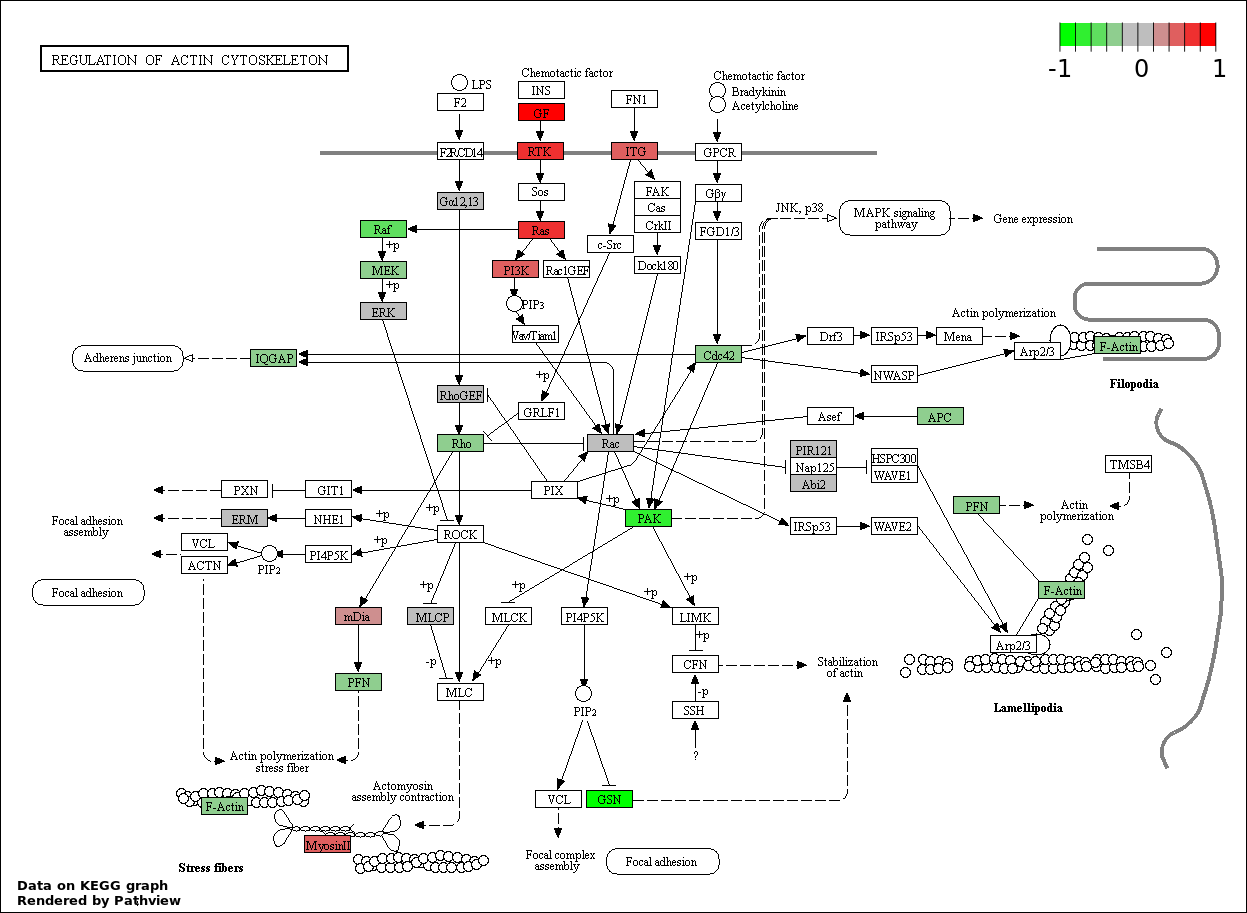
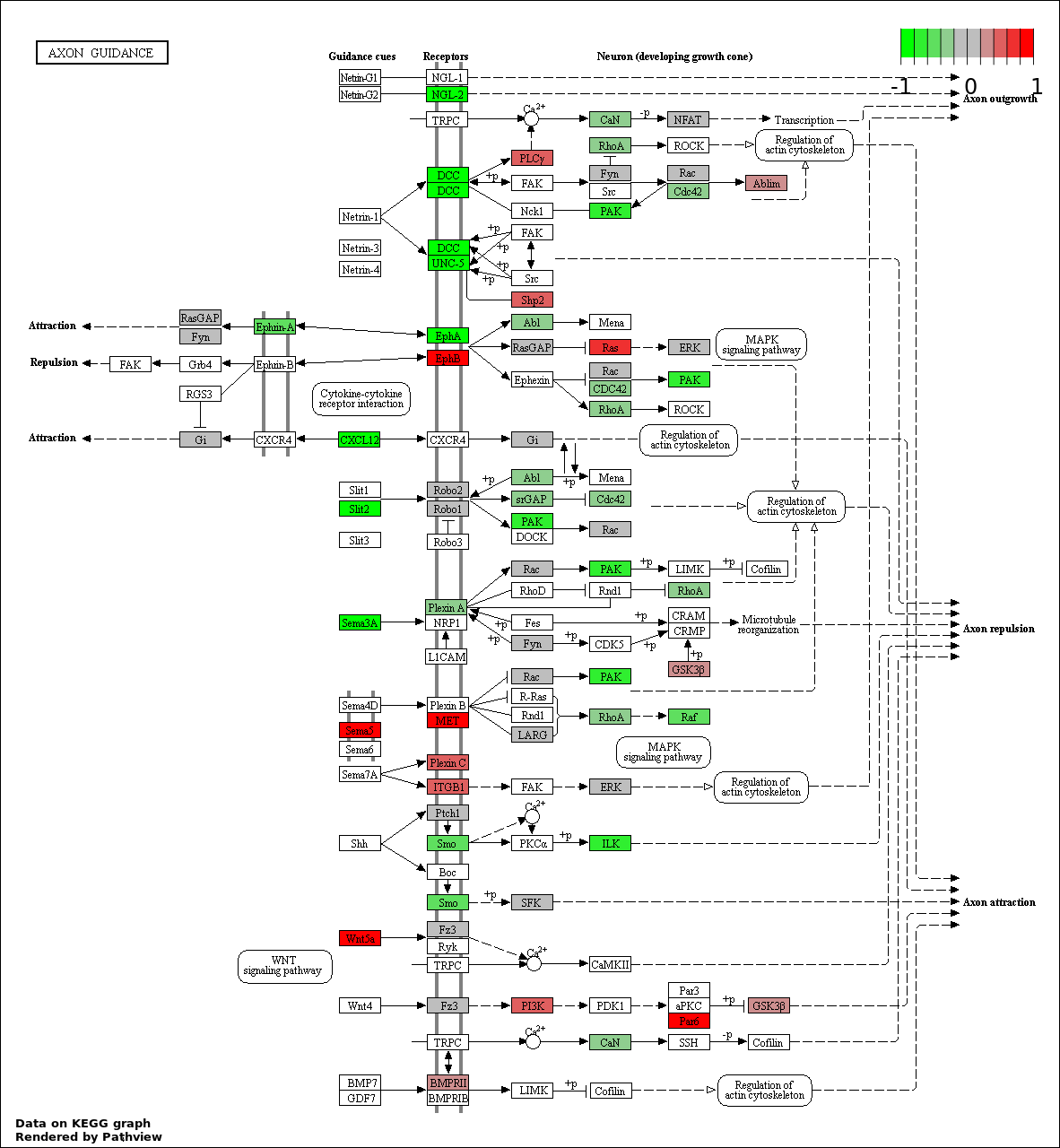
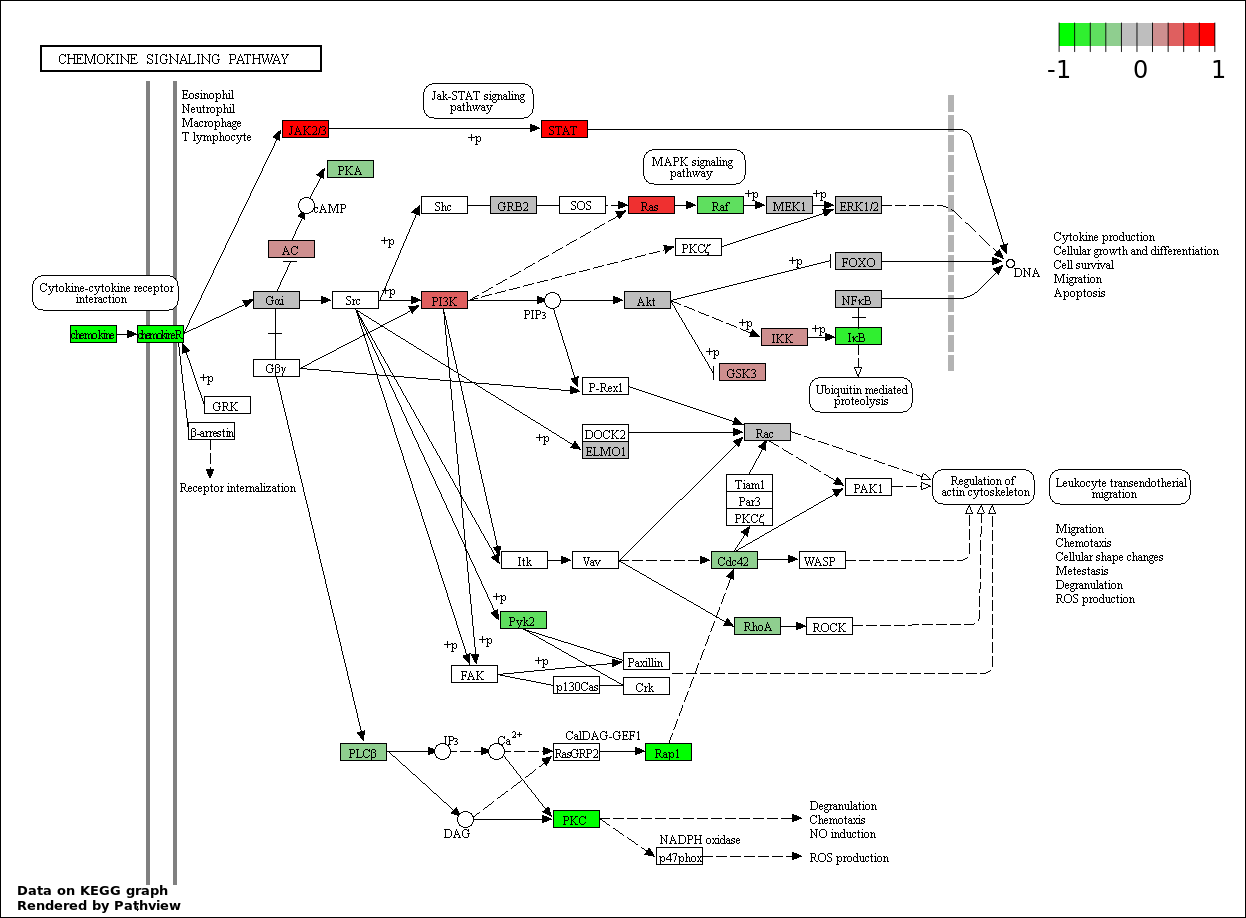
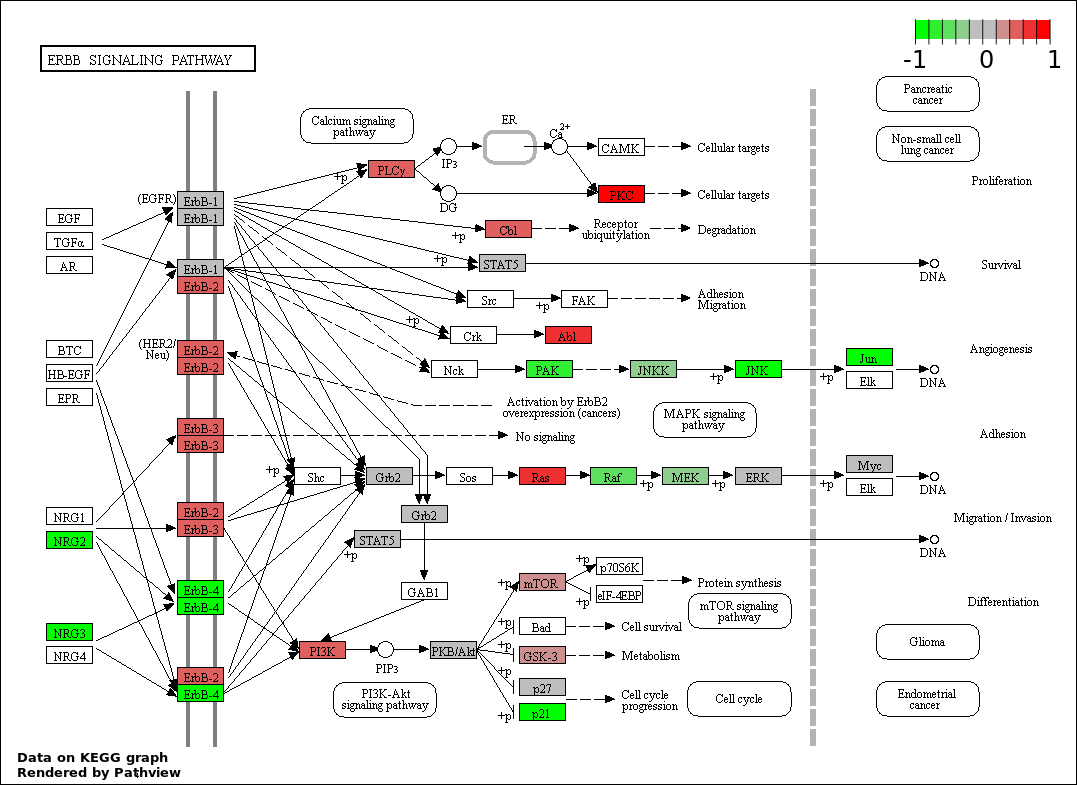
This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-324-5p | GLI1 | 0.8 | 0.00101 | 0.94 | 0.00099 | mir2Disease; miRNAWalker2 validate; miRTarBase | -0.2 | 0.00636 | ||
| 2 | hsa-let-7a-3p | TRIM33 | 0.28 | 0.12701 | 0.4 | 7.0E-5 | miRNAWalker2 validate | -0.15 | 0 | ||
| 3 | hsa-let-7a-5p | CDKN1A | 0.3 | 0.00963 | -1.27 | 0 | miRNAWalker2 validate; MirTarget; TargetScan | -0.36 | 0.00039 | ||
| 4 | hsa-let-7a-5p | GADD45GIP1 | 0.3 | 0.00963 | -0.11 | 0.32137 | miRNAWalker2 validate | -0.2 | 0.00086 | ||
| 5 | hsa-let-7a-5p | HRAS | 0.3 | 0.00963 | 0 | 0.98399 | miRNAWalker2 validate; miRTarBase | -0.16 | 0.00583 | ||
| 6 | hsa-let-7a-5p | PNRC1 | 0.3 | 0.00963 | -0.65 | 0 | miRNAWalker2 validate | -0.13 | 0.03661 | ||
| 7 | hsa-let-7a-5p | TUSC2 | 0.3 | 0.00963 | -0.44 | 0 | miRNAWalker2 validate; TargetScan; miRNATAP | -0.18 | 0.00023 | ||
| 8 | hsa-let-7a-5p | ZFP36L1 | 0.3 | 0.00963 | -0.51 | 8.0E-5 | miRNAWalker2 validate | -0.21 | 0.00223 | ||
| 9 | hsa-let-7b-3p | SYT4 | 0.38 | 0.00617 | -3.21 | 0 | miRNAWalker2 validate | -1.61 | 0 | ||
| 10 | hsa-let-7b-5p | ABL1 | 0.17 | 0.1606 | -0.21 | 0.07804 | miRNAWalker2 validate | -0.18 | 0.00351 | ||
| 11 | hsa-let-7b-5p | AMPH | 0.17 | 0.1606 | -0.92 | 5.0E-5 | miRNAWalker2 validate | -0.24 | 0.04099 | ||
| 12 | hsa-let-7b-5p | AR | 0.17 | 0.1606 | -1.55 | 0 | miRNAWalker2 validate | -0.48 | 0.00744 | ||
| 13 | hsa-let-7b-5p | DMD | 0.17 | 0.1606 | -1.51 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.57 | 0.00107 | ||
| 14 | hsa-let-7b-5p | ERC1 | 0.17 | 0.1606 | -0.11 | 0.38794 | miRNAWalker2 validate | -0.15 | 0.02169 | ||
| 15 | hsa-let-7b-5p | FLNA | 0.17 | 0.1606 | -0.77 | 0.01377 | miRNAWalker2 validate | -0.46 | 0.00493 | ||
| 16 | hsa-let-7b-5p | GLTSCR2 | 0.17 | 0.1606 | -0.67 | 0 | miRNAWalker2 validate | -0.15 | 0.02223 | ||
| 17 | hsa-let-7b-5p | HMGB1 | 0.17 | 0.1606 | 0.21 | 0.02228 | miRNAWalker2 validate | -0.1 | 0.04547 | ||
| 18 | hsa-let-7b-5p | MAP2K2 | 0.17 | 0.1606 | -0.5 | 0 | miRNAWalker2 validate | -0.12 | 0.0194 | ||
| 19 | hsa-let-7b-5p | MLLT1 | 0.17 | 0.1606 | 0.04 | 0.66424 | miRNAWalker2 validate | -0.12 | 0.00518 | ||
| 20 | hsa-let-7b-5p | NEDD4 | 0.17 | 0.1606 | 0.02 | 0.89834 | miRNAWalker2 validate | -0.22 | 0.00727 | ||
| 21 | hsa-let-7b-5p | PRKAA2 | 0.17 | 0.1606 | -2.46 | 0 | miRNAWalker2 validate; MirTarget | -0.83 | 0.00199 | ||
| 22 | hsa-let-7b-5p | SALL2 | 0.17 | 0.1606 | -1.4 | 2.0E-5 | miRNAWalker2 validate | -0.6 | 0.00051 | ||
| 23 | hsa-let-7b-5p | ST13 | 0.17 | 0.1606 | -0.21 | 0.01092 | miRNAWalker2 validate | -0.09 | 0.02928 | ||
| 24 | hsa-let-7b-5p | TES | 0.17 | 0.1606 | -0.35 | 0.00639 | miRNAWalker2 validate | -0.16 | 0.01773 | ||
| 25 | hsa-let-7c-5p | ACVR1B | -0.15 | 0.54808 | 0.22 | 0.12477 | miRNAWalker2 validate | -0.07 | 0.03766 | ||
| 26 | hsa-let-7c-5p | ATP2A2 | -0.15 | 0.54808 | 0.6 | 0 | miRNAWalker2 validate | -0.09 | 0.00012 | ||
| 27 | hsa-let-7c-5p | CCNB2 | -0.15 | 0.54808 | 1.86 | 0 | miRNAWalker2 validate | -0.36 | 0 | ||
| 28 | hsa-let-7c-5p | CCNC | -0.15 | 0.54808 | -0.22 | 0.03806 | miRNAWalker2 validate | -0.18 | 0 | ||
| 29 | hsa-let-7c-5p | CNOT3 | -0.15 | 0.54808 | 0.2 | 0.03181 | miRNAWalker2 validate | -0.08 | 0.00054 | ||
| 30 | hsa-let-7c-5p | DOT1L | -0.15 | 0.54808 | 0.32 | 0.00907 | miRNAWalker2 validate | -0.18 | 0 | ||
| 31 | hsa-let-7c-5p | EIF3A | -0.15 | 0.54808 | 0.32 | 0.00012 | miRNAWalker2 validate | -0.08 | 4.0E-5 | ||
| 32 | hsa-let-7c-5p | EWSR1 | -0.15 | 0.54808 | 0.19 | 0.00219 | miRNAWalker2 validate | -0.12 | 0 | ||
| 33 | hsa-let-7c-5p | LTA4H | -0.15 | 0.54808 | -0.26 | 0.01046 | miRNAWalker2 validate | -0.05 | 0.0396 | ||
| 34 | hsa-let-7c-5p | MYC | -0.15 | 0.54808 | -0.03 | 0.88917 | miRNAWalker2 validate; miRTarBase | -0.12 | 0.00982 | ||
| 35 | hsa-let-7c-5p | NRAS | -0.15 | 0.54808 | 0.47 | 1.0E-5 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.12 | 2.0E-5 | ||
| 36 | hsa-let-7c-5p | SMARCC1 | -0.15 | 0.54808 | 0.54 | 0 | miRNAWalker2 validate | -0.1 | 8.0E-5 | ||
| 37 | hsa-let-7c-5p | SMARCD1 | -0.15 | 0.54808 | 0.45 | 0 | miRNAWalker2 validate | -0.08 | 0.00022 | ||
| 38 | hsa-let-7d-5p | DICER1 | 0.69 | 0 | 0.04 | 0.674 | miRNAWalker2 validate; miRTarBase | -0.14 | 0.00143 | ||
| 39 | hsa-let-7d-5p | DLC1 | 0.69 | 0 | -0.38 | 0.05038 | miRNAWalker2 validate | -0.54 | 0 | ||
| 40 | hsa-let-7d-5p | ERC1 | 0.69 | 0 | -0.11 | 0.38794 | miRNAWalker2 validate | -0.31 | 0 | ||
| 41 | hsa-let-7d-5p | MPL | 0.69 | 0 | -0.62 | 0.00133 | miRNAWalker2 validate | -0.7 | 0 | ||
| 42 | hsa-let-7d-5p | NCOA1 | 0.69 | 0 | -0.43 | 0 | miRNAWalker2 validate; miRNATAP | -0.29 | 0 | ||
| 43 | hsa-let-7d-5p | THBS1 | 0.69 | 0 | 0.74 | 0.00386 | miRNAWalker2 validate | -0.51 | 0 | ||
| 44 | hsa-let-7d-5p | TLE4 | 0.69 | 0 | -1.16 | 0 | miRNAWalker2 validate | -0.62 | 0 | ||
| 45 | hsa-let-7d-5p | TMTC3 | 0.69 | 0 | 0.12 | 0.22189 | miRNAWalker2 validate | -0.18 | 3.0E-5 | ||
| 46 | hsa-let-7e-5p | CNBP | 0.23 | 0.16229 | -0.39 | 0 | miRNAWalker2 validate | -0.06 | 0.03061 | ||
| 47 | hsa-let-7e-5p | DIAPH1 | 0.23 | 0.16229 | 0.27 | 0.0326 | miRNAWalker2 validate | -0.2 | 3.0E-5 | ||
| 48 | hsa-let-7e-5p | NCKIPSD | 0.23 | 0.16229 | 0.15 | 0.10057 | miRNAWalker2 validate | -0.1 | 0.00431 | ||
| 49 | hsa-let-7e-5p | RPA1 | 0.23 | 0.16229 | 0.04 | 0.57415 | miRNAWalker2 validate | -0.06 | 0.04715 | ||
| 50 | hsa-let-7e-5p | RPL10 | 0.23 | 0.16229 | -0.6 | 0 | miRNAWalker2 validate | -0.16 | 0.00142 | ||
| 51 | hsa-let-7e-5p | SRSF2 | 0.23 | 0.16229 | 0.27 | 0.00028 | miRNAWalker2 validate | -0.08 | 0.00564 | ||
| 52 | hsa-let-7e-5p | YWHAE | 0.23 | 0.16229 | 0 | 0.98905 | miRNAWalker2 validate | -0.15 | 0.00017 | ||
| 53 | hsa-let-7f-5p | CDKN1A | 0.3 | 0.0221 | -1.27 | 0 | miRNAWalker2 validate; MirTarget | -0.25 | 0.00539 | ||
| 54 | hsa-let-7f-5p | MPL | 0.3 | 0.0221 | -0.62 | 0.00133 | miRNAWalker2 validate | -0.35 | 0.00016 | ||
| 55 | hsa-let-7f-5p | ROR2 | 0.3 | 0.0221 | -1.09 | 0.0008 | miRNAWalker2 validate | -0.73 | 0 | ||
| 56 | hsa-let-7g-5p | AKAP11 | 0.3 | 0.02968 | 0.2 | 0.09825 | miRNAWalker2 validate | -0.25 | 0 | ||
| 57 | hsa-let-7g-5p | COL1A2 | 0.3 | 0.02968 | 1.99 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.31 | 0.00943 | ||
| 58 | hsa-let-7i-5p | EGLN3 | 0.91 | 0 | -0.95 | 0 | miRNAWalker2 validate | -0.36 | 0.00083 | ||
| 59 | hsa-miR-100-5p | ACSL3 | -0.34 | 0.134 | -0.14 | 0.20403 | miRNAWalker2 validate | -0.17 | 0 | ||
| 60 | hsa-miR-100-5p | ANAPC1 | -0.34 | 0.134 | 0.7 | 0 | miRNAWalker2 validate | -0.17 | 0 | ||
| 61 | hsa-miR-100-5p | ATP2A2 | -0.34 | 0.134 | 0.6 | 0 | miRNAWalker2 validate | -0.13 | 0 | ||
| 62 | hsa-miR-100-5p | BCL10 | -0.34 | 0.134 | -0.19 | 0.11901 | miRNAWalker2 validate | -0.18 | 0 | ||
| 63 | hsa-miR-100-5p | BECN1 | -0.34 | 0.134 | -0.25 | 0.00015 | miRNAWalker2 validate | -0.06 | 0.00199 | ||
| 64 | hsa-miR-100-5p | CDC73 | -0.34 | 0.134 | 0.09 | 0.20463 | miRNAWalker2 validate | -0.07 | 0.00033 | ||
| 65 | hsa-miR-100-5p | CUL2 | -0.34 | 0.134 | 0.2 | 0.01264 | miRNAWalker2 validate | -0.06 | 0.00581 | ||
| 66 | hsa-miR-100-5p | DNMT1 | -0.34 | 0.134 | 1.19 | 0 | miRNAWalker2 validate | -0.19 | 0 | ||
| 67 | hsa-miR-100-5p | EWSR1 | -0.34 | 0.134 | 0.19 | 0.00219 | miRNAWalker2 validate | -0.11 | 0 | ||
| 68 | hsa-miR-100-5p | FGFR3 | -0.34 | 0.134 | 0.41 | 0.23565 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.24 | 0.00862 | 25344675; 25344675; 23778527; 23778527; 23778527; 23778527; 23778488 | The predicted target of miR-100 fibroblast growth factor receptor 3 FGFR3 was downregulated by siRNA to examine its effect on pancreatic cancer cells;FGFR3 was significantly downregulated by overexpressing miR-100 in pancreatic cancer cells and knocking down FGFR3 by siRNA exerted similar effect as miR-100;Hypoxia regulates FGFR3 expression via HIF-1α and miR-100 and contributes to cell survival in non-muscle invasive bladder cancer;In this study we investigated the effects of hypoxia on miR-100 and FGFR3 expression and the link between miR-100 and FGFR3 in hypoxia;Bladder cancer cell lines were exposed to normoxic or hypoxic conditions and examined for the expression of FGFR3 by quantitative PCR qPCR and western blotting and miR-100 by qPCR;Hypoxia in part via suppression of miR-100 induces FGFR3 expression in bladder cancer both of which have an important role in maintaining cell viability under conditions of stress;We demonstrate that miR-100 is a context-dependent miRNA controlling BAZ2 mTOR FGFR3 SMARCA5 and THAP2 that might be involved in PC progression |
| 69 | hsa-miR-100-5p | FXN | -0.34 | 0.134 | 0.17 | 0.13008 | miRNAWalker2 validate | -0.11 | 0.00015 | ||
| 70 | hsa-miR-100-5p | GRB2 | -0.34 | 0.134 | -0.05 | 0.48951 | miRNAWalker2 validate | -0.05 | 0.0027 | ||
| 71 | hsa-miR-100-5p | HMGB1 | -0.34 | 0.134 | 0.21 | 0.02228 | miRNAWalker2 validate | -0.09 | 0.00055 | ||
| 72 | hsa-miR-100-5p | MTOR | -0.34 | 0.134 | 0.22 | 0.02719 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.07 | 0.01018 | 22926517; 23778488; 23778488; 23778488; 20081105; 20081105 | MiR-100 has also been shown to suppress other proteins of the IGF/mammalian target of rapamycin mTOR signaling cascade in different human tumors;There was reduction in mTOR p=0.025 THAP2 p=0.038 SMARCA5 p=0.001 and BAZ2A p=0.006 mRNA expression in DU145 cells after exposure to miR-100;In PC3 cells mTOR expression was decreased by miR-100 p=0.01;We demonstrate that miR-100 is a context-dependent miRNA controlling BAZ2 mTOR FGFR3 SMARCA5 and THAP2 that might be involved in PC progression;Because of the critical roles of the phosphatidylinositol 3-kinase/v-akt murine thymoma viral oncogene homolog 1/mammalian target of rapamycin mTOR pathway in clear cell ovarian cancer we focused on mir-100 a putative tumor suppressor that was the most down-regulated miRNA in our cancer cell lines and its up-regulated target FRAP1/mTOR;Overexpression of mir-100 inhibited mTOR signaling and enhanced sensitivity to the rapamycin analog RAD001 everolimus confirming the key relationship between mir-100 and the mTOR pathway |
| 73 | hsa-miR-100-5p | OXA1L | -0.34 | 0.134 | -0.29 | 0.0006 | miRNAWalker2 validate | -0.09 | 5.0E-5 | ||
| 74 | hsa-miR-100-5p | PLK1 | -0.34 | 0.134 | 2 | 0 | miRNAWalker2 validate; miRTarBase | -0.45 | 0 | 21636267; 21636267; 21636267; 21636267; 21636267; 21636267; 23842624; 23842624; 23842624; 23151088; 23151088; 23151088; 23151088; 23151088; 22246341; 22246341; 22120675; 22120675; 22120675 | Reduced miR-100 expression in cervical cancer and precursors and its carcinogenic effect through targeting PLK1 protein;Through modulating miR-100 expression using miR-100 inhibitor or mimic in vitro cell growth cycle and apoptosis were tested separately by MTT or flow cytometry and meanwhile Polo-like kinase1 PLK1 mRNA and protein expressions were detected by qRT-PCR and immunoblotting;The expression of PLK1 in 125 cervical tissues was also examined by immunohistochemical staining and the correlation between miR-100 and PLK1 expression in the same tissues was analysed;The modulation of miR-100 expression remarkably influenced cell proliferation cycle and apoptosis as well as the level of PLK1 protein but not mRNA in vitro experiments;PLK1 expression was negatively correlated with miR-100 expression in CIN3 and cervical cancer tissues;The reduced miR-100 expression participates in the development of cervical cancer at least partly through loss of inhibition to target gene PLK1 which probably occurs in a relative late phase of carcinogenesis;In addition we found that upregulation of miR-100 could inhibit growth and increase apoptosis of HCC cells by downregulating polo-like kinase 1 plk1;In HCC tissues miR-100 expression was inversely correlated with the expression of plk1 protein r = -0.418; P = 0.029;Therefore downregulation of miR-100 was correlated with progressive pathological feature and poor prognosis in HCC patients and miR-100 could function as a tumor suppressor by targeting plk1;MicroRNA-100 is a potential molecular marker of non-small cell lung cancer and functions as a tumor suppressor by targeting polo-like kinase 1;Finally the effects of miR-100 expression on growth apoptosis and cell cycle of NSCLC cells by posttranscriptionally regulating PLK1 expression were determined;Meanwhile miR-100 mimics could significantly inhibit PLK1 mRNA and protein expression and reduce the luciferase activity of a PLK1 3' untranslated region-based reporter construct in A549 cells;Furthermore small interfering RNA siRNA-mediated PLK1 downregulation could mimic the effects of miR-100 mimics while PLK1 overexpression could partially rescue the phenotypical changes of NSCLC cells induced by miR-100 mimics;Our findings indicate that low miR-100 may be a poor prognostic factor for NSCLC patients and functions as a tumor suppressor by posttranscriptionally regulating PLK1 expression;Additionally miR-100 could affect the growth of EOC cells by post-transcriptionally regulating polo-like kinase 1 PLK1 expression;Together these results suggest that low miR-100 expression may be an independent poor prognostic factor and miR-100 can function as a tumor suppressor by targeting PLK1 in human EOCs;Knock-down of Plk1 which was a direct target of miR-100 yielded similar effects as that of ectopic miR-100 expression;The inverse correlation between miR-100 and Plk1 expression was also detected in nude mice SPC-A1/DTX tumor xenografts and clinical lung adenocarcinoma tissues and was proved to be related with the in vivo response to docetaxel;Thus our results suggested that down-regulation of miR-100 could lead to Plk1 over-expression and eventually to docetaxel chemoresistance of human lung adenocarcinoma |
| 75 | hsa-miR-100-5p | RB1 | -0.34 | 0.134 | 0.38 | 0.0017 | miRNAWalker2 validate | -0.09 | 0.00504 | ||
| 76 | hsa-miR-100-5p | SET | -0.34 | 0.134 | 0.47 | 0 | miRNAWalker2 validate | -0.14 | 0 | ||
| 77 | hsa-miR-100-5p | UBB | -0.34 | 0.134 | -0.43 | 4.0E-5 | miRNAWalker2 validate | -0.07 | 0.0183 | ||
| 78 | hsa-miR-100-5p | XRCC5 | -0.34 | 0.134 | 0.2 | 0.00319 | miRNAWalker2 validate | -0.11 | 0 | ||
| 79 | hsa-miR-100-5p | YWHAE | -0.34 | 0.134 | 0 | 0.98905 | miRNAWalker2 validate | -0.17 | 0 | ||
| 80 | hsa-miR-101-3p | E2F3 | -0.07 | 0.54416 | 1.29 | 0 | miRNAWalker2 validate | -0.2 | 0.00606 | ||
| 81 | hsa-miR-101-3p | GPAM | -0.07 | 0.54416 | 0.14 | 0.29732 | miRNAWalker2 validate; mirMAP | -0.15 | 0.04309 | ||
| 82 | hsa-miR-101-3p | MAP2K1 | -0.07 | 0.54416 | 0.11 | 0.188 | miRNAWalker2 validate | -0.12 | 0.01272 | ||
| 83 | hsa-miR-101-3p | MSH2 | -0.07 | 0.54416 | 1.04 | 0 | miRNAWalker2 validate | -0.17 | 0.03146 | ||
| 84 | hsa-miR-101-3p | RAC1 | -0.07 | 0.54416 | 0.08 | 0.39234 | miRNAWalker2 validate; MirTarget | -0.12 | 0.01667 | ||
| 85 | hsa-miR-101-3p | SUZ12 | -0.07 | 0.54416 | 0.62 | 0 | miRNAWalker2 validate | -0.19 | 0.00128 | ||
| 86 | hsa-miR-101-3p | TMTC3 | -0.07 | 0.54416 | 0.12 | 0.22189 | miRNAWalker2 validate | -0.14 | 0.01004 | ||
| 87 | hsa-miR-101-3p | VEGFA | -0.07 | 0.54416 | 0.63 | 0.00062 | miRNAWalker2 validate | -0.23 | 0.02392 | ||
| 88 | hsa-miR-101-5p | ATM | -0.37 | 0.04353 | 0.18 | 0.21568 | miRNAWalker2 validate | -0.1 | 0.04098 | ||
| 89 | hsa-miR-103a-3p | BCL2 | 1.01 | 0 | -0.9 | 0 | miRNAWalker2 validate | -0.34 | 1.0E-5 | ||
| 90 | hsa-miR-103a-3p | CAV1 | 1.01 | 0 | -1.01 | 6.0E-5 | miRNAWalker2 validate; miRTarBase | -0.59 | 0 | ||
| 91 | hsa-miR-103a-3p | CITED2 | 1.01 | 0 | -1.54 | 0 | miRNAWalker2 validate | -0.57 | 0 | ||
| 92 | hsa-miR-103a-3p | DNAJB4 | 1.01 | 0 | -0.7 | 1.0E-5 | miRNAWalker2 validate | -0.26 | 4.0E-5 | ||
| 93 | hsa-miR-103a-3p | JAKMIP2 | 1.01 | 0 | -1.39 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.77 | 0 | ||
| 94 | hsa-miR-103a-3p | KLF4 | 1.01 | 0 | -1.89 | 0 | miRNAWalker2 validate; miRTarBase | -0.26 | 0.0044 | ||
| 95 | hsa-miR-103a-3p | PLSCR4 | 1.01 | 0 | -0.47 | 0.03833 | miRNAWalker2 validate | -0.54 | 0 | ||
| 96 | hsa-miR-103a-3p | RECK | 1.01 | 0 | -1.04 | 0 | miRNAWalker2 validate | -0.57 | 0 | ||
| 97 | hsa-miR-103a-3p | RUNX1T1 | 1.01 | 0 | -0.34 | 0.2374 | miRNAWalker2 validate; miRNATAP | -0.64 | 0 | ||
| 98 | hsa-miR-103a-3p | SIRT4 | 1.01 | 0 | -0.99 | 0 | miRNAWalker2 validate | -0.36 | 0 | ||
| 99 | hsa-miR-103a-3p | TIMP3 | 1.01 | 0 | -0.55 | 0.02307 | miRNAWalker2 validate; miRTarBase | -0.58 | 0 | ||
| 100 | hsa-miR-103a-3p | TLE4 | 1.01 | 0 | -1.16 | 0 | miRNAWalker2 validate | -0.34 | 0 | ||
| 101 | hsa-miR-103a-3p | ZYX | 1.01 | 0 | 0.02 | 0.89763 | miRNAWalker2 validate; miRNATAP | -0.33 | 0 | ||
| 102 | hsa-miR-106a-5p | CDKN1A | 0.59 | 0.00839 | -1.27 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.17 | 0.00069 | ||
| 103 | hsa-miR-106a-5p | NOTCH2 | 0.59 | 0.00839 | 0.14 | 0.35163 | miRNAWalker2 validate | -0.14 | 0.00052 | ||
| 104 | hsa-miR-106b-3p | FOXO1 | 1.36 | 0 | -0.14 | 0.33874 | miRNAWalker2 validate | -0.16 | 0.00052 | ||
| 105 | hsa-miR-106b-3p | NFE2L2 | 1.36 | 0 | -0.86 | 0 | miRNAWalker2 validate | -0.18 | 0 | ||
| 106 | hsa-miR-106b-3p | PRDM2 | 1.36 | 0 | -0.26 | 0.00721 | miRNAWalker2 validate | -0.19 | 0 | ||
| 107 | hsa-miR-106b-3p | SALL2 | 1.36 | 0 | -1.4 | 2.0E-5 | miRNAWalker2 validate | -0.76 | 0 | ||
| 108 | hsa-miR-106b-5p | AKAP11 | 1.09 | 0 | 0.2 | 0.09825 | miRNAWalker2 validate | -0.12 | 0.00117 | ||
| 109 | hsa-miR-106b-5p | APC | 1.09 | 0 | -0.26 | 0.04051 | miRNAWalker2 validate; miRTarBase | -0.3 | 0 | 23087084 | miR-106b downregulates adenomatous polyposis coli and promotes cell proliferation in human hepatocellular carcinoma |
| 110 | hsa-miR-106b-5p | CCND2 | 1.09 | 0 | -0.27 | 0.3112 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.58 | 0 | ||
| 111 | hsa-miR-106b-5p | CDC42 | 1.09 | 0 | -0.34 | 1.0E-5 | miRNAWalker2 validate | -0.06 | 0.02563 | ||
| 112 | hsa-miR-106b-5p | EEF1A1 | 1.09 | 0 | -0.6 | 0 | miRNAWalker2 validate | -0.08 | 0.03819 | ||
| 113 | hsa-miR-106b-5p | EIF3F | 1.09 | 0 | -0.59 | 0 | miRNAWalker2 validate | -0.07 | 0.01743 | ||
| 114 | hsa-miR-106b-5p | FNBP1 | 1.09 | 0 | -0.97 | 4.0E-5 | miRNAWalker2 validate | -0.76 | 0 | ||
| 115 | hsa-miR-106b-5p | JAK1 | 1.09 | 0 | 0 | 0.98982 | miRNAWalker2 validate; MirTarget | -0.12 | 4.0E-5 | ||
| 116 | hsa-miR-106b-5p | MXI1 | 1.09 | 0 | -0.99 | 0 | miRNAWalker2 validate | -0.38 | 0 | ||
| 117 | hsa-miR-106b-5p | PTEN | 1.09 | 0 | -0.19 | 0.07748 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.16 | 0 | 24842611; 24842611 | MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN;Here we have shown that miR-106b is up-regulated in cancer associated fibroblasts compared with normal fibroblasts established from patients with gastric cancer the expression level of miR-106b is associated with poor prognosis of patients and CAFs with down-regulated miR-106b could significantly inhibit gastric cancer cell migration and invasion by targeting PTEN |
| 118 | hsa-miR-106b-5p | RBL2 | 1.09 | 0 | -0.28 | 0.00254 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.22 | 0 | ||
| 119 | hsa-miR-106b-5p | RHOBTB2 | 1.09 | 0 | -0.17 | 0.19499 | miRNAWalker2 validate | -0.13 | 0.00145 | ||
| 120 | hsa-miR-106b-5p | RUNX1T1 | 1.09 | 0 | -0.34 | 0.2374 | miRNAWalker2 validate | -0.95 | 0 | ||
| 121 | hsa-miR-106b-5p | SETD2 | 1.09 | 0 | 0.03 | 0.73068 | miRNAWalker2 validate; miRNATAP | -0.06 | 0.03157 | ||
| 122 | hsa-miR-106b-5p | SYT4 | 1.09 | 0 | -3.21 | 0 | miRNAWalker2 validate | -1.35 | 0 | ||
| 123 | hsa-miR-106b-5p | TSC1 | 1.09 | 0 | 0.1 | 0.26071 | miRNAWalker2 validate | -0.06 | 0.04723 | ||
| 124 | hsa-miR-107 | ARNT | 0.85 | 0 | -0.08 | 0.26073 | miRNAWalker2 validate; miRTarBase; miRanda | -0.1 | 0.00248 | ||
| 125 | hsa-miR-107 | DAPK1 | 0.85 | 0 | -0.08 | 0.65985 | miRNAWalker2 validate; miRTarBase | -0.27 | 0.0032 | ||
| 126 | hsa-miR-107 | NINJ1 | 0.85 | 0 | -0.14 | 0.22595 | miRNAWalker2 validate | -0.17 | 0.00205 | ||
| 127 | hsa-miR-107 | NOTCH2 | 0.85 | 0 | 0.14 | 0.35163 | miRNAWalker2 validate; MirTarget; miRanda; miRNATAP | -0.35 | 0 | ||
| 128 | hsa-miR-10a-5p | AJUBA | 0.19 | 0.25421 | 1.84 | 0 | miRNAWalker2 validate | -0.23 | 0.00591 | ||
| 129 | hsa-miR-10a-5p | CHFR | 0.19 | 0.25421 | -0.68 | 7.0E-5 | miRNAWalker2 validate | -0.2 | 0.00126 | ||
| 130 | hsa-miR-10a-5p | CTNND1 | 0.19 | 0.25421 | -0.18 | 0.08478 | miRNAWalker2 validate | -0.09 | 0.02177 | ||
| 131 | hsa-miR-10a-5p | FAT2 | 0.19 | 0.25421 | -0.28 | 0.44877 | miRNAWalker2 validate | -0.41 | 0.00188 | ||
| 132 | hsa-miR-10a-5p | KLHL6 | 0.19 | 0.25421 | 0 | 0.99199 | miRNAWalker2 validate | -0.21 | 0.03094 | ||
| 133 | hsa-miR-10a-5p | SPECC1 | 0.19 | 0.25421 | -0.08 | 0.57179 | miRNAWalker2 validate | -0.19 | 0.00027 | ||
| 134 | hsa-miR-10b-5p | INHBA | -0.25 | 0.14496 | 3.97 | 0 | miRNAWalker2 validate | -0.3 | 0.02078 | ||
| 135 | hsa-miR-10b-5p | NCOR2 | -0.25 | 0.14496 | 0.55 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.07 | 0.04495 | ||
| 136 | hsa-miR-10b-5p | NF1 | -0.25 | 0.14496 | 0.51 | 0 | miRNAWalker2 validate; miRTarBase | -0.14 | 0.00019 | 24096486 | miR-10b also enhanced the stimulatory effects of EGF and TGF-β on cell migration and epithelial-mesenchymal transition EMT and decreased the expression of RAP2A EPHB2 KLF4 and NF1 |
| 137 | hsa-miR-10b-5p | PABPC1 | -0.25 | 0.14496 | 0.44 | 0.00029 | miRNAWalker2 validate | -0.13 | 0.00413 | ||
| 138 | hsa-miR-10b-5p | SMCHD1 | -0.25 | 0.14496 | 0.14 | 0.18254 | miRNAWalker2 validate | -0.11 | 0.00335 | ||
| 139 | hsa-miR-125a-5p | MAP3K1 | -0.26 | 0.11611 | 0.57 | 4.0E-5 | miRNAWalker2 validate | -0.17 | 0.00088 | ||
| 140 | hsa-miR-125a-5p | MAT2A | -0.26 | 0.11611 | -0.07 | 0.36195 | miRNAWalker2 validate | -0.09 | 0.00322 | ||
| 141 | hsa-miR-125a-5p | NPM1 | -0.26 | 0.11611 | 0.19 | 0.13186 | miRNAWalker2 validate | -0.2 | 1.0E-5 | ||
| 142 | hsa-miR-125a-5p | TDG | -0.26 | 0.11611 | 0.45 | 2.0E-5 | miRNAWalker2 validate; miRanda | -0.12 | 0.0028 | ||
| 143 | hsa-miR-125a-5p | TFRC | -0.26 | 0.11611 | 0.82 | 2.0E-5 | miRNAWalker2 validate | -0.19 | 0.00958 | ||
| 144 | hsa-miR-125a-5p | THRAP3 | -0.26 | 0.11611 | 0.14 | 0.0313 | miRNAWalker2 validate | -0.1 | 1.0E-5 | ||
| 145 | hsa-miR-125a-5p | TP53 | -0.26 | 0.11611 | 0.51 | 0.00486 | miRNAWalker2 validate; miRTarBase; miRanda | -0.24 | 0.00029 | 23079745 | Furthermore treatment of HCC cells with 5-aza-2'-deoxycytidine or ectopic expression of wildtype but not mutated p53 restored miR-125a-5p and miR-125b expression and inhibited tumor cell growth suggesting their regulation by promoter methylation and p53 activity |
| 146 | hsa-miR-125a-5p | XPO1 | -0.26 | 0.11611 | 0.76 | 0 | miRNAWalker2 validate | -0.16 | 2.0E-5 | ||
| 147 | hsa-miR-125b-5p | BBC3 | -0.18 | 0.43772 | 1.36 | 0 | miRNAWalker2 validate; miRTarBase | -0.12 | 0.0199 | ||
| 148 | hsa-miR-125b-5p | CBFB | -0.18 | 0.43772 | 1.06 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.15 | 1.0E-5 | ||
| 149 | hsa-miR-125b-5p | CCNE1 | -0.18 | 0.43772 | 1.64 | 0 | miRNAWalker2 validate | -0.33 | 5.0E-5 | ||
| 150 | hsa-miR-125b-5p | E2F2 | -0.18 | 0.43772 | 1.36 | 0 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.44 | 0 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | POSITIVE REGULATION OF GENE EXPRESSION | 421 | 1733 | 2.271e-105 | 1.057e-101 |
| 2 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 422 | 1784 | 1.847e-101 | 4.297e-98 |
| 3 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 409 | 1805 | 1.566e-91 | 2.429e-88 |
| 4 | REGULATION OF CELL PROLIFERATION | 360 | 1496 | 2.944e-87 | 3.425e-84 |
| 5 | REGULATION OF CELL DIFFERENTIATION | 353 | 1492 | 3.365e-83 | 3.131e-80 |
| 6 | REGULATION OF CELL DEATH | 345 | 1472 | 5.921e-80 | 4.592e-77 |
| 7 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 371 | 1672 | 2.521e-79 | 1.676e-76 |
| 8 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 274 | 1004 | 5.589e-78 | 3.251e-75 |
| 9 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 395 | 1929 | 4.223e-74 | 2.183e-71 |
| 10 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 337 | 1517 | 1.673e-71 | 7.786e-69 |
| 11 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 351 | 1656 | 4.682e-69 | 1.981e-66 |
| 12 | POSITIVE REGULATION OF CELL COMMUNICATION | 334 | 1532 | 1.26e-68 | 4.886e-66 |
| 13 | REGULATION OF PROTEIN MODIFICATION PROCESS | 354 | 1710 | 7.014e-67 | 2.51e-64 |
| 14 | NEGATIVE REGULATION OF GENE EXPRESSION | 324 | 1493 | 7.051e-66 | 2.343e-63 |
| 15 | TISSUE DEVELOPMENT | 325 | 1518 | 1.36e-64 | 4.219e-62 |
| 16 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 335 | 1618 | 5.757e-63 | 1.674e-60 |
| 17 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 270 | 1142 | 2.846e-62 | 7.79e-60 |
| 18 | REGULATION OF CELL CYCLE | 242 | 949 | 3.448e-62 | 8.913e-60 |
| 19 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 314 | 1492 | 2.101e-60 | 5.146e-58 |
| 20 | INTRACELLULAR SIGNAL TRANSDUCTION | 324 | 1572 | 3.164e-60 | 7.362e-58 |
| 21 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 207 | 740 | 3.379e-60 | 7.487e-58 |
| 22 | NEGATIVE REGULATION OF CELL PROLIFERATION | 190 | 643 | 2.916e-59 | 6.167e-57 |
| 23 | NEUROGENESIS | 298 | 1402 | 4.334e-58 | 8.767e-56 |
| 24 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 210 | 788 | 3.618e-57 | 6.734e-55 |
| 25 | CIRCULATORY SYSTEM DEVELOPMENT | 210 | 788 | 3.618e-57 | 6.734e-55 |
| 26 | EMBRYO DEVELOPMENT | 224 | 894 | 6.297e-56 | 1.127e-53 |
| 27 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 213 | 823 | 1.182e-55 | 2.037e-53 |
| 28 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 284 | 1360 | 1.702e-53 | 2.829e-51 |
| 29 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 288 | 1395 | 3.291e-53 | 5.281e-51 |
| 30 | CELL DEVELOPMENT | 289 | 1426 | 1.189e-51 | 1.844e-49 |
| 31 | POSITIVE REGULATION OF CELL DEATH | 173 | 605 | 1.281e-51 | 1.923e-49 |
| 32 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 235 | 1021 | 1.331e-51 | 1.935e-49 |
| 33 | CELL DEATH | 231 | 1001 | 6.681e-51 | 9.421e-49 |
| 34 | NEGATIVE REGULATION OF CELL COMMUNICATION | 257 | 1192 | 7.151e-51 | 9.787e-49 |
| 35 | RESPONSE TO ENDOGENOUS STIMULUS | 290 | 1450 | 1.348e-50 | 1.792e-48 |
| 36 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 331 | 1791 | 4.456e-50 | 5.759e-48 |
| 37 | REGULATION OF KINASE ACTIVITY | 197 | 776 | 5.902e-50 | 7.423e-48 |
| 38 | REGULATION OF CELL DEVELOPMENT | 204 | 836 | 7.993e-49 | 9.787e-47 |
| 39 | EPITHELIUM DEVELOPMENT | 219 | 945 | 1.719e-48 | 2.051e-46 |
| 40 | TUBE DEVELOPMENT | 159 | 552 | 7.31e-48 | 8.503e-46 |
| 41 | NEGATIVE REGULATION OF CELL DEATH | 207 | 872 | 1.626e-47 | 1.845e-45 |
| 42 | REGULATION OF TRANSFERASE ACTIVITY | 216 | 946 | 1.097e-46 | 1.215e-44 |
| 43 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 205 | 872 | 2.437e-46 | 2.637e-44 |
| 44 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 241 | 1135 | 2.498e-46 | 2.642e-44 |
| 45 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 329 | 1848 | 5.062e-46 | 5.234e-44 |
| 46 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 205 | 876 | 5.221e-46 | 5.281e-44 |
| 47 | RESPONSE TO ABIOTIC STIMULUS | 225 | 1024 | 1.003e-45 | 9.929e-44 |
| 48 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 189 | 771 | 1.796e-45 | 1.741e-43 |
| 49 | CELL CYCLE | 262 | 1316 | 4.783e-45 | 4.542e-43 |
| 50 | NEGATIVE REGULATION OF CELL CYCLE | 135 | 433 | 6.128e-45 | 5.703e-43 |
| 51 | ORGAN MORPHOGENESIS | 198 | 841 | 8.005e-45 | 7.303e-43 |
| 52 | CELL PROLIFERATION | 173 | 672 | 1.132e-44 | 1.013e-42 |
| 53 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 224 | 1036 | 2.689e-44 | 2.317e-42 |
| 54 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 224 | 1036 | 2.689e-44 | 2.317e-42 |
| 55 | TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 180 | 724 | 3.399e-44 | 2.876e-42 |
| 56 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 217 | 1008 | 1.488e-42 | 1.236e-40 |
| 57 | PROTEIN PHOSPHORYLATION | 208 | 944 | 2.28e-42 | 1.861e-40 |
| 58 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 188 | 801 | 2.585e-42 | 2.074e-40 |
| 59 | NEURON DIFFERENTIATION | 198 | 874 | 3.641e-42 | 2.871e-40 |
| 60 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 208 | 957 | 2.079e-41 | 1.612e-39 |
| 61 | REGULATION OF GROWTH | 161 | 633 | 7.782e-41 | 5.936e-39 |
| 62 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 178 | 750 | 9.847e-41 | 7.39e-39 |
| 63 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 169 | 689 | 1.211e-40 | 8.945e-39 |
| 64 | TISSUE MORPHOGENESIS | 144 | 533 | 8.951e-40 | 6.507e-38 |
| 65 | REGULATION OF MAPK CASCADE | 163 | 660 | 1.349e-39 | 9.655e-38 |
| 66 | REGULATION OF CELL ADHESION | 158 | 629 | 2.37e-39 | 1.671e-37 |
| 67 | CELLULAR RESPONSE TO STRESS | 280 | 1565 | 4.221e-39 | 2.932e-37 |
| 68 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 274 | 1518 | 6.253e-39 | 4.279e-37 |
| 69 | HEAD DEVELOPMENT | 169 | 709 | 6.449e-39 | 4.349e-37 |
| 70 | HEART DEVELOPMENT | 132 | 466 | 6.931e-39 | 4.607e-37 |
| 71 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 227 | 1152 | 4.717e-38 | 3.091e-36 |
| 72 | VASCULATURE DEVELOPMENT | 131 | 469 | 6.988e-38 | 4.516e-36 |
| 73 | IMMUNE SYSTEM DEVELOPMENT | 148 | 582 | 1.57e-37 | 1.001e-35 |
| 74 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 143 | 554 | 4.678e-37 | 2.941e-35 |
| 75 | LOCOMOTION | 220 | 1114 | 5.349e-37 | 3.318e-35 |
| 76 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 240 | 1275 | 6.612e-37 | 4.048e-35 |
| 77 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 194 | 917 | 7.89e-37 | 4.768e-35 |
| 78 | POSITIVE REGULATION OF CELL PROLIFERATION | 180 | 814 | 8.542e-37 | 5.096e-35 |
| 79 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 201 | 983 | 5.914e-36 | 3.483e-34 |
| 80 | RESPONSE TO HORMONE | 189 | 893 | 6.817e-36 | 3.965e-34 |
| 81 | MORPHOGENESIS OF AN EPITHELIUM | 116 | 400 | 2.639e-35 | 1.516e-33 |
| 82 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 148 | 609 | 4.205e-35 | 2.386e-33 |
| 83 | PHOSPHORYLATION | 230 | 1228 | 5.753e-35 | 3.225e-33 |
| 84 | POSITIVE REGULATION OF KINASE ACTIVITY | 128 | 482 | 1.449e-34 | 8.025e-33 |
| 85 | POSITIVE REGULATION OF CELL DEVELOPMENT | 126 | 472 | 2.791e-34 | 1.528e-32 |
| 86 | EMBRYONIC MORPHOGENESIS | 136 | 539 | 3.997e-34 | 2.162e-32 |
| 87 | CELL CYCLE PROCESS | 209 | 1081 | 1.008e-33 | 5.392e-32 |
| 88 | RESPONSE TO GROWTH FACTOR | 125 | 475 | 2.413e-33 | 1.262e-31 |
| 89 | BIOLOGICAL ADHESION | 202 | 1032 | 2.408e-33 | 1.262e-31 |
| 90 | IMMUNE SYSTEM PROCESS | 314 | 1984 | 2.496e-33 | 1.291e-31 |
| 91 | PEPTIDYL AMINO ACID MODIFICATION | 177 | 841 | 2.853e-33 | 1.459e-31 |
| 92 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 93 | 285 | 6.349e-33 | 3.211e-31 |
| 93 | GLAND DEVELOPMENT | 111 | 395 | 1.88e-32 | 9.405e-31 |
| 94 | NEURON PROJECTION DEVELOPMENT | 133 | 545 | 8.794e-32 | 4.329e-30 |
| 95 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 126 | 498 | 8.838e-32 | 4.329e-30 |
| 96 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 241 | 1381 | 1.129e-31 | 5.473e-30 |
| 97 | NEURON DEVELOPMENT | 153 | 687 | 1.213e-31 | 5.82e-30 |
| 98 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 143 | 616 | 1.262e-31 | 5.94e-30 |
| 99 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 141 | 602 | 1.264e-31 | 5.94e-30 |
| 100 | GROWTH | 112 | 410 | 1.598e-31 | 7.436e-30 |
| 101 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 121 | 470 | 2.583e-31 | 1.19e-29 |
| 102 | RESPONSE TO LIPID | 179 | 888 | 3.639e-31 | 1.66e-29 |
| 103 | UROGENITAL SYSTEM DEVELOPMENT | 93 | 299 | 4.408e-31 | 1.991e-29 |
| 104 | TUBE MORPHOGENESIS | 97 | 323 | 5.299e-31 | 2.371e-29 |
| 105 | REGULATION OF RESPONSE TO STRESS | 249 | 1468 | 9.246e-31 | 4.097e-29 |
| 106 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 103 | 363 | 1.466e-30 | 6.437e-29 |
| 107 | REGULATION OF CELLULAR RESPONSE TO STRESS | 151 | 691 | 2.847e-30 | 1.238e-28 |
| 108 | REGULATION OF CELL GROWTH | 107 | 391 | 3.175e-30 | 1.368e-28 |
| 109 | CHROMOSOME ORGANIZATION | 192 | 1009 | 5.231e-30 | 2.233e-28 |
| 110 | CHROMATIN MODIFICATION | 129 | 539 | 5.931e-30 | 2.509e-28 |
| 111 | REGULATION OF NEURON DIFFERENTIATION | 131 | 554 | 7.46e-30 | 3.127e-28 |
| 112 | CELL ACTIVATION | 133 | 568 | 7.594e-30 | 3.155e-28 |
| 113 | REPRODUCTIVE SYSTEM DEVELOPMENT | 109 | 408 | 8.76e-30 | 3.607e-28 |
| 114 | CELL MOTILITY | 169 | 835 | 1.215e-29 | 4.916e-28 |
| 115 | LOCALIZATION OF CELL | 169 | 835 | 1.215e-29 | 4.916e-28 |
| 116 | RESPONSE TO EXTERNAL STIMULUS | 286 | 1821 | 1.381e-29 | 5.541e-28 |
| 117 | CELL CELL ADHESION | 138 | 608 | 1.815e-29 | 7.218e-28 |
| 118 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 124 | 513 | 2.985e-29 | 1.177e-27 |
| 119 | REGULATION OF TRANSPORT | 283 | 1804 | 3.586e-29 | 1.402e-27 |
| 120 | NEURON PROJECTION MORPHOGENESIS | 107 | 402 | 4.185e-29 | 1.623e-27 |
| 121 | NEGATIVE REGULATION OF PHOSPHORYLATION | 110 | 422 | 4.845e-29 | 1.863e-27 |
| 122 | CELL CYCLE ARREST | 63 | 154 | 5.412e-29 | 2.064e-27 |
| 123 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 129 | 552 | 7.045e-29 | 2.665e-27 |
| 124 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 112 | 437 | 7.406e-29 | 2.779e-27 |
| 125 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 199 | 1087 | 8.279e-29 | 3.082e-27 |
| 126 | APOPTOTIC SIGNALING PATHWAY | 87 | 289 | 5.373e-28 | 1.984e-26 |
| 127 | DEVELOPMENTAL GROWTH | 94 | 333 | 8.718e-28 | 3.194e-26 |
| 128 | REGULATION OF CELL MORPHOGENESIS | 127 | 552 | 9.339e-28 | 3.395e-26 |
| 129 | REGULATION OF CELLULAR LOCALIZATION | 219 | 1277 | 1.249e-27 | 4.504e-26 |
| 130 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 114 | 465 | 1.581e-27 | 5.658e-26 |
| 131 | BLOOD VESSEL MORPHOGENESIS | 98 | 364 | 3.716e-27 | 1.32e-25 |
| 132 | RESPONSE TO STEROID HORMONE | 118 | 497 | 3.965e-27 | 1.397e-25 |
| 133 | CELLULAR COMPONENT MORPHOGENESIS | 172 | 900 | 4.181e-27 | 1.463e-25 |
| 134 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 159 | 799 | 4.696e-27 | 1.631e-25 |
| 135 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 109 | 437 | 4.948e-27 | 1.705e-25 |
| 136 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 64 | 171 | 8.115e-27 | 2.776e-25 |
| 137 | REPRODUCTION | 219 | 1297 | 1.103e-26 | 3.747e-25 |
| 138 | CELL PROJECTION ORGANIZATION | 171 | 902 | 1.564e-26 | 5.273e-25 |
| 139 | CHROMATIN ORGANIZATION | 140 | 663 | 1.604e-26 | 5.37e-25 |
| 140 | REGULATION OF ORGANELLE ORGANIZATION | 204 | 1178 | 2.854e-26 | 9.486e-25 |
| 141 | RESPONSE TO RADIATION | 104 | 413 | 3.422e-26 | 1.121e-24 |
| 142 | REGULATION OF CELL CYCLE PROCESS | 125 | 558 | 3.398e-26 | 1.121e-24 |
| 143 | REGULATION OF MITOTIC CELL CYCLE | 112 | 468 | 4.202e-26 | 1.367e-24 |
| 144 | LEUKOCYTE DIFFERENTIATION | 84 | 292 | 1.46e-25 | 4.717e-24 |
| 145 | POSITIVE REGULATION OF MAPK CASCADE | 111 | 470 | 2.303e-25 | 7.39e-24 |
| 146 | POSITIVE REGULATION OF CELL CYCLE | 90 | 332 | 2.943e-25 | 9.379e-24 |
| 147 | PEPTIDYL TYROSINE MODIFICATION | 65 | 186 | 2.975e-25 | 9.417e-24 |
| 148 | REGULATION OF DEVELOPMENTAL GROWTH | 83 | 289 | 3.224e-25 | 1.014e-23 |
| 149 | EMBRYONIC ORGAN DEVELOPMENT | 101 | 406 | 5.07e-25 | 1.583e-23 |
| 150 | LEUKOCYTE ACTIVATION | 102 | 414 | 6.607e-25 | 2.049e-23 |
| 151 | RESPONSE TO NITROGEN COMPOUND | 162 | 859 | 6.759e-25 | 2.083e-23 |
| 152 | LYMPHOCYTE ACTIVATION | 91 | 342 | 6.861e-25 | 2.1e-23 |
| 153 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 95 | 368 | 7.134e-25 | 2.169e-23 |
| 154 | SINGLE ORGANISM CELL ADHESION | 108 | 459 | 1.454e-24 | 4.392e-23 |
| 155 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 143 | 720 | 2.714e-24 | 8.148e-23 |
| 156 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 149 | 767 | 2.85e-24 | 8.5e-23 |
| 157 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 119 | 541 | 3.007e-24 | 8.855e-23 |
| 158 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 119 | 541 | 3.007e-24 | 8.855e-23 |
| 159 | RESPONSE TO ALCOHOL | 93 | 362 | 3.306e-24 | 9.675e-23 |
| 160 | REGULATION OF HYDROLASE ACTIVITY | 216 | 1327 | 3.628e-24 | 1.055e-22 |
| 161 | CELLULAR RESPONSE TO LIPID | 107 | 457 | 3.666e-24 | 1.059e-22 |
| 162 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 129 | 616 | 3.832e-24 | 1.101e-22 |
| 163 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 60 | 167 | 3.914e-24 | 1.117e-22 |
| 164 | REGULATION OF CATABOLIC PROCESS | 144 | 731 | 4.299e-24 | 1.22e-22 |
| 165 | MUSCLE STRUCTURE DEVELOPMENT | 103 | 432 | 6.025e-24 | 1.699e-22 |
| 166 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 93 | 365 | 6.301e-24 | 1.766e-22 |
| 167 | RESPONSE TO WOUNDING | 121 | 563 | 1.027e-23 | 2.86e-22 |
| 168 | IN UTERO EMBRYONIC DEVELOPMENT | 84 | 311 | 1.559e-23 | 4.319e-22 |
| 169 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 88 | 337 | 1.715e-23 | 4.723e-22 |
| 170 | CELLULAR RESPONSE TO HORMONE STIMULUS | 119 | 552 | 1.861e-23 | 5.094e-22 |
| 171 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 285 | 1977 | 2.262e-23 | 6.155e-22 |
| 172 | REGULATION OF INTRACELLULAR TRANSPORT | 128 | 621 | 2.601e-23 | 7.036e-22 |
| 173 | REGULATION OF PROTEIN LOCALIZATION | 169 | 950 | 4.547e-23 | 1.223e-21 |
| 174 | RESPONSE TO OXYGEN LEVELS | 83 | 311 | 6.791e-23 | 1.816e-21 |
| 175 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 68 | 220 | 7.264e-23 | 1.931e-21 |
| 176 | FOREBRAIN DEVELOPMENT | 90 | 357 | 7.631e-23 | 2.017e-21 |
| 177 | REGULATION OF IMMUNE SYSTEM PROCESS | 221 | 1403 | 7.83e-23 | 2.047e-21 |
| 178 | REGULATION OF CYTOPLASMIC TRANSPORT | 108 | 481 | 7.795e-23 | 2.047e-21 |
| 179 | ANGIOGENESIS | 80 | 293 | 8.523e-23 | 2.215e-21 |
| 180 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 82 | 306 | 9.295e-23 | 2.403e-21 |
| 181 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 135 | 684 | 1.042e-22 | 2.679e-21 |
| 182 | SKELETAL SYSTEM DEVELOPMENT | 104 | 455 | 1.182e-22 | 3.023e-21 |
| 183 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 64 | 199 | 1.232e-22 | 3.134e-21 |
| 184 | POSITIVE REGULATION OF CELL ADHESION | 92 | 376 | 2.424e-22 | 6.129e-21 |
| 185 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 182 | 1079 | 4.103e-22 | 1.028e-20 |
| 186 | REGULATION OF MAP KINASE ACTIVITY | 83 | 319 | 4.108e-22 | 1.028e-20 |
| 187 | LYMPHOCYTE DIFFERENTIATION | 65 | 209 | 4.472e-22 | 1.113e-20 |
| 188 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 161 | 905 | 5.348e-22 | 1.324e-20 |
| 189 | RESPONSE TO MECHANICAL STIMULUS | 65 | 210 | 5.976e-22 | 1.471e-20 |
| 190 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 76 | 279 | 1.193e-21 | 2.921e-20 |
| 191 | REGULATION OF DNA METABOLIC PROCESS | 85 | 340 | 2.209e-21 | 5.381e-20 |
| 192 | PROTEIN AUTOPHOSPHORYLATION | 61 | 192 | 2.495e-21 | 6.047e-20 |
| 193 | REGULATION OF NEURON DEATH | 71 | 252 | 3.202e-21 | 7.72e-20 |
| 194 | REGULATION OF WNT SIGNALING PATHWAY | 80 | 310 | 4.076e-21 | 9.775e-20 |
| 195 | CELL PART MORPHOGENESIS | 125 | 633 | 4.177e-21 | 9.966e-20 |
| 196 | CELL CYCLE CHECKPOINT | 61 | 194 | 4.529e-21 | 1.075e-19 |
| 197 | RHYTHMIC PROCESS | 78 | 298 | 5.03e-21 | 1.188e-19 |
| 198 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 94 | 408 | 7.832e-21 | 1.84e-19 |
| 199 | BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE | 49 | 131 | 7.884e-21 | 1.843e-19 |
| 200 | INTRINSIC APOPTOTIC SIGNALING PATHWAY | 53 | 152 | 8.766e-21 | 2.039e-19 |
| 201 | NEGATIVE REGULATION OF CELL CYCLE PROCESS | 64 | 214 | 9.481e-21 | 2.195e-19 |
| 202 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 76 | 289 | 1.194e-20 | 2.751e-19 |
| 203 | REGULATION OF VASCULATURE DEVELOPMENT | 67 | 233 | 1.258e-20 | 2.884e-19 |
| 204 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 93 | 404 | 1.35e-20 | 3.079e-19 |
| 205 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 153 | 867 | 1.394e-20 | 3.164e-19 |
| 206 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 50 | 138 | 1.609e-20 | 3.634e-19 |
| 207 | POSITIVE REGULATION OF TRANSPORT | 161 | 936 | 1.784e-20 | 4.01e-19 |
| 208 | POSITIVE REGULATION OF LOCOMOTION | 95 | 420 | 1.898e-20 | 4.245e-19 |
| 209 | REGULATION OF ORGAN MORPHOGENESIS | 68 | 242 | 2.618e-20 | 5.829e-19 |
| 210 | NEGATIVE REGULATION OF GROWTH | 67 | 236 | 2.704e-20 | 5.992e-19 |
| 211 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 64 | 218 | 2.79e-20 | 6.153e-19 |
| 212 | EPITHELIAL CELL DIFFERENTIATION | 105 | 495 | 2.951e-20 | 6.478e-19 |
| 213 | NEGATIVE REGULATION OF MAPK CASCADE | 51 | 145 | 3.05e-20 | 6.663e-19 |
| 214 | SENSORY ORGAN DEVELOPMENT | 104 | 493 | 6.893e-20 | 1.499e-18 |
| 215 | SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 47 | 127 | 8.214e-20 | 1.778e-18 |
| 216 | RESPONSE TO ESTROGEN | 63 | 218 | 1.351e-19 | 2.91e-18 |
| 217 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 105 | 505 | 1.397e-19 | 2.994e-18 |
| 218 | REGULATION OF PROTEIN IMPORT | 57 | 183 | 1.477e-19 | 3.153e-18 |
| 219 | RESPONSE TO IONIZING RADIATION | 50 | 145 | 1.91e-19 | 4.058e-18 |
| 220 | WOUND HEALING | 100 | 470 | 1.929e-19 | 4.08e-18 |
| 221 | NEGATIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 44 | 115 | 2.712e-19 | 5.71e-18 |
| 222 | RESPIRATORY SYSTEM DEVELOPMENT | 59 | 197 | 2.889e-19 | 6.054e-18 |
| 223 | REGULATION OF CELL CELL ADHESION | 87 | 380 | 3.514e-19 | 7.331e-18 |
| 224 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 67 | 247 | 3.945e-19 | 8.194e-18 |
| 225 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 64 | 229 | 4.66e-19 | 9.637e-18 |
| 226 | NEURON PROJECTION GUIDANCE | 60 | 205 | 5.037e-19 | 1.037e-17 |
| 227 | REGULATION OF HEMOPOIESIS | 77 | 314 | 5.922e-19 | 1.214e-17 |
| 228 | POSITIVE REGULATION OF PROTEOLYSIS | 84 | 363 | 7.24e-19 | 1.478e-17 |
| 229 | TAXIS | 98 | 464 | 7.673e-19 | 1.559e-17 |
| 230 | REGULATION OF ERK1 AND ERK2 CASCADE | 65 | 238 | 9.139e-19 | 1.849e-17 |
| 231 | CELLULAR RESPONSE TO EXTERNAL STIMULUS | 69 | 264 | 1.043e-18 | 2.092e-17 |
| 232 | AGING | 69 | 264 | 1.043e-18 | 2.092e-17 |
| 233 | STEM CELL DIFFERENTIATION | 57 | 190 | 1.071e-18 | 2.138e-17 |
| 234 | REGULATION OF CELL PROJECTION ORGANIZATION | 110 | 558 | 1.251e-18 | 2.487e-17 |
| 235 | RESPONSE TO ESTRADIOL | 49 | 146 | 1.611e-18 | 3.189e-17 |
| 236 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 50 | 153 | 2.605e-18 | 5.135e-17 |
| 237 | MAINTENANCE OF CELL NUMBER | 46 | 132 | 3.293e-18 | 6.465e-17 |
| 238 | NEGATIVE REGULATION OF LOCOMOTION | 68 | 263 | 3.47e-18 | 6.785e-17 |
| 239 | REGULATION OF BINDING | 71 | 283 | 3.727e-18 | 7.255e-17 |
| 240 | POSITIVE REGULATION OF GROWTH | 64 | 238 | 4.003e-18 | 7.761e-17 |
| 241 | POSITIVE REGULATION OF NEURON PROJECTION DEVELOPMENT | 63 | 232 | 4.299e-18 | 8.299e-17 |
| 242 | REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 85 | 381 | 5.021e-18 | 9.623e-17 |
| 243 | REGULATION OF OSSIFICATION | 54 | 178 | 5.025e-18 | 9.623e-17 |
| 244 | CELL FATE COMMITMENT | 62 | 227 | 5.808e-18 | 1.108e-16 |
| 245 | DNA INTEGRITY CHECKPOINT | 48 | 146 | 9.386e-18 | 1.783e-16 |
| 246 | HEART MORPHOGENESIS | 59 | 212 | 1.405e-17 | 2.657e-16 |
| 247 | NEGATIVE REGULATION OF KINASE ACTIVITY | 65 | 250 | 1.418e-17 | 2.661e-16 |
| 248 | CELLULAR RESPONSE TO ABIOTIC STIMULUS | 67 | 263 | 1.413e-17 | 2.661e-16 |
| 249 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 59 | 213 | 1.792e-17 | 3.349e-16 |
| 250 | POSITIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 49 | 154 | 1.975e-17 | 3.677e-16 |
| 251 | RESPONSE TO INORGANIC SUBSTANCE | 97 | 479 | 2.298e-17 | 4.26e-16 |
| 252 | REGULATION OF EPITHELIAL CELL MIGRATION | 51 | 166 | 2.33e-17 | 4.302e-16 |
| 253 | GLAND MORPHOGENESIS | 38 | 97 | 2.921e-17 | 5.372e-16 |
| 254 | TELENCEPHALON DEVELOPMENT | 61 | 228 | 3.2e-17 | 5.862e-16 |
| 255 | REGULATION OF CELL SUBSTRATE ADHESION | 52 | 173 | 3.235e-17 | 5.902e-16 |
| 256 | T CELL DIFFERENTIATION | 43 | 123 | 3.592e-17 | 6.529e-16 |
| 257 | POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 50 | 162 | 3.873e-17 | 7.011e-16 |
| 258 | LEUKOCYTE CELL CELL ADHESION | 65 | 255 | 4.183e-17 | 7.544e-16 |
| 259 | MUSCLE TISSUE DEVELOPMENT | 68 | 275 | 4.326e-17 | 7.771e-16 |
| 260 | NEGATIVE REGULATION OF CELL ADHESION | 60 | 223 | 4.352e-17 | 7.789e-16 |
| 261 | NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 38 | 98 | 4.401e-17 | 7.846e-16 |
| 262 | REGULATION OF LIPID METABOLIC PROCESS | 69 | 282 | 4.595e-17 | 8.16e-16 |
| 263 | REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 62 | 236 | 4.617e-17 | 8.168e-16 |
| 264 | REGULATION OF CELL ACTIVATION | 97 | 484 | 4.732e-17 | 8.34e-16 |
| 265 | REGULATION OF ENDOTHELIAL CELL MIGRATION | 41 | 114 | 6.075e-17 | 1.067e-15 |
| 266 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 80 | 360 | 6.212e-17 | 1.087e-15 |
| 267 | KIDNEY EPITHELIUM DEVELOPMENT | 43 | 125 | 7.133e-17 | 1.243e-15 |
| 268 | REGULATION OF PROTEOLYSIS | 125 | 711 | 7.84e-17 | 1.361e-15 |
| 269 | MESONEPHROS DEVELOPMENT | 36 | 90 | 9.013e-17 | 1.559e-15 |
| 270 | SEX DIFFERENTIATION | 66 | 266 | 1.045e-16 | 1.801e-15 |
| 271 | POSITIVE REGULATION OF PEPTIDASE ACTIVITY | 48 | 154 | 1.064e-16 | 1.827e-15 |
| 272 | NEGATIVE REGULATION OF IMMUNE SYSTEM PROCESS | 81 | 372 | 1.39e-16 | 2.378e-15 |
| 273 | SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE | 37 | 96 | 1.439e-16 | 2.453e-15 |
| 274 | COVALENT CHROMATIN MODIFICATION | 77 | 345 | 1.859e-16 | 3.157e-15 |
| 275 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 71 | 303 | 1.898e-16 | 3.211e-15 |
| 276 | REGULATION OF OSTEOBLAST DIFFERENTIATION | 40 | 112 | 1.927e-16 | 3.248e-15 |
| 277 | REGULATION OF CYTOSKELETON ORGANIZATION | 98 | 502 | 2.002e-16 | 3.362e-15 |
| 278 | REGULATION OF NEURON APOPTOTIC PROCESS | 54 | 192 | 2.013e-16 | 3.369e-15 |
| 279 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 107 | 573 | 2.078e-16 | 3.466e-15 |
| 280 | REGULATION OF STEM CELL DIFFERENTIATION | 40 | 113 | 2.751e-16 | 4.572e-15 |
| 281 | REGULATION OF ACTIN FILAMENT BASED PROCESS | 72 | 312 | 2.769e-16 | 4.584e-15 |
| 282 | NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 46 | 146 | 2.902e-16 | 4.789e-15 |
| 283 | MITOTIC CELL CYCLE | 130 | 766 | 3.339e-16 | 5.489e-15 |
| 284 | NEGATIVE REGULATION OF CELL GROWTH | 50 | 170 | 3.537e-16 | 5.795e-15 |
| 285 | REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 46 | 147 | 3.897e-16 | 6.362e-15 |
| 286 | RESPONSE TO DRUG | 88 | 431 | 4.323e-16 | 7.033e-15 |
| 287 | RESPONSE TO LIGHT STIMULUS | 67 | 280 | 4.402e-16 | 7.137e-15 |
| 288 | REGULATION OF MYELOID CELL DIFFERENTIATION | 52 | 183 | 4.522e-16 | 7.294e-15 |
| 289 | EPITHELIAL CELL PROLIFERATION | 35 | 89 | 4.53e-16 | 7.294e-15 |
| 290 | REGULATION OF HOMEOSTATIC PROCESS | 90 | 447 | 4.907e-16 | 7.874e-15 |
| 291 | INTERSPECIES INTERACTION BETWEEN ORGANISMS | 117 | 662 | 5.425e-16 | 8.645e-15 |
| 292 | SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM | 117 | 662 | 5.425e-16 | 8.645e-15 |
| 293 | MESENCHYME DEVELOPMENT | 53 | 190 | 5.613e-16 | 8.913e-15 |
| 294 | PATTERN SPECIFICATION PROCESS | 86 | 418 | 5.845e-16 | 9.25e-15 |
| 295 | RESPONSE TO PEPTIDE | 84 | 404 | 6.739e-16 | 1.063e-14 |
| 296 | NEGATIVE REGULATION OF WNT SIGNALING PATHWAY | 54 | 197 | 6.803e-16 | 1.066e-14 |
| 297 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 70 | 303 | 6.791e-16 | 1.066e-14 |
| 298 | POSITIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 41 | 121 | 6.865e-16 | 1.072e-14 |
| 299 | REGULATION OF PROTEIN COMPLEX ASSEMBLY | 80 | 375 | 7.343e-16 | 1.143e-14 |
| 300 | RESPONSE TO TRANSFORMING GROWTH FACTOR BETA | 45 | 144 | 8.628e-16 | 1.338e-14 |
| 301 | POSITIVE REGULATION OF DEVELOPMENTAL GROWTH | 47 | 156 | 9.724e-16 | 1.503e-14 |
| 302 | INTRACELLULAR RECEPTOR SIGNALING PATHWAY | 49 | 168 | 1.012e-15 | 1.56e-14 |
| 303 | MITOTIC CELL CYCLE CHECKPOINT | 44 | 139 | 1.059e-15 | 1.626e-14 |
| 304 | APPENDAGE DEVELOPMENT | 49 | 169 | 1.315e-15 | 2.006e-14 |
| 305 | LIMB DEVELOPMENT | 49 | 169 | 1.315e-15 | 2.006e-14 |
| 306 | REGULATION OF PROTEIN TARGETING | 70 | 307 | 1.393e-15 | 2.118e-14 |
| 307 | REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 55 | 207 | 1.608e-15 | 2.438e-14 |
| 308 | REGULATION OF MUSCLE CELL DIFFERENTIATION | 46 | 152 | 1.625e-15 | 2.455e-14 |
| 309 | POSITIVE REGULATION OF CATABOLIC PROCESS | 82 | 395 | 1.652e-15 | 2.488e-14 |
| 310 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 76 | 351 | 1.689e-15 | 2.536e-14 |
| 311 | INOSITOL LIPID MEDIATED SIGNALING | 41 | 124 | 1.819e-15 | 2.722e-14 |
| 312 | MYELOID CELL DIFFERENTIATION | 52 | 189 | 1.981e-15 | 2.955e-14 |
| 313 | REGULATION OF LIPID KINASE ACTIVITY | 25 | 48 | 2.337e-15 | 3.474e-14 |
| 314 | CELLULAR RESPONSE TO OXIDATIVE STRESS | 51 | 184 | 2.653e-15 | 3.931e-14 |
| 315 | CYTOSKELETON ORGANIZATION | 136 | 838 | 2.718e-15 | 4.015e-14 |
| 316 | DEVELOPMENT OF PRIMARY SEXUAL CHARACTERISTICS | 56 | 216 | 2.858e-15 | 4.209e-14 |
| 317 | CELLULAR RESPONSE TO OXYGEN LEVELS | 44 | 143 | 3.402e-15 | 4.993e-14 |
| 318 | RESPONSE TO UV | 41 | 126 | 3.416e-15 | 4.998e-14 |
| 319 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 26 | 53 | 4.267e-15 | 6.224e-14 |
| 320 | CELLULAR RESPONSE TO STEROID HORMONE STIMULUS | 56 | 218 | 4.412e-15 | 6.415e-14 |
| 321 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 69 | 307 | 4.809e-15 | 6.97e-14 |
| 322 | REGULATION OF CELL MATRIX ADHESION | 34 | 90 | 4.955e-15 | 7.159e-14 |
| 323 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 30 | 71 | 5.557e-15 | 8.006e-14 |
| 324 | POSITIVE REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 28 | 62 | 5.582e-15 | 8.016e-14 |
| 325 | HOMEOSTASIS OF NUMBER OF CELLS | 49 | 175 | 6.019e-15 | 8.617e-14 |
| 326 | MAMMARY GLAND DEVELOPMENT | 39 | 117 | 6.503e-15 | 9.282e-14 |
| 327 | REGULATION OF DNA TEMPLATED TRANSCRIPTION IN RESPONSE TO STRESS | 29 | 67 | 7.292e-15 | 1.038e-13 |
| 328 | REGULATION OF IMMUNE RESPONSE | 137 | 858 | 7.998e-15 | 1.135e-13 |
| 329 | POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS | 82 | 406 | 8.442e-15 | 1.194e-13 |
| 330 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 77 | 370 | 1.06e-14 | 1.495e-13 |
| 331 | RESPONSE TO ACID CHEMICAL | 70 | 319 | 1.098e-14 | 1.544e-13 |
| 332 | REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 52 | 197 | 1.271e-14 | 1.781e-13 |
| 333 | DIGESTIVE SYSTEM DEVELOPMENT | 44 | 148 | 1.366e-14 | 1.909e-13 |
| 334 | REGULATION OF CELL CYCLE PHASE TRANSITION | 70 | 321 | 1.53e-14 | 2.132e-13 |
| 335 | OSSIFICATION | 60 | 251 | 1.6e-14 | 2.222e-13 |
| 336 | REGULATION OF LEUKOCYTE DIFFERENTIATION | 57 | 232 | 2.052e-14 | 2.842e-13 |
| 337 | PROTEIN COMPLEX SUBUNIT ORGANIZATION | 208 | 1527 | 2.416e-14 | 3.336e-13 |
| 338 | NEGATIVE REGULATION OF ERK1 AND ERK2 CASCADE | 25 | 52 | 2.618e-14 | 3.603e-13 |
| 339 | GLIOGENESIS | 48 | 175 | 2.707e-14 | 3.715e-13 |
| 340 | EYE DEVELOPMENT | 70 | 326 | 3.453e-14 | 4.726e-13 |
| 341 | POSITIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 56 | 228 | 3.524e-14 | 4.808e-13 |
| 342 | MUSCLE ORGAN DEVELOPMENT | 63 | 277 | 4.189e-14 | 5.699e-13 |
| 343 | RESPONSE TO OXIDATIVE STRESS | 73 | 352 | 6.335e-14 | 8.593e-13 |
| 344 | GLIAL CELL DIFFERENTIATION | 41 | 136 | 6.415e-14 | 8.678e-13 |
| 345 | AMEBOIDAL TYPE CELL MIGRATION | 44 | 154 | 6.598e-14 | 8.899e-13 |
| 346 | POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION | 35 | 103 | 8.195e-14 | 1.102e-12 |
| 347 | REGENERATION | 45 | 161 | 8.235e-14 | 1.104e-12 |
| 348 | NEURAL TUBE DEVELOPMENT | 43 | 149 | 8.636e-14 | 1.155e-12 |
| 349 | RESPONSE TO GAMMA RADIATION | 24 | 50 | 9.022e-14 | 1.203e-12 |
| 350 | REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 53 | 213 | 9.712e-14 | 1.291e-12 |
| 351 | REGULATION OF PROTEIN CATABOLIC PROCESS | 78 | 393 | 1.016e-13 | 1.347e-12 |
| 352 | REGULATION OF DNA BINDING | 33 | 93 | 1.02e-13 | 1.349e-12 |
| 353 | REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 45 | 162 | 1.052e-13 | 1.387e-12 |
| 354 | POSITIVE REGULATION OF MAP KINASE ACTIVITY | 52 | 207 | 1.104e-13 | 1.451e-12 |
| 355 | CARDIAC CHAMBER DEVELOPMENT | 42 | 144 | 1.123e-13 | 1.472e-12 |
| 356 | PEPTIDYL LYSINE MODIFICATION | 67 | 312 | 1.222e-13 | 1.597e-12 |
| 357 | RESPONSE TO EXTRACELLULAR STIMULUS | 84 | 441 | 1.244e-13 | 1.621e-12 |
| 358 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 93 | 514 | 1.451e-13 | 1.886e-12 |
| 359 | REGULATION OF CHROMOSOME ORGANIZATION | 62 | 278 | 1.706e-13 | 2.212e-12 |
| 360 | REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 34 | 100 | 1.818e-13 | 2.35e-12 |
| 361 | EMBRYONIC ORGAN MORPHOGENESIS | 62 | 279 | 2.024e-13 | 2.608e-12 |
| 362 | ENDOCRINE SYSTEM DEVELOPMENT | 38 | 123 | 2.234e-13 | 2.872e-12 |
| 363 | PALLIUM DEVELOPMENT | 43 | 153 | 2.378e-13 | 3.048e-12 |
| 364 | CENTRAL NERVOUS SYSTEM NEURON DIFFERENTIATION | 45 | 166 | 2.736e-13 | 3.497e-12 |
| 365 | REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 26 | 61 | 2.813e-13 | 3.586e-12 |
| 366 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 21 | 40 | 3.318e-13 | 4.218e-12 |
| 367 | DNA METABOLIC PROCESS | 121 | 758 | 3.339e-13 | 4.233e-12 |
| 368 | POSITIVE REGULATION OF IMMUNE RESPONSE | 98 | 563 | 3.649e-13 | 4.614e-12 |
| 369 | REGULATION OF FIBROBLAST PROLIFERATION | 30 | 81 | 3.66e-13 | 4.615e-12 |
| 370 | MULTICELLULAR ORGANISM REPRODUCTION | 122 | 768 | 3.754e-13 | 4.721e-12 |
| 371 | NEGATIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 50 | 200 | 3.998e-13 | 5.014e-12 |
| 372 | REGULATION OF CELL CYCLE ARREST | 35 | 108 | 4.067e-13 | 5.087e-12 |
| 373 | CELLULAR RESPONSE TO RADIATION | 40 | 137 | 4.15e-13 | 5.177e-12 |
| 374 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 59 | 262 | 4.244e-13 | 5.28e-12 |
| 375 | REGULATION OF AXONOGENESIS | 45 | 168 | 4.353e-13 | 5.401e-12 |
| 376 | NEGATIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 44 | 162 | 4.67e-13 | 5.779e-12 |
| 377 | POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 27 | 67 | 5.074e-13 | 6.246e-12 |
| 378 | CELL AGING | 27 | 67 | 5.074e-13 | 6.246e-12 |
| 379 | B CELL ACTIVATION | 39 | 132 | 5.354e-13 | 6.574e-12 |
| 380 | PLACENTA DEVELOPMENT | 40 | 138 | 5.392e-13 | 6.602e-12 |
| 381 | REGULATION OF CELLULAR COMPONENT SIZE | 69 | 337 | 5.896e-13 | 7.201e-12 |
| 382 | POSITIVE REGULATION OF CELL CELL ADHESION | 56 | 243 | 6.118e-13 | 7.432e-12 |
| 383 | NEPHRON DEVELOPMENT | 36 | 115 | 6.115e-13 | 7.432e-12 |
| 384 | PROSTATE GLAND DEVELOPMENT | 21 | 41 | 6.281e-13 | 7.611e-12 |
| 385 | POSITIVE REGULATION OF PROTEIN IMPORT | 34 | 104 | 6.609e-13 | 7.987e-12 |
| 386 | REGULATION OF STEM CELL PROLIFERATION | 31 | 88 | 7.116e-13 | 8.578e-12 |
| 387 | MUSCLE CELL DIFFERENTIATION | 55 | 237 | 7.276e-13 | 8.748e-12 |
| 388 | FORMATION OF PRIMARY GERM LAYER | 35 | 110 | 7.48e-13 | 8.97e-12 |
| 389 | G1 DNA DAMAGE CHECKPOINT | 28 | 73 | 8.099e-13 | 9.688e-12 |
| 390 | REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 42 | 152 | 8.429e-13 | 1.006e-11 |
| 391 | MORPHOGENESIS OF EMBRYONIC EPITHELIUM | 39 | 134 | 9.096e-13 | 1.08e-11 |
| 392 | MESENCHYMAL CELL DIFFERENTIATION | 39 | 134 | 9.096e-13 | 1.08e-11 |
| 393 | POSITIVE REGULATION OF CELL ACTIVATION | 65 | 311 | 1.048e-12 | 1.241e-11 |
| 394 | REGULATION OF CELL SIZE | 45 | 172 | 1.074e-12 | 1.268e-11 |
| 395 | TUBE FORMATION | 38 | 129 | 1.176e-12 | 1.385e-11 |
| 396 | ACTIVATION OF IMMUNE RESPONSE | 80 | 427 | 1.184e-12 | 1.391e-11 |
| 397 | POSITIVE REGULATION OF CELL GROWTH | 41 | 148 | 1.435e-12 | 1.682e-11 |
| 398 | PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 132 | 873 | 1.559e-12 | 1.823e-11 |
| 399 | POSITIVE REGULATION OF CELL CYCLE ARREST | 30 | 85 | 1.576e-12 | 1.838e-11 |
| 400 | GASTRULATION | 42 | 155 | 1.721e-12 | 2.001e-11 |
| 401 | CONNECTIVE TISSUE DEVELOPMENT | 48 | 194 | 1.771e-12 | 2.055e-11 |
| 402 | REGULATION OF CELL DIVISION | 59 | 272 | 2.289e-12 | 2.649e-11 |
| 403 | RESPONSE TO HYDROGEN PEROXIDE | 34 | 109 | 2.992e-12 | 3.455e-11 |
| 404 | REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 32 | 98 | 3.252e-12 | 3.746e-11 |
| 405 | NEGATIVE REGULATION OF NEURON DEATH | 44 | 171 | 3.547e-12 | 4.075e-11 |
| 406 | POSITIVE REGULATION OF NEURON DEATH | 26 | 67 | 3.863e-12 | 4.427e-11 |
| 407 | REGULATION OF MEMBRANE POTENTIAL | 68 | 343 | 4.16e-12 | 4.756e-11 |
| 408 | POSITIVE REGULATION OF DNA METABOLIC PROCESS | 46 | 185 | 4.251e-12 | 4.848e-11 |
| 409 | REGULATION OF JNK CASCADE | 42 | 159 | 4.31e-12 | 4.903e-11 |
| 410 | REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 36 | 122 | 4.34e-12 | 4.925e-11 |
| 411 | REGULATION OF I KAPPAB KINASE NF KAPPAB SIGNALING | 53 | 233 | 4.36e-12 | 4.936e-11 |
| 412 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 32 | 99 | 4.435e-12 | 5.009e-11 |
| 413 | NEGATIVE REGULATION OF DNA METABOLIC PROCESS | 34 | 111 | 5.316e-12 | 5.989e-11 |
| 414 | MITOTIC DNA INTEGRITY CHECKPOINT | 32 | 100 | 6.018e-12 | 6.764e-11 |
| 415 | CARDIAC VENTRICLE DEVELOPMENT | 33 | 106 | 6.618e-12 | 7.42e-11 |
| 416 | ACTIVATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 31 | 95 | 7.216e-12 | 8.071e-11 |
| 417 | ERBB SIGNALING PATHWAY | 28 | 79 | 7.729e-12 | 8.624e-11 |
| 418 | CELLULAR RESPONSE TO EXTRACELLULAR STIMULUS | 46 | 188 | 7.785e-12 | 8.666e-11 |
| 419 | REGULATION OF INNATE IMMUNE RESPONSE | 69 | 357 | 9.84e-12 | 1.093e-10 |
| 420 | MACROMOLECULAR COMPLEX ASSEMBLY | 185 | 1398 | 1.052e-11 | 1.165e-10 |
| 421 | POSITIVE REGULATION OF CYTOPLASMIC TRANSPORT | 59 | 282 | 1.115e-11 | 1.232e-10 |
| 422 | TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 46 | 190 | 1.155e-11 | 1.274e-10 |
| 423 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 135 | 926 | 1.233e-11 | 1.356e-10 |
| 424 | REGULATION OF ANATOMICAL STRUCTURE SIZE | 83 | 472 | 1.415e-11 | 1.553e-10 |
| 425 | HORMONE MEDIATED SIGNALING PATHWAY | 41 | 158 | 1.436e-11 | 1.572e-10 |
| 426 | POSITIVE REGULATION OF VASCULATURE DEVELOPMENT | 37 | 133 | 1.533e-11 | 1.675e-10 |
| 427 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 35 | 121 | 1.611e-11 | 1.752e-10 |
| 428 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 124 | 829 | 1.611e-11 | 1.752e-10 |
| 429 | CELL JUNCTION ORGANIZATION | 45 | 185 | 1.627e-11 | 1.765e-10 |
| 430 | IMMUNE RESPONSE REGULATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 64 | 323 | 1.828e-11 | 1.979e-10 |
| 431 | CELLULAR RESPONSE TO HYDROGEN PEROXIDE | 24 | 61 | 1.833e-11 | 1.979e-10 |
| 432 | HOMOPHILIC CELL ADHESION VIA PLASMA MEMBRANE ADHESION MOLECULES | 40 | 153 | 1.973e-11 | 2.125e-10 |
| 433 | NEGATIVE REGULATION OF HEMOPOIESIS | 36 | 128 | 2.037e-11 | 2.189e-10 |
| 434 | DEVELOPMENTAL MATURATION | 46 | 193 | 2.063e-11 | 2.212e-10 |
| 435 | KIDNEY MORPHOGENESIS | 28 | 82 | 2.17e-11 | 2.321e-10 |
| 436 | NEGATIVE REGULATION OF ORGANELLE ORGANIZATION | 72 | 387 | 2.259e-11 | 2.411e-10 |
| 437 | REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 31 | 99 | 2.419e-11 | 2.57e-10 |
| 438 | REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 31 | 99 | 2.419e-11 | 2.57e-10 |
| 439 | ANTERIOR POSTERIOR PATTERN SPECIFICATION | 46 | 194 | 2.495e-11 | 2.644e-10 |
| 440 | RESPONSE TO METAL ION | 65 | 333 | 2.517e-11 | 2.662e-10 |
| 441 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 36 | 129 | 2.607e-11 | 2.751e-10 |
| 442 | PEPTIDYL SERINE MODIFICATION | 39 | 148 | 2.703e-11 | 2.845e-10 |
| 443 | REGULATION OF DNA REPLICATION | 41 | 161 | 2.741e-11 | 2.878e-10 |
| 444 | REGIONALIZATION | 62 | 311 | 2.892e-11 | 3.031e-10 |
| 445 | EAR DEVELOPMENT | 46 | 195 | 3.012e-11 | 3.15e-10 |
| 446 | RENAL TUBULE DEVELOPMENT | 27 | 78 | 3.435e-11 | 3.584e-10 |
| 447 | WNT SIGNALING PATHWAY | 67 | 351 | 3.496e-11 | 3.639e-10 |
| 448 | MESODERM DEVELOPMENT | 34 | 118 | 3.525e-11 | 3.661e-10 |
| 449 | INTRINSIC APOPTOTIC SIGNALING PATHWAY BY P53 CLASS MEDIATOR | 22 | 53 | 3.601e-11 | 3.732e-10 |
| 450 | BONE DEVELOPMENT | 40 | 156 | 3.794e-11 | 3.923e-10 |
| 451 | REGULATION OF BODY FLUID LEVELS | 86 | 506 | 3.808e-11 | 3.929e-10 |
| 452 | CIRCADIAN RHYTHM | 37 | 137 | 3.98e-11 | 4.097e-10 |
| 453 | ORGAN GROWTH | 25 | 68 | 4.037e-11 | 4.147e-10 |
| 454 | SINGLE ORGANISM BEHAVIOR | 71 | 384 | 4.181e-11 | 4.273e-10 |
| 455 | REGULATION OF PEPTIDASE ACTIVITY | 72 | 392 | 4.187e-11 | 4.273e-10 |
| 456 | POSITIVE REGULATION OF OSSIFICATION | 28 | 84 | 4.185e-11 | 4.273e-10 |
| 457 | POSITIVE REGULATION OF PROTEIN COMPLEX ASSEMBLY | 46 | 197 | 4.37e-11 | 4.449e-10 |
| 458 | RESPONSE TO NUTRIENT | 45 | 191 | 5.154e-11 | 5.236e-10 |
| 459 | NEGATIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 35 | 126 | 5.661e-11 | 5.739e-10 |
| 460 | REGULATION OF STRIATED MUSCLE CELL DIFFERENTIATION | 28 | 85 | 5.761e-11 | 5.827e-10 |
| 461 | REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 38 | 145 | 5.783e-11 | 5.837e-10 |
| 462 | REGULATION OF CHROMATIN ORGANIZATION | 39 | 152 | 6.502e-11 | 6.549e-10 |
| 463 | REGULATION OF CYTOKINE PRODUCTION | 92 | 563 | 7.06e-11 | 7.095e-10 |
| 464 | EPITHELIAL CELL DEVELOPMENT | 44 | 186 | 7.31e-11 | 7.33e-10 |
| 465 | CELLULAR RESPONSE TO ALCOHOL | 33 | 115 | 7.707e-11 | 7.712e-10 |
| 466 | REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 57 | 280 | 7.738e-11 | 7.726e-10 |
| 467 | CARDIAC MUSCLE TISSUE DEVELOPMENT | 37 | 140 | 7.913e-11 | 7.884e-10 |
| 468 | TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER | 39 | 153 | 8.052e-11 | 8.005e-10 |
| 469 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 22 | 55 | 8.637e-11 | 8.551e-10 |
| 470 | REGULATION OF KIDNEY DEVELOPMENT | 22 | 55 | 8.637e-11 | 8.551e-10 |
| 471 | POSITIVE REGULATION OF OSTEOBLAST DIFFERENTIATION | 23 | 60 | 9.026e-11 | 8.916e-10 |
| 472 | PROTEIN LOCALIZATION | 222 | 1805 | 9.341e-11 | 9.208e-10 |
| 473 | RESPONSE TO VITAMIN | 30 | 98 | 9.516e-11 | 9.361e-10 |
| 474 | CARDIAC CHAMBER MORPHOGENESIS | 31 | 104 | 9.917e-11 | 9.735e-10 |
| 475 | MULTICELLULAR ORGANISM GROWTH | 26 | 76 | 1.07e-10 | 1.048e-09 |
| 476 | MALE SEX DIFFERENTIATION | 38 | 148 | 1.114e-10 | 1.089e-09 |
| 477 | NEGATIVE REGULATION OF TRANSPORT | 79 | 458 | 1.162e-10 | 1.133e-09 |
| 478 | NEPHRON EPITHELIUM DEVELOPMENT | 29 | 93 | 1.185e-10 | 1.153e-09 |
| 479 | LIPID PHOSPHORYLATION | 30 | 99 | 1.26e-10 | 1.224e-09 |
| 480 | SOMATIC STEM CELL POPULATION MAINTENANCE | 24 | 66 | 1.292e-10 | 1.253e-09 |
| 481 | CEREBRAL CORTEX DEVELOPMENT | 31 | 105 | 1.299e-10 | 1.256e-09 |
| 482 | COLUMNAR CUBOIDAL EPITHELIAL CELL DIFFERENTIATION | 32 | 111 | 1.303e-10 | 1.258e-09 |
| 483 | NEGATIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 36 | 136 | 1.354e-10 | 1.304e-09 |
| 484 | SKIN DEVELOPMENT | 47 | 211 | 1.51e-10 | 1.452e-09 |
| 485 | PLATELET DERIVED GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 17 | 34 | 1.672e-10 | 1.601e-09 |
| 486 | REGULATION OF MESENCHYMAL CELL PROLIFERATION | 17 | 34 | 1.672e-10 | 1.601e-09 |
| 487 | HINDBRAIN DEVELOPMENT | 36 | 137 | 1.694e-10 | 1.619e-09 |
| 488 | POSITIVE REGULATION OF CHROMOSOME ORGANIZATION | 38 | 150 | 1.705e-10 | 1.626e-09 |
| 489 | HOMEOSTATIC PROCESS | 174 | 1337 | 1.792e-10 | 1.705e-09 |
| 490 | RESPONSE TO CYTOKINE | 108 | 714 | 1.8e-10 | 1.709e-09 |
| 491 | RESPONSE TO REACTIVE OXYGEN SPECIES | 44 | 191 | 1.838e-10 | 1.742e-09 |
| 492 | REGULATION OF METAL ION TRANSPORT | 62 | 325 | 1.937e-10 | 1.831e-09 |
| 493 | ENDOTHELIAL CELL MIGRATION | 22 | 57 | 1.976e-10 | 1.864e-09 |
| 494 | STEROID HORMONE MEDIATED SIGNALING PATHWAY | 34 | 125 | 1.979e-10 | 1.864e-09 |
| 495 | REGULATION OF METANEPHROS DEVELOPMENT | 14 | 23 | 2.117e-10 | 1.99e-09 |
| 496 | REGULATION OF EXTENT OF CELL GROWTH | 30 | 101 | 2.182e-10 | 2.046e-09 |
| 497 | REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS | 37 | 145 | 2.365e-10 | 2.214e-09 |
| 498 | POSITIVE REGULATION OF MUSCLE CELL DIFFERENTIATION | 27 | 84 | 2.381e-10 | 2.225e-09 |
| 499 | POSITIVE REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 53 | 258 | 2.516e-10 | 2.346e-09 |
| 500 | POSITIVE REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 24 | 68 | 2.653e-10 | 2.469e-09 |
| 501 | MAMMARY GLAND EPITHELIUM DEVELOPMENT | 21 | 53 | 2.859e-10 | 2.645e-09 |
| 502 | MESONEPHRIC TUBULE MORPHOGENESIS | 21 | 53 | 2.859e-10 | 2.645e-09 |
| 503 | REGULATION OF MYELOID LEUKOCYTE DIFFERENTIATION | 31 | 108 | 2.849e-10 | 2.645e-09 |
| 504 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 11 | 14 | 2.873e-10 | 2.653e-09 |
| 505 | CARDIAC SEPTUM DEVELOPMENT | 27 | 85 | 3.221e-10 | 2.968e-09 |
| 506 | BRANCHING INVOLVED IN URETERIC BUD MORPHOGENESIS | 19 | 44 | 3.446e-10 | 3.169e-09 |
| 507 | CARTILAGE DEVELOPMENT | 37 | 147 | 3.605e-10 | 3.308e-09 |
| 508 | CELLULAR RESPONSE TO MECHANICAL STIMULUS | 26 | 80 | 3.886e-10 | 3.559e-09 |
| 509 | PHOSPHATIDYLINOSITOL 3 PHOSPHATE BIOSYNTHETIC PROCESS | 20 | 49 | 4.033e-10 | 3.687e-09 |
| 510 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 27 | 86 | 4.334e-10 | 3.954e-09 |
| 511 | RESPONSE TO KETONE | 42 | 182 | 4.474e-10 | 4.074e-09 |
| 512 | NEGATIVE REGULATION OF NEURON APOPTOTIC PROCESS | 35 | 135 | 4.512e-10 | 4.1e-09 |
| 513 | FEMALE SEX DIFFERENTIATION | 32 | 116 | 4.532e-10 | 4.111e-09 |
| 514 | CELLULAR RESPONSE TO REACTIVE OXYGEN SPECIES | 30 | 104 | 4.819e-10 | 4.362e-09 |
| 515 | POSITIVE REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 39 | 162 | 5.008e-10 | 4.525e-09 |
| 516 | REGULATION OF GTPASE ACTIVITY | 102 | 673 | 5.254e-10 | 4.738e-09 |
| 517 | METANEPHROS DEVELOPMENT | 26 | 81 | 5.283e-10 | 4.755e-09 |
| 518 | SPROUTING ANGIOGENESIS | 19 | 45 | 5.511e-10 | 4.95e-09 |
| 519 | REGULATION OF SYSTEM PROCESS | 83 | 507 | 5.588e-10 | 5.01e-09 |
| 520 | PROTEIN COMPLEX BIOGENESIS | 151 | 1132 | 5.73e-10 | 5.118e-09 |
| 521 | PROTEIN COMPLEX ASSEMBLY | 151 | 1132 | 5.73e-10 | 5.118e-09 |
| 522 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER IN RESPONSE TO HYPOXIA | 16 | 32 | 5.807e-10 | 5.176e-09 |
| 523 | BEHAVIOR | 84 | 516 | 5.822e-10 | 5.18e-09 |
| 524 | POSITIVE REGULATION OF HEMOPOIESIS | 39 | 163 | 6.074e-10 | 5.394e-09 |
| 525 | POSITIVE REGULATION OF STEM CELL DIFFERENTIATION | 20 | 50 | 6.209e-10 | 5.503e-09 |
| 526 | EPIDERMAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 21 | 55 | 6.457e-10 | 5.712e-09 |
| 527 | ACTIN FILAMENT BASED PROCESS | 76 | 450 | 7.125e-10 | 6.291e-09 |
| 528 | ENDODERM DEVELOPMENT | 24 | 71 | 7.362e-10 | 6.488e-09 |
| 529 | CELL MATURATION | 34 | 131 | 7.729e-10 | 6.786e-09 |
| 530 | ZYMOGEN ACTIVATION | 31 | 112 | 7.718e-10 | 6.786e-09 |
| 531 | REGULATION OF PROTEIN STABILITY | 47 | 221 | 7.8e-10 | 6.835e-09 |
| 532 | CELLULAR RESPONSE TO UV | 23 | 66 | 8.342e-10 | 7.282e-09 |
| 533 | LENS DEVELOPMENT IN CAMERA TYPE EYE | 23 | 66 | 8.342e-10 | 7.282e-09 |
| 534 | POSITIVE REGULATION OF STEM CELL PROLIFERATION | 22 | 61 | 9.122e-10 | 7.948e-09 |
| 535 | EMBRYONIC PLACENTA DEVELOPMENT | 26 | 83 | 9.602e-10 | 8.351e-09 |
| 536 | SKELETAL SYSTEM MORPHOGENESIS | 44 | 201 | 1.037e-09 | 9e-09 |
| 537 | TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 28 | 95 | 1.054e-09 | 9.135e-09 |
| 538 | NEGATIVE REGULATION OF MYELOID CELL DIFFERENTIATION | 26 | 84 | 1.284e-09 | 1.11e-08 |
| 539 | REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS | 41 | 181 | 1.303e-09 | 1.125e-08 |
| 540 | POSITIVE REGULATION OF NEURON APOPTOTIC PROCESS | 19 | 47 | 1.345e-09 | 1.159e-08 |
| 541 | REGULATION OF SYNAPSE STRUCTURE OR ACTIVITY | 48 | 232 | 1.363e-09 | 1.172e-08 |
| 542 | CELLULAR RESPONSE TO IONIZING RADIATION | 20 | 52 | 1.417e-09 | 1.212e-08 |
| 543 | POSITIVE REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 20 | 52 | 1.417e-09 | 1.212e-08 |
| 544 | ACTIVATION OF MAPKK ACTIVITY | 20 | 52 | 1.417e-09 | 1.212e-08 |
| 545 | REGULATION OF DEFENSE RESPONSE | 110 | 759 | 1.522e-09 | 1.3e-08 |
| 546 | CELLULAR RESPONSE TO ACID CHEMICAL | 40 | 175 | 1.56e-09 | 1.329e-08 |
| 547 | ACTIVATION OF INNATE IMMUNE RESPONSE | 44 | 204 | 1.694e-09 | 1.439e-08 |
| 548 | CELL CELL ADHESION VIA PLASMA MEMBRANE ADHESION MOLECULES | 44 | 204 | 1.694e-09 | 1.439e-08 |
| 549 | REGULATION OF PROTEASOMAL UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 36 | 148 | 1.704e-09 | 1.444e-08 |
| 550 | RESPONSE TO X RAY | 15 | 30 | 2.02e-09 | 1.709e-08 |
| 551 | EXTRACELLULAR STRUCTURE ORGANIZATION | 57 | 304 | 2.034e-09 | 1.717e-08 |
| 552 | REGULATION OF MORPHOGENESIS OF A BRANCHING STRUCTURE | 20 | 53 | 2.102e-09 | 1.772e-08 |
| 553 | POSITIVE REGULATION OF MITOTIC CELL CYCLE | 32 | 123 | 2.268e-09 | 1.905e-08 |
| 554 | POSITIVE REGULATION OF AXONOGENESIS | 23 | 69 | 2.268e-09 | 1.905e-08 |
| 555 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 28 | 98 | 2.304e-09 | 1.932e-08 |
| 556 | REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 53 | 274 | 2.407e-09 | 2.011e-08 |
| 557 | CELLULAR RESPONSE TO PEPTIDE | 53 | 274 | 2.407e-09 | 2.011e-08 |
| 558 | NEGATIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 31 | 117 | 2.487e-09 | 2.074e-08 |
| 559 | REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 63 | 354 | 2.544e-09 | 2.117e-08 |
| 560 | EPIDERMIS DEVELOPMENT | 50 | 253 | 3.205e-09 | 2.663e-08 |
| 561 | SYNAPSE ORGANIZATION | 35 | 145 | 3.552e-09 | 2.946e-08 |
| 562 | POSITIVE REGULATION OF MESENCHYMAL CELL PROLIFERATION | 14 | 27 | 3.804e-09 | 3.144e-08 |
| 563 | POSITIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 28 | 100 | 3.803e-09 | 3.144e-08 |
| 564 | MAMMARY GLAND MORPHOGENESIS | 17 | 40 | 3.964e-09 | 3.27e-08 |
| 565 | CELL CYCLE PHASE TRANSITION | 50 | 255 | 4.225e-09 | 3.479e-08 |
| 566 | NEGATIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 23 | 71 | 4.261e-09 | 3.503e-08 |
| 567 | NEGATIVE REGULATION OF CATABOLIC PROCESS | 43 | 203 | 4.617e-09 | 3.789e-08 |
| 568 | REGULATION OF ION TRANSPORT | 90 | 592 | 4.804e-09 | 3.935e-08 |
| 569 | SEGMENTATION | 26 | 89 | 5.087e-09 | 4.16e-08 |
| 570 | IMMUNE RESPONSE | 144 | 1100 | 5.214e-09 | 4.256e-08 |
| 571 | POSITIVE REGULATION OF BINDING | 32 | 127 | 5.343e-09 | 4.354e-08 |
| 572 | REGULATION OF PROTEIN BINDING | 38 | 168 | 5.412e-09 | 4.403e-08 |
| 573 | DNA REPAIR | 77 | 480 | 5.953e-09 | 4.834e-08 |
| 574 | NEGATIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 28 | 102 | 6.181e-09 | 5.011e-08 |
| 575 | NEGATIVE REGULATION OF STRESS ACTIVATED MAPK CASCADE | 17 | 41 | 6.259e-09 | 5.056e-08 |
| 576 | NEGATIVE REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 17 | 41 | 6.259e-09 | 5.056e-08 |
| 577 | TISSUE MIGRATION | 25 | 84 | 6.556e-09 | 5.287e-08 |
| 578 | POSITIVE REGULATION OF LIPID METABOLIC PROCESS | 32 | 128 | 6.575e-09 | 5.293e-08 |
| 579 | REGULATION OF MICROTUBULE BASED PROCESS | 48 | 243 | 6.695e-09 | 5.38e-08 |
| 580 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 18 | 46 | 6.798e-09 | 5.454e-08 |
| 581 | REGULATION OF EPITHELIAL TO MESENCHYMAL TRANSITION | 22 | 67 | 6.939e-09 | 5.557e-08 |
| 582 | NEGATIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 33 | 135 | 7.039e-09 | 5.627e-08 |
| 583 | REGULATION OF LEUKOCYTE PROLIFERATION | 43 | 206 | 7.348e-09 | 5.854e-08 |
| 584 | FC RECEPTOR SIGNALING PATHWAY | 43 | 206 | 7.348e-09 | 5.854e-08 |
| 585 | PLATELET ACTIVATION | 34 | 142 | 7.385e-09 | 5.874e-08 |
| 586 | REGULATION OF ORGAN GROWTH | 23 | 73 | 7.797e-09 | 6.191e-08 |
| 587 | REGULATION OF CIRCADIAN RHYTHM | 28 | 103 | 7.837e-09 | 6.212e-08 |
| 588 | PROTEOLYSIS | 154 | 1208 | 9.175e-09 | 7.26e-08 |
| 589 | POSITIVE REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 17 | 42 | 9.72e-09 | 7.679e-08 |
| 590 | POSITIVE REGULATION OF INNATE IMMUNE RESPONSE | 48 | 246 | 1.011e-08 | 7.975e-08 |
| 591 | IMMUNE EFFECTOR PROCESS | 77 | 486 | 1.038e-08 | 8.176e-08 |
| 592 | CELL DIVISION | 74 | 460 | 1.042e-08 | 8.193e-08 |
| 593 | POSITIVE REGULATION OF I KAPPAB KINASE NF KAPPAB SIGNALING | 39 | 179 | 1.045e-08 | 8.198e-08 |
| 594 | REGULATION OF RESPONSE TO CYTOKINE STIMULUS | 34 | 144 | 1.08e-08 | 8.462e-08 |
| 595 | SENSORY ORGAN MORPHOGENESIS | 47 | 239 | 1.111e-08 | 8.69e-08 |
| 596 | NEGATIVE REGULATION OF BINDING | 32 | 131 | 1.207e-08 | 9.422e-08 |
| 597 | NEGATIVE REGULATION OF CELL CELL ADHESION | 33 | 138 | 1.265e-08 | 9.849e-08 |
| 598 | HEMOSTASIS | 56 | 311 | 1.266e-08 | 9.849e-08 |
| 599 | PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 13 | 25 | 1.327e-08 | 1.031e-07 |
| 600 | POSITIVE REGULATION OF CELL SUBSTRATE ADHESION | 27 | 99 | 1.338e-08 | 1.038e-07 |
| 601 | NEURAL CREST CELL DIFFERENTIATION | 23 | 75 | 1.392e-08 | 1.078e-07 |
| 602 | NEGATIVE REGULATION OF CELL SUBSTRATE ADHESION | 19 | 53 | 1.43e-08 | 1.105e-07 |
| 603 | REGULATION OF AUTOPHAGY | 48 | 249 | 1.514e-08 | 1.168e-07 |
| 604 | POSITIVE REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS | 21 | 64 | 1.544e-08 | 1.19e-07 |
| 605 | REGULATION OF FAT CELL DIFFERENTIATION | 28 | 106 | 1.564e-08 | 1.203e-07 |
| 606 | LIMBIC SYSTEM DEVELOPMENT | 27 | 100 | 1.693e-08 | 1.3e-07 |
| 607 | POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION | 72 | 448 | 1.716e-08 | 1.316e-07 |
| 608 | CENTRAL NERVOUS SYSTEM NEURON DEVELOPMENT | 22 | 70 | 1.731e-08 | 1.324e-07 |
| 609 | LUNG EPITHELIUM DEVELOPMENT | 15 | 34 | 1.768e-08 | 1.346e-07 |
| 610 | REGULATION OF SYNAPSE ORGANIZATION | 29 | 113 | 1.765e-08 | 1.346e-07 |
| 611 | HEART VALVE DEVELOPMENT | 15 | 34 | 1.768e-08 | 1.346e-07 |
| 612 | NEURAL TUBE FORMATION | 26 | 94 | 1.797e-08 | 1.367e-07 |
| 613 | NEGATIVE REGULATION OF LEUKOCYTE DIFFERENTIATION | 24 | 82 | 1.885e-08 | 1.431e-07 |
| 614 | RESPONSE TO CORTICOSTEROID | 38 | 176 | 2.084e-08 | 1.579e-07 |
| 615 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE BY P53 CLASS MEDIATOR | 14 | 30 | 2.183e-08 | 1.651e-07 |
| 616 | REGULATION OF EMBRYONIC DEVELOPMENT | 29 | 114 | 2.185e-08 | 1.651e-07 |
| 617 | NEGATIVE REGULATION OF NEURON DIFFERENTIATION | 40 | 191 | 2.208e-08 | 1.665e-07 |
| 618 | CELLULAR MACROMOLECULE LOCALIZATION | 155 | 1234 | 2.218e-08 | 1.67e-07 |
| 619 | CANONICAL WNT SIGNALING PATHWAY | 26 | 95 | 2.285e-08 | 1.718e-07 |
| 620 | REGULATION OF PROTEIN KINASE B SIGNALING | 30 | 121 | 2.375e-08 | 1.782e-07 |
| 621 | ORGAN REGENERATION | 24 | 83 | 2.45e-08 | 1.835e-07 |
| 622 | HEPATICOBILIARY SYSTEM DEVELOPMENT | 31 | 128 | 2.521e-08 | 1.884e-07 |
| 623 | DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION RESULTING IN TRANSCRIPTION | 10 | 15 | 2.523e-08 | 1.884e-07 |
| 624 | CELL GROWTH | 32 | 135 | 2.622e-08 | 1.955e-07 |
| 625 | PROTEIN LOCALIZATION TO NUCLEUS | 35 | 156 | 2.675e-08 | 1.992e-07 |
| 626 | REGULATION OF VESICLE MEDIATED TRANSPORT | 73 | 462 | 2.808e-08 | 2.087e-07 |
| 627 | MESODERM MORPHOGENESIS | 21 | 66 | 2.856e-08 | 2.119e-07 |
| 628 | NEGATIVE REGULATION OF EPITHELIAL CELL APOPTOTIC PROCESS | 15 | 35 | 2.861e-08 | 2.12e-07 |
| 629 | MYELOID LEUKOCYTE DIFFERENTIATION | 26 | 96 | 2.894e-08 | 2.141e-07 |
| 630 | AXIS SPECIFICATION | 25 | 90 | 3.064e-08 | 2.263e-07 |
| 631 | RAS PROTEIN SIGNAL TRANSDUCTION | 33 | 143 | 3.214e-08 | 2.368e-07 |
| 632 | CHROMATIN REMODELING | 34 | 150 | 3.217e-08 | 2.368e-07 |
| 633 | THYMOCYTE AGGREGATION | 17 | 45 | 3.326e-08 | 2.441e-07 |
| 634 | T CELL DIFFERENTIATION IN THYMUS | 17 | 45 | 3.326e-08 | 2.441e-07 |
| 635 | POSITIVE REGULATION OF PROTEIN CATABOLIC PROCESS | 49 | 263 | 3.352e-08 | 2.452e-07 |
| 636 | REGULATION OF MUSCLE TISSUE DEVELOPMENT | 27 | 103 | 3.351e-08 | 2.452e-07 |
| 637 | POSITIVE REGULATION OF ERK1 AND ERK2 CASCADE | 37 | 172 | 3.526e-08 | 2.576e-07 |
| 638 | REGULATION OF ADAPTIVE IMMUNE RESPONSE | 30 | 123 | 3.55e-08 | 2.589e-07 |
| 639 | REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 26 | 97 | 3.65e-08 | 2.658e-07 |
| 640 | NEGATIVE REGULATION OF CELL ACTIVATION | 35 | 158 | 3.767e-08 | 2.739e-07 |
| 641 | REGULATION OF CHEMOTAXIS | 38 | 180 | 3.937e-08 | 2.858e-07 |
| 642 | MULTI ORGANISM REPRODUCTIVE PROCESS | 119 | 891 | 3.977e-08 | 2.883e-07 |
| 643 | NEGATIVE REGULATION OF CYTOPLASMIC TRANSPORT | 29 | 117 | 4.076e-08 | 2.945e-07 |
| 644 | REGULATION OF NEURAL PRECURSOR CELL PROLIFERATION | 22 | 73 | 4.074e-08 | 2.945e-07 |
| 645 | POSITIVE REGULATION OF HEART GROWTH | 13 | 27 | 4.384e-08 | 3.163e-07 |
| 646 | NEGATIVE REGULATION OF I KAPPAB KINASE NF KAPPAB SIGNALING | 18 | 51 | 4.538e-08 | 3.264e-07 |
| 647 | REGULATION OF TRANSCRIPTION REGULATORY REGION DNA BINDING | 15 | 36 | 4.536e-08 | 3.264e-07 |
| 648 | POSITIVE REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS | 26 | 98 | 4.588e-08 | 3.294e-07 |
| 649 | CARDIAC VENTRICLE MORPHOGENESIS | 20 | 62 | 4.692e-08 | 3.364e-07 |
| 650 | REGULATION OF CELL JUNCTION ASSEMBLY | 21 | 68 | 5.137e-08 | 3.677e-07 |
| 651 | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 22 | 74 | 5.355e-08 | 3.828e-07 |
| 652 | COGNITION | 47 | 251 | 5.417e-08 | 3.86e-07 |
| 653 | POSITIVE REGULATION OF NF KAPPAB TRANSCRIPTION FACTOR ACTIVITY | 31 | 132 | 5.416e-08 | 3.86e-07 |
| 654 | POSITIVE REGULATION OF LIPID KINASE ACTIVITY | 14 | 32 | 6.058e-08 | 4.31e-07 |
| 655 | CELLULAR RESPONSE TO CYTOKINE STIMULUS | 88 | 606 | 6.266e-08 | 4.451e-07 |
| 656 | REGULATION OF GENE EXPRESSION EPIGENETIC | 44 | 229 | 6.585e-08 | 4.671e-07 |
| 657 | OVULATION CYCLE | 28 | 113 | 6.995e-08 | 4.954e-07 |
| 658 | THYMUS DEVELOPMENT | 17 | 47 | 7.064e-08 | 4.995e-07 |
| 659 | REGULATION OF DNA BIOSYNTHETIC PROCESS | 25 | 94 | 7.86e-08 | 5.55e-07 |
| 660 | ACTIVATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 9 | 13 | 8.038e-08 | 5.666e-07 |
| 661 | EMBRYONIC HEMOPOIESIS | 11 | 20 | 8.367e-08 | 5.872e-07 |
| 662 | GENETIC IMPRINTING | 11 | 20 | 8.367e-08 | 5.872e-07 |
| 663 | POSITIVE REGULATION OF CYTOKINE PRODUCTION | 61 | 370 | 8.35e-08 | 5.872e-07 |
| 664 | REGULATION OF NUCLEAR DIVISION | 35 | 163 | 8.593e-08 | 5.996e-07 |
| 665 | POSITIVE REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 12 | 24 | 8.595e-08 | 5.996e-07 |
| 666 | BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 12 | 24 | 8.595e-08 | 5.996e-07 |
| 667 | POSITIVE REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 12 | 24 | 8.595e-08 | 5.996e-07 |
| 668 | PROTEIN UBIQUITINATION | 90 | 629 | 8.837e-08 | 6.156e-07 |
| 669 | POSITIVE REGULATION OF FIBROBLAST PROLIFERATION | 18 | 53 | 8.979e-08 | 6.236e-07 |
| 670 | SKELETAL MUSCLE CELL DIFFERENTIATION | 18 | 53 | 8.979e-08 | 6.236e-07 |
| 671 | CELLULAR SENESCENCE | 14 | 33 | 9.734e-08 | 6.75e-07 |
| 672 | DIGESTIVE TRACT MORPHOGENESIS | 17 | 48 | 1.011e-07 | 7.002e-07 |
| 673 | PROTEIN SUMOYLATION | 28 | 115 | 1.045e-07 | 7.224e-07 |
| 674 | POSITIVE REGULATION OF ORGAN GROWTH | 15 | 38 | 1.078e-07 | 7.444e-07 |
| 675 | DNA TEMPLATED TRANSCRIPTION INITIATION | 40 | 202 | 1.108e-07 | 7.639e-07 |
| 676 | REGULATION OF REPRODUCTIVE PROCESS | 30 | 129 | 1.115e-07 | 7.676e-07 |
| 677 | REGULATION OF BMP SIGNALING PATHWAY | 22 | 77 | 1.177e-07 | 8.091e-07 |
| 678 | ADHERENS JUNCTION ORGANIZATION | 21 | 71 | 1.181e-07 | 8.107e-07 |
| 679 | REGULATION OF MUSCLE SYSTEM PROCESS | 39 | 195 | 1.208e-07 | 8.281e-07 |
| 680 | NEGATIVE REGULATION OF CHROMOSOME ORGANIZATION | 25 | 96 | 1.23e-07 | 8.405e-07 |
| 681 | CARDIOCYTE DIFFERENTIATION | 25 | 96 | 1.23e-07 | 8.405e-07 |
| 682 | REGULATION OF SECRETION | 97 | 699 | 1.238e-07 | 8.445e-07 |
| 683 | VENTRICULAR SEPTUM DEVELOPMENT | 18 | 54 | 1.245e-07 | 8.481e-07 |
| 684 | SKELETAL MUSCLE ORGAN DEVELOPMENT | 31 | 137 | 1.338e-07 | 9.09e-07 |
| 685 | ACTIVATION OF MAPK ACTIVITY | 31 | 137 | 1.338e-07 | 9.09e-07 |
| 686 | REGULATION OF MUSCLE ORGAN DEVELOPMENT | 26 | 103 | 1.365e-07 | 9.257e-07 |
| 687 | CELL CELL SIGNALING | 104 | 767 | 1.39e-07 | 9.416e-07 |
| 688 | POSITIVE REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 10 | 17 | 1.403e-07 | 9.476e-07 |
| 689 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 10 | 17 | 1.403e-07 | 9.476e-07 |
| 690 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 19 | 60 | 1.414e-07 | 9.533e-07 |
| 691 | CARDIAC SEPTUM MORPHOGENESIS | 17 | 49 | 1.432e-07 | 9.64e-07 |
| 692 | NEGATIVE REGULATION OF DEVELOPMENTAL GROWTH | 23 | 84 | 1.446e-07 | 9.711e-07 |
| 693 | LEUKOCYTE MIGRATION | 47 | 259 | 1.446e-07 | 9.711e-07 |
| 694 | NEURON MIGRATION | 27 | 110 | 1.47e-07 | 9.853e-07 |
| 695 | RESPONSE TO FLUID SHEAR STRESS | 14 | 34 | 1.531e-07 | 1.024e-06 |
| 696 | LUNG CELL DIFFERENTIATION | 12 | 25 | 1.53e-07 | 1.024e-06 |
| 697 | NEGATIVE REGULATION OF MYELOID LEUKOCYTE DIFFERENTIATION | 16 | 44 | 1.572e-07 | 1.049e-06 |
| 698 | PROTEIN STABILIZATION | 30 | 131 | 1.603e-07 | 1.068e-06 |
| 699 | NEGATIVE REGULATION OF TRANSCRIPTION FACTOR IMPORT INTO NUCLEUS | 15 | 39 | 1.621e-07 | 1.074e-06 |
| 700 | ASTROCYTE DIFFERENTIATION | 15 | 39 | 1.621e-07 | 1.074e-06 |
| 701 | CELLULAR RESPONSE TO NUTRIENT | 15 | 39 | 1.621e-07 | 1.074e-06 |
| 702 | GLANDULAR EPITHELIAL CELL DIFFERENTIATION | 15 | 39 | 1.621e-07 | 1.074e-06 |
| 703 | DEVELOPMENTAL GROWTH INVOLVED IN MORPHOGENESIS | 26 | 104 | 1.68e-07 | 1.112e-06 |
| 704 | CELLULAR RESPONSE TO LIGHT STIMULUS | 24 | 91 | 1.69e-07 | 1.117e-06 |
| 705 | EPHRIN RECEPTOR SIGNALING PATHWAY | 23 | 85 | 1.831e-07 | 1.207e-06 |
| 706 | POSITIVE REGULATION OF CHROMATIN MODIFICATION | 23 | 85 | 1.831e-07 | 1.207e-06 |
| 707 | REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 28 | 118 | 1.868e-07 | 1.23e-06 |
| 708 | REGULATION OF LYMPHOCYTE DIFFERENTIATION | 30 | 132 | 1.915e-07 | 1.258e-06 |
| 709 | REGULATION OF LEUKOCYTE APOPTOTIC PROCESS | 22 | 79 | 1.939e-07 | 1.273e-06 |
| 710 | SINGLE ORGANISM CELLULAR LOCALIZATION | 117 | 898 | 1.975e-07 | 1.294e-06 |
| 711 | HIPPOCAMPUS DEVELOPMENT | 21 | 73 | 1.998e-07 | 1.308e-06 |
| 712 | REGULATION OF ADHERENS JUNCTION ORGANIZATION | 17 | 50 | 2.005e-07 | 1.311e-06 |
| 713 | RESPONSE TO EPIDERMAL GROWTH FACTOR | 13 | 30 | 2.075e-07 | 1.352e-06 |
| 714 | CELLULAR RESPONSE TO VASCULAR ENDOTHELIAL GROWTH FACTOR STIMULUS | 13 | 30 | 2.075e-07 | 1.352e-06 |
| 715 | VASCULAR ENDOTHELIAL GROWTH FACTOR SIGNALING PATHWAY | 9 | 14 | 2.087e-07 | 1.358e-06 |
| 716 | OSTEOBLAST DIFFERENTIATION | 29 | 126 | 2.288e-07 | 1.487e-06 |
| 717 | POSITIVE REGULATION OF DNA REPLICATION | 23 | 86 | 2.309e-07 | 1.498e-06 |
| 718 | POSITIVE REGULATION OF MUSCLE TISSUE DEVELOPMENT | 18 | 56 | 2.33e-07 | 1.51e-06 |
| 719 | POSITIVE REGULATION OF CELL MATRIX ADHESION | 15 | 40 | 2.399e-07 | 1.552e-06 |
| 720 | POSITIVE REGULATION OF DEFENSE RESPONSE | 59 | 364 | 2.466e-07 | 1.594e-06 |
| 721 | PHOSPHATIDYLINOSITOL METABOLIC PROCESS | 38 | 193 | 2.66e-07 | 1.717e-06 |
| 722 | CYTOKINE PRODUCTION | 28 | 120 | 2.717e-07 | 1.749e-06 |
| 723 | REGULATION OF DENDRITE DEVELOPMENT | 28 | 120 | 2.717e-07 | 1.749e-06 |
| 724 | TISSUE REMODELING | 23 | 87 | 2.899e-07 | 1.863e-06 |
| 725 | RESPONSE TO TOXIC SUBSTANCE | 44 | 241 | 3.002e-07 | 1.927e-06 |
| 726 | POSITIVE REGULATION OF MYELOID CELL DIFFERENTIATION | 22 | 81 | 3.134e-07 | 2.009e-06 |
| 727 | CIRCADIAN REGULATION OF GENE EXPRESSION | 18 | 57 | 3.148e-07 | 2.015e-06 |
| 728 | NEGATIVE REGULATION OF DNA BINDING | 16 | 46 | 3.198e-07 | 2.041e-06 |
| 729 | NEGATIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 48 | 274 | 3.197e-07 | 2.041e-06 |
| 730 | NOTCH SIGNALING PATHWAY | 27 | 114 | 3.203e-07 | 2.042e-06 |
| 731 | MACROMOLECULE CATABOLIC PROCESS | 119 | 926 | 3.217e-07 | 2.048e-06 |
| 732 | ARTERY DEVELOPMENT | 21 | 75 | 3.309e-07 | 2.1e-06 |
| 733 | POSITIVE REGULATION OF AUTOPHAGY | 21 | 75 | 3.309e-07 | 2.1e-06 |
| 734 | NEGATIVE REGULATION OF OSSIFICATION | 20 | 69 | 3.39e-07 | 2.149e-06 |
| 735 | NEGATIVE REGULATION OF FAT CELL DIFFERENTIATION | 15 | 41 | 3.499e-07 | 2.213e-06 |
| 736 | ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 38 | 195 | 3.5e-07 | 2.213e-06 |
| 737 | REGULATION OF ERYTHROCYTE DIFFERENTIATION | 14 | 36 | 3.575e-07 | 2.254e-06 |
| 738 | POSITIVE REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 14 | 36 | 3.575e-07 | 2.254e-06 |
| 739 | MYELOID CELL HOMEOSTASIS | 23 | 88 | 3.625e-07 | 2.283e-06 |
| 740 | TISSUE REGENERATION | 17 | 52 | 3.82e-07 | 2.402e-06 |
| 741 | CELL JUNCTION ASSEMBLY | 29 | 129 | 3.892e-07 | 2.444e-06 |
| 742 | STRIATED MUSCLE CELL DIFFERENTIATION | 35 | 173 | 3.958e-07 | 2.482e-06 |
| 743 | MODULATION OF SYNAPTIC TRANSMISSION | 51 | 301 | 4.022e-07 | 2.519e-06 |
| 744 | NEGATIVE REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 24 | 95 | 4.029e-07 | 2.52e-06 |
| 745 | REGULATION OF RESPONSE TO WOUNDING | 64 | 413 | 4.068e-07 | 2.541e-06 |
| 746 | EMBRYONIC PATTERN SPECIFICATION | 18 | 58 | 4.22e-07 | 2.632e-06 |
| 747 | REGULATION OF CATENIN IMPORT INTO NUCLEUS | 12 | 27 | 4.386e-07 | 2.728e-06 |
| 748 | AXIS ELONGATION | 12 | 27 | 4.386e-07 | 2.728e-06 |
| 749 | REGULATION OF PHOSPHOLIPASE ACTIVITY | 19 | 64 | 4.399e-07 | 2.733e-06 |
| 750 | B CELL DIFFERENTIATION | 23 | 89 | 4.516e-07 | 2.798e-06 |
| 751 | POST EMBRYONIC DEVELOPMENT | 23 | 89 | 4.516e-07 | 2.798e-06 |
| 752 | PATTERN RECOGNITION RECEPTOR SIGNALING PATHWAY | 26 | 109 | 4.541e-07 | 2.81e-06 |
| 753 | CELL MIGRATION INVOLVED IN SPROUTING ANGIOGENESIS | 9 | 15 | 4.84e-07 | 2.99e-06 |
| 754 | POSITIVE REGULATION OF WNT SIGNALING PATHWAY | 32 | 152 | 4.851e-07 | 2.994e-06 |
| 755 | POSITIVE REGULATION OF DNA BINDING | 15 | 42 | 5.035e-07 | 3.095e-06 |
| 756 | AUTONOMIC NERVOUS SYSTEM DEVELOPMENT | 15 | 42 | 5.035e-07 | 3.095e-06 |
| 757 | REGULATION OF HEART GROWTH | 15 | 42 | 5.035e-07 | 3.095e-06 |
| 758 | ENDOCARDIAL CUSHION DEVELOPMENT | 13 | 32 | 5.157e-07 | 3.166e-06 |
| 759 | PROSTATE GLAND MORPHOGENESIS | 11 | 23 | 5.354e-07 | 3.274e-06 |
| 760 | RESPONSE TO INCREASED OXYGEN LEVELS | 11 | 23 | 5.354e-07 | 3.274e-06 |
| 761 | RESPONSE TO HYPEROXIA | 11 | 23 | 5.354e-07 | 3.274e-06 |
| 762 | CELLULAR RESPONSE TO STARVATION | 27 | 117 | 5.585e-07 | 3.41e-06 |
| 763 | REGULATION OF EPITHELIAL CELL APOPTOTIC PROCESS | 18 | 59 | 5.614e-07 | 3.419e-06 |
| 764 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 18 | 59 | 5.614e-07 | 3.419e-06 |
| 765 | REGULATION OF NON CANONICAL WNT SIGNALING PATHWAY | 10 | 19 | 5.726e-07 | 3.478e-06 |
| 766 | NEURONAL STEM CELL POPULATION MAINTENANCE | 10 | 19 | 5.726e-07 | 3.478e-06 |
| 767 | REGULATION OF TRANSMEMBRANE TRANSPORT | 65 | 426 | 5.811e-07 | 3.525e-06 |
| 768 | ADAPTIVE IMMUNE RESPONSE | 49 | 288 | 5.95e-07 | 3.605e-06 |
| 769 | REGULATION OF INTERLEUKIN 2 PRODUCTION | 16 | 48 | 6.22e-07 | 3.764e-06 |
| 770 | POSITIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 37 | 192 | 6.619e-07 | 3.987e-06 |
| 771 | POSITIVE REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 26 | 111 | 6.624e-07 | 3.987e-06 |
| 772 | CELL CYCLE G1 S PHASE TRANSITION | 26 | 111 | 6.624e-07 | 3.987e-06 |
| 773 | G1 S TRANSITION OF MITOTIC CELL CYCLE | 26 | 111 | 6.624e-07 | 3.987e-06 |
| 774 | SOMITE DEVELOPMENT | 21 | 78 | 6.786e-07 | 4.08e-06 |
| 775 | CHONDROCYTE DIFFERENTIATION | 18 | 60 | 7.414e-07 | 4.446e-06 |
| 776 | LEUKOCYTE HOMEOSTASIS | 18 | 60 | 7.414e-07 | 4.446e-06 |
| 777 | TOLL LIKE RECEPTOR SIGNALING PATHWAY | 22 | 85 | 7.762e-07 | 4.648e-06 |
| 778 | EMBRYONIC AXIS SPECIFICATION | 13 | 33 | 7.876e-07 | 4.686e-06 |
| 779 | SIGNAL TRANSDUCTION IN ABSENCE OF LIGAND | 13 | 33 | 7.876e-07 | 4.686e-06 |
| 780 | NEGATIVE REGULATION OF JNK CASCADE | 13 | 33 | 7.876e-07 | 4.686e-06 |
| 781 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 13 | 33 | 7.876e-07 | 4.686e-06 |
| 782 | REGULATION OF CELL AGING | 13 | 33 | 7.876e-07 | 4.686e-06 |
| 783 | GAMETE GENERATION | 83 | 595 | 8.159e-07 | 4.848e-06 |
| 784 | CELLULAR RESPONSE TO INORGANIC SUBSTANCE | 32 | 156 | 8.915e-07 | 5.291e-06 |
| 785 | PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 29 | 134 | 9.03e-07 | 5.353e-06 |
| 786 | REGULATION OF PLASMA MEMBRANE ORGANIZATION | 20 | 73 | 9.191e-07 | 5.441e-06 |
| 787 | EMBRYONIC DIGIT MORPHOGENESIS | 18 | 61 | 9.723e-07 | 5.749e-06 |
| 788 | PROTEIN CATABOLIC PROCESS | 81 | 579 | 9.851e-07 | 5.817e-06 |
| 789 | FC EPSILON RECEPTOR SIGNALING PATHWAY | 30 | 142 | 1.011e-06 | 5.957e-06 |
| 790 | REGULATION OF CARBOHYDRATE METABOLIC PROCESS | 34 | 172 | 1.011e-06 | 5.957e-06 |
| 791 | NEGATIVE REGULATION OF T CELL RECEPTOR SIGNALING PATHWAY | 9 | 16 | 1.026e-06 | 6.036e-06 |
| 792 | POSITIVE REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 29 | 135 | 1.062e-06 | 6.239e-06 |
| 793 | NEGATIVE REGULATION OF VASCULATURE DEVELOPMENT | 21 | 80 | 1.07e-06 | 6.279e-06 |
| 794 | REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 12 | 29 | 1.123e-06 | 6.562e-06 |
| 795 | POSITIVE REGULATION OF PROTEIN INSERTION INTO MITOCHONDRIAL MEMBRANE INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 12 | 29 | 1.123e-06 | 6.562e-06 |
| 796 | POSITIVE REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT | 12 | 29 | 1.123e-06 | 6.562e-06 |
| 797 | PEPTIDYL TYROSINE AUTOPHOSPHORYLATION | 14 | 39 | 1.125e-06 | 6.562e-06 |
| 798 | NEGATIVE CHEMOTAXIS | 14 | 39 | 1.125e-06 | 6.562e-06 |
| 799 | RESPONSE TO RETINOIC ACID | 25 | 107 | 1.135e-06 | 6.611e-06 |
| 800 | ENDODERM FORMATION | 16 | 50 | 1.162e-06 | 6.757e-06 |
| 801 | POSITIVE REGULATION OF EPITHELIAL TO MESENCHYMAL TRANSITION | 13 | 34 | 1.18e-06 | 6.84e-06 |
| 802 | PROTEIN KINASE B SIGNALING | 13 | 34 | 1.18e-06 | 6.84e-06 |
| 803 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 13 | 34 | 1.18e-06 | 6.84e-06 |
| 804 | RESPONSE TO PURINE CONTAINING COMPOUND | 32 | 158 | 1.197e-06 | 6.925e-06 |
| 805 | REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 47 | 278 | 1.206e-06 | 6.974e-06 |
| 806 | REGULATION OF SMOOTHENED SIGNALING PATHWAY | 18 | 62 | 1.267e-06 | 7.312e-06 |
| 807 | REGULATION OF SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM | 38 | 205 | 1.284e-06 | 7.401e-06 |
| 808 | DNA GEOMETRIC CHANGE | 21 | 81 | 1.335e-06 | 7.688e-06 |
| 809 | EMBRYONIC SKELETAL SYSTEM DEVELOPMENT | 27 | 122 | 1.342e-06 | 7.718e-06 |
| 810 | MICROTUBULE CYTOSKELETON ORGANIZATION | 55 | 348 | 1.406e-06 | 8.079e-06 |
| 811 | CHEMICAL HOMEOSTASIS | 111 | 874 | 1.438e-06 | 8.251e-06 |
| 812 | CELLULAR RESPONSE TO EPIDERMAL GROWTH FACTOR STIMULUS | 11 | 25 | 1.515e-06 | 8.67e-06 |
| 813 | EPITHELIAL CELL APOPTOTIC PROCESS | 11 | 25 | 1.515e-06 | 8.67e-06 |
| 814 | REGULATION OF TRANSCRIPTION FACTOR IMPORT INTO NUCLEUS | 23 | 95 | 1.562e-06 | 8.926e-06 |
| 815 | REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 16 | 51 | 1.566e-06 | 8.939e-06 |
| 816 | NEGATIVE REGULATION OF PROTEIN CATABOLIC PROCESS | 25 | 109 | 1.635e-06 | 9.314e-06 |
| 817 | REGULATION OF ENDOCYTOSIS | 37 | 199 | 1.634e-06 | 9.314e-06 |
| 818 | REGULATION OF CARTILAGE DEVELOPMENT | 18 | 63 | 1.639e-06 | 9.314e-06 |
| 819 | REGULATION OF MUSCLE ADAPTATION | 18 | 63 | 1.639e-06 | 9.314e-06 |
| 820 | RECEPTOR METABOLIC PROCESS | 21 | 82 | 1.658e-06 | 9.41e-06 |
| 821 | MICROTUBULE BASED PROCESS | 74 | 522 | 1.757e-06 | 9.96e-06 |
| 822 | REGULATION OF GENERATION OF PRECURSOR METABOLITES AND ENERGY | 22 | 89 | 1.802e-06 | 1.02e-05 |
| 823 | GLIAL CELL DEVELOPMENT | 20 | 76 | 1.84e-06 | 1.04e-05 |
| 824 | POSITIVE REGULATION OF HOMEOSTATIC PROCESS | 39 | 216 | 1.846e-06 | 1.043e-05 |
| 825 | SYMPATHETIC NERVOUS SYSTEM DEVELOPMENT | 10 | 21 | 1.879e-06 | 1.059e-05 |
| 826 | PEPTIDYL THREONINE MODIFICATION | 15 | 46 | 1.913e-06 | 1.077e-05 |
| 827 | STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 24 | 103 | 1.946e-06 | 1.095e-05 |
| 828 | REGULATION OF CELL SHAPE | 29 | 139 | 1.991e-06 | 1.119e-05 |
| 829 | BRANCH ELONGATION OF AN EPITHELIUM | 9 | 17 | 2.022e-06 | 1.132e-05 |
| 830 | REGULATION OF STEM CELL POPULATION MAINTENANCE | 9 | 17 | 2.022e-06 | 1.132e-05 |
| 831 | RECEPTOR CATABOLIC PROCESS | 9 | 17 | 2.022e-06 | 1.132e-05 |
| 832 | REGULATION OF LIPASE ACTIVITY | 21 | 83 | 2.052e-06 | 1.147e-05 |
| 833 | REGULATION OF ION HOMEOSTASIS | 37 | 201 | 2.093e-06 | 1.168e-05 |
| 834 | REGULATION OF GENE SILENCING | 16 | 52 | 2.092e-06 | 1.168e-05 |
| 835 | POSITIVE REGULATION OF CELL DIVISION | 28 | 132 | 2.108e-06 | 1.174e-05 |
| 836 | POSITIVE REGULATION OF DENDRITE DEVELOPMENT | 18 | 64 | 2.109e-06 | 1.174e-05 |
| 837 | REGULATION OF ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 14 | 41 | 2.252e-06 | 1.247e-05 |
| 838 | POSITIVE REGULATION OF KIDNEY DEVELOPMENT | 14 | 41 | 2.252e-06 | 1.247e-05 |
| 839 | REGULATION OF POTASSIUM ION TRANSMEMBRANE TRANSPORTER ACTIVITY | 14 | 41 | 2.252e-06 | 1.247e-05 |
| 840 | LACTATION | 14 | 41 | 2.252e-06 | 1.247e-05 |
| 841 | REGULATION OF CELLULAR SENESCENCE | 11 | 26 | 2.433e-06 | 1.343e-05 |
| 842 | REGULATION OF P38MAPK CASCADE | 11 | 26 | 2.433e-06 | 1.343e-05 |
| 843 | CELLULAR RESPONSE TO VITAMIN | 11 | 26 | 2.433e-06 | 1.343e-05 |
| 844 | T CELL SELECTION | 13 | 36 | 2.519e-06 | 1.389e-05 |
| 845 | REGULATION OF DNA TEMPLATED TRANSCRIPTION INITIATION | 12 | 31 | 2.618e-06 | 1.442e-05 |
| 846 | CELLULAR PROCESS INVOLVED IN REPRODUCTION IN MULTICELLULAR ORGANISM | 43 | 252 | 2.64e-06 | 1.452e-05 |
| 847 | NEGATIVE REGULATION OF CELLULAR CATABOLIC PROCESS | 31 | 156 | 2.661e-06 | 1.462e-05 |
| 848 | NEGATIVE REGULATION OF AXONOGENESIS | 18 | 65 | 2.696e-06 | 1.48e-05 |
| 849 | HEMATOPOIETIC PROGENITOR CELL DIFFERENTIATION | 23 | 98 | 2.774e-06 | 1.517e-05 |
| 850 | POSITIVE REGULATION OF PHOSPHOLIPASE ACTIVITY | 16 | 53 | 2.772e-06 | 1.517e-05 |
| 851 | MYELOID LEUKOCYTE ACTIVATION | 23 | 98 | 2.774e-06 | 1.517e-05 |
| 852 | REGULATION OF INTRACELLULAR STEROID HORMONE RECEPTOR SIGNALING PATHWAY | 17 | 59 | 2.792e-06 | 1.521e-05 |
| 853 | CELL SUBSTRATE ADHESION | 32 | 164 | 2.788e-06 | 1.521e-05 |
| 854 | VASCULOGENESIS | 17 | 59 | 2.792e-06 | 1.521e-05 |
| 855 | ODONTOGENESIS | 24 | 105 | 2.799e-06 | 1.523e-05 |
| 856 | PALATE DEVELOPMENT | 21 | 85 | 3.103e-06 | 1.684e-05 |
| 857 | REGULATION OF CHROMOSOME SEGREGATION | 21 | 85 | 3.103e-06 | 1.684e-05 |
| 858 | POSITIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 48 | 296 | 3.131e-06 | 1.694e-05 |
| 859 | EPIDERMAL CELL DIFFERENTIATION | 29 | 142 | 3.128e-06 | 1.694e-05 |
| 860 | NEGATIVE REGULATION OF BMP SIGNALING PATHWAY | 14 | 42 | 3.126e-06 | 1.694e-05 |
| 861 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 10 | 22 | 3.193e-06 | 1.723e-05 |
| 862 | REGULATION OF RESPONSE TO INTERFERON GAMMA | 10 | 22 | 3.193e-06 | 1.723e-05 |
| 863 | INNER EAR MORPHOGENESIS | 22 | 92 | 3.261e-06 | 1.758e-05 |
| 864 | SPINAL CORD DEVELOPMENT | 24 | 106 | 3.343e-06 | 1.8e-05 |
| 865 | SECRETION | 80 | 588 | 3.42e-06 | 1.839e-05 |
| 866 | FOREBRAIN GENERATION OF NEURONS | 18 | 66 | 3.428e-06 | 1.842e-05 |
| 867 | REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT | 15 | 48 | 3.498e-06 | 1.875e-05 |
| 868 | COLUMNAR CUBOIDAL EPITHELIAL CELL DEVELOPMENT | 15 | 48 | 3.498e-06 | 1.875e-05 |
| 869 | BONE MORPHOGENESIS | 20 | 79 | 3.534e-06 | 1.892e-05 |
| 870 | REGULATION OF LIPID BIOSYNTHETIC PROCESS | 27 | 128 | 3.563e-06 | 1.905e-05 |
| 871 | HINDLIMB MORPHOGENESIS | 13 | 37 | 3.595e-06 | 1.921e-05 |
| 872 | POSITIVE REGULATION OF METANEPHROS DEVELOPMENT | 8 | 14 | 3.609e-06 | 1.921e-05 |
| 873 | STEM CELL PROLIFERATION | 17 | 60 | 3.603e-06 | 1.921e-05 |
| 874 | POSITIVE REGULATION OF HISTONE DEACETYLATION | 8 | 14 | 3.609e-06 | 1.921e-05 |
| 875 | NEGATIVE REGULATION OF INTRACELLULAR TRANSPORT | 29 | 143 | 3.623e-06 | 1.927e-05 |
| 876 | REGULATION OF LYMPHOCYTE APOPTOTIC PROCESS | 16 | 54 | 3.643e-06 | 1.935e-05 |
| 877 | REGULATION OF CELL MATURATION | 9 | 18 | 3.752e-06 | 1.986e-05 |
| 878 | REGULATION OF B CELL ACTIVATION | 26 | 121 | 3.745e-06 | 1.986e-05 |
| 879 | POSITIVE REGULATION OF PROTEIN DEACETYLATION | 9 | 18 | 3.752e-06 | 1.986e-05 |
| 880 | REGULATION OF CARDIAC MUSCLE CELL CONTRACTION | 11 | 27 | 3.804e-06 | 2.007e-05 |
| 881 | NEGATIVE REGULATION OF FIBROBLAST PROLIFERATION | 11 | 27 | 3.804e-06 | 2.007e-05 |
| 882 | NEGATIVE REGULATION OF ENDOTHELIAL CELL APOPTOTIC PROCESS | 11 | 27 | 3.804e-06 | 2.007e-05 |
| 883 | RESPONSE TO ETHANOL | 28 | 136 | 3.9e-06 | 2.055e-05 |
| 884 | NEGATIVE REGULATION OF MAP KINASE ACTIVITY | 19 | 73 | 3.956e-06 | 2.075e-05 |
| 885 | EMBRYONIC SKELETAL SYSTEM MORPHOGENESIS | 22 | 93 | 3.947e-06 | 2.075e-05 |
| 886 | PANCREAS DEVELOPMENT | 19 | 73 | 3.956e-06 | 2.075e-05 |
| 887 | CELLULAR RESPONSE TO KETONE | 19 | 73 | 3.956e-06 | 2.075e-05 |
| 888 | REGULATION OF T CELL DIFFERENTIATION | 24 | 107 | 3.981e-06 | 2.086e-05 |
| 889 | METENCEPHALON DEVELOPMENT | 23 | 100 | 4.005e-06 | 2.096e-05 |
| 890 | NEGATIVE REGULATION OF STEM CELL DIFFERENTIATION | 14 | 43 | 4.289e-06 | 2.237e-05 |
| 891 | CELL FATE DETERMINATION | 14 | 43 | 4.289e-06 | 2.237e-05 |
| 892 | REGULATION OF MITOCHONDRIAL OUTER MEMBRANE PERMEABILIZATION INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 14 | 43 | 4.289e-06 | 2.237e-05 |
| 893 | GLOMERULUS DEVELOPMENT | 15 | 49 | 4.664e-06 | 2.43e-05 |
| 894 | CRANIAL SKELETAL SYSTEM DEVELOPMENT | 16 | 55 | 4.752e-06 | 2.468e-05 |
| 895 | REGULATION OF MYOTUBE DIFFERENTIATION | 16 | 55 | 4.752e-06 | 2.468e-05 |
| 896 | NEGATIVE REGULATION OF DNA REPLICATION | 16 | 55 | 4.752e-06 | 2.468e-05 |
| 897 | CARDIAC MUSCLE CELL DIFFERENTIATION | 19 | 74 | 4.921e-06 | 2.552e-05 |
| 898 | POSITIVE REGULATION OF ERYTHROCYTE DIFFERENTIATION | 10 | 23 | 5.237e-06 | 2.71e-05 |
| 899 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER IN RESPONSE TO STRESS | 10 | 23 | 5.237e-06 | 2.71e-05 |
| 900 | NEGATIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 37 | 209 | 5.398e-06 | 2.791e-05 |
| 901 | REGULATION OF ALPHA BETA T CELL ACTIVATION | 18 | 68 | 5.45e-06 | 2.811e-05 |
| 902 | NERVE DEVELOPMENT | 18 | 68 | 5.45e-06 | 2.811e-05 |
| 903 | LEUKOCYTE PROLIFERATION | 21 | 88 | 5.61e-06 | 2.887e-05 |
| 904 | POSITIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 21 | 88 | 5.61e-06 | 2.887e-05 |
| 905 | REGULATION OF PROTEIN EXPORT FROM NUCLEUS | 12 | 33 | 5.647e-06 | 2.894e-05 |
| 906 | NEGATIVE REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 27 | 131 | 5.643e-06 | 2.894e-05 |
| 907 | POSITIVE REGULATION OF LEUKOCYTE DIFFERENTIATION | 27 | 131 | 5.643e-06 | 2.894e-05 |
| 908 | G2 DNA DAMAGE CHECKPOINT | 12 | 33 | 5.647e-06 | 2.894e-05 |
| 909 | PROSTATE GLANDULAR ACINUS DEVELOPMENT | 7 | 11 | 5.709e-06 | 2.922e-05 |
| 910 | NEGATIVE REGULATION OF HOMOTYPIC CELL CELL ADHESION | 23 | 102 | 5.716e-06 | 2.923e-05 |
| 911 | REGULATION OF T CELL RECEPTOR SIGNALING PATHWAY | 11 | 28 | 5.806e-06 | 2.959e-05 |
| 912 | REGULATION OF NEUROBLAST PROLIFERATION | 11 | 28 | 5.806e-06 | 2.959e-05 |
| 913 | DOPAMINERGIC NEURON DIFFERENTIATION | 11 | 28 | 5.806e-06 | 2.959e-05 |
| 914 | SOMITOGENESIS | 17 | 62 | 5.886e-06 | 2.997e-05 |
| 915 | CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 28 | 139 | 6.065e-06 | 3.084e-05 |
| 916 | REGULATION OF HISTONE METHYLATION | 16 | 56 | 6.155e-06 | 3.123e-05 |
| 917 | EPITHELIAL TO MESENCHYMAL TRANSITION | 16 | 56 | 6.155e-06 | 3.123e-05 |
| 918 | LYMPHOCYTE HOMEOSTASIS | 15 | 50 | 6.164e-06 | 3.125e-05 |
| 919 | INTEGRIN MEDIATED SIGNALING PATHWAY | 20 | 82 | 6.535e-06 | 3.309e-05 |
| 920 | REGULATION OF CARDIAC MUSCLE CELL ACTION POTENTIAL | 9 | 19 | 6.614e-06 | 3.338e-05 |
| 921 | CARTILAGE DEVELOPMENT INVOLVED IN ENDOCHONDRAL BONE MORPHOGENESIS | 9 | 19 | 6.614e-06 | 3.338e-05 |
| 922 | REGULATION OF LONG TERM SYNAPTIC POTENTIATION | 9 | 19 | 6.614e-06 | 3.338e-05 |
| 923 | NEGATIVE REGULATION OF CYTOKINE PRODUCTION | 37 | 211 | 6.77e-06 | 3.413e-05 |
| 924 | PERIPHERAL NERVOUS SYSTEM DEVELOPMENT | 18 | 69 | 6.818e-06 | 3.433e-05 |
| 925 | REGULATION OF AXON GUIDANCE | 13 | 39 | 7.026e-06 | 3.527e-05 |
| 926 | ERBB2 SIGNALING PATHWAY | 13 | 39 | 7.026e-06 | 3.527e-05 |
| 927 | NEGATIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 13 | 39 | 7.026e-06 | 3.527e-05 |
| 928 | REGULATION OF EPITHELIAL CELL DIFFERENTIATION INVOLVED IN KIDNEY DEVELOPMENT | 8 | 15 | 7.179e-06 | 3.592e-05 |
| 929 | JAK STAT CASCADE INVOLVED IN GROWTH HORMONE SIGNALING PATHWAY | 8 | 15 | 7.179e-06 | 3.592e-05 |
| 930 | T CELL APOPTOTIC PROCESS | 8 | 15 | 7.179e-06 | 3.592e-05 |
| 931 | ALPHA BETA T CELL DIFFERENTIATION | 14 | 45 | 7.823e-06 | 3.901e-05 |
| 932 | LUNG MORPHOGENESIS | 14 | 45 | 7.823e-06 | 3.901e-05 |
| 933 | ENDOCHONDRAL BONE MORPHOGENESIS | 14 | 45 | 7.823e-06 | 3.901e-05 |
| 934 | POSITIVE REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 16 | 57 | 7.915e-06 | 3.943e-05 |
| 935 | RESPONSE TO FATTY ACID | 20 | 83 | 7.958e-06 | 3.956e-05 |
| 936 | REGULATION OF POTASSIUM ION TRANSPORT | 20 | 83 | 7.958e-06 | 3.956e-05 |
| 937 | ORGAN FORMATION | 12 | 34 | 8.083e-06 | 4.01e-05 |
| 938 | NEGATIVE REGULATION OF CHEMOTAXIS | 15 | 51 | 8.081e-06 | 4.01e-05 |
| 939 | ESTABLISHMENT OF PROTEIN LOCALIZATION | 161 | 1423 | 8.15e-06 | 4.039e-05 |
| 940 | NEGATIVE REGULATION OF CYTOSKELETON ORGANIZATION | 38 | 221 | 8.209e-06 | 4.063e-05 |
| 941 | MULTI MULTICELLULAR ORGANISM PROCESS | 37 | 213 | 8.457e-06 | 4.182e-05 |
| 942 | STEM CELL DIVISION | 11 | 29 | 8.667e-06 | 4.269e-05 |
| 943 | REGULATION OF HEART MORPHOGENESIS | 11 | 29 | 8.667e-06 | 4.269e-05 |
| 944 | RESPONSE TO INSULIN | 36 | 205 | 8.669e-06 | 4.269e-05 |
| 945 | EMBRYONIC HINDLIMB MORPHOGENESIS | 11 | 29 | 8.667e-06 | 4.269e-05 |
| 946 | REGULATION OF PROTEIN POLYMERIZATION | 32 | 173 | 8.994e-06 | 4.424e-05 |
| 947 | RESPONSE TO AMINO ACID | 24 | 112 | 9.175e-06 | 4.508e-05 |
| 948 | SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 53 | 352 | 9.216e-06 | 4.523e-05 |
| 949 | PROTEIN LOCALIZATION TO ORGANELLE | 75 | 556 | 9.347e-06 | 4.583e-05 |
| 950 | ENDOCRINE PANCREAS DEVELOPMENT | 13 | 40 | 9.638e-06 | 4.72e-05 |
| 951 | PROTEIN DEPHOSPHORYLATION | 34 | 190 | 1.007e-05 | 4.926e-05 |
| 952 | PHOSPHATIDYLINOSITOL BIOSYNTHETIC PROCESS | 25 | 120 | 1.016e-05 | 4.965e-05 |
| 953 | REGULATION OF CHONDROCYTE DIFFERENTIATION | 14 | 46 | 1.041e-05 | 5.071e-05 |
| 954 | REGULATION OF ALPHA BETA T CELL DIFFERENTIATION | 14 | 46 | 1.041e-05 | 5.071e-05 |
| 955 | EMBRYONIC CRANIAL SKELETON MORPHOGENESIS | 14 | 46 | 1.041e-05 | 5.071e-05 |
| 956 | SKIN EPIDERMIS DEVELOPMENT | 18 | 71 | 1.051e-05 | 5.106e-05 |
| 957 | CELL FATE SPECIFICATION | 18 | 71 | 1.051e-05 | 5.106e-05 |
| 958 | POSITIVE REGULATION OF STRIATED MUSCLE CELL DIFFERENTIATION | 15 | 52 | 1.051e-05 | 5.106e-05 |
| 959 | CELLULAR RESPONSE TO GROWTH HORMONE STIMULUS | 9 | 20 | 1.116e-05 | 5.402e-05 |
| 960 | TRACHEA DEVELOPMENT | 9 | 20 | 1.116e-05 | 5.402e-05 |
| 961 | NEGATIVE REGULATION OF ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 9 | 20 | 1.116e-05 | 5.402e-05 |
| 962 | NEURON APOPTOTIC PROCESS | 12 | 35 | 1.139e-05 | 5.511e-05 |
| 963 | EYE MORPHOGENESIS | 27 | 136 | 1.169e-05 | 5.647e-05 |
| 964 | REGULATION OF CATION TRANSMEMBRANE TRANSPORT | 36 | 208 | 1.21e-05 | 5.839e-05 |
| 965 | NEGATIVE REGULATION OF CELL PROJECTION ORGANIZATION | 28 | 144 | 1.222e-05 | 5.893e-05 |
| 966 | PROTEIN EXPORT FROM NUCLEUS | 11 | 30 | 1.268e-05 | 6.065e-05 |
| 967 | SMOOTH MUSCLE CELL DIFFERENTIATION | 11 | 30 | 1.268e-05 | 6.065e-05 |
| 968 | NEGATIVE REGULATION OF MUSCLE CELL APOPTOTIC PROCESS | 11 | 30 | 1.268e-05 | 6.065e-05 |
| 969 | NEGATIVE REGULATION OF CELL MATRIX ADHESION | 11 | 30 | 1.268e-05 | 6.065e-05 |
| 970 | NEGATIVE REGULATION OF TOR SIGNALING | 11 | 30 | 1.268e-05 | 6.065e-05 |
| 971 | REGULATION OF LYMPHOCYTE MEDIATED IMMUNITY | 24 | 114 | 1.26e-05 | 6.065e-05 |
| 972 | MODULATION OF EXCITATORY POSTSYNAPTIC POTENTIAL | 11 | 30 | 1.268e-05 | 6.065e-05 |
| 973 | REGULATION OF MEMBRANE REPOLARIZATION | 11 | 30 | 1.268e-05 | 6.065e-05 |
| 974 | POSITIVE REGULATION OF CATENIN IMPORT INTO NUCLEUS | 7 | 12 | 1.274e-05 | 6.071e-05 |
| 975 | MAMMARY GLAND EPITHELIAL CELL PROLIFERATION | 7 | 12 | 1.274e-05 | 6.071e-05 |
| 976 | REGULATION OF WNT SIGNALING PATHWAY PLANAR CELL POLARITY PATHWAY | 7 | 12 | 1.274e-05 | 6.071e-05 |
| 977 | POSITIVE REGULATION OF DNA BIOSYNTHETIC PROCESS | 16 | 59 | 1.284e-05 | 6.113e-05 |
| 978 | HISTONE PHOSPHORYLATION | 10 | 25 | 1.286e-05 | 6.119e-05 |
| 979 | RESPONSE TO CARBOHYDRATE | 31 | 168 | 1.297e-05 | 6.166e-05 |
| 980 | REGULATION OF MEMBRANE DEPOLARIZATION | 13 | 41 | 1.307e-05 | 6.198e-05 |
| 981 | ANDROGEN RECEPTOR SIGNALING PATHWAY | 13 | 41 | 1.307e-05 | 6.198e-05 |
| 982 | REGULATION OF SMAD PROTEIN IMPORT INTO NUCLEUS | 8 | 16 | 1.333e-05 | 6.311e-05 |
| 983 | REGULATION OF GENE EXPRESSION BY GENETIC IMPRINTING | 8 | 16 | 1.333e-05 | 6.311e-05 |
| 984 | REGULATION OF CALCIUM ION TRANSPORT | 36 | 209 | 1.349e-05 | 6.38e-05 |
| 985 | PEPTIDYL TYROSINE DEPHOSPHORYLATION | 22 | 100 | 1.376e-05 | 6.502e-05 |
| 986 | REGULATION OF MULTICELLULAR ORGANISM GROWTH | 17 | 66 | 1.465e-05 | 6.907e-05 |
| 987 | POSITIVE REGULATION OF LIPASE ACTIVITY | 17 | 66 | 1.465e-05 | 6.907e-05 |
| 988 | LIPID MODIFICATION | 36 | 210 | 1.503e-05 | 7.079e-05 |
| 989 | CELL CYCLE G2 M PHASE TRANSITION | 27 | 138 | 1.544e-05 | 7.264e-05 |
| 990 | SEXUAL REPRODUCTION | 92 | 730 | 1.56e-05 | 7.333e-05 |
| 991 | NEGATIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 12 | 36 | 1.583e-05 | 7.418e-05 |
| 992 | NEGATIVE REGULATION OF PROTEIN KINASE B SIGNALING | 12 | 36 | 1.583e-05 | 7.418e-05 |
| 993 | POSITIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 12 | 36 | 1.583e-05 | 7.418e-05 |
| 994 | POSITIVE REGULATION OF PROTEIN BINDING | 18 | 73 | 1.591e-05 | 7.44e-05 |
| 995 | POSITIVE REGULATION OF ADAPTIVE IMMUNE RESPONSE | 18 | 73 | 1.591e-05 | 7.44e-05 |
| 996 | CAMERA TYPE EYE MORPHOGENESIS | 22 | 101 | 1.626e-05 | 7.597e-05 |
| 997 | RESPONSE TO STARVATION | 29 | 154 | 1.635e-05 | 7.63e-05 |
| 998 | REGULATION OF MULTI ORGANISM PROCESS | 65 | 470 | 1.65e-05 | 7.694e-05 |
| 999 | ESTABLISHMENT OF LOCALIZATION IN CELL | 183 | 1676 | 1.654e-05 | 7.703e-05 |
| 1000 | ACTION POTENTIAL | 21 | 94 | 1.672e-05 | 7.782e-05 |
| 1001 | LEARNING | 26 | 131 | 1.693e-05 | 7.871e-05 |
| 1002 | CARDIAC MUSCLE TISSUE MORPHOGENESIS | 15 | 54 | 1.738e-05 | 8.046e-05 |
| 1003 | ALPHA BETA T CELL ACTIVATION | 15 | 54 | 1.738e-05 | 8.046e-05 |
| 1004 | REGULATION OF MITOCHONDRIAL MEMBRANE POTENTIAL | 15 | 54 | 1.738e-05 | 8.046e-05 |
| 1005 | B CELL RECEPTOR SIGNALING PATHWAY | 15 | 54 | 1.738e-05 | 8.046e-05 |
| 1006 | REGULATION OF MYOBLAST DIFFERENTIATION | 14 | 48 | 1.793e-05 | 8.294e-05 |
| 1007 | REGULATION OF GENE SILENCING BY RNA | 9 | 21 | 1.812e-05 | 8.329e-05 |
| 1008 | REGULATION OF PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION | 9 | 21 | 1.812e-05 | 8.329e-05 |
| 1009 | MAST CELL ACTIVATION | 9 | 21 | 1.812e-05 | 8.329e-05 |
| 1010 | REGULATION OF GLIAL CELL PROLIFERATION | 9 | 21 | 1.812e-05 | 8.329e-05 |
| 1011 | NEGATIVE REGULATION OF NEURAL PRECURSOR CELL PROLIFERATION | 9 | 21 | 1.812e-05 | 8.329e-05 |
| 1012 | REGULATION OF POSTTRANSCRIPTIONAL GENE SILENCING | 9 | 21 | 1.812e-05 | 8.329e-05 |
| 1013 | MISMATCH REPAIR | 11 | 31 | 1.822e-05 | 8.351e-05 |
| 1014 | SCHWANN CELL DIFFERENTIATION | 11 | 31 | 1.822e-05 | 8.351e-05 |
| 1015 | POSITIVE REGULATION OF INTERLEUKIN 2 PRODUCTION | 11 | 31 | 1.822e-05 | 8.351e-05 |
| 1016 | CELLULAR RESPONSE TO BIOTIC STIMULUS | 30 | 163 | 1.871e-05 | 8.571e-05 |
| 1017 | REGULATION OF MESONEPHROS DEVELOPMENT | 10 | 26 | 1.938e-05 | 8.867e-05 |
| 1018 | POSITIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 34 | 196 | 1.988e-05 | 9.086e-05 |
| 1019 | OVULATION CYCLE PROCESS | 20 | 88 | 2.019e-05 | 9.219e-05 |
| 1020 | REGULATION OF SYNAPTIC PLASTICITY | 27 | 140 | 2.025e-05 | 9.24e-05 |
| 1021 | RESPONSE TO TEMPERATURE STIMULUS | 28 | 148 | 2.078e-05 | 9.469e-05 |
| 1022 | POSITIVE REGULATION OF SYNAPTIC TRANSMISSION | 23 | 110 | 2.13e-05 | 9.698e-05 |
| 1023 | MYOBLAST DIFFERENTIATION | 12 | 37 | 2.17e-05 | 9.862e-05 |
| 1024 | NEGATIVE REGULATION OF EPITHELIAL CELL DIFFERENTIATION | 12 | 37 | 2.17e-05 | 9.862e-05 |
| 1025 | REGULATION OF B CELL PROLIFERATION | 15 | 55 | 2.211e-05 | 0.0001004 |
| 1026 | PROTEIN MATURATION | 42 | 265 | 2.223e-05 | 0.0001008 |
| 1027 | REGULATION OF TOR SIGNALING | 17 | 68 | 2.24e-05 | 0.0001014 |
| 1028 | POSITIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 17 | 68 | 2.24e-05 | 0.0001014 |
| 1029 | REGULATION OF CALCIUM ION IMPORT | 22 | 103 | 2.252e-05 | 0.0001018 |
| 1030 | ESTABLISHMENT OR MAINTENANCE OF CELL POLARITY | 27 | 141 | 2.314e-05 | 0.0001045 |
| 1031 | REGULATION OF MUSCLE CELL APOPTOTIC PROCESS | 13 | 43 | 2.326e-05 | 0.0001047 |
| 1032 | REGULATION OF POLYSACCHARIDE METABOLIC PROCESS | 13 | 43 | 2.326e-05 | 0.0001047 |
| 1033 | BETA CATENIN TCF COMPLEX ASSEMBLY | 13 | 43 | 2.326e-05 | 0.0001047 |
| 1034 | REGULATION OF TELOMERASE ACTIVITY | 13 | 43 | 2.326e-05 | 0.0001047 |
| 1035 | NEGATIVE REGULATION OF KIDNEY DEVELOPMENT | 8 | 17 | 2.339e-05 | 0.0001049 |
| 1036 | REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 8 | 17 | 2.339e-05 | 0.0001049 |
| 1037 | REGULATION OF ENDOTHELIAL CELL CHEMOTAXIS | 8 | 17 | 2.339e-05 | 0.0001049 |
| 1038 | MAINTENANCE OF LOCATION IN CELL | 21 | 96 | 2.348e-05 | 0.0001053 |
| 1039 | NEGATIVE REGULATION OF VIRAL PROCESS | 20 | 89 | 2.407e-05 | 0.0001078 |
| 1040 | JNK CASCADE | 19 | 82 | 2.418e-05 | 0.0001082 |
| 1041 | REGULATION OF WOUND HEALING | 25 | 126 | 2.455e-05 | 0.0001097 |
| 1042 | REGULATION OF TRANSPORTER ACTIVITY | 34 | 198 | 2.473e-05 | 0.0001104 |
| 1043 | REGULATION OF TYPE I INTERFERON PRODUCTION | 23 | 111 | 2.483e-05 | 0.0001108 |
| 1044 | FOREBRAIN CELL MIGRATION | 16 | 62 | 2.532e-05 | 0.0001126 |
| 1045 | NEGATIVE REGULATION OF SYNAPTIC TRANSMISSION | 16 | 62 | 2.532e-05 | 0.0001126 |
| 1046 | BLASTOCYST DEVELOPMENT | 16 | 62 | 2.532e-05 | 0.0001126 |
| 1047 | PHAGOCYTOSIS | 33 | 190 | 2.549e-05 | 0.0001133 |
| 1048 | SECRETION BY CELL | 66 | 486 | 2.555e-05 | 0.0001134 |
| 1049 | MITOTIC CELL CYCLE ARREST | 7 | 13 | 2.565e-05 | 0.0001136 |
| 1050 | POSITIVE REGULATION OF PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION | 7 | 13 | 2.565e-05 | 0.0001136 |
| 1051 | POSITIVE REGULATION OF ENDOPLASMIC RETICULUM UNFOLDED PROTEIN RESPONSE | 7 | 13 | 2.565e-05 | 0.0001136 |
| 1052 | NEGATIVE REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 11 | 32 | 2.573e-05 | 0.0001136 |
| 1053 | REGULATION OF ORGAN FORMATION | 11 | 32 | 2.573e-05 | 0.0001136 |
| 1054 | REGULATION OF ACTIN FILAMENT BASED MOVEMENT | 11 | 32 | 2.573e-05 | 0.0001136 |
| 1055 | RESPONSE TO CAMP | 22 | 104 | 2.64e-05 | 0.0001164 |
| 1056 | ACTIN FILAMENT ORGANIZATION | 31 | 174 | 2.661e-05 | 0.0001173 |
| 1057 | CELL MATRIX ADHESION | 24 | 119 | 2.674e-05 | 0.0001177 |
| 1058 | RESPONSE TO ACTIVITY | 17 | 69 | 2.749e-05 | 0.0001209 |
| 1059 | NEUROMUSCULAR PROCESS | 21 | 97 | 2.77e-05 | 0.0001217 |
| 1060 | OUTFLOW TRACT MORPHOGENESIS | 15 | 56 | 2.795e-05 | 0.0001227 |
| 1061 | BONE CELL DEVELOPMENT | 9 | 22 | 2.845e-05 | 0.0001246 |
| 1062 | POSITIVE REGULATION OF MESONEPHROS DEVELOPMENT | 9 | 22 | 2.845e-05 | 0.0001246 |
| 1063 | REGULATION OF OXIDOREDUCTASE ACTIVITY | 20 | 90 | 2.861e-05 | 0.0001247 |
| 1064 | NEGATIVE REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 20 | 90 | 2.861e-05 | 0.0001247 |
| 1065 | REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 10 | 27 | 2.854e-05 | 0.0001247 |
| 1066 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER INVOLVED IN CELLULAR RESPONSE TO CHEMICAL STIMULUS | 10 | 27 | 2.854e-05 | 0.0001247 |
| 1067 | REGULATION OF GLIOGENESIS | 20 | 90 | 2.861e-05 | 0.0001247 |
| 1068 | CENTRAL NERVOUS SYSTEM NEURON AXONOGENESIS | 10 | 27 | 2.854e-05 | 0.0001247 |
| 1069 | EAR MORPHOGENESIS | 23 | 112 | 2.887e-05 | 0.0001257 |
| 1070 | COLLAGEN FIBRIL ORGANIZATION | 12 | 38 | 2.939e-05 | 0.0001277 |
| 1071 | REGULATION OF ACTION POTENTIAL | 12 | 38 | 2.939e-05 | 0.0001277 |
| 1072 | POSITIVE REGULATION OF CYTOSKELETON ORGANIZATION | 31 | 175 | 2.986e-05 | 0.0001293 |
| 1073 | FACE DEVELOPMENT | 14 | 50 | 2.988e-05 | 0.0001293 |
| 1074 | STAT CASCADE | 14 | 50 | 2.988e-05 | 0.0001293 |
| 1075 | JAK STAT CASCADE | 14 | 50 | 2.988e-05 | 0.0001293 |
| 1076 | NEGATIVE REGULATION OF MULTI ORGANISM PROCESS | 28 | 151 | 3.043e-05 | 0.0001316 |
| 1077 | POSITIVE REGULATION OF CHEMOTAXIS | 24 | 120 | 3.088e-05 | 0.0001334 |
| 1078 | PROTEIN POLYUBIQUITINATION | 39 | 243 | 3.225e-05 | 0.0001392 |
| 1079 | POSITIVE REGULATION OF LEUKOCYTE PROLIFERATION | 26 | 136 | 3.351e-05 | 0.0001444 |
| 1080 | MUSCLE ORGAN MORPHOGENESIS | 17 | 70 | 3.358e-05 | 0.0001444 |
| 1081 | INTERFERON GAMMA MEDIATED SIGNALING PATHWAY | 17 | 70 | 3.358e-05 | 0.0001444 |
| 1082 | REGULATION OF MEMBRANE PERMEABILITY | 17 | 70 | 3.358e-05 | 0.0001444 |
| 1083 | SECOND MESSENGER MEDIATED SIGNALING | 29 | 160 | 3.441e-05 | 0.0001478 |
| 1084 | REGULATION OF SODIUM ION TRANSPORT | 18 | 77 | 3.46e-05 | 0.0001484 |
| 1085 | REGULATION OF ACTIN FILAMENT BUNDLE ASSEMBLY | 18 | 77 | 3.46e-05 | 0.0001484 |
| 1086 | HISTONE METHYLATION | 19 | 84 | 3.464e-05 | 0.0001484 |
| 1087 | REGULATION OF CYTOKINE PRODUCTION INVOLVED IN IMMUNE RESPONSE | 15 | 57 | 3.511e-05 | 0.0001503 |
| 1088 | SOMATIC CELL DNA RECOMBINATION | 11 | 33 | 3.577e-05 | 0.0001527 |
| 1089 | SOMATIC DIVERSIFICATION OF IMMUNE RECEPTORS VIA GERMLINE RECOMBINATION WITHIN A SINGLE LOCUS | 11 | 33 | 3.577e-05 | 0.0001527 |
| 1090 | RESPONSE TO VITAMIN D | 11 | 33 | 3.577e-05 | 0.0001527 |
| 1091 | FAT CELL DIFFERENTIATION | 22 | 106 | 3.6e-05 | 0.0001535 |
| 1092 | NUCLEAR IMPORT | 25 | 129 | 3.715e-05 | 0.0001583 |
| 1093 | POSITIVE REGULATION OF ION TRANSPORT | 38 | 236 | 3.762e-05 | 0.0001602 |
| 1094 | MECHANORECEPTOR DIFFERENTIATION | 14 | 51 | 3.813e-05 | 0.0001622 |
| 1095 | MEMBRANE ORGANIZATION | 107 | 899 | 3.834e-05 | 0.0001629 |
| 1096 | POSITIVE REGULATION OF ENDOCYTOSIS | 23 | 114 | 3.879e-05 | 0.0001647 |
| 1097 | ORGAN MATURATION | 8 | 18 | 3.909e-05 | 0.0001654 |
| 1098 | REGULATION OF PROTEIN SECRETION | 55 | 389 | 3.907e-05 | 0.0001654 |
| 1099 | NOTOCHORD DEVELOPMENT | 8 | 18 | 3.909e-05 | 0.0001654 |
| 1100 | LYMPHOCYTE APOPTOTIC PROCESS | 8 | 18 | 3.909e-05 | 0.0001654 |
| 1101 | REGULATION OF PHOSPHOLIPASE C ACTIVITY | 12 | 39 | 3.932e-05 | 0.0001659 |
| 1102 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY VIA DEATH DOMAIN RECEPTORS | 12 | 39 | 3.932e-05 | 0.0001659 |
| 1103 | POSITIVE REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 12 | 39 | 3.932e-05 | 0.0001659 |
| 1104 | VENTRICULAR CARDIAC MUSCLE TISSUE DEVELOPMENT | 13 | 45 | 3.981e-05 | 0.0001678 |
| 1105 | INTRACELLULAR STEROID HORMONE RECEPTOR SIGNALING PATHWAY | 17 | 71 | 4.085e-05 | 0.000172 |
| 1106 | REGULATION OF MACROPHAGE DERIVED FOAM CELL DIFFERENTIATION | 10 | 28 | 4.117e-05 | 0.0001731 |
| 1107 | MAMMARY GLAND DUCT MORPHOGENESIS | 10 | 28 | 4.117e-05 | 0.0001731 |
| 1108 | LYMPHOCYTE COSTIMULATION | 18 | 78 | 4.16e-05 | 0.0001747 |
| 1109 | NEGATIVE REGULATION OF PROTEIN COMPLEX ASSEMBLY | 22 | 107 | 4.188e-05 | 0.0001757 |
| 1110 | GLUCOSE HOMEOSTASIS | 30 | 170 | 4.29e-05 | 0.0001797 |
| 1111 | CARBOHYDRATE HOMEOSTASIS | 30 | 170 | 4.29e-05 | 0.0001797 |
| 1112 | ADRENAL GLAND DEVELOPMENT | 9 | 23 | 4.337e-05 | 0.0001813 |
| 1113 | RESPONSE TO AUDITORY STIMULUS | 9 | 23 | 4.337e-05 | 0.0001813 |
| 1114 | PROTEIN DNA COMPLEX SUBUNIT ORGANIZATION | 37 | 229 | 4.387e-05 | 0.0001832 |
| 1115 | T CELL RECEPTOR SIGNALING PATHWAY | 27 | 146 | 4.396e-05 | 0.0001835 |
| 1116 | MULTICELLULAR ORGANISMAL SIGNALING | 24 | 123 | 4.701e-05 | 0.000196 |
| 1117 | CELLULAR RESPONSE TO RETINOIC ACID | 16 | 65 | 4.755e-05 | 0.0001979 |
| 1118 | PRODUCTION OF MOLECULAR MEDIATOR OF IMMUNE RESPONSE | 16 | 65 | 4.755e-05 | 0.0001979 |
| 1119 | DETERMINATION OF ADULT LIFESPAN | 7 | 14 | 4.769e-05 | 0.0001981 |
| 1120 | MIDGUT DEVELOPMENT | 7 | 14 | 4.769e-05 | 0.0001981 |
| 1121 | REGULATION OF PROTEIN DEACETYLATION | 11 | 34 | 4.901e-05 | 0.0002034 |
| 1122 | NEGATIVE REGULATION OF PROTEIN BINDING | 18 | 79 | 4.983e-05 | 0.0002063 |
| 1123 | REGULATION OF SYNAPSE ASSEMBLY | 18 | 79 | 4.983e-05 | 0.0002063 |
| 1124 | REGULATION OF T CELL PROLIFERATION | 27 | 147 | 4.975e-05 | 0.0002063 |
| 1125 | NEGATIVE REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 12 | 40 | 5.205e-05 | 0.0002149 |
| 1126 | ENDODERMAL CELL DIFFERENTIATION | 12 | 40 | 5.205e-05 | 0.0002149 |
| 1127 | NEGATIVE REGULATION OF OSTEOBLAST DIFFERENTIATION | 12 | 40 | 5.205e-05 | 0.0002149 |
| 1128 | REGULATION OF CELL CYCLE G2 M PHASE TRANSITION | 15 | 59 | 5.439e-05 | 0.0002244 |
| 1129 | RESPONSE TO BMP | 20 | 94 | 5.541e-05 | 0.0002282 |
| 1130 | CELLULAR RESPONSE TO BMP STIMULUS | 20 | 94 | 5.541e-05 | 0.0002282 |
| 1131 | REGULATION OF ACTIN FILAMENT LENGTH | 28 | 156 | 5.582e-05 | 0.0002297 |
| 1132 | ION HOMEOSTASIS | 74 | 576 | 5.593e-05 | 0.0002299 |
| 1133 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 16 | 66 | 5.808e-05 | 0.0002381 |
| 1134 | NEGATIVE REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 16 | 66 | 5.808e-05 | 0.0002381 |
| 1135 | REGULATION OF CARDIAC MUSCLE CONTRACTION | 16 | 66 | 5.808e-05 | 0.0002381 |
| 1136 | EPIDERMIS MORPHOGENESIS | 10 | 29 | 5.828e-05 | 0.0002387 |
| 1137 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 47 | 321 | 5.881e-05 | 0.0002407 |
| 1138 | ENDOCYTOSIS | 67 | 509 | 5.934e-05 | 0.0002426 |
| 1139 | NEGATIVE REGULATION OF LIPID METABOLIC PROCESS | 18 | 80 | 5.947e-05 | 0.0002429 |
| 1140 | ERYTHROCYTE HOMEOSTASIS | 17 | 73 | 5.965e-05 | 0.0002433 |
| 1141 | EMBRYONIC HEART TUBE DEVELOPMENT | 17 | 73 | 5.965e-05 | 0.0002433 |
| 1142 | REGULATION OF NITRIC OXIDE BIOSYNTHETIC PROCESS | 14 | 53 | 6.077e-05 | 0.0002474 |
| 1143 | NEGATIVE REGULATION OF EPITHELIAL CELL MIGRATION | 14 | 53 | 6.077e-05 | 0.0002474 |
| 1144 | CELLULAR RESPONSE TO FLUID SHEAR STRESS | 8 | 19 | 6.272e-05 | 0.0002551 |
| 1145 | REGULATION OF HISTONE DEACETYLATION | 9 | 24 | 6.44e-05 | 0.0002608 |
| 1146 | CELLULAR RESPONSE TO PROSTAGLANDIN STIMULUS | 9 | 24 | 6.44e-05 | 0.0002608 |
| 1147 | REGULATION OF ANOIKIS | 9 | 24 | 6.44e-05 | 0.0002608 |
| 1148 | NEGATIVE REGULATION OF VIRAL TRANSCRIPTION | 9 | 24 | 6.44e-05 | 0.0002608 |
| 1149 | REGULATION OF EXECUTION PHASE OF APOPTOSIS | 9 | 24 | 6.44e-05 | 0.0002608 |
| 1150 | FC GAMMA RECEPTOR SIGNALING PATHWAY | 20 | 95 | 6.489e-05 | 0.0002623 |
| 1151 | REGULATION OF LIPID TRANSPORT | 20 | 95 | 6.489e-05 | 0.0002623 |
| 1152 | REGULATION OF ESTABLISHMENT OF PLANAR POLARITY | 22 | 110 | 6.505e-05 | 0.0002627 |
| 1153 | NEURON DEATH | 13 | 47 | 6.58e-05 | 0.0002653 |
| 1154 | POSITIVE REGULATION OF DEPHOSPHORYLATION | 13 | 47 | 6.58e-05 | 0.0002653 |
| 1155 | RESPONSE TO IRON ION | 11 | 35 | 6.625e-05 | 0.0002664 |
| 1156 | RESPONSE TO MONOAMINE | 11 | 35 | 6.625e-05 | 0.0002664 |
| 1157 | B CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 11 | 35 | 6.625e-05 | 0.0002664 |
| 1158 | NEGATIVE REGULATION OF CELL DIVISION | 15 | 60 | 6.712e-05 | 0.0002697 |
| 1159 | REGULATION OF LIPID STORAGE | 12 | 41 | 6.818e-05 | 0.0002735 |
| 1160 | AORTA DEVELOPMENT | 12 | 41 | 6.818e-05 | 0.0002735 |
| 1161 | MACROMOLECULE DEACYLATION | 16 | 67 | 7.061e-05 | 0.0002827 |
| 1162 | REGULATION OF SISTER CHROMATID SEGREGATION | 16 | 67 | 7.061e-05 | 0.0002827 |
| 1163 | POSITIVE REGULATION OF PROTEIN KINASE B SIGNALING | 18 | 81 | 7.072e-05 | 0.0002829 |
| 1164 | REGULATION OF DENDRITE MORPHOGENESIS | 17 | 74 | 7.164e-05 | 0.0002861 |
| 1165 | CELL CELL JUNCTION ASSEMBLY | 17 | 74 | 7.164e-05 | 0.0002861 |
| 1166 | DIVALENT INORGANIC CATION HOMEOSTASIS | 49 | 343 | 7.776e-05 | 0.0003103 |
| 1167 | REGULATION OF RAS PROTEIN SIGNAL TRANSDUCTION | 31 | 184 | 8e-05 | 0.000319 |
| 1168 | ADULT BEHAVIOR | 25 | 135 | 8.109e-05 | 0.0003217 |
| 1169 | POSITIVE REGULATION OF CYTOKINE PRODUCTION INVOLVED IN IMMUNE RESPONSE | 10 | 30 | 8.108e-05 | 0.0003217 |
| 1170 | NEGATIVE REGULATION OF INTRACELLULAR STEROID HORMONE RECEPTOR SIGNALING PATHWAY | 10 | 30 | 8.108e-05 | 0.0003217 |
| 1171 | RESPONSE TO GROWTH HORMONE | 10 | 30 | 8.108e-05 | 0.0003217 |
| 1172 | REGULATION OF HEART RATE BY CARDIAC CONDUCTION | 10 | 30 | 8.108e-05 | 0.0003217 |
| 1173 | MYD88 INDEPENDENT TOLL LIKE RECEPTOR SIGNALING PATHWAY | 10 | 30 | 8.108e-05 | 0.0003217 |
| 1174 | REGULATION OF POTASSIUM ION TRANSMEMBRANE TRANSPORT | 15 | 61 | 8.239e-05 | 0.0003265 |
| 1175 | OLFACTORY BULB INTERNEURON DIFFERENTIATION | 7 | 15 | 8.314e-05 | 0.0003289 |
| 1176 | RESPONSE TO VITAMIN E | 7 | 15 | 8.314e-05 | 0.0003289 |
| 1177 | POSITIVE REGULATION OF REACTIVE OXYGEN SPECIES BIOSYNTHETIC PROCESS | 13 | 48 | 8.358e-05 | 0.0003301 |
| 1178 | POSITIVE REGULATION OF WOUND HEALING | 13 | 48 | 8.358e-05 | 0.0003301 |
| 1179 | NEGATIVE REGULATION OF NF KAPPAB TRANSCRIPTION FACTOR ACTIVITY | 16 | 68 | 8.544e-05 | 0.0003372 |
| 1180 | REGULATION OF BIOMINERAL TISSUE DEVELOPMENT | 17 | 75 | 8.571e-05 | 0.0003374 |
| 1181 | ODONTOGENESIS OF DENTIN CONTAINING TOOTH | 17 | 75 | 8.571e-05 | 0.0003374 |
| 1182 | MULTI ORGANISM BEHAVIOR | 17 | 75 | 8.571e-05 | 0.0003374 |
| 1183 | REGULATION OF ENDOTHELIAL CELL APOPTOTIC PROCESS | 12 | 42 | 8.844e-05 | 0.0003467 |
| 1184 | REGULATION OF NUCLEOTIDE CATABOLIC PROCESS | 11 | 36 | 8.843e-05 | 0.0003467 |
| 1185 | CELL DIFFERENTIATION INVOLVED IN KIDNEY DEVELOPMENT | 11 | 36 | 8.843e-05 | 0.0003467 |
| 1186 | POSITIVE REGULATION OF AXON EXTENSION | 11 | 36 | 8.843e-05 | 0.0003467 |
| 1187 | POSITIVE REGULATION OF GLUCOSE TRANSPORT | 12 | 42 | 8.844e-05 | 0.0003467 |
| 1188 | POSITIVE REGULATION OF ENDOTHELIAL CELL CHEMOTAXIS | 6 | 11 | 9.241e-05 | 0.0003598 |
| 1189 | REGULATION OF VENTRICULAR CARDIAC MUSCLE CELL ACTION POTENTIAL | 6 | 11 | 9.241e-05 | 0.0003598 |
| 1190 | NEGATIVE REGULATION OF NEUROTRANSMITTER SECRETION | 6 | 11 | 9.241e-05 | 0.0003598 |
| 1191 | POSITIVE REGULATION OF NON CANONICAL WNT SIGNALING PATHWAY | 6 | 11 | 9.241e-05 | 0.0003598 |
| 1192 | REGULATION OF ATTACHMENT OF SPINDLE MICROTUBULES TO KINETOCHORE | 6 | 11 | 9.241e-05 | 0.0003598 |
| 1193 | PROSTATE GLAND GROWTH | 6 | 11 | 9.241e-05 | 0.0003598 |
| 1194 | EPITHELIAL CELL DIFFERENTIATION INVOLVED IN PROSTATE GLAND DEVELOPMENT | 6 | 11 | 9.241e-05 | 0.0003598 |
| 1195 | OLFACTORY BULB INTERNEURON DEVELOPMENT | 6 | 11 | 9.241e-05 | 0.0003598 |
| 1196 | LENS FIBER CELL DIFFERENTIATION | 9 | 25 | 9.338e-05 | 0.0003624 |
| 1197 | EPITHELIAL TUBE BRANCHING INVOLVED IN LUNG MORPHOGENESIS | 9 | 25 | 9.338e-05 | 0.0003624 |
| 1198 | MEMBRANE ASSEMBLY | 9 | 25 | 9.338e-05 | 0.0003624 |
| 1199 | POSITIVE REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 9 | 25 | 9.338e-05 | 0.0003624 |
| 1200 | ENDOTHELIUM DEVELOPMENT | 19 | 90 | 9.418e-05 | 0.0003652 |
| 1201 | NEGATIVE REGULATION OF MUSCLE CELL DIFFERENTIATION | 14 | 55 | 9.435e-05 | 0.0003655 |
| 1202 | BONE GROWTH | 8 | 20 | 9.709e-05 | 0.000374 |
| 1203 | RESPONSE TO VITAMIN A | 8 | 20 | 9.709e-05 | 0.000374 |
| 1204 | REGULATION OF NEURON PROJECTION REGENERATION | 8 | 20 | 9.709e-05 | 0.000374 |
| 1205 | MEGAKARYOCYTE DIFFERENTIATION | 8 | 20 | 9.709e-05 | 0.000374 |
| 1206 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT5 PROTEIN | 8 | 20 | 9.709e-05 | 0.000374 |
| 1207 | LYMPH VESSEL DEVELOPMENT | 8 | 20 | 9.709e-05 | 0.000374 |
| 1208 | BRANCHING INVOLVED IN MAMMARY GLAND DUCT MORPHOGENESIS | 8 | 20 | 9.709e-05 | 0.000374 |
| 1209 | MEIOTIC CELL CYCLE | 31 | 186 | 9.838e-05 | 0.0003786 |
| 1210 | HAIR CYCLE | 18 | 83 | 9.899e-05 | 0.0003804 |
| 1211 | MOLTING CYCLE | 18 | 83 | 9.899e-05 | 0.0003804 |
| 1212 | POSITIVE REGULATION OF NUCLEAR DIVISION | 15 | 62 | 0.0001006 | 0.0003862 |
| 1213 | REGULATION OF MICROTUBULE POLYMERIZATION OR DEPOLYMERIZATION | 30 | 178 | 0.0001028 | 0.0003942 |
| 1214 | PROTEIN ACETYLATION | 24 | 129 | 0.0001035 | 0.0003967 |
| 1215 | REGULATION OF GLUCOSE METABOLIC PROCESS | 21 | 106 | 0.0001089 | 0.0004172 |
| 1216 | ENSHEATHMENT OF NEURONS | 19 | 91 | 0.0001101 | 0.0004209 |
| 1217 | AXON ENSHEATHMENT | 19 | 91 | 0.0001101 | 0.0004209 |
| 1218 | CELL CHEMOTAXIS | 28 | 162 | 0.0001105 | 0.000422 |
| 1219 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 10 | 31 | 0.000111 | 0.0004231 |
| 1220 | CARDIAC ATRIUM DEVELOPMENT | 10 | 31 | 0.000111 | 0.0004231 |
| 1221 | CELLULAR RESPONSE TO GLUCOSE STARVATION | 10 | 31 | 0.000111 | 0.0004231 |
| 1222 | CYTOKINE MEDIATED SIGNALING PATHWAY | 60 | 452 | 0.0001118 | 0.0004258 |
| 1223 | ADAPTIVE IMMUNE RESPONSE BASED ON SOMATIC RECOMBINATION OF IMMUNE RECEPTORS BUILT FROM IMMUNOGLOBULIN SUPERFAMILY DOMAINS | 27 | 154 | 0.0001135 | 0.0004314 |
| 1224 | CRANIAL NERVE DEVELOPMENT | 12 | 43 | 0.0001137 | 0.0004314 |
| 1225 | MORPHOGENESIS OF AN EPITHELIAL SHEET | 12 | 43 | 0.0001137 | 0.0004314 |
| 1226 | CEREBRAL CORTEX CELL MIGRATION | 12 | 43 | 0.0001137 | 0.0004314 |
| 1227 | CORONARY VASCULATURE DEVELOPMENT | 11 | 37 | 0.0001167 | 0.0004406 |
| 1228 | RESPONSE TO NERVE GROWTH FACTOR | 11 | 37 | 0.0001167 | 0.0004406 |
| 1229 | CELL COMMUNICATION INVOLVED IN CARDIAC CONDUCTION | 11 | 37 | 0.0001167 | 0.0004406 |
| 1230 | REGULATION OF MUSCLE HYPERTROPHY | 11 | 37 | 0.0001167 | 0.0004406 |
| 1231 | MICROTUBULE ORGANIZING CENTER ORGANIZATION | 18 | 84 | 0.0001165 | 0.0004406 |
| 1232 | REGULATION OF PROTEIN AUTOPHOSPHORYLATION | 11 | 37 | 0.0001167 | 0.0004406 |
| 1233 | ORGANELLE LOCALIZATION | 56 | 415 | 0.0001208 | 0.000456 |
| 1234 | REGULATION OF OSTEOCLAST DIFFERENTIATION | 15 | 63 | 0.0001222 | 0.0004609 |
| 1235 | POSITIVE REGULATION OF TYPE I INTERFERON PRODUCTION | 16 | 70 | 0.0001235 | 0.0004653 |
| 1236 | SENSORY PERCEPTION OF MECHANICAL STIMULUS | 27 | 155 | 0.000127 | 0.0004781 |
| 1237 | REGULATION OF CALCIUM ION TRANSPORT INTO CYTOSOL | 19 | 92 | 0.0001283 | 0.0004826 |
| 1238 | RESPONSE TO CALCIUM ION | 22 | 115 | 0.0001297 | 0.0004873 |
| 1239 | DEVELOPMENTAL PROGRAMMED CELL DEATH | 9 | 26 | 0.0001326 | 0.0004962 |
| 1240 | NEGATIVE REGULATION OF EMBRYONIC DEVELOPMENT | 9 | 26 | 0.0001326 | 0.0004962 |
| 1241 | MESODERMAL CELL DIFFERENTIATION | 9 | 26 | 0.0001326 | 0.0004962 |
| 1242 | STRAND DISPLACEMENT | 9 | 26 | 0.0001326 | 0.0004962 |
| 1243 | SCHWANN CELL DEVELOPMENT | 9 | 26 | 0.0001326 | 0.0004962 |
| 1244 | REGULATION OF MAMMARY GLAND EPITHELIAL CELL PROLIFERATION | 7 | 16 | 0.0001374 | 0.0005091 |
| 1245 | MAMMARY GLAND EPITHELIAL CELL DIFFERENTIATION | 7 | 16 | 0.0001374 | 0.0005091 |
| 1246 | MODIFICATION OF MORPHOLOGY OR PHYSIOLOGY OF OTHER ORGANISM | 20 | 100 | 0.0001374 | 0.0005091 |
| 1247 | MEGAKARYOCYTE DEVELOPMENT | 7 | 16 | 0.0001374 | 0.0005091 |
| 1248 | NEURON CELL CELL ADHESION | 7 | 16 | 0.0001374 | 0.0005091 |
| 1249 | REGULATION OF PROTEIN HOMOOLIGOMERIZATION | 7 | 16 | 0.0001374 | 0.0005091 |
| 1250 | HEART PROCESS | 18 | 85 | 0.0001368 | 0.0005091 |
| 1251 | PROTEIN LOCALIZATION TO SYNAPSE | 7 | 16 | 0.0001374 | 0.0005091 |
| 1252 | REGULATION OF GLUCOSE TRANSPORT | 20 | 100 | 0.0001374 | 0.0005091 |
| 1253 | POSITIVE REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 7 | 16 | 0.0001374 | 0.0005091 |
| 1254 | ATRIOVENTRICULAR VALVE MORPHOGENESIS | 7 | 16 | 0.0001374 | 0.0005091 |
| 1255 | MONOCYTE DIFFERENTIATION | 7 | 16 | 0.0001374 | 0.0005091 |
| 1256 | REGULATION OF DEVELOPMENTAL PIGMENTATION | 7 | 16 | 0.0001374 | 0.0005091 |
| 1257 | REGULATION OF LEUKOCYTE MEDIATED IMMUNITY | 27 | 156 | 0.0001419 | 0.0005252 |
| 1258 | LABYRINTHINE LAYER DEVELOPMENT | 12 | 44 | 0.0001448 | 0.0005348 |
| 1259 | NEGATIVE REGULATION OF ERBB SIGNALING PATHWAY | 12 | 44 | 0.0001448 | 0.0005348 |
| 1260 | INTERSTRAND CROSS LINK REPAIR | 12 | 44 | 0.0001448 | 0.0005348 |
| 1261 | POSITIVE REGULATION OF NEUROBLAST PROLIFERATION | 8 | 21 | 0.0001457 | 0.0005367 |
| 1262 | POSITIVE REGULATION OF EXCITATORY POSTSYNAPTIC POTENTIAL | 8 | 21 | 0.0001457 | 0.0005367 |
| 1263 | SOMATIC RECOMBINATION OF IMMUNOGLOBULIN GENE SEGMENTS | 8 | 21 | 0.0001457 | 0.0005367 |
| 1264 | PROXIMAL DISTAL PATTERN FORMATION | 10 | 32 | 0.0001498 | 0.0005503 |
| 1265 | NEGATIVE REGULATION OF DNA BIOSYNTHETIC PROCESS | 10 | 32 | 0.0001498 | 0.0005503 |
| 1266 | NEGATIVE REGULATION OF HOMEOSTATIC PROCESS | 23 | 124 | 0.0001498 | 0.0005503 |
| 1267 | POSITIVE REGULATION OF DENDRITE MORPHOGENESIS | 10 | 32 | 0.0001498 | 0.0005503 |
| 1268 | ORGANELLE FISSION | 64 | 496 | 0.0001533 | 0.0005624 |
| 1269 | DNA REPLICATION | 33 | 208 | 0.0001579 | 0.0005788 |
| 1270 | REGULATION OF TUMOR NECROSIS FACTOR SUPERFAMILY CYTOKINE PRODUCTION | 20 | 101 | 0.0001584 | 0.0005799 |
| 1271 | KERATINOCYTE DIFFERENTIATION | 20 | 101 | 0.0001584 | 0.0005799 |
| 1272 | REGULATION OF HEART RATE | 18 | 86 | 0.00016 | 0.0005849 |
| 1273 | POSITIVE REGULATION OF B CELL ACTIVATION | 18 | 86 | 0.00016 | 0.0005849 |
| 1274 | POSITIVE REGULATION OF MITOTIC NUCLEAR DIVISION | 13 | 51 | 0.000164 | 0.000597 |
| 1275 | NEURAL CREST CELL MIGRATION | 13 | 51 | 0.000164 | 0.000597 |
| 1276 | ARTERY MORPHOGENESIS | 13 | 51 | 0.000164 | 0.000597 |
| 1277 | CELLULAR RESPONSE TO FATTY ACID | 13 | 51 | 0.000164 | 0.000597 |
| 1278 | HOMOTYPIC CELL CELL ADHESION | 13 | 51 | 0.000164 | 0.000597 |
| 1279 | CELLULAR MACROMOLECULAR COMPLEX ASSEMBLY | 87 | 727 | 0.0001666 | 0.0006059 |
| 1280 | PROTEIN METHYLATION | 22 | 117 | 0.0001684 | 0.0006117 |
| 1281 | PROTEIN ALKYLATION | 22 | 117 | 0.0001684 | 0.0006117 |
| 1282 | VESICLE MEDIATED TRANSPORT | 136 | 1239 | 0.0001689 | 0.0006131 |
| 1283 | NEGATIVE REGULATION OF ANDROGEN RECEPTOR SIGNALING PATHWAY | 6 | 12 | 0.0001721 | 0.0006212 |
| 1284 | POSITIVE REGULATION OF SMAD PROTEIN IMPORT INTO NUCLEUS | 6 | 12 | 0.0001721 | 0.0006212 |
| 1285 | TRACHEA MORPHOGENESIS | 6 | 12 | 0.0001721 | 0.0006212 |
| 1286 | REPLICATIVE SENESCENCE | 6 | 12 | 0.0001721 | 0.0006212 |
| 1287 | ACTIVATION OF TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE ACTIVITY | 6 | 12 | 0.0001721 | 0.0006212 |
| 1288 | POSITIVE REGULATION OF INTERLEUKIN 2 BIOSYNTHETIC PROCESS | 6 | 12 | 0.0001721 | 0.0006212 |
| 1289 | POSITIVE REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 6 | 12 | 0.0001721 | 0.0006212 |
| 1290 | REGULATION OF CYTOKINE BIOSYNTHETIC PROCESS | 19 | 94 | 0.0001729 | 0.0006236 |
| 1291 | GERM CELL DEVELOPMENT | 33 | 209 | 0.0001731 | 0.000624 |
| 1292 | CELLULAR RESPONSE TO CORTICOSTEROID STIMULUS | 14 | 58 | 0.0001745 | 0.0006285 |
| 1293 | REGULATION OF MITOCHONDRION ORGANIZATION | 34 | 218 | 0.0001772 | 0.0006376 |
| 1294 | REGULATION OF REACTIVE OXYGEN SPECIES BIOSYNTHETIC PROCESS | 15 | 65 | 0.0001778 | 0.0006395 |
| 1295 | NUCLEAR TRANSPORT | 49 | 355 | 0.0001819 | 0.0006537 |
| 1296 | CENTROSOME CYCLE | 12 | 45 | 0.000183 | 0.0006561 |
| 1297 | EXOCRINE SYSTEM DEVELOPMENT | 12 | 45 | 0.000183 | 0.0006561 |
| 1298 | NEGATIVE REGULATION OF CHROMATIN MODIFICATION | 12 | 45 | 0.000183 | 0.0006561 |
| 1299 | HIPPO SIGNALING | 9 | 27 | 0.0001846 | 0.0006596 |
| 1300 | MOTOR NEURON AXON GUIDANCE | 9 | 27 | 0.0001846 | 0.0006596 |
| 1301 | REGULATION OF HISTONE H3 K4 METHYLATION | 9 | 27 | 0.0001846 | 0.0006596 |
| 1302 | HETEROTYPIC CELL CELL ADHESION | 9 | 27 | 0.0001846 | 0.0006596 |
| 1303 | REGULATION OF FATTY ACID METABOLIC PROCESS | 18 | 87 | 0.0001866 | 0.0006665 |
| 1304 | POSITIVE REGULATION OF MITOCHONDRION ORGANIZATION | 28 | 167 | 0.0001884 | 0.0006724 |
| 1305 | INTERACTION WITH HOST | 24 | 134 | 0.0001904 | 0.0006788 |
| 1306 | LONG TERM SYNAPTIC POTENTIATION | 11 | 39 | 0.0001966 | 0.0007003 |
| 1307 | DNA CONFORMATION CHANGE | 40 | 273 | 0.0001989 | 0.000708 |
| 1308 | REGULATION OF MYELINATION | 10 | 33 | 0.0001994 | 0.0007092 |
| 1309 | CIRCULATORY SYSTEM PROCESS | 50 | 366 | 0.0002023 | 0.0007187 |
| 1310 | CELLULAR CHEMICAL HOMEOSTASIS | 71 | 570 | 0.0002023 | 0.0007187 |
| 1311 | ASSOCIATIVE LEARNING | 16 | 73 | 0.0002081 | 0.0007385 |
| 1312 | POSITIVE REGULATION OF PROTEIN OLIGOMERIZATION | 8 | 22 | 0.0002127 | 0.0007508 |
| 1313 | REGULATION OF ANDROGEN RECEPTOR SIGNALING PATHWAY | 8 | 22 | 0.0002127 | 0.0007508 |
| 1314 | MODULATION OF TRANSCRIPTION IN OTHER ORGANISM INVOLVED IN SYMBIOTIC INTERACTION | 8 | 22 | 0.0002127 | 0.0007508 |
| 1315 | REGULATION OF B CELL DIFFERENTIATION | 8 | 22 | 0.0002127 | 0.0007508 |
| 1316 | CENTRAL NERVOUS SYSTEM PROJECTION NEURON AXONOGENESIS | 8 | 22 | 0.0002127 | 0.0007508 |
| 1317 | DNA METHYLATION OR DEMETHYLATION | 14 | 59 | 0.0002119 | 0.0007508 |
| 1318 | REGULATION OF ESTABLISHMENT OR MAINTENANCE OF CELL POLARITY | 8 | 22 | 0.0002127 | 0.0007508 |
| 1319 | NUCLEOSOME DISASSEMBLY | 7 | 17 | 0.0002173 | 0.0007607 |
| 1320 | PROTEIN DNA COMPLEX DISASSEMBLY | 7 | 17 | 0.0002173 | 0.0007607 |
| 1321 | NEGATIVE REGULATION OF CELL AGING | 7 | 17 | 0.0002173 | 0.0007607 |
| 1322 | NEGATIVE REGULATION OF STEM CELL PROLIFERATION | 7 | 17 | 0.0002173 | 0.0007607 |
| 1323 | POSITIVE REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 7 | 17 | 0.0002173 | 0.0007607 |
| 1324 | CHROMATIN DISASSEMBLY | 7 | 17 | 0.0002173 | 0.0007607 |
| 1325 | REGULATION OF HISTONE H3 K9 METHYLATION | 7 | 17 | 0.0002173 | 0.0007607 |
| 1326 | NEGATIVE REGULATION OF ANOIKIS | 7 | 17 | 0.0002173 | 0.0007607 |
| 1327 | NEGATIVE REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 18 | 88 | 0.0002171 | 0.0007607 |
| 1328 | REGULATION OF SISTER CHROMATID COHESION | 7 | 17 | 0.0002173 | 0.0007607 |
| 1329 | REGULATION OF PRI MIRNA TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 7 | 17 | 0.0002173 | 0.0007607 |
| 1330 | INFLAMMATORY RESPONSE | 59 | 454 | 0.0002256 | 0.0007891 |
| 1331 | REGULATION OF OXIDATIVE STRESS INDUCED CELL DEATH | 12 | 46 | 0.0002295 | 0.0008016 |
| 1332 | NEGATIVE REGULATION OF NUCLEAR DIVISION | 12 | 46 | 0.0002295 | 0.0008016 |
| 1333 | REGULATION OF JUN KINASE ACTIVITY | 17 | 81 | 0.0002328 | 0.0008126 |
| 1334 | PEPTIDYL LYSINE METHYLATION | 16 | 74 | 0.0002457 | 0.0008571 |
| 1335 | FORELIMB MORPHOGENESIS | 11 | 40 | 0.0002514 | 0.0008749 |
| 1336 | HINDBRAIN MORPHOGENESIS | 11 | 40 | 0.0002514 | 0.0008749 |
| 1337 | NEGATIVE REGULATION OF LYMPHOCYTE DIFFERENTIATION | 11 | 40 | 0.0002514 | 0.0008749 |
| 1338 | VENTRICULAR SEPTUM MORPHOGENESIS | 9 | 28 | 0.0002525 | 0.0008761 |
| 1339 | POSITIVE REGULATION OF LEUKOCYTE APOPTOTIC PROCESS | 9 | 28 | 0.0002525 | 0.0008761 |
| 1340 | REGULATION OF FATTY ACID OXIDATION | 9 | 28 | 0.0002525 | 0.0008761 |
| 1341 | REGULATION OF SPROUTING ANGIOGENESIS | 9 | 28 | 0.0002525 | 0.0008761 |
| 1342 | REGULATION OF TELOMERE MAINTENANCE | 15 | 67 | 0.0002542 | 0.0008799 |
| 1343 | REGULATION OF NOTCH SIGNALING PATHWAY | 15 | 67 | 0.0002542 | 0.0008799 |
| 1344 | NEURON FATE COMMITMENT | 15 | 67 | 0.0002542 | 0.0008799 |
| 1345 | REGULATION OF GLUCOSE IMPORT | 14 | 60 | 0.000256 | 0.0008841 |
| 1346 | MATERNAL PROCESS INVOLVED IN FEMALE PREGNANCY | 14 | 60 | 0.000256 | 0.0008841 |
| 1347 | REGULATION OF PHOSPHOPROTEIN PHOSPHATASE ACTIVITY | 14 | 60 | 0.000256 | 0.0008841 |
| 1348 | FOREBRAIN NEURON DEVELOPMENT | 10 | 34 | 0.000262 | 0.0009024 |
| 1349 | T CELL HOMEOSTASIS | 10 | 34 | 0.000262 | 0.0009024 |
| 1350 | RESPONSE TO PROSTAGLANDIN | 10 | 34 | 0.000262 | 0.0009024 |
| 1351 | BRAIN MORPHOGENESIS | 10 | 34 | 0.000262 | 0.0009024 |
| 1352 | MALE GAMETE GENERATION | 62 | 486 | 0.0002635 | 0.0009069 |
| 1353 | MITOTIC NUCLEAR DIVISION | 49 | 361 | 0.0002716 | 0.0009338 |
| 1354 | CARDIAC CONDUCTION | 17 | 82 | 0.0002717 | 0.0009338 |
| 1355 | RESPONSE TO ANTIBIOTIC | 12 | 47 | 0.0002856 | 0.0009807 |
| 1356 | REGULATION OF DNA REPAIR | 16 | 75 | 0.0002891 | 0.0009913 |
| 1357 | POSITIVE REGULATION OF CARBOHYDRATE METABOLIC PROCESS | 16 | 75 | 0.0002891 | 0.0009913 |
| 1358 | CALCIUM MEDIATED SIGNALING | 18 | 90 | 0.0002911 | 0.0009967 |
| 1359 | MIDBRAIN DEVELOPMENT | 18 | 90 | 0.0002911 | 0.0009967 |
| 1360 | REGULATION OF CELL PROLIFERATION INVOLVED IN KIDNEY DEVELOPMENT | 6 | 13 | 0.0002976 | 0.001015 |
| 1361 | EYELID DEVELOPMENT IN CAMERA TYPE EYE | 6 | 13 | 0.0002976 | 0.001015 |
| 1362 | HEART VALVE FORMATION | 6 | 13 | 0.0002976 | 0.001015 |
| 1363 | LYMPH VESSEL MORPHOGENESIS | 6 | 13 | 0.0002976 | 0.001015 |
| 1364 | POSITIVE REGULATION OF CARDIAC MUSCLE CELL DIFFERENTIATION | 6 | 13 | 0.0002976 | 0.001015 |
| 1365 | MAINTENANCE OF LOCATION | 24 | 138 | 0.0003011 | 0.001027 |
| 1366 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT PROTEIN | 15 | 68 | 0.0003019 | 0.001028 |
| 1367 | REGULATION OF NUCLEASE ACTIVITY | 8 | 23 | 0.000303 | 0.001028 |
| 1368 | REGULATION OF OSTEOBLAST PROLIFERATION | 8 | 23 | 0.000303 | 0.001028 |
| 1369 | POSITIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 8 | 23 | 0.000303 | 0.001028 |
| 1370 | LEUKOCYTE APOPTOTIC PROCESS | 8 | 23 | 0.000303 | 0.001028 |
| 1371 | NEUROTROPHIN SIGNALING PATHWAY | 8 | 23 | 0.000303 | 0.001028 |
| 1372 | REGULATION OF TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER | 8 | 23 | 0.000303 | 0.001028 |
| 1373 | DEMETHYLATION | 13 | 54 | 0.0003033 | 0.001028 |
| 1374 | LYMPHOCYTE ACTIVATION INVOLVED IN IMMUNE RESPONSE | 19 | 98 | 0.0003043 | 0.00103 |
| 1375 | REGULATION OF PROTEIN TYROSINE KINASE ACTIVITY | 14 | 61 | 0.0003077 | 0.00104 |
| 1376 | REGULATION OF VIRAL TRANSCRIPTION | 14 | 61 | 0.0003077 | 0.00104 |
| 1377 | PROTEIN IMPORT | 26 | 155 | 0.0003118 | 0.001053 |
| 1378 | INNATE IMMUNE RESPONSE ACTIVATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 20 | 106 | 0.0003118 | 0.001053 |
| 1379 | REGULATION OF MITOCHONDRIAL DEPOLARIZATION | 7 | 18 | 0.0003306 | 0.001111 |
| 1380 | POSITIVE REGULATION OF SYNAPTIC TRANSMISSION GLUTAMATERGIC | 7 | 18 | 0.0003306 | 0.001111 |
| 1381 | POSITIVE REGULATION OF CELL CYCLE G2 M PHASE TRANSITION | 7 | 18 | 0.0003306 | 0.001111 |
| 1382 | MAST CELL MEDIATED IMMUNITY | 7 | 18 | 0.0003306 | 0.001111 |
| 1383 | POST ANAL TAIL MORPHOGENESIS | 7 | 18 | 0.0003306 | 0.001111 |
| 1384 | RETINOIC ACID RECEPTOR SIGNALING PATHWAY | 7 | 18 | 0.0003306 | 0.001111 |
| 1385 | UTERUS DEVELOPMENT | 7 | 18 | 0.0003306 | 0.001111 |
| 1386 | RESPONSE TO ORGANOPHOSPHORUS | 24 | 139 | 0.0003365 | 0.00113 |
| 1387 | REGULATION OF PROTEIN OLIGOMERIZATION | 10 | 35 | 0.0003403 | 0.001135 |
| 1388 | NEGATIVE REGULATION OF RESPONSE TO OXIDATIVE STRESS | 10 | 35 | 0.0003403 | 0.001135 |
| 1389 | REGULATION OF CARDIAC MUSCLE CELL PROLIFERATION | 9 | 29 | 0.0003399 | 0.001135 |
| 1390 | REGULATION OF CELLULAR RESPONSE TO HEAT | 16 | 76 | 0.000339 | 0.001135 |
| 1391 | REGULATION OF VACUOLAR TRANSPORT | 9 | 29 | 0.0003399 | 0.001135 |
| 1392 | REGULATION OF OLIGODENDROCYTE DIFFERENTIATION | 9 | 29 | 0.0003399 | 0.001135 |
| 1393 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO OXIDATIVE STRESS | 10 | 35 | 0.0003403 | 0.001135 |
| 1394 | EMBRYONIC CAMERA TYPE EYE DEVELOPMENT | 10 | 35 | 0.0003403 | 0.001135 |
| 1395 | REGULATION OF OXIDATIVE STRESS INDUCED INTRINSIC APOPTOTIC SIGNALING PATHWAY | 9 | 29 | 0.0003399 | 0.001135 |
| 1396 | PROTEASOMAL PROTEIN CATABOLIC PROCESS | 39 | 271 | 0.0003438 | 0.001146 |
| 1397 | IMMUNOGLOBULIN PRODUCTION | 12 | 48 | 0.0003529 | 0.001174 |
| 1398 | POSITIVE REGULATION OF RESPONSE TO EXTRACELLULAR STIMULUS | 12 | 48 | 0.0003529 | 0.001174 |
| 1399 | POSITIVE REGULATION OF RESPONSE TO NUTRIENT LEVELS | 12 | 48 | 0.0003529 | 0.001174 |
| 1400 | POSITIVE REGULATION OF LYMPHOCYTE MEDIATED IMMUNITY | 15 | 69 | 0.0003572 | 0.001187 |
| 1401 | CYTOKINESIS | 17 | 84 | 0.0003669 | 0.001219 |
| 1402 | REGULATION OF TISSUE REMODELING | 14 | 62 | 0.000368 | 0.001221 |
| 1403 | CHROMOSOME SEGREGATION | 39 | 272 | 0.0003706 | 0.001228 |
| 1404 | MULTICELLULAR ORGANISMAL HOMEOSTASIS | 39 | 272 | 0.0003706 | 0.001228 |
| 1405 | DIENCEPHALON DEVELOPMENT | 16 | 77 | 0.0003961 | 0.001312 |
| 1406 | DETECTION OF MECHANICAL STIMULUS | 11 | 42 | 0.0004002 | 0.001319 |
| 1407 | DENDRITE MORPHOGENESIS | 11 | 42 | 0.0004002 | 0.001319 |
| 1408 | SOMATIC DIVERSIFICATION OF IMMUNE RECEPTORS | 11 | 42 | 0.0004002 | 0.001319 |
| 1409 | REGULATION OF NIK NF KAPPAB SIGNALING | 11 | 42 | 0.0004002 | 0.001319 |
| 1410 | REGULATION OF CELLULAR CARBOHYDRATE CATABOLIC PROCESS | 11 | 42 | 0.0004002 | 0.001319 |
| 1411 | GENITALIA DEVELOPMENT | 11 | 42 | 0.0004002 | 0.001319 |
| 1412 | REGULATION OF CARBOHYDRATE CATABOLIC PROCESS | 11 | 42 | 0.0004002 | 0.001319 |
| 1413 | RESPONSE TO FIBROBLAST GROWTH FACTOR | 21 | 116 | 0.0004018 | 0.001323 |
| 1414 | POSITIVE REGULATION OF DNA TEMPLATED TRANSCRIPTION INITIATION | 8 | 24 | 0.0004223 | 0.001385 |
| 1415 | EMBRYONIC CAMERA TYPE EYE MORPHOGENESIS | 8 | 24 | 0.0004223 | 0.001385 |
| 1416 | POSITIVE REGULATION OF CELL JUNCTION ASSEMBLY | 8 | 24 | 0.0004223 | 0.001385 |
| 1417 | NEGATIVE REGULATION OF MYOBLAST DIFFERENTIATION | 8 | 24 | 0.0004223 | 0.001385 |
| 1418 | NON RECOMBINATIONAL REPAIR | 15 | 70 | 0.0004209 | 0.001385 |
| 1419 | EPITHELIAL CELL DIFFERENTIATION INVOLVED IN KIDNEY DEVELOPMENT | 8 | 24 | 0.0004223 | 0.001385 |
| 1420 | ACTIN FILAMENT BUNDLE ORGANIZATION | 12 | 49 | 0.0004332 | 0.00142 |
| 1421 | NEGATIVE REGULATION OF HISTONE MODIFICATION | 10 | 36 | 0.0004372 | 0.00143 |
| 1422 | SEMAPHORIN PLEXIN SIGNALING PATHWAY | 10 | 36 | 0.0004372 | 0.00143 |
| 1423 | POSITIVE CHEMOTAXIS | 10 | 36 | 0.0004372 | 0.00143 |
| 1424 | MEMBRANE BIOGENESIS | 9 | 30 | 0.0004508 | 0.00147 |
| 1425 | CEREBELLAR CORTEX MORPHOGENESIS | 9 | 30 | 0.0004508 | 0.00147 |
| 1426 | REGULATION OF ENDOPLASMIC RETICULUM STRESS INDUCED INTRINSIC APOPTOTIC SIGNALING PATHWAY | 9 | 30 | 0.0004508 | 0.00147 |
| 1427 | REGULATION OF NEURON MIGRATION | 9 | 30 | 0.0004508 | 0.00147 |
| 1428 | REGULATION OF RECEPTOR MEDIATED ENDOCYTOSIS | 16 | 78 | 0.0004613 | 0.001503 |
| 1429 | PROTEIN ACYLATION | 26 | 159 | 0.000468 | 0.001524 |
| 1430 | RESPONSE TO MUSCLE STRETCH | 7 | 19 | 0.0004869 | 0.001562 |
| 1431 | CELLULAR RESPONSE TO GAMMA RADIATION | 7 | 19 | 0.0004869 | 0.001562 |
| 1432 | FIBRIL ORGANIZATION | 7 | 19 | 0.0004869 | 0.001562 |
| 1433 | REGULATION OF CARDIAC MUSCLE CELL DIFFERENTIATION | 7 | 19 | 0.0004869 | 0.001562 |
| 1434 | MUSCLE CELL PROLIFERATION | 7 | 19 | 0.0004869 | 0.001562 |
| 1435 | REGULATION OF FIBROBLAST APOPTOTIC PROCESS | 6 | 14 | 0.000485 | 0.001562 |
| 1436 | INSULIN LIKE GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 6 | 14 | 0.000485 | 0.001562 |
| 1437 | NEGATIVE REGULATION OF ACTIN FILAMENT BUNDLE ASSEMBLY | 7 | 19 | 0.0004869 | 0.001562 |
| 1438 | POSITIVE REGULATION OF P38MAPK CASCADE | 6 | 14 | 0.000485 | 0.001562 |
| 1439 | POSITIVE REGULATION OF LONG TERM SYNAPTIC POTENTIATION | 6 | 14 | 0.000485 | 0.001562 |
| 1440 | ENTEROENDOCRINE CELL DIFFERENTIATION | 7 | 19 | 0.0004869 | 0.001562 |
| 1441 | POSITIVE REGULATION OF CARDIAC MUSCLE CELL PROLIFERATION | 7 | 19 | 0.0004869 | 0.001562 |
| 1442 | POSITIVE REGULATION OF TRANSCRIPTION REGULATORY REGION DNA BINDING | 6 | 14 | 0.000485 | 0.001562 |
| 1443 | POSITIVE REGULATION OF CELL SIZE | 6 | 14 | 0.000485 | 0.001562 |
| 1444 | CELLULAR RESPONSE TO VITAMIN D | 6 | 14 | 0.000485 | 0.001562 |
| 1445 | POSITIVE REGULATION OF PROTEIN EXPORT FROM NUCLEUS | 7 | 19 | 0.0004869 | 0.001562 |
| 1446 | ATRIOVENTRICULAR VALVE DEVELOPMENT | 7 | 19 | 0.0004869 | 0.001562 |
| 1447 | CELLULAR RESPONSE TO REACTIVE NITROGEN SPECIES | 7 | 19 | 0.0004869 | 0.001562 |
| 1448 | NEGATIVE REGULATION OF GENE SILENCING | 7 | 19 | 0.0004869 | 0.001562 |
| 1449 | RESPONSE TO LAMINAR FLUID SHEAR STRESS | 6 | 14 | 0.000485 | 0.001562 |
| 1450 | ANATOMICAL STRUCTURE HOMEOSTASIS | 40 | 285 | 0.0004871 | 0.001562 |
| 1451 | ESTROUS CYCLE | 7 | 19 | 0.0004869 | 0.001562 |
| 1452 | REGULATION OF HEART CONTRACTION | 33 | 221 | 0.0004905 | 0.001572 |
| 1453 | REGULATION OF INSULIN RECEPTOR SIGNALING PATHWAY | 11 | 43 | 0.0004987 | 0.001597 |
| 1454 | INNATE IMMUNE RESPONSE | 74 | 619 | 0.0005132 | 0.001642 |
| 1455 | POSITIVE REGULATION OF PRODUCTION OF MOLECULAR MEDIATOR OF IMMUNE RESPONSE | 14 | 64 | 0.0005196 | 0.001662 |
| 1456 | VISUAL BEHAVIOR | 12 | 50 | 0.0005284 | 0.001689 |
| 1457 | APOPTOTIC MITOCHONDRIAL CHANGES | 13 | 57 | 0.000533 | 0.001702 |
| 1458 | REGULATION OF STRIATED MUSCLE CONTRACTION | 16 | 79 | 0.0005354 | 0.001709 |
| 1459 | POSITIVE REGULATION OF B CELL PROLIFERATION | 10 | 37 | 0.0005559 | 0.001772 |
| 1460 | CARDIAC MUSCLE CELL ACTION POTENTIAL | 10 | 37 | 0.0005559 | 0.001772 |
| 1461 | REGULATION OF CARBOHYDRATE BIOSYNTHETIC PROCESS | 17 | 87 | 0.0005632 | 0.001794 |
| 1462 | POSITIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 21 | 119 | 0.0005721 | 0.001821 |
| 1463 | REGULATION OF JAK STAT CASCADE | 24 | 144 | 0.0005732 | 0.001822 |
| 1464 | REGULATION OF STAT CASCADE | 24 | 144 | 0.0005732 | 0.001822 |
| 1465 | NEGATIVE REGULATION OF SMOOTHENED SIGNALING PATHWAY | 8 | 25 | 0.000577 | 0.00183 |
| 1466 | THYROID GLAND DEVELOPMENT | 8 | 25 | 0.000577 | 0.00183 |
| 1467 | NEGATIVE REGULATION OF OSTEOCLAST DIFFERENTIATION | 8 | 25 | 0.000577 | 0.00183 |
| 1468 | POSITIVE REGULATION OF T CELL PROLIFERATION | 18 | 95 | 0.0005789 | 0.001835 |
| 1469 | GLYCEROPHOSPHOLIPID METABOLIC PROCESS | 41 | 297 | 0.0005861 | 0.001857 |
| 1470 | MULTICELLULAR ORGANISM AGING | 9 | 31 | 0.0005898 | 0.001864 |
| 1471 | REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR PRODUCTION | 9 | 31 | 0.0005898 | 0.001864 |
| 1472 | HOMEOSTASIS OF NUMBER OF CELLS WITHIN A TISSUE | 9 | 31 | 0.0005898 | 0.001864 |
| 1473 | MYELOID CELL DEVELOPMENT | 11 | 44 | 0.0006167 | 0.001945 |
| 1474 | REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 11 | 44 | 0.0006167 | 0.001945 |
| 1475 | NEGATIVE REGULATION OF RESPONSE TO CYTOKINE STIMULUS | 11 | 44 | 0.0006167 | 0.001945 |
| 1476 | POSITIVE REGULATION OF LYMPHOCYTE DIFFERENTIATION | 16 | 80 | 0.0006196 | 0.001953 |
| 1477 | RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 34 | 233 | 0.0006209 | 0.001955 |
| 1478 | RESPONSE TO TUMOR NECROSIS FACTOR | 34 | 233 | 0.0006209 | 0.001955 |
| 1479 | REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION TO MITOCHONDRION | 22 | 128 | 0.0006223 | 0.001958 |
| 1480 | REGULATION OF INTERLEUKIN 1 PRODUCTION | 13 | 58 | 0.0006366 | 0.002 |
| 1481 | POSITIVE REGULATION OF CYTOKINE BIOSYNTHETIC PROCESS | 13 | 58 | 0.0006366 | 0.002 |
| 1482 | POSITIVE REGULATION OF FAT CELL DIFFERENTIATION | 12 | 51 | 0.0006405 | 0.002008 |
| 1483 | REGULATION OF NEUROTRANSMITTER SECRETION | 12 | 51 | 0.0006405 | 0.002008 |
| 1484 | NEUROMUSCULAR PROCESS CONTROLLING BALANCE | 12 | 51 | 0.0006405 | 0.002008 |
| 1485 | REGULATION OF RESPONSE TO EXTRACELLULAR STIMULUS | 27 | 171 | 0.000646 | 0.002023 |
| 1486 | REGULATION OF RESPONSE TO NUTRIENT LEVELS | 27 | 171 | 0.000646 | 0.002023 |
| 1487 | GLYCEROLIPID METABOLIC PROCESS | 47 | 356 | 0.0006644 | 0.002079 |
| 1488 | RESPONSE TO ALKALOID | 23 | 137 | 0.0006657 | 0.002082 |
| 1489 | ENDOPLASMIC RETICULUM CALCIUM ION HOMEOSTASIS | 7 | 20 | 0.0006968 | 0.002175 |
| 1490 | REGULATION OF MACROPHAGE DIFFERENTIATION | 7 | 20 | 0.0006968 | 0.002175 |
| 1491 | DORSAL VENTRAL AXIS SPECIFICATION | 7 | 20 | 0.0006968 | 0.002175 |
| 1492 | B CELL PROLIFERATION | 10 | 38 | 0.0007001 | 0.002176 |
| 1493 | POSITIVE REGULATION OF POTASSIUM ION TRANSPORT | 10 | 38 | 0.0007001 | 0.002176 |
| 1494 | BONE MINERALIZATION | 10 | 38 | 0.0007001 | 0.002176 |
| 1495 | POSITIVE REGULATION OF DNA REPAIR | 10 | 38 | 0.0007001 | 0.002176 |
| 1496 | MESENCHYME MORPHOGENESIS | 10 | 38 | 0.0007001 | 0.002176 |
| 1497 | REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 10 | 38 | 0.0007001 | 0.002176 |
| 1498 | CELLULAR RESPONSE TO INSULIN STIMULUS | 24 | 146 | 0.0007026 | 0.002182 |
| 1499 | NEGATIVE REGULATION OF PROTEOLYSIS | 44 | 329 | 0.0007419 | 0.002303 |
| 1500 | POSITIVE REGULATION OF ENDOTHELIAL CELL DIFFERENTIATION | 6 | 15 | 0.0007529 | 0.002317 |
| 1501 | MESENCHYMAL TO EPITHELIAL TRANSITION | 6 | 15 | 0.0007529 | 0.002317 |
| 1502 | POSITIVE REGULATION OF CREB TRANSCRIPTION FACTOR ACTIVITY | 6 | 15 | 0.0007529 | 0.002317 |
| 1503 | RESPONSE TO MURAMYL DIPEPTIDE | 6 | 15 | 0.0007529 | 0.002317 |
| 1504 | REGULATION OF MESODERM DEVELOPMENT | 6 | 15 | 0.0007529 | 0.002317 |
| 1505 | ECTODERMAL PLACODE DEVELOPMENT | 6 | 15 | 0.0007529 | 0.002317 |
| 1506 | BLASTODERM SEGMENTATION | 6 | 15 | 0.0007529 | 0.002317 |
| 1507 | REGULATION OF CELL PROLIFERATION INVOLVED IN HEART MORPHOGENESIS | 6 | 15 | 0.0007529 | 0.002317 |
| 1508 | STRIATED MUSCLE CELL PROLIFERATION | 6 | 15 | 0.0007529 | 0.002317 |
| 1509 | RESPONSE TO HYDROPEROXIDE | 6 | 15 | 0.0007529 | 0.002317 |
| 1510 | ECTODERMAL PLACODE MORPHOGENESIS | 6 | 15 | 0.0007529 | 0.002317 |
| 1511 | ECTODERMAL PLACODE FORMATION | 6 | 15 | 0.0007529 | 0.002317 |
| 1512 | MRNA TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 6 | 15 | 0.0007529 | 0.002317 |
| 1513 | SALIVARY GLAND DEVELOPMENT | 9 | 32 | 0.000762 | 0.002334 |
| 1514 | PATTERNING OF BLOOD VESSELS | 9 | 32 | 0.000762 | 0.002334 |
| 1515 | EMBRYONIC FORELIMB MORPHOGENESIS | 9 | 32 | 0.000762 | 0.002334 |
| 1516 | METANEPHRIC NEPHRON DEVELOPMENT | 9 | 32 | 0.000762 | 0.002334 |
| 1517 | NEGATIVE REGULATION OF T CELL DIFFERENTIATION | 9 | 32 | 0.000762 | 0.002334 |
| 1518 | REGULATION OF ACTIN CYTOSKELETON REORGANIZATION | 9 | 32 | 0.000762 | 0.002334 |
| 1519 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 9 | 32 | 0.000762 | 0.002334 |
| 1520 | POSITIVE REGULATION OF REPRODUCTIVE PROCESS | 12 | 52 | 0.0007719 | 0.002358 |
| 1521 | REGULATION OF T CELL MEDIATED IMMUNITY | 12 | 52 | 0.0007719 | 0.002358 |
| 1522 | INTERACTION WITH SYMBIONT | 12 | 52 | 0.0007719 | 0.002358 |
| 1523 | REGULATION OF LIPID CATABOLIC PROCESS | 12 | 52 | 0.0007719 | 0.002358 |
| 1524 | ENTRAINMENT OF CIRCADIAN CLOCK | 8 | 26 | 0.0007746 | 0.002363 |
| 1525 | POSITIVE REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR PRODUCTION | 8 | 26 | 0.0007746 | 0.002363 |
| 1526 | CELLULAR RESPONSE TO INTERFERON GAMMA | 21 | 122 | 0.0008018 | 0.002445 |
| 1527 | DOUBLE STRAND BREAK REPAIR | 26 | 165 | 0.0008307 | 0.002531 |
| 1528 | RESPONSE TO TRANSITION METAL NANOPARTICLE | 24 | 148 | 0.0008566 | 0.002608 |
| 1529 | NEGATIVE REGULATION OF PEPTIDYL TYROSINE PHOSPHORYLATION | 10 | 39 | 0.0008737 | 0.002652 |
| 1530 | ANATOMICAL STRUCTURE MATURATION | 10 | 39 | 0.0008737 | 0.002652 |
| 1531 | TRABECULA MORPHOGENESIS | 10 | 39 | 0.0008737 | 0.002652 |
| 1532 | PLATELET AGGREGATION | 10 | 39 | 0.0008737 | 0.002652 |
| 1533 | SPLEEN DEVELOPMENT | 10 | 39 | 0.0008737 | 0.002652 |
| 1534 | REGULATION OF PEPTIDE TRANSPORT | 36 | 256 | 0.0008753 | 0.002655 |
| 1535 | REGULATION OF PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 13 | 60 | 0.0008951 | 0.002711 |
| 1536 | OLIGODENDROCYTE DIFFERENTIATION | 13 | 60 | 0.0008951 | 0.002711 |
| 1537 | NON CANONICAL WNT SIGNALING PATHWAY | 23 | 140 | 0.0009055 | 0.00274 |
| 1538 | REGULATION OF RELEASE OF SEQUESTERED CALCIUM ION INTO CYTOSOL | 15 | 75 | 0.0009058 | 0.00274 |
| 1539 | CEREBELLAR CORTEX DEVELOPMENT | 11 | 46 | 0.0009228 | 0.00279 |
| 1540 | REGULATION OF ERBB SIGNALING PATHWAY | 16 | 83 | 0.0009431 | 0.002849 |
| 1541 | METANEPHRIC NEPHRON MORPHOGENESIS | 7 | 21 | 0.0009724 | 0.002915 |
| 1542 | NEURON RECOGNITION | 9 | 33 | 0.000973 | 0.002915 |
| 1543 | B CELL HOMEOSTASIS | 7 | 21 | 0.0009724 | 0.002915 |
| 1544 | REGULATION OF CARDIAC MUSCLE CELL MEMBRANE REPOLARIZATION | 7 | 21 | 0.0009724 | 0.002915 |
| 1545 | REGULATION OF T CELL APOPTOTIC PROCESS | 9 | 33 | 0.000973 | 0.002915 |
| 1546 | REGULATION OF CHROMATIN SILENCING | 7 | 21 | 0.0009724 | 0.002915 |
| 1547 | REGULATION OF RECEPTOR BIOSYNTHETIC PROCESS | 7 | 21 | 0.0009724 | 0.002915 |
| 1548 | CELL DIFFERENTIATION IN HINDBRAIN | 7 | 21 | 0.0009724 | 0.002915 |
| 1549 | MRNA TRANSCRIPTION | 7 | 21 | 0.0009724 | 0.002915 |
| 1550 | POSITIVE REGULATION OF ADHERENS JUNCTION ORGANIZATION | 7 | 21 | 0.0009724 | 0.002915 |
| 1551 | POSITIVE REGULATION OF HISTONE METHYLATION | 9 | 33 | 0.000973 | 0.002915 |
| 1552 | EMBRYONIC EYE MORPHOGENESIS | 9 | 33 | 0.000973 | 0.002915 |
| 1553 | NEGATIVE REGULATION OF OXIDATIVE STRESS INDUCED INTRINSIC APOPTOTIC SIGNALING PATHWAY | 7 | 21 | 0.0009724 | 0.002915 |
| 1554 | REGULATION OF DEPHOSPHORYLATION | 25 | 158 | 0.0009742 | 0.002917 |
| 1555 | SKELETAL MUSCLE TISSUE REGENERATION | 8 | 27 | 0.001023 | 0.003055 |
| 1556 | DEVELOPMENTAL INDUCTION | 8 | 27 | 0.001023 | 0.003055 |
| 1557 | NEGATIVE REGULATION OF AXON GUIDANCE | 8 | 27 | 0.001023 | 0.003055 |
| 1558 | NEGATIVE REGULATION OF LYMPHOCYTE APOPTOTIC PROCESS | 8 | 27 | 0.001023 | 0.003055 |
| 1559 | PROTEIN SECRETION | 20 | 116 | 0.001034 | 0.003086 |
| 1560 | POSITIVE REGULATION OF SYNAPSE ASSEMBLY | 13 | 61 | 0.001054 | 0.003142 |
| 1561 | OVARIAN FOLLICLE DEVELOPMENT | 13 | 61 | 0.001054 | 0.003142 |
| 1562 | RESPONSE TO UV C | 5 | 11 | 0.001075 | 0.003188 |
| 1563 | POSITIVE REGULATION OF MEMBRANE DEPOLARIZATION | 5 | 11 | 0.001075 | 0.003188 |
| 1564 | POSITIVE REGULATION OF SYNAPSE MATURATION | 5 | 11 | 0.001075 | 0.003188 |
| 1565 | POSITIVE REGULATION OF PRI MIRNA TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 5 | 11 | 0.001075 | 0.003188 |
| 1566 | NEGATIVE REGULATION OF CELLULAR SENESCENCE | 5 | 11 | 0.001075 | 0.003188 |
| 1567 | POSITIVE REGULATION OF MYELOID LEUKOCYTE CYTOKINE PRODUCTION INVOLVED IN IMMUNE RESPONSE | 5 | 11 | 0.001075 | 0.003188 |
| 1568 | REGULATION OF OXIDATIVE STRESS INDUCED NEURON DEATH | 5 | 11 | 0.001075 | 0.003188 |
| 1569 | LOCOMOTORY EXPLORATION BEHAVIOR | 5 | 11 | 0.001075 | 0.003188 |
| 1570 | REGULATION OF MEIOTIC CELL CYCLE | 10 | 40 | 0.001081 | 0.003202 |
| 1571 | REGULATION OF ACTIVATED T CELL PROLIFERATION | 10 | 40 | 0.001081 | 0.003202 |
| 1572 | MITOCHONDRIAL MEMBRANE ORGANIZATION | 17 | 92 | 0.00109 | 0.003227 |
| 1573 | DEFENSE RESPONSE | 130 | 1231 | 0.001099 | 0.003252 |
| 1574 | SOMATIC RECOMBINATION OF IMMUNOGLOBULIN GENES INVOLVED IN IMMUNE RESPONSE | 6 | 16 | 0.001122 | 0.003292 |
| 1575 | POSITIVE REGULATION OF HISTONE H3 K4 METHYLATION | 6 | 16 | 0.001122 | 0.003292 |
| 1576 | POSITIVE REGULATION OF RECEPTOR MEDIATED ENDOCYTOSIS | 11 | 47 | 0.001118 | 0.003292 |
| 1577 | MORPHOGENESIS OF AN ENDOTHELIUM | 6 | 16 | 0.001122 | 0.003292 |
| 1578 | NEGATIVE REGULATION OF NEUROTRANSMITTER TRANSPORT | 6 | 16 | 0.001122 | 0.003292 |
| 1579 | NEGATIVE REGULATION OF HISTONE METHYLATION | 6 | 16 | 0.001122 | 0.003292 |
| 1580 | REGULATION OF EARLY ENDOSOME TO LATE ENDOSOME TRANSPORT | 6 | 16 | 0.001122 | 0.003292 |
| 1581 | ORGAN INDUCTION | 6 | 16 | 0.001122 | 0.003292 |
| 1582 | ISOTYPE SWITCHING | 6 | 16 | 0.001122 | 0.003292 |
| 1583 | SOMATIC DIVERSIFICATION OF IMMUNOGLOBULINS INVOLVED IN IMMUNE RESPONSE | 6 | 16 | 0.001122 | 0.003292 |
| 1584 | POSITIVE REGULATION OF TELOMERE MAINTENANCE | 11 | 47 | 0.001118 | 0.003292 |
| 1585 | PARAXIAL MESODERM DEVELOPMENT | 6 | 16 | 0.001122 | 0.003292 |
| 1586 | CELL SURFACE RECEPTOR SIGNALING PATHWAY INVOLVED IN HEART DEVELOPMENT | 6 | 16 | 0.001122 | 0.003292 |
| 1587 | REGULATION OF RECEPTOR ACTIVITY | 20 | 117 | 0.001154 | 0.003383 |
| 1588 | MACROMOLECULE METHYLATION | 30 | 205 | 0.0012 | 0.003516 |
| 1589 | REGULATION OF PRODUCTION OF MOLECULAR MEDIATOR OF IMMUNE RESPONSE | 18 | 101 | 0.001221 | 0.003575 |
| 1590 | POSITIVE REGULATION OF SODIUM ION TRANSPORT | 9 | 34 | 0.001229 | 0.003594 |
| 1591 | REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 9 | 34 | 0.001229 | 0.003594 |
| 1592 | POSITIVE REGULATION OF LEUKOCYTE MEDIATED IMMUNITY | 16 | 85 | 0.00123 | 0.003596 |
| 1593 | EMBRYONIC HEART TUBE MORPHOGENESIS | 13 | 62 | 0.001236 | 0.003611 |
| 1594 | REGULATED EXOCYTOSIS | 32 | 224 | 0.001265 | 0.003692 |
| 1595 | DNA RECOMBINATION | 31 | 215 | 0.001283 | 0.003744 |
| 1596 | NEGATIVE REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 26 | 170 | 0.001299 | 0.003788 |
| 1597 | ACTIN CYTOSKELETON REORGANIZATION | 12 | 55 | 0.001308 | 0.00381 |
| 1598 | MYELINATION IN PERIPHERAL NERVOUS SYSTEM | 7 | 22 | 0.001327 | 0.003821 |
| 1599 | NECROTIC CELL DEATH | 8 | 28 | 0.001331 | 0.003821 |
| 1600 | REGULATION OF FIBROBLAST MIGRATION | 8 | 28 | 0.001331 | 0.003821 |
| 1601 | ENDOCARDIAL CUSHION MORPHOGENESIS | 7 | 22 | 0.001327 | 0.003821 |
| 1602 | REGULATION OF DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 8 | 28 | 0.001331 | 0.003821 |
| 1603 | POSITIVE REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 8 | 28 | 0.001331 | 0.003821 |
| 1604 | MODULATION BY SYMBIONT OF HOST CELLULAR PROCESS | 8 | 28 | 0.001331 | 0.003821 |
| 1605 | POSITIVE REGULATION OF TELOMERASE ACTIVITY | 8 | 28 | 0.001331 | 0.003821 |
| 1606 | PLACENTA BLOOD VESSEL DEVELOPMENT | 8 | 28 | 0.001331 | 0.003821 |
| 1607 | CELL FATE COMMITMENT INVOLVED IN FORMATION OF PRIMARY GERM LAYER | 8 | 28 | 0.001331 | 0.003821 |
| 1608 | POSITIVE REGULATION OF MUSCLE HYPERTROPHY | 7 | 22 | 0.001327 | 0.003821 |
| 1609 | RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 7 | 22 | 0.001327 | 0.003821 |
| 1610 | POSITIVE REGULATION OF MYOBLAST DIFFERENTIATION | 7 | 22 | 0.001327 | 0.003821 |
| 1611 | ACTIVIN RECEPTOR SIGNALING PATHWAY | 7 | 22 | 0.001327 | 0.003821 |
| 1612 | PERIPHERAL NERVOUS SYSTEM AXON ENSHEATHMENT | 7 | 22 | 0.001327 | 0.003821 |
| 1613 | SOMATIC STEM CELL DIVISION | 7 | 22 | 0.001327 | 0.003821 |
| 1614 | MYELOID CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 10 | 41 | 0.001327 | 0.003821 |
| 1615 | POSITIVE REGULATION OF CARDIAC MUSCLE HYPERTROPHY | 7 | 22 | 0.001327 | 0.003821 |
| 1616 | CEREBELLAR CORTEX FORMATION | 7 | 22 | 0.001327 | 0.003821 |
| 1617 | EMBRYONIC PLACENTA MORPHOGENESIS | 7 | 22 | 0.001327 | 0.003821 |
| 1618 | ERK1 AND ERK2 CASCADE | 7 | 22 | 0.001327 | 0.003821 |
| 1619 | METANEPHROS MORPHOGENESIS | 8 | 28 | 0.001331 | 0.003821 |
| 1620 | HISTONE H3 DEACETYLATION | 7 | 22 | 0.001327 | 0.003821 |
| 1621 | LUNG ALVEOLUS DEVELOPMENT | 10 | 41 | 0.001327 | 0.003821 |
| 1622 | NEGATIVE REGULATION OF DEFENSE RESPONSE | 23 | 144 | 0.001339 | 0.003841 |
| 1623 | REGULATION OF INTERLEUKIN 1 BETA PRODUCTION | 11 | 48 | 0.001345 | 0.003851 |
| 1624 | ANTERIOR POSTERIOR AXIS SPECIFICATION | 11 | 48 | 0.001345 | 0.003851 |
| 1625 | POSITIVE REGULATION OF PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 11 | 48 | 0.001345 | 0.003851 |
| 1626 | REGULATION OF HORMONE LEVELS | 58 | 478 | 0.001393 | 0.003986 |
| 1627 | POSITIVE REGULATION OF RESPONSE TO WOUNDING | 25 | 162 | 0.001401 | 0.004006 |
| 1628 | TISSUE HOMEOSTASIS | 26 | 171 | 0.001417 | 0.004049 |
| 1629 | NEUROEPITHELIAL CELL DIFFERENTIATION | 13 | 63 | 0.001444 | 0.004124 |
| 1630 | REGULATION OF PROTEIN COMPLEX DISASSEMBLY | 31 | 217 | 0.001493 | 0.004261 |
| 1631 | POSITIVE REGULATION OF SECRETION | 47 | 370 | 0.001508 | 0.004301 |
| 1632 | BODY FLUID SECRETION | 14 | 71 | 0.001531 | 0.004364 |
| 1633 | ACTIVATION OF JUN KINASE ACTIVITY | 9 | 35 | 0.001536 | 0.004373 |
| 1634 | REGULATION OF GLYCOGEN METABOLIC PROCESS | 9 | 35 | 0.001536 | 0.004373 |
| 1635 | POSITIVE REGULATION OF DENDRITIC SPINE DEVELOPMENT | 9 | 35 | 0.001536 | 0.004373 |
| 1636 | REGULATION OF DENDRITIC SPINE DEVELOPMENT | 12 | 56 | 0.001543 | 0.004387 |
| 1637 | RESPONSE TO BIOTIC STIMULUS | 97 | 886 | 0.001543 | 0.004387 |
| 1638 | SYSTEM PROCESS | 179 | 1785 | 0.001577 | 0.004481 |
| 1639 | CELLULAR HOMEOSTASIS | 77 | 676 | 0.001594 | 0.004526 |
| 1640 | NEGATIVE REGULATION OF PLATELET ACTIVATION | 6 | 17 | 0.001616 | 0.004559 |
| 1641 | PITUITARY GLAND DEVELOPMENT | 10 | 42 | 0.001617 | 0.004559 |
| 1642 | REGULATION OF ENERGY HOMEOSTASIS | 6 | 17 | 0.001616 | 0.004559 |
| 1643 | REGULATION OF NUCLEOSIDE METABOLIC PROCESS | 11 | 49 | 0.001609 | 0.004559 |
| 1644 | POSITIVE REGULATION OF NEURAL PRECURSOR CELL PROLIFERATION | 10 | 42 | 0.001617 | 0.004559 |
| 1645 | NEGATIVE REGULATION OF LIPID STORAGE | 6 | 17 | 0.001616 | 0.004559 |
| 1646 | MAMMARY GLAND ALVEOLUS DEVELOPMENT | 6 | 17 | 0.001616 | 0.004559 |
| 1647 | MAMMARY GLAND LOBULE DEVELOPMENT | 6 | 17 | 0.001616 | 0.004559 |
| 1648 | EPITHELIAL CELL MORPHOGENESIS | 10 | 42 | 0.001617 | 0.004559 |
| 1649 | REGULATION OF ATP METABOLIC PROCESS | 11 | 49 | 0.001609 | 0.004559 |
| 1650 | DEFINITIVE HEMOPOIESIS | 6 | 17 | 0.001616 | 0.004559 |
| 1651 | NEGATIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 13 | 64 | 0.001679 | 0.004724 |
| 1652 | REGULATION OF CELLULAR KETONE METABOLIC PROCESS | 26 | 173 | 0.001679 | 0.004724 |
| 1653 | REGULATION OF NEUROTRANSMITTER TRANSPORT | 13 | 64 | 0.001679 | 0.004724 |
| 1654 | REGULATION OF PROTEIN ACETYLATION | 13 | 64 | 0.001679 | 0.004724 |
| 1655 | NUCLEAR CHROMOSOME SEGREGATION | 32 | 228 | 0.001695 | 0.004765 |
| 1656 | REGULATION OF GOLGI ORGANIZATION | 5 | 12 | 0.00172 | 0.00478 |
| 1657 | ANTERIOR POSTERIOR AXIS SPECIFICATION EMBRYO | 5 | 12 | 0.00172 | 0.00478 |
| 1658 | SOMATIC DIVERSIFICATION OF IMMUNOGLOBULINS | 8 | 29 | 0.001709 | 0.00478 |
| 1659 | HEMATOPOIETIC STEM CELL DIFFERENTIATION | 5 | 12 | 0.00172 | 0.00478 |
| 1660 | REGULATION OF DEFENSE RESPONSE TO VIRUS BY VIRUS | 8 | 29 | 0.001709 | 0.00478 |
| 1661 | RESPONSE TO PAIN | 8 | 29 | 0.001709 | 0.00478 |
| 1662 | REGULATION OF THYMOCYTE APOPTOTIC PROCESS | 5 | 12 | 0.00172 | 0.00478 |
| 1663 | NUCLEOTIDE EXCISION REPAIR PREINCISION COMPLEX ASSEMBLY | 8 | 29 | 0.001709 | 0.00478 |
| 1664 | POSITIVE REGULATION OF CARTILAGE DEVELOPMENT | 8 | 29 | 0.001709 | 0.00478 |
| 1665 | LATERAL VENTRICLE DEVELOPMENT | 5 | 12 | 0.00172 | 0.00478 |
| 1666 | TRIPARTITE REGIONAL SUBDIVISION | 5 | 12 | 0.00172 | 0.00478 |
| 1667 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL METABOLIC PROCESS | 5 | 12 | 0.00172 | 0.00478 |
| 1668 | MYELIN MAINTENANCE | 5 | 12 | 0.00172 | 0.00478 |
| 1669 | MECHANOSENSORY BEHAVIOR | 5 | 12 | 0.00172 | 0.00478 |
| 1670 | CEREBRAL CORTEX RADIALLY ORIENTED CELL MIGRATION | 8 | 29 | 0.001709 | 0.00478 |
| 1671 | GROWTH PLATE CARTILAGE DEVELOPMENT | 5 | 12 | 0.00172 | 0.00478 |
| 1672 | EMBRYONIC CAMERA TYPE EYE FORMATION | 5 | 12 | 0.00172 | 0.00478 |
| 1673 | POSITIVE REGULATION OF EXECUTION PHASE OF APOPTOSIS | 5 | 12 | 0.00172 | 0.00478 |
| 1674 | POSITIVE REGULATION OF INSULIN LIKE GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 5 | 12 | 0.00172 | 0.00478 |
| 1675 | ENDOTHELIAL CELL DIFFERENTIATION | 14 | 72 | 0.001761 | 0.004891 |
| 1676 | REGULATION OF MUSCLE CONTRACTION | 23 | 147 | 0.001771 | 0.004916 |
| 1677 | NUCLEOTIDE EXCISION REPAIR DNA DAMAGE RECOGNITION | 7 | 23 | 0.001775 | 0.004919 |
| 1678 | LYSOSOME LOCALIZATION | 7 | 23 | 0.001775 | 0.004919 |
| 1679 | EPIBOLY | 7 | 23 | 0.001775 | 0.004919 |
| 1680 | REGULATION OF BLOOD CIRCULATION | 39 | 295 | 0.001778 | 0.004923 |
| 1681 | POSITIVE REGULATION OF IMMUNE EFFECTOR PROCESS | 24 | 156 | 0.001801 | 0.004982 |
| 1682 | NEGATIVE REGULATION OF RESPONSE TO WOUNDING | 24 | 156 | 0.001801 | 0.004982 |
| 1683 | DEPHOSPHORYLATION | 38 | 286 | 0.001846 | 0.005103 |
| 1684 | NEUROMUSCULAR JUNCTION DEVELOPMENT | 9 | 36 | 0.001903 | 0.005239 |
| 1685 | T CELL PROLIFERATION | 9 | 36 | 0.001903 | 0.005239 |
| 1686 | NEGATIVE REGULATION OF BLOOD CIRCULATION | 9 | 36 | 0.001903 | 0.005239 |
| 1687 | OLFACTORY LOBE DEVELOPMENT | 9 | 36 | 0.001903 | 0.005239 |
| 1688 | NEGATIVE REGULATION OF MUSCLE TISSUE DEVELOPMENT | 9 | 36 | 0.001903 | 0.005239 |
| 1689 | REGULATION OF NUCLEOTIDE METABOLIC PROCESS | 30 | 211 | 0.001898 | 0.005239 |
| 1690 | POSITIVE REGULATION OF GLUCOSE METABOLIC PROCESS | 9 | 36 | 0.001903 | 0.005239 |
| 1691 | REGULATION OF SKELETAL MUSCLE TISSUE DEVELOPMENT | 11 | 50 | 0.001913 | 0.005256 |
| 1692 | REGULATION OF SYNAPTIC TRANSMISSION GLUTAMATERGIC | 11 | 50 | 0.001913 | 0.005256 |
| 1693 | RECEPTOR INTERNALIZATION | 11 | 50 | 0.001913 | 0.005256 |
| 1694 | HISTONE H4 ACETYLATION | 11 | 50 | 0.001913 | 0.005256 |
| 1695 | REGULATION OF RESPONSE TO OXIDATIVE STRESS | 13 | 65 | 0.001946 | 0.005341 |
| 1696 | NEGATIVE REGULATION OF LEUKOCYTE APOPTOTIC PROCESS | 10 | 43 | 0.001956 | 0.005365 |
| 1697 | REGULATION OF RNA SPLICING | 17 | 97 | 0.001987 | 0.005449 |
| 1698 | POSITIVE REGULATION OF STAT CASCADE | 14 | 73 | 0.002019 | 0.005528 |
| 1699 | POSITIVE REGULATION OF JAK STAT CASCADE | 14 | 73 | 0.002019 | 0.005528 |
| 1700 | POSITIVE REGULATION OF TRANSMEMBRANE TRANSPORT | 21 | 131 | 0.002024 | 0.005537 |
| 1701 | RETINA DEVELOPMENT IN CAMERA TYPE EYE | 21 | 131 | 0.002024 | 0.005537 |
| 1702 | RESPONSE TO HEAT | 16 | 89 | 0.002029 | 0.005548 |
| 1703 | PLASMA MEMBRANE ORGANIZATION | 29 | 203 | 0.00208 | 0.005684 |
| 1704 | REGULATION OF CYTOKINE SECRETION | 23 | 149 | 0.002121 | 0.005791 |
| 1705 | NEURON MATURATION | 8 | 30 | 0.002166 | 0.005909 |
| 1706 | NEGATIVE REGULATION OF B CELL ACTIVATION | 8 | 30 | 0.002166 | 0.005909 |
| 1707 | RETINAL GANGLION CELL AXON GUIDANCE | 6 | 18 | 0.002258 | 0.006106 |
| 1708 | GLANDULAR EPITHELIAL CELL DEVELOPMENT | 6 | 18 | 0.002258 | 0.006106 |
| 1709 | ATRIAL SEPTUM DEVELOPMENT | 6 | 18 | 0.002258 | 0.006106 |
| 1710 | SMOOTH MUSCLE TISSUE DEVELOPMENT | 6 | 18 | 0.002258 | 0.006106 |
| 1711 | REGULATION OF INTERLEUKIN 2 BIOSYNTHETIC PROCESS | 6 | 18 | 0.002258 | 0.006106 |
| 1712 | CELL DEATH IN RESPONSE TO OXIDATIVE STRESS | 6 | 18 | 0.002258 | 0.006106 |
| 1713 | RESPONSE TO PLATELET DERIVED GROWTH FACTOR | 6 | 18 | 0.002258 | 0.006106 |
| 1714 | REGULATION OF B CELL APOPTOTIC PROCESS | 6 | 18 | 0.002258 | 0.006106 |
| 1715 | CELLULAR RESPONSE TO PROSTAGLANDIN E STIMULUS | 6 | 18 | 0.002258 | 0.006106 |
| 1716 | RESPONSE TO CAFFEINE | 6 | 18 | 0.002258 | 0.006106 |
| 1717 | PERICARDIUM DEVELOPMENT | 6 | 18 | 0.002258 | 0.006106 |
| 1718 | NEGATIVE REGULATION OF NF KAPPAB IMPORT INTO NUCLEUS | 6 | 18 | 0.002258 | 0.006106 |
| 1719 | NEPHRON TUBULE FORMATION | 6 | 18 | 0.002258 | 0.006106 |
| 1720 | MUSCLE CELL MIGRATION | 6 | 18 | 0.002258 | 0.006106 |
| 1721 | CENTROSOME LOCALIZATION | 6 | 18 | 0.002258 | 0.006106 |
| 1722 | RESPONSE TO AMMONIUM ION | 11 | 51 | 0.002264 | 0.006109 |
| 1723 | PROTEIN MONOUBIQUITINATION | 11 | 51 | 0.002264 | 0.006109 |
| 1724 | POSITIVE REGULATION OF ALPHA BETA T CELL ACTIVATION | 11 | 51 | 0.002264 | 0.006109 |
| 1725 | CELLULAR CATABOLIC PROCESS | 136 | 1322 | 0.002303 | 0.006213 |
| 1726 | NEGATIVE REGULATION OF GLIOGENESIS | 9 | 37 | 0.002336 | 0.006243 |
| 1727 | NEGATIVE REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 7 | 24 | 0.002332 | 0.006243 |
| 1728 | REGULATION OF MYELOID CELL APOPTOTIC PROCESS | 7 | 24 | 0.002332 | 0.006243 |
| 1729 | POSITIVE REGULATION OF NUCLEOSIDE METABOLIC PROCESS | 7 | 24 | 0.002332 | 0.006243 |
| 1730 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO CELL PERIPHERY | 9 | 37 | 0.002336 | 0.006243 |
| 1731 | POSITIVE REGULATION OF PROTEIN TYROSINE KINASE ACTIVITY | 9 | 37 | 0.002336 | 0.006243 |
| 1732 | TRICARBOXYLIC ACID METABOLIC PROCESS | 9 | 37 | 0.002336 | 0.006243 |
| 1733 | POSITIVE REGULATION OF ATP METABOLIC PROCESS | 7 | 24 | 0.002332 | 0.006243 |
| 1734 | MODULATION BY VIRUS OF HOST MORPHOLOGY OR PHYSIOLOGY | 9 | 37 | 0.002336 | 0.006243 |
| 1735 | IMMUNOGLOBULIN PRODUCTION INVOLVED IN IMMUNOGLOBULIN MEDIATED IMMUNE RESPONSE | 7 | 24 | 0.002332 | 0.006243 |
| 1736 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO PLASMA MEMBRANE | 9 | 37 | 0.002336 | 0.006243 |
| 1737 | CARDIAC EPITHELIAL TO MESENCHYMAL TRANSITION | 7 | 24 | 0.002332 | 0.006243 |
| 1738 | PROTEIN K63 LINKED DEUBIQUITINATION | 7 | 24 | 0.002332 | 0.006243 |
| 1739 | POSITIVE REGULATION OF ALPHA BETA T CELL DIFFERENTIATION | 9 | 37 | 0.002336 | 0.006243 |
| 1740 | POSITIVE REGULATION OF SMOOTHENED SIGNALING PATHWAY | 7 | 24 | 0.002332 | 0.006243 |
| 1741 | REGULATION OF PROTEIN PHOSPHATASE TYPE 2A ACTIVITY | 7 | 24 | 0.002332 | 0.006243 |
| 1742 | NEGATIVE REGULATION OF LIPID BIOSYNTHETIC PROCESS | 10 | 44 | 0.002349 | 0.006272 |
| 1743 | REGULATION OF TYROSINE PHOSPHORYLATION OF STAT3 PROTEIN | 10 | 44 | 0.002349 | 0.006272 |
| 1744 | PLATELET DEGRANULATION | 18 | 107 | 0.002388 | 0.006371 |
| 1745 | REGULATION OF GLIAL CELL DIFFERENTIATION | 12 | 59 | 0.002467 | 0.006579 |
| 1746 | REGULATION OF HORMONE SECRETION | 35 | 262 | 0.002491 | 0.006637 |
| 1747 | REGULATION OF CALCIUM ION TRANSMEMBRANE TRANSPORT | 19 | 116 | 0.002516 | 0.006702 |
| 1748 | REGULATION OF HEPATOCYTE PROLIFERATION | 5 | 13 | 0.002608 | 0.006877 |
| 1749 | POSITIVE REGULATION OF DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 5 | 13 | 0.002608 | 0.006877 |
| 1750 | NEGATIVE REGULATION OF OLIGODENDROCYTE DIFFERENTIATION | 5 | 13 | 0.002608 | 0.006877 |
| 1751 | WHITE FAT CELL DIFFERENTIATION | 5 | 13 | 0.002608 | 0.006877 |
| 1752 | POSITIVE REGULATION OF INTRACELLULAR STEROID HORMONE RECEPTOR SIGNALING PATHWAY | 5 | 13 | 0.002608 | 0.006877 |
| 1753 | REGULATION OF IRE1 MEDIATED UNFOLDED PROTEIN RESPONSE | 5 | 13 | 0.002608 | 0.006877 |
| 1754 | LIPOPROTEIN TRANSPORT | 5 | 13 | 0.002608 | 0.006877 |
| 1755 | HEMATOPOIETIC STEM CELL PROLIFERATION | 5 | 13 | 0.002608 | 0.006877 |
| 1756 | HEPATOCYTE APOPTOTIC PROCESS | 5 | 13 | 0.002608 | 0.006877 |
| 1757 | CELLULAR RESPONSE TO HEPATOCYTE GROWTH FACTOR STIMULUS | 5 | 13 | 0.002608 | 0.006877 |
| 1758 | RESPONSE TO HEPATOCYTE GROWTH FACTOR | 5 | 13 | 0.002608 | 0.006877 |
| 1759 | LIPOPROTEIN LOCALIZATION | 5 | 13 | 0.002608 | 0.006877 |
| 1760 | NEGATIVE REGULATION OF MACROPHAGE DERIVED FOAM CELL DIFFERENTIATION | 5 | 13 | 0.002608 | 0.006877 |
| 1761 | GLIAL CELL FATE COMMITMENT | 5 | 13 | 0.002608 | 0.006877 |
| 1762 | REGULATION OF CELL FATE SPECIFICATION | 5 | 13 | 0.002608 | 0.006877 |
| 1763 | RESPONSE TO COBALT ION | 5 | 13 | 0.002608 | 0.006877 |
| 1764 | REGULATION OF PROTEIN POLYUBIQUITINATION | 5 | 13 | 0.002608 | 0.006877 |
| 1765 | ENDODERMAL CELL FATE COMMITMENT | 5 | 13 | 0.002608 | 0.006877 |
| 1766 | BIOMINERAL TISSUE DEVELOPMENT | 14 | 75 | 0.002629 | 0.006922 |
| 1767 | REGULATION OF VIRAL GENOME REPLICATION | 14 | 75 | 0.002629 | 0.006922 |
| 1768 | NEGATIVE REGULATION OF RESPONSE TO DNA DAMAGE STIMULUS | 11 | 52 | 0.002664 | 0.00701 |
| 1769 | CELLULAR LIPID METABOLIC PROCESS | 98 | 913 | 0.002684 | 0.00706 |
| 1770 | ESTABLISHMENT OF PROTEIN LOCALIZATION TO ORGANELLE | 45 | 361 | 0.002709 | 0.007121 |
| 1771 | RNA STABILIZATION | 8 | 31 | 0.002715 | 0.007125 |
| 1772 | MACROPHAGE ACTIVATION | 8 | 31 | 0.002715 | 0.007125 |
| 1773 | REGULATION OF PLATELET ACTIVATION | 8 | 31 | 0.002715 | 0.007125 |
| 1774 | MEIOTIC CELL CYCLE PROCESS | 23 | 152 | 0.002755 | 0.007226 |
| 1775 | DNA METHYLATION | 10 | 45 | 0.002804 | 0.007335 |
| 1776 | MODIFICATION BY SYMBIONT OF HOST MORPHOLOGY OR PHYSIOLOGY | 10 | 45 | 0.002804 | 0.007335 |
| 1777 | PROTEIN LOCALIZATION TO CHROMOSOME | 10 | 45 | 0.002804 | 0.007335 |
| 1778 | HETEROPHILIC CELL CELL ADHESION VIA PLASMA MEMBRANE CELL ADHESION MOLECULES | 10 | 45 | 0.002804 | 0.007335 |
| 1779 | DNA ALKYLATION | 10 | 45 | 0.002804 | 0.007335 |
| 1780 | NEGATIVE REGULATION OF AXON EXTENSION | 9 | 38 | 0.002844 | 0.007416 |
| 1781 | POSITIVE REGULATION OF BIOMINERAL TISSUE DEVELOPMENT | 9 | 38 | 0.002844 | 0.007416 |
| 1782 | REGULATION OF CD4 POSITIVE ALPHA BETA T CELL ACTIVATION | 9 | 38 | 0.002844 | 0.007416 |
| 1783 | CYTOKINE SECRETION | 9 | 38 | 0.002844 | 0.007416 |
| 1784 | REGULATION OF LYMPHOCYTE MIGRATION | 9 | 38 | 0.002844 | 0.007416 |
| 1785 | REGULATION OF MONOOXYGENASE ACTIVITY | 12 | 60 | 0.002861 | 0.007457 |
| 1786 | LEUKOCYTE MEDIATED IMMUNITY | 27 | 189 | 0.002904 | 0.007566 |
| 1787 | RESPONSE TO INTERFERON GAMMA | 22 | 144 | 0.002981 | 0.007763 |
| 1788 | REGULATION OF CALCIUM MEDIATED SIGNALING | 14 | 76 | 0.002986 | 0.007771 |
| 1789 | REGULATION OF INFLAMMATORY RESPONSE | 38 | 294 | 0.003002 | 0.007808 |
| 1790 | RESPONSE TO PROSTAGLANDIN E | 7 | 25 | 0.003014 | 0.00781 |
| 1791 | DETECTION OF MECHANICAL STIMULUS INVOLVED IN SENSORY PERCEPTION | 7 | 25 | 0.003014 | 0.00781 |
| 1792 | CELLULAR RESPONSE TO TOXIC SUBSTANCE | 7 | 25 | 0.003014 | 0.00781 |
| 1793 | REGULATION OF FIBROBLAST GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 7 | 25 | 0.003014 | 0.00781 |
| 1794 | POSTSYNAPTIC MEMBRANE ORGANIZATION | 7 | 25 | 0.003014 | 0.00781 |
| 1795 | REGULATION OF ACTIVIN RECEPTOR SIGNALING PATHWAY | 7 | 25 | 0.003014 | 0.00781 |
| 1796 | POSITIVE REGULATION OF CHROMOSOME SEGREGATION | 7 | 25 | 0.003014 | 0.00781 |
| 1797 | THYMIC T CELL SELECTION | 6 | 19 | 0.003076 | 0.007925 |
| 1798 | ENTRAINMENT OF CIRCADIAN CLOCK BY PHOTOPERIOD | 6 | 19 | 0.003076 | 0.007925 |
| 1799 | KIDNEY VASCULATURE DEVELOPMENT | 6 | 19 | 0.003076 | 0.007925 |
| 1800 | NEGATIVE REGULATION OF MEIOTIC CELL CYCLE | 6 | 19 | 0.003076 | 0.007925 |
| 1801 | REGULATION OF CELL MIGRATION INVOLVED IN SPROUTING ANGIOGENESIS | 6 | 19 | 0.003076 | 0.007925 |
| 1802 | RENAL SYSTEM VASCULATURE DEVELOPMENT | 6 | 19 | 0.003076 | 0.007925 |
| 1803 | POSITIVE REGULATION OF DENDRITE EXTENSION | 6 | 19 | 0.003076 | 0.007925 |
| 1804 | REGULATION OF DENDRITE EXTENSION | 6 | 19 | 0.003076 | 0.007925 |
| 1805 | POSITIVE REGULATION OF CHONDROCYTE DIFFERENTIATION | 6 | 19 | 0.003076 | 0.007925 |
| 1806 | ASTROCYTE DEVELOPMENT | 6 | 19 | 0.003076 | 0.007925 |
| 1807 | PROTEIN OLIGOMERIZATION | 52 | 434 | 0.00311 | 0.008007 |
| 1808 | CELLULAR RESPONSE TO AMINO ACID STIMULUS | 11 | 53 | 0.003119 | 0.008027 |
| 1809 | REGULATION OF IMMUNE EFFECTOR PROCESS | 51 | 424 | 0.003129 | 0.008048 |
| 1810 | NEGATIVE REGULATION OF ION TRANSPORT | 20 | 127 | 0.003158 | 0.008119 |
| 1811 | PHOSPHOLIPID METABOLIC PROCESS | 45 | 364 | 0.003173 | 0.008151 |
| 1812 | ACTIN FILAMENT BASED MOVEMENT | 16 | 93 | 0.003222 | 0.008273 |
| 1813 | NEGATIVE REGULATION OF SECRETION | 28 | 200 | 0.003297 | 0.008461 |
| 1814 | MEMBRANE DEPOLARIZATION | 12 | 61 | 0.003303 | 0.008472 |
| 1815 | VENTRAL SPINAL CORD DEVELOPMENT | 10 | 46 | 0.003327 | 0.008515 |
| 1816 | INTRASPECIES INTERACTION BETWEEN ORGANISMS | 10 | 46 | 0.003327 | 0.008515 |
| 1817 | SOCIAL BEHAVIOR | 10 | 46 | 0.003327 | 0.008515 |
| 1818 | HISTONE H3 ACETYLATION | 10 | 46 | 0.003327 | 0.008515 |
| 1819 | GLOBAL GENOME NUCLEOTIDE EXCISION REPAIR | 8 | 32 | 0.003366 | 0.008595 |
| 1820 | BLOOD VESSEL REMODELING | 8 | 32 | 0.003366 | 0.008595 |
| 1821 | NEGATIVE REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 8 | 32 | 0.003366 | 0.008595 |
| 1822 | POSITIVE REGULATION OF MULTICELLULAR ORGANISM GROWTH | 8 | 32 | 0.003366 | 0.008595 |
| 1823 | NEGATIVE REGULATION OF LEUKOCYTE PROLIFERATION | 13 | 69 | 0.003383 | 0.008629 |
| 1824 | ACTOMYOSIN STRUCTURE ORGANIZATION | 14 | 77 | 0.003382 | 0.008629 |
| 1825 | MEMBRANE DEPOLARIZATION DURING ACTION POTENTIAL | 9 | 39 | 0.003434 | 0.008752 |
| 1826 | BASE EXCISION REPAIR | 9 | 39 | 0.003434 | 0.008752 |
| 1827 | CELLULAR PROTEIN COMPLEX ASSEMBLY | 43 | 346 | 0.003508 | 0.008934 |
| 1828 | SISTER CHROMATID COHESION | 18 | 111 | 0.003598 | 0.009158 |
| 1829 | NEGATIVE REGULATION OF REPRODUCTIVE PROCESS | 11 | 54 | 0.003635 | 0.009242 |
| 1830 | CELL DIFFERENTIATION IN SPINAL CORD | 11 | 54 | 0.003635 | 0.009242 |
| 1831 | NEGATIVE REGULATION OF HYDROLASE ACTIVITY | 48 | 397 | 0.003674 | 0.009337 |
| 1832 | POSITIVE REGULATION OF PROTEIN SECRETION | 29 | 211 | 0.003697 | 0.009389 |
| 1833 | CELL MIGRATION INVOLVED IN HEART DEVELOPMENT | 5 | 14 | 0.003788 | 0.009528 |
| 1834 | REGULATION OF PODOSOME ASSEMBLY | 5 | 14 | 0.003788 | 0.009528 |
| 1835 | POSITIVE REGULATION OF RHO PROTEIN SIGNAL TRANSDUCTION | 5 | 14 | 0.003788 | 0.009528 |
| 1836 | LAYER FORMATION IN CEREBRAL CORTEX | 5 | 14 | 0.003788 | 0.009528 |
| 1837 | SPECIFICATION OF ORGAN IDENTITY | 5 | 14 | 0.003788 | 0.009528 |
| 1838 | NEGATIVE REGULATION OF RHO PROTEIN SIGNAL TRANSDUCTION | 5 | 14 | 0.003788 | 0.009528 |
| 1839 | PROTEIN LOCALIZATION TO CHROMOSOME TELOMERIC REGION | 5 | 14 | 0.003788 | 0.009528 |
| 1840 | POSITIVE REGULATION OF T CELL CYTOKINE PRODUCTION | 5 | 14 | 0.003788 | 0.009528 |
| 1841 | VOCALIZATION BEHAVIOR | 5 | 14 | 0.003788 | 0.009528 |
| 1842 | RESPONSE TO LOW DENSITY LIPOPROTEIN PARTICLE | 5 | 14 | 0.003788 | 0.009528 |
| 1843 | METANEPHRIC MESENCHYME DEVELOPMENT | 5 | 14 | 0.003788 | 0.009528 |
| 1844 | REGULATION OF MONOCYTE DIFFERENTIATION | 5 | 14 | 0.003788 | 0.009528 |
| 1845 | POSITIVE REGULATION OF SPROUTING ANGIOGENESIS | 5 | 14 | 0.003788 | 0.009528 |
| 1846 | POSITIVE REGULATION OF FATTY ACID OXIDATION | 5 | 14 | 0.003788 | 0.009528 |
| 1847 | NEGATIVE REGULATION OF NUCLEOTIDE CATABOLIC PROCESS | 5 | 14 | 0.003788 | 0.009528 |
| 1848 | REGULATION OF SYNAPSE MATURATION | 5 | 14 | 0.003788 | 0.009528 |
| 1849 | BONE MATURATION | 5 | 14 | 0.003788 | 0.009528 |
| 1850 | POST EMBRYONIC ORGAN DEVELOPMENT | 5 | 14 | 0.003788 | 0.009528 |
| 1851 | NEGATIVE REGULATION OF STRIATED MUSCLE CELL DIFFERENTIATION | 7 | 26 | 0.00384 | 0.009616 |
| 1852 | HEART TRABECULA MORPHOGENESIS | 7 | 26 | 0.00384 | 0.009616 |
| 1853 | POSITIVE REGULATION OF LYMPHOCYTE MIGRATION | 7 | 26 | 0.00384 | 0.009616 |
| 1854 | REGULATION OF T HELPER CELL DIFFERENTIATION | 7 | 26 | 0.00384 | 0.009616 |
| 1855 | VENTRICULAR SYSTEM DEVELOPMENT | 7 | 26 | 0.00384 | 0.009616 |
| 1856 | NEGATIVE REGULATION OF LIPID TRANSPORT | 7 | 26 | 0.00384 | 0.009616 |
| 1857 | NEGATIVE REGULATION OF TELOMERE MAINTENANCE | 7 | 26 | 0.00384 | 0.009616 |
| 1858 | REGULATION OF HORMONE METABOLIC PROCESS | 7 | 26 | 0.00384 | 0.009616 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | ENZYME BINDING | 353 | 1737 | 1.71e-64 | 1.588e-61 |
| 2 | REGULATORY REGION NUCLEIC ACID BINDING | 210 | 818 | 3.268e-54 | 1.518e-51 |
| 3 | MACROMOLECULAR COMPLEX BINDING | 287 | 1399 | 2.06e-52 | 6.38e-50 |
| 4 | DOUBLE STRANDED DNA BINDING | 190 | 764 | 1.039e-46 | 2.414e-44 |
| 5 | SEQUENCE SPECIFIC DNA BINDING | 228 | 1037 | 2.095e-46 | 3.893e-44 |
| 6 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 167 | 629 | 4.91e-45 | 7.603e-43 |
| 7 | KINASE BINDING | 162 | 606 | 4.251e-44 | 5.032e-42 |
| 8 | TRANSCRIPTION FACTOR BINDING | 149 | 524 | 4.333e-44 | 5.032e-42 |
| 9 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 238 | 1199 | 1.624e-40 | 1.676e-38 |
| 10 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 102 | 315 | 1.079e-35 | 1.003e-33 |
| 11 | PROTEIN KINASE ACTIVITY | 150 | 640 | 1.175e-33 | 9.921e-32 |
| 12 | KINASE ACTIVITY | 177 | 842 | 3.341e-33 | 2.586e-31 |
| 13 | TRANSCRIPTION FACTOR ACTIVITY PROTEIN BINDING | 140 | 588 | 3.302e-32 | 2.36e-30 |
| 14 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 99 | 328 | 8.019e-32 | 5.321e-30 |
| 15 | PROTEIN COMPLEX BINDING | 186 | 935 | 1.292e-31 | 7.999e-30 |
| 16 | CHROMATIN BINDING | 115 | 435 | 6.294e-31 | 3.654e-29 |
| 17 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 78 | 226 | 1.218e-29 | 6.655e-28 |
| 18 | RECEPTOR BINDING | 245 | 1476 | 8.763e-29 | 4.523e-27 |
| 19 | CORE PROMOTER PROXIMAL REGION DNA BINDING | 96 | 371 | 3.308e-25 | 1.618e-23 |
| 20 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 178 | 992 | 1.007e-24 | 4.68e-23 |
| 21 | PROTEIN DOMAIN SPECIFIC BINDING | 131 | 624 | 1.305e-24 | 5.773e-23 |
| 22 | PROTEIN TYROSINE KINASE ACTIVITY | 61 | 176 | 1.463e-23 | 6.178e-22 |
| 23 | IDENTICAL PROTEIN BINDING | 200 | 1209 | 3.117e-23 | 1.259e-21 |
| 24 | CORE PROMOTER BINDING | 55 | 152 | 2.098e-22 | 8.121e-21 |
| 25 | ADENYL NUCLEOTIDE BINDING | 230 | 1514 | 1.154e-21 | 4.288e-20 |
| 26 | TRANSMEMBRANE RECEPTOR PROTEIN KINASE ACTIVITY | 38 | 81 | 1.468e-20 | 5.245e-19 |
| 27 | GROWTH FACTOR BINDING | 47 | 123 | 1.736e-20 | 5.973e-19 |
| 28 | RIBONUCLEOTIDE BINDING | 261 | 1860 | 1.063e-19 | 3.526e-18 |
| 29 | TRANSCRIPTION COACTIVATOR ACTIVITY | 75 | 296 | 2.253e-19 | 7.218e-18 |
| 30 | PROTEIN DIMERIZATION ACTIVITY | 179 | 1149 | 5.071e-18 | 1.57e-16 |
| 31 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 93 | 445 | 1.268e-17 | 3.799e-16 |
| 32 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE ACTIVITY | 31 | 64 | 1.564e-17 | 4.54e-16 |
| 33 | CELL ADHESION MOLECULE BINDING | 53 | 186 | 2.069e-16 | 5.826e-15 |
| 34 | PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 31 | 70 | 3.879e-16 | 1.06e-14 |
| 35 | ZINC ION BINDING | 173 | 1155 | 9.183e-16 | 2.437e-14 |
| 36 | MOLECULAR FUNCTION REGULATOR | 194 | 1353 | 1.279e-15 | 3.3e-14 |
| 37 | SMAD BINDING | 30 | 72 | 8.792e-15 | 2.207e-13 |
| 38 | RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 36 | 104 | 1.883e-14 | 4.604e-13 |
| 39 | BETA CATENIN BINDING | 32 | 84 | 2.377e-14 | 5.663e-13 |
| 40 | CORE PROMOTER SEQUENCE SPECIFIC DNA BINDING | 35 | 101 | 4.174e-14 | 9.695e-13 |
| 41 | TRANSITION METAL ION BINDING | 193 | 1400 | 7.58e-14 | 1.717e-12 |
| 42 | KINASE REGULATOR ACTIVITY | 48 | 186 | 3.309e-13 | 7.32e-12 |
| 43 | RECEPTOR SIGNALING PROTEIN ACTIVITY | 45 | 172 | 1.074e-12 | 2.32e-11 |
| 44 | PROTEIN HOMODIMERIZATION ACTIVITY | 115 | 722 | 1.54e-12 | 3.251e-11 |
| 45 | TRANSCRIPTION COREPRESSOR ACTIVITY | 52 | 221 | 1.742e-12 | 3.596e-11 |
| 46 | ENHANCER BINDING | 31 | 93 | 3.825e-12 | 7.726e-11 |
| 47 | PHOSPHATASE BINDING | 42 | 162 | 8.381e-12 | 1.657e-10 |
| 48 | PROTEIN PHOSPHATASE BINDING | 35 | 120 | 1.241e-11 | 2.402e-10 |
| 49 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 37 | 133 | 1.533e-11 | 2.907e-10 |
| 50 | UBIQUITIN LIKE PROTEIN LIGASE BINDING | 56 | 264 | 2.107e-11 | 3.915e-10 |
| 51 | HISTONE DEACETYLASE BINDING | 32 | 105 | 2.589e-11 | 4.65e-10 |
| 52 | ACTIVATING TRANSCRIPTION FACTOR BINDING | 23 | 57 | 2.603e-11 | 4.65e-10 |
| 53 | STRUCTURE SPECIFIC DNA BINDING | 34 | 118 | 3.525e-11 | 6.179e-10 |
| 54 | SIGNAL TRANSDUCER ACTIVITY | 216 | 1731 | 4.627e-11 | 7.96e-10 |
| 55 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 28 | 86 | 7.884e-11 | 1.332e-09 |
| 56 | HORMONE RECEPTOR BINDING | 41 | 168 | 1.152e-10 | 1.912e-09 |
| 57 | ENZYME REGULATOR ACTIVITY | 134 | 959 | 2.785e-10 | 4.539e-09 |
| 58 | RECEPTOR SIGNALING PROTEIN SERINE THREONINE KINASE ACTIVITY | 28 | 92 | 4.639e-10 | 7.43e-09 |
| 59 | STEROID HORMONE RECEPTOR BINDING | 26 | 81 | 5.283e-10 | 8.319e-09 |
| 60 | CYTOSKELETAL PROTEIN BINDING | 118 | 819 | 5.406e-10 | 8.371e-09 |
| 61 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 39 | 168 | 1.55e-09 | 2.361e-08 |
| 62 | X1 PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 18 | 43 | 1.859e-09 | 2.785e-08 |
| 63 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II ACTIVATING TRANSCRIPTION FACTOR BINDING | 20 | 53 | 2.102e-09 | 3.1e-08 |
| 64 | UBIQUITIN LIKE PROTEIN TRANSFERASE ACTIVITY | 71 | 420 | 2.489e-09 | 3.613e-08 |
| 65 | RNA POLYMERASE II DISTAL ENHANCER SEQUENCE SPECIFIC DNA BINDING | 22 | 65 | 3.642e-09 | 5.205e-08 |
| 66 | RNA POLYMERASE II ACTIVATING TRANSCRIPTION FACTOR BINDING | 16 | 36 | 5.158e-09 | 7.26e-08 |
| 67 | PHOSPHATIDYLINOSITOL KINASE ACTIVITY | 19 | 51 | 6.816e-09 | 9.451e-08 |
| 68 | KINASE ACTIVATOR ACTIVITY | 21 | 62 | 8.106e-09 | 1.107e-07 |
| 69 | PROTEIN HETERODIMERIZATION ACTIVITY | 75 | 468 | 9.804e-09 | 1.32e-07 |
| 70 | INTEGRIN BINDING | 28 | 105 | 1.246e-08 | 1.654e-07 |
| 71 | TRANSCRIPTION FACTOR ACTIVITY DIRECT LIGAND REGULATED SEQUENCE SPECIFIC DNA BINDING | 18 | 48 | 1.506e-08 | 1.97e-07 |
| 72 | CHROMATIN DNA BINDING | 23 | 80 | 5.371e-08 | 6.931e-07 |
| 73 | DAMAGED DNA BINDING | 20 | 63 | 6.352e-08 | 8.083e-07 |
| 74 | UBIQUITIN LIKE PROTEIN LIGASE ACTIVITY | 40 | 199 | 7.248e-08 | 9.099e-07 |
| 75 | FIBRONECTIN BINDING | 13 | 28 | 7.574e-08 | 9.382e-07 |
| 76 | GLYCOPROTEIN BINDING | 26 | 101 | 8.915e-08 | 1.09e-06 |
| 77 | STEROID HORMONE RECEPTOR ACTIVITY | 19 | 59 | 1.045e-07 | 1.261e-06 |
| 78 | SH3 DOMAIN BINDING | 28 | 116 | 1.272e-07 | 1.515e-06 |
| 79 | ENZYME ACTIVATOR ACTIVITY | 72 | 471 | 1.346e-07 | 1.582e-06 |
| 80 | INSULIN LIKE GROWTH FACTOR BINDING | 12 | 25 | 1.53e-07 | 1.777e-06 |
| 81 | PROTEIN SERINE THREONINE TYROSINE KINASE ACTIVITY | 15 | 39 | 1.621e-07 | 1.859e-06 |
| 82 | P53 BINDING | 20 | 67 | 1.99e-07 | 2.255e-06 |
| 83 | PHOSPHATIDYLINOSITOL 3 KINASE BINDING | 13 | 30 | 2.075e-07 | 2.323e-06 |
| 84 | PROTEIN C TERMINUS BINDING | 37 | 186 | 2.909e-07 | 3.217e-06 |
| 85 | REPRESSING TRANSCRIPTION FACTOR BINDING | 18 | 57 | 3.148e-07 | 3.441e-06 |
| 86 | PLATELET DERIVED GROWTH FACTOR RECEPTOR BINDING | 9 | 15 | 4.84e-07 | 5.228e-06 |
| 87 | PROTEIN KINASE A BINDING | 15 | 42 | 5.035e-07 | 5.376e-06 |
| 88 | RNA POLYMERASE II CORE PROMOTER SEQUENCE SPECIFIC DNA BINDING | 17 | 53 | 5.2e-07 | 5.49e-06 |
| 89 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II DISTAL ENHANCER SEQUENCE SPECIFIC BINDING | 23 | 90 | 5.604e-07 | 5.849e-06 |
| 90 | HISTONE BINDING | 35 | 177 | 6.994e-07 | 6.936e-06 |
| 91 | MISMATCHED DNA BINDING | 8 | 12 | 6.901e-07 | 6.936e-06 |
| 92 | PROTEIN TYROSINE KINASE BINDING | 17 | 54 | 7.018e-07 | 6.936e-06 |
| 93 | RNA POLYMERASE II TRANSCRIPTION COFACTOR ACTIVITY | 23 | 91 | 6.928e-07 | 6.936e-06 |
| 94 | GAMMA CATENIN BINDING | 8 | 12 | 6.901e-07 | 6.936e-06 |
| 95 | TRANSCRIPTIONAL REPRESSOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 25 | 105 | 7.793e-07 | 7.621e-06 |
| 96 | ANDROGEN RECEPTOR BINDING | 14 | 39 | 1.125e-06 | 1.089e-05 |
| 97 | HEAT SHOCK PROTEIN BINDING | 22 | 89 | 1.802e-06 | 1.726e-05 |
| 98 | PHOSPHOPROTEIN PHOSPHATASE ACTIVITY | 34 | 178 | 2.278e-06 | 2.16e-05 |
| 99 | RNA POLYMERASE II TRANSCRIPTION COACTIVATOR ACTIVITY | 13 | 36 | 2.519e-06 | 2.364e-05 |
| 100 | GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 49 | 303 | 2.688e-06 | 2.497e-05 |
| 101 | GTPASE BINDING | 48 | 295 | 2.843e-06 | 2.615e-05 |
| 102 | MAP KINASE KINASE KINASE ACTIVITY | 10 | 22 | 3.193e-06 | 2.908e-05 |
| 103 | ACTIN BINDING | 59 | 393 | 3.228e-06 | 2.911e-05 |
| 104 | DNA DEPENDENT ATPASE ACTIVITY | 20 | 79 | 3.534e-06 | 3.157e-05 |
| 105 | BHLH TRANSCRIPTION FACTOR BINDING | 11 | 28 | 5.806e-06 | 4.948e-05 |
| 106 | PLATELET DERIVED GROWTH FACTOR BINDING | 7 | 11 | 5.709e-06 | 4.948e-05 |
| 107 | INSULIN RECEPTOR SUBSTRATE BINDING | 7 | 11 | 5.709e-06 | 4.948e-05 |
| 108 | HISTONE ACETYLTRANSFERASE BINDING | 11 | 28 | 5.806e-06 | 4.948e-05 |
| 109 | CADHERIN BINDING | 11 | 28 | 5.806e-06 | 4.948e-05 |
| 110 | CYCLIN BINDING | 9 | 19 | 6.614e-06 | 5.586e-05 |
| 111 | PROTEIN TYROSINE PHOSPHATASE ACTIVITY | 23 | 103 | 6.8e-06 | 5.641e-05 |
| 112 | PROTEIN N TERMINUS BINDING | 23 | 103 | 6.8e-06 | 5.641e-05 |
| 113 | PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR BINDING | 8 | 15 | 7.179e-06 | 5.902e-05 |
| 114 | CALCIUM ION BINDING | 90 | 697 | 7.433e-06 | 6.057e-05 |
| 115 | ION CHANNEL BINDING | 24 | 111 | 7.803e-06 | 6.303e-05 |
| 116 | MITOGEN ACTIVATED PROTEIN KINASE BINDING | 10 | 24 | 8.323e-06 | 6.666e-05 |
| 117 | MAP KINASE KINASE ACTIVITY | 7 | 12 | 1.274e-05 | 0.0001011 |
| 118 | HYDROLASE ACTIVITY ACTING ON ACID ANHYDRIDES | 101 | 820 | 1.589e-05 | 0.0001251 |
| 119 | LIGASE ACTIVITY | 58 | 406 | 1.791e-05 | 0.0001399 |
| 120 | RETINOID X RECEPTOR BINDING | 8 | 17 | 2.339e-05 | 0.0001811 |
| 121 | BINDING BRIDGING | 31 | 173 | 2.368e-05 | 0.0001818 |
| 122 | RECEPTOR ACTIVATOR ACTIVITY | 11 | 32 | 2.573e-05 | 0.0001959 |
| 123 | ACTIN FILAMENT BINDING | 24 | 121 | 3.56e-05 | 0.0002689 |
| 124 | CYTOKINE RECEPTOR BINDING | 42 | 271 | 3.814e-05 | 0.0002858 |
| 125 | HELICASE ACTIVITY | 28 | 153 | 3.896e-05 | 0.0002895 |
| 126 | RAS GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 37 | 228 | 3.984e-05 | 0.0002937 |
| 127 | RHO GTPASE BINDING | 18 | 78 | 4.16e-05 | 0.0003043 |
| 128 | R SMAD BINDING | 9 | 23 | 4.337e-05 | 0.0003148 |
| 129 | GLUCOCORTICOID RECEPTOR BINDING | 7 | 14 | 4.769e-05 | 0.0003408 |
| 130 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II CORE PROMOTER SEQUENCE SPECIFIC | 7 | 14 | 4.769e-05 | 0.0003408 |
| 131 | E BOX BINDING | 11 | 34 | 4.901e-05 | 0.0003475 |
| 132 | NON MEMBRANE SPANNING PROTEIN TYROSINE KINASE ACTIVITY | 13 | 46 | 5.139e-05 | 0.0003617 |
| 133 | PROLINE RICH REGION BINDING | 8 | 19 | 6.272e-05 | 0.0004348 |
| 134 | EPHRIN RECEPTOR ACTIVITY | 8 | 19 | 6.272e-05 | 0.0004348 |
| 135 | PROTEIN TYROSINE KINASE ACTIVATOR ACTIVITY | 7 | 15 | 8.314e-05 | 0.0005721 |
| 136 | PROTEIN KINASE C BINDING | 13 | 50 | 0.0001319 | 0.0009009 |
| 137 | G PROTEIN COUPLED RECEPTOR BINDING | 39 | 259 | 0.0001331 | 0.0009028 |
| 138 | PROTEIN KINASE A REGULATORY SUBUNIT BINDING | 7 | 16 | 0.0001374 | 0.0009185 |
| 139 | PHOSPHATIDYLINOSITOL PHOSPHATE KINASE ACTIVITY | 7 | 16 | 0.0001374 | 0.0009185 |
| 140 | ATPASE ACTIVITY | 57 | 427 | 0.0001413 | 0.0009374 |
| 141 | SINGLE STRANDED DNA BINDING | 19 | 93 | 0.0001491 | 0.0009826 |
| 142 | RECEPTOR REGULATOR ACTIVITY | 12 | 45 | 0.000183 | 0.001197 |
| 143 | TUBULIN BINDING | 40 | 273 | 0.0001989 | 0.001292 |
| 144 | METHYLATED HISTONE BINDING | 13 | 52 | 0.0002025 | 0.001307 |
| 145 | TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE ACTIVITY | 7 | 17 | 0.0002173 | 0.001392 |
| 146 | PROTEASE BINDING | 20 | 104 | 0.0002394 | 0.001523 |
| 147 | ESTROGEN RECEPTOR BINDING | 11 | 40 | 0.0002514 | 0.001585 |
| 148 | CYCLIN DEPENDENT PROTEIN SERINE THREONINE KINASE REGULATOR ACTIVITY | 9 | 28 | 0.0002525 | 0.001585 |
| 149 | PHOSPHOPROTEIN BINDING | 14 | 60 | 0.000256 | 0.001596 |
| 150 | ATP DEPENDENT DNA HELICASE ACTIVITY | 10 | 34 | 0.000262 | 0.001623 |
| 151 | LIGAND DEPENDENT NUCLEAR RECEPTOR BINDING | 8 | 23 | 0.000303 | 0.001864 |
| 152 | HMG BOX DOMAIN BINDING | 7 | 18 | 0.0003306 | 0.002008 |
| 153 | MITOGEN ACTIVATED PROTEIN KINASE KINASE KINASE BINDING | 7 | 18 | 0.0003306 | 0.002008 |
| 154 | CYTOKINE BINDING | 18 | 92 | 0.0003862 | 0.00233 |
| 155 | TRANSCRIPTION COFACTOR BINDING | 8 | 24 | 0.0004223 | 0.002515 |
| 156 | EPHRIN RECEPTOR BINDING | 8 | 24 | 0.0004223 | 0.002515 |
| 157 | HSP70 PROTEIN BINDING | 9 | 30 | 0.0004508 | 0.002651 |
| 158 | NF KAPPAB BINDING | 9 | 30 | 0.0004508 | 0.002651 |
| 159 | PROTEIN SERINE THREONINE KINASE ACTIVATOR ACTIVITY | 7 | 19 | 0.0004869 | 0.002827 |
| 160 | NEUROTROPHIN RECEPTOR BINDING | 6 | 14 | 0.000485 | 0.002827 |
| 161 | GLUTAMATE RECEPTOR BINDING | 10 | 37 | 0.0005559 | 0.003208 |
| 162 | WNT PROTEIN BINDING | 9 | 31 | 0.0005898 | 0.003382 |
| 163 | PROTEIN SELF ASSOCIATION | 11 | 44 | 0.0006167 | 0.003515 |
| 164 | ANKYRIN BINDING | 7 | 20 | 0.0006968 | 0.003947 |
| 165 | UBIQUITIN LIKE PROTEIN BINDING | 21 | 121 | 0.0007177 | 0.004041 |
| 166 | SCAFFOLD PROTEIN BINDING | 11 | 45 | 0.000757 | 0.004186 |
| 167 | PROTEIN KINASE A CATALYTIC SUBUNIT BINDING | 6 | 15 | 0.0007529 | 0.004186 |
| 168 | RETINOIC ACID RECEPTOR BINDING | 11 | 45 | 0.000757 | 0.004186 |
| 169 | PHOSPHATASE ACTIVITY | 38 | 275 | 0.000895 | 0.00492 |
| 170 | DNA HELICASE ACTIVITY | 12 | 53 | 0.0009251 | 0.005055 |
| 171 | PROTEIN PHOSPHATASE TYPE 2A REGULATOR ACTIVITY | 7 | 21 | 0.0009724 | 0.005283 |
| 172 | RNA POLYMERASE II REPRESSING TRANSCRIPTION FACTOR BINDING | 8 | 27 | 0.001023 | 0.005494 |
| 173 | CHEMOREPELLENT ACTIVITY | 8 | 27 | 0.001023 | 0.005494 |
| 174 | TRANSFORMING GROWTH FACTOR BETA BINDING | 6 | 16 | 0.001122 | 0.005957 |
| 175 | TUMOR NECROSIS FACTOR RECEPTOR SUPERFAMILY BINDING | 11 | 47 | 0.001118 | 0.005957 |
| 176 | LYSINE ACETYLATED HISTONE BINDING | 6 | 17 | 0.001616 | 0.008481 |
| 177 | LRR DOMAIN BINDING | 6 | 17 | 0.001616 | 0.008481 |
| 178 | MRNA BINDING | 24 | 155 | 0.001648 | 0.008602 |
| 179 | GROWTH FACTOR RECEPTOR BINDING | 21 | 129 | 0.001665 | 0.008643 |
| 180 | ACTININ BINDING | 8 | 29 | 0.001709 | 0.008778 |
| 181 | ACTIVIN BINDING | 5 | 12 | 0.00172 | 0.008778 |
| 182 | DNA POLYMERASE BINDING | 5 | 12 | 0.00172 | 0.008778 |
| 183 | IONOTROPIC GLUTAMATE RECEPTOR BINDING | 7 | 23 | 0.001775 | 0.008962 |
| 184 | FIBROBLAST GROWTH FACTOR BINDING | 7 | 23 | 0.001775 | 0.008962 |
| 185 | MICROTUBULE BINDING | 29 | 201 | 0.001788 | 0.008978 |
| 186 | PROTEIN DEACETYLASE ACTIVITY | 10 | 43 | 0.001956 | 0.009768 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | ANCHORING JUNCTION | 122 | 489 | 3.588e-30 | 2.095e-27 |
| 2 | CHROMOSOME | 170 | 880 | 2.379e-27 | 6.945e-25 |
| 3 | CELL JUNCTION | 198 | 1151 | 4.004e-25 | 7.795e-23 |
| 4 | NUCLEOPLASM PART | 138 | 708 | 1.147e-22 | 1.675e-20 |
| 5 | CHROMATIN | 101 | 441 | 4.25e-22 | 4.964e-20 |
| 6 | CELL SUBSTRATE JUNCTION | 93 | 398 | 4.494e-21 | 4.375e-19 |
| 7 | NUCLEAR CHROMOSOME | 110 | 523 | 7.178e-21 | 5.988e-19 |
| 8 | NUCLEAR BODY | 84 | 349 | 5.181e-20 | 3.782e-18 |
| 9 | NEURON PART | 197 | 1265 | 8.847e-20 | 5.167e-18 |
| 10 | TRANSCRIPTION FACTOR COMPLEX | 76 | 298 | 8.547e-20 | 5.167e-18 |
| 11 | MEMBRANE REGION | 182 | 1134 | 1.058e-19 | 5.619e-18 |
| 12 | CELL PROJECTION | 252 | 1786 | 2.53e-19 | 1.231e-17 |
| 13 | NUCLEAR CHROMATIN | 73 | 291 | 1.255e-18 | 5.639e-17 |
| 14 | MEMBRANE MICRODOMAIN | 72 | 288 | 2.661e-18 | 1.11e-16 |
| 15 | NEURON PROJECTION | 156 | 942 | 2.989e-18 | 1.164e-16 |
| 16 | CATALYTIC COMPLEX | 158 | 1038 | 4.481e-15 | 1.636e-13 |
| 17 | RECEPTOR COMPLEX | 70 | 327 | 4.053e-14 | 1.319e-12 |
| 18 | PLASMA MEMBRANE REGION | 143 | 929 | 4.065e-14 | 1.319e-12 |
| 19 | TRANSFERASE COMPLEX | 117 | 703 | 4.517e-14 | 1.364e-12 |
| 20 | CELL LEADING EDGE | 73 | 350 | 4.673e-14 | 1.364e-12 |
| 21 | PML BODY | 34 | 97 | 6.549e-14 | 1.821e-12 |
| 22 | CELL SURFACE | 120 | 757 | 6.857e-13 | 1.82e-11 |
| 23 | PROTEIN KINASE COMPLEX | 31 | 90 | 1.419e-12 | 3.602e-11 |
| 24 | EXTRINSIC COMPONENT OF MEMBRANE | 56 | 252 | 2.962e-12 | 7.208e-11 |
| 25 | CELL PROJECTION PART | 139 | 946 | 3.348e-12 | 7.821e-11 |
| 26 | CYTOSKELETON | 241 | 1967 | 1.836e-11 | 4.124e-10 |
| 27 | SIDE OF MEMBRANE | 77 | 428 | 2.509e-11 | 5.427e-10 |
| 28 | CELL CELL JUNCTION | 71 | 383 | 3.695e-11 | 7.706e-10 |
| 29 | CYTOPLASMIC SIDE OF MEMBRANE | 42 | 170 | 4.504e-11 | 9.069e-10 |
| 30 | CYTOPLASMIC REGION | 58 | 287 | 7.157e-11 | 1.393e-09 |
| 31 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 51 | 237 | 9.555e-11 | 1.8e-09 |
| 32 | CELL CORTEX | 51 | 238 | 1.122e-10 | 2.047e-09 |
| 33 | PHOSPHATIDYLINOSITOL 3 KINASE COMPLEX | 13 | 20 | 2.917e-10 | 5.161e-09 |
| 34 | PERINUCLEAR REGION OF CYTOPLASM | 99 | 642 | 3.66e-10 | 6.287e-09 |
| 35 | NUCLEAR SPECK | 43 | 194 | 1.065e-09 | 1.777e-08 |
| 36 | SOMATODENDRITIC COMPARTMENT | 98 | 650 | 1.536e-09 | 2.491e-08 |
| 37 | SWI SNF SUPERFAMILY TYPE COMPLEX | 23 | 71 | 4.261e-09 | 6.725e-08 |
| 38 | DENDRITE | 74 | 451 | 4.379e-09 | 6.73e-08 |
| 39 | LEADING EDGE MEMBRANE | 33 | 134 | 5.762e-09 | 8.629e-08 |
| 40 | CYTOSKELETAL PART | 176 | 1436 | 1.381e-08 | 2.016e-07 |
| 41 | SYNAPSE | 106 | 754 | 1.6e-08 | 2.225e-07 |
| 42 | NUCLEOLUS | 116 | 848 | 1.577e-08 | 2.225e-07 |
| 43 | CHROMOSOMAL REGION | 58 | 330 | 1.729e-08 | 2.348e-07 |
| 44 | LATERAL PLASMA MEMBRANE | 18 | 50 | 3.177e-08 | 4.217e-07 |
| 45 | CELL BODY | 76 | 494 | 4.615e-08 | 5.989e-07 |
| 46 | PLASMA MEMBRANE RAFT | 24 | 86 | 5.221e-08 | 6.629e-07 |
| 47 | RUFFLE MEMBRANE | 23 | 80 | 5.371e-08 | 6.674e-07 |
| 48 | MICROTUBULE CYTOSKELETON | 136 | 1068 | 7.677e-08 | 9.164e-07 |
| 49 | NUCLEAR TRANSCRIPTION FACTOR COMPLEX | 30 | 127 | 7.689e-08 | 9.164e-07 |
| 50 | RNA POLYMERASE II TRANSCRIPTION FACTOR COMPLEX | 26 | 101 | 8.915e-08 | 1.041e-06 |
| 51 | EXTRINSIC COMPONENT OF PLASMA MEMBRANE | 31 | 136 | 1.122e-07 | 1.284e-06 |
| 52 | EXTRACELLULAR MATRIX | 67 | 426 | 1.243e-07 | 1.397e-06 |
| 53 | ACTIN CYTOSKELETON | 69 | 444 | 1.294e-07 | 1.426e-06 |
| 54 | EXCITATORY SYNAPSE | 39 | 197 | 1.602e-07 | 1.732e-06 |
| 55 | TRANSCRIPTIONAL REPRESSOR COMPLEX | 21 | 74 | 2.578e-07 | 2.738e-06 |
| 56 | AXON | 65 | 418 | 2.912e-07 | 3.037e-06 |
| 57 | CYCLIN DEPENDENT PROTEIN KINASE HOLOENZYME COMPLEX | 13 | 31 | 3.308e-07 | 3.389e-06 |
| 58 | POSTSYNAPSE | 60 | 378 | 4.078e-07 | 4.106e-06 |
| 59 | BAF TYPE COMPLEX | 11 | 23 | 5.354e-07 | 5.3e-06 |
| 60 | PROTEINACEOUS EXTRACELLULAR MATRIX | 57 | 356 | 5.921e-07 | 5.763e-06 |
| 61 | CELL CELL ADHERENS JUNCTION | 17 | 54 | 7.018e-07 | 6.719e-06 |
| 62 | RUFFLE | 32 | 156 | 8.915e-07 | 8.397e-06 |
| 63 | CHROMOSOME CENTROMERIC REGION | 34 | 174 | 1.333e-06 | 1.236e-05 |
| 64 | PLASMA MEMBRANE PROTEIN COMPLEX | 73 | 510 | 1.438e-06 | 1.312e-05 |
| 65 | PLASMA MEMBRANE RECEPTOR COMPLEX | 34 | 175 | 1.527e-06 | 1.372e-05 |
| 66 | LAMELLIPODIUM | 33 | 172 | 2.885e-06 | 2.553e-05 |
| 67 | BETA CATENIN DESTRUCTION COMPLEX | 8 | 14 | 3.609e-06 | 3.145e-05 |
| 68 | MICROTUBULE ORGANIZING CENTER | 83 | 623 | 5.131e-06 | 4.407e-05 |
| 69 | EXTRACELLULAR MATRIX COMPONENT | 26 | 125 | 7.019e-06 | 5.94e-05 |
| 70 | SYNAPSE PART | 81 | 610 | 7.536e-06 | 6.287e-05 |
| 71 | SITE OF POLARIZED GROWTH | 29 | 149 | 8.443e-06 | 6.878e-05 |
| 72 | CELL PROJECTION MEMBRANE | 47 | 298 | 8.48e-06 | 6.878e-05 |
| 73 | CELL CELL CONTACT ZONE | 17 | 64 | 9.39e-06 | 7.512e-05 |
| 74 | EXTRINSIC COMPONENT OF CYTOPLASMIC SIDE OF PLASMA MEMBRANE | 22 | 98 | 9.777e-06 | 7.716e-05 |
| 75 | HETEROCHROMATIN | 17 | 67 | 1.816e-05 | 0.000138 |
| 76 | SPINDLE | 45 | 289 | 1.82e-05 | 0.000138 |
| 77 | MAST CELL GRANULE | 9 | 21 | 1.812e-05 | 0.000138 |
| 78 | GOLGI APPARATUS | 161 | 1445 | 1.899e-05 | 0.0001422 |
| 79 | CORTICAL CYTOSKELETON | 19 | 81 | 2.009e-05 | 0.0001482 |
| 80 | HISTONE DEACETYLASE COMPLEX | 16 | 61 | 2.031e-05 | 0.0001482 |
| 81 | DNA REPAIR COMPLEX | 13 | 43 | 2.326e-05 | 0.0001677 |
| 82 | INCLUSION BODY | 17 | 70 | 3.358e-05 | 0.0002392 |
| 83 | NUCLEAR PERIPHERY | 24 | 121 | 3.56e-05 | 0.0002505 |
| 84 | CONTRACTILE FIBER | 35 | 211 | 4.004e-05 | 0.0002784 |
| 85 | MISMATCH REPAIR COMPLEX | 7 | 14 | 4.769e-05 | 0.0003239 |
| 86 | NBAF COMPLEX | 7 | 14 | 4.769e-05 | 0.0003239 |
| 87 | INTRINSIC COMPONENT OF PLASMA MEMBRANE | 177 | 1649 | 5.949e-05 | 0.0003994 |
| 88 | COSTAMERE | 8 | 19 | 6.272e-05 | 0.0004162 |
| 89 | CELL CORTEX PART | 23 | 119 | 7.814e-05 | 0.0005128 |
| 90 | SWI SNF COMPLEX | 7 | 15 | 8.314e-05 | 0.0005395 |
| 91 | CENTROSOME | 64 | 487 | 9.026e-05 | 0.0005793 |
| 92 | VACUOLE | 132 | 1180 | 9.219e-05 | 0.0005852 |
| 93 | PCG PROTEIN COMPLEX | 12 | 43 | 0.0001137 | 0.0007138 |
| 94 | DESMOSOME | 9 | 26 | 0.0001326 | 0.0008236 |
| 95 | ACTIN BASED CELL PROJECTION | 30 | 181 | 0.00014 | 0.0008604 |
| 96 | APICAL PART OF CELL | 50 | 361 | 0.0001444 | 0.0008781 |
| 97 | HISTONE METHYLTRANSFERASE COMPLEX | 16 | 71 | 0.0001475 | 0.0008882 |
| 98 | NUCLEAR HETEROCHROMATIN | 10 | 32 | 0.0001498 | 0.0008926 |
| 99 | INTERCALATED DISC | 13 | 51 | 0.000164 | 0.0009672 |
| 100 | FILOPODIUM | 19 | 94 | 0.0001729 | 0.0009895 |
| 101 | NPBAF COMPLEX | 6 | 12 | 0.0001721 | 0.0009895 |
| 102 | CORTICAL ACTIN CYTOSKELETON | 14 | 58 | 0.0001745 | 0.0009895 |
| 103 | BANDED COLLAGEN FIBRIL | 6 | 12 | 0.0001721 | 0.0009895 |
| 104 | BASOLATERAL PLASMA MEMBRANE | 33 | 211 | 0.0002077 | 0.001166 |
| 105 | NUCLEAR ENVELOPE | 55 | 416 | 0.0002347 | 0.001305 |
| 106 | NEURONAL POSTSYNAPTIC DENSITY | 13 | 53 | 0.0002486 | 0.00137 |
| 107 | CHROMOSOME TELOMERIC REGION | 27 | 162 | 0.0002687 | 0.001467 |
| 108 | COMPLEX OF COLLAGEN TRIMERS | 8 | 23 | 0.000303 | 0.00163 |
| 109 | NUCLEAR MATRIX | 19 | 98 | 0.0003043 | 0.00163 |
| 110 | UBIQUITIN LIGASE COMPLEX | 38 | 262 | 0.000348 | 0.001847 |
| 111 | MICROTUBULE | 53 | 405 | 0.0003878 | 0.002022 |
| 112 | MIDBODY | 23 | 132 | 0.0003873 | 0.002022 |
| 113 | BASEMENT MEMBRANE | 18 | 93 | 0.0004431 | 0.00229 |
| 114 | EXTERNAL SIDE OF PLASMA MEMBRANE | 35 | 238 | 0.0004503 | 0.002307 |
| 115 | SYNAPTIC MEMBRANE | 37 | 261 | 0.0006424 | 0.003262 |
| 116 | APICAL DENDRITE | 6 | 15 | 0.0007529 | 0.003779 |
| 117 | T TUBULE | 11 | 45 | 0.000757 | 0.003779 |
| 118 | EARLY ENDOSOME | 41 | 301 | 0.0007677 | 0.003799 |
| 119 | APICAL PLASMA MEMBRANE | 40 | 292 | 0.0007909 | 0.003881 |
| 120 | METHYLTRANSFERASE COMPLEX | 17 | 90 | 0.0008435 | 0.004105 |
| 121 | AXON PART | 32 | 219 | 0.0008628 | 0.004164 |
| 122 | IMMUNOLOGICAL SYNAPSE | 9 | 33 | 0.000973 | 0.004657 |
| 123 | SEX CHROMOSOME | 8 | 27 | 0.001023 | 0.004857 |
| 124 | WNT SIGNALOSOME | 5 | 11 | 0.001075 | 0.005062 |
| 125 | SARCOLEMMA | 21 | 125 | 0.001107 | 0.005172 |
| 126 | ESC E Z COMPLEX | 6 | 16 | 0.001122 | 0.005201 |
| 127 | POSTSYNAPTIC MEMBRANE | 30 | 205 | 0.0012 | 0.005518 |
| 128 | CONDENSED NUCLEAR CHROMOSOME | 16 | 85 | 0.00123 | 0.005613 |
| 129 | NUCLEAR MEMBRANE | 38 | 280 | 0.001254 | 0.005677 |
| 130 | MITOTIC SPINDLE | 12 | 55 | 0.001308 | 0.005875 |
| 131 | KINETOCHORE | 20 | 120 | 0.001587 | 0.007074 |
| 132 | CHD TYPE COMPLEX | 6 | 17 | 0.001616 | 0.007149 |
| 133 | ENDOCYTIC VESICLE | 35 | 256 | 0.00168 | 0.007376 |
| 134 | PARANODE REGION OF AXON | 5 | 12 | 0.00172 | 0.007439 |
| 135 | PRESYNAPTIC ACTIVE ZONE | 8 | 29 | 0.001709 | 0.007439 |
| 136 | DYNEIN COMPLEX | 10 | 43 | 0.001956 | 0.008398 |
| 137 | MAIN AXON | 12 | 58 | 0.002119 | 0.009035 |
| 138 | PROTEIN COMPLEX INVOLVED IN CELL ADHESION | 8 | 30 | 0.002166 | 0.009168 |
| 139 | NUCLEAR CHROMOSOME TELOMERIC REGION | 21 | 132 | 0.002227 | 0.009357 |
| 140 | CONDENSED CHROMOSOME | 28 | 195 | 0.002279 | 0.009506 |
| 141 | NUCLEAR EUCHROMATIN | 7 | 24 | 0.002332 | 0.009659 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04010_MAPK_signaling_pathway | 71 | 387 | 6.039e-11 | 1.069e-08 | |
| 2 | hsa04510_Focal_adhesion | 68 | 387 | 1.038e-09 | 9.189e-08 | |
| 3 | hsa04722_Neurotrophin_signaling_pathway | 49 | 387 | 0.0013 | 0.0767 | |
| 4 | hsa04110_Cell_cycle | 48 | 387 | 0.002214 | 0.09797 | |
| 5 | hsa04310_Wnt_signaling_pathway | 47 | 387 | 0.003688 | 0.1306 | |
| 6 | hsa04810_Regulation_of_actin_cytoskeleton | 46 | 387 | 0.006007 | 0.1772 | |
| 7 | hsa04360_Axon_guidance | 43 | 387 | 0.02264 | 0.5724 | |
| 8 | hsa04062_Chemokine_signaling_pathway | 42 | 387 | 0.03363 | 0.7441 | |
| 9 | hsa04012_ErbB_signaling_pathway | 40 | 387 | 0.06921 | 1 | |
| 10 | hsa04144_Endocytosis | 39 | 387 | 0.09583 | 1 | |
| 11 | hsa04630_Jak.STAT_signaling_pathway | 39 | 387 | 0.09583 | 1 | |
| 12 | hsa04660_T_cell_receptor_signaling_pathway | 39 | 387 | 0.09583 | 1 | |
| 13 | hsa04115_p53_signaling_pathway | 36 | 387 | 0.2209 | 1 | |
| 14 | hsa04120_Ubiquitin_mediated_proteolysis | 35 | 387 | 0.2785 | 1 | |
| 15 | hsa04910_Insulin_signaling_pathway | 35 | 387 | 0.2785 | 1 | |
| 16 | hsa04380_Osteoclast_differentiation | 34 | 387 | 0.3431 | 1 | |
| 17 | hsa04020_Calcium_signaling_pathway | 32 | 387 | 0.4871 | 1 | |
| 18 | hsa04662_B_cell_receptor_signaling_pathway | 32 | 387 | 0.4871 | 1 | |
| 19 | hsa04210_Apoptosis | 30 | 387 | 0.6356 | 1 | |
| 20 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 30 | 387 | 0.6356 | 1 | |
| 21 | hsa04670_Leukocyte_transendothelial_migration | 30 | 387 | 0.6356 | 1 | |
| 22 | hsa04912_GnRH_signaling_pathway | 30 | 387 | 0.6356 | 1 | |
| 23 | hsa04914_Progesterone.mediated_oocyte_maturation | 30 | 387 | 0.6356 | 1 | |
| 24 | hsa04916_Melanogenesis | 30 | 387 | 0.6356 | 1 | |
| 25 | hsa04114_Oocyte_meiosis | 29 | 387 | 0.705 | 1 | |
| 26 | hsa04350_TGF.beta_signaling_pathway | 28 | 387 | 0.7683 | 1 | |
| 27 | hsa04530_Tight_junction | 28 | 387 | 0.7683 | 1 | |
| 28 | hsa04620_Toll.like_receptor_signaling_pathway | 28 | 387 | 0.7683 | 1 | |
| 29 | hsa04664_Fc_epsilon_RI_signaling_pathway | 28 | 387 | 0.7683 | 1 | |
| 30 | hsa04540_Gap_junction | 27 | 387 | 0.8237 | 1 | |
| 31 | hsa04270_Vascular_smooth_muscle_contraction | 26 | 387 | 0.8704 | 1 | |
| 32 | hsa04720_Long.term_potentiation | 25 | 387 | 0.9081 | 1 | |
| 33 | hsa04514_Cell_adhesion_molecules_.CAMs. | 24 | 387 | 0.9373 | 1 | |
| 34 | hsa04520_Adherens_junction | 24 | 387 | 0.9373 | 1 | |
| 35 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 24 | 387 | 0.9373 | 1 | |
| 36 | hsa04370_VEGF_signaling_pathway | 23 | 387 | 0.9589 | 1 | |
| 37 | hsa04730_Long.term_depression | 23 | 387 | 0.9589 | 1 | |
| 38 | hsa04150_mTOR_signaling_pathway | 22 | 387 | 0.9742 | 1 | |
| 39 | hsa04512_ECM.receptor_interaction | 22 | 387 | 0.9742 | 1 | |
| 40 | hsa04070_Phosphatidylinositol_signaling_system | 21 | 387 | 0.9846 | 1 | |
| 41 | hsa04920_Adipocytokine_signaling_pathway | 19 | 387 | 0.9952 | 1 | |
| 42 | hsa00562_Inositol_phosphate_metabolism | 17 | 387 | 0.9988 | 1 | |
| 43 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 17 | 387 | 0.9988 | 1 | |
| 44 | hsa04145_Phagosome | 16 | 387 | 0.9995 | 1 | |
| 45 | hsa04622_RIG.I.like_receptor_signaling_pathway | 16 | 387 | 0.9995 | 1 | |
| 46 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 16 | 387 | 0.9995 | 1 | |
| 47 | hsa04971_Gastric_acid_secretion | 15 | 387 | 0.9998 | 1 | |
| 48 | hsa04972_Pancreatic_secretion | 15 | 387 | 0.9998 | 1 | |
| 49 | hsa03013_RNA_transport | 14 | 387 | 0.9999 | 1 | |
| 50 | hsa00010_Glycolysis_._Gluconeogenesis | 4 | 387 | 1 | 1 | |
| 51 | hsa00020_Citrate_cycle_.TCA_cycle. | 6 | 387 | 1 | 1 | |
| 52 | hsa00051_Fructose_and_mannose_metabolism | 3 | 387 | 1 | 1 | |
| 53 | hsa00071_Fatty_acid_metabolism | 4 | 387 | 1 | 1 | |
| 54 | hsa00190_Oxidative_phosphorylation | 4 | 387 | 1 | 1 | |
| 55 | hsa00230_Purine_metabolism | 7 | 387 | 1 | 1 | |
| 56 | hsa00240_Pyrimidine_metabolism | 3 | 387 | 1 | 1 | |
| 57 | hsa00270_Cysteine_and_methionine_metabolism | 7 | 387 | 1 | 1 | |
| 58 | hsa00310_Lysine_degradation | 4 | 387 | 1 | 1 | |
| 59 | hsa00330_Arginine_and_proline_metabolism | 2 | 387 | 1 | 1 | |
| 60 | hsa00340_Histidine_metabolism | 2 | 387 | 1 | 1 | |
| 61 | hsa00350_Tyrosine_metabolism | 2 | 387 | 1 | 1 | |
| 62 | hsa00380_Tryptophan_metabolism | 2 | 387 | 1 | 1 | |
| 63 | hsa00410_beta.Alanine_metabolism | 2 | 387 | 1 | 1 | |
| 64 | hsa00480_Glutathione_metabolism | 4 | 387 | 1 | 1 | |
| 65 | hsa00510_N.Glycan_biosynthesis | 2 | 387 | 1 | 1 | |
| 66 | hsa00514_Other_types_of_O.glycan_biosynthesis | 2 | 387 | 1 | 1 | |
| 67 | hsa00534_Glycosaminoglycan_biosynthesis_._heparan_sulfate | 3 | 387 | 1 | 1 | |
| 68 | hsa00561_Glycerolipid_metabolism | 2 | 387 | 1 | 1 | |
| 69 | hsa00564_Glycerophospholipid_metabolism | 2 | 387 | 1 | 1 | |
| 70 | hsa00565_Ether_lipid_metabolism | 3 | 387 | 1 | 1 | |
| 71 | hsa00590_Arachidonic_acid_metabolism | 6 | 387 | 1 | 1 | |
| 72 | hsa00591_Linoleic_acid_metabolism | 4 | 387 | 1 | 1 | |
| 73 | hsa00600_Sphingolipid_metabolism | 2 | 387 | 1 | 1 | |
| 74 | hsa00620_Pyruvate_metabolism | 2 | 387 | 1 | 1 | |
| 75 | hsa00650_Butanoate_metabolism | 2 | 387 | 1 | 1 | |
| 76 | hsa00670_One_carbon_pool_by_folate | 2 | 387 | 1 | 1 | |
| 77 | hsa00830_Retinol_metabolism | 3 | 387 | 1 | 1 | |
| 78 | hsa00980_Metabolism_of_xenobiotics_by_cytochrome_P450 | 3 | 387 | 1 | 1 | |
| 79 | hsa00982_Drug_metabolism_._cytochrome_P450 | 3 | 387 | 1 | 1 | |
| 80 | hsa00983_Drug_metabolism_._other_enzymes | 2 | 387 | 1 | 1 | |
| 81 | hsa02010_ABC_transporters | 6 | 387 | 1 | 1 | |
| 82 | hsa03008_Ribosome_biogenesis_in_eukaryotes | 5 | 387 | 1 | 1 | |
| 83 | hsa03010_Ribosome | 4 | 387 | 1 | 1 | |
| 84 | hsa03015_mRNA_surveillance_pathway | 12 | 387 | 1 | 1 | |
| 85 | hsa03018_RNA_degradation | 8 | 387 | 1 | 1 | |
| 86 | hsa03022_Basal_transcription_factors | 2 | 387 | 1 | 1 | |
| 87 | hsa03040_Spliceosome | 5 | 387 | 1 | 1 | |
| 88 | hsa03320_PPAR_signaling_pathway | 9 | 387 | 1 | 1 | |
| 89 | hsa03410_Base_excision_repair | 3 | 387 | 1 | 1 | |
| 90 | hsa03420_Nucleotide_excision_repair | 8 | 387 | 1 | 1 | |
| 91 | hsa03430_Mismatch_repair | 6 | 387 | 1 | 1 | |
| 92 | hsa03440_Homologous_recombination | 4 | 387 | 1 | 1 | |
| 93 | hsa04140_Regulation_of_autophagy | 4 | 387 | 1 | 1 | |
| 94 | hsa04142_Lysosome | 3 | 387 | 1 | 1 | |
| 95 | hsa04146_Peroxisome | 6 | 387 | 1 | 1 | |
| 96 | hsa04260_Cardiac_muscle_contraction | 8 | 387 | 1 | 1 | |
| 97 | hsa04320_Dorso.ventral_axis_formation | 11 | 387 | 1 | 1 | |
| 98 | hsa04330_Notch_signaling_pathway | 11 | 387 | 1 | 1 | |
| 99 | hsa04340_Hedgehog_signaling_pathway | 13 | 387 | 1 | 1 | |
| 100 | hsa04610_Complement_and_coagulation_cascades | 4 | 387 | 1 | 1 | |
| 101 | hsa04612_Antigen_processing_and_presentation | 6 | 387 | 1 | 1 | |
| 102 | hsa04614_Renin.angiotensin_system | 3 | 387 | 1 | 1 | |
| 103 | hsa04621_NOD.like_receptor_signaling_pathway | 13 | 387 | 1 | 1 | |
| 104 | hsa04623_Cytosolic_DNA.sensing_pathway | 6 | 387 | 1 | 1 | |
| 105 | hsa04640_Hematopoietic_cell_lineage | 12 | 387 | 1 | 1 | |
| 106 | hsa04672_Intestinal_immune_network_for_IgA_production | 3 | 387 | 1 | 1 | |
| 107 | hsa04710_Circadian_rhythm_._mammal | 6 | 387 | 1 | 1 | |
| 108 | hsa04740_Olfactory_transduction | 2 | 388 | 1 | 1 | |
| 109 | hsa04742_Taste_transduction | 3 | 387 | 1 | 1 | |
| 110 | hsa04962_Vasopressin.regulated_water_reabsorption | 7 | 387 | 1 | 1 | |
| 111 | hsa04964_Proximal_tubule_bicarbonate_reclamation | 2 | 387 | 1 | 1 | |
| 112 | hsa04970_Salivary_secretion | 12 | 387 | 1 | 1 | |
| 113 | hsa04973_Carbohydrate_digestion_and_absorption | 11 | 387 | 1 | 1 | |
| 114 | hsa04974_Protein_digestion_and_absorption | 9 | 387 | 1 | 1 | |
| 115 | hsa04976_Bile_secretion | 8 | 387 | 1 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MYLK-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p | 11 | MRVI1 | Sponge network | -1.331 | 3.0E-5 | -1.051 | 0.00022 | 0.939 |
| 2 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p | 12 | SYNM | Sponge network | -1.331 | 3.0E-5 | -2.309 | 1.0E-5 | 0.934 |
| 3 | MYLK-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-340-5p | 16 | SYNPO2 | Sponge network | -1.331 | 3.0E-5 | -2.342 | 0 | 0.931 |
| 4 | MYLK-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-484;hsa-miR-7-5p | 11 | FLNA | Sponge network | -1.331 | 3.0E-5 | -0.771 | 0.01377 | 0.917 |
| 5 | MIR143HG |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 24 | MRVI1 | Sponge network | -0.739 | 0.0574 | -1.051 | 0.00022 | 0.909 |
| 6 | MRVI1-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p | 17 | MRVI1 | Sponge network | -1.092 | 4.0E-5 | -1.051 | 0.00022 | 0.903 |
| 7 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 30 | RASSF8 | Sponge network | 0.335 | 0.20596 | -0.122 | 0.65591 | 0.897 |
| 8 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p | 17 | AKAP6 | Sponge network | -1.331 | 3.0E-5 | -1.586 | 3.0E-5 | 0.893 |
| 9 | MIR143HG |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-769-5p | 34 | SYNPO2 | Sponge network | -0.739 | 0.0574 | -2.342 | 0 | 0.891 |
| 10 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | ABCC9 | Sponge network | -0.576 | 0.10121 | -0.466 | 0.14121 | 0.885 |
| 11 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 58 | BNC2 | Sponge network | -0.129 | 0.57177 | -0.491 | 0.09988 | 0.881 |
| 12 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ABCC9 | Sponge network | -0.129 | 0.57177 | -0.466 | 0.14121 | 0.881 |
| 13 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | ABCC9 | Sponge network | -0.739 | 0.0574 | -0.466 | 0.14121 | 0.88 |
| 14 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p | 15 | PGR | Sponge network | -1.331 | 3.0E-5 | -1.704 | 0 | 0.877 |
| 15 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 54 | BNC2 | Sponge network | -0.576 | 0.10121 | -0.491 | 0.09988 | 0.876 |
| 16 | ADAMTS9-AS2 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 12 | FHL1 | Sponge network | -2.452 | 0 | -2.687 | 0 | 0.873 |
| 17 | MIR143HG |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | FLNA | Sponge network | -0.739 | 0.0574 | -0.771 | 0.01377 | 0.872 |
| 18 | MIR143HG |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -0.739 | 0.0574 | -2.633 | 0 | 0.871 |
| 19 | LINC00702 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 31 | SYNPO2 | Sponge network | -0.737 | 0.06182 | -2.342 | 0 | 0.868 |
| 20 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-539-5p;hsa-miR-542-3p | 14 | SYNPO2 | Sponge network | -1.092 | 4.0E-5 | -2.342 | 0 | 0.865 |
| 21 | MIR143HG |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 23 | SYNM | Sponge network | -0.739 | 0.0574 | -2.309 | 1.0E-5 | 0.863 |
| 22 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | PEG3 | Sponge network | -0.976 | 0.00025 | -1.74 | 0 | 0.863 |
| 23 | RP11-166D19.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | MRVI1 | Sponge network | -0.576 | 0.10121 | -1.051 | 0.00022 | 0.862 |
| 24 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-935 | 23 | DMD | Sponge network | -1.331 | 3.0E-5 | -1.506 | 0 | 0.855 |
| 25 | LINC00702 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | -0.737 | 0.06182 | -2.633 | 0 | 0.854 |
| 26 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 56 | RBMS3 | Sponge network | -0.129 | 0.57177 | -0.519 | 0.03551 | 0.853 |
| 27 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-340-5p | 10 | PRUNE2 | Sponge network | -1.331 | 3.0E-5 | -1.733 | 0.00022 | 0.852 |
| 28 | MIR143HG |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-98-5p | 11 | HSPB7 | Sponge network | -0.739 | 0.0574 | -2.268 | 1.0E-5 | 0.852 |
| 29 | MIR143HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 38 | AKAP6 | Sponge network | -0.739 | 0.0574 | -1.586 | 3.0E-5 | 0.851 |
| 30 | LINC00702 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | MRVI1 | Sponge network | -0.737 | 0.06182 | -1.051 | 0.00022 | 0.85 |
| 31 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 62 | BNC2 | Sponge network | -0.739 | 0.0574 | -0.491 | 0.09988 | 0.848 |
| 32 | MAGI2-AS3 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 13 | PRKD1 | Sponge network | -0.129 | 0.57177 | -0.466 | 0.03583 | 0.848 |
| 33 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-935 | 35 | FAT3 | Sponge network | -1.331 | 3.0E-5 | -1.601 | 0.00051 | 0.846 |
| 34 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-370-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | JAZF1 | Sponge network | -0.769 | 0.00035 | -0.377 | 0.02677 | 0.844 |
| 35 | RP11-166D19.1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -0.576 | 0.10121 | -2.633 | 0 | 0.844 |
| 36 | MIR143HG |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 13 | FHL1 | Sponge network | -0.739 | 0.0574 | -2.687 | 0 | 0.843 |
| 37 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLNA | Sponge network | -0.576 | 0.10121 | -0.771 | 0.01377 | 0.842 |
| 38 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p | 21 | ANK2 | Sponge network | -1.331 | 3.0E-5 | -1.603 | 1.0E-5 | 0.842 |
| 39 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p | 36 | PGR | Sponge network | -0.576 | 0.10121 | -1.704 | 0 | 0.841 |
| 40 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 67 | RUNX1T1 | Sponge network | -0.576 | 0.10121 | -0.344 | 0.2374 | 0.841 |
| 41 | RP11-554A11.4 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-5p | 14 | MRVI1 | Sponge network | -0.583 | 0.14589 | -1.051 | 0.00022 | 0.841 |
| 42 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | PEG3 | Sponge network | -0.576 | 0.10121 | -1.74 | 0 | 0.839 |
| 43 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 42 | PGR | Sponge network | -0.739 | 0.0574 | -1.704 | 0 | 0.839 |
| 44 | MAGI2-AS3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | MRVI1 | Sponge network | -0.129 | 0.57177 | -1.051 | 0.00022 | 0.839 |
| 45 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 32 | SYNPO2 | Sponge network | -2.452 | 0 | -2.342 | 0 | 0.839 |
| 46 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-590-5p | 10 | MYH11 | Sponge network | -2.452 | 0 | -2.895 | 0 | 0.839 |
| 47 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 70 | RUNX1T1 | Sponge network | -0.129 | 0.57177 | -0.344 | 0.2374 | 0.838 |
| 48 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-664a-5p;hsa-miR-7-5p;hsa-miR-935 | 55 | LPP | Sponge network | -1.331 | 3.0E-5 | -0.459 | 0.04475 | 0.837 |
| 49 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 51 | RBMS3 | Sponge network | -0.576 | 0.10121 | -0.519 | 0.03551 | 0.836 |
| 50 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 38 | PGR | Sponge network | -2.452 | 0 | -1.704 | 0 | 0.836 |
| 51 | HAND2-AS1 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-874-3p;hsa-miR-98-5p | 11 | HSPB7 | Sponge network | -1.697 | 0.00889 | -2.268 | 1.0E-5 | 0.836 |
| 52 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 33 | SYNPO2 | Sponge network | -0.576 | 0.10121 | -2.342 | 0 | 0.836 |
| 53 | RP11-166D19.1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 10 | PRKD1 | Sponge network | -0.576 | 0.10121 | -0.466 | 0.03583 | 0.836 |
| 54 | RP11-166D19.1 |
hsa-let-7f-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 10 | ROR2 | Sponge network | -0.576 | 0.10121 | -1.091 | 0.0008 | 0.835 |
| 55 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-671-5p;hsa-miR-7-5p | 22 | RBMS3 | Sponge network | -1.331 | 3.0E-5 | -0.519 | 0.03551 | 0.835 |
| 56 | HAND2-AS1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -1.697 | 0.00889 | -2.633 | 0 | 0.835 |
| 57 | MIR143HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 30 | NRXN3 | Sponge network | -0.739 | 0.0574 | -0.893 | 0.04186 | 0.835 |
| 58 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 42 | TSHZ3 | Sponge network | -0.129 | 0.57177 | -0.556 | 0.01516 | 0.835 |
| 59 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-550a-5p;hsa-miR-935 | 20 | TSHZ3 | Sponge network | -1.331 | 3.0E-5 | -0.556 | 0.01516 | 0.834 |
| 60 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 34 | PCDH10 | Sponge network | -0.576 | 0.10121 | -1.644 | 0.0078 | 0.834 |
| 61 | MYLK-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-671-5p | 28 | BNC2 | Sponge network | -1.331 | 3.0E-5 | -0.491 | 0.09988 | 0.832 |
| 62 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-424-5p | 15 | SYNE1 | Sponge network | -1.331 | 3.0E-5 | -0.738 | 0.00316 | 0.831 |
| 63 | MIR143HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 74 | RUNX1T1 | Sponge network | -0.739 | 0.0574 | -0.344 | 0.2374 | 0.831 |
| 64 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 21 | GLI3 | Sponge network | -0.129 | 0.57177 | -0.615 | 0.01482 | 0.83 |
| 65 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 59 | RBMS3 | Sponge network | -0.739 | 0.0574 | -0.519 | 0.03551 | 0.83 |
| 66 | HAND2-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-769-5p | 33 | SYNPO2 | Sponge network | -1.697 | 0.00889 | -2.342 | 0 | 0.829 |
| 67 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | RECK | Sponge network | -0.129 | 0.57177 | -1.043 | 0 | 0.829 |
| 68 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 41 | PGR | Sponge network | -0.129 | 0.57177 | -1.704 | 0 | 0.828 |
| 69 | LINC00702 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | FLNA | Sponge network | -0.737 | 0.06182 | -0.771 | 0.01377 | 0.828 |
| 70 | MYLK-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p | 37 | AR | Sponge network | -1.331 | 3.0E-5 | -1.554 | 0 | 0.827 |
| 71 | MRVI1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-542-3p | 16 | SYNM | Sponge network | -1.092 | 4.0E-5 | -2.309 | 1.0E-5 | 0.827 |
| 72 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | ABCC9 | Sponge network | -0.737 | 0.06182 | -0.466 | 0.14121 | 0.827 |
| 73 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-935;hsa-miR-940 | 13 | MAPK10 | Sponge network | -1.331 | 3.0E-5 | -1.774 | 0 | 0.827 |
| 74 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 53 | RBMS3 | Sponge network | -2.452 | 0 | -0.519 | 0.03551 | 0.826 |
| 75 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | -0.976 | 0.00025 | -1.398 | 2.0E-5 | 0.825 |
| 76 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92b-3p | 18 | AKAP12 | Sponge network | -0.576 | 0.10121 | -0.932 | 0.0028 | 0.825 |
| 77 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 61 | AR | Sponge network | -0.576 | 0.10121 | -1.554 | 0 | 0.825 |
| 78 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 31 | SETBP1 | Sponge network | -2.452 | 0 | -1.302 | 1.0E-5 | 0.825 |
| 79 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | SYNE1 | Sponge network | -0.129 | 0.57177 | -0.738 | 0.00316 | 0.824 |
| 80 | ADAMTS9-AS2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 18 | MAPK10 | Sponge network | -2.452 | 0 | -1.774 | 0 | 0.823 |
| 81 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ABCC9 | Sponge network | -1.697 | 0.00889 | -0.466 | 0.14121 | 0.822 |
| 82 | MIR143HG |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | PEG3 | Sponge network | -0.739 | 0.0574 | -1.74 | 0 | 0.821 |
| 83 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | PEG3 | Sponge network | -0.129 | 0.57177 | -1.74 | 0 | 0.821 |
| 84 | FENDRR |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-98-5p | 11 | ADAMTS8 | Sponge network | -1.281 | 0.00012 | -1.476 | 0.00069 | 0.82 |
| 85 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 35 | ANK2 | Sponge network | -0.576 | 0.10121 | -1.603 | 1.0E-5 | 0.819 |
| 86 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 40 | TSHZ3 | Sponge network | -0.576 | 0.10121 | -0.556 | 0.01516 | 0.819 |
| 87 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-96-5p | 62 | FAT3 | Sponge network | -0.576 | 0.10121 | -1.601 | 0.00051 | 0.818 |
| 88 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 26 | PRUNE2 | Sponge network | -0.739 | 0.0574 | -1.733 | 0.00022 | 0.818 |
| 89 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 25 | BNC2 | Sponge network | -0.583 | 0.14589 | -0.491 | 0.09988 | 0.818 |
| 90 | MIR143HG |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-3913-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 112 | LPP | Sponge network | -0.739 | 0.0574 | -0.459 | 0.04475 | 0.818 |
| 91 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 24 | AKAP6 | Sponge network | -1.092 | 4.0E-5 | -1.586 | 3.0E-5 | 0.817 |
| 92 | LINC00702 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-532-3p;hsa-miR-542-3p | 18 | SYNM | Sponge network | -0.737 | 0.06182 | -2.309 | 1.0E-5 | 0.817 |
| 93 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 37 | CNTN1 | Sponge network | -0.576 | 0.10121 | -2.594 | 0 | 0.817 |
| 94 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 15 | ADAMTSL3 | Sponge network | -1.331 | 3.0E-5 | -1.559 | 0 | 0.816 |
| 95 | FENDRR |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | MRVI1 | Sponge network | -1.281 | 0.00012 | -1.051 | 0.00022 | 0.816 |
| 96 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 33 | ANK2 | Sponge network | -2.452 | 0 | -1.603 | 1.0E-5 | 0.815 |
| 97 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 47 | TSHZ3 | Sponge network | -0.739 | 0.0574 | -0.556 | 0.01516 | 0.815 |
| 98 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | -1.331 | 3.0E-5 | 0.358 | 0.13727 | 0.815 |
| 99 | RP11-166D19.1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 14 | FHL1 | Sponge network | -0.576 | 0.10121 | -2.687 | 0 | 0.814 |
| 100 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 15 | MN1 | Sponge network | -0.576 | 0.10121 | -0.358 | 0.24172 | 0.814 |
| 101 | ADAMTS9-AS2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | MRVI1 | Sponge network | -2.452 | 0 | -1.051 | 0.00022 | 0.814 |
| 102 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-484;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | NRXN3 | Sponge network | -0.576 | 0.10121 | -0.893 | 0.04186 | 0.813 |
| 103 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-374b-3p;hsa-miR-452-5p;hsa-miR-539-5p | 15 | PGR | Sponge network | -1.092 | 4.0E-5 | -1.704 | 0 | 0.813 |
| 104 | PCA3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | -0.709 | 0.18168 | -1.733 | 0.00022 | 0.813 |
| 105 | SNHG14 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | RNF180 | Sponge network | -1.111 | 0.0003 | -1.127 | 1.0E-5 | 0.812 |
| 106 | LINC00702 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 12 | FHL1 | Sponge network | -0.737 | 0.06182 | -2.687 | 0 | 0.812 |
| 107 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 59 | BNC2 | Sponge network | -0.737 | 0.06182 | -0.491 | 0.09988 | 0.81 |
| 108 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 18 | CNTNAP1 | Sponge network | -0.739 | 0.0574 | 0.358 | 0.13727 | 0.81 |
| 109 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | PLSCR4 | Sponge network | -0.576 | 0.10121 | -0.474 | 0.03833 | 0.81 |
| 110 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-424-5p | 40 | RUNX1T1 | Sponge network | -1.331 | 3.0E-5 | -0.344 | 0.2374 | 0.81 |
| 111 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-424-5p | 13 | RECK | Sponge network | -1.331 | 3.0E-5 | -1.043 | 0 | 0.809 |
| 112 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | RECK | Sponge network | -0.576 | 0.10121 | -1.043 | 0 | 0.809 |
| 113 | HAND2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -1.697 | 0.00889 | -2.046 | 0.00019 | 0.809 |
| 114 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 19 | GLI3 | Sponge network | -0.576 | 0.10121 | -0.615 | 0.01482 | 0.808 |
| 115 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-769-5p | 10 | SYNPO2 | Sponge network | -0.583 | 0.14589 | -2.342 | 0 | 0.808 |
| 116 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 73 | RUNX1T1 | Sponge network | -1.697 | 0.00889 | -0.344 | 0.2374 | 0.808 |
| 117 | FENDRR |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 56 | EPHA7 | Sponge network | -1.281 | 0.00012 | -2.197 | 3.0E-5 | 0.808 |
| 118 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 41 | RASSF8 | Sponge network | -0.129 | 0.57177 | -0.122 | 0.65591 | 0.807 |
| 119 | MYLK-AS1 |
hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-370-3p;hsa-miR-935 | 13 | PCDH10 | Sponge network | -1.331 | 3.0E-5 | -1.644 | 0.0078 | 0.807 |
| 120 | MYLK-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-34a-5p | 11 | ITPKB | Sponge network | -1.331 | 3.0E-5 | -0.704 | 0.00106 | 0.807 |
| 121 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-940 | 13 | MAPK10 | Sponge network | -1.439 | 4.0E-5 | -1.774 | 0 | 0.806 |
| 122 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 19 | MAPK10 | Sponge network | -0.576 | 0.10121 | -1.774 | 0 | 0.806 |
| 123 | MAGI2-AS3 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | RNF180 | Sponge network | -0.129 | 0.57177 | -1.127 | 1.0E-5 | 0.805 |
| 124 | HAND2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 23 | SYNM | Sponge network | -1.697 | 0.00889 | -2.309 | 1.0E-5 | 0.805 |
| 125 | RP11-166D19.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 20 | SYNM | Sponge network | -0.576 | 0.10121 | -2.309 | 1.0E-5 | 0.805 |
| 126 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | -0.129 | 0.57177 | -0.474 | 0.03833 | 0.804 |
| 127 | MIR143HG |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 69 | AR | Sponge network | -0.739 | 0.0574 | -1.554 | 0 | 0.803 |
| 128 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 61 | BNC2 | Sponge network | -1.697 | 0.00889 | -0.491 | 0.09988 | 0.803 |
| 129 | MRVI1-AS1 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-452-5p;hsa-miR-7-1-3p | 19 | ANK2 | Sponge network | -1.092 | 4.0E-5 | -1.603 | 1.0E-5 | 0.803 |
| 130 | NR2F1-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 21 | MRVI1 | Sponge network | -0.49 | 0.04946 | -1.051 | 0.00022 | 0.803 |
| 131 | MAGI2-AS3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 33 | DLC1 | Sponge network | -0.129 | 0.57177 | -0.382 | 0.05038 | 0.803 |
| 132 | MIR143HG |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 22 | GLI3 | Sponge network | -0.739 | 0.0574 | -0.615 | 0.01482 | 0.803 |
| 133 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 35 | PGR | Sponge network | -0.49 | 0.04946 | -1.704 | 0 | 0.802 |
| 134 | MIR497HG |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p | 18 | MRVI1 | Sponge network | -1.439 | 4.0E-5 | -1.051 | 0.00022 | 0.802 |
| 135 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-935 | 22 | FBXO32 | Sponge network | -1.331 | 3.0E-5 | -0.495 | 0.07561 | 0.801 |
| 136 | ADAMTS9-AS2 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-98-5p | 11 | HSPB7 | Sponge network | -2.452 | 0 | -2.268 | 1.0E-5 | 0.801 |
| 137 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 12 | ABCC9 | Sponge network | -1.092 | 4.0E-5 | -0.466 | 0.14121 | 0.801 |
| 138 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-664a-3p | 23 | GREM1 | Sponge network | -0.576 | 0.10121 | 0.902 | 0.01691 | 0.8 |
| 139 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | ADAMTSL3 | Sponge network | -0.576 | 0.10121 | -1.559 | 0 | 0.8 |
| 140 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | PLSCR4 | Sponge network | -0.49 | 0.04946 | -0.474 | 0.03833 | 0.8 |
| 141 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | SYNE1 | Sponge network | -0.739 | 0.0574 | -0.738 | 0.00316 | 0.8 |
| 142 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | LHFP | Sponge network | -0.129 | 0.57177 | -0.167 | 0.41371 | 0.8 |
| 143 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-1-3p | 24 | ADAMTSL3 | Sponge network | -1.439 | 4.0E-5 | -1.559 | 0 | 0.8 |
| 144 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | SYNE1 | Sponge network | -1.111 | 0.0003 | -0.738 | 0.00316 | 0.799 |
| 145 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | -0.129 | 0.57177 | 0.809 | 0.00544 | 0.799 |
| 146 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | ZNF521 | Sponge network | -0.129 | 0.57177 | -0.111 | 0.58068 | 0.798 |
| 147 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | ADAMTSL3 | Sponge network | -2.452 | 0 | -1.559 | 0 | 0.798 |
| 148 | MIR143HG |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-935 | 41 | PCDH10 | Sponge network | -0.739 | 0.0574 | -1.644 | 0.0078 | 0.798 |
| 149 | ADAMTS9-AS2 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | RNF180 | Sponge network | -2.452 | 0 | -1.127 | 1.0E-5 | 0.797 |
| 150 | ADAMTS9-AS2 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 22 | SFRP1 | Sponge network | -2.452 | 0 | -3.566 | 0 | 0.797 |
| 151 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 62 | AR | Sponge network | -0.49 | 0.04946 | -1.554 | 0 | 0.797 |
| 152 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 36 | ANK2 | Sponge network | -0.739 | 0.0574 | -1.603 | 1.0E-5 | 0.797 |
| 153 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 53 | RBMS3 | Sponge network | -0.49 | 0.04946 | -0.519 | 0.03551 | 0.796 |
| 154 | MRVI1-AS1 |
hsa-let-7b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-7-1-3p | 33 | AR | Sponge network | -1.092 | 4.0E-5 | -1.554 | 0 | 0.796 |
| 155 | MIR143HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935 | 47 | FBXO32 | Sponge network | -0.739 | 0.0574 | -0.495 | 0.07561 | 0.796 |
| 156 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 67 | FAT3 | Sponge network | -0.739 | 0.0574 | -1.601 | 0.00051 | 0.796 |
| 157 | ZNF667-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-511-5p | 17 | MRVI1 | Sponge network | -0.976 | 0.00025 | -1.051 | 0.00022 | 0.796 |
| 158 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 48 | DMD | Sponge network | -2.452 | 0 | -1.506 | 0 | 0.796 |
| 159 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 11 | PEG3 | Sponge network | -1.092 | 4.0E-5 | -1.74 | 0 | 0.796 |
| 160 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 16 | CNTNAP1 | Sponge network | -0.576 | 0.10121 | 0.358 | 0.13727 | 0.796 |
| 161 | HAND2-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 23 | MRVI1 | Sponge network | -1.697 | 0.00889 | -1.051 | 0.00022 | 0.795 |
| 162 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LHFP | Sponge network | -0.576 | 0.10121 | -0.167 | 0.41371 | 0.795 |
| 163 | MAGI2-AS3 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLNA | Sponge network | -0.129 | 0.57177 | -0.771 | 0.01377 | 0.794 |
| 164 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 33 | AKAP6 | Sponge network | -0.576 | 0.10121 | -1.586 | 3.0E-5 | 0.794 |
| 165 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 77 | TCF4 | Sponge network | -0.129 | 0.57177 | 0.016 | 0.92271 | 0.794 |
| 166 | MAGI2-AS3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 34 | NAV3 | Sponge network | -0.129 | 0.57177 | 0.212 | 0.47729 | 0.794 |
| 167 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 38 | PGR | Sponge network | -1.697 | 0.00889 | -1.704 | 0 | 0.794 |
| 168 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 41 | PGR | Sponge network | -0.737 | 0.06182 | -1.704 | 0 | 0.794 |
| 169 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 61 | RBMS3 | Sponge network | -1.111 | 0.0003 | -0.519 | 0.03551 | 0.794 |
| 170 | USP3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-495-3p | 19 | SYNM | Sponge network | -0.162 | 0.47687 | -2.309 | 1.0E-5 | 0.793 |
| 171 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 54 | BNC2 | Sponge network | -0.49 | 0.04946 | -0.491 | 0.09988 | 0.793 |
| 172 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-7-5p | 10 | FNBP1 | Sponge network | -1.331 | 3.0E-5 | -0.969 | 4.0E-5 | 0.793 |
| 173 | ZNF667-AS1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-484;hsa-miR-96-5p | 11 | FHL1 | Sponge network | -0.976 | 0.00025 | -2.687 | 0 | 0.792 |
| 174 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 36 | ANK2 | Sponge network | -0.129 | 0.57177 | -1.603 | 1.0E-5 | 0.792 |
| 175 | LINC00702 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 36 | AKAP6 | Sponge network | -0.737 | 0.06182 | -1.586 | 3.0E-5 | 0.792 |
| 176 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | BNC2 | Sponge network | -0.976 | 0.00025 | -0.491 | 0.09988 | 0.792 |
| 177 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | SETBP1 | Sponge network | -0.576 | 0.10121 | -1.302 | 1.0E-5 | 0.792 |
| 178 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | BNC2 | Sponge network | 0.572 | 0.01722 | -0.491 | 0.09988 | 0.792 |
| 179 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 66 | AR | Sponge network | -0.129 | 0.57177 | -1.554 | 0 | 0.792 |
| 180 | SNHG14 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 39 | SETBP1 | Sponge network | -1.111 | 0.0003 | -1.302 | 1.0E-5 | 0.792 |
| 181 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | -0.576 | 0.10121 | -1.398 | 2.0E-5 | 0.792 |
| 182 | SNHG14 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | PEG3 | Sponge network | -1.111 | 0.0003 | -1.74 | 0 | 0.792 |
| 183 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 16 | SETBP1 | Sponge network | -1.092 | 4.0E-5 | -1.302 | 1.0E-5 | 0.791 |
| 184 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 28 | NRXN3 | Sponge network | -1.697 | 0.00889 | -0.893 | 0.04186 | 0.79 |
| 185 | MIR497HG |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p | 22 | SYNPO2 | Sponge network | -1.439 | 4.0E-5 | -2.342 | 0 | 0.79 |
| 186 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-92b-3p | 19 | AKAP12 | Sponge network | -0.129 | 0.57177 | -0.932 | 0.0028 | 0.79 |
| 187 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | PEG3 | Sponge network | -2.452 | 0 | -1.74 | 0 | 0.789 |
| 188 | MIR143HG |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | CTNNA3 | Sponge network | -0.739 | 0.0574 | -2.046 | 0.00019 | 0.789 |
| 189 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 41 | RBMS3 | Sponge network | -0.976 | 0.00025 | -0.519 | 0.03551 | 0.789 |
| 190 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-940 | 28 | HLF | Sponge network | -1.331 | 3.0E-5 | -2.443 | 0 | 0.789 |
| 191 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | RECK | Sponge network | -2.452 | 0 | -1.043 | 0 | 0.788 |
| 192 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | TET1 | Sponge network | 0.225 | 0.30644 | -0.135 | 0.52138 | 0.788 |
| 193 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-5p | 11 | LHFP | Sponge network | -0.976 | 0.00025 | -0.167 | 0.41371 | 0.788 |
| 194 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-7-5p | 22 | ZBTB4 | Sponge network | -1.331 | 3.0E-5 | -0.739 | 0 | 0.788 |
| 195 | MIR497HG |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p | 10 | HSPB7 | Sponge network | -1.439 | 4.0E-5 | -2.268 | 1.0E-5 | 0.788 |
| 196 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-424-5p | 28 | EPHA7 | Sponge network | -1.331 | 3.0E-5 | -2.197 | 3.0E-5 | 0.788 |
| 197 | HAND2-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLNA | Sponge network | -1.697 | 0.00889 | -0.771 | 0.01377 | 0.788 |
| 198 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 64 | FAT3 | Sponge network | -1.697 | 0.00889 | -1.601 | 0.00051 | 0.787 |
| 199 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | RECK | Sponge network | -0.976 | 0.00025 | -1.043 | 0 | 0.787 |
| 200 | MYLK-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-7-5p;hsa-miR-940 | 11 | GLI3 | Sponge network | -1.331 | 3.0E-5 | -0.615 | 0.01482 | 0.787 |
| 201 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 61 | AR | Sponge network | -2.452 | 0 | -1.554 | 0 | 0.787 |
| 202 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-92b-3p | 17 | AKAP12 | Sponge network | -0.49 | 0.04946 | -0.932 | 0.0028 | 0.787 |
| 203 | RP11-166D19.1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | RNF180 | Sponge network | -0.576 | 0.10121 | -1.127 | 1.0E-5 | 0.786 |
| 204 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | RASSF8 | Sponge network | -0.576 | 0.10121 | -0.122 | 0.65591 | 0.786 |
| 205 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 15 | CNTNAP1 | Sponge network | -0.129 | 0.57177 | 0.358 | 0.13727 | 0.786 |
| 206 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 32 | RBMS3 | Sponge network | -1.092 | 4.0E-5 | -0.519 | 0.03551 | 0.785 |
| 207 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-671-5p | 19 | DLC1 | Sponge network | -1.331 | 3.0E-5 | -0.382 | 0.05038 | 0.785 |
| 208 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 33 | AKAP6 | Sponge network | -2.452 | 0 | -1.586 | 3.0E-5 | 0.785 |
| 209 | ZNF667-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 14 | MAPK10 | Sponge network | -0.976 | 0.00025 | -1.774 | 0 | 0.785 |
| 210 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-511-5p;hsa-miR-590-3p | 19 | ADAMTSL3 | Sponge network | -0.358 | 0.17255 | -1.559 | 0 | 0.785 |
| 211 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 36 | PCDH10 | Sponge network | -1.697 | 0.00889 | -1.644 | 0.0078 | 0.784 |
| 212 | NR2F1-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p | 12 | PRKD1 | Sponge network | -0.49 | 0.04946 | -0.466 | 0.03583 | 0.784 |
| 213 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | -1.439 | 4.0E-5 | -1.603 | 1.0E-5 | 0.784 |
| 214 | FENDRR |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | FAT4 | Sponge network | -1.281 | 0.00012 | -0.354 | 0.14694 | 0.784 |
| 215 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | ATRX | Sponge network | 0.782 | 0.00016 | 0.261 | 0.04408 | 0.784 |
| 216 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-505-3p | 18 | TSHZ3 | Sponge network | -0.583 | 0.14589 | -0.556 | 0.01516 | 0.784 |
| 217 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | -0.976 | 0.00025 | -0.466 | 0.14121 | 0.783 |
| 218 | HAND2-AS1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 13 | FHL1 | Sponge network | -1.697 | 0.00889 | -2.687 | 0 | 0.783 |
| 219 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | -0.576 | 0.10121 | -1.324 | 0.00014 | 0.783 |
| 220 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-877-5p | 25 | NRK | Sponge network | -0.576 | 0.10121 | 0.656 | 0.19217 | 0.782 |
| 221 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ABCC9 | Sponge network | -0.49 | 0.04946 | -0.466 | 0.14121 | 0.782 |
| 222 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 54 | AR | Sponge network | -0.976 | 0.00025 | -1.554 | 0 | 0.782 |
| 223 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 38 | ANK2 | Sponge network | -1.111 | 0.0003 | -1.603 | 1.0E-5 | 0.782 |
| 224 | MIR143HG |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 35 | SETBP1 | Sponge network | -0.739 | 0.0574 | -1.302 | 1.0E-5 | 0.782 |
| 225 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | ABCC9 | Sponge network | -2.452 | 0 | -0.466 | 0.14121 | 0.781 |
| 226 | USP3-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-92a-1-5p | 31 | AKAP6 | Sponge network | -0.162 | 0.47687 | -1.586 | 3.0E-5 | 0.781 |
| 227 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-664a-3p;hsa-miR-769-5p | 20 | SYNPO2 | Sponge network | -2.69 | 0.0001 | -2.342 | 0 | 0.78 |
| 228 | HAND2-AS1 |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 10 | DPP6 | Sponge network | -1.697 | 0.00889 | -3.777 | 0 | 0.78 |
| 229 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | PEG3 | Sponge network | -1.697 | 0.00889 | -1.74 | 0 | 0.78 |
| 230 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | PARK2 | Sponge network | -2.452 | 0 | -1.892 | 0 | 0.78 |
| 231 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 71 | RUNX1T1 | Sponge network | -0.49 | 0.04946 | -0.344 | 0.2374 | 0.78 |
| 232 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 10 | NRXN3 | Sponge network | -1.092 | 4.0E-5 | -0.893 | 0.04186 | 0.78 |
| 233 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-421 | 17 | RCAN2 | Sponge network | -0.162 | 0.47687 | -0.33 | 0.15172 | 0.779 |
| 234 | BVES-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-576-5p | 22 | SYNPO2 | Sponge network | -2.62 | 0 | -2.342 | 0 | 0.779 |
| 235 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 18 | MAPK10 | Sponge network | -0.49 | 0.04946 | -1.774 | 0 | 0.779 |
| 236 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 36 | SETBP1 | Sponge network | -0.129 | 0.57177 | -1.302 | 1.0E-5 | 0.779 |
| 237 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | SYNE1 | Sponge network | -0.576 | 0.10121 | -0.738 | 0.00316 | 0.779 |
| 238 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | SYNE1 | Sponge network | -2.452 | 0 | -0.738 | 0.00316 | 0.778 |
| 239 | RP11-166D19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 31 | NAV3 | Sponge network | -0.576 | 0.10121 | 0.212 | 0.47729 | 0.777 |
| 240 | SNHG14 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-935;hsa-miR-98-5p | 53 | DMD | Sponge network | -1.111 | 0.0003 | -1.506 | 0 | 0.777 |
| 241 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 63 | FAT3 | Sponge network | -0.49 | 0.04946 | -1.601 | 0.00051 | 0.777 |
| 242 | MIR497HG |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | SFRP1 | Sponge network | -1.439 | 4.0E-5 | -3.566 | 0 | 0.777 |
| 243 | NR2F1-AS1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | RNF180 | Sponge network | -0.49 | 0.04946 | -1.127 | 1.0E-5 | 0.777 |
| 244 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 16 | RECK | Sponge network | -1.092 | 4.0E-5 | -1.043 | 0 | 0.777 |
| 245 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p | 21 | FGFR1 | Sponge network | -0.576 | 0.10121 | -0.509 | 0.03957 | 0.777 |
| 246 | MYLK-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p | 32 | DTNA | Sponge network | -1.331 | 3.0E-5 | -1.648 | 1.0E-5 | 0.777 |
| 247 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 30 | BNC2 | Sponge network | -1.092 | 4.0E-5 | -0.491 | 0.09988 | 0.776 |
| 248 | LINC00702 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 35 | PCDH10 | Sponge network | -0.737 | 0.06182 | -1.644 | 0.0078 | 0.776 |
| 249 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 21 | ADAMTSL3 | Sponge network | -1.092 | 4.0E-5 | -1.559 | 0 | 0.776 |
| 250 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 16 | MN1 | Sponge network | -0.129 | 0.57177 | -0.358 | 0.24172 | 0.776 |
| 251 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 14 | SYNE1 | Sponge network | -1.092 | 4.0E-5 | -0.738 | 0.00316 | 0.776 |
| 252 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | TIMP3 | Sponge network | -0.576 | 0.10121 | -0.546 | 0.02307 | 0.776 |
| 253 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | PCDH9 | Sponge network | -0.576 | 0.10121 | -1.823 | 4.0E-5 | 0.776 |
| 254 | MIR143HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-98-5p | 51 | DMD | Sponge network | -0.739 | 0.0574 | -1.506 | 0 | 0.776 |
| 255 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 39 | AKAP6 | Sponge network | -1.697 | 0.00889 | -1.586 | 3.0E-5 | 0.775 |
| 256 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 32 | ADAMTSL3 | Sponge network | -0.129 | 0.57177 | -1.559 | 0 | 0.775 |
| 257 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 26 | PRUNE2 | Sponge network | -1.697 | 0.00889 | -1.733 | 0.00022 | 0.775 |
| 258 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 21 | MAPK10 | Sponge network | -1.111 | 0.0003 | -1.774 | 0 | 0.775 |
| 259 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | -0.576 | 0.10121 | -1.047 | 0.00364 | 0.775 |
| 260 | USP3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935 | 89 | LPP | Sponge network | -0.162 | 0.47687 | -0.459 | 0.04475 | 0.775 |
| 261 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | PRUNE2 | Sponge network | -0.737 | 0.06182 | -1.733 | 0.00022 | 0.775 |
| 262 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | RECK | Sponge network | -0.49 | 0.04946 | -1.043 | 0 | 0.774 |
| 263 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 51 | ZFHX4 | Sponge network | -0.576 | 0.10121 | 0.018 | 0.95574 | 0.774 |
| 264 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p | 14 | FGFR1 | Sponge network | -1.331 | 3.0E-5 | -0.509 | 0.03957 | 0.774 |
| 265 | MYLK-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-424-5p | 15 | PCDH9 | Sponge network | -1.331 | 3.0E-5 | -1.823 | 4.0E-5 | 0.773 |
| 266 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-452-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-590-3p | 29 | RBMS3 | Sponge network | -0.358 | 0.17255 | -0.519 | 0.03551 | 0.773 |
| 267 | LINC00702 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | PEG3 | Sponge network | -0.737 | 0.06182 | -1.74 | 0 | 0.773 |
| 268 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 32 | TIMP3 | Sponge network | -0.129 | 0.57177 | -0.546 | 0.02307 | 0.773 |
| 269 | USP3-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-511-5p | 18 | MRVI1 | Sponge network | -0.162 | 0.47687 | -1.051 | 0.00022 | 0.773 |
| 270 | LINC00702 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 12 | PRKD1 | Sponge network | -0.737 | 0.06182 | -0.466 | 0.03583 | 0.773 |
| 271 | MIR497HG |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-5p | 10 | LHFP | Sponge network | -1.439 | 4.0E-5 | -0.167 | 0.41371 | 0.773 |
| 272 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-628-5p | 42 | RUNX1T1 | Sponge network | -0.583 | 0.14589 | -0.344 | 0.2374 | 0.773 |
| 273 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 54 | RBMS3 | Sponge network | -0.737 | 0.06182 | -0.519 | 0.03551 | 0.772 |
| 274 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-769-5p | 36 | SYNPO2 | Sponge network | -0.129 | 0.57177 | -2.342 | 0 | 0.772 |
| 275 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-340-5p;hsa-miR-424-5p | 15 | DYNC1I1 | Sponge network | -1.331 | 3.0E-5 | -1.12 | 0.00107 | 0.772 |
| 276 | ADAMTS9-AS2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 21 | SYNM | Sponge network | -2.452 | 0 | -2.309 | 1.0E-5 | 0.772 |
| 277 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-7-1-3p | 27 | TSHZ3 | Sponge network | -1.092 | 4.0E-5 | -0.556 | 0.01516 | 0.772 |
| 278 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 39 | EPHA3 | Sponge network | -0.129 | 0.57177 | 0.185 | 0.53588 | 0.771 |
| 279 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 66 | QKI | Sponge network | -0.129 | 0.57177 | -0.333 | 0.04111 | 0.771 |
| 280 | RP11-166D19.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 31 | WWTR1 | Sponge network | -0.576 | 0.10121 | -0.345 | 0.11997 | 0.771 |
| 281 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 22 | RCAN2 | Sponge network | -0.739 | 0.0574 | -0.33 | 0.15172 | 0.771 |
| 282 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 20 | DCLK1 | Sponge network | -0.49 | 0.04946 | -1.324 | 0.00014 | 0.771 |
| 283 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92b-3p | 21 | AKAP12 | Sponge network | -0.739 | 0.0574 | -0.932 | 0.0028 | 0.771 |
| 284 | AC011526.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-96-5p | 10 | ERG | Sponge network | 0.342 | 0.03129 | 0.002 | 0.99011 | 0.771 |
| 285 | NR2F1-AS1 |
hsa-let-7f-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | ROR2 | Sponge network | -0.49 | 0.04946 | -1.091 | 0.0008 | 0.77 |
| 286 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-98-5p | 32 | DMD | Sponge network | -1.092 | 4.0E-5 | -1.506 | 0 | 0.77 |
| 287 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | SYNE1 | Sponge network | -0.737 | 0.06182 | -0.738 | 0.00316 | 0.77 |
| 288 | ZNF667-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 17 | SFRP1 | Sponge network | -0.976 | 0.00025 | -3.566 | 0 | 0.77 |
| 289 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b | 11 | SYNM | Sponge network | -0.583 | 0.14589 | -2.309 | 1.0E-5 | 0.77 |
| 290 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 44 | PGR | Sponge network | -1.111 | 0.0003 | -1.704 | 0 | 0.77 |
| 291 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | RECK | Sponge network | -1.439 | 4.0E-5 | -1.043 | 0 | 0.77 |
| 292 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 21 | FNBP1 | Sponge network | -1.111 | 0.0003 | -0.969 | 4.0E-5 | 0.769 |
| 293 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 37 | ANK2 | Sponge network | -0.49 | 0.04946 | -1.603 | 1.0E-5 | 0.769 |
| 294 | LINC00702 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 20 | GLI3 | Sponge network | -0.737 | 0.06182 | -0.615 | 0.01482 | 0.769 |
| 295 | SNHG14 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-574-3p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | FHL1 | Sponge network | -1.111 | 0.0003 | -2.687 | 0 | 0.769 |
| 296 | MAGI2-AS3 |
hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | ROR2 | Sponge network | -0.129 | 0.57177 | -1.091 | 0.0008 | 0.769 |
| 297 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | -0.576 | 0.10121 | 0.809 | 0.00544 | 0.769 |
| 298 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 22 | PRUNE2 | Sponge network | -0.576 | 0.10121 | -1.733 | 0.00022 | 0.769 |
| 299 | MIR143HG |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | RNF180 | Sponge network | -0.739 | 0.0574 | -1.127 | 1.0E-5 | 0.769 |
| 300 | MIR497HG |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 31 | SETBP1 | Sponge network | -1.439 | 4.0E-5 | -1.302 | 1.0E-5 | 0.769 |
| 301 | MYLK-AS1 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-940 | 11 | SFRP1 | Sponge network | -1.331 | 3.0E-5 | -3.566 | 0 | 0.768 |
| 302 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p | 30 | PGR | Sponge network | -0.976 | 0.00025 | -1.704 | 0 | 0.768 |
| 303 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 19 | MAPK10 | Sponge network | -0.129 | 0.57177 | -1.774 | 0 | 0.768 |
| 304 | MIR143HG |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-542-3p | 11 | PRKD1 | Sponge network | -0.739 | 0.0574 | -0.466 | 0.03583 | 0.767 |
| 305 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DCN | Sponge network | -0.576 | 0.10121 | -1.288 | 0 | 0.767 |
| 306 | FENDRR |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 12 | FHL1 | Sponge network | -1.281 | 0.00012 | -2.687 | 0 | 0.767 |
| 307 | LINC00702 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 72 | RUNX1T1 | Sponge network | -0.737 | 0.06182 | -0.344 | 0.2374 | 0.767 |
| 308 | ADAMTS9-AS2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-940 | 20 | SORCS1 | Sponge network | -2.452 | 0 | -3.492 | 0 | 0.766 |
| 309 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 22 | PTGFR | Sponge network | -0.129 | 0.57177 | -0.233 | 0.45421 | 0.766 |
| 310 | LINC00702 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 65 | DTNA | Sponge network | -0.737 | 0.06182 | -1.648 | 1.0E-5 | 0.766 |
| 311 | RP11-166D19.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 22 | SFRP1 | Sponge network | -0.576 | 0.10121 | -3.566 | 0 | 0.766 |
| 312 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | PEG3 | Sponge network | -0.49 | 0.04946 | -1.74 | 0 | 0.766 |
| 313 | ZNF667-AS1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | -0.976 | 0.00025 | -2.633 | 0 | 0.765 |
| 314 | LINC00702 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 66 | AR | Sponge network | -0.737 | 0.06182 | -1.554 | 0 | 0.765 |
| 315 | MRVI1-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 13 | PRUNE2 | Sponge network | -1.092 | 4.0E-5 | -1.733 | 0.00022 | 0.765 |
| 316 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 21 | SETBP1 | Sponge network | -0.976 | 0.00025 | -1.302 | 1.0E-5 | 0.764 |
| 317 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 53 | RUNX1T1 | Sponge network | 0.572 | 0.01722 | -0.344 | 0.2374 | 0.764 |
| 318 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 45 | RBMS3 | Sponge network | -1.439 | 4.0E-5 | -0.519 | 0.03551 | 0.764 |
| 319 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | ADAMTSL3 | Sponge network | -1.111 | 0.0003 | -1.559 | 0 | 0.764 |
| 320 | USP3-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p | 29 | SYNPO2 | Sponge network | -0.162 | 0.47687 | -2.342 | 0 | 0.764 |
| 321 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-590-3p | 23 | EPHA3 | Sponge network | -0.358 | 0.17255 | 0.185 | 0.53588 | 0.764 |
| 322 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | ADAMTSL3 | Sponge network | -0.49 | 0.04946 | -1.559 | 0 | 0.764 |
| 323 | PGM5-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421 | 13 | SYNPO2 | Sponge network | -4.546 | 0 | -2.342 | 0 | 0.764 |
| 324 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p | 20 | AKAP12 | Sponge network | -2.452 | 0 | -0.932 | 0.0028 | 0.764 |
| 325 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | DCLK1 | Sponge network | -0.129 | 0.57177 | -1.324 | 0.00014 | 0.764 |
| 326 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 40 | RASSF8 | Sponge network | -0.49 | 0.04946 | -0.122 | 0.65591 | 0.764 |
| 327 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | PDZRN3 | Sponge network | -0.576 | 0.10121 | -1.548 | 0 | 0.763 |
| 328 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 13 | HMCN1 | Sponge network | 0.997 | 0.00066 | 0.809 | 0.00544 | 0.763 |
| 329 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p | 27 | PGR | Sponge network | -1.439 | 4.0E-5 | -1.704 | 0 | 0.763 |
| 330 | MIR143HG |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 17 | MN1 | Sponge network | -0.739 | 0.0574 | -0.358 | 0.24172 | 0.763 |
| 331 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 20 | MAPK10 | Sponge network | -0.739 | 0.0574 | -1.774 | 0 | 0.762 |
| 332 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 23 | FGFR1 | Sponge network | -0.129 | 0.57177 | -0.509 | 0.03957 | 0.762 |
| 333 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 56 | GAS7 | Sponge network | -0.129 | 0.57177 | -0.555 | 0.01602 | 0.762 |
| 334 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p | 12 | GLI3 | Sponge network | -1.092 | 4.0E-5 | -0.615 | 0.01482 | 0.761 |
| 335 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935;hsa-miR-96-5p | 43 | FBXO32 | Sponge network | -0.576 | 0.10121 | -0.495 | 0.07561 | 0.761 |
| 336 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-664a-3p | 20 | SYNPO2 | Sponge network | -0.976 | 0.00025 | -2.342 | 0 | 0.761 |
| 337 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | ABCC9 | Sponge network | -1.111 | 0.0003 | -0.466 | 0.14121 | 0.761 |
| 338 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 25 | ADAMTSL3 | Sponge network | -0.976 | 0.00025 | -1.559 | 0 | 0.761 |
| 339 | NR2F1-AS1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p | 13 | FHL1 | Sponge network | -0.49 | 0.04946 | -2.687 | 0 | 0.761 |
| 340 | RP11-166D19.1 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | -0.576 | 0.10121 | -1.628 | 0 | 0.761 |
| 341 | SNHG14 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 24 | MRVI1 | Sponge network | -1.111 | 0.0003 | -1.051 | 0.00022 | 0.761 |
| 342 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 14 | DCN | Sponge network | -0.129 | 0.57177 | -1.288 | 0 | 0.76 |
| 343 | FENDRR |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 28 | SYNPO2 | Sponge network | -1.281 | 0.00012 | -2.342 | 0 | 0.76 |
| 344 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | NCAM2 | Sponge network | -0.576 | 0.10121 | -0.482 | 0.22565 | 0.76 |
| 345 | PWAR6 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | RNF180 | Sponge network | -1.575 | 6.0E-5 | -1.127 | 1.0E-5 | 0.76 |
| 346 | LINC00702 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 27 | NRXN3 | Sponge network | -0.737 | 0.06182 | -0.893 | 0.04186 | 0.76 |
| 347 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p | 13 | THSD7A | Sponge network | -0.129 | 0.57177 | 0.242 | 0.25154 | 0.76 |
| 348 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 35 | TSHZ3 | Sponge network | -0.976 | 0.00025 | -0.556 | 0.01516 | 0.759 |
| 349 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | TSHZ3 | Sponge network | -0.737 | 0.06182 | -0.556 | 0.01516 | 0.759 |
| 350 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 40 | DTNA | Sponge network | -1.092 | 4.0E-5 | -1.648 | 1.0E-5 | 0.758 |
| 351 | MIR497HG |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 11 | DCN | Sponge network | -1.439 | 4.0E-5 | -1.288 | 0 | 0.758 |
| 352 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | CNTNAP1 | Sponge network | -0.737 | 0.06182 | 0.358 | 0.13727 | 0.758 |
| 353 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-877-5p | 17 | CDH19 | Sponge network | -2.62 | 0 | -3.434 | 0 | 0.758 |
| 354 | SNHG14 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-769-5p | 36 | SYNPO2 | Sponge network | -1.111 | 0.0003 | -2.342 | 0 | 0.758 |
| 355 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | RECK | Sponge network | -0.739 | 0.0574 | -1.043 | 0 | 0.758 |
| 356 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | RASSF8 | Sponge network | -1.111 | 0.0003 | -0.122 | 0.65591 | 0.757 |
| 357 | BVES-AS1 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-628-5p | 46 | DTNA | Sponge network | -2.62 | 0 | -1.648 | 1.0E-5 | 0.757 |
| 358 | ADAMTS9-AS2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PRUNE2 | Sponge network | -2.452 | 0 | -1.733 | 0.00022 | 0.757 |
| 359 | MIR143HG |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | -0.739 | 0.0574 | -1.398 | 2.0E-5 | 0.757 |
| 360 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | TLL1 | Sponge network | -2.452 | 0 | -1.195 | 3.0E-5 | 0.757 |
| 361 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 12 | LHFP | Sponge network | -0.49 | 0.04946 | -0.167 | 0.41371 | 0.757 |
| 362 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-629-3p | 37 | AR | Sponge network | -0.583 | 0.14589 | -1.554 | 0 | 0.757 |
| 363 | NR2F1-AS1 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-874-3p;hsa-miR-98-5p | 10 | HSPB7 | Sponge network | -0.49 | 0.04946 | -2.268 | 1.0E-5 | 0.756 |
| 364 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 59 | SSBP2 | Sponge network | -2.452 | 0 | -1.58 | 0 | 0.756 |
| 365 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 10 | CTNNA3 | Sponge network | -0.576 | 0.10121 | -2.046 | 0.00019 | 0.756 |
| 366 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-651-5p | 14 | WWTR1 | Sponge network | -1.331 | 3.0E-5 | -0.345 | 0.11997 | 0.756 |
| 367 | RP11-166D19.1 |
hsa-miR-144-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | FMN2 | Sponge network | -0.576 | 0.10121 | -1.746 | 0.00047 | 0.755 |
| 368 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ABCC9 | Sponge network | 0.572 | 0.01722 | -0.466 | 0.14121 | 0.755 |
| 369 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 58 | DTNA | Sponge network | -0.576 | 0.10121 | -1.648 | 1.0E-5 | 0.755 |
| 370 | MYLK-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-766-3p | 15 | EZH1 | Sponge network | -1.331 | 3.0E-5 | -0.564 | 1.0E-5 | 0.755 |
| 371 | USP3-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-7-5p | 14 | FLNA | Sponge network | -0.162 | 0.47687 | -0.771 | 0.01377 | 0.755 |
| 372 | SNHG14 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 71 | AR | Sponge network | -1.111 | 0.0003 | -1.554 | 0 | 0.754 |
| 373 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p | 24 | RBMS3 | Sponge network | -0.583 | 0.14589 | -0.519 | 0.03551 | 0.754 |
| 374 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | -0.49 | 0.04946 | -1.398 | 2.0E-5 | 0.754 |
| 375 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-34a-5p;hsa-miR-664a-5p;hsa-miR-671-5p | 34 | ADARB1 | Sponge network | -1.331 | 3.0E-5 | -0.335 | 0.06631 | 0.754 |
| 376 | C20orf166-AS1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484 | 11 | FHL1 | Sponge network | -2.69 | 0.0001 | -2.687 | 0 | 0.754 |
| 377 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-628-5p | 40 | RUNX1T1 | Sponge network | -1.092 | 4.0E-5 | -0.344 | 0.2374 | 0.754 |
| 378 | MAGI2-AS3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 37 | WWTR1 | Sponge network | -0.129 | 0.57177 | -0.345 | 0.11997 | 0.753 |
| 379 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | ANK2 | Sponge network | -1.697 | 0.00889 | -1.603 | 1.0E-5 | 0.753 |
| 380 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-421 | 11 | NRXN3 | Sponge network | -0.583 | 0.14589 | -0.893 | 0.04186 | 0.753 |
| 381 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | PDZRN3 | Sponge network | -0.739 | 0.0574 | -1.548 | 0 | 0.753 |
| 382 | BVES-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p | 22 | SYNM | Sponge network | -2.62 | 0 | -2.309 | 1.0E-5 | 0.753 |
| 383 | MIR143HG |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | PCDH9 | Sponge network | -0.739 | 0.0574 | -1.823 | 4.0E-5 | 0.752 |
| 384 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-7-5p | 32 | ZBTB4 | Sponge network | -2.452 | 0 | -0.739 | 0 | 0.752 |
| 385 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ABCC9 | Sponge network | -1.439 | 4.0E-5 | -0.466 | 0.14121 | 0.752 |
| 386 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-532-3p | 21 | CADM3 | Sponge network | -2.452 | 0 | -3.663 | 0 | 0.751 |
| 387 | MIR143HG |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ITPKB | Sponge network | -0.739 | 0.0574 | -0.704 | 0.00106 | 0.751 |
| 388 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 15 | MN1 | Sponge network | -0.49 | 0.04946 | -0.358 | 0.24172 | 0.751 |
| 389 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-450b-5p | 15 | DLC1 | Sponge network | -0.583 | 0.14589 | -0.382 | 0.05038 | 0.751 |
| 390 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-486-5p | 14 | NRXN3 | Sponge network | -0.162 | 0.47687 | -0.893 | 0.04186 | 0.75 |
| 391 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | NRXN3 | Sponge network | -0.49 | 0.04946 | -0.893 | 0.04186 | 0.75 |
| 392 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 28 | NRXN3 | Sponge network | -0.129 | 0.57177 | -0.893 | 0.04186 | 0.75 |
| 393 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-421 | 16 | PGR | Sponge network | -0.583 | 0.14589 | -1.704 | 0 | 0.75 |
| 394 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | -0.129 | 0.57177 | -1.047 | 0.00364 | 0.749 |
| 395 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | ABCC9 | Sponge network | -0.358 | 0.17255 | -0.466 | 0.14121 | 0.749 |
| 396 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p | 19 | SYNM | Sponge network | -2.69 | 0.0001 | -2.309 | 1.0E-5 | 0.749 |
| 397 | MAGI2-AS3 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 13 | FHL1 | Sponge network | -0.129 | 0.57177 | -2.687 | 0 | 0.749 |
| 398 | DNM3OS |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 19 | GLI3 | Sponge network | 0.572 | 0.01722 | -0.615 | 0.01482 | 0.748 |
| 399 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | ADAMTSL3 | Sponge network | -0.739 | 0.0574 | -1.559 | 0 | 0.748 |
| 400 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | AFF3 | Sponge network | -2.452 | 0 | -2.373 | 0 | 0.748 |
| 401 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH11 | Sponge network | 0.572 | 0.01722 | 1.427 | 0 | 0.748 |
| 402 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 33 | ANK2 | Sponge network | -0.737 | 0.06182 | -1.603 | 1.0E-5 | 0.748 |
| 403 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 49 | RBMS3 | Sponge network | -1.575 | 6.0E-5 | -0.519 | 0.03551 | 0.748 |
| 404 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 21 | GLI3 | Sponge network | -0.49 | 0.04946 | -0.615 | 0.01482 | 0.748 |
| 405 | SNHG14 |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | -1.111 | 0.0003 | -1.398 | 2.0E-5 | 0.748 |
| 406 | NR2F1-AS1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -0.49 | 0.04946 | -2.633 | 0 | 0.748 |
| 407 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | PDZRN3 | Sponge network | -2.452 | 0 | -1.548 | 0 | 0.747 |
| 408 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | FLNA | Sponge network | -0.49 | 0.04946 | -0.771 | 0.01377 | 0.747 |
| 409 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-508-3p;hsa-miR-590-3p | 17 | SYNE1 | Sponge network | -0.358 | 0.17255 | -0.738 | 0.00316 | 0.747 |
| 410 | PGM5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 14 | CDH19 | Sponge network | -4.546 | 0 | -3.434 | 0 | 0.747 |
| 411 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 39 | RASSF8 | Sponge network | -0.739 | 0.0574 | -0.122 | 0.65591 | 0.747 |
| 412 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | PRRX1 | Sponge network | 0.572 | 0.01722 | 1.316 | 0 | 0.747 |
| 413 | PWAR6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | PEG3 | Sponge network | -1.575 | 6.0E-5 | -1.74 | 0 | 0.747 |
| 414 | SNHG14 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | SPG20 | Sponge network | -1.111 | 0.0003 | -1.16 | 0 | 0.746 |
| 415 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | SETBP1 | Sponge network | -0.49 | 0.04946 | -1.302 | 1.0E-5 | 0.746 |
| 416 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 54 | ZFHX4 | Sponge network | -0.49 | 0.04946 | 0.018 | 0.95574 | 0.746 |
| 417 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | TSHZ3 | Sponge network | -0.49 | 0.04946 | -0.556 | 0.01516 | 0.746 |
| 418 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 41 | NRXN1 | Sponge network | -2.452 | 0 | -3.778 | 0 | 0.746 |
| 419 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 16 | GLI3 | Sponge network | -0.976 | 0.00025 | -0.615 | 0.01482 | 0.746 |
| 420 | MAGI2-AS3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 31 | NRK | Sponge network | -0.129 | 0.57177 | 0.656 | 0.19217 | 0.745 |
| 421 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 32 | ANK2 | Sponge network | -0.976 | 0.00025 | -1.603 | 1.0E-5 | 0.745 |
| 422 | LINC00702 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | RNF180 | Sponge network | -0.737 | 0.06182 | -1.127 | 1.0E-5 | 0.744 |
| 423 | MRVI1-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 13 | SFRP1 | Sponge network | -1.092 | 4.0E-5 | -3.566 | 0 | 0.744 |
| 424 | MIR143HG |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 37 | WWTR1 | Sponge network | -0.739 | 0.0574 | -0.345 | 0.11997 | 0.743 |
| 425 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | SPG20 | Sponge network | -0.576 | 0.10121 | -1.16 | 0 | 0.743 |
| 426 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 24 | SPG20 | Sponge network | -0.129 | 0.57177 | -1.16 | 0 | 0.743 |
| 427 | MIR497HG |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 20 | CADM3 | Sponge network | -1.439 | 4.0E-5 | -3.663 | 0 | 0.743 |
| 428 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 66 | FAT3 | Sponge network | -0.129 | 0.57177 | -1.601 | 0.00051 | 0.743 |
| 429 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 61 | DTNA | Sponge network | -2.452 | 0 | -1.648 | 1.0E-5 | 0.743 |
| 430 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 13 | RECK | Sponge network | -0.583 | 0.14589 | -1.043 | 0 | 0.743 |
| 431 | SNHG14 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-590-5p | 10 | MYH11 | Sponge network | -1.111 | 0.0003 | -2.895 | 0 | 0.742 |
| 432 | HAND2-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | SFRP1 | Sponge network | -1.697 | 0.00889 | -3.566 | 0 | 0.742 |
| 433 | BVES-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p | 20 | MRVI1 | Sponge network | -2.62 | 0 | -1.051 | 0.00022 | 0.742 |
| 434 | MIR143HG |
hsa-let-7f-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | ROR2 | Sponge network | -0.739 | 0.0574 | -1.091 | 0.0008 | 0.742 |
| 435 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-484;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | NRXN3 | Sponge network | -0.976 | 0.00025 | -0.893 | 0.04186 | 0.742 |
| 436 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-508-3p;hsa-miR-628-5p;hsa-miR-935 | 41 | FBXO32 | Sponge network | -0.162 | 0.47687 | -0.495 | 0.07561 | 0.742 |
| 437 | USP3-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-93-5p | 18 | PRUNE2 | Sponge network | -0.162 | 0.47687 | -1.733 | 0.00022 | 0.742 |
| 438 | MIR143HG |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 67 | DTNA | Sponge network | -0.739 | 0.0574 | -1.648 | 1.0E-5 | 0.742 |
| 439 | MIR143HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 58 | EPHA7 | Sponge network | -0.739 | 0.0574 | -2.197 | 3.0E-5 | 0.741 |
| 440 | SNHG14 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 70 | DTNA | Sponge network | -1.111 | 0.0003 | -1.648 | 1.0E-5 | 0.741 |
| 441 | MIR143HG |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 22 | MACF1 | Sponge network | -0.739 | 0.0574 | 0.019 | 0.90335 | 0.741 |
| 442 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | SYNE1 | Sponge network | -0.976 | 0.00025 | -0.738 | 0.00316 | 0.741 |
| 443 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 30 | SYNPO2 | Sponge network | -0.49 | 0.04946 | -2.342 | 0 | 0.74 |
| 444 | HAND2-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 31 | NRK | Sponge network | -1.697 | 0.00889 | 0.656 | 0.19217 | 0.74 |
| 445 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p | 10 | SPG20 | Sponge network | -1.331 | 3.0E-5 | -1.16 | 0 | 0.74 |
| 446 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | RECK | Sponge network | -1.111 | 0.0003 | -1.043 | 0 | 0.74 |
| 447 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-98-5p | 13 | DCN | Sponge network | -0.976 | 0.00025 | -1.288 | 0 | 0.74 |
| 448 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 24 | GREM1 | Sponge network | -0.129 | 0.57177 | 0.902 | 0.01691 | 0.74 |
| 449 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-744-5p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | -0.976 | 0.00025 | -0.509 | 0.03957 | 0.74 |
| 450 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-628-5p;hsa-miR-93-3p | 54 | RUNX1T1 | Sponge network | -0.976 | 0.00025 | -0.344 | 0.2374 | 0.74 |
| 451 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 26 | NRXN1 | Sponge network | -4.546 | 0 | -3.778 | 0 | 0.74 |
| 452 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 11 | CNTNAP1 | Sponge network | -1.092 | 4.0E-5 | 0.358 | 0.13727 | 0.739 |
| 453 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 54 | RBMS3 | Sponge network | -1.697 | 0.00889 | -0.519 | 0.03551 | 0.739 |
| 454 | MAGI2-AS3 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -0.129 | 0.57177 | -2.633 | 0 | 0.739 |
| 455 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | PLSCR4 | Sponge network | -0.739 | 0.0574 | -0.474 | 0.03833 | 0.739 |
| 456 | SNHG14 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 31 | NRXN3 | Sponge network | -1.111 | 0.0003 | -0.893 | 0.04186 | 0.739 |
| 457 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-98-5p | 35 | THBS1 | Sponge network | -0.129 | 0.57177 | 0.741 | 0.00386 | 0.739 |
| 458 | LINC00702 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | PCDH9 | Sponge network | -0.737 | 0.06182 | -1.823 | 4.0E-5 | 0.739 |
| 459 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | PCDH9 | Sponge network | -2.452 | 0 | -1.823 | 4.0E-5 | 0.739 |
| 460 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 18 | SYNE1 | Sponge network | -0.583 | 0.14589 | -0.738 | 0.00316 | 0.739 |
| 461 | MYLK-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27b-5p | 11 | CNTN1 | Sponge network | -1.331 | 3.0E-5 | -2.594 | 0 | 0.739 |
| 462 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p | 21 | ANK2 | Sponge network | -0.583 | 0.14589 | -1.603 | 1.0E-5 | 0.738 |
| 463 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-590-3p | 16 | SETBP1 | Sponge network | -0.358 | 0.17255 | -1.302 | 1.0E-5 | 0.738 |
| 464 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 52 | ZFHX4 | Sponge network | -0.129 | 0.57177 | 0.018 | 0.95574 | 0.738 |
| 465 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p | 10 | DNAJB4 | Sponge network | -1.331 | 3.0E-5 | -0.7 | 1.0E-5 | 0.738 |
| 466 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-628-5p;hsa-miR-7-1-3p | 51 | DTNA | Sponge network | -0.976 | 0.00025 | -1.648 | 1.0E-5 | 0.738 |
| 467 | LINC00702 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-598-3p;hsa-miR-664a-3p | 15 | MN1 | Sponge network | -0.737 | 0.06182 | -0.358 | 0.24172 | 0.738 |
| 468 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MDM4 | Sponge network | 0.931 | 1.0E-5 | 0.345 | 0.00762 | 0.738 |
| 469 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-508-3p;hsa-miR-590-3p | 16 | RECK | Sponge network | -0.358 | 0.17255 | -1.043 | 0 | 0.738 |
| 470 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p | 20 | FNBP1 | Sponge network | -0.739 | 0.0574 | -0.969 | 4.0E-5 | 0.738 |
| 471 | PWAR6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 45 | DMD | Sponge network | -1.575 | 6.0E-5 | -1.506 | 0 | 0.737 |
| 472 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p | 18 | PLSCR4 | Sponge network | -1.331 | 3.0E-5 | -0.474 | 0.03833 | 0.737 |
| 473 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 35 | ZBTB4 | Sponge network | -0.739 | 0.0574 | -0.739 | 0 | 0.737 |
| 474 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-935 | 19 | DIP2C | Sponge network | -1.331 | 3.0E-5 | -0.436 | 0.01713 | 0.737 |
| 475 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-424-5p | 16 | RASSF8 | Sponge network | -1.331 | 3.0E-5 | -0.122 | 0.65591 | 0.737 |
| 476 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | -0.737 | 0.06182 | -1.324 | 0.00014 | 0.737 |
| 477 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-532-5p;hsa-miR-582-5p | 11 | PRICKLE1 | Sponge network | -0.129 | 0.57177 | -0.112 | 0.67088 | 0.736 |
| 478 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-877-5p | 24 | PGR | Sponge network | -2.62 | 0 | -1.704 | 0 | 0.736 |
| 479 | MAGI2-AS3 |
hsa-miR-182-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-769-5p | 11 | TCEAL7 | Sponge network | -0.129 | 0.57177 | -1.445 | 0 | 0.736 |
| 480 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 19 | MCC | Sponge network | -1.092 | 4.0E-5 | -1.005 | 0 | 0.736 |
| 481 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 31 | EPHA3 | Sponge network | 0.572 | 0.01722 | 0.185 | 0.53588 | 0.736 |
| 482 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 54 | BNC2 | Sponge network | -2.452 | 0 | -0.491 | 0.09988 | 0.736 |
| 483 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 23 | FGFR1 | Sponge network | -0.49 | 0.04946 | -0.509 | 0.03957 | 0.736 |
| 484 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 41 | TSHZ3 | Sponge network | -2.452 | 0 | -0.556 | 0.01516 | 0.736 |
| 485 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-3p | 17 | ABCC9 | Sponge network | -0.162 | 0.47687 | -0.466 | 0.14121 | 0.736 |
| 486 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-340-5p | 17 | NRXN1 | Sponge network | -1.331 | 3.0E-5 | -3.778 | 0 | 0.736 |
| 487 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 23 | CDH19 | Sponge network | -2.452 | 0 | -3.434 | 0 | 0.736 |
| 488 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 73 | NTRK3 | Sponge network | -0.576 | 0.10121 | -1.77 | 3.0E-5 | 0.736 |
| 489 | DNM3OS |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 20 | PTGFR | Sponge network | 0.572 | 0.01722 | -0.233 | 0.45421 | 0.735 |
| 490 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 16 | ADAMTSL3 | Sponge network | -1.886 | 0 | -1.559 | 0 | 0.735 |
| 491 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 62 | BNC2 | Sponge network | -1.111 | 0.0003 | -0.491 | 0.09988 | 0.735 |
| 492 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | FLNA | Sponge network | -2.452 | 0 | -0.771 | 0.01377 | 0.735 |
| 493 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 58 | ADARB1 | Sponge network | -0.739 | 0.0574 | -0.335 | 0.06631 | 0.735 |
| 494 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 43 | MCC | Sponge network | -2.452 | 0 | -1.005 | 0 | 0.735 |
| 495 | MIR143HG |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 21 | EZH1 | Sponge network | -0.739 | 0.0574 | -0.564 | 1.0E-5 | 0.735 |
| 496 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 11 | AFF3 | Sponge network | -1.886 | 0 | -2.373 | 0 | 0.734 |
| 497 | MIR143HG |
hsa-miR-126-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p | 10 | TNC | Sponge network | -0.739 | 0.0574 | 0.313 | 0.33705 | 0.734 |
| 498 | BVES-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-98-5p | 27 | AKAP6 | Sponge network | -2.62 | 0 | -1.586 | 3.0E-5 | 0.734 |
| 499 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CNTN1 | Sponge network | -0.976 | 0.00025 | -2.594 | 0 | 0.734 |
| 500 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-98-5p | 44 | PRKAA2 | Sponge network | -2.62 | 0 | -2.462 | 0 | 0.733 |
| 501 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | FAT4 | Sponge network | -0.129 | 0.57177 | -0.354 | 0.14694 | 0.733 |
| 502 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | CNTNAP1 | Sponge network | -0.49 | 0.04946 | 0.358 | 0.13727 | 0.733 |
| 503 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 73 | CADM2 | Sponge network | -2.452 | 0 | -3.782 | 0 | 0.733 |
| 504 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 22 | SYNE1 | Sponge network | -0.896 | 0.00099 | -0.738 | 0.00316 | 0.733 |
| 505 | RP11-736K20.5 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-624-5p | 26 | ANK2 | Sponge network | -0.358 | 0.17255 | -1.603 | 1.0E-5 | 0.733 |
| 506 | C20orf166-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p | 19 | MRVI1 | Sponge network | -2.69 | 0.0001 | -1.051 | 0.00022 | 0.733 |
| 507 | LINC00702 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -0.737 | 0.06182 | -2.046 | 0.00019 | 0.732 |
| 508 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | SYNE1 | Sponge network | -0.49 | 0.04946 | -0.738 | 0.00316 | 0.732 |
| 509 | MIR143HG |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 32 | DLC1 | Sponge network | -0.739 | 0.0574 | -0.382 | 0.05038 | 0.732 |
| 510 | SNHG14 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 25 | SORCS1 | Sponge network | -1.111 | 0.0003 | -3.492 | 0 | 0.732 |
| 511 | LINC00702 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 32 | SETBP1 | Sponge network | -0.737 | 0.06182 | -1.302 | 1.0E-5 | 0.732 |
| 512 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-935 | 39 | PCDH10 | Sponge network | -0.129 | 0.57177 | -1.644 | 0.0078 | 0.732 |
| 513 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 45 | MCC | Sponge network | -0.129 | 0.57177 | -1.005 | 0 | 0.732 |
| 514 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | ANK2 | Sponge network | -1.575 | 6.0E-5 | -1.603 | 1.0E-5 | 0.731 |
| 515 | LINC00578 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 21 | BNC2 | Sponge network | 0.811 | 0.03228 | -0.491 | 0.09988 | 0.731 |
| 516 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-628-5p | 28 | FBXO32 | Sponge network | -1.092 | 4.0E-5 | -0.495 | 0.07561 | 0.731 |
| 517 | FENDRR |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 16 | MAPK10 | Sponge network | -1.281 | 0.00012 | -1.774 | 0 | 0.731 |
| 518 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 20 | FNBP1 | Sponge network | -2.452 | 0 | -0.969 | 4.0E-5 | 0.731 |
| 519 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p | 14 | FLNA | Sponge network | -0.976 | 0.00025 | -0.771 | 0.01377 | 0.731 |
| 520 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p | 11 | DCLK1 | Sponge network | -1.331 | 3.0E-5 | -1.324 | 0.00014 | 0.731 |
| 521 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p | 19 | SPG20 | Sponge network | -0.976 | 0.00025 | -1.16 | 0 | 0.731 |
| 522 | USP3-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-935 | 40 | DMD | Sponge network | -0.162 | 0.47687 | -1.506 | 0 | 0.731 |
| 523 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 42 | RBMS3 | Sponge network | 0.572 | 0.01722 | -0.519 | 0.03551 | 0.731 |
| 524 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | CNTN1 | Sponge network | -1.697 | 0.00889 | -2.594 | 0 | 0.731 |
| 525 | PGM5-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-421;hsa-miR-940 | 12 | SORCS1 | Sponge network | -4.546 | 0 | -3.492 | 0 | 0.73 |
| 526 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -0.129 | 0.57177 | -2.046 | 0.00019 | 0.73 |
| 527 | RP11-166D19.1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | -0.576 | 0.10121 | 0.154 | 0.74723 | 0.73 |
| 528 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-940;hsa-miR-96-5p | 55 | GAS7 | Sponge network | -0.576 | 0.10121 | -0.555 | 0.01602 | 0.73 |
| 529 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 23 | GREM1 | Sponge network | -0.737 | 0.06182 | 0.902 | 0.01691 | 0.73 |
| 530 | MIR497HG |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | CXCL12 | Sponge network | -1.439 | 4.0E-5 | -1.421 | 0 | 0.73 |
| 531 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | SPG20 | Sponge network | -2.452 | 0 | -1.16 | 0 | 0.73 |
| 532 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p | 36 | RUNX1T1 | Sponge network | -0.358 | 0.17255 | -0.344 | 0.2374 | 0.73 |
| 533 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ABCC9 | Sponge network | 0.194 | 0.64278 | -0.466 | 0.14121 | 0.73 |
| 534 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | ANK2 | Sponge network | -1.281 | 0.00012 | -1.603 | 1.0E-5 | 0.73 |
| 535 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 43 | MCC | Sponge network | -0.576 | 0.10121 | -1.005 | 0 | 0.729 |
| 536 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p | 24 | ADAMTSL3 | Sponge network | -0.896 | 0.00099 | -1.559 | 0 | 0.729 |
| 537 | MALAT1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 63 | MDM4 | Sponge network | 0.782 | 0.00016 | 0.345 | 0.00762 | 0.729 |
| 538 | MAGI2-AS3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 23 | SYNM | Sponge network | -0.129 | 0.57177 | -2.309 | 1.0E-5 | 0.729 |
| 539 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 56 | QKI | Sponge network | -0.576 | 0.10121 | -0.333 | 0.04111 | 0.729 |
| 540 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 59 | EPHA7 | Sponge network | -1.697 | 0.00889 | -2.197 | 3.0E-5 | 0.729 |
| 541 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-628-5p | 25 | FBXO32 | Sponge network | -0.583 | 0.14589 | -0.495 | 0.07561 | 0.729 |
| 542 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-450b-5p | 11 | AMPH | Sponge network | -0.583 | 0.14589 | -0.916 | 5.0E-5 | 0.728 |
| 543 | RP11-867G23.10 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p | 12 | MRVI1 | Sponge network | -2.531 | 0 | -1.051 | 0.00022 | 0.728 |
| 544 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 21 | AKAP6 | Sponge network | -0.583 | 0.14589 | -1.586 | 3.0E-5 | 0.728 |
| 545 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-664a-3p | 11 | MN1 | Sponge network | -0.976 | 0.00025 | -0.358 | 0.24172 | 0.728 |
| 546 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 71 | RUNX1T1 | Sponge network | -2.452 | 0 | -0.344 | 0.2374 | 0.728 |
| 547 | LINC00702 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 21 | SFRP1 | Sponge network | -0.737 | 0.06182 | -3.566 | 0 | 0.728 |
| 548 | BVES-AS1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p | 13 | PRUNE2 | Sponge network | -2.62 | 0 | -1.733 | 0.00022 | 0.728 |
| 549 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | RASSF8 | Sponge network | -0.976 | 0.00025 | -0.122 | 0.65591 | 0.728 |
| 550 | ADAMTS9-AS2 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ADH1B | Sponge network | -2.452 | 0 | -3.716 | 0 | 0.728 |
| 551 | HAND2-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 20 | GLI3 | Sponge network | -1.697 | 0.00889 | -0.615 | 0.01482 | 0.727 |
| 552 | SNHG14 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | ZDBF2 | Sponge network | -1.111 | 0.0003 | -0.998 | 0.00025 | 0.727 |
| 553 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | SYNE1 | Sponge network | -1.697 | 0.00889 | -0.738 | 0.00316 | 0.727 |
| 554 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 39 | CNTN1 | Sponge network | -0.129 | 0.57177 | -2.594 | 0 | 0.727 |
| 555 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | -0.739 | 0.0574 | -1.324 | 0.00014 | 0.727 |
| 556 | RP11-166D19.1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 39 | IGFBP5 | Sponge network | -0.576 | 0.10121 | -0.806 | 0.00105 | 0.727 |
| 557 | SNHG14 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 24 | SYNM | Sponge network | -1.111 | 0.0003 | -2.309 | 1.0E-5 | 0.726 |
| 558 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 78 | NTRK3 | Sponge network | -2.452 | 0 | -1.77 | 3.0E-5 | 0.726 |
| 559 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | DCLK1 | Sponge network | -2.452 | 0 | -1.324 | 0.00014 | 0.726 |
| 560 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | ADAMTSL3 | Sponge network | -1.697 | 0.00889 | -1.559 | 0 | 0.726 |
| 561 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-424-5p | 21 | TACC1 | Sponge network | -1.331 | 3.0E-5 | -0.362 | 0.05406 | 0.726 |
| 562 | AC003090.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | SYNPO2 | Sponge network | -2.117 | 0.00161 | -2.342 | 0 | 0.726 |
| 563 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-429;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 30 | EBF3 | Sponge network | -0.129 | 0.57177 | -0.058 | 0.81437 | 0.725 |
| 564 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | PCDH9 | Sponge network | -0.49 | 0.04946 | -1.823 | 4.0E-5 | 0.725 |
| 565 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-539-5p | 33 | FAT3 | Sponge network | -1.092 | 4.0E-5 | -1.601 | 0.00051 | 0.725 |
| 566 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 52 | QKI | Sponge network | -0.976 | 0.00025 | -0.333 | 0.04111 | 0.725 |
| 567 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | SYNE1 | Sponge network | -1.439 | 4.0E-5 | -0.738 | 0.00316 | 0.725 |
| 568 | MIR497HG |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-671-5p | 29 | DLC1 | Sponge network | -1.439 | 4.0E-5 | -0.382 | 0.05038 | 0.725 |
| 569 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 31 | ADAMTSL3 | Sponge network | -1.281 | 0.00012 | -1.559 | 0 | 0.724 |
| 570 | PWAR6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 32 | SETBP1 | Sponge network | -1.575 | 6.0E-5 | -1.302 | 1.0E-5 | 0.724 |
| 571 | NR2F1-AS1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-532-5p;hsa-miR-582-5p | 10 | PRICKLE1 | Sponge network | -0.49 | 0.04946 | -0.112 | 0.67088 | 0.724 |
| 572 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 36 | EPHA3 | Sponge network | -0.739 | 0.0574 | 0.185 | 0.53588 | 0.724 |
| 573 | HAND2-AS1 |
hsa-let-7f-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 11 | ROR2 | Sponge network | -1.697 | 0.00889 | -1.091 | 0.0008 | 0.724 |
| 574 | DNM3OS |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 13 | HMCN1 | Sponge network | 0.572 | 0.01722 | 0.809 | 0.00544 | 0.724 |
| 575 | RP11-166D19.1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 14 | NGFR | Sponge network | -0.576 | 0.10121 | -1.271 | 0.01058 | 0.724 |
| 576 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p | 15 | FAT4 | Sponge network | -0.583 | 0.14589 | -0.354 | 0.14694 | 0.724 |
| 577 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 18 | MAPK10 | Sponge network | -1.697 | 0.00889 | -1.774 | 0 | 0.724 |
| 578 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | RECK | Sponge network | -0.737 | 0.06182 | -1.043 | 0 | 0.724 |
| 579 | MIR497HG |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 49 | AR | Sponge network | -1.439 | 4.0E-5 | -1.554 | 0 | 0.724 |
| 580 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 41 | CNTN1 | Sponge network | -1.111 | 0.0003 | -2.594 | 0 | 0.724 |
| 581 | ACTA2-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-362-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | 0.194 | 0.64278 | -1.051 | 0.00022 | 0.723 |
| 582 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | BNC2 | Sponge network | 0.249 | 0.35902 | -0.491 | 0.09988 | 0.723 |
| 583 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-589-3p;hsa-miR-769-5p | 11 | SYNPO2 | Sponge network | -2.531 | 0 | -2.342 | 0 | 0.723 |
| 584 | MRVI1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 56 | LPP | Sponge network | -1.092 | 4.0E-5 | -0.459 | 0.04475 | 0.723 |
| 585 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 40 | FBXO32 | Sponge network | -0.737 | 0.06182 | -0.495 | 0.07561 | 0.723 |
| 586 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-590-3p | 11 | MAPK10 | Sponge network | -0.358 | 0.17255 | -1.774 | 0 | 0.722 |
| 587 | BVES-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-935;hsa-miR-98-5p | 39 | DMD | Sponge network | -2.62 | 0 | -1.506 | 0 | 0.722 |
| 588 | RP11-736K20.5 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-590-3p | 21 | DLC1 | Sponge network | -0.358 | 0.17255 | -0.382 | 0.05038 | 0.722 |
| 589 | MIR497HG |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-625-5p | 12 | NGFR | Sponge network | -1.439 | 4.0E-5 | -1.271 | 0.01058 | 0.722 |
| 590 | DNM3OS |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | NAV3 | Sponge network | 0.572 | 0.01722 | 0.212 | 0.47729 | 0.722 |
| 591 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p | 13 | DCLK1 | Sponge network | -1.092 | 4.0E-5 | -1.324 | 0.00014 | 0.722 |
| 592 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-590-5p;hsa-miR-664a-3p | 13 | AKAP12 | Sponge network | -0.976 | 0.00025 | -0.932 | 0.0028 | 0.722 |
| 593 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 61 | DTNA | Sponge network | -2.46 | 0 | -1.648 | 1.0E-5 | 0.721 |
| 594 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | RASSF8 | Sponge network | 0.572 | 0.01722 | -0.122 | 0.65591 | 0.721 |
| 595 | LINC00092 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-671-5p | 14 | CADM3 | Sponge network | -1.886 | 0 | -3.663 | 0 | 0.721 |
| 596 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p | 31 | BNC2 | Sponge network | -0.358 | 0.17255 | -0.491 | 0.09988 | 0.721 |
| 597 | LINC00702 |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | -0.737 | 0.06182 | -1.398 | 2.0E-5 | 0.721 |
| 598 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | PDZRN3 | Sponge network | -0.129 | 0.57177 | -1.548 | 0 | 0.721 |
| 599 | MIR143HG |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 10 | DPP6 | Sponge network | -0.739 | 0.0574 | -3.777 | 0 | 0.721 |
| 600 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 55 | MEF2C | Sponge network | -1.111 | 0.0003 | -0.499 | 0.01448 | 0.721 |
| 601 | ZNF667-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-493-5p | 18 | SYNM | Sponge network | -0.976 | 0.00025 | -2.309 | 1.0E-5 | 0.721 |
| 602 | MIR497HG |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | PEG3 | Sponge network | -1.439 | 4.0E-5 | -1.74 | 0 | 0.721 |
| 603 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | PTGFR | Sponge network | -0.576 | 0.10121 | -0.233 | 0.45421 | 0.721 |
| 604 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | SYNE1 | Sponge network | -1.575 | 6.0E-5 | -0.738 | 0.00316 | 0.721 |
| 605 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 37 | PCDH9 | Sponge network | -0.129 | 0.57177 | -1.823 | 4.0E-5 | 0.72 |
| 606 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92b-3p | 20 | AKAP12 | Sponge network | -0.737 | 0.06182 | -0.932 | 0.0028 | 0.72 |
| 607 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-98-5p | 11 | ADAMTS8 | Sponge network | -2.452 | 0 | -1.476 | 0.00069 | 0.72 |
| 608 | SNHG14 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 76 | RUNX1T1 | Sponge network | -1.111 | 0.0003 | -0.344 | 0.2374 | 0.72 |
| 609 | LINC00702 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 107 | LPP | Sponge network | -0.737 | 0.06182 | -0.459 | 0.04475 | 0.72 |
| 610 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 11 | FAT4 | Sponge network | -1.092 | 4.0E-5 | -0.354 | 0.14694 | 0.72 |
| 611 | HAND2-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | PCDH9 | Sponge network | -1.697 | 0.00889 | -1.823 | 4.0E-5 | 0.72 |
| 612 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | PGR | Sponge network | -0.358 | 0.17255 | -1.704 | 0 | 0.72 |
| 613 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 35 | AKAP6 | Sponge network | -0.129 | 0.57177 | -1.586 | 3.0E-5 | 0.72 |
| 614 | SNHG14 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 38 | AKAP6 | Sponge network | -1.111 | 0.0003 | -1.586 | 3.0E-5 | 0.72 |
| 615 | MIR143HG |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | SFRP1 | Sponge network | -0.739 | 0.0574 | -3.566 | 0 | 0.719 |
| 616 | NR2F1-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 34 | WWTR1 | Sponge network | -0.49 | 0.04946 | -0.345 | 0.11997 | 0.719 |
| 617 | SNHG14 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 42 | PCDH9 | Sponge network | -1.111 | 0.0003 | -1.823 | 4.0E-5 | 0.719 |
| 618 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | LRFN5 | Sponge network | -0.576 | 0.10121 | -1.483 | 0 | 0.719 |
| 619 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 69 | FAT3 | Sponge network | -1.111 | 0.0003 | -1.601 | 0.00051 | 0.719 |
| 620 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | CNTN1 | Sponge network | -0.49 | 0.04946 | -2.594 | 0 | 0.719 |
| 621 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | DNAJB4 | Sponge network | -0.576 | 0.10121 | -0.7 | 1.0E-5 | 0.718 |
| 622 | FENDRR |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-98-5p | 45 | DMD | Sponge network | -1.281 | 0.00012 | -1.506 | 0 | 0.718 |
| 623 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-877-5p | 22 | ANK2 | Sponge network | -2.62 | 0 | -1.603 | 1.0E-5 | 0.718 |
| 624 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | -0.49 | 0.04946 | -1.628 | 0 | 0.718 |
| 625 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 61 | FAT3 | Sponge network | -2.452 | 0 | -1.601 | 0.00051 | 0.718 |
| 626 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-671-5p | 16 | FAT4 | Sponge network | -1.331 | 3.0E-5 | -0.354 | 0.14694 | 0.718 |
| 627 | LINC00702 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 45 | DMD | Sponge network | -0.737 | 0.06182 | -1.506 | 0 | 0.718 |
| 628 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 41 | PGR | Sponge network | -1.575 | 6.0E-5 | -1.704 | 0 | 0.717 |
| 629 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 47 | MEF2C | Sponge network | -0.129 | 0.57177 | -0.499 | 0.01448 | 0.717 |
| 630 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p | 27 | DMD | Sponge network | -0.583 | 0.14589 | -1.506 | 0 | 0.717 |
| 631 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 28 | PRUNE2 | Sponge network | -1.111 | 0.0003 | -1.733 | 0.00022 | 0.717 |
| 632 | RP11-244O19.1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | MACF1 | Sponge network | -0.096 | 0.67958 | 0.019 | 0.90335 | 0.717 |
| 633 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | PLSCR4 | Sponge network | -2.452 | 0 | -0.474 | 0.03833 | 0.717 |
| 634 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935 | 41 | FBXO32 | Sponge network | -0.129 | 0.57177 | -0.495 | 0.07561 | 0.717 |
| 635 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-628-5p | 34 | DTNA | Sponge network | -0.583 | 0.14589 | -1.648 | 1.0E-5 | 0.717 |
| 636 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | DCN | Sponge network | -0.49 | 0.04946 | -1.288 | 0 | 0.717 |
| 637 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-664a-3p;hsa-miR-92a-1-5p | 28 | AKAP6 | Sponge network | -2.69 | 0.0001 | -1.586 | 3.0E-5 | 0.717 |
| 638 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 54 | ZFHX4 | Sponge network | -0.739 | 0.0574 | 0.018 | 0.95574 | 0.717 |
| 639 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | TSHZ3 | Sponge network | -1.439 | 4.0E-5 | -0.556 | 0.01516 | 0.717 |
| 640 | MYLK-AS1 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-7-5p | 17 | EPHA3 | Sponge network | -1.331 | 3.0E-5 | 0.185 | 0.53588 | 0.717 |
| 641 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 22 | DYNC1I1 | Sponge network | -2.452 | 0 | -1.12 | 0.00107 | 0.717 |
| 642 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-940 | 24 | MCC | Sponge network | -1.331 | 3.0E-5 | -1.005 | 0 | 0.717 |
| 643 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-93-3p;hsa-miR-940 | 14 | CNTNAP1 | Sponge network | -0.162 | 0.47687 | 0.358 | 0.13727 | 0.717 |
| 644 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | PLSCR4 | Sponge network | -0.976 | 0.00025 | -0.474 | 0.03833 | 0.716 |
| 645 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-664a-3p;hsa-miR-769-5p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | -2.69 | 0.0001 | -1.733 | 0.00022 | 0.716 |
| 646 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 35 | EPHA7 | Sponge network | -1.092 | 4.0E-5 | -2.197 | 3.0E-5 | 0.716 |
| 647 | HAND2-AS1 |
hsa-miR-182-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-769-5p | 11 | TCEAL7 | Sponge network | -1.697 | 0.00889 | -1.445 | 0 | 0.716 |
| 648 | ARHGEF26-AS1 |
hsa-miR-144-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | FMN2 | Sponge network | -2.46 | 0 | -1.746 | 0.00047 | 0.716 |
| 649 | RP11-166D19.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 33 | DLC1 | Sponge network | -0.576 | 0.10121 | -0.382 | 0.05038 | 0.716 |
| 650 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-942-5p | 27 | NRXN1 | Sponge network | -2.62 | 0 | -3.778 | 0 | 0.716 |
| 651 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | ADAMTSL3 | Sponge network | -0.737 | 0.06182 | -1.559 | 0 | 0.716 |
| 652 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 23 | FGFR1 | Sponge network | -0.739 | 0.0574 | -0.509 | 0.03957 | 0.716 |
| 653 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p | 10 | MN1 | Sponge network | -1.092 | 4.0E-5 | -0.358 | 0.24172 | 0.716 |
| 654 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-590-5p | 13 | AKAP12 | Sponge network | -1.439 | 4.0E-5 | -0.932 | 0.0028 | 0.716 |
| 655 | MIR497HG |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | TLL1 | Sponge network | -1.439 | 4.0E-5 | -1.195 | 3.0E-5 | 0.716 |
| 656 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 11 | NALCN | Sponge network | -0.576 | 0.10121 | 1.068 | 0.00237 | 0.716 |
| 657 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-7-1-3p | 16 | PLSCR4 | Sponge network | -1.092 | 4.0E-5 | -0.474 | 0.03833 | 0.716 |
| 658 | MIR497HG |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p | 11 | ADH1B | Sponge network | -1.439 | 4.0E-5 | -3.716 | 0 | 0.715 |
| 659 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 58 | TCF4 | Sponge network | 0.572 | 0.01722 | 0.016 | 0.92271 | 0.715 |
| 660 | NR2F1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p | 21 | SYNM | Sponge network | -0.49 | 0.04946 | -2.309 | 1.0E-5 | 0.715 |
| 661 | MIR497HG |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-532-3p;hsa-miR-542-3p | 17 | SYNM | Sponge network | -1.439 | 4.0E-5 | -2.309 | 1.0E-5 | 0.715 |
| 662 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 24 | DYNC1I1 | Sponge network | -0.49 | 0.04946 | -1.12 | 0.00107 | 0.715 |
| 663 | NR2F1-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 33 | DLC1 | Sponge network | -0.49 | 0.04946 | -0.382 | 0.05038 | 0.715 |
| 664 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | LHFP | Sponge network | -2.452 | 0 | -0.167 | 0.41371 | 0.715 |
| 665 | FENDRR |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p | 12 | AMPH | Sponge network | -1.281 | 0.00012 | -0.916 | 5.0E-5 | 0.715 |
| 666 | AC011526.1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 13 | PCDH17 | Sponge network | 0.342 | 0.03129 | 0.731 | 2.0E-5 | 0.715 |
| 667 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | PDZRN3 | Sponge network | -1.697 | 0.00889 | -1.548 | 0 | 0.715 |
| 668 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-628-5p;hsa-miR-629-3p | 18 | RASSF8 | Sponge network | -0.583 | 0.14589 | -0.122 | 0.65591 | 0.715 |
| 669 | MRVI1-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-7-1-3p | 12 | WWTR1 | Sponge network | -1.092 | 4.0E-5 | -0.345 | 0.11997 | 0.715 |
| 670 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 31 | NCAM2 | Sponge network | -0.129 | 0.57177 | -0.482 | 0.22565 | 0.715 |
| 671 | MIR143HG |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LHFP | Sponge network | -0.739 | 0.0574 | -0.167 | 0.41371 | 0.714 |
| 672 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 16 | PCDH10 | Sponge network | -1.092 | 4.0E-5 | -1.644 | 0.0078 | 0.714 |
| 673 | RP11-532F6.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-511-5p;hsa-miR-590-3p | 15 | MRVI1 | Sponge network | -0.896 | 0.00099 | -1.051 | 0.00022 | 0.714 |
| 674 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 20 | SYNPO2 | Sponge network | -0.896 | 0.00099 | -2.342 | 0 | 0.714 |
| 675 | LINC00578 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 14 | GREM1 | Sponge network | 0.811 | 0.03228 | 0.902 | 0.01691 | 0.714 |
| 676 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | PREX2 | Sponge network | -0.129 | 0.57177 | -0.272 | 0.25382 | 0.714 |
| 677 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 44 | TSHZ3 | Sponge network | -1.697 | 0.00889 | -0.556 | 0.01516 | 0.714 |
| 678 | HAND2-AS1 |
hsa-miR-144-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | FMN2 | Sponge network | -1.697 | 0.00889 | -1.746 | 0.00047 | 0.714 |
| 679 | MAGI2-AS3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | SFRP1 | Sponge network | -0.129 | 0.57177 | -3.566 | 0 | 0.714 |
| 680 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | CNTNAP1 | Sponge network | -1.697 | 0.00889 | 0.358 | 0.13727 | 0.714 |
| 681 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 28 | PGR | Sponge network | -0.896 | 0.00099 | -1.704 | 0 | 0.714 |
| 682 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-542-3p | 10 | LRRK2 | Sponge network | -1.092 | 4.0E-5 | -1.136 | 1.0E-5 | 0.714 |
| 683 | MIR143HG |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-942-5p | 14 | ZBTB20 | Sponge network | -0.739 | 0.0574 | -0.122 | 0.57569 | 0.713 |
| 684 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 60 | EPHA7 | Sponge network | -1.111 | 0.0003 | -2.197 | 3.0E-5 | 0.713 |
| 685 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-93-5p | 21 | PGR | Sponge network | -2.69 | 0.0001 | -1.704 | 0 | 0.713 |
| 686 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 19 | GLI3 | Sponge network | -2.452 | 0 | -0.615 | 0.01482 | 0.713 |
| 687 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421 | 12 | PEG3 | Sponge network | -2.62 | 0 | -1.74 | 0 | 0.713 |
| 688 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 12 | RADIL | Sponge network | -2.452 | 0 | -1.628 | 0 | 0.713 |
| 689 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | CNTN1 | Sponge network | -0.739 | 0.0574 | -2.594 | 0 | 0.713 |
| 690 | MYLK-AS1 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-7-5p | 10 | PTGFR | Sponge network | -1.331 | 3.0E-5 | -0.233 | 0.45421 | 0.713 |
| 691 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 36 | IGF1 | Sponge network | -0.129 | 0.57177 | -0.204 | 0.5898 | 0.713 |
| 692 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 35 | EPHA3 | Sponge network | -0.576 | 0.10121 | 0.185 | 0.53588 | 0.713 |
| 693 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 31 | NCAM2 | Sponge network | -1.697 | 0.00889 | -0.482 | 0.22565 | 0.712 |
| 694 | SNHG14 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 23 | SFRP1 | Sponge network | -1.111 | 0.0003 | -3.566 | 0 | 0.712 |
| 695 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | CDH19 | Sponge network | -0.576 | 0.10121 | -3.434 | 0 | 0.712 |
| 696 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p | 31 | FAT3 | Sponge network | -0.583 | 0.14589 | -1.601 | 0.00051 | 0.712 |
| 697 | ADAMTS9-AS2 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 20 | CXCL12 | Sponge network | -2.452 | 0 | -1.421 | 0 | 0.712 |
| 698 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p | 16 | SETBP1 | Sponge network | -2.62 | 0 | -1.302 | 1.0E-5 | 0.712 |
| 699 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 54 | ZFHX4 | Sponge network | -1.697 | 0.00889 | 0.018 | 0.95574 | 0.711 |
| 700 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 25 | GREM1 | Sponge network | -0.739 | 0.0574 | 0.902 | 0.01691 | 0.711 |
| 701 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-96-5p | 20 | FNBP1 | Sponge network | -0.576 | 0.10121 | -0.969 | 4.0E-5 | 0.711 |
| 702 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-500a-3p | 18 | AFF3 | Sponge network | -1.092 | 4.0E-5 | -2.373 | 0 | 0.711 |
| 703 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-505-3p | 18 | TIMP3 | Sponge network | -0.583 | 0.14589 | -0.546 | 0.02307 | 0.711 |
| 704 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-935;hsa-miR-940 | 22 | GRIK3 | Sponge network | -1.331 | 3.0E-5 | -3.208 | 0 | 0.711 |
| 705 | ADAMTS9-AS2 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-99b-3p | 48 | GRIK3 | Sponge network | -2.452 | 0 | -3.208 | 0 | 0.711 |
| 706 | USP3-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 16 | EZH1 | Sponge network | -0.162 | 0.47687 | -0.564 | 1.0E-5 | 0.711 |
| 707 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 14 | PEG3 | Sponge network | -0.358 | 0.17255 | -1.74 | 0 | 0.711 |
| 708 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p | 32 | PGR | Sponge network | -0.162 | 0.47687 | -1.704 | 0 | 0.711 |
| 709 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-5p | 12 | THSD7A | Sponge network | -0.576 | 0.10121 | 0.242 | 0.25154 | 0.711 |
| 710 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 54 | BNC2 | Sponge network | 0.997 | 0.00066 | -0.491 | 0.09988 | 0.711 |
| 711 | MRVI1-AS1 |
hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p | 13 | FNBP1 | Sponge network | -1.092 | 4.0E-5 | -0.969 | 4.0E-5 | 0.71 |
| 712 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 16 | FLNA | Sponge network | 0.194 | 0.64278 | -0.771 | 0.01377 | 0.71 |
| 713 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p | 41 | FBXO32 | Sponge network | -1.697 | 0.00889 | -0.495 | 0.07561 | 0.71 |
| 714 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | ERG | Sponge network | -0.358 | 0.17255 | 0.002 | 0.99011 | 0.71 |
| 715 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | RCAN2 | Sponge network | -0.576 | 0.10121 | -0.33 | 0.15172 | 0.71 |
| 716 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-7-5p | 19 | FNBP1 | Sponge network | -0.129 | 0.57177 | -0.969 | 4.0E-5 | 0.71 |
| 717 | PWAR6 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | FHL1 | Sponge network | -1.575 | 6.0E-5 | -2.687 | 0 | 0.71 |
| 718 | HAND2-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 23 | SORCS1 | Sponge network | -1.697 | 0.00889 | -3.492 | 0 | 0.71 |
| 719 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | -1.439 | 4.0E-5 | -0.509 | 0.03957 | 0.71 |
| 720 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 15 | CNTNAP1 | Sponge network | 0.572 | 0.01722 | 0.358 | 0.13727 | 0.71 |
| 721 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | -1.697 | 0.00889 | -1.324 | 0.00014 | 0.71 |
| 722 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 10 | CTNNA3 | Sponge network | -2.452 | 0 | -2.046 | 0.00019 | 0.71 |
| 723 | NR2F1-AS1 |
hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-502-5p | 11 | ELN | Sponge network | -0.49 | 0.04946 | 0.451 | 0.13752 | 0.71 |
| 724 | SNHG14 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 14 | ZNF382 | Sponge network | -1.111 | 0.0003 | -0.494 | 0.03831 | 0.71 |
| 725 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 24 | DYNC1I1 | Sponge network | -0.576 | 0.10121 | -1.12 | 0.00107 | 0.709 |
| 726 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-671-5p;hsa-miR-940 | 11 | SORCS1 | Sponge network | -1.331 | 3.0E-5 | -3.492 | 0 | 0.709 |
| 727 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 42 | NRXN1 | Sponge network | -1.697 | 0.00889 | -3.778 | 0 | 0.709 |
| 728 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 21 | CNTN1 | Sponge network | -1.092 | 4.0E-5 | -2.594 | 0 | 0.709 |
| 729 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 34 | IGF1 | Sponge network | -0.576 | 0.10121 | -0.204 | 0.5898 | 0.709 |
| 730 | DNM3OS |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 19 | NRK | Sponge network | 0.572 | 0.01722 | 0.656 | 0.19217 | 0.709 |
| 731 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | DCN | Sponge network | -2.452 | 0 | -1.288 | 0 | 0.709 |
| 732 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 21 | ANK2 | Sponge network | -4.546 | 0 | -1.603 | 1.0E-5 | 0.709 |
| 733 | FENDRR |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p | 21 | SYNM | Sponge network | -1.281 | 0.00012 | -2.309 | 1.0E-5 | 0.709 |
| 734 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p | 13 | RYR2 | Sponge network | -1.331 | 3.0E-5 | -1.466 | 0.00031 | 0.709 |
| 735 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 40 | TSHZ3 | Sponge network | 0.572 | 0.01722 | -0.556 | 0.01516 | 0.709 |
| 736 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-625-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -0.129 | 0.57177 | 0.024 | 0.89889 | 0.709 |
| 737 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ERG | Sponge network | -0.129 | 0.57177 | 0.002 | 0.99011 | 0.708 |
| 738 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 28 | PCDH10 | Sponge network | -0.976 | 0.00025 | -1.644 | 0.0078 | 0.708 |
| 739 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 18 | MAPK10 | Sponge network | -0.737 | 0.06182 | -1.774 | 0 | 0.708 |
| 740 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 28 | CDH19 | Sponge network | -1.697 | 0.00889 | -3.434 | 0 | 0.708 |
| 741 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-2355-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 11 | NUDT11 | Sponge network | -0.129 | 0.57177 | -0.921 | 0.00376 | 0.708 |
| 742 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-942-5p | 26 | HLF | Sponge network | -1.092 | 4.0E-5 | -2.443 | 0 | 0.708 |
| 743 | ZNF667-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-511-5p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | WWTR1 | Sponge network | -0.976 | 0.00025 | -0.345 | 0.11997 | 0.708 |
| 744 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-93-5p | 28 | ANK2 | Sponge network | -2.69 | 0.0001 | -1.603 | 1.0E-5 | 0.708 |
| 745 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | PDGFC | Sponge network | -0.49 | 0.04946 | -0.33 | 0.06483 | 0.708 |
| 746 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 13 | HMCN1 | Sponge network | -0.49 | 0.04946 | 0.809 | 0.00544 | 0.708 |
| 747 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 35 | AKAP6 | Sponge network | -0.49 | 0.04946 | -1.586 | 3.0E-5 | 0.708 |
| 748 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 20 | SPG20 | Sponge network | -0.49 | 0.04946 | -1.16 | 0 | 0.708 |
| 749 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 35 | PGR | Sponge network | -1.281 | 0.00012 | -1.704 | 0 | 0.707 |
| 750 | RP11-400K9.4 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-542-3p | 16 | MRVI1 | Sponge network | -0.611 | 0.09607 | -1.051 | 0.00022 | 0.707 |
| 751 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 49 | MEF2C | Sponge network | -2.452 | 0 | -0.499 | 0.01448 | 0.707 |
| 752 | HAND2-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 67 | AR | Sponge network | -1.697 | 0.00889 | -1.554 | 0 | 0.707 |
| 753 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-935 | 35 | SSBP2 | Sponge network | -1.331 | 3.0E-5 | -1.58 | 0 | 0.707 |
| 754 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-935 | 54 | FAT3 | Sponge network | -0.162 | 0.47687 | -1.601 | 0.00051 | 0.707 |
| 755 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-421 | 13 | AKAP6 | Sponge network | -4.463 | 0.00025 | -1.586 | 3.0E-5 | 0.707 |
| 756 | DNM3OS |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | 0.572 | 0.01722 | -0.382 | 0.05038 | 0.707 |
| 757 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 12 | GAS1 | Sponge network | -1.697 | 0.00889 | -1.047 | 0.00364 | 0.706 |
| 758 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | BNC2 | Sponge network | -1.439 | 4.0E-5 | -0.491 | 0.09988 | 0.706 |
| 759 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | FAT4 | Sponge network | -0.739 | 0.0574 | -0.354 | 0.14694 | 0.706 |
| 760 | MAGI2-AS3 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 42 | IGFBP5 | Sponge network | -0.129 | 0.57177 | -0.806 | 0.00105 | 0.706 |
| 761 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 61 | DTNA | Sponge network | -0.49 | 0.04946 | -1.648 | 1.0E-5 | 0.706 |
| 762 | SERTAD4-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-576-5p | 11 | SYNPO2 | Sponge network | -0.932 | 0.00329 | -2.342 | 0 | 0.706 |
| 763 | ADAMTS9-AS2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p | 32 | DLC1 | Sponge network | -2.452 | 0 | -0.382 | 0.05038 | 0.706 |
| 764 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 32 | GRIN2A | Sponge network | -2.452 | 0 | -2.161 | 0 | 0.706 |
| 765 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 33 | ST6GAL2 | Sponge network | -0.576 | 0.10121 | -1.201 | 0.00597 | 0.706 |
| 766 | FGF14-AS2 |
hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | SETBP1 | Sponge network | -1.582 | 8.0E-5 | -1.302 | 1.0E-5 | 0.706 |
| 767 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | CNTNAP1 | Sponge network | -1.439 | 4.0E-5 | 0.358 | 0.13727 | 0.706 |
| 768 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-98-5p | 38 | DMD | Sponge network | -0.976 | 0.00025 | -1.506 | 0 | 0.706 |
| 769 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 14 | FAT4 | Sponge network | -0.358 | 0.17255 | -0.354 | 0.14694 | 0.706 |
| 770 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 32 | PCDH10 | Sponge network | -0.49 | 0.04946 | -1.644 | 0.0078 | 0.706 |
| 771 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 51 | BNC2 | Sponge network | 0.194 | 0.64278 | -0.491 | 0.09988 | 0.706 |
| 772 | FGF14-AS2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 29 | DTNA | Sponge network | -1.582 | 8.0E-5 | -1.648 | 1.0E-5 | 0.705 |
| 773 | USP3-AS1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-375;hsa-miR-455-5p | 17 | ITPKB | Sponge network | -0.162 | 0.47687 | -0.704 | 0.00106 | 0.705 |
| 774 | HAND2-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 13 | PRKD1 | Sponge network | -1.697 | 0.00889 | -0.466 | 0.03583 | 0.705 |
| 775 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 16 | CACNA2D1 | Sponge network | -1.092 | 4.0E-5 | -0.846 | 0.00675 | 0.705 |
| 776 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 10 | DNAJB4 | Sponge network | -1.092 | 4.0E-5 | -0.7 | 1.0E-5 | 0.705 |
| 777 | RP11-166D19.1 |
hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-502-5p | 11 | ELN | Sponge network | -0.576 | 0.10121 | 0.451 | 0.13752 | 0.705 |
| 778 | AC003090.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | SORCS1 | Sponge network | -2.117 | 0.00161 | -3.492 | 0 | 0.705 |
| 779 | LINC00578 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577 | 17 | NRK | Sponge network | 0.811 | 0.03228 | 0.656 | 0.19217 | 0.705 |
| 780 | LINC00092 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 13 | SORCS1 | Sponge network | -1.886 | 0 | -3.492 | 0 | 0.705 |
| 781 | MIR497HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-93-3p | 51 | RUNX1T1 | Sponge network | -1.439 | 4.0E-5 | -0.344 | 0.2374 | 0.704 |
| 782 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 36 | AKAP6 | Sponge network | -2.46 | 0 | -1.586 | 3.0E-5 | 0.704 |
| 783 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 21 | JAKMIP2 | Sponge network | -1.697 | 0.00889 | -1.388 | 0 | 0.704 |
| 784 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 22 | NR2C2 | Sponge network | 0.537 | 0.00139 | 0.21 | 0.03224 | 0.704 |
| 785 | LINC00702 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LHFP | Sponge network | -0.737 | 0.06182 | -0.167 | 0.41371 | 0.704 |
| 786 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-98-5p | 49 | DMD | Sponge network | -0.129 | 0.57177 | -1.506 | 0 | 0.704 |
| 787 | CTC-296K1.3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-342-3p | 11 | SYNPO2 | Sponge network | -2.095 | 0.00112 | -2.342 | 0 | 0.704 |
| 788 | MEG3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | 0.249 | 0.35902 | -0.615 | 0.01482 | 0.704 |
| 789 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | DNAJB4 | Sponge network | -0.129 | 0.57177 | -0.7 | 1.0E-5 | 0.704 |
| 790 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429 | 14 | SPG20 | Sponge network | -2.62 | 0 | -1.16 | 0 | 0.704 |
| 791 | LINC00702 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | -0.737 | 0.06182 | 0.809 | 0.00544 | 0.704 |
| 792 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | TIMP3 | Sponge network | -0.49 | 0.04946 | -0.546 | 0.02307 | 0.703 |
| 793 | PWAR6 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 19 | MAPK10 | Sponge network | -1.575 | 6.0E-5 | -1.774 | 0 | 0.703 |
| 794 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | PDZRN3 | Sponge network | -1.111 | 0.0003 | -1.548 | 0 | 0.703 |
| 795 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 20 | DCLK1 | Sponge network | -1.111 | 0.0003 | -1.324 | 0.00014 | 0.703 |
| 796 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 15 | NALCN | Sponge network | -0.129 | 0.57177 | 1.068 | 0.00237 | 0.703 |
| 797 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | -0.737 | 0.06182 | -0.474 | 0.03833 | 0.703 |
| 798 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-93-5p | 46 | AR | Sponge network | -2.69 | 0.0001 | -1.554 | 0 | 0.703 |
| 799 | SNHG14 |
hsa-miR-144-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | FMN2 | Sponge network | -1.111 | 0.0003 | -1.746 | 0.00047 | 0.703 |
| 800 | MIR143HG |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 24 | PTGFR | Sponge network | -0.739 | 0.0574 | -0.233 | 0.45421 | 0.703 |
| 801 | NR2F1-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | SFRP1 | Sponge network | -0.49 | 0.04946 | -3.566 | 0 | 0.703 |
| 802 | RP11-554A11.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-629-3p | 24 | IGFBP5 | Sponge network | -0.583 | 0.14589 | -0.806 | 0.00105 | 0.703 |
| 803 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 35 | NRXN1 | Sponge network | -1.439 | 4.0E-5 | -3.778 | 0 | 0.702 |
| 804 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | ANK2 | Sponge network | -2.117 | 0.00161 | -1.603 | 1.0E-5 | 0.702 |
| 805 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | THBS1 | Sponge network | 0.572 | 0.01722 | 0.741 | 0.00386 | 0.702 |
| 806 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 18 | PCDH9 | Sponge network | -1.092 | 4.0E-5 | -1.823 | 4.0E-5 | 0.702 |
| 807 | LINC00578 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-589-3p | 11 | ABCC9 | Sponge network | 0.811 | 0.03228 | -0.466 | 0.14121 | 0.702 |
| 808 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | TIMP3 | Sponge network | 0.572 | 0.01722 | -0.546 | 0.02307 | 0.702 |
| 809 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | RECK | Sponge network | 0.572 | 0.01722 | -1.043 | 0 | 0.702 |
| 810 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 48 | RBMS3 | Sponge network | 0.194 | 0.64278 | -0.519 | 0.03551 | 0.702 |
| 811 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p | 11 | PGR | Sponge network | -0.932 | 0.00329 | -1.704 | 0 | 0.702 |
| 812 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-629-3p | 39 | EPHA7 | Sponge network | -2.62 | 0 | -2.197 | 3.0E-5 | 0.701 |
| 813 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 50 | DIP2C | Sponge network | -0.739 | 0.0574 | -0.436 | 0.01713 | 0.701 |
| 814 | ZNF667-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-625-5p | 11 | NGFR | Sponge network | -0.976 | 0.00025 | -1.271 | 0.01058 | 0.701 |
| 815 | MIR143HG |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 33 | NRK | Sponge network | -0.739 | 0.0574 | 0.656 | 0.19217 | 0.701 |
| 816 | PWAR6 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 62 | AR | Sponge network | -1.575 | 6.0E-5 | -1.554 | 0 | 0.701 |
| 817 | MIR143HG |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 15 | AMPH | Sponge network | -0.739 | 0.0574 | -0.916 | 5.0E-5 | 0.701 |
| 818 | SNHG14 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | HSPB7 | Sponge network | -1.111 | 0.0003 | -2.268 | 1.0E-5 | 0.701 |
| 819 | MIR497HG |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | FNBP1 | Sponge network | -1.439 | 4.0E-5 | -0.969 | 4.0E-5 | 0.701 |
| 820 | RP11-736K20.5 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-511-5p;hsa-miR-590-3p | 12 | MRVI1 | Sponge network | -0.358 | 0.17255 | -1.051 | 0.00022 | 0.701 |
| 821 | BVES-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-766-3p;hsa-miR-98-5p | 44 | CADM2 | Sponge network | -2.62 | 0 | -3.782 | 0 | 0.701 |
| 822 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 17 | FNBP1 | Sponge network | -0.976 | 0.00025 | -0.969 | 4.0E-5 | 0.701 |
| 823 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 37 | EBF1 | Sponge network | -0.129 | 0.57177 | -0.384 | 0.08856 | 0.701 |
| 824 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 36 | CNTN1 | Sponge network | -0.737 | 0.06182 | -2.594 | 0 | 0.701 |
| 825 | MYLK-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484 | 28 | TGFBR3 | Sponge network | -1.331 | 3.0E-5 | -1.173 | 1.0E-5 | 0.701 |
| 826 | MRVI1-AS1 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p | 16 | ZBTB4 | Sponge network | -1.092 | 4.0E-5 | -0.739 | 0 | 0.701 |
| 827 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-484 | 13 | FLNA | Sponge network | -2.69 | 0.0001 | -0.771 | 0.01377 | 0.701 |
| 828 | ACTA2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-96-5p | 87 | LPP | Sponge network | 0.194 | 0.64278 | -0.459 | 0.04475 | 0.701 |
| 829 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 61 | NTRK3 | Sponge network | -1.439 | 4.0E-5 | -1.77 | 3.0E-5 | 0.7 |
| 830 | PWAR6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | SPG20 | Sponge network | -1.575 | 6.0E-5 | -1.16 | 0 | 0.7 |
| 831 | MIR497HG |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 16 | SORCS1 | Sponge network | -1.439 | 4.0E-5 | -3.492 | 0 | 0.7 |
| 832 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-940 | 33 | CADM2 | Sponge network | -1.331 | 3.0E-5 | -3.782 | 0 | 0.7 |
| 833 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-769-5p | 12 | SETBP1 | Sponge network | -0.583 | 0.14589 | -1.302 | 1.0E-5 | 0.7 |
| 834 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p | 19 | EZH1 | Sponge network | -2.452 | 0 | -0.564 | 1.0E-5 | 0.7 |
| 835 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p | 15 | DCLK1 | Sponge network | -0.976 | 0.00025 | -1.324 | 0.00014 | 0.7 |
| 836 | NR2F1-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 32 | NAV3 | Sponge network | -0.49 | 0.04946 | 0.212 | 0.47729 | 0.7 |
| 837 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | ZNF521 | Sponge network | -0.576 | 0.10121 | -0.111 | 0.58068 | 0.7 |
| 838 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-629-3p | 18 | EBF3 | Sponge network | -0.358 | 0.17255 | -0.058 | 0.81437 | 0.7 |
| 839 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 35 | CNTN1 | Sponge network | -2.452 | 0 | -2.594 | 0 | 0.7 |
| 840 | SNHG14 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 114 | LPP | Sponge network | -1.111 | 0.0003 | -0.459 | 0.04475 | 0.7 |
| 841 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 29 | SYNPO2 | Sponge network | 0.194 | 0.64278 | -2.342 | 0 | 0.7 |
| 842 | AC003090.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 16 | MAPK10 | Sponge network | -2.117 | 0.00161 | -1.774 | 0 | 0.7 |
| 843 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-532-3p | 12 | SYNM | Sponge network | -2.531 | 0 | -2.309 | 1.0E-5 | 0.7 |
| 844 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 47 | MCC | Sponge network | -0.739 | 0.0574 | -1.005 | 0 | 0.699 |
| 845 | MRVI1-AS1 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-744-5p | 10 | FGFR1 | Sponge network | -1.092 | 4.0E-5 | -0.509 | 0.03957 | 0.699 |
| 846 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-628-5p;hsa-miR-664a-5p | 56 | LPP | Sponge network | -0.583 | 0.14589 | -0.459 | 0.04475 | 0.699 |
| 847 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 28 | AKAP6 | Sponge network | -0.976 | 0.00025 | -1.586 | 3.0E-5 | 0.699 |
| 848 | SNHG14 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ITPKB | Sponge network | -1.111 | 0.0003 | -0.704 | 0.00106 | 0.699 |
| 849 | ADAMTS9-AS2 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 14 | NGFR | Sponge network | -2.452 | 0 | -1.271 | 0.01058 | 0.699 |
| 850 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 24 | GREM1 | Sponge network | -1.697 | 0.00889 | 0.902 | 0.01691 | 0.699 |
| 851 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-577 | 12 | AMPH | Sponge network | -2.452 | 0 | -0.916 | 5.0E-5 | 0.699 |
| 852 | SNHG14 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 17 | STAT5B | Sponge network | -1.111 | 0.0003 | -0.182 | 0.1082 | 0.699 |
| 853 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-769-5p | 20 | SYNPO2 | Sponge network | -0.611 | 0.09607 | -2.342 | 0 | 0.699 |
| 854 | SNHG14 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | KL | Sponge network | -1.111 | 0.0003 | -1.294 | 0 | 0.699 |
| 855 | MIR143HG |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 34 | NAV3 | Sponge network | -0.739 | 0.0574 | 0.212 | 0.47729 | 0.699 |
| 856 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-590-3p | 23 | AFF3 | Sponge network | -1.281 | 0.00012 | -2.373 | 0 | 0.699 |
| 857 | CTC-444N24.11 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-409-5p;hsa-miR-590-3p | 11 | TPR | Sponge network | 1.153 | 0 | 0.582 | 0 | 0.699 |
| 858 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935 | 47 | DMD | Sponge network | -0.576 | 0.10121 | -1.506 | 0 | 0.698 |
| 859 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-501-3p | 12 | SLITRK3 | Sponge network | -1.092 | 4.0E-5 | -3.328 | 0 | 0.698 |
| 860 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 41 | EBF1 | Sponge network | -1.111 | 0.0003 | -0.384 | 0.08856 | 0.698 |
| 861 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 25 | PRUNE2 | Sponge network | -0.129 | 0.57177 | -1.733 | 0.00022 | 0.698 |
| 862 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | FNBP1 | Sponge network | -1.575 | 6.0E-5 | -0.969 | 4.0E-5 | 0.698 |
| 863 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 64 | FAT3 | Sponge network | -0.737 | 0.06182 | -1.601 | 0.00051 | 0.698 |
| 864 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b | 19 | PLSCR4 | Sponge network | -0.583 | 0.14589 | -0.474 | 0.03833 | 0.698 |
| 865 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-493-5p | 13 | NRXN3 | Sponge network | -2.62 | 0 | -0.893 | 0.04186 | 0.698 |
| 866 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 22 | MACF1 | Sponge network | -0.129 | 0.57177 | 0.019 | 0.90335 | 0.698 |
| 867 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 36 | MEF2C | Sponge network | -1.439 | 4.0E-5 | -0.499 | 0.01448 | 0.698 |
| 868 | PWAR6 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 30 | SYNPO2 | Sponge network | -1.575 | 6.0E-5 | -2.342 | 0 | 0.698 |
| 869 | RP11-166D19.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 23 | SORCS1 | Sponge network | -0.576 | 0.10121 | -3.492 | 0 | 0.698 |
| 870 | MRVI1-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-671-5p | 22 | DLC1 | Sponge network | -1.092 | 4.0E-5 | -0.382 | 0.05038 | 0.697 |
| 871 | HAND2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 16 | MN1 | Sponge network | -1.697 | 0.00889 | -0.358 | 0.24172 | 0.697 |
| 872 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 17 | CADM3 | Sponge network | -1.092 | 4.0E-5 | -3.663 | 0 | 0.697 |
| 873 | MYLK-AS1 |
hsa-miR-107;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p | 13 | GRIN2A | Sponge network | -1.331 | 3.0E-5 | -2.161 | 0 | 0.697 |
| 874 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 35 | EPHA3 | Sponge network | -0.49 | 0.04946 | 0.185 | 0.53588 | 0.697 |
| 875 | HAND2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 102 | LPP | Sponge network | -1.697 | 0.00889 | -0.459 | 0.04475 | 0.697 |
| 876 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 53 | EPHA7 | Sponge network | -0.576 | 0.10121 | -2.197 | 3.0E-5 | 0.697 |
| 877 | DNM3OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | LHFP | Sponge network | 0.572 | 0.01722 | -0.167 | 0.41371 | 0.697 |
| 878 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 46 | GAS7 | Sponge network | -0.976 | 0.00025 | -0.555 | 0.01602 | 0.697 |
| 879 | MIR497HG |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-7-5p | 15 | FLNA | Sponge network | -1.439 | 4.0E-5 | -0.771 | 0.01377 | 0.697 |
| 880 | RP11-736K20.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p | 24 | NAV3 | Sponge network | -0.358 | 0.17255 | 0.212 | 0.47729 | 0.696 |
| 881 | MIR143HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 27 | CDH19 | Sponge network | -0.739 | 0.0574 | -3.434 | 0 | 0.696 |
| 882 | LINC00702 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 32 | NRK | Sponge network | -0.737 | 0.06182 | 0.656 | 0.19217 | 0.696 |
| 883 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | RASSF8 | Sponge network | -0.737 | 0.06182 | -0.122 | 0.65591 | 0.696 |
| 884 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 38 | NRXN1 | Sponge network | -0.737 | 0.06182 | -3.778 | 0 | 0.696 |
| 885 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 23 | GREM1 | Sponge network | 0.572 | 0.01722 | 0.902 | 0.01691 | 0.696 |
| 886 | MIR497HG |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-577 | 10 | AMPH | Sponge network | -1.439 | 4.0E-5 | -0.916 | 5.0E-5 | 0.696 |
| 887 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 81 | IL6ST | Sponge network | -0.129 | 0.57177 | -0.402 | 0.00676 | 0.696 |
| 888 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 67 | PRKAA2 | Sponge network | -2.46 | 0 | -2.462 | 0 | 0.696 |
| 889 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CDH11 | Sponge network | -0.129 | 0.57177 | 1.427 | 0 | 0.696 |
| 890 | MIR497HG |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 43 | GRIK3 | Sponge network | -1.439 | 4.0E-5 | -3.208 | 0 | 0.696 |
| 891 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | CAV1 | Sponge network | -0.739 | 0.0574 | -1.013 | 6.0E-5 | 0.696 |
| 892 | MYLK-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-671-5p | 10 | CADM3 | Sponge network | -1.331 | 3.0E-5 | -3.663 | 0 | 0.696 |
| 893 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 15 | LRRK2 | Sponge network | -0.129 | 0.57177 | -1.136 | 1.0E-5 | 0.696 |
| 894 | MAGI2-AS3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 110 | LPP | Sponge network | -0.129 | 0.57177 | -0.459 | 0.04475 | 0.696 |
| 895 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 26 | GREM1 | Sponge network | 0.997 | 0.00066 | 0.902 | 0.01691 | 0.696 |
| 896 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p | 12 | CACNA2D1 | Sponge network | -1.331 | 3.0E-5 | -0.846 | 0.00675 | 0.695 |
| 897 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ABCC9 | Sponge network | 0.997 | 0.00066 | -0.466 | 0.14121 | 0.695 |
| 898 | DNM3OS |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 20 | MRVI1 | Sponge network | 0.572 | 0.01722 | -1.051 | 0.00022 | 0.695 |
| 899 | LINC00702 |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 11 | DPP6 | Sponge network | -0.737 | 0.06182 | -3.777 | 0 | 0.695 |
| 900 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | RECK | Sponge network | -1.281 | 0.00012 | -1.043 | 0 | 0.695 |
| 901 | ZNF667-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-671-5p | 29 | DLC1 | Sponge network | -0.976 | 0.00025 | -0.382 | 0.05038 | 0.695 |
| 902 | SNHG14 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | FLNA | Sponge network | -1.111 | 0.0003 | -0.771 | 0.01377 | 0.695 |
| 903 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-93-5p | 37 | AR | Sponge network | -2.531 | 0 | -1.554 | 0 | 0.695 |
| 904 | LINC00702 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 34 | NAV3 | Sponge network | -0.737 | 0.06182 | 0.212 | 0.47729 | 0.695 |
| 905 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 10 | LRFN5 | Sponge network | -0.129 | 0.57177 | -1.483 | 0 | 0.695 |
| 906 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 19 | ADAMTSL3 | Sponge network | -0.611 | 0.09607 | -1.559 | 0 | 0.695 |
| 907 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-708-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-942-5p | 48 | HLF | Sponge network | -2.452 | 0 | -2.443 | 0 | 0.695 |
| 908 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p | 25 | KIT | Sponge network | -2.452 | 0 | -2.184 | 0 | 0.695 |
| 909 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | CACNA2D1 | Sponge network | -0.129 | 0.57177 | -0.846 | 0.00675 | 0.695 |
| 910 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-505-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 60 | TCF4 | Sponge network | -0.976 | 0.00025 | 0.016 | 0.92271 | 0.695 |
| 911 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 31 | AKAP6 | Sponge network | 0.194 | 0.64278 | -1.586 | 3.0E-5 | 0.694 |
| 912 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 55 | EPHA7 | Sponge network | -0.49 | 0.04946 | -2.197 | 3.0E-5 | 0.694 |
| 913 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p | 11 | STARD13 | Sponge network | -1.331 | 3.0E-5 | -0.187 | 0.27383 | 0.694 |
| 914 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | ANK2 | Sponge network | -1.886 | 0 | -1.603 | 1.0E-5 | 0.694 |
| 915 | PGM5-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 13 | SETBP1 | Sponge network | -4.546 | 0 | -1.302 | 1.0E-5 | 0.694 |
| 916 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 40 | TCF4 | Sponge network | -0.358 | 0.17255 | 0.016 | 0.92271 | 0.694 |
| 917 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 13 | ZNF382 | Sponge network | -0.129 | 0.57177 | -0.494 | 0.03831 | 0.694 |
| 918 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-624-5p | 10 | HMCN1 | Sponge network | -0.358 | 0.17255 | 0.809 | 0.00544 | 0.694 |
| 919 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p | 13 | TIMP3 | Sponge network | -1.331 | 3.0E-5 | -0.546 | 0.02307 | 0.694 |
| 920 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 14 | CNTNAP1 | Sponge network | -0.976 | 0.00025 | 0.358 | 0.13727 | 0.694 |
| 921 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 24 | ADAMTSL3 | Sponge network | -2.117 | 0.00161 | -1.559 | 0 | 0.693 |
| 922 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 36 | PCDH10 | Sponge network | -2.452 | 0 | -1.644 | 0.0078 | 0.693 |
| 923 | MIR143HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 53 | HLF | Sponge network | -0.739 | 0.0574 | -2.443 | 0 | 0.693 |
| 924 | PGM5-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-660-5p;hsa-miR-7-1-3p | 12 | PCDH9 | Sponge network | -4.546 | 0 | -1.823 | 4.0E-5 | 0.693 |
| 925 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 17 | JAKMIP2 | Sponge network | -0.576 | 0.10121 | -1.388 | 0 | 0.693 |
| 926 | SERTAD4-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p | 10 | SYNM | Sponge network | -0.932 | 0.00329 | -2.309 | 1.0E-5 | 0.693 |
| 927 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p | 25 | DYNC1I1 | Sponge network | -0.739 | 0.0574 | -1.12 | 0.00107 | 0.693 |
| 928 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p | 53 | DTNA | Sponge network | -2.69 | 0.0001 | -1.648 | 1.0E-5 | 0.693 |
| 929 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 56 | ADARB1 | Sponge network | -0.129 | 0.57177 | -0.335 | 0.06631 | 0.693 |
| 930 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 44 | NRXN1 | Sponge network | -0.739 | 0.0574 | -3.778 | 0 | 0.693 |
| 931 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | SYNE1 | Sponge network | 0.572 | 0.01722 | -0.738 | 0.00316 | 0.693 |
| 932 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 20 | CDH19 | Sponge network | -0.976 | 0.00025 | -3.434 | 0 | 0.693 |
| 933 | LINC00702 |
hsa-miR-144-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | FMN2 | Sponge network | -0.737 | 0.06182 | -1.746 | 0.00047 | 0.693 |
| 934 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 53 | EPHA7 | Sponge network | -2.452 | 0 | -2.197 | 3.0E-5 | 0.693 |
| 935 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 13 | AMPH | Sponge network | -0.129 | 0.57177 | -0.916 | 5.0E-5 | 0.693 |
| 936 | BVES-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-874-3p | 16 | SORCS1 | Sponge network | -2.62 | 0 | -3.492 | 0 | 0.693 |
| 937 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-577 | 19 | AFF3 | Sponge network | -1.439 | 4.0E-5 | -2.373 | 0 | 0.693 |
| 938 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-744-3p | 21 | ADAMTSL3 | Sponge network | -2.62 | 0 | -1.559 | 0 | 0.693 |
| 939 | C20orf166-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-664a-3p | 16 | CDH19 | Sponge network | -2.69 | 0.0001 | -3.434 | 0 | 0.692 |
| 940 | BVES-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-877-5p | 43 | AR | Sponge network | -2.62 | 0 | -1.554 | 0 | 0.692 |
| 941 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 14 | ANK2 | Sponge network | -1.582 | 8.0E-5 | -1.603 | 1.0E-5 | 0.692 |
| 942 | ADAMTS9-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 100 | LPP | Sponge network | -2.452 | 0 | -0.459 | 0.04475 | 0.692 |
| 943 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 85 | IL6ST | Sponge network | -1.111 | 0.0003 | -0.402 | 0.00676 | 0.692 |
| 944 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | EBF1 | Sponge network | -2.452 | 0 | -0.384 | 0.08856 | 0.692 |
| 945 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 21 | CADM3 | Sponge network | -0.576 | 0.10121 | -3.663 | 0 | 0.692 |
| 946 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | SLITRK3 | Sponge network | -0.737 | 0.06182 | -3.328 | 0 | 0.692 |
| 947 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-92b-3p | 20 | AKAP12 | Sponge network | -1.697 | 0.00889 | -0.932 | 0.0028 | 0.692 |
| 948 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 37 | MCC | Sponge network | -0.976 | 0.00025 | -1.005 | 0 | 0.692 |
| 949 | SNHG14 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -1.111 | 0.0003 | -2.633 | 0 | 0.692 |
| 950 | MIR497HG |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | ERG | Sponge network | -1.439 | 4.0E-5 | 0.002 | 0.99011 | 0.692 |
| 951 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 12 | DCN | Sponge network | -0.583 | 0.14589 | -1.288 | 0 | 0.691 |
| 952 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 53 | ADARB1 | Sponge network | -0.576 | 0.10121 | -0.335 | 0.06631 | 0.691 |
| 953 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 31 | SSBP2 | Sponge network | -1.092 | 4.0E-5 | -1.58 | 0 | 0.691 |
| 954 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p | 19 | SLITRK3 | Sponge network | -0.576 | 0.10121 | -3.328 | 0 | 0.691 |
| 955 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 71 | PRKAA2 | Sponge network | -1.111 | 0.0003 | -2.462 | 0 | 0.691 |
| 956 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p | 14 | FAT4 | Sponge network | -0.469 | 0.08721 | -0.354 | 0.14694 | 0.691 |
| 957 | PWAR6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | AKAP6 | Sponge network | -1.575 | 6.0E-5 | -1.586 | 3.0E-5 | 0.691 |
| 958 | SNHG14 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 17 | MN1 | Sponge network | -1.111 | 0.0003 | -0.358 | 0.24172 | 0.691 |
| 959 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | CNTNAP1 | Sponge network | 0.997 | 0.00066 | 0.358 | 0.13727 | 0.691 |
| 960 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-550a-3p | 24 | DTNA | Sponge network | -2.531 | 0 | -1.648 | 1.0E-5 | 0.691 |
| 961 | LINC00702 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 22 | CDH19 | Sponge network | -0.737 | 0.06182 | -3.434 | 0 | 0.691 |
| 962 | SNHG14 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 22 | GLI3 | Sponge network | -1.111 | 0.0003 | -0.615 | 0.01482 | 0.691 |
| 963 | FENDRR |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 31 | SETBP1 | Sponge network | -1.281 | 0.00012 | -1.302 | 1.0E-5 | 0.691 |
| 964 | RP11-244O19.1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLNA | Sponge network | -0.096 | 0.67958 | -0.771 | 0.01377 | 0.691 |
| 965 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-370-3p | 26 | JAZF1 | Sponge network | -1.331 | 3.0E-5 | -0.377 | 0.02677 | 0.69 |
| 966 | MAGI2-AS3 |
hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-502-5p | 10 | ELN | Sponge network | -0.129 | 0.57177 | 0.451 | 0.13752 | 0.69 |
| 967 | MYLK-AS1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-940 | 21 | MEF2C | Sponge network | -1.331 | 3.0E-5 | -0.499 | 0.01448 | 0.69 |
| 968 | CTD-3064M3.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-429;hsa-miR-505-5p | 12 | MRVI1 | Sponge network | -0.469 | 0.08721 | -1.051 | 0.00022 | 0.69 |
| 969 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | ZFHX4 | Sponge network | -0.976 | 0.00025 | 0.018 | 0.95574 | 0.69 |
| 970 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-429 | 10 | ABCC9 | Sponge network | -2.62 | 0 | -0.466 | 0.14121 | 0.69 |
| 971 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 50 | FAT3 | Sponge network | -0.976 | 0.00025 | -1.601 | 0.00051 | 0.69 |
| 972 | RP11-532F6.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-493-5p;hsa-miR-495-3p | 15 | SYNM | Sponge network | -0.896 | 0.00099 | -2.309 | 1.0E-5 | 0.69 |
| 973 | MIR497HG |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-5p | 17 | SPG20 | Sponge network | -1.439 | 4.0E-5 | -1.16 | 0 | 0.69 |
| 974 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 45 | MCC | Sponge network | -0.737 | 0.06182 | -1.005 | 0 | 0.69 |
| 975 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | ZNF521 | Sponge network | -0.49 | 0.04946 | -0.111 | 0.58068 | 0.69 |
| 976 | LINC00702 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 35 | WWTR1 | Sponge network | -0.737 | 0.06182 | -0.345 | 0.11997 | 0.69 |
| 977 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 34 | SETBP1 | Sponge network | -1.697 | 0.00889 | -1.302 | 1.0E-5 | 0.69 |
| 978 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p | 13 | DCLK1 | Sponge network | -1.439 | 4.0E-5 | -1.324 | 0.00014 | 0.69 |
| 979 | PGM5-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-940 | 14 | SFRP1 | Sponge network | -4.546 | 0 | -3.566 | 0 | 0.69 |
| 980 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 28 | NRXN3 | Sponge network | -2.452 | 0 | -0.893 | 0.04186 | 0.689 |
| 981 | FENDRR |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | PCDH18 | Sponge network | -1.281 | 0.00012 | 0.216 | 0.24645 | 0.689 |
| 982 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-98-5p | 28 | PRRX1 | Sponge network | -0.129 | 0.57177 | 1.316 | 0 | 0.689 |
| 983 | ADAMTS9-AS2 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ITPKB | Sponge network | -2.452 | 0 | -0.704 | 0.00106 | 0.689 |
| 984 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 72 | IL6ST | Sponge network | -2.452 | 0 | -0.402 | 0.00676 | 0.689 |
| 985 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 49 | MCC | Sponge network | -1.111 | 0.0003 | -1.005 | 0 | 0.689 |
| 986 | MYLK-AS1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-940 | 26 | QKI | Sponge network | -1.331 | 3.0E-5 | -0.333 | 0.04111 | 0.689 |
| 987 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 51 | DMD | Sponge network | -1.697 | 0.00889 | -1.506 | 0 | 0.689 |
| 988 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ATRX | Sponge network | 1.308 | 0 | 0.261 | 0.04408 | 0.689 |
| 989 | ACTA2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-532-3p;hsa-miR-542-3p | 18 | SYNM | Sponge network | 0.194 | 0.64278 | -2.309 | 1.0E-5 | 0.689 |
| 990 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p | 12 | SPG20 | Sponge network | -1.092 | 4.0E-5 | -1.16 | 0 | 0.689 |
| 991 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-93-3p | 21 | PCDH10 | Sponge network | -2.69 | 0.0001 | -1.644 | 0.0078 | 0.689 |
| 992 | RP11-554A11.4 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-212-3p;hsa-miR-27a-5p | 12 | ITPKB | Sponge network | -0.583 | 0.14589 | -0.704 | 0.00106 | 0.688 |
| 993 | MYLK-AS1 |
hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p | 11 | AFF3 | Sponge network | -1.331 | 3.0E-5 | -2.373 | 0 | 0.688 |
| 994 | PGM5-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 38 | DTNA | Sponge network | -4.546 | 0 | -1.648 | 1.0E-5 | 0.688 |
| 995 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 43 | HLF | Sponge network | -0.162 | 0.47687 | -2.443 | 0 | 0.688 |
| 996 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DNAJB4 | Sponge network | -0.737 | 0.06182 | -0.7 | 1.0E-5 | 0.688 |
| 997 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 69 | QKI | Sponge network | -1.111 | 0.0003 | -0.333 | 0.04111 | 0.688 |
| 998 | PGM5-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p | 13 | MRVI1 | Sponge network | -4.546 | 0 | -1.051 | 0.00022 | 0.688 |
| 999 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | RECK | Sponge network | -1.697 | 0.00889 | -1.043 | 0 | 0.688 |
| 1000 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429 | 13 | ZNF521 | Sponge network | -0.976 | 0.00025 | -0.111 | 0.58068 | 0.688 |
| 1001 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 48 | RBMS3 | Sponge network | 0.335 | 0.20596 | -0.519 | 0.03551 | 0.688 |
| 1002 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 47 | RBMS3 | Sponge network | -0.323 | 0.01372 | -0.519 | 0.03551 | 0.688 |
| 1003 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | -0.162 | 0.47687 | -1.548 | 0 | 0.688 |
| 1004 | AC003090.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | -2.117 | 0.00161 | -1.733 | 0.00022 | 0.688 |
| 1005 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 83 | CADM2 | Sponge network | -1.111 | 0.0003 | -3.782 | 0 | 0.688 |
| 1006 | MIR497HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | CDH19 | Sponge network | -1.439 | 4.0E-5 | -3.434 | 0 | 0.688 |
| 1007 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92b-3p | 22 | AKAP12 | Sponge network | -1.111 | 0.0003 | -0.932 | 0.0028 | 0.688 |
| 1008 | SNHG14 |
hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | ROR2 | Sponge network | -1.111 | 0.0003 | -1.091 | 0.0008 | 0.688 |
| 1009 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | PLSCR4 | Sponge network | -1.697 | 0.00889 | -0.474 | 0.03833 | 0.688 |
| 1010 | PGM5-AS1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 13 | PCDH10 | Sponge network | -4.546 | 0 | -1.644 | 0.0078 | 0.688 |
| 1011 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 41 | EPHA3 | Sponge network | -1.111 | 0.0003 | 0.185 | 0.53588 | 0.688 |
| 1012 | ARHGEF26-AS1 |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 10 | DPP6 | Sponge network | -2.46 | 0 | -3.777 | 0 | 0.688 |
| 1013 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 22 | AKAP6 | Sponge network | -0.096 | 0.67958 | -1.586 | 3.0E-5 | 0.687 |
| 1014 | GNG12-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | -0.793 | 7.0E-5 | -1.051 | 0.00022 | 0.687 |
| 1015 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-455-3p;hsa-miR-708-5p;hsa-miR-766-3p | 11 | EZH1 | Sponge network | -1.092 | 4.0E-5 | -0.564 | 1.0E-5 | 0.687 |
| 1016 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-935 | 37 | TSHZ3 | Sponge network | -0.162 | 0.47687 | -0.556 | 0.01516 | 0.687 |
| 1017 | PWAR6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 60 | DTNA | Sponge network | -1.575 | 6.0E-5 | -1.648 | 1.0E-5 | 0.687 |
| 1018 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 68 | DTNA | Sponge network | -0.129 | 0.57177 | -1.648 | 1.0E-5 | 0.687 |
| 1019 | GNG12-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p | 11 | PARK2 | Sponge network | -0.793 | 7.0E-5 | -1.892 | 0 | 0.687 |
| 1020 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | ZNF521 | Sponge network | -0.737 | 0.06182 | -0.111 | 0.58068 | 0.687 |
| 1021 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | RASSF8 | Sponge network | -1.575 | 6.0E-5 | -0.122 | 0.65591 | 0.687 |
| 1022 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-589-3p | 13 | SYNPO2 | Sponge network | -4.463 | 0.00025 | -2.342 | 0 | 0.687 |
| 1023 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | CNTNAP1 | Sponge network | -2.452 | 0 | 0.358 | 0.13727 | 0.687 |
| 1024 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 24 | NRXN1 | Sponge network | -1.092 | 4.0E-5 | -3.778 | 0 | 0.687 |
| 1025 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 26 | NR2C2 | Sponge network | -0.222 | 0.32445 | 0.21 | 0.03224 | 0.687 |
| 1026 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-577 | 25 | NRXN1 | Sponge network | -2.69 | 0.0001 | -3.778 | 0 | 0.687 |
| 1027 | GNG12-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 40 | DMD | Sponge network | -0.793 | 7.0E-5 | -1.506 | 0 | 0.687 |
| 1028 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 44 | SSBP2 | Sponge network | -1.439 | 4.0E-5 | -1.58 | 0 | 0.687 |
| 1029 | C20orf166-AS1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577 | 10 | ABCC9 | Sponge network | -2.69 | 0.0001 | -0.466 | 0.14121 | 0.687 |
| 1030 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | DKK3 | Sponge network | -0.49 | 0.04946 | -0.052 | 0.77783 | 0.687 |
| 1031 | MIR143HG |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | -0.739 | 0.0574 | 0.809 | 0.00544 | 0.686 |
| 1032 | LINC00092 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-769-5p | 14 | SETBP1 | Sponge network | -1.886 | 0 | -1.302 | 1.0E-5 | 0.686 |
| 1033 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-590-5p | 11 | PTPN13 | Sponge network | -0.976 | 0.00025 | -1.208 | 0 | 0.686 |
| 1034 | ARHGEF26-AS1 |
hsa-miR-146b-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | FHL1 | Sponge network | -2.46 | 0 | -2.687 | 0 | 0.686 |
| 1035 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-493-5p;hsa-miR-590-3p | 23 | ANK2 | Sponge network | -0.896 | 0.00099 | -1.603 | 1.0E-5 | 0.686 |
| 1036 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p | 13 | AKAP6 | Sponge network | -4.546 | 0 | -1.586 | 3.0E-5 | 0.686 |
| 1037 | FAM66C |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | RNF180 | Sponge network | -0.901 | 0.00019 | -1.127 | 1.0E-5 | 0.686 |
| 1038 | RP11-400K9.4 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-7-5p | 10 | FLNA | Sponge network | -0.611 | 0.09607 | -0.771 | 0.01377 | 0.686 |
| 1039 | AC127904.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | MDM4 | Sponge network | 1.308 | 0 | 0.345 | 0.00762 | 0.686 |
| 1040 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | CNTNAP1 | Sponge network | -1.111 | 0.0003 | 0.358 | 0.13727 | 0.686 |
| 1041 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 35 | EPHA3 | Sponge network | -0.737 | 0.06182 | 0.185 | 0.53588 | 0.686 |
| 1042 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 16 | CNTNAP1 | Sponge network | 0.194 | 0.64278 | 0.358 | 0.13727 | 0.685 |
| 1043 | AC003090.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 49 | DTNA | Sponge network | -2.117 | 0.00161 | -1.648 | 1.0E-5 | 0.685 |
| 1044 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | PDZRN3 | Sponge network | -0.49 | 0.04946 | -1.548 | 0 | 0.685 |
| 1045 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 35 | EPHA3 | Sponge network | -1.697 | 0.00889 | 0.185 | 0.53588 | 0.685 |
| 1046 | MIR497HG |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-1-3p | 48 | DTNA | Sponge network | -1.439 | 4.0E-5 | -1.648 | 1.0E-5 | 0.685 |
| 1047 | MIR497HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-5p | 38 | DMD | Sponge network | -1.439 | 4.0E-5 | -1.506 | 0 | 0.685 |
| 1048 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p | 12 | PTGFR | Sponge network | -0.583 | 0.14589 | -0.233 | 0.45421 | 0.685 |
| 1049 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p | 29 | QKI | Sponge network | -0.473 | 0.07602 | -0.333 | 0.04111 | 0.685 |
| 1050 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-424-5p | 26 | EPHA7 | Sponge network | -0.932 | 0.00329 | -2.197 | 3.0E-5 | 0.685 |
| 1051 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 10 | PEG3 | Sponge network | -1.582 | 8.0E-5 | -1.74 | 0 | 0.685 |
| 1052 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | ABCC9 | Sponge network | -1.575 | 6.0E-5 | -0.466 | 0.14121 | 0.685 |
| 1053 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-598-3p;hsa-miR-664a-3p | 14 | MN1 | Sponge network | 0.997 | 0.00066 | -0.358 | 0.24172 | 0.685 |
| 1054 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | LSAMP | Sponge network | -0.129 | 0.57177 | -0.066 | 0.81708 | 0.684 |
| 1055 | ADAMTS9-AS2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | KCTD8 | Sponge network | -2.452 | 0 | -3.359 | 0 | 0.684 |
| 1056 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 14 | PARK2 | Sponge network | -0.739 | 0.0574 | -1.892 | 0 | 0.684 |
| 1057 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | FAT4 | Sponge network | -2.452 | 0 | -0.354 | 0.14694 | 0.684 |
| 1058 | ADAMTS9-AS2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-7-1-3p | 32 | WWTR1 | Sponge network | -2.452 | 0 | -0.345 | 0.11997 | 0.684 |
| 1059 | MYLK-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-34a-5p | 14 | NAV3 | Sponge network | -1.331 | 3.0E-5 | 0.212 | 0.47729 | 0.684 |
| 1060 | RP11-554A11.4 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p | 12 | SFRP1 | Sponge network | -0.583 | 0.14589 | -3.566 | 0 | 0.684 |
| 1061 | FENDRR |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | -1.281 | 0.00012 | -3.143 | 0 | 0.684 |
| 1062 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-7-1-3p | 12 | CDH19 | Sponge network | -1.092 | 4.0E-5 | -3.434 | 0 | 0.684 |
| 1063 | SNHG14 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | TLL1 | Sponge network | -1.111 | 0.0003 | -1.195 | 3.0E-5 | 0.684 |
| 1064 | RP11-819C21.1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p | 18 | ATM | Sponge network | 0.537 | 0.00139 | 0.178 | 0.21568 | 0.684 |
| 1065 | MIR143HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-98-5p | 11 | ADAMTS8 | Sponge network | -0.739 | 0.0574 | -1.476 | 0.00069 | 0.684 |
| 1066 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 48 | ADARB1 | Sponge network | -0.096 | 0.67958 | -0.335 | 0.06631 | 0.683 |
| 1067 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-505-3p;hsa-miR-590-3p | 21 | JAKMIP2 | Sponge network | -0.739 | 0.0574 | -1.388 | 0 | 0.683 |
| 1068 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ADAMTSL3 | Sponge network | -1.575 | 6.0E-5 | -1.559 | 0 | 0.683 |
| 1069 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | PLSCR4 | Sponge network | -1.111 | 0.0003 | -0.474 | 0.03833 | 0.683 |
| 1070 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TLL1 | Sponge network | -0.129 | 0.57177 | -1.195 | 3.0E-5 | 0.683 |
| 1071 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940;hsa-miR-98-5p | 45 | PBX1 | Sponge network | -2.452 | 0 | -1.206 | 0 | 0.683 |
| 1072 | ARHGEF26-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p | 20 | SYNM | Sponge network | -2.46 | 0 | -2.309 | 1.0E-5 | 0.683 |
| 1073 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-629-3p | 29 | EPHA7 | Sponge network | -0.583 | 0.14589 | -2.197 | 3.0E-5 | 0.683 |
| 1074 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 56 | AR | Sponge network | -0.901 | 0.00019 | -1.554 | 0 | 0.683 |
| 1075 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 68 | NTRK3 | Sponge network | -0.976 | 0.00025 | -1.77 | 3.0E-5 | 0.683 |
| 1076 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-628-5p | 10 | CAV1 | Sponge network | -0.583 | 0.14589 | -1.013 | 6.0E-5 | 0.683 |
| 1077 | PWAR6 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 22 | SORCS1 | Sponge network | -1.575 | 6.0E-5 | -3.492 | 0 | 0.683 |
| 1078 | ADAMTS9-AS2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-942-5p;hsa-miR-96-5p | 16 | ZBTB20 | Sponge network | -2.452 | 0 | -0.122 | 0.57569 | 0.683 |
| 1079 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 38 | MYO9A | Sponge network | -0.222 | 0.32445 | -0.147 | 0.32373 | 0.683 |
| 1080 | MIR497HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -1.439 | 4.0E-5 | 0.024 | 0.89889 | 0.683 |
| 1081 | BVES-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p | 13 | SFRP1 | Sponge network | -2.62 | 0 | -3.566 | 0 | 0.683 |
| 1082 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-335-3p;hsa-miR-34a-5p | 11 | AKAP6 | Sponge network | -0.691 | 0.00146 | -1.586 | 3.0E-5 | 0.683 |
| 1083 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b | 15 | ADAMTSL3 | Sponge network | -0.583 | 0.14589 | -1.559 | 0 | 0.682 |
| 1084 | RP11-554A11.4 |
hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-450b-5p | 14 | PCDH10 | Sponge network | -0.583 | 0.14589 | -1.644 | 0.0078 | 0.682 |
| 1085 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-940 | 16 | PEG3 | Sponge network | -0.162 | 0.47687 | -1.74 | 0 | 0.682 |
| 1086 | LINC00843 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p | 29 | MDM4 | Sponge network | 1.106 | 0 | 0.345 | 0.00762 | 0.682 |
| 1087 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p | 27 | ZFHX4 | Sponge network | -0.583 | 0.14589 | 0.018 | 0.95574 | 0.682 |
| 1088 | MYLK-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-484;hsa-miR-7-5p | 14 | PRKCB | Sponge network | -1.331 | 3.0E-5 | -1.313 | 2.0E-5 | 0.682 |
| 1089 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 54 | ZFHX4 | Sponge network | -0.737 | 0.06182 | 0.018 | 0.95574 | 0.682 |
| 1090 | CTC-510F12.2 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-935 | 27 | LPP | Sponge network | -0.691 | 0.00146 | -0.459 | 0.04475 | 0.682 |
| 1091 | LINC00702 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 34 | DLC1 | Sponge network | -0.737 | 0.06182 | -0.382 | 0.05038 | 0.682 |
| 1092 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 23 | RCAN2 | Sponge network | -1.697 | 0.00889 | -0.33 | 0.15172 | 0.682 |
| 1093 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 35 | SCN9A | Sponge network | -0.129 | 0.57177 | -0.525 | 0.10593 | 0.682 |
| 1094 | RP5-1198O20.4 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p | 10 | PRKD1 | Sponge network | 0.997 | 0.00066 | -0.466 | 0.03583 | 0.682 |
| 1095 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-590-3p | 13 | TLL1 | Sponge network | -0.358 | 0.17255 | -1.195 | 3.0E-5 | 0.681 |
| 1096 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-550a-3p | 21 | PGR | Sponge network | -4.546 | 0 | -1.704 | 0 | 0.681 |
| 1097 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-629-3p | 31 | ZBTB4 | Sponge network | -0.576 | 0.10121 | -0.739 | 0 | 0.681 |
| 1098 | PWAR6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 97 | LPP | Sponge network | -1.575 | 6.0E-5 | -0.459 | 0.04475 | 0.681 |
| 1099 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 12 | ABCC9 | Sponge network | -0.611 | 0.09607 | -0.466 | 0.14121 | 0.681 |
| 1100 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLNA | Sponge network | 0.997 | 0.00066 | -0.771 | 0.01377 | 0.681 |
| 1101 | MIR143HG |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 17 | STAT5B | Sponge network | -0.739 | 0.0574 | -0.182 | 0.1082 | 0.681 |
| 1102 | SNHG14 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 28 | CDH19 | Sponge network | -1.111 | 0.0003 | -3.434 | 0 | 0.681 |
| 1103 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-935 | 43 | FAT3 | Sponge network | -2.62 | 0 | -1.601 | 0.00051 | 0.681 |
| 1104 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-628-5p | 12 | TRIM33 | Sponge network | 1.923 | 0 | 0.402 | 7.0E-5 | 0.681 |
| 1105 | MRVI1-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-7-1-3p | 11 | ITPKB | Sponge network | -1.092 | 4.0E-5 | -0.704 | 0.00106 | 0.681 |
| 1106 | RP11-389C8.2 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 15 | FMNL3 | Sponge network | 0.727 | 3.0E-5 | 0.555 | 4.0E-5 | 0.681 |
| 1107 | FENDRR |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | PRUNE2 | Sponge network | -1.281 | 0.00012 | -1.733 | 0.00022 | 0.681 |
| 1108 | LINC00702 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-625-5p | 11 | NGFR | Sponge network | -0.737 | 0.06182 | -1.271 | 0.01058 | 0.681 |
| 1109 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-2355-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | NUDT11 | Sponge network | -0.49 | 0.04946 | -0.921 | 0.00376 | 0.68 |
| 1110 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 33 | ANK2 | Sponge network | -0.901 | 0.00019 | -1.603 | 1.0E-5 | 0.68 |
| 1111 | LINC00641 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 92 | LPP | Sponge network | -0.222 | 0.32445 | -0.459 | 0.04475 | 0.68 |
| 1112 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | JAKMIP2 | Sponge network | -0.49 | 0.04946 | -1.388 | 0 | 0.68 |
| 1113 | PWAR6 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 14 | ZNF382 | Sponge network | -1.575 | 6.0E-5 | -0.494 | 0.03831 | 0.68 |
| 1114 | FENDRR |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 22 | NRXN3 | Sponge network | -1.281 | 0.00012 | -0.893 | 0.04186 | 0.68 |
| 1115 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-577 | 14 | AMPH | Sponge network | -0.49 | 0.04946 | -0.916 | 5.0E-5 | 0.68 |
| 1116 | HAND2-AS1 |
hsa-miR-185-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-542-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-940 | 10 | ILK | Sponge network | -1.697 | 0.00889 | -0.74 | 0 | 0.68 |
| 1117 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 58 | RUNX1T1 | Sponge network | 0.194 | 0.64278 | -0.344 | 0.2374 | 0.68 |
| 1118 | RP11-166D19.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-96-5p | 101 | LPP | Sponge network | -0.576 | 0.10121 | -0.459 | 0.04475 | 0.68 |
| 1119 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 44 | RBMS3 | Sponge network | -0.901 | 0.00019 | -0.519 | 0.03551 | 0.68 |
| 1120 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 14 | NRK | Sponge network | -0.583 | 0.14589 | 0.656 | 0.19217 | 0.68 |
| 1121 | MYLK-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-940 | 27 | PBX1 | Sponge network | -1.331 | 3.0E-5 | -1.206 | 0 | 0.68 |
| 1122 | HAND2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | NAV3 | Sponge network | -1.697 | 0.00889 | 0.212 | 0.47729 | 0.68 |
| 1123 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | RCAN2 | Sponge network | -0.612 | 0.00094 | -0.33 | 0.15172 | 0.679 |
| 1124 | ADAMTS9-AS2 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 13 | PRKD1 | Sponge network | -2.452 | 0 | -0.466 | 0.03583 | 0.679 |
| 1125 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 32 | GPC6 | Sponge network | -0.129 | 0.57177 | -0.054 | 0.81465 | 0.679 |
| 1126 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 17 | DKK3 | Sponge network | -0.976 | 0.00025 | -0.052 | 0.77783 | 0.679 |
| 1127 | MIR497HG |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-542-3p | 10 | PRKD1 | Sponge network | -1.439 | 4.0E-5 | -0.466 | 0.03583 | 0.679 |
| 1128 | PWAR6 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p | 20 | MRVI1 | Sponge network | -1.575 | 6.0E-5 | -1.051 | 0.00022 | 0.679 |
| 1129 | RP11-166D19.1 |
hsa-miR-185-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-940 | 10 | ILK | Sponge network | -0.576 | 0.10121 | -0.74 | 0 | 0.679 |
| 1130 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 31 | THBS1 | Sponge network | -0.576 | 0.10121 | 0.741 | 0.00386 | 0.679 |
| 1131 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 28 | PGR | Sponge network | 0.572 | 0.01722 | -1.704 | 0 | 0.679 |
| 1132 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-93-5p | 43 | EPHA7 | Sponge network | -0.162 | 0.47687 | -2.197 | 3.0E-5 | 0.679 |
| 1133 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p | 13 | AMPH | Sponge network | -0.576 | 0.10121 | -0.916 | 5.0E-5 | 0.679 |
| 1134 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429 | 20 | DYNC1I1 | Sponge network | -0.976 | 0.00025 | -1.12 | 0.00107 | 0.679 |
| 1135 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | PCDH9 | Sponge network | -0.896 | 0.00099 | -1.823 | 4.0E-5 | 0.679 |
| 1136 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 45 | NRXN1 | Sponge network | -1.111 | 0.0003 | -3.778 | 0 | 0.678 |
| 1137 | BVES-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429 | 14 | PCDH9 | Sponge network | -2.62 | 0 | -1.823 | 4.0E-5 | 0.678 |
| 1138 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | PRUNE2 | Sponge network | -0.49 | 0.04946 | -1.733 | 0.00022 | 0.678 |
| 1139 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -0.49 | 0.04946 | -2.046 | 0.00019 | 0.678 |
| 1140 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577 | 25 | CNTN1 | Sponge network | -2.69 | 0.0001 | -2.594 | 0 | 0.678 |
| 1141 | MEG3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | RASSF8 | Sponge network | 0.249 | 0.35902 | -0.122 | 0.65591 | 0.678 |
| 1142 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | AFF3 | Sponge network | -1.111 | 0.0003 | -2.373 | 0 | 0.678 |
| 1143 | LINC00578 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-664a-3p | 14 | NRXN3 | Sponge network | 0.811 | 0.03228 | -0.893 | 0.04186 | 0.678 |
| 1144 | MIR143HG |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 23 | SPG20 | Sponge network | -0.739 | 0.0574 | -1.16 | 0 | 0.678 |
| 1145 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 81 | NTRK3 | Sponge network | -0.129 | 0.57177 | -1.77 | 3.0E-5 | 0.678 |
| 1146 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 33 | TIMP3 | Sponge network | -0.739 | 0.0574 | -0.546 | 0.02307 | 0.678 |
| 1147 | LINC00702 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-361-5p;hsa-miR-532-5p;hsa-miR-582-5p | 10 | PRICKLE1 | Sponge network | -0.737 | 0.06182 | -0.112 | 0.67088 | 0.678 |
| 1148 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | NRXN3 | Sponge network | 0.194 | 0.64278 | -0.893 | 0.04186 | 0.678 |
| 1149 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p | 27 | ZFHX4 | Sponge network | -1.331 | 3.0E-5 | 0.018 | 0.95574 | 0.678 |
| 1150 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 26 | NRK | Sponge network | 0.997 | 0.00066 | 0.656 | 0.19217 | 0.677 |
| 1151 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 12 | SPG20 | Sponge network | -1.582 | 8.0E-5 | -1.16 | 0 | 0.677 |
| 1152 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-486-5p | 20 | DYNC1I1 | Sponge network | -0.162 | 0.47687 | -1.12 | 0.00107 | 0.677 |
| 1153 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | CAV1 | Sponge network | -2.452 | 0 | -1.013 | 6.0E-5 | 0.677 |
| 1154 | BZRAP1-AS1 |
hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-589-5p;hsa-miR-7-5p | 21 | PRKCB | Sponge network | -0.465 | 0.04703 | -1.313 | 2.0E-5 | 0.677 |
| 1155 | LINC00702 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 13 | AMPH | Sponge network | -0.737 | 0.06182 | -0.916 | 5.0E-5 | 0.677 |
| 1156 | MIR143HG |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 22 | SORCS1 | Sponge network | -0.739 | 0.0574 | -3.492 | 0 | 0.677 |
| 1157 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | DNAJB4 | Sponge network | -0.739 | 0.0574 | -0.7 | 1.0E-5 | 0.677 |
| 1158 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 42 | MCC | Sponge network | -0.49 | 0.04946 | -1.005 | 0 | 0.677 |
| 1159 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-744-3p;hsa-miR-935 | 41 | DIP2C | Sponge network | -0.162 | 0.47687 | -0.436 | 0.01713 | 0.677 |
| 1160 | MIR497HG |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | PRKCB | Sponge network | -1.439 | 4.0E-5 | -1.313 | 2.0E-5 | 0.677 |
| 1161 | MRVI1-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-671-5p | 32 | GAS7 | Sponge network | -1.092 | 4.0E-5 | -0.555 | 0.01602 | 0.677 |
| 1162 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-628-5p | 38 | RUNX1T1 | Sponge network | 0.811 | 0.03228 | -0.344 | 0.2374 | 0.676 |
| 1163 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-5p | 34 | BCL2 | Sponge network | -0.465 | 0.04703 | -0.899 | 0 | 0.676 |
| 1164 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 31 | EPHA3 | Sponge network | -0.976 | 0.00025 | 0.185 | 0.53588 | 0.676 |
| 1165 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p | 28 | PCDH9 | Sponge network | -0.976 | 0.00025 | -1.823 | 4.0E-5 | 0.676 |
| 1166 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p | 21 | MCC | Sponge network | -0.583 | 0.14589 | -1.005 | 0 | 0.676 |
| 1167 | LINC00702 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | TLL1 | Sponge network | -0.737 | 0.06182 | -1.195 | 3.0E-5 | 0.676 |
| 1168 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-769-5p | 11 | RCAN2 | Sponge network | -0.583 | 0.14589 | -0.33 | 0.15172 | 0.676 |
| 1169 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | ZNF521 | Sponge network | -0.739 | 0.0574 | -0.111 | 0.58068 | 0.676 |
| 1170 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 13 | ABCC9 | Sponge network | -0.896 | 0.00099 | -0.466 | 0.14121 | 0.676 |
| 1171 | FENDRR |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-874-3p;hsa-miR-98-5p | 10 | HSPB7 | Sponge network | -1.281 | 0.00012 | -2.268 | 1.0E-5 | 0.676 |
| 1172 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 18 | GLI3 | Sponge network | 0.997 | 0.00066 | -0.615 | 0.01482 | 0.676 |
| 1173 | RP11-532F6.3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-590-3p | 20 | PRKCB | Sponge network | -0.896 | 0.00099 | -1.313 | 2.0E-5 | 0.676 |
| 1174 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | PGR | Sponge network | -0.901 | 0.00019 | -1.704 | 0 | 0.676 |
| 1175 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 40 | JAZF1 | Sponge network | -0.129 | 0.57177 | -0.377 | 0.02677 | 0.676 |
| 1176 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 40 | NRXN1 | Sponge network | -0.576 | 0.10121 | -3.778 | 0 | 0.675 |
| 1177 | MYLK-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-671-5p;hsa-miR-940 | 33 | GAS7 | Sponge network | -1.331 | 3.0E-5 | -0.555 | 0.01602 | 0.675 |
| 1178 | MAGI2-AS3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ITPKB | Sponge network | -0.129 | 0.57177 | -0.704 | 0.00106 | 0.675 |
| 1179 | GNG12-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p | 30 | AKAP6 | Sponge network | -0.793 | 7.0E-5 | -1.586 | 3.0E-5 | 0.675 |
| 1180 | ZNF667-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 29 | NAV3 | Sponge network | -0.976 | 0.00025 | 0.212 | 0.47729 | 0.675 |
| 1181 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-98-5p | 13 | SCN11A | Sponge network | -2.452 | 0 | -1.15 | 0.00299 | 0.675 |
| 1182 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 45 | DMD | Sponge network | -0.49 | 0.04946 | -1.506 | 0 | 0.675 |
| 1183 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-452-5p;hsa-miR-590-3p | 29 | MEF2C | Sponge network | -0.358 | 0.17255 | -0.499 | 0.01448 | 0.675 |
| 1184 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p | 46 | BNC2 | Sponge network | -0.162 | 0.47687 | -0.491 | 0.09988 | 0.675 |
| 1185 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-93-5p | 15 | PGR | Sponge network | -2.531 | 0 | -1.704 | 0 | 0.675 |
| 1186 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 13 | PARK2 | Sponge network | -2.46 | 0 | -1.892 | 0 | 0.675 |
| 1187 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | PRUNE2 | Sponge network | -1.439 | 4.0E-5 | -1.733 | 0.00022 | 0.675 |
| 1188 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p | 17 | FGFR1 | Sponge network | -0.739 | 0.00044 | -0.509 | 0.03957 | 0.675 |
| 1189 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 12 | GAS1 | Sponge network | -0.739 | 0.0574 | -1.047 | 0.00364 | 0.675 |
| 1190 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 25 | EBF3 | Sponge network | 0.572 | 0.01722 | -0.058 | 0.81437 | 0.675 |
| 1191 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 41 | FBXO32 | Sponge network | -2.452 | 0 | -0.495 | 0.07561 | 0.675 |
| 1192 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940 | 47 | HLF | Sponge network | -2.46 | 0 | -2.443 | 0 | 0.674 |
| 1193 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 12 | CNTN1 | Sponge network | -0.583 | 0.14589 | -2.594 | 0 | 0.674 |
| 1194 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 32 | PGR | Sponge network | 0.194 | 0.64278 | -1.704 | 0 | 0.674 |
| 1195 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 20 | JAKMIP2 | Sponge network | -0.129 | 0.57177 | -1.388 | 0 | 0.674 |
| 1196 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-671-5p | 19 | CADM3 | Sponge network | -0.976 | 0.00025 | -3.663 | 0 | 0.674 |
| 1197 | MIR143HG |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-5p | 14 | THSD7A | Sponge network | -0.739 | 0.0574 | 0.242 | 0.25154 | 0.674 |
| 1198 | HAND2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 65 | DTNA | Sponge network | -1.697 | 0.00889 | -1.648 | 1.0E-5 | 0.674 |
| 1199 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | CAV1 | Sponge network | -0.576 | 0.10121 | -1.013 | 6.0E-5 | 0.674 |
| 1200 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-769-5p | 32 | SYNPO2 | Sponge network | -2.46 | 0 | -2.342 | 0 | 0.674 |
| 1201 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | PLSCR4 | Sponge network | -1.439 | 4.0E-5 | -0.474 | 0.03833 | 0.674 |
| 1202 | PWAR6 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | ZDBF2 | Sponge network | -1.575 | 6.0E-5 | -0.998 | 0.00025 | 0.674 |
| 1203 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-542-3p | 20 | PCDH9 | Sponge network | -2.69 | 0.0001 | -1.823 | 4.0E-5 | 0.674 |
| 1204 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | DKK3 | Sponge network | -0.129 | 0.57177 | -0.052 | 0.77783 | 0.674 |
| 1205 | RP11-401P9.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-877-5p;hsa-miR-93-5p | 33 | IL17RD | Sponge network | -0.384 | 0.40652 | -0.019 | 0.94873 | 0.674 |
| 1206 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 11 | SYNE1 | Sponge network | -0.473 | 0.07602 | -0.738 | 0.00316 | 0.674 |
| 1207 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 67 | RUNX1T1 | Sponge network | 0.997 | 0.00066 | -0.344 | 0.2374 | 0.674 |
| 1208 | MIR497HG |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-3913-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-942-5p | 35 | IGFBP5 | Sponge network | -1.439 | 4.0E-5 | -0.806 | 0.00105 | 0.674 |
| 1209 | LINC00578 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-511-5p | 14 | MRVI1 | Sponge network | 0.811 | 0.03228 | -1.051 | 0.00022 | 0.674 |
| 1210 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-590-3p | 15 | PLSCR4 | Sponge network | -0.358 | 0.17255 | -0.474 | 0.03833 | 0.674 |
| 1211 | LINC00702 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 21 | CADM3 | Sponge network | -0.737 | 0.06182 | -3.663 | 0 | 0.674 |
| 1212 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 19 | SLITRK3 | Sponge network | -2.452 | 0 | -3.328 | 0 | 0.674 |
| 1213 | MIR497HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 25 | AKAP6 | Sponge network | -1.439 | 4.0E-5 | -1.586 | 3.0E-5 | 0.673 |
| 1214 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-34a-5p | 37 | PRKAA2 | Sponge network | -1.331 | 3.0E-5 | -2.462 | 0 | 0.673 |
| 1215 | SNHG1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-195-5p;hsa-miR-23b-3p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-497-5p;hsa-miR-654-3p | 10 | HNRNPA2B1 | Sponge network | 1.265 | 0 | 0.55 | 0 | 0.673 |
| 1216 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 58 | BNC2 | Sponge network | -1.575 | 6.0E-5 | -0.491 | 0.09988 | 0.673 |
| 1217 | RP5-1198O20.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | NAV3 | Sponge network | 0.997 | 0.00066 | 0.212 | 0.47729 | 0.673 |
| 1218 | FAM66C |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | PEG3 | Sponge network | -0.901 | 0.00019 | -1.74 | 0 | 0.673 |
| 1219 | RP11-894P9.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-582-5p | 11 | NR2C2 | Sponge network | 0.993 | 0 | 0.21 | 0.03224 | 0.673 |
| 1220 | RP11-166D19.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-877-5p;hsa-miR-96-5p | 34 | ASTN1 | Sponge network | -0.576 | 0.10121 | -2.389 | 2.0E-5 | 0.673 |
| 1221 | MIR497HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | EPHA7 | Sponge network | -1.439 | 4.0E-5 | -2.197 | 3.0E-5 | 0.673 |
| 1222 | C20orf166-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-484;hsa-miR-542-3p;hsa-miR-625-5p | 15 | SFRP1 | Sponge network | -2.69 | 0.0001 | -3.566 | 0 | 0.673 |
| 1223 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940 | 49 | MEF2C | Sponge network | -0.739 | 0.0574 | -0.499 | 0.01448 | 0.673 |
| 1224 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 25 | NCAM2 | Sponge network | -0.49 | 0.04946 | -0.482 | 0.22565 | 0.673 |
| 1225 | MIR143HG |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | TPO | Sponge network | -0.739 | 0.0574 | -1.87 | 2.0E-5 | 0.673 |
| 1226 | ADAMTS9-AS2 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | TPO | Sponge network | -2.452 | 0 | -1.87 | 2.0E-5 | 0.673 |
| 1227 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | TLL1 | Sponge network | -0.976 | 0.00025 | -1.195 | 3.0E-5 | 0.673 |
| 1228 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | RECK | Sponge network | 0.335 | 0.20596 | -1.043 | 0 | 0.673 |
| 1229 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | TIMP3 | Sponge network | -0.976 | 0.00025 | -0.546 | 0.02307 | 0.672 |
| 1230 | BVES-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429 | 10 | PARK2 | Sponge network | -2.62 | 0 | -1.892 | 0 | 0.672 |
| 1231 | MEG3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | PEG3 | Sponge network | 0.249 | 0.35902 | -1.74 | 0 | 0.672 |
| 1232 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | TIMP3 | Sponge network | -0.737 | 0.06182 | -0.546 | 0.02307 | 0.672 |
| 1233 | LINC00578 |
hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577 | 19 | TSHZ3 | Sponge network | 0.811 | 0.03228 | -0.556 | 0.01516 | 0.672 |
| 1234 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-511-5p;hsa-miR-7-1-3p | 15 | ADAMTSL3 | Sponge network | -4.546 | 0 | -1.559 | 0 | 0.672 |
| 1235 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 42 | MEF2C | Sponge network | -0.976 | 0.00025 | -0.499 | 0.01448 | 0.672 |
| 1236 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | SLITRK3 | Sponge network | -0.739 | 0.0574 | -3.328 | 0 | 0.672 |
| 1237 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | DCN | Sponge network | -1.697 | 0.00889 | -1.288 | 0 | 0.672 |
| 1238 | RP5-1198O20.4 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 20 | MRVI1 | Sponge network | 0.997 | 0.00066 | -1.051 | 0.00022 | 0.672 |
| 1239 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 58 | QKI | Sponge network | -0.49 | 0.04946 | -0.333 | 0.04111 | 0.672 |
| 1240 | SNHG14 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 14 | PRKD1 | Sponge network | -1.111 | 0.0003 | -0.466 | 0.03583 | 0.672 |
| 1241 | RP4-717I23.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 48 | MDM4 | Sponge network | 0.637 | 0.00069 | 0.345 | 0.00762 | 0.672 |
| 1242 | ADAMTS9-AS2 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-744-3p;hsa-miR-940 | 16 | RHOB | Sponge network | -2.452 | 0 | -1.052 | 0 | 0.671 |
| 1243 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-98-5p | 15 | SCN11A | Sponge network | -1.111 | 0.0003 | -1.15 | 0.00299 | 0.671 |
| 1244 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-877-5p | 34 | RBMS3 | Sponge network | -2.62 | 0 | -0.519 | 0.03551 | 0.671 |
| 1245 | RP11-736K20.5 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p | 13 | SFRP1 | Sponge network | -0.358 | 0.17255 | -3.566 | 0 | 0.671 |
| 1246 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | FAT4 | Sponge network | -0.49 | 0.04946 | -0.354 | 0.14694 | 0.671 |
| 1247 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p | 16 | TXNIP | Sponge network | -2.452 | 0 | -1.277 | 0 | 0.671 |
| 1248 | MIR143HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 59 | GAS7 | Sponge network | -0.739 | 0.0574 | -0.555 | 0.01602 | 0.671 |
| 1249 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 45 | FBXO32 | Sponge network | -0.49 | 0.04946 | -0.495 | 0.07561 | 0.671 |
| 1250 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-935 | 56 | SSBP2 | Sponge network | -0.576 | 0.10121 | -1.58 | 0 | 0.671 |
| 1251 | FAM66C |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 31 | SETBP1 | Sponge network | -0.901 | 0.00019 | -1.302 | 1.0E-5 | 0.671 |
| 1252 | MIR497HG |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 16 | GLI3 | Sponge network | -1.439 | 4.0E-5 | -0.615 | 0.01482 | 0.671 |
| 1253 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | DKK3 | Sponge network | -0.576 | 0.10121 | -0.052 | 0.77783 | 0.671 |
| 1254 | MRVI1-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-671-5p;hsa-miR-874-3p | 14 | SORCS1 | Sponge network | -1.092 | 4.0E-5 | -3.492 | 0 | 0.671 |
| 1255 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 21 | FNBP1 | Sponge network | -0.737 | 0.06182 | -0.969 | 4.0E-5 | 0.671 |
| 1256 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 49 | TSHZ3 | Sponge network | -1.111 | 0.0003 | -0.556 | 0.01516 | 0.671 |
| 1257 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p | 16 | DMD | Sponge network | -4.463 | 0.00025 | -1.506 | 0 | 0.671 |
| 1258 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | PLSCR4 | Sponge network | 0.572 | 0.01722 | -0.474 | 0.03833 | 0.671 |
| 1259 | RP11-244O19.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-93-5p | 18 | PRUNE2 | Sponge network | -0.096 | 0.67958 | -1.733 | 0.00022 | 0.671 |
| 1260 | MRVI1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-98-5p | 11 | ITGB3 | Sponge network | -1.092 | 4.0E-5 | -0.011 | 0.95751 | 0.671 |
| 1261 | MIR143HG |
hsa-miR-185-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-940 | 10 | ILK | Sponge network | -0.739 | 0.0574 | -0.74 | 0 | 0.67 |
| 1262 | PWAR6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | NRXN3 | Sponge network | -1.575 | 6.0E-5 | -0.893 | 0.04186 | 0.67 |
| 1263 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-940 | 29 | MDM4 | Sponge network | 0.993 | 0 | 0.345 | 0.00762 | 0.67 |
| 1264 | PWAR6 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-532-3p | 19 | SYNM | Sponge network | -1.575 | 6.0E-5 | -2.309 | 1.0E-5 | 0.67 |
| 1265 | SNHG14 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 23 | RELN | Sponge network | -1.111 | 0.0003 | -2.403 | 0 | 0.67 |
| 1266 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-577;hsa-miR-940 | 28 | IGF1 | Sponge network | -1.439 | 4.0E-5 | -0.204 | 0.5898 | 0.67 |
| 1267 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 12 | HIC1 | Sponge network | -0.576 | 0.10121 | 0.024 | 0.89889 | 0.67 |
| 1268 | RP11-244O19.1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-935 | 14 | STAT5B | Sponge network | -0.096 | 0.67958 | -0.182 | 0.1082 | 0.67 |
| 1269 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-96-5p | 26 | SETBP1 | Sponge network | -0.563 | 0.02545 | -1.302 | 1.0E-5 | 0.67 |
| 1270 | ADAMTS9-AS2 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | PRKCB | Sponge network | -2.452 | 0 | -1.313 | 2.0E-5 | 0.67 |
| 1271 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 40 | JAZF1 | Sponge network | -0.49 | 0.04946 | -0.377 | 0.02677 | 0.67 |
| 1272 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-766-3p | 39 | IL6ST | Sponge network | -1.331 | 3.0E-5 | -0.402 | 0.00676 | 0.67 |
| 1273 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 78 | TCF4 | Sponge network | -0.739 | 0.0574 | 0.016 | 0.92271 | 0.67 |
| 1274 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 41 | MYO9A | Sponge network | -0.739 | 0.0574 | -0.147 | 0.32373 | 0.669 |
| 1275 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 36 | PGR | Sponge network | -2.46 | 0 | -1.704 | 0 | 0.669 |
| 1276 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | NRXN3 | Sponge network | -0.896 | 0.00099 | -0.893 | 0.04186 | 0.669 |
| 1277 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 21 | SYNPO2 | Sponge network | -0.358 | 0.17255 | -2.342 | 0 | 0.669 |
| 1278 | MALAT1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RBBP6 | Sponge network | 0.782 | 0.00016 | 0.163 | 0.05101 | 0.669 |
| 1279 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-96-5p | 40 | FBXO32 | Sponge network | 0.194 | 0.64278 | -0.495 | 0.07561 | 0.669 |
| 1280 | FENDRR |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 34 | AKAP6 | Sponge network | -1.281 | 0.00012 | -1.586 | 3.0E-5 | 0.669 |
| 1281 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | ANK2 | Sponge network | -2.46 | 0 | -1.603 | 1.0E-5 | 0.669 |
| 1282 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-96-5p | 33 | NTRK3 | Sponge network | -1.582 | 8.0E-5 | -1.77 | 3.0E-5 | 0.669 |
| 1283 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-93-5p | 28 | PGR | Sponge network | -0.611 | 0.09607 | -1.704 | 0 | 0.669 |
| 1284 | SNHG14 |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 11 | DPP6 | Sponge network | -1.111 | 0.0003 | -3.777 | 0 | 0.669 |
| 1285 | EMX2OS |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-664a-3p | 13 | MN1 | Sponge network | -0.777 | 0.1134 | -0.358 | 0.24172 | 0.669 |
| 1286 | SNHG14 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | -1.111 | 0.0003 | -1.628 | 0 | 0.669 |
| 1287 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | -0.737 | 0.06182 | -1.047 | 0.00364 | 0.669 |
| 1288 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 70 | CADM2 | Sponge network | -1.575 | 6.0E-5 | -3.782 | 0 | 0.669 |
| 1289 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 12 | GAS1 | Sponge network | -0.49 | 0.04946 | -1.047 | 0.00364 | 0.669 |
| 1290 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-501-5p | 14 | MN1 | Sponge network | -2.452 | 0 | -0.358 | 0.24172 | 0.669 |
| 1291 | FENDRR |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PREX2 | Sponge network | -1.281 | 0.00012 | -0.272 | 0.25382 | 0.669 |
| 1292 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 22 | TRIM33 | Sponge network | 1.153 | 0 | 0.402 | 7.0E-5 | 0.669 |
| 1293 | RP11-159D12.2 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p | 10 | ZNF638 | Sponge network | 1.254 | 0 | 0.22 | 0.01214 | 0.669 |
| 1294 | PWAR6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-766-3p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -1.575 | 6.0E-5 | -0.564 | 1.0E-5 | 0.669 |
| 1295 | ZNF667-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 18 | CXCL12 | Sponge network | -0.976 | 0.00025 | -1.421 | 0 | 0.668 |
| 1296 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 18 | EZH1 | Sponge network | -0.096 | 0.67958 | -0.564 | 1.0E-5 | 0.668 |
| 1297 | RP11-554A11.4 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-628-5p | 19 | EPHA3 | Sponge network | -0.583 | 0.14589 | 0.185 | 0.53588 | 0.668 |
| 1298 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-98-5p | 11 | ADAMTS8 | Sponge network | -0.49 | 0.04946 | -1.476 | 0.00069 | 0.668 |
| 1299 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p | 48 | RBMS3 | Sponge network | -0.162 | 0.47687 | -0.519 | 0.03551 | 0.668 |
| 1300 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | CAV1 | Sponge network | -0.49 | 0.04946 | -1.013 | 6.0E-5 | 0.668 |
| 1301 | GNG12-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 58 | DTNA | Sponge network | -0.793 | 7.0E-5 | -1.648 | 1.0E-5 | 0.668 |
| 1302 | RP11-244O19.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-532-3p | 15 | SYNM | Sponge network | -0.096 | 0.67958 | -2.309 | 1.0E-5 | 0.668 |
| 1303 | LINC00702 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 12 | THSD7A | Sponge network | -0.737 | 0.06182 | 0.242 | 0.25154 | 0.668 |
| 1304 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b | 10 | DCLK1 | Sponge network | -0.583 | 0.14589 | -1.324 | 0.00014 | 0.668 |
| 1305 | MYLK-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 12 | NRK | Sponge network | -1.331 | 3.0E-5 | 0.656 | 0.19217 | 0.668 |
| 1306 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | SYNE1 | Sponge network | -0.611 | 0.09607 | -0.738 | 0.00316 | 0.668 |
| 1307 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | -2.46 | 0 | -1.548 | 0 | 0.668 |
| 1308 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | LRRK2 | Sponge network | -1.111 | 0.0003 | -1.136 | 1.0E-5 | 0.668 |
| 1309 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-629-3p | 13 | ITGA5 | Sponge network | -0.583 | 0.14589 | 0.452 | 0.03524 | 0.668 |
| 1310 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | PEG3 | Sponge network | 0.194 | 0.64278 | -1.74 | 0 | 0.668 |
| 1311 | MYLK-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-340-5p | 12 | KLF10 | Sponge network | -1.331 | 3.0E-5 | -0.783 | 0 | 0.668 |
| 1312 | AC003090.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | -2.117 | 0.00161 | -1.051 | 0.00022 | 0.668 |
| 1313 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 71 | TCF4 | Sponge network | -0.576 | 0.10121 | 0.016 | 0.92271 | 0.668 |
| 1314 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-505-3p;hsa-miR-590-3p | 24 | JAKMIP2 | Sponge network | -1.111 | 0.0003 | -1.388 | 0 | 0.668 |
| 1315 | MYLK-AS1 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-484 | 15 | ABL1 | Sponge network | -1.331 | 3.0E-5 | -0.215 | 0.07804 | 0.668 |
| 1316 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-93-5p | 43 | FAT3 | Sponge network | -2.69 | 0.0001 | -1.601 | 0.00051 | 0.668 |
| 1317 | DNM3OS |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | 0.572 | 0.01722 | -1.74 | 0 | 0.667 |
| 1318 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p | 38 | FBXO32 | Sponge network | -0.612 | 0.00094 | -0.495 | 0.07561 | 0.667 |
| 1319 | PWAR6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 17 | FAM172A | Sponge network | -1.575 | 6.0E-5 | -0.57 | 0 | 0.667 |
| 1320 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-582-5p | 14 | MAPK10 | Sponge network | -2.69 | 0.0001 | -1.774 | 0 | 0.667 |
| 1321 | MAGI2-AS3 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | THSD7B | Sponge network | -0.129 | 0.57177 | 0.154 | 0.74723 | 0.667 |
| 1322 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-93-5p | 39 | EPHA7 | Sponge network | -2.69 | 0.0001 | -2.197 | 3.0E-5 | 0.667 |
| 1323 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 15 | PARK2 | Sponge network | -1.111 | 0.0003 | -1.892 | 0 | 0.667 |
| 1324 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p | 48 | IL6ST | Sponge network | -0.358 | 0.17255 | -0.402 | 0.00676 | 0.667 |
| 1325 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 18 | CAV1 | Sponge network | -0.737 | 0.06182 | -1.013 | 6.0E-5 | 0.667 |
| 1326 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-505-3p | 10 | JAKMIP2 | Sponge network | -1.092 | 4.0E-5 | -1.388 | 0 | 0.667 |
| 1327 | BVES-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-452-5p | 12 | FNBP1 | Sponge network | -2.62 | 0 | -0.969 | 4.0E-5 | 0.667 |
| 1328 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 29 | PGR | Sponge network | -2.117 | 0.00161 | -1.704 | 0 | 0.667 |
| 1329 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 20 | PCDH17 | Sponge network | 0.727 | 3.0E-5 | 0.731 | 2.0E-5 | 0.667 |
| 1330 | MAGI2-AS3 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 14 | NGFR | Sponge network | -0.129 | 0.57177 | -1.271 | 0.01058 | 0.667 |
| 1331 | HAND2-AS1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | RNF180 | Sponge network | -1.697 | 0.00889 | -1.127 | 1.0E-5 | 0.667 |
| 1332 | SNHG14 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 21 | EZH1 | Sponge network | -1.111 | 0.0003 | -0.564 | 1.0E-5 | 0.667 |
| 1333 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-501-3p | 22 | TSHZ3 | Sponge network | -0.154 | 0.53954 | -0.556 | 0.01516 | 0.667 |
| 1334 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-7-5p | 34 | TCF4 | Sponge network | -1.331 | 3.0E-5 | 0.016 | 0.92271 | 0.667 |
| 1335 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | CNTN1 | Sponge network | -1.439 | 4.0E-5 | -2.594 | 0 | 0.667 |
| 1336 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 22 | FGFR1 | Sponge network | -0.737 | 0.06182 | -0.509 | 0.03957 | 0.667 |
| 1337 | PWAR6 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | PCDH9 | Sponge network | -1.575 | 6.0E-5 | -1.823 | 4.0E-5 | 0.667 |
| 1338 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 23 | IGF1 | Sponge network | -0.358 | 0.17255 | -0.204 | 0.5898 | 0.666 |
| 1339 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-542-3p | 18 | DYNC1I1 | Sponge network | -1.439 | 4.0E-5 | -1.12 | 0.00107 | 0.666 |
| 1340 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 18 | RASSF8 | Sponge network | -1.092 | 4.0E-5 | -0.122 | 0.65591 | 0.666 |
| 1341 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | FAT4 | Sponge network | -1.439 | 4.0E-5 | -0.354 | 0.14694 | 0.666 |
| 1342 | MIR497HG |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 23 | PCDH10 | Sponge network | -1.439 | 4.0E-5 | -1.644 | 0.0078 | 0.666 |
| 1343 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-542-3p | 19 | AKAP6 | Sponge network | -0.517 | 0.02935 | -1.586 | 3.0E-5 | 0.666 |
| 1344 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-3p | 32 | AR | Sponge network | -0.693 | 0.05629 | -1.554 | 0 | 0.666 |
| 1345 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | TSHZ3 | Sponge network | 0.194 | 0.64278 | -0.556 | 0.01516 | 0.666 |
| 1346 | USP3-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 57 | RUNX1T1 | Sponge network | -0.162 | 0.47687 | -0.344 | 0.2374 | 0.666 |
| 1347 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 82 | TCF4 | Sponge network | -1.111 | 0.0003 | 0.016 | 0.92271 | 0.666 |
| 1348 | ARHGEF26-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 23 | PRUNE2 | Sponge network | -2.46 | 0 | -1.733 | 0.00022 | 0.666 |
| 1349 | ZNF667-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 31 | IGFBP5 | Sponge network | -0.976 | 0.00025 | -0.806 | 0.00105 | 0.666 |
| 1350 | HAND2-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 34 | DLC1 | Sponge network | -1.697 | 0.00889 | -0.382 | 0.05038 | 0.665 |
| 1351 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | RECK | Sponge network | -0.901 | 0.00019 | -1.043 | 0 | 0.665 |
| 1352 | RP11-166D19.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ITPKB | Sponge network | -0.576 | 0.10121 | -0.704 | 0.00106 | 0.665 |
| 1353 | NR2F1-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 44 | IGFBP5 | Sponge network | -0.49 | 0.04946 | -0.806 | 0.00105 | 0.665 |
| 1354 | MEG3 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p | 11 | PRKD1 | Sponge network | 0.249 | 0.35902 | -0.466 | 0.03583 | 0.665 |
| 1355 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | FAT3 | Sponge network | -1.439 | 4.0E-5 | -1.601 | 0.00051 | 0.665 |
| 1356 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-940 | 39 | PBX1 | Sponge network | -0.793 | 7.0E-5 | -1.206 | 0 | 0.665 |
| 1357 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-505-3p | 14 | FNBP1 | Sponge network | -0.583 | 0.14589 | -0.969 | 4.0E-5 | 0.665 |
| 1358 | MIR143HG |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 29 | RYR2 | Sponge network | -0.739 | 0.0574 | -1.466 | 0.00031 | 0.665 |
| 1359 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484 | 10 | FLNA | Sponge network | -2.531 | 0 | -0.771 | 0.01377 | 0.665 |
| 1360 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-93-5p | 15 | TSHZ3 | Sponge network | -2.531 | 0 | -0.556 | 0.01516 | 0.665 |
| 1361 | MYLK-AS1 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-940 | 17 | MYO9A | Sponge network | -1.331 | 3.0E-5 | -0.147 | 0.32373 | 0.665 |
| 1362 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-629-3p | 31 | GAS7 | Sponge network | -0.583 | 0.14589 | -0.555 | 0.01602 | 0.665 |
| 1363 | C20orf166-AS1 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-664a-3p | 15 | PEG3 | Sponge network | -2.69 | 0.0001 | -1.74 | 0 | 0.665 |
| 1364 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 28 | PRRX1 | Sponge network | 0.249 | 0.35902 | 1.316 | 0 | 0.665 |
| 1365 | ADAMTS9-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-96-5p | 31 | ASTN1 | Sponge network | -2.452 | 0 | -2.389 | 2.0E-5 | 0.665 |
| 1366 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | GPC6 | Sponge network | 0.572 | 0.01722 | -0.054 | 0.81465 | 0.665 |
| 1367 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-877-5p | 26 | NRK | Sponge network | -0.976 | 0.00025 | 0.656 | 0.19217 | 0.665 |
| 1368 | MALAT1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ZNF638 | Sponge network | 0.782 | 0.00016 | 0.22 | 0.01214 | 0.665 |
| 1369 | MAGI2-AS3 |
hsa-miR-144-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | FMN2 | Sponge network | -0.129 | 0.57177 | -1.746 | 0.00047 | 0.665 |
| 1370 | DNM3OS |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 10 | FLNA | Sponge network | 0.572 | 0.01722 | -0.771 | 0.01377 | 0.665 |
| 1371 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 16 | STAT5B | Sponge network | -0.129 | 0.57177 | -0.182 | 0.1082 | 0.664 |
| 1372 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p | 12 | THSD7A | Sponge network | -0.49 | 0.04946 | 0.242 | 0.25154 | 0.664 |
| 1373 | RP11-867G23.10 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-625-5p | 13 | SFRP1 | Sponge network | -2.531 | 0 | -3.566 | 0 | 0.664 |
| 1374 | RP11-389C8.2 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ERG | Sponge network | 0.727 | 3.0E-5 | 0.002 | 0.99011 | 0.664 |
| 1375 | GNG12-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 25 | SYNPO2 | Sponge network | -0.793 | 7.0E-5 | -2.342 | 0 | 0.664 |
| 1376 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-486-5p;hsa-miR-542-3p | 20 | DYNC1I1 | Sponge network | -2.69 | 0.0001 | -1.12 | 0.00107 | 0.664 |
| 1377 | TRAF3IP2-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-942-5p | 13 | ZBTB20 | Sponge network | -0.323 | 0.01372 | -0.122 | 0.57569 | 0.664 |
| 1378 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | RECK | Sponge network | -1.575 | 6.0E-5 | -1.043 | 0 | 0.664 |
| 1379 | LINC00702 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 19 | MACF1 | Sponge network | -0.737 | 0.06182 | 0.019 | 0.90335 | 0.664 |
| 1380 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | NRXN1 | Sponge network | -2.117 | 0.00161 | -3.778 | 0 | 0.664 |
| 1381 | SERTAD4-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-369-3p;hsa-miR-500a-5p | 18 | DMD | Sponge network | -0.932 | 0.00329 | -1.506 | 0 | 0.664 |
| 1382 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | DACT1 | Sponge network | -0.129 | 0.57177 | 0.783 | 0.00072 | 0.664 |
| 1383 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 20 | RCAN2 | Sponge network | -0.129 | 0.57177 | -0.33 | 0.15172 | 0.664 |
| 1384 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-877-5p;hsa-miR-93-5p | 29 | ANK2 | Sponge network | -0.162 | 0.47687 | -1.603 | 1.0E-5 | 0.664 |
| 1385 | SNHG14 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 19 | FAM172A | Sponge network | -1.111 | 0.0003 | -0.57 | 0 | 0.663 |
| 1386 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 13 | PGR | Sponge network | -1.582 | 8.0E-5 | -1.704 | 0 | 0.663 |
| 1387 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | CADM2 | Sponge network | -4.546 | 0 | -3.782 | 0 | 0.663 |
| 1388 | RP11-244O19.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 16 | ITPKB | Sponge network | -0.096 | 0.67958 | -0.704 | 0.00106 | 0.663 |
| 1389 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | CACNA2D1 | Sponge network | -1.111 | 0.0003 | -0.846 | 0.00675 | 0.663 |
| 1390 | ZNF667-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-96-5p | 30 | ASTN1 | Sponge network | -0.976 | 0.00025 | -2.389 | 2.0E-5 | 0.663 |
| 1391 | HAND2-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | TPO | Sponge network | -1.697 | 0.00889 | -1.87 | 2.0E-5 | 0.663 |
| 1392 | ARHGEF26-AS1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p | 10 | CNN1 | Sponge network | -2.46 | 0 | -2.633 | 0 | 0.663 |
| 1393 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | RCAN2 | Sponge network | -0.49 | 0.04946 | -0.33 | 0.15172 | 0.663 |
| 1394 | RP11-554A11.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3913-5p;hsa-miR-450b-5p | 15 | WWTR1 | Sponge network | -0.583 | 0.14589 | -0.345 | 0.11997 | 0.663 |
| 1395 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 57 | GAS7 | Sponge network | -0.49 | 0.04946 | -0.555 | 0.01602 | 0.663 |
| 1396 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | SETBP1 | Sponge network | -2.117 | 0.00161 | -1.302 | 1.0E-5 | 0.663 |
| 1397 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p | 32 | EPHA7 | Sponge network | -0.469 | 0.08721 | -2.197 | 3.0E-5 | 0.663 |
| 1398 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | LRRK2 | Sponge network | -2.452 | 0 | -1.136 | 1.0E-5 | 0.663 |
| 1399 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | PTGFR | Sponge network | -0.358 | 0.17255 | -0.233 | 0.45421 | 0.663 |
| 1400 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | ATRX | Sponge network | 0.537 | 0.00139 | 0.261 | 0.04408 | 0.662 |
| 1401 | PWAR6 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | PRUNE2 | Sponge network | -1.575 | 6.0E-5 | -1.733 | 0.00022 | 0.662 |
| 1402 | HAND2-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 16 | KCTD8 | Sponge network | -1.697 | 0.00889 | -3.359 | 0 | 0.662 |
| 1403 | RP11-532F6.3 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p | 10 | FLNA | Sponge network | -0.896 | 0.00099 | -0.771 | 0.01377 | 0.662 |
| 1404 | RP11-819C21.1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-590-3p | 10 | ZNF638 | Sponge network | 0.537 | 0.00139 | 0.22 | 0.01214 | 0.662 |
| 1405 | ZNF667-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -0.976 | 0.00025 | -0.704 | 0.00106 | 0.662 |
| 1406 | LINC00092 |
hsa-miR-1301-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-940 | 11 | SFRP1 | Sponge network | -1.886 | 0 | -3.566 | 0 | 0.662 |
| 1407 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p | 22 | PTGFR | Sponge network | -0.49 | 0.04946 | -0.233 | 0.45421 | 0.662 |
| 1408 | HAND2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 28 | RYR2 | Sponge network | -1.697 | 0.00889 | -1.466 | 0.00031 | 0.662 |
| 1409 | SNHG14 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 61 | GAS7 | Sponge network | -1.111 | 0.0003 | -0.555 | 0.01602 | 0.662 |
| 1410 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 48 | TGFBR3 | Sponge network | -2.452 | 0 | -1.173 | 1.0E-5 | 0.662 |
| 1411 | FGF14-AS2 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-590-3p | 13 | MRVI1 | Sponge network | -1.582 | 8.0E-5 | -1.051 | 0.00022 | 0.662 |
| 1412 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-744-5p;hsa-miR-93-5p | 15 | FGFR1 | Sponge network | -2.531 | 0 | -0.509 | 0.03957 | 0.662 |
| 1413 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | FAT4 | Sponge network | 0.572 | 0.01722 | -0.354 | 0.14694 | 0.662 |
| 1414 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DNAJB4 | Sponge network | -2.452 | 0 | -0.7 | 1.0E-5 | 0.662 |
| 1415 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | NRXN1 | Sponge network | -1.886 | 0 | -3.778 | 0 | 0.662 |
| 1416 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 23 | DIP2C | Sponge network | -1.092 | 4.0E-5 | -0.436 | 0.01713 | 0.662 |
| 1417 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 15 | PCDH9 | Sponge network | -0.583 | 0.14589 | -1.823 | 4.0E-5 | 0.661 |
| 1418 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-935 | 21 | PCDH10 | Sponge network | -2.62 | 0 | -1.644 | 0.0078 | 0.661 |
| 1419 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | CNTN1 | Sponge network | -2.117 | 0.00161 | -2.594 | 0 | 0.661 |
| 1420 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 50 | TACC1 | Sponge network | -0.739 | 0.0574 | -0.362 | 0.05406 | 0.661 |
| 1421 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p | 37 | AR | Sponge network | -0.358 | 0.17255 | -1.554 | 0 | 0.661 |
| 1422 | LINC00578 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-484 | 10 | FLNA | Sponge network | 0.811 | 0.03228 | -0.771 | 0.01377 | 0.661 |
| 1423 | LINC00702 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 19 | ITGA5 | Sponge network | -0.737 | 0.06182 | 0.452 | 0.03524 | 0.661 |
| 1424 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 48 | ZFHX4 | Sponge network | 0.572 | 0.01722 | 0.018 | 0.95574 | 0.661 |
| 1425 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 15 | NALCN | Sponge network | 0.997 | 0.00066 | 1.068 | 0.00237 | 0.661 |
| 1426 | SNHG14 |
hsa-miR-182-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-769-5p | 11 | TCEAL7 | Sponge network | -1.111 | 0.0003 | -1.445 | 0 | 0.661 |
| 1427 | LINC00641 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p | 16 | MACF1 | Sponge network | -0.222 | 0.32445 | 0.019 | 0.90335 | 0.661 |
| 1428 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 28 | NRK | Sponge network | -0.49 | 0.04946 | 0.656 | 0.19217 | 0.661 |
| 1429 | RP11-121C2.2 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-409-5p | 11 | TPR | Sponge network | 1.147 | 0 | 0.582 | 0 | 0.661 |
| 1430 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | ERG | Sponge network | -2.452 | 0 | 0.002 | 0.99011 | 0.661 |
| 1431 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-577 | 23 | ADAMTSL3 | Sponge network | -2.69 | 0.0001 | -1.559 | 0 | 0.661 |
| 1432 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | MYO9A | Sponge network | -0.487 | 0.02157 | -0.147 | 0.32373 | 0.661 |
| 1433 | CTC-296K1.4 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-539-5p;hsa-miR-576-5p | 11 | SYNPO2 | Sponge network | -2.076 | 0.00548 | -2.342 | 0 | 0.661 |
| 1434 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 17 | RCAN2 | Sponge network | 0.194 | 0.64278 | -0.33 | 0.15172 | 0.661 |
| 1435 | PCA3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | AKAP6 | Sponge network | -0.709 | 0.18168 | -1.586 | 3.0E-5 | 0.66 |
| 1436 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p | 16 | DYNC1I1 | Sponge network | -2.62 | 0 | -1.12 | 0.00107 | 0.66 |
| 1437 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 35 | IGF1 | Sponge network | -0.49 | 0.04946 | -0.204 | 0.5898 | 0.66 |
| 1438 | MIR143HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 23 | STARD13 | Sponge network | -0.739 | 0.0574 | -0.187 | 0.27383 | 0.66 |
| 1439 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 56 | SOX5 | Sponge network | -1.111 | 0.0003 | -1.091 | 4.0E-5 | 0.66 |
| 1440 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 39 | NTRK3 | Sponge network | -1.092 | 4.0E-5 | -1.77 | 3.0E-5 | 0.66 |
| 1441 | MAGI2-AS3 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-589-5p | 11 | SORCS2 | Sponge network | -0.129 | 0.57177 | -0.223 | 0.4253 | 0.66 |
| 1442 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-429;hsa-miR-505-3p | 16 | JAKMIP2 | Sponge network | -0.976 | 0.00025 | -1.388 | 0 | 0.66 |
| 1443 | PWAR6 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 20 | SFRP1 | Sponge network | -1.575 | 6.0E-5 | -3.566 | 0 | 0.66 |
| 1444 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | BNC2 | Sponge network | 0.335 | 0.20596 | -0.491 | 0.09988 | 0.66 |
| 1445 | RP11-532F6.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-505-3p | 11 | FNBP1 | Sponge network | -0.896 | 0.00099 | -0.969 | 4.0E-5 | 0.66 |
| 1446 | SNHG14 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -1.111 | 0.0003 | -2.046 | 0.00019 | 0.66 |
| 1447 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | RASSF8 | Sponge network | -2.452 | 0 | -0.122 | 0.65591 | 0.66 |
| 1448 | SERTAD4-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-576-5p | 28 | AR | Sponge network | -0.932 | 0.00329 | -1.554 | 0 | 0.66 |
| 1449 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 73 | NTRK3 | Sponge network | -0.49 | 0.04946 | -1.77 | 3.0E-5 | 0.66 |
| 1450 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 49 | MEF2C | Sponge network | -1.575 | 6.0E-5 | -0.499 | 0.01448 | 0.66 |
| 1451 | MRVI1-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-664a-5p | 38 | DST | Sponge network | -1.092 | 4.0E-5 | -0.713 | 0.00096 | 0.66 |
| 1452 | ZNF667-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 37 | GRIK3 | Sponge network | -0.976 | 0.00025 | -3.208 | 0 | 0.66 |
| 1453 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | SYNE1 | Sponge network | -1.281 | 0.00012 | -0.738 | 0.00316 | 0.659 |
| 1454 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p | 11 | JAKMIP2 | Sponge network | -2.62 | 0 | -1.388 | 0 | 0.659 |
| 1455 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 39 | EBF1 | Sponge network | -0.739 | 0.0574 | -0.384 | 0.08856 | 0.659 |
| 1456 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-550a-3p;hsa-miR-7-1-3p | 27 | EPHA7 | Sponge network | -4.546 | 0 | -2.197 | 3.0E-5 | 0.659 |
| 1457 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 47 | ADARB1 | Sponge network | -0.162 | 0.47687 | -0.335 | 0.06631 | 0.659 |
| 1458 | C20orf166-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-664a-3p | 17 | NRXN3 | Sponge network | -2.69 | 0.0001 | -0.893 | 0.04186 | 0.659 |
| 1459 | HAND2-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 38 | WWTR1 | Sponge network | -1.697 | 0.00889 | -0.345 | 0.11997 | 0.659 |
| 1460 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 30 | SCN9A | Sponge network | -0.576 | 0.10121 | -0.525 | 0.10593 | 0.659 |
| 1461 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | CNTN1 | Sponge network | -1.575 | 6.0E-5 | -2.594 | 0 | 0.659 |
| 1462 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 43 | RBMS3 | Sponge network | -0.487 | 0.02157 | -0.519 | 0.03551 | 0.659 |
| 1463 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-628-5p;hsa-miR-935;hsa-miR-96-5p | 32 | FBXO32 | Sponge network | -0.976 | 0.00025 | -0.495 | 0.07561 | 0.659 |
| 1464 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-369-3p | 19 | FBXO32 | Sponge network | -0.932 | 0.00329 | -0.495 | 0.07561 | 0.659 |
| 1465 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p;hsa-miR-98-5p | 67 | PRKAA2 | Sponge network | -2.452 | 0 | -2.462 | 0 | 0.659 |
| 1466 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | PEG3 | Sponge network | -2.46 | 0 | -1.74 | 0 | 0.659 |
| 1467 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | EBF1 | Sponge network | -0.323 | 0.01372 | -0.384 | 0.08856 | 0.659 |
| 1468 | MEG3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p | 13 | MN1 | Sponge network | 0.249 | 0.35902 | -0.358 | 0.24172 | 0.659 |
| 1469 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 73 | CADM2 | Sponge network | -1.697 | 0.00889 | -3.782 | 0 | 0.659 |
| 1470 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p | 10 | DKK3 | Sponge network | -1.331 | 3.0E-5 | -0.052 | 0.77783 | 0.659 |
| 1471 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-935 | 36 | DMD | Sponge network | -0.096 | 0.67958 | -1.506 | 0 | 0.659 |
| 1472 | RP11-166D19.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | RYR2 | Sponge network | -0.576 | 0.10121 | -1.466 | 0.00031 | 0.659 |
| 1473 | MIR143HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 22 | ITGB3 | Sponge network | -0.739 | 0.0574 | -0.011 | 0.95751 | 0.659 |
| 1474 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-93-3p;hsa-miR-935 | 26 | PCDH10 | Sponge network | -0.162 | 0.47687 | -1.644 | 0.0078 | 0.658 |
| 1475 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | LRRK2 | Sponge network | -0.739 | 0.0574 | -1.136 | 1.0E-5 | 0.658 |
| 1476 | RP11-400K9.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | WWTR1 | Sponge network | -0.611 | 0.09607 | -0.345 | 0.11997 | 0.658 |
| 1477 | MIR497HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | NRXN3 | Sponge network | -1.439 | 4.0E-5 | -0.893 | 0.04186 | 0.658 |
| 1478 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 14 | CDH19 | Sponge network | -1.582 | 8.0E-5 | -3.434 | 0 | 0.658 |
| 1479 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | PLSCR4 | Sponge network | 0.335 | 0.20596 | -0.474 | 0.03833 | 0.658 |
| 1480 | MAGI2-AS3 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 23 | CXCL12 | Sponge network | -0.129 | 0.57177 | -1.421 | 0 | 0.658 |
| 1481 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 26 | NR2C2 | Sponge network | 0.782 | 0.00016 | 0.21 | 0.03224 | 0.658 |
| 1482 | LINC00702 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | ERG | Sponge network | -0.737 | 0.06182 | 0.002 | 0.99011 | 0.658 |
| 1483 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-940 | 10 | CTNNA3 | Sponge network | -0.976 | 0.00025 | -2.046 | 0.00019 | 0.658 |
| 1484 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 42 | ST6GAL2 | Sponge network | -0.129 | 0.57177 | -1.201 | 0.00597 | 0.658 |
| 1485 | PWAR6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 64 | RUNX1T1 | Sponge network | -1.575 | 6.0E-5 | -0.344 | 0.2374 | 0.658 |
| 1486 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-590-3p | 11 | HERC2 | Sponge network | 0.083 | 0.66405 | 0.114 | 0.30638 | 0.657 |
| 1487 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p | 11 | TXNIP | Sponge network | -1.331 | 3.0E-5 | -1.277 | 0 | 0.657 |
| 1488 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | ANK2 | Sponge network | 0.194 | 0.64278 | -1.603 | 1.0E-5 | 0.657 |
| 1489 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 21 | PTGFR | Sponge network | -2.452 | 0 | -0.233 | 0.45421 | 0.657 |
| 1490 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-98-5p | 20 | ITGB3 | Sponge network | -0.129 | 0.57177 | -0.011 | 0.95751 | 0.657 |
| 1491 | ADAMTS9-AS2 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-7-5p;hsa-miR-940 | 10 | PHOX2B | Sponge network | -2.452 | 0 | -2.68 | 0 | 0.657 |
| 1492 | USP3-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-455-5p | 12 | MACF1 | Sponge network | -0.162 | 0.47687 | 0.019 | 0.90335 | 0.657 |
| 1493 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-769-5p | 14 | SETBP1 | Sponge network | -2.531 | 0 | -1.302 | 1.0E-5 | 0.657 |
| 1494 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 31 | THBS1 | Sponge network | 0.249 | 0.35902 | 0.741 | 0.00386 | 0.657 |
| 1495 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p | 12 | SCN11A | Sponge network | -1.575 | 6.0E-5 | -1.15 | 0.00299 | 0.657 |
| 1496 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-7-5p | 18 | FNBP1 | Sponge network | -1.697 | 0.00889 | -0.969 | 4.0E-5 | 0.657 |
| 1497 | LINC00702 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 56 | GAS7 | Sponge network | -0.737 | 0.06182 | -0.555 | 0.01602 | 0.657 |
| 1498 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-590-3p | 10 | NALCN | Sponge network | -0.358 | 0.17255 | 1.068 | 0.00237 | 0.657 |
| 1499 | DNM3OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-664a-3p | 14 | MN1 | Sponge network | 0.572 | 0.01722 | -0.358 | 0.24172 | 0.657 |
| 1500 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-671-5p | 32 | RBMS3 | Sponge network | -2.69 | 0.0001 | -0.519 | 0.03551 | 0.657 |
| 1501 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 50 | RUNX1T1 | Sponge network | 0.335 | 0.20596 | -0.344 | 0.2374 | 0.657 |
| 1502 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 19 | EPHA3 | Sponge network | -1.092 | 4.0E-5 | 0.185 | 0.53588 | 0.657 |
| 1503 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | ABCC9 | Sponge network | 0.249 | 0.35902 | -0.466 | 0.14121 | 0.657 |
| 1504 | SNHG14 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 25 | PTGFR | Sponge network | -1.111 | 0.0003 | -0.233 | 0.45421 | 0.656 |
| 1505 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 45 | RBMS3 | Sponge network | -1.281 | 0.00012 | -0.519 | 0.03551 | 0.656 |
| 1506 | ACTA2-AS1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ITPKB | Sponge network | 0.194 | 0.64278 | -0.704 | 0.00106 | 0.656 |
| 1507 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 50 | PRKAA2 | Sponge network | -1.249 | 7.0E-5 | -2.462 | 0 | 0.656 |
| 1508 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-532-3p | 11 | CADM3 | Sponge network | -2.531 | 0 | -3.663 | 0 | 0.656 |
| 1509 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-455-5p;hsa-miR-590-3p | 15 | RECK | Sponge network | -0.896 | 0.00099 | -1.043 | 0 | 0.656 |
| 1510 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 36 | AR | Sponge network | -4.546 | 0 | -1.554 | 0 | 0.656 |
| 1511 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 53 | AR | Sponge network | 0.194 | 0.64278 | -1.554 | 0 | 0.656 |
| 1512 | SNHG14 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 41 | PCDH10 | Sponge network | -1.111 | 0.0003 | -1.644 | 0.0078 | 0.656 |
| 1513 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3913-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 30 | ZBTB4 | Sponge network | -1.439 | 4.0E-5 | -0.739 | 0 | 0.656 |
| 1514 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 11 | LRFN5 | Sponge network | -1.111 | 0.0003 | -1.483 | 0 | 0.656 |
| 1515 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-629-5p;hsa-miR-940 | 28 | IGF1 | Sponge network | -0.976 | 0.00025 | -0.204 | 0.5898 | 0.656 |
| 1516 | RP11-244O19.1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-503-5p | 25 | ABL1 | Sponge network | -0.096 | 0.67958 | -0.215 | 0.07804 | 0.656 |
| 1517 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p | 43 | RBMS3 | Sponge network | -0.793 | 7.0E-5 | -0.519 | 0.03551 | 0.656 |
| 1518 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-421 | 24 | DTNA | Sponge network | -4.463 | 0.00025 | -1.648 | 1.0E-5 | 0.656 |
| 1519 | SERTAD4-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-424-5p | 12 | PLSCR4 | Sponge network | -0.932 | 0.00329 | -0.474 | 0.03833 | 0.656 |
| 1520 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 10 | TSLP | Sponge network | -2.452 | 0 | -2.209 | 0 | 0.656 |
| 1521 | ADAMTS9-AS2 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | RPS6KA6 | Sponge network | -2.452 | 0 | -2.989 | 0 | 0.655 |
| 1522 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 20 | RASSF2 | Sponge network | 0.693 | 0.00076 | -0.273 | 0.21961 | 0.655 |
| 1523 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-576-5p | 13 | AKAP12 | Sponge network | -2.62 | 0 | -0.932 | 0.0028 | 0.655 |
| 1524 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 51 | EPHA7 | Sponge network | -1.575 | 6.0E-5 | -2.197 | 3.0E-5 | 0.655 |
| 1525 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 31 | TIMP3 | Sponge network | -1.697 | 0.00889 | -0.546 | 0.02307 | 0.655 |
| 1526 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 65 | FAT3 | Sponge network | -2.46 | 0 | -1.601 | 0.00051 | 0.655 |
| 1527 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 30 | EBF3 | Sponge network | -0.739 | 0.0574 | -0.058 | 0.81437 | 0.655 |
| 1528 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 24 | SETBP1 | Sponge network | -0.611 | 0.09607 | -1.302 | 1.0E-5 | 0.655 |
| 1529 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-93-5p | 22 | FAT3 | Sponge network | -2.531 | 0 | -1.601 | 0.00051 | 0.655 |
| 1530 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 40 | TSHZ3 | Sponge network | 0.249 | 0.35902 | -0.556 | 0.01516 | 0.655 |
| 1531 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DCN | Sponge network | 0.572 | 0.01722 | -1.288 | 0 | 0.655 |
| 1532 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | ADAMTSL3 | Sponge network | -0.901 | 0.00019 | -1.559 | 0 | 0.655 |
| 1533 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 30 | NRXN1 | Sponge network | -0.976 | 0.00025 | -3.778 | 0 | 0.655 |
| 1534 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-577;hsa-miR-664a-3p | 23 | GREM1 | Sponge network | -0.49 | 0.04946 | 0.902 | 0.01691 | 0.655 |
| 1535 | ZNF582-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-940 | 13 | MAPK10 | Sponge network | -1.349 | 0.00017 | -1.774 | 0 | 0.655 |
| 1536 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-5p | 11 | ZNF208 | Sponge network | -0.976 | 0.00025 | -0.4 | 0.22381 | 0.655 |
| 1537 | HAND2-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-940 | 12 | PHOX2B | Sponge network | -1.697 | 0.00889 | -2.68 | 0 | 0.655 |
| 1538 | SNHG14 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 15 | PTPN13 | Sponge network | -1.111 | 0.0003 | -1.208 | 0 | 0.655 |
| 1539 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-628-5p | 31 | TCF4 | Sponge network | -0.583 | 0.14589 | 0.016 | 0.92271 | 0.655 |
| 1540 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | CNTN1 | Sponge network | -1.582 | 8.0E-5 | -2.594 | 0 | 0.655 |
| 1541 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 17 | JAKMIP2 | Sponge network | -0.737 | 0.06182 | -1.388 | 0 | 0.654 |
| 1542 | GNG12-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-5p | 81 | LPP | Sponge network | -0.793 | 7.0E-5 | -0.459 | 0.04475 | 0.654 |
| 1543 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-582-5p | 29 | FBXO32 | Sponge network | -2.69 | 0.0001 | -0.495 | 0.07561 | 0.654 |
| 1544 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | AKAP6 | Sponge network | -2.117 | 0.00161 | -1.586 | 3.0E-5 | 0.654 |
| 1545 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 32 | PGR | Sponge network | -0.793 | 7.0E-5 | -1.704 | 0 | 0.654 |
| 1546 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | EBF1 | Sponge network | -1.575 | 6.0E-5 | -0.384 | 0.08856 | 0.654 |
| 1547 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 25 | DYNC1I1 | Sponge network | -2.46 | 0 | -1.12 | 0.00107 | 0.654 |
| 1548 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-452-5p;hsa-miR-590-3p | 26 | DMD | Sponge network | -0.358 | 0.17255 | -1.506 | 0 | 0.654 |
| 1549 | MIR497HG |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3913-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | WWTR1 | Sponge network | -1.439 | 4.0E-5 | -0.345 | 0.11997 | 0.654 |
| 1550 | MIR143HG |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | KCTD8 | Sponge network | -0.739 | 0.0574 | -3.359 | 0 | 0.654 |
| 1551 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-542-3p | 16 | DYNC1I1 | Sponge network | -1.092 | 4.0E-5 | -1.12 | 0.00107 | 0.654 |
| 1552 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | FAT4 | Sponge network | -0.737 | 0.06182 | -0.354 | 0.14694 | 0.654 |
| 1553 | C20orf166-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-874-3p | 17 | SORCS1 | Sponge network | -2.69 | 0.0001 | -3.492 | 0 | 0.654 |
| 1554 | MIR143HG |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 16 | RHOB | Sponge network | -0.739 | 0.0574 | -1.052 | 0 | 0.654 |
| 1555 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 76 | CADM2 | Sponge network | -2.46 | 0 | -3.782 | 0 | 0.654 |
| 1556 | LINC00865 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-450b-5p | 11 | PRDM5 | Sponge network | -1.249 | 7.0E-5 | -1.09 | 0.00014 | 0.654 |
| 1557 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | TMEFF2 | Sponge network | -2.452 | 0 | -2.426 | 0 | 0.654 |
| 1558 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 53 | GAS7 | Sponge network | 0.997 | 0.00066 | -0.555 | 0.01602 | 0.654 |
| 1559 | MIR143HG |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 15 | DCN | Sponge network | -0.739 | 0.0574 | -1.288 | 0 | 0.654 |
| 1560 | DNM3OS |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | NCAM2 | Sponge network | 0.572 | 0.01722 | -0.482 | 0.22565 | 0.654 |
| 1561 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 19 | JAKMIP2 | Sponge network | -2.452 | 0 | -1.388 | 0 | 0.654 |
| 1562 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | ROBO1 | Sponge network | -0.129 | 0.57177 | -0.009 | 0.96154 | 0.654 |
| 1563 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p | 14 | NR2C2 | Sponge network | 1.106 | 0 | 0.21 | 0.03224 | 0.654 |
| 1564 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-96-5p | 29 | FAT3 | Sponge network | -1.582 | 8.0E-5 | -1.601 | 0.00051 | 0.654 |
| 1565 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-769-5p | 23 | SETBP1 | Sponge network | -2.69 | 0.0001 | -1.302 | 1.0E-5 | 0.654 |
| 1566 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | DNAJB4 | Sponge network | -0.49 | 0.04946 | -0.7 | 1.0E-5 | 0.653 |
| 1567 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 65 | QKI | Sponge network | -0.739 | 0.0574 | -0.333 | 0.04111 | 0.653 |
| 1568 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 43 | RBMS3 | Sponge network | 0.249 | 0.35902 | -0.519 | 0.03551 | 0.653 |
| 1569 | MYLK-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-484 | 11 | GREM1 | Sponge network | -1.331 | 3.0E-5 | 0.902 | 0.01691 | 0.653 |
| 1570 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 33 | ZBTB4 | Sponge network | -1.697 | 0.00889 | -0.739 | 0 | 0.653 |
| 1571 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 35 | MCC | Sponge network | -1.439 | 4.0E-5 | -1.005 | 0 | 0.653 |
| 1572 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-576-5p | 10 | AKAP12 | Sponge network | -0.932 | 0.00329 | -0.932 | 0.0028 | 0.653 |
| 1573 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-940 | 13 | PEG3 | Sponge network | -1.349 | 0.00017 | -1.74 | 0 | 0.653 |
| 1574 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 34 | QKI | Sponge network | -1.092 | 4.0E-5 | -0.333 | 0.04111 | 0.653 |
| 1575 | LINC00702 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | KCTD8 | Sponge network | -0.737 | 0.06182 | -3.359 | 0 | 0.653 |
| 1576 | AC003090.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | RIMS1 | Sponge network | -2.117 | 0.00161 | -3.143 | 0 | 0.653 |
| 1577 | MIR497HG |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | TMEFF2 | Sponge network | -1.439 | 4.0E-5 | -2.426 | 0 | 0.653 |
| 1578 | FENDRR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-98-5p | 18 | ITGB3 | Sponge network | -1.281 | 0.00012 | -0.011 | 0.95751 | 0.653 |
| 1579 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | LSAMP | Sponge network | -0.576 | 0.10121 | -0.066 | 0.81708 | 0.653 |
| 1580 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-374b-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 11 | CNTNAP1 | Sponge network | 0.335 | 0.20596 | 0.358 | 0.13727 | 0.653 |
| 1581 | AC006116.24 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577 | 34 | DTNA | Sponge network | -0.473 | 0.07602 | -1.648 | 1.0E-5 | 0.653 |
| 1582 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 30 | ANK2 | Sponge network | -0.323 | 0.01372 | -1.603 | 1.0E-5 | 0.653 |
| 1583 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-629-3p | 28 | GAS7 | Sponge network | 0.811 | 0.03228 | -0.555 | 0.01602 | 0.653 |
| 1584 | FAM66C |
hsa-miR-146b-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p | 10 | FHL1 | Sponge network | -0.901 | 0.00019 | -2.687 | 0 | 0.653 |
| 1585 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-5p | 16 | FOXO3 | Sponge network | 0.537 | 0.0006 | -0.04 | 0.72028 | 0.653 |
| 1586 | NR2F1-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | THSD7B | Sponge network | -0.49 | 0.04946 | 0.154 | 0.74723 | 0.653 |
| 1587 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 11 | ADAMTSL3 | Sponge network | -1.582 | 8.0E-5 | -1.559 | 0 | 0.653 |
| 1588 | ACTA2-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | MACF1 | Sponge network | 0.194 | 0.64278 | 0.019 | 0.90335 | 0.653 |
| 1589 | MIR143HG |
hsa-let-7a-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 12 | RADIL | Sponge network | -0.739 | 0.0574 | -1.628 | 0 | 0.653 |
| 1590 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-7-5p | 17 | FNBP1 | Sponge network | -0.49 | 0.04946 | -0.969 | 4.0E-5 | 0.653 |
| 1591 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 30 | RBMS3 | Sponge network | -4.546 | 0 | -0.519 | 0.03551 | 0.653 |
| 1592 | RP11-244O19.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-382-5p;hsa-miR-3913-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 85 | LPP | Sponge network | -0.096 | 0.67958 | -0.459 | 0.04475 | 0.653 |
| 1593 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3913-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 32 | ZBTB4 | Sponge network | -0.737 | 0.06182 | -0.739 | 0 | 0.652 |
| 1594 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 11 | MN1 | Sponge network | -2.69 | 0.0001 | -0.358 | 0.24172 | 0.652 |
| 1595 | EMX2OS |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 26 | NRK | Sponge network | -0.777 | 0.1134 | 0.656 | 0.19217 | 0.652 |
| 1596 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 35 | IGF1 | Sponge network | -2.452 | 0 | -0.204 | 0.5898 | 0.652 |
| 1597 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | 0.249 | 0.35902 | -1.047 | 0.00364 | 0.652 |
| 1598 | FENDRR |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 62 | DTNA | Sponge network | -1.281 | 0.00012 | -1.648 | 1.0E-5 | 0.652 |
| 1599 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 14 | PARK2 | Sponge network | -0.576 | 0.10121 | -1.892 | 0 | 0.652 |
| 1600 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | EDA2R | Sponge network | -0.129 | 0.57177 | -0.587 | 0.01598 | 0.652 |
| 1601 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | PRDM5 | Sponge network | -0.49 | 0.04946 | -1.09 | 0.00014 | 0.652 |
| 1602 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 77 | IL6ST | Sponge network | -1.575 | 6.0E-5 | -0.402 | 0.00676 | 0.652 |
| 1603 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-629-3p | 34 | GAS7 | Sponge network | -0.358 | 0.17255 | -0.555 | 0.01602 | 0.652 |
| 1604 | CTD-3064M3.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-582-5p | 10 | NRXN3 | Sponge network | -0.469 | 0.08721 | -0.893 | 0.04186 | 0.652 |
| 1605 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-660-5p | 28 | RUNX1T1 | Sponge network | -1.582 | 8.0E-5 | -0.344 | 0.2374 | 0.652 |
| 1606 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | 0.335 | 0.20596 | 0.809 | 0.00544 | 0.652 |
| 1607 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p | 12 | TIMP3 | Sponge network | 0.811 | 0.03228 | -0.546 | 0.02307 | 0.652 |
| 1608 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-493-5p | 19 | SYNE1 | Sponge network | -2.62 | 0 | -0.738 | 0.00316 | 0.652 |
| 1609 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-3p;hsa-miR-93-5p | 45 | SSBP2 | Sponge network | -2.69 | 0.0001 | -1.58 | 0 | 0.652 |
| 1610 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | PCDH9 | Sponge network | -0.358 | 0.17255 | -1.823 | 4.0E-5 | 0.652 |
| 1611 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-769-5p | 31 | SYNPO2 | Sponge network | 0.997 | 0.00066 | -2.342 | 0 | 0.652 |
| 1612 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p | 10 | PDZD2 | Sponge network | -1.092 | 4.0E-5 | -0.909 | 3.0E-5 | 0.652 |
| 1613 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CACNA2D1 | Sponge network | -0.737 | 0.06182 | -0.846 | 0.00675 | 0.651 |
| 1614 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | EPHA7 | Sponge network | -2.117 | 0.00161 | -2.197 | 3.0E-5 | 0.651 |
| 1615 | FENDRR |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-671-5p | 31 | DLC1 | Sponge network | -1.281 | 0.00012 | -0.382 | 0.05038 | 0.651 |
| 1616 | DNM3OS |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 12 | AMPH | Sponge network | 0.572 | 0.01722 | -0.916 | 5.0E-5 | 0.651 |
| 1617 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | -0.162 | 0.47687 | -0.509 | 0.03957 | 0.651 |
| 1618 | PCA3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 28 | SYNPO2 | Sponge network | -0.709 | 0.18168 | -2.342 | 0 | 0.651 |
| 1619 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-935 | 37 | SSBP2 | Sponge network | -2.62 | 0 | -1.58 | 0 | 0.651 |
| 1620 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-590-3p | 20 | TSHZ3 | Sponge network | -0.358 | 0.17255 | -0.556 | 0.01516 | 0.651 |
| 1621 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-421;hsa-miR-532-3p | 13 | CADM3 | Sponge network | -2.62 | 0 | -3.663 | 0 | 0.651 |
| 1622 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | ANK2 | Sponge network | -0.611 | 0.09607 | -1.603 | 1.0E-5 | 0.651 |
| 1623 | MIR143HG |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 13 | ZNF382 | Sponge network | -0.739 | 0.0574 | -0.494 | 0.03831 | 0.651 |
| 1624 | SNHG14 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | LHFP | Sponge network | -1.111 | 0.0003 | -0.167 | 0.41371 | 0.651 |
| 1625 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 14 | ABCC9 | Sponge network | -0.096 | 0.67958 | -0.466 | 0.14121 | 0.651 |
| 1626 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 13 | NALCN | Sponge network | -0.49 | 0.04946 | 1.068 | 0.00237 | 0.651 |
| 1627 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 35 | SETBP1 | Sponge network | -2.46 | 0 | -1.302 | 1.0E-5 | 0.651 |
| 1628 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | EBF1 | Sponge network | -1.439 | 4.0E-5 | -0.384 | 0.08856 | 0.651 |
| 1629 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 65 | SSBP2 | Sponge network | -1.111 | 0.0003 | -1.58 | 0 | 0.651 |
| 1630 | BVES-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-378a-3p | 12 | FLNA | Sponge network | -2.62 | 0 | -0.771 | 0.01377 | 0.651 |
| 1631 | ACTA2-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 16 | PRUNE2 | Sponge network | 0.194 | 0.64278 | -1.733 | 0.00022 | 0.651 |
| 1632 | USP3-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421 | 19 | STARD13 | Sponge network | -0.162 | 0.47687 | -0.187 | 0.27383 | 0.65 |
| 1633 | DNM3OS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 13 | THSD7A | Sponge network | 0.572 | 0.01722 | 0.242 | 0.25154 | 0.65 |
| 1634 | FAM66C |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 13 | MAPK10 | Sponge network | -0.901 | 0.00019 | -1.774 | 0 | 0.65 |
| 1635 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CDH19 | Sponge network | -2.117 | 0.00161 | -3.434 | 0 | 0.65 |
| 1636 | DNM3OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DACT1 | Sponge network | 0.572 | 0.01722 | 0.783 | 0.00072 | 0.65 |
| 1637 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 24 | TIMP3 | Sponge network | 0.249 | 0.35902 | -0.546 | 0.02307 | 0.65 |
| 1638 | AC053503.6 | hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p | 13 | LPP | Sponge network | -4.017 | 0.00205 | -0.459 | 0.04475 | 0.65 |
| 1639 | HAND2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | LHFP | Sponge network | -1.697 | 0.00889 | -0.167 | 0.41371 | 0.65 |
| 1640 | AC006116.24 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 12 | SPG20 | Sponge network | -0.473 | 0.07602 | -1.16 | 0 | 0.65 |
| 1641 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | RECK | Sponge network | -0.739 | 0.00044 | -1.043 | 0 | 0.65 |
| 1642 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 49 | MEF2C | Sponge network | -0.576 | 0.10121 | -0.499 | 0.01448 | 0.65 |
| 1643 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-589-5p | 11 | SLITRK3 | Sponge network | -4.546 | 0 | -3.328 | 0 | 0.65 |
| 1644 | MIR143HG |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-2355-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | NUDT11 | Sponge network | -0.739 | 0.0574 | -0.921 | 0.00376 | 0.65 |
| 1645 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-5p | 26 | RBMS3 | Sponge network | -1.582 | 8.0E-5 | -0.519 | 0.03551 | 0.65 |
| 1646 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | BNC2 | Sponge network | -1.349 | 0.00017 | -0.491 | 0.09988 | 0.649 |
| 1647 | MYLK-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-7-5p | 25 | IGFBP5 | Sponge network | -1.331 | 3.0E-5 | -0.806 | 0.00105 | 0.649 |
| 1648 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | RELN | Sponge network | -2.452 | 0 | -2.403 | 0 | 0.649 |
| 1649 | MIR497HG |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-96-5p | 27 | ASTN1 | Sponge network | -1.439 | 4.0E-5 | -2.389 | 2.0E-5 | 0.649 |
| 1650 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | TAL1 | Sponge network | -0.358 | 0.17255 | -0.462 | 0.00574 | 0.649 |
| 1651 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-93-5p | 34 | RASSF2 | Sponge network | -0.465 | 0.04703 | -0.273 | 0.21961 | 0.649 |
| 1652 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 59 | FAT3 | Sponge network | -1.575 | 6.0E-5 | -1.601 | 0.00051 | 0.649 |
| 1653 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 41 | ESR1 | Sponge network | -0.129 | 0.57177 | -1.035 | 3.0E-5 | 0.649 |
| 1654 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | PRUNE2 | Sponge network | -0.896 | 0.00099 | -1.733 | 0.00022 | 0.649 |
| 1655 | LINC00092 |
hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | PRKCB | Sponge network | -1.886 | 0 | -1.313 | 2.0E-5 | 0.649 |
| 1656 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-744-3p;hsa-miR-942-5p | 36 | HLF | Sponge network | -2.62 | 0 | -2.443 | 0 | 0.649 |
| 1657 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 36 | EPHA3 | Sponge network | 0.997 | 0.00066 | 0.185 | 0.53588 | 0.649 |
| 1658 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 16 | AKAP12 | Sponge network | 0.572 | 0.01722 | -0.932 | 0.0028 | 0.649 |
| 1659 | RP11-166D19.1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 15 | RHOB | Sponge network | -0.576 | 0.10121 | -1.052 | 0 | 0.649 |
| 1660 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 25 | EPHA3 | Sponge network | -0.896 | 0.00099 | 0.185 | 0.53588 | 0.649 |
| 1661 | LINC00702 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | ITGB3 | Sponge network | -0.737 | 0.06182 | -0.011 | 0.95751 | 0.649 |
| 1662 | LINC00702 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ITPKB | Sponge network | -0.737 | 0.06182 | -0.704 | 0.00106 | 0.649 |
| 1663 | BVES-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-628-5p | 45 | RUNX1T1 | Sponge network | -2.62 | 0 | -0.344 | 0.2374 | 0.649 |
| 1664 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-935 | 46 | FAT3 | Sponge network | -0.896 | 0.00099 | -1.601 | 0.00051 | 0.649 |
| 1665 | MIR143HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 54 | TGFBR3 | Sponge network | -0.739 | 0.0574 | -1.173 | 1.0E-5 | 0.649 |
| 1666 | LINC00702 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 22 | SORCS1 | Sponge network | -0.737 | 0.06182 | -3.492 | 0 | 0.649 |
| 1667 | SNHG14 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 34 | DLC1 | Sponge network | -1.111 | 0.0003 | -0.382 | 0.05038 | 0.648 |
| 1668 | AC003090.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 26 | ASTN1 | Sponge network | -2.117 | 0.00161 | -2.389 | 2.0E-5 | 0.648 |
| 1669 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 17 | DAB2 | Sponge network | -0.129 | 0.57177 | 0.431 | 0.0113 | 0.648 |
| 1670 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | EBF1 | Sponge network | -0.358 | 0.17255 | -0.384 | 0.08856 | 0.648 |
| 1671 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 45 | SSBP2 | Sponge network | -0.976 | 0.00025 | -1.58 | 0 | 0.648 |
| 1672 | FAM66C |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 23 | MRVI1 | Sponge network | -0.901 | 0.00019 | -1.051 | 0.00022 | 0.648 |
| 1673 | HOXB-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-378a-3p | 10 | FLNA | Sponge network | -0.154 | 0.53954 | -0.771 | 0.01377 | 0.648 |
| 1674 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -0.612 | 0.00094 | -0.564 | 1.0E-5 | 0.648 |
| 1675 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 38 | TCF4 | Sponge network | -1.092 | 4.0E-5 | 0.016 | 0.92271 | 0.648 |
| 1676 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | ERG | Sponge network | -0.976 | 0.00025 | 0.002 | 0.99011 | 0.648 |
| 1677 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | LRRK2 | Sponge network | -0.576 | 0.10121 | -1.136 | 1.0E-5 | 0.648 |
| 1678 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 48 | PRKAA2 | Sponge network | -0.563 | 0.02545 | -2.462 | 0 | 0.648 |
| 1679 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 61 | QKI | Sponge network | -0.737 | 0.06182 | -0.333 | 0.04111 | 0.648 |
| 1680 | EMX2OS |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 25 | PCDH10 | Sponge network | -0.777 | 0.1134 | -1.644 | 0.0078 | 0.648 |
| 1681 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 37 | IGF1 | Sponge network | -1.111 | 0.0003 | -0.204 | 0.5898 | 0.648 |
| 1682 | GNG12-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | -0.793 | 7.0E-5 | -1.733 | 0.00022 | 0.648 |
| 1683 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-671-5p | 22 | CADM3 | Sponge network | -1.697 | 0.00889 | -3.663 | 0 | 0.648 |
| 1684 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 17 | DCLK1 | Sponge network | -1.575 | 6.0E-5 | -1.324 | 0.00014 | 0.648 |
| 1685 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-93-5p | 25 | ANK2 | Sponge network | -2.531 | 0 | -1.603 | 1.0E-5 | 0.648 |
| 1686 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 21 | DMD | Sponge network | -4.546 | 0 | -1.506 | 0 | 0.648 |
| 1687 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | TLL1 | Sponge network | -0.576 | 0.10121 | -1.195 | 3.0E-5 | 0.648 |
| 1688 | RP11-110I1.11 |
hsa-let-7d-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 20 | MDM4 | Sponge network | 1.034 | 0 | 0.345 | 0.00762 | 0.648 |
| 1689 | TP73-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | ZDBF2 | Sponge network | -0.563 | 0.02545 | -0.998 | 0.00025 | 0.648 |
| 1690 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 30 | ADARB1 | Sponge network | -1.092 | 4.0E-5 | -0.335 | 0.06631 | 0.648 |
| 1691 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | PTPN13 | Sponge network | -0.129 | 0.57177 | -1.208 | 0 | 0.648 |
| 1692 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 64 | SSBP2 | Sponge network | -0.739 | 0.0574 | -1.58 | 0 | 0.648 |
| 1693 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 55 | EPHA7 | Sponge network | -0.737 | 0.06182 | -2.197 | 3.0E-5 | 0.648 |
| 1694 | PWAR6 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ITPKB | Sponge network | -1.575 | 6.0E-5 | -0.704 | 0.00106 | 0.648 |
| 1695 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 29 | AKAP6 | Sponge network | -0.901 | 0.00019 | -1.586 | 3.0E-5 | 0.648 |
| 1696 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 17 | DCLK1 | Sponge network | 0.572 | 0.01722 | -1.324 | 0.00014 | 0.647 |
| 1697 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 68 | PRKAA2 | Sponge network | -1.575 | 6.0E-5 | -2.462 | 0 | 0.647 |
| 1698 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PDGFC | Sponge network | -0.576 | 0.10121 | -0.33 | 0.06483 | 0.647 |
| 1699 | HAND2-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ITPKB | Sponge network | -1.697 | 0.00889 | -0.704 | 0.00106 | 0.647 |
| 1700 | LINC00702 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | RADIL | Sponge network | -0.737 | 0.06182 | -1.628 | 0 | 0.647 |
| 1701 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | LSAMP | Sponge network | -0.49 | 0.04946 | -0.066 | 0.81708 | 0.647 |
| 1702 | MIR497HG |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -1.439 | 4.0E-5 | -0.704 | 0.00106 | 0.647 |
| 1703 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 32 | ZBTB4 | Sponge network | -0.129 | 0.57177 | -0.739 | 0 | 0.647 |
| 1704 | LINC00641 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | FAM172A | Sponge network | -0.222 | 0.32445 | -0.57 | 0 | 0.647 |
| 1705 | FGF14-AS2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-874-3p | 14 | SORCS1 | Sponge network | -1.582 | 8.0E-5 | -3.492 | 0 | 0.647 |
| 1706 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 58 | MDM4 | Sponge network | 0.537 | 0.00139 | 0.345 | 0.00762 | 0.647 |
| 1707 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 34 | IGF1 | Sponge network | -0.739 | 0.0574 | -0.204 | 0.5898 | 0.647 |
| 1708 | MIR497HG |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-5p | 16 | PTGFR | Sponge network | -1.439 | 4.0E-5 | -0.233 | 0.45421 | 0.647 |
| 1709 | USP3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-628-5p | 54 | DTNA | Sponge network | -0.162 | 0.47687 | -1.648 | 1.0E-5 | 0.647 |
| 1710 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | FAT4 | Sponge network | -1.111 | 0.0003 | -0.354 | 0.14694 | 0.647 |
| 1711 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-589-3p;hsa-miR-590-3p | 31 | QKI | Sponge network | -0.358 | 0.17255 | -0.333 | 0.04111 | 0.647 |
| 1712 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p;hsa-miR-624-5p | 14 | SMARCA1 | Sponge network | -0.49 | 0.04946 | -0.684 | 0.00159 | 0.647 |
| 1713 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-93-5p | 36 | TCF4 | Sponge network | -0.473 | 0.07602 | 0.016 | 0.92271 | 0.647 |
| 1714 | LINC00092 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-424-5p | 17 | CBFA2T3 | Sponge network | -1.886 | 0 | -1.221 | 1.0E-5 | 0.647 |
| 1715 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 77 | IL6ST | Sponge network | -0.739 | 0.0574 | -0.402 | 0.00676 | 0.647 |
| 1716 | MIR143HG |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 16 | NGFR | Sponge network | -0.739 | 0.0574 | -1.271 | 0.01058 | 0.647 |
| 1717 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 16 | ADAMTSL3 | Sponge network | -0.932 | 0.00329 | -1.559 | 0 | 0.647 |
| 1718 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 32 | MEF2C | Sponge network | -1.092 | 4.0E-5 | -0.499 | 0.01448 | 0.646 |
| 1719 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-940 | 34 | MYO9A | Sponge network | -0.162 | 0.47687 | -0.147 | 0.32373 | 0.646 |
| 1720 | MRVI1-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-98-5p | 35 | CADM2 | Sponge network | -1.092 | 4.0E-5 | -3.782 | 0 | 0.646 |
| 1721 | ZNF667-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | PRUNE2 | Sponge network | -0.976 | 0.00025 | -1.733 | 0.00022 | 0.646 |
| 1722 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-337-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 31 | RAP1A | Sponge network | -0.612 | 0.00094 | -0.929 | 0 | 0.646 |
| 1723 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | CAV1 | Sponge network | -0.129 | 0.57177 | -1.013 | 6.0E-5 | 0.646 |
| 1724 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | RECK | Sponge network | -1.886 | 0 | -1.043 | 0 | 0.646 |
| 1725 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 30 | SCN9A | Sponge network | -0.49 | 0.04946 | -0.525 | 0.10593 | 0.646 |
| 1726 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 24 | NR2C2 | Sponge network | 0.083 | 0.66405 | 0.21 | 0.03224 | 0.646 |
| 1727 | MYLK-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-550a-5p;hsa-miR-664a-5p | 37 | DST | Sponge network | -1.331 | 3.0E-5 | -0.713 | 0.00096 | 0.646 |
| 1728 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | DYNC1I1 | Sponge network | -0.739 | 0.00044 | -1.12 | 0.00107 | 0.646 |
| 1729 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | CACNA2D1 | Sponge network | -0.739 | 0.0574 | -0.846 | 0.00675 | 0.646 |
| 1730 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 35 | IGF1 | Sponge network | -0.737 | 0.06182 | -0.204 | 0.5898 | 0.646 |
| 1731 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 39 | ATRX | Sponge network | 0.311 | 0.10344 | 0.261 | 0.04408 | 0.646 |
| 1732 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | SETBP1 | Sponge network | -0.896 | 0.00099 | -1.302 | 1.0E-5 | 0.646 |
| 1733 | ADAMTS9-AS2 |
hsa-miR-185-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-542-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-940 | 11 | ILK | Sponge network | -2.452 | 0 | -0.74 | 0 | 0.645 |
| 1734 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | -2.69 | 0.0001 | -1.398 | 2.0E-5 | 0.645 |
| 1735 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | CNTN1 | Sponge network | -0.901 | 0.00019 | -2.594 | 0 | 0.645 |
| 1736 | DNM3OS |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-7-5p | 11 | COL1A2 | Sponge network | 0.572 | 0.01722 | 1.991 | 0 | 0.645 |
| 1737 | LINC00578 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p | 10 | GLI3 | Sponge network | 0.811 | 0.03228 | -0.615 | 0.01482 | 0.645 |
| 1738 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p | 25 | DYNC1I1 | Sponge network | -0.129 | 0.57177 | -1.12 | 0.00107 | 0.645 |
| 1739 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 52 | DIP2C | Sponge network | -0.129 | 0.57177 | -0.436 | 0.01713 | 0.645 |
| 1740 | LINC00702 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | SPG20 | Sponge network | -0.737 | 0.06182 | -1.16 | 0 | 0.645 |
| 1741 | TRAF3IP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 92 | LPP | Sponge network | -0.323 | 0.01372 | -0.459 | 0.04475 | 0.645 |
| 1742 | ACTA2-AS1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 10 | FHL1 | Sponge network | 0.194 | 0.64278 | -2.687 | 0 | 0.645 |
| 1743 | RP11-774O3.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p | 87 | LPP | Sponge network | -0.487 | 0.02157 | -0.459 | 0.04475 | 0.645 |
| 1744 | ARHGEF26-AS1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | RNF180 | Sponge network | -2.46 | 0 | -1.127 | 1.0E-5 | 0.645 |
| 1745 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-429 | 13 | RECK | Sponge network | -2.62 | 0 | -1.043 | 0 | 0.645 |
| 1746 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 69 | TCF4 | Sponge network | -0.49 | 0.04946 | 0.016 | 0.92271 | 0.645 |
| 1747 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 50 | QKI | Sponge network | 0.572 | 0.01722 | -0.333 | 0.04111 | 0.645 |
| 1748 | MIR143HG |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 31 | NCAM2 | Sponge network | -0.739 | 0.0574 | -0.482 | 0.22565 | 0.645 |
| 1749 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | NRXN1 | Sponge network | -1.575 | 6.0E-5 | -3.778 | 0 | 0.645 |
| 1750 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-93-5p | 28 | BNC2 | Sponge network | -2.531 | 0 | -0.491 | 0.09988 | 0.645 |
| 1751 | PGM5-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-2110;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | -4.546 | 0 | -1.74 | 0 | 0.645 |
| 1752 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | SYNE1 | Sponge network | 0.194 | 0.64278 | -0.738 | 0.00316 | 0.645 |
| 1753 | SNHG14 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 24 | CADM3 | Sponge network | -1.111 | 0.0003 | -3.663 | 0 | 0.645 |
| 1754 | LINC00702 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | TPO | Sponge network | -0.737 | 0.06182 | -1.87 | 2.0E-5 | 0.645 |
| 1755 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 14 | DKK3 | Sponge network | 0.572 | 0.01722 | -0.052 | 0.77783 | 0.645 |
| 1756 | RP11-774O3.3 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 15 | MACF1 | Sponge network | -0.487 | 0.02157 | 0.019 | 0.90335 | 0.645 |
| 1757 | SNHG14 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 35 | NAV3 | Sponge network | -1.111 | 0.0003 | 0.212 | 0.47729 | 0.645 |
| 1758 | MEG3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p | 11 | HMCN1 | Sponge network | 0.249 | 0.35902 | 0.809 | 0.00544 | 0.645 |
| 1759 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 11 | FGFR1 | Sponge network | -0.473 | 0.07602 | -0.509 | 0.03957 | 0.645 |
| 1760 | SNHG14 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 36 | PRKCB | Sponge network | -1.111 | 0.0003 | -1.313 | 2.0E-5 | 0.645 |
| 1761 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 35 | RBMS3 | Sponge network | -0.896 | 0.00099 | -0.519 | 0.03551 | 0.644 |
| 1762 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | SYNE1 | Sponge network | -0.769 | 0.00035 | -0.738 | 0.00316 | 0.644 |
| 1763 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | CNTNAP1 | Sponge network | 0.889 | 9.0E-5 | 0.358 | 0.13727 | 0.644 |
| 1764 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p | 23 | CNTN1 | Sponge network | -2.62 | 0 | -2.594 | 0 | 0.644 |
| 1765 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 50 | RBMS3 | Sponge network | 0.997 | 0.00066 | -0.519 | 0.03551 | 0.644 |
| 1766 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | RASSF8 | Sponge network | 0.997 | 0.00066 | -0.122 | 0.65591 | 0.644 |
| 1767 | FAM66C |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 54 | DTNA | Sponge network | -0.901 | 0.00019 | -1.648 | 1.0E-5 | 0.644 |
| 1768 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-5p | 33 | ESR1 | Sponge network | -1.439 | 4.0E-5 | -1.035 | 3.0E-5 | 0.644 |
| 1769 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 38 | EPHA7 | Sponge network | -0.896 | 0.00099 | -2.197 | 3.0E-5 | 0.644 |
| 1770 | HAND2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-2355-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 11 | NUDT11 | Sponge network | -1.697 | 0.00889 | -0.921 | 0.00376 | 0.644 |
| 1771 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-423-5p | 12 | PTGFR | Sponge network | -1.092 | 4.0E-5 | -0.233 | 0.45421 | 0.644 |
| 1772 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 32 | DDX6 | Sponge network | 0.537 | 0.00139 | 0.091 | 0.36428 | 0.644 |
| 1773 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429 | 10 | PARK2 | Sponge network | -0.976 | 0.00025 | -1.892 | 0 | 0.644 |
| 1774 | SERTAD4-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-502-5p;hsa-miR-576-5p | 21 | DTNA | Sponge network | -0.932 | 0.00329 | -1.648 | 1.0E-5 | 0.644 |
| 1775 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 20 | RBMS3 | Sponge network | -0.932 | 0.00329 | -0.519 | 0.03551 | 0.644 |
| 1776 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 54 | EPHA7 | Sponge network | -0.129 | 0.57177 | -2.197 | 3.0E-5 | 0.644 |
| 1777 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CACNA2D1 | Sponge network | -2.452 | 0 | -0.846 | 0.00675 | 0.644 |
| 1778 | LINC00702 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DCN | Sponge network | -0.737 | 0.06182 | -1.288 | 0 | 0.644 |
| 1779 | FENDRR |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 92 | LPP | Sponge network | -1.281 | 0.00012 | -0.459 | 0.04475 | 0.644 |
| 1780 | MIR143HG |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 78 | CADM2 | Sponge network | -0.739 | 0.0574 | -3.782 | 0 | 0.644 |
| 1781 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-484 | 11 | EBF3 | Sponge network | -1.331 | 3.0E-5 | -0.058 | 0.81437 | 0.643 |
| 1782 | RP11-774O3.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | MRVI1 | Sponge network | -0.487 | 0.02157 | -1.051 | 0.00022 | 0.643 |
| 1783 | FENDRR |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | TMEFF2 | Sponge network | -1.281 | 0.00012 | -2.426 | 0 | 0.643 |
| 1784 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 56 | SSBP2 | Sponge network | -0.49 | 0.04946 | -1.58 | 0 | 0.643 |
| 1785 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-664a-5p | 36 | ADARB1 | Sponge network | -0.583 | 0.14589 | -0.335 | 0.06631 | 0.643 |
| 1786 | USP3-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-877-5p;hsa-miR-93-5p | 54 | AR | Sponge network | -0.162 | 0.47687 | -1.554 | 0 | 0.643 |
| 1787 | MIR143HG |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 23 | CADM3 | Sponge network | -0.739 | 0.0574 | -3.663 | 0 | 0.643 |
| 1788 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 30 | EBF1 | Sponge network | -0.976 | 0.00025 | -0.384 | 0.08856 | 0.643 |
| 1789 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 39 | MCC | Sponge network | -0.901 | 0.00019 | -1.005 | 0 | 0.643 |
| 1790 | RAD51-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-766-3p | 30 | MDM4 | Sponge network | 0.768 | 7.0E-5 | 0.345 | 0.00762 | 0.643 |
| 1791 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 35 | GRIN2A | Sponge network | -1.111 | 0.0003 | -2.161 | 0 | 0.643 |
| 1792 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | SPG20 | Sponge network | -0.358 | 0.17255 | -1.16 | 0 | 0.643 |
| 1793 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 59 | AR | Sponge network | -2.46 | 0 | -1.554 | 0 | 0.643 |
| 1794 | PART1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 21 | PRUNE2 | Sponge network | -2.925 | 0 | -1.733 | 0.00022 | 0.643 |
| 1795 | MIR497HG |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-5p | 11 | MN1 | Sponge network | -1.439 | 4.0E-5 | -0.358 | 0.24172 | 0.643 |
| 1796 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 29 | EPHA3 | Sponge network | -1.439 | 4.0E-5 | 0.185 | 0.53588 | 0.643 |
| 1797 | RP11-488L18.10 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p | 13 | MDM4 | Sponge network | 1.392 | 0 | 0.345 | 0.00762 | 0.643 |
| 1798 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DACT1 | Sponge network | -0.576 | 0.10121 | 0.783 | 0.00072 | 0.643 |
| 1799 | HAND2-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 43 | IGFBP5 | Sponge network | -1.697 | 0.00889 | -0.806 | 0.00105 | 0.643 |
| 1800 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 50 | QKI | Sponge network | 0.335 | 0.20596 | -0.333 | 0.04111 | 0.643 |
| 1801 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 14 | MAPK10 | Sponge network | -0.896 | 0.00099 | -1.774 | 0 | 0.643 |
| 1802 | RP11-774O3.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-708-5p;hsa-miR-766-3p | 16 | EZH1 | Sponge network | -0.487 | 0.02157 | -0.564 | 1.0E-5 | 0.643 |
| 1803 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p | 20 | FGFR1 | Sponge network | -2.452 | 0 | -0.509 | 0.03957 | 0.643 |
| 1804 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 23 | RUNX1T1 | Sponge network | -0.376 | 0.00337 | -0.344 | 0.2374 | 0.643 |
| 1805 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 37 | JAZF1 | Sponge network | -0.576 | 0.10121 | -0.377 | 0.02677 | 0.642 |
| 1806 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-935 | 10 | DMD | Sponge network | -0.691 | 0.00146 | -1.506 | 0 | 0.642 |
| 1807 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 25 | AKAP6 | Sponge network | -0.487 | 0.02157 | -1.586 | 3.0E-5 | 0.642 |
| 1808 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940;hsa-miR-98-5p | 45 | PBX1 | Sponge network | -1.111 | 0.0003 | -1.206 | 0 | 0.642 |
| 1809 | MRVI1-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-942-5p | 23 | IGFBP5 | Sponge network | -1.092 | 4.0E-5 | -0.806 | 0.00105 | 0.642 |
| 1810 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-5p | 32 | TSHZ3 | Sponge network | -2.69 | 0.0001 | -0.556 | 0.01516 | 0.642 |
| 1811 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 81 | NTRK3 | Sponge network | -0.737 | 0.06182 | -1.77 | 3.0E-5 | 0.642 |
| 1812 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 13 | PTPN13 | Sponge network | -0.576 | 0.10121 | -1.208 | 0 | 0.642 |
| 1813 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | FAM172A | Sponge network | -2.452 | 0 | -0.57 | 0 | 0.641 |
| 1814 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 84 | NTRK3 | Sponge network | -0.739 | 0.0574 | -1.77 | 3.0E-5 | 0.641 |
| 1815 | LINC00578 |
hsa-let-7d-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-664a-3p | 16 | THBS1 | Sponge network | 0.811 | 0.03228 | 0.741 | 0.00386 | 0.641 |
| 1816 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 10 | ABCC9 | Sponge network | -4.546 | 0 | -0.466 | 0.14121 | 0.641 |
| 1817 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-942-5p | 16 | NRXN1 | Sponge network | -2.531 | 0 | -3.778 | 0 | 0.641 |
| 1818 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-598-3p | 11 | MN1 | Sponge network | 0.194 | 0.64278 | -0.358 | 0.24172 | 0.641 |
| 1819 | ARHGEF26-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 20 | MAPK10 | Sponge network | -2.46 | 0 | -1.774 | 0 | 0.641 |
| 1820 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-628-5p | 28 | ERC1 | Sponge network | 0.064 | 0.72934 | -0.108 | 0.38794 | 0.641 |
| 1821 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 25 | SYNPO2 | Sponge network | -0.096 | 0.67958 | -2.342 | 0 | 0.641 |
| 1822 | MIR497HG |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 22 | PCDH9 | Sponge network | -1.439 | 4.0E-5 | -1.823 | 4.0E-5 | 0.641 |
| 1823 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | NALCN | Sponge network | 0.572 | 0.01722 | 1.068 | 0.00237 | 0.641 |
| 1824 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 20 | RCAN2 | Sponge network | -0.737 | 0.06182 | -0.33 | 0.15172 | 0.641 |
| 1825 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 46 | DIP2C | Sponge network | -0.576 | 0.10121 | -0.436 | 0.01713 | 0.641 |
| 1826 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | JAZF1 | Sponge network | -1.111 | 0.0003 | -0.377 | 0.02677 | 0.641 |
| 1827 | LINC00092 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-345-5p;hsa-miR-423-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-769-5p;hsa-miR-940 | 36 | NTRK3 | Sponge network | -1.886 | 0 | -1.77 | 3.0E-5 | 0.641 |
| 1828 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-421 | 11 | PEG3 | Sponge network | -1.654 | 0.00144 | -1.74 | 0 | 0.64 |
| 1829 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-629-3p | 25 | RASSF2 | Sponge network | -0.358 | 0.17255 | -0.273 | 0.21961 | 0.64 |
| 1830 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 13 | HIC1 | Sponge network | -0.49 | 0.04946 | 0.024 | 0.89889 | 0.64 |
| 1831 | NR2F1-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 15 | NGFR | Sponge network | -0.49 | 0.04946 | -1.271 | 0.01058 | 0.64 |
| 1832 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 39 | MYO9A | Sponge network | 0.101 | 0.60919 | -0.147 | 0.32373 | 0.64 |
| 1833 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 55 | ADARB1 | Sponge network | -0.737 | 0.06182 | -0.335 | 0.06631 | 0.64 |
| 1834 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 37 | EBF1 | Sponge network | -0.576 | 0.10121 | -0.384 | 0.08856 | 0.64 |
| 1835 | ACTA2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p | 16 | EZH1 | Sponge network | 0.194 | 0.64278 | -0.564 | 1.0E-5 | 0.64 |
| 1836 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 53 | RUNX1T1 | Sponge network | -0.896 | 0.00099 | -0.344 | 0.2374 | 0.64 |
| 1837 | FGF14-AS2 |
hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-96-5p | 10 | RECK | Sponge network | -1.582 | 8.0E-5 | -1.043 | 0 | 0.64 |
| 1838 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 56 | ADARB1 | Sponge network | -1.697 | 0.00889 | -0.335 | 0.06631 | 0.64 |
| 1839 | HAND2-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | -1.697 | 0.00889 | -0.564 | 1.0E-5 | 0.64 |
| 1840 | LINC00472 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 21 | SETBP1 | Sponge network | -0.842 | 0.01361 | -1.302 | 1.0E-5 | 0.64 |
| 1841 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | PEG3 | Sponge network | -0.896 | 0.00099 | -1.74 | 0 | 0.64 |
| 1842 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PEG3 | Sponge network | 0.335 | 0.20596 | -1.74 | 0 | 0.64 |
| 1843 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-671-5p | 21 | RBMS3 | Sponge network | -1.886 | 0 | -0.519 | 0.03551 | 0.64 |
| 1844 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-425-5p | 12 | ITGB3 | Sponge network | -0.583 | 0.14589 | -0.011 | 0.95751 | 0.64 |
| 1845 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-3p | 23 | RASSF8 | Sponge network | -0.358 | 0.17255 | -0.122 | 0.65591 | 0.64 |
| 1846 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-93-3p | 56 | RUNX1T1 | Sponge network | 0.249 | 0.35902 | -0.344 | 0.2374 | 0.64 |
| 1847 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p | 15 | AKAP12 | Sponge network | -0.162 | 0.47687 | -0.932 | 0.0028 | 0.64 |
| 1848 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 24 | PTGFR | Sponge network | -1.697 | 0.00889 | -0.233 | 0.45421 | 0.64 |
| 1849 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 59 | QKI | Sponge network | -2.452 | 0 | -0.333 | 0.04111 | 0.64 |
| 1850 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-508-3p;hsa-miR-590-3p | 21 | CACNA2D1 | Sponge network | -0.358 | 0.17255 | -0.846 | 0.00675 | 0.64 |
| 1851 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 15 | DAB2 | Sponge network | 0.572 | 0.01722 | 0.431 | 0.0113 | 0.64 |
| 1852 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-935;hsa-miR-98-5p | 49 | DMD | Sponge network | -2.46 | 0 | -1.506 | 0 | 0.639 |
| 1853 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-550a-3p;hsa-miR-660-5p | 34 | RUNX1T1 | Sponge network | -4.546 | 0 | -0.344 | 0.2374 | 0.639 |
| 1854 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | PREX2 | Sponge network | -2.452 | 0 | -0.272 | 0.25382 | 0.639 |
| 1855 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-98-5p | 29 | PBX1 | Sponge network | -1.092 | 4.0E-5 | -1.206 | 0 | 0.639 |
| 1856 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-589-3p | 29 | ZFHX4 | Sponge network | 0.811 | 0.03228 | 0.018 | 0.95574 | 0.639 |
| 1857 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | CACNA2D1 | Sponge network | -1.281 | 0.00012 | -0.846 | 0.00675 | 0.639 |
| 1858 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p | 19 | AFF3 | Sponge network | -2.62 | 0 | -2.373 | 0 | 0.639 |
| 1859 | SNHG14 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-96-5p | 37 | ASTN1 | Sponge network | -1.111 | 0.0003 | -2.389 | 2.0E-5 | 0.639 |
| 1860 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 54 | TACC1 | Sponge network | -0.129 | 0.57177 | -0.362 | 0.05406 | 0.639 |
| 1861 | RP11-693J15.4 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | PRKCB | Sponge network | -0.72 | 0.24239 | -1.313 | 2.0E-5 | 0.639 |
| 1862 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 16 | NR2C2 | Sponge network | 1.601 | 0 | 0.21 | 0.03224 | 0.639 |
| 1863 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-324-3p;hsa-miR-3913-5p;hsa-miR-629-3p | 20 | ZBTB4 | Sponge network | -0.583 | 0.14589 | -0.739 | 0 | 0.639 |
| 1864 | MIR143HG |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 81 | DST | Sponge network | -0.739 | 0.0574 | -0.713 | 0.00096 | 0.639 |
| 1865 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ABCC9 | Sponge network | 0.335 | 0.20596 | -0.466 | 0.14121 | 0.639 |
| 1866 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-874-3p | 18 | NR2C2 | Sponge network | 1.254 | 0 | 0.21 | 0.03224 | 0.639 |
| 1867 | AC003090.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-532-3p | 17 | SYNM | Sponge network | -2.117 | 0.00161 | -2.309 | 1.0E-5 | 0.639 |
| 1868 | RP11-359E10.1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-484;hsa-miR-7-5p | 11 | FLNA | Sponge network | 0.299 | 0.57722 | -0.771 | 0.01377 | 0.639 |
| 1869 | RP11-736K20.5 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p | 14 | FGFR1 | Sponge network | -0.358 | 0.17255 | -0.509 | 0.03957 | 0.639 |
| 1870 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 47 | DIP2C | Sponge network | -2.452 | 0 | -0.436 | 0.01713 | 0.638 |
| 1871 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p | 32 | AR | Sponge network | -1.582 | 8.0E-5 | -1.554 | 0 | 0.638 |
| 1872 | LINC01018 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 48 | PRKAA2 | Sponge network | -2.915 | 0.00015 | -2.462 | 0 | 0.638 |
| 1873 | BZRAP1-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-7-5p | 11 | FNBP1 | Sponge network | -0.465 | 0.04703 | -0.969 | 4.0E-5 | 0.638 |
| 1874 | HAND2-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 16 | NGFR | Sponge network | -1.697 | 0.00889 | -1.271 | 0.01058 | 0.638 |
| 1875 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-877-5p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | -0.096 | 0.67958 | -1.603 | 1.0E-5 | 0.638 |
| 1876 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 14 | GLI3 | Sponge network | 0.194 | 0.64278 | -0.615 | 0.01482 | 0.638 |
| 1877 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-744-5p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | -0.563 | 0.02545 | -0.509 | 0.03957 | 0.638 |
| 1878 | LINC00641 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-766-3p | 17 | EZH1 | Sponge network | -0.222 | 0.32445 | -0.564 | 1.0E-5 | 0.638 |
| 1879 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 43 | RBMS3 | Sponge network | -2.117 | 0.00161 | -0.519 | 0.03551 | 0.638 |
| 1880 | MRVI1-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 18 | PRKCB | Sponge network | -1.092 | 4.0E-5 | -1.313 | 2.0E-5 | 0.638 |
| 1881 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p | 24 | ANK2 | Sponge network | -0.739 | 0.00044 | -1.603 | 1.0E-5 | 0.638 |
| 1882 | AC009120.5 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 19 | MDM4 | Sponge network | 1.383 | 0 | 0.345 | 0.00762 | 0.638 |
| 1883 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | PGR | Sponge network | -0.376 | 0.00337 | -1.704 | 0 | 0.638 |
| 1884 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 29 | GPC6 | Sponge network | -0.576 | 0.10121 | -0.054 | 0.81465 | 0.638 |
| 1885 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-5p | 10 | MN1 | Sponge network | 0.335 | 0.20596 | -0.358 | 0.24172 | 0.638 |
| 1886 | LINC00092 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-940 | 25 | GRIK3 | Sponge network | -1.886 | 0 | -3.208 | 0 | 0.638 |
| 1887 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 22 | BNC2 | Sponge network | -0.376 | 0.00337 | -0.491 | 0.09988 | 0.638 |
| 1888 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-500a-3p | 16 | ADAMTSL3 | Sponge network | -0.469 | 0.08721 | -1.559 | 0 | 0.638 |
| 1889 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | SLITRK3 | Sponge network | -2.46 | 0 | -3.328 | 0 | 0.638 |
| 1890 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 44 | BNC2 | Sponge network | -0.611 | 0.09607 | -0.491 | 0.09988 | 0.638 |
| 1891 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | PDGFC | Sponge network | -0.129 | 0.57177 | -0.33 | 0.06483 | 0.638 |
| 1892 | ARHGEF26-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | MRVI1 | Sponge network | -2.46 | 0 | -1.051 | 0.00022 | 0.637 |
| 1893 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 14 | CDH19 | Sponge network | -0.583 | 0.14589 | -3.434 | 0 | 0.637 |
| 1894 | PCA3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-96-5p | 86 | LPP | Sponge network | -0.709 | 0.18168 | -0.459 | 0.04475 | 0.637 |
| 1895 | MIR497HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 47 | GAS7 | Sponge network | -1.439 | 4.0E-5 | -0.555 | 0.01602 | 0.637 |
| 1896 | RP11-774O3.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | FAM172A | Sponge network | -0.487 | 0.02157 | -0.57 | 0 | 0.637 |
| 1897 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-7-1-3p | 11 | DCN | Sponge network | -1.349 | 0.00017 | -1.288 | 0 | 0.637 |
| 1898 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-940 | 14 | CNTNAP1 | Sponge network | -0.096 | 0.67958 | 0.358 | 0.13727 | 0.637 |
| 1899 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 20 | ITGA5 | Sponge network | -0.129 | 0.57177 | 0.452 | 0.03524 | 0.637 |
| 1900 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 28 | SETBP1 | Sponge network | -0.323 | 0.01372 | -1.302 | 1.0E-5 | 0.637 |
| 1901 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 50 | GAS7 | Sponge network | 0.572 | 0.01722 | -0.555 | 0.01602 | 0.637 |
| 1902 | USP3-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-342-3p;hsa-miR-361-5p | 17 | ABL1 | Sponge network | -0.162 | 0.47687 | -0.215 | 0.07804 | 0.637 |
| 1903 | AC006116.24 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p | 11 | RASSF8 | Sponge network | -0.473 | 0.07602 | -0.122 | 0.65591 | 0.637 |
| 1904 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 59 | ZFHX4 | Sponge network | -1.111 | 0.0003 | 0.018 | 0.95574 | 0.637 |
| 1905 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | PDZRN3 | Sponge network | -1.575 | 6.0E-5 | -1.548 | 0 | 0.637 |
| 1906 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p | 15 | CADM3 | Sponge network | -0.583 | 0.14589 | -3.663 | 0 | 0.637 |
| 1907 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-576-5p | 25 | FAT3 | Sponge network | -0.932 | 0.00329 | -1.601 | 0.00051 | 0.637 |
| 1908 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-589-5p | 16 | SLITRK3 | Sponge network | -2.62 | 0 | -3.328 | 0 | 0.637 |
| 1909 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-511-5p | 13 | ADAMTSL3 | Sponge network | -2.531 | 0 | -1.559 | 0 | 0.637 |
| 1910 | HCG18 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | TPR | Sponge network | 1.063 | 0 | 0.582 | 0 | 0.637 |
| 1911 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | JAZF1 | Sponge network | -0.976 | 0.00025 | -0.377 | 0.02677 | 0.637 |
| 1912 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | ZNF521 | Sponge network | -2.452 | 0 | -0.111 | 0.58068 | 0.637 |
| 1913 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | NRXN3 | Sponge network | -2.46 | 0 | -0.893 | 0.04186 | 0.637 |
| 1914 | MAGI2-AS3 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | MSN | Sponge network | -0.129 | 0.57177 | -0.023 | 0.88422 | 0.637 |
| 1915 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 36 | ZBTB4 | Sponge network | -1.111 | 0.0003 | -0.739 | 0 | 0.637 |
| 1916 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-3p | 32 | BNC2 | Sponge network | -0.154 | 0.53954 | -0.491 | 0.09988 | 0.637 |
| 1917 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p | 37 | ZFHX4 | Sponge network | -1.092 | 4.0E-5 | 0.018 | 0.95574 | 0.637 |
| 1918 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 22 | FAT4 | Sponge network | -0.576 | 0.10121 | -0.354 | 0.14694 | 0.636 |
| 1919 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-671-5p | 14 | KIT | Sponge network | -1.092 | 4.0E-5 | -2.184 | 0 | 0.636 |
| 1920 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 11 | CAV1 | Sponge network | -1.439 | 4.0E-5 | -1.013 | 6.0E-5 | 0.636 |
| 1921 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | HOOK3 | Sponge network | 0.225 | 0.30644 | 0.273 | 0.09957 | 0.636 |
| 1922 | MIR143HG |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 14 | PHOX2B | Sponge network | -0.739 | 0.0574 | -2.68 | 0 | 0.636 |
| 1923 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | ABI2 | Sponge network | 0.532 | 0.00505 | 0.175 | 0.14787 | 0.636 |
| 1924 | ZNF667-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 21 | SORCS1 | Sponge network | -0.976 | 0.00025 | -3.492 | 0 | 0.636 |
| 1925 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | TACC1 | Sponge network | -0.576 | 0.10121 | -0.362 | 0.05406 | 0.636 |
| 1926 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 10 | JAKMIP2 | Sponge network | -0.896 | 0.00099 | -1.388 | 0 | 0.636 |
| 1927 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 30 | EPHA3 | Sponge network | -2.452 | 0 | 0.185 | 0.53588 | 0.636 |
| 1928 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | PRRX1 | Sponge network | -0.576 | 0.10121 | 1.316 | 0 | 0.636 |
| 1929 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CDH11 | Sponge network | -0.576 | 0.10121 | 1.427 | 0 | 0.636 |
| 1930 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 75 | TCF4 | Sponge network | -0.737 | 0.06182 | 0.016 | 0.92271 | 0.636 |
| 1931 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 56 | ZFHX4 | Sponge network | 0.997 | 0.00066 | 0.018 | 0.95574 | 0.636 |
| 1932 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 42 | JAZF1 | Sponge network | -0.739 | 0.0574 | -0.377 | 0.02677 | 0.636 |
| 1933 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p | 37 | ESR1 | Sponge network | -2.452 | 0 | -1.035 | 3.0E-5 | 0.636 |
| 1934 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 54 | FAT3 | Sponge network | 0.194 | 0.64278 | -1.601 | 0.00051 | 0.636 |
| 1935 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | CACNA2D1 | Sponge network | -0.576 | 0.10121 | -0.846 | 0.00675 | 0.636 |
| 1936 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 68 | TCF4 | Sponge network | 0.249 | 0.35902 | 0.016 | 0.92271 | 0.636 |
| 1937 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | EBF3 | Sponge network | -0.576 | 0.10121 | -0.058 | 0.81437 | 0.636 |
| 1938 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | ZNF521 | Sponge network | -1.111 | 0.0003 | -0.111 | 0.58068 | 0.636 |
| 1939 | MIR143HG |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | THSD7B | Sponge network | -0.739 | 0.0574 | 0.154 | 0.74723 | 0.636 |
| 1940 | RP11-855A2.3 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 25 | MDM4 | Sponge network | 2.094 | 0 | 0.345 | 0.00762 | 0.636 |
| 1941 | EMX2OS |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-625-5p | 14 | NGFR | Sponge network | -0.777 | 0.1134 | -1.271 | 0.01058 | 0.636 |
| 1942 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 16 | FNBP1 | Sponge network | -0.769 | 0.00035 | -0.969 | 4.0E-5 | 0.636 |
| 1943 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | EBF3 | Sponge network | -0.976 | 0.00025 | -0.058 | 0.81437 | 0.636 |
| 1944 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 25 | CDH19 | Sponge network | -2.46 | 0 | -3.434 | 0 | 0.635 |
| 1945 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | CDH19 | Sponge network | -0.49 | 0.04946 | -3.434 | 0 | 0.635 |
| 1946 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-34a-5p | 11 | AHNAK | Sponge network | -1.331 | 3.0E-5 | -1.207 | 0 | 0.635 |
| 1947 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p | 17 | AKAP12 | Sponge network | -0.901 | 0.00019 | -0.932 | 0.0028 | 0.635 |
| 1948 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-550a-3p | 16 | PGR | Sponge network | -2.095 | 0.00112 | -1.704 | 0 | 0.635 |
| 1949 | FENDRR |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | -1.281 | 0.00012 | -1.398 | 2.0E-5 | 0.635 |
| 1950 | RP11-774O3.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 19 | PRUNE2 | Sponge network | -0.487 | 0.02157 | -1.733 | 0.00022 | 0.635 |
| 1951 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | GRIN2A | Sponge network | -1.439 | 4.0E-5 | -2.161 | 0 | 0.635 |
| 1952 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | FAT4 | Sponge network | -0.976 | 0.00025 | -0.354 | 0.14694 | 0.635 |
| 1953 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-5p | 29 | ESR1 | Sponge network | -0.465 | 0.04703 | -1.035 | 3.0E-5 | 0.635 |
| 1954 | AC006116.24 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-582-3p;hsa-miR-93-5p | 28 | GAS7 | Sponge network | -0.473 | 0.07602 | -0.555 | 0.01602 | 0.635 |
| 1955 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | CNTNAP1 | Sponge network | 0.299 | 0.57722 | 0.358 | 0.13727 | 0.635 |
| 1956 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 39 | RASSF8 | Sponge network | -1.697 | 0.00889 | -0.122 | 0.65591 | 0.635 |
| 1957 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-7-1-3p | 21 | CNTN1 | Sponge network | -4.546 | 0 | -2.594 | 0 | 0.635 |
| 1958 | FENDRR |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-93-3p | 67 | RUNX1T1 | Sponge network | -1.281 | 0.00012 | -0.344 | 0.2374 | 0.635 |
| 1959 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-3p | 39 | BNC2 | Sponge network | -0.896 | 0.00099 | -0.491 | 0.09988 | 0.635 |
| 1960 | MIR143HG |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-582-5p | 12 | PRICKLE1 | Sponge network | -0.739 | 0.0574 | -0.112 | 0.67088 | 0.635 |
| 1961 | RP11-166D19.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p | 17 | EZH1 | Sponge network | -0.576 | 0.10121 | -0.564 | 1.0E-5 | 0.635 |
| 1962 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 21 | CADM3 | Sponge network | -2.69 | 0.0001 | -3.663 | 0 | 0.635 |
| 1963 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 31 | ZBTB4 | Sponge network | -0.976 | 0.00025 | -0.739 | 0 | 0.635 |
| 1964 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 39 | JAZF1 | Sponge network | -2.452 | 0 | -0.377 | 0.02677 | 0.635 |
| 1965 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 34 | MITF | Sponge network | -0.739 | 0.0574 | -0.769 | 0.00022 | 0.635 |
| 1966 | FAM66C |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-501-5p | 13 | MN1 | Sponge network | -0.901 | 0.00019 | -0.358 | 0.24172 | 0.634 |
| 1967 | CTD-3064M3.3 |
hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429 | 11 | SYNE1 | Sponge network | -0.469 | 0.08721 | -0.738 | 0.00316 | 0.634 |
| 1968 | MIR143HG |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | FAM172A | Sponge network | -0.739 | 0.0574 | -0.57 | 0 | 0.634 |
| 1969 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 51 | AR | Sponge network | 0.335 | 0.20596 | -1.554 | 0 | 0.634 |
| 1970 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 59 | WDFY3 | Sponge network | -0.942 | 0 | -0.201 | 0.0967 | 0.634 |
| 1971 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-96-5p | 27 | BNC2 | Sponge network | -1.582 | 8.0E-5 | -0.491 | 0.09988 | 0.634 |
| 1972 | FENDRR |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 20 | SORCS1 | Sponge network | -1.281 | 0.00012 | -3.492 | 0 | 0.634 |
| 1973 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 47 | EPHA7 | Sponge network | -0.976 | 0.00025 | -2.197 | 3.0E-5 | 0.634 |
| 1974 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-582-5p | 11 | MAPK10 | Sponge network | -0.469 | 0.08721 | -1.774 | 0 | 0.634 |
| 1975 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 23 | SYNE1 | Sponge network | -0.096 | 0.67958 | -0.738 | 0.00316 | 0.634 |
| 1976 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-33a-3p;hsa-miR-577 | 12 | ESR1 | Sponge network | -0.31 | 0.33871 | -1.035 | 3.0E-5 | 0.634 |
| 1977 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 38 | MCC | Sponge network | -0.487 | 0.02157 | -1.005 | 0 | 0.634 |
| 1978 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-501-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | EBF3 | Sponge network | -1.439 | 4.0E-5 | -0.058 | 0.81437 | 0.634 |
| 1979 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-550a-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 35 | MEF2C | Sponge network | 0.4 | 0.14611 | -0.499 | 0.01448 | 0.634 |
| 1980 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3913-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | ESR1 | Sponge network | -1.886 | 0 | -1.035 | 3.0E-5 | 0.634 |
| 1981 | MRVI1-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 18 | NAV3 | Sponge network | -1.092 | 4.0E-5 | 0.212 | 0.47729 | 0.634 |
| 1982 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p | 41 | IL6ST | Sponge network | -1.092 | 4.0E-5 | -0.402 | 0.00676 | 0.634 |
| 1983 | MRVI1-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p | 30 | GRIK3 | Sponge network | -1.092 | 4.0E-5 | -3.208 | 0 | 0.633 |
| 1984 | PWAR6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CDH19 | Sponge network | -1.575 | 6.0E-5 | -3.434 | 0 | 0.633 |
| 1985 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p | 10 | ABCC9 | Sponge network | -0.154 | 0.53954 | -0.466 | 0.14121 | 0.633 |
| 1986 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-624-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-940 | 38 | NTRK3 | Sponge network | -4.546 | 0 | -1.77 | 3.0E-5 | 0.633 |
| 1987 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-582-3p;hsa-miR-590-3p | 49 | ADARB1 | Sponge network | -2.452 | 0 | -0.335 | 0.06631 | 0.633 |
| 1988 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 18 | GRIN2A | Sponge network | -1.092 | 4.0E-5 | -2.161 | 0 | 0.633 |
| 1989 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 55 | AR | Sponge network | -0.487 | 0.02157 | -1.554 | 0 | 0.633 |
| 1990 | MEG3 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | RECK | Sponge network | 0.249 | 0.35902 | -1.043 | 0 | 0.633 |
| 1991 | RP5-1198O20.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 30 | WWTR1 | Sponge network | 0.997 | 0.00066 | -0.345 | 0.11997 | 0.633 |
| 1992 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | CAV1 | Sponge network | -0.976 | 0.00025 | -1.013 | 6.0E-5 | 0.633 |
| 1993 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 47 | AR | Sponge network | 0.572 | 0.01722 | -1.554 | 0 | 0.633 |
| 1994 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 46 | MCC | Sponge network | -1.575 | 6.0E-5 | -1.005 | 0 | 0.633 |
| 1995 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 80 | NTRK3 | Sponge network | -1.697 | 0.00889 | -1.77 | 3.0E-5 | 0.633 |
| 1996 | SERTAD4-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-424-5p | 10 | DYNC1I1 | Sponge network | -0.932 | 0.00329 | -1.12 | 0.00107 | 0.633 |
| 1997 | BVES-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-874-3p;hsa-miR-935 | 26 | GRIK3 | Sponge network | -2.62 | 0 | -3.208 | 0 | 0.633 |
| 1998 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-508-3p;hsa-miR-628-5p | 24 | FBXO32 | Sponge network | -0.154 | 0.53954 | -0.495 | 0.07561 | 0.633 |
| 1999 | SERTAD4-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-335-3p | 12 | AKAP6 | Sponge network | -0.932 | 0.00329 | -1.586 | 3.0E-5 | 0.633 |
| 2000 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | NRXN3 | Sponge network | -0.096 | 0.67958 | -0.893 | 0.04186 | 0.633 |
| 2001 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-374b-5p | 14 | CBL | Sponge network | 0.542 | 0.00054 | 0.291 | 0.01267 | 0.633 |
| 2002 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 31 | PRKAA2 | Sponge network | -1.092 | 4.0E-5 | -2.462 | 0 | 0.633 |
| 2003 | LINC00578 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 15 | NCAM2 | Sponge network | 0.811 | 0.03228 | -0.482 | 0.22565 | 0.633 |
| 2004 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-505-3p | 24 | DIP2C | Sponge network | -0.583 | 0.14589 | -0.436 | 0.01713 | 0.633 |
| 2005 | RP11-400K9.4 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-542-3p | 12 | SYNM | Sponge network | -0.611 | 0.09607 | -2.309 | 1.0E-5 | 0.633 |
| 2006 | NR2F1-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 25 | SORCS1 | Sponge network | -0.49 | 0.04946 | -3.492 | 0 | 0.633 |
| 2007 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | RASSF8 | Sponge network | 0.194 | 0.64278 | -0.122 | 0.65591 | 0.633 |
| 2008 | CTD-3064M3.3 |
hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-502-5p | 10 | ELN | Sponge network | -0.469 | 0.08721 | 0.451 | 0.13752 | 0.633 |
| 2009 | LINC00702 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 73 | CADM2 | Sponge network | -0.737 | 0.06182 | -3.782 | 0 | 0.633 |
| 2010 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p | 11 | PARK2 | Sponge network | -0.162 | 0.47687 | -1.892 | 0 | 0.633 |
| 2011 | TRAF3IP2-AS1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | RNF180 | Sponge network | -0.323 | 0.01372 | -1.127 | 1.0E-5 | 0.632 |
| 2012 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 16 | PRDM5 | Sponge network | -0.576 | 0.10121 | -1.09 | 0.00014 | 0.632 |
| 2013 | ZNF667-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | ZDBF2 | Sponge network | -0.976 | 0.00025 | -0.998 | 0.00025 | 0.632 |
| 2014 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | AFF3 | Sponge network | -2.117 | 0.00161 | -2.373 | 0 | 0.632 |
| 2015 | MIR497HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-625-5p | 28 | RSPO2 | Sponge network | -1.439 | 4.0E-5 | -3.038 | 0 | 0.632 |
| 2016 | PGM5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-484;hsa-miR-7-1-3p | 12 | NRXN3 | Sponge network | -4.546 | 0 | -0.893 | 0.04186 | 0.632 |
| 2017 | ADAMTS9-AS2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 28 | RYR2 | Sponge network | -2.452 | 0 | -1.466 | 0.00031 | 0.632 |
| 2018 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | RECK | Sponge network | -1.349 | 0.00017 | -1.043 | 0 | 0.632 |
| 2019 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-502-5p | 14 | ANK2 | Sponge network | -0.932 | 0.00329 | -1.603 | 1.0E-5 | 0.632 |
| 2020 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | SYNE1 | Sponge network | 0.335 | 0.20596 | -0.738 | 0.00316 | 0.632 |
| 2021 | RP11-166D19.1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 15 | STAT5B | Sponge network | -0.576 | 0.10121 | -0.182 | 0.1082 | 0.632 |
| 2022 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-577 | 16 | ADAMTSL3 | Sponge network | -0.473 | 0.07602 | -1.559 | 0 | 0.632 |
| 2023 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-5p | 11 | THSD7A | Sponge network | -0.976 | 0.00025 | 0.242 | 0.25154 | 0.632 |
| 2024 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 39 | EPHA3 | Sponge network | -1.575 | 6.0E-5 | 0.185 | 0.53588 | 0.632 |
| 2025 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | SYNE1 | Sponge network | -0.563 | 0.02545 | -0.738 | 0.00316 | 0.632 |
| 2026 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 57 | RUNX1T1 | Sponge network | -0.901 | 0.00019 | -0.344 | 0.2374 | 0.632 |
| 2027 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 59 | NTRK3 | Sponge network | -1.349 | 0.00017 | -1.77 | 3.0E-5 | 0.632 |
| 2028 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 51 | BNC2 | Sponge network | -0.777 | 0.1134 | -0.491 | 0.09988 | 0.632 |
| 2029 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 44 | RASSF2 | Sponge network | -1.111 | 0.0003 | -0.273 | 0.21961 | 0.632 |
| 2030 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p | 21 | NRK | Sponge network | 0.194 | 0.64278 | 0.656 | 0.19217 | 0.632 |
| 2031 | PART1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -2.925 | 0 | -2.633 | 0 | 0.632 |
| 2032 | MEG3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | NAV3 | Sponge network | 0.249 | 0.35902 | 0.212 | 0.47729 | 0.632 |
| 2033 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p | 34 | DMD | Sponge network | -0.842 | 0.01361 | -1.506 | 0 | 0.631 |
| 2034 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | NRXN3 | Sponge network | -2.117 | 0.00161 | -0.893 | 0.04186 | 0.631 |
| 2035 | USP3-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 16 | GLI3 | Sponge network | -0.162 | 0.47687 | -0.615 | 0.01482 | 0.631 |
| 2036 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | ZNF382 | Sponge network | -0.576 | 0.10121 | -0.494 | 0.03831 | 0.631 |
| 2037 | RP11-736K20.5 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3614-5p | 11 | NGFR | Sponge network | -0.358 | 0.17255 | -1.271 | 0.01058 | 0.631 |
| 2038 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 28 | AFF3 | Sponge network | -1.575 | 6.0E-5 | -2.373 | 0 | 0.631 |
| 2039 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | CNTNAP1 | Sponge network | -1.281 | 0.00012 | 0.358 | 0.13727 | 0.631 |
| 2040 | CTC-296K1.3 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-452-5p;hsa-miR-493-5p | 13 | SYNM | Sponge network | -2.095 | 0.00112 | -2.309 | 1.0E-5 | 0.631 |
| 2041 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 59 | RUNX1T1 | Sponge network | 0.246 | 0.59912 | -0.344 | 0.2374 | 0.631 |
| 2042 | LINC00865 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 29 | SETBP1 | Sponge network | -1.249 | 7.0E-5 | -1.302 | 1.0E-5 | 0.631 |
| 2043 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 11 | DCLK1 | Sponge network | -0.611 | 0.09607 | -1.324 | 0.00014 | 0.631 |
| 2044 | MIR143HG |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 57 | GRIK3 | Sponge network | -0.739 | 0.0574 | -3.208 | 0 | 0.631 |
| 2045 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 33 | GRIN2A | Sponge network | -0.739 | 0.0574 | -2.161 | 0 | 0.631 |
| 2046 | LINC00702 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | NCAM2 | Sponge network | -0.737 | 0.06182 | -0.482 | 0.22565 | 0.631 |
| 2047 | DNM3OS |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 29 | WWTR1 | Sponge network | 0.572 | 0.01722 | -0.345 | 0.11997 | 0.631 |
| 2048 | MYLK-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-424-5p | 20 | ASTN1 | Sponge network | -1.331 | 3.0E-5 | -2.389 | 2.0E-5 | 0.631 |
| 2049 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ABCC9 | Sponge network | -0.777 | 0.1134 | -0.466 | 0.14121 | 0.631 |
| 2050 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 38 | DTNA | Sponge network | -0.739 | 0.00044 | -1.648 | 1.0E-5 | 0.631 |
| 2051 | LINC00578 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-664a-3p | 16 | SYNPO2 | Sponge network | 0.811 | 0.03228 | -2.342 | 0 | 0.631 |
| 2052 | FENDRR |
hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p | 10 | ELN | Sponge network | -1.281 | 0.00012 | 0.451 | 0.13752 | 0.631 |
| 2053 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 28 | KIT | Sponge network | -1.111 | 0.0003 | -2.184 | 0 | 0.631 |
| 2054 | SNHG14 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-96-5p | 24 | RPS6KA6 | Sponge network | -1.111 | 0.0003 | -2.989 | 0 | 0.631 |
| 2055 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 23 | SPG20 | Sponge network | -1.697 | 0.00889 | -1.16 | 0 | 0.631 |
| 2056 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940;hsa-miR-96-5p | 71 | CADM2 | Sponge network | -0.576 | 0.10121 | -3.782 | 0 | 0.631 |
| 2057 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-3p | 22 | AFF3 | Sponge network | -0.976 | 0.00025 | -2.373 | 0 | 0.631 |
| 2058 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 44 | TACC1 | Sponge network | -2.452 | 0 | -0.362 | 0.05406 | 0.631 |
| 2059 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-421 | 10 | SPG20 | Sponge network | -0.583 | 0.14589 | -1.16 | 0 | 0.631 |
| 2060 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-671-5p;hsa-miR-93-5p | 47 | ADARB1 | Sponge network | -1.439 | 4.0E-5 | -0.335 | 0.06631 | 0.631 |
| 2061 | PART1 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p | 11 | HSPB7 | Sponge network | -2.925 | 0 | -2.268 | 1.0E-5 | 0.63 |
| 2062 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | -2.117 | 0.00161 | -1.74 | 0 | 0.63 |
| 2063 | AC006116.24 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-582-3p | 12 | SETBP1 | Sponge network | -0.473 | 0.07602 | -1.302 | 1.0E-5 | 0.63 |
| 2064 | CTD-2002H8.2 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | ATM | Sponge network | 0.931 | 1.0E-5 | 0.178 | 0.21568 | 0.63 |
| 2065 | CTD-3064M3.3 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p | 22 | ANK2 | Sponge network | -0.469 | 0.08721 | -1.603 | 1.0E-5 | 0.63 |
| 2066 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 61 | RUNX1T1 | Sponge network | -0.323 | 0.01372 | -0.344 | 0.2374 | 0.63 |
| 2067 | AC006116.24 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-539-5p;hsa-miR-98-5p | 22 | DMD | Sponge network | -0.473 | 0.07602 | -1.506 | 0 | 0.63 |
| 2068 | RP11-166D19.1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | MACF1 | Sponge network | -0.576 | 0.10121 | 0.019 | 0.90335 | 0.63 |
| 2069 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p | 15 | GLI3 | Sponge network | 0.335 | 0.20596 | -0.615 | 0.01482 | 0.63 |
| 2070 | SNHG14 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 13 | HMCN1 | Sponge network | -1.111 | 0.0003 | 0.809 | 0.00544 | 0.63 |
| 2071 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 31 | GRIN2A | Sponge network | -2.46 | 0 | -2.161 | 0 | 0.63 |
| 2072 | USP3-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-744-3p;hsa-miR-940 | 11 | RHOB | Sponge network | -0.162 | 0.47687 | -1.052 | 0 | 0.63 |
| 2073 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 11 | SCN11A | Sponge network | -0.576 | 0.10121 | -1.15 | 0.00299 | 0.63 |
| 2074 | HAND2-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 12 | STAT5B | Sponge network | -1.697 | 0.00889 | -0.182 | 0.1082 | 0.63 |
| 2075 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-576-5p | 40 | BNC2 | Sponge network | -2.62 | 0 | -0.491 | 0.09988 | 0.63 |
| 2076 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-625-5p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -0.976 | 0.00025 | 0.024 | 0.89889 | 0.63 |
| 2077 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 47 | ADARB1 | Sponge network | 0.194 | 0.64278 | -0.335 | 0.06631 | 0.63 |
| 2078 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p | 12 | DNAJB4 | Sponge network | -0.583 | 0.14589 | -0.7 | 1.0E-5 | 0.63 |
| 2079 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 18 | DMD | Sponge network | -1.582 | 8.0E-5 | -1.506 | 0 | 0.63 |
| 2080 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 49 | SOX5 | Sponge network | -0.49 | 0.04946 | -1.091 | 4.0E-5 | 0.63 |
| 2081 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-93-3p | 33 | RUNX1T1 | Sponge network | -2.531 | 0 | -0.344 | 0.2374 | 0.63 |
| 2082 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-3p;hsa-miR-629-3p | 41 | NTRK3 | Sponge network | -0.358 | 0.17255 | -1.77 | 3.0E-5 | 0.63 |
| 2083 | MIR143HG |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 35 | PRKCB | Sponge network | -0.739 | 0.0574 | -1.313 | 2.0E-5 | 0.63 |
| 2084 | MIR497HG |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 11 | RHOB | Sponge network | -1.439 | 4.0E-5 | -1.052 | 0 | 0.63 |
| 2085 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-338-3p;hsa-miR-93-5p | 10 | SLITRK3 | Sponge network | -2.531 | 0 | -3.328 | 0 | 0.63 |
| 2086 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-576-5p | 10 | PREX2 | Sponge network | 0.889 | 9.0E-5 | -0.272 | 0.25382 | 0.629 |
| 2087 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | PREX2 | Sponge network | -0.513 | 0.04703 | -0.272 | 0.25382 | 0.629 |
| 2088 | C20orf166-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 13 | NGFR | Sponge network | -2.69 | 0.0001 | -1.271 | 0.01058 | 0.629 |
| 2089 | BVES-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-486-5p;hsa-miR-708-5p;hsa-miR-766-3p | 11 | EZH1 | Sponge network | -2.62 | 0 | -0.564 | 1.0E-5 | 0.629 |
| 2090 | PCA3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-532-3p | 16 | SYNM | Sponge network | -0.709 | 0.18168 | -2.309 | 1.0E-5 | 0.629 |
| 2091 | MIR143HG |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 46 | IGFBP5 | Sponge network | -0.739 | 0.0574 | -0.806 | 0.00105 | 0.629 |
| 2092 | FGF14-AS2 |
hsa-miR-103a-2-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b | 11 | SYNM | Sponge network | -1.582 | 8.0E-5 | -2.309 | 1.0E-5 | 0.629 |
| 2093 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484 | 23 | SOX5 | Sponge network | -1.331 | 3.0E-5 | -1.091 | 4.0E-5 | 0.629 |
| 2094 | FGF14-AS2 |
hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-24-2-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-590-3p;hsa-miR-660-5p | 12 | PCDH9 | Sponge network | -1.582 | 8.0E-5 | -1.823 | 4.0E-5 | 0.629 |
| 2095 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-511-5p | 22 | RBMS3 | Sponge network | -2.531 | 0 | -0.519 | 0.03551 | 0.629 |
| 2096 | MEG3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-671-5p | 27 | DLC1 | Sponge network | 0.249 | 0.35902 | -0.382 | 0.05038 | 0.629 |
| 2097 | MIR143HG |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 22 | ITGA5 | Sponge network | -0.739 | 0.0574 | 0.452 | 0.03524 | 0.629 |
| 2098 | ACTA2-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | WWTR1 | Sponge network | 0.194 | 0.64278 | -0.345 | 0.11997 | 0.629 |
| 2099 | DNM3OS |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | RNF180 | Sponge network | 0.572 | 0.01722 | -1.127 | 1.0E-5 | 0.629 |
| 2100 | RP11-532F6.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 26 | DLC1 | Sponge network | -0.896 | 0.00099 | -0.382 | 0.05038 | 0.629 |
| 2101 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | ADAMTSL3 | Sponge network | -0.563 | 0.02545 | -1.559 | 0 | 0.629 |
| 2102 | LINC00702 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 21 | PTGFR | Sponge network | -0.737 | 0.06182 | -0.233 | 0.45421 | 0.629 |
| 2103 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 61 | NTRK3 | Sponge network | -2.117 | 0.00161 | -1.77 | 3.0E-5 | 0.629 |
| 2104 | LINC00702 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | THSD7B | Sponge network | -0.737 | 0.06182 | 0.154 | 0.74723 | 0.629 |
| 2105 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 23 | CADM3 | Sponge network | -0.49 | 0.04946 | -3.663 | 0 | 0.629 |
| 2106 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 63 | TCF4 | Sponge network | 0.335 | 0.20596 | 0.016 | 0.92271 | 0.629 |
| 2107 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-532-5p;hsa-miR-590-3p | 11 | DCLK1 | Sponge network | -0.896 | 0.00099 | -1.324 | 0.00014 | 0.629 |
| 2108 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | ZNF292 | Sponge network | 0.782 | 0.00016 | 0.276 | 0.04148 | 0.629 |
| 2109 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 14 | PARK2 | Sponge network | -1.697 | 0.00889 | -1.892 | 0 | 0.629 |
| 2110 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-93-5p | 19 | FGFR1 | Sponge network | -2.69 | 0.0001 | -0.509 | 0.03957 | 0.629 |
| 2111 | MYLK-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-484 | 13 | ITGA5 | Sponge network | -1.331 | 3.0E-5 | 0.452 | 0.03524 | 0.629 |
| 2112 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p | 25 | DYNC1I1 | Sponge network | -1.111 | 0.0003 | -1.12 | 0.00107 | 0.629 |
| 2113 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-7-5p;hsa-miR-93-5p | 28 | ZBTB4 | Sponge network | -0.162 | 0.47687 | -0.739 | 0 | 0.629 |
| 2114 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | IGSF10 | Sponge network | -1.111 | 0.0003 | -1.751 | 1.0E-5 | 0.629 |
| 2115 | SERTAD4-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-93-3p | 10 | WWTR1 | Sponge network | -0.932 | 0.00329 | -0.345 | 0.11997 | 0.629 |
| 2116 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 53 | QKI | Sponge network | 0.532 | 0.00505 | -0.333 | 0.04111 | 0.628 |
| 2117 | AC006116.24 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-454-3p | 21 | DLC1 | Sponge network | -0.473 | 0.07602 | -0.382 | 0.05038 | 0.628 |
| 2118 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 27 | BNC2 | Sponge network | -0.473 | 0.07602 | -0.491 | 0.09988 | 0.628 |
| 2119 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 51 | HLF | Sponge network | -1.697 | 0.00889 | -2.443 | 0 | 0.628 |
| 2120 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | ZFHX3 | Sponge network | -0.323 | 0.01372 | 0.389 | 0.01363 | 0.628 |
| 2121 | MIR143HG |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 21 | CXCL12 | Sponge network | -0.739 | 0.0574 | -1.421 | 0 | 0.628 |
| 2122 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 29 | EBF3 | Sponge network | -0.49 | 0.04946 | -0.058 | 0.81437 | 0.628 |
| 2123 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-93-5p | 44 | AR | Sponge network | -1.654 | 0.00144 | -1.554 | 0 | 0.628 |
| 2124 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | DNAJB4 | Sponge network | -1.697 | 0.00889 | -0.7 | 1.0E-5 | 0.628 |
| 2125 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p | 11 | GREM1 | Sponge network | -1.092 | 4.0E-5 | 0.902 | 0.01691 | 0.628 |
| 2126 | LINC00865 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 53 | DTNA | Sponge network | -1.249 | 7.0E-5 | -1.648 | 1.0E-5 | 0.628 |
| 2127 | NR2F1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 102 | LPP | Sponge network | -0.49 | 0.04946 | -0.459 | 0.04475 | 0.628 |
| 2128 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-671-5p | 19 | CADM3 | Sponge network | -0.129 | 0.57177 | -3.663 | 0 | 0.628 |
| 2129 | SNHG14 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | THSD7B | Sponge network | -1.111 | 0.0003 | 0.154 | 0.74723 | 0.628 |
| 2130 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-92a-1-5p | 18 | AKAP6 | Sponge network | -2.531 | 0 | -1.586 | 3.0E-5 | 0.628 |
| 2131 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 18 | RCAN2 | Sponge network | -0.323 | 0.01372 | -0.33 | 0.15172 | 0.628 |
| 2132 | SNHG14 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 37 | WWTR1 | Sponge network | -1.111 | 0.0003 | -0.345 | 0.11997 | 0.628 |
| 2133 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p | 13 | LRRK2 | Sponge network | -0.976 | 0.00025 | -1.136 | 1.0E-5 | 0.628 |
| 2134 | ZNF582-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 48 | AR | Sponge network | -1.349 | 0.00017 | -1.554 | 0 | 0.628 |
| 2135 | USP3-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 15 | STAT5B | Sponge network | -0.162 | 0.47687 | -0.182 | 0.1082 | 0.628 |
| 2136 | ZNF582-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 17 | SFRP1 | Sponge network | -1.349 | 0.00017 | -3.566 | 0 | 0.628 |
| 2137 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 33 | RASSF8 | Sponge network | -0.901 | 0.00019 | -0.122 | 0.65591 | 0.628 |
| 2138 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 33 | EPHA3 | Sponge network | 0.194 | 0.64278 | 0.185 | 0.53588 | 0.628 |
| 2139 | LINC00578 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-628-5p | 18 | EPHA3 | Sponge network | 0.811 | 0.03228 | 0.185 | 0.53588 | 0.628 |
| 2140 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 48 | SOX5 | Sponge network | -2.452 | 0 | -1.091 | 4.0E-5 | 0.628 |
| 2141 | NR2F1-AS1 |
hsa-miR-185-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-940 | 10 | ILK | Sponge network | -0.49 | 0.04946 | -0.74 | 0 | 0.628 |
| 2142 | USP3-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-671-5p | 29 | DLC1 | Sponge network | -0.162 | 0.47687 | -0.382 | 0.05038 | 0.627 |
| 2143 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-935 | 17 | RELN | Sponge network | -0.976 | 0.00025 | -2.403 | 0 | 0.627 |
| 2144 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-340-5p | 15 | PLSCR4 | Sponge network | -0.693 | 0.05629 | -0.474 | 0.03833 | 0.627 |
| 2145 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | GLI3 | Sponge network | -0.358 | 0.17255 | -0.615 | 0.01482 | 0.627 |
| 2146 | RP11-736K20.5 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 25 | GRIK3 | Sponge network | -0.358 | 0.17255 | -3.208 | 0 | 0.627 |
| 2147 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-940 | 37 | NTRK3 | Sponge network | -1.331 | 3.0E-5 | -1.77 | 3.0E-5 | 0.627 |
| 2148 | RP11-554A11.4 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-628-5p | 27 | MEF2C | Sponge network | -0.583 | 0.14589 | -0.499 | 0.01448 | 0.627 |
| 2149 | AC003090.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 19 | SFRP1 | Sponge network | -2.117 | 0.00161 | -3.566 | 0 | 0.627 |
| 2150 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 61 | QKI | Sponge network | -1.575 | 6.0E-5 | -0.333 | 0.04111 | 0.627 |
| 2151 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | AFF3 | Sponge network | -0.769 | 0.00035 | -2.373 | 0 | 0.627 |
| 2152 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-5p | 17 | PTGFR | Sponge network | -0.976 | 0.00025 | -0.233 | 0.45421 | 0.627 |
| 2153 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 33 | EBF3 | Sponge network | -1.111 | 0.0003 | -0.058 | 0.81437 | 0.627 |
| 2154 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | DNAJB4 | Sponge network | -1.111 | 0.0003 | -0.7 | 1.0E-5 | 0.627 |
| 2155 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-93-5p | 15 | RASSF2 | Sponge network | -0.31 | 0.33871 | -0.273 | 0.21961 | 0.627 |
| 2156 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-93-3p | 10 | CNTNAP1 | Sponge network | -0.469 | 0.08721 | 0.358 | 0.13727 | 0.627 |
| 2157 | PWAR6 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p | 16 | MN1 | Sponge network | -1.575 | 6.0E-5 | -0.358 | 0.24172 | 0.627 |
| 2158 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | LRRC7 | Sponge network | -1.697 | 0.00889 | -1.341 | 0.01193 | 0.627 |
| 2159 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 39 | DMD | Sponge network | 0.194 | 0.64278 | -1.506 | 0 | 0.627 |
| 2160 | MIR143HG |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | PREX2 | Sponge network | -0.739 | 0.0574 | -0.272 | 0.25382 | 0.627 |
| 2161 | USP3-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p | 10 | PRKD1 | Sponge network | -0.162 | 0.47687 | -0.466 | 0.03583 | 0.627 |
| 2162 | LINC00702 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 77 | DST | Sponge network | -0.737 | 0.06182 | -0.713 | 0.00096 | 0.627 |
| 2163 | LINC00702 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3913-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 43 | IGFBP5 | Sponge network | -0.737 | 0.06182 | -0.806 | 0.00105 | 0.627 |
| 2164 | RASSF8-AS1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-582-5p | 11 | PRICKLE1 | Sponge network | 0.335 | 0.20596 | -0.112 | 0.67088 | 0.627 |
| 2165 | HAND2-AS1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-582-5p | 12 | PRICKLE1 | Sponge network | -1.697 | 0.00889 | -0.112 | 0.67088 | 0.627 |
| 2166 | AC009948.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | RASSF8 | Sponge network | 0.532 | 0.00505 | -0.122 | 0.65591 | 0.627 |
| 2167 | FENDRR |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | PEG3 | Sponge network | -1.281 | 0.00012 | -1.74 | 0 | 0.627 |
| 2168 | DNM3OS |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 19 | GLIPR1 | Sponge network | 0.572 | 0.01722 | 0.146 | 0.3276 | 0.627 |
| 2169 | SERTAD4-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-93-3p | 33 | RUNX1T1 | Sponge network | -0.932 | 0.00329 | -0.344 | 0.2374 | 0.627 |
| 2170 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p | 26 | DYNC1I1 | Sponge network | -1.697 | 0.00889 | -1.12 | 0.00107 | 0.627 |
| 2171 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | PGR | Sponge network | -1.886 | 0 | -1.704 | 0 | 0.627 |
| 2172 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | DYNC1I1 | Sponge network | -0.737 | 0.06182 | -1.12 | 0.00107 | 0.626 |
| 2173 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p | 35 | ZFHX4 | Sponge network | -0.358 | 0.17255 | 0.018 | 0.95574 | 0.626 |
| 2174 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 19 | PEG3 | Sponge network | -0.487 | 0.02157 | -1.74 | 0 | 0.626 |
| 2175 | RP11-159D12.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 44 | MDM4 | Sponge network | 1.254 | 0 | 0.345 | 0.00762 | 0.626 |
| 2176 | RP11-67L2.2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | PIK3CA | Sponge network | 0.72 | 0.0001 | 0.195 | 0.06908 | 0.626 |
| 2177 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-532-3p;hsa-miR-93-3p | 10 | CNTNAP1 | Sponge network | -2.69 | 0.0001 | 0.358 | 0.13727 | 0.626 |
| 2178 | RP11-736K20.5 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-629-3p;hsa-miR-942-5p | 21 | IGFBP5 | Sponge network | -0.358 | 0.17255 | -0.806 | 0.00105 | 0.626 |
| 2179 | SERTAD4-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-369-3p | 12 | DIP2C | Sponge network | -0.932 | 0.00329 | -0.436 | 0.01713 | 0.626 |
| 2180 | MEG3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577 | 23 | NRK | Sponge network | 0.249 | 0.35902 | 0.656 | 0.19217 | 0.626 |
| 2181 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 70 | IL6ST | Sponge network | -0.576 | 0.10121 | -0.402 | 0.00676 | 0.626 |
| 2182 | PWAR6 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 21 | GLI3 | Sponge network | -1.575 | 6.0E-5 | -0.615 | 0.01482 | 0.626 |
| 2183 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-628-5p;hsa-miR-629-3p | 14 | RASSF8 | Sponge network | 0.811 | 0.03228 | -0.122 | 0.65591 | 0.626 |
| 2184 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 25 | DMD | Sponge network | -0.469 | 0.08721 | -1.506 | 0 | 0.626 |
| 2185 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-361-5p;hsa-miR-450b-5p | 17 | DLC1 | Sponge network | -2.531 | 0 | -0.382 | 0.05038 | 0.626 |
| 2186 | MAGI2-AS3 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CTGF | Sponge network | -0.129 | 0.57177 | 0.321 | 0.11682 | 0.626 |
| 2187 | ANKRD10-IT1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-7-1-3p | 22 | MDM4 | Sponge network | 1.739 | 0 | 0.345 | 0.00762 | 0.626 |
| 2188 | MIR143HG |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | TLL1 | Sponge network | -0.739 | 0.0574 | -1.195 | 3.0E-5 | 0.626 |
| 2189 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-93-5p | 26 | SYNE1 | Sponge network | -0.162 | 0.47687 | -0.738 | 0.00316 | 0.626 |
| 2190 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 16 | SYNE1 | Sponge network | -4.546 | 0 | -0.738 | 0.00316 | 0.626 |
| 2191 | MAGI2-AS3 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | ZDBF2 | Sponge network | -0.129 | 0.57177 | -0.998 | 0.00025 | 0.626 |
| 2192 | USP3-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-5p | 12 | MN1 | Sponge network | -0.162 | 0.47687 | -0.358 | 0.24172 | 0.626 |
| 2193 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 26 | CDH19 | Sponge network | -0.129 | 0.57177 | -3.434 | 0 | 0.626 |
| 2194 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p | 19 | IGF1 | Sponge network | -1.092 | 4.0E-5 | -0.204 | 0.5898 | 0.626 |
| 2195 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | THBS1 | Sponge network | 0.889 | 9.0E-5 | 0.741 | 0.00386 | 0.626 |
| 2196 | RP4-717I23.3 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-766-3p | 13 | ATM | Sponge network | 0.637 | 0.00069 | 0.178 | 0.21568 | 0.626 |
| 2197 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | AHNAK | Sponge network | -2.452 | 0 | -1.207 | 0 | 0.626 |
| 2198 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p | 11 | ZNF382 | Sponge network | -2.452 | 0 | -0.494 | 0.03831 | 0.626 |
| 2199 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-532-5p | 11 | PARK2 | Sponge network | -2.69 | 0.0001 | -1.892 | 0 | 0.626 |
| 2200 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 31 | DIP2C | Sponge network | -0.096 | 0.67958 | -0.436 | 0.01713 | 0.626 |
| 2201 | LINC00702 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | -0.737 | 0.06182 | -0.564 | 1.0E-5 | 0.626 |
| 2202 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | PCDH9 | Sponge network | -2.46 | 0 | -1.823 | 4.0E-5 | 0.626 |
| 2203 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 47 | HLF | Sponge network | -0.737 | 0.06182 | -2.443 | 0 | 0.625 |
| 2204 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-671-5p | 21 | KIT | Sponge network | -1.439 | 4.0E-5 | -2.184 | 0 | 0.625 |
| 2205 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | 0.194 | 0.64278 | 0.809 | 0.00544 | 0.625 |
| 2206 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-335-3p;hsa-miR-935 | 10 | FBXO32 | Sponge network | -0.691 | 0.00146 | -0.495 | 0.07561 | 0.625 |
| 2207 | RP11-166D19.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | KCTD8 | Sponge network | -0.576 | 0.10121 | -3.359 | 0 | 0.625 |
| 2208 | NR2F1-AS1 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-5p | 11 | MLLT11 | Sponge network | -0.49 | 0.04946 | -0.661 | 0.01733 | 0.625 |
| 2209 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940;hsa-miR-98-5p | 47 | PBX1 | Sponge network | -0.739 | 0.0574 | -1.206 | 0 | 0.625 |
| 2210 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 22 | CHD2 | Sponge network | 0.782 | 0.00016 | 0.049 | 0.55371 | 0.625 |
| 2211 | LINC00702 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 28 | RYR2 | Sponge network | -0.737 | 0.06182 | -1.466 | 0.00031 | 0.625 |
| 2212 | HAND2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 55 | GAS7 | Sponge network | -1.697 | 0.00889 | -0.555 | 0.01602 | 0.625 |
| 2213 | AC003090.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PCDH9 | Sponge network | -2.117 | 0.00161 | -1.823 | 4.0E-5 | 0.625 |
| 2214 | AC006116.24 |
hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p | 10 | MRVI1 | Sponge network | -0.473 | 0.07602 | -1.051 | 0.00022 | 0.625 |
| 2215 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 49 | MEF2C | Sponge network | -0.737 | 0.06182 | -0.499 | 0.01448 | 0.625 |
| 2216 | SNHG14 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 16 | NGFR | Sponge network | -1.111 | 0.0003 | -1.271 | 0.01058 | 0.625 |
| 2217 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 23 | KIT | Sponge network | -1.281 | 0.00012 | -2.184 | 0 | 0.625 |
| 2218 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 15 | SYNE1 | Sponge network | -1.582 | 8.0E-5 | -0.738 | 0.00316 | 0.625 |
| 2219 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-590-3p | 21 | AKAP6 | Sponge network | -0.896 | 0.00099 | -1.586 | 3.0E-5 | 0.625 |
| 2220 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 16 | AKAP12 | Sponge network | -1.575 | 6.0E-5 | -0.932 | 0.0028 | 0.625 |
| 2221 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p | 20 | RECK | Sponge network | -2.69 | 0.0001 | -1.043 | 0 | 0.625 |
| 2222 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | RECK | Sponge network | -0.769 | 0.00035 | -1.043 | 0 | 0.625 |
| 2223 | LINC00702 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-96-5p | 34 | ASTN1 | Sponge network | -0.737 | 0.06182 | -2.389 | 2.0E-5 | 0.625 |
| 2224 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-7-5p | 26 | RBMS3 | Sponge network | -0.693 | 0.05629 | -0.519 | 0.03551 | 0.624 |
| 2225 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | TSHZ3 | Sponge network | -0.611 | 0.09607 | -0.556 | 0.01516 | 0.624 |
| 2226 | RP11-464F9.1 |
hsa-miR-125a-3p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 10 | TAF1 | Sponge network | 0.959 | 0 | 0.39 | 3.0E-5 | 0.624 |
| 2227 | MAGI2-AS3 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-940 | 15 | RHOB | Sponge network | -0.129 | 0.57177 | -1.052 | 0 | 0.624 |
| 2228 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | ERG | Sponge network | -0.576 | 0.10121 | 0.002 | 0.99011 | 0.624 |
| 2229 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 18 | KIT | Sponge network | -0.358 | 0.17255 | -2.184 | 0 | 0.624 |
| 2230 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b | 16 | PLSCR4 | Sponge network | 0.811 | 0.03228 | -0.474 | 0.03833 | 0.624 |
| 2231 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 26 | SYNPO2 | Sponge network | -0.487 | 0.02157 | -2.342 | 0 | 0.624 |
| 2232 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 11 | RIMS1 | Sponge network | -2.452 | 0 | -3.143 | 0 | 0.624 |
| 2233 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 53 | CADM2 | Sponge network | -0.976 | 0.00025 | -3.782 | 0 | 0.624 |
| 2234 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-940 | 17 | IGF1 | Sponge network | -1.331 | 3.0E-5 | -0.204 | 0.5898 | 0.624 |
| 2235 | NR2F1-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ITPKB | Sponge network | -0.49 | 0.04946 | -0.704 | 0.00106 | 0.624 |
| 2236 | LINC00092 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 12 | SYNPO2 | Sponge network | -1.886 | 0 | -2.342 | 0 | 0.624 |
| 2237 | MYLK-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p | 12 | ERG | Sponge network | -1.331 | 3.0E-5 | 0.002 | 0.99011 | 0.624 |
| 2238 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940 | 47 | MEF2C | Sponge network | -1.697 | 0.00889 | -0.499 | 0.01448 | 0.624 |
| 2239 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | TSHZ3 | Sponge network | 0.997 | 0.00066 | -0.556 | 0.01516 | 0.624 |
| 2240 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-655-3p | 14 | LATS1 | Sponge network | 0.542 | 0.00054 | 0.109 | 0.34208 | 0.624 |
| 2241 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-582-3p | 20 | RBMS3 | Sponge network | -0.473 | 0.07602 | -0.519 | 0.03551 | 0.624 |
| 2242 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 32 | SETBP1 | Sponge network | 0.194 | 0.64278 | -1.302 | 1.0E-5 | 0.624 |
| 2243 | LINC00641 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | HERC1 | Sponge network | -0.222 | 0.32445 | -0.196 | 0.08544 | 0.624 |
| 2244 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p | 12 | DKK3 | Sponge network | -0.932 | 0.00329 | -0.052 | 0.77783 | 0.624 |
| 2245 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-342-5p;hsa-miR-539-5p | 15 | SYNPO2 | Sponge network | -0.469 | 0.08721 | -2.342 | 0 | 0.623 |
| 2246 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 16 | RCAN2 | Sponge network | 0.572 | 0.01722 | -0.33 | 0.15172 | 0.623 |
| 2247 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429 | 11 | THSD7A | Sponge network | 0.335 | 0.20596 | 0.242 | 0.25154 | 0.623 |
| 2248 | ZNF667-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 16 | RHOB | Sponge network | -0.976 | 0.00025 | -1.052 | 0 | 0.623 |
| 2249 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 36 | MITF | Sponge network | -0.129 | 0.57177 | -0.769 | 0.00022 | 0.623 |
| 2250 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935;hsa-miR-96-5p | 33 | FBXO32 | Sponge network | 0.572 | 0.01722 | -0.495 | 0.07561 | 0.623 |
| 2251 | CTD-2528L19.6 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 42 | MDM4 | Sponge network | 0.953 | 0 | 0.345 | 0.00762 | 0.623 |
| 2252 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-590-3p | 11 | ADAMTSL3 | Sponge network | -1.375 | 0.08642 | -1.559 | 0 | 0.623 |
| 2253 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | MCC | Sponge network | -0.358 | 0.17255 | -1.005 | 0 | 0.623 |
| 2254 | PWAR6 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-7-5p | 12 | FLNA | Sponge network | -1.575 | 6.0E-5 | -0.771 | 0.01377 | 0.623 |
| 2255 | ZNF582-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 21 | SETBP1 | Sponge network | -1.349 | 0.00017 | -1.302 | 1.0E-5 | 0.623 |
| 2256 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 17 | PLSCR4 | Sponge network | -1.582 | 8.0E-5 | -0.474 | 0.03833 | 0.623 |
| 2257 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-98-5p | 13 | SCN11A | Sponge network | -0.129 | 0.57177 | -1.15 | 0.00299 | 0.623 |
| 2258 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-766-3p | 12 | EZH1 | Sponge network | -0.583 | 0.14589 | -0.564 | 1.0E-5 | 0.623 |
| 2259 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p | 14 | MYO9A | Sponge network | 0.178 | 0.29106 | -0.147 | 0.32373 | 0.623 |
| 2260 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 14 | PARK2 | Sponge network | -1.575 | 6.0E-5 | -1.892 | 0 | 0.623 |
| 2261 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-93-5p | 16 | PLSCR4 | Sponge network | -2.531 | 0 | -0.474 | 0.03833 | 0.623 |
| 2262 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 53 | RUNX1T1 | Sponge network | -2.117 | 0.00161 | -0.344 | 0.2374 | 0.623 |
| 2263 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 39 | FBXO32 | Sponge network | -0.793 | 7.0E-5 | -0.495 | 0.07561 | 0.623 |
| 2264 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 55 | TACC1 | Sponge network | -1.111 | 0.0003 | -0.362 | 0.05406 | 0.623 |
| 2265 | SERTAD4-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-576-5p | 34 | LPP | Sponge network | -0.932 | 0.00329 | -0.459 | 0.04475 | 0.623 |
| 2266 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940 | 48 | MEF2C | Sponge network | -0.49 | 0.04946 | -0.499 | 0.01448 | 0.623 |
| 2267 | CTD-2528L19.6 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ZNF638 | Sponge network | 0.953 | 0 | 0.22 | 0.01214 | 0.623 |
| 2268 | MIR4435-1HG |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 10 | CBFB | Sponge network | 2.275 | 0 | 1.061 | 0 | 0.623 |
| 2269 | RP11-159D12.2 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-486-5p;hsa-miR-590-3p | 12 | TPR | Sponge network | 1.254 | 0 | 0.582 | 0 | 0.622 |
| 2270 | HAND2-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | PRKCB | Sponge network | -1.697 | 0.00889 | -1.313 | 2.0E-5 | 0.622 |
| 2271 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-98-5p | 37 | PBX1 | Sponge network | -2.62 | 0 | -1.206 | 0 | 0.622 |
| 2272 | MIR497HG |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 30 | NAV3 | Sponge network | -1.439 | 4.0E-5 | 0.212 | 0.47729 | 0.622 |
| 2273 | FENDRR |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ZNF536 | Sponge network | -1.281 | 0.00012 | -3.115 | 0 | 0.622 |
| 2274 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | AFF3 | Sponge network | -0.739 | 0.0574 | -2.373 | 0 | 0.622 |
| 2275 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935 | 32 | FBXO32 | Sponge network | -0.096 | 0.67958 | -0.495 | 0.07561 | 0.622 |
| 2276 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ABCC9 | Sponge network | -0.709 | 0.18168 | -0.466 | 0.14121 | 0.622 |
| 2277 | RP11-57H14.4 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | HERC1 | Sponge network | 0.083 | 0.66405 | -0.196 | 0.08544 | 0.622 |
| 2278 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 34 | ST6GAL2 | Sponge network | -0.777 | 0.1134 | -1.201 | 0.00597 | 0.622 |
| 2279 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 41 | PBX1 | Sponge network | -1.575 | 6.0E-5 | -1.206 | 0 | 0.622 |
| 2280 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 42 | DMD | Sponge network | -0.901 | 0.00019 | -1.506 | 0 | 0.622 |
| 2281 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TLL1 | Sponge network | -1.697 | 0.00889 | -1.195 | 3.0E-5 | 0.622 |
| 2282 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 41 | MYO9A | Sponge network | -0.129 | 0.57177 | -0.147 | 0.32373 | 0.622 |
| 2283 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 26 | AKAP6 | Sponge network | -0.323 | 0.01372 | -1.586 | 3.0E-5 | 0.622 |
| 2284 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | SYNE1 | Sponge network | -1.886 | 0 | -0.738 | 0.00316 | 0.622 |
| 2285 | MIR143HG |
hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-502-5p | 10 | ELN | Sponge network | -0.739 | 0.0574 | 0.451 | 0.13752 | 0.621 |
| 2286 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | SLITRK3 | Sponge network | -0.49 | 0.04946 | -3.328 | 0 | 0.621 |
| 2287 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-93-5p | 16 | SYNE1 | Sponge network | -2.531 | 0 | -0.738 | 0.00316 | 0.621 |
| 2288 | CTD-2554C21.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-505-5p;hsa-miR-532-3p | 14 | MRVI1 | Sponge network | -1.654 | 0.00144 | -1.051 | 0.00022 | 0.621 |
| 2289 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-93-5p | 35 | GAS7 | Sponge network | -0.611 | 0.09607 | -0.555 | 0.01602 | 0.621 |
| 2290 | ADAMTS9-AS2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | NAV3 | Sponge network | -2.452 | 0 | 0.212 | 0.47729 | 0.621 |
| 2291 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p | 22 | ADAMTSL3 | Sponge network | -1.349 | 0.00017 | -1.559 | 0 | 0.621 |
| 2292 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p | 10 | PREX2 | Sponge network | -0.976 | 0.00025 | -0.272 | 0.25382 | 0.621 |
| 2293 | DNM3OS |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | NRXN3 | Sponge network | 0.572 | 0.01722 | -0.893 | 0.04186 | 0.621 |
| 2294 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | IGSF10 | Sponge network | -2.452 | 0 | -1.751 | 1.0E-5 | 0.621 |
| 2295 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p | 17 | EDA2R | Sponge network | -0.49 | 0.04946 | -0.587 | 0.01598 | 0.621 |
| 2296 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 52 | PRKAA2 | Sponge network | -0.976 | 0.00025 | -2.462 | 0 | 0.621 |
| 2297 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p | 11 | FGFR1 | Sponge network | -0.154 | 0.53954 | -0.509 | 0.03957 | 0.621 |
| 2298 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 29 | AKAP6 | Sponge network | -0.612 | 0.00094 | -1.586 | 3.0E-5 | 0.621 |
| 2299 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p | 18 | WWTR1 | Sponge network | -0.358 | 0.17255 | -0.345 | 0.11997 | 0.621 |
| 2300 | MAGI2-AS3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-877-5p | 32 | ASTN1 | Sponge network | -0.129 | 0.57177 | -2.389 | 2.0E-5 | 0.621 |
| 2301 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 26 | LATS2 | Sponge network | -0.129 | 0.57177 | 0.159 | 0.2304 | 0.621 |
| 2302 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 35 | RBMS3 | Sponge network | -0.611 | 0.09607 | -0.519 | 0.03551 | 0.621 |
| 2303 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | NRXN1 | Sponge network | -2.46 | 0 | -3.778 | 0 | 0.621 |
| 2304 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-671-5p;hsa-miR-93-5p | 45 | ADARB1 | Sponge network | -0.976 | 0.00025 | -0.335 | 0.06631 | 0.621 |
| 2305 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 44 | EPHA7 | Sponge network | -0.793 | 7.0E-5 | -2.197 | 3.0E-5 | 0.621 |
| 2306 | BVES-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-877-5p | 22 | ASTN1 | Sponge network | -2.62 | 0 | -2.389 | 2.0E-5 | 0.621 |
| 2307 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 16 | ITCH | Sponge network | 0.91 | 0 | 0.084 | 0.34058 | 0.621 |
| 2308 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p | 10 | NUAK1 | Sponge network | 1.344 | 0 | 0.904 | 0 | 0.621 |
| 2309 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-330-5p | 10 | CNTN1 | Sponge network | -2.531 | 0 | -2.594 | 0 | 0.621 |
| 2310 | ADAMTS9-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p | 21 | CDH23 | Sponge network | -2.452 | 0 | -0.974 | 0.00034 | 0.621 |
| 2311 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | ZFHX4 | Sponge network | 0.335 | 0.20596 | 0.018 | 0.95574 | 0.621 |
| 2312 | RP11-798M19.6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p | 80 | LPP | Sponge network | -0.612 | 0.00094 | -0.459 | 0.04475 | 0.621 |
| 2313 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 10 | LRFN5 | Sponge network | -1.697 | 0.00889 | -1.483 | 0 | 0.621 |
| 2314 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-7-1-3p | 12 | RCAN2 | Sponge network | -1.092 | 4.0E-5 | -0.33 | 0.15172 | 0.621 |
| 2315 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-542-3p;hsa-miR-590-3p | 26 | BCL2 | Sponge network | -1.886 | 0 | -0.899 | 0 | 0.621 |
| 2316 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-542-3p | 10 | PEG3 | Sponge network | -0.517 | 0.02935 | -1.74 | 0 | 0.62 |
| 2317 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 19 | NRXN1 | Sponge network | -0.583 | 0.14589 | -3.778 | 0 | 0.62 |
| 2318 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ABCC9 | Sponge network | -1.281 | 0.00012 | -0.466 | 0.14121 | 0.62 |
| 2319 | SNHG14 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | PREX2 | Sponge network | -1.111 | 0.0003 | -0.272 | 0.25382 | 0.62 |
| 2320 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 42 | NRXN1 | Sponge network | -0.49 | 0.04946 | -3.778 | 0 | 0.62 |
| 2321 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | SYNE1 | Sponge network | -0.376 | 0.00337 | -0.738 | 0.00316 | 0.62 |
| 2322 | LINC00702 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 22 | CXCL12 | Sponge network | -0.737 | 0.06182 | -1.421 | 0 | 0.62 |
| 2323 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-5p | 20 | SETBP1 | Sponge network | -0.469 | 0.08721 | -1.302 | 1.0E-5 | 0.62 |
| 2324 | RP11-166D19.1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | TPO | Sponge network | -0.576 | 0.10121 | -1.87 | 2.0E-5 | 0.62 |
| 2325 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-93-3p | 11 | CNTNAP1 | Sponge network | -2.531 | 0 | 0.358 | 0.13727 | 0.62 |
| 2326 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | PEG3 | Sponge network | -0.323 | 0.01372 | -1.74 | 0 | 0.62 |
| 2327 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | -1.575 | 6.0E-5 | -0.474 | 0.03833 | 0.62 |
| 2328 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | SYNE1 | Sponge network | 0.693 | 0.00076 | -0.738 | 0.00316 | 0.62 |
| 2329 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 12 | CACNA2D1 | Sponge network | -0.583 | 0.14589 | -0.846 | 0.00675 | 0.62 |
| 2330 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-590-3p | 11 | AKAP12 | Sponge network | -1.582 | 8.0E-5 | -0.932 | 0.0028 | 0.62 |
| 2331 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 34 | SOX5 | Sponge network | -0.513 | 0.04703 | -1.091 | 4.0E-5 | 0.62 |
| 2332 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-3p | 56 | RUNX1T1 | Sponge network | -2.69 | 0.0001 | -0.344 | 0.2374 | 0.62 |
| 2333 | SERTAD4-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-93-3p | 10 | CNTNAP1 | Sponge network | -0.932 | 0.00329 | 0.358 | 0.13727 | 0.62 |
| 2334 | ZNF667-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 84 | LPP | Sponge network | -0.976 | 0.00025 | -0.459 | 0.04475 | 0.62 |
| 2335 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p | 33 | SSBP2 | Sponge network | -0.583 | 0.14589 | -1.58 | 0 | 0.62 |
| 2336 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-501-5p | 10 | MN1 | Sponge network | -0.246 | 0.35034 | -0.358 | 0.24172 | 0.62 |
| 2337 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | ZNF521 | Sponge network | -1.697 | 0.00889 | -0.111 | 0.58068 | 0.62 |
| 2338 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | RASSF8 | Sponge network | -0.739 | 0.00044 | -0.122 | 0.65591 | 0.62 |
| 2339 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 44 | EPHA7 | Sponge network | 0.246 | 0.59912 | -2.197 | 3.0E-5 | 0.62 |
| 2340 | RP11-244O19.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-362-5p;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 19 | MRVI1 | Sponge network | -0.096 | 0.67958 | -1.051 | 0.00022 | 0.62 |
| 2341 | MAGI2-AS3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | -0.129 | 0.57177 | -0.564 | 1.0E-5 | 0.619 |
| 2342 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 36 | FBXO32 | Sponge network | 0.997 | 0.00066 | -0.495 | 0.07561 | 0.619 |
| 2343 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 54 | FAT3 | Sponge network | 0.572 | 0.01722 | -1.601 | 0.00051 | 0.619 |
| 2344 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 13 | NCAM2 | Sponge network | -0.583 | 0.14589 | -0.482 | 0.22565 | 0.619 |
| 2345 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p | 32 | PGR | Sponge network | -0.323 | 0.01372 | -1.704 | 0 | 0.619 |
| 2346 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-628-5p | 21 | FBXO32 | Sponge network | 0.811 | 0.03228 | -0.495 | 0.07561 | 0.619 |
| 2347 | PWAR6 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 15 | STAT5B | Sponge network | -1.575 | 6.0E-5 | -0.182 | 0.1082 | 0.619 |
| 2348 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | -0.096 | 0.67958 | -1.892 | 0 | 0.619 |
| 2349 | PWAR6 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -1.575 | 6.0E-5 | -2.046 | 0.00019 | 0.619 |
| 2350 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 34 | THBS1 | Sponge network | 0.997 | 0.00066 | 0.741 | 0.00386 | 0.619 |
| 2351 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-3913-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 43 | HLF | Sponge network | -0.793 | 7.0E-5 | -2.443 | 0 | 0.619 |
| 2352 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 15 | AFF3 | Sponge network | -0.358 | 0.17255 | -2.373 | 0 | 0.619 |
| 2353 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 32 | ZFHX3 | Sponge network | 0.225 | 0.30644 | 0.389 | 0.01363 | 0.619 |
| 2354 | LINC00702 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | PREX2 | Sponge network | -0.737 | 0.06182 | -0.272 | 0.25382 | 0.619 |
| 2355 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-589-3p | 12 | PGR | Sponge network | -4.463 | 0.00025 | -1.704 | 0 | 0.619 |
| 2356 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 48 | QKI | Sponge network | -1.439 | 4.0E-5 | -0.333 | 0.04111 | 0.619 |
| 2357 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 62 | SSBP2 | Sponge network | -2.46 | 0 | -1.58 | 0 | 0.619 |
| 2358 | MEG3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | WWTR1 | Sponge network | 0.249 | 0.35902 | -0.345 | 0.11997 | 0.619 |
| 2359 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 41 | RICTOR | Sponge network | 0.537 | 0.00139 | 0.388 | 0.00188 | 0.619 |
| 2360 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 38 | CNTN1 | Sponge network | -2.46 | 0 | -2.594 | 0 | 0.619 |
| 2361 | FENDRR |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-7-5p;hsa-miR-96-5p | 17 | FNBP1 | Sponge network | -1.281 | 0.00012 | -0.969 | 4.0E-5 | 0.619 |
| 2362 | EMX2OS |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | PEG3 | Sponge network | -0.777 | 0.1134 | -1.74 | 0 | 0.619 |
| 2363 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-532-3p | 14 | CADM3 | Sponge network | -2.117 | 0.00161 | -3.663 | 0 | 0.619 |
| 2364 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | FGF5 | Sponge network | -0.576 | 0.10121 | 0.549 | 0.28659 | 0.619 |
| 2365 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | PEG3 | Sponge network | -0.739 | 0.00044 | -1.74 | 0 | 0.619 |
| 2366 | RP11-999E24.3 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-425-5p | 34 | DTNA | Sponge network | -0.693 | 0.05629 | -1.648 | 1.0E-5 | 0.619 |
| 2367 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-877-5p;hsa-miR-93-5p | 26 | PGR | Sponge network | -1.349 | 0.00017 | -1.704 | 0 | 0.619 |
| 2368 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-93-3p | 13 | PCDH10 | Sponge network | -2.531 | 0 | -1.644 | 0.0078 | 0.618 |
| 2369 | PART1 |
hsa-miR-146b-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 12 | FHL1 | Sponge network | -2.925 | 0 | -2.687 | 0 | 0.618 |
| 2370 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | SYNE1 | Sponge network | -0.901 | 0.00019 | -0.738 | 0.00316 | 0.618 |
| 2371 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-96-5p | 28 | PRKAA2 | Sponge network | -1.582 | 8.0E-5 | -2.462 | 0 | 0.618 |
| 2372 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-424-5p | 19 | FAT3 | Sponge network | -4.463 | 0.00025 | -1.601 | 0.00051 | 0.618 |
| 2373 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 16 | FGFR1 | Sponge network | 0.335 | 0.20596 | -0.509 | 0.03957 | 0.618 |
| 2374 | RP11-356J5.12 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p | 12 | PARK2 | Sponge network | -0.899 | 6.0E-5 | -1.892 | 0 | 0.618 |
| 2375 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-93-5p | 19 | BNC2 | Sponge network | -1.255 | 0.00981 | -0.491 | 0.09988 | 0.618 |
| 2376 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 37 | SCN9A | Sponge network | -0.739 | 0.0574 | -0.525 | 0.10593 | 0.618 |
| 2377 | AC006116.24 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-93-5p | 26 | AR | Sponge network | -0.473 | 0.07602 | -1.554 | 0 | 0.618 |
| 2378 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 32 | EPHA3 | Sponge network | 0.246 | 0.59912 | 0.185 | 0.53588 | 0.618 |
| 2379 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 45 | DIP2C | Sponge network | -0.49 | 0.04946 | -0.436 | 0.01713 | 0.618 |
| 2380 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-576-5p | 13 | HIP1 | Sponge network | 0.889 | 9.0E-5 | 0.774 | 0 | 0.618 |
| 2381 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | -0.323 | 0.01372 | -0.57 | 0 | 0.618 |
| 2382 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 26 | ESR1 | Sponge network | -0.358 | 0.17255 | -1.035 | 3.0E-5 | 0.618 |
| 2383 | FAM66C |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-769-5p | 24 | SYNPO2 | Sponge network | -0.901 | 0.00019 | -2.342 | 0 | 0.618 |
| 2384 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429 | 17 | SPG20 | Sponge network | -0.563 | 0.02545 | -1.16 | 0 | 0.618 |
| 2385 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-7-5p | 10 | PDGFC | Sponge network | -0.976 | 0.00025 | -0.33 | 0.06483 | 0.618 |
| 2386 | MAGI2-AS3 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 22 | SORCS1 | Sponge network | -0.129 | 0.57177 | -3.492 | 0 | 0.618 |
| 2387 | FENDRR |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | PARK2 | Sponge network | -1.281 | 0.00012 | -1.892 | 0 | 0.618 |
| 2388 | FAM66C |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 20 | SPG20 | Sponge network | -0.901 | 0.00019 | -1.16 | 0 | 0.618 |
| 2389 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-374b-3p;hsa-miR-450b-5p | 31 | FAT3 | Sponge network | -4.546 | 0 | -1.601 | 0.00051 | 0.618 |
| 2390 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p | 10 | JAKMIP2 | Sponge network | -0.583 | 0.14589 | -1.388 | 0 | 0.618 |
| 2391 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 24 | RCAN2 | Sponge network | -1.111 | 0.0003 | -0.33 | 0.15172 | 0.618 |
| 2392 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-744-5p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | -1.349 | 0.00017 | -0.509 | 0.03957 | 0.618 |
| 2393 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 49 | QKI | Sponge network | -0.563 | 0.02545 | -0.333 | 0.04111 | 0.618 |
| 2394 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-942-5p | 15 | ZBTB20 | Sponge network | -0.129 | 0.57177 | -0.122 | 0.57569 | 0.618 |
| 2395 | SNHG14 |
hsa-miR-10b-3p;hsa-miR-193b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p | 10 | LRRTM1 | Sponge network | -1.111 | 0.0003 | -3.361 | 0 | 0.618 |
| 2396 | RP11-166D19.1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 20 | GLIPR1 | Sponge network | -0.576 | 0.10121 | 0.146 | 0.3276 | 0.618 |
| 2397 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 37 | DIP2C | Sponge network | -0.487 | 0.02157 | -0.436 | 0.01713 | 0.618 |
| 2398 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-93-5p | 30 | MCC | Sponge network | -2.69 | 0.0001 | -1.005 | 0 | 0.618 |
| 2399 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-93-5p | 13 | ANK2 | Sponge network | -0.473 | 0.07602 | -1.603 | 1.0E-5 | 0.618 |
| 2400 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-424-5p | 22 | EPHA7 | Sponge network | -4.463 | 0.00025 | -2.197 | 3.0E-5 | 0.618 |
| 2401 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -2.46 | 0 | -2.046 | 0.00019 | 0.618 |
| 2402 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 33 | MYO9A | Sponge network | 0.194 | 0.64278 | -0.147 | 0.32373 | 0.618 |
| 2403 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-589-3p | 15 | PCDH9 | Sponge network | -2.531 | 0 | -1.823 | 4.0E-5 | 0.618 |
| 2404 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-589-5p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -0.976 | 0.00025 | -3.328 | 0 | 0.618 |
| 2405 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-671-5p | 21 | DLC1 | Sponge network | -0.469 | 0.08721 | -0.382 | 0.05038 | 0.617 |
| 2406 | AC003090.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | PARK2 | Sponge network | -2.117 | 0.00161 | -1.892 | 0 | 0.617 |
| 2407 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-744-3p | 26 | ADAMTSL3 | Sponge network | -0.162 | 0.47687 | -1.559 | 0 | 0.617 |
| 2408 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-154-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 16 | GRIN2A | Sponge network | -4.546 | 0 | -2.161 | 0 | 0.617 |
| 2409 | LINC00472 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-511-5p | 18 | MRVI1 | Sponge network | -0.842 | 0.01361 | -1.051 | 0.00022 | 0.617 |
| 2410 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-590-5p | 13 | THSD7A | Sponge network | -2.452 | 0 | 0.242 | 0.25154 | 0.617 |
| 2411 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-589-5p;hsa-miR-93-5p | 14 | SLITRK3 | Sponge network | -1.439 | 4.0E-5 | -3.328 | 0 | 0.617 |
| 2412 | FENDRR |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 61 | AR | Sponge network | -1.281 | 0.00012 | -1.554 | 0 | 0.617 |
| 2413 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | ADAMTSL3 | Sponge network | -0.769 | 0.00035 | -1.559 | 0 | 0.617 |
| 2414 | ARHGEF26-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-940 | 22 | SORCS1 | Sponge network | -2.46 | 0 | -3.492 | 0 | 0.617 |
| 2415 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 34 | RSPO2 | Sponge network | -2.452 | 0 | -3.038 | 0 | 0.617 |
| 2416 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-671-5p | 10 | KIT | Sponge network | -1.886 | 0 | -2.184 | 0 | 0.617 |
| 2417 | RP11-307B6.3 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-5p | 27 | AR | Sponge network | -4.463 | 0.00025 | -1.554 | 0 | 0.617 |
| 2418 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | PCDH10 | Sponge network | 0.194 | 0.64278 | -1.644 | 0.0078 | 0.617 |
| 2419 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TLL1 | Sponge network | -0.49 | 0.04946 | -1.195 | 3.0E-5 | 0.617 |
| 2420 | GNG12-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | MAPK10 | Sponge network | -0.793 | 7.0E-5 | -1.774 | 0 | 0.617 |
| 2421 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 24 | GRIN2A | Sponge network | -0.976 | 0.00025 | -2.161 | 0 | 0.617 |
| 2422 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 28 | TET1 | Sponge network | -0.323 | 0.01372 | -0.135 | 0.52138 | 0.617 |
| 2423 | MRVI1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-539-5p | 13 | RYR2 | Sponge network | -1.092 | 4.0E-5 | -1.466 | 0.00031 | 0.617 |
| 2424 | MIR497HG |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p | 15 | RELN | Sponge network | -1.439 | 4.0E-5 | -2.403 | 0 | 0.617 |
| 2425 | CTD-2002H8.2 |
hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ATRX | Sponge network | 0.931 | 1.0E-5 | 0.261 | 0.04408 | 0.617 |
| 2426 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-877-5p | 12 | PGR | Sponge network | -0.309 | 0.1145 | -1.704 | 0 | 0.617 |
| 2427 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-330-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-550a-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 31 | CHD5 | Sponge network | -1.331 | 3.0E-5 | -0.922 | 0.01521 | 0.617 |
| 2428 | MIR143HG |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 26 | ABL1 | Sponge network | -0.739 | 0.0574 | -0.215 | 0.07804 | 0.617 |
| 2429 | PWAR6 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-96-5p | 21 | RPS6KA6 | Sponge network | -1.575 | 6.0E-5 | -2.989 | 0 | 0.617 |
| 2430 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-424-5p | 10 | DYNC1I1 | Sponge network | -2.531 | 0 | -1.12 | 0.00107 | 0.617 |
| 2431 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 14 | PEG3 | Sponge network | -0.611 | 0.09607 | -1.74 | 0 | 0.617 |
| 2432 | NR2F1-AS1 |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 10 | DPP6 | Sponge network | -0.49 | 0.04946 | -3.777 | 0 | 0.617 |
| 2433 | FAM66C |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PLSCR4 | Sponge network | -0.901 | 0.00019 | -0.474 | 0.03833 | 0.617 |
| 2434 | RP11-532F6.3 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 13 | SORCS1 | Sponge network | -0.896 | 0.00099 | -3.492 | 0 | 0.617 |
| 2435 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p | 18 | AFF3 | Sponge network | -4.546 | 0 | -2.373 | 0 | 0.617 |
| 2436 | MIR143HG |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 61 | TRPS1 | Sponge network | -0.739 | 0.0574 | -0.191 | 0.44109 | 0.617 |
| 2437 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | PEG3 | Sponge network | -1.216 | 0 | -1.74 | 0 | 0.617 |
| 2438 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | TAL1 | Sponge network | -2.452 | 0 | -0.462 | 0.00574 | 0.617 |
| 2439 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 40 | NRXN1 | Sponge network | -0.129 | 0.57177 | -3.778 | 0 | 0.617 |
| 2440 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-940;hsa-miR-98-5p | 34 | PBX1 | Sponge network | -0.976 | 0.00025 | -1.206 | 0 | 0.617 |
| 2441 | MYLK-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-935 | 10 | LRRC7 | Sponge network | -1.331 | 3.0E-5 | -1.341 | 0.01193 | 0.617 |
| 2442 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 15 | NRXN3 | Sponge network | -0.611 | 0.09607 | -0.893 | 0.04186 | 0.617 |
| 2443 | LINC00473 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | -1.203 | 0.03881 | -0.382 | 0.05038 | 0.617 |
| 2444 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | AKAP12 | Sponge network | 0.335 | 0.20596 | -0.932 | 0.0028 | 0.616 |
| 2445 | PGM5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421 | 11 | SPG20 | Sponge network | -4.546 | 0 | -1.16 | 0 | 0.616 |
| 2446 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-935 | 12 | GJA1 | Sponge network | -1.331 | 3.0E-5 | -0.485 | 0.02179 | 0.616 |
| 2447 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-629-3p | 11 | CDH23 | Sponge network | -0.358 | 0.17255 | -0.974 | 0.00034 | 0.616 |
| 2448 | LINC00578 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 12 | NAV3 | Sponge network | 0.811 | 0.03228 | 0.212 | 0.47729 | 0.616 |
| 2449 | SNHG14 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | KCTD8 | Sponge network | -1.111 | 0.0003 | -3.359 | 0 | 0.616 |
| 2450 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935 | 34 | DMD | Sponge network | -0.896 | 0.00099 | -1.506 | 0 | 0.616 |
| 2451 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-486-5p;hsa-miR-532-3p | 10 | DMTF1 | Sponge network | 0.637 | 0.00069 | 0.248 | 0.02726 | 0.616 |
| 2452 | TRAF3IP2-AS1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ITPKB | Sponge network | -0.323 | 0.01372 | -0.704 | 0.00106 | 0.616 |
| 2453 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | TSHZ3 | Sponge network | 0.889 | 9.0E-5 | -0.556 | 0.01516 | 0.616 |
| 2454 | MAGI2-AS3 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 22 | GLIPR1 | Sponge network | -0.129 | 0.57177 | 0.146 | 0.3276 | 0.616 |
| 2455 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 30 | RSPO2 | Sponge network | -0.576 | 0.10121 | -3.038 | 0 | 0.616 |
| 2456 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 39 | FAT3 | Sponge network | -0.358 | 0.17255 | -1.601 | 0.00051 | 0.616 |
| 2457 | MRVI1-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 10 | TPO | Sponge network | -1.092 | 4.0E-5 | -1.87 | 2.0E-5 | 0.616 |
| 2458 | SNHG14 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ADH1B | Sponge network | -1.111 | 0.0003 | -3.716 | 0 | 0.616 |
| 2459 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-576-5p | 11 | AKAP12 | Sponge network | -0.611 | 0.09607 | -0.932 | 0.0028 | 0.616 |
| 2460 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | CACNA2D1 | Sponge network | -0.49 | 0.04946 | -0.846 | 0.00675 | 0.616 |
| 2461 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 74 | TCF4 | Sponge network | -2.452 | 0 | 0.016 | 0.92271 | 0.616 |
| 2462 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | TCF4 | Sponge network | 0.342 | 0.03129 | 0.016 | 0.92271 | 0.616 |
| 2463 | FENDRR |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-98-5p | 29 | RSPO2 | Sponge network | -1.281 | 0.00012 | -3.038 | 0 | 0.616 |
| 2464 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 39 | MEF2C | Sponge network | 0.572 | 0.01722 | -0.499 | 0.01448 | 0.616 |
| 2465 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 32 | IGF1 | Sponge network | 0.572 | 0.01722 | -0.204 | 0.5898 | 0.616 |
| 2466 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p;hsa-miR-874-3p | 21 | TACC1 | Sponge network | -1.092 | 4.0E-5 | -0.362 | 0.05406 | 0.616 |
| 2467 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-7-5p | 21 | SETBP1 | Sponge network | -0.162 | 0.47687 | -1.302 | 1.0E-5 | 0.616 |
| 2468 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 56 | PRKAA2 | Sponge network | -0.576 | 0.10121 | -2.462 | 0 | 0.616 |
| 2469 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 36 | IGF1 | Sponge network | -1.697 | 0.00889 | -0.204 | 0.5898 | 0.616 |
| 2470 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-671-5p | 14 | KIT | Sponge network | -1.331 | 3.0E-5 | -2.184 | 0 | 0.616 |
| 2471 | FENDRR |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p | 15 | FLNA | Sponge network | -1.281 | 0.00012 | -0.771 | 0.01377 | 0.615 |
| 2472 | AC006116.24 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-577;hsa-miR-93-5p | 10 | GLI3 | Sponge network | -0.473 | 0.07602 | -0.615 | 0.01482 | 0.615 |
| 2473 | GNG12-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-3p;hsa-miR-486-5p;hsa-miR-542-3p | 19 | SYNM | Sponge network | -0.793 | 7.0E-5 | -2.309 | 1.0E-5 | 0.615 |
| 2474 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p | 13 | GRIN2A | Sponge network | -0.932 | 0.00329 | -2.161 | 0 | 0.615 |
| 2475 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | PCDH9 | Sponge network | -0.739 | 0.00044 | -1.823 | 4.0E-5 | 0.615 |
| 2476 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | ZFHX4 | Sponge network | 0.249 | 0.35902 | 0.018 | 0.95574 | 0.615 |
| 2477 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-582-3p | 22 | ABL2 | Sponge network | 0.993 | 0 | 0.82 | 0 | 0.615 |
| 2478 | AC003090.1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 50 | AR | Sponge network | -2.117 | 0.00161 | -1.554 | 0 | 0.615 |
| 2479 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-744-3p;hsa-miR-940 | 32 | MEF2C | Sponge network | -0.465 | 0.04703 | -0.499 | 0.01448 | 0.615 |
| 2480 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | LSAMP | Sponge network | -1.111 | 0.0003 | -0.066 | 0.81708 | 0.615 |
| 2481 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 24 | TSHZ3 | Sponge network | -0.473 | 0.07602 | -0.556 | 0.01516 | 0.615 |
| 2482 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-877-5p | 19 | ANK2 | Sponge network | -0.309 | 0.1145 | -1.603 | 1.0E-5 | 0.615 |
| 2483 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | -0.709 | 0.18168 | -1.603 | 1.0E-5 | 0.615 |
| 2484 | AC003090.1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | RNF180 | Sponge network | -2.117 | 0.00161 | -1.127 | 1.0E-5 | 0.615 |
| 2485 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -0.976 | 0.00025 | -0.7 | 1.0E-5 | 0.615 |
| 2486 | ACTA2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | NAV3 | Sponge network | 0.194 | 0.64278 | 0.212 | 0.47729 | 0.615 |
| 2487 | HAND2-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | THSD7B | Sponge network | -1.697 | 0.00889 | 0.154 | 0.74723 | 0.615 |
| 2488 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 65 | TCF4 | Sponge network | 0.532 | 0.00505 | 0.016 | 0.92271 | 0.615 |
| 2489 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 22 | AFF3 | Sponge network | -1.057 | 0.00029 | -2.373 | 0 | 0.615 |
| 2490 | ZNF582-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-671-5p | 16 | CADM3 | Sponge network | -1.349 | 0.00017 | -3.663 | 0 | 0.615 |
| 2491 | GNG12-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 48 | AR | Sponge network | -0.793 | 7.0E-5 | -1.554 | 0 | 0.615 |
| 2492 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 48 | BNC2 | Sponge network | -0.487 | 0.02157 | -0.491 | 0.09988 | 0.615 |
| 2493 | DNM3OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | ERG | Sponge network | 0.572 | 0.01722 | 0.002 | 0.99011 | 0.615 |
| 2494 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p | 39 | BNC2 | Sponge network | -2.69 | 0.0001 | -0.491 | 0.09988 | 0.615 |
| 2495 | ARHGEF26-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 21 | SFRP1 | Sponge network | -2.46 | 0 | -3.566 | 0 | 0.615 |
| 2496 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 44 | RASSF2 | Sponge network | -0.129 | 0.57177 | -0.273 | 0.21961 | 0.614 |
| 2497 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | LRP1B | Sponge network | -2.452 | 0 | -2.163 | 0 | 0.614 |
| 2498 | GNG12-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 14 | RHOB | Sponge network | -0.793 | 7.0E-5 | -1.052 | 0 | 0.614 |
| 2499 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 14 | PARK2 | Sponge network | -0.49 | 0.04946 | -1.892 | 0 | 0.614 |
| 2500 | AC003090.1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TPO | Sponge network | -2.117 | 0.00161 | -1.87 | 2.0E-5 | 0.614 |
| 2501 | HOXB-AS1 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p | 10 | LHFP | Sponge network | -0.154 | 0.53954 | -0.167 | 0.41371 | 0.614 |
| 2502 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 24 | FGFR1 | Sponge network | -1.111 | 0.0003 | -0.509 | 0.03957 | 0.614 |
| 2503 | RP5-1198O20.4 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | THSD7B | Sponge network | 0.997 | 0.00066 | 0.154 | 0.74723 | 0.614 |
| 2504 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 12 | MAPK10 | Sponge network | -0.769 | 0.00035 | -1.774 | 0 | 0.614 |
| 2505 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 37 | DMD | Sponge network | -0.323 | 0.01372 | -1.506 | 0 | 0.614 |
| 2506 | LINC00957 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SETBP1 | Sponge network | -0.421 | 0.0114 | -1.302 | 1.0E-5 | 0.614 |
| 2507 | CTC-510F12.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-935 | 14 | FAT3 | Sponge network | -0.691 | 0.00146 | -1.601 | 0.00051 | 0.614 |
| 2508 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 30 | MYO9A | Sponge network | 0.083 | 0.66405 | -0.147 | 0.32373 | 0.614 |
| 2509 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 42 | ST6GAL2 | Sponge network | -1.697 | 0.00889 | -1.201 | 0.00597 | 0.614 |
| 2510 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 34 | PGR | Sponge network | -0.487 | 0.02157 | -1.704 | 0 | 0.614 |
| 2511 | RP11-554A11.4 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-874-3p | 12 | SORCS1 | Sponge network | -0.583 | 0.14589 | -3.492 | 0 | 0.614 |
| 2512 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | JAZF1 | Sponge network | -1.439 | 4.0E-5 | -0.377 | 0.02677 | 0.614 |
| 2513 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-629-3p;hsa-miR-7-5p | 29 | ZBTB4 | Sponge network | -0.487 | 0.02157 | -0.739 | 0 | 0.614 |
| 2514 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 33 | EBF1 | Sponge network | 0.572 | 0.01722 | -0.384 | 0.08856 | 0.614 |
| 2515 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 53 | ZFHX4 | Sponge network | -0.777 | 0.1134 | 0.018 | 0.95574 | 0.614 |
| 2516 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 30 | GRIN2A | Sponge network | -0.576 | 0.10121 | -2.161 | 0 | 0.614 |
| 2517 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 44 | AR | Sponge network | -0.739 | 0.00044 | -1.554 | 0 | 0.614 |
| 2518 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 26 | ANK2 | Sponge network | -0.793 | 7.0E-5 | -1.603 | 1.0E-5 | 0.614 |
| 2519 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 20 | MEF2C | Sponge network | 0.693 | 0.00076 | -0.499 | 0.01448 | 0.614 |
| 2520 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-421;hsa-miR-429 | 16 | KIT | Sponge network | -2.62 | 0 | -2.184 | 0 | 0.614 |
| 2521 | MEG3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-5p | 11 | LHFP | Sponge network | 0.249 | 0.35902 | -0.167 | 0.41371 | 0.614 |
| 2522 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | ANK2 | Sponge network | 0.572 | 0.01722 | -1.603 | 1.0E-5 | 0.614 |
| 2523 | PGM5-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-378a-3p;hsa-miR-484 | 10 | FLNA | Sponge network | -4.546 | 0 | -0.771 | 0.01377 | 0.614 |
| 2524 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-500a-5p | 23 | DMD | Sponge network | -2.531 | 0 | -1.506 | 0 | 0.614 |
| 2525 | JAZF1-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | PRKCB | Sponge network | -0.769 | 0.00035 | -1.313 | 2.0E-5 | 0.614 |
| 2526 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | SYNE1 | Sponge network | 0.532 | 0.00505 | -0.738 | 0.00316 | 0.614 |
| 2527 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-877-5p | 23 | GRIN2A | Sponge network | -2.62 | 0 | -2.161 | 0 | 0.614 |
| 2528 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 16 | EZH1 | Sponge network | -2.46 | 0 | -0.564 | 1.0E-5 | 0.614 |
| 2529 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 43 | QKI | Sponge network | -0.611 | 0.09607 | -0.333 | 0.04111 | 0.614 |
| 2530 | USP3-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-651-5p;hsa-miR-93-3p | 26 | WWTR1 | Sponge network | -0.162 | 0.47687 | -0.345 | 0.11997 | 0.614 |
| 2531 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-744-3p;hsa-miR-93-5p | 44 | FAT3 | Sponge network | -0.611 | 0.09607 | -1.601 | 0.00051 | 0.614 |
| 2532 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 20 | TMEFF2 | Sponge network | -0.576 | 0.10121 | -2.426 | 0 | 0.614 |
| 2533 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | AKAP6 | Sponge network | 0.997 | 0.00066 | -1.586 | 3.0E-5 | 0.614 |
| 2534 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 15 | PARK2 | Sponge network | -0.129 | 0.57177 | -1.892 | 0 | 0.613 |
| 2535 | ADAMTS9-AS2 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-532-5p;hsa-miR-582-5p | 11 | PRICKLE1 | Sponge network | -2.452 | 0 | -0.112 | 0.67088 | 0.613 |
| 2536 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | PARK2 | Sponge network | -0.737 | 0.06182 | -1.892 | 0 | 0.613 |
| 2537 | RP11-554A11.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-769-5p;hsa-miR-874-3p | 27 | GRIK3 | Sponge network | -0.583 | 0.14589 | -3.208 | 0 | 0.613 |
| 2538 | HAND2-AS1 |
hsa-let-7a-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | -1.697 | 0.00889 | -1.628 | 0 | 0.613 |
| 2539 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-744-5p | 10 | FGFR1 | Sponge network | -0.309 | 0.1145 | -0.509 | 0.03957 | 0.613 |
| 2540 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p | 14 | DCLK1 | Sponge network | -1.349 | 0.00017 | -1.324 | 0.00014 | 0.613 |
| 2541 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 25 | SSBP2 | Sponge network | -1.582 | 8.0E-5 | -1.58 | 0 | 0.613 |
| 2542 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p | 20 | RBMS3 | Sponge network | -0.376 | 0.00337 | -0.519 | 0.03551 | 0.613 |
| 2543 | EMX2OS |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | SFRP1 | Sponge network | -0.777 | 0.1134 | -3.566 | 0 | 0.613 |
| 2544 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-542-3p;hsa-miR-582-5p | 33 | DMD | Sponge network | -2.69 | 0.0001 | -1.506 | 0 | 0.613 |
| 2545 | RP11-166D19.1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 12 | SORCS2 | Sponge network | -0.576 | 0.10121 | -0.223 | 0.4253 | 0.613 |
| 2546 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-940 | 34 | MEF2C | Sponge network | -0.896 | 0.00099 | -0.499 | 0.01448 | 0.613 |
| 2547 | SNHG14 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 15 | AMPH | Sponge network | -1.111 | 0.0003 | -0.916 | 5.0E-5 | 0.613 |
| 2548 | DNM3OS |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p | 16 | ITGA5 | Sponge network | 0.572 | 0.01722 | 0.452 | 0.03524 | 0.613 |
| 2549 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-577;hsa-miR-93-5p | 13 | GLI3 | Sponge network | -2.69 | 0.0001 | -0.615 | 0.01482 | 0.613 |
| 2550 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 10 | LRFN5 | Sponge network | -0.737 | 0.06182 | -1.483 | 0 | 0.613 |
| 2551 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-877-5p;hsa-miR-93-5p | 19 | PGR | Sponge network | -1.654 | 0.00144 | -1.704 | 0 | 0.613 |
| 2552 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 13 | PTPN13 | Sponge network | -2.452 | 0 | -1.208 | 0 | 0.613 |
| 2553 | HAND2-AS1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 14 | AMPH | Sponge network | -1.697 | 0.00889 | -0.916 | 5.0E-5 | 0.613 |
| 2554 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-942-5p | 43 | HLF | Sponge network | -0.576 | 0.10121 | -2.443 | 0 | 0.613 |
| 2555 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 59 | SSBP2 | Sponge network | -0.129 | 0.57177 | -1.58 | 0 | 0.613 |
| 2556 | PWAR6 |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 10 | DPP6 | Sponge network | -1.575 | 6.0E-5 | -3.777 | 0 | 0.613 |
| 2557 | ZNF582-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-505-5p | 13 | MRVI1 | Sponge network | -1.349 | 0.00017 | -1.051 | 0.00022 | 0.613 |
| 2558 | RP11-307B6.3 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p | 15 | ANK2 | Sponge network | -4.463 | 0.00025 | -1.603 | 1.0E-5 | 0.613 |
| 2559 | RP5-1198O20.4 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-532-3p | 18 | SYNM | Sponge network | 0.997 | 0.00066 | -2.309 | 1.0E-5 | 0.612 |
| 2560 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | ANK2 | Sponge network | -0.376 | 0.00337 | -1.603 | 1.0E-5 | 0.612 |
| 2561 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | TACC1 | Sponge network | -0.737 | 0.06182 | -0.362 | 0.05406 | 0.612 |
| 2562 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CNTN1 | Sponge network | -2.915 | 0.00015 | -2.594 | 0 | 0.612 |
| 2563 | SNHG14 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 59 | GRIK3 | Sponge network | -1.111 | 0.0003 | -3.208 | 0 | 0.612 |
| 2564 | MIR497HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-589-5p;hsa-miR-590-5p | 12 | ITGB3 | Sponge network | -1.439 | 4.0E-5 | -0.011 | 0.95751 | 0.612 |
| 2565 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577 | 27 | RBMS3 | Sponge network | 0.811 | 0.03228 | -0.519 | 0.03551 | 0.612 |
| 2566 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | EBF1 | Sponge network | -0.737 | 0.06182 | -0.384 | 0.08856 | 0.612 |
| 2567 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p | 11 | RECK | Sponge network | -4.546 | 0 | -1.043 | 0 | 0.612 |
| 2568 | LINC00702 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 13 | SCN11A | Sponge network | -0.737 | 0.06182 | -1.15 | 0.00299 | 0.612 |
| 2569 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | TSHZ3 | Sponge network | 0.335 | 0.20596 | -0.556 | 0.01516 | 0.612 |
| 2570 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p | 17 | RSPO2 | Sponge network | -1.331 | 3.0E-5 | -3.038 | 0 | 0.612 |
| 2571 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 12 | DYNC1I1 | Sponge network | -0.583 | 0.14589 | -1.12 | 0.00107 | 0.612 |
| 2572 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-877-5p | 37 | RBMS3 | Sponge network | -0.096 | 0.67958 | -0.519 | 0.03551 | 0.612 |
| 2573 | FAM66C |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 13 | PARK2 | Sponge network | -0.901 | 0.00019 | -1.892 | 0 | 0.612 |
| 2574 | JAZF1-AS1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 12 | FHL1 | Sponge network | -0.769 | 0.00035 | -2.687 | 0 | 0.612 |
| 2575 | LINC00702 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 16 | STAT5B | Sponge network | -0.737 | 0.06182 | -0.182 | 0.1082 | 0.612 |
| 2576 | TPTEP1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-455-5p | 11 | AMPH | Sponge network | -1.216 | 0 | -0.916 | 5.0E-5 | 0.612 |
| 2577 | SERTAD4-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-24-3p;hsa-miR-335-3p | 16 | DLC1 | Sponge network | -0.932 | 0.00329 | -0.382 | 0.05038 | 0.612 |
| 2578 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 49 | HLF | Sponge network | -1.111 | 0.0003 | -2.443 | 0 | 0.612 |
| 2579 | TRAF3IP2-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | MAPK10 | Sponge network | -0.323 | 0.01372 | -1.774 | 0 | 0.612 |
| 2580 | RP11-736K20.5 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-452-5p | 12 | SYNM | Sponge network | -0.358 | 0.17255 | -2.309 | 1.0E-5 | 0.612 |
| 2581 | AC009948.5 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZDBF2 | Sponge network | 0.532 | 0.00505 | -0.998 | 0.00025 | 0.612 |
| 2582 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 28 | ANK2 | Sponge network | 0.335 | 0.20596 | -1.603 | 1.0E-5 | 0.612 |
| 2583 | SNHG14 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | ZNF208 | Sponge network | -1.111 | 0.0003 | -0.4 | 0.22381 | 0.612 |
| 2584 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-874-3p | 13 | NR2C2 | Sponge network | 1.162 | 5.0E-5 | 0.21 | 0.03224 | 0.612 |
| 2585 | MIR143HG |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 32 | PTPN14 | Sponge network | -0.739 | 0.0574 | 0.144 | 0.34595 | 0.612 |
| 2586 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-7-5p;hsa-miR-93-5p | 26 | ZBTB4 | Sponge network | -0.611 | 0.09607 | -0.739 | 0 | 0.612 |
| 2587 | FGF14-AS2 |
hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | PCDH10 | Sponge network | -1.582 | 8.0E-5 | -1.644 | 0.0078 | 0.612 |
| 2588 | LINC00702 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | ZNF382 | Sponge network | -0.737 | 0.06182 | -0.494 | 0.03831 | 0.612 |
| 2589 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 46 | QKI | Sponge network | -0.769 | 0.00035 | -0.333 | 0.04111 | 0.612 |
| 2590 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | PDZRN3 | Sponge network | -0.793 | 7.0E-5 | -1.548 | 0 | 0.612 |
| 2591 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | ZNF770 | Sponge network | 1.153 | 0 | 0.206 | 0.06751 | 0.612 |
| 2592 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-3913-5p | 10 | RIMS1 | Sponge network | -2.69 | 0.0001 | -3.143 | 0 | 0.612 |
| 2593 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 43 | AR | Sponge network | -0.896 | 0.00099 | -1.554 | 0 | 0.611 |
| 2594 | ACTA2-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 14 | STAT5B | Sponge network | 0.194 | 0.64278 | -0.182 | 0.1082 | 0.611 |
| 2595 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-769-5p | 12 | RCAN2 | Sponge network | -0.096 | 0.67958 | -0.33 | 0.15172 | 0.611 |
| 2596 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-744-5p | 14 | FGFR1 | Sponge network | -0.246 | 0.35034 | -0.509 | 0.03957 | 0.611 |
| 2597 | MIR143HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-98-5p | 66 | PRKAA2 | Sponge network | -0.739 | 0.0574 | -2.462 | 0 | 0.611 |
| 2598 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p | 15 | FNBP1 | Sponge network | -0.323 | 0.01372 | -0.969 | 4.0E-5 | 0.611 |
| 2599 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | PEG3 | Sponge network | 0.997 | 0.00066 | -1.74 | 0 | 0.611 |
| 2600 | ACTA2-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 31 | DLC1 | Sponge network | 0.194 | 0.64278 | -0.382 | 0.05038 | 0.611 |
| 2601 | RP11-166D19.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-942-5p;hsa-miR-96-5p | 13 | ZBTB20 | Sponge network | -0.576 | 0.10121 | -0.122 | 0.57569 | 0.611 |
| 2602 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 58 | ADARB1 | Sponge network | -0.49 | 0.04946 | -0.335 | 0.06631 | 0.611 |
| 2603 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 46 | EPHA5 | Sponge network | -0.576 | 0.10121 | -0.957 | 0.01733 | 0.611 |
| 2604 | ADAMTS9-AS2 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 42 | IGFBP5 | Sponge network | -2.452 | 0 | -0.806 | 0.00105 | 0.611 |
| 2605 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | NRXN3 | Sponge network | 0.299 | 0.57722 | -0.893 | 0.04186 | 0.611 |
| 2606 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 32 | EPHA3 | Sponge network | -0.162 | 0.47687 | 0.185 | 0.53588 | 0.611 |
| 2607 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | DKK3 | Sponge network | -0.737 | 0.06182 | -0.052 | 0.77783 | 0.611 |
| 2608 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | PRDM5 | Sponge network | -2.452 | 0 | -1.09 | 0.00014 | 0.611 |
| 2609 | PCA3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-96-5p | 38 | FBXO32 | Sponge network | -0.709 | 0.18168 | -0.495 | 0.07561 | 0.611 |
| 2610 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p | 14 | THSD7A | Sponge network | 0.997 | 0.00066 | 0.242 | 0.25154 | 0.611 |
| 2611 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | CACNA2D1 | Sponge network | -1.439 | 4.0E-5 | -0.846 | 0.00675 | 0.611 |
| 2612 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p | 18 | KIT | Sponge network | -0.842 | 0.01361 | -2.184 | 0 | 0.611 |
| 2613 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 76 | IL6ST | Sponge network | -0.737 | 0.06182 | -0.402 | 0.00676 | 0.611 |
| 2614 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p | 21 | CNTN1 | Sponge network | -0.358 | 0.17255 | -2.594 | 0 | 0.611 |
| 2615 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 29 | LRRC7 | Sponge network | -0.739 | 0.0574 | -1.341 | 0.01193 | 0.611 |
| 2616 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 27 | SETBP1 | Sponge network | -0.769 | 0.00035 | -1.302 | 1.0E-5 | 0.611 |
| 2617 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 50 | SSBP2 | Sponge network | -0.769 | 0.00035 | -1.58 | 0 | 0.611 |
| 2618 | RP11-166D19.1 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CTGF | Sponge network | -0.576 | 0.10121 | 0.321 | 0.11682 | 0.611 |
| 2619 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 90 | NTRK3 | Sponge network | -1.111 | 0.0003 | -1.77 | 3.0E-5 | 0.611 |
| 2620 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-98-5p | 35 | ZFHX3 | Sponge network | 0.494 | 0.00339 | 0.389 | 0.01363 | 0.611 |
| 2621 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | ANK2 | Sponge network | -0.421 | 0.0114 | -1.603 | 1.0E-5 | 0.611 |
| 2622 | USP3-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-935;hsa-miR-940 | 16 | MAPK10 | Sponge network | -0.162 | 0.47687 | -1.774 | 0 | 0.611 |
| 2623 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 34 | ZBTB4 | Sponge network | -0.49 | 0.04946 | -0.739 | 0 | 0.611 |
| 2624 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-484;hsa-miR-93-5p | 14 | EDA2R | Sponge network | -0.976 | 0.00025 | -0.587 | 0.01598 | 0.61 |
| 2625 | FGF14-AS2 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-5p;hsa-miR-96-5p | 10 | FNBP1 | Sponge network | -1.582 | 8.0E-5 | -0.969 | 4.0E-5 | 0.61 |
| 2626 | MIR143HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-98-5p | 14 | SCN11A | Sponge network | -0.739 | 0.0574 | -1.15 | 0.00299 | 0.61 |
| 2627 | RP11-390K5.6 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 22 | MDM4 | Sponge network | 1.148 | 2.0E-5 | 0.345 | 0.00762 | 0.61 |
| 2628 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 14 | DCN | Sponge network | -1.111 | 0.0003 | -1.288 | 0 | 0.61 |
| 2629 | USP3-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-940 | 16 | FAM172A | Sponge network | -0.162 | 0.47687 | -0.57 | 0 | 0.61 |
| 2630 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 30 | ANK2 | Sponge network | -0.487 | 0.02157 | -1.603 | 1.0E-5 | 0.61 |
| 2631 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-3614-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | NRXN3 | Sponge network | -1.349 | 0.00017 | -0.893 | 0.04186 | 0.61 |
| 2632 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | LHFP | Sponge network | 0.335 | 0.20596 | -0.167 | 0.41371 | 0.61 |
| 2633 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-874-3p | 20 | NR2C2 | Sponge network | 0.817 | 3.0E-5 | 0.21 | 0.03224 | 0.61 |
| 2634 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 29 | MYO9A | Sponge network | -0.096 | 0.67958 | -0.147 | 0.32373 | 0.61 |
| 2635 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 16 | PDZD2 | Sponge network | -2.452 | 0 | -0.909 | 3.0E-5 | 0.61 |
| 2636 | HAND2-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-942-5p | 15 | ZBTB20 | Sponge network | -1.697 | 0.00889 | -0.122 | 0.57569 | 0.61 |
| 2637 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 53 | FAT3 | Sponge network | -0.096 | 0.67958 | -1.601 | 0.00051 | 0.61 |
| 2638 | LINC00854 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-625-5p;hsa-miR-940 | 24 | MDM4 | Sponge network | 1.18 | 0 | 0.345 | 0.00762 | 0.61 |
| 2639 | CTC-510F12.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-484 | 10 | NRXN3 | Sponge network | -0.691 | 0.00146 | -0.893 | 0.04186 | 0.61 |
| 2640 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 30 | GRIN2A | Sponge network | -1.575 | 6.0E-5 | -2.161 | 0 | 0.61 |
| 2641 | RP11-798M19.6 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | -0.612 | 0.00094 | -1.733 | 0.00022 | 0.61 |
| 2642 | RP11-400K9.4 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | SFRP1 | Sponge network | -0.611 | 0.09607 | -3.566 | 0 | 0.61 |
| 2643 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-429 | 11 | ZNF521 | Sponge network | 0.335 | 0.20596 | -0.111 | 0.58068 | 0.61 |
| 2644 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 30 | KCNA6 | Sponge network | -2.452 | 0 | -1.531 | 0.00013 | 0.61 |
| 2645 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 40 | HLF | Sponge network | -0.976 | 0.00025 | -2.443 | 0 | 0.61 |
| 2646 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-421 | 23 | PRKAA2 | Sponge network | -4.463 | 0.00025 | -2.462 | 0 | 0.61 |
| 2647 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940;hsa-miR-98-5p | 43 | PBX1 | Sponge network | -0.49 | 0.04946 | -1.206 | 0 | 0.61 |
| 2648 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | 0.572 | 0.01722 | -0.509 | 0.03957 | 0.61 |
| 2649 | RP11-867G23.10 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-942-5p | 23 | IGFBP5 | Sponge network | -2.531 | 0 | -0.806 | 0.00105 | 0.61 |
| 2650 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ZNF536 | Sponge network | -2.452 | 0 | -3.115 | 0 | 0.61 |
| 2651 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-374a-5p;hsa-miR-93-3p | 34 | RUNX1T1 | Sponge network | -0.693 | 0.05629 | -0.344 | 0.2374 | 0.61 |
| 2652 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | DYNC1I1 | Sponge network | -0.793 | 7.0E-5 | -1.12 | 0.00107 | 0.61 |
| 2653 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p | 13 | AKAP12 | Sponge network | 0.249 | 0.35902 | -0.932 | 0.0028 | 0.61 |
| 2654 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 58 | AR | Sponge network | 0.997 | 0.00066 | -1.554 | 0 | 0.61 |
| 2655 | CTC-296K1.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-582-5p | 33 | DTNA | Sponge network | -2.095 | 0.00112 | -1.648 | 1.0E-5 | 0.61 |
| 2656 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 63 | BMPR2 | Sponge network | 0.532 | 0.00505 | 0.206 | 0.0635 | 0.61 |
| 2657 | SNHG14 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 54 | TGFBR3 | Sponge network | -1.111 | 0.0003 | -1.173 | 1.0E-5 | 0.61 |
| 2658 | BZRAP1-AS1 |
hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-5p | 14 | ITPKB | Sponge network | -0.465 | 0.04703 | -0.704 | 0.00106 | 0.61 |
| 2659 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | PARK2 | Sponge network | -0.487 | 0.02157 | -1.892 | 0 | 0.61 |
| 2660 | SNHG14 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-532-5p;hsa-miR-582-5p | 13 | PRICKLE1 | Sponge network | -1.111 | 0.0003 | -0.112 | 0.67088 | 0.61 |
| 2661 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 38 | PBX1 | Sponge network | -0.323 | 0.01372 | -1.206 | 0 | 0.609 |
| 2662 | MAGI2-AS3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 33 | PRKCB | Sponge network | -0.129 | 0.57177 | -1.313 | 2.0E-5 | 0.609 |
| 2663 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | -2.117 | 0.00161 | -0.466 | 0.14121 | 0.609 |
| 2664 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-429;hsa-miR-502-5p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 17 | SETBP1 | Sponge network | -1.654 | 0.00144 | -1.302 | 1.0E-5 | 0.609 |
| 2665 | LINC00865 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | ZDBF2 | Sponge network | -1.249 | 7.0E-5 | -0.998 | 0.00025 | 0.609 |
| 2666 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | RECK | Sponge network | -2.117 | 0.00161 | -1.043 | 0 | 0.609 |
| 2667 | SNHG14 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-2355-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | NUDT11 | Sponge network | -1.111 | 0.0003 | -0.921 | 0.00376 | 0.609 |
| 2668 | HAND2-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 18 | RHOB | Sponge network | -1.697 | 0.00889 | -1.052 | 0 | 0.609 |
| 2669 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 19 | RCAN2 | Sponge network | -0.222 | 0.32445 | -0.33 | 0.15172 | 0.609 |
| 2670 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 25 | SETBP1 | Sponge network | 0.335 | 0.20596 | -1.302 | 1.0E-5 | 0.609 |
| 2671 | LINC00578 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 18 | PRRX1 | Sponge network | 0.811 | 0.03228 | 1.316 | 0 | 0.609 |
| 2672 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 29 | EBF3 | Sponge network | -2.452 | 0 | -0.058 | 0.81437 | 0.609 |
| 2673 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 17 | IGF1 | Sponge network | -0.583 | 0.14589 | -0.204 | 0.5898 | 0.609 |
| 2674 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 23 | PRUNE2 | Sponge network | 0.997 | 0.00066 | -1.733 | 0.00022 | 0.609 |
| 2675 | RP11-175O19.4 |
hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-363-3p;hsa-miR-497-5p | 10 | E2F3 | Sponge network | 0.917 | 0 | 1.289 | 0 | 0.609 |
| 2676 | RASSF8-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 24 | NAV3 | Sponge network | 0.335 | 0.20596 | 0.212 | 0.47729 | 0.609 |
| 2677 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 25 | BNC2 | Sponge network | -4.546 | 0 | -0.491 | 0.09988 | 0.609 |
| 2678 | TP73-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-935;hsa-miR-940 | 16 | MAPK10 | Sponge network | -0.563 | 0.02545 | -1.774 | 0 | 0.609 |
| 2679 | MRVI1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-92a-1-5p | 24 | SNX29 | Sponge network | -1.092 | 4.0E-5 | -0.34 | 0.01371 | 0.609 |
| 2680 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-5p | 11 | LRRK2 | Sponge network | -1.439 | 4.0E-5 | -1.136 | 1.0E-5 | 0.609 |
| 2681 | FAM66C |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p | 11 | AMPH | Sponge network | -0.901 | 0.00019 | -0.916 | 5.0E-5 | 0.609 |
| 2682 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | LRRK2 | Sponge network | -0.358 | 0.17255 | -1.136 | 1.0E-5 | 0.609 |
| 2683 | LINC00092 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | RSPO2 | Sponge network | -1.886 | 0 | -3.038 | 0 | 0.609 |
| 2684 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 51 | ZFHX3 | Sponge network | -0.739 | 0.0574 | 0.389 | 0.01363 | 0.609 |
| 2685 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-744-3p | 23 | ADAMTSL3 | Sponge network | -0.777 | 0.16667 | -1.559 | 0 | 0.609 |
| 2686 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | SYNE1 | Sponge network | -0.323 | 0.01372 | -0.738 | 0.00316 | 0.609 |
| 2687 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 12 | ERG | Sponge network | -0.583 | 0.14589 | 0.002 | 0.99011 | 0.609 |
| 2688 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 56 | IL6ST | Sponge network | -0.611 | 0.09607 | -0.402 | 0.00676 | 0.609 |
| 2689 | FENDRR |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p | 18 | PDZD2 | Sponge network | -1.281 | 0.00012 | -0.909 | 3.0E-5 | 0.609 |
| 2690 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | LRRK2 | Sponge network | -0.737 | 0.06182 | -1.136 | 1.0E-5 | 0.609 |
| 2691 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-98-5p | 20 | RSPO2 | Sponge network | -1.092 | 4.0E-5 | -3.038 | 0 | 0.609 |
| 2692 | RP11-166D19.1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 18 | CXCL12 | Sponge network | -0.576 | 0.10121 | -1.421 | 0 | 0.609 |
| 2693 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p | 10 | SYNM | Sponge network | 0.299 | 0.57722 | -2.309 | 1.0E-5 | 0.609 |
| 2694 | RP11-305E6.4 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | KIF5B | Sponge network | 1.317 | 0 | 0.362 | 0.00066 | 0.609 |
| 2695 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-98-5p | 13 | SCN11A | Sponge network | -0.976 | 0.00025 | -1.15 | 0.00299 | 0.609 |
| 2696 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-93-5p | 25 | SYNE1 | Sponge network | -2.69 | 0.0001 | -0.738 | 0.00316 | 0.609 |
| 2697 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | ZNF521 | Sponge network | 0.997 | 0.00066 | -0.111 | 0.58068 | 0.609 |
| 2698 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p | 13 | PREX2 | Sponge network | -0.611 | 0.09607 | -0.272 | 0.25382 | 0.609 |
| 2699 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-590-3p | 13 | JAKMIP2 | Sponge network | -0.739 | 0.00044 | -1.388 | 0 | 0.608 |
| 2700 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-424-5p | 17 | LRIG1 | Sponge network | -1.331 | 3.0E-5 | -0.537 | 0.00588 | 0.608 |
| 2701 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 15 | LRRK2 | Sponge network | -1.575 | 6.0E-5 | -1.136 | 1.0E-5 | 0.608 |
| 2702 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CDH11 | Sponge network | 0.997 | 0.00066 | 1.427 | 0 | 0.608 |
| 2703 | LINC00641 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ZMYND11 | Sponge network | -0.222 | 0.32445 | -0.2 | 0.05449 | 0.608 |
| 2704 | RP11-867G23.10 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 29 | GRIK3 | Sponge network | -2.531 | 0 | -3.208 | 0 | 0.608 |
| 2705 | MYLK-AS1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-935 | 12 | RELN | Sponge network | -1.331 | 3.0E-5 | -2.403 | 0 | 0.608 |
| 2706 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 56 | TCF4 | Sponge network | -1.439 | 4.0E-5 | 0.016 | 0.92271 | 0.608 |
| 2707 | MIR143HG |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-877-5p | 34 | ASTN1 | Sponge network | -0.739 | 0.0574 | -2.389 | 2.0E-5 | 0.608 |
| 2708 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 56 | FAT3 | Sponge network | -0.901 | 0.00019 | -1.601 | 0.00051 | 0.608 |
| 2709 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p | 18 | RASSF2 | Sponge network | -0.164 | 0.45106 | -0.273 | 0.21961 | 0.608 |
| 2710 | RP5-1198O20.4 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p | 10 | CNN1 | Sponge network | 0.997 | 0.00066 | -2.633 | 0 | 0.608 |
| 2711 | ADAMTS9-AS2 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | NOV | Sponge network | -2.452 | 0 | -0.58 | 0.04676 | 0.608 |
| 2712 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p | 10 | AKAP12 | Sponge network | -0.693 | 0.05629 | -0.932 | 0.0028 | 0.608 |
| 2713 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-589-5p;hsa-miR-7-1-3p | 34 | PRKAA2 | Sponge network | -4.546 | 0 | -2.462 | 0 | 0.608 |
| 2714 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | PLSCR4 | Sponge network | 0.249 | 0.35902 | -0.474 | 0.03833 | 0.608 |
| 2715 | RP11-774O3.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 20 | SYNM | Sponge network | -0.487 | 0.02157 | -2.309 | 1.0E-5 | 0.608 |
| 2716 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 13 | BCLAF1 | Sponge network | 1.147 | 0 | 0.211 | 0.00684 | 0.608 |
| 2717 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ABCC9 | Sponge network | -0.323 | 0.01372 | -0.466 | 0.14121 | 0.608 |
| 2718 | RP11-819C21.1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-590-3p | 11 | TPR | Sponge network | 0.537 | 0.00139 | 0.582 | 0 | 0.608 |
| 2719 | ARHGEF26-AS1 |
hsa-miR-182-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-769-5p | 10 | TCEAL7 | Sponge network | -2.46 | 0 | -1.445 | 0 | 0.608 |
| 2720 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 18 | TIMP3 | Sponge network | -1.092 | 4.0E-5 | -0.546 | 0.02307 | 0.608 |
| 2721 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-370-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-935 | 20 | PCDH10 | Sponge network | -0.896 | 0.00099 | -1.644 | 0.0078 | 0.608 |
| 2722 | MIR143HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | ZNF208 | Sponge network | -0.739 | 0.0574 | -0.4 | 0.22381 | 0.608 |
| 2723 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p | 48 | FAT3 | Sponge network | -0.612 | 0.00094 | -1.601 | 0.00051 | 0.608 |
| 2724 | RP5-1198O20.4 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | MACF1 | Sponge network | 0.997 | 0.00066 | 0.019 | 0.90335 | 0.608 |
| 2725 | RP11-57H14.4 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 13 | MACF1 | Sponge network | 0.083 | 0.66405 | 0.019 | 0.90335 | 0.608 |
| 2726 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | ZNF208 | Sponge network | -0.129 | 0.57177 | -0.4 | 0.22381 | 0.608 |
| 2727 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 33 | GRIN2A | Sponge network | -0.737 | 0.06182 | -2.161 | 0 | 0.608 |
| 2728 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 52 | RBMS3 | Sponge network | -2.46 | 0 | -0.519 | 0.03551 | 0.608 |
| 2729 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p | 11 | FGFR1 | Sponge network | -2.62 | 0 | -0.509 | 0.03957 | 0.608 |
| 2730 | SNHG14 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 17 | ZBTB20 | Sponge network | -1.111 | 0.0003 | -0.122 | 0.57569 | 0.607 |
| 2731 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3913-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 29 | ZBTB4 | Sponge network | -0.769 | 0.00035 | -0.739 | 0 | 0.607 |
| 2732 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-628-5p;hsa-miR-935 | 32 | FBXO32 | Sponge network | -2.62 | 0 | -0.495 | 0.07561 | 0.607 |
| 2733 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-671-5p | 33 | ADARB1 | Sponge network | -0.154 | 0.53954 | -0.335 | 0.06631 | 0.607 |
| 2734 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 32 | SSBP2 | Sponge network | -4.546 | 0 | -1.58 | 0 | 0.607 |
| 2735 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 59 | IL6ST | Sponge network | -1.439 | 4.0E-5 | -0.402 | 0.00676 | 0.607 |
| 2736 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-508-3p;hsa-miR-582-3p;hsa-miR-671-5p | 28 | RBMS3 | Sponge network | -0.154 | 0.53954 | -0.519 | 0.03551 | 0.607 |
| 2737 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-98-5p | 15 | SCN11A | Sponge network | -1.697 | 0.00889 | -1.15 | 0.00299 | 0.607 |
| 2738 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 54 | EPHA7 | Sponge network | -2.46 | 0 | -2.197 | 3.0E-5 | 0.607 |
| 2739 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-340-5p | 16 | SCN9A | Sponge network | -1.331 | 3.0E-5 | -0.525 | 0.10593 | 0.607 |
| 2740 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 50 | DTNA | Sponge network | 0.335 | 0.20596 | -1.648 | 1.0E-5 | 0.607 |
| 2741 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 20 | IGSF10 | Sponge network | -1.249 | 7.0E-5 | -1.751 | 1.0E-5 | 0.607 |
| 2742 | JAZF1-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | MRVI1 | Sponge network | -0.769 | 0.00035 | -1.051 | 0.00022 | 0.607 |
| 2743 | EMX2OS |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLNA | Sponge network | -0.777 | 0.1134 | -0.771 | 0.01377 | 0.607 |
| 2744 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-93-3p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | -0.896 | 0.00099 | 0.358 | 0.13727 | 0.607 |
| 2745 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p | 15 | JAZF1 | Sponge network | -0.932 | 0.00329 | -0.377 | 0.02677 | 0.607 |
| 2746 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p | 43 | ZFHX4 | Sponge network | -0.162 | 0.47687 | 0.018 | 0.95574 | 0.607 |
| 2747 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-505-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 31 | CADM2 | Sponge network | -1.582 | 8.0E-5 | -3.782 | 0 | 0.607 |
| 2748 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | SYNE1 | Sponge network | -0.842 | 0.01361 | -0.738 | 0.00316 | 0.607 |
| 2749 | PWAR6 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PTPN13 | Sponge network | -1.575 | 6.0E-5 | -1.208 | 0 | 0.607 |
| 2750 | MEG3 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 13 | CDH11 | Sponge network | 0.249 | 0.35902 | 1.427 | 0 | 0.607 |
| 2751 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 16 | RECK | Sponge network | -0.611 | 0.09607 | -1.043 | 0 | 0.607 |
| 2752 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-582-5p | 38 | DTNA | Sponge network | -0.469 | 0.08721 | -1.648 | 1.0E-5 | 0.607 |
| 2753 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 41 | PBX1 | Sponge network | -0.576 | 0.10121 | -1.206 | 0 | 0.607 |
| 2754 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-629-3p | 23 | ZBTB4 | Sponge network | -2.62 | 0 | -0.739 | 0 | 0.607 |
| 2755 | HAND2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p | 34 | ASTN1 | Sponge network | -1.697 | 0.00889 | -2.389 | 2.0E-5 | 0.607 |
| 2756 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 17 | AKAP6 | Sponge network | -0.611 | 0.09607 | -1.586 | 3.0E-5 | 0.606 |
| 2757 | MYLK-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-484 | 14 | CTIF | Sponge network | -1.331 | 3.0E-5 | -0.389 | 0.00549 | 0.606 |
| 2758 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p | 14 | FGFR1 | Sponge network | -0.517 | 0.02935 | -0.509 | 0.03957 | 0.606 |
| 2759 | RP11-736K20.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 17 | NRK | Sponge network | -0.358 | 0.17255 | 0.656 | 0.19217 | 0.606 |
| 2760 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p | 14 | ATRX | Sponge network | 1.106 | 0 | 0.261 | 0.04408 | 0.606 |
| 2761 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-93-3p | 34 | BNC2 | Sponge network | -0.693 | 0.05629 | -0.491 | 0.09988 | 0.606 |
| 2762 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-23a-3p | 13 | NRXN1 | Sponge network | -2.095 | 0.00112 | -3.778 | 0 | 0.606 |
| 2763 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 14 | AKAP12 | Sponge network | -0.793 | 7.0E-5 | -0.932 | 0.0028 | 0.606 |
| 2764 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | TMEFF2 | Sponge network | -0.49 | 0.04946 | -2.426 | 0 | 0.606 |
| 2765 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p | 21 | PLSCR4 | Sponge network | -0.739 | 0.00044 | -0.474 | 0.03833 | 0.606 |
| 2766 | ARHGEF26-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-96-5p | 32 | ASTN1 | Sponge network | -2.46 | 0 | -2.389 | 2.0E-5 | 0.606 |
| 2767 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 11 | NRXN3 | Sponge network | -0.358 | 0.17255 | -0.893 | 0.04186 | 0.606 |
| 2768 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-502-3p | 23 | JAZF1 | Sponge network | -1.092 | 4.0E-5 | -0.377 | 0.02677 | 0.606 |
| 2769 | BVES-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-935 | 67 | LPP | Sponge network | -2.62 | 0 | -0.459 | 0.04475 | 0.606 |
| 2770 | PCA3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | NRXN3 | Sponge network | -0.709 | 0.18168 | -0.893 | 0.04186 | 0.606 |
| 2771 | RP11-464F9.1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | ZNF638 | Sponge network | 0.959 | 0 | 0.22 | 0.01214 | 0.606 |
| 2772 | AC010226.4 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-409-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-5p | 13 | PRKCB | Sponge network | -0.811 | 2.0E-5 | -1.313 | 2.0E-5 | 0.606 |
| 2773 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | DKK3 | Sponge network | -0.739 | 0.0574 | -0.052 | 0.77783 | 0.606 |
| 2774 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-628-5p;hsa-miR-744-3p | 35 | MEF2C | Sponge network | -2.62 | 0 | -0.499 | 0.01448 | 0.606 |
| 2775 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-7-1-3p | 11 | DKK3 | Sponge network | 0.889 | 9.0E-5 | -0.052 | 0.77783 | 0.606 |
| 2776 | TPTEP1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-542-3p | 10 | PRKD1 | Sponge network | -1.216 | 0 | -0.466 | 0.03583 | 0.606 |
| 2777 | AC159540.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 25 | MDM4 | Sponge network | 1.122 | 0 | 0.345 | 0.00762 | 0.606 |
| 2778 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-590-3p | 10 | NR2C2 | Sponge network | 0.768 | 7.0E-5 | 0.21 | 0.03224 | 0.606 |
| 2779 | MALAT1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 12 | FBXO11 | Sponge network | 0.782 | 0.00016 | 0.142 | 0.04938 | 0.606 |
| 2780 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 21 | AFF3 | Sponge network | -0.896 | 0.00099 | -2.373 | 0 | 0.606 |
| 2781 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | NCAM2 | Sponge network | -0.976 | 0.00025 | -0.482 | 0.22565 | 0.606 |
| 2782 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p;hsa-miR-98-5p | 10 | THBS1 | Sponge network | 1.344 | 0 | 0.741 | 0.00386 | 0.606 |
| 2783 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | -2.452 | 0 | -1.047 | 0.00364 | 0.606 |
| 2784 | USP3-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 13 | PHOX2B | Sponge network | -0.162 | 0.47687 | -2.68 | 0 | 0.606 |
| 2785 | FENDRR |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -1.281 | 0.00012 | -2.046 | 0.00019 | 0.606 |
| 2786 | BVES-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-502-5p | 12 | FAM172A | Sponge network | -2.62 | 0 | -0.57 | 0 | 0.606 |
| 2787 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 13 | HMCN1 | Sponge network | -1.697 | 0.00889 | 0.809 | 0.00544 | 0.606 |
| 2788 | BZRAP1-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-577 | 15 | FMNL3 | Sponge network | -0.465 | 0.04703 | 0.555 | 4.0E-5 | 0.606 |
| 2789 | CTD-3064M3.3 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-539-5p | 12 | PGR | Sponge network | -0.469 | 0.08721 | -1.704 | 0 | 0.606 |
| 2790 | SERTAD4-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-502-5p | 10 | SETBP1 | Sponge network | -0.932 | 0.00329 | -1.302 | 1.0E-5 | 0.605 |
| 2791 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577 | 47 | RUNX1T1 | Sponge network | -1.349 | 0.00017 | -0.344 | 0.2374 | 0.605 |
| 2792 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 33 | CBL | Sponge network | 0.083 | 0.66405 | 0.291 | 0.01267 | 0.605 |
| 2793 | PWAR6 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p | 18 | RELN | Sponge network | -1.575 | 6.0E-5 | -2.403 | 0 | 0.605 |
| 2794 | MIR143HG |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | ERG | Sponge network | -0.739 | 0.0574 | 0.002 | 0.99011 | 0.605 |
| 2795 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 25 | MEF2C | Sponge network | -1.886 | 0 | -0.499 | 0.01448 | 0.605 |
| 2796 | RP11-798M19.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | -0.612 | 0.00094 | -0.57 | 0 | 0.605 |
| 2797 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | 0.311 | 0.10344 | 0.273 | 0.09957 | 0.605 |
| 2798 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PLSCR4 | Sponge network | 0.194 | 0.64278 | -0.474 | 0.03833 | 0.605 |
| 2799 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-7-5p | 18 | FNBP1 | Sponge network | -0.162 | 0.47687 | -0.969 | 4.0E-5 | 0.605 |
| 2800 | NR2F1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 46 | IL17RD | Sponge network | -0.49 | 0.04946 | -0.019 | 0.94873 | 0.605 |
| 2801 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429 | 12 | DNAJB4 | Sponge network | -2.62 | 0 | -0.7 | 1.0E-5 | 0.605 |
| 2802 | SNHG14 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | MACF1 | Sponge network | -1.111 | 0.0003 | 0.019 | 0.90335 | 0.605 |
| 2803 | FAM66C |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 18 | GLI3 | Sponge network | -0.901 | 0.00019 | -0.615 | 0.01482 | 0.605 |
| 2804 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | TMEFF2 | Sponge network | -2.117 | 0.00161 | -2.426 | 0 | 0.605 |
| 2805 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-590-3p | 35 | SCN9A | Sponge network | -1.697 | 0.00889 | -0.525 | 0.10593 | 0.605 |
| 2806 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 38 | CLASP2 | Sponge network | -0.222 | 0.32445 | -0.038 | 0.71 | 0.605 |
| 2807 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 16 | RCAN2 | Sponge network | -0.976 | 0.00025 | -0.33 | 0.15172 | 0.605 |
| 2808 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-625-5p;hsa-miR-93-5p | 10 | LHX6 | Sponge network | 0.342 | 0.03129 | -0.087 | 0.69193 | 0.605 |
| 2809 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-338-5p | 15 | EBF3 | Sponge network | -1.092 | 4.0E-5 | -0.058 | 0.81437 | 0.605 |
| 2810 | RP11-67L2.2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-369-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | MACF1 | Sponge network | 0.72 | 0.0001 | 0.019 | 0.90335 | 0.605 |
| 2811 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | RCAN2 | Sponge network | -2.452 | 0 | -0.33 | 0.15172 | 0.605 |
| 2812 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 33 | QKI | Sponge network | -1.582 | 8.0E-5 | -0.333 | 0.04111 | 0.605 |
| 2813 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 65 | QKI | Sponge network | -1.697 | 0.00889 | -0.333 | 0.04111 | 0.605 |
| 2814 | SNHG14 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 17 | PHOX2B | Sponge network | -1.111 | 0.0003 | -2.68 | 0 | 0.605 |
| 2815 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | FGF5 | Sponge network | -0.737 | 0.06182 | 0.549 | 0.28659 | 0.605 |
| 2816 | RP11-244O19.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | -0.096 | 0.67958 | -0.382 | 0.05038 | 0.604 |
| 2817 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 35 | TSHZ3 | Sponge network | -0.901 | 0.00019 | -0.556 | 0.01516 | 0.604 |
| 2818 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | RECK | Sponge network | -0.793 | 7.0E-5 | -1.043 | 0 | 0.604 |
| 2819 | CTC-429P9.1 |
hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p | 13 | MYO9A | Sponge network | 0.497 | 0.01451 | -0.147 | 0.32373 | 0.604 |
| 2820 | RP11-166D19.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 43 | IL17RD | Sponge network | -0.576 | 0.10121 | -0.019 | 0.94873 | 0.604 |
| 2821 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 16 | FAM172A | Sponge network | -2.46 | 0 | -0.57 | 0 | 0.604 |
| 2822 | PWAR6 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 33 | PCDH10 | Sponge network | -1.575 | 6.0E-5 | -1.644 | 0.0078 | 0.604 |
| 2823 | LINC00843 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p | 11 | TRIM33 | Sponge network | 1.106 | 0 | 0.402 | 7.0E-5 | 0.604 |
| 2824 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 30 | PGR | Sponge network | 0.997 | 0.00066 | -1.704 | 0 | 0.604 |
| 2825 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-7-1-3p | 14 | ERG | Sponge network | -1.092 | 4.0E-5 | 0.002 | 0.99011 | 0.604 |
| 2826 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | ST6GAL2 | Sponge network | -0.976 | 0.00025 | -1.201 | 0.00597 | 0.604 |
| 2827 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-502-5p;hsa-miR-590-3p | 19 | SETBP1 | Sponge network | -0.739 | 0.00044 | -1.302 | 1.0E-5 | 0.604 |
| 2828 | DNM3OS |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 11 | MACF1 | Sponge network | 0.572 | 0.01722 | 0.019 | 0.90335 | 0.604 |
| 2829 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-452-5p | 10 | TMEFF2 | Sponge network | -2.62 | 0 | -2.426 | 0 | 0.604 |
| 2830 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-874-3p | 10 | NR2C2 | Sponge network | 0.93 | 0 | 0.21 | 0.03224 | 0.604 |
| 2831 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 26 | IL6ST | Sponge network | -0.376 | 0.00337 | -0.402 | 0.00676 | 0.604 |
| 2832 | RP11-244O19.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 16 | FAM172A | Sponge network | -0.096 | 0.67958 | -0.57 | 0 | 0.604 |
| 2833 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 18 | ITGA5 | Sponge network | -0.576 | 0.10121 | 0.452 | 0.03524 | 0.604 |
| 2834 | H19 |
hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-23a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-484 | 10 | IGF2 | Sponge network | 2.439 | 0 | 0.848 | 0.03842 | 0.604 |
| 2835 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 35 | EBF1 | Sponge network | -0.49 | 0.04946 | -0.384 | 0.08856 | 0.604 |
| 2836 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 32 | PCDH10 | Sponge network | -2.46 | 0 | -1.644 | 0.0078 | 0.604 |
| 2837 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-532-5p;hsa-miR-93-3p;hsa-miR-940 | 11 | POU2F2 | Sponge network | -0.465 | 0.04703 | -0.233 | 0.32214 | 0.604 |
| 2838 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-589-5p | 37 | ZFHX3 | Sponge network | -0.162 | 0.47687 | 0.389 | 0.01363 | 0.604 |
| 2839 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 12 | GAS1 | Sponge network | -0.777 | 0.1134 | -1.047 | 0.00364 | 0.604 |
| 2840 | LINC00702 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 15 | RHOB | Sponge network | -0.737 | 0.06182 | -1.052 | 0 | 0.604 |
| 2841 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-7-1-3p | 17 | MYO9A | Sponge network | -1.092 | 4.0E-5 | -0.147 | 0.32373 | 0.604 |
| 2842 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 18 | SPG20 | Sponge network | -0.769 | 0.00035 | -1.16 | 0 | 0.603 |
| 2843 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 52 | TRPS1 | Sponge network | -0.576 | 0.10121 | -0.191 | 0.44109 | 0.603 |
| 2844 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 30 | EBF3 | Sponge network | -0.737 | 0.06182 | -0.058 | 0.81437 | 0.603 |
| 2845 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-874-3p | 27 | TACC1 | Sponge network | -0.583 | 0.14589 | -0.362 | 0.05406 | 0.603 |
| 2846 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | IGSF10 | Sponge network | -0.49 | 0.04946 | -1.751 | 1.0E-5 | 0.603 |
| 2847 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-590-3p | 16 | STARD13 | Sponge network | -0.096 | 0.67958 | -0.187 | 0.27383 | 0.603 |
| 2848 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-744-3p | 14 | ADAMTSL3 | Sponge network | -2.095 | 0.00112 | -1.559 | 0 | 0.603 |
| 2849 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-576-5p | 17 | TSHZ3 | Sponge network | -0.932 | 0.00329 | -0.556 | 0.01516 | 0.603 |
| 2850 | FENDRR |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 58 | PRKAA2 | Sponge network | -1.281 | 0.00012 | -2.462 | 0 | 0.603 |
| 2851 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 17 | EDA2R | Sponge network | -0.576 | 0.10121 | -0.587 | 0.01598 | 0.603 |
| 2852 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-223-3p;hsa-miR-455-5p;hsa-miR-577 | 14 | EPHA3 | Sponge network | -0.473 | 0.07602 | 0.185 | 0.53588 | 0.603 |
| 2853 | SERTAD4-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p | 11 | FAT4 | Sponge network | -0.932 | 0.00329 | -0.354 | 0.14694 | 0.603 |
| 2854 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 41 | TSHZ3 | Sponge network | -1.281 | 0.00012 | -0.556 | 0.01516 | 0.603 |
| 2855 | MEG3 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 16 | CNTNAP1 | Sponge network | 0.249 | 0.35902 | 0.358 | 0.13727 | 0.603 |
| 2856 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | SYNE1 | Sponge network | -0.811 | 2.0E-5 | -0.738 | 0.00316 | 0.603 |
| 2857 | FENDRR |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 61 | CADM2 | Sponge network | -1.281 | 0.00012 | -3.782 | 0 | 0.603 |
| 2858 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | FGF5 | Sponge network | -0.739 | 0.0574 | 0.549 | 0.28659 | 0.603 |
| 2859 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-590-3p | 10 | DCN | Sponge network | -0.739 | 0.00044 | -1.288 | 0 | 0.603 |
| 2860 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 13 | SLITRK3 | Sponge network | -1.582 | 8.0E-5 | -3.328 | 0 | 0.603 |
| 2861 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 40 | RBMS3 | Sponge network | -1.349 | 0.00017 | -0.519 | 0.03551 | 0.603 |
| 2862 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-335-3p;hsa-miR-424-5p | 10 | FGFR1 | Sponge network | -0.932 | 0.00329 | -0.509 | 0.03957 | 0.603 |
| 2863 | ACTA2-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-542-3p | 10 | PRKD1 | Sponge network | 0.194 | 0.64278 | -0.466 | 0.03583 | 0.603 |
| 2864 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | NRXN3 | Sponge network | 0.997 | 0.00066 | -0.893 | 0.04186 | 0.603 |
| 2865 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p | 30 | ESR1 | Sponge network | -1.349 | 0.00017 | -1.035 | 3.0E-5 | 0.603 |
| 2866 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 19 | CRTC3 | Sponge network | -0.162 | 0.47687 | 0.164 | 0.16786 | 0.603 |
| 2867 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 10 | ZNF382 | Sponge network | 0.335 | 0.20596 | -0.494 | 0.03831 | 0.603 |
| 2868 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 48 | ADARB1 | Sponge network | -0.323 | 0.01372 | -0.335 | 0.06631 | 0.603 |
| 2869 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | SLITRK3 | Sponge network | -0.129 | 0.57177 | -3.328 | 0 | 0.603 |
| 2870 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | ZFHX4 | Sponge network | -1.439 | 4.0E-5 | 0.018 | 0.95574 | 0.603 |
| 2871 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | ABCC9 | Sponge network | -2.46 | 0 | -0.466 | 0.14121 | 0.603 |
| 2872 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940 | 28 | CRTC3 | Sponge network | -0.129 | 0.57177 | 0.164 | 0.16786 | 0.603 |
| 2873 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-503-5p;hsa-miR-624-5p | 10 | SMARCA1 | Sponge network | -0.976 | 0.00025 | -0.684 | 0.00159 | 0.603 |
| 2874 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-625-5p | 19 | ANK2 | Sponge network | -2.095 | 0.00112 | -1.603 | 1.0E-5 | 0.603 |
| 2875 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 41 | RBMS3 | Sponge network | -0.709 | 0.18168 | -0.519 | 0.03551 | 0.603 |
| 2876 | RP5-1198O20.4 |
hsa-miR-144-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-502-5p | 10 | ELN | Sponge network | 0.997 | 0.00066 | 0.451 | 0.13752 | 0.603 |
| 2877 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 22 | FGFR1 | Sponge network | -1.697 | 0.00889 | -0.509 | 0.03957 | 0.603 |
| 2878 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 49 | GAS7 | Sponge network | 0.249 | 0.35902 | -0.555 | 0.01602 | 0.603 |
| 2879 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | MCC | Sponge network | -4.546 | 0 | -1.005 | 0 | 0.603 |
| 2880 | ZNF582-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 47 | GAS7 | Sponge network | -1.349 | 0.00017 | -0.555 | 0.01602 | 0.603 |
| 2881 | LINC00472 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421 | 17 | SPG20 | Sponge network | -0.842 | 0.01361 | -1.16 | 0 | 0.603 |
| 2882 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 49 | BNC2 | Sponge network | -0.901 | 0.00019 | -0.491 | 0.09988 | 0.602 |
| 2883 | RP11-307B6.3 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-378a-3p | 11 | FLNA | Sponge network | -4.463 | 0.00025 | -0.771 | 0.01377 | 0.602 |
| 2884 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | ANK2 | Sponge network | 0.997 | 0.00066 | -1.603 | 1.0E-5 | 0.602 |
| 2885 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | DCLK1 | Sponge network | -0.901 | 0.00019 | -1.324 | 0.00014 | 0.602 |
| 2886 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 31 | IGF1 | Sponge network | 0.335 | 0.20596 | -0.204 | 0.5898 | 0.602 |
| 2887 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | -0.323 | 0.01372 | -1.324 | 0.00014 | 0.602 |
| 2888 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 41 | RBMS3 | Sponge network | -0.842 | 0.01361 | -0.519 | 0.03551 | 0.602 |
| 2889 | SNHG14 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | PRDM5 | Sponge network | -1.111 | 0.0003 | -1.09 | 0.00014 | 0.602 |
| 2890 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 17 | EBF1 | Sponge network | -0.583 | 0.14589 | -0.384 | 0.08856 | 0.602 |
| 2891 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-331-3p;hsa-miR-629-3p;hsa-miR-93-5p | 16 | ZBTB4 | Sponge network | -2.531 | 0 | -0.739 | 0 | 0.602 |
| 2892 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-505-3p | 17 | JAKMIP2 | Sponge network | -0.162 | 0.47687 | -1.388 | 0 | 0.602 |
| 2893 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p | 32 | PBX1 | Sponge network | -2.69 | 0.0001 | -1.206 | 0 | 0.602 |
| 2894 | LINC00578 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 18 | WWTR1 | Sponge network | 0.811 | 0.03228 | -0.345 | 0.11997 | 0.602 |
| 2895 | RP11-156P1.3 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 18 | ABL1 | Sponge network | 0.214 | 0.18246 | -0.215 | 0.07804 | 0.602 |
| 2896 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 28 | PCDH10 | Sponge network | 0.997 | 0.00066 | -1.644 | 0.0078 | 0.602 |
| 2897 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | IGSF10 | Sponge network | -2.46 | 0 | -1.751 | 1.0E-5 | 0.602 |
| 2898 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | SETBP1 | Sponge network | -0.487 | 0.02157 | -1.302 | 1.0E-5 | 0.602 |
| 2899 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 26 | LRRC7 | Sponge network | -0.576 | 0.10121 | -1.341 | 0.01193 | 0.602 |
| 2900 | TP73-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-501-5p | 12 | MN1 | Sponge network | -0.563 | 0.02545 | -0.358 | 0.24172 | 0.602 |
| 2901 | CTD-3092A11.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 44 | MDM4 | Sponge network | 0.873 | 0 | 0.345 | 0.00762 | 0.602 |
| 2902 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-331-3p;hsa-miR-7-1-3p | 19 | QKI | Sponge network | 0.693 | 0.00076 | -0.333 | 0.04111 | 0.602 |
| 2903 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 33 | MITF | Sponge network | -0.576 | 0.10121 | -0.769 | 0.00022 | 0.602 |
| 2904 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | AKAP12 | Sponge network | 0.194 | 0.64278 | -0.932 | 0.0028 | 0.602 |
| 2905 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 48 | DIP2C | Sponge network | -0.737 | 0.06182 | -0.436 | 0.01713 | 0.602 |
| 2906 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | -0.769 | 0.00035 | -1.603 | 1.0E-5 | 0.602 |
| 2907 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 72 | TCF4 | Sponge network | -1.575 | 6.0E-5 | 0.016 | 0.92271 | 0.602 |
| 2908 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p | 12 | SYNM | Sponge network | -2.076 | 0.00548 | -2.309 | 1.0E-5 | 0.602 |
| 2909 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-942-5p | 10 | CREBBP | Sponge network | 0.542 | 0.00054 | 0.126 | 0.15186 | 0.602 |
| 2910 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-589-3p | 17 | TIMP3 | Sponge network | -0.154 | 0.53954 | -0.546 | 0.02307 | 0.602 |
| 2911 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p | 14 | DCLK1 | Sponge network | 0.335 | 0.20596 | -1.324 | 0.00014 | 0.602 |
| 2912 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 17 | TXNIP | Sponge network | -0.739 | 0.0574 | -1.277 | 0 | 0.602 |
| 2913 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | PEG3 | Sponge network | -0.563 | 0.02545 | -1.74 | 0 | 0.602 |
| 2914 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | CAV1 | Sponge network | -1.697 | 0.00889 | -1.013 | 6.0E-5 | 0.602 |
| 2915 | LINC00578 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-511-5p | 12 | PCDH10 | Sponge network | 0.811 | 0.03228 | -1.644 | 0.0078 | 0.602 |
| 2916 | MAGI2-AS3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 18 | STARD13 | Sponge network | -0.129 | 0.57177 | -0.187 | 0.27383 | 0.601 |
| 2917 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 39 | ABI2 | Sponge network | -0.323 | 0.01372 | 0.175 | 0.14787 | 0.601 |
| 2918 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | PCDH9 | Sponge network | 0.335 | 0.20596 | -1.823 | 4.0E-5 | 0.601 |
| 2919 | HOXB-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-429;hsa-miR-505-5p | 13 | MRVI1 | Sponge network | -0.154 | 0.53954 | -1.051 | 0.00022 | 0.601 |
| 2920 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | HOOK3 | Sponge network | -0.323 | 0.01372 | 0.273 | 0.09957 | 0.601 |
| 2921 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-98-5p | 42 | PBX1 | Sponge network | -2.46 | 0 | -1.206 | 0 | 0.601 |
| 2922 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | AFF3 | Sponge network | -0.129 | 0.57177 | -2.373 | 0 | 0.601 |
| 2923 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-7-1-3p | 10 | TMEFF2 | Sponge network | -1.092 | 4.0E-5 | -2.426 | 0 | 0.601 |
| 2924 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 16 | NAV3 | Sponge network | -0.583 | 0.14589 | 0.212 | 0.47729 | 0.601 |
| 2925 | ZNF667-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | -0.976 | 0.00025 | -0.564 | 1.0E-5 | 0.601 |
| 2926 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | RECK | Sponge network | 0.194 | 0.64278 | -1.043 | 0 | 0.601 |
| 2927 | RP5-1198O20.4 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484 | 11 | LZTS1 | Sponge network | 0.997 | 0.00066 | 1.664 | 0 | 0.601 |
| 2928 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 37 | NRXN1 | Sponge network | -1.281 | 0.00012 | -3.778 | 0 | 0.601 |
| 2929 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p | 19 | CNTNAP3 | Sponge network | -2.452 | 0 | -1.465 | 0.00033 | 0.601 |
| 2930 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | EBF1 | Sponge network | 0.4 | 0.14611 | -0.384 | 0.08856 | 0.601 |
| 2931 | RP11-736K20.5 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | SORCS1 | Sponge network | -0.358 | 0.17255 | -3.492 | 0 | 0.601 |
| 2932 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935;hsa-miR-96-5p | 42 | FBXO32 | Sponge network | -2.46 | 0 | -0.495 | 0.07561 | 0.601 |
| 2933 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-940;hsa-miR-96-5p | 55 | GAS7 | Sponge network | -2.452 | 0 | -0.555 | 0.01602 | 0.601 |
| 2934 | LINC00702 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 53 | GRIK3 | Sponge network | -0.737 | 0.06182 | -3.208 | 0 | 0.601 |
| 2935 | RP5-1198O20.4 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p | 10 | SORCS2 | Sponge network | 0.997 | 0.00066 | -0.223 | 0.4253 | 0.601 |
| 2936 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 56 | FAT3 | Sponge network | -0.323 | 0.01372 | -1.601 | 0.00051 | 0.601 |
| 2937 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 58 | SSBP2 | Sponge network | -0.737 | 0.06182 | -1.58 | 0 | 0.601 |
| 2938 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | PLSCR4 | Sponge network | -1.349 | 0.00017 | -0.474 | 0.03833 | 0.601 |
| 2939 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-589-5p | 11 | MN1 | Sponge network | -0.154 | 0.78939 | -0.358 | 0.24172 | 0.601 |
| 2940 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 12 | BCLAF1 | Sponge network | 1.153 | 0 | 0.211 | 0.00684 | 0.601 |
| 2941 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 30 | GPC6 | Sponge network | -0.49 | 0.04946 | -0.054 | 0.81465 | 0.601 |
| 2942 | RP11-400K9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 22 | NAV3 | Sponge network | -0.611 | 0.09607 | 0.212 | 0.47729 | 0.601 |
| 2943 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 46 | BNC2 | Sponge network | -0.323 | 0.01372 | -0.491 | 0.09988 | 0.601 |
| 2944 | PGM5-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p | 16 | ASTN1 | Sponge network | -4.546 | 0 | -2.389 | 2.0E-5 | 0.601 |
| 2945 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-654-3p;hsa-miR-874-3p | 23 | NR2C2 | Sponge network | 0.637 | 0.00069 | 0.21 | 0.03224 | 0.601 |
| 2946 | LINC00957 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 47 | DTNA | Sponge network | -0.421 | 0.0114 | -1.648 | 1.0E-5 | 0.601 |
| 2947 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-337-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | HOOK3 | Sponge network | 0.782 | 0.00016 | 0.273 | 0.09957 | 0.601 |
| 2948 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 19 | JAKMIP2 | Sponge network | -1.575 | 6.0E-5 | -1.388 | 0 | 0.6 |
| 2949 | PWAR6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ZNF208 | Sponge network | -1.575 | 6.0E-5 | -0.4 | 0.22381 | 0.6 |
| 2950 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935 | 32 | TSHZ3 | Sponge network | -0.896 | 0.00099 | -0.556 | 0.01516 | 0.6 |
| 2951 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-590-3p | 11 | NR2C2 | Sponge network | 1.148 | 2.0E-5 | 0.21 | 0.03224 | 0.6 |
| 2952 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-3p | 12 | SYNM | Sponge network | 0.811 | 0.03228 | -2.309 | 1.0E-5 | 0.6 |
| 2953 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 44 | AR | Sponge network | -0.611 | 0.09607 | -1.554 | 0 | 0.6 |
| 2954 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 30 | NRXN1 | Sponge network | -0.896 | 0.00099 | -3.778 | 0 | 0.6 |
| 2955 | PGM5-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 10 | TMEFF2 | Sponge network | -4.546 | 0 | -2.426 | 0 | 0.6 |
| 2956 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 44 | PHC3 | Sponge network | -0.222 | 0.32445 | 0.212 | 0.0499 | 0.6 |
| 2957 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | TIMP3 | Sponge network | -0.358 | 0.17255 | -0.546 | 0.02307 | 0.6 |
| 2958 | PWAR6 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | TLL1 | Sponge network | -1.575 | 6.0E-5 | -1.195 | 3.0E-5 | 0.6 |
| 2959 | DNM3OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p | 23 | SYNPO2 | Sponge network | 0.572 | 0.01722 | -2.342 | 0 | 0.6 |
| 2960 | LINC00641 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RBM6 | Sponge network | -0.222 | 0.32445 | -0.137 | 0.15663 | 0.6 |
| 2961 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | CDH19 | Sponge network | -0.896 | 0.00099 | -3.434 | 0 | 0.6 |
| 2962 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 43 | HLF | Sponge network | -1.575 | 6.0E-5 | -2.443 | 0 | 0.6 |
| 2963 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p | 14 | ITGB3 | Sponge network | 0.889 | 9.0E-5 | -0.011 | 0.95751 | 0.6 |
| 2964 | ADAMTS9-AS2 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ID4 | Sponge network | -2.452 | 0 | -1.644 | 0 | 0.6 |
| 2965 | RP11-774O3.3 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-7-5p | 13 | FLNA | Sponge network | -0.487 | 0.02157 | -0.771 | 0.01377 | 0.6 |
| 2966 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | GRIN2A | Sponge network | -0.901 | 0.00019 | -2.161 | 0 | 0.6 |
| 2967 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 11 | NALCN | Sponge network | 0.335 | 0.20596 | 1.068 | 0.00237 | 0.6 |
| 2968 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 42 | ESR1 | Sponge network | -1.111 | 0.0003 | -1.035 | 3.0E-5 | 0.6 |
| 2969 | AC003090.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 58 | CADM2 | Sponge network | -2.117 | 0.00161 | -3.782 | 0 | 0.6 |
| 2970 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 16 | GLI3 | Sponge network | -0.487 | 0.02157 | -0.615 | 0.01482 | 0.6 |
| 2971 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | AKAP12 | Sponge network | -0.739 | 0.00044 | -0.932 | 0.0028 | 0.6 |
| 2972 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 20 | SYNE1 | Sponge network | -0.465 | 0.04703 | -0.738 | 0.00316 | 0.6 |
| 2973 | USP3-AS1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-940 | 13 | SPOP | Sponge network | -0.162 | 0.47687 | -0.419 | 1.0E-5 | 0.6 |
| 2974 | EMX2OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 11 | LHFP | Sponge network | -0.777 | 0.1134 | -0.167 | 0.41371 | 0.6 |
| 2975 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 17 | DCLK1 | Sponge network | 0.997 | 0.00066 | -1.324 | 0.00014 | 0.6 |
| 2976 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | RCAN2 | Sponge network | -1.575 | 6.0E-5 | -0.33 | 0.15172 | 0.6 |
| 2977 | C20orf166-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577 | 12 | TLL1 | Sponge network | -2.69 | 0.0001 | -1.195 | 3.0E-5 | 0.6 |
| 2978 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 14 | CNTNAP1 | Sponge network | -1.575 | 6.0E-5 | 0.358 | 0.13727 | 0.6 |
| 2979 | RP11-774O3.3 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -0.487 | 0.02157 | -0.704 | 0.00106 | 0.6 |
| 2980 | PWAR6 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-96-5p | 15 | ZBTB20 | Sponge network | -1.575 | 6.0E-5 | -0.122 | 0.57569 | 0.6 |
| 2981 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p | 16 | CBFA2T3 | Sponge network | -0.72 | 0.24239 | -1.221 | 1.0E-5 | 0.6 |
| 2982 | NR2F1-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 15 | RHOB | Sponge network | -0.49 | 0.04946 | -1.052 | 0 | 0.6 |
| 2983 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-505-3p;hsa-miR-629-3p;hsa-miR-769-5p | 36 | NTRK3 | Sponge network | -0.583 | 0.14589 | -1.77 | 3.0E-5 | 0.6 |
| 2984 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p | 24 | CNTN1 | Sponge network | -0.739 | 0.00044 | -2.594 | 0 | 0.6 |
| 2985 | RP11-25K19.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | MAPK10 | Sponge network | -1.057 | 0.00029 | -1.774 | 0 | 0.6 |
| 2986 | SNHG14 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | TPO | Sponge network | -1.111 | 0.0003 | -1.87 | 2.0E-5 | 0.6 |
| 2987 | SNHG14 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-577;hsa-miR-96-5p;hsa-miR-98-5p | 12 | PLAGL1 | Sponge network | -1.111 | 0.0003 | -0.588 | 0.00912 | 0.6 |
| 2988 | MEG3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | NCAM2 | Sponge network | 0.249 | 0.35902 | -0.482 | 0.22565 | 0.6 |
| 2989 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 34 | IGF1 | Sponge network | -1.575 | 6.0E-5 | -0.204 | 0.5898 | 0.6 |
| 2990 | BVES-AS1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-375;hsa-miR-429 | 16 | ITPKB | Sponge network | -2.62 | 0 | -0.704 | 0.00106 | 0.6 |
| 2991 | RP11-166D19.1 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-542-3p | 10 | MLLT11 | Sponge network | -0.576 | 0.10121 | -0.661 | 0.01733 | 0.6 |
| 2992 | PGM5-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-940 | 27 | GRIK3 | Sponge network | -4.546 | 0 | -3.208 | 0 | 0.599 |
| 2993 | LINC00657 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 35 | ABL2 | Sponge network | 0.91 | 0 | 0.82 | 0 | 0.599 |
| 2994 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 28 | EPHA3 | Sponge network | -0.096 | 0.67958 | 0.185 | 0.53588 | 0.599 |
| 2995 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p | 19 | MCC | Sponge network | -2.531 | 0 | -1.005 | 0 | 0.599 |
| 2996 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 52 | SSBP2 | Sponge network | -0.901 | 0.00019 | -1.58 | 0 | 0.599 |
| 2997 | RP11-999E24.3 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p | 14 | RASSF8 | Sponge network | -0.693 | 0.05629 | -0.122 | 0.65591 | 0.599 |
| 2998 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 25 | SEMA3C | Sponge network | -0.49 | 0.04946 | -0.272 | 0.31153 | 0.599 |
| 2999 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 23 | ADAMTSL3 | Sponge network | -0.842 | 0.01361 | -1.559 | 0 | 0.599 |
| 3000 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CNTN1 | Sponge network | 0.194 | 0.64278 | -2.594 | 0 | 0.599 |
| 3001 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-93-5p | 26 | PRKAA2 | Sponge network | -2.531 | 0 | -2.462 | 0 | 0.599 |
| 3002 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 21 | LRRC4 | Sponge network | -2.452 | 0 | -1.774 | 0 | 0.599 |
| 3003 | LINC00472 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p | 13 | PRDM5 | Sponge network | -0.842 | 0.01361 | -1.09 | 0.00014 | 0.599 |
| 3004 | RP11-540B6.6 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p | 10 | TPR | Sponge network | 0.93 | 0 | 0.582 | 0 | 0.599 |
| 3005 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935;hsa-miR-96-5p | 45 | FBXO32 | Sponge network | -1.111 | 0.0003 | -0.495 | 0.07561 | 0.599 |
| 3006 | FGF14-AS2 |
hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | GLI3 | Sponge network | -1.582 | 8.0E-5 | -0.615 | 0.01482 | 0.599 |
| 3007 | ADAMTS9-AS2 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | STAT5B | Sponge network | -2.452 | 0 | -0.182 | 0.1082 | 0.599 |
| 3008 | RP11-774O3.3 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | HERC1 | Sponge network | -0.487 | 0.02157 | -0.196 | 0.08544 | 0.599 |
| 3009 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-744-3p | 18 | ADAMTSL3 | Sponge network | -0.739 | 0.00044 | -1.559 | 0 | 0.599 |
| 3010 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 34 | ADARB1 | Sponge network | -0.896 | 0.00099 | -0.335 | 0.06631 | 0.599 |
| 3011 | RP11-10N23.2 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p | 13 | ABL2 | Sponge network | 1.134 | 0 | 0.82 | 0 | 0.599 |
| 3012 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | TIMP3 | Sponge network | -1.439 | 4.0E-5 | -0.546 | 0.02307 | 0.599 |
| 3013 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-454-3p;hsa-miR-93-5p | 21 | MCC | Sponge network | -0.473 | 0.07602 | -1.005 | 0 | 0.599 |
| 3014 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-629-5p | 20 | GRIN2A | Sponge network | -2.69 | 0.0001 | -2.161 | 0 | 0.599 |
| 3015 | TP73-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-92a-3p | 10 | PTPN13 | Sponge network | -0.563 | 0.02545 | -1.208 | 0 | 0.599 |
| 3016 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-7-1-3p | 29 | ANK2 | Sponge network | -1.249 | 7.0E-5 | -1.603 | 1.0E-5 | 0.599 |
| 3017 | LINC00865 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429 | 16 | SPG20 | Sponge network | -1.249 | 7.0E-5 | -1.16 | 0 | 0.599 |
| 3018 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-877-5p | 21 | FAT3 | Sponge network | -0.309 | 0.1145 | -1.601 | 0.00051 | 0.599 |
| 3019 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-7-5p;hsa-miR-940 | 20 | SNX29 | Sponge network | -1.331 | 3.0E-5 | -0.34 | 0.01371 | 0.599 |
| 3020 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-93-5p | 49 | NTRK3 | Sponge network | -2.69 | 0.0001 | -1.77 | 3.0E-5 | 0.599 |
| 3021 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p | 18 | ZFHX3 | Sponge network | 0.178 | 0.29106 | 0.389 | 0.01363 | 0.599 |
| 3022 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | FAT3 | Sponge network | -2.117 | 0.00161 | -1.601 | 0.00051 | 0.599 |
| 3023 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 20 | RELN | Sponge network | -0.576 | 0.10121 | -2.403 | 0 | 0.599 |
| 3024 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | -2.117 | 0.00161 | 0.358 | 0.13727 | 0.599 |
| 3025 | ADAMTS9-AS2 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | THSD7B | Sponge network | -2.452 | 0 | 0.154 | 0.74723 | 0.599 |
| 3026 | RP11-244O19.1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | HERC1 | Sponge network | -0.096 | 0.67958 | -0.196 | 0.08544 | 0.599 |
| 3027 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-582-3p;hsa-miR-629-3p | 38 | AR | Sponge network | -0.154 | 0.53954 | -1.554 | 0 | 0.599 |
| 3028 | SERTAD4-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-576-5p | 26 | CADM2 | Sponge network | -0.932 | 0.00329 | -3.782 | 0 | 0.599 |
| 3029 | ADAMTS9-AS2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 67 | DST | Sponge network | -2.452 | 0 | -0.713 | 0.00096 | 0.599 |
| 3030 | MEG3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-5p | 20 | PTGFR | Sponge network | 0.249 | 0.35902 | -0.233 | 0.45421 | 0.598 |
| 3031 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-664a-5p;hsa-miR-940 | 21 | TRPS1 | Sponge network | -1.331 | 3.0E-5 | -0.191 | 0.44109 | 0.598 |
| 3032 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 40 | MCC | Sponge network | -1.281 | 0.00012 | -1.005 | 0 | 0.598 |
| 3033 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 15 | NALCN | Sponge network | -1.697 | 0.00889 | 1.068 | 0.00237 | 0.598 |
| 3034 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-7-5p;hsa-miR-93-3p | 24 | SNX29 | Sponge network | -0.611 | 0.09607 | -0.34 | 0.01371 | 0.598 |
| 3035 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 11 | PRDM5 | Sponge network | -0.563 | 0.02545 | -1.09 | 0.00014 | 0.598 |
| 3036 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | LSAMP | Sponge network | -0.739 | 0.0574 | -0.066 | 0.81708 | 0.598 |
| 3037 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | ATRX | Sponge network | 0.559 | 0.00026 | 0.261 | 0.04408 | 0.598 |
| 3038 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | NRXN1 | Sponge network | -1.582 | 8.0E-5 | -3.778 | 0 | 0.598 |
| 3039 | BVES-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-744-3p;hsa-miR-935 | 29 | DIP2C | Sponge network | -2.62 | 0 | -0.436 | 0.01713 | 0.598 |
| 3040 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 13 | RBMS3 | Sponge network | -2.039 | 0.03447 | -0.519 | 0.03551 | 0.598 |
| 3041 | ADAMTS9-AS2 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZDBF2 | Sponge network | -2.452 | 0 | -0.998 | 0.00025 | 0.598 |
| 3042 | PCA3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | -0.709 | 0.18168 | -1.051 | 0.00022 | 0.598 |
| 3043 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 36 | ESR1 | Sponge network | -0.576 | 0.10121 | -1.035 | 3.0E-5 | 0.598 |
| 3044 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-484;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 41 | SOX5 | Sponge network | -0.976 | 0.00025 | -1.091 | 4.0E-5 | 0.598 |
| 3045 | PCA3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p | 57 | RUNX1T1 | Sponge network | -0.709 | 0.18168 | -0.344 | 0.2374 | 0.598 |
| 3046 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | PGR | Sponge network | -0.769 | 0.00035 | -1.704 | 0 | 0.598 |
| 3047 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 60 | FAT3 | Sponge network | -0.777 | 0.1134 | -1.601 | 0.00051 | 0.598 |
| 3048 | RP11-166D19.1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 18 | KLF10 | Sponge network | -0.576 | 0.10121 | -0.783 | 0 | 0.598 |
| 3049 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 43 | TACC1 | Sponge network | 0.194 | 0.64278 | -0.362 | 0.05406 | 0.598 |
| 3050 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 32 | MYO9A | Sponge network | 0.72 | 0.0001 | -0.147 | 0.32373 | 0.598 |
| 3051 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | -0.842 | 0.01361 | -1.603 | 1.0E-5 | 0.598 |
| 3052 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 23 | PLSCR4 | Sponge network | -1.249 | 7.0E-5 | -0.474 | 0.03833 | 0.598 |
| 3053 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940 | 21 | CRTC3 | Sponge network | 0.214 | 0.18246 | 0.164 | 0.16786 | 0.598 |
| 3054 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | SOX5 | Sponge network | -0.563 | 0.02545 | -1.091 | 4.0E-5 | 0.598 |
| 3055 | CTD-3092A11.2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 11 | ZNF638 | Sponge network | 0.873 | 0 | 0.22 | 0.01214 | 0.598 |
| 3056 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p | 14 | AFF3 | Sponge network | -0.72 | 0.24239 | -2.373 | 0 | 0.598 |
| 3057 | EMX2OS |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | RADIL | Sponge network | -0.777 | 0.1134 | -1.628 | 0 | 0.598 |
| 3058 | DNM3OS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 84 | LPP | Sponge network | 0.572 | 0.01722 | -0.459 | 0.04475 | 0.598 |
| 3059 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3614-5p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-7-5p | 25 | RBMS3 | Sponge network | -0.469 | 0.08721 | -0.519 | 0.03551 | 0.598 |
| 3060 | MIR497HG |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 51 | CADM2 | Sponge network | -1.439 | 4.0E-5 | -3.782 | 0 | 0.598 |
| 3061 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 67 | IL6ST | Sponge network | -1.281 | 0.00012 | -0.402 | 0.00676 | 0.598 |
| 3062 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 19 | PTGFR | Sponge network | 0.335 | 0.20596 | -0.233 | 0.45421 | 0.598 |
| 3063 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 72 | CADM2 | Sponge network | -0.49 | 0.04946 | -3.782 | 0 | 0.597 |
| 3064 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | SLITRK3 | Sponge network | -1.697 | 0.00889 | -3.328 | 0 | 0.597 |
| 3065 | C20orf166-AS1 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-3913-5p;hsa-miR-582-5p;hsa-miR-664a-3p;hsa-miR-93-3p | 22 | WWTR1 | Sponge network | -2.69 | 0.0001 | -0.345 | 0.11997 | 0.597 |
| 3066 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | DACT1 | Sponge network | 0.997 | 0.00066 | 0.783 | 0.00072 | 0.597 |
| 3067 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-629-3p | 22 | EPHA7 | Sponge network | -1.582 | 8.0E-5 | -2.197 | 3.0E-5 | 0.597 |
| 3068 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 30 | SOX5 | Sponge network | -0.739 | 0.00044 | -1.091 | 4.0E-5 | 0.597 |
| 3069 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-940 | 28 | IGF1 | Sponge network | -1.349 | 0.00017 | -0.204 | 0.5898 | 0.597 |
| 3070 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | SYNE1 | Sponge network | -1.349 | 0.00017 | -0.738 | 0.00316 | 0.597 |
| 3071 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 23 | ANK2 | Sponge network | -1.349 | 0.00017 | -1.603 | 1.0E-5 | 0.597 |
| 3072 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | DCLK1 | Sponge network | -0.739 | 0.00044 | -1.324 | 0.00014 | 0.597 |
| 3073 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 62 | TRPS1 | Sponge network | -0.129 | 0.57177 | -0.191 | 0.44109 | 0.597 |
| 3074 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p | 11 | ELMO1 | Sponge network | -0.465 | 0.04703 | 0.04 | 0.83147 | 0.597 |
| 3075 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-369-3p | 15 | CNTN1 | Sponge network | -0.932 | 0.00329 | -2.594 | 0 | 0.597 |
| 3076 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p | 32 | PDGFRA | Sponge network | -0.358 | 0.17255 | -0.372 | 0.0037 | 0.597 |
| 3077 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | CNTN1 | Sponge network | -0.777 | 0.1134 | -2.594 | 0 | 0.597 |
| 3078 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 17 | PRDM5 | Sponge network | -0.129 | 0.57177 | -1.09 | 0.00014 | 0.597 |
| 3079 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-421 | 10 | TLL1 | Sponge network | -0.583 | 0.14589 | -1.195 | 3.0E-5 | 0.597 |
| 3080 | SNHG14 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 15 | THSD7A | Sponge network | -1.111 | 0.0003 | 0.242 | 0.25154 | 0.597 |
| 3081 | RP11-400K9.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 18 | PRUNE2 | Sponge network | -0.611 | 0.09607 | -1.733 | 0.00022 | 0.597 |
| 3082 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 51 | AR | Sponge network | 0.249 | 0.35902 | -1.554 | 0 | 0.597 |
| 3083 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | MCC | Sponge network | 0.335 | 0.20596 | -1.005 | 0 | 0.597 |
| 3084 | TP73-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 50 | DTNA | Sponge network | -0.563 | 0.02545 | -1.648 | 1.0E-5 | 0.597 |
| 3085 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 30 | RSPO2 | Sponge network | -2.117 | 0.00161 | -3.038 | 0 | 0.597 |
| 3086 | NR2F1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-877-5p | 34 | ASTN1 | Sponge network | -0.49 | 0.04946 | -2.389 | 2.0E-5 | 0.597 |
| 3087 | SNX29P2 |
hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-96-5p | 10 | ELMO1 | Sponge network | 0.4 | 0.14611 | 0.04 | 0.83147 | 0.597 |
| 3088 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 30 | TRPS1 | Sponge network | -1.092 | 4.0E-5 | -0.191 | 0.44109 | 0.597 |
| 3089 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | -1.439 | 4.0E-5 | -1.047 | 0.00364 | 0.597 |
| 3090 | RP5-1198O20.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 91 | LPP | Sponge network | 0.997 | 0.00066 | -0.459 | 0.04475 | 0.597 |
| 3091 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 13 | FAT4 | Sponge network | 0.889 | 9.0E-5 | -0.354 | 0.14694 | 0.597 |
| 3092 | PART1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 33 | SYNPO2 | Sponge network | -2.925 | 0 | -2.342 | 0 | 0.597 |
| 3093 | RP11-244O19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 39 | TGFBR3 | Sponge network | -0.096 | 0.67958 | -1.173 | 1.0E-5 | 0.597 |
| 3094 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 35 | RASSF2 | Sponge network | 0.4 | 0.14611 | -0.273 | 0.21961 | 0.597 |
| 3095 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 25 | SOX5 | Sponge network | -1.092 | 4.0E-5 | -1.091 | 4.0E-5 | 0.597 |
| 3096 | LINC00957 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CNTN1 | Sponge network | -0.421 | 0.0114 | -2.594 | 0 | 0.597 |
| 3097 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | PRRX1 | Sponge network | 0.997 | 0.00066 | 1.316 | 0 | 0.596 |
| 3098 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 37 | CNTN1 | Sponge network | 0.997 | 0.00066 | -2.594 | 0 | 0.596 |
| 3099 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | 0.997 | 0.00066 | -0.474 | 0.03833 | 0.596 |
| 3100 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 37 | DMD | Sponge network | -2.117 | 0.00161 | -1.506 | 0 | 0.596 |
| 3101 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 16 | FAM172A | Sponge network | -0.576 | 0.10121 | -0.57 | 0 | 0.596 |
| 3102 | PART1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-542-3p | 20 | SYNM | Sponge network | -2.925 | 0 | -2.309 | 1.0E-5 | 0.596 |
| 3103 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p | 13 | PDZD2 | Sponge network | -0.487 | 0.02157 | -0.909 | 3.0E-5 | 0.596 |
| 3104 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-486-5p;hsa-miR-93-5p | 21 | LRIG1 | Sponge network | -0.162 | 0.47687 | -0.537 | 0.00588 | 0.596 |
| 3105 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 63 | IL6ST | Sponge network | -0.976 | 0.00025 | -0.402 | 0.00676 | 0.596 |
| 3106 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 45 | MYO9A | Sponge network | -1.111 | 0.0003 | -0.147 | 0.32373 | 0.596 |
| 3107 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 15 | DKK3 | Sponge network | -1.092 | 4.0E-5 | -0.052 | 0.77783 | 0.596 |
| 3108 | HAND2-AS1 |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 12 | GLIPR2 | Sponge network | -1.697 | 0.00889 | -0.996 | 0 | 0.596 |
| 3109 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TBX18 | Sponge network | -0.576 | 0.10121 | 0.843 | 0.02945 | 0.596 |
| 3110 | MYLK-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-7-5p | 21 | ESR1 | Sponge network | -1.331 | 3.0E-5 | -1.035 | 3.0E-5 | 0.596 |
| 3111 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-582-5p;hsa-miR-629-5p | 24 | RASSF8 | Sponge network | -2.69 | 0.0001 | -0.122 | 0.65591 | 0.596 |
| 3112 | PWAR6 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p | 12 | PRKD1 | Sponge network | -1.575 | 6.0E-5 | -0.466 | 0.03583 | 0.596 |
| 3113 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ZNF208 | Sponge network | -0.576 | 0.10121 | -0.4 | 0.22381 | 0.596 |
| 3114 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | PDGFRA | Sponge network | -1.281 | 0.00012 | -0.372 | 0.0037 | 0.596 |
| 3115 | SNHG14 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 24 | CXCL12 | Sponge network | -1.111 | 0.0003 | -1.421 | 0 | 0.596 |
| 3116 | FENDRR |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ADH1B | Sponge network | -1.281 | 0.00012 | -3.716 | 0 | 0.596 |
| 3117 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-769-5p | 15 | SYNPO2 | Sponge network | -1.349 | 0.00017 | -2.342 | 0 | 0.596 |
| 3118 | FENDRR |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 33 | PCDH9 | Sponge network | -1.281 | 0.00012 | -1.823 | 4.0E-5 | 0.596 |
| 3119 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 14 | FNBP1 | Sponge network | -0.563 | 0.02545 | -0.969 | 4.0E-5 | 0.596 |
| 3120 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 34 | THBS1 | Sponge network | -0.737 | 0.06182 | 0.741 | 0.00386 | 0.596 |
| 3121 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | SYNE1 | Sponge network | 0.997 | 0.00066 | -0.738 | 0.00316 | 0.596 |
| 3122 | LINC00578 |
hsa-miR-107;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-629-3p | 11 | ITGA5 | Sponge network | 0.811 | 0.03228 | 0.452 | 0.03524 | 0.596 |
| 3123 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-744-3p | 19 | ADAMTSL3 | Sponge network | -1.654 | 0.00144 | -1.559 | 0 | 0.596 |
| 3124 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-27a-5p;hsa-miR-3913-5p | 11 | HLF | Sponge network | -0.691 | 0.00146 | -2.443 | 0 | 0.596 |
| 3125 | MIR497HG |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | TAL1 | Sponge network | -1.439 | 4.0E-5 | -0.462 | 0.00574 | 0.596 |
| 3126 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 38 | ST6GAL2 | Sponge network | -0.737 | 0.06182 | -1.201 | 0.00597 | 0.596 |
| 3127 | RP11-819C21.1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PHF3 | Sponge network | 0.537 | 0.00139 | 0.126 | 0.19652 | 0.596 |
| 3128 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | 0.532 | 0.00505 | 0.809 | 0.00544 | 0.595 |
| 3129 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-338-5p | 10 | AKAP12 | Sponge network | -0.842 | 0.01361 | -0.932 | 0.0028 | 0.595 |
| 3130 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ERG | Sponge network | -0.49 | 0.04946 | 0.002 | 0.99011 | 0.595 |
| 3131 | HCG18 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ZNF638 | Sponge network | 1.063 | 0 | 0.22 | 0.01214 | 0.595 |
| 3132 | MAGI2-AS3 |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 12 | GLIPR2 | Sponge network | -0.129 | 0.57177 | -0.996 | 0 | 0.595 |
| 3133 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 29 | KCNA6 | Sponge network | -0.976 | 0.00025 | -1.531 | 0.00013 | 0.595 |
| 3134 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-23b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 15 | BCLAF1 | Sponge network | 1.254 | 0 | 0.211 | 0.00684 | 0.595 |
| 3135 | FENDRR |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | -1.281 | 0.00012 | -0.474 | 0.03833 | 0.595 |
| 3136 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 45 | TGFBR3 | Sponge network | -0.576 | 0.10121 | -1.173 | 1.0E-5 | 0.595 |
| 3137 | NR2F1-AS1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 12 | SORCS2 | Sponge network | -0.49 | 0.04946 | -0.223 | 0.4253 | 0.595 |
| 3138 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | -1.439 | 4.0E-5 | -1.277 | 0 | 0.595 |
| 3139 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 12 | CNTNAP1 | Sponge network | -0.611 | 0.09607 | 0.358 | 0.13727 | 0.595 |
| 3140 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 16 | FAM172A | Sponge network | -0.793 | 7.0E-5 | -0.57 | 0 | 0.595 |
| 3141 | LINC00472 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-450b-5p | 11 | MAPK10 | Sponge network | -0.842 | 0.01361 | -1.774 | 0 | 0.595 |
| 3142 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p | 28 | GRIN2A | Sponge network | -0.793 | 7.0E-5 | -2.161 | 0 | 0.595 |
| 3143 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-590-3p | 25 | ST6GALNAC3 | Sponge network | -0.358 | 0.17255 | -0.987 | 0 | 0.595 |
| 3144 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | LATS2 | Sponge network | 0.572 | 0.01722 | 0.159 | 0.2304 | 0.595 |
| 3145 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p | 47 | GAS7 | Sponge network | 0.335 | 0.20596 | -0.555 | 0.01602 | 0.595 |
| 3146 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | AHNAK | Sponge network | -0.739 | 0.0574 | -1.207 | 0 | 0.595 |
| 3147 | LINC00672 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | -0.227 | 0.31657 | -0.564 | 1.0E-5 | 0.595 |
| 3148 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | CACNA2D1 | Sponge network | -1.697 | 0.00889 | -0.846 | 0.00675 | 0.595 |
| 3149 | MIR497HG |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-590-5p | 11 | THSD7A | Sponge network | -1.439 | 4.0E-5 | 0.242 | 0.25154 | 0.595 |
| 3150 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | DYNC1I1 | Sponge network | -0.769 | 0.00035 | -1.12 | 0.00107 | 0.595 |
| 3151 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 69 | BMPR2 | Sponge network | 0.72 | 0.0001 | 0.206 | 0.0635 | 0.595 |
| 3152 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-424-5p | 12 | ADAMTSL3 | Sponge network | -4.463 | 0.00025 | -1.559 | 0 | 0.595 |
| 3153 | SNHG14 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 33 | KCNA6 | Sponge network | -1.111 | 0.0003 | -1.531 | 0.00013 | 0.595 |
| 3154 | RASSF8-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | ZDBF2 | Sponge network | 0.335 | 0.20596 | -0.998 | 0.00025 | 0.595 |
| 3155 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p | 11 | FAM172A | Sponge network | -0.583 | 0.14589 | -0.57 | 0 | 0.594 |
| 3156 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-98-5p | 61 | PRKAA2 | Sponge network | -0.49 | 0.04946 | -2.462 | 0 | 0.594 |
| 3157 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | TCF4 | Sponge network | 0.727 | 3.0E-5 | 0.016 | 0.92271 | 0.594 |
| 3158 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | 0.889 | 9.0E-5 | 0.783 | 0.00072 | 0.594 |
| 3159 | AC011526.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-502-5p;hsa-miR-590-3p | 10 | FMNL3 | Sponge network | 0.342 | 0.03129 | 0.555 | 4.0E-5 | 0.594 |
| 3160 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p | 26 | EPHA7 | Sponge network | -0.358 | 0.17255 | -2.197 | 3.0E-5 | 0.594 |
| 3161 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | FNBP1 | Sponge network | 0.194 | 0.64278 | -0.969 | 4.0E-5 | 0.594 |
| 3162 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 33 | CBL | Sponge network | 0.683 | 1.0E-5 | 0.291 | 0.01267 | 0.594 |
| 3163 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p | 22 | SYNE1 | Sponge network | -0.739 | 0.00044 | -0.738 | 0.00316 | 0.594 |
| 3164 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-93-5p | 43 | PRKAA2 | Sponge network | -2.69 | 0.0001 | -2.462 | 0 | 0.594 |
| 3165 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 20 | MITF | Sponge network | -1.092 | 4.0E-5 | -0.769 | 0.00022 | 0.594 |
| 3166 | PWAR6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 45 | TGFBR3 | Sponge network | -1.575 | 6.0E-5 | -1.173 | 1.0E-5 | 0.594 |
| 3167 | LINC00578 |
hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 16 | PGR | Sponge network | 0.811 | 0.03228 | -1.704 | 0 | 0.594 |
| 3168 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 22 | CBL | Sponge network | 0.673 | 0 | 0.291 | 0.01267 | 0.594 |
| 3169 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | BNC2 | Sponge network | -1.216 | 0 | -0.491 | 0.09988 | 0.594 |
| 3170 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-625-5p;hsa-miR-671-5p | 12 | HIC1 | Sponge network | -1.697 | 0.00889 | 0.024 | 0.89889 | 0.594 |
| 3171 | RP11-554A11.4 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-629-3p;hsa-miR-664a-5p | 38 | DST | Sponge network | -0.583 | 0.14589 | -0.713 | 0.00096 | 0.594 |
| 3172 | RP11-166D19.1 |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-590-3p | 10 | GLIPR2 | Sponge network | -0.576 | 0.10121 | -0.996 | 0 | 0.594 |
| 3173 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | CNTN1 | Sponge network | 0.335 | 0.20596 | -2.594 | 0 | 0.594 |
| 3174 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PEG3 | Sponge network | -1.161 | 0.00591 | -1.74 | 0 | 0.594 |
| 3175 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | TACC1 | Sponge network | -1.575 | 6.0E-5 | -0.362 | 0.05406 | 0.594 |
| 3176 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 40 | RASSF2 | Sponge network | -0.769 | 0.00035 | -0.273 | 0.21961 | 0.594 |
| 3177 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p | 24 | ZFHX4 | Sponge network | -2.531 | 0 | 0.018 | 0.95574 | 0.594 |
| 3178 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | CACNA2D1 | Sponge network | -0.976 | 0.00025 | -0.846 | 0.00675 | 0.594 |
| 3179 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 71 | IL6ST | Sponge network | -0.49 | 0.04946 | -0.402 | 0.00676 | 0.594 |
| 3180 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p | 12 | PLSCR4 | Sponge network | -4.463 | 0.00025 | -0.474 | 0.03833 | 0.594 |
| 3181 | AP001347.6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 47 | DTNA | Sponge network | -1.161 | 0.00591 | -1.648 | 1.0E-5 | 0.594 |
| 3182 | RP11-307B6.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27b-5p;hsa-miR-421 | 12 | CNTN1 | Sponge network | -4.463 | 0.00025 | -2.594 | 0 | 0.594 |
| 3183 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 53 | RBMS3 | Sponge network | -0.777 | 0.1134 | -0.519 | 0.03551 | 0.594 |
| 3184 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p | 21 | DYNC1I1 | Sponge network | -0.563 | 0.02545 | -1.12 | 0.00107 | 0.594 |
| 3185 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 33 | PGR | Sponge network | -0.612 | 0.00094 | -1.704 | 0 | 0.594 |
| 3186 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 32 | IGF1 | Sponge network | 0.997 | 0.00066 | -0.204 | 0.5898 | 0.594 |
| 3187 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | SETBP1 | Sponge network | -1.161 | 0.00591 | -1.302 | 1.0E-5 | 0.594 |
| 3188 | EMX2OS |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 32 | NAV3 | Sponge network | -0.777 | 0.1134 | 0.212 | 0.47729 | 0.593 |
| 3189 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | RCAN2 | Sponge network | 0.064 | 0.72934 | -0.33 | 0.15172 | 0.593 |
| 3190 | MYLK-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-940 | 13 | CRTC3 | Sponge network | -1.331 | 3.0E-5 | 0.164 | 0.16786 | 0.593 |
| 3191 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 44 | NOTCH2 | Sponge network | 0.997 | 0.00066 | 0.138 | 0.35163 | 0.593 |
| 3192 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 20 | STARD13 | Sponge network | -1.697 | 0.00889 | -0.187 | 0.27383 | 0.593 |
| 3193 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-424-5p;hsa-miR-429 | 17 | DYNC1I1 | Sponge network | -0.469 | 0.08721 | -1.12 | 0.00107 | 0.593 |
| 3194 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-421 | 13 | DYNC1I1 | Sponge network | -4.546 | 0 | -1.12 | 0.00107 | 0.593 |
| 3195 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p | 33 | ADARB1 | Sponge network | -0.358 | 0.17255 | -0.335 | 0.06631 | 0.593 |
| 3196 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | NALCN | Sponge network | -0.737 | 0.06182 | 1.068 | 0.00237 | 0.593 |
| 3197 | FAM215B |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | MDM4 | Sponge network | 0.817 | 3.0E-5 | 0.345 | 0.00762 | 0.593 |
| 3198 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | -0.487 | 0.02157 | -0.466 | 0.14121 | 0.593 |
| 3199 | MEG3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 14 | THSD7A | Sponge network | 0.249 | 0.35902 | 0.242 | 0.25154 | 0.593 |
| 3200 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | RASSF2 | Sponge network | -1.439 | 4.0E-5 | -0.273 | 0.21961 | 0.593 |
| 3201 | LINC00702 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -0.737 | 0.06182 | 0.024 | 0.89889 | 0.593 |
| 3202 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-455-3p | 13 | EZH1 | Sponge network | -0.896 | 0.00099 | -0.564 | 1.0E-5 | 0.593 |
| 3203 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | ABCA10 | Sponge network | -2.452 | 0 | -0.571 | 0.03674 | 0.593 |
| 3204 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-378a-3p;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-93-5p | 36 | NTRK3 | Sponge network | -2.531 | 0 | -1.77 | 3.0E-5 | 0.593 |
| 3205 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-7-5p | 29 | BCL2 | Sponge network | -1.331 | 3.0E-5 | -0.899 | 0 | 0.593 |
| 3206 | LINC00865 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-935;hsa-miR-940 | 13 | MAPK10 | Sponge network | -1.249 | 7.0E-5 | -1.774 | 0 | 0.593 |
| 3207 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 27 | GRIN2A | Sponge network | -1.281 | 0.00012 | -2.161 | 0 | 0.593 |
| 3208 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-542-3p | 13 | SYNM | Sponge network | -0.517 | 0.02935 | -2.309 | 1.0E-5 | 0.593 |
| 3209 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | PLSCR4 | Sponge network | -0.563 | 0.02545 | -0.474 | 0.03833 | 0.593 |
| 3210 | BZRAP1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-671-5p | 17 | CYLD | Sponge network | -0.465 | 0.04703 | -0.367 | 0.00171 | 0.593 |
| 3211 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-335-3p | 14 | PBX1 | Sponge network | -0.691 | 0.00146 | -1.206 | 0 | 0.593 |
| 3212 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 26 | RUNX1T1 | Sponge network | -1.375 | 0.08642 | -0.344 | 0.2374 | 0.593 |
| 3213 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 13 | AKAP6 | Sponge network | -1.582 | 8.0E-5 | -1.586 | 3.0E-5 | 0.593 |
| 3214 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 33 | ST6GAL2 | Sponge network | -0.49 | 0.04946 | -1.201 | 0.00597 | 0.593 |
| 3215 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p | 20 | ADAMTSL3 | Sponge network | -0.811 | 2.0E-5 | -1.559 | 0 | 0.593 |
| 3216 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 12 | WWTR1 | Sponge network | -1.582 | 8.0E-5 | -0.345 | 0.11997 | 0.593 |
| 3217 | ACTA2-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | MAPK10 | Sponge network | 0.194 | 0.64278 | -1.774 | 0 | 0.593 |
| 3218 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-589-5p;hsa-miR-671-5p | 21 | ADARB1 | Sponge network | -0.309 | 0.1145 | -0.335 | 0.06631 | 0.593 |
| 3219 | RASSF8-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-877-5p | 23 | NRK | Sponge network | 0.335 | 0.20596 | 0.656 | 0.19217 | 0.593 |
| 3220 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 31 | ESR1 | Sponge network | -0.769 | 0.00035 | -1.035 | 3.0E-5 | 0.593 |
| 3221 | MIR497HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3913-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 41 | HLF | Sponge network | -1.439 | 4.0E-5 | -2.443 | 0 | 0.593 |
| 3222 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 45 | DIP2C | Sponge network | -1.697 | 0.00889 | -0.436 | 0.01713 | 0.593 |
| 3223 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 20 | ITGA5 | Sponge network | 0.997 | 0.00066 | 0.452 | 0.03524 | 0.593 |
| 3224 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NR4A3 | Sponge network | -1.203 | 0.03881 | -1.385 | 0 | 0.593 |
| 3225 | ZRANB2-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-590-3p | 15 | DLC1 | Sponge network | -0.376 | 0.00337 | -0.382 | 0.05038 | 0.593 |
| 3226 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-454-3p;hsa-miR-577 | 21 | SOX5 | Sponge network | -0.473 | 0.07602 | -1.091 | 4.0E-5 | 0.593 |
| 3227 | PWAR6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | NAV3 | Sponge network | -1.575 | 6.0E-5 | 0.212 | 0.47729 | 0.593 |
| 3228 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | IGF1 | Sponge network | -1.886 | 0 | -0.204 | 0.5898 | 0.593 |
| 3229 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 35 | MITF | Sponge network | -1.697 | 0.00889 | -0.769 | 0.00022 | 0.593 |
| 3230 | MRVI1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-874-3p | 21 | ASTN1 | Sponge network | -1.092 | 4.0E-5 | -2.389 | 2.0E-5 | 0.592 |
| 3231 | TP73-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 10 | PRKD1 | Sponge network | -0.563 | 0.02545 | -0.466 | 0.03583 | 0.592 |
| 3232 | RP11-244O19.1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | -0.096 | 0.67958 | -2.633 | 0 | 0.592 |
| 3233 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | RELN | Sponge network | -0.49 | 0.04946 | -2.403 | 0 | 0.592 |
| 3234 | LINC00641 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 41 | DMD | Sponge network | -0.222 | 0.32445 | -1.506 | 0 | 0.592 |
| 3235 | RP11-166D19.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 72 | DST | Sponge network | -0.576 | 0.10121 | -0.713 | 0.00096 | 0.592 |
| 3236 | RP11-156P1.3 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | MACF1 | Sponge network | 0.214 | 0.18246 | 0.019 | 0.90335 | 0.592 |
| 3237 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | AKAP6 | Sponge network | -0.777 | 0.1134 | -1.586 | 3.0E-5 | 0.592 |
| 3238 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ATRX | Sponge network | 1.153 | 0 | 0.261 | 0.04408 | 0.592 |
| 3239 | HOXB-AS1 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-940 | 11 | PEG3 | Sponge network | -0.154 | 0.53954 | -1.74 | 0 | 0.592 |
| 3240 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-744-3p | 29 | FAT3 | Sponge network | 0.811 | 0.03228 | -1.601 | 0.00051 | 0.592 |
| 3241 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-493-5p | 20 | NR2C2 | Sponge network | 0.614 | 1.0E-5 | 0.21 | 0.03224 | 0.592 |
| 3242 | RP11-47A8.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-625-5p | 12 | SUFU | Sponge network | 0.103 | 0.43169 | -0.04 | 0.65489 | 0.592 |
| 3243 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-222-3p;hsa-miR-590-3p | 10 | RECK | Sponge network | -1.375 | 0.08642 | -1.043 | 0 | 0.592 |
| 3244 | RP11-166D19.1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | ZDBF2 | Sponge network | -0.576 | 0.10121 | -0.998 | 0.00025 | 0.592 |
| 3245 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-93-5p | 16 | AHNAK | Sponge network | -1.645 | 0 | -1.207 | 0 | 0.592 |
| 3246 | MIR497HG |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 15 | ID4 | Sponge network | -1.439 | 4.0E-5 | -1.644 | 0 | 0.592 |
| 3247 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | HOOK3 | Sponge network | 0.559 | 0.00026 | 0.273 | 0.09957 | 0.592 |
| 3248 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 46 | ADARB1 | Sponge network | 0.572 | 0.01722 | -0.335 | 0.06631 | 0.592 |
| 3249 | LINC00702 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | FAM172A | Sponge network | -0.737 | 0.06182 | -0.57 | 0 | 0.592 |
| 3250 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | PHC3 | Sponge network | 0.537 | 0.00139 | 0.212 | 0.0499 | 0.592 |
| 3251 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 37 | SOX5 | Sponge network | 0.335 | 0.20596 | -1.091 | 4.0E-5 | 0.592 |
| 3252 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 30 | SCN9A | Sponge network | 0.572 | 0.01722 | -0.525 | 0.10593 | 0.592 |
| 3253 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | SYNE1 | Sponge network | -0.487 | 0.02157 | -0.738 | 0.00316 | 0.592 |
| 3254 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-34a-5p | 25 | EPHA5 | Sponge network | -1.331 | 3.0E-5 | -0.957 | 0.01733 | 0.592 |
| 3255 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -2.69 | 0.0001 | -3.328 | 0 | 0.592 |
| 3256 | RP1-59D14.5 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 36 | MDM4 | Sponge network | 1.162 | 5.0E-5 | 0.345 | 0.00762 | 0.592 |
| 3257 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 28 | EBF1 | Sponge network | 0.335 | 0.20596 | -0.384 | 0.08856 | 0.592 |
| 3258 | HNRNPU-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 44 | MDM4 | Sponge network | 1.601 | 0 | 0.345 | 0.00762 | 0.592 |
| 3259 | MAGI2-AS3 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 19 | FMNL3 | Sponge network | -0.129 | 0.57177 | 0.555 | 4.0E-5 | 0.592 |
| 3260 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 17 | PTPN14 | Sponge network | -1.331 | 3.0E-5 | 0.144 | 0.34595 | 0.592 |
| 3261 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-550a-3p | 26 | PGR | Sponge network | -1.249 | 7.0E-5 | -1.704 | 0 | 0.592 |
| 3262 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 40 | AKAP11 | Sponge network | -0.222 | 0.32445 | 0.196 | 0.09825 | 0.592 |
| 3263 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ZNF292 | Sponge network | 0.537 | 0.00139 | 0.276 | 0.04148 | 0.592 |
| 3264 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-629-3p | 15 | GPC6 | Sponge network | 0.811 | 0.03228 | -0.054 | 0.81465 | 0.592 |
| 3265 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-7-1-3p | 14 | EBF1 | Sponge network | 0.693 | 0.00076 | -0.384 | 0.08856 | 0.591 |
| 3266 | HAND2-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 22 | CXCL12 | Sponge network | -1.697 | 0.00889 | -1.421 | 0 | 0.591 |
| 3267 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 30 | CADM1 | Sponge network | -0.563 | 0.02545 | -0.9 | 0.00084 | 0.591 |
| 3268 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-429;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 31 | EBF3 | Sponge network | -1.697 | 0.00889 | -0.058 | 0.81437 | 0.591 |
| 3269 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | DNAJB4 | Sponge network | 0.572 | 0.01722 | -0.7 | 1.0E-5 | 0.591 |
| 3270 | CTD-2554C21.2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429 | 10 | MAPK10 | Sponge network | -1.654 | 0.00144 | -1.774 | 0 | 0.591 |
| 3271 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | DCLK1 | Sponge network | -0.421 | 0.0114 | -1.324 | 0.00014 | 0.591 |
| 3272 | ZRANB2-AS1 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p | 10 | MRVI1 | Sponge network | -0.376 | 0.00337 | -1.051 | 0.00022 | 0.591 |
| 3273 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 46 | RBMS3 | Sponge network | 0.532 | 0.00505 | -0.519 | 0.03551 | 0.591 |
| 3274 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-629-3p | 11 | THBS1 | Sponge network | -0.583 | 0.14589 | 0.741 | 0.00386 | 0.591 |
| 3275 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-5p | 18 | PLSCR4 | Sponge network | -2.62 | 0 | -0.474 | 0.03833 | 0.591 |
| 3276 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | TSHZ3 | Sponge network | -0.769 | 0.00035 | -0.556 | 0.01516 | 0.591 |
| 3277 | RP11-867G23.10 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-589-3p;hsa-miR-874-3p | 12 | SORCS1 | Sponge network | -2.531 | 0 | -3.492 | 0 | 0.591 |
| 3278 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | TAL1 | Sponge network | -0.129 | 0.57177 | -0.462 | 0.00574 | 0.591 |
| 3279 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | SPG20 | Sponge network | -2.46 | 0 | -1.16 | 0 | 0.591 |
| 3280 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | ZFHX4 | Sponge network | -1.582 | 8.0E-5 | 0.018 | 0.95574 | 0.591 |
| 3281 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | LSAMP | Sponge network | -1.697 | 0.00889 | -0.066 | 0.81708 | 0.591 |
| 3282 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | PCDH10 | Sponge network | -2.117 | 0.00161 | -1.644 | 0.0078 | 0.591 |
| 3283 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 39 | DMD | Sponge network | -0.899 | 6.0E-5 | -1.506 | 0 | 0.591 |
| 3284 | C20orf166-AS1 |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p | 20 | AFF3 | Sponge network | -2.69 | 0.0001 | -2.373 | 0 | 0.591 |
| 3285 | LINC00641 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ITPKB | Sponge network | -0.222 | 0.32445 | -0.704 | 0.00106 | 0.591 |
| 3286 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | TLE4 | Sponge network | -2.452 | 0 | -1.157 | 0 | 0.591 |
| 3287 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 21 | MCC | Sponge network | -1.582 | 8.0E-5 | -1.005 | 0 | 0.591 |
| 3288 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-30d-3p | 11 | CACNA2D1 | Sponge network | -0.473 | 0.07602 | -0.846 | 0.00675 | 0.591 |
| 3289 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 30 | GRIN2A | Sponge network | -2.117 | 0.00161 | -2.161 | 0 | 0.591 |
| 3290 | MRVI1-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p | 19 | NRK | Sponge network | -1.092 | 4.0E-5 | 0.656 | 0.19217 | 0.591 |
| 3291 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CDH11 | Sponge network | -0.49 | 0.04946 | 1.427 | 0 | 0.591 |
| 3292 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | ZFHX4 | Sponge network | -1.349 | 0.00017 | 0.018 | 0.95574 | 0.591 |
| 3293 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 31 | AKAP11 | Sponge network | 0.083 | 0.66405 | 0.196 | 0.09825 | 0.591 |
| 3294 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | PGR | Sponge network | -0.811 | 2.0E-5 | -1.704 | 0 | 0.591 |
| 3295 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PTGFR | Sponge network | -0.896 | 0.00099 | -0.233 | 0.45421 | 0.591 |
| 3296 | FENDRR |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 19 | PTGFR | Sponge network | -1.281 | 0.00012 | -0.233 | 0.45421 | 0.591 |
| 3297 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-93-3p | 23 | BNC2 | Sponge network | -0.932 | 0.00329 | -0.491 | 0.09988 | 0.591 |
| 3298 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-5p | 13 | ZNF521 | Sponge network | -1.281 | 0.00012 | -0.111 | 0.58068 | 0.591 |
| 3299 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 36 | RICTOR | Sponge network | 1.153 | 0 | 0.388 | 0.00188 | 0.591 |
| 3300 | FAM66C |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | SFRP1 | Sponge network | -0.901 | 0.00019 | -3.566 | 0 | 0.591 |
| 3301 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 44 | SOX5 | Sponge network | -0.901 | 0.00019 | -1.091 | 4.0E-5 | 0.591 |
| 3302 | AC006116.24 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-577 | 13 | SYNPO2 | Sponge network | -0.473 | 0.07602 | -2.342 | 0 | 0.591 |
| 3303 | RP11-798M19.6 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-484 | 11 | FLNA | Sponge network | -0.612 | 0.00094 | -0.771 | 0.01377 | 0.591 |
| 3304 | LINC00865 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-5p | 13 | MN1 | Sponge network | -1.249 | 7.0E-5 | -0.358 | 0.24172 | 0.591 |
| 3305 | LINC00092 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | MRVI1 | Sponge network | -1.886 | 0 | -1.051 | 0.00022 | 0.591 |
| 3306 | RP11-819C21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 38 | CREB1 | Sponge network | 0.537 | 0.00139 | 0.203 | 0.00537 | 0.591 |
| 3307 | RASSF8-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 20 | MRVI1 | Sponge network | 0.335 | 0.20596 | -1.051 | 0.00022 | 0.591 |
| 3308 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-5p | 27 | ATRX | Sponge network | 0.637 | 0.00069 | 0.261 | 0.04408 | 0.59 |
| 3309 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | IGSF10 | Sponge network | -1.575 | 6.0E-5 | -1.751 | 1.0E-5 | 0.59 |
| 3310 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429 | 14 | RECK | Sponge network | -0.469 | 0.08721 | -1.043 | 0 | 0.59 |
| 3311 | RP11-434H6.7 |
hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-374b-3p;hsa-miR-409-3p | 10 | ATRX | Sponge network | 0.358 | 0.14302 | 0.261 | 0.04408 | 0.59 |
| 3312 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-339-5p;hsa-miR-421 | 13 | STARD13 | Sponge network | -0.583 | 0.14589 | -0.187 | 0.27383 | 0.59 |
| 3313 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | ABCC9 | Sponge network | -0.901 | 0.00019 | -0.466 | 0.14121 | 0.59 |
| 3314 | JAZF1-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -0.769 | 0.00035 | -0.704 | 0.00106 | 0.59 |
| 3315 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TBX18 | Sponge network | -0.129 | 0.57177 | 0.843 | 0.02945 | 0.59 |
| 3316 | CTC-559E9.5 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-576-5p | 33 | MDM4 | Sponge network | 0.594 | 0.00064 | 0.345 | 0.00762 | 0.59 |
| 3317 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-30d-5p | 12 | ITGA5 | Sponge network | 0.889 | 9.0E-5 | 0.452 | 0.03524 | 0.59 |
| 3318 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-93-5p | 24 | PLSCR4 | Sponge network | -0.777 | 0.16667 | -0.474 | 0.03833 | 0.59 |
| 3319 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 36 | EBF1 | Sponge network | 0.249 | 0.35902 | -0.384 | 0.08856 | 0.59 |
| 3320 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-589-5p;hsa-miR-664a-3p | 12 | MN1 | Sponge network | -0.487 | 0.02157 | -0.358 | 0.24172 | 0.59 |
| 3321 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 15 | GRIN2A | Sponge network | -1.582 | 8.0E-5 | -2.161 | 0 | 0.59 |
| 3322 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-423-5p | 12 | ABL1 | Sponge network | -0.583 | 0.14589 | -0.215 | 0.07804 | 0.59 |
| 3323 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 10 | ADAMTSL3 | Sponge network | -0.376 | 0.00337 | -1.559 | 0 | 0.59 |
| 3324 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-454-3p;hsa-miR-652-3p;hsa-miR-7-5p | 27 | ZBTB4 | Sponge network | 0.194 | 0.64278 | -0.739 | 0 | 0.59 |
| 3325 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-940 | 12 | FAM172A | Sponge network | -0.976 | 0.00025 | -0.57 | 0 | 0.59 |
| 3326 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 37 | MCC | Sponge network | 0.997 | 0.00066 | -1.005 | 0 | 0.59 |
| 3327 | RP4-639F20.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p | 12 | MRVI1 | Sponge network | -0.517 | 0.02935 | -1.051 | 0.00022 | 0.59 |
| 3328 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | -0.563 | 0.02545 | -1.603 | 1.0E-5 | 0.59 |
| 3329 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 53 | QKI | Sponge network | 0.249 | 0.35902 | -0.333 | 0.04111 | 0.59 |
| 3330 | DNM3OS |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | PCDH17 | Sponge network | 0.572 | 0.01722 | 0.731 | 2.0E-5 | 0.59 |
| 3331 | FAM66C |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | PCDH9 | Sponge network | -0.901 | 0.00019 | -1.823 | 4.0E-5 | 0.59 |
| 3332 | PART1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | AKAP6 | Sponge network | -2.925 | 0 | -1.586 | 3.0E-5 | 0.59 |
| 3333 | RP11-359B12.2 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 21 | ABL1 | Sponge network | 0.064 | 0.72934 | -0.215 | 0.07804 | 0.59 |
| 3334 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | 0.194 | 0.64278 | -1.548 | 0 | 0.59 |
| 3335 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | TACC1 | Sponge network | -0.976 | 0.00025 | -0.362 | 0.05406 | 0.59 |
| 3336 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p | 15 | DYNC1I1 | Sponge network | -0.517 | 0.02935 | -1.12 | 0.00107 | 0.589 |
| 3337 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-629-3p;hsa-miR-93-5p | 30 | EPHA7 | Sponge network | -2.531 | 0 | -2.197 | 3.0E-5 | 0.589 |
| 3338 | EMX2OS |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 13 | PRKD1 | Sponge network | -0.777 | 0.1134 | -0.466 | 0.03583 | 0.589 |
| 3339 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 35 | CBL | Sponge network | 0.537 | 0.00139 | 0.291 | 0.01267 | 0.589 |
| 3340 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CACNA2D1 | Sponge network | -1.575 | 6.0E-5 | -0.846 | 0.00675 | 0.589 |
| 3341 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3913-5p | 13 | FNBP1 | Sponge network | -2.69 | 0.0001 | -0.969 | 4.0E-5 | 0.589 |
| 3342 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | JAZF1 | Sponge network | -1.575 | 6.0E-5 | -0.377 | 0.02677 | 0.589 |
| 3343 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p | 49 | FAT3 | Sponge network | -0.793 | 7.0E-5 | -1.601 | 0.00051 | 0.589 |
| 3344 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 57 | EPHA5 | Sponge network | -0.129 | 0.57177 | -0.957 | 0.01733 | 0.589 |
| 3345 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | RASSF8 | Sponge network | 0.889 | 9.0E-5 | -0.122 | 0.65591 | 0.589 |
| 3346 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-576-5p | 29 | MCC | Sponge network | -2.62 | 0 | -1.005 | 0 | 0.589 |
| 3347 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | NRXN3 | Sponge network | 0.335 | 0.20596 | -0.893 | 0.04186 | 0.589 |
| 3348 | RP5-1198O20.4 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | RNF180 | Sponge network | 0.997 | 0.00066 | -1.127 | 1.0E-5 | 0.589 |
| 3349 | ZNF667-AS1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-589-5p | 11 | SORCS2 | Sponge network | -0.976 | 0.00025 | -0.223 | 0.4253 | 0.589 |
| 3350 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 23 | LRRC55 | Sponge network | -0.129 | 0.57177 | -0.026 | 0.93163 | 0.589 |
| 3351 | NR2F1-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | MACF1 | Sponge network | -0.49 | 0.04946 | 0.019 | 0.90335 | 0.589 |
| 3352 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 34 | SOX5 | Sponge network | -0.358 | 0.17255 | -1.091 | 4.0E-5 | 0.589 |
| 3353 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | ADAMTSL3 | Sponge network | -2.46 | 0 | -1.559 | 0 | 0.589 |
| 3354 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | CACNA2D1 | Sponge network | -0.611 | 0.09607 | -0.846 | 0.00675 | 0.589 |
| 3355 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 33 | EPHA7 | Sponge network | -1.057 | 0.00029 | -2.197 | 3.0E-5 | 0.589 |
| 3356 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-450b-5p | 10 | CNTNAP1 | Sponge network | -0.517 | 0.02935 | 0.358 | 0.13727 | 0.589 |
| 3357 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p | 11 | SCN11A | Sponge network | -0.323 | 0.01372 | -1.15 | 0.00299 | 0.589 |
| 3358 | ANKRD10-IT1 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-378a-3p;hsa-miR-378c | 18 | ABL2 | Sponge network | 1.739 | 0 | 0.82 | 0 | 0.589 |
| 3359 | LINC00641 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ZNF638 | Sponge network | -0.222 | 0.32445 | 0.22 | 0.01214 | 0.589 |
| 3360 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-93-5p | 20 | PLSCR4 | Sponge network | -2.69 | 0.0001 | -0.474 | 0.03833 | 0.589 |
| 3361 | LINC00641 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 55 | MDM4 | Sponge network | -0.222 | 0.32445 | 0.345 | 0.00762 | 0.589 |
| 3362 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-671-5p | 27 | DLC1 | Sponge network | -2.69 | 0.0001 | -0.382 | 0.05038 | 0.589 |
| 3363 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | ZFHX3 | Sponge network | 0.064 | 0.72934 | 0.389 | 0.01363 | 0.589 |
| 3364 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 31 | MYO9A | Sponge network | 0.537 | 0.00139 | -0.147 | 0.32373 | 0.589 |
| 3365 | RP11-620J15.3 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 37 | FAT3 | Sponge network | -0.739 | 0.00044 | -1.601 | 0.00051 | 0.589 |
| 3366 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 69 | IL6ST | Sponge network | -0.487 | 0.02157 | -0.402 | 0.00676 | 0.589 |
| 3367 | ZRANB2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | NAV3 | Sponge network | -0.376 | 0.00337 | 0.212 | 0.47729 | 0.589 |
| 3368 | AC009948.5 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p | 12 | ZNF382 | Sponge network | 0.532 | 0.00505 | -0.494 | 0.03831 | 0.589 |
| 3369 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p | 15 | FSTL5 | Sponge network | -0.49 | 0.04946 | -1.3 | 0.01619 | 0.589 |
| 3370 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-744-5p;hsa-miR-93-5p | 21 | FGFR1 | Sponge network | 0.249 | 0.35902 | -0.509 | 0.03957 | 0.589 |
| 3371 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 13 | LHX6 | Sponge network | -0.513 | 0.04703 | -0.087 | 0.69193 | 0.589 |
| 3372 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-7-5p;hsa-miR-93-5p | 11 | GLI3 | Sponge network | -0.611 | 0.09607 | -0.615 | 0.01482 | 0.589 |
| 3373 | SNHG14 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | NOV | Sponge network | -1.111 | 0.0003 | -0.58 | 0.04676 | 0.589 |
| 3374 | RP11-532F6.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-455-5p;hsa-miR-590-3p | 15 | ITPKB | Sponge network | -0.896 | 0.00099 | -0.704 | 0.00106 | 0.589 |
| 3375 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 40 | MOB1B | Sponge network | -0.222 | 0.32445 | 0.197 | 0.15266 | 0.589 |
| 3376 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 17 | FAM172A | Sponge network | -0.49 | 0.04946 | -0.57 | 0 | 0.589 |
| 3377 | NR2F1-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | ZDBF2 | Sponge network | -0.49 | 0.04946 | -0.998 | 0.00025 | 0.589 |
| 3378 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-590-3p | 14 | SPG20 | Sponge network | -0.739 | 0.00044 | -1.16 | 0 | 0.589 |
| 3379 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-493-5p | 11 | SYNE1 | Sponge network | -2.095 | 0.00112 | -0.738 | 0.00316 | 0.588 |
| 3380 | ADAMTS9-AS2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | MACF1 | Sponge network | -2.452 | 0 | 0.019 | 0.90335 | 0.588 |
| 3381 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-450b-5p | 29 | TGFBR3 | Sponge network | -0.583 | 0.14589 | -1.173 | 1.0E-5 | 0.588 |
| 3382 | RP11-798M19.6 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-532-3p | 15 | SYNM | Sponge network | -0.612 | 0.00094 | -2.309 | 1.0E-5 | 0.588 |
| 3383 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 48 | MDM4 | Sponge network | 1.153 | 0 | 0.345 | 0.00762 | 0.588 |
| 3384 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-542-3p | 30 | DMD | Sponge network | -0.517 | 0.02935 | -1.506 | 0 | 0.588 |
| 3385 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p | 10 | BCL6 | Sponge network | -1.331 | 3.0E-5 | -0.211 | 0.19489 | 0.588 |
| 3386 | GNG12-AS1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 13 | AMPH | Sponge network | -0.793 | 7.0E-5 | -0.916 | 5.0E-5 | 0.588 |
| 3387 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-769-5p | 17 | SYNPO2 | Sponge network | -1.654 | 0.00144 | -2.342 | 0 | 0.588 |
| 3388 | LINC00578 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577 | 15 | SYNE1 | Sponge network | 0.811 | 0.03228 | -0.738 | 0.00316 | 0.588 |
| 3389 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p | 12 | DKK3 | Sponge network | -0.473 | 0.07602 | -0.052 | 0.77783 | 0.588 |
| 3390 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-766-3p | 10 | EZH1 | Sponge network | -0.187 | 0.23787 | -0.564 | 1.0E-5 | 0.588 |
| 3391 | PCA3 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 17 | SORCS1 | Sponge network | -0.709 | 0.18168 | -3.492 | 0 | 0.588 |
| 3392 | RP11-307B6.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p | 33 | LPP | Sponge network | -4.463 | 0.00025 | -0.459 | 0.04475 | 0.588 |
| 3393 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 27 | PRKAA2 | Sponge network | -0.932 | 0.00329 | -2.462 | 0 | 0.588 |
| 3394 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p | 19 | ESR1 | Sponge network | -1.092 | 4.0E-5 | -1.035 | 3.0E-5 | 0.588 |
| 3395 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 20 | RSPO2 | Sponge network | -4.546 | 0 | -3.038 | 0 | 0.588 |
| 3396 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-539-5p;hsa-miR-576-5p | 11 | NRXN1 | Sponge network | -2.076 | 0.00548 | -3.778 | 0 | 0.588 |
| 3397 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-942-5p | 35 | NRXN1 | Sponge network | -0.162 | 0.47687 | -3.778 | 0 | 0.588 |
| 3398 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | PRDM5 | Sponge network | -0.942 | 0 | -1.09 | 0.00014 | 0.588 |
| 3399 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 14 | MAPK10 | Sponge network | 0.335 | 0.20596 | -1.774 | 0 | 0.588 |
| 3400 | DNM3OS |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 25 | SETBP1 | Sponge network | 0.572 | 0.01722 | -1.302 | 1.0E-5 | 0.588 |
| 3401 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ANK2 | Sponge network | -1.161 | 0.00591 | -1.603 | 1.0E-5 | 0.588 |
| 3402 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 40 | PRKAA2 | Sponge network | -0.739 | 0.00044 | -2.462 | 0 | 0.588 |
| 3403 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 18 | MAPK10 | Sponge network | 0.572 | 0.01722 | -1.774 | 0 | 0.588 |
| 3404 | PWAR6 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 33 | PRKCB | Sponge network | -1.575 | 6.0E-5 | -1.313 | 2.0E-5 | 0.588 |
| 3405 | LINC00472 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | -0.842 | 0.01361 | -0.998 | 0.00025 | 0.588 |
| 3406 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 27 | RBMS3 | Sponge network | -0.739 | 0.00044 | -0.519 | 0.03551 | 0.588 |
| 3407 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | GRIN2A | Sponge network | -1.886 | 0 | -2.161 | 0 | 0.588 |
| 3408 | BVES-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429 | 10 | NGFR | Sponge network | -2.62 | 0 | -1.271 | 0.01058 | 0.588 |
| 3409 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 37 | DTNA | Sponge network | -0.358 | 0.17255 | -1.648 | 1.0E-5 | 0.588 |
| 3410 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p | 51 | ADARB1 | Sponge network | -0.222 | 0.32445 | -0.335 | 0.06631 | 0.588 |
| 3411 | RP11-554A11.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-874-3p | 21 | ASTN1 | Sponge network | -0.583 | 0.14589 | -2.389 | 2.0E-5 | 0.588 |
| 3412 | TRAF3IP2-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 15 | STAT5B | Sponge network | -0.323 | 0.01372 | -0.182 | 0.1082 | 0.588 |
| 3413 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-3913-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 40 | HLF | Sponge network | -0.096 | 0.67958 | -2.443 | 0 | 0.587 |
| 3414 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 25 | LATS2 | Sponge network | -0.739 | 0.0574 | 0.159 | 0.2304 | 0.587 |
| 3415 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p | 11 | RECK | Sponge network | 0.811 | 0.03228 | -1.043 | 0 | 0.587 |
| 3416 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-3p | 27 | ZFHX4 | Sponge network | -0.693 | 0.05629 | 0.018 | 0.95574 | 0.587 |
| 3417 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-93-3p | 23 | SSBP2 | Sponge network | -0.932 | 0.00329 | -1.58 | 0 | 0.587 |
| 3418 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 30 | GRIN2A | Sponge network | -0.49 | 0.04946 | -2.161 | 0 | 0.587 |
| 3419 | HAND2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p | 13 | THSD7A | Sponge network | -1.697 | 0.00889 | 0.242 | 0.25154 | 0.587 |
| 3420 | LINC00702 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 15 | ZBTB20 | Sponge network | -0.737 | 0.06182 | -0.122 | 0.57569 | 0.587 |
| 3421 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | SYNE1 | Sponge network | 0.249 | 0.35902 | -0.738 | 0.00316 | 0.587 |
| 3422 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-7-1-3p | 26 | RASSF2 | Sponge network | -0.72 | 0.24239 | -0.273 | 0.21961 | 0.587 |
| 3423 | MIR497HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-96-5p | 31 | FBXO32 | Sponge network | -1.439 | 4.0E-5 | -0.495 | 0.07561 | 0.587 |
| 3424 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p | 10 | CTNNA3 | Sponge network | -0.487 | 0.02157 | -2.046 | 0.00019 | 0.587 |
| 3425 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-93-3p;hsa-miR-940 | 10 | POU2F2 | Sponge network | -0.72 | 0.24239 | -0.233 | 0.32214 | 0.587 |
| 3426 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-96-5p | 19 | RECK | Sponge network | -1.654 | 0.00144 | -1.043 | 0 | 0.587 |
| 3427 | AP001055.6 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p | 25 | GAS7 | Sponge network | 0.693 | 0.00076 | -0.555 | 0.01602 | 0.587 |
| 3428 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p | 19 | RECK | Sponge network | -0.777 | 0.16667 | -1.043 | 0 | 0.587 |
| 3429 | FAM66C |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-542-3p | 10 | PRKD1 | Sponge network | -0.901 | 0.00019 | -0.466 | 0.03583 | 0.587 |
| 3430 | PWAR6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 21 | PTGFR | Sponge network | -1.575 | 6.0E-5 | -0.233 | 0.45421 | 0.587 |
| 3431 | RP11-999E24.3 |
hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-362-3p | 10 | RECK | Sponge network | -0.693 | 0.05629 | -1.043 | 0 | 0.587 |
| 3432 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 27 | ANK2 | Sponge network | -1.203 | 0.03881 | -1.603 | 1.0E-5 | 0.587 |
| 3433 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 55 | FAT3 | Sponge network | -1.281 | 0.00012 | -1.601 | 0.00051 | 0.587 |
| 3434 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | AFF3 | Sponge network | -0.576 | 0.10121 | -2.373 | 0 | 0.587 |
| 3435 | SERTAD4-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-576-5p | 10 | CDH19 | Sponge network | -0.932 | 0.00329 | -3.434 | 0 | 0.587 |
| 3436 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 60 | ADARB1 | Sponge network | -1.111 | 0.0003 | -0.335 | 0.06631 | 0.587 |
| 3437 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 41 | ST6GAL2 | Sponge network | -0.739 | 0.0574 | -1.201 | 0.00597 | 0.587 |
| 3438 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-577 | 11 | DNAJB4 | Sponge network | -2.69 | 0.0001 | -0.7 | 1.0E-5 | 0.587 |
| 3439 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | NRXN1 | Sponge network | -0.709 | 0.18168 | -3.778 | 0 | 0.587 |
| 3440 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-654-3p;hsa-miR-874-3p | 12 | NR2C2 | Sponge network | 1.923 | 0 | 0.21 | 0.03224 | 0.587 |
| 3441 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | PTPN13 | Sponge network | -0.49 | 0.04946 | -1.208 | 0 | 0.587 |
| 3442 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ADAMTSL3 | Sponge network | -2.915 | 0.00015 | -1.559 | 0 | 0.587 |
| 3443 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 56 | DTNA | Sponge network | 0.194 | 0.64278 | -1.648 | 1.0E-5 | 0.587 |
| 3444 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | AKAP11 | Sponge network | 0.537 | 0.00139 | 0.196 | 0.09825 | 0.587 |
| 3445 | RP5-1074L1.4 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 16 | PHF6 | Sponge network | 1.923 | 0 | 0.785 | 0 | 0.587 |
| 3446 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-625-5p | 15 | DCLK1 | Sponge network | -2.69 | 0.0001 | -1.324 | 0.00014 | 0.587 |
| 3447 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 37 | SCN9A | Sponge network | -1.111 | 0.0003 | -0.525 | 0.10593 | 0.587 |
| 3448 | LINC00657 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 11 | TPR | Sponge network | 0.91 | 0 | 0.582 | 0 | 0.587 |
| 3449 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-3p | 14 | TIMP3 | Sponge network | -2.531 | 0 | -0.546 | 0.02307 | 0.587 |
| 3450 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 50 | ZFHX4 | Sponge network | -1.575 | 6.0E-5 | 0.018 | 0.95574 | 0.587 |
| 3451 | MAGI2-AS3 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 72 | DST | Sponge network | -0.129 | 0.57177 | -0.713 | 0.00096 | 0.587 |
| 3452 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-590-3p | 10 | NRXN3 | Sponge network | -1.582 | 8.0E-5 | -0.893 | 0.04186 | 0.587 |
| 3453 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 15 | ESR1 | Sponge network | 0.693 | 0.00076 | -1.035 | 3.0E-5 | 0.587 |
| 3454 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-532-5p | 16 | FBXO32 | Sponge network | -4.463 | 0.00025 | -0.495 | 0.07561 | 0.587 |
| 3455 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | RECK | Sponge network | -0.323 | 0.01372 | -1.043 | 0 | 0.587 |
| 3456 | USP3-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-589-3p | 23 | PCDH9 | Sponge network | -0.162 | 0.47687 | -1.823 | 4.0E-5 | 0.587 |
| 3457 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | ZNF208 | Sponge network | -0.49 | 0.04946 | -0.4 | 0.22381 | 0.587 |
| 3458 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p | 28 | ZBTB4 | Sponge network | -0.901 | 0.00019 | -0.739 | 0 | 0.587 |
| 3459 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-628-5p | 28 | QKI | Sponge network | -0.583 | 0.14589 | -0.333 | 0.04111 | 0.587 |
| 3460 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | SYNE1 | Sponge network | -2.117 | 0.00161 | -0.738 | 0.00316 | 0.587 |
| 3461 | PCA3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 50 | DTNA | Sponge network | -0.709 | 0.18168 | -1.648 | 1.0E-5 | 0.587 |
| 3462 | NR2F1-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 50 | GRIK3 | Sponge network | -0.49 | 0.04946 | -3.208 | 0 | 0.587 |
| 3463 | DNM3OS |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | PCDH9 | Sponge network | 0.572 | 0.01722 | -1.823 | 4.0E-5 | 0.587 |
| 3464 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-455-3p;hsa-miR-590-3p | 20 | PLSCR4 | Sponge network | -0.896 | 0.00099 | -0.474 | 0.03833 | 0.587 |
| 3465 | EMX2OS |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -0.777 | 0.1134 | -2.633 | 0 | 0.587 |
| 3466 | C20orf166-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-505-5p;hsa-miR-664a-3p;hsa-miR-874-3p | 24 | ASTN1 | Sponge network | -2.69 | 0.0001 | -2.389 | 2.0E-5 | 0.587 |
| 3467 | RP11-434B12.1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | MDM4 | Sponge network | -0.031 | 0.89063 | 0.345 | 0.00762 | 0.587 |
| 3468 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-671-5p | 17 | FAT4 | Sponge network | -0.896 | 0.00099 | -0.354 | 0.14694 | 0.587 |
| 3469 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 52 | SOX5 | Sponge network | -1.575 | 6.0E-5 | -1.091 | 4.0E-5 | 0.586 |
| 3470 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-744-3p | 26 | MEF2C | Sponge network | -0.739 | 0.00044 | -0.499 | 0.01448 | 0.586 |
| 3471 | TPTEP1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | MRVI1 | Sponge network | -1.216 | 0 | -1.051 | 0.00022 | 0.586 |
| 3472 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | LRRK2 | Sponge network | -0.769 | 0.00035 | -1.136 | 1.0E-5 | 0.586 |
| 3473 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-424-5p | 13 | DYNC1I1 | Sponge network | -4.463 | 0.00025 | -1.12 | 0.00107 | 0.586 |
| 3474 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-766-3p;hsa-miR-96-5p | 24 | IL17RD | Sponge network | -0.517 | 0.02935 | -0.019 | 0.94873 | 0.586 |
| 3475 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-429 | 13 | SYNM | Sponge network | -0.469 | 0.08721 | -2.309 | 1.0E-5 | 0.586 |
| 3476 | FAM66C |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | FAT4 | Sponge network | -0.901 | 0.00019 | -0.354 | 0.14694 | 0.586 |
| 3477 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 51 | ZFHX4 | Sponge network | -2.452 | 0 | 0.018 | 0.95574 | 0.586 |
| 3478 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | MYO9A | Sponge network | -0.323 | 0.01372 | -0.147 | 0.32373 | 0.586 |
| 3479 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p | 14 | SPG20 | Sponge network | -2.69 | 0.0001 | -1.16 | 0 | 0.586 |
| 3480 | ARHGEF26-AS1 |
hsa-miR-10b-3p;hsa-miR-193b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p | 10 | LRRTM1 | Sponge network | -2.46 | 0 | -3.361 | 0 | 0.586 |
| 3481 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 21 | GPC6 | Sponge network | -0.358 | 0.17255 | -0.054 | 0.81465 | 0.586 |
| 3482 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-93-3p | 45 | RUNX1T1 | Sponge network | -0.611 | 0.09607 | -0.344 | 0.2374 | 0.586 |
| 3483 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | RECK | Sponge network | -1.216 | 0 | -1.043 | 0 | 0.586 |
| 3484 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940 | 44 | MEF2C | Sponge network | -0.323 | 0.01372 | -0.499 | 0.01448 | 0.586 |
| 3485 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 34 | ZBTB4 | Sponge network | -1.575 | 6.0E-5 | -0.739 | 0 | 0.586 |
| 3486 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-582-5p | 14 | DLC1 | Sponge network | 0.889 | 9.0E-5 | -0.382 | 0.05038 | 0.586 |
| 3487 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-629-5p;hsa-miR-940 | 31 | IGF1 | Sponge network | 0.249 | 0.35902 | -0.204 | 0.5898 | 0.586 |
| 3488 | MIR143HG |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 23 | KLF10 | Sponge network | -0.739 | 0.0574 | -0.783 | 0 | 0.586 |
| 3489 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 40 | SSBP2 | Sponge network | -0.421 | 0.0114 | -1.58 | 0 | 0.586 |
| 3490 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 20 | FAT4 | Sponge network | 0.194 | 0.64278 | -0.354 | 0.14694 | 0.586 |
| 3491 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-629-3p | 24 | ZBTB4 | Sponge network | -0.896 | 0.00099 | -0.739 | 0 | 0.586 |
| 3492 | LINC00578 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-502-5p | 16 | ANK2 | Sponge network | 0.811 | 0.03228 | -1.603 | 1.0E-5 | 0.586 |
| 3493 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 37 | DMD | Sponge network | -0.487 | 0.02157 | -1.506 | 0 | 0.586 |
| 3494 | RP11-389C8.2 |
hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TAL1 | Sponge network | 0.727 | 3.0E-5 | -0.462 | 0.00574 | 0.586 |
| 3495 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | AHNAK | Sponge network | -0.737 | 0.06182 | -1.207 | 0 | 0.586 |
| 3496 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 22 | MITF | Sponge network | -0.896 | 0.00099 | -0.769 | 0.00022 | 0.586 |
| 3497 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | DCLK1 | Sponge network | 0.194 | 0.64278 | -1.324 | 0.00014 | 0.586 |
| 3498 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 21 | SETBP1 | Sponge network | -0.793 | 7.0E-5 | -1.302 | 1.0E-5 | 0.586 |
| 3499 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-942-5p | 37 | HLF | Sponge network | -0.323 | 0.01372 | -2.443 | 0 | 0.586 |
| 3500 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | IGSF10 | Sponge network | -0.901 | 0.00019 | -1.751 | 1.0E-5 | 0.586 |
| 3501 | LINC00578 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-374a-3p;hsa-miR-577 | 15 | CNTN1 | Sponge network | 0.811 | 0.03228 | -2.594 | 0 | 0.586 |
| 3502 | HAND2-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 22 | MACF1 | Sponge network | -1.697 | 0.00889 | 0.019 | 0.90335 | 0.586 |
| 3503 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | EPHA7 | Sponge network | -1.375 | 0.08642 | -2.197 | 3.0E-5 | 0.586 |
| 3504 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-3065-3p;hsa-miR-7-1-3p | 19 | BNC2 | Sponge network | 0.693 | 0.00076 | -0.491 | 0.09988 | 0.586 |
| 3505 | MIR143HG |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 46 | IL17RD | Sponge network | -0.739 | 0.0574 | -0.019 | 0.94873 | 0.586 |
| 3506 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-532-3p | 20 | RPS6KA2 | Sponge network | -2.452 | 0 | -0.57 | 0.00013 | 0.586 |
| 3507 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-3p | 15 | CACNA2D1 | Sponge network | -2.69 | 0.0001 | -0.846 | 0.00675 | 0.586 |
| 3508 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 23 | ADAMTSL3 | Sponge network | 0.246 | 0.59912 | -1.559 | 0 | 0.586 |
| 3509 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 17 | ITGB3 | Sponge network | -2.452 | 0 | -0.011 | 0.95751 | 0.586 |
| 3510 | RP11-159D12.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p | 11 | FBXO11 | Sponge network | 1.254 | 0 | 0.142 | 0.04938 | 0.586 |
| 3511 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 24 | CDH23 | Sponge network | -1.111 | 0.0003 | -0.974 | 0.00034 | 0.586 |
| 3512 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | MLLT10 | Sponge network | 0.537 | 0.00139 | 0.105 | 0.33292 | 0.586 |
| 3513 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 58 | AR | Sponge network | -0.777 | 0.1134 | -1.554 | 0 | 0.586 |
| 3514 | MAGI2-AS3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 19 | FAM172A | Sponge network | -0.129 | 0.57177 | -0.57 | 0 | 0.586 |
| 3515 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 57 | DTNA | Sponge network | -0.323 | 0.01372 | -1.648 | 1.0E-5 | 0.586 |
| 3516 | ARHGEF26-AS1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | SPOP | Sponge network | -2.46 | 0 | -0.419 | 1.0E-5 | 0.586 |
| 3517 | HNRNPU-AS1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 11 | TAF1 | Sponge network | 1.601 | 0 | 0.39 | 3.0E-5 | 0.585 |
| 3518 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 31 | PGR | Sponge network | -0.096 | 0.67958 | -1.704 | 0 | 0.585 |
| 3519 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-370-3p | 13 | RPS6KA2 | Sponge network | -1.331 | 3.0E-5 | -0.57 | 0.00013 | 0.585 |
| 3520 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | IGSF10 | Sponge network | -1.439 | 4.0E-5 | -1.751 | 1.0E-5 | 0.585 |
| 3521 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 56 | EPHA5 | Sponge network | -0.739 | 0.0574 | -0.957 | 0.01733 | 0.585 |
| 3522 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | -0.612 | 0.00094 | -1.12 | 0.00107 | 0.585 |
| 3523 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-7-1-3p | 19 | AKAP6 | Sponge network | 0.299 | 0.57722 | -1.586 | 3.0E-5 | 0.585 |
| 3524 | HCG18 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-590-3p | 29 | ABL2 | Sponge network | 1.063 | 0 | 0.82 | 0 | 0.585 |
| 3525 | MRVI1-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 28 | TGFBR3 | Sponge network | -1.092 | 4.0E-5 | -1.173 | 1.0E-5 | 0.585 |
| 3526 | ARHGEF26-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 108 | LPP | Sponge network | -2.46 | 0 | -0.459 | 0.04475 | 0.585 |
| 3527 | USP3-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 36 | GRIK3 | Sponge network | -0.162 | 0.47687 | -3.208 | 0 | 0.585 |
| 3528 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-877-5p;hsa-miR-93-5p | 24 | ANK2 | Sponge network | -1.654 | 0.00144 | -1.603 | 1.0E-5 | 0.585 |
| 3529 | NR2F1-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 18 | CXCL12 | Sponge network | -0.49 | 0.04946 | -1.421 | 0 | 0.585 |
| 3530 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p | 20 | ZFHX3 | Sponge network | -1.331 | 3.0E-5 | 0.389 | 0.01363 | 0.585 |
| 3531 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | ADAMTSL3 | Sponge network | -0.096 | 0.67958 | -1.559 | 0 | 0.585 |
| 3532 | NR2F1-AS1 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | NOV | Sponge network | -0.49 | 0.04946 | -0.58 | 0.04676 | 0.585 |
| 3533 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | ST6GAL2 | Sponge network | 0.572 | 0.01722 | -1.201 | 0.00597 | 0.585 |
| 3534 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-452-5p;hsa-miR-455-3p | 12 | RELN | Sponge network | -1.092 | 4.0E-5 | -2.403 | 0 | 0.585 |
| 3535 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 33 | MCC | Sponge network | 0.249 | 0.35902 | -1.005 | 0 | 0.585 |
| 3536 | LINC00092 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | ASTN1 | Sponge network | -1.886 | 0 | -2.389 | 2.0E-5 | 0.585 |
| 3537 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-590-3p | 29 | SCN9A | Sponge network | 0.335 | 0.20596 | -0.525 | 0.10593 | 0.585 |
| 3538 | TPTEP1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | RNF180 | Sponge network | -1.216 | 0 | -1.127 | 1.0E-5 | 0.585 |
| 3539 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | RASSF8 | Sponge network | -1.439 | 4.0E-5 | -0.122 | 0.65591 | 0.585 |
| 3540 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p | 19 | SLITRK3 | Sponge network | -0.901 | 0.00019 | -3.328 | 0 | 0.585 |
| 3541 | FAM66C |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | PRDM5 | Sponge network | -0.901 | 0.00019 | -1.09 | 0.00014 | 0.585 |
| 3542 | FENDRR |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | TLL1 | Sponge network | -1.281 | 0.00012 | -1.195 | 3.0E-5 | 0.585 |
| 3543 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 30 | GJA1 | Sponge network | -0.129 | 0.57177 | -0.485 | 0.02179 | 0.585 |
| 3544 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 27 | TCF4 | Sponge network | 0.693 | 0.00076 | 0.016 | 0.92271 | 0.585 |
| 3545 | PCA3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | PARK2 | Sponge network | -0.709 | 0.18168 | -1.892 | 0 | 0.585 |
| 3546 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-664a-3p | 19 | GREM1 | Sponge network | -0.976 | 0.00025 | 0.902 | 0.01691 | 0.585 |
| 3547 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | EPHA7 | Sponge network | 0.194 | 0.64278 | -2.197 | 3.0E-5 | 0.585 |
| 3548 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 38 | ATRX | Sponge network | -0.222 | 0.32445 | 0.261 | 0.04408 | 0.585 |
| 3549 | USP3-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-625-5p | 38 | TGFBR3 | Sponge network | -0.162 | 0.47687 | -1.173 | 1.0E-5 | 0.585 |
| 3550 | RP11-554A11.4 |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-450b-5p | 10 | AFF3 | Sponge network | -0.583 | 0.14589 | -2.373 | 0 | 0.585 |
| 3551 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p | 15 | PCDH9 | Sponge network | -4.463 | 0.00025 | -1.823 | 4.0E-5 | 0.585 |
| 3552 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-5p | 30 | ESR1 | Sponge network | -0.976 | 0.00025 | -1.035 | 3.0E-5 | 0.585 |
| 3553 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 22 | PCDH9 | Sponge network | -0.611 | 0.09607 | -1.823 | 4.0E-5 | 0.585 |
| 3554 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 33 | PGR | Sponge network | 0.335 | 0.20596 | -1.704 | 0 | 0.585 |
| 3555 | RP11-822E23.8 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-511-5p;hsa-miR-532-3p | 19 | MRVI1 | Sponge network | -0.777 | 0.16667 | -1.051 | 0.00022 | 0.585 |
| 3556 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 24 | EBF1 | Sponge network | -1.092 | 4.0E-5 | -0.384 | 0.08856 | 0.585 |
| 3557 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-98-5p | 22 | RSPO2 | Sponge network | -2.62 | 0 | -3.038 | 0 | 0.585 |
| 3558 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-5p | 30 | SASH1 | Sponge network | -1.645 | 0 | -0.502 | 0.00175 | 0.585 |
| 3559 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | SLITRK3 | Sponge network | -2.117 | 0.00161 | -3.328 | 0 | 0.584 |
| 3560 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-582-5p | 15 | GRIN2A | Sponge network | -2.095 | 0.00112 | -2.161 | 0 | 0.584 |
| 3561 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-766-3p | 24 | IL17RD | Sponge network | -1.331 | 3.0E-5 | -0.019 | 0.94873 | 0.584 |
| 3562 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | RASSF8 | Sponge network | -0.563 | 0.02545 | -0.122 | 0.65591 | 0.584 |
| 3563 | FAM66C |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-5p;hsa-miR-7-5p | 17 | FNBP1 | Sponge network | -0.901 | 0.00019 | -0.969 | 4.0E-5 | 0.584 |
| 3564 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | -0.777 | 0.1134 | -1.548 | 0 | 0.584 |
| 3565 | RP11-359E10.1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | 0.299 | 0.57722 | -2.633 | 0 | 0.584 |
| 3566 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421 | 17 | RBMS3 | Sponge network | -4.463 | 0.00025 | -0.519 | 0.03551 | 0.584 |
| 3567 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-338-3p | 12 | AKAP12 | Sponge network | -1.249 | 7.0E-5 | -0.932 | 0.0028 | 0.584 |
| 3568 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-942-5p | 17 | HLF | Sponge network | -4.463 | 0.00025 | -2.443 | 0 | 0.584 |
| 3569 | HCG18 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 28 | TBL1XR1 | Sponge network | 1.063 | 0 | 0.578 | 0 | 0.584 |
| 3570 | LINC00641 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 22 | ATM | Sponge network | -0.222 | 0.32445 | 0.178 | 0.21568 | 0.584 |
| 3571 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 20 | FGFR1 | Sponge network | -0.096 | 0.67958 | -0.509 | 0.03957 | 0.584 |
| 3572 | SNHG14 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | KCNAB1 | Sponge network | -1.111 | 0.0003 | -0.755 | 2.0E-5 | 0.584 |
| 3573 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | PCDH9 | Sponge network | -0.323 | 0.01372 | -1.823 | 4.0E-5 | 0.584 |
| 3574 | LINC00092 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-542-3p;hsa-miR-590-3p | 27 | DTNA | Sponge network | -1.886 | 0 | -1.648 | 1.0E-5 | 0.584 |
| 3575 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 34 | SNX29 | Sponge network | -0.769 | 0.00035 | -0.34 | 0.01371 | 0.584 |
| 3576 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 23 | CNTN1 | Sponge network | -0.611 | 0.09607 | -2.594 | 0 | 0.584 |
| 3577 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 13 | ERCC4 | Sponge network | 0.542 | 0.00054 | 0.126 | 0.15266 | 0.584 |
| 3578 | FAM66C |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-542-3p | 18 | SYNM | Sponge network | -0.901 | 0.00019 | -2.309 | 1.0E-5 | 0.584 |
| 3579 | LINC01018 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 48 | DTNA | Sponge network | -2.915 | 0.00015 | -1.648 | 1.0E-5 | 0.584 |
| 3580 | RASSF8-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p | 22 | ASTN1 | Sponge network | 0.335 | 0.20596 | -2.389 | 2.0E-5 | 0.584 |
| 3581 | CTC-296K1.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-582-5p | 22 | DMD | Sponge network | -2.095 | 0.00112 | -1.506 | 0 | 0.584 |
| 3582 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 23 | MCC | Sponge network | -1.886 | 0 | -1.005 | 0 | 0.584 |
| 3583 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-671-5p | 19 | GAS7 | Sponge network | 1.344 | 0 | -0.555 | 0.01602 | 0.584 |
| 3584 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 44 | MCC | Sponge network | -1.697 | 0.00889 | -1.005 | 0 | 0.584 |
| 3585 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 39 | SNX29 | Sponge network | -0.129 | 0.57177 | -0.34 | 0.01371 | 0.584 |
| 3586 | LINC00641 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 31 | AKAP6 | Sponge network | -0.222 | 0.32445 | -1.586 | 3.0E-5 | 0.584 |
| 3587 | RP11-389C8.2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | DOCK4 | Sponge network | 0.727 | 3.0E-5 | 0.502 | 0.00108 | 0.584 |
| 3588 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 16 | PTGFR | Sponge network | 0.194 | 0.64278 | -0.233 | 0.45421 | 0.584 |
| 3589 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 42 | TGFBR3 | Sponge network | -0.976 | 0.00025 | -1.173 | 1.0E-5 | 0.584 |
| 3590 | MAGI2-AS3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 30 | RYR2 | Sponge network | -0.129 | 0.57177 | -1.466 | 0.00031 | 0.584 |
| 3591 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p | 19 | FGFR1 | Sponge network | 0.194 | 0.64278 | -0.509 | 0.03957 | 0.584 |
| 3592 | NR2F1-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 19 | KLF10 | Sponge network | -0.49 | 0.04946 | -0.783 | 0 | 0.584 |
| 3593 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-98-5p | 37 | PRKAA2 | Sponge network | -0.473 | 0.07602 | -2.462 | 0 | 0.584 |
| 3594 | LINC00672 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 73 | LPP | Sponge network | -0.227 | 0.31657 | -0.459 | 0.04475 | 0.584 |
| 3595 | CTC-296K1.4 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-378a-5p;hsa-miR-576-5p | 19 | DTNA | Sponge network | -2.076 | 0.00548 | -1.648 | 1.0E-5 | 0.584 |
| 3596 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p | 48 | FAT3 | Sponge network | 0.335 | 0.20596 | -1.601 | 0.00051 | 0.584 |
| 3597 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DNAJB4 | Sponge network | -1.575 | 6.0E-5 | -0.7 | 1.0E-5 | 0.584 |
| 3598 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 31 | AR | Sponge network | -1.886 | 0 | -1.554 | 0 | 0.584 |
| 3599 | RP11-307B6.3 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-93-3p | 16 | BNC2 | Sponge network | -4.463 | 0.00025 | -0.491 | 0.09988 | 0.584 |
| 3600 | RP11-672A2.4 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-671-5p | 10 | BNC2 | Sponge network | 1.344 | 0 | -0.491 | 0.09988 | 0.584 |
| 3601 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-671-5p | 23 | KIT | Sponge network | -0.563 | 0.02545 | -2.184 | 0 | 0.584 |
| 3602 | DNM3OS |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 28 | PCDH10 | Sponge network | 0.572 | 0.01722 | -1.644 | 0.0078 | 0.583 |
| 3603 | MIR497HG |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 10 | TPO | Sponge network | -1.439 | 4.0E-5 | -1.87 | 2.0E-5 | 0.583 |
| 3604 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 22 | DYNC1I1 | Sponge network | -0.901 | 0.00019 | -1.12 | 0.00107 | 0.583 |
| 3605 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 62 | PRKAA2 | Sponge network | -0.737 | 0.06182 | -2.462 | 0 | 0.583 |
| 3606 | LINC00472 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 36 | DIP2C | Sponge network | -0.842 | 0.01361 | -0.436 | 0.01713 | 0.583 |
| 3607 | CTC-296K1.3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-625-5p;hsa-miR-940 | 10 | SFRP1 | Sponge network | -2.095 | 0.00112 | -3.566 | 0 | 0.583 |
| 3608 | PWAR6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 19 | CADM3 | Sponge network | -1.575 | 6.0E-5 | -3.663 | 0 | 0.583 |
| 3609 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | MCC | Sponge network | -0.611 | 0.09607 | -1.005 | 0 | 0.583 |
| 3610 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 26 | SEMA3C | Sponge network | -0.129 | 0.57177 | -0.272 | 0.31153 | 0.583 |
| 3611 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 16 | TMEFF2 | Sponge network | -0.976 | 0.00025 | -2.426 | 0 | 0.583 |
| 3612 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 30 | RAP1A | Sponge network | -0.793 | 7.0E-5 | -0.929 | 0 | 0.583 |
| 3613 | TRAF3IP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-766-3p | 15 | EZH1 | Sponge network | -0.323 | 0.01372 | -0.564 | 1.0E-5 | 0.583 |
| 3614 | RP11-977G19.11 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-940 | 11 | MDM4 | Sponge network | 0.957 | 0 | 0.345 | 0.00762 | 0.583 |
| 3615 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-3p | 28 | ZFHX4 | Sponge network | -0.154 | 0.53954 | 0.018 | 0.95574 | 0.583 |
| 3616 | RP11-822E23.8 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-940 | 12 | MAPK10 | Sponge network | -0.777 | 0.16667 | -1.774 | 0 | 0.583 |
| 3617 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 56 | SSBP2 | Sponge network | -1.575 | 6.0E-5 | -1.58 | 0 | 0.583 |
| 3618 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-576-5p | 24 | ZFHX4 | Sponge network | -0.932 | 0.00329 | 0.018 | 0.95574 | 0.583 |
| 3619 | RP11-819C21.1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 12 | TAF1 | Sponge network | 0.537 | 0.00139 | 0.39 | 3.0E-5 | 0.583 |
| 3620 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 35 | EPHA3 | Sponge network | -1.281 | 0.00012 | 0.185 | 0.53588 | 0.583 |
| 3621 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PCDH9 | Sponge network | 0.246 | 0.59912 | -1.823 | 4.0E-5 | 0.583 |
| 3622 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-942-5p | 25 | NRXN1 | Sponge network | -0.358 | 0.17255 | -3.778 | 0 | 0.583 |
| 3623 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 14 | NALCN | Sponge network | -0.739 | 0.0574 | 1.068 | 0.00237 | 0.583 |
| 3624 | FGF14-AS2 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-629-3p;hsa-miR-7-5p | 23 | IGFBP5 | Sponge network | -1.582 | 8.0E-5 | -0.806 | 0.00105 | 0.583 |
| 3625 | RP11-736K20.5 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | FMNL3 | Sponge network | -0.358 | 0.17255 | 0.555 | 4.0E-5 | 0.583 |
| 3626 | MEG3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | LATS2 | Sponge network | 0.249 | 0.35902 | 0.159 | 0.2304 | 0.583 |
| 3627 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 46 | EPHA7 | Sponge network | -0.901 | 0.00019 | -2.197 | 3.0E-5 | 0.583 |
| 3628 | CTC-510F12.2 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-3614-5p | 11 | RBMS3 | Sponge network | -0.691 | 0.00146 | -0.519 | 0.03551 | 0.583 |
| 3629 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ABCC9 | Sponge network | -1.349 | 0.00017 | -0.466 | 0.14121 | 0.583 |
| 3630 | MIR497HG |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p | 10 | HMCN1 | Sponge network | -1.439 | 4.0E-5 | 0.809 | 0.00544 | 0.583 |
| 3631 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | LRRK2 | Sponge network | -1.281 | 0.00012 | -1.136 | 1.0E-5 | 0.583 |
| 3632 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 64 | RUNX1T1 | Sponge network | -0.777 | 0.1134 | -0.344 | 0.2374 | 0.583 |
| 3633 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-935 | 29 | TSHZ3 | Sponge network | -2.62 | 0 | -0.556 | 0.01516 | 0.583 |
| 3634 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 16 | ANK2 | Sponge network | -1.375 | 0.08642 | -1.603 | 1.0E-5 | 0.583 |
| 3635 | CTA-363E19.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p | 10 | NR2C2 | Sponge network | 1.055 | 0 | 0.21 | 0.03224 | 0.583 |
| 3636 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 47 | PDGFRA | Sponge network | -0.129 | 0.57177 | -0.372 | 0.0037 | 0.583 |
| 3637 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | LRRC7 | Sponge network | -0.737 | 0.06182 | -1.341 | 0.01193 | 0.583 |
| 3638 | LINC00578 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-744-3p | 30 | IGFBP5 | Sponge network | 0.811 | 0.03228 | -0.806 | 0.00105 | 0.583 |
| 3639 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-200c-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p | 12 | NRK | Sponge network | -0.517 | 0.02935 | 0.656 | 0.19217 | 0.583 |
| 3640 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | PCDH9 | Sponge network | 0.194 | 0.64278 | -1.823 | 4.0E-5 | 0.583 |
| 3641 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 16 | GRIN2A | Sponge network | -0.358 | 0.17255 | -2.161 | 0 | 0.582 |
| 3642 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-655-3p;hsa-miR-935;hsa-miR-98-5p | 42 | DMD | Sponge network | -0.405 | 0.22013 | -1.506 | 0 | 0.582 |
| 3643 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-5p | 33 | NTRK2 | Sponge network | -0.384 | 0.40652 | -0.736 | 0.06495 | 0.582 |
| 3644 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 31 | IGF1 | Sponge network | -2.117 | 0.00161 | -0.204 | 0.5898 | 0.582 |
| 3645 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935 | 34 | FBXO32 | Sponge network | -0.896 | 0.00099 | -0.495 | 0.07561 | 0.582 |
| 3646 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 41 | TACC1 | Sponge network | -0.096 | 0.67958 | -0.362 | 0.05406 | 0.582 |
| 3647 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 64 | IL6ST | Sponge network | 0.572 | 0.01722 | -0.402 | 0.00676 | 0.582 |
| 3648 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | MYO9A | Sponge network | 0.559 | 0.00026 | -0.147 | 0.32373 | 0.582 |
| 3649 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-33a-5p;hsa-miR-3607-3p | 18 | CNTN1 | Sponge network | -0.693 | 0.05629 | -2.594 | 0 | 0.582 |
| 3650 | RP11-166D19.1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 20 | ID4 | Sponge network | -0.576 | 0.10121 | -1.644 | 0 | 0.582 |
| 3651 | TPTEP1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 13 | FHL1 | Sponge network | -1.216 | 0 | -2.687 | 0 | 0.582 |
| 3652 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-23a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-96-5p | 13 | CNTN1 | Sponge network | -0.517 | 0.02935 | -2.594 | 0 | 0.582 |
| 3653 | LINC00957 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 16 | MAPK10 | Sponge network | -0.421 | 0.0114 | -1.774 | 0 | 0.582 |
| 3654 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-93-5p | 42 | TACC1 | Sponge network | -0.162 | 0.47687 | -0.362 | 0.05406 | 0.582 |
| 3655 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | FAT3 | Sponge network | -0.709 | 0.18168 | -1.601 | 0.00051 | 0.582 |
| 3656 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-92b-3p | 19 | AKAP12 | Sponge network | 0.997 | 0.00066 | -0.932 | 0.0028 | 0.582 |
| 3657 | NR2F1-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 12 | KCTD8 | Sponge network | -0.49 | 0.04946 | -3.359 | 0 | 0.582 |
| 3658 | TPTEP1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 14 | PTPN13 | Sponge network | -1.216 | 0 | -1.208 | 0 | 0.582 |
| 3659 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-542-3p | 30 | DMD | Sponge network | -0.611 | 0.09607 | -1.506 | 0 | 0.582 |
| 3660 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 16 | PTPN11 | Sponge network | 1.153 | 0 | 0.431 | 2.0E-5 | 0.582 |
| 3661 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | PREX2 | Sponge network | -0.576 | 0.10121 | -0.272 | 0.25382 | 0.582 |
| 3662 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 60 | TCF4 | Sponge network | 0.194 | 0.64278 | 0.016 | 0.92271 | 0.582 |
| 3663 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-424-5p | 20 | PLSCR4 | Sponge network | 0.176 | 0.37402 | -0.474 | 0.03833 | 0.582 |
| 3664 | RP4-613B23.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p | 10 | ITPKB | Sponge network | -0.309 | 0.1145 | -0.704 | 0.00106 | 0.582 |
| 3665 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | RECK | Sponge network | -0.563 | 0.02545 | -1.043 | 0 | 0.582 |
| 3666 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 18 | ANK2 | Sponge network | -1.057 | 0.00029 | -1.603 | 1.0E-5 | 0.582 |
| 3667 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-5p;hsa-miR-409-3p;hsa-miR-576-5p | 24 | ATRX | Sponge network | 0.594 | 0.00064 | 0.261 | 0.04408 | 0.582 |
| 3668 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | PEG3 | Sponge network | -0.793 | 7.0E-5 | -1.74 | 0 | 0.582 |
| 3669 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-429;hsa-miR-576-5p | 34 | ZFHX4 | Sponge network | -2.62 | 0 | 0.018 | 0.95574 | 0.582 |
| 3670 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p | 13 | TGFBR2 | Sponge network | -0.358 | 0.17255 | -0.243 | 0.11408 | 0.582 |
| 3671 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 50 | SOX5 | Sponge network | -0.129 | 0.57177 | -1.091 | 4.0E-5 | 0.582 |
| 3672 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 41 | RBMS3 | Sponge network | -0.769 | 0.00035 | -0.519 | 0.03551 | 0.582 |
| 3673 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 51 | PRKAA2 | Sponge network | -0.842 | 0.01361 | -2.462 | 0 | 0.582 |
| 3674 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 23 | ADAMTSL3 | Sponge network | -1.249 | 7.0E-5 | -1.559 | 0 | 0.582 |
| 3675 | AC006116.24 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-93-5p | 15 | PLSCR4 | Sponge network | -0.473 | 0.07602 | -0.474 | 0.03833 | 0.582 |
| 3676 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | PCDH10 | Sponge network | -0.611 | 0.09607 | -1.644 | 0.0078 | 0.582 |
| 3677 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-7-5p;hsa-miR-96-5p | 17 | SETBP1 | Sponge network | -0.517 | 0.02935 | -1.302 | 1.0E-5 | 0.582 |
| 3678 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DCN | Sponge network | -0.901 | 0.00019 | -1.288 | 0 | 0.582 |
| 3679 | RP11-999E24.3 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p | 10 | SFRP1 | Sponge network | -0.693 | 0.05629 | -3.566 | 0 | 0.582 |
| 3680 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | LRRK2 | Sponge network | -1.697 | 0.00889 | -1.136 | 1.0E-5 | 0.582 |
| 3681 | PCA3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | FAM172A | Sponge network | -0.709 | 0.18168 | -0.57 | 0 | 0.582 |
| 3682 | HOXB-AS1 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-93-3p | 13 | WWTR1 | Sponge network | -0.154 | 0.53954 | -0.345 | 0.11997 | 0.581 |
| 3683 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 21 | TRIM33 | Sponge network | 1.254 | 0 | 0.402 | 7.0E-5 | 0.581 |
| 3684 | RP11-798M19.6 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ITPKB | Sponge network | -0.612 | 0.00094 | -0.704 | 0.00106 | 0.581 |
| 3685 | RP11-999E24.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-3614-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 21 | IGFBP5 | Sponge network | -0.693 | 0.05629 | -0.806 | 0.00105 | 0.581 |
| 3686 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 49 | ZFHX4 | Sponge network | 0.194 | 0.64278 | 0.018 | 0.95574 | 0.581 |
| 3687 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | WWTR1 | Sponge network | 0.335 | 0.20596 | -0.345 | 0.11997 | 0.581 |
| 3688 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 25 | EBF3 | Sponge network | 0.249 | 0.35902 | -0.058 | 0.81437 | 0.581 |
| 3689 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p | 10 | LHFP | Sponge network | -0.611 | 0.09607 | -0.167 | 0.41371 | 0.581 |
| 3690 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-16-2-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-493-5p;hsa-miR-582-5p | 10 | AFF3 | Sponge network | -2.095 | 0.00112 | -2.373 | 0 | 0.581 |
| 3691 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | ZNF292 | Sponge network | 1.254 | 0 | 0.276 | 0.04148 | 0.581 |
| 3692 | MAGI2-AS3 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PCDH18 | Sponge network | -0.129 | 0.57177 | 0.216 | 0.24645 | 0.581 |
| 3693 | ZNF582-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | PCDH10 | Sponge network | -1.349 | 0.00017 | -1.644 | 0.0078 | 0.581 |
| 3694 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-92a-1-5p | 15 | AKAP6 | Sponge network | -0.309 | 0.1145 | -1.586 | 3.0E-5 | 0.581 |
| 3695 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-503-5p | 11 | SMARCA1 | Sponge network | -0.563 | 0.02545 | -0.684 | 0.00159 | 0.581 |
| 3696 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | HOOK3 | Sponge network | -0.739 | 0.0574 | 0.273 | 0.09957 | 0.581 |
| 3697 | AC003090.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-590-3p | 10 | SCN11A | Sponge network | -2.117 | 0.00161 | -1.15 | 0.00299 | 0.581 |
| 3698 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 18 | NR2C2 | Sponge network | 1.153 | 0 | 0.21 | 0.03224 | 0.581 |
| 3699 | RP11-166D19.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | PRKCB | Sponge network | -0.576 | 0.10121 | -1.313 | 2.0E-5 | 0.581 |
| 3700 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | SASH1 | Sponge network | -0.487 | 0.02157 | -0.502 | 0.00175 | 0.581 |
| 3701 | PCA3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 37 | DMD | Sponge network | -0.709 | 0.18168 | -1.506 | 0 | 0.581 |
| 3702 | FENDRR |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | PRKCB | Sponge network | -1.281 | 0.00012 | -1.313 | 2.0E-5 | 0.581 |
| 3703 | C20orf166-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-582-5p | 10 | THSD7B | Sponge network | -2.69 | 0.0001 | 0.154 | 0.74723 | 0.581 |
| 3704 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | PTGFR | Sponge network | -0.323 | 0.01372 | -0.233 | 0.45421 | 0.581 |
| 3705 | MEG3 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 16 | DCLK1 | Sponge network | 0.249 | 0.35902 | -1.324 | 0.00014 | 0.581 |
| 3706 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-577 | 13 | AKAP12 | Sponge network | -0.473 | 0.07602 | -0.932 | 0.0028 | 0.581 |
| 3707 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | RELN | Sponge network | -0.358 | 0.17255 | -2.403 | 0 | 0.581 |
| 3708 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-671-5p;hsa-miR-877-5p | 12 | RBMS3 | Sponge network | -0.309 | 0.1145 | -0.519 | 0.03551 | 0.581 |
| 3709 | ZNF667-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-484;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | PRKCB | Sponge network | -0.976 | 0.00025 | -1.313 | 2.0E-5 | 0.581 |
| 3710 | ZNF582-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-671-5p;hsa-miR-940 | 12 | SORCS1 | Sponge network | -1.349 | 0.00017 | -3.492 | 0 | 0.581 |
| 3711 | FAM66C |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p | 15 | EZH1 | Sponge network | -0.901 | 0.00019 | -0.564 | 1.0E-5 | 0.581 |
| 3712 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 37 | RICTOR | Sponge network | 1.063 | 0 | 0.388 | 0.00188 | 0.581 |
| 3713 | BVES-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429 | 13 | GLI3 | Sponge network | -2.62 | 0 | -0.615 | 0.01482 | 0.581 |
| 3714 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-3913-5p | 11 | FNBP1 | Sponge network | -0.096 | 0.67958 | -0.969 | 4.0E-5 | 0.58 |
| 3715 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 49 | SOX5 | Sponge network | -0.576 | 0.10121 | -1.091 | 4.0E-5 | 0.58 |
| 3716 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | ROBO1 | Sponge network | -0.576 | 0.10121 | -0.009 | 0.96154 | 0.58 |
| 3717 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-550a-5p;hsa-miR-93-5p | 31 | TSHZ3 | Sponge network | -0.777 | 0.16667 | -0.556 | 0.01516 | 0.58 |
| 3718 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 41 | SOX5 | Sponge network | -1.249 | 7.0E-5 | -1.091 | 4.0E-5 | 0.58 |
| 3719 | JAZF1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | MSN | Sponge network | -0.769 | 0.00035 | -0.023 | 0.88422 | 0.58 |
| 3720 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 59 | SSBP2 | Sponge network | -1.697 | 0.00889 | -1.58 | 0 | 0.58 |
| 3721 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-628-5p;hsa-miR-629-3p | 16 | RASSF8 | Sponge network | -0.154 | 0.53954 | -0.122 | 0.65591 | 0.58 |
| 3722 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 28 | ADAMTSL3 | Sponge network | 0.572 | 0.01722 | -1.559 | 0 | 0.58 |
| 3723 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 19 | CHD2 | Sponge network | 0.537 | 0.00139 | 0.049 | 0.55371 | 0.58 |
| 3724 | LINC00702 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 48 | TGFBR3 | Sponge network | -0.737 | 0.06182 | -1.173 | 1.0E-5 | 0.58 |
| 3725 | LINC00473 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-935 | 12 | MAPK10 | Sponge network | -1.203 | 0.03881 | -1.774 | 0 | 0.58 |
| 3726 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p | 11 | ABCC9 | Sponge network | -0.693 | 0.05629 | -0.466 | 0.14121 | 0.58 |
| 3727 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 44 | CHD5 | Sponge network | -0.096 | 0.67958 | -0.922 | 0.01521 | 0.58 |
| 3728 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 35 | FBXO32 | Sponge network | -0.487 | 0.02157 | -0.495 | 0.07561 | 0.58 |
| 3729 | FAM66C |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | WWTR1 | Sponge network | -0.901 | 0.00019 | -0.345 | 0.11997 | 0.58 |
| 3730 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-664a-3p | 18 | AKAP6 | Sponge network | -0.358 | 0.17255 | -1.586 | 3.0E-5 | 0.58 |
| 3731 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-22-5p;hsa-miR-23b-3p;hsa-miR-26a-2-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p | 10 | XPO1 | Sponge network | 1.254 | 0 | 0.759 | 0 | 0.58 |
| 3732 | ZNF667-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | IL17RD | Sponge network | -0.976 | 0.00025 | -0.019 | 0.94873 | 0.58 |
| 3733 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-7-5p | 24 | ESR1 | Sponge network | -0.611 | 0.09607 | -1.035 | 3.0E-5 | 0.58 |
| 3734 | RASSF8-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 15 | FLNA | Sponge network | 0.335 | 0.20596 | -0.771 | 0.01377 | 0.58 |
| 3735 | HAND2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 52 | TGFBR3 | Sponge network | -1.697 | 0.00889 | -1.173 | 1.0E-5 | 0.58 |
| 3736 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | CNTN1 | Sponge network | 0.572 | 0.01722 | -2.594 | 0 | 0.58 |
| 3737 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 15 | MEF2C | Sponge network | 0.342 | 0.03129 | -0.499 | 0.01448 | 0.58 |
| 3738 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | RASSF2 | Sponge network | 0.727 | 3.0E-5 | -0.273 | 0.21961 | 0.58 |
| 3739 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-577;hsa-miR-93-5p | 19 | NIN | Sponge network | -0.465 | 0.04703 | -0.253 | 0.13794 | 0.58 |
| 3740 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p | 13 | DYNC1I1 | Sponge network | -0.896 | 0.00099 | -1.12 | 0.00107 | 0.58 |
| 3741 | FENDRR |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | PRDM5 | Sponge network | -1.281 | 0.00012 | -1.09 | 0.00014 | 0.58 |
| 3742 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | TIMP3 | Sponge network | 0.997 | 0.00066 | -0.546 | 0.02307 | 0.58 |
| 3743 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 35 | MITF | Sponge network | -2.452 | 0 | -0.769 | 0.00022 | 0.58 |
| 3744 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 26 | ADAMTSL3 | Sponge network | 0.194 | 0.64278 | -1.559 | 0 | 0.58 |
| 3745 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | -1.281 | 0.00012 | -1.548 | 0 | 0.58 |
| 3746 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | PRKAA2 | Sponge network | -1.161 | 0.00591 | -2.462 | 0 | 0.58 |
| 3747 | FAM66C |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -0.901 | 0.00019 | -2.633 | 0 | 0.58 |
| 3748 | BVES-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p | 12 | ID4 | Sponge network | -2.62 | 0 | -1.644 | 0 | 0.58 |
| 3749 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 22 | FGFR1 | Sponge network | -0.777 | 0.1134 | -0.509 | 0.03957 | 0.58 |
| 3750 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | PCDH9 | Sponge network | 0.997 | 0.00066 | -1.823 | 4.0E-5 | 0.58 |
| 3751 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 28 | RASSF8 | Sponge network | -0.323 | 0.01372 | -0.122 | 0.65591 | 0.58 |
| 3752 | FAM66C |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | -0.901 | 0.00019 | 0.154 | 0.74723 | 0.58 |
| 3753 | MIR143HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 13 | HIC1 | Sponge network | -0.739 | 0.0574 | 0.024 | 0.89889 | 0.58 |
| 3754 | PWAR6 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 14 | KCNAB1 | Sponge network | -1.575 | 6.0E-5 | -0.755 | 2.0E-5 | 0.58 |
| 3755 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 43 | TSHZ3 | Sponge network | -1.575 | 6.0E-5 | -0.556 | 0.01516 | 0.58 |
| 3756 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-628-5p | 31 | RASSF8 | Sponge network | -0.162 | 0.47687 | -0.122 | 0.65591 | 0.58 |
| 3757 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 19 | ZNF770 | Sponge network | 0.421 | 0.00084 | 0.206 | 0.06751 | 0.58 |
| 3758 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-93-5p | 28 | ZBTB4 | Sponge network | -2.69 | 0.0001 | -0.739 | 0 | 0.58 |
| 3759 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | FGF5 | Sponge network | -0.129 | 0.57177 | 0.549 | 0.28659 | 0.58 |
| 3760 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 16 | TXNIP | Sponge network | -0.769 | 0.00035 | -1.277 | 0 | 0.58 |
| 3761 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 24 | KCNA6 | Sponge network | -1.092 | 4.0E-5 | -1.531 | 0.00013 | 0.58 |
| 3762 | EMX2OS |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 19 | GLI3 | Sponge network | -0.777 | 0.1134 | -0.615 | 0.01482 | 0.58 |
| 3763 | ARHGEF26-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | KCTD8 | Sponge network | -2.46 | 0 | -3.359 | 0 | 0.58 |
| 3764 | MEG3 |
hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 12 | FLNA | Sponge network | 0.249 | 0.35902 | -0.771 | 0.01377 | 0.58 |
| 3765 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 20 | STARD13 | Sponge network | -0.576 | 0.10121 | -0.187 | 0.27383 | 0.58 |
| 3766 | RASSF8-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-7-5p | 12 | FNBP1 | Sponge network | 0.335 | 0.20596 | -0.969 | 4.0E-5 | 0.58 |
| 3767 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-450b-5p | 20 | FBXO32 | Sponge network | -2.531 | 0 | -0.495 | 0.07561 | 0.58 |
| 3768 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92b-3p | 21 | AKAP12 | Sponge network | -2.46 | 0 | -0.932 | 0.0028 | 0.58 |
| 3769 | MRVI1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-671-5p | 16 | CDH23 | Sponge network | -1.092 | 4.0E-5 | -0.974 | 0.00034 | 0.58 |
| 3770 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 40 | PBX1 | Sponge network | -0.162 | 0.47687 | -1.206 | 0 | 0.579 |
| 3771 | LINC00865 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p | 19 | MRVI1 | Sponge network | -1.249 | 7.0E-5 | -1.051 | 0.00022 | 0.579 |
| 3772 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p | 14 | TXNIP | Sponge network | -0.576 | 0.10121 | -1.277 | 0 | 0.579 |
| 3773 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p | 16 | RCAN2 | Sponge network | -0.793 | 7.0E-5 | -0.33 | 0.15172 | 0.579 |
| 3774 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 49 | BCL2 | Sponge network | -2.452 | 0 | -0.899 | 0 | 0.579 |
| 3775 | MIR143HG |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | TMEFF2 | Sponge network | -0.739 | 0.0574 | -2.426 | 0 | 0.579 |
| 3776 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 50 | AR | Sponge network | -0.563 | 0.02545 | -1.554 | 0 | 0.579 |
| 3777 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-5p | 55 | PRKAA2 | Sponge network | -0.793 | 7.0E-5 | -2.462 | 0 | 0.579 |
| 3778 | ZNF582-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 16 | FNBP1 | Sponge network | -1.349 | 0.00017 | -0.969 | 4.0E-5 | 0.579 |
| 3779 | TUG1 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-582-3p | 22 | ABL2 | Sponge network | 0.972 | 0 | 0.82 | 0 | 0.579 |
| 3780 | MEG3 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-577 | 15 | MACF1 | Sponge network | 0.249 | 0.35902 | 0.019 | 0.90335 | 0.579 |
| 3781 | MALAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 23 | TRIM33 | Sponge network | 0.782 | 0.00016 | 0.402 | 7.0E-5 | 0.579 |
| 3782 | MEG3 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-7-5p | 11 | COL1A2 | Sponge network | 0.249 | 0.35902 | 1.991 | 0 | 0.579 |
| 3783 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 34 | PGR | Sponge network | -1.216 | 0 | -1.704 | 0 | 0.579 |
| 3784 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 17 | CDH19 | Sponge network | -1.349 | 0.00017 | -3.434 | 0 | 0.579 |
| 3785 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 22 | GPC6 | Sponge network | 0.335 | 0.20596 | -0.054 | 0.81465 | 0.579 |
| 3786 | HAND2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-3p | 12 | ZNF382 | Sponge network | -1.697 | 0.00889 | -0.494 | 0.03831 | 0.579 |
| 3787 | CTC-296K1.3 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-671-5p;hsa-miR-940 | 20 | GRIK3 | Sponge network | -2.095 | 0.00112 | -3.208 | 0 | 0.579 |
| 3788 | LINC00702 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p | 20 | STARD13 | Sponge network | -0.737 | 0.06182 | -0.187 | 0.27383 | 0.579 |
| 3789 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 18 | EZH1 | Sponge network | -2.69 | 0.0001 | -0.564 | 1.0E-5 | 0.579 |
| 3790 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-7-5p | 14 | FNBP1 | Sponge network | -0.487 | 0.02157 | -0.969 | 4.0E-5 | 0.579 |
| 3791 | RP11-890B15.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.101 | 0.60919 | -0.57 | 0 | 0.579 |
| 3792 | TPTEP1 |
hsa-let-7a-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 12 | RADIL | Sponge network | -1.216 | 0 | -1.628 | 0 | 0.579 |
| 3793 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 27 | EPHA3 | Sponge network | -0.611 | 0.09607 | 0.185 | 0.53588 | 0.579 |
| 3794 | LINC00957 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | -0.421 | 0.0114 | -0.974 | 0.00034 | 0.579 |
| 3795 | LINC00578 |
hsa-let-7b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-629-3p | 31 | AR | Sponge network | 0.811 | 0.03228 | -1.554 | 0 | 0.579 |
| 3796 | LINC00657 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p | 18 | NF1 | Sponge network | 0.91 | 0 | 0.51 | 0 | 0.579 |
| 3797 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-424-5p | 19 | RUNX1T1 | Sponge network | 1.344 | 0 | -0.344 | 0.2374 | 0.579 |
| 3798 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-590-3p | 16 | ATRX | Sponge network | 0.178 | 0.29106 | 0.261 | 0.04408 | 0.579 |
| 3799 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-93-3p | 43 | RUNX1T1 | Sponge network | -0.469 | 0.08721 | -0.344 | 0.2374 | 0.579 |
| 3800 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-96-5p | 51 | FAT3 | Sponge network | 0.246 | 0.59912 | -1.601 | 0.00051 | 0.579 |
| 3801 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 22 | FGFR1 | Sponge network | 0.997 | 0.00066 | -0.509 | 0.03957 | 0.579 |
| 3802 | ANKRD10-IT1 |
hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p | 14 | RICTOR | Sponge network | 1.739 | 0 | 0.388 | 0.00188 | 0.579 |
| 3803 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | EDA2R | Sponge network | -1.111 | 0.0003 | -0.587 | 0.01598 | 0.579 |
| 3804 | MAGI2-AS3 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | TPO | Sponge network | -0.129 | 0.57177 | -1.87 | 2.0E-5 | 0.579 |
| 3805 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TIMP3 | Sponge network | 0.335 | 0.20596 | -0.546 | 0.02307 | 0.579 |
| 3806 | HOXB-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-671-5p | 24 | DLC1 | Sponge network | -0.154 | 0.53954 | -0.382 | 0.05038 | 0.579 |
| 3807 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 73 | LPP | Sponge network | -2.69 | 0.0001 | -0.459 | 0.04475 | 0.579 |
| 3808 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 32 | GRIN2A | Sponge network | -2.925 | 0 | -2.161 | 0 | 0.579 |
| 3809 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 17 | RCAN2 | Sponge network | -0.709 | 0.18168 | -0.33 | 0.15172 | 0.578 |
| 3810 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429 | 14 | RCAN2 | Sponge network | -2.62 | 0 | -0.33 | 0.15172 | 0.578 |
| 3811 | CTD-2554C21.2 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-96-5p | 13 | SFRP1 | Sponge network | -1.654 | 0.00144 | -3.566 | 0 | 0.578 |
| 3812 | MAGI2-AS3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 22 | KLF10 | Sponge network | -0.129 | 0.57177 | -0.783 | 0 | 0.578 |
| 3813 | PWAR6 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429 | 12 | NGFR | Sponge network | -1.575 | 6.0E-5 | -1.271 | 0.01058 | 0.578 |
| 3814 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-542-3p | 29 | DIP2C | Sponge network | -2.69 | 0.0001 | -0.436 | 0.01713 | 0.578 |
| 3815 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-96-5p | 11 | CRTC3 | Sponge network | -0.187 | 0.23787 | 0.164 | 0.16786 | 0.578 |
| 3816 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-577 | 19 | CNTN1 | Sponge network | -0.473 | 0.07602 | -2.594 | 0 | 0.578 |
| 3817 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | -0.096 | 0.67958 | -1.74 | 0 | 0.578 |
| 3818 | CTA-204B4.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-766-3p | 35 | MDM4 | Sponge network | 0.765 | 7.0E-5 | 0.345 | 0.00762 | 0.578 |
| 3819 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-664a-3p | 18 | AKAP6 | Sponge network | 0.811 | 0.03228 | -1.586 | 3.0E-5 | 0.578 |
| 3820 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-485-3p;hsa-miR-590-3p | 10 | LRRK2 | Sponge network | -0.896 | 0.00099 | -1.136 | 1.0E-5 | 0.578 |
| 3821 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-769-5p | 23 | SETBP1 | Sponge network | -0.096 | 0.67958 | -1.302 | 1.0E-5 | 0.578 |
| 3822 | LINC00641 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p | 22 | CSDE1 | Sponge network | -0.222 | 0.32445 | -0.493 | 0 | 0.578 |
| 3823 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 44 | TGFBR3 | Sponge network | -0.129 | 0.57177 | -1.173 | 1.0E-5 | 0.578 |
| 3824 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 24 | CBFA2T3 | Sponge network | -2.452 | 0 | -1.221 | 1.0E-5 | 0.578 |
| 3825 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 57 | EPHA5 | Sponge network | -1.111 | 0.0003 | -0.957 | 0.01733 | 0.578 |
| 3826 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | PREX2 | Sponge network | -0.49 | 0.04946 | -0.272 | 0.25382 | 0.578 |
| 3827 | BVES-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-935 | 13 | RELN | Sponge network | -2.62 | 0 | -2.403 | 0 | 0.578 |
| 3828 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | TSHZ3 | Sponge network | -1.349 | 0.00017 | -0.556 | 0.01516 | 0.578 |
| 3829 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 57 | DTNA | Sponge network | 0.997 | 0.00066 | -1.648 | 1.0E-5 | 0.578 |
| 3830 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-98-5p | 41 | PBX1 | Sponge network | -1.281 | 0.00012 | -1.206 | 0 | 0.578 |
| 3831 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 39 | ZFHX3 | Sponge network | 0.537 | 0.00139 | 0.389 | 0.01363 | 0.578 |
| 3832 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-629-3p;hsa-miR-942-5p | 28 | GPC6 | Sponge network | 0.249 | 0.35902 | -0.054 | 0.81465 | 0.578 |
| 3833 | CTC-429P9.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-744-3p | 41 | LPP | Sponge network | 0.178 | 0.29106 | -0.459 | 0.04475 | 0.578 |
| 3834 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p | 21 | NR2C2 | Sponge network | 0.75 | 0.00054 | 0.21 | 0.03224 | 0.578 |
| 3835 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p | 35 | DTNA | Sponge network | -1.654 | 0.00144 | -1.648 | 1.0E-5 | 0.578 |
| 3836 | PWAR6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 53 | GAS7 | Sponge network | -1.575 | 6.0E-5 | -0.555 | 0.01602 | 0.578 |
| 3837 | MIR143HG |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 29 | GJA1 | Sponge network | -0.739 | 0.0574 | -0.485 | 0.02179 | 0.578 |
| 3838 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 11 | SYNE1 | Sponge network | -2.039 | 0.03447 | -0.738 | 0.00316 | 0.578 |
| 3839 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-877-5p;hsa-miR-93-5p | 29 | PGR | Sponge network | -0.842 | 0.01361 | -1.704 | 0 | 0.578 |
| 3840 | EMX2OS |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-877-5p | 34 | ASTN1 | Sponge network | -0.777 | 0.1134 | -2.389 | 2.0E-5 | 0.578 |
| 3841 | MIR497HG |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | DIP2C | Sponge network | -1.439 | 4.0E-5 | -0.436 | 0.01713 | 0.578 |
| 3842 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-508-3p | 16 | SYNE1 | Sponge network | -0.154 | 0.53954 | -0.738 | 0.00316 | 0.578 |
| 3843 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-93-5p | 13 | LHX6 | Sponge network | 0.727 | 3.0E-5 | -0.087 | 0.69193 | 0.578 |
| 3844 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-324-3p | 16 | ZBTB4 | Sponge network | -4.546 | 0 | -0.739 | 0 | 0.578 |
| 3845 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-93-5p | 21 | RASSF2 | Sponge network | -0.473 | 0.07602 | -0.273 | 0.21961 | 0.578 |
| 3846 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 20 | TMEFF2 | Sponge network | -2.46 | 0 | -2.426 | 0 | 0.578 |
| 3847 | USP3-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-877-5p | 22 | NRK | Sponge network | -0.162 | 0.47687 | 0.656 | 0.19217 | 0.578 |
| 3848 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 17 | TXNIP | Sponge network | -1.111 | 0.0003 | -1.277 | 0 | 0.578 |
| 3849 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p | 10 | HMCN1 | Sponge network | -0.611 | 0.09607 | 0.809 | 0.00544 | 0.578 |
| 3850 | MRVI1-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 11 | GJA1 | Sponge network | -1.092 | 4.0E-5 | -0.485 | 0.02179 | 0.578 |
| 3851 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-93-5p | 19 | PGR | Sponge network | -0.473 | 0.07602 | -1.704 | 0 | 0.578 |
| 3852 | RP5-1198O20.4 |
hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3127-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-664a-3p | 10 | PDGFRB | Sponge network | 0.997 | 0.00066 | 1.588 | 0 | 0.578 |
| 3853 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 35 | LIFR | Sponge network | -2.452 | 0 | -1.794 | 0 | 0.578 |
| 3854 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92b-3p | 17 | AKAP12 | Sponge network | -0.323 | 0.01372 | -0.932 | 0.0028 | 0.578 |
| 3855 | FAM66C |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LHFP | Sponge network | -0.901 | 0.00019 | -0.167 | 0.41371 | 0.577 |
| 3856 | RP11-400K9.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-671-5p | 25 | DLC1 | Sponge network | -0.611 | 0.09607 | -0.382 | 0.05038 | 0.577 |
| 3857 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 23 | RASSF8 | Sponge network | -0.896 | 0.00099 | -0.122 | 0.65591 | 0.577 |
| 3858 | LINC00865 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429 | 11 | PTPN13 | Sponge network | -1.249 | 7.0E-5 | -1.208 | 0 | 0.577 |
| 3859 | MAGI2-AS3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 49 | GRIK3 | Sponge network | -0.129 | 0.57177 | -3.208 | 0 | 0.577 |
| 3860 | MEG3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p | 18 | MRVI1 | Sponge network | 0.249 | 0.35902 | -1.051 | 0.00022 | 0.577 |
| 3861 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 26 | LATS1 | Sponge network | 0.683 | 1.0E-5 | 0.109 | 0.34208 | 0.577 |
| 3862 | CTD-2314G24.2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 30 | DLC1 | Sponge network | 0.246 | 0.59912 | -0.382 | 0.05038 | 0.577 |
| 3863 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-93-3p | 61 | IL6ST | Sponge network | -0.896 | 0.00099 | -0.402 | 0.00676 | 0.577 |
| 3864 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 65 | NTRK3 | Sponge network | -1.575 | 6.0E-5 | -1.77 | 3.0E-5 | 0.577 |
| 3865 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-486-5p | 26 | CNTN1 | Sponge network | -0.162 | 0.47687 | -2.594 | 0 | 0.577 |
| 3866 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 29 | SOX5 | Sponge network | -0.246 | 0.35034 | -1.091 | 4.0E-5 | 0.577 |
| 3867 | OIP5-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 46 | CREB1 | Sponge network | 0.421 | 0.00084 | 0.203 | 0.00537 | 0.577 |
| 3868 | LINC00476 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 28 | AKAP6 | Sponge network | -0.615 | 0.00015 | -1.586 | 3.0E-5 | 0.577 |
| 3869 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | FAT3 | Sponge network | -2.915 | 0.00015 | -1.601 | 0.00051 | 0.577 |
| 3870 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | KCNAB1 | Sponge network | -2.452 | 0 | -0.755 | 2.0E-5 | 0.577 |
| 3871 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ITGB3 | Sponge network | 0.572 | 0.01722 | -0.011 | 0.95751 | 0.577 |
| 3872 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-940 | 21 | IGF1 | Sponge network | -4.546 | 0 | -0.204 | 0.5898 | 0.577 |
| 3873 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 14 | GRIN2A | Sponge network | -0.583 | 0.14589 | -2.161 | 0 | 0.577 |
| 3874 | FENDRR |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 51 | GAS7 | Sponge network | -1.281 | 0.00012 | -0.555 | 0.01602 | 0.577 |
| 3875 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 24 | GRIN2A | Sponge network | -0.842 | 0.01361 | -2.161 | 0 | 0.577 |
| 3876 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 12 | NR2C2 | Sponge network | 0.997 | 0 | 0.21 | 0.03224 | 0.577 |
| 3877 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | NR2C2 | Sponge network | 0.673 | 0 | 0.21 | 0.03224 | 0.577 |
| 3878 | DLEU2 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-486-5p | 11 | PHF6 | Sponge network | 1.831 | 0 | 0.785 | 0 | 0.577 |
| 3879 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-361-5p | 12 | SYNE1 | Sponge network | -0.309 | 0.1145 | -0.738 | 0.00316 | 0.577 |
| 3880 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p | 10 | PDZD2 | Sponge network | -0.976 | 0.00025 | -0.909 | 3.0E-5 | 0.577 |
| 3881 | PWAR6 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | -1.575 | 6.0E-5 | 0.154 | 0.74723 | 0.577 |
| 3882 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 18 | TRIM33 | Sponge network | 1.601 | 0 | 0.402 | 7.0E-5 | 0.577 |
| 3883 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 48 | RUNX1T1 | Sponge network | 0.532 | 0.00505 | -0.344 | 0.2374 | 0.577 |
| 3884 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 16 | CNTNAP1 | Sponge network | 0.246 | 0.59912 | 0.358 | 0.13727 | 0.577 |
| 3885 | LINC00702 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 14 | PHOX2B | Sponge network | -0.737 | 0.06182 | -2.68 | 0 | 0.577 |
| 3886 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-96-5p | 31 | SETBP1 | Sponge network | 0.997 | 0.00066 | -1.302 | 1.0E-5 | 0.577 |
| 3887 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-5p | 17 | RASSF8 | Sponge network | 0.176 | 0.37402 | -0.122 | 0.65591 | 0.577 |
| 3888 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 19 | ABCC9 | Sponge network | -0.793 | 7.0E-5 | -0.466 | 0.14121 | 0.577 |
| 3889 | RP11-166D19.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 47 | GRIK3 | Sponge network | -0.576 | 0.10121 | -3.208 | 0 | 0.577 |
| 3890 | FGF14-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-96-5p | 19 | ASTN1 | Sponge network | -1.582 | 8.0E-5 | -2.389 | 2.0E-5 | 0.577 |
| 3891 | MYLK-AS1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-424-5p | 14 | RPS6KA6 | Sponge network | -1.331 | 3.0E-5 | -2.989 | 0 | 0.577 |
| 3892 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 15 | CAV1 | Sponge network | -0.793 | 7.0E-5 | -1.013 | 6.0E-5 | 0.577 |
| 3893 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 50 | AR | Sponge network | -1.249 | 7.0E-5 | -1.554 | 0 | 0.577 |
| 3894 | TPTEP1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 18 | MAPK10 | Sponge network | -1.216 | 0 | -1.774 | 0 | 0.577 |
| 3895 | ADAMTS9-AS2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 19 | KLF10 | Sponge network | -2.452 | 0 | -0.783 | 0 | 0.577 |
| 3896 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-5p | 36 | TSHZ3 | Sponge network | -0.793 | 7.0E-5 | -0.556 | 0.01516 | 0.577 |
| 3897 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-3p | 10 | CNTNAP1 | Sponge network | -0.739 | 0.00044 | 0.358 | 0.13727 | 0.577 |
| 3898 | ZNF582-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-96-5p | 25 | ASTN1 | Sponge network | -1.349 | 0.00017 | -2.389 | 2.0E-5 | 0.577 |
| 3899 | PWAR6 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ADH1B | Sponge network | -1.575 | 6.0E-5 | -3.716 | 0 | 0.576 |
| 3900 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | LRFN5 | Sponge network | -2.46 | 0 | -1.483 | 0 | 0.576 |
| 3901 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | RECK | Sponge network | 0.997 | 0.00066 | -1.043 | 0 | 0.576 |
| 3902 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | PLSCR4 | Sponge network | -0.942 | 0 | -0.474 | 0.03833 | 0.576 |
| 3903 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 38 | PRKAA2 | Sponge network | -0.246 | 0.35034 | -2.462 | 0 | 0.576 |
| 3904 | RP11-25K19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-429 | 12 | CBFA2T3 | Sponge network | -1.057 | 0.00029 | -1.221 | 1.0E-5 | 0.576 |
| 3905 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-940;hsa-miR-942-5p | 27 | HLF | Sponge network | -4.546 | 0 | -2.443 | 0 | 0.576 |
| 3906 | ZNF582-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 16 | CXCL12 | Sponge network | -1.349 | 0.00017 | -1.421 | 0 | 0.576 |
| 3907 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-628-5p;hsa-miR-93-3p | 35 | RUNX1T1 | Sponge network | -0.154 | 0.53954 | -0.344 | 0.2374 | 0.576 |
| 3908 | GNG12-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 10 | ADAMTS8 | Sponge network | -0.793 | 7.0E-5 | -1.476 | 0.00069 | 0.576 |
| 3909 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 50 | GAS7 | Sponge network | -0.563 | 0.02545 | -0.555 | 0.01602 | 0.576 |
| 3910 | MRVI1-AS1 |
hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-374b-3p | 10 | RPS6KA6 | Sponge network | -1.092 | 4.0E-5 | -2.989 | 0 | 0.576 |
| 3911 | RP5-1198O20.4 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | 0.997 | 0.00066 | 0.524 | 0.00017 | 0.576 |
| 3912 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p | 26 | FBXO32 | Sponge network | -0.611 | 0.09607 | -0.495 | 0.07561 | 0.576 |
| 3913 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | HOOK3 | Sponge network | -0.222 | 0.32445 | 0.273 | 0.09957 | 0.576 |
| 3914 | RP11-532F6.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | -0.896 | 0.00099 | -1.892 | 0 | 0.576 |
| 3915 | JAZF1-AS1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | RNF180 | Sponge network | -0.769 | 0.00035 | -1.127 | 1.0E-5 | 0.576 |
| 3916 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 33 | RSPO2 | Sponge network | -0.49 | 0.04946 | -3.038 | 0 | 0.576 |
| 3917 | CTC-429P9.5 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | ATM | Sponge network | 0.178 | 0.29106 | 0.178 | 0.21568 | 0.576 |
| 3918 | RP11-166D19.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 24 | GJA1 | Sponge network | -0.576 | 0.10121 | -0.485 | 0.02179 | 0.576 |
| 3919 | BVES-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-589-5p | 19 | PRKCB | Sponge network | -2.62 | 0 | -1.313 | 2.0E-5 | 0.576 |
| 3920 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | NRXN3 | Sponge network | -0.323 | 0.01372 | -0.893 | 0.04186 | 0.576 |
| 3921 | FAM66C |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-5p | 14 | PDZD2 | Sponge network | -0.901 | 0.00019 | -0.909 | 3.0E-5 | 0.576 |
| 3922 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 10 | DCN | Sponge network | -0.376 | 0.00337 | -1.288 | 0 | 0.576 |
| 3923 | CTC-296K1.3 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-454-3p;hsa-miR-493-5p | 30 | AR | Sponge network | -2.095 | 0.00112 | -1.554 | 0 | 0.576 |
| 3924 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 51 | CHD5 | Sponge network | -0.739 | 0.0574 | -0.922 | 0.01521 | 0.576 |
| 3925 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-502-5p;hsa-miR-624-5p | 20 | ANK2 | Sponge network | -0.517 | 0.02935 | -1.603 | 1.0E-5 | 0.576 |
| 3926 | GNG12-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 57 | CADM2 | Sponge network | -0.793 | 7.0E-5 | -3.782 | 0 | 0.576 |
| 3927 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | SYNE1 | Sponge network | -0.421 | 0.0114 | -0.738 | 0.00316 | 0.576 |
| 3928 | RP11-867G23.10 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-874-3p | 22 | ASTN1 | Sponge network | -2.531 | 0 | -2.389 | 2.0E-5 | 0.576 |
| 3929 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-93-5p | 20 | SYNE1 | Sponge network | -1.654 | 0.00144 | -0.738 | 0.00316 | 0.576 |
| 3930 | LINC00641 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 43 | ABI2 | Sponge network | -0.222 | 0.32445 | 0.175 | 0.14787 | 0.576 |
| 3931 | CTC-559E9.5 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-409-3p;hsa-miR-576-5p | 13 | ATM | Sponge network | 0.594 | 0.00064 | 0.178 | 0.21568 | 0.576 |
| 3932 | FENDRR |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 25 | CDH19 | Sponge network | -1.281 | 0.00012 | -3.434 | 0 | 0.576 |
| 3933 | SNHG14 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 20 | PAK3 | Sponge network | -1.111 | 0.0003 | -0.628 | 0.07253 | 0.576 |
| 3934 | RP11-307B6.3 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-424-5p | 10 | SYNE1 | Sponge network | -4.463 | 0.00025 | -0.738 | 0.00316 | 0.576 |
| 3935 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 38 | MYO9A | Sponge network | -2.452 | 0 | -0.147 | 0.32373 | 0.575 |
| 3936 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-625-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | 0.246 | 0.59912 | 0.024 | 0.89889 | 0.575 |
| 3937 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | FGF5 | Sponge network | 0.572 | 0.01722 | 0.549 | 0.28659 | 0.575 |
| 3938 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | FAT4 | Sponge network | 0.997 | 0.00066 | -0.354 | 0.14694 | 0.575 |
| 3939 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-455-3p | 13 | TSHZ3 | Sponge network | -0.309 | 0.1145 | -0.556 | 0.01516 | 0.575 |
| 3940 | LINC00578 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-24-3p;hsa-miR-362-3p | 13 | DLC1 | Sponge network | 0.811 | 0.03228 | -0.382 | 0.05038 | 0.575 |
| 3941 | LINC01018 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | SETBP1 | Sponge network | -2.915 | 0.00015 | -1.302 | 1.0E-5 | 0.575 |
| 3942 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | SEMA3C | Sponge network | -0.576 | 0.10121 | -0.272 | 0.31153 | 0.575 |
| 3943 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 41 | RUNX1T1 | Sponge network | -0.739 | 0.00044 | -0.344 | 0.2374 | 0.575 |
| 3944 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-664a-3p | 10 | PRUNE2 | Sponge network | 0.811 | 0.03228 | -1.733 | 0.00022 | 0.575 |
| 3945 | SNX29P2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 15 | POU2F2 | Sponge network | 0.4 | 0.14611 | -0.233 | 0.32214 | 0.575 |
| 3946 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-98-5p | 47 | NTRK3 | Sponge network | -2.62 | 0 | -1.77 | 3.0E-5 | 0.575 |
| 3947 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | SLITRK3 | Sponge network | -1.216 | 0 | -3.328 | 0 | 0.575 |
| 3948 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | FAT4 | Sponge network | -1.575 | 6.0E-5 | -0.354 | 0.14694 | 0.575 |
| 3949 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 33 | KCNA6 | Sponge network | -1.697 | 0.00889 | -1.531 | 0.00013 | 0.575 |
| 3950 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 36 | DIP2C | Sponge network | -0.976 | 0.00025 | -0.436 | 0.01713 | 0.575 |
| 3951 | RP11-25K19.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-874-3p | 16 | SORCS1 | Sponge network | -1.057 | 0.00029 | -3.492 | 0 | 0.575 |
| 3952 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-576-5p | 19 | GREM1 | Sponge network | 0.194 | 0.64278 | 0.902 | 0.01691 | 0.575 |
| 3953 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | AHNAK | Sponge network | -0.487 | 0.02157 | -1.207 | 0 | 0.575 |
| 3954 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | RASSF8 | Sponge network | -1.281 | 0.00012 | -0.122 | 0.65591 | 0.575 |
| 3955 | RP11-1112J20.2 |
hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p | 12 | PHF6 | Sponge network | 1.416 | 0 | 0.785 | 0 | 0.575 |
| 3956 | SNHG14 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 32 | RYR2 | Sponge network | -1.111 | 0.0003 | -1.466 | 0.00031 | 0.575 |
| 3957 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 27 | AKAP6 | Sponge network | -0.405 | 0.22013 | -1.586 | 3.0E-5 | 0.575 |
| 3958 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-505-3p;hsa-miR-671-5p | 21 | KIT | Sponge network | -0.976 | 0.00025 | -2.184 | 0 | 0.575 |
| 3959 | RP11-156E6.1 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p | 18 | PHF6 | Sponge network | 1.266 | 0 | 0.785 | 0 | 0.575 |
| 3960 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | PLSCR4 | Sponge network | -0.777 | 0.1134 | -0.474 | 0.03833 | 0.575 |
| 3961 | AC006116.24 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | FAT4 | Sponge network | -0.473 | 0.07602 | -0.354 | 0.14694 | 0.575 |
| 3962 | MIR143HG |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 15 | PTPN13 | Sponge network | -0.739 | 0.0574 | -1.208 | 0 | 0.575 |
| 3963 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-98-5p | 10 | ADAMTS8 | Sponge network | -1.697 | 0.00889 | -1.476 | 0.00069 | 0.575 |
| 3964 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | -0.793 | 7.0E-5 | 0.358 | 0.13727 | 0.575 |
| 3965 | RP11-819C21.1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 15 | SHPRH | Sponge network | 0.537 | 0.00139 | 0.098 | 0.40994 | 0.575 |
| 3966 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-501-3p;hsa-miR-502-3p | 19 | RUNX1T1 | Sponge network | -1.255 | 0.00981 | -0.344 | 0.2374 | 0.575 |
| 3967 | RP11-620J15.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p | 24 | ASTN1 | Sponge network | -0.739 | 0.00044 | -2.389 | 2.0E-5 | 0.575 |
| 3968 | PWAR6 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | LHFP | Sponge network | -1.575 | 6.0E-5 | -0.167 | 0.41371 | 0.575 |
| 3969 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-582-5p | 21 | TSHZ3 | Sponge network | -0.469 | 0.08721 | -0.556 | 0.01516 | 0.575 |
| 3970 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 59 | IL6ST | Sponge network | -0.769 | 0.00035 | -0.402 | 0.00676 | 0.575 |
| 3971 | MAGI2-AS3 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ITGB1 | Sponge network | -0.129 | 0.57177 | 0.524 | 0.00017 | 0.575 |
| 3972 | CTC-296K1.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p | 13 | AKAP6 | Sponge network | -2.095 | 0.00112 | -1.586 | 3.0E-5 | 0.575 |
| 3973 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-3p | 11 | MITF | Sponge network | -0.932 | 0.00329 | -0.769 | 0.00022 | 0.575 |
| 3974 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-361-5p;hsa-miR-671-5p | 12 | DLC1 | Sponge network | -0.309 | 0.1145 | -0.382 | 0.05038 | 0.574 |
| 3975 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-7-1-3p | 14 | EPHA3 | Sponge network | 0.693 | 0.00076 | 0.185 | 0.53588 | 0.574 |
| 3976 | ZNF667-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 13 | ID4 | Sponge network | -0.976 | 0.00025 | -1.644 | 0 | 0.574 |
| 3977 | AC005154.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 55 | MDM4 | Sponge network | 0.848 | 0 | 0.345 | 0.00762 | 0.574 |
| 3978 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940;hsa-miR-98-5p | 38 | PBX1 | Sponge network | -0.129 | 0.57177 | -1.206 | 0 | 0.574 |
| 3979 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 53 | RUNX1T1 | Sponge network | -0.096 | 0.67958 | -0.344 | 0.2374 | 0.574 |
| 3980 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | GPC6 | Sponge network | -1.375 | 0.08642 | -0.054 | 0.81465 | 0.574 |
| 3981 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 28 | ABI2 | Sponge network | 0.225 | 0.30644 | 0.175 | 0.14787 | 0.574 |
| 3982 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 35 | ZFHX3 | Sponge network | 0.75 | 0.00054 | 0.389 | 0.01363 | 0.574 |
| 3983 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | TSHZ3 | Sponge network | -0.777 | 0.1134 | -0.556 | 0.01516 | 0.574 |
| 3984 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 34 | TET1 | Sponge network | -1.111 | 0.0003 | -0.135 | 0.52138 | 0.574 |
| 3985 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-589-3p | 18 | PGR | Sponge network | -0.154 | 0.53954 | -1.704 | 0 | 0.574 |
| 3986 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-7-1-3p | 11 | TLL1 | Sponge network | -0.611 | 0.09607 | -1.195 | 3.0E-5 | 0.574 |
| 3987 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | AFF3 | Sponge network | -0.737 | 0.06182 | -2.373 | 0 | 0.574 |
| 3988 | GNG12-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 13 | DCN | Sponge network | -0.793 | 7.0E-5 | -1.288 | 0 | 0.574 |
| 3989 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | PGR | Sponge network | -0.739 | 0.00044 | -1.704 | 0 | 0.574 |
| 3990 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-495-3p | 10 | TXNIP | Sponge network | -0.896 | 0.00099 | -1.277 | 0 | 0.574 |
| 3991 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-429 | 24 | DMD | Sponge network | -1.654 | 0.00144 | -1.506 | 0 | 0.574 |
| 3992 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3913-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-93-5p | 32 | ZBTB4 | Sponge network | -0.096 | 0.67958 | -0.739 | 0 | 0.574 |
| 3993 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 35 | CNTN1 | Sponge network | -1.216 | 0 | -2.594 | 0 | 0.574 |
| 3994 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 12 | PRUNE2 | Sponge network | -0.358 | 0.17255 | -1.733 | 0.00022 | 0.574 |
| 3995 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | AKAP6 | Sponge network | 0.064 | 0.72934 | -1.586 | 3.0E-5 | 0.574 |
| 3996 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 13 | RASSF8 | Sponge network | -2.531 | 0 | -0.122 | 0.65591 | 0.574 |
| 3997 | SNHG14 |
hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-502-5p | 11 | ELN | Sponge network | -1.111 | 0.0003 | 0.451 | 0.13752 | 0.574 |
| 3998 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 12 | HERC2 | Sponge network | -0.222 | 0.32445 | 0.114 | 0.30638 | 0.574 |
| 3999 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 44 | RBMS3 | Sponge network | -0.563 | 0.02545 | -0.519 | 0.03551 | 0.574 |
| 4000 | FENDRR |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 15 | RHOB | Sponge network | -1.281 | 0.00012 | -1.052 | 0 | 0.574 |
| 4001 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-590-3p | 14 | NR2C2 | Sponge network | 0.539 | 0.03722 | 0.21 | 0.03224 | 0.574 |
| 4002 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 28 | SYNPO2 | Sponge network | -0.323 | 0.01372 | -2.342 | 0 | 0.574 |
| 4003 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | GRIN2A | Sponge network | -2.915 | 0.00015 | -2.161 | 0 | 0.574 |
| 4004 | RP4-613B23.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-539-5p | 11 | DIP2C | Sponge network | -0.309 | 0.1145 | -0.436 | 0.01713 | 0.574 |
| 4005 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-450b-5p;hsa-miR-502-5p | 34 | AR | Sponge network | -0.517 | 0.02935 | -1.554 | 0 | 0.574 |
| 4006 | LINC00702 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 13 | PTPN13 | Sponge network | -0.737 | 0.06182 | -1.208 | 0 | 0.574 |
| 4007 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 40 | MCC | Sponge network | -1.216 | 0 | -1.005 | 0 | 0.574 |
| 4008 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 37 | RBMS3 | Sponge network | -1.161 | 0.00591 | -0.519 | 0.03551 | 0.574 |
| 4009 | RP5-1074L1.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 25 | MDM4 | Sponge network | 1.923 | 0 | 0.345 | 0.00762 | 0.574 |
| 4010 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 31 | ZBTB4 | Sponge network | -2.46 | 0 | -0.739 | 0 | 0.574 |
| 4011 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 24 | ADAMTSL3 | Sponge network | -1.203 | 0.03881 | -1.559 | 0 | 0.574 |
| 4012 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p | 18 | FGFR1 | Sponge network | -0.901 | 0.00019 | -0.509 | 0.03957 | 0.574 |
| 4013 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 14 | AKAP12 | Sponge network | -0.896 | 0.00099 | -0.932 | 0.0028 | 0.574 |
| 4014 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | PTPRB | Sponge network | -0.358 | 0.17255 | 0.105 | 0.47122 | 0.574 |
| 4015 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-654-3p;hsa-miR-874-3p | 19 | NR2C2 | Sponge network | 0.946 | 0 | 0.21 | 0.03224 | 0.574 |
| 4016 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | ABCB5 | Sponge network | -1.697 | 0.00889 | -0.956 | 0.04077 | 0.574 |
| 4017 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-3p | 11 | FBXO32 | Sponge network | -0.309 | 0.1145 | -0.495 | 0.07561 | 0.574 |
| 4018 | FENDRR |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ITPKB | Sponge network | -1.281 | 0.00012 | -0.704 | 0.00106 | 0.574 |
| 4019 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | EDA2R | Sponge network | 0.335 | 0.20596 | -0.587 | 0.01598 | 0.574 |
| 4020 | BVES-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-429 | 24 | DLC1 | Sponge network | -2.62 | 0 | -0.382 | 0.05038 | 0.573 |
| 4021 | BVES-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-629-3p | 17 | RYR2 | Sponge network | -2.62 | 0 | -1.466 | 0.00031 | 0.573 |
| 4022 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | RECK | Sponge network | -0.811 | 2.0E-5 | -1.043 | 0 | 0.573 |
| 4023 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b | 10 | SLITRK3 | Sponge network | -0.583 | 0.14589 | -3.328 | 0 | 0.573 |
| 4024 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 40 | PBX1 | Sponge network | -0.737 | 0.06182 | -1.206 | 0 | 0.573 |
| 4025 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | TACC1 | Sponge network | -1.439 | 4.0E-5 | -0.362 | 0.05406 | 0.573 |
| 4026 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-339-5p | 13 | STARD13 | Sponge network | -0.517 | 0.02935 | -0.187 | 0.27383 | 0.573 |
| 4027 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 25 | JAZF1 | Sponge network | -0.739 | 0.00044 | -0.377 | 0.02677 | 0.573 |
| 4028 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-96-5p | 29 | BNC2 | Sponge network | -0.517 | 0.02935 | -0.491 | 0.09988 | 0.573 |
| 4029 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-671-5p | 24 | RBMS3 | Sponge network | -2.095 | 0.00112 | -0.519 | 0.03551 | 0.573 |
| 4030 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 16 | TXNIP | Sponge network | -0.129 | 0.57177 | -1.277 | 0 | 0.573 |
| 4031 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-577 | 10 | CAV1 | Sponge network | -2.69 | 0.0001 | -1.013 | 6.0E-5 | 0.573 |
| 4032 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 42 | TCF4 | Sponge network | 0.889 | 9.0E-5 | 0.016 | 0.92271 | 0.573 |
| 4033 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-324-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | TSHZ3 | Sponge network | -0.739 | 0.00044 | -0.556 | 0.01516 | 0.573 |
| 4034 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 28 | CNTN1 | Sponge network | -1.249 | 7.0E-5 | -2.594 | 0 | 0.573 |
| 4035 | RP11-736K20.5 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p | 23 | ASTN1 | Sponge network | -0.358 | 0.17255 | -2.389 | 2.0E-5 | 0.573 |
| 4036 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | PLSCR4 | Sponge network | -0.611 | 0.09607 | -0.474 | 0.03833 | 0.573 |
| 4037 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -1.203 | 0.03881 | -0.354 | 0.14694 | 0.573 |
| 4038 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 69 | TCF4 | Sponge network | 0.997 | 0.00066 | 0.016 | 0.92271 | 0.573 |
| 4039 | RASSF8-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 26 | DLC1 | Sponge network | 0.335 | 0.20596 | -0.382 | 0.05038 | 0.573 |
| 4040 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p | 44 | ADARB1 | Sponge network | -0.487 | 0.02157 | -0.335 | 0.06631 | 0.573 |
| 4041 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | PDGFC | Sponge network | 0.572 | 0.01722 | -0.33 | 0.06483 | 0.573 |
| 4042 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | TIMP3 | Sponge network | -1.216 | 0 | -0.546 | 0.02307 | 0.573 |
| 4043 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-576-5p | 15 | GREM1 | Sponge network | -0.611 | 0.09607 | 0.902 | 0.01691 | 0.573 |
| 4044 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-362-3p | 19 | TIMP3 | Sponge network | -0.693 | 0.05629 | -0.546 | 0.02307 | 0.573 |
| 4045 | GNG12-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-93-5p | 60 | DST | Sponge network | -0.793 | 7.0E-5 | -0.713 | 0.00096 | 0.573 |
| 4046 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | TSHZ3 | Sponge network | 0.299 | 0.57722 | -0.556 | 0.01516 | 0.573 |
| 4047 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | FAT4 | Sponge network | -1.697 | 0.00889 | -0.354 | 0.14694 | 0.573 |
| 4048 | RP11-244O19.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 26 | PTPN14 | Sponge network | -0.096 | 0.67958 | 0.144 | 0.34595 | 0.573 |
| 4049 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-942-5p | 39 | HLF | Sponge network | -0.901 | 0.00019 | -2.443 | 0 | 0.573 |
| 4050 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-421 | 12 | NRXN3 | Sponge network | -1.654 | 0.00144 | -0.893 | 0.04186 | 0.573 |
| 4051 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 27 | SNX29 | Sponge network | -0.421 | 0.0114 | -0.34 | 0.01371 | 0.573 |
| 4052 | MYLK-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-940 | 11 | TAL1 | Sponge network | -1.331 | 3.0E-5 | -0.462 | 0.00574 | 0.573 |
| 4053 | NR2F1-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-942-5p | 15 | ZBTB20 | Sponge network | -0.49 | 0.04946 | -0.122 | 0.57569 | 0.573 |
| 4054 | GNG12-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 60 | RUNX1T1 | Sponge network | -0.793 | 7.0E-5 | -0.344 | 0.2374 | 0.573 |
| 4055 | LINC00092 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | RELN | Sponge network | -1.886 | 0 | -2.403 | 0 | 0.573 |
| 4056 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 39 | QKI | Sponge network | -0.739 | 0.00044 | -0.333 | 0.04111 | 0.573 |
| 4057 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-3p | 24 | ATRX | Sponge network | 1.254 | 0 | 0.261 | 0.04408 | 0.573 |
| 4058 | PCA3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 47 | EPHA7 | Sponge network | -0.709 | 0.18168 | -2.197 | 3.0E-5 | 0.573 |
| 4059 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p | 17 | ANK2 | Sponge network | -0.693 | 0.05629 | -1.603 | 1.0E-5 | 0.573 |
| 4060 | PART1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 64 | DTNA | Sponge network | -2.925 | 0 | -1.648 | 1.0E-5 | 0.573 |
| 4061 | RP11-894P9.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-339-5p | 11 | SON | Sponge network | 0.993 | 0 | 0.168 | 0.04406 | 0.573 |
| 4062 | MIR143HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 32 | RSPO2 | Sponge network | -0.739 | 0.0574 | -3.038 | 0 | 0.573 |
| 4063 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 41 | MYO9A | Sponge network | -1.575 | 6.0E-5 | -0.147 | 0.32373 | 0.573 |
| 4064 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | SLITRK3 | Sponge network | -2.915 | 0.00015 | -3.328 | 0 | 0.573 |
| 4065 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p | 20 | DYNC1I1 | Sponge network | -0.842 | 0.01361 | -1.12 | 0.00107 | 0.573 |
| 4066 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CACNA2D1 | Sponge network | 0.997 | 0.00066 | -0.846 | 0.00675 | 0.573 |
| 4067 | PART1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-935 | 48 | DMD | Sponge network | -2.925 | 0 | -1.506 | 0 | 0.573 |
| 4068 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | LRFN5 | Sponge network | -1.575 | 6.0E-5 | -1.483 | 0 | 0.573 |
| 4069 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 11 | LHFP | Sponge network | 0.194 | 0.64278 | -0.167 | 0.41371 | 0.573 |
| 4070 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-98-5p | 35 | THBS1 | Sponge network | -0.739 | 0.0574 | 0.741 | 0.00386 | 0.573 |
| 4071 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p | 11 | AKAP12 | Sponge network | -0.563 | 0.02545 | -0.932 | 0.0028 | 0.573 |
| 4072 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | ATRX | Sponge network | -0.031 | 0.89063 | 0.261 | 0.04408 | 0.572 |
| 4073 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 11 | FGFR1 | Sponge network | -1.582 | 8.0E-5 | -0.509 | 0.03957 | 0.572 |
| 4074 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | TRIM33 | Sponge network | 0.953 | 0 | 0.402 | 7.0E-5 | 0.572 |
| 4075 | SNHG14 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | ERG | Sponge network | -1.111 | 0.0003 | 0.002 | 0.99011 | 0.572 |
| 4076 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-93-5p | 16 | FGFR1 | Sponge network | 0.299 | 0.57722 | -0.509 | 0.03957 | 0.572 |
| 4077 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | 0.537 | 0.00139 | 0.273 | 0.09957 | 0.572 |
| 4078 | C20orf166-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p | 17 | ITPKB | Sponge network | -2.69 | 0.0001 | -0.704 | 0.00106 | 0.572 |
| 4079 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 21 | TSHZ3 | Sponge network | -0.376 | 0.00337 | -0.556 | 0.01516 | 0.572 |
| 4080 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-625-5p;hsa-miR-93-5p | 31 | JAZF1 | Sponge network | -0.842 | 0.01361 | -0.377 | 0.02677 | 0.572 |
| 4081 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 27 | TET1 | Sponge network | 0.532 | 0.00505 | -0.135 | 0.52138 | 0.572 |
| 4082 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-508-3p;hsa-miR-96-5p | 16 | CACNA2D1 | Sponge network | -1.654 | 0.00144 | -0.846 | 0.00675 | 0.572 |
| 4083 | CTD-2554C21.2 |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429 | 14 | AFF3 | Sponge network | -1.654 | 0.00144 | -2.373 | 0 | 0.572 |
| 4084 | RP11-999E24.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p | 14 | MRVI1 | Sponge network | -0.693 | 0.05629 | -1.051 | 0.00022 | 0.572 |
| 4085 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | -0.612 | 0.00094 | -1.892 | 0 | 0.572 |
| 4086 | LINC01018 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-5p | 13 | MN1 | Sponge network | -2.915 | 0.00015 | -0.358 | 0.24172 | 0.572 |
| 4087 | RP11-359E10.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-505-5p | 14 | MRVI1 | Sponge network | 0.299 | 0.57722 | -1.051 | 0.00022 | 0.572 |
| 4088 | PCA3 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -0.709 | 0.18168 | -0.564 | 1.0E-5 | 0.572 |
| 4089 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-98-5p | 13 | SCN11A | Sponge network | -0.49 | 0.04946 | -1.15 | 0.00299 | 0.572 |
| 4090 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-590-3p | 20 | RBMS3 | Sponge network | 0.178 | 0.29106 | -0.519 | 0.03551 | 0.572 |
| 4091 | ZRANB2-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 10 | SYNPO2 | Sponge network | -0.376 | 0.00337 | -2.342 | 0 | 0.572 |
| 4092 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 53 | SSBP2 | Sponge network | -1.281 | 0.00012 | -1.58 | 0 | 0.572 |
| 4093 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 32 | ABI2 | Sponge network | 0.214 | 0.18246 | 0.175 | 0.14787 | 0.572 |
| 4094 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-424-5p;hsa-miR-486-5p | 12 | DYNC1I1 | Sponge network | 0.176 | 0.37402 | -1.12 | 0.00107 | 0.572 |
| 4095 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-590-3p | 16 | SPG20 | Sponge network | -0.323 | 0.01372 | -1.16 | 0 | 0.572 |
| 4096 | FENDRR |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-590-3p | 20 | SPG20 | Sponge network | -1.281 | 0.00012 | -1.16 | 0 | 0.572 |
| 4097 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 33 | IGF1 | Sponge network | -0.777 | 0.1134 | -0.204 | 0.5898 | 0.572 |
| 4098 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p | 25 | ABL2 | Sponge network | 0.72 | 0.0001 | 0.82 | 0 | 0.572 |
| 4099 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-744-3p;hsa-miR-93-3p | 38 | NTRK3 | Sponge network | -0.693 | 0.05629 | -1.77 | 3.0E-5 | 0.572 |
| 4100 | PWAR6 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 32 | WWTR1 | Sponge network | -1.575 | 6.0E-5 | -0.345 | 0.11997 | 0.572 |
| 4101 | MYLK-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-7-5p | 19 | ERC1 | Sponge network | -1.331 | 3.0E-5 | -0.108 | 0.38794 | 0.572 |
| 4102 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 35 | PBX1 | Sponge network | -0.096 | 0.67958 | -1.206 | 0 | 0.572 |
| 4103 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-940 | 64 | NTRK3 | Sponge network | -0.901 | 0.00019 | -1.77 | 3.0E-5 | 0.572 |
| 4104 | LINC00472 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p | 24 | SYNPO2 | Sponge network | -0.842 | 0.01361 | -2.342 | 0 | 0.572 |
| 4105 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DNAJB4 | Sponge network | 0.194 | 0.64278 | -0.7 | 1.0E-5 | 0.572 |
| 4106 | ZNF582-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-940 | 30 | GRIK3 | Sponge network | -1.349 | 0.00017 | -3.208 | 0 | 0.572 |
| 4107 | RP11-166D19.1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 12 | PHOX2B | Sponge network | -0.576 | 0.10121 | -2.68 | 0 | 0.572 |
| 4108 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | TIMP3 | Sponge network | -1.349 | 0.00017 | -0.546 | 0.02307 | 0.572 |
| 4109 | AC011526.1 |
hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 10 | CXCL12 | Sponge network | 0.342 | 0.03129 | -1.421 | 0 | 0.572 |
| 4110 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 22 | FGFR1 | Sponge network | -1.281 | 0.00012 | -0.509 | 0.03957 | 0.572 |
| 4111 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-877-5p | 27 | GRIN2A | Sponge network | -0.162 | 0.47687 | -2.161 | 0 | 0.572 |
| 4112 | GNG12-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | KCTD8 | Sponge network | -0.793 | 7.0E-5 | -3.359 | 0 | 0.572 |
| 4113 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | TLL1 | Sponge network | -2.117 | 0.00161 | -1.195 | 3.0E-5 | 0.572 |
| 4114 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 30 | EPHA3 | Sponge network | -0.487 | 0.02157 | 0.185 | 0.53588 | 0.572 |
| 4115 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 41 | DIP2C | Sponge network | 0.194 | 0.64278 | -0.436 | 0.01713 | 0.572 |
| 4116 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 16 | CHD1 | Sponge network | 1.254 | 0 | 0.322 | 0.00215 | 0.572 |
| 4117 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 71 | IL6ST | Sponge network | 0.194 | 0.64278 | -0.402 | 0.00676 | 0.572 |
| 4118 | MYLK-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-935 | 15 | RAP1A | Sponge network | -1.331 | 3.0E-5 | -0.929 | 0 | 0.572 |
| 4119 | ADAMTS9-AS2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p | 11 | DUSP1 | Sponge network | -2.452 | 0 | -1.754 | 0 | 0.572 |
| 4120 | RP11-890B15.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-96-5p | 85 | LPP | Sponge network | 0.101 | 0.60919 | -0.459 | 0.04475 | 0.572 |
| 4121 | ZNF667-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 14 | STAT5B | Sponge network | -0.976 | 0.00025 | -0.182 | 0.1082 | 0.572 |
| 4122 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 52 | DTNA | Sponge network | -0.405 | 0.22013 | -1.648 | 1.0E-5 | 0.572 |
| 4123 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-625-5p | 19 | RSPO2 | Sponge network | -2.69 | 0.0001 | -3.038 | 0 | 0.572 |
| 4124 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-539-5p | 19 | DMD | Sponge network | -0.309 | 0.1145 | -1.506 | 0 | 0.572 |
| 4125 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-486-5p | 10 | ADAMTSL3 | Sponge network | -0.309 | 0.1145 | -1.559 | 0 | 0.572 |
| 4126 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-495-3p | 14 | DICER1 | Sponge network | 0.993 | 0 | 0.042 | 0.674 | 0.572 |
| 4127 | LINC00865 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 14 | SEMA3E | Sponge network | -1.249 | 7.0E-5 | -1.717 | 0.00137 | 0.572 |
| 4128 | TRAF3IP2-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | MACF1 | Sponge network | -0.323 | 0.01372 | 0.019 | 0.90335 | 0.571 |
| 4129 | CTC-444N24.11 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p | 33 | ABL2 | Sponge network | 1.153 | 0 | 0.82 | 0 | 0.571 |
| 4130 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 70 | RUNX1T1 | Sponge network | -2.46 | 0 | -0.344 | 0.2374 | 0.571 |
| 4131 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-577 | 13 | AMPH | Sponge network | -2.69 | 0.0001 | -0.916 | 5.0E-5 | 0.571 |
| 4132 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 31 | PGR | Sponge network | -0.709 | 0.18168 | -1.704 | 0 | 0.571 |
| 4133 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | NALCN | Sponge network | 0.246 | 0.59912 | 1.068 | 0.00237 | 0.571 |
| 4134 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 22 | LATS2 | Sponge network | -0.576 | 0.10121 | 0.159 | 0.2304 | 0.571 |
| 4135 | PWAR6 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 32 | DLC1 | Sponge network | -1.575 | 6.0E-5 | -0.382 | 0.05038 | 0.571 |
| 4136 | PCA3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 36 | TGFBR3 | Sponge network | -0.709 | 0.18168 | -1.173 | 1.0E-5 | 0.571 |
| 4137 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | NUAK1 | Sponge network | 0.572 | 0.01722 | 0.904 | 0 | 0.571 |
| 4138 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 17 | NR2C2 | Sponge network | 0.959 | 0 | 0.21 | 0.03224 | 0.571 |
| 4139 | MIR143HG |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 12 | SORCS2 | Sponge network | -0.739 | 0.0574 | -0.223 | 0.4253 | 0.571 |
| 4140 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 14 | JAKMIP2 | Sponge network | -2.117 | 0.00161 | -1.388 | 0 | 0.571 |
| 4141 | MIR143HG |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 26 | SEMA3C | Sponge network | -0.739 | 0.0574 | -0.272 | 0.31153 | 0.571 |
| 4142 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-942-5p | 41 | HLF | Sponge network | 0.194 | 0.64278 | -2.443 | 0 | 0.571 |
| 4143 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p | 64 | IL6ST | Sponge network | 0.532 | 0.00505 | -0.402 | 0.00676 | 0.571 |
| 4144 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 32 | SCN9A | Sponge network | 0.532 | 0.00505 | -0.525 | 0.10593 | 0.571 |
| 4145 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 27 | KIT | Sponge network | -0.129 | 0.57177 | -2.184 | 0 | 0.571 |
| 4146 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-455-3p;hsa-miR-671-5p | 12 | CADM3 | Sponge network | -0.896 | 0.00099 | -3.663 | 0 | 0.571 |
| 4147 | SNHG14 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p;hsa-miR-624-5p | 14 | SMARCA1 | Sponge network | -1.111 | 0.0003 | -0.684 | 0.00159 | 0.571 |
| 4148 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p | 13 | ITGB3 | Sponge network | -0.611 | 0.09607 | -0.011 | 0.95751 | 0.571 |
| 4149 | AP001347.6 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-590-3p | 12 | MAPK10 | Sponge network | -1.161 | 0.00591 | -1.774 | 0 | 0.571 |
| 4150 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-708-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 32 | GRIA3 | Sponge network | -2.452 | 0 | -1.956 | 0 | 0.571 |
| 4151 | MIR143HG |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 18 | PDZD2 | Sponge network | -0.739 | 0.0574 | -0.909 | 3.0E-5 | 0.571 |
| 4152 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940 | 30 | CRTC3 | Sponge network | -0.739 | 0.0574 | 0.164 | 0.16786 | 0.571 |
| 4153 | TPTEP1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | DCN | Sponge network | -1.216 | 0 | -1.288 | 0 | 0.571 |
| 4154 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 55 | BNC2 | Sponge network | -2.46 | 0 | -0.491 | 0.09988 | 0.571 |
| 4155 | TPTEP1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 18 | GLI3 | Sponge network | -1.216 | 0 | -0.615 | 0.01482 | 0.571 |
| 4156 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 46 | DIP2C | Sponge network | -2.46 | 0 | -0.436 | 0.01713 | 0.571 |
| 4157 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | LRRC7 | Sponge network | -2.452 | 0 | -1.341 | 0.01193 | 0.571 |
| 4158 | BVES-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-424-3p;hsa-miR-511-5p;hsa-miR-576-5p | 17 | WWTR1 | Sponge network | -2.62 | 0 | -0.345 | 0.11997 | 0.571 |
| 4159 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p | 11 | CACNA2D1 | Sponge network | -0.376 | 0.00337 | -0.846 | 0.00675 | 0.571 |
| 4160 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-484 | 13 | NRXN3 | Sponge network | -0.777 | 0.16667 | -0.893 | 0.04186 | 0.571 |
| 4161 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | SYNPO2 | Sponge network | -0.612 | 0.00094 | -2.342 | 0 | 0.57 |
| 4162 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | JAKMIP2 | Sponge network | -1.281 | 0.00012 | -1.388 | 0 | 0.57 |
| 4163 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p | 15 | MN1 | Sponge network | -0.323 | 0.01372 | -0.358 | 0.24172 | 0.57 |
| 4164 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 42 | TACC1 | Sponge network | -0.323 | 0.01372 | -0.362 | 0.05406 | 0.57 |
| 4165 | RP11-554A11.4 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-450b-5p | 13 | PRKCB | Sponge network | -0.583 | 0.14589 | -1.313 | 2.0E-5 | 0.57 |
| 4166 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | MEF2C | Sponge network | -0.811 | 2.0E-5 | -0.499 | 0.01448 | 0.57 |
| 4167 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | PLSCR4 | Sponge network | -0.793 | 7.0E-5 | -0.474 | 0.03833 | 0.57 |
| 4168 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-424-5p;hsa-miR-590-3p | 17 | DYNC1I1 | Sponge network | -0.096 | 0.67958 | -1.12 | 0.00107 | 0.57 |
| 4169 | RP4-639F20.1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-96-5p | 10 | PCDH10 | Sponge network | -0.517 | 0.02935 | -1.644 | 0.0078 | 0.57 |
| 4170 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 34 | MITF | Sponge network | -0.737 | 0.06182 | -0.769 | 0.00022 | 0.57 |
| 4171 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-370-3p;hsa-miR-532-3p | 10 | RPS6KA2 | Sponge network | -1.092 | 4.0E-5 | -0.57 | 0.00013 | 0.57 |
| 4172 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ATRX | Sponge network | 2.094 | 0 | 0.261 | 0.04408 | 0.57 |
| 4173 | PWAR6 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-940 | 45 | GRIK3 | Sponge network | -1.575 | 6.0E-5 | -3.208 | 0 | 0.57 |
| 4174 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-2355-3p;hsa-miR-26b-3p | 10 | EPHA7 | Sponge network | -0.691 | 0.00146 | -2.197 | 3.0E-5 | 0.57 |
| 4175 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | BCLAF1 | Sponge network | 0.537 | 0.00139 | 0.211 | 0.00684 | 0.57 |
| 4176 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-93-5p | 51 | SSBP2 | Sponge network | -0.793 | 7.0E-5 | -1.58 | 0 | 0.57 |
| 4177 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-429 | 10 | NALCN | Sponge network | -0.976 | 0.00025 | 1.068 | 0.00237 | 0.57 |
| 4178 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-671-5p | 18 | BNC2 | Sponge network | -0.309 | 0.1145 | -0.491 | 0.09988 | 0.57 |
| 4179 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-93-5p | 54 | PRKAA2 | Sponge network | -0.162 | 0.47687 | -2.462 | 0 | 0.57 |
| 4180 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p | 27 | PGR | Sponge network | 0.249 | 0.35902 | -1.704 | 0 | 0.57 |
| 4181 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | ROBO1 | Sponge network | -0.739 | 0.0574 | -0.009 | 0.96154 | 0.57 |
| 4182 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 19 | AKAP12 | Sponge network | -0.942 | 0 | -0.932 | 0.0028 | 0.57 |
| 4183 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | RBMS3 | Sponge network | 0.225 | 0.30644 | -0.519 | 0.03551 | 0.57 |
| 4184 | CTC-429P9.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p | 12 | EZH1 | Sponge network | 0.178 | 0.29106 | -0.564 | 1.0E-5 | 0.57 |
| 4185 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-378a-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | FLNA | Sponge network | -2.46 | 0 | -0.771 | 0.01377 | 0.57 |
| 4186 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 49 | ZFHX4 | Sponge network | 0.246 | 0.59912 | 0.018 | 0.95574 | 0.57 |
| 4187 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-93-5p | 14 | FGFR1 | Sponge network | -0.611 | 0.09607 | -0.509 | 0.03957 | 0.57 |
| 4188 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 14 | PARK2 | Sponge network | -0.942 | 0 | -1.892 | 0 | 0.57 |
| 4189 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 22 | DYNC1I1 | Sponge network | -1.575 | 6.0E-5 | -1.12 | 0.00107 | 0.57 |
| 4190 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-675-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 39 | MEF2C | Sponge network | -0.769 | 0.00035 | -0.499 | 0.01448 | 0.57 |
| 4191 | PWAR6 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 29 | ASTN1 | Sponge network | -1.575 | 6.0E-5 | -2.389 | 2.0E-5 | 0.57 |
| 4192 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -2.531 | 0 | -0.354 | 0.14694 | 0.57 |
| 4193 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-629-3p | 31 | AR | Sponge network | -0.469 | 0.08721 | -1.554 | 0 | 0.57 |
| 4194 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-589-3p | 14 | DIP2C | Sponge network | -4.463 | 0.00025 | -0.436 | 0.01713 | 0.57 |
| 4195 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | -2.915 | 0.00015 | -1.603 | 1.0E-5 | 0.57 |
| 4196 | RP11-25K19.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PRUNE2 | Sponge network | -1.057 | 0.00029 | -1.733 | 0.00022 | 0.57 |
| 4197 | EMX2OS |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p | 16 | SYNM | Sponge network | -0.777 | 0.1134 | -2.309 | 1.0E-5 | 0.57 |
| 4198 | RP11-532F6.3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-940 | 15 | SFRP1 | Sponge network | -0.896 | 0.00099 | -3.566 | 0 | 0.57 |
| 4199 | DNM3OS |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 10 | FHL1 | Sponge network | 0.572 | 0.01722 | -2.687 | 0 | 0.57 |
| 4200 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-590-5p | 17 | SPG20 | Sponge network | -1.349 | 0.00017 | -1.16 | 0 | 0.57 |
| 4201 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 13 | ABCC9 | Sponge network | -0.842 | 0.01361 | -0.466 | 0.14121 | 0.57 |
| 4202 | ZNF667-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-5p | 16 | ROBO1 | Sponge network | -0.976 | 0.00025 | -0.009 | 0.96154 | 0.57 |
| 4203 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH11 | Sponge network | 0.335 | 0.20596 | 1.427 | 0 | 0.57 |
| 4204 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | TSHZ3 | Sponge network | -0.487 | 0.02157 | -0.556 | 0.01516 | 0.569 |
| 4205 | DLEU2 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p | 15 | CBFB | Sponge network | 1.831 | 0 | 1.061 | 0 | 0.569 |
| 4206 | LINC00092 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-24-2-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p | 13 | PCDH9 | Sponge network | -1.886 | 0 | -1.823 | 4.0E-5 | 0.569 |
| 4207 | RP11-504P24.4 |
hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | MDM4 | Sponge network | 0.624 | 0.0004 | 0.345 | 0.00762 | 0.569 |
| 4208 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 51 | DTNA | Sponge network | -0.096 | 0.67958 | -1.648 | 1.0E-5 | 0.569 |
| 4209 | RP11-307B6.3 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p | 10 | ZBTB4 | Sponge network | -4.463 | 0.00025 | -0.739 | 0 | 0.569 |
| 4210 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-7-1-3p | 13 | PLSCR4 | Sponge network | -4.546 | 0 | -0.474 | 0.03833 | 0.569 |
| 4211 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-576-5p | 14 | NRXN1 | Sponge network | -0.932 | 0.00329 | -3.778 | 0 | 0.569 |
| 4212 | LINC00476 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-93-5p | 20 | PRUNE2 | Sponge network | -0.615 | 0.00015 | -1.733 | 0.00022 | 0.569 |
| 4213 | CTD-2528L19.6 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RBBP6 | Sponge network | 0.953 | 0 | 0.163 | 0.05101 | 0.569 |
| 4214 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | LRRK2 | Sponge network | -0.487 | 0.02157 | -1.136 | 1.0E-5 | 0.569 |
| 4215 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 55 | DIP2C | Sponge network | -1.111 | 0.0003 | -0.436 | 0.01713 | 0.569 |
| 4216 | TP73-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-96-5p | 14 | PAK3 | Sponge network | -0.563 | 0.02545 | -0.628 | 0.07253 | 0.569 |
| 4217 | MIR497HG |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-3p | 12 | STAT5B | Sponge network | -1.439 | 4.0E-5 | -0.182 | 0.1082 | 0.569 |
| 4218 | PWAR6 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p | 20 | MACF1 | Sponge network | -1.575 | 6.0E-5 | 0.019 | 0.90335 | 0.569 |
| 4219 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-5p | 31 | RBMS3 | Sponge network | 0.176 | 0.37402 | -0.519 | 0.03551 | 0.569 |
| 4220 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 12 | TAL1 | Sponge network | -1.092 | 4.0E-5 | -0.462 | 0.00574 | 0.569 |
| 4221 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 34 | SSBP2 | Sponge network | -0.739 | 0.00044 | -1.58 | 0 | 0.569 |
| 4222 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-500a-3p | 18 | TSHZ3 | Sponge network | -0.517 | 0.02935 | -0.556 | 0.01516 | 0.569 |
| 4223 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p | 15 | FGFR1 | Sponge network | 0.176 | 0.37402 | -0.509 | 0.03957 | 0.569 |
| 4224 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-1266-5p;hsa-miR-129-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-590-3p | 10 | MLH3 | Sponge network | 1.254 | 0 | 0.608 | 0 | 0.569 |
| 4225 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-940 | 32 | CHD5 | Sponge network | -0.246 | 0.35034 | -0.922 | 0.01521 | 0.569 |
| 4226 | BVES-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p;hsa-miR-744-3p | 11 | RHOB | Sponge network | -2.62 | 0 | -1.052 | 0 | 0.569 |
| 4227 | PCA3 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ITPKB | Sponge network | -0.709 | 0.18168 | -0.704 | 0.00106 | 0.569 |
| 4228 | ADAMTS9-AS2 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | FAM117A | Sponge network | -2.452 | 0 | -1.379 | 0 | 0.569 |
| 4229 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-532-3p | 13 | SYNM | Sponge network | -1.654 | 0.00144 | -2.309 | 1.0E-5 | 0.569 |
| 4230 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p | 11 | JAKMIP2 | Sponge network | -0.769 | 0.00035 | -1.388 | 0 | 0.569 |
| 4231 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 36 | SNX29 | Sponge network | -0.576 | 0.10121 | -0.34 | 0.01371 | 0.569 |
| 4232 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p | 12 | CNTNAP3 | Sponge network | -1.331 | 3.0E-5 | -1.465 | 0.00033 | 0.569 |
| 4233 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-664a-5p;hsa-miR-766-3p | 31 | CADM2 | Sponge network | -0.583 | 0.14589 | -3.782 | 0 | 0.569 |
| 4234 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p | 24 | PGR | Sponge network | -0.777 | 0.16667 | -1.704 | 0 | 0.569 |
| 4235 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 38 | DDX6 | Sponge network | -0.222 | 0.32445 | 0.091 | 0.36428 | 0.569 |
| 4236 | RP11-532F6.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-935 | 69 | LPP | Sponge network | -0.896 | 0.00099 | -0.459 | 0.04475 | 0.569 |
| 4237 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-93-5p | 65 | NTRK3 | Sponge network | 0.335 | 0.20596 | -1.77 | 3.0E-5 | 0.569 |
| 4238 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 16 | SPG20 | Sponge network | -0.896 | 0.00099 | -1.16 | 0 | 0.569 |
| 4239 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 37 | EBF1 | Sponge network | -1.697 | 0.00889 | -0.384 | 0.08856 | 0.569 |
| 4240 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | SLITRK3 | Sponge network | -1.111 | 0.0003 | -3.328 | 0 | 0.569 |
| 4241 | BZRAP1-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-877-5p;hsa-miR-93-3p | 12 | STAT5B | Sponge network | -0.465 | 0.04703 | -0.182 | 0.1082 | 0.568 |
| 4242 | HOXB-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-3p | 19 | NAV3 | Sponge network | -0.154 | 0.53954 | 0.212 | 0.47729 | 0.568 |
| 4243 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | SPG20 | Sponge network | -1.216 | 0 | -1.16 | 0 | 0.568 |
| 4244 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TAL1 | Sponge network | -0.513 | 0.04703 | -0.462 | 0.00574 | 0.568 |
| 4245 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-935 | 34 | DMD | Sponge network | -0.563 | 0.02545 | -1.506 | 0 | 0.568 |
| 4246 | RP11-159D12.2 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | RBBP6 | Sponge network | 1.254 | 0 | 0.163 | 0.05101 | 0.568 |
| 4247 | MAGI2-AS3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 44 | IL17RD | Sponge network | -0.129 | 0.57177 | -0.019 | 0.94873 | 0.568 |
| 4248 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p;hsa-miR-874-3p | 21 | NR2C2 | Sponge network | 0.267 | 0.2937 | 0.21 | 0.03224 | 0.568 |
| 4249 | RP11-736K20.5 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | IGSF10 | Sponge network | -0.358 | 0.17255 | -1.751 | 1.0E-5 | 0.568 |
| 4250 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 34 | USP12 | Sponge network | -0.222 | 0.32445 | -0.102 | 0.40768 | 0.568 |
| 4251 | FENDRR |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 11 | PRKD1 | Sponge network | -1.281 | 0.00012 | -0.466 | 0.03583 | 0.568 |
| 4252 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-5p | 28 | SOX5 | Sponge network | -2.62 | 0 | -1.091 | 4.0E-5 | 0.568 |
| 4253 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | MCC | Sponge network | 0.194 | 0.64278 | -1.005 | 0 | 0.568 |
| 4254 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 35 | PBX1 | Sponge network | -0.901 | 0.00019 | -1.206 | 0 | 0.568 |
| 4255 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DNAJB4 | Sponge network | -0.612 | 0.00094 | -0.7 | 1.0E-5 | 0.568 |
| 4256 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | AR | Sponge network | -0.376 | 0.00337 | -1.554 | 0 | 0.568 |
| 4257 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 18 | TMEFF2 | Sponge network | -0.793 | 7.0E-5 | -2.426 | 0 | 0.568 |
| 4258 | RP11-248G5.8 |
hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-363-3p;hsa-miR-497-5p | 10 | E2F3 | Sponge network | 1.294 | 0 | 1.289 | 0 | 0.568 |
| 4259 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 64 | IL6ST | Sponge network | -0.323 | 0.01372 | -0.402 | 0.00676 | 0.568 |
| 4260 | GNG12-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-744-3p | 41 | DIP2C | Sponge network | -0.793 | 7.0E-5 | -0.436 | 0.01713 | 0.568 |
| 4261 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | EPHA3 | Sponge network | 0.532 | 0.00505 | 0.185 | 0.53588 | 0.568 |
| 4262 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-98-5p | 18 | ITGB3 | Sponge network | -0.976 | 0.00025 | -0.011 | 0.95751 | 0.568 |
| 4263 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-429 | 17 | AKAP6 | Sponge network | -0.469 | 0.08721 | -1.586 | 3.0E-5 | 0.568 |
| 4264 | NR2F1-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 75 | DST | Sponge network | -0.49 | 0.04946 | -0.713 | 0.00096 | 0.568 |
| 4265 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 29 | EPHA3 | Sponge network | -0.901 | 0.00019 | 0.185 | 0.53588 | 0.568 |
| 4266 | RP11-620J15.3 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 16 | MRVI1 | Sponge network | -0.739 | 0.00044 | -1.051 | 0.00022 | 0.568 |
| 4267 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | AKAP6 | Sponge network | -1.161 | 0.00591 | -1.586 | 3.0E-5 | 0.568 |
| 4268 | LINC01018 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | MAPK10 | Sponge network | -2.915 | 0.00015 | -1.774 | 0 | 0.568 |
| 4269 | RP11-400K9.4 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-7-5p | 12 | FNBP1 | Sponge network | -0.611 | 0.09607 | -0.969 | 4.0E-5 | 0.568 |
| 4270 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 22 | ADAMTSL3 | Sponge network | -0.421 | 0.0114 | -1.559 | 0 | 0.568 |
| 4271 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 49 | EPHA5 | Sponge network | -0.49 | 0.04946 | -0.957 | 0.01733 | 0.568 |
| 4272 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | PCDH9 | Sponge network | -0.096 | 0.67958 | -1.823 | 4.0E-5 | 0.568 |
| 4273 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | RASSF8 | Sponge network | 0.225 | 0.30644 | -0.122 | 0.65591 | 0.568 |
| 4274 | EMX2OS |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | THSD7B | Sponge network | -0.777 | 0.1134 | 0.154 | 0.74723 | 0.568 |
| 4275 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 17 | TXNIP | Sponge network | -1.697 | 0.00889 | -1.277 | 0 | 0.568 |
| 4276 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | SYNE1 | Sponge network | 0.246 | 0.59912 | -0.738 | 0.00316 | 0.568 |
| 4277 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ADAMTSL3 | Sponge network | -1.057 | 0.00029 | -1.559 | 0 | 0.568 |
| 4278 | ARHGEF26-AS1 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | RIMS2 | Sponge network | -2.46 | 0 | -1.365 | 0.00208 | 0.568 |
| 4279 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-576-5p | 19 | SYNPO2 | Sponge network | 0.299 | 0.57722 | -2.342 | 0 | 0.568 |
| 4280 | RP11-750H9.5 |
hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-671-5p;hsa-miR-877-5p | 11 | GAS7 | Sponge network | 0.349 | 0.13152 | -0.555 | 0.01602 | 0.568 |
| 4281 | LINC00702 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | DACT1 | Sponge network | -0.737 | 0.06182 | 0.783 | 0.00072 | 0.568 |
| 4282 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | AFF3 | Sponge network | -0.49 | 0.04946 | -2.373 | 0 | 0.568 |
| 4283 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | EBF3 | Sponge network | 0.194 | 0.64278 | -0.058 | 0.81437 | 0.568 |
| 4284 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | 0.335 | 0.20596 | -0.684 | 0.00159 | 0.568 |
| 4285 | RP11-798M19.6 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | -0.612 | 0.00094 | -1.051 | 0.00022 | 0.568 |
| 4286 | RP5-1198O20.4 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 12 | STAT5B | Sponge network | 0.997 | 0.00066 | -0.182 | 0.1082 | 0.568 |
| 4287 | TUG1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-495-3p;hsa-miR-654-3p | 16 | PHF6 | Sponge network | 0.972 | 0 | 0.785 | 0 | 0.568 |
| 4288 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 12 | LHFP | Sponge network | 0.997 | 0.00066 | -0.167 | 0.41371 | 0.568 |
| 4289 | JAZF1-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | FAM117A | Sponge network | -0.769 | 0.00035 | -1.379 | 0 | 0.568 |
| 4290 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3913-5p | 13 | ADARB1 | Sponge network | -0.691 | 0.00146 | -0.335 | 0.06631 | 0.568 |
| 4291 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-502-5p;hsa-miR-877-5p | 25 | AR | Sponge network | -0.309 | 0.1145 | -1.554 | 0 | 0.568 |
| 4292 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | -0.976 | 0.00025 | -1.751 | 1.0E-5 | 0.568 |
| 4293 | JAZF1-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | FLNA | Sponge network | -0.769 | 0.00035 | -0.771 | 0.01377 | 0.568 |
| 4294 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 17 | MN1 | Sponge network | -2.46 | 0 | -0.358 | 0.24172 | 0.568 |
| 4295 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-577;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -1.439 | 4.0E-5 | -0.7 | 1.0E-5 | 0.568 |
| 4296 | MIR143HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 34 | KCNA6 | Sponge network | -0.739 | 0.0574 | -1.531 | 0.00013 | 0.568 |
| 4297 | SNHG1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 14 | PHF6 | Sponge network | 1.265 | 0 | 0.785 | 0 | 0.568 |
| 4298 | PGM5-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-345-5p;hsa-miR-942-5p | 19 | IGFBP5 | Sponge network | -4.546 | 0 | -0.806 | 0.00105 | 0.568 |
| 4299 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 49 | AR | Sponge network | -0.842 | 0.01361 | -1.554 | 0 | 0.568 |
| 4300 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | ROBO1 | Sponge network | -0.49 | 0.04946 | -0.009 | 0.96154 | 0.568 |
| 4301 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-577;hsa-miR-98-5p | 10 | PLAGL1 | Sponge network | -0.49 | 0.04946 | -0.588 | 0.00912 | 0.568 |
| 4302 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 27 | LRRC7 | Sponge network | -2.46 | 0 | -1.341 | 0.01193 | 0.568 |
| 4303 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-93-5p | 39 | FAT3 | Sponge network | 0.299 | 0.57722 | -1.601 | 0.00051 | 0.568 |
| 4304 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-93-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | -0.777 | 0.16667 | 0.358 | 0.13727 | 0.568 |
| 4305 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p | 31 | IL17RD | Sponge network | -0.246 | 0.35034 | -0.019 | 0.94873 | 0.567 |
| 4306 | MIR143HG |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | ZDBF2 | Sponge network | -0.739 | 0.0574 | -0.998 | 0.00025 | 0.567 |
| 4307 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940;hsa-miR-96-5p | 49 | GAS7 | Sponge network | 0.194 | 0.64278 | -0.555 | 0.01602 | 0.567 |
| 4308 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 20 | RELN | Sponge network | -0.129 | 0.57177 | -2.403 | 0 | 0.567 |
| 4309 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 22 | AKAP6 | Sponge network | 0.331 | 0.57247 | -1.586 | 3.0E-5 | 0.567 |
| 4310 | RP11-244O19.1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-93-5p | 53 | AR | Sponge network | -0.096 | 0.67958 | -1.554 | 0 | 0.567 |
| 4311 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | IGSF10 | Sponge network | -0.129 | 0.57177 | -1.751 | 1.0E-5 | 0.567 |
| 4312 | ACTA2-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | FAM172A | Sponge network | 0.194 | 0.64278 | -0.57 | 0 | 0.567 |
| 4313 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | FLNA | Sponge network | -0.323 | 0.01372 | -0.771 | 0.01377 | 0.567 |
| 4314 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-335-5p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-96-5p | 10 | TP53INP1 | Sponge network | -0.31 | 0.33871 | -0.496 | 0.00064 | 0.567 |
| 4315 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | ST6GALNAC3 | Sponge network | -0.976 | 0.00025 | -0.987 | 0 | 0.567 |
| 4316 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-451a;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 52 | BCL2 | Sponge network | -1.111 | 0.0003 | -0.899 | 0 | 0.567 |
| 4317 | RP11-473I1.9 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | ERCC4 | Sponge network | 0.683 | 1.0E-5 | 0.126 | 0.15266 | 0.567 |
| 4318 | GNG12-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-940 | 16 | SFRP1 | Sponge network | -0.793 | 7.0E-5 | -3.566 | 0 | 0.567 |
| 4319 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-93-5p | 46 | CADM2 | Sponge network | -2.69 | 0.0001 | -3.782 | 0 | 0.567 |
| 4320 | BVES-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-98-5p | 13 | DCN | Sponge network | -2.62 | 0 | -1.288 | 0 | 0.567 |
| 4321 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-93-3p | 14 | PCDH10 | Sponge network | -0.154 | 0.53954 | -1.644 | 0.0078 | 0.567 |
| 4322 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | PLSCR4 | Sponge network | -2.46 | 0 | -0.474 | 0.03833 | 0.567 |
| 4323 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 12 | GLI3 | Sponge network | -0.896 | 0.00099 | -0.615 | 0.01482 | 0.567 |
| 4324 | RP11-672A2.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-671-5p | 14 | FAT4 | Sponge network | 1.344 | 0 | -0.354 | 0.14694 | 0.567 |
| 4325 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | ZNF521 | Sponge network | -0.901 | 0.00019 | -0.111 | 0.58068 | 0.567 |
| 4326 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3913-5p | 23 | HLF | Sponge network | -0.583 | 0.14589 | -2.443 | 0 | 0.567 |
| 4327 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-369-3p | 13 | JAKMIP2 | Sponge network | -0.611 | 0.09607 | -1.388 | 0 | 0.567 |
| 4328 | LINC00476 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-590-3p | 15 | FAM172A | Sponge network | -0.615 | 0.00015 | -0.57 | 0 | 0.567 |
| 4329 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | NRXN3 | Sponge network | -0.487 | 0.02157 | -0.893 | 0.04186 | 0.567 |
| 4330 | DNM3OS |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ITGB1 | Sponge network | 0.572 | 0.01722 | 0.524 | 0.00017 | 0.567 |
| 4331 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 21 | FAT4 | Sponge network | -0.793 | 7.0E-5 | -0.354 | 0.14694 | 0.567 |
| 4332 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-7-5p | 12 | FLNA | Sponge network | -0.469 | 0.08721 | -0.771 | 0.01377 | 0.567 |
| 4333 | LINC01018 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PCDH9 | Sponge network | -2.915 | 0.00015 | -1.823 | 4.0E-5 | 0.567 |
| 4334 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 11 | SEPT6 | Sponge network | -0.896 | 0.00099 | -0.598 | 0.00181 | 0.567 |
| 4335 | AC006116.24 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-577 | 12 | CADM1 | Sponge network | -0.473 | 0.07602 | -0.9 | 0.00084 | 0.567 |
| 4336 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-590-3p | 15 | KIT | Sponge network | -1.582 | 8.0E-5 | -2.184 | 0 | 0.567 |
| 4337 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 51 | AR | Sponge network | -0.769 | 0.00035 | -1.554 | 0 | 0.567 |
| 4338 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-92b-3p | 19 | AKAP12 | Sponge network | -0.777 | 0.1134 | -0.932 | 0.0028 | 0.567 |
| 4339 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 18 | IGF1 | Sponge network | -1.582 | 8.0E-5 | -0.204 | 0.5898 | 0.567 |
| 4340 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 34 | MEF2C | Sponge network | -1.349 | 0.00017 | -0.499 | 0.01448 | 0.567 |
| 4341 | CTD-2006H14.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MDM4 | Sponge network | 1.378 | 0 | 0.345 | 0.00762 | 0.567 |
| 4342 | ACTA2-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 15 | ZBTB20 | Sponge network | 0.194 | 0.64278 | -0.122 | 0.57569 | 0.567 |
| 4343 | MIR497HG |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 77 | LPP | Sponge network | -1.439 | 4.0E-5 | -0.459 | 0.04475 | 0.567 |
| 4344 | DNM3OS |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p | 33 | IGFBP5 | Sponge network | 0.572 | 0.01722 | -0.806 | 0.00105 | 0.567 |
| 4345 | HCG18 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 13 | TAF1 | Sponge network | 1.063 | 0 | 0.39 | 3.0E-5 | 0.566 |
| 4346 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 26 | EBF3 | Sponge network | 0.335 | 0.20596 | -0.058 | 0.81437 | 0.566 |
| 4347 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 26 | ADAMTSL3 | Sponge network | -0.487 | 0.02157 | -1.559 | 0 | 0.566 |
| 4348 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | -0.563 | 0.02545 | -1.324 | 0.00014 | 0.566 |
| 4349 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 36 | ADARB1 | Sponge network | -0.612 | 0.00094 | -0.335 | 0.06631 | 0.566 |
| 4350 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-505-5p | 24 | KCNA6 | Sponge network | -0.583 | 0.14589 | -1.531 | 0.00013 | 0.566 |
| 4351 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p | 22 | STARD13 | Sponge network | -2.452 | 0 | -0.187 | 0.27383 | 0.566 |
| 4352 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p | 12 | DCLK1 | Sponge network | 0.811 | 0.03228 | -1.324 | 0.00014 | 0.566 |
| 4353 | RP11-166D19.1 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | MSN | Sponge network | -0.576 | 0.10121 | -0.023 | 0.88422 | 0.566 |
| 4354 | LINC00702 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p | 11 | SORCS2 | Sponge network | -0.737 | 0.06182 | -0.223 | 0.4253 | 0.566 |
| 4355 | EMX2OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 29 | SYNPO2 | Sponge network | -0.777 | 0.1134 | -2.342 | 0 | 0.566 |
| 4356 | TPTEP1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 60 | DTNA | Sponge network | -1.216 | 0 | -1.648 | 1.0E-5 | 0.566 |
| 4357 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | ANK2 | Sponge network | -0.811 | 2.0E-5 | -1.603 | 1.0E-5 | 0.566 |
| 4358 | RP11-736K20.5 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-511-5p;hsa-miR-590-3p | 12 | PCDH10 | Sponge network | -0.358 | 0.17255 | -1.644 | 0.0078 | 0.566 |
| 4359 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-508-3p;hsa-miR-7-5p;hsa-miR-877-5p | 30 | RBMS3 | Sponge network | -1.654 | 0.00144 | -0.519 | 0.03551 | 0.566 |
| 4360 | LINC00578 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577 | 10 | AMPH | Sponge network | 0.811 | 0.03228 | -0.916 | 5.0E-5 | 0.566 |
| 4361 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 46 | EPHA7 | Sponge network | -0.096 | 0.67958 | -2.197 | 3.0E-5 | 0.566 |
| 4362 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 32 | IGF1 | Sponge network | -0.323 | 0.01372 | -0.204 | 0.5898 | 0.566 |
| 4363 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-452-5p;hsa-miR-590-3p | 26 | JAZF1 | Sponge network | -0.358 | 0.17255 | -0.377 | 0.02677 | 0.566 |
| 4364 | MIR497HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-96-5p | 24 | CBFA2T3 | Sponge network | -1.439 | 4.0E-5 | -1.221 | 1.0E-5 | 0.566 |
| 4365 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p | 14 | ZBTB4 | Sponge network | -0.932 | 0.00329 | -0.739 | 0 | 0.566 |
| 4366 | AC003090.1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-96-5p | 18 | FNBP1 | Sponge network | -2.117 | 0.00161 | -0.969 | 4.0E-5 | 0.566 |
| 4367 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-485-3p | 10 | LRRK2 | Sponge network | -0.465 | 0.04703 | -1.136 | 1.0E-5 | 0.566 |
| 4368 | TRAF3IP2-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | MRVI1 | Sponge network | -0.323 | 0.01372 | -1.051 | 0.00022 | 0.566 |
| 4369 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-942-5p | 24 | HLF | Sponge network | -0.517 | 0.02935 | -2.443 | 0 | 0.566 |
| 4370 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 30 | EPHA3 | Sponge network | 0.249 | 0.35902 | 0.185 | 0.53588 | 0.566 |
| 4371 | MIR143HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | ABCA10 | Sponge network | -0.739 | 0.0574 | -0.571 | 0.03674 | 0.566 |
| 4372 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-652-3p | 26 | ZBTB4 | Sponge network | -0.323 | 0.01372 | -0.739 | 0 | 0.566 |
| 4373 | RP11-6O2.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-96-5p | 68 | LPP | Sponge network | 0.331 | 0.57247 | -0.459 | 0.04475 | 0.566 |
| 4374 | RP11-356J5.12 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | FHL1 | Sponge network | -0.899 | 6.0E-5 | -2.687 | 0 | 0.566 |
| 4375 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p | 18 | PDZD2 | Sponge network | -0.49 | 0.04946 | -0.909 | 3.0E-5 | 0.566 |
| 4376 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-33a-3p;hsa-miR-33a-5p | 12 | AKAP11 | Sponge network | 0.497 | 0.01451 | 0.196 | 0.09825 | 0.566 |
| 4377 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-5p | 29 | JAZF1 | Sponge network | 0.335 | 0.20596 | -0.377 | 0.02677 | 0.566 |
| 4378 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 53 | AR | Sponge network | -0.323 | 0.01372 | -1.554 | 0 | 0.566 |
| 4379 | RP11-166D19.1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 10 | PEA15 | Sponge network | -0.576 | 0.10121 | 0.009 | 0.93318 | 0.566 |
| 4380 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | DLC1 | Sponge network | 0.342 | 0.03129 | -0.382 | 0.05038 | 0.566 |
| 4381 | TRAF3IP2-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 30 | DLC1 | Sponge network | -0.323 | 0.01372 | -0.382 | 0.05038 | 0.566 |
| 4382 | CTC-429P9.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-940 | 11 | FAM172A | Sponge network | 0.082 | 0.55397 | -0.57 | 0 | 0.566 |
| 4383 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 14 | NR2C2 | Sponge network | 2.094 | 0 | 0.21 | 0.03224 | 0.566 |
| 4384 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-93-5p | 29 | CADM2 | Sponge network | -2.531 | 0 | -3.782 | 0 | 0.566 |
| 4385 | LINC00654 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 13 | HMCN1 | Sponge network | 0.374 | 0.20054 | 0.809 | 0.00544 | 0.566 |
| 4386 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-93-5p | 26 | TCF4 | Sponge network | -0.376 | 0.00337 | 0.016 | 0.92271 | 0.566 |
| 4387 | ADAMTS9-AS2 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | SPOP | Sponge network | -2.452 | 0 | -0.419 | 1.0E-5 | 0.566 |
| 4388 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | IGSF10 | Sponge network | -0.576 | 0.10121 | -1.751 | 1.0E-5 | 0.566 |
| 4389 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-7-5p | 17 | CAV1 | Sponge network | -0.162 | 0.47687 | -1.013 | 6.0E-5 | 0.566 |
| 4390 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-577 | 20 | IGF1 | Sponge network | 0.811 | 0.03228 | -0.204 | 0.5898 | 0.566 |
| 4391 | MIR143HG |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | SPOP | Sponge network | -0.739 | 0.0574 | -0.419 | 1.0E-5 | 0.566 |
| 4392 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 54 | TRPS1 | Sponge network | -0.49 | 0.04946 | -0.191 | 0.44109 | 0.566 |
| 4393 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 16 | DCLK1 | Sponge network | -2.46 | 0 | -1.324 | 0.00014 | 0.566 |
| 4394 | TUG1 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-664a-3p | 15 | RASAL2 | Sponge network | 0.972 | 0 | 0.869 | 0 | 0.566 |
| 4395 | RP11-736K20.5 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | PRKCB | Sponge network | -0.358 | 0.17255 | -1.313 | 2.0E-5 | 0.565 |
| 4396 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | EYA4 | Sponge network | -0.49 | 0.04946 | 0.3 | 0.36876 | 0.565 |
| 4397 | RP11-890B15.3 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 16 | MACF1 | Sponge network | 0.101 | 0.60919 | 0.019 | 0.90335 | 0.565 |
| 4398 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-590-3p | 10 | DCN | Sponge network | -0.896 | 0.00099 | -1.288 | 0 | 0.565 |
| 4399 | LINC00843 |
hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-429 | 17 | ABL2 | Sponge network | 1.106 | 0 | 0.82 | 0 | 0.565 |
| 4400 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-7-1-3p | 14 | NCAM2 | Sponge network | -1.092 | 4.0E-5 | -0.482 | 0.22565 | 0.565 |
| 4401 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | 0.532 | 0.00505 | 0.273 | 0.09957 | 0.565 |
| 4402 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 30 | KCNA6 | Sponge network | -0.129 | 0.57177 | -1.531 | 0.00013 | 0.565 |
| 4403 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 78 | BMPR2 | Sponge network | -0.129 | 0.57177 | 0.206 | 0.0635 | 0.565 |
| 4404 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 18 | CNTNAP3 | Sponge network | -0.901 | 0.00019 | -1.465 | 0.00033 | 0.565 |
| 4405 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 52 | QKI | Sponge network | -0.901 | 0.00019 | -0.333 | 0.04111 | 0.565 |
| 4406 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 53 | EPHA5 | Sponge network | -1.575 | 6.0E-5 | -0.957 | 0.01733 | 0.565 |
| 4407 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-940 | 15 | LIFR | Sponge network | -1.886 | 0 | -1.794 | 0 | 0.565 |
| 4408 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-671-5p | 11 | CADM3 | Sponge network | -0.611 | 0.09607 | -3.663 | 0 | 0.565 |
| 4409 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-590-3p | 14 | NR2C2 | Sponge network | 0.765 | 7.0E-5 | 0.21 | 0.03224 | 0.565 |
| 4410 | FAM66C |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | FLNA | Sponge network | -0.901 | 0.00019 | -0.771 | 0.01377 | 0.565 |
| 4411 | MALAT1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 18 | BAZ2B | Sponge network | 0.782 | 0.00016 | -0.276 | 0.02833 | 0.565 |
| 4412 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | LIFR | Sponge network | -0.358 | 0.17255 | -1.794 | 0 | 0.565 |
| 4413 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | NR2C2 | Sponge network | 0.826 | 1.0E-5 | 0.21 | 0.03224 | 0.565 |
| 4414 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | MYO9A | Sponge network | 0.421 | 0.00084 | -0.147 | 0.32373 | 0.565 |
| 4415 | RP11-890B15.3 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | HERC1 | Sponge network | 0.101 | 0.60919 | -0.196 | 0.08544 | 0.565 |
| 4416 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-3p | 23 | AR | Sponge network | -2.076 | 0.00548 | -1.554 | 0 | 0.565 |
| 4417 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-30d-3p | 10 | SLITRK3 | Sponge network | -0.932 | 0.00329 | -3.328 | 0 | 0.565 |
| 4418 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-940;hsa-miR-96-5p | 38 | SNX29 | Sponge network | -2.452 | 0 | -0.34 | 0.01371 | 0.565 |
| 4419 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 28 | TIMP3 | Sponge network | -2.452 | 0 | -0.546 | 0.02307 | 0.565 |
| 4420 | HOXB-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429 | 15 | ITPKB | Sponge network | -0.154 | 0.53954 | -0.704 | 0.00106 | 0.565 |
| 4421 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | LHFP | Sponge network | -0.769 | 0.00035 | -0.167 | 0.41371 | 0.565 |
| 4422 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 20 | KIT | Sponge network | -0.896 | 0.00099 | -2.184 | 0 | 0.565 |
| 4423 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p | 21 | SOX5 | Sponge network | -1.886 | 0 | -1.091 | 4.0E-5 | 0.565 |
| 4424 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | JAZF1 | Sponge network | 0.572 | 0.01722 | -0.377 | 0.02677 | 0.565 |
| 4425 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 31 | CNTN1 | Sponge network | -0.563 | 0.02545 | -2.594 | 0 | 0.565 |
| 4426 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | NRXN3 | Sponge network | -0.793 | 7.0E-5 | -0.893 | 0.04186 | 0.565 |
| 4427 | TP73-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p | 11 | LHFP | Sponge network | -0.563 | 0.02545 | -0.167 | 0.41371 | 0.565 |
| 4428 | RP11-999E24.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-7-5p | 15 | SETBP1 | Sponge network | -0.693 | 0.05629 | -1.302 | 1.0E-5 | 0.565 |
| 4429 | SNHG14 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 21 | PDZD2 | Sponge network | -1.111 | 0.0003 | -0.909 | 3.0E-5 | 0.565 |
| 4430 | PCA3 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TPO | Sponge network | -0.709 | 0.18168 | -1.87 | 2.0E-5 | 0.565 |
| 4431 | HOXB-AS1 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-429;hsa-miR-629-3p | 13 | ITGA5 | Sponge network | -0.154 | 0.53954 | 0.452 | 0.03524 | 0.565 |
| 4432 | DNM3OS |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-532-3p | 16 | SYNM | Sponge network | 0.572 | 0.01722 | -2.309 | 1.0E-5 | 0.565 |
| 4433 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | SLITRK3 | Sponge network | -0.793 | 7.0E-5 | -3.328 | 0 | 0.565 |
| 4434 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | CAV1 | Sponge network | 0.572 | 0.01722 | -1.013 | 6.0E-5 | 0.565 |
| 4435 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-96-5p | 17 | DAB2 | Sponge network | -0.576 | 0.10121 | 0.431 | 0.0113 | 0.565 |
| 4436 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-628-5p | 12 | CAV1 | Sponge network | -2.62 | 0 | -1.013 | 6.0E-5 | 0.565 |
| 4437 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ZMYND11 | Sponge network | -0.612 | 0.00094 | -0.2 | 0.05449 | 0.565 |
| 4438 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | NCAM2 | Sponge network | 0.997 | 0.00066 | -0.482 | 0.22565 | 0.565 |
| 4439 | FENDRR |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 52 | GRIK3 | Sponge network | -1.281 | 0.00012 | -3.208 | 0 | 0.565 |
| 4440 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-629-3p | 15 | GPC6 | Sponge network | -0.583 | 0.14589 | -0.054 | 0.81465 | 0.565 |
| 4441 | RP11-894P9.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-331-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p | 13 | ATRX | Sponge network | 0.993 | 0 | 0.261 | 0.04408 | 0.565 |
| 4442 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-590-3p | 12 | ITGB3 | Sponge network | -0.096 | 0.67958 | -0.011 | 0.95751 | 0.565 |
| 4443 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 24 | RICTOR | Sponge network | 1.205 | 0 | 0.388 | 0.00188 | 0.565 |
| 4444 | RP11-464F9.1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | ERCC6 | Sponge network | 0.959 | 0 | 0.23 | 0.015 | 0.565 |
| 4445 | TP73-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p | 20 | MRVI1 | Sponge network | -0.563 | 0.02545 | -1.051 | 0.00022 | 0.565 |
| 4446 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | ABCC9 | Sponge network | 0.889 | 9.0E-5 | -0.466 | 0.14121 | 0.564 |
| 4447 | RP11-532F6.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 33 | GRIK3 | Sponge network | -0.896 | 0.00099 | -3.208 | 0 | 0.564 |
| 4448 | NR2F1-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 22 | GLIPR1 | Sponge network | -0.49 | 0.04946 | 0.146 | 0.3276 | 0.564 |
| 4449 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | BNC2 | Sponge network | -0.709 | 0.18168 | -0.491 | 0.09988 | 0.564 |
| 4450 | MEG3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | 0.249 | 0.35902 | 0.783 | 0.00072 | 0.564 |
| 4451 | AC090587.5 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-93-3p | 15 | SNX29 | Sponge network | -0.088 | 0.65011 | -0.34 | 0.01371 | 0.564 |
| 4452 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 20 | CNTNAP3 | Sponge network | -0.737 | 0.06182 | -1.465 | 0.00033 | 0.564 |
| 4453 | MYLK-AS1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-340-5p;hsa-miR-424-5p | 10 | CSDE1 | Sponge network | -1.331 | 3.0E-5 | -0.493 | 0 | 0.564 |
| 4454 | FENDRR |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 19 | SFRP1 | Sponge network | -1.281 | 0.00012 | -3.566 | 0 | 0.564 |
| 4455 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 29 | CRTC3 | Sponge network | -0.576 | 0.10121 | 0.164 | 0.16786 | 0.564 |
| 4456 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 15 | DCN | Sponge network | -0.769 | 0.00035 | -1.288 | 0 | 0.564 |
| 4457 | RP11-620J15.3 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | ZDBF2 | Sponge network | -0.739 | 0.00044 | -0.998 | 0.00025 | 0.564 |
| 4458 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-589-5p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 49 | ADARB1 | Sponge network | -2.69 | 0.0001 | -0.335 | 0.06631 | 0.564 |
| 4459 | AGAP11 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-708-5p;hsa-miR-93-3p | 25 | TOM1L2 | Sponge network | -1.645 | 0 | -0.994 | 0 | 0.564 |
| 4460 | PCA3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | RYR2 | Sponge network | -0.709 | 0.18168 | -1.466 | 0.00031 | 0.564 |
| 4461 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | FSTL5 | Sponge network | -0.576 | 0.10121 | -1.3 | 0.01619 | 0.564 |
| 4462 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 20 | EZH1 | Sponge network | -0.49 | 0.04946 | -0.564 | 1.0E-5 | 0.564 |
| 4463 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 18 | HOOK3 | Sponge network | -1.575 | 6.0E-5 | 0.273 | 0.09957 | 0.564 |
| 4464 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92b-3p | 14 | AKAP12 | Sponge network | -0.096 | 0.67958 | -0.932 | 0.0028 | 0.564 |
| 4465 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 47 | BNC2 | Sponge network | -0.096 | 0.67958 | -0.491 | 0.09988 | 0.564 |
| 4466 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | 0.246 | 0.59912 | -0.466 | 0.14121 | 0.564 |
| 4467 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-429 | 15 | AKAP6 | Sponge network | -0.154 | 0.53954 | -1.586 | 3.0E-5 | 0.564 |
| 4468 | RP11-400K9.4 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | PRKCB | Sponge network | -0.611 | 0.09607 | -1.313 | 2.0E-5 | 0.564 |
| 4469 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-542-3p | 11 | SYNPO2 | Sponge network | -0.517 | 0.02935 | -2.342 | 0 | 0.564 |
| 4470 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p | 21 | DYNC1I1 | Sponge network | -2.915 | 0.00015 | -1.12 | 0.00107 | 0.564 |
| 4471 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 58 | AR | Sponge network | -1.216 | 0 | -1.554 | 0 | 0.564 |
| 4472 | MIR143HG |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-486-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 24 | GLIPR1 | Sponge network | -0.739 | 0.0574 | 0.146 | 0.3276 | 0.564 |
| 4473 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 16 | DKK3 | Sponge network | -1.439 | 4.0E-5 | -0.052 | 0.77783 | 0.564 |
| 4474 | MIR143HG |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 11 | GLIPR2 | Sponge network | -0.739 | 0.0574 | -0.996 | 0 | 0.564 |
| 4475 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-7-1-3p | 19 | TBX18 | Sponge network | 0.249 | 0.35902 | 0.843 | 0.02945 | 0.564 |
| 4476 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | ST6GAL2 | Sponge network | 0.249 | 0.35902 | -1.201 | 0.00597 | 0.564 |
| 4477 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577 | 23 | GREM1 | Sponge network | 0.249 | 0.35902 | 0.902 | 0.01691 | 0.564 |
| 4478 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | FAT4 | Sponge network | -0.769 | 0.00035 | -0.354 | 0.14694 | 0.564 |
| 4479 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-93-5p | 31 | ADARB1 | Sponge network | 0.299 | 0.57722 | -0.335 | 0.06631 | 0.564 |
| 4480 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | ATRX | Sponge network | 0.225 | 0.30644 | 0.261 | 0.04408 | 0.564 |
| 4481 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 20 | TET1 | Sponge network | -0.513 | 0.04703 | -0.135 | 0.52138 | 0.564 |
| 4482 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 35 | SCN9A | Sponge network | -0.737 | 0.06182 | -0.525 | 0.10593 | 0.564 |
| 4483 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-590-3p | 11 | DACT1 | Sponge network | -0.358 | 0.17255 | 0.783 | 0.00072 | 0.564 |
| 4484 | CTD-3064M3.3 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-744-5p | 10 | FGFR1 | Sponge network | -0.469 | 0.08721 | -0.509 | 0.03957 | 0.564 |
| 4485 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-940 | 41 | QKI | Sponge network | -0.896 | 0.00099 | -0.333 | 0.04111 | 0.564 |
| 4486 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-624-5p | 14 | WWTR1 | Sponge network | -0.517 | 0.02935 | -0.345 | 0.11997 | 0.564 |
| 4487 | RP11-774O3.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-589-5p;hsa-miR-590-3p | 15 | ROBO1 | Sponge network | -0.487 | 0.02157 | -0.009 | 0.96154 | 0.564 |
| 4488 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | LSAMP | Sponge network | -2.452 | 0 | -0.066 | 0.81708 | 0.564 |
| 4489 | SNHG14 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 62 | TRPS1 | Sponge network | -1.111 | 0.0003 | -0.191 | 0.44109 | 0.564 |
| 4490 | ZNF582-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p | 14 | SYNM | Sponge network | -1.349 | 0.00017 | -2.309 | 1.0E-5 | 0.563 |
| 4491 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-935 | 21 | DMD | Sponge network | -0.376 | 0.00337 | -1.506 | 0 | 0.563 |
| 4492 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | MLLT10 | Sponge network | 1.317 | 0 | 0.105 | 0.33292 | 0.563 |
| 4493 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-424-5p | 11 | SYNE1 | Sponge network | -0.932 | 0.00329 | -0.738 | 0.00316 | 0.563 |
| 4494 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 36 | MCC | Sponge network | -0.563 | 0.02545 | -1.005 | 0 | 0.563 |
| 4495 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-589-5p | 34 | ADARB1 | Sponge network | -2.62 | 0 | -0.335 | 0.06631 | 0.563 |
| 4496 | LINC00472 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p | 11 | PDZD2 | Sponge network | -0.842 | 0.01361 | -0.909 | 3.0E-5 | 0.563 |
| 4497 | CTD-3064M3.3 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-550a-5p | 10 | PCDH18 | Sponge network | -0.469 | 0.08721 | 0.216 | 0.24645 | 0.563 |
| 4498 | ARHGEF26-AS1 |
hsa-let-7f-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 10 | ROR2 | Sponge network | -2.46 | 0 | -1.091 | 0.0008 | 0.563 |
| 4499 | FGF14-AS2 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 13 | RASSF8 | Sponge network | -1.582 | 8.0E-5 | -0.122 | 0.65591 | 0.563 |
| 4500 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | 0.246 | 0.59912 | -1.603 | 1.0E-5 | 0.563 |
| 4501 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-3p | 16 | EPHA3 | Sponge network | 0.178 | 0.29106 | 0.185 | 0.53588 | 0.563 |
| 4502 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -1.654 | 0.00144 | -0.354 | 0.14694 | 0.563 |
| 4503 | LINC00578 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-664a-3p | 10 | RCAN2 | Sponge network | 0.811 | 0.03228 | -0.33 | 0.15172 | 0.563 |
| 4504 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-625-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -1.349 | 0.00017 | 0.024 | 0.89889 | 0.563 |
| 4505 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | TRIM33 | Sponge network | 0.768 | 7.0E-5 | 0.402 | 7.0E-5 | 0.563 |
| 4506 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 31 | ZFHX4 | Sponge network | -0.739 | 0.00044 | 0.018 | 0.95574 | 0.563 |
| 4507 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 45 | EPHA7 | Sponge network | -0.323 | 0.01372 | -2.197 | 3.0E-5 | 0.563 |
| 4508 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-98-5p | 18 | ITGA5 | Sponge network | -0.976 | 0.00025 | 0.452 | 0.03524 | 0.563 |
| 4509 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | EBF1 | Sponge network | 0.194 | 0.64278 | -0.384 | 0.08856 | 0.563 |
| 4510 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 60 | TCF4 | Sponge network | -0.563 | 0.02545 | 0.016 | 0.92271 | 0.563 |
| 4511 | FENDRR |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 16 | UNC5C | Sponge network | -1.281 | 0.00012 | -0.178 | 0.40214 | 0.563 |
| 4512 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ANK2 | Sponge network | -0.942 | 0 | -1.603 | 1.0E-5 | 0.563 |
| 4513 | RP11-488L18.10 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p | 10 | PHF6 | Sponge network | 1.392 | 0 | 0.785 | 0 | 0.563 |
| 4514 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-940 | 28 | MYO9A | Sponge network | 0.214 | 0.18246 | -0.147 | 0.32373 | 0.563 |
| 4515 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p | 11 | AKAP12 | Sponge network | -0.777 | 0.16667 | -0.932 | 0.0028 | 0.563 |
| 4516 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 16 | CNTNAP1 | Sponge network | -0.901 | 0.00019 | 0.358 | 0.13727 | 0.563 |
| 4517 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-93-5p | 20 | SLITRK3 | Sponge network | -0.162 | 0.47687 | -3.328 | 0 | 0.563 |
| 4518 | RP11-244O19.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | KCTD8 | Sponge network | -0.096 | 0.67958 | -3.359 | 0 | 0.563 |
| 4519 | AC006116.24 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577 | 15 | NRK | Sponge network | -0.473 | 0.07602 | 0.656 | 0.19217 | 0.563 |
| 4520 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p | 20 | GREM1 | Sponge network | 0.335 | 0.20596 | 0.902 | 0.01691 | 0.563 |
| 4521 | LINC00865 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-935 | 36 | DMD | Sponge network | -1.249 | 7.0E-5 | -1.506 | 0 | 0.563 |
| 4522 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p | 21 | CNTNAP3 | Sponge network | -0.576 | 0.10121 | -1.465 | 0.00033 | 0.563 |
| 4523 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | DNAJB4 | Sponge network | -2.46 | 0 | -0.7 | 1.0E-5 | 0.563 |
| 4524 | RP11-532F6.3 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-590-3p | 10 | TPO | Sponge network | -0.896 | 0.00099 | -1.87 | 2.0E-5 | 0.563 |
| 4525 | RP11-53O19.1 |
hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3613-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-7-1-3p | 23 | ANK2 | Sponge network | -0.405 | 0.22013 | -1.603 | 1.0E-5 | 0.563 |
| 4526 | USP3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p | 37 | IL17RD | Sponge network | -0.162 | 0.47687 | -0.019 | 0.94873 | 0.563 |
| 4527 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | RICTOR | Sponge network | 0.931 | 1.0E-5 | 0.388 | 0.00188 | 0.562 |
| 4528 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-940 | 37 | PBX1 | Sponge network | -1.439 | 4.0E-5 | -1.206 | 0 | 0.562 |
| 4529 | FAM66C |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 19 | PRUNE2 | Sponge network | -0.901 | 0.00019 | -1.733 | 0.00022 | 0.562 |
| 4530 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 11 | SYNE1 | Sponge network | -1.375 | 0.08642 | -0.738 | 0.00316 | 0.562 |
| 4531 | MEG3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | NRXN3 | Sponge network | 0.249 | 0.35902 | -0.893 | 0.04186 | 0.562 |
| 4532 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p | 20 | MAFB | Sponge network | -0.129 | 0.57177 | -0.023 | 0.89865 | 0.562 |
| 4533 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 30 | DDX6 | Sponge network | 1.153 | 0 | 0.091 | 0.36428 | 0.562 |
| 4534 | PWAR6 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 14 | KCTD8 | Sponge network | -1.575 | 6.0E-5 | -3.359 | 0 | 0.562 |
| 4535 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | FGF5 | Sponge network | -1.697 | 0.00889 | 0.549 | 0.28659 | 0.562 |
| 4536 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-98-5p | 14 | ITGA5 | Sponge network | -1.092 | 4.0E-5 | 0.452 | 0.03524 | 0.562 |
| 4537 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 25 | SON | Sponge network | 0.782 | 0.00016 | 0.168 | 0.04406 | 0.562 |
| 4538 | HOXB-AS1 |
hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-508-3p | 13 | RECK | Sponge network | -0.154 | 0.53954 | -1.043 | 0 | 0.562 |
| 4539 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 49 | PRKAA2 | Sponge network | -0.901 | 0.00019 | -2.462 | 0 | 0.562 |
| 4540 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 12 | ITGB3 | Sponge network | 0.997 | 0.00066 | -0.011 | 0.95751 | 0.562 |
| 4541 | CTA-941F9.9 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-576-5p | 15 | RUNX1T1 | Sponge network | -0.344 | 0.49624 | -0.344 | 0.2374 | 0.562 |
| 4542 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 18 | NR2C2 | Sponge network | 1.063 | 0 | 0.21 | 0.03224 | 0.562 |
| 4543 | USP3-AS1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p | 11 | AMPH | Sponge network | -0.162 | 0.47687 | -0.916 | 5.0E-5 | 0.562 |
| 4544 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-33a-3p;hsa-miR-361-5p | 10 | RASSF8 | Sponge network | -0.309 | 0.1145 | -0.122 | 0.65591 | 0.562 |
| 4545 | FAM66C |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | NRXN3 | Sponge network | -0.901 | 0.00019 | -0.893 | 0.04186 | 0.562 |
| 4546 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | CNTN1 | Sponge network | -1.349 | 0.00017 | -2.594 | 0 | 0.562 |
| 4547 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ITGB3 | Sponge network | -0.576 | 0.10121 | -0.011 | 0.95751 | 0.562 |
| 4548 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | CNTN1 | Sponge network | -1.281 | 0.00012 | -2.594 | 0 | 0.562 |
| 4549 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 29 | NR2C2 | Sponge network | 0.421 | 0.00084 | 0.21 | 0.03224 | 0.562 |
| 4550 | FENDRR |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 37 | IGFBP5 | Sponge network | -1.281 | 0.00012 | -0.806 | 0.00105 | 0.562 |
| 4551 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p | 63 | IL6ST | Sponge network | -0.096 | 0.67958 | -0.402 | 0.00676 | 0.562 |
| 4552 | ARHGEF26-AS1 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 11 | MLLT11 | Sponge network | -2.46 | 0 | -0.661 | 0.01733 | 0.562 |
| 4553 | RP11-774O3.3 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | MAPK10 | Sponge network | -0.487 | 0.02157 | -1.774 | 0 | 0.562 |
| 4554 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | SETBP1 | Sponge network | 0.532 | 0.00505 | -1.302 | 1.0E-5 | 0.562 |
| 4555 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-93-5p | 40 | ADARB1 | Sponge network | -0.611 | 0.09607 | -0.335 | 0.06631 | 0.562 |
| 4556 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-539-5p;hsa-miR-550a-3p | 24 | MEF2C | Sponge network | -0.473 | 0.07602 | -0.499 | 0.01448 | 0.562 |
| 4557 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | NR2C2 | Sponge network | 0.178 | 0.29106 | 0.21 | 0.03224 | 0.562 |
| 4558 | AC005154.6 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p | 13 | ZNF638 | Sponge network | 0.848 | 0 | 0.22 | 0.01214 | 0.562 |
| 4559 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p | 15 | ZNF208 | Sponge network | 0.194 | 0.64278 | -0.4 | 0.22381 | 0.562 |
| 4560 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-98-5p | 13 | PRRX1 | Sponge network | 1.344 | 0 | 1.316 | 0 | 0.562 |
| 4561 | LINC00092 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-942-5p | 24 | IGFBP5 | Sponge network | -1.886 | 0 | -0.806 | 0.00105 | 0.562 |
| 4562 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 21 | TRIM33 | Sponge network | 0.537 | 0.00139 | 0.402 | 7.0E-5 | 0.562 |
| 4563 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-93-5p;hsa-miR-940 | 19 | GAS7 | Sponge network | -1.255 | 0.00981 | -0.555 | 0.01602 | 0.562 |
| 4564 | MYLK-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p | 13 | ST6GAL2 | Sponge network | -1.331 | 3.0E-5 | -1.201 | 0.00597 | 0.562 |
| 4565 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | PDGFC | Sponge network | -0.739 | 0.0574 | -0.33 | 0.06483 | 0.562 |
| 4566 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | ANK2 | Sponge network | 0.249 | 0.35902 | -1.603 | 1.0E-5 | 0.562 |
| 4567 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 20 | RECK | Sponge network | -0.842 | 0.01361 | -1.043 | 0 | 0.562 |
| 4568 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p | 24 | PBX1 | Sponge network | -0.517 | 0.02935 | -1.206 | 0 | 0.562 |
| 4569 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-576-5p | 25 | IGF1 | Sponge network | -0.611 | 0.09607 | -0.204 | 0.5898 | 0.562 |
| 4570 | USP3-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-589-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | -0.162 | 0.47687 | -0.366 | 0.01036 | 0.562 |
| 4571 | CTD-2314G24.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | 0.246 | 0.59912 | -1.051 | 0.00022 | 0.562 |
| 4572 | EMX2OS |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-484;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | NRXN3 | Sponge network | -0.777 | 0.1134 | -0.893 | 0.04186 | 0.562 |
| 4573 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-590-3p | 10 | NRXN3 | Sponge network | -0.739 | 0.00044 | -0.893 | 0.04186 | 0.562 |
| 4574 | EMX2OS |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 39 | NTRK2 | Sponge network | -0.777 | 0.1134 | -0.736 | 0.06495 | 0.562 |
| 4575 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-629-5p | 34 | SOX5 | Sponge network | -2.69 | 0.0001 | -1.091 | 4.0E-5 | 0.562 |
| 4576 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | ROBO1 | Sponge network | -1.111 | 0.0003 | -0.009 | 0.96154 | 0.562 |
| 4577 | NDUFA6-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 18 | EZH1 | Sponge network | -0.355 | 0.0077 | -0.564 | 1.0E-5 | 0.562 |
| 4578 | THAP9-AS1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-195-5p;hsa-miR-23b-3p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-497-5p;hsa-miR-654-3p | 10 | HNRNPA2B1 | Sponge network | 1.079 | 0 | 0.55 | 0 | 0.562 |
| 4579 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-542-3p | 30 | DTNA | Sponge network | -0.517 | 0.02935 | -1.648 | 1.0E-5 | 0.561 |
| 4580 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 31 | TSHZ3 | Sponge network | -0.096 | 0.67958 | -0.556 | 0.01516 | 0.561 |
| 4581 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 34 | ANK2 | Sponge network | -0.777 | 0.1134 | -1.603 | 1.0E-5 | 0.561 |
| 4582 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-576-5p | 26 | CACNA2D1 | Sponge network | -0.162 | 0.47687 | -0.846 | 0.00675 | 0.561 |
| 4583 | AC010226.4 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-7-5p | 24 | BTG2 | Sponge network | -0.811 | 2.0E-5 | -1.763 | 0 | 0.561 |
| 4584 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | PRUNE2 | Sponge network | 0.572 | 0.01722 | -1.733 | 0.00022 | 0.561 |
| 4585 | CTC-296K1.3 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-3913-5p;hsa-miR-452-5p | 10 | FNBP1 | Sponge network | -2.095 | 0.00112 | -0.969 | 4.0E-5 | 0.561 |
| 4586 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 39 | MYO9A | Sponge network | -0.737 | 0.06182 | -0.147 | 0.32373 | 0.561 |
| 4587 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 49 | DTNA | Sponge network | -0.896 | 0.00099 | -1.648 | 1.0E-5 | 0.561 |
| 4588 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | ROBO1 | Sponge network | 0.997 | 0.00066 | -0.009 | 0.96154 | 0.561 |
| 4589 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | EDA2R | Sponge network | 0.572 | 0.01722 | -0.587 | 0.01598 | 0.561 |
| 4590 | LINC00702 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 58 | TRPS1 | Sponge network | -0.737 | 0.06182 | -0.191 | 0.44109 | 0.561 |
| 4591 | CTD-2349P21.10 |
hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 13 | MDM4 | Sponge network | 1.341 | 0.00014 | 0.345 | 0.00762 | 0.561 |
| 4592 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | FAT3 | Sponge network | -1.349 | 0.00017 | -1.601 | 0.00051 | 0.561 |
| 4593 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | PGR | Sponge network | -0.421 | 0.0114 | -1.704 | 0 | 0.561 |
| 4594 | JAZF1-AS1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-93-3p | 10 | PEA15 | Sponge network | -0.769 | 0.00035 | 0.009 | 0.93318 | 0.561 |
| 4595 | SNHG14 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 45 | IGFBP5 | Sponge network | -1.111 | 0.0003 | -0.806 | 0.00105 | 0.561 |
| 4596 | ARHGEF26-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 32 | WWTR1 | Sponge network | -2.46 | 0 | -0.345 | 0.11997 | 0.561 |
| 4597 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 18 | FGFR1 | Sponge network | 0.214 | 0.18246 | -0.509 | 0.03957 | 0.561 |
| 4598 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | ABCC9 | Sponge network | -0.612 | 0.00094 | -0.466 | 0.14121 | 0.561 |
| 4599 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p | 34 | ZFHX4 | Sponge network | -2.69 | 0.0001 | 0.018 | 0.95574 | 0.561 |
| 4600 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b | 10 | DCN | Sponge network | -2.69 | 0.0001 | -1.288 | 0 | 0.561 |
| 4601 | RP11-156P1.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-590-3p | 16 | ITPKB | Sponge network | 0.214 | 0.18246 | -0.704 | 0.00106 | 0.561 |
| 4602 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-660-5p | 18 | ESR1 | Sponge network | -0.72 | 0.24239 | -1.035 | 3.0E-5 | 0.561 |
| 4603 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | EPHA3 | Sponge network | 0.335 | 0.20596 | 0.185 | 0.53588 | 0.561 |
| 4604 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-577;hsa-miR-664a-3p | 17 | GREM1 | Sponge network | -2.69 | 0.0001 | 0.902 | 0.01691 | 0.561 |
| 4605 | CTC-444N24.11 |
hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-940 | 11 | TAF1 | Sponge network | 1.153 | 0 | 0.39 | 3.0E-5 | 0.561 |
| 4606 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 15 | STARD13 | Sponge network | 0.194 | 0.64278 | -0.187 | 0.27383 | 0.561 |
| 4607 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 21 | TSHZ3 | Sponge network | -1.255 | 0.00981 | -0.556 | 0.01516 | 0.561 |
| 4608 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 38 | TSHZ3 | Sponge network | -0.563 | 0.02545 | -0.556 | 0.01516 | 0.561 |
| 4609 | RP11-736K20.5 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-590-3p | 14 | ITPKB | Sponge network | -0.358 | 0.17255 | -0.704 | 0.00106 | 0.561 |
| 4610 | RP11-400K9.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-671-5p | 10 | SORCS1 | Sponge network | -0.611 | 0.09607 | -3.492 | 0 | 0.561 |
| 4611 | LINC00094 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-576-5p | 19 | HIP1 | Sponge network | 0.253 | 0.09565 | 0.774 | 0 | 0.561 |
| 4612 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 14 | UVRAG | Sponge network | -0.323 | 0.01372 | -0.017 | 0.83865 | 0.561 |
| 4613 | JAZF1-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3913-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | WWTR1 | Sponge network | -0.769 | 0.00035 | -0.345 | 0.11997 | 0.561 |
| 4614 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -0.611 | 0.09607 | -0.354 | 0.14694 | 0.561 |
| 4615 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 22 | EBF1 | Sponge network | -1.582 | 8.0E-5 | -0.384 | 0.08856 | 0.561 |
| 4616 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 73 | CADM2 | Sponge network | -0.129 | 0.57177 | -3.782 | 0 | 0.561 |
| 4617 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-93-5p | 21 | ST6GALNAC3 | Sponge network | -0.473 | 0.07602 | -0.987 | 0 | 0.561 |
| 4618 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 28 | NR2C2 | Sponge network | 0.72 | 0.0001 | 0.21 | 0.03224 | 0.561 |
| 4619 | RP11-774O3.3 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | -0.487 | 0.02157 | -2.633 | 0 | 0.561 |
| 4620 | LINC00092 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | DMD | Sponge network | -1.886 | 0 | -1.506 | 0 | 0.56 |
| 4621 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 22 | NR2C2 | Sponge network | 0.848 | 0 | 0.21 | 0.03224 | 0.56 |
| 4622 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 32 | PGR | Sponge network | 0.246 | 0.59912 | -1.704 | 0 | 0.56 |
| 4623 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 29 | CBFA2T3 | Sponge network | -1.111 | 0.0003 | -1.221 | 1.0E-5 | 0.56 |
| 4624 | CTC-296K1.3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-582-5p | 16 | PCDH10 | Sponge network | -2.095 | 0.00112 | -1.644 | 0.0078 | 0.56 |
| 4625 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-590-3p | 23 | CACNA2D1 | Sponge network | -0.096 | 0.67958 | -0.846 | 0.00675 | 0.56 |
| 4626 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 28 | ZFHX4 | Sponge network | -4.546 | 0 | 0.018 | 0.95574 | 0.56 |
| 4627 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -1.092 | 4.0E-5 | 0.024 | 0.89889 | 0.56 |
| 4628 | LINC00092 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-769-5p | 11 | PRUNE2 | Sponge network | -1.886 | 0 | -1.733 | 0.00022 | 0.56 |
| 4629 | ZNF667-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | GJA1 | Sponge network | -0.976 | 0.00025 | -0.485 | 0.02179 | 0.56 |
| 4630 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | -0.49 | 0.04946 | 0.783 | 0.00072 | 0.56 |
| 4631 | TPTEP1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-769-5p | 31 | SYNPO2 | Sponge network | -1.216 | 0 | -2.342 | 0 | 0.56 |
| 4632 | HAND2-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 18 | FAM172A | Sponge network | -1.697 | 0.00889 | -0.57 | 0 | 0.56 |
| 4633 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-590-3p | 31 | SSBP2 | Sponge network | -1.886 | 0 | -1.58 | 0 | 0.56 |
| 4634 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-3p | 10 | GRIN2A | Sponge network | -4.463 | 0.00025 | -2.161 | 0 | 0.56 |
| 4635 | C20orf166-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 39 | GRIK3 | Sponge network | -2.69 | 0.0001 | -3.208 | 0 | 0.56 |
| 4636 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-5p | 25 | KCNA6 | Sponge network | -1.331 | 3.0E-5 | -1.531 | 0.00013 | 0.56 |
| 4637 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 16 | PLSCR4 | Sponge network | -0.517 | 0.02935 | -0.474 | 0.03833 | 0.56 |
| 4638 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 15 | AFF3 | Sponge network | -0.811 | 2.0E-5 | -2.373 | 0 | 0.56 |
| 4639 | MEG3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 12 | HIC1 | Sponge network | 0.249 | 0.35902 | 0.024 | 0.89889 | 0.56 |
| 4640 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CACNA2D1 | Sponge network | -0.901 | 0.00019 | -0.846 | 0.00675 | 0.56 |
| 4641 | TRAF3IP2-AS1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | SPOP | Sponge network | -0.323 | 0.01372 | -0.419 | 1.0E-5 | 0.56 |
| 4642 | CTD-2089N3.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p | 16 | EPHA7 | Sponge network | -0.314 | 0.62033 | -2.197 | 3.0E-5 | 0.56 |
| 4643 | HAND2-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-93-3p;hsa-miR-940 | 49 | GRIK3 | Sponge network | -1.697 | 0.00889 | -3.208 | 0 | 0.56 |
| 4644 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 45 | HLF | Sponge network | -0.129 | 0.57177 | -2.443 | 0 | 0.56 |
| 4645 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | EYA4 | Sponge network | 0.335 | 0.20596 | 0.3 | 0.36876 | 0.56 |
| 4646 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | SON | Sponge network | 1.106 | 0 | 0.168 | 0.04406 | 0.56 |
| 4647 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 16 | AKAP12 | Sponge network | -1.281 | 0.00012 | -0.932 | 0.0028 | 0.56 |
| 4648 | RP11-244O19.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3913-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-651-5p | 22 | WWTR1 | Sponge network | -0.096 | 0.67958 | -0.345 | 0.11997 | 0.56 |
| 4649 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-503-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 41 | RICTOR | Sponge network | 0.782 | 0.00016 | 0.388 | 0.00188 | 0.56 |
| 4650 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p | 17 | SPG20 | Sponge network | -0.611 | 0.09607 | -1.16 | 0 | 0.56 |
| 4651 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-98-5p | 62 | PRKAA2 | Sponge network | -0.129 | 0.57177 | -2.462 | 0 | 0.56 |
| 4652 | RP11-620J15.3 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p | 11 | SFRP1 | Sponge network | -0.739 | 0.00044 | -3.566 | 0 | 0.56 |
| 4653 | ZNF667-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p | 13 | KCTD8 | Sponge network | -0.976 | 0.00025 | -3.359 | 0 | 0.56 |
| 4654 | TRAF3IP2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-877-5p | 27 | ASTN1 | Sponge network | -0.323 | 0.01372 | -2.389 | 2.0E-5 | 0.56 |
| 4655 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-455-3p;hsa-miR-93-5p | 23 | MCC | Sponge network | -1.654 | 0.00144 | -1.005 | 0 | 0.56 |
| 4656 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-942-5p | 34 | NRXN1 | Sponge network | -0.793 | 7.0E-5 | -3.778 | 0 | 0.56 |
| 4657 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 21 | CNTNAP3 | Sponge network | -0.739 | 0.0574 | -1.465 | 0.00033 | 0.56 |
| 4658 | LINC00472 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-942-5p | 11 | ZBTB20 | Sponge network | -0.842 | 0.01361 | -0.122 | 0.57569 | 0.56 |
| 4659 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | CACNA2D1 | Sponge network | 0.572 | 0.01722 | -0.846 | 0.00675 | 0.56 |
| 4660 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-93-3p | 33 | SSBP2 | Sponge network | -0.693 | 0.05629 | -1.58 | 0 | 0.56 |
| 4661 | CTD-3064M3.3 |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p | 15 | AFF3 | Sponge network | -0.469 | 0.08721 | -2.373 | 0 | 0.56 |
| 4662 | FENDRR |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 20 | CADM3 | Sponge network | -1.281 | 0.00012 | -3.663 | 0 | 0.56 |
| 4663 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | GLI3 | Sponge network | -0.739 | 0.00044 | -0.615 | 0.01482 | 0.56 |
| 4664 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | RECK | Sponge network | -1.203 | 0.03881 | -1.043 | 0 | 0.56 |
| 4665 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | EBF1 | Sponge network | -0.739 | 0.00044 | -0.384 | 0.08856 | 0.56 |
| 4666 | NR2F1-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 16 | STAT5B | Sponge network | -0.49 | 0.04946 | -0.182 | 0.1082 | 0.56 |
| 4667 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | FSTL5 | Sponge network | -1.697 | 0.00889 | -1.3 | 0.01619 | 0.56 |
| 4668 | PWAR6 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TPO | Sponge network | -1.575 | 6.0E-5 | -1.87 | 2.0E-5 | 0.56 |
| 4669 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | FAT4 | Sponge network | 0.249 | 0.35902 | -0.354 | 0.14694 | 0.56 |
| 4670 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 23 | TBL1XR1 | Sponge network | 1.923 | 0 | 0.578 | 0 | 0.56 |
| 4671 | PART1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 23 | MRVI1 | Sponge network | -2.925 | 0 | -1.051 | 0.00022 | 0.56 |
| 4672 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 27 | PCDH10 | Sponge network | -0.793 | 7.0E-5 | -1.644 | 0.0078 | 0.56 |
| 4673 | LINC00957 |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-590-3p | 17 | AFF3 | Sponge network | -0.421 | 0.0114 | -2.373 | 0 | 0.559 |
| 4674 | C20orf166-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-542-3p | 10 | ID4 | Sponge network | -2.69 | 0.0001 | -1.644 | 0 | 0.559 |
| 4675 | CTD-2002H8.2 |
hsa-miR-125a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PHC3 | Sponge network | 0.931 | 1.0E-5 | 0.212 | 0.0499 | 0.559 |
| 4676 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-450b-5p | 11 | CNTNAP1 | Sponge network | 0.176 | 0.37402 | 0.358 | 0.13727 | 0.559 |
| 4677 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-339-5p | 13 | STARD13 | Sponge network | -1.092 | 4.0E-5 | -0.187 | 0.27383 | 0.559 |
| 4678 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-450b-5p | 20 | JAZF1 | Sponge network | -0.583 | 0.14589 | -0.377 | 0.02677 | 0.559 |
| 4679 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-628-5p;hsa-miR-629-3p | 21 | RASSF8 | Sponge network | -2.62 | 0 | -0.122 | 0.65591 | 0.559 |
| 4680 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-93-3p | 56 | RUNX1T1 | Sponge network | -0.487 | 0.02157 | -0.344 | 0.2374 | 0.559 |
| 4681 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 38 | MITF | Sponge network | -1.111 | 0.0003 | -0.769 | 0.00022 | 0.559 |
| 4682 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-98-5p | 42 | PBX1 | Sponge network | -0.942 | 0 | -1.206 | 0 | 0.559 |
| 4683 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | PTGFR | Sponge network | 0.997 | 0.00066 | -0.233 | 0.45421 | 0.559 |
| 4684 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-511-5p;hsa-miR-590-3p | 20 | PCDH10 | Sponge network | -0.739 | 0.00044 | -1.644 | 0.0078 | 0.559 |
| 4685 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-582-5p | 22 | FBXO32 | Sponge network | 0.299 | 0.57722 | -0.495 | 0.07561 | 0.559 |
| 4686 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-577 | 17 | GREM1 | Sponge network | -1.439 | 4.0E-5 | 0.902 | 0.01691 | 0.559 |
| 4687 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | RCAN2 | Sponge network | -0.896 | 0.00099 | -0.33 | 0.15172 | 0.559 |
| 4688 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-582-5p | 26 | EPHA7 | Sponge network | -2.095 | 0.00112 | -2.197 | 3.0E-5 | 0.559 |
| 4689 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | EPHA3 | Sponge network | 0.727 | 3.0E-5 | 0.185 | 0.53588 | 0.559 |
| 4690 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p | 43 | ADARB1 | Sponge network | 0.064 | 0.72934 | -0.335 | 0.06631 | 0.559 |
| 4691 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | NRXN3 | Sponge network | -0.612 | 0.00094 | -0.893 | 0.04186 | 0.559 |
| 4692 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 25 | BCL2 | Sponge network | -1.092 | 4.0E-5 | -0.899 | 0 | 0.559 |
| 4693 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | AHNAK | Sponge network | -0.611 | 0.09607 | -1.207 | 0 | 0.559 |
| 4694 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | RBMS3 | Sponge network | -0.021 | 0.93598 | -0.519 | 0.03551 | 0.559 |
| 4695 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p | 25 | ZFHX4 | Sponge network | -0.473 | 0.07602 | 0.018 | 0.95574 | 0.559 |
| 4696 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 30 | RICTOR | Sponge network | 0.91 | 0 | 0.388 | 0.00188 | 0.559 |
| 4697 | LINC00702 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 30 | CTIF | Sponge network | -0.737 | 0.06182 | -0.389 | 0.00549 | 0.559 |
| 4698 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 46 | GAS7 | Sponge network | -0.901 | 0.00019 | -0.555 | 0.01602 | 0.559 |
| 4699 | LINC00702 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 41 | SNX29 | Sponge network | -0.737 | 0.06182 | -0.34 | 0.01371 | 0.559 |
| 4700 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p | 31 | DMD | Sponge network | -1.161 | 0.00591 | -1.506 | 0 | 0.559 |
| 4701 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 36 | MCC | Sponge network | -2.117 | 0.00161 | -1.005 | 0 | 0.559 |
| 4702 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 28 | ANK2 | Sponge network | -0.513 | 0.04703 | -1.603 | 1.0E-5 | 0.559 |
| 4703 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-589-3p;hsa-miR-590-3p | 22 | DIP2C | Sponge network | -0.358 | 0.17255 | -0.436 | 0.01713 | 0.559 |
| 4704 | MIR497HG |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-577;hsa-miR-877-5p | 20 | NRK | Sponge network | -1.439 | 4.0E-5 | 0.656 | 0.19217 | 0.559 |
| 4705 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | WWTR1 | Sponge network | -0.739 | 0.00044 | -0.345 | 0.11997 | 0.559 |
| 4706 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 46 | TACC1 | Sponge network | -0.49 | 0.04946 | -0.362 | 0.05406 | 0.559 |
| 4707 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-424-5p | 14 | ANK2 | Sponge network | -2.076 | 0.00548 | -1.603 | 1.0E-5 | 0.559 |
| 4708 | FENDRR |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-2355-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p | 10 | NUDT11 | Sponge network | -1.281 | 0.00012 | -0.921 | 0.00376 | 0.559 |
| 4709 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 41 | ESR1 | Sponge network | -0.739 | 0.0574 | -1.035 | 3.0E-5 | 0.559 |
| 4710 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | EDA2R | Sponge network | -0.739 | 0.00044 | -0.587 | 0.01598 | 0.559 |
| 4711 | PGM5-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-589-5p;hsa-miR-7-1-3p | 19 | PRKCB | Sponge network | -4.546 | 0 | -1.313 | 2.0E-5 | 0.559 |
| 4712 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 22 | CADM3 | Sponge network | -0.769 | 0.00035 | -3.663 | 0 | 0.559 |
| 4713 | RP11-119F7.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-502-5p;hsa-miR-92a-3p | 28 | IL17RD | Sponge network | 0.225 | 0.30644 | -0.019 | 0.94873 | 0.559 |
| 4714 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 44 | TSHZ3 | Sponge network | -1.216 | 0 | -0.556 | 0.01516 | 0.559 |
| 4715 | SNHG14 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | TLE4 | Sponge network | -1.111 | 0.0003 | -1.157 | 0 | 0.559 |
| 4716 | RP11-10N23.2 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 21 | MDM4 | Sponge network | 1.134 | 0 | 0.345 | 0.00762 | 0.559 |
| 4717 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | PLSCR4 | Sponge network | -0.842 | 0.01361 | -0.474 | 0.03833 | 0.559 |
| 4718 | MIR497HG |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 19 | EZH1 | Sponge network | -1.439 | 4.0E-5 | -0.564 | 1.0E-5 | 0.559 |
| 4719 | LINC00654 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-5p;hsa-miR-589-5p | 11 | MN1 | Sponge network | 0.374 | 0.20054 | -0.358 | 0.24172 | 0.558 |
| 4720 | OIP5-AS1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p | 14 | ZNF638 | Sponge network | 0.421 | 0.00084 | 0.22 | 0.01214 | 0.558 |
| 4721 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p | 15 | CADM3 | Sponge network | -2.46 | 0 | -3.663 | 0 | 0.558 |
| 4722 | MYLK-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-340-5p | 12 | CLIP1 | Sponge network | -1.331 | 3.0E-5 | -0.215 | 0.08684 | 0.558 |
| 4723 | AC003090.1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | RPS6KA6 | Sponge network | -2.117 | 0.00161 | -2.989 | 0 | 0.558 |
| 4724 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-508-3p | 21 | RECK | Sponge network | -0.162 | 0.47687 | -1.043 | 0 | 0.558 |
| 4725 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | MEF2C | Sponge network | -4.546 | 0 | -0.499 | 0.01448 | 0.558 |
| 4726 | DNM3OS |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 17 | FMNL3 | Sponge network | 0.572 | 0.01722 | 0.555 | 4.0E-5 | 0.558 |
| 4727 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-451a;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 48 | BCL2 | Sponge network | -0.129 | 0.57177 | -0.899 | 0 | 0.558 |
| 4728 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 22 | ADAMTSL3 | Sponge network | 0.335 | 0.20596 | -1.559 | 0 | 0.558 |
| 4729 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p | 12 | ZBTB4 | Sponge network | -0.309 | 0.1145 | -0.739 | 0 | 0.558 |
| 4730 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 17 | PAK3 | Sponge network | -2.452 | 0 | -0.628 | 0.07253 | 0.558 |
| 4731 | GNG12-AS1 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | RTN4 | Sponge network | -0.793 | 7.0E-5 | -0.412 | 0.00014 | 0.558 |
| 4732 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 15 | CNTNAP1 | Sponge network | -0.323 | 0.01372 | 0.358 | 0.13727 | 0.558 |
| 4733 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 29 | AKAP11 | Sponge network | 0.614 | 1.0E-5 | 0.196 | 0.09825 | 0.558 |
| 4734 | ACTA2-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 19 | SFRP1 | Sponge network | 0.194 | 0.64278 | -3.566 | 0 | 0.558 |
| 4735 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 57 | QKI | Sponge network | 0.997 | 0.00066 | -0.333 | 0.04111 | 0.558 |
| 4736 | MAGI2-AS3 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-503-5p | 24 | ABL1 | Sponge network | -0.129 | 0.57177 | -0.215 | 0.07804 | 0.558 |
| 4737 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-629-3p | 19 | RASSF8 | Sponge network | -0.469 | 0.08721 | -0.122 | 0.65591 | 0.558 |
| 4738 | HAND2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | RELN | Sponge network | -1.697 | 0.00889 | -2.403 | 0 | 0.558 |
| 4739 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | RECK | Sponge network | -0.777 | 0.1134 | -1.043 | 0 | 0.558 |
| 4740 | PGM5-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-940 | 10 | GLI3 | Sponge network | -4.546 | 0 | -0.615 | 0.01482 | 0.558 |
| 4741 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 15 | EBF3 | Sponge network | 0.727 | 3.0E-5 | -0.058 | 0.81437 | 0.558 |
| 4742 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429 | 18 | AKAP6 | Sponge network | -1.654 | 0.00144 | -1.586 | 3.0E-5 | 0.558 |
| 4743 | PART1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | KCTD8 | Sponge network | -2.925 | 0 | -3.359 | 0 | 0.558 |
| 4744 | ZNF582-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-7-5p | 12 | FLNA | Sponge network | -1.349 | 0.00017 | -0.771 | 0.01377 | 0.558 |
| 4745 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | ADAMTSL3 | Sponge network | -1.216 | 0 | -1.559 | 0 | 0.558 |
| 4746 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 48 | ZFHX4 | Sponge network | -0.901 | 0.00019 | 0.018 | 0.95574 | 0.558 |
| 4747 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-874-3p | 19 | CRTC3 | Sponge network | -1.092 | 4.0E-5 | 0.164 | 0.16786 | 0.558 |
| 4748 | SH3RF3-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-5p | 13 | MRVI1 | Sponge network | 0.889 | 9.0E-5 | -1.051 | 0.00022 | 0.558 |
| 4749 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | JAZF1 | Sponge network | -0.737 | 0.06182 | -0.377 | 0.02677 | 0.558 |
| 4750 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 39 | BNC2 | Sponge network | -0.739 | 0.00044 | -0.491 | 0.09988 | 0.558 |
| 4751 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | IGSF10 | Sponge network | -0.739 | 0.00044 | -1.751 | 1.0E-5 | 0.558 |
| 4752 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-511-5p;hsa-miR-590-3p | 23 | ZFHX3 | Sponge network | 1.04 | 0.00023 | 0.389 | 0.01363 | 0.558 |
| 4753 | C20orf166-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p | 35 | IGFBP5 | Sponge network | -2.69 | 0.0001 | -0.806 | 0.00105 | 0.558 |
| 4754 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 27 | SCN9A | Sponge network | -0.976 | 0.00025 | -0.525 | 0.10593 | 0.558 |
| 4755 | GNG12-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 18 | EZH1 | Sponge network | -0.793 | 7.0E-5 | -0.564 | 1.0E-5 | 0.558 |
| 4756 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | 0.532 | 0.00505 | -0.466 | 0.14121 | 0.558 |
| 4757 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p | 19 | PGR | Sponge network | -0.517 | 0.02935 | -1.704 | 0 | 0.558 |
| 4758 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 30 | RNF144A | Sponge network | -0.129 | 0.57177 | 0.594 | 0.0009 | 0.558 |
| 4759 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-93-3p | 23 | SSBP2 | Sponge network | -4.463 | 0.00025 | -1.58 | 0 | 0.558 |
| 4760 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 33 | GPC6 | Sponge network | -1.111 | 0.0003 | -0.054 | 0.81465 | 0.558 |
| 4761 | RP11-326G21.1 |
hsa-let-7b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 25 | RUNX1T1 | Sponge network | -0.021 | 0.93598 | -0.344 | 0.2374 | 0.558 |
| 4762 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | -0.323 | 0.01372 | -1.892 | 0 | 0.558 |
| 4763 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 36 | CLASP2 | Sponge network | 0.537 | 0.00139 | -0.038 | 0.71 | 0.558 |
| 4764 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-940 | 39 | QKI | Sponge network | 0.199 | 0.38015 | -0.333 | 0.04111 | 0.558 |
| 4765 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 74 | NTRK3 | Sponge network | -0.777 | 0.1134 | -1.77 | 3.0E-5 | 0.558 |
| 4766 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-935;hsa-miR-96-5p | 28 | FBXO32 | Sponge network | 0.249 | 0.35902 | -0.495 | 0.07561 | 0.558 |
| 4767 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 39 | TRPS1 | Sponge network | -0.976 | 0.00025 | -0.191 | 0.44109 | 0.558 |
| 4768 | RP11-867G23.10 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-34a-5p;hsa-miR-361-3p | 11 | ITPKB | Sponge network | -2.531 | 0 | -0.704 | 0.00106 | 0.558 |
| 4769 | TRAF3IP2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 27 | NAV3 | Sponge network | -0.323 | 0.01372 | 0.212 | 0.47729 | 0.558 |
| 4770 | PCA3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940 | 44 | HLF | Sponge network | -0.709 | 0.18168 | -2.443 | 0 | 0.557 |
| 4771 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | SLITRK3 | Sponge network | -0.777 | 0.1134 | -3.328 | 0 | 0.557 |
| 4772 | USP3-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-940 | 16 | SFRP1 | Sponge network | -0.162 | 0.47687 | -3.566 | 0 | 0.557 |
| 4773 | EMX2OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | PCDH9 | Sponge network | -0.777 | 0.1134 | -1.823 | 4.0E-5 | 0.557 |
| 4774 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 49 | PRKAA2 | Sponge network | -0.405 | 0.22013 | -2.462 | 0 | 0.557 |
| 4775 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 22 | FAT3 | Sponge network | -0.376 | 0.00337 | -1.601 | 0.00051 | 0.557 |
| 4776 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 31 | GRIN2A | Sponge network | -0.899 | 6.0E-5 | -2.161 | 0 | 0.557 |
| 4777 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-511-5p;hsa-miR-628-5p;hsa-miR-940 | 12 | TRIM33 | Sponge network | 1.162 | 5.0E-5 | 0.402 | 7.0E-5 | 0.557 |
| 4778 | LINC01018 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-532-3p | 15 | SYNM | Sponge network | -2.915 | 0.00015 | -2.309 | 1.0E-5 | 0.557 |
| 4779 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 19 | MITF | Sponge network | -0.611 | 0.09607 | -0.769 | 0.00022 | 0.557 |
| 4780 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CACNA2D1 | Sponge network | -0.421 | 0.0114 | -0.846 | 0.00675 | 0.557 |
| 4781 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | CACNA2D1 | Sponge network | -0.896 | 0.00099 | -0.846 | 0.00675 | 0.557 |
| 4782 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p | 29 | ADAMTSL3 | Sponge network | -0.793 | 7.0E-5 | -1.559 | 0 | 0.557 |
| 4783 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | EBF1 | Sponge network | 0.342 | 0.03129 | -0.384 | 0.08856 | 0.557 |
| 4784 | HOXB-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-429 | 13 | SYNM | Sponge network | -0.154 | 0.53954 | -2.309 | 1.0E-5 | 0.557 |
| 4785 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-590-3p | 14 | RBM5 | Sponge network | -0.222 | 0.32445 | -0.205 | 0.0129 | 0.557 |
| 4786 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 23 | RCAN2 | Sponge network | -2.46 | 0 | -0.33 | 0.15172 | 0.557 |
| 4787 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-3p | 22 | SLITRK3 | Sponge network | -0.942 | 0 | -3.328 | 0 | 0.557 |
| 4788 | CTD-2002H8.2 |
hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | TRIM33 | Sponge network | 0.931 | 1.0E-5 | 0.402 | 7.0E-5 | 0.557 |
| 4789 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | ZNF770 | Sponge network | 0.34 | 0.00273 | 0.206 | 0.06751 | 0.557 |
| 4790 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 43 | CBX5 | Sponge network | 0.083 | 0.66405 | 0.165 | 0.24446 | 0.557 |
| 4791 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-874-3p | 13 | NR2C2 | Sponge network | 0.924 | 0 | 0.21 | 0.03224 | 0.557 |
| 4792 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 35 | ANK2 | Sponge network | -1.216 | 0 | -1.603 | 1.0E-5 | 0.557 |
| 4793 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | DYNC1I1 | Sponge network | -2.117 | 0.00161 | -1.12 | 0.00107 | 0.557 |
| 4794 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-451a;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 37 | BCL2 | Sponge network | -0.896 | 0.00099 | -0.899 | 0 | 0.557 |
| 4795 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 35 | AKAP6 | Sponge network | -0.942 | 0 | -1.586 | 3.0E-5 | 0.557 |
| 4796 | RP11-53O19.3 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 15 | PHF6 | Sponge network | 1.205 | 0 | 0.785 | 0 | 0.557 |
| 4797 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 52 | PIK3R1 | Sponge network | -0.739 | 0.0574 | -0.133 | 0.36854 | 0.557 |
| 4798 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | PREX2 | Sponge network | 0.997 | 0.00066 | -0.272 | 0.25382 | 0.557 |
| 4799 | MEG3 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p | 10 | RADIL | Sponge network | 0.249 | 0.35902 | -1.628 | 0 | 0.557 |
| 4800 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | -0.576 | 0.10121 | -2.417 | 0 | 0.557 |
| 4801 | AC009948.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 25 | NAV3 | Sponge network | 0.532 | 0.00505 | 0.212 | 0.47729 | 0.557 |
| 4802 | LINC00702 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | PRKCB | Sponge network | -0.737 | 0.06182 | -1.313 | 2.0E-5 | 0.557 |
| 4803 | SNHG14 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 28 | TMEFF2 | Sponge network | -1.111 | 0.0003 | -2.426 | 0 | 0.557 |
| 4804 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-93-5p | 13 | EZH1 | Sponge network | -0.611 | 0.09607 | -0.564 | 1.0E-5 | 0.557 |
| 4805 | AP001347.6 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 46 | AR | Sponge network | -1.161 | 0.00591 | -1.554 | 0 | 0.557 |
| 4806 | PWAR6 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-940 | 14 | PHOX2B | Sponge network | -1.575 | 6.0E-5 | -2.68 | 0 | 0.557 |
| 4807 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-577;hsa-miR-590-3p | 15 | DNAJB4 | Sponge network | -0.793 | 7.0E-5 | -0.7 | 1.0E-5 | 0.557 |
| 4808 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 20 | SON | Sponge network | 0.537 | 0.00139 | 0.168 | 0.04406 | 0.557 |
| 4809 | FAM66C |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 35 | DIP2C | Sponge network | -0.901 | 0.00019 | -0.436 | 0.01713 | 0.557 |
| 4810 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-590-3p | 14 | JAKMIP2 | Sponge network | -1.216 | 0 | -1.388 | 0 | 0.557 |
| 4811 | HNRNPU-AS1 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p | 27 | ABL2 | Sponge network | 1.601 | 0 | 0.82 | 0 | 0.557 |
| 4812 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | FNBP1 | Sponge network | 0.572 | 0.01722 | -0.969 | 4.0E-5 | 0.557 |
| 4813 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p | 19 | FGFR1 | Sponge network | -0.896 | 0.00099 | -0.509 | 0.03957 | 0.557 |
| 4814 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 12 | AKAP12 | Sponge network | -0.405 | 0.22013 | -0.932 | 0.0028 | 0.557 |
| 4815 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p | 20 | PLSCR4 | Sponge network | -0.773 | 0.04073 | -0.474 | 0.03833 | 0.557 |
| 4816 | RP11-503E24.2 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-495-3p | 16 | MDM4 | Sponge network | 2.218 | 0 | 0.345 | 0.00762 | 0.557 |
| 4817 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-424-5p | 20 | LRP1B | Sponge network | -1.331 | 3.0E-5 | -2.163 | 0 | 0.556 |
| 4818 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | FYN | Sponge network | -0.563 | 0.02545 | -0.131 | 0.37051 | 0.556 |
| 4819 | ARHGEF26-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 12 | PHOX2B | Sponge network | -2.46 | 0 | -2.68 | 0 | 0.556 |
| 4820 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-98-5p | 33 | THBS1 | Sponge network | -0.49 | 0.04946 | 0.741 | 0.00386 | 0.556 |
| 4821 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 45 | BNC2 | Sponge network | -0.793 | 7.0E-5 | -0.491 | 0.09988 | 0.556 |
| 4822 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-96-5p | 12 | CBFA2T3 | Sponge network | -0.351 | 0.11358 | -1.221 | 1.0E-5 | 0.556 |
| 4823 | LINC00472 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 52 | DTNA | Sponge network | -0.842 | 0.01361 | -1.648 | 1.0E-5 | 0.556 |
| 4824 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-508-3p;hsa-miR-590-3p | 14 | MITF | Sponge network | -0.376 | 0.00337 | -0.769 | 0.00022 | 0.556 |
| 4825 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p | 14 | RB1CC1 | Sponge network | 0.75 | 0.00054 | 0.175 | 0.12559 | 0.556 |
| 4826 | JAZF1-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-671-5p | 29 | DLC1 | Sponge network | -0.769 | 0.00035 | -0.382 | 0.05038 | 0.556 |
| 4827 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 21 | SON | Sponge network | 0.959 | 0 | 0.168 | 0.04406 | 0.556 |
| 4828 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | PIK3CA | Sponge network | 0.683 | 1.0E-5 | 0.195 | 0.06908 | 0.556 |
| 4829 | LINC00654 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 22 | HIP1 | Sponge network | 0.374 | 0.20054 | 0.774 | 0 | 0.556 |
| 4830 | FENDRR |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | KCTD8 | Sponge network | -1.281 | 0.00012 | -3.359 | 0 | 0.556 |
| 4831 | RP11-389C8.2 |
hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PCDH18 | Sponge network | 0.727 | 3.0E-5 | 0.216 | 0.24645 | 0.556 |
| 4832 | USP3-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-338-3p | 13 | ITGA5 | Sponge network | -0.162 | 0.47687 | 0.452 | 0.03524 | 0.556 |
| 4833 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-374a-5p | 14 | SPG20 | Sponge network | -0.693 | 0.05629 | -1.16 | 0 | 0.556 |
| 4834 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-3p | 11 | PLSCR4 | Sponge network | -0.469 | 0.08721 | -0.474 | 0.03833 | 0.556 |
| 4835 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577 | 24 | NRK | Sponge network | -2.69 | 0.0001 | 0.656 | 0.19217 | 0.556 |
| 4836 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421 | 17 | SPG20 | Sponge network | -0.162 | 0.47687 | -1.16 | 0 | 0.556 |
| 4837 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p | 28 | ESR1 | Sponge network | -0.896 | 0.00099 | -1.035 | 3.0E-5 | 0.556 |
| 4838 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -0.777 | 0.16667 | 0.024 | 0.89889 | 0.556 |
| 4839 | FENDRR |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-935 | 13 | STAT5B | Sponge network | -1.281 | 0.00012 | -0.182 | 0.1082 | 0.556 |
| 4840 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | DACT1 | Sponge network | -1.697 | 0.00889 | 0.783 | 0.00072 | 0.556 |
| 4841 | RP11-283I3.6 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-766-3p | 16 | ATM | Sponge network | 0.614 | 1.0E-5 | 0.178 | 0.21568 | 0.556 |
| 4842 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-378a-3p;hsa-miR-7-5p | 11 | FLNA | Sponge network | -1.654 | 0.00144 | -0.771 | 0.01377 | 0.556 |
| 4843 | LINC00473 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 41 | EPHA7 | Sponge network | -1.203 | 0.03881 | -2.197 | 3.0E-5 | 0.556 |
| 4844 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | DMTF1 | Sponge network | 0.782 | 0.00016 | 0.248 | 0.02726 | 0.556 |
| 4845 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-93-3p | 30 | RUNX1T1 | Sponge network | -4.463 | 0.00025 | -0.344 | 0.2374 | 0.556 |
| 4846 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-455-5p | 13 | THSD7A | Sponge network | 0.194 | 0.64278 | 0.242 | 0.25154 | 0.556 |
| 4847 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-625-5p | 23 | ANK2 | Sponge network | -0.154 | 0.53954 | -1.603 | 1.0E-5 | 0.556 |
| 4848 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MEF2C | Sponge network | 0.727 | 3.0E-5 | -0.499 | 0.01448 | 0.556 |
| 4849 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | LSAMP | Sponge network | 0.997 | 0.00066 | -0.066 | 0.81708 | 0.556 |
| 4850 | LINC00472 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-493-5p | 16 | SYNM | Sponge network | -0.842 | 0.01361 | -2.309 | 1.0E-5 | 0.556 |
| 4851 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-370-3p;hsa-miR-455-5p;hsa-miR-590-3p | 28 | JAZF1 | Sponge network | -0.896 | 0.00099 | -0.377 | 0.02677 | 0.556 |
| 4852 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | TAL1 | Sponge network | -0.976 | 0.00025 | -0.462 | 0.00574 | 0.556 |
| 4853 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TBX18 | Sponge network | -0.777 | 0.1134 | 0.843 | 0.02945 | 0.556 |
| 4854 | FENDRR |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-664a-3p | 13 | MN1 | Sponge network | -1.281 | 0.00012 | -0.358 | 0.24172 | 0.556 |
| 4855 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | RECK | Sponge network | -0.487 | 0.02157 | -1.043 | 0 | 0.556 |
| 4856 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-590-3p | 17 | ROBO1 | Sponge network | 0.572 | 0.01722 | -0.009 | 0.96154 | 0.556 |
| 4857 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-1266-5p;hsa-miR-129-5p;hsa-miR-1468-5p;hsa-miR-326;hsa-miR-484;hsa-miR-590-3p | 10 | MLH3 | Sponge network | 1.153 | 0 | 0.608 | 0 | 0.556 |
| 4858 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 46 | RBMS3 | Sponge network | 0.101 | 0.60919 | -0.519 | 0.03551 | 0.556 |
| 4859 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-5p | 11 | PLSCR4 | Sponge network | -0.154 | 0.53954 | -0.474 | 0.03833 | 0.556 |
| 4860 | RP11-672A2.4 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 14 | ZFHX4 | Sponge network | 1.344 | 0 | 0.018 | 0.95574 | 0.556 |
| 4861 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-628-5p | 32 | QKI | Sponge network | 0.811 | 0.03228 | -0.333 | 0.04111 | 0.556 |
| 4862 | TRAF3IP2-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 21 | PRUNE2 | Sponge network | -0.323 | 0.01372 | -1.733 | 0.00022 | 0.556 |
| 4863 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-582-5p | 21 | RICTOR | Sponge network | 0.993 | 0 | 0.388 | 0.00188 | 0.556 |
| 4864 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 36 | MYO9A | Sponge network | -0.576 | 0.10121 | -0.147 | 0.32373 | 0.556 |
| 4865 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | AKAP6 | Sponge network | 0.572 | 0.01722 | -1.586 | 3.0E-5 | 0.556 |
| 4866 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p | 20 | PBX1 | Sponge network | -0.932 | 0.00329 | -1.206 | 0 | 0.556 |
| 4867 | MIR143HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 40 | SNX29 | Sponge network | -0.739 | 0.0574 | -0.34 | 0.01371 | 0.556 |
| 4868 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 15 | LRRK2 | Sponge network | -0.49 | 0.04946 | -1.136 | 1.0E-5 | 0.556 |
| 4869 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | CACNA2D1 | Sponge network | -1.582 | 8.0E-5 | -0.846 | 0.00675 | 0.556 |
| 4870 | RP11-248G5.8 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p | 17 | PHF6 | Sponge network | 1.294 | 0 | 0.785 | 0 | 0.556 |
| 4871 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 36 | ABI2 | Sponge network | 0.082 | 0.55654 | 0.175 | 0.14787 | 0.556 |
| 4872 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 32 | FAT3 | Sponge network | -0.517 | 0.02935 | -1.601 | 0.00051 | 0.556 |
| 4873 | AC127904.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | TRIM33 | Sponge network | 1.308 | 0 | 0.402 | 7.0E-5 | 0.556 |
| 4874 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 35 | MCC | Sponge network | 0.572 | 0.01722 | -1.005 | 0 | 0.556 |
| 4875 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | RABEP1 | Sponge network | -0.222 | 0.32445 | -0.066 | 0.43142 | 0.556 |
| 4876 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p | 10 | TSLP | Sponge network | -2.46 | 0 | -2.209 | 0 | 0.556 |
| 4877 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -2.69 | 0.0001 | -0.354 | 0.14694 | 0.556 |
| 4878 | RP11-819C21.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-940 | 21 | SIRT1 | Sponge network | 0.537 | 0.00139 | 0.109 | 0.2593 | 0.556 |
| 4879 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 55 | PIK3R1 | Sponge network | -1.111 | 0.0003 | -0.133 | 0.36854 | 0.556 |
| 4880 | CKMT2-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-590-3p | 19 | ITPKB | Sponge network | 0.082 | 0.55654 | -0.704 | 0.00106 | 0.555 |
| 4881 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p | 22 | PGR | Sponge network | -0.693 | 0.05629 | -1.704 | 0 | 0.555 |
| 4882 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-542-3p | 16 | DYNC1I1 | Sponge network | -0.611 | 0.09607 | -1.12 | 0.00107 | 0.555 |
| 4883 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 46 | NOTCH2 | Sponge network | -0.129 | 0.57177 | 0.138 | 0.35163 | 0.555 |
| 4884 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-335-3p | 13 | DTNA | Sponge network | -0.691 | 0.00146 | -1.648 | 1.0E-5 | 0.555 |
| 4885 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-550a-3p | 13 | NRK | Sponge network | -2.531 | 0 | 0.656 | 0.19217 | 0.555 |
| 4886 | MIR497HG |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 11 | FLI1 | Sponge network | -1.439 | 4.0E-5 | -0.294 | 0.10905 | 0.555 |
| 4887 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 16 | EPHA3 | Sponge network | -0.376 | 0.00337 | 0.185 | 0.53588 | 0.555 |
| 4888 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-96-5p | 31 | PRKAA2 | Sponge network | -0.517 | 0.02935 | -2.462 | 0 | 0.555 |
| 4889 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-429 | 12 | MLLT10 | Sponge network | 1.106 | 0 | 0.105 | 0.33292 | 0.555 |
| 4890 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p | 12 | FSTL5 | Sponge network | -0.976 | 0.00025 | -1.3 | 0.01619 | 0.555 |
| 4891 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p | 24 | RICTOR | Sponge network | 1.106 | 0 | 0.388 | 0.00188 | 0.555 |
| 4892 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SEMA3E | Sponge network | -2.452 | 0 | -1.717 | 0.00137 | 0.555 |
| 4893 | PGM5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-450b-5p | 13 | RYR2 | Sponge network | -4.546 | 0 | -1.466 | 0.00031 | 0.555 |
| 4894 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 31 | ANK2 | Sponge network | -0.615 | 0.00015 | -1.603 | 1.0E-5 | 0.555 |
| 4895 | ARHGEF26-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-429 | 11 | NGFR | Sponge network | -2.46 | 0 | -1.271 | 0.01058 | 0.555 |
| 4896 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 25 | TBL1XR1 | Sponge network | 1.147 | 0 | 0.578 | 0 | 0.555 |
| 4897 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ERG | Sponge network | 0.335 | 0.20596 | 0.002 | 0.99011 | 0.555 |
| 4898 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 34 | ABL2 | Sponge network | 0.537 | 0.00139 | 0.82 | 0 | 0.555 |
| 4899 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | DCLK1 | Sponge network | -1.886 | 0 | -1.324 | 0.00014 | 0.555 |
| 4900 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935 | 34 | DMD | Sponge network | 0.572 | 0.01722 | -1.506 | 0 | 0.555 |
| 4901 | MAGI2-AS3 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-625-5p;hsa-miR-7-5p | 11 | LZTS1 | Sponge network | -0.129 | 0.57177 | 1.664 | 0 | 0.555 |
| 4902 | MEG3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 11 | AMPH | Sponge network | 0.249 | 0.35902 | -0.916 | 5.0E-5 | 0.555 |
| 4903 | LINC00641 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 22 | ABL1 | Sponge network | -0.222 | 0.32445 | -0.215 | 0.07804 | 0.555 |
| 4904 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-345-5p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-940 | 31 | NTRK3 | Sponge network | -2.095 | 0.00112 | -1.77 | 3.0E-5 | 0.555 |
| 4905 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 34 | ABI2 | Sponge network | 0.537 | 0.00139 | 0.175 | 0.14787 | 0.555 |
| 4906 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 57 | EPHA5 | Sponge network | -1.697 | 0.00889 | -0.957 | 0.01733 | 0.555 |
| 4907 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | ATRX | Sponge network | 0.953 | 0 | 0.261 | 0.04408 | 0.555 |
| 4908 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-34a-5p;hsa-miR-370-3p | 13 | ZMYND11 | Sponge network | -1.331 | 3.0E-5 | -0.2 | 0.05449 | 0.555 |
| 4909 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -0.162 | 0.47687 | -0.354 | 0.14694 | 0.555 |
| 4910 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | 0.214 | 0.18246 | 0.358 | 0.13727 | 0.555 |
| 4911 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 41 | DMD | Sponge network | -0.612 | 0.00094 | -1.506 | 0 | 0.555 |
| 4912 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-452-5p | 29 | JAZF1 | Sponge network | -2.62 | 0 | -0.377 | 0.02677 | 0.555 |
| 4913 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-590-3p | 13 | DLC1 | Sponge network | -1.375 | 0.08642 | -0.382 | 0.05038 | 0.555 |
| 4914 | ARHGEF26-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 26 | RYR2 | Sponge network | -2.46 | 0 | -1.466 | 0.00031 | 0.555 |
| 4915 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | FNBP1 | Sponge network | -2.46 | 0 | -0.969 | 4.0E-5 | 0.555 |
| 4916 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 12 | ZNF292 | Sponge network | 1.317 | 0 | 0.276 | 0.04148 | 0.555 |
| 4917 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-196a-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-362-5p;hsa-miR-500b-5p | 15 | RSPO2 | Sponge network | -0.932 | 0.00329 | -3.038 | 0 | 0.555 |
| 4918 | MAGI2-AS3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 16 | KCTD8 | Sponge network | -0.129 | 0.57177 | -3.359 | 0 | 0.555 |
| 4919 | RP11-35G9.3 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 13 | ATM | Sponge network | 0.657 | 4.0E-5 | 0.178 | 0.21568 | 0.555 |
| 4920 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 13 | AKAP6 | Sponge network | -0.187 | 0.23787 | -1.586 | 3.0E-5 | 0.555 |
| 4921 | CTC-429P9.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-542-3p | 22 | LPP | Sponge network | 0.043 | 0.81628 | -0.459 | 0.04475 | 0.555 |
| 4922 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 39 | CHD5 | Sponge network | -2.46 | 0 | -0.922 | 0.01521 | 0.555 |
| 4923 | MIR143HG |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 36 | CTIF | Sponge network | -0.739 | 0.0574 | -0.389 | 0.00549 | 0.555 |
| 4924 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 20 | KIT | Sponge network | -1.203 | 0.03881 | -2.184 | 0 | 0.555 |
| 4925 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 38 | RBMS3 | Sponge network | -0.612 | 0.00094 | -0.519 | 0.03551 | 0.555 |
| 4926 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-338-3p;hsa-miR-429 | 18 | CADM1 | Sponge network | -0.246 | 0.35034 | -0.9 | 0.00084 | 0.554 |
| 4927 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-93-3p;hsa-miR-93-5p | 24 | SSBP2 | Sponge network | -2.531 | 0 | -1.58 | 0 | 0.554 |
| 4928 | AC006116.24 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-539-5p | 13 | GRIK3 | Sponge network | -0.473 | 0.07602 | -3.208 | 0 | 0.554 |
| 4929 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-744-3p | 36 | HLF | Sponge network | -0.487 | 0.02157 | -2.443 | 0 | 0.554 |
| 4930 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-361-5p | 12 | DNAJB4 | Sponge network | -2.531 | 0 | -0.7 | 1.0E-5 | 0.554 |
| 4931 | RP11-736K20.5 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-629-3p | 20 | ZBTB4 | Sponge network | -0.358 | 0.17255 | -0.739 | 0 | 0.554 |
| 4932 | RP11-225B17.2 |
hsa-miR-101-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | KIF5B | Sponge network | 1.055 | 0 | 0.362 | 0.00066 | 0.554 |
| 4933 | RP11-67L2.2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITGB1 | Sponge network | 0.72 | 0.0001 | 0.524 | 0.00017 | 0.554 |
| 4934 | C20orf166-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 55 | DST | Sponge network | -2.69 | 0.0001 | -0.713 | 0.00096 | 0.554 |
| 4935 | MYLK-AS1 |
hsa-let-7d-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p;hsa-miR-940 | 13 | THBS1 | Sponge network | -1.331 | 3.0E-5 | 0.741 | 0.00386 | 0.554 |
| 4936 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 36 | SCN9A | Sponge network | -1.575 | 6.0E-5 | -0.525 | 0.10593 | 0.554 |
| 4937 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-590-3p;hsa-miR-96-5p | 27 | MEF2C | Sponge network | -1.582 | 8.0E-5 | -0.499 | 0.01448 | 0.554 |
| 4938 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-7-5p;hsa-miR-93-3p | 20 | EPHA3 | Sponge network | -0.469 | 0.08721 | 0.185 | 0.53588 | 0.554 |
| 4939 | MEG3 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | RNF180 | Sponge network | 0.249 | 0.35902 | -1.127 | 1.0E-5 | 0.554 |
| 4940 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-338-3p;hsa-miR-542-3p;hsa-miR-590-3p | 11 | LRRK2 | Sponge network | 0.194 | 0.64278 | -1.136 | 1.0E-5 | 0.554 |
| 4941 | FAM66C |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 15 | SORCS1 | Sponge network | -0.901 | 0.00019 | -3.492 | 0 | 0.554 |
| 4942 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429 | 12 | DCLK1 | Sponge network | -1.654 | 0.00144 | -1.324 | 0.00014 | 0.554 |
| 4943 | RP11-359E10.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | BNC2 | Sponge network | 0.299 | 0.57722 | -0.491 | 0.09988 | 0.554 |
| 4944 | TRAF3IP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-542-3p | 17 | SYNM | Sponge network | -0.323 | 0.01372 | -2.309 | 1.0E-5 | 0.554 |
| 4945 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p | 25 | RASSF2 | Sponge network | -1.886 | 0 | -0.273 | 0.21961 | 0.554 |
| 4946 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-589-5p | 14 | SOX5 | Sponge network | -0.309 | 0.1145 | -1.091 | 4.0E-5 | 0.554 |
| 4947 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 20 | RASSF8 | Sponge network | 0.299 | 0.57722 | -0.122 | 0.65591 | 0.554 |
| 4948 | TRAF3IP2-AS1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | RBM6 | Sponge network | -0.323 | 0.01372 | -0.137 | 0.15663 | 0.554 |
| 4949 | LINC00957 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 31 | TSHZ3 | Sponge network | -0.421 | 0.0114 | -0.556 | 0.01516 | 0.554 |
| 4950 | BVES-AS1 |
hsa-miR-136-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-629-3p | 10 | CXCL12 | Sponge network | -2.62 | 0 | -1.421 | 0 | 0.554 |
| 4951 | RP11-554A11.4 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-629-3p | 14 | CDH23 | Sponge network | -0.583 | 0.14589 | -0.974 | 0.00034 | 0.554 |
| 4952 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 25 | RICTOR | Sponge network | 0.959 | 0 | 0.388 | 0.00188 | 0.554 |
| 4953 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 16 | CNTNAP1 | Sponge network | -0.563 | 0.02545 | 0.358 | 0.13727 | 0.554 |
| 4954 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | ZNF208 | Sponge network | -2.452 | 0 | -0.4 | 0.22381 | 0.554 |
| 4955 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 11 | LHFP | Sponge network | -0.323 | 0.01372 | -0.167 | 0.41371 | 0.554 |
| 4956 | RP11-166D19.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 32 | PTPN14 | Sponge network | -0.576 | 0.10121 | 0.144 | 0.34595 | 0.554 |
| 4957 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | SYNE1 | Sponge network | -1.216 | 0 | -0.738 | 0.00316 | 0.554 |
| 4958 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 19 | TRIM33 | Sponge network | 0.873 | 0 | 0.402 | 7.0E-5 | 0.554 |
| 4959 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p | 31 | FAT3 | Sponge network | -0.154 | 0.53954 | -1.601 | 0.00051 | 0.554 |
| 4960 | NR2F1-AS1 |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-590-3p | 10 | GLIPR2 | Sponge network | -0.49 | 0.04946 | -0.996 | 0 | 0.554 |
| 4961 | FAM66C |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-942-5p | 13 | ZBTB20 | Sponge network | -0.901 | 0.00019 | -0.122 | 0.57569 | 0.554 |
| 4962 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 45 | RBMS3 | Sponge network | 0.246 | 0.59912 | -0.519 | 0.03551 | 0.554 |
| 4963 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-590-5p;hsa-miR-769-5p | 14 | RCAN2 | Sponge network | 0.331 | 0.57247 | -0.33 | 0.15172 | 0.554 |
| 4964 | MEG3 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 11 | SORCS2 | Sponge network | 0.249 | 0.35902 | -0.223 | 0.4253 | 0.554 |
| 4965 | AC011526.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-98-5p | 10 | ITGB3 | Sponge network | 0.342 | 0.03129 | -0.011 | 0.95751 | 0.554 |
| 4966 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 60 | TCF4 | Sponge network | -0.323 | 0.01372 | 0.016 | 0.92271 | 0.554 |
| 4967 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 16 | LHX6 | Sponge network | -0.976 | 0.00025 | -0.087 | 0.69193 | 0.554 |
| 4968 | RP11-359E10.1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 14 | PCDH10 | Sponge network | 0.299 | 0.57722 | -1.644 | 0.0078 | 0.554 |
| 4969 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | TIMP3 | Sponge network | -0.739 | 0.00044 | -0.546 | 0.02307 | 0.554 |
| 4970 | JAZF1-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 47 | GRIK3 | Sponge network | -0.769 | 0.00035 | -3.208 | 0 | 0.554 |
| 4971 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 10 | CAV1 | Sponge network | -0.612 | 0.00094 | -1.013 | 6.0E-5 | 0.554 |
| 4972 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 15 | EPHA3 | Sponge network | -1.375 | 0.08642 | 0.185 | 0.53588 | 0.554 |
| 4973 | RP11-159D12.2 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | TAF1 | Sponge network | 1.254 | 0 | 0.39 | 3.0E-5 | 0.554 |
| 4974 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-671-5p | 12 | CDH23 | Sponge network | -1.331 | 3.0E-5 | -0.974 | 0.00034 | 0.554 |
| 4975 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 28 | ZBTB4 | Sponge network | -1.281 | 0.00012 | -0.739 | 0 | 0.554 |
| 4976 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 40 | ADARB1 | Sponge network | 0.335 | 0.20596 | -0.335 | 0.06631 | 0.554 |
| 4977 | USP3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-589-5p | 13 | ITGB3 | Sponge network | -0.162 | 0.47687 | -0.011 | 0.95751 | 0.553 |
| 4978 | BCYRN1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RBBP6 | Sponge network | 0.311 | 0.10344 | 0.163 | 0.05101 | 0.553 |
| 4979 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | BNC2 | Sponge network | 0.532 | 0.00505 | -0.491 | 0.09988 | 0.553 |
| 4980 | RASSF8-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-942-5p | 12 | ZBTB20 | Sponge network | 0.335 | 0.20596 | -0.122 | 0.57569 | 0.553 |
| 4981 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 39 | DTNA | Sponge network | -0.246 | 0.35034 | -1.648 | 1.0E-5 | 0.553 |
| 4982 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | ST6GALNAC3 | Sponge network | -1.281 | 0.00012 | -0.987 | 0 | 0.553 |
| 4983 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 25 | SYNPO2 | Sponge network | -0.769 | 0.00035 | -2.342 | 0 | 0.553 |
| 4984 | LINC00641 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p | 14 | FLNA | Sponge network | -0.222 | 0.32445 | -0.771 | 0.01377 | 0.553 |
| 4985 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-675-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | MEF2C | Sponge network | -0.72 | 0.24239 | -0.499 | 0.01448 | 0.553 |
| 4986 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-409-3p;hsa-miR-590-3p | 20 | ATRX | Sponge network | 0.765 | 7.0E-5 | 0.261 | 0.04408 | 0.553 |
| 4987 | GS1-124K5.11 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-369-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 14 | MACF1 | Sponge network | 0.494 | 0.00339 | 0.019 | 0.90335 | 0.553 |
| 4988 | LINC00578 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-589-3p | 11 | PCDH9 | Sponge network | 0.811 | 0.03228 | -1.823 | 4.0E-5 | 0.553 |
| 4989 | PCA3 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | PCDH10 | Sponge network | -0.709 | 0.18168 | -1.644 | 0.0078 | 0.553 |
| 4990 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-577 | 14 | EBF3 | Sponge network | -0.473 | 0.07602 | -0.058 | 0.81437 | 0.553 |
| 4991 | TRAF3IP2-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZDBF2 | Sponge network | -0.323 | 0.01372 | -0.998 | 0.00025 | 0.553 |
| 4992 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | ERG | Sponge network | 0.997 | 0.00066 | 0.002 | 0.99011 | 0.553 |
| 4993 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-675-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 46 | MEF2C | Sponge network | -1.281 | 0.00012 | -0.499 | 0.01448 | 0.553 |
| 4994 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | TSC1 | Sponge network | 0.083 | 0.66405 | 0.103 | 0.26071 | 0.553 |
| 4995 | TPTEP1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-590-5p | 10 | MYH11 | Sponge network | -1.216 | 0 | -2.895 | 0 | 0.553 |
| 4996 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | NCAM2 | Sponge network | -1.349 | 0.00017 | -0.482 | 0.22565 | 0.553 |
| 4997 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-96-5p | 15 | TLL1 | Sponge network | -1.654 | 0.00144 | -1.195 | 3.0E-5 | 0.553 |
| 4998 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 33 | GPC6 | Sponge network | -1.697 | 0.00889 | -0.054 | 0.81465 | 0.553 |
| 4999 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p | 38 | IL6ST | Sponge network | -0.583 | 0.14589 | -0.402 | 0.00676 | 0.553 |
| 5000 | PCA3 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-7-5p | 13 | FLNA | Sponge network | -0.709 | 0.18168 | -0.771 | 0.01377 | 0.553 |
| 5001 | SH3RF3-AS1 |
hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | DOCK4 | Sponge network | 0.889 | 9.0E-5 | 0.502 | 0.00108 | 0.553 |
| 5002 | RP11-121C2.2 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p;hsa-miR-654-3p | 20 | PHF6 | Sponge network | 1.147 | 0 | 0.785 | 0 | 0.553 |
| 5003 | LINC01018 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-501-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | SFRP1 | Sponge network | -2.915 | 0.00015 | -3.566 | 0 | 0.553 |
| 5004 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 10 | SCN11A | Sponge network | 0.335 | 0.20596 | -1.15 | 0.00299 | 0.553 |
| 5005 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p | 13 | FLNA | Sponge network | -0.739 | 0.00044 | -0.771 | 0.01377 | 0.553 |
| 5006 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | SYNE1 | Sponge network | 0.214 | 0.18246 | -0.738 | 0.00316 | 0.553 |
| 5007 | FAM66C |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | FAM172A | Sponge network | -0.901 | 0.00019 | -0.57 | 0 | 0.553 |
| 5008 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 54 | BNC2 | Sponge network | -1.281 | 0.00012 | -0.491 | 0.09988 | 0.553 |
| 5009 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 15 | FAT4 | Sponge network | -1.375 | 0.08642 | -0.354 | 0.14694 | 0.553 |
| 5010 | MAGI2-AS3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 32 | PTPN14 | Sponge network | -0.129 | 0.57177 | 0.144 | 0.34595 | 0.553 |
| 5011 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 22 | ITGB3 | Sponge network | -1.111 | 0.0003 | -0.011 | 0.95751 | 0.553 |
| 5012 | USP3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p | 21 | RYR2 | Sponge network | -0.162 | 0.47687 | -1.466 | 0.00031 | 0.553 |
| 5013 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | RASSF2 | Sponge network | -0.351 | 0.11358 | -0.273 | 0.21961 | 0.553 |
| 5014 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 55 | QKI | Sponge network | 0.194 | 0.64278 | -0.333 | 0.04111 | 0.553 |
| 5015 | RP11-121C2.2 |
hsa-miR-101-3p;hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-497-5p | 10 | E2F3 | Sponge network | 1.147 | 0 | 1.289 | 0 | 0.553 |
| 5016 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 23 | MCC | Sponge network | -0.693 | 0.05629 | -1.005 | 0 | 0.553 |
| 5017 | LINC00473 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | PREX2 | Sponge network | -1.203 | 0.03881 | -0.272 | 0.25382 | 0.553 |
| 5018 | LINC00957 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 38 | BNC2 | Sponge network | -0.421 | 0.0114 | -0.491 | 0.09988 | 0.553 |
| 5019 | PART1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 56 | EPHA7 | Sponge network | -2.925 | 0 | -2.197 | 3.0E-5 | 0.553 |
| 5020 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 53 | TCF4 | Sponge network | 0.4 | 0.14611 | 0.016 | 0.92271 | 0.553 |
| 5021 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | MCC | Sponge network | -0.842 | 0.01361 | -1.005 | 0 | 0.553 |
| 5022 | MIR143HG |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-98-5p | 32 | LRIG1 | Sponge network | -0.739 | 0.0574 | -0.537 | 0.00588 | 0.553 |
| 5023 | USP3-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-942-5p | 12 | ZBTB20 | Sponge network | -0.162 | 0.47687 | -0.122 | 0.57569 | 0.553 |
| 5024 | ARHGEF26-AS1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-889-3p;hsa-miR-96-5p | 23 | RPS6KA6 | Sponge network | -2.46 | 0 | -2.989 | 0 | 0.553 |
| 5025 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 44 | MDM4 | Sponge network | -0.323 | 0.01372 | 0.345 | 0.00762 | 0.553 |
| 5026 | MIR143HG |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-942-5p | 13 | BMPR1A | Sponge network | -0.739 | 0.0574 | -0.366 | 0.01036 | 0.552 |
| 5027 | HOXB-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429 | 16 | NRK | Sponge network | -0.154 | 0.53954 | 0.656 | 0.19217 | 0.552 |
| 5028 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-5p | 27 | ADARB1 | Sponge network | -2.531 | 0 | -0.335 | 0.06631 | 0.552 |
| 5029 | PCA3 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-500a-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-940 | 10 | PHOX2B | Sponge network | -0.709 | 0.18168 | -2.68 | 0 | 0.552 |
| 5030 | RP11-356J5.12 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 18 | PRUNE2 | Sponge network | -0.899 | 6.0E-5 | -1.733 | 0.00022 | 0.552 |
| 5031 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 32 | GRIN2A | Sponge network | -0.129 | 0.57177 | -2.161 | 0 | 0.552 |
| 5032 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 43 | DTNA | Sponge network | -1.349 | 0.00017 | -1.648 | 1.0E-5 | 0.552 |
| 5033 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 19 | ITGA5 | Sponge network | -0.49 | 0.04946 | 0.452 | 0.03524 | 0.552 |
| 5034 | LINC00957 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | RADIL | Sponge network | -0.421 | 0.0114 | -1.628 | 0 | 0.552 |
| 5035 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p | 50 | RUNX1T1 | Sponge network | -0.421 | 0.0114 | -0.344 | 0.2374 | 0.552 |
| 5036 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-940 | 45 | CHD5 | Sponge network | -0.576 | 0.10121 | -0.922 | 0.01521 | 0.552 |
| 5037 | RP11-307B6.3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-93-3p | 11 | WWTR1 | Sponge network | -4.463 | 0.00025 | -0.345 | 0.11997 | 0.552 |
| 5038 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-769-5p | 12 | RCAN2 | Sponge network | 0.082 | 0.55397 | -0.33 | 0.15172 | 0.552 |
| 5039 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p | 14 | ST6GAL2 | Sponge network | -0.583 | 0.14589 | -1.201 | 0.00597 | 0.552 |
| 5040 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 44 | EPHA7 | Sponge network | -1.249 | 7.0E-5 | -2.197 | 3.0E-5 | 0.552 |
| 5041 | FENDRR |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | DCN | Sponge network | -1.281 | 0.00012 | -1.288 | 0 | 0.552 |
| 5042 | EMX2OS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | GALNT13 | Sponge network | -0.777 | 0.1134 | -0.857 | 0.07439 | 0.552 |
| 5043 | SNX29P2 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-500a-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 11 | ATM | Sponge network | 0.4 | 0.14611 | 0.178 | 0.21568 | 0.552 |
| 5044 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 21 | PGR | Sponge network | -1.161 | 0.00591 | -1.704 | 0 | 0.552 |
| 5045 | DNM3OS |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ZNF208 | Sponge network | 0.572 | 0.01722 | -0.4 | 0.22381 | 0.552 |
| 5046 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 56 | PRKAA2 | Sponge network | -2.117 | 0.00161 | -2.462 | 0 | 0.552 |
| 5047 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 25 | LATS2 | Sponge network | -0.737 | 0.06182 | 0.159 | 0.2304 | 0.552 |
| 5048 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-486-5p;hsa-miR-590-3p | 17 | NR2C2 | Sponge network | 0.919 | 0 | 0.21 | 0.03224 | 0.552 |
| 5049 | FAM66C |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 14 | NGFR | Sponge network | -0.901 | 0.00019 | -1.271 | 0.01058 | 0.552 |
| 5050 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FSTL5 | Sponge network | -0.739 | 0.0574 | -1.3 | 0.01619 | 0.552 |
| 5051 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 54 | EPHA5 | Sponge network | -0.737 | 0.06182 | -0.957 | 0.01733 | 0.552 |
| 5052 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p | 11 | TSLP | Sponge network | -0.769 | 0.00035 | -2.209 | 0 | 0.552 |
| 5053 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-93-5p | 16 | SLITRK3 | Sponge network | -1.349 | 0.00017 | -3.328 | 0 | 0.552 |
| 5054 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-744-3p | 20 | CUL5 | Sponge network | 0.537 | 0.00139 | 0.026 | 0.77301 | 0.552 |
| 5055 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p | 24 | SSBP2 | Sponge network | -0.309 | 0.1145 | -1.58 | 0 | 0.552 |
| 5056 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 34 | MOB1B | Sponge network | 0.559 | 0.00026 | 0.197 | 0.15266 | 0.552 |
| 5057 | MIR497HG |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 19 | RPS6KA2 | Sponge network | -1.439 | 4.0E-5 | -0.57 | 0.00013 | 0.552 |
| 5058 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 24 | SNX29 | Sponge network | -0.358 | 0.17255 | -0.34 | 0.01371 | 0.552 |
| 5059 | LINC00472 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 14 | TMEFF2 | Sponge network | -0.842 | 0.01361 | -2.426 | 0 | 0.552 |
| 5060 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 11 | GLI3 | Sponge network | -1.349 | 0.00017 | -0.615 | 0.01482 | 0.552 |
| 5061 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 12 | LATS2 | Sponge network | -1.331 | 3.0E-5 | 0.159 | 0.2304 | 0.552 |
| 5062 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | CITED2 | Sponge network | -2.452 | 0 | -1.545 | 0 | 0.552 |
| 5063 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-940 | 38 | PBX1 | Sponge network | -1.249 | 7.0E-5 | -1.206 | 0 | 0.552 |
| 5064 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 27 | SEMA3C | Sponge network | -1.697 | 0.00889 | -0.272 | 0.31153 | 0.552 |
| 5065 | ZNF582-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p | 33 | IGFBP5 | Sponge network | -1.349 | 0.00017 | -0.806 | 0.00105 | 0.552 |
| 5066 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | PTPN11 | Sponge network | 0.683 | 1.0E-5 | 0.431 | 2.0E-5 | 0.552 |
| 5067 | LINC01018 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 17 | SPG20 | Sponge network | -2.915 | 0.00015 | -1.16 | 0 | 0.552 |
| 5068 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PCDH10 | Sponge network | 0.335 | 0.20596 | -1.644 | 0.0078 | 0.552 |
| 5069 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 13 | AKAP12 | Sponge network | -0.487 | 0.02157 | -0.932 | 0.0028 | 0.552 |
| 5070 | MEG3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 38 | IGFBP5 | Sponge network | 0.249 | 0.35902 | -0.806 | 0.00105 | 0.552 |
| 5071 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-942-5p | 25 | GPC6 | Sponge network | -0.976 | 0.00025 | -0.054 | 0.81465 | 0.552 |
| 5072 | RP11-2B6.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 39 | MDM4 | Sponge network | 0.539 | 0.03722 | 0.345 | 0.00762 | 0.552 |
| 5073 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 72 | IL6ST | Sponge network | -0.222 | 0.32445 | -0.402 | 0.00676 | 0.552 |
| 5074 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | -1.645 | 0 | -1.465 | 0.00033 | 0.551 |
| 5075 | SNHG14 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 34 | NRK | Sponge network | -1.111 | 0.0003 | 0.656 | 0.19217 | 0.551 |
| 5076 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 29 | EBF3 | Sponge network | -1.575 | 6.0E-5 | -0.058 | 0.81437 | 0.551 |
| 5077 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p | 30 | PRDM2 | Sponge network | -0.323 | 0.01372 | -0.258 | 0.00721 | 0.551 |
| 5078 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p | 19 | AFF3 | Sponge network | -0.611 | 0.09607 | -2.373 | 0 | 0.551 |
| 5079 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 20 | AKAP6 | Sponge network | -0.421 | 0.0114 | -1.586 | 3.0E-5 | 0.551 |
| 5080 | CTC-296K1.3 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p | 15 | ASTN1 | Sponge network | -2.095 | 0.00112 | -2.389 | 2.0E-5 | 0.551 |
| 5081 | FAM66C |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | NAV3 | Sponge network | -0.901 | 0.00019 | 0.212 | 0.47729 | 0.551 |
| 5082 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | RASSF2 | Sponge network | -0.976 | 0.00025 | -0.273 | 0.21961 | 0.551 |
| 5083 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429 | 19 | DYNC1I1 | Sponge network | -1.249 | 7.0E-5 | -1.12 | 0.00107 | 0.551 |
| 5084 | MEG3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-98-5p | 17 | ITGA5 | Sponge network | 0.249 | 0.35902 | 0.452 | 0.03524 | 0.551 |
| 5085 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 20 | RARB | Sponge network | -0.976 | 0.00025 | -0.825 | 2.0E-5 | 0.551 |
| 5086 | AC003090.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-501-5p | 12 | MN1 | Sponge network | -2.117 | 0.00161 | -0.358 | 0.24172 | 0.551 |
| 5087 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p | 16 | DKK3 | Sponge network | -0.162 | 0.47687 | -0.052 | 0.77783 | 0.551 |
| 5088 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 63 | BMPR2 | Sponge network | 0.997 | 0.00066 | 0.206 | 0.0635 | 0.551 |
| 5089 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 39 | ESR1 | Sponge network | -0.49 | 0.04946 | -1.035 | 3.0E-5 | 0.551 |
| 5090 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | RECK | Sponge network | -2.46 | 0 | -1.043 | 0 | 0.551 |
| 5091 | MIR143HG |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ADH1B | Sponge network | -0.739 | 0.0574 | -3.716 | 0 | 0.551 |
| 5092 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | DAB2 | Sponge network | -0.49 | 0.04946 | 0.431 | 0.0113 | 0.551 |
| 5093 | FENDRR |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-758-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 48 | HLF | Sponge network | -1.281 | 0.00012 | -2.443 | 0 | 0.551 |
| 5094 | RP11-307B6.3 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-744-5p | 10 | FGFR1 | Sponge network | -4.463 | 0.00025 | -0.509 | 0.03957 | 0.551 |
| 5095 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p | 20 | DYNC1I1 | Sponge network | -0.899 | 6.0E-5 | -1.12 | 0.00107 | 0.551 |
| 5096 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 46 | PIK3R1 | Sponge network | -0.129 | 0.57177 | -0.133 | 0.36854 | 0.551 |
| 5097 | DNM3OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 28 | RNF144A | Sponge network | 0.572 | 0.01722 | 0.594 | 0.0009 | 0.551 |
| 5098 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 10 | TSLP | Sponge network | -1.111 | 0.0003 | -2.209 | 0 | 0.551 |
| 5099 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 46 | AR | Sponge network | -0.421 | 0.0114 | -1.554 | 0 | 0.551 |
| 5100 | EMX2OS |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 22 | NCAM2 | Sponge network | -0.777 | 0.1134 | -0.482 | 0.22565 | 0.551 |
| 5101 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-370-3p;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 39 | CHD5 | Sponge network | -0.162 | 0.47687 | -0.922 | 0.01521 | 0.551 |
| 5102 | RP11-307B6.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-766-3p | 22 | CADM2 | Sponge network | -4.463 | 0.00025 | -3.782 | 0 | 0.551 |
| 5103 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940;hsa-miR-98-5p | 41 | PBX1 | Sponge network | -0.899 | 6.0E-5 | -1.206 | 0 | 0.551 |
| 5104 | TRAF3IP2-AS1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 18 | ATM | Sponge network | -0.323 | 0.01372 | 0.178 | 0.21568 | 0.551 |
| 5105 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 29 | GRIN2A | Sponge network | -1.697 | 0.00889 | -2.161 | 0 | 0.551 |
| 5106 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | MLLT10 | Sponge network | -0.031 | 0.89063 | 0.105 | 0.33292 | 0.551 |
| 5107 | DNM3OS |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-96-5p | 13 | ZBTB20 | Sponge network | 0.572 | 0.01722 | -0.122 | 0.57569 | 0.551 |
| 5108 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-590-3p | 14 | RSPO2 | Sponge network | -1.582 | 8.0E-5 | -3.038 | 0 | 0.551 |
| 5109 | DNM3OS |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | EYA4 | Sponge network | 0.572 | 0.01722 | 0.3 | 0.36876 | 0.551 |
| 5110 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 18 | PTGFR | Sponge network | 0.246 | 0.59912 | -0.233 | 0.45421 | 0.551 |
| 5111 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p | 11 | FAT2 | Sponge network | -1.645 | 0 | -0.278 | 0.44877 | 0.551 |
| 5112 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-503-5p;hsa-miR-590-3p | 11 | SMARCA1 | Sponge network | -0.129 | 0.57177 | -0.684 | 0.00159 | 0.551 |
| 5113 | HOXB-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-589-3p | 16 | SYNPO2 | Sponge network | -0.154 | 0.53954 | -2.342 | 0 | 0.551 |
| 5114 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-5p | 50 | AR | Sponge network | -0.154 | 0.78939 | -1.554 | 0 | 0.551 |
| 5115 | LINC00957 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | -0.421 | 0.0114 | -1.74 | 0 | 0.551 |
| 5116 | RP11-384K6.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-378c;hsa-miR-424-5p | 22 | ABL2 | Sponge network | 0.946 | 0 | 0.82 | 0 | 0.551 |
| 5117 | RP11-400K9.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-93-3p | 30 | GRIK3 | Sponge network | -0.611 | 0.09607 | -3.208 | 0 | 0.551 |
| 5118 | RP11-307B6.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p | 21 | PBX1 | Sponge network | -4.463 | 0.00025 | -1.206 | 0 | 0.551 |
| 5119 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | 0.335 | 0.20596 | 0.024 | 0.89889 | 0.551 |
| 5120 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 21 | RECK | Sponge network | -1.249 | 7.0E-5 | -1.043 | 0 | 0.551 |
| 5121 | RP11-736K20.5 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | TPO | Sponge network | -0.358 | 0.17255 | -1.87 | 2.0E-5 | 0.551 |
| 5122 | AC006116.24 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-582-3p | 11 | NAV3 | Sponge network | -0.473 | 0.07602 | 0.212 | 0.47729 | 0.551 |
| 5123 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 31 | EPHA3 | Sponge network | -0.323 | 0.01372 | 0.185 | 0.53588 | 0.551 |
| 5124 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p | 40 | BNC2 | Sponge network | -0.777 | 0.16667 | -0.491 | 0.09988 | 0.551 |
| 5125 | DNM3OS |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | PCDH18 | Sponge network | 0.572 | 0.01722 | 0.216 | 0.24645 | 0.551 |
| 5126 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 18 | ABCB5 | Sponge network | -2.452 | 0 | -0.956 | 0.04077 | 0.551 |
| 5127 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-484 | 19 | TGFBR3 | Sponge network | -0.691 | 0.00146 | -1.173 | 1.0E-5 | 0.551 |
| 5128 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 38 | PBX1 | Sponge network | -2.117 | 0.00161 | -1.206 | 0 | 0.551 |
| 5129 | LINC00957 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-362-5p;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | -0.421 | 0.0114 | -1.051 | 0.00022 | 0.551 |
| 5130 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421 | 10 | RELN | Sponge network | -0.583 | 0.14589 | -2.403 | 0 | 0.551 |
| 5131 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 38 | GAS7 | Sponge network | -0.896 | 0.00099 | -0.555 | 0.01602 | 0.551 |
| 5132 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 26 | AKAP6 | Sponge network | -0.899 | 6.0E-5 | -1.586 | 3.0E-5 | 0.55 |
| 5133 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 49 | ADARB1 | Sponge network | 0.249 | 0.35902 | -0.335 | 0.06631 | 0.55 |
| 5134 | LINC00472 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 23 | PCDH9 | Sponge network | -0.842 | 0.01361 | -1.823 | 4.0E-5 | 0.55 |
| 5135 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-590-3p | 10 | IGSF10 | Sponge network | -1.886 | 0 | -1.751 | 1.0E-5 | 0.55 |
| 5136 | CTD-2314G24.2 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CDH11 | Sponge network | 0.246 | 0.59912 | 1.427 | 0 | 0.55 |
| 5137 | AC006116.24 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577 | 34 | RUNX1T1 | Sponge network | -0.473 | 0.07602 | -0.344 | 0.2374 | 0.55 |
| 5138 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-486-5p | 13 | EZH1 | Sponge network | -0.309 | 0.1145 | -0.564 | 1.0E-5 | 0.55 |
| 5139 | LINC00578 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-628-5p | 36 | DTNA | Sponge network | 0.811 | 0.03228 | -1.648 | 1.0E-5 | 0.55 |
| 5140 | CTC-296K1.4 |
hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-30d-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p | 22 | CADM2 | Sponge network | -2.076 | 0.00548 | -3.782 | 0 | 0.55 |
| 5141 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-744-3p | 19 | ADAMTSL3 | Sponge network | 0.811 | 0.03228 | -1.559 | 0 | 0.55 |
| 5142 | RP11-819C21.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | 0.537 | 0.00139 | 0.142 | 0.04938 | 0.55 |
| 5143 | LINC00476 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 12 | FHL1 | Sponge network | -0.615 | 0.00015 | -2.687 | 0 | 0.55 |
| 5144 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p | 28 | SNX29 | Sponge network | -0.487 | 0.02157 | -0.34 | 0.01371 | 0.55 |
| 5145 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3614-5p | 14 | RICTOR | Sponge network | 0.793 | 0 | 0.388 | 0.00188 | 0.55 |
| 5146 | ACTA2-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | 0.194 | 0.64278 | 0.154 | 0.74723 | 0.55 |
| 5147 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 68 | IL6ST | Sponge network | 0.997 | 0.00066 | -0.402 | 0.00676 | 0.55 |
| 5148 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-5p | 16 | TCF4 | Sponge network | 1.344 | 0 | 0.016 | 0.92271 | 0.55 |
| 5149 | LINC00092 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 25 | PTPRT | Sponge network | -1.886 | 0 | -0.907 | 0.03727 | 0.55 |
| 5150 | HOXB-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-625-5p | 12 | NGFR | Sponge network | -0.154 | 0.53954 | -1.271 | 0.01058 | 0.55 |
| 5151 | LINC00641 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 14 | STAT5B | Sponge network | -0.222 | 0.32445 | -0.182 | 0.1082 | 0.55 |
| 5152 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | MITF | Sponge network | -1.439 | 4.0E-5 | -0.769 | 0.00022 | 0.55 |
| 5153 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 23 | LATS2 | Sponge network | 0.997 | 0.00066 | 0.159 | 0.2304 | 0.55 |
| 5154 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 15 | IGSF10 | Sponge network | -4.546 | 0 | -1.751 | 1.0E-5 | 0.55 |
| 5155 | AP001055.6 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-301a-3p | 11 | KCNJ5 | Sponge network | 0.693 | 0.00076 | -0.281 | 0.3339 | 0.55 |
| 5156 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | EPHA5 | Sponge network | 0.572 | 0.01722 | -0.957 | 0.01733 | 0.55 |
| 5157 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 18 | SPG20 | Sponge network | 0.335 | 0.20596 | -1.16 | 0 | 0.55 |
| 5158 | CTA-941F9.9 |
hsa-let-7d-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-576-5p | 11 | GAS7 | Sponge network | -0.344 | 0.49624 | -0.555 | 0.01602 | 0.55 |
| 5159 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 34 | ABI2 | Sponge network | 0.222 | 0.29133 | 0.175 | 0.14787 | 0.55 |
| 5160 | LINC00957 |
hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484 | 11 | FLNA | Sponge network | -0.421 | 0.0114 | -0.771 | 0.01377 | 0.55 |
| 5161 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 31 | FAT3 | Sponge network | -0.469 | 0.08721 | -1.601 | 0.00051 | 0.55 |
| 5162 | LINC00473 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-935 | 33 | DMD | Sponge network | -1.203 | 0.03881 | -1.506 | 0 | 0.55 |
| 5163 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 36 | MEF2C | Sponge network | 0.335 | 0.20596 | -0.499 | 0.01448 | 0.55 |
| 5164 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 74 | IL6ST | Sponge network | -1.697 | 0.00889 | -0.402 | 0.00676 | 0.55 |
| 5165 | EMX2OS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 39 | IL17RD | Sponge network | -0.777 | 0.1134 | -0.019 | 0.94873 | 0.55 |
| 5166 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | DTNA | Sponge network | -0.513 | 0.04703 | -1.648 | 1.0E-5 | 0.55 |
| 5167 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 46 | TRPS1 | Sponge network | -0.162 | 0.47687 | -0.191 | 0.44109 | 0.55 |
| 5168 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | PHC3 | Sponge network | 0.72 | 0.0001 | 0.212 | 0.0499 | 0.55 |
| 5169 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | PRKAA2 | Sponge network | -0.513 | 0.04703 | -2.462 | 0 | 0.55 |
| 5170 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-744-3p | 50 | LPP | Sponge network | 0.811 | 0.03228 | -0.459 | 0.04475 | 0.55 |
| 5171 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 63 | CADM2 | Sponge network | -0.323 | 0.01372 | -3.782 | 0 | 0.55 |
| 5172 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-93-5p | 11 | EDA2R | Sponge network | -1.439 | 4.0E-5 | -0.587 | 0.01598 | 0.55 |
| 5173 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 18 | DKK3 | Sponge network | 0.249 | 0.35902 | -0.052 | 0.77783 | 0.55 |
| 5174 | MIR143HG |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 17 | PRDM5 | Sponge network | -0.739 | 0.0574 | -1.09 | 0.00014 | 0.55 |
| 5175 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-378a-3p;hsa-miR-590-3p | 16 | RBMS3 | Sponge network | -1.375 | 0.08642 | -0.519 | 0.03551 | 0.55 |
| 5176 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 55 | NTRK3 | Sponge network | -0.421 | 0.0114 | -1.77 | 3.0E-5 | 0.55 |
| 5177 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | SOX5 | Sponge network | -1.281 | 0.00012 | -1.091 | 4.0E-5 | 0.55 |
| 5178 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 35 | MITF | Sponge network | -0.49 | 0.04946 | -0.769 | 0.00022 | 0.549 |
| 5179 | SNHG14 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | TAL1 | Sponge network | -1.111 | 0.0003 | -0.462 | 0.00574 | 0.549 |
| 5180 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-429 | 16 | CNTN1 | Sponge network | -0.154 | 0.53954 | -2.594 | 0 | 0.549 |
| 5181 | RP11-244O19.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 15 | MAPK10 | Sponge network | -0.096 | 0.67958 | -1.774 | 0 | 0.549 |
| 5182 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -0.777 | 0.16667 | -0.354 | 0.14694 | 0.549 |
| 5183 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p | 11 | PTGFR | Sponge network | -2.62 | 0 | -0.233 | 0.45421 | 0.549 |
| 5184 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p | 10 | ZNF292 | Sponge network | 0.594 | 0.00064 | 0.276 | 0.04148 | 0.549 |
| 5185 | AC009948.5 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | PIK3CA | Sponge network | 0.532 | 0.00505 | 0.195 | 0.06908 | 0.549 |
| 5186 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-590-3p | 17 | SPG20 | Sponge network | -0.096 | 0.67958 | -1.16 | 0 | 0.549 |
| 5187 | MRVI1-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-455-3p | 12 | ABL1 | Sponge network | -1.092 | 4.0E-5 | -0.215 | 0.07804 | 0.549 |
| 5188 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 43 | QKI | Sponge network | -1.349 | 0.00017 | -0.333 | 0.04111 | 0.549 |
| 5189 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3913-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-93-5p | 30 | ZBTB4 | Sponge network | -0.793 | 7.0E-5 | -0.739 | 0 | 0.549 |
| 5190 | MIR143HG |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | HERC1 | Sponge network | -0.739 | 0.0574 | -0.196 | 0.08544 | 0.549 |
| 5191 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 32 | KCNA6 | Sponge network | -0.576 | 0.10121 | -1.531 | 0.00013 | 0.549 |
| 5192 | CTD-2314G24.2 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-874-3p;hsa-miR-98-5p | 10 | HSPB7 | Sponge network | 0.246 | 0.59912 | -2.268 | 1.0E-5 | 0.549 |
| 5193 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 11 | ATRX | Sponge network | 0.545 | 0.05789 | 0.261 | 0.04408 | 0.549 |
| 5194 | LINC00957 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 20 | CADM3 | Sponge network | -0.421 | 0.0114 | -3.663 | 0 | 0.549 |
| 5195 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-590-3p | 13 | AKAP12 | Sponge network | -1.203 | 0.03881 | -0.932 | 0.0028 | 0.549 |
| 5196 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 20 | TCF4 | Sponge network | -2.039 | 0.03447 | 0.016 | 0.92271 | 0.549 |
| 5197 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CLIP1 | Sponge network | -0.739 | 0.0574 | -0.215 | 0.08684 | 0.549 |
| 5198 | RP11-531A24.5 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-655-3p;hsa-miR-940 | 13 | TAF1 | Sponge network | 0.75 | 0.00054 | 0.39 | 3.0E-5 | 0.549 |
| 5199 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577 | 36 | IL6ST | Sponge network | -0.473 | 0.07602 | -0.402 | 0.00676 | 0.549 |
| 5200 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 27 | BCL6 | Sponge network | -0.739 | 0.0574 | -0.211 | 0.19489 | 0.549 |
| 5201 | ACTA2-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-744-3p | 28 | PTPN14 | Sponge network | 0.194 | 0.64278 | 0.144 | 0.34595 | 0.549 |
| 5202 | RP1-59D14.5 |
hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-511-5p | 13 | ATRX | Sponge network | 1.162 | 5.0E-5 | 0.261 | 0.04408 | 0.549 |
| 5203 | PART1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 13 | PARK2 | Sponge network | -2.925 | 0 | -1.892 | 0 | 0.549 |
| 5204 | MEG3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 30 | SETBP1 | Sponge network | 0.249 | 0.35902 | -1.302 | 1.0E-5 | 0.549 |
| 5205 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p | 15 | KIT | Sponge network | -4.546 | 0 | -2.184 | 0 | 0.549 |
| 5206 | CTC-296K1.4 |
hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p | 10 | SFRP1 | Sponge network | -2.076 | 0.00548 | -3.566 | 0 | 0.549 |
| 5207 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-98-5p | 37 | DMD | Sponge network | 0.101 | 0.60919 | -1.506 | 0 | 0.549 |
| 5208 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 10 | ATRX | Sponge network | 1.055 | 0 | 0.261 | 0.04408 | 0.549 |
| 5209 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 36 | JAZF1 | Sponge network | -0.323 | 0.01372 | -0.377 | 0.02677 | 0.549 |
| 5210 | FAM66C |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 63 | DST | Sponge network | -0.901 | 0.00019 | -0.713 | 0.00096 | 0.549 |
| 5211 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 38 | DIP2C | Sponge network | -0.323 | 0.01372 | -0.436 | 0.01713 | 0.549 |
| 5212 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-576-5p | 12 | MCC | Sponge network | -0.932 | 0.00329 | -1.005 | 0 | 0.549 |
| 5213 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-671-5p | 25 | KIT | Sponge network | -0.576 | 0.10121 | -2.184 | 0 | 0.549 |
| 5214 | RP11-440L14.4 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-5p;hsa-miR-590-3p | 15 | MDM4 | Sponge network | 0.545 | 0.05789 | 0.345 | 0.00762 | 0.549 |
| 5215 | CTD-3092A11.2 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-940 | 10 | TAF1 | Sponge network | 0.873 | 0 | 0.39 | 3.0E-5 | 0.549 |
| 5216 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 21 | TSHZ3 | Sponge network | -4.546 | 0 | -0.556 | 0.01516 | 0.549 |
| 5217 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 18 | HOOK3 | Sponge network | 0.194 | 0.64278 | 0.273 | 0.09957 | 0.549 |
| 5218 | RP11-554A11.4 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 11 | ROBO1 | Sponge network | -0.583 | 0.14589 | -0.009 | 0.96154 | 0.549 |
| 5219 | CTD-2089N3.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-455-5p | 18 | RUNX1T1 | Sponge network | -0.314 | 0.62033 | -0.344 | 0.2374 | 0.549 |
| 5220 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | NR2C2 | Sponge network | 1.308 | 0 | 0.21 | 0.03224 | 0.549 |
| 5221 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 45 | BNC2 | Sponge network | -0.612 | 0.00094 | -0.491 | 0.09988 | 0.549 |
| 5222 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-92a-3p;hsa-miR-93-5p | 19 | RASSF2 | Sponge network | 0.342 | 0.03129 | -0.273 | 0.21961 | 0.549 |
| 5223 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 27 | KIT | Sponge network | -1.575 | 6.0E-5 | -2.184 | 0 | 0.549 |
| 5224 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | DCLK1 | Sponge network | -0.777 | 0.1134 | -1.324 | 0.00014 | 0.549 |
| 5225 | ARHGEF26-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 17 | RHOB | Sponge network | -2.46 | 0 | -1.052 | 0 | 0.549 |
| 5226 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 23 | ABL1 | Sponge network | -0.576 | 0.10121 | -0.215 | 0.07804 | 0.549 |
| 5227 | NEAT1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ATRX | Sponge network | 0.705 | 0.00014 | 0.261 | 0.04408 | 0.549 |
| 5228 | LINC00092 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 24 | PBX1 | Sponge network | -1.886 | 0 | -1.206 | 0 | 0.549 |
| 5229 | AC003090.1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ZNF536 | Sponge network | -2.117 | 0.00161 | -3.115 | 0 | 0.549 |
| 5230 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | NALCN | Sponge network | 0.194 | 0.64278 | 1.068 | 0.00237 | 0.549 |
| 5231 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | CADM1 | Sponge network | -0.739 | 0.00044 | -0.9 | 0.00084 | 0.549 |
| 5232 | NR2F1-AS1 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ADH1B | Sponge network | -0.49 | 0.04946 | -3.716 | 0 | 0.549 |
| 5233 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 40 | MOB1B | Sponge network | 0.537 | 0.00139 | 0.197 | 0.15266 | 0.549 |
| 5234 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | -0.358 | 0.17255 | -1.12 | 0.00107 | 0.549 |
| 5235 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 11 | ITCH | Sponge network | 0.826 | 1.0E-5 | 0.084 | 0.34058 | 0.548 |
| 5236 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p | 11 | RBM5 | Sponge network | 0.267 | 0.2937 | -0.205 | 0.0129 | 0.548 |
| 5237 | AC010226.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-455-3p | 10 | CADM3 | Sponge network | -0.811 | 2.0E-5 | -3.663 | 0 | 0.548 |
| 5238 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | ADAMTSL3 | Sponge network | -0.323 | 0.01372 | -1.559 | 0 | 0.548 |
| 5239 | LINC00641 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 15 | SHPRH | Sponge network | -0.222 | 0.32445 | 0.098 | 0.40994 | 0.548 |
| 5240 | RP11-53O19.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-940 | 14 | FAM172A | Sponge network | -0.405 | 0.22013 | -0.57 | 0 | 0.548 |
| 5241 | PART1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 28 | CDH19 | Sponge network | -2.925 | 0 | -3.434 | 0 | 0.548 |
| 5242 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 21 | FGFR1 | Sponge network | -1.216 | 0 | -0.509 | 0.03957 | 0.548 |
| 5243 | PCA3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | -0.709 | 0.18168 | 0.358 | 0.13727 | 0.548 |
| 5244 | MRVI1-AS1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p | 30 | CHD5 | Sponge network | -1.092 | 4.0E-5 | -0.922 | 0.01521 | 0.548 |
| 5245 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p | 24 | FBXO32 | Sponge network | -0.693 | 0.05629 | -0.495 | 0.07561 | 0.548 |
| 5246 | HHIP-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DCN | Sponge network | -0.737 | 0.10822 | -1.288 | 0 | 0.548 |
| 5247 | LINC00092 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | ID4 | Sponge network | -1.886 | 0 | -1.644 | 0 | 0.548 |
| 5248 | HAND2-AS1 |
hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-502-5p | 11 | ELN | Sponge network | -1.697 | 0.00889 | 0.451 | 0.13752 | 0.548 |
| 5249 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | FAT4 | Sponge network | -0.487 | 0.02157 | -0.354 | 0.14694 | 0.548 |
| 5250 | HCG18 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 20 | TRIM33 | Sponge network | 1.063 | 0 | 0.402 | 7.0E-5 | 0.548 |
| 5251 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-577;hsa-miR-93-5p | 17 | TACC1 | Sponge network | -0.473 | 0.07602 | -0.362 | 0.05406 | 0.548 |
| 5252 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 22 | TBL1XR1 | Sponge network | 0.972 | 0 | 0.578 | 0 | 0.548 |
| 5253 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 21 | CNTNAP3 | Sponge network | -0.129 | 0.57177 | -1.465 | 0.00033 | 0.548 |
| 5254 | AC006547.13 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-766-3p;hsa-miR-940 | 22 | MDM4 | Sponge network | 1.159 | 0 | 0.345 | 0.00762 | 0.548 |
| 5255 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p | 18 | ZNF208 | Sponge network | -1.697 | 0.00889 | -0.4 | 0.22381 | 0.548 |
| 5256 | DNM3OS |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 15 | CXCL12 | Sponge network | 0.572 | 0.01722 | -1.421 | 0 | 0.548 |
| 5257 | BCYRN1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 23 | BAZ2B | Sponge network | 0.311 | 0.10344 | -0.276 | 0.02833 | 0.548 |
| 5258 | LINC00578 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 20 | ST6GAL2 | Sponge network | 0.811 | 0.03228 | -1.201 | 0.00597 | 0.548 |
| 5259 | GABPB1-AS1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 48 | MDM4 | Sponge network | 1.11 | 0 | 0.345 | 0.00762 | 0.548 |
| 5260 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-940 | 46 | CADM2 | Sponge network | -0.896 | 0.00099 | -3.782 | 0 | 0.548 |
| 5261 | CTA-204B4.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 21 | ABL2 | Sponge network | 0.765 | 7.0E-5 | 0.82 | 0 | 0.548 |
| 5262 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p | 26 | PIK3C2A | Sponge network | 0.537 | 0.00139 | 0.061 | 0.53776 | 0.548 |
| 5263 | RP11-798M19.6 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 14 | RHOB | Sponge network | -0.612 | 0.00094 | -1.052 | 0 | 0.548 |
| 5264 | AP001347.6 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-362-5p;hsa-miR-590-3p | 13 | MRVI1 | Sponge network | -1.161 | 0.00591 | -1.051 | 0.00022 | 0.548 |
| 5265 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | MCC | Sponge network | -1.161 | 0.00591 | -1.005 | 0 | 0.548 |
| 5266 | FAM66C |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | -0.901 | 0.00019 | 0.809 | 0.00544 | 0.548 |
| 5267 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429 | 10 | SPG20 | Sponge network | -0.469 | 0.08721 | -1.16 | 0 | 0.548 |
| 5268 | CTC-510F12.2 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-181a-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-3913-5p | 10 | ZBTB4 | Sponge network | -0.691 | 0.00146 | -0.739 | 0 | 0.548 |
| 5269 | AC003090.1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p | 12 | FLNA | Sponge network | -2.117 | 0.00161 | -0.771 | 0.01377 | 0.548 |
| 5270 | USP3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 63 | CADM2 | Sponge network | -0.162 | 0.47687 | -3.782 | 0 | 0.548 |
| 5271 | RP11-473I1.9 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | DICER1 | Sponge network | 0.683 | 1.0E-5 | 0.042 | 0.674 | 0.548 |
| 5272 | GABPB1-AS1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-486-5p;hsa-miR-590-3p | 12 | TPR | Sponge network | 1.11 | 0 | 0.582 | 0 | 0.548 |
| 5273 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-3615;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p | 13 | DMTF1 | Sponge network | 0.537 | 0.00139 | 0.248 | 0.02726 | 0.548 |
| 5274 | LINC00472 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 20 | WWTR1 | Sponge network | -0.842 | 0.01361 | -0.345 | 0.11997 | 0.548 |
| 5275 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-455-5p | 21 | ZMYND11 | Sponge network | -0.162 | 0.47687 | -0.2 | 0.05449 | 0.548 |
| 5276 | RP11-67L2.2 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 16 | TAF1 | Sponge network | 0.72 | 0.0001 | 0.39 | 3.0E-5 | 0.548 |
| 5277 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 48 | SSBP2 | Sponge network | -0.563 | 0.02545 | -1.58 | 0 | 0.548 |
| 5278 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 68 | TCF4 | Sponge network | -1.281 | 0.00012 | 0.016 | 0.92271 | 0.548 |
| 5279 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-7-1-3p | 20 | PCDH9 | Sponge network | -0.405 | 0.22013 | -1.823 | 4.0E-5 | 0.548 |
| 5280 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 27 | THBS1 | Sponge network | -0.976 | 0.00025 | 0.741 | 0.00386 | 0.547 |
| 5281 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 29 | IGF1 | Sponge network | -0.901 | 0.00019 | -0.204 | 0.5898 | 0.547 |
| 5282 | AC009948.5 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p | 15 | MACF1 | Sponge network | 0.532 | 0.00505 | 0.019 | 0.90335 | 0.547 |
| 5283 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-542-3p | 14 | ADAMTSL3 | Sponge network | -0.517 | 0.02935 | -1.559 | 0 | 0.547 |
| 5284 | RP11-890B15.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | 0.101 | 0.60919 | -0.704 | 0.00106 | 0.547 |
| 5285 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-429 | 12 | LRRK2 | Sponge network | -2.62 | 0 | -1.136 | 1.0E-5 | 0.547 |
| 5286 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 18 | RCAN2 | Sponge network | 0.249 | 0.35902 | -0.33 | 0.15172 | 0.547 |
| 5287 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 13 | HMCN1 | Sponge network | -2.452 | 0 | 0.809 | 0.00544 | 0.547 |
| 5288 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-576-5p | 12 | PGR | Sponge network | -0.733 | 0.19469 | -1.704 | 0 | 0.547 |
| 5289 | LINC00473 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-935 | 38 | GRIK3 | Sponge network | -1.203 | 0.03881 | -3.208 | 0 | 0.547 |
| 5290 | GABPB1-AS1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p | 13 | ZNF638 | Sponge network | 1.11 | 0 | 0.22 | 0.01214 | 0.547 |
| 5291 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 25 | GPC6 | Sponge network | 0.246 | 0.59912 | -0.054 | 0.81465 | 0.547 |
| 5292 | CTC-429P9.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-766-3p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | 0.082 | 0.55397 | -0.564 | 1.0E-5 | 0.547 |
| 5293 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935 | 46 | ABI2 | Sponge network | -0.129 | 0.57177 | 0.175 | 0.14787 | 0.547 |
| 5294 | LINC00657 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-766-3p | 22 | TBL1XR1 | Sponge network | 0.91 | 0 | 0.578 | 0 | 0.547 |
| 5295 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-93-5p | 23 | PGR | Sponge network | -0.563 | 0.02545 | -1.704 | 0 | 0.547 |
| 5296 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-628-5p | 40 | TCF4 | Sponge network | -0.154 | 0.53954 | 0.016 | 0.92271 | 0.547 |
| 5297 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 28 | PRKAA2 | Sponge network | -0.469 | 0.08721 | -2.462 | 0 | 0.547 |
| 5298 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-576-5p | 17 | CACNA2D1 | Sponge network | -2.62 | 0 | -0.846 | 0.00675 | 0.547 |
| 5299 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p | 40 | RBMS3 | Sponge network | -0.777 | 0.16667 | -0.519 | 0.03551 | 0.547 |
| 5300 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 20 | CDH19 | Sponge network | -0.793 | 7.0E-5 | -3.434 | 0 | 0.547 |
| 5301 | DNM3OS |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p | 11 | PXDN | Sponge network | 0.572 | 0.01722 | 1.038 | 0 | 0.547 |
| 5302 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-532-3p | 14 | SYNM | Sponge network | -0.246 | 0.35034 | -2.309 | 1.0E-5 | 0.547 |
| 5303 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | DAB2 | Sponge network | 0.727 | 3.0E-5 | 0.431 | 0.0113 | 0.547 |
| 5304 | LINC00865 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p | 13 | PDZD2 | Sponge network | -1.249 | 7.0E-5 | -0.909 | 3.0E-5 | 0.547 |
| 5305 | AP001055.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 13 | PRRX1 | Sponge network | 0.693 | 0.00076 | 1.316 | 0 | 0.547 |
| 5306 | RP11-434B12.1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-654-3p | 18 | BAZ2B | Sponge network | -0.031 | 0.89063 | -0.276 | 0.02833 | 0.547 |
| 5307 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p | 25 | RASSF8 | Sponge network | -0.096 | 0.67958 | -0.122 | 0.65591 | 0.547 |
| 5308 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 37 | NRXN1 | Sponge network | -0.323 | 0.01372 | -3.778 | 0 | 0.547 |
| 5309 | RP11-359B12.2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 11 | MACF1 | Sponge network | 0.064 | 0.72934 | 0.019 | 0.90335 | 0.547 |
| 5310 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-539-5p | 23 | EPHA7 | Sponge network | -0.309 | 0.1145 | -2.197 | 3.0E-5 | 0.547 |
| 5311 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 41 | TCF4 | Sponge network | -0.739 | 0.00044 | 0.016 | 0.92271 | 0.547 |
| 5312 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-877-5p | 20 | CDH19 | Sponge network | -0.162 | 0.47687 | -3.434 | 0 | 0.547 |
| 5313 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-486-5p;hsa-miR-582-3p;hsa-miR-590-3p | 38 | ADARB1 | Sponge network | 0.214 | 0.18246 | -0.335 | 0.06631 | 0.547 |
| 5314 | LINC00702 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | NOV | Sponge network | -0.737 | 0.06182 | -0.58 | 0.04676 | 0.547 |
| 5315 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 28 | PBX1 | Sponge network | -0.583 | 0.14589 | -1.206 | 0 | 0.547 |
| 5316 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p | 11 | IGSF10 | Sponge network | -1.331 | 3.0E-5 | -1.751 | 1.0E-5 | 0.547 |
| 5317 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 16 | LHX6 | Sponge network | -1.439 | 4.0E-5 | -0.087 | 0.69193 | 0.547 |
| 5318 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 16 | ABCC9 | Sponge network | -0.563 | 0.02545 | -0.466 | 0.14121 | 0.547 |
| 5319 | BVES-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-550a-3p | 19 | SYT4 | Sponge network | -2.62 | 0 | -3.207 | 0 | 0.547 |
| 5320 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 41 | MEF2C | Sponge network | 0.194 | 0.64278 | -0.499 | 0.01448 | 0.547 |
| 5321 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-455-3p | 22 | RUNX1T1 | Sponge network | -0.309 | 0.1145 | -0.344 | 0.2374 | 0.547 |
| 5322 | HOXB-AS1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-423-5p | 11 | ABL1 | Sponge network | -0.154 | 0.53954 | -0.215 | 0.07804 | 0.547 |
| 5323 | CTC-296K1.3 |
hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-5p;hsa-miR-940 | 11 | PEG3 | Sponge network | -2.095 | 0.00112 | -1.74 | 0 | 0.547 |
| 5324 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | EYA4 | Sponge network | -0.576 | 0.10121 | 0.3 | 0.36876 | 0.547 |
| 5325 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | EBF1 | Sponge network | -1.886 | 0 | -0.384 | 0.08856 | 0.547 |
| 5326 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-421 | 27 | PRKAA2 | Sponge network | -0.583 | 0.14589 | -2.462 | 0 | 0.547 |
| 5327 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-500a-3p;hsa-miR-542-3p | 33 | RUNX1T1 | Sponge network | -0.517 | 0.02935 | -0.344 | 0.2374 | 0.547 |
| 5328 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 14 | HERC2 | Sponge network | 0.683 | 1.0E-5 | 0.114 | 0.30638 | 0.547 |
| 5329 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 13 | THBS1 | Sponge network | 0.342 | 0.03129 | 0.741 | 0.00386 | 0.547 |
| 5330 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 24 | TSHZ3 | Sponge network | -1.582 | 8.0E-5 | -0.556 | 0.01516 | 0.547 |
| 5331 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-339-5p | 11 | ZNF521 | Sponge network | -0.611 | 0.09607 | -0.111 | 0.58068 | 0.547 |
| 5332 | GS1-124K5.11 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-766-3p | 15 | ATM | Sponge network | 0.494 | 0.00339 | 0.178 | 0.21568 | 0.547 |
| 5333 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-370-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-655-3p | 23 | ZMYND11 | Sponge network | -0.096 | 0.67958 | -0.2 | 0.05449 | 0.547 |
| 5334 | ACTA2-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TPO | Sponge network | 0.194 | 0.64278 | -1.87 | 2.0E-5 | 0.547 |
| 5335 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 16 | RBMS3 | Sponge network | 0.693 | 0.00076 | -0.519 | 0.03551 | 0.547 |
| 5336 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p | 42 | GAS7 | Sponge network | -0.487 | 0.02157 | -0.555 | 0.01602 | 0.547 |
| 5337 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | ABCA10 | Sponge network | -0.323 | 0.01372 | -0.571 | 0.03674 | 0.547 |
| 5338 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 38 | NRXN1 | Sponge network | -1.216 | 0 | -3.778 | 0 | 0.547 |
| 5339 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 16 | TXNIP | Sponge network | -1.575 | 6.0E-5 | -1.277 | 0 | 0.547 |
| 5340 | FAM66C |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ADH1B | Sponge network | -0.901 | 0.00019 | -3.716 | 0 | 0.547 |
| 5341 | AC006116.24 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-455-5p;hsa-miR-98-5p | 14 | AKAP6 | Sponge network | -0.473 | 0.07602 | -1.586 | 3.0E-5 | 0.547 |
| 5342 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-539-5p | 29 | MITF | Sponge network | -0.162 | 0.47687 | -0.769 | 0.00022 | 0.547 |
| 5343 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ATRX | Sponge network | 0.768 | 7.0E-5 | 0.261 | 0.04408 | 0.546 |
| 5344 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-320b;hsa-miR-424-5p | 11 | DYNC1I1 | Sponge network | -1.349 | 0.00017 | -1.12 | 0.00107 | 0.546 |
| 5345 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 17 | ESR1 | Sponge network | -0.811 | 2.0E-5 | -1.035 | 3.0E-5 | 0.546 |
| 5346 | LINC01018 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p | 18 | SYNPO2 | Sponge network | -2.915 | 0.00015 | -2.342 | 0 | 0.546 |
| 5347 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 20 | ATRX | Sponge network | 0.541 | 0.00125 | 0.261 | 0.04408 | 0.546 |
| 5348 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | -0.318 | 0.65847 | -0.509 | 0.03957 | 0.546 |
| 5349 | NR2F1-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | PRKCB | Sponge network | -0.49 | 0.04946 | -1.313 | 2.0E-5 | 0.546 |
| 5350 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p | 15 | ZNF521 | Sponge network | -1.575 | 6.0E-5 | -0.111 | 0.58068 | 0.546 |
| 5351 | MIR143HG |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | INPP4A | Sponge network | -0.739 | 0.0574 | 0.07 | 0.53563 | 0.546 |
| 5352 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | ST6GALNAC3 | Sponge network | -1.439 | 4.0E-5 | -0.987 | 0 | 0.546 |
| 5353 | PCA3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 25 | SETBP1 | Sponge network | -0.709 | 0.18168 | -1.302 | 1.0E-5 | 0.546 |
| 5354 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p | 21 | LRP1B | Sponge network | -2.62 | 0 | -2.163 | 0 | 0.546 |
| 5355 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 46 | SOX5 | Sponge network | -0.323 | 0.01372 | -1.091 | 4.0E-5 | 0.546 |
| 5356 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 49 | SOX5 | Sponge network | -0.739 | 0.0574 | -1.091 | 4.0E-5 | 0.546 |
| 5357 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 21 | CDH23 | Sponge network | -0.129 | 0.57177 | -0.974 | 0.00034 | 0.546 |
| 5358 | PCA3 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 15 | MAPK10 | Sponge network | -0.709 | 0.18168 | -1.774 | 0 | 0.546 |
| 5359 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | BNC2 | Sponge network | -2.117 | 0.00161 | -0.491 | 0.09988 | 0.546 |
| 5360 | BOLA3-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | -0.246 | 0.35034 | -0.998 | 0.00025 | 0.546 |
| 5361 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30d-5p;hsa-miR-3614-5p | 10 | NRXN3 | Sponge network | -0.693 | 0.05629 | -0.893 | 0.04186 | 0.546 |
| 5362 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 29 | PCDH17 | Sponge network | -0.129 | 0.57177 | 0.731 | 2.0E-5 | 0.546 |
| 5363 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-1-5p | 40 | PRKAB2 | Sponge network | -0.222 | 0.32445 | -0.367 | 0.01371 | 0.546 |
| 5364 | PART1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | NRXN3 | Sponge network | -2.925 | 0 | -0.893 | 0.04186 | 0.546 |
| 5365 | CTC-296K1.4 |
hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-324-5p;hsa-miR-582-3p;hsa-miR-590-5p | 10 | PCDH10 | Sponge network | -2.076 | 0.00548 | -1.644 | 0.0078 | 0.546 |
| 5366 | TP73-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-92a-3p | 11 | PREX2 | Sponge network | -0.563 | 0.02545 | -0.272 | 0.25382 | 0.546 |
| 5367 | NR2F1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 28 | RYR2 | Sponge network | -0.49 | 0.04946 | -1.466 | 0.00031 | 0.546 |
| 5368 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | WWTR1 | Sponge network | 0.299 | 0.57722 | -0.345 | 0.11997 | 0.546 |
| 5369 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 28 | ZFHX3 | Sponge network | 0.559 | 0.00026 | 0.389 | 0.01363 | 0.546 |
| 5370 | FENDRR |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | TLE4 | Sponge network | -1.281 | 0.00012 | -1.157 | 0 | 0.546 |
| 5371 | HCG11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | 0.385 | 0.10067 | 0.809 | 0.00544 | 0.546 |
| 5372 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-93-5p | 14 | TXNIP | Sponge network | -0.976 | 0.00025 | -1.277 | 0 | 0.546 |
| 5373 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | ATRX | Sponge network | 0.848 | 0 | 0.261 | 0.04408 | 0.546 |
| 5374 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-362-3p | 10 | TIMP3 | Sponge network | -1.255 | 0.00981 | -0.546 | 0.02307 | 0.546 |
| 5375 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ABCC9 | Sponge network | 0.101 | 0.60919 | -0.466 | 0.14121 | 0.546 |
| 5376 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | FGF5 | Sponge network | 0.997 | 0.00066 | 0.549 | 0.28659 | 0.546 |
| 5377 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | AFF3 | Sponge network | -1.216 | 0 | -2.373 | 0 | 0.546 |
| 5378 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | IGSF10 | Sponge network | -0.942 | 0 | -1.751 | 1.0E-5 | 0.546 |
| 5379 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -0.487 | 0.02157 | -0.7 | 1.0E-5 | 0.546 |
| 5380 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 77 | NTRK3 | Sponge network | -2.46 | 0 | -1.77 | 3.0E-5 | 0.546 |
| 5381 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 30 | EPHA3 | Sponge network | 0.101 | 0.60919 | 0.185 | 0.53588 | 0.546 |
| 5382 | GNG12-AS1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-889-3p | 17 | RPS6KA6 | Sponge network | -0.793 | 7.0E-5 | -2.989 | 0 | 0.546 |
| 5383 | RP11-532F6.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | ASTN1 | Sponge network | -0.896 | 0.00099 | -2.389 | 2.0E-5 | 0.546 |
| 5384 | MIR143HG |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | MSN | Sponge network | -0.739 | 0.0574 | -0.023 | 0.88422 | 0.546 |
| 5385 | AC010226.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-769-5p | 14 | SYNPO2 | Sponge network | -0.811 | 2.0E-5 | -2.342 | 0 | 0.546 |
| 5386 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | DCLK1 | Sponge network | -2.117 | 0.00161 | -1.324 | 0.00014 | 0.546 |
| 5387 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | TACC1 | Sponge network | -0.901 | 0.00019 | -0.362 | 0.05406 | 0.546 |
| 5388 | RP11-1006G14.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | BRD3 | Sponge network | 0.559 | 0.00026 | 0.37 | 0.00013 | 0.546 |
| 5389 | LINC00094 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-625-5p | 37 | ABI2 | Sponge network | 0.253 | 0.09565 | 0.175 | 0.14787 | 0.546 |
| 5390 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-93-5p | 24 | PLSCR4 | Sponge network | -0.162 | 0.47687 | -0.474 | 0.03833 | 0.546 |
| 5391 | RP11-822E23.8 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p | 12 | DCN | Sponge network | -0.777 | 0.16667 | -1.288 | 0 | 0.545 |
| 5392 | RP11-620J15.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-590-3p | 13 | ITPKB | Sponge network | -0.739 | 0.00044 | -0.704 | 0.00106 | 0.545 |
| 5393 | ARHGEF26-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 46 | GRIK3 | Sponge network | -2.46 | 0 | -3.208 | 0 | 0.545 |
| 5394 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | CDKN1C | Sponge network | -2.46 | 0 | -0.929 | 0.00033 | 0.545 |
| 5395 | SH3RF3-AS1 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | PTGFR | Sponge network | 0.889 | 9.0E-5 | -0.233 | 0.45421 | 0.545 |
| 5396 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | TSHZ3 | Sponge network | -2.117 | 0.00161 | -0.556 | 0.01516 | 0.545 |
| 5397 | RP11-736K20.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | STARD13 | Sponge network | -0.358 | 0.17255 | -0.187 | 0.27383 | 0.545 |
| 5398 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 32 | EYA4 | Sponge network | -0.129 | 0.57177 | 0.3 | 0.36876 | 0.545 |
| 5399 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 19 | HOOK3 | Sponge network | -1.111 | 0.0003 | 0.273 | 0.09957 | 0.545 |
| 5400 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 39 | EPHA7 | Sponge network | -0.227 | 0.31657 | -2.197 | 3.0E-5 | 0.545 |
| 5401 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DCN | Sponge network | 0.335 | 0.20596 | -1.288 | 0 | 0.545 |
| 5402 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-93-5p | 41 | TACC1 | Sponge network | -0.793 | 7.0E-5 | -0.362 | 0.05406 | 0.545 |
| 5403 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-5p | 32 | MCC | Sponge network | -1.645 | 0 | -1.005 | 0 | 0.545 |
| 5404 | FAM66C |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 13 | PTPN13 | Sponge network | -0.901 | 0.00019 | -1.208 | 0 | 0.545 |
| 5405 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | FAT4 | Sponge network | 0.246 | 0.59912 | -0.354 | 0.14694 | 0.545 |
| 5406 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-629-3p;hsa-miR-93-5p | 33 | EPHA7 | Sponge network | -1.654 | 0.00144 | -2.197 | 3.0E-5 | 0.545 |
| 5407 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 42 | DDX6 | Sponge network | 0.421 | 0.00084 | 0.091 | 0.36428 | 0.545 |
| 5408 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | CDH19 | Sponge network | -1.161 | 0.00591 | -3.434 | 0 | 0.545 |
| 5409 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 31 | IGF1 | Sponge network | 0.194 | 0.64278 | -0.204 | 0.5898 | 0.545 |
| 5410 | MEG3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | ZNF208 | Sponge network | 0.249 | 0.35902 | -0.4 | 0.22381 | 0.545 |
| 5411 | AC009948.5 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p | 20 | ATM | Sponge network | 0.532 | 0.00505 | 0.178 | 0.21568 | 0.545 |
| 5412 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 50 | TRPS1 | Sponge network | 0.997 | 0.00066 | -0.191 | 0.44109 | 0.545 |
| 5413 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 28 | SCN9A | Sponge network | 0.249 | 0.35902 | -0.525 | 0.10593 | 0.545 |
| 5414 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-429 | 10 | THSD7A | Sponge network | -0.154 | 0.53954 | 0.242 | 0.25154 | 0.545 |
| 5415 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-450b-5p | 16 | DLC1 | Sponge network | -0.517 | 0.02935 | -0.382 | 0.05038 | 0.545 |
| 5416 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 27 | MYO9A | Sponge network | 0.494 | 0.00339 | -0.147 | 0.32373 | 0.545 |
| 5417 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 44 | QKI | Sponge network | -0.842 | 0.01361 | -0.333 | 0.04111 | 0.545 |
| 5418 | MEG3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 16 | ZBTB20 | Sponge network | 0.249 | 0.35902 | -0.122 | 0.57569 | 0.545 |
| 5419 | MIR143HG |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | TAL1 | Sponge network | -0.739 | 0.0574 | -0.462 | 0.00574 | 0.545 |
| 5420 | CTD-2314G24.2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-940 | 16 | MAPK10 | Sponge network | 0.246 | 0.59912 | -1.774 | 0 | 0.545 |
| 5421 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p | 26 | TACC1 | Sponge network | -0.358 | 0.17255 | -0.362 | 0.05406 | 0.545 |
| 5422 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b | 11 | TXNIP | Sponge network | -0.583 | 0.14589 | -1.277 | 0 | 0.545 |
| 5423 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | BNC2 | Sponge network | -0.021 | 0.93598 | -0.491 | 0.09988 | 0.545 |
| 5424 | PCA3 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | RNF180 | Sponge network | -0.709 | 0.18168 | -1.127 | 1.0E-5 | 0.545 |
| 5425 | FAM66C |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-590-3p | 17 | RELN | Sponge network | -0.901 | 0.00019 | -2.403 | 0 | 0.545 |
| 5426 | BZRAP1-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 10 | PIK3IP1 | Sponge network | -0.465 | 0.04703 | -0.187 | 0.19945 | 0.545 |
| 5427 | ARHGEF26-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | -2.46 | 0 | 0.154 | 0.74723 | 0.545 |
| 5428 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 34 | TIMP3 | Sponge network | -1.111 | 0.0003 | -0.546 | 0.02307 | 0.545 |
| 5429 | CTD-2002H8.2 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CREB1 | Sponge network | 0.931 | 1.0E-5 | 0.203 | 0.00537 | 0.545 |
| 5430 | LINC00657 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 18 | RASAL2 | Sponge network | 0.91 | 0 | 0.869 | 0 | 0.545 |
| 5431 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 43 | AKAP11 | Sponge network | 0.421 | 0.00084 | 0.196 | 0.09825 | 0.545 |
| 5432 | ACTA2-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-744-3p | 64 | DST | Sponge network | 0.194 | 0.64278 | -0.713 | 0.00096 | 0.545 |
| 5433 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | RECK | Sponge network | 0.246 | 0.59912 | -1.043 | 0 | 0.545 |
| 5434 | FENDRR |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 42 | DIP2C | Sponge network | -1.281 | 0.00012 | -0.436 | 0.01713 | 0.545 |
| 5435 | WDFY3-AS2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 29 | WWTR1 | Sponge network | -0.942 | 0 | -0.345 | 0.11997 | 0.545 |
| 5436 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p | 11 | HMCN1 | Sponge network | -0.777 | 0.16667 | 0.809 | 0.00544 | 0.545 |
| 5437 | PWAR6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | KCNA6 | Sponge network | -1.575 | 6.0E-5 | -1.531 | 0.00013 | 0.545 |
| 5438 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ABCC9 | Sponge network | -1.203 | 0.03881 | -0.466 | 0.14121 | 0.545 |
| 5439 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 29 | RASSF8 | Sponge network | -0.487 | 0.02157 | -0.122 | 0.65591 | 0.545 |
| 5440 | PCA3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PCDH9 | Sponge network | -0.709 | 0.18168 | -1.823 | 4.0E-5 | 0.545 |
| 5441 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 36 | MYO9A | Sponge network | 0.064 | 0.72934 | -0.147 | 0.32373 | 0.545 |
| 5442 | SNHG14 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | NCAM2 | Sponge network | -1.111 | 0.0003 | -0.482 | 0.22565 | 0.545 |
| 5443 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p | 10 | DKK3 | Sponge network | -0.739 | 0.00044 | -0.052 | 0.77783 | 0.545 |
| 5444 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-629-3p | 19 | EBF1 | Sponge network | -0.154 | 0.53954 | -0.384 | 0.08856 | 0.545 |
| 5445 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p | 15 | CDH19 | Sponge network | -0.358 | 0.17255 | -3.434 | 0 | 0.544 |
| 5446 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-502-5p | 20 | DTNA | Sponge network | -0.309 | 0.1145 | -1.648 | 1.0E-5 | 0.544 |
| 5447 | AC009120.6 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-590-3p | 28 | ABL2 | Sponge network | 0.919 | 0 | 0.82 | 0 | 0.544 |
| 5448 | SNX29P2 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 14 | ZBTB20 | Sponge network | 0.4 | 0.14611 | -0.122 | 0.57569 | 0.544 |
| 5449 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 16 | TXNIP | Sponge network | -0.49 | 0.04946 | -1.277 | 0 | 0.544 |
| 5450 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p | 21 | AKAP6 | Sponge network | -0.246 | 0.35034 | -1.586 | 3.0E-5 | 0.544 |
| 5451 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | -2.117 | 0.00161 | -0.474 | 0.03833 | 0.544 |
| 5452 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 49 | SSBP2 | Sponge network | -0.162 | 0.47687 | -1.58 | 0 | 0.544 |
| 5453 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 31 | SNX29 | Sponge network | -0.563 | 0.02545 | -0.34 | 0.01371 | 0.544 |
| 5454 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-874-3p;hsa-miR-98-5p | 11 | ST5 | Sponge network | -2.452 | 0 | -0.78 | 0 | 0.544 |
| 5455 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-577 | 17 | AFF3 | Sponge network | -0.473 | 0.07602 | -2.373 | 0 | 0.544 |
| 5456 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 32 | SCN9A | Sponge network | -2.452 | 0 | -0.525 | 0.10593 | 0.544 |
| 5457 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 12 | SEMA3C | Sponge network | -0.583 | 0.14589 | -0.272 | 0.31153 | 0.544 |
| 5458 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 22 | CNTNAP3 | Sponge network | -2.46 | 0 | -1.465 | 0.00033 | 0.544 |
| 5459 | RP5-1198O20.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 33 | DLC1 | Sponge network | 0.997 | 0.00066 | -0.382 | 0.05038 | 0.544 |
| 5460 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 36 | FBXO32 | Sponge network | -0.222 | 0.32445 | -0.495 | 0.07561 | 0.544 |
| 5461 | BVES-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-98-5p | 28 | TGFBR3 | Sponge network | -2.62 | 0 | -1.173 | 1.0E-5 | 0.544 |
| 5462 | RP11-758N13.1 |
hsa-let-7d-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-375 | 10 | THBS1 | Sponge network | 2.939 | 3.0E-5 | 0.741 | 0.00386 | 0.544 |
| 5463 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 44 | ZFHX3 | Sponge network | 0.311 | 0.10344 | 0.389 | 0.01363 | 0.544 |
| 5464 | USP3-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-877-5p | 24 | NAV3 | Sponge network | -0.162 | 0.47687 | 0.212 | 0.47729 | 0.544 |
| 5465 | AGAP11 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-5p | 13 | PDZD2 | Sponge network | -1.645 | 0 | -0.909 | 3.0E-5 | 0.544 |
| 5466 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-98-5p | 46 | DMD | Sponge network | -0.942 | 0 | -1.506 | 0 | 0.544 |
| 5467 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-589-3p | 19 | SOX5 | Sponge network | -2.531 | 0 | -1.091 | 4.0E-5 | 0.544 |
| 5468 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-503-5p;hsa-miR-590-3p | 12 | SMARCA1 | Sponge network | -0.576 | 0.10121 | -0.684 | 0.00159 | 0.544 |
| 5469 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | AKAP6 | Sponge network | -1.349 | 0.00017 | -1.586 | 3.0E-5 | 0.544 |
| 5470 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | DYNC1I1 | Sponge network | -0.323 | 0.01372 | -1.12 | 0.00107 | 0.544 |
| 5471 | PART1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 64 | PRKAA2 | Sponge network | -2.925 | 0 | -2.462 | 0 | 0.544 |
| 5472 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 24 | AKAP6 | Sponge network | -0.154 | 0.78939 | -1.586 | 3.0E-5 | 0.544 |
| 5473 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 19 | MAPK10 | Sponge network | 0.997 | 0.00066 | -1.774 | 0 | 0.544 |
| 5474 | AP001347.6 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 18 | SYNPO2 | Sponge network | -1.161 | 0.00591 | -2.342 | 0 | 0.544 |
| 5475 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 41 | NRXN1 | Sponge network | -2.925 | 0 | -3.778 | 0 | 0.544 |
| 5476 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 25 | DYNC1I1 | Sponge network | -0.942 | 0 | -1.12 | 0.00107 | 0.544 |
| 5477 | PWAR6 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 21 | CXCL12 | Sponge network | -1.575 | 6.0E-5 | -1.421 | 0 | 0.544 |
| 5478 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | NRXN1 | Sponge network | -1.057 | 0.00029 | -3.778 | 0 | 0.544 |
| 5479 | RP11-6O2.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-452-5p;hsa-miR-495-3p | 17 | SYNM | Sponge network | 0.331 | 0.57247 | -2.309 | 1.0E-5 | 0.544 |
| 5480 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-940 | 18 | CREB1 | Sponge network | 0.993 | 0 | 0.203 | 0.00537 | 0.544 |
| 5481 | TP73-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-940 | 18 | GLI3 | Sponge network | -0.563 | 0.02545 | -0.615 | 0.01482 | 0.544 |
| 5482 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-542-3p | 10 | ALDH1A2 | Sponge network | -2.452 | 0 | -2.411 | 0 | 0.544 |
| 5483 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | PREX2 | Sponge network | -0.096 | 0.67958 | -0.272 | 0.25382 | 0.544 |
| 5484 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | ABCC9 | Sponge network | -1.216 | 0 | -0.466 | 0.14121 | 0.544 |
| 5485 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 23 | CDH23 | Sponge network | -0.739 | 0.0574 | -0.974 | 0.00034 | 0.544 |
| 5486 | RP11-473I1.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-369-3p | 14 | AKAP11 | Sponge network | 0.542 | 0.00054 | 0.196 | 0.09825 | 0.544 |
| 5487 | RP11-356J5.12 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 15 | FAM172A | Sponge network | -0.899 | 6.0E-5 | -0.57 | 0 | 0.544 |
| 5488 | LINC00957 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | SFRP1 | Sponge network | -0.421 | 0.0114 | -3.566 | 0 | 0.544 |
| 5489 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-589-3p | 23 | TIMP3 | Sponge network | -0.162 | 0.47687 | -0.546 | 0.02307 | 0.544 |
| 5490 | PWAR6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-577;hsa-miR-96-5p | 10 | PLAGL1 | Sponge network | -1.575 | 6.0E-5 | -0.588 | 0.00912 | 0.544 |
| 5491 | PGM5-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 20 | WWTR1 | Sponge network | -4.546 | 0 | -0.345 | 0.11997 | 0.544 |
| 5492 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p | 23 | RASSF8 | Sponge network | -0.246 | 0.35034 | -0.122 | 0.65591 | 0.544 |
| 5493 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-98-5p | 19 | PNRC1 | Sponge network | -2.452 | 0 | -0.646 | 0 | 0.544 |
| 5494 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-361-5p;hsa-miR-362-5p | 11 | IGF1 | Sponge network | -2.531 | 0 | -0.204 | 0.5898 | 0.544 |
| 5495 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | RAP1A | Sponge network | -2.452 | 0 | -0.929 | 0 | 0.544 |
| 5496 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p;hsa-miR-98-5p | 27 | MITF | Sponge network | -1.281 | 0.00012 | -0.769 | 0.00022 | 0.544 |
| 5497 | MIR497HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 51 | PRKAA2 | Sponge network | -1.439 | 4.0E-5 | -2.462 | 0 | 0.544 |
| 5498 | RP11-326G21.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | DTNA | Sponge network | -0.021 | 0.93598 | -1.648 | 1.0E-5 | 0.544 |
| 5499 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 20 | FGFR1 | Sponge network | -0.769 | 0.00035 | -0.509 | 0.03957 | 0.544 |
| 5500 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-362-3p | 16 | NRK | Sponge network | -0.693 | 0.05629 | 0.656 | 0.19217 | 0.543 |
| 5501 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | MLLT10 | Sponge network | 0.953 | 0 | 0.105 | 0.33292 | 0.543 |
| 5502 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 10 | CTNNA3 | Sponge network | -0.323 | 0.01372 | -2.046 | 0.00019 | 0.543 |
| 5503 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 10 | CAV1 | Sponge network | -0.896 | 0.00099 | -1.013 | 6.0E-5 | 0.543 |
| 5504 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | HOOK3 | Sponge network | -0.129 | 0.57177 | 0.273 | 0.09957 | 0.543 |
| 5505 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 26 | KIT | Sponge network | -0.739 | 0.0574 | -2.184 | 0 | 0.543 |
| 5506 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-505-3p | 13 | JAKMIP2 | Sponge network | -1.349 | 0.00017 | -1.388 | 0 | 0.543 |
| 5507 | LINC01018 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PEG3 | Sponge network | -2.915 | 0.00015 | -1.74 | 0 | 0.543 |
| 5508 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLI1 | Sponge network | -0.769 | 0.00035 | -0.294 | 0.10905 | 0.543 |
| 5509 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-671-5p | 14 | KIT | Sponge network | -2.095 | 0.00112 | -2.184 | 0 | 0.543 |
| 5510 | CTC-559E9.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 24 | CREB1 | Sponge network | 0.594 | 0.00064 | 0.203 | 0.00537 | 0.543 |
| 5511 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-539-5p | 17 | PDZD2 | Sponge network | -0.129 | 0.57177 | -0.909 | 3.0E-5 | 0.543 |
| 5512 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p | 43 | RUNX1T1 | Sponge network | -1.654 | 0.00144 | -0.344 | 0.2374 | 0.543 |
| 5513 | RP11-156E6.1 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326 | 14 | CBFB | Sponge network | 1.266 | 0 | 1.061 | 0 | 0.543 |
| 5514 | CTD-2002H8.2 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | MYO9A | Sponge network | 0.931 | 1.0E-5 | -0.147 | 0.32373 | 0.543 |
| 5515 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | AKAP12 | Sponge network | -2.117 | 0.00161 | -0.932 | 0.0028 | 0.543 |
| 5516 | RP11-774O3.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 31 | WWTR1 | Sponge network | -0.487 | 0.02157 | -0.345 | 0.11997 | 0.543 |
| 5517 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p | 35 | JAZF1 | Sponge network | -0.901 | 0.00019 | -0.377 | 0.02677 | 0.543 |
| 5518 | MIR497HG |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-584-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | PCDH18 | Sponge network | -1.439 | 4.0E-5 | 0.216 | 0.24645 | 0.543 |
| 5519 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 21 | SASH1 | Sponge network | -1.092 | 4.0E-5 | -0.502 | 0.00175 | 0.543 |
| 5520 | SH3BP5-AS1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 54 | MDM4 | Sponge network | 0.267 | 0.2937 | 0.345 | 0.00762 | 0.543 |
| 5521 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p | 25 | TBL1XR1 | Sponge network | 0.959 | 0 | 0.578 | 0 | 0.543 |
| 5522 | SNHG14 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 12 | SORCS2 | Sponge network | -1.111 | 0.0003 | -0.223 | 0.4253 | 0.543 |
| 5523 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | RASSF2 | Sponge network | -1.575 | 6.0E-5 | -0.273 | 0.21961 | 0.543 |
| 5524 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 43 | SNX29 | Sponge network | -1.111 | 0.0003 | -0.34 | 0.01371 | 0.543 |
| 5525 | PWAR6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | -1.575 | 6.0E-5 | 0.809 | 0.00544 | 0.543 |
| 5526 | NR2F1-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | TPO | Sponge network | -0.49 | 0.04946 | -1.87 | 2.0E-5 | 0.543 |
| 5527 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 18 | CDH23 | Sponge network | -1.439 | 4.0E-5 | -0.974 | 0.00034 | 0.543 |
| 5528 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 39 | RASSF2 | Sponge network | -2.452 | 0 | -0.273 | 0.21961 | 0.543 |
| 5529 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3913-5p;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-708-3p;hsa-miR-744-3p;hsa-miR-758-3p;hsa-miR-93-5p;hsa-miR-942-5p | 45 | HLF | Sponge network | -0.615 | 0.00015 | -2.443 | 0 | 0.543 |
| 5530 | LINC00472 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p | 10 | PARK2 | Sponge network | -0.842 | 0.01361 | -1.892 | 0 | 0.543 |
| 5531 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 13 | CNTNAP1 | Sponge network | -0.842 | 0.01361 | 0.358 | 0.13727 | 0.543 |
| 5532 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-421 | 11 | AHNAK | Sponge network | -0.583 | 0.14589 | -1.207 | 0 | 0.543 |
| 5533 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 17 | PDZD2 | Sponge network | -0.576 | 0.10121 | -0.909 | 3.0E-5 | 0.543 |
| 5534 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 18 | RSPO2 | Sponge network | -0.811 | 2.0E-5 | -3.038 | 0 | 0.543 |
| 5535 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 32 | LATS1 | Sponge network | 0.421 | 0.00084 | 0.109 | 0.34208 | 0.543 |
| 5536 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 33 | ZFHX3 | Sponge network | 0.194 | 0.64278 | 0.389 | 0.01363 | 0.543 |
| 5537 | MYLK-AS1 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-424-5p | 35 | THRB | Sponge network | -1.331 | 3.0E-5 | -1.117 | 0.0004 | 0.543 |
| 5538 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-576-5p | 18 | IL6ST | Sponge network | -2.039 | 0.03447 | -0.402 | 0.00676 | 0.543 |
| 5539 | CTA-204B4.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 15 | BRD3 | Sponge network | 0.765 | 7.0E-5 | 0.37 | 0.00013 | 0.543 |
| 5540 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 75 | TCF4 | Sponge network | -1.697 | 0.00889 | 0.016 | 0.92271 | 0.543 |
| 5541 | ARHGEF26-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-96-5p | 14 | ZBTB20 | Sponge network | -2.46 | 0 | -0.122 | 0.57569 | 0.543 |
| 5542 | AC005154.6 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-486-5p;hsa-miR-590-3p | 12 | TPR | Sponge network | 0.848 | 0 | 0.582 | 0 | 0.543 |
| 5543 | RP11-798M19.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 35 | TGFBR3 | Sponge network | -0.612 | 0.00094 | -1.173 | 1.0E-5 | 0.543 |
| 5544 | AC010226.4 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-590-3p | 12 | ITPKB | Sponge network | -0.811 | 2.0E-5 | -0.704 | 0.00106 | 0.543 |
| 5545 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 20 | FGFR1 | Sponge network | -1.575 | 6.0E-5 | -0.509 | 0.03957 | 0.543 |
| 5546 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-629-5p;hsa-miR-96-5p | 20 | RASSF8 | Sponge network | -0.517 | 0.02935 | -0.122 | 0.65591 | 0.543 |
| 5547 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | LRRC7 | Sponge network | -0.709 | 0.18168 | -1.341 | 0.01193 | 0.543 |
| 5548 | FENDRR |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | ZDBF2 | Sponge network | -1.281 | 0.00012 | -0.998 | 0.00025 | 0.543 |
| 5549 | RP11-890B15.3 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-877-5p | 12 | STAT5B | Sponge network | 0.101 | 0.60919 | -0.182 | 0.1082 | 0.543 |
| 5550 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-766-3p | 25 | CHD5 | Sponge network | -2.62 | 0 | -0.922 | 0.01521 | 0.543 |
| 5551 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 32 | RSPO2 | Sponge network | -1.111 | 0.0003 | -3.038 | 0 | 0.542 |
| 5552 | FAM66C |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | PAK3 | Sponge network | -0.901 | 0.00019 | -0.628 | 0.07253 | 0.542 |
| 5553 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 24 | KIT | Sponge network | -0.769 | 0.00035 | -2.184 | 0 | 0.542 |
| 5554 | EMX2OS |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | PTPN13 | Sponge network | -0.777 | 0.1134 | -1.208 | 0 | 0.542 |
| 5555 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | FSTL5 | Sponge network | -1.111 | 0.0003 | -1.3 | 0.01619 | 0.542 |
| 5556 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | JAZF1 | Sponge network | -1.281 | 0.00012 | -0.377 | 0.02677 | 0.542 |
| 5557 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-484 | 22 | ABL1 | Sponge network | 0.194 | 0.64278 | -0.215 | 0.07804 | 0.542 |
| 5558 | ANKRD10-IT1 |
hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-7-1-3p | 18 | CREB1 | Sponge network | 1.739 | 0 | 0.203 | 0.00537 | 0.542 |
| 5559 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 13 | TAF1 | Sponge network | 0.683 | 1.0E-5 | 0.39 | 3.0E-5 | 0.542 |
| 5560 | DNM3OS |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-323a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-935 | 10 | STAT5B | Sponge network | 0.572 | 0.01722 | -0.182 | 0.1082 | 0.542 |
| 5561 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p | 12 | WWTR1 | Sponge network | 0.176 | 0.37402 | -0.345 | 0.11997 | 0.542 |
| 5562 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-508-3p;hsa-miR-590-3p | 14 | SCN9A | Sponge network | -0.376 | 0.00337 | -0.525 | 0.10593 | 0.542 |
| 5563 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-590-3p | 16 | FGFR1 | Sponge network | -1.161 | 0.00591 | -0.509 | 0.03957 | 0.542 |
| 5564 | DNM3OS |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ITPKB | Sponge network | 0.572 | 0.01722 | -0.704 | 0.00106 | 0.542 |
| 5565 | SNHG14 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 20 | RHOB | Sponge network | -1.111 | 0.0003 | -1.052 | 0 | 0.542 |
| 5566 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 39 | JAZF1 | Sponge network | 0.194 | 0.64278 | -0.377 | 0.02677 | 0.542 |
| 5567 | ZNF582-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-877-5p | 21 | NRK | Sponge network | -1.349 | 0.00017 | 0.656 | 0.19217 | 0.542 |
| 5568 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 49 | RBMS3 | Sponge network | -1.216 | 0 | -0.519 | 0.03551 | 0.542 |
| 5569 | PCA3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | PEG3 | Sponge network | -0.709 | 0.18168 | -1.74 | 0 | 0.542 |
| 5570 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-935 | 34 | ABI2 | Sponge network | -0.096 | 0.67958 | 0.175 | 0.14787 | 0.542 |
| 5571 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-93-5p | 15 | SLITRK3 | Sponge network | -0.611 | 0.09607 | -3.328 | 0 | 0.542 |
| 5572 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-877-5p | 16 | CRTC3 | Sponge network | 0.889 | 9.0E-5 | 0.164 | 0.16786 | 0.542 |
| 5573 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-98-5p | 24 | MITF | Sponge network | -2.62 | 0 | -0.769 | 0.00022 | 0.542 |
| 5574 | MRVI1-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-542-3p | 10 | ID4 | Sponge network | -1.092 | 4.0E-5 | -1.644 | 0 | 0.542 |
| 5575 | LINC01018 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 51 | AR | Sponge network | -2.915 | 0.00015 | -1.554 | 0 | 0.542 |
| 5576 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-338-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | DYNC1I1 | Sponge network | 0.194 | 0.64278 | -1.12 | 0.00107 | 0.542 |
| 5577 | GNG12-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | -0.793 | 7.0E-5 | -0.382 | 0.05038 | 0.542 |
| 5578 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-5p | 14 | DMD | Sponge network | -2.076 | 0.00548 | -1.506 | 0 | 0.542 |
| 5579 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-26b-5p | 11 | DMD | Sponge network | -0.187 | 0.23787 | -1.506 | 0 | 0.542 |
| 5580 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | ST6GALNAC3 | Sponge network | -1.111 | 0.0003 | -0.987 | 0 | 0.542 |
| 5581 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 52 | SSBP2 | Sponge network | -2.117 | 0.00161 | -1.58 | 0 | 0.542 |
| 5582 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-577 | 18 | GREM1 | Sponge network | -0.896 | 0.00099 | 0.902 | 0.01691 | 0.542 |
| 5583 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 26 | MCC | Sponge network | -0.739 | 0.00044 | -1.005 | 0 | 0.542 |
| 5584 | FGF14-AS2 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-590-3p | 10 | PRUNE2 | Sponge network | -1.582 | 8.0E-5 | -1.733 | 0.00022 | 0.542 |
| 5585 | RP5-1198O20.4 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-574-3p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | FHL1 | Sponge network | 0.997 | 0.00066 | -2.687 | 0 | 0.542 |
| 5586 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | FGFR1 | Sponge network | -1.203 | 0.03881 | -0.509 | 0.03957 | 0.542 |
| 5587 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-508-3p;hsa-miR-590-3p | 18 | MITF | Sponge network | -0.358 | 0.17255 | -0.769 | 0.00022 | 0.542 |
| 5588 | CTC-296K1.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-582-5p | 33 | RUNX1T1 | Sponge network | -2.095 | 0.00112 | -0.344 | 0.2374 | 0.542 |
| 5589 | LINC00472 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p | 11 | MN1 | Sponge network | -0.842 | 0.01361 | -0.358 | 0.24172 | 0.542 |
| 5590 | ZRANB2-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-942-5p | 18 | IGFBP5 | Sponge network | -0.376 | 0.00337 | -0.806 | 0.00105 | 0.542 |
| 5591 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | PRDM5 | Sponge network | -2.46 | 0 | -1.09 | 0.00014 | 0.542 |
| 5592 | RP11-175O19.4 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p | 17 | PHF6 | Sponge network | 0.917 | 0 | 0.785 | 0 | 0.542 |
| 5593 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-590-3p | 18 | AKAP6 | Sponge network | -0.739 | 0.00044 | -1.586 | 3.0E-5 | 0.542 |
| 5594 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | TACC1 | Sponge network | 0.572 | 0.01722 | -0.362 | 0.05406 | 0.542 |
| 5595 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 13 | JAZF1 | Sponge network | -0.309 | 0.1145 | -0.377 | 0.02677 | 0.542 |
| 5596 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-590-5p | 12 | PTPN11 | Sponge network | 0.706 | 0 | 0.431 | 2.0E-5 | 0.542 |
| 5597 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-744-3p;hsa-miR-940 | 20 | MEF2C | Sponge network | -2.095 | 0.00112 | -0.499 | 0.01448 | 0.542 |
| 5598 | RP11-359B12.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-96-5p | 87 | LPP | Sponge network | 0.064 | 0.72934 | -0.459 | 0.04475 | 0.542 |
| 5599 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 16 | CNTNAP1 | Sponge network | -0.769 | 0.00035 | 0.358 | 0.13727 | 0.542 |
| 5600 | BVES-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-942-5p | 30 | IGFBP5 | Sponge network | -2.62 | 0 | -0.806 | 0.00105 | 0.542 |
| 5601 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-769-5p | 19 | SETBP1 | Sponge network | -0.777 | 0.16667 | -1.302 | 1.0E-5 | 0.542 |
| 5602 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 39 | FBXO32 | Sponge network | -1.575 | 6.0E-5 | -0.495 | 0.07561 | 0.542 |
| 5603 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 60 | CADM2 | Sponge network | -0.901 | 0.00019 | -3.782 | 0 | 0.542 |
| 5604 | CTD-2002H8.2 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-582-3p;hsa-miR-590-3p | 11 | ABL2 | Sponge network | 0.931 | 1.0E-5 | 0.82 | 0 | 0.542 |
| 5605 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p | 25 | ADARB1 | Sponge network | -0.517 | 0.02935 | -0.335 | 0.06631 | 0.542 |
| 5606 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | SYNE1 | Sponge network | 0.727 | 3.0E-5 | -0.738 | 0.00316 | 0.542 |
| 5607 | RP11-504P24.8 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 19 | MDM4 | Sponge network | 1.178 | 0 | 0.345 | 0.00762 | 0.542 |
| 5608 | MIR17HG |
hsa-miR-143-5p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-497-5p | 10 | MDM4 | Sponge network | 1.699 | 0 | 0.345 | 0.00762 | 0.542 |
| 5609 | RP11-473I1.10 |
hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-331-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | DDX6 | Sponge network | 0.673 | 0 | 0.091 | 0.36428 | 0.542 |
| 5610 | RP11-532F6.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p | 16 | WWTR1 | Sponge network | -0.896 | 0.00099 | -0.345 | 0.11997 | 0.542 |
| 5611 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b | 18 | NR2C2 | Sponge network | 0.594 | 0.00064 | 0.21 | 0.03224 | 0.542 |
| 5612 | PCA3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-500a-5p;hsa-miR-96-5p | 11 | ZBTB20 | Sponge network | -0.709 | 0.18168 | -0.122 | 0.57569 | 0.542 |
| 5613 | SNHG14 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | ABCA10 | Sponge network | -1.111 | 0.0003 | -0.571 | 0.03674 | 0.542 |
| 5614 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | MITF | Sponge network | -0.487 | 0.02157 | -0.769 | 0.00022 | 0.542 |
| 5615 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-629-3p;hsa-miR-93-3p | 13 | EBF3 | Sponge network | -2.531 | 0 | -0.058 | 0.81437 | 0.542 |
| 5616 | RP11-203M5.7 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-5p | 19 | MDM4 | Sponge network | 1.684 | 1.0E-5 | 0.345 | 0.00762 | 0.542 |
| 5617 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 32 | JAZF1 | Sponge network | -0.793 | 7.0E-5 | -0.377 | 0.02677 | 0.542 |
| 5618 | RP11-244O19.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 38 | IL17RD | Sponge network | -0.096 | 0.67958 | -0.019 | 0.94873 | 0.542 |
| 5619 | RAMP2-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-671-5p | 22 | DLC1 | Sponge network | -0.513 | 0.04703 | -0.382 | 0.05038 | 0.541 |
| 5620 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-2110;hsa-miR-331-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 13 | SNX29 | Sponge network | -0.295 | 0.02604 | -0.34 | 0.01371 | 0.541 |
| 5621 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | PDGFC | Sponge network | -0.737 | 0.06182 | -0.33 | 0.06483 | 0.541 |
| 5622 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 29 | JAZF1 | Sponge network | -0.612 | 0.00094 | -0.377 | 0.02677 | 0.541 |
| 5623 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | NRXN1 | Sponge network | -1.349 | 0.00017 | -3.778 | 0 | 0.541 |
| 5624 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | JAKMIP2 | Sponge network | -2.46 | 0 | -1.388 | 0 | 0.541 |
| 5625 | AC159540.2 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | MDM4 | Sponge network | 1.634 | 0 | 0.345 | 0.00762 | 0.541 |
| 5626 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | GRIN2A | Sponge network | -0.487 | 0.02157 | -2.161 | 0 | 0.541 |
| 5627 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 34 | MYO9A | Sponge network | 0.997 | 0.00066 | -0.147 | 0.32373 | 0.541 |
| 5628 | RP11-359B12.2 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-484 | 12 | FLNA | Sponge network | 0.064 | 0.72934 | -0.771 | 0.01377 | 0.541 |
| 5629 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p | 17 | GPC6 | Sponge network | 0.889 | 9.0E-5 | -0.054 | 0.81465 | 0.541 |
| 5630 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-577 | 18 | AFF3 | Sponge network | -0.465 | 0.04703 | -2.373 | 0 | 0.541 |
| 5631 | LINC00473 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p | 13 | PAK3 | Sponge network | -1.203 | 0.03881 | -0.628 | 0.07253 | 0.541 |
| 5632 | RP11-532F6.3 |
hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | CXCL12 | Sponge network | -0.896 | 0.00099 | -1.421 | 0 | 0.541 |
| 5633 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 38 | ERC1 | Sponge network | 0.997 | 0.00066 | -0.108 | 0.38794 | 0.541 |
| 5634 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | DCLK1 | Sponge network | -0.769 | 0.00035 | -1.324 | 0.00014 | 0.541 |
| 5635 | RP11-248G5.8 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-495-3p | 12 | STAG2 | Sponge network | 1.294 | 0 | 0.252 | 0.00438 | 0.541 |
| 5636 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | YAP1 | Sponge network | 0.222 | 0.29133 | 0.22 | 0.11836 | 0.541 |
| 5637 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | EPHA7 | Sponge network | 0.572 | 0.01722 | -2.197 | 3.0E-5 | 0.541 |
| 5638 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | TET1 | Sponge network | -1.575 | 6.0E-5 | -0.135 | 0.52138 | 0.541 |
| 5639 | RP11-672A2.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p | 10 | PLSCR4 | Sponge network | 1.344 | 0 | -0.474 | 0.03833 | 0.541 |
| 5640 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 32 | MCC | Sponge network | -0.421 | 0.0114 | -1.005 | 0 | 0.541 |
| 5641 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-590-5p | 13 | NRXN3 | Sponge network | 0.176 | 0.37402 | -0.893 | 0.04186 | 0.541 |
| 5642 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | NUAK1 | Sponge network | -0.129 | 0.57177 | 0.904 | 0 | 0.541 |
| 5643 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-766-3p | 24 | MAPK1 | Sponge network | 0.34 | 0.00273 | -0.017 | 0.81465 | 0.541 |
| 5644 | AP001347.6 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-7-1-3p | 14 | SFRP1 | Sponge network | -1.161 | 0.00591 | -3.566 | 0 | 0.541 |
| 5645 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-409-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 45 | TRPS1 | Sponge network | -0.487 | 0.02157 | -0.191 | 0.44109 | 0.541 |
| 5646 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 21 | TBL1XR1 | Sponge network | 1.4 | 0 | 0.578 | 0 | 0.541 |
| 5647 | TPTEP1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | PCDH9 | Sponge network | -1.216 | 0 | -1.823 | 4.0E-5 | 0.541 |
| 5648 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-34a-5p | 13 | AR | Sponge network | -0.691 | 0.00146 | -1.554 | 0 | 0.541 |
| 5649 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-425-5p | 43 | DTNA | Sponge network | -0.777 | 0.16667 | -1.648 | 1.0E-5 | 0.541 |
| 5650 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-7-1-3p | 21 | PDGFRA | Sponge network | -1.092 | 4.0E-5 | -0.372 | 0.0037 | 0.541 |
| 5651 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p | 12 | PTPN13 | Sponge network | 0.335 | 0.20596 | -1.208 | 0 | 0.541 |
| 5652 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 30 | TIMP3 | Sponge network | -0.777 | 0.1134 | -0.546 | 0.02307 | 0.541 |
| 5653 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TBX18 | Sponge network | 0.572 | 0.01722 | 0.843 | 0.02945 | 0.541 |
| 5654 | TRAF3IP2-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | SFRP1 | Sponge network | -0.323 | 0.01372 | -3.566 | 0 | 0.541 |
| 5655 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PTPRB | Sponge network | -0.129 | 0.57177 | 0.105 | 0.47122 | 0.541 |
| 5656 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ATRX | Sponge network | -0.323 | 0.01372 | 0.261 | 0.04408 | 0.541 |
| 5657 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-361-3p | 20 | RUNX1T1 | Sponge network | -0.691 | 0.00146 | -0.344 | 0.2374 | 0.541 |
| 5658 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-96-5p | 14 | DAB2 | Sponge network | 1.302 | 7.0E-5 | 0.431 | 0.0113 | 0.541 |
| 5659 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 24 | RSPO2 | Sponge network | -0.976 | 0.00025 | -3.038 | 0 | 0.541 |
| 5660 | ACTA2-AS1 |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 10 | DPP6 | Sponge network | 0.194 | 0.64278 | -3.777 | 0 | 0.541 |
| 5661 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | SLITRK3 | Sponge network | 0.997 | 0.00066 | -3.328 | 0 | 0.541 |
| 5662 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 35 | DMD | Sponge network | 0.997 | 0.00066 | -1.506 | 0 | 0.541 |
| 5663 | RP11-2B6.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TSC1 | Sponge network | 0.539 | 0.03722 | 0.103 | 0.26071 | 0.541 |
| 5664 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-590-3p | 13 | DCLK1 | Sponge network | -1.161 | 0.00591 | -1.324 | 0.00014 | 0.541 |
| 5665 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-628-5p | 33 | TCF4 | Sponge network | 0.811 | 0.03228 | 0.016 | 0.92271 | 0.541 |
| 5666 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-589-3p | 19 | ZFHX4 | Sponge network | -4.463 | 0.00025 | 0.018 | 0.95574 | 0.541 |
| 5667 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-590-5p | 17 | AKAP6 | Sponge network | -0.384 | 0.40652 | -1.586 | 3.0E-5 | 0.541 |
| 5668 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -2.117 | 0.00161 | -0.354 | 0.14694 | 0.541 |
| 5669 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 50 | SSBP2 | Sponge network | -0.487 | 0.02157 | -1.58 | 0 | 0.541 |
| 5670 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p | 22 | AFF3 | Sponge network | -0.842 | 0.01361 | -2.373 | 0 | 0.541 |
| 5671 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 26 | MITF | Sponge network | -0.769 | 0.00035 | -0.769 | 0.00022 | 0.54 |
| 5672 | HAND2-AS1 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ADH1B | Sponge network | -1.697 | 0.00889 | -3.716 | 0 | 0.54 |
| 5673 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | -0.777 | 0.16667 | -0.509 | 0.03957 | 0.54 |
| 5674 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | ZFHX3 | Sponge network | 0.083 | 0.66405 | 0.389 | 0.01363 | 0.54 |
| 5675 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-576-5p | 20 | GREM1 | Sponge network | -0.162 | 0.47687 | 0.902 | 0.01691 | 0.54 |
| 5676 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-340-5p;hsa-miR-486-5p;hsa-miR-590-3p | 17 | DYNC1I1 | Sponge network | -1.161 | 0.00591 | -1.12 | 0.00107 | 0.54 |
| 5677 | PWAR6 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 24 | ATM | Sponge network | -1.575 | 6.0E-5 | 0.178 | 0.21568 | 0.54 |
| 5678 | TP73-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | -0.563 | 0.02545 | -0.974 | 0.00034 | 0.54 |
| 5679 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-629-3p | 25 | RASSF8 | Sponge network | -0.777 | 0.16667 | -0.122 | 0.65591 | 0.54 |
| 5680 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940 | 26 | SNX29 | Sponge network | -0.465 | 0.04703 | -0.34 | 0.01371 | 0.54 |
| 5681 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 33 | PGR | Sponge network | -0.777 | 0.1134 | -1.704 | 0 | 0.54 |
| 5682 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 25 | LRIG1 | Sponge network | -0.096 | 0.67958 | -0.537 | 0.00588 | 0.54 |
| 5683 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | PDGFRA | Sponge network | -1.439 | 4.0E-5 | -0.372 | 0.0037 | 0.54 |
| 5684 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-7-1-3p | 12 | NRXN3 | Sponge network | -0.246 | 0.35034 | -0.893 | 0.04186 | 0.54 |
| 5685 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 18 | LHX6 | Sponge network | -0.129 | 0.57177 | -0.087 | 0.69193 | 0.54 |
| 5686 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-625-5p | 22 | ZMYM2 | Sponge network | 1.294 | 0 | 0.279 | 0.01273 | 0.54 |
| 5687 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | ST6GALNAC3 | Sponge network | -2.452 | 0 | -0.987 | 0 | 0.54 |
| 5688 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | SYNE1 | Sponge network | 0.101 | 0.60919 | -0.738 | 0.00316 | 0.54 |
| 5689 | OIP5-AS1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-590-3p | 12 | TPR | Sponge network | 0.421 | 0.00084 | 0.582 | 0 | 0.54 |
| 5690 | FAM66C |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 85 | LPP | Sponge network | -0.901 | 0.00019 | -0.459 | 0.04475 | 0.54 |
| 5691 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 13 | NR4A3 | Sponge network | -1.439 | 4.0E-5 | -1.385 | 0 | 0.54 |
| 5692 | DNM3OS |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CTGF | Sponge network | 0.572 | 0.01722 | 0.321 | 0.11682 | 0.54 |
| 5693 | ZNF667-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-93-5p;hsa-miR-940 | 10 | FAM117A | Sponge network | -0.976 | 0.00025 | -1.379 | 0 | 0.54 |
| 5694 | LINC00865 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | PEG3 | Sponge network | -1.249 | 7.0E-5 | -1.74 | 0 | 0.54 |
| 5695 | LINC00702 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | TMEFF2 | Sponge network | -0.737 | 0.06182 | -2.426 | 0 | 0.54 |
| 5696 | RP11-156P1.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p | 16 | EZH1 | Sponge network | 0.214 | 0.18246 | -0.564 | 1.0E-5 | 0.54 |
| 5697 | RP11-307B6.3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-942-5p | 17 | NRXN1 | Sponge network | -4.463 | 0.00025 | -3.778 | 0 | 0.54 |
| 5698 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-660-5p;hsa-miR-940 | 32 | GAS7 | Sponge network | -4.546 | 0 | -0.555 | 0.01602 | 0.54 |
| 5699 | ZNF667-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-484;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | -0.976 | 0.00025 | -0.974 | 0.00034 | 0.54 |
| 5700 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-671-5p | 21 | KIT | Sponge network | -1.249 | 7.0E-5 | -2.184 | 0 | 0.54 |
| 5701 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | MYO9A | Sponge network | 0.75 | 0.00054 | -0.147 | 0.32373 | 0.54 |
| 5702 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | EPHA3 | Sponge network | -0.187 | 0.23787 | 0.185 | 0.53588 | 0.54 |
| 5703 | ZNF667-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-98-5p | 15 | KLF10 | Sponge network | -0.976 | 0.00025 | -0.783 | 0 | 0.54 |
| 5704 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 44 | MEF2C | Sponge network | -0.563 | 0.02545 | -0.499 | 0.01448 | 0.54 |
| 5705 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-7-1-3p | 16 | SON | Sponge network | 0.817 | 3.0E-5 | 0.168 | 0.04406 | 0.54 |
| 5706 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | NIN | Sponge network | -0.129 | 0.57177 | -0.253 | 0.13794 | 0.54 |
| 5707 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 51 | FOXP1 | Sponge network | -0.222 | 0.32445 | 0.042 | 0.74368 | 0.54 |
| 5708 | LINC01018 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 14 | SORCS1 | Sponge network | -2.915 | 0.00015 | -3.492 | 0 | 0.54 |
| 5709 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-582-5p | 20 | RYR2 | Sponge network | -2.69 | 0.0001 | -1.466 | 0.00031 | 0.54 |
| 5710 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-629-3p;hsa-miR-935 | 27 | EBF1 | Sponge network | -2.62 | 0 | -0.384 | 0.08856 | 0.54 |
| 5711 | RP11-736K20.5 |
hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-335-5p | 10 | RPS6KA2 | Sponge network | -0.358 | 0.17255 | -0.57 | 0.00013 | 0.54 |
| 5712 | HHIP-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 41 | RUNX1T1 | Sponge network | -0.737 | 0.10822 | -0.344 | 0.2374 | 0.54 |
| 5713 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 42 | AR | Sponge network | -0.318 | 0.65847 | -1.554 | 0 | 0.54 |
| 5714 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92b-3p | 20 | AKAP12 | Sponge network | -1.216 | 0 | -0.932 | 0.0028 | 0.54 |
| 5715 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DNAJB4 | Sponge network | 0.997 | 0.00066 | -0.7 | 1.0E-5 | 0.54 |
| 5716 | MIR143HG |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-590-3p | 12 | RIMS1 | Sponge network | -0.739 | 0.0574 | -3.143 | 0 | 0.54 |
| 5717 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 29 | ABI2 | Sponge network | 0.559 | 0.00026 | 0.175 | 0.14787 | 0.54 |
| 5718 | MEG3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 82 | LPP | Sponge network | 0.249 | 0.35902 | -0.459 | 0.04475 | 0.539 |
| 5719 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | -0.612 | 0.00094 | -1.74 | 0 | 0.539 |
| 5720 | CTB-31O20.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 19 | MDM4 | Sponge network | 1.619 | 0 | 0.345 | 0.00762 | 0.539 |
| 5721 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-654-3p | 13 | ITCH | Sponge network | 0.959 | 0 | 0.084 | 0.34058 | 0.539 |
| 5722 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-628-5p | 35 | DTNA | Sponge network | -0.154 | 0.53954 | -1.648 | 1.0E-5 | 0.539 |
| 5723 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 57 | DTNA | Sponge network | -0.487 | 0.02157 | -1.648 | 1.0E-5 | 0.539 |
| 5724 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 33 | TRPS1 | Sponge network | -0.611 | 0.09607 | -0.191 | 0.44109 | 0.539 |
| 5725 | TRAF3IP2-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-940 | 19 | SORCS1 | Sponge network | -0.323 | 0.01372 | -3.492 | 0 | 0.539 |
| 5726 | RP11-244O19.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 12 | INPP4A | Sponge network | -0.096 | 0.67958 | 0.07 | 0.53563 | 0.539 |
| 5727 | NEAT1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 47 | MDM4 | Sponge network | 0.705 | 0.00014 | 0.345 | 0.00762 | 0.539 |
| 5728 | LINC00092 |
hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | RPS6KA6 | Sponge network | -1.886 | 0 | -2.989 | 0 | 0.539 |
| 5729 | LINC00641 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 43 | TGFBR3 | Sponge network | -0.222 | 0.32445 | -1.173 | 1.0E-5 | 0.539 |
| 5730 | RP11-53O19.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 20 | PRUNE2 | Sponge network | -0.405 | 0.22013 | -1.733 | 0.00022 | 0.539 |
| 5731 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-542-3p | 20 | SYNE1 | Sponge network | -0.517 | 0.02935 | -0.738 | 0.00316 | 0.539 |
| 5732 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p | 19 | SSBP2 | Sponge network | -2.076 | 0.00548 | -1.58 | 0 | 0.539 |
| 5733 | RP11-251G23.5 |
hsa-miR-101-3p;hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-363-3p | 10 | E2F3 | Sponge network | 1.344 | 0 | 1.289 | 0 | 0.539 |
| 5734 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | CNTN1 | Sponge network | -0.709 | 0.18168 | -2.594 | 0 | 0.539 |
| 5735 | ARHGEF26-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 74 | DST | Sponge network | -2.46 | 0 | -0.713 | 0.00096 | 0.539 |
| 5736 | MRVI1-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-628-5p | 23 | ERC1 | Sponge network | -1.092 | 4.0E-5 | -0.108 | 0.38794 | 0.539 |
| 5737 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | TSC1 | Sponge network | -0.222 | 0.32445 | 0.103 | 0.26071 | 0.539 |
| 5738 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-424-5p | 20 | ANK2 | Sponge network | 0.176 | 0.37402 | -1.603 | 1.0E-5 | 0.539 |
| 5739 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 43 | LRP1B | Sponge network | -1.111 | 0.0003 | -2.163 | 0 | 0.539 |
| 5740 | LINC00702 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 28 | PRRX1 | Sponge network | -0.737 | 0.06182 | 1.316 | 0 | 0.539 |
| 5741 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-539-5p;hsa-miR-625-5p | 17 | RSPO2 | Sponge network | -0.469 | 0.08721 | -3.038 | 0 | 0.539 |
| 5742 | LENG8-AS1 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-766-3p | 19 | MDM4 | Sponge network | 1.471 | 0 | 0.345 | 0.00762 | 0.539 |
| 5743 | LINC00654 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | DACT1 | Sponge network | 0.374 | 0.20054 | 0.783 | 0.00072 | 0.539 |
| 5744 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 45 | TACC1 | Sponge network | -1.697 | 0.00889 | -0.362 | 0.05406 | 0.539 |
| 5745 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 22 | TIMP3 | Sponge network | -0.611 | 0.09607 | -0.546 | 0.02307 | 0.539 |
| 5746 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 38 | EPHA7 | Sponge network | -0.777 | 0.16667 | -2.197 | 3.0E-5 | 0.539 |
| 5747 | LINC01018 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | -2.915 | 0.00015 | 0.154 | 0.74723 | 0.539 |
| 5748 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 57 | AR | Sponge network | -0.942 | 0 | -1.554 | 0 | 0.539 |
| 5749 | LINC00865 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | NRXN3 | Sponge network | -1.249 | 7.0E-5 | -0.893 | 0.04186 | 0.539 |
| 5750 | HAND2-AS1 |
hsa-miR-10b-3p;hsa-miR-193b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p | 10 | LRRTM1 | Sponge network | -1.697 | 0.00889 | -3.361 | 0 | 0.539 |
| 5751 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b | 17 | PRRX1 | Sponge network | -0.583 | 0.14589 | 1.316 | 0 | 0.539 |
| 5752 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | CNTNAP1 | Sponge network | -0.777 | 0.1134 | 0.358 | 0.13727 | 0.539 |
| 5753 | RP11-305E6.4 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 12 | PHF6 | Sponge network | 1.317 | 0 | 0.785 | 0 | 0.539 |
| 5754 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-338-3p | 10 | CACNA2D1 | Sponge network | -4.463 | 0.00025 | -0.846 | 0.00675 | 0.539 |
| 5755 | RP11-798M19.6 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 52 | AR | Sponge network | -0.612 | 0.00094 | -1.554 | 0 | 0.539 |
| 5756 | AC138035.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3614-5p | 17 | RICTOR | Sponge network | 1.073 | 0 | 0.388 | 0.00188 | 0.539 |
| 5757 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ATRX | Sponge network | 1.601 | 0 | 0.261 | 0.04408 | 0.539 |
| 5758 | MIR497HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-339-5p | 15 | STARD13 | Sponge network | -1.439 | 4.0E-5 | -0.187 | 0.27383 | 0.539 |
| 5759 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-7-1-3p | 22 | AKAP6 | Sponge network | -0.842 | 0.01361 | -1.586 | 3.0E-5 | 0.539 |
| 5760 | AC003090.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 32 | IGFBP5 | Sponge network | -2.117 | 0.00161 | -0.806 | 0.00105 | 0.539 |
| 5761 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-93-5p | 29 | ANK2 | Sponge network | -0.777 | 0.16667 | -1.603 | 1.0E-5 | 0.539 |
| 5762 | RP11-620J15.3 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | SORCS1 | Sponge network | -0.739 | 0.00044 | -3.492 | 0 | 0.539 |
| 5763 | AC003090.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 26 | RYR2 | Sponge network | -2.117 | 0.00161 | -1.466 | 0.00031 | 0.539 |
| 5764 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 37 | MAF | Sponge network | -2.452 | 0 | -1.257 | 0 | 0.539 |
| 5765 | CTD-3064M3.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-429 | 12 | ITPKB | Sponge network | -0.469 | 0.08721 | -0.704 | 0.00106 | 0.539 |
| 5766 | RASSF8-AS1 |
hsa-miR-146b-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-484;hsa-miR-574-3p;hsa-miR-590-3p | 11 | FHL1 | Sponge network | 0.335 | 0.20596 | -2.687 | 0 | 0.539 |
| 5767 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-590-3p | 16 | PAK3 | Sponge network | -0.49 | 0.04946 | -0.628 | 0.07253 | 0.539 |
| 5768 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-940 | 14 | HOOK3 | Sponge network | 0.75 | 0.00054 | 0.273 | 0.09957 | 0.539 |
| 5769 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p | 25 | CNTN1 | Sponge network | -0.777 | 0.16667 | -2.594 | 0 | 0.539 |
| 5770 | RP11-159D12.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p | 12 | SHPRH | Sponge network | 1.254 | 0 | 0.098 | 0.40994 | 0.539 |
| 5771 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 21 | CADM3 | Sponge network | -0.563 | 0.02545 | -3.663 | 0 | 0.539 |
| 5772 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 14 | NCAM2 | Sponge network | -0.896 | 0.00099 | -0.482 | 0.22565 | 0.539 |
| 5773 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p | 19 | PCDH9 | Sponge network | -1.654 | 0.00144 | -1.823 | 4.0E-5 | 0.539 |
| 5774 | FENDRR |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p | 10 | SORCS2 | Sponge network | -1.281 | 0.00012 | -0.223 | 0.4253 | 0.539 |
| 5775 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-93-3p | 25 | EPHA3 | Sponge network | -2.69 | 0.0001 | 0.185 | 0.53588 | 0.539 |
| 5776 | HOXB-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-628-5p | 54 | LPP | Sponge network | -0.154 | 0.53954 | -0.459 | 0.04475 | 0.539 |
| 5777 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-664a-3p | 14 | MN1 | Sponge network | -0.405 | 0.22013 | -0.358 | 0.24172 | 0.539 |
| 5778 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | MYO9A | Sponge network | 0.349 | 0.01695 | -0.147 | 0.32373 | 0.539 |
| 5779 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | DCLK1 | Sponge network | -0.487 | 0.02157 | -1.324 | 0.00014 | 0.538 |
| 5780 | RP11-554A11.4 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-33a-3p;hsa-miR-421 | 10 | GJA1 | Sponge network | -0.583 | 0.14589 | -0.485 | 0.02179 | 0.538 |
| 5781 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 47 | ZFHX4 | Sponge network | -0.563 | 0.02545 | 0.018 | 0.95574 | 0.538 |
| 5782 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 27 | AKAP6 | Sponge network | 0.101 | 0.60919 | -1.586 | 3.0E-5 | 0.538 |
| 5783 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-940;hsa-miR-96-5p | 21 | MEF2C | Sponge network | -0.31 | 0.33871 | -0.499 | 0.01448 | 0.538 |
| 5784 | LINC00472 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p | 13 | FNBP1 | Sponge network | -0.842 | 0.01361 | -0.969 | 4.0E-5 | 0.538 |
| 5785 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-96-5p | 30 | GAS7 | Sponge network | -1.582 | 8.0E-5 | -0.555 | 0.01602 | 0.538 |
| 5786 | RP11-701H24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-33a-3p | 15 | DMD | Sponge network | -2.039 | 0.03447 | -1.506 | 0 | 0.538 |
| 5787 | CTC-444N24.11 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 34 | CREB1 | Sponge network | 1.153 | 0 | 0.203 | 0.00537 | 0.538 |
| 5788 | LINC00476 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p | 21 | SYNM | Sponge network | -0.615 | 0.00015 | -2.309 | 1.0E-5 | 0.538 |
| 5789 | FAM66C |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 10 | DPP6 | Sponge network | -0.901 | 0.00019 | -3.777 | 0 | 0.538 |
| 5790 | RP11-326G21.1 |
hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | GLIPR1 | Sponge network | -0.021 | 0.93598 | 0.146 | 0.3276 | 0.538 |
| 5791 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 27 | RICTOR | Sponge network | 0.946 | 0 | 0.388 | 0.00188 | 0.538 |
| 5792 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-590-3p | 12 | ITGB3 | Sponge network | 0.194 | 0.64278 | -0.011 | 0.95751 | 0.538 |
| 5793 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p | 31 | CTIF | Sponge network | -2.452 | 0 | -0.389 | 0.00549 | 0.538 |
| 5794 | PGM5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 50 | LPP | Sponge network | -4.546 | 0 | -0.459 | 0.04475 | 0.538 |
| 5795 | NR2F1-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 14 | PHOX2B | Sponge network | -0.49 | 0.04946 | -2.68 | 0 | 0.538 |
| 5796 | LINC00472 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 16 | PEG3 | Sponge network | -0.842 | 0.01361 | -1.74 | 0 | 0.538 |
| 5797 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | FSTL5 | Sponge network | -2.46 | 0 | -1.3 | 0.01619 | 0.538 |
| 5798 | ZRANB2-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-628-5p | 21 | DTNA | Sponge network | -0.376 | 0.00337 | -1.648 | 1.0E-5 | 0.538 |
| 5799 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 20 | LRRC7 | Sponge network | -1.092 | 4.0E-5 | -1.341 | 0.01193 | 0.538 |
| 5800 | EMX2OS |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | MRVI1 | Sponge network | -0.777 | 0.1134 | -1.051 | 0.00022 | 0.538 |
| 5801 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-92a-3p | 22 | AFF3 | Sponge network | -0.563 | 0.02545 | -2.373 | 0 | 0.538 |
| 5802 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-93-5p | 14 | TXNIP | Sponge network | -0.611 | 0.09607 | -1.277 | 0 | 0.538 |
| 5803 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-98-5p | 17 | ITGB3 | Sponge network | -0.49 | 0.04946 | -0.011 | 0.95751 | 0.538 |
| 5804 | WDFY3-AS2 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | RNF180 | Sponge network | -0.942 | 0 | -1.127 | 1.0E-5 | 0.538 |
| 5805 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-877-5p | 29 | GAS7 | Sponge network | 0.889 | 9.0E-5 | -0.555 | 0.01602 | 0.538 |
| 5806 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-874-3p | 22 | NR2C2 | Sponge network | 0.683 | 1.0E-5 | 0.21 | 0.03224 | 0.538 |
| 5807 | RP11-822E23.8 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-93-5p | 45 | AR | Sponge network | -0.777 | 0.16667 | -1.554 | 0 | 0.538 |
| 5808 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-429 | 23 | DLC1 | Sponge network | -1.654 | 0.00144 | -0.382 | 0.05038 | 0.538 |
| 5809 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-582-3p;hsa-miR-590-3p | 44 | ADARB1 | Sponge network | -0.901 | 0.00019 | -0.335 | 0.06631 | 0.538 |
| 5810 | ZNF667-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429 | 12 | MACF1 | Sponge network | -0.976 | 0.00025 | 0.019 | 0.90335 | 0.538 |
| 5811 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 13 | CNTNAP1 | Sponge network | -0.487 | 0.02157 | 0.358 | 0.13727 | 0.538 |
| 5812 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | PDZRN3 | Sponge network | 0.997 | 0.00066 | -1.548 | 0 | 0.538 |
| 5813 | AC010226.4 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-7-5p;hsa-miR-96-5p | 11 | FNBP1 | Sponge network | -0.811 | 2.0E-5 | -0.969 | 4.0E-5 | 0.538 |
| 5814 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-96-5p;hsa-miR-98-5p | 25 | MITF | Sponge network | -0.976 | 0.00025 | -0.769 | 0.00022 | 0.538 |
| 5815 | RP11-894P9.1 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-495-3p | 15 | TSC1 | Sponge network | 0.993 | 0 | 0.103 | 0.26071 | 0.538 |
| 5816 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | ADAMTSL3 | Sponge network | 0.997 | 0.00066 | -1.559 | 0 | 0.538 |
| 5817 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-582-3p | 19 | CBL | Sponge network | 0.993 | 0 | 0.291 | 0.01267 | 0.538 |
| 5818 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | SOX5 | Sponge network | -1.439 | 4.0E-5 | -1.091 | 4.0E-5 | 0.538 |
| 5819 | LINC00702 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 29 | GJA1 | Sponge network | -0.737 | 0.06182 | -0.485 | 0.02179 | 0.538 |
| 5820 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 31 | DMD | Sponge network | -0.769 | 0.00035 | -1.506 | 0 | 0.538 |
| 5821 | LINC00092 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 28 | CADM2 | Sponge network | -1.886 | 0 | -3.782 | 0 | 0.538 |
| 5822 | HAND2-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-5p;hsa-miR-486-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-942-5p | 22 | GLIPR1 | Sponge network | -1.697 | 0.00889 | 0.146 | 0.3276 | 0.538 |
| 5823 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-455-3p | 15 | PLSCR4 | Sponge network | -0.309 | 0.1145 | -0.474 | 0.03833 | 0.538 |
| 5824 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-98-5p | 35 | THBS1 | Sponge network | -1.697 | 0.00889 | 0.741 | 0.00386 | 0.538 |
| 5825 | RP11-736K20.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 54 | LPP | Sponge network | -0.358 | 0.17255 | -0.459 | 0.04475 | 0.538 |
| 5826 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-654-3p | 16 | RICTOR | Sponge network | 1.134 | 0 | 0.388 | 0.00188 | 0.538 |
| 5827 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429 | 14 | DCLK1 | Sponge network | -1.249 | 7.0E-5 | -1.324 | 0.00014 | 0.538 |
| 5828 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p | 19 | SPG20 | Sponge network | -0.487 | 0.02157 | -1.16 | 0 | 0.538 |
| 5829 | AC010226.4 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -0.811 | 2.0E-5 | -0.564 | 1.0E-5 | 0.538 |
| 5830 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-577 | 17 | IGF1 | Sponge network | -0.473 | 0.07602 | -0.204 | 0.5898 | 0.538 |
| 5831 | MIR497HG |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-33a-3p | 12 | KLF10 | Sponge network | -1.439 | 4.0E-5 | -0.783 | 0 | 0.538 |
| 5832 | LINC00957 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 10 | FHL1 | Sponge network | -0.421 | 0.0114 | -2.687 | 0 | 0.538 |
| 5833 | AP001055.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-455-3p | 26 | RUNX1T1 | Sponge network | 0.693 | 0.00076 | -0.344 | 0.2374 | 0.538 |
| 5834 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p | 21 | TIMP3 | Sponge network | -0.896 | 0.00099 | -0.546 | 0.02307 | 0.538 |
| 5835 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 53 | QKI | Sponge network | -0.487 | 0.02157 | -0.333 | 0.04111 | 0.538 |
| 5836 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | -0.246 | 0.35034 | -1.74 | 0 | 0.538 |
| 5837 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 42 | ZFHX4 | Sponge network | -1.249 | 7.0E-5 | 0.018 | 0.95574 | 0.538 |
| 5838 | RP11-359B12.2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | 0.064 | 0.72934 | -0.704 | 0.00106 | 0.537 |
| 5839 | LINC00957 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | PRUNE2 | Sponge network | -0.421 | 0.0114 | -1.733 | 0.00022 | 0.537 |
| 5840 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 18 | DYNC1I1 | Sponge network | 0.335 | 0.20596 | -1.12 | 0.00107 | 0.537 |
| 5841 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 27 | THBS1 | Sponge network | 0.335 | 0.20596 | 0.741 | 0.00386 | 0.537 |
| 5842 | LINC00641 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 24 | SIRT1 | Sponge network | -0.222 | 0.32445 | 0.109 | 0.2593 | 0.537 |
| 5843 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 31 | RICTOR | Sponge network | 0.799 | 1.0E-5 | 0.388 | 0.00188 | 0.537 |
| 5844 | CTA-363E19.2 |
hsa-let-7d-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 13 | RICTOR | Sponge network | 1.055 | 0 | 0.388 | 0.00188 | 0.537 |
| 5845 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-3p | 17 | PAK3 | Sponge network | -0.129 | 0.57177 | -0.628 | 0.07253 | 0.537 |
| 5846 | AC006116.24 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-454-3p;hsa-miR-455-5p | 12 | ITPKB | Sponge network | -0.473 | 0.07602 | -0.704 | 0.00106 | 0.537 |
| 5847 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-590-3p | 32 | DMD | Sponge network | -0.739 | 0.00044 | -1.506 | 0 | 0.537 |
| 5848 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 19 | KIT | Sponge network | -1.057 | 0.00029 | -2.184 | 0 | 0.537 |
| 5849 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p | 11 | HIC1 | Sponge network | -2.452 | 0 | 0.024 | 0.89889 | 0.537 |
| 5850 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-455-5p | 17 | GPC6 | Sponge network | -0.473 | 0.07602 | -0.054 | 0.81465 | 0.537 |
| 5851 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 34 | MCC | Sponge network | -1.349 | 0.00017 | -1.005 | 0 | 0.537 |
| 5852 | LINC00094 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p | 26 | BRD3 | Sponge network | 0.253 | 0.09565 | 0.37 | 0.00013 | 0.537 |
| 5853 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p | 37 | PBX1 | Sponge network | -0.487 | 0.02157 | -1.206 | 0 | 0.537 |
| 5854 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 15 | DCN | Sponge network | 0.246 | 0.59912 | -1.288 | 0 | 0.537 |
| 5855 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | AR | Sponge network | 0.299 | 0.57722 | -1.554 | 0 | 0.537 |
| 5856 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 53 | ZFHX4 | Sponge network | -0.323 | 0.01372 | 0.018 | 0.95574 | 0.537 |
| 5857 | RP11-400K9.3 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-3p | 16 | BNC2 | Sponge network | -0.733 | 0.19469 | -0.491 | 0.09988 | 0.537 |
| 5858 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-96-5p | 11 | LRRC4 | Sponge network | -1.439 | 4.0E-5 | -1.774 | 0 | 0.537 |
| 5859 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 52 | DTNA | Sponge network | -0.612 | 0.00094 | -1.648 | 1.0E-5 | 0.537 |
| 5860 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 27 | RICTOR | Sponge network | 0.594 | 0.00064 | 0.388 | 0.00188 | 0.537 |
| 5861 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 15 | GLI3 | Sponge network | -0.096 | 0.67958 | -0.615 | 0.01482 | 0.537 |
| 5862 | RP11-822E23.8 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-93-3p | 50 | RUNX1T1 | Sponge network | -0.777 | 0.16667 | -0.344 | 0.2374 | 0.537 |
| 5863 | RP11-532F6.3 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 10 | RHOB | Sponge network | -0.896 | 0.00099 | -1.052 | 0 | 0.537 |
| 5864 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 14 | HIP1 | Sponge network | 1.757 | 0 | 0.774 | 0 | 0.537 |
| 5865 | FAM66C |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | LSAMP | Sponge network | -0.901 | 0.00019 | -0.066 | 0.81708 | 0.537 |
| 5866 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 12 | DICER1 | Sponge network | 0.542 | 0.00054 | 0.042 | 0.674 | 0.537 |
| 5867 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-423-5p | 15 | CBFA2T3 | Sponge network | -4.546 | 0 | -1.221 | 1.0E-5 | 0.537 |
| 5868 | CTC-429P9.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 63 | LPP | Sponge network | 0.082 | 0.55397 | -0.459 | 0.04475 | 0.537 |
| 5869 | FENDRR |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-532-3p;hsa-miR-93-3p | 17 | RPS6KA2 | Sponge network | -1.281 | 0.00012 | -0.57 | 0.00013 | 0.537 |
| 5870 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-5p | 33 | GAS7 | Sponge network | -2.531 | 0 | -0.555 | 0.01602 | 0.537 |
| 5871 | RP11-890B15.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 16 | EZH1 | Sponge network | 0.101 | 0.60919 | -0.564 | 1.0E-5 | 0.537 |
| 5872 | LINC00957 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | SLITRK3 | Sponge network | -0.421 | 0.0114 | -3.328 | 0 | 0.537 |
| 5873 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-654-3p | 22 | RICTOR | Sponge network | 1.923 | 0 | 0.388 | 0.00188 | 0.537 |
| 5874 | CTC-429P9.2 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-485-3p;hsa-miR-542-3p | 10 | PHC3 | Sponge network | 0.043 | 0.81628 | 0.212 | 0.0499 | 0.537 |
| 5875 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PLSCR4 | Sponge network | -0.487 | 0.02157 | -0.474 | 0.03833 | 0.537 |
| 5876 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-486-5p | 20 | ADAMTSL3 | Sponge network | 0.176 | 0.37402 | -1.559 | 0 | 0.537 |
| 5877 | FAM66C |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | TPO | Sponge network | -0.901 | 0.00019 | -1.87 | 2.0E-5 | 0.537 |
| 5878 | WDFY3-AS2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p | 68 | DST | Sponge network | -0.942 | 0 | -0.713 | 0.00096 | 0.537 |
| 5879 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-7-5p | 23 | RBMS3 | Sponge network | -0.517 | 0.02935 | -0.519 | 0.03551 | 0.537 |
| 5880 | RP11-822E23.8 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-671-5p | 26 | DLC1 | Sponge network | -0.777 | 0.16667 | -0.382 | 0.05038 | 0.537 |
| 5881 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 22 | SON | Sponge network | 1.601 | 0 | 0.168 | 0.04406 | 0.537 |
| 5882 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 21 | QKI | Sponge network | -0.376 | 0.00337 | -0.333 | 0.04111 | 0.537 |
| 5883 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | BNC2 | Sponge network | -1.654 | 0.00144 | -0.491 | 0.09988 | 0.537 |
| 5884 | MIR143HG |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | DACT1 | Sponge network | -0.739 | 0.0574 | 0.783 | 0.00072 | 0.537 |
| 5885 | RP11-2B6.2 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p | 12 | ATM | Sponge network | 0.539 | 0.03722 | 0.178 | 0.21568 | 0.537 |
| 5886 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | -0.769 | 0.00035 | -0.615 | 0.01482 | 0.537 |
| 5887 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | ZFHX4 | Sponge network | -2.915 | 0.00015 | 0.018 | 0.95574 | 0.537 |
| 5888 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 50 | GAS7 | Sponge network | -1.216 | 0 | -0.555 | 0.01602 | 0.537 |
| 5889 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 19 | RARB | Sponge network | -0.769 | 0.00035 | -0.825 | 2.0E-5 | 0.537 |
| 5890 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-874-3p | 14 | NR2C2 | Sponge network | 1.093 | 0 | 0.21 | 0.03224 | 0.537 |
| 5891 | RP11-359B12.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p | 16 | EZH1 | Sponge network | 0.064 | 0.72934 | -0.564 | 1.0E-5 | 0.537 |
| 5892 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TIMP3 | Sponge network | 0.194 | 0.64278 | -0.546 | 0.02307 | 0.537 |
| 5893 | TRAF3IP2-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | CXCL12 | Sponge network | -0.323 | 0.01372 | -1.421 | 0 | 0.537 |
| 5894 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | ZMYND11 | Sponge network | 0.064 | 0.72934 | -0.2 | 0.05449 | 0.537 |
| 5895 | AC010226.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p | 32 | RUNX1T1 | Sponge network | -0.811 | 2.0E-5 | -0.344 | 0.2374 | 0.537 |
| 5896 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 37 | GAS7 | Sponge network | -0.154 | 0.53954 | -0.555 | 0.01602 | 0.537 |
| 5897 | RP11-822E23.8 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 32 | IGFBP5 | Sponge network | -0.777 | 0.16667 | -0.806 | 0.00105 | 0.537 |
| 5898 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 22 | ZFHX3 | Sponge network | 0.765 | 7.0E-5 | 0.389 | 0.01363 | 0.537 |
| 5899 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | 0.335 | 0.20596 | -0.354 | 0.14694 | 0.537 |
| 5900 | TRAF3IP2-AS1 |
hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-877-5p;hsa-miR-940 | 13 | PHOX2B | Sponge network | -0.323 | 0.01372 | -2.68 | 0 | 0.537 |
| 5901 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-340-5p | 22 | TSHZ3 | Sponge network | -0.693 | 0.05629 | -0.556 | 0.01516 | 0.537 |
| 5902 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-589-3p | 15 | PEG3 | Sponge network | -0.154 | 0.78939 | -1.74 | 0 | 0.537 |
| 5903 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 34 | MCC | Sponge network | -1.249 | 7.0E-5 | -1.005 | 0 | 0.537 |
| 5904 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 17 | NR2C2 | Sponge network | 0.873 | 0 | 0.21 | 0.03224 | 0.536 |
| 5905 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 12 | GAS1 | Sponge network | -1.111 | 0.0003 | -1.047 | 0.00364 | 0.536 |
| 5906 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | PEG3 | Sponge network | 0.299 | 0.57722 | -1.74 | 0 | 0.536 |
| 5907 | MEG3 |
hsa-miR-144-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-502-5p | 10 | ELN | Sponge network | 0.249 | 0.35902 | 0.451 | 0.13752 | 0.536 |
| 5908 | MIR143HG |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 23 | RPS6KA6 | Sponge network | -0.739 | 0.0574 | -2.989 | 0 | 0.536 |
| 5909 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | TLL1 | Sponge network | -0.769 | 0.00035 | -1.195 | 3.0E-5 | 0.536 |
| 5910 | RP11-244O19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-378c;hsa-miR-628-5p | 29 | ERC1 | Sponge network | -0.096 | 0.67958 | -0.108 | 0.38794 | 0.536 |
| 5911 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 11 | FSTL5 | Sponge network | -0.162 | 0.47687 | -1.3 | 0.01619 | 0.536 |
| 5912 | RP11-554A11.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 13 | SCN9A | Sponge network | -0.583 | 0.14589 | -0.525 | 0.10593 | 0.536 |
| 5913 | SNX29P2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | FLI1 | Sponge network | 0.4 | 0.14611 | -0.294 | 0.10905 | 0.536 |
| 5914 | RP5-1085F17.3 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | RBBP6 | Sponge network | 0.541 | 0.00125 | 0.163 | 0.05101 | 0.536 |
| 5915 | RP11-473I1.10 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | AKAP11 | Sponge network | 0.673 | 0 | 0.196 | 0.09825 | 0.536 |
| 5916 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | DYNC1I1 | Sponge network | -1.281 | 0.00012 | -1.12 | 0.00107 | 0.536 |
| 5917 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-96-5p | 44 | PIK3R1 | Sponge network | 0.194 | 0.64278 | -0.133 | 0.36854 | 0.536 |
| 5918 | FENDRR |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 15 | GLI3 | Sponge network | -1.281 | 0.00012 | -0.615 | 0.01482 | 0.536 |
| 5919 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p | 16 | DNAJB4 | Sponge network | -0.162 | 0.47687 | -0.7 | 1.0E-5 | 0.536 |
| 5920 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-7-1-3p | 20 | PCDH9 | Sponge network | -1.349 | 0.00017 | -1.823 | 4.0E-5 | 0.536 |
| 5921 | RP11-182J1.1 |
hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | ANK2 | Sponge network | -0.187 | 0.23787 | -1.603 | 1.0E-5 | 0.536 |
| 5922 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | -0.421 | 0.0114 | -1.751 | 1.0E-5 | 0.536 |
| 5923 | LINC00702 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | IL17RD | Sponge network | -0.737 | 0.06182 | -0.019 | 0.94873 | 0.536 |
| 5924 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 16 | DDX5 | Sponge network | -0.222 | 0.32445 | -0.099 | 0.17428 | 0.536 |
| 5925 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | RICTOR | Sponge network | 1.378 | 0 | 0.388 | 0.00188 | 0.536 |
| 5926 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 41 | ABI2 | Sponge network | -0.576 | 0.10121 | 0.175 | 0.14787 | 0.536 |
| 5927 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-7-1-3p | 15 | DKK3 | Sponge network | 0.335 | 0.20596 | -0.052 | 0.77783 | 0.536 |
| 5928 | AC009948.5 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 17 | INPP4A | Sponge network | 0.532 | 0.00505 | 0.07 | 0.53563 | 0.536 |
| 5929 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 48 | BCL2 | Sponge network | -1.575 | 6.0E-5 | -0.899 | 0 | 0.536 |
| 5930 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | TGFBR3 | Sponge network | 0.194 | 0.64278 | -1.173 | 1.0E-5 | 0.536 |
| 5931 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p | 33 | ZFHX4 | Sponge network | 0.176 | 0.37402 | 0.018 | 0.95574 | 0.536 |
| 5932 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | -0.318 | 0.65847 | 0.358 | 0.13727 | 0.536 |
| 5933 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 28 | RASSF8 | Sponge network | -0.842 | 0.01361 | -0.122 | 0.65591 | 0.536 |
| 5934 | RP11-867G23.10 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-550a-3p | 19 | MEF2C | Sponge network | -2.531 | 0 | -0.499 | 0.01448 | 0.536 |
| 5935 | RP11-473I1.9 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | USP9X | Sponge network | 0.683 | 1.0E-5 | 0.222 | 0.01689 | 0.536 |
| 5936 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 20 | TACC1 | Sponge network | -0.932 | 0.00329 | -0.362 | 0.05406 | 0.536 |
| 5937 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-653-5p | 32 | DMXL1 | Sponge network | -0.222 | 0.32445 | -0.426 | 0.00056 | 0.536 |
| 5938 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 40 | ESR1 | Sponge network | -1.697 | 0.00889 | -1.035 | 3.0E-5 | 0.536 |
| 5939 | BCYRN1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 62 | MDM4 | Sponge network | 0.311 | 0.10344 | 0.345 | 0.00762 | 0.536 |
| 5940 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p | 38 | ZFHX4 | Sponge network | -0.896 | 0.00099 | 0.018 | 0.95574 | 0.536 |
| 5941 | WDFY3-AS2 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | FHL1 | Sponge network | -0.942 | 0 | -2.687 | 0 | 0.536 |
| 5942 | AC009948.5 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | 0.532 | 0.00505 | 0.358 | 0.13727 | 0.536 |
| 5943 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 26 | ZBTB4 | Sponge network | -0.612 | 0.00094 | -0.739 | 0 | 0.536 |
| 5944 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 12 | HMCN1 | Sponge network | -0.487 | 0.02157 | 0.809 | 0.00544 | 0.536 |
| 5945 | TPTEP1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 21 | SFRP1 | Sponge network | -1.216 | 0 | -3.566 | 0 | 0.536 |
| 5946 | MIR22HG |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-940 | 13 | CSRNP1 | Sponge network | -1.047 | 0 | -1.448 | 0 | 0.536 |
| 5947 | LINC00702 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ZNF208 | Sponge network | -0.737 | 0.06182 | -0.4 | 0.22381 | 0.536 |
| 5948 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | SOX5 | Sponge network | -1.582 | 8.0E-5 | -1.091 | 4.0E-5 | 0.536 |
| 5949 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | PAK3 | Sponge network | -0.769 | 0.00035 | -0.628 | 0.07253 | 0.536 |
| 5950 | TRAF3IP2-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | HERC1 | Sponge network | -0.323 | 0.01372 | -0.196 | 0.08544 | 0.536 |
| 5951 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 25 | DDX6 | Sponge network | 0.873 | 0 | 0.091 | 0.36428 | 0.536 |
| 5952 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p | 13 | PRUNE2 | Sponge network | -0.154 | 0.53954 | -1.733 | 0.00022 | 0.536 |
| 5953 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-671-5p | 29 | ADARB1 | Sponge network | -0.469 | 0.08721 | -0.335 | 0.06631 | 0.536 |
| 5954 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 38 | MCC | Sponge network | -0.942 | 0 | -1.005 | 0 | 0.536 |
| 5955 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p | 11 | EDA2R | Sponge network | -0.583 | 0.14589 | -0.587 | 0.01598 | 0.536 |
| 5956 | LINC00472 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-7-1-3p | 15 | SEMA3E | Sponge network | -0.842 | 0.01361 | -1.717 | 0.00137 | 0.536 |
| 5957 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | LRRC4 | Sponge network | -0.739 | 0.0574 | -1.774 | 0 | 0.536 |
| 5958 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | AKAP11 | Sponge network | 0.683 | 1.0E-5 | 0.196 | 0.09825 | 0.536 |
| 5959 | LINC01018 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | AKAP6 | Sponge network | -2.915 | 0.00015 | -1.586 | 3.0E-5 | 0.536 |
| 5960 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-590-3p | 13 | TRIM33 | Sponge network | 1.378 | 0 | 0.402 | 7.0E-5 | 0.536 |
| 5961 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 23 | BCL6 | Sponge network | -0.576 | 0.10121 | -0.211 | 0.19489 | 0.536 |
| 5962 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-877-5p | 10 | ANK2 | Sponge network | 0.043 | 0.81628 | -1.603 | 1.0E-5 | 0.536 |
| 5963 | MIR143HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 42 | ERC1 | Sponge network | -0.739 | 0.0574 | -0.108 | 0.38794 | 0.536 |
| 5964 | LINC01018 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SEMA3E | Sponge network | -2.915 | 0.00015 | -1.717 | 0.00137 | 0.536 |
| 5965 | RP11-244O19.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | RYR2 | Sponge network | -0.096 | 0.67958 | -1.466 | 0.00031 | 0.535 |
| 5966 | SNHG14 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | SYT4 | Sponge network | -1.111 | 0.0003 | -3.207 | 0 | 0.535 |
| 5967 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-7-5p | 20 | EPHA3 | Sponge network | 0.176 | 0.37402 | 0.185 | 0.53588 | 0.535 |
| 5968 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p | 11 | GAS1 | Sponge network | 0.997 | 0.00066 | -1.047 | 0.00364 | 0.535 |
| 5969 | CTD-3064M3.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-93-3p | 34 | GRIK3 | Sponge network | -0.469 | 0.08721 | -3.208 | 0 | 0.535 |
| 5970 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 35 | TCF4 | Sponge network | -1.582 | 8.0E-5 | 0.016 | 0.92271 | 0.535 |
| 5971 | RP11-166D19.1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 37 | RIMS2 | Sponge network | -0.576 | 0.10121 | -1.365 | 0.00208 | 0.535 |
| 5972 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 11 | CHD1 | Sponge network | 0.873 | 0 | 0.322 | 0.00215 | 0.535 |
| 5973 | PWAR6 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | RYR2 | Sponge network | -1.575 | 6.0E-5 | -1.466 | 0.00031 | 0.535 |
| 5974 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-940 | 31 | QKI | Sponge network | -0.154 | 0.53954 | -0.333 | 0.04111 | 0.535 |
| 5975 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | RECK | Sponge network | -1.161 | 0.00591 | -1.043 | 0 | 0.535 |
| 5976 | RP11-53O19.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p | 18 | SYNM | Sponge network | -0.405 | 0.22013 | -2.309 | 1.0E-5 | 0.535 |
| 5977 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 32 | GPC6 | Sponge network | 0.997 | 0.00066 | -0.054 | 0.81465 | 0.535 |
| 5978 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | DMTF1 | Sponge network | 1.153 | 0 | 0.248 | 0.02726 | 0.535 |
| 5979 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | SYNE1 | Sponge network | 0.4 | 0.14611 | -0.738 | 0.00316 | 0.535 |
| 5980 | CTC-444N24.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 1.153 | 0 | 0.943 | 0 | 0.535 |
| 5981 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 63 | IL6ST | Sponge network | -0.901 | 0.00019 | -0.402 | 0.00676 | 0.535 |
| 5982 | LINC00702 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | KCNA6 | Sponge network | -0.737 | 0.06182 | -1.531 | 0.00013 | 0.535 |
| 5983 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 57 | KIF1B | Sponge network | 0.083 | 0.66405 | -0.118 | 0.32258 | 0.535 |
| 5984 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-744-3p | 28 | FAT3 | Sponge network | -0.693 | 0.05629 | -1.601 | 0.00051 | 0.535 |
| 5985 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | GALNT13 | Sponge network | -0.976 | 0.00025 | -0.857 | 0.07439 | 0.535 |
| 5986 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-450b-5p | 16 | SCN9A | Sponge network | -1.092 | 4.0E-5 | -0.525 | 0.10593 | 0.535 |
| 5987 | LINC00641 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 20 | PRUNE2 | Sponge network | -0.222 | 0.32445 | -1.733 | 0.00022 | 0.535 |
| 5988 | LPP-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 71 | LPP | Sponge network | -0.18 | 0.20343 | -0.459 | 0.04475 | 0.535 |
| 5989 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-577;hsa-miR-590-3p | 13 | AKAP12 | Sponge network | -1.161 | 0.00591 | -0.932 | 0.0028 | 0.535 |
| 5990 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | SCN9A | Sponge network | -0.739 | 0.00044 | -0.525 | 0.10593 | 0.535 |
| 5991 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SON | Sponge network | 1.153 | 0 | 0.168 | 0.04406 | 0.535 |
| 5992 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-5p | 17 | IGF1 | Sponge network | -0.376 | 0.00337 | -0.204 | 0.5898 | 0.535 |
| 5993 | LINC00092 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-323a-3p;hsa-miR-590-3p | 11 | SLITRK3 | Sponge network | -1.886 | 0 | -3.328 | 0 | 0.535 |
| 5994 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | RNF144A | Sponge network | 0.997 | 0.00066 | 0.594 | 0.0009 | 0.535 |
| 5995 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | SYNE1 | Sponge network | -0.709 | 0.18168 | -0.738 | 0.00316 | 0.535 |
| 5996 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p | 10 | HMCN1 | Sponge network | -0.563 | 0.02545 | 0.809 | 0.00544 | 0.535 |
| 5997 | RP11-400K9.4 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 25 | IGFBP5 | Sponge network | -0.611 | 0.09607 | -0.806 | 0.00105 | 0.535 |
| 5998 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 11 | MCC | Sponge network | -2.039 | 0.03447 | -1.005 | 0 | 0.535 |
| 5999 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 42 | MEF2C | Sponge network | -0.842 | 0.01361 | -0.499 | 0.01448 | 0.535 |
| 6000 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CDH19 | Sponge network | 0.194 | 0.64278 | -3.434 | 0 | 0.535 |
| 6001 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-664a-3p;hsa-miR-769-5p | 12 | RCAN2 | Sponge network | -2.69 | 0.0001 | -0.33 | 0.15172 | 0.535 |
| 6002 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | CACNA2D1 | Sponge network | 0.101 | 0.60919 | -0.846 | 0.00675 | 0.535 |
| 6003 | RP11-488L18.10 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p | 12 | TBL1XR1 | Sponge network | 1.392 | 0 | 0.578 | 0 | 0.535 |
| 6004 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-485-3p;hsa-miR-542-3p | 13 | PIK3R1 | Sponge network | 0.043 | 0.81628 | -0.133 | 0.36854 | 0.535 |
| 6005 | RP11-48B3.4 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p | 26 | ABL2 | Sponge network | 1.757 | 0 | 0.82 | 0 | 0.535 |
| 6006 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-942-5p | 34 | HLF | Sponge network | -1.161 | 0.00591 | -2.443 | 0 | 0.535 |
| 6007 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | 0.532 | 0.00505 | -1.603 | 1.0E-5 | 0.535 |
| 6008 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p | 12 | RECK | Sponge network | -4.463 | 0.00025 | -1.043 | 0 | 0.535 |
| 6009 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | CDH19 | Sponge network | -0.611 | 0.09607 | -3.434 | 0 | 0.535 |
| 6010 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 10 | DCLK1 | Sponge network | -0.517 | 0.02935 | -1.324 | 0.00014 | 0.535 |
| 6011 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | MED12L | Sponge network | -0.513 | 0.04703 | -0.194 | 0.55299 | 0.535 |
| 6012 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p | 25 | EBF3 | Sponge network | -0.323 | 0.01372 | -0.058 | 0.81437 | 0.535 |
| 6013 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 30 | LIFR | Sponge network | -0.769 | 0.00035 | -1.794 | 0 | 0.535 |
| 6014 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-361-5p;hsa-miR-455-3p | 10 | PCDH9 | Sponge network | -0.309 | 0.1145 | -1.823 | 4.0E-5 | 0.535 |
| 6015 | LINC00702 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 15 | PRDM5 | Sponge network | -0.737 | 0.06182 | -1.09 | 0.00014 | 0.535 |
| 6016 | SH3RF3-AS1 |
hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | SYNE1 | Sponge network | 0.889 | 9.0E-5 | -0.738 | 0.00316 | 0.535 |
| 6017 | TPTEP1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LHFP | Sponge network | -1.216 | 0 | -0.167 | 0.41371 | 0.535 |
| 6018 | PCA3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 18 | GLI3 | Sponge network | -0.709 | 0.18168 | -0.615 | 0.01482 | 0.534 |
| 6019 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-3p | 21 | ZFHX4 | Sponge network | -0.376 | 0.00337 | 0.018 | 0.95574 | 0.534 |
| 6020 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 46 | DIP2C | Sponge network | -1.575 | 6.0E-5 | -0.436 | 0.01713 | 0.534 |
| 6021 | DNM3OS |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | MSN | Sponge network | 0.572 | 0.01722 | -0.023 | 0.88422 | 0.534 |
| 6022 | ARHGEF26-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 10 | PRKD1 | Sponge network | -2.46 | 0 | -0.466 | 0.03583 | 0.534 |
| 6023 | EMX2OS |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-624-5p | 11 | HMCN1 | Sponge network | -0.777 | 0.1134 | 0.809 | 0.00544 | 0.534 |
| 6024 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577 | 13 | NALCN | Sponge network | 0.249 | 0.35902 | 1.068 | 0.00237 | 0.534 |
| 6025 | RP11-13K12.5 |
hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | PEG3 | Sponge network | -0.318 | 0.65847 | -1.74 | 0 | 0.534 |
| 6026 | RP11-554A11.4 |
hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 17 | MYO9A | Sponge network | -0.583 | 0.14589 | -0.147 | 0.32373 | 0.534 |
| 6027 | FAM66C |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | TMEFF2 | Sponge network | -0.901 | 0.00019 | -2.426 | 0 | 0.534 |
| 6028 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-424-5p | 24 | PIK3R1 | Sponge network | -1.331 | 3.0E-5 | -0.133 | 0.36854 | 0.534 |
| 6029 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | EPHA7 | Sponge network | -0.376 | 0.00337 | -2.197 | 3.0E-5 | 0.534 |
| 6030 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-708-3p;hsa-miR-940 | 37 | HLF | Sponge network | -0.222 | 0.32445 | -2.443 | 0 | 0.534 |
| 6031 | LINC00854 |
hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-7-5p | 11 | ATRX | Sponge network | 1.18 | 0 | 0.261 | 0.04408 | 0.534 |
| 6032 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 19 | AKAP6 | Sponge network | 0.335 | 0.20596 | -1.586 | 3.0E-5 | 0.534 |
| 6033 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 27 | EPHA3 | Sponge network | 0.214 | 0.18246 | 0.185 | 0.53588 | 0.534 |
| 6034 | MRVI1-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-7-1-3p | 24 | RASSF2 | Sponge network | -1.092 | 4.0E-5 | -0.273 | 0.21961 | 0.534 |
| 6035 | RP11-326G21.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RASSF8 | Sponge network | -0.021 | 0.93598 | -0.122 | 0.65591 | 0.534 |
| 6036 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 14 | KCNAB1 | Sponge network | -0.487 | 0.02157 | -0.755 | 2.0E-5 | 0.534 |
| 6037 | LINC00092 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | PTGFR | Sponge network | -1.886 | 0 | -0.233 | 0.45421 | 0.534 |
| 6038 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 48 | PIK3R1 | Sponge network | -1.575 | 6.0E-5 | -0.133 | 0.36854 | 0.534 |
| 6039 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 31 | NRXN1 | Sponge network | -2.915 | 0.00015 | -3.778 | 0 | 0.534 |
| 6040 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 29 | NRXN1 | Sponge network | -0.611 | 0.09607 | -3.778 | 0 | 0.534 |
| 6041 | RP11-620J15.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-532-3p | 14 | SYNM | Sponge network | -0.739 | 0.00044 | -2.309 | 1.0E-5 | 0.534 |
| 6042 | ARHGEF26-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | TPO | Sponge network | -2.46 | 0 | -1.87 | 2.0E-5 | 0.534 |
| 6043 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | PIK3R1 | Sponge network | 0.178 | 0.29106 | -0.133 | 0.36854 | 0.534 |
| 6044 | LINC00957 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 20 | SYNPO2 | Sponge network | -0.421 | 0.0114 | -2.342 | 0 | 0.534 |
| 6045 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | SLITRK3 | Sponge network | -1.281 | 0.00012 | -3.328 | 0 | 0.534 |
| 6046 | FAM66C |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-532-3p | 16 | CADM3 | Sponge network | -0.901 | 0.00019 | -3.663 | 0 | 0.534 |
| 6047 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 29 | DOCK4 | Sponge network | -0.129 | 0.57177 | 0.502 | 0.00108 | 0.534 |
| 6048 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 20 | ARHGAP29 | Sponge network | 0.572 | 0.01722 | 0.131 | 0.42953 | 0.534 |
| 6049 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | LATS2 | Sponge network | 0.194 | 0.64278 | 0.159 | 0.2304 | 0.534 |
| 6050 | RP11-400K9.4 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p | 10 | CXCL12 | Sponge network | -0.611 | 0.09607 | -1.421 | 0 | 0.534 |
| 6051 | ADAMTS9-AS2 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | SYT4 | Sponge network | -2.452 | 0 | -3.207 | 0 | 0.534 |
| 6052 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NRXN3 | Sponge network | -0.021 | 0.93598 | -0.893 | 0.04186 | 0.534 |
| 6053 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 41 | MCC | Sponge network | -2.46 | 0 | -1.005 | 0 | 0.534 |
| 6054 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p | 12 | PRUNE2 | Sponge network | -0.469 | 0.08721 | -1.733 | 0.00022 | 0.534 |
| 6055 | ZNF582-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | PRKCB | Sponge network | -1.349 | 0.00017 | -1.313 | 2.0E-5 | 0.534 |
| 6056 | AC003090.1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p | 11 | AMPH | Sponge network | -2.117 | 0.00161 | -0.916 | 5.0E-5 | 0.534 |
| 6057 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 41 | TSHZ3 | Sponge network | -2.46 | 0 | -0.556 | 0.01516 | 0.534 |
| 6058 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | DKK3 | Sponge network | -1.111 | 0.0003 | -0.052 | 0.77783 | 0.534 |
| 6059 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DICER1 | Sponge network | 0.673 | 0 | 0.042 | 0.674 | 0.534 |
| 6060 | DNM3OS |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | SPG20 | Sponge network | 0.572 | 0.01722 | -1.16 | 0 | 0.534 |
| 6061 | CTD-2089N3.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-590-3p | 21 | FAT3 | Sponge network | -1.375 | 0.08642 | -1.601 | 0.00051 | 0.534 |
| 6062 | LINC00476 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935 | 18 | MAPK10 | Sponge network | -0.615 | 0.00015 | -1.774 | 0 | 0.534 |
| 6063 | RP11-473I1.9 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 40 | ARHGEF12 | Sponge network | 0.683 | 1.0E-5 | 0.09 | 0.35693 | 0.534 |
| 6064 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 38 | MCC | Sponge network | -0.777 | 0.1134 | -1.005 | 0 | 0.534 |
| 6065 | CTC-429P9.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p | 13 | BNC2 | Sponge network | 0.043 | 0.81628 | -0.491 | 0.09988 | 0.534 |
| 6066 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | TLL1 | Sponge network | -0.563 | 0.02545 | -1.195 | 3.0E-5 | 0.534 |
| 6067 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p | 37 | ZFHX3 | Sponge network | -0.096 | 0.67958 | 0.389 | 0.01363 | 0.534 |
| 6068 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-429 | 14 | PIK3C2A | Sponge network | 1.106 | 0 | 0.061 | 0.53776 | 0.534 |
| 6069 | RP11-736K20.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-942-5p | 10 | ZBTB20 | Sponge network | -0.358 | 0.17255 | -0.122 | 0.57569 | 0.534 |
| 6070 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p | 12 | LATS2 | Sponge network | 0.811 | 0.03228 | 0.159 | 0.2304 | 0.534 |
| 6071 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 26 | KIT | Sponge network | -0.737 | 0.06182 | -2.184 | 0 | 0.534 |
| 6072 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-935 | 17 | RARB | Sponge network | -1.331 | 3.0E-5 | -0.825 | 2.0E-5 | 0.534 |
| 6073 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | SON | Sponge network | 0.826 | 1.0E-5 | 0.168 | 0.04406 | 0.534 |
| 6074 | LINC00473 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | -1.203 | 0.03881 | -1.051 | 0.00022 | 0.534 |
| 6075 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 25 | NR2C2 | Sponge network | 0.349 | 0.01695 | 0.21 | 0.03224 | 0.534 |
| 6076 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-942-5p | 35 | UNC80 | Sponge network | -2.62 | 0 | -1.934 | 0 | 0.534 |
| 6077 | PART1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 17 | MAPK10 | Sponge network | -2.925 | 0 | -1.774 | 0 | 0.534 |
| 6078 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 69 | AFF4 | Sponge network | -0.222 | 0.32445 | -0.266 | 0.01565 | 0.534 |
| 6079 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -0.49 | 0.04946 | -2.417 | 0 | 0.534 |
| 6080 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-940 | 10 | MAPK10 | Sponge network | -0.154 | 0.53954 | -1.774 | 0 | 0.534 |
| 6081 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 54 | GAS7 | Sponge network | -0.777 | 0.1134 | -0.555 | 0.01602 | 0.533 |
| 6082 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 17 | RYR2 | Sponge network | -0.583 | 0.14589 | -1.466 | 0.00031 | 0.533 |
| 6083 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p | 20 | FGFR1 | Sponge network | -0.323 | 0.01372 | -0.509 | 0.03957 | 0.533 |
| 6084 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-590-3p | 15 | JAKMIP2 | Sponge network | -0.901 | 0.00019 | -1.388 | 0 | 0.533 |
| 6085 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-935 | 15 | LRRC7 | Sponge network | -2.62 | 0 | -1.341 | 0.01193 | 0.533 |
| 6086 | LINC00092 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 33 | RUNX1T1 | Sponge network | -1.886 | 0 | -0.344 | 0.2374 | 0.533 |
| 6087 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 41 | HLF | Sponge network | -0.612 | 0.00094 | -2.443 | 0 | 0.533 |
| 6088 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 39 | DTNA | Sponge network | -0.318 | 0.65847 | -1.648 | 1.0E-5 | 0.533 |
| 6089 | HCG18 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p | 15 | PHF6 | Sponge network | 1.063 | 0 | 0.785 | 0 | 0.533 |
| 6090 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | AFF3 | Sponge network | -0.323 | 0.01372 | -2.373 | 0 | 0.533 |
| 6091 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 55 | ADARB1 | Sponge network | -1.575 | 6.0E-5 | -0.335 | 0.06631 | 0.533 |
| 6092 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | KL | Sponge network | -2.46 | 0 | -1.294 | 0 | 0.533 |
| 6093 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 14 | PGR | Sponge network | -2.039 | 0.03447 | -1.704 | 0 | 0.533 |
| 6094 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-769-5p | 23 | SYNPO2 | Sponge network | 0.335 | 0.20596 | -2.342 | 0 | 0.533 |
| 6095 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-877-5p | 20 | GAS7 | Sponge network | 1.122 | 0.02989 | -0.555 | 0.01602 | 0.533 |
| 6096 | MIR497HG |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-96-5p | 12 | ELMO1 | Sponge network | -1.439 | 4.0E-5 | 0.04 | 0.83147 | 0.533 |
| 6097 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | KAT6B | Sponge network | -0.222 | 0.32445 | -0.478 | 0.00018 | 0.533 |
| 6098 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-629-3p | 13 | EBF3 | Sponge network | 0.811 | 0.03228 | -0.058 | 0.81437 | 0.533 |
| 6099 | RP11-574K11.29 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 29 | MDM4 | Sponge network | 0.997 | 0 | 0.345 | 0.00762 | 0.533 |
| 6100 | HAND2-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 24 | KLF10 | Sponge network | -1.697 | 0.00889 | -0.783 | 0 | 0.533 |
| 6101 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 17 | RCAN2 | Sponge network | 0.101 | 0.60919 | -0.33 | 0.15172 | 0.533 |
| 6102 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-664a-3p;hsa-miR-93-5p | 20 | CNTNAP3 | Sponge network | -0.976 | 0.00025 | -1.465 | 0.00033 | 0.533 |
| 6103 | MALAT1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RBM6 | Sponge network | 0.782 | 0.00016 | -0.137 | 0.15663 | 0.533 |
| 6104 | LINC01018 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 22 | PCDH10 | Sponge network | -2.915 | 0.00015 | -1.644 | 0.0078 | 0.533 |
| 6105 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 29 | ANK2 | Sponge network | -0.612 | 0.00094 | -1.603 | 1.0E-5 | 0.533 |
| 6106 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | PCDH10 | Sponge network | -0.612 | 0.00094 | -1.644 | 0.0078 | 0.533 |
| 6107 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 12 | AMPH | Sponge network | 0.997 | 0.00066 | -0.916 | 5.0E-5 | 0.533 |
| 6108 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-935 | 32 | FBXO32 | Sponge network | -0.303 | 0.21002 | -0.495 | 0.07561 | 0.533 |
| 6109 | RP11-473I1.9 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-590-3p | 12 | TPR | Sponge network | 0.683 | 1.0E-5 | 0.582 | 0 | 0.533 |
| 6110 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 19 | FGFR1 | Sponge network | -0.487 | 0.02157 | -0.509 | 0.03957 | 0.533 |
| 6111 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | AKAP12 | Sponge network | -0.612 | 0.00094 | -0.932 | 0.0028 | 0.533 |
| 6112 | AC009948.5 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 13 | DCLK1 | Sponge network | 0.532 | 0.00505 | -1.324 | 0.00014 | 0.533 |
| 6113 | FAM66C |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p | 17 | CDH23 | Sponge network | -0.901 | 0.00019 | -0.974 | 0.00034 | 0.533 |
| 6114 | CTC-296K1.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-940 | 35 | CADM2 | Sponge network | -2.095 | 0.00112 | -3.782 | 0 | 0.533 |
| 6115 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | RBMS3 | Sponge network | 0.299 | 0.57722 | -0.519 | 0.03551 | 0.533 |
| 6116 | LINC00473 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 13 | SFRP1 | Sponge network | -1.203 | 0.03881 | -3.566 | 0 | 0.533 |
| 6117 | RP11-166D19.1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 23 | RPS6KA6 | Sponge network | -0.576 | 0.10121 | -2.989 | 0 | 0.533 |
| 6118 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-539-5p | 31 | LPP | Sponge network | -0.309 | 0.1145 | -0.459 | 0.04475 | 0.533 |
| 6119 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 48 | ADARB1 | Sponge network | -0.709 | 0.18168 | -0.335 | 0.06631 | 0.533 |
| 6120 | CTA-363E19.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-625-5p | 22 | MDM4 | Sponge network | 1.055 | 0 | 0.345 | 0.00762 | 0.533 |
| 6121 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-940 | 22 | MCC | Sponge network | -2.095 | 0.00112 | -1.005 | 0 | 0.533 |
| 6122 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-93-3p | 50 | RUNX1T1 | Sponge network | -0.612 | 0.00094 | -0.344 | 0.2374 | 0.533 |
| 6123 | GNG12-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p | 12 | FLNA | Sponge network | -0.793 | 7.0E-5 | -0.771 | 0.01377 | 0.533 |
| 6124 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-628-5p | 20 | MEF2C | Sponge network | -0.376 | 0.00337 | -0.499 | 0.01448 | 0.533 |
| 6125 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p | 15 | RBMS3 | Sponge network | -2.076 | 0.00548 | -0.519 | 0.03551 | 0.533 |
| 6126 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | GLI3 | Sponge network | -0.323 | 0.01372 | -0.615 | 0.01482 | 0.533 |
| 6127 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | FOXP1 | Sponge network | -0.709 | 0.18168 | 0.042 | 0.74368 | 0.533 |
| 6128 | TPTEP1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 22 | RPS6KA6 | Sponge network | -1.216 | 0 | -2.989 | 0 | 0.533 |
| 6129 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-532-3p;hsa-miR-93-5p | 20 | PIK3R1 | Sponge network | 0.497 | 0.01451 | -0.133 | 0.36854 | 0.533 |
| 6130 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | CAV1 | Sponge network | -0.769 | 0.00035 | -1.013 | 6.0E-5 | 0.533 |
| 6131 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | JAZF1 | Sponge network | 0.532 | 0.00505 | -0.377 | 0.02677 | 0.533 |
| 6132 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | DMTF1 | Sponge network | 1.254 | 0 | 0.248 | 0.02726 | 0.533 |
| 6133 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | LRRC7 | Sponge network | -0.129 | 0.57177 | -1.341 | 0.01193 | 0.533 |
| 6134 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | LHFP | Sponge network | -0.096 | 0.67958 | -0.167 | 0.41371 | 0.533 |
| 6135 | CTC-378H22.2 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-501-5p;hsa-miR-744-3p | 13 | MEF2C | Sponge network | 0.001 | 0.99663 | -0.499 | 0.01448 | 0.533 |
| 6136 | NR2F1-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PCDH18 | Sponge network | -0.49 | 0.04946 | 0.216 | 0.24645 | 0.533 |
| 6137 | RP11-359B12.2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-532-3p | 19 | SYNM | Sponge network | 0.064 | 0.72934 | -2.309 | 1.0E-5 | 0.533 |
| 6138 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | ABCC9 | Sponge network | -0.769 | 0.00035 | -0.466 | 0.14121 | 0.533 |
| 6139 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 20 | ATRX | Sponge network | 0.817 | 3.0E-5 | 0.261 | 0.04408 | 0.533 |
| 6140 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 60 | IL6ST | Sponge network | 0.335 | 0.20596 | -0.402 | 0.00676 | 0.533 |
| 6141 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-7-1-3p | 26 | EPHA3 | Sponge network | -0.842 | 0.01361 | 0.185 | 0.53588 | 0.532 |
| 6142 | CTB-31O20.4 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-378c;hsa-miR-424-5p | 14 | ABL2 | Sponge network | 1.619 | 0 | 0.82 | 0 | 0.532 |
| 6143 | GNG12-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-3913-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 25 | WWTR1 | Sponge network | -0.793 | 7.0E-5 | -0.345 | 0.11997 | 0.532 |
| 6144 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 26 | PGR | Sponge network | -2.915 | 0.00015 | -1.704 | 0 | 0.532 |
| 6145 | MEG3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 31 | RNF144A | Sponge network | 0.249 | 0.35902 | 0.594 | 0.0009 | 0.532 |
| 6146 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 16 | EDA2R | Sponge network | -0.901 | 0.00019 | -0.587 | 0.01598 | 0.532 |
| 6147 | RP11-159D12.2 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p | 13 | STAG2 | Sponge network | 1.254 | 0 | 0.252 | 0.00438 | 0.532 |
| 6148 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-5p | 26 | RBMS3 | Sponge network | -0.811 | 2.0E-5 | -0.519 | 0.03551 | 0.532 |
| 6149 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-3p | 20 | MAFB | Sponge network | -0.576 | 0.10121 | -0.023 | 0.89865 | 0.532 |
| 6150 | RP11-356J5.12 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p | 21 | SYNM | Sponge network | -0.899 | 6.0E-5 | -2.309 | 1.0E-5 | 0.532 |
| 6151 | RP11-206L10.2 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3614-5p | 11 | ABL2 | Sponge network | 0.793 | 0 | 0.82 | 0 | 0.532 |
| 6152 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 33 | IL6ST | Sponge network | -1.886 | 0 | -0.402 | 0.00676 | 0.532 |
| 6153 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 13 | HMCN1 | Sponge network | 0.214 | 0.18246 | 0.809 | 0.00544 | 0.532 |
| 6154 | EMX2OS |
hsa-miR-146b-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 12 | FHL1 | Sponge network | -0.777 | 0.1134 | -2.687 | 0 | 0.532 |
| 6155 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3913-5p;hsa-miR-744-3p;hsa-miR-940 | 30 | HLF | Sponge network | -2.095 | 0.00112 | -2.443 | 0 | 0.532 |
| 6156 | LINC01018 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | ZDBF2 | Sponge network | -2.915 | 0.00015 | -0.998 | 0.00025 | 0.532 |
| 6157 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-96-5p | 12 | RECK | Sponge network | -0.517 | 0.02935 | -1.043 | 0 | 0.532 |
| 6158 | LINC00702 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | KLF10 | Sponge network | -0.737 | 0.06182 | -0.783 | 0 | 0.532 |
| 6159 | TRAF3IP2-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PRKCB | Sponge network | -0.323 | 0.01372 | -1.313 | 2.0E-5 | 0.532 |
| 6160 | LINC00702 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | MSN | Sponge network | -0.737 | 0.06182 | -0.023 | 0.88422 | 0.532 |
| 6161 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p | 36 | EPHA7 | Sponge network | -0.739 | 0.00044 | -2.197 | 3.0E-5 | 0.532 |
| 6162 | MAGI2-AS3 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | NOV | Sponge network | -0.129 | 0.57177 | -0.58 | 0.04676 | 0.532 |
| 6163 | LINC00702 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 19 | GLIPR1 | Sponge network | -0.737 | 0.06182 | 0.146 | 0.3276 | 0.532 |
| 6164 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-539-5p | 14 | PBX1 | Sponge network | -0.309 | 0.1145 | -1.206 | 0 | 0.532 |
| 6165 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-590-3p | 23 | CNTN1 | Sponge network | -0.896 | 0.00099 | -2.594 | 0 | 0.532 |
| 6166 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | MCC | Sponge network | -0.376 | 0.00337 | -1.005 | 0 | 0.532 |
| 6167 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p | 10 | SYNE1 | Sponge network | 1.344 | 0 | -0.738 | 0.00316 | 0.532 |
| 6168 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p | 31 | RBMS3 | Sponge network | 0.082 | 0.55397 | -0.519 | 0.03551 | 0.532 |
| 6169 | PWAR6 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 12 | NOV | Sponge network | -1.575 | 6.0E-5 | -0.58 | 0.04676 | 0.532 |
| 6170 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-935 | 31 | DMD | Sponge network | -0.384 | 0.40652 | -1.506 | 0 | 0.532 |
| 6171 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-3p;hsa-miR-96-5p | 11 | FNBP1 | Sponge network | 0.532 | 0.00505 | -0.969 | 4.0E-5 | 0.532 |
| 6172 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-93-5p | 24 | TSHZ3 | Sponge network | -1.654 | 0.00144 | -0.556 | 0.01516 | 0.532 |
| 6173 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | CACNA2D1 | Sponge network | 0.889 | 9.0E-5 | -0.846 | 0.00675 | 0.532 |
| 6174 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 31 | SETBP1 | Sponge network | -1.216 | 0 | -1.302 | 1.0E-5 | 0.532 |
| 6175 | RP11-166D19.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | ITGB1 | Sponge network | -0.576 | 0.10121 | 0.524 | 0.00017 | 0.532 |
| 6176 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-532-5p;hsa-miR-590-3p | 14 | PARK2 | Sponge network | -0.615 | 0.00015 | -1.892 | 0 | 0.532 |
| 6177 | RP11-554A11.4 |
hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-874-3p | 13 | CRTC3 | Sponge network | -0.583 | 0.14589 | 0.164 | 0.16786 | 0.532 |
| 6178 | RP11-400K9.4 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 14 | ITPKB | Sponge network | -0.611 | 0.09607 | -0.704 | 0.00106 | 0.532 |
| 6179 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p | 11 | FAT4 | Sponge network | 0.811 | 0.03228 | -0.354 | 0.14694 | 0.532 |
| 6180 | RP11-400K9.4 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-369-3p;hsa-miR-576-5p | 16 | KLF10 | Sponge network | -0.611 | 0.09607 | -0.783 | 0 | 0.532 |
| 6181 | PART1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 70 | CADM2 | Sponge network | -2.925 | 0 | -3.782 | 0 | 0.532 |
| 6182 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 38 | DIP2C | Sponge network | -0.222 | 0.32445 | -0.436 | 0.01713 | 0.532 |
| 6183 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.532 | 0.00505 | 0.943 | 0 | 0.532 |
| 6184 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | NAV3 | Sponge network | 0.299 | 0.57722 | 0.212 | 0.47729 | 0.532 |
| 6185 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-342-3p | 11 | ZNRF3 | Sponge network | 1.294 | 0 | 0.637 | 0.00172 | 0.532 |
| 6186 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | MOB1B | Sponge network | 0.225 | 0.30644 | 0.197 | 0.15266 | 0.532 |
| 6187 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | CNTNAP1 | Sponge network | -0.246 | 0.35034 | 0.358 | 0.13727 | 0.532 |
| 6188 | USP3-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p | 12 | TPO | Sponge network | -0.162 | 0.47687 | -1.87 | 2.0E-5 | 0.532 |
| 6189 | MAGI2-AS3 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | ID4 | Sponge network | -0.129 | 0.57177 | -1.644 | 0 | 0.532 |
| 6190 | TRAF3IP2-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-92b-3p;hsa-miR-940 | 22 | BAZ2B | Sponge network | -0.323 | 0.01372 | -0.276 | 0.02833 | 0.532 |
| 6191 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 12 | CNTNAP1 | Sponge network | -1.203 | 0.03881 | 0.358 | 0.13727 | 0.532 |
| 6192 | BVES-AS1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-628-5p | 23 | RIMS2 | Sponge network | -2.62 | 0 | -1.365 | 0.00208 | 0.532 |
| 6193 | SNHG14 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 34 | LRIG1 | Sponge network | -1.111 | 0.0003 | -0.537 | 0.00588 | 0.532 |
| 6194 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 12 | AKAP12 | Sponge network | -0.421 | 0.0114 | -0.932 | 0.0028 | 0.532 |
| 6195 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-589-3p;hsa-miR-769-5p | 19 | SYNPO2 | Sponge network | -0.777 | 0.16667 | -2.342 | 0 | 0.532 |
| 6196 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | DMD | Sponge network | 0.178 | 0.29106 | -1.506 | 0 | 0.532 |
| 6197 | FGF14-AS2 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-874-3p | 16 | GRIK3 | Sponge network | -1.582 | 8.0E-5 | -3.208 | 0 | 0.532 |
| 6198 | RP11-798M19.6 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 23 | RYR2 | Sponge network | -0.612 | 0.00094 | -1.466 | 0.00031 | 0.532 |
| 6199 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p | 10 | SETBP1 | Sponge network | 0.178 | 0.29106 | -1.302 | 1.0E-5 | 0.531 |
| 6200 | RP11-819C21.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | PIK3CA | Sponge network | 0.537 | 0.00139 | 0.195 | 0.06908 | 0.531 |
| 6201 | SNHG14 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 22 | STARD13 | Sponge network | -1.111 | 0.0003 | -0.187 | 0.27383 | 0.531 |
| 6202 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-532-3p | 23 | PBX1 | Sponge network | -2.531 | 0 | -1.206 | 0 | 0.531 |
| 6203 | DNM3OS |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 20 | SFRP1 | Sponge network | 0.572 | 0.01722 | -3.566 | 0 | 0.531 |
| 6204 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 46 | DTNA | Sponge network | -0.611 | 0.09607 | -1.648 | 1.0E-5 | 0.531 |
| 6205 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | -0.513 | 0.04703 | -1.751 | 1.0E-5 | 0.531 |
| 6206 | FAM66C |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | CAV1 | Sponge network | -0.901 | 0.00019 | -1.013 | 6.0E-5 | 0.531 |
| 6207 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | EPHA3 | Sponge network | 0.342 | 0.03129 | 0.185 | 0.53588 | 0.531 |
| 6208 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | NF1 | Sponge network | 0.826 | 1.0E-5 | 0.51 | 0 | 0.531 |
| 6209 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p | 12 | GREM1 | Sponge network | -0.932 | 0.00329 | 0.902 | 0.01691 | 0.531 |
| 6210 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-23b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 10 | BCLAF1 | Sponge network | 1.923 | 0 | 0.211 | 0.00684 | 0.531 |
| 6211 | SNHG1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p | 15 | CBFB | Sponge network | 1.265 | 0 | 1.061 | 0 | 0.531 |
| 6212 | TPTEP1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 23 | PRUNE2 | Sponge network | -1.216 | 0 | -1.733 | 0.00022 | 0.531 |
| 6213 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-98-5p | 10 | ADAMTS8 | Sponge network | -2.46 | 0 | -1.476 | 0.00069 | 0.531 |
| 6214 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-98-5p | 64 | PRKAA2 | Sponge network | -1.697 | 0.00889 | -2.462 | 0 | 0.531 |
| 6215 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 22 | AKAP6 | Sponge network | 0.056 | 0.81195 | -1.586 | 3.0E-5 | 0.531 |
| 6216 | LINC00654 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | PREX2 | Sponge network | 0.374 | 0.20054 | -0.272 | 0.25382 | 0.531 |
| 6217 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-654-3p | 24 | RICTOR | Sponge network | 0.997 | 0 | 0.388 | 0.00188 | 0.531 |
| 6218 | TRAF3IP2-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-942-5p | 11 | BMPR1A | Sponge network | -0.323 | 0.01372 | -0.366 | 0.01036 | 0.531 |
| 6219 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | NCAM2 | Sponge network | -2.452 | 0 | -0.482 | 0.22565 | 0.531 |
| 6220 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DCN | Sponge network | -1.575 | 6.0E-5 | -1.288 | 0 | 0.531 |
| 6221 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 14 | RCAN2 | Sponge network | -0.303 | 0.21002 | -0.33 | 0.15172 | 0.531 |
| 6222 | CKMT2-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 12 | STAT5B | Sponge network | 0.082 | 0.55654 | -0.182 | 0.1082 | 0.531 |
| 6223 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-93-5p | 47 | EPHA5 | Sponge network | -0.162 | 0.47687 | -0.957 | 0.01733 | 0.531 |
| 6224 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | MITF | Sponge network | -1.575 | 6.0E-5 | -0.769 | 0.00022 | 0.531 |
| 6225 | RP11-33B1.4 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 12 | RICTOR | Sponge network | 0.826 | 0 | 0.388 | 0.00188 | 0.531 |
| 6226 | RP11-517B11.7 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 13 | GSK3B | Sponge network | 0.706 | 0 | 0.266 | 0.00045 | 0.531 |
| 6227 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | MITF | Sponge network | -0.612 | 0.00094 | -0.769 | 0.00022 | 0.531 |
| 6228 | PART1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 14 | RIMS1 | Sponge network | -2.925 | 0 | -3.143 | 0 | 0.531 |
| 6229 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-628-5p | 47 | TCF4 | Sponge network | -2.62 | 0 | 0.016 | 0.92271 | 0.531 |
| 6230 | DNM3OS |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 13 | STARD13 | Sponge network | 0.572 | 0.01722 | -0.187 | 0.27383 | 0.531 |
| 6231 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p | 15 | SETBP1 | Sponge network | 0.811 | 0.03228 | -1.302 | 1.0E-5 | 0.531 |
| 6232 | LINC00843 |
hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-495-3p | 14 | CREB1 | Sponge network | 1.106 | 0 | 0.203 | 0.00537 | 0.531 |
| 6233 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p | 12 | CACNA2D1 | Sponge network | 0.811 | 0.03228 | -0.846 | 0.00675 | 0.531 |
| 6234 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | ABCB5 | Sponge network | -0.777 | 0.1134 | -0.956 | 0.04077 | 0.531 |
| 6235 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | EPHA7 | Sponge network | -0.611 | 0.09607 | -2.197 | 3.0E-5 | 0.531 |
| 6236 | AC003090.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-93-5p | 14 | EZH1 | Sponge network | -2.117 | 0.00161 | -0.564 | 1.0E-5 | 0.531 |
| 6237 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-98-5p | 13 | PRRX1 | Sponge network | 1.122 | 0.02989 | 1.316 | 0 | 0.531 |
| 6238 | MYLK-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-935 | 21 | ABI2 | Sponge network | -1.331 | 3.0E-5 | 0.175 | 0.14787 | 0.531 |
| 6239 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | ABCC9 | Sponge network | -0.222 | 0.32445 | -0.466 | 0.14121 | 0.531 |
| 6240 | WDFY3-AS2 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 20 | SFRP1 | Sponge network | -0.942 | 0 | -3.566 | 0 | 0.531 |
| 6241 | RP1-59D14.5 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p;hsa-miR-629-3p | 10 | CHD2 | Sponge network | 1.162 | 5.0E-5 | 0.049 | 0.55371 | 0.531 |
| 6242 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | SCN9A | Sponge network | -0.358 | 0.17255 | -0.525 | 0.10593 | 0.531 |
| 6243 | RP11-53O19.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-502-5p | 27 | IL17RD | Sponge network | -0.405 | 0.22013 | -0.019 | 0.94873 | 0.531 |
| 6244 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-33a-3p | 12 | RBMS3 | Sponge network | -1.255 | 0.00981 | -0.519 | 0.03551 | 0.531 |
| 6245 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 18 | ST6GAL2 | Sponge network | -1.092 | 4.0E-5 | -1.201 | 0.00597 | 0.531 |
| 6246 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 32 | RASSF8 | Sponge network | -1.249 | 7.0E-5 | -0.122 | 0.65591 | 0.531 |
| 6247 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 20 | RELN | Sponge network | -2.46 | 0 | -2.403 | 0 | 0.531 |
| 6248 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-93-5p | 14 | RASSF2 | Sponge network | -0.295 | 0.02604 | -0.273 | 0.21961 | 0.531 |
| 6249 | RP11-473I1.9 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p | 34 | ABL2 | Sponge network | 0.683 | 1.0E-5 | 0.82 | 0 | 0.531 |
| 6250 | CTD-3092A11.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | 0.873 | 0 | 0.142 | 0.04938 | 0.531 |
| 6251 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 14 | PHF6 | Sponge network | 1.153 | 0 | 0.785 | 0 | 0.531 |
| 6252 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 31 | GPC6 | Sponge network | -0.739 | 0.0574 | -0.054 | 0.81465 | 0.531 |
| 6253 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 39 | ESR1 | Sponge network | -0.737 | 0.06182 | -1.035 | 3.0E-5 | 0.531 |
| 6254 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 31 | PCDH9 | Sponge network | -0.487 | 0.02157 | -1.823 | 4.0E-5 | 0.531 |
| 6255 | FAM66C |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | TLL1 | Sponge network | -0.901 | 0.00019 | -1.195 | 3.0E-5 | 0.531 |
| 6256 | RP11-244O19.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | PDE4DIP | Sponge network | -0.096 | 0.67958 | -0.282 | 0.0257 | 0.531 |
| 6257 | RP11-307B6.3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-93-3p | 14 | GRIK3 | Sponge network | -4.463 | 0.00025 | -3.208 | 0 | 0.531 |
| 6258 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 20 | DKK3 | Sponge network | -0.563 | 0.02545 | -0.052 | 0.77783 | 0.531 |
| 6259 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 28 | SYNPO2 | Sponge network | -0.899 | 6.0E-5 | -2.342 | 0 | 0.531 |
| 6260 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ATRX | Sponge network | 0.873 | 0 | 0.261 | 0.04408 | 0.531 |
| 6261 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 52 | ZFHX4 | Sponge network | -2.46 | 0 | 0.018 | 0.95574 | 0.531 |
| 6262 | RP11-822E23.8 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p | 11 | AMPH | Sponge network | -0.777 | 0.16667 | -0.916 | 5.0E-5 | 0.531 |
| 6263 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-589-3p | 11 | RELN | Sponge network | -2.531 | 0 | -2.403 | 0 | 0.531 |
| 6264 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-576-5p | 12 | GREM1 | Sponge network | 0.299 | 0.57722 | 0.902 | 0.01691 | 0.531 |
| 6265 | LINC00702 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 26 | ABL1 | Sponge network | -0.737 | 0.06182 | -0.215 | 0.07804 | 0.531 |
| 6266 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 26 | IGF1 | Sponge network | -0.896 | 0.00099 | -0.204 | 0.5898 | 0.53 |
| 6267 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | DACT1 | Sponge network | 0.335 | 0.20596 | 0.783 | 0.00072 | 0.53 |
| 6268 | RP5-1198O20.4 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429 | 15 | NGFR | Sponge network | 0.997 | 0.00066 | -1.271 | 0.01058 | 0.53 |
| 6269 | SNHG14 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 17 | SEPT6 | Sponge network | -1.111 | 0.0003 | -0.598 | 0.00181 | 0.53 |
| 6270 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-93-5p | 26 | HBP1 | Sponge network | -1.645 | 0 | -0.454 | 0 | 0.53 |
| 6271 | RP4-613B23.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-378a-5p | 10 | PTGFR | Sponge network | -0.309 | 0.1145 | -0.233 | 0.45421 | 0.53 |
| 6272 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p | 13 | TRIM33 | Sponge network | 1.055 | 0 | 0.402 | 7.0E-5 | 0.53 |
| 6273 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b | 12 | TAL1 | Sponge network | -0.583 | 0.14589 | -0.462 | 0.00574 | 0.53 |
| 6274 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SLC6A15 | Sponge network | -2.46 | 0 | -2.373 | 2.0E-5 | 0.53 |
| 6275 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | AFF3 | Sponge network | -0.901 | 0.00019 | -2.373 | 0 | 0.53 |
| 6276 | FZD10-AS1 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p | 10 | DIRAS1 | Sponge network | -0.727 | 0.04726 | -2.518 | 0 | 0.53 |
| 6277 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p | 17 | DCLK1 | Sponge network | -1.281 | 0.00012 | -1.324 | 0.00014 | 0.53 |
| 6278 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 11 | UBE2QL1 | Sponge network | -0.563 | 0.02545 | -2.417 | 0 | 0.53 |
| 6279 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | CDH19 | Sponge network | -0.739 | 0.00044 | -3.434 | 0 | 0.53 |
| 6280 | CTC-296K1.3 |
hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-3913-5p;hsa-miR-582-5p | 10 | WWTR1 | Sponge network | -2.095 | 0.00112 | -0.345 | 0.11997 | 0.53 |
| 6281 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 37 | PBX1 | Sponge network | -0.612 | 0.00094 | -1.206 | 0 | 0.53 |
| 6282 | RP5-1198O20.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-744-3p | 28 | PTPN14 | Sponge network | 0.997 | 0.00066 | 0.144 | 0.34595 | 0.53 |
| 6283 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 36 | DIP2C | Sponge network | 0.101 | 0.60919 | -0.436 | 0.01713 | 0.53 |
| 6284 | TPTEP1 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | HSPB7 | Sponge network | -1.216 | 0 | -2.268 | 1.0E-5 | 0.53 |
| 6285 | RP11-890B15.3 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | RNF180 | Sponge network | 0.101 | 0.60919 | -1.127 | 1.0E-5 | 0.53 |
| 6286 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 45 | ABI2 | Sponge network | -0.49 | 0.04946 | 0.175 | 0.14787 | 0.53 |
| 6287 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-628-5p | 24 | EPHA3 | Sponge network | -2.62 | 0 | 0.185 | 0.53588 | 0.53 |
| 6288 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-532-5p;hsa-miR-93-5p | 14 | FGFR1 | Sponge network | -0.384 | 0.40652 | -0.509 | 0.03957 | 0.53 |
| 6289 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429 | 21 | SCN9A | Sponge network | -2.62 | 0 | -0.525 | 0.10593 | 0.53 |
| 6290 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-7-1-3p | 18 | AKAP6 | Sponge network | -0.318 | 0.65847 | -1.586 | 3.0E-5 | 0.53 |
| 6291 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-7-5p;hsa-miR-93-5p | 11 | GLI3 | Sponge network | -1.654 | 0.00144 | -0.615 | 0.01482 | 0.53 |
| 6292 | DNM3OS |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | TAL1 | Sponge network | 0.572 | 0.01722 | -0.462 | 0.00574 | 0.53 |
| 6293 | RP11-347P5.1 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-940 | 14 | MDM4 | Sponge network | 1.364 | 0 | 0.345 | 0.00762 | 0.53 |
| 6294 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 63 | BMPR2 | Sponge network | 0.572 | 0.01722 | 0.206 | 0.0635 | 0.53 |
| 6295 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 45 | HLF | Sponge network | -0.49 | 0.04946 | -2.443 | 0 | 0.53 |
| 6296 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | PDGFRA | Sponge network | -0.976 | 0.00025 | -0.372 | 0.0037 | 0.53 |
| 6297 | RP4-613B23.1 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-486-5p | 11 | EPHA3 | Sponge network | -0.309 | 0.1145 | 0.185 | 0.53588 | 0.53 |
| 6298 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 36 | NRXN1 | Sponge network | 0.194 | 0.64278 | -3.778 | 0 | 0.53 |
| 6299 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | -0.842 | 0.01361 | -1.465 | 0.00033 | 0.53 |
| 6300 | RP11-701H24.3 |
hsa-let-7g-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 10 | EBF1 | Sponge network | -1.425 | 0.04204 | -0.384 | 0.08856 | 0.53 |
| 6301 | SNHG14 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 53 | NTRK2 | Sponge network | -1.111 | 0.0003 | -0.736 | 0.06495 | 0.53 |
| 6302 | FAM66C |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 29 | PCDH10 | Sponge network | -0.901 | 0.00019 | -1.644 | 0.0078 | 0.53 |
| 6303 | EMX2OS |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | WWTR1 | Sponge network | -0.777 | 0.1134 | -0.345 | 0.11997 | 0.53 |
| 6304 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | JAZF1 | Sponge network | -1.582 | 8.0E-5 | -0.377 | 0.02677 | 0.53 |
| 6305 | USP3-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p | 10 | HERC1 | Sponge network | -0.162 | 0.47687 | -0.196 | 0.08544 | 0.53 |
| 6306 | RP11-244O19.1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | CSDE1 | Sponge network | -0.096 | 0.67958 | -0.493 | 0 | 0.53 |
| 6307 | AC138035.2 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p | 16 | ABL2 | Sponge network | 1.073 | 0 | 0.82 | 0 | 0.53 |
| 6308 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | HECA | Sponge network | -0.222 | 0.32445 | -0.217 | 0.03463 | 0.53 |
| 6309 | RP11-999E24.3 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-30d-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-3p | 10 | PRUNE2 | Sponge network | -0.693 | 0.05629 | -1.733 | 0.00022 | 0.53 |
| 6310 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 44 | GAS7 | Sponge network | -0.421 | 0.0114 | -0.555 | 0.01602 | 0.53 |
| 6311 | TP73-AS1 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-874-3p | 10 | HSPB7 | Sponge network | -0.563 | 0.02545 | -2.268 | 1.0E-5 | 0.53 |
| 6312 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 30 | SSBP2 | Sponge network | -0.358 | 0.17255 | -1.58 | 0 | 0.53 |
| 6313 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | JAKMIP2 | Sponge network | 0.572 | 0.01722 | -1.388 | 0 | 0.53 |
| 6314 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 23 | BNC2 | Sponge network | -1.886 | 0 | -0.491 | 0.09988 | 0.53 |
| 6315 | RP3-430N8.11 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-378a-3p;hsa-miR-378c | 11 | ABL2 | Sponge network | 0.973 | 0 | 0.82 | 0 | 0.53 |
| 6316 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 27 | ABL2 | Sponge network | 0.559 | 0.00026 | 0.82 | 0 | 0.53 |
| 6317 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p | 10 | GAS1 | Sponge network | 0.194 | 0.64278 | -1.047 | 0.00364 | 0.53 |
| 6318 | GABPB1-AS1 |
hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-940 | 14 | TAF1 | Sponge network | 1.11 | 0 | 0.39 | 3.0E-5 | 0.53 |
| 6319 | AP001347.6 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-493-5p | 14 | SYNM | Sponge network | -1.161 | 0.00591 | -2.309 | 1.0E-5 | 0.53 |
| 6320 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-542-3p | 13 | RUNX1T1 | Sponge network | 0.043 | 0.81628 | -0.344 | 0.2374 | 0.53 |
| 6321 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92a-1-5p | 23 | AKAP6 | Sponge network | 0.214 | 0.18246 | -1.586 | 3.0E-5 | 0.53 |
| 6322 | ZNF667-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-629-3p | 19 | RYR2 | Sponge network | -0.976 | 0.00025 | -1.466 | 0.00031 | 0.53 |
| 6323 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-576-5p | 10 | ZNF208 | Sponge network | -2.62 | 0 | -0.4 | 0.22381 | 0.53 |
| 6324 | RP11-927P21.1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-940 | 38 | MDM4 | Sponge network | 0.921 | 0.00118 | 0.345 | 0.00762 | 0.53 |
| 6325 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | BNC2 | Sponge network | -0.563 | 0.02545 | -0.491 | 0.09988 | 0.53 |
| 6326 | RP11-589P10.7 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-671-5p;hsa-miR-93-3p | 11 | GRIK3 | Sponge network | -2.044 | 0.00066 | -3.208 | 0 | 0.53 |
| 6327 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-629-5p | 25 | IGF1 | Sponge network | -2.69 | 0.0001 | -0.204 | 0.5898 | 0.53 |
| 6328 | LINC00641 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-584-5p;hsa-miR-589-5p | 11 | BMPR1A | Sponge network | -0.222 | 0.32445 | -0.366 | 0.01036 | 0.53 |
| 6329 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 46 | ZFHX3 | Sponge network | -0.222 | 0.32445 | 0.389 | 0.01363 | 0.53 |
| 6330 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429 | 16 | DYNC1I1 | Sponge network | -0.246 | 0.35034 | -1.12 | 0.00107 | 0.53 |
| 6331 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | -2.117 | 0.00161 | 0.809 | 0.00544 | 0.53 |
| 6332 | RP5-1198O20.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 39 | IGFBP5 | Sponge network | 0.997 | 0.00066 | -0.806 | 0.00105 | 0.53 |
| 6333 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 42 | RBMS3 | Sponge network | -1.249 | 7.0E-5 | -0.519 | 0.03551 | 0.53 |
| 6334 | RP11-589P10.7 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-671-5p | 11 | CADM3 | Sponge network | -2.044 | 0.00066 | -3.663 | 0 | 0.53 |
| 6335 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CLIP1 | Sponge network | -0.737 | 0.06182 | -0.215 | 0.08684 | 0.53 |
| 6336 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 34 | FBXO32 | Sponge network | -2.117 | 0.00161 | -0.495 | 0.07561 | 0.53 |
| 6337 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374a-5p | 15 | AKAP6 | Sponge network | -0.693 | 0.05629 | -1.586 | 3.0E-5 | 0.53 |
| 6338 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 16 | DAB2 | Sponge network | 0.249 | 0.35902 | 0.431 | 0.0113 | 0.529 |
| 6339 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-539-5p | 12 | PDZD2 | Sponge network | -0.793 | 7.0E-5 | -0.909 | 3.0E-5 | 0.529 |
| 6340 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 28 | BCL6 | Sponge network | -0.129 | 0.57177 | -0.211 | 0.19489 | 0.529 |
| 6341 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | ABI2 | Sponge network | 0.335 | 0.20596 | 0.175 | 0.14787 | 0.529 |
| 6342 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PLSCR4 | Sponge network | -1.161 | 0.00591 | -0.474 | 0.03833 | 0.529 |
| 6343 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 18 | PTPN11 | Sponge network | 0.537 | 0.00139 | 0.431 | 2.0E-5 | 0.529 |
| 6344 | PWAR6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | ABCA10 | Sponge network | -1.575 | 6.0E-5 | -0.571 | 0.03674 | 0.529 |
| 6345 | MRVI1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-455-3p | 16 | MAF | Sponge network | -1.092 | 4.0E-5 | -1.257 | 0 | 0.529 |
| 6346 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | SYNE1 | Sponge network | 0.178 | 0.29106 | -0.738 | 0.00316 | 0.529 |
| 6347 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | NALCN | Sponge network | -0.777 | 0.1134 | 1.068 | 0.00237 | 0.529 |
| 6348 | RP11-156P1.3 |
hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | HERC1 | Sponge network | 0.214 | 0.18246 | -0.196 | 0.08544 | 0.529 |
| 6349 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | AFF3 | Sponge network | 0.4 | 0.14611 | -2.373 | 0 | 0.529 |
| 6350 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 20 | TBL1XR1 | Sponge network | 0.826 | 1.0E-5 | 0.578 | 0 | 0.529 |
| 6351 | RP11-13K12.5 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p | 15 | MRVI1 | Sponge network | -0.318 | 0.65847 | -1.051 | 0.00022 | 0.529 |
| 6352 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p | 43 | CHD5 | Sponge network | -2.69 | 0.0001 | -0.922 | 0.01521 | 0.529 |
| 6353 | AC016747.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 36 | GAS7 | Sponge network | 0.199 | 0.38015 | -0.555 | 0.01602 | 0.529 |
| 6354 | RP11-400K9.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p | 66 | LPP | Sponge network | -0.611 | 0.09607 | -0.459 | 0.04475 | 0.529 |
| 6355 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | AKAP12 | Sponge network | 0.176 | 0.37402 | -0.932 | 0.0028 | 0.529 |
| 6356 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | 0.335 | 0.20596 | -1.09 | 0.00014 | 0.529 |
| 6357 | RP4-639F20.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 18 | ZFHX3 | Sponge network | -0.517 | 0.02935 | 0.389 | 0.01363 | 0.529 |
| 6358 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p | 19 | DCLK1 | Sponge network | -0.942 | 0 | -1.324 | 0.00014 | 0.529 |
| 6359 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-874-3p | 16 | NR2C2 | Sponge network | 0.556 | 0.00298 | 0.21 | 0.03224 | 0.529 |
| 6360 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-628-5p | 21 | GRIA3 | Sponge network | -2.62 | 0 | -1.956 | 0 | 0.529 |
| 6361 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | FGFR1 | Sponge network | -0.513 | 0.04703 | -0.509 | 0.03957 | 0.529 |
| 6362 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-590-3p | 30 | CADM2 | Sponge network | -0.358 | 0.17255 | -3.782 | 0 | 0.529 |
| 6363 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-96-5p | 13 | FNBP1 | Sponge network | 0.997 | 0.00066 | -0.969 | 4.0E-5 | 0.529 |
| 6364 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-361-5p;hsa-miR-424-5p | 11 | RELN | Sponge network | -1.349 | 0.00017 | -2.403 | 0 | 0.529 |
| 6365 | LINC00641 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 32 | RASSF8 | Sponge network | -0.222 | 0.32445 | -0.122 | 0.65591 | 0.529 |
| 6366 | TP73-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-935 | 18 | RELN | Sponge network | -0.563 | 0.02545 | -2.403 | 0 | 0.529 |
| 6367 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-582-5p | 14 | RELN | Sponge network | -2.69 | 0.0001 | -2.403 | 0 | 0.529 |
| 6368 | LINC00702 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | -0.737 | 0.06182 | -3.143 | 0 | 0.529 |
| 6369 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | SOX5 | Sponge network | -2.117 | 0.00161 | -1.091 | 4.0E-5 | 0.529 |
| 6370 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p | 22 | SOX5 | Sponge network | -0.469 | 0.08721 | -1.091 | 4.0E-5 | 0.529 |
| 6371 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-451a;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 50 | BCL2 | Sponge network | -0.739 | 0.0574 | -0.899 | 0 | 0.529 |
| 6372 | EMX2OS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 60 | DTNA | Sponge network | -0.777 | 0.1134 | -1.648 | 1.0E-5 | 0.529 |
| 6373 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | MDM4 | Sponge network | 0.225 | 0.30644 | 0.345 | 0.00762 | 0.529 |
| 6374 | ARHGEF26-AS1 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ADH1B | Sponge network | -2.46 | 0 | -3.716 | 0 | 0.529 |
| 6375 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p | 17 | FBXO32 | Sponge network | -0.473 | 0.07602 | -0.495 | 0.07561 | 0.529 |
| 6376 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p | 14 | CNTN1 | Sponge network | -0.376 | 0.00337 | -2.594 | 0 | 0.529 |
| 6377 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-590-3p | 19 | SPG20 | Sponge network | -0.793 | 7.0E-5 | -1.16 | 0 | 0.529 |
| 6378 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 52 | PAFAH1B1 | Sponge network | -0.222 | 0.32445 | -0.429 | 0 | 0.529 |
| 6379 | WDFY3-AS2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 19 | MAPK10 | Sponge network | -0.942 | 0 | -1.774 | 0 | 0.529 |
| 6380 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 18 | GLIPR1 | Sponge network | -0.323 | 0.01372 | 0.146 | 0.3276 | 0.529 |
| 6381 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-589-5p;hsa-miR-590-3p | 18 | SLITRK3 | Sponge network | -0.487 | 0.02157 | -3.328 | 0 | 0.529 |
| 6382 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 60 | FAT3 | Sponge network | 0.997 | 0.00066 | -1.601 | 0.00051 | 0.529 |
| 6383 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484 | 21 | ABL1 | Sponge network | 0.083 | 0.66405 | -0.215 | 0.07804 | 0.529 |
| 6384 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p | 10 | AKAP12 | Sponge network | -0.246 | 0.35034 | -0.932 | 0.0028 | 0.529 |
| 6385 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-429 | 14 | IGSF10 | Sponge network | -2.62 | 0 | -1.751 | 1.0E-5 | 0.529 |
| 6386 | MIR143HG |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 23 | RPS6KA2 | Sponge network | -0.739 | 0.0574 | -0.57 | 0.00013 | 0.529 |
| 6387 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-485-3p;hsa-miR-590-3p | 21 | MLLT10 | Sponge network | 1.254 | 0 | 0.105 | 0.33292 | 0.529 |
| 6388 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 56 | QKI | Sponge network | -0.323 | 0.01372 | -0.333 | 0.04111 | 0.529 |
| 6389 | AC009948.5 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 13 | MN1 | Sponge network | 0.532 | 0.00505 | -0.358 | 0.24172 | 0.529 |
| 6390 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429 | 10 | RCAN2 | Sponge network | -0.154 | 0.53954 | -0.33 | 0.15172 | 0.529 |
| 6391 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 26 | GRIN2A | Sponge network | -0.096 | 0.67958 | -2.161 | 0 | 0.529 |
| 6392 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | 0.997 | 0.00066 | 0.024 | 0.89889 | 0.529 |
| 6393 | RP11-819C21.1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p | 12 | MACF1 | Sponge network | 0.537 | 0.00139 | 0.019 | 0.90335 | 0.529 |
| 6394 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-374a-5p;hsa-miR-744-3p | 13 | ADAMTSL3 | Sponge network | -0.693 | 0.05629 | -1.559 | 0 | 0.529 |
| 6395 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 22 | RET | Sponge network | -0.49 | 0.04946 | -0.833 | 0.03517 | 0.529 |
| 6396 | DNM3OS |
hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3065-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-664a-3p | 11 | LOX | Sponge network | 0.572 | 0.01722 | 1.164 | 0 | 0.529 |
| 6397 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p | 10 | ITGB3 | Sponge network | -0.896 | 0.00099 | -0.011 | 0.95751 | 0.529 |
| 6398 | CTC-296K1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-671-5p | 13 | FAT4 | Sponge network | -2.095 | 0.00112 | -0.354 | 0.14694 | 0.529 |
| 6399 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-93-5p | 13 | AHNAK | Sponge network | -1.047 | 0 | -1.207 | 0 | 0.529 |
| 6400 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | PRRX1 | Sponge network | 0.335 | 0.20596 | 1.316 | 0 | 0.529 |
| 6401 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 14 | CREBBP | Sponge network | 0.683 | 1.0E-5 | 0.126 | 0.15186 | 0.529 |
| 6402 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 15 | RCAN2 | Sponge network | -0.405 | 0.22013 | -0.33 | 0.15172 | 0.529 |
| 6403 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 38 | PBX1 | Sponge network | 0.194 | 0.64278 | -1.206 | 0 | 0.529 |
| 6404 | RP11-532F6.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 22 | NRK | Sponge network | -0.896 | 0.00099 | 0.656 | 0.19217 | 0.529 |
| 6405 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p | 13 | PLSCR4 | Sponge network | -1.375 | 0.08642 | -0.474 | 0.03833 | 0.529 |
| 6406 | OIP5-AS1 |
hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p | 11 | TTL | Sponge network | 0.421 | 0.00084 | 0.494 | 1.0E-5 | 0.529 |
| 6407 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-940 | 14 | PEG3 | Sponge network | -0.777 | 0.16667 | -1.74 | 0 | 0.529 |
| 6408 | FENDRR |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-935 | 19 | RELN | Sponge network | -1.281 | 0.00012 | -2.403 | 0 | 0.529 |
| 6409 | MIR497HG |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 12 | PDZD2 | Sponge network | -1.439 | 4.0E-5 | -0.909 | 3.0E-5 | 0.528 |
| 6410 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 57 | DTNA | Sponge network | -0.942 | 0 | -1.648 | 1.0E-5 | 0.528 |
| 6411 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | IGSF10 | Sponge network | -2.117 | 0.00161 | -1.751 | 1.0E-5 | 0.528 |
| 6412 | PWAR6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-590-5p | 18 | PDZD2 | Sponge network | -1.575 | 6.0E-5 | -0.909 | 3.0E-5 | 0.528 |
| 6413 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-3065-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 26 | CRTC3 | Sponge network | 0.997 | 0.00066 | 0.164 | 0.16786 | 0.528 |
| 6414 | DNM3OS |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 38 | ABI2 | Sponge network | 0.572 | 0.01722 | 0.175 | 0.14787 | 0.528 |
| 6415 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 21 | CAV1 | Sponge network | -1.111 | 0.0003 | -1.013 | 6.0E-5 | 0.528 |
| 6416 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TRIM58 | Sponge network | -2.452 | 0 | -1.692 | 0 | 0.528 |
| 6417 | RP11-620J15.3 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | CXCL12 | Sponge network | -0.739 | 0.00044 | -1.421 | 0 | 0.528 |
| 6418 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 10 | LRRC7 | Sponge network | -4.546 | 0 | -1.341 | 0.01193 | 0.528 |
| 6419 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-29a-5p | 12 | CNTNAP3 | Sponge network | -2.095 | 0.00112 | -1.465 | 0.00033 | 0.528 |
| 6420 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | NRXN3 | Sponge network | -1.161 | 0.00591 | -0.893 | 0.04186 | 0.528 |
| 6421 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DNAJB4 | Sponge network | -0.901 | 0.00019 | -0.7 | 1.0E-5 | 0.528 |
| 6422 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 49 | BCL2 | Sponge network | -1.697 | 0.00889 | -0.899 | 0 | 0.528 |
| 6423 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-590-3p | 12 | HOOK3 | Sponge network | 0.335 | 0.20596 | 0.273 | 0.09957 | 0.528 |
| 6424 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 17 | LRRC55 | Sponge network | -0.976 | 0.00025 | -0.026 | 0.93163 | 0.528 |
| 6425 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 50 | RBMS3 | Sponge network | -0.222 | 0.32445 | -0.519 | 0.03551 | 0.528 |
| 6426 | LINC00702 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 22 | RPS6KA6 | Sponge network | -0.737 | 0.06182 | -2.989 | 0 | 0.528 |
| 6427 | RP11-473I1.5 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p | 16 | MYO9A | Sponge network | 0.542 | 0.00054 | -0.147 | 0.32373 | 0.528 |
| 6428 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p | 16 | FGFR1 | Sponge network | -1.886 | 0 | -0.509 | 0.03957 | 0.528 |
| 6429 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | FAM172A | Sponge network | -2.117 | 0.00161 | -0.57 | 0 | 0.528 |
| 6430 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 31 | GPC6 | Sponge network | -0.737 | 0.06182 | -0.054 | 0.81465 | 0.528 |
| 6431 | MIR497HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | KCNA6 | Sponge network | -1.439 | 4.0E-5 | -1.531 | 0.00013 | 0.528 |
| 6432 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 23 | NRXN1 | Sponge network | -0.739 | 0.00044 | -3.778 | 0 | 0.528 |
| 6433 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 37 | BTG2 | Sponge network | -2.452 | 0 | -1.763 | 0 | 0.528 |
| 6434 | BOLA3-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p | 19 | HIP1 | Sponge network | -0.246 | 0.35034 | 0.774 | 0 | 0.528 |
| 6435 | PWAR6 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 21 | TLE4 | Sponge network | -1.575 | 6.0E-5 | -1.157 | 0 | 0.528 |
| 6436 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 41 | BCL2 | Sponge network | -1.439 | 4.0E-5 | -0.899 | 0 | 0.528 |
| 6437 | LINC00476 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-769-5p | 32 | SYNPO2 | Sponge network | -0.615 | 0.00015 | -2.342 | 0 | 0.528 |
| 6438 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 39 | RICTOR | Sponge network | 0.848 | 0 | 0.388 | 0.00188 | 0.528 |
| 6439 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 22 | THBS1 | Sponge network | -0.154 | 0.53954 | 0.741 | 0.00386 | 0.528 |
| 6440 | RP5-1198O20.4 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 21 | SFRP1 | Sponge network | 0.997 | 0.00066 | -3.566 | 0 | 0.528 |
| 6441 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | ERG | Sponge network | 0.889 | 9.0E-5 | 0.002 | 0.99011 | 0.528 |
| 6442 | MALAT1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 17 | ATM | Sponge network | 0.782 | 0.00016 | 0.178 | 0.21568 | 0.528 |
| 6443 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 46 | GAS7 | Sponge network | -0.769 | 0.00035 | -0.555 | 0.01602 | 0.528 |
| 6444 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | PLSCR4 | Sponge network | -1.216 | 0 | -0.474 | 0.03833 | 0.528 |
| 6445 | MIR497HG |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p | 10 | DUSP1 | Sponge network | -1.439 | 4.0E-5 | -1.754 | 0 | 0.528 |
| 6446 | AP001347.6 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | RELN | Sponge network | -1.161 | 0.00591 | -2.403 | 0 | 0.528 |
| 6447 | LINC00657 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | KIF5B | Sponge network | 0.91 | 0 | 0.362 | 0.00066 | 0.528 |
| 6448 | PWAR6 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 14 | AMPH | Sponge network | -1.575 | 6.0E-5 | -0.916 | 5.0E-5 | 0.528 |
| 6449 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-769-5p | 24 | SETBP1 | Sponge network | -0.154 | 0.78939 | -1.302 | 1.0E-5 | 0.528 |
| 6450 | RP11-890B15.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-424-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p | 20 | SYNM | Sponge network | 0.101 | 0.60919 | -2.309 | 1.0E-5 | 0.528 |
| 6451 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-7-1-3p | 16 | ERG | Sponge network | -0.611 | 0.09607 | 0.002 | 0.99011 | 0.528 |
| 6452 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-590-3p | 10 | SLITRK3 | Sponge network | -0.739 | 0.00044 | -3.328 | 0 | 0.528 |
| 6453 | RP11-503E24.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | TRIM33 | Sponge network | 2.218 | 0 | 0.402 | 7.0E-5 | 0.528 |
| 6454 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p | 23 | EBF3 | Sponge network | -0.096 | 0.67958 | -0.058 | 0.81437 | 0.528 |
| 6455 | RP11-356J5.12 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -0.899 | 6.0E-5 | -2.633 | 0 | 0.528 |
| 6456 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | TSHZ3 | Sponge network | -0.709 | 0.18168 | -0.556 | 0.01516 | 0.528 |
| 6457 | RP11-504P24.4 |
hsa-miR-10b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p | 10 | NR2C2 | Sponge network | 0.624 | 0.0004 | 0.21 | 0.03224 | 0.528 |
| 6458 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 28 | ANK2 | Sponge network | 0.101 | 0.60919 | -1.603 | 1.0E-5 | 0.528 |
| 6459 | RP11-53O19.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | WWTR1 | Sponge network | -0.405 | 0.22013 | -0.345 | 0.11997 | 0.528 |
| 6460 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-590-3p | 16 | EPHA3 | Sponge network | -1.886 | 0 | 0.185 | 0.53588 | 0.528 |
| 6461 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-5p | 10 | SYNE1 | Sponge network | -1.255 | 0.00981 | -0.738 | 0.00316 | 0.528 |
| 6462 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SEMA3E | Sponge network | -2.46 | 0 | -1.717 | 0.00137 | 0.528 |
| 6463 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 37 | PIK3R1 | Sponge network | -0.096 | 0.67958 | -0.133 | 0.36854 | 0.528 |
| 6464 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 31 | PGR | Sponge network | -0.942 | 0 | -1.704 | 0 | 0.528 |
| 6465 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 31 | FBXO32 | Sponge network | -0.777 | 0.1134 | -0.495 | 0.07561 | 0.528 |
| 6466 | RP11-25K19.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PRKCB | Sponge network | -1.057 | 0.00029 | -1.313 | 2.0E-5 | 0.528 |
| 6467 | CTC-510F12.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-146b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-193b-3p | 10 | TACC1 | Sponge network | -0.691 | 0.00146 | -0.362 | 0.05406 | 0.528 |
| 6468 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 38 | RBMS3 | Sponge network | -1.203 | 0.03881 | -0.519 | 0.03551 | 0.528 |
| 6469 | RP11-57H14.4 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p | 18 | ATM | Sponge network | 0.083 | 0.66405 | 0.178 | 0.21568 | 0.528 |
| 6470 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 32 | MCC | Sponge network | -0.769 | 0.00035 | -1.005 | 0 | 0.528 |
| 6471 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 31 | SSBP2 | Sponge network | -0.811 | 2.0E-5 | -1.58 | 0 | 0.528 |
| 6472 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-425-5p | 12 | ATRX | Sponge network | 0.37 | 0.27614 | 0.261 | 0.04408 | 0.528 |
| 6473 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | EPHA3 | Sponge network | 0.889 | 9.0E-5 | 0.185 | 0.53588 | 0.528 |
| 6474 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-339-5p | 11 | STARD13 | Sponge network | -0.611 | 0.09607 | -0.187 | 0.27383 | 0.528 |
| 6475 | MIR143HG |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 21 | RELN | Sponge network | -0.739 | 0.0574 | -2.403 | 0 | 0.528 |
| 6476 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 50 | TCF4 | Sponge network | -0.896 | 0.00099 | 0.016 | 0.92271 | 0.528 |
| 6477 | SNHG14 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | MSN | Sponge network | -1.111 | 0.0003 | -0.023 | 0.88422 | 0.528 |
| 6478 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | FAT4 | Sponge network | -1.057 | 0.00029 | -0.354 | 0.14694 | 0.528 |
| 6479 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-96-5p | 28 | CADM1 | Sponge network | -0.976 | 0.00025 | -0.9 | 0.00084 | 0.528 |
| 6480 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | TSHZ3 | Sponge network | -0.842 | 0.01361 | -0.556 | 0.01516 | 0.528 |
| 6481 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | RADIL | Sponge network | -0.769 | 0.00035 | -1.628 | 0 | 0.528 |
| 6482 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 23 | ABCB5 | Sponge network | -1.111 | 0.0003 | -0.956 | 0.04077 | 0.528 |
| 6483 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -0.842 | 0.01361 | -0.354 | 0.14694 | 0.528 |
| 6484 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 13 | PTPN13 | Sponge network | -2.46 | 0 | -1.208 | 0 | 0.528 |
| 6485 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-589-3p | 14 | ABCC9 | Sponge network | -0.154 | 0.78939 | -0.466 | 0.14121 | 0.527 |
| 6486 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | DMTF1 | Sponge network | 0.848 | 0 | 0.248 | 0.02726 | 0.527 |
| 6487 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 17 | TXNIP | Sponge network | -0.737 | 0.06182 | -1.277 | 0 | 0.527 |
| 6488 | EMX2OS |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | PRUNE2 | Sponge network | -0.777 | 0.1134 | -1.733 | 0.00022 | 0.527 |
| 6489 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p | 15 | CADM3 | Sponge network | -1.654 | 0.00144 | -3.663 | 0 | 0.527 |
| 6490 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | ADAMTSL3 | Sponge network | -0.187 | 0.23787 | -1.559 | 0 | 0.527 |
| 6491 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | 0.249 | 0.35902 | 0.273 | 0.09957 | 0.527 |
| 6492 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ZMYND11 | Sponge network | -0.323 | 0.01372 | -0.2 | 0.05449 | 0.527 |
| 6493 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 51 | SSBP2 | Sponge network | -0.709 | 0.18168 | -1.58 | 0 | 0.527 |
| 6494 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RCAN2 | Sponge network | -0.021 | 0.93598 | -0.33 | 0.15172 | 0.527 |
| 6495 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 34 | ST6GAL2 | Sponge network | -1.216 | 0 | -1.201 | 0.00597 | 0.527 |
| 6496 | LINC00702 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ADH1B | Sponge network | -0.737 | 0.06182 | -3.716 | 0 | 0.527 |
| 6497 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 26 | PCDH10 | Sponge network | -0.096 | 0.67958 | -1.644 | 0.0078 | 0.527 |
| 6498 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | PREX2 | Sponge network | 0.194 | 0.64278 | -0.272 | 0.25382 | 0.527 |
| 6499 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 18 | SYNPO2 | Sponge network | -0.739 | 0.00044 | -2.342 | 0 | 0.527 |
| 6500 | CTC-510F12.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-484 | 16 | DST | Sponge network | -0.691 | 0.00146 | -0.713 | 0.00096 | 0.527 |
| 6501 | FENDRR |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 14 | PHOX2B | Sponge network | -1.281 | 0.00012 | -2.68 | 0 | 0.527 |
| 6502 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 22 | LATS2 | Sponge network | -0.49 | 0.04946 | 0.159 | 0.2304 | 0.527 |
| 6503 | OIP5-AS1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 17 | SHPRH | Sponge network | 0.421 | 0.00084 | 0.098 | 0.40994 | 0.527 |
| 6504 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p | 19 | PCDH9 | Sponge network | 0.299 | 0.57722 | -1.823 | 4.0E-5 | 0.527 |
| 6505 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | AFF3 | Sponge network | -0.487 | 0.02157 | -2.373 | 0 | 0.527 |
| 6506 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-590-3p | 10 | PTPN13 | Sponge network | -0.487 | 0.02157 | -1.208 | 0 | 0.527 |
| 6507 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-590-3p | 16 | SPG20 | Sponge network | 0.532 | 0.00505 | -1.16 | 0 | 0.527 |
| 6508 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 64 | NTRK3 | Sponge network | 0.194 | 0.64278 | -1.77 | 3.0E-5 | 0.527 |
| 6509 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 27 | KIT | Sponge network | -0.49 | 0.04946 | -2.184 | 0 | 0.527 |
| 6510 | MAGI2-AS3 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ADH1B | Sponge network | -0.129 | 0.57177 | -3.716 | 0 | 0.527 |
| 6511 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-93-5p | 33 | HLF | Sponge network | -2.69 | 0.0001 | -2.443 | 0 | 0.527 |
| 6512 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | RCAN2 | Sponge network | -0.487 | 0.02157 | -0.33 | 0.15172 | 0.527 |
| 6513 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-582-5p | 23 | TSHZ3 | Sponge network | -2.095 | 0.00112 | -0.556 | 0.01516 | 0.527 |
| 6514 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | DKK3 | Sponge network | 0.997 | 0.00066 | -0.052 | 0.77783 | 0.527 |
| 6515 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | SYNE1 | Sponge network | -2.46 | 0 | -0.738 | 0.00316 | 0.527 |
| 6516 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | MLLT10 | Sponge network | 0.311 | 0.10344 | 0.105 | 0.33292 | 0.527 |
| 6517 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p | 17 | DYNC1I1 | Sponge network | -0.405 | 0.22013 | -1.12 | 0.00107 | 0.527 |
| 6518 | RP11-254F7.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p | 28 | IL17RD | Sponge network | 0.176 | 0.37402 | -0.019 | 0.94873 | 0.527 |
| 6519 | USP3-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-576-5p | 17 | KLF10 | Sponge network | -0.162 | 0.47687 | -0.783 | 0 | 0.527 |
| 6520 | AC025171.1 |
hsa-let-7g-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 27 | RICTOR | Sponge network | 0.998 | 0 | 0.388 | 0.00188 | 0.527 |
| 6521 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p | 10 | IGF1 | Sponge network | 0.693 | 0.00076 | -0.204 | 0.5898 | 0.527 |
| 6522 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 18 | SPG20 | Sponge network | 0.194 | 0.64278 | -1.16 | 0 | 0.527 |
| 6523 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940 | 34 | HLF | Sponge network | -0.896 | 0.00099 | -2.443 | 0 | 0.527 |
| 6524 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | ERG | Sponge network | -0.769 | 0.00035 | 0.002 | 0.99011 | 0.527 |
| 6525 | PWAR6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 58 | TRPS1 | Sponge network | -1.575 | 6.0E-5 | -0.191 | 0.44109 | 0.527 |
| 6526 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-5p | 18 | PTGFR | Sponge network | -1.349 | 0.00017 | -0.233 | 0.45421 | 0.527 |
| 6527 | RP11-774O3.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p | 55 | DST | Sponge network | -0.487 | 0.02157 | -0.713 | 0.00096 | 0.527 |
| 6528 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-629-3p;hsa-miR-7-5p | 21 | ZBTB4 | Sponge network | -1.582 | 8.0E-5 | -0.739 | 0 | 0.527 |
| 6529 | MIR143HG |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 12 | MLLT11 | Sponge network | -0.739 | 0.0574 | -0.661 | 0.01733 | 0.527 |
| 6530 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 10 | SETD2 | Sponge network | 0.537 | 0.00139 | 0.031 | 0.73068 | 0.527 |
| 6531 | LINC00641 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | NRXN3 | Sponge network | -0.222 | 0.32445 | -0.893 | 0.04186 | 0.527 |
| 6532 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | PGR | Sponge network | -0.021 | 0.93598 | -1.704 | 0 | 0.527 |
| 6533 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | JAZF1 | Sponge network | -0.811 | 2.0E-5 | -0.377 | 0.02677 | 0.527 |
| 6534 | RP11-999E24.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-93-3p | 16 | WWTR1 | Sponge network | -0.693 | 0.05629 | -0.345 | 0.11997 | 0.527 |
| 6535 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-98-5p | 31 | PRRX1 | Sponge network | -1.697 | 0.00889 | 1.316 | 0 | 0.527 |
| 6536 | GNG12-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 41 | TGFBR3 | Sponge network | -0.793 | 7.0E-5 | -1.173 | 1.0E-5 | 0.527 |
| 6537 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | AHNAK | Sponge network | -0.129 | 0.57177 | -1.207 | 0 | 0.527 |
| 6538 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | RELN | Sponge network | -0.739 | 0.00044 | -2.403 | 0 | 0.527 |
| 6539 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 40 | TGFBR3 | Sponge network | -0.323 | 0.01372 | -1.173 | 1.0E-5 | 0.527 |
| 6540 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 29 | DMD | Sponge network | 0.335 | 0.20596 | -1.506 | 0 | 0.527 |
| 6541 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | PLSCR4 | Sponge network | -0.421 | 0.0114 | -0.474 | 0.03833 | 0.527 |
| 6542 | HHIP-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | IGFBP5 | Sponge network | -0.737 | 0.10822 | -0.806 | 0.00105 | 0.527 |
| 6543 | LINC01018 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | -2.915 | 0.00015 | -1.733 | 0.00022 | 0.526 |
| 6544 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | STARD13 | Sponge network | -0.896 | 0.00099 | -0.187 | 0.27383 | 0.526 |
| 6545 | SNX29P2 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-502-5p | 16 | FMNL3 | Sponge network | 0.4 | 0.14611 | 0.555 | 4.0E-5 | 0.526 |
| 6546 | TP73-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 14 | NGFR | Sponge network | -0.563 | 0.02545 | -1.271 | 0.01058 | 0.526 |
| 6547 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p | 15 | FGFR1 | Sponge network | -1.249 | 7.0E-5 | -0.509 | 0.03957 | 0.526 |
| 6548 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 23 | ST6GAL2 | Sponge network | -1.349 | 0.00017 | -1.201 | 0.00597 | 0.526 |
| 6549 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 23 | PGR | Sponge network | -1.203 | 0.03881 | -1.704 | 0 | 0.526 |
| 6550 | DNM3OS |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SEMA3C | Sponge network | 0.572 | 0.01722 | -0.272 | 0.31153 | 0.526 |
| 6551 | JAZF1-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-940 | 15 | RHOB | Sponge network | -0.769 | 0.00035 | -1.052 | 0 | 0.526 |
| 6552 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | RNF144A | Sponge network | 0.335 | 0.20596 | 0.594 | 0.0009 | 0.526 |
| 6553 | ANKRD10-IT1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-7-1-3p | 13 | ATRX | Sponge network | 1.739 | 0 | 0.261 | 0.04408 | 0.526 |
| 6554 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-421 | 24 | CNTN1 | Sponge network | -0.154 | 0.78939 | -2.594 | 0 | 0.526 |
| 6555 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 18 | ABCB5 | Sponge network | -0.576 | 0.10121 | -0.956 | 0.04077 | 0.526 |
| 6556 | FENDRR |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | RPS6KA6 | Sponge network | -1.281 | 0.00012 | -2.989 | 0 | 0.526 |
| 6557 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-93-3p | 16 | EPHA3 | Sponge network | -0.932 | 0.00329 | 0.185 | 0.53588 | 0.526 |
| 6558 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p | 25 | BCL6 | Sponge network | -2.452 | 0 | -0.211 | 0.19489 | 0.526 |
| 6559 | SH3RF3-AS1 |
hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-339-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | NAV3 | Sponge network | 0.889 | 9.0E-5 | 0.212 | 0.47729 | 0.526 |
| 6560 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 20 | PTPN11 | Sponge network | -0.222 | 0.32445 | 0.431 | 2.0E-5 | 0.526 |
| 6561 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-93-5p | 29 | ANK2 | Sponge network | -0.154 | 0.78939 | -1.603 | 1.0E-5 | 0.526 |
| 6562 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 19 | FGFR1 | Sponge network | -0.793 | 7.0E-5 | -0.509 | 0.03957 | 0.526 |
| 6563 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p | 10 | FGFR1 | Sponge network | -4.546 | 0 | -0.509 | 0.03957 | 0.526 |
| 6564 | MIR143HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-98-5p | 31 | PRRX1 | Sponge network | -0.739 | 0.0574 | 1.316 | 0 | 0.526 |
| 6565 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | SON | Sponge network | 1.162 | 5.0E-5 | 0.168 | 0.04406 | 0.526 |
| 6566 | CTD-3138B18.6 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | MDM4 | Sponge network | 2.248 | 0 | 0.345 | 0.00762 | 0.526 |
| 6567 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 64 | TCF4 | Sponge network | -0.487 | 0.02157 | 0.016 | 0.92271 | 0.526 |
| 6568 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | NRXN3 | Sponge network | -0.563 | 0.02545 | -0.893 | 0.04186 | 0.526 |
| 6569 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-671-5p | 14 | FAT4 | Sponge network | -0.309 | 0.1145 | -0.354 | 0.14694 | 0.526 |
| 6570 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-942-5p | 25 | NOTCH2 | Sponge network | -1.092 | 4.0E-5 | 0.138 | 0.35163 | 0.526 |
| 6571 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | ANK2 | Sponge network | 0.299 | 0.57722 | -1.603 | 1.0E-5 | 0.526 |
| 6572 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 39 | ESR1 | Sponge network | -1.575 | 6.0E-5 | -1.035 | 3.0E-5 | 0.526 |
| 6573 | RP11-13K12.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-93-5p | 34 | IL17RD | Sponge network | -0.154 | 0.78939 | -0.019 | 0.94873 | 0.526 |
| 6574 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 42 | DTNA | Sponge network | -1.057 | 0.00029 | -1.648 | 1.0E-5 | 0.526 |
| 6575 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | NALCN | Sponge network | 0.532 | 0.00505 | 1.068 | 0.00237 | 0.526 |
| 6576 | MIR143HG |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-98-5p | 11 | MPL | Sponge network | -0.739 | 0.0574 | -0.624 | 0.00133 | 0.526 |
| 6577 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 25 | EBF3 | Sponge network | -0.777 | 0.1134 | -0.058 | 0.81437 | 0.526 |
| 6578 | FGF14-AS2 |
hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 11 | CXCL12 | Sponge network | -1.582 | 8.0E-5 | -1.421 | 0 | 0.526 |
| 6579 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-429;hsa-miR-7-1-3p | 11 | DKK3 | Sponge network | -0.246 | 0.35034 | -0.052 | 0.77783 | 0.526 |
| 6580 | RASSF8-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p | 74 | LPP | Sponge network | 0.335 | 0.20596 | -0.459 | 0.04475 | 0.526 |
| 6581 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p | 29 | RASSF8 | Sponge network | -0.793 | 7.0E-5 | -0.122 | 0.65591 | 0.526 |
| 6582 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | FAT3 | Sponge network | -1.654 | 0.00144 | -1.601 | 0.00051 | 0.526 |
| 6583 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 25 | GREM1 | Sponge network | -1.111 | 0.0003 | 0.902 | 0.01691 | 0.526 |
| 6584 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 51 | TRPS1 | Sponge network | 0.194 | 0.64278 | -0.191 | 0.44109 | 0.526 |
| 6585 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | LRRC7 | Sponge network | -0.323 | 0.01372 | -1.341 | 0.01193 | 0.526 |
| 6586 | PWAR6 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | PREX2 | Sponge network | -1.575 | 6.0E-5 | -0.272 | 0.25382 | 0.526 |
| 6587 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | PCDH17 | Sponge network | -0.358 | 0.17255 | 0.731 | 2.0E-5 | 0.526 |
| 6588 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 45 | PIK3R1 | Sponge network | -0.222 | 0.32445 | -0.133 | 0.36854 | 0.526 |
| 6589 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | ADAMTSL3 | Sponge network | -0.737 | 0.10822 | -1.559 | 0 | 0.526 |
| 6590 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-532-3p | 14 | SYNM | Sponge network | -0.421 | 0.0114 | -2.309 | 1.0E-5 | 0.526 |
| 6591 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p | 14 | SPG20 | Sponge network | -0.777 | 0.16667 | -1.16 | 0 | 0.526 |
| 6592 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | LRP1B | Sponge network | -2.117 | 0.00161 | -2.163 | 0 | 0.526 |
| 6593 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MLLT10 | Sponge network | 1.153 | 0 | 0.105 | 0.33292 | 0.526 |
| 6594 | RP11-251G23.5 |
hsa-miR-101-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p;hsa-miR-654-3p | 17 | PHF6 | Sponge network | 1.344 | 0 | 0.785 | 0 | 0.526 |
| 6595 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | SON | Sponge network | 0.946 | 0 | 0.168 | 0.04406 | 0.526 |
| 6596 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CNTN1 | Sponge network | 0.249 | 0.35902 | -2.594 | 0 | 0.526 |
| 6597 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940 | 27 | CRTC3 | Sponge network | -0.49 | 0.04946 | 0.164 | 0.16786 | 0.526 |
| 6598 | PWAR6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 13 | THSD7A | Sponge network | -1.575 | 6.0E-5 | 0.242 | 0.25154 | 0.526 |
| 6599 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | BNC2 | Sponge network | 0.889 | 9.0E-5 | -0.491 | 0.09988 | 0.526 |
| 6600 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 21 | GLIPR1 | Sponge network | 0.335 | 0.20596 | 0.146 | 0.3276 | 0.526 |
| 6601 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CACNA2D1 | Sponge network | -1.057 | 0.00029 | -0.846 | 0.00675 | 0.526 |
| 6602 | RASSF8-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p | 13 | MACF1 | Sponge network | 0.335 | 0.20596 | 0.019 | 0.90335 | 0.526 |
| 6603 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-7-1-3p | 14 | SEMA3E | Sponge network | -0.563 | 0.02545 | -1.717 | 0.00137 | 0.526 |
| 6604 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | JAZF1 | Sponge network | -0.563 | 0.02545 | -0.377 | 0.02677 | 0.526 |
| 6605 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 28 | NRK | Sponge network | -2.452 | 0 | 0.656 | 0.19217 | 0.526 |
| 6606 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 24 | RARB | Sponge network | -0.129 | 0.57177 | -0.825 | 2.0E-5 | 0.526 |
| 6607 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | ZFHX4 | Sponge network | -2.117 | 0.00161 | 0.018 | 0.95574 | 0.526 |
| 6608 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-93-5p | 53 | NTRK3 | Sponge network | -0.611 | 0.09607 | -1.77 | 3.0E-5 | 0.525 |
| 6609 | FENDRR |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | LRP1B | Sponge network | -1.281 | 0.00012 | -2.163 | 0 | 0.525 |
| 6610 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 20 | PHC3 | Sponge network | 0.178 | 0.29106 | 0.212 | 0.0499 | 0.525 |
| 6611 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-93-5p | 10 | LRRC55 | Sponge network | 0.342 | 0.03129 | -0.026 | 0.93163 | 0.525 |
| 6612 | RP11-400K9.4 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326 | 18 | NRK | Sponge network | -0.611 | 0.09607 | 0.656 | 0.19217 | 0.525 |
| 6613 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 34 | ST6GAL2 | Sponge network | 0.997 | 0.00066 | -1.201 | 0.00597 | 0.525 |
| 6614 | RP11-672A2.4 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-30b-3p | 11 | IGFBP5 | Sponge network | 1.344 | 0 | -0.806 | 0.00105 | 0.525 |
| 6615 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-3913-5p;hsa-miR-454-3p | 22 | ZBTB4 | Sponge network | -2.095 | 0.00112 | -0.739 | 0 | 0.525 |
| 6616 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p | 24 | PBX1 | Sponge network | -0.358 | 0.17255 | -1.206 | 0 | 0.525 |
| 6617 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 16 | ZMYND11 | Sponge network | -0.031 | 0.89063 | -0.2 | 0.05449 | 0.525 |
| 6618 | FAM66C |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 30 | DLC1 | Sponge network | -0.901 | 0.00019 | -0.382 | 0.05038 | 0.525 |
| 6619 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 27 | TBL1XR1 | Sponge network | 1.254 | 0 | 0.578 | 0 | 0.525 |
| 6620 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-5p | 19 | SETBP1 | Sponge network | 0.176 | 0.37402 | -1.302 | 1.0E-5 | 0.525 |
| 6621 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-505-3p | 12 | PTPN11 | Sponge network | 1.147 | 0 | 0.431 | 2.0E-5 | 0.525 |
| 6622 | MIR497HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 40 | TGFBR3 | Sponge network | -1.439 | 4.0E-5 | -1.173 | 1.0E-5 | 0.525 |
| 6623 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 24 | CDH23 | Sponge network | -0.576 | 0.10121 | -0.974 | 0.00034 | 0.525 |
| 6624 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-93-5p | 19 | PLSCR4 | Sponge network | -1.654 | 0.00144 | -0.474 | 0.03833 | 0.525 |
| 6625 | ZNF667-AS1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-96-5p | 17 | RPS6KA6 | Sponge network | -0.976 | 0.00025 | -2.989 | 0 | 0.525 |
| 6626 | CTD-3092A11.2 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p | 11 | ATM | Sponge network | 0.873 | 0 | 0.178 | 0.21568 | 0.525 |
| 6627 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-362-3p | 11 | DKK3 | Sponge network | -0.358 | 0.17255 | -0.052 | 0.77783 | 0.525 |
| 6628 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 16 | PTPN11 | Sponge network | 0.91 | 0 | 0.431 | 2.0E-5 | 0.525 |
| 6629 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | RICTOR | Sponge network | 0.826 | 1.0E-5 | 0.388 | 0.00188 | 0.525 |
| 6630 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-577;hsa-miR-96-5p;hsa-miR-98-5p | 11 | PLAGL1 | Sponge network | -2.452 | 0 | -0.588 | 0.00912 | 0.525 |
| 6631 | NR2F1-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | ID4 | Sponge network | -0.49 | 0.04946 | -1.644 | 0 | 0.525 |
| 6632 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3E | Sponge network | -0.576 | 0.10121 | -1.717 | 0.00137 | 0.525 |
| 6633 | TPTEP1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-3913-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | FNBP1 | Sponge network | -1.216 | 0 | -0.969 | 4.0E-5 | 0.525 |
| 6634 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | CCND2 | Sponge network | -0.709 | 0.18168 | -0.267 | 0.3112 | 0.525 |
| 6635 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 40 | RASSF2 | Sponge network | -0.563 | 0.02545 | -0.273 | 0.21961 | 0.525 |
| 6636 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-455-5p;hsa-miR-582-3p;hsa-miR-93-5p | 21 | JAZF1 | Sponge network | -0.473 | 0.07602 | -0.377 | 0.02677 | 0.525 |
| 6637 | LINC01018 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | NRXN3 | Sponge network | -2.915 | 0.00015 | -0.893 | 0.04186 | 0.525 |
| 6638 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 21 | PCDH9 | Sponge network | -0.612 | 0.00094 | -1.823 | 4.0E-5 | 0.525 |
| 6639 | JAZF1-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-542-3p | 10 | PRKD1 | Sponge network | -0.769 | 0.00035 | -0.466 | 0.03583 | 0.525 |
| 6640 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-577;hsa-miR-590-3p | 14 | DNAJB4 | Sponge network | -0.896 | 0.00099 | -0.7 | 1.0E-5 | 0.525 |
| 6641 | RP11-244O19.1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-625-5p | 11 | NGFR | Sponge network | -0.096 | 0.67958 | -1.271 | 0.01058 | 0.525 |
| 6642 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-93-3p | 10 | ZNF382 | Sponge network | -0.611 | 0.09607 | -0.494 | 0.03831 | 0.525 |
| 6643 | JAZF1-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 14 | CXCL12 | Sponge network | -0.769 | 0.00035 | -1.421 | 0 | 0.525 |
| 6644 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p | 16 | AKAP12 | Sponge network | -0.769 | 0.00035 | -0.932 | 0.0028 | 0.525 |
| 6645 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | GPC6 | Sponge network | -0.376 | 0.00337 | -0.054 | 0.81465 | 0.525 |
| 6646 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935 | 28 | DMD | Sponge network | 0.225 | 0.30644 | -1.506 | 0 | 0.525 |
| 6647 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-625-5p | 14 | DCLK1 | Sponge network | -0.162 | 0.47687 | -1.324 | 0.00014 | 0.525 |
| 6648 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-214-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 52 | PPM1A | Sponge network | -0.222 | 0.32445 | -0.623 | 0 | 0.525 |
| 6649 | LINC00865 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-582-5p | 10 | THSD7B | Sponge network | -1.249 | 7.0E-5 | 0.154 | 0.74723 | 0.525 |
| 6650 | ZNF582-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 20 | NAV3 | Sponge network | -1.349 | 0.00017 | 0.212 | 0.47729 | 0.525 |
| 6651 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | LRRC55 | Sponge network | 0.727 | 3.0E-5 | -0.026 | 0.93163 | 0.525 |
| 6652 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | DAB2 | Sponge network | 0.997 | 0.00066 | 0.431 | 0.0113 | 0.525 |
| 6653 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 11 | TIMP3 | Sponge network | -0.376 | 0.00337 | -0.546 | 0.02307 | 0.525 |
| 6654 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-484 | 16 | GREM1 | Sponge network | -0.777 | 0.16667 | 0.902 | 0.01691 | 0.525 |
| 6655 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 23 | RARB | Sponge network | -2.452 | 0 | -0.825 | 2.0E-5 | 0.525 |
| 6656 | LINC00641 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-532-3p | 22 | SYNM | Sponge network | -0.222 | 0.32445 | -2.309 | 1.0E-5 | 0.525 |
| 6657 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-935 | 41 | ABI2 | Sponge network | -0.162 | 0.47687 | 0.175 | 0.14787 | 0.525 |
| 6658 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-577 | 13 | TIMP3 | Sponge network | -0.473 | 0.07602 | -0.546 | 0.02307 | 0.525 |
| 6659 | SERTAD4-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-502-5p | 14 | IGFBP5 | Sponge network | -0.932 | 0.00329 | -0.806 | 0.00105 | 0.525 |
| 6660 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-29a-5p | 10 | FBXO32 | Sponge network | -2.076 | 0.00548 | -0.495 | 0.07561 | 0.525 |
| 6661 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 22 | CNTNAP3 | Sponge network | -1.111 | 0.0003 | -1.465 | 0.00033 | 0.525 |
| 6662 | PWAR6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | -1.575 | 6.0E-5 | -0.974 | 0.00034 | 0.525 |
| 6663 | MAGI2-AS3 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 12 | PHOX2B | Sponge network | -0.129 | 0.57177 | -2.68 | 0 | 0.525 |
| 6664 | CTD-3138B18.6 |
hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 12 | ABL2 | Sponge network | 2.248 | 0 | 0.82 | 0 | 0.525 |
| 6665 | LINC00702 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 19 | ID4 | Sponge network | -0.737 | 0.06182 | -1.644 | 0 | 0.525 |
| 6666 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | ST5 | Sponge network | -0.739 | 0.0574 | -0.78 | 0 | 0.525 |
| 6667 | AC010226.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 13 | SETBP1 | Sponge network | -0.811 | 2.0E-5 | -1.302 | 1.0E-5 | 0.525 |
| 6668 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | GALNT13 | Sponge network | -0.576 | 0.10121 | -0.857 | 0.07439 | 0.525 |
| 6669 | LINC00702 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | RELN | Sponge network | -0.737 | 0.06182 | -2.403 | 0 | 0.525 |
| 6670 | LINC00957 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-935 | 11 | STAT5B | Sponge network | -0.421 | 0.0114 | -0.182 | 0.1082 | 0.525 |
| 6671 | PCA3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | FNBP1 | Sponge network | -0.709 | 0.18168 | -0.969 | 4.0E-5 | 0.525 |
| 6672 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-654-3p | 15 | PSIP1 | Sponge network | -0.323 | 0.01372 | -0.005 | 0.96897 | 0.525 |
| 6673 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-34a-5p;hsa-miR-424-5p | 14 | SYT4 | Sponge network | -1.331 | 3.0E-5 | -3.207 | 0 | 0.525 |
| 6674 | LINC00702 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 29 | PTPN14 | Sponge network | -0.737 | 0.06182 | 0.144 | 0.34595 | 0.525 |
| 6675 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | CACNA2D1 | Sponge network | -0.487 | 0.02157 | -0.846 | 0.00675 | 0.525 |
| 6676 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p | 15 | PDS5B | Sponge network | 1.294 | 0 | 0.259 | 0.00962 | 0.525 |
| 6677 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 20 | NR2C2 | Sponge network | 0.559 | 0.00026 | 0.21 | 0.03224 | 0.525 |
| 6678 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 36 | DDX6 | Sponge network | 0.782 | 0.00016 | 0.091 | 0.36428 | 0.524 |
| 6679 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-337-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PBRM1 | Sponge network | 0.537 | 0.00139 | -0.029 | 0.78601 | 0.524 |
| 6680 | OIP5-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | PIK3CA | Sponge network | 0.421 | 0.00084 | 0.195 | 0.06908 | 0.524 |
| 6681 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | CAV1 | Sponge network | 0.194 | 0.64278 | -1.013 | 6.0E-5 | 0.524 |
| 6682 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 48 | SSBP2 | Sponge network | -0.612 | 0.00094 | -1.58 | 0 | 0.524 |
| 6683 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 22 | CDH19 | Sponge network | -0.612 | 0.00094 | -3.434 | 0 | 0.524 |
| 6684 | MIR497HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 36 | SNX29 | Sponge network | -1.439 | 4.0E-5 | -0.34 | 0.01371 | 0.524 |
| 6685 | TRAF3IP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | RYR2 | Sponge network | -0.323 | 0.01372 | -1.466 | 0.00031 | 0.524 |
| 6686 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | 0.194 | 0.64278 | -1.892 | 0 | 0.524 |
| 6687 | LINC00672 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-98-5p | 33 | DMD | Sponge network | -0.227 | 0.31657 | -1.506 | 0 | 0.524 |
| 6688 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p | 19 | PRKAA2 | Sponge network | -2.076 | 0.00548 | -2.462 | 0 | 0.524 |
| 6689 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 25 | SYNPO2 | Sponge network | 0.246 | 0.59912 | -2.342 | 0 | 0.524 |
| 6690 | ACTA2-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3614-5p | 12 | NGFR | Sponge network | 0.194 | 0.64278 | -1.271 | 0.01058 | 0.524 |
| 6691 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 42 | BNC2 | Sponge network | -1.161 | 0.00591 | -0.491 | 0.09988 | 0.524 |
| 6692 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-93-5p | 24 | PGR | Sponge network | 0.299 | 0.57722 | -1.704 | 0 | 0.524 |
| 6693 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | SON | Sponge network | 0.997 | 0 | 0.168 | 0.04406 | 0.524 |
| 6694 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500b-5p | 29 | SCN9A | Sponge network | -0.405 | 0.22013 | -0.525 | 0.10593 | 0.524 |
| 6695 | MALAT1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-590-3p | 11 | TPR | Sponge network | 0.782 | 0.00016 | 0.582 | 0 | 0.524 |
| 6696 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | PEG3 | Sponge network | -0.769 | 0.00035 | -1.74 | 0 | 0.524 |
| 6697 | RP11-159D12.2 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-653-5p | 26 | UBR3 | Sponge network | 1.254 | 0 | -0.021 | 0.82774 | 0.524 |
| 6698 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-589-3p | 23 | TIMP3 | Sponge network | -0.777 | 0.16667 | -0.546 | 0.02307 | 0.524 |
| 6699 | ZNF582-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-671-5p | 25 | DLC1 | Sponge network | -1.349 | 0.00017 | -0.382 | 0.05038 | 0.524 |
| 6700 | HCG18 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | PHF3 | Sponge network | 1.063 | 0 | 0.126 | 0.19652 | 0.524 |
| 6701 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | CNTNAP1 | Sponge network | -2.46 | 0 | 0.358 | 0.13727 | 0.524 |
| 6702 | GNG12-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-93-5p | 33 | IL17RD | Sponge network | -0.793 | 7.0E-5 | -0.019 | 0.94873 | 0.524 |
| 6703 | ACTA2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p | 24 | RYR2 | Sponge network | 0.194 | 0.64278 | -1.466 | 0.00031 | 0.524 |
| 6704 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-590-3p | 10 | PTCH1 | Sponge network | -1.886 | 0 | -0.1 | 0.6179 | 0.524 |
| 6705 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-744-3p | 30 | FAT3 | Sponge network | -2.095 | 0.00112 | -1.601 | 0.00051 | 0.524 |
| 6706 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 36 | DIP2C | Sponge network | 0.997 | 0.00066 | -0.436 | 0.01713 | 0.524 |
| 6707 | AC108488.4 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 27 | ABI2 | Sponge network | 0.259 | 0.12542 | 0.175 | 0.14787 | 0.524 |
| 6708 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | LSAMP | Sponge network | -0.323 | 0.01372 | -0.066 | 0.81708 | 0.524 |
| 6709 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 34 | KCNA6 | Sponge network | -0.49 | 0.04946 | -1.531 | 0.00013 | 0.524 |
| 6710 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-628-5p | 41 | QKI | Sponge network | -2.62 | 0 | -0.333 | 0.04111 | 0.524 |
| 6711 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 20 | SCN9A | Sponge network | -0.896 | 0.00099 | -0.525 | 0.10593 | 0.524 |
| 6712 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-429 | 15 | GREM1 | Sponge network | -0.154 | 0.53954 | 0.902 | 0.01691 | 0.524 |
| 6713 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | RBMS3 | Sponge network | -0.737 | 0.10822 | -0.519 | 0.03551 | 0.524 |
| 6714 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | FYN | Sponge network | 0.335 | 0.20596 | -0.131 | 0.37051 | 0.524 |
| 6715 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 27 | ADAMTSL3 | Sponge network | -0.942 | 0 | -1.559 | 0 | 0.524 |
| 6716 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | CDH10 | Sponge network | -2.452 | 0 | -2.397 | 0 | 0.524 |
| 6717 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p | 35 | AR | Sponge network | 0.176 | 0.37402 | -1.554 | 0 | 0.524 |
| 6718 | ZNF667-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-5p | 12 | EHD3 | Sponge network | -0.976 | 0.00025 | -0.484 | 0.01747 | 0.524 |
| 6719 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 44 | SSBP2 | Sponge network | -0.842 | 0.01361 | -1.58 | 0 | 0.524 |
| 6720 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92a-3p | 22 | NRXN3 | Sponge network | 0.214 | 0.18246 | -0.893 | 0.04186 | 0.524 |
| 6721 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | UBE2QL1 | Sponge network | -2.452 | 0 | -2.417 | 0 | 0.524 |
| 6722 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p | 11 | SPTBN4 | Sponge network | -1.331 | 3.0E-5 | -0.786 | 0.01281 | 0.524 |
| 6723 | RASSF8-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 25 | TET1 | Sponge network | 0.335 | 0.20596 | -0.135 | 0.52138 | 0.524 |
| 6724 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 47 | CHD5 | Sponge network | -0.49 | 0.04946 | -0.922 | 0.01521 | 0.524 |
| 6725 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 30 | SNX29 | Sponge network | -0.976 | 0.00025 | -0.34 | 0.01371 | 0.524 |
| 6726 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 30 | RBMS3 | Sponge network | -0.318 | 0.65847 | -0.519 | 0.03551 | 0.524 |
| 6727 | MIR143HG |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ITGB1 | Sponge network | -0.739 | 0.0574 | 0.524 | 0.00017 | 0.524 |
| 6728 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 41 | MYO9A | Sponge network | -2.46 | 0 | -0.147 | 0.32373 | 0.524 |
| 6729 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-98-5p | 14 | SCN11A | Sponge network | -2.46 | 0 | -1.15 | 0.00299 | 0.524 |
| 6730 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 42 | MEF2C | Sponge network | 0.249 | 0.35902 | -0.499 | 0.01448 | 0.524 |
| 6731 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p | 11 | JAKMIP2 | Sponge network | -0.777 | 0.16667 | -1.388 | 0 | 0.524 |
| 6732 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p | 19 | BCL6 | Sponge network | -0.769 | 0.00035 | -0.211 | 0.19489 | 0.524 |
| 6733 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | SYNE1 | Sponge network | -0.295 | 0.02604 | -0.738 | 0.00316 | 0.524 |
| 6734 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-93-5p | 24 | TCF4 | Sponge network | -2.531 | 0 | 0.016 | 0.92271 | 0.524 |
| 6735 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | CDH19 | Sponge network | -0.323 | 0.01372 | -3.434 | 0 | 0.524 |
| 6736 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | EBF1 | Sponge network | -0.021 | 0.93598 | -0.384 | 0.08856 | 0.524 |
| 6737 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | TMEFF2 | Sponge network | -0.129 | 0.57177 | -2.426 | 0 | 0.524 |
| 6738 | WDFY3-AS2 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | NOV | Sponge network | -0.942 | 0 | -0.58 | 0.04676 | 0.524 |
| 6739 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 29 | RASSF2 | Sponge network | -1.331 | 3.0E-5 | -0.273 | 0.21961 | 0.524 |
| 6740 | LINC00641 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 29 | DLC1 | Sponge network | -0.222 | 0.32445 | -0.382 | 0.05038 | 0.523 |
| 6741 | SNHG14 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | INPP4A | Sponge network | -1.111 | 0.0003 | 0.07 | 0.53563 | 0.523 |
| 6742 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 47 | NTRK3 | Sponge network | -1.161 | 0.00591 | -1.77 | 3.0E-5 | 0.523 |
| 6743 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p | 15 | RPS6KA2 | Sponge network | -2.62 | 0 | -0.57 | 0.00013 | 0.523 |
| 6744 | RP11-798M19.6 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 19 | WWTR1 | Sponge network | -0.612 | 0.00094 | -0.345 | 0.11997 | 0.523 |
| 6745 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | LPP | Sponge network | -0.021 | 0.93598 | -0.459 | 0.04475 | 0.523 |
| 6746 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 38 | PBX1 | Sponge network | -0.709 | 0.18168 | -1.206 | 0 | 0.523 |
| 6747 | MIR143HG |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935 | 45 | ABI2 | Sponge network | -0.739 | 0.0574 | 0.175 | 0.14787 | 0.523 |
| 6748 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-625-5p | 13 | DCLK1 | Sponge network | -0.842 | 0.01361 | -1.324 | 0.00014 | 0.523 |
| 6749 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 51 | SOX5 | Sponge network | -2.46 | 0 | -1.091 | 4.0E-5 | 0.523 |
| 6750 | MIR143HG |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 19 | ID4 | Sponge network | -0.739 | 0.0574 | -1.644 | 0 | 0.523 |
| 6751 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-425-5p | 20 | ATRX | Sponge network | 0.082 | 0.55397 | 0.261 | 0.04408 | 0.523 |
| 6752 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PTGFR | Sponge network | 0.532 | 0.00505 | -0.233 | 0.45421 | 0.523 |
| 6753 | TPTEP1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 20 | SORCS1 | Sponge network | -1.216 | 0 | -3.492 | 0 | 0.523 |
| 6754 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 39 | PHC3 | Sponge network | 0.782 | 0.00016 | 0.212 | 0.0499 | 0.523 |
| 6755 | MIR497HG |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3913-5p | 10 | RIMS1 | Sponge network | -1.439 | 4.0E-5 | -3.143 | 0 | 0.523 |
| 6756 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p | 27 | EBF1 | Sponge network | -0.465 | 0.04703 | -0.384 | 0.08856 | 0.523 |
| 6757 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | SSBP2 | Sponge network | -1.161 | 0.00591 | -1.58 | 0 | 0.523 |
| 6758 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-654-3p | 21 | NR2C2 | Sponge network | 0.101 | 0.60919 | 0.21 | 0.03224 | 0.523 |
| 6759 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | ATRX | Sponge network | 0.101 | 0.60919 | 0.261 | 0.04408 | 0.523 |
| 6760 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 32 | GAS7 | Sponge network | -0.739 | 0.00044 | -0.555 | 0.01602 | 0.523 |
| 6761 | RP11-359E10.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p | 16 | NRK | Sponge network | 0.299 | 0.57722 | 0.656 | 0.19217 | 0.523 |
| 6762 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 28 | RSPO2 | Sponge network | -0.129 | 0.57177 | -3.038 | 0 | 0.523 |
| 6763 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 58 | THRB | Sponge network | -1.249 | 7.0E-5 | -1.117 | 0.0004 | 0.523 |
| 6764 | RP11-620J15.3 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-744-3p | 23 | IGFBP5 | Sponge network | -0.739 | 0.00044 | -0.806 | 0.00105 | 0.523 |
| 6765 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | KCNA6 | Sponge network | -1.349 | 0.00017 | -1.531 | 0.00013 | 0.523 |
| 6766 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-486-5p;hsa-miR-590-3p | 14 | NR2C2 | Sponge network | 0.986 | 0.01022 | 0.21 | 0.03224 | 0.523 |
| 6767 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | BNC2 | Sponge network | -0.769 | 0.00035 | -0.491 | 0.09988 | 0.523 |
| 6768 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-628-5p;hsa-miR-93-3p | 50 | RUNX1T1 | Sponge network | -0.154 | 0.78939 | -0.344 | 0.2374 | 0.523 |
| 6769 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 15 | LHX6 | Sponge network | -0.737 | 0.06182 | -0.087 | 0.69193 | 0.523 |
| 6770 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-5p | 12 | RCAN2 | Sponge network | -0.384 | 0.40652 | -0.33 | 0.15172 | 0.523 |
| 6771 | TPTEP1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 10 | CTNNA3 | Sponge network | -1.216 | 0 | -2.046 | 0.00019 | 0.523 |
| 6772 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | 0.374 | 0.20054 | -0.509 | 0.03957 | 0.523 |
| 6773 | RP11-166D19.1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-93-3p | 31 | CTIF | Sponge network | -0.576 | 0.10121 | -0.389 | 0.00549 | 0.523 |
| 6774 | CTD-2089N3.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-629-3p | 15 | IGFBP5 | Sponge network | -1.375 | 0.08642 | -0.806 | 0.00105 | 0.523 |
| 6775 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p | 12 | TAL1 | Sponge network | -0.469 | 0.08721 | -0.462 | 0.00574 | 0.523 |
| 6776 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | RASSF8 | Sponge network | 0.101 | 0.60919 | -0.122 | 0.65591 | 0.523 |
| 6777 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 53 | FAT3 | Sponge network | 0.249 | 0.35902 | -1.601 | 0.00051 | 0.523 |
| 6778 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p | 36 | FBXO32 | Sponge network | -0.901 | 0.00019 | -0.495 | 0.07561 | 0.523 |
| 6779 | AC003090.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 38 | GRIK3 | Sponge network | -2.117 | 0.00161 | -3.208 | 0 | 0.523 |
| 6780 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-374b-5p;hsa-miR-424-5p | 10 | KCNAB1 | Sponge network | 0.178 | 0.29106 | -0.755 | 2.0E-5 | 0.523 |
| 6781 | RP11-359E10.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | PRUNE2 | Sponge network | 0.299 | 0.57722 | -1.733 | 0.00022 | 0.523 |
| 6782 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b | 11 | CHD2 | Sponge network | 0.594 | 0.00064 | 0.049 | 0.55371 | 0.523 |
| 6783 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 37 | SOX5 | Sponge network | -0.842 | 0.01361 | -1.091 | 4.0E-5 | 0.523 |
| 6784 | HAND2-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 21 | ITGA5 | Sponge network | -1.697 | 0.00889 | 0.452 | 0.03524 | 0.523 |
| 6785 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 31 | JAZF1 | Sponge network | -0.096 | 0.67958 | -0.377 | 0.02677 | 0.523 |
| 6786 | MAGI2-AS3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 20 | INPP4A | Sponge network | -0.129 | 0.57177 | 0.07 | 0.53563 | 0.523 |
| 6787 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p | 17 | HIP1 | Sponge network | -0.384 | 0.40652 | 0.774 | 0 | 0.523 |
| 6788 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 18 | HOOK3 | Sponge network | -0.576 | 0.10121 | 0.273 | 0.09957 | 0.523 |
| 6789 | CTC-429P9.3 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p | 12 | HERC1 | Sponge network | 0.082 | 0.55397 | -0.196 | 0.08544 | 0.523 |
| 6790 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 13 | DNAJB4 | Sponge network | -0.611 | 0.09607 | -0.7 | 1.0E-5 | 0.523 |
| 6791 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 10 | RARB | Sponge network | -1.092 | 4.0E-5 | -0.825 | 2.0E-5 | 0.523 |
| 6792 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 17 | TXNIP | Sponge network | -1.281 | 0.00012 | -1.277 | 0 | 0.523 |
| 6793 | RP11-473I1.10 |
hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | ABL2 | Sponge network | 0.673 | 0 | 0.82 | 0 | 0.523 |
| 6794 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | PLSCR4 | Sponge network | -0.376 | 0.00337 | -0.474 | 0.03833 | 0.523 |
| 6795 | CTD-2554C21.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-96-5p | 27 | ASTN1 | Sponge network | -1.654 | 0.00144 | -2.389 | 2.0E-5 | 0.523 |
| 6796 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TMEFF2 | Sponge network | -1.057 | 0.00029 | -2.426 | 0 | 0.523 |
| 6797 | LINC00702 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | TAL1 | Sponge network | -0.737 | 0.06182 | -0.462 | 0.00574 | 0.523 |
| 6798 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 24 | ARHGAP29 | Sponge network | -0.129 | 0.57177 | 0.131 | 0.42953 | 0.523 |
| 6799 | CTD-2314G24.2 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p | 20 | NRK | Sponge network | 0.246 | 0.59912 | 0.656 | 0.19217 | 0.523 |
| 6800 | AC003090.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DCN | Sponge network | -2.117 | 0.00161 | -1.288 | 0 | 0.523 |
| 6801 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 36 | MCC | Sponge network | -0.793 | 7.0E-5 | -1.005 | 0 | 0.523 |
| 6802 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | -0.323 | 0.01372 | -0.7 | 1.0E-5 | 0.523 |
| 6803 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 33 | MYO9A | Sponge network | 0.532 | 0.00505 | -0.147 | 0.32373 | 0.523 |
| 6804 | RP11-182J1.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-24-3p | 10 | MRVI1 | Sponge network | -0.187 | 0.23787 | -1.051 | 0.00022 | 0.523 |
| 6805 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | CACNA2D1 | Sponge network | -0.769 | 0.00035 | -0.846 | 0.00675 | 0.523 |
| 6806 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | UBE2QL1 | Sponge network | -1.697 | 0.00889 | -2.417 | 0 | 0.523 |
| 6807 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 51 | RBMS3 | Sponge network | -0.942 | 0 | -0.519 | 0.03551 | 0.522 |
| 6808 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | PTPN11 | Sponge network | 1.055 | 0 | 0.431 | 2.0E-5 | 0.522 |
| 6809 | MYLK-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p | 10 | TLE4 | Sponge network | -1.331 | 3.0E-5 | -1.157 | 0 | 0.522 |
| 6810 | CTD-2006H14.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | KIF5B | Sponge network | 1.378 | 0 | 0.362 | 0.00066 | 0.522 |
| 6811 | EMX2OS |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | MAPK10 | Sponge network | -0.777 | 0.1134 | -1.774 | 0 | 0.522 |
| 6812 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-550a-3p;hsa-miR-708-5p;hsa-miR-877-5p;hsa-miR-935 | 28 | RAP1A | Sponge network | -2.62 | 0 | -0.929 | 0 | 0.522 |
| 6813 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 30 | ANK2 | Sponge network | -0.222 | 0.32445 | -1.603 | 1.0E-5 | 0.522 |
| 6814 | RP11-867G23.10 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-5p | 34 | DST | Sponge network | -2.531 | 0 | -0.713 | 0.00096 | 0.522 |
| 6815 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-654-3p;hsa-miR-874-3p | 14 | NR2C2 | Sponge network | 0.856 | 0 | 0.21 | 0.03224 | 0.522 |
| 6816 | LINC00865 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 53 | CADM2 | Sponge network | -1.249 | 7.0E-5 | -3.782 | 0 | 0.522 |
| 6817 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-629-5p;hsa-miR-877-5p | 14 | GRIN2A | Sponge network | -1.654 | 0.00144 | -2.161 | 0 | 0.522 |
| 6818 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429 | 11 | PDZD2 | Sponge network | -2.62 | 0 | -0.909 | 3.0E-5 | 0.522 |
| 6819 | LINC00472 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 15 | TLL1 | Sponge network | -0.842 | 0.01361 | -1.195 | 3.0E-5 | 0.522 |
| 6820 | PCA3 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 54 | AR | Sponge network | -0.709 | 0.18168 | -1.554 | 0 | 0.522 |
| 6821 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | SETBP1 | Sponge network | -0.513 | 0.04703 | -1.302 | 1.0E-5 | 0.522 |
| 6822 | LINC00854 |
hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p | 13 | ABL2 | Sponge network | 1.18 | 0 | 0.82 | 0 | 0.522 |
| 6823 | MIR497HG |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | PAK3 | Sponge network | -1.439 | 4.0E-5 | -0.628 | 0.07253 | 0.522 |
| 6824 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-96-5p | 16 | FBXO32 | Sponge network | -1.582 | 8.0E-5 | -0.495 | 0.07561 | 0.522 |
| 6825 | RP11-356J5.12 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | RNF180 | Sponge network | -0.899 | 6.0E-5 | -1.127 | 1.0E-5 | 0.522 |
| 6826 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 14 | TLL1 | Sponge network | -0.896 | 0.00099 | -1.195 | 3.0E-5 | 0.522 |
| 6827 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-935 | 26 | DMD | Sponge network | -0.513 | 0.04703 | -1.506 | 0 | 0.522 |
| 6828 | TP73-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 30 | PCDH9 | Sponge network | -0.563 | 0.02545 | -1.823 | 4.0E-5 | 0.522 |
| 6829 | RP11-701H24.3 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 18 | MEF2C | Sponge network | -1.425 | 0.04204 | -0.499 | 0.01448 | 0.522 |
| 6830 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-589-5p | 33 | ZFHX3 | Sponge network | -0.384 | 0.40652 | 0.389 | 0.01363 | 0.522 |
| 6831 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p | 14 | RARB | Sponge network | -0.358 | 0.17255 | -0.825 | 2.0E-5 | 0.522 |
| 6832 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-877-5p | 50 | RBMS3 | Sponge network | -0.615 | 0.00015 | -0.519 | 0.03551 | 0.522 |
| 6833 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | DKK3 | Sponge network | -1.697 | 0.00889 | -0.052 | 0.77783 | 0.522 |
| 6834 | LINC00702 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | SEMA3C | Sponge network | -0.737 | 0.06182 | -0.272 | 0.31153 | 0.522 |
| 6835 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-7-5p | 51 | LPP | Sponge network | -0.469 | 0.08721 | -0.459 | 0.04475 | 0.522 |
| 6836 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 36 | ZFHX4 | Sponge network | 0.299 | 0.57722 | 0.018 | 0.95574 | 0.522 |
| 6837 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | RECK | Sponge network | -0.737 | 0.10822 | -1.043 | 0 | 0.522 |
| 6838 | RP11-13K12.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-532-3p | 16 | MRVI1 | Sponge network | -0.154 | 0.78939 | -1.051 | 0.00022 | 0.522 |
| 6839 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429 | 10 | SCN5A | Sponge network | -0.246 | 0.35034 | -0.259 | 0.48224 | 0.522 |
| 6840 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | SON | Sponge network | 1.254 | 0 | 0.168 | 0.04406 | 0.522 |
| 6841 | PCA3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | RASSF8 | Sponge network | -0.709 | 0.18168 | -0.122 | 0.65591 | 0.522 |
| 6842 | RP11-620J15.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | GRIK3 | Sponge network | -0.739 | 0.00044 | -3.208 | 0 | 0.522 |
| 6843 | GNG12-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p | 25 | PCDH9 | Sponge network | -0.793 | 7.0E-5 | -1.823 | 4.0E-5 | 0.522 |
| 6844 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-25-3p;hsa-miR-362-3p | 10 | DKK3 | Sponge network | -0.693 | 0.05629 | -0.052 | 0.77783 | 0.522 |
| 6845 | ADAMTS9-AS2 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NRG3 | Sponge network | -2.452 | 0 | -1.836 | 4.0E-5 | 0.522 |
| 6846 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 28 | PGR | Sponge network | -0.899 | 6.0E-5 | -1.704 | 0 | 0.522 |
| 6847 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 22 | NF1 | Sponge network | 0.959 | 0 | 0.51 | 0 | 0.522 |
| 6848 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | LRRC55 | Sponge network | -1.439 | 4.0E-5 | -0.026 | 0.93163 | 0.522 |
| 6849 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 16 | HOOK3 | Sponge network | 0.572 | 0.01722 | 0.273 | 0.09957 | 0.522 |
| 6850 | HOXB-AS1 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p | 22 | DIP2C | Sponge network | -0.154 | 0.53954 | -0.436 | 0.01713 | 0.522 |
| 6851 | THAP9-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p;hsa-miR-654-3p | 18 | PHF6 | Sponge network | 1.079 | 0 | 0.785 | 0 | 0.522 |
| 6852 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-96-5p | 13 | LRRC4 | Sponge network | -0.976 | 0.00025 | -1.774 | 0 | 0.522 |
| 6853 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-625-5p | 18 | RSPO2 | Sponge network | -2.531 | 0 | -3.038 | 0 | 0.522 |
| 6854 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | SPG20 | Sponge network | -2.117 | 0.00161 | -1.16 | 0 | 0.522 |
| 6855 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 27 | AKAP6 | Sponge network | 0.083 | 0.66405 | -1.586 | 3.0E-5 | 0.522 |
| 6856 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 21 | LRRC7 | Sponge network | -0.976 | 0.00025 | -1.341 | 0.01193 | 0.522 |
| 6857 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p | 12 | LATS2 | Sponge network | -1.092 | 4.0E-5 | 0.159 | 0.2304 | 0.522 |
| 6858 | LINC00578 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-589-3p | 20 | MCC | Sponge network | 0.811 | 0.03228 | -1.005 | 0 | 0.522 |
| 6859 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-590-3p | 21 | ADARB1 | Sponge network | 0.178 | 0.29106 | -0.335 | 0.06631 | 0.522 |
| 6860 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 40 | MEF2C | Sponge network | 0.532 | 0.00505 | -0.499 | 0.01448 | 0.522 |
| 6861 | RP3-337H4.8 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p | 10 | CBFB | Sponge network | 1.527 | 0 | 1.061 | 0 | 0.522 |
| 6862 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | RICTOR | Sponge network | 0.768 | 7.0E-5 | 0.388 | 0.00188 | 0.522 |
| 6863 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 69 | THRB | Sponge network | -2.46 | 0 | -1.117 | 0.0004 | 0.522 |
| 6864 | BVES-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-877-5p | 17 | CBFA2T3 | Sponge network | -2.62 | 0 | -1.221 | 1.0E-5 | 0.522 |
| 6865 | JAZF1-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 20 | SFRP1 | Sponge network | -0.769 | 0.00035 | -3.566 | 0 | 0.522 |
| 6866 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 13 | GLI3 | Sponge network | -0.738 | 0.21233 | -0.615 | 0.01482 | 0.522 |
| 6867 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940 | 22 | CRTC3 | Sponge network | -0.096 | 0.67958 | 0.164 | 0.16786 | 0.521 |
| 6868 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | LHFP | Sponge network | -0.487 | 0.02157 | -0.167 | 0.41371 | 0.521 |
| 6869 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | PDGFC | Sponge network | -1.697 | 0.00889 | -0.33 | 0.06483 | 0.521 |
| 6870 | RP1-228H13.5 |
hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-654-3p | 14 | PHF6 | Sponge network | 1.397 | 0 | 0.785 | 0 | 0.521 |
| 6871 | CTD-2089N3.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 19 | ZFHX4 | Sponge network | -1.375 | 0.08642 | 0.018 | 0.95574 | 0.521 |
| 6872 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | CDH19 | Sponge network | -0.376 | 0.00337 | -3.434 | 0 | 0.521 |
| 6873 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 24 | EBF3 | Sponge network | -0.487 | 0.02157 | -0.058 | 0.81437 | 0.521 |
| 6874 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | -0.976 | 0.00025 | 0.783 | 0.00072 | 0.521 |
| 6875 | CTC-429P9.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p | 24 | RUNX1T1 | Sponge network | 0.178 | 0.29106 | -0.344 | 0.2374 | 0.521 |
| 6876 | THAP9-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326 | 16 | CBFB | Sponge network | 1.079 | 0 | 1.061 | 0 | 0.521 |
| 6877 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 18 | THBS1 | Sponge network | -1.092 | 4.0E-5 | 0.741 | 0.00386 | 0.521 |
| 6878 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | SLITRK3 | Sponge network | -1.575 | 6.0E-5 | -3.328 | 0 | 0.521 |
| 6879 | FENDRR |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-3p | 15 | MACF1 | Sponge network | -1.281 | 0.00012 | 0.019 | 0.90335 | 0.521 |
| 6880 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 13 | ZNF382 | Sponge network | -0.487 | 0.02157 | -0.494 | 0.03831 | 0.521 |
| 6881 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3E | Sponge network | -0.49 | 0.04946 | -1.717 | 0.00137 | 0.521 |
| 6882 | LINC00092 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 10 | TAL1 | Sponge network | -1.886 | 0 | -0.462 | 0.00574 | 0.521 |
| 6883 | THAP9-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 20 | TBL1XR1 | Sponge network | 1.079 | 0 | 0.578 | 0 | 0.521 |
| 6884 | RP11-166D19.1 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | SPOP | Sponge network | -0.576 | 0.10121 | -0.419 | 1.0E-5 | 0.521 |
| 6885 | ZNF667-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-942-5p;hsa-miR-96-5p | 11 | ZBTB20 | Sponge network | -0.976 | 0.00025 | -0.122 | 0.57569 | 0.521 |
| 6886 | MIR497HG |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-942-5p;hsa-miR-96-5p | 14 | ZBTB20 | Sponge network | -1.439 | 4.0E-5 | -0.122 | 0.57569 | 0.521 |
| 6887 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 34 | ANK2 | Sponge network | -0.899 | 6.0E-5 | -1.603 | 1.0E-5 | 0.521 |
| 6888 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 27 | ATRX | Sponge network | 0.75 | 0.00054 | 0.261 | 0.04408 | 0.521 |
| 6889 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-96-5p | 18 | ABI2 | Sponge network | -0.517 | 0.02935 | 0.175 | 0.14787 | 0.521 |
| 6890 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 27 | NRXN1 | Sponge network | -0.421 | 0.0114 | -3.778 | 0 | 0.521 |
| 6891 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 12 | AFF3 | Sponge network | -0.376 | 0.00337 | -2.373 | 0 | 0.521 |
| 6892 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 10 | SETD2 | Sponge network | 0.683 | 1.0E-5 | 0.031 | 0.73068 | 0.521 |
| 6893 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-582-5p | 25 | RICTOR | Sponge network | 1.093 | 0 | 0.388 | 0.00188 | 0.521 |
| 6894 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | PLSCR4 | Sponge network | 0.532 | 0.00505 | -0.474 | 0.03833 | 0.521 |
| 6895 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-664a-5p | 22 | TRPS1 | Sponge network | -0.583 | 0.14589 | -0.191 | 0.44109 | 0.521 |
| 6896 | USP3-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-744-3p | 26 | PTPN14 | Sponge network | -0.162 | 0.47687 | 0.144 | 0.34595 | 0.521 |
| 6897 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-935 | 19 | LRRC7 | Sponge network | -0.162 | 0.47687 | -1.341 | 0.01193 | 0.521 |
| 6898 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | HOOK3 | Sponge network | -0.021 | 0.93598 | 0.273 | 0.09957 | 0.521 |
| 6899 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-766-3p | 24 | CHD5 | Sponge network | -0.517 | 0.02935 | -0.922 | 0.01521 | 0.521 |
| 6900 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | FAT4 | Sponge network | 0.727 | 3.0E-5 | -0.354 | 0.14694 | 0.521 |
| 6901 | FGF14-AS2 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-5p | 13 | PRKCB | Sponge network | -1.582 | 8.0E-5 | -1.313 | 2.0E-5 | 0.521 |
| 6902 | FAM13A-AS1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | NR3C2 | Sponge network | -0.83 | 0 | -1.56 | 0 | 0.521 |
| 6903 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-589-3p | 25 | SCN9A | Sponge network | -0.162 | 0.47687 | -0.525 | 0.10593 | 0.521 |
| 6904 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | IGSF10 | Sponge network | -0.563 | 0.02545 | -1.751 | 1.0E-5 | 0.521 |
| 6905 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | ABI2 | Sponge network | 0.194 | 0.64278 | 0.175 | 0.14787 | 0.521 |
| 6906 | AC010226.4 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-96-5p | 13 | SFRP1 | Sponge network | -0.811 | 2.0E-5 | -3.566 | 0 | 0.521 |
| 6907 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-625-5p | 17 | ST6GALNAC3 | Sponge network | -0.469 | 0.08721 | -0.987 | 0 | 0.521 |
| 6908 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 47 | NTRK2 | Sponge network | -0.576 | 0.10121 | -0.736 | 0.06495 | 0.521 |
| 6909 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-874-3p;hsa-miR-98-5p | 10 | ST5 | Sponge network | -1.281 | 0.00012 | -0.78 | 0 | 0.521 |
| 6910 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p | 30 | RICTOR | Sponge network | 1.601 | 0 | 0.388 | 0.00188 | 0.521 |
| 6911 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-539-5p | 22 | DDX6 | Sponge network | 0.594 | 0.00064 | 0.091 | 0.36428 | 0.521 |
| 6912 | FAM157B |
hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3614-5p | 18 | ABL2 | Sponge network | 0.92 | 0 | 0.82 | 0 | 0.521 |
| 6913 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-96-5p | 47 | PIK3R1 | Sponge network | 0.064 | 0.72934 | -0.133 | 0.36854 | 0.521 |
| 6914 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 26 | GRIN2A | Sponge network | -1.203 | 0.03881 | -2.161 | 0 | 0.521 |
| 6915 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | ABCA10 | Sponge network | -2.46 | 0 | -0.571 | 0.03674 | 0.521 |
| 6916 | LINC01018 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 33 | DMD | Sponge network | -2.915 | 0.00015 | -1.506 | 0 | 0.521 |
| 6917 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 80 | NTRK3 | Sponge network | -1.216 | 0 | -1.77 | 3.0E-5 | 0.521 |
| 6918 | RP11-307B6.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p | 14 | DLC1 | Sponge network | -4.463 | 0.00025 | -0.382 | 0.05038 | 0.521 |
| 6919 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | TACC1 | Sponge network | 0.335 | 0.20596 | -0.362 | 0.05406 | 0.521 |
| 6920 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 23 | IL6ST | Sponge network | 0.693 | 0.00076 | -0.402 | 0.00676 | 0.521 |
| 6921 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-874-3p | 35 | TACC1 | Sponge network | -2.62 | 0 | -0.362 | 0.05406 | 0.521 |
| 6922 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-5p | 26 | IGF1 | Sponge network | -1.203 | 0.03881 | -0.204 | 0.5898 | 0.521 |
| 6923 | FAM66C |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 45 | TRPS1 | Sponge network | -0.901 | 0.00019 | -0.191 | 0.44109 | 0.521 |
| 6924 | RP11-434B12.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | -0.031 | 0.89063 | 0.142 | 0.04938 | 0.521 |
| 6925 | LINC00472 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p | 79 | LPP | Sponge network | -0.842 | 0.01361 | -0.459 | 0.04475 | 0.521 |
| 6926 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | EPHA7 | Sponge network | -1.161 | 0.00591 | -2.197 | 3.0E-5 | 0.521 |
| 6927 | AC009948.5 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | RECK | Sponge network | 0.532 | 0.00505 | -1.043 | 0 | 0.521 |
| 6928 | CTD-2528L19.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | CREB1 | Sponge network | 0.953 | 0 | 0.203 | 0.00537 | 0.521 |
| 6929 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 15 | CITED2 | Sponge network | -0.769 | 0.00035 | -1.545 | 0 | 0.521 |
| 6930 | RP11-890B15.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 30 | DLC1 | Sponge network | 0.101 | 0.60919 | -0.382 | 0.05038 | 0.521 |
| 6931 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 40 | JAZF1 | Sponge network | -1.697 | 0.00889 | -0.377 | 0.02677 | 0.521 |
| 6932 | LINC00957 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | -0.421 | 0.0114 | -0.704 | 0.00106 | 0.521 |
| 6933 | ZNF667-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-484;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 28 | CTIF | Sponge network | -0.976 | 0.00025 | -0.389 | 0.00549 | 0.521 |
| 6934 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | TACC1 | Sponge network | -1.281 | 0.00012 | -0.362 | 0.05406 | 0.521 |
| 6935 | HAND2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ERG | Sponge network | -1.697 | 0.00889 | 0.002 | 0.99011 | 0.521 |
| 6936 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 56 | DTNA | Sponge network | -0.899 | 6.0E-5 | -1.648 | 1.0E-5 | 0.521 |
| 6937 | RP11-25K19.1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-450b-5p | 13 | FNBP1 | Sponge network | -1.057 | 0.00029 | -0.969 | 4.0E-5 | 0.521 |
| 6938 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | RCAN2 | Sponge network | 0.174 | 0.30034 | -0.33 | 0.15172 | 0.52 |
| 6939 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-942-5p | 42 | THRB | Sponge network | -2.62 | 0 | -1.117 | 0.0004 | 0.52 |
| 6940 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | PCDH9 | Sponge network | -0.769 | 0.00035 | -1.823 | 4.0E-5 | 0.52 |
| 6941 | LINC00641 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | NIN | Sponge network | -0.222 | 0.32445 | -0.253 | 0.13794 | 0.52 |
| 6942 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-96-5p | 24 | RASSF8 | Sponge network | -1.654 | 0.00144 | -0.122 | 0.65591 | 0.52 |
| 6943 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | IGSF10 | Sponge network | -0.323 | 0.01372 | -1.751 | 1.0E-5 | 0.52 |
| 6944 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-7-5p | 24 | BTG2 | Sponge network | -1.331 | 3.0E-5 | -1.763 | 0 | 0.52 |
| 6945 | LINC00641 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -0.222 | 0.32445 | -2.046 | 0.00019 | 0.52 |
| 6946 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-501-5p | 11 | MN1 | Sponge network | -0.777 | 0.16667 | -0.358 | 0.24172 | 0.52 |
| 6947 | CTD-2006H14.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | ABL2 | Sponge network | 1.378 | 0 | 0.82 | 0 | 0.52 |
| 6948 | RP11-531A24.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 25 | BRD3 | Sponge network | 0.75 | 0.00054 | 0.37 | 0.00013 | 0.52 |
| 6949 | RP11-473I1.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p | 25 | AFF4 | Sponge network | 0.542 | 0.00054 | -0.266 | 0.01565 | 0.52 |
| 6950 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | APC | Sponge network | -0.222 | 0.32445 | -0.255 | 0.04051 | 0.52 |
| 6951 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 16 | ZNF292 | Sponge network | 0.848 | 0 | 0.276 | 0.04148 | 0.52 |
| 6952 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-874-3p;hsa-miR-93-5p | 31 | TACC1 | Sponge network | -0.384 | 0.40652 | -0.362 | 0.05406 | 0.52 |
| 6953 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-940 | 28 | SNX29 | Sponge network | 0.214 | 0.18246 | -0.34 | 0.01371 | 0.52 |
| 6954 | LINC00476 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935 | 40 | DMD | Sponge network | -0.615 | 0.00015 | -1.506 | 0 | 0.52 |
| 6955 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | HACE1 | Sponge network | -0.222 | 0.32445 | -0.072 | 0.55239 | 0.52 |
| 6956 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 14 | ITGB3 | Sponge network | -0.769 | 0.00035 | -0.011 | 0.95751 | 0.52 |
| 6957 | LINC00865 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-7-1-3p | 27 | EYA4 | Sponge network | -1.249 | 7.0E-5 | 0.3 | 0.36876 | 0.52 |
| 6958 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 22 | KCNA6 | Sponge network | -2.117 | 0.00161 | -1.531 | 0.00013 | 0.52 |
| 6959 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DNAJB4 | Sponge network | -1.281 | 0.00012 | -0.7 | 1.0E-5 | 0.52 |
| 6960 | RP11-589P10.7 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-671-5p | 12 | RBMS3 | Sponge network | -2.044 | 0.00066 | -0.519 | 0.03551 | 0.52 |
| 6961 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p | 51 | IL6ST | Sponge network | -2.62 | 0 | -0.402 | 0.00676 | 0.52 |
| 6962 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 38 | ZFHX4 | Sponge network | -0.611 | 0.09607 | 0.018 | 0.95574 | 0.52 |
| 6963 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p | 17 | STARD13 | Sponge network | -0.323 | 0.01372 | -0.187 | 0.27383 | 0.52 |
| 6964 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | LRRC7 | Sponge network | 0.194 | 0.64278 | -1.341 | 0.01193 | 0.52 |
| 6965 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 42 | MEF2C | Sponge network | -2.117 | 0.00161 | -0.499 | 0.01448 | 0.52 |
| 6966 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 21 | ADAMTSL3 | Sponge network | -0.513 | 0.04703 | -1.559 | 0 | 0.52 |
| 6967 | RP11-819C21.1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | RBBP6 | Sponge network | 0.537 | 0.00139 | 0.163 | 0.05101 | 0.52 |
| 6968 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-421 | 11 | ZNF521 | Sponge network | -0.487 | 0.02157 | -0.111 | 0.58068 | 0.52 |
| 6969 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 32 | RASSF8 | Sponge network | -0.777 | 0.1134 | -0.122 | 0.65591 | 0.52 |
| 6970 | MEG3 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 18 | MAPK10 | Sponge network | 0.249 | 0.35902 | -1.774 | 0 | 0.52 |
| 6971 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-539-5p | 34 | PBX1 | Sponge network | -0.842 | 0.01361 | -1.206 | 0 | 0.52 |
| 6972 | AC010226.4 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 32 | AR | Sponge network | -0.811 | 2.0E-5 | -1.554 | 0 | 0.52 |
| 6973 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-378a-5p | 32 | PBX1 | Sponge network | -1.161 | 0.00591 | -1.206 | 0 | 0.52 |
| 6974 | RP11-307B6.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-589-3p | 14 | MCC | Sponge network | -4.463 | 0.00025 | -1.005 | 0 | 0.52 |
| 6975 | MIR143HG |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | ZMYND11 | Sponge network | -0.739 | 0.0574 | -0.2 | 0.05449 | 0.52 |
| 6976 | CTD-3064M3.3 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-5p | 13 | PTGFR | Sponge network | -0.469 | 0.08721 | -0.233 | 0.45421 | 0.52 |
| 6977 | ZRANB2-AS1 |
hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p | 10 | RASSF8 | Sponge network | -0.376 | 0.00337 | -0.122 | 0.65591 | 0.52 |
| 6978 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 20 | TGFBR2 | Sponge network | -2.452 | 0 | -0.243 | 0.11408 | 0.52 |
| 6979 | RP4-639F20.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-96-5p | 21 | ASTN1 | Sponge network | -0.517 | 0.02935 | -2.389 | 2.0E-5 | 0.52 |
| 6980 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | ST6GALNAC3 | Sponge network | -0.513 | 0.04703 | -0.987 | 0 | 0.52 |
| 6981 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-532-3p | 15 | CADM3 | Sponge network | -1.057 | 0.00029 | -3.663 | 0 | 0.52 |
| 6982 | FENDRR |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 17 | CAV1 | Sponge network | -1.281 | 0.00012 | -1.013 | 6.0E-5 | 0.52 |
| 6983 | ZNF667-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 12 | PHOX2B | Sponge network | -0.976 | 0.00025 | -2.68 | 0 | 0.52 |
| 6984 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-326;hsa-miR-330-5p | 17 | CNTN1 | Sponge network | -2.095 | 0.00112 | -2.594 | 0 | 0.52 |
| 6985 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 53 | ZFHX4 | Sponge network | -1.216 | 0 | 0.018 | 0.95574 | 0.52 |
| 6986 | PWAR6 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | PRDM5 | Sponge network | -1.575 | 6.0E-5 | -1.09 | 0.00014 | 0.52 |
| 6987 | TP73-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-935 | 13 | STAT5B | Sponge network | -0.563 | 0.02545 | -0.182 | 0.1082 | 0.52 |
| 6988 | LINC00473 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-769-5p | 19 | SYNPO2 | Sponge network | -1.203 | 0.03881 | -2.342 | 0 | 0.52 |
| 6989 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | TLL1 | Sponge network | -1.349 | 0.00017 | -1.195 | 3.0E-5 | 0.52 |
| 6990 | TPTEP1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 43 | IGFBP5 | Sponge network | -1.216 | 0 | -0.806 | 0.00105 | 0.52 |
| 6991 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-29b-1-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-582-5p | 15 | PHC3 | Sponge network | 0.993 | 0 | 0.212 | 0.0499 | 0.52 |
| 6992 | RP11-532F6.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | RYR2 | Sponge network | -0.896 | 0.00099 | -1.466 | 0.00031 | 0.52 |
| 6993 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-361-5p;hsa-miR-590-3p | 23 | DLC1 | Sponge network | -0.739 | 0.00044 | -0.382 | 0.05038 | 0.52 |
| 6994 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-93-5p | 16 | EDA2R | Sponge network | 0.249 | 0.35902 | -0.587 | 0.01598 | 0.52 |
| 6995 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 44 | EPHA7 | Sponge network | -0.842 | 0.01361 | -2.197 | 3.0E-5 | 0.52 |
| 6996 | TP73-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-378a-3p;hsa-miR-92a-3p | 13 | FLNA | Sponge network | -0.563 | 0.02545 | -0.771 | 0.01377 | 0.52 |
| 6997 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p | 14 | TXNIP | Sponge network | -0.487 | 0.02157 | -1.277 | 0 | 0.52 |
| 6998 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 27 | ARHGAP28 | Sponge network | -1.281 | 0.00012 | -0.911 | 0.00013 | 0.52 |
| 6999 | FAM157C |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-511-5p | 10 | TRIM33 | Sponge network | 0.91 | 0 | 0.402 | 7.0E-5 | 0.52 |
| 7000 | RP11-119F7.5 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 10 | RNF180 | Sponge network | 0.225 | 0.30644 | -1.127 | 1.0E-5 | 0.52 |
| 7001 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-576-5p | 17 | QKI | Sponge network | -2.039 | 0.03447 | -0.333 | 0.04111 | 0.52 |
| 7002 | ACTA2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-96-5p | 35 | IL17RD | Sponge network | 0.194 | 0.64278 | -0.019 | 0.94873 | 0.52 |
| 7003 | MRVI1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-423-5p | 16 | CBFA2T3 | Sponge network | -1.092 | 4.0E-5 | -1.221 | 1.0E-5 | 0.52 |
| 7004 | PWAR6 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | LRIG1 | Sponge network | -1.575 | 6.0E-5 | -0.537 | 0.00588 | 0.52 |
| 7005 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-3p | 13 | SETBP1 | Sponge network | -0.154 | 0.53954 | -1.302 | 1.0E-5 | 0.519 |
| 7006 | HCG18 |
hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p | 12 | SHPRH | Sponge network | 1.063 | 0 | 0.098 | 0.40994 | 0.519 |
| 7007 | SNHG14 |
hsa-miR-185-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-542-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-940 | 11 | ILK | Sponge network | -1.111 | 0.0003 | -0.74 | 0 | 0.519 |
| 7008 | ARHGEF26-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | ZDBF2 | Sponge network | -2.46 | 0 | -0.998 | 0.00025 | 0.519 |
| 7009 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | IGSF10 | Sponge network | -0.351 | 0.11358 | -1.751 | 1.0E-5 | 0.519 |
| 7010 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-942-5p | 33 | HLF | Sponge network | -0.842 | 0.01361 | -2.443 | 0 | 0.519 |
| 7011 | RP11-536O18.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-3065-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-502-5p | 16 | BNC2 | Sponge network | -0.074 | 0.90758 | -0.491 | 0.09988 | 0.519 |
| 7012 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-628-5p | 13 | TRIM33 | Sponge network | 0.93 | 0 | 0.402 | 7.0E-5 | 0.519 |
| 7013 | EMX2OS |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 19 | SPG20 | Sponge network | -0.777 | 0.1134 | -1.16 | 0 | 0.519 |
| 7014 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | PDGFC | Sponge network | 0.335 | 0.20596 | -0.33 | 0.06483 | 0.519 |
| 7015 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | BNC2 | Sponge network | -0.811 | 2.0E-5 | -0.491 | 0.09988 | 0.519 |
| 7016 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 17 | PAK3 | Sponge network | -0.576 | 0.10121 | -0.628 | 0.07253 | 0.519 |
| 7017 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | RECK | Sponge network | -1.057 | 0.00029 | -1.043 | 0 | 0.519 |
| 7018 | CTC-559E9.6 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-940 | 26 | MDM4 | Sponge network | 0.601 | 1.0E-5 | 0.345 | 0.00762 | 0.519 |
| 7019 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p | 22 | RASSF8 | Sponge network | -0.738 | 0.21233 | -0.122 | 0.65591 | 0.519 |
| 7020 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 27 | ZFHX3 | Sponge network | 0.389 | 0.04293 | 0.389 | 0.01363 | 0.519 |
| 7021 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 20 | EBF1 | Sponge network | -4.546 | 0 | -0.384 | 0.08856 | 0.519 |
| 7022 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | NCAM2 | Sponge network | 0.335 | 0.20596 | -0.482 | 0.22565 | 0.519 |
| 7023 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-940;hsa-miR-96-5p | 36 | MYO9A | Sponge network | 0.082 | 0.55654 | -0.147 | 0.32373 | 0.519 |
| 7024 | PWAR6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 11 | SMARCA1 | Sponge network | -1.575 | 6.0E-5 | -0.684 | 0.00159 | 0.519 |
| 7025 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-590-3p | 13 | JAKMIP2 | Sponge network | -0.421 | 0.0114 | -1.388 | 0 | 0.519 |
| 7026 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | EDA2R | Sponge network | -0.739 | 0.0574 | -0.587 | 0.01598 | 0.519 |
| 7027 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 46 | RUNX1T1 | Sponge network | -1.161 | 0.00591 | -0.344 | 0.2374 | 0.519 |
| 7028 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 20 | LATS2 | Sponge network | -0.976 | 0.00025 | 0.159 | 0.2304 | 0.519 |
| 7029 | LINC00957 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | NRXN3 | Sponge network | -0.421 | 0.0114 | -0.893 | 0.04186 | 0.519 |
| 7030 | LINC00092 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 10 | ERG | Sponge network | -1.886 | 0 | 0.002 | 0.99011 | 0.519 |
| 7031 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 21 | GRIN2A | Sponge network | -0.896 | 0.00099 | -2.161 | 0 | 0.519 |
| 7032 | ZNF582-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-940 | 10 | FAM117A | Sponge network | -1.349 | 0.00017 | -1.379 | 0 | 0.519 |
| 7033 | BOLA3-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | TET1 | Sponge network | -0.246 | 0.35034 | -0.135 | 0.52138 | 0.519 |
| 7034 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p | 11 | LATS2 | Sponge network | -0.583 | 0.14589 | 0.159 | 0.2304 | 0.519 |
| 7035 | LENG8-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-625-3p | 14 | ABL2 | Sponge network | 1.471 | 0 | 0.82 | 0 | 0.519 |
| 7036 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 59 | PTPRD | Sponge network | -0.129 | 0.57177 | -1.005 | 0.00064 | 0.519 |
| 7037 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-508-3p;hsa-miR-590-3p | 11 | RIMS1 | Sponge network | -2.46 | 0 | -3.143 | 0 | 0.519 |
| 7038 | OIP5-AS1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 25 | ATM | Sponge network | 0.421 | 0.00084 | 0.178 | 0.21568 | 0.519 |
| 7039 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p | 12 | PHC3 | Sponge network | 0.858 | 3.0E-5 | 0.212 | 0.0499 | 0.519 |
| 7040 | TPTEP1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 54 | GRIK3 | Sponge network | -1.216 | 0 | -3.208 | 0 | 0.519 |
| 7041 | SNHG14 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | FYN | Sponge network | -1.111 | 0.0003 | -0.131 | 0.37051 | 0.519 |
| 7042 | LINC01001 |
hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-326 | 14 | RICTOR | Sponge network | 1.055 | 0 | 0.388 | 0.00188 | 0.519 |
| 7043 | MEG3 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | 0.249 | 0.35902 | -0.998 | 0.00025 | 0.519 |
| 7044 | FENDRR |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TPO | Sponge network | -1.281 | 0.00012 | -1.87 | 2.0E-5 | 0.519 |
| 7045 | LINC00702 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p | 18 | MAFB | Sponge network | -0.737 | 0.06182 | -0.023 | 0.89865 | 0.519 |
| 7046 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 44 | CHD5 | Sponge network | -1.697 | 0.00889 | -0.922 | 0.01521 | 0.519 |
| 7047 | LINC00578 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 11 | PTGFR | Sponge network | 0.811 | 0.03228 | -0.233 | 0.45421 | 0.519 |
| 7048 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-7-1-3p | 10 | MLF1 | Sponge network | -0.563 | 0.02545 | -0.893 | 0.00727 | 0.519 |
| 7049 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | LRP1B | Sponge network | -2.46 | 0 | -2.163 | 0 | 0.519 |
| 7050 | ACTA2-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | INPP4A | Sponge network | 0.194 | 0.64278 | 0.07 | 0.53563 | 0.519 |
| 7051 | LINC00957 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 34 | RBMS3 | Sponge network | -0.421 | 0.0114 | -0.519 | 0.03551 | 0.519 |
| 7052 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-940 | 39 | NTRK2 | Sponge network | -0.976 | 0.00025 | -0.736 | 0.06495 | 0.519 |
| 7053 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 24 | TBL1XR1 | Sponge network | 0.683 | 1.0E-5 | 0.578 | 0 | 0.519 |
| 7054 | LINC00265 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p | 11 | RASAL2 | Sponge network | 1.07 | 0 | 0.869 | 0 | 0.519 |
| 7055 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | ABCB5 | Sponge network | -0.129 | 0.57177 | -0.956 | 0.04077 | 0.519 |
| 7056 | RP11-473I1.9 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p | 22 | DDX3X | Sponge network | 0.683 | 1.0E-5 | -0.152 | 0.07686 | 0.519 |
| 7057 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-582-3p;hsa-miR-93-5p | 20 | ADARB1 | Sponge network | -0.473 | 0.07602 | -0.335 | 0.06631 | 0.519 |
| 7058 | LINC00092 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | AKAP6 | Sponge network | -1.886 | 0 | -1.586 | 3.0E-5 | 0.519 |
| 7059 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | RECK | Sponge network | -0.421 | 0.0114 | -1.043 | 0 | 0.519 |
| 7060 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p | 27 | ATRX | Sponge network | 0.921 | 0.00118 | 0.261 | 0.04408 | 0.519 |
| 7061 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 34 | DIP2C | Sponge network | 0.572 | 0.01722 | -0.436 | 0.01713 | 0.519 |
| 7062 | PWAR6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 50 | NTRK2 | Sponge network | -1.575 | 6.0E-5 | -0.736 | 0.06495 | 0.519 |
| 7063 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | -0.738 | 0.21233 | 0.358 | 0.13727 | 0.519 |
| 7064 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TLL1 | Sponge network | 0.335 | 0.20596 | -1.195 | 3.0E-5 | 0.519 |
| 7065 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 22 | LRRC55 | Sponge network | -0.563 | 0.02545 | -0.026 | 0.93163 | 0.519 |
| 7066 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 53 | TCF4 | Sponge network | -0.842 | 0.01361 | 0.016 | 0.92271 | 0.519 |
| 7067 | RP11-13K12.5 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-940 | 11 | MAPK10 | Sponge network | -0.318 | 0.65847 | -1.774 | 0 | 0.519 |
| 7068 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 30 | MCC | Sponge network | -0.896 | 0.00099 | -1.005 | 0 | 0.519 |
| 7069 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ARHGEF12 | Sponge network | 0.673 | 0 | 0.09 | 0.35693 | 0.519 |
| 7070 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p | 11 | KCNAB1 | Sponge network | 0.082 | 0.55397 | -0.755 | 2.0E-5 | 0.519 |
| 7071 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | DAB2 | Sponge network | 0.335 | 0.20596 | 0.431 | 0.0113 | 0.519 |
| 7072 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 69 | NTRK3 | Sponge network | 0.249 | 0.35902 | -1.77 | 3.0E-5 | 0.519 |
| 7073 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 38 | RUNX1T1 | Sponge network | 0.176 | 0.37402 | -0.344 | 0.2374 | 0.519 |
| 7074 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | NTRK3 | Sponge network | -0.811 | 2.0E-5 | -1.77 | 3.0E-5 | 0.519 |
| 7075 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 34 | DDX6 | Sponge network | 0.683 | 1.0E-5 | 0.091 | 0.36428 | 0.519 |
| 7076 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | 1.063 | 0 | 0.323 | 0.00792 | 0.519 |
| 7077 | PCA3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | KCNA6 | Sponge network | -0.709 | 0.18168 | -1.531 | 0.00013 | 0.519 |
| 7078 | LINC00092 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-532-3p;hsa-miR-542-3p | 12 | SYNM | Sponge network | -1.886 | 0 | -2.309 | 1.0E-5 | 0.519 |
| 7079 | RP11-819C21.1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p | 13 | STAG2 | Sponge network | 0.537 | 0.00139 | 0.252 | 0.00438 | 0.519 |
| 7080 | LINC01018 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 26 | RIMS2 | Sponge network | -2.915 | 0.00015 | -1.365 | 0.00208 | 0.519 |
| 7081 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p | 12 | FGFR1 | Sponge network | 1.344 | 0 | -0.509 | 0.03957 | 0.519 |
| 7082 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 32 | SETBP1 | Sponge network | -0.942 | 0 | -1.302 | 1.0E-5 | 0.519 |
| 7083 | MEG3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | IL17RD | Sponge network | 0.249 | 0.35902 | -0.019 | 0.94873 | 0.519 |
| 7084 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 12 | ABCB5 | Sponge network | -0.358 | 0.17255 | -0.956 | 0.04077 | 0.519 |
| 7085 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | EBF1 | Sponge network | -1.349 | 0.00017 | -0.384 | 0.08856 | 0.519 |
| 7086 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-96-5p | 34 | FBXO32 | Sponge network | 0.331 | 0.57247 | -0.495 | 0.07561 | 0.519 |
| 7087 | RP11-867G23.10 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-93-3p | 14 | WWTR1 | Sponge network | -2.531 | 0 | -0.345 | 0.11997 | 0.519 |
| 7088 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | ABCB5 | Sponge network | -0.739 | 0.0574 | -0.956 | 0.04077 | 0.519 |
| 7089 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | PAK3 | Sponge network | -0.976 | 0.00025 | -0.628 | 0.07253 | 0.519 |
| 7090 | GNG12-AS1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-590-3p | 17 | ITPKB | Sponge network | -0.793 | 7.0E-5 | -0.704 | 0.00106 | 0.519 |
| 7091 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 26 | BCL2 | Sponge network | -0.811 | 2.0E-5 | -0.899 | 0 | 0.518 |
| 7092 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p | 17 | ABCA10 | Sponge network | -0.49 | 0.04946 | -0.571 | 0.03674 | 0.518 |
| 7093 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 15 | RCAN2 | Sponge network | 0.082 | 0.55654 | -0.33 | 0.15172 | 0.518 |
| 7094 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-7-5p;hsa-miR-874-3p | 20 | CRTC3 | Sponge network | -0.487 | 0.02157 | 0.164 | 0.16786 | 0.518 |
| 7095 | KB-1572G7.2 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 25 | MDM4 | Sponge network | 0.924 | 0 | 0.345 | 0.00762 | 0.518 |
| 7096 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | MXI1 | Sponge network | -0.612 | 0.00094 | -0.987 | 0 | 0.518 |
| 7097 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 24 | RUNX1T1 | Sponge network | -2.076 | 0.00548 | -0.344 | 0.2374 | 0.518 |
| 7098 | CTD-2554C21.2 |
hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429 | 13 | KIT | Sponge network | -1.654 | 0.00144 | -2.184 | 0 | 0.518 |
| 7099 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-503-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 49 | RICTOR | Sponge network | 0.421 | 0.00084 | 0.388 | 0.00188 | 0.518 |
| 7100 | LINC00702 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | ITGB1 | Sponge network | -0.737 | 0.06182 | 0.524 | 0.00017 | 0.518 |
| 7101 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-744-3p;hsa-miR-935 | 51 | FAT3 | Sponge network | -1.249 | 7.0E-5 | -1.601 | 0.00051 | 0.518 |
| 7102 | TUG1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 16 | NF1 | Sponge network | 0.972 | 0 | 0.51 | 0 | 0.518 |
| 7103 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | SYNE1 | Sponge network | -0.222 | 0.32445 | -0.738 | 0.00316 | 0.518 |
| 7104 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 20 | ZBTB4 | Sponge network | -0.811 | 2.0E-5 | -0.739 | 0 | 0.518 |
| 7105 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | ANK2 | Sponge network | -2.039 | 0.03447 | -1.603 | 1.0E-5 | 0.518 |
| 7106 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-93-5p | 24 | SYNE1 | Sponge network | -0.777 | 0.16667 | -0.738 | 0.00316 | 0.518 |
| 7107 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 26 | TBL1XR1 | Sponge network | 1.153 | 0 | 0.578 | 0 | 0.518 |
| 7108 | LINC00865 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-7-1-3p | 12 | DCN | Sponge network | -1.249 | 7.0E-5 | -1.288 | 0 | 0.518 |
| 7109 | SNX29P2 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | PRKCB | Sponge network | 0.4 | 0.14611 | -1.313 | 2.0E-5 | 0.518 |
| 7110 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 42 | AR | Sponge network | -0.246 | 0.35034 | -1.554 | 0 | 0.518 |
| 7111 | FENDRR |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | TAL1 | Sponge network | -1.281 | 0.00012 | -0.462 | 0.00574 | 0.518 |
| 7112 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-3p | 18 | SLITRK3 | Sponge network | 0.194 | 0.64278 | -3.328 | 0 | 0.518 |
| 7113 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-590-5p | 18 | PDZD2 | Sponge network | -2.46 | 0 | -0.909 | 3.0E-5 | 0.518 |
| 7114 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | PDGFC | Sponge network | -1.216 | 0 | -0.33 | 0.06483 | 0.518 |
| 7115 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | TIMP3 | Sponge network | 0.889 | 9.0E-5 | -0.546 | 0.02307 | 0.518 |
| 7116 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-362-3p;hsa-miR-93-5p | 15 | AR | Sponge network | -1.255 | 0.00981 | -1.554 | 0 | 0.518 |
| 7117 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-508-3p;hsa-miR-590-3p | 13 | ROBO1 | Sponge network | -0.358 | 0.17255 | -0.009 | 0.96154 | 0.518 |
| 7118 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 41 | BCL2 | Sponge network | -0.615 | 0.00015 | -0.899 | 0 | 0.518 |
| 7119 | MEG3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | ERG | Sponge network | 0.249 | 0.35902 | 0.002 | 0.99011 | 0.518 |
| 7120 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 57 | RUNX1T1 | Sponge network | -0.563 | 0.02545 | -0.344 | 0.2374 | 0.518 |
| 7121 | RP11-999E24.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p | 14 | NAV3 | Sponge network | -0.693 | 0.05629 | 0.212 | 0.47729 | 0.518 |
| 7122 | MRVI1-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p | 12 | TLE4 | Sponge network | -1.092 | 4.0E-5 | -1.157 | 0 | 0.518 |
| 7123 | NEAT1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | ZNF638 | Sponge network | 0.705 | 0.00014 | 0.22 | 0.01214 | 0.518 |
| 7124 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-664a-3p;hsa-miR-93-5p | 16 | CNTNAP3 | Sponge network | -2.69 | 0.0001 | -1.465 | 0.00033 | 0.518 |
| 7125 | RP11-999E24.3 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-362-3p | 10 | CADM3 | Sponge network | -0.693 | 0.05629 | -3.663 | 0 | 0.518 |
| 7126 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | ST6GALNAC3 | Sponge network | -0.563 | 0.02545 | -0.987 | 0 | 0.518 |
| 7127 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | PDGFRA | Sponge network | 0.572 | 0.01722 | -0.372 | 0.0037 | 0.518 |
| 7128 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | CACNA2D1 | Sponge network | 0.194 | 0.64278 | -0.846 | 0.00675 | 0.518 |
| 7129 | FAM66C |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | PREX2 | Sponge network | -0.901 | 0.00019 | -0.272 | 0.25382 | 0.518 |
| 7130 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p | 22 | THBS1 | Sponge network | -0.358 | 0.17255 | 0.741 | 0.00386 | 0.518 |
| 7131 | TP73-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-769-5p | 22 | SYNPO2 | Sponge network | -0.563 | 0.02545 | -2.342 | 0 | 0.518 |
| 7132 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 24 | CADM1 | Sponge network | -0.513 | 0.04703 | -0.9 | 0.00084 | 0.518 |
| 7133 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 17 | DKK3 | Sponge network | -1.281 | 0.00012 | -0.052 | 0.77783 | 0.518 |
| 7134 | TRAF3IP2-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | TPO | Sponge network | -0.323 | 0.01372 | -1.87 | 2.0E-5 | 0.518 |
| 7135 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 19 | RCAN2 | Sponge network | 0.997 | 0.00066 | -0.33 | 0.15172 | 0.518 |
| 7136 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 13 | LRRK2 | Sponge network | 0.997 | 0.00066 | -1.136 | 1.0E-5 | 0.518 |
| 7137 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 35 | ANK2 | Sponge network | -2.925 | 0 | -1.603 | 1.0E-5 | 0.518 |
| 7138 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-577 | 18 | AFF3 | Sponge network | -1.349 | 0.00017 | -2.373 | 0 | 0.518 |
| 7139 | MIR497HG |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-629-3p | 12 | ITGA5 | Sponge network | -1.439 | 4.0E-5 | 0.452 | 0.03524 | 0.518 |
| 7140 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-335-3p;hsa-miR-34a-5p | 16 | PRKAA2 | Sponge network | -0.691 | 0.00146 | -2.462 | 0 | 0.518 |
| 7141 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-486-5p | 21 | FGF5 | Sponge network | -0.162 | 0.47687 | 0.549 | 0.28659 | 0.518 |
| 7142 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-660-5p | 22 | ESR1 | Sponge network | -4.546 | 0 | -1.035 | 3.0E-5 | 0.518 |
| 7143 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 37 | NOTCH2 | Sponge network | -0.487 | 0.02157 | 0.138 | 0.35163 | 0.518 |
| 7144 | RP11-33B1.4 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-424-5p | 12 | ABL2 | Sponge network | 0.826 | 0 | 0.82 | 0 | 0.518 |
| 7145 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 15 | GLI3 | Sponge network | -0.777 | 0.16667 | -0.615 | 0.01482 | 0.518 |
| 7146 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | TRIM33 | Sponge network | 1.055 | 0 | 0.402 | 7.0E-5 | 0.518 |
| 7147 | GNG12-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | -0.793 | 7.0E-5 | -0.615 | 0.01482 | 0.518 |
| 7148 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TBX18 | Sponge network | 0.997 | 0.00066 | 0.843 | 0.02945 | 0.518 |
| 7149 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 22 | CDH23 | Sponge network | -0.737 | 0.06182 | -0.974 | 0.00034 | 0.518 |
| 7150 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p | 12 | DKK3 | Sponge network | -0.469 | 0.08721 | -0.052 | 0.77783 | 0.518 |
| 7151 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-625-5p;hsa-miR-93-5p | 36 | JAZF1 | Sponge network | -0.162 | 0.47687 | -0.377 | 0.02677 | 0.518 |
| 7152 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-628-5p;hsa-miR-629-3p | 16 | CHD2 | Sponge network | 0.637 | 0.00069 | 0.049 | 0.55371 | 0.518 |
| 7153 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | RECK | Sponge network | 0.889 | 9.0E-5 | -1.043 | 0 | 0.518 |
| 7154 | SNHG14 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | FAM117A | Sponge network | -1.111 | 0.0003 | -1.379 | 0 | 0.518 |
| 7155 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-539-5p | 20 | EPHA7 | Sponge network | -2.076 | 0.00548 | -2.197 | 3.0E-5 | 0.518 |
| 7156 | LINC00672 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-590-3p | 13 | NRXN3 | Sponge network | -0.227 | 0.31657 | -0.893 | 0.04186 | 0.518 |
| 7157 | LINC00472 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-766-3p;hsa-miR-93-5p | 16 | EZH1 | Sponge network | -0.842 | 0.01361 | -0.564 | 1.0E-5 | 0.518 |
| 7158 | RP11-774O3.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p | 10 | ZBTB20 | Sponge network | -0.487 | 0.02157 | -0.122 | 0.57569 | 0.518 |
| 7159 | LINC00472 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | -0.842 | 0.01361 | -2.633 | 0 | 0.518 |
| 7160 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | DCLK1 | Sponge network | -0.811 | 2.0E-5 | -1.324 | 0.00014 | 0.518 |
| 7161 | LINC00094 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-330-5p | 16 | ABL1 | Sponge network | 0.253 | 0.09565 | -0.215 | 0.07804 | 0.518 |
| 7162 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p | 30 | ADARB1 | Sponge network | -0.739 | 0.00044 | -0.335 | 0.06631 | 0.518 |
| 7163 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-935 | 48 | FAT3 | Sponge network | -0.405 | 0.22013 | -1.601 | 0.00051 | 0.518 |
| 7164 | MEG3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 21 | SFRP1 | Sponge network | 0.249 | 0.35902 | -3.566 | 0 | 0.518 |
| 7165 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 50 | PTPRD | Sponge network | -0.576 | 0.10121 | -1.005 | 0.00064 | 0.518 |
| 7166 | PART1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 19 | ID4 | Sponge network | -2.925 | 0 | -1.644 | 0 | 0.518 |
| 7167 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 12 | NCAM2 | Sponge network | -0.376 | 0.00337 | -0.482 | 0.22565 | 0.518 |
| 7168 | FGF14-AS2 |
hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | ERG | Sponge network | -1.582 | 8.0E-5 | 0.002 | 0.99011 | 0.517 |
| 7169 | LINC00641 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | PEG3 | Sponge network | -0.222 | 0.32445 | -1.74 | 0 | 0.517 |
| 7170 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SON | Sponge network | 0.673 | 0 | 0.168 | 0.04406 | 0.517 |
| 7171 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SYNE1 | Sponge network | -1.057 | 0.00029 | -0.738 | 0.00316 | 0.517 |
| 7172 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p | 19 | RCAN2 | Sponge network | -0.615 | 0.00015 | -0.33 | 0.15172 | 0.517 |
| 7173 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | PLSCR4 | Sponge network | 0.299 | 0.57722 | -0.474 | 0.03833 | 0.517 |
| 7174 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-625-3p | 34 | BMPR2 | Sponge network | -0.358 | 0.17255 | 0.206 | 0.0635 | 0.517 |
| 7175 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 32 | DDX6 | Sponge network | 0.614 | 1.0E-5 | 0.091 | 0.36428 | 0.517 |
| 7176 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 29 | ZMYM2 | Sponge network | 1.063 | 0 | 0.279 | 0.01273 | 0.517 |
| 7177 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 45 | TGFBR3 | Sponge network | -2.46 | 0 | -1.173 | 1.0E-5 | 0.517 |
| 7178 | CTD-2089N3.2 |
hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-590-3p | 17 | BNC2 | Sponge network | -1.375 | 0.08642 | -0.491 | 0.09988 | 0.517 |
| 7179 | RP11-999E24.3 |
hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p | 13 | PCDH9 | Sponge network | -0.693 | 0.05629 | -1.823 | 4.0E-5 | 0.517 |
| 7180 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-589-5p;hsa-miR-93-5p | 47 | PRKAA2 | Sponge network | -0.384 | 0.40652 | -2.462 | 0 | 0.517 |
| 7181 | FAM66C |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | ZDBF2 | Sponge network | -0.901 | 0.00019 | -0.998 | 0.00025 | 0.517 |
| 7182 | FAM157A |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326 | 14 | RICTOR | Sponge network | 0.813 | 1.0E-5 | 0.388 | 0.00188 | 0.517 |
| 7183 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-576-5p | 12 | CACNA2D1 | Sponge network | -0.932 | 0.00329 | -0.846 | 0.00675 | 0.517 |
| 7184 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-421;hsa-miR-424-5p | 13 | RASSF8 | Sponge network | -4.463 | 0.00025 | -0.122 | 0.65591 | 0.517 |
| 7185 | RP11-166D19.1 |
hsa-miR-10b-3p;hsa-miR-193b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p | 10 | LRRTM1 | Sponge network | -0.576 | 0.10121 | -3.361 | 0 | 0.517 |
| 7186 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 27 | SON | Sponge network | 0.683 | 1.0E-5 | 0.168 | 0.04406 | 0.517 |
| 7187 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CREB1 | Sponge network | 0.673 | 0 | 0.203 | 0.00537 | 0.517 |
| 7188 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-7-1-3p | 26 | TGFBR3 | Sponge network | -4.546 | 0 | -1.173 | 1.0E-5 | 0.517 |
| 7189 | LINC00472 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-7-1-3p | 14 | NRXN3 | Sponge network | -0.842 | 0.01361 | -0.893 | 0.04186 | 0.517 |
| 7190 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-589-5p;hsa-miR-98-5p | 29 | ZFHX3 | Sponge network | 0.082 | 0.55397 | 0.389 | 0.01363 | 0.517 |
| 7191 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 36 | SNX29 | Sponge network | 0.997 | 0.00066 | -0.34 | 0.01371 | 0.517 |
| 7192 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-877-5p | 18 | CBFA2T3 | Sponge network | -0.465 | 0.04703 | -1.221 | 1.0E-5 | 0.517 |
| 7193 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 24 | TGFBR3 | Sponge network | -0.517 | 0.02935 | -1.173 | 1.0E-5 | 0.517 |
| 7194 | AGAP11 |
hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-590-5p | 11 | SLC12A6 | Sponge network | -1.645 | 0 | -0.343 | 0.00043 | 0.517 |
| 7195 | AC106786.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-629-3p;hsa-miR-942-5p | 18 | IGFBP5 | Sponge network | 1.122 | 0.02989 | -0.806 | 0.00105 | 0.517 |
| 7196 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | TRIM33 | Sponge network | 1.205 | 0 | 0.402 | 7.0E-5 | 0.517 |
| 7197 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | DCLK1 | Sponge network | -1.216 | 0 | -1.324 | 0.00014 | 0.517 |
| 7198 | ATP1B3-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p | 17 | MDM4 | Sponge network | 1.2 | 0.01813 | 0.345 | 0.00762 | 0.517 |
| 7199 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-421;hsa-miR-429;hsa-miR-96-5p | 21 | CNTN1 | Sponge network | -1.654 | 0.00144 | -2.594 | 0 | 0.517 |
| 7200 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | NUAK1 | Sponge network | 0.249 | 0.35902 | 0.904 | 0 | 0.517 |
| 7201 | HCG18 |
hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-664a-3p | 19 | RASAL2 | Sponge network | 1.063 | 0 | 0.869 | 0 | 0.517 |
| 7202 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | TSHZ3 | Sponge network | -0.612 | 0.00094 | -0.556 | 0.01516 | 0.517 |
| 7203 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-98-5p | 28 | PRRX1 | Sponge network | -0.49 | 0.04946 | 1.316 | 0 | 0.517 |
| 7204 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 15 | EDA2R | Sponge network | -2.452 | 0 | -0.587 | 0.01598 | 0.517 |
| 7205 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 26 | GAS7 | Sponge network | -0.376 | 0.00337 | -0.555 | 0.01602 | 0.517 |
| 7206 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 34 | NRXN1 | Sponge network | -0.901 | 0.00019 | -3.778 | 0 | 0.517 |
| 7207 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 28 | MYO9A | Sponge network | 0.331 | 0.57247 | -0.147 | 0.32373 | 0.517 |
| 7208 | AC138035.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-766-3p | 20 | MDM4 | Sponge network | 1.073 | 0 | 0.345 | 0.00762 | 0.517 |
| 7209 | FAM66C |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | TET1 | Sponge network | -0.901 | 0.00019 | -0.135 | 0.52138 | 0.517 |
| 7210 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 13 | MITF | Sponge network | -0.583 | 0.14589 | -0.769 | 0.00022 | 0.517 |
| 7211 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p | 11 | CNTNAP1 | Sponge network | -1.654 | 0.00144 | 0.358 | 0.13727 | 0.517 |
| 7212 | SNHG14 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 12 | HIC1 | Sponge network | -1.111 | 0.0003 | 0.024 | 0.89889 | 0.517 |
| 7213 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-429 | 12 | SYNM | Sponge network | -0.738 | 0.21233 | -2.309 | 1.0E-5 | 0.517 |
| 7214 | RP11-345P4.7 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-33a-5p;hsa-miR-424-5p | 15 | ABL2 | Sponge network | 1.597 | 0 | 0.82 | 0 | 0.517 |
| 7215 | FGF14-AS2 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 11 | SYT4 | Sponge network | -1.582 | 8.0E-5 | -3.207 | 0 | 0.517 |
| 7216 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 13 | DCLK1 | Sponge network | 0.299 | 0.57722 | -1.324 | 0.00014 | 0.517 |
| 7217 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | 0.374 | 0.20054 | -0.474 | 0.03833 | 0.517 |
| 7218 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | RB1CC1 | Sponge network | 0.91 | 0 | 0.175 | 0.12559 | 0.517 |
| 7219 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p | 12 | ABL1 | Sponge network | -0.517 | 0.02935 | -0.215 | 0.07804 | 0.517 |
| 7220 | SNHG14 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-942-5p | 12 | BMPR1A | Sponge network | -1.111 | 0.0003 | -0.366 | 0.01036 | 0.517 |
| 7221 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | KCNA6 | Sponge network | 0.335 | 0.20596 | -1.531 | 0.00013 | 0.517 |
| 7222 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 18 | LRP1B | Sponge network | -1.092 | 4.0E-5 | -2.163 | 0 | 0.517 |
| 7223 | LINC01018 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | KCTD8 | Sponge network | -2.915 | 0.00015 | -3.359 | 0 | 0.517 |
| 7224 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-98-5p | 18 | AKAP6 | Sponge network | 0.082 | 0.55397 | -1.586 | 3.0E-5 | 0.517 |
| 7225 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p;hsa-miR-98-5p | 57 | PRKAA2 | Sponge network | -0.942 | 0 | -2.462 | 0 | 0.517 |
| 7226 | TP73-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 20 | SFRP1 | Sponge network | -0.563 | 0.02545 | -3.566 | 0 | 0.517 |
| 7227 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 43 | PIK3R1 | Sponge network | -0.487 | 0.02157 | -0.133 | 0.36854 | 0.517 |
| 7228 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 53 | TCF4 | Sponge network | 0.225 | 0.30644 | 0.016 | 0.92271 | 0.517 |
| 7229 | LINC00702 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-93-3p | 19 | RPS6KA2 | Sponge network | -0.737 | 0.06182 | -0.57 | 0.00013 | 0.517 |
| 7230 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-629-3p | 23 | ZBTB4 | Sponge network | -0.154 | 0.53954 | -0.739 | 0 | 0.517 |
| 7231 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 21 | TGFBR2 | Sponge network | -0.769 | 0.00035 | -0.243 | 0.11408 | 0.517 |
| 7232 | RP5-1198O20.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | IL17RD | Sponge network | 0.997 | 0.00066 | -0.019 | 0.94873 | 0.517 |
| 7233 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -0.563 | 0.02545 | 0.024 | 0.89889 | 0.517 |
| 7234 | HAND2-AS1 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 12 | MLLT11 | Sponge network | -1.697 | 0.00889 | -0.661 | 0.01733 | 0.517 |
| 7235 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935 | 41 | FAT3 | Sponge network | -0.421 | 0.0114 | -1.601 | 0.00051 | 0.517 |
| 7236 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | TCF4 | Sponge network | 1.153 | 0.00114 | 0.016 | 0.92271 | 0.517 |
| 7237 | RP11-532F6.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-935 | 30 | DIP2C | Sponge network | -0.896 | 0.00099 | -0.436 | 0.01713 | 0.517 |
| 7238 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-935 | 16 | SSBP2 | Sponge network | -0.691 | 0.00146 | -1.58 | 0 | 0.517 |
| 7239 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -0.769 | 0.00035 | 0.024 | 0.89889 | 0.517 |
| 7240 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 46 | EPHA5 | Sponge network | 0.194 | 0.64278 | -0.957 | 0.01733 | 0.517 |
| 7241 | RP11-464F9.1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | ATM | Sponge network | 0.959 | 0 | 0.178 | 0.21568 | 0.516 |
| 7242 | AC003090.1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | PAK3 | Sponge network | -2.117 | 0.00161 | -0.628 | 0.07253 | 0.516 |
| 7243 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | NRXN3 | Sponge network | 0.246 | 0.59912 | -0.893 | 0.04186 | 0.516 |
| 7244 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | PDGFRA | Sponge network | -0.376 | 0.00337 | -0.372 | 0.0037 | 0.516 |
| 7245 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 45 | DIP2C | Sponge network | -0.942 | 0 | -0.436 | 0.01713 | 0.516 |
| 7246 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 10 | DMTF1 | Sponge network | 0.267 | 0.2937 | 0.248 | 0.02726 | 0.516 |
| 7247 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 22 | GLI3 | Sponge network | -2.46 | 0 | -0.615 | 0.01482 | 0.516 |
| 7248 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-664a-3p | 19 | GREM1 | Sponge network | -0.358 | 0.17255 | 0.902 | 0.01691 | 0.516 |
| 7249 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 74 | BMPR2 | Sponge network | -0.739 | 0.0574 | 0.206 | 0.0635 | 0.516 |
| 7250 | GAS6-AS2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-664a-3p;hsa-miR-7-1-3p | 14 | PRUNE2 | Sponge network | 0.16 | 0.50785 | -1.733 | 0.00022 | 0.516 |
| 7251 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | -0.942 | 0 | -0.684 | 0.00159 | 0.516 |
| 7252 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | LRRC7 | Sponge network | -0.49 | 0.04946 | -1.341 | 0.01193 | 0.516 |
| 7253 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-429;hsa-miR-629-5p;hsa-miR-96-5p | 27 | SOX5 | Sponge network | -1.654 | 0.00144 | -1.091 | 4.0E-5 | 0.516 |
| 7254 | RP11-400K9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-940 | 19 | GAS7 | Sponge network | -0.733 | 0.19469 | -0.555 | 0.01602 | 0.516 |
| 7255 | RP11-434H6.7 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-32-5p | 20 | MDM4 | Sponge network | 0.358 | 0.14302 | 0.345 | 0.00762 | 0.516 |
| 7256 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-624-5p | 14 | LATS1 | Sponge network | 0.673 | 0 | 0.109 | 0.34208 | 0.516 |
| 7257 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 17 | ZMYM2 | Sponge network | 1.923 | 0 | 0.279 | 0.01273 | 0.516 |
| 7258 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 47 | CHD5 | Sponge network | -0.737 | 0.06182 | -0.922 | 0.01521 | 0.516 |
| 7259 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 45 | EPHA7 | Sponge network | -0.615 | 0.00015 | -2.197 | 3.0E-5 | 0.516 |
| 7260 | C20orf166-AS1 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-455-5p | 12 | PTGFR | Sponge network | -2.69 | 0.0001 | -0.233 | 0.45421 | 0.516 |
| 7261 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 20 | FAT4 | Sponge network | -0.096 | 0.67958 | -0.354 | 0.14694 | 0.516 |
| 7262 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | CAV1 | Sponge network | -2.46 | 0 | -1.013 | 6.0E-5 | 0.516 |
| 7263 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | ZNF521 | Sponge network | -0.769 | 0.00035 | -0.111 | 0.58068 | 0.516 |
| 7264 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 32 | RSPO2 | Sponge network | -1.697 | 0.00889 | -3.038 | 0 | 0.516 |
| 7265 | SNHG14 |
hsa-miR-126-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p | 10 | TNC | Sponge network | -1.111 | 0.0003 | 0.313 | 0.33705 | 0.516 |
| 7266 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-96-5p | 26 | CBFA2T3 | Sponge network | -0.563 | 0.02545 | -1.221 | 1.0E-5 | 0.516 |
| 7267 | RP11-144G6.12 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 23 | ARHGEF12 | Sponge network | 0.826 | 1.0E-5 | 0.09 | 0.35693 | 0.516 |
| 7268 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-429 | 13 | ELMO1 | Sponge network | -0.129 | 0.57177 | 0.04 | 0.83147 | 0.516 |
| 7269 | LINC00865 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-7-1-3p | 25 | MED12L | Sponge network | -1.249 | 7.0E-5 | -0.194 | 0.55299 | 0.516 |
| 7270 | AC003090.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | KCTD8 | Sponge network | -2.117 | 0.00161 | -3.359 | 0 | 0.516 |
| 7271 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-452-5p;hsa-miR-671-5p | 25 | BNC2 | Sponge network | -2.095 | 0.00112 | -0.491 | 0.09988 | 0.516 |
| 7272 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-93-5p | 14 | SLITRK3 | Sponge network | -1.654 | 0.00144 | -3.328 | 0 | 0.516 |
| 7273 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-455-3p;hsa-miR-550a-5p;hsa-miR-7-1-3p | 18 | TSHZ3 | Sponge network | 0.693 | 0.00076 | -0.556 | 0.01516 | 0.516 |
| 7274 | FENDRR |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 35 | WWTR1 | Sponge network | -1.281 | 0.00012 | -0.345 | 0.11997 | 0.516 |
| 7275 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-511-5p | 17 | ATRX | Sponge network | 0.497 | 0.01451 | 0.261 | 0.04408 | 0.516 |
| 7276 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 28 | SCN9A | Sponge network | 0.194 | 0.64278 | -0.525 | 0.10593 | 0.516 |
| 7277 | RP11-774O3.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | NAV3 | Sponge network | -0.487 | 0.02157 | 0.212 | 0.47729 | 0.516 |
| 7278 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-409-3p | 12 | PHC3 | Sponge network | 0.497 | 0.01451 | 0.212 | 0.0499 | 0.516 |
| 7279 | AC016747.3 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 10 | MSN | Sponge network | 0.199 | 0.38015 | -0.023 | 0.88422 | 0.516 |
| 7280 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | PBRM1 | Sponge network | -0.222 | 0.32445 | -0.029 | 0.78601 | 0.516 |
| 7281 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-628-5p | 21 | BCL6 | Sponge network | -0.976 | 0.00025 | -0.211 | 0.19489 | 0.516 |
| 7282 | RP11-819C21.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ARHGEF12 | Sponge network | 0.537 | 0.00139 | 0.09 | 0.35693 | 0.516 |
| 7283 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | 0.246 | 0.59912 | 0.783 | 0.00072 | 0.516 |
| 7284 | LINC00092 |
hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 29 | EPHA7 | Sponge network | -1.886 | 0 | -2.197 | 3.0E-5 | 0.516 |
| 7285 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 21 | KIT | Sponge network | -0.901 | 0.00019 | -2.184 | 0 | 0.516 |
| 7286 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 12 | RB1CC1 | Sponge network | 1.153 | 0 | 0.175 | 0.12559 | 0.516 |
| 7287 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | BCL2 | Sponge network | -0.72 | 0.24239 | -0.899 | 0 | 0.516 |
| 7288 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-935 | 34 | DMD | Sponge network | -0.246 | 0.35034 | -1.506 | 0 | 0.516 |
| 7289 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 34 | PGR | Sponge network | 0.532 | 0.00505 | -1.704 | 0 | 0.516 |
| 7290 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | CNTN1 | Sponge network | -0.942 | 0 | -2.594 | 0 | 0.516 |
| 7291 | C20orf166-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-664a-3p | 15 | NCAM2 | Sponge network | -2.69 | 0.0001 | -0.482 | 0.22565 | 0.516 |
| 7292 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940;hsa-miR-98-5p | 42 | PBX1 | Sponge network | -1.697 | 0.00889 | -1.206 | 0 | 0.516 |
| 7293 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-508-3p;hsa-miR-590-3p | 24 | FBXO32 | Sponge network | -0.358 | 0.17255 | -0.495 | 0.07561 | 0.516 |
| 7294 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-495-3p | 11 | BCLAF1 | Sponge network | 1.294 | 0 | 0.211 | 0.00684 | 0.516 |
| 7295 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p | 12 | CRTC3 | Sponge network | -0.358 | 0.17255 | 0.164 | 0.16786 | 0.516 |
| 7296 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | SON | Sponge network | 1.148 | 2.0E-5 | 0.168 | 0.04406 | 0.516 |
| 7297 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 43 | PBX1 | Sponge network | -2.925 | 0 | -1.206 | 0 | 0.516 |
| 7298 | RP11-53O19.3 |
hsa-miR-101-3p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | KIF5B | Sponge network | 1.205 | 0 | 0.362 | 0.00066 | 0.516 |
| 7299 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | ST6GALNAC3 | Sponge network | -1.203 | 0.03881 | -0.987 | 0 | 0.516 |
| 7300 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | ABI2 | Sponge network | 0.064 | 0.72934 | 0.175 | 0.14787 | 0.516 |
| 7301 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 30 | NRXN1 | Sponge network | -1.203 | 0.03881 | -3.778 | 0 | 0.516 |
| 7302 | RP11-983P16.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 23 | BRD3 | Sponge network | 0.41 | 0.02214 | 0.37 | 0.00013 | 0.516 |
| 7303 | RP11-356J5.12 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 88 | LPP | Sponge network | -0.899 | 6.0E-5 | -0.459 | 0.04475 | 0.516 |
| 7304 | CTC-296K1.4 |
hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-629-5p | 12 | GRIN2A | Sponge network | -2.076 | 0.00548 | -2.161 | 0 | 0.516 |
| 7305 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-3p | 13 | RB1CC1 | Sponge network | 0.537 | 0.00139 | 0.175 | 0.12559 | 0.516 |
| 7306 | ARHGEF26-AS1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ITPKB | Sponge network | -2.46 | 0 | -0.704 | 0.00106 | 0.515 |
| 7307 | RP5-1198O20.4 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ITPKB | Sponge network | 0.997 | 0.00066 | -0.704 | 0.00106 | 0.515 |
| 7308 | AC009948.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | CREB1 | Sponge network | 0.532 | 0.00505 | 0.203 | 0.00537 | 0.515 |
| 7309 | LINC00672 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | MRVI1 | Sponge network | -0.227 | 0.31657 | -1.051 | 0.00022 | 0.515 |
| 7310 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | AFF3 | Sponge network | -2.46 | 0 | -2.373 | 0 | 0.515 |
| 7311 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-452-5p | 28 | BCL2 | Sponge network | -2.62 | 0 | -0.899 | 0 | 0.515 |
| 7312 | RP11-526I2.5 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-495-3p | 13 | MDM4 | Sponge network | 1.054 | 1.0E-5 | 0.345 | 0.00762 | 0.515 |
| 7313 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DCN | Sponge network | -0.777 | 0.1134 | -1.288 | 0 | 0.515 |
| 7314 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -0.896 | 0.00099 | 0.024 | 0.89889 | 0.515 |
| 7315 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | PAK3 | Sponge network | 0.246 | 0.59912 | -0.628 | 0.07253 | 0.515 |
| 7316 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | EBF1 | Sponge network | 0.532 | 0.00505 | -0.384 | 0.08856 | 0.515 |
| 7317 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p | 33 | RBMS3 | Sponge network | -0.227 | 0.31657 | -0.519 | 0.03551 | 0.515 |
| 7318 | CKMT2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 18 | EZH1 | Sponge network | 0.082 | 0.55654 | -0.564 | 1.0E-5 | 0.515 |
| 7319 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p | 20 | FAT3 | Sponge network | -2.076 | 0.00548 | -1.601 | 0.00051 | 0.515 |
| 7320 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | -0.899 | 6.0E-5 | -1.548 | 0 | 0.515 |
| 7321 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p | 10 | TIMP3 | Sponge network | -0.309 | 0.1145 | -0.546 | 0.02307 | 0.515 |
| 7322 | TP73-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-671-5p | 29 | DLC1 | Sponge network | -0.563 | 0.02545 | -0.382 | 0.05038 | 0.515 |
| 7323 | LINC00865 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p | 11 | AMPH | Sponge network | -1.249 | 7.0E-5 | -0.916 | 5.0E-5 | 0.515 |
| 7324 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p | 22 | CTIF | Sponge network | -1.092 | 4.0E-5 | -0.389 | 0.00549 | 0.515 |
| 7325 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-542-3p | 17 | PCDH9 | Sponge network | -0.517 | 0.02935 | -1.823 | 4.0E-5 | 0.515 |
| 7326 | RP11-254F7.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-5p | 24 | DIP2C | Sponge network | 0.176 | 0.37402 | -0.436 | 0.01713 | 0.515 |
| 7327 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | BNC2 | Sponge network | 0.246 | 0.59912 | -0.491 | 0.09988 | 0.515 |
| 7328 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TBX18 | Sponge network | 0.335 | 0.20596 | 0.843 | 0.02945 | 0.515 |
| 7329 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-34a-5p | 15 | ZFHX4 | Sponge network | -0.309 | 0.1145 | 0.018 | 0.95574 | 0.515 |
| 7330 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-7-1-3p | 12 | DCN | Sponge network | -0.563 | 0.02545 | -1.288 | 0 | 0.515 |
| 7331 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-93-3p | 16 | CTIF | Sponge network | -0.611 | 0.09607 | -0.389 | 0.00549 | 0.515 |
| 7332 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p | 15 | AFF3 | Sponge network | -0.739 | 0.00044 | -2.373 | 0 | 0.515 |
| 7333 | RP11-119F7.5 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 15 | ATM | Sponge network | 0.225 | 0.30644 | 0.178 | 0.21568 | 0.515 |
| 7334 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 27 | GRIN2A | Sponge network | -0.942 | 0 | -2.161 | 0 | 0.515 |
| 7335 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 23 | EBF3 | Sponge network | -0.901 | 0.00019 | -0.058 | 0.81437 | 0.515 |
| 7336 | CTD-3064M3.3 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-874-3p | 15 | SORCS1 | Sponge network | -0.469 | 0.08721 | -3.492 | 0 | 0.515 |
| 7337 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | RCAN2 | Sponge network | -1.439 | 4.0E-5 | -0.33 | 0.15172 | 0.515 |
| 7338 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 42 | RUNX1T1 | Sponge network | 0.225 | 0.30644 | -0.344 | 0.2374 | 0.515 |
| 7339 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-3p | 19 | EBF3 | Sponge network | -0.154 | 0.53954 | -0.058 | 0.81437 | 0.515 |
| 7340 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | SYNE1 | Sponge network | 0.342 | 0.03129 | -0.738 | 0.00316 | 0.515 |
| 7341 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | PREX2 | Sponge network | -0.769 | 0.00035 | -0.272 | 0.25382 | 0.515 |
| 7342 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 38 | MCC | Sponge network | -0.709 | 0.18168 | -1.005 | 0 | 0.515 |
| 7343 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 35 | NRXN1 | Sponge network | -0.612 | 0.00094 | -3.778 | 0 | 0.515 |
| 7344 | CTC-429P9.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-769-5p;hsa-miR-93-5p | 14 | PRUNE2 | Sponge network | 0.082 | 0.55397 | -1.733 | 0.00022 | 0.515 |
| 7345 | HAND2-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 70 | DST | Sponge network | -1.697 | 0.00889 | -0.713 | 0.00096 | 0.515 |
| 7346 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 71 | TCF4 | Sponge network | -1.216 | 0 | 0.016 | 0.92271 | 0.515 |
| 7347 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | EXOC8 | Sponge network | 0.683 | 1.0E-5 | 0.216 | 0.00598 | 0.515 |
| 7348 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | AKAP11 | Sponge network | 1.294 | 0 | 0.196 | 0.09825 | 0.515 |
| 7349 | FENDRR |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p | 17 | ROBO1 | Sponge network | -1.281 | 0.00012 | -0.009 | 0.96154 | 0.515 |
| 7350 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | DYNC1I1 | Sponge network | -0.777 | 0.1134 | -1.12 | 0.00107 | 0.515 |
| 7351 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | KAT6B | Sponge network | -0.323 | 0.01372 | -0.478 | 0.00018 | 0.515 |
| 7352 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 40 | TGFBR3 | Sponge network | -0.487 | 0.02157 | -1.173 | 1.0E-5 | 0.515 |
| 7353 | HAND2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 11 | RIMS1 | Sponge network | -1.697 | 0.00889 | -3.143 | 0 | 0.515 |
| 7354 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-34a-5p | 15 | GPC6 | Sponge network | -1.331 | 3.0E-5 | -0.054 | 0.81465 | 0.515 |
| 7355 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | GALNT13 | Sponge network | -0.129 | 0.57177 | -0.857 | 0.07439 | 0.515 |
| 7356 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | RNF144A | Sponge network | 0.889 | 9.0E-5 | 0.594 | 0.0009 | 0.515 |
| 7357 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p | 35 | RUNX1T1 | Sponge network | 0.889 | 9.0E-5 | -0.344 | 0.2374 | 0.515 |
| 7358 | CTD-3064M3.3 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p | 25 | MEF2C | Sponge network | -0.469 | 0.08721 | -0.499 | 0.01448 | 0.515 |
| 7359 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | AHNAK | Sponge network | -1.281 | 0.00012 | -1.207 | 0 | 0.515 |
| 7360 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | ZNF208 | Sponge network | -0.021 | 0.93598 | -0.4 | 0.22381 | 0.515 |
| 7361 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | GAS7 | Sponge network | -1.654 | 0.00144 | -0.555 | 0.01602 | 0.515 |
| 7362 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-628-5p | 28 | FBXO32 | Sponge network | -1.161 | 0.00591 | -0.495 | 0.07561 | 0.515 |
| 7363 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-335-3p | 10 | GRIN2A | Sponge network | -0.691 | 0.00146 | -2.161 | 0 | 0.515 |
| 7364 | GABPB1-AS1 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 14 | ATM | Sponge network | 1.11 | 0 | 0.178 | 0.21568 | 0.515 |
| 7365 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 18 | RASSF8 | Sponge network | -0.611 | 0.09607 | -0.122 | 0.65591 | 0.515 |
| 7366 | MAGI2-AS3 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | HERC1 | Sponge network | -0.129 | 0.57177 | -0.196 | 0.08544 | 0.515 |
| 7367 | MIR497HG |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-577;hsa-miR-7-1-3p | 11 | CTGF | Sponge network | -1.439 | 4.0E-5 | 0.321 | 0.11682 | 0.515 |
| 7368 | RP11-305E6.4 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | BRD3 | Sponge network | 1.317 | 0 | 0.37 | 0.00013 | 0.515 |
| 7369 | NR2F1-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 39 | RIMS2 | Sponge network | -0.49 | 0.04946 | -1.365 | 0.00208 | 0.515 |
| 7370 | H19 |
hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p | 13 | NUAK1 | Sponge network | 2.439 | 0 | 0.904 | 0 | 0.515 |
| 7371 | FGF14-AS2 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 15 | RIMS2 | Sponge network | -1.582 | 8.0E-5 | -1.365 | 0.00208 | 0.514 |
| 7372 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-96-5p | 12 | PCDH8 | Sponge network | -0.563 | 0.02545 | -0.997 | 0.05366 | 0.514 |
| 7373 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 26 | ADAMTSL3 | Sponge network | 0.532 | 0.00505 | -1.559 | 0 | 0.514 |
| 7374 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-370-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-935 | 42 | SSBP2 | Sponge network | -0.896 | 0.00099 | -1.58 | 0 | 0.514 |
| 7375 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | -0.777 | 0.1134 | -0.7 | 1.0E-5 | 0.514 |
| 7376 | MIR497HG |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 13 | DACT1 | Sponge network | -1.439 | 4.0E-5 | 0.783 | 0.00072 | 0.514 |
| 7377 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | AFF3 | Sponge network | -1.697 | 0.00889 | -2.373 | 0 | 0.514 |
| 7378 | RP11-532F6.3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | KLF10 | Sponge network | -0.896 | 0.00099 | -0.783 | 0 | 0.514 |
| 7379 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-96-5p | 23 | FBXO32 | Sponge network | -1.654 | 0.00144 | -0.495 | 0.07561 | 0.514 |
| 7380 | RP11-798M19.6 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | CSDE1 | Sponge network | -0.612 | 0.00094 | -0.493 | 0 | 0.514 |
| 7381 | RP11-504P24.4 |
hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | RICTOR | Sponge network | 0.624 | 0.0004 | 0.388 | 0.00188 | 0.514 |
| 7382 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p | 12 | JAZF1 | Sponge network | 0.693 | 0.00076 | -0.377 | 0.02677 | 0.514 |
| 7383 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 17 | RSPO2 | Sponge network | -0.583 | 0.14589 | -3.038 | 0 | 0.514 |
| 7384 | PGM5-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-7-1-3p | 18 | RIMS2 | Sponge network | -4.546 | 0 | -1.365 | 0.00208 | 0.514 |
| 7385 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | ERG | Sponge network | -1.349 | 0.00017 | 0.002 | 0.99011 | 0.514 |
| 7386 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | PRKAA2 | Sponge network | 0.225 | 0.30644 | -2.462 | 0 | 0.514 |
| 7387 | LINC00672 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 22 | AKAP6 | Sponge network | -0.227 | 0.31657 | -1.586 | 3.0E-5 | 0.514 |
| 7388 | HOXB-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p | 23 | IGFBP5 | Sponge network | -0.154 | 0.53954 | -0.806 | 0.00105 | 0.514 |
| 7389 | DNM3OS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 47 | DTNA | Sponge network | 0.572 | 0.01722 | -1.648 | 1.0E-5 | 0.514 |
| 7390 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p | 12 | DACT1 | Sponge network | -0.693 | 0.05629 | 0.783 | 0.00072 | 0.514 |
| 7391 | DNM3OS |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | TLL1 | Sponge network | 0.572 | 0.01722 | -1.195 | 3.0E-5 | 0.514 |
| 7392 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p | 17 | PHC3 | Sponge network | 1.106 | 0 | 0.212 | 0.0499 | 0.514 |
| 7393 | LINC00641 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-125a-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-653-5p | 33 | UBR3 | Sponge network | -0.222 | 0.32445 | -0.021 | 0.82774 | 0.514 |
| 7394 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b | 11 | TXNIP | Sponge network | -2.62 | 0 | -1.277 | 0 | 0.514 |
| 7395 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-629-3p | 30 | RASSF2 | Sponge network | -0.896 | 0.00099 | -0.273 | 0.21961 | 0.514 |
| 7396 | FAM66C |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-5p | 11 | THSD7A | Sponge network | -0.901 | 0.00019 | 0.242 | 0.25154 | 0.514 |
| 7397 | RP11-53O19.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-629-5p | 52 | DST | Sponge network | -0.405 | 0.22013 | -0.713 | 0.00096 | 0.514 |
| 7398 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 16 | EZH1 | Sponge network | -0.421 | 0.0114 | -0.564 | 1.0E-5 | 0.514 |
| 7399 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p | 26 | PBX1 | Sponge network | -0.693 | 0.05629 | -1.206 | 0 | 0.514 |
| 7400 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 20 | NRK | Sponge network | -2.117 | 0.00161 | 0.656 | 0.19217 | 0.514 |
| 7401 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 14 | DKK3 | Sponge network | -1.349 | 0.00017 | -0.052 | 0.77783 | 0.514 |
| 7402 | LINC00473 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ERG | Sponge network | -1.203 | 0.03881 | 0.002 | 0.99011 | 0.514 |
| 7403 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 42 | ST6GAL2 | Sponge network | -1.111 | 0.0003 | -1.201 | 0.00597 | 0.514 |
| 7404 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | NR4A3 | Sponge network | -0.769 | 0.00035 | -1.385 | 0 | 0.514 |
| 7405 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 24 | CADM3 | Sponge network | -1.216 | 0 | -3.663 | 0 | 0.514 |
| 7406 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-671-5p | 16 | CADM3 | Sponge network | -0.777 | 0.16667 | -3.663 | 0 | 0.514 |
| 7407 | ACTA2-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITGB1 | Sponge network | 0.194 | 0.64278 | 0.524 | 0.00017 | 0.514 |
| 7408 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935;hsa-miR-96-5p | 41 | FBXO32 | Sponge network | -1.216 | 0 | -0.495 | 0.07561 | 0.514 |
| 7409 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-93-5p | 25 | PDGFRA | Sponge network | -0.473 | 0.07602 | -0.372 | 0.0037 | 0.514 |
| 7410 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p | 18 | CACNA2D1 | Sponge network | 0.056 | 0.81195 | -0.846 | 0.00675 | 0.514 |
| 7411 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p | 20 | CDH23 | Sponge network | -1.697 | 0.00889 | -0.974 | 0.00034 | 0.514 |
| 7412 | MIR497HG |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-940 | 13 | FAM172A | Sponge network | -1.439 | 4.0E-5 | -0.57 | 0 | 0.514 |
| 7413 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 30 | FAT3 | Sponge network | -0.811 | 2.0E-5 | -1.601 | 0.00051 | 0.514 |
| 7414 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 28 | PIK3C2A | Sponge network | 0.782 | 0.00016 | 0.061 | 0.53776 | 0.514 |
| 7415 | HCG11 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-664a-3p | 13 | MN1 | Sponge network | 0.385 | 0.10067 | -0.358 | 0.24172 | 0.514 |
| 7416 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 37 | ABI2 | Sponge network | 0.083 | 0.66405 | 0.175 | 0.14787 | 0.514 |
| 7417 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 14 | AMPH | Sponge network | -2.46 | 0 | -0.916 | 5.0E-5 | 0.514 |
| 7418 | MALAT1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 17 | SHPRH | Sponge network | 0.782 | 0.00016 | 0.098 | 0.40994 | 0.514 |
| 7419 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-7-5p | 37 | TCF4 | Sponge network | -0.469 | 0.08721 | 0.016 | 0.92271 | 0.514 |
| 7420 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-940 | 54 | NTRK3 | Sponge network | -1.249 | 7.0E-5 | -1.77 | 3.0E-5 | 0.514 |
| 7421 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | SLITRK6 | Sponge network | -1.281 | 0.00012 | -2.121 | 0 | 0.514 |
| 7422 | TPTEP1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 71 | RUNX1T1 | Sponge network | -1.216 | 0 | -0.344 | 0.2374 | 0.514 |
| 7423 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 12 | BCLAF1 | Sponge network | 1.063 | 0 | 0.211 | 0.00684 | 0.514 |
| 7424 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p | 24 | NAV3 | Sponge network | -2.69 | 0.0001 | 0.212 | 0.47729 | 0.514 |
| 7425 | LINC00657 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ITGB1 | Sponge network | 0.91 | 0 | 0.524 | 0.00017 | 0.514 |
| 7426 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CDH11 | Sponge network | -1.697 | 0.00889 | 1.427 | 0 | 0.514 |
| 7427 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PTGFR | Sponge network | -0.739 | 0.00044 | -0.233 | 0.45421 | 0.514 |
| 7428 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | FSTL5 | Sponge network | -2.452 | 0 | -1.3 | 0.01619 | 0.514 |
| 7429 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-424-5p;hsa-miR-532-5p | 10 | DCLK1 | Sponge network | -4.463 | 0.00025 | -1.324 | 0.00014 | 0.514 |
| 7430 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 21 | NUAK1 | Sponge network | 0.335 | 0.20596 | 0.904 | 0 | 0.514 |
| 7431 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-93-5p | 32 | FAT3 | Sponge network | -0.473 | 0.07602 | -1.601 | 0.00051 | 0.514 |
| 7432 | BVES-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-877-5p | 19 | NAV3 | Sponge network | -2.62 | 0 | 0.212 | 0.47729 | 0.514 |
| 7433 | HAND2-AS1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | RPS6KA6 | Sponge network | -1.697 | 0.00889 | -2.989 | 0 | 0.514 |
| 7434 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-3614-5p | 22 | EPHA3 | Sponge network | 0.082 | 0.55397 | 0.185 | 0.53588 | 0.514 |
| 7435 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 28 | PCDH10 | Sponge network | -0.323 | 0.01372 | -1.644 | 0.0078 | 0.514 |
| 7436 | NEAT1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 21 | NR2C2 | Sponge network | 0.705 | 0.00014 | 0.21 | 0.03224 | 0.514 |
| 7437 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | CHD2 | Sponge network | 0.848 | 0 | 0.049 | 0.55371 | 0.514 |
| 7438 | LINC00957 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | WWTR1 | Sponge network | -0.421 | 0.0114 | -0.345 | 0.11997 | 0.514 |
| 7439 | LINC00578 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-629-3p | 25 | EPHA7 | Sponge network | 0.811 | 0.03228 | -2.197 | 3.0E-5 | 0.514 |
| 7440 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-92a-1-5p | 29 | PRKAB2 | Sponge network | -0.162 | 0.47687 | -0.367 | 0.01371 | 0.514 |
| 7441 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-940 | 26 | MCC | Sponge network | -0.154 | 0.53954 | -1.005 | 0 | 0.514 |
| 7442 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-5p | 12 | RECK | Sponge network | 0.176 | 0.37402 | -1.043 | 0 | 0.513 |
| 7443 | RASSF8-AS1 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | SFRP1 | Sponge network | 0.335 | 0.20596 | -3.566 | 0 | 0.513 |
| 7444 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 30 | ERC1 | Sponge network | -0.487 | 0.02157 | -0.108 | 0.38794 | 0.513 |
| 7445 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | UNC5C | Sponge network | -0.358 | 0.17255 | -0.178 | 0.40214 | 0.513 |
| 7446 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-671-5p | 36 | RBMS3 | Sponge network | -0.154 | 0.78939 | -0.519 | 0.03551 | 0.513 |
| 7447 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-93-5p | 10 | TXNIP | Sponge network | -0.612 | 0.00094 | -1.277 | 0 | 0.513 |
| 7448 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 27 | SSBP2 | Sponge network | -0.376 | 0.00337 | -1.58 | 0 | 0.513 |
| 7449 | HAND2-AS1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | SPOP | Sponge network | -1.697 | 0.00889 | -0.419 | 1.0E-5 | 0.513 |
| 7450 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 18 | EPHA3 | Sponge network | 0.174 | 0.30034 | 0.185 | 0.53588 | 0.513 |
| 7451 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500b-5p | 16 | GRIA3 | Sponge network | -2.531 | 0 | -1.956 | 0 | 0.513 |
| 7452 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | NCAM2 | Sponge network | 0.395 | 0.15451 | -0.482 | 0.22565 | 0.513 |
| 7453 | LINC00865 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 13 | TMEFF2 | Sponge network | -1.249 | 7.0E-5 | -2.426 | 0 | 0.513 |
| 7454 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | RECK | Sponge network | -0.942 | 0 | -1.043 | 0 | 0.513 |
| 7455 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-582-5p | 37 | PRKAA2 | Sponge network | -2.095 | 0.00112 | -2.462 | 0 | 0.513 |
| 7456 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-542-3p | 18 | DIP2C | Sponge network | -0.517 | 0.02935 | -0.436 | 0.01713 | 0.513 |
| 7457 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-935 | 35 | FBXO32 | Sponge network | -0.405 | 0.22013 | -0.495 | 0.07561 | 0.513 |
| 7458 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | RASSF2 | Sponge network | 0.572 | 0.01722 | -0.273 | 0.21961 | 0.513 |
| 7459 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-323a-3p | 13 | CADM2 | Sponge network | -7.551 | 0 | -3.782 | 0 | 0.513 |
| 7460 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 18 | SETBP1 | Sponge network | -0.318 | 0.65847 | -1.302 | 1.0E-5 | 0.513 |
| 7461 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-628-5p | 11 | BCL6 | Sponge network | -1.092 | 4.0E-5 | -0.211 | 0.19489 | 0.513 |
| 7462 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577 | 16 | GREM1 | Sponge network | -1.349 | 0.00017 | 0.902 | 0.01691 | 0.513 |
| 7463 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ZFHX4 | Sponge network | -0.021 | 0.93598 | 0.018 | 0.95574 | 0.513 |
| 7464 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-3607-3p | 21 | IL17RD | Sponge network | -0.693 | 0.05629 | -0.019 | 0.94873 | 0.513 |
| 7465 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 29 | SCN9A | Sponge network | -0.323 | 0.01372 | -0.525 | 0.10593 | 0.513 |
| 7466 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-429 | 15 | CNTN1 | Sponge network | -0.469 | 0.08721 | -2.594 | 0 | 0.513 |
| 7467 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p | 19 | FAT4 | Sponge network | -1.249 | 7.0E-5 | -0.354 | 0.14694 | 0.513 |
| 7468 | RP11-356J5.12 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-766-3p | 18 | EZH1 | Sponge network | -0.899 | 6.0E-5 | -0.564 | 1.0E-5 | 0.513 |
| 7469 | PCA3 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | STAT5B | Sponge network | -0.709 | 0.18168 | -0.182 | 0.1082 | 0.513 |
| 7470 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 53 | TCF4 | Sponge network | -0.513 | 0.04703 | 0.016 | 0.92271 | 0.513 |
| 7471 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | MOB1B | Sponge network | 0.782 | 0.00016 | 0.197 | 0.15266 | 0.513 |
| 7472 | RP11-48B3.4 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p | 19 | RASSF8 | Sponge network | 1.757 | 0 | -0.122 | 0.65591 | 0.513 |
| 7473 | RP11-244O19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-877-5p | 26 | NAV3 | Sponge network | -0.096 | 0.67958 | 0.212 | 0.47729 | 0.513 |
| 7474 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | KCNAB1 | Sponge network | -0.129 | 0.57177 | -0.755 | 2.0E-5 | 0.513 |
| 7475 | HCG18 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 13 | KIAA1024 | Sponge network | 1.063 | 0 | 1.083 | 0 | 0.513 |
| 7476 | SERTAD4-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-424-5p | 10 | EZH1 | Sponge network | -0.932 | 0.00329 | -0.564 | 1.0E-5 | 0.513 |
| 7477 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 12 | FYN | Sponge network | -0.513 | 0.04703 | -0.131 | 0.37051 | 0.513 |
| 7478 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-625-3p | 32 | ABL2 | Sponge network | 0.75 | 0.00054 | 0.82 | 0 | 0.513 |
| 7479 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p | 44 | BNC2 | Sponge network | -0.154 | 0.78939 | -0.491 | 0.09988 | 0.513 |
| 7480 | SNHG14 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 14 | RIMS1 | Sponge network | -1.111 | 0.0003 | -3.143 | 0 | 0.513 |
| 7481 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-5p;hsa-miR-93-5p | 14 | ZBTB4 | Sponge network | -0.376 | 0.00337 | -0.739 | 0 | 0.513 |
| 7482 | RP11-1006G14.4 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 44 | MDM4 | Sponge network | 0.559 | 0.00026 | 0.345 | 0.00762 | 0.513 |
| 7483 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-590-3p | 12 | RBM5 | Sponge network | -0.323 | 0.01372 | -0.205 | 0.0129 | 0.513 |
| 7484 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-511-5p | 14 | TRIM33 | Sponge network | 0.997 | 0 | 0.402 | 7.0E-5 | 0.513 |
| 7485 | MIR497HG |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | FYN | Sponge network | -1.439 | 4.0E-5 | -0.131 | 0.37051 | 0.513 |
| 7486 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 53 | PRKAA2 | Sponge network | -0.323 | 0.01372 | -2.462 | 0 | 0.513 |
| 7487 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SMARCA2 | Sponge network | -0.487 | 0.02157 | -0.529 | 0.00014 | 0.513 |
| 7488 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 15 | AKAP12 | Sponge network | -0.709 | 0.18168 | -0.932 | 0.0028 | 0.513 |
| 7489 | AC006116.24 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-539-5p;hsa-miR-93-5p | 10 | PRUNE2 | Sponge network | -0.473 | 0.07602 | -1.733 | 0.00022 | 0.513 |
| 7490 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 39 | PRKAA2 | Sponge network | -1.654 | 0.00144 | -2.462 | 0 | 0.513 |
| 7491 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 25 | GJA1 | Sponge network | -0.49 | 0.04946 | -0.485 | 0.02179 | 0.513 |
| 7492 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p | 11 | LHX6 | Sponge network | -0.583 | 0.14589 | -0.087 | 0.69193 | 0.513 |
| 7493 | LINC00957 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | GLI3 | Sponge network | -0.421 | 0.0114 | -0.615 | 0.01482 | 0.513 |
| 7494 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-582-3p | 12 | PIK3CA | Sponge network | 0.993 | 0 | 0.195 | 0.06908 | 0.513 |
| 7495 | GNG12-AS1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p | 10 | MN1 | Sponge network | -0.793 | 7.0E-5 | -0.358 | 0.24172 | 0.513 |
| 7496 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | DCLK1 | Sponge network | -0.096 | 0.67958 | -1.324 | 0.00014 | 0.513 |
| 7497 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | MEF2C | Sponge network | 1.153 | 0.00114 | -0.499 | 0.01448 | 0.513 |
| 7498 | LINC00472 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-421 | 11 | MACF1 | Sponge network | -0.842 | 0.01361 | 0.019 | 0.90335 | 0.513 |
| 7499 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-96-5p | 21 | EBF3 | Sponge network | -1.349 | 0.00017 | -0.058 | 0.81437 | 0.513 |
| 7500 | MIR143HG |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | KCNAB1 | Sponge network | -0.739 | 0.0574 | -0.755 | 2.0E-5 | 0.513 |
| 7501 | RP11-254F7.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-362-5p | 12 | MRVI1 | Sponge network | 0.176 | 0.37402 | -1.051 | 0.00022 | 0.513 |
| 7502 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-590-3p | 10 | HOOK3 | Sponge network | -0.031 | 0.89063 | 0.273 | 0.09957 | 0.513 |
| 7503 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 26 | GRIN2A | Sponge network | -1.249 | 7.0E-5 | -2.161 | 0 | 0.513 |
| 7504 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484 | 15 | FLNA | Sponge network | 0.082 | 0.55654 | -0.771 | 0.01377 | 0.513 |
| 7505 | LINC00641 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | KCNAB1 | Sponge network | -0.222 | 0.32445 | -0.755 | 2.0E-5 | 0.513 |
| 7506 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p | 10 | ST5 | Sponge network | -0.487 | 0.02157 | -0.78 | 0 | 0.513 |
| 7507 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | DAB2 | Sponge network | -0.737 | 0.06182 | 0.431 | 0.0113 | 0.513 |
| 7508 | SNHG14 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 27 | ABL1 | Sponge network | -1.111 | 0.0003 | -0.215 | 0.07804 | 0.513 |
| 7509 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-935 | 28 | FBXO32 | Sponge network | 0.16 | 0.50785 | -0.495 | 0.07561 | 0.513 |
| 7510 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-93-3p | 43 | RUNX1T1 | Sponge network | -0.318 | 0.65847 | -0.344 | 0.2374 | 0.513 |
| 7511 | RP11-6O2.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-590-5p | 13 | NRXN3 | Sponge network | 0.331 | 0.57247 | -0.893 | 0.04186 | 0.513 |
| 7512 | TP73-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 21 | SORCS1 | Sponge network | -0.563 | 0.02545 | -3.492 | 0 | 0.513 |
| 7513 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-93-3p | 18 | PCDH10 | Sponge network | -0.777 | 0.16667 | -1.644 | 0.0078 | 0.513 |
| 7514 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 28 | AKAP6 | Sponge network | -0.355 | 0.0077 | -1.586 | 3.0E-5 | 0.513 |
| 7515 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p | 21 | ANK2 | Sponge network | 0.178 | 0.29106 | -1.603 | 1.0E-5 | 0.513 |
| 7516 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-576-5p | 26 | IGF1 | Sponge network | -2.62 | 0 | -0.204 | 0.5898 | 0.513 |
| 7517 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | PDGFRA | Sponge network | -2.452 | 0 | -0.372 | 0.0037 | 0.512 |
| 7518 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | RARB | Sponge network | -1.886 | 0 | -0.825 | 2.0E-5 | 0.512 |
| 7519 | RP11-305E6.4 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-625-3p | 23 | ABL2 | Sponge network | 1.317 | 0 | 0.82 | 0 | 0.512 |
| 7520 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 51 | NTRK3 | Sponge network | -0.896 | 0.00099 | -1.77 | 3.0E-5 | 0.512 |
| 7521 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b | 14 | ST6GAL2 | Sponge network | -1.582 | 8.0E-5 | -1.201 | 0.00597 | 0.512 |
| 7522 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | ROBO1 | Sponge network | -0.737 | 0.06182 | -0.009 | 0.96154 | 0.512 |
| 7523 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 15 | NALCN | Sponge network | -1.111 | 0.0003 | 1.068 | 0.00237 | 0.512 |
| 7524 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-3p | 13 | WWTR1 | Sponge network | -0.376 | 0.00337 | -0.345 | 0.11997 | 0.512 |
| 7525 | MAGI2-AS3 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 12 | MLLT11 | Sponge network | -0.129 | 0.57177 | -0.661 | 0.01733 | 0.512 |
| 7526 | LINC00672 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 17 | ZMYND11 | Sponge network | -0.227 | 0.31657 | -0.2 | 0.05449 | 0.512 |
| 7527 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 25 | TSHZ3 | Sponge network | -0.811 | 2.0E-5 | -0.556 | 0.01516 | 0.512 |
| 7528 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 12 | FGFR1 | Sponge network | -0.376 | 0.00337 | -0.509 | 0.03957 | 0.512 |
| 7529 | EMX2OS |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | SETBP1 | Sponge network | -0.777 | 0.1134 | -1.302 | 1.0E-5 | 0.512 |
| 7530 | RP11-326G21.1 |
hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 13 | ANK2 | Sponge network | -0.021 | 0.93598 | -1.603 | 1.0E-5 | 0.512 |
| 7531 | TPTEP1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 19 | SYNM | Sponge network | -1.216 | 0 | -2.309 | 1.0E-5 | 0.512 |
| 7532 | RAMP2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-877-5p | 25 | ASTN1 | Sponge network | -0.513 | 0.04703 | -2.389 | 2.0E-5 | 0.512 |
| 7533 | RP11-620J15.3 |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PAIP2 | Sponge network | -0.739 | 0.00044 | -0.515 | 0 | 0.512 |
| 7534 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 29 | CNTN1 | Sponge network | -0.793 | 7.0E-5 | -2.594 | 0 | 0.512 |
| 7535 | LINC00702 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 40 | ERC1 | Sponge network | -0.737 | 0.06182 | -0.108 | 0.38794 | 0.512 |
| 7536 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 21 | STARD13 | Sponge network | -0.49 | 0.04946 | -0.187 | 0.27383 | 0.512 |
| 7537 | RP11-822E23.8 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 45 | GAS7 | Sponge network | -0.777 | 0.16667 | -0.555 | 0.01602 | 0.512 |
| 7538 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p | 11 | TSLP | Sponge network | -0.49 | 0.04946 | -2.209 | 0 | 0.512 |
| 7539 | LINC00473 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | PRKAA2 | Sponge network | -1.203 | 0.03881 | -2.462 | 0 | 0.512 |
| 7540 | PVT1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-486-5p | 11 | CBFB | Sponge network | 1.985 | 0 | 1.061 | 0 | 0.512 |
| 7541 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | BCL2 | Sponge network | -0.576 | 0.10121 | -0.899 | 0 | 0.512 |
| 7542 | RP4-669L17.10 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-625-5p;hsa-miR-766-3p | 24 | MDM4 | Sponge network | 1.093 | 0 | 0.345 | 0.00762 | 0.512 |
| 7543 | NR2F1-AS1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 11 | PEA15 | Sponge network | -0.49 | 0.04946 | 0.009 | 0.93318 | 0.512 |
| 7544 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | RASSF2 | Sponge network | 1.153 | 0.00114 | -0.273 | 0.21961 | 0.512 |
| 7545 | AC006116.24 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-98-5p | 15 | KCNA6 | Sponge network | -0.473 | 0.07602 | -1.531 | 0.00013 | 0.512 |
| 7546 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 18 | SYNPO2 | Sponge network | -1.057 | 0.00029 | -2.342 | 0 | 0.512 |
| 7547 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p | 37 | DTNA | Sponge network | 0.176 | 0.37402 | -1.648 | 1.0E-5 | 0.512 |
| 7548 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-628-5p | 23 | FBXO32 | Sponge network | -0.726 | 0.00132 | -0.495 | 0.07561 | 0.512 |
| 7549 | USP3-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-940 | 19 | SORCS1 | Sponge network | -0.162 | 0.47687 | -3.492 | 0 | 0.512 |
| 7550 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 11 | NALCN | Sponge network | 1.302 | 7.0E-5 | 1.068 | 0.00237 | 0.512 |
| 7551 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-93-3p | 23 | BNC2 | Sponge network | -0.469 | 0.08721 | -0.491 | 0.09988 | 0.512 |
| 7552 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p;hsa-miR-98-5p | 26 | LRIG1 | Sponge network | -2.452 | 0 | -0.537 | 0.00588 | 0.512 |
| 7553 | PGM5-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-450b-5p | 10 | STAT5B | Sponge network | -4.546 | 0 | -0.182 | 0.1082 | 0.512 |
| 7554 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-7-1-3p | 14 | CLIP1 | Sponge network | -1.092 | 4.0E-5 | -0.215 | 0.08684 | 0.512 |
| 7555 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 37 | ADARB1 | Sponge network | 0.082 | 0.55397 | -0.335 | 0.06631 | 0.512 |
| 7556 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PBRM1 | Sponge network | 0.083 | 0.66405 | -0.029 | 0.78601 | 0.512 |
| 7557 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | GRIN2A | Sponge network | -0.769 | 0.00035 | -2.161 | 0 | 0.512 |
| 7558 | AP001347.6 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 12 | GLI3 | Sponge network | -1.161 | 0.00591 | -0.615 | 0.01482 | 0.512 |
| 7559 | GUSBP11 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 25 | MDM4 | Sponge network | 0.856 | 0 | 0.345 | 0.00762 | 0.512 |
| 7560 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 65 | THRB | Sponge network | -2.452 | 0 | -1.117 | 0.0004 | 0.512 |
| 7561 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CDH11 | Sponge network | -0.737 | 0.06182 | 1.427 | 0 | 0.512 |
| 7562 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 21 | ESR1 | Sponge network | -0.583 | 0.14589 | -1.035 | 3.0E-5 | 0.512 |
| 7563 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p | 12 | TRIM33 | Sponge network | 1.147 | 0 | 0.402 | 7.0E-5 | 0.512 |
| 7564 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-582-5p | 16 | EBF3 | Sponge network | 0.889 | 9.0E-5 | -0.058 | 0.81437 | 0.512 |
| 7565 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | NR2C2 | Sponge network | 1.378 | 0 | 0.21 | 0.03224 | 0.512 |
| 7566 | RP11-144G6.12 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 22 | CCDC6 | Sponge network | 0.826 | 1.0E-5 | 0.27 | 0.02555 | 0.512 |
| 7567 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p | 11 | PDZD2 | Sponge network | -0.611 | 0.09607 | -0.909 | 3.0E-5 | 0.512 |
| 7568 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 34 | RBMS3 | Sponge network | 0.4 | 0.14611 | -0.519 | 0.03551 | 0.512 |
| 7569 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | ZNF292 | Sponge network | 1.153 | 0 | 0.276 | 0.04148 | 0.512 |
| 7570 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 17 | HOOK3 | Sponge network | 0.997 | 0.00066 | 0.273 | 0.09957 | 0.512 |
| 7571 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 21 | RET | Sponge network | -1.281 | 0.00012 | -0.833 | 0.03517 | 0.512 |
| 7572 | EMX2OS |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | RNF180 | Sponge network | -0.777 | 0.1134 | -1.127 | 1.0E-5 | 0.512 |
| 7573 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 23 | CBFA2T3 | Sponge network | 0.4 | 0.14611 | -1.221 | 1.0E-5 | 0.512 |
| 7574 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 28 | EPHA3 | Sponge network | -0.563 | 0.02545 | 0.185 | 0.53588 | 0.512 |
| 7575 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 22 | PCDH10 | Sponge network | 0.246 | 0.59912 | -1.644 | 0.0078 | 0.512 |
| 7576 | CTC-429P9.2 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-766-3p | 18 | IL6ST | Sponge network | 0.043 | 0.81628 | -0.402 | 0.00676 | 0.512 |
| 7577 | AC009948.5 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PREX2 | Sponge network | 0.532 | 0.00505 | -0.272 | 0.25382 | 0.512 |
| 7578 | NR2F1-AS1 |
hsa-miR-10b-3p;hsa-miR-193b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p | 10 | LRRTM1 | Sponge network | -0.49 | 0.04946 | -3.361 | 0 | 0.512 |
| 7579 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-3p | 15 | GREM1 | Sponge network | -0.693 | 0.05629 | 0.902 | 0.01691 | 0.512 |
| 7580 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 17 | JAKMIP2 | Sponge network | 0.335 | 0.20596 | -1.388 | 0 | 0.512 |
| 7581 | LINC00957 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-874-3p | 28 | ASTN1 | Sponge network | -0.421 | 0.0114 | -2.389 | 2.0E-5 | 0.512 |
| 7582 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-424-5p | 14 | DYNC1I1 | Sponge network | 0.299 | 0.57722 | -1.12 | 0.00107 | 0.512 |
| 7583 | PWAR6 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | HERC1 | Sponge network | -1.575 | 6.0E-5 | -0.196 | 0.08544 | 0.512 |
| 7584 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MYO9A | Sponge network | 0.385 | 0.10067 | -0.147 | 0.32373 | 0.512 |
| 7585 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p | 16 | FAT4 | Sponge network | -1.886 | 0 | -0.354 | 0.14694 | 0.512 |
| 7586 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-369-3p;hsa-miR-590-3p | 12 | TMEFF2 | Sponge network | -0.739 | 0.00044 | -2.426 | 0 | 0.512 |
| 7587 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 11 | HERC2 | Sponge network | 0.537 | 0.00139 | 0.114 | 0.30638 | 0.512 |
| 7588 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | CNTN1 | Sponge network | -0.323 | 0.01372 | -2.594 | 0 | 0.512 |
| 7589 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | QKI | Sponge network | 0.342 | 0.03129 | -0.333 | 0.04111 | 0.512 |
| 7590 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | IGF1 | Sponge network | -0.739 | 0.00044 | -0.204 | 0.5898 | 0.512 |
| 7591 | PCA3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 61 | CADM2 | Sponge network | -0.709 | 0.18168 | -3.782 | 0 | 0.512 |
| 7592 | ADAMTS9-AS2 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 28 | GJA1 | Sponge network | -2.452 | 0 | -0.485 | 0.02179 | 0.512 |
| 7593 | RP11-774O3.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p | 27 | DLC1 | Sponge network | -0.487 | 0.02157 | -0.382 | 0.05038 | 0.512 |
| 7594 | RP11-999E24.3 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-93-3p | 16 | PCDH10 | Sponge network | -0.693 | 0.05629 | -1.644 | 0.0078 | 0.512 |
| 7595 | LINC00957 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PCDH9 | Sponge network | -0.421 | 0.0114 | -1.823 | 4.0E-5 | 0.512 |
| 7596 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TBX18 | Sponge network | -0.49 | 0.04946 | 0.843 | 0.02945 | 0.512 |
| 7597 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-629-3p;hsa-miR-942-5p | 23 | GPC6 | Sponge network | -0.777 | 0.16667 | -0.054 | 0.81465 | 0.512 |
| 7598 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 10 | ZNF208 | Sponge network | -1.582 | 8.0E-5 | -0.4 | 0.22381 | 0.512 |
| 7599 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-664a-3p | 15 | MN1 | Sponge network | -0.942 | 0 | -0.358 | 0.24172 | 0.512 |
| 7600 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p | 16 | PCDH9 | Sponge network | -0.469 | 0.08721 | -1.823 | 4.0E-5 | 0.511 |
| 7601 | RP11-890B15.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 16 | PRUNE2 | Sponge network | 0.101 | 0.60919 | -1.733 | 0.00022 | 0.511 |
| 7602 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ABCC9 | Sponge network | -0.021 | 0.93598 | -0.466 | 0.14121 | 0.511 |
| 7603 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 21 | FBXO32 | Sponge network | -0.517 | 0.02935 | -0.495 | 0.07561 | 0.511 |
| 7604 | AP001347.6 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-505-3p;hsa-miR-7-5p | 11 | FNBP1 | Sponge network | -1.161 | 0.00591 | -0.969 | 4.0E-5 | 0.511 |
| 7605 | TPTEP1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 10 | LRFN5 | Sponge network | -1.216 | 0 | -1.483 | 0 | 0.511 |
| 7606 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 36 | NRXN1 | Sponge network | -0.777 | 0.1134 | -3.778 | 0 | 0.511 |
| 7607 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 24 | FAT3 | Sponge network | -0.021 | 0.93598 | -1.601 | 0.00051 | 0.511 |
| 7608 | MEG3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 30 | PTPN14 | Sponge network | 0.249 | 0.35902 | 0.144 | 0.34595 | 0.511 |
| 7609 | RP11-400K9.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-362-5p;hsa-miR-671-5p | 18 | CYLD | Sponge network | -0.611 | 0.09607 | -0.367 | 0.00171 | 0.511 |
| 7610 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-98-5p | 18 | PBX1 | Sponge network | -0.473 | 0.07602 | -1.206 | 0 | 0.511 |
| 7611 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 31 | KCNA6 | Sponge network | -2.46 | 0 | -1.531 | 0.00013 | 0.511 |
| 7612 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p | 10 | LRRC4 | Sponge network | -2.62 | 0 | -1.774 | 0 | 0.511 |
| 7613 | SH3RF3-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | NRXN3 | Sponge network | 0.889 | 9.0E-5 | -0.893 | 0.04186 | 0.511 |
| 7614 | PWAR6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | TMEFF2 | Sponge network | -1.575 | 6.0E-5 | -2.426 | 0 | 0.511 |
| 7615 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DACT1 | Sponge network | -0.737 | 0.10822 | 0.783 | 0.00072 | 0.511 |
| 7616 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 12 | GLI3 | Sponge network | 0.299 | 0.57722 | -0.615 | 0.01482 | 0.511 |
| 7617 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 12 | FGFR1 | Sponge network | -0.726 | 0.00132 | -0.509 | 0.03957 | 0.511 |
| 7618 | U91328.19 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 19 | AKAP6 | Sponge network | -0.303 | 0.21002 | -1.586 | 3.0E-5 | 0.511 |
| 7619 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p | 51 | BMPR2 | Sponge network | 0.494 | 0.00339 | 0.206 | 0.0635 | 0.511 |
| 7620 | RP4-717I23.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-654-3p;hsa-miR-940 | 17 | BAZ2B | Sponge network | 0.637 | 0.00069 | -0.276 | 0.02833 | 0.511 |
| 7621 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 35 | NRXN1 | Sponge network | 0.246 | 0.59912 | -3.778 | 0 | 0.511 |
| 7622 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-511-5p | 15 | TRIM33 | Sponge network | 1.093 | 0 | 0.402 | 7.0E-5 | 0.511 |
| 7623 | AC009948.5 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 16 | FMNL3 | Sponge network | 0.532 | 0.00505 | 0.555 | 4.0E-5 | 0.511 |
| 7624 | EMX2OS |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 28 | PTPN14 | Sponge network | -0.777 | 0.1134 | 0.144 | 0.34595 | 0.511 |
| 7625 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 45 | FOXP1 | Sponge network | -0.323 | 0.01372 | 0.042 | 0.74368 | 0.511 |
| 7626 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 38 | ZFHX4 | Sponge network | -1.161 | 0.00591 | 0.018 | 0.95574 | 0.511 |
| 7627 | RP11-798M19.6 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RTN4 | Sponge network | -0.612 | 0.00094 | -0.412 | 0.00014 | 0.511 |
| 7628 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | 0.335 | 0.20596 | -0.7 | 1.0E-5 | 0.511 |
| 7629 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-425-5p | 13 | LRIG1 | Sponge network | -0.583 | 0.14589 | -0.537 | 0.00588 | 0.511 |
| 7630 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 23 | RASSF2 | Sponge network | -0.811 | 2.0E-5 | -0.273 | 0.21961 | 0.511 |
| 7631 | HAND2-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | SYT4 | Sponge network | -1.697 | 0.00889 | -3.207 | 0 | 0.511 |
| 7632 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-629-3p | 25 | RASSF2 | Sponge network | -0.583 | 0.14589 | -0.273 | 0.21961 | 0.511 |
| 7633 | RASSF8-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 33 | IGFBP5 | Sponge network | 0.335 | 0.20596 | -0.806 | 0.00105 | 0.511 |
| 7634 | H19 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-576-5p | 20 | BRD3 | Sponge network | 2.439 | 0 | 0.37 | 0.00013 | 0.511 |
| 7635 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p | 10 | TBL1XR1 | Sponge network | 0.673 | 0 | 0.578 | 0 | 0.511 |
| 7636 | MIR497HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-423-5p;hsa-miR-450b-5p | 12 | PNRC1 | Sponge network | -1.439 | 4.0E-5 | -0.646 | 0 | 0.511 |
| 7637 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 18 | CADM3 | Sponge network | 0.335 | 0.20596 | -3.663 | 0 | 0.511 |
| 7638 | SNHG14 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 19 | HERC1 | Sponge network | -1.111 | 0.0003 | -0.196 | 0.08544 | 0.511 |
| 7639 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-5p | 26 | DMD | Sponge network | 0.176 | 0.37402 | -1.506 | 0 | 0.511 |
| 7640 | AC003090.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-96-5p | 77 | LPP | Sponge network | -2.117 | 0.00161 | -0.459 | 0.04475 | 0.511 |
| 7641 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p | 20 | CACNA2D1 | Sponge network | -0.777 | 0.16667 | -0.846 | 0.00675 | 0.511 |
| 7642 | C20orf166-AS1 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p | 11 | PDZD2 | Sponge network | -2.69 | 0.0001 | -0.909 | 3.0E-5 | 0.511 |
| 7643 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-342-3p;hsa-miR-93-5p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | -0.162 | 0.47687 | -0.085 | 0.42389 | 0.511 |
| 7644 | FAM157C |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p | 13 | RICTOR | Sponge network | 0.91 | 0 | 0.388 | 0.00188 | 0.511 |
| 7645 | RP11-345P4.7 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 23 | MDM4 | Sponge network | 1.597 | 0 | 0.345 | 0.00762 | 0.511 |
| 7646 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | ZFHX4 | Sponge network | -0.737 | 0.10822 | 0.018 | 0.95574 | 0.511 |
| 7647 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | TIMP3 | Sponge network | -0.769 | 0.00035 | -0.546 | 0.02307 | 0.511 |
| 7648 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 18 | HOOK3 | Sponge network | 0.101 | 0.60919 | 0.273 | 0.09957 | 0.511 |
| 7649 | BZRAP1-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-625-5p | 13 | CXCL12 | Sponge network | -0.465 | 0.04703 | -1.421 | 0 | 0.511 |
| 7650 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | TMEM132B | Sponge network | -0.487 | 0.02157 | -0.83 | 0.01084 | 0.511 |
| 7651 | RP11-473I1.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374a-3p;hsa-miR-942-5p | 19 | ARHGEF12 | Sponge network | 0.542 | 0.00054 | 0.09 | 0.35693 | 0.511 |
| 7652 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-589-3p | 40 | ZFHX4 | Sponge network | -0.777 | 0.16667 | 0.018 | 0.95574 | 0.511 |
| 7653 | LINC00578 |
hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-484 | 11 | CRTC3 | Sponge network | 0.811 | 0.03228 | 0.164 | 0.16786 | 0.511 |
| 7654 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-624-5p | 14 | CDH19 | Sponge network | -0.517 | 0.02935 | -3.434 | 0 | 0.511 |
| 7655 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -0.129 | 0.57177 | -2.417 | 0 | 0.511 |
| 7656 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 31 | MEF2C | Sponge network | -0.611 | 0.09607 | -0.499 | 0.01448 | 0.511 |
| 7657 | MIR497HG |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-590-5p | 10 | CITED2 | Sponge network | -1.439 | 4.0E-5 | -1.545 | 0 | 0.511 |
| 7658 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | DKK3 | Sponge network | -2.452 | 0 | -0.052 | 0.77783 | 0.511 |
| 7659 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | TCF4 | Sponge network | -1.654 | 0.00144 | 0.016 | 0.92271 | 0.511 |
| 7660 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 32 | MYO9A | Sponge network | 0.572 | 0.01722 | -0.147 | 0.32373 | 0.511 |
| 7661 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | TRIM33 | Sponge network | 0.959 | 0 | 0.402 | 7.0E-5 | 0.511 |
| 7662 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 29 | RBMS3 | Sponge network | 0.889 | 9.0E-5 | -0.519 | 0.03551 | 0.511 |
| 7663 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p | 10 | EBF3 | Sponge network | 0.693 | 0.00076 | -0.058 | 0.81437 | 0.511 |
| 7664 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p | 17 | DKK3 | Sponge network | -0.793 | 7.0E-5 | -0.052 | 0.77783 | 0.511 |
| 7665 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CLIP1 | Sponge network | -0.222 | 0.32445 | -0.215 | 0.08684 | 0.511 |
| 7666 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 30 | RSPO2 | Sponge network | -0.737 | 0.06182 | -3.038 | 0 | 0.511 |
| 7667 | AC003090.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITPKB | Sponge network | -2.117 | 0.00161 | -0.704 | 0.00106 | 0.511 |
| 7668 | HOXB-AS1 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-3p | 14 | PCDH9 | Sponge network | -0.154 | 0.53954 | -1.823 | 4.0E-5 | 0.511 |
| 7669 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-7-5p | 18 | PTGFR | Sponge network | -0.162 | 0.47687 | -0.233 | 0.45421 | 0.511 |
| 7670 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-429 | 13 | FAT4 | Sponge network | -2.62 | 0 | -0.354 | 0.14694 | 0.511 |
| 7671 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-628-5p | 22 | ERC1 | Sponge network | -0.583 | 0.14589 | -0.108 | 0.38794 | 0.511 |
| 7672 | RASSF8-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-877-5p | 10 | STAT5B | Sponge network | 0.335 | 0.20596 | -0.182 | 0.1082 | 0.511 |
| 7673 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-576-5p | 13 | SEMA3E | Sponge network | -2.62 | 0 | -1.717 | 0.00137 | 0.511 |
| 7674 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 23 | BNC2 | Sponge network | 0.342 | 0.03129 | -0.491 | 0.09988 | 0.511 |
| 7675 | PCA3 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-342-3p | 16 | ABL1 | Sponge network | -0.709 | 0.18168 | -0.215 | 0.07804 | 0.511 |
| 7676 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-652-3p;hsa-miR-940 | 18 | QKI | Sponge network | -1.255 | 0.00981 | -0.333 | 0.04111 | 0.511 |
| 7677 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 34 | PHC3 | Sponge network | 0.75 | 0.00054 | 0.212 | 0.0499 | 0.511 |
| 7678 | LINC00476 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p | 20 | ITPKB | Sponge network | -0.615 | 0.00015 | -0.704 | 0.00106 | 0.511 |
| 7679 | USP3-AS1 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-508-3p | 14 | RASSF3 | Sponge network | -0.162 | 0.47687 | -0.226 | 0.11691 | 0.511 |
| 7680 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | SPG20 | Sponge network | -1.161 | 0.00591 | -1.16 | 0 | 0.511 |
| 7681 | CTC-429P9.5 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p | 22 | MDM4 | Sponge network | 0.178 | 0.29106 | 0.345 | 0.00762 | 0.511 |
| 7682 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-744-3p;hsa-miR-93-5p | 29 | HLF | Sponge network | -1.654 | 0.00144 | -2.443 | 0 | 0.51 |
| 7683 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-93-5p | 15 | FGFR1 | Sponge network | -1.654 | 0.00144 | -0.509 | 0.03957 | 0.51 |
| 7684 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | ZNF770 | Sponge network | 1.317 | 0 | 0.206 | 0.06751 | 0.51 |
| 7685 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 11 | EDA2R | Sponge network | -1.582 | 8.0E-5 | -0.587 | 0.01598 | 0.51 |
| 7686 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-942-5p | 37 | HLF | Sponge network | -0.611 | 0.09607 | -2.443 | 0 | 0.51 |
| 7687 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-654-3p;hsa-miR-874-3p | 15 | NR2C2 | Sponge network | 1.147 | 0 | 0.21 | 0.03224 | 0.51 |
| 7688 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-7-5p | 23 | ZBTB4 | Sponge network | -1.161 | 0.00591 | -0.739 | 0 | 0.51 |
| 7689 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CNTN1 | Sponge network | -1.161 | 0.00591 | -2.594 | 0 | 0.51 |
| 7690 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | TLL1 | Sponge network | -0.739 | 0.00044 | -1.195 | 3.0E-5 | 0.51 |
| 7691 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p | 13 | LRRK2 | Sponge network | -0.563 | 0.02545 | -1.136 | 1.0E-5 | 0.51 |
| 7692 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | CBL | Sponge network | 0.537 | 0.0006 | 0.291 | 0.01267 | 0.51 |
| 7693 | CTC-296K1.4 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-590-5p | 10 | CNTN1 | Sponge network | -2.076 | 0.00548 | -2.594 | 0 | 0.51 |
| 7694 | LINC00578 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p | 15 | SFRP1 | Sponge network | 0.811 | 0.03228 | -3.566 | 0 | 0.51 |
| 7695 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ABCC9 | Sponge network | 0.214 | 0.18246 | -0.466 | 0.14121 | 0.51 |
| 7696 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | MED12L | Sponge network | 0.225 | 0.30644 | -0.194 | 0.55299 | 0.51 |
| 7697 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-629-3p | 12 | ITGA5 | Sponge network | -2.531 | 0 | 0.452 | 0.03524 | 0.51 |
| 7698 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p | 35 | PRKAA2 | Sponge network | -0.693 | 0.05629 | -2.462 | 0 | 0.51 |
| 7699 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | NCAM2 | Sponge network | -0.611 | 0.09607 | -0.482 | 0.22565 | 0.51 |
| 7700 | ARHGEF26-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 20 | CELF4 | Sponge network | -2.46 | 0 | -1.135 | 0.00344 | 0.51 |
| 7701 | BZRAP1-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-877-5p | 10 | INPP4A | Sponge network | -0.465 | 0.04703 | 0.07 | 0.53563 | 0.51 |
| 7702 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-624-5p | 12 | HMCN1 | Sponge network | -0.246 | 0.35034 | 0.809 | 0.00544 | 0.51 |
| 7703 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p | 10 | SON | Sponge network | 1.597 | 0 | 0.168 | 0.04406 | 0.51 |
| 7704 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | MYO9A | Sponge network | -0.901 | 0.00019 | -0.147 | 0.32373 | 0.51 |
| 7705 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 24 | THBS1 | Sponge network | -0.777 | 0.16667 | 0.741 | 0.00386 | 0.51 |
| 7706 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p | 16 | CBFA2T3 | Sponge network | -0.358 | 0.17255 | -1.221 | 1.0E-5 | 0.51 |
| 7707 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | NRXN3 | Sponge network | 0.064 | 0.72934 | -0.893 | 0.04186 | 0.51 |
| 7708 | PCA3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DNAJB4 | Sponge network | -0.709 | 0.18168 | -0.7 | 1.0E-5 | 0.51 |
| 7709 | RP11-540B6.6 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 31 | MDM4 | Sponge network | 0.93 | 0 | 0.345 | 0.00762 | 0.51 |
| 7710 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | PDGFC | Sponge network | -0.793 | 7.0E-5 | -0.33 | 0.06483 | 0.51 |
| 7711 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-7-1-3p | 10 | DCN | Sponge network | -0.611 | 0.09607 | -1.288 | 0 | 0.51 |
| 7712 | TP73-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 48 | GRIK3 | Sponge network | -0.563 | 0.02545 | -3.208 | 0 | 0.51 |
| 7713 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421 | 25 | PDGFRA | Sponge network | -0.583 | 0.14589 | -0.372 | 0.0037 | 0.51 |
| 7714 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 57 | RUNX1T1 | Sponge network | -0.769 | 0.00035 | -0.344 | 0.2374 | 0.51 |
| 7715 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-93-5p | 47 | TCF4 | Sponge network | -0.465 | 0.04703 | 0.016 | 0.92271 | 0.51 |
| 7716 | GNG12-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | ABCA10 | Sponge network | -0.793 | 7.0E-5 | -0.571 | 0.03674 | 0.51 |
| 7717 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 28 | PGR | Sponge network | -0.227 | 0.31657 | -1.704 | 0 | 0.51 |
| 7718 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p | 12 | ZNF292 | Sponge network | 0.637 | 0.00069 | 0.276 | 0.04148 | 0.51 |
| 7719 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-935 | 25 | FBXO32 | Sponge network | -0.421 | 0.0114 | -0.495 | 0.07561 | 0.51 |
| 7720 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | HACE1 | Sponge network | 0.537 | 0.00139 | -0.072 | 0.55239 | 0.51 |
| 7721 | FENDRR |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | PAK3 | Sponge network | -1.281 | 0.00012 | -0.628 | 0.07253 | 0.51 |
| 7722 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-5p | 25 | ATRX | Sponge network | 0.494 | 0.00339 | 0.261 | 0.04408 | 0.51 |
| 7723 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p | 10 | ABCB5 | Sponge network | -1.331 | 3.0E-5 | -0.956 | 0.04077 | 0.51 |
| 7724 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-940 | 27 | IGF1 | Sponge network | -0.777 | 0.16667 | -0.204 | 0.5898 | 0.51 |
| 7725 | AC009948.5 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-582-5p | 11 | PRICKLE1 | Sponge network | 0.532 | 0.00505 | -0.112 | 0.67088 | 0.51 |
| 7726 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-576-5p;hsa-miR-590-5p | 15 | TSHZ3 | Sponge network | -2.076 | 0.00548 | -0.556 | 0.01516 | 0.51 |
| 7727 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-935 | 28 | FBXO32 | Sponge network | -0.738 | 0.21233 | -0.495 | 0.07561 | 0.51 |
| 7728 | RP4-639F20.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | CRTC3 | Sponge network | -0.517 | 0.02935 | 0.164 | 0.16786 | 0.51 |
| 7729 | ERVH48-1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 35 | MDM4 | Sponge network | 1.04 | 0.00023 | 0.345 | 0.00762 | 0.51 |
| 7730 | FENDRR |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 29 | LRIG1 | Sponge network | -1.281 | 0.00012 | -0.537 | 0.00588 | 0.51 |
| 7731 | FENDRR |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | -1.281 | 0.00012 | -0.57 | 0 | 0.51 |
| 7732 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | PREX2 | Sponge network | 0.335 | 0.20596 | -0.272 | 0.25382 | 0.51 |
| 7733 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | NR4A3 | Sponge network | -2.452 | 0 | -1.385 | 0 | 0.51 |
| 7734 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-3p | 21 | ZFHX3 | Sponge network | 0.174 | 0.30034 | 0.389 | 0.01363 | 0.51 |
| 7735 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p | 10 | LHX6 | Sponge network | -1.331 | 3.0E-5 | -0.087 | 0.69193 | 0.51 |
| 7736 | HAND2-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | ID4 | Sponge network | -1.697 | 0.00889 | -1.644 | 0 | 0.51 |
| 7737 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | TAL1 | Sponge network | -0.323 | 0.01372 | -0.462 | 0.00574 | 0.51 |
| 7738 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p | 27 | TRPS1 | Sponge network | 0.811 | 0.03228 | -0.191 | 0.44109 | 0.51 |
| 7739 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 31 | GRIN2A | Sponge network | -0.323 | 0.01372 | -2.161 | 0 | 0.51 |
| 7740 | AP001347.6 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 42 | CADM2 | Sponge network | -1.161 | 0.00591 | -3.782 | 0 | 0.51 |
| 7741 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-940 | 28 | PBX1 | Sponge network | -4.546 | 0 | -1.206 | 0 | 0.51 |
| 7742 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 26 | ST6GALNAC3 | Sponge network | -1.057 | 0.00029 | -0.987 | 0 | 0.51 |
| 7743 | AC010226.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-93-5p | 10 | PRUNE2 | Sponge network | -0.811 | 2.0E-5 | -1.733 | 0.00022 | 0.51 |
| 7744 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | NRXN1 | Sponge network | -0.376 | 0.00337 | -3.778 | 0 | 0.51 |
| 7745 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 15 | CDH19 | Sponge network | -0.021 | 0.93598 | -3.434 | 0 | 0.51 |
| 7746 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | LHFP | Sponge network | -0.942 | 0 | -0.167 | 0.41371 | 0.51 |
| 7747 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 10 | LHFP | Sponge network | 0.246 | 0.59912 | -0.167 | 0.41371 | 0.51 |
| 7748 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 40 | EPHA7 | Sponge network | -0.405 | 0.22013 | -2.197 | 3.0E-5 | 0.51 |
| 7749 | HAND2-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-423-5p | 22 | ABL1 | Sponge network | -1.697 | 0.00889 | -0.215 | 0.07804 | 0.51 |
| 7750 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-450b-5p | 12 | TIMP3 | Sponge network | -0.517 | 0.02935 | -0.546 | 0.02307 | 0.51 |
| 7751 | LINC00473 |
hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | CXCL12 | Sponge network | -1.203 | 0.03881 | -1.421 | 0 | 0.51 |
| 7752 | AC003090.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | LHFP | Sponge network | -2.117 | 0.00161 | -0.167 | 0.41371 | 0.51 |
| 7753 | CTD-2089N3.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 11 | SYT4 | Sponge network | -1.375 | 0.08642 | -3.207 | 0 | 0.51 |
| 7754 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-93-3p | 25 | QKI | Sponge network | -0.932 | 0.00329 | -0.333 | 0.04111 | 0.51 |
| 7755 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-455-3p;hsa-miR-93-5p | 13 | EDA2R | Sponge network | -0.563 | 0.02545 | -0.587 | 0.01598 | 0.51 |
| 7756 | LINC00578 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b | 11 | ITGB3 | Sponge network | 0.811 | 0.03228 | -0.011 | 0.95751 | 0.51 |
| 7757 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 47 | EPHA5 | Sponge network | -2.452 | 0 | -0.957 | 0.01733 | 0.51 |
| 7758 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 11 | DNAJB4 | Sponge network | -0.358 | 0.17255 | -0.7 | 1.0E-5 | 0.51 |
| 7759 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p | 48 | NTRK3 | Sponge network | -0.739 | 0.00044 | -1.77 | 3.0E-5 | 0.51 |
| 7760 | SNHG16 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p | 22 | TBL1XR1 | Sponge network | 0.961 | 0 | 0.578 | 0 | 0.51 |
| 7761 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-93-5p;hsa-miR-935 | 21 | ARNT2 | Sponge network | -0.563 | 0.02545 | -0.332 | 0.25851 | 0.51 |
| 7762 | DIO3OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-501-5p | 10 | MN1 | Sponge network | -0.773 | 0.04073 | -0.358 | 0.24172 | 0.509 |
| 7763 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | FSTL5 | Sponge network | -0.129 | 0.57177 | -1.3 | 0.01619 | 0.509 |
| 7764 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | ADARB1 | Sponge network | -0.376 | 0.00337 | -0.335 | 0.06631 | 0.509 |
| 7765 | ZNF667-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | PCDH18 | Sponge network | -0.976 | 0.00025 | 0.216 | 0.24645 | 0.509 |
| 7766 | FAM66C |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CDH19 | Sponge network | -0.901 | 0.00019 | -3.434 | 0 | 0.509 |
| 7767 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374b-5p | 13 | DNAJB4 | Sponge network | -1.645 | 0 | -0.7 | 1.0E-5 | 0.509 |
| 7768 | RP3-402G11.28 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 11 | MDM4 | Sponge network | 1.119 | 5.0E-5 | 0.345 | 0.00762 | 0.509 |
| 7769 | AC009948.5 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | PCDH9 | Sponge network | 0.532 | 0.00505 | -1.823 | 4.0E-5 | 0.509 |
| 7770 | CTD-3138B18.6 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-331-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p | 14 | ATRX | Sponge network | 2.248 | 0 | 0.261 | 0.04408 | 0.509 |
| 7771 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p | 13 | DCLK1 | Sponge network | -0.777 | 0.16667 | -1.324 | 0.00014 | 0.509 |
| 7772 | LINC00865 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p | 24 | CADM1 | Sponge network | -1.249 | 7.0E-5 | -0.9 | 0.00084 | 0.509 |
| 7773 | AP001055.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-331-3p | 14 | SNX29 | Sponge network | 0.693 | 0.00076 | -0.34 | 0.01371 | 0.509 |
| 7774 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | FAT3 | Sponge network | -0.769 | 0.00035 | -1.601 | 0.00051 | 0.509 |
| 7775 | MRVI1-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p | 11 | HERC1 | Sponge network | -1.092 | 4.0E-5 | -0.196 | 0.08544 | 0.509 |
| 7776 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-5p | 15 | SOX5 | Sponge network | -0.376 | 0.00337 | -1.091 | 4.0E-5 | 0.509 |
| 7777 | LINC00473 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | PCDH9 | Sponge network | -1.203 | 0.03881 | -1.823 | 4.0E-5 | 0.509 |
| 7778 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-582-3p | 14 | ATRX | Sponge network | 0.858 | 3.0E-5 | 0.261 | 0.04408 | 0.509 |
| 7779 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 29 | RSPO2 | Sponge network | -1.575 | 6.0E-5 | -3.038 | 0 | 0.509 |
| 7780 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 15 | NF1 | Sponge network | 0.946 | 0 | 0.51 | 0 | 0.509 |
| 7781 | LINC00472 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 18 | TAL1 | Sponge network | -0.842 | 0.01361 | -0.462 | 0.00574 | 0.509 |
| 7782 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | WWTR1 | Sponge network | -1.161 | 0.00591 | -0.345 | 0.11997 | 0.509 |
| 7783 | SERTAD4-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-502-5p | 24 | DST | Sponge network | -0.932 | 0.00329 | -0.713 | 0.00096 | 0.509 |
| 7784 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p | 25 | AFF3 | Sponge network | -0.709 | 0.18168 | -2.373 | 0 | 0.509 |
| 7785 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p | 20 | MYO9A | Sponge network | -0.358 | 0.17255 | -0.147 | 0.32373 | 0.509 |
| 7786 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484 | 16 | ITGA5 | Sponge network | 0.194 | 0.64278 | 0.452 | 0.03524 | 0.509 |
| 7787 | RP11-195F19.9 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 19 | DIP2C | Sponge network | -0.726 | 0.00132 | -0.436 | 0.01713 | 0.509 |
| 7788 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p | 18 | AKAP11 | Sponge network | 0.178 | 0.29106 | 0.196 | 0.09825 | 0.509 |
| 7789 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 27 | ADAMTSL3 | Sponge network | -0.709 | 0.18168 | -1.559 | 0 | 0.509 |
| 7790 | LINC00861 |
hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-675-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 14 | MEF2C | Sponge network | 0.911 | 0.00854 | -0.499 | 0.01448 | 0.509 |
| 7791 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-942-5p | 18 | DDX6 | Sponge network | 1.106 | 0 | 0.091 | 0.36428 | 0.509 |
| 7792 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 31 | RICTOR | Sponge network | 0.559 | 0.00026 | 0.388 | 0.00188 | 0.509 |
| 7793 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-505-5p;hsa-miR-874-3p | 19 | CTIF | Sponge network | -0.583 | 0.14589 | -0.389 | 0.00549 | 0.509 |
| 7794 | LINC00957 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TLL1 | Sponge network | -0.421 | 0.0114 | -1.195 | 3.0E-5 | 0.509 |
| 7795 | CTD-3092A11.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.873 | 0 | 0.943 | 0 | 0.509 |
| 7796 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p | 24 | TACC1 | Sponge network | -0.154 | 0.53954 | -0.362 | 0.05406 | 0.509 |
| 7797 | RP4-717I23.3 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-940 | 10 | TAF1 | Sponge network | 0.637 | 0.00069 | 0.39 | 3.0E-5 | 0.509 |
| 7798 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | DYNC1I1 | Sponge network | -1.886 | 0 | -1.12 | 0.00107 | 0.509 |
| 7799 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 18 | GRIN2A | Sponge network | -0.611 | 0.09607 | -2.161 | 0 | 0.509 |
| 7800 | HOXB-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-940 | 11 | RHOB | Sponge network | -0.154 | 0.53954 | -1.052 | 0 | 0.509 |
| 7801 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p | 10 | AMPH | Sponge network | 0.194 | 0.64278 | -0.916 | 5.0E-5 | 0.509 |
| 7802 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 46 | SSBP2 | Sponge network | -1.249 | 7.0E-5 | -1.58 | 0 | 0.509 |
| 7803 | RAMP2-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-935 | 11 | MAPK10 | Sponge network | -0.513 | 0.04703 | -1.774 | 0 | 0.509 |
| 7804 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p | 19 | CNTNAP3 | Sponge network | -0.487 | 0.02157 | -1.465 | 0.00033 | 0.509 |
| 7805 | LINC00092 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 31 | GAS7 | Sponge network | -1.886 | 0 | -0.555 | 0.01602 | 0.509 |
| 7806 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-590-3p | 10 | PAK3 | Sponge network | -1.057 | 0.00029 | -0.628 | 0.07253 | 0.509 |
| 7807 | MEG3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | 0.249 | 0.35902 | -0.7 | 1.0E-5 | 0.509 |
| 7808 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | MED12L | Sponge network | -1.575 | 6.0E-5 | -0.194 | 0.55299 | 0.509 |
| 7809 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-93-3p | 18 | ST6GAL2 | Sponge network | -0.154 | 0.53954 | -1.201 | 0.00597 | 0.509 |
| 7810 | AC005154.6 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-940 | 16 | TAF1 | Sponge network | 0.848 | 0 | 0.39 | 3.0E-5 | 0.509 |
| 7811 | RP11-774O3.3 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-589-5p | 10 | SORCS2 | Sponge network | -0.487 | 0.02157 | -0.223 | 0.4253 | 0.509 |
| 7812 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 20 | CHD2 | Sponge network | 1.254 | 0 | 0.049 | 0.55371 | 0.509 |
| 7813 | RP11-182J1.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-96-5p | 11 | SCN9A | Sponge network | -0.187 | 0.23787 | -0.525 | 0.10593 | 0.509 |
| 7814 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | MED12L | Sponge network | -1.111 | 0.0003 | -0.194 | 0.55299 | 0.509 |
| 7815 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-3p | 41 | QKI | Sponge network | -2.69 | 0.0001 | -0.333 | 0.04111 | 0.509 |
| 7816 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | CADM2 | Sponge network | -0.376 | 0.00337 | -3.782 | 0 | 0.509 |
| 7817 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ZMYND11 | Sponge network | 0.194 | 0.64278 | -0.2 | 0.05449 | 0.509 |
| 7818 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | THBS1 | Sponge network | 0.194 | 0.64278 | 0.741 | 0.00386 | 0.509 |
| 7819 | MIR497HG |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 10 | PEA15 | Sponge network | -1.439 | 4.0E-5 | 0.009 | 0.93318 | 0.509 |
| 7820 | MEG3 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 16 | NGFR | Sponge network | 0.249 | 0.35902 | -1.271 | 0.01058 | 0.509 |
| 7821 | RP11-253E3.3 |
hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p | 11 | ABL2 | Sponge network | 1.764 | 0 | 0.82 | 0 | 0.509 |
| 7822 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-378a-5p;hsa-miR-539-5p | 23 | PBX1 | Sponge network | -0.469 | 0.08721 | -1.206 | 0 | 0.509 |
| 7823 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | EPHA7 | Sponge network | -0.612 | 0.00094 | -2.197 | 3.0E-5 | 0.509 |
| 7824 | RP11-13K12.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-455-5p | 13 | MAPK10 | Sponge network | -0.154 | 0.78939 | -1.774 | 0 | 0.508 |
| 7825 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p | 17 | PTGFR | Sponge network | -0.096 | 0.67958 | -0.233 | 0.45421 | 0.508 |
| 7826 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-589-3p | 23 | KCNA6 | Sponge network | -2.531 | 0 | -1.531 | 0.00013 | 0.508 |
| 7827 | CTC-510F12.2 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-766-3p | 11 | IL17RD | Sponge network | -0.691 | 0.00146 | -0.019 | 0.94873 | 0.508 |
| 7828 | SNHG16 |
hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-363-3p;hsa-miR-497-5p | 10 | E2F3 | Sponge network | 0.961 | 0 | 1.289 | 0 | 0.508 |
| 7829 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-29a-5p;hsa-miR-342-3p | 18 | EPHA3 | Sponge network | -2.095 | 0.00112 | 0.185 | 0.53588 | 0.508 |
| 7830 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-93-5p | 15 | AHNAK | Sponge network | -2.69 | 0.0001 | -1.207 | 0 | 0.508 |
| 7831 | MRVI1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-7-1-3p | 20 | NCOA1 | Sponge network | -1.092 | 4.0E-5 | -0.432 | 0 | 0.508 |
| 7832 | BZRAP1-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-93-3p;hsa-miR-940 | 37 | GRIK3 | Sponge network | -0.465 | 0.04703 | -3.208 | 0 | 0.508 |
| 7833 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p | 29 | FBXO32 | Sponge network | 0.335 | 0.20596 | -0.495 | 0.07561 | 0.508 |
| 7834 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-96-5p | 23 | TACC1 | Sponge network | -0.517 | 0.02935 | -0.362 | 0.05406 | 0.508 |
| 7835 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-877-5p | 12 | CDH19 | Sponge network | -1.654 | 0.00144 | -3.434 | 0 | 0.508 |
| 7836 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PLSCR4 | Sponge network | 1.302 | 7.0E-5 | -0.474 | 0.03833 | 0.508 |
| 7837 | TP73-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-542-3p | 10 | PCLO | Sponge network | -0.563 | 0.02545 | -1.843 | 0.00064 | 0.508 |
| 7838 | AC006116.24 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-577 | 19 | IGFBP5 | Sponge network | -0.473 | 0.07602 | -0.806 | 0.00105 | 0.508 |
| 7839 | C20orf166-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-625-5p | 15 | CXCL12 | Sponge network | -2.69 | 0.0001 | -1.421 | 0 | 0.508 |
| 7840 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PREX2 | Sponge network | -0.487 | 0.02157 | -0.272 | 0.25382 | 0.508 |
| 7841 | MIR143HG |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | NOV | Sponge network | -0.739 | 0.0574 | -0.58 | 0.04676 | 0.508 |
| 7842 | FGF14-AS2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | NAV3 | Sponge network | -1.582 | 8.0E-5 | 0.212 | 0.47729 | 0.508 |
| 7843 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-940 | 23 | MYO9A | Sponge network | 0.082 | 0.55397 | -0.147 | 0.32373 | 0.508 |
| 7844 | AC010226.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-5p | 14 | PTGFR | Sponge network | -0.811 | 2.0E-5 | -0.233 | 0.45421 | 0.508 |
| 7845 | AP000254.8 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 13 | MDM4 | Sponge network | 0.555 | 0.00012 | 0.345 | 0.00762 | 0.508 |
| 7846 | RP11-13K12.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-589-3p | 24 | TET1 | Sponge network | -0.154 | 0.78939 | -0.135 | 0.52138 | 0.508 |
| 7847 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | RASSF8 | Sponge network | -2.117 | 0.00161 | -0.122 | 0.65591 | 0.508 |
| 7848 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | LRRK2 | Sponge network | -1.216 | 0 | -1.136 | 1.0E-5 | 0.508 |
| 7849 | RASSF8-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | 0.335 | 0.20596 | -0.704 | 0.00106 | 0.508 |
| 7850 | FENDRR |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 33 | PCDH10 | Sponge network | -1.281 | 0.00012 | -1.644 | 0.0078 | 0.508 |
| 7851 | MEG3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-5p | 17 | ROBO1 | Sponge network | 0.249 | 0.35902 | -0.009 | 0.96154 | 0.508 |
| 7852 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 33 | RICTOR | Sponge network | 0.614 | 1.0E-5 | 0.388 | 0.00188 | 0.508 |
| 7853 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | EPAS1 | Sponge network | -1.439 | 4.0E-5 | -0.184 | 0.14418 | 0.508 |
| 7854 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-576-5p | 12 | GREM1 | Sponge network | 0.889 | 9.0E-5 | 0.902 | 0.01691 | 0.508 |
| 7855 | FAM66C |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 18 | CXCL12 | Sponge network | -0.901 | 0.00019 | -1.421 | 0 | 0.508 |
| 7856 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | CHD2 | Sponge network | 1.308 | 0 | 0.049 | 0.55371 | 0.508 |
| 7857 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | MYO9A | Sponge network | 0.817 | 3.0E-5 | -0.147 | 0.32373 | 0.508 |
| 7858 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | KAT6B | Sponge network | -1.575 | 6.0E-5 | -0.478 | 0.00018 | 0.508 |
| 7859 | LINC00473 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p | 15 | SORCS1 | Sponge network | -1.203 | 0.03881 | -3.492 | 0 | 0.508 |
| 7860 | RP11-774O3.3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | SFRP1 | Sponge network | -0.487 | 0.02157 | -3.566 | 0 | 0.508 |
| 7861 | RP11-473I1.9 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-200b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | UBR3 | Sponge network | 0.683 | 1.0E-5 | -0.021 | 0.82774 | 0.508 |
| 7862 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | TSHZ3 | Sponge network | -0.733 | 0.19469 | -0.556 | 0.01516 | 0.508 |
| 7863 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 20 | FGFR1 | Sponge network | -0.421 | 0.0114 | -0.509 | 0.03957 | 0.508 |
| 7864 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 53 | ADARB1 | Sponge network | -0.777 | 0.1134 | -0.335 | 0.06631 | 0.508 |
| 7865 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935 | 36 | ABI2 | Sponge network | 0.75 | 0.00054 | 0.175 | 0.14787 | 0.508 |
| 7866 | LINC01018 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 20 | MRVI1 | Sponge network | -2.915 | 0.00015 | -1.051 | 0.00022 | 0.508 |
| 7867 | RP11-13K12.1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 10 | SORCS2 | Sponge network | -0.154 | 0.78939 | -0.223 | 0.4253 | 0.508 |
| 7868 | ZNF582-AS1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-577 | 11 | AMPH | Sponge network | -1.349 | 0.00017 | -0.916 | 5.0E-5 | 0.508 |
| 7869 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | EDA2R | Sponge network | -1.575 | 6.0E-5 | -0.587 | 0.01598 | 0.508 |
| 7870 | FENDRR |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 21 | EZH1 | Sponge network | -1.281 | 0.00012 | -0.564 | 1.0E-5 | 0.508 |
| 7871 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 15 | CHD1 | Sponge network | 0.537 | 0.00139 | 0.322 | 0.00215 | 0.508 |
| 7872 | RP5-1085F17.3 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 52 | MDM4 | Sponge network | 0.541 | 0.00125 | 0.345 | 0.00762 | 0.508 |
| 7873 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p | 26 | EPHA7 | Sponge network | -0.517 | 0.02935 | -2.197 | 3.0E-5 | 0.508 |
| 7874 | MRVI1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 21 | AFF1 | Sponge network | -1.092 | 4.0E-5 | -0.384 | 0.00053 | 0.508 |
| 7875 | CTC-378H22.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-744-3p;hsa-miR-877-5p | 13 | PTPRT | Sponge network | 0.001 | 0.99663 | -0.907 | 0.03727 | 0.508 |
| 7876 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-493-5p | 17 | ZFHX3 | Sponge network | 0.368 | 0.20093 | 0.389 | 0.01363 | 0.508 |
| 7877 | HNRNPU-AS1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | ATM | Sponge network | 1.601 | 0 | 0.178 | 0.21568 | 0.508 |
| 7878 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | RB1CC1 | Sponge network | 0.765 | 7.0E-5 | 0.175 | 0.12559 | 0.508 |
| 7879 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378c;hsa-miR-576-5p | 21 | ERC1 | Sponge network | 0.889 | 9.0E-5 | -0.108 | 0.38794 | 0.508 |
| 7880 | LINC00473 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 19 | PRUNE2 | Sponge network | -1.203 | 0.03881 | -1.733 | 0.00022 | 0.508 |
| 7881 | LINC00672 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 25 | SYNPO2 | Sponge network | -0.227 | 0.31657 | -2.342 | 0 | 0.508 |
| 7882 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 45 | NTRK3 | Sponge network | -0.737 | 0.10822 | -1.77 | 3.0E-5 | 0.508 |
| 7883 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | JAKMIP2 | Sponge network | -0.323 | 0.01372 | -1.388 | 0 | 0.508 |
| 7884 | LINC00865 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 23 | AKAP6 | Sponge network | -1.249 | 7.0E-5 | -1.586 | 3.0E-5 | 0.508 |
| 7885 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-766-3p | 18 | TBL1XR1 | Sponge network | 1.317 | 0 | 0.578 | 0 | 0.508 |
| 7886 | RP11-774O3.3 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | CSDE1 | Sponge network | -0.487 | 0.02157 | -0.493 | 0 | 0.508 |
| 7887 | JAZF1-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NAV3 | Sponge network | -0.769 | 0.00035 | 0.212 | 0.47729 | 0.508 |
| 7888 | RP11-2H8.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | NR2C2 | Sponge network | 1.089 | 0 | 0.21 | 0.03224 | 0.508 |
| 7889 | PCA3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-501-5p;hsa-miR-589-5p | 12 | MN1 | Sponge network | -0.709 | 0.18168 | -0.358 | 0.24172 | 0.508 |
| 7890 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-96-5p | 28 | ZFHX4 | Sponge network | -0.517 | 0.02935 | 0.018 | 0.95574 | 0.508 |
| 7891 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 26 | RARB | Sponge network | -1.111 | 0.0003 | -0.825 | 2.0E-5 | 0.508 |
| 7892 | MIR143HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 19 | SPTBN4 | Sponge network | -0.739 | 0.0574 | -0.786 | 0.01281 | 0.508 |
| 7893 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 41 | TACC1 | Sponge network | -0.222 | 0.32445 | -0.362 | 0.05406 | 0.508 |
| 7894 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 61 | SSBP2 | Sponge network | -2.925 | 0 | -1.58 | 0 | 0.508 |
| 7895 | LINC00641 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 11 | PCM1 | Sponge network | -0.222 | 0.32445 | 0.164 | 0.20307 | 0.508 |
| 7896 | FENDRR |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935;hsa-miR-96-5p | 42 | FBXO32 | Sponge network | -1.281 | 0.00012 | -0.495 | 0.07561 | 0.508 |
| 7897 | ACTA2-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 19 | SORCS1 | Sponge network | 0.194 | 0.64278 | -3.492 | 0 | 0.508 |
| 7898 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-96-5p | 24 | CRTC3 | Sponge network | 0.194 | 0.64278 | 0.164 | 0.16786 | 0.508 |
| 7899 | RP11-57H14.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p | 17 | EZH1 | Sponge network | 0.083 | 0.66405 | -0.564 | 1.0E-5 | 0.508 |
| 7900 | MEG3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-5p | 15 | SPG20 | Sponge network | 0.249 | 0.35902 | -1.16 | 0 | 0.508 |
| 7901 | LINC00578 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-324-5p;hsa-miR-342-3p | 10 | STARD13 | Sponge network | 0.811 | 0.03228 | -0.187 | 0.27383 | 0.507 |
| 7902 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-5p | 36 | BMPR2 | Sponge network | -1.331 | 3.0E-5 | 0.206 | 0.0635 | 0.507 |
| 7903 | RP11-531A24.5 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 58 | MDM4 | Sponge network | 0.75 | 0.00054 | 0.345 | 0.00762 | 0.507 |
| 7904 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 16 | ATRX | Sponge network | 1.04 | 0.00023 | 0.261 | 0.04408 | 0.507 |
| 7905 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-7-5p | 27 | ERC1 | Sponge network | 0.214 | 0.18246 | -0.108 | 0.38794 | 0.507 |
| 7906 | ARHGEF26-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 18 | ID4 | Sponge network | -2.46 | 0 | -1.644 | 0 | 0.507 |
| 7907 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SEMA3E | Sponge network | -0.942 | 0 | -1.717 | 0.00137 | 0.507 |
| 7908 | CTA-363E19.2 |
hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-409-3p;hsa-miR-495-3p | 10 | ATRX | Sponge network | 1.055 | 0 | 0.261 | 0.04408 | 0.507 |
| 7909 | RP11-25K19.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-493-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | -1.057 | 0.00029 | -1.051 | 0.00022 | 0.507 |
| 7910 | LINC00969 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 27 | TBL1XR1 | Sponge network | 0.683 | 1.0E-5 | 0.578 | 0 | 0.507 |
| 7911 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-96-5p | 37 | FBXO32 | Sponge network | 0.101 | 0.60919 | -0.495 | 0.07561 | 0.507 |
| 7912 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | TCF4 | Sponge network | -1.349 | 0.00017 | 0.016 | 0.92271 | 0.507 |
| 7913 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-98-5p | 38 | PBX1 | Sponge network | -0.405 | 0.22013 | -1.206 | 0 | 0.507 |
| 7914 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-451a;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 42 | BCL2 | Sponge network | -0.323 | 0.01372 | -0.899 | 0 | 0.507 |
| 7915 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 39 | AKAP11 | Sponge network | 0.72 | 0.0001 | 0.196 | 0.09825 | 0.507 |
| 7916 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-654-3p | 34 | RICTOR | Sponge network | 0.637 | 0.00069 | 0.388 | 0.00188 | 0.507 |
| 7917 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-455-5p | 10 | THSD7A | Sponge network | -0.323 | 0.01372 | 0.242 | 0.25154 | 0.507 |
| 7918 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 28 | LIFR | Sponge network | -1.439 | 4.0E-5 | -1.794 | 0 | 0.507 |
| 7919 | TRAF3IP2-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 18 | INPP4A | Sponge network | -0.323 | 0.01372 | 0.07 | 0.53563 | 0.507 |
| 7920 | CTA-941F9.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-576-5p | 14 | AR | Sponge network | -0.344 | 0.49624 | -1.554 | 0 | 0.507 |
| 7921 | RP11-774O3.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p | 34 | IL17RD | Sponge network | -0.487 | 0.02157 | -0.019 | 0.94873 | 0.507 |
| 7922 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-98-5p | 20 | ITGB3 | Sponge network | -1.697 | 0.00889 | -0.011 | 0.95751 | 0.507 |
| 7923 | PCA3 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | RPS6KA6 | Sponge network | -0.709 | 0.18168 | -2.989 | 0 | 0.507 |
| 7924 | PART1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-889-3p | 22 | RPS6KA6 | Sponge network | -2.925 | 0 | -2.989 | 0 | 0.507 |
| 7925 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | TSHZ3 | Sponge network | -1.161 | 0.00591 | -0.556 | 0.01516 | 0.507 |
| 7926 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-497-5p | 20 | CBL | Sponge network | 0.946 | 0 | 0.291 | 0.01267 | 0.507 |
| 7927 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-98-5p | 38 | DMD | Sponge network | 0.056 | 0.81195 | -1.506 | 0 | 0.507 |
| 7928 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 51 | PDGFRA | Sponge network | -1.111 | 0.0003 | -0.372 | 0.0037 | 0.507 |
| 7929 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | KL | Sponge network | -0.576 | 0.10121 | -1.294 | 0 | 0.507 |
| 7930 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 13 | BCLAF1 | Sponge network | 0.782 | 0.00016 | 0.211 | 0.00684 | 0.507 |
| 7931 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | FLI1 | Sponge network | -0.129 | 0.57177 | -0.294 | 0.10905 | 0.507 |
| 7932 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-582-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 38 | GAS7 | Sponge network | -0.465 | 0.04703 | -0.555 | 0.01602 | 0.507 |
| 7933 | RP11-822E23.8 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 15 | FLNA | Sponge network | -0.777 | 0.16667 | -0.771 | 0.01377 | 0.507 |
| 7934 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-576-5p | 19 | GREM1 | Sponge network | -2.117 | 0.00161 | 0.902 | 0.01691 | 0.507 |
| 7935 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | 0.101 | 0.60919 | 0.358 | 0.13727 | 0.507 |
| 7936 | LINC00473 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | -1.203 | 0.03881 | -0.974 | 0.00034 | 0.507 |
| 7937 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-940 | 45 | QKI | Sponge network | -0.777 | 0.16667 | -0.333 | 0.04111 | 0.507 |
| 7938 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 65 | BMPR2 | Sponge network | 0.194 | 0.64278 | 0.206 | 0.0635 | 0.507 |
| 7939 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | NCAM2 | Sponge network | 0.194 | 0.64278 | -0.482 | 0.22565 | 0.507 |
| 7940 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 20 | SPG20 | Sponge network | -0.942 | 0 | -1.16 | 0 | 0.507 |
| 7941 | TRAF3IP2-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 11 | PRKD1 | Sponge network | -0.323 | 0.01372 | -0.466 | 0.03583 | 0.507 |
| 7942 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 56 | LPP | Sponge network | -0.031 | 0.89063 | -0.459 | 0.04475 | 0.507 |
| 7943 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p | 37 | BCL2 | Sponge network | -0.096 | 0.67958 | -0.899 | 0 | 0.507 |
| 7944 | LINC00473 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TAL1 | Sponge network | -1.203 | 0.03881 | -0.462 | 0.00574 | 0.507 |
| 7945 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-625-5p;hsa-miR-93-5p | 33 | JAZF1 | Sponge network | -2.69 | 0.0001 | -0.377 | 0.02677 | 0.507 |
| 7946 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p | 22 | DLC1 | Sponge network | -0.246 | 0.35034 | -0.382 | 0.05038 | 0.507 |
| 7947 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | PLSCR4 | Sponge network | -0.769 | 0.00035 | -0.474 | 0.03833 | 0.507 |
| 7948 | RP11-283I3.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 34 | CREB1 | Sponge network | 0.614 | 1.0E-5 | 0.203 | 0.00537 | 0.507 |
| 7949 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ABCC9 | Sponge network | -1.161 | 0.00591 | -0.466 | 0.14121 | 0.507 |
| 7950 | HAND2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 21 | ABCA10 | Sponge network | -1.697 | 0.00889 | -0.571 | 0.03674 | 0.507 |
| 7951 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | TSHZ3 | Sponge network | -0.318 | 0.65847 | -0.556 | 0.01516 | 0.507 |
| 7952 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-362-3p | 12 | CACNA2D1 | Sponge network | -0.469 | 0.08721 | -0.846 | 0.00675 | 0.507 |
| 7953 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-93-3p | 31 | NTRK3 | Sponge network | -0.932 | 0.00329 | -1.77 | 3.0E-5 | 0.507 |
| 7954 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 19 | ITCH | Sponge network | 1.063 | 0 | 0.084 | 0.34058 | 0.507 |
| 7955 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | ABCB5 | Sponge network | -0.737 | 0.06182 | -0.956 | 0.04077 | 0.507 |
| 7956 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 34 | MYO9A | Sponge network | -0.709 | 0.18168 | -0.147 | 0.32373 | 0.507 |
| 7957 | ADAMTS9-AS2 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | LIMCH1 | Sponge network | -2.452 | 0 | -0.915 | 1.0E-5 | 0.507 |
| 7958 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | ZFHX3 | Sponge network | 0.219 | 0.1516 | 0.389 | 0.01363 | 0.507 |
| 7959 | SNHG14 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRIM58 | Sponge network | -1.111 | 0.0003 | -1.692 | 0 | 0.507 |
| 7960 | AC003090.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | PREX2 | Sponge network | -2.117 | 0.00161 | -0.272 | 0.25382 | 0.507 |
| 7961 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 59 | BMPR2 | Sponge network | -0.222 | 0.32445 | 0.206 | 0.0635 | 0.507 |
| 7962 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p | 12 | EPHA3 | Sponge network | 0.497 | 0.01451 | 0.185 | 0.53588 | 0.507 |
| 7963 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | SNX29 | Sponge network | -0.351 | 0.11358 | -0.34 | 0.01371 | 0.507 |
| 7964 | RP11-166D19.1 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | RTN4 | Sponge network | -0.576 | 0.10121 | -0.412 | 0.00014 | 0.507 |
| 7965 | EMX2OS |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | CDH19 | Sponge network | -0.777 | 0.1134 | -3.434 | 0 | 0.507 |
| 7966 | CTC-429P9.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p | 14 | ITPKB | Sponge network | 0.082 | 0.55397 | -0.704 | 0.00106 | 0.507 |
| 7967 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-493-5p;hsa-miR-7-1-3p | 12 | SEMA3E | Sponge network | -0.976 | 0.00025 | -1.717 | 0.00137 | 0.507 |
| 7968 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p | 14 | PRRX1 | Sponge network | -0.154 | 0.53954 | 1.316 | 0 | 0.507 |
| 7969 | RP11-307B6.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-589-3p | 24 | TGFBR3 | Sponge network | -4.463 | 0.00025 | -1.173 | 1.0E-5 | 0.507 |
| 7970 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 26 | SYNE1 | Sponge network | -1.249 | 7.0E-5 | -0.738 | 0.00316 | 0.507 |
| 7971 | SNHG14 |
hsa-let-7d-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-708-5p;hsa-miR-98-5p | 11 | MPL | Sponge network | -1.111 | 0.0003 | -0.624 | 0.00133 | 0.507 |
| 7972 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p | 22 | DMD | Sponge network | -0.154 | 0.53954 | -1.506 | 0 | 0.507 |
| 7973 | ANKRD10-IT1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 11 | PHF6 | Sponge network | 1.739 | 0 | 0.785 | 0 | 0.507 |
| 7974 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-7-5p | 13 | PTGFR | Sponge network | -0.611 | 0.09607 | -0.233 | 0.45421 | 0.507 |
| 7975 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-337-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | MOB1B | Sponge network | 1.378 | 0 | 0.197 | 0.15266 | 0.507 |
| 7976 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-532-3p | 24 | PBX1 | Sponge network | -0.737 | 0.10822 | -1.206 | 0 | 0.507 |
| 7977 | LINC00865 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-93-3p | 51 | RUNX1T1 | Sponge network | -1.249 | 7.0E-5 | -0.344 | 0.2374 | 0.507 |
| 7978 | TP73-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-542-3p | 19 | SYNM | Sponge network | -0.563 | 0.02545 | -2.309 | 1.0E-5 | 0.506 |
| 7979 | WDFY3-AS2 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429 | 15 | NGFR | Sponge network | -0.942 | 0 | -1.271 | 0.01058 | 0.506 |
| 7980 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 16 | PDE4DIP | Sponge network | -0.358 | 0.17255 | -0.282 | 0.0257 | 0.506 |
| 7981 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 15 | AKAP12 | Sponge network | 0.101 | 0.60919 | -0.932 | 0.0028 | 0.506 |
| 7982 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 54 | ADARB1 | Sponge network | 0.997 | 0.00066 | -0.335 | 0.06631 | 0.506 |
| 7983 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-450b-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 28 | CREB3L2 | Sponge network | -1.092 | 4.0E-5 | -0.525 | 2.0E-5 | 0.506 |
| 7984 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-92a-3p | 11 | DKK3 | Sponge network | 0.199 | 0.38015 | -0.052 | 0.77783 | 0.506 |
| 7985 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 16 | EPHA3 | Sponge network | -1.582 | 8.0E-5 | 0.185 | 0.53588 | 0.506 |
| 7986 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p | 10 | ZFHX3 | Sponge network | 1.134 | 0 | 0.389 | 0.01363 | 0.506 |
| 7987 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-452-5p;hsa-miR-590-3p | 30 | BCL2 | Sponge network | -0.358 | 0.17255 | -0.899 | 0 | 0.506 |
| 7988 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-500a-5p | 11 | PREX2 | Sponge network | -0.777 | 0.16667 | -0.272 | 0.25382 | 0.506 |
| 7989 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 18 | PDS5B | Sponge network | 0.537 | 0.00139 | 0.259 | 0.00962 | 0.506 |
| 7990 | PCA3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 37 | DIP2C | Sponge network | -0.709 | 0.18168 | -0.436 | 0.01713 | 0.506 |
| 7991 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-628-5p | 51 | DTNA | Sponge network | -0.154 | 0.78939 | -1.648 | 1.0E-5 | 0.506 |
| 7992 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p | 10 | SYNM | Sponge network | -0.187 | 0.23787 | -2.309 | 1.0E-5 | 0.506 |
| 7993 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 49 | ZFHX3 | Sponge network | -0.129 | 0.57177 | 0.389 | 0.01363 | 0.506 |
| 7994 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 43 | ZFHX4 | Sponge network | -0.096 | 0.67958 | 0.018 | 0.95574 | 0.506 |
| 7995 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-589-3p | 13 | RELN | Sponge network | -0.777 | 0.16667 | -2.403 | 0 | 0.506 |
| 7996 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 35 | PGR | Sponge network | -0.615 | 0.00015 | -1.704 | 0 | 0.506 |
| 7997 | LINC00473 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 12 | UNC5C | Sponge network | -1.203 | 0.03881 | -0.178 | 0.40214 | 0.506 |
| 7998 | CTD-2314G24.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 34 | IGFBP5 | Sponge network | 0.246 | 0.59912 | -0.806 | 0.00105 | 0.506 |
| 7999 | MIR497HG |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | NCAM2 | Sponge network | -1.439 | 4.0E-5 | -0.482 | 0.22565 | 0.506 |
| 8000 | RP11-867G23.10 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-589-3p | 17 | DIP2C | Sponge network | -2.531 | 0 | -0.436 | 0.01713 | 0.506 |
| 8001 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-582-5p | 12 | EBF3 | Sponge network | -2.095 | 0.00112 | -0.058 | 0.81437 | 0.506 |
| 8002 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | ABCA10 | Sponge network | -0.129 | 0.57177 | -0.571 | 0.03674 | 0.506 |
| 8003 | CTC-296K1.4 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p | 16 | GRIK3 | Sponge network | -2.076 | 0.00548 | -3.208 | 0 | 0.506 |
| 8004 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 10 | PGR | Sponge network | -1.255 | 0.00981 | -1.704 | 0 | 0.506 |
| 8005 | AC006116.24 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-455-5p | 13 | GJA1 | Sponge network | -0.473 | 0.07602 | -0.485 | 0.02179 | 0.506 |
| 8006 | FENDRR |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 20 | CDH23 | Sponge network | -1.281 | 0.00012 | -0.974 | 0.00034 | 0.506 |
| 8007 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-550a-3p;hsa-miR-576-5p | 22 | HECA | Sponge network | -1.645 | 0 | -0.217 | 0.03463 | 0.506 |
| 8008 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 22 | CNTNAP3 | Sponge network | -0.49 | 0.04946 | -1.465 | 0.00033 | 0.506 |
| 8009 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 37 | SNX29 | Sponge network | -0.49 | 0.04946 | -0.34 | 0.01371 | 0.506 |
| 8010 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 37 | ABI2 | Sponge network | -0.405 | 0.22013 | 0.175 | 0.14787 | 0.506 |
| 8011 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | DCN | Sponge network | -2.46 | 0 | -1.288 | 0 | 0.506 |
| 8012 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-874-3p | 12 | GIGYF2 | Sponge network | 1.254 | 0 | 0.136 | 0.04403 | 0.506 |
| 8013 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-877-5p | 10 | GRIN2A | Sponge network | -0.309 | 0.1145 | -2.161 | 0 | 0.506 |
| 8014 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p | 29 | ZFHX3 | Sponge network | 1.757 | 0 | 0.389 | 0.01363 | 0.506 |
| 8015 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b | 10 | CITED2 | Sponge network | -1.092 | 4.0E-5 | -1.545 | 0 | 0.506 |
| 8016 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-424-5p | 10 | PLSCR4 | Sponge network | -2.076 | 0.00548 | -0.474 | 0.03833 | 0.506 |
| 8017 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-495-3p | 10 | TRIM33 | Sponge network | 1.178 | 0 | 0.402 | 7.0E-5 | 0.506 |
| 8018 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 37 | PTPRT | Sponge network | -2.117 | 0.00161 | -0.907 | 0.03727 | 0.506 |
| 8019 | RP4-669L17.10 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c | 23 | ABL2 | Sponge network | 1.093 | 0 | 0.82 | 0 | 0.506 |
| 8020 | CTC-429P9.5 |
hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-590-3p | 10 | ITPKB | Sponge network | 0.178 | 0.29106 | -0.704 | 0.00106 | 0.506 |
| 8021 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p | 34 | PRKAA2 | Sponge network | 0.176 | 0.37402 | -2.462 | 0 | 0.506 |
| 8022 | RP11-815I9.4 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-625-3p | 16 | ABL2 | Sponge network | 0.537 | 0.0006 | 0.82 | 0 | 0.506 |
| 8023 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-93-3p | 18 | RPS6KA2 | Sponge network | -0.769 | 0.00035 | -0.57 | 0.00013 | 0.506 |
| 8024 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-98-5p | 30 | LRIG1 | Sponge network | -1.697 | 0.00889 | -0.537 | 0.00588 | 0.506 |
| 8025 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-503-5p | 24 | ABL1 | Sponge network | -0.976 | 0.00025 | -0.215 | 0.07804 | 0.506 |
| 8026 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-500a-5p | 21 | HLF | Sponge network | -0.932 | 0.00329 | -2.443 | 0 | 0.506 |
| 8027 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | FGF5 | Sponge network | 0.194 | 0.64278 | 0.549 | 0.28659 | 0.506 |
| 8028 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-550a-5p;hsa-miR-940 | 20 | TOM1L2 | Sponge network | -1.331 | 3.0E-5 | -0.994 | 0 | 0.506 |
| 8029 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-654-3p | 26 | DDX6 | Sponge network | 0.946 | 0 | 0.091 | 0.36428 | 0.506 |
| 8030 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 62 | TCF4 | Sponge network | -0.901 | 0.00019 | 0.016 | 0.92271 | 0.506 |
| 8031 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 15 | EBF3 | Sponge network | -1.582 | 8.0E-5 | -0.058 | 0.81437 | 0.506 |
| 8032 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | RCAN2 | Sponge network | -0.227 | 0.31657 | -0.33 | 0.15172 | 0.506 |
| 8033 | RP11-589P10.7 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-324-3p | 12 | MEF2C | Sponge network | -2.044 | 0.00066 | -0.499 | 0.01448 | 0.506 |
| 8034 | RP11-867G23.10 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-424-5p | 12 | PTGFR | Sponge network | -2.531 | 0 | -0.233 | 0.45421 | 0.506 |
| 8035 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DCN | Sponge network | 0.997 | 0.00066 | -1.288 | 0 | 0.506 |
| 8036 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 14 | PTPN11 | Sponge network | 1.317 | 0 | 0.431 | 2.0E-5 | 0.506 |
| 8037 | AC003090.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p | 25 | WWTR1 | Sponge network | -2.117 | 0.00161 | -0.345 | 0.11997 | 0.506 |
| 8038 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-337-3p;hsa-miR-590-3p;hsa-miR-940 | 10 | HOOK3 | Sponge network | 1.153 | 0 | 0.273 | 0.09957 | 0.506 |
| 8039 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | AKAP11 | Sponge network | 1.153 | 0 | 0.196 | 0.09825 | 0.506 |
| 8040 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | MCC | Sponge network | -0.323 | 0.01372 | -1.005 | 0 | 0.506 |
| 8041 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 22 | NAV3 | Sponge network | 0.214 | 0.18246 | 0.212 | 0.47729 | 0.506 |
| 8042 | LINC00473 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 20 | CADM3 | Sponge network | -1.203 | 0.03881 | -3.663 | 0 | 0.506 |
| 8043 | CTC-510F12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-766-3p | 14 | CADM2 | Sponge network | -0.691 | 0.00146 | -3.782 | 0 | 0.506 |
| 8044 | GS1-124K5.11 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-766-3p | 15 | EZH1 | Sponge network | 0.494 | 0.00339 | -0.564 | 1.0E-5 | 0.506 |
| 8045 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-660-5p | 16 | TIMP3 | Sponge network | -1.582 | 8.0E-5 | -0.546 | 0.02307 | 0.506 |
| 8046 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 26 | CNTN1 | Sponge network | -0.405 | 0.22013 | -2.594 | 0 | 0.506 |
| 8047 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | CHD2 | Sponge network | -0.031 | 0.89063 | 0.049 | 0.55371 | 0.506 |
| 8048 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p | 45 | FAT3 | Sponge network | -0.777 | 0.16667 | -1.601 | 0.00051 | 0.506 |
| 8049 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-940 | 41 | MEF2C | Sponge network | -0.162 | 0.47687 | -0.499 | 0.01448 | 0.506 |
| 8050 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-429;hsa-miR-532-3p;hsa-miR-671-5p;hsa-miR-93-5p | 43 | GAS7 | Sponge network | -2.69 | 0.0001 | -0.555 | 0.01602 | 0.506 |
| 8051 | CTD-3064M3.3 |
hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-671-5p | 11 | KIT | Sponge network | -0.469 | 0.08721 | -2.184 | 0 | 0.506 |
| 8052 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p | 15 | RET | Sponge network | -0.469 | 0.08721 | -0.833 | 0.03517 | 0.506 |
| 8053 | MYLK-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-330-3p | 22 | AFF1 | Sponge network | -1.331 | 3.0E-5 | -0.384 | 0.00053 | 0.506 |
| 8054 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 54 | QKI | Sponge network | -0.777 | 0.1134 | -0.333 | 0.04111 | 0.505 |
| 8055 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 26 | TSC1 | Sponge network | 0.75 | 0.00054 | 0.103 | 0.26071 | 0.505 |
| 8056 | NR2F1-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | SYT4 | Sponge network | -0.49 | 0.04946 | -3.207 | 0 | 0.505 |
| 8057 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-624-5p | 11 | SMARCA1 | Sponge network | -2.452 | 0 | -0.684 | 0.00159 | 0.505 |
| 8058 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | TSHZ3 | Sponge network | 0.342 | 0.03129 | -0.556 | 0.01516 | 0.505 |
| 8059 | AC009948.5 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-877-5p | 20 | GLIPR1 | Sponge network | 0.532 | 0.00505 | 0.146 | 0.3276 | 0.505 |
| 8060 | LINC00702 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 16 | PDZD2 | Sponge network | -0.737 | 0.06182 | -0.909 | 3.0E-5 | 0.505 |
| 8061 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 16 | CHD1 | Sponge network | 0.782 | 0.00016 | 0.322 | 0.00215 | 0.505 |
| 8062 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-484;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p | 37 | SOX5 | Sponge network | -0.096 | 0.67958 | -1.091 | 4.0E-5 | 0.505 |
| 8063 | MIR143HG |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | RTN4 | Sponge network | -0.739 | 0.0574 | -0.412 | 0.00014 | 0.505 |
| 8064 | FGF14-AS2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-590-3p | 15 | DLC1 | Sponge network | -1.582 | 8.0E-5 | -0.382 | 0.05038 | 0.505 |
| 8065 | RP11-283I3.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-582-3p;hsa-miR-625-3p | 29 | ABL2 | Sponge network | 0.614 | 1.0E-5 | 0.82 | 0 | 0.505 |
| 8066 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 44 | ZFHX3 | Sponge network | 0.997 | 0.00066 | 0.389 | 0.01363 | 0.505 |
| 8067 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 43 | MOB1B | Sponge network | 0.421 | 0.00084 | 0.197 | 0.15266 | 0.505 |
| 8068 | TP73-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | ERG | Sponge network | -0.563 | 0.02545 | 0.002 | 0.99011 | 0.505 |
| 8069 | RAMP2-AS1 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | RADIL | Sponge network | -0.513 | 0.04703 | -1.628 | 0 | 0.505 |
| 8070 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | WWTR1 | Sponge network | -0.563 | 0.02545 | -0.345 | 0.11997 | 0.505 |
| 8071 | RP11-122K13.12 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 12 | ABL2 | Sponge network | 1.219 | 0 | 0.82 | 0 | 0.505 |
| 8072 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-940 | 12 | HOOK3 | Sponge network | 0.389 | 0.04293 | 0.273 | 0.09957 | 0.505 |
| 8073 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 16 | ADAMTSL3 | Sponge network | 0.693 | 0.00076 | -1.559 | 0 | 0.505 |
| 8074 | CTA-363E19.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | SHPRH | Sponge network | 1.055 | 0 | 0.098 | 0.40994 | 0.505 |
| 8075 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p | 13 | RCAN2 | Sponge network | 0.75 | 0.00054 | -0.33 | 0.15172 | 0.505 |
| 8076 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 21 | IGF1 | Sponge network | -0.737 | 0.10822 | -0.204 | 0.5898 | 0.505 |
| 8077 | LINC01018 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 11 | PARK2 | Sponge network | -2.915 | 0.00015 | -1.892 | 0 | 0.505 |
| 8078 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 24 | GJA1 | Sponge network | 0.997 | 0.00066 | -0.485 | 0.02179 | 0.505 |
| 8079 | MYLK-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-484 | 14 | GRIA3 | Sponge network | -1.331 | 3.0E-5 | -1.956 | 0 | 0.505 |
| 8080 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | DOCK4 | Sponge network | 0.532 | 0.00505 | 0.502 | 0.00108 | 0.505 |
| 8081 | C20orf166-AS1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429 | 27 | MEF2C | Sponge network | -2.69 | 0.0001 | -0.499 | 0.01448 | 0.505 |
| 8082 | MAGI2-AS3 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-7-5p | 11 | COL1A2 | Sponge network | -0.129 | 0.57177 | 1.991 | 0 | 0.505 |
| 8083 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p | 16 | ZNF208 | Sponge network | 0.335 | 0.20596 | -0.4 | 0.22381 | 0.505 |
| 8084 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | TMEFF2 | Sponge network | -0.769 | 0.00035 | -2.426 | 0 | 0.505 |
| 8085 | AC010226.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 21 | DMD | Sponge network | -0.811 | 2.0E-5 | -1.506 | 0 | 0.505 |
| 8086 | USP3-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375 | 14 | ITGB1 | Sponge network | -0.162 | 0.47687 | 0.524 | 0.00017 | 0.505 |
| 8087 | LINC00657 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | ERCC6 | Sponge network | 0.91 | 0 | 0.23 | 0.015 | 0.505 |
| 8088 | RP11-244O19.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-92a-1-5p;hsa-miR-935;hsa-miR-940 | 43 | GRIK3 | Sponge network | -0.096 | 0.67958 | -3.208 | 0 | 0.505 |
| 8089 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p | 10 | TXNIP | Sponge network | -4.546 | 0 | -1.277 | 0 | 0.505 |
| 8090 | LINC00578 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 18 | CDH19 | Sponge network | 0.811 | 0.03228 | -3.434 | 0 | 0.505 |
| 8091 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-22-5p | 10 | PRKAA2 | Sponge network | -0.326 | 0.28809 | -2.462 | 0 | 0.505 |
| 8092 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b | 10 | DCN | Sponge network | -0.517 | 0.02935 | -1.288 | 0 | 0.505 |
| 8093 | HCG11 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-590-3p | 11 | MACF1 | Sponge network | 0.385 | 0.10067 | 0.019 | 0.90335 | 0.505 |
| 8094 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 21 | EBF3 | Sponge network | -0.896 | 0.00099 | -0.058 | 0.81437 | 0.505 |
| 8095 | TRAF3IP2-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 42 | GRIK3 | Sponge network | -0.323 | 0.01372 | -3.208 | 0 | 0.505 |
| 8096 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p | 14 | NR2C2 | Sponge network | 1.044 | 2.0E-5 | 0.21 | 0.03224 | 0.505 |
| 8097 | RASSF8-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-532-3p | 17 | SYNM | Sponge network | 0.335 | 0.20596 | -2.309 | 1.0E-5 | 0.505 |
| 8098 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 31 | MYO9A | Sponge network | 0.249 | 0.35902 | -0.147 | 0.32373 | 0.505 |
| 8099 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935 | 47 | FAT3 | Sponge network | -1.203 | 0.03881 | -1.601 | 0.00051 | 0.505 |
| 8100 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | NRXN1 | Sponge network | -0.487 | 0.02157 | -3.778 | 0 | 0.505 |
| 8101 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-671-5p;hsa-miR-7-5p | 16 | CRTC3 | Sponge network | -0.611 | 0.09607 | 0.164 | 0.16786 | 0.505 |
| 8102 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 20 | PRUNE2 | Sponge network | 0.335 | 0.20596 | -1.733 | 0.00022 | 0.505 |
| 8103 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | SYNE1 | Sponge network | -0.72 | 0.24239 | -0.738 | 0.00316 | 0.505 |
| 8104 | AC006116.24 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-577 | 12 | TET1 | Sponge network | -0.473 | 0.07602 | -0.135 | 0.52138 | 0.505 |
| 8105 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | AR | Sponge network | -0.726 | 0.00132 | -1.554 | 0 | 0.505 |
| 8106 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -0.842 | 0.01361 | -0.243 | 0.11408 | 0.505 |
| 8107 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-425-5p;hsa-miR-589-5p | 20 | ADARB1 | Sponge network | -0.187 | 0.23787 | -0.335 | 0.06631 | 0.505 |
| 8108 | ZNF582-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-7-1-3p | 13 | ITPKB | Sponge network | -1.349 | 0.00017 | -0.704 | 0.00106 | 0.505 |
| 8109 | RP11-6O2.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-766-3p | 13 | EZH1 | Sponge network | 0.331 | 0.57247 | -0.564 | 1.0E-5 | 0.505 |
| 8110 | MRVI1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-628-5p;hsa-miR-708-5p | 22 | TOM1L2 | Sponge network | -1.092 | 4.0E-5 | -0.994 | 0 | 0.505 |
| 8111 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-628-5p | 48 | RUNX1T1 | Sponge network | -0.842 | 0.01361 | -0.344 | 0.2374 | 0.505 |
| 8112 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p | 13 | RET | Sponge network | -1.092 | 4.0E-5 | -0.833 | 0.03517 | 0.505 |
| 8113 | RP11-359B12.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PRUNE2 | Sponge network | 0.064 | 0.72934 | -1.733 | 0.00022 | 0.505 |
| 8114 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p | 36 | FBXO32 | Sponge network | -0.323 | 0.01372 | -0.495 | 0.07561 | 0.505 |
| 8115 | U91328.19 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p | 36 | IL17RD | Sponge network | -0.303 | 0.21002 | -0.019 | 0.94873 | 0.505 |
| 8116 | RP11-774O3.3 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 12 | NGFR | Sponge network | -0.487 | 0.02157 | -1.271 | 0.01058 | 0.505 |
| 8117 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 41 | RASSF2 | Sponge network | -0.576 | 0.10121 | -0.273 | 0.21961 | 0.505 |
| 8118 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 34 | MYO9A | Sponge network | -0.49 | 0.04946 | -0.147 | 0.32373 | 0.505 |
| 8119 | MIR143HG |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PCDH18 | Sponge network | -0.739 | 0.0574 | 0.216 | 0.24645 | 0.505 |
| 8120 | LINC00641 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FBXO11 | Sponge network | -0.222 | 0.32445 | 0.142 | 0.04938 | 0.505 |
| 8121 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p | 15 | MYCBP2 | Sponge network | -0.222 | 0.32445 | -0.027 | 0.82016 | 0.505 |
| 8122 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 13 | PDZD2 | Sponge network | -2.117 | 0.00161 | -0.909 | 3.0E-5 | 0.505 |
| 8123 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 21 | NRXN3 | Sponge network | -0.899 | 6.0E-5 | -0.893 | 0.04186 | 0.504 |
| 8124 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 18 | NAV3 | Sponge network | -0.739 | 0.00044 | 0.212 | 0.47729 | 0.504 |
| 8125 | HOXB-AS1 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-940 | 12 | SFRP1 | Sponge network | -0.154 | 0.53954 | -3.566 | 0 | 0.504 |
| 8126 | GAS6-AS2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-429 | 14 | MRVI1 | Sponge network | 0.16 | 0.50785 | -1.051 | 0.00022 | 0.504 |
| 8127 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | FAM172A | Sponge network | -1.161 | 0.00591 | -0.57 | 0 | 0.504 |
| 8128 | CTC-296K1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p | 10 | PLSCR4 | Sponge network | -2.095 | 0.00112 | -0.474 | 0.03833 | 0.504 |
| 8129 | LINC00865 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 19 | CDH19 | Sponge network | -1.249 | 7.0E-5 | -3.434 | 0 | 0.504 |
| 8130 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-590-3p | 22 | HLF | Sponge network | -1.582 | 8.0E-5 | -2.443 | 0 | 0.504 |
| 8131 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-455-3p;hsa-miR-582-5p | 13 | KCNAB1 | Sponge network | -0.793 | 7.0E-5 | -0.755 | 2.0E-5 | 0.504 |
| 8132 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 25 | EPHA3 | Sponge network | 1.302 | 7.0E-5 | 0.185 | 0.53588 | 0.504 |
| 8133 | RP11-774O3.3 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 13 | STAT5B | Sponge network | -0.487 | 0.02157 | -0.182 | 0.1082 | 0.504 |
| 8134 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-93-3p | 22 | EBF3 | Sponge network | -0.162 | 0.47687 | -0.058 | 0.81437 | 0.504 |
| 8135 | JAZF1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-532-3p;hsa-miR-542-3p | 16 | SYNM | Sponge network | -0.769 | 0.00035 | -2.309 | 1.0E-5 | 0.504 |
| 8136 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p | 13 | SYNM | Sponge network | -0.318 | 0.65847 | -2.309 | 1.0E-5 | 0.504 |
| 8137 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-577 | 18 | TIMP3 | Sponge network | -2.69 | 0.0001 | -0.546 | 0.02307 | 0.504 |
| 8138 | LINC00174 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 17 | RICTOR | Sponge network | 1.253 | 0 | 0.388 | 0.00188 | 0.504 |
| 8139 | AC005154.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 23 | TRIM33 | Sponge network | 0.848 | 0 | 0.402 | 7.0E-5 | 0.504 |
| 8140 | WDFY3-AS2 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 23 | MRVI1 | Sponge network | -0.942 | 0 | -1.051 | 0.00022 | 0.504 |
| 8141 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p | 10 | MGA | Sponge network | 0.663 | 0.00073 | 0.323 | 0.00792 | 0.504 |
| 8142 | RP5-1198O20.4 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | 0.997 | 0.00066 | -1.628 | 0 | 0.504 |
| 8143 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-744-3p | 25 | PTPN14 | Sponge network | -0.323 | 0.01372 | 0.144 | 0.34595 | 0.504 |
| 8144 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | PHC3 | Sponge network | 0.659 | 3.0E-5 | 0.212 | 0.0499 | 0.504 |
| 8145 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-93-3p | 20 | EBF3 | Sponge network | -0.611 | 0.09607 | -0.058 | 0.81437 | 0.504 |
| 8146 | USP3-AS1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-493-3p | 18 | CSDE1 | Sponge network | -0.162 | 0.47687 | -0.493 | 0 | 0.504 |
| 8147 | SNHG14 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | IL17RD | Sponge network | -1.111 | 0.0003 | -0.019 | 0.94873 | 0.504 |
| 8148 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429 | 13 | ERG | Sponge network | -0.469 | 0.08721 | 0.002 | 0.99011 | 0.504 |
| 8149 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | MYO9A | Sponge network | -1.697 | 0.00889 | -0.147 | 0.32373 | 0.504 |
| 8150 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-93-5p | 15 | CNTNAP3 | Sponge network | -1.439 | 4.0E-5 | -1.465 | 0.00033 | 0.504 |
| 8151 | DNM3OS |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 13 | RHOB | Sponge network | 0.572 | 0.01722 | -1.052 | 0 | 0.504 |
| 8152 | TUG1 |
hsa-miR-125a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | ERCC6 | Sponge network | 0.972 | 0 | 0.23 | 0.015 | 0.504 |
| 8153 | RP11-307B6.3 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-93-3p | 12 | EPHA3 | Sponge network | -4.463 | 0.00025 | 0.185 | 0.53588 | 0.504 |
| 8154 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-628-5p | 23 | GRIA3 | Sponge network | -0.793 | 7.0E-5 | -1.956 | 0 | 0.504 |
| 8155 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | CLASP2 | Sponge network | -0.323 | 0.01372 | -0.038 | 0.71 | 0.504 |
| 8156 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ADAMTSL3 | Sponge network | -1.161 | 0.00591 | -1.559 | 0 | 0.504 |
| 8157 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | PIK3C2A | Sponge network | 0.826 | 1.0E-5 | 0.061 | 0.53776 | 0.504 |
| 8158 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ABCA10 | Sponge network | -0.576 | 0.10121 | -0.571 | 0.03674 | 0.504 |
| 8159 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-744-5p;hsa-miR-766-3p | 15 | CHD5 | Sponge network | -4.463 | 0.00025 | -0.922 | 0.01521 | 0.504 |
| 8160 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3913-5p;hsa-miR-93-5p | 27 | ZBTB4 | Sponge network | -0.421 | 0.0114 | -0.739 | 0 | 0.504 |
| 8161 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-940;hsa-miR-96-5p | 30 | CRTC3 | Sponge network | -0.976 | 0.00025 | 0.164 | 0.16786 | 0.504 |
| 8162 | RP11-156P1.3 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | FMNL3 | Sponge network | 0.214 | 0.18246 | 0.555 | 4.0E-5 | 0.504 |
| 8163 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 22 | NR2C2 | Sponge network | 1.11 | 0 | 0.21 | 0.03224 | 0.504 |
| 8164 | WDFY3-AS2 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-98-5p | 11 | HSPB7 | Sponge network | -0.942 | 0 | -2.268 | 1.0E-5 | 0.504 |
| 8165 | LINC00473 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-532-3p | 14 | SYNM | Sponge network | -1.203 | 0.03881 | -2.309 | 1.0E-5 | 0.504 |
| 8166 | PCA3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | RECK | Sponge network | -0.709 | 0.18168 | -1.043 | 0 | 0.504 |
| 8167 | RP11-532F6.3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | INPP4A | Sponge network | -0.896 | 0.00099 | 0.07 | 0.53563 | 0.504 |
| 8168 | RP11-216B9.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 13 | BRD3 | Sponge network | 0.174 | 0.30034 | 0.37 | 0.00013 | 0.504 |
| 8169 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | SOX5 | Sponge network | 0.225 | 0.30644 | -1.091 | 4.0E-5 | 0.504 |
| 8170 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | DOCK4 | Sponge network | 0.572 | 0.01722 | 0.502 | 0.00108 | 0.504 |
| 8171 | RP11-182J1.1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 11 | ITPKB | Sponge network | -0.187 | 0.23787 | -0.704 | 0.00106 | 0.504 |
| 8172 | RP11-384K6.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p;hsa-miR-940 | 32 | MDM4 | Sponge network | 0.946 | 0 | 0.345 | 0.00762 | 0.504 |
| 8173 | LINC00578 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p | 16 | SCN9A | Sponge network | 0.811 | 0.03228 | -0.525 | 0.10593 | 0.504 |
| 8174 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | CACNA2D1 | Sponge network | -0.709 | 0.18168 | -0.846 | 0.00675 | 0.504 |
| 8175 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-127-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-539-5p;hsa-miR-877-5p | 11 | PGR | Sponge network | 0.043 | 0.81628 | -1.704 | 0 | 0.504 |
| 8176 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-93-3p | 13 | CNTNAP1 | Sponge network | -0.154 | 0.78939 | 0.358 | 0.13727 | 0.504 |
| 8177 | PCA3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 19 | SFRP1 | Sponge network | -0.709 | 0.18168 | -3.566 | 0 | 0.504 |
| 8178 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-500b-5p | 13 | SCN9A | Sponge network | -0.932 | 0.00329 | -0.525 | 0.10593 | 0.504 |
| 8179 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421 | 20 | CADM3 | Sponge network | -0.323 | 0.01372 | -3.663 | 0 | 0.504 |
| 8180 | RP11-344B5.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b | 20 | GAS7 | Sponge network | -0.055 | 0.88482 | -0.555 | 0.01602 | 0.504 |
| 8181 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-93-5p | 19 | BNC2 | Sponge network | -2.039 | 0.03447 | -0.491 | 0.09988 | 0.504 |
| 8182 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-766-3p | 21 | IL17RD | Sponge network | -0.583 | 0.14589 | -0.019 | 0.94873 | 0.504 |
| 8183 | RP11-774O3.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | AFF1 | Sponge network | -0.487 | 0.02157 | -0.384 | 0.00053 | 0.504 |
| 8184 | RP4-639F20.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-500a-3p | 16 | GRIK3 | Sponge network | -0.517 | 0.02935 | -3.208 | 0 | 0.504 |
| 8185 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | TACC1 | Sponge network | 0.997 | 0.00066 | -0.362 | 0.05406 | 0.504 |
| 8186 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | IGSF10 | Sponge network | -0.842 | 0.01361 | -1.751 | 1.0E-5 | 0.504 |
| 8187 | LINC00092 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 14 | CDH23 | Sponge network | -1.886 | 0 | -0.974 | 0.00034 | 0.504 |
| 8188 | HCG11 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 19 | HIP1 | Sponge network | 0.385 | 0.10067 | 0.774 | 0 | 0.504 |
| 8189 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-539-5p | 21 | BTG2 | Sponge network | -1.057 | 0.00029 | -1.763 | 0 | 0.504 |
| 8190 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p | 13 | MLLT10 | Sponge network | 1.294 | 0 | 0.105 | 0.33292 | 0.504 |
| 8191 | FAM66C |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 35 | IL17RD | Sponge network | -0.901 | 0.00019 | -0.019 | 0.94873 | 0.504 |
| 8192 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 44 | MEF2C | Sponge network | -0.901 | 0.00019 | -0.499 | 0.01448 | 0.504 |
| 8193 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | PEG3 | Sponge network | -0.942 | 0 | -1.74 | 0 | 0.504 |
| 8194 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-577;hsa-miR-93-5p | 27 | HLF | Sponge network | -0.473 | 0.07602 | -2.443 | 0 | 0.504 |
| 8195 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-421 | 22 | EPHA5 | Sponge network | -0.583 | 0.14589 | -0.957 | 0.01733 | 0.504 |
| 8196 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 40 | DMD | Sponge network | 0.064 | 0.72934 | -1.506 | 0 | 0.504 |
| 8197 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CACNA2D1 | Sponge network | -1.203 | 0.03881 | -0.846 | 0.00675 | 0.504 |
| 8198 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p | 24 | ANK2 | Sponge network | 0.214 | 0.18246 | -1.603 | 1.0E-5 | 0.504 |
| 8199 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ANK2 | Sponge network | -0.737 | 0.10822 | -1.603 | 1.0E-5 | 0.504 |
| 8200 | HOXB-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-629-3p | 20 | ASTN1 | Sponge network | -0.154 | 0.53954 | -2.389 | 2.0E-5 | 0.504 |
| 8201 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | BCLAF1 | Sponge network | 0.953 | 0 | 0.211 | 0.00684 | 0.504 |
| 8202 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | PCDH8 | Sponge network | -2.915 | 0.00015 | -0.997 | 0.05366 | 0.504 |
| 8203 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p | 34 | DMD | Sponge network | 0.532 | 0.00505 | -1.506 | 0 | 0.504 |
| 8204 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-93-5p | 12 | LHX6 | Sponge network | -1.203 | 0.03881 | -0.087 | 0.69193 | 0.504 |
| 8205 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PLSCR4 | Sponge network | -0.737 | 0.10822 | -0.474 | 0.03833 | 0.504 |
| 8206 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 34 | SCN9A | Sponge network | -2.46 | 0 | -0.525 | 0.10593 | 0.504 |
| 8207 | H19 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-664a-3p | 11 | SOX11 | Sponge network | 2.439 | 0 | 2.68 | 0 | 0.504 |
| 8208 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | ST6GALNAC3 | Sponge network | -1.582 | 8.0E-5 | -0.987 | 0 | 0.504 |
| 8209 | LINC00473 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 34 | IGFBP5 | Sponge network | -1.203 | 0.03881 | -0.806 | 0.00105 | 0.504 |
| 8210 | AC010226.4 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-486-5p | 13 | SYNM | Sponge network | -0.811 | 2.0E-5 | -2.309 | 1.0E-5 | 0.504 |
| 8211 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | TSHZ3 | Sponge network | -0.323 | 0.01372 | -0.556 | 0.01516 | 0.504 |
| 8212 | AP001347.6 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | SORCS1 | Sponge network | -1.161 | 0.00591 | -3.492 | 0 | 0.504 |
| 8213 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 33 | ZFHX4 | Sponge network | -0.246 | 0.35034 | 0.018 | 0.95574 | 0.504 |
| 8214 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | MYO9A | Sponge network | 0.919 | 0 | -0.147 | 0.32373 | 0.504 |
| 8215 | LINC00578 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-589-3p | 23 | ADARB1 | Sponge network | 0.811 | 0.03228 | -0.335 | 0.06631 | 0.504 |
| 8216 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | EPHA3 | Sponge network | 0.064 | 0.72934 | 0.185 | 0.53588 | 0.503 |
| 8217 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-7-1-3p | 12 | TIMP3 | Sponge network | 0.693 | 0.00076 | -0.546 | 0.02307 | 0.503 |
| 8218 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 20 | EPHA3 | Sponge network | 0.4 | 0.14611 | 0.185 | 0.53588 | 0.503 |
| 8219 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-590-3p | 12 | RB1CC1 | Sponge network | 0.559 | 0.00026 | 0.175 | 0.12559 | 0.503 |
| 8220 | RP11-244O19.1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-590-3p | 16 | KLF10 | Sponge network | -0.096 | 0.67958 | -0.783 | 0 | 0.503 |
| 8221 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | RASSF8 | Sponge network | -1.375 | 0.08642 | -0.122 | 0.65591 | 0.503 |
| 8222 | C20orf166-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577 | 13 | MACF1 | Sponge network | -2.69 | 0.0001 | 0.019 | 0.90335 | 0.503 |
| 8223 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-940 | 28 | MEF2C | Sponge network | -0.154 | 0.53954 | -0.499 | 0.01448 | 0.503 |
| 8224 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p | 55 | KIF1B | Sponge network | -0.096 | 0.67958 | -0.118 | 0.32258 | 0.503 |
| 8225 | LINC00969 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-874-3p | 14 | NR2C2 | Sponge network | 0.683 | 1.0E-5 | 0.21 | 0.03224 | 0.503 |
| 8226 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | PIK3R1 | Sponge network | 0.572 | 0.01722 | -0.133 | 0.36854 | 0.503 |
| 8227 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | PEG3 | Sponge network | 0.101 | 0.60919 | -1.74 | 0 | 0.503 |
| 8228 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 22 | FGFR1 | Sponge network | -2.46 | 0 | -0.509 | 0.03957 | 0.503 |
| 8229 | DNAJC3-AS1 |
hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-582-5p | 13 | ZMYM2 | Sponge network | 0.753 | 0.00014 | 0.279 | 0.01273 | 0.503 |
| 8230 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 27 | SOX5 | Sponge network | -0.318 | 0.65847 | -1.091 | 4.0E-5 | 0.503 |
| 8231 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | TLL1 | Sponge network | -2.46 | 0 | -1.195 | 3.0E-5 | 0.503 |
| 8232 | SNHG14 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 79 | DST | Sponge network | -1.111 | 0.0003 | -0.713 | 0.00096 | 0.503 |
| 8233 | CTD-2314G24.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 19 | PRUNE2 | Sponge network | 0.246 | 0.59912 | -1.733 | 0.00022 | 0.503 |
| 8234 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | DCLK1 | Sponge network | -2.915 | 0.00015 | -1.324 | 0.00014 | 0.503 |
| 8235 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SON | Sponge network | -0.222 | 0.32445 | 0.168 | 0.04406 | 0.503 |
| 8236 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-942-5p | 25 | UNC80 | Sponge network | -1.886 | 0 | -1.934 | 0 | 0.503 |
| 8237 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | RASSF8 | Sponge network | 0.246 | 0.59912 | -0.122 | 0.65591 | 0.503 |
| 8238 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 19 | FGFR1 | Sponge network | 0.246 | 0.59912 | -0.509 | 0.03957 | 0.503 |
| 8239 | RP11-404E16.1 |
hsa-miR-103a-2-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 20 | LPP | Sponge network | 0.097 | 0.91311 | -0.459 | 0.04475 | 0.503 |
| 8240 | SH3BP5-AS1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 10 | RBM6 | Sponge network | 0.267 | 0.2937 | -0.137 | 0.15663 | 0.503 |
| 8241 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | UBE4B | Sponge network | 0.083 | 0.66405 | -0.235 | 0.007 | 0.503 |
| 8242 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 56 | QKI | Sponge network | -1.216 | 0 | -0.333 | 0.04111 | 0.503 |
| 8243 | RP11-473I1.10 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | UBR3 | Sponge network | 0.673 | 0 | -0.021 | 0.82774 | 0.503 |
| 8244 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-582-5p;hsa-miR-671-5p | 23 | DLC1 | Sponge network | 0.299 | 0.57722 | -0.382 | 0.05038 | 0.503 |
| 8245 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 23 | EBF3 | Sponge network | 0.101 | 0.60919 | -0.058 | 0.81437 | 0.503 |
| 8246 | ARHGEF26-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | IL17RD | Sponge network | -2.46 | 0 | -0.019 | 0.94873 | 0.503 |
| 8247 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | ZFHX3 | Sponge network | 0.335 | 0.20596 | 0.389 | 0.01363 | 0.503 |
| 8248 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | CNTN1 | Sponge network | -0.612 | 0.00094 | -2.594 | 0 | 0.503 |
| 8249 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p | 37 | PRKAA2 | Sponge network | -0.358 | 0.17255 | -2.462 | 0 | 0.503 |
| 8250 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | NCOA1 | Sponge network | -0.487 | 0.02157 | -0.432 | 0 | 0.503 |
| 8251 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-629-3p | 21 | SNX29 | Sponge network | -0.583 | 0.14589 | -0.34 | 0.01371 | 0.503 |
| 8252 | BVES-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-766-3p;hsa-miR-877-5p | 25 | IL17RD | Sponge network | -2.62 | 0 | -0.019 | 0.94873 | 0.503 |
| 8253 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-455-3p | 28 | FBXO32 | Sponge network | -1.645 | 0 | -0.495 | 0.07561 | 0.503 |
| 8254 | WDFY3-AS2 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | -0.942 | 0 | 0.154 | 0.74723 | 0.503 |
| 8255 | RP11-356J5.12 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | KCTD8 | Sponge network | -0.899 | 6.0E-5 | -3.359 | 0 | 0.503 |
| 8256 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 37 | CBL | Sponge network | -0.222 | 0.32445 | 0.291 | 0.01267 | 0.503 |
| 8257 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | FSTL5 | Sponge network | -1.575 | 6.0E-5 | -1.3 | 0.01619 | 0.503 |
| 8258 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p | 24 | MDM4 | Sponge network | 0.497 | 0.01451 | 0.345 | 0.00762 | 0.503 |
| 8259 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 19 | RCAN2 | Sponge network | 0.335 | 0.20596 | -0.33 | 0.15172 | 0.503 |
| 8260 | MIR497HG |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-96-5p | 17 | RPS6KA6 | Sponge network | -1.439 | 4.0E-5 | -2.989 | 0 | 0.503 |
| 8261 | RP11-532F6.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | NAV3 | Sponge network | -0.896 | 0.00099 | 0.212 | 0.47729 | 0.503 |
| 8262 | C20orf166-AS1 |
hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-671-5p | 17 | KIT | Sponge network | -2.69 | 0.0001 | -2.184 | 0 | 0.503 |
| 8263 | RP11-532F6.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-744-3p | 28 | IGFBP5 | Sponge network | -0.896 | 0.00099 | -0.806 | 0.00105 | 0.503 |
| 8264 | AC009948.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | NIN | Sponge network | 0.532 | 0.00505 | -0.253 | 0.13794 | 0.503 |
| 8265 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | SCN9A | Sponge network | -0.612 | 0.00094 | -0.525 | 0.10593 | 0.503 |
| 8266 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p | 45 | RUNX1T1 | Sponge network | 0.299 | 0.57722 | -0.344 | 0.2374 | 0.503 |
| 8267 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-503-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 42 | RICTOR | Sponge network | -0.222 | 0.32445 | 0.388 | 0.00188 | 0.503 |
| 8268 | LINC00657 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p | 17 | PHF6 | Sponge network | 0.91 | 0 | 0.785 | 0 | 0.503 |
| 8269 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | SYNE1 | Sponge network | 0.299 | 0.57722 | -0.738 | 0.00316 | 0.503 |
| 8270 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-940 | 13 | MCC | Sponge network | -0.733 | 0.19469 | -1.005 | 0 | 0.503 |
| 8271 | RP11-244O19.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 24 | NRK | Sponge network | -0.096 | 0.67958 | 0.656 | 0.19217 | 0.503 |
| 8272 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | MOB1B | Sponge network | 0.614 | 1.0E-5 | 0.197 | 0.15266 | 0.503 |
| 8273 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 22 | CACNA2D1 | Sponge network | -0.793 | 7.0E-5 | -0.846 | 0.00675 | 0.503 |
| 8274 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | ZNF208 | Sponge network | -0.896 | 0.00099 | -0.4 | 0.22381 | 0.503 |
| 8275 | GNG12-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p | 21 | STARD13 | Sponge network | -0.793 | 7.0E-5 | -0.187 | 0.27383 | 0.503 |
| 8276 | FGF14-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-505-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 50 | LPP | Sponge network | -1.582 | 8.0E-5 | -0.459 | 0.04475 | 0.503 |
| 8277 | RP11-356J5.12 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ITPKB | Sponge network | -0.899 | 6.0E-5 | -0.704 | 0.00106 | 0.503 |
| 8278 | EMX2OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p | 10 | SCN11A | Sponge network | -0.777 | 0.1134 | -1.15 | 0.00299 | 0.503 |
| 8279 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 39 | ERC1 | Sponge network | -0.129 | 0.57177 | -0.108 | 0.38794 | 0.503 |
| 8280 | AC009948.5 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-590-3p | 17 | MRVI1 | Sponge network | 0.532 | 0.00505 | -1.051 | 0.00022 | 0.503 |
| 8281 | LINC00473 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 19 | SETBP1 | Sponge network | -1.203 | 0.03881 | -1.302 | 1.0E-5 | 0.503 |
| 8282 | LINC00473 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PEG3 | Sponge network | -1.203 | 0.03881 | -1.74 | 0 | 0.503 |
| 8283 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | DAB2 | Sponge network | -1.697 | 0.00889 | 0.431 | 0.0113 | 0.503 |
| 8284 | RP11-890B15.3 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 20 | CSDE1 | Sponge network | 0.101 | 0.60919 | -0.493 | 0 | 0.503 |
| 8285 | RP11-589P10.7 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-505-3p | 10 | AR | Sponge network | -2.044 | 0.00066 | -1.554 | 0 | 0.503 |
| 8286 | FENDRR |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | ERG | Sponge network | -1.281 | 0.00012 | 0.002 | 0.99011 | 0.503 |
| 8287 | U91328.19 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-484 | 11 | WNK2 | Sponge network | -0.303 | 0.21002 | -0.352 | 0.31066 | 0.503 |
| 8288 | MEG3 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-942-5p | 16 | GLIPR1 | Sponge network | 0.249 | 0.35902 | 0.146 | 0.3276 | 0.503 |
| 8289 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | AKAP12 | Sponge network | -0.738 | 0.21233 | -0.932 | 0.0028 | 0.503 |
| 8290 | MIR497HG |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 16 | GJA1 | Sponge network | -1.439 | 4.0E-5 | -0.485 | 0.02179 | 0.503 |
| 8291 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p | 11 | SMARCA1 | Sponge network | -2.46 | 0 | -0.684 | 0.00159 | 0.503 |
| 8292 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 13 | FAT4 | Sponge network | -4.546 | 0 | -0.354 | 0.14694 | 0.503 |
| 8293 | LINC00472 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-378a-3p | 12 | FLNA | Sponge network | -0.842 | 0.01361 | -0.771 | 0.01377 | 0.503 |
| 8294 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-93-5p | 19 | FGFR1 | Sponge network | -0.154 | 0.78939 | -0.509 | 0.03957 | 0.503 |
| 8295 | RP11-389C8.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | PTGFR | Sponge network | 0.727 | 3.0E-5 | -0.233 | 0.45421 | 0.503 |
| 8296 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 15 | FYN | Sponge network | -0.129 | 0.57177 | -0.131 | 0.37051 | 0.503 |
| 8297 | MEG3 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-98-5p | 11 | HSPB7 | Sponge network | 0.249 | 0.35902 | -2.268 | 1.0E-5 | 0.502 |
| 8298 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 18 | ABCB5 | Sponge network | -0.49 | 0.04946 | -0.956 | 0.04077 | 0.502 |
| 8299 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-484 | 24 | ABL1 | Sponge network | -2.452 | 0 | -0.215 | 0.07804 | 0.502 |
| 8300 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-7-1-3p | 23 | EPHA3 | Sponge network | -4.546 | 0 | 0.185 | 0.53588 | 0.502 |
| 8301 | SH3RF3-AS1 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | NUAK1 | Sponge network | 0.889 | 9.0E-5 | 0.904 | 0 | 0.502 |
| 8302 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p | 16 | ZNF208 | Sponge network | -0.323 | 0.01372 | -0.4 | 0.22381 | 0.502 |
| 8303 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | EBF1 | Sponge network | -2.117 | 0.00161 | -0.384 | 0.08856 | 0.502 |
| 8304 | RP11-672A2.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-424-5p | 10 | RASSF8 | Sponge network | 1.344 | 0 | -0.122 | 0.65591 | 0.502 |
| 8305 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SEMA3C | Sponge network | -0.021 | 0.93598 | -0.272 | 0.31153 | 0.502 |
| 8306 | LINC00957 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 18 | SORCS1 | Sponge network | -0.421 | 0.0114 | -3.492 | 0 | 0.502 |
| 8307 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 16 | NAV3 | Sponge network | -0.517 | 0.02935 | 0.212 | 0.47729 | 0.502 |
| 8308 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | PDZRN3 | Sponge network | -0.942 | 0 | -1.548 | 0 | 0.502 |
| 8309 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PCDH9 | Sponge network | -1.057 | 0.00029 | -1.823 | 4.0E-5 | 0.502 |
| 8310 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | RELN | Sponge network | -0.323 | 0.01372 | -2.403 | 0 | 0.502 |
| 8311 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-330-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 15 | RASSF8 | Sponge network | 0.381 | 0.25967 | -0.122 | 0.65591 | 0.502 |
| 8312 | FAM66C |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p | 29 | ASTN1 | Sponge network | -0.901 | 0.00019 | -2.389 | 2.0E-5 | 0.502 |
| 8313 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | ST6GAL2 | Sponge network | 0.194 | 0.64278 | -1.201 | 0.00597 | 0.502 |
| 8314 | ZRANB2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p | 11 | SYNM | Sponge network | -0.376 | 0.00337 | -2.309 | 1.0E-5 | 0.502 |
| 8315 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | AKAP6 | Sponge network | -0.376 | 0.00337 | -1.586 | 3.0E-5 | 0.502 |
| 8316 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 16 | RASSF8 | Sponge network | -4.546 | 0 | -0.122 | 0.65591 | 0.502 |
| 8317 | ADAMTS9-AS2 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | MSN | Sponge network | -2.452 | 0 | -0.023 | 0.88422 | 0.502 |
| 8318 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-625-5p | 16 | LHX6 | Sponge network | -2.452 | 0 | -0.087 | 0.69193 | 0.502 |
| 8319 | RP11-720L2.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 10 | NAV3 | Sponge network | -1.255 | 0.00981 | 0.212 | 0.47729 | 0.502 |
| 8320 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p | 15 | NCAM2 | Sponge network | -0.358 | 0.17255 | -0.482 | 0.22565 | 0.502 |
| 8321 | CTD-2314G24.2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 20 | SORCS1 | Sponge network | 0.246 | 0.59912 | -3.492 | 0 | 0.502 |
| 8322 | FAM157A |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | NR2C2 | Sponge network | 0.813 | 1.0E-5 | 0.21 | 0.03224 | 0.502 |
| 8323 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 48 | EPHA7 | Sponge network | -0.222 | 0.32445 | -2.197 | 3.0E-5 | 0.502 |
| 8324 | TP73-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-92a-3p | 19 | ROBO1 | Sponge network | -0.563 | 0.02545 | -0.009 | 0.96154 | 0.502 |
| 8325 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 39 | CADM2 | Sponge network | -0.739 | 0.00044 | -3.782 | 0 | 0.502 |
| 8326 | LINC01018 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | CADM2 | Sponge network | -2.915 | 0.00015 | -3.782 | 0 | 0.502 |
| 8327 | RP11-473I1.5 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 17 | ABL2 | Sponge network | 0.542 | 0.00054 | 0.82 | 0 | 0.502 |
| 8328 | RP11-244O19.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-940 | 15 | SFRP1 | Sponge network | -0.096 | 0.67958 | -3.566 | 0 | 0.502 |
| 8329 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p | 12 | JAKMIP2 | Sponge network | -0.563 | 0.02545 | -1.388 | 0 | 0.502 |
| 8330 | RP11-67L2.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-625-5p | 25 | PTPN14 | Sponge network | 0.72 | 0.0001 | 0.144 | 0.34595 | 0.502 |
| 8331 | RP5-1198O20.4 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 16 | ZBTB20 | Sponge network | 0.997 | 0.00066 | -0.122 | 0.57569 | 0.502 |
| 8332 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-590-3p | 11 | KIT | Sponge network | -0.811 | 2.0E-5 | -2.184 | 0 | 0.502 |
| 8333 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 11 | TMEFF2 | Sponge network | -0.358 | 0.17255 | -2.426 | 0 | 0.502 |
| 8334 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-93-5p | 32 | CHD5 | Sponge network | -0.384 | 0.40652 | -0.922 | 0.01521 | 0.502 |
| 8335 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-576-5p | 20 | NCAM2 | Sponge network | -0.162 | 0.47687 | -0.482 | 0.22565 | 0.502 |
| 8336 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 14 | LATS1 | Sponge network | 0.959 | 0 | 0.109 | 0.34208 | 0.502 |
| 8337 | RP11-672A2.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-651-5p;hsa-miR-671-5p | 19 | BMPR2 | Sponge network | 1.344 | 0 | 0.206 | 0.0635 | 0.502 |
| 8338 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 17 | PCDH10 | Sponge network | -0.318 | 0.65847 | -1.644 | 0.0078 | 0.502 |
| 8339 | ADAMTS9-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 40 | IL17RD | Sponge network | -2.452 | 0 | -0.019 | 0.94873 | 0.502 |
| 8340 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p | 18 | EBF1 | Sponge network | -0.693 | 0.05629 | -0.384 | 0.08856 | 0.502 |
| 8341 | LINC01018 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | SYT4 | Sponge network | -2.915 | 0.00015 | -3.207 | 0 | 0.502 |
| 8342 | OIP5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | RBL2 | Sponge network | 0.421 | 0.00084 | -0.282 | 0.00254 | 0.502 |
| 8343 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p | 14 | ITGB3 | Sponge network | -2.69 | 0.0001 | -0.011 | 0.95751 | 0.502 |
| 8344 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-590-5p | 19 | CNTN1 | Sponge network | 0.176 | 0.37402 | -2.594 | 0 | 0.502 |
| 8345 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | -1.349 | 0.00017 | 0.358 | 0.13727 | 0.502 |
| 8346 | LINC00672 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-424-3p;hsa-miR-532-3p;hsa-miR-542-3p | 18 | SYNM | Sponge network | -0.227 | 0.31657 | -2.309 | 1.0E-5 | 0.502 |
| 8347 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 65 | NTRK3 | Sponge network | -0.563 | 0.02545 | -1.77 | 3.0E-5 | 0.502 |
| 8348 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 25 | AFF3 | Sponge network | -0.793 | 7.0E-5 | -2.373 | 0 | 0.502 |
| 8349 | MIR497HG |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 10 | DIRAS1 | Sponge network | -1.439 | 4.0E-5 | -2.518 | 0 | 0.502 |
| 8350 | MAPKAPK5-AS1 |
hsa-miR-101-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-654-3p | 16 | PHF6 | Sponge network | 0.765 | 0 | 0.785 | 0 | 0.502 |
| 8351 | MRVI1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-766-3p | 20 | IL17RD | Sponge network | -1.092 | 4.0E-5 | -0.019 | 0.94873 | 0.502 |
| 8352 | MIR497HG |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-629-3p | 24 | RYR2 | Sponge network | -1.439 | 4.0E-5 | -1.466 | 0.00031 | 0.502 |
| 8353 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 44 | TRPS1 | Sponge network | 0.335 | 0.20596 | -0.191 | 0.44109 | 0.502 |
| 8354 | RP11-798M19.6 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 11 | KCTD8 | Sponge network | -0.612 | 0.00094 | -3.359 | 0 | 0.502 |
| 8355 | RP11-819C21.1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-96-5p | 68 | LPP | Sponge network | 0.537 | 0.00139 | -0.459 | 0.04475 | 0.502 |
| 8356 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p | 15 | AKAP12 | Sponge network | -1.349 | 0.00017 | -0.932 | 0.0028 | 0.502 |
| 8357 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 33 | RICTOR | Sponge network | 1.254 | 0 | 0.388 | 0.00188 | 0.502 |
| 8358 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 41 | ZFHX4 | Sponge network | 0.374 | 0.20054 | 0.018 | 0.95574 | 0.502 |
| 8359 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 43 | RASSF2 | Sponge network | -0.737 | 0.06182 | -0.273 | 0.21961 | 0.502 |
| 8360 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 31 | ZBTB4 | Sponge network | -0.777 | 0.1134 | -0.739 | 0 | 0.502 |
| 8361 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 26 | MYO9A | Sponge network | -0.611 | 0.09607 | -0.147 | 0.32373 | 0.502 |
| 8362 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 38 | BCL2 | Sponge network | -0.976 | 0.00025 | -0.899 | 0 | 0.502 |
| 8363 | C20orf166-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-582-5p | 11 | KLF10 | Sponge network | -2.69 | 0.0001 | -0.783 | 0 | 0.502 |
| 8364 | RP3-430N8.11 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | TBL1XR1 | Sponge network | 0.973 | 0 | 0.578 | 0 | 0.502 |
| 8365 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-5p | 55 | TCF4 | Sponge network | -0.162 | 0.47687 | 0.016 | 0.92271 | 0.502 |
| 8366 | RP11-283I3.6 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-7-1-3p | 12 | PHF3 | Sponge network | 0.614 | 1.0E-5 | 0.126 | 0.19652 | 0.502 |
| 8367 | MAP3K14 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-93-5p | 10 | EZH1 | Sponge network | -0.295 | 0.02604 | -0.564 | 1.0E-5 | 0.502 |
| 8368 | LINC01001 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-766-3p | 18 | MDM4 | Sponge network | 1.055 | 0 | 0.345 | 0.00762 | 0.502 |
| 8369 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-625-5p;hsa-miR-766-3p | 31 | MDM4 | Sponge network | 0.858 | 3.0E-5 | 0.345 | 0.00762 | 0.502 |
| 8370 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | NRXN3 | Sponge network | -1.216 | 0 | -0.893 | 0.04186 | 0.502 |
| 8371 | SNX29P2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 41 | MDM4 | Sponge network | 0.4 | 0.14611 | 0.345 | 0.00762 | 0.502 |
| 8372 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | IGSF10 | Sponge network | -0.739 | 0.0574 | -1.751 | 1.0E-5 | 0.501 |
| 8373 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 41 | THRB | Sponge network | -1.092 | 4.0E-5 | -1.117 | 0.0004 | 0.501 |
| 8374 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 25 | EPHA3 | Sponge network | 0.374 | 0.20054 | 0.185 | 0.53588 | 0.501 |
| 8375 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-625-5p;hsa-miR-93-5p | 10 | ANK2 | Sponge network | -1.255 | 0.00981 | -1.603 | 1.0E-5 | 0.501 |
| 8376 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 55 | FAT3 | Sponge network | -0.899 | 6.0E-5 | -1.601 | 0.00051 | 0.501 |
| 8377 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 41 | CHD5 | Sponge network | 0.083 | 0.66405 | -0.922 | 0.01521 | 0.501 |
| 8378 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-511-5p | 13 | ATRX | Sponge network | 1.122 | 0 | 0.261 | 0.04408 | 0.501 |
| 8379 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p | 10 | MGA | Sponge network | 1.093 | 0 | 0.323 | 0.00792 | 0.501 |
| 8380 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CLIP1 | Sponge network | 0.194 | 0.64278 | -0.215 | 0.08684 | 0.501 |
| 8381 | RP11-356J5.12 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | HSPB7 | Sponge network | -0.899 | 6.0E-5 | -2.268 | 1.0E-5 | 0.501 |
| 8382 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CDH11 | Sponge network | -0.739 | 0.0574 | 1.427 | 0 | 0.501 |
| 8383 | RP11-35G9.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 42 | MDM4 | Sponge network | 0.657 | 4.0E-5 | 0.345 | 0.00762 | 0.501 |
| 8384 | TP73-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-96-5p | 14 | ELMO1 | Sponge network | -0.563 | 0.02545 | 0.04 | 0.83147 | 0.501 |
| 8385 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 34 | EBF1 | Sponge network | -0.901 | 0.00019 | -0.384 | 0.08856 | 0.501 |
| 8386 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | CDH10 | Sponge network | -0.49 | 0.04946 | -2.397 | 0 | 0.501 |
| 8387 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p | 43 | SSBP2 | Sponge network | -1.645 | 0 | -1.58 | 0 | 0.501 |
| 8388 | PWAR6 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | TAL1 | Sponge network | -1.575 | 6.0E-5 | -0.462 | 0.00574 | 0.501 |
| 8389 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-874-3p;hsa-miR-93-5p | 27 | TACC1 | Sponge network | -2.531 | 0 | -0.362 | 0.05406 | 0.501 |
| 8390 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | EPHA3 | Sponge network | 0.225 | 0.30644 | 0.185 | 0.53588 | 0.501 |
| 8391 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-628-5p | 12 | TRIM33 | Sponge network | 2.094 | 0 | 0.402 | 7.0E-5 | 0.501 |
| 8392 | RP11-307B6.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-5p | 24 | DST | Sponge network | -4.463 | 0.00025 | -0.713 | 0.00096 | 0.501 |
| 8393 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 13 | RASSF2 | Sponge network | -2.039 | 0.03447 | -0.273 | 0.21961 | 0.501 |
| 8394 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-942-5p | 57 | THRB | Sponge network | -0.793 | 7.0E-5 | -1.117 | 0.0004 | 0.501 |
| 8395 | LINC01018 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 46 | EPHA7 | Sponge network | -2.915 | 0.00015 | -2.197 | 3.0E-5 | 0.501 |
| 8396 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | FAT4 | Sponge network | -0.563 | 0.02545 | -0.354 | 0.14694 | 0.501 |
| 8397 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577 | 23 | GREM1 | Sponge network | 0.246 | 0.59912 | 0.902 | 0.01691 | 0.501 |
| 8398 | OIP5-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 10 | SETD2 | Sponge network | 0.421 | 0.00084 | 0.031 | 0.73068 | 0.501 |
| 8399 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 25 | PBX1 | Sponge network | 0.225 | 0.30644 | -1.206 | 0 | 0.501 |
| 8400 | GNG12-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | KLF10 | Sponge network | -0.793 | 7.0E-5 | -0.783 | 0 | 0.501 |
| 8401 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | -1.216 | 0 | -2.417 | 0 | 0.501 |
| 8402 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | EPHA5 | Sponge network | -0.376 | 0.00337 | -0.957 | 0.01733 | 0.501 |
| 8403 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3913-5p;hsa-miR-629-3p;hsa-miR-652-3p | 22 | ZBTB4 | Sponge network | -1.886 | 0 | -0.739 | 0 | 0.501 |
| 8404 | PART1 |
hsa-miR-144-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | FMN2 | Sponge network | -2.925 | 0 | -1.746 | 0.00047 | 0.501 |
| 8405 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p | 30 | PGR | Sponge network | -0.405 | 0.22013 | -1.704 | 0 | 0.501 |
| 8406 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p | 14 | ITGA5 | Sponge network | 0.335 | 0.20596 | 0.452 | 0.03524 | 0.501 |
| 8407 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 18 | AKAP6 | Sponge network | 0.16 | 0.50785 | -1.586 | 3.0E-5 | 0.501 |
| 8408 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 24 | PTPRT | Sponge network | -1.057 | 0.00029 | -0.907 | 0.03727 | 0.501 |
| 8409 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | CNTN1 | Sponge network | -0.096 | 0.67958 | -2.594 | 0 | 0.501 |
| 8410 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | PTPRB | Sponge network | -0.737 | 0.06182 | 0.105 | 0.47122 | 0.501 |
| 8411 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p | 20 | KIT | Sponge network | -2.117 | 0.00161 | -2.184 | 0 | 0.501 |
| 8412 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | EPHA5 | Sponge network | -0.976 | 0.00025 | -0.957 | 0.01733 | 0.501 |
| 8413 | PWAR6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 28 | ZMYND11 | Sponge network | -1.575 | 6.0E-5 | -0.2 | 0.05449 | 0.501 |
| 8414 | FENDRR |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-93-5p | 62 | DST | Sponge network | -1.281 | 0.00012 | -0.713 | 0.00096 | 0.501 |
| 8415 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | GALNT13 | Sponge network | -2.46 | 0 | -0.857 | 0.07439 | 0.501 |
| 8416 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 26 | KIT | Sponge network | -1.697 | 0.00889 | -2.184 | 0 | 0.501 |
| 8417 | LINC00702 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 18 | PAK3 | Sponge network | -0.737 | 0.06182 | -0.628 | 0.07253 | 0.501 |
| 8418 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p | 31 | LIFR | Sponge network | -0.563 | 0.02545 | -1.794 | 0 | 0.501 |
| 8419 | RP11-701H24.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-590-3p | 16 | RBMS3 | Sponge network | -1.425 | 0.04204 | -0.519 | 0.03551 | 0.501 |
| 8420 | LINC00092 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-708-5p | 15 | EZH1 | Sponge network | -1.886 | 0 | -0.564 | 1.0E-5 | 0.501 |
| 8421 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 22 | TBL1XR1 | Sponge network | 1.266 | 0 | 0.578 | 0 | 0.501 |
| 8422 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CACNA2D1 | Sponge network | 0.335 | 0.20596 | -0.846 | 0.00675 | 0.501 |
| 8423 | RP11-159D12.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 34 | CREB1 | Sponge network | 1.254 | 0 | 0.203 | 0.00537 | 0.501 |
| 8424 | RP11-499P20.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-766-3p;hsa-miR-940 | 22 | MDM4 | Sponge network | 0.187 | 0.44121 | 0.345 | 0.00762 | 0.501 |
| 8425 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-98-5p | 18 | THBS1 | Sponge network | -0.473 | 0.07602 | 0.741 | 0.00386 | 0.501 |
| 8426 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-629-5p | 24 | MYO9A | Sponge network | -0.154 | 0.78939 | -0.147 | 0.32373 | 0.501 |
| 8427 | BVES-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-421;hsa-miR-429 | 15 | STARD13 | Sponge network | -2.62 | 0 | -0.187 | 0.27383 | 0.501 |
| 8428 | LINC00957 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-590-3p | 15 | SPG20 | Sponge network | -0.421 | 0.0114 | -1.16 | 0 | 0.501 |
| 8429 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p | 16 | ZNF208 | Sponge network | 0.532 | 0.00505 | -0.4 | 0.22381 | 0.501 |
| 8430 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-485-3p | 11 | AKAP11 | Sponge network | 1.055 | 0 | 0.196 | 0.09825 | 0.501 |
| 8431 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-486-5p;hsa-miR-590-3p | 12 | ZNF208 | Sponge network | -1.161 | 0.00591 | -0.4 | 0.22381 | 0.501 |
| 8432 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 45 | PRKAA2 | Sponge network | -0.896 | 0.00099 | -2.462 | 0 | 0.501 |
| 8433 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 18 | NR2C2 | Sponge network | 0.225 | 0.30644 | 0.21 | 0.03224 | 0.501 |
| 8434 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-874-3p | 14 | NR2C2 | Sponge network | 0.594 | 0.41522 | 0.21 | 0.03224 | 0.501 |
| 8435 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p | 19 | PLSCR4 | Sponge network | -0.246 | 0.35034 | -0.474 | 0.03833 | 0.501 |
| 8436 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 31 | CHD5 | Sponge network | 0.299 | 0.57722 | -0.922 | 0.01521 | 0.501 |
| 8437 | AC003090.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | PRKCB | Sponge network | -2.117 | 0.00161 | -1.313 | 2.0E-5 | 0.501 |
| 8438 | TPTEP1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | PARK2 | Sponge network | -1.216 | 0 | -1.892 | 0 | 0.501 |
| 8439 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | PCDH9 | Sponge network | -0.942 | 0 | -1.823 | 4.0E-5 | 0.501 |
| 8440 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-766-3p | 43 | MDM4 | Sponge network | 1.089 | 0 | 0.345 | 0.00762 | 0.501 |
| 8441 | ZNF667-AS1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | SPOP | Sponge network | -0.976 | 0.00025 | -0.419 | 1.0E-5 | 0.501 |
| 8442 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-424-5p | 14 | NR2C2 | Sponge network | 0.082 | 0.55397 | 0.21 | 0.03224 | 0.501 |
| 8443 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 22 | RPS6KA2 | Sponge network | -0.129 | 0.57177 | -0.57 | 0.00013 | 0.501 |
| 8444 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | CNTN1 | Sponge network | -0.811 | 2.0E-5 | -2.594 | 0 | 0.501 |
| 8445 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 34 | IGF1 | Sponge network | -1.281 | 0.00012 | -0.204 | 0.5898 | 0.501 |
| 8446 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 23 | EPHA5 | Sponge network | -1.092 | 4.0E-5 | -0.957 | 0.01733 | 0.501 |
| 8447 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | TLL1 | Sponge network | 0.997 | 0.00066 | -1.195 | 3.0E-5 | 0.501 |
| 8448 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-503-5p | 19 | ABL1 | Sponge network | -0.323 | 0.01372 | -0.215 | 0.07804 | 0.501 |
| 8449 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | HECA | Sponge network | 0.101 | 0.60919 | -0.217 | 0.03463 | 0.501 |
| 8450 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | 0.056 | 0.81195 | -1.751 | 1.0E-5 | 0.501 |
| 8451 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 14 | CNTNAP1 | Sponge network | -0.421 | 0.0114 | 0.358 | 0.13727 | 0.501 |
| 8452 | RP11-159D12.2 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-766-3p | 14 | ATM | Sponge network | 1.254 | 0 | 0.178 | 0.21568 | 0.501 |
| 8453 | HAND2-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | PREX2 | Sponge network | -1.697 | 0.00889 | -0.272 | 0.25382 | 0.501 |
| 8454 | RP11-384O8.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 14 | PRUNE2 | Sponge network | -0.738 | 0.21233 | -1.733 | 0.00022 | 0.501 |
| 8455 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | NRXN1 | Sponge network | -0.811 | 2.0E-5 | -3.778 | 0 | 0.501 |
| 8456 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-454-3p;hsa-miR-582-5p | 27 | SSBP2 | Sponge network | -2.095 | 0.00112 | -1.58 | 0 | 0.501 |
| 8457 | RP11-57H14.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | FAM172A | Sponge network | 0.083 | 0.66405 | -0.57 | 0 | 0.501 |
| 8458 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-331-3p | 15 | ZBTB4 | Sponge network | -2.076 | 0.00548 | -0.739 | 0 | 0.5 |
| 8459 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p | 23 | PGR | Sponge network | 0.176 | 0.37402 | -1.704 | 0 | 0.5 |
| 8460 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 11 | MGA | Sponge network | 0.91 | 0 | 0.323 | 0.00792 | 0.5 |
| 8461 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 21 | SOX11 | Sponge network | 1.308 | 0 | 2.68 | 0 | 0.5 |
| 8462 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | MITF | Sponge network | 0.572 | 0.01722 | -0.769 | 0.00022 | 0.5 |
| 8463 | CKMT2-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 21 | PIK3CA | Sponge network | 0.082 | 0.55654 | 0.195 | 0.06908 | 0.5 |
| 8464 | CTC-296K1.3 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p | 11 | PRKCB | Sponge network | -2.095 | 0.00112 | -1.313 | 2.0E-5 | 0.5 |
| 8465 | BDNF-AS |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | LIMCH1 | Sponge network | -0.668 | 3.0E-5 | -0.915 | 1.0E-5 | 0.5 |
| 8466 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 23 | HECA | Sponge network | 0.537 | 0.00139 | -0.217 | 0.03463 | 0.5 |
| 8467 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | CHD2 | Sponge network | 1.106 | 0 | 0.049 | 0.55371 | 0.5 |
| 8468 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 39 | LIFR | Sponge network | -1.111 | 0.0003 | -1.794 | 0 | 0.5 |
| 8469 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | PEG3 | Sponge network | 0.532 | 0.00505 | -1.74 | 0 | 0.5 |
| 8470 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 30 | ABI2 | Sponge network | -0.246 | 0.35034 | 0.175 | 0.14787 | 0.5 |
| 8471 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 31 | CRTC3 | Sponge network | -0.737 | 0.06182 | 0.164 | 0.16786 | 0.5 |
| 8472 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-625-5p | 33 | JAZF1 | Sponge network | -0.487 | 0.02157 | -0.377 | 0.02677 | 0.5 |
| 8473 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | -0.405 | 0.22013 | -0.7 | 1.0E-5 | 0.5 |
| 8474 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ZNF536 | Sponge network | -2.46 | 0 | -3.115 | 0 | 0.5 |
| 8475 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-654-3p | 17 | NR2C2 | Sponge network | -0.031 | 0.89063 | 0.21 | 0.03224 | 0.5 |
| 8476 | TP73-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | CXCL12 | Sponge network | -0.563 | 0.02545 | -1.421 | 0 | 0.5 |
| 8477 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-3p | 22 | PBX1 | Sponge network | -1.582 | 8.0E-5 | -1.206 | 0 | 0.5 |
| 8478 | U91328.19 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-935 | 39 | DIP2C | Sponge network | -0.303 | 0.21002 | -0.436 | 0.01713 | 0.5 |
| 8479 | RP11-822E23.8 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-940 | 15 | SFRP1 | Sponge network | -0.777 | 0.16667 | -3.566 | 0 | 0.5 |
| 8480 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | CACNA2D1 | Sponge network | -2.117 | 0.00161 | -0.846 | 0.00675 | 0.5 |
| 8481 | BZRAP1-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-625-5p | 12 | NGFR | Sponge network | -0.465 | 0.04703 | -1.271 | 0.01058 | 0.5 |
| 8482 | LINC00641 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p | 14 | UVRAG | Sponge network | -0.222 | 0.32445 | -0.017 | 0.83865 | 0.5 |
| 8483 | PWAR6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | IL17RD | Sponge network | -1.575 | 6.0E-5 | -0.019 | 0.94873 | 0.5 |
| 8484 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | PTPRB | Sponge network | -2.452 | 0 | 0.105 | 0.47122 | 0.5 |
| 8485 | USP3-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-93-5p | 61 | DST | Sponge network | -0.162 | 0.47687 | -0.713 | 0.00096 | 0.5 |
| 8486 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p | 51 | FAT3 | Sponge network | -0.842 | 0.01361 | -1.601 | 0.00051 | 0.5 |
| 8487 | RP11-359E10.1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p | 10 | NGFR | Sponge network | 0.299 | 0.57722 | -1.271 | 0.01058 | 0.5 |
| 8488 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 19 | FBXO32 | Sponge network | -4.546 | 0 | -0.495 | 0.07561 | 0.5 |
| 8489 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | LATS1 | Sponge network | -0.222 | 0.32445 | 0.109 | 0.34208 | 0.5 |
| 8490 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577 | 18 | GRIN2A | Sponge network | -0.18 | 0.20343 | -2.161 | 0 | 0.5 |
| 8491 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-539-5p | 22 | RSPO2 | Sponge network | -0.611 | 0.09607 | -3.038 | 0 | 0.5 |
| 8492 | AC003090.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p | 16 | CDH23 | Sponge network | -2.117 | 0.00161 | -0.974 | 0.00034 | 0.5 |
| 8493 | RP11-774O3.3 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-423-5p | 19 | ABL1 | Sponge network | -0.487 | 0.02157 | -0.215 | 0.07804 | 0.5 |
| 8494 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 59 | TCF4 | Sponge network | -0.769 | 0.00035 | 0.016 | 0.92271 | 0.5 |
| 8495 | LINC00702 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | ZDBF2 | Sponge network | -0.737 | 0.06182 | -0.998 | 0.00025 | 0.5 |
| 8496 | LINC00641 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | RNF180 | Sponge network | -0.222 | 0.32445 | -1.127 | 1.0E-5 | 0.5 |
| 8497 | USP3-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-877-5p | 14 | INPP4A | Sponge network | -0.162 | 0.47687 | 0.07 | 0.53563 | 0.5 |
| 8498 | RP11-473I1.9 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ZNF638 | Sponge network | 0.683 | 1.0E-5 | 0.22 | 0.01214 | 0.5 |
| 8499 | RP11-597D13.9 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 31 | ABL2 | Sponge network | 1.302 | 7.0E-5 | 0.82 | 0 | 0.5 |
| 8500 | FENDRR |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 19 | LUZP2 | Sponge network | -1.281 | 0.00012 | -0.056 | 0.89416 | 0.5 |
| 8501 | AC010226.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-92a-1-5p | 20 | GRIK3 | Sponge network | -0.811 | 2.0E-5 | -3.208 | 0 | 0.5 |
| 8502 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | EBF1 | Sponge network | -1.161 | 0.00591 | -0.384 | 0.08856 | 0.5 |
| 8503 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-98-5p | 16 | PNRC1 | Sponge network | -0.769 | 0.00035 | -0.646 | 0 | 0.5 |
| 8504 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429 | 16 | ABCB5 | Sponge network | -0.976 | 0.00025 | -0.956 | 0.04077 | 0.5 |
| 8505 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 38 | ZFHX3 | Sponge network | 0.572 | 0.01722 | 0.389 | 0.01363 | 0.5 |
| 8506 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 39 | QKI | Sponge network | -1.161 | 0.00591 | -0.333 | 0.04111 | 0.5 |
| 8507 | LINC00473 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TMEFF2 | Sponge network | -1.203 | 0.03881 | -2.426 | 0 | 0.5 |
| 8508 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 12 | PAK3 | Sponge network | -0.896 | 0.00099 | -0.628 | 0.07253 | 0.5 |
| 8509 | RP11-701H24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 19 | CADM2 | Sponge network | -2.039 | 0.03447 | -3.782 | 0 | 0.5 |
| 8510 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-532-3p | 25 | PBX1 | Sponge network | -1.654 | 0.00144 | -1.206 | 0 | 0.5 |
| 8511 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 13 | JAKMIP2 | Sponge network | 0.246 | 0.59912 | -1.388 | 0 | 0.5 |
| 8512 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 27 | ANK2 | Sponge network | -0.318 | 0.65847 | -1.603 | 1.0E-5 | 0.5 |
| 8513 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 63 | TCF4 | Sponge network | 0.101 | 0.60919 | 0.016 | 0.92271 | 0.5 |
| 8514 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | PLSCR4 | Sponge network | -2.915 | 0.00015 | -0.474 | 0.03833 | 0.5 |
| 8515 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | PDGFRA | Sponge network | 0.889 | 9.0E-5 | -0.372 | 0.0037 | 0.5 |
| 8516 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-98-5p | 31 | DMD | Sponge network | 0.249 | 0.35902 | -1.506 | 0 | 0.5 |
| 8517 | MEG3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 35 | DIP2C | Sponge network | 0.249 | 0.35902 | -0.436 | 0.01713 | 0.5 |
| 8518 | MIR497HG |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577 | 10 | MACF1 | Sponge network | -1.439 | 4.0E-5 | 0.019 | 0.90335 | 0.5 |
| 8519 | RP11-384K6.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p | 11 | PIK3CA | Sponge network | 0.946 | 0 | 0.195 | 0.06908 | 0.5 |
| 8520 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 28 | JAZF1 | Sponge network | -0.18 | 0.20343 | -0.377 | 0.02677 | 0.5 |
| 8521 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 12 | CHD1 | Sponge network | 1.153 | 0 | 0.322 | 0.00215 | 0.5 |
| 8522 | MEG3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 19 | ITPKB | Sponge network | 0.249 | 0.35902 | -0.704 | 0.00106 | 0.5 |
| 8523 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-7-1-3p | 11 | KCNAB1 | Sponge network | -1.092 | 4.0E-5 | -0.755 | 2.0E-5 | 0.5 |
| 8524 | MEG3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-5p | 11 | PTPN13 | Sponge network | 0.249 | 0.35902 | -1.208 | 0 | 0.5 |
| 8525 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | AKAP11 | Sponge network | 0.101 | 0.60919 | 0.196 | 0.09825 | 0.5 |
| 8526 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 10 | LRRK2 | Sponge network | -0.842 | 0.01361 | -1.136 | 1.0E-5 | 0.5 |
| 8527 | RP11-384O8.1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-484;hsa-miR-92a-3p | 10 | FLNA | Sponge network | -0.738 | 0.21233 | -0.771 | 0.01377 | 0.5 |
| 8528 | SNHG14 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | UBE2QL1 | Sponge network | -1.111 | 0.0003 | -2.417 | 0 | 0.5 |
| 8529 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-590-3p | 11 | PAK3 | Sponge network | 0.335 | 0.20596 | -0.628 | 0.07253 | 0.5 |
| 8530 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 45 | FAT3 | Sponge network | 0.214 | 0.18246 | -1.601 | 0.00051 | 0.5 |
| 8531 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-664a-3p | 16 | LRRC7 | Sponge network | -2.69 | 0.0001 | -1.341 | 0.01193 | 0.5 |
| 8532 | RP4-613B23.1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p | 16 | GRIK3 | Sponge network | -0.309 | 0.1145 | -3.208 | 0 | 0.499 |
| 8533 | MEG3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-769-5p | 25 | SYNPO2 | Sponge network | 0.249 | 0.35902 | -2.342 | 0 | 0.499 |
| 8534 | RP11-67L2.2 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-590-3p | 10 | TPR | Sponge network | 0.72 | 0.0001 | 0.582 | 0 | 0.499 |
| 8535 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-590-3p | 13 | SYNPO2 | Sponge network | 0.178 | 0.29106 | -2.342 | 0 | 0.499 |
| 8536 | RP11-359E10.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p | 16 | SCN9A | Sponge network | 0.299 | 0.57722 | -0.525 | 0.10593 | 0.499 |
| 8537 | MIR497HG |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 28 | CTIF | Sponge network | -1.439 | 4.0E-5 | -0.389 | 0.00549 | 0.499 |
| 8538 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | MED12L | Sponge network | -0.563 | 0.02545 | -0.194 | 0.55299 | 0.499 |
| 8539 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 53 | PRKAA2 | Sponge network | -0.487 | 0.02157 | -2.462 | 0 | 0.499 |
| 8540 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | KCNAB1 | Sponge network | -2.46 | 0 | -0.755 | 2.0E-5 | 0.499 |
| 8541 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | AKAP12 | Sponge network | -2.915 | 0.00015 | -0.932 | 0.0028 | 0.499 |
| 8542 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | RECK | Sponge network | -0.096 | 0.67958 | -1.043 | 0 | 0.499 |
| 8543 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p | 14 | FSTL5 | Sponge network | 0.246 | 0.59912 | -1.3 | 0.01619 | 0.499 |
| 8544 | MALAT1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 44 | CREB1 | Sponge network | 0.782 | 0.00016 | 0.203 | 0.00537 | 0.499 |
| 8545 | ZRANB2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-378a-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-5p;hsa-miR-935 | 37 | LPP | Sponge network | -0.376 | 0.00337 | -0.459 | 0.04475 | 0.499 |
| 8546 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | KAT6B | Sponge network | -0.487 | 0.02157 | -0.478 | 0.00018 | 0.499 |
| 8547 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-93-5p | 24 | EPHA7 | Sponge network | -0.473 | 0.07602 | -2.197 | 3.0E-5 | 0.499 |
| 8548 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-5p | 12 | ABCC9 | Sponge network | 0.176 | 0.37402 | -0.466 | 0.14121 | 0.499 |
| 8549 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 17 | ZNF292 | Sponge network | 0.311 | 0.10344 | 0.276 | 0.04148 | 0.499 |
| 8550 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-338-3p;hsa-miR-590-3p | 14 | SLITRK3 | Sponge network | -1.161 | 0.00591 | -3.328 | 0 | 0.499 |
| 8551 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-577 | 15 | KIT | Sponge network | -0.473 | 0.07602 | -2.184 | 0 | 0.499 |
| 8552 | LINC00578 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-744-3p | 34 | NTRK3 | Sponge network | 0.811 | 0.03228 | -1.77 | 3.0E-5 | 0.499 |
| 8553 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | LRRC7 | Sponge network | -1.575 | 6.0E-5 | -1.341 | 0.01193 | 0.499 |
| 8554 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-484 | 14 | NRXN3 | Sponge network | -0.154 | 0.78939 | -0.893 | 0.04186 | 0.499 |
| 8555 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-539-5p | 19 | NRXN1 | Sponge network | -0.469 | 0.08721 | -3.778 | 0 | 0.499 |
| 8556 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | -0.842 | 0.01361 | -0.509 | 0.03957 | 0.499 |
| 8557 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 21 | CNTNAP3 | Sponge network | -1.575 | 6.0E-5 | -1.465 | 0.00033 | 0.499 |
| 8558 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | SLITRK3 | Sponge network | -2.925 | 0 | -3.328 | 0 | 0.499 |
| 8559 | LINC00641 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | AFF1 | Sponge network | -0.222 | 0.32445 | -0.384 | 0.00053 | 0.499 |
| 8560 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-589-3p | 11 | PEG3 | Sponge network | 0.176 | 0.37402 | -1.74 | 0 | 0.499 |
| 8561 | RP11-473I1.9 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | ERCC6 | Sponge network | 0.683 | 1.0E-5 | 0.23 | 0.015 | 0.499 |
| 8562 | MEG3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 25 | PCDH10 | Sponge network | 0.249 | 0.35902 | -1.644 | 0.0078 | 0.499 |
| 8563 | ADAMTS9-AS2 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-421;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | HERC1 | Sponge network | -2.452 | 0 | -0.196 | 0.08544 | 0.499 |
| 8564 | RP3-475N16.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c | 16 | ABL2 | Sponge network | 1.056 | 0 | 0.82 | 0 | 0.499 |
| 8565 | GAS6-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-5p | 11 | LHFP | Sponge network | 0.16 | 0.50785 | -0.167 | 0.41371 | 0.499 |
| 8566 | CTC-296K1.3 |
hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-454-3p | 11 | ITPKB | Sponge network | -2.095 | 0.00112 | -0.704 | 0.00106 | 0.499 |
| 8567 | LINC00957 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PDE4DIP | Sponge network | -0.421 | 0.0114 | -0.282 | 0.0257 | 0.499 |
| 8568 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-92b-3p | 16 | AKAP12 | Sponge network | 0.056 | 0.81195 | -0.932 | 0.0028 | 0.499 |
| 8569 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 40 | CHD5 | Sponge network | 0.194 | 0.64278 | -0.922 | 0.01521 | 0.499 |
| 8570 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | RCAN2 | Sponge network | 0.559 | 0.00026 | -0.33 | 0.15172 | 0.499 |
| 8571 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | EDA2R | Sponge network | 0.199 | 0.38015 | -0.587 | 0.01598 | 0.499 |
| 8572 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-3p | 24 | BNC2 | Sponge network | 0.178 | 0.29106 | -0.491 | 0.09988 | 0.499 |
| 8573 | RP11-620J15.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p | 22 | SYT4 | Sponge network | -0.739 | 0.00044 | -3.207 | 0 | 0.499 |
| 8574 | FGF14-AS2 |
hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | PAK3 | Sponge network | -1.582 | 8.0E-5 | -0.628 | 0.07253 | 0.499 |
| 8575 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | JAKMIP2 | Sponge network | 0.194 | 0.64278 | -1.388 | 0 | 0.499 |
| 8576 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p | 51 | CADM2 | Sponge network | -0.842 | 0.01361 | -3.782 | 0 | 0.499 |
| 8577 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 41 | ZFHX3 | Sponge network | -0.576 | 0.10121 | 0.389 | 0.01363 | 0.499 |
| 8578 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | ADAMTSL3 | Sponge network | -0.777 | 0.1134 | -1.559 | 0 | 0.499 |
| 8579 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 17 | LRRC4 | Sponge network | -0.769 | 0.00035 | -1.774 | 0 | 0.499 |
| 8580 | CTD-2089N3.2 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-194-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | PRKCB | Sponge network | -1.375 | 0.08642 | -1.313 | 2.0E-5 | 0.499 |
| 8581 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | MCC | Sponge network | -0.727 | 0.04726 | -1.005 | 0 | 0.499 |
| 8582 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-376c-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | PHC3 | Sponge network | 0.101 | 0.60919 | 0.212 | 0.0499 | 0.499 |
| 8583 | NR2F1-AS1 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 12 | RTN4 | Sponge network | -0.49 | 0.04946 | -0.412 | 0.00014 | 0.499 |
| 8584 | RP5-1198O20.4 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-942-5p | 21 | GLIPR1 | Sponge network | 0.997 | 0.00066 | 0.146 | 0.3276 | 0.499 |
| 8585 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 61 | NTRK3 | Sponge network | 0.572 | 0.01722 | -1.77 | 3.0E-5 | 0.499 |
| 8586 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ZNF292 | Sponge network | 0.559 | 0.00026 | 0.276 | 0.04148 | 0.499 |
| 8587 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940 | 38 | HLF | Sponge network | -2.117 | 0.00161 | -2.443 | 0 | 0.499 |
| 8588 | RP1-59D14.5 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-940 | 10 | MYO9A | Sponge network | 1.162 | 5.0E-5 | -0.147 | 0.32373 | 0.499 |
| 8589 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-93-5p | 32 | ZBTB4 | Sponge network | 0.997 | 0.00066 | -0.739 | 0 | 0.499 |
| 8590 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-539-5p;hsa-miR-542-3p | 10 | DMD | Sponge network | 0.043 | 0.81628 | -1.506 | 0 | 0.499 |
| 8591 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | WWTR1 | Sponge network | -1.349 | 0.00017 | -0.345 | 0.11997 | 0.499 |
| 8592 | TRAF3IP2-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 11 | NGFR | Sponge network | -0.323 | 0.01372 | -1.271 | 0.01058 | 0.499 |
| 8593 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 16 | DYNC1I1 | Sponge network | -0.206 | 0.12942 | -1.12 | 0.00107 | 0.499 |
| 8594 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 59 | BMPR2 | Sponge network | 0.537 | 0.00139 | 0.206 | 0.0635 | 0.499 |
| 8595 | RP11-67L2.2 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | ZNF638 | Sponge network | 0.72 | 0.0001 | 0.22 | 0.01214 | 0.499 |
| 8596 | PCA3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 19 | PTGFR | Sponge network | -0.709 | 0.18168 | -0.233 | 0.45421 | 0.499 |
| 8597 | RP11-822E23.8 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-769-5p;hsa-miR-93-5p | 14 | PRUNE2 | Sponge network | -0.777 | 0.16667 | -1.733 | 0.00022 | 0.499 |
| 8598 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 13 | SEPT6 | Sponge network | -0.769 | 0.00035 | -0.598 | 0.00181 | 0.499 |
| 8599 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-5p | 13 | ABCC9 | Sponge network | 0.331 | 0.57247 | -0.466 | 0.14121 | 0.499 |
| 8600 | LINC00865 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 22 | PCDH9 | Sponge network | -1.249 | 7.0E-5 | -1.823 | 4.0E-5 | 0.499 |
| 8601 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | TACC1 | Sponge network | -1.349 | 0.00017 | -0.362 | 0.05406 | 0.499 |
| 8602 | GNG12-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 18 | SORCS1 | Sponge network | -0.793 | 7.0E-5 | -3.492 | 0 | 0.499 |
| 8603 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PTPRB | Sponge network | -1.281 | 0.00012 | 0.105 | 0.47122 | 0.499 |
| 8604 | RP11-620J15.3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p | 25 | PRKCB | Sponge network | -0.739 | 0.00044 | -1.313 | 2.0E-5 | 0.499 |
| 8605 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 65 | KIF1B | Sponge network | 0.72 | 0.0001 | -0.118 | 0.32258 | 0.499 |
| 8606 | RP11-554A11.4 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p | 12 | KIT | Sponge network | -0.583 | 0.14589 | -2.184 | 0 | 0.499 |
| 8607 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-93-3p | 22 | ST6GAL2 | Sponge network | -0.693 | 0.05629 | -1.201 | 0.00597 | 0.499 |
| 8608 | AC009120.6 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | ATM | Sponge network | 0.919 | 0 | 0.178 | 0.21568 | 0.499 |
| 8609 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p | 27 | MYO9A | Sponge network | -2.62 | 0 | -0.147 | 0.32373 | 0.498 |
| 8610 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 31 | TGFBR3 | Sponge network | -0.896 | 0.00099 | -1.173 | 1.0E-5 | 0.498 |
| 8611 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 21 | PTGFR | Sponge network | -0.769 | 0.00035 | -0.233 | 0.45421 | 0.498 |
| 8612 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 12 | ZNF292 | Sponge network | 1.063 | 0 | 0.276 | 0.04148 | 0.498 |
| 8613 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-93-3p | 19 | PTPN11 | Sponge network | 0.75 | 0.00054 | 0.431 | 2.0E-5 | 0.498 |
| 8614 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484 | 14 | ITGA5 | Sponge network | 0.299 | 0.57722 | 0.452 | 0.03524 | 0.498 |
| 8615 | LINC00473 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-935 | 33 | PTPRT | Sponge network | -1.203 | 0.03881 | -0.907 | 0.03727 | 0.498 |
| 8616 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-629-3p | 17 | EBF1 | Sponge network | -2.531 | 0 | -0.384 | 0.08856 | 0.498 |
| 8617 | ARHGEF26-AS1 |
hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-660-5p | 10 | DNER | Sponge network | -2.46 | 0 | -3.787 | 0 | 0.498 |
| 8618 | AC006116.24 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p | 11 | CDH19 | Sponge network | -0.473 | 0.07602 | -3.434 | 0 | 0.498 |
| 8619 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p | 42 | RICTOR | Sponge network | 0.72 | 0.0001 | 0.388 | 0.00188 | 0.498 |
| 8620 | LINC00473 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-877-5p | 27 | ASTN1 | Sponge network | -1.203 | 0.03881 | -2.389 | 2.0E-5 | 0.498 |
| 8621 | PWAR6 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 17 | PAK3 | Sponge network | -1.575 | 6.0E-5 | -0.628 | 0.07253 | 0.498 |
| 8622 | LINC00472 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 14 | GLI3 | Sponge network | -0.842 | 0.01361 | -0.615 | 0.01482 | 0.498 |
| 8623 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | AR | Sponge network | -1.375 | 0.08642 | -1.554 | 0 | 0.498 |
| 8624 | LINC00865 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p | 56 | DST | Sponge network | -1.249 | 7.0E-5 | -0.713 | 0.00096 | 0.498 |
| 8625 | RP11-473I1.9 |
hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p | 10 | TTL | Sponge network | 0.683 | 1.0E-5 | 0.494 | 1.0E-5 | 0.498 |
| 8626 | TPTEP1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 32 | DLC1 | Sponge network | -1.216 | 0 | -0.382 | 0.05038 | 0.498 |
| 8627 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p | 23 | EBF1 | Sponge network | -0.473 | 0.07602 | -0.384 | 0.08856 | 0.498 |
| 8628 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 48 | PRKAA2 | Sponge network | 0.335 | 0.20596 | -2.462 | 0 | 0.498 |
| 8629 | HOXB-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-501-3p | 11 | ZBTB20 | Sponge network | -0.154 | 0.53954 | -0.122 | 0.57569 | 0.498 |
| 8630 | TRAF3IP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 38 | IL17RD | Sponge network | -0.323 | 0.01372 | -0.019 | 0.94873 | 0.498 |
| 8631 | LINC00641 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 29 | SYNPO2 | Sponge network | -0.222 | 0.32445 | -2.342 | 0 | 0.498 |
| 8632 | GS1-124K5.11 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p | 14 | ZBTB20 | Sponge network | 0.494 | 0.00339 | -0.122 | 0.57569 | 0.498 |
| 8633 | SNHG14 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-96-5p | 15 | ELMO1 | Sponge network | -1.111 | 0.0003 | 0.04 | 0.83147 | 0.498 |
| 8634 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 62 | IL6ST | Sponge network | 0.249 | 0.35902 | -0.402 | 0.00676 | 0.498 |
| 8635 | NR2F1-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-942-5p | 11 | BMPR1A | Sponge network | -0.49 | 0.04946 | -0.366 | 0.01036 | 0.498 |
| 8636 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 24 | ABL1 | Sponge network | -0.49 | 0.04946 | -0.215 | 0.07804 | 0.498 |
| 8637 | RP11-37B2.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-625-5p | 19 | ABI2 | Sponge network | 1.03 | 0 | 0.175 | 0.14787 | 0.498 |
| 8638 | AC127904.2 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 10 | TPR | Sponge network | 1.308 | 0 | 0.582 | 0 | 0.498 |
| 8639 | RP11-798M19.6 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 12 | MAPK10 | Sponge network | -0.612 | 0.00094 | -1.774 | 0 | 0.498 |
| 8640 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 25 | TMEFF2 | Sponge network | -1.697 | 0.00889 | -2.426 | 0 | 0.498 |
| 8641 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 38 | PGR | Sponge network | -2.925 | 0 | -1.704 | 0 | 0.498 |
| 8642 | RP11-359E10.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-7-1-3p | 15 | ITPKB | Sponge network | 0.299 | 0.57722 | -0.704 | 0.00106 | 0.498 |
| 8643 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-660-5p | 35 | ESR1 | Sponge network | -0.563 | 0.02545 | -1.035 | 3.0E-5 | 0.498 |
| 8644 | LINC00672 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-590-3p | 12 | FAM172A | Sponge network | -0.227 | 0.31657 | -0.57 | 0 | 0.498 |
| 8645 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-7-5p;hsa-miR-96-5p | 16 | FNBP1 | Sponge network | 0.101 | 0.60919 | -0.969 | 4.0E-5 | 0.498 |
| 8646 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-500a-5p;hsa-miR-93-5p;hsa-miR-942-5p | 23 | HLF | Sponge network | -2.531 | 0 | -2.443 | 0 | 0.498 |
| 8647 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 10 | TMEFF2 | Sponge network | -0.896 | 0.00099 | -2.426 | 0 | 0.498 |
| 8648 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | TACC1 | Sponge network | -0.612 | 0.00094 | -0.362 | 0.05406 | 0.498 |
| 8649 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 17 | TRIM33 | Sponge network | 0.559 | 0.00026 | 0.402 | 7.0E-5 | 0.498 |
| 8650 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | -1.203 | 0.03881 | -1.12 | 0.00107 | 0.498 |
| 8651 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 16 | ZMYND11 | Sponge network | 0.537 | 0.00139 | -0.2 | 0.05449 | 0.498 |
| 8652 | BOLA3-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-935 | 30 | DIP2C | Sponge network | -0.246 | 0.35034 | -0.436 | 0.01713 | 0.498 |
| 8653 | PCA3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-590-3p | 17 | STARD13 | Sponge network | -0.709 | 0.18168 | -0.187 | 0.27383 | 0.498 |
| 8654 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-7-1-3p | 18 | PLSCR4 | Sponge network | -0.405 | 0.22013 | -0.474 | 0.03833 | 0.498 |
| 8655 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | TACC1 | Sponge network | -2.46 | 0 | -0.362 | 0.05406 | 0.498 |
| 8656 | BCYRN1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p | 12 | ZNF638 | Sponge network | 0.311 | 0.10344 | 0.22 | 0.01214 | 0.498 |
| 8657 | RP11-175O19.4 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-664a-3p | 17 | RASAL2 | Sponge network | 0.917 | 0 | 0.869 | 0 | 0.498 |
| 8658 | LINC00473 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 12 | ITGB3 | Sponge network | -1.203 | 0.03881 | -0.011 | 0.95751 | 0.498 |
| 8659 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | ZFHX4 | Sponge network | 0.532 | 0.00505 | 0.018 | 0.95574 | 0.498 |
| 8660 | TRAF3IP2-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | WWTR1 | Sponge network | -0.323 | 0.01372 | -0.345 | 0.11997 | 0.498 |
| 8661 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | JAZF1 | Sponge network | -1.886 | 0 | -0.377 | 0.02677 | 0.498 |
| 8662 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-766-3p;hsa-miR-93-3p | 51 | IL6ST | Sponge network | -2.69 | 0.0001 | -0.402 | 0.00676 | 0.498 |
| 8663 | LINC00476 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-590-5p | 10 | MYH11 | Sponge network | -0.615 | 0.00015 | -2.895 | 0 | 0.498 |
| 8664 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 24 | NR2C2 | Sponge network | 0.401 | 0.00066 | 0.21 | 0.03224 | 0.498 |
| 8665 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p | 10 | GAS1 | Sponge network | -0.612 | 0.00094 | -1.047 | 0.00364 | 0.498 |
| 8666 | SNHG14 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 25 | ATM | Sponge network | -1.111 | 0.0003 | 0.178 | 0.21568 | 0.498 |
| 8667 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 25 | CACNA2D1 | Sponge network | -0.405 | 0.22013 | -0.846 | 0.00675 | 0.498 |
| 8668 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 41 | RASSF8 | Sponge network | -2.46 | 0 | -0.122 | 0.65591 | 0.498 |
| 8669 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | EBF3 | Sponge network | 0.997 | 0.00066 | -0.058 | 0.81437 | 0.498 |
| 8670 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p | 12 | THSD7A | Sponge network | -2.69 | 0.0001 | 0.242 | 0.25154 | 0.498 |
| 8671 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 31 | ABI2 | Sponge network | 0.494 | 0.00339 | 0.175 | 0.14787 | 0.498 |
| 8672 | RP11-767N6.7 |
hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | ATRX | Sponge network | 0.466 | 0.00155 | 0.261 | 0.04408 | 0.498 |
| 8673 | FAM66C |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | CDKN1C | Sponge network | -0.901 | 0.00019 | -0.929 | 0.00033 | 0.498 |
| 8674 | CTC-296K1.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-424-5p | 15 | ASTN1 | Sponge network | -2.076 | 0.00548 | -2.389 | 2.0E-5 | 0.498 |
| 8675 | AC003090.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-590-5p | 10 | THSD7A | Sponge network | -2.117 | 0.00161 | 0.242 | 0.25154 | 0.498 |
| 8676 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p | 18 | FNBP1 | Sponge network | -0.222 | 0.32445 | -0.969 | 4.0E-5 | 0.498 |
| 8677 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-532-3p;hsa-miR-671-5p | 22 | GAS7 | Sponge network | -0.164 | 0.45106 | -0.555 | 0.01602 | 0.498 |
| 8678 | FAM66C |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 21 | PTGFR | Sponge network | -0.901 | 0.00019 | -0.233 | 0.45421 | 0.498 |
| 8679 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-550a-5p | 28 | PDGFRA | Sponge network | -1.331 | 3.0E-5 | -0.372 | 0.0037 | 0.498 |
| 8680 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 22 | ATRX | Sponge network | 0.389 | 0.04293 | 0.261 | 0.04408 | 0.498 |
| 8681 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-374a-5p | 12 | DNAJB4 | Sponge network | -0.693 | 0.05629 | -0.7 | 1.0E-5 | 0.498 |
| 8682 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-421 | 12 | ZNF521 | Sponge network | 0.532 | 0.00505 | -0.111 | 0.58068 | 0.498 |
| 8683 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | TACC1 | Sponge network | 0.249 | 0.35902 | -0.362 | 0.05406 | 0.498 |
| 8684 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | NOTCH2 | Sponge network | 0.889 | 9.0E-5 | 0.138 | 0.35163 | 0.498 |
| 8685 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-7-5p;hsa-miR-93-3p | 24 | EPHA3 | Sponge network | -0.777 | 0.16667 | 0.185 | 0.53588 | 0.498 |
| 8686 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 22 | TCF4 | Sponge network | -1.886 | 0 | 0.016 | 0.92271 | 0.498 |
| 8687 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-93-5p | 27 | FAT3 | Sponge network | -0.726 | 0.00132 | -1.601 | 0.00051 | 0.498 |
| 8688 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | JAZF1 | Sponge network | -0.376 | 0.00337 | -0.377 | 0.02677 | 0.498 |
| 8689 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | PDGFRA | Sponge network | 0.249 | 0.35902 | -0.372 | 0.0037 | 0.498 |
| 8690 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 15 | ZNF770 | Sponge network | 1.147 | 0 | 0.206 | 0.06751 | 0.498 |
| 8691 | LINC00092 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-539-5p | 19 | BTG2 | Sponge network | -1.886 | 0 | -1.763 | 0 | 0.498 |
| 8692 | LINC00702 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CTGF | Sponge network | -0.737 | 0.06182 | 0.321 | 0.11682 | 0.498 |
| 8693 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-5p | 19 | RET | Sponge network | -1.439 | 4.0E-5 | -0.833 | 0.03517 | 0.497 |
| 8694 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 33 | AR | Sponge network | 0.889 | 9.0E-5 | -1.554 | 0 | 0.497 |
| 8695 | RP11-967K21.1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | RNF180 | Sponge network | 0.056 | 0.81195 | -1.127 | 1.0E-5 | 0.497 |
| 8696 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-766-3p | 13 | EZH1 | Sponge network | -0.517 | 0.02935 | -0.564 | 1.0E-5 | 0.497 |
| 8697 | RP11-822E23.8 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-532-3p | 16 | SYNM | Sponge network | -0.777 | 0.16667 | -2.309 | 1.0E-5 | 0.497 |
| 8698 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | LRRC7 | Sponge network | 0.997 | 0.00066 | -1.341 | 0.01193 | 0.497 |
| 8699 | MIR497HG |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p | 27 | SYT4 | Sponge network | -1.439 | 4.0E-5 | -3.207 | 0 | 0.497 |
| 8700 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-628-5p | 19 | BCL6 | Sponge network | -0.154 | 0.53954 | -0.211 | 0.19489 | 0.497 |
| 8701 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 23 | ERCC4 | Sponge network | 0.532 | 0.00505 | 0.126 | 0.15266 | 0.497 |
| 8702 | PART1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935 | 41 | FBXO32 | Sponge network | -2.925 | 0 | -0.495 | 0.07561 | 0.497 |
| 8703 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | TACC1 | Sponge network | -0.563 | 0.02545 | -0.362 | 0.05406 | 0.497 |
| 8704 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-590-3p | 25 | AFF3 | Sponge network | 0.194 | 0.64278 | -2.373 | 0 | 0.497 |
| 8705 | CTC-429P9.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-542-3p | 10 | FOXP1 | Sponge network | 0.043 | 0.81628 | 0.042 | 0.74368 | 0.497 |
| 8706 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | TRPS1 | Sponge network | -1.161 | 0.00591 | -0.191 | 0.44109 | 0.497 |
| 8707 | FENDRR |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 44 | TGFBR3 | Sponge network | -1.281 | 0.00012 | -1.173 | 1.0E-5 | 0.497 |
| 8708 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | MOB1B | Sponge network | 0.72 | 0.0001 | 0.197 | 0.15266 | 0.497 |
| 8709 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NCAM2 | Sponge network | -1.216 | 0 | -0.482 | 0.22565 | 0.497 |
| 8710 | MIR143HG |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | RASSF3 | Sponge network | -0.739 | 0.0574 | -0.226 | 0.11691 | 0.497 |
| 8711 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-7-5p | 12 | FLNA | Sponge network | -0.693 | 0.05629 | -0.771 | 0.01377 | 0.497 |
| 8712 | RP11-13K12.5 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p | 11 | NGFR | Sponge network | -0.318 | 0.65847 | -1.271 | 0.01058 | 0.497 |
| 8713 | RP11-890B15.3 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p | 20 | ATM | Sponge network | 0.101 | 0.60919 | 0.178 | 0.21568 | 0.497 |
| 8714 | CTD-2089N3.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 15 | NRXN1 | Sponge network | -1.375 | 0.08642 | -3.778 | 0 | 0.497 |
| 8715 | MIR497HG |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-7-1-3p | 11 | SEMA3E | Sponge network | -1.439 | 4.0E-5 | -1.717 | 0.00137 | 0.497 |
| 8716 | HAND2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | GALNT13 | Sponge network | -1.697 | 0.00889 | -0.857 | 0.07439 | 0.497 |
| 8717 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 33 | BNC2 | Sponge network | 0.16 | 0.50785 | -0.491 | 0.09988 | 0.497 |
| 8718 | NR2F1-AS1 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CTGF | Sponge network | -0.49 | 0.04946 | 0.321 | 0.11682 | 0.497 |
| 8719 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-450b-5p | 24 | JAZF1 | Sponge network | -4.546 | 0 | -0.377 | 0.02677 | 0.497 |
| 8720 | MEG3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-7-1-3p | 52 | DTNA | Sponge network | 0.249 | 0.35902 | -1.648 | 1.0E-5 | 0.497 |
| 8721 | RP11-13K12.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p | 18 | NRK | Sponge network | -0.154 | 0.78939 | 0.656 | 0.19217 | 0.497 |
| 8722 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-93-3p | 17 | EBF3 | Sponge network | -0.469 | 0.08721 | -0.058 | 0.81437 | 0.497 |
| 8723 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | EBF3 | Sponge network | -1.281 | 0.00012 | -0.058 | 0.81437 | 0.497 |
| 8724 | RP11-400K9.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-93-3p | 17 | RUNX1T1 | Sponge network | -0.733 | 0.19469 | -0.344 | 0.2374 | 0.497 |
| 8725 | LINC00641 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | WAC | Sponge network | -0.222 | 0.32445 | -0.054 | 0.43688 | 0.497 |
| 8726 | AP001347.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | NAV3 | Sponge network | -1.161 | 0.00591 | 0.212 | 0.47729 | 0.497 |
| 8727 | NEAT1 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | CHD2 | Sponge network | 0.705 | 0.00014 | 0.049 | 0.55371 | 0.497 |
| 8728 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-589-3p | 15 | SOX5 | Sponge network | -4.463 | 0.00025 | -1.091 | 4.0E-5 | 0.497 |
| 8729 | RP5-1198O20.4 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | TPO | Sponge network | 0.997 | 0.00066 | -1.87 | 2.0E-5 | 0.497 |
| 8730 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 29 | PBX1 | Sponge network | -0.896 | 0.00099 | -1.206 | 0 | 0.497 |
| 8731 | MYLK-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-940 | 25 | NTRK2 | Sponge network | -1.331 | 3.0E-5 | -0.736 | 0.06495 | 0.497 |
| 8732 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | FAM172A | Sponge network | -0.358 | 0.17255 | -0.57 | 0 | 0.497 |
| 8733 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 19 | RICTOR | Sponge network | 1.178 | 0 | 0.388 | 0.00188 | 0.497 |
| 8734 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | PLSCR4 | Sponge network | -0.612 | 0.00094 | -0.474 | 0.03833 | 0.497 |
| 8735 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ZMYND11 | Sponge network | 0.101 | 0.60919 | -0.2 | 0.05449 | 0.497 |
| 8736 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-874-3p | 13 | NR2C2 | Sponge network | 1.122 | 0 | 0.21 | 0.03224 | 0.497 |
| 8737 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | ERG | Sponge network | 0.194 | 0.64278 | 0.002 | 0.99011 | 0.497 |
| 8738 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | LRP1B | Sponge network | -0.323 | 0.01372 | -2.163 | 0 | 0.497 |
| 8739 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-450b-5p;hsa-miR-589-5p | 15 | ROBO1 | Sponge network | -1.439 | 4.0E-5 | -0.009 | 0.96154 | 0.497 |
| 8740 | RP11-967K21.1 |
hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-92a-3p | 13 | MACF1 | Sponge network | 0.056 | 0.81195 | 0.019 | 0.90335 | 0.497 |
| 8741 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | RCAN2 | Sponge network | 0.083 | 0.66405 | -0.33 | 0.15172 | 0.497 |
| 8742 | HOXB-AS1 |
hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-429 | 11 | KLF10 | Sponge network | -0.154 | 0.53954 | -0.783 | 0 | 0.497 |
| 8743 | PCA3 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-409-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p | 13 | MACF1 | Sponge network | -0.709 | 0.18168 | 0.019 | 0.90335 | 0.497 |
| 8744 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p | 15 | CDH19 | Sponge network | -0.693 | 0.05629 | -3.434 | 0 | 0.497 |
| 8745 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 18 | LATS2 | Sponge network | 0.335 | 0.20596 | 0.159 | 0.2304 | 0.497 |
| 8746 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p | 21 | JAZF1 | Sponge network | 0.176 | 0.37402 | -0.377 | 0.02677 | 0.497 |
| 8747 | AGAP11 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-214-5p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-874-3p | 22 | MXI1 | Sponge network | -1.645 | 0 | -0.987 | 0 | 0.497 |
| 8748 | AC009948.5 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 18 | GLI3 | Sponge network | 0.532 | 0.00505 | -0.615 | 0.01482 | 0.497 |
| 8749 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 46 | PDGFRA | Sponge network | -0.49 | 0.04946 | -0.372 | 0.0037 | 0.497 |
| 8750 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | THBS1 | Sponge network | 0.299 | 0.57722 | 0.741 | 0.00386 | 0.497 |
| 8751 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 18 | ABCB5 | Sponge network | -1.575 | 6.0E-5 | -0.956 | 0.04077 | 0.497 |
| 8752 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-369-3p | 24 | MEF2C | Sponge network | -0.932 | 0.00329 | -0.499 | 0.01448 | 0.497 |
| 8753 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 25 | HIP1 | Sponge network | 0.997 | 0.00066 | 0.774 | 0 | 0.497 |
| 8754 | AC010226.4 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p | 14 | PCDH9 | Sponge network | -0.811 | 2.0E-5 | -1.823 | 4.0E-5 | 0.497 |
| 8755 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 48 | PRKAA2 | Sponge network | -0.899 | 6.0E-5 | -2.462 | 0 | 0.497 |
| 8756 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | PDGFC | Sponge network | 0.249 | 0.35902 | -0.33 | 0.06483 | 0.497 |
| 8757 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-429 | 15 | CNTNAP3 | Sponge network | -2.62 | 0 | -1.465 | 0.00033 | 0.497 |
| 8758 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-671-5p;hsa-miR-93-5p | 37 | ADARB1 | Sponge network | -1.349 | 0.00017 | -0.335 | 0.06631 | 0.497 |
| 8759 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-340-5p | 12 | TGFBR2 | Sponge network | -1.331 | 3.0E-5 | -0.243 | 0.11408 | 0.497 |
| 8760 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-671-5p | 16 | KIT | Sponge network | -0.611 | 0.09607 | -2.184 | 0 | 0.497 |
| 8761 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | CACNA2D1 | Sponge network | -0.563 | 0.02545 | -0.846 | 0.00675 | 0.497 |
| 8762 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | ZFHX4 | Sponge network | -0.709 | 0.18168 | 0.018 | 0.95574 | 0.497 |
| 8763 | EMX2OS |
hsa-miR-126-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 16 | CXCL12 | Sponge network | -0.777 | 0.1134 | -1.421 | 0 | 0.497 |
| 8764 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 54 | SSBP2 | Sponge network | -1.216 | 0 | -1.58 | 0 | 0.497 |
| 8765 | CTD-2554C21.2 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-629-3p;hsa-miR-96-5p | 11 | CXCL12 | Sponge network | -1.654 | 0.00144 | -1.421 | 0 | 0.497 |
| 8766 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-29a-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-582-5p | 21 | FBXO32 | Sponge network | -2.095 | 0.00112 | -0.495 | 0.07561 | 0.497 |
| 8767 | SERTAD4-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-93-3p | 19 | TRPS1 | Sponge network | -0.932 | 0.00329 | -0.191 | 0.44109 | 0.497 |
| 8768 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 30 | BNC2 | Sponge network | -0.318 | 0.65847 | -0.491 | 0.09988 | 0.497 |
| 8769 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | CAV1 | Sponge network | -0.739 | 0.00044 | -1.013 | 6.0E-5 | 0.497 |
| 8770 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-590-3p | 21 | PLSCR4 | Sponge network | -0.738 | 0.21233 | -0.474 | 0.03833 | 0.497 |
| 8771 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 17 | TGFBR3 | Sponge network | -0.376 | 0.00337 | -1.173 | 1.0E-5 | 0.497 |
| 8772 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | 0.064 | 0.72934 | 0.358 | 0.13727 | 0.497 |
| 8773 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 35 | EPHA3 | Sponge network | -0.222 | 0.32445 | 0.185 | 0.53588 | 0.497 |
| 8774 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | TACC1 | Sponge network | -1.161 | 0.00591 | -0.362 | 0.05406 | 0.497 |
| 8775 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 29 | PRKAB2 | Sponge network | -0.096 | 0.67958 | -0.367 | 0.01371 | 0.497 |
| 8776 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 11 | CNTNAP3 | Sponge network | -1.582 | 8.0E-5 | -1.465 | 0.00033 | 0.497 |
| 8777 | SH3RF3-AS1 |
hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p | 14 | ABL1 | Sponge network | 0.889 | 9.0E-5 | -0.215 | 0.07804 | 0.496 |
| 8778 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 20 | CNTN1 | Sponge network | -0.246 | 0.35034 | -2.594 | 0 | 0.496 |
| 8779 | CTD-2314G24.2 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | SYT4 | Sponge network | 0.246 | 0.59912 | -3.207 | 0 | 0.496 |
| 8780 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-629-3p | 23 | DDX6 | Sponge network | 1.254 | 0 | 0.091 | 0.36428 | 0.496 |
| 8781 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | ZNF208 | Sponge network | -2.46 | 0 | -0.4 | 0.22381 | 0.496 |
| 8782 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TSC1 | Sponge network | 0.537 | 0.00139 | 0.103 | 0.26071 | 0.496 |
| 8783 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 17 | RCAN2 | Sponge network | -0.899 | 6.0E-5 | -0.33 | 0.15172 | 0.496 |
| 8784 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p | 19 | TACC1 | Sponge network | -4.463 | 0.00025 | -0.362 | 0.05406 | 0.496 |
| 8785 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | RASSF8 | Sponge network | -1.349 | 0.00017 | -0.122 | 0.65591 | 0.496 |
| 8786 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | SYNE1 | Sponge network | -1.203 | 0.03881 | -0.738 | 0.00316 | 0.496 |
| 8787 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 44 | QKI | Sponge network | -0.096 | 0.67958 | -0.333 | 0.04111 | 0.496 |
| 8788 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | PIK3R1 | Sponge network | 0.997 | 0.00066 | -0.133 | 0.36854 | 0.496 |
| 8789 | HOXB-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-429 | 10 | DYNC1I1 | Sponge network | -0.154 | 0.53954 | -1.12 | 0.00107 | 0.496 |
| 8790 | TP73-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-96-5p | 28 | ASTN1 | Sponge network | -0.563 | 0.02545 | -2.389 | 2.0E-5 | 0.496 |
| 8791 | C20orf166-AS1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-532-5p | 15 | RPS6KA6 | Sponge network | -2.69 | 0.0001 | -2.989 | 0 | 0.496 |
| 8792 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | MOB1B | Sponge network | 1.153 | 0 | 0.197 | 0.15266 | 0.496 |
| 8793 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935 | 18 | LRRC7 | Sponge network | -0.896 | 0.00099 | -1.341 | 0.01193 | 0.496 |
| 8794 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-96-5p | 12 | NCAM2 | Sponge network | -1.582 | 8.0E-5 | -0.482 | 0.22565 | 0.496 |
| 8795 | RP1-302G2.5 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-935;hsa-miR-940 | 13 | PTPRT | Sponge network | -1.483 | 9.0E-5 | -0.907 | 0.03727 | 0.496 |
| 8796 | FAM66C |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ITPKB | Sponge network | -0.901 | 0.00019 | -0.704 | 0.00106 | 0.496 |
| 8797 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-942-5p | 40 | UNC80 | Sponge network | -1.203 | 0.03881 | -1.934 | 0 | 0.496 |
| 8798 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p | 12 | FAT4 | Sponge network | -1.054 | 0.0706 | -0.354 | 0.14694 | 0.496 |
| 8799 | ACTA2-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | KLF10 | Sponge network | 0.194 | 0.64278 | -0.783 | 0 | 0.496 |
| 8800 | RP11-473I1.9 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 14 | SHPRH | Sponge network | 0.683 | 1.0E-5 | 0.098 | 0.40994 | 0.496 |
| 8801 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | HOOK3 | Sponge network | 0.541 | 0.00125 | 0.273 | 0.09957 | 0.496 |
| 8802 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 23 | BCL2 | Sponge network | -4.546 | 0 | -0.899 | 0 | 0.496 |
| 8803 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-940 | 19 | MYO9A | Sponge network | 0.921 | 0.00118 | -0.147 | 0.32373 | 0.496 |
| 8804 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 34 | SOX5 | Sponge network | -0.896 | 0.00099 | -1.091 | 4.0E-5 | 0.496 |
| 8805 | PART1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | TMEFF2 | Sponge network | -2.925 | 0 | -2.426 | 0 | 0.496 |
| 8806 | RP5-1198O20.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-96-5p | 34 | ASTN1 | Sponge network | 0.997 | 0.00066 | -2.389 | 2.0E-5 | 0.496 |
| 8807 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 45 | ZFHX3 | Sponge network | -0.737 | 0.06182 | 0.389 | 0.01363 | 0.496 |
| 8808 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p | 55 | IL6ST | Sponge network | -0.842 | 0.01361 | -0.402 | 0.00676 | 0.496 |
| 8809 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-671-5p | 15 | CADM3 | Sponge network | -0.469 | 0.08721 | -3.663 | 0 | 0.496 |
| 8810 | MAP3K14 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p | 12 | BCL2 | Sponge network | -0.295 | 0.02604 | -0.899 | 0 | 0.496 |
| 8811 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-629-3p;hsa-miR-942-5p | 26 | GPC6 | Sponge network | -1.439 | 4.0E-5 | -0.054 | 0.81465 | 0.496 |
| 8812 | MALAT1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-940 | 14 | TAF1 | Sponge network | 0.782 | 0.00016 | 0.39 | 3.0E-5 | 0.496 |
| 8813 | LINC00578 |
hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-744-3p | 19 | DIP2C | Sponge network | 0.811 | 0.03228 | -0.436 | 0.01713 | 0.496 |
| 8814 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 32 | IGF1 | Sponge network | 0.532 | 0.00505 | -0.204 | 0.5898 | 0.496 |
| 8815 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-484 | 13 | GREM1 | Sponge network | -4.546 | 0 | 0.902 | 0.01691 | 0.496 |
| 8816 | RP11-693J15.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-671-5p | 11 | CADM3 | Sponge network | -0.72 | 0.24239 | -3.663 | 0 | 0.496 |
| 8817 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429 | 19 | PTGFR | Sponge network | -0.563 | 0.02545 | -0.233 | 0.45421 | 0.496 |
| 8818 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 24 | CACNA2D1 | Sponge network | -0.842 | 0.01361 | -0.846 | 0.00675 | 0.496 |
| 8819 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | RASSF2 | Sponge network | -1.281 | 0.00012 | -0.273 | 0.21961 | 0.496 |
| 8820 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-590-3p | 13 | FGFR1 | Sponge network | -1.375 | 0.08642 | -0.509 | 0.03957 | 0.496 |
| 8821 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 26 | SETBP1 | Sponge network | -0.405 | 0.22013 | -1.302 | 1.0E-5 | 0.496 |
| 8822 | ACTA2-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | CXCL12 | Sponge network | 0.194 | 0.64278 | -1.421 | 0 | 0.496 |
| 8823 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-5p;hsa-miR-652-3p;hsa-miR-93-5p | 15 | ZBTB4 | Sponge network | -1.255 | 0.00981 | -0.739 | 0 | 0.496 |
| 8824 | LINC00092 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-940 | 23 | SNX29 | Sponge network | -1.886 | 0 | -0.34 | 0.01371 | 0.496 |
| 8825 | MEG3 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-577;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | CAV1 | Sponge network | 0.249 | 0.35902 | -1.013 | 6.0E-5 | 0.496 |
| 8826 | MIR143HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-503-5p;hsa-miR-590-3p | 11 | SMARCA1 | Sponge network | -0.739 | 0.0574 | -0.684 | 0.00159 | 0.496 |
| 8827 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 22 | PGR | Sponge network | -1.057 | 0.00029 | -1.704 | 0 | 0.496 |
| 8828 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-671-5p | 22 | KIT | Sponge network | -2.915 | 0.00015 | -2.184 | 0 | 0.496 |
| 8829 | RP11-1246C19.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | BRD3 | Sponge network | 1.352 | 0 | 0.37 | 0.00013 | 0.496 |
| 8830 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-5p | 35 | ADARB1 | Sponge network | 0.16 | 0.50785 | -0.335 | 0.06631 | 0.496 |
| 8831 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 11 | RELN | Sponge network | -1.057 | 0.00029 | -2.403 | 0 | 0.496 |
| 8832 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RB1CC1 | Sponge network | 1.063 | 0 | 0.175 | 0.12559 | 0.496 |
| 8833 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | ABCA10 | Sponge network | 0.194 | 0.64278 | -0.571 | 0.03674 | 0.496 |
| 8834 | SNHG12 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p;hsa-miR-664a-5p | 17 | CBFB | Sponge network | 1.103 | 0 | 1.061 | 0 | 0.496 |
| 8835 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | KCNA6 | Sponge network | 0.194 | 0.64278 | -1.531 | 0.00013 | 0.496 |
| 8836 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | NEDD4 | Sponge network | -0.129 | 0.57177 | 0.02 | 0.89834 | 0.496 |
| 8837 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940;hsa-miR-96-5p | 24 | CRTC3 | Sponge network | 0.101 | 0.60919 | 0.164 | 0.16786 | 0.496 |
| 8838 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 33 | AKAP11 | Sponge network | 0.75 | 0.00054 | 0.196 | 0.09825 | 0.496 |
| 8839 | JAZF1-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p | 10 | DUSP1 | Sponge network | -0.769 | 0.00035 | -1.754 | 0 | 0.496 |
| 8840 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-93-5p | 18 | PLSCR4 | Sponge network | 0.056 | 0.81195 | -0.474 | 0.03833 | 0.496 |
| 8841 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | SON | Sponge network | 0.93 | 0 | 0.168 | 0.04406 | 0.496 |
| 8842 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-374a-3p | 11 | MOB1B | Sponge network | 1.134 | 0 | 0.197 | 0.15266 | 0.496 |
| 8843 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-33a-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 15 | MCC | Sponge network | -1.255 | 0.00981 | -1.005 | 0 | 0.496 |
| 8844 | RP11-473I1.9 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 14 | GSK3B | Sponge network | 0.683 | 1.0E-5 | 0.266 | 0.00045 | 0.496 |
| 8845 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 38 | RBMS3 | Sponge network | -0.405 | 0.22013 | -0.519 | 0.03551 | 0.496 |
| 8846 | SNX29P2 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 22 | SETBP1 | Sponge network | 0.4 | 0.14611 | -1.302 | 1.0E-5 | 0.496 |
| 8847 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484 | 25 | PPM1A | Sponge network | -1.331 | 3.0E-5 | -0.623 | 0 | 0.496 |
| 8848 | AC009948.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-96-5p | 82 | LPP | Sponge network | 0.532 | 0.00505 | -0.459 | 0.04475 | 0.495 |
| 8849 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p | 28 | ZFHX3 | Sponge network | 0.214 | 0.18246 | 0.389 | 0.01363 | 0.495 |
| 8850 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 27 | SYNPO2 | Sponge network | -0.405 | 0.22013 | -2.342 | 0 | 0.495 |
| 8851 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-590-3p | 10 | PAK3 | Sponge network | -0.739 | 0.00044 | -0.628 | 0.07253 | 0.495 |
| 8852 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484 | 15 | RET | Sponge network | -1.331 | 3.0E-5 | -0.833 | 0.03517 | 0.495 |
| 8853 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 28 | ABL2 | Sponge network | 1.089 | 0 | 0.82 | 0 | 0.495 |
| 8854 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-550a-3p | 17 | EBF1 | Sponge network | -2.095 | 0.00112 | -0.384 | 0.08856 | 0.495 |
| 8855 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p | 22 | MAFB | Sponge network | 0.997 | 0.00066 | -0.023 | 0.89865 | 0.495 |
| 8856 | PCA3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 60 | PRKAA2 | Sponge network | -0.709 | 0.18168 | -2.462 | 0 | 0.495 |
| 8857 | CTC-429P9.3 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-500a-5p;hsa-miR-766-3p | 12 | ATM | Sponge network | 0.082 | 0.55397 | 0.178 | 0.21568 | 0.495 |
| 8858 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-590-5p | 15 | AKAP6 | Sponge network | 0.176 | 0.37402 | -1.586 | 3.0E-5 | 0.495 |
| 8859 | CTD-2314G24.2 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-577 | 11 | AMPH | Sponge network | 0.246 | 0.59912 | -0.916 | 5.0E-5 | 0.495 |
| 8860 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | RNF144A | Sponge network | -0.576 | 0.10121 | 0.594 | 0.0009 | 0.495 |
| 8861 | RP11-182J1.1 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-652-3p;hsa-miR-7-5p | 18 | ZBTB4 | Sponge network | -0.187 | 0.23787 | -0.739 | 0 | 0.495 |
| 8862 | RP11-254F7.2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-450b-5p;hsa-miR-454-3p | 20 | DLC1 | Sponge network | 0.176 | 0.37402 | -0.382 | 0.05038 | 0.495 |
| 8863 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | GRIN2A | Sponge network | -0.563 | 0.02545 | -2.161 | 0 | 0.495 |
| 8864 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | LRRC7 | Sponge network | -2.117 | 0.00161 | -1.341 | 0.01193 | 0.495 |
| 8865 | SNX29P2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | FNBP1 | Sponge network | 0.4 | 0.14611 | -0.969 | 4.0E-5 | 0.495 |
| 8866 | SNHG14 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | CITED2 | Sponge network | -1.111 | 0.0003 | -1.545 | 0 | 0.495 |
| 8867 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b | 15 | WWTR1 | Sponge network | -1.654 | 0.00144 | -0.345 | 0.11997 | 0.495 |
| 8868 | BCYRN1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PTPN12 | Sponge network | 0.311 | 0.10344 | 0.768 | 0 | 0.495 |
| 8869 | RAD51-AS1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-766-3p | 11 | ATM | Sponge network | 0.768 | 7.0E-5 | 0.178 | 0.21568 | 0.495 |
| 8870 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p | 10 | ADAMTSL3 | Sponge network | -0.154 | 0.53954 | -1.559 | 0 | 0.495 |
| 8871 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p | 22 | PBX1 | Sponge network | 0.176 | 0.37402 | -1.206 | 0 | 0.495 |
| 8872 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p | 16 | SSBP2 | Sponge network | -7.551 | 0 | -1.58 | 0 | 0.495 |
| 8873 | BOLA3-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-935;hsa-miR-940 | 11 | MAPK10 | Sponge network | -0.246 | 0.35034 | -1.774 | 0 | 0.495 |
| 8874 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-508-3p;hsa-miR-590-3p | 11 | RIMS1 | Sponge network | -0.576 | 0.10121 | -3.143 | 0 | 0.495 |
| 8875 | C20orf166-AS1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 12 | SORCS2 | Sponge network | -2.69 | 0.0001 | -0.223 | 0.4253 | 0.495 |
| 8876 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 69 | AFF4 | Sponge network | 0.72 | 0.0001 | -0.266 | 0.01565 | 0.495 |
| 8877 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-370-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-940 | 41 | CHD5 | Sponge network | -2.452 | 0 | -0.922 | 0.01521 | 0.495 |
| 8878 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | PLSCR4 | Sponge network | 0.199 | 0.38015 | -0.474 | 0.03833 | 0.495 |
| 8879 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 13 | NR4A3 | Sponge network | -0.976 | 0.00025 | -1.385 | 0 | 0.495 |
| 8880 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-369-3p | 12 | AFF3 | Sponge network | -0.932 | 0.00329 | -2.373 | 0 | 0.495 |
| 8881 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | -0.769 | 0.00035 | -1.09 | 0.00014 | 0.495 |
| 8882 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-5p | 23 | FAT3 | Sponge network | -1.255 | 0.00981 | -1.601 | 0.00051 | 0.495 |
| 8883 | LINC00265 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p | 11 | PHF6 | Sponge network | 1.07 | 0 | 0.785 | 0 | 0.495 |
| 8884 | MRVI1-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-450b-5p | 20 | PRDM2 | Sponge network | -1.092 | 4.0E-5 | -0.258 | 0.00721 | 0.495 |
| 8885 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p | 19 | EPHA3 | Sponge network | -0.739 | 0.00044 | 0.185 | 0.53588 | 0.495 |
| 8886 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p | 22 | FGFR1 | Sponge network | -0.942 | 0 | -0.509 | 0.03957 | 0.495 |
| 8887 | RP11-620J15.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p | 25 | IL17RD | Sponge network | -0.739 | 0.00044 | -0.019 | 0.94873 | 0.495 |
| 8888 | AP001347.6 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-942-5p | 10 | ZBTB20 | Sponge network | -1.161 | 0.00591 | -0.122 | 0.57569 | 0.495 |
| 8889 | CTC-296K1.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-582-5p;hsa-miR-625-5p | 22 | TGFBR3 | Sponge network | -2.095 | 0.00112 | -1.173 | 1.0E-5 | 0.495 |
| 8890 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p | 20 | RET | Sponge network | -0.563 | 0.02545 | -0.833 | 0.03517 | 0.495 |
| 8891 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 46 | ADARB1 | Sponge network | -2.46 | 0 | -0.335 | 0.06631 | 0.495 |
| 8892 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | SYNE1 | Sponge network | -0.777 | 0.1134 | -0.738 | 0.00316 | 0.495 |
| 8893 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 24 | RARB | Sponge network | -0.576 | 0.10121 | -0.825 | 2.0E-5 | 0.495 |
| 8894 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-629-3p | 16 | GPC6 | Sponge network | -0.469 | 0.08721 | -0.054 | 0.81465 | 0.495 |
| 8895 | ACTA2-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | GJA1 | Sponge network | 0.194 | 0.64278 | -0.485 | 0.02179 | 0.495 |
| 8896 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-708-5p;hsa-miR-98-5p | 11 | MPL | Sponge network | -2.452 | 0 | -0.624 | 0.00133 | 0.495 |
| 8897 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | ZNF208 | Sponge network | 0.225 | 0.30644 | -0.4 | 0.22381 | 0.495 |
| 8898 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-671-5p | 20 | KIT | Sponge network | -1.349 | 0.00017 | -2.184 | 0 | 0.495 |
| 8899 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 32 | LRRC7 | Sponge network | -1.111 | 0.0003 | -1.341 | 0.01193 | 0.495 |
| 8900 | MEG3 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-7-1-3p | 10 | CTGF | Sponge network | 0.249 | 0.35902 | 0.321 | 0.11682 | 0.495 |
| 8901 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p | 26 | ABL2 | Sponge network | 0.083 | 0.66405 | 0.82 | 0 | 0.495 |
| 8902 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-93-5p | 27 | LRP1B | Sponge network | -2.69 | 0.0001 | -2.163 | 0 | 0.495 |
| 8903 | AC009948.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 50 | DTNA | Sponge network | 0.532 | 0.00505 | -1.648 | 1.0E-5 | 0.495 |
| 8904 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-96-5p | 22 | CBFA2T3 | Sponge network | -1.575 | 6.0E-5 | -1.221 | 1.0E-5 | 0.495 |
| 8905 | CTD-2554C21.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-769-5p;hsa-miR-93-5p | 15 | PRUNE2 | Sponge network | -1.654 | 0.00144 | -1.733 | 0.00022 | 0.495 |
| 8906 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-93-5p | 15 | LATS2 | Sponge network | -0.611 | 0.09607 | 0.159 | 0.2304 | 0.495 |
| 8907 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 65 | CADM2 | Sponge network | -0.942 | 0 | -3.782 | 0 | 0.495 |
| 8908 | AP001347.6 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-590-3p | 25 | ASTN1 | Sponge network | -1.161 | 0.00591 | -2.389 | 2.0E-5 | 0.495 |
| 8909 | PGM5-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-550a-3p;hsa-miR-7-1-3p | 17 | SYT4 | Sponge network | -4.546 | 0 | -3.207 | 0 | 0.495 |
| 8910 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | DYNC1I1 | Sponge network | -0.487 | 0.02157 | -1.12 | 0.00107 | 0.495 |
| 8911 | CTD-3064M3.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-500a-3p | 10 | CDH19 | Sponge network | -0.469 | 0.08721 | -3.434 | 0 | 0.495 |
| 8912 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-361-5p | 14 | QKI | Sponge network | -0.309 | 0.1145 | -0.333 | 0.04111 | 0.495 |
| 8913 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | HECA | Sponge network | -0.487 | 0.02157 | -0.217 | 0.03463 | 0.495 |
| 8914 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 41 | RASSF2 | Sponge network | -0.739 | 0.0574 | -0.273 | 0.21961 | 0.495 |
| 8915 | MRVI1-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-671-5p | 10 | EHD3 | Sponge network | -1.092 | 4.0E-5 | -0.484 | 0.01747 | 0.495 |
| 8916 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-877-5p | 13 | NRK | Sponge network | -0.309 | 0.1145 | 0.656 | 0.19217 | 0.495 |
| 8917 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p | 11 | KCNA6 | Sponge network | -1.255 | 0.00981 | -1.531 | 0.00013 | 0.495 |
| 8918 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-7-5p | 10 | PDGFC | Sponge network | -0.777 | 0.16667 | -0.33 | 0.06483 | 0.495 |
| 8919 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-93-3p | 10 | RPS6KA2 | Sponge network | -2.531 | 0 | -0.57 | 0.00013 | 0.495 |
| 8920 | LINC00865 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429 | 10 | SCN5A | Sponge network | -1.249 | 7.0E-5 | -0.259 | 0.48224 | 0.495 |
| 8921 | FGF14-AS2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 13 | ITPKB | Sponge network | -1.582 | 8.0E-5 | -0.704 | 0.00106 | 0.495 |
| 8922 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p | 31 | TACC1 | Sponge network | 0.176 | 0.37402 | -0.362 | 0.05406 | 0.495 |
| 8923 | CTD-3092A11.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | CREB1 | Sponge network | 0.873 | 0 | 0.203 | 0.00537 | 0.495 |
| 8924 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 25 | ZBTB4 | Sponge network | -1.349 | 0.00017 | -0.739 | 0 | 0.495 |
| 8925 | RP11-774O3.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-93-3p | 40 | GRIK3 | Sponge network | -0.487 | 0.02157 | -3.208 | 0 | 0.494 |
| 8926 | LINC00865 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-940 | 15 | SORCS1 | Sponge network | -1.249 | 7.0E-5 | -3.492 | 0 | 0.494 |
| 8927 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LHFP | Sponge network | -2.46 | 0 | -0.167 | 0.41371 | 0.494 |
| 8928 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-378a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 23 | NTRK3 | Sponge network | -2.076 | 0.00548 | -1.77 | 3.0E-5 | 0.494 |
| 8929 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 36 | DIP2C | Sponge network | -0.612 | 0.00094 | -0.436 | 0.01713 | 0.494 |
| 8930 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p | 24 | ADARB1 | Sponge network | 0.889 | 9.0E-5 | -0.335 | 0.06631 | 0.494 |
| 8931 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 28 | EPHA3 | Sponge network | -1.349 | 0.00017 | 0.185 | 0.53588 | 0.494 |
| 8932 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-625-5p | 18 | RSPO2 | Sponge network | -2.095 | 0.00112 | -3.038 | 0 | 0.494 |
| 8933 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-98-5p | 32 | DMD | Sponge network | -0.154 | 0.78939 | -1.506 | 0 | 0.494 |
| 8934 | NR2F1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p | 10 | MSN | Sponge network | -0.49 | 0.04946 | -0.023 | 0.88422 | 0.494 |
| 8935 | RP11-819C21.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | 0.537 | 0.00139 | 0.524 | 0.00017 | 0.494 |
| 8936 | CKMT2-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 13 | MACF1 | Sponge network | 0.082 | 0.55654 | 0.019 | 0.90335 | 0.494 |
| 8937 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 22 | EPHA3 | Sponge network | -0.513 | 0.04703 | 0.185 | 0.53588 | 0.494 |
| 8938 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-450b-5p | 12 | DKK3 | Sponge network | 0.176 | 0.37402 | -0.052 | 0.77783 | 0.494 |
| 8939 | LINC00472 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-550a-3p | 13 | PAK3 | Sponge network | -0.842 | 0.01361 | -0.628 | 0.07253 | 0.494 |
| 8940 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | ZMYM2 | Sponge network | 1.308 | 0 | 0.279 | 0.01273 | 0.494 |
| 8941 | RP1-59D14.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-3614-5p | 11 | TSC1 | Sponge network | 1.162 | 5.0E-5 | 0.103 | 0.26071 | 0.494 |
| 8942 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 27 | PTPN14 | Sponge network | -0.487 | 0.02157 | 0.144 | 0.34595 | 0.494 |
| 8943 | RP11-999E24.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p | 14 | PRRX1 | Sponge network | -0.693 | 0.05629 | 1.316 | 0 | 0.494 |
| 8944 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 28 | SETBP1 | Sponge network | 0.246 | 0.59912 | -1.302 | 1.0E-5 | 0.494 |
| 8945 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 23 | TRIM33 | Sponge network | 1.11 | 0 | 0.402 | 7.0E-5 | 0.494 |
| 8946 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 49 | SOX5 | Sponge network | -0.737 | 0.06182 | -1.091 | 4.0E-5 | 0.494 |
| 8947 | PWAR6 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-590-3p | 18 | ROBO1 | Sponge network | -1.575 | 6.0E-5 | -0.009 | 0.96154 | 0.494 |
| 8948 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-93-3p | 17 | PTPN11 | Sponge network | 0.401 | 0.00066 | 0.431 | 2.0E-5 | 0.494 |
| 8949 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 26 | PGR | Sponge network | -0.222 | 0.32445 | -1.704 | 0 | 0.494 |
| 8950 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 43 | NOTCH2 | Sponge network | -0.576 | 0.10121 | 0.138 | 0.35163 | 0.494 |
| 8951 | LINC00473 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | SYT4 | Sponge network | -1.203 | 0.03881 | -3.207 | 0 | 0.494 |
| 8952 | CTD-3064M3.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p | 25 | IGFBP5 | Sponge network | -0.469 | 0.08721 | -0.806 | 0.00105 | 0.494 |
| 8953 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 29 | SYNPO2 | Sponge network | -0.942 | 0 | -2.342 | 0 | 0.494 |
| 8954 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 21 | CHD2 | Sponge network | -0.222 | 0.32445 | 0.049 | 0.55371 | 0.494 |
| 8955 | RP11-620J15.3 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p | 11 | PRUNE2 | Sponge network | -0.739 | 0.00044 | -1.733 | 0.00022 | 0.494 |
| 8956 | RP11-798M19.6 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | SFRP1 | Sponge network | -0.612 | 0.00094 | -3.566 | 0 | 0.494 |
| 8957 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 22 | NRK | Sponge network | -0.323 | 0.01372 | 0.656 | 0.19217 | 0.494 |
| 8958 | PART1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-935;hsa-miR-940;hsa-miR-99b-3p | 54 | GRIK3 | Sponge network | -2.925 | 0 | -3.208 | 0 | 0.494 |
| 8959 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | -2.117 | 0.00161 | -0.7 | 1.0E-5 | 0.494 |
| 8960 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | SOX5 | Sponge network | -0.769 | 0.00035 | -1.091 | 4.0E-5 | 0.494 |
| 8961 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LRRK2 | Sponge network | -2.117 | 0.00161 | -1.136 | 1.0E-5 | 0.494 |
| 8962 | FGF14-AS2 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-629-3p | 10 | CDH23 | Sponge network | -1.582 | 8.0E-5 | -0.974 | 0.00034 | 0.494 |
| 8963 | PCA3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | KCTD8 | Sponge network | -0.709 | 0.18168 | -3.359 | 0 | 0.494 |
| 8964 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 24 | DLC1 | Sponge network | -1.057 | 0.00029 | -0.382 | 0.05038 | 0.494 |
| 8965 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | AHNAK | Sponge network | -0.576 | 0.10121 | -1.207 | 0 | 0.494 |
| 8966 | ACTA2-AS1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p | 10 | SORCS2 | Sponge network | 0.194 | 0.64278 | -0.223 | 0.4253 | 0.494 |
| 8967 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | MGA | Sponge network | 0.421 | 0.00084 | 0.323 | 0.00792 | 0.494 |
| 8968 | FGD5-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PIK3CA | Sponge network | 0.401 | 0.00066 | 0.195 | 0.06908 | 0.494 |
| 8969 | LINC00092 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-590-3p | 15 | GRIA3 | Sponge network | -1.886 | 0 | -1.956 | 0 | 0.494 |
| 8970 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 14 | GLI3 | Sponge network | -0.612 | 0.00094 | -0.615 | 0.01482 | 0.494 |
| 8971 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-421;hsa-miR-429;hsa-miR-92a-3p | 10 | ZNF521 | Sponge network | -0.563 | 0.02545 | -0.111 | 0.58068 | 0.494 |
| 8972 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 16 | SPG20 | Sponge network | -1.057 | 0.00029 | -1.16 | 0 | 0.494 |
| 8973 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 22 | LRRC4 | Sponge network | -1.111 | 0.0003 | -1.774 | 0 | 0.494 |
| 8974 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | PTGFR | Sponge network | -1.582 | 8.0E-5 | -0.233 | 0.45421 | 0.494 |
| 8975 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421 | 12 | CNTNAP3 | Sponge network | -4.546 | 0 | -1.465 | 0.00033 | 0.494 |
| 8976 | H19 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 19 | ABL2 | Sponge network | 2.439 | 0 | 0.82 | 0 | 0.494 |
| 8977 | DNM3OS |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | INPP4A | Sponge network | 0.572 | 0.01722 | 0.07 | 0.53563 | 0.494 |
| 8978 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-671-5p | 16 | CADM3 | Sponge network | -0.793 | 7.0E-5 | -3.663 | 0 | 0.494 |
| 8979 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | PIK3R1 | Sponge network | -0.358 | 0.17255 | -0.133 | 0.36854 | 0.494 |
| 8980 | LINC00641 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | CREB1 | Sponge network | -0.222 | 0.32445 | 0.203 | 0.00537 | 0.494 |
| 8981 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 53 | FAT3 | Sponge network | -0.222 | 0.32445 | -1.601 | 0.00051 | 0.494 |
| 8982 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-664a-3p | 22 | GREM1 | Sponge network | -1.281 | 0.00012 | 0.902 | 0.01691 | 0.494 |
| 8983 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | -2.69 | 0.0001 | -0.974 | 0.00034 | 0.494 |
| 8984 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PDS5B | Sponge network | 0.083 | 0.66405 | 0.259 | 0.00962 | 0.494 |
| 8985 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p | 18 | SYNE1 | Sponge network | -0.693 | 0.05629 | -0.738 | 0.00316 | 0.494 |
| 8986 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-7-5p | 10 | PTGFR | Sponge network | -0.693 | 0.05629 | -0.233 | 0.45421 | 0.494 |
| 8987 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p | 13 | DCLK1 | Sponge network | -0.405 | 0.22013 | -1.324 | 0.00014 | 0.494 |
| 8988 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3613-5p;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 44 | NTRK3 | Sponge network | -0.318 | 0.65847 | -1.77 | 3.0E-5 | 0.494 |
| 8989 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p | 30 | ZBTB4 | Sponge network | -0.222 | 0.32445 | -0.739 | 0 | 0.494 |
| 8990 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 27 | JAZF1 | Sponge network | -0.611 | 0.09607 | -0.377 | 0.02677 | 0.494 |
| 8991 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 29 | AKAP11 | Sponge network | 0.385 | 0.10067 | 0.196 | 0.09825 | 0.494 |
| 8992 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-361-5p | 21 | IL6ST | Sponge network | -0.309 | 0.1145 | -0.402 | 0.00676 | 0.494 |
| 8993 | WDFY3-AS2 |
hsa-let-7f-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 10 | ROR2 | Sponge network | -0.942 | 0 | -1.091 | 0.0008 | 0.494 |
| 8994 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-365a-3p;hsa-miR-590-3p | 17 | RICTOR | Sponge network | 1.148 | 2.0E-5 | 0.388 | 0.00188 | 0.494 |
| 8995 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-96-5p | 20 | NRXN1 | Sponge network | -1.654 | 0.00144 | -3.778 | 0 | 0.494 |
| 8996 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-577;hsa-miR-590-3p | 14 | DNAJB4 | Sponge network | -0.096 | 0.67958 | -0.7 | 1.0E-5 | 0.493 |
| 8997 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | RCAN2 | Sponge network | -0.031 | 0.89063 | -0.33 | 0.15172 | 0.493 |
| 8998 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | EBF3 | Sponge network | -0.563 | 0.02545 | -0.058 | 0.81437 | 0.493 |
| 8999 | RP11-57H14.4 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 12 | TAF1 | Sponge network | 0.083 | 0.66405 | 0.39 | 3.0E-5 | 0.493 |
| 9000 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429 | 18 | CBFA2T3 | Sponge network | -0.469 | 0.08721 | -1.221 | 1.0E-5 | 0.493 |
| 9001 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | ZNF382 | Sponge network | -2.46 | 0 | -0.494 | 0.03831 | 0.493 |
| 9002 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 29 | ZFHX3 | Sponge network | 0.41 | 0.02214 | 0.389 | 0.01363 | 0.493 |
| 9003 | LINC00641 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | PIK3CA | Sponge network | -0.222 | 0.32445 | 0.195 | 0.06908 | 0.493 |
| 9004 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | ZMYM2 | Sponge network | 1.106 | 0 | 0.279 | 0.01273 | 0.493 |
| 9005 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 45 | SASH1 | Sponge network | -0.129 | 0.57177 | -0.502 | 0.00175 | 0.493 |
| 9006 | FAM157B |
hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-3614-5p | 17 | RICTOR | Sponge network | 0.92 | 0 | 0.388 | 0.00188 | 0.493 |
| 9007 | HCG18 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 36 | CREB1 | Sponge network | 1.063 | 0 | 0.203 | 0.00537 | 0.493 |
| 9008 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 25 | LRRC55 | Sponge network | -0.739 | 0.0574 | -0.026 | 0.93163 | 0.493 |
| 9009 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | MYO9A | Sponge network | -0.405 | 0.22013 | -0.147 | 0.32373 | 0.493 |
| 9010 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-935 | 31 | DIP2C | Sponge network | -0.738 | 0.21233 | -0.436 | 0.01713 | 0.493 |
| 9011 | RP11-166D19.1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-942-5p | 12 | BMPR1A | Sponge network | -0.576 | 0.10121 | -0.366 | 0.01036 | 0.493 |
| 9012 | GNG12-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-940 | 10 | PHOX2B | Sponge network | -0.793 | 7.0E-5 | -2.68 | 0 | 0.493 |
| 9013 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 16 | PGR | Sponge network | -0.737 | 0.10822 | -1.704 | 0 | 0.493 |
| 9014 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 29 | SETBP1 | Sponge network | 0.101 | 0.60919 | -1.302 | 1.0E-5 | 0.493 |
| 9015 | TP73-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-671-5p | 13 | EHD3 | Sponge network | -0.563 | 0.02545 | -0.484 | 0.01747 | 0.493 |
| 9016 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-590-3p | 15 | RICTOR | Sponge network | 1.055 | 0 | 0.388 | 0.00188 | 0.493 |
| 9017 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | ABCC9 | Sponge network | -2.915 | 0.00015 | -0.466 | 0.14121 | 0.493 |
| 9018 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | PLSCR4 | Sponge network | 0.889 | 9.0E-5 | -0.474 | 0.03833 | 0.493 |
| 9019 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 11 | FYN | Sponge network | -0.465 | 0.04703 | -0.131 | 0.37051 | 0.493 |
| 9020 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 23 | TBL1XR1 | Sponge network | 0.93 | 0 | 0.578 | 0 | 0.493 |
| 9021 | ZRANB2-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | NRK | Sponge network | -0.376 | 0.00337 | 0.656 | 0.19217 | 0.493 |
| 9022 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 20 | STARD13 | Sponge network | 0.997 | 0.00066 | -0.187 | 0.27383 | 0.493 |
| 9023 | DNM3OS |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-744-3p | 26 | PTPN14 | Sponge network | 0.572 | 0.01722 | 0.144 | 0.34595 | 0.493 |
| 9024 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p | 28 | RASSF8 | Sponge network | -0.154 | 0.78939 | -0.122 | 0.65591 | 0.493 |
| 9025 | FENDRR |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 10 | LHFP | Sponge network | -1.281 | 0.00012 | -0.167 | 0.41371 | 0.493 |
| 9026 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-511-5p | 10 | LRRC4 | Sponge network | -0.842 | 0.01361 | -1.774 | 0 | 0.493 |
| 9027 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | CAV1 | Sponge network | -0.942 | 0 | -1.013 | 6.0E-5 | 0.493 |
| 9028 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 36 | QKI | Sponge network | 0.889 | 9.0E-5 | -0.333 | 0.04111 | 0.493 |
| 9029 | LINC01001 |
hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 14 | ABL2 | Sponge network | 1.055 | 0 | 0.82 | 0 | 0.493 |
| 9030 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 31 | PTPRD | Sponge network | -0.358 | 0.17255 | -1.005 | 0.00064 | 0.493 |
| 9031 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-589-3p | 20 | PCDH9 | Sponge network | -0.777 | 0.16667 | -1.823 | 4.0E-5 | 0.493 |
| 9032 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 44 | SASH1 | Sponge network | -2.452 | 0 | -0.502 | 0.00175 | 0.493 |
| 9033 | RP5-1198O20.4 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CTGF | Sponge network | 0.997 | 0.00066 | 0.321 | 0.11682 | 0.493 |
| 9034 | LINC00865 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-769-5p | 21 | SYNPO2 | Sponge network | -1.249 | 7.0E-5 | -2.342 | 0 | 0.493 |
| 9035 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | RB1CC1 | Sponge network | 1.317 | 0 | 0.175 | 0.12559 | 0.493 |
| 9036 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | PSIP1 | Sponge network | -0.222 | 0.32445 | -0.005 | 0.96897 | 0.493 |
| 9037 | PWAR6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | ABI2 | Sponge network | -1.575 | 6.0E-5 | 0.175 | 0.14787 | 0.493 |
| 9038 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | PCDH17 | Sponge network | 0.889 | 9.0E-5 | 0.731 | 2.0E-5 | 0.493 |
| 9039 | PART1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | RNF180 | Sponge network | -2.925 | 0 | -1.127 | 1.0E-5 | 0.493 |
| 9040 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | ZFHX3 | Sponge network | 0.222 | 0.29133 | 0.389 | 0.01363 | 0.493 |
| 9041 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | FGFR1 | Sponge network | -2.117 | 0.00161 | -0.509 | 0.03957 | 0.493 |
| 9042 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-5p | 15 | ESR1 | Sponge network | -0.351 | 0.11358 | -1.035 | 3.0E-5 | 0.493 |
| 9043 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 55 | CHD5 | Sponge network | -1.111 | 0.0003 | -0.922 | 0.01521 | 0.493 |
| 9044 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | PLSCR4 | Sponge network | 0.246 | 0.59912 | -0.474 | 0.03833 | 0.493 |
| 9045 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 22 | LATS1 | Sponge network | 0.401 | 0.00066 | 0.109 | 0.34208 | 0.493 |
| 9046 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 17 | TRIM33 | Sponge network | 1.317 | 0 | 0.402 | 7.0E-5 | 0.493 |
| 9047 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 44 | CHD5 | Sponge network | -0.976 | 0.00025 | -0.922 | 0.01521 | 0.493 |
| 9048 | RP11-13K12.1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484 | 12 | FLNA | Sponge network | -0.154 | 0.78939 | -0.771 | 0.01377 | 0.493 |
| 9049 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-423-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 44 | NTRK3 | Sponge network | -0.154 | 0.53954 | -1.77 | 3.0E-5 | 0.493 |
| 9050 | LINC00957 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | -0.421 | 0.0114 | 0.809 | 0.00544 | 0.493 |
| 9051 | PCA3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ABCA10 | Sponge network | -0.709 | 0.18168 | -0.571 | 0.03674 | 0.493 |
| 9052 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p | 23 | ZMYM2 | Sponge network | 1.147 | 0 | 0.279 | 0.01273 | 0.493 |
| 9053 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | BCL2 | Sponge network | -0.421 | 0.0114 | -0.899 | 0 | 0.493 |
| 9054 | HAND2-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-942-5p | 12 | BMPR1A | Sponge network | -1.697 | 0.00889 | -0.366 | 0.01036 | 0.493 |
| 9055 | BVES-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-744-3p | 47 | DST | Sponge network | -2.62 | 0 | -0.713 | 0.00096 | 0.493 |
| 9056 | PART1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 22 | SORCS1 | Sponge network | -2.925 | 0 | -3.492 | 0 | 0.493 |
| 9057 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-877-5p | 44 | AR | Sponge network | -0.773 | 0.04073 | -1.554 | 0 | 0.493 |
| 9058 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 13 | LRRK2 | Sponge network | 0.572 | 0.01722 | -1.136 | 1.0E-5 | 0.493 |
| 9059 | BVES-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-877-5p | 23 | NRK | Sponge network | -2.62 | 0 | 0.656 | 0.19217 | 0.493 |
| 9060 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | EBF1 | Sponge network | -0.769 | 0.00035 | -0.384 | 0.08856 | 0.493 |
| 9061 | SNHG14 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SEMA3E | Sponge network | -1.111 | 0.0003 | -1.717 | 0.00137 | 0.493 |
| 9062 | RP11-359B12.2 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | STAT5B | Sponge network | 0.064 | 0.72934 | -0.182 | 0.1082 | 0.493 |
| 9063 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | RET | Sponge network | -2.452 | 0 | -0.833 | 0.03517 | 0.493 |
| 9064 | RASSF8-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p | 16 | FMNL3 | Sponge network | 0.335 | 0.20596 | 0.555 | 4.0E-5 | 0.493 |
| 9065 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | ESR1 | Sponge network | -0.376 | 0.00337 | -1.035 | 3.0E-5 | 0.493 |
| 9066 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | EBF3 | Sponge network | -1.203 | 0.03881 | -0.058 | 0.81437 | 0.493 |
| 9067 | ZNF667-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-877-5p;hsa-miR-942-5p | 15 | GLIPR1 | Sponge network | -0.976 | 0.00025 | 0.146 | 0.3276 | 0.493 |
| 9068 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-429;hsa-miR-7-5p | 10 | GLI3 | Sponge network | -0.469 | 0.08721 | -0.615 | 0.01482 | 0.493 |
| 9069 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 10 | LRRC7 | Sponge network | -0.583 | 0.14589 | -1.341 | 0.01193 | 0.493 |
| 9070 | RP11-400K9.3 |
hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-3p | 22 | IL6ST | Sponge network | -0.733 | 0.19469 | -0.402 | 0.00676 | 0.493 |
| 9071 | LINC00641 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 23 | UBE4B | Sponge network | -0.222 | 0.32445 | -0.235 | 0.007 | 0.493 |
| 9072 | DNM3OS |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p | 18 | MAFB | Sponge network | 0.572 | 0.01722 | -0.023 | 0.89865 | 0.493 |
| 9073 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | -1.161 | 0.00591 | 0.809 | 0.00544 | 0.493 |
| 9074 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | IGSF10 | Sponge network | 0.335 | 0.20596 | -1.751 | 1.0E-5 | 0.493 |
| 9075 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 54 | BMPR2 | Sponge network | 0.385 | 0.10067 | 0.206 | 0.0635 | 0.493 |
| 9076 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-7-1-3p | 38 | IL6ST | Sponge network | -4.546 | 0 | -0.402 | 0.00676 | 0.493 |
| 9077 | TP73-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 28 | SYT4 | Sponge network | -0.563 | 0.02545 | -3.207 | 0 | 0.493 |
| 9078 | GAS6-AS2 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | FLNA | Sponge network | 0.16 | 0.50785 | -0.771 | 0.01377 | 0.493 |
| 9079 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 23 | ANK2 | Sponge network | 0.225 | 0.30644 | -1.603 | 1.0E-5 | 0.493 |
| 9080 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b | 10 | SLITRK3 | Sponge network | -0.517 | 0.02935 | -3.328 | 0 | 0.492 |
| 9081 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-576-5p | 17 | GREM1 | Sponge network | -2.62 | 0 | 0.902 | 0.01691 | 0.492 |
| 9082 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 25 | GRIN2A | Sponge network | 0.194 | 0.64278 | -2.161 | 0 | 0.492 |
| 9083 | TRAF3IP2-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 18 | CELF4 | Sponge network | -0.323 | 0.01372 | -1.135 | 0.00344 | 0.492 |
| 9084 | SNHG14 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 41 | RIMS2 | Sponge network | -1.111 | 0.0003 | -1.365 | 0.00208 | 0.492 |
| 9085 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-502-5p;hsa-miR-877-5p | 12 | IL17RD | Sponge network | -0.309 | 0.1145 | -0.019 | 0.94873 | 0.492 |
| 9086 | RP11-999E24.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 14 | ASTN1 | Sponge network | -0.693 | 0.05629 | -2.389 | 2.0E-5 | 0.492 |
| 9087 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-589-3p;hsa-miR-590-3p | 16 | TET1 | Sponge network | -0.358 | 0.17255 | -0.135 | 0.52138 | 0.492 |
| 9088 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-628-5p;hsa-miR-940 | 18 | TRIM33 | Sponge network | 0.637 | 0.00069 | 0.402 | 7.0E-5 | 0.492 |
| 9089 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 45 | DTNA | Sponge network | -0.342 | 0.02288 | -1.648 | 1.0E-5 | 0.492 |
| 9090 | AC006116.24 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-539-5p | 11 | PRKCB | Sponge network | -0.473 | 0.07602 | -1.313 | 2.0E-5 | 0.492 |
| 9091 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 28 | ABI2 | Sponge network | 0.299 | 0.57722 | 0.175 | 0.14787 | 0.492 |
| 9092 | AC003090.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | RELN | Sponge network | -2.117 | 0.00161 | -2.403 | 0 | 0.492 |
| 9093 | CTD-3110H11.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-150-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 15 | MDM4 | Sponge network | 1.735 | 0 | 0.345 | 0.00762 | 0.492 |
| 9094 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-577 | 11 | NALCN | Sponge network | -1.439 | 4.0E-5 | 1.068 | 0.00237 | 0.492 |
| 9095 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | EDA2R | Sponge network | -0.737 | 0.06182 | -0.587 | 0.01598 | 0.492 |
| 9096 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 35 | TSHZ3 | Sponge network | -1.203 | 0.03881 | -0.556 | 0.01516 | 0.492 |
| 9097 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-550a-3p | 11 | PAK3 | Sponge network | -2.531 | 0 | -0.628 | 0.07253 | 0.492 |
| 9098 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 36 | CBL | Sponge network | 0.72 | 0.0001 | 0.291 | 0.01267 | 0.492 |
| 9099 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-93-5p | 27 | PIK3R1 | Sponge network | 0.082 | 0.55397 | -0.133 | 0.36854 | 0.492 |
| 9100 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | -0.709 | 0.18168 | -1.465 | 0.00033 | 0.492 |
| 9101 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 20 | MLLT10 | Sponge network | 0.594 | 0.00064 | 0.105 | 0.33292 | 0.492 |
| 9102 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 14 | KCNAB1 | Sponge network | -0.323 | 0.01372 | -0.755 | 2.0E-5 | 0.492 |
| 9103 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p | 24 | KIT | Sponge network | -2.46 | 0 | -2.184 | 0 | 0.492 |
| 9104 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-576-5p | 17 | GREM1 | Sponge network | 0.395 | 0.15451 | 0.902 | 0.01691 | 0.492 |
| 9105 | TP73-AS1 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 10 | DIRAS1 | Sponge network | -0.563 | 0.02545 | -2.518 | 0 | 0.492 |
| 9106 | RP11-967K21.1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | FLNA | Sponge network | 0.056 | 0.81195 | -0.771 | 0.01377 | 0.492 |
| 9107 | RP11-389C8.2 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-335-5p;hsa-miR-429 | 14 | PLXNC1 | Sponge network | 0.727 | 3.0E-5 | 0.569 | 0.00329 | 0.492 |
| 9108 | LINC00702 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 11 | PEA15 | Sponge network | -0.737 | 0.06182 | 0.009 | 0.93318 | 0.492 |
| 9109 | FAM13A-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-940 | 51 | NFIB | Sponge network | -0.83 | 0 | 0.023 | 0.90795 | 0.492 |
| 9110 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | ZNF521 | Sponge network | -0.323 | 0.01372 | -0.111 | 0.58068 | 0.492 |
| 9111 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PTPN11 | Sponge network | 1.205 | 0 | 0.431 | 2.0E-5 | 0.492 |
| 9112 | LINC00641 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | INPP4A | Sponge network | -0.222 | 0.32445 | 0.07 | 0.53563 | 0.492 |
| 9113 | FENDRR |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 12 | HMCN1 | Sponge network | -1.281 | 0.00012 | 0.809 | 0.00544 | 0.492 |
| 9114 | RP11-326G21.1 |
hsa-let-7b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-590-3p | 15 | NRK | Sponge network | -0.021 | 0.93598 | 0.656 | 0.19217 | 0.492 |
| 9115 | CTA-204B4.2 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-766-3p | 10 | ATM | Sponge network | 0.765 | 7.0E-5 | 0.178 | 0.21568 | 0.492 |
| 9116 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-942-5p | 23 | ARHGAP28 | Sponge network | -1.203 | 0.03881 | -0.911 | 0.00013 | 0.492 |
| 9117 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-576-5p | 21 | ERC1 | Sponge network | 0.614 | 1.0E-5 | -0.108 | 0.38794 | 0.492 |
| 9118 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 23 | LPP | Sponge network | -2.039 | 0.03447 | -0.459 | 0.04475 | 0.492 |
| 9119 | LINC00473 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-5p | 19 | PTGFR | Sponge network | -1.203 | 0.03881 | -0.233 | 0.45421 | 0.492 |
| 9120 | CTC-550B14.7 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374a-3p;hsa-miR-766-3p | 11 | MDM4 | Sponge network | 1.161 | 0 | 0.345 | 0.00762 | 0.492 |
| 9121 | CTC-444N24.11 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 17 | KIF5B | Sponge network | 1.153 | 0 | 0.362 | 0.00066 | 0.492 |
| 9122 | RP11-464F9.1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-654-3p | 17 | PHF6 | Sponge network | 0.959 | 0 | 0.785 | 0 | 0.492 |
| 9123 | LINC00641 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 28 | RYR2 | Sponge network | -0.222 | 0.32445 | -1.466 | 0.00031 | 0.492 |
| 9124 | NR2F1-AS1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | SPOP | Sponge network | -0.49 | 0.04946 | -0.419 | 1.0E-5 | 0.492 |
| 9125 | HAND2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 10 | MSN | Sponge network | -1.697 | 0.00889 | -0.023 | 0.88422 | 0.492 |
| 9126 | RP11-589P10.7 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-93-3p | 15 | RUNX1T1 | Sponge network | -2.044 | 0.00066 | -0.344 | 0.2374 | 0.492 |
| 9127 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MLLT10 | Sponge network | 0.873 | 0 | 0.105 | 0.33292 | 0.492 |
| 9128 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 33 | EBF1 | Sponge network | -0.777 | 0.1134 | -0.384 | 0.08856 | 0.492 |
| 9129 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | TIMP3 | Sponge network | -0.901 | 0.00019 | -0.546 | 0.02307 | 0.492 |
| 9130 | SERTAD4-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-505-5p | 14 | ASTN1 | Sponge network | -0.932 | 0.00329 | -2.389 | 2.0E-5 | 0.492 |
| 9131 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p | 11 | JAKMIP2 | Sponge network | 0.214 | 0.18246 | -1.388 | 0 | 0.492 |
| 9132 | WDFY3-AS2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 19 | SYNM | Sponge network | -0.942 | 0 | -2.309 | 1.0E-5 | 0.492 |
| 9133 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p | 19 | HIP1 | Sponge network | 0.176 | 0.37402 | 0.774 | 0 | 0.492 |
| 9134 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | ANK2 | Sponge network | -0.246 | 0.35034 | -1.603 | 1.0E-5 | 0.492 |
| 9135 | FENDRR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-96-5p | 24 | CBFA2T3 | Sponge network | -1.281 | 0.00012 | -1.221 | 1.0E-5 | 0.492 |
| 9136 | ARHGEF26-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 20 | KLF10 | Sponge network | -2.46 | 0 | -0.783 | 0 | 0.492 |
| 9137 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-652-3p;hsa-miR-7-5p | 27 | ZBTB4 | Sponge network | 0.214 | 0.18246 | -0.739 | 0 | 0.492 |
| 9138 | RP11-890B15.3 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-484;hsa-miR-7-5p | 11 | FLNA | Sponge network | 0.101 | 0.60919 | -0.771 | 0.01377 | 0.492 |
| 9139 | CTD-2528L19.6 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | FBXO11 | Sponge network | 0.953 | 0 | 0.142 | 0.04938 | 0.492 |
| 9140 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p | 14 | ZNF770 | Sponge network | 0.93 | 0 | 0.206 | 0.06751 | 0.492 |
| 9141 | PART1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-708-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 50 | HLF | Sponge network | -2.925 | 0 | -2.443 | 0 | 0.492 |
| 9142 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-505-3p | 11 | KIT | Sponge network | -4.477 | 0 | -2.184 | 0 | 0.492 |
| 9143 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | MLLT10 | Sponge network | 0.782 | 0.00016 | 0.105 | 0.33292 | 0.492 |
| 9144 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-495-3p | 18 | CLIP1 | Sponge network | -0.162 | 0.47687 | -0.215 | 0.08684 | 0.492 |
| 9145 | RASSF8-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-877-5p;hsa-miR-93-5p | 32 | IL17RD | Sponge network | 0.335 | 0.20596 | -0.019 | 0.94873 | 0.492 |
| 9146 | LINC00092 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 24 | PRKAA2 | Sponge network | -1.886 | 0 | -2.462 | 0 | 0.492 |
| 9147 | MIR143HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 51 | NTRK2 | Sponge network | -0.739 | 0.0574 | -0.736 | 0.06495 | 0.492 |
| 9148 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 13 | NCAM2 | Sponge network | -0.517 | 0.02935 | -0.482 | 0.22565 | 0.492 |
| 9149 | USP3-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 23 | CTIF | Sponge network | -0.162 | 0.47687 | -0.389 | 0.00549 | 0.492 |
| 9150 | MIR143HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | -0.739 | 0.0574 | -0.001 | 0.99669 | 0.492 |
| 9151 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-409-3p;hsa-miR-576-5p | 23 | PHC3 | Sponge network | 0.594 | 0.00064 | 0.212 | 0.0499 | 0.492 |
| 9152 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | NR4A3 | Sponge network | -0.129 | 0.57177 | -1.385 | 0 | 0.492 |
| 9153 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-96-5p | 26 | SCN9A | Sponge network | -0.563 | 0.02545 | -0.525 | 0.10593 | 0.492 |
| 9154 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-590-5p | 30 | DMD | Sponge network | 0.331 | 0.57247 | -1.506 | 0 | 0.492 |
| 9155 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 51 | AR | Sponge network | 0.101 | 0.60919 | -1.554 | 0 | 0.492 |
| 9156 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 29 | ANK2 | Sponge network | 0.4 | 0.14611 | -1.603 | 1.0E-5 | 0.492 |
| 9157 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | RBMS3 | Sponge network | 0.342 | 0.03129 | -0.519 | 0.03551 | 0.492 |
| 9158 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p | 20 | AKAP6 | Sponge network | -0.738 | 0.21233 | -1.586 | 3.0E-5 | 0.492 |
| 9159 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 50 | SSBP2 | Sponge network | -0.899 | 6.0E-5 | -1.58 | 0 | 0.492 |
| 9160 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 31 | TACC1 | Sponge network | -0.896 | 0.00099 | -0.362 | 0.05406 | 0.492 |
| 9161 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 28 | BCL6 | Sponge network | -1.111 | 0.0003 | -0.211 | 0.19489 | 0.492 |
| 9162 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 42 | CBL | Sponge network | 0.421 | 0.00084 | 0.291 | 0.01267 | 0.492 |
| 9163 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 14 | CEP170 | Sponge network | 0.537 | 0.00139 | 0.943 | 0 | 0.492 |
| 9164 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 50 | NTRK2 | Sponge network | -2.452 | 0 | -0.736 | 0.06495 | 0.492 |
| 9165 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-769-5p | 26 | SYNPO2 | Sponge network | 0.101 | 0.60919 | -2.342 | 0 | 0.492 |
| 9166 | LINC01018 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 54 | RUNX1T1 | Sponge network | -2.915 | 0.00015 | -0.344 | 0.2374 | 0.492 |
| 9167 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-582-5p | 31 | CADM2 | Sponge network | -0.469 | 0.08721 | -3.782 | 0 | 0.492 |
| 9168 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 60 | IL6ST | Sponge network | -2.117 | 0.00161 | -0.402 | 0.00676 | 0.492 |
| 9169 | MIR497HG |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | LRIG1 | Sponge network | -1.439 | 4.0E-5 | -0.537 | 0.00588 | 0.492 |
| 9170 | ACTA2-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | HERC1 | Sponge network | 0.194 | 0.64278 | -0.196 | 0.08544 | 0.492 |
| 9171 | RP11-894P9.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-940 | 19 | AKAP11 | Sponge network | 0.993 | 0 | 0.196 | 0.09825 | 0.492 |
| 9172 | SERTAD4-AS1 |
hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-424-5p | 10 | RPS6KA6 | Sponge network | -0.932 | 0.00329 | -2.989 | 0 | 0.492 |
| 9173 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-7-5p | 25 | KCNA6 | Sponge network | -1.654 | 0.00144 | -1.531 | 0.00013 | 0.492 |
| 9174 | LINC00657 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 0.91 | 0 | 0.321 | 0.00267 | 0.492 |
| 9175 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p | 32 | PBX1 | Sponge network | -1.203 | 0.03881 | -1.206 | 0 | 0.492 |
| 9176 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DCN | Sponge network | 0.194 | 0.64278 | -1.288 | 0 | 0.492 |
| 9177 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 22 | CRTC3 | Sponge network | 0.064 | 0.72934 | 0.164 | 0.16786 | 0.491 |
| 9178 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | FAT4 | Sponge network | -0.323 | 0.01372 | -0.354 | 0.14694 | 0.491 |
| 9179 | AC009120.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 18 | ZFHX3 | Sponge network | 0.919 | 0 | 0.389 | 0.01363 | 0.491 |
| 9180 | WDFY3-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-96-5p | 40 | IL17RD | Sponge network | -0.942 | 0 | -0.019 | 0.94873 | 0.491 |
| 9181 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 12 | TMEFF2 | Sponge network | -0.563 | 0.02545 | -2.426 | 0 | 0.491 |
| 9182 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 13 | PARK2 | Sponge network | -0.222 | 0.32445 | -1.892 | 0 | 0.491 |
| 9183 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | CDHR3 | Sponge network | -0.793 | 7.0E-5 | -0.977 | 4.0E-5 | 0.491 |
| 9184 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-940 | 15 | HOOK3 | Sponge network | -0.162 | 0.47687 | 0.273 | 0.09957 | 0.491 |
| 9185 | RP11-400K9.4 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-485-3p;hsa-miR-539-5p | 16 | RYR2 | Sponge network | -0.611 | 0.09607 | -1.466 | 0.00031 | 0.491 |
| 9186 | RP11-927P21.1 |
hsa-miR-125a-3p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p | 14 | ATM | Sponge network | 0.921 | 0.00118 | 0.178 | 0.21568 | 0.491 |
| 9187 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-3p | 39 | BNC2 | Sponge network | -0.773 | 0.04073 | -0.491 | 0.09988 | 0.491 |
| 9188 | AGAP11 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 18 | EZH1 | Sponge network | -1.645 | 0 | -0.564 | 1.0E-5 | 0.491 |
| 9189 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | QKI | Sponge network | -4.546 | 0 | -0.333 | 0.04111 | 0.491 |
| 9190 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | PLSCR4 | Sponge network | -1.886 | 0 | -0.474 | 0.03833 | 0.491 |
| 9191 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 32 | TRPS1 | Sponge network | -0.154 | 0.53954 | -0.191 | 0.44109 | 0.491 |
| 9192 | RP11-464F9.1 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | ABL2 | Sponge network | 0.959 | 0 | 0.82 | 0 | 0.491 |
| 9193 | PGM5-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 11 | TLE4 | Sponge network | -4.546 | 0 | -1.157 | 0 | 0.491 |
| 9194 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p | 41 | GAS7 | Sponge network | -0.154 | 0.78939 | -0.555 | 0.01602 | 0.491 |
| 9195 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | FGFR1 | Sponge network | -0.206 | 0.12942 | -0.509 | 0.03957 | 0.491 |
| 9196 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-93-5p | 27 | SSBP2 | Sponge network | -0.473 | 0.07602 | -1.58 | 0 | 0.491 |
| 9197 | RP1-59D14.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3614-5p;hsa-miR-424-5p | 22 | ABL2 | Sponge network | 1.162 | 5.0E-5 | 0.82 | 0 | 0.491 |
| 9198 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 23 | ADAMTSL3 | Sponge network | 0.199 | 0.38015 | -1.559 | 0 | 0.491 |
| 9199 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | ADAMTSL3 | Sponge network | 0.056 | 0.81195 | -1.559 | 0 | 0.491 |
| 9200 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-7-5p | 38 | TCF4 | Sponge network | -0.693 | 0.05629 | 0.016 | 0.92271 | 0.491 |
| 9201 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b | 15 | CLIP1 | Sponge network | -1.645 | 0 | -0.215 | 0.08684 | 0.491 |
| 9202 | RP11-284N8.3 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-96-5p | 13 | BCL2 | Sponge network | -0.31 | 0.33871 | -0.899 | 0 | 0.491 |
| 9203 | MEG3 |
hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p | 10 | THSD7B | Sponge network | 0.249 | 0.35902 | 0.154 | 0.74723 | 0.491 |
| 9204 | RP11-159D12.2 |
hsa-miR-125a-3p;hsa-miR-149-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | CDC27 | Sponge network | 1.254 | 0 | 0.384 | 0 | 0.491 |
| 9205 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 17 | HECA | Sponge network | 0.178 | 0.29106 | -0.217 | 0.03463 | 0.491 |
| 9206 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p | 14 | AKAP12 | Sponge network | 0.532 | 0.00505 | -0.932 | 0.0028 | 0.491 |
| 9207 | LINC00963 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 29 | BRD3 | Sponge network | 0.523 | 9.0E-5 | 0.37 | 0.00013 | 0.491 |
| 9208 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-374a-5p | 28 | DMD | Sponge network | -0.693 | 0.05629 | -1.506 | 0 | 0.491 |
| 9209 | BZRAP1-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 14 | SORCS1 | Sponge network | -0.465 | 0.04703 | -3.492 | 0 | 0.491 |
| 9210 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | RASSF8 | Sponge network | 0.064 | 0.72934 | -0.122 | 0.65591 | 0.491 |
| 9211 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | LRIG1 | Sponge network | -0.487 | 0.02157 | -0.537 | 0.00588 | 0.491 |
| 9212 | CTC-510F12.2 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-3614-5p | 12 | EPHA3 | Sponge network | -0.691 | 0.00146 | 0.185 | 0.53588 | 0.491 |
| 9213 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-376c-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p | 30 | PHC3 | Sponge network | 0.637 | 0.00069 | 0.212 | 0.0499 | 0.491 |
| 9214 | MYLK-AS1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-484 | 19 | RIMS2 | Sponge network | -1.331 | 3.0E-5 | -1.365 | 0.00208 | 0.491 |
| 9215 | CTC-429P9.5 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-376c-3p | 10 | SYNM | Sponge network | 0.178 | 0.29106 | -2.309 | 1.0E-5 | 0.491 |
| 9216 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 35 | MEF2C | Sponge network | -1.057 | 0.00029 | -0.499 | 0.01448 | 0.491 |
| 9217 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 31 | RASSF8 | Sponge network | -0.612 | 0.00094 | -0.122 | 0.65591 | 0.491 |
| 9218 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 20 | FOXP1 | Sponge network | 0.178 | 0.29106 | 0.042 | 0.74368 | 0.491 |
| 9219 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429 | 12 | EDA2R | Sponge network | -1.249 | 7.0E-5 | -0.587 | 0.01598 | 0.491 |
| 9220 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 48 | BCL2 | Sponge network | -0.737 | 0.06182 | -0.899 | 0 | 0.491 |
| 9221 | SH3RF3-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | MCC | Sponge network | 0.889 | 9.0E-5 | -1.005 | 0 | 0.491 |
| 9222 | AC016747.3 |
hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p | 15 | FMNL3 | Sponge network | 0.199 | 0.38015 | 0.555 | 4.0E-5 | 0.491 |
| 9223 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p | 18 | SPG20 | Sponge network | -0.405 | 0.22013 | -1.16 | 0 | 0.491 |
| 9224 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-7-1-3p | 18 | DDX6 | Sponge network | 0.545 | 0.00064 | 0.091 | 0.36428 | 0.491 |
| 9225 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-7-1-3p | 22 | ADAMTSL3 | Sponge network | 0.889 | 9.0E-5 | -1.559 | 0 | 0.491 |
| 9226 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 30 | PGR | Sponge network | 0.101 | 0.60919 | -1.704 | 0 | 0.491 |
| 9227 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-429;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-3p | 21 | EBF3 | Sponge network | -2.69 | 0.0001 | -0.058 | 0.81437 | 0.491 |
| 9228 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 25 | CTIF | Sponge network | -2.531 | 0 | -0.389 | 0.00549 | 0.491 |
| 9229 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 24 | TET1 | Sponge network | -0.976 | 0.00025 | -0.135 | 0.52138 | 0.491 |
| 9230 | TP73-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p | 11 | AMPH | Sponge network | -0.563 | 0.02545 | -0.916 | 5.0E-5 | 0.491 |
| 9231 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p | 12 | DKK3 | Sponge network | -0.777 | 0.16667 | -0.052 | 0.77783 | 0.491 |
| 9232 | CTA-363E19.2 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-625-3p | 18 | ABL2 | Sponge network | 1.055 | 0 | 0.82 | 0 | 0.491 |
| 9233 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | EPHA5 | Sponge network | -0.323 | 0.01372 | -0.957 | 0.01733 | 0.491 |
| 9234 | RP4-669P10.16 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-766-3p | 10 | EZH1 | Sponge network | -0.301 | 0.06597 | -0.564 | 1.0E-5 | 0.491 |
| 9235 | RP11-464F9.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-940 | 35 | MDM4 | Sponge network | 0.959 | 0 | 0.345 | 0.00762 | 0.491 |
| 9236 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ITGAV | Sponge network | 0.572 | 0.01722 | 0.337 | 0.01002 | 0.491 |
| 9237 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | NRXN3 | Sponge network | 0.532 | 0.00505 | -0.893 | 0.04186 | 0.491 |
| 9238 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-582-5p | 21 | SETBP1 | Sponge network | 0.056 | 0.81195 | -1.302 | 1.0E-5 | 0.491 |
| 9239 | KB-1460A1.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | BRD3 | Sponge network | 0.603 | 0.00127 | 0.37 | 0.00013 | 0.491 |
| 9240 | RP11-819C21.1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 18 | KIF5B | Sponge network | 0.537 | 0.00139 | 0.362 | 0.00066 | 0.491 |
| 9241 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | LRP1B | Sponge network | -0.739 | 0.0574 | -2.163 | 0 | 0.491 |
| 9242 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-7-1-3p | 26 | LIFR | Sponge network | -0.842 | 0.01361 | -1.794 | 0 | 0.491 |
| 9243 | MYLK-AS1 |
hsa-let-7b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-940 | 11 | NEDD4 | Sponge network | -1.331 | 3.0E-5 | 0.02 | 0.89834 | 0.491 |
| 9244 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 15 | RET | Sponge network | -0.583 | 0.14589 | -0.833 | 0.03517 | 0.491 |
| 9245 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 25 | KCNA6 | Sponge network | -0.323 | 0.01372 | -1.531 | 0.00013 | 0.491 |
| 9246 | FAM66C |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 16 | KLF10 | Sponge network | -0.901 | 0.00019 | -0.783 | 0 | 0.491 |
| 9247 | RP11-166D19.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | INPP4A | Sponge network | -0.576 | 0.10121 | 0.07 | 0.53563 | 0.491 |
| 9248 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-450b-5p | 13 | FNBP1 | Sponge network | -0.421 | 0.0114 | -0.969 | 4.0E-5 | 0.491 |
| 9249 | LINC00702 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | PCDH18 | Sponge network | -0.737 | 0.06182 | 0.216 | 0.24645 | 0.491 |
| 9250 | CTD-2002H8.2 |
hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MOB1B | Sponge network | 0.931 | 1.0E-5 | 0.197 | 0.15266 | 0.491 |
| 9251 | LINC00578 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 12 | ERG | Sponge network | 0.811 | 0.03228 | 0.002 | 0.99011 | 0.491 |
| 9252 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | LRRC7 | Sponge network | -0.612 | 0.00094 | -1.341 | 0.01193 | 0.491 |
| 9253 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-590-3p | 25 | RICTOR | Sponge network | 0.765 | 7.0E-5 | 0.388 | 0.00188 | 0.491 |
| 9254 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-5p | 29 | DMD | Sponge network | -1.349 | 0.00017 | -1.506 | 0 | 0.491 |
| 9255 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | FAM172A | Sponge network | -0.942 | 0 | -0.57 | 0 | 0.491 |
| 9256 | FAM66C |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 37 | SNX29 | Sponge network | -0.901 | 0.00019 | -0.34 | 0.01371 | 0.491 |
| 9257 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 32 | CBL | Sponge network | 0.614 | 1.0E-5 | 0.291 | 0.01267 | 0.491 |
| 9258 | LINC00476 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p | 25 | SETBP1 | Sponge network | -0.615 | 0.00015 | -1.302 | 1.0E-5 | 0.491 |
| 9259 | FAM157A |
hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 15 | ABL2 | Sponge network | 0.813 | 1.0E-5 | 0.82 | 0 | 0.491 |
| 9260 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-769-5p | 18 | SYNPO2 | Sponge network | -0.318 | 0.65847 | -2.342 | 0 | 0.491 |
| 9261 | CTC-296K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-5p | 12 | CBFA2T3 | Sponge network | -2.095 | 0.00112 | -1.221 | 1.0E-5 | 0.491 |
| 9262 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-96-5p | 28 | PIK3R1 | Sponge network | 0.614 | 1.0E-5 | -0.133 | 0.36854 | 0.491 |
| 9263 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SETBP1 | Sponge network | -1.057 | 0.00029 | -1.302 | 1.0E-5 | 0.49 |
| 9264 | RASSF8-AS1 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 12 | NOV | Sponge network | 0.335 | 0.20596 | -0.58 | 0.04676 | 0.49 |
| 9265 | RP11-13K12.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-532-3p | 15 | SYNM | Sponge network | -0.154 | 0.78939 | -2.309 | 1.0E-5 | 0.49 |
| 9266 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | PTPN13 | Sponge network | 0.997 | 0.00066 | -1.208 | 0 | 0.49 |
| 9267 | RP11-326G21.1 |
hsa-let-7b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | AR | Sponge network | -0.021 | 0.93598 | -1.554 | 0 | 0.49 |
| 9268 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-590-3p | 13 | TRIM33 | Sponge network | 0.826 | 1.0E-5 | 0.402 | 7.0E-5 | 0.49 |
| 9269 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | PREX2 | Sponge network | 0.214 | 0.18246 | -0.272 | 0.25382 | 0.49 |
| 9270 | RP11-326G21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SETBP1 | Sponge network | -0.021 | 0.93598 | -1.302 | 1.0E-5 | 0.49 |
| 9271 | AC010226.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-590-3p | 19 | DLC1 | Sponge network | -0.811 | 2.0E-5 | -0.382 | 0.05038 | 0.49 |
| 9272 | RP11-620J15.3 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-3p | 18 | RIMS2 | Sponge network | -0.739 | 0.00044 | -1.365 | 0.00208 | 0.49 |
| 9273 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-7-5p;hsa-miR-96-5p | 22 | EPHA3 | Sponge network | -1.654 | 0.00144 | 0.185 | 0.53588 | 0.49 |
| 9274 | MRVI1-AS1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-423-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p | 13 | CSDE1 | Sponge network | -1.092 | 4.0E-5 | -0.493 | 0 | 0.49 |
| 9275 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | RBL2 | Sponge network | -0.222 | 0.32445 | -0.282 | 0.00254 | 0.49 |
| 9276 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 27 | PRKAB2 | Sponge network | -0.303 | 0.21002 | -0.367 | 0.01371 | 0.49 |
| 9277 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 22 | RARB | Sponge network | -1.439 | 4.0E-5 | -0.825 | 2.0E-5 | 0.49 |
| 9278 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 58 | BMPR2 | Sponge network | -0.096 | 0.67958 | 0.206 | 0.0635 | 0.49 |
| 9279 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-629-5p | 18 | IGF1 | Sponge network | -1.654 | 0.00144 | -0.204 | 0.5898 | 0.49 |
| 9280 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-874-3p | 12 | GIGYF2 | Sponge network | 0.782 | 0.00016 | 0.136 | 0.04403 | 0.49 |
| 9281 | RP11-574K11.29 |
hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-424-5p | 13 | ABL2 | Sponge network | 0.997 | 0 | 0.82 | 0 | 0.49 |
| 9282 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | PLSCR4 | Sponge network | -0.187 | 0.23787 | -0.474 | 0.03833 | 0.49 |
| 9283 | MEG3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 35 | ABI2 | Sponge network | 0.249 | 0.35902 | 0.175 | 0.14787 | 0.49 |
| 9284 | RP11-532F6.3 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-590-3p | 14 | GLIPR1 | Sponge network | -0.896 | 0.00099 | 0.146 | 0.3276 | 0.49 |
| 9285 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | PTPRB | Sponge network | -0.739 | 0.0574 | 0.105 | 0.47122 | 0.49 |
| 9286 | CTC-429P9.5 |
hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p | 13 | PRDM2 | Sponge network | 0.178 | 0.29106 | -0.258 | 0.00721 | 0.49 |
| 9287 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p | 11 | CADM3 | Sponge network | -1.161 | 0.00591 | -3.663 | 0 | 0.49 |
| 9288 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 50 | TGFBR3 | Sponge network | -0.49 | 0.04946 | -1.173 | 1.0E-5 | 0.49 |
| 9289 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 61 | THRB | Sponge network | -1.281 | 0.00012 | -1.117 | 0.0004 | 0.49 |
| 9290 | BVES-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-505-5p;hsa-miR-98-5p | 24 | KCNA6 | Sponge network | -2.62 | 0 | -1.531 | 0.00013 | 0.49 |
| 9291 | RP11-13K12.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-3p | 25 | MED12L | Sponge network | -0.154 | 0.78939 | -0.194 | 0.55299 | 0.49 |
| 9292 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 35 | BCL2 | Sponge network | -1.349 | 0.00017 | -0.899 | 0 | 0.49 |
| 9293 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 25 | ESR1 | Sponge network | -1.582 | 8.0E-5 | -1.035 | 3.0E-5 | 0.49 |
| 9294 | RP11-620J15.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-744-3p | 25 | DIP2C | Sponge network | -0.739 | 0.00044 | -0.436 | 0.01713 | 0.49 |
| 9295 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 51 | FAT3 | Sponge network | -0.563 | 0.02545 | -1.601 | 0.00051 | 0.49 |
| 9296 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 45 | ADARB1 | Sponge network | 0.082 | 0.55654 | -0.335 | 0.06631 | 0.49 |
| 9297 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 49 | GAS7 | Sponge network | -2.117 | 0.00161 | -0.555 | 0.01602 | 0.49 |
| 9298 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p | 10 | TSLP | Sponge network | -1.161 | 0.00591 | -2.209 | 0 | 0.49 |
| 9299 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-93-5p | 14 | CNTNAP3 | Sponge network | -0.611 | 0.09607 | -1.465 | 0.00033 | 0.49 |
| 9300 | LINC00472 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-450b-5p | 27 | DLC1 | Sponge network | -0.842 | 0.01361 | -0.382 | 0.05038 | 0.49 |
| 9301 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | ST6GAL2 | Sponge network | -2.452 | 0 | -1.201 | 0.00597 | 0.49 |
| 9302 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TBX18 | Sponge network | 0.532 | 0.00505 | 0.843 | 0.02945 | 0.49 |
| 9303 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-582-5p | 18 | NR2C2 | Sponge network | 0.921 | 0.00118 | 0.21 | 0.03224 | 0.49 |
| 9304 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-940 | 52 | QKI | Sponge network | -0.162 | 0.47687 | -0.333 | 0.04111 | 0.49 |
| 9305 | RP11-13K12.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 13 | PRUNE2 | Sponge network | -0.318 | 0.65847 | -1.733 | 0.00022 | 0.49 |
| 9306 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 20 | SPG20 | Sponge network | -0.612 | 0.00094 | -1.16 | 0 | 0.49 |
| 9307 | RP11-21A7A.2 |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-93-3p | 18 | GRIK3 | Sponge network | -1.116 | 0.23686 | -3.208 | 0 | 0.49 |
| 9308 | LINC01018 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | RELN | Sponge network | -2.915 | 0.00015 | -2.403 | 0 | 0.49 |
| 9309 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 30 | TET1 | Sponge network | -0.49 | 0.04946 | -0.135 | 0.52138 | 0.49 |
| 9310 | RP11-326G21.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | SYNPO2 | Sponge network | -0.021 | 0.93598 | -2.342 | 0 | 0.49 |
| 9311 | PCA3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | SPG20 | Sponge network | -0.709 | 0.18168 | -1.16 | 0 | 0.49 |
| 9312 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429 | 17 | DYNC1I1 | Sponge network | -1.654 | 0.00144 | -1.12 | 0.00107 | 0.49 |
| 9313 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 19 | DYNC1I1 | Sponge network | 0.572 | 0.01722 | -1.12 | 0.00107 | 0.49 |
| 9314 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p | 17 | SEMA3E | Sponge network | -0.793 | 7.0E-5 | -1.717 | 0.00137 | 0.49 |
| 9315 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | MOB1B | Sponge network | 0.532 | 0.00505 | 0.197 | 0.15266 | 0.49 |
| 9316 | AC009948.5 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH11 | Sponge network | 0.532 | 0.00505 | 1.427 | 0 | 0.49 |
| 9317 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-7-5p;hsa-miR-96-5p | 47 | LPP | Sponge network | -0.517 | 0.02935 | -0.459 | 0.04475 | 0.49 |
| 9318 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p | 51 | LPP | Sponge network | -2.531 | 0 | -0.459 | 0.04475 | 0.49 |
| 9319 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p | 16 | AKAP11 | Sponge network | 1.106 | 0 | 0.196 | 0.09825 | 0.49 |
| 9320 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 15 | CNTNAP1 | Sponge network | 0.374 | 0.20054 | 0.358 | 0.13727 | 0.49 |
| 9321 | DNM3OS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | IL17RD | Sponge network | 0.572 | 0.01722 | -0.019 | 0.94873 | 0.49 |
| 9322 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | FLI1 | Sponge network | -2.452 | 0 | -0.294 | 0.10905 | 0.49 |
| 9323 | MEG3 |
hsa-miR-126-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | CXCL12 | Sponge network | 0.249 | 0.35902 | -1.421 | 0 | 0.49 |
| 9324 | EMX2OS |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 31 | DLC1 | Sponge network | -0.777 | 0.1134 | -0.382 | 0.05038 | 0.49 |
| 9325 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-744-3p | 25 | ADAMTSL3 | Sponge network | 0.249 | 0.35902 | -1.559 | 0 | 0.49 |
| 9326 | RP11-156P1.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | MRVI1 | Sponge network | 0.214 | 0.18246 | -1.051 | 0.00022 | 0.49 |
| 9327 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 35 | EPHA7 | Sponge network | -0.811 | 2.0E-5 | -2.197 | 3.0E-5 | 0.49 |
| 9328 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 16 | HERC2 | Sponge network | 0.421 | 0.00084 | 0.114 | 0.30638 | 0.49 |
| 9329 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 27 | BHLHE41 | Sponge network | 0.335 | 0.20596 | 0.353 | 0.12753 | 0.49 |
| 9330 | RP11-888D10.4 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 20 | MDM4 | Sponge network | 0.702 | 7.0E-5 | 0.345 | 0.00762 | 0.49 |
| 9331 | AC006116.24 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-98-5p | 20 | TGFBR3 | Sponge network | -0.473 | 0.07602 | -1.173 | 1.0E-5 | 0.49 |
| 9332 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 22 | NF1 | Sponge network | 0.683 | 1.0E-5 | 0.51 | 0 | 0.49 |
| 9333 | RP11-736K20.5 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 20 | PTPRT | Sponge network | -0.358 | 0.17255 | -0.907 | 0.03727 | 0.49 |
| 9334 | AC010226.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 16 | AKAP6 | Sponge network | -0.811 | 2.0E-5 | -1.586 | 3.0E-5 | 0.49 |
| 9335 | HAND2-AS1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 11 | SORCS2 | Sponge network | -1.697 | 0.00889 | -0.223 | 0.4253 | 0.49 |
| 9336 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-7-1-3p | 11 | CNTNAP1 | Sponge network | -1.161 | 0.00591 | 0.358 | 0.13727 | 0.49 |
| 9337 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429 | 12 | ABCB5 | Sponge network | -2.62 | 0 | -0.956 | 0.04077 | 0.49 |
| 9338 | RP11-53O19.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-935;hsa-miR-940 | 13 | MAPK10 | Sponge network | -0.405 | 0.22013 | -1.774 | 0 | 0.49 |
| 9339 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-424-5p | 22 | UNC80 | Sponge network | -1.331 | 3.0E-5 | -1.934 | 0 | 0.49 |
| 9340 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 17 | SPTBN4 | Sponge network | -0.576 | 0.10121 | -0.786 | 0.01281 | 0.49 |
| 9341 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 24 | CDH23 | Sponge network | -0.49 | 0.04946 | -0.974 | 0.00034 | 0.49 |
| 9342 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-940 | 22 | MYO9A | Sponge network | 0.637 | 0.00069 | -0.147 | 0.32373 | 0.49 |
| 9343 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 36 | GAS7 | Sponge network | -1.161 | 0.00591 | -0.555 | 0.01602 | 0.49 |
| 9344 | RP11-390K5.6 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | ABL2 | Sponge network | 1.148 | 2.0E-5 | 0.82 | 0 | 0.49 |
| 9345 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 18 | ST6GALNAC3 | Sponge network | -1.331 | 3.0E-5 | -0.987 | 0 | 0.49 |
| 9346 | MIR497HG |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-671-5p | 10 | SEPT6 | Sponge network | -1.439 | 4.0E-5 | -0.598 | 0.00181 | 0.49 |
| 9347 | RP11-206L10.11 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-664a-5p;hsa-miR-671-5p | 10 | ADARB1 | Sponge network | 0.315 | 0.01647 | -0.335 | 0.06631 | 0.49 |
| 9348 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 26 | ZBTB4 | Sponge network | 0.572 | 0.01722 | -0.739 | 0 | 0.49 |
| 9349 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -1.439 | 4.0E-5 | -0.243 | 0.11408 | 0.49 |
| 9350 | ACTA2-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | GLIPR1 | Sponge network | 0.194 | 0.64278 | 0.146 | 0.3276 | 0.49 |
| 9351 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 43 | ADARB1 | Sponge network | -0.769 | 0.00035 | -0.335 | 0.06631 | 0.49 |
| 9352 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 21 | ST6GALNAC3 | Sponge network | -1.886 | 0 | -0.987 | 0 | 0.49 |
| 9353 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p | 16 | ZFX | Sponge network | 0.542 | 0.00054 | 0.09 | 0.42563 | 0.49 |
| 9354 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p | 13 | PRDM5 | Sponge network | -0.405 | 0.22013 | -1.09 | 0.00014 | 0.489 |
| 9355 | HOXB-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-429 | 13 | ERG | Sponge network | -0.154 | 0.53954 | 0.002 | 0.99011 | 0.489 |
| 9356 | RP11-156P1.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | PIK3CA | Sponge network | 0.214 | 0.18246 | 0.195 | 0.06908 | 0.489 |
| 9357 | NDUFA6-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-590-3p | 11 | PARK2 | Sponge network | -0.355 | 0.0077 | -1.892 | 0 | 0.489 |
| 9358 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 15 | FGFR1 | Sponge network | 0.199 | 0.38015 | -0.509 | 0.03957 | 0.489 |
| 9359 | RP11-13K12.5 |
hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-877-5p | 15 | NRK | Sponge network | -0.318 | 0.65847 | 0.656 | 0.19217 | 0.489 |
| 9360 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | PRDM5 | Sponge network | -0.793 | 7.0E-5 | -1.09 | 0.00014 | 0.489 |
| 9361 | LINC01018 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-96-5p | 29 | ASTN1 | Sponge network | -2.915 | 0.00015 | -2.389 | 2.0E-5 | 0.489 |
| 9362 | MEG3 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | 0.249 | 0.35902 | -2.633 | 0 | 0.489 |
| 9363 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | PEG3 | Sponge network | 0.225 | 0.30644 | -1.74 | 0 | 0.489 |
| 9364 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | GALNT13 | Sponge network | -0.49 | 0.04946 | -0.857 | 0.07439 | 0.489 |
| 9365 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 24 | TIMP3 | Sponge network | -0.612 | 0.00094 | -0.546 | 0.02307 | 0.489 |
| 9366 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 15 | RB1CC1 | Sponge network | 0.782 | 0.00016 | 0.175 | 0.12559 | 0.489 |
| 9367 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 27 | MITF | Sponge network | -0.793 | 7.0E-5 | -0.769 | 0.00022 | 0.489 |
| 9368 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 70 | NTRK3 | Sponge network | -0.942 | 0 | -1.77 | 3.0E-5 | 0.489 |
| 9369 | RP11-25K19.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p | 13 | CDH23 | Sponge network | -1.057 | 0.00029 | -0.974 | 0.00034 | 0.489 |
| 9370 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 40 | DTNA | Sponge network | 0.299 | 0.57722 | -1.648 | 1.0E-5 | 0.489 |
| 9371 | RP11-400K9.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 30 | DIP2C | Sponge network | -0.611 | 0.09607 | -0.436 | 0.01713 | 0.489 |
| 9372 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | DAB2 | Sponge network | 0.246 | 0.59912 | 0.431 | 0.0113 | 0.489 |
| 9373 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | RBL2 | Sponge network | -0.487 | 0.02157 | -0.282 | 0.00254 | 0.489 |
| 9374 | CTC-378H22.2 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-455-5p | 10 | ESR1 | Sponge network | 0.001 | 0.99663 | -1.035 | 3.0E-5 | 0.489 |
| 9375 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 73 | BMPR2 | Sponge network | -0.737 | 0.06182 | 0.206 | 0.0635 | 0.489 |
| 9376 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | MEF2C | Sponge network | -0.021 | 0.93598 | -0.499 | 0.01448 | 0.489 |
| 9377 | PWAR6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 35 | PRDM2 | Sponge network | -1.575 | 6.0E-5 | -0.258 | 0.00721 | 0.489 |
| 9378 | MIR497HG |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | SPOP | Sponge network | -1.439 | 4.0E-5 | -0.419 | 1.0E-5 | 0.489 |
| 9379 | MEG3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | TAL1 | Sponge network | 0.249 | 0.35902 | -0.462 | 0.00574 | 0.489 |
| 9380 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-940 | 20 | MYO9A | Sponge network | -0.154 | 0.53954 | -0.147 | 0.32373 | 0.489 |
| 9381 | RP5-1074L1.4 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p | 21 | CREB1 | Sponge network | 1.923 | 0 | 0.203 | 0.00537 | 0.489 |
| 9382 | BVES-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429 | 14 | TLE4 | Sponge network | -2.62 | 0 | -1.157 | 0 | 0.489 |
| 9383 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 31 | SNX29 | Sponge network | -0.096 | 0.67958 | -0.34 | 0.01371 | 0.489 |
| 9384 | RP5-1198O20.4 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 70 | DST | Sponge network | 0.997 | 0.00066 | -0.713 | 0.00096 | 0.489 |
| 9385 | LINC00473 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | -1.203 | 0.03881 | -1.892 | 0 | 0.489 |
| 9386 | CTD-3099C6.9 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-3p | 19 | ABL2 | Sponge network | 1.361 | 0 | 0.82 | 0 | 0.489 |
| 9387 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-425-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | SOX5 | Sponge network | 0.889 | 9.0E-5 | -1.091 | 4.0E-5 | 0.489 |
| 9388 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-455-3p;hsa-miR-671-5p | 24 | KCNA6 | Sponge network | -0.896 | 0.00099 | -1.531 | 0.00013 | 0.489 |
| 9389 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | DCN | Sponge network | -0.942 | 0 | -1.288 | 0 | 0.489 |
| 9390 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | RCAN2 | Sponge network | 0.537 | 0.00139 | -0.33 | 0.15172 | 0.489 |
| 9391 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p | 36 | MDM4 | Sponge network | 0.919 | 0 | 0.345 | 0.00762 | 0.489 |
| 9392 | TUG1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-654-3p | 13 | NR2C2 | Sponge network | 0.972 | 0 | 0.21 | 0.03224 | 0.489 |
| 9393 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 37 | MOB1B | Sponge network | 0.75 | 0.00054 | 0.197 | 0.15266 | 0.489 |
| 9394 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 35 | MCC | Sponge network | -0.162 | 0.47687 | -1.005 | 0 | 0.489 |
| 9395 | AP001347.6 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -1.161 | 0.00591 | -0.704 | 0.00106 | 0.489 |
| 9396 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | -0.162 | 0.47687 | -1.277 | 0 | 0.489 |
| 9397 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-3p | 45 | RUNX1T1 | Sponge network | 0.082 | 0.55397 | -0.344 | 0.2374 | 0.489 |
| 9398 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | THBS1 | Sponge network | -1.439 | 4.0E-5 | 0.741 | 0.00386 | 0.489 |
| 9399 | RP11-389C8.2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p | 23 | DLC1 | Sponge network | 0.727 | 3.0E-5 | -0.382 | 0.05038 | 0.489 |
| 9400 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-660-5p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | ZHX2 | Sponge network | -1.111 | 0.0003 | -0.192 | 0.18459 | 0.489 |
| 9401 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-628-5p;hsa-miR-940 | 14 | TRIM33 | Sponge network | 0.946 | 0 | 0.402 | 7.0E-5 | 0.489 |
| 9402 | RP11-867G23.10 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p | 15 | PRKCB | Sponge network | -2.531 | 0 | -1.313 | 2.0E-5 | 0.489 |
| 9403 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | ABCB5 | Sponge network | -2.46 | 0 | -0.956 | 0.04077 | 0.489 |
| 9404 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-93-5p | 14 | CNTNAP3 | Sponge network | -2.531 | 0 | -1.465 | 0.00033 | 0.489 |
| 9405 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-93-5p | 41 | AR | Sponge network | 0.056 | 0.81195 | -1.554 | 0 | 0.489 |
| 9406 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 17 | TET1 | Sponge network | 1.757 | 0 | -0.135 | 0.52138 | 0.489 |
| 9407 | SNHG14 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 37 | PRDM2 | Sponge network | -1.111 | 0.0003 | -0.258 | 0.00721 | 0.489 |
| 9408 | SH3BP5-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-766-3p | 15 | ATM | Sponge network | 0.267 | 0.2937 | 0.178 | 0.21568 | 0.489 |
| 9409 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p | 11 | TSLP | Sponge network | -0.739 | 0.0574 | -2.209 | 0 | 0.489 |
| 9410 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | CADM2 | Sponge network | -1.654 | 0.00144 | -3.782 | 0 | 0.489 |
| 9411 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p | 24 | ST6GALNAC3 | Sponge network | -2.62 | 0 | -0.987 | 0 | 0.489 |
| 9412 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CLASP2 | Sponge network | 0.083 | 0.66405 | -0.038 | 0.71 | 0.489 |
| 9413 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | CTNNA3 | Sponge network | 0.101 | 0.60919 | -2.046 | 0.00019 | 0.489 |
| 9414 | TPTEP1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLNA | Sponge network | -1.216 | 0 | -0.771 | 0.01377 | 0.489 |
| 9415 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 45 | DTNA | Sponge network | 0.225 | 0.30644 | -1.648 | 1.0E-5 | 0.489 |
| 9416 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 65 | KIF1B | Sponge network | -0.222 | 0.32445 | -0.118 | 0.32258 | 0.489 |
| 9417 | ARHGEF26-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | SYT4 | Sponge network | -2.46 | 0 | -3.207 | 0 | 0.489 |
| 9418 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-577 | 12 | SCN9A | Sponge network | -0.473 | 0.07602 | -0.525 | 0.10593 | 0.489 |
| 9419 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-5p | 12 | FSTL5 | Sponge network | -1.439 | 4.0E-5 | -1.3 | 0.01619 | 0.489 |
| 9420 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-7-1-3p | 31 | TGFBR3 | Sponge network | -0.611 | 0.09607 | -1.173 | 1.0E-5 | 0.489 |
| 9421 | LINC00473 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 47 | DTNA | Sponge network | -1.203 | 0.03881 | -1.648 | 1.0E-5 | 0.489 |
| 9422 | AC006116.24 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-577 | 15 | SNX29 | Sponge network | -0.473 | 0.07602 | -0.34 | 0.01371 | 0.489 |
| 9423 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | PDGFRA | Sponge network | -0.576 | 0.10121 | -0.372 | 0.0037 | 0.489 |
| 9424 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-590-3p | 23 | ABI2 | Sponge network | 0.174 | 0.30034 | 0.175 | 0.14787 | 0.489 |
| 9425 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | LRRC4 | Sponge network | -0.901 | 0.00019 | -1.774 | 0 | 0.489 |
| 9426 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p;hsa-miR-766-3p | 42 | IL6ST | Sponge network | -1.654 | 0.00144 | -0.402 | 0.00676 | 0.489 |
| 9427 | CTD-3064M3.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-625-5p | 21 | JAZF1 | Sponge network | -0.469 | 0.08721 | -0.377 | 0.02677 | 0.489 |
| 9428 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | PRRX1 | Sponge network | 0.194 | 0.64278 | 1.316 | 0 | 0.489 |
| 9429 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-93-3p | 13 | UNC5C | Sponge network | -0.469 | 0.08721 | -0.178 | 0.40214 | 0.489 |
| 9430 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-589-5p;hsa-miR-7-1-3p | 27 | SOX5 | Sponge network | -4.546 | 0 | -1.091 | 4.0E-5 | 0.489 |
| 9431 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | SYNE1 | Sponge network | -0.612 | 0.00094 | -0.738 | 0.00316 | 0.489 |
| 9432 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 27 | TBL1XR1 | Sponge network | 1.294 | 0 | 0.578 | 0 | 0.489 |
| 9433 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-7-5p | 14 | FNBP1 | Sponge network | -0.777 | 0.1134 | -0.969 | 4.0E-5 | 0.489 |
| 9434 | RP11-401P9.4 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 15 | SYNM | Sponge network | -0.384 | 0.40652 | -2.309 | 1.0E-5 | 0.489 |
| 9435 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 25 | ZMYM2 | Sponge network | 1.153 | 0 | 0.279 | 0.01273 | 0.489 |
| 9436 | BOLA3-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-629-3p | 42 | DST | Sponge network | -0.246 | 0.35034 | -0.713 | 0.00096 | 0.489 |
| 9437 | SNHG14 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 48 | ABI2 | Sponge network | -1.111 | 0.0003 | 0.175 | 0.14787 | 0.489 |
| 9438 | CTD-2554C21.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-629-3p | 13 | CDH23 | Sponge network | -1.654 | 0.00144 | -0.974 | 0.00034 | 0.489 |
| 9439 | ACTA2-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-590-3p | 16 | ROBO1 | Sponge network | 0.194 | 0.64278 | -0.009 | 0.96154 | 0.489 |
| 9440 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 14 | CNTNAP1 | Sponge network | -0.405 | 0.22013 | 0.358 | 0.13727 | 0.488 |
| 9441 | FAM66C |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 13 | KCNAB1 | Sponge network | -0.901 | 0.00019 | -0.755 | 2.0E-5 | 0.488 |
| 9442 | PART1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -2.925 | 0 | -2.046 | 0.00019 | 0.488 |
| 9443 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 15 | DAB2 | Sponge network | 0.374 | 0.20054 | 0.431 | 0.0113 | 0.488 |
| 9444 | RP11-488L18.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p | 11 | RICTOR | Sponge network | 1.392 | 0 | 0.388 | 0.00188 | 0.488 |
| 9445 | RP11-458F8.4 |
hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p | 12 | RICTOR | Sponge network | 1.518 | 0 | 0.388 | 0.00188 | 0.488 |
| 9446 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | PCDH10 | Sponge network | -0.487 | 0.02157 | -1.644 | 0.0078 | 0.488 |
| 9447 | BZRAP1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -0.465 | 0.04703 | -0.564 | 1.0E-5 | 0.488 |
| 9448 | RP4-791M13.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 23 | ABI2 | Sponge network | 0.419 | 0.0319 | 0.175 | 0.14787 | 0.488 |
| 9449 | TPTEP1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-98-5p | 42 | DMD | Sponge network | -1.216 | 0 | -1.506 | 0 | 0.488 |
| 9450 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | LRRK2 | Sponge network | -0.901 | 0.00019 | -1.136 | 1.0E-5 | 0.488 |
| 9451 | PWAR6 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | ERG | Sponge network | -1.575 | 6.0E-5 | 0.002 | 0.99011 | 0.488 |
| 9452 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 45 | ZFHX4 | Sponge network | -0.487 | 0.02157 | 0.018 | 0.95574 | 0.488 |
| 9453 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-5p | 23 | GRIA3 | Sponge network | -1.439 | 4.0E-5 | -1.956 | 0 | 0.488 |
| 9454 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 14 | CHD2 | Sponge network | 0.559 | 0.00026 | 0.049 | 0.55371 | 0.488 |
| 9455 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-766-3p;hsa-miR-940 | 34 | MDM4 | Sponge network | 0.082 | 0.55397 | 0.345 | 0.00762 | 0.488 |
| 9456 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-582-5p | 14 | ZMYM2 | Sponge network | 0.993 | 0 | 0.279 | 0.01273 | 0.488 |
| 9457 | RP11-307B6.3 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-421 | 12 | AFF3 | Sponge network | -4.463 | 0.00025 | -2.373 | 0 | 0.488 |
| 9458 | GNG12-AS1 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p | 20 | LIMCH1 | Sponge network | -0.793 | 7.0E-5 | -0.915 | 1.0E-5 | 0.488 |
| 9459 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 22 | DDX6 | Sponge network | 0.953 | 0 | 0.091 | 0.36428 | 0.488 |
| 9460 | TUG1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 18 | SLC5A3 | Sponge network | 0.972 | 0 | 0.493 | 5.0E-5 | 0.488 |
| 9461 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p | 14 | NR2C2 | Sponge network | 1.317 | 0 | 0.21 | 0.03224 | 0.488 |
| 9462 | LINC00473 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TLL1 | Sponge network | -1.203 | 0.03881 | -1.195 | 3.0E-5 | 0.488 |
| 9463 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p | 11 | ABCC9 | Sponge network | 0.082 | 0.55397 | -0.466 | 0.14121 | 0.488 |
| 9464 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-590-3p | 13 | NR2C2 | Sponge network | 1.04 | 0.00023 | 0.21 | 0.03224 | 0.488 |
| 9465 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-93-3p | 19 | EPHA3 | Sponge network | -0.154 | 0.53954 | 0.185 | 0.53588 | 0.488 |
| 9466 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p | 10 | AMPH | Sponge network | -0.487 | 0.02157 | -0.916 | 5.0E-5 | 0.488 |
| 9467 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 28 | SCN9A | Sponge network | -1.439 | 4.0E-5 | -0.525 | 0.10593 | 0.488 |
| 9468 | RP11-400K9.3 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-576-5p | 17 | AR | Sponge network | -0.733 | 0.19469 | -1.554 | 0 | 0.488 |
| 9469 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-7-5p | 14 | TET1 | Sponge network | -0.517 | 0.02935 | -0.135 | 0.52138 | 0.488 |
| 9470 | MEG3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | FNBP1 | Sponge network | 0.249 | 0.35902 | -0.969 | 4.0E-5 | 0.488 |
| 9471 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p | 13 | TLL1 | Sponge network | -0.777 | 0.16667 | -1.195 | 3.0E-5 | 0.488 |
| 9472 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-577;hsa-miR-874-3p;hsa-miR-93-5p | 33 | TACC1 | Sponge network | -2.69 | 0.0001 | -0.362 | 0.05406 | 0.488 |
| 9473 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 26 | AKAP11 | Sponge network | 0.559 | 0.00026 | 0.196 | 0.09825 | 0.488 |
| 9474 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 35 | EPHA3 | Sponge network | -1.216 | 0 | 0.185 | 0.53588 | 0.488 |
| 9475 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | NIN | Sponge network | 0.421 | 0.00084 | -0.253 | 0.13794 | 0.488 |
| 9476 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 14 | FYN | Sponge network | -0.576 | 0.10121 | -0.131 | 0.37051 | 0.488 |
| 9477 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 21 | RASSF8 | Sponge network | 0.056 | 0.81195 | -0.122 | 0.65591 | 0.488 |
| 9478 | H19 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-576-5p | 11 | HIP1 | Sponge network | 2.439 | 0 | 0.774 | 0 | 0.488 |
| 9479 | RP1-59D14.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-497-5p | 27 | RICTOR | Sponge network | 1.162 | 5.0E-5 | 0.388 | 0.00188 | 0.488 |
| 9480 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 50 | TRPS1 | Sponge network | -2.452 | 0 | -0.191 | 0.44109 | 0.488 |
| 9481 | RP11-400K9.4 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 13 | INPP4A | Sponge network | -0.611 | 0.09607 | 0.07 | 0.53563 | 0.488 |
| 9482 | NEAT1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | RBBP6 | Sponge network | 0.705 | 0.00014 | 0.163 | 0.05101 | 0.488 |
| 9483 | LINC00865 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | WWTR1 | Sponge network | -1.249 | 7.0E-5 | -0.345 | 0.11997 | 0.488 |
| 9484 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 72 | NTRK3 | Sponge network | 0.997 | 0.00066 | -1.77 | 3.0E-5 | 0.488 |
| 9485 | LINC00473 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | CDH19 | Sponge network | -1.203 | 0.03881 | -3.434 | 0 | 0.488 |
| 9486 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 41 | IL6ST | Sponge network | 0.727 | 3.0E-5 | -0.402 | 0.00676 | 0.488 |
| 9487 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-877-5p;hsa-miR-93-5p | 35 | FAT3 | Sponge network | -0.318 | 0.65847 | -1.601 | 0.00051 | 0.488 |
| 9488 | RAMP2-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | CELF4 | Sponge network | -0.513 | 0.04703 | -1.135 | 0.00344 | 0.488 |
| 9489 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 42 | GAS7 | Sponge network | 0.532 | 0.00505 | -0.555 | 0.01602 | 0.488 |
| 9490 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 21 | RASSF8 | Sponge network | -1.161 | 0.00591 | -0.122 | 0.65591 | 0.488 |
| 9491 | WAC-AS1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | KIF5B | Sponge network | 0.821 | 0 | 0.362 | 0.00066 | 0.488 |
| 9492 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | FSTL5 | Sponge network | -0.737 | 0.06182 | -1.3 | 0.01619 | 0.488 |
| 9493 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | FAT4 | Sponge network | 0.342 | 0.03129 | -0.354 | 0.14694 | 0.488 |
| 9494 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 47 | EPHA7 | Sponge network | -0.899 | 6.0E-5 | -2.197 | 3.0E-5 | 0.488 |
| 9495 | HOXA-AS2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 15 | RCAN2 | Sponge network | 0.61 | 0.01593 | -0.33 | 0.15172 | 0.488 |
| 9496 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-93-3p | 18 | RPS6KA2 | Sponge network | -0.976 | 0.00025 | -0.57 | 0.00013 | 0.488 |
| 9497 | BVES-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-98-5p | 13 | KLF10 | Sponge network | -2.62 | 0 | -0.783 | 0 | 0.488 |
| 9498 | RASSF8-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TPO | Sponge network | 0.335 | 0.20596 | -1.87 | 2.0E-5 | 0.488 |
| 9499 | RP11-890B15.3 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | 0.101 | 0.60919 | -0.998 | 0.00025 | 0.488 |
| 9500 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | EPHA3 | Sponge network | -2.117 | 0.00161 | 0.185 | 0.53588 | 0.488 |
| 9501 | PGM5-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-450b-5p | 21 | DLC1 | Sponge network | -4.546 | 0 | -0.382 | 0.05038 | 0.488 |
| 9502 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | MLLT10 | Sponge network | 1.923 | 0 | 0.105 | 0.33292 | 0.488 |
| 9503 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | ST6GALNAC3 | Sponge network | -0.769 | 0.00035 | -0.987 | 0 | 0.488 |
| 9504 | CTD-3064M3.3 |
hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p | 12 | LHX6 | Sponge network | -0.469 | 0.08721 | -0.087 | 0.69193 | 0.488 |
| 9505 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | CNTN1 | Sponge network | -0.487 | 0.02157 | -2.594 | 0 | 0.488 |
| 9506 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | AHNAK | Sponge network | -1.439 | 4.0E-5 | -1.207 | 0 | 0.488 |
| 9507 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CLIP1 | Sponge network | 0.997 | 0.00066 | -0.215 | 0.08684 | 0.488 |
| 9508 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 32 | TACC1 | Sponge network | -0.246 | 0.35034 | -0.362 | 0.05406 | 0.488 |
| 9509 | CTC-429P9.2 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-542-3p | 12 | DIP2C | Sponge network | 0.043 | 0.81628 | -0.436 | 0.01713 | 0.488 |
| 9510 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 48 | TCF4 | Sponge network | -1.161 | 0.00591 | 0.016 | 0.92271 | 0.488 |
| 9511 | EMX2OS |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 17 | SORCS1 | Sponge network | -0.777 | 0.1134 | -3.492 | 0 | 0.488 |
| 9512 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-590-5p | 10 | MYH11 | Sponge network | -0.942 | 0 | -2.895 | 0 | 0.488 |
| 9513 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CREB1 | Sponge network | 0.67 | 0.00048 | 0.203 | 0.00537 | 0.488 |
| 9514 | LINC00969 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-625-5p;hsa-miR-766-3p | 35 | MDM4 | Sponge network | 0.683 | 1.0E-5 | 0.345 | 0.00762 | 0.488 |
| 9515 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p | 28 | ABL2 | Sponge network | 0.494 | 0.00339 | 0.82 | 0 | 0.488 |
| 9516 | USP3-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 52 | GAS7 | Sponge network | -0.162 | 0.47687 | -0.555 | 0.01602 | 0.488 |
| 9517 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-628-5p | 20 | BCL6 | Sponge network | -0.162 | 0.47687 | -0.211 | 0.19489 | 0.487 |
| 9518 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 15 | DKK3 | Sponge network | -0.842 | 0.01361 | -0.052 | 0.77783 | 0.487 |
| 9519 | BOLA3-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-532-3p | 19 | MRVI1 | Sponge network | -0.246 | 0.35034 | -1.051 | 0.00022 | 0.487 |
| 9520 | LINC00865 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429 | 15 | SYNM | Sponge network | -1.249 | 7.0E-5 | -2.309 | 1.0E-5 | 0.487 |
| 9521 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | FSTL5 | Sponge network | -1.281 | 0.00012 | -1.3 | 0.01619 | 0.487 |
| 9522 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-382-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | AFF3 | Sponge network | -0.096 | 0.67958 | -2.373 | 0 | 0.487 |
| 9523 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 49 | ADARB1 | Sponge network | 0.101 | 0.60919 | -0.335 | 0.06631 | 0.487 |
| 9524 | LINC00641 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 29 | PTPN14 | Sponge network | -0.222 | 0.32445 | 0.144 | 0.34595 | 0.487 |
| 9525 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-766-3p | 22 | CHD5 | Sponge network | -0.583 | 0.14589 | -0.922 | 0.01521 | 0.487 |
| 9526 | LINC00657 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 27 | ARHGEF12 | Sponge network | 0.91 | 0 | 0.09 | 0.35693 | 0.487 |
| 9527 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 24 | ZFP36L1 | Sponge network | -2.452 | 0 | -0.505 | 8.0E-5 | 0.487 |
| 9528 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TET1 | Sponge network | 0.385 | 0.10067 | -0.135 | 0.52138 | 0.487 |
| 9529 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-671-5p | 21 | KIT | Sponge network | -0.777 | 0.16667 | -2.184 | 0 | 0.487 |
| 9530 | RP11-473I1.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p | 28 | BMPR2 | Sponge network | 0.542 | 0.00054 | 0.206 | 0.0635 | 0.487 |
| 9531 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-98-5p | 14 | ARHGAP28 | Sponge network | -0.473 | 0.07602 | -0.911 | 0.00013 | 0.487 |
| 9532 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p | 12 | ATRX | Sponge network | 1.2 | 0.01813 | 0.261 | 0.04408 | 0.487 |
| 9533 | RP11-326G21.1 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | RYR2 | Sponge network | -0.021 | 0.93598 | -1.466 | 0.00031 | 0.487 |
| 9534 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | LRRK2 | Sponge network | -2.46 | 0 | -1.136 | 1.0E-5 | 0.487 |
| 9535 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | EPHA5 | Sponge network | -0.612 | 0.00094 | -0.957 | 0.01733 | 0.487 |
| 9536 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 14 | RCAN2 | Sponge network | 0.214 | 0.18246 | -0.33 | 0.15172 | 0.487 |
| 9537 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-429 | 12 | LRRC55 | Sponge network | -0.154 | 0.53954 | -0.026 | 0.93163 | 0.487 |
| 9538 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 22 | NR2C2 | Sponge network | -0.323 | 0.01372 | 0.21 | 0.03224 | 0.487 |
| 9539 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 48 | ADARB1 | Sponge network | -0.154 | 0.78939 | -0.335 | 0.06631 | 0.487 |
| 9540 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-505-5p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 15 | SNX29 | Sponge network | 0.889 | 9.0E-5 | -0.34 | 0.01371 | 0.487 |
| 9541 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p | 15 | AKAP12 | Sponge network | -0.899 | 6.0E-5 | -0.932 | 0.0028 | 0.487 |
| 9542 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-485-3p;hsa-miR-493-5p | 21 | MLLT10 | Sponge network | 0.637 | 0.00069 | 0.105 | 0.33292 | 0.487 |
| 9543 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 12 | NALCN | Sponge network | -0.901 | 0.00019 | 1.068 | 0.00237 | 0.487 |
| 9544 | RP11-283I3.6 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 16 | SHPRH | Sponge network | 0.614 | 1.0E-5 | 0.098 | 0.40994 | 0.487 |
| 9545 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 33 | DIP2C | Sponge network | 0.214 | 0.18246 | -0.436 | 0.01713 | 0.487 |
| 9546 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | SSBP2 | Sponge network | -1.057 | 0.00029 | -1.58 | 0 | 0.487 |
| 9547 | RP11-473I1.9 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | CREB1 | Sponge network | 0.683 | 1.0E-5 | 0.203 | 0.00537 | 0.487 |
| 9548 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-93-5p | 55 | PRKAA2 | Sponge network | -0.096 | 0.67958 | -2.462 | 0 | 0.487 |
| 9549 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | LRP1B | Sponge network | -1.575 | 6.0E-5 | -2.163 | 0 | 0.487 |
| 9550 | LINC00957 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 18 | PCDH10 | Sponge network | -0.421 | 0.0114 | -1.644 | 0.0078 | 0.487 |
| 9551 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | SSBP2 | Sponge network | 0.335 | 0.20596 | -1.58 | 0 | 0.487 |
| 9552 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DCN | Sponge network | -0.421 | 0.0114 | -1.288 | 0 | 0.487 |
| 9553 | LINC00473 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935 | 32 | FBXO32 | Sponge network | -1.203 | 0.03881 | -0.495 | 0.07561 | 0.487 |
| 9554 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p;hsa-miR-654-3p | 16 | PHF6 | Sponge network | 0.421 | 0.00084 | 0.785 | 0 | 0.487 |
| 9555 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429 | 13 | HERC2 | Sponge network | 0.75 | 0.00054 | 0.114 | 0.30638 | 0.487 |
| 9556 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | SEMA3C | Sponge network | -0.896 | 0.00099 | -0.272 | 0.31153 | 0.487 |
| 9557 | TRAF3IP2-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | THSD7B | Sponge network | -0.323 | 0.01372 | 0.154 | 0.74723 | 0.487 |
| 9558 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p | 12 | DCLK1 | Sponge network | -0.469 | 0.08721 | -1.324 | 0.00014 | 0.487 |
| 9559 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 32 | ESR1 | Sponge network | 0.572 | 0.01722 | -1.035 | 3.0E-5 | 0.487 |
| 9560 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 15 | GRIN2A | Sponge network | -0.693 | 0.05629 | -2.161 | 0 | 0.487 |
| 9561 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -0.129 | 0.57177 | 0.309 | 0.17079 | 0.487 |
| 9562 | LINC00472 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p | 15 | CADM3 | Sponge network | -0.842 | 0.01361 | -3.663 | 0 | 0.487 |
| 9563 | TP73-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-429 | 11 | THSD7A | Sponge network | -0.563 | 0.02545 | 0.242 | 0.25154 | 0.487 |
| 9564 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 51 | TCF4 | Sponge network | -0.611 | 0.09607 | 0.016 | 0.92271 | 0.487 |
| 9565 | RP11-57H14.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | BRD3 | Sponge network | 0.083 | 0.66405 | 0.37 | 0.00013 | 0.487 |
| 9566 | RP11-110I1.11 |
hsa-miR-10b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 11 | NR2C2 | Sponge network | 1.034 | 0 | 0.21 | 0.03224 | 0.487 |
| 9567 | LINC00641 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 19 | STARD13 | Sponge network | -0.222 | 0.32445 | -0.187 | 0.27383 | 0.487 |
| 9568 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -0.421 | 0.0114 | -0.7 | 1.0E-5 | 0.487 |
| 9569 | RP11-98I9.4 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p | 10 | ZNF638 | Sponge network | 0.67 | 0.00048 | 0.22 | 0.01214 | 0.487 |
| 9570 | RP11-356J5.12 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 20 | MRVI1 | Sponge network | -0.899 | 6.0E-5 | -1.051 | 0.00022 | 0.487 |
| 9571 | CTD-2089N3.2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 21 | DTNA | Sponge network | -1.375 | 0.08642 | -1.648 | 1.0E-5 | 0.487 |
| 9572 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-96-5p | 61 | LPP | Sponge network | -1.654 | 0.00144 | -0.459 | 0.04475 | 0.487 |
| 9573 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p | 28 | ZFHX4 | Sponge network | -0.469 | 0.08721 | 0.018 | 0.95574 | 0.487 |
| 9574 | OIP5-AS1 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | MAP3K4 | Sponge network | 0.421 | 0.00084 | 0.058 | 0.54229 | 0.487 |
| 9575 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 41 | TACC1 | Sponge network | -0.487 | 0.02157 | -0.362 | 0.05406 | 0.487 |
| 9576 | BVES-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-589-5p;hsa-miR-98-5p | 14 | ITGB3 | Sponge network | -2.62 | 0 | -0.011 | 0.95751 | 0.487 |
| 9577 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | CDH19 | Sponge network | -1.216 | 0 | -3.434 | 0 | 0.487 |
| 9578 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-3p | 18 | ITGB3 | Sponge network | -0.487 | 0.02157 | -0.011 | 0.95751 | 0.487 |
| 9579 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 18 | TBX18 | Sponge network | -0.976 | 0.00025 | 0.843 | 0.02945 | 0.487 |
| 9580 | RP11-497H16.9 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 24 | MDM4 | Sponge network | 0.545 | 0.00064 | 0.345 | 0.00762 | 0.487 |
| 9581 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p | 31 | MCC | Sponge network | -0.154 | 0.78939 | -1.005 | 0 | 0.487 |
| 9582 | LINC00265 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p | 12 | NR2C2 | Sponge network | 1.07 | 0 | 0.21 | 0.03224 | 0.487 |
| 9583 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-93-5p | 35 | TSHZ3 | Sponge network | -0.154 | 0.78939 | -0.556 | 0.01516 | 0.487 |
| 9584 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | RCAN2 | Sponge network | 0.222 | 0.29133 | -0.33 | 0.15172 | 0.487 |
| 9585 | AC009948.5 |
hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | PAK3 | Sponge network | 0.532 | 0.00505 | -0.628 | 0.07253 | 0.487 |
| 9586 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-576-5p;hsa-miR-590-5p | 17 | BNC2 | Sponge network | -2.076 | 0.00548 | -0.491 | 0.09988 | 0.487 |
| 9587 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | -0.318 | 0.65847 | -1.751 | 1.0E-5 | 0.487 |
| 9588 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 39 | TACC1 | Sponge network | -0.842 | 0.01361 | -0.362 | 0.05406 | 0.487 |
| 9589 | RP11-156P1.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p | 15 | SYNM | Sponge network | 0.214 | 0.18246 | -2.309 | 1.0E-5 | 0.487 |
| 9590 | RP11-822E23.8 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-877-5p | 24 | NAV3 | Sponge network | -0.777 | 0.16667 | 0.212 | 0.47729 | 0.487 |
| 9591 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -1.645 | 0 | -3.328 | 0 | 0.487 |
| 9592 | RP11-736K20.5 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-590-3p | 12 | PTCH1 | Sponge network | -0.358 | 0.17255 | -0.1 | 0.6179 | 0.487 |
| 9593 | LINC00472 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-7-1-3p | 17 | ERG | Sponge network | -0.842 | 0.01361 | 0.002 | 0.99011 | 0.487 |
| 9594 | CTC-296K1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-454-3p;hsa-miR-493-5p | 12 | LRP1B | Sponge network | -2.095 | 0.00112 | -2.163 | 0 | 0.487 |
| 9595 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-505-5p | 22 | AR | Sponge network | 1.344 | 0 | -1.554 | 0 | 0.487 |
| 9596 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 22 | LRRC55 | Sponge network | 0.572 | 0.01722 | -0.026 | 0.93163 | 0.487 |
| 9597 | RP11-400K9.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-485-3p;hsa-miR-7-1-3p | 14 | SAMHD1 | Sponge network | -0.611 | 0.09607 | 0.072 | 0.62895 | 0.487 |
| 9598 | SNHG14 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 22 | GLIPR1 | Sponge network | -1.111 | 0.0003 | 0.146 | 0.3276 | 0.487 |
| 9599 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 17 | PKNOX1 | Sponge network | -0.222 | 0.32445 | 0.085 | 0.28917 | 0.487 |
| 9600 | RP11-254F7.2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-5p | 11 | MAPK10 | Sponge network | 0.176 | 0.37402 | -1.774 | 0 | 0.487 |
| 9601 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | JAZF1 | Sponge network | -2.46 | 0 | -0.377 | 0.02677 | 0.487 |
| 9602 | PWAR6 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 31 | NRK | Sponge network | -1.575 | 6.0E-5 | 0.656 | 0.19217 | 0.487 |
| 9603 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CACNA2D1 | Sponge network | -0.942 | 0 | -0.846 | 0.00675 | 0.487 |
| 9604 | RP5-1198O20.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | FAM172A | Sponge network | 0.997 | 0.00066 | -0.57 | 0 | 0.487 |
| 9605 | FAM66C |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | MACF1 | Sponge network | -0.901 | 0.00019 | 0.019 | 0.90335 | 0.487 |
| 9606 | LINC00657 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PIK3CA | Sponge network | 0.91 | 0 | 0.195 | 0.06908 | 0.487 |
| 9607 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 37 | LIFR | Sponge network | -1.575 | 6.0E-5 | -1.794 | 0 | 0.487 |
| 9608 | NR2F1-AS1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | RPS6KA6 | Sponge network | -0.49 | 0.04946 | -2.989 | 0 | 0.487 |
| 9609 | FAM66C |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SEMA3E | Sponge network | -0.901 | 0.00019 | -1.717 | 0.00137 | 0.487 |
| 9610 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | ZNF208 | Sponge network | 0.997 | 0.00066 | -0.4 | 0.22381 | 0.487 |
| 9611 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | NIN | Sponge network | -0.487 | 0.02157 | -0.253 | 0.13794 | 0.486 |
| 9612 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NUAK1 | Sponge network | 0.997 | 0.00066 | 0.904 | 0 | 0.486 |
| 9613 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | NF1 | Sponge network | 0.997 | 0 | 0.51 | 0 | 0.486 |
| 9614 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-96-5p | 13 | ELMO1 | Sponge network | -0.976 | 0.00025 | 0.04 | 0.83147 | 0.486 |
| 9615 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 32 | ZBTB4 | Sponge network | -0.709 | 0.18168 | -0.739 | 0 | 0.486 |
| 9616 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | DKK3 | Sponge network | -0.769 | 0.00035 | -0.052 | 0.77783 | 0.486 |
| 9617 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-940 | 18 | IGF1 | Sponge network | -0.154 | 0.53954 | -0.204 | 0.5898 | 0.486 |
| 9618 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-93-5p | 46 | FAT3 | Sponge network | -0.154 | 0.78939 | -1.601 | 0.00051 | 0.486 |
| 9619 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 73 | NTRK3 | Sponge network | -1.281 | 0.00012 | -1.77 | 3.0E-5 | 0.486 |
| 9620 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p | 15 | PSIP1 | Sponge network | 0.537 | 0.00139 | -0.005 | 0.96897 | 0.486 |
| 9621 | PGM5-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484 | 10 | CDH23 | Sponge network | -4.546 | 0 | -0.974 | 0.00034 | 0.486 |
| 9622 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 17 | TXNIP | Sponge network | -2.46 | 0 | -1.277 | 0 | 0.486 |
| 9623 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 19 | RASSF8 | Sponge network | 0.199 | 0.38015 | -0.122 | 0.65591 | 0.486 |
| 9624 | PWAR6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | NCAM2 | Sponge network | -1.575 | 6.0E-5 | -0.482 | 0.22565 | 0.486 |
| 9625 | PCA3 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 40 | BCL2 | Sponge network | -0.709 | 0.18168 | -0.899 | 0 | 0.486 |
| 9626 | ACTA2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-96-5p | 27 | ASTN1 | Sponge network | 0.194 | 0.64278 | -2.389 | 2.0E-5 | 0.486 |
| 9627 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | GRIA3 | Sponge network | -2.46 | 0 | -1.956 | 0 | 0.486 |
| 9628 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-5p | 16 | SYNE1 | Sponge network | 0.176 | 0.37402 | -0.738 | 0.00316 | 0.486 |
| 9629 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p | 19 | RECK | Sponge network | -0.154 | 0.78939 | -1.043 | 0 | 0.486 |
| 9630 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-425-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 35 | SOX5 | Sponge network | -1.203 | 0.03881 | -1.091 | 4.0E-5 | 0.486 |
| 9631 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 19 | RCAN2 | Sponge network | -0.901 | 0.00019 | -0.33 | 0.15172 | 0.486 |
| 9632 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | CHD2 | Sponge network | 0.765 | 7.0E-5 | 0.049 | 0.55371 | 0.486 |
| 9633 | HOTAIRM1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 42 | DST | Sponge network | -0.053 | 0.80685 | -0.713 | 0.00096 | 0.486 |
| 9634 | LINC00092 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p | 32 | FAT3 | Sponge network | -1.886 | 0 | -1.601 | 0.00051 | 0.486 |
| 9635 | CTD-2089N3.2 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 16 | MEF2C | Sponge network | -1.375 | 0.08642 | -0.499 | 0.01448 | 0.486 |
| 9636 | CTD-2002H8.2 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | AKAP11 | Sponge network | 0.931 | 1.0E-5 | 0.196 | 0.09825 | 0.486 |
| 9637 | RP11-67L2.2 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 21 | ATM | Sponge network | 0.72 | 0.0001 | 0.178 | 0.21568 | 0.486 |
| 9638 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | EPHA5 | Sponge network | -0.901 | 0.00019 | -0.957 | 0.01733 | 0.486 |
| 9639 | HOXA-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p | 10 | MN1 | Sponge network | 0.61 | 0.01593 | -0.358 | 0.24172 | 0.486 |
| 9640 | MEG3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 21 | SEMA3C | Sponge network | 0.249 | 0.35902 | -0.272 | 0.31153 | 0.486 |
| 9641 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-942-5p | 44 | HLF | Sponge network | 0.064 | 0.72934 | -2.443 | 0 | 0.486 |
| 9642 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | NRXN3 | Sponge network | 0.225 | 0.30644 | -0.893 | 0.04186 | 0.486 |
| 9643 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-671-5p | 28 | CREB3L2 | Sponge network | -1.331 | 3.0E-5 | -0.525 | 2.0E-5 | 0.486 |
| 9644 | MIR22HG |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-582-5p | 12 | KLF10 | Sponge network | -1.047 | 0 | -0.783 | 0 | 0.486 |
| 9645 | CTC-444N24.11 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | BRD3 | Sponge network | 1.153 | 0 | 0.37 | 0.00013 | 0.486 |
| 9646 | RP11-244H3.1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 42 | MDM4 | Sponge network | 0.659 | 3.0E-5 | 0.345 | 0.00762 | 0.486 |
| 9647 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | RASSF8 | Sponge network | -0.421 | 0.0114 | -0.122 | 0.65591 | 0.486 |
| 9648 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 20 | RASSF8 | Sponge network | -0.318 | 0.65847 | -0.122 | 0.65591 | 0.486 |
| 9649 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | RECK | Sponge network | -0.513 | 0.04703 | -1.043 | 0 | 0.486 |
| 9650 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 42 | ADARB1 | Sponge network | -2.117 | 0.00161 | -0.335 | 0.06631 | 0.486 |
| 9651 | AGAP11 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-574-3p | 10 | FHL1 | Sponge network | -1.645 | 0 | -2.687 | 0 | 0.486 |
| 9652 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 55 | SSBP2 | Sponge network | -0.323 | 0.01372 | -1.58 | 0 | 0.486 |
| 9653 | AGAP11 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-455-5p;hsa-miR-590-5p | 10 | PTPN13 | Sponge network | -1.645 | 0 | -1.208 | 0 | 0.486 |
| 9654 | RP11-284N8.3 |
hsa-miR-135b-5p;hsa-miR-151a-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577 | 10 | FMNL3 | Sponge network | -0.31 | 0.33871 | 0.555 | 4.0E-5 | 0.486 |
| 9655 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-625-5p;hsa-miR-93-5p | 22 | JAZF1 | Sponge network | -2.531 | 0 | -0.377 | 0.02677 | 0.486 |
| 9656 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 43 | RBMS3 | Sponge network | -2.915 | 0.00015 | -0.519 | 0.03551 | 0.486 |
| 9657 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-93-5p | 25 | CHD5 | Sponge network | -2.531 | 0 | -0.922 | 0.01521 | 0.486 |
| 9658 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 63 | THRB | Sponge network | -0.901 | 0.00019 | -1.117 | 0.0004 | 0.486 |
| 9659 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 10 | ABCC9 | Sponge network | -0.811 | 2.0E-5 | -0.466 | 0.14121 | 0.486 |
| 9660 | WDFY3-AS2 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 11 | PRKD1 | Sponge network | -0.942 | 0 | -0.466 | 0.03583 | 0.486 |
| 9661 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 29 | TSHZ3 | Sponge network | -0.738 | 0.21233 | -0.556 | 0.01516 | 0.486 |
| 9662 | RP5-855D21.1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30a-3p | 13 | MDM4 | Sponge network | 1.115 | 0.00188 | 0.345 | 0.00762 | 0.486 |
| 9663 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | PDZRN3 | Sponge network | -0.222 | 0.32445 | -1.548 | 0 | 0.486 |
| 9664 | AC093110.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-766-3p | 31 | MDM4 | Sponge network | 1.044 | 2.0E-5 | 0.345 | 0.00762 | 0.486 |
| 9665 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-502-5p | 11 | TCF4 | Sponge network | 0.043 | 0.81628 | 0.016 | 0.92271 | 0.486 |
| 9666 | RP11-251G23.5 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-664a-3p | 12 | RASAL2 | Sponge network | 1.344 | 0 | 0.869 | 0 | 0.486 |
| 9667 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | PAFAH1B1 | Sponge network | 0.101 | 0.60919 | -0.429 | 0 | 0.486 |
| 9668 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 24 | DDX6 | Sponge network | 1.601 | 0 | 0.091 | 0.36428 | 0.486 |
| 9669 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p | 17 | ROBO1 | Sponge network | 0.335 | 0.20596 | -0.009 | 0.96154 | 0.486 |
| 9670 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-93-5p | 15 | LHX6 | Sponge network | 0.335 | 0.20596 | -0.087 | 0.69193 | 0.486 |
| 9671 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 16 | SASH1 | Sponge network | -1.331 | 3.0E-5 | -0.502 | 0.00175 | 0.486 |
| 9672 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | BCL2 | Sponge network | -1.057 | 0.00029 | -0.899 | 0 | 0.486 |
| 9673 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p | 10 | PDE4DIP | Sponge network | -1.331 | 3.0E-5 | -0.282 | 0.0257 | 0.486 |
| 9674 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 44 | ZFHX3 | Sponge network | -1.575 | 6.0E-5 | 0.389 | 0.01363 | 0.486 |
| 9675 | RP11-798M19.6 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 54 | CADM2 | Sponge network | -0.612 | 0.00094 | -3.782 | 0 | 0.486 |
| 9676 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 32 | WDFY3 | Sponge network | -1.092 | 4.0E-5 | -0.201 | 0.0967 | 0.486 |
| 9677 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p | 23 | BMPR2 | Sponge network | 0.993 | 0 | 0.206 | 0.0635 | 0.486 |
| 9678 | RP11-121C2.2 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p | 13 | STAG2 | Sponge network | 1.147 | 0 | 0.252 | 0.00438 | 0.486 |
| 9679 | LINC00654 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | 0.374 | 0.20054 | -0.382 | 0.05038 | 0.486 |
| 9680 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p | 12 | MAFB | Sponge network | -0.611 | 0.09607 | -0.023 | 0.89865 | 0.486 |
| 9681 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | CNTN1 | Sponge network | -0.737 | 0.10822 | -2.594 | 0 | 0.486 |
| 9682 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-942-5p | 38 | NOTCH2 | Sponge network | 0.194 | 0.64278 | 0.138 | 0.35163 | 0.486 |
| 9683 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | ZNF208 | Sponge network | -1.349 | 0.00017 | -0.4 | 0.22381 | 0.486 |
| 9684 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 19 | ITGA5 | Sponge network | -0.096 | 0.67958 | 0.452 | 0.03524 | 0.486 |
| 9685 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 49 | BMPR2 | Sponge network | 0.225 | 0.30644 | 0.206 | 0.0635 | 0.486 |
| 9686 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-940 | 42 | CHD5 | Sponge network | -0.323 | 0.01372 | -0.922 | 0.01521 | 0.486 |
| 9687 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-429;hsa-miR-629-3p | 19 | EBF3 | Sponge network | -2.62 | 0 | -0.058 | 0.81437 | 0.486 |
| 9688 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-532-3p;hsa-miR-539-5p | 32 | PBX1 | Sponge network | -0.384 | 0.40652 | -1.206 | 0 | 0.486 |
| 9689 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 32 | TET1 | Sponge network | -0.129 | 0.57177 | -0.135 | 0.52138 | 0.486 |
| 9690 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p | 12 | FGFR1 | Sponge network | -1.057 | 0.00029 | -0.509 | 0.03957 | 0.486 |
| 9691 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-942-5p | 45 | UNC80 | Sponge network | -2.452 | 0 | -1.934 | 0 | 0.486 |
| 9692 | RP11-119F7.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | NAV3 | Sponge network | 0.225 | 0.30644 | 0.212 | 0.47729 | 0.486 |
| 9693 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | IGSF10 | Sponge network | -1.349 | 0.00017 | -1.751 | 1.0E-5 | 0.486 |
| 9694 | TPTEP1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ITPKB | Sponge network | -1.216 | 0 | -0.704 | 0.00106 | 0.486 |
| 9695 | SNHG14 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | ZNF536 | Sponge network | -1.111 | 0.0003 | -3.115 | 0 | 0.486 |
| 9696 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-5p | 18 | MYO9A | Sponge network | -0.376 | 0.00337 | -0.147 | 0.32373 | 0.486 |
| 9697 | LINC00472 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p | 14 | ROBO1 | Sponge network | -0.842 | 0.01361 | -0.009 | 0.96154 | 0.486 |
| 9698 | PCA3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CDH19 | Sponge network | -0.709 | 0.18168 | -3.434 | 0 | 0.486 |
| 9699 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-500a-5p | 23 | DMD | Sponge network | 0.811 | 0.03228 | -1.506 | 0 | 0.486 |
| 9700 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 20 | TMEFF2 | Sponge network | -0.942 | 0 | -2.426 | 0 | 0.486 |
| 9701 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 21 | EPHA3 | Sponge network | -0.811 | 2.0E-5 | 0.185 | 0.53588 | 0.485 |
| 9702 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 41 | PTPRT | Sponge network | -2.452 | 0 | -0.907 | 0.03727 | 0.485 |
| 9703 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | MOB1B | Sponge network | 0.673 | 0 | 0.197 | 0.15266 | 0.485 |
| 9704 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-874-3p | 11 | BCLAF1 | Sponge network | 0.959 | 0 | 0.211 | 0.00684 | 0.485 |
| 9705 | HAND2-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | ZDBF2 | Sponge network | -1.697 | 0.00889 | -0.998 | 0.00025 | 0.485 |
| 9706 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | PCDH9 | Sponge network | -0.187 | 0.23787 | -1.823 | 4.0E-5 | 0.485 |
| 9707 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-92a-1-5p | 26 | CTIF | Sponge network | -0.096 | 0.67958 | -0.389 | 0.00549 | 0.485 |
| 9708 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-590-3p | 11 | PNRC1 | Sponge network | -0.811 | 2.0E-5 | -0.646 | 0 | 0.485 |
| 9709 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | HOOK3 | Sponge network | 0.494 | 0.00339 | 0.273 | 0.09957 | 0.485 |
| 9710 | CTC-296K1.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-744-3p | 51 | LPP | Sponge network | -2.095 | 0.00112 | -0.459 | 0.04475 | 0.485 |
| 9711 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 16 | PGR | Sponge network | 0.178 | 0.29106 | -1.704 | 0 | 0.485 |
| 9712 | RP11-57H14.4 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 16 | SHPRH | Sponge network | 0.083 | 0.66405 | 0.098 | 0.40994 | 0.485 |
| 9713 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-93-3p | 14 | TRPS1 | Sponge network | -4.463 | 0.00025 | -0.191 | 0.44109 | 0.485 |
| 9714 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-let-7i-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-539-5p;hsa-miR-590-5p | 10 | RSPO2 | Sponge network | -2.076 | 0.00548 | -3.038 | 0 | 0.485 |
| 9715 | WDFY3-AS2 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p | 10 | CNN1 | Sponge network | -0.942 | 0 | -2.633 | 0 | 0.485 |
| 9716 | SNHG14 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 22 | KLF10 | Sponge network | -1.111 | 0.0003 | -0.783 | 0 | 0.485 |
| 9717 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 27 | ARHGAP28 | Sponge network | -2.452 | 0 | -0.911 | 0.00013 | 0.485 |
| 9718 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | ZNF536 | Sponge network | -1.057 | 0.00029 | -3.115 | 0 | 0.485 |
| 9719 | MYLK-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-484;hsa-miR-940 | 14 | ZFP36L1 | Sponge network | -1.331 | 3.0E-5 | -0.505 | 8.0E-5 | 0.485 |
| 9720 | AC011526.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 15 | PRRX1 | Sponge network | 0.342 | 0.03129 | 1.316 | 0 | 0.485 |
| 9721 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | ANK2 | Sponge network | -0.227 | 0.31657 | -1.603 | 1.0E-5 | 0.485 |
| 9722 | LINC00657 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PHF3 | Sponge network | 0.91 | 0 | 0.126 | 0.19652 | 0.485 |
| 9723 | MIR17HG |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-654-3p | 11 | PHF6 | Sponge network | 1.699 | 0 | 0.785 | 0 | 0.485 |
| 9724 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | MYO9A | Sponge network | -0.227 | 0.31657 | -0.147 | 0.32373 | 0.485 |
| 9725 | RP11-283I3.6 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | PIK3CA | Sponge network | 0.614 | 1.0E-5 | 0.195 | 0.06908 | 0.485 |
| 9726 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-362-3p;hsa-miR-429 | 11 | DKK3 | Sponge network | -0.154 | 0.53954 | -0.052 | 0.77783 | 0.485 |
| 9727 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 13 | KCNAB1 | Sponge network | -0.976 | 0.00025 | -0.755 | 2.0E-5 | 0.485 |
| 9728 | RP11-244H3.1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-766-3p | 14 | ATM | Sponge network | 0.659 | 3.0E-5 | 0.178 | 0.21568 | 0.485 |
| 9729 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | ATRX | Sponge network | 0.411 | 0.1335 | 0.261 | 0.04408 | 0.485 |
| 9730 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-550a-3p;hsa-miR-590-3p | 13 | HIVEP1 | Sponge network | 0.683 | 1.0E-5 | 0.368 | 0.00064 | 0.485 |
| 9731 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 19 | JAZF1 | Sponge network | -0.021 | 0.93598 | -0.377 | 0.02677 | 0.485 |
| 9732 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | FYN | Sponge network | -0.976 | 0.00025 | -0.131 | 0.37051 | 0.485 |
| 9733 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-500a-5p;hsa-miR-98-5p | 26 | DMD | Sponge network | 0.082 | 0.55397 | -1.506 | 0 | 0.485 |
| 9734 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | ZNF521 | Sponge network | -1.216 | 0 | -0.111 | 0.58068 | 0.485 |
| 9735 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 44 | TMEM132B | Sponge network | -0.901 | 0.00019 | -0.83 | 0.01084 | 0.485 |
| 9736 | CTC-444N24.11 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-940 | 16 | SIRT1 | Sponge network | 1.153 | 0 | 0.109 | 0.2593 | 0.485 |
| 9737 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | HOOK3 | Sponge network | -0.487 | 0.02157 | 0.273 | 0.09957 | 0.485 |
| 9738 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-874-3p | 19 | TACC1 | Sponge network | -0.469 | 0.08721 | -0.362 | 0.05406 | 0.485 |
| 9739 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-708-5p | 11 | EZH1 | Sponge network | -2.076 | 0.00548 | -0.564 | 1.0E-5 | 0.485 |
| 9740 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-3p | 35 | PRKAB2 | Sponge network | -0.899 | 6.0E-5 | -0.367 | 0.01371 | 0.485 |
| 9741 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p | 51 | FAT3 | Sponge network | -0.487 | 0.02157 | -1.601 | 0.00051 | 0.485 |
| 9742 | SNX29P2 |
hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-940 | 10 | MAPK10 | Sponge network | 0.4 | 0.14611 | -1.774 | 0 | 0.485 |
| 9743 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | RECK | Sponge network | -0.318 | 0.65847 | -1.043 | 0 | 0.485 |
| 9744 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-877-5p;hsa-miR-935 | 39 | FAT3 | Sponge network | -0.246 | 0.35034 | -1.601 | 0.00051 | 0.485 |
| 9745 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 23 | TACC1 | Sponge network | 0.225 | 0.30644 | -0.362 | 0.05406 | 0.485 |
| 9746 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-93-5p | 23 | PLSCR4 | Sponge network | -0.154 | 0.78939 | -0.474 | 0.03833 | 0.485 |
| 9747 | SNHG14 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NRG3 | Sponge network | -1.111 | 0.0003 | -1.836 | 4.0E-5 | 0.485 |
| 9748 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 16 | HOOK3 | Sponge network | 0.222 | 0.29133 | 0.273 | 0.09957 | 0.485 |
| 9749 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-577;hsa-miR-590-3p | 13 | MYCBP2 | Sponge network | 0.537 | 0.00139 | -0.027 | 0.82016 | 0.485 |
| 9750 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | EBF3 | Sponge network | -0.376 | 0.00337 | -0.058 | 0.81437 | 0.485 |
| 9751 | SNHG14 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 16 | UVRAG | Sponge network | -1.111 | 0.0003 | -0.017 | 0.83865 | 0.485 |
| 9752 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 17 | IGF1 | Sponge network | -0.693 | 0.05629 | -0.204 | 0.5898 | 0.485 |
| 9753 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-625-5p | 21 | RSPO2 | Sponge network | -0.421 | 0.0114 | -3.038 | 0 | 0.485 |
| 9754 | LINC00641 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DNAJB4 | Sponge network | -0.222 | 0.32445 | -0.7 | 1.0E-5 | 0.485 |
| 9755 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | RBMS3 | Sponge network | -0.187 | 0.23787 | -0.519 | 0.03551 | 0.485 |
| 9756 | RP11-307B6.3 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-532-5p | 11 | RYR2 | Sponge network | -4.463 | 0.00025 | -1.466 | 0.00031 | 0.485 |
| 9757 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | SYNE1 | Sponge network | 0.064 | 0.72934 | -0.738 | 0.00316 | 0.485 |
| 9758 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-940 | 14 | CRTC3 | Sponge network | -0.154 | 0.53954 | 0.164 | 0.16786 | 0.485 |
| 9759 | PCA3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 15 | DCLK1 | Sponge network | -0.709 | 0.18168 | -1.324 | 0.00014 | 0.485 |
| 9760 | CKMT2-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.082 | 0.55654 | -0.57 | 0 | 0.485 |
| 9761 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-629-5p;hsa-miR-93-5p | 52 | TCF4 | Sponge network | -2.69 | 0.0001 | 0.016 | 0.92271 | 0.485 |
| 9762 | C20orf166-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577 | 17 | SCN9A | Sponge network | -2.69 | 0.0001 | -0.525 | 0.10593 | 0.485 |
| 9763 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-22-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-652-3p | 12 | QKI | Sponge network | -0.326 | 0.28809 | -0.333 | 0.04111 | 0.485 |
| 9764 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | BNC2 | Sponge network | 0.101 | 0.60919 | -0.491 | 0.09988 | 0.485 |
| 9765 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-935 | 30 | EBF1 | Sponge network | -0.896 | 0.00099 | -0.384 | 0.08856 | 0.485 |
| 9766 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-93-3p | 26 | QKI | Sponge network | -2.531 | 0 | -0.333 | 0.04111 | 0.485 |
| 9767 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-744-3p;hsa-miR-96-5p | 23 | HIP1 | Sponge network | 0.41 | 0.02214 | 0.774 | 0 | 0.485 |
| 9768 | RP11-540B6.6 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 16 | PHF6 | Sponge network | 0.93 | 0 | 0.785 | 0 | 0.485 |
| 9769 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 32 | CHD5 | Sponge network | -0.318 | 0.65847 | -0.922 | 0.01521 | 0.485 |
| 9770 | CTD-2314G24.2 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | STAT5B | Sponge network | 0.246 | 0.59912 | -0.182 | 0.1082 | 0.485 |
| 9771 | EMX2OS |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ITPKB | Sponge network | -0.777 | 0.1134 | -0.704 | 0.00106 | 0.485 |
| 9772 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-424-5p | 15 | LATS2 | Sponge network | 0.889 | 9.0E-5 | 0.159 | 0.2304 | 0.485 |
| 9773 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-505-3p;hsa-miR-590-3p | 15 | JAKMIP2 | Sponge network | -2.915 | 0.00015 | -1.388 | 0 | 0.485 |
| 9774 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 24 | TCF12 | Sponge network | 0.421 | 0.00084 | 0.174 | 0.13271 | 0.485 |
| 9775 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-455-3p | 17 | CREB1 | Sponge network | 0.858 | 3.0E-5 | 0.203 | 0.00537 | 0.485 |
| 9776 | RP11-672A2.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-330-5p;hsa-miR-424-5p | 16 | NOTCH2 | Sponge network | 1.344 | 0 | 0.138 | 0.35163 | 0.485 |
| 9777 | GNG12-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | ZNF536 | Sponge network | -0.793 | 7.0E-5 | -3.115 | 0 | 0.485 |
| 9778 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-589-3p | 10 | MAFB | Sponge network | 0.811 | 0.03228 | -0.023 | 0.89865 | 0.485 |
| 9779 | RP11-774O3.3 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TPO | Sponge network | -0.487 | 0.02157 | -1.87 | 2.0E-5 | 0.485 |
| 9780 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-486-5p;hsa-miR-590-3p | 10 | NALCN | Sponge network | -2.915 | 0.00015 | 1.068 | 0.00237 | 0.485 |
| 9781 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 17 | ABI2 | Sponge network | 0.178 | 0.29106 | 0.175 | 0.14787 | 0.485 |
| 9782 | DNM3OS |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-484 | 17 | ABL1 | Sponge network | 0.572 | 0.01722 | -0.215 | 0.07804 | 0.485 |
| 9783 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-744-3p | 21 | ADAMTSL3 | Sponge network | -0.465 | 0.04703 | -1.559 | 0 | 0.484 |
| 9784 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 41 | RBMS3 | Sponge network | 0.064 | 0.72934 | -0.519 | 0.03551 | 0.484 |
| 9785 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZFHX3 | Sponge network | 1.378 | 0 | 0.389 | 0.01363 | 0.484 |
| 9786 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | CACNA2D1 | Sponge network | -2.46 | 0 | -0.846 | 0.00675 | 0.484 |
| 9787 | CTD-2017D11.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-940 | 33 | MDM4 | Sponge network | 0.792 | 0.0049 | 0.345 | 0.00762 | 0.484 |
| 9788 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 31 | IGF1 | Sponge network | -0.487 | 0.02157 | -0.204 | 0.5898 | 0.484 |
| 9789 | PGM5-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 14 | ERG | Sponge network | -4.546 | 0 | 0.002 | 0.99011 | 0.484 |
| 9790 | RP11-156P1.3 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 19 | HIP1 | Sponge network | 0.214 | 0.18246 | 0.774 | 0 | 0.484 |
| 9791 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p | 28 | EPHA3 | Sponge network | -0.227 | 0.31657 | 0.185 | 0.53588 | 0.484 |
| 9792 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-5p | 12 | AKAP12 | Sponge network | -0.154 | 0.78939 | -0.932 | 0.0028 | 0.484 |
| 9793 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-93-3p;hsa-miR-940 | 34 | QKI | Sponge network | -0.465 | 0.04703 | -0.333 | 0.04111 | 0.484 |
| 9794 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-590-5p | 25 | CBL | Sponge network | 0.214 | 0.18246 | 0.291 | 0.01267 | 0.484 |
| 9795 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-5p | 27 | TSHZ3 | Sponge network | 0.176 | 0.37402 | -0.556 | 0.01516 | 0.484 |
| 9796 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-652-3p;hsa-miR-940 | 22 | QKI | Sponge network | -1.886 | 0 | -0.333 | 0.04111 | 0.484 |
| 9797 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-542-3p;hsa-miR-708-3p;hsa-miR-93-5p | 32 | HLF | Sponge network | -0.384 | 0.40652 | -2.443 | 0 | 0.484 |
| 9798 | LINC00641 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 15 | CNTNAP1 | Sponge network | -0.222 | 0.32445 | 0.358 | 0.13727 | 0.484 |
| 9799 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-7-5p;hsa-miR-93-3p | 21 | EPHA3 | Sponge network | -0.693 | 0.05629 | 0.185 | 0.53588 | 0.484 |
| 9800 | LINC00957 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 11 | LHFP | Sponge network | -0.421 | 0.0114 | -0.167 | 0.41371 | 0.484 |
| 9801 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | EDA2R | Sponge network | -1.216 | 0 | -0.587 | 0.01598 | 0.484 |
| 9802 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | AKAP6 | Sponge network | 0.225 | 0.30644 | -1.586 | 3.0E-5 | 0.484 |
| 9803 | RP11-57H14.4 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ITPKB | Sponge network | 0.083 | 0.66405 | -0.704 | 0.00106 | 0.484 |
| 9804 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-655-3p | 22 | LATS1 | Sponge network | 0.083 | 0.66405 | 0.109 | 0.34208 | 0.484 |
| 9805 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-93-3p | 34 | NTRK3 | Sponge network | -0.469 | 0.08721 | -1.77 | 3.0E-5 | 0.484 |
| 9806 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 12 | LRRK2 | Sponge network | 0.532 | 0.00505 | -1.136 | 1.0E-5 | 0.484 |
| 9807 | LINC01018 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PTPN13 | Sponge network | -2.915 | 0.00015 | -1.208 | 0 | 0.484 |
| 9808 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | ABCC9 | Sponge network | 0.064 | 0.72934 | -0.466 | 0.14121 | 0.484 |
| 9809 | SNHG14 |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 12 | GLIPR2 | Sponge network | -1.111 | 0.0003 | -0.996 | 0 | 0.484 |
| 9810 | SNHG14 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | BTG2 | Sponge network | -1.111 | 0.0003 | -1.763 | 0 | 0.484 |
| 9811 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | GPC6 | Sponge network | -1.582 | 8.0E-5 | -0.054 | 0.81465 | 0.484 |
| 9812 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | PCDH9 | Sponge network | 0.101 | 0.60919 | -1.823 | 4.0E-5 | 0.484 |
| 9813 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-96-5p | 57 | FAT3 | Sponge network | -0.942 | 0 | -1.601 | 0.00051 | 0.484 |
| 9814 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 25 | EBF3 | Sponge network | 0.532 | 0.00505 | -0.058 | 0.81437 | 0.484 |
| 9815 | RP11-384O8.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-590-3p | 23 | SCN9A | Sponge network | -0.738 | 0.21233 | -0.525 | 0.10593 | 0.484 |
| 9816 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 80 | BMPR2 | Sponge network | -1.111 | 0.0003 | 0.206 | 0.0635 | 0.484 |
| 9817 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | RADIL | Sponge network | -0.096 | 0.67958 | -1.628 | 0 | 0.484 |
| 9818 | SNHG14 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 42 | ERC1 | Sponge network | -1.111 | 0.0003 | -0.108 | 0.38794 | 0.484 |
| 9819 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-940 | 27 | NOTCH2 | Sponge network | -1.331 | 3.0E-5 | 0.138 | 0.35163 | 0.484 |
| 9820 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | PREX2 | Sponge network | 0.101 | 0.60919 | -0.272 | 0.25382 | 0.484 |
| 9821 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-335-5p;hsa-miR-589-3p | 19 | KCNA6 | Sponge network | -0.358 | 0.17255 | -1.531 | 0.00013 | 0.484 |
| 9822 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 29 | ZFHX3 | Sponge network | 0.614 | 1.0E-5 | 0.389 | 0.01363 | 0.484 |
| 9823 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p | 10 | THSD7A | Sponge network | -0.487 | 0.02157 | 0.242 | 0.25154 | 0.484 |
| 9824 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-652-3p;hsa-miR-96-5p | 32 | QKI | Sponge network | -1.654 | 0.00144 | -0.333 | 0.04111 | 0.484 |
| 9825 | LINC00957 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-625-5p | 10 | NGFR | Sponge network | -0.421 | 0.0114 | -1.271 | 0.01058 | 0.484 |
| 9826 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-539-5p | 25 | NRXN1 | Sponge network | -0.154 | 0.53954 | -3.778 | 0 | 0.484 |
| 9827 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | IGSF10 | Sponge network | -0.793 | 7.0E-5 | -1.751 | 1.0E-5 | 0.484 |
| 9828 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | MYO9A | Sponge network | 0.683 | 1.0E-5 | -0.147 | 0.32373 | 0.484 |
| 9829 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940;hsa-miR-98-5p | 40 | PBX1 | Sponge network | -0.222 | 0.32445 | -1.206 | 0 | 0.484 |
| 9830 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p | 15 | TMEFF2 | Sponge network | -0.162 | 0.47687 | -2.426 | 0 | 0.484 |
| 9831 | C20orf166-AS1 |
hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-629-5p | 24 | MYO9A | Sponge network | -2.69 | 0.0001 | -0.147 | 0.32373 | 0.484 |
| 9832 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SEMA3E | Sponge network | -0.129 | 0.57177 | -1.717 | 0.00137 | 0.484 |
| 9833 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-484 | 10 | SOX5 | Sponge network | -0.326 | 0.28809 | -1.091 | 4.0E-5 | 0.484 |
| 9834 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 43 | NOTCH2 | Sponge network | -0.737 | 0.06182 | 0.138 | 0.35163 | 0.484 |
| 9835 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-589-5p | 30 | ADARB1 | Sponge network | -4.546 | 0 | -0.335 | 0.06631 | 0.484 |
| 9836 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | CAV1 | Sponge network | 0.335 | 0.20596 | -1.013 | 6.0E-5 | 0.484 |
| 9837 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 12 | HMCN1 | Sponge network | -1.216 | 0 | 0.809 | 0.00544 | 0.484 |
| 9838 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-484;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NRXN3 | Sponge network | 0.61 | 0.01593 | -0.893 | 0.04186 | 0.484 |
| 9839 | LINC00657 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 13 | GSK3B | Sponge network | 0.91 | 0 | 0.266 | 0.00045 | 0.484 |
| 9840 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 10 | TXNIP | Sponge network | -2.531 | 0 | -1.277 | 0 | 0.484 |
| 9841 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-5p;hsa-miR-98-5p | 47 | PRKAA2 | Sponge network | 0.056 | 0.81195 | -2.462 | 0 | 0.484 |
| 9842 | LINC01018 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-96-5p | 16 | FNBP1 | Sponge network | -2.915 | 0.00015 | -0.969 | 4.0E-5 | 0.484 |
| 9843 | SNHG14 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | FLI1 | Sponge network | -1.111 | 0.0003 | -0.294 | 0.10905 | 0.484 |
| 9844 | PART1 |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | -2.925 | 0 | -1.398 | 2.0E-5 | 0.484 |
| 9845 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | MAF | Sponge network | -1.886 | 0 | -1.257 | 0 | 0.484 |
| 9846 | RASSF8-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p | 16 | SORCS1 | Sponge network | 0.335 | 0.20596 | -3.492 | 0 | 0.484 |
| 9847 | RP11-400K9.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-744-3p | 15 | FAT3 | Sponge network | -0.733 | 0.19469 | -1.601 | 0.00051 | 0.484 |
| 9848 | RP11-13K12.5 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-7-5p | 11 | FLNA | Sponge network | -0.318 | 0.65847 | -0.771 | 0.01377 | 0.484 |
| 9849 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 18 | HOOK3 | Sponge network | -0.737 | 0.06182 | 0.273 | 0.09957 | 0.484 |
| 9850 | RP11-693J15.4 |
hsa-miR-130b-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 10 | SORCS1 | Sponge network | -0.72 | 0.24239 | -3.492 | 0 | 0.484 |
| 9851 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-590-3p | 14 | JAKMIP2 | Sponge network | -1.057 | 0.00029 | -1.388 | 0 | 0.484 |
| 9852 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-582-5p | 17 | DDX6 | Sponge network | 0.826 | 1.0E-5 | 0.091 | 0.36428 | 0.484 |
| 9853 | USP3-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-576-5p | 11 | PREX2 | Sponge network | -0.162 | 0.47687 | -0.272 | 0.25382 | 0.484 |
| 9854 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 15 | ABCB5 | Sponge network | -0.323 | 0.01372 | -0.956 | 0.04077 | 0.484 |
| 9855 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 22 | NTRK3 | Sponge network | -0.021 | 0.93598 | -1.77 | 3.0E-5 | 0.484 |
| 9856 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 26 | SYNE1 | Sponge network | -0.793 | 7.0E-5 | -0.738 | 0.00316 | 0.484 |
| 9857 | RP11-701H24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 12 | RUNX1T1 | Sponge network | -2.039 | 0.03447 | -0.344 | 0.2374 | 0.484 |
| 9858 | AC009948.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p | 16 | ROBO1 | Sponge network | 0.532 | 0.00505 | -0.009 | 0.96154 | 0.484 |
| 9859 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | AHNAK | Sponge network | -0.096 | 0.67958 | -1.207 | 0 | 0.484 |
| 9860 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 18 | CAV1 | Sponge network | -1.575 | 6.0E-5 | -1.013 | 6.0E-5 | 0.484 |
| 9861 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | HECA | Sponge network | 0.083 | 0.66405 | -0.217 | 0.03463 | 0.484 |
| 9862 | RAMP2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 18 | CDH23 | Sponge network | -0.513 | 0.04703 | -0.974 | 0.00034 | 0.484 |
| 9863 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 12 | FYN | Sponge network | -0.323 | 0.01372 | -0.131 | 0.37051 | 0.484 |
| 9864 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | KAT6B | Sponge network | -1.111 | 0.0003 | -0.478 | 0.00018 | 0.484 |
| 9865 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 49 | PRKAA2 | Sponge network | -0.303 | 0.21002 | -2.462 | 0 | 0.483 |
| 9866 | GNG12-AS1 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374a-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577 | 10 | MLLT11 | Sponge network | -0.793 | 7.0E-5 | -0.661 | 0.01733 | 0.483 |
| 9867 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 49 | TRPS1 | Sponge network | -1.216 | 0 | -0.191 | 0.44109 | 0.483 |
| 9868 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-511-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | ST6GALNAC3 | Sponge network | -0.842 | 0.01361 | -0.987 | 0 | 0.483 |
| 9869 | RP11-13K12.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-769-5p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | -0.154 | 0.78939 | -1.733 | 0.00022 | 0.483 |
| 9870 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | RBMS3 | Sponge network | -1.057 | 0.00029 | -0.519 | 0.03551 | 0.483 |
| 9871 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p | 22 | AFF3 | Sponge network | -0.162 | 0.47687 | -2.373 | 0 | 0.483 |
| 9872 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 12 | CNTNAP3 | Sponge network | -0.358 | 0.17255 | -1.465 | 0.00033 | 0.483 |
| 9873 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-576-5p | 25 | PBX1 | Sponge network | -0.739 | 0.00044 | -1.206 | 0 | 0.483 |
| 9874 | LINC00641 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ITGB1 | Sponge network | -0.222 | 0.32445 | 0.524 | 0.00017 | 0.483 |
| 9875 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | NOTCH2 | Sponge network | 0.532 | 0.00505 | 0.138 | 0.35163 | 0.483 |
| 9876 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577 | 17 | ESR1 | Sponge network | -0.473 | 0.07602 | -1.035 | 3.0E-5 | 0.483 |
| 9877 | RAMP2-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-590-3p | 15 | SPG20 | Sponge network | -0.513 | 0.04703 | -1.16 | 0 | 0.483 |
| 9878 | HOTAIRM1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-766-3p;hsa-miR-93-5p | 28 | IL17RD | Sponge network | -0.053 | 0.80685 | -0.019 | 0.94873 | 0.483 |
| 9879 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-940 | 30 | MCC | Sponge network | -0.777 | 0.16667 | -1.005 | 0 | 0.483 |
| 9880 | ZNF582-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 15 | PRUNE2 | Sponge network | -1.349 | 0.00017 | -1.733 | 0.00022 | 0.483 |
| 9881 | WDFY3-AS2 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ADH1B | Sponge network | -0.942 | 0 | -3.716 | 0 | 0.483 |
| 9882 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CACNA2D1 | Sponge network | -0.513 | 0.04703 | -0.846 | 0.00675 | 0.483 |
| 9883 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 20 | RARB | Sponge network | -1.249 | 7.0E-5 | -0.825 | 2.0E-5 | 0.483 |
| 9884 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | 0.61 | 0.01593 | -0.474 | 0.03833 | 0.483 |
| 9885 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-93-3p | 17 | PTPN11 | Sponge network | 0.559 | 0.00026 | 0.431 | 2.0E-5 | 0.483 |
| 9886 | LINC00174 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-625-5p;hsa-miR-766-3p | 24 | MDM4 | Sponge network | 1.253 | 0 | 0.345 | 0.00762 | 0.483 |
| 9887 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ZMYND11 | Sponge network | -0.487 | 0.02157 | -0.2 | 0.05449 | 0.483 |
| 9888 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ZNF208 | Sponge network | -2.117 | 0.00161 | -0.4 | 0.22381 | 0.483 |
| 9889 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FSTL5 | Sponge network | -2.117 | 0.00161 | -1.3 | 0.01619 | 0.483 |
| 9890 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-935 | 10 | LRRN3 | Sponge network | -0.576 | 0.10121 | -0.393 | 0.16293 | 0.483 |
| 9891 | FENDRR |
hsa-miR-10b-3p;hsa-miR-193b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p | 10 | LRRTM1 | Sponge network | -1.281 | 0.00012 | -3.361 | 0 | 0.483 |
| 9892 | LINC00654 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | IL17RD | Sponge network | 0.374 | 0.20054 | -0.019 | 0.94873 | 0.483 |
| 9893 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-340-5p | 15 | MXI1 | Sponge network | -1.331 | 3.0E-5 | -0.987 | 0 | 0.483 |
| 9894 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | AHNAK | Sponge network | -1.111 | 0.0003 | -1.207 | 0 | 0.483 |
| 9895 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CLIP1 | Sponge network | -0.487 | 0.02157 | -0.215 | 0.08684 | 0.483 |
| 9896 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 36 | NOTCH2 | Sponge network | 0.214 | 0.18246 | 0.138 | 0.35163 | 0.483 |
| 9897 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | MYO9A | Sponge network | -0.303 | 0.21002 | -0.147 | 0.32373 | 0.483 |
| 9898 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p | 12 | RCAN2 | Sponge network | -0.246 | 0.35034 | -0.33 | 0.15172 | 0.483 |
| 9899 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 48 | NTRK2 | Sponge network | -0.49 | 0.04946 | -0.736 | 0.06495 | 0.483 |
| 9900 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p | 28 | IL17RD | Sponge network | -0.318 | 0.65847 | -0.019 | 0.94873 | 0.483 |
| 9901 | RP11-473I1.10 |
hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p | 12 | DDX3X | Sponge network | 0.673 | 0 | -0.152 | 0.07686 | 0.483 |
| 9902 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-877-5p | 27 | PGR | Sponge network | -0.246 | 0.35034 | -1.704 | 0 | 0.483 |
| 9903 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940;hsa-miR-98-5p | 61 | CADM2 | Sponge network | -0.899 | 6.0E-5 | -3.782 | 0 | 0.483 |
| 9904 | PART1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 24 | DYNC1I1 | Sponge network | -2.925 | 0 | -1.12 | 0.00107 | 0.483 |
| 9905 | WDFY3-AS2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | PRUNE2 | Sponge network | -0.942 | 0 | -1.733 | 0.00022 | 0.483 |
| 9906 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-486-5p;hsa-miR-590-3p | 15 | NR2C2 | Sponge network | 0.545 | 0.05789 | 0.21 | 0.03224 | 0.483 |
| 9907 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | RAP1A | Sponge network | -0.323 | 0.01372 | -0.929 | 0 | 0.483 |
| 9908 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 31 | NTRK3 | Sponge network | -0.376 | 0.00337 | -1.77 | 3.0E-5 | 0.483 |
| 9909 | LINC00865 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-429 | 11 | PARK2 | Sponge network | -1.249 | 7.0E-5 | -1.892 | 0 | 0.483 |
| 9910 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-98-5p | 17 | LRIG1 | Sponge network | -2.62 | 0 | -0.537 | 0.00588 | 0.483 |
| 9911 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | EBF1 | Sponge network | -0.709 | 0.18168 | -0.384 | 0.08856 | 0.483 |
| 9912 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-339-5p | 11 | FBXO31 | Sponge network | -0.583 | 0.14589 | -0.085 | 0.42389 | 0.483 |
| 9913 | SNHG16 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p | 11 | PAWR | Sponge network | 0.961 | 0 | 0.636 | 0 | 0.483 |
| 9914 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 14 | LRIG1 | Sponge network | -0.358 | 0.17255 | -0.537 | 0.00588 | 0.483 |
| 9915 | CTC-296K1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-671-5p | 22 | DLC1 | Sponge network | -2.095 | 0.00112 | -0.382 | 0.05038 | 0.483 |
| 9916 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-33a-3p | 12 | CLASP2 | Sponge network | 0.542 | 0.00054 | -0.038 | 0.71 | 0.483 |
| 9917 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-96-5p | 13 | CBFA2T3 | Sponge network | -1.582 | 8.0E-5 | -1.221 | 1.0E-5 | 0.483 |
| 9918 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 13 | DCLK1 | Sponge network | -0.612 | 0.00094 | -1.324 | 0.00014 | 0.483 |
| 9919 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | UVRAG | Sponge network | 0.225 | 0.30644 | -0.017 | 0.83865 | 0.483 |
| 9920 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p | 23 | ZMYND11 | Sponge network | -0.793 | 7.0E-5 | -0.2 | 0.05449 | 0.483 |
| 9921 | RP11-144G6.12 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 25 | MDM4 | Sponge network | 0.826 | 1.0E-5 | 0.345 | 0.00762 | 0.483 |
| 9922 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-93-5p;hsa-miR-96-5p | 25 | MAF | Sponge network | -1.349 | 0.00017 | -1.257 | 0 | 0.483 |
| 9923 | BOLA3-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-877-5p | 16 | NRK | Sponge network | -0.246 | 0.35034 | 0.656 | 0.19217 | 0.483 |
| 9924 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 24 | PTPN11 | Sponge network | 0.421 | 0.00084 | 0.431 | 2.0E-5 | 0.483 |
| 9925 | RP11-531A24.5 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429 | 10 | MACF1 | Sponge network | 0.75 | 0.00054 | 0.019 | 0.90335 | 0.483 |
| 9926 | ADAMTS9-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-708-5p;hsa-miR-744-3p;hsa-miR-940 | 30 | TOM1L2 | Sponge network | -2.452 | 0 | -0.994 | 0 | 0.483 |
| 9927 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ZMYM2 | Sponge network | 0.953 | 0 | 0.279 | 0.01273 | 0.483 |
| 9928 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 44 | RBMS3 | Sponge network | -0.899 | 6.0E-5 | -0.519 | 0.03551 | 0.483 |
| 9929 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | RECK | Sponge network | -0.612 | 0.00094 | -1.043 | 0 | 0.483 |
| 9930 | RP11-166D19.1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | SYT4 | Sponge network | -0.576 | 0.10121 | -3.207 | 0 | 0.483 |
| 9931 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 14 | GIGYF2 | Sponge network | 1.601 | 0 | 0.136 | 0.04403 | 0.483 |
| 9932 | MIR497HG |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p | 16 | TLE4 | Sponge network | -1.439 | 4.0E-5 | -1.157 | 0 | 0.483 |
| 9933 | FENDRR |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-590-3p | 20 | STARD13 | Sponge network | -1.281 | 0.00012 | -0.187 | 0.27383 | 0.483 |
| 9934 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-96-5p | 14 | EBF3 | Sponge network | -1.654 | 0.00144 | -0.058 | 0.81437 | 0.483 |
| 9935 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 47 | NOTCH2 | Sponge network | -0.739 | 0.0574 | 0.138 | 0.35163 | 0.483 |
| 9936 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 13 | BCLAF1 | Sponge network | 1.601 | 0 | 0.211 | 0.00684 | 0.483 |
| 9937 | RP11-867G23.10 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-942-5p | 31 | THRB | Sponge network | -2.531 | 0 | -1.117 | 0.0004 | 0.483 |
| 9938 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 30 | TSHZ3 | Sponge network | 0.16 | 0.50785 | -0.556 | 0.01516 | 0.483 |
| 9939 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-625-5p;hsa-miR-93-5p | 22 | ANK2 | Sponge network | 0.056 | 0.81195 | -1.603 | 1.0E-5 | 0.483 |
| 9940 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 13 | GJA1 | Sponge network | 0.811 | 0.03228 | -0.485 | 0.02179 | 0.483 |
| 9941 | RP11-503E24.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-495-3p | 13 | RICTOR | Sponge network | 2.218 | 0 | 0.388 | 0.00188 | 0.483 |
| 9942 | RP11-554A11.4 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-423-5p | 30 | CCND2 | Sponge network | -0.583 | 0.14589 | -0.267 | 0.3112 | 0.483 |
| 9943 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 35 | PBX1 | Sponge network | -0.563 | 0.02545 | -1.206 | 0 | 0.483 |
| 9944 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 27 | RSPO2 | Sponge network | -0.901 | 0.00019 | -3.038 | 0 | 0.483 |
| 9945 | RP11-166D19.1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | PCDH18 | Sponge network | -0.576 | 0.10121 | 0.216 | 0.24645 | 0.483 |
| 9946 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-769-5p | 21 | SYNPO2 | Sponge network | 0.331 | 0.57247 | -2.342 | 0 | 0.483 |
| 9947 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-935 | 11 | ABI2 | Sponge network | -0.691 | 0.00146 | 0.175 | 0.14787 | 0.483 |
| 9948 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | MYO9A | Sponge network | 0.225 | 0.30644 | -0.147 | 0.32373 | 0.483 |
| 9949 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | RNF144A | Sponge network | -0.49 | 0.04946 | 0.594 | 0.0009 | 0.483 |
| 9950 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-7-5p | 15 | TET1 | Sponge network | -1.331 | 3.0E-5 | -0.135 | 0.52138 | 0.483 |
| 9951 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 0.349 | 0.01695 | 0.114 | 0.30638 | 0.483 |
| 9952 | CTC-429P9.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-625-3p | 15 | ZFHX3 | Sponge network | 0.497 | 0.01451 | 0.389 | 0.01363 | 0.483 |
| 9953 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-942-5p | 15 | GPC6 | Sponge network | -1.092 | 4.0E-5 | -0.054 | 0.81465 | 0.483 |
| 9954 | PART1 |
hsa-miR-182-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-769-5p | 10 | TCEAL7 | Sponge network | -2.925 | 0 | -1.445 | 0 | 0.483 |
| 9955 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 48 | DTNA | Sponge network | -0.769 | 0.00035 | -1.648 | 1.0E-5 | 0.483 |
| 9956 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 42 | BNC2 | Sponge network | -1.249 | 7.0E-5 | -0.491 | 0.09988 | 0.483 |
| 9957 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 19 | SPTBN4 | Sponge network | -1.697 | 0.00889 | -0.786 | 0.01281 | 0.483 |
| 9958 | LINC00578 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-374a-3p | 10 | GLIPR1 | Sponge network | 0.811 | 0.03228 | 0.146 | 0.3276 | 0.483 |
| 9959 | AP001347.6 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-7-5p | 10 | FLNA | Sponge network | -1.161 | 0.00591 | -0.771 | 0.01377 | 0.483 |
| 9960 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 39 | MEF2C | Sponge network | -0.487 | 0.02157 | -0.499 | 0.01448 | 0.483 |
| 9961 | PART1 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | NOV | Sponge network | -2.925 | 0 | -0.58 | 0.04676 | 0.483 |
| 9962 | PWAR6 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 19 | STARD13 | Sponge network | -1.575 | 6.0E-5 | -0.187 | 0.27383 | 0.483 |
| 9963 | RP11-6O2.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-511-5p | 17 | MRVI1 | Sponge network | 0.331 | 0.57247 | -1.051 | 0.00022 | 0.483 |
| 9964 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | SYNE1 | Sponge network | -0.513 | 0.04703 | -0.738 | 0.00316 | 0.483 |
| 9965 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 42 | SASH1 | Sponge network | -0.737 | 0.06182 | -0.502 | 0.00175 | 0.483 |
| 9966 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PLSCR4 | Sponge network | -0.323 | 0.01372 | -0.474 | 0.03833 | 0.483 |
| 9967 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 24 | CRTC3 | Sponge network | -0.563 | 0.02545 | 0.164 | 0.16786 | 0.483 |
| 9968 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-484 | 14 | ITGA5 | Sponge network | -2.69 | 0.0001 | 0.452 | 0.03524 | 0.482 |
| 9969 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 20 | CAV1 | Sponge network | -1.216 | 0 | -1.013 | 6.0E-5 | 0.482 |
| 9970 | RP11-400K9.3 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-22-5p;hsa-miR-576-5p;hsa-miR-93-3p;hsa-miR-940 | 10 | QKI | Sponge network | -0.733 | 0.19469 | -0.333 | 0.04111 | 0.482 |
| 9971 | FAM157C |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-766-3p | 17 | MDM4 | Sponge network | 0.91 | 0 | 0.345 | 0.00762 | 0.482 |
| 9972 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-940 | 16 | IGF1 | Sponge network | -2.095 | 0.00112 | -0.204 | 0.5898 | 0.482 |
| 9973 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-590-3p | 13 | TSHZ3 | Sponge network | -1.375 | 0.08642 | -0.556 | 0.01516 | 0.482 |
| 9974 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-590-5p | 17 | FNBP1 | Sponge network | -0.615 | 0.00015 | -0.969 | 4.0E-5 | 0.482 |
| 9975 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | -1.216 | 0 | -1.047 | 0.00364 | 0.482 |
| 9976 | MEG3 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 13 | LZTS1 | Sponge network | 0.249 | 0.35902 | 1.664 | 0 | 0.482 |
| 9977 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 27 | ZBTB4 | Sponge network | -1.654 | 0.00144 | -0.739 | 0 | 0.482 |
| 9978 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 25 | TIMP3 | Sponge network | -0.563 | 0.02545 | -0.546 | 0.02307 | 0.482 |
| 9979 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | LRRC7 | Sponge network | -0.777 | 0.1134 | -1.341 | 0.01193 | 0.482 |
| 9980 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | DMTF1 | Sponge network | 0.311 | 0.10344 | 0.248 | 0.02726 | 0.482 |
| 9981 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-590-3p | 17 | CACNA2D1 | Sponge network | 0.214 | 0.18246 | -0.846 | 0.00675 | 0.482 |
| 9982 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 12 | THSD7A | Sponge network | -2.46 | 0 | 0.242 | 0.25154 | 0.482 |
| 9983 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 27 | TGFBR3 | Sponge network | -0.358 | 0.17255 | -1.173 | 1.0E-5 | 0.482 |
| 9984 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | NOTCH2 | Sponge network | -0.611 | 0.09607 | 0.138 | 0.35163 | 0.482 |
| 9985 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-98-5p | 58 | NTRK3 | Sponge network | -0.154 | 0.78939 | -1.77 | 3.0E-5 | 0.482 |
| 9986 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 29 | SCN9A | Sponge network | 0.246 | 0.59912 | -0.525 | 0.10593 | 0.482 |
| 9987 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 25 | NTRK3 | Sponge network | -1.255 | 0.00981 | -1.77 | 3.0E-5 | 0.482 |
| 9988 | THAP9-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-23b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 12 | BCLAF1 | Sponge network | 1.079 | 0 | 0.211 | 0.00684 | 0.482 |
| 9989 | EMX2OS |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 66 | DST | Sponge network | -0.777 | 0.1134 | -0.713 | 0.00096 | 0.482 |
| 9990 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-5p | 17 | PDZD2 | Sponge network | -1.216 | 0 | -0.909 | 3.0E-5 | 0.482 |
| 9991 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | CNTN1 | Sponge network | -1.203 | 0.03881 | -2.594 | 0 | 0.482 |
| 9992 | AP001055.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-455-3p | 10 | NRK | Sponge network | 0.693 | 0.00076 | 0.656 | 0.19217 | 0.482 |
| 9993 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p | 34 | ST6GAL2 | Sponge network | -1.575 | 6.0E-5 | -1.201 | 0.00597 | 0.482 |
| 9994 | AC005154.6 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 19 | ATM | Sponge network | 0.848 | 0 | 0.178 | 0.21568 | 0.482 |
| 9995 | RP11-774O3.3 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | THSD7B | Sponge network | -0.487 | 0.02157 | 0.154 | 0.74723 | 0.482 |
| 9996 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-550a-5p | 24 | PDGFRA | Sponge network | -0.469 | 0.08721 | -0.372 | 0.0037 | 0.482 |
| 9997 | LINC01018 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | -2.915 | 0.00015 | -1.09 | 0.00014 | 0.482 |
| 9998 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 10 | FAT4 | Sponge network | -1.582 | 8.0E-5 | -0.354 | 0.14694 | 0.482 |
| 9999 | AC106786.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-576-5p | 16 | ZFHX4 | Sponge network | 1.122 | 0.02989 | 0.018 | 0.95574 | 0.482 |
| 10000 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ABCC9 | Sponge network | 0.16 | 0.50785 | -0.466 | 0.14121 | 0.482 |
| 10001 | LINC00641 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 20 | KLF10 | Sponge network | -0.222 | 0.32445 | -0.783 | 0 | 0.482 |
| 10002 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-96-5p | 42 | FAT3 | Sponge network | 0.331 | 0.57247 | -1.601 | 0.00051 | 0.482 |
| 10003 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p | 20 | CBFA2T3 | Sponge network | -0.421 | 0.0114 | -1.221 | 1.0E-5 | 0.482 |
| 10004 | RP11-307B6.3 |
hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-424-5p | 12 | LRIG1 | Sponge network | -4.463 | 0.00025 | -0.537 | 0.00588 | 0.482 |
| 10005 | TP73-AS1 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-96-5p | 10 | MSN | Sponge network | -0.563 | 0.02545 | -0.023 | 0.88422 | 0.482 |
| 10006 | RAMP2-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 10 | NGFR | Sponge network | -0.513 | 0.04703 | -1.271 | 0.01058 | 0.482 |
| 10007 | ACTA2-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-940 | 10 | PHOX2B | Sponge network | 0.194 | 0.64278 | -2.68 | 0 | 0.482 |
| 10008 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CACNA2D1 | Sponge network | -0.222 | 0.32445 | -0.846 | 0.00675 | 0.482 |
| 10009 | NR2F1-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 17 | INPP4A | Sponge network | -0.49 | 0.04946 | 0.07 | 0.53563 | 0.482 |
| 10010 | AC003090.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 31 | DLC1 | Sponge network | -2.117 | 0.00161 | -0.382 | 0.05038 | 0.482 |
| 10011 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-421;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-940 | 43 | GAS7 | Sponge network | 0.214 | 0.18246 | -0.555 | 0.01602 | 0.482 |
| 10012 | RP11-244O19.1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | TPO | Sponge network | -0.096 | 0.67958 | -1.87 | 2.0E-5 | 0.482 |
| 10013 | RP11-356J5.12 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 14 | MAPK10 | Sponge network | -0.899 | 6.0E-5 | -1.774 | 0 | 0.482 |
| 10014 | LINC00865 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p | 10 | CNN1 | Sponge network | -1.249 | 7.0E-5 | -2.633 | 0 | 0.482 |
| 10015 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 21 | EBF3 | Sponge network | -0.513 | 0.04703 | -0.058 | 0.81437 | 0.482 |
| 10016 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | -1.886 | 0 | 0.358 | 0.13727 | 0.482 |
| 10017 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | PDGFRA | Sponge network | -1.203 | 0.03881 | -0.372 | 0.0037 | 0.482 |
| 10018 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 33 | MCC | Sponge network | -0.096 | 0.67958 | -1.005 | 0 | 0.482 |
| 10019 | MEG3 |
hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-96-5p | 12 | FHL1 | Sponge network | 0.249 | 0.35902 | -2.687 | 0 | 0.482 |
| 10020 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 34 | FOXP1 | Sponge network | 0.782 | 0.00016 | 0.042 | 0.74368 | 0.482 |
| 10021 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 26 | RICTOR | Sponge network | 1.317 | 0 | 0.388 | 0.00188 | 0.482 |
| 10022 | LINC00657 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | HIP1 | Sponge network | 0.91 | 0 | 0.774 | 0 | 0.482 |
| 10023 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 18 | CNTN1 | Sponge network | -0.318 | 0.65847 | -2.594 | 0 | 0.482 |
| 10024 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-5p;hsa-miR-590-5p | 35 | SOX5 | Sponge network | -0.384 | 0.40652 | -1.091 | 4.0E-5 | 0.482 |
| 10025 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | RET | Sponge network | -0.739 | 0.0574 | -0.833 | 0.03517 | 0.482 |
| 10026 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 46 | ADARB1 | Sponge network | 0.083 | 0.66405 | -0.335 | 0.06631 | 0.482 |
| 10027 | DNM3OS |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-429;hsa-miR-455-5p | 14 | PLXNC1 | Sponge network | 0.572 | 0.01722 | 0.569 | 0.00329 | 0.482 |
| 10028 | FAM66C |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 45 | NTRK2 | Sponge network | -0.901 | 0.00019 | -0.736 | 0.06495 | 0.482 |
| 10029 | RP11-53O19.3 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | BRD3 | Sponge network | 1.205 | 0 | 0.37 | 0.00013 | 0.482 |
| 10030 | RP11-25K19.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-99b-3p | 30 | GRIK3 | Sponge network | -1.057 | 0.00029 | -3.208 | 0 | 0.482 |
| 10031 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-374b-5p | 29 | KIF1B | Sponge network | 0.542 | 0.00054 | -0.118 | 0.32258 | 0.482 |
| 10032 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | -0.769 | 0.00035 | -0.7 | 1.0E-5 | 0.482 |
| 10033 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p | 15 | NCAM2 | Sponge network | -0.777 | 0.16667 | -0.482 | 0.22565 | 0.482 |
| 10034 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 22 | CACNA2D1 | Sponge network | -1.249 | 7.0E-5 | -0.846 | 0.00675 | 0.482 |
| 10035 | PART1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | SFRP1 | Sponge network | -2.925 | 0 | -3.566 | 0 | 0.482 |
| 10036 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 12 | CHD2 | Sponge network | 2.094 | 0 | 0.049 | 0.55371 | 0.482 |
| 10037 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | ZFHX3 | Sponge network | 0.392 | 0.0278 | 0.389 | 0.01363 | 0.482 |
| 10038 | RP11-822E23.8 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p | 10 | SORCS2 | Sponge network | -0.777 | 0.16667 | -0.223 | 0.4253 | 0.482 |
| 10039 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-7-5p | 13 | PTGFR | Sponge network | -0.465 | 0.04703 | -0.233 | 0.45421 | 0.482 |
| 10040 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 48 | CREB3L2 | Sponge network | -1.111 | 0.0003 | -0.525 | 2.0E-5 | 0.482 |
| 10041 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 33 | PHC3 | Sponge network | 0.848 | 0 | 0.212 | 0.0499 | 0.482 |
| 10042 | FAM13A-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-3913-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 37 | AFF1 | Sponge network | -0.83 | 0 | -0.384 | 0.00053 | 0.482 |
| 10043 | MALAT1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 10 | PTPN12 | Sponge network | 0.782 | 0.00016 | 0.768 | 0 | 0.482 |
| 10044 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CLIP1 | Sponge network | -0.576 | 0.10121 | -0.215 | 0.08684 | 0.482 |
| 10045 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577 | 21 | GREM1 | Sponge network | -2.452 | 0 | 0.902 | 0.01691 | 0.482 |
| 10046 | RP11-554A11.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 22 | ZFHX3 | Sponge network | -0.583 | 0.14589 | 0.389 | 0.01363 | 0.482 |
| 10047 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429 | 17 | STARD13 | Sponge network | -0.976 | 0.00025 | -0.187 | 0.27383 | 0.482 |
| 10048 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | RCAN2 | Sponge network | -1.161 | 0.00591 | -0.33 | 0.15172 | 0.482 |
| 10049 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 15 | RB1CC1 | Sponge network | 0.953 | 0 | 0.175 | 0.12559 | 0.482 |
| 10050 | RP11-531A24.5 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-655-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 14 | SHPRH | Sponge network | 0.75 | 0.00054 | 0.098 | 0.40994 | 0.482 |
| 10051 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 14 | AMPH | Sponge network | -0.323 | 0.01372 | -0.916 | 5.0E-5 | 0.482 |
| 10052 | RP11-121C2.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-582-5p | 11 | PHF3 | Sponge network | 1.147 | 0 | 0.126 | 0.19652 | 0.482 |
| 10053 | PWAR6 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 65 | DST | Sponge network | -1.575 | 6.0E-5 | -0.713 | 0.00096 | 0.482 |
| 10054 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 49 | AR | Sponge network | -0.738 | 0.21233 | -1.554 | 0 | 0.482 |
| 10055 | LINC00476 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-590-3p | 18 | STARD13 | Sponge network | -0.615 | 0.00015 | -0.187 | 0.27383 | 0.482 |
| 10056 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-93-5p | 15 | FGFR1 | Sponge network | 0.253 | 0.09565 | -0.509 | 0.03957 | 0.482 |
| 10057 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 25 | EBF3 | Sponge network | -2.117 | 0.00161 | -0.058 | 0.81437 | 0.482 |
| 10058 | PCA3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | DYNC1I1 | Sponge network | -0.709 | 0.18168 | -1.12 | 0.00107 | 0.482 |
| 10059 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-629-5p | 36 | BCL2 | Sponge network | -2.69 | 0.0001 | -0.899 | 0 | 0.482 |
| 10060 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | CUL5 | Sponge network | 0.953 | 0 | 0.026 | 0.77301 | 0.482 |
| 10061 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-93-5p | 29 | ZBTB4 | Sponge network | 0.101 | 0.60919 | -0.739 | 0 | 0.482 |
| 10062 | LINC00476 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935 | 92 | LPP | Sponge network | -0.615 | 0.00015 | -0.459 | 0.04475 | 0.482 |
| 10063 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-582-5p | 13 | GRIN2A | Sponge network | -0.469 | 0.08721 | -2.161 | 0 | 0.481 |
| 10064 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-33a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 39 | SSBP2 | Sponge network | -0.611 | 0.09607 | -1.58 | 0 | 0.481 |
| 10065 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 19 | NRK | Sponge network | -0.739 | 0.00044 | 0.656 | 0.19217 | 0.481 |
| 10066 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p | 11 | FBXO32 | Sponge network | -0.285 | 0.2172 | -0.495 | 0.07561 | 0.481 |
| 10067 | HHIP-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 10 | SFRP1 | Sponge network | -0.737 | 0.10822 | -3.566 | 0 | 0.481 |
| 10068 | RP4-639F20.1 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | EPHA3 | Sponge network | -0.517 | 0.02935 | 0.185 | 0.53588 | 0.481 |
| 10069 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-629-5p;hsa-miR-940 | 29 | IGF1 | Sponge network | -0.563 | 0.02545 | -0.204 | 0.5898 | 0.481 |
| 10070 | AP001347.6 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | DIP2C | Sponge network | -1.161 | 0.00591 | -0.436 | 0.01713 | 0.481 |
| 10071 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-96-5p | 14 | MITF | Sponge network | -1.582 | 8.0E-5 | -0.769 | 0.00022 | 0.481 |
| 10072 | RAMP2-AS1 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 12 | PEG3 | Sponge network | -0.513 | 0.04703 | -1.74 | 0 | 0.481 |
| 10073 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | SCN9A | Sponge network | -1.582 | 8.0E-5 | -0.525 | 0.10593 | 0.481 |
| 10074 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-93-5p | 11 | EDA2R | Sponge network | -0.611 | 0.09607 | -0.587 | 0.01598 | 0.481 |
| 10075 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 18 | CDH19 | Sponge network | -0.563 | 0.02545 | -3.434 | 0 | 0.481 |
| 10076 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 44 | RUNX1T1 | Sponge network | -0.405 | 0.22013 | -0.344 | 0.2374 | 0.481 |
| 10077 | RP11-48B3.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 20 | ABI2 | Sponge network | 1.757 | 0 | 0.175 | 0.14787 | 0.481 |
| 10078 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-93-3p | 16 | ST6GAL2 | Sponge network | -0.376 | 0.00337 | -1.201 | 0.00597 | 0.481 |
| 10079 | H19 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-511-5p | 16 | ZFHX3 | Sponge network | 2.439 | 0 | 0.389 | 0.01363 | 0.481 |
| 10080 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-421;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 15 | TLL1 | Sponge network | -0.793 | 7.0E-5 | -1.195 | 3.0E-5 | 0.481 |
| 10081 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PHC3 | Sponge network | 1.308 | 0 | 0.212 | 0.0499 | 0.481 |
| 10082 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 24 | CDH19 | Sponge network | -0.942 | 0 | -3.434 | 0 | 0.481 |
| 10083 | CTA-204B4.2 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 10 | PHF3 | Sponge network | 0.765 | 7.0E-5 | 0.126 | 0.19652 | 0.481 |
| 10084 | RP11-57H14.4 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-935 | 10 | STAT5B | Sponge network | 0.083 | 0.66405 | -0.182 | 0.1082 | 0.481 |
| 10085 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-93-5p | 20 | IL17RD | Sponge network | -2.531 | 0 | -0.019 | 0.94873 | 0.481 |
| 10086 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | PCDH17 | Sponge network | 1.253 | 0 | 0.731 | 2.0E-5 | 0.481 |
| 10087 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | EDA2R | Sponge network | 0.532 | 0.00505 | -0.587 | 0.01598 | 0.481 |
| 10088 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 16 | CSRNP1 | Sponge network | -0.769 | 0.00035 | -1.448 | 0 | 0.481 |
| 10089 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p | 25 | PTPN14 | Sponge network | 0.214 | 0.18246 | 0.144 | 0.34595 | 0.481 |
| 10090 | LINC00672 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | PRUNE2 | Sponge network | -0.227 | 0.31657 | -1.733 | 0.00022 | 0.481 |
| 10091 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 53 | ADARB1 | Sponge network | -0.615 | 0.00015 | -0.335 | 0.06631 | 0.481 |
| 10092 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 15 | TIMP3 | Sponge network | -1.375 | 0.08642 | -0.546 | 0.02307 | 0.481 |
| 10093 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 30 | RICTOR | Sponge network | 0.225 | 0.30644 | 0.388 | 0.00188 | 0.481 |
| 10094 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-495-3p | 18 | CLIP1 | Sponge network | 0.331 | 0.57247 | -0.215 | 0.08684 | 0.481 |
| 10095 | CTC-444N24.11 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-409-5p;hsa-miR-495-3p | 11 | STAG2 | Sponge network | 1.153 | 0 | 0.252 | 0.00438 | 0.481 |
| 10096 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-223-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-935;hsa-miR-96-5p | 30 | LPP | Sponge network | 0.37 | 0.27614 | -0.459 | 0.04475 | 0.481 |
| 10097 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-3p | 17 | PTPN11 | Sponge network | 0.083 | 0.66405 | 0.431 | 2.0E-5 | 0.481 |
| 10098 | NR2F1-AS1 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | DIRAS1 | Sponge network | -0.49 | 0.04946 | -2.518 | 0 | 0.481 |
| 10099 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 18 | GPAM | Sponge network | 0.91 | 0 | 0.142 | 0.29732 | 0.481 |
| 10100 | PCA3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-940 | 38 | GRIK3 | Sponge network | -0.709 | 0.18168 | -3.208 | 0 | 0.481 |
| 10101 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-92a-3p | 18 | SPTBN4 | Sponge network | -2.452 | 0 | -0.786 | 0.01281 | 0.481 |
| 10102 | PCA3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | FAT4 | Sponge network | -0.709 | 0.18168 | -0.354 | 0.14694 | 0.481 |
| 10103 | RP11-473I1.10 |
hsa-miR-125a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | ERCC6 | Sponge network | 0.673 | 0 | 0.23 | 0.015 | 0.481 |
| 10104 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p | 10 | THSD7A | Sponge network | -0.777 | 0.16667 | 0.242 | 0.25154 | 0.481 |
| 10105 | RP11-554A11.4 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p | 12 | PCDH17 | Sponge network | -0.583 | 0.14589 | 0.731 | 2.0E-5 | 0.481 |
| 10106 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-7-1-3p | 12 | KCNAB1 | Sponge network | 0.101 | 0.60919 | -0.755 | 2.0E-5 | 0.481 |
| 10107 | AC003090.1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | STAT5B | Sponge network | -2.117 | 0.00161 | -0.182 | 0.1082 | 0.481 |
| 10108 | RP11-434B12.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | SIRT1 | Sponge network | -0.031 | 0.89063 | 0.109 | 0.2593 | 0.481 |
| 10109 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-199b-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | ZMYND11 | Sponge network | 0.75 | 0.00054 | -0.2 | 0.05449 | 0.481 |
| 10110 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 16 | HOOK3 | Sponge network | 0.064 | 0.72934 | 0.273 | 0.09957 | 0.481 |
| 10111 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PHC3 | Sponge network | 0.873 | 0 | 0.212 | 0.0499 | 0.481 |
| 10112 | HOXA-AS2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | WWTR1 | Sponge network | 0.61 | 0.01593 | -0.345 | 0.11997 | 0.481 |
| 10113 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 23 | TRPS1 | Sponge network | -2.531 | 0 | -0.191 | 0.44109 | 0.481 |
| 10114 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | PTGFR | Sponge network | -2.117 | 0.00161 | -0.233 | 0.45421 | 0.481 |
| 10115 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p | 17 | PTGFR | Sponge network | -0.793 | 7.0E-5 | -0.233 | 0.45421 | 0.481 |
| 10116 | ANKRD10-IT1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-32-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 10 | KIF5B | Sponge network | 1.739 | 0 | 0.362 | 0.00066 | 0.481 |
| 10117 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 32 | CRTC3 | Sponge network | -1.111 | 0.0003 | 0.164 | 0.16786 | 0.481 |
| 10118 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 12 | NALCN | Sponge network | 0.374 | 0.20054 | 1.068 | 0.00237 | 0.481 |
| 10119 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 23 | TET1 | Sponge network | 0.214 | 0.18246 | -0.135 | 0.52138 | 0.481 |
| 10120 | RP11-25K19.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ITPKB | Sponge network | -1.057 | 0.00029 | -0.704 | 0.00106 | 0.481 |
| 10121 | RP5-1092A3.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-766-3p;hsa-miR-940 | 15 | MDM4 | Sponge network | 1.495 | 0 | 0.345 | 0.00762 | 0.481 |
| 10122 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | TAL1 | Sponge network | -0.49 | 0.04946 | -0.462 | 0.00574 | 0.481 |
| 10123 | PWAR6 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NRG3 | Sponge network | -1.575 | 6.0E-5 | -1.836 | 4.0E-5 | 0.481 |
| 10124 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | PLSCR4 | Sponge network | -0.318 | 0.65847 | -0.474 | 0.03833 | 0.481 |
| 10125 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-96-5p | 58 | CADM2 | Sponge network | 0.194 | 0.64278 | -3.782 | 0 | 0.481 |
| 10126 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 22 | AKAP6 | Sponge network | 0.494 | 0.00339 | -1.586 | 3.0E-5 | 0.481 |
| 10127 | AC004893.11 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 24 | ABL2 | Sponge network | 1.317 | 0 | 0.82 | 0 | 0.481 |
| 10128 | LINC00865 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p | 10 | SCN11A | Sponge network | -1.249 | 7.0E-5 | -1.15 | 0.00299 | 0.481 |
| 10129 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NCAM2 | Sponge network | -0.021 | 0.93598 | -0.482 | 0.22565 | 0.481 |
| 10130 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-324-3p;hsa-miR-629-3p | 13 | PTPN14 | Sponge network | -0.583 | 0.14589 | 0.144 | 0.34595 | 0.481 |
| 10131 | TP73-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-542-3p | 10 | ALDH1A2 | Sponge network | -0.563 | 0.02545 | -2.411 | 0 | 0.481 |
| 10132 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 30 | NRXN1 | Sponge network | -0.096 | 0.67958 | -3.778 | 0 | 0.481 |
| 10133 | AC003090.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PTPN13 | Sponge network | -2.117 | 0.00161 | -1.208 | 0 | 0.481 |
| 10134 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 37 | FBXO32 | Sponge network | -0.899 | 6.0E-5 | -0.495 | 0.07561 | 0.481 |
| 10135 | USP3-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-589-3p;hsa-miR-653-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 42 | PPM1L | Sponge network | -0.162 | 0.47687 | -0.537 | 0.00616 | 0.481 |
| 10136 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-660-5p | 42 | RUNX1T1 | Sponge network | -0.246 | 0.35034 | -0.344 | 0.2374 | 0.481 |
| 10137 | SNHG14 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 20 | ID4 | Sponge network | -1.111 | 0.0003 | -1.644 | 0 | 0.481 |
| 10138 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p | 18 | SCN9A | Sponge network | -0.693 | 0.05629 | -0.525 | 0.10593 | 0.481 |
| 10139 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 31 | LRIG1 | Sponge network | -0.576 | 0.10121 | -0.537 | 0.00588 | 0.481 |
| 10140 | RP11-540B6.6 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 18 | ABL2 | Sponge network | 0.93 | 0 | 0.82 | 0 | 0.481 |
| 10141 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 25 | SNX29 | Sponge network | -2.69 | 0.0001 | -0.34 | 0.01371 | 0.481 |
| 10142 | CTD-2314G24.2 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p | 15 | FLNA | Sponge network | 0.246 | 0.59912 | -0.771 | 0.01377 | 0.481 |
| 10143 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 62 | NTRK3 | Sponge network | -0.709 | 0.18168 | -1.77 | 3.0E-5 | 0.481 |
| 10144 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-5p | 28 | DIP2C | Sponge network | 0.331 | 0.57247 | -0.436 | 0.01713 | 0.481 |
| 10145 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-93-5p | 22 | ANK2 | Sponge network | 0.082 | 0.55397 | -1.603 | 1.0E-5 | 0.481 |
| 10146 | RAD51-AS1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p | 13 | SHPRH | Sponge network | 0.768 | 7.0E-5 | 0.098 | 0.40994 | 0.481 |
| 10147 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p | 16 | TXNIP | Sponge network | -0.901 | 0.00019 | -1.277 | 0 | 0.481 |
| 10148 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 46 | EPHA7 | Sponge network | -0.487 | 0.02157 | -2.197 | 3.0E-5 | 0.481 |
| 10149 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 17 | ZMYM2 | Sponge network | 1.4 | 0 | 0.279 | 0.01273 | 0.481 |
| 10150 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | FAT4 | Sponge network | -0.811 | 2.0E-5 | -0.354 | 0.14694 | 0.481 |
| 10151 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | CREBBP | Sponge network | 0.494 | 0.00339 | 0.126 | 0.15186 | 0.481 |
| 10152 | LINC00957 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | EPHA7 | Sponge network | -0.421 | 0.0114 | -2.197 | 3.0E-5 | 0.481 |
| 10153 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-486-5p;hsa-miR-495-3p | 18 | PHF6 | Sponge network | 1.254 | 0 | 0.785 | 0 | 0.481 |
| 10154 | RP5-1198O20.4 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 21 | FMNL3 | Sponge network | 0.997 | 0.00066 | 0.555 | 4.0E-5 | 0.481 |
| 10155 | LINC00957 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 16 | CRTC3 | Sponge network | -0.421 | 0.0114 | 0.164 | 0.16786 | 0.481 |
| 10156 | RP11-57H14.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 25 | PTPN14 | Sponge network | 0.083 | 0.66405 | 0.144 | 0.34595 | 0.481 |
| 10157 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-424-5p | 20 | PHC3 | Sponge network | 0.082 | 0.55397 | 0.212 | 0.0499 | 0.481 |
| 10158 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-744-3p | 20 | CUL5 | Sponge network | 1.153 | 0 | 0.026 | 0.77301 | 0.481 |
| 10159 | FAM157B |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326 | 10 | NF1 | Sponge network | 0.92 | 0 | 0.51 | 0 | 0.481 |
| 10160 | U91328.19 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 20 | PRUNE2 | Sponge network | -0.303 | 0.21002 | -1.733 | 0.00022 | 0.481 |
| 10161 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-940 | 35 | PTPRT | Sponge network | -0.563 | 0.02545 | -0.907 | 0.03727 | 0.481 |
| 10162 | RP11-774O3.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p | 18 | CDH23 | Sponge network | -0.487 | 0.02157 | -0.974 | 0.00034 | 0.481 |
| 10163 | RP5-1198O20.4 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | MSN | Sponge network | 0.997 | 0.00066 | -0.023 | 0.88422 | 0.481 |
| 10164 | HOXB-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p | 12 | CXCL12 | Sponge network | -0.154 | 0.53954 | -1.421 | 0 | 0.481 |
| 10165 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p | 19 | IL17RD | Sponge network | 0.811 | 0.03228 | -0.019 | 0.94873 | 0.481 |
| 10166 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | TLL1 | Sponge network | 0.194 | 0.64278 | -1.195 | 3.0E-5 | 0.481 |
| 10167 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 18 | WWTR1 | Sponge network | -0.246 | 0.35034 | -0.345 | 0.11997 | 0.481 |
| 10168 | DNM3OS |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 50 | TRPS1 | Sponge network | 0.572 | 0.01722 | -0.191 | 0.44109 | 0.481 |
| 10169 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-93-3p | 18 | WWTR1 | Sponge network | 0.056 | 0.81195 | -0.345 | 0.11997 | 0.481 |
| 10170 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-7-1-3p | 29 | ST6GAL2 | Sponge network | 0.335 | 0.20596 | -1.201 | 0.00597 | 0.481 |
| 10171 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p | 19 | PNRC1 | Sponge network | -0.576 | 0.10121 | -0.646 | 0 | 0.481 |
| 10172 | RP4-613B23.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-877-5p | 14 | ASTN1 | Sponge network | -0.309 | 0.1145 | -2.389 | 2.0E-5 | 0.481 |
| 10173 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-629-3p | 13 | RYR2 | Sponge network | -2.531 | 0 | -1.466 | 0.00031 | 0.481 |
| 10174 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-375;hsa-miR-590-3p | 13 | MLLT10 | Sponge network | 0.768 | 7.0E-5 | 0.105 | 0.33292 | 0.481 |
| 10175 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-7-5p | 14 | PDGFC | Sponge network | -0.162 | 0.47687 | -0.33 | 0.06483 | 0.481 |
| 10176 | PGM5-AS1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | SPOP | Sponge network | -4.546 | 0 | -0.419 | 1.0E-5 | 0.48 |
| 10177 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 42 | ADARB1 | Sponge network | -0.421 | 0.0114 | -0.335 | 0.06631 | 0.48 |
| 10178 | CTC-510F12.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-335-3p | 10 | CNTN1 | Sponge network | -0.691 | 0.00146 | -2.594 | 0 | 0.48 |
| 10179 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | MITF | Sponge network | -1.886 | 0 | -0.769 | 0.00022 | 0.48 |
| 10180 | RP11-473I1.5 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p | 14 | UBR3 | Sponge network | 0.542 | 0.00054 | -0.021 | 0.82774 | 0.48 |
| 10181 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-629-3p;hsa-miR-93-5p | 27 | ZBTB4 | Sponge network | -2.117 | 0.00161 | -0.739 | 0 | 0.48 |
| 10182 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p | 15 | LRRC55 | Sponge network | -0.739 | 0.00044 | -0.026 | 0.93163 | 0.48 |
| 10183 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p | 16 | ZFHX3 | Sponge network | 0.37 | 0.27614 | 0.389 | 0.01363 | 0.48 |
| 10184 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 32 | AKAP11 | Sponge network | -0.096 | 0.67958 | 0.196 | 0.09825 | 0.48 |
| 10185 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-375;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-96-5p | 27 | RASSF8 | Sponge network | 0.082 | 0.55654 | -0.122 | 0.65591 | 0.48 |
| 10186 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 35 | NRXN1 | Sponge network | -0.769 | 0.00035 | -3.778 | 0 | 0.48 |
| 10187 | RP11-597D13.9 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-374b-3p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 11 | CNTNAP1 | Sponge network | 1.302 | 7.0E-5 | 0.358 | 0.13727 | 0.48 |
| 10188 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | PRKAA2 | Sponge network | 0.374 | 0.20054 | -2.462 | 0 | 0.48 |
| 10189 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-29a-5p | 11 | PGR | Sponge network | -7.551 | 0 | -1.704 | 0 | 0.48 |
| 10190 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | RABEP1 | Sponge network | -0.096 | 0.67958 | -0.066 | 0.43142 | 0.48 |
| 10191 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 62 | BMPR2 | Sponge network | -0.323 | 0.01372 | 0.206 | 0.0635 | 0.48 |
| 10192 | AC009948.5 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p | 13 | STAT5B | Sponge network | 0.532 | 0.00505 | -0.182 | 0.1082 | 0.48 |
| 10193 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 29 | RSPO2 | Sponge network | -0.769 | 0.00035 | -3.038 | 0 | 0.48 |
| 10194 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | FSTL5 | Sponge network | -0.793 | 7.0E-5 | -1.3 | 0.01619 | 0.48 |
| 10195 | RP11-166D19.1 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p | 12 | LZTS1 | Sponge network | -0.576 | 0.10121 | 1.664 | 0 | 0.48 |
| 10196 | CTD-2002H8.2 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 12 | DDX6 | Sponge network | 0.931 | 1.0E-5 | 0.091 | 0.36428 | 0.48 |
| 10197 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-93-3p | 14 | EPHA3 | Sponge network | -2.531 | 0 | 0.185 | 0.53588 | 0.48 |
| 10198 | ZNF667-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-429 | 12 | DUSP1 | Sponge network | -0.976 | 0.00025 | -1.754 | 0 | 0.48 |
| 10199 | RP11-159D12.2 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p | 18 | TSC1 | Sponge network | 1.254 | 0 | 0.103 | 0.26071 | 0.48 |
| 10200 | RP11-967K21.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 19 | TET1 | Sponge network | 0.056 | 0.81195 | -0.135 | 0.52138 | 0.48 |
| 10201 | RP11-589P10.7 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-338-3p | 12 | DTNA | Sponge network | -2.044 | 0.00066 | -1.648 | 1.0E-5 | 0.48 |
| 10202 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p | 27 | MOB1B | Sponge network | 1.757 | 0 | 0.197 | 0.15266 | 0.48 |
| 10203 | PCA3 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-577 | 13 | AMPH | Sponge network | -0.709 | 0.18168 | -0.916 | 5.0E-5 | 0.48 |
| 10204 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-940;hsa-miR-96-5p | 23 | CRTC3 | Sponge network | 0.572 | 0.01722 | 0.164 | 0.16786 | 0.48 |
| 10205 | RP11-736K20.5 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-375;hsa-miR-590-3p | 12 | DOCK4 | Sponge network | -0.358 | 0.17255 | 0.502 | 0.00108 | 0.48 |
| 10206 | JAZF1-AS1 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-542-3p | 10 | ZNF331 | Sponge network | -0.769 | 0.00035 | -1.251 | 0 | 0.48 |
| 10207 | LINC00473 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-935 | 16 | RELN | Sponge network | -1.203 | 0.03881 | -2.403 | 0 | 0.48 |
| 10208 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | BNC2 | Sponge network | -0.842 | 0.01361 | -0.491 | 0.09988 | 0.48 |
| 10209 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | CNTNAP3 | Sponge network | -2.117 | 0.00161 | -1.465 | 0.00033 | 0.48 |
| 10210 | LINC01018 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | -2.915 | 0.00015 | -0.684 | 0.00159 | 0.48 |
| 10211 | LINC00472 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-3127-5p;hsa-miR-378a-3p | 13 | RPS6KA2 | Sponge network | -0.842 | 0.01361 | -0.57 | 0.00013 | 0.48 |
| 10212 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 51 | PTPRD | Sponge network | 0.249 | 0.35902 | -1.005 | 0.00064 | 0.48 |
| 10213 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 25 | RUNX1T1 | Sponge network | -0.187 | 0.23787 | -0.344 | 0.2374 | 0.48 |
| 10214 | RP11-144G6.12 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-330-3p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | CREB1 | Sponge network | 0.826 | 1.0E-5 | 0.203 | 0.00537 | 0.48 |
| 10215 | NEAT1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p | 12 | FBXO11 | Sponge network | 0.705 | 0.00014 | 0.142 | 0.04938 | 0.48 |
| 10216 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 53 | PRKAA2 | Sponge network | 0.194 | 0.64278 | -2.462 | 0 | 0.48 |
| 10217 | HOXB-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-335-5p | 10 | CDH19 | Sponge network | -0.154 | 0.53954 | -3.434 | 0 | 0.48 |
| 10218 | PART1 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 14 | AMPH | Sponge network | -2.925 | 0 | -0.916 | 5.0E-5 | 0.48 |
| 10219 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-744-3p | 23 | PTPN14 | Sponge network | 0.335 | 0.20596 | 0.144 | 0.34595 | 0.48 |
| 10220 | RP11-822E23.8 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-500a-5p | 29 | DMD | Sponge network | -0.777 | 0.16667 | -1.506 | 0 | 0.48 |
| 10221 | ACTA2-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | PRKCB | Sponge network | 0.194 | 0.64278 | -1.313 | 2.0E-5 | 0.48 |
| 10222 | AP001347.6 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p | 44 | DST | Sponge network | -1.161 | 0.00591 | -0.713 | 0.00096 | 0.48 |
| 10223 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-935 | 38 | DMD | Sponge network | 0.083 | 0.66405 | -1.506 | 0 | 0.48 |
| 10224 | AC108488.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-424-5p | 15 | DYNC1I1 | Sponge network | 0.259 | 0.12542 | -1.12 | 0.00107 | 0.48 |
| 10225 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-495-3p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 37 | MYO9A | Sponge network | -0.615 | 0.00015 | -0.147 | 0.32373 | 0.48 |
| 10226 | RP1-151F17.1 |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-330-3p | 14 | WDFY3 | Sponge network | -0.321 | 0.22761 | -0.201 | 0.0967 | 0.48 |
| 10227 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-766-3p | 49 | MDM4 | Sponge network | 0.614 | 1.0E-5 | 0.345 | 0.00762 | 0.48 |
| 10228 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-628-5p | 18 | GRIA3 | Sponge network | -1.092 | 4.0E-5 | -1.956 | 0 | 0.48 |
| 10229 | HAND2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-93-5p | 40 | IL17RD | Sponge network | -1.697 | 0.00889 | -0.019 | 0.94873 | 0.48 |
| 10230 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935 | 16 | DAB2 | Sponge network | -0.739 | 0.0574 | 0.431 | 0.0113 | 0.48 |
| 10231 | AC106786.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p | 11 | PLSCR4 | Sponge network | 1.122 | 0.02989 | -0.474 | 0.03833 | 0.48 |
| 10232 | AC009948.5 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | MAPK10 | Sponge network | 0.532 | 0.00505 | -1.774 | 0 | 0.48 |
| 10233 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | PEG3 | Sponge network | 0.246 | 0.59912 | -1.74 | 0 | 0.48 |
| 10234 | LINC00641 |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | -0.222 | 0.32445 | -1.398 | 2.0E-5 | 0.48 |
| 10235 | BVES-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429 | 15 | ERG | Sponge network | -2.62 | 0 | 0.002 | 0.99011 | 0.48 |
| 10236 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 17 | DKK3 | Sponge network | -1.249 | 7.0E-5 | -0.052 | 0.77783 | 0.48 |
| 10237 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-625-5p | 27 | TGFBR3 | Sponge network | -0.154 | 0.53954 | -1.173 | 1.0E-5 | 0.48 |
| 10238 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-935 | 15 | LRRC55 | Sponge network | -1.331 | 3.0E-5 | -0.026 | 0.93163 | 0.48 |
| 10239 | RP11-1006G14.4 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-590-3p | 11 | MACF1 | Sponge network | 0.559 | 0.00026 | 0.019 | 0.90335 | 0.48 |
| 10240 | EMX2OS |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | TPO | Sponge network | -0.777 | 0.1134 | -1.87 | 2.0E-5 | 0.48 |
| 10241 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 22 | LRRC7 | Sponge network | -0.487 | 0.02157 | -1.341 | 0.01193 | 0.48 |
| 10242 | AP001347.6 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 11 | CXCL12 | Sponge network | -1.161 | 0.00591 | -1.421 | 0 | 0.48 |
| 10243 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | NIN | Sponge network | -0.611 | 0.09607 | -0.253 | 0.13794 | 0.48 |
| 10244 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 20 | FGFR1 | Sponge network | -2.915 | 0.00015 | -0.509 | 0.03957 | 0.48 |
| 10245 | LINC00473 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 28 | DIP2C | Sponge network | -1.203 | 0.03881 | -0.436 | 0.01713 | 0.48 |
| 10246 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 31 | ESR1 | Sponge network | 0.335 | 0.20596 | -1.035 | 3.0E-5 | 0.48 |
| 10247 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-455-3p | 13 | MCC | Sponge network | -0.309 | 0.1145 | -1.005 | 0 | 0.48 |
| 10248 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-877-5p | 22 | NTRK3 | Sponge network | -0.309 | 0.1145 | -1.77 | 3.0E-5 | 0.48 |
| 10249 | RP11-264B17.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 10 | PHF6 | Sponge network | 1.82 | 0 | 0.785 | 0 | 0.48 |
| 10250 | GS1-124K5.11 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | HERC1 | Sponge network | 0.494 | 0.00339 | -0.196 | 0.08544 | 0.48 |
| 10251 | MAGI2-AS3 |
hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | GUCY1A2 | Sponge network | -0.129 | 0.57177 | 1.213 | 0 | 0.48 |
| 10252 | RP5-1198O20.4 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | PIK3CA | Sponge network | 0.997 | 0.00066 | 0.195 | 0.06908 | 0.48 |
| 10253 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-577;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 36 | TRPS1 | Sponge network | -2.69 | 0.0001 | -0.191 | 0.44109 | 0.48 |
| 10254 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | AHNAK | Sponge network | 0.194 | 0.64278 | -1.207 | 0 | 0.48 |
| 10255 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-96-5p | 39 | FBXO32 | Sponge network | 0.064 | 0.72934 | -0.495 | 0.07561 | 0.48 |
| 10256 | LINC01018 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | RPS6KA6 | Sponge network | -2.915 | 0.00015 | -2.989 | 0 | 0.48 |
| 10257 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 11 | PRDM5 | Sponge network | -1.161 | 0.00591 | -1.09 | 0.00014 | 0.48 |
| 10258 | JAZF1-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p | 11 | NGFR | Sponge network | -0.769 | 0.00035 | -1.271 | 0.01058 | 0.48 |
| 10259 | U91328.19 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p | 14 | SYNM | Sponge network | -0.303 | 0.21002 | -2.309 | 1.0E-5 | 0.48 |
| 10260 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-3p | 66 | NTRK3 | Sponge network | -0.487 | 0.02157 | -1.77 | 3.0E-5 | 0.48 |
| 10261 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 33 | PIK3R1 | Sponge network | 0.537 | 0.00139 | -0.133 | 0.36854 | 0.48 |
| 10262 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 11 | IGSF10 | Sponge network | -0.583 | 0.14589 | -1.751 | 1.0E-5 | 0.48 |
| 10263 | MAGI2-AS3 |
hsa-miR-10b-3p;hsa-miR-193b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p | 10 | LRRTM1 | Sponge network | -0.129 | 0.57177 | -3.361 | 0 | 0.48 |
| 10264 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | AHNAK | Sponge network | -0.769 | 0.00035 | -1.207 | 0 | 0.48 |
| 10265 | RP11-67L2.2 |
hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | HERC1 | Sponge network | 0.72 | 0.0001 | -0.196 | 0.08544 | 0.48 |
| 10266 | CTC-457L16.2 |
hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | NIN | Sponge network | 1.153 | 0.00114 | -0.253 | 0.13794 | 0.48 |
| 10267 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | BNC2 | Sponge network | -0.187 | 0.23787 | -0.491 | 0.09988 | 0.48 |
| 10268 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p | 12 | SMARCA1 | Sponge network | -0.323 | 0.01372 | -0.684 | 0.00159 | 0.479 |
| 10269 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | EBF1 | Sponge network | 0.056 | 0.8494 | -0.384 | 0.08856 | 0.479 |
| 10270 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | SETBP1 | Sponge network | 0.299 | 0.57722 | -1.302 | 1.0E-5 | 0.479 |
| 10271 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-301a-3p;hsa-miR-539-5p;hsa-miR-940 | 12 | PBX1 | Sponge network | 0.043 | 0.81628 | -1.206 | 0 | 0.479 |
| 10272 | LINC00702 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | KCNAB1 | Sponge network | -0.737 | 0.06182 | -0.755 | 2.0E-5 | 0.479 |
| 10273 | LINC00578 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 14 | NUAK1 | Sponge network | 0.811 | 0.03228 | 0.904 | 0 | 0.479 |
| 10274 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p | 29 | RASSF2 | Sponge network | -1.057 | 0.00029 | -0.273 | 0.21961 | 0.479 |
| 10275 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | DLC1 | Sponge network | 0.214 | 0.18246 | -0.382 | 0.05038 | 0.479 |
| 10276 | RP11-359B12.2 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | RNF180 | Sponge network | 0.064 | 0.72934 | -1.127 | 1.0E-5 | 0.479 |
| 10277 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | GRIN2A | Sponge network | -0.737 | 0.10822 | -2.161 | 0 | 0.479 |
| 10278 | AC003090.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | ERG | Sponge network | -2.117 | 0.00161 | 0.002 | 0.99011 | 0.479 |
| 10279 | RP11-536O18.1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-502-5p;hsa-miR-942-5p | 10 | IGFBP5 | Sponge network | -0.074 | 0.90758 | -0.806 | 0.00105 | 0.479 |
| 10280 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | LRP1B | Sponge network | -1.439 | 4.0E-5 | -2.163 | 0 | 0.479 |
| 10281 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-425-5p;hsa-miR-7-1-3p | 12 | DIP2C | Sponge network | -0.187 | 0.23787 | -0.436 | 0.01713 | 0.479 |
| 10282 | CTD-2528L19.6 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | SIRT1 | Sponge network | 0.953 | 0 | 0.109 | 0.2593 | 0.479 |
| 10283 | LINC00672 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-590-3p | 17 | ITPKB | Sponge network | -0.227 | 0.31657 | -0.704 | 0.00106 | 0.479 |
| 10284 | RP11-53O19.1 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 15 | IGSF10 | Sponge network | -0.405 | 0.22013 | -1.751 | 1.0E-5 | 0.479 |
| 10285 | MIR497HG |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 39 | TRPS1 | Sponge network | -1.439 | 4.0E-5 | -0.191 | 0.44109 | 0.479 |
| 10286 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 48 | EPHA5 | Sponge network | 0.997 | 0.00066 | -0.957 | 0.01733 | 0.479 |
| 10287 | AC006116.24 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-577 | 21 | TRPS1 | Sponge network | -0.473 | 0.07602 | -0.191 | 0.44109 | 0.479 |
| 10288 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 15 | AKAP12 | Sponge network | -0.222 | 0.32445 | -0.932 | 0.0028 | 0.479 |
| 10289 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421 | 10 | DAB2 | Sponge network | -0.583 | 0.14589 | 0.431 | 0.0113 | 0.479 |
| 10290 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PEG3 | Sponge network | -0.737 | 0.10822 | -1.74 | 0 | 0.479 |
| 10291 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p | 22 | SCN9A | Sponge network | -0.154 | 0.53954 | -0.525 | 0.10593 | 0.479 |
| 10292 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | -0.421 | 0.0114 | -1.465 | 0.00033 | 0.479 |
| 10293 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 20 | RPS6KA2 | Sponge network | -0.49 | 0.04946 | -0.57 | 0.00013 | 0.479 |
| 10294 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-590-5p | 11 | LRRK2 | Sponge network | -1.349 | 0.00017 | -1.136 | 1.0E-5 | 0.479 |
| 10295 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 24 | ESR1 | Sponge network | -0.739 | 0.00044 | -1.035 | 3.0E-5 | 0.479 |
| 10296 | RP11-119F7.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935 | 60 | LPP | Sponge network | 0.225 | 0.30644 | -0.459 | 0.04475 | 0.479 |
| 10297 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | NALCN | Sponge network | -2.117 | 0.00161 | 1.068 | 0.00237 | 0.479 |
| 10298 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 33 | NOTCH2 | Sponge network | 0.72 | 0.0001 | 0.138 | 0.35163 | 0.479 |
| 10299 | HCG11 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 10 | CNTNAP1 | Sponge network | 0.385 | 0.10067 | 0.358 | 0.13727 | 0.479 |
| 10300 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | PIK3R1 | Sponge network | -0.737 | 0.06182 | -0.133 | 0.36854 | 0.479 |
| 10301 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p | 10 | PRDM5 | Sponge network | -0.693 | 0.05629 | -1.09 | 0.00014 | 0.479 |
| 10302 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | CDH10 | Sponge network | -0.576 | 0.10121 | -2.397 | 0 | 0.479 |
| 10303 | A1BG-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-589-3p | 14 | ESR1 | Sponge network | -0.164 | 0.45106 | -1.035 | 3.0E-5 | 0.479 |
| 10304 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 14 | RCAN2 | Sponge network | 0.16 | 0.50785 | -0.33 | 0.15172 | 0.479 |
| 10305 | GAS5 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 12 | CBFB | Sponge network | 0.728 | 0 | 1.061 | 0 | 0.479 |
| 10306 | PCA3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-590-3p | 13 | JAKMIP2 | Sponge network | -0.709 | 0.18168 | -1.388 | 0 | 0.479 |
| 10307 | AC009948.5 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-744-3p | 26 | PTPN14 | Sponge network | 0.532 | 0.00505 | 0.144 | 0.34595 | 0.479 |
| 10308 | FAM66C |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 42 | GRIK3 | Sponge network | -0.901 | 0.00019 | -3.208 | 0 | 0.479 |
| 10309 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-935 | 59 | FAT3 | Sponge network | -0.615 | 0.00015 | -1.601 | 0.00051 | 0.479 |
| 10310 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 15 | RET | Sponge network | -0.513 | 0.04703 | -0.833 | 0.03517 | 0.479 |
| 10311 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 55 | FAT3 | Sponge network | 0.101 | 0.60919 | -1.601 | 0.00051 | 0.479 |
| 10312 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 35 | CLASP2 | Sponge network | 0.421 | 0.00084 | -0.038 | 0.71 | 0.479 |
| 10313 | MIR143HG |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-940;hsa-miR-98-5p | 26 | ZFP36L1 | Sponge network | -0.739 | 0.0574 | -0.505 | 8.0E-5 | 0.479 |
| 10314 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p | 20 | RET | Sponge network | -0.129 | 0.57177 | -0.833 | 0.03517 | 0.479 |
| 10315 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-708-3p;hsa-miR-940;hsa-miR-942-5p | 38 | HLF | Sponge network | -0.899 | 6.0E-5 | -2.443 | 0 | 0.479 |
| 10316 | RP11-736K20.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-589-3p | 20 | IL17RD | Sponge network | -0.358 | 0.17255 | -0.019 | 0.94873 | 0.479 |
| 10317 | RP11-736K20.5 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-589-3p | 17 | KCNJ5 | Sponge network | -0.358 | 0.17255 | -0.281 | 0.3339 | 0.479 |
| 10318 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-542-3p | 10 | ALDH1A2 | Sponge network | -1.092 | 4.0E-5 | -2.411 | 0 | 0.479 |
| 10319 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 19 | RPS6KA2 | Sponge network | -2.69 | 0.0001 | -0.57 | 0.00013 | 0.479 |
| 10320 | LINC00641 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | SETBP1 | Sponge network | -0.222 | 0.32445 | -1.302 | 1.0E-5 | 0.479 |
| 10321 | PGM5-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-550a-3p | 12 | NRK | Sponge network | -4.546 | 0 | 0.656 | 0.19217 | 0.479 |
| 10322 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | ST6GAL2 | Sponge network | -1.439 | 4.0E-5 | -1.201 | 0.00597 | 0.479 |
| 10323 | MEG3 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-96-5p | 11 | MSN | Sponge network | 0.249 | 0.35902 | -0.023 | 0.88422 | 0.479 |
| 10324 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | CACNA2D1 | Sponge network | -0.323 | 0.01372 | -0.846 | 0.00675 | 0.479 |
| 10325 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | RASSF8 | Sponge network | -1.216 | 0 | -0.122 | 0.65591 | 0.479 |
| 10326 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-93-5p | 35 | LPP | Sponge network | 0.497 | 0.01451 | -0.459 | 0.04475 | 0.479 |
| 10327 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 14 | NRK | Sponge network | -1.582 | 8.0E-5 | 0.656 | 0.19217 | 0.479 |
| 10328 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 33 | EPHA3 | Sponge network | -0.709 | 0.18168 | 0.185 | 0.53588 | 0.479 |
| 10329 | RP11-359B12.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 29 | PTPN14 | Sponge network | 0.064 | 0.72934 | 0.144 | 0.34595 | 0.479 |
| 10330 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 26 | IL6ST | Sponge network | -0.187 | 0.23787 | -0.402 | 0.00676 | 0.479 |
| 10331 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 16 | PTPN11 | Sponge network | 1.254 | 0 | 0.431 | 2.0E-5 | 0.479 |
| 10332 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 63 | IL6ST | Sponge network | 0.101 | 0.60919 | -0.402 | 0.00676 | 0.479 |
| 10333 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 42 | TGFBR3 | Sponge network | 0.064 | 0.72934 | -1.173 | 1.0E-5 | 0.479 |
| 10334 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-624-5p;hsa-miR-96-5p | 33 | NTRK3 | Sponge network | -0.517 | 0.02935 | -1.77 | 3.0E-5 | 0.479 |
| 10335 | AC083843.1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p;hsa-miR-495-3p | 15 | PHF6 | Sponge network | 1.4 | 0 | 0.785 | 0 | 0.479 |
| 10336 | BCYRN1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-942-5p;hsa-miR-96-5p | 12 | ZBTB20 | Sponge network | 0.311 | 0.10344 | -0.122 | 0.57569 | 0.479 |
| 10337 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-664a-5p | 30 | PPM1L | Sponge network | -1.331 | 3.0E-5 | -0.537 | 0.00616 | 0.479 |
| 10338 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 32 | EBF1 | Sponge network | -0.563 | 0.02545 | -0.384 | 0.08856 | 0.479 |
| 10339 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 28 | PIK3R1 | Sponge network | -0.583 | 0.14589 | -0.133 | 0.36854 | 0.479 |
| 10340 | LINC00702 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | RTN4 | Sponge network | -0.737 | 0.06182 | -0.412 | 0.00014 | 0.479 |
| 10341 | U91328.19 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-935 | 34 | DMD | Sponge network | -0.303 | 0.21002 | -1.506 | 0 | 0.479 |
| 10342 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 24 | ADAMTSL3 | Sponge network | 0.214 | 0.18246 | -1.559 | 0 | 0.479 |
| 10343 | LINC00865 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-671-5p | 17 | CADM3 | Sponge network | -1.249 | 7.0E-5 | -3.663 | 0 | 0.479 |
| 10344 | MALAT1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 23 | SIRT1 | Sponge network | 0.782 | 0.00016 | 0.109 | 0.2593 | 0.479 |
| 10345 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 16 | DCLK1 | Sponge network | -0.154 | 0.78939 | -1.324 | 0.00014 | 0.479 |
| 10346 | LINC00472 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 15 | SFRP1 | Sponge network | -0.842 | 0.01361 | -3.566 | 0 | 0.479 |
| 10347 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 25 | AKAP6 | Sponge network | 0.249 | 0.35902 | -1.586 | 3.0E-5 | 0.479 |
| 10348 | ZSCAN16-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | MAPK10 | Sponge network | -0.342 | 0.02288 | -1.774 | 0 | 0.479 |
| 10349 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-429 | 12 | GJA1 | Sponge network | -0.154 | 0.53954 | -0.485 | 0.02179 | 0.479 |
| 10350 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | DAB2 | Sponge network | -1.439 | 4.0E-5 | 0.431 | 0.0113 | 0.479 |
| 10351 | LINC01018 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 19 | CADM3 | Sponge network | -2.915 | 0.00015 | -3.663 | 0 | 0.479 |
| 10352 | PGM5-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 21 | NAV3 | Sponge network | -4.546 | 0 | 0.212 | 0.47729 | 0.479 |
| 10353 | FAM66C |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | ERG | Sponge network | -0.901 | 0.00019 | 0.002 | 0.99011 | 0.479 |
| 10354 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | PCDH9 | Sponge network | -0.899 | 6.0E-5 | -1.823 | 4.0E-5 | 0.479 |
| 10355 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 18 | RUNX1T1 | Sponge network | 1.122 | 0.02989 | -0.344 | 0.2374 | 0.479 |
| 10356 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | CDHR3 | Sponge network | -0.612 | 0.00094 | -0.977 | 4.0E-5 | 0.479 |
| 10357 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 14 | RECK | Sponge network | 0.199 | 0.38015 | -1.043 | 0 | 0.479 |
| 10358 | RP11-244O19.1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-940 | 13 | RHOB | Sponge network | -0.096 | 0.67958 | -1.052 | 0 | 0.479 |
| 10359 | RP11-774O3.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p | 15 | STARD13 | Sponge network | -0.487 | 0.02157 | -0.187 | 0.27383 | 0.479 |
| 10360 | GS1-124K5.11 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 12 | STAT5B | Sponge network | 0.494 | 0.00339 | -0.182 | 0.1082 | 0.479 |
| 10361 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | PDGFC | Sponge network | -1.111 | 0.0003 | -0.33 | 0.06483 | 0.479 |
| 10362 | RP11-968O1.5 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p | 33 | DST | Sponge network | -0.829 | 0.01343 | -0.713 | 0.00096 | 0.478 |
| 10363 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-98-5p | 39 | DMD | Sponge network | 0.246 | 0.59912 | -1.506 | 0 | 0.478 |
| 10364 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 18 | GJA1 | Sponge network | -0.769 | 0.00035 | -0.485 | 0.02179 | 0.478 |
| 10365 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 36 | RBMS3 | Sponge network | -0.513 | 0.04703 | -0.519 | 0.03551 | 0.478 |
| 10366 | RP11-720L2.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-625-5p | 14 | IGFBP5 | Sponge network | -1.255 | 0.00981 | -0.806 | 0.00105 | 0.478 |
| 10367 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-429 | 10 | DNAJB4 | Sponge network | -1.654 | 0.00144 | -0.7 | 1.0E-5 | 0.478 |
| 10368 | RP4-717I23.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-940 | 32 | CREB1 | Sponge network | 0.637 | 0.00069 | 0.203 | 0.00537 | 0.478 |
| 10369 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | KCNAB1 | Sponge network | -0.49 | 0.04946 | -0.755 | 2.0E-5 | 0.478 |
| 10370 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 63 | NTRK3 | Sponge network | -0.162 | 0.47687 | -1.77 | 3.0E-5 | 0.478 |
| 10371 | DNM3OS |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | 0.572 | 0.01722 | -0.998 | 0.00025 | 0.478 |
| 10372 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | LRRC55 | Sponge network | 0.335 | 0.20596 | -0.026 | 0.93163 | 0.478 |
| 10373 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 22 | FGFR1 | Sponge network | 0.101 | 0.60919 | -0.509 | 0.03957 | 0.478 |
| 10374 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 18 | ABCC9 | Sponge network | -0.405 | 0.22013 | -0.466 | 0.14121 | 0.478 |
| 10375 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p | 10 | DKK3 | Sponge network | -1.582 | 8.0E-5 | -0.052 | 0.77783 | 0.478 |
| 10376 | LINC00641 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 21 | MRVI1 | Sponge network | -0.222 | 0.32445 | -1.051 | 0.00022 | 0.478 |
| 10377 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 32 | PRKAB2 | Sponge network | 0.083 | 0.66405 | -0.367 | 0.01371 | 0.478 |
| 10378 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 27 | IGF1 | Sponge network | -1.161 | 0.00591 | -0.204 | 0.5898 | 0.478 |
| 10379 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 21 | MOB1B | Sponge network | 0.799 | 1.0E-5 | 0.197 | 0.15266 | 0.478 |
| 10380 | USP3-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 31 | ERC1 | Sponge network | -0.162 | 0.47687 | -0.108 | 0.38794 | 0.478 |
| 10381 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 26 | CXXC4 | Sponge network | 0.374 | 0.20054 | -0.433 | 0.21055 | 0.478 |
| 10382 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | SETBP1 | Sponge network | 0.889 | 9.0E-5 | -1.302 | 1.0E-5 | 0.478 |
| 10383 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-93-5p;hsa-miR-98-5p | 23 | LRIG1 | Sponge network | 0.082 | 0.55397 | -0.537 | 0.00588 | 0.478 |
| 10384 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 27 | GRIN2A | Sponge network | -0.709 | 0.18168 | -2.161 | 0 | 0.478 |
| 10385 | MAGI2-AS3 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p | 10 | PXDN | Sponge network | -0.129 | 0.57177 | 1.038 | 0 | 0.478 |
| 10386 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-3p | 37 | IL6ST | Sponge network | 0.178 | 0.29106 | -0.402 | 0.00676 | 0.478 |
| 10387 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 25 | PRRX1 | Sponge network | -0.976 | 0.00025 | 1.316 | 0 | 0.478 |
| 10388 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-628-5p | 11 | TRIM33 | Sponge network | 1.597 | 0 | 0.402 | 7.0E-5 | 0.478 |
| 10389 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-940 | 67 | NTRK3 | Sponge network | -0.323 | 0.01372 | -1.77 | 3.0E-5 | 0.478 |
| 10390 | DNAJC27-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p | 12 | PARK2 | Sponge network | -0.969 | 2.0E-5 | -1.892 | 0 | 0.478 |
| 10391 | ARHGEF26-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-935 | 14 | STAT5B | Sponge network | -2.46 | 0 | -0.182 | 0.1082 | 0.478 |
| 10392 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 26 | LRRC55 | Sponge network | -1.111 | 0.0003 | -0.026 | 0.93163 | 0.478 |
| 10393 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | NRXN3 | Sponge network | 0.16 | 0.50785 | -0.893 | 0.04186 | 0.478 |
| 10394 | CTC-444N24.11 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-766-3p | 11 | ATM | Sponge network | 1.153 | 0 | 0.178 | 0.21568 | 0.478 |
| 10395 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 42 | ZFHX4 | Sponge network | -0.793 | 7.0E-5 | 0.018 | 0.95574 | 0.478 |
| 10396 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 37 | HLF | Sponge network | -0.355 | 0.0077 | -2.443 | 0 | 0.478 |
| 10397 | LINC01018 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p | 11 | FLNA | Sponge network | -2.915 | 0.00015 | -0.771 | 0.01377 | 0.478 |
| 10398 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | NAV3 | Sponge network | 0.342 | 0.03129 | 0.212 | 0.47729 | 0.478 |
| 10399 | RP11-967K21.1 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-542-3p | 15 | SYNM | Sponge network | 0.056 | 0.81195 | -2.309 | 1.0E-5 | 0.478 |
| 10400 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 26 | PTPN14 | Sponge network | -0.976 | 0.00025 | 0.144 | 0.34595 | 0.478 |
| 10401 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-323a-3p;hsa-miR-33a-5p | 13 | SLITRK3 | Sponge network | -0.693 | 0.05629 | -3.328 | 0 | 0.478 |
| 10402 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-942-5p;hsa-miR-96-5p | 17 | NRXN1 | Sponge network | -0.517 | 0.02935 | -3.778 | 0 | 0.478 |
| 10403 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-429;hsa-miR-542-3p;hsa-miR-582-5p | 18 | MITF | Sponge network | -2.69 | 0.0001 | -0.769 | 0.00022 | 0.478 |
| 10404 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-940 | 17 | CRTC3 | Sponge network | -0.896 | 0.00099 | 0.164 | 0.16786 | 0.478 |
| 10405 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421 | 12 | RET | Sponge network | -2.62 | 0 | -0.833 | 0.03517 | 0.478 |
| 10406 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 48 | SOX5 | Sponge network | -1.697 | 0.00889 | -1.091 | 4.0E-5 | 0.478 |
| 10407 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-7-1-3p | 25 | PRKAA2 | Sponge network | -4.477 | 0 | -2.462 | 0 | 0.478 |
| 10408 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-93-5p | 13 | TXNIP | Sponge network | -0.096 | 0.67958 | -1.277 | 0 | 0.478 |
| 10409 | RP11-597D13.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 25 | NRK | Sponge network | 1.302 | 7.0E-5 | 0.656 | 0.19217 | 0.478 |
| 10410 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | FNBP1 | Sponge network | -1.654 | 0.00144 | -0.969 | 4.0E-5 | 0.478 |
| 10411 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | IGSF10 | Sponge network | -0.777 | 0.1134 | -1.751 | 1.0E-5 | 0.478 |
| 10412 | CTD-3064M3.3 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-7-5p | 17 | PRKCB | Sponge network | -0.469 | 0.08721 | -1.313 | 2.0E-5 | 0.478 |
| 10413 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 12 | FSTL5 | Sponge network | 0.335 | 0.20596 | -1.3 | 0.01619 | 0.478 |
| 10414 | CTD-3064M3.3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-625-5p | 10 | SFRP1 | Sponge network | -0.469 | 0.08721 | -3.566 | 0 | 0.478 |
| 10415 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 47 | SSBP2 | Sponge network | 0.194 | 0.64278 | -1.58 | 0 | 0.478 |
| 10416 | TPTEP1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 19 | ID4 | Sponge network | -1.216 | 0 | -1.644 | 0 | 0.478 |
| 10417 | AC006116.24 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-93-5p | 39 | LPP | Sponge network | -0.473 | 0.07602 | -0.459 | 0.04475 | 0.478 |
| 10418 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | NRXN3 | Sponge network | -0.738 | 0.21233 | -0.893 | 0.04186 | 0.478 |
| 10419 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p | 19 | MAF | Sponge network | -1.331 | 3.0E-5 | -1.257 | 0 | 0.478 |
| 10420 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-93-5p | 31 | SSBP2 | Sponge network | -1.654 | 0.00144 | -1.58 | 0 | 0.478 |
| 10421 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-93-3p | 39 | IL6ST | Sponge network | -0.469 | 0.08721 | -0.402 | 0.00676 | 0.478 |
| 10422 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-93-3p | 28 | SSBP2 | Sponge network | -0.469 | 0.08721 | -1.58 | 0 | 0.478 |
| 10423 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-7-5p;hsa-miR-93-5p | 17 | ZBTB4 | Sponge network | -0.726 | 0.00132 | -0.739 | 0 | 0.478 |
| 10424 | LINC00092 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3913-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-942-5p | 26 | HLF | Sponge network | -1.886 | 0 | -2.443 | 0 | 0.478 |
| 10425 | FAM66C |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p | 11 | SCN11A | Sponge network | -0.901 | 0.00019 | -1.15 | 0.00299 | 0.478 |
| 10426 | FAM66C |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | ROBO1 | Sponge network | -0.901 | 0.00019 | -0.009 | 0.96154 | 0.478 |
| 10427 | RP11-774O3.3 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-874-3p | 19 | SORCS1 | Sponge network | -0.487 | 0.02157 | -3.492 | 0 | 0.478 |
| 10428 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 22 | HIP1 | Sponge network | -0.129 | 0.57177 | 0.774 | 0 | 0.478 |
| 10429 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 31 | ESR1 | Sponge network | -0.901 | 0.00019 | -1.035 | 3.0E-5 | 0.478 |
| 10430 | NEAT1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | TAF1 | Sponge network | 0.705 | 0.00014 | 0.39 | 3.0E-5 | 0.478 |
| 10431 | USP3-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-625-5p | 14 | CXCL12 | Sponge network | -0.162 | 0.47687 | -1.421 | 0 | 0.478 |
| 10432 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-424-5p;hsa-miR-500a-5p | 12 | LRP1B | Sponge network | -2.076 | 0.00548 | -2.163 | 0 | 0.478 |
| 10433 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 20 | ZFHX3 | Sponge network | 0.419 | 0.0319 | 0.389 | 0.01363 | 0.478 |
| 10434 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 50 | ZFHX3 | Sponge network | -1.111 | 0.0003 | 0.389 | 0.01363 | 0.478 |
| 10435 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 14 | CHD2 | Sponge network | 1.153 | 0 | 0.049 | 0.55371 | 0.478 |
| 10436 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-629-3p | 16 | RYR2 | Sponge network | 0.811 | 0.03228 | -1.466 | 0.00031 | 0.478 |
| 10437 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p | 10 | ALDH1A2 | Sponge network | -0.129 | 0.57177 | -2.411 | 0 | 0.478 |
| 10438 | PWAR6 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | SYT4 | Sponge network | -1.575 | 6.0E-5 | -3.207 | 0 | 0.478 |
| 10439 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | QKI | Sponge network | 0.727 | 3.0E-5 | -0.333 | 0.04111 | 0.478 |
| 10440 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-590-3p | 17 | SLITRK3 | Sponge network | -0.896 | 0.00099 | -3.328 | 0 | 0.478 |
| 10441 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ITGB3 | Sponge network | 0.214 | 0.18246 | -0.011 | 0.95751 | 0.478 |
| 10442 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429 | 15 | ERG | Sponge network | -2.69 | 0.0001 | 0.002 | 0.99011 | 0.478 |
| 10443 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 32 | PCDH10 | Sponge network | -1.216 | 0 | -1.644 | 0.0078 | 0.478 |
| 10444 | LINC00702 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | SYT4 | Sponge network | -0.737 | 0.06182 | -3.207 | 0 | 0.478 |
| 10445 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | FGF5 | Sponge network | -0.49 | 0.04946 | 0.549 | 0.28659 | 0.478 |
| 10446 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | SUFU | Sponge network | 0.214 | 0.18246 | -0.04 | 0.65489 | 0.478 |
| 10447 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 47 | NTRK2 | Sponge network | -2.46 | 0 | -0.736 | 0.06495 | 0.478 |
| 10448 | RP11-359E10.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-582-5p;hsa-miR-940 | 10 | MAPK10 | Sponge network | 0.299 | 0.57722 | -1.774 | 0 | 0.478 |
| 10449 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-7-1-3p | 23 | AKAP6 | Sponge network | -0.563 | 0.02545 | -1.586 | 3.0E-5 | 0.478 |
| 10450 | FAM66C |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 39 | IGFBP5 | Sponge network | -0.901 | 0.00019 | -0.806 | 0.00105 | 0.478 |
| 10451 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 12 | PSIP1 | Sponge network | -0.021 | 0.93598 | -0.005 | 0.96897 | 0.478 |
| 10452 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | ROBO1 | Sponge network | -1.697 | 0.00889 | -0.009 | 0.96154 | 0.478 |
| 10453 | RP11-359B12.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 17 | FAM172A | Sponge network | 0.064 | 0.72934 | -0.57 | 0 | 0.478 |
| 10454 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-96-5p | 16 | LRRC4 | Sponge network | -1.281 | 0.00012 | -1.774 | 0 | 0.478 |
| 10455 | AC010226.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-96-5p | 15 | CBFA2T3 | Sponge network | -0.811 | 2.0E-5 | -1.221 | 1.0E-5 | 0.478 |
| 10456 | AC009948.5 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITGB1 | Sponge network | 0.532 | 0.00505 | 0.524 | 0.00017 | 0.478 |
| 10457 | RP4-639F20.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-629-5p | 35 | DST | Sponge network | -0.517 | 0.02935 | -0.713 | 0.00096 | 0.478 |
| 10458 | EMX2OS |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 47 | TRPS1 | Sponge network | -0.777 | 0.1134 | -0.191 | 0.44109 | 0.478 |
| 10459 | FENDRR |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-98-5p | 16 | KLF10 | Sponge network | -1.281 | 0.00012 | -0.783 | 0 | 0.478 |
| 10460 | CTD-3020H12.4 |
hsa-let-7d-5p;hsa-miR-127-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-5p;hsa-miR-671-5p | 10 | MDM4 | Sponge network | 1.874 | 0.00389 | 0.345 | 0.00762 | 0.478 |
| 10461 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 35 | TCF4 | Sponge network | -0.72 | 0.24239 | 0.016 | 0.92271 | 0.478 |
| 10462 | SNHG14 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | PCDH18 | Sponge network | -1.111 | 0.0003 | 0.216 | 0.24645 | 0.478 |
| 10463 | ZRANB2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 13 | ASTN1 | Sponge network | -0.376 | 0.00337 | -2.389 | 2.0E-5 | 0.478 |
| 10464 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-96-5p;hsa-miR-98-5p | 10 | PLAGL1 | Sponge network | -0.942 | 0 | -0.588 | 0.00912 | 0.478 |
| 10465 | LINC00843 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p | 16 | TBL1XR1 | Sponge network | 1.106 | 0 | 0.578 | 0 | 0.478 |
| 10466 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 45 | CHD5 | Sponge network | -1.575 | 6.0E-5 | -0.922 | 0.01521 | 0.478 |
| 10467 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | LRFN5 | Sponge network | -0.942 | 0 | -1.483 | 0 | 0.478 |
| 10468 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 38 | MOB1B | Sponge network | -0.405 | 0.22013 | 0.197 | 0.15266 | 0.478 |
| 10469 | CKMT2-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-877-5p | 15 | INPP4A | Sponge network | 0.082 | 0.55654 | 0.07 | 0.53563 | 0.478 |
| 10470 | RP11-822E23.8 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-92a-1-5p | 21 | AKAP6 | Sponge network | -0.777 | 0.16667 | -1.586 | 3.0E-5 | 0.478 |
| 10471 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | GRIN2A | Sponge network | -1.349 | 0.00017 | -2.161 | 0 | 0.478 |
| 10472 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p | 20 | EYA4 | Sponge network | -0.739 | 0.00044 | 0.3 | 0.36876 | 0.478 |
| 10473 | CTC-444N24.11 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 13 | SHPRH | Sponge network | 1.153 | 0 | 0.098 | 0.40994 | 0.478 |
| 10474 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-7-5p;hsa-miR-93-5p | 51 | TCF4 | Sponge network | -0.777 | 0.16667 | 0.016 | 0.92271 | 0.478 |
| 10475 | C20orf166-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-375 | 10 | ZBTB20 | Sponge network | -2.69 | 0.0001 | -0.122 | 0.57569 | 0.478 |
| 10476 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-484 | 15 | EBF3 | Sponge network | -4.546 | 0 | -0.058 | 0.81437 | 0.478 |
| 10477 | AC011526.1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-96-5p | 12 | RECK | Sponge network | 0.342 | 0.03129 | -1.043 | 0 | 0.478 |
| 10478 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-424-5p | 17 | KCNA6 | Sponge network | -2.076 | 0.00548 | -1.531 | 0.00013 | 0.478 |
| 10479 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-423-5p | 14 | PAK3 | Sponge network | -0.777 | 0.16667 | -0.628 | 0.07253 | 0.478 |
| 10480 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TIMP3 | Sponge network | -0.421 | 0.0114 | -0.546 | 0.02307 | 0.477 |
| 10481 | WDFY3-AS2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-98-5p | 19 | KLF10 | Sponge network | -0.942 | 0 | -0.783 | 0 | 0.477 |
| 10482 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 25 | CRTC3 | Sponge network | -2.452 | 0 | 0.164 | 0.16786 | 0.477 |
| 10483 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-935 | 34 | GRIK3 | Sponge network | -0.513 | 0.04703 | -3.208 | 0 | 0.477 |
| 10484 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | PTPRB | Sponge network | 0.572 | 0.01722 | 0.105 | 0.47122 | 0.477 |
| 10485 | RP11-798M19.6 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 12 | SORCS1 | Sponge network | -0.612 | 0.00094 | -3.492 | 0 | 0.477 |
| 10486 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | FAT4 | Sponge network | 0.532 | 0.00505 | -0.354 | 0.14694 | 0.477 |
| 10487 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SEMA3C | Sponge network | -0.612 | 0.00094 | -0.272 | 0.31153 | 0.477 |
| 10488 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | PEG3 | Sponge network | -0.899 | 6.0E-5 | -1.74 | 0 | 0.477 |
| 10489 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-93-3p | 31 | EPHA3 | Sponge network | -0.615 | 0.00015 | 0.185 | 0.53588 | 0.477 |
| 10490 | DNM3OS |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TPO | Sponge network | 0.572 | 0.01722 | -1.87 | 2.0E-5 | 0.477 |
| 10491 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -0.49 | 0.04946 | -0.252 | 0.15511 | 0.477 |
| 10492 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-671-5p | 32 | GAS7 | Sponge network | -0.469 | 0.08721 | -0.555 | 0.01602 | 0.477 |
| 10493 | PGM5-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-423-5p;hsa-miR-550a-3p | 10 | PAK3 | Sponge network | -4.546 | 0 | -0.628 | 0.07253 | 0.477 |
| 10494 | RP11-254F7.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p | 19 | ASTN1 | Sponge network | 0.176 | 0.37402 | -2.389 | 2.0E-5 | 0.477 |
| 10495 | FAM66C |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p | 23 | NRK | Sponge network | -0.901 | 0.00019 | 0.656 | 0.19217 | 0.477 |
| 10496 | RP11-867G23.10 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-93-3p | 11 | ST6GAL2 | Sponge network | -2.531 | 0 | -1.201 | 0.00597 | 0.477 |
| 10497 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 22 | BCL6 | Sponge network | 0.194 | 0.64278 | -0.211 | 0.19489 | 0.477 |
| 10498 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | SETBP1 | Sponge network | -0.612 | 0.00094 | -1.302 | 1.0E-5 | 0.477 |
| 10499 | CTD-3025N20.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 16 | MDM4 | Sponge network | 1.046 | 0 | 0.345 | 0.00762 | 0.477 |
| 10500 | SNHG16 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 12 | ZNF770 | Sponge network | 0.961 | 0 | 0.206 | 0.06751 | 0.477 |
| 10501 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-940 | 47 | TRPS1 | Sponge network | 0.532 | 0.00505 | -0.191 | 0.44109 | 0.477 |
| 10502 | RP11-2H8.2 |
hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-511-5p;hsa-miR-590-3p | 18 | ATRX | Sponge network | 1.089 | 0 | 0.261 | 0.04408 | 0.477 |
| 10503 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 27 | PBX1 | Sponge network | -0.318 | 0.65847 | -1.206 | 0 | 0.477 |
| 10504 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-93-5p | 25 | ZBTB4 | Sponge network | -0.842 | 0.01361 | -0.739 | 0 | 0.477 |
| 10505 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-576-5p | 19 | ST6GAL2 | Sponge network | -0.739 | 0.00044 | -1.201 | 0.00597 | 0.477 |
| 10506 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | NEDD4 | Sponge network | -0.49 | 0.04946 | 0.02 | 0.89834 | 0.477 |
| 10507 | HHIP-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 21 | SETBP1 | Sponge network | -0.737 | 0.10822 | -1.302 | 1.0E-5 | 0.477 |
| 10508 | HAND2-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 15 | SEPT6 | Sponge network | -1.697 | 0.00889 | -0.598 | 0.00181 | 0.477 |
| 10509 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-425-5p | 13 | ITGB3 | Sponge network | -0.842 | 0.01361 | -0.011 | 0.95751 | 0.477 |
| 10510 | RP11-307B6.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p | 16 | ASTN1 | Sponge network | -4.463 | 0.00025 | -2.389 | 2.0E-5 | 0.477 |
| 10511 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | SETBP1 | Sponge network | 0.225 | 0.30644 | -1.302 | 1.0E-5 | 0.477 |
| 10512 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 28 | AKAP6 | Sponge network | 0.082 | 0.55654 | -1.586 | 3.0E-5 | 0.477 |
| 10513 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 29 | PBX1 | Sponge network | -0.246 | 0.35034 | -1.206 | 0 | 0.477 |
| 10514 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | TAL1 | Sponge network | -0.576 | 0.10121 | -0.462 | 0.00574 | 0.477 |
| 10515 | RP11-33B1.4 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 0.826 | 0 | 0.51 | 0 | 0.477 |
| 10516 | AC003090.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-96-5p | 21 | CBFA2T3 | Sponge network | -2.117 | 0.00161 | -1.221 | 1.0E-5 | 0.477 |
| 10517 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-655-3p | 15 | SON | Sponge network | 0.542 | 0.00054 | 0.168 | 0.04406 | 0.477 |
| 10518 | RP11-967K21.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-664a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 52 | DST | Sponge network | 0.056 | 0.81195 | -0.713 | 0.00096 | 0.477 |
| 10519 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-940 | 26 | CRTC3 | Sponge network | -0.901 | 0.00019 | 0.164 | 0.16786 | 0.477 |
| 10520 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-98-5p | 29 | PBX1 | Sponge network | 0.056 | 0.81195 | -1.206 | 0 | 0.477 |
| 10521 | GNG12-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p | 25 | ASTN1 | Sponge network | -0.793 | 7.0E-5 | -2.389 | 2.0E-5 | 0.477 |
| 10522 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | LRRC55 | Sponge network | 0.194 | 0.64278 | -0.026 | 0.93163 | 0.477 |
| 10523 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 27 | TACC1 | Sponge network | -0.739 | 0.00044 | -0.362 | 0.05406 | 0.477 |
| 10524 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-93-5p | 13 | TXNIP | Sponge network | -0.465 | 0.04703 | -1.277 | 0 | 0.477 |
| 10525 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | CACNA2D1 | Sponge network | 0.246 | 0.59912 | -0.846 | 0.00675 | 0.477 |
| 10526 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-940 | 11 | HOOK3 | Sponge network | 0.082 | 0.55397 | 0.273 | 0.09957 | 0.477 |
| 10527 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 10 | IGF1 | Sponge network | -2.039 | 0.03447 | -0.204 | 0.5898 | 0.477 |
| 10528 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DICER1 | Sponge network | 0.931 | 1.0E-5 | 0.042 | 0.674 | 0.477 |
| 10529 | LINC00641 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-940 | 14 | PHOX2B | Sponge network | -0.222 | 0.32445 | -2.68 | 0 | 0.477 |
| 10530 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 44 | TCF4 | Sponge network | -0.318 | 0.65847 | 0.016 | 0.92271 | 0.477 |
| 10531 | RAMP2-AS1 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | -0.513 | 0.04703 | -1.09 | 0.00014 | 0.477 |
| 10532 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p | 37 | RICTOR | Sponge network | 0.75 | 0.00054 | 0.388 | 0.00188 | 0.477 |
| 10533 | RP11-473I1.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ZFX | Sponge network | 0.683 | 1.0E-5 | 0.09 | 0.42563 | 0.477 |
| 10534 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-493-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 18 | NRXN3 | Sponge network | -0.405 | 0.22013 | -0.893 | 0.04186 | 0.477 |
| 10535 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 36 | MCC | Sponge network | 0.101 | 0.60919 | -1.005 | 0 | 0.477 |
| 10536 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | PCDH9 | Sponge network | -0.738 | 0.21233 | -1.823 | 4.0E-5 | 0.477 |
| 10537 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 41 | MEF2C | Sponge network | 0.246 | 0.59912 | -0.499 | 0.01448 | 0.477 |
| 10538 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-7-1-3p | 12 | TMEFF2 | Sponge network | -0.611 | 0.09607 | -2.426 | 0 | 0.477 |
| 10539 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p | 44 | PBX1 | Sponge network | -0.615 | 0.00015 | -1.206 | 0 | 0.477 |
| 10540 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | FAT4 | Sponge network | -0.737 | 0.10822 | -0.354 | 0.14694 | 0.477 |
| 10541 | EMX2OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | -0.777 | 0.1134 | 0.783 | 0.00072 | 0.477 |
| 10542 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | PDGFC | Sponge network | -0.769 | 0.00035 | -0.33 | 0.06483 | 0.477 |
| 10543 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | RARB | Sponge network | -0.563 | 0.02545 | -0.825 | 2.0E-5 | 0.477 |
| 10544 | HCG18 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | PIK3CA | Sponge network | 1.063 | 0 | 0.195 | 0.06908 | 0.477 |
| 10545 | LINC00865 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-550a-3p | 14 | PAK3 | Sponge network | -1.249 | 7.0E-5 | -0.628 | 0.07253 | 0.477 |
| 10546 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | PAK3 | Sponge network | -0.513 | 0.04703 | -0.628 | 0.07253 | 0.477 |
| 10547 | TRAF3IP2-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 64 | DST | Sponge network | -0.323 | 0.01372 | -0.713 | 0.00096 | 0.477 |
| 10548 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421 | 12 | LRRK2 | Sponge network | -0.162 | 0.47687 | -1.136 | 1.0E-5 | 0.477 |
| 10549 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-744-5p | 16 | FGFR1 | Sponge network | -0.738 | 0.21233 | -0.509 | 0.03957 | 0.477 |
| 10550 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 12 | SLITRK6 | Sponge network | -1.057 | 0.00029 | -2.121 | 0 | 0.477 |
| 10551 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-629-3p | 26 | ESR1 | Sponge network | -2.62 | 0 | -1.035 | 3.0E-5 | 0.477 |
| 10552 | RP11-326G21.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PRUNE2 | Sponge network | -0.021 | 0.93598 | -1.733 | 0.00022 | 0.477 |
| 10553 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 38 | SNX29 | Sponge network | -1.575 | 6.0E-5 | -0.34 | 0.01371 | 0.477 |
| 10554 | RP11-736K20.5 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | ID4 | Sponge network | -0.358 | 0.17255 | -1.644 | 0 | 0.477 |
| 10555 | TPTEP1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 13 | NGFR | Sponge network | -1.216 | 0 | -1.271 | 0.01058 | 0.477 |
| 10556 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-7-1-3p | 13 | CHD2 | Sponge network | 0.817 | 3.0E-5 | 0.049 | 0.55371 | 0.477 |
| 10557 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 36 | CHD5 | Sponge network | -0.709 | 0.18168 | -0.922 | 0.01521 | 0.477 |
| 10558 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-5p | 17 | CRTC3 | Sponge network | 0.176 | 0.37402 | 0.164 | 0.16786 | 0.477 |
| 10559 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 35 | MAF | Sponge network | -0.576 | 0.10121 | -1.257 | 0 | 0.477 |
| 10560 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 16 | JAZF1 | Sponge network | -2.039 | 0.03447 | -0.377 | 0.02677 | 0.477 |
| 10561 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | USP12 | Sponge network | 0.537 | 0.00139 | -0.102 | 0.40768 | 0.477 |
| 10562 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | DCLK1 | Sponge network | -0.318 | 0.65847 | -1.324 | 0.00014 | 0.477 |
| 10563 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | TLL1 | Sponge network | -1.216 | 0 | -1.195 | 3.0E-5 | 0.477 |
| 10564 | USP3-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-7-5p | 25 | PRKCB | Sponge network | -0.162 | 0.47687 | -1.313 | 2.0E-5 | 0.477 |
| 10565 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 40 | NTRK2 | Sponge network | -0.096 | 0.67958 | -0.736 | 0.06495 | 0.477 |
| 10566 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p | 19 | IL17RD | Sponge network | 1.757 | 0 | -0.019 | 0.94873 | 0.477 |
| 10567 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 33 | SCN9A | Sponge network | 0.997 | 0.00066 | -0.525 | 0.10593 | 0.477 |
| 10568 | FAM66C |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 12 | SPTBN4 | Sponge network | -0.901 | 0.00019 | -0.786 | 0.01281 | 0.477 |
| 10569 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | ABCC9 | Sponge network | -0.615 | 0.00015 | -0.466 | 0.14121 | 0.477 |
| 10570 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -1.281 | 0.00012 | -0.087 | 0.69193 | 0.477 |
| 10571 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-625-5p | 22 | PTPN14 | Sponge network | 0.299 | 0.57722 | 0.144 | 0.34595 | 0.477 |
| 10572 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | CXXC4 | Sponge network | -0.513 | 0.04703 | -0.433 | 0.21055 | 0.477 |
| 10573 | CTD-2528L19.6 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 14 | ATM | Sponge network | 0.953 | 0 | 0.178 | 0.21568 | 0.477 |
| 10574 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 23 | ST6GALNAC3 | Sponge network | -0.739 | 0.00044 | -0.987 | 0 | 0.477 |
| 10575 | LINC00702 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | ABCA10 | Sponge network | -0.737 | 0.06182 | -0.571 | 0.03674 | 0.477 |
| 10576 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 38 | ETV1 | Sponge network | -0.49 | 0.04946 | -0.096 | 0.67674 | 0.477 |
| 10577 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-671-5p | 11 | CADM3 | Sponge network | -0.154 | 0.53954 | -3.663 | 0 | 0.477 |
| 10578 | SNX29P2 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-3p | 12 | STAT5B | Sponge network | 0.4 | 0.14611 | -0.182 | 0.1082 | 0.477 |
| 10579 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 20 | LRRC4 | Sponge network | -2.46 | 0 | -1.774 | 0 | 0.477 |
| 10580 | RP11-359B12.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 20 | MRVI1 | Sponge network | 0.064 | 0.72934 | -1.051 | 0.00022 | 0.477 |
| 10581 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-7-5p | 16 | PTGFR | Sponge network | -0.777 | 0.16667 | -0.233 | 0.45421 | 0.477 |
| 10582 | RP11-2H8.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 23 | AKAP11 | Sponge network | 1.089 | 0 | 0.196 | 0.09825 | 0.476 |
| 10583 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-454-3p | 18 | DDX6 | Sponge network | 0.178 | 0.29106 | 0.091 | 0.36428 | 0.476 |
| 10584 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | PRDM5 | Sponge network | -2.117 | 0.00161 | -1.09 | 0.00014 | 0.476 |
| 10585 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 18 | RPS6KA2 | Sponge network | -0.576 | 0.10121 | -0.57 | 0.00013 | 0.476 |
| 10586 | LINC00473 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 10 | CDH10 | Sponge network | -1.203 | 0.03881 | -2.397 | 0 | 0.476 |
| 10587 | HAND2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-577;hsa-miR-98-5p | 11 | PLAGL1 | Sponge network | -1.697 | 0.00889 | -0.588 | 0.00912 | 0.476 |
| 10588 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-7-1-3p | 12 | TSHZ3 | Sponge network | -0.187 | 0.23787 | -0.556 | 0.01516 | 0.476 |
| 10589 | SERTAD4-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p | 11 | NRK | Sponge network | -0.932 | 0.00329 | 0.656 | 0.19217 | 0.476 |
| 10590 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 43 | ADARB1 | Sponge network | 0.532 | 0.00505 | -0.335 | 0.06631 | 0.476 |
| 10591 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | RCAN2 | Sponge network | -1.281 | 0.00012 | -0.33 | 0.15172 | 0.476 |
| 10592 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | ARHGAP28 | Sponge network | -0.358 | 0.17255 | -0.911 | 0.00013 | 0.476 |
| 10593 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-98-5p | 37 | PBX1 | Sponge network | -0.355 | 0.0077 | -1.206 | 0 | 0.476 |
| 10594 | GNG12-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | ID4 | Sponge network | -0.793 | 7.0E-5 | -1.644 | 0 | 0.476 |
| 10595 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p | 10 | GAS1 | Sponge network | -0.777 | 0.16667 | -1.047 | 0.00364 | 0.476 |
| 10596 | SNHG14 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 32 | GJA1 | Sponge network | -1.111 | 0.0003 | -0.485 | 0.02179 | 0.476 |
| 10597 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 22 | PHC3 | Sponge network | 0.817 | 3.0E-5 | 0.212 | 0.0499 | 0.476 |
| 10598 | KB-1572G7.3 |
hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p;hsa-miR-940 | 13 | MDM4 | Sponge network | 0.786 | 1.0E-5 | 0.345 | 0.00762 | 0.476 |
| 10599 | ADAMTS9-AS2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | ROBO1 | Sponge network | -2.452 | 0 | -0.009 | 0.96154 | 0.476 |
| 10600 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 23 | ADAMTSL3 | Sponge network | -0.227 | 0.31657 | -1.559 | 0 | 0.476 |
| 10601 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-429;hsa-miR-96-5p | 15 | PCDH10 | Sponge network | -1.654 | 0.00144 | -1.644 | 0.0078 | 0.476 |
| 10602 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-96-5p | 20 | HIP1 | Sponge network | -0.563 | 0.02545 | 0.774 | 0 | 0.476 |
| 10603 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-375;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | PSIP1 | Sponge network | 1.254 | 0 | -0.005 | 0.96897 | 0.476 |
| 10604 | RP11-400K9.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-374b-3p | 11 | ROBO1 | Sponge network | -0.611 | 0.09607 | -0.009 | 0.96154 | 0.476 |
| 10605 | CTC-510F12.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-3614-5p | 10 | JAZF1 | Sponge network | -0.691 | 0.00146 | -0.377 | 0.02677 | 0.476 |
| 10606 | RP11-554A11.4 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-629-3p | 10 | ID4 | Sponge network | -0.583 | 0.14589 | -1.644 | 0 | 0.476 |
| 10607 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-3p | 18 | LRIG1 | Sponge network | 0.178 | 0.29106 | -0.537 | 0.00588 | 0.476 |
| 10608 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | NEDD4 | Sponge network | -0.739 | 0.0574 | 0.02 | 0.89834 | 0.476 |
| 10609 | KB-1410C5.5 |
hsa-miR-130b-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-7-1-3p | 11 | ZFHX3 | Sponge network | 0.959 | 0 | 0.389 | 0.01363 | 0.476 |
| 10610 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 37 | RAP1A | Sponge network | -2.46 | 0 | -0.929 | 0 | 0.476 |
| 10611 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-628-5p;hsa-miR-940 | 21 | TRIM33 | Sponge network | 0.75 | 0.00054 | 0.402 | 7.0E-5 | 0.476 |
| 10612 | USP3-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-935 | 22 | GJA1 | Sponge network | -0.162 | 0.47687 | -0.485 | 0.02179 | 0.476 |
| 10613 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | CDKN1C | Sponge network | -2.452 | 0 | -0.929 | 0.00033 | 0.476 |
| 10614 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PRKAB2 | Sponge network | -0.031 | 0.89063 | -0.367 | 0.01371 | 0.476 |
| 10615 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-582-3p | 18 | ABL2 | Sponge network | 0.858 | 3.0E-5 | 0.82 | 0 | 0.476 |
| 10616 | SNHG14 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 32 | PTPN14 | Sponge network | -1.111 | 0.0003 | 0.144 | 0.34595 | 0.476 |
| 10617 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 25 | SEMA3C | Sponge network | -2.452 | 0 | -0.272 | 0.31153 | 0.476 |
| 10618 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-576-5p | 20 | SEMA3C | Sponge network | -0.162 | 0.47687 | -0.272 | 0.31153 | 0.476 |
| 10619 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-455-3p;hsa-miR-582-5p | 19 | FBXO32 | Sponge network | -0.469 | 0.08721 | -0.495 | 0.07561 | 0.476 |
| 10620 | AP001055.6 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-7-1-3p | 17 | BCL2 | Sponge network | 0.693 | 0.00076 | -0.899 | 0 | 0.476 |
| 10621 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 22 | ZFP36L1 | Sponge network | -0.576 | 0.10121 | -0.505 | 8.0E-5 | 0.476 |
| 10622 | AC007879.7 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p | 12 | RASAL2 | Sponge network | 4.3 | 0 | 0.869 | 0 | 0.476 |
| 10623 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3913-5p;hsa-miR-629-3p;hsa-miR-93-5p | 29 | ZBTB4 | Sponge network | -1.645 | 0 | -0.739 | 0 | 0.476 |
| 10624 | LINC00865 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-7-1-3p | 20 | TET1 | Sponge network | -1.249 | 7.0E-5 | -0.135 | 0.52138 | 0.476 |
| 10625 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 14 | DKK3 | Sponge network | 0.194 | 0.64278 | -0.052 | 0.77783 | 0.476 |
| 10626 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-93-5p;hsa-miR-96-5p | 30 | MAF | Sponge network | -1.439 | 4.0E-5 | -1.257 | 0 | 0.476 |
| 10627 | HCG18 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | ERCC6 | Sponge network | 1.063 | 0 | 0.23 | 0.015 | 0.476 |
| 10628 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 16 | STARD13 | Sponge network | -0.612 | 0.00094 | -0.187 | 0.27383 | 0.476 |
| 10629 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | RECK | Sponge network | 0.299 | 0.57722 | -1.043 | 0 | 0.476 |
| 10630 | RP11-554A11.4 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p | 36 | THRB | Sponge network | -0.583 | 0.14589 | -1.117 | 0.0004 | 0.476 |
| 10631 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | TGFBR3 | Sponge network | 0.178 | 0.29106 | -1.173 | 1.0E-5 | 0.476 |
| 10632 | EMX2OS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-455-5p | 12 | THSD7A | Sponge network | -0.777 | 0.1134 | 0.242 | 0.25154 | 0.476 |
| 10633 | EMX2OS |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TLL1 | Sponge network | -0.777 | 0.1134 | -1.195 | 3.0E-5 | 0.476 |
| 10634 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p | 45 | DTNA | Sponge network | 0.056 | 0.81195 | -1.648 | 1.0E-5 | 0.476 |
| 10635 | AC006116.24 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-582-3p | 13 | PCDH10 | Sponge network | -0.473 | 0.07602 | -1.644 | 0.0078 | 0.476 |
| 10636 | RP11-21A7A.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-576-5p;hsa-miR-877-5p | 16 | FAT3 | Sponge network | -1.116 | 0.23686 | -1.601 | 0.00051 | 0.476 |
| 10637 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-421;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 34 | BNC2 | Sponge network | 0.214 | 0.18246 | -0.491 | 0.09988 | 0.476 |
| 10638 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | RELN | Sponge network | -0.769 | 0.00035 | -2.403 | 0 | 0.476 |
| 10639 | RP11-1006G14.4 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITGB1 | Sponge network | 0.559 | 0.00026 | 0.524 | 0.00017 | 0.476 |
| 10640 | RP11-672A2.4 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-424-5p | 10 | ABL2 | Sponge network | 1.344 | 0 | 0.82 | 0 | 0.476 |
| 10641 | PCA3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-671-5p | 16 | CADM3 | Sponge network | -0.709 | 0.18168 | -3.663 | 0 | 0.476 |
| 10642 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 31 | CNTN1 | Sponge network | -0.769 | 0.00035 | -2.594 | 0 | 0.476 |
| 10643 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 29 | RASSF8 | Sponge network | -1.203 | 0.03881 | -0.122 | 0.65591 | 0.476 |
| 10644 | LINC00094 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 35 | IL17RD | Sponge network | 0.253 | 0.09565 | -0.019 | 0.94873 | 0.476 |
| 10645 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 43 | RASSF2 | Sponge network | -0.49 | 0.04946 | -0.273 | 0.21961 | 0.476 |
| 10646 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 29 | EBF1 | Sponge network | -0.842 | 0.01361 | -0.384 | 0.08856 | 0.476 |
| 10647 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-5p | 33 | EPHA5 | Sponge network | -2.69 | 0.0001 | -0.957 | 0.01733 | 0.476 |
| 10648 | RP11-774O3.3 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 16 | CXCL12 | Sponge network | -0.487 | 0.02157 | -1.421 | 0 | 0.476 |
| 10649 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | ABCB5 | Sponge network | -2.117 | 0.00161 | -0.956 | 0.04077 | 0.476 |
| 10650 | RP5-1198O20.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 20 | EZH1 | Sponge network | 0.997 | 0.00066 | -0.564 | 1.0E-5 | 0.476 |
| 10651 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 27 | LRIG1 | Sponge network | -0.976 | 0.00025 | -0.537 | 0.00588 | 0.476 |
| 10652 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-935 | 12 | SEPT6 | Sponge network | -0.976 | 0.00025 | -0.598 | 0.00181 | 0.476 |
| 10653 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | CHD2 | Sponge network | 0.267 | 0.2937 | 0.049 | 0.55371 | 0.476 |
| 10654 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-671-5p | 20 | CADM3 | Sponge network | -0.942 | 0 | -3.663 | 0 | 0.476 |
| 10655 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | DKK3 | Sponge network | -0.901 | 0.00019 | -0.052 | 0.77783 | 0.476 |
| 10656 | RP11-736K20.5 |
hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-629-3p | 11 | RHOB | Sponge network | -0.358 | 0.17255 | -1.052 | 0 | 0.476 |
| 10657 | RP11-536O18.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-362-5p | 12 | IGF1 | Sponge network | -0.074 | 0.90758 | -0.204 | 0.5898 | 0.476 |
| 10658 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-877-5p;hsa-miR-93-5p | 26 | ANK2 | Sponge network | -0.465 | 0.04703 | -1.603 | 1.0E-5 | 0.476 |
| 10659 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | 0.16 | 0.50785 | 0.358 | 0.13727 | 0.476 |
| 10660 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 59 | NTRK3 | Sponge network | -0.777 | 0.16667 | -1.77 | 3.0E-5 | 0.476 |
| 10661 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-664a-3p | 10 | SCN11A | Sponge network | 0.532 | 0.00505 | -1.15 | 0.00299 | 0.476 |
| 10662 | RP11-464F9.1 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 12 | RASAL2 | Sponge network | 0.959 | 0 | 0.869 | 0 | 0.476 |
| 10663 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 53 | CADM2 | Sponge network | -0.563 | 0.02545 | -3.782 | 0 | 0.476 |
| 10664 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ABI2 | Sponge network | -0.612 | 0.00094 | 0.175 | 0.14787 | 0.476 |
| 10665 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 10 | FGF5 | Sponge network | 0.299 | 0.57722 | 0.549 | 0.28659 | 0.476 |
| 10666 | GAS6-AS2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-493-5p | 15 | SYNM | Sponge network | 0.16 | 0.50785 | -2.309 | 1.0E-5 | 0.476 |
| 10667 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | PDGFC | Sponge network | -1.439 | 4.0E-5 | -0.33 | 0.06483 | 0.476 |
| 10668 | LINC00641 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 61 | DST | Sponge network | -0.222 | 0.32445 | -0.713 | 0.00096 | 0.476 |
| 10669 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | MITF | Sponge network | -0.811 | 2.0E-5 | -0.769 | 0.00022 | 0.476 |
| 10670 | RP11-67L2.2 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 36 | ARHGEF12 | Sponge network | 0.72 | 0.0001 | 0.09 | 0.35693 | 0.476 |
| 10671 | CTD-2314G24.2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-532-3p | 19 | SYNM | Sponge network | 0.246 | 0.59912 | -2.309 | 1.0E-5 | 0.476 |
| 10672 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p | 13 | GPC6 | Sponge network | 0.693 | 0.00076 | -0.054 | 0.81465 | 0.476 |
| 10673 | RP11-6O2.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-340-5p;hsa-miR-539-5p;hsa-miR-769-5p | 11 | PRUNE2 | Sponge network | 0.331 | 0.57247 | -1.733 | 0.00022 | 0.476 |
| 10674 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | LRRC7 | Sponge network | -1.439 | 4.0E-5 | -1.341 | 0.01193 | 0.476 |
| 10675 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | ST6GALNAC3 | Sponge network | -1.575 | 6.0E-5 | -0.987 | 0 | 0.475 |
| 10676 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-93-5p | 26 | PGR | Sponge network | -0.154 | 0.78939 | -1.704 | 0 | 0.475 |
| 10677 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-5p | 15 | CITED2 | Sponge network | -0.976 | 0.00025 | -1.545 | 0 | 0.475 |
| 10678 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-501-5p | 12 | DACT1 | Sponge network | -0.777 | 0.16667 | 0.783 | 0.00072 | 0.475 |
| 10679 | RP5-1198O20.4 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | PCDH18 | Sponge network | 0.997 | 0.00066 | 0.216 | 0.24645 | 0.475 |
| 10680 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p | 16 | PCDH9 | Sponge network | 0.176 | 0.37402 | -1.823 | 4.0E-5 | 0.475 |
| 10681 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | CNTN1 | Sponge network | 0.056 | 0.81195 | -2.594 | 0 | 0.475 |
| 10682 | RP11-166D19.1 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 10 | DIRAS1 | Sponge network | -0.576 | 0.10121 | -2.518 | 0 | 0.475 |
| 10683 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 26 | TSHZ3 | Sponge network | -0.246 | 0.35034 | -0.556 | 0.01516 | 0.475 |
| 10684 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | PARK2 | Sponge network | 0.101 | 0.60919 | -1.892 | 0 | 0.475 |
| 10685 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 25 | RBMS3 | Sponge network | 0.727 | 3.0E-5 | -0.519 | 0.03551 | 0.475 |
| 10686 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-424-5p | 16 | ANK2 | Sponge network | 1.344 | 0 | -1.603 | 1.0E-5 | 0.475 |
| 10687 | USP3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-455-5p | 11 | THSD7A | Sponge network | -0.162 | 0.47687 | 0.242 | 0.25154 | 0.475 |
| 10688 | LINC00473 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | -1.203 | 0.03881 | -1.09 | 0.00014 | 0.475 |
| 10689 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 53 | AFF4 | Sponge network | 0.683 | 1.0E-5 | -0.266 | 0.01565 | 0.475 |
| 10690 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | CAV1 | Sponge network | -1.349 | 0.00017 | -1.013 | 6.0E-5 | 0.475 |
| 10691 | LINC00957 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-590-3p | 10 | PTPN13 | Sponge network | -0.421 | 0.0114 | -1.208 | 0 | 0.475 |
| 10692 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-874-3p | 21 | NR2C2 | Sponge network | 0.494 | 0.00339 | 0.21 | 0.03224 | 0.475 |
| 10693 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-582-5p;hsa-miR-7-5p | 29 | MOB1B | Sponge network | 0.637 | 0.00069 | 0.197 | 0.15266 | 0.475 |
| 10694 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-98-5p | 34 | PBX1 | Sponge network | -0.154 | 0.78939 | -1.206 | 0 | 0.475 |
| 10695 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | RARB | Sponge network | -1.281 | 0.00012 | -0.825 | 2.0E-5 | 0.475 |
| 10696 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 20 | LRRC4 | Sponge network | -0.737 | 0.06182 | -1.774 | 0 | 0.475 |
| 10697 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 36 | ADARB1 | Sponge network | -0.318 | 0.65847 | -0.335 | 0.06631 | 0.475 |
| 10698 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 23 | ABL1 | Sponge network | 0.997 | 0.00066 | -0.215 | 0.07804 | 0.475 |
| 10699 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 24 | NUAK1 | Sponge network | -0.49 | 0.04946 | 0.904 | 0 | 0.475 |
| 10700 | AC093110.3 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p | 10 | ZNF638 | Sponge network | 1.044 | 2.0E-5 | 0.22 | 0.01214 | 0.475 |
| 10701 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 49 | QKI | Sponge network | -1.249 | 7.0E-5 | -0.333 | 0.04111 | 0.475 |
| 10702 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 14 | CSRNP1 | Sponge network | -2.452 | 0 | -1.448 | 0 | 0.475 |
| 10703 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-940 | 11 | CRTC3 | Sponge network | 0.056 | 0.81195 | 0.164 | 0.16786 | 0.475 |
| 10704 | RP11-464F9.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p | 25 | ARHGEF12 | Sponge network | 0.959 | 0 | 0.09 | 0.35693 | 0.475 |
| 10705 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CLASP2 | Sponge network | 0.673 | 0 | -0.038 | 0.71 | 0.475 |
| 10706 | LINC00957 |
hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | MYO9A | Sponge network | -0.421 | 0.0114 | -0.147 | 0.32373 | 0.475 |
| 10707 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 26 | BCL6 | Sponge network | -0.737 | 0.06182 | -0.211 | 0.19489 | 0.475 |
| 10708 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-577 | 10 | PLAGL1 | Sponge network | -0.323 | 0.01372 | -0.588 | 0.00912 | 0.475 |
| 10709 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-708-5p | 23 | RAP1A | Sponge network | -1.092 | 4.0E-5 | -0.929 | 0 | 0.475 |
| 10710 | PWAR6 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 16 | SEPT6 | Sponge network | -1.575 | 6.0E-5 | -0.598 | 0.00181 | 0.475 |
| 10711 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | AFF3 | Sponge network | -1.203 | 0.03881 | -2.373 | 0 | 0.475 |
| 10712 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 0.559 | 0.00026 | 0.943 | 0 | 0.475 |
| 10713 | FAM66C |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | FAM117A | Sponge network | -0.901 | 0.00019 | -1.379 | 0 | 0.475 |
| 10714 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | PPM1A | Sponge network | -0.612 | 0.00094 | -0.623 | 0 | 0.475 |
| 10715 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 44 | PAFAH1B1 | Sponge network | -1.047 | 0 | -0.429 | 0 | 0.475 |
| 10716 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p | 34 | EPHA5 | Sponge network | -2.62 | 0 | -0.957 | 0.01733 | 0.475 |
| 10717 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-629-5p;hsa-miR-96-5p | 23 | SOX5 | Sponge network | -0.517 | 0.02935 | -1.091 | 4.0E-5 | 0.475 |
| 10718 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | ROBO1 | Sponge network | -0.769 | 0.00035 | -0.009 | 0.96154 | 0.475 |
| 10719 | AC006116.24 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-93-5p | 12 | NIN | Sponge network | -0.473 | 0.07602 | -0.253 | 0.13794 | 0.475 |
| 10720 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-450b-5p | 26 | BCL2 | Sponge network | -0.583 | 0.14589 | -0.899 | 0 | 0.475 |
| 10721 | MIR143HG |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | TLE4 | Sponge network | -0.739 | 0.0574 | -1.157 | 0 | 0.475 |
| 10722 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-940 | 36 | NOTCH2 | Sponge network | -0.096 | 0.67958 | 0.138 | 0.35163 | 0.475 |
| 10723 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 14 | BCLAF1 | Sponge network | 0.421 | 0.00084 | 0.211 | 0.00684 | 0.475 |
| 10724 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | RASSF2 | Sponge network | -0.611 | 0.09607 | -0.273 | 0.21961 | 0.475 |
| 10725 | MEG3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 28 | EYA4 | Sponge network | 0.249 | 0.35902 | 0.3 | 0.36876 | 0.475 |
| 10726 | CTD-2554C21.2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p | 13 | SORCS1 | Sponge network | -1.654 | 0.00144 | -3.492 | 0 | 0.475 |
| 10727 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | TET1 | Sponge network | -0.222 | 0.32445 | -0.135 | 0.52138 | 0.475 |
| 10728 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 36 | IGF1 | Sponge network | -2.46 | 0 | -0.204 | 0.5898 | 0.475 |
| 10729 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | JAKMIP2 | Sponge network | -0.612 | 0.00094 | -1.388 | 0 | 0.475 |
| 10730 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 29 | SCN9A | Sponge network | -0.901 | 0.00019 | -0.525 | 0.10593 | 0.475 |
| 10731 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-424-5p | 16 | PRKAB2 | Sponge network | -1.331 | 3.0E-5 | -0.367 | 0.01371 | 0.475 |
| 10732 | SNHG14 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | NIN | Sponge network | -1.111 | 0.0003 | -0.253 | 0.13794 | 0.475 |
| 10733 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940 | 61 | NTRK3 | Sponge network | -0.793 | 7.0E-5 | -1.77 | 3.0E-5 | 0.475 |
| 10734 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 40 | ZFHX3 | Sponge network | 1.302 | 7.0E-5 | 0.389 | 0.01363 | 0.475 |
| 10735 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-193b-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-500a-5p;hsa-miR-98-5p | 15 | DMD | Sponge network | -1.116 | 0.23686 | -1.506 | 0 | 0.475 |
| 10736 | MEG3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | PCDH9 | Sponge network | 0.249 | 0.35902 | -1.823 | 4.0E-5 | 0.475 |
| 10737 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p | 23 | TBL1XR1 | Sponge network | 0.401 | 0.00066 | 0.578 | 0 | 0.475 |
| 10738 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p;hsa-miR-935;hsa-miR-98-5p | 25 | LIFR | Sponge network | -2.62 | 0 | -1.794 | 0 | 0.475 |
| 10739 | RP11-53O19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 25 | NAV3 | Sponge network | -0.405 | 0.22013 | 0.212 | 0.47729 | 0.475 |
| 10740 | LINC00578 |
hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 17 | RNF144A | Sponge network | 0.811 | 0.03228 | 0.594 | 0.0009 | 0.475 |
| 10741 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p | 30 | ADARB1 | Sponge network | 0.176 | 0.37402 | -0.335 | 0.06631 | 0.475 |
| 10742 | LINC00865 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p | 10 | ZNF536 | Sponge network | -1.249 | 7.0E-5 | -3.115 | 0 | 0.475 |
| 10743 | TINCR |
hsa-miR-141-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-940 | 10 | RANBP9 | Sponge network | -0.664 | 0.14543 | -0.191 | 0.04628 | 0.475 |
| 10744 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-590-5p | 12 | RCAN2 | Sponge network | 0.176 | 0.37402 | -0.33 | 0.15172 | 0.475 |
| 10745 | RP11-21A7A.2 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-877-5p | 15 | ANK2 | Sponge network | -1.116 | 0.23686 | -1.603 | 1.0E-5 | 0.475 |
| 10746 | RP11-244O19.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 11 | ITGB1 | Sponge network | -0.096 | 0.67958 | 0.524 | 0.00017 | 0.475 |
| 10747 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 23 | SYNE1 | Sponge network | 0.082 | 0.55654 | -0.738 | 0.00316 | 0.475 |
| 10748 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | ADAMTSL3 | Sponge network | -0.899 | 6.0E-5 | -1.559 | 0 | 0.475 |
| 10749 | RP11-403I13.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p | 10 | HMCN1 | Sponge network | 0.395 | 0.15451 | 0.809 | 0.00544 | 0.475 |
| 10750 | LINC00957 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-942-5p | 32 | IGFBP5 | Sponge network | -0.421 | 0.0114 | -0.806 | 0.00105 | 0.475 |
| 10751 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-5p | 15 | BCL2 | Sponge network | -0.376 | 0.00337 | -0.899 | 0 | 0.475 |
| 10752 | BVES-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p | 11 | CELF4 | Sponge network | -2.62 | 0 | -1.135 | 0.00344 | 0.475 |
| 10753 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p | 29 | MYO9A | Sponge network | -1.645 | 0 | -0.147 | 0.32373 | 0.475 |
| 10754 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-96-5p | 10 | ZYX | Sponge network | -0.576 | 0.10121 | 0.019 | 0.89763 | 0.475 |
| 10755 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | PEG3 | Sponge network | -0.405 | 0.22013 | -1.74 | 0 | 0.475 |
| 10756 | AC016747.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-877-5p | 15 | NAV3 | Sponge network | 0.199 | 0.38015 | 0.212 | 0.47729 | 0.475 |
| 10757 | MEG3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p | 11 | SMARCA1 | Sponge network | 0.249 | 0.35902 | -0.684 | 0.00159 | 0.475 |
| 10758 | FENDRR |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 14 | CITED2 | Sponge network | -1.281 | 0.00012 | -1.545 | 0 | 0.475 |
| 10759 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p | 40 | FAT3 | Sponge network | -1.161 | 0.00591 | -1.601 | 0.00051 | 0.475 |
| 10760 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-940 | 24 | IGF1 | Sponge network | 0.225 | 0.30644 | -0.204 | 0.5898 | 0.475 |
| 10761 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 49 | PIK3R1 | Sponge network | -1.697 | 0.00889 | -0.133 | 0.36854 | 0.475 |
| 10762 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-576-5p | 17 | NCAM2 | Sponge network | -2.62 | 0 | -0.482 | 0.22565 | 0.475 |
| 10763 | RP11-254F7.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-539-5p | 10 | PRUNE2 | Sponge network | 0.176 | 0.37402 | -1.733 | 0.00022 | 0.475 |
| 10764 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | LRP1B | Sponge network | -1.697 | 0.00889 | -2.163 | 0 | 0.475 |
| 10765 | CTD-3064M3.3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-582-5p;hsa-miR-93-3p | 14 | WWTR1 | Sponge network | -0.469 | 0.08721 | -0.345 | 0.11997 | 0.475 |
| 10766 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-93-5p | 14 | TXNIP | Sponge network | -0.793 | 7.0E-5 | -1.277 | 0 | 0.475 |
| 10767 | DNM3OS |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | CDH19 | Sponge network | 0.572 | 0.01722 | -3.434 | 0 | 0.475 |
| 10768 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-7-1-3p | 12 | DKK3 | Sponge network | -0.611 | 0.09607 | -0.052 | 0.77783 | 0.475 |
| 10769 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 20 | CBL | Sponge network | 1.601 | 0 | 0.291 | 0.01267 | 0.475 |
| 10770 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 20 | RARB | Sponge network | -0.842 | 0.01361 | -0.825 | 2.0E-5 | 0.475 |
| 10771 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 22 | CDH19 | Sponge network | 0.335 | 0.20596 | -3.434 | 0 | 0.475 |
| 10772 | RP11-967K21.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-542-3p | 17 | MRVI1 | Sponge network | 0.056 | 0.81195 | -1.051 | 0.00022 | 0.475 |
| 10773 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 10 | UVRAG | Sponge network | 0.082 | 0.55654 | -0.017 | 0.83865 | 0.475 |
| 10774 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p | 18 | DYNC1I1 | Sponge network | -0.18 | 0.20343 | -1.12 | 0.00107 | 0.475 |
| 10775 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | PAK3 | Sponge network | -0.323 | 0.01372 | -0.628 | 0.07253 | 0.475 |
| 10776 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 19 | ZFX | Sponge network | 0.673 | 0 | 0.09 | 0.42563 | 0.475 |
| 10777 | FENDRR |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | NR3C2 | Sponge network | -1.281 | 0.00012 | -1.56 | 0 | 0.475 |
| 10778 | MIR497HG |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-503-5p | 20 | ABL1 | Sponge network | -1.439 | 4.0E-5 | -0.215 | 0.07804 | 0.475 |
| 10779 | LINC01018 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | TMEFF2 | Sponge network | -2.915 | 0.00015 | -2.426 | 0 | 0.474 |
| 10780 | LINC00861 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-93-3p | 14 | IL6ST | Sponge network | 0.911 | 0.00854 | -0.402 | 0.00676 | 0.474 |
| 10781 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-3614-5p | 26 | GAS7 | Sponge network | -0.693 | 0.05629 | -0.555 | 0.01602 | 0.474 |
| 10782 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NRXN3 | Sponge network | 0.101 | 0.60919 | -0.893 | 0.04186 | 0.474 |
| 10783 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 52 | BMPR2 | Sponge network | 0.056 | 0.81195 | 0.206 | 0.0635 | 0.474 |
| 10784 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 30 | EPHA3 | Sponge network | -0.793 | 7.0E-5 | 0.185 | 0.53588 | 0.474 |
| 10785 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CLIP1 | Sponge network | -0.129 | 0.57177 | -0.215 | 0.08684 | 0.474 |
| 10786 | LENG8-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-338-5p | 17 | RICTOR | Sponge network | 1.471 | 0 | 0.388 | 0.00188 | 0.474 |
| 10787 | RP11-195B17.1 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-96-5p | 15 | MYO9A | Sponge network | 0.37 | 0.27614 | -0.147 | 0.32373 | 0.474 |
| 10788 | H19 |
hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 13 | PRRX1 | Sponge network | 2.439 | 0 | 1.316 | 0 | 0.474 |
| 10789 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-590-3p | 13 | STARD13 | Sponge network | 0.083 | 0.66405 | -0.187 | 0.27383 | 0.474 |
| 10790 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 15 | DAB2 | Sponge network | -0.976 | 0.00025 | 0.431 | 0.0113 | 0.474 |
| 10791 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | ST6GALNAC3 | Sponge network | -2.117 | 0.00161 | -0.987 | 0 | 0.474 |
| 10792 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 15 | CNTNAP1 | Sponge network | -0.612 | 0.00094 | 0.358 | 0.13727 | 0.474 |
| 10793 | RP11-867G23.10 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-589-3p | 16 | SYT4 | Sponge network | -2.531 | 0 | -3.207 | 0 | 0.474 |
| 10794 | AC016747.3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p | 11 | MN1 | Sponge network | 0.199 | 0.38015 | -0.358 | 0.24172 | 0.474 |
| 10795 | MIR143HG |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-98-5p | 22 | PNRC1 | Sponge network | -0.739 | 0.0574 | -0.646 | 0 | 0.474 |
| 10796 | RP1-151F17.2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | 0.222 | 0.29133 | 0.524 | 0.00017 | 0.474 |
| 10797 | MEG3 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 27 | PCDH17 | Sponge network | 0.249 | 0.35902 | 0.731 | 2.0E-5 | 0.474 |
| 10798 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 35 | BNC2 | Sponge network | -0.738 | 0.21233 | -0.491 | 0.09988 | 0.474 |
| 10799 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 39 | TGFBR3 | Sponge network | -2.117 | 0.00161 | -1.173 | 1.0E-5 | 0.474 |
| 10800 | JAZF1-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | KCTD8 | Sponge network | -0.769 | 0.00035 | -3.359 | 0 | 0.474 |
| 10801 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | -0.162 | 0.47687 | -1.465 | 0.00033 | 0.474 |
| 10802 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 20 | SETBP1 | Sponge network | -0.246 | 0.35034 | -1.302 | 1.0E-5 | 0.474 |
| 10803 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-590-3p | 28 | PHC3 | Sponge network | 0.494 | 0.00339 | 0.212 | 0.0499 | 0.474 |
| 10804 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p | 27 | ABI2 | Sponge network | 0.176 | 0.37402 | 0.175 | 0.14787 | 0.474 |
| 10805 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | EGR2 | Sponge network | -0.576 | 0.10121 | 0.309 | 0.17079 | 0.474 |
| 10806 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-98-5p | 37 | THBS1 | Sponge network | -1.111 | 0.0003 | 0.741 | 0.00386 | 0.474 |
| 10807 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 36 | TACC1 | Sponge network | 1.302 | 7.0E-5 | -0.362 | 0.05406 | 0.474 |
| 10808 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 10 | RIMS1 | Sponge network | -0.611 | 0.09607 | -3.143 | 0 | 0.474 |
| 10809 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | ZFHX4 | Sponge network | -1.886 | 0 | 0.018 | 0.95574 | 0.474 |
| 10810 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 18 | GLI3 | Sponge network | 0.101 | 0.60919 | -0.615 | 0.01482 | 0.474 |
| 10811 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | PRDM5 | Sponge network | -1.216 | 0 | -1.09 | 0.00014 | 0.474 |
| 10812 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | ZNF521 | Sponge network | -0.777 | 0.1134 | -0.111 | 0.58068 | 0.474 |
| 10813 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-589-3p | 14 | ADARB1 | Sponge network | -4.463 | 0.00025 | -0.335 | 0.06631 | 0.474 |
| 10814 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p | 17 | ABI2 | Sponge network | 0.497 | 0.01451 | 0.175 | 0.14787 | 0.474 |
| 10815 | SNHG14 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 16 | EHD3 | Sponge network | -1.111 | 0.0003 | -0.484 | 0.01747 | 0.474 |
| 10816 | FAM157A |
hsa-miR-130a-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p | 11 | RASAL2 | Sponge network | 0.813 | 1.0E-5 | 0.869 | 0 | 0.474 |
| 10817 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 20 | LATS2 | Sponge network | -2.452 | 0 | 0.159 | 0.2304 | 0.474 |
| 10818 | LINC00473 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 12 | RIMS1 | Sponge network | -1.203 | 0.03881 | -3.143 | 0 | 0.474 |
| 10819 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 29 | GPC6 | Sponge network | -1.281 | 0.00012 | -0.054 | 0.81465 | 0.474 |
| 10820 | RP11-774O3.3 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | ZDBF2 | Sponge network | -0.487 | 0.02157 | -0.998 | 0.00025 | 0.474 |
| 10821 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 32 | BTG2 | Sponge network | -0.769 | 0.00035 | -1.763 | 0 | 0.474 |
| 10822 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429 | 13 | SPG20 | Sponge network | -0.154 | 0.53954 | -1.16 | 0 | 0.474 |
| 10823 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p | 20 | PGR | Sponge network | -0.318 | 0.65847 | -1.704 | 0 | 0.474 |
| 10824 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-769-5p | 25 | SYNPO2 | Sponge network | -0.154 | 0.78939 | -2.342 | 0 | 0.474 |
| 10825 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | SON | Sponge network | 0.72 | 0.0001 | 0.168 | 0.04406 | 0.474 |
| 10826 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | 0.873 | 0 | 0.206 | 0.06751 | 0.474 |
| 10827 | NDUFA6-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-493-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 20 | PRUNE2 | Sponge network | -0.355 | 0.0077 | -1.733 | 0.00022 | 0.474 |
| 10828 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 18 | USP12 | Sponge network | 0.614 | 1.0E-5 | -0.102 | 0.40768 | 0.474 |
| 10829 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | SASH1 | Sponge network | -1.161 | 0.00591 | -0.502 | 0.00175 | 0.474 |
| 10830 | TUG1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-654-3p;hsa-miR-664a-3p | 12 | ITCH | Sponge network | 0.972 | 0 | 0.084 | 0.34058 | 0.474 |
| 10831 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-93-3p | 18 | RPS6KA2 | Sponge network | -0.563 | 0.02545 | -0.57 | 0.00013 | 0.474 |
| 10832 | AP001347.6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-378a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 64 | LPP | Sponge network | -1.161 | 0.00591 | -0.459 | 0.04475 | 0.474 |
| 10833 | RP11-156E6.1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-495-3p | 11 | STAG2 | Sponge network | 1.266 | 0 | 0.252 | 0.00438 | 0.474 |
| 10834 | RP11-13K12.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 21 | MED12L | Sponge network | -0.318 | 0.65847 | -0.194 | 0.55299 | 0.474 |
| 10835 | CTD-2554C21.2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-429 | 12 | ITPKB | Sponge network | -1.654 | 0.00144 | -0.704 | 0.00106 | 0.474 |
| 10836 | OIP5-AS1 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 25 | DICER1 | Sponge network | 0.421 | 0.00084 | 0.042 | 0.674 | 0.474 |
| 10837 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-340-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 10 | KCNAB1 | Sponge network | -1.161 | 0.00591 | -0.755 | 2.0E-5 | 0.474 |
| 10838 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 68 | THRB | Sponge network | -0.49 | 0.04946 | -1.117 | 0.0004 | 0.474 |
| 10839 | HHIP-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-532-3p;hsa-miR-590-3p | 12 | MRVI1 | Sponge network | -0.737 | 0.10822 | -1.051 | 0.00022 | 0.474 |
| 10840 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-7-5p | 10 | FNBP1 | Sponge network | -0.342 | 0.02288 | -0.969 | 4.0E-5 | 0.474 |
| 10841 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-493-3p;hsa-miR-590-3p | 14 | JAKMIP2 | Sponge network | -0.096 | 0.67958 | -1.388 | 0 | 0.474 |
| 10842 | RP11-53O19.3 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 17 | ABL2 | Sponge network | 1.205 | 0 | 0.82 | 0 | 0.474 |
| 10843 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -2.531 | 0 | -0.564 | 1.0E-5 | 0.474 |
| 10844 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 48 | RUNX1T1 | Sponge network | 0.16 | 0.50785 | -0.344 | 0.2374 | 0.474 |
| 10845 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-93-5p | 16 | SLITRK3 | Sponge network | -0.842 | 0.01361 | -3.328 | 0 | 0.474 |
| 10846 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 30 | ZFHX4 | Sponge network | -0.318 | 0.65847 | 0.018 | 0.95574 | 0.474 |
| 10847 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | CHD2 | Sponge network | 1.601 | 0 | 0.049 | 0.55371 | 0.474 |
| 10848 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | ANK2 | Sponge network | 0.693 | 0.00076 | -1.603 | 1.0E-5 | 0.474 |
| 10849 | BCYRN1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FBXO11 | Sponge network | 0.311 | 0.10344 | 0.142 | 0.04938 | 0.474 |
| 10850 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TAL1 | Sponge network | 0.335 | 0.20596 | -0.462 | 0.00574 | 0.474 |
| 10851 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | CEP170 | Sponge network | 0.421 | 0.00084 | 0.943 | 0 | 0.474 |
| 10852 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p | 10 | ZFHX4 | Sponge network | -0.326 | 0.28809 | 0.018 | 0.95574 | 0.474 |
| 10853 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 38 | BCL2 | Sponge network | -0.487 | 0.02157 | -0.899 | 0 | 0.474 |
| 10854 | LINC00174 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-625-5p | 15 | ZMYM2 | Sponge network | 1.253 | 0 | 0.279 | 0.01273 | 0.474 |
| 10855 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p | 24 | SOX5 | Sponge network | -0.583 | 0.14589 | -1.091 | 4.0E-5 | 0.474 |
| 10856 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | TBX18 | Sponge network | 0.889 | 9.0E-5 | 0.843 | 0.02945 | 0.474 |
| 10857 | FENDRR |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 50 | TRPS1 | Sponge network | -1.281 | 0.00012 | -0.191 | 0.44109 | 0.474 |
| 10858 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | SYNE1 | Sponge network | -2.915 | 0.00015 | -0.738 | 0.00316 | 0.474 |
| 10859 | FGF14-AS2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | RYR2 | Sponge network | -1.582 | 8.0E-5 | -1.466 | 0.00031 | 0.474 |
| 10860 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 63 | LPP | Sponge network | 0.299 | 0.57722 | -0.459 | 0.04475 | 0.474 |
| 10861 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | TSHZ3 | Sponge network | 0.214 | 0.18246 | -0.556 | 0.01516 | 0.474 |
| 10862 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 49 | BMPR2 | Sponge network | 0.683 | 1.0E-5 | 0.206 | 0.0635 | 0.474 |
| 10863 | AC010226.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-590-3p | 14 | CDH19 | Sponge network | -0.811 | 2.0E-5 | -3.434 | 0 | 0.474 |
| 10864 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-96-5p | 20 | FAT3 | Sponge network | -0.187 | 0.23787 | -1.601 | 0.00051 | 0.474 |
| 10865 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p | 12 | SLITRK6 | Sponge network | -1.092 | 4.0E-5 | -2.121 | 0 | 0.474 |
| 10866 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | PDGFC | Sponge network | -0.901 | 0.00019 | -0.33 | 0.06483 | 0.474 |
| 10867 | FAM66C |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 14 | RHOB | Sponge network | -0.901 | 0.00019 | -1.052 | 0 | 0.474 |
| 10868 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p | 15 | RELN | Sponge network | -1.654 | 0.00144 | -2.403 | 0 | 0.474 |
| 10869 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p | 13 | CRTC3 | Sponge network | 0.385 | 0.10067 | 0.164 | 0.16786 | 0.474 |
| 10870 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 58 | BMPR2 | Sponge network | 0.249 | 0.35902 | 0.206 | 0.0635 | 0.474 |
| 10871 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | SASH1 | Sponge network | -0.611 | 0.09607 | -0.502 | 0.00175 | 0.474 |
| 10872 | RP11-156P1.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 16 | PRUNE2 | Sponge network | 0.214 | 0.18246 | -1.733 | 0.00022 | 0.474 |
| 10873 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | -0.738 | 0.21233 | -1.74 | 0 | 0.474 |
| 10874 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 62 | NTRK3 | Sponge network | -2.915 | 0.00015 | -1.77 | 3.0E-5 | 0.474 |
| 10875 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-5p;hsa-miR-301a-3p | 12 | ZBTB4 | Sponge network | -0.733 | 0.19469 | -0.739 | 0 | 0.474 |
| 10876 | RP11-473I1.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p | 22 | CBX5 | Sponge network | 0.542 | 0.00054 | 0.165 | 0.24446 | 0.474 |
| 10877 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-590-3p | 22 | LRIG1 | Sponge network | -0.896 | 0.00099 | -0.537 | 0.00588 | 0.474 |
| 10878 | RP11-119F7.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | CREB1 | Sponge network | 0.225 | 0.30644 | 0.203 | 0.00537 | 0.474 |
| 10879 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 27 | ZBTB4 | Sponge network | -0.563 | 0.02545 | -0.739 | 0 | 0.474 |
| 10880 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-664a-3p | 18 | ST6GAL2 | Sponge network | -0.358 | 0.17255 | -1.201 | 0.00597 | 0.474 |
| 10881 | TP73-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 23 | PRKCB | Sponge network | -0.563 | 0.02545 | -1.313 | 2.0E-5 | 0.474 |
| 10882 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p | 15 | DYNC1I1 | Sponge network | 0.056 | 0.81195 | -1.12 | 0.00107 | 0.474 |
| 10883 | RP13-516M14.1 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577 | 10 | MLLT11 | Sponge network | 0.389 | 0.04293 | -0.661 | 0.01733 | 0.474 |
| 10884 | LINC00702 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 13 | LZTS1 | Sponge network | -0.737 | 0.06182 | 1.664 | 0 | 0.473 |
| 10885 | RP11-53O19.3 |
hsa-miR-101-3p;hsa-miR-139-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-497-5p | 10 | SUZ12 | Sponge network | 1.205 | 0 | 0.619 | 0 | 0.473 |
| 10886 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-590-3p | 15 | JAKMIP2 | Sponge network | -0.487 | 0.02157 | -1.388 | 0 | 0.473 |
| 10887 | RP11-819C21.1 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-629-5p | 28 | UBR3 | Sponge network | 0.537 | 0.00139 | -0.021 | 0.82774 | 0.473 |
| 10888 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | HOOK3 | Sponge network | -0.49 | 0.04946 | 0.273 | 0.09957 | 0.473 |
| 10889 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p | 10 | GAS1 | Sponge network | -0.611 | 0.09607 | -1.047 | 0.00364 | 0.473 |
| 10890 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p | 14 | TXNIP | Sponge network | 0.194 | 0.64278 | -1.277 | 0 | 0.473 |
| 10891 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 16 | JAKMIP2 | Sponge network | 0.997 | 0.00066 | -1.388 | 0 | 0.473 |
| 10892 | MALAT1 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | UBR3 | Sponge network | 0.782 | 0.00016 | -0.021 | 0.82774 | 0.473 |
| 10893 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 19 | PCDH9 | Sponge network | -0.246 | 0.35034 | -1.823 | 4.0E-5 | 0.473 |
| 10894 | CTD-2006H14.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | BRD3 | Sponge network | 1.378 | 0 | 0.37 | 0.00013 | 0.473 |
| 10895 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-500a-5p | 15 | HLF | Sponge network | -2.076 | 0.00548 | -2.443 | 0 | 0.473 |
| 10896 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p | 21 | FBXO32 | Sponge network | -0.318 | 0.65847 | -0.495 | 0.07561 | 0.473 |
| 10897 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-628-5p;hsa-miR-629-3p | 12 | CHD2 | Sponge network | 1.122 | 0 | 0.049 | 0.55371 | 0.473 |
| 10898 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 14 | LATS2 | Sponge network | -0.358 | 0.17255 | 0.159 | 0.2304 | 0.473 |
| 10899 | PWAR6 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-3p | 22 | ABL1 | Sponge network | -1.575 | 6.0E-5 | -0.215 | 0.07804 | 0.473 |
| 10900 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p | 24 | GPC6 | Sponge network | -0.896 | 0.00099 | -0.054 | 0.81465 | 0.473 |
| 10901 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 37 | ZFHX4 | Sponge network | -0.842 | 0.01361 | 0.018 | 0.95574 | 0.473 |
| 10902 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-450b-5p | 13 | NAV3 | Sponge network | -2.531 | 0 | 0.212 | 0.47729 | 0.473 |
| 10903 | ARHGEF26-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | -2.46 | 0 | -1.379 | 0 | 0.473 |
| 10904 | MIR143HG |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 11 | PEA15 | Sponge network | -0.739 | 0.0574 | 0.009 | 0.93318 | 0.473 |
| 10905 | AC159540.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-628-5p | 13 | TRIM33 | Sponge network | 1.122 | 0 | 0.402 | 7.0E-5 | 0.473 |
| 10906 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 17 | KCNAB1 | Sponge network | -0.576 | 0.10121 | -0.755 | 2.0E-5 | 0.473 |
| 10907 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 22 | LRRC55 | Sponge network | -0.576 | 0.10121 | -0.026 | 0.93163 | 0.473 |
| 10908 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 16 | ABCB5 | Sponge network | 0.572 | 0.01722 | -0.956 | 0.04077 | 0.473 |
| 10909 | FENDRR |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-96-5p | 31 | ASTN1 | Sponge network | -1.281 | 0.00012 | -2.389 | 2.0E-5 | 0.473 |
| 10910 | RP11-597D13.9 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-664a-3p | 11 | MN1 | Sponge network | 1.302 | 7.0E-5 | -0.358 | 0.24172 | 0.473 |
| 10911 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PTPN13 | Sponge network | -0.942 | 0 | -1.208 | 0 | 0.473 |
| 10912 | RP11-2B6.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 10 | SHPRH | Sponge network | 0.539 | 0.03722 | 0.098 | 0.40994 | 0.473 |
| 10913 | PWAR6 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p | 19 | CSDE1 | Sponge network | -1.575 | 6.0E-5 | -0.493 | 0 | 0.473 |
| 10914 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-502-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | SETBP1 | Sponge network | -0.351 | 0.11358 | -1.302 | 1.0E-5 | 0.473 |
| 10915 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | 0.826 | 1.0E-5 | 0.206 | 0.06751 | 0.473 |
| 10916 | U91328.19 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-7-1-3p | 18 | NRXN3 | Sponge network | -0.303 | 0.21002 | -0.893 | 0.04186 | 0.473 |
| 10917 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-98-5p | 37 | NTRK3 | Sponge network | -0.473 | 0.07602 | -1.77 | 3.0E-5 | 0.473 |
| 10918 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | -2.69 | 0.0001 | -1.277 | 0 | 0.473 |
| 10919 | PWAR6 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | SPOP | Sponge network | -1.575 | 6.0E-5 | -0.419 | 1.0E-5 | 0.473 |
| 10920 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 57 | BMPR2 | Sponge network | 0.083 | 0.66405 | 0.206 | 0.0635 | 0.473 |
| 10921 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 44 | CHD5 | Sponge network | -0.222 | 0.32445 | -0.922 | 0.01521 | 0.473 |
| 10922 | LINC00641 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | ERCC4 | Sponge network | -0.222 | 0.32445 | 0.126 | 0.15266 | 0.473 |
| 10923 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-629-3p | 15 | ESR1 | Sponge network | -2.531 | 0 | -1.035 | 3.0E-5 | 0.473 |
| 10924 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | TAL1 | Sponge network | 0.194 | 0.64278 | -0.462 | 0.00574 | 0.473 |
| 10925 | RP11-400K9.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 23 | NTRK3 | Sponge network | -0.733 | 0.19469 | -1.77 | 3.0E-5 | 0.473 |
| 10926 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-576-5p | 29 | PBX1 | Sponge network | 0.335 | 0.20596 | -1.206 | 0 | 0.473 |
| 10927 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 27 | HECA | Sponge network | 0.72 | 0.0001 | -0.217 | 0.03463 | 0.473 |
| 10928 | RP11-254F7.2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-486-5p | 12 | SYNM | Sponge network | 0.176 | 0.37402 | -2.309 | 1.0E-5 | 0.473 |
| 10929 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421 | 16 | CNTNAP3 | Sponge network | -0.583 | 0.14589 | -1.465 | 0.00033 | 0.473 |
| 10930 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | ZFHX3 | Sponge network | 0.532 | 0.00505 | 0.389 | 0.01363 | 0.473 |
| 10931 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | ERG | Sponge network | -0.896 | 0.00099 | 0.002 | 0.99011 | 0.473 |
| 10932 | RP11-774O3.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-590-3p | 22 | CYLD | Sponge network | -0.487 | 0.02157 | -0.367 | 0.00171 | 0.473 |
| 10933 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 24 | GJA1 | Sponge network | 0.572 | 0.01722 | -0.485 | 0.02179 | 0.473 |
| 10934 | DNM3OS |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | PRKCB | Sponge network | 0.572 | 0.01722 | -1.313 | 2.0E-5 | 0.473 |
| 10935 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | MYO9A | Sponge network | -0.612 | 0.00094 | -0.147 | 0.32373 | 0.473 |
| 10936 | RP11-307B6.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 15 | JAZF1 | Sponge network | -4.463 | 0.00025 | -0.377 | 0.02677 | 0.473 |
| 10937 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-330-5p;hsa-miR-7-5p;hsa-miR-96-5p | 10 | ZYX | Sponge network | -0.737 | 0.06182 | 0.019 | 0.89763 | 0.473 |
| 10938 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-98-5p | 33 | LRIG1 | Sponge network | -0.129 | 0.57177 | -0.537 | 0.00588 | 0.473 |
| 10939 | CTA-363E19.2 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-485-3p | 14 | PIK3C2A | Sponge network | 1.055 | 0 | 0.061 | 0.53776 | 0.473 |
| 10940 | JAZF1-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3913-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 38 | IGFBP5 | Sponge network | -0.769 | 0.00035 | -0.806 | 0.00105 | 0.473 |
| 10941 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | LRRC55 | Sponge network | -0.49 | 0.04946 | -0.026 | 0.93163 | 0.473 |
| 10942 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SYT4 | Sponge network | -1.057 | 0.00029 | -3.207 | 0 | 0.473 |
| 10943 | LINC00641 |
hsa-miR-140-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | CHD6 | Sponge network | -0.222 | 0.32445 | 0.32 | 0.01326 | 0.473 |
| 10944 | LINC00957 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | EPHA3 | Sponge network | -0.421 | 0.0114 | 0.185 | 0.53588 | 0.473 |
| 10945 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-7-1-3p | 25 | MYO9A | Sponge network | -0.842 | 0.01361 | -0.147 | 0.32373 | 0.473 |
| 10946 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 30 | SCN9A | Sponge network | -0.222 | 0.32445 | -0.525 | 0.10593 | 0.473 |
| 10947 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940 | 32 | MEF2C | Sponge network | 0.199 | 0.38015 | -0.499 | 0.01448 | 0.473 |
| 10948 | LINC00957 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 10 | TMEFF2 | Sponge network | -0.421 | 0.0114 | -2.426 | 0 | 0.473 |
| 10949 | RASSF8-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 16 | CXCL12 | Sponge network | 0.335 | 0.20596 | -1.421 | 0 | 0.473 |
| 10950 | RP11-464F9.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 28 | CREB1 | Sponge network | 0.959 | 0 | 0.203 | 0.00537 | 0.473 |
| 10951 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-708-5p;hsa-miR-98-5p | 11 | MPL | Sponge network | -0.942 | 0 | -0.624 | 0.00133 | 0.473 |
| 10952 | CTD-2303H24.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 54 | MDM4 | Sponge network | 0.415 | 0.14657 | 0.345 | 0.00762 | 0.473 |
| 10953 | RP11-175O19.4 |
hsa-miR-100-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-363-3p | 10 | ATP2A2 | Sponge network | 0.917 | 0 | 0.604 | 0 | 0.473 |
| 10954 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 10 | MLLT10 | Sponge network | 1.055 | 0 | 0.105 | 0.33292 | 0.473 |
| 10955 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p | 45 | CHD5 | Sponge network | -0.615 | 0.00015 | -0.922 | 0.01521 | 0.473 |
| 10956 | AP001347.6 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PCDH9 | Sponge network | -1.161 | 0.00591 | -1.823 | 4.0E-5 | 0.473 |
| 10957 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 32 | IL6ST | Sponge network | -0.811 | 2.0E-5 | -0.402 | 0.00676 | 0.473 |
| 10958 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-766-3p;hsa-miR-93-5p | 26 | CHD5 | Sponge network | -1.654 | 0.00144 | -0.922 | 0.01521 | 0.473 |
| 10959 | MALAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | EXOC8 | Sponge network | 0.782 | 0.00016 | 0.216 | 0.00598 | 0.473 |
| 10960 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 11 | IGSF10 | Sponge network | -2.039 | 0.03447 | -1.751 | 1.0E-5 | 0.473 |
| 10961 | RP11-244O19.1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 17 | CXCL12 | Sponge network | -0.096 | 0.67958 | -1.421 | 0 | 0.473 |
| 10962 | EMX2OS |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-503-5p | 20 | ABL1 | Sponge network | -0.777 | 0.1134 | -0.215 | 0.07804 | 0.473 |
| 10963 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | LRRC55 | Sponge network | -0.737 | 0.06182 | -0.026 | 0.93163 | 0.473 |
| 10964 | DNM3OS |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 16 | KLF10 | Sponge network | 0.572 | 0.01722 | -0.783 | 0 | 0.473 |
| 10965 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | GALNT13 | Sponge network | -0.739 | 0.00044 | -0.857 | 0.07439 | 0.473 |
| 10966 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 38 | MAF | Sponge network | -0.129 | 0.57177 | -1.257 | 0 | 0.473 |
| 10967 | RP11-326G21.1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PCDH10 | Sponge network | -0.021 | 0.93598 | -1.644 | 0.0078 | 0.473 |
| 10968 | TP73-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429 | 15 | FMNL3 | Sponge network | -0.563 | 0.02545 | 0.555 | 4.0E-5 | 0.473 |
| 10969 | TPTEP1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | ZDBF2 | Sponge network | -1.216 | 0 | -0.998 | 0.00025 | 0.473 |
| 10970 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 47 | CREB3L2 | Sponge network | -2.452 | 0 | -0.525 | 2.0E-5 | 0.473 |
| 10971 | LINC00173 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | PRKCB | Sponge network | 0.056 | 0.8494 | -1.313 | 2.0E-5 | 0.473 |
| 10972 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p | 25 | RSPO2 | Sponge network | -1.249 | 7.0E-5 | -3.038 | 0 | 0.473 |
| 10973 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 51 | FAT3 | Sponge network | 0.532 | 0.00505 | -1.601 | 0.00051 | 0.473 |
| 10974 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-7-5p;hsa-miR-96-5p | 18 | SNX29 | Sponge network | -0.187 | 0.23787 | -0.34 | 0.01371 | 0.473 |
| 10975 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 34 | EPHA7 | Sponge network | -0.737 | 0.10822 | -2.197 | 3.0E-5 | 0.473 |
| 10976 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 49 | NTRK2 | Sponge network | -0.129 | 0.57177 | -0.736 | 0.06495 | 0.473 |
| 10977 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | PLSCR4 | Sponge network | 0.101 | 0.60919 | -0.474 | 0.03833 | 0.473 |
| 10978 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-497-5p | 11 | CBL | Sponge network | 0.959 | 0 | 0.291 | 0.01267 | 0.472 |
| 10979 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 11 | CAV1 | Sponge network | -0.096 | 0.67958 | -1.013 | 6.0E-5 | 0.472 |
| 10980 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-654-3p;hsa-miR-940;hsa-miR-942-5p | 36 | HLF | Sponge network | -0.405 | 0.22013 | -2.443 | 0 | 0.472 |
| 10981 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-93-3p | 29 | SSBP2 | Sponge network | -0.154 | 0.53954 | -1.58 | 0 | 0.472 |
| 10982 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p | 17 | FGF5 | Sponge network | 0.249 | 0.35902 | 0.549 | 0.28659 | 0.472 |
| 10983 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-766-3p;hsa-miR-93-3p | 64 | IL6ST | Sponge network | -0.162 | 0.47687 | -0.402 | 0.00676 | 0.472 |
| 10984 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 23 | EBF3 | Sponge network | -0.709 | 0.18168 | -0.058 | 0.81437 | 0.472 |
| 10985 | AC006116.24 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-93-5p | 22 | IL17RD | Sponge network | -0.473 | 0.07602 | -0.019 | 0.94873 | 0.472 |
| 10986 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 28 | SETBP1 | Sponge network | -0.899 | 6.0E-5 | -1.302 | 1.0E-5 | 0.472 |
| 10987 | MIR497HG |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-5p | 32 | BTG2 | Sponge network | -1.439 | 4.0E-5 | -1.763 | 0 | 0.472 |
| 10988 | MALAT1 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p | 31 | ABL2 | Sponge network | 0.782 | 0.00016 | 0.82 | 0 | 0.472 |
| 10989 | ANKRD10-IT1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 12 | MOB1B | Sponge network | 1.739 | 0 | 0.197 | 0.15266 | 0.472 |
| 10990 | HAND2-AS1 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | NOV | Sponge network | -1.697 | 0.00889 | -0.58 | 0.04676 | 0.472 |
| 10991 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-625-5p | 35 | TGFBR3 | Sponge network | -2.69 | 0.0001 | -1.173 | 1.0E-5 | 0.472 |
| 10992 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 19 | NF1 | Sponge network | 0.72 | 0.0001 | 0.51 | 0 | 0.472 |
| 10993 | EMX2OS |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 20 | CDH23 | Sponge network | -0.777 | 0.1134 | -0.974 | 0.00034 | 0.472 |
| 10994 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-7-1-3p | 32 | PAFAH1B1 | Sponge network | -1.092 | 4.0E-5 | -0.429 | 0 | 0.472 |
| 10995 | DNM3OS |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-96-5p | 24 | ASTN1 | Sponge network | 0.572 | 0.01722 | -2.389 | 2.0E-5 | 0.472 |
| 10996 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p | 13 | DDX6 | Sponge network | 1.2 | 0.01813 | 0.091 | 0.36428 | 0.472 |
| 10997 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 22 | RICTOR | Sponge network | 0.93 | 0 | 0.388 | 0.00188 | 0.472 |
| 10998 | LPP-AS2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-96-5p | 13 | ZBTB20 | Sponge network | -0.18 | 0.20343 | -0.122 | 0.57569 | 0.472 |
| 10999 | GNG12-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | TPO | Sponge network | -0.793 | 7.0E-5 | -1.87 | 2.0E-5 | 0.472 |
| 11000 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 19 | NCAM2 | Sponge network | -0.612 | 0.00094 | -0.482 | 0.22565 | 0.472 |
| 11001 | OIP5-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 31 | UBR3 | Sponge network | 0.421 | 0.00084 | -0.021 | 0.82774 | 0.472 |
| 11002 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 48 | EPHA7 | Sponge network | -0.942 | 0 | -2.197 | 3.0E-5 | 0.472 |
| 11003 | MIR143HG |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | CSDE1 | Sponge network | -0.739 | 0.0574 | -0.493 | 0 | 0.472 |
| 11004 | HOTAIRM1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | DTNA | Sponge network | -0.053 | 0.80685 | -1.648 | 1.0E-5 | 0.472 |
| 11005 | FENDRR |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-455-5p | 11 | THSD7A | Sponge network | -1.281 | 0.00012 | 0.242 | 0.25154 | 0.472 |
| 11006 | FGF14-AS2 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | TRPS1 | Sponge network | -1.582 | 8.0E-5 | -0.191 | 0.44109 | 0.472 |
| 11007 | RP11-400K9.4 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-324-3p;hsa-miR-502-5p | 11 | FMNL3 | Sponge network | -0.611 | 0.09607 | 0.555 | 4.0E-5 | 0.472 |
| 11008 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 18 | HOOK3 | Sponge network | -2.46 | 0 | 0.273 | 0.09957 | 0.472 |
| 11009 | RP11-356J5.12 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | SPOP | Sponge network | -0.899 | 6.0E-5 | -0.419 | 1.0E-5 | 0.472 |
| 11010 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | CACNA2D1 | Sponge network | 0.532 | 0.00505 | -0.846 | 0.00675 | 0.472 |
| 11011 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-98-5p | 21 | PNRC1 | Sponge network | -1.697 | 0.00889 | -0.646 | 0 | 0.472 |
| 11012 | EMX2OS |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-93-3p;hsa-miR-940 | 45 | GRIK3 | Sponge network | -0.777 | 0.1134 | -3.208 | 0 | 0.472 |
| 11013 | CTD-2554C21.2 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-769-5p | 26 | GRIK3 | Sponge network | -1.654 | 0.00144 | -3.208 | 0 | 0.472 |
| 11014 | FAM66C |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | KCTD8 | Sponge network | -0.901 | 0.00019 | -3.359 | 0 | 0.472 |
| 11015 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-590-3p | 14 | DMD | Sponge network | -1.375 | 0.08642 | -1.506 | 0 | 0.472 |
| 11016 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | DNAJB4 | Sponge network | -1.216 | 0 | -0.7 | 1.0E-5 | 0.472 |
| 11017 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | EPHA7 | Sponge network | -1.349 | 0.00017 | -2.197 | 3.0E-5 | 0.472 |
| 11018 | FENDRR |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CDH10 | Sponge network | -1.281 | 0.00012 | -2.397 | 0 | 0.472 |
| 11019 | FZD10-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 12 | NGFR | Sponge network | -0.727 | 0.04726 | -1.271 | 0.01058 | 0.472 |
| 11020 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | TIMP3 | Sponge network | 0.246 | 0.59912 | -0.546 | 0.02307 | 0.472 |
| 11021 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | FAT4 | Sponge network | -0.513 | 0.04703 | -0.354 | 0.14694 | 0.472 |
| 11022 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-424-3p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 17 | HOOK3 | Sponge network | 0.083 | 0.66405 | 0.273 | 0.09957 | 0.472 |
| 11023 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 29 | EBF1 | Sponge network | -0.487 | 0.02157 | -0.384 | 0.08856 | 0.472 |
| 11024 | RP11-620J15.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | KCTD8 | Sponge network | -0.739 | 0.00044 | -3.359 | 0 | 0.472 |
| 11025 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-5p | 20 | ADAMTSL3 | Sponge network | -0.154 | 0.78939 | -1.559 | 0 | 0.472 |
| 11026 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-539-5p | 19 | DIP2C | Sponge network | -0.469 | 0.08721 | -0.436 | 0.01713 | 0.472 |
| 11027 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 33 | PTPN14 | Sponge network | -0.49 | 0.04946 | 0.144 | 0.34595 | 0.472 |
| 11028 | RP11-305E6.4 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-495-3p | 10 | STAG2 | Sponge network | 1.317 | 0 | 0.252 | 0.00438 | 0.472 |
| 11029 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 29 | ABL2 | Sponge network | 0.349 | 0.01695 | 0.82 | 0 | 0.472 |
| 11030 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PEG3 | Sponge network | -0.021 | 0.93598 | -1.74 | 0 | 0.472 |
| 11031 | ZRANB2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-935 | 17 | DIP2C | Sponge network | -0.376 | 0.00337 | -0.436 | 0.01713 | 0.472 |
| 11032 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 45 | BMPR2 | Sponge network | -0.611 | 0.09607 | 0.206 | 0.0635 | 0.472 |
| 11033 | RP3-475N16.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-378a-3p;hsa-miR-378c | 10 | BRD3 | Sponge network | 1.056 | 0 | 0.37 | 0.00013 | 0.472 |
| 11034 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | CDH10 | Sponge network | -0.323 | 0.01372 | -2.397 | 0 | 0.472 |
| 11035 | LINC01004 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 16 | MDM4 | Sponge network | 1.115 | 0 | 0.345 | 0.00762 | 0.472 |
| 11036 | LINC00641 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p | 11 | FHL1 | Sponge network | -0.222 | 0.32445 | -2.687 | 0 | 0.472 |
| 11037 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 39 | RAP1A | Sponge network | -0.739 | 0.0574 | -0.929 | 0 | 0.472 |
| 11038 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 33 | IGF1 | Sponge network | 0.246 | 0.59912 | -0.204 | 0.5898 | 0.472 |
| 11039 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 35 | TOM1L2 | Sponge network | -0.739 | 0.0574 | -0.994 | 0 | 0.472 |
| 11040 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | AKAP12 | Sponge network | 0.374 | 0.20054 | -0.932 | 0.0028 | 0.472 |
| 11041 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 14 | ITCH | Sponge network | 1.153 | 0 | 0.084 | 0.34058 | 0.472 |
| 11042 | RP11-2H8.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | PTPN11 | Sponge network | 1.089 | 0 | 0.431 | 2.0E-5 | 0.472 |
| 11043 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-5p | 11 | SYNE1 | Sponge network | -2.076 | 0.00548 | -0.738 | 0.00316 | 0.472 |
| 11044 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-96-5p | 13 | ELMO1 | Sponge network | -0.769 | 0.00035 | 0.04 | 0.83147 | 0.472 |
| 11045 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 37 | NRXN1 | Sponge network | -0.615 | 0.00015 | -3.778 | 0 | 0.472 |
| 11046 | RP11-326G21.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-511-5p;hsa-miR-590-3p | 11 | MRVI1 | Sponge network | -0.021 | 0.93598 | -1.051 | 0.00022 | 0.472 |
| 11047 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-5p | 28 | ZBTB4 | Sponge network | -0.942 | 0 | -0.739 | 0 | 0.472 |
| 11048 | PWAR6 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p | 34 | IGFBP5 | Sponge network | -1.575 | 6.0E-5 | -0.806 | 0.00105 | 0.472 |
| 11049 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-5p | 13 | EYA4 | Sponge network | -0.693 | 0.05629 | 0.3 | 0.36876 | 0.472 |
| 11050 | CTD-2089N3.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p | 12 | NRK | Sponge network | -1.375 | 0.08642 | 0.656 | 0.19217 | 0.472 |
| 11051 | ZNF582-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 45 | CADM2 | Sponge network | -1.349 | 0.00017 | -3.782 | 0 | 0.472 |
| 11052 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-423-5p | 10 | PAK3 | Sponge network | -1.092 | 4.0E-5 | -0.628 | 0.07253 | 0.472 |
| 11053 | U91328.19 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-7-1-3p | 25 | MXI1 | Sponge network | -0.303 | 0.21002 | -0.987 | 0 | 0.472 |
| 11054 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-450b-5p | 13 | JAKMIP2 | Sponge network | -1.249 | 7.0E-5 | -1.388 | 0 | 0.472 |
| 11055 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-98-5p | 13 | AKAP6 | Sponge network | -1.116 | 0.23686 | -1.586 | 3.0E-5 | 0.472 |
| 11056 | SERTAD4-AS1 |
hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-502-5p | 11 | PRKCB | Sponge network | -0.932 | 0.00329 | -1.313 | 2.0E-5 | 0.472 |
| 11057 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 29 | ARHGAP28 | Sponge network | -1.111 | 0.0003 | -0.911 | 0.00013 | 0.472 |
| 11058 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-320b;hsa-miR-493-5p | 10 | MYCBP2 | Sponge network | 0.614 | 1.0E-5 | -0.027 | 0.82016 | 0.472 |
| 11059 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SON | Sponge network | 1.308 | 0 | 0.168 | 0.04406 | 0.472 |
| 11060 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 44 | ZFHX3 | Sponge network | 0.249 | 0.35902 | 0.389 | 0.01363 | 0.472 |
| 11061 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p | 20 | PBX1 | Sponge network | -2.076 | 0.00548 | -1.206 | 0 | 0.472 |
| 11062 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 27 | PCDH10 | Sponge network | -0.405 | 0.22013 | -1.644 | 0.0078 | 0.472 |
| 11063 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 32 | RASSF8 | Sponge network | -0.405 | 0.22013 | -0.122 | 0.65591 | 0.472 |
| 11064 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-421 | 12 | LRRC55 | Sponge network | -0.583 | 0.14589 | -0.026 | 0.93163 | 0.472 |
| 11065 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-625-3p | 25 | ABL2 | Sponge network | 0.225 | 0.30644 | 0.82 | 0 | 0.472 |
| 11066 | LINC00641 |
hsa-miR-126-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p | 10 | TNC | Sponge network | -0.222 | 0.32445 | 0.313 | 0.33705 | 0.472 |
| 11067 | LINC00963 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 31 | ZFHX3 | Sponge network | 0.523 | 9.0E-5 | 0.389 | 0.01363 | 0.472 |
| 11068 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-589-5p | 19 | SLITRK3 | Sponge network | -1.249 | 7.0E-5 | -3.328 | 0 | 0.472 |
| 11069 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-7-1-3p | 18 | ADAMTSL3 | Sponge network | 0.299 | 0.57722 | -1.559 | 0 | 0.472 |
| 11070 | FENDRR |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -1.281 | 0.00012 | 0.024 | 0.89889 | 0.472 |
| 11071 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRIM58 | Sponge network | -1.216 | 0 | -1.692 | 0 | 0.472 |
| 11072 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 48 | EPHA7 | Sponge network | 0.997 | 0.00066 | -2.197 | 3.0E-5 | 0.472 |
| 11073 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-628-5p;hsa-miR-93-3p | 27 | EPHA3 | Sponge network | -0.154 | 0.78939 | 0.185 | 0.53588 | 0.472 |
| 11074 | AC006116.24 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-93-5p;hsa-miR-98-5p | 39 | CADM2 | Sponge network | -0.473 | 0.07602 | -3.782 | 0 | 0.472 |
| 11075 | RP11-701H24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-93-5p | 15 | GAS7 | Sponge network | -2.039 | 0.03447 | -0.555 | 0.01602 | 0.472 |
| 11076 | RP11-890B15.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | 0.101 | 0.60919 | -1.051 | 0.00022 | 0.472 |
| 11077 | RP11-774O3.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-708-5p;hsa-miR-744-3p;hsa-miR-93-3p | 26 | TOM1L2 | Sponge network | -0.487 | 0.02157 | -0.994 | 0 | 0.472 |
| 11078 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | CNTN1 | Sponge network | 0.246 | 0.59912 | -2.594 | 0 | 0.472 |
| 11079 | RP11-244O19.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-484;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 65 | DST | Sponge network | -0.096 | 0.67958 | -0.713 | 0.00096 | 0.472 |
| 11080 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-582-3p | 28 | PBX1 | Sponge network | -0.513 | 0.04703 | -1.206 | 0 | 0.472 |
| 11081 | RP11-244O19.1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | -0.096 | 0.67958 | 0.154 | 0.74723 | 0.471 |
| 11082 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 46 | TRPS1 | Sponge network | -0.942 | 0 | -0.191 | 0.44109 | 0.471 |
| 11083 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 35 | PRKAB2 | Sponge network | 0.101 | 0.60919 | -0.367 | 0.01371 | 0.471 |
| 11084 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | NAV3 | Sponge network | -0.187 | 0.23787 | 0.212 | 0.47729 | 0.471 |
| 11085 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-23a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-484;hsa-miR-629-3p | 19 | DDX6 | Sponge network | 1.148 | 2.0E-5 | 0.091 | 0.36428 | 0.471 |
| 11086 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | RASSF2 | Sponge network | 0.532 | 0.00505 | -0.273 | 0.21961 | 0.471 |
| 11087 | BZRAP1-AS1 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-940 | 12 | MAPK10 | Sponge network | -0.465 | 0.04703 | -1.774 | 0 | 0.471 |
| 11088 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | LATS1 | Sponge network | 0.826 | 1.0E-5 | 0.109 | 0.34208 | 0.471 |
| 11089 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 60 | FAT3 | Sponge network | -1.216 | 0 | -1.601 | 0.00051 | 0.471 |
| 11090 | BVES-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p | 12 | TAL1 | Sponge network | -2.62 | 0 | -0.462 | 0.00574 | 0.471 |
| 11091 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | PHC3 | Sponge network | 0.194 | 0.64278 | 0.212 | 0.0499 | 0.471 |
| 11092 | PWAR6 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 17 | CELF4 | Sponge network | -1.575 | 6.0E-5 | -1.135 | 0.00344 | 0.471 |
| 11093 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-590-3p | 12 | DNAJB4 | Sponge network | -0.739 | 0.00044 | -0.7 | 1.0E-5 | 0.471 |
| 11094 | LINC00473 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 48 | CADM2 | Sponge network | -1.203 | 0.03881 | -3.782 | 0 | 0.471 |
| 11095 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-93-5p | 33 | ADARB1 | Sponge network | -1.654 | 0.00144 | -0.335 | 0.06631 | 0.471 |
| 11096 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-675-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 49 | MEF2C | Sponge network | -1.216 | 0 | -0.499 | 0.01448 | 0.471 |
| 11097 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | MLLT10 | Sponge network | -0.222 | 0.32445 | 0.105 | 0.33292 | 0.471 |
| 11098 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 20 | RET | Sponge network | -0.727 | 0.04726 | -0.833 | 0.03517 | 0.471 |
| 11099 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-940 | 26 | HLF | Sponge network | -0.154 | 0.53954 | -2.443 | 0 | 0.471 |
| 11100 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | PTPRB | Sponge network | -1.439 | 4.0E-5 | 0.105 | 0.47122 | 0.471 |
| 11101 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-424-5p | 11 | ABCB5 | Sponge network | -2.531 | 0 | -0.956 | 0.04077 | 0.471 |
| 11102 | CTA-941F9.9 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-576-5p | 12 | ZFHX4 | Sponge network | -0.344 | 0.49624 | 0.018 | 0.95574 | 0.471 |
| 11103 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FAM133A | Sponge network | -0.576 | 0.10121 | 0.549 | 0.29009 | 0.471 |
| 11104 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 59 | QKI | Sponge network | -1.281 | 0.00012 | -0.333 | 0.04111 | 0.471 |
| 11105 | RP11-400K9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-940 | 12 | SNX29 | Sponge network | -0.733 | 0.19469 | -0.34 | 0.01371 | 0.471 |
| 11106 | RP11-798M19.6 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TPO | Sponge network | -0.612 | 0.00094 | -1.87 | 2.0E-5 | 0.471 |
| 11107 | RP11-894P9.1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-582-5p | 21 | DDX6 | Sponge network | 0.993 | 0 | 0.091 | 0.36428 | 0.471 |
| 11108 | LINC00641 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | DICER1 | Sponge network | -0.222 | 0.32445 | 0.042 | 0.674 | 0.471 |
| 11109 | LINC01018 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | -2.915 | 0.00015 | -0.564 | 1.0E-5 | 0.471 |
| 11110 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | PBRM1 | Sponge network | 0.683 | 1.0E-5 | -0.029 | 0.78601 | 0.471 |
| 11111 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-455-5p | 10 | ZNF521 | Sponge network | -0.793 | 7.0E-5 | -0.111 | 0.58068 | 0.471 |
| 11112 | MAGI2-AS3 |
hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-7-5p | 10 | CMKLR1 | Sponge network | -0.129 | 0.57177 | 0.288 | 0.17621 | 0.471 |
| 11113 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-766-3p | 26 | IL17RD | Sponge network | 0.75 | 0.00054 | -0.019 | 0.94873 | 0.471 |
| 11114 | HOTAIRM1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | WWTR1 | Sponge network | -0.053 | 0.80685 | -0.345 | 0.11997 | 0.471 |
| 11115 | BZRAP1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-7-5p | 22 | SETBP1 | Sponge network | -0.465 | 0.04703 | -1.302 | 1.0E-5 | 0.471 |
| 11116 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-935 | 35 | DIP2C | Sponge network | -0.405 | 0.22013 | -0.436 | 0.01713 | 0.471 |
| 11117 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 26 | RSPO2 | Sponge network | -0.793 | 7.0E-5 | -3.038 | 0 | 0.471 |
| 11118 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p | 12 | ERC1 | Sponge network | 0.542 | 0.00054 | -0.108 | 0.38794 | 0.471 |
| 11119 | MAGI2-AS3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PIK3CA | Sponge network | -0.129 | 0.57177 | 0.195 | 0.06908 | 0.471 |
| 11120 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-664a-3p | 17 | GREM1 | Sponge network | 0.16 | 0.50785 | 0.902 | 0.01691 | 0.471 |
| 11121 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-5p | 13 | RET | Sponge network | 0.889 | 9.0E-5 | -0.833 | 0.03517 | 0.471 |
| 11122 | CTD-2554C21.2 |
hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-629-5p;hsa-miR-96-5p | 20 | MYO9A | Sponge network | -1.654 | 0.00144 | -0.147 | 0.32373 | 0.471 |
| 11123 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | -0.738 | 0.21233 | -0.382 | 0.05038 | 0.471 |
| 11124 | RP11-13K12.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 21 | CDH23 | Sponge network | -0.154 | 0.78939 | -0.974 | 0.00034 | 0.471 |
| 11125 | LINC00957 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | NAV3 | Sponge network | -0.421 | 0.0114 | 0.212 | 0.47729 | 0.471 |
| 11126 | PWAR6 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | INPP4A | Sponge network | -1.575 | 6.0E-5 | 0.07 | 0.53563 | 0.471 |
| 11127 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -1.161 | 0.00591 | -0.7 | 1.0E-5 | 0.471 |
| 11128 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 29 | GPC6 | Sponge network | -0.901 | 0.00019 | -0.054 | 0.81465 | 0.471 |
| 11129 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | CHD2 | Sponge network | 0.178 | 0.29106 | 0.049 | 0.55371 | 0.471 |
| 11130 | RP4-714D9.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-497-5p;hsa-miR-766-3p | 13 | TBL1XR1 | Sponge network | 1.015 | 0 | 0.578 | 0 | 0.471 |
| 11131 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | EPAS1 | Sponge network | -2.452 | 0 | -0.184 | 0.14418 | 0.471 |
| 11132 | SNHG14 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | DIRAS1 | Sponge network | -1.111 | 0.0003 | -2.518 | 0 | 0.471 |
| 11133 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | ST6GAL2 | Sponge network | -0.611 | 0.09607 | -1.201 | 0.00597 | 0.471 |
| 11134 | RP11-13K12.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-454-3p;hsa-miR-589-5p | 14 | ROBO1 | Sponge network | -0.154 | 0.78939 | -0.009 | 0.96154 | 0.471 |
| 11135 | FAM66C |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | LIMCH1 | Sponge network | -0.901 | 0.00019 | -0.915 | 1.0E-5 | 0.471 |
| 11136 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 28 | ATRX | Sponge network | 0.614 | 1.0E-5 | 0.261 | 0.04408 | 0.471 |
| 11137 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 48 | SSBP2 | Sponge network | -2.915 | 0.00015 | -1.58 | 0 | 0.471 |
| 11138 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-590-3p | 26 | AFF3 | Sponge network | -0.615 | 0.00015 | -2.373 | 0 | 0.471 |
| 11139 | SNHG16 |
hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | SLC5A3 | Sponge network | 0.961 | 0 | 0.493 | 5.0E-5 | 0.471 |
| 11140 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | SLITRK6 | Sponge network | -2.452 | 0 | -2.121 | 0 | 0.471 |
| 11141 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-374b-5p | 13 | PHC3 | Sponge network | 0.542 | 0.00054 | 0.212 | 0.0499 | 0.471 |
| 11142 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p | 13 | ZNF292 | Sponge network | 0.75 | 0.00054 | 0.276 | 0.04148 | 0.471 |
| 11143 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429 | 29 | NOTCH2 | Sponge network | -1.654 | 0.00144 | 0.138 | 0.35163 | 0.471 |
| 11144 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-940;hsa-miR-96-5p | 25 | CRTC3 | Sponge network | -1.439 | 4.0E-5 | 0.164 | 0.16786 | 0.471 |
| 11145 | AC083843.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 22 | MDM4 | Sponge network | 1.4 | 0 | 0.345 | 0.00762 | 0.471 |
| 11146 | PCA3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-93-5p | 40 | UNC80 | Sponge network | -0.709 | 0.18168 | -1.934 | 0 | 0.471 |
| 11147 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 35 | MCC | Sponge network | 0.532 | 0.00505 | -1.005 | 0 | 0.471 |
| 11148 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-624-5p | 11 | SMARCA1 | Sponge network | -0.769 | 0.00035 | -0.684 | 0.00159 | 0.471 |
| 11149 | USP3-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 12 | DCN | Sponge network | -0.162 | 0.47687 | -1.288 | 0 | 0.471 |
| 11150 | TPTEP1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 34 | AKAP6 | Sponge network | -1.216 | 0 | -1.586 | 3.0E-5 | 0.471 |
| 11151 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-935 | 27 | EBF1 | Sponge network | -0.162 | 0.47687 | -0.384 | 0.08856 | 0.471 |
| 11152 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p | 23 | PGR | Sponge network | 0.331 | 0.57247 | -1.704 | 0 | 0.471 |
| 11153 | AC010761.8 |
hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-766-3p | 14 | TBL1XR1 | Sponge network | 1.638 | 0 | 0.578 | 0 | 0.471 |
| 11154 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-484 | 12 | TRIM3 | Sponge network | -1.331 | 3.0E-5 | -0.577 | 0 | 0.471 |
| 11155 | MYLK-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-651-5p | 17 | PRDM2 | Sponge network | -1.331 | 3.0E-5 | -0.258 | 0.00721 | 0.471 |
| 11156 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 10 | ZHX2 | Sponge network | -0.49 | 0.04946 | -0.192 | 0.18459 | 0.471 |
| 11157 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-940 | 20 | SNX29 | Sponge network | -4.546 | 0 | -0.34 | 0.01371 | 0.471 |
| 11158 | LINC00654 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 10 | PCLO | Sponge network | 0.374 | 0.20054 | -1.843 | 0.00064 | 0.471 |
| 11159 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-452-5p;hsa-miR-671-5p | 28 | ADARB1 | Sponge network | -2.095 | 0.00112 | -0.335 | 0.06631 | 0.471 |
| 11160 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-582-3p;hsa-miR-625-5p | 30 | JAZF1 | Sponge network | -0.154 | 0.53954 | -0.377 | 0.02677 | 0.471 |
| 11161 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-590-3p | 11 | CAV1 | Sponge network | -0.738 | 0.21233 | -1.013 | 6.0E-5 | 0.471 |
| 11162 | RP11-464F9.1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | PHF3 | Sponge network | 0.959 | 0 | 0.126 | 0.19652 | 0.471 |
| 11163 | LINC00957 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-935 | 16 | RELN | Sponge network | -0.421 | 0.0114 | -2.403 | 0 | 0.471 |
| 11164 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 20 | SLC6A15 | Sponge network | -0.976 | 0.00025 | -2.373 | 2.0E-5 | 0.471 |
| 11165 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429 | 10 | LHFP | Sponge network | -1.654 | 0.00144 | -0.167 | 0.41371 | 0.471 |
| 11166 | GNG12-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-505-3p | 16 | FNBP1 | Sponge network | -0.793 | 7.0E-5 | -0.969 | 4.0E-5 | 0.471 |
| 11167 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 25 | BCL6 | Sponge network | -0.49 | 0.04946 | -0.211 | 0.19489 | 0.471 |
| 11168 | RP11-156P1.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-92a-3p | 33 | IL17RD | Sponge network | 0.214 | 0.18246 | -0.019 | 0.94873 | 0.471 |
| 11169 | LINC00957 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-455-5p | 11 | THSD7A | Sponge network | -0.421 | 0.0114 | 0.242 | 0.25154 | 0.471 |
| 11170 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-7-5p | 10 | PTGFR | Sponge network | -0.517 | 0.02935 | -0.233 | 0.45421 | 0.471 |
| 11171 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 55 | THRB | Sponge network | -0.976 | 0.00025 | -1.117 | 0.0004 | 0.471 |
| 11172 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 21 | SYNE1 | Sponge network | 0.199 | 0.38015 | -0.738 | 0.00316 | 0.471 |
| 11173 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 24 | AFF3 | Sponge network | 0.335 | 0.20596 | -2.373 | 0 | 0.471 |
| 11174 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-362-3p;hsa-miR-7-1-3p | 11 | DACT1 | Sponge network | -1.349 | 0.00017 | 0.783 | 0.00072 | 0.471 |
| 11175 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-935 | 31 | DMD | Sponge network | -0.738 | 0.21233 | -1.506 | 0 | 0.471 |
| 11176 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 25 | ADAMTSL3 | Sponge network | 0.374 | 0.20054 | -1.559 | 0 | 0.471 |
| 11177 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | SEMA3C | Sponge network | -0.376 | 0.00337 | -0.272 | 0.31153 | 0.471 |
| 11178 | ACTA2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-671-5p | 17 | CDH23 | Sponge network | 0.194 | 0.64278 | -0.974 | 0.00034 | 0.471 |
| 11179 | PCA3 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 13 | RHOB | Sponge network | -0.709 | 0.18168 | -1.052 | 0 | 0.471 |
| 11180 | RP11-672A2.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p | 11 | EYA4 | Sponge network | 1.344 | 0 | 0.3 | 0.36876 | 0.471 |
| 11181 | RP11-212P7.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 31 | DIP2C | Sponge network | 0.219 | 0.1516 | -0.436 | 0.01713 | 0.471 |
| 11182 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-501-5p | 10 | UBE2QL1 | Sponge network | -0.777 | 0.16667 | -2.417 | 0 | 0.471 |
| 11183 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p | 28 | GPC6 | Sponge network | -1.575 | 6.0E-5 | -0.054 | 0.81465 | 0.471 |
| 11184 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-7-5p | 21 | ZBTB4 | Sponge network | -0.517 | 0.02935 | -0.739 | 0 | 0.471 |
| 11185 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-744-3p | 27 | PTPN14 | Sponge network | -2.452 | 0 | 0.144 | 0.34595 | 0.471 |
| 11186 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 51 | TCF4 | Sponge network | 0.199 | 0.38015 | 0.016 | 0.92271 | 0.471 |
| 11187 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484 | 18 | ABL1 | Sponge network | 0.082 | 0.55654 | -0.215 | 0.07804 | 0.471 |
| 11188 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-361-5p;hsa-miR-590-3p | 17 | ERG | Sponge network | -0.739 | 0.00044 | 0.002 | 0.99011 | 0.471 |
| 11189 | CTA-363E19.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-485-3p;hsa-miR-495-3p | 14 | PHC3 | Sponge network | 1.055 | 0 | 0.212 | 0.0499 | 0.471 |
| 11190 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 49 | TMEM132B | Sponge network | -2.46 | 0 | -0.83 | 0.01084 | 0.471 |
| 11191 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-628-5p | 28 | FBXO32 | Sponge network | -0.154 | 0.78939 | -0.495 | 0.07561 | 0.471 |
| 11192 | TPTEP1 |
hsa-miR-126-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | CXCL12 | Sponge network | -1.216 | 0 | -1.421 | 0 | 0.471 |
| 11193 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 18 | LHX6 | Sponge network | -0.739 | 0.0574 | -0.087 | 0.69193 | 0.471 |
| 11194 | SERTAD4-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-576-5p | 24 | GAS7 | Sponge network | -0.932 | 0.00329 | -0.555 | 0.01602 | 0.471 |
| 11195 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 40 | NOTCH2 | Sponge network | -0.976 | 0.00025 | 0.138 | 0.35163 | 0.471 |
| 11196 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p | 23 | ZFHX3 | Sponge network | 0.176 | 0.37402 | 0.389 | 0.01363 | 0.471 |
| 11197 | RP11-13K12.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-629-3p;hsa-miR-671-5p | 14 | CDH23 | Sponge network | -0.318 | 0.65847 | -0.974 | 0.00034 | 0.471 |
| 11198 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-589-5p;hsa-miR-93-5p | 14 | SLITRK3 | Sponge network | -0.318 | 0.65847 | -3.328 | 0 | 0.471 |
| 11199 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SON | Sponge network | 0.873 | 0 | 0.168 | 0.04406 | 0.47 |
| 11200 | RP11-672A2.4 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p | 12 | QKI | Sponge network | 1.344 | 0 | -0.333 | 0.04111 | 0.47 |
| 11201 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-500a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 35 | HLF | Sponge network | -1.645 | 0 | -2.443 | 0 | 0.47 |
| 11202 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | FYN | Sponge network | 0.246 | 0.59912 | -0.131 | 0.37051 | 0.47 |
| 11203 | PCA3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | PRKCB | Sponge network | -0.709 | 0.18168 | -1.313 | 2.0E-5 | 0.47 |
| 11204 | TPTEP1 |
hsa-miR-144-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | FMN2 | Sponge network | -1.216 | 0 | -1.746 | 0.00047 | 0.47 |
| 11205 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 36 | BMPR2 | Sponge network | 0.889 | 9.0E-5 | 0.206 | 0.0635 | 0.47 |
| 11206 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 32 | ST6GAL2 | Sponge network | -0.901 | 0.00019 | -1.201 | 0.00597 | 0.47 |
| 11207 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-151a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-7-1-3p | 11 | PTPRB | Sponge network | -1.092 | 4.0E-5 | 0.105 | 0.47122 | 0.47 |
| 11208 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-424-5p | 11 | FAT4 | Sponge network | -4.463 | 0.00025 | -0.354 | 0.14694 | 0.47 |
| 11209 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-432-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-889-3p;hsa-miR-93-5p | 55 | UNC80 | Sponge network | -2.46 | 0 | -1.934 | 0 | 0.47 |
| 11210 | PCA3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 25 | ASTN1 | Sponge network | -0.709 | 0.18168 | -2.389 | 2.0E-5 | 0.47 |
| 11211 | RP11-848P1.3 |
hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | DOCK4 | Sponge network | 1.253 | 0 | 0.502 | 0.00108 | 0.47 |
| 11212 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-589-3p;hsa-miR-93-5p | 41 | PIK3R1 | Sponge network | -0.162 | 0.47687 | -0.133 | 0.36854 | 0.47 |
| 11213 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 20 | PLSCR4 | Sponge network | 0.16 | 0.50785 | -0.474 | 0.03833 | 0.47 |
| 11214 | MIR143HG |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PAK3 | Sponge network | -0.739 | 0.0574 | -0.628 | 0.07253 | 0.47 |
| 11215 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-940 | 25 | AKAP11 | Sponge network | 1.254 | 0 | 0.196 | 0.09825 | 0.47 |
| 11216 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | 1.302 | 7.0E-5 | 0.783 | 0.00072 | 0.47 |
| 11217 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 53 | TRPS1 | Sponge network | -2.46 | 0 | -0.191 | 0.44109 | 0.47 |
| 11218 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 44 | EPHA7 | Sponge network | -0.563 | 0.02545 | -2.197 | 3.0E-5 | 0.47 |
| 11219 | RP11-248G5.8 |
hsa-miR-125a-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | ERCC6 | Sponge network | 1.294 | 0 | 0.23 | 0.015 | 0.47 |
| 11220 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | CNTN1 | Sponge network | -2.925 | 0 | -2.594 | 0 | 0.47 |
| 11221 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 30 | NRXN1 | Sponge network | -1.161 | 0.00591 | -3.778 | 0 | 0.47 |
| 11222 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 11 | SPG20 | Sponge network | -0.021 | 0.93598 | -1.16 | 0 | 0.47 |
| 11223 | MYLK-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p | 15 | NIN | Sponge network | -1.331 | 3.0E-5 | -0.253 | 0.13794 | 0.47 |
| 11224 | LINC00641 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PHF3 | Sponge network | -0.222 | 0.32445 | 0.126 | 0.19652 | 0.47 |
| 11225 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 27 | KCNA6 | Sponge network | 0.249 | 0.35902 | -1.531 | 0.00013 | 0.47 |
| 11226 | PWAR6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | NIN | Sponge network | -1.575 | 6.0E-5 | -0.253 | 0.13794 | 0.47 |
| 11227 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-93-5p | 24 | IL17RD | Sponge network | 0.051 | 0.83388 | -0.019 | 0.94873 | 0.47 |
| 11228 | RP11-400K9.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-185-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-940 | 10 | GRIK3 | Sponge network | -0.733 | 0.19469 | -3.208 | 0 | 0.47 |
| 11229 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-484;hsa-miR-576-5p | 21 | CBL | Sponge network | 0.919 | 0 | 0.291 | 0.01267 | 0.47 |
| 11230 | CTD-2314G24.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-96-5p | 28 | ASTN1 | Sponge network | 0.246 | 0.59912 | -2.389 | 2.0E-5 | 0.47 |
| 11231 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 57 | RUNX1T1 | Sponge network | 0.101 | 0.60919 | -0.344 | 0.2374 | 0.47 |
| 11232 | RAMP2-AS1 |
hsa-miR-141-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 12 | STAT5B | Sponge network | -0.513 | 0.04703 | -0.182 | 0.1082 | 0.47 |
| 11233 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 30 | CLASP2 | Sponge network | 0.75 | 0.00054 | -0.038 | 0.71 | 0.47 |
| 11234 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 45 | TRPS1 | Sponge network | -0.323 | 0.01372 | -0.191 | 0.44109 | 0.47 |
| 11235 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | FAT4 | Sponge network | -1.216 | 0 | -0.354 | 0.14694 | 0.47 |
| 11236 | MEG3 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 12 | TPO | Sponge network | 0.249 | 0.35902 | -1.87 | 2.0E-5 | 0.47 |
| 11237 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-339-5p;hsa-miR-421 | 11 | STARD13 | Sponge network | -4.463 | 0.00025 | -0.187 | 0.27383 | 0.47 |
| 11238 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 22 | LIFR | Sponge network | -0.611 | 0.09607 | -1.794 | 0 | 0.47 |
| 11239 | EMX2OS |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 39 | IGFBP5 | Sponge network | -0.777 | 0.1134 | -0.806 | 0.00105 | 0.47 |
| 11240 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-874-3p | 11 | NR2C2 | Sponge network | 0.79 | 0 | 0.21 | 0.03224 | 0.47 |
| 11241 | JAZF1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-96-5p | 28 | ASTN1 | Sponge network | -0.769 | 0.00035 | -2.389 | 2.0E-5 | 0.47 |
| 11242 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p | 35 | IL6ST | Sponge network | -1.582 | 8.0E-5 | -0.402 | 0.00676 | 0.47 |
| 11243 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | CNTNAP1 | Sponge network | -0.187 | 0.23787 | 0.358 | 0.13727 | 0.47 |
| 11244 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-7-1-3p | 14 | CAV1 | Sponge network | -0.842 | 0.01361 | -1.013 | 6.0E-5 | 0.47 |
| 11245 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 52 | SSBP2 | Sponge network | -0.942 | 0 | -1.58 | 0 | 0.47 |
| 11246 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-577 | 19 | GREM1 | Sponge network | -0.709 | 0.18168 | 0.902 | 0.01691 | 0.47 |
| 11247 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 15 | PBX1 | Sponge network | -0.187 | 0.23787 | -1.206 | 0 | 0.47 |
| 11248 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | FNBP1 | Sponge network | -0.517 | 0.02935 | -0.969 | 4.0E-5 | 0.47 |
| 11249 | LINC00476 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-99b-3p | 50 | GRIK3 | Sponge network | -0.615 | 0.00015 | -3.208 | 0 | 0.47 |
| 11250 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | PREX2 | Sponge network | -1.057 | 0.00029 | -0.272 | 0.25382 | 0.47 |
| 11251 | MIR497HG |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 10 | MLLT11 | Sponge network | -1.439 | 4.0E-5 | -0.661 | 0.01733 | 0.47 |
| 11252 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 46 | PDGFRA | Sponge network | -0.739 | 0.0574 | -0.372 | 0.0037 | 0.47 |
| 11253 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-590-3p | 11 | PSIP1 | Sponge network | -0.342 | 0.02288 | -0.005 | 0.96897 | 0.47 |
| 11254 | RP11-531A24.5 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-766-3p | 18 | ATM | Sponge network | 0.75 | 0.00054 | 0.178 | 0.21568 | 0.47 |
| 11255 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-7-1-3p | 19 | ADAMTSL3 | Sponge network | -0.405 | 0.22013 | -1.559 | 0 | 0.47 |
| 11256 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-423-5p | 10 | PAK3 | Sponge network | -0.583 | 0.14589 | -0.628 | 0.07253 | 0.47 |
| 11257 | CTC-510F12.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-935 | 11 | EBF1 | Sponge network | -0.691 | 0.00146 | -0.384 | 0.08856 | 0.47 |
| 11258 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | JAZF1 | Sponge network | -0.295 | 0.02604 | -0.377 | 0.02677 | 0.47 |
| 11259 | RP11-119F7.5 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-590-3p | 11 | DNMT3A | Sponge network | 0.225 | 0.30644 | 0.302 | 0.0262 | 0.47 |
| 11260 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-590-5p | 12 | PDZD2 | Sponge network | -0.769 | 0.00035 | -0.909 | 3.0E-5 | 0.47 |
| 11261 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 21 | NF1 | Sponge network | 0.537 | 0.00139 | 0.51 | 0 | 0.47 |
| 11262 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 29 | PBX1 | Sponge network | 0.374 | 0.20054 | -1.206 | 0 | 0.47 |
| 11263 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | EPAS1 | Sponge network | -0.129 | 0.57177 | -0.184 | 0.14418 | 0.47 |
| 11264 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | RNF144A | Sponge network | -0.358 | 0.17255 | 0.594 | 0.0009 | 0.47 |
| 11265 | AC016747.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429 | 10 | ELMO1 | Sponge network | 0.199 | 0.38015 | 0.04 | 0.83147 | 0.47 |
| 11266 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 66 | RUNX1T1 | Sponge network | -0.942 | 0 | -0.344 | 0.2374 | 0.47 |
| 11267 | LINC00265 |
hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p | 16 | ABL2 | Sponge network | 1.07 | 0 | 0.82 | 0 | 0.47 |
| 11268 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | EDA2R | Sponge network | 0.997 | 0.00066 | -0.587 | 0.01598 | 0.47 |
| 11269 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 34 | ZFHX4 | Sponge network | 0.889 | 9.0E-5 | 0.018 | 0.95574 | 0.47 |
| 11270 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | DMTF1 | Sponge network | 0.541 | 0.00125 | 0.248 | 0.02726 | 0.47 |
| 11271 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | PLSCR4 | Sponge network | -0.053 | 0.80685 | -0.474 | 0.03833 | 0.47 |
| 11272 | MEG3 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | 0.249 | 0.35902 | 0.524 | 0.00017 | 0.47 |
| 11273 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 41 | NRXN1 | Sponge network | -0.942 | 0 | -3.778 | 0 | 0.47 |
| 11274 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 12 | AKAP12 | Sponge network | 0.214 | 0.18246 | -0.932 | 0.0028 | 0.47 |
| 11275 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | LRRC55 | Sponge network | -2.452 | 0 | -0.026 | 0.93163 | 0.47 |
| 11276 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-7-1-3p | 18 | CNTN1 | Sponge network | 0.299 | 0.57722 | -2.594 | 0 | 0.47 |
| 11277 | LINC00473 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 17 | SPG20 | Sponge network | -1.203 | 0.03881 | -1.16 | 0 | 0.47 |
| 11278 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | EPHA5 | Sponge network | 0.335 | 0.20596 | -0.957 | 0.01733 | 0.47 |
| 11279 | RP11-254F7.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-486-5p | 14 | EZH1 | Sponge network | 0.176 | 0.37402 | -0.564 | 1.0E-5 | 0.47 |
| 11280 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 71 | THRB | Sponge network | -0.739 | 0.0574 | -1.117 | 0.0004 | 0.47 |
| 11281 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 45 | MDM4 | Sponge network | 0.494 | 0.00339 | 0.345 | 0.00762 | 0.47 |
| 11282 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p;hsa-miR-877-5p | 22 | GAS7 | Sponge network | -0.309 | 0.1145 | -0.555 | 0.01602 | 0.47 |
| 11283 | HCG18 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 21 | NF1 | Sponge network | 1.063 | 0 | 0.51 | 0 | 0.47 |
| 11284 | AC006116.24 |
hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-2-5p;hsa-miR-455-5p;hsa-miR-577 | 10 | DOCK4 | Sponge network | -0.473 | 0.07602 | 0.502 | 0.00108 | 0.47 |
| 11285 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -0.612 | 0.00094 | -3.328 | 0 | 0.47 |
| 11286 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 38 | ZFHX4 | Sponge network | 0.214 | 0.18246 | 0.018 | 0.95574 | 0.47 |
| 11287 | CTC-296K1.3 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | SPG20 | Sponge network | -2.095 | 0.00112 | -1.16 | 0 | 0.47 |
| 11288 | LINC00865 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | SFRP1 | Sponge network | -1.249 | 7.0E-5 | -3.566 | 0 | 0.47 |
| 11289 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-744-3p | 25 | DIP2C | Sponge network | -1.654 | 0.00144 | -0.436 | 0.01713 | 0.47 |
| 11290 | RP11-67L2.2 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-376c-3p | 12 | RASAL2 | Sponge network | 0.72 | 0.0001 | 0.869 | 0 | 0.47 |
| 11291 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 28 | HLF | Sponge network | -0.358 | 0.17255 | -2.443 | 0 | 0.47 |
| 11292 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p | 12 | SCN11A | Sponge network | -0.487 | 0.02157 | -1.15 | 0.00299 | 0.47 |
| 11293 | C20orf166-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 18 | GJA1 | Sponge network | -2.69 | 0.0001 | -0.485 | 0.02179 | 0.47 |
| 11294 | LINC00865 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-589-5p | 10 | SORCS2 | Sponge network | -1.249 | 7.0E-5 | -0.223 | 0.4253 | 0.47 |
| 11295 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 24 | DYNC1I1 | Sponge network | -0.615 | 0.00015 | -1.12 | 0.00107 | 0.47 |
| 11296 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CHD2 | Sponge network | 0.953 | 0 | 0.049 | 0.55371 | 0.47 |
| 11297 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-766-3p | 22 | TBL1XR1 | Sponge network | 0.706 | 0 | 0.578 | 0 | 0.469 |
| 11298 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-93-5p | 12 | TXNIP | Sponge network | -0.421 | 0.0114 | -1.277 | 0 | 0.469 |
| 11299 | GS1-124K5.11 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-5p;hsa-miR-744-3p | 72 | LPP | Sponge network | 0.494 | 0.00339 | -0.459 | 0.04475 | 0.469 |
| 11300 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-495-3p | 11 | MYO9A | Sponge network | 1.106 | 0 | -0.147 | 0.32373 | 0.469 |
| 11301 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 24 | MYO9A | Sponge network | -0.896 | 0.00099 | -0.147 | 0.32373 | 0.469 |
| 11302 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 41 | SOX5 | Sponge network | -2.915 | 0.00015 | -1.091 | 4.0E-5 | 0.469 |
| 11303 | SH3RF3-AS1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-625-3p | 23 | ABL2 | Sponge network | 0.889 | 9.0E-5 | 0.82 | 0 | 0.469 |
| 11304 | LINC00472 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | PRUNE2 | Sponge network | -0.842 | 0.01361 | -1.733 | 0.00022 | 0.469 |
| 11305 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-3p | 34 | BMPR2 | Sponge network | 0.178 | 0.29106 | 0.206 | 0.0635 | 0.469 |
| 11306 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 16 | BCL2 | Sponge network | -0.187 | 0.23787 | -0.899 | 0 | 0.469 |
| 11307 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 27 | CRTC3 | Sponge network | 0.374 | 0.20054 | 0.164 | 0.16786 | 0.469 |
| 11308 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-93-5p | 16 | TP53INP1 | Sponge network | -0.465 | 0.04703 | -0.496 | 0.00064 | 0.469 |
| 11309 | PCA3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | -0.709 | 0.18168 | -0.382 | 0.05038 | 0.469 |
| 11310 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 23 | DMD | Sponge network | 0.174 | 0.30034 | -1.506 | 0 | 0.469 |
| 11311 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | MED12L | Sponge network | -2.46 | 0 | -0.194 | 0.55299 | 0.469 |
| 11312 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 48 | BNC2 | Sponge network | -0.222 | 0.32445 | -0.491 | 0.09988 | 0.469 |
| 11313 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | SYNE1 | Sponge network | -1.161 | 0.00591 | -0.738 | 0.00316 | 0.469 |
| 11314 | AC003090.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NAV3 | Sponge network | -2.117 | 0.00161 | 0.212 | 0.47729 | 0.469 |
| 11315 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | TACC1 | Sponge network | -0.769 | 0.00035 | -0.362 | 0.05406 | 0.469 |
| 11316 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | IGSF10 | Sponge network | -0.769 | 0.00035 | -1.751 | 1.0E-5 | 0.469 |
| 11317 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TLL1 | Sponge network | -0.323 | 0.01372 | -1.195 | 3.0E-5 | 0.469 |
| 11318 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | ITGAV | Sponge network | -0.129 | 0.57177 | 0.337 | 0.01002 | 0.469 |
| 11319 | ZNF667-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-5p | 14 | FMNL3 | Sponge network | -0.976 | 0.00025 | 0.555 | 4.0E-5 | 0.469 |
| 11320 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 31 | PRKAB2 | Sponge network | -0.323 | 0.01372 | -0.367 | 0.01371 | 0.469 |
| 11321 | GNG12-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 35 | GRIK3 | Sponge network | -0.793 | 7.0E-5 | -3.208 | 0 | 0.469 |
| 11322 | LINC00476 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 18 | EZH1 | Sponge network | -0.615 | 0.00015 | -0.564 | 1.0E-5 | 0.469 |
| 11323 | RP11-359B12.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 16 | STARD13 | Sponge network | 0.064 | 0.72934 | -0.187 | 0.27383 | 0.469 |
| 11324 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-590-3p | 15 | AKAP6 | Sponge network | 0.174 | 0.30034 | -1.586 | 3.0E-5 | 0.469 |
| 11325 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p | 14 | AHNAK | Sponge network | -0.358 | 0.17255 | -1.207 | 0 | 0.469 |
| 11326 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | RELN | Sponge network | 0.335 | 0.20596 | -2.403 | 0 | 0.469 |
| 11327 | LINC00265 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | ERCC6 | Sponge network | 1.07 | 0 | 0.23 | 0.015 | 0.469 |
| 11328 | OIP5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 26 | TBL1XR1 | Sponge network | 0.421 | 0.00084 | 0.578 | 0 | 0.469 |
| 11329 | C20orf166-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-5p | 21 | PRKCB | Sponge network | -2.69 | 0.0001 | -1.313 | 2.0E-5 | 0.469 |
| 11330 | RP4-717I23.3 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-495-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 11 | SHPRH | Sponge network | 0.637 | 0.00069 | 0.098 | 0.40994 | 0.469 |
| 11331 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | RCAN2 | Sponge network | -0.777 | 0.1134 | -0.33 | 0.15172 | 0.469 |
| 11332 | RP11-119F7.5 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | PCDH9 | Sponge network | 0.225 | 0.30644 | -1.823 | 4.0E-5 | 0.469 |
| 11333 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 17 | TXNIP | Sponge network | -1.216 | 0 | -1.277 | 0 | 0.469 |
| 11334 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 14 | LRRC7 | Sponge network | -0.358 | 0.17255 | -1.341 | 0.01193 | 0.469 |
| 11335 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 29 | RICTOR | Sponge network | 1.089 | 0 | 0.388 | 0.00188 | 0.469 |
| 11336 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-532-3p | 14 | CBL | Sponge network | 0.497 | 0.01451 | 0.291 | 0.01267 | 0.469 |
| 11337 | BVES-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-942-5p | 11 | BMPR1A | Sponge network | -2.62 | 0 | -0.366 | 0.01036 | 0.469 |
| 11338 | HOXB-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-93-3p | 19 | CTIF | Sponge network | -0.154 | 0.53954 | -0.389 | 0.00549 | 0.469 |
| 11339 | MIR143HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p | 33 | PRDM2 | Sponge network | -0.739 | 0.0574 | -0.258 | 0.00721 | 0.469 |
| 11340 | LINC00702 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | GALNT13 | Sponge network | -0.737 | 0.06182 | -0.857 | 0.07439 | 0.469 |
| 11341 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ABCC9 | Sponge network | -0.513 | 0.04703 | -0.466 | 0.14121 | 0.469 |
| 11342 | PCA3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | LHFP | Sponge network | -0.709 | 0.18168 | -0.167 | 0.41371 | 0.469 |
| 11343 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 17 | ABCC9 | Sponge network | -0.227 | 0.31657 | -0.466 | 0.14121 | 0.469 |
| 11344 | RP11-571M6.17 |
hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | RICTOR | Sponge network | 1.013 | 0 | 0.388 | 0.00188 | 0.469 |
| 11345 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p | 11 | ZYX | Sponge network | -0.129 | 0.57177 | 0.019 | 0.89763 | 0.469 |
| 11346 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-495-3p | 10 | CUL5 | Sponge network | 1.147 | 0 | 0.026 | 0.77301 | 0.469 |
| 11347 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | EPHA3 | Sponge network | -1.161 | 0.00591 | 0.185 | 0.53588 | 0.469 |
| 11348 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-93-5p | 14 | EZH1 | Sponge network | -1.349 | 0.00017 | -0.564 | 1.0E-5 | 0.469 |
| 11349 | LINC00654 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p | 11 | SMARCA1 | Sponge network | 0.374 | 0.20054 | -0.684 | 0.00159 | 0.469 |
| 11350 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 28 | MCC | Sponge network | -0.318 | 0.65847 | -1.005 | 0 | 0.469 |
| 11351 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | -0.563 | 0.02545 | -1.465 | 0.00033 | 0.469 |
| 11352 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CNTN1 | Sponge network | -1.057 | 0.00029 | -2.594 | 0 | 0.469 |
| 11353 | RP11-244O19.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | -0.096 | 0.67958 | -0.974 | 0.00034 | 0.469 |
| 11354 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 19 | CACNA2D1 | Sponge network | -4.546 | 0 | -0.846 | 0.00675 | 0.469 |
| 11355 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 14 | AKAP12 | Sponge network | 0.246 | 0.59912 | -0.932 | 0.0028 | 0.469 |
| 11356 | SNHG14 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 11 | MLLT11 | Sponge network | -1.111 | 0.0003 | -0.661 | 0.01733 | 0.469 |
| 11357 | AGAP11 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-651-5p;hsa-miR-93-3p | 27 | WWTR1 | Sponge network | -1.645 | 0 | -0.345 | 0.11997 | 0.469 |
| 11358 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-942-5p | 25 | HLF | Sponge network | -0.376 | 0.00337 | -2.443 | 0 | 0.469 |
| 11359 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92b-3p | 14 | AKAP12 | Sponge network | 0.16 | 0.50785 | -0.932 | 0.0028 | 0.469 |
| 11360 | LINC00473 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-532-3p | 12 | RPS6KA2 | Sponge network | -1.203 | 0.03881 | -0.57 | 0.00013 | 0.469 |
| 11361 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 22 | NAV3 | Sponge network | -0.318 | 0.65847 | 0.212 | 0.47729 | 0.469 |
| 11362 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | ZFHX4 | Sponge network | 0.395 | 0.15451 | 0.018 | 0.95574 | 0.469 |
| 11363 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-766-3p | 39 | CHD5 | Sponge network | -0.487 | 0.02157 | -0.922 | 0.01521 | 0.469 |
| 11364 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | IGSF10 | Sponge network | -1.281 | 0.00012 | -1.751 | 1.0E-5 | 0.469 |
| 11365 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429 | 11 | PARK2 | Sponge network | -0.563 | 0.02545 | -1.892 | 0 | 0.469 |
| 11366 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | EYA4 | Sponge network | 0.246 | 0.59912 | 0.3 | 0.36876 | 0.469 |
| 11367 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-629-3p | 21 | ZBTB4 | Sponge network | -0.739 | 0.00044 | -0.739 | 0 | 0.469 |
| 11368 | LINC00641 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 14 | TAF1 | Sponge network | -0.222 | 0.32445 | 0.39 | 3.0E-5 | 0.469 |
| 11369 | GNG12-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | GJA1 | Sponge network | -0.793 | 7.0E-5 | -0.485 | 0.02179 | 0.469 |
| 11370 | LINC00094 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-7-5p | 11 | MLLT11 | Sponge network | 0.253 | 0.09565 | -0.661 | 0.01733 | 0.469 |
| 11371 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 17 | PRKAB2 | Sponge network | 0.178 | 0.29106 | -0.367 | 0.01371 | 0.469 |
| 11372 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | ZNF292 | Sponge network | 0.083 | 0.66405 | 0.276 | 0.04148 | 0.469 |
| 11373 | MIR497HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-940 | 34 | NTRK2 | Sponge network | -1.439 | 4.0E-5 | -0.736 | 0.06495 | 0.469 |
| 11374 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 53 | ERBB4 | Sponge network | -2.46 | 0 | -1.975 | 2.0E-5 | 0.469 |
| 11375 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429 | 10 | ZNF521 | Sponge network | -2.62 | 0 | -0.111 | 0.58068 | 0.469 |
| 11376 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | FGFR1 | Sponge network | -0.811 | 2.0E-5 | -0.509 | 0.03957 | 0.469 |
| 11377 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | -0.513 | 0.04703 | -1.12 | 0.00107 | 0.469 |
| 11378 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 70 | BMPR2 | Sponge network | -1.575 | 6.0E-5 | 0.206 | 0.0635 | 0.469 |
| 11379 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-542-3p | 10 | ALDH1A2 | Sponge network | -0.576 | 0.10121 | -2.411 | 0 | 0.469 |
| 11380 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577 | 25 | EBF1 | Sponge network | -2.69 | 0.0001 | -0.384 | 0.08856 | 0.469 |
| 11381 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-502-5p;hsa-miR-744-3p | 22 | PTPN14 | Sponge network | -0.611 | 0.09607 | 0.144 | 0.34595 | 0.469 |
| 11382 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | ADAMTSL3 | Sponge network | -2.925 | 0 | -1.559 | 0 | 0.469 |
| 11383 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-942-5p | 17 | FBXO31 | Sponge network | -0.739 | 0.0574 | -0.085 | 0.42389 | 0.469 |
| 11384 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 32 | BNC2 | Sponge network | 0.176 | 0.37402 | -0.491 | 0.09988 | 0.469 |
| 11385 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 27 | GAS7 | Sponge network | -0.517 | 0.02935 | -0.555 | 0.01602 | 0.469 |
| 11386 | FAM66C |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | TAL1 | Sponge network | -0.901 | 0.00019 | -0.462 | 0.00574 | 0.469 |
| 11387 | JAZF1-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-98-5p | 16 | KLF10 | Sponge network | -0.769 | 0.00035 | -0.783 | 0 | 0.469 |
| 11388 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 15 | CNTNAP1 | Sponge network | -1.249 | 7.0E-5 | 0.358 | 0.13727 | 0.469 |
| 11389 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p | 17 | CITED2 | Sponge network | -0.49 | 0.04946 | -1.545 | 0 | 0.469 |
| 11390 | RP11-400K9.4 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-542-3p | 10 | KCTD8 | Sponge network | -0.611 | 0.09607 | -3.359 | 0 | 0.469 |
| 11391 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-96-5p | 13 | EBF1 | Sponge network | 0.622 | 0.00015 | -0.384 | 0.08856 | 0.469 |
| 11392 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | ATRX | Sponge network | -1.575 | 6.0E-5 | 0.261 | 0.04408 | 0.469 |
| 11393 | RP11-894P9.1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-940 | 10 | BAZ2B | Sponge network | 0.993 | 0 | -0.276 | 0.02833 | 0.469 |
| 11394 | MEG3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 49 | TRPS1 | Sponge network | 0.249 | 0.35902 | -0.191 | 0.44109 | 0.469 |
| 11395 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | ST6GALNAC3 | Sponge network | -0.129 | 0.57177 | -0.987 | 0 | 0.469 |
| 11396 | NR2F1-AS1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p | 18 | IRS1 | Sponge network | -0.49 | 0.04946 | -0.026 | 0.88855 | 0.469 |
| 11397 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | QKI | Sponge network | 0.225 | 0.30644 | -0.333 | 0.04111 | 0.469 |
| 11398 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 46 | CHD5 | Sponge network | -1.281 | 0.00012 | -0.922 | 0.01521 | 0.469 |
| 11399 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 36 | ST6GAL2 | Sponge network | -2.46 | 0 | -1.201 | 0.00597 | 0.469 |
| 11400 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-484;hsa-miR-629-3p;hsa-miR-93-3p | 22 | EBF3 | Sponge network | -0.154 | 0.78939 | -0.058 | 0.81437 | 0.469 |
| 11401 | RP11-532F6.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | PDE4DIP | Sponge network | -0.896 | 0.00099 | -0.282 | 0.0257 | 0.469 |
| 11402 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-940 | 16 | IGF1 | Sponge network | -0.72 | 0.24239 | -0.204 | 0.5898 | 0.469 |
| 11403 | RP11-400K9.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-340-5p;hsa-miR-590-5p | 17 | DMD | Sponge network | -0.733 | 0.19469 | -1.506 | 0 | 0.469 |
| 11404 | RP11-53O19.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-493-3p;hsa-miR-532-3p | 19 | MRVI1 | Sponge network | -0.405 | 0.22013 | -1.051 | 0.00022 | 0.469 |
| 11405 | RP11-359E10.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-629-5p;hsa-miR-93-5p | 44 | DST | Sponge network | 0.299 | 0.57722 | -0.713 | 0.00096 | 0.469 |
| 11406 | MEG3 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-93-5p | 66 | DST | Sponge network | 0.249 | 0.35902 | -0.713 | 0.00096 | 0.469 |
| 11407 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 46 | SASH1 | Sponge network | -0.739 | 0.0574 | -0.502 | 0.00175 | 0.469 |
| 11408 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-362-5p;hsa-miR-576-5p | 14 | IGF1 | Sponge network | -0.932 | 0.00329 | -0.204 | 0.5898 | 0.469 |
| 11409 | JAZF1-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-96-5p | 12 | PRKACA | Sponge network | -0.769 | 0.00035 | -0.255 | 0.0012 | 0.469 |
| 11410 | AC083843.1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 11 | KIF5B | Sponge network | 1.4 | 0 | 0.362 | 0.00066 | 0.469 |
| 11411 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 32 | ANK2 | Sponge network | 0.374 | 0.20054 | -1.603 | 1.0E-5 | 0.469 |
| 11412 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 35 | PIK3R1 | Sponge network | 0.214 | 0.18246 | -0.133 | 0.36854 | 0.469 |
| 11413 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-7-1-3p | 19 | LRP1B | Sponge network | -4.546 | 0 | -2.163 | 0 | 0.469 |
| 11414 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-589-5p;hsa-miR-96-5p | 18 | ARHGAP29 | Sponge network | 0.249 | 0.35902 | 0.131 | 0.42953 | 0.469 |
| 11415 | AP001347.6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-486-5p | 12 | EZH1 | Sponge network | -1.161 | 0.00591 | -0.564 | 1.0E-5 | 0.469 |
| 11416 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-582-3p | 32 | CBL | Sponge network | 0.494 | 0.00339 | 0.291 | 0.01267 | 0.469 |
| 11417 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 37 | GAS7 | Sponge network | -0.318 | 0.65847 | -0.555 | 0.01602 | 0.469 |
| 11418 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 19 | DKK3 | Sponge network | -1.575 | 6.0E-5 | -0.052 | 0.77783 | 0.469 |
| 11419 | LINC01018 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-942-5p | 36 | HLF | Sponge network | -2.915 | 0.00015 | -2.443 | 0 | 0.469 |
| 11420 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 15 | RCAN2 | Sponge network | 0.494 | 0.00339 | -0.33 | 0.15172 | 0.468 |
| 11421 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-766-3p | 24 | IL17RD | Sponge network | 0.559 | 0.00026 | -0.019 | 0.94873 | 0.468 |
| 11422 | RP11-13K12.1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-625-5p | 14 | NGFR | Sponge network | -0.154 | 0.78939 | -1.271 | 0.01058 | 0.468 |
| 11423 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | EXOC8 | Sponge network | 0.959 | 0 | 0.216 | 0.00598 | 0.468 |
| 11424 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 55 | PRKAA2 | Sponge network | 0.532 | 0.00505 | -2.462 | 0 | 0.468 |
| 11425 | USP3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-589-3p | 15 | ABCA10 | Sponge network | -0.162 | 0.47687 | -0.571 | 0.03674 | 0.468 |
| 11426 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-542-3p | 12 | ABCB5 | Sponge network | -1.092 | 4.0E-5 | -0.956 | 0.04077 | 0.468 |
| 11427 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-628-5p | 12 | TRIM33 | Sponge network | 1.206 | 0 | 0.402 | 7.0E-5 | 0.468 |
| 11428 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | SASH1 | Sponge network | -0.976 | 0.00025 | -0.502 | 0.00175 | 0.468 |
| 11429 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p | 13 | LRRK2 | Sponge network | 0.249 | 0.35902 | -1.136 | 1.0E-5 | 0.468 |
| 11430 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | TCF4 | Sponge network | 1.253 | 0 | 0.016 | 0.92271 | 0.468 |
| 11431 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-550a-3p;hsa-miR-589-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 42 | TCF4 | Sponge network | -4.546 | 0 | 0.016 | 0.92271 | 0.468 |
| 11432 | RP11-403I13.8 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-590-3p | 20 | NRK | Sponge network | 0.619 | 0.00157 | 0.656 | 0.19217 | 0.468 |
| 11433 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | TIMP3 | Sponge network | -1.575 | 6.0E-5 | -0.546 | 0.02307 | 0.468 |
| 11434 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-511-5p | 12 | CREB1 | Sponge network | 0.497 | 0.01451 | 0.203 | 0.00537 | 0.468 |
| 11435 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940 | 13 | NR4A3 | Sponge network | -1.047 | 0 | -1.385 | 0 | 0.468 |
| 11436 | MIR497HG |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 21 | JDP2 | Sponge network | -1.439 | 4.0E-5 | -0.967 | 0 | 0.468 |
| 11437 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-511-5p;hsa-miR-590-3p | 13 | BCL6 | Sponge network | -0.739 | 0.00044 | -0.211 | 0.19489 | 0.468 |
| 11438 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 20 | PDZD2 | Sponge network | -0.942 | 0 | -0.909 | 3.0E-5 | 0.468 |
| 11439 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-93-3p | 18 | PCDH10 | Sponge network | -0.469 | 0.08721 | -1.644 | 0.0078 | 0.468 |
| 11440 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p | 10 | LHFP | Sponge network | -0.154 | 0.78939 | -0.167 | 0.41371 | 0.468 |
| 11441 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 63 | IL6ST | Sponge network | -0.709 | 0.18168 | -0.402 | 0.00676 | 0.468 |
| 11442 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-942-5p | 55 | THRB | Sponge network | -0.162 | 0.47687 | -1.117 | 0.0004 | 0.468 |
| 11443 | AC011526.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-92a-3p | 13 | IGFBP5 | Sponge network | 0.342 | 0.03129 | -0.806 | 0.00105 | 0.468 |
| 11444 | RP11-121C2.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p | 30 | CREB1 | Sponge network | 1.147 | 0 | 0.203 | 0.00537 | 0.468 |
| 11445 | RP5-1198O20.4 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 24 | RYR2 | Sponge network | 0.997 | 0.00066 | -1.466 | 0.00031 | 0.468 |
| 11446 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | TSHZ3 | Sponge network | 0.246 | 0.59912 | -0.556 | 0.01516 | 0.468 |
| 11447 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p | 20 | PLXNC1 | Sponge network | -0.129 | 0.57177 | 0.569 | 0.00329 | 0.468 |
| 11448 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 20 | CDH23 | Sponge network | 0.997 | 0.00066 | -0.974 | 0.00034 | 0.468 |
| 11449 | RP1-151F17.2 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 21 | ATM | Sponge network | 0.222 | 0.29133 | 0.178 | 0.21568 | 0.468 |
| 11450 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 39 | TGFBR3 | Sponge network | -0.901 | 0.00019 | -1.173 | 1.0E-5 | 0.468 |
| 11451 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-493-5p | 16 | DMD | Sponge network | -0.829 | 0.01343 | -1.506 | 0 | 0.468 |
| 11452 | RP11-307B6.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-532-5p;hsa-miR-942-5p | 18 | IGFBP5 | Sponge network | -4.463 | 0.00025 | -0.806 | 0.00105 | 0.468 |
| 11453 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-629-5p | 10 | UVRAG | Sponge network | -0.096 | 0.67958 | -0.017 | 0.83865 | 0.468 |
| 11454 | AC010761.8 |
hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-766-3p | 14 | MDM4 | Sponge network | 1.638 | 0 | 0.345 | 0.00762 | 0.468 |
| 11455 | LINC00476 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 30 | PCDH9 | Sponge network | -0.615 | 0.00015 | -1.823 | 4.0E-5 | 0.468 |
| 11456 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-625-5p | 24 | ZMYM2 | Sponge network | 0.537 | 0.00139 | 0.279 | 0.01273 | 0.468 |
| 11457 | FENDRR |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-96-5p;hsa-miR-98-5p | 43 | CREB3L2 | Sponge network | -1.281 | 0.00012 | -0.525 | 2.0E-5 | 0.468 |
| 11458 | RP11-25K19.1 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | NR3C2 | Sponge network | -1.057 | 0.00029 | -1.56 | 0 | 0.468 |
| 11459 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | ZNF292 | Sponge network | 0.953 | 0 | 0.276 | 0.04148 | 0.468 |
| 11460 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | DKK3 | Sponge network | 0.16 | 0.50785 | -0.052 | 0.77783 | 0.468 |
| 11461 | CTC-429P9.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-744-3p | 23 | DIP2C | Sponge network | 0.178 | 0.29106 | -0.436 | 0.01713 | 0.468 |
| 11462 | LINC01018 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | GALNT13 | Sponge network | -2.915 | 0.00015 | -0.857 | 0.07439 | 0.468 |
| 11463 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-7-5p;hsa-miR-96-5p | 12 | CAV1 | Sponge network | -1.654 | 0.00144 | -1.013 | 6.0E-5 | 0.468 |
| 11464 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-576-5p | 16 | ST6GALNAC3 | Sponge network | -0.932 | 0.00329 | -0.987 | 0 | 0.468 |
| 11465 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | ITCH | Sponge network | 0.34 | 0.00273 | 0.084 | 0.34058 | 0.468 |
| 11466 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 43 | PHC3 | Sponge network | -0.739 | 0.0574 | 0.212 | 0.0499 | 0.468 |
| 11467 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p | 13 | RBM5 | Sponge network | 0.082 | 0.55397 | -0.205 | 0.0129 | 0.468 |
| 11468 | LINC00265 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 13 | NF1 | Sponge network | 1.07 | 0 | 0.51 | 0 | 0.468 |
| 11469 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-590-3p | 13 | BNC2 | Sponge network | -1.425 | 0.04204 | -0.491 | 0.09988 | 0.468 |
| 11470 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-93-5p | 24 | SYNE1 | Sponge network | -0.154 | 0.78939 | -0.738 | 0.00316 | 0.468 |
| 11471 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p | 18 | CITED2 | Sponge network | -2.62 | 0 | -1.545 | 0 | 0.468 |
| 11472 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 52 | PTPRD | Sponge network | -0.49 | 0.04946 | -1.005 | 0.00064 | 0.468 |
| 11473 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | LIFR | Sponge network | -0.901 | 0.00019 | -1.794 | 0 | 0.468 |
| 11474 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 28 | ZFHX3 | Sponge network | 0.889 | 9.0E-5 | 0.389 | 0.01363 | 0.468 |
| 11475 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 23 | RIMBP2 | Sponge network | -2.46 | 0 | -1.968 | 5.0E-5 | 0.468 |
| 11476 | CTD-2303H24.2 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p | 17 | ATM | Sponge network | 0.415 | 0.14657 | 0.178 | 0.21568 | 0.468 |
| 11477 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-671-5p | 24 | KIT | Sponge network | -1.216 | 0 | -2.184 | 0 | 0.468 |
| 11478 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 17 | DYNC1I1 | Sponge network | -0.421 | 0.0114 | -1.12 | 0.00107 | 0.468 |
| 11479 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | EBF3 | Sponge network | 0.225 | 0.30644 | -0.058 | 0.81437 | 0.468 |
| 11480 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-942-5p | 45 | UNC80 | Sponge network | -0.323 | 0.01372 | -1.934 | 0 | 0.468 |
| 11481 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-629-3p;hsa-miR-92a-3p | 15 | SPTBN4 | Sponge network | -0.096 | 0.67958 | -0.786 | 0.01281 | 0.468 |
| 11482 | SERTAD4-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-424-5p | 11 | LRIG1 | Sponge network | -0.932 | 0.00329 | -0.537 | 0.00588 | 0.468 |
| 11483 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p | 15 | HOOK3 | Sponge network | 0.385 | 0.10067 | 0.273 | 0.09957 | 0.468 |
| 11484 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | 0.101 | 0.60919 | -0.354 | 0.14694 | 0.468 |
| 11485 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 0.919 | 0 | 0.51 | 0 | 0.468 |
| 11486 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 27 | LRRC7 | Sponge network | 0.572 | 0.01722 | -1.341 | 0.01193 | 0.468 |
| 11487 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-589-5p | 19 | ADARB1 | Sponge network | 0.693 | 0.00076 | -0.335 | 0.06631 | 0.468 |
| 11488 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-3p | 12 | JAKMIP2 | Sponge network | -0.842 | 0.01361 | -1.388 | 0 | 0.468 |
| 11489 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 30 | MYO9A | Sponge network | 0.782 | 0.00016 | -0.147 | 0.32373 | 0.468 |
| 11490 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p | 11 | AKAP6 | Sponge network | 0.37 | 0.27614 | -1.586 | 3.0E-5 | 0.468 |
| 11491 | MAGI2-AS3 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 22 | RPS6KA6 | Sponge network | -0.129 | 0.57177 | -2.989 | 0 | 0.468 |
| 11492 | AC010226.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-5p | 24 | KCNA6 | Sponge network | -0.811 | 2.0E-5 | -1.531 | 0.00013 | 0.468 |
| 11493 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 48 | PIK3R1 | Sponge network | -0.576 | 0.10121 | -0.133 | 0.36854 | 0.468 |
| 11494 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | CDH19 | Sponge network | 0.997 | 0.00066 | -3.434 | 0 | 0.468 |
| 11495 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-629-5p | 40 | SOX5 | Sponge network | -0.154 | 0.78939 | -1.091 | 4.0E-5 | 0.468 |
| 11496 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | SOX5 | Sponge network | 0.532 | 0.00505 | -1.091 | 4.0E-5 | 0.468 |
| 11497 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | ARHGAP28 | Sponge network | -2.117 | 0.00161 | -0.911 | 0.00013 | 0.468 |
| 11498 | LINC00641 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | ZFX | Sponge network | -0.222 | 0.32445 | 0.09 | 0.42563 | 0.468 |
| 11499 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | ZNF521 | Sponge network | 0.4 | 0.14611 | -0.111 | 0.58068 | 0.468 |
| 11500 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | NIN | Sponge network | -0.376 | 0.00337 | -0.253 | 0.13794 | 0.468 |
| 11501 | BOLA3-AS1 |
hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 11 | DACT1 | Sponge network | -0.246 | 0.35034 | 0.783 | 0.00072 | 0.468 |
| 11502 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 20 | RARB | Sponge network | -0.901 | 0.00019 | -0.825 | 2.0E-5 | 0.468 |
| 11503 | AC010226.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | GAS7 | Sponge network | -0.811 | 2.0E-5 | -0.555 | 0.01602 | 0.468 |
| 11504 | RP11-389C8.2 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 10 | CNTNAP1 | Sponge network | 0.727 | 3.0E-5 | 0.358 | 0.13727 | 0.468 |
| 11505 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 32 | LPP | Sponge network | -0.187 | 0.23787 | -0.459 | 0.04475 | 0.468 |
| 11506 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 45 | ZFHX3 | Sponge network | -0.49 | 0.04946 | 0.389 | 0.01363 | 0.468 |
| 11507 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | -0.096 | 0.67958 | 0.809 | 0.00544 | 0.468 |
| 11508 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-484;hsa-miR-92a-3p;hsa-miR-98-5p | 17 | ITGA5 | Sponge network | -2.452 | 0 | 0.452 | 0.03524 | 0.468 |
| 11509 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p | 13 | IL17RD | Sponge network | -4.463 | 0.00025 | -0.019 | 0.94873 | 0.468 |
| 11510 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 19 | SON | Sponge network | 0.083 | 0.66405 | 0.168 | 0.04406 | 0.468 |
| 11511 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 10 | FSTL5 | Sponge network | -4.546 | 0 | -1.3 | 0.01619 | 0.468 |
| 11512 | CTD-3064M3.3 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p | 12 | CXCL12 | Sponge network | -0.469 | 0.08721 | -1.421 | 0 | 0.468 |
| 11513 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 32 | SOX5 | Sponge network | -0.611 | 0.09607 | -1.091 | 4.0E-5 | 0.468 |
| 11514 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-629-3p | 37 | IL6ST | Sponge network | 0.811 | 0.03228 | -0.402 | 0.00676 | 0.468 |
| 11515 | LINC00865 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-942-5p | 34 | IGFBP5 | Sponge network | -1.249 | 7.0E-5 | -0.806 | 0.00105 | 0.468 |
| 11516 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 40 | ZFHX3 | Sponge network | 0.72 | 0.0001 | 0.389 | 0.01363 | 0.468 |
| 11517 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 26 | SCN9A | Sponge network | 0.082 | 0.55654 | -0.525 | 0.10593 | 0.468 |
| 11518 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | 0.374 | 0.20054 | -1.751 | 1.0E-5 | 0.468 |
| 11519 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-539-5p;hsa-miR-577 | 15 | GRIN2A | Sponge network | -0.473 | 0.07602 | -2.161 | 0 | 0.468 |
| 11520 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-590-3p | 19 | DYNC1I1 | Sponge network | 0.246 | 0.59912 | -1.12 | 0.00107 | 0.468 |
| 11521 | LINC00473 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DCN | Sponge network | -1.203 | 0.03881 | -1.288 | 0 | 0.468 |
| 11522 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 21 | EBF1 | Sponge network | 0.727 | 3.0E-5 | -0.384 | 0.08856 | 0.468 |
| 11523 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | PDGFC | Sponge network | -1.281 | 0.00012 | -0.33 | 0.06483 | 0.468 |
| 11524 | CTC-444N24.11 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 19 | RASAL2 | Sponge network | 1.153 | 0 | 0.869 | 0 | 0.468 |
| 11525 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 16 | NEDD4 | Sponge network | -1.092 | 4.0E-5 | 0.02 | 0.89834 | 0.468 |
| 11526 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 49 | SASH1 | Sponge network | -1.111 | 0.0003 | -0.502 | 0.00175 | 0.468 |
| 11527 | AC006116.24 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p;hsa-miR-577 | 10 | INPP4A | Sponge network | -0.473 | 0.07602 | 0.07 | 0.53563 | 0.468 |
| 11528 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | EPG5 | Sponge network | 0.683 | 1.0E-5 | 0.101 | 0.3563 | 0.468 |
| 11529 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-577 | 25 | NOTCH2 | Sponge network | 0.811 | 0.03228 | 0.138 | 0.35163 | 0.468 |
| 11530 | GAS6-AS2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429 | 14 | NRK | Sponge network | 0.16 | 0.50785 | 0.656 | 0.19217 | 0.468 |
| 11531 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | PRDM5 | Sponge network | -0.896 | 0.00099 | -1.09 | 0.00014 | 0.468 |
| 11532 | H19 |
hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-589-3p | 12 | IL17RD | Sponge network | 2.439 | 0 | -0.019 | 0.94873 | 0.468 |
| 11533 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 27 | SCN9A | Sponge network | -0.096 | 0.67958 | -0.525 | 0.10593 | 0.468 |
| 11534 | PGM5-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 11 | NCAM2 | Sponge network | -4.546 | 0 | -0.482 | 0.22565 | 0.468 |
| 11535 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | HACE1 | Sponge network | 1.254 | 0 | -0.072 | 0.55239 | 0.468 |
| 11536 | FAM157A |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326 | 12 | NF1 | Sponge network | 0.813 | 1.0E-5 | 0.51 | 0 | 0.468 |
| 11537 | RP11-532F6.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | FAM172A | Sponge network | -0.896 | 0.00099 | -0.57 | 0 | 0.468 |
| 11538 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | NR2C2 | Sponge network | 1.205 | 0 | 0.21 | 0.03224 | 0.468 |
| 11539 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-93-5p | 13 | EDA2R | Sponge network | -1.654 | 0.00144 | -0.587 | 0.01598 | 0.468 |
| 11540 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | PTPRB | Sponge network | -1.111 | 0.0003 | 0.105 | 0.47122 | 0.468 |
| 11541 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | CDKN1C | Sponge network | -0.49 | 0.04946 | -0.929 | 0.00033 | 0.467 |
| 11542 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-3p;hsa-miR-362-3p;hsa-miR-590-3p | 11 | CACNA2D1 | Sponge network | -1.886 | 0 | -0.846 | 0.00675 | 0.467 |
| 11543 | DIO3OS |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-940 | 12 | GLI3 | Sponge network | -0.773 | 0.04073 | -0.615 | 0.01482 | 0.467 |
| 11544 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 45 | AR | Sponge network | -0.405 | 0.22013 | -1.554 | 0 | 0.467 |
| 11545 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-671-5p | 17 | CYLD | Sponge network | -0.896 | 0.00099 | -0.367 | 0.00171 | 0.467 |
| 11546 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-93-5p | 13 | GLI3 | Sponge network | -0.154 | 0.78939 | -0.615 | 0.01482 | 0.467 |
| 11547 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p | 42 | CHD5 | Sponge network | -0.154 | 0.78939 | -0.922 | 0.01521 | 0.467 |
| 11548 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 33 | CTIF | Sponge network | -0.769 | 0.00035 | -0.389 | 0.00549 | 0.467 |
| 11549 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-766-3p;hsa-miR-940 | 10 | MDM4 | Sponge network | 0.043 | 0.81628 | 0.345 | 0.00762 | 0.467 |
| 11550 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-935 | 22 | FBXO32 | Sponge network | -0.246 | 0.35034 | -0.495 | 0.07561 | 0.467 |
| 11551 | FENDRR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-98-5p | 13 | SCN11A | Sponge network | -1.281 | 0.00012 | -1.15 | 0.00299 | 0.467 |
| 11552 | RP11-532F6.3 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 15 | CSDE1 | Sponge network | -0.896 | 0.00099 | -0.493 | 0 | 0.467 |
| 11553 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MYO9A | Sponge network | -1.161 | 0.00591 | -0.147 | 0.32373 | 0.467 |
| 11554 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 30 | MCC | Sponge network | 0.199 | 0.38015 | -1.005 | 0 | 0.467 |
| 11555 | SNHG14 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | DACT1 | Sponge network | -1.111 | 0.0003 | 0.783 | 0.00072 | 0.467 |
| 11556 | RP11-400K9.4 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-744-3p;hsa-miR-93-5p | 44 | DST | Sponge network | -0.611 | 0.09607 | -0.713 | 0.00096 | 0.467 |
| 11557 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 24 | SCN9A | Sponge network | 0.331 | 0.57247 | -0.525 | 0.10593 | 0.467 |
| 11558 | PART1 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | DIRAS1 | Sponge network | -2.925 | 0 | -2.518 | 0 | 0.467 |
| 11559 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-7-1-3p | 14 | TIMP3 | Sponge network | 0.299 | 0.57722 | -0.546 | 0.02307 | 0.467 |
| 11560 | TPTEP1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 32 | WWTR1 | Sponge network | -1.216 | 0 | -0.345 | 0.11997 | 0.467 |
| 11561 | MAGI2-AS3 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 29 | CTIF | Sponge network | -0.129 | 0.57177 | -0.389 | 0.00549 | 0.467 |
| 11562 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-5p | 11 | GREM1 | Sponge network | -2.095 | 0.00112 | 0.902 | 0.01691 | 0.467 |
| 11563 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 11 | CTNNA3 | Sponge network | -0.942 | 0 | -2.046 | 0.00019 | 0.467 |
| 11564 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | BNC2 | Sponge network | -2.915 | 0.00015 | -0.491 | 0.09988 | 0.467 |
| 11565 | RP11-244O19.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 15 | BCL9 | Sponge network | -0.096 | 0.67958 | 0.321 | 0.00267 | 0.467 |
| 11566 | PART1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 60 | AR | Sponge network | -2.925 | 0 | -1.554 | 0 | 0.467 |
| 11567 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FSTL5 | Sponge network | -0.709 | 0.18168 | -1.3 | 0.01619 | 0.467 |
| 11568 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-590-3p | 27 | PHC3 | Sponge network | -0.096 | 0.67958 | 0.212 | 0.0499 | 0.467 |
| 11569 | RP5-1085F17.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | BRD3 | Sponge network | 0.541 | 0.00125 | 0.37 | 0.00013 | 0.467 |
| 11570 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 10 | IGSF10 | Sponge network | -0.517 | 0.02935 | -1.751 | 1.0E-5 | 0.467 |
| 11571 | TRAF3IP2-AS1 |
hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | RPS6KA6 | Sponge network | -0.323 | 0.01372 | -2.989 | 0 | 0.467 |
| 11572 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 23 | TBL1XR1 | Sponge network | 1.601 | 0 | 0.578 | 0 | 0.467 |
| 11573 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-424-5p | 11 | LRRC7 | Sponge network | -2.531 | 0 | -1.341 | 0.01193 | 0.467 |
| 11574 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 22 | PRKAB2 | Sponge network | -1.092 | 4.0E-5 | -0.367 | 0.01371 | 0.467 |
| 11575 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 21 | RSPO2 | Sponge network | -0.896 | 0.00099 | -3.038 | 0 | 0.467 |
| 11576 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | FAM133A | Sponge network | 0.335 | 0.20596 | 0.549 | 0.29009 | 0.467 |
| 11577 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 10 | PREX2 | Sponge network | -0.154 | 0.78939 | -0.272 | 0.25382 | 0.467 |
| 11578 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | TIMP3 | Sponge network | -0.487 | 0.02157 | -0.546 | 0.02307 | 0.467 |
| 11579 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 28 | RICTOR | Sponge network | 0.67 | 0.00048 | 0.388 | 0.00188 | 0.467 |
| 11580 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | PDE4DIP | Sponge network | 0.214 | 0.18246 | -0.282 | 0.0257 | 0.467 |
| 11581 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 17 | CNTNAP3 | Sponge network | 0.194 | 0.64278 | -1.465 | 0.00033 | 0.467 |
| 11582 | AC116366.6 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-940 | 22 | MDM4 | Sponge network | 0.329 | 0.11122 | 0.345 | 0.00762 | 0.467 |
| 11583 | FENDRR |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | PDE4DIP | Sponge network | -1.281 | 0.00012 | -0.282 | 0.0257 | 0.467 |
| 11584 | AC016747.3 |
hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-93-3p | 18 | WWTR1 | Sponge network | 0.199 | 0.38015 | -0.345 | 0.11997 | 0.467 |
| 11585 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | ZMYND11 | Sponge network | -0.129 | 0.57177 | -0.2 | 0.05449 | 0.467 |
| 11586 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421 | 12 | PTPRB | Sponge network | -0.583 | 0.14589 | 0.105 | 0.47122 | 0.467 |
| 11587 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-500a-5p | 23 | DMD | Sponge network | -0.318 | 0.65847 | -1.506 | 0 | 0.467 |
| 11588 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 50 | RUNX1T1 | Sponge network | 1.302 | 7.0E-5 | -0.344 | 0.2374 | 0.467 |
| 11589 | RP1-151F17.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 39 | CREB1 | Sponge network | 0.222 | 0.29133 | 0.203 | 0.00537 | 0.467 |
| 11590 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CLASP2 | Sponge network | 0.683 | 1.0E-5 | -0.038 | 0.71 | 0.467 |
| 11591 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 22 | ADAMTSL3 | Sponge network | -0.612 | 0.00094 | -1.559 | 0 | 0.467 |
| 11592 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 26 | ARHGAP28 | Sponge network | -0.901 | 0.00019 | -0.911 | 0.00013 | 0.467 |
| 11593 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-429 | 15 | ARHGAP28 | Sponge network | -1.654 | 0.00144 | -0.911 | 0.00013 | 0.467 |
| 11594 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 16 | ABCC9 | Sponge network | -1.249 | 7.0E-5 | -0.466 | 0.14121 | 0.467 |
| 11595 | RP11-61L19.3 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-374b-5p | 10 | UBE2D1 | Sponge network | 1.411 | 0 | 0.468 | 0 | 0.467 |
| 11596 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-590-5p | 10 | PAK3 | Sponge network | 0.176 | 0.37402 | -0.628 | 0.07253 | 0.467 |
| 11597 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 31 | RASSF2 | Sponge network | 0.199 | 0.38015 | -0.273 | 0.21961 | 0.467 |
| 11598 | RP11-35G9.3 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 13 | FMNL3 | Sponge network | 0.657 | 4.0E-5 | 0.555 | 4.0E-5 | 0.467 |
| 11599 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | RCAN2 | Sponge network | 0.225 | 0.30644 | -0.33 | 0.15172 | 0.467 |
| 11600 | ZNF667-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-93-5p | 53 | DST | Sponge network | -0.976 | 0.00025 | -0.713 | 0.00096 | 0.467 |
| 11601 | LINC01018 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | LHFP | Sponge network | -2.915 | 0.00015 | -0.167 | 0.41371 | 0.467 |
| 11602 | OIP5-AS1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-940 | 18 | TAF1 | Sponge network | 0.421 | 0.00084 | 0.39 | 3.0E-5 | 0.467 |
| 11603 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-92a-3p | 47 | BMPR2 | Sponge network | 0.214 | 0.18246 | 0.206 | 0.0635 | 0.467 |
| 11604 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-7-1-3p | 10 | DKK3 | Sponge network | 0.299 | 0.57722 | -0.052 | 0.77783 | 0.467 |
| 11605 | RP11-248G5.8 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 11 | KIF5B | Sponge network | 1.294 | 0 | 0.362 | 0.00066 | 0.467 |
| 11606 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | EBF1 | Sponge network | 0.657 | 4.0E-5 | -0.384 | 0.08856 | 0.467 |
| 11607 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 27 | ZMYM2 | Sponge network | 0.782 | 0.00016 | 0.279 | 0.01273 | 0.467 |
| 11608 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-505-5p | 15 | TBL1XR1 | Sponge network | 0.757 | 0 | 0.578 | 0 | 0.467 |
| 11609 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-505-3p;hsa-miR-7-1-3p | 14 | ST6GALNAC3 | Sponge network | -1.092 | 4.0E-5 | -0.987 | 0 | 0.467 |
| 11610 | RP11-119F7.5 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p | 10 | DYNC1I1 | Sponge network | 0.225 | 0.30644 | -1.12 | 0.00107 | 0.467 |
| 11611 | CKMT2-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-532-5p | 23 | PTPN14 | Sponge network | 0.082 | 0.55654 | 0.144 | 0.34595 | 0.467 |
| 11612 | BZRAP1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-671-5p | 14 | CADM3 | Sponge network | -0.465 | 0.04703 | -3.663 | 0 | 0.467 |
| 11613 | LINC00865 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-582-5p | 13 | ZNF208 | Sponge network | -1.249 | 7.0E-5 | -0.4 | 0.22381 | 0.467 |
| 11614 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | CDKN1C | Sponge network | -0.576 | 0.10121 | -0.929 | 0.00033 | 0.467 |
| 11615 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | MLLT10 | Sponge network | 0.545 | 0.00064 | 0.105 | 0.33292 | 0.467 |
| 11616 | RP11-867G23.10 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p | 13 | CDH23 | Sponge network | -2.531 | 0 | -0.974 | 0.00034 | 0.467 |
| 11617 | PWAR6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 15 | UVRAG | Sponge network | -1.575 | 6.0E-5 | -0.017 | 0.83865 | 0.467 |
| 11618 | SH3RF3-AS1 |
hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-877-5p | 16 | NRK | Sponge network | 0.889 | 9.0E-5 | 0.656 | 0.19217 | 0.467 |
| 11619 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p | 14 | TRIM33 | Sponge network | 1.4 | 0 | 0.402 | 7.0E-5 | 0.467 |
| 11620 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | RCAN2 | Sponge network | 0.392 | 0.0278 | -0.33 | 0.15172 | 0.467 |
| 11621 | RP11-597D13.9 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 27 | NAV3 | Sponge network | 1.302 | 7.0E-5 | 0.212 | 0.47729 | 0.467 |
| 11622 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p | 15 | LRRC4 | Sponge network | -0.487 | 0.02157 | -1.774 | 0 | 0.467 |
| 11623 | LINC00702 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | -0.737 | 0.06182 | -2.417 | 0 | 0.467 |
| 11624 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 25 | SYNPO2 | Sponge network | 0.064 | 0.72934 | -2.342 | 0 | 0.467 |
| 11625 | LINC00641 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p | 13 | ZBTB20 | Sponge network | -0.222 | 0.32445 | -0.122 | 0.57569 | 0.467 |
| 11626 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-877-5p;hsa-miR-935 | 30 | RAP1A | Sponge network | -0.162 | 0.47687 | -0.929 | 0 | 0.467 |
| 11627 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | MITF | Sponge network | -0.901 | 0.00019 | -0.769 | 0.00022 | 0.467 |
| 11628 | HCG11 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-590-3p;hsa-miR-625-3p | 28 | ABL2 | Sponge network | 0.385 | 0.10067 | 0.82 | 0 | 0.467 |
| 11629 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -0.739 | 0.0574 | -0.252 | 0.15511 | 0.467 |
| 11630 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 27 | SON | Sponge network | 0.421 | 0.00084 | 0.168 | 0.04406 | 0.467 |
| 11631 | ADAMTS9-AS2 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | RTN4 | Sponge network | -2.452 | 0 | -0.412 | 0.00014 | 0.467 |
| 11632 | CTC-510F12.2 |
hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-3913-5p;hsa-miR-484;hsa-miR-935 | 11 | GRIK3 | Sponge network | -0.691 | 0.00146 | -3.208 | 0 | 0.467 |
| 11633 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-655-3p | 25 | LATS1 | Sponge network | 0.537 | 0.00139 | 0.109 | 0.34208 | 0.467 |
| 11634 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | SOX5 | Sponge network | 0.374 | 0.20054 | -1.091 | 4.0E-5 | 0.467 |
| 11635 | AGAP11 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 38 | TRPS1 | Sponge network | -1.645 | 0 | -0.191 | 0.44109 | 0.467 |
| 11636 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-96-5p | 18 | GAS7 | Sponge network | -0.187 | 0.23787 | -0.555 | 0.01602 | 0.467 |
| 11637 | DNM3OS |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 0.572 | 0.01722 | 0.943 | 0 | 0.467 |
| 11638 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p | 22 | IGF1 | Sponge network | 0.176 | 0.37402 | -0.204 | 0.5898 | 0.467 |
| 11639 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-429 | 10 | JAKMIP2 | Sponge network | -0.246 | 0.35034 | -1.388 | 0 | 0.467 |
| 11640 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | ST6GALNAC3 | Sponge network | 0.342 | 0.03129 | -0.987 | 0 | 0.467 |
| 11641 | AC009948.5 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | LATS2 | Sponge network | 0.532 | 0.00505 | 0.159 | 0.2304 | 0.467 |
| 11642 | LINC00476 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-5p | 12 | SPOP | Sponge network | -0.615 | 0.00015 | -0.419 | 1.0E-5 | 0.467 |
| 11643 | AP001347.6 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p | 27 | GRIK3 | Sponge network | -1.161 | 0.00591 | -3.208 | 0 | 0.466 |
| 11644 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | RB1CC1 | Sponge network | 0.222 | 0.29133 | 0.175 | 0.12559 | 0.466 |
| 11645 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 22 | ZFHX4 | Sponge network | 0.693 | 0.00076 | 0.018 | 0.95574 | 0.466 |
| 11646 | CTC-510F12.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-219a-1-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-376c-3p | 13 | EPHA5 | Sponge network | -0.691 | 0.00146 | -0.957 | 0.01733 | 0.466 |
| 11647 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-424-5p | 10 | DYNC1I1 | Sponge network | -0.187 | 0.23787 | -1.12 | 0.00107 | 0.466 |
| 11648 | LINC00174 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-582-5p | 12 | NR2C2 | Sponge network | 1.253 | 0 | 0.21 | 0.03224 | 0.466 |
| 11649 | AC006547.13 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-628-5p | 10 | CHD2 | Sponge network | 1.159 | 0 | 0.049 | 0.55371 | 0.466 |
| 11650 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ERG | Sponge network | -0.487 | 0.02157 | 0.002 | 0.99011 | 0.466 |
| 11651 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | MYO9A | Sponge network | 0.659 | 3.0E-5 | -0.147 | 0.32373 | 0.466 |
| 11652 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | WWTR1 | Sponge network | 0.532 | 0.00505 | -0.345 | 0.11997 | 0.466 |
| 11653 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 31 | DDX6 | Sponge network | 0.75 | 0.00054 | 0.091 | 0.36428 | 0.466 |
| 11654 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p | 28 | ZBTB4 | Sponge network | -0.899 | 6.0E-5 | -0.739 | 0 | 0.466 |
| 11655 | RP11-359E10.1 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 22 | ZFHX3 | Sponge network | 0.299 | 0.57722 | 0.389 | 0.01363 | 0.466 |
| 11656 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-96-5p;hsa-miR-98-5p | 36 | CREB3L2 | Sponge network | -0.976 | 0.00025 | -0.525 | 2.0E-5 | 0.466 |
| 11657 | RP11-967K21.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577 | 19 | PIK3CA | Sponge network | 0.056 | 0.81195 | 0.195 | 0.06908 | 0.466 |
| 11658 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-766-3p;hsa-miR-96-5p | 24 | IL17RD | Sponge network | 0.41 | 0.02214 | -0.019 | 0.94873 | 0.466 |
| 11659 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 21 | HIP1 | Sponge network | 0.532 | 0.00505 | 0.774 | 0 | 0.466 |
| 11660 | RP11-701H24.7 |
hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 13 | DTNA | Sponge network | -2.039 | 0.03447 | -1.648 | 1.0E-5 | 0.466 |
| 11661 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 45 | CHD5 | Sponge network | -0.129 | 0.57177 | -0.922 | 0.01521 | 0.466 |
| 11662 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 49 | ZFHX3 | Sponge network | -1.697 | 0.00889 | 0.389 | 0.01363 | 0.466 |
| 11663 | OIP5-AS1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | ERCC6 | Sponge network | 0.421 | 0.00084 | 0.23 | 0.015 | 0.466 |
| 11664 | RP11-815I9.4 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-664a-3p | 11 | RASAL2 | Sponge network | 0.537 | 0.0006 | 0.869 | 0 | 0.466 |
| 11665 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | LRIG1 | Sponge network | -0.793 | 7.0E-5 | -0.537 | 0.00588 | 0.466 |
| 11666 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 19 | MYO9A | Sponge network | 0.765 | 7.0E-5 | -0.147 | 0.32373 | 0.466 |
| 11667 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 17 | CAV1 | Sponge network | -0.323 | 0.01372 | -1.013 | 6.0E-5 | 0.466 |
| 11668 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | FBXO32 | Sponge network | -0.021 | 0.93598 | -0.495 | 0.07561 | 0.466 |
| 11669 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-5p | 29 | ERC1 | Sponge network | 0.335 | 0.20596 | -0.108 | 0.38794 | 0.466 |
| 11670 | RP11-57H14.4 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-590-3p | 10 | RBM6 | Sponge network | 0.083 | 0.66405 | -0.137 | 0.15663 | 0.466 |
| 11671 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-505-3p | 18 | PTPN11 | Sponge network | 0.614 | 1.0E-5 | 0.431 | 2.0E-5 | 0.466 |
| 11672 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 23 | RARB | Sponge network | -0.49 | 0.04946 | -0.825 | 2.0E-5 | 0.466 |
| 11673 | BVES-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-550a-3p | 13 | PAK3 | Sponge network | -2.62 | 0 | -0.628 | 0.07253 | 0.466 |
| 11674 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | LHFP | Sponge network | -0.612 | 0.00094 | -0.167 | 0.41371 | 0.466 |
| 11675 | RP11-400K9.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-223-5p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 12 | DTNA | Sponge network | -0.733 | 0.19469 | -1.648 | 1.0E-5 | 0.466 |
| 11676 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | HOOK3 | Sponge network | 0.174 | 0.30034 | 0.273 | 0.09957 | 0.466 |
| 11677 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 41 | EPHA5 | Sponge network | 0.082 | 0.55654 | -0.957 | 0.01733 | 0.466 |
| 11678 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-452-5p | 18 | TIMP3 | Sponge network | -2.62 | 0 | -0.546 | 0.02307 | 0.466 |
| 11679 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | ZFX | Sponge network | 0.421 | 0.00084 | 0.09 | 0.42563 | 0.466 |
| 11680 | RP11-307B6.3 |
hsa-let-7g-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-421 | 13 | MEF2C | Sponge network | -4.463 | 0.00025 | -0.499 | 0.01448 | 0.466 |
| 11681 | AC083843.1 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-582-3p | 19 | ABL2 | Sponge network | 1.4 | 0 | 0.82 | 0 | 0.466 |
| 11682 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PHC3 | Sponge network | 0.673 | 0 | 0.212 | 0.0499 | 0.466 |
| 11683 | AC005562.1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | DNMT3A | Sponge network | 0.488 | 0.00084 | 0.302 | 0.0262 | 0.466 |
| 11684 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | SOX5 | Sponge network | -1.161 | 0.00591 | -1.091 | 4.0E-5 | 0.466 |
| 11685 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-5p | 12 | MN1 | Sponge network | 0.101 | 0.60919 | -0.358 | 0.24172 | 0.466 |
| 11686 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-532-3p | 27 | CBL | Sponge network | 0.594 | 0.41522 | 0.291 | 0.01267 | 0.466 |
| 11687 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 22 | TGFBR2 | Sponge network | -0.129 | 0.57177 | -0.243 | 0.11408 | 0.466 |
| 11688 | RP11-48B3.4 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | MED12L | Sponge network | 1.757 | 0 | -0.194 | 0.55299 | 0.466 |
| 11689 | CTD-2528L19.6 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PHF3 | Sponge network | 0.953 | 0 | 0.126 | 0.19652 | 0.466 |
| 11690 | RP11-359B12.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 16 | CSDE1 | Sponge network | 0.064 | 0.72934 | -0.493 | 0 | 0.466 |
| 11691 | RP11-53O19.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-708-5p | 10 | EZH1 | Sponge network | -0.405 | 0.22013 | -0.564 | 1.0E-5 | 0.466 |
| 11692 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | TGFBR3 | Sponge network | 0.572 | 0.01722 | -1.173 | 1.0E-5 | 0.466 |
| 11693 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p | 14 | NRXN3 | Sponge network | 0.082 | 0.55397 | -0.893 | 0.04186 | 0.466 |
| 11694 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 15 | SEPT6 | Sponge network | -0.129 | 0.57177 | -0.598 | 0.00181 | 0.466 |
| 11695 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-424-3p;hsa-miR-493-5p;hsa-miR-96-5p | 11 | HOOK3 | Sponge network | 0.614 | 1.0E-5 | 0.273 | 0.09957 | 0.466 |
| 11696 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 25 | LRRC55 | Sponge network | 0.249 | 0.35902 | -0.026 | 0.93163 | 0.466 |
| 11697 | BVES-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-877-5p | 43 | GAS7 | Sponge network | -2.62 | 0 | -0.555 | 0.01602 | 0.466 |
| 11698 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-96-5p | 81 | LPP | Sponge network | 0.311 | 0.10344 | -0.459 | 0.04475 | 0.466 |
| 11699 | TP73-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 35 | IGFBP5 | Sponge network | -0.563 | 0.02545 | -0.806 | 0.00105 | 0.466 |
| 11700 | RP11-53O19.1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | -0.405 | 0.22013 | -0.366 | 0.01036 | 0.466 |
| 11701 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-409-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 14 | RCAN2 | Sponge network | 0.177 | 0.16681 | -0.33 | 0.15172 | 0.466 |
| 11702 | AC005154.6 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FBXO11 | Sponge network | 0.848 | 0 | 0.142 | 0.04938 | 0.466 |
| 11703 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p | 15 | MAFB | Sponge network | -0.976 | 0.00025 | -0.023 | 0.89865 | 0.466 |
| 11704 | GAS6-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-664a-3p | 18 | SYNPO2 | Sponge network | 0.16 | 0.50785 | -2.342 | 0 | 0.466 |
| 11705 | RP11-57H14.4 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 11 | ZNF638 | Sponge network | 0.083 | 0.66405 | 0.22 | 0.01214 | 0.466 |
| 11706 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 25 | PHC3 | Sponge network | 1.254 | 0 | 0.212 | 0.0499 | 0.466 |
| 11707 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p | 11 | GAS1 | Sponge network | -1.575 | 6.0E-5 | -1.047 | 0.00364 | 0.466 |
| 11708 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | FLI1 | Sponge network | -0.976 | 0.00025 | -0.294 | 0.10905 | 0.466 |
| 11709 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 30 | THRB | Sponge network | -0.932 | 0.00329 | -1.117 | 0.0004 | 0.466 |
| 11710 | RP11-464F9.1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 28 | CCDC6 | Sponge network | 0.959 | 0 | 0.27 | 0.02555 | 0.466 |
| 11711 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p | 24 | SYNE1 | Sponge network | -0.738 | 0.21233 | -0.738 | 0.00316 | 0.466 |
| 11712 | FAM66C |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | LRP1B | Sponge network | -0.901 | 0.00019 | -2.163 | 0 | 0.466 |
| 11713 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | TACC1 | Sponge network | 0.064 | 0.72934 | -0.362 | 0.05406 | 0.466 |
| 11714 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-625-3p | 22 | TMTC3 | Sponge network | 1.153 | 0 | 0.123 | 0.22189 | 0.466 |
| 11715 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940 | 28 | CRTC3 | Sponge network | -1.697 | 0.00889 | 0.164 | 0.16786 | 0.466 |
| 11716 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-7-1-3p | 19 | PLSCR4 | Sponge network | 0.395 | 0.15451 | -0.474 | 0.03833 | 0.466 |
| 11717 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 26 | NRK | Sponge network | -2.46 | 0 | 0.656 | 0.19217 | 0.466 |
| 11718 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | MLLT10 | Sponge network | 0.222 | 0.29133 | 0.105 | 0.33292 | 0.466 |
| 11719 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p | 10 | MGA | Sponge network | 0.75 | 0.00054 | 0.323 | 0.00792 | 0.466 |
| 11720 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p | 12 | CADM3 | Sponge network | -0.517 | 0.02935 | -3.663 | 0 | 0.466 |
| 11721 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-625-3p | 28 | BMPR2 | Sponge network | 0.497 | 0.01451 | 0.206 | 0.0635 | 0.466 |
| 11722 | RP11-359E10.1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | MYO9A | Sponge network | 0.299 | 0.57722 | -0.147 | 0.32373 | 0.466 |
| 11723 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | IGSF10 | Sponge network | -0.737 | 0.06182 | -1.751 | 1.0E-5 | 0.466 |
| 11724 | ADAMTS9-AS2 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | PCDH18 | Sponge network | -2.452 | 0 | 0.216 | 0.24645 | 0.466 |
| 11725 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 57 | IL6ST | Sponge network | -0.563 | 0.02545 | -0.402 | 0.00676 | 0.466 |
| 11726 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-7-1-3p | 13 | CNTN1 | Sponge network | -0.726 | 0.00132 | -2.594 | 0 | 0.466 |
| 11727 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 11 | DOCK4 | Sponge network | 0.542 | 0.00054 | 0.502 | 0.00108 | 0.466 |
| 11728 | AC006116.24 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-577;hsa-miR-98-5p | 23 | CREB3L2 | Sponge network | -0.473 | 0.07602 | -0.525 | 2.0E-5 | 0.466 |
| 11729 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-532-5p;hsa-miR-93-5p | 30 | BNC2 | Sponge network | 0.082 | 0.55397 | -0.491 | 0.09988 | 0.466 |
| 11730 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p | 11 | ZNF208 | Sponge network | -0.693 | 0.05629 | -0.4 | 0.22381 | 0.466 |
| 11731 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PDE4DIP | Sponge network | -1.057 | 0.00029 | -0.282 | 0.0257 | 0.466 |
| 11732 | HOXB-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-589-3p | 23 | IL17RD | Sponge network | -0.154 | 0.53954 | -0.019 | 0.94873 | 0.466 |
| 11733 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | NIN | Sponge network | 0.101 | 0.60919 | -0.253 | 0.13794 | 0.466 |
| 11734 | WDFY3-AS2 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-935;hsa-miR-940 | 49 | GRIK3 | Sponge network | -0.942 | 0 | -3.208 | 0 | 0.466 |
| 11735 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-1301-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | ZNF770 | Sponge network | 1.254 | 0 | 0.206 | 0.06751 | 0.466 |
| 11736 | RP11-110I1.11 |
hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 11 | ABL2 | Sponge network | 1.034 | 0 | 0.82 | 0 | 0.466 |
| 11737 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-493-5p;hsa-miR-577 | 10 | AFF3 | Sponge network | -0.31 | 0.33871 | -2.373 | 0 | 0.466 |
| 11738 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 27 | SCN9A | Sponge network | -0.777 | 0.1134 | -0.525 | 0.10593 | 0.466 |
| 11739 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 40 | PDGFRA | Sponge network | -0.901 | 0.00019 | -0.372 | 0.0037 | 0.466 |
| 11740 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 14 | PSIP1 | Sponge network | -0.612 | 0.00094 | -0.005 | 0.96897 | 0.466 |
| 11741 | RP11-1112J20.2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-486-5p;hsa-miR-664a-5p | 11 | CBFB | Sponge network | 1.416 | 0 | 1.061 | 0 | 0.465 |
| 11742 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-5p | 11 | LATS1 | Sponge network | 0.706 | 0 | 0.109 | 0.34208 | 0.465 |
| 11743 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | CNTN1 | Sponge network | -0.187 | 0.23787 | -2.594 | 0 | 0.465 |
| 11744 | RP11-693J15.4 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | PTPRT | Sponge network | -0.72 | 0.24239 | -0.907 | 0.03727 | 0.465 |
| 11745 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p | 12 | PDZD2 | Sponge network | -0.563 | 0.02545 | -0.909 | 3.0E-5 | 0.465 |
| 11746 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 68 | CADM2 | Sponge network | -1.216 | 0 | -3.782 | 0 | 0.465 |
| 11747 | RP1-151F17.2 |
hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | MACF1 | Sponge network | 0.222 | 0.29133 | 0.019 | 0.90335 | 0.465 |
| 11748 | ZRANB2-AS1 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | PRKCB | Sponge network | -0.376 | 0.00337 | -1.313 | 2.0E-5 | 0.465 |
| 11749 | RP11-589P10.7 |
hsa-miR-1296-5p;hsa-miR-146b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-324-3p;hsa-miR-331-3p | 11 | ZBTB4 | Sponge network | -2.044 | 0.00066 | -0.739 | 0 | 0.465 |
| 11750 | AC006116.24 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-550a-3p | 19 | DIP2C | Sponge network | -0.473 | 0.07602 | -0.436 | 0.01713 | 0.465 |
| 11751 | LINC00957 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 26 | ESR1 | Sponge network | -0.421 | 0.0114 | -1.035 | 3.0E-5 | 0.465 |
| 11752 | MEG3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429 | 16 | STARD13 | Sponge network | 0.249 | 0.35902 | -0.187 | 0.27383 | 0.465 |
| 11753 | AC009948.5 |
hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 10 | HERC1 | Sponge network | 0.532 | 0.00505 | -0.196 | 0.08544 | 0.465 |
| 11754 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 24 | PIK3C2A | Sponge network | 1.153 | 0 | 0.061 | 0.53776 | 0.465 |
| 11755 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 32 | TET1 | Sponge network | -0.739 | 0.0574 | -0.135 | 0.52138 | 0.465 |
| 11756 | MEG3 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | TLL1 | Sponge network | 0.249 | 0.35902 | -1.195 | 3.0E-5 | 0.465 |
| 11757 | MIR143HG |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 47 | AFF1 | Sponge network | -0.739 | 0.0574 | -0.384 | 0.00053 | 0.465 |
| 11758 | MAGI2-AS3 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | CSDE1 | Sponge network | -0.129 | 0.57177 | -0.493 | 0 | 0.465 |
| 11759 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-424-5p | 13 | DYNC1I1 | Sponge network | -0.318 | 0.65847 | -1.12 | 0.00107 | 0.465 |
| 11760 | LINC00472 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 21 | PCDH10 | Sponge network | -0.842 | 0.01361 | -1.644 | 0.0078 | 0.465 |
| 11761 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-429 | 17 | MAF | Sponge network | -2.62 | 0 | -1.257 | 0 | 0.465 |
| 11762 | AC010226.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-590-3p | 28 | DTNA | Sponge network | -0.811 | 2.0E-5 | -1.648 | 1.0E-5 | 0.465 |
| 11763 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | AR | Sponge network | -0.737 | 0.10822 | -1.554 | 0 | 0.465 |
| 11764 | FENDRR |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 12 | ZNF382 | Sponge network | -1.281 | 0.00012 | -0.494 | 0.03831 | 0.465 |
| 11765 | RP11-736K20.5 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-335-5p | 12 | PLXNC1 | Sponge network | -0.358 | 0.17255 | 0.569 | 0.00329 | 0.465 |
| 11766 | RP5-1061H20.4 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 10 | TBL1XR1 | Sponge network | 1.921 | 0 | 0.578 | 0 | 0.465 |
| 11767 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-539-5p | 13 | ABI2 | Sponge network | -0.309 | 0.1145 | 0.175 | 0.14787 | 0.465 |
| 11768 | RP11-254F7.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-486-5p | 14 | NRK | Sponge network | 0.176 | 0.37402 | 0.656 | 0.19217 | 0.465 |
| 11769 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | PHC3 | Sponge network | 0.683 | 1.0E-5 | 0.212 | 0.0499 | 0.465 |
| 11770 | RP11-999E24.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-744-3p | 38 | DST | Sponge network | -0.693 | 0.05629 | -0.713 | 0.00096 | 0.465 |
| 11771 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 26 | PCDH10 | Sponge network | -0.942 | 0 | -1.644 | 0.0078 | 0.465 |
| 11772 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | 0.72 | 0.0001 | 0.126 | 0.15266 | 0.465 |
| 11773 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-484;hsa-miR-590-3p | 14 | PKNOX1 | Sponge network | 0.083 | 0.66405 | 0.085 | 0.28917 | 0.465 |
| 11774 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-5p | 18 | PTGFR | Sponge network | -1.654 | 0.00144 | -0.233 | 0.45421 | 0.465 |
| 11775 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PTGFR | Sponge network | 0.225 | 0.30644 | -0.233 | 0.45421 | 0.465 |
| 11776 | RP11-59H7.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 22 | MDM4 | Sponge network | 2.163 | 0 | 0.345 | 0.00762 | 0.465 |
| 11777 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429 | 10 | PTPN13 | Sponge network | -0.246 | 0.35034 | -1.208 | 0 | 0.465 |
| 11778 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | EDA2R | Sponge network | -0.769 | 0.00035 | -0.587 | 0.01598 | 0.465 |
| 11779 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PRDM11 | Sponge network | 0.374 | 0.20054 | -0.252 | 0.15511 | 0.465 |
| 11780 | RP11-175O19.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-409-5p;hsa-miR-497-5p;hsa-miR-664a-3p | 28 | CCDC6 | Sponge network | 0.917 | 0 | 0.27 | 0.02555 | 0.465 |
| 11781 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-5p | 15 | PTGFR | Sponge network | 0.299 | 0.57722 | -0.233 | 0.45421 | 0.465 |
| 11782 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-590-3p | 15 | TSHZ3 | Sponge network | -1.886 | 0 | -0.556 | 0.01516 | 0.465 |
| 11783 | LINC01018 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -2.915 | 0.00015 | -2.417 | 0 | 0.465 |
| 11784 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p | 23 | TIMP3 | Sponge network | 0.214 | 0.18246 | -0.546 | 0.02307 | 0.465 |
| 11785 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | MYO9A | Sponge network | -0.355 | 0.0077 | -0.147 | 0.32373 | 0.465 |
| 11786 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 22 | IGF1 | Sponge network | 0.199 | 0.38015 | -0.204 | 0.5898 | 0.465 |
| 11787 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | -0.154 | 0.78939 | -1.751 | 1.0E-5 | 0.465 |
| 11788 | AC003090.1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | ZDBF2 | Sponge network | -2.117 | 0.00161 | -0.998 | 0.00025 | 0.465 |
| 11789 | RP11-307B6.3 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 10 | NRK | Sponge network | -4.463 | 0.00025 | 0.656 | 0.19217 | 0.465 |
| 11790 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-576-5p | 11 | BNC2 | Sponge network | 1.122 | 0.02989 | -0.491 | 0.09988 | 0.465 |
| 11791 | RP11-13K12.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p | 25 | NAV3 | Sponge network | -0.154 | 0.78939 | 0.212 | 0.47729 | 0.465 |
| 11792 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 14 | FYN | Sponge network | -0.49 | 0.04946 | -0.131 | 0.37051 | 0.465 |
| 11793 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ERG | Sponge network | -0.513 | 0.04703 | 0.002 | 0.99011 | 0.465 |
| 11794 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p | 13 | CRTC3 | Sponge network | -0.739 | 0.00044 | 0.164 | 0.16786 | 0.465 |
| 11795 | RP11-400K9.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-378a-5p | 21 | ASTN1 | Sponge network | -0.611 | 0.09607 | -2.389 | 2.0E-5 | 0.465 |
| 11796 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-93-5p | 23 | ZBTB4 | Sponge network | -0.465 | 0.04703 | -0.739 | 0 | 0.465 |
| 11797 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | SYNPO2 | Sponge network | -0.738 | 0.21233 | -2.342 | 0 | 0.465 |
| 11798 | RP11-967K21.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 35 | IL17RD | Sponge network | 0.056 | 0.81195 | -0.019 | 0.94873 | 0.465 |
| 11799 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-505-3p | 16 | SON | Sponge network | 0.637 | 0.00069 | 0.168 | 0.04406 | 0.465 |
| 11800 | RP11-254F7.2 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 11 | FLNA | Sponge network | 0.176 | 0.37402 | -0.771 | 0.01377 | 0.465 |
| 11801 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 42 | EPHA7 | Sponge network | -0.769 | 0.00035 | -2.197 | 3.0E-5 | 0.465 |
| 11802 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 56 | FOXP1 | Sponge network | -0.739 | 0.0574 | 0.042 | 0.74368 | 0.465 |
| 11803 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | MOB1B | Sponge network | 0.683 | 1.0E-5 | 0.197 | 0.15266 | 0.465 |
| 11804 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 34 | ST6GALNAC3 | Sponge network | -0.901 | 0.00019 | -0.987 | 0 | 0.465 |
| 11805 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 19 | PTGFR | Sponge network | -0.487 | 0.02157 | -0.233 | 0.45421 | 0.465 |
| 11806 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | PCDH9 | Sponge network | -0.513 | 0.04703 | -1.823 | 4.0E-5 | 0.465 |
| 11807 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 17 | LHX6 | Sponge network | -0.49 | 0.04946 | -0.087 | 0.69193 | 0.465 |
| 11808 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-93-3p | 21 | EPHA3 | Sponge network | -0.465 | 0.04703 | 0.185 | 0.53588 | 0.465 |
| 11809 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 21 | KCNA6 | Sponge network | -1.057 | 0.00029 | -1.531 | 0.00013 | 0.465 |
| 11810 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 48 | AR | Sponge network | -0.727 | 0.04726 | -1.554 | 0 | 0.465 |
| 11811 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 67 | AFF4 | Sponge network | 0.101 | 0.60919 | -0.266 | 0.01565 | 0.465 |
| 11812 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | SEMA3C | Sponge network | -1.092 | 4.0E-5 | -0.272 | 0.31153 | 0.465 |
| 11813 | ADAMTS9-AS2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p | 12 | PIK3IP1 | Sponge network | -2.452 | 0 | -0.187 | 0.19945 | 0.465 |
| 11814 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-590-3p | 10 | PTPN13 | Sponge network | 0.194 | 0.64278 | -1.208 | 0 | 0.465 |
| 11815 | LINC00473 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-935 | 12 | PHOX2B | Sponge network | -1.203 | 0.03881 | -2.68 | 0 | 0.465 |
| 11816 | CTD-2002H8.2 |
hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MLLT10 | Sponge network | 0.931 | 1.0E-5 | 0.105 | 0.33292 | 0.465 |
| 11817 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | SCN9A | Sponge network | 0.214 | 0.18246 | -0.525 | 0.10593 | 0.465 |
| 11818 | SNHG14 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 35 | PDE4DIP | Sponge network | -1.111 | 0.0003 | -0.282 | 0.0257 | 0.465 |
| 11819 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-577;hsa-miR-590-3p | 12 | AKAP12 | Sponge network | -0.227 | 0.31657 | -0.932 | 0.0028 | 0.465 |
| 11820 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-942-5p | 17 | NOTCH2 | Sponge network | 0.542 | 0.00054 | 0.138 | 0.35163 | 0.465 |
| 11821 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 47 | PPM1A | Sponge network | -0.615 | 0.00015 | -0.623 | 0 | 0.465 |
| 11822 | MIR143HG |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 16 | SEPT6 | Sponge network | -0.739 | 0.0574 | -0.598 | 0.00181 | 0.465 |
| 11823 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | RCAN2 | Sponge network | -1.582 | 8.0E-5 | -0.33 | 0.15172 | 0.465 |
| 11824 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 24 | PRDM2 | Sponge network | -0.487 | 0.02157 | -0.258 | 0.00721 | 0.465 |
| 11825 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p | 12 | EDA2R | Sponge network | -0.487 | 0.02157 | -0.587 | 0.01598 | 0.465 |
| 11826 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 32 | MYO9A | Sponge network | -0.976 | 0.00025 | -0.147 | 0.32373 | 0.465 |
| 11827 | MRVI1-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-942-5p | 13 | GLIPR1 | Sponge network | -1.092 | 4.0E-5 | 0.146 | 0.3276 | 0.465 |
| 11828 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935 | 45 | SSBP2 | Sponge network | -1.203 | 0.03881 | -1.58 | 0 | 0.465 |
| 11829 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 13 | HERC2 | Sponge network | 0.72 | 0.0001 | 0.114 | 0.30638 | 0.465 |
| 11830 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 25 | SNX29 | Sponge network | -0.896 | 0.00099 | -0.34 | 0.01371 | 0.465 |
| 11831 | LINC00092 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-940 | 11 | POU2F2 | Sponge network | -1.886 | 0 | -0.233 | 0.32214 | 0.465 |
| 11832 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | CLASP2 | Sponge network | 0.064 | 0.72934 | -0.038 | 0.71 | 0.465 |
| 11833 | PCA3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-5p | 10 | THSD7A | Sponge network | -0.709 | 0.18168 | 0.242 | 0.25154 | 0.465 |
| 11834 | BVES-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-429 | 11 | ABCA10 | Sponge network | -2.62 | 0 | -0.571 | 0.03674 | 0.464 |
| 11835 | LINC00476 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p | 57 | DTNA | Sponge network | -0.615 | 0.00015 | -1.648 | 1.0E-5 | 0.464 |
| 11836 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | HDAC9 | Sponge network | -0.769 | 0.00035 | -0.755 | 0.00495 | 0.464 |
| 11837 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-485-3p;hsa-miR-590-3p | 10 | LRRK2 | Sponge network | -0.096 | 0.67958 | -1.136 | 1.0E-5 | 0.464 |
| 11838 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | NTRK3 | Sponge network | -1.654 | 0.00144 | -1.77 | 3.0E-5 | 0.464 |
| 11839 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-655-3p | 22 | LATS1 | Sponge network | 0.614 | 1.0E-5 | 0.109 | 0.34208 | 0.464 |
| 11840 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-589-5p;hsa-miR-874-3p | 10 | ST5 | Sponge network | -1.439 | 4.0E-5 | -0.78 | 0 | 0.464 |
| 11841 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 58 | BMPR2 | Sponge network | -0.487 | 0.02157 | 0.206 | 0.0635 | 0.464 |
| 11842 | SH3RF3-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ITGB1 | Sponge network | 0.889 | 9.0E-5 | 0.524 | 0.00017 | 0.464 |
| 11843 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 23 | RSPO2 | Sponge network | -1.057 | 0.00029 | -3.038 | 0 | 0.464 |
| 11844 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p | 13 | PTPN11 | Sponge network | 0.873 | 0 | 0.431 | 2.0E-5 | 0.464 |
| 11845 | FGD5-AS1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | TPR | Sponge network | 0.401 | 0.00066 | 0.582 | 0 | 0.464 |
| 11846 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 60 | QKI | Sponge network | -2.46 | 0 | -0.333 | 0.04111 | 0.464 |
| 11847 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 65 | BMPR2 | Sponge network | 1.302 | 7.0E-5 | 0.206 | 0.0635 | 0.464 |
| 11848 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | -0.49 | 0.04946 | -3.143 | 0 | 0.464 |
| 11849 | LINC00843 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-942-5p | 21 | ARHGEF12 | Sponge network | 1.106 | 0 | 0.09 | 0.35693 | 0.464 |
| 11850 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-7-1-3p | 22 | DTNA | Sponge network | -0.187 | 0.23787 | -1.648 | 1.0E-5 | 0.464 |
| 11851 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-629-5p | 23 | IGF1 | Sponge network | -0.513 | 0.04703 | -0.204 | 0.5898 | 0.464 |
| 11852 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | EPHA7 | Sponge network | 0.101 | 0.60919 | -2.197 | 3.0E-5 | 0.464 |
| 11853 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 34 | MEF2C | Sponge network | -1.161 | 0.00591 | -0.499 | 0.01448 | 0.464 |
| 11854 | HCG11 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PIK3CA | Sponge network | 0.385 | 0.10067 | 0.195 | 0.06908 | 0.464 |
| 11855 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-940 | 12 | TRIM33 | Sponge network | 1.134 | 0 | 0.402 | 7.0E-5 | 0.464 |
| 11856 | NDUFA6-AS1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ITPKB | Sponge network | -0.355 | 0.0077 | -0.704 | 0.00106 | 0.464 |
| 11857 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-98-5p | 30 | TGFBR3 | Sponge network | 0.082 | 0.55397 | -1.173 | 1.0E-5 | 0.464 |
| 11858 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 23 | EPHA7 | Sponge network | 0.178 | 0.29106 | -2.197 | 3.0E-5 | 0.464 |
| 11859 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-942-5p | 44 | HLF | Sponge network | -0.942 | 0 | -2.443 | 0 | 0.464 |
| 11860 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 26 | MOB1B | Sponge network | 0.765 | 7.0E-5 | 0.197 | 0.15266 | 0.464 |
| 11861 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p | 25 | SNX29 | Sponge network | -0.727 | 0.04726 | -0.34 | 0.01371 | 0.464 |
| 11862 | EMX2OS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | -0.777 | 0.1134 | -0.564 | 1.0E-5 | 0.464 |
| 11863 | AGAP11 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-769-5p | 22 | SYNPO2 | Sponge network | -1.645 | 0 | -2.342 | 0 | 0.464 |
| 11864 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 18 | SLITRK6 | Sponge network | -2.117 | 0.00161 | -2.121 | 0 | 0.464 |
| 11865 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p | 30 | RICTOR | Sponge network | 0.953 | 0 | 0.388 | 0.00188 | 0.464 |
| 11866 | SNHG1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 15 | TBL1XR1 | Sponge network | 1.265 | 0 | 0.578 | 0 | 0.464 |
| 11867 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 29 | ZFHX3 | Sponge network | 0.541 | 0.00125 | 0.389 | 0.01363 | 0.464 |
| 11868 | CTC-444N24.11 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | DICER1 | Sponge network | 1.153 | 0 | 0.042 | 0.674 | 0.464 |
| 11869 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 10 | RCAN2 | Sponge network | -0.611 | 0.09607 | -0.33 | 0.15172 | 0.464 |
| 11870 | LINC00702 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 10 | EPB41L3 | Sponge network | -0.737 | 0.06182 | -1.193 | 0 | 0.464 |
| 11871 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-429 | 12 | DACT1 | Sponge network | -0.154 | 0.53954 | 0.783 | 0.00072 | 0.464 |
| 11872 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | PHC3 | Sponge network | -0.323 | 0.01372 | 0.212 | 0.0499 | 0.464 |
| 11873 | JAZF1-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 18 | SORCS1 | Sponge network | -0.769 | 0.00035 | -3.492 | 0 | 0.464 |
| 11874 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-629-3p;hsa-miR-671-5p | 14 | CDH23 | Sponge network | -0.469 | 0.08721 | -0.974 | 0.00034 | 0.464 |
| 11875 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484 | 13 | FLNA | Sponge network | 0.083 | 0.66405 | -0.771 | 0.01377 | 0.464 |
| 11876 | RP11-57H14.4 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p | 58 | DST | Sponge network | 0.083 | 0.66405 | -0.713 | 0.00096 | 0.464 |
| 11877 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 24 | LPP | Sponge network | 0.959 | 0 | -0.459 | 0.04475 | 0.464 |
| 11878 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-532-3p | 34 | PBX1 | Sponge network | -2.915 | 0.00015 | -1.206 | 0 | 0.464 |
| 11879 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 45 | BCL2 | Sponge network | -0.49 | 0.04946 | -0.899 | 0 | 0.464 |
| 11880 | CTD-3092A11.2 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | SIRT1 | Sponge network | 0.873 | 0 | 0.109 | 0.2593 | 0.464 |
| 11881 | LINC00472 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | CDH19 | Sponge network | -0.842 | 0.01361 | -3.434 | 0 | 0.464 |
| 11882 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 27 | TET1 | Sponge network | -0.576 | 0.10121 | -0.135 | 0.52138 | 0.464 |
| 11883 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 35 | RICTOR | Sponge network | 0.532 | 0.00505 | 0.388 | 0.00188 | 0.464 |
| 11884 | RP11-1006G14.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-625-5p;hsa-miR-744-3p | 20 | PTPN14 | Sponge network | 0.559 | 0.00026 | 0.144 | 0.34595 | 0.464 |
| 11885 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-590-3p | 13 | NAV3 | Sponge network | -1.886 | 0 | 0.212 | 0.47729 | 0.464 |
| 11886 | RP11-458D21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-375;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 25 | ABI2 | Sponge network | 0.663 | 0.00073 | 0.175 | 0.14787 | 0.464 |
| 11887 | CTD-2554C21.2 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-744-3p;hsa-miR-96-5p | 24 | MEF2C | Sponge network | -1.654 | 0.00144 | -0.499 | 0.01448 | 0.464 |
| 11888 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 19 | ZFHX3 | Sponge network | 0.594 | 0.00064 | 0.389 | 0.01363 | 0.464 |
| 11889 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 21 | LIFR | Sponge network | -1.582 | 8.0E-5 | -1.794 | 0 | 0.464 |
| 11890 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | TACC1 | Sponge network | 0.299 | 0.57722 | -0.362 | 0.05406 | 0.464 |
| 11891 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-671-5p;hsa-miR-93-5p | 40 | ADARB1 | Sponge network | -0.465 | 0.04703 | -0.335 | 0.06631 | 0.464 |
| 11892 | FAM66C |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 18 | RPS6KA6 | Sponge network | -0.901 | 0.00019 | -2.989 | 0 | 0.464 |
| 11893 | ACTA2-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 34 | IGFBP5 | Sponge network | 0.194 | 0.64278 | -0.806 | 0.00105 | 0.464 |
| 11894 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | JAZF1 | Sponge network | -2.117 | 0.00161 | -0.377 | 0.02677 | 0.464 |
| 11895 | EMX2OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-625-5p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -0.777 | 0.1134 | 0.024 | 0.89889 | 0.464 |
| 11896 | RP11-21A7A.2 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-505-5p;hsa-miR-877-5p | 11 | ASTN1 | Sponge network | -1.116 | 0.23686 | -2.389 | 2.0E-5 | 0.464 |
| 11897 | RP11-822E23.8 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-940 | 17 | SORCS1 | Sponge network | -0.777 | 0.16667 | -3.492 | 0 | 0.464 |
| 11898 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NUAK1 | Sponge network | -0.576 | 0.10121 | 0.904 | 0 | 0.464 |
| 11899 | AC016747.3 |
hsa-let-7a-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-92a-3p | 14 | ZNF521 | Sponge network | 0.199 | 0.38015 | -0.111 | 0.58068 | 0.464 |
| 11900 | ZRANB2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 14 | RYR2 | Sponge network | -0.376 | 0.00337 | -1.466 | 0.00031 | 0.464 |
| 11901 | U91328.19 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-7-1-3p | 48 | DTNA | Sponge network | -0.303 | 0.21002 | -1.648 | 1.0E-5 | 0.464 |
| 11902 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-486-5p;hsa-miR-576-5p | 10 | NALCN | Sponge network | -0.162 | 0.47687 | 1.068 | 0.00237 | 0.464 |
| 11903 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-539-5p | 27 | EPHA7 | Sponge network | 0.176 | 0.37402 | -2.197 | 3.0E-5 | 0.464 |
| 11904 | SH3RF3-AS1 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 17 | ANK2 | Sponge network | 0.889 | 9.0E-5 | -1.603 | 1.0E-5 | 0.464 |
| 11905 | RASSF8-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 31 | DIP2C | Sponge network | 0.335 | 0.20596 | -0.436 | 0.01713 | 0.464 |
| 11906 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | RET | Sponge network | -1.111 | 0.0003 | -0.833 | 0.03517 | 0.464 |
| 11907 | MIR497HG |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 52 | DST | Sponge network | -1.439 | 4.0E-5 | -0.713 | 0.00096 | 0.464 |
| 11908 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p | 26 | SCN9A | Sponge network | -1.249 | 7.0E-5 | -0.525 | 0.10593 | 0.464 |
| 11909 | LINC00092 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 29 | TGFBR3 | Sponge network | -1.886 | 0 | -1.173 | 1.0E-5 | 0.464 |
| 11910 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 32 | MXI1 | Sponge network | -2.46 | 0 | -0.987 | 0 | 0.464 |
| 11911 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-582-5p | 15 | STARD13 | Sponge network | -0.469 | 0.08721 | -0.187 | 0.27383 | 0.464 |
| 11912 | CTC-296K1.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-452-5p;hsa-miR-671-5p;hsa-miR-940 | 29 | GAS7 | Sponge network | -2.095 | 0.00112 | -0.555 | 0.01602 | 0.464 |
| 11913 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935 | 25 | FBXO32 | Sponge network | -0.376 | 0.00337 | -0.495 | 0.07561 | 0.464 |
| 11914 | AC074289.1 |
hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-96-5p | 10 | ELMO1 | Sponge network | -0.326 | 0.14314 | 0.04 | 0.83147 | 0.464 |
| 11915 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-582-5p | 27 | RICTOR | Sponge network | 0.817 | 3.0E-5 | 0.388 | 0.00188 | 0.464 |
| 11916 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TSHZ3 | Sponge network | -0.021 | 0.93598 | -0.556 | 0.01516 | 0.464 |
| 11917 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | SYNE1 | Sponge network | 1.153 | 0.00114 | -0.738 | 0.00316 | 0.464 |
| 11918 | MIR497HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-584-5p;hsa-miR-590-5p | 13 | ZNF208 | Sponge network | -1.439 | 4.0E-5 | -0.4 | 0.22381 | 0.464 |
| 11919 | RP11-166D19.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | FMNL3 | Sponge network | -0.576 | 0.10121 | 0.555 | 4.0E-5 | 0.464 |
| 11920 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | TRIM33 | Sponge network | 1.266 | 0 | 0.402 | 7.0E-5 | 0.464 |
| 11921 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 35 | PBX1 | Sponge network | -0.777 | 0.1134 | -1.206 | 0 | 0.464 |
| 11922 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 17 | MLLT10 | Sponge network | 0.541 | 0.00125 | 0.105 | 0.33292 | 0.464 |
| 11923 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p | 36 | RBMS3 | Sponge network | 0.199 | 0.38015 | -0.519 | 0.03551 | 0.464 |
| 11924 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-429;hsa-miR-576-5p | 21 | ST6GAL2 | Sponge network | -2.62 | 0 | -1.201 | 0.00597 | 0.464 |
| 11925 | RP11-156P1.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 20 | NRK | Sponge network | 0.214 | 0.18246 | 0.656 | 0.19217 | 0.464 |
| 11926 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 14 | RCAN2 | Sponge network | -0.358 | 0.17255 | -0.33 | 0.15172 | 0.464 |
| 11927 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-502-5p | 19 | TCF4 | Sponge network | 0.598 | 0.38481 | 0.016 | 0.92271 | 0.464 |
| 11928 | HHIP-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | DTNA | Sponge network | -0.737 | 0.10822 | -1.648 | 1.0E-5 | 0.464 |
| 11929 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 13 | EBF1 | Sponge network | 1.153 | 0.00114 | -0.384 | 0.08856 | 0.464 |
| 11930 | RP11-46C24.7 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-766-3p | 32 | IL17RD | Sponge network | 0.392 | 0.0278 | -0.019 | 0.94873 | 0.464 |
| 11931 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-935 | 31 | TSHZ3 | Sponge network | -1.249 | 7.0E-5 | -0.556 | 0.01516 | 0.464 |
| 11932 | RP11-13K12.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-629-3p | 28 | ASTN1 | Sponge network | -0.154 | 0.78939 | -2.389 | 2.0E-5 | 0.464 |
| 11933 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ZMYM2 | Sponge network | 0.225 | 0.30644 | 0.279 | 0.01273 | 0.464 |
| 11934 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-940 | 31 | IGF1 | Sponge network | -0.162 | 0.47687 | -0.204 | 0.5898 | 0.464 |
| 11935 | TP73-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | NAV3 | Sponge network | -0.563 | 0.02545 | 0.212 | 0.47729 | 0.464 |
| 11936 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p | 16 | LHX6 | Sponge network | -0.576 | 0.10121 | -0.087 | 0.69193 | 0.464 |
| 11937 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p | 12 | PDZD2 | Sponge network | 0.194 | 0.64278 | -0.909 | 3.0E-5 | 0.464 |
| 11938 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | BCLAF1 | Sponge network | 1.4 | 0 | 0.211 | 0.00684 | 0.464 |
| 11939 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | FYN | Sponge network | -2.452 | 0 | -0.131 | 0.37051 | 0.464 |
| 11940 | DNM3OS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | 0.572 | 0.01722 | -0.564 | 1.0E-5 | 0.464 |
| 11941 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | MEF2C | Sponge network | 1.253 | 0 | -0.499 | 0.01448 | 0.464 |
| 11942 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-589-5p;hsa-miR-93-5p;hsa-miR-98-5p | 50 | PRKAA2 | Sponge network | -0.154 | 0.78939 | -2.462 | 0 | 0.464 |
| 11943 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 23 | ZMYND11 | Sponge network | 0.72 | 0.0001 | -0.2 | 0.05449 | 0.464 |
| 11944 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | NRXN3 | Sponge network | -0.355 | 0.0077 | -0.893 | 0.04186 | 0.464 |
| 11945 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-624-5p | 11 | HMCN1 | Sponge network | 0.225 | 0.30644 | 0.809 | 0.00544 | 0.464 |
| 11946 | AGAP11 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-576-5p | 47 | DTNA | Sponge network | -1.645 | 0 | -1.648 | 1.0E-5 | 0.463 |
| 11947 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | EDA2R | Sponge network | -0.777 | 0.1134 | -0.587 | 0.01598 | 0.463 |
| 11948 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-766-3p | 20 | TBL1XR1 | Sponge network | 1.206 | 0 | 0.578 | 0 | 0.463 |
| 11949 | RP11-25K19.1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | TLE4 | Sponge network | -1.057 | 0.00029 | -1.157 | 0 | 0.463 |
| 11950 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 52 | TMEM132B | Sponge network | -1.111 | 0.0003 | -0.83 | 0.01084 | 0.463 |
| 11951 | RP11-384K6.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-940 | 24 | CREB1 | Sponge network | 0.946 | 0 | 0.203 | 0.00537 | 0.463 |
| 11952 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-744-3p | 19 | HLF | Sponge network | 0.178 | 0.29106 | -2.443 | 0 | 0.463 |
| 11953 | RP11-819C21.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | BCL9 | Sponge network | 0.537 | 0.00139 | 0.321 | 0.00267 | 0.463 |
| 11954 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-577;hsa-miR-744-3p;hsa-miR-96-5p | 20 | HIP1 | Sponge network | 0.853 | 0.15352 | 0.774 | 0 | 0.463 |
| 11955 | RP11-736K20.5 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-629-3p | 16 | RYR2 | Sponge network | -0.358 | 0.17255 | -1.466 | 0.00031 | 0.463 |
| 11956 | PART1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 47 | DIP2C | Sponge network | -2.925 | 0 | -0.436 | 0.01713 | 0.463 |
| 11957 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-340-5p;hsa-miR-495-3p;hsa-miR-940 | 29 | HLF | Sponge network | 0.331 | 0.57247 | -2.443 | 0 | 0.463 |
| 11958 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 53 | THRB | Sponge network | -1.161 | 0.00591 | -1.117 | 0.0004 | 0.463 |
| 11959 | HNRNPU-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PHF3 | Sponge network | 1.601 | 0 | 0.126 | 0.19652 | 0.463 |
| 11960 | CTC-378H22.2 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-502-5p | 12 | TCF4 | Sponge network | 0.001 | 0.99663 | 0.016 | 0.92271 | 0.463 |
| 11961 | RP4-639F20.1 |
hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-629-5p;hsa-miR-96-5p | 18 | MYO9A | Sponge network | -0.517 | 0.02935 | -0.147 | 0.32373 | 0.463 |
| 11962 | LINC00865 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-940 | 14 | GLI3 | Sponge network | -1.249 | 7.0E-5 | -0.615 | 0.01482 | 0.463 |
| 11963 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p | 15 | RET | Sponge network | -0.358 | 0.17255 | -0.833 | 0.03517 | 0.463 |
| 11964 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-660-5p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | ZHX2 | Sponge network | -0.563 | 0.02545 | -0.192 | 0.18459 | 0.463 |
| 11965 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | EPHA7 | Sponge network | 0.335 | 0.20596 | -2.197 | 3.0E-5 | 0.463 |
| 11966 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | MAF | Sponge network | -0.976 | 0.00025 | -1.257 | 0 | 0.463 |
| 11967 | AC006116.24 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-93-5p | 11 | LATS2 | Sponge network | -0.473 | 0.07602 | 0.159 | 0.2304 | 0.463 |
| 11968 | BCYRN1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 19 | KIF5B | Sponge network | 0.311 | 0.10344 | 0.362 | 0.00066 | 0.463 |
| 11969 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | 0.225 | 0.30644 | -1.548 | 0 | 0.463 |
| 11970 | H19 |
hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-664a-3p | 10 | THBS1 | Sponge network | 2.439 | 0 | 0.741 | 0.00386 | 0.463 |
| 11971 | LINC00641 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ABCA10 | Sponge network | -0.222 | 0.32445 | -0.571 | 0.03674 | 0.463 |
| 11972 | LINC00476 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | RNF180 | Sponge network | -0.615 | 0.00015 | -1.127 | 1.0E-5 | 0.463 |
| 11973 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-625-5p | 11 | LHX6 | Sponge network | -2.095 | 0.00112 | -0.087 | 0.69193 | 0.463 |
| 11974 | RP11-326G21.1 |
hsa-let-7b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | KCNA6 | Sponge network | -0.021 | 0.93598 | -1.531 | 0.00013 | 0.463 |
| 11975 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p | 13 | RAP1A | Sponge network | -0.932 | 0.00329 | -0.929 | 0 | 0.463 |
| 11976 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-93-5p | 19 | ARNT2 | Sponge network | -0.842 | 0.01361 | -0.332 | 0.25851 | 0.463 |
| 11977 | LINC00672 |
hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | STAT5B | Sponge network | -0.227 | 0.31657 | -0.182 | 0.1082 | 0.463 |
| 11978 | SNHG14 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 13 | ALDH1A2 | Sponge network | -1.111 | 0.0003 | -2.411 | 0 | 0.463 |
| 11979 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | ABCC9 | Sponge network | -0.942 | 0 | -0.466 | 0.14121 | 0.463 |
| 11980 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PCDH10 | Sponge network | -1.161 | 0.00591 | -1.644 | 0.0078 | 0.463 |
| 11981 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CDH19 | Sponge network | -1.057 | 0.00029 | -3.434 | 0 | 0.463 |
| 11982 | AC074289.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-539-5p | 17 | NIN | Sponge network | -0.326 | 0.14314 | -0.253 | 0.13794 | 0.463 |
| 11983 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-96-5p | 16 | ERG | Sponge network | -1.654 | 0.00144 | 0.002 | 0.99011 | 0.463 |
| 11984 | CKMT2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-93-5p;hsa-miR-96-5p | 76 | LPP | Sponge network | 0.082 | 0.55654 | -0.459 | 0.04475 | 0.463 |
| 11985 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p | 19 | SLC6A15 | Sponge network | -1.645 | 0 | -2.373 | 2.0E-5 | 0.463 |
| 11986 | RP11-384O8.1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-935 | 10 | STAT5B | Sponge network | -0.738 | 0.21233 | -0.182 | 0.1082 | 0.463 |
| 11987 | RP11-400K9.4 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-7-1-3p | 30 | AFF1 | Sponge network | -0.611 | 0.09607 | -0.384 | 0.00053 | 0.463 |
| 11988 | HOXB-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-629-3p | 19 | RYR2 | Sponge network | -0.154 | 0.53954 | -1.466 | 0.00031 | 0.463 |
| 11989 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-429 | 13 | ITGB3 | Sponge network | -0.563 | 0.02545 | -0.011 | 0.95751 | 0.463 |
| 11990 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-378a-5p | 14 | PBX1 | Sponge network | 0.178 | 0.29106 | -1.206 | 0 | 0.463 |
| 11991 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 49 | WDFY3 | Sponge network | 0.72 | 0.0001 | -0.201 | 0.0967 | 0.463 |
| 11992 | TP73-AS1 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-92a-3p | 10 | CDKN1C | Sponge network | -0.563 | 0.02545 | -0.929 | 0.00033 | 0.463 |
| 11993 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-92a-3p | 15 | NRXN3 | Sponge network | 0.056 | 0.81195 | -0.893 | 0.04186 | 0.463 |
| 11994 | RP11-389C8.2 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 12 | CXCL12 | Sponge network | 0.727 | 3.0E-5 | -1.421 | 0 | 0.463 |
| 11995 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p | 15 | SYNPO2 | Sponge network | 0.176 | 0.37402 | -2.342 | 0 | 0.463 |
| 11996 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CHD2 | Sponge network | 0.873 | 0 | 0.049 | 0.55371 | 0.463 |
| 11997 | AC003090.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-96-5p | 13 | ZBTB20 | Sponge network | -2.117 | 0.00161 | -0.122 | 0.57569 | 0.463 |
| 11998 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-655-3p | 18 | SON | Sponge network | 1.147 | 0 | 0.168 | 0.04406 | 0.463 |
| 11999 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-874-3p | 10 | BCLAF1 | Sponge network | 1.344 | 0 | 0.211 | 0.00684 | 0.463 |
| 12000 | MAGI2-AS3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 14 | EHD3 | Sponge network | -0.129 | 0.57177 | -0.484 | 0.01747 | 0.463 |
| 12001 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-93-3p | 12 | EPHA3 | Sponge network | 0.598 | 0.38481 | 0.185 | 0.53588 | 0.463 |
| 12002 | LINC00578 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-629-3p | 16 | ZBTB4 | Sponge network | 0.811 | 0.03228 | -0.739 | 0 | 0.463 |
| 12003 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 19 | EBF1 | Sponge network | -0.811 | 2.0E-5 | -0.384 | 0.08856 | 0.463 |
| 12004 | EMX2OS |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-671-5p | 20 | CADM3 | Sponge network | -0.777 | 0.1134 | -3.663 | 0 | 0.463 |
| 12005 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p | 12 | KIT | Sponge network | -0.739 | 0.00044 | -2.184 | 0 | 0.463 |
| 12006 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 36 | MAF | Sponge network | -0.49 | 0.04946 | -1.257 | 0 | 0.463 |
| 12007 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-7-1-3p | 15 | RECK | Sponge network | -0.246 | 0.35034 | -1.043 | 0 | 0.463 |
| 12008 | BCYRN1 |
hsa-miR-130b-5p;hsa-miR-197-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 10 | RPL22 | Sponge network | 0.311 | 0.10344 | -0.065 | 0.5828 | 0.463 |
| 12009 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-940 | 18 | PEG3 | Sponge network | 0.214 | 0.18246 | -1.74 | 0 | 0.463 |
| 12010 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 33 | RSPO2 | Sponge network | -2.46 | 0 | -3.038 | 0 | 0.463 |
| 12011 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p | 17 | CBFA2T3 | Sponge network | -1.331 | 3.0E-5 | -1.221 | 1.0E-5 | 0.463 |
| 12012 | RP11-6O2.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-370-3p | 16 | ZMYND11 | Sponge network | 0.331 | 0.57247 | -0.2 | 0.05449 | 0.463 |
| 12013 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | EBF3 | Sponge network | -0.769 | 0.00035 | -0.058 | 0.81437 | 0.463 |
| 12014 | RP11-401P9.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-5p | 16 | KANK1 | Sponge network | -0.384 | 0.40652 | -0.55 | 0.00134 | 0.463 |
| 12015 | RP11-554A11.4 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-423-5p | 15 | CSDE1 | Sponge network | -0.583 | 0.14589 | -0.493 | 0 | 0.463 |
| 12016 | LINC00473 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 50 | AR | Sponge network | -1.203 | 0.03881 | -1.554 | 0 | 0.463 |
| 12017 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p | 11 | HMCN1 | Sponge network | -0.405 | 0.22013 | 0.809 | 0.00544 | 0.463 |
| 12018 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 28 | ADAMTSL3 | Sponge network | 0.101 | 0.60919 | -1.559 | 0 | 0.463 |
| 12019 | RP11-379F4.7 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-424-5p | 16 | ABL2 | Sponge network | 1.316 | 0 | 0.82 | 0 | 0.463 |
| 12020 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-744-3p | 28 | ADAMTSL3 | Sponge network | -0.615 | 0.00015 | -1.559 | 0 | 0.463 |
| 12021 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-93-5p | 14 | AHNAK | Sponge network | -0.162 | 0.47687 | -1.207 | 0 | 0.463 |
| 12022 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | MGA | Sponge network | 0.349 | 0.01695 | 0.323 | 0.00792 | 0.463 |
| 12023 | RP5-994D16.9 |
hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 11 | NR2C2 | Sponge network | 0.252 | 0.08508 | 0.21 | 0.03224 | 0.463 |
| 12024 | MIR143HG |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CTGF | Sponge network | -0.739 | 0.0574 | 0.321 | 0.11682 | 0.463 |
| 12025 | AC127904.2 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 18 | ABL2 | Sponge network | 1.308 | 0 | 0.82 | 0 | 0.463 |
| 12026 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p | 11 | ZYX | Sponge network | -0.739 | 0.0574 | 0.019 | 0.89763 | 0.463 |
| 12027 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p | 31 | JAZF1 | Sponge network | -1.249 | 7.0E-5 | -0.377 | 0.02677 | 0.463 |
| 12028 | AC003090.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 16 | GLI3 | Sponge network | -2.117 | 0.00161 | -0.615 | 0.01482 | 0.463 |
| 12029 | RP11-48B3.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | BRD3 | Sponge network | 1.757 | 0 | 0.37 | 0.00013 | 0.463 |
| 12030 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-577;hsa-miR-96-5p;hsa-miR-98-5p | 10 | PLAGL1 | Sponge network | -2.46 | 0 | -0.588 | 0.00912 | 0.463 |
| 12031 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | SLITRK3 | Sponge network | -0.709 | 0.18168 | -3.328 | 0 | 0.463 |
| 12032 | PART1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 35 | PCDH10 | Sponge network | -2.925 | 0 | -1.644 | 0.0078 | 0.463 |
| 12033 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | LRIG1 | Sponge network | -1.092 | 4.0E-5 | -0.537 | 0.00588 | 0.463 |
| 12034 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 1.262 | 0 | 0.578 | 0 | 0.463 |
| 12035 | RP11-33B1.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | SON | Sponge network | 0.826 | 0 | 0.168 | 0.04406 | 0.463 |
| 12036 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | 0.246 | 0.59912 | -1.324 | 0.00014 | 0.463 |
| 12037 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-651-5p | 32 | PRDM2 | Sponge network | -2.452 | 0 | -0.258 | 0.00721 | 0.463 |
| 12038 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-590-3p | 15 | NRXN3 | Sponge network | 0.494 | 0.00339 | -0.893 | 0.04186 | 0.463 |
| 12039 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-582-5p | 31 | TCF4 | Sponge network | -2.095 | 0.00112 | 0.016 | 0.92271 | 0.463 |
| 12040 | DNM3OS |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | FAM172A | Sponge network | 0.572 | 0.01722 | -0.57 | 0 | 0.463 |
| 12041 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | RASSF8 | Sponge network | -0.769 | 0.00035 | -0.122 | 0.65591 | 0.463 |
| 12042 | RP11-119F7.5 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | BCL9 | Sponge network | 0.225 | 0.30644 | 0.321 | 0.00267 | 0.463 |
| 12043 | RP11-196G18.22 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-582-3p | 16 | ABL2 | Sponge network | 1.206 | 0 | 0.82 | 0 | 0.463 |
| 12044 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-542-3p | 14 | SEMA3C | Sponge network | -0.517 | 0.02935 | -0.272 | 0.31153 | 0.463 |
| 12045 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | DAB2 | Sponge network | 0.199 | 0.38015 | 0.431 | 0.0113 | 0.463 |
| 12046 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | -0.709 | 0.18168 | -0.509 | 0.03957 | 0.463 |
| 12047 | LINC00265 |
hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | RICTOR | Sponge network | 1.07 | 0 | 0.388 | 0.00188 | 0.463 |
| 12048 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 42 | CADM2 | Sponge network | -0.421 | 0.0114 | -3.782 | 0 | 0.463 |
| 12049 | RP4-613B23.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-502-5p | 15 | IGFBP5 | Sponge network | -0.309 | 0.1145 | -0.806 | 0.00105 | 0.463 |
| 12050 | RP11-894P9.1 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-369-3p;hsa-miR-495-3p | 10 | NF1 | Sponge network | 0.993 | 0 | 0.51 | 0 | 0.463 |
| 12051 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 19 | SPG20 | Sponge network | 0.997 | 0.00066 | -1.16 | 0 | 0.463 |
| 12052 | LINC00578 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p | 12 | SEMA3C | Sponge network | 0.811 | 0.03228 | -0.272 | 0.31153 | 0.463 |
| 12053 | RP11-53O19.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-375;hsa-miR-7-1-3p | 12 | ITPKB | Sponge network | -0.405 | 0.22013 | -0.704 | 0.00106 | 0.463 |
| 12054 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | ABI2 | Sponge network | 0.72 | 0.0001 | 0.175 | 0.14787 | 0.463 |
| 12055 | FZD10-AS1 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | -0.727 | 0.04726 | 0.809 | 0.00544 | 0.463 |
| 12056 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-940 | 15 | CRTC3 | Sponge network | -0.246 | 0.35034 | 0.164 | 0.16786 | 0.463 |
| 12057 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 68 | BMPR2 | Sponge network | -0.576 | 0.10121 | 0.206 | 0.0635 | 0.463 |
| 12058 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-7-1-3p | 32 | EPHA5 | Sponge network | -4.546 | 0 | -0.957 | 0.01733 | 0.463 |
| 12059 | CTD-2647L4.4 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-495-3p;hsa-miR-590-5p | 14 | ARHGEF12 | Sponge network | 0.555 | 0.00197 | 0.09 | 0.35693 | 0.463 |
| 12060 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-495-3p | 21 | BMPR2 | Sponge network | 1.106 | 0 | 0.206 | 0.0635 | 0.463 |
| 12061 | TP73-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-92a-3p | 14 | MACF1 | Sponge network | -0.563 | 0.02545 | 0.019 | 0.90335 | 0.462 |
| 12062 | AC005562.1 |
hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p | 10 | TTL | Sponge network | 0.488 | 0.00084 | 0.494 | 1.0E-5 | 0.462 |
| 12063 | FENDRR |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | DIRAS1 | Sponge network | -1.281 | 0.00012 | -2.518 | 0 | 0.462 |
| 12064 | BVES-AS1 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-493-3p | 13 | CSDE1 | Sponge network | -2.62 | 0 | -0.493 | 0 | 0.462 |
| 12065 | FZD10-AS1 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | -0.727 | 0.04726 | -1.628 | 0 | 0.462 |
| 12066 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | FGF5 | Sponge network | -0.777 | 0.1134 | 0.549 | 0.28659 | 0.462 |
| 12067 | PCA3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | WWTR1 | Sponge network | -0.709 | 0.18168 | -0.345 | 0.11997 | 0.462 |
| 12068 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 16 | CADM3 | Sponge network | 0.194 | 0.64278 | -3.663 | 0 | 0.462 |
| 12069 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 31 | TCF4 | Sponge network | 0.178 | 0.29106 | 0.016 | 0.92271 | 0.462 |
| 12070 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p | 17 | ERG | Sponge network | -0.777 | 0.16667 | 0.002 | 0.99011 | 0.462 |
| 12071 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-5p | 47 | ADARB1 | Sponge network | -0.563 | 0.02545 | -0.335 | 0.06631 | 0.462 |
| 12072 | HCG18 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | CEP170 | Sponge network | 1.063 | 0 | 0.943 | 0 | 0.462 |
| 12073 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-93-5p | 13 | IGSF10 | Sponge network | -0.473 | 0.07602 | -1.751 | 1.0E-5 | 0.462 |
| 12074 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-429 | 11 | DCLK1 | Sponge network | 0.16 | 0.50785 | -1.324 | 0.00014 | 0.462 |
| 12075 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p | 17 | CBFA2T3 | Sponge network | -2.531 | 0 | -1.221 | 1.0E-5 | 0.462 |
| 12076 | AC003090.1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | TLE4 | Sponge network | -2.117 | 0.00161 | -1.157 | 0 | 0.462 |
| 12077 | RP11-48B3.4 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p | 14 | BCL9 | Sponge network | 1.757 | 0 | 0.321 | 0.00267 | 0.462 |
| 12078 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | CREBBP | Sponge network | 0.72 | 0.0001 | 0.126 | 0.15186 | 0.462 |
| 12079 | GNG12-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3913-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-942-5p | 33 | IGFBP5 | Sponge network | -0.793 | 7.0E-5 | -0.806 | 0.00105 | 0.462 |
| 12080 | USP3-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 32 | IGFBP5 | Sponge network | -0.162 | 0.47687 | -0.806 | 0.00105 | 0.462 |
| 12081 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | NEDD4 | Sponge network | -0.576 | 0.10121 | 0.02 | 0.89834 | 0.462 |
| 12082 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p | 21 | CNTNAP3 | Sponge network | -0.942 | 0 | -1.465 | 0.00033 | 0.462 |
| 12083 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-511-5p | 17 | RBMS3 | Sponge network | 0.497 | 0.01451 | -0.519 | 0.03551 | 0.462 |
| 12084 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 30 | UNC80 | Sponge network | -1.057 | 0.00029 | -1.934 | 0 | 0.462 |
| 12085 | RP11-473I1.5 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-655-3p | 13 | NF1 | Sponge network | 0.542 | 0.00054 | 0.51 | 0 | 0.462 |
| 12086 | AC108488.4 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 11 | CNTNAP1 | Sponge network | 0.259 | 0.12542 | 0.358 | 0.13727 | 0.462 |
| 12087 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-361-5p;hsa-miR-590-3p | 19 | RSPO2 | Sponge network | -0.739 | 0.00044 | -3.038 | 0 | 0.462 |
| 12088 | LINC00865 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 43 | GAS7 | Sponge network | -1.249 | 7.0E-5 | -0.555 | 0.01602 | 0.462 |
| 12089 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p | 14 | EZH1 | Sponge network | 0.299 | 0.57722 | -0.564 | 1.0E-5 | 0.462 |
| 12090 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | EBF1 | Sponge network | 0.997 | 0.00066 | -0.384 | 0.08856 | 0.462 |
| 12091 | MIR143HG |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | UBE2QL1 | Sponge network | -0.739 | 0.0574 | -2.417 | 0 | 0.462 |
| 12092 | LINC01018 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 18 | CDH19 | Sponge network | -2.915 | 0.00015 | -3.434 | 0 | 0.462 |
| 12093 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 14 | FBXO31 | Sponge network | -0.096 | 0.67958 | -0.085 | 0.42389 | 0.462 |
| 12094 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DCN | Sponge network | -0.323 | 0.01372 | -1.288 | 0 | 0.462 |
| 12095 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-940 | 10 | HOOK3 | Sponge network | 0.921 | 0.00118 | 0.273 | 0.09957 | 0.462 |
| 12096 | RP11-400K9.3 |
hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-340-5p;hsa-miR-576-5p | 10 | ZFHX4 | Sponge network | -0.733 | 0.19469 | 0.018 | 0.95574 | 0.462 |
| 12097 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 40 | PAFAH1B1 | Sponge network | -0.487 | 0.02157 | -0.429 | 0 | 0.462 |
| 12098 | MIR497HG |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-5p | 16 | FMNL3 | Sponge network | -1.439 | 4.0E-5 | 0.555 | 4.0E-5 | 0.462 |
| 12099 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-505-5p | 14 | TBL1XR1 | Sponge network | 0.993 | 0 | 0.578 | 0 | 0.462 |
| 12100 | ADAMTS9-AS2 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | CSDE1 | Sponge network | -2.452 | 0 | -0.493 | 0 | 0.462 |
| 12101 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-196b-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-577 | 15 | NRXN1 | Sponge network | -0.473 | 0.07602 | -3.778 | 0 | 0.462 |
| 12102 | LINC00969 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-628-5p | 17 | TRIM33 | Sponge network | 0.683 | 1.0E-5 | 0.402 | 7.0E-5 | 0.462 |
| 12103 | CTC-510F12.2 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-361-3p | 14 | THRB | Sponge network | -0.691 | 0.00146 | -1.117 | 0.0004 | 0.462 |
| 12104 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | PDGFC | Sponge network | -2.452 | 0 | -0.33 | 0.06483 | 0.462 |
| 12105 | OIP5-AS1 |
hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | HERC1 | Sponge network | 0.421 | 0.00084 | -0.196 | 0.08544 | 0.462 |
| 12106 | MEG3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | NIN | Sponge network | 0.249 | 0.35902 | -0.253 | 0.13794 | 0.462 |
| 12107 | MYLK-AS1 |
hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-935;hsa-miR-940 | 14 | LIFR | Sponge network | -1.331 | 3.0E-5 | -1.794 | 0 | 0.462 |
| 12108 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 25 | HECA | Sponge network | 0.349 | 0.01695 | -0.217 | 0.03463 | 0.462 |
| 12109 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-935 | 65 | LPP | Sponge network | 0.75 | 0.00054 | -0.459 | 0.04475 | 0.462 |
| 12110 | TUG1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-654-3p | 25 | RICTOR | Sponge network | 0.972 | 0 | 0.388 | 0.00188 | 0.462 |
| 12111 | RP11-226L15.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | BRD3 | Sponge network | 0.8 | 0 | 0.37 | 0.00013 | 0.462 |
| 12112 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 51 | MEF2C | Sponge network | -2.46 | 0 | -0.499 | 0.01448 | 0.462 |
| 12113 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 22 | AKAP11 | Sponge network | 0.765 | 7.0E-5 | 0.196 | 0.09825 | 0.462 |
| 12114 | RP11-33B1.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 18 | MDM4 | Sponge network | 0.826 | 0 | 0.345 | 0.00762 | 0.462 |
| 12115 | MIR143HG |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-935 | 10 | LRRN3 | Sponge network | -0.739 | 0.0574 | -0.393 | 0.16293 | 0.462 |
| 12116 | TRAF3IP2-AS1 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | RIMS2 | Sponge network | -0.323 | 0.01372 | -1.365 | 0.00208 | 0.462 |
| 12117 | PCA3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | LRP1B | Sponge network | -0.709 | 0.18168 | -2.163 | 0 | 0.462 |
| 12118 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-330-3p;hsa-miR-576-5p | 16 | ATRX | Sponge network | 2.163 | 0 | 0.261 | 0.04408 | 0.462 |
| 12119 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-93-5p | 15 | AR | Sponge network | -2.039 | 0.03447 | -1.554 | 0 | 0.462 |
| 12120 | CTC-429P9.5 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 10 | PTGFR | Sponge network | 0.178 | 0.29106 | -0.233 | 0.45421 | 0.462 |
| 12121 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 38 | EBF1 | Sponge network | -2.46 | 0 | -0.384 | 0.08856 | 0.462 |
| 12122 | KB-431C1.4 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p;hsa-miR-654-3p | 15 | PHF6 | Sponge network | 0.909 | 0 | 0.785 | 0 | 0.462 |
| 12123 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | QKI | Sponge network | 0.299 | 0.57722 | -0.333 | 0.04111 | 0.462 |
| 12124 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 37 | RICTOR | Sponge network | 1.11 | 0 | 0.388 | 0.00188 | 0.462 |
| 12125 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | NCAM2 | Sponge network | 0.246 | 0.59912 | -0.482 | 0.22565 | 0.462 |
| 12126 | GHRLOS |
hsa-let-7d-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p | 14 | CBFA2T3 | Sponge network | -1.942 | 0 | -1.221 | 1.0E-5 | 0.462 |
| 12127 | RP11-819C21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | BRD3 | Sponge network | 0.537 | 0.00139 | 0.37 | 0.00013 | 0.462 |
| 12128 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-93-5p | 21 | CDHR3 | Sponge network | -0.162 | 0.47687 | -0.977 | 4.0E-5 | 0.462 |
| 12129 | FENDRR |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-940 | 37 | PTPRT | Sponge network | -1.281 | 0.00012 | -0.907 | 0.03727 | 0.462 |
| 12130 | HOXB-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-629-3p | 41 | DST | Sponge network | -0.154 | 0.53954 | -0.713 | 0.00096 | 0.462 |
| 12131 | LINC00957 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TPO | Sponge network | -0.421 | 0.0114 | -1.87 | 2.0E-5 | 0.462 |
| 12132 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 19 | DOCK4 | Sponge network | -0.096 | 0.67958 | 0.502 | 0.00108 | 0.462 |
| 12133 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | PRKAA2 | Sponge network | -0.318 | 0.65847 | -2.462 | 0 | 0.462 |
| 12134 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p | 65 | IL6ST | Sponge network | -0.793 | 7.0E-5 | -0.402 | 0.00676 | 0.462 |
| 12135 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 34 | THBS1 | Sponge network | -0.777 | 0.1134 | 0.741 | 0.00386 | 0.462 |
| 12136 | TP73-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | -0.563 | 0.02545 | -0.704 | 0.00106 | 0.462 |
| 12137 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | CDH10 | Sponge network | -1.697 | 0.00889 | -2.397 | 0 | 0.462 |
| 12138 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-484 | 13 | CBL | Sponge network | 1.056 | 0 | 0.291 | 0.01267 | 0.462 |
| 12139 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 58 | THRB | Sponge network | -1.439 | 4.0E-5 | -1.117 | 0.0004 | 0.462 |
| 12140 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | DIP2C | Sponge network | 0.889 | 9.0E-5 | -0.436 | 0.01713 | 0.462 |
| 12141 | RP11-121C2.2 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | ERCC6 | Sponge network | 1.147 | 0 | 0.23 | 0.015 | 0.462 |
| 12142 | ADAMTS9-AS2 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374a-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 11 | MLLT11 | Sponge network | -2.452 | 0 | -0.661 | 0.01733 | 0.462 |
| 12143 | AC009948.5 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | EYA4 | Sponge network | 0.532 | 0.00505 | 0.3 | 0.36876 | 0.462 |
| 12144 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-93-5p | 22 | NTRK3 | Sponge network | -2.039 | 0.03447 | -1.77 | 3.0E-5 | 0.462 |
| 12145 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | MITF | Sponge network | 0.194 | 0.64278 | -0.769 | 0.00022 | 0.462 |
| 12146 | MEG3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-625-3p | 30 | ABL2 | Sponge network | 0.249 | 0.35902 | 0.82 | 0 | 0.462 |
| 12147 | CTC-429P9.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-877-5p | 17 | BMPR2 | Sponge network | 0.043 | 0.81628 | 0.206 | 0.0635 | 0.462 |
| 12148 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | NR4A3 | Sponge network | -0.576 | 0.10121 | -1.385 | 0 | 0.462 |
| 12149 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-589-5p;hsa-miR-935 | 10 | RARB | Sponge network | -2.62 | 0 | -0.825 | 2.0E-5 | 0.462 |
| 12150 | PGM5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-940 | 23 | TRPS1 | Sponge network | -4.546 | 0 | -0.191 | 0.44109 | 0.462 |
| 12151 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 22 | BCL6 | Sponge network | -2.46 | 0 | -0.211 | 0.19489 | 0.462 |
| 12152 | RP11-401P9.4 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-93-5p | 52 | DST | Sponge network | -0.384 | 0.40652 | -0.713 | 0.00096 | 0.462 |
| 12153 | LINC00641 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | BRD3 | Sponge network | -0.222 | 0.32445 | 0.37 | 0.00013 | 0.462 |
| 12154 | MIR143HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | NIN | Sponge network | -0.739 | 0.0574 | -0.253 | 0.13794 | 0.462 |
| 12155 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-93-3p | 14 | EBF3 | Sponge network | -0.693 | 0.05629 | -0.058 | 0.81437 | 0.462 |
| 12156 | LINC00957 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 20 | EBF1 | Sponge network | -0.421 | 0.0114 | -0.384 | 0.08856 | 0.462 |
| 12157 | RP11-384K6.6 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-539-5p | 10 | PHF3 | Sponge network | 0.946 | 0 | 0.126 | 0.19652 | 0.462 |
| 12158 | LINC00473 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 12 | TPO | Sponge network | -1.203 | 0.03881 | -1.87 | 2.0E-5 | 0.462 |
| 12159 | RP11-473I1.10 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 0.673 | 0 | 0.51 | 0 | 0.461 |
| 12160 | RP11-13K12.5 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | -0.318 | 0.65847 | -2.633 | 0 | 0.461 |
| 12161 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-93-5p | 26 | IL17RD | Sponge network | 0.299 | 0.57722 | -0.019 | 0.94873 | 0.461 |
| 12162 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | CUL5 | Sponge network | 0.873 | 0 | 0.026 | 0.77301 | 0.461 |
| 12163 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-7-1-3p | 15 | KCNA6 | Sponge network | -4.546 | 0 | -1.531 | 0.00013 | 0.461 |
| 12164 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-935 | 34 | DIP2C | Sponge network | -0.563 | 0.02545 | -0.436 | 0.01713 | 0.461 |
| 12165 | AP001347.6 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-7-5p | 11 | EHD3 | Sponge network | -1.161 | 0.00591 | -0.484 | 0.01747 | 0.461 |
| 12166 | LINC00476 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | PRKCB | Sponge network | -0.615 | 0.00015 | -1.313 | 2.0E-5 | 0.461 |
| 12167 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484 | 14 | TRIM3 | Sponge network | -2.452 | 0 | -0.577 | 0 | 0.461 |
| 12168 | RP11-83A24.2 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-654-3p | 11 | ZNF638 | Sponge network | 0.417 | 0.00445 | 0.22 | 0.01214 | 0.461 |
| 12169 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ATRX | Sponge network | -0.021 | 0.93598 | 0.261 | 0.04408 | 0.461 |
| 12170 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | MED12L | Sponge network | 0.532 | 0.00505 | -0.194 | 0.55299 | 0.461 |
| 12171 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 33 | ZFHX4 | Sponge network | 0.225 | 0.30644 | 0.018 | 0.95574 | 0.461 |
| 12172 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-877-5p;hsa-miR-935 | 34 | RAP1A | Sponge network | -0.615 | 0.00015 | -0.929 | 0 | 0.461 |
| 12173 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ZNF844 | Sponge network | -1.111 | 0.0003 | -0.966 | 0.00035 | 0.461 |
| 12174 | RP11-356J5.12 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | DIP2C | Sponge network | -0.899 | 6.0E-5 | -0.436 | 0.01713 | 0.461 |
| 12175 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 29 | TACC1 | Sponge network | 0.889 | 9.0E-5 | -0.362 | 0.05406 | 0.461 |
| 12176 | FZD10-AS1 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p | 10 | PDZD2 | Sponge network | -0.727 | 0.04726 | -0.909 | 3.0E-5 | 0.461 |
| 12177 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | QKI | Sponge network | -0.021 | 0.93598 | -0.333 | 0.04111 | 0.461 |
| 12178 | ADAMTS9-AS2 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | NR3C2 | Sponge network | -2.452 | 0 | -1.56 | 0 | 0.461 |
| 12179 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | ABCC9 | Sponge network | 0.225 | 0.30644 | -0.466 | 0.14121 | 0.461 |
| 12180 | RP5-855D21.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 16 | TBL1XR1 | Sponge network | 1.728 | 0 | 0.578 | 0 | 0.461 |
| 12181 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 33 | BMPR2 | Sponge network | -1.092 | 4.0E-5 | 0.206 | 0.0635 | 0.461 |
| 12182 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-940 | 19 | IGF1 | Sponge network | 0.4 | 0.14611 | -0.204 | 0.5898 | 0.461 |
| 12183 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | TLE4 | Sponge network | -0.769 | 0.00035 | -1.157 | 0 | 0.461 |
| 12184 | RP11-473I1.5 |
hsa-miR-1266-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 14 | AFF1 | Sponge network | 0.542 | 0.00054 | -0.384 | 0.00053 | 0.461 |
| 12185 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | PIK3R1 | Sponge network | -0.376 | 0.00337 | -0.133 | 0.36854 | 0.461 |
| 12186 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | LRP1B | Sponge network | -0.49 | 0.04946 | -2.163 | 0 | 0.461 |
| 12187 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | NRXN3 | Sponge network | -0.769 | 0.00035 | -0.893 | 0.04186 | 0.461 |
| 12188 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-338-3p;hsa-miR-590-3p | 10 | LRRK2 | Sponge network | -1.161 | 0.00591 | -1.136 | 1.0E-5 | 0.461 |
| 12189 | RP11-798M19.6 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 29 | DLC1 | Sponge network | -0.612 | 0.00094 | -0.382 | 0.05038 | 0.461 |
| 12190 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | THBS1 | Sponge network | 0.693 | 0.00076 | 0.741 | 0.00386 | 0.461 |
| 12191 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | EPHA3 | Sponge network | 0.385 | 0.10067 | 0.185 | 0.53588 | 0.461 |
| 12192 | LINC00672 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-484 | 12 | FLNA | Sponge network | -0.227 | 0.31657 | -0.771 | 0.01377 | 0.461 |
| 12193 | RP3-337H4.8 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30c-5p | 10 | TBL1XR1 | Sponge network | 1.527 | 0 | 0.578 | 0 | 0.461 |
| 12194 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 23 | MAF | Sponge network | -4.546 | 0 | -1.257 | 0 | 0.461 |
| 12195 | RP5-1198O20.4 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p | 11 | COL1A2 | Sponge network | 0.997 | 0.00066 | 1.991 | 0 | 0.461 |
| 12196 | RP11-225B17.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 17 | ABL2 | Sponge network | 1.055 | 0 | 0.82 | 0 | 0.461 |
| 12197 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 73 | THRB | Sponge network | -1.111 | 0.0003 | -1.117 | 0.0004 | 0.461 |
| 12198 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 37 | MAF | Sponge network | -1.697 | 0.00889 | -1.257 | 0 | 0.461 |
| 12199 | LINC01018 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 10 | ADAMTS8 | Sponge network | -2.915 | 0.00015 | -1.476 | 0.00069 | 0.461 |
| 12200 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 11 | NALCN | Sponge network | 0.199 | 0.38015 | 1.068 | 0.00237 | 0.461 |
| 12201 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-502-5p;hsa-miR-576-5p | 23 | TCF4 | Sponge network | -0.932 | 0.00329 | 0.016 | 0.92271 | 0.461 |
| 12202 | LINC00578 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 10 | ZNF208 | Sponge network | 0.811 | 0.03228 | -0.4 | 0.22381 | 0.461 |
| 12203 | CTC-429P9.5 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 16 | BCL2 | Sponge network | 0.178 | 0.29106 | -0.899 | 0 | 0.461 |
| 12204 | TP73-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-766-3p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | -0.563 | 0.02545 | -0.564 | 1.0E-5 | 0.461 |
| 12205 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-98-5p | 14 | ITGB3 | Sponge network | 0.249 | 0.35902 | -0.011 | 0.95751 | 0.461 |
| 12206 | HHIP-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-532-3p | 12 | CADM3 | Sponge network | -0.737 | 0.10822 | -3.663 | 0 | 0.461 |
| 12207 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-935 | 17 | PCDH10 | Sponge network | -0.246 | 0.35034 | -1.644 | 0.0078 | 0.461 |
| 12208 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | PRKAA2 | Sponge network | -0.187 | 0.23787 | -2.462 | 0 | 0.461 |
| 12209 | EMX2OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | PRRX1 | Sponge network | -0.777 | 0.1134 | 1.316 | 0 | 0.461 |
| 12210 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | TACC1 | Sponge network | 0.532 | 0.00505 | -0.362 | 0.05406 | 0.461 |
| 12211 | RP11-25K19.1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | ZDBF2 | Sponge network | -1.057 | 0.00029 | -0.998 | 0.00025 | 0.461 |
| 12212 | ADAMTS9-AS2 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | GATA2 | Sponge network | -2.452 | 0 | -0.654 | 0.0016 | 0.461 |
| 12213 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | DKK3 | Sponge network | -1.216 | 0 | -0.052 | 0.77783 | 0.461 |
| 12214 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-421;hsa-miR-511-5p | 11 | LRRC4 | Sponge network | -0.162 | 0.47687 | -1.774 | 0 | 0.461 |
| 12215 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p | 17 | PPM1L | Sponge network | 0.542 | 0.00054 | -0.537 | 0.00616 | 0.461 |
| 12216 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-577;hsa-miR-590-3p | 11 | FAM133A | Sponge network | -0.49 | 0.04946 | 0.549 | 0.29009 | 0.461 |
| 12217 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 34 | BCL2 | Sponge network | -0.611 | 0.09607 | -0.899 | 0 | 0.461 |
| 12218 | LIPE-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-96-5p | 16 | PIK3CA | Sponge network | 0.078 | 0.55803 | 0.195 | 0.06908 | 0.461 |
| 12219 | RP11-536O18.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 14 | RUNX1T1 | Sponge network | -0.074 | 0.90758 | -0.344 | 0.2374 | 0.461 |
| 12220 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | THBS1 | Sponge network | 0.532 | 0.00505 | 0.741 | 0.00386 | 0.461 |
| 12221 | ADAMTS9-AS2 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | RIMS2 | Sponge network | -2.452 | 0 | -1.365 | 0.00208 | 0.461 |
| 12222 | CTD-2647L4.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p | 14 | KIF1B | Sponge network | 0.555 | 0.00197 | -0.118 | 0.32258 | 0.461 |
| 12223 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 51 | DTNA | Sponge network | 0.246 | 0.59912 | -1.648 | 1.0E-5 | 0.461 |
| 12224 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | RBL2 | Sponge network | 0.101 | 0.60919 | -0.282 | 0.00254 | 0.461 |
| 12225 | AC009948.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-96-5p | 11 | ZBTB20 | Sponge network | 0.532 | 0.00505 | -0.122 | 0.57569 | 0.461 |
| 12226 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-935 | 18 | LRRC7 | Sponge network | -0.096 | 0.67958 | -1.341 | 0.01193 | 0.461 |
| 12227 | RP11-25K19.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p | 15 | ASTN1 | Sponge network | -1.057 | 0.00029 | -2.389 | 2.0E-5 | 0.461 |
| 12228 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | LATS2 | Sponge network | -1.111 | 0.0003 | 0.159 | 0.2304 | 0.461 |
| 12229 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-744-3p;hsa-miR-940 | 33 | MEF2C | Sponge network | -0.777 | 0.16667 | -0.499 | 0.01448 | 0.461 |
| 12230 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-361-3p;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-744-5p | 18 | CHD5 | Sponge network | -0.309 | 0.1145 | -0.922 | 0.01521 | 0.461 |
| 12231 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-590-5p | 13 | FNBP1 | Sponge network | -0.612 | 0.00094 | -0.969 | 4.0E-5 | 0.461 |
| 12232 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p | 18 | RECK | Sponge network | -0.773 | 0.04073 | -1.043 | 0 | 0.461 |
| 12233 | RP11-159D12.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-940 | 20 | SIRT1 | Sponge network | 1.254 | 0 | 0.109 | 0.2593 | 0.461 |
| 12234 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 20 | ADARB1 | Sponge network | -1.255 | 0.00981 | -0.335 | 0.06631 | 0.461 |
| 12235 | LINC00957 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | -0.421 | 0.0114 | 0.154 | 0.74723 | 0.461 |
| 12236 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | IGSF10 | Sponge network | -1.697 | 0.00889 | -1.751 | 1.0E-5 | 0.461 |
| 12237 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 53 | ADARB1 | Sponge network | -1.281 | 0.00012 | -0.335 | 0.06631 | 0.461 |
| 12238 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-874-3p;hsa-miR-98-5p | 10 | ST5 | Sponge network | -0.769 | 0.00035 | -0.78 | 0 | 0.461 |
| 12239 | LINC00578 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-576-5p | 24 | ABI2 | Sponge network | 0.811 | 0.03228 | 0.175 | 0.14787 | 0.461 |
| 12240 | RP11-384O8.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p | 17 | MRVI1 | Sponge network | -0.738 | 0.21233 | -1.051 | 0.00022 | 0.461 |
| 12241 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p | 31 | CHD5 | Sponge network | -0.513 | 0.04703 | -0.922 | 0.01521 | 0.461 |
| 12242 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 13 | LRRK2 | Sponge network | -0.323 | 0.01372 | -1.136 | 1.0E-5 | 0.461 |
| 12243 | HAND2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | PTPN13 | Sponge network | -1.697 | 0.00889 | -1.208 | 0 | 0.461 |
| 12244 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 21 | RET | Sponge network | -0.576 | 0.10121 | -0.833 | 0.03517 | 0.461 |
| 12245 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 12 | GLI3 | Sponge network | -0.318 | 0.65847 | -0.615 | 0.01482 | 0.461 |
| 12246 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p | 16 | SPG20 | Sponge network | -0.246 | 0.35034 | -1.16 | 0 | 0.461 |
| 12247 | LENG8-AS1 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-511-5p | 10 | ATRX | Sponge network | 1.471 | 0 | 0.261 | 0.04408 | 0.461 |
| 12248 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | MCC | Sponge network | -1.203 | 0.03881 | -1.005 | 0 | 0.461 |
| 12249 | BOLA3-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-940 | 10 | CELF4 | Sponge network | -0.246 | 0.35034 | -1.135 | 0.00344 | 0.461 |
| 12250 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-708-5p | 13 | EZH1 | Sponge network | -0.469 | 0.08721 | -0.564 | 1.0E-5 | 0.461 |
| 12251 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p | 30 | PGR | Sponge network | 0.056 | 0.81195 | -1.704 | 0 | 0.461 |
| 12252 | HCG11 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | PREX2 | Sponge network | 0.385 | 0.10067 | -0.272 | 0.25382 | 0.461 |
| 12253 | FAM66C |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p | 11 | HIC1 | Sponge network | -0.901 | 0.00019 | 0.024 | 0.89889 | 0.461 |
| 12254 | RP11-554A11.4 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 14 | RNF144A | Sponge network | -0.583 | 0.14589 | 0.594 | 0.0009 | 0.461 |
| 12255 | RP11-307B6.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-93-3p | 22 | NTRK3 | Sponge network | -4.463 | 0.00025 | -1.77 | 3.0E-5 | 0.461 |
| 12256 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-7-1-3p | 13 | RECK | Sponge network | -0.726 | 0.00132 | -1.043 | 0 | 0.461 |
| 12257 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 55 | TRPS1 | Sponge network | -1.697 | 0.00889 | -0.191 | 0.44109 | 0.461 |
| 12258 | RP11-6O2.3 |
hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-335-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-940 | 13 | PEG3 | Sponge network | 0.331 | 0.57247 | -1.74 | 0 | 0.461 |
| 12259 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-625-5p | 31 | TGFBR3 | Sponge network | -2.531 | 0 | -1.173 | 1.0E-5 | 0.461 |
| 12260 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | SOX5 | Sponge network | -0.421 | 0.0114 | -1.091 | 4.0E-5 | 0.461 |
| 12261 | LINC00472 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p | 11 | ZNF208 | Sponge network | -0.842 | 0.01361 | -0.4 | 0.22381 | 0.461 |
| 12262 | RP11-535M15.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-589-5p | 15 | PRKAA2 | Sponge network | -1.14 | 0.03513 | -2.462 | 0 | 0.461 |
| 12263 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 20 | ZFP36L1 | Sponge network | -0.976 | 0.00025 | -0.505 | 8.0E-5 | 0.461 |
| 12264 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421 | 27 | IGF1 | Sponge network | -0.842 | 0.01361 | -0.204 | 0.5898 | 0.461 |
| 12265 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-98-5p | 31 | PBX1 | Sponge network | -0.727 | 0.04726 | -1.206 | 0 | 0.461 |
| 12266 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | SOX5 | Sponge network | -0.187 | 0.23787 | -1.091 | 4.0E-5 | 0.46 |
| 12267 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-877-5p | 18 | AR | Sponge network | 0.043 | 0.81628 | -1.554 | 0 | 0.46 |
| 12268 | RAMP2-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 16 | CXCL12 | Sponge network | -0.513 | 0.04703 | -1.421 | 0 | 0.46 |
| 12269 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-378a-5p;hsa-miR-582-3p | 17 | ADARB1 | Sponge network | -2.076 | 0.00548 | -0.335 | 0.06631 | 0.46 |
| 12270 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p | 24 | CREB1 | Sponge network | 0.542 | 0.00054 | 0.203 | 0.00537 | 0.46 |
| 12271 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 16 | ZNF292 | Sponge network | -0.222 | 0.32445 | 0.276 | 0.04148 | 0.46 |
| 12272 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 27 | TET1 | Sponge network | 0.374 | 0.20054 | -0.135 | 0.52138 | 0.46 |
| 12273 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p | 13 | ZFHX3 | Sponge network | 0.858 | 3.0E-5 | 0.389 | 0.01363 | 0.46 |
| 12274 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-409-3p;hsa-miR-450b-5p | 12 | IGSF10 | Sponge network | 0.176 | 0.37402 | -1.751 | 1.0E-5 | 0.46 |
| 12275 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-493-5p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | -0.976 | 0.00025 | 0.273 | 0.09957 | 0.46 |
| 12276 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 34 | DDX6 | Sponge network | 0.72 | 0.0001 | 0.091 | 0.36428 | 0.46 |
| 12277 | CTD-2002H8.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TSC1 | Sponge network | 0.931 | 1.0E-5 | 0.103 | 0.26071 | 0.46 |
| 12278 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | RASSF2 | Sponge network | 0.335 | 0.20596 | -0.273 | 0.21961 | 0.46 |
| 12279 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-92a-3p | 12 | ZNF521 | Sponge network | -0.513 | 0.04703 | -0.111 | 0.58068 | 0.46 |
| 12280 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | ZMYM2 | Sponge network | 1.378 | 0 | 0.279 | 0.01273 | 0.46 |
| 12281 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | BNC2 | Sponge network | -0.737 | 0.10822 | -0.491 | 0.09988 | 0.46 |
| 12282 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 19 | RYR2 | Sponge network | -0.421 | 0.0114 | -1.466 | 0.00031 | 0.46 |
| 12283 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 17 | RB1CC1 | Sponge network | 0.311 | 0.10344 | 0.175 | 0.12559 | 0.46 |
| 12284 | RP11-305E6.4 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | SHPRH | Sponge network | 1.317 | 0 | 0.098 | 0.40994 | 0.46 |
| 12285 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-455-3p | 16 | ABL1 | Sponge network | -0.469 | 0.08721 | -0.215 | 0.07804 | 0.46 |
| 12286 | RP11-305E6.4 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-766-3p | 29 | MDM4 | Sponge network | 1.317 | 0 | 0.345 | 0.00762 | 0.46 |
| 12287 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-93-5p | 20 | ZBTB4 | Sponge network | -0.473 | 0.07602 | -0.739 | 0 | 0.46 |
| 12288 | RP11-67L2.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 76 | LPP | Sponge network | 0.72 | 0.0001 | -0.459 | 0.04475 | 0.46 |
| 12289 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p | 17 | CBFA2T3 | Sponge network | -0.896 | 0.00099 | -1.221 | 1.0E-5 | 0.46 |
| 12290 | LINC00092 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | SYT4 | Sponge network | -1.886 | 0 | -3.207 | 0 | 0.46 |
| 12291 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | NRXN3 | Sponge network | -0.513 | 0.04703 | -0.893 | 0.04186 | 0.46 |
| 12292 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 32 | SOX5 | Sponge network | 0.176 | 0.37402 | -1.091 | 4.0E-5 | 0.46 |
| 12293 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-429 | 15 | BMPR2 | Sponge network | 2.364 | 0.00134 | 0.206 | 0.0635 | 0.46 |
| 12294 | ARHGEF26-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 18 | CXCL12 | Sponge network | -2.46 | 0 | -1.421 | 0 | 0.46 |
| 12295 | MIR143HG |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SEMA3E | Sponge network | -0.739 | 0.0574 | -1.717 | 0.00137 | 0.46 |
| 12296 | FENDRR |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 15 | ZBTB20 | Sponge network | -1.281 | 0.00012 | -0.122 | 0.57569 | 0.46 |
| 12297 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | -0.563 | 0.02545 | -1.047 | 0.00364 | 0.46 |
| 12298 | FAM66C |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | NCAM2 | Sponge network | -0.901 | 0.00019 | -0.482 | 0.22565 | 0.46 |
| 12299 | RP11-999E24.3 |
hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-33a-3p | 12 | ERG | Sponge network | -0.693 | 0.05629 | 0.002 | 0.99011 | 0.46 |
| 12300 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | DOCK4 | Sponge network | -0.513 | 0.04703 | 0.502 | 0.00108 | 0.46 |
| 12301 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p | 20 | PTPN14 | Sponge network | -1.092 | 4.0E-5 | 0.144 | 0.34595 | 0.46 |
| 12302 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-590-3p | 17 | EYA4 | Sponge network | -0.358 | 0.17255 | 0.3 | 0.36876 | 0.46 |
| 12303 | SNX29P2 |
hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-7-1-3p | 12 | ITPKB | Sponge network | 0.4 | 0.14611 | -0.704 | 0.00106 | 0.46 |
| 12304 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p | 24 | GRIA3 | Sponge network | -1.203 | 0.03881 | -1.956 | 0 | 0.46 |
| 12305 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 21 | DYNC1I1 | Sponge network | 0.997 | 0.00066 | -1.12 | 0.00107 | 0.46 |
| 12306 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-29a-5p | 10 | RBMS3 | Sponge network | -0.326 | 0.28809 | -0.519 | 0.03551 | 0.46 |
| 12307 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p | 16 | ZNF208 | Sponge network | -0.793 | 7.0E-5 | -0.4 | 0.22381 | 0.46 |
| 12308 | LINC00641 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 35 | ERC1 | Sponge network | -0.222 | 0.32445 | -0.108 | 0.38794 | 0.46 |
| 12309 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-942-5p | 28 | NOTCH2 | Sponge network | -0.358 | 0.17255 | 0.138 | 0.35163 | 0.46 |
| 12310 | NDUFA6-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 12 | STAT5B | Sponge network | -0.355 | 0.0077 | -0.182 | 0.1082 | 0.46 |
| 12311 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 55 | DTNA | Sponge network | 0.101 | 0.60919 | -1.648 | 1.0E-5 | 0.46 |
| 12312 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-429 | 10 | TAL1 | Sponge network | -1.654 | 0.00144 | -0.462 | 0.00574 | 0.46 |
| 12313 | AC009120.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p | 10 | RICTOR | Sponge network | 1.383 | 0 | 0.388 | 0.00188 | 0.46 |
| 12314 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-501-5p | 14 | RET | Sponge network | -0.246 | 0.35034 | -0.833 | 0.03517 | 0.46 |
| 12315 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | SOX5 | Sponge network | -1.057 | 0.00029 | -1.091 | 4.0E-5 | 0.46 |
| 12316 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-942-5p;hsa-miR-98-5p | 24 | ARHGAP28 | Sponge network | -0.976 | 0.00025 | -0.911 | 0.00013 | 0.46 |
| 12317 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p | 27 | NOTCH2 | Sponge network | -0.583 | 0.14589 | 0.138 | 0.35163 | 0.46 |
| 12318 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | DCLK1 | Sponge network | -0.513 | 0.04703 | -1.324 | 0.00014 | 0.46 |
| 12319 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | KCNA6 | Sponge network | 0.572 | 0.01722 | -1.531 | 0.00013 | 0.46 |
| 12320 | AC011526.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 19 | RUNX1T1 | Sponge network | 0.342 | 0.03129 | -0.344 | 0.2374 | 0.46 |
| 12321 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 55 | CCND2 | Sponge network | -1.697 | 0.00889 | -0.267 | 0.3112 | 0.46 |
| 12322 | LINC00473 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | AKAP6 | Sponge network | -1.203 | 0.03881 | -1.586 | 3.0E-5 | 0.46 |
| 12323 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-382-5p;hsa-miR-455-3p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 31 | ACSL6 | Sponge network | -2.452 | 0 | -1.593 | 0 | 0.46 |
| 12324 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 59 | BMPR2 | Sponge network | 0.222 | 0.29133 | 0.206 | 0.0635 | 0.46 |
| 12325 | PWAR6 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 18 | RHOB | Sponge network | -1.575 | 6.0E-5 | -1.052 | 0 | 0.46 |
| 12326 | RP11-283I3.6 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-500a-5p | 10 | MACF1 | Sponge network | 0.614 | 1.0E-5 | 0.019 | 0.90335 | 0.46 |
| 12327 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | USP6 | Sponge network | 1.308 | 0 | 0.188 | 0.3871 | 0.46 |
| 12328 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-629-3p | 16 | THBS1 | Sponge network | -2.531 | 0 | 0.741 | 0.00386 | 0.46 |
| 12329 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 17 | ADAMTSL3 | Sponge network | -0.318 | 0.65847 | -1.559 | 0 | 0.46 |
| 12330 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-96-5p | 45 | KIF1B | Sponge network | 0.331 | 0.57247 | -0.118 | 0.32258 | 0.46 |
| 12331 | PWAR6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-629-3p | 26 | PTPN14 | Sponge network | -1.575 | 6.0E-5 | 0.144 | 0.34595 | 0.46 |
| 12332 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-93-5p;hsa-miR-940 | 45 | CHD5 | Sponge network | -0.777 | 0.1134 | -0.922 | 0.01521 | 0.46 |
| 12333 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-5p | 41 | TCF4 | Sponge network | 0.176 | 0.37402 | 0.016 | 0.92271 | 0.46 |
| 12334 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p;hsa-miR-98-5p | 35 | CLASP2 | Sponge network | 0.101 | 0.60919 | -0.038 | 0.71 | 0.46 |
| 12335 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 32 | TGFBR3 | Sponge network | -0.842 | 0.01361 | -1.173 | 1.0E-5 | 0.46 |
| 12336 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 25 | TBL1XR1 | Sponge network | 0.917 | 0 | 0.578 | 0 | 0.46 |
| 12337 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | ANK2 | Sponge network | -0.726 | 0.00132 | -1.603 | 1.0E-5 | 0.46 |
| 12338 | SH3RF3-AS1 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 20 | WWTR1 | Sponge network | 0.889 | 9.0E-5 | -0.345 | 0.11997 | 0.46 |
| 12339 | CKMT2-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ITGB1 | Sponge network | 0.082 | 0.55654 | 0.524 | 0.00017 | 0.46 |
| 12340 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p | 11 | CREBBP | Sponge network | 0.889 | 9.0E-5 | 0.126 | 0.15186 | 0.46 |
| 12341 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-935 | 26 | DMD | Sponge network | -0.421 | 0.0114 | -1.506 | 0 | 0.46 |
| 12342 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-942-5p;hsa-miR-98-5p | 20 | ARHGAP28 | Sponge network | -1.092 | 4.0E-5 | -0.911 | 0.00013 | 0.46 |
| 12343 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-93-3p | 21 | QKI | Sponge network | -4.463 | 0.00025 | -0.333 | 0.04111 | 0.46 |
| 12344 | FAM66C |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ZNF536 | Sponge network | -0.901 | 0.00019 | -3.115 | 0 | 0.46 |
| 12345 | CTC-429P9.1 |
hsa-miR-126-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 14 | MOB1B | Sponge network | 0.497 | 0.01451 | 0.197 | 0.15266 | 0.46 |
| 12346 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p | 16 | LRP1B | Sponge network | -4.463 | 0.00025 | -2.163 | 0 | 0.46 |
| 12347 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | NIN | Sponge network | -0.576 | 0.10121 | -0.253 | 0.13794 | 0.46 |
| 12348 | FENDRR |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-7-5p | 32 | BTG2 | Sponge network | -1.281 | 0.00012 | -1.763 | 0 | 0.46 |
| 12349 | GABPB1-AS1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p | 14 | SHPRH | Sponge network | 1.11 | 0 | 0.098 | 0.40994 | 0.46 |
| 12350 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | ZMYM2 | Sponge network | 0.768 | 7.0E-5 | 0.279 | 0.01273 | 0.46 |
| 12351 | RP11-254F7.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p | 16 | NAV3 | Sponge network | 0.176 | 0.37402 | 0.212 | 0.47729 | 0.46 |
| 12352 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 19 | TRIM33 | Sponge network | 0.222 | 0.29133 | 0.402 | 7.0E-5 | 0.46 |
| 12353 | TTC25 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-5p | 10 | DMD | Sponge network | -0.208 | 0.38261 | -1.506 | 0 | 0.46 |
| 12354 | RP11-13K12.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-625-5p | 15 | SFRP1 | Sponge network | -0.154 | 0.78939 | -3.566 | 0 | 0.46 |
| 12355 | LINC00702 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 10 | MLLT11 | Sponge network | -0.737 | 0.06182 | -0.661 | 0.01733 | 0.46 |
| 12356 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | DAB2 | Sponge network | -1.349 | 0.00017 | 0.431 | 0.0113 | 0.46 |
| 12357 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 13 | UVRAG | Sponge network | -2.452 | 0 | -0.017 | 0.83865 | 0.46 |
| 12358 | MAGI2-AS3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 32 | PDE4DIP | Sponge network | -0.129 | 0.57177 | -0.282 | 0.0257 | 0.46 |
| 12359 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 31 | ANK2 | Sponge network | 0.064 | 0.72934 | -1.603 | 1.0E-5 | 0.46 |
| 12360 | CTC-296K1.3 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-582-5p | 11 | RYR2 | Sponge network | -2.095 | 0.00112 | -1.466 | 0.00031 | 0.46 |
| 12361 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 40 | TRPS1 | Sponge network | -0.769 | 0.00035 | -0.191 | 0.44109 | 0.46 |
| 12362 | SOX2-OT |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 28 | RIMS2 | Sponge network | -1.264 | 6.0E-5 | -1.365 | 0.00208 | 0.46 |
| 12363 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | ARHGAP29 | Sponge network | 0.532 | 0.00505 | 0.131 | 0.42953 | 0.46 |
| 12364 | TPTEP1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-96-5p | 34 | ASTN1 | Sponge network | -1.216 | 0 | -2.389 | 2.0E-5 | 0.46 |
| 12365 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-339-5p;hsa-miR-342-3p | 12 | STARD13 | Sponge network | 0.082 | 0.55397 | -0.187 | 0.27383 | 0.46 |
| 12366 | RP11-13K12.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-589-5p | 11 | ROBO1 | Sponge network | -0.318 | 0.65847 | -0.009 | 0.96154 | 0.46 |
| 12367 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-655-3p | 34 | DMD | Sponge network | -1.057 | 0.00029 | -1.506 | 0 | 0.459 |
| 12368 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-429 | 11 | THSD7A | Sponge network | -0.469 | 0.08721 | 0.242 | 0.25154 | 0.459 |
| 12369 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | CACNA2D1 | Sponge network | -0.738 | 0.21233 | -0.846 | 0.00675 | 0.459 |
| 12370 | LINC00641 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | ZDBF2 | Sponge network | -0.222 | 0.32445 | -0.998 | 0.00025 | 0.459 |
| 12371 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-93-5p | 37 | CHD5 | Sponge network | -2.915 | 0.00015 | -0.922 | 0.01521 | 0.459 |
| 12372 | RP11-57H14.4 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | ARHGEF12 | Sponge network | 0.083 | 0.66405 | 0.09 | 0.35693 | 0.459 |
| 12373 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 50 | AR | Sponge network | 0.532 | 0.00505 | -1.554 | 0 | 0.459 |
| 12374 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p | 41 | RBMS3 | Sponge network | 0.494 | 0.00339 | -0.519 | 0.03551 | 0.459 |
| 12375 | RP11-119F7.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | BRD3 | Sponge network | 0.225 | 0.30644 | 0.37 | 0.00013 | 0.459 |
| 12376 | RP11-359E10.1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-484 | 13 | ABL1 | Sponge network | 0.299 | 0.57722 | -0.215 | 0.07804 | 0.459 |
| 12377 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-331-3p;hsa-miR-424-5p | 15 | CHD5 | Sponge network | -2.076 | 0.00548 | -0.922 | 0.01521 | 0.459 |
| 12378 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-935 | 43 | FAT3 | Sponge network | -0.384 | 0.40652 | -1.601 | 0.00051 | 0.459 |
| 12379 | CTD-2314G24.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 17 | UNC5C | Sponge network | 0.246 | 0.59912 | -0.178 | 0.40214 | 0.459 |
| 12380 | MIR143HG |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 43 | RIMS2 | Sponge network | -0.739 | 0.0574 | -1.365 | 0.00208 | 0.459 |
| 12381 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | PCDH9 | Sponge network | 0.056 | 0.81195 | -1.823 | 4.0E-5 | 0.459 |
| 12382 | RP11-894P9.1 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p | 14 | UBR3 | Sponge network | 0.993 | 0 | -0.021 | 0.82774 | 0.459 |
| 12383 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p | 34 | QKI | Sponge network | 0.176 | 0.37402 | -0.333 | 0.04111 | 0.459 |
| 12384 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 51 | AR | Sponge network | -0.899 | 6.0E-5 | -1.554 | 0 | 0.459 |
| 12385 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | RET | Sponge network | -1.203 | 0.03881 | -0.833 | 0.03517 | 0.459 |
| 12386 | LINC00654 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 28 | SETBP1 | Sponge network | 0.374 | 0.20054 | -1.302 | 1.0E-5 | 0.459 |
| 12387 | LINC01018 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 14 | ID4 | Sponge network | -2.915 | 0.00015 | -1.644 | 0 | 0.459 |
| 12388 | FAM157C |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-378a-3p;hsa-miR-378c | 14 | ABL2 | Sponge network | 0.91 | 0 | 0.82 | 0 | 0.459 |
| 12389 | RP11-344B5.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-27a-5p | 12 | SNX29 | Sponge network | -0.055 | 0.88482 | -0.34 | 0.01371 | 0.459 |
| 12390 | ZNF667-AS1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-429;hsa-miR-96-5p | 17 | PAIP2 | Sponge network | -0.976 | 0.00025 | -0.515 | 0 | 0.459 |
| 12391 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 58 | TCF4 | Sponge network | -0.096 | 0.67958 | 0.016 | 0.92271 | 0.459 |
| 12392 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 42 | BCL2 | Sponge network | 0.101 | 0.60919 | -0.899 | 0 | 0.459 |
| 12393 | HAND2-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-93-3p | 33 | CTIF | Sponge network | -1.697 | 0.00889 | -0.389 | 0.00549 | 0.459 |
| 12394 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 46 | NOTCH2 | Sponge network | -0.49 | 0.04946 | 0.138 | 0.35163 | 0.459 |
| 12395 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | RABEP1 | Sponge network | 0.101 | 0.60919 | -0.066 | 0.43142 | 0.459 |
| 12396 | LINC00449 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 12 | PHF6 | Sponge network | 1.525 | 0 | 0.785 | 0 | 0.459 |
| 12397 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | LRP1B | Sponge network | -0.576 | 0.10121 | -2.163 | 0 | 0.459 |
| 12398 | CTD-2089N3.1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-455-5p | 10 | IGFBP5 | Sponge network | -0.314 | 0.62033 | -0.806 | 0.00105 | 0.459 |
| 12399 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 14 | HOOK3 | Sponge network | -0.709 | 0.18168 | 0.273 | 0.09957 | 0.459 |
| 12400 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | CAV1 | Sponge network | -0.487 | 0.02157 | -1.013 | 6.0E-5 | 0.459 |
| 12401 | FGD5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 40 | ARHGEF12 | Sponge network | 0.401 | 0.00066 | 0.09 | 0.35693 | 0.459 |
| 12402 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-935 | 15 | RARB | Sponge network | -0.896 | 0.00099 | -0.825 | 2.0E-5 | 0.459 |
| 12403 | A1BG-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-589-3p | 11 | KCNJ5 | Sponge network | -0.164 | 0.45106 | -0.281 | 0.3339 | 0.459 |
| 12404 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SEMA3C | Sponge network | 0.335 | 0.20596 | -0.272 | 0.31153 | 0.459 |
| 12405 | CTC-429P9.5 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-744-3p | 16 | PTPN14 | Sponge network | 0.178 | 0.29106 | 0.144 | 0.34595 | 0.459 |
| 12406 | RP11-774O3.3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-7-5p | 12 | EHD3 | Sponge network | -0.487 | 0.02157 | -0.484 | 0.01747 | 0.459 |
| 12407 | AC006116.24 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-577;hsa-miR-582-3p | 10 | PIK3CA | Sponge network | -0.473 | 0.07602 | 0.195 | 0.06908 | 0.459 |
| 12408 | IQCH-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.929 | 0 | 1.083 | 0 | 0.459 |
| 12409 | RP11-389C8.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TLL1 | Sponge network | 0.727 | 3.0E-5 | -1.195 | 3.0E-5 | 0.459 |
| 12410 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 19 | NRXN1 | Sponge network | -0.72 | 0.24239 | -3.778 | 0 | 0.459 |
| 12411 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p | 26 | RICTOR | Sponge network | 1.294 | 0 | 0.388 | 0.00188 | 0.459 |
| 12412 | CTD-2314G24.2 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | PRKCB | Sponge network | 0.246 | 0.59912 | -1.313 | 2.0E-5 | 0.459 |
| 12413 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 22 | SON | Sponge network | 0.75 | 0.00054 | 0.168 | 0.04406 | 0.459 |
| 12414 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p | 11 | SLITRK3 | Sponge network | -0.376 | 0.00337 | -3.328 | 0 | 0.459 |
| 12415 | LINC00476 |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p | 12 | CTNNA2 | Sponge network | -0.615 | 0.00015 | -2.547 | 0 | 0.459 |
| 12416 | LINC00865 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-629-3p | 28 | ASTN1 | Sponge network | -1.249 | 7.0E-5 | -2.389 | 2.0E-5 | 0.459 |
| 12417 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p | 13 | CHD2 | Sponge network | 0.082 | 0.55397 | 0.049 | 0.55371 | 0.459 |
| 12418 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 43 | RUNX1T1 | Sponge network | -1.057 | 0.00029 | -0.344 | 0.2374 | 0.459 |
| 12419 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-493-5p;hsa-miR-550a-5p | 12 | HOOK3 | Sponge network | -0.384 | 0.40652 | 0.273 | 0.09957 | 0.459 |
| 12420 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 22 | HIP1 | Sponge network | 0.72 | 0.0001 | 0.774 | 0 | 0.459 |
| 12421 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429 | 11 | NALCN | Sponge network | -0.563 | 0.02545 | 1.068 | 0.00237 | 0.459 |
| 12422 | KB-1572G7.2 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-378c;hsa-miR-424-5p | 20 | ABL2 | Sponge network | 0.924 | 0 | 0.82 | 0 | 0.459 |
| 12423 | PCA3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | PIK3R1 | Sponge network | -0.709 | 0.18168 | -0.133 | 0.36854 | 0.459 |
| 12424 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | NCAM2 | Sponge network | -0.739 | 0.00044 | -0.482 | 0.22565 | 0.459 |
| 12425 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | PLSCR4 | Sponge network | -0.811 | 2.0E-5 | -0.474 | 0.03833 | 0.459 |
| 12426 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-361-5p | 19 | HLF | Sponge network | -0.309 | 0.1145 | -2.443 | 0 | 0.459 |
| 12427 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 27 | GRIN2A | Sponge network | -0.612 | 0.00094 | -2.161 | 0 | 0.459 |
| 12428 | DIO3OS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-940 | 31 | NTRK2 | Sponge network | -0.773 | 0.04073 | -0.736 | 0.06495 | 0.459 |
| 12429 | AC006116.24 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-5p | 16 | ERC1 | Sponge network | -0.473 | 0.07602 | -0.108 | 0.38794 | 0.459 |
| 12430 | LINC01018 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-93-3p | 38 | GRIK3 | Sponge network | -2.915 | 0.00015 | -3.208 | 0 | 0.459 |
| 12431 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-877-5p | 21 | GRIN2A | Sponge network | -0.384 | 0.40652 | -2.161 | 0 | 0.459 |
| 12432 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 16 | MYCBP2 | Sponge network | 0.083 | 0.66405 | -0.027 | 0.82016 | 0.459 |
| 12433 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p | 19 | DYNC1I1 | Sponge network | -0.154 | 0.78939 | -1.12 | 0.00107 | 0.459 |
| 12434 | LINC00476 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-942-5p | 14 | ZBTB20 | Sponge network | -0.615 | 0.00015 | -0.122 | 0.57569 | 0.459 |
| 12435 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 44 | CREB3L2 | Sponge network | -0.129 | 0.57177 | -0.525 | 2.0E-5 | 0.459 |
| 12436 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-484;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-628-5p | 24 | GRIA3 | Sponge network | -0.976 | 0.00025 | -1.956 | 0 | 0.459 |
| 12437 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-7-1-3p | 16 | FGF5 | Sponge network | -0.976 | 0.00025 | 0.549 | 0.28659 | 0.459 |
| 12438 | RP1-302G2.5 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-26b-3p;hsa-miR-423-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 11 | GRIK3 | Sponge network | -1.483 | 9.0E-5 | -3.208 | 0 | 0.459 |
| 12439 | JAZF1-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 18 | FMNL3 | Sponge network | -0.769 | 0.00035 | 0.555 | 4.0E-5 | 0.459 |
| 12440 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-628-5p | 33 | FBXO32 | Sponge network | -0.842 | 0.01361 | -0.495 | 0.07561 | 0.459 |
| 12441 | JAZF1-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | STAT5B | Sponge network | -0.769 | 0.00035 | -0.182 | 0.1082 | 0.459 |
| 12442 | AP001347.6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p | 30 | IL17RD | Sponge network | -1.161 | 0.00591 | -0.019 | 0.94873 | 0.459 |
| 12443 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 27 | TACC1 | Sponge network | 0.811 | 0.03228 | -0.362 | 0.05406 | 0.459 |
| 12444 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-96-5p | 28 | ZFHX4 | Sponge network | -1.654 | 0.00144 | 0.018 | 0.95574 | 0.459 |
| 12445 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p | 22 | MOB1B | Sponge network | 0.594 | 0.00064 | 0.197 | 0.15266 | 0.459 |
| 12446 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-505-5p;hsa-miR-7-5p | 14 | KCNA6 | Sponge network | -1.582 | 8.0E-5 | -1.531 | 0.00013 | 0.459 |
| 12447 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 17 | ITGB3 | Sponge network | 0.335 | 0.20596 | -0.011 | 0.95751 | 0.459 |
| 12448 | EMX2OS |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 24 | GJA1 | Sponge network | -0.777 | 0.1134 | -0.485 | 0.02179 | 0.459 |
| 12449 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | -2.915 | 0.00015 | -1.751 | 1.0E-5 | 0.459 |
| 12450 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 62 | PTPRD | Sponge network | -1.111 | 0.0003 | -1.005 | 0.00064 | 0.459 |
| 12451 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | RASSF2 | Sponge network | -1.349 | 0.00017 | -0.273 | 0.21961 | 0.459 |
| 12452 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p | 42 | PIK3R1 | Sponge network | -0.323 | 0.01372 | -0.133 | 0.36854 | 0.459 |
| 12453 | MRVI1-AS1 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 18 | RIMS2 | Sponge network | -1.092 | 4.0E-5 | -1.365 | 0.00208 | 0.459 |
| 12454 | RP11-867G23.10 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p | 12 | RIMS2 | Sponge network | -2.531 | 0 | -1.365 | 0.00208 | 0.459 |
| 12455 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-589-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-93-5p | 58 | TCF4 | Sponge network | -0.154 | 0.78939 | 0.016 | 0.92271 | 0.459 |
| 12456 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-7-1-3p | 14 | ZMYND11 | Sponge network | -1.092 | 4.0E-5 | -0.2 | 0.05449 | 0.459 |
| 12457 | HCG11 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 24 | RASSF8 | Sponge network | 0.385 | 0.10067 | -0.122 | 0.65591 | 0.459 |
| 12458 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 41 | FOXP1 | Sponge network | -0.612 | 0.00094 | 0.042 | 0.74368 | 0.459 |
| 12459 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | TGFBR2 | Sponge network | -1.281 | 0.00012 | -0.243 | 0.11408 | 0.459 |
| 12460 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-590-3p | 11 | ARHGAP29 | Sponge network | -0.358 | 0.17255 | 0.131 | 0.42953 | 0.459 |
| 12461 | BZRAP1-AS1 |
hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-484;hsa-miR-7-5p | 10 | FLNA | Sponge network | -0.465 | 0.04703 | -0.771 | 0.01377 | 0.459 |
| 12462 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 28 | ADARB1 | Sponge network | -1.886 | 0 | -0.335 | 0.06631 | 0.459 |
| 12463 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p | 14 | MCC | Sponge network | 1.344 | 0 | -1.005 | 0 | 0.459 |
| 12464 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 35 | SNX29 | Sponge network | 0.194 | 0.64278 | -0.34 | 0.01371 | 0.459 |
| 12465 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 31 | PHC3 | Sponge network | 0.614 | 1.0E-5 | 0.212 | 0.0499 | 0.459 |
| 12466 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | DCLK1 | Sponge network | -0.727 | 0.04726 | -1.324 | 0.00014 | 0.459 |
| 12467 | RP5-1074L1.4 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 17 | ABL2 | Sponge network | 1.923 | 0 | 0.82 | 0 | 0.459 |
| 12468 | RP11-597D13.9 |
hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | GUCY1A2 | Sponge network | 1.302 | 7.0E-5 | 1.213 | 0 | 0.459 |
| 12469 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p | 12 | ADAMTSL3 | Sponge network | -0.351 | 0.11358 | -1.559 | 0 | 0.459 |
| 12470 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-497-5p | 16 | CBL | Sponge network | 1.106 | 0 | 0.291 | 0.01267 | 0.459 |
| 12471 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | PAFAH1B1 | Sponge network | -1.575 | 6.0E-5 | -0.429 | 0 | 0.459 |
| 12472 | HAND2-AS1 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | RTN4 | Sponge network | -1.697 | 0.00889 | -0.412 | 0.00014 | 0.459 |
| 12473 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PTPRB | Sponge network | 0.246 | 0.59912 | 0.105 | 0.47122 | 0.459 |
| 12474 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 13 | ZNF770 | Sponge network | 1.344 | 0 | 0.206 | 0.06751 | 0.458 |
| 12475 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p | 12 | MAFB | Sponge network | -1.349 | 0.00017 | -0.023 | 0.89865 | 0.458 |
| 12476 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429 | 16 | CNTNAP3 | Sponge network | -1.249 | 7.0E-5 | -1.465 | 0.00033 | 0.458 |
| 12477 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p | 12 | CHD2 | Sponge network | 0.93 | 0 | 0.049 | 0.55371 | 0.458 |
| 12478 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 46 | SOX5 | Sponge network | -0.942 | 0 | -1.091 | 4.0E-5 | 0.458 |
| 12479 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p | 25 | USP6 | Sponge network | 0.782 | 0.00016 | 0.188 | 0.3871 | 0.458 |
| 12480 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-769-5p | 13 | SETBP1 | Sponge network | 0.082 | 0.55397 | -1.302 | 1.0E-5 | 0.458 |
| 12481 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | SPG20 | Sponge network | -0.899 | 6.0E-5 | -1.16 | 0 | 0.458 |
| 12482 | RP11-967K21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 17 | NAV3 | Sponge network | 0.056 | 0.81195 | 0.212 | 0.47729 | 0.458 |
| 12483 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 28 | ANK2 | Sponge network | -0.342 | 0.02288 | -1.603 | 1.0E-5 | 0.458 |
| 12484 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-93-5p | 22 | ADARB1 | Sponge network | -0.726 | 0.00132 | -0.335 | 0.06631 | 0.458 |
| 12485 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | FAT3 | Sponge network | 0.082 | 0.55654 | -1.601 | 0.00051 | 0.458 |
| 12486 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 11 | HERC2 | Sponge network | 1.601 | 0 | 0.114 | 0.30638 | 0.458 |
| 12487 | LINC00957 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-3p | 16 | EBF3 | Sponge network | -0.421 | 0.0114 | -0.058 | 0.81437 | 0.458 |
| 12488 | RP11-13K12.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 19 | EZH1 | Sponge network | -0.154 | 0.78939 | -0.564 | 1.0E-5 | 0.458 |
| 12489 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 12 | DACT1 | Sponge network | 0.16 | 0.50785 | 0.783 | 0.00072 | 0.458 |
| 12490 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p | 16 | CRTC3 | Sponge network | -0.469 | 0.08721 | 0.164 | 0.16786 | 0.458 |
| 12491 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 20 | AKAP6 | Sponge network | 0.385 | 0.10067 | -1.586 | 3.0E-5 | 0.458 |
| 12492 | RP11-620J15.3 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p | 11 | CDH23 | Sponge network | -0.739 | 0.00044 | -0.974 | 0.00034 | 0.458 |
| 12493 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 48 | EPHA5 | Sponge network | -1.281 | 0.00012 | -0.957 | 0.01733 | 0.458 |
| 12494 | SNHG14 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | CDH10 | Sponge network | -1.111 | 0.0003 | -2.397 | 0 | 0.458 |
| 12495 | EMX2OS |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-942-5p | 14 | ZBTB20 | Sponge network | -0.777 | 0.1134 | -0.122 | 0.57569 | 0.458 |
| 12496 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 46 | TRPS1 | Sponge network | -0.405 | 0.22013 | -0.191 | 0.44109 | 0.458 |
| 12497 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 53 | BMPR2 | Sponge network | 0.349 | 0.01695 | 0.206 | 0.0635 | 0.458 |
| 12498 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 16 | LRRC4 | Sponge network | -1.575 | 6.0E-5 | -1.774 | 0 | 0.458 |
| 12499 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 22 | PGR | Sponge network | 0.214 | 0.18246 | -1.704 | 0 | 0.458 |
| 12500 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | ZNF292 | Sponge network | -0.031 | 0.89063 | 0.276 | 0.04148 | 0.458 |
| 12501 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-455-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | PSIP1 | Sponge network | 0.782 | 0.00016 | -0.005 | 0.96897 | 0.458 |
| 12502 | RP11-57H14.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 75 | LPP | Sponge network | 0.083 | 0.66405 | -0.459 | 0.04475 | 0.458 |
| 12503 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 11 | CACNA2D1 | Sponge network | -2.039 | 0.03447 | -0.846 | 0.00675 | 0.458 |
| 12504 | BVES-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p | 15 | CDH23 | Sponge network | -2.62 | 0 | -0.974 | 0.00034 | 0.458 |
| 12505 | RP11-890B15.3 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484 | 18 | ABL1 | Sponge network | 0.101 | 0.60919 | -0.215 | 0.07804 | 0.458 |
| 12506 | RP11-13K12.5 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 37 | DST | Sponge network | -0.318 | 0.65847 | -0.713 | 0.00096 | 0.458 |
| 12507 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-542-3p | 10 | ALDH1A2 | Sponge network | -0.942 | 0 | -2.411 | 0 | 0.458 |
| 12508 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-93-5p | 33 | IL17RD | Sponge network | -2.69 | 0.0001 | -0.019 | 0.94873 | 0.458 |
| 12509 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | PLSCR4 | Sponge network | -0.096 | 0.67958 | -0.474 | 0.03833 | 0.458 |
| 12510 | RP11-195F19.9 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-93-5p | 12 | EZH1 | Sponge network | -0.726 | 0.00132 | -0.564 | 1.0E-5 | 0.458 |
| 12511 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | MAF | Sponge network | -1.582 | 8.0E-5 | -1.257 | 0 | 0.458 |
| 12512 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-582-5p | 18 | ZFHX4 | Sponge network | -2.095 | 0.00112 | 0.018 | 0.95574 | 0.458 |
| 12513 | PWAR6 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 11 | RIMS1 | Sponge network | -1.575 | 6.0E-5 | -3.143 | 0 | 0.458 |
| 12514 | LINC00578 |
hsa-let-7b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p | 12 | NEDD4 | Sponge network | 0.811 | 0.03228 | 0.02 | 0.89834 | 0.458 |
| 12515 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 17 | WWTR1 | Sponge network | 0.214 | 0.18246 | -0.345 | 0.11997 | 0.458 |
| 12516 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-501-5p | 10 | MN1 | Sponge network | -0.769 | 0.00035 | -0.358 | 0.24172 | 0.458 |
| 12517 | RP11-540B6.6 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | ERCC6 | Sponge network | 0.93 | 0 | 0.23 | 0.015 | 0.458 |
| 12518 | FENDRR |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 25 | LIMCH1 | Sponge network | -1.281 | 0.00012 | -0.915 | 1.0E-5 | 0.458 |
| 12519 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 35 | SASH1 | Sponge network | -0.222 | 0.32445 | -0.502 | 0.00175 | 0.458 |
| 12520 | RP11-999E24.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p | 25 | PTPRD | Sponge network | -0.693 | 0.05629 | -1.005 | 0.00064 | 0.458 |
| 12521 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | PHC3 | Sponge network | -0.487 | 0.02157 | 0.212 | 0.0499 | 0.458 |
| 12522 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-500a-3p | 22 | AFF3 | Sponge network | -1.645 | 0 | -2.373 | 0 | 0.458 |
| 12523 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | IGSF10 | Sponge network | -0.737 | 0.10822 | -1.751 | 1.0E-5 | 0.458 |
| 12524 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | NRXN1 | Sponge network | -0.021 | 0.93598 | -3.778 | 0 | 0.458 |
| 12525 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p | 15 | NCAM2 | Sponge network | -0.693 | 0.05629 | -0.482 | 0.22565 | 0.458 |
| 12526 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p | 10 | THSD7A | Sponge network | -0.318 | 0.65847 | 0.242 | 0.25154 | 0.458 |
| 12527 | RP11-156P1.3 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-664a-3p | 42 | PPM1L | Sponge network | 0.214 | 0.18246 | -0.537 | 0.00616 | 0.458 |
| 12528 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 12 | RASSF8 | Sponge network | 0.178 | 0.29106 | -0.122 | 0.65591 | 0.458 |
| 12529 | LINC00957 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-590-3p | 19 | SCN9A | Sponge network | -0.421 | 0.0114 | -0.525 | 0.10593 | 0.458 |
| 12530 | LINC00094 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-3p | 16 | BCL9 | Sponge network | 0.253 | 0.09565 | 0.321 | 0.00267 | 0.458 |
| 12531 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p;hsa-miR-539-5p | 12 | LIMD1 | Sponge network | 0.683 | 1.0E-5 | 0.103 | 0.28978 | 0.458 |
| 12532 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 14 | AKAP11 | Sponge network | 1.147 | 0 | 0.196 | 0.09825 | 0.458 |
| 12533 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-940 | 32 | NTRK2 | Sponge network | -1.349 | 0.00017 | -0.736 | 0.06495 | 0.458 |
| 12534 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-766-3p;hsa-miR-93-5p | 16 | EZH1 | Sponge network | -1.654 | 0.00144 | -0.564 | 1.0E-5 | 0.458 |
| 12535 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p | 38 | JAZF1 | Sponge network | -0.222 | 0.32445 | -0.377 | 0.02677 | 0.458 |
| 12536 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 12 | GAS1 | Sponge network | -0.901 | 0.00019 | -1.047 | 0.00364 | 0.458 |
| 12537 | LINC00957 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p | 11 | MACF1 | Sponge network | -0.421 | 0.0114 | 0.019 | 0.90335 | 0.458 |
| 12538 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 38 | TGFBR3 | Sponge network | 0.249 | 0.35902 | -1.173 | 1.0E-5 | 0.458 |
| 12539 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 52 | FAT3 | Sponge network | 0.064 | 0.72934 | -1.601 | 0.00051 | 0.458 |
| 12540 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | EPHA3 | Sponge network | -1.203 | 0.03881 | 0.185 | 0.53588 | 0.458 |
| 12541 | GHRLOS |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-493-5p | 10 | AFF3 | Sponge network | -1.942 | 0 | -2.373 | 0 | 0.458 |
| 12542 | GABPB1-AS1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RBBP6 | Sponge network | 1.11 | 0 | 0.163 | 0.05101 | 0.458 |
| 12543 | AC010226.4 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | LRIG1 | Sponge network | -0.811 | 2.0E-5 | -0.537 | 0.00588 | 0.458 |
| 12544 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 24 | MLLT10 | Sponge network | 0.75 | 0.00054 | 0.105 | 0.33292 | 0.458 |
| 12545 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 36 | KIF1B | Sponge network | 0.556 | 0.00298 | -0.118 | 0.32258 | 0.458 |
| 12546 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 25 | MOB1B | Sponge network | 1.089 | 0 | 0.197 | 0.15266 | 0.458 |
| 12547 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | KCNA6 | Sponge network | -0.901 | 0.00019 | -1.531 | 0.00013 | 0.458 |
| 12548 | RP11-774O3.3 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | ID4 | Sponge network | -0.487 | 0.02157 | -1.644 | 0 | 0.458 |
| 12549 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 11 | CACNA2D1 | Sponge network | 0.178 | 0.29106 | -0.846 | 0.00675 | 0.458 |
| 12550 | AGAP11 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-5p | 16 | UBE4B | Sponge network | -1.645 | 0 | -0.235 | 0.007 | 0.458 |
| 12551 | RP11-517B11.7 |
hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-495-3p | 11 | TMEM30A | Sponge network | 0.706 | 0 | 0.096 | 0.31031 | 0.458 |
| 12552 | RP11-25K19.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | SFRP1 | Sponge network | -1.057 | 0.00029 | -3.566 | 0 | 0.458 |
| 12553 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | ABI2 | Sponge network | 0.101 | 0.60919 | 0.175 | 0.14787 | 0.458 |
| 12554 | MEG3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-96-5p | 28 | ASTN1 | Sponge network | 0.249 | 0.35902 | -2.389 | 2.0E-5 | 0.458 |
| 12555 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | EPHA5 | Sponge network | 0.249 | 0.35902 | -0.957 | 0.01733 | 0.458 |
| 12556 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | SYNE1 | Sponge network | 0.225 | 0.30644 | -0.738 | 0.00316 | 0.458 |
| 12557 | SNHG14 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | SPOP | Sponge network | -1.111 | 0.0003 | -0.419 | 1.0E-5 | 0.458 |
| 12558 | RP11-589P10.7 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-93-3p | 15 | NTRK3 | Sponge network | -2.044 | 0.00066 | -1.77 | 3.0E-5 | 0.458 |
| 12559 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p | 11 | MGA | Sponge network | 0.614 | 1.0E-5 | 0.323 | 0.00792 | 0.458 |
| 12560 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | SEPT6 | Sponge network | -2.452 | 0 | -0.598 | 0.00181 | 0.458 |
| 12561 | LINC00092 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-671-5p | 17 | DLC1 | Sponge network | -1.886 | 0 | -0.382 | 0.05038 | 0.458 |
| 12562 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | EBF1 | Sponge network | 0.101 | 0.60919 | -0.384 | 0.08856 | 0.458 |
| 12563 | CTB-31O20.4 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-497-5p | 16 | RICTOR | Sponge network | 1.619 | 0 | 0.388 | 0.00188 | 0.458 |
| 12564 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-5p;hsa-miR-7-5p | 12 | FNBP1 | Sponge network | 0.214 | 0.18246 | -0.969 | 4.0E-5 | 0.458 |
| 12565 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-589-5p;hsa-miR-590-3p | 16 | ITGB3 | Sponge network | 0.532 | 0.00505 | -0.011 | 0.95751 | 0.458 |
| 12566 | CTD-3064M3.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-874-3p | 21 | ASTN1 | Sponge network | -0.469 | 0.08721 | -2.389 | 2.0E-5 | 0.458 |
| 12567 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-3p | 23 | EPHA3 | Sponge network | 0.199 | 0.38015 | 0.185 | 0.53588 | 0.458 |
| 12568 | ZSCAN16-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ITPKB | Sponge network | -0.342 | 0.02288 | -0.704 | 0.00106 | 0.458 |
| 12569 | RP5-1198O20.4 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-93-3p | 11 | PEA15 | Sponge network | 0.997 | 0.00066 | 0.009 | 0.93318 | 0.458 |
| 12570 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 10 | ZNF292 | Sponge network | 1.601 | 0 | 0.276 | 0.04148 | 0.458 |
| 12571 | RP11-25K19.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 45 | CADM2 | Sponge network | -1.057 | 0.00029 | -3.782 | 0 | 0.458 |
| 12572 | FAM66C |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p | 10 | PRDM11 | Sponge network | -0.901 | 0.00019 | -0.252 | 0.15511 | 0.458 |
| 12573 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 45 | PRKAA2 | Sponge network | -1.057 | 0.00029 | -2.462 | 0 | 0.458 |
| 12574 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 16 | HIP1 | Sponge network | -0.358 | 0.17255 | 0.774 | 0 | 0.458 |
| 12575 | RP11-798M19.6 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-361-5p;hsa-miR-484 | 13 | ABL1 | Sponge network | -0.612 | 0.00094 | -0.215 | 0.07804 | 0.458 |
| 12576 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-942-5p | 20 | ARHGAP28 | Sponge network | -1.439 | 4.0E-5 | -0.911 | 0.00013 | 0.458 |
| 12577 | CTC-296K1.4 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-299-5p;hsa-miR-539-5p;hsa-miR-590-5p | 13 | DIP2C | Sponge network | -2.076 | 0.00548 | -0.436 | 0.01713 | 0.458 |
| 12578 | AC009948.5 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | ERG | Sponge network | 0.532 | 0.00505 | 0.002 | 0.99011 | 0.458 |
| 12579 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | -0.777 | 0.1134 | -1.892 | 0 | 0.458 |
| 12580 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 24 | NTRK2 | Sponge network | -1.582 | 8.0E-5 | -0.736 | 0.06495 | 0.458 |
| 12581 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-500a-5p | 24 | FOXP1 | Sponge network | 0.082 | 0.55397 | 0.042 | 0.74368 | 0.458 |
| 12582 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TBX18 | Sponge network | -0.739 | 0.0574 | 0.843 | 0.02945 | 0.458 |
| 12583 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-942-5p | 53 | THRB | Sponge network | -0.371 | 0.24604 | -1.117 | 0.0004 | 0.458 |
| 12584 | CTC-444N24.11 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PHF3 | Sponge network | 1.153 | 0 | 0.126 | 0.19652 | 0.458 |
| 12585 | RP11-359E10.1 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | EPHA3 | Sponge network | 0.299 | 0.57722 | 0.185 | 0.53588 | 0.458 |
| 12586 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-629-5p | 16 | IGF1 | Sponge network | -0.517 | 0.02935 | -0.204 | 0.5898 | 0.458 |
| 12587 | RP11-798M19.6 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | TES | Sponge network | -0.612 | 0.00094 | -0.348 | 0.00639 | 0.458 |
| 12588 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | RBMS3 | Sponge network | 0.16 | 0.50785 | -0.519 | 0.03551 | 0.458 |
| 12589 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DCN | Sponge network | -0.612 | 0.00094 | -1.288 | 0 | 0.457 |
| 12590 | LINC00702 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | SPOP | Sponge network | -0.737 | 0.06182 | -0.419 | 1.0E-5 | 0.457 |
| 12591 | MEG3 |
hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-5p | 10 | MLLT11 | Sponge network | 0.249 | 0.35902 | -0.661 | 0.01733 | 0.457 |
| 12592 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | ST5 | Sponge network | -2.62 | 0 | -0.78 | 0 | 0.457 |
| 12593 | RP11-434H6.7 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-132-3p;hsa-miR-150-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-409-3p | 13 | FOXP1 | Sponge network | 0.358 | 0.14302 | 0.042 | 0.74368 | 0.457 |
| 12594 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | LRP1B | Sponge network | -0.129 | 0.57177 | -2.163 | 0 | 0.457 |
| 12595 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | CDH19 | Sponge network | 0.299 | 0.57722 | -3.434 | 0 | 0.457 |
| 12596 | HAND2-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 32 | PTPN14 | Sponge network | -1.697 | 0.00889 | 0.144 | 0.34595 | 0.457 |
| 12597 | LINC00957 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | FAM172A | Sponge network | -0.421 | 0.0114 | -0.57 | 0 | 0.457 |
| 12598 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ZMYND11 | Sponge network | -0.576 | 0.10121 | -0.2 | 0.05449 | 0.457 |
| 12599 | FENDRR |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 30 | KCNA6 | Sponge network | -1.281 | 0.00012 | -1.531 | 0.00013 | 0.457 |
| 12600 | MRVI1-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 16 | SYT4 | Sponge network | -1.092 | 4.0E-5 | -3.207 | 0 | 0.457 |
| 12601 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-455-3p | 10 | RELN | Sponge network | -0.309 | 0.1145 | -2.403 | 0 | 0.457 |
| 12602 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 18 | GREM1 | Sponge network | 0.532 | 0.00505 | 0.902 | 0.01691 | 0.457 |
| 12603 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-93-5p | 34 | EPHA7 | Sponge network | -0.384 | 0.40652 | -2.197 | 3.0E-5 | 0.457 |
| 12604 | BVES-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-935 | 11 | SEPT6 | Sponge network | -2.62 | 0 | -0.598 | 0.00181 | 0.457 |
| 12605 | H1FX-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 12 | LHFP | Sponge network | -0.206 | 0.12942 | -0.167 | 0.41371 | 0.457 |
| 12606 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-5p | 20 | EYA4 | Sponge network | 0.176 | 0.37402 | 0.3 | 0.36876 | 0.457 |
| 12607 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | HOOK3 | Sponge network | -0.096 | 0.67958 | 0.273 | 0.09957 | 0.457 |
| 12608 | RP11-473I1.9 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 18 | ATM | Sponge network | 0.683 | 1.0E-5 | 0.178 | 0.21568 | 0.457 |
| 12609 | LINC00702 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | INPP4A | Sponge network | -0.737 | 0.06182 | 0.07 | 0.53563 | 0.457 |
| 12610 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-539-5p | 12 | NRXN1 | Sponge network | -0.309 | 0.1145 | -3.778 | 0 | 0.457 |
| 12611 | GAS6-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-5p | 11 | THSD7A | Sponge network | 0.16 | 0.50785 | 0.242 | 0.25154 | 0.457 |
| 12612 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 32 | MYO9A | Sponge network | -0.793 | 7.0E-5 | -0.147 | 0.32373 | 0.457 |
| 12613 | LINC01018 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-96-5p | 28 | CADM1 | Sponge network | -2.915 | 0.00015 | -0.9 | 0.00084 | 0.457 |
| 12614 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | EPAS1 | Sponge network | 0.727 | 3.0E-5 | -0.184 | 0.14418 | 0.457 |
| 12615 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | CADM2 | Sponge network | -0.611 | 0.09607 | -3.782 | 0 | 0.457 |
| 12616 | LINC00472 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 34 | TRPS1 | Sponge network | -0.842 | 0.01361 | -0.191 | 0.44109 | 0.457 |
| 12617 | OIP5-AS1 |
hsa-miR-125a-3p;hsa-miR-149-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | CDC27 | Sponge network | 0.421 | 0.00084 | 0.384 | 0 | 0.457 |
| 12618 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | SCN9A | Sponge network | 0.225 | 0.30644 | -0.525 | 0.10593 | 0.457 |
| 12619 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 23 | NR2C2 | Sponge network | 0.659 | 3.0E-5 | 0.21 | 0.03224 | 0.457 |
| 12620 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-590-3p | 15 | EBF1 | Sponge network | 0.178 | 0.29106 | -0.384 | 0.08856 | 0.457 |
| 12621 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 11 | PTPN13 | Sponge network | -0.769 | 0.00035 | -1.208 | 0 | 0.457 |
| 12622 | RP11-531A24.5 |
hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-576-5p | 10 | HERC1 | Sponge network | 0.75 | 0.00054 | -0.196 | 0.08544 | 0.457 |
| 12623 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 16 | SEPT6 | Sponge network | -0.576 | 0.10121 | -0.598 | 0.00181 | 0.457 |
| 12624 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | BCLAF1 | Sponge network | 0.768 | 7.0E-5 | 0.211 | 0.00684 | 0.457 |
| 12625 | PCA3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-940 | 56 | NFIB | Sponge network | -0.709 | 0.18168 | 0.023 | 0.90795 | 0.457 |
| 12626 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p | 32 | SOX5 | Sponge network | 0.214 | 0.18246 | -1.091 | 4.0E-5 | 0.457 |
| 12627 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PHC3 | Sponge network | 1.601 | 0 | 0.212 | 0.0499 | 0.457 |
| 12628 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p | 31 | ZFHX4 | Sponge network | 1.757 | 0 | 0.018 | 0.95574 | 0.457 |
| 12629 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p | 11 | TBX18 | Sponge network | -0.693 | 0.05629 | 0.843 | 0.02945 | 0.457 |
| 12630 | RP11-983P16.4 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 0.41 | 0.02214 | 0.321 | 0.00267 | 0.457 |
| 12631 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | LRIG1 | Sponge network | -0.323 | 0.01372 | -0.537 | 0.00588 | 0.457 |
| 12632 | RP11-13K12.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 57 | DST | Sponge network | -0.154 | 0.78939 | -0.713 | 0.00096 | 0.457 |
| 12633 | RP11-819C21.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 10 | UVRAG | Sponge network | 0.537 | 0.00139 | -0.017 | 0.83865 | 0.457 |
| 12634 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 22 | SYNE1 | Sponge network | -0.246 | 0.35034 | -0.738 | 0.00316 | 0.457 |
| 12635 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | NALCN | Sponge network | 0.385 | 0.10067 | 1.068 | 0.00237 | 0.457 |
| 12636 | ZNF252P-AS1 |
hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 11 | MDM4 | Sponge network | 1.076 | 0 | 0.345 | 0.00762 | 0.457 |
| 12637 | AP001055.6 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-7-1-3p | 11 | PCDH17 | Sponge network | 0.693 | 0.00076 | 0.731 | 2.0E-5 | 0.457 |
| 12638 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-5p;hsa-miR-96-5p | 28 | SCN9A | Sponge network | 0.395 | 0.15451 | -0.525 | 0.10593 | 0.457 |
| 12639 | USP3-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-877-5p | 25 | ASTN1 | Sponge network | -0.162 | 0.47687 | -2.389 | 2.0E-5 | 0.457 |
| 12640 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | GRIN2A | Sponge network | -1.161 | 0.00591 | -2.161 | 0 | 0.457 |
| 12641 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 34 | AKAP11 | Sponge network | 0.782 | 0.00016 | 0.196 | 0.09825 | 0.457 |
| 12642 | LINC00957 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 44 | GRIK3 | Sponge network | -0.421 | 0.0114 | -3.208 | 0 | 0.457 |
| 12643 | RP5-1074L1.4 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p | 14 | RASAL2 | Sponge network | 1.923 | 0 | 0.869 | 0 | 0.457 |
| 12644 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 46 | ERBB4 | Sponge network | -0.942 | 0 | -1.975 | 2.0E-5 | 0.457 |
| 12645 | RP11-244O19.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-3913-5p;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 38 | AFF1 | Sponge network | -0.096 | 0.67958 | -0.384 | 0.00053 | 0.457 |
| 12646 | RP4-613B23.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-539-5p | 24 | DST | Sponge network | -0.309 | 0.1145 | -0.713 | 0.00096 | 0.457 |
| 12647 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-326 | 11 | ITGA5 | Sponge network | -0.611 | 0.09607 | 0.452 | 0.03524 | 0.457 |
| 12648 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | PIK3R1 | Sponge network | 0.532 | 0.00505 | -0.133 | 0.36854 | 0.457 |
| 12649 | RP11-57H14.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PRUNE2 | Sponge network | 0.083 | 0.66405 | -1.733 | 0.00022 | 0.457 |
| 12650 | SNHG14 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-940 | 45 | PTPRT | Sponge network | -1.111 | 0.0003 | -0.907 | 0.03727 | 0.457 |
| 12651 | RP11-148O21.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-362-5p;hsa-miR-501-5p | 13 | PTPRT | Sponge network | 0.233 | 0.81029 | -0.907 | 0.03727 | 0.457 |
| 12652 | ZSCAN16-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | SYT4 | Sponge network | -0.342 | 0.02288 | -3.207 | 0 | 0.457 |
| 12653 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 30 | DDX6 | Sponge network | 0.222 | 0.29133 | 0.091 | 0.36428 | 0.457 |
| 12654 | LINC00473 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p | 12 | NGFR | Sponge network | -1.203 | 0.03881 | -1.271 | 0.01058 | 0.457 |
| 12655 | LINC00473 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-92a-3p | 58 | RUNX1T1 | Sponge network | -1.203 | 0.03881 | -0.344 | 0.2374 | 0.457 |
| 12656 | SERTAD4-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-93-3p | 16 | GRIK3 | Sponge network | -0.932 | 0.00329 | -3.208 | 0 | 0.457 |
| 12657 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-501-3p | 26 | ABI2 | Sponge network | 0.082 | 0.55397 | 0.175 | 0.14787 | 0.457 |
| 12658 | AC010761.8 |
hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 10 | PHF6 | Sponge network | 1.638 | 0 | 0.785 | 0 | 0.457 |
| 12659 | RAMP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 32 | IL17RD | Sponge network | -0.513 | 0.04703 | -0.019 | 0.94873 | 0.457 |
| 12660 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | EPHA5 | Sponge network | -1.439 | 4.0E-5 | -0.957 | 0.01733 | 0.457 |
| 12661 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 42 | RASSF2 | Sponge network | -1.697 | 0.00889 | -0.273 | 0.21961 | 0.457 |
| 12662 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 24 | RASSF8 | Sponge network | -0.513 | 0.04703 | -0.122 | 0.65591 | 0.457 |
| 12663 | LINC00473 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p | 11 | RHOB | Sponge network | -1.203 | 0.03881 | -1.052 | 0 | 0.457 |
| 12664 | RP11-464F9.1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 13 | CDK6 | Sponge network | 0.959 | 0 | 0.991 | 0 | 0.457 |
| 12665 | RP11-2H8.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | SON | Sponge network | 1.089 | 0 | 0.168 | 0.04406 | 0.457 |
| 12666 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 25 | RECK | Sponge network | -0.615 | 0.00015 | -1.043 | 0 | 0.457 |
| 12667 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 48 | EPHA5 | Sponge network | 0.532 | 0.00505 | -0.957 | 0.01733 | 0.457 |
| 12668 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 49 | SSBP2 | Sponge network | -0.096 | 0.67958 | -1.58 | 0 | 0.457 |
| 12669 | LINC00657 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | EXOC8 | Sponge network | 0.91 | 0 | 0.216 | 0.00598 | 0.457 |
| 12670 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 35 | ESR1 | Sponge network | -0.487 | 0.02157 | -1.035 | 3.0E-5 | 0.457 |
| 12671 | LINC00702 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | LRIG1 | Sponge network | -0.737 | 0.06182 | -0.537 | 0.00588 | 0.457 |
| 12672 | HOXA-AS2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | 0.61 | 0.01593 | 0.809 | 0.00544 | 0.457 |
| 12673 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940;hsa-miR-96-5p | 27 | SNX29 | Sponge network | 0.532 | 0.00505 | -0.34 | 0.01371 | 0.457 |
| 12674 | RP11-401P9.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-877-5p | 23 | TET1 | Sponge network | -0.384 | 0.40652 | -0.135 | 0.52138 | 0.457 |
| 12675 | AC025171.1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ATM | Sponge network | 0.998 | 0 | 0.178 | 0.21568 | 0.457 |
| 12676 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 21 | MYO9A | Sponge network | 0.174 | 0.30034 | -0.147 | 0.32373 | 0.457 |
| 12677 | LINC00578 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-744-3p | 16 | PTPN14 | Sponge network | 0.811 | 0.03228 | 0.144 | 0.34595 | 0.457 |
| 12678 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | TMEFF2 | Sponge network | -1.349 | 0.00017 | -2.426 | 0 | 0.457 |
| 12679 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 47 | TMEM132B | Sponge network | -2.452 | 0 | -0.83 | 0.01084 | 0.457 |
| 12680 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | -0.709 | 0.18168 | -1.277 | 0 | 0.457 |
| 12681 | LINC00865 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p | 10 | PRDM11 | Sponge network | -1.249 | 7.0E-5 | -0.252 | 0.15511 | 0.457 |
| 12682 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-582-5p | 11 | MOB1B | Sponge network | 0.993 | 0 | 0.197 | 0.15266 | 0.457 |
| 12683 | RP11-206L10.2 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 16 | CREB1 | Sponge network | 0.793 | 0 | 0.203 | 0.00537 | 0.457 |
| 12684 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RB1CC1 | Sponge network | 1.205 | 0 | 0.175 | 0.12559 | 0.457 |
| 12685 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | 0.374 | 0.20054 | -0.466 | 0.14121 | 0.457 |
| 12686 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-590-3p | 17 | TCF4 | Sponge network | -1.375 | 0.08642 | 0.016 | 0.92271 | 0.457 |
| 12687 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | RASSF8 | Sponge network | -0.187 | 0.23787 | -0.122 | 0.65591 | 0.457 |
| 12688 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | ABCC9 | Sponge network | -0.899 | 6.0E-5 | -0.466 | 0.14121 | 0.457 |
| 12689 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-500a-3p | 22 | HLF | Sponge network | -0.469 | 0.08721 | -2.443 | 0 | 0.457 |
| 12690 | CTD-3064M3.3 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-582-5p | 38 | THRB | Sponge network | -0.469 | 0.08721 | -1.117 | 0.0004 | 0.457 |
| 12691 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | PTPN13 | Sponge network | -0.513 | 0.04703 | -1.208 | 0 | 0.457 |
| 12692 | RP11-890B15.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 14 | ZBTB20 | Sponge network | 0.101 | 0.60919 | -0.122 | 0.57569 | 0.457 |
| 12693 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-455-5p | 16 | RECK | Sponge network | -0.465 | 0.04703 | -1.043 | 0 | 0.457 |
| 12694 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-93-3p | 27 | ST6GAL2 | Sponge network | -0.162 | 0.47687 | -1.201 | 0.00597 | 0.457 |
| 12695 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 36 | CTIF | Sponge network | -0.49 | 0.04946 | -0.389 | 0.00549 | 0.457 |
| 12696 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-5p | 25 | RUNX1T1 | Sponge network | 0.497 | 0.01451 | -0.344 | 0.2374 | 0.457 |
| 12697 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-429 | 13 | ZFHX3 | Sponge network | 1.106 | 0 | 0.389 | 0.01363 | 0.457 |
| 12698 | RP5-1085F17.3 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | KIF5B | Sponge network | 0.541 | 0.00125 | 0.362 | 0.00066 | 0.457 |
| 12699 | TRAF3IP2-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | KLF10 | Sponge network | -0.323 | 0.01372 | -0.783 | 0 | 0.456 |
| 12700 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-671-5p | 32 | ADARB1 | Sponge network | 0.75 | 0.00054 | -0.335 | 0.06631 | 0.456 |
| 12701 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 61 | CCND2 | Sponge network | -0.739 | 0.0574 | -0.267 | 0.3112 | 0.456 |
| 12702 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 21 | TET1 | Sponge network | -0.405 | 0.22013 | -0.135 | 0.52138 | 0.456 |
| 12703 | RP11-707P17.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 18 | MDM4 | Sponge network | 1.877 | 0 | 0.345 | 0.00762 | 0.456 |
| 12704 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | MYO9A | Sponge network | 0.272 | 0.44692 | -0.147 | 0.32373 | 0.456 |
| 12705 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | MOB1B | Sponge network | 0.349 | 0.01695 | 0.197 | 0.15266 | 0.456 |
| 12706 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 30 | EPHA7 | Sponge network | 0.056 | 0.81195 | -2.197 | 3.0E-5 | 0.456 |
| 12707 | LINC00092 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-590-3p | 14 | LRIG1 | Sponge network | -1.886 | 0 | -0.537 | 0.00588 | 0.456 |
| 12708 | ARHGEF26-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | NAV3 | Sponge network | -2.46 | 0 | 0.212 | 0.47729 | 0.456 |
| 12709 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-655-3p | 22 | SON | Sponge network | 1.063 | 0 | 0.168 | 0.04406 | 0.456 |
| 12710 | AC005154.6 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 15 | SHPRH | Sponge network | 0.848 | 0 | 0.098 | 0.40994 | 0.456 |
| 12711 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-590-3p | 16 | PTPN11 | Sponge network | 1.601 | 0 | 0.431 | 2.0E-5 | 0.456 |
| 12712 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p | 39 | SOX5 | Sponge network | -0.793 | 7.0E-5 | -1.091 | 4.0E-5 | 0.456 |
| 12713 | MRVI1-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 16 | PDE4DIP | Sponge network | -1.092 | 4.0E-5 | -0.282 | 0.0257 | 0.456 |
| 12714 | MEG3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | TET1 | Sponge network | 0.249 | 0.35902 | -0.135 | 0.52138 | 0.456 |
| 12715 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 25 | KCNA6 | Sponge network | -0.421 | 0.0114 | -1.531 | 0.00013 | 0.456 |
| 12716 | NR2F1-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 19 | FMNL3 | Sponge network | -0.49 | 0.04946 | 0.555 | 4.0E-5 | 0.456 |
| 12717 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 40 | TSHZ3 | Sponge network | -0.942 | 0 | -0.556 | 0.01516 | 0.456 |
| 12718 | LINC00641 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 34 | PRDM2 | Sponge network | -0.222 | 0.32445 | -0.258 | 0.00721 | 0.456 |
| 12719 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-26b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 10 | DMD | Sponge network | -1.425 | 0.04204 | -1.506 | 0 | 0.456 |
| 12720 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 67 | THRB | Sponge network | -0.576 | 0.10121 | -1.117 | 0.0004 | 0.456 |
| 12721 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p | 13 | ARNT2 | Sponge network | 0.176 | 0.37402 | -0.332 | 0.25851 | 0.456 |
| 12722 | LINC00472 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | NAV3 | Sponge network | -0.842 | 0.01361 | 0.212 | 0.47729 | 0.456 |
| 12723 | RP11-620J15.3 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | MED12L | Sponge network | -0.739 | 0.00044 | -0.194 | 0.55299 | 0.456 |
| 12724 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 43 | DTNA | Sponge network | -0.738 | 0.21233 | -1.648 | 1.0E-5 | 0.456 |
| 12725 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 20 | TIMP3 | Sponge network | -4.546 | 0 | -0.546 | 0.02307 | 0.456 |
| 12726 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-532-3p;hsa-miR-93-5p | 12 | DAB2 | Sponge network | -0.777 | 0.16667 | 0.431 | 0.0113 | 0.456 |
| 12727 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-93-5p | 43 | EPHA5 | Sponge network | -0.096 | 0.67958 | -0.957 | 0.01733 | 0.456 |
| 12728 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-5p | 33 | ERC1 | Sponge network | 0.101 | 0.60919 | -0.108 | 0.38794 | 0.456 |
| 12729 | EMX2OS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 91 | LPP | Sponge network | -0.777 | 0.1134 | -0.459 | 0.04475 | 0.456 |
| 12730 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | ITGB3 | Sponge network | -1.575 | 6.0E-5 | -0.011 | 0.95751 | 0.456 |
| 12731 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 29 | DDX6 | Sponge network | 0.559 | 0.00026 | 0.091 | 0.36428 | 0.456 |
| 12732 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p | 15 | IGF2 | Sponge network | 0.572 | 0.01722 | 0.848 | 0.03842 | 0.456 |
| 12733 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 32 | RBMS3 | Sponge network | -0.246 | 0.35034 | -0.519 | 0.03551 | 0.456 |
| 12734 | PART1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 21 | EZH1 | Sponge network | -2.925 | 0 | -0.564 | 1.0E-5 | 0.456 |
| 12735 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 32 | AKAP11 | Sponge network | 0.603 | 0.00127 | 0.196 | 0.09825 | 0.456 |
| 12736 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-93-3p | 18 | PTPN11 | Sponge network | 0.72 | 0.0001 | 0.431 | 2.0E-5 | 0.456 |
| 12737 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 13 | SYNE1 | Sponge network | 0.497 | 0.01451 | -0.738 | 0.00316 | 0.456 |
| 12738 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | NCOA1 | Sponge network | -2.452 | 0 | -0.432 | 0 | 0.456 |
| 12739 | CTD-2002H8.2 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | BMPR2 | Sponge network | 0.931 | 1.0E-5 | 0.206 | 0.0635 | 0.456 |
| 12740 | WDFY3-AS2 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-7-5p | 10 | MLLT11 | Sponge network | -0.942 | 0 | -0.661 | 0.01733 | 0.456 |
| 12741 | HAND2-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | INPP4A | Sponge network | -1.697 | 0.00889 | 0.07 | 0.53563 | 0.456 |
| 12742 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 52 | CADM2 | Sponge network | -0.405 | 0.22013 | -3.782 | 0 | 0.456 |
| 12743 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 32 | IGF1 | Sponge network | -0.709 | 0.18168 | -0.204 | 0.5898 | 0.456 |
| 12744 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | ATRX | Sponge network | 0.72 | 0.0001 | 0.261 | 0.04408 | 0.456 |
| 12745 | BOLA3-AS1 |
hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 33 | EPHA7 | Sponge network | -0.246 | 0.35034 | -2.197 | 3.0E-5 | 0.456 |
| 12746 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-429;hsa-miR-484;hsa-miR-98-5p | 15 | PNRC1 | Sponge network | -0.976 | 0.00025 | -0.646 | 0 | 0.456 |
| 12747 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-33a-3p | 14 | ESR1 | Sponge network | -1.255 | 0.00981 | -1.035 | 3.0E-5 | 0.456 |
| 12748 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TLL1 | Sponge network | -1.161 | 0.00591 | -1.195 | 3.0E-5 | 0.456 |
| 12749 | TRAF3IP2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | CREB1 | Sponge network | -0.323 | 0.01372 | 0.203 | 0.00537 | 0.456 |
| 12750 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935 | 15 | RELN | Sponge network | -0.896 | 0.00099 | -2.403 | 0 | 0.456 |
| 12751 | RP11-175O19.4 |
hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p | 17 | ABL2 | Sponge network | 0.917 | 0 | 0.82 | 0 | 0.456 |
| 12752 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 33 | DDX6 | Sponge network | 0.083 | 0.66405 | 0.091 | 0.36428 | 0.456 |
| 12753 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-495-3p | 11 | TRIM33 | Sponge network | 0.924 | 0 | 0.402 | 7.0E-5 | 0.456 |
| 12754 | RP11-359E10.1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-576-5p;hsa-miR-582-5p | 12 | KLF10 | Sponge network | 0.299 | 0.57722 | -0.783 | 0 | 0.456 |
| 12755 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 38 | BMPR2 | Sponge network | 0.91 | 0 | 0.206 | 0.0635 | 0.456 |
| 12756 | MEG3 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-935 | 13 | STAT5B | Sponge network | 0.249 | 0.35902 | -0.182 | 0.1082 | 0.456 |
| 12757 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 11 | NEDD4 | Sponge network | -0.517 | 0.02935 | 0.02 | 0.89834 | 0.456 |
| 12758 | CALML3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-766-3p | 31 | IL17RD | Sponge network | 0.95 | 0.15943 | -0.019 | 0.94873 | 0.456 |
| 12759 | LINC01018 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 13 | STAT5B | Sponge network | -2.915 | 0.00015 | -0.182 | 0.1082 | 0.456 |
| 12760 | RP11-159D12.2 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | KIF5B | Sponge network | 1.254 | 0 | 0.362 | 0.00066 | 0.456 |
| 12761 | RP11-532F6.3 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | HERC1 | Sponge network | -0.896 | 0.00099 | -0.196 | 0.08544 | 0.456 |
| 12762 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-935 | 13 | EBF1 | Sponge network | -0.376 | 0.00337 | -0.384 | 0.08856 | 0.456 |
| 12763 | FLNB-AS1 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-9-5p | 20 | CCDC6 | Sponge network | 1.163 | 0.00018 | 0.27 | 0.02555 | 0.456 |
| 12764 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 24 | EPHA3 | Sponge network | 0.494 | 0.00339 | 0.185 | 0.53588 | 0.456 |
| 12765 | PWAR6 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 18 | GLIPR1 | Sponge network | -1.575 | 6.0E-5 | 0.146 | 0.3276 | 0.456 |
| 12766 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 40 | ZFHX3 | Sponge network | -0.709 | 0.18168 | 0.389 | 0.01363 | 0.456 |
| 12767 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | TSHZ3 | Sponge network | 0.101 | 0.60919 | -0.556 | 0.01516 | 0.456 |
| 12768 | LINC00702 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | HERC1 | Sponge network | -0.737 | 0.06182 | -0.196 | 0.08544 | 0.456 |
| 12769 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p | 16 | TACC1 | Sponge network | -0.309 | 0.1145 | -0.362 | 0.05406 | 0.456 |
| 12770 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | MYO9A | Sponge network | 0.614 | 1.0E-5 | -0.147 | 0.32373 | 0.456 |
| 12771 | TUG1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-628-5p;hsa-miR-766-3p | 11 | KIAA1024 | Sponge network | 0.972 | 0 | 1.083 | 0 | 0.456 |
| 12772 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | CNTNAP1 | Sponge network | -1.216 | 0 | 0.358 | 0.13727 | 0.456 |
| 12773 | GNG12-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | RYR2 | Sponge network | -0.793 | 7.0E-5 | -1.466 | 0.00031 | 0.456 |
| 12774 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-96-5p | 20 | SNX29 | Sponge network | -1.654 | 0.00144 | -0.34 | 0.01371 | 0.456 |
| 12775 | DNM3OS |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | NIN | Sponge network | 0.572 | 0.01722 | -0.253 | 0.13794 | 0.456 |
| 12776 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p | 12 | RELN | Sponge network | -0.611 | 0.09607 | -2.403 | 0 | 0.456 |
| 12777 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-93-3p | 49 | RUNX1T1 | Sponge network | -0.773 | 0.04073 | -0.344 | 0.2374 | 0.456 |
| 12778 | AP001347.6 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PRUNE2 | Sponge network | -1.161 | 0.00591 | -1.733 | 0.00022 | 0.456 |
| 12779 | DNM3OS |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | RYR2 | Sponge network | 0.572 | 0.01722 | -1.466 | 0.00031 | 0.456 |
| 12780 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | CDH10 | Sponge network | -2.46 | 0 | -2.397 | 0 | 0.456 |
| 12781 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b | 10 | NR2C2 | Sponge network | 0.37 | 0.27614 | 0.21 | 0.03224 | 0.456 |
| 12782 | TUG1 |
hsa-miR-139-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-495-3p | 11 | GSK3B | Sponge network | 0.972 | 0 | 0.266 | 0.00045 | 0.456 |
| 12783 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-93-5p | 16 | FAT4 | Sponge network | -0.18 | 0.20343 | -0.354 | 0.14694 | 0.456 |
| 12784 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | ATRX | Sponge network | 0.267 | 0.2937 | 0.261 | 0.04408 | 0.456 |
| 12785 | RP11-156P1.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p | 12 | STARD13 | Sponge network | 0.214 | 0.18246 | -0.187 | 0.27383 | 0.456 |
| 12786 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | FSTL5 | Sponge network | 0.214 | 0.18246 | -1.3 | 0.01619 | 0.456 |
| 12787 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-590-3p | 20 | HIP1 | Sponge network | -0.096 | 0.67958 | 0.774 | 0 | 0.456 |
| 12788 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 19 | DIP2C | Sponge network | 0.299 | 0.57722 | -0.436 | 0.01713 | 0.456 |
| 12789 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | -0.896 | 0.00099 | -1.3 | 0.01619 | 0.456 |
| 12790 | PART1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | -2.925 | 0 | -0.57 | 0 | 0.456 |
| 12791 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-590-3p | 15 | TIMP3 | Sponge network | -0.811 | 2.0E-5 | -0.546 | 0.02307 | 0.456 |
| 12792 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 71 | BMPR2 | Sponge network | 0.421 | 0.00084 | 0.206 | 0.0635 | 0.456 |
| 12793 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 14 | NALCN | Sponge network | -1.575 | 6.0E-5 | 1.068 | 0.00237 | 0.456 |
| 12794 | ZNF32-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-766-3p | 19 | MDM4 | Sponge network | 1.552 | 7.0E-5 | 0.345 | 0.00762 | 0.456 |
| 12795 | RP11-400K9.4 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | PPM1L | Sponge network | -0.611 | 0.09607 | -0.537 | 0.00616 | 0.456 |
| 12796 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p | 11 | MLLT10 | Sponge network | 0.793 | 0 | 0.105 | 0.33292 | 0.456 |
| 12797 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | NEDD4 | Sponge network | 0.572 | 0.01722 | 0.02 | 0.89834 | 0.456 |
| 12798 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 11 | CLASP2 | Sponge network | 0.178 | 0.29106 | -0.038 | 0.71 | 0.456 |
| 12799 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p | 29 | BCL6 | Sponge network | -1.697 | 0.00889 | -0.211 | 0.19489 | 0.456 |
| 12800 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 54 | RUNX1T1 | Sponge network | 0.214 | 0.18246 | -0.344 | 0.2374 | 0.456 |
| 12801 | RP11-693J15.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ERG | Sponge network | -0.72 | 0.24239 | 0.002 | 0.99011 | 0.456 |
| 12802 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 38 | EPHA7 | Sponge network | -0.513 | 0.04703 | -2.197 | 3.0E-5 | 0.456 |
| 12803 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-5p | 18 | RET | Sponge network | -1.249 | 7.0E-5 | -0.833 | 0.03517 | 0.456 |
| 12804 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | NF1 | Sponge network | 0.93 | 0 | 0.51 | 0 | 0.456 |
| 12805 | RP11-35G9.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-375 | 12 | NIN | Sponge network | 0.622 | 0.00015 | -0.253 | 0.13794 | 0.456 |
| 12806 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | CACNA2D1 | Sponge network | -1.216 | 0 | -0.846 | 0.00675 | 0.456 |
| 12807 | RP11-384O8.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 18 | ITPKB | Sponge network | -0.738 | 0.21233 | -0.704 | 0.00106 | 0.456 |
| 12808 | RP11-401P9.1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-511-5p | 13 | ZFHX3 | Sponge network | 1.524 | 0.1098 | 0.389 | 0.01363 | 0.456 |
| 12809 | AC011526.1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | DCLK1 | Sponge network | 0.342 | 0.03129 | -1.324 | 0.00014 | 0.456 |
| 12810 | MEG3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | 0.249 | 0.35902 | -1.733 | 0.00022 | 0.456 |
| 12811 | LINC00092 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p | 22 | KCNA6 | Sponge network | -1.886 | 0 | -1.531 | 0.00013 | 0.456 |
| 12812 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-625-5p | 10 | ZMYM2 | Sponge network | 1.134 | 0 | 0.279 | 0.01273 | 0.456 |
| 12813 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 11 | NALCN | Sponge network | -0.896 | 0.00099 | 1.068 | 0.00237 | 0.455 |
| 12814 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-542-3p | 13 | ST6GAL2 | Sponge network | -0.517 | 0.02935 | -1.201 | 0.00597 | 0.455 |
| 12815 | RP11-248G5.8 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p | 24 | CREB1 | Sponge network | 1.294 | 0 | 0.203 | 0.00537 | 0.455 |
| 12816 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 15 | AHNAK | Sponge network | -1.654 | 0.00144 | -1.207 | 0 | 0.455 |
| 12817 | LINC00654 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p | 12 | THSD7A | Sponge network | 0.374 | 0.20054 | 0.242 | 0.25154 | 0.455 |
| 12818 | TRAF3IP2-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 11 | KCTD8 | Sponge network | -0.323 | 0.01372 | -3.359 | 0 | 0.455 |
| 12819 | RP11-401P9.4 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p | 13 | FLNA | Sponge network | -0.384 | 0.40652 | -0.771 | 0.01377 | 0.455 |
| 12820 | CTC-490E21.10 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-93-3p | 24 | BMPR2 | Sponge network | 1.46 | 1.0E-5 | 0.206 | 0.0635 | 0.455 |
| 12821 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 36 | DMD | Sponge network | -0.355 | 0.0077 | -1.506 | 0 | 0.455 |
| 12822 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 16 | PIK3C2A | Sponge network | 1.308 | 0 | 0.061 | 0.53776 | 0.455 |
| 12823 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | -2.117 | 0.00161 | -1.277 | 0 | 0.455 |
| 12824 | BOLA3-AS1 |
hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 34 | BNC2 | Sponge network | -0.246 | 0.35034 | -0.491 | 0.09988 | 0.455 |
| 12825 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ZMYND11 | Sponge network | 0.222 | 0.29133 | -0.2 | 0.05449 | 0.455 |
| 12826 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-5p | 14 | RNF144A | Sponge network | -0.693 | 0.05629 | 0.594 | 0.0009 | 0.455 |
| 12827 | MIR143HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | CBFA2T3 | Sponge network | -0.739 | 0.0574 | -1.221 | 1.0E-5 | 0.455 |
| 12828 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p | 13 | PDZD2 | Sponge network | 0.997 | 0.00066 | -0.909 | 3.0E-5 | 0.455 |
| 12829 | FENDRR |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3E | Sponge network | -1.281 | 0.00012 | -1.717 | 0.00137 | 0.455 |
| 12830 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-7-1-3p | 25 | MITF | Sponge network | -0.842 | 0.01361 | -0.769 | 0.00022 | 0.455 |
| 12831 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 24 | BCL6 | Sponge network | -0.777 | 0.1134 | -0.211 | 0.19489 | 0.455 |
| 12832 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.225 | 0.30644 | -0.57 | 0 | 0.455 |
| 12833 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 54 | CADM2 | Sponge network | -0.769 | 0.00035 | -3.782 | 0 | 0.455 |
| 12834 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p | 10 | SLC5A3 | Sponge network | 0.673 | 0 | 0.493 | 5.0E-5 | 0.455 |
| 12835 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p | 16 | RET | Sponge network | -2.531 | 0 | -0.833 | 0.03517 | 0.455 |
| 12836 | RP5-994D16.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | AKAP11 | Sponge network | 0.252 | 0.08508 | 0.196 | 0.09825 | 0.455 |
| 12837 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | PIK3C2A | Sponge network | -0.222 | 0.32445 | 0.061 | 0.53776 | 0.455 |
| 12838 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-577 | 16 | ST6GAL2 | Sponge network | -0.473 | 0.07602 | -1.201 | 0.00597 | 0.455 |
| 12839 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450a-5p;hsa-miR-497-5p;hsa-miR-628-5p | 10 | CHD2 | Sponge network | 1.923 | 0 | 0.049 | 0.55371 | 0.455 |
| 12840 | U91328.19 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | -0.303 | 0.21002 | -0.564 | 1.0E-5 | 0.455 |
| 12841 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 10 | ZHX2 | Sponge network | -0.976 | 0.00025 | -0.192 | 0.18459 | 0.455 |
| 12842 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429 | 16 | TAL1 | Sponge network | -2.69 | 0.0001 | -0.462 | 0.00574 | 0.455 |
| 12843 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | ZNF208 | Sponge network | -0.487 | 0.02157 | -0.4 | 0.22381 | 0.455 |
| 12844 | HHIP-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | PCDH9 | Sponge network | -0.737 | 0.10822 | -1.823 | 4.0E-5 | 0.455 |
| 12845 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p | 12 | DLC1 | Sponge network | -0.187 | 0.23787 | -0.382 | 0.05038 | 0.455 |
| 12846 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -0.154 | 0.78939 | -0.354 | 0.14694 | 0.455 |
| 12847 | RP11-535M15.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-532-3p | 15 | PBX1 | Sponge network | -1.14 | 0.03513 | -1.206 | 0 | 0.455 |
| 12848 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | PIK3C2A | Sponge network | 1.055 | 0 | 0.061 | 0.53776 | 0.455 |
| 12849 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | SCN9A | Sponge network | -0.021 | 0.93598 | -0.525 | 0.10593 | 0.455 |
| 12850 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 21 | TRIM33 | Sponge network | 0.705 | 0.00014 | 0.402 | 7.0E-5 | 0.455 |
| 12851 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CLASP2 | Sponge network | 0.931 | 1.0E-5 | -0.038 | 0.71 | 0.455 |
| 12852 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | PNRC1 | Sponge network | -0.323 | 0.01372 | -0.646 | 0 | 0.455 |
| 12853 | RP11-283I3.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | ABI2 | Sponge network | 0.614 | 1.0E-5 | 0.175 | 0.14787 | 0.455 |
| 12854 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-93-3p | 30 | IL6ST | Sponge network | -4.463 | 0.00025 | -0.402 | 0.00676 | 0.455 |
| 12855 | RP11-473I1.9 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p | 13 | MACF1 | Sponge network | 0.683 | 1.0E-5 | 0.019 | 0.90335 | 0.455 |
| 12856 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-3p | 12 | RYR2 | Sponge network | -2.076 | 0.00548 | -1.466 | 0.00031 | 0.455 |
| 12857 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | KAT6B | Sponge network | -2.452 | 0 | -0.478 | 0.00018 | 0.455 |
| 12858 | RP11-175O19.4 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-497-5p | 13 | ERCC6 | Sponge network | 0.917 | 0 | 0.23 | 0.015 | 0.455 |
| 12859 | GHRLOS |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-361-5p | 12 | FAM135B | Sponge network | -1.942 | 0 | -3.978 | 0 | 0.455 |
| 12860 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | MLLT10 | Sponge network | 0.826 | 1.0E-5 | 0.105 | 0.33292 | 0.455 |
| 12861 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 21 | KALRN | Sponge network | -1.281 | 0.00012 | -0.819 | 1.0E-5 | 0.455 |
| 12862 | LINC00641 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 57 | DTNA | Sponge network | -0.222 | 0.32445 | -1.648 | 1.0E-5 | 0.455 |
| 12863 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | LATS2 | Sponge network | -0.769 | 0.00035 | 0.159 | 0.2304 | 0.455 |
| 12864 | LINC00957 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p | 52 | DST | Sponge network | -0.421 | 0.0114 | -0.713 | 0.00096 | 0.455 |
| 12865 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-874-3p | 30 | TACC1 | Sponge network | 0.214 | 0.18246 | -0.362 | 0.05406 | 0.455 |
| 12866 | RP11-620J15.3 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | ID4 | Sponge network | -0.739 | 0.00044 | -1.644 | 0 | 0.455 |
| 12867 | RP11-999E24.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-744-3p | 19 | DIP2C | Sponge network | -0.693 | 0.05629 | -0.436 | 0.01713 | 0.455 |
| 12868 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 28 | PGR | Sponge network | -0.738 | 0.21233 | -1.704 | 0 | 0.455 |
| 12869 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-940 | 13 | PBX1 | Sponge network | -1.483 | 9.0E-5 | -1.206 | 0 | 0.455 |
| 12870 | RP4-613B23.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p | 16 | MEF2C | Sponge network | -0.309 | 0.1145 | -0.499 | 0.01448 | 0.455 |
| 12871 | CTD-3099C6.9 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-429 | 11 | DOCK4 | Sponge network | 1.361 | 0 | 0.502 | 0.00108 | 0.455 |
| 12872 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-7-1-3p | 20 | ANK2 | Sponge network | -0.72 | 0.24239 | -1.603 | 1.0E-5 | 0.455 |
| 12873 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 29 | MYO9A | Sponge network | -0.031 | 0.89063 | -0.147 | 0.32373 | 0.455 |
| 12874 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | SLC6A15 | Sponge network | -1.575 | 6.0E-5 | -2.373 | 2.0E-5 | 0.455 |
| 12875 | WDFY3-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 21 | CDH23 | Sponge network | -0.942 | 0 | -0.974 | 0.00034 | 0.455 |
| 12876 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-539-5p | 19 | CADM2 | Sponge network | -0.309 | 0.1145 | -3.782 | 0 | 0.455 |
| 12877 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 20 | CDH19 | Sponge network | -0.769 | 0.00035 | -3.434 | 0 | 0.455 |
| 12878 | AC004540.4 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-484 | 13 | SOX5 | Sponge network | -2.549 | 0.00528 | -1.091 | 4.0E-5 | 0.455 |
| 12879 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 19 | CNTNAP3 | Sponge network | -1.216 | 0 | -1.465 | 0.00033 | 0.455 |
| 12880 | PWAR6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | FYN | Sponge network | -1.575 | 6.0E-5 | -0.131 | 0.37051 | 0.455 |
| 12881 | SNHG14 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 30 | ZMYND11 | Sponge network | -1.111 | 0.0003 | -0.2 | 0.05449 | 0.455 |
| 12882 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 39 | SASH1 | Sponge network | 0.997 | 0.00066 | -0.502 | 0.00175 | 0.455 |
| 12883 | JAZF1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 76 | LPP | Sponge network | -0.769 | 0.00035 | -0.459 | 0.04475 | 0.455 |
| 12884 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-629-3p | 26 | EPHA7 | Sponge network | -0.154 | 0.53954 | -2.197 | 3.0E-5 | 0.455 |
| 12885 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | TSHZ3 | Sponge network | 0.532 | 0.00505 | -0.556 | 0.01516 | 0.455 |
| 12886 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 48 | AR | Sponge network | -0.513 | 0.04703 | -1.554 | 0 | 0.455 |
| 12887 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 25 | ZFHX3 | Sponge network | -1.092 | 4.0E-5 | 0.389 | 0.01363 | 0.455 |
| 12888 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PDGFC | Sponge network | -0.612 | 0.00094 | -0.33 | 0.06483 | 0.455 |
| 12889 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-582-5p | 34 | IL6ST | Sponge network | -2.095 | 0.00112 | -0.402 | 0.00676 | 0.455 |
| 12890 | ARHGEF26-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p | 18 | CDH23 | Sponge network | -2.46 | 0 | -0.974 | 0.00034 | 0.455 |
| 12891 | RP11-536O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-502-5p | 14 | AR | Sponge network | -0.074 | 0.90758 | -1.554 | 0 | 0.455 |
| 12892 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | ZMYM2 | Sponge network | 0.959 | 0 | 0.279 | 0.01273 | 0.455 |
| 12893 | AC003090.1 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | RIMS2 | Sponge network | -2.117 | 0.00161 | -1.365 | 0.00208 | 0.455 |
| 12894 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 14 | SPG20 | Sponge network | -0.342 | 0.02288 | -1.16 | 0 | 0.455 |
| 12895 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 12 | PLA2R1 | Sponge network | -0.129 | 0.57177 | -0.265 | 0.24676 | 0.455 |
| 12896 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PDGFC | Sponge network | 0.997 | 0.00066 | -0.33 | 0.06483 | 0.455 |
| 12897 | LINC00473 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-454-3p | 10 | THSD7A | Sponge network | -1.203 | 0.03881 | 0.242 | 0.25154 | 0.455 |
| 12898 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 32 | BCL2 | Sponge network | 0.4 | 0.14611 | -0.899 | 0 | 0.455 |
| 12899 | RP11-13K12.1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-455-5p | 10 | MACF1 | Sponge network | -0.154 | 0.78939 | 0.019 | 0.90335 | 0.455 |
| 12900 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 13 | TRIM3 | Sponge network | -1.281 | 0.00012 | -0.577 | 0 | 0.455 |
| 12901 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | SPG20 | Sponge network | 0.225 | 0.30644 | -1.16 | 0 | 0.455 |
| 12902 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-628-5p | 14 | TRIM33 | Sponge network | 0.972 | 0 | 0.402 | 7.0E-5 | 0.455 |
| 12903 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 35 | NRXN1 | Sponge network | -0.899 | 6.0E-5 | -3.778 | 0 | 0.455 |
| 12904 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | SASH1 | Sponge network | -1.575 | 6.0E-5 | -0.502 | 0.00175 | 0.455 |
| 12905 | MEG3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 22 | CADM3 | Sponge network | 0.249 | 0.35902 | -3.663 | 0 | 0.455 |
| 12906 | HOXB-AS2 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-576-5p | 14 | MYO9A | Sponge network | 1.316 | 0.01106 | -0.147 | 0.32373 | 0.455 |
| 12907 | LINC00472 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | LRIG1 | Sponge network | -0.842 | 0.01361 | -0.537 | 0.00588 | 0.455 |
| 12908 | RP11-968O1.5 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-505-5p;hsa-miR-532-3p | 12 | MRVI1 | Sponge network | -0.829 | 0.01343 | -1.051 | 0.00022 | 0.455 |
| 12909 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-34a-5p | 16 | BCL6 | Sponge network | -0.611 | 0.09607 | -0.211 | 0.19489 | 0.455 |
| 12910 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p | 13 | ADARB1 | Sponge network | -0.932 | 0.00329 | -0.335 | 0.06631 | 0.455 |
| 12911 | RP11-244O19.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-590-3p | 24 | BRD3 | Sponge network | -0.096 | 0.67958 | 0.37 | 0.00013 | 0.455 |
| 12912 | PWAR6 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 20 | KLF10 | Sponge network | -1.575 | 6.0E-5 | -0.783 | 0 | 0.455 |
| 12913 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-374a-5p | 11 | FAT4 | Sponge network | -0.693 | 0.05629 | -0.354 | 0.14694 | 0.455 |
| 12914 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-98-5p | 16 | NTRK3 | Sponge network | 1.344 | 0 | -1.77 | 3.0E-5 | 0.455 |
| 12915 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 25 | KIT | Sponge network | -0.942 | 0 | -2.184 | 0 | 0.455 |
| 12916 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p | 28 | RICTOR | Sponge network | 1.308 | 0 | 0.388 | 0.00188 | 0.455 |
| 12917 | RASSF8-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | PRKCB | Sponge network | 0.335 | 0.20596 | -1.313 | 2.0E-5 | 0.455 |
| 12918 | RP11-326G21.1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ITPKB | Sponge network | -0.021 | 0.93598 | -0.704 | 0.00106 | 0.455 |
| 12919 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-98-5p | 40 | PBX1 | Sponge network | -1.216 | 0 | -1.206 | 0 | 0.455 |
| 12920 | NDUFA6-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | HERC1 | Sponge network | -0.355 | 0.0077 | -0.196 | 0.08544 | 0.455 |
| 12921 | PWAR6 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 35 | RIMS2 | Sponge network | -1.575 | 6.0E-5 | -1.365 | 0.00208 | 0.455 |
| 12922 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 19 | EPHA7 | Sponge network | -0.187 | 0.23787 | -2.197 | 3.0E-5 | 0.455 |
| 12923 | RP11-819C21.1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 18 | BAZ2B | Sponge network | 0.537 | 0.00139 | -0.276 | 0.02833 | 0.455 |
| 12924 | RP5-1198O20.4 |
hsa-miR-126-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 16 | CXCL12 | Sponge network | 0.997 | 0.00066 | -1.421 | 0 | 0.455 |
| 12925 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-935 | 19 | PCDH10 | Sponge network | -0.384 | 0.40652 | -1.644 | 0.0078 | 0.455 |
| 12926 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-671-5p | 27 | KCNA6 | Sponge network | -2.69 | 0.0001 | -1.531 | 0.00013 | 0.455 |
| 12927 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | TAL1 | Sponge network | -0.096 | 0.67958 | -0.462 | 0.00574 | 0.454 |
| 12928 | BZRAP1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-7-5p | 20 | KCNJ5 | Sponge network | -0.465 | 0.04703 | -0.281 | 0.3339 | 0.454 |
| 12929 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-96-5p | 15 | LRIG1 | Sponge network | -0.517 | 0.02935 | -0.537 | 0.00588 | 0.454 |
| 12930 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 46 | RUNX1T1 | Sponge network | 0.4 | 0.14611 | -0.344 | 0.2374 | 0.454 |
| 12931 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 34 | ATRX | Sponge network | 0.222 | 0.29133 | 0.261 | 0.04408 | 0.454 |
| 12932 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p | 23 | PGR | Sponge network | -0.18 | 0.20343 | -1.704 | 0 | 0.454 |
| 12933 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-577;hsa-miR-590-3p | 30 | PRKAB2 | Sponge network | -0.793 | 7.0E-5 | -0.367 | 0.01371 | 0.454 |
| 12934 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | PCDH9 | Sponge network | 0.178 | 0.29106 | -1.823 | 4.0E-5 | 0.454 |
| 12935 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-532-3p;hsa-miR-93-3p | 11 | CNTNAP1 | Sponge network | -0.384 | 0.40652 | 0.358 | 0.13727 | 0.454 |
| 12936 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 56 | PRKAA2 | Sponge network | -1.216 | 0 | -2.462 | 0 | 0.454 |
| 12937 | C20orf166-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-664a-3p;hsa-miR-93-3p | 24 | ST6GAL2 | Sponge network | -2.69 | 0.0001 | -1.201 | 0.00597 | 0.454 |
| 12938 | FENDRR |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | RYR2 | Sponge network | -1.281 | 0.00012 | -1.466 | 0.00031 | 0.454 |
| 12939 | RP11-244O19.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 17 | SORCS1 | Sponge network | -0.096 | 0.67958 | -3.492 | 0 | 0.454 |
| 12940 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | NR2C2 | Sponge network | 0.272 | 0.44692 | 0.21 | 0.03224 | 0.454 |
| 12941 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 23 | NR2C2 | Sponge network | 0.415 | 0.14657 | 0.21 | 0.03224 | 0.454 |
| 12942 | LINC00672 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-590-3p | 22 | DLC1 | Sponge network | -0.227 | 0.31657 | -0.382 | 0.05038 | 0.454 |
| 12943 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | PEG3 | Sponge network | 0.064 | 0.72934 | -1.74 | 0 | 0.454 |
| 12944 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 41 | HLF | Sponge network | -0.777 | 0.1134 | -2.443 | 0 | 0.454 |
| 12945 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 20 | CRTC3 | Sponge network | -0.303 | 0.21002 | 0.164 | 0.16786 | 0.454 |
| 12946 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 28 | EPHA3 | Sponge network | -0.769 | 0.00035 | 0.185 | 0.53588 | 0.454 |
| 12947 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-93-5p | 23 | LRP1B | Sponge network | -1.654 | 0.00144 | -2.163 | 0 | 0.454 |
| 12948 | MAPKAPK5-AS1 |
hsa-miR-125b-2-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p | 11 | PAWR | Sponge network | 0.765 | 0 | 0.636 | 0 | 0.454 |
| 12949 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-655-3p | 28 | LATS1 | Sponge network | 0.72 | 0.0001 | 0.109 | 0.34208 | 0.454 |
| 12950 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 27 | SASH1 | Sponge network | -0.096 | 0.67958 | -0.502 | 0.00175 | 0.454 |
| 12951 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | ETV1 | Sponge network | -0.576 | 0.10121 | -0.096 | 0.67674 | 0.454 |
| 12952 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | AHNAK | Sponge network | -0.421 | 0.0114 | -1.207 | 0 | 0.454 |
| 12953 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-495-3p | 11 | ATRX | Sponge network | 1.597 | 0 | 0.261 | 0.04408 | 0.454 |
| 12954 | RP11-119F7.5 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 15 | CELF4 | Sponge network | 0.225 | 0.30644 | -1.135 | 0.00344 | 0.454 |
| 12955 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH11 | Sponge network | 1.302 | 7.0E-5 | 1.427 | 0 | 0.454 |
| 12956 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-590-5p | 21 | MYO9A | Sponge network | -0.384 | 0.40652 | -0.147 | 0.32373 | 0.454 |
| 12957 | SERTAD4-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-502-5p | 17 | IL17RD | Sponge network | -0.932 | 0.00329 | -0.019 | 0.94873 | 0.454 |
| 12958 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 43 | SOX5 | Sponge network | -0.899 | 6.0E-5 | -1.091 | 4.0E-5 | 0.454 |
| 12959 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p | 51 | THRB | Sponge network | -0.18 | 0.20343 | -1.117 | 0.0004 | 0.454 |
| 12960 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 31 | PRKAB2 | Sponge network | 0.75 | 0.00054 | -0.367 | 0.01371 | 0.454 |
| 12961 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-590-3p | 11 | ZNF292 | Sponge network | 0.873 | 0 | 0.276 | 0.04148 | 0.454 |
| 12962 | RP11-473I1.9 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | 0.683 | 1.0E-5 | 0.524 | 0.00017 | 0.454 |
| 12963 | CTD-2089N3.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p | 11 | ZFHX4 | Sponge network | -0.314 | 0.62033 | 0.018 | 0.95574 | 0.454 |
| 12964 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 10 | FAM133A | Sponge network | -0.129 | 0.57177 | 0.549 | 0.29009 | 0.454 |
| 12965 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 29 | DMD | Sponge network | 0.214 | 0.18246 | -1.506 | 0 | 0.454 |
| 12966 | MIR497HG |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 16 | CSRNP1 | Sponge network | -1.439 | 4.0E-5 | -1.448 | 0 | 0.454 |
| 12967 | LINC00578 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-501-5p | 15 | JAZF1 | Sponge network | 0.811 | 0.03228 | -0.377 | 0.02677 | 0.454 |
| 12968 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | ABCC9 | Sponge network | -2.925 | 0 | -0.466 | 0.14121 | 0.454 |
| 12969 | RP11-497H16.9 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-323a-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CREB1 | Sponge network | 0.545 | 0.00064 | 0.203 | 0.00537 | 0.454 |
| 12970 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | SLITRK6 | Sponge network | -0.942 | 0 | -2.121 | 0 | 0.454 |
| 12971 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | -0.342 | 0.02288 | -1.12 | 0.00107 | 0.454 |
| 12972 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-424-3p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 18 | HOOK3 | Sponge network | 0.72 | 0.0001 | 0.273 | 0.09957 | 0.454 |
| 12973 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | RAP1A | Sponge network | -0.021 | 0.93598 | -0.929 | 0 | 0.454 |
| 12974 | NR2F1-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITGB1 | Sponge network | -0.49 | 0.04946 | 0.524 | 0.00017 | 0.454 |
| 12975 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-590-3p | 30 | JAZF1 | Sponge network | -1.161 | 0.00591 | -0.377 | 0.02677 | 0.454 |
| 12976 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 26 | CRTC3 | Sponge network | 0.72 | 0.0001 | 0.164 | 0.16786 | 0.454 |
| 12977 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p | 26 | NTRK2 | Sponge network | -0.358 | 0.17255 | -0.736 | 0.06495 | 0.454 |
| 12978 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 30 | ST6GAL2 | Sponge network | -0.487 | 0.02157 | -1.201 | 0.00597 | 0.454 |
| 12979 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-655-3p;hsa-miR-7-1-3p | 16 | UBE4B | Sponge network | 0.75 | 0.00054 | -0.235 | 0.007 | 0.454 |
| 12980 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-361-3p;hsa-miR-370-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 29 | CHD5 | Sponge network | 0.331 | 0.57247 | -0.922 | 0.01521 | 0.454 |
| 12981 | TPTEP1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 19 | EZH1 | Sponge network | -1.216 | 0 | -0.564 | 1.0E-5 | 0.454 |
| 12982 | ZNF582-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 11 | RHOB | Sponge network | -1.349 | 0.00017 | -1.052 | 0 | 0.454 |
| 12983 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-98-5p | 29 | ZFHX3 | Sponge network | 0.056 | 0.81195 | 0.389 | 0.01363 | 0.454 |
| 12984 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-629-3p | 19 | EBF1 | Sponge network | -0.469 | 0.08721 | -0.384 | 0.08856 | 0.454 |
| 12985 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 12 | SUFU | Sponge network | 0.889 | 9.0E-5 | -0.04 | 0.65489 | 0.454 |
| 12986 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-654-3p;hsa-miR-874-3p | 10 | GIGYF2 | Sponge network | 1.147 | 0 | 0.136 | 0.04403 | 0.454 |
| 12987 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | CAV1 | Sponge network | -0.777 | 0.1134 | -1.013 | 6.0E-5 | 0.454 |
| 12988 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p | 59 | LPP | Sponge network | 0.559 | 0.00026 | -0.459 | 0.04475 | 0.454 |
| 12989 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-7-5p | 12 | PTGFR | Sponge network | 0.4 | 0.14611 | -0.233 | 0.45421 | 0.454 |
| 12990 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-874-3p | 10 | BCLAF1 | Sponge network | -0.222 | 0.32445 | 0.211 | 0.00684 | 0.454 |
| 12991 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p | 13 | HECA | Sponge network | 0.497 | 0.01451 | -0.217 | 0.03463 | 0.454 |
| 12992 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-5p | 43 | FAT3 | Sponge network | -0.227 | 0.31657 | -1.601 | 0.00051 | 0.454 |
| 12993 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-324-5p | 16 | NTRK3 | Sponge network | -7.551 | 0 | -1.77 | 3.0E-5 | 0.454 |
| 12994 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-3p | 13 | JAKMIP2 | Sponge network | -0.405 | 0.22013 | -1.388 | 0 | 0.454 |
| 12995 | ADAMTS9-AS2 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-942-5p | 20 | GLIPR1 | Sponge network | -2.452 | 0 | 0.146 | 0.3276 | 0.454 |
| 12996 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-590-3p | 12 | HERC2 | Sponge network | 0.101 | 0.60919 | 0.114 | 0.30638 | 0.454 |
| 12997 | AGAP11 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-501-5p;hsa-miR-576-5p | 12 | SFRP1 | Sponge network | -1.645 | 0 | -3.566 | 0 | 0.454 |
| 12998 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-590-3p | 14 | DNAJB4 | Sponge network | -0.615 | 0.00015 | -0.7 | 1.0E-5 | 0.454 |
| 12999 | LINC00954 |
hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429 | 17 | MDM4 | Sponge network | 1.157 | 0.00455 | 0.345 | 0.00762 | 0.454 |
| 13000 | RP11-244O19.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p | 25 | PRKCB | Sponge network | -0.096 | 0.67958 | -1.313 | 2.0E-5 | 0.454 |
| 13001 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | MCC | Sponge network | -2.076 | 0.00548 | -1.005 | 0 | 0.454 |
| 13002 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3913-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 38 | ESR1 | Sponge network | -1.216 | 0 | -1.035 | 3.0E-5 | 0.454 |
| 13003 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-342-3p;hsa-miR-582-5p | 11 | LRRC7 | Sponge network | -2.095 | 0.00112 | -1.341 | 0.01193 | 0.454 |
| 13004 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 50 | MDM4 | Sponge network | 0.101 | 0.60919 | 0.345 | 0.00762 | 0.454 |
| 13005 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-590-3p | 15 | NR2C2 | Sponge network | 0.91 | 0 | 0.21 | 0.03224 | 0.454 |
| 13006 | HAND2-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-940;hsa-miR-98-5p | 24 | ZFP36L1 | Sponge network | -1.697 | 0.00889 | -0.505 | 8.0E-5 | 0.454 |
| 13007 | BCYRN1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 22 | TRIM33 | Sponge network | 0.311 | 0.10344 | 0.402 | 7.0E-5 | 0.454 |
| 13008 | AC016747.3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 16 | GLI3 | Sponge network | 0.199 | 0.38015 | -0.615 | 0.01482 | 0.454 |
| 13009 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-378a-5p | 29 | BMPR2 | Sponge network | 0.368 | 0.20093 | 0.206 | 0.0635 | 0.454 |
| 13010 | SH3RF3-AS1 |
hsa-miR-130b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378a-3p | 12 | RPS6KA2 | Sponge network | 0.889 | 9.0E-5 | -0.57 | 0.00013 | 0.454 |
| 13011 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | RABEP1 | Sponge network | -0.323 | 0.01372 | -0.066 | 0.43142 | 0.454 |
| 13012 | CTD-3092A11.2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PIK3CA | Sponge network | 0.873 | 0 | 0.195 | 0.06908 | 0.454 |
| 13013 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p | 17 | FGFR1 | Sponge network | -0.342 | 0.02288 | -0.509 | 0.03957 | 0.454 |
| 13014 | HNRNPU-AS1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 12 | SHPRH | Sponge network | 1.601 | 0 | 0.098 | 0.40994 | 0.454 |
| 13015 | OIP5-AS1 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-224-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 11 | PCM1 | Sponge network | 0.421 | 0.00084 | 0.164 | 0.20307 | 0.454 |
| 13016 | RAMP2-AS1 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 14 | SFRP1 | Sponge network | -0.513 | 0.04703 | -3.566 | 0 | 0.454 |
| 13017 | RP11-46C24.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 31 | ABI2 | Sponge network | 0.392 | 0.0278 | 0.175 | 0.14787 | 0.454 |
| 13018 | RP11-532F6.3 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p | 14 | ABL1 | Sponge network | -0.896 | 0.00099 | -0.215 | 0.07804 | 0.454 |
| 13019 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p | 16 | PRDM5 | Sponge network | -0.162 | 0.47687 | -1.09 | 0.00014 | 0.454 |
| 13020 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | PIK3R1 | Sponge network | 0.249 | 0.35902 | -0.133 | 0.36854 | 0.454 |
| 13021 | FZD10-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-5p;hsa-miR-589-5p | 11 | MN1 | Sponge network | -0.727 | 0.04726 | -0.358 | 0.24172 | 0.454 |
| 13022 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | RARB | Sponge network | -2.46 | 0 | -0.825 | 2.0E-5 | 0.454 |
| 13023 | EMX2OS |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 12 | PHOX2B | Sponge network | -0.777 | 0.1134 | -2.68 | 0 | 0.454 |
| 13024 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | PRKAA2 | Sponge network | 0.299 | 0.57722 | -2.462 | 0 | 0.454 |
| 13025 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429 | 32 | TGFBR3 | Sponge network | -1.654 | 0.00144 | -1.173 | 1.0E-5 | 0.453 |
| 13026 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | NRXN3 | Sponge network | -0.942 | 0 | -0.893 | 0.04186 | 0.453 |
| 13027 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | PIK3R1 | Sponge network | 0.101 | 0.60919 | -0.133 | 0.36854 | 0.453 |
| 13028 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 47 | TCF4 | Sponge network | -0.246 | 0.35034 | 0.016 | 0.92271 | 0.453 |
| 13029 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | AHNAK | Sponge network | -0.976 | 0.00025 | -1.207 | 0 | 0.453 |
| 13030 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 19 | RARB | Sponge network | -0.611 | 0.09607 | -0.825 | 2.0E-5 | 0.453 |
| 13031 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 17 | JAKMIP2 | Sponge network | -0.793 | 7.0E-5 | -1.388 | 0 | 0.453 |
| 13032 | EMX2OS |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | CDKN1C | Sponge network | -0.777 | 0.1134 | -0.929 | 0.00033 | 0.453 |
| 13033 | CTD-2528L19.6 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | 0.953 | 0 | 0.098 | 0.40994 | 0.453 |
| 13034 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-940;hsa-miR-98-5p | 22 | ZFP36L1 | Sponge network | -0.49 | 0.04946 | -0.505 | 8.0E-5 | 0.453 |
| 13035 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 19 | RCAN2 | Sponge network | -2.925 | 0 | -0.33 | 0.15172 | 0.453 |
| 13036 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-3913-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | WWTR1 | Sponge network | -0.738 | 0.21233 | -0.345 | 0.11997 | 0.453 |
| 13037 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-424-3p;hsa-miR-629-5p;hsa-miR-877-5p | 35 | BMPR2 | Sponge network | 0.594 | 0.00064 | 0.206 | 0.0635 | 0.453 |
| 13038 | RP11-212P7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 21 | AKAP6 | Sponge network | 0.219 | 0.1516 | -1.586 | 3.0E-5 | 0.453 |
| 13039 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | EPG5 | Sponge network | 0.421 | 0.00084 | 0.101 | 0.3563 | 0.453 |
| 13040 | AC003090.1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | LIMCH1 | Sponge network | -2.117 | 0.00161 | -0.915 | 1.0E-5 | 0.453 |
| 13041 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-660-5p;hsa-miR-93-5p | 10 | ZHX2 | Sponge network | -0.129 | 0.57177 | -0.192 | 0.18459 | 0.453 |
| 13042 | RP11-102N12.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-7-5p | 17 | KCNJ5 | Sponge network | -0.351 | 0.11358 | -0.281 | 0.3339 | 0.453 |
| 13043 | SNHG14 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | CSDE1 | Sponge network | -1.111 | 0.0003 | -0.493 | 0 | 0.453 |
| 13044 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-93-5p | 10 | CNTNAP3 | Sponge network | -0.473 | 0.07602 | -1.465 | 0.00033 | 0.453 |
| 13045 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-590-3p | 13 | RBM5 | Sponge network | -0.031 | 0.89063 | -0.205 | 0.0129 | 0.453 |
| 13046 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | NRXN1 | Sponge network | 0.572 | 0.01722 | -3.778 | 0 | 0.453 |
| 13047 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p | 39 | SOX5 | Sponge network | -0.162 | 0.47687 | -1.091 | 4.0E-5 | 0.453 |
| 13048 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | AHNAK | Sponge network | -1.575 | 6.0E-5 | -1.207 | 0 | 0.453 |
| 13049 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-493-5p;hsa-miR-877-5p;hsa-miR-93-5p | 26 | ANK2 | Sponge network | -0.384 | 0.40652 | -1.603 | 1.0E-5 | 0.453 |
| 13050 | LINC00865 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p | 11 | PREX2 | Sponge network | -1.249 | 7.0E-5 | -0.272 | 0.25382 | 0.453 |
| 13051 | LINC00702 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 12 | ALDH1A2 | Sponge network | -0.737 | 0.06182 | -2.411 | 0 | 0.453 |
| 13052 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p | 28 | AFF3 | Sponge network | -0.942 | 0 | -2.373 | 0 | 0.453 |
| 13053 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | NRXN3 | Sponge network | 0.082 | 0.55654 | -0.893 | 0.04186 | 0.453 |
| 13054 | RP11-159D12.2 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | DICER1 | Sponge network | 1.254 | 0 | 0.042 | 0.674 | 0.453 |
| 13055 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p | 14 | PDZD2 | Sponge network | -0.154 | 0.78939 | -0.909 | 3.0E-5 | 0.453 |
| 13056 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | ABCA10 | Sponge network | -0.358 | 0.17255 | -0.571 | 0.03674 | 0.453 |
| 13057 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TBX18 | Sponge network | -0.737 | 0.06182 | 0.843 | 0.02945 | 0.453 |
| 13058 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577 | 11 | NALCN | Sponge network | 0.056 | 0.81195 | 1.068 | 0.00237 | 0.453 |
| 13059 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 20 | BMPR2 | Sponge network | 0.673 | 0 | 0.206 | 0.0635 | 0.453 |
| 13060 | FAM157C |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | RASAL2 | Sponge network | 0.91 | 0 | 0.869 | 0 | 0.453 |
| 13061 | RP11-400K9.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p | 11 | FBXO32 | Sponge network | -0.733 | 0.19469 | -0.495 | 0.07561 | 0.453 |
| 13062 | LINC00702 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3E | Sponge network | -0.737 | 0.06182 | -1.717 | 0.00137 | 0.453 |
| 13063 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 25 | MCC | Sponge network | 0.299 | 0.57722 | -1.005 | 0 | 0.453 |
| 13064 | LINC00476 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 65 | RUNX1T1 | Sponge network | -0.615 | 0.00015 | -0.344 | 0.2374 | 0.453 |
| 13065 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-629-3p | 34 | EPHA7 | Sponge network | 0.331 | 0.57247 | -2.197 | 3.0E-5 | 0.453 |
| 13066 | TPTEP1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-877-5p | 25 | NRK | Sponge network | -1.216 | 0 | 0.656 | 0.19217 | 0.453 |
| 13067 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-582-3p;hsa-miR-590-3p | 26 | ADARB1 | Sponge network | -1.582 | 8.0E-5 | -0.335 | 0.06631 | 0.453 |
| 13068 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-3p | 27 | TGFBR3 | Sponge network | -0.693 | 0.05629 | -1.173 | 1.0E-5 | 0.453 |
| 13069 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 34 | JAZF1 | Sponge network | -0.899 | 6.0E-5 | -0.377 | 0.02677 | 0.453 |
| 13070 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p | 13 | MN1 | Sponge network | 0.219 | 0.1516 | -0.358 | 0.24172 | 0.453 |
| 13071 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 26 | BCL6 | Sponge network | 0.997 | 0.00066 | -0.211 | 0.19489 | 0.453 |
| 13072 | RP11-395G23.3 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-651-5p | 12 | WWTR1 | Sponge network | 0.381 | 0.25967 | -0.345 | 0.11997 | 0.453 |
| 13073 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | HECA | Sponge network | 0.683 | 1.0E-5 | -0.217 | 0.03463 | 0.453 |
| 13074 | MEG3 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484 | 21 | ABL1 | Sponge network | 0.249 | 0.35902 | -0.215 | 0.07804 | 0.453 |
| 13075 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | TLE4 | Sponge network | -2.46 | 0 | -1.157 | 0 | 0.453 |
| 13076 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-589-5p | 18 | TCF4 | Sponge network | -0.309 | 0.1145 | 0.016 | 0.92271 | 0.453 |
| 13077 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-935;hsa-miR-940 | 26 | LIFR | Sponge network | -0.896 | 0.00099 | -1.794 | 0 | 0.453 |
| 13078 | RP11-890B15.3 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | MAPK10 | Sponge network | 0.101 | 0.60919 | -1.774 | 0 | 0.453 |
| 13079 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 53 | PRKAA2 | Sponge network | -0.777 | 0.1134 | -2.462 | 0 | 0.453 |
| 13080 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | ZFHX4 | Sponge network | -0.421 | 0.0114 | 0.018 | 0.95574 | 0.453 |
| 13081 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | ABCB5 | Sponge network | 0.194 | 0.64278 | -0.956 | 0.04077 | 0.453 |
| 13082 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 35 | CHD5 | Sponge network | 0.41 | 0.02214 | -0.922 | 0.01521 | 0.453 |
| 13083 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | ST6GALNAC3 | Sponge network | -0.49 | 0.04946 | -0.987 | 0 | 0.453 |
| 13084 | PCA3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 13 | KCNAB1 | Sponge network | -0.709 | 0.18168 | -0.755 | 2.0E-5 | 0.453 |
| 13085 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-96-5p | 24 | CBFA2T3 | Sponge network | -0.976 | 0.00025 | -1.221 | 1.0E-5 | 0.453 |
| 13086 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | RAP1A | Sponge network | -1.575 | 6.0E-5 | -0.929 | 0 | 0.453 |
| 13087 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | EPHA5 | Sponge network | -0.709 | 0.18168 | -0.957 | 0.01733 | 0.453 |
| 13088 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 13 | AKAP12 | Sponge network | -0.513 | 0.04703 | -0.932 | 0.0028 | 0.453 |
| 13089 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-93-3p | 25 | EPHA3 | Sponge network | 0.056 | 0.81195 | 0.185 | 0.53588 | 0.453 |
| 13090 | RP11-283I3.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.614 | 1.0E-5 | 0.943 | 0 | 0.453 |
| 13091 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-98-5p | 36 | PBX1 | Sponge network | -0.227 | 0.31657 | -1.206 | 0 | 0.453 |
| 13092 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p | 19 | DYNC1I1 | Sponge network | -0.777 | 0.16667 | -1.12 | 0.00107 | 0.453 |
| 13093 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-708-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | GRIA3 | Sponge network | -2.925 | 0 | -1.956 | 0 | 0.453 |
| 13094 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-935 | 19 | RELN | Sponge network | -0.942 | 0 | -2.403 | 0 | 0.453 |
| 13095 | CTA-204B4.2 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-590-3p | 16 | TSC1 | Sponge network | 0.765 | 7.0E-5 | 0.103 | 0.26071 | 0.453 |
| 13096 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-935 | 34 | RAP1A | Sponge network | -1.281 | 0.00012 | -0.929 | 0 | 0.453 |
| 13097 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-362-3p;hsa-miR-942-5p | 17 | GPC6 | Sponge network | -0.693 | 0.05629 | -0.054 | 0.81465 | 0.453 |
| 13098 | USP3-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p | 10 | ZDBF2 | Sponge network | -0.162 | 0.47687 | -0.998 | 0.00025 | 0.453 |
| 13099 | RP11-13K12.1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421 | 11 | PRKD1 | Sponge network | -0.154 | 0.78939 | -0.466 | 0.03583 | 0.453 |
| 13100 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p | 27 | ANK2 | Sponge network | -0.738 | 0.21233 | -1.603 | 1.0E-5 | 0.453 |
| 13101 | HCG11 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-93-5p | 51 | DST | Sponge network | 0.385 | 0.10067 | -0.713 | 0.00096 | 0.453 |
| 13102 | FZD10-AS1 |
hsa-miR-107;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 23 | RIMBP2 | Sponge network | -0.727 | 0.04726 | -1.968 | 5.0E-5 | 0.453 |
| 13103 | LINC01018 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | -2.915 | 0.00015 | -0.974 | 0.00034 | 0.453 |
| 13104 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | TAL1 | Sponge network | -0.896 | 0.00099 | -0.462 | 0.00574 | 0.453 |
| 13105 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 13 | PDS5B | Sponge network | 0.765 | 7.0E-5 | 0.259 | 0.00962 | 0.453 |
| 13106 | FENDRR |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | NOV | Sponge network | -1.281 | 0.00012 | -0.58 | 0.04676 | 0.453 |
| 13107 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 16 | TET1 | Sponge network | 0.131 | 0.8436 | -0.135 | 0.52138 | 0.453 |
| 13108 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | AKAP11 | Sponge network | 0.225 | 0.30644 | 0.196 | 0.09825 | 0.453 |
| 13109 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 33 | GAS7 | Sponge network | 0.299 | 0.57722 | -0.555 | 0.01602 | 0.453 |
| 13110 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 43 | ZFHX4 | Sponge network | -1.203 | 0.03881 | 0.018 | 0.95574 | 0.453 |
| 13111 | LINC00957 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-590-3p | 10 | CITED2 | Sponge network | -0.421 | 0.0114 | -1.545 | 0 | 0.453 |
| 13112 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-92a-3p | 20 | CRTC3 | Sponge network | 0.253 | 0.09565 | 0.164 | 0.16786 | 0.453 |
| 13113 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-935 | 48 | FAT3 | Sponge network | -0.738 | 0.21233 | -1.601 | 0.00051 | 0.453 |
| 13114 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-5p | 22 | CADM1 | Sponge network | -0.384 | 0.40652 | -0.9 | 0.00084 | 0.453 |
| 13115 | RP11-359E10.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-582-5p | 14 | EBF3 | Sponge network | 0.299 | 0.57722 | -0.058 | 0.81437 | 0.453 |
| 13116 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 33 | JAZF1 | Sponge network | -0.421 | 0.0114 | -0.377 | 0.02677 | 0.453 |
| 13117 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | IGSF10 | Sponge network | -0.487 | 0.02157 | -1.751 | 1.0E-5 | 0.453 |
| 13118 | RP11-359E10.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p | 18 | HIP1 | Sponge network | 0.299 | 0.57722 | 0.774 | 0 | 0.453 |
| 13119 | CTD-2314G24.2 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-940 | 11 | PHOX2B | Sponge network | 0.246 | 0.59912 | -2.68 | 0 | 0.453 |
| 13120 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-7-1-3p | 15 | DKK3 | Sponge network | -1.161 | 0.00591 | -0.052 | 0.77783 | 0.453 |
| 13121 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 58 | CADM2 | Sponge network | -0.777 | 0.1134 | -3.782 | 0 | 0.453 |
| 13122 | AP001347.6 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-590-3p | 25 | DLC1 | Sponge network | -1.161 | 0.00591 | -0.382 | 0.05038 | 0.453 |
| 13123 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-5p | 25 | RBMS3 | Sponge network | 0.174 | 0.30034 | -0.519 | 0.03551 | 0.453 |
| 13124 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-320a;hsa-miR-320b | 11 | DYNC1I1 | Sponge network | 0.811 | 0.03228 | -1.12 | 0.00107 | 0.453 |
| 13125 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 37 | ERC1 | Sponge network | 0.194 | 0.64278 | -0.108 | 0.38794 | 0.453 |
| 13126 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p | 17 | RYR2 | Sponge network | 0.299 | 0.57722 | -1.466 | 0.00031 | 0.453 |
| 13127 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p | 19 | CRTC3 | Sponge network | 0.335 | 0.20596 | 0.164 | 0.16786 | 0.453 |
| 13128 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TLL1 | Sponge network | -0.487 | 0.02157 | -1.195 | 3.0E-5 | 0.453 |
| 13129 | FENDRR |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 25 | ABL1 | Sponge network | -1.281 | 0.00012 | -0.215 | 0.07804 | 0.452 |
| 13130 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 32 | ADARB1 | Sponge network | 0.225 | 0.30644 | -0.335 | 0.06631 | 0.452 |
| 13131 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p | 10 | ZBTB4 | Sponge network | -7.551 | 0 | -0.739 | 0 | 0.452 |
| 13132 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-409-3p | 11 | AKAP6 | Sponge network | 0.497 | 0.01451 | -1.586 | 3.0E-5 | 0.452 |
| 13133 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | AKAP6 | Sponge network | 0.532 | 0.00505 | -1.586 | 3.0E-5 | 0.452 |
| 13134 | LINC00476 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 43 | TGFBR3 | Sponge network | -0.615 | 0.00015 | -1.173 | 1.0E-5 | 0.452 |
| 13135 | RP11-434B12.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p | 12 | FAM172A | Sponge network | -0.031 | 0.89063 | -0.57 | 0 | 0.452 |
| 13136 | PART1 |
hsa-miR-185-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-542-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-940 | 10 | ILK | Sponge network | -2.925 | 0 | -0.74 | 0 | 0.452 |
| 13137 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 20 | THBS1 | Sponge network | -0.611 | 0.09607 | 0.741 | 0.00386 | 0.452 |
| 13138 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 35 | CHD5 | Sponge network | -0.793 | 7.0E-5 | -0.922 | 0.01521 | 0.452 |
| 13139 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | RASSF8 | Sponge network | 1.302 | 7.0E-5 | -0.122 | 0.65591 | 0.452 |
| 13140 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p;hsa-miR-98-5p | 24 | MITF | Sponge network | 0.249 | 0.35902 | -0.769 | 0.00022 | 0.452 |
| 13141 | AC010226.4 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-345-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | PCDH10 | Sponge network | -0.811 | 2.0E-5 | -1.644 | 0.0078 | 0.452 |
| 13142 | CTD-2089N3.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | ASTN1 | Sponge network | -1.375 | 0.08642 | -2.389 | 2.0E-5 | 0.452 |
| 13143 | HHIP-AS1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | RPS6KA6 | Sponge network | -0.737 | 0.10822 | -2.989 | 0 | 0.452 |
| 13144 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 22 | NIN | Sponge network | -0.096 | 0.67958 | -0.253 | 0.13794 | 0.452 |
| 13145 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-654-3p | 31 | FBXO32 | Sponge network | -0.227 | 0.31657 | -0.495 | 0.07561 | 0.452 |
| 13146 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 11 | SNX29 | Sponge network | 0.542 | 0.00054 | -0.34 | 0.01371 | 0.452 |
| 13147 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NRXN3 | Sponge network | -1.057 | 0.00029 | -0.893 | 0.04186 | 0.452 |
| 13148 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-501-5p | 19 | EBF3 | Sponge network | 0.176 | 0.37402 | -0.058 | 0.81437 | 0.452 |
| 13149 | CTD-3092A11.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-654-3p | 10 | PHF6 | Sponge network | 0.873 | 0 | 0.785 | 0 | 0.452 |
| 13150 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p | 11 | GPC6 | Sponge network | -0.932 | 0.00329 | -0.054 | 0.81465 | 0.452 |
| 13151 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-511-5p | 27 | ZFHX3 | Sponge network | 0.331 | 0.57247 | 0.389 | 0.01363 | 0.452 |
| 13152 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-542-3p | 14 | ABCB5 | Sponge network | -1.439 | 4.0E-5 | -0.956 | 0.04077 | 0.452 |
| 13153 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p | 19 | DMD | Sponge network | -2.549 | 0.00528 | -1.506 | 0 | 0.452 |
| 13154 | RP11-686D22.8 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 18 | DOCK4 | Sponge network | 0.898 | 1.0E-5 | 0.502 | 0.00108 | 0.452 |
| 13155 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-654-3p;hsa-miR-940 | 11 | AKAP11 | Sponge network | 1.134 | 0 | 0.196 | 0.09825 | 0.452 |
| 13156 | CTB-31O20.4 |
hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p;hsa-miR-629-3p | 11 | CHD2 | Sponge network | 1.619 | 0 | 0.049 | 0.55371 | 0.452 |
| 13157 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | BNC2 | Sponge network | 0.225 | 0.30644 | -0.491 | 0.09988 | 0.452 |
| 13158 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-5p | 13 | SPG20 | Sponge network | 0.176 | 0.37402 | -1.16 | 0 | 0.452 |
| 13159 | CTD-2528L19.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CEP170 | Sponge network | 0.953 | 0 | 0.943 | 0 | 0.452 |
| 13160 | AP000487.6 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-625-3p | 11 | ABL2 | Sponge network | 2.208 | 0 | 0.82 | 0 | 0.452 |
| 13161 | CTD-2291D10.4 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p | 12 | ABL2 | Sponge network | 3.21 | 0 | 0.82 | 0 | 0.452 |
| 13162 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-30b-3p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-877-5p | 19 | AR | Sponge network | -1.116 | 0.23686 | -1.554 | 0 | 0.452 |
| 13163 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-625-5p | 24 | RSPO2 | Sponge network | -1.349 | 0.00017 | -3.038 | 0 | 0.452 |
| 13164 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | PREX2 | Sponge network | 0.056 | 0.81195 | -0.272 | 0.25382 | 0.452 |
| 13165 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 14 | ZMYM2 | Sponge network | 0.997 | 0 | 0.279 | 0.01273 | 0.452 |
| 13166 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p | 18 | PTPN14 | Sponge network | -0.154 | 0.53954 | 0.144 | 0.34595 | 0.452 |
| 13167 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 22 | BCL6 | Sponge network | -0.323 | 0.01372 | -0.211 | 0.19489 | 0.452 |
| 13168 | RP11-13K12.1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-574-3p | 11 | FHL1 | Sponge network | -0.154 | 0.78939 | -2.687 | 0 | 0.452 |
| 13169 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | PDGFRA | Sponge network | -1.654 | 0.00144 | -0.372 | 0.0037 | 0.452 |
| 13170 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-424-5p | 16 | RARB | Sponge network | -0.739 | 0.00044 | -0.825 | 2.0E-5 | 0.452 |
| 13171 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-629-3p | 13 | ITGA5 | Sponge network | -0.469 | 0.08721 | 0.452 | 0.03524 | 0.452 |
| 13172 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.683 | 1.0E-5 | 0.323 | 0.00792 | 0.452 |
| 13173 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 42 | RASSF2 | Sponge network | 0.997 | 0.00066 | -0.273 | 0.21961 | 0.452 |
| 13174 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 39 | ZFHX4 | Sponge network | -0.738 | 0.21233 | 0.018 | 0.95574 | 0.452 |
| 13175 | LINC00865 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-935 | 17 | RELN | Sponge network | -1.249 | 7.0E-5 | -2.403 | 0 | 0.452 |
| 13176 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PTPRB | Sponge network | 0.194 | 0.64278 | 0.105 | 0.47122 | 0.452 |
| 13177 | RP13-516M14.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 23 | ABI2 | Sponge network | 0.389 | 0.04293 | 0.175 | 0.14787 | 0.452 |
| 13178 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p | 17 | RARB | Sponge network | -0.18 | 0.20343 | -0.825 | 2.0E-5 | 0.452 |
| 13179 | RP11-6O2.3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-629-3p | 24 | PTPN14 | Sponge network | 0.331 | 0.57247 | 0.144 | 0.34595 | 0.452 |
| 13180 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | NEDD4 | Sponge network | 0.374 | 0.20054 | 0.02 | 0.89834 | 0.452 |
| 13181 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-3p | 16 | ADAMTSL3 | Sponge network | 0.178 | 0.29106 | -1.559 | 0 | 0.452 |
| 13182 | RP11-53O19.1 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-98-5p | 18 | KLF10 | Sponge network | -0.405 | 0.22013 | -0.783 | 0 | 0.452 |
| 13183 | PART1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | PCDH9 | Sponge network | -2.925 | 0 | -1.823 | 4.0E-5 | 0.452 |
| 13184 | SNHG14 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 48 | AFF1 | Sponge network | -1.111 | 0.0003 | -0.384 | 0.00053 | 0.452 |
| 13185 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 64 | THRB | Sponge network | -0.942 | 0 | -1.117 | 0.0004 | 0.452 |
| 13186 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-503-5p | 16 | ABL1 | Sponge network | 0.082 | 0.55397 | -0.215 | 0.07804 | 0.452 |
| 13187 | FAM157B |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 12 | RASAL2 | Sponge network | 0.92 | 0 | 0.869 | 0 | 0.452 |
| 13188 | LINC00472 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p | 16 | PTGFR | Sponge network | -0.842 | 0.01361 | -0.233 | 0.45421 | 0.452 |
| 13189 | RP11-458D21.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | TET1 | Sponge network | 0.663 | 0.00073 | -0.135 | 0.52138 | 0.452 |
| 13190 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | CBFA2T3 | Sponge network | -0.49 | 0.04946 | -1.221 | 1.0E-5 | 0.452 |
| 13191 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 15 | CHD1 | Sponge network | 1.601 | 0 | 0.322 | 0.00215 | 0.452 |
| 13192 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p | 33 | RBMS3 | Sponge network | -0.773 | 0.04073 | -0.519 | 0.03551 | 0.452 |
| 13193 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 40 | PRKAB2 | Sponge network | -0.739 | 0.0574 | -0.367 | 0.01371 | 0.452 |
| 13194 | LINC00654 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 58 | DST | Sponge network | 0.374 | 0.20054 | -0.713 | 0.00096 | 0.452 |
| 13195 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p | 17 | ABCC9 | Sponge network | 0.056 | 0.81195 | -0.466 | 0.14121 | 0.452 |
| 13196 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 58 | TCF4 | Sponge network | -2.117 | 0.00161 | 0.016 | 0.92271 | 0.452 |
| 13197 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p | 24 | DMD | Sponge network | -0.031 | 0.89063 | -1.506 | 0 | 0.452 |
| 13198 | FAM157C |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p | 10 | ZMYM2 | Sponge network | 0.91 | 0 | 0.279 | 0.01273 | 0.452 |
| 13199 | LINC00865 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429 | 10 | LHFP | Sponge network | -1.249 | 7.0E-5 | -0.167 | 0.41371 | 0.452 |
| 13200 | AP001055.6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-3p | 17 | PIK3R1 | Sponge network | 0.693 | 0.00076 | -0.133 | 0.36854 | 0.452 |
| 13201 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p | 28 | FBXO32 | Sponge network | 0.61 | 0.01593 | -0.495 | 0.07561 | 0.452 |
| 13202 | FZD10-AS1 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | -0.727 | 0.04726 | -1.09 | 0.00014 | 0.452 |
| 13203 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p | 37 | RBMS3 | Sponge network | 0.056 | 0.81195 | -0.519 | 0.03551 | 0.452 |
| 13204 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-96-5p | 21 | NAV3 | Sponge network | -1.654 | 0.00144 | 0.212 | 0.47729 | 0.452 |
| 13205 | LPP-AS2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-505-5p;hsa-miR-532-3p | 15 | MRVI1 | Sponge network | -0.18 | 0.20343 | -1.051 | 0.00022 | 0.452 |
| 13206 | AC005154.6 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RBBP6 | Sponge network | 0.848 | 0 | 0.163 | 0.05101 | 0.452 |
| 13207 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-5p | 10 | CDH23 | Sponge network | -4.463 | 0.00025 | -0.974 | 0.00034 | 0.452 |
| 13208 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | PDGFRA | Sponge network | -0.737 | 0.10822 | -0.372 | 0.0037 | 0.452 |
| 13209 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 28 | HLF | Sponge network | -0.811 | 2.0E-5 | -2.443 | 0 | 0.452 |
| 13210 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 22 | SYNE1 | Sponge network | -0.227 | 0.31657 | -0.738 | 0.00316 | 0.452 |
| 13211 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-7-5p;hsa-miR-744-3p | 54 | LPP | Sponge network | -0.693 | 0.05629 | -0.459 | 0.04475 | 0.452 |
| 13212 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 21 | NF1 | Sponge network | 1.601 | 0 | 0.51 | 0 | 0.452 |
| 13213 | CTD-3064M3.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-625-5p | 15 | MCC | Sponge network | -0.469 | 0.08721 | -1.005 | 0 | 0.452 |
| 13214 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | RAP1A | Sponge network | -1.697 | 0.00889 | -0.929 | 0 | 0.452 |
| 13215 | MALAT1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-409-5p;hsa-miR-495-3p | 12 | STAG2 | Sponge network | 0.782 | 0.00016 | 0.252 | 0.00438 | 0.452 |
| 13216 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 41 | MEF2C | Sponge network | 0.997 | 0.00066 | -0.499 | 0.01448 | 0.452 |
| 13217 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 20 | PAK3 | Sponge network | -0.942 | 0 | -0.628 | 0.07253 | 0.452 |
| 13218 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 33 | IGF1 | Sponge network | -0.942 | 0 | -0.204 | 0.5898 | 0.452 |
| 13219 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 37 | PBX1 | Sponge network | 0.997 | 0.00066 | -1.206 | 0 | 0.452 |
| 13220 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | AHNAK | Sponge network | -0.896 | 0.00099 | -1.207 | 0 | 0.452 |
| 13221 | RP11-13K12.5 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | SFRP1 | Sponge network | -0.318 | 0.65847 | -3.566 | 0 | 0.452 |
| 13222 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | RCAN2 | Sponge network | -0.376 | 0.00337 | -0.33 | 0.15172 | 0.452 |
| 13223 | EMX2OS |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | RYR2 | Sponge network | -0.777 | 0.1134 | -1.466 | 0.00031 | 0.452 |
| 13224 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 10 | RIMS1 | Sponge network | -1.057 | 0.00029 | -3.143 | 0 | 0.452 |
| 13225 | FENDRR |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 20 | CXCL12 | Sponge network | -1.281 | 0.00012 | -1.421 | 0 | 0.452 |
| 13226 | PGM5-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | TAL1 | Sponge network | -4.546 | 0 | -0.462 | 0.00574 | 0.452 |
| 13227 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 13 | FREM2 | Sponge network | -1.249 | 7.0E-5 | -0.437 | 0.46353 | 0.452 |
| 13228 | AC009948.5 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | MSN | Sponge network | 0.532 | 0.00505 | -0.023 | 0.88422 | 0.452 |
| 13229 | RP11-13K12.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | -0.154 | 0.78939 | -0.382 | 0.05038 | 0.452 |
| 13230 | RP11-144G6.12 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | KIF5B | Sponge network | 0.826 | 1.0E-5 | 0.362 | 0.00066 | 0.452 |
| 13231 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-93-5p | 15 | LATS2 | Sponge network | -0.842 | 0.01361 | 0.159 | 0.2304 | 0.452 |
| 13232 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p | 24 | PAFAH1B1 | Sponge network | -1.331 | 3.0E-5 | -0.429 | 0 | 0.452 |
| 13233 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 30 | EPHA3 | Sponge network | -0.405 | 0.22013 | 0.185 | 0.53588 | 0.452 |
| 13234 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-590-5p | 16 | GRIN2A | Sponge network | 0.176 | 0.37402 | -2.161 | 0 | 0.452 |
| 13235 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 20 | CREBBP | Sponge network | -0.739 | 0.0574 | 0.126 | 0.15186 | 0.452 |
| 13236 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-424-5p;hsa-miR-629-3p | 14 | EPHA7 | Sponge network | -1.054 | 0.0706 | -2.197 | 3.0E-5 | 0.452 |
| 13237 | RP11-736K20.5 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-664a-3p | 14 | NUAK1 | Sponge network | -0.358 | 0.17255 | 0.904 | 0 | 0.452 |
| 13238 | DNAJC27-AS1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | SPOP | Sponge network | -0.969 | 2.0E-5 | -0.419 | 1.0E-5 | 0.452 |
| 13239 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-7-1-3p | 14 | DKK3 | Sponge network | -0.487 | 0.02157 | -0.052 | 0.77783 | 0.452 |
| 13240 | RP11-359E10.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-582-5p | 15 | STARD13 | Sponge network | 0.299 | 0.57722 | -0.187 | 0.27383 | 0.452 |
| 13241 | RP11-37B2.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-625-3p | 32 | ABL2 | Sponge network | 1.03 | 0 | 0.82 | 0 | 0.452 |
| 13242 | ACTA2-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 48 | GRIK3 | Sponge network | 0.194 | 0.64278 | -3.208 | 0 | 0.452 |
| 13243 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p | 17 | SYNPO2 | Sponge network | -0.246 | 0.35034 | -2.342 | 0 | 0.452 |
| 13244 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-3p | 27 | MOB1B | Sponge network | 1.254 | 0 | 0.197 | 0.15266 | 0.452 |
| 13245 | RP11-620J15.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | TET1 | Sponge network | -0.739 | 0.00044 | -0.135 | 0.52138 | 0.452 |
| 13246 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-940 | 52 | NFIB | Sponge network | -0.793 | 7.0E-5 | 0.023 | 0.90795 | 0.452 |
| 13247 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 29 | ERC1 | Sponge network | -0.976 | 0.00025 | -0.108 | 0.38794 | 0.452 |
| 13248 | CTD-2245F17.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-484;hsa-miR-505-3p | 12 | SOX5 | Sponge network | -0.421 | 0.12556 | -1.091 | 4.0E-5 | 0.451 |
| 13249 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | SYNE1 | Sponge network | -0.899 | 6.0E-5 | -0.738 | 0.00316 | 0.451 |
| 13250 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 54 | EPHA7 | Sponge network | -1.216 | 0 | -2.197 | 3.0E-5 | 0.451 |
| 13251 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | SYNE1 | Sponge network | -0.318 | 0.65847 | -0.738 | 0.00316 | 0.451 |
| 13252 | DNM3OS |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 17 | SORCS1 | Sponge network | 0.572 | 0.01722 | -3.492 | 0 | 0.451 |
| 13253 | FENDRR |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-455-5p;hsa-miR-590-3p | 11 | PTPN13 | Sponge network | -1.281 | 0.00012 | -1.208 | 0 | 0.451 |
| 13254 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-93-5p | 18 | SYNE1 | Sponge network | 0.082 | 0.55397 | -0.738 | 0.00316 | 0.451 |
| 13255 | AC003090.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 39 | DIP2C | Sponge network | -2.117 | 0.00161 | -0.436 | 0.01713 | 0.451 |
| 13256 | FAM66C |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | LUZP2 | Sponge network | -0.901 | 0.00019 | -0.056 | 0.89416 | 0.451 |
| 13257 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p | 30 | EPHA5 | Sponge network | -0.358 | 0.17255 | -0.957 | 0.01733 | 0.451 |
| 13258 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 49 | PAFAH1B1 | Sponge network | -2.452 | 0 | -0.429 | 0 | 0.451 |
| 13259 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-660-5p | 11 | LRRC4 | Sponge network | -4.546 | 0 | -1.774 | 0 | 0.451 |
| 13260 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-532-3p;hsa-miR-93-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | 0.082 | 0.55654 | 0.358 | 0.13727 | 0.451 |
| 13261 | MIR497HG |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-940 | 35 | PTPRT | Sponge network | -1.439 | 4.0E-5 | -0.907 | 0.03727 | 0.451 |
| 13262 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-5p | 11 | SETBP1 | Sponge network | -1.116 | 0.23686 | -1.302 | 1.0E-5 | 0.451 |
| 13263 | AGAP11 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p | 19 | CSDE1 | Sponge network | -1.645 | 0 | -0.493 | 0 | 0.451 |
| 13264 | LINC00641 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p | 63 | RUNX1T1 | Sponge network | -0.222 | 0.32445 | -0.344 | 0.2374 | 0.451 |
| 13265 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-93-5p;hsa-miR-940 | 28 | HLF | Sponge network | 0.299 | 0.57722 | -2.443 | 0 | 0.451 |
| 13266 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-877-5p | 35 | RBMS3 | Sponge network | -0.384 | 0.40652 | -0.519 | 0.03551 | 0.451 |
| 13267 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | 0.919 | 0 | 0.323 | 0.00792 | 0.451 |
| 13268 | RP11-774O3.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | KCTD8 | Sponge network | -0.487 | 0.02157 | -3.359 | 0 | 0.451 |
| 13269 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-455-5p | 12 | PAK3 | Sponge network | -2.69 | 0.0001 | -0.628 | 0.07253 | 0.451 |
| 13270 | HCG11 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-93-5p | 30 | IL17RD | Sponge network | 0.385 | 0.10067 | -0.019 | 0.94873 | 0.451 |
| 13271 | GAS6-AS2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-92a-3p | 10 | MACF1 | Sponge network | 0.16 | 0.50785 | 0.019 | 0.90335 | 0.451 |
| 13272 | RP11-758N13.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 14 | PRRX1 | Sponge network | 2.939 | 3.0E-5 | 1.316 | 0 | 0.451 |
| 13273 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-664a-3p | 12 | CNTNAP3 | Sponge network | -0.405 | 0.22013 | -1.465 | 0.00033 | 0.451 |
| 13274 | RASSF8-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | MED12L | Sponge network | 0.335 | 0.20596 | -0.194 | 0.55299 | 0.451 |
| 13275 | BZRAP1-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-577 | 11 | ATM | Sponge network | -0.465 | 0.04703 | 0.178 | 0.21568 | 0.451 |
| 13276 | RP11-589P10.7 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-324-3p;hsa-miR-671-5p | 11 | ADARB1 | Sponge network | -2.044 | 0.00066 | -0.335 | 0.06631 | 0.451 |
| 13277 | LINC00865 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-671-5p | 30 | DLC1 | Sponge network | -1.249 | 7.0E-5 | -0.382 | 0.05038 | 0.451 |
| 13278 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | MOB1B | Sponge network | 0.953 | 0 | 0.197 | 0.15266 | 0.451 |
| 13279 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | NEDD4 | Sponge network | 0.997 | 0.00066 | 0.02 | 0.89834 | 0.451 |
| 13280 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p | 19 | NCOA1 | Sponge network | -1.331 | 3.0E-5 | -0.432 | 0 | 0.451 |
| 13281 | CTD-2314G24.2 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | PCDH18 | Sponge network | 0.246 | 0.59912 | 0.216 | 0.24645 | 0.451 |
| 13282 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | FAT4 | Sponge network | -1.161 | 0.00591 | -0.354 | 0.14694 | 0.451 |
| 13283 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 60 | THRB | Sponge network | -0.563 | 0.02545 | -1.117 | 0.0004 | 0.451 |
| 13284 | LINC01018 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 33 | FBXO32 | Sponge network | -2.915 | 0.00015 | -0.495 | 0.07561 | 0.451 |
| 13285 | LINC00865 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 27 | RIMS2 | Sponge network | -1.249 | 7.0E-5 | -1.365 | 0.00208 | 0.451 |
| 13286 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-96-5p | 28 | IL17RD | Sponge network | 0.853 | 0.15352 | -0.019 | 0.94873 | 0.451 |
| 13287 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | SASH1 | Sponge network | 0.194 | 0.64278 | -0.502 | 0.00175 | 0.451 |
| 13288 | C20orf166-AS1 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-664a-3p | 25 | RIMS2 | Sponge network | -2.69 | 0.0001 | -1.365 | 0.00208 | 0.451 |
| 13289 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 22 | NRK | Sponge network | -1.161 | 0.00591 | 0.656 | 0.19217 | 0.451 |
| 13290 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-589-5p | 21 | PRKAA2 | Sponge network | -0.309 | 0.1145 | -2.462 | 0 | 0.451 |
| 13291 | RP11-53O19.1 |
hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 77 | LPP | Sponge network | -0.405 | 0.22013 | -0.459 | 0.04475 | 0.451 |
| 13292 | PWAR6 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-550a-5p;hsa-miR-589-5p | 11 | SORCS2 | Sponge network | -1.575 | 6.0E-5 | -0.223 | 0.4253 | 0.451 |
| 13293 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 37 | RICTOR | Sponge network | 0.683 | 1.0E-5 | 0.388 | 0.00188 | 0.451 |
| 13294 | RP11-1246C19.1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-625-3p | 19 | ABL2 | Sponge network | 1.352 | 0 | 0.82 | 0 | 0.451 |
| 13295 | AC005154.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 41 | CREB1 | Sponge network | 0.848 | 0 | 0.203 | 0.00537 | 0.451 |
| 13296 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-940 | 19 | MEF2C | Sponge network | -1.255 | 0.00981 | -0.499 | 0.01448 | 0.451 |
| 13297 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 20 | LATS1 | Sponge network | 1.063 | 0 | 0.109 | 0.34208 | 0.451 |
| 13298 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | KL | Sponge network | -0.942 | 0 | -1.294 | 0 | 0.451 |
| 13299 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-92a-3p | 15 | ITGA5 | Sponge network | -0.738 | 0.21233 | 0.452 | 0.03524 | 0.451 |
| 13300 | PWAR6 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | -1.575 | 6.0E-5 | -1.379 | 0 | 0.451 |
| 13301 | RP11-574K11.29 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-495-3p | 10 | TSC1 | Sponge network | 0.997 | 0 | 0.103 | 0.26071 | 0.451 |
| 13302 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p | 10 | ROBO1 | Sponge network | 0.727 | 3.0E-5 | -0.009 | 0.96154 | 0.451 |
| 13303 | SERTAD4-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 10 | LATS2 | Sponge network | -0.932 | 0.00329 | 0.159 | 0.2304 | 0.451 |
| 13304 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 34 | CBL | Sponge network | 0.75 | 0.00054 | 0.291 | 0.01267 | 0.451 |
| 13305 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | ZFHX4 | Sponge network | 0.61 | 0.01593 | 0.018 | 0.95574 | 0.451 |
| 13306 | LINC00476 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | ABCA10 | Sponge network | -0.615 | 0.00015 | -0.571 | 0.03674 | 0.451 |
| 13307 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 29 | PBX1 | Sponge network | -0.421 | 0.0114 | -1.206 | 0 | 0.451 |
| 13308 | RP11-384O8.1 |
hsa-miR-146b-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p | 10 | FHL1 | Sponge network | -0.738 | 0.21233 | -2.687 | 0 | 0.451 |
| 13309 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p | 12 | RCAN2 | Sponge network | -0.738 | 0.21233 | -0.33 | 0.15172 | 0.451 |
| 13310 | LINC00578 |
hsa-let-7b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-342-3p;hsa-miR-484 | 10 | ABL1 | Sponge network | 0.811 | 0.03228 | -0.215 | 0.07804 | 0.451 |
| 13311 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-589-5p | 10 | SOX5 | Sponge network | -1.14 | 0.03513 | -1.091 | 4.0E-5 | 0.451 |
| 13312 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | PDGFC | Sponge network | 0.246 | 0.59912 | -0.33 | 0.06483 | 0.451 |
| 13313 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-940 | 32 | AKAP11 | Sponge network | 0.494 | 0.00339 | 0.196 | 0.09825 | 0.451 |
| 13314 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 32 | CHD5 | Sponge network | 0.75 | 0.00054 | -0.922 | 0.01521 | 0.451 |
| 13315 | RP11-999E24.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p | 20 | DLC1 | Sponge network | -0.693 | 0.05629 | -0.382 | 0.05038 | 0.451 |
| 13316 | RP11-503E24.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-505-5p | 10 | TBL1XR1 | Sponge network | 2.218 | 0 | 0.578 | 0 | 0.451 |
| 13317 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DACT1 | Sponge network | 0.194 | 0.64278 | 0.783 | 0.00072 | 0.451 |
| 13318 | BMS1P20 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-3p | 11 | RASAL2 | Sponge network | 0.34 | 0.00273 | 0.869 | 0 | 0.451 |
| 13319 | RP11-244O19.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | ROBO1 | Sponge network | -0.096 | 0.67958 | -0.009 | 0.96154 | 0.451 |
| 13320 | PSMG3-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-7-5p | 18 | KCNJ5 | Sponge network | -0.125 | 0.49414 | -0.281 | 0.3339 | 0.451 |
| 13321 | LINC00957 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-93-3p | 16 | RPS6KA2 | Sponge network | -0.421 | 0.0114 | -0.57 | 0.00013 | 0.451 |
| 13322 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | CREBBP | Sponge network | 0.214 | 0.18246 | 0.126 | 0.15186 | 0.451 |
| 13323 | CTD-2314G24.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | PDE4DIP | Sponge network | 0.246 | 0.59912 | -0.282 | 0.0257 | 0.451 |
| 13324 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-744-3p | 22 | DIP2C | Sponge network | 0.051 | 0.83388 | -0.436 | 0.01713 | 0.451 |
| 13325 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 11 | ERCC4 | Sponge network | 0.497 | 0.01451 | 0.126 | 0.15266 | 0.451 |
| 13326 | RP11-35G9.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-940 | 32 | MDM4 | Sponge network | 0.622 | 0.00015 | 0.345 | 0.00762 | 0.451 |
| 13327 | DNM3OS |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-671-5p | 14 | CADM3 | Sponge network | 0.572 | 0.01722 | -3.663 | 0 | 0.451 |
| 13328 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-3p | 37 | ZFHX4 | Sponge network | -0.773 | 0.04073 | 0.018 | 0.95574 | 0.451 |
| 13329 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 42 | BNC2 | Sponge network | 1.302 | 7.0E-5 | -0.491 | 0.09988 | 0.451 |
| 13330 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | TSHZ3 | Sponge network | -0.726 | 0.00132 | -0.556 | 0.01516 | 0.451 |
| 13331 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 14 | IGSF10 | Sponge network | -0.693 | 0.05629 | -1.751 | 1.0E-5 | 0.451 |
| 13332 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | RECK | Sponge network | -0.187 | 0.23787 | -1.043 | 0 | 0.451 |
| 13333 | PCA3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-93-5p | 61 | DST | Sponge network | -0.709 | 0.18168 | -0.713 | 0.00096 | 0.451 |
| 13334 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | PEG3 | Sponge network | 0.082 | 0.55654 | -1.74 | 0 | 0.451 |
| 13335 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | SEMA3C | Sponge network | 0.194 | 0.64278 | -0.272 | 0.31153 | 0.451 |
| 13336 | RP11-144G6.12 |
hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p | 10 | PHF3 | Sponge network | 0.826 | 1.0E-5 | 0.126 | 0.19652 | 0.451 |
| 13337 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429 | 10 | NALCN | Sponge network | -1.249 | 7.0E-5 | 1.068 | 0.00237 | 0.451 |
| 13338 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 12 | ST6GAL2 | Sponge network | -0.557 | 0.11135 | -1.201 | 0.00597 | 0.451 |
| 13339 | PCA3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | NCAM2 | Sponge network | -0.709 | 0.18168 | -0.482 | 0.22565 | 0.451 |
| 13340 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-96-5p | 14 | ELMO1 | Sponge network | -2.452 | 0 | 0.04 | 0.83147 | 0.451 |
| 13341 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-590-5p | 31 | TGFBR3 | Sponge network | -0.384 | 0.40652 | -1.173 | 1.0E-5 | 0.451 |
| 13342 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 12 | CNTNAP1 | Sponge network | -0.513 | 0.04703 | 0.358 | 0.13727 | 0.451 |
| 13343 | AC009948.5 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 26 | SYNPO2 | Sponge network | 0.532 | 0.00505 | -2.342 | 0 | 0.451 |
| 13344 | RP11-701H24.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-511-5p;hsa-miR-590-3p | 18 | CADM2 | Sponge network | -1.425 | 0.04204 | -3.782 | 0 | 0.451 |
| 13345 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 21 | TMEM132B | Sponge network | -1.331 | 3.0E-5 | -0.83 | 0.01084 | 0.451 |
| 13346 | FAM215B |
hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | 0.817 | 3.0E-5 | 0.098 | 0.40994 | 0.451 |
| 13347 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | CLASP2 | Sponge network | 0.225 | 0.30644 | -0.038 | 0.71 | 0.451 |
| 13348 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-889-3p | 36 | UNC80 | Sponge network | -0.896 | 0.00099 | -1.934 | 0 | 0.451 |
| 13349 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-576-5p;hsa-miR-582-5p | 24 | RICTOR | Sponge network | 0.921 | 0.00118 | 0.388 | 0.00188 | 0.451 |
| 13350 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 37 | ZFHX3 | Sponge network | 0.385 | 0.10067 | 0.389 | 0.01363 | 0.451 |
| 13351 | RP11-597D13.9 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | ITGB1 | Sponge network | 1.302 | 7.0E-5 | 0.524 | 0.00017 | 0.451 |
| 13352 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p | 15 | LRRC55 | Sponge network | 0.176 | 0.37402 | -0.026 | 0.93163 | 0.451 |
| 13353 | LINC00472 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-874-3p | 14 | SORCS1 | Sponge network | -0.842 | 0.01361 | -3.492 | 0 | 0.451 |
| 13354 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -0.08 | 0.90092 | -0.354 | 0.14694 | 0.451 |
| 13355 | AGAP11 |
hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-374b-5p;hsa-miR-590-5p | 11 | TP63 | Sponge network | -1.645 | 0 | -0.363 | 0.38191 | 0.451 |
| 13356 | LINC00578 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p | 16 | MYO9A | Sponge network | 0.811 | 0.03228 | -0.147 | 0.32373 | 0.451 |
| 13357 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | FGF5 | Sponge network | 0.335 | 0.20596 | 0.549 | 0.28659 | 0.451 |
| 13358 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-96-5p | 15 | PIK3R1 | Sponge network | -0.187 | 0.23787 | -0.133 | 0.36854 | 0.451 |
| 13359 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | DCLK1 | Sponge network | -0.793 | 7.0E-5 | -1.324 | 0.00014 | 0.451 |
| 13360 | LINC00657 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-766-3p | 13 | KIAA1024 | Sponge network | 0.91 | 0 | 1.083 | 0 | 0.451 |
| 13361 | RP11-798M19.6 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-877-5p | 21 | NRK | Sponge network | -0.612 | 0.00094 | 0.656 | 0.19217 | 0.451 |
| 13362 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | JAZF1 | Sponge network | -1.349 | 0.00017 | -0.377 | 0.02677 | 0.451 |
| 13363 | RP11-1006G14.4 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 14 | BCL9 | Sponge network | 0.559 | 0.00026 | 0.321 | 0.00267 | 0.451 |
| 13364 | SNHG14 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | GALNT13 | Sponge network | -1.111 | 0.0003 | -0.857 | 0.07439 | 0.451 |
| 13365 | LINC00265 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p | 12 | CBL | Sponge network | 1.07 | 0 | 0.291 | 0.01267 | 0.451 |
| 13366 | LINC00957 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | PAK3 | Sponge network | -0.421 | 0.0114 | -0.628 | 0.07253 | 0.451 |
| 13367 | RP11-968O1.5 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-493-5p | 11 | CACNA2D1 | Sponge network | -0.829 | 0.01343 | -0.846 | 0.00675 | 0.451 |
| 13368 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-335-5p | 14 | ZBTB4 | Sponge network | 0.043 | 0.81628 | -0.739 | 0 | 0.451 |
| 13369 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 13 | NIN | Sponge network | -0.187 | 0.23787 | -0.253 | 0.13794 | 0.451 |
| 13370 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 12 | NR2C2 | Sponge network | 1.294 | 0 | 0.21 | 0.03224 | 0.451 |
| 13371 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-421;hsa-miR-429 | 14 | DKK3 | Sponge network | -2.62 | 0 | -0.052 | 0.77783 | 0.45 |
| 13372 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-942-5p | 43 | TMEM132B | Sponge network | -0.793 | 7.0E-5 | -0.83 | 0.01084 | 0.45 |
| 13373 | LINC00094 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-93-3p | 13 | CNTNAP1 | Sponge network | 0.253 | 0.09565 | 0.358 | 0.13727 | 0.45 |
| 13374 | RP11-13K12.5 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-877-5p | 23 | ASTN1 | Sponge network | -0.318 | 0.65847 | -2.389 | 2.0E-5 | 0.45 |
| 13375 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | CBL | Sponge network | 0.826 | 1.0E-5 | 0.291 | 0.01267 | 0.45 |
| 13376 | MIR497HG |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | CDH10 | Sponge network | -1.439 | 4.0E-5 | -2.397 | 0 | 0.45 |
| 13377 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-93-5p;hsa-miR-940 | 34 | HLF | Sponge network | -0.563 | 0.02545 | -2.443 | 0 | 0.45 |
| 13378 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-940 | 43 | MEF2C | Sponge network | -0.793 | 7.0E-5 | -0.499 | 0.01448 | 0.45 |
| 13379 | LINC00578 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-628-5p;hsa-miR-744-3p | 25 | MEF2C | Sponge network | 0.811 | 0.03228 | -0.499 | 0.01448 | 0.45 |
| 13380 | LINC00957 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 22 | RIMS2 | Sponge network | -0.421 | 0.0114 | -1.365 | 0.00208 | 0.45 |
| 13381 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-935 | 35 | EBF1 | Sponge network | -0.615 | 0.00015 | -0.384 | 0.08856 | 0.45 |
| 13382 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-877-5p | 20 | PGR | Sponge network | -0.773 | 0.04073 | -1.704 | 0 | 0.45 |
| 13383 | RP11-531A24.5 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-576-5p | 21 | UBR3 | Sponge network | 0.75 | 0.00054 | -0.021 | 0.82774 | 0.45 |
| 13384 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-942-5p | 26 | GPC6 | Sponge network | -1.249 | 7.0E-5 | -0.054 | 0.81465 | 0.45 |
| 13385 | LINC00957 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-664a-3p | 16 | GREM1 | Sponge network | -0.421 | 0.0114 | 0.902 | 0.01691 | 0.45 |
| 13386 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-93-5p | 24 | ACSL6 | Sponge network | -0.465 | 0.04703 | -1.593 | 0 | 0.45 |
| 13387 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | BCLAF1 | Sponge network | 1.266 | 0 | 0.211 | 0.00684 | 0.45 |
| 13388 | OIP5-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FBXO11 | Sponge network | 0.421 | 0.00084 | 0.142 | 0.04938 | 0.45 |
| 13389 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | RET | Sponge network | -0.739 | 0.00044 | -0.833 | 0.03517 | 0.45 |
| 13390 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-935 | 20 | PCDH10 | Sponge network | -0.738 | 0.21233 | -1.644 | 0.0078 | 0.45 |
| 13391 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | BMPR2 | Sponge network | -0.187 | 0.23787 | 0.206 | 0.0635 | 0.45 |
| 13392 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-7-1-3p | 24 | TACC1 | Sponge network | -4.546 | 0 | -0.362 | 0.05406 | 0.45 |
| 13393 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | -1.203 | 0.03881 | -0.474 | 0.03833 | 0.45 |
| 13394 | CTA-941F9.9 |
hsa-let-7d-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-151a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-502-5p;hsa-miR-576-5p | 15 | NTRK3 | Sponge network | -0.344 | 0.49624 | -1.77 | 3.0E-5 | 0.45 |
| 13395 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-98-5p | 17 | PRKAA2 | Sponge network | -1.116 | 0.23686 | -2.462 | 0 | 0.45 |
| 13396 | RP11-458D21.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | BCL9 | Sponge network | 0.663 | 0.00073 | 0.321 | 0.00267 | 0.45 |
| 13397 | RP11-182J1.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 25 | AR | Sponge network | -0.187 | 0.23787 | -1.554 | 0 | 0.45 |
| 13398 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-660-5p | 46 | AR | Sponge network | 0.331 | 0.57247 | -1.554 | 0 | 0.45 |
| 13399 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 41 | NTRK3 | Sponge network | 0.176 | 0.37402 | -1.77 | 3.0E-5 | 0.45 |
| 13400 | LINC00654 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | FAM133A | Sponge network | 0.374 | 0.20054 | 0.549 | 0.29009 | 0.45 |
| 13401 | LINC00641 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 18 | ITGB3 | Sponge network | -0.222 | 0.32445 | -0.011 | 0.95751 | 0.45 |
| 13402 | RP11-678G14.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-940 | 17 | GRIK3 | Sponge network | -4.477 | 0 | -3.208 | 0 | 0.45 |
| 13403 | RP11-305E6.4 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 18 | RASAL2 | Sponge network | 1.317 | 0 | 0.869 | 0 | 0.45 |
| 13404 | LINC00861 |
hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-96-5p | 10 | BCL2 | Sponge network | 0.911 | 0.00854 | -0.899 | 0 | 0.45 |
| 13405 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | PDS5B | Sponge network | 1.063 | 0 | 0.259 | 0.00962 | 0.45 |
| 13406 | RP11-890B15.3 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 10 | FHL1 | Sponge network | 0.101 | 0.60919 | -2.687 | 0 | 0.45 |
| 13407 | LINC00476 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | MRVI1 | Sponge network | -0.615 | 0.00015 | -1.051 | 0.00022 | 0.45 |
| 13408 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | CITED2 | Sponge network | -2.46 | 0 | -1.545 | 0 | 0.45 |
| 13409 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p | 20 | CXXC4 | Sponge network | 0.176 | 0.37402 | -0.433 | 0.21055 | 0.45 |
| 13410 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-874-3p | 30 | CREB3L2 | Sponge network | -0.583 | 0.14589 | -0.525 | 2.0E-5 | 0.45 |
| 13411 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-93-5p | 22 | SASH1 | Sponge network | -0.473 | 0.07602 | -0.502 | 0.00175 | 0.45 |
| 13412 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-590-3p | 23 | FBXO32 | Sponge network | -0.739 | 0.00044 | -0.495 | 0.07561 | 0.45 |
| 13413 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | -0.323 | 0.01372 | 0.809 | 0.00544 | 0.45 |
| 13414 | AC009948.5 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 25 | DLC1 | Sponge network | 0.532 | 0.00505 | -0.382 | 0.05038 | 0.45 |
| 13415 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 30 | EPHA3 | Sponge network | -0.612 | 0.00094 | 0.185 | 0.53588 | 0.45 |
| 13416 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 19 | CNTNAP3 | Sponge network | -1.281 | 0.00012 | -1.465 | 0.00033 | 0.45 |
| 13417 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-769-5p | 14 | RCAN2 | Sponge network | -0.154 | 0.78939 | -0.33 | 0.15172 | 0.45 |
| 13418 | MIR497HG |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-542-3p | 11 | ALDH1A2 | Sponge network | -1.439 | 4.0E-5 | -2.411 | 0 | 0.45 |
| 13419 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p | 30 | NTRK2 | Sponge network | 0.176 | 0.37402 | -0.736 | 0.06495 | 0.45 |
| 13420 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-486-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 26 | ADARB1 | Sponge network | -0.811 | 2.0E-5 | -0.335 | 0.06631 | 0.45 |
| 13421 | CTC-429P9.2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-877-5p | 13 | FAT3 | Sponge network | 0.043 | 0.81628 | -1.601 | 0.00051 | 0.45 |
| 13422 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | CUL5 | Sponge network | -0.222 | 0.32445 | 0.026 | 0.77301 | 0.45 |
| 13423 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-744-3p | 35 | HLF | Sponge network | -0.739 | 0.00044 | -2.443 | 0 | 0.45 |
| 13424 | DNM3OS |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 22 | HIP1 | Sponge network | 0.572 | 0.01722 | 0.774 | 0 | 0.45 |
| 13425 | RP11-597D13.8 |
hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p | 12 | ZFHX3 | Sponge network | 0.936 | 0.03889 | 0.389 | 0.01363 | 0.45 |
| 13426 | BZRAP1-AS1 |
hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-574-3p | 10 | FHL1 | Sponge network | -0.465 | 0.04703 | -2.687 | 0 | 0.45 |
| 13427 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-486-5p;hsa-miR-582-5p | 12 | NR2C2 | Sponge network | 0.792 | 0.0049 | 0.21 | 0.03224 | 0.45 |
| 13428 | USP3-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p | 28 | MXI1 | Sponge network | -0.162 | 0.47687 | -0.987 | 0 | 0.45 |
| 13429 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | CREM | Sponge network | -0.769 | 0.00035 | -0.345 | 0.00258 | 0.45 |
| 13430 | LINC00657 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | CREB1 | Sponge network | 0.91 | 0 | 0.203 | 0.00537 | 0.45 |
| 13431 | OIP5-AS1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 67 | MDM4 | Sponge network | 0.421 | 0.00084 | 0.345 | 0.00762 | 0.45 |
| 13432 | LINC00174 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-375 | 10 | MLLT10 | Sponge network | 1.253 | 0 | 0.105 | 0.33292 | 0.45 |
| 13433 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | CACNA2D1 | Sponge network | -2.915 | 0.00015 | -0.846 | 0.00675 | 0.45 |
| 13434 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 22 | SASH1 | Sponge network | -1.654 | 0.00144 | -0.502 | 0.00175 | 0.45 |
| 13435 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-98-5p | 21 | LIFR | Sponge network | -0.473 | 0.07602 | -1.794 | 0 | 0.45 |
| 13436 | LINC00472 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-874-3p | 32 | GRIK3 | Sponge network | -0.842 | 0.01361 | -3.208 | 0 | 0.45 |
| 13437 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429 | 12 | ELMO1 | Sponge network | 0.335 | 0.20596 | 0.04 | 0.83147 | 0.45 |
| 13438 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-590-5p | 12 | AKAP12 | Sponge network | -0.384 | 0.40652 | -0.932 | 0.0028 | 0.45 |
| 13439 | PCA3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | TMEFF2 | Sponge network | -0.709 | 0.18168 | -2.426 | 0 | 0.45 |
| 13440 | HAND2-AS1 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CTGF | Sponge network | -1.697 | 0.00889 | 0.321 | 0.11682 | 0.45 |
| 13441 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-539-5p | 14 | PDZD2 | Sponge network | -0.162 | 0.47687 | -0.909 | 3.0E-5 | 0.45 |
| 13442 | ARHGEF26-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p | 33 | DLC1 | Sponge network | -2.46 | 0 | -0.382 | 0.05038 | 0.45 |
| 13443 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 45 | TGFBR3 | Sponge network | 0.997 | 0.00066 | -1.173 | 1.0E-5 | 0.45 |
| 13444 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-3p | 15 | EBF3 | Sponge network | 0.178 | 0.29106 | -0.058 | 0.81437 | 0.45 |
| 13445 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 30 | CBL | Sponge network | 1.153 | 0 | 0.291 | 0.01267 | 0.45 |
| 13446 | RP11-720L2.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p | 13 | DLC1 | Sponge network | -1.255 | 0.00981 | -0.382 | 0.05038 | 0.45 |
| 13447 | LINC00654 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | NRXN3 | Sponge network | 0.374 | 0.20054 | -0.893 | 0.04186 | 0.45 |
| 13448 | MIR143HG |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ZNF536 | Sponge network | -0.739 | 0.0574 | -3.115 | 0 | 0.45 |
| 13449 | RP11-774O3.3 |
hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p | 10 | ELN | Sponge network | -0.487 | 0.02157 | 0.451 | 0.13752 | 0.45 |
| 13450 | FENDRR |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | SYT4 | Sponge network | -1.281 | 0.00012 | -3.207 | 0 | 0.45 |
| 13451 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ABCC9 | Sponge network | 0.082 | 0.55654 | -0.466 | 0.14121 | 0.45 |
| 13452 | RP11-195F19.9 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p | 15 | DLC1 | Sponge network | -0.726 | 0.00132 | -0.382 | 0.05038 | 0.45 |
| 13453 | TUG1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-582-5p | 10 | PHF3 | Sponge network | 0.972 | 0 | 0.126 | 0.19652 | 0.45 |
| 13454 | RP11-384K6.6 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p | 13 | RASAL2 | Sponge network | 0.946 | 0 | 0.869 | 0 | 0.45 |
| 13455 | RP11-360F5.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | TCF4 | Sponge network | 1.867 | 0 | 0.016 | 0.92271 | 0.45 |
| 13456 | CTD-2002H8.2 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | PIK3C2A | Sponge network | 0.931 | 1.0E-5 | 0.061 | 0.53776 | 0.45 |
| 13457 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 70 | BMPR2 | Sponge network | -0.49 | 0.04946 | 0.206 | 0.0635 | 0.45 |
| 13458 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-877-5p | 24 | TET1 | Sponge network | 0.494 | 0.00339 | -0.135 | 0.52138 | 0.45 |
| 13459 | RP11-113K21.4 |
hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-766-3p | 14 | MDM4 | Sponge network | 0.676 | 0 | 0.345 | 0.00762 | 0.45 |
| 13460 | FGD5-AS1 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 12 | ITGB1 | Sponge network | 0.401 | 0.00066 | 0.524 | 0.00017 | 0.45 |
| 13461 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-744-3p | 42 | MEF2C | Sponge network | -0.615 | 0.00015 | -0.499 | 0.01448 | 0.45 |
| 13462 | RP11-119F7.5 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-502-5p | 20 | PTPN14 | Sponge network | 0.225 | 0.30644 | 0.144 | 0.34595 | 0.45 |
| 13463 | SNHG14 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 30 | LIMCH1 | Sponge network | -1.111 | 0.0003 | -0.915 | 1.0E-5 | 0.45 |
| 13464 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p | 12 | STARD13 | Sponge network | 0.331 | 0.57247 | -0.187 | 0.27383 | 0.45 |
| 13465 | LINC00092 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-532-3p | 10 | RPS6KA2 | Sponge network | -1.886 | 0 | -0.57 | 0.00013 | 0.45 |
| 13466 | MIR7-3HG |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-942-5p | 10 | UNC80 | Sponge network | -2.688 | 0.00397 | -1.934 | 0 | 0.45 |
| 13467 | SNX29P2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 34 | PTPRT | Sponge network | 0.4 | 0.14611 | -0.907 | 0.03727 | 0.45 |
| 13468 | GS1-124K5.11 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-93-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | 0.494 | 0.00339 | 0.358 | 0.13727 | 0.45 |
| 13469 | RP11-307B6.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421 | 21 | GAS7 | Sponge network | -4.463 | 0.00025 | -0.555 | 0.01602 | 0.45 |
| 13470 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 13 | JAKMIP2 | Sponge network | -0.738 | 0.21233 | -1.388 | 0 | 0.45 |
| 13471 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-98-5p | 29 | DMD | Sponge network | -0.18 | 0.20343 | -1.506 | 0 | 0.45 |
| 13472 | CTC-444N24.11 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | ERCC6 | Sponge network | 1.153 | 0 | 0.23 | 0.015 | 0.45 |
| 13473 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | PRRX1 | Sponge network | 0.889 | 9.0E-5 | 1.316 | 0 | 0.45 |
| 13474 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 33 | EBF1 | Sponge network | -0.222 | 0.32445 | -0.384 | 0.08856 | 0.45 |
| 13475 | RP11-212P7.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | IL17RD | Sponge network | 0.219 | 0.1516 | -0.019 | 0.94873 | 0.45 |
| 13476 | LINC00472 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 14 | KCNAB1 | Sponge network | -0.842 | 0.01361 | -0.755 | 2.0E-5 | 0.45 |
| 13477 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 44 | LPP | Sponge network | -0.285 | 0.2172 | -0.459 | 0.04475 | 0.45 |
| 13478 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-96-5p | 13 | ELMO1 | Sponge network | -0.576 | 0.10121 | 0.04 | 0.83147 | 0.45 |
| 13479 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-500b-5p;hsa-miR-590-5p | 18 | ACSL6 | Sponge network | -0.72 | 0.24239 | -1.593 | 0 | 0.45 |
| 13480 | GNG12-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-671-5p | 26 | KCNA6 | Sponge network | -0.793 | 7.0E-5 | -1.531 | 0.00013 | 0.45 |
| 13481 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p | 20 | CNTN1 | Sponge network | -0.773 | 0.04073 | -2.594 | 0 | 0.45 |
| 13482 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-34a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | LRRC7 | Sponge network | -0.611 | 0.09607 | -1.341 | 0.01193 | 0.45 |
| 13483 | RP11-6O2.3 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-96-5p | 22 | PCDH10 | Sponge network | 0.331 | 0.57247 | -1.644 | 0.0078 | 0.45 |
| 13484 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | RAP1A | Sponge network | -0.342 | 0.02288 | -0.929 | 0 | 0.45 |
| 13485 | LINC01001 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | ZMYM2 | Sponge network | 1.055 | 0 | 0.279 | 0.01273 | 0.45 |
| 13486 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 23 | RARB | Sponge network | -1.575 | 6.0E-5 | -0.825 | 2.0E-5 | 0.45 |
| 13487 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 18 | PRKAA2 | Sponge network | -2.549 | 0.00528 | -2.462 | 0 | 0.45 |
| 13488 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 21 | ZFHX3 | Sponge network | -0.246 | 0.35034 | 0.389 | 0.01363 | 0.45 |
| 13489 | WDFY3-AS2 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-378a-3p;hsa-miR-7-5p | 13 | FLNA | Sponge network | -0.942 | 0 | -0.771 | 0.01377 | 0.45 |
| 13490 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-942-5p | 28 | NRXN1 | Sponge network | -0.777 | 0.16667 | -3.778 | 0 | 0.45 |
| 13491 | LINC00472 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 23 | PRKCB | Sponge network | -0.842 | 0.01361 | -1.313 | 2.0E-5 | 0.45 |
| 13492 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-7-1-3p | 20 | ST6GAL2 | Sponge network | -1.161 | 0.00591 | -1.201 | 0.00597 | 0.45 |
| 13493 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 32 | THRB | Sponge network | -1.582 | 8.0E-5 | -1.117 | 0.0004 | 0.45 |
| 13494 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-93-5p | 17 | RASSF2 | Sponge network | -0.376 | 0.00337 | -0.273 | 0.21961 | 0.45 |
| 13495 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-93-3p | 29 | IL6ST | Sponge network | -0.932 | 0.00329 | -0.402 | 0.00676 | 0.449 |
| 13496 | PART1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 18 | JAKMIP2 | Sponge network | -2.925 | 0 | -1.388 | 0 | 0.449 |
| 13497 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484 | 11 | EDA2R | Sponge network | -0.773 | 0.04073 | -0.587 | 0.01598 | 0.449 |
| 13498 | AC011526.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | GAS7 | Sponge network | 0.342 | 0.03129 | -0.555 | 0.01602 | 0.449 |
| 13499 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | SNX29 | Sponge network | -1.582 | 8.0E-5 | -0.34 | 0.01371 | 0.449 |
| 13500 | RP5-1139B12.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-5p | 20 | AR | Sponge network | 0.598 | 0.38481 | -1.554 | 0 | 0.449 |
| 13501 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 36 | SNX29 | Sponge network | -1.216 | 0 | -0.34 | 0.01371 | 0.449 |
| 13502 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | 0.101 | 0.60919 | -0.7 | 1.0E-5 | 0.449 |
| 13503 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | 0.727 | 3.0E-5 | -0.243 | 0.11408 | 0.449 |
| 13504 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 13 | PDE4DIP | Sponge network | 0.419 | 0.0319 | -0.282 | 0.0257 | 0.449 |
| 13505 | RP11-67L2.2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 19 | BCL9 | Sponge network | 0.72 | 0.0001 | 0.321 | 0.00267 | 0.449 |
| 13506 | RP11-6O2.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-340-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-96-5p | 30 | ABI2 | Sponge network | 0.331 | 0.57247 | 0.175 | 0.14787 | 0.449 |
| 13507 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-93-3p | 27 | RUNX1T1 | Sponge network | 0.598 | 0.38481 | -0.344 | 0.2374 | 0.449 |
| 13508 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-629-3p | 29 | RASSF2 | Sponge network | -0.739 | 0.00044 | -0.273 | 0.21961 | 0.449 |
| 13509 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-93-5p | 15 | PRRX1 | Sponge network | -1.255 | 0.00981 | 1.316 | 0 | 0.449 |
| 13510 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | PRKAB2 | Sponge network | 0.349 | 0.01695 | -0.367 | 0.01371 | 0.449 |
| 13511 | CTA-204B4.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | BAZ2B | Sponge network | 0.765 | 7.0E-5 | -0.276 | 0.02833 | 0.449 |
| 13512 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | PPM1A | Sponge network | -0.709 | 0.18168 | -0.623 | 0 | 0.449 |
| 13513 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-5p | 14 | PCDH10 | Sponge network | 0.176 | 0.37402 | -1.644 | 0.0078 | 0.449 |
| 13514 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | PTPRB | Sponge network | -0.576 | 0.10121 | 0.105 | 0.47122 | 0.449 |
| 13515 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-671-5p | 16 | CADM3 | Sponge network | -0.162 | 0.47687 | -3.663 | 0 | 0.449 |
| 13516 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p | 34 | BNC2 | Sponge network | -0.465 | 0.04703 | -0.491 | 0.09988 | 0.449 |
| 13517 | BVES-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-935;hsa-miR-942-5p | 17 | OPCML | Sponge network | -2.62 | 0 | -2.498 | 0 | 0.449 |
| 13518 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -1.349 | 0.00017 | -0.354 | 0.14694 | 0.449 |
| 13519 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | MOB1B | Sponge network | 1.063 | 0 | 0.197 | 0.15266 | 0.449 |
| 13520 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-532-5p | 26 | FBXO32 | Sponge network | 0.082 | 0.55397 | -0.495 | 0.07561 | 0.449 |
| 13521 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-5p;hsa-miR-411-5p;hsa-miR-590-3p | 14 | USP6 | Sponge network | 2.094 | 0 | 0.188 | 0.3871 | 0.449 |
| 13522 | PCA3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-590-3p | 10 | SCN11A | Sponge network | -0.709 | 0.18168 | -1.15 | 0.00299 | 0.449 |
| 13523 | CTA-941F9.9 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-576-5p;hsa-miR-935 | 13 | FAT3 | Sponge network | -0.344 | 0.49624 | -1.601 | 0.00051 | 0.449 |
| 13524 | MAGI2-AS3 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | SYT4 | Sponge network | -0.129 | 0.57177 | -3.207 | 0 | 0.449 |
| 13525 | RP11-158K1.3 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | HERC1 | Sponge network | 0.349 | 0.01695 | -0.196 | 0.08544 | 0.449 |
| 13526 | FAM66C |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | MED12L | Sponge network | -0.901 | 0.00019 | -0.194 | 0.55299 | 0.449 |
| 13527 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | EYA4 | Sponge network | 0.997 | 0.00066 | 0.3 | 0.36876 | 0.449 |
| 13528 | CTD-2554C21.2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-429 | 10 | KLF10 | Sponge network | -1.654 | 0.00144 | -0.783 | 0 | 0.449 |
| 13529 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-940 | 44 | TRPS1 | Sponge network | -0.793 | 7.0E-5 | -0.191 | 0.44109 | 0.449 |
| 13530 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-93-5p | 10 | RBL2 | Sponge network | 0.497 | 0.01451 | -0.282 | 0.00254 | 0.449 |
| 13531 | CTD-2314G24.2 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 52 | AR | Sponge network | 0.246 | 0.59912 | -1.554 | 0 | 0.449 |
| 13532 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p | 12 | ZFHX3 | Sponge network | 1.056 | 0 | 0.389 | 0.01363 | 0.449 |
| 13533 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 24 | LIFR | Sponge network | -0.513 | 0.04703 | -1.794 | 0 | 0.449 |
| 13534 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | ST6GALNAC3 | Sponge network | -0.376 | 0.00337 | -0.987 | 0 | 0.449 |
| 13535 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-425-3p;hsa-miR-454-3p;hsa-miR-582-5p | 25 | SOX5 | Sponge network | -2.095 | 0.00112 | -1.091 | 4.0E-5 | 0.449 |
| 13536 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | JAZF1 | Sponge network | 0.199 | 0.38015 | -0.377 | 0.02677 | 0.449 |
| 13537 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | TLL1 | Sponge network | -0.942 | 0 | -1.195 | 3.0E-5 | 0.449 |
| 13538 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 15 | RICTOR | Sponge network | 0.702 | 7.0E-5 | 0.388 | 0.00188 | 0.449 |
| 13539 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 12 | KIAA1024 | Sponge network | 1.153 | 0 | 1.083 | 0 | 0.449 |
| 13540 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 40 | ZFHX3 | Sponge network | 0.101 | 0.60919 | 0.389 | 0.01363 | 0.449 |
| 13541 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 24 | SNX29 | Sponge network | -0.739 | 0.00044 | -0.34 | 0.01371 | 0.449 |
| 13542 | LINC00476 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | CDH10 | Sponge network | -0.615 | 0.00015 | -2.397 | 0 | 0.449 |
| 13543 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LRRK2 | Sponge network | -0.709 | 0.18168 | -1.136 | 1.0E-5 | 0.449 |
| 13544 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | -1.111 | 0.0003 | -0.001 | 0.99669 | 0.449 |
| 13545 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-497-5p | 15 | NF1 | Sponge network | 0.917 | 0 | 0.51 | 0 | 0.449 |
| 13546 | RP11-400K9.4 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326 | 15 | ABL1 | Sponge network | -0.611 | 0.09607 | -0.215 | 0.07804 | 0.449 |
| 13547 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 36 | MOB1B | Sponge network | -0.303 | 0.21002 | 0.197 | 0.15266 | 0.449 |
| 13548 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | ZYX | Sponge network | -1.439 | 4.0E-5 | 0.019 | 0.89763 | 0.449 |
| 13549 | SNHG14 |
hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-660-5p | 10 | DNER | Sponge network | -1.111 | 0.0003 | -3.787 | 0 | 0.449 |
| 13550 | RP11-57H14.4 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p | 21 | SYNM | Sponge network | 0.083 | 0.66405 | -2.309 | 1.0E-5 | 0.449 |
| 13551 | LINC00654 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 49 | DTNA | Sponge network | 0.374 | 0.20054 | -1.648 | 1.0E-5 | 0.449 |
| 13552 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | ARNT2 | Sponge network | -0.739 | 0.00044 | -0.332 | 0.25851 | 0.449 |
| 13553 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 46 | ADARB1 | Sponge network | -0.793 | 7.0E-5 | -0.335 | 0.06631 | 0.449 |
| 13554 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p | 16 | CBL | Sponge network | 0.972 | 0 | 0.291 | 0.01267 | 0.449 |
| 13555 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-590-3p | 26 | BCL2 | Sponge network | -0.739 | 0.00044 | -0.899 | 0 | 0.449 |
| 13556 | RP11-517B11.7 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-493-5p | 17 | DDX3X | Sponge network | 0.706 | 0 | -0.152 | 0.07686 | 0.449 |
| 13557 | RP11-479G22.8 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 26 | ABL2 | Sponge network | 1.308 | 0 | 0.82 | 0 | 0.449 |
| 13558 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 19 | SLC5A3 | Sponge network | 0.683 | 1.0E-5 | 0.493 | 5.0E-5 | 0.449 |
| 13559 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-582-5p;hsa-miR-654-3p | 18 | DDX6 | Sponge network | 0.959 | 0 | 0.091 | 0.36428 | 0.449 |
| 13560 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p | 22 | SCN9A | Sponge network | -0.842 | 0.01361 | -0.525 | 0.10593 | 0.449 |
| 13561 | TRAF3IP2-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | SYT4 | Sponge network | -0.323 | 0.01372 | -3.207 | 0 | 0.449 |
| 13562 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-874-3p | 10 | ST5 | Sponge network | -0.737 | 0.06182 | -0.78 | 0 | 0.449 |
| 13563 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 30 | HECA | Sponge network | -1.575 | 6.0E-5 | -0.217 | 0.03463 | 0.449 |
| 13564 | HCG18 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-495-3p | 14 | STAG2 | Sponge network | 1.063 | 0 | 0.252 | 0.00438 | 0.449 |
| 13565 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-3614-5p | 23 | JAZF1 | Sponge network | -0.693 | 0.05629 | -0.377 | 0.02677 | 0.449 |
| 13566 | CTC-429P9.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b | 14 | SYNM | Sponge network | 0.082 | 0.55397 | -2.309 | 1.0E-5 | 0.449 |
| 13567 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | FYN | Sponge network | 0.4 | 0.14611 | -0.131 | 0.37051 | 0.449 |
| 13568 | MAGI2-AS3 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 39 | RIMS2 | Sponge network | -0.129 | 0.57177 | -1.365 | 0.00208 | 0.449 |
| 13569 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-629-3p | 21 | TRPS1 | Sponge network | -1.654 | 0.00144 | -0.191 | 0.44109 | 0.449 |
| 13570 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 23 | AKAP6 | Sponge network | -1.645 | 0 | -1.586 | 3.0E-5 | 0.449 |
| 13571 | RP11-384K6.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-550a-5p | 22 | ARHGEF12 | Sponge network | 0.946 | 0 | 0.09 | 0.35693 | 0.449 |
| 13572 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | ZNF521 | Sponge network | -0.096 | 0.67958 | -0.111 | 0.58068 | 0.449 |
| 13573 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-484;hsa-miR-582-3p | 17 | CBL | Sponge network | 0.858 | 3.0E-5 | 0.291 | 0.01267 | 0.449 |
| 13574 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | CNTN1 | Sponge network | 0.199 | 0.38015 | -2.594 | 0 | 0.449 |
| 13575 | ANKRD10-IT1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-505-3p | 10 | PTPN11 | Sponge network | 1.739 | 0 | 0.431 | 2.0E-5 | 0.449 |
| 13576 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | PDE4DIP | Sponge network | -0.563 | 0.02545 | -0.282 | 0.0257 | 0.449 |
| 13577 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | HACE1 | Sponge network | 0.421 | 0.00084 | -0.072 | 0.55239 | 0.449 |
| 13578 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 36 | MITF | Sponge network | -2.46 | 0 | -0.769 | 0.00022 | 0.449 |
| 13579 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-212-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 20 | RUNX1T1 | Sponge network | -1.425 | 0.04204 | -0.344 | 0.2374 | 0.449 |
| 13580 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-758-3p;hsa-miR-93-5p | 36 | HLF | Sponge network | -0.227 | 0.31657 | -2.443 | 0 | 0.449 |
| 13581 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 15 | DPYD | Sponge network | -0.129 | 0.57177 | -0.736 | 0.00076 | 0.449 |
| 13582 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-501-5p;hsa-miR-589-5p | 12 | MN1 | Sponge network | 0.395 | 0.15451 | -0.358 | 0.24172 | 0.449 |
| 13583 | RP11-701H24.3 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-590-3p | 26 | LPP | Sponge network | -1.425 | 0.04204 | -0.459 | 0.04475 | 0.449 |
| 13584 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 67 | THRB | Sponge network | -0.129 | 0.57177 | -1.117 | 0.0004 | 0.449 |
| 13585 | LINC00865 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 16 | LUZP2 | Sponge network | -1.249 | 7.0E-5 | -0.056 | 0.89416 | 0.449 |
| 13586 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RB1CC1 | Sponge network | 0.225 | 0.30644 | 0.175 | 0.12559 | 0.449 |
| 13587 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-7-1-3p | 12 | PTPRB | Sponge network | 0.4 | 0.14611 | 0.105 | 0.47122 | 0.449 |
| 13588 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | RECK | Sponge network | -2.915 | 0.00015 | -1.043 | 0 | 0.449 |
| 13589 | AP001347.6 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | RPS6KA6 | Sponge network | -1.161 | 0.00591 | -2.989 | 0 | 0.449 |
| 13590 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | IGSF10 | Sponge network | -0.899 | 6.0E-5 | -1.751 | 1.0E-5 | 0.449 |
| 13591 | GNG12-AS1 |
hsa-miR-107;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p | 11 | RNF103 | Sponge network | -0.793 | 7.0E-5 | -0.341 | 1.0E-5 | 0.449 |
| 13592 | CTC-429P9.5 |
hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 12 | BAZ2B | Sponge network | 0.178 | 0.29106 | -0.276 | 0.02833 | 0.449 |
| 13593 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p | 22 | FOXO3 | Sponge network | 0.91 | 0 | -0.04 | 0.72028 | 0.449 |
| 13594 | USP3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-940 | 37 | NTRK2 | Sponge network | -0.162 | 0.47687 | -0.736 | 0.06495 | 0.449 |
| 13595 | PART1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 10 | ADAMTS8 | Sponge network | -2.925 | 0 | -1.476 | 0.00069 | 0.449 |
| 13596 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-589-3p | 34 | SOX5 | Sponge network | -0.777 | 0.16667 | -1.091 | 4.0E-5 | 0.449 |
| 13597 | SNX29P2 |
hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-92a-3p | 10 | HERC1 | Sponge network | 0.4 | 0.14611 | -0.196 | 0.08544 | 0.449 |
| 13598 | CTD-3064M3.3 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-5p | 19 | ZBTB4 | Sponge network | -0.469 | 0.08721 | -0.739 | 0 | 0.449 |
| 13599 | CROCCP2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p | 14 | PHF6 | Sponge network | 0.79 | 0 | 0.785 | 0 | 0.449 |
| 13600 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | 0.532 | 0.00505 | -0.509 | 0.03957 | 0.449 |
| 13601 | MEG3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 19 | EZH1 | Sponge network | 0.249 | 0.35902 | -0.564 | 1.0E-5 | 0.449 |
| 13602 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | EPHA7 | Sponge network | -0.021 | 0.93598 | -2.197 | 3.0E-5 | 0.449 |
| 13603 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -0.611 | 0.09607 | -0.243 | 0.11408 | 0.449 |
| 13604 | HNRNPU-AS1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | CREB1 | Sponge network | 1.601 | 0 | 0.203 | 0.00537 | 0.449 |
| 13605 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p | 17 | DCLK1 | Sponge network | -1.203 | 0.03881 | -1.324 | 0.00014 | 0.449 |
| 13606 | AP001347.6 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | STAT5B | Sponge network | -1.161 | 0.00591 | -0.182 | 0.1082 | 0.449 |
| 13607 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-429;hsa-miR-589-5p | 10 | ITGB3 | Sponge network | -0.154 | 0.53954 | -0.011 | 0.95751 | 0.449 |
| 13608 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 14 | HIP1 | Sponge network | 0.727 | 3.0E-5 | 0.774 | 0 | 0.449 |
| 13609 | CTA-363E19.2 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-485-3p | 14 | UBR3 | Sponge network | 1.055 | 0 | -0.021 | 0.82774 | 0.449 |
| 13610 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p | 27 | EPHA7 | Sponge network | -0.693 | 0.05629 | -2.197 | 3.0E-5 | 0.449 |
| 13611 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-625-5p | 26 | SNX29 | Sponge network | -0.842 | 0.01361 | -0.34 | 0.01371 | 0.449 |
| 13612 | RP5-994D16.9 |
hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | MOB1B | Sponge network | 0.252 | 0.08508 | 0.197 | 0.15266 | 0.449 |
| 13613 | RP11-589P10.7 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-338-3p;hsa-miR-671-5p | 14 | GAS7 | Sponge network | -2.044 | 0.00066 | -0.555 | 0.01602 | 0.449 |
| 13614 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 0.953 | 0 | 0.322 | 0.00215 | 0.449 |
| 13615 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | KCNAB1 | Sponge network | -1.697 | 0.00889 | -0.755 | 2.0E-5 | 0.449 |
| 13616 | RP11-196G18.24 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-424-5p | 16 | ABL2 | Sponge network | 1.262 | 0 | 0.82 | 0 | 0.449 |
| 13617 | RP11-760H22.2 |
hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-655-3p | 11 | ZMYND11 | Sponge network | 0.001 | 0.99753 | -0.2 | 0.05449 | 0.449 |
| 13618 | CTC-429P9.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 10 | TET1 | Sponge network | 0.178 | 0.29106 | -0.135 | 0.52138 | 0.448 |
| 13619 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 18 | CACNA2D1 | Sponge network | -0.726 | 0.00132 | -0.846 | 0.00675 | 0.448 |
| 13620 | CTD-2002H8.2 |
hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PIK3CA | Sponge network | 0.931 | 1.0E-5 | 0.195 | 0.06908 | 0.448 |
| 13621 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-744-5p;hsa-miR-93-5p | 22 | FGFR1 | Sponge network | -0.465 | 0.04703 | -0.509 | 0.03957 | 0.448 |
| 13622 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 21 | RNF144A | Sponge network | -0.976 | 0.00025 | 0.594 | 0.0009 | 0.448 |
| 13623 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 42 | MEF2C | Sponge network | 0.101 | 0.60919 | -0.499 | 0.01448 | 0.448 |
| 13624 | RP11-890B15.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | IL17RD | Sponge network | 0.101 | 0.60919 | -0.019 | 0.94873 | 0.448 |
| 13625 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 17 | RICTOR | Sponge network | 0.673 | 0 | 0.388 | 0.00188 | 0.448 |
| 13626 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 22 | CADM1 | Sponge network | 0.225 | 0.30644 | -0.9 | 0.00084 | 0.448 |
| 13627 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-940 | 26 | MYO9A | Sponge network | 0.056 | 0.81195 | -0.147 | 0.32373 | 0.448 |
| 13628 | AC005154.6 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PTPN12 | Sponge network | 0.848 | 0 | 0.768 | 0 | 0.448 |
| 13629 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 46 | BMPR2 | Sponge network | 0.614 | 1.0E-5 | 0.206 | 0.0635 | 0.448 |
| 13630 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-93-5p | 14 | LHX6 | Sponge network | 0.572 | 0.01722 | -0.087 | 0.69193 | 0.448 |
| 13631 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484 | 10 | EZH1 | Sponge network | 0.811 | 0.03228 | -0.564 | 1.0E-5 | 0.448 |
| 13632 | A1BG-AS1 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-589-3p;hsa-miR-93-3p | 13 | SNX29 | Sponge network | -0.164 | 0.45106 | -0.34 | 0.01371 | 0.448 |
| 13633 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-653-5p | 27 | DMXL1 | Sponge network | -0.487 | 0.02157 | -0.426 | 0.00056 | 0.448 |
| 13634 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PTPN11 | Sponge network | 1.063 | 0 | 0.431 | 2.0E-5 | 0.448 |
| 13635 | TPTEP1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 12 | SORCS2 | Sponge network | -1.216 | 0 | -0.223 | 0.4253 | 0.448 |
| 13636 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 18 | IGF1 | Sponge network | -0.021 | 0.93598 | -0.204 | 0.5898 | 0.448 |
| 13637 | LINC01018 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 29 | RSPO2 | Sponge network | -2.915 | 0.00015 | -3.038 | 0 | 0.448 |
| 13638 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-93-3p | 16 | PCDH10 | Sponge network | -0.154 | 0.78939 | -1.644 | 0.0078 | 0.448 |
| 13639 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | EBF3 | Sponge network | -0.739 | 0.00044 | -0.058 | 0.81437 | 0.448 |
| 13640 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-5p | 16 | PTGFR | Sponge network | 0.176 | 0.37402 | -0.233 | 0.45421 | 0.448 |
| 13641 | RP11-815I9.4 |
hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-365a-3p | 10 | ERCC6 | Sponge network | 0.537 | 0.0006 | 0.23 | 0.015 | 0.448 |
| 13642 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-429 | 14 | SPG20 | Sponge network | -1.654 | 0.00144 | -1.16 | 0 | 0.448 |
| 13643 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 28 | SCN9A | Sponge network | -1.216 | 0 | -0.525 | 0.10593 | 0.448 |
| 13644 | RP11-35G9.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | NIN | Sponge network | 0.657 | 4.0E-5 | -0.253 | 0.13794 | 0.448 |
| 13645 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-5p | 16 | GRIN2A | Sponge network | -0.376 | 0.00337 | -2.161 | 0 | 0.448 |
| 13646 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p | 21 | MAFB | Sponge network | -0.49 | 0.04946 | -0.023 | 0.89865 | 0.448 |
| 13647 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 43 | TGFBR3 | Sponge network | -0.355 | 0.0077 | -1.173 | 1.0E-5 | 0.448 |
| 13648 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CLIP1 | Sponge network | -2.46 | 0 | -0.215 | 0.08684 | 0.448 |
| 13649 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-429 | 12 | CADM1 | Sponge network | -2.62 | 0 | -0.9 | 0.00084 | 0.448 |
| 13650 | LINC00473 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | FYN | Sponge network | -1.203 | 0.03881 | -0.131 | 0.37051 | 0.448 |
| 13651 | FAM215B |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | CREB1 | Sponge network | 0.817 | 3.0E-5 | 0.203 | 0.00537 | 0.448 |
| 13652 | PCA3 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 20 | NRK | Sponge network | -0.709 | 0.18168 | 0.656 | 0.19217 | 0.448 |
| 13653 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 43 | BNC2 | Sponge network | -0.405 | 0.22013 | -0.491 | 0.09988 | 0.448 |
| 13654 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH10 | Sponge network | -2.117 | 0.00161 | -2.397 | 0 | 0.448 |
| 13655 | RP11-819C21.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.537 | 0.00139 | -0.57 | 0 | 0.448 |
| 13656 | AC012360.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-92a-3p;hsa-miR-940 | 20 | NFIB | Sponge network | 0.624 | 0.02176 | 0.023 | 0.90795 | 0.448 |
| 13657 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | ZMYM2 | Sponge network | 0.765 | 7.0E-5 | 0.279 | 0.01273 | 0.448 |
| 13658 | RP11-21A7A.2 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-625-5p | 12 | CHD5 | Sponge network | -1.116 | 0.23686 | -0.922 | 0.01521 | 0.448 |
| 13659 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | PTPRB | Sponge network | -2.117 | 0.00161 | 0.105 | 0.47122 | 0.448 |
| 13660 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-5p | 14 | SLITRK3 | Sponge network | -0.154 | 0.53954 | -3.328 | 0 | 0.448 |
| 13661 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | PRKAA2 | Sponge network | -0.053 | 0.80685 | -2.462 | 0 | 0.448 |
| 13662 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-362-3p | 11 | GPC6 | Sponge network | -1.255 | 0.00981 | -0.054 | 0.81465 | 0.448 |
| 13663 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 24 | AKAP6 | Sponge network | 0.246 | 0.59912 | -1.586 | 3.0E-5 | 0.448 |
| 13664 | CTC-378H22.2 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-532-3p | 10 | CADM3 | Sponge network | 0.001 | 0.99663 | -3.663 | 0 | 0.448 |
| 13665 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | 0.101 | 0.60919 | -1.548 | 0 | 0.448 |
| 13666 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | AHNAK | Sponge network | 0.997 | 0.00066 | -1.207 | 0 | 0.448 |
| 13667 | AC009948.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 22 | NRK | Sponge network | 0.532 | 0.00505 | 0.656 | 0.19217 | 0.448 |
| 13668 | RP11-720L2.4 |
hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-362-3p | 14 | DTNA | Sponge network | -1.255 | 0.00981 | -1.648 | 1.0E-5 | 0.448 |
| 13669 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-3p | 16 | RNF144A | Sponge network | -0.154 | 0.53954 | 0.594 | 0.0009 | 0.448 |
| 13670 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 11 | PREX2 | Sponge network | -0.246 | 0.35034 | -0.272 | 0.25382 | 0.448 |
| 13671 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-421 | 10 | SEMA3E | Sponge network | -1.654 | 0.00144 | -1.717 | 0.00137 | 0.448 |
| 13672 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | AR | Sponge network | 0.225 | 0.30644 | -1.554 | 0 | 0.448 |
| 13673 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | JAZF1 | Sponge network | 0.997 | 0.00066 | -0.377 | 0.02677 | 0.448 |
| 13674 | HOXA-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p | 33 | IL17RD | Sponge network | 0.61 | 0.01593 | -0.019 | 0.94873 | 0.448 |
| 13675 | AC005154.6 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 31 | ABL2 | Sponge network | 0.848 | 0 | 0.82 | 0 | 0.448 |
| 13676 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-143-3p;hsa-miR-154-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-409-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p | 17 | LIMD1 | Sponge network | 1.254 | 0 | 0.103 | 0.28978 | 0.448 |
| 13677 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | LHFP | Sponge network | -0.342 | 0.02288 | -0.167 | 0.41371 | 0.448 |
| 13678 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 31 | SYNE1 | Sponge network | -0.615 | 0.00015 | -0.738 | 0.00316 | 0.448 |
| 13679 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 20 | CNTNAP3 | Sponge network | 0.997 | 0.00066 | -1.465 | 0.00033 | 0.448 |
| 13680 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 31 | LIFR | Sponge network | -0.976 | 0.00025 | -1.794 | 0 | 0.448 |
| 13681 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-589-3p;hsa-miR-590-3p | 25 | TACC1 | Sponge network | 0.174 | 0.30034 | -0.362 | 0.05406 | 0.448 |
| 13682 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | SLC6A15 | Sponge network | -1.111 | 0.0003 | -2.373 | 2.0E-5 | 0.448 |
| 13683 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 16 | NR4A3 | Sponge network | -1.281 | 0.00012 | -1.385 | 0 | 0.448 |
| 13684 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-671-5p | 14 | SEPT6 | Sponge network | -0.49 | 0.04946 | -0.598 | 0.00181 | 0.448 |
| 13685 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p | 12 | FSTL5 | Sponge network | -1.249 | 7.0E-5 | -1.3 | 0.01619 | 0.448 |
| 13686 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 10 | RCAN2 | Sponge network | -0.726 | 0.00132 | -0.33 | 0.15172 | 0.448 |
| 13687 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 43 | TCF4 | Sponge network | 0.299 | 0.57722 | 0.016 | 0.92271 | 0.448 |
| 13688 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-629-3p | 22 | RASSF2 | Sponge network | -1.582 | 8.0E-5 | -0.273 | 0.21961 | 0.448 |
| 13689 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 21 | LRP1B | Sponge network | -2.531 | 0 | -2.163 | 0 | 0.448 |
| 13690 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | MOB1B | Sponge network | 0.67 | 0.00048 | 0.197 | 0.15266 | 0.448 |
| 13691 | AC108488.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-1-3p | 26 | ZFHX3 | Sponge network | 0.259 | 0.12542 | 0.389 | 0.01363 | 0.448 |
| 13692 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 28 | PRRX1 | Sponge network | 0.532 | 0.00505 | 1.316 | 0 | 0.448 |
| 13693 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p | 10 | RBM5 | Sponge network | 0.594 | 0.41522 | -0.205 | 0.0129 | 0.448 |
| 13694 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-744-3p | 23 | PTPN14 | Sponge network | 0.537 | 0.00139 | 0.144 | 0.34595 | 0.448 |
| 13695 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p | 13 | NR2C2 | Sponge network | 0.983 | 0.00048 | 0.21 | 0.03224 | 0.448 |
| 13696 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p | 14 | PDGFRA | Sponge network | 1.344 | 0 | -0.372 | 0.0037 | 0.448 |
| 13697 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PCDH9 | Sponge network | -0.342 | 0.02288 | -1.823 | 4.0E-5 | 0.448 |
| 13698 | RP11-206L10.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-766-3p | 19 | MDM4 | Sponge network | 0.793 | 0 | 0.345 | 0.00762 | 0.448 |
| 13699 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-98-5p | 35 | PBX1 | Sponge network | -0.769 | 0.00035 | -1.206 | 0 | 0.448 |
| 13700 | TP73-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-7-1-3p | 12 | NRG3 | Sponge network | -0.563 | 0.02545 | -1.836 | 4.0E-5 | 0.448 |
| 13701 | BMS1P20 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | ERCC6 | Sponge network | 0.34 | 0.00273 | 0.23 | 0.015 | 0.448 |
| 13702 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | ERBB4 | Sponge network | -2.915 | 0.00015 | -1.975 | 2.0E-5 | 0.448 |
| 13703 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-625-5p;hsa-miR-93-5p | 11 | LHX6 | Sponge network | -2.531 | 0 | -0.087 | 0.69193 | 0.448 |
| 13704 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ZMYND11 | Sponge network | -0.899 | 6.0E-5 | -0.2 | 0.05449 | 0.448 |
| 13705 | RP11-156P1.3 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 10 | RNF180 | Sponge network | 0.214 | 0.18246 | -1.127 | 1.0E-5 | 0.448 |
| 13706 | RP11-57H14.4 |
hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-874-3p | 11 | ARID1B | Sponge network | 0.083 | 0.66405 | 0.003 | 0.97503 | 0.448 |
| 13707 | MYLK-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-340-5p;hsa-miR-34a-5p | 12 | TES | Sponge network | -1.331 | 3.0E-5 | -0.348 | 0.00639 | 0.448 |
| 13708 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | NRXN1 | Sponge network | -0.842 | 0.01361 | -3.778 | 0 | 0.448 |
| 13709 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-625-3p | 13 | ABL2 | Sponge network | 0.497 | 0.01451 | 0.82 | 0 | 0.448 |
| 13710 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p | 17 | BCL6 | Sponge network | -0.18 | 0.20343 | -0.211 | 0.19489 | 0.448 |
| 13711 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | TLL1 | Sponge network | 0.4 | 0.14611 | -1.195 | 3.0E-5 | 0.448 |
| 13712 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-378a-5p;hsa-miR-532-3p | 27 | PBX1 | Sponge network | -0.376 | 0.00337 | -1.206 | 0 | 0.448 |
| 13713 | MAGI2-AS3 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | RTN4 | Sponge network | -0.129 | 0.57177 | -0.412 | 0.00014 | 0.448 |
| 13714 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-576-5p | 23 | IGF1 | Sponge network | 0.889 | 9.0E-5 | -0.204 | 0.5898 | 0.448 |
| 13715 | HCG18 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 19 | ATM | Sponge network | 1.063 | 0 | 0.178 | 0.21568 | 0.448 |
| 13716 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-93-3p | 22 | WWTR1 | Sponge network | -0.777 | 0.16667 | -0.345 | 0.11997 | 0.448 |
| 13717 | RP5-1139B12.3 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-505-5p | 11 | MRVI1 | Sponge network | 0.598 | 0.38481 | -1.051 | 0.00022 | 0.448 |
| 13718 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-590-3p | 11 | HERC2 | Sponge network | -0.096 | 0.67958 | 0.114 | 0.30638 | 0.448 |
| 13719 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-409-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | RCAN2 | Sponge network | 0.41 | 0.02214 | -0.33 | 0.15172 | 0.448 |
| 13720 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 33 | RICTOR | Sponge network | 0.222 | 0.29133 | 0.388 | 0.00188 | 0.448 |
| 13721 | TUG1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p | 14 | SON | Sponge network | 0.972 | 0 | 0.168 | 0.04406 | 0.447 |
| 13722 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-589-3p | 25 | KCNA6 | Sponge network | -0.739 | 0.00044 | -1.531 | 0.00013 | 0.447 |
| 13723 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | KAT6B | Sponge network | 0.101 | 0.60919 | -0.478 | 0.00018 | 0.447 |
| 13724 | RP11-686D22.8 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-5p | 13 | PLXNC1 | Sponge network | 0.898 | 1.0E-5 | 0.569 | 0.00329 | 0.447 |
| 13725 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | 0.331 | 0.57247 | 0.358 | 0.13727 | 0.447 |
| 13726 | LINC00641 |
hsa-let-7b-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 55 | AR | Sponge network | -0.222 | 0.32445 | -1.554 | 0 | 0.447 |
| 13727 | AC108488.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-7-1-3p | 12 | NRXN3 | Sponge network | 0.259 | 0.12542 | -0.893 | 0.04186 | 0.447 |
| 13728 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | GALNT13 | Sponge network | -0.246 | 0.35034 | -0.857 | 0.07439 | 0.447 |
| 13729 | MIR497HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-5p | 14 | ABCA10 | Sponge network | -1.439 | 4.0E-5 | -0.571 | 0.03674 | 0.447 |
| 13730 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ZNF208 | Sponge network | -1.216 | 0 | -0.4 | 0.22381 | 0.447 |
| 13731 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-92a-3p | 14 | DKK3 | Sponge network | 0.253 | 0.09565 | -0.052 | 0.77783 | 0.447 |
| 13732 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-511-5p;hsa-miR-582-5p | 11 | ZFHX3 | Sponge network | 0.993 | 0 | 0.389 | 0.01363 | 0.447 |
| 13733 | MEG3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-532-3p | 17 | SYNM | Sponge network | 0.249 | 0.35902 | -2.309 | 1.0E-5 | 0.447 |
| 13734 | RP11-144G6.12 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | ABL2 | Sponge network | 0.826 | 1.0E-5 | 0.82 | 0 | 0.447 |
| 13735 | RP11-110I1.11 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-3607-3p;hsa-miR-484 | 11 | DDX6 | Sponge network | 1.034 | 0 | 0.091 | 0.36428 | 0.447 |
| 13736 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-93-5p | 12 | EDA2R | Sponge network | -0.842 | 0.01361 | -0.587 | 0.01598 | 0.447 |
| 13737 | RP11-53O19.1 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p | 12 | FGFR1 | Sponge network | -0.405 | 0.22013 | -0.509 | 0.03957 | 0.447 |
| 13738 | RP11-226L15.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 22 | ABL2 | Sponge network | 0.8 | 0 | 0.82 | 0 | 0.447 |
| 13739 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-375 | 14 | TET1 | Sponge network | 0.858 | 3.0E-5 | -0.135 | 0.52138 | 0.447 |
| 13740 | RP1-302G2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p | 10 | ANK2 | Sponge network | -1.483 | 9.0E-5 | -1.603 | 1.0E-5 | 0.447 |
| 13741 | RP11-282O18.3 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-5p | 15 | ABL2 | Sponge network | 0.757 | 0 | 0.82 | 0 | 0.447 |
| 13742 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ATRX | Sponge network | 0.083 | 0.66405 | 0.261 | 0.04408 | 0.447 |
| 13743 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-582-3p;hsa-miR-590-3p | 27 | PRDM2 | Sponge network | 0.083 | 0.66405 | -0.258 | 0.00721 | 0.447 |
| 13744 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-93-5p | 40 | UNC80 | Sponge network | -2.117 | 0.00161 | -1.934 | 0 | 0.447 |
| 13745 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p | 16 | NR2C2 | Sponge network | 0.953 | 0 | 0.21 | 0.03224 | 0.447 |
| 13746 | DNM3OS |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | FYN | Sponge network | 0.572 | 0.01722 | -0.131 | 0.37051 | 0.447 |
| 13747 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-935 | 29 | PTPRT | Sponge network | -2.62 | 0 | -0.907 | 0.03727 | 0.447 |
| 13748 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | ZFHX3 | Sponge network | 0.374 | 0.20054 | 0.389 | 0.01363 | 0.447 |
| 13749 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | FOXP1 | Sponge network | 0.537 | 0.00139 | 0.042 | 0.74368 | 0.447 |
| 13750 | RP11-156P1.3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | KLF10 | Sponge network | 0.214 | 0.18246 | -0.783 | 0 | 0.447 |
| 13751 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 10 | PTPN12 | Sponge network | 1.601 | 0 | 0.768 | 0 | 0.447 |
| 13752 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-429;hsa-miR-455-3p;hsa-miR-877-5p | 17 | NRK | Sponge network | -1.654 | 0.00144 | 0.656 | 0.19217 | 0.447 |
| 13753 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-93-5p | 11 | EDA2R | Sponge network | -1.349 | 0.00017 | -0.587 | 0.01598 | 0.447 |
| 13754 | HOXA-AS2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-93-5p | 56 | DST | Sponge network | 0.61 | 0.01593 | -0.713 | 0.00096 | 0.447 |
| 13755 | RP5-994D16.9 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-766-3p | 10 | ATM | Sponge network | 0.252 | 0.08508 | 0.178 | 0.21568 | 0.447 |
| 13756 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | DMTF1 | Sponge network | 1.11 | 0 | 0.248 | 0.02726 | 0.447 |
| 13757 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p | 34 | KIF1B | Sponge network | 0.178 | 0.29106 | -0.118 | 0.32258 | 0.447 |
| 13758 | LINC00641 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | PCDH9 | Sponge network | -0.222 | 0.32445 | -1.823 | 4.0E-5 | 0.447 |
| 13759 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-485-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | MLLT10 | Sponge network | 0.267 | 0.2937 | 0.105 | 0.33292 | 0.447 |
| 13760 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CDH19 | Sponge network | -0.737 | 0.10822 | -3.434 | 0 | 0.447 |
| 13761 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 12 | MITF | Sponge network | 0.811 | 0.03228 | -0.769 | 0.00022 | 0.447 |
| 13762 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | CNTN1 | Sponge network | -0.899 | 6.0E-5 | -2.594 | 0 | 0.447 |
| 13763 | AGAP11 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-576-5p | 11 | ZNF536 | Sponge network | -1.645 | 0 | -3.115 | 0 | 0.447 |
| 13764 | RP11-53O19.3 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RBBP6 | Sponge network | 1.205 | 0 | 0.163 | 0.05101 | 0.447 |
| 13765 | RP11-400K9.4 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-7-5p | 16 | KCNJ5 | Sponge network | -0.611 | 0.09607 | -0.281 | 0.3339 | 0.447 |
| 13766 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-3p | 17 | TRPS1 | Sponge network | 0.598 | 0.38481 | -0.191 | 0.44109 | 0.447 |
| 13767 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-532-3p;hsa-miR-940 | 20 | PBX1 | Sponge network | -0.829 | 0.01343 | -1.206 | 0 | 0.447 |
| 13768 | LINC01018 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 38 | NTRK2 | Sponge network | -2.915 | 0.00015 | -0.736 | 0.06495 | 0.447 |
| 13769 | CTC-457L16.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 15 | NAV3 | Sponge network | 1.153 | 0.00114 | 0.212 | 0.47729 | 0.447 |
| 13770 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p | 10 | TXNIP | Sponge network | -1.582 | 8.0E-5 | -1.277 | 0 | 0.447 |
| 13771 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-493-5p | 12 | SLITRK6 | Sponge network | -2.62 | 0 | -2.121 | 0 | 0.447 |
| 13772 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TBX18 | Sponge network | 0.194 | 0.64278 | 0.843 | 0.02945 | 0.447 |
| 13773 | RP11-67L2.2 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | ERCC6 | Sponge network | 0.72 | 0.0001 | 0.23 | 0.015 | 0.447 |
| 13774 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-361-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 53 | RUNX1T1 | Sponge network | 0.064 | 0.72934 | -0.344 | 0.2374 | 0.447 |
| 13775 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-98-5p | 32 | ZFHX3 | Sponge network | -0.154 | 0.78939 | 0.389 | 0.01363 | 0.447 |
| 13776 | MEG3 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-5p | 20 | FMNL3 | Sponge network | 0.249 | 0.35902 | 0.555 | 4.0E-5 | 0.447 |
| 13777 | EMX2OS |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 15 | STAT5B | Sponge network | -0.777 | 0.1134 | -0.182 | 0.1082 | 0.447 |
| 13778 | RP11-326G21.1 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PRKCB | Sponge network | -0.021 | 0.93598 | -1.313 | 2.0E-5 | 0.447 |
| 13779 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 29 | SCN9A | Sponge network | -0.487 | 0.02157 | -0.525 | 0.10593 | 0.447 |
| 13780 | HAND2-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ITGB1 | Sponge network | -1.697 | 0.00889 | 0.524 | 0.00017 | 0.447 |
| 13781 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | UBE4B | Sponge network | 0.064 | 0.72934 | -0.235 | 0.007 | 0.447 |
| 13782 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | DOCK4 | Sponge network | 0.997 | 0.00066 | 0.502 | 0.00108 | 0.447 |
| 13783 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-96-5p | 11 | ELMO1 | Sponge network | -1.349 | 0.00017 | 0.04 | 0.83147 | 0.447 |
| 13784 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | NRXN1 | Sponge network | -0.563 | 0.02545 | -3.778 | 0 | 0.447 |
| 13785 | SNHG14 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | EDNRB | Sponge network | -1.111 | 0.0003 | -0.682 | 0.00138 | 0.447 |
| 13786 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-651-5p;hsa-miR-93-3p | 18 | WWTR1 | Sponge network | -0.154 | 0.78939 | -0.345 | 0.11997 | 0.447 |
| 13787 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | TMEM132B | Sponge network | -1.092 | 4.0E-5 | -0.83 | 0.01084 | 0.447 |
| 13788 | TPTEP1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 13 | MN1 | Sponge network | -1.216 | 0 | -0.358 | 0.24172 | 0.447 |
| 13789 | LINC00641 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 15 | MN1 | Sponge network | -0.222 | 0.32445 | -0.358 | 0.24172 | 0.447 |
| 13790 | FZD10-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 11 | LHFP | Sponge network | -0.727 | 0.04726 | -0.167 | 0.41371 | 0.447 |
| 13791 | RP11-774O3.3 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p | 15 | RHOB | Sponge network | -0.487 | 0.02157 | -1.052 | 0 | 0.447 |
| 13792 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 28 | KCNA6 | Sponge network | 0.246 | 0.59912 | -1.531 | 0.00013 | 0.447 |
| 13793 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 15 | JAKMIP2 | Sponge network | -0.777 | 0.1134 | -1.388 | 0 | 0.447 |
| 13794 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p | 17 | BCL6 | Sponge network | 0.532 | 0.00505 | -0.211 | 0.19489 | 0.447 |
| 13795 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PTPRB | Sponge network | 0.997 | 0.00066 | 0.105 | 0.47122 | 0.447 |
| 13796 | PART1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3E | Sponge network | -2.925 | 0 | -1.717 | 0.00137 | 0.447 |
| 13797 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 13 | LRRK2 | Sponge network | 0.335 | 0.20596 | -1.136 | 1.0E-5 | 0.447 |
| 13798 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-93-5p | 30 | ZBTB4 | Sponge network | -0.154 | 0.78939 | -0.739 | 0 | 0.447 |
| 13799 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 28 | PCDH17 | Sponge network | 0.997 | 0.00066 | 0.731 | 2.0E-5 | 0.447 |
| 13800 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-708-5p;hsa-miR-766-3p | 17 | EZH1 | Sponge network | -0.942 | 0 | -0.564 | 1.0E-5 | 0.447 |
| 13801 | RP11-6O2.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-629-3p | 19 | RYR2 | Sponge network | 0.331 | 0.57247 | -1.466 | 0.00031 | 0.447 |
| 13802 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-411-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 50 | SMAD2 | Sponge network | 0.421 | 0.00084 | -0.128 | 0.12732 | 0.447 |
| 13803 | HAND2-AS1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | RASSF3 | Sponge network | -1.697 | 0.00889 | -0.226 | 0.11691 | 0.447 |
| 13804 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | KAT6B | Sponge network | -0.739 | 0.0574 | -0.478 | 0.00018 | 0.447 |
| 13805 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 20 | EPHA3 | Sponge network | -0.318 | 0.65847 | 0.185 | 0.53588 | 0.447 |
| 13806 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 19 | FGFR1 | Sponge network | -0.727 | 0.04726 | -0.509 | 0.03957 | 0.447 |
| 13807 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-935 | 41 | FAT3 | Sponge network | 0.225 | 0.30644 | -1.601 | 0.00051 | 0.447 |
| 13808 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | TLE4 | Sponge network | -0.323 | 0.01372 | -1.157 | 0 | 0.447 |
| 13809 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | RCAN2 | Sponge network | 0.311 | 0.10344 | -0.33 | 0.15172 | 0.447 |
| 13810 | DIO3OS |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p | 18 | MRVI1 | Sponge network | -0.773 | 0.04073 | -1.051 | 0.00022 | 0.447 |
| 13811 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PREX2 | Sponge network | -0.942 | 0 | -0.272 | 0.25382 | 0.447 |
| 13812 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p | 17 | MITF | Sponge network | -4.546 | 0 | -0.769 | 0.00022 | 0.447 |
| 13813 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-769-5p | 15 | SYNPO2 | Sponge network | 0.082 | 0.55397 | -2.342 | 0 | 0.447 |
| 13814 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 19 | NR4A3 | Sponge network | -1.111 | 0.0003 | -1.385 | 0 | 0.447 |
| 13815 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 30 | MCC | Sponge network | -0.738 | 0.21233 | -1.005 | 0 | 0.447 |
| 13816 | LINC00672 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-93-3p | 10 | CNTNAP1 | Sponge network | -0.227 | 0.31657 | 0.358 | 0.13727 | 0.447 |
| 13817 | CTC-429P9.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-505-5p | 13 | MRVI1 | Sponge network | 0.082 | 0.55397 | -1.051 | 0.00022 | 0.447 |
| 13818 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 24 | PPM1A | Sponge network | -1.092 | 4.0E-5 | -0.623 | 0 | 0.447 |
| 13819 | CTD-3064M3.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-539-5p | 12 | PDE4DIP | Sponge network | -0.469 | 0.08721 | -0.282 | 0.0257 | 0.447 |
| 13820 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | PRKAB2 | Sponge network | 0.537 | 0.00139 | -0.367 | 0.01371 | 0.447 |
| 13821 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 36 | ERC1 | Sponge network | -0.576 | 0.10121 | -0.108 | 0.38794 | 0.447 |
| 13822 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 35 | BMPR2 | Sponge network | 0.727 | 3.0E-5 | 0.206 | 0.0635 | 0.447 |
| 13823 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p | 18 | BCL6 | Sponge network | 0.335 | 0.20596 | -0.211 | 0.19489 | 0.447 |
| 13824 | GABPB1-AS1 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 30 | ABL2 | Sponge network | 1.11 | 0 | 0.82 | 0 | 0.447 |
| 13825 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-625-5p | 17 | ZMYM2 | Sponge network | 1.093 | 0 | 0.279 | 0.01273 | 0.447 |
| 13826 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-495-3p | 13 | NF1 | Sponge network | 0.706 | 0 | 0.51 | 0 | 0.447 |
| 13827 | CTC-444N24.11 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM13 | Sponge network | 1.153 | 0 | 0.183 | 0.06968 | 0.447 |
| 13828 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-877-5p;hsa-miR-93-5p | 43 | GAS7 | Sponge network | -0.842 | 0.01361 | -0.555 | 0.01602 | 0.447 |
| 13829 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | FSTL5 | Sponge network | 0.194 | 0.64278 | -1.3 | 0.01619 | 0.447 |
| 13830 | RP11-182J1.1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-484 | 16 | GRIK3 | Sponge network | -0.187 | 0.23787 | -3.208 | 0 | 0.447 |
| 13831 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | ABCB5 | Sponge network | -0.793 | 7.0E-5 | -0.956 | 0.04077 | 0.447 |
| 13832 | RP11-156E6.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 15 | SLC5A3 | Sponge network | 1.266 | 0 | 0.493 | 5.0E-5 | 0.447 |
| 13833 | MAGI2-AS3 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 13 | SPTBN4 | Sponge network | -0.129 | 0.57177 | -0.786 | 0.01281 | 0.446 |
| 13834 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 26 | PTPN14 | Sponge network | 0.494 | 0.00339 | 0.144 | 0.34595 | 0.446 |
| 13835 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-424-5p | 19 | CBFA2T3 | Sponge network | -0.739 | 0.00044 | -1.221 | 1.0E-5 | 0.446 |
| 13836 | CTC-429P9.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-485-3p;hsa-miR-539-5p | 13 | EPHA7 | Sponge network | 0.043 | 0.81628 | -2.197 | 3.0E-5 | 0.446 |
| 13837 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | LATS2 | Sponge network | -1.439 | 4.0E-5 | 0.159 | 0.2304 | 0.446 |
| 13838 | AC006116.24 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-455-5p;hsa-miR-577 | 19 | NOTCH2 | Sponge network | -0.473 | 0.07602 | 0.138 | 0.35163 | 0.446 |
| 13839 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 21 | NR2C2 | Sponge network | -0.487 | 0.02157 | 0.21 | 0.03224 | 0.446 |
| 13840 | CTA-941F9.9 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-576-5p | 10 | QKI | Sponge network | -0.344 | 0.49624 | -0.333 | 0.04111 | 0.446 |
| 13841 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | PRKAA2 | Sponge network | -1.349 | 0.00017 | -2.462 | 0 | 0.446 |
| 13842 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | AKAP12 | Sponge network | 0.082 | 0.55654 | -0.932 | 0.0028 | 0.446 |
| 13843 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 19 | CHD2 | Sponge network | 1.11 | 0 | 0.049 | 0.55371 | 0.446 |
| 13844 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-143-5p;hsa-miR-151a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 12 | CLASP2 | Sponge network | 0.497 | 0.01451 | -0.038 | 0.71 | 0.446 |
| 13845 | LINC00472 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 29 | PPP1R9A | Sponge network | -0.842 | 0.01361 | -0.903 | 0.04254 | 0.446 |
| 13846 | ZNF667-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 10 | NRG3 | Sponge network | -0.976 | 0.00025 | -1.836 | 4.0E-5 | 0.446 |
| 13847 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935 | 38 | FBXO32 | Sponge network | -0.615 | 0.00015 | -0.495 | 0.07561 | 0.446 |
| 13848 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | TLE4 | Sponge network | -0.487 | 0.02157 | -1.157 | 0 | 0.446 |
| 13849 | PWAR6 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | ZNF536 | Sponge network | -1.575 | 6.0E-5 | -3.115 | 0 | 0.446 |
| 13850 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 53 | BNC2 | Sponge network | -0.942 | 0 | -0.491 | 0.09988 | 0.446 |
| 13851 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | MED12L | Sponge network | 0.374 | 0.20054 | -0.194 | 0.55299 | 0.446 |
| 13852 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | 0.064 | 0.72934 | -1.548 | 0 | 0.446 |
| 13853 | RP11-356J5.12 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-940 | 19 | SORCS1 | Sponge network | -0.899 | 6.0E-5 | -3.492 | 0 | 0.446 |
| 13854 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-455-5p | 11 | MN1 | Sponge network | 0.083 | 0.66405 | -0.358 | 0.24172 | 0.446 |
| 13855 | RP4-639F20.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-502-5p;hsa-miR-7-5p;hsa-miR-942-5p | 19 | IGFBP5 | Sponge network | -0.517 | 0.02935 | -0.806 | 0.00105 | 0.446 |
| 13856 | RP11-531A24.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-940 | 22 | SIRT1 | Sponge network | 0.75 | 0.00054 | 0.109 | 0.2593 | 0.446 |
| 13857 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TRIM58 | Sponge network | -0.576 | 0.10121 | -1.692 | 0 | 0.446 |
| 13858 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | PTPN11 | Sponge network | 0.959 | 0 | 0.431 | 2.0E-5 | 0.446 |
| 13859 | RP11-359B12.2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | ITGB1 | Sponge network | 0.064 | 0.72934 | 0.524 | 0.00017 | 0.446 |
| 13860 | RP11-401P9.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-935 | 32 | DIP2C | Sponge network | -0.384 | 0.40652 | -0.436 | 0.01713 | 0.446 |
| 13861 | RP11-890B15.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-590-3p | 18 | ROBO1 | Sponge network | 0.101 | 0.60919 | -0.009 | 0.96154 | 0.446 |
| 13862 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 45 | BCL2 | Sponge network | 0.194 | 0.64278 | -0.899 | 0 | 0.446 |
| 13863 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p | 18 | PTGFR | Sponge network | -0.154 | 0.78939 | -0.233 | 0.45421 | 0.446 |
| 13864 | LINC00865 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 34 | DIP2C | Sponge network | -1.249 | 7.0E-5 | -0.436 | 0.01713 | 0.446 |
| 13865 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-589-3p | 22 | SCN9A | Sponge network | -0.777 | 0.16667 | -0.525 | 0.10593 | 0.446 |
| 13866 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-429 | 11 | LHX6 | Sponge network | -0.246 | 0.35034 | -0.087 | 0.69193 | 0.446 |
| 13867 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-424-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577 | 19 | NRXN1 | Sponge network | 0.811 | 0.03228 | -3.778 | 0 | 0.446 |
| 13868 | RP11-326G21.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | ADARB1 | Sponge network | -0.021 | 0.93598 | -0.335 | 0.06631 | 0.446 |
| 13869 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 15 | RB1CC1 | Sponge network | 0.848 | 0 | 0.175 | 0.12559 | 0.446 |
| 13870 | RP11-166D19.1 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 10 | EPB41L3 | Sponge network | -0.576 | 0.10121 | -1.193 | 0 | 0.446 |
| 13871 | RP11-774O3.3 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-582-5p | 11 | PRICKLE1 | Sponge network | -0.487 | 0.02157 | -0.112 | 0.67088 | 0.446 |
| 13872 | RAB30-AS1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-766-3p | 23 | MDM4 | Sponge network | 0.752 | 0 | 0.345 | 0.00762 | 0.446 |
| 13873 | DLEU2 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-497-5p | 12 | TBL1XR1 | Sponge network | 1.831 | 0 | 0.578 | 0 | 0.446 |
| 13874 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | SON | Sponge network | 1.923 | 0 | 0.168 | 0.04406 | 0.446 |
| 13875 | ZRANB2-AS1 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p | 10 | TAL1 | Sponge network | -0.376 | 0.00337 | -0.462 | 0.00574 | 0.446 |
| 13876 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 69 | THRB | Sponge network | -0.737 | 0.06182 | -1.117 | 0.0004 | 0.446 |
| 13877 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p | 34 | MOB1B | Sponge network | 0.056 | 0.81195 | 0.197 | 0.15266 | 0.446 |
| 13878 | MAGI2-AS3 |
hsa-miR-148b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | GNAI2 | Sponge network | -0.129 | 0.57177 | -0.063 | 0.54532 | 0.446 |
| 13879 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-629-3p;hsa-miR-93-3p | 14 | EBF3 | Sponge network | -0.318 | 0.65847 | -0.058 | 0.81437 | 0.446 |
| 13880 | PWAR6 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CDH10 | Sponge network | -1.575 | 6.0E-5 | -2.397 | 0 | 0.446 |
| 13881 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 32 | PRKAB2 | Sponge network | 0.72 | 0.0001 | -0.367 | 0.01371 | 0.446 |
| 13882 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-590-3p | 12 | JAKMIP2 | Sponge network | -1.161 | 0.00591 | -1.388 | 0 | 0.446 |
| 13883 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p | 22 | KCNA6 | Sponge network | -4.463 | 0.00025 | -1.531 | 0.00013 | 0.446 |
| 13884 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 51 | PTPRD | Sponge network | -2.452 | 0 | -1.005 | 0.00064 | 0.446 |
| 13885 | AC009948.5 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ITPKB | Sponge network | 0.532 | 0.00505 | -0.704 | 0.00106 | 0.446 |
| 13886 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 11 | SEMA3E | Sponge network | -1.349 | 0.00017 | -1.717 | 0.00137 | 0.446 |
| 13887 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p | 20 | HIP1 | Sponge network | 0.199 | 0.38015 | 0.774 | 0 | 0.446 |
| 13888 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 19 | PIK3C2A | Sponge network | 0.953 | 0 | 0.061 | 0.53776 | 0.446 |
| 13889 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p | 18 | PNRC1 | Sponge network | -0.612 | 0.00094 | -0.646 | 0 | 0.446 |
| 13890 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 67 | NTRK3 | Sponge network | -0.769 | 0.00035 | -1.77 | 3.0E-5 | 0.446 |
| 13891 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p | 14 | ABCB5 | Sponge network | -2.69 | 0.0001 | -0.956 | 0.04077 | 0.446 |
| 13892 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 26 | EBF3 | Sponge network | -0.222 | 0.32445 | -0.058 | 0.81437 | 0.446 |
| 13893 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | GRIA3 | Sponge network | -1.216 | 0 | -1.956 | 0 | 0.446 |
| 13894 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577 | 18 | ADAMTSL3 | Sponge network | -0.18 | 0.20343 | -1.559 | 0 | 0.446 |
| 13895 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-942-5p | 20 | UNC80 | Sponge network | -0.376 | 0.00337 | -1.934 | 0 | 0.446 |
| 13896 | RP11-6O2.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-629-3p | 44 | DST | Sponge network | 0.331 | 0.57247 | -0.713 | 0.00096 | 0.446 |
| 13897 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-655-3p | 10 | NF1 | Sponge network | 0.757 | 0 | 0.51 | 0 | 0.446 |
| 13898 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 13 | FAT4 | Sponge network | -0.187 | 0.23787 | -0.354 | 0.14694 | 0.446 |
| 13899 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MOB1B | Sponge network | 0.873 | 0 | 0.197 | 0.15266 | 0.446 |
| 13900 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | AKAP11 | Sponge network | 1.317 | 0 | 0.196 | 0.09825 | 0.446 |
| 13901 | RP11-822E23.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-5p | 40 | PTPRD | Sponge network | -0.777 | 0.16667 | -1.005 | 0.00064 | 0.446 |
| 13902 | MIR143HG |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 30 | PCDH17 | Sponge network | -0.739 | 0.0574 | 0.731 | 2.0E-5 | 0.446 |
| 13903 | RP11-181G12.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-424-5p | 18 | ABL2 | Sponge network | 0.556 | 0.00298 | 0.82 | 0 | 0.446 |
| 13904 | ZNF667-AS1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p | 15 | CSDE1 | Sponge network | -0.976 | 0.00025 | -0.493 | 0 | 0.446 |
| 13905 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | LRP1B | Sponge network | -0.737 | 0.06182 | -2.163 | 0 | 0.446 |
| 13906 | AC005154.6 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 13 | RAPGEF5 | Sponge network | 0.848 | 0 | 0.536 | 4.0E-5 | 0.446 |
| 13907 | FENDRR |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-93-5p | 26 | RNASEL | Sponge network | -1.281 | 0.00012 | -0.272 | 0.01796 | 0.446 |
| 13908 | RP11-774O3.3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | PRKCB | Sponge network | -0.487 | 0.02157 | -1.313 | 2.0E-5 | 0.446 |
| 13909 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 41 | ADARB1 | Sponge network | -0.227 | 0.31657 | -0.335 | 0.06631 | 0.446 |
| 13910 | HCG11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ABI2 | Sponge network | 0.385 | 0.10067 | 0.175 | 0.14787 | 0.446 |
| 13911 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-5p | 21 | ZFHX3 | Sponge network | -0.154 | 0.53954 | 0.389 | 0.01363 | 0.446 |
| 13912 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-93-5p | 46 | PRKAA2 | Sponge network | -0.777 | 0.16667 | -2.462 | 0 | 0.446 |
| 13913 | USP3-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-589-5p | 18 | ROBO1 | Sponge network | -0.162 | 0.47687 | -0.009 | 0.96154 | 0.446 |
| 13914 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | ABI2 | Sponge network | -0.739 | 0.00044 | 0.175 | 0.14787 | 0.446 |
| 13915 | LINC00654 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 20 | CDH23 | Sponge network | 0.374 | 0.20054 | -0.974 | 0.00034 | 0.446 |
| 13916 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-199b-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 28 | KIF1B | Sponge network | 0.673 | 0 | -0.118 | 0.32258 | 0.446 |
| 13917 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-532-3p | 20 | FOXP1 | Sponge network | 0.497 | 0.01451 | 0.042 | 0.74368 | 0.446 |
| 13918 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | FLI1 | Sponge network | -0.563 | 0.02545 | -0.294 | 0.10905 | 0.446 |
| 13919 | LINC00654 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PTCH1 | Sponge network | 0.374 | 0.20054 | -0.1 | 0.6179 | 0.446 |
| 13920 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | RABEP1 | Sponge network | -1.575 | 6.0E-5 | -0.066 | 0.43142 | 0.446 |
| 13921 | RP11-403I13.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | 0.619 | 0.00157 | 0.809 | 0.00544 | 0.446 |
| 13922 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-671-5p | 17 | CADM3 | Sponge network | 0.246 | 0.59912 | -3.663 | 0 | 0.446 |
| 13923 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | LRP1B | Sponge network | -0.842 | 0.01361 | -2.163 | 0 | 0.446 |
| 13924 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | ABCC9 | Sponge network | 1.302 | 7.0E-5 | -0.466 | 0.14121 | 0.446 |
| 13925 | FZD10-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 46 | DTNA | Sponge network | -0.727 | 0.04726 | -1.648 | 1.0E-5 | 0.446 |
| 13926 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 16 | GRIN2A | Sponge network | -0.811 | 2.0E-5 | -2.161 | 0 | 0.446 |
| 13927 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-339-5p;hsa-miR-421 | 11 | ABCB5 | Sponge network | -0.583 | 0.14589 | -0.956 | 0.04077 | 0.446 |
| 13928 | FAM215B |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-378a-3p | 18 | ABL2 | Sponge network | 0.817 | 3.0E-5 | 0.82 | 0 | 0.446 |
| 13929 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | EPHA3 | Sponge network | -0.08 | 0.90092 | 0.185 | 0.53588 | 0.446 |
| 13930 | C1RL-AS1 |
hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-3p | 11 | MDM4 | Sponge network | 1.26 | 0 | 0.345 | 0.00762 | 0.446 |
| 13931 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 34 | LIFR | Sponge network | -1.281 | 0.00012 | -1.794 | 0 | 0.446 |
| 13932 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-331-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ABI2 | Sponge network | 0.673 | 0 | 0.175 | 0.14787 | 0.446 |
| 13933 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-7-1-3p | 28 | CNTN1 | Sponge network | -0.842 | 0.01361 | -2.594 | 0 | 0.446 |
| 13934 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 32 | ARID5B | Sponge network | 0.997 | 0.00066 | 0.342 | 0.01676 | 0.446 |
| 13935 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | ZNF770 | Sponge network | 0.683 | 1.0E-5 | 0.206 | 0.06751 | 0.446 |
| 13936 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 26 | SON | Sponge network | 0.848 | 0 | 0.168 | 0.04406 | 0.446 |
| 13937 | AC093110.3 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 12 | KIF5B | Sponge network | 1.044 | 2.0E-5 | 0.362 | 0.00066 | 0.446 |
| 13938 | RP11-119F7.5 |
hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | RECK | Sponge network | 0.225 | 0.30644 | -1.043 | 0 | 0.446 |
| 13939 | LINC00265 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | ZMYM2 | Sponge network | 1.07 | 0 | 0.279 | 0.01273 | 0.446 |
| 13940 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ZMYND11 | Sponge network | -0.49 | 0.04946 | -0.2 | 0.05449 | 0.446 |
| 13941 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-629-5p | 27 | IGF1 | Sponge network | -0.154 | 0.78939 | -0.204 | 0.5898 | 0.446 |
| 13942 | RP11-967K21.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-542-3p | 11 | ITGB1 | Sponge network | 0.056 | 0.81195 | 0.524 | 0.00017 | 0.446 |
| 13943 | RP11-248G5.8 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 16 | BRD3 | Sponge network | 1.294 | 0 | 0.37 | 0.00013 | 0.446 |
| 13944 | TP73-AS1 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 10 | ADH1B | Sponge network | -0.563 | 0.02545 | -3.716 | 0 | 0.446 |
| 13945 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 11 | WWTR1 | Sponge network | -0.021 | 0.93598 | -0.345 | 0.11997 | 0.446 |
| 13946 | RP11-527J8.1 |
hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p | 12 | MDM4 | Sponge network | 1.324 | 0 | 0.345 | 0.00762 | 0.446 |
| 13947 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PLSCR4 | Sponge network | 0.619 | 0.00157 | -0.474 | 0.03833 | 0.446 |
| 13948 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-654-3p | 21 | NR2C2 | Sponge network | 0.657 | 4.0E-5 | 0.21 | 0.03224 | 0.445 |
| 13949 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 36 | EPHA3 | Sponge network | -2.46 | 0 | 0.185 | 0.53588 | 0.445 |
| 13950 | RP11-254F7.2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-5p | 13 | FNBP1 | Sponge network | 0.176 | 0.37402 | -0.969 | 4.0E-5 | 0.445 |
| 13951 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | RECK | Sponge network | -0.72 | 0.24239 | -1.043 | 0 | 0.445 |
| 13952 | DNM3OS |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-93-5p | 62 | DST | Sponge network | 0.572 | 0.01722 | -0.713 | 0.00096 | 0.445 |
| 13953 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-940 | 16 | ZFP36L1 | Sponge network | -0.612 | 0.00094 | -0.505 | 8.0E-5 | 0.445 |
| 13954 | RP11-298J20.4 |
hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3607-3p | 17 | KIF1B | Sponge network | 0.002 | 0.99057 | -0.118 | 0.32258 | 0.445 |
| 13955 | TUG1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | USP9X | Sponge network | 0.972 | 0 | 0.222 | 0.01689 | 0.445 |
| 13956 | FAM83H-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p | 10 | CBFB | Sponge network | 3.083 | 0 | 1.061 | 0 | 0.445 |
| 13957 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | HOOK3 | Sponge network | -0.777 | 0.1134 | 0.273 | 0.09957 | 0.445 |
| 13958 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 40 | DIP2C | Sponge network | -0.777 | 0.1134 | -0.436 | 0.01713 | 0.445 |
| 13959 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | PPM1A | Sponge network | 0.101 | 0.60919 | -0.623 | 0 | 0.445 |
| 13960 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | GRIA3 | Sponge network | -0.576 | 0.10121 | -1.956 | 0 | 0.445 |
| 13961 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 64 | FAT3 | Sponge network | -2.925 | 0 | -1.601 | 0.00051 | 0.445 |
| 13962 | RP11-359E10.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-940 | 15 | CRTC3 | Sponge network | 0.299 | 0.57722 | 0.164 | 0.16786 | 0.445 |
| 13963 | BVES-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-511-5p | 24 | BTG2 | Sponge network | -2.62 | 0 | -1.763 | 0 | 0.445 |
| 13964 | SH3RF3-AS1 |
hsa-miR-126-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-576-5p;hsa-miR-877-5p | 17 | PGR | Sponge network | 0.889 | 9.0E-5 | -1.704 | 0 | 0.445 |
| 13965 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 17 | RIMBP2 | Sponge network | -0.513 | 0.04703 | -1.968 | 5.0E-5 | 0.445 |
| 13966 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 73 | IL6ST | Sponge network | -2.46 | 0 | -0.402 | 0.00676 | 0.445 |
| 13967 | LINC00472 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-7-1-3p | 10 | MLF1 | Sponge network | -0.842 | 0.01361 | -0.893 | 0.00727 | 0.445 |
| 13968 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-93-5p | 49 | AR | Sponge network | -1.645 | 0 | -1.554 | 0 | 0.445 |
| 13969 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-511-5p | 12 | TRIM33 | Sponge network | 1.294 | 0 | 0.402 | 7.0E-5 | 0.445 |
| 13970 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -0.096 | 0.67958 | -3.328 | 0 | 0.445 |
| 13971 | RP11-156P1.3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | BCL9 | Sponge network | 0.214 | 0.18246 | 0.321 | 0.00267 | 0.445 |
| 13972 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | AHNAK | Sponge network | -1.697 | 0.00889 | -1.207 | 0 | 0.445 |
| 13973 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 54 | WDFY3 | Sponge network | -0.096 | 0.67958 | -0.201 | 0.0967 | 0.445 |
| 13974 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 39 | CBX5 | Sponge network | 0.614 | 1.0E-5 | 0.165 | 0.24446 | 0.445 |
| 13975 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ERCC4 | Sponge network | 0.931 | 1.0E-5 | 0.126 | 0.15266 | 0.445 |
| 13976 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 28 | ANK2 | Sponge network | 0.083 | 0.66405 | -1.603 | 1.0E-5 | 0.445 |
| 13977 | SNX29P2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-671-5p | 16 | CDH23 | Sponge network | 0.4 | 0.14611 | -0.974 | 0.00034 | 0.445 |
| 13978 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | RASSF8 | Sponge network | -0.811 | 2.0E-5 | -0.122 | 0.65591 | 0.445 |
| 13979 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | SYNE1 | Sponge network | -0.351 | 0.11358 | -0.738 | 0.00316 | 0.445 |
| 13980 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | NRXN1 | Sponge network | 0.335 | 0.20596 | -3.778 | 0 | 0.445 |
| 13981 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | TET1 | Sponge network | 0.572 | 0.01722 | -0.135 | 0.52138 | 0.445 |
| 13982 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-454-3p | 18 | ZBTB4 | Sponge network | 0.178 | 0.29106 | -0.739 | 0 | 0.445 |
| 13983 | EIF3J-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | ABI2 | Sponge network | 0.331 | 0.00509 | 0.175 | 0.14787 | 0.445 |
| 13984 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450a-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 20 | CHD2 | Sponge network | 0.494 | 0.00339 | 0.049 | 0.55371 | 0.445 |
| 13985 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p | 13 | DCLK1 | Sponge network | 0.176 | 0.37402 | -1.324 | 0.00014 | 0.445 |
| 13986 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 25 | HECA | Sponge network | 0.782 | 0.00016 | -0.217 | 0.03463 | 0.445 |
| 13987 | H19 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-200b-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-625-5p | 13 | ABI2 | Sponge network | 2.439 | 0 | 0.175 | 0.14787 | 0.445 |
| 13988 | LINC00473 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | NRXN3 | Sponge network | -1.203 | 0.03881 | -0.893 | 0.04186 | 0.445 |
| 13989 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 18 | CHD2 | Sponge network | -0.323 | 0.01372 | 0.049 | 0.55371 | 0.445 |
| 13990 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 23 | GREM1 | Sponge network | -0.777 | 0.1134 | 0.902 | 0.01691 | 0.445 |
| 13991 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-532-3p;hsa-miR-93-3p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | 0.199 | 0.38015 | 0.358 | 0.13727 | 0.445 |
| 13992 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-93-5p | 59 | TCF4 | Sponge network | -0.793 | 7.0E-5 | 0.016 | 0.92271 | 0.445 |
| 13993 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 46 | ZFHX4 | Sponge network | -1.281 | 0.00012 | 0.018 | 0.95574 | 0.445 |
| 13994 | RP11-473I1.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CEP170 | Sponge network | 0.683 | 1.0E-5 | 0.943 | 0 | 0.445 |
| 13995 | EMX2OS |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 18 | ITGA5 | Sponge network | -0.777 | 0.1134 | 0.452 | 0.03524 | 0.445 |
| 13996 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 18 | RICTOR | Sponge network | 1.597 | 0 | 0.388 | 0.00188 | 0.445 |
| 13997 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | BNC2 | Sponge network | -1.203 | 0.03881 | -0.491 | 0.09988 | 0.445 |
| 13998 | SNHG12 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p;hsa-miR-495-3p | 16 | PHF6 | Sponge network | 1.103 | 0 | 0.785 | 0 | 0.445 |
| 13999 | RP11-434H6.7 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-409-3p | 13 | MOB1B | Sponge network | 0.358 | 0.14302 | 0.197 | 0.15266 | 0.445 |
| 14000 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-576-5p | 36 | FAT3 | Sponge network | 0.176 | 0.37402 | -1.601 | 0.00051 | 0.445 |
| 14001 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | HACE1 | Sponge network | 1.153 | 0 | -0.072 | 0.55239 | 0.445 |
| 14002 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 12 | ADAMTSL3 | Sponge network | -0.726 | 0.00132 | -1.559 | 0 | 0.445 |
| 14003 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | RB1CC1 | Sponge network | 0.417 | 0.00445 | 0.175 | 0.12559 | 0.445 |
| 14004 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | TRIM33 | Sponge network | 0.545 | 0.00064 | 0.402 | 7.0E-5 | 0.445 |
| 14005 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | PBRM1 | Sponge network | 0.421 | 0.00084 | -0.029 | 0.78601 | 0.445 |
| 14006 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 25 | GRIN2A | Sponge network | -0.777 | 0.1134 | -2.161 | 0 | 0.445 |
| 14007 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 20 | ATRX | Sponge network | 1.757 | 0 | 0.261 | 0.04408 | 0.445 |
| 14008 | LINC00657 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-495-3p | 13 | STAG2 | Sponge network | 0.91 | 0 | 0.252 | 0.00438 | 0.445 |
| 14009 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940 | 23 | CRTC3 | Sponge network | -0.355 | 0.0077 | 0.164 | 0.16786 | 0.445 |
| 14010 | AC127904.2 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 10 | BAZ2B | Sponge network | 1.308 | 0 | -0.276 | 0.02833 | 0.445 |
| 14011 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | USP12 | Sponge network | 0.72 | 0.0001 | -0.102 | 0.40768 | 0.445 |
| 14012 | SERTAD4-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-424-5p;hsa-miR-505-5p | 21 | KCNA6 | Sponge network | -0.932 | 0.00329 | -1.531 | 0.00013 | 0.445 |
| 14013 | DNM3OS |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-5p | 12 | MLLT11 | Sponge network | 0.572 | 0.01722 | -0.661 | 0.01733 | 0.445 |
| 14014 | AP001347.6 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | TET1 | Sponge network | -1.161 | 0.00591 | -0.135 | 0.52138 | 0.445 |
| 14015 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 37 | ETV1 | Sponge network | -0.129 | 0.57177 | -0.096 | 0.67674 | 0.445 |
| 14016 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-576-5p | 12 | NUAK1 | Sponge network | 1.122 | 0.02989 | 0.904 | 0 | 0.445 |
| 14017 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | SEMA3C | Sponge network | 0.997 | 0.00066 | -0.272 | 0.31153 | 0.445 |
| 14018 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3E | Sponge network | -2.117 | 0.00161 | -1.717 | 0.00137 | 0.445 |
| 14019 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-340-5p | 14 | DYNC1I1 | Sponge network | 0.331 | 0.57247 | -1.12 | 0.00107 | 0.445 |
| 14020 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-450b-5p | 21 | KAT6B | Sponge network | -0.162 | 0.47687 | -0.478 | 0.00018 | 0.445 |
| 14021 | RP11-535M15.1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-362-3p | 12 | AR | Sponge network | -1.14 | 0.03513 | -1.554 | 0 | 0.445 |
| 14022 | RP11-175O19.4 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p | 14 | GSK3B | Sponge network | 0.917 | 0 | 0.266 | 0.00045 | 0.445 |
| 14023 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | PHC3 | Sponge network | 0.532 | 0.00505 | 0.212 | 0.0499 | 0.445 |
| 14024 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-577;hsa-miR-590-3p | 20 | SCN9A | Sponge network | -0.342 | 0.02288 | -0.525 | 0.10593 | 0.445 |
| 14025 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | -0.563 | 0.02545 | 0.783 | 0.00072 | 0.445 |
| 14026 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p | 10 | SCN9A | Sponge network | 0.693 | 0.00076 | -0.525 | 0.10593 | 0.445 |
| 14027 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 36 | ESR1 | Sponge network | 0.249 | 0.35902 | -1.035 | 3.0E-5 | 0.445 |
| 14028 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 25 | RARB | Sponge network | -0.739 | 0.0574 | -0.825 | 2.0E-5 | 0.445 |
| 14029 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-532-3p | 12 | CADM3 | Sponge network | -0.342 | 0.02288 | -3.663 | 0 | 0.445 |
| 14030 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-589-3p | 31 | ADARB1 | Sponge network | -0.246 | 0.35034 | -0.335 | 0.06631 | 0.445 |
| 14031 | ZSCAN16-AS1 |
hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | PAIP2 | Sponge network | -0.342 | 0.02288 | -0.515 | 0 | 0.445 |
| 14032 | ACTA2-AS1 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-7-1-3p | 14 | RASSF3 | Sponge network | 0.194 | 0.64278 | -0.226 | 0.11691 | 0.445 |
| 14033 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-589-5p | 21 | GRIA3 | Sponge network | -2.69 | 0.0001 | -1.956 | 0 | 0.445 |
| 14034 | ZNF582-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 10 | TPO | Sponge network | -1.349 | 0.00017 | -1.87 | 2.0E-5 | 0.445 |
| 14035 | RP11-981G7.6 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 37 | MDM4 | Sponge network | 0.986 | 0.01022 | 0.345 | 0.00762 | 0.445 |
| 14036 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | HIP1 | Sponge network | 0.488 | 0.00084 | 0.774 | 0 | 0.445 |
| 14037 | HCG11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PDE4DIP | Sponge network | 0.385 | 0.10067 | -0.282 | 0.0257 | 0.445 |
| 14038 | LINC00641 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p | 38 | IL17RD | Sponge network | -0.222 | 0.32445 | -0.019 | 0.94873 | 0.445 |
| 14039 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | AKAP11 | Sponge network | 0.91 | 0 | 0.196 | 0.09825 | 0.445 |
| 14040 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p | 16 | FAT4 | Sponge network | 0.176 | 0.37402 | -0.354 | 0.14694 | 0.445 |
| 14041 | TP73-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-629-3p | 13 | ID4 | Sponge network | -0.563 | 0.02545 | -1.644 | 0 | 0.445 |
| 14042 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-98-5p | 30 | DMD | Sponge network | 0.494 | 0.00339 | -1.506 | 0 | 0.445 |
| 14043 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-423-5p | 12 | TIMP3 | Sponge network | -0.469 | 0.08721 | -0.546 | 0.02307 | 0.445 |
| 14044 | EMX2OS |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | MACF1 | Sponge network | -0.777 | 0.1134 | 0.019 | 0.90335 | 0.445 |
| 14045 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 16 | ABCA10 | Sponge network | 0.101 | 0.60919 | -0.571 | 0.03674 | 0.445 |
| 14046 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | BCLAF1 | Sponge network | 0.614 | 1.0E-5 | 0.211 | 0.00684 | 0.445 |
| 14047 | WDFY3-AS2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-671-5p | 31 | DLC1 | Sponge network | -0.942 | 0 | -0.382 | 0.05038 | 0.445 |
| 14048 | TINCR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p | 20 | SASH1 | Sponge network | -0.664 | 0.14543 | -0.502 | 0.00175 | 0.445 |
| 14049 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | PLSCR4 | Sponge network | 0.385 | 0.10067 | -0.474 | 0.03833 | 0.445 |
| 14050 | CTC-490E21.10 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p | 28 | LPP | Sponge network | 1.46 | 1.0E-5 | -0.459 | 0.04475 | 0.445 |
| 14051 | RP4-639F20.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 14 | SCN9A | Sponge network | -0.517 | 0.02935 | -0.525 | 0.10593 | 0.445 |
| 14052 | NUTM2A-AS1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p | 21 | ABL2 | Sponge network | 0.643 | 0 | 0.82 | 0 | 0.445 |
| 14053 | AC009948.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | 0.532 | 0.00505 | -0.7 | 1.0E-5 | 0.445 |
| 14054 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | GALNT13 | Sponge network | -0.358 | 0.17255 | -0.857 | 0.07439 | 0.445 |
| 14055 | CTC-296K1.4 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | PTGFR | Sponge network | -2.076 | 0.00548 | -0.233 | 0.45421 | 0.445 |
| 14056 | PWAR6 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 18 | ID4 | Sponge network | -1.575 | 6.0E-5 | -1.644 | 0 | 0.445 |
| 14057 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 34 | ZFHX3 | Sponge network | 0.782 | 0.00016 | 0.389 | 0.01363 | 0.445 |
| 14058 | WDFY3-AS2 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 19 | CXCL12 | Sponge network | -0.942 | 0 | -1.421 | 0 | 0.445 |
| 14059 | AC003090.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | LUZP2 | Sponge network | -2.117 | 0.00161 | -0.056 | 0.89416 | 0.445 |
| 14060 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 23 | AFF3 | Sponge network | -1.249 | 7.0E-5 | -2.373 | 0 | 0.445 |
| 14061 | RP11-736K20.5 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | SYT4 | Sponge network | -0.358 | 0.17255 | -3.207 | 0 | 0.445 |
| 14062 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3C | Sponge network | 1.302 | 7.0E-5 | -0.272 | 0.31153 | 0.445 |
| 14063 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 38 | QKI | Sponge network | -0.421 | 0.0114 | -0.333 | 0.04111 | 0.445 |
| 14064 | CTC-444N24.11 |
hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TMEM30A | Sponge network | 1.153 | 0 | 0.096 | 0.31031 | 0.445 |
| 14065 | RP11-181G12.2 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-497-5p | 20 | BRD3 | Sponge network | 0.556 | 0.00298 | 0.37 | 0.00013 | 0.445 |
| 14066 | EMX2OS |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 10 | PEA15 | Sponge network | -0.777 | 0.1134 | 0.009 | 0.93318 | 0.445 |
| 14067 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 66 | THRB | Sponge network | -1.697 | 0.00889 | -1.117 | 0.0004 | 0.445 |
| 14068 | RP11-798M19.6 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 17 | KLF10 | Sponge network | -0.612 | 0.00094 | -0.783 | 0 | 0.445 |
| 14069 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 28 | ZMYM2 | Sponge network | 0.848 | 0 | 0.279 | 0.01273 | 0.445 |
| 14070 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p | 23 | NIN | Sponge network | -0.738 | 0.21233 | -0.253 | 0.13794 | 0.445 |
| 14071 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | ZFX | Sponge network | 0.537 | 0.00139 | 0.09 | 0.42563 | 0.445 |
| 14072 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-940 | 11 | MCC | Sponge network | 0.043 | 0.81628 | -1.005 | 0 | 0.445 |
| 14073 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-577 | 13 | AKAP12 | Sponge network | -0.303 | 0.21002 | -0.932 | 0.0028 | 0.445 |
| 14074 | AC007566.10 |
hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p | 19 | MDM4 | Sponge network | 2.368 | 2.0E-5 | 0.345 | 0.00762 | 0.445 |
| 14075 | LINC00476 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-651-5p | 31 | PRDM2 | Sponge network | -0.615 | 0.00015 | -0.258 | 0.00721 | 0.445 |
| 14076 | LINC00843 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | NF1 | Sponge network | 1.106 | 0 | 0.51 | 0 | 0.445 |
| 14077 | SNX29P2 |
hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | PREX2 | Sponge network | 0.4 | 0.14611 | -0.272 | 0.25382 | 0.445 |
| 14078 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 32 | DMD | Sponge network | -0.777 | 0.1134 | -1.506 | 0 | 0.445 |
| 14079 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374a-3p;hsa-miR-589-3p;hsa-miR-629-3p | 21 | PIK3R1 | Sponge network | 0.811 | 0.03228 | -0.133 | 0.36854 | 0.445 |
| 14080 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 19 | FBXO32 | Sponge network | -0.053 | 0.80685 | -0.495 | 0.07561 | 0.445 |
| 14081 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 25 | AKAP6 | Sponge network | -0.769 | 0.00035 | -1.586 | 3.0E-5 | 0.444 |
| 14082 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-628-5p | 25 | BCL6 | Sponge network | -1.575 | 6.0E-5 | -0.211 | 0.19489 | 0.444 |
| 14083 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ATRX | Sponge network | 0.545 | 0.00064 | 0.261 | 0.04408 | 0.444 |
| 14084 | RP11-282O18.3 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-374a-3p;hsa-miR-495-3p | 11 | SLC5A3 | Sponge network | 0.757 | 0 | 0.493 | 5.0E-5 | 0.444 |
| 14085 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PLSCR4 | Sponge network | 0.225 | 0.30644 | -0.474 | 0.03833 | 0.444 |
| 14086 | LINC00963 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 23 | HIP1 | Sponge network | 0.523 | 9.0E-5 | 0.774 | 0 | 0.444 |
| 14087 | FAM66C |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | ABCA10 | Sponge network | -0.901 | 0.00019 | -0.571 | 0.03674 | 0.444 |
| 14088 | LINC00865 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 34 | PPP1R9A | Sponge network | -1.249 | 7.0E-5 | -0.903 | 0.04254 | 0.444 |
| 14089 | NEAT1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 35 | RICTOR | Sponge network | 0.705 | 0.00014 | 0.388 | 0.00188 | 0.444 |
| 14090 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 22 | CACNA2D1 | Sponge network | -0.154 | 0.78939 | -0.846 | 0.00675 | 0.444 |
| 14091 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-590-3p | 15 | ERCC4 | Sponge network | 0.214 | 0.18246 | 0.126 | 0.15266 | 0.444 |
| 14092 | CTC-510F12.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-34a-5p | 13 | ZFHX4 | Sponge network | -0.691 | 0.00146 | 0.018 | 0.95574 | 0.444 |
| 14093 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p | 17 | KCNA6 | Sponge network | -1.375 | 0.08642 | -1.531 | 0.00013 | 0.444 |
| 14094 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | NCAM2 | Sponge network | 0.299 | 0.57722 | -0.482 | 0.22565 | 0.444 |
| 14095 | LINC00578 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577 | 25 | EPHA5 | Sponge network | 0.811 | 0.03228 | -0.957 | 0.01733 | 0.444 |
| 14096 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-671-5p | 15 | RBMS3 | Sponge network | 0.598 | 0.38481 | -0.519 | 0.03551 | 0.444 |
| 14097 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-484;hsa-miR-93-5p | 10 | EDA2R | Sponge network | -2.531 | 0 | -0.587 | 0.01598 | 0.444 |
| 14098 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p | 27 | PBX1 | Sponge network | -1.057 | 0.00029 | -1.206 | 0 | 0.444 |
| 14099 | RP11-35G9.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-96-5p | 18 | NAV3 | Sponge network | 0.622 | 0.00015 | 0.212 | 0.47729 | 0.444 |
| 14100 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 11 | PARK2 | Sponge network | 0.064 | 0.72934 | -1.892 | 0 | 0.444 |
| 14101 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | ZFHX4 | Sponge network | 0.619 | 0.00157 | 0.018 | 0.95574 | 0.444 |
| 14102 | RP11-10N23.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-940 | 16 | CREB1 | Sponge network | 1.134 | 0 | 0.203 | 0.00537 | 0.444 |
| 14103 | CTD-2089N3.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27b-5p | 15 | KCNA6 | Sponge network | -0.314 | 0.62033 | -1.531 | 0.00013 | 0.444 |
| 14104 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | NEDD4 | Sponge network | 0.72 | 0.0001 | 0.02 | 0.89834 | 0.444 |
| 14105 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | CBFA2T3 | Sponge network | -0.129 | 0.57177 | -1.221 | 1.0E-5 | 0.444 |
| 14106 | RP11-1006G14.4 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p | 16 | ATM | Sponge network | 0.559 | 0.00026 | 0.178 | 0.21568 | 0.444 |
| 14107 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | LIFR | Sponge network | -0.323 | 0.01372 | -1.794 | 0 | 0.444 |
| 14108 | RP11-359B12.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | BRD3 | Sponge network | 0.064 | 0.72934 | 0.37 | 0.00013 | 0.444 |
| 14109 | RP11-774O3.3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZNF844 | Sponge network | -0.487 | 0.02157 | -0.966 | 0.00035 | 0.444 |
| 14110 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 36 | PHC3 | Sponge network | -1.575 | 6.0E-5 | 0.212 | 0.0499 | 0.444 |
| 14111 | AC016747.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-7-5p | 16 | KCNJ5 | Sponge network | 0.199 | 0.38015 | -0.281 | 0.3339 | 0.444 |
| 14112 | AC010226.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p | 12 | RELN | Sponge network | -0.811 | 2.0E-5 | -2.403 | 0 | 0.444 |
| 14113 | JAZF1-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | MACF1 | Sponge network | -0.769 | 0.00035 | 0.019 | 0.90335 | 0.444 |
| 14114 | RP11-359E10.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-505-5p | 18 | ASTN1 | Sponge network | 0.299 | 0.57722 | -2.389 | 2.0E-5 | 0.444 |
| 14115 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-5p | 48 | EPHA5 | Sponge network | -0.793 | 7.0E-5 | -0.957 | 0.01733 | 0.444 |
| 14116 | ARHGEF26-AS1 |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | PACRG | Sponge network | -2.46 | 0 | -2.248 | 0 | 0.444 |
| 14117 | RP11-251G23.5 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-628-5p | 13 | SLC5A3 | Sponge network | 1.344 | 0 | 0.493 | 5.0E-5 | 0.444 |
| 14118 | RP11-981G7.6 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-590-3p | 10 | TPR | Sponge network | 0.986 | 0.01022 | 0.582 | 0 | 0.444 |
| 14119 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 32 | CBL | Sponge network | 0.385 | 0.10067 | 0.291 | 0.01267 | 0.444 |
| 14120 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 19 | HIP1 | Sponge network | 0.335 | 0.20596 | 0.774 | 0 | 0.444 |
| 14121 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | 0.194 | 0.64278 | -0.001 | 0.99669 | 0.444 |
| 14122 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-590-3p | 13 | LRRC4 | Sponge network | -0.793 | 7.0E-5 | -1.774 | 0 | 0.444 |
| 14123 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-495-3p | 13 | NF1 | Sponge network | 1.4 | 0 | 0.51 | 0 | 0.444 |
| 14124 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 39 | ZFHX4 | Sponge network | 0.385 | 0.10067 | 0.018 | 0.95574 | 0.444 |
| 14125 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 58 | NTRK3 | Sponge network | -1.203 | 0.03881 | -1.77 | 3.0E-5 | 0.444 |
| 14126 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-7-1-3p | 18 | CBL | Sponge network | 0.817 | 3.0E-5 | 0.291 | 0.01267 | 0.444 |
| 14127 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLIP1 | Sponge network | 0.222 | 0.29133 | -0.215 | 0.08684 | 0.444 |
| 14128 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 45 | SOX5 | Sponge network | -0.777 | 0.1134 | -1.091 | 4.0E-5 | 0.444 |
| 14129 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-935 | 24 | RAP1A | Sponge network | -0.896 | 0.00099 | -0.929 | 0 | 0.444 |
| 14130 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 42 | MOB1B | Sponge network | -1.575 | 6.0E-5 | 0.197 | 0.15266 | 0.444 |
| 14131 | AC009948.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-590-3p | 31 | ABL2 | Sponge network | 0.532 | 0.00505 | 0.82 | 0 | 0.444 |
| 14132 | LINC00672 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | -0.227 | 0.31657 | -1.12 | 0.00107 | 0.444 |
| 14133 | GNG12-AS1 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 14 | MACF1 | Sponge network | -0.793 | 7.0E-5 | 0.019 | 0.90335 | 0.444 |
| 14134 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-590-3p | 17 | ERG | Sponge network | -0.096 | 0.67958 | 0.002 | 0.99011 | 0.444 |
| 14135 | OIP5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 26 | ERCC4 | Sponge network | 0.421 | 0.00084 | 0.126 | 0.15266 | 0.444 |
| 14136 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-654-3p | 19 | DDX6 | Sponge network | 1.147 | 0 | 0.091 | 0.36428 | 0.444 |
| 14137 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | LRIG1 | Sponge network | 0.194 | 0.64278 | -0.537 | 0.00588 | 0.444 |
| 14138 | GAS6-AS2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-940 | 11 | GLI3 | Sponge network | 0.16 | 0.50785 | -0.615 | 0.01482 | 0.444 |
| 14139 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ATRX | Sponge network | 0.476 | 0.19203 | 0.261 | 0.04408 | 0.444 |
| 14140 | PART1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | SPOP | Sponge network | -2.925 | 0 | -0.419 | 1.0E-5 | 0.444 |
| 14141 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-590-3p | 12 | RBM5 | Sponge network | 0.083 | 0.66405 | -0.205 | 0.0129 | 0.444 |
| 14142 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | MITF | Sponge network | 0.532 | 0.00505 | -0.769 | 0.00022 | 0.444 |
| 14143 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-375 | 15 | BCL6 | Sponge network | -0.465 | 0.04703 | -0.211 | 0.19489 | 0.444 |
| 14144 | AGAP11 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p | 12 | KCNAB1 | Sponge network | -1.645 | 0 | -0.755 | 2.0E-5 | 0.444 |
| 14145 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 64 | WDFY3 | Sponge network | -0.129 | 0.57177 | -0.201 | 0.0967 | 0.444 |
| 14146 | FAM66C |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | RYR2 | Sponge network | -0.901 | 0.00019 | -1.466 | 0.00031 | 0.444 |
| 14147 | CTC-559E9.5 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p | 15 | PIK3R1 | Sponge network | 0.594 | 0.00064 | -0.133 | 0.36854 | 0.444 |
| 14148 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | EPAS1 | Sponge network | -0.976 | 0.00025 | -0.184 | 0.14418 | 0.444 |
| 14149 | RP11-34P13.14 |
hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 12 | MDM4 | Sponge network | 1.888 | 0 | 0.345 | 0.00762 | 0.444 |
| 14150 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-590-3p | 18 | PDGFRA | Sponge network | -1.375 | 0.08642 | -0.372 | 0.0037 | 0.444 |
| 14151 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 17 | RASSF8 | Sponge network | -0.726 | 0.00132 | -0.122 | 0.65591 | 0.444 |
| 14152 | LINC00702 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 16 | SPTBN4 | Sponge network | -0.737 | 0.06182 | -0.786 | 0.01281 | 0.444 |
| 14153 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | MOB1B | Sponge network | -0.031 | 0.89063 | 0.197 | 0.15266 | 0.444 |
| 14154 | CTD-2314G24.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 58 | CADM2 | Sponge network | 0.246 | 0.59912 | -3.782 | 0 | 0.444 |
| 14155 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 18 | AKAP6 | Sponge network | -0.513 | 0.04703 | -1.586 | 3.0E-5 | 0.444 |
| 14156 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 17 | ERRFI1 | Sponge network | -0.576 | 0.10121 | -0.798 | 1.0E-5 | 0.444 |
| 14157 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p | 13 | PDS5B | Sponge network | 0.614 | 1.0E-5 | 0.259 | 0.00962 | 0.444 |
| 14158 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-450b-5p | 20 | FBXO32 | Sponge network | 0.176 | 0.37402 | -0.495 | 0.07561 | 0.444 |
| 14159 | LINC00672 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | RASSF8 | Sponge network | -0.227 | 0.31657 | -0.122 | 0.65591 | 0.444 |
| 14160 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | PTPRB | Sponge network | 0.889 | 9.0E-5 | 0.105 | 0.47122 | 0.444 |
| 14161 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-590-5p | 13 | PTPN11 | Sponge network | 0.331 | 0.57247 | 0.431 | 2.0E-5 | 0.444 |
| 14162 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-374a-3p;hsa-miR-654-3p | 10 | ITCH | Sponge network | 0.997 | 0 | 0.084 | 0.34058 | 0.444 |
| 14163 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DOCK4 | Sponge network | 0.225 | 0.30644 | 0.502 | 0.00108 | 0.444 |
| 14164 | LINC00657 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 13 | SHPRH | Sponge network | 0.91 | 0 | 0.098 | 0.40994 | 0.444 |
| 14165 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-539-5p | 10 | SYNPO2 | Sponge network | -0.726 | 0.00132 | -2.342 | 0 | 0.444 |
| 14166 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 23 | IL6ST | Sponge network | -1.255 | 0.00981 | -0.402 | 0.00676 | 0.444 |
| 14167 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 15 | RCAN2 | Sponge network | 0.532 | 0.00505 | -0.33 | 0.15172 | 0.444 |
| 14168 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p | 18 | RICTOR | Sponge network | 1.11 | 1.0E-5 | 0.388 | 0.00188 | 0.444 |
| 14169 | LINC00476 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | CXCL12 | Sponge network | -0.615 | 0.00015 | -1.421 | 0 | 0.444 |
| 14170 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | DMTF1 | Sponge network | 0.494 | 0.00339 | 0.248 | 0.02726 | 0.444 |
| 14171 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b | 11 | PTPN11 | Sponge network | 0.946 | 0 | 0.431 | 2.0E-5 | 0.444 |
| 14172 | AC003090.1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | -2.117 | 0.00161 | -1.379 | 0 | 0.444 |
| 14173 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 59 | PAFAH1B1 | Sponge network | -1.111 | 0.0003 | -0.429 | 0 | 0.444 |
| 14174 | LINC01018 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | DIP2C | Sponge network | -2.915 | 0.00015 | -0.436 | 0.01713 | 0.444 |
| 14175 | LINC00702 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | CSDE1 | Sponge network | -0.737 | 0.06182 | -0.493 | 0 | 0.444 |
| 14176 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | -0.899 | 6.0E-5 | 0.358 | 0.13727 | 0.444 |
| 14177 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | RASSF2 | Sponge network | -0.842 | 0.01361 | -0.273 | 0.21961 | 0.444 |
| 14178 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | EDA2R | Sponge network | -1.697 | 0.00889 | -0.587 | 0.01598 | 0.444 |
| 14179 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 30 | EPHA7 | Sponge network | 0.225 | 0.30644 | -2.197 | 3.0E-5 | 0.444 |
| 14180 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | LRP1B | Sponge network | -0.899 | 6.0E-5 | -2.163 | 0 | 0.444 |
| 14181 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-671-5p | 11 | FAT4 | Sponge network | -0.154 | 0.53954 | -0.354 | 0.14694 | 0.444 |
| 14182 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 26 | JAZF1 | Sponge network | -0.227 | 0.31657 | -0.377 | 0.02677 | 0.444 |
| 14183 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 47 | ZFHX4 | Sponge network | -0.769 | 0.00035 | 0.018 | 0.95574 | 0.444 |
| 14184 | LINC00476 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-484 | 20 | ABL1 | Sponge network | -0.615 | 0.00015 | -0.215 | 0.07804 | 0.444 |
| 14185 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | DIP2C | Sponge network | -0.769 | 0.00035 | -0.436 | 0.01713 | 0.444 |
| 14186 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | NR2C2 | Sponge network | 1.163 | 0.00018 | 0.21 | 0.03224 | 0.444 |
| 14187 | MEG3 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 40 | NOTCH2 | Sponge network | 0.249 | 0.35902 | 0.138 | 0.35163 | 0.444 |
| 14188 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-590-3p;hsa-miR-96-5p | 29 | QKI | Sponge network | -0.811 | 2.0E-5 | -0.333 | 0.04111 | 0.444 |
| 14189 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 33 | FBXO32 | Sponge network | 0.082 | 0.55654 | -0.495 | 0.07561 | 0.444 |
| 14190 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-590-3p | 13 | RSPO2 | Sponge network | -1.375 | 0.08642 | -3.038 | 0 | 0.444 |
| 14191 | RP11-254F19.2 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 15 | LPP | Sponge network | -1.428 | 0.07715 | -0.459 | 0.04475 | 0.444 |
| 14192 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | MGA | Sponge network | 0.765 | 7.0E-5 | 0.323 | 0.00792 | 0.444 |
| 14193 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-940 | 14 | HOOK3 | Sponge network | -0.405 | 0.22013 | 0.273 | 0.09957 | 0.444 |
| 14194 | RP5-1198O20.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 20 | SORCS1 | Sponge network | 0.997 | 0.00066 | -3.492 | 0 | 0.444 |
| 14195 | RP11-166D19.1 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 17 | IRS1 | Sponge network | -0.576 | 0.10121 | -0.026 | 0.88855 | 0.444 |
| 14196 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-654-3p | 10 | ITCH | Sponge network | 1.923 | 0 | 0.084 | 0.34058 | 0.444 |
| 14197 | RP11-968O1.5 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-532-5p | 15 | PDE4DIP | Sponge network | -0.829 | 0.01343 | -0.282 | 0.0257 | 0.444 |
| 14198 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | GIGYF2 | Sponge network | 0.537 | 0.00139 | 0.136 | 0.04403 | 0.444 |
| 14199 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p | 27 | ADARB1 | Sponge network | -0.693 | 0.05629 | -0.335 | 0.06631 | 0.444 |
| 14200 | SNHG16 |
hsa-miR-125a-5p;hsa-miR-150-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-411-5p;hsa-miR-497-5p | 14 | RAD23B | Sponge network | 0.961 | 0 | 0.108 | 0.17414 | 0.444 |
| 14201 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-629-3p | 17 | EBF1 | Sponge network | 0.811 | 0.03228 | -0.384 | 0.08856 | 0.444 |
| 14202 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DNAJB4 | Sponge network | -0.942 | 0 | -0.7 | 1.0E-5 | 0.444 |
| 14203 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-625-5p | 20 | ST6GALNAC3 | Sponge network | -2.095 | 0.00112 | -0.987 | 0 | 0.444 |
| 14204 | ADAMTS9-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 41 | AFF1 | Sponge network | -2.452 | 0 | -0.384 | 0.00053 | 0.444 |
| 14205 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 16 | ARHGAP29 | Sponge network | 0.335 | 0.20596 | 0.131 | 0.42953 | 0.444 |
| 14206 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-576-5p | 18 | EPHA5 | Sponge network | -0.932 | 0.00329 | -0.957 | 0.01733 | 0.444 |
| 14207 | TPTEP1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 31 | KCNA6 | Sponge network | -1.216 | 0 | -1.531 | 0.00013 | 0.444 |
| 14208 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | PIK3R1 | Sponge network | 0.082 | 0.55654 | -0.133 | 0.36854 | 0.444 |
| 14209 | TRAF3IP2-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 19 | SIRT1 | Sponge network | -0.323 | 0.01372 | 0.109 | 0.2593 | 0.444 |
| 14210 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-5p | 22 | SETBP1 | Sponge network | 0.494 | 0.00339 | -1.302 | 1.0E-5 | 0.444 |
| 14211 | RP11-102N12.3 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | MEF2C | Sponge network | -0.351 | 0.11358 | -0.499 | 0.01448 | 0.444 |
| 14212 | DNM3OS |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 25 | SOX11 | Sponge network | 0.572 | 0.01722 | 2.68 | 0 | 0.444 |
| 14213 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PRDM11 | Sponge network | -0.976 | 0.00025 | -0.252 | 0.15511 | 0.444 |
| 14214 | TP73-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 18 | PRUNE2 | Sponge network | -0.563 | 0.02545 | -1.733 | 0.00022 | 0.444 |
| 14215 | RP11-464F9.1 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | DICER1 | Sponge network | 0.959 | 0 | 0.042 | 0.674 | 0.444 |
| 14216 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-550a-3p;hsa-miR-93-5p | 17 | PGR | Sponge network | 0.082 | 0.55397 | -1.704 | 0 | 0.444 |
| 14217 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ABCA10 | Sponge network | -0.612 | 0.00094 | -0.571 | 0.03674 | 0.444 |
| 14218 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | FAM172A | Sponge network | -0.739 | 0.00044 | -0.57 | 0 | 0.443 |
| 14219 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 40 | PAFAH1B1 | Sponge network | -0.096 | 0.67958 | -0.429 | 0 | 0.443 |
| 14220 | AP001347.6 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DCN | Sponge network | -1.161 | 0.00591 | -1.288 | 0 | 0.443 |
| 14221 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 31 | MTSS1 | Sponge network | -0.727 | 0.04726 | -0.81 | 0.00035 | 0.443 |
| 14222 | CTC-429P9.5 |
hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-93-3p | 12 | ERC1 | Sponge network | 0.178 | 0.29106 | -0.108 | 0.38794 | 0.443 |
| 14223 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p | 30 | SCN9A | Sponge network | 0.056 | 0.81195 | -0.525 | 0.10593 | 0.443 |
| 14224 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | EYA4 | Sponge network | -0.976 | 0.00025 | 0.3 | 0.36876 | 0.443 |
| 14225 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p | 25 | TET1 | Sponge network | -0.096 | 0.67958 | -0.135 | 0.52138 | 0.443 |
| 14226 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | TCF4 | Sponge network | -0.811 | 2.0E-5 | 0.016 | 0.92271 | 0.443 |
| 14227 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p | 12 | HLF | Sponge network | -0.187 | 0.23787 | -2.443 | 0 | 0.443 |
| 14228 | AGAP11 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-93-5p | 52 | DST | Sponge network | -1.645 | 0 | -0.713 | 0.00096 | 0.443 |
| 14229 | MIR497HG |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | IL17RD | Sponge network | -1.439 | 4.0E-5 | -0.019 | 0.94873 | 0.443 |
| 14230 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 19 | RARB | Sponge network | 0.532 | 0.00505 | -0.825 | 2.0E-5 | 0.443 |
| 14231 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PRDM11 | Sponge network | -1.111 | 0.0003 | -0.252 | 0.15511 | 0.443 |
| 14232 | DNM3OS |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | PAK3 | Sponge network | 0.572 | 0.01722 | -0.628 | 0.07253 | 0.443 |
| 14233 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SLC6A15 | Sponge network | -2.452 | 0 | -2.373 | 2.0E-5 | 0.443 |
| 14234 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 11 | ZNF382 | Sponge network | 0.064 | 0.72934 | -0.494 | 0.03831 | 0.443 |
| 14235 | AGAP11 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421 | 14 | PEG3 | Sponge network | -1.645 | 0 | -1.74 | 0 | 0.443 |
| 14236 | LINC00654 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | 0.374 | 0.20054 | 0.154 | 0.74723 | 0.443 |
| 14237 | RP11-121C2.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 30 | MDM4 | Sponge network | 1.147 | 0 | 0.345 | 0.00762 | 0.443 |
| 14238 | RP11-701H24.3 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-590-3p | 10 | NRXN1 | Sponge network | -1.425 | 0.04204 | -3.778 | 0 | 0.443 |
| 14239 | RP11-356J5.12 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | FLNA | Sponge network | -0.899 | 6.0E-5 | -0.771 | 0.01377 | 0.443 |
| 14240 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 20 | TBL1XR1 | Sponge network | 0.946 | 0 | 0.578 | 0 | 0.443 |
| 14241 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-577 | 13 | GREM1 | Sponge network | -0.473 | 0.07602 | 0.902 | 0.01691 | 0.443 |
| 14242 | LINC00702 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 27 | CBFA2T3 | Sponge network | -0.737 | 0.06182 | -1.221 | 1.0E-5 | 0.443 |
| 14243 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-7-1-3p | 17 | HDAC9 | Sponge network | -0.842 | 0.01361 | -0.755 | 0.00495 | 0.443 |
| 14244 | HNRNPU-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 21 | UBR3 | Sponge network | 1.601 | 0 | -0.021 | 0.82774 | 0.443 |
| 14245 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-200c-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-550a-3p;hsa-miR-590-3p | 10 | HIVEP1 | Sponge network | 0.72 | 0.0001 | 0.368 | 0.00064 | 0.443 |
| 14246 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | HOOK3 | Sponge network | -0.612 | 0.00094 | 0.273 | 0.09957 | 0.443 |
| 14247 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p | 13 | MYO9A | Sponge network | -0.473 | 0.07602 | -0.147 | 0.32373 | 0.443 |
| 14248 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-940 | 10 | HOOK3 | Sponge network | -0.154 | 0.53954 | 0.273 | 0.09957 | 0.443 |
| 14249 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429 | 12 | CITED2 | Sponge network | -2.69 | 0.0001 | -1.545 | 0 | 0.443 |
| 14250 | LINC00865 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p | 13 | FNBP1 | Sponge network | -1.249 | 7.0E-5 | -0.969 | 4.0E-5 | 0.443 |
| 14251 | LINC00094 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-369-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-92a-3p | 12 | MACF1 | Sponge network | 0.253 | 0.09565 | 0.019 | 0.90335 | 0.443 |
| 14252 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | DYNC1I1 | Sponge network | 0.214 | 0.18246 | -1.12 | 0.00107 | 0.443 |
| 14253 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | MED12L | Sponge network | -0.323 | 0.01372 | -0.194 | 0.55299 | 0.443 |
| 14254 | ARHGEF26-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PCLO | Sponge network | -2.46 | 0 | -1.843 | 0.00064 | 0.443 |
| 14255 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 23 | ZFP36L1 | Sponge network | -2.46 | 0 | -0.505 | 8.0E-5 | 0.443 |
| 14256 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 40 | NTRK2 | Sponge network | -0.487 | 0.02157 | -0.736 | 0.06495 | 0.443 |
| 14257 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | NTRK3 | Sponge network | -0.187 | 0.23787 | -1.77 | 3.0E-5 | 0.443 |
| 14258 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | GPC6 | Sponge network | -0.739 | 0.00044 | -0.054 | 0.81465 | 0.443 |
| 14259 | H1FX-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 24 | SOX5 | Sponge network | -0.206 | 0.12942 | -1.091 | 4.0E-5 | 0.443 |
| 14260 | SNHG14 |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | CTNNA2 | Sponge network | -1.111 | 0.0003 | -2.547 | 0 | 0.443 |
| 14261 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 17 | TGFBR2 | Sponge network | 0.199 | 0.38015 | -0.243 | 0.11408 | 0.443 |
| 14262 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | SOX5 | Sponge network | 0.246 | 0.59912 | -1.091 | 4.0E-5 | 0.443 |
| 14263 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 29 | LRRC7 | Sponge network | -1.216 | 0 | -1.341 | 0.01193 | 0.443 |
| 14264 | RP11-57H14.4 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | BCL9 | Sponge network | 0.083 | 0.66405 | 0.321 | 0.00267 | 0.443 |
| 14265 | SOS1-IT1 |
hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 10 | MDM4 | Sponge network | 0.892 | 0 | 0.345 | 0.00762 | 0.443 |
| 14266 | RP11-867G23.10 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p | 10 | FAM172A | Sponge network | -2.531 | 0 | -0.57 | 0 | 0.443 |
| 14267 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-940 | 10 | PEG3 | Sponge network | 0.082 | 0.55397 | -1.74 | 0 | 0.443 |
| 14268 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p | 19 | CUL5 | Sponge network | 1.254 | 0 | 0.026 | 0.77301 | 0.443 |
| 14269 | RP11-196G18.24 |
hsa-miR-101-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | KIF5B | Sponge network | 1.262 | 0 | 0.362 | 0.00066 | 0.443 |
| 14270 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-744-3p | 24 | MEF2C | Sponge network | -0.693 | 0.05629 | -0.499 | 0.01448 | 0.443 |
| 14271 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-7-1-3p | 32 | CCND2 | Sponge network | -1.092 | 4.0E-5 | -0.267 | 0.3112 | 0.443 |
| 14272 | RP11-693J15.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | SETBP1 | Sponge network | -0.72 | 0.24239 | -1.302 | 1.0E-5 | 0.443 |
| 14273 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | JAZF1 | Sponge network | 0.249 | 0.35902 | -0.377 | 0.02677 | 0.443 |
| 14274 | SERTAD4-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-502-5p | 14 | SYT4 | Sponge network | -0.932 | 0.00329 | -3.207 | 0 | 0.443 |
| 14275 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 33 | HECA | Sponge network | -1.111 | 0.0003 | -0.217 | 0.03463 | 0.443 |
| 14276 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | 1.317 | 0 | 0.323 | 0.00792 | 0.443 |
| 14277 | EMX2OS |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | -0.777 | 0.1134 | -0.998 | 0.00025 | 0.443 |
| 14278 | RP11-356J5.12 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | TPO | Sponge network | -0.899 | 6.0E-5 | -1.87 | 2.0E-5 | 0.443 |
| 14279 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | PREX2 | Sponge network | -0.323 | 0.01372 | -0.272 | 0.25382 | 0.443 |
| 14280 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935 | 28 | DMD | Sponge network | 1.302 | 7.0E-5 | -1.506 | 0 | 0.443 |
| 14281 | ZNF582-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-96-5p | 21 | CBFA2T3 | Sponge network | -1.349 | 0.00017 | -1.221 | 1.0E-5 | 0.443 |
| 14282 | GAS6-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 43 | DTNA | Sponge network | 0.16 | 0.50785 | -1.648 | 1.0E-5 | 0.443 |
| 14283 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-429 | 20 | CBFA2T3 | Sponge network | -2.69 | 0.0001 | -1.221 | 1.0E-5 | 0.443 |
| 14284 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-5p | 19 | EBF3 | Sponge network | -0.842 | 0.01361 | -0.058 | 0.81437 | 0.443 |
| 14285 | RP11-439A17.10 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-5p | 15 | DST | Sponge network | 0.051 | 0.95756 | -0.713 | 0.00096 | 0.443 |
| 14286 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-576-5p | 17 | SOX5 | Sponge network | -0.932 | 0.00329 | -1.091 | 4.0E-5 | 0.443 |
| 14287 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PIK3CA | Sponge network | 1.153 | 0 | 0.195 | 0.06908 | 0.443 |
| 14288 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p | 18 | CITED2 | Sponge network | -1.697 | 0.00889 | -1.545 | 0 | 0.443 |
| 14289 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 25 | QKI | Sponge network | -0.72 | 0.24239 | -0.333 | 0.04111 | 0.443 |
| 14290 | RP11-384O8.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 15 | MAPK10 | Sponge network | -0.738 | 0.21233 | -1.774 | 0 | 0.443 |
| 14291 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-96-5p | 39 | PIK3R1 | Sponge network | 0.72 | 0.0001 | -0.133 | 0.36854 | 0.443 |
| 14292 | FZD10-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | PTPN13 | Sponge network | -0.727 | 0.04726 | -1.208 | 0 | 0.443 |
| 14293 | BVES-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429 | 24 | PRDM2 | Sponge network | -2.62 | 0 | -0.258 | 0.00721 | 0.443 |
| 14294 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 14 | BCLAF1 | Sponge network | 0.683 | 1.0E-5 | 0.211 | 0.00684 | 0.443 |
| 14295 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-577;hsa-miR-93-5p | 20 | SYNE1 | Sponge network | 0.056 | 0.81195 | -0.738 | 0.00316 | 0.443 |
| 14296 | HAND2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | TAL1 | Sponge network | -1.697 | 0.00889 | -0.462 | 0.00574 | 0.443 |
| 14297 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-93-5p | 12 | IGSF10 | Sponge network | -1.654 | 0.00144 | -1.751 | 1.0E-5 | 0.443 |
| 14298 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | AFF3 | Sponge network | -0.737 | 0.10822 | -2.373 | 0 | 0.443 |
| 14299 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | MAF | Sponge network | -1.111 | 0.0003 | -1.257 | 0 | 0.443 |
| 14300 | C20orf166-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577 | 22 | OPCML | Sponge network | -2.69 | 0.0001 | -2.498 | 0 | 0.443 |
| 14301 | MIR143HG |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-340-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-940 | 68 | NFIB | Sponge network | -0.739 | 0.0574 | 0.023 | 0.90795 | 0.443 |
| 14302 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 23 | EPHA3 | Sponge network | -1.057 | 0.00029 | 0.185 | 0.53588 | 0.443 |
| 14303 | DOCK9-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-940 | 18 | NTRK2 | Sponge network | -0.231 | 0.28214 | -0.736 | 0.06495 | 0.443 |
| 14304 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | EBF1 | Sponge network | -0.342 | 0.02288 | -0.384 | 0.08856 | 0.443 |
| 14305 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-629-3p | 11 | EBF3 | Sponge network | -1.886 | 0 | -0.058 | 0.81437 | 0.443 |
| 14306 | PCA3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 30 | ERC1 | Sponge network | -0.709 | 0.18168 | -0.108 | 0.38794 | 0.443 |
| 14307 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-766-3p | 65 | AFF4 | Sponge network | -0.487 | 0.02157 | -0.266 | 0.01565 | 0.443 |
| 14308 | RP11-389C8.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | NAV3 | Sponge network | 0.727 | 3.0E-5 | 0.212 | 0.47729 | 0.443 |
| 14309 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 22 | PTPRT | Sponge network | -1.092 | 4.0E-5 | -0.907 | 0.03727 | 0.443 |
| 14310 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-98-5p | 15 | AKAP6 | Sponge network | -0.301 | 0.06597 | -1.586 | 3.0E-5 | 0.443 |
| 14311 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | LRP1B | Sponge network | -1.057 | 0.00029 | -2.163 | 0 | 0.443 |
| 14312 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-25-3p;hsa-miR-590-3p | 10 | MYCBP2 | Sponge network | 0.178 | 0.29106 | -0.027 | 0.82016 | 0.443 |
| 14313 | RP4-613B23.1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-92a-1-5p | 15 | CTIF | Sponge network | -0.309 | 0.1145 | -0.389 | 0.00549 | 0.443 |
| 14314 | CTD-2554C21.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-93-5p | 45 | DST | Sponge network | -1.654 | 0.00144 | -0.713 | 0.00096 | 0.443 |
| 14315 | MALAT1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 20 | KIF5B | Sponge network | 0.782 | 0.00016 | 0.362 | 0.00066 | 0.443 |
| 14316 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-766-3p | 16 | TBL1XR1 | Sponge network | 0.953 | 0 | 0.578 | 0 | 0.443 |
| 14317 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p | 14 | RPS6KA2 | Sponge network | -0.793 | 7.0E-5 | -0.57 | 0.00013 | 0.443 |
| 14318 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | NEDD4 | Sponge network | -0.737 | 0.06182 | 0.02 | 0.89834 | 0.443 |
| 14319 | SNHG14 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 22 | FMNL3 | Sponge network | -1.111 | 0.0003 | 0.555 | 4.0E-5 | 0.443 |
| 14320 | SNHG14 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 17 | SPTBN4 | Sponge network | -1.111 | 0.0003 | -0.786 | 0.01281 | 0.443 |
| 14321 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-628-5p | 14 | TRIM33 | Sponge network | 0.856 | 0 | 0.402 | 7.0E-5 | 0.443 |
| 14322 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 14 | SPG20 | Sponge network | -0.737 | 0.10822 | -1.16 | 0 | 0.443 |
| 14323 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 23 | SETBP1 | Sponge network | 0.214 | 0.18246 | -1.302 | 1.0E-5 | 0.443 |
| 14324 | RP11-574K11.29 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-374a-3p | 10 | PHF3 | Sponge network | 0.997 | 0 | 0.126 | 0.19652 | 0.443 |
| 14325 | LINC00473 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | ZNF536 | Sponge network | -1.203 | 0.03881 | -3.115 | 0 | 0.443 |
| 14326 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | NRXN1 | Sponge network | -1.249 | 7.0E-5 | -3.778 | 0 | 0.443 |
| 14327 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 16 | PRRX1 | Sponge network | -0.358 | 0.17255 | 1.316 | 0 | 0.443 |
| 14328 | CTA-363E19.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-550a-5p | 16 | ARHGEF12 | Sponge network | 1.055 | 0 | 0.09 | 0.35693 | 0.443 |
| 14329 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-93-5p | 31 | EPHA7 | Sponge network | 0.082 | 0.55397 | -2.197 | 3.0E-5 | 0.443 |
| 14330 | LINC00476 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-877-5p;hsa-miR-935 | 14 | PHOX2B | Sponge network | -0.615 | 0.00015 | -2.68 | 0 | 0.443 |
| 14331 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | BMPR2 | Sponge network | 0.959 | 0 | 0.206 | 0.0635 | 0.443 |
| 14332 | RP11-701H24.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 10 | PGR | Sponge network | -1.425 | 0.04204 | -1.704 | 0 | 0.442 |
| 14333 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ITGAV | Sponge network | 0.532 | 0.00505 | 0.337 | 0.01002 | 0.442 |
| 14334 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-625-5p | 10 | DCLK1 | Sponge network | 0.056 | 0.81195 | -1.324 | 0.00014 | 0.442 |
| 14335 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p | 18 | CDH23 | Sponge network | 0.335 | 0.20596 | -0.974 | 0.00034 | 0.442 |
| 14336 | MEG3 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | GJA1 | Sponge network | 0.249 | 0.35902 | -0.485 | 0.02179 | 0.442 |
| 14337 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 39 | HLF | Sponge network | 0.101 | 0.60919 | -2.443 | 0 | 0.442 |
| 14338 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-539-5p | 15 | MITF | Sponge network | -0.154 | 0.53954 | -0.769 | 0.00022 | 0.442 |
| 14339 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 25 | CBFA2T3 | Sponge network | -0.769 | 0.00035 | -1.221 | 1.0E-5 | 0.442 |
| 14340 | RP11-13K12.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-589-3p | 34 | DIP2C | Sponge network | -0.154 | 0.78939 | -0.436 | 0.01713 | 0.442 |
| 14341 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-589-3p | 18 | MAFB | Sponge network | -0.769 | 0.00035 | -0.023 | 0.89865 | 0.442 |
| 14342 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | FAT4 | Sponge network | 0.4 | 0.14611 | -0.354 | 0.14694 | 0.442 |
| 14343 | MIR143HG |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-3p | 20 | MAFB | Sponge network | -0.739 | 0.0574 | -0.023 | 0.89865 | 0.442 |
| 14344 | AP001347.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-628-5p | 11 | CDH23 | Sponge network | -1.161 | 0.00591 | -0.974 | 0.00034 | 0.442 |
| 14345 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | CAV1 | Sponge network | 0.997 | 0.00066 | -1.013 | 6.0E-5 | 0.442 |
| 14346 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-654-3p | 26 | RICTOR | Sponge network | 1.147 | 0 | 0.388 | 0.00188 | 0.442 |
| 14347 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | -0.727 | 0.04726 | -1.603 | 1.0E-5 | 0.442 |
| 14348 | A1BG-AS1 |
hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-93-3p | 16 | QKI | Sponge network | -0.164 | 0.45106 | -0.333 | 0.04111 | 0.442 |
| 14349 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-940 | 34 | HLF | Sponge network | -1.349 | 0.00017 | -2.443 | 0 | 0.442 |
| 14350 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p | 15 | IGF1 | Sponge network | -0.469 | 0.08721 | -0.204 | 0.5898 | 0.442 |
| 14351 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484 | 20 | ABL1 | Sponge network | -2.69 | 0.0001 | -0.215 | 0.07804 | 0.442 |
| 14352 | RP5-1198O20.4 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | INPP4A | Sponge network | 0.997 | 0.00066 | 0.07 | 0.53563 | 0.442 |
| 14353 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 24 | TGFBR2 | Sponge network | -1.111 | 0.0003 | -0.243 | 0.11408 | 0.442 |
| 14354 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-93-3p | 29 | TOM1L2 | Sponge network | -2.69 | 0.0001 | -0.994 | 0 | 0.442 |
| 14355 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 19 | SYNPO2 | Sponge network | 0.214 | 0.18246 | -2.342 | 0 | 0.442 |
| 14356 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 12 | TBX18 | Sponge network | -1.582 | 8.0E-5 | 0.843 | 0.02945 | 0.442 |
| 14357 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -1.092 | 4.0E-5 | -1.692 | 0 | 0.442 |
| 14358 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-942-5p | 26 | NRXN1 | Sponge network | -0.693 | 0.05629 | -3.778 | 0 | 0.442 |
| 14359 | DIO3OS |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | -0.773 | 0.04073 | -1.74 | 0 | 0.442 |
| 14360 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | FYN | Sponge network | -0.896 | 0.00099 | -0.131 | 0.37051 | 0.442 |
| 14361 | RP11-403I13.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | PRRX1 | Sponge network | 0.619 | 0.00157 | 1.316 | 0 | 0.442 |
| 14362 | RP11-110I1.11 |
hsa-let-7d-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 15 | RICTOR | Sponge network | 1.034 | 0 | 0.388 | 0.00188 | 0.442 |
| 14363 | ARHGEF26-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 33 | PRKCB | Sponge network | -2.46 | 0 | -1.313 | 2.0E-5 | 0.442 |
| 14364 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p | 24 | SCN9A | Sponge network | -0.611 | 0.09607 | -0.525 | 0.10593 | 0.442 |
| 14365 | SNHG14 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | CDKN1C | Sponge network | -1.111 | 0.0003 | -0.929 | 0.00033 | 0.442 |
| 14366 | RP11-21A7A.2 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-628-5p | 24 | DTNA | Sponge network | -1.116 | 0.23686 | -1.648 | 1.0E-5 | 0.442 |
| 14367 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p | 18 | PIK3C2A | Sponge network | 0.959 | 0 | 0.061 | 0.53776 | 0.442 |
| 14368 | RP11-248G5.8 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-539-5p | 13 | TSC1 | Sponge network | 1.294 | 0 | 0.103 | 0.26071 | 0.442 |
| 14369 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 21 | TET1 | Sponge network | -0.563 | 0.02545 | -0.135 | 0.52138 | 0.442 |
| 14370 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 25 | SNX29 | Sponge network | -2.531 | 0 | -0.34 | 0.01371 | 0.442 |
| 14371 | LINC00654 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-935 | 34 | DMD | Sponge network | 0.374 | 0.20054 | -1.506 | 0 | 0.442 |
| 14372 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-582-5p | 11 | KCNAB1 | Sponge network | -2.69 | 0.0001 | -0.755 | 2.0E-5 | 0.442 |
| 14373 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | ZNF292 | Sponge network | 0.421 | 0.00084 | 0.276 | 0.04148 | 0.442 |
| 14374 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | -0.612 | 0.00094 | -0.509 | 0.03957 | 0.442 |
| 14375 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-590-3p | 18 | CADM2 | Sponge network | -1.375 | 0.08642 | -3.782 | 0 | 0.442 |
| 14376 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -0.154 | 0.78939 | -3.328 | 0 | 0.442 |
| 14377 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-5p | 16 | PCDH9 | Sponge network | -0.465 | 0.04703 | -1.823 | 4.0E-5 | 0.442 |
| 14378 | GS1-124K5.11 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-501-5p | 11 | MN1 | Sponge network | 0.494 | 0.00339 | -0.358 | 0.24172 | 0.442 |
| 14379 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-331-3p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 13 | ZBTB4 | Sponge network | -0.295 | 0.02604 | -0.739 | 0 | 0.442 |
| 14380 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 28 | EPHA3 | Sponge network | 0.083 | 0.66405 | 0.185 | 0.53588 | 0.442 |
| 14381 | RP11-473I1.9 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FBXO11 | Sponge network | 0.683 | 1.0E-5 | 0.142 | 0.04938 | 0.442 |
| 14382 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p | 18 | DCLK1 | Sponge network | -0.615 | 0.00015 | -1.324 | 0.00014 | 0.442 |
| 14383 | LINC00578 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577 | 25 | ZFHX3 | Sponge network | 0.811 | 0.03228 | 0.389 | 0.01363 | 0.442 |
| 14384 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p | 10 | GAS1 | Sponge network | -2.46 | 0 | -1.047 | 0.00364 | 0.442 |
| 14385 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-628-5p;hsa-miR-654-3p | 26 | BMPR2 | Sponge network | 0.946 | 0 | 0.206 | 0.0635 | 0.442 |
| 14386 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-500a-3p | 11 | HLF | Sponge network | -7.551 | 0 | -2.443 | 0 | 0.442 |
| 14387 | AP001347.6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p | 33 | NTRK2 | Sponge network | -1.161 | 0.00591 | -0.736 | 0.06495 | 0.442 |
| 14388 | RP11-216B9.6 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 10 | DYNC1I1 | Sponge network | 0.174 | 0.30034 | -1.12 | 0.00107 | 0.442 |
| 14389 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 12 | FYN | Sponge network | 0.199 | 0.38015 | -0.131 | 0.37051 | 0.442 |
| 14390 | SNHG14 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 23 | CELF4 | Sponge network | -1.111 | 0.0003 | -1.135 | 0.00344 | 0.442 |
| 14391 | MEG3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 22 | CDH23 | Sponge network | 0.249 | 0.35902 | -0.974 | 0.00034 | 0.442 |
| 14392 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429 | 18 | ZMYND11 | Sponge network | -2.62 | 0 | -0.2 | 0.05449 | 0.442 |
| 14393 | MIR497HG |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-589-3p | 16 | MAFB | Sponge network | -1.439 | 4.0E-5 | -0.023 | 0.89865 | 0.442 |
| 14394 | CTD-2006H14.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CREB1 | Sponge network | 1.378 | 0 | 0.203 | 0.00537 | 0.442 |
| 14395 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TRIM58 | Sponge network | -2.117 | 0.00161 | -1.692 | 0 | 0.442 |
| 14396 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 19 | LRRC4 | Sponge network | -1.216 | 0 | -1.774 | 0 | 0.442 |
| 14397 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 21 | TBL1XR1 | Sponge network | 0.79 | 0 | 0.578 | 0 | 0.442 |
| 14398 | CTA-204B4.2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | BCL9 | Sponge network | 0.765 | 7.0E-5 | 0.321 | 0.00267 | 0.442 |
| 14399 | RP11-855A2.3 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CREB1 | Sponge network | 2.094 | 0 | 0.203 | 0.00537 | 0.442 |
| 14400 | LINC00641 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 23 | DYNC1I1 | Sponge network | -0.222 | 0.32445 | -1.12 | 0.00107 | 0.442 |
| 14401 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-664a-3p | 21 | GREM1 | Sponge network | -1.216 | 0 | 0.902 | 0.01691 | 0.442 |
| 14402 | PCA3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 16 | CAV1 | Sponge network | -0.709 | 0.18168 | -1.013 | 6.0E-5 | 0.442 |
| 14403 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PRDM11 | Sponge network | -0.129 | 0.57177 | -0.252 | 0.15511 | 0.442 |
| 14404 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 34 | RUNX1T1 | Sponge network | 0.727 | 3.0E-5 | -0.344 | 0.2374 | 0.442 |
| 14405 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | KAT6B | Sponge network | -0.709 | 0.18168 | -0.478 | 0.00018 | 0.442 |
| 14406 | LINC00641 |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 10 | DPP6 | Sponge network | -0.222 | 0.32445 | -3.777 | 0 | 0.442 |
| 14407 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 53 | IL6ST | Sponge network | 0.225 | 0.30644 | -0.402 | 0.00676 | 0.442 |
| 14408 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-423-5p | 16 | CBFA2T3 | Sponge network | -0.583 | 0.14589 | -1.221 | 1.0E-5 | 0.442 |
| 14409 | RP11-119F7.5 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | LHFP | Sponge network | 0.225 | 0.30644 | -0.167 | 0.41371 | 0.442 |
| 14410 | RP11-102N12.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 14 | PTPRT | Sponge network | -0.351 | 0.11358 | -0.907 | 0.03727 | 0.442 |
| 14411 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-93-5p | 17 | EPHA7 | Sponge network | -2.039 | 0.03447 | -2.197 | 3.0E-5 | 0.442 |
| 14412 | RP11-119F7.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-940 | 20 | SIRT1 | Sponge network | 0.225 | 0.30644 | 0.109 | 0.2593 | 0.442 |
| 14413 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-93-5p | 32 | HLF | Sponge network | -0.154 | 0.78939 | -2.443 | 0 | 0.442 |
| 14414 | RP11-102N12.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-671-5p | 10 | CDH23 | Sponge network | -0.351 | 0.11358 | -0.974 | 0.00034 | 0.442 |
| 14415 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 63 | KIF1B | Sponge network | 0.064 | 0.72934 | -0.118 | 0.32258 | 0.442 |
| 14416 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 24 | SNX29 | Sponge network | -0.154 | 0.53954 | -0.34 | 0.01371 | 0.442 |
| 14417 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 14 | PIK3IP1 | Sponge network | -0.576 | 0.10121 | -0.187 | 0.19945 | 0.442 |
| 14418 | ZNF667-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | INPP4A | Sponge network | -0.976 | 0.00025 | 0.07 | 0.53563 | 0.442 |
| 14419 | LINC00657 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-200b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | UBR3 | Sponge network | 0.91 | 0 | -0.021 | 0.82774 | 0.442 |
| 14420 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 28 | SCN9A | Sponge network | -0.709 | 0.18168 | -0.525 | 0.10593 | 0.442 |
| 14421 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-5p | 19 | RET | Sponge network | -0.976 | 0.00025 | -0.833 | 0.03517 | 0.442 |
| 14422 | MIR497HG |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-877-5p | 25 | CRTC1 | Sponge network | -1.439 | 4.0E-5 | -0.116 | 0.28412 | 0.442 |
| 14423 | H1FX-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-629-3p | 20 | ASTN1 | Sponge network | -0.206 | 0.12942 | -2.389 | 2.0E-5 | 0.442 |
| 14424 | AC016747.3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 11 | AKAP12 | Sponge network | 0.199 | 0.38015 | -0.932 | 0.0028 | 0.442 |
| 14425 | LINC00865 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 16 | TLL1 | Sponge network | -1.249 | 7.0E-5 | -1.195 | 3.0E-5 | 0.442 |
| 14426 | MIR497HG |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 17 | SPTBN4 | Sponge network | -1.439 | 4.0E-5 | -0.786 | 0.01281 | 0.442 |
| 14427 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429 | 12 | DCLK1 | Sponge network | -0.246 | 0.35034 | -1.324 | 0.00014 | 0.442 |
| 14428 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-877-5p;hsa-miR-93-5p | 22 | PGR | Sponge network | -0.465 | 0.04703 | -1.704 | 0 | 0.442 |
| 14429 | AC108488.4 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-331-3p;hsa-miR-484 | 11 | FLNA | Sponge network | 0.259 | 0.12542 | -0.771 | 0.01377 | 0.442 |
| 14430 | RP11-473I1.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30c-5p;hsa-miR-374a-3p | 10 | TBL1XR1 | Sponge network | 0.542 | 0.00054 | 0.578 | 0 | 0.442 |
| 14431 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940 | 38 | SNX29 | Sponge network | -1.697 | 0.00889 | -0.34 | 0.01371 | 0.442 |
| 14432 | HOXB-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429 | 10 | STARD13 | Sponge network | -0.154 | 0.53954 | -0.187 | 0.27383 | 0.442 |
| 14433 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 39 | MOB1B | Sponge network | 0.101 | 0.60919 | 0.197 | 0.15266 | 0.442 |
| 14434 | RP11-720L2.4 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p | 10 | SYNM | Sponge network | -1.255 | 0.00981 | -2.309 | 1.0E-5 | 0.442 |
| 14435 | LINC00476 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p | 20 | SORCS1 | Sponge network | -0.615 | 0.00015 | -3.492 | 0 | 0.442 |
| 14436 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-511-5p;hsa-miR-940 | 14 | TRIM33 | Sponge network | 0.921 | 0.00118 | 0.402 | 7.0E-5 | 0.442 |
| 14437 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | FAM172A | Sponge network | -1.057 | 0.00029 | -0.57 | 0 | 0.442 |
| 14438 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SEMA3E | Sponge network | -1.697 | 0.00889 | -1.717 | 0.00137 | 0.442 |
| 14439 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p | 21 | BCL6 | Sponge network | -1.439 | 4.0E-5 | -0.211 | 0.19489 | 0.442 |
| 14440 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | PHC3 | Sponge network | 0.083 | 0.66405 | 0.212 | 0.0499 | 0.442 |
| 14441 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-96-5p;hsa-miR-98-5p | 10 | PLAGL1 | Sponge network | 0.101 | 0.60919 | -0.588 | 0.00912 | 0.442 |
| 14442 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | TMEFF2 | Sponge network | -1.216 | 0 | -2.426 | 0 | 0.442 |
| 14443 | WDFY3-AS2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | NAV3 | Sponge network | -0.942 | 0 | 0.212 | 0.47729 | 0.442 |
| 14444 | TINCR |
hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 12 | PPP2R2C | Sponge network | -0.664 | 0.14543 | -0.959 | 0.07428 | 0.442 |
| 14445 | GNG12-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | TLE4 | Sponge network | -0.793 | 7.0E-5 | -1.157 | 0 | 0.442 |
| 14446 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-5p | 10 | FSTL5 | Sponge network | 0.176 | 0.37402 | -1.3 | 0.01619 | 0.442 |
| 14447 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-590-3p | 14 | NR2C2 | Sponge network | 0.67 | 0.00048 | 0.21 | 0.03224 | 0.442 |
| 14448 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 11 | KCNAB1 | Sponge network | 0.335 | 0.20596 | -0.755 | 2.0E-5 | 0.442 |
| 14449 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | PPM1A | Sponge network | -0.83 | 0 | -0.623 | 0 | 0.442 |
| 14450 | RP11-736K20.5 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p | 15 | ABL1 | Sponge network | -0.358 | 0.17255 | -0.215 | 0.07804 | 0.442 |
| 14451 | RP11-356J5.12 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 24 | RYR2 | Sponge network | -0.899 | 6.0E-5 | -1.466 | 0.00031 | 0.442 |
| 14452 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577 | 14 | SEMA3E | Sponge network | 0.056 | 0.81195 | -1.717 | 0.00137 | 0.442 |
| 14453 | RP11-705C15.3 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | CREB1 | Sponge network | 0.796 | 4.0E-5 | 0.203 | 0.00537 | 0.442 |
| 14454 | DIO3OS |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p | 10 | HMCN1 | Sponge network | -0.773 | 0.04073 | 0.809 | 0.00544 | 0.442 |
| 14455 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 10 | TSLP | Sponge network | -0.942 | 0 | -2.209 | 0 | 0.442 |
| 14456 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-877-5p | 33 | FAT3 | Sponge network | 0.889 | 9.0E-5 | -1.601 | 0.00051 | 0.442 |
| 14457 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-342-3p | 11 | STARD13 | Sponge network | 0.259 | 0.12542 | -0.187 | 0.27383 | 0.442 |
| 14458 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p | 11 | ZYX | Sponge network | -1.697 | 0.00889 | 0.019 | 0.89763 | 0.442 |
| 14459 | RP11-298J20.4 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-5p | 10 | ERC1 | Sponge network | 0.002 | 0.99057 | -0.108 | 0.38794 | 0.442 |
| 14460 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 17 | SUFU | Sponge network | -0.611 | 0.09607 | -0.04 | 0.65489 | 0.442 |
| 14461 | LINC00473 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-503-5p | 15 | ABL1 | Sponge network | -1.203 | 0.03881 | -0.215 | 0.07804 | 0.442 |
| 14462 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-7-1-3p | 16 | RCAN2 | Sponge network | -0.842 | 0.01361 | -0.33 | 0.15172 | 0.442 |
| 14463 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p | 14 | CBL | Sponge network | 0.178 | 0.29106 | 0.291 | 0.01267 | 0.441 |
| 14464 | MIR600HG |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p | 11 | ATM | Sponge network | 0.594 | 0.41522 | 0.178 | 0.21568 | 0.441 |
| 14465 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 31 | GPC6 | Sponge network | -1.216 | 0 | -0.054 | 0.81465 | 0.441 |
| 14466 | RP11-35G9.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.657 | 4.0E-5 | 0.943 | 0 | 0.441 |
| 14467 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 45 | SSBP2 | Sponge network | -0.405 | 0.22013 | -1.58 | 0 | 0.441 |
| 14468 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | CACNA2D1 | Sponge network | 0.374 | 0.20054 | -0.846 | 0.00675 | 0.441 |
| 14469 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | DKK3 | Sponge network | 0.374 | 0.20054 | -0.052 | 0.77783 | 0.441 |
| 14470 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 18 | AR | Sponge network | -1.425 | 0.04204 | -1.554 | 0 | 0.441 |
| 14471 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 38 | PRKAB2 | Sponge network | -2.46 | 0 | -0.367 | 0.01371 | 0.441 |
| 14472 | AP001055.6 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p | 14 | DLC1 | Sponge network | 0.693 | 0.00076 | -0.382 | 0.05038 | 0.441 |
| 14473 | AP001055.6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-7-1-3p | 11 | NIN | Sponge network | 0.693 | 0.00076 | -0.253 | 0.13794 | 0.441 |
| 14474 | LINC00641 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | WWTR1 | Sponge network | -0.222 | 0.32445 | -0.345 | 0.11997 | 0.441 |
| 14475 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p | 32 | ABL2 | Sponge network | 0.997 | 0.00066 | 0.82 | 0 | 0.441 |
| 14476 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-940 | 16 | TRIM33 | Sponge network | 0.541 | 0.00125 | 0.402 | 7.0E-5 | 0.441 |
| 14477 | AC002117.1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MDM4 | Sponge network | 0.476 | 0.19203 | 0.345 | 0.00762 | 0.441 |
| 14478 | HCG18 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-766-3p | 48 | MDM4 | Sponge network | 1.063 | 0 | 0.345 | 0.00762 | 0.441 |
| 14479 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 19 | AKAP11 | Sponge network | 1.063 | 0 | 0.196 | 0.09825 | 0.441 |
| 14480 | RP11-225B17.2 |
hsa-let-7d-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | MDM4 | Sponge network | 1.055 | 0 | 0.345 | 0.00762 | 0.441 |
| 14481 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-655-3p | 28 | DMD | Sponge network | 0.385 | 0.10067 | -1.506 | 0 | 0.441 |
| 14482 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 21 | SON | Sponge network | 0.614 | 1.0E-5 | 0.168 | 0.04406 | 0.441 |
| 14483 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 34 | NRXN1 | Sponge network | -0.405 | 0.22013 | -3.778 | 0 | 0.441 |
| 14484 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 28 | IGF1 | Sponge network | -0.096 | 0.67958 | -0.204 | 0.5898 | 0.441 |
| 14485 | RP11-102N12.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | SOX5 | Sponge network | -0.351 | 0.11358 | -1.091 | 4.0E-5 | 0.441 |
| 14486 | BOLA3-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-877-5p | 20 | ASTN1 | Sponge network | -0.246 | 0.35034 | -2.389 | 2.0E-5 | 0.441 |
| 14487 | LINC00702 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p | 11 | SMARCA1 | Sponge network | -0.737 | 0.06182 | -0.684 | 0.00159 | 0.441 |
| 14488 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 28 | IGF1 | Sponge network | -0.769 | 0.00035 | -0.204 | 0.5898 | 0.441 |
| 14489 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 29 | PRKAB2 | Sponge network | 0.219 | 0.1516 | -0.367 | 0.01371 | 0.441 |
| 14490 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 10 | ZNF292 | Sponge network | 0.91 | 0 | 0.276 | 0.04148 | 0.441 |
| 14491 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p | 18 | ZFHX4 | Sponge network | -1.255 | 0.00981 | 0.018 | 0.95574 | 0.441 |
| 14492 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-484 | 12 | EZH1 | Sponge network | -0.738 | 0.21233 | -0.564 | 1.0E-5 | 0.441 |
| 14493 | LINC00578 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 15 | LRRC7 | Sponge network | 0.811 | 0.03228 | -1.341 | 0.01193 | 0.441 |
| 14494 | TPTEP1 |
hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p | 10 | ELN | Sponge network | -1.216 | 0 | 0.451 | 0.13752 | 0.441 |
| 14495 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-7-1-3p | 19 | EPHA3 | Sponge network | -0.246 | 0.35034 | 0.185 | 0.53588 | 0.441 |
| 14496 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DCN | Sponge network | -0.487 | 0.02157 | -1.288 | 0 | 0.441 |
| 14497 | LINC00957 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 46 | TCF4 | Sponge network | -0.421 | 0.0114 | 0.016 | 0.92271 | 0.441 |
| 14498 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 17 | SPG20 | Sponge network | 0.101 | 0.60919 | -1.16 | 0 | 0.441 |
| 14499 | RP11-13K12.5 |
hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | MYO9A | Sponge network | -0.318 | 0.65847 | -0.147 | 0.32373 | 0.441 |
| 14500 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 29 | PRKAB2 | Sponge network | -0.612 | 0.00094 | -0.367 | 0.01371 | 0.441 |
| 14501 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-940 | 17 | PEG3 | Sponge network | 0.056 | 0.81195 | -1.74 | 0 | 0.441 |
| 14502 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | SOX5 | Sponge network | 0.572 | 0.01722 | -1.091 | 4.0E-5 | 0.441 |
| 14503 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p | 13 | DMD | Sponge network | -4.477 | 0 | -1.506 | 0 | 0.441 |
| 14504 | AC004540.4 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-629-3p | 13 | EPHA7 | Sponge network | -2.549 | 0.00528 | -2.197 | 3.0E-5 | 0.441 |
| 14505 | RP11-536O18.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p | 16 | GAS7 | Sponge network | -0.074 | 0.90758 | -0.555 | 0.01602 | 0.441 |
| 14506 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | EBF1 | Sponge network | -0.72 | 0.24239 | -0.384 | 0.08856 | 0.441 |
| 14507 | LINC00657 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-495-3p | 16 | SLC5A3 | Sponge network | 0.91 | 0 | 0.493 | 5.0E-5 | 0.441 |
| 14508 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-5p | 15 | ABCB5 | Sponge network | -0.162 | 0.47687 | -0.956 | 0.04077 | 0.441 |
| 14509 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-877-5p;hsa-miR-93-5p | 26 | PGR | Sponge network | -0.384 | 0.40652 | -1.704 | 0 | 0.441 |
| 14510 | RP11-156P1.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 71 | LPP | Sponge network | 0.214 | 0.18246 | -0.459 | 0.04475 | 0.441 |
| 14511 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-625-5p | 10 | MPL | Sponge network | -2.69 | 0.0001 | -0.624 | 0.00133 | 0.441 |
| 14512 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 29 | SOX5 | Sponge network | 0.299 | 0.57722 | -1.091 | 4.0E-5 | 0.441 |
| 14513 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-651-5p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 58 | AFF4 | Sponge network | -1.047 | 0 | -0.266 | 0.01565 | 0.441 |
| 14514 | PGM5-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-940 | 24 | NTRK2 | Sponge network | -4.546 | 0 | -0.736 | 0.06495 | 0.441 |
| 14515 | RP11-356J5.12 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 61 | DST | Sponge network | -0.899 | 6.0E-5 | -0.713 | 0.00096 | 0.441 |
| 14516 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PHC3 | Sponge network | 0.539 | 0.03722 | 0.212 | 0.0499 | 0.441 |
| 14517 | NEAT1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-486-5p;hsa-miR-590-3p | 10 | TPR | Sponge network | 0.705 | 0.00014 | 0.582 | 0 | 0.441 |
| 14518 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | 0.16 | 0.50785 | -1.74 | 0 | 0.441 |
| 14519 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-429 | 11 | THSD7A | Sponge network | -1.654 | 0.00144 | 0.242 | 0.25154 | 0.441 |
| 14520 | LINC00472 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 23 | CADM1 | Sponge network | -0.842 | 0.01361 | -0.9 | 0.00084 | 0.441 |
| 14521 | RP11-67L2.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-708-5p;hsa-miR-766-3p | 17 | EZH1 | Sponge network | 0.72 | 0.0001 | -0.564 | 1.0E-5 | 0.441 |
| 14522 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-3p | 16 | SNX29 | Sponge network | -0.376 | 0.00337 | -0.34 | 0.01371 | 0.441 |
| 14523 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-486-5p | 15 | EPHA5 | Sponge network | -0.309 | 0.1145 | -0.957 | 0.01733 | 0.441 |
| 14524 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-539-5p | 11 | PDZD2 | Sponge network | 0.056 | 0.81195 | -0.909 | 3.0E-5 | 0.441 |
| 14525 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-582-5p | 16 | ABI2 | Sponge network | 0.993 | 0 | 0.175 | 0.14787 | 0.441 |
| 14526 | PART1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 16 | NGFR | Sponge network | -2.925 | 0 | -1.271 | 0.01058 | 0.441 |
| 14527 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-877-5p | 16 | CDH19 | Sponge network | -0.096 | 0.67958 | -3.434 | 0 | 0.441 |
| 14528 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-96-5p | 10 | LRRC4 | Sponge network | -1.582 | 8.0E-5 | -1.774 | 0 | 0.441 |
| 14529 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-424-5p | 13 | EZH1 | Sponge network | -0.739 | 0.00044 | -0.564 | 1.0E-5 | 0.441 |
| 14530 | HCG11 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | HERC1 | Sponge network | 0.385 | 0.10067 | -0.196 | 0.08544 | 0.441 |
| 14531 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p | 27 | ZBTB4 | Sponge network | -1.057 | 0.00029 | -0.739 | 0 | 0.441 |
| 14532 | RP11-359B12.2 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 20 | ATM | Sponge network | 0.064 | 0.72934 | 0.178 | 0.21568 | 0.441 |
| 14533 | MYLK-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-940 | 33 | WDFY3 | Sponge network | -1.331 | 3.0E-5 | -0.201 | 0.0967 | 0.441 |
| 14534 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p | 35 | LPP | Sponge network | -2.076 | 0.00548 | -0.459 | 0.04475 | 0.441 |
| 14535 | RP11-693J15.4 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-502-5p;hsa-miR-590-5p | 12 | FMNL3 | Sponge network | -0.72 | 0.24239 | 0.555 | 4.0E-5 | 0.441 |
| 14536 | CTD-2089N3.2 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p | 13 | GRIK3 | Sponge network | -1.375 | 0.08642 | -3.208 | 0 | 0.441 |
| 14537 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-590-3p | 15 | PHC3 | Sponge network | 1.2 | 0.01813 | 0.212 | 0.0499 | 0.441 |
| 14538 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | EDA2R | Sponge network | -0.896 | 0.00099 | -0.587 | 0.01598 | 0.441 |
| 14539 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 33 | TACC1 | Sponge network | -0.227 | 0.31657 | -0.362 | 0.05406 | 0.441 |
| 14540 | RP11-119F7.5 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | DLC1 | Sponge network | 0.225 | 0.30644 | -0.382 | 0.05038 | 0.441 |
| 14541 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | CNTNAP1 | Sponge network | -0.942 | 0 | 0.358 | 0.13727 | 0.441 |
| 14542 | LINC00092 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | CTIF | Sponge network | -1.886 | 0 | -0.389 | 0.00549 | 0.441 |
| 14543 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-93-5p | 13 | MAF | Sponge network | -2.531 | 0 | -1.257 | 0 | 0.441 |
| 14544 | RP1-59D14.5 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-331-3p;hsa-miR-629-3p;hsa-miR-942-5p | 19 | DDX6 | Sponge network | 1.162 | 5.0E-5 | 0.091 | 0.36428 | 0.441 |
| 14545 | RP11-400K9.3 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-940 | 13 | MEF2C | Sponge network | -0.733 | 0.19469 | -0.499 | 0.01448 | 0.441 |
| 14546 | TPTEP1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 15 | PHOX2B | Sponge network | -1.216 | 0 | -2.68 | 0 | 0.441 |
| 14547 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-375;hsa-miR-582-3p | 25 | BMPR2 | Sponge network | 0.858 | 3.0E-5 | 0.206 | 0.0635 | 0.441 |
| 14548 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | PHC3 | Sponge network | 0.919 | 0 | 0.212 | 0.0499 | 0.441 |
| 14549 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 40 | NTRK2 | Sponge network | 0.194 | 0.64278 | -0.736 | 0.06495 | 0.441 |
| 14550 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p | 23 | TBL1XR1 | Sponge network | 0.997 | 0 | 0.578 | 0 | 0.441 |
| 14551 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | NCAM2 | Sponge network | 0.619 | 0.00157 | -0.482 | 0.22565 | 0.441 |
| 14552 | MEG3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-935 | 18 | RELN | Sponge network | 0.249 | 0.35902 | -2.403 | 0 | 0.441 |
| 14553 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-629-3p | 17 | ESR1 | Sponge network | 1.153 | 0.00114 | -1.035 | 3.0E-5 | 0.441 |
| 14554 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | IGSF10 | Sponge network | -1.161 | 0.00591 | -1.751 | 1.0E-5 | 0.441 |
| 14555 | RP11-21A7A.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-505-5p | 10 | MRVI1 | Sponge network | -1.116 | 0.23686 | -1.051 | 0.00022 | 0.441 |
| 14556 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 10 | FLI1 | Sponge network | -1.057 | 0.00029 | -0.294 | 0.10905 | 0.441 |
| 14557 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 34 | PBX1 | Sponge network | -0.303 | 0.21002 | -1.206 | 0 | 0.441 |
| 14558 | RP11-166D19.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-429 | 11 | DUSP1 | Sponge network | -0.576 | 0.10121 | -1.754 | 0 | 0.441 |
| 14559 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 36 | NOTCH2 | Sponge network | -0.421 | 0.0114 | 0.138 | 0.35163 | 0.441 |
| 14560 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 37 | BNC2 | Sponge network | 0.61 | 0.01593 | -0.491 | 0.09988 | 0.441 |
| 14561 | BOLA3-AS1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-429 | 11 | DNMT3A | Sponge network | -0.246 | 0.35034 | 0.302 | 0.0262 | 0.441 |
| 14562 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | ABI2 | Sponge network | 0.997 | 0.00066 | 0.175 | 0.14787 | 0.441 |
| 14563 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MLF1 | Sponge network | -2.46 | 0 | -0.893 | 0.00727 | 0.441 |
| 14564 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 15 | ST6GALNAC3 | Sponge network | -2.039 | 0.03447 | -0.987 | 0 | 0.441 |
| 14565 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 39 | ZFHX3 | Sponge network | -0.487 | 0.02157 | 0.389 | 0.01363 | 0.441 |
| 14566 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | TGFBR2 | Sponge network | -1.886 | 0 | -0.243 | 0.11408 | 0.441 |
| 14567 | RP11-983P16.4 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-542-3p | 12 | DYNC1I1 | Sponge network | 0.41 | 0.02214 | -1.12 | 0.00107 | 0.441 |
| 14568 | CTD-2089N3.1 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p | 12 | RASSF2 | Sponge network | -0.314 | 0.62033 | -0.273 | 0.21961 | 0.441 |
| 14569 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-375;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p | 30 | TRPS1 | Sponge network | -2.62 | 0 | -0.191 | 0.44109 | 0.441 |
| 14570 | FAM66C |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-542-3p | 11 | ALDH1A2 | Sponge network | -0.901 | 0.00019 | -2.411 | 0 | 0.441 |
| 14571 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | TET1 | Sponge network | 0.083 | 0.66405 | -0.135 | 0.52138 | 0.441 |
| 14572 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 18 | FAT4 | Sponge network | -0.222 | 0.32445 | -0.354 | 0.14694 | 0.441 |
| 14573 | AGAP11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-576-5p | 19 | CDH19 | Sponge network | -1.645 | 0 | -3.434 | 0 | 0.441 |
| 14574 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 12 | PTPN11 | Sponge network | -0.096 | 0.67958 | 0.431 | 2.0E-5 | 0.441 |
| 14575 | MEG3 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | DOCK4 | Sponge network | 0.249 | 0.35902 | 0.502 | 0.00108 | 0.441 |
| 14576 | GS1-124K5.11 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 11 | UVRAG | Sponge network | 0.494 | 0.00339 | -0.017 | 0.83865 | 0.441 |
| 14577 | RP11-122K13.12 |
hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p | 10 | MDM4 | Sponge network | 1.219 | 0 | 0.345 | 0.00762 | 0.441 |
| 14578 | RP11-2B6.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 20 | ABL2 | Sponge network | 0.539 | 0.03722 | 0.82 | 0 | 0.441 |
| 14579 | RP11-195F19.9 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-7-1-3p | 10 | ITPKB | Sponge network | -0.726 | 0.00132 | -0.704 | 0.00106 | 0.441 |
| 14580 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-660-5p;hsa-miR-96-5p | 40 | PPM1L | Sponge network | 0.078 | 0.55803 | -0.537 | 0.00616 | 0.441 |
| 14581 | EMX2OS |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 12 | AMPH | Sponge network | -0.777 | 0.1134 | -0.916 | 5.0E-5 | 0.441 |
| 14582 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 24 | JAZF1 | Sponge network | 0.225 | 0.30644 | -0.377 | 0.02677 | 0.441 |
| 14583 | FAM215B |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-940 | 14 | TRIM33 | Sponge network | 0.817 | 3.0E-5 | 0.402 | 7.0E-5 | 0.441 |
| 14584 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 17 | BNC2 | Sponge network | 0.497 | 0.01451 | -0.491 | 0.09988 | 0.441 |
| 14585 | RP11-736K20.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-3p | 31 | DST | Sponge network | -0.358 | 0.17255 | -0.713 | 0.00096 | 0.441 |
| 14586 | RP11-121C2.2 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-424-5p | 20 | ABL2 | Sponge network | 1.147 | 0 | 0.82 | 0 | 0.441 |
| 14587 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | PLSCR4 | Sponge network | -0.513 | 0.04703 | -0.474 | 0.03833 | 0.44 |
| 14588 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-628-5p | 17 | BCL6 | Sponge network | -0.842 | 0.01361 | -0.211 | 0.19489 | 0.44 |
| 14589 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-590-3p | 12 | PTPN11 | Sponge network | 0.826 | 1.0E-5 | 0.431 | 2.0E-5 | 0.44 |
| 14590 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-93-3p | 14 | BNC2 | Sponge network | 0.598 | 0.38481 | -0.491 | 0.09988 | 0.44 |
| 14591 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-424-5p | 13 | FGFR1 | Sponge network | 0.16 | 0.50785 | -0.509 | 0.03957 | 0.44 |
| 14592 | RP11-166D19.1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | CSDE1 | Sponge network | -0.576 | 0.10121 | -0.493 | 0 | 0.44 |
| 14593 | MIR17HG |
hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-497-5p | 11 | TBL1XR1 | Sponge network | 1.699 | 0 | 0.578 | 0 | 0.44 |
| 14594 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-5p | 34 | ERC1 | Sponge network | -2.452 | 0 | -0.108 | 0.38794 | 0.44 |
| 14595 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-654-3p | 18 | RICTOR | Sponge network | 2.094 | 0 | 0.388 | 0.00188 | 0.44 |
| 14596 | RP11-536O18.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p | 12 | RBMS3 | Sponge network | -0.074 | 0.90758 | -0.519 | 0.03551 | 0.44 |
| 14597 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-744-5p | 28 | CHD5 | Sponge network | -0.739 | 0.00044 | -0.922 | 0.01521 | 0.44 |
| 14598 | GNG12-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-625-5p | 10 | NGFR | Sponge network | -0.793 | 7.0E-5 | -1.271 | 0.01058 | 0.44 |
| 14599 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p | 10 | THBS1 | Sponge network | -0.517 | 0.02935 | 0.741 | 0.00386 | 0.44 |
| 14600 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 27 | EBF1 | Sponge network | -0.612 | 0.00094 | -0.384 | 0.08856 | 0.44 |
| 14601 | CTC-444N24.11 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-200b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 22 | UBR3 | Sponge network | 1.153 | 0 | -0.021 | 0.82774 | 0.44 |
| 14602 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p | 59 | NTRK3 | Sponge network | -0.513 | 0.04703 | -1.77 | 3.0E-5 | 0.44 |
| 14603 | LINC00641 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-874-3p | 14 | ARID1B | Sponge network | -0.222 | 0.32445 | 0.003 | 0.97503 | 0.44 |
| 14604 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 55 | NTRK3 | Sponge network | 0.4 | 0.14611 | -1.77 | 3.0E-5 | 0.44 |
| 14605 | AC003090.1 |
hsa-miR-126-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | CXCL12 | Sponge network | -2.117 | 0.00161 | -1.421 | 0 | 0.44 |
| 14606 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | ATRX | Sponge network | 0.532 | 0.00505 | 0.261 | 0.04408 | 0.44 |
| 14607 | HAND2-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p | 20 | MAFB | Sponge network | -1.697 | 0.00889 | -0.023 | 0.89865 | 0.44 |
| 14608 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 35 | TGFBR3 | Sponge network | 0.101 | 0.60919 | -1.173 | 1.0E-5 | 0.44 |
| 14609 | TRAF3IP2-AS1 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ADH1B | Sponge network | -0.323 | 0.01372 | -3.716 | 0 | 0.44 |
| 14610 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 41 | TACC1 | Sponge network | -0.777 | 0.1134 | -0.362 | 0.05406 | 0.44 |
| 14611 | RP11-890B15.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p | 24 | WWTR1 | Sponge network | 0.101 | 0.60919 | -0.345 | 0.11997 | 0.44 |
| 14612 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 24 | CNTN1 | Sponge network | -0.738 | 0.21233 | -2.594 | 0 | 0.44 |
| 14613 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-940 | 12 | IGF1 | Sponge network | -1.255 | 0.00981 | -0.204 | 0.5898 | 0.44 |
| 14614 | RP11-21A7A.2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-942-5p | 10 | NRXN1 | Sponge network | -1.116 | 0.23686 | -3.778 | 0 | 0.44 |
| 14615 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 41 | NOTCH2 | Sponge network | 0.572 | 0.01722 | 0.138 | 0.35163 | 0.44 |
| 14616 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3913-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 31 | ZBTB4 | Sponge network | -1.216 | 0 | -0.739 | 0 | 0.44 |
| 14617 | GAS6-AS2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429 | 14 | STARD13 | Sponge network | 0.16 | 0.50785 | -0.187 | 0.27383 | 0.44 |
| 14618 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | ADAMTSL3 | Sponge network | -0.125 | 0.49414 | -1.559 | 0 | 0.44 |
| 14619 | USP3-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-877-5p | 40 | AFF1 | Sponge network | -0.162 | 0.47687 | -0.384 | 0.00053 | 0.44 |
| 14620 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-671-5p | 15 | KIT | Sponge network | -0.72 | 0.24239 | -2.184 | 0 | 0.44 |
| 14621 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-495-3p | 10 | ATRX | Sponge network | 1.923 | 0 | 0.261 | 0.04408 | 0.44 |
| 14622 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 35 | PHC3 | Sponge network | 0.267 | 0.2937 | 0.212 | 0.0499 | 0.44 |
| 14623 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92b-3p | 13 | AKAP12 | Sponge network | -0.737 | 0.10822 | -0.932 | 0.0028 | 0.44 |
| 14624 | SNHG16 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-664a-3p | 25 | CCDC6 | Sponge network | 0.961 | 0 | 0.27 | 0.02555 | 0.44 |
| 14625 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-940 | 12 | SNX29 | Sponge network | 0.043 | 0.81628 | -0.34 | 0.01371 | 0.44 |
| 14626 | USP3-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-450b-5p | 12 | SPTBN4 | Sponge network | -0.162 | 0.47687 | -0.786 | 0.01281 | 0.44 |
| 14627 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-96-5p | 66 | BMPR2 | Sponge network | 0.101 | 0.60919 | 0.206 | 0.0635 | 0.44 |
| 14628 | RP11-244O19.1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-92a-3p | 12 | RASSF3 | Sponge network | -0.096 | 0.67958 | -0.226 | 0.11691 | 0.44 |
| 14629 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | HACE1 | Sponge network | 0.782 | 0.00016 | -0.072 | 0.55239 | 0.44 |
| 14630 | RP11-400K9.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-93-3p;hsa-miR-940 | 11 | TRPS1 | Sponge network | -0.733 | 0.19469 | -0.191 | 0.44109 | 0.44 |
| 14631 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 25 | DMXL1 | Sponge network | 0.537 | 0.00139 | -0.426 | 0.00056 | 0.44 |
| 14632 | RP11-254F7.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 16 | TET1 | Sponge network | 0.176 | 0.37402 | -0.135 | 0.52138 | 0.44 |
| 14633 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-664a-3p | 10 | ITCH | Sponge network | 0.917 | 0 | 0.084 | 0.34058 | 0.44 |
| 14634 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 31 | MYO9A | Sponge network | 0.335 | 0.20596 | -0.147 | 0.32373 | 0.44 |
| 14635 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-942-5p | 28 | GPC6 | Sponge network | 0.194 | 0.64278 | -0.054 | 0.81465 | 0.44 |
| 14636 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | AKAP6 | Sponge network | -0.031 | 0.89063 | -1.586 | 3.0E-5 | 0.44 |
| 14637 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-655-3p | 10 | HECA | Sponge network | 0.542 | 0.00054 | -0.217 | 0.03463 | 0.44 |
| 14638 | RP13-516M14.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 38 | MDM4 | Sponge network | 0.389 | 0.04293 | 0.345 | 0.00762 | 0.44 |
| 14639 | BZRAP1-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-582-3p;hsa-miR-877-5p | 24 | NAV3 | Sponge network | -0.465 | 0.04703 | 0.212 | 0.47729 | 0.44 |
| 14640 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 12 | CHD2 | Sponge network | 0.389 | 0.04293 | 0.049 | 0.55371 | 0.44 |
| 14641 | DNM3OS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | GALNT13 | Sponge network | 0.572 | 0.01722 | -0.857 | 0.07439 | 0.44 |
| 14642 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | SEMA3C | Sponge network | -0.793 | 7.0E-5 | -0.272 | 0.31153 | 0.44 |
| 14643 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 26 | TSHZ3 | Sponge network | 0.199 | 0.38015 | -0.556 | 0.01516 | 0.44 |
| 14644 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | PRKAA2 | Sponge network | -0.421 | 0.0114 | -2.462 | 0 | 0.44 |
| 14645 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 69 | TCF4 | Sponge network | -0.777 | 0.1134 | 0.016 | 0.92271 | 0.44 |
| 14646 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | TAL1 | Sponge network | -0.769 | 0.00035 | -0.462 | 0.00574 | 0.44 |
| 14647 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p | 17 | ADAMTSL3 | Sponge network | -0.246 | 0.35034 | -1.559 | 0 | 0.44 |
| 14648 | CTD-2314G24.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-96-5p | 79 | LPP | Sponge network | 0.246 | 0.59912 | -0.459 | 0.04475 | 0.44 |
| 14649 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 44 | NTRK3 | Sponge network | 0.299 | 0.57722 | -1.77 | 3.0E-5 | 0.44 |
| 14650 | RP11-67L2.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PHF3 | Sponge network | 0.72 | 0.0001 | 0.126 | 0.19652 | 0.44 |
| 14651 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p | 13 | AMPH | Sponge network | -0.942 | 0 | -0.916 | 5.0E-5 | 0.44 |
| 14652 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | TET1 | Sponge network | 0.064 | 0.72934 | -0.135 | 0.52138 | 0.44 |
| 14653 | CTD-3138B18.6 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-331-3p;hsa-miR-495-3p | 11 | DDX6 | Sponge network | 2.248 | 0 | 0.091 | 0.36428 | 0.44 |
| 14654 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DCN | Sponge network | -1.057 | 0.00029 | -1.288 | 0 | 0.44 |
| 14655 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | IGSF10 | Sponge network | -1.057 | 0.00029 | -1.751 | 1.0E-5 | 0.44 |
| 14656 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 14 | AKAP6 | Sponge network | -0.726 | 0.00132 | -1.586 | 3.0E-5 | 0.44 |
| 14657 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-7-1-3p | 36 | PDGFRA | Sponge network | -1.249 | 7.0E-5 | -0.372 | 0.0037 | 0.44 |
| 14658 | U91328.19 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577 | 16 | CSDE1 | Sponge network | -0.303 | 0.21002 | -0.493 | 0 | 0.44 |
| 14659 | WDFY3-AS2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 22 | SORCS1 | Sponge network | -0.942 | 0 | -3.492 | 0 | 0.44 |
| 14660 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 35 | IGF1 | Sponge network | -1.216 | 0 | -0.204 | 0.5898 | 0.44 |
| 14661 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | SOX5 | Sponge network | 0.101 | 0.60919 | -1.091 | 4.0E-5 | 0.44 |
| 14662 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | -0.323 | 0.01372 | -1.047 | 0.00364 | 0.44 |
| 14663 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 42 | FOXP1 | Sponge network | -0.487 | 0.02157 | 0.042 | 0.74368 | 0.44 |
| 14664 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-96-5p | 37 | FBXO32 | Sponge network | -0.942 | 0 | -0.495 | 0.07561 | 0.44 |
| 14665 | RP11-727A23.5 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-625-5p;hsa-miR-766-3p | 29 | MDM4 | Sponge network | 1.117 | 0 | 0.345 | 0.00762 | 0.44 |
| 14666 | RP11-110I1.11 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 11 | TSC1 | Sponge network | 1.034 | 0 | 0.103 | 0.26071 | 0.44 |
| 14667 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | CDHR3 | Sponge network | -0.899 | 6.0E-5 | -0.977 | 4.0E-5 | 0.44 |
| 14668 | CTD-2314G24.2 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | INPP4A | Sponge network | 0.246 | 0.59912 | 0.07 | 0.53563 | 0.44 |
| 14669 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-455-3p;hsa-miR-542-3p | 15 | TLE4 | Sponge network | -2.69 | 0.0001 | -1.157 | 0 | 0.44 |
| 14670 | AC003090.1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | SYT4 | Sponge network | -2.117 | 0.00161 | -3.207 | 0 | 0.44 |
| 14671 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 17 | AFF3 | Sponge network | -1.161 | 0.00591 | -2.373 | 0 | 0.44 |
| 14672 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SON | Sponge network | 2.094 | 0 | 0.168 | 0.04406 | 0.44 |
| 14673 | HNRNPU-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ARHGEF12 | Sponge network | 1.601 | 0 | 0.09 | 0.35693 | 0.44 |
| 14674 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | ERBB4 | Sponge network | -1.886 | 0 | -1.975 | 2.0E-5 | 0.44 |
| 14675 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-654-3p | 29 | RICTOR | Sponge network | 1.03 | 0 | 0.388 | 0.00188 | 0.44 |
| 14676 | BVES-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-874-3p | 21 | CTIF | Sponge network | -2.62 | 0 | -0.389 | 0.00549 | 0.44 |
| 14677 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 18 | DYNC1I1 | Sponge network | -0.738 | 0.21233 | -1.12 | 0.00107 | 0.44 |
| 14678 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 16 | LHX6 | Sponge network | -0.563 | 0.02545 | -0.087 | 0.69193 | 0.44 |
| 14679 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | TET1 | Sponge network | -0.487 | 0.02157 | -0.135 | 0.52138 | 0.44 |
| 14680 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p | 12 | DKK3 | Sponge network | -2.69 | 0.0001 | -0.052 | 0.77783 | 0.44 |
| 14681 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-708-3p;hsa-miR-92a-1-5p | 32 | PRKAB2 | Sponge network | -0.615 | 0.00015 | -0.367 | 0.01371 | 0.439 |
| 14682 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | CDH19 | Sponge network | -0.246 | 0.35034 | -3.434 | 0 | 0.439 |
| 14683 | FENDRR |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | HERC1 | Sponge network | -1.281 | 0.00012 | -0.196 | 0.08544 | 0.439 |
| 14684 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429 | 14 | TLE4 | Sponge network | -0.976 | 0.00025 | -1.157 | 0 | 0.439 |
| 14685 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-3614-5p | 15 | MOB1B | Sponge network | 1.162 | 5.0E-5 | 0.197 | 0.15266 | 0.439 |
| 14686 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | EBF1 | Sponge network | 0.225 | 0.30644 | -0.384 | 0.08856 | 0.439 |
| 14687 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-345-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 41 | FAT3 | Sponge network | -0.08 | 0.90092 | -1.601 | 0.00051 | 0.439 |
| 14688 | RP11-57H14.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | CREB1 | Sponge network | 0.083 | 0.66405 | 0.203 | 0.00537 | 0.439 |
| 14689 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-940 | 19 | NEDD4 | Sponge network | -0.162 | 0.47687 | 0.02 | 0.89834 | 0.439 |
| 14690 | FAM66C |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | PRKCB | Sponge network | -0.901 | 0.00019 | -1.313 | 2.0E-5 | 0.439 |
| 14691 | RP11-244O19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 46 | GAS7 | Sponge network | -0.096 | 0.67958 | -0.555 | 0.01602 | 0.439 |
| 14692 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 11 | FSTL5 | Sponge network | -0.842 | 0.01361 | -1.3 | 0.01619 | 0.439 |
| 14693 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-493-3p | 12 | JAKMIP2 | Sponge network | -0.465 | 0.04703 | -1.388 | 0 | 0.439 |
| 14694 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | FYN | Sponge network | -0.769 | 0.00035 | -0.131 | 0.37051 | 0.439 |
| 14695 | AP000692.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | MDM4 | Sponge network | 1.617 | 0.00159 | 0.345 | 0.00762 | 0.439 |
| 14696 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 44 | AR | Sponge network | 0.16 | 0.50785 | -1.554 | 0 | 0.439 |
| 14697 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-590-3p | 12 | DNAJB4 | Sponge network | -0.811 | 2.0E-5 | -0.7 | 1.0E-5 | 0.439 |
| 14698 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 34 | NTRK3 | Sponge network | -0.72 | 0.24239 | -1.77 | 3.0E-5 | 0.439 |
| 14699 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-940 | 34 | MEF2C | Sponge network | -0.096 | 0.67958 | -0.499 | 0.01448 | 0.439 |
| 14700 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 20 | CDH23 | Sponge network | -0.769 | 0.00035 | -0.974 | 0.00034 | 0.439 |
| 14701 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PHF3 | Sponge network | 0.683 | 1.0E-5 | 0.126 | 0.19652 | 0.439 |
| 14702 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 34 | RASSF2 | Sponge network | -1.654 | 0.00144 | -0.273 | 0.21961 | 0.439 |
| 14703 | LINC00702 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 50 | NTRK2 | Sponge network | -0.737 | 0.06182 | -0.736 | 0.06495 | 0.439 |
| 14704 | RP11-384O8.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 49 | DST | Sponge network | -0.738 | 0.21233 | -0.713 | 0.00096 | 0.439 |
| 14705 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | AHNAK | Sponge network | -0.222 | 0.32445 | -1.207 | 0 | 0.439 |
| 14706 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-940 | 21 | IGF1 | Sponge network | -0.465 | 0.04703 | -0.204 | 0.5898 | 0.439 |
| 14707 | RP11-360F5.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p | 20 | LPP | Sponge network | 1.867 | 0 | -0.459 | 0.04475 | 0.439 |
| 14708 | AC009948.5 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p | 20 | DYNC1I1 | Sponge network | 0.532 | 0.00505 | -1.12 | 0.00107 | 0.439 |
| 14709 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p | 13 | DKK3 | Sponge network | -0.384 | 0.40652 | -0.052 | 0.77783 | 0.439 |
| 14710 | RP11-98I9.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p | 15 | TBL1XR1 | Sponge network | 0.67 | 0.00048 | 0.578 | 0 | 0.439 |
| 14711 | FAM13A-AS1 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | ACVR2A | Sponge network | -0.83 | 0 | -0.409 | 0.00039 | 0.439 |
| 14712 | RP11-597D13.9 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p | 56 | DST | Sponge network | 1.302 | 7.0E-5 | -0.713 | 0.00096 | 0.439 |
| 14713 | MIR143HG |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 22 | FMNL3 | Sponge network | -0.739 | 0.0574 | 0.555 | 4.0E-5 | 0.439 |
| 14714 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-424-5p | 12 | PTGFR | Sponge network | 0.082 | 0.55397 | -0.233 | 0.45421 | 0.439 |
| 14715 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 34 | USP12 | Sponge network | -1.111 | 0.0003 | -0.102 | 0.40768 | 0.439 |
| 14716 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-5p | 11 | MAPK10 | Sponge network | 0.889 | 9.0E-5 | -1.774 | 0 | 0.439 |
| 14717 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TMEFF2 | Sponge network | 0.194 | 0.64278 | -2.426 | 0 | 0.439 |
| 14718 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-590-3p | 19 | BCL6 | Sponge network | -0.487 | 0.02157 | -0.211 | 0.19489 | 0.439 |
| 14719 | USP3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-450b-5p | 10 | UVRAG | Sponge network | -0.162 | 0.47687 | -0.017 | 0.83865 | 0.439 |
| 14720 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-411-5p | 18 | USP6 | Sponge network | 0.594 | 0.00064 | 0.188 | 0.3871 | 0.439 |
| 14721 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | TET1 | Sponge network | 0.537 | 0.00139 | -0.135 | 0.52138 | 0.439 |
| 14722 | LINC00702 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 10 | DSE | Sponge network | -0.737 | 0.06182 | -0.231 | 0.05347 | 0.439 |
| 14723 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | PLSCR4 | Sponge network | 0.381 | 0.25967 | -0.474 | 0.03833 | 0.439 |
| 14724 | LINC00578 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-3p | 12 | CADM3 | Sponge network | 0.811 | 0.03228 | -3.663 | 0 | 0.439 |
| 14725 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | NALCN | Sponge network | -2.452 | 0 | 1.068 | 0.00237 | 0.439 |
| 14726 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | -0.129 | 0.57177 | -0.001 | 0.99669 | 0.439 |
| 14727 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 45 | PTPRD | Sponge network | -0.976 | 0.00025 | -1.005 | 0.00064 | 0.439 |
| 14728 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-93-5p | 16 | PGR | Sponge network | -0.726 | 0.00132 | -1.704 | 0 | 0.439 |
| 14729 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 36 | MOB1B | Sponge network | 0.222 | 0.29133 | 0.197 | 0.15266 | 0.439 |
| 14730 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 15 | GPAM | Sponge network | 0.083 | 0.66405 | 0.142 | 0.29732 | 0.439 |
| 14731 | PART1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | PEG3 | Sponge network | -2.925 | 0 | -1.74 | 0 | 0.439 |
| 14732 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-590-3p | 20 | GPC6 | Sponge network | 0.532 | 0.00505 | -0.054 | 0.81465 | 0.439 |
| 14733 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-589-5p | 15 | ROBO1 | Sponge network | -2.69 | 0.0001 | -0.009 | 0.96154 | 0.439 |
| 14734 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-582-5p | 12 | PIK3C2A | Sponge network | 0.993 | 0 | 0.061 | 0.53776 | 0.439 |
| 14735 | FENDRR |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | THSD7B | Sponge network | -1.281 | 0.00012 | 0.154 | 0.74723 | 0.439 |
| 14736 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-590-5p | 15 | MEF2C | Sponge network | -2.076 | 0.00548 | -0.499 | 0.01448 | 0.439 |
| 14737 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-542-3p | 15 | TXNIP | Sponge network | -0.323 | 0.01372 | -1.277 | 0 | 0.439 |
| 14738 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 54 | RBMS3 | Sponge network | -2.925 | 0 | -0.519 | 0.03551 | 0.439 |
| 14739 | CTD-3064M3.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429 | 14 | SCN9A | Sponge network | -0.469 | 0.08721 | -0.525 | 0.10593 | 0.439 |
| 14740 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-766-3p | 11 | EZH1 | Sponge network | 0.75 | 0.00054 | -0.564 | 1.0E-5 | 0.439 |
| 14741 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-429;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p | 32 | GAS7 | Sponge network | 0.727 | 3.0E-5 | -0.555 | 0.01602 | 0.439 |
| 14742 | RP11-244H3.1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | MACF1 | Sponge network | 0.659 | 3.0E-5 | 0.019 | 0.90335 | 0.439 |
| 14743 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 26 | LRIG1 | Sponge network | 0.246 | 0.59912 | -0.537 | 0.00588 | 0.439 |
| 14744 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CXXC4 | Sponge network | -1.161 | 0.00591 | -0.433 | 0.21055 | 0.439 |
| 14745 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-370-3p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 18 | RPS6KA2 | Sponge network | -0.487 | 0.02157 | -0.57 | 0.00013 | 0.439 |
| 14746 | RP11-983P16.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-935 | 29 | DMD | Sponge network | 0.41 | 0.02214 | -1.506 | 0 | 0.439 |
| 14747 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | MOB1B | Sponge network | 0.385 | 0.10067 | 0.197 | 0.15266 | 0.439 |
| 14748 | FAM66C |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | CBFA2T3 | Sponge network | -0.901 | 0.00019 | -1.221 | 1.0E-5 | 0.439 |
| 14749 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 12 | SOX5 | Sponge network | -2.039 | 0.03447 | -1.091 | 4.0E-5 | 0.439 |
| 14750 | LINC00957 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PRKCB | Sponge network | -0.421 | 0.0114 | -1.313 | 2.0E-5 | 0.439 |
| 14751 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | IGSF10 | Sponge network | 0.225 | 0.30644 | -1.751 | 1.0E-5 | 0.439 |
| 14752 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p | 44 | SOX5 | Sponge network | -0.615 | 0.00015 | -1.091 | 4.0E-5 | 0.439 |
| 14753 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 54 | ERBB4 | Sponge network | -1.111 | 0.0003 | -1.975 | 2.0E-5 | 0.439 |
| 14754 | RP11-400K9.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-340-5p | 12 | EPHA7 | Sponge network | -0.733 | 0.19469 | -2.197 | 3.0E-5 | 0.439 |
| 14755 | RP11-589P10.7 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-26b-3p;hsa-miR-324-3p;hsa-miR-331-3p | 14 | EPHA7 | Sponge network | -2.044 | 0.00066 | -2.197 | 3.0E-5 | 0.439 |
| 14756 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p | 11 | SPTBN4 | Sponge network | -0.611 | 0.09607 | -0.786 | 0.01281 | 0.439 |
| 14757 | RP11-6O2.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-96-5p | 30 | IL17RD | Sponge network | 0.331 | 0.57247 | -0.019 | 0.94873 | 0.439 |
| 14758 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CLIP1 | Sponge network | 0.537 | 0.00139 | -0.215 | 0.08684 | 0.439 |
| 14759 | RP11-264B14.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p | 14 | MDM4 | Sponge network | 0.557 | 0.00564 | 0.345 | 0.00762 | 0.439 |
| 14760 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 16 | SYT4 | Sponge network | -0.583 | 0.14589 | -3.207 | 0 | 0.439 |
| 14761 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-454-3p | 16 | CHD5 | Sponge network | 0.178 | 0.29106 | -0.922 | 0.01521 | 0.439 |
| 14762 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | TIMP3 | Sponge network | 0.342 | 0.03129 | -0.546 | 0.02307 | 0.439 |
| 14763 | HCG11 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | NRK | Sponge network | 0.385 | 0.10067 | 0.656 | 0.19217 | 0.439 |
| 14764 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-421;hsa-miR-486-5p | 11 | PDGFC | Sponge network | -2.62 | 0 | -0.33 | 0.06483 | 0.439 |
| 14765 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DACT1 | Sponge network | -2.452 | 0 | 0.783 | 0.00072 | 0.439 |
| 14766 | PGM5-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 25 | DIP2C | Sponge network | -4.546 | 0 | -0.436 | 0.01713 | 0.439 |
| 14767 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | AFF4 | Sponge network | 0.673 | 0 | -0.266 | 0.01565 | 0.439 |
| 14768 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-96-5p | 11 | LRRC7 | Sponge network | -0.517 | 0.02935 | -1.341 | 0.01193 | 0.439 |
| 14769 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-495-3p;hsa-miR-654-3p | 15 | PHF6 | Sponge network | 1.601 | 0 | 0.785 | 0 | 0.439 |
| 14770 | CTA-363E19.2 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-369-3p;hsa-miR-495-3p | 10 | NF1 | Sponge network | 1.055 | 0 | 0.51 | 0 | 0.439 |
| 14771 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 23 | GRIN2A | Sponge network | -0.227 | 0.31657 | -2.161 | 0 | 0.439 |
| 14772 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -0.227 | 0.31657 | -0.354 | 0.14694 | 0.439 |
| 14773 | H19 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-3p | 10 | BCL9 | Sponge network | 2.439 | 0 | 0.321 | 0.00267 | 0.439 |
| 14774 | ANKRD10-IT1 |
hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-191-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 11 | DDX6 | Sponge network | 1.739 | 0 | 0.091 | 0.36428 | 0.439 |
| 14775 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p | 14 | UNC5C | Sponge network | 0.727 | 3.0E-5 | -0.178 | 0.40214 | 0.439 |
| 14776 | LINC00963 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-484 | 18 | ABL1 | Sponge network | 0.523 | 9.0E-5 | -0.215 | 0.07804 | 0.439 |
| 14777 | RP11-102N12.3 |
hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-671-5p | 12 | KIT | Sponge network | -0.351 | 0.11358 | -2.184 | 0 | 0.439 |
| 14778 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 11 | NRXN3 | Sponge network | 1.757 | 0 | -0.893 | 0.04186 | 0.439 |
| 14779 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 19 | SON | Sponge network | 0.91 | 0 | 0.168 | 0.04406 | 0.439 |
| 14780 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 17 | PRKAA2 | Sponge network | -1.425 | 0.04204 | -2.462 | 0 | 0.439 |
| 14781 | PCA3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-629-3p | 13 | SPTBN4 | Sponge network | -0.709 | 0.18168 | -0.786 | 0.01281 | 0.439 |
| 14782 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 13 | GRIN2A | Sponge network | -4.477 | 0 | -2.161 | 0 | 0.439 |
| 14783 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 16 | RELN | Sponge network | -0.513 | 0.04703 | -2.403 | 0 | 0.439 |
| 14784 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-495-3p | 19 | DDX6 | Sponge network | 0.706 | 0 | 0.091 | 0.36428 | 0.439 |
| 14785 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-93-5p | 31 | LRP1B | Sponge network | -0.162 | 0.47687 | -2.163 | 0 | 0.439 |
| 14786 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CACNA2D1 | Sponge network | 0.385 | 0.10067 | -0.846 | 0.00675 | 0.439 |
| 14787 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p | 30 | DMD | Sponge network | 0.299 | 0.57722 | -1.506 | 0 | 0.439 |
| 14788 | LINC00476 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-484 | 14 | FLNA | Sponge network | -0.615 | 0.00015 | -0.771 | 0.01377 | 0.439 |
| 14789 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p | 10 | AKAP12 | Sponge network | 0.082 | 0.55397 | -0.932 | 0.0028 | 0.439 |
| 14790 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-577;hsa-miR-590-3p | 16 | MAP3K12 | Sponge network | -2.452 | 0 | -0.057 | 0.59369 | 0.439 |
| 14791 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 21 | SYNE1 | Sponge network | -0.405 | 0.22013 | -0.738 | 0.00316 | 0.439 |
| 14792 | RP11-359B12.2 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | INPP4A | Sponge network | 0.064 | 0.72934 | 0.07 | 0.53563 | 0.438 |
| 14793 | SERTAD4-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-500a-5p | 15 | LRP1B | Sponge network | -0.932 | 0.00329 | -2.163 | 0 | 0.438 |
| 14794 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 42 | RUNX1T1 | Sponge network | 0.385 | 0.10067 | -0.344 | 0.2374 | 0.438 |
| 14795 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 18 | ERCC4 | Sponge network | -0.096 | 0.67958 | 0.126 | 0.15266 | 0.438 |
| 14796 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p | 11 | ZMYM2 | Sponge network | 0.813 | 1.0E-5 | 0.279 | 0.01273 | 0.438 |
| 14797 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-671-5p | 19 | DLC1 | Sponge network | -0.318 | 0.65847 | -0.382 | 0.05038 | 0.438 |
| 14798 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | TACC1 | Sponge network | -2.117 | 0.00161 | -0.362 | 0.05406 | 0.438 |
| 14799 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | FAT4 | Sponge network | 0.374 | 0.20054 | -0.354 | 0.14694 | 0.438 |
| 14800 | RP11-359E10.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | SFRP1 | Sponge network | 0.299 | 0.57722 | -3.566 | 0 | 0.438 |
| 14801 | AP001056.1 |
hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-96-5p | 12 | TCF4 | Sponge network | 1.062 | 0.00912 | 0.016 | 0.92271 | 0.438 |
| 14802 | LINC01018 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-942-5p | 31 | IGFBP5 | Sponge network | -2.915 | 0.00015 | -0.806 | 0.00105 | 0.438 |
| 14803 | DNM3OS |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PTPN13 | Sponge network | 0.572 | 0.01722 | -1.208 | 0 | 0.438 |
| 14804 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 65 | LPP | Sponge network | 0.782 | 0.00016 | -0.459 | 0.04475 | 0.438 |
| 14805 | AGAP11 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-3p | 12 | ID4 | Sponge network | -1.645 | 0 | -1.644 | 0 | 0.438 |
| 14806 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | HECA | Sponge network | 0.614 | 1.0E-5 | -0.217 | 0.03463 | 0.438 |
| 14807 | SNHG14 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MLF1 | Sponge network | -1.111 | 0.0003 | -0.893 | 0.00727 | 0.438 |
| 14808 | AC004893.11 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p | 11 | ZNF638 | Sponge network | 1.317 | 0 | 0.22 | 0.01214 | 0.438 |
| 14809 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 23 | ESR1 | Sponge network | 0.727 | 3.0E-5 | -1.035 | 3.0E-5 | 0.438 |
| 14810 | RP11-356J5.12 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | STARD13 | Sponge network | -0.899 | 6.0E-5 | -0.187 | 0.27383 | 0.438 |
| 14811 | RP11-356J5.12 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | SFRP1 | Sponge network | -0.899 | 6.0E-5 | -3.566 | 0 | 0.438 |
| 14812 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 25 | RSPO2 | Sponge network | 0.194 | 0.64278 | -3.038 | 0 | 0.438 |
| 14813 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-671-5p | 24 | DLC1 | Sponge network | -0.421 | 0.0114 | -0.382 | 0.05038 | 0.438 |
| 14814 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 17 | RET | Sponge network | 0.374 | 0.20054 | -0.833 | 0.03517 | 0.438 |
| 14815 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PTGFR | Sponge network | -0.612 | 0.00094 | -0.233 | 0.45421 | 0.438 |
| 14816 | ZNF667-AS1 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 12 | LZTS1 | Sponge network | -0.976 | 0.00025 | 1.664 | 0 | 0.438 |
| 14817 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ABCC9 | Sponge network | 0.385 | 0.10067 | -0.466 | 0.14121 | 0.438 |
| 14818 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | CACNA2D1 | Sponge network | -0.739 | 0.00044 | -0.846 | 0.00675 | 0.438 |
| 14819 | RP11-13J8.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-495-3p;hsa-miR-766-3p;hsa-miR-940 | 19 | MDM4 | Sponge network | 3.03 | 0 | 0.345 | 0.00762 | 0.438 |
| 14820 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | FYN | Sponge network | -1.057 | 0.00029 | -0.131 | 0.37051 | 0.438 |
| 14821 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 42 | CBX5 | Sponge network | 0.683 | 1.0E-5 | 0.165 | 0.24446 | 0.438 |
| 14822 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-98-5p | 11 | ZFP36L1 | Sponge network | -1.092 | 4.0E-5 | -0.505 | 8.0E-5 | 0.438 |
| 14823 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-942-5p | 17 | FBXO31 | Sponge network | -0.737 | 0.06182 | -0.085 | 0.42389 | 0.438 |
| 14824 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 31 | IGF1 | Sponge network | -0.793 | 7.0E-5 | -0.204 | 0.5898 | 0.438 |
| 14825 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 20 | RICTOR | Sponge network | 0.924 | 0 | 0.388 | 0.00188 | 0.438 |
| 14826 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 26 | SYNPO2 | Sponge network | 0.056 | 0.81195 | -2.342 | 0 | 0.438 |
| 14827 | CTC-296K1.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-132-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-940 | 16 | TRPS1 | Sponge network | -2.095 | 0.00112 | -0.191 | 0.44109 | 0.438 |
| 14828 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | DOCK4 | Sponge network | 0.72 | 0.0001 | 0.502 | 0.00108 | 0.438 |
| 14829 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 11 | NRXN1 | Sponge network | -2.039 | 0.03447 | -3.778 | 0 | 0.438 |
| 14830 | RP11-484D2.5 | hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p | 10 | MDM4 | Sponge network | 2.365 | 0.01134 | 0.345 | 0.00762 | 0.438 |
| 14831 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-93-3p | 31 | ERC1 | Sponge network | 0.72 | 0.0001 | -0.108 | 0.38794 | 0.438 |
| 14832 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | 0.494 | 0.00339 | 0.809 | 0.00544 | 0.438 |
| 14833 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | GJA1 | Sponge network | -1.697 | 0.00889 | -0.485 | 0.02179 | 0.438 |
| 14834 | LINC00657 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p | 16 | TRIM33 | Sponge network | 0.91 | 0 | 0.402 | 7.0E-5 | 0.438 |
| 14835 | CTD-2089N3.2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 11 | ERG | Sponge network | -1.375 | 0.08642 | 0.002 | 0.99011 | 0.438 |
| 14836 | RP11-401P9.4 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p | 16 | MRVI1 | Sponge network | -0.384 | 0.40652 | -1.051 | 0.00022 | 0.438 |
| 14837 | SNX29P2 |
hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | TAL1 | Sponge network | 0.4 | 0.14611 | -0.462 | 0.00574 | 0.438 |
| 14838 | ADAMTS9-AS2 |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | PACRG | Sponge network | -2.452 | 0 | -2.248 | 0 | 0.438 |
| 14839 | TPTEP1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 14 | RHOB | Sponge network | -1.216 | 0 | -1.052 | 0 | 0.438 |
| 14840 | LINC00473 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | TGFBR3 | Sponge network | -1.203 | 0.03881 | -1.173 | 1.0E-5 | 0.438 |
| 14841 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | PTPN11 | Sponge network | 1.294 | 0 | 0.431 | 2.0E-5 | 0.438 |
| 14842 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | MOB1B | Sponge network | 0.663 | 0.00073 | 0.197 | 0.15266 | 0.438 |
| 14843 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-511-5p | 23 | BTG2 | Sponge network | -2.531 | 0 | -1.763 | 0 | 0.438 |
| 14844 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577 | 16 | TLL1 | Sponge network | -0.465 | 0.04703 | -1.195 | 3.0E-5 | 0.438 |
| 14845 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | RASSF8 | Sponge network | 0.16 | 0.50785 | -0.122 | 0.65591 | 0.438 |
| 14846 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-495-3p | 12 | PHF6 | Sponge network | 0.769 | 0 | 0.785 | 0 | 0.438 |
| 14847 | EMX2OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ERG | Sponge network | -0.777 | 0.1134 | 0.002 | 0.99011 | 0.438 |
| 14848 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 53 | KIF1B | Sponge network | 0.683 | 1.0E-5 | -0.118 | 0.32258 | 0.438 |
| 14849 | HHIP-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | EYA4 | Sponge network | -0.737 | 0.10822 | 0.3 | 0.36876 | 0.438 |
| 14850 | MAGI2-AS3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 47 | AFF1 | Sponge network | -0.129 | 0.57177 | -0.384 | 0.00053 | 0.438 |
| 14851 | CTC-490E21.10 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p | 11 | MYO9A | Sponge network | 1.46 | 1.0E-5 | -0.147 | 0.32373 | 0.438 |
| 14852 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 22 | EPHA3 | Sponge network | -0.738 | 0.21233 | 0.185 | 0.53588 | 0.438 |
| 14853 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-92a-3p | 33 | IL17RD | Sponge network | -0.738 | 0.21233 | -0.019 | 0.94873 | 0.438 |
| 14854 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-542-3p | 19 | RARB | Sponge network | -0.793 | 7.0E-5 | -0.825 | 2.0E-5 | 0.438 |
| 14855 | RP11-554A11.4 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p | 10 | RASSF3 | Sponge network | -0.583 | 0.14589 | -0.226 | 0.11691 | 0.438 |
| 14856 | ZNF667-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-7-5p | 28 | BTG2 | Sponge network | -0.976 | 0.00025 | -1.763 | 0 | 0.438 |
| 14857 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 37 | TMEM132B | Sponge network | -0.976 | 0.00025 | -0.83 | 0.01084 | 0.438 |
| 14858 | DNM3OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FAM133A | Sponge network | 0.572 | 0.01722 | 0.549 | 0.29009 | 0.438 |
| 14859 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p | 19 | HIP1 | Sponge network | -0.154 | 0.78939 | 0.774 | 0 | 0.438 |
| 14860 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | USP12 | Sponge network | -0.096 | 0.67958 | -0.102 | 0.40768 | 0.438 |
| 14861 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 63 | BMPR2 | Sponge network | 0.064 | 0.72934 | 0.206 | 0.0635 | 0.438 |
| 14862 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | EPG5 | Sponge network | 0.72 | 0.0001 | 0.101 | 0.3563 | 0.438 |
| 14863 | FAM66C |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | EYA4 | Sponge network | -0.901 | 0.00019 | 0.3 | 0.36876 | 0.438 |
| 14864 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-98-5p | 12 | SCN11A | Sponge network | -0.899 | 6.0E-5 | -1.15 | 0.00299 | 0.438 |
| 14865 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 15 | ST6GAL2 | Sponge network | -4.546 | 0 | -1.201 | 0.00597 | 0.438 |
| 14866 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 32 | BHLHE41 | Sponge network | -0.129 | 0.57177 | 0.353 | 0.12753 | 0.438 |
| 14867 | RP11-119F7.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 18 | NRK | Sponge network | 0.225 | 0.30644 | 0.656 | 0.19217 | 0.438 |
| 14868 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p | 10 | EYA4 | Sponge network | -0.376 | 0.00337 | 0.3 | 0.36876 | 0.438 |
| 14869 | TINCR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-589-3p | 18 | HECA | Sponge network | -0.664 | 0.14543 | -0.217 | 0.03463 | 0.438 |
| 14870 | RP11-119F7.5 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | TAL1 | Sponge network | 0.225 | 0.30644 | -0.462 | 0.00574 | 0.438 |
| 14871 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p | 11 | ITGA5 | Sponge network | -0.358 | 0.17255 | 0.452 | 0.03524 | 0.438 |
| 14872 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-503-5p;hsa-miR-590-3p | 33 | RICTOR | Sponge network | 0.494 | 0.00339 | 0.388 | 0.00188 | 0.438 |
| 14873 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | NR2C2 | Sponge network | 0.532 | 0.00505 | 0.21 | 0.03224 | 0.438 |
| 14874 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 30 | ESR1 | Sponge network | 0.532 | 0.00505 | -1.035 | 3.0E-5 | 0.438 |
| 14875 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-93-3p | 37 | TRPS1 | Sponge network | -0.384 | 0.40652 | -0.191 | 0.44109 | 0.438 |
| 14876 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-7-5p | 35 | RBMS3 | Sponge network | -0.18 | 0.20343 | -0.519 | 0.03551 | 0.438 |
| 14877 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | PRKAA1 | Sponge network | 1.205 | 0 | 0.317 | 0.0031 | 0.438 |
| 14878 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 14 | MXI1 | Sponge network | -1.092 | 4.0E-5 | -0.987 | 0 | 0.438 |
| 14879 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | MCC | Sponge network | -0.405 | 0.22013 | -1.005 | 0 | 0.438 |
| 14880 | PCA3 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | TLL1 | Sponge network | -0.709 | 0.18168 | -1.195 | 3.0E-5 | 0.438 |
| 14881 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-940 | 38 | GAS7 | Sponge network | -0.773 | 0.04073 | -0.555 | 0.01602 | 0.438 |
| 14882 | SNHG14 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 28 | SEMA3C | Sponge network | -1.111 | 0.0003 | -0.272 | 0.31153 | 0.438 |
| 14883 | RP11-37B2.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-625-5p | 10 | CEP170 | Sponge network | 1.03 | 0 | 0.943 | 0 | 0.438 |
| 14884 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 31 | PRKAA2 | Sponge network | 1.757 | 0 | -2.462 | 0 | 0.438 |
| 14885 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 12 | PLA2R1 | Sponge network | 0.997 | 0.00066 | -0.265 | 0.24676 | 0.438 |
| 14886 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 30 | DIP2C | Sponge network | 0.374 | 0.20054 | -0.436 | 0.01713 | 0.438 |
| 14887 | RP11-464F9.1 |
hsa-miR-142-5p;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | SIRT1 | Sponge network | 0.959 | 0 | 0.109 | 0.2593 | 0.438 |
| 14888 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | RABEP1 | Sponge network | -1.111 | 0.0003 | -0.066 | 0.43142 | 0.438 |
| 14889 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | FGFR1 | Sponge network | 0.051 | 0.83388 | -0.509 | 0.03957 | 0.438 |
| 14890 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 23 | PGR | Sponge network | 0.225 | 0.30644 | -1.704 | 0 | 0.438 |
| 14891 | FAM66C |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p | 30 | CADM1 | Sponge network | -0.901 | 0.00019 | -0.9 | 0.00084 | 0.438 |
| 14892 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 30 | LRP1B | Sponge network | -0.793 | 7.0E-5 | -2.163 | 0 | 0.438 |
| 14893 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | MITF | Sponge network | -0.323 | 0.01372 | -0.769 | 0.00022 | 0.438 |
| 14894 | CTC-296K1.3 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-625-5p;hsa-miR-744-3p | 20 | IGFBP5 | Sponge network | -2.095 | 0.00112 | -0.806 | 0.00105 | 0.438 |
| 14895 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-93-3p | 14 | EBF3 | Sponge network | -0.932 | 0.00329 | -0.058 | 0.81437 | 0.438 |
| 14896 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 57 | PRKAA2 | Sponge network | -0.222 | 0.32445 | -2.462 | 0 | 0.438 |
| 14897 | ANKRD10-IT1 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-7-1-3p | 11 | CBL | Sponge network | 1.739 | 0 | 0.291 | 0.01267 | 0.438 |
| 14898 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 14 | PIK3IP1 | Sponge network | -0.129 | 0.57177 | -0.187 | 0.19945 | 0.438 |
| 14899 | RP11-890B15.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-92a-1-5p;hsa-miR-940 | 46 | GRIK3 | Sponge network | 0.101 | 0.60919 | -3.208 | 0 | 0.438 |
| 14900 | LINC00672 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p | 49 | RUNX1T1 | Sponge network | -0.227 | 0.31657 | -0.344 | 0.2374 | 0.438 |
| 14901 | TRAF3IP2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-628-5p | 18 | CDH23 | Sponge network | -0.323 | 0.01372 | -0.974 | 0.00034 | 0.438 |
| 14902 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 38 | MYO9A | Sponge network | -1.281 | 0.00012 | -0.147 | 0.32373 | 0.438 |
| 14903 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-450b-5p | 12 | SPTBN4 | Sponge network | -1.092 | 4.0E-5 | -0.786 | 0.01281 | 0.438 |
| 14904 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 22 | TBL1XR1 | Sponge network | 1.344 | 0 | 0.578 | 0 | 0.438 |
| 14905 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p | 32 | CHD5 | Sponge network | -0.469 | 0.08721 | -0.922 | 0.01521 | 0.438 |
| 14906 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 41 | TGFBR3 | Sponge network | -0.777 | 0.1134 | -1.173 | 1.0E-5 | 0.438 |
| 14907 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 13 | HMCN1 | Sponge network | -2.46 | 0 | 0.809 | 0.00544 | 0.438 |
| 14908 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-590-3p | 13 | CREBBP | Sponge network | 0.919 | 0 | 0.126 | 0.15186 | 0.438 |
| 14909 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-940 | 13 | IGF1 | Sponge network | 1.153 | 0.00114 | -0.204 | 0.5898 | 0.438 |
| 14910 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-629-5p | 18 | MYO9A | Sponge network | 0.594 | 0.00064 | -0.147 | 0.32373 | 0.438 |
| 14911 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-5p | 13 | CHD5 | Sponge network | -0.473 | 0.07602 | -0.922 | 0.01521 | 0.438 |
| 14912 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-93-5p | 25 | PTPRD | Sponge network | -2.531 | 0 | -1.005 | 0.00064 | 0.438 |
| 14913 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | MITF | Sponge network | -2.117 | 0.00161 | -0.769 | 0.00022 | 0.438 |
| 14914 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 18 | KIT | Sponge network | -0.513 | 0.04703 | -2.184 | 0 | 0.438 |
| 14915 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 30 | IGF1 | Sponge network | 0.374 | 0.20054 | -0.204 | 0.5898 | 0.438 |
| 14916 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | PDS5B | Sponge network | 1.153 | 0 | 0.259 | 0.00962 | 0.438 |
| 14917 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | RASSF8 | Sponge network | -0.08 | 0.90092 | -0.122 | 0.65591 | 0.438 |
| 14918 | WDFY3-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-96-5p | 35 | ASTN1 | Sponge network | -0.942 | 0 | -2.389 | 2.0E-5 | 0.438 |
| 14919 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-486-5p | 13 | FGF5 | Sponge network | -2.62 | 0 | 0.549 | 0.28659 | 0.438 |
| 14920 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 46 | WDFY3 | Sponge network | 0.222 | 0.29133 | -0.201 | 0.0967 | 0.438 |
| 14921 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b | 17 | EYA4 | Sponge network | -0.583 | 0.14589 | 0.3 | 0.36876 | 0.438 |
| 14922 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MLLT10 | Sponge network | -0.323 | 0.01372 | 0.105 | 0.33292 | 0.437 |
| 14923 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 19 | MEF2C | Sponge network | 0.622 | 0.00015 | -0.499 | 0.01448 | 0.437 |
| 14924 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | PRDM5 | Sponge network | -1.697 | 0.00889 | -1.09 | 0.00014 | 0.437 |
| 14925 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | IL6ST | Sponge network | -1.375 | 0.08642 | -0.402 | 0.00676 | 0.437 |
| 14926 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-455-5p | 13 | THSD7A | Sponge network | -0.154 | 0.78939 | 0.242 | 0.25154 | 0.437 |
| 14927 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-374b-5p | 11 | NIN | Sponge network | 0.542 | 0.00054 | -0.253 | 0.13794 | 0.437 |
| 14928 | LINC00094 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 13 | FLNA | Sponge network | 0.253 | 0.09565 | -0.771 | 0.01377 | 0.437 |
| 14929 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-93-3p | 10 | EBF3 | Sponge network | 0.598 | 0.38481 | -0.058 | 0.81437 | 0.437 |
| 14930 | RP11-798G7.7 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-539-5p | 19 | DDX6 | Sponge network | 0.858 | 3.0E-5 | 0.091 | 0.36428 | 0.437 |
| 14931 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 23 | PCDH9 | Sponge network | 0.214 | 0.18246 | -1.823 | 4.0E-5 | 0.437 |
| 14932 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 32 | PRDM2 | Sponge network | -2.46 | 0 | -0.258 | 0.00721 | 0.437 |
| 14933 | LINC00865 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 18 | CDH23 | Sponge network | -1.249 | 7.0E-5 | -0.974 | 0.00034 | 0.437 |
| 14934 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | DCLK1 | Sponge network | -0.738 | 0.21233 | -1.324 | 0.00014 | 0.437 |
| 14935 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | RBMS3 | Sponge network | 0.385 | 0.10067 | -0.519 | 0.03551 | 0.437 |
| 14936 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p | 17 | RASSF8 | Sponge network | 0.082 | 0.55397 | -0.122 | 0.65591 | 0.437 |
| 14937 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | GALNT13 | Sponge network | -1.349 | 0.00017 | -0.857 | 0.07439 | 0.437 |
| 14938 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p | 25 | LRP1B | Sponge network | -0.896 | 0.00099 | -2.163 | 0 | 0.437 |
| 14939 | AC006116.24 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p | 17 | MED12L | Sponge network | -0.473 | 0.07602 | -0.194 | 0.55299 | 0.437 |
| 14940 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 52 | FOXP1 | Sponge network | -1.575 | 6.0E-5 | 0.042 | 0.74368 | 0.437 |
| 14941 | CTD-2089N3.2 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | PDE4DIP | Sponge network | -1.375 | 0.08642 | -0.282 | 0.0257 | 0.437 |
| 14942 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 29 | DIP2C | Sponge network | -0.227 | 0.31657 | -0.436 | 0.01713 | 0.437 |
| 14943 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | GALNT13 | Sponge network | 0.335 | 0.20596 | -0.857 | 0.07439 | 0.437 |
| 14944 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 55 | NTRK3 | Sponge network | -0.612 | 0.00094 | -1.77 | 3.0E-5 | 0.437 |
| 14945 | FENDRR |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | FAM117A | Sponge network | -1.281 | 0.00012 | -1.379 | 0 | 0.437 |
| 14946 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-628-5p;hsa-miR-93-3p | 29 | ERC1 | Sponge network | -0.154 | 0.78939 | -0.108 | 0.38794 | 0.437 |
| 14947 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-320b;hsa-miR-484 | 10 | MYCBP2 | Sponge network | 0.594 | 0.41522 | -0.027 | 0.82016 | 0.437 |
| 14948 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-940;hsa-miR-96-5p | 18 | CRTC3 | Sponge network | 0.532 | 0.00505 | 0.164 | 0.16786 | 0.437 |
| 14949 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-7-5p | 16 | KCNA6 | Sponge network | -0.517 | 0.02935 | -1.531 | 0.00013 | 0.437 |
| 14950 | RP11-822E23.8 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-7-5p | 13 | FNBP1 | Sponge network | -0.777 | 0.16667 | -0.969 | 4.0E-5 | 0.437 |
| 14951 | RP11-389C8.2 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PRKCB | Sponge network | 0.727 | 3.0E-5 | -1.313 | 2.0E-5 | 0.437 |
| 14952 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-98-5p | 11 | SCN11A | Sponge network | -0.769 | 0.00035 | -1.15 | 0.00299 | 0.437 |
| 14953 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 16 | SON | Sponge network | 0.919 | 0 | 0.168 | 0.04406 | 0.437 |
| 14954 | LINC00641 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 22 | BAZ2B | Sponge network | -0.222 | 0.32445 | -0.276 | 0.02833 | 0.437 |
| 14955 | TUG1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 14 | KIF5B | Sponge network | 0.972 | 0 | 0.362 | 0.00066 | 0.437 |
| 14956 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-375;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | CREM | Sponge network | -2.452 | 0 | -0.345 | 0.00258 | 0.437 |
| 14957 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30a-3p | 11 | NR2C2 | Sponge network | 2.163 | 0 | 0.21 | 0.03224 | 0.437 |
| 14958 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | USP12 | Sponge network | 1.089 | 0 | -0.102 | 0.40768 | 0.437 |
| 14959 | LINC01018 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | IL17RD | Sponge network | -2.915 | 0.00015 | -0.019 | 0.94873 | 0.437 |
| 14960 | RP11-401P9.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-361-3p | 17 | BRD3 | Sponge network | -0.384 | 0.40652 | 0.37 | 0.00013 | 0.437 |
| 14961 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-877-5p | 26 | RAP1A | Sponge network | -1.439 | 4.0E-5 | -0.929 | 0 | 0.437 |
| 14962 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 43 | FAT3 | Sponge network | 0.056 | 0.81195 | -1.601 | 0.00051 | 0.437 |
| 14963 | OIP5-AS1 |
hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p | 11 | RNF111 | Sponge network | 0.421 | 0.00084 | 0.098 | 0.21093 | 0.437 |
| 14964 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | PDGFRA | Sponge network | -0.769 | 0.00035 | -0.372 | 0.0037 | 0.437 |
| 14965 | BVES-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-505-5p;hsa-miR-629-3p | 22 | SNX29 | Sponge network | -2.62 | 0 | -0.34 | 0.01371 | 0.437 |
| 14966 | CTD-2528L19.6 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 14 | BAZ2B | Sponge network | 0.953 | 0 | -0.276 | 0.02833 | 0.437 |
| 14967 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 37 | DDX6 | Sponge network | 0.064 | 0.72934 | 0.091 | 0.36428 | 0.437 |
| 14968 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 26 | PRKAA2 | Sponge network | -0.376 | 0.00337 | -2.462 | 0 | 0.437 |
| 14969 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-577 | 23 | SYT4 | Sponge network | -2.69 | 0.0001 | -3.207 | 0 | 0.437 |
| 14970 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | MLLT10 | Sponge network | 0.67 | 0.00048 | 0.105 | 0.33292 | 0.437 |
| 14971 | LINC00473 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p | 14 | GLI3 | Sponge network | -1.203 | 0.03881 | -0.615 | 0.01482 | 0.437 |
| 14972 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-590-3p | 13 | ROBO1 | Sponge network | -0.896 | 0.00099 | -0.009 | 0.96154 | 0.437 |
| 14973 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 26 | SCN9A | Sponge network | 0.619 | 0.00157 | -0.525 | 0.10593 | 0.437 |
| 14974 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | NR4A3 | Sponge network | -0.49 | 0.04946 | -1.385 | 0 | 0.437 |
| 14975 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 47 | WDFY3 | Sponge network | -0.611 | 0.09607 | -0.201 | 0.0967 | 0.437 |
| 14976 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-33a-3p | 11 | ZMYM2 | Sponge network | 0.594 | 0.00064 | 0.279 | 0.01273 | 0.437 |
| 14977 | RP11-159D12.2 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-9-5p | 42 | CCDC6 | Sponge network | 1.254 | 0 | 0.27 | 0.02555 | 0.437 |
| 14978 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | LRRC55 | Sponge network | 0.4 | 0.14611 | -0.026 | 0.93163 | 0.437 |
| 14979 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 56 | PRKAA2 | Sponge network | 0.101 | 0.60919 | -2.462 | 0 | 0.437 |
| 14980 | LINC00672 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-590-3p;hsa-miR-93-3p | 19 | WWTR1 | Sponge network | -0.227 | 0.31657 | -0.345 | 0.11997 | 0.437 |
| 14981 | AC009948.5 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 50 | MDM4 | Sponge network | 0.532 | 0.00505 | 0.345 | 0.00762 | 0.437 |
| 14982 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-210-3p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p | 12 | CLASP2 | Sponge network | 0.993 | 0 | -0.038 | 0.71 | 0.437 |
| 14983 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SEMA3E | Sponge network | -0.899 | 6.0E-5 | -1.717 | 0.00137 | 0.437 |
| 14984 | AP001347.6 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-590-3p | 13 | ROBO1 | Sponge network | -1.161 | 0.00591 | -0.009 | 0.96154 | 0.437 |
| 14985 | RP1-302G2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p | 10 | FGFR1 | Sponge network | -1.483 | 9.0E-5 | -0.509 | 0.03957 | 0.437 |
| 14986 | RP11-379F4.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | TBL1XR1 | Sponge network | 1.316 | 0 | 0.578 | 0 | 0.437 |
| 14987 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 20 | PDE4DIP | Sponge network | -0.513 | 0.04703 | -0.282 | 0.0257 | 0.437 |
| 14988 | RP11-736K20.5 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | RPS6KA6 | Sponge network | -0.358 | 0.17255 | -2.989 | 0 | 0.437 |
| 14989 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-582-5p | 16 | STARD13 | Sponge network | -2.69 | 0.0001 | -0.187 | 0.27383 | 0.437 |
| 14990 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | PDS5B | Sponge network | 1.317 | 0 | 0.259 | 0.00962 | 0.437 |
| 14991 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-93-3p;hsa-miR-93-5p | 41 | SSBP2 | Sponge network | -0.777 | 0.16667 | -1.58 | 0 | 0.437 |
| 14992 | CTD-2089N3.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 15 | ST6GALNAC3 | Sponge network | -1.375 | 0.08642 | -0.987 | 0 | 0.437 |
| 14993 | RP11-927P21.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-424-5p | 10 | MDM4 | Sponge network | 1.272 | 0.00021 | 0.345 | 0.00762 | 0.437 |
| 14994 | LINC00847 |
hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-7-5p | 10 | KCNJ5 | Sponge network | 0.035 | 0.80563 | -0.281 | 0.3339 | 0.437 |
| 14995 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 55 | CCND2 | Sponge network | -0.129 | 0.57177 | -0.267 | 0.3112 | 0.437 |
| 14996 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-940 | 24 | AKAP11 | Sponge network | 0.082 | 0.55397 | 0.196 | 0.09825 | 0.437 |
| 14997 | CTC-296K1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-582-5p | 13 | RASSF8 | Sponge network | -2.095 | 0.00112 | -0.122 | 0.65591 | 0.437 |
| 14998 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-424-5p | 19 | RASSF2 | Sponge network | 1.344 | 0 | -0.273 | 0.21961 | 0.437 |
| 14999 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-3127-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-93-3p | 14 | RPS6KA2 | Sponge network | -0.469 | 0.08721 | -0.57 | 0.00013 | 0.437 |
| 15000 | PCA3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | LRIG1 | Sponge network | -0.709 | 0.18168 | -0.537 | 0.00588 | 0.437 |
| 15001 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 13 | ABCC9 | Sponge network | 0.174 | 0.30034 | -0.466 | 0.14121 | 0.437 |
| 15002 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 30 | THBS1 | Sponge network | 1.302 | 7.0E-5 | 0.741 | 0.00386 | 0.437 |
| 15003 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 18 | NIN | Sponge network | -0.896 | 0.00099 | -0.253 | 0.13794 | 0.437 |
| 15004 | LINC00654 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935 | 51 | PTPRD | Sponge network | 0.374 | 0.20054 | -1.005 | 0.00064 | 0.437 |
| 15005 | RP11-497H16.9 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZFX | Sponge network | 0.545 | 0.00064 | 0.09 | 0.42563 | 0.437 |
| 15006 | RP11-13K12.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-671-5p | 13 | EHD3 | Sponge network | -0.154 | 0.78939 | -0.484 | 0.01747 | 0.437 |
| 15007 | RP11-540B6.6 |
hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-511-5p | 24 | CREB1 | Sponge network | 0.93 | 0 | 0.203 | 0.00537 | 0.437 |
| 15008 | RP1-59D14.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-331-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | PHC3 | Sponge network | 1.162 | 5.0E-5 | 0.212 | 0.0499 | 0.437 |
| 15009 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-940;hsa-miR-98-5p | 28 | PBX1 | Sponge network | 0.082 | 0.55397 | -1.206 | 0 | 0.437 |
| 15010 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-590-3p | 26 | CLASP2 | Sponge network | -0.096 | 0.67958 | -0.038 | 0.71 | 0.437 |
| 15011 | RP11-33B1.4 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-582-5p | 16 | CREB1 | Sponge network | 0.826 | 0 | 0.203 | 0.00537 | 0.437 |
| 15012 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-940 | 45 | TRPS1 | Sponge network | -0.096 | 0.67958 | -0.191 | 0.44109 | 0.437 |
| 15013 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3127-5p;hsa-miR-532-3p | 10 | RPS6KA2 | Sponge network | -0.829 | 0.01343 | -0.57 | 0.00013 | 0.437 |
| 15014 | RP11-305E6.4 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ERCC6 | Sponge network | 1.317 | 0 | 0.23 | 0.015 | 0.437 |
| 15015 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p | 22 | ERC1 | Sponge network | 0.811 | 0.03228 | -0.108 | 0.38794 | 0.437 |
| 15016 | CTD-2089N3.2 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-30b-3p;hsa-miR-590-3p | 13 | SOX5 | Sponge network | -1.375 | 0.08642 | -1.091 | 4.0E-5 | 0.437 |
| 15017 | RP11-356J5.12 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 24 | WWTR1 | Sponge network | -0.899 | 6.0E-5 | -0.345 | 0.11997 | 0.437 |
| 15018 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-7-1-3p | 17 | AKAP6 | Sponge network | 0.259 | 0.12542 | -1.586 | 3.0E-5 | 0.437 |
| 15019 | HOXB-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-940 | 27 | GRIK3 | Sponge network | -0.154 | 0.53954 | -3.208 | 0 | 0.437 |
| 15020 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | -1.575 | 6.0E-5 | -0.001 | 0.99669 | 0.437 |
| 15021 | LINC00865 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 10 | UBE2QL1 | Sponge network | -1.249 | 7.0E-5 | -2.417 | 0 | 0.437 |
| 15022 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-940 | 25 | SNX29 | Sponge network | 0.299 | 0.57722 | -0.34 | 0.01371 | 0.437 |
| 15023 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 11 | PIK3IP1 | Sponge network | -0.896 | 0.00099 | -0.187 | 0.19945 | 0.437 |
| 15024 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 12 | ZNF292 | Sponge network | 1.11 | 0 | 0.276 | 0.04148 | 0.437 |
| 15025 | RP11-13K12.1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-93-3p | 10 | STAT5B | Sponge network | -0.154 | 0.78939 | -0.182 | 0.1082 | 0.437 |
| 15026 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-940 | 22 | CHD5 | Sponge network | -4.546 | 0 | -0.922 | 0.01521 | 0.437 |
| 15027 | ZRANB2-AS1 |
hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-3p | 11 | CTIF | Sponge network | -0.376 | 0.00337 | -0.389 | 0.00549 | 0.437 |
| 15028 | RP11-736K20.5 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 16 | CTIF | Sponge network | -0.358 | 0.17255 | -0.389 | 0.00549 | 0.437 |
| 15029 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 19 | RARB | Sponge network | -2.117 | 0.00161 | -0.825 | 2.0E-5 | 0.437 |
| 15030 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | LIFR | Sponge network | -0.739 | 0.00044 | -1.794 | 0 | 0.437 |
| 15031 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p | 32 | EBF1 | Sponge network | -0.793 | 7.0E-5 | -0.384 | 0.08856 | 0.437 |
| 15032 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | CNTN1 | Sponge network | 0.532 | 0.00505 | -2.594 | 0 | 0.437 |
| 15033 | PGM5-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p | 10 | SEMA3E | Sponge network | -4.546 | 0 | -1.717 | 0.00137 | 0.437 |
| 15034 | RP11-774O3.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-539-5p | 12 | PIK3IP1 | Sponge network | -0.487 | 0.02157 | -0.187 | 0.19945 | 0.437 |
| 15035 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | ZNF208 | Sponge network | -0.739 | 0.00044 | -0.4 | 0.22381 | 0.437 |
| 15036 | NR2F1-AS1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | CSDE1 | Sponge network | -0.49 | 0.04946 | -0.493 | 0 | 0.437 |
| 15037 | RP11-473I1.10 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 20 | CBX5 | Sponge network | 0.673 | 0 | 0.165 | 0.24446 | 0.437 |
| 15038 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-96-5p | 12 | NUAK1 | Sponge network | 0.342 | 0.03129 | 0.904 | 0 | 0.437 |
| 15039 | CTC-444N24.11 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ARHGEF12 | Sponge network | 1.153 | 0 | 0.09 | 0.35693 | 0.437 |
| 15040 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TRIM58 | Sponge network | -0.129 | 0.57177 | -1.692 | 0 | 0.437 |
| 15041 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | ZMYM2 | Sponge network | 0.93 | 0 | 0.279 | 0.01273 | 0.437 |
| 15042 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | TET1 | Sponge network | 0.75 | 0.00054 | -0.135 | 0.52138 | 0.437 |
| 15043 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 16 | ABCB5 | Sponge network | -0.901 | 0.00019 | -0.956 | 0.04077 | 0.437 |
| 15044 | RP11-798G7.7 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p | 13 | AKAP11 | Sponge network | 0.858 | 3.0E-5 | 0.196 | 0.09825 | 0.437 |
| 15045 | DNM3OS |
hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 10 | PHOX2B | Sponge network | 0.572 | 0.01722 | -2.68 | 0 | 0.437 |
| 15046 | LINC00702 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | RIMS2 | Sponge network | -0.737 | 0.06182 | -1.365 | 0.00208 | 0.437 |
| 15047 | PSMD5-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | TAF1 | Sponge network | 0.569 | 0.00067 | 0.39 | 3.0E-5 | 0.437 |
| 15048 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 11 | KCNAB1 | Sponge network | 0.194 | 0.64278 | -0.755 | 2.0E-5 | 0.437 |
| 15049 | AC011526.1 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | DOCK4 | Sponge network | 0.342 | 0.03129 | 0.502 | 0.00108 | 0.437 |
| 15050 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | HOOK3 | Sponge network | -1.697 | 0.00889 | 0.273 | 0.09957 | 0.437 |
| 15051 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-429 | 16 | LRP1B | Sponge network | -0.469 | 0.08721 | -2.163 | 0 | 0.437 |
| 15052 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-7-1-3p | 12 | TET1 | Sponge network | -1.092 | 4.0E-5 | -0.135 | 0.52138 | 0.437 |
| 15053 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 19 | RET | Sponge network | 0.335 | 0.20596 | -0.833 | 0.03517 | 0.437 |
| 15054 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | ZMYM2 | Sponge network | 1.178 | 0 | 0.279 | 0.01273 | 0.436 |
| 15055 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 17 | NF1 | Sponge network | 1.294 | 0 | 0.51 | 0 | 0.436 |
| 15056 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p | 13 | RELN | Sponge network | -0.465 | 0.04703 | -2.403 | 0 | 0.436 |
| 15057 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 41 | RBMS3 | Sponge network | 1.302 | 7.0E-5 | -0.519 | 0.03551 | 0.436 |
| 15058 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-940 | 35 | CHD5 | Sponge network | 0.214 | 0.18246 | -0.922 | 0.01521 | 0.436 |
| 15059 | LINC00472 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-3p | 36 | NTRK2 | Sponge network | -0.842 | 0.01361 | -0.736 | 0.06495 | 0.436 |
| 15060 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-338-5p | 12 | MAFB | Sponge network | -1.092 | 4.0E-5 | -0.023 | 0.89865 | 0.436 |
| 15061 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | TACC1 | Sponge network | -0.611 | 0.09607 | -0.362 | 0.05406 | 0.436 |
| 15062 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 23 | RBMS3 | Sponge network | -0.72 | 0.24239 | -0.519 | 0.03551 | 0.436 |
| 15063 | RP11-503E24.2 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-374b-5p | 12 | ABL2 | Sponge network | 2.218 | 0 | 0.82 | 0 | 0.436 |
| 15064 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | MYO9A | Sponge network | 0.889 | 9.0E-5 | -0.147 | 0.32373 | 0.436 |
| 15065 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ARNT | Sponge network | -0.942 | 0 | -0.08 | 0.26073 | 0.436 |
| 15066 | MAGI2-AS3 |
hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-664a-3p | 12 | LOX | Sponge network | -0.129 | 0.57177 | 1.164 | 0 | 0.436 |
| 15067 | RP11-156P1.3 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p | 10 | HIC1 | Sponge network | 0.214 | 0.18246 | 0.024 | 0.89889 | 0.436 |
| 15068 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 20 | CDC73 | Sponge network | 1.153 | 0 | 0.094 | 0.20463 | 0.436 |
| 15069 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | 0.246 | 0.59912 | 0.809 | 0.00544 | 0.436 |
| 15070 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ZFHX3 | Sponge network | -0.031 | 0.89063 | 0.389 | 0.01363 | 0.436 |
| 15071 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p | 11 | PCDH8 | Sponge network | -0.513 | 0.04703 | -0.997 | 0.05366 | 0.436 |
| 15072 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | -1.161 | 0.00591 | -1.3 | 0.01619 | 0.436 |
| 15073 | RP11-156P1.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 28 | AFF1 | Sponge network | 0.214 | 0.18246 | -0.384 | 0.00053 | 0.436 |
| 15074 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-940 | 25 | AKAP11 | Sponge network | 0.214 | 0.18246 | 0.196 | 0.09825 | 0.436 |
| 15075 | SH3RF3-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | PDE4DIP | Sponge network | 0.889 | 9.0E-5 | -0.282 | 0.0257 | 0.436 |
| 15076 | EMX2OS |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | RIMS2 | Sponge network | -0.777 | 0.1134 | -1.365 | 0.00208 | 0.436 |
| 15077 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 22 | CADM3 | Sponge network | 0.997 | 0.00066 | -3.663 | 0 | 0.436 |
| 15078 | ASH1L-AS1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-495-3p | 12 | PHF6 | Sponge network | 0.948 | 0 | 0.785 | 0 | 0.436 |
| 15079 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PLSCR4 | Sponge network | -0.899 | 6.0E-5 | -0.474 | 0.03833 | 0.436 |
| 15080 | LINC00092 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-3913-5p;hsa-miR-590-3p | 10 | WWTR1 | Sponge network | -1.886 | 0 | -0.345 | 0.11997 | 0.436 |
| 15081 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-766-3p | 25 | IL17RD | Sponge network | 0.389 | 0.04293 | -0.019 | 0.94873 | 0.436 |
| 15082 | AC010226.4 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-590-3p | 16 | SYT4 | Sponge network | -0.811 | 2.0E-5 | -3.207 | 0 | 0.436 |
| 15083 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 38 | RBMS3 | Sponge network | 0.374 | 0.20054 | -0.519 | 0.03551 | 0.436 |
| 15084 | SNX29P2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 12 | SORCS1 | Sponge network | 0.4 | 0.14611 | -3.492 | 0 | 0.436 |
| 15085 | RAMP2-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TLL1 | Sponge network | -0.513 | 0.04703 | -1.195 | 3.0E-5 | 0.436 |
| 15086 | RP11-967K21.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p | 31 | DIP2C | Sponge network | 0.056 | 0.81195 | -0.436 | 0.01713 | 0.436 |
| 15087 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | PDZRN3 | Sponge network | 0.246 | 0.59912 | -1.548 | 0 | 0.436 |
| 15088 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | LRP1B | Sponge network | -0.769 | 0.00035 | -2.163 | 0 | 0.436 |
| 15089 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-655-3p | 19 | KANK1 | Sponge network | -0.096 | 0.67958 | -0.55 | 0.00134 | 0.436 |
| 15090 | LINC01018 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-877-5p | 23 | NRK | Sponge network | -2.915 | 0.00015 | 0.656 | 0.19217 | 0.436 |
| 15091 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | NRXN3 | Sponge network | 0.083 | 0.66405 | -0.893 | 0.04186 | 0.436 |
| 15092 | RP11-488L18.10 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p | 11 | CREB1 | Sponge network | 1.392 | 0 | 0.203 | 0.00537 | 0.436 |
| 15093 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 31 | QKI | Sponge network | 0.381 | 0.25967 | -0.333 | 0.04111 | 0.436 |
| 15094 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 11 | HERC2 | Sponge network | 1.153 | 0 | 0.114 | 0.30638 | 0.436 |
| 15095 | ZNF667-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 25 | SYT4 | Sponge network | -0.976 | 0.00025 | -3.207 | 0 | 0.436 |
| 15096 | AC011526.1 |
hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p | 10 | WWTR1 | Sponge network | 0.342 | 0.03129 | -0.345 | 0.11997 | 0.436 |
| 15097 | FENDRR |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | PPP1R9A | Sponge network | -1.281 | 0.00012 | -0.903 | 0.04254 | 0.436 |
| 15098 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | EPHA7 | Sponge network | -0.318 | 0.65847 | -2.197 | 3.0E-5 | 0.436 |
| 15099 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-7-5p | 11 | FNBP1 | Sponge network | 0.299 | 0.57722 | -0.969 | 4.0E-5 | 0.436 |
| 15100 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-96-5p | 27 | CRTC3 | Sponge network | 0.249 | 0.35902 | 0.164 | 0.16786 | 0.436 |
| 15101 | RP11-21A7A.2 |
hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-877-5p | 12 | IL17RD | Sponge network | -1.116 | 0.23686 | -0.019 | 0.94873 | 0.436 |
| 15102 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-628-5p | 11 | CHD2 | Sponge network | 1.597 | 0 | 0.049 | 0.55371 | 0.436 |
| 15103 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | KDSR | Sponge network | 0.559 | 0.00026 | 0.239 | 0.02216 | 0.436 |
| 15104 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-495-3p | 11 | ZNF292 | Sponge network | 0.817 | 3.0E-5 | 0.276 | 0.04148 | 0.436 |
| 15105 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-590-5p;hsa-miR-654-3p | 17 | BMPR2 | Sponge network | 1.134 | 0 | 0.206 | 0.0635 | 0.436 |
| 15106 | RP11-967K21.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 12 | EZH1 | Sponge network | 0.056 | 0.81195 | -0.564 | 1.0E-5 | 0.436 |
| 15107 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | NRXN3 | Sponge network | 0.395 | 0.15451 | -0.893 | 0.04186 | 0.436 |
| 15108 | LINC00472 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-628-5p | 15 | CDH23 | Sponge network | -0.842 | 0.01361 | -0.974 | 0.00034 | 0.436 |
| 15109 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | LRP1B | Sponge network | -0.976 | 0.00025 | -2.163 | 0 | 0.436 |
| 15110 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | GJA1 | Sponge network | 0.335 | 0.20596 | -0.485 | 0.02179 | 0.436 |
| 15111 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 46 | FAT3 | Sponge network | -0.342 | 0.02288 | -1.601 | 0.00051 | 0.436 |
| 15112 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-92a-1-5p | 14 | TRPS1 | Sponge network | -0.309 | 0.1145 | -0.191 | 0.44109 | 0.436 |
| 15113 | SH3RF3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-486-5p | 14 | SYNM | Sponge network | 0.889 | 9.0E-5 | -2.309 | 1.0E-5 | 0.436 |
| 15114 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-33a-3p | 20 | PBX1 | Sponge network | -0.811 | 2.0E-5 | -1.206 | 0 | 0.436 |
| 15115 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-93-3p | 16 | RPS6KA2 | Sponge network | -0.777 | 0.16667 | -0.57 | 0.00013 | 0.436 |
| 15116 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 33 | PBX1 | Sponge network | 0.064 | 0.72934 | -1.206 | 0 | 0.436 |
| 15117 | DIO3OS |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-940 | 13 | MAPK10 | Sponge network | -0.773 | 0.04073 | -1.774 | 0 | 0.436 |
| 15118 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | EPHA3 | Sponge network | -1.249 | 7.0E-5 | 0.185 | 0.53588 | 0.436 |
| 15119 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 19 | RNF144A | Sponge network | 0.176 | 0.37402 | 0.594 | 0.0009 | 0.436 |
| 15120 | LINC00654 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 52 | RUNX1T1 | Sponge network | 0.374 | 0.20054 | -0.344 | 0.2374 | 0.436 |
| 15121 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-940 | 44 | CHD5 | Sponge network | 0.064 | 0.72934 | -0.922 | 0.01521 | 0.436 |
| 15122 | CTA-363E19.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 12 | RPS6KA3 | Sponge network | 1.055 | 0 | 0.256 | 0.02419 | 0.436 |
| 15123 | FENDRR |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-758-3p | 21 | CSDE1 | Sponge network | -1.281 | 0.00012 | -0.493 | 0 | 0.436 |
| 15124 | LINC00865 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-378a-3p | 12 | FLNA | Sponge network | -1.249 | 7.0E-5 | -0.771 | 0.01377 | 0.436 |
| 15125 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CLIP1 | Sponge network | 0.72 | 0.0001 | -0.215 | 0.08684 | 0.436 |
| 15126 | RP11-356J5.12 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p | 28 | DLC1 | Sponge network | -0.899 | 6.0E-5 | -0.382 | 0.05038 | 0.436 |
| 15127 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | CACNA2D1 | Sponge network | 0.249 | 0.35902 | -0.846 | 0.00675 | 0.436 |
| 15128 | RP11-798M19.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | NAV3 | Sponge network | -0.612 | 0.00094 | 0.212 | 0.47729 | 0.436 |
| 15129 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | MLLT10 | Sponge network | 0.848 | 0 | 0.105 | 0.33292 | 0.436 |
| 15130 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 62 | WDFY3 | Sponge network | -0.737 | 0.06182 | -0.201 | 0.0967 | 0.436 |
| 15131 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | TACC1 | Sponge network | 0.082 | 0.55654 | -0.362 | 0.05406 | 0.436 |
| 15132 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p | 13 | CHD1 | Sponge network | 0.93 | 0 | 0.322 | 0.00215 | 0.436 |
| 15133 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577 | 23 | GREM1 | Sponge network | -1.575 | 6.0E-5 | 0.902 | 0.01691 | 0.436 |
| 15134 | RP11-144G6.12 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 12 | PHF6 | Sponge network | 0.826 | 1.0E-5 | 0.785 | 0 | 0.436 |
| 15135 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | SYNE1 | Sponge network | 0.385 | 0.10067 | -0.738 | 0.00316 | 0.436 |
| 15136 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | MYO9A | Sponge network | 0.392 | 0.0278 | -0.147 | 0.32373 | 0.436 |
| 15137 | PWAR6 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 54 | MDM4 | Sponge network | -1.575 | 6.0E-5 | 0.345 | 0.00762 | 0.436 |
| 15138 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p | 31 | ST6GALNAC3 | Sponge network | -1.249 | 7.0E-5 | -0.987 | 0 | 0.436 |
| 15139 | LINC00865 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-629-3p | 11 | ID4 | Sponge network | -1.249 | 7.0E-5 | -1.644 | 0 | 0.436 |
| 15140 | RP11-389C8.2 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SETBP1 | Sponge network | 0.727 | 3.0E-5 | -1.302 | 1.0E-5 | 0.436 |
| 15141 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-424-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-942-5p | 32 | NRXN1 | Sponge network | -0.465 | 0.04703 | -3.778 | 0 | 0.436 |
| 15142 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p | 36 | RICTOR | Sponge network | 0.873 | 0 | 0.388 | 0.00188 | 0.436 |
| 15143 | ZNF582-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 38 | TGFBR3 | Sponge network | -1.349 | 0.00017 | -1.173 | 1.0E-5 | 0.436 |
| 15144 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 30 | SNX29 | Sponge network | 0.4 | 0.14611 | -0.34 | 0.01371 | 0.436 |
| 15145 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 15 | TACC1 | Sponge network | -2.039 | 0.03447 | -0.362 | 0.05406 | 0.436 |
| 15146 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-940;hsa-miR-98-5p | 22 | ZFP36L1 | Sponge network | -0.129 | 0.57177 | -0.505 | 8.0E-5 | 0.436 |
| 15147 | MRVI1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 27 | PTPRD | Sponge network | -1.092 | 4.0E-5 | -1.005 | 0.00064 | 0.436 |
| 15148 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 37 | TACC1 | Sponge network | -1.249 | 7.0E-5 | -0.362 | 0.05406 | 0.436 |
| 15149 | RP11-25K19.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-532-3p;hsa-miR-542-3p | 15 | SYNM | Sponge network | -1.057 | 0.00029 | -2.309 | 1.0E-5 | 0.436 |
| 15150 | FENDRR |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 26 | GJA1 | Sponge network | -1.281 | 0.00012 | -0.485 | 0.02179 | 0.436 |
| 15151 | LINC00342 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p | 16 | MDM4 | Sponge network | 0.297 | 0.19933 | 0.345 | 0.00762 | 0.436 |
| 15152 | RP11-326G21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ABI2 | Sponge network | -0.021 | 0.93598 | 0.175 | 0.14787 | 0.436 |
| 15153 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-940 | 17 | LIFR | Sponge network | -4.546 | 0 | -1.794 | 0 | 0.436 |
| 15154 | C20orf166-AS1 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 11 | RTN4 | Sponge network | -2.69 | 0.0001 | -0.412 | 0.00014 | 0.436 |
| 15155 | CTC-429P9.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-877-5p | 17 | IL17RD | Sponge network | 0.043 | 0.81628 | -0.019 | 0.94873 | 0.436 |
| 15156 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 20 | MYO9A | Sponge network | 0.4 | 0.14611 | -0.147 | 0.32373 | 0.436 |
| 15157 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-493-5p | 18 | ANK2 | Sponge network | -0.829 | 0.01343 | -1.603 | 1.0E-5 | 0.436 |
| 15158 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | KAT6B | Sponge network | 0.225 | 0.30644 | -0.478 | 0.00018 | 0.436 |
| 15159 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-421 | 11 | IGF1 | Sponge network | -4.463 | 0.00025 | -0.204 | 0.5898 | 0.436 |
| 15160 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | DOCK4 | Sponge network | 0.101 | 0.60919 | 0.502 | 0.00108 | 0.436 |
| 15161 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 44 | HLF | Sponge network | 0.997 | 0.00066 | -2.443 | 0 | 0.436 |
| 15162 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-93-3p | 30 | BMPR2 | Sponge network | -0.376 | 0.00337 | 0.206 | 0.0635 | 0.436 |
| 15163 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-424-3p;hsa-miR-493-5p;hsa-miR-940 | 11 | HOOK3 | Sponge network | 0.637 | 0.00069 | 0.273 | 0.09957 | 0.436 |
| 15164 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 19 | PRKAB2 | Sponge network | 0.556 | 0.00298 | -0.367 | 0.01371 | 0.436 |
| 15165 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | CDH19 | Sponge network | -0.405 | 0.22013 | -3.434 | 0 | 0.436 |
| 15166 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | MYO9A | Sponge network | 0.873 | 0 | -0.147 | 0.32373 | 0.436 |
| 15167 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 45 | PRKAA2 | Sponge network | -0.738 | 0.21233 | -2.462 | 0 | 0.436 |
| 15168 | RP11-517B11.7 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-5p | 24 | ARHGEF12 | Sponge network | 0.706 | 0 | 0.09 | 0.35693 | 0.436 |
| 15169 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | ZFHX3 | Sponge network | -0.777 | 0.1134 | 0.389 | 0.01363 | 0.436 |
| 15170 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-185-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-450a-5p | 12 | PBX1 | Sponge network | -7.551 | 0 | -1.206 | 0 | 0.436 |
| 15171 | SOS1-IT1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-378a-5p;hsa-miR-495-3p | 12 | CREB1 | Sponge network | 0.892 | 0 | 0.203 | 0.00537 | 0.436 |
| 15172 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 41 | TMEM132B | Sponge network | -0.842 | 0.01361 | -0.83 | 0.01084 | 0.436 |
| 15173 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-532-3p;hsa-miR-576-5p | 35 | PBX1 | Sponge network | -1.645 | 0 | -1.206 | 0 | 0.436 |
| 15174 | RP11-701H24.7 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p | 12 | DLC1 | Sponge network | -2.039 | 0.03447 | -0.382 | 0.05038 | 0.436 |
| 15175 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-93-3p | 17 | IL6ST | Sponge network | -0.31 | 0.33871 | -0.402 | 0.00676 | 0.436 |
| 15176 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | -0.096 | 0.67958 | -1.3 | 0.01619 | 0.436 |
| 15177 | CTC-457L16.2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | DOCK4 | Sponge network | 1.153 | 0.00114 | 0.502 | 0.00108 | 0.436 |
| 15178 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-576-5p | 17 | PDGFRA | Sponge network | -0.932 | 0.00329 | -0.372 | 0.0037 | 0.436 |
| 15179 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-370-3p;hsa-miR-452-5p;hsa-miR-625-5p | 24 | JAZF1 | Sponge network | -2.095 | 0.00112 | -0.377 | 0.02677 | 0.436 |
| 15180 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p | 13 | FSTL5 | Sponge network | -0.563 | 0.02545 | -1.3 | 0.01619 | 0.436 |
| 15181 | WDFY3-AS2 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | NRG3 | Sponge network | -0.942 | 0 | -1.836 | 4.0E-5 | 0.436 |
| 15182 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-96-5p | 27 | FBXO32 | Sponge network | -0.811 | 2.0E-5 | -0.495 | 0.07561 | 0.436 |
| 15183 | HNRNPU-AS1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | KIF5B | Sponge network | 1.601 | 0 | 0.362 | 0.00066 | 0.436 |
| 15184 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 27 | MEF2C | Sponge network | 0.056 | 0.8494 | -0.499 | 0.01448 | 0.436 |
| 15185 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c | 14 | RBMS3 | Sponge network | -1.14 | 0.03513 | -0.519 | 0.03551 | 0.435 |
| 15186 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-33a-3p | 19 | AKAP11 | Sponge network | 0.594 | 0.00064 | 0.196 | 0.09825 | 0.435 |
| 15187 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-940;hsa-miR-98-5p | 32 | PBX1 | Sponge network | 0.494 | 0.00339 | -1.206 | 0 | 0.435 |
| 15188 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | FNBP1 | Sponge network | 0.082 | 0.55654 | -0.969 | 4.0E-5 | 0.435 |
| 15189 | RP11-57H14.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-96-5p | 30 | IL17RD | Sponge network | 0.083 | 0.66405 | -0.019 | 0.94873 | 0.435 |
| 15190 | MEG3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-5p | 15 | ABCA10 | Sponge network | 0.249 | 0.35902 | -0.571 | 0.03674 | 0.435 |
| 15191 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-486-5p;hsa-miR-576-5p | 13 | ZNF208 | Sponge network | -0.162 | 0.47687 | -0.4 | 0.22381 | 0.435 |
| 15192 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | RELN | Sponge network | 0.246 | 0.59912 | -2.403 | 0 | 0.435 |
| 15193 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-590-3p | 16 | GRIA3 | Sponge network | -0.739 | 0.00044 | -1.956 | 0 | 0.435 |
| 15194 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 25 | CREB3L2 | Sponge network | -0.358 | 0.17255 | -0.525 | 2.0E-5 | 0.435 |
| 15195 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-96-5p | 41 | PIK3R1 | Sponge network | 0.083 | 0.66405 | -0.133 | 0.36854 | 0.435 |
| 15196 | ARHGEF26-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | GJA1 | Sponge network | -2.46 | 0 | -0.485 | 0.02179 | 0.435 |
| 15197 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-93-5p | 12 | TRIM3 | Sponge network | -1.439 | 4.0E-5 | -0.577 | 0 | 0.435 |
| 15198 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-542-3p | 27 | PIK3R1 | Sponge network | -1.092 | 4.0E-5 | -0.133 | 0.36854 | 0.435 |
| 15199 | GNG12-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-590-3p | 10 | SCN11A | Sponge network | -0.793 | 7.0E-5 | -1.15 | 0.00299 | 0.435 |
| 15200 | LINC00865 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-766-3p | 33 | IL17RD | Sponge network | -1.249 | 7.0E-5 | -0.019 | 0.94873 | 0.435 |
| 15201 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 10 | CBL | Sponge network | 0.931 | 1.0E-5 | 0.291 | 0.01267 | 0.435 |
| 15202 | RP11-999E24.3 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-942-5p | 17 | OPCML | Sponge network | -0.693 | 0.05629 | -2.498 | 0 | 0.435 |
| 15203 | U91328.19 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-93-5p | 52 | DST | Sponge network | -0.303 | 0.21002 | -0.713 | 0.00096 | 0.435 |
| 15204 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | PDGFC | Sponge network | -1.575 | 6.0E-5 | -0.33 | 0.06483 | 0.435 |
| 15205 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-671-5p;hsa-miR-93-5p | 46 | ADARB1 | Sponge network | 0.253 | 0.09565 | -0.335 | 0.06631 | 0.435 |
| 15206 | SNX29P2 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | 0.4 | 0.14611 | 0.358 | 0.13727 | 0.435 |
| 15207 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-5p | 21 | CADM1 | Sponge network | 0.176 | 0.37402 | -0.9 | 0.00084 | 0.435 |
| 15208 | RASSF8-AS1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-93-5p | 56 | DST | Sponge network | 0.335 | 0.20596 | -0.713 | 0.00096 | 0.435 |
| 15209 | CTC-296K1.3 |
hsa-let-7a-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-625-5p | 11 | CTIF | Sponge network | -2.095 | 0.00112 | -0.389 | 0.00549 | 0.435 |
| 15210 | TTC28-AS1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-486-5p | 10 | TPR | Sponge network | 0.373 | 0.00068 | 0.582 | 0 | 0.435 |
| 15211 | PWAR6 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-96-5p | 14 | ELMO1 | Sponge network | -1.575 | 6.0E-5 | 0.04 | 0.83147 | 0.435 |
| 15212 | MEG3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 19 | CDH19 | Sponge network | 0.249 | 0.35902 | -3.434 | 0 | 0.435 |
| 15213 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-542-3p | 41 | DTNA | Sponge network | -0.384 | 0.40652 | -1.648 | 1.0E-5 | 0.435 |
| 15214 | HCG11 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PCDH9 | Sponge network | 0.385 | 0.10067 | -1.823 | 4.0E-5 | 0.435 |
| 15215 | RP11-535M15.1 |
hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-3p | 13 | DTNA | Sponge network | -1.14 | 0.03513 | -1.648 | 1.0E-5 | 0.435 |
| 15216 | ZNF667-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -0.976 | 0.00025 | -0.966 | 0.00035 | 0.435 |
| 15217 | TUG1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p;hsa-miR-582-5p | 20 | BRD3 | Sponge network | 0.972 | 0 | 0.37 | 0.00013 | 0.435 |
| 15218 | CTC-296K1.4 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p | 11 | ITPKB | Sponge network | -2.076 | 0.00548 | -0.704 | 0.00106 | 0.435 |
| 15219 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p | 10 | CUL5 | Sponge network | 1.106 | 0 | 0.026 | 0.77301 | 0.435 |
| 15220 | LINC00702 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 20 | FMNL3 | Sponge network | -0.737 | 0.06182 | 0.555 | 4.0E-5 | 0.435 |
| 15221 | RP11-822E23.8 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-550a-5p | 11 | PCDH18 | Sponge network | -0.777 | 0.16667 | 0.216 | 0.24645 | 0.435 |
| 15222 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3913-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-93-5p | 33 | ZBTB4 | Sponge network | -0.615 | 0.00015 | -0.739 | 0 | 0.435 |
| 15223 | DNAJC27-AS1 |
hsa-let-7d-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-98-5p | 10 | MPL | Sponge network | -0.969 | 2.0E-5 | -0.624 | 0.00133 | 0.435 |
| 15224 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | TET1 | Sponge network | -2.46 | 0 | -0.135 | 0.52138 | 0.435 |
| 15225 | LINC00641 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 19 | PTGFR | Sponge network | -0.222 | 0.32445 | -0.233 | 0.45421 | 0.435 |
| 15226 | LINC00641 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | TAL1 | Sponge network | -0.222 | 0.32445 | -0.462 | 0.00574 | 0.435 |
| 15227 | RP11-384O8.1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | MACF1 | Sponge network | -0.738 | 0.21233 | 0.019 | 0.90335 | 0.435 |
| 15228 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 23 | ARHGAP29 | Sponge network | -0.576 | 0.10121 | 0.131 | 0.42953 | 0.435 |
| 15229 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | -2.452 | 0 | 0.273 | 0.09957 | 0.435 |
| 15230 | MAGI2-AS3 |
hsa-miR-10b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 10 | EXTL2 | Sponge network | -0.129 | 0.57177 | 0.109 | 0.37375 | 0.435 |
| 15231 | LINC00672 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 11 | PEG3 | Sponge network | -0.227 | 0.31657 | -1.74 | 0 | 0.435 |
| 15232 | HCG11 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 13 | PEG3 | Sponge network | 0.385 | 0.10067 | -1.74 | 0 | 0.435 |
| 15233 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p | 19 | MOB1B | Sponge network | 1.147 | 0 | 0.197 | 0.15266 | 0.435 |
| 15234 | AC141928.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | BCL9 | Sponge network | 0.853 | 0.15352 | 0.321 | 0.00267 | 0.435 |
| 15235 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p | 12 | CUL5 | Sponge network | 0.545 | 0.00064 | 0.026 | 0.77301 | 0.435 |
| 15236 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 31 | DMD | Sponge network | 0.082 | 0.55654 | -1.506 | 0 | 0.435 |
| 15237 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | NIN | Sponge network | -0.323 | 0.01372 | -0.253 | 0.13794 | 0.435 |
| 15238 | FAM66C |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | GALNT13 | Sponge network | -0.901 | 0.00019 | -0.857 | 0.07439 | 0.435 |
| 15239 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p | 12 | ZNF521 | Sponge network | -0.942 | 0 | -0.111 | 0.58068 | 0.435 |
| 15240 | PWAR6 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ZNF844 | Sponge network | -1.575 | 6.0E-5 | -0.966 | 0.00035 | 0.435 |
| 15241 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-5p | 15 | NRXN3 | Sponge network | -0.384 | 0.40652 | -0.893 | 0.04186 | 0.435 |
| 15242 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-940 | 24 | PBX1 | Sponge network | -2.095 | 0.00112 | -1.206 | 0 | 0.435 |
| 15243 | RASSF8-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | RIMS2 | Sponge network | 0.335 | 0.20596 | -1.365 | 0.00208 | 0.435 |
| 15244 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 16 | MYCBP2 | Sponge network | 0.421 | 0.00084 | -0.027 | 0.82016 | 0.435 |
| 15245 | RP11-597D13.9 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | PREX2 | Sponge network | 1.302 | 7.0E-5 | -0.272 | 0.25382 | 0.435 |
| 15246 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-590-3p | 14 | TLE4 | Sponge network | -0.896 | 0.00099 | -1.157 | 0 | 0.435 |
| 15247 | RP11-123M6.2 |
hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 11 | NTRK3 | Sponge network | 0.543 | 0.12356 | -1.77 | 3.0E-5 | 0.435 |
| 15248 | LINC00702 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | RNF144A | Sponge network | -0.737 | 0.06182 | 0.594 | 0.0009 | 0.435 |
| 15249 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 23 | PRKAB2 | Sponge network | 0.559 | 0.00026 | -0.367 | 0.01371 | 0.435 |
| 15250 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-628-5p | 12 | FBXO32 | Sponge network | -0.557 | 0.11135 | -0.495 | 0.07561 | 0.435 |
| 15251 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 19 | RECK | Sponge network | -0.738 | 0.21233 | -1.043 | 0 | 0.435 |
| 15252 | RP11-822E23.8 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-3p | 11 | STAT5B | Sponge network | -0.777 | 0.16667 | -0.182 | 0.1082 | 0.435 |
| 15253 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 12 | GAS1 | Sponge network | 0.246 | 0.59912 | -1.047 | 0.00364 | 0.435 |
| 15254 | LINC00174 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p | 12 | RASAL2 | Sponge network | 1.253 | 0 | 0.869 | 0 | 0.435 |
| 15255 | RP11-13K12.1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -0.154 | 0.78939 | -2.633 | 0 | 0.435 |
| 15256 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | PTPN13 | Sponge network | -0.323 | 0.01372 | -1.208 | 0 | 0.435 |
| 15257 | TPTEP1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | PREX2 | Sponge network | -1.216 | 0 | -0.272 | 0.25382 | 0.435 |
| 15258 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | EPHA3 | Sponge network | 0.16 | 0.50785 | 0.185 | 0.53588 | 0.435 |
| 15259 | PART1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 17 | RHOB | Sponge network | -2.925 | 0 | -1.052 | 0 | 0.435 |
| 15260 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-365a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 30 | RICTOR | Sponge network | 0.919 | 0 | 0.388 | 0.00188 | 0.435 |
| 15261 | ERVH48-1 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 25 | ABL2 | Sponge network | 1.04 | 0.00023 | 0.82 | 0 | 0.435 |
| 15262 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | TRIM33 | Sponge network | 0.765 | 7.0E-5 | 0.402 | 7.0E-5 | 0.435 |
| 15263 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | YAP1 | Sponge network | 0.72 | 0.0001 | 0.22 | 0.11836 | 0.435 |
| 15264 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 39 | SOX5 | Sponge network | -0.405 | 0.22013 | -1.091 | 4.0E-5 | 0.435 |
| 15265 | RASSF8-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-584-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | 0.335 | 0.20596 | -0.366 | 0.01036 | 0.435 |
| 15266 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 24 | RARB | Sponge network | -0.737 | 0.06182 | -0.825 | 2.0E-5 | 0.435 |
| 15267 | CTD-3092A11.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ZFX | Sponge network | 0.873 | 0 | 0.09 | 0.42563 | 0.435 |
| 15268 | HCG11 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-532-3p | 16 | SYNM | Sponge network | 0.385 | 0.10067 | -2.309 | 1.0E-5 | 0.435 |
| 15269 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 14 | CITED2 | Sponge network | -1.057 | 0.00029 | -1.545 | 0 | 0.435 |
| 15270 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | MITF | Sponge network | -0.096 | 0.67958 | -0.769 | 0.00022 | 0.435 |
| 15271 | LINC00476 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-889-3p | 19 | RPS6KA6 | Sponge network | -0.615 | 0.00015 | -2.989 | 0 | 0.435 |
| 15272 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p | 10 | ZNRF3 | Sponge network | 1.266 | 0 | 0.637 | 0.00172 | 0.435 |
| 15273 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-625-5p | 13 | ZMYM2 | Sponge network | 1.055 | 0 | 0.279 | 0.01273 | 0.435 |
| 15274 | RP11-156P1.3 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ITGB1 | Sponge network | 0.214 | 0.18246 | 0.524 | 0.00017 | 0.435 |
| 15275 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-629-5p | 16 | CTIF | Sponge network | -0.517 | 0.02935 | -0.389 | 0.00549 | 0.435 |
| 15276 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-484 | 11 | ABL1 | Sponge network | -2.531 | 0 | -0.215 | 0.07804 | 0.435 |
| 15277 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | FGFR1 | Sponge network | -0.08 | 0.90092 | -0.509 | 0.03957 | 0.435 |
| 15278 | FZD10-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 35 | IL17RD | Sponge network | -0.727 | 0.04726 | -0.019 | 0.94873 | 0.435 |
| 15279 | LINC00672 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | SETBP1 | Sponge network | -0.227 | 0.31657 | -1.302 | 1.0E-5 | 0.435 |
| 15280 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-576-5p | 51 | KIF1B | Sponge network | -0.162 | 0.47687 | -0.118 | 0.32258 | 0.435 |
| 15281 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ADAMTSL3 | Sponge network | 0.727 | 3.0E-5 | -1.559 | 0 | 0.435 |
| 15282 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 27 | CBFA2T3 | Sponge network | -0.576 | 0.10121 | -1.221 | 1.0E-5 | 0.435 |
| 15283 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-671-5p | 22 | GAS7 | Sponge network | 0.598 | 0.38481 | -0.555 | 0.01602 | 0.435 |
| 15284 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-584-5p;hsa-miR-93-5p | 32 | PIK3R1 | Sponge network | -0.465 | 0.04703 | -0.133 | 0.36854 | 0.435 |
| 15285 | TUG1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 20 | ARHGEF12 | Sponge network | 0.972 | 0 | 0.09 | 0.35693 | 0.435 |
| 15286 | GHRLOS |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-421;hsa-miR-455-3p | 10 | CADM3 | Sponge network | -1.942 | 0 | -3.663 | 0 | 0.435 |
| 15287 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 22 | TGFBR2 | Sponge network | -0.737 | 0.06182 | -0.243 | 0.11408 | 0.435 |
| 15288 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p | 25 | RASSF8 | Sponge network | 0.214 | 0.18246 | -0.122 | 0.65591 | 0.435 |
| 15289 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 45 | EPHA5 | Sponge network | -0.487 | 0.02157 | -0.957 | 0.01733 | 0.435 |
| 15290 | RP11-678G14.3 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-500a-3p;hsa-miR-7-1-3p | 11 | CDH19 | Sponge network | -4.477 | 0 | -3.434 | 0 | 0.435 |
| 15291 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | PRKAB2 | Sponge network | 0.603 | 0.00127 | -0.367 | 0.01371 | 0.435 |
| 15292 | LINC00473 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-3913-5p;hsa-miR-7-5p | 15 | FNBP1 | Sponge network | -1.203 | 0.03881 | -0.969 | 4.0E-5 | 0.435 |
| 15293 | WDFY3-AS2 |
hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | PACRG | Sponge network | -0.942 | 0 | -2.248 | 0 | 0.435 |
| 15294 | GNG12-AS1 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 19 | CSDE1 | Sponge network | -0.793 | 7.0E-5 | -0.493 | 0 | 0.435 |
| 15295 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-542-3p | 23 | DYNC1I1 | Sponge network | -0.969 | 2.0E-5 | -1.12 | 0.00107 | 0.435 |
| 15296 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 11 | RCAN2 | Sponge network | -0.342 | 0.02288 | -0.33 | 0.15172 | 0.435 |
| 15297 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 51 | ADARB1 | Sponge network | 0.246 | 0.59912 | -0.335 | 0.06631 | 0.435 |
| 15298 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-590-3p | 18 | TIMP3 | Sponge network | -0.738 | 0.21233 | -0.546 | 0.02307 | 0.435 |
| 15299 | RP11-216B9.6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-5p | 52 | LPP | Sponge network | 0.174 | 0.30034 | -0.459 | 0.04475 | 0.435 |
| 15300 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | ZMYND11 | Sponge network | -1.697 | 0.00889 | -0.2 | 0.05449 | 0.435 |
| 15301 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 47 | IL6ST | Sponge network | -1.057 | 0.00029 | -0.402 | 0.00676 | 0.435 |
| 15302 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SLC6A15 | Sponge network | -0.777 | 0.1134 | -2.373 | 2.0E-5 | 0.435 |
| 15303 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-96-5p | 25 | FBXO32 | Sponge network | -0.08 | 0.90092 | -0.495 | 0.07561 | 0.435 |
| 15304 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-942-5p | 35 | NOTCH2 | Sponge network | 0.335 | 0.20596 | 0.138 | 0.35163 | 0.435 |
| 15305 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 38 | ZFHX3 | Sponge network | -0.976 | 0.00025 | 0.389 | 0.01363 | 0.435 |
| 15306 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-940 | 19 | IGF1 | Sponge network | 0.299 | 0.57722 | -0.204 | 0.5898 | 0.435 |
| 15307 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-361-5p | 15 | LRP1B | Sponge network | -0.309 | 0.1145 | -2.163 | 0 | 0.435 |
| 15308 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-550a-3p | 20 | EBF1 | Sponge network | 0.082 | 0.55397 | -0.384 | 0.08856 | 0.435 |
| 15309 | CTD-2554C21.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p | 12 | ROBO1 | Sponge network | -1.654 | 0.00144 | -0.009 | 0.96154 | 0.435 |
| 15310 | RP11-536O18.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p | 10 | PRRX1 | Sponge network | -0.074 | 0.90758 | 1.316 | 0 | 0.435 |
| 15311 | PART1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p | 15 | MN1 | Sponge network | -2.925 | 0 | -0.358 | 0.24172 | 0.435 |
| 15312 | RP11-531A24.5 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 17 | PHF3 | Sponge network | 0.75 | 0.00054 | 0.126 | 0.19652 | 0.435 |
| 15313 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 15 | NCAM2 | Sponge network | 0.214 | 0.18246 | -0.482 | 0.22565 | 0.435 |
| 15314 | TPT1-AS1 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-3p;hsa-miR-625-3p | 24 | ABL2 | Sponge network | 0.903 | 0 | 0.82 | 0 | 0.435 |
| 15315 | LINC00641 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | EPS15 | Sponge network | -0.222 | 0.32445 | -0.052 | 0.53998 | 0.435 |
| 15316 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | RASSF2 | Sponge network | -0.323 | 0.01372 | -0.273 | 0.21961 | 0.434 |
| 15317 | AC083843.1 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 16 | RASAL2 | Sponge network | 1.4 | 0 | 0.869 | 0 | 0.434 |
| 15318 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-628-5p | 12 | TRIM33 | Sponge network | 1.344 | 0 | 0.402 | 7.0E-5 | 0.434 |
| 15319 | AC009120.6 |
hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 10 | SHPRH | Sponge network | 0.919 | 0 | 0.098 | 0.40994 | 0.434 |
| 15320 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-93-5p | 38 | PDGFRA | Sponge network | -0.777 | 0.16667 | -0.372 | 0.0037 | 0.434 |
| 15321 | AF131215.2 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-576-5p | 12 | ATM | Sponge network | -0.176 | 0.59692 | 0.178 | 0.21568 | 0.434 |
| 15322 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 14 | PIK3IP1 | Sponge network | -0.739 | 0.0574 | -0.187 | 0.19945 | 0.434 |
| 15323 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-33a-3p;hsa-miR-3613-5p | 17 | DMD | Sponge network | 0.497 | 0.01451 | -1.506 | 0 | 0.434 |
| 15324 | RP11-344B5.2 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b | 15 | QKI | Sponge network | -0.055 | 0.88482 | -0.333 | 0.04111 | 0.434 |
| 15325 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | 0.532 | 0.00505 | -1.09 | 0.00014 | 0.434 |
| 15326 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | SSBP2 | Sponge network | -0.726 | 0.00132 | -1.58 | 0 | 0.434 |
| 15327 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-655-3p | 12 | SON | Sponge network | 0.768 | 7.0E-5 | 0.168 | 0.04406 | 0.434 |
| 15328 | RP11-497H16.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | EPS15 | Sponge network | 0.545 | 0.00064 | -0.052 | 0.53998 | 0.434 |
| 15329 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ATRX | Sponge network | 0.539 | 0.03722 | 0.261 | 0.04408 | 0.434 |
| 15330 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 24 | SCN9A | Sponge network | 0.101 | 0.60919 | -0.525 | 0.10593 | 0.434 |
| 15331 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-5p | 25 | ESR1 | Sponge network | 0.4 | 0.14611 | -1.035 | 3.0E-5 | 0.434 |
| 15332 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | GRIA3 | Sponge network | -0.49 | 0.04946 | -1.956 | 0 | 0.434 |
| 15333 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | STARD13 | Sponge network | -0.355 | 0.0077 | -0.187 | 0.27383 | 0.434 |
| 15334 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-429;hsa-miR-590-3p | 10 | RBL2 | Sponge network | 0.537 | 0.00139 | -0.282 | 0.00254 | 0.434 |
| 15335 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-93-5p | 29 | SASH1 | Sponge network | -0.154 | 0.78939 | -0.502 | 0.00175 | 0.434 |
| 15336 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 15 | CACNA2D1 | Sponge network | -0.693 | 0.05629 | -0.846 | 0.00675 | 0.434 |
| 15337 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-374a-3p | 10 | SLC5A3 | Sponge network | 0.542 | 0.00054 | 0.493 | 5.0E-5 | 0.434 |
| 15338 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-33a-3p | 11 | EBF1 | Sponge network | -1.255 | 0.00981 | -0.384 | 0.08856 | 0.434 |
| 15339 | RP11-356J5.12 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 16 | ID4 | Sponge network | -0.899 | 6.0E-5 | -1.644 | 0 | 0.434 |
| 15340 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | MGA | Sponge network | 0.8 | 0 | 0.323 | 0.00792 | 0.434 |
| 15341 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 21 | TBL1XR1 | Sponge network | 1.205 | 0 | 0.578 | 0 | 0.434 |
| 15342 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p | 11 | ABCB5 | Sponge network | -4.463 | 0.00025 | -0.956 | 0.04077 | 0.434 |
| 15343 | SH3RF3-AS1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-582-5p | 10 | STARD13 | Sponge network | 0.889 | 9.0E-5 | -0.187 | 0.27383 | 0.434 |
| 15344 | SNX29P2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | RECK | Sponge network | 0.4 | 0.14611 | -1.043 | 0 | 0.434 |
| 15345 | RP11-102N12.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | MED12L | Sponge network | -0.351 | 0.11358 | -0.194 | 0.55299 | 0.434 |
| 15346 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | CDHR3 | Sponge network | 0.246 | 0.59912 | -0.977 | 4.0E-5 | 0.434 |
| 15347 | ZNF582-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-940;hsa-miR-96-5p | 31 | SNX29 | Sponge network | -1.349 | 0.00017 | -0.34 | 0.01371 | 0.434 |
| 15348 | NNT-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-3677-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p | 14 | PRKAA1 | Sponge network | 0.799 | 1.0E-5 | 0.317 | 0.0031 | 0.434 |
| 15349 | HCG11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | WWTR1 | Sponge network | 0.385 | 0.10067 | -0.345 | 0.11997 | 0.434 |
| 15350 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RBMS3 | Sponge network | 1.253 | 0 | -0.519 | 0.03551 | 0.434 |
| 15351 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-429 | 14 | MOB1B | Sponge network | 1.106 | 0 | 0.197 | 0.15266 | 0.434 |
| 15352 | LINC00641 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 18 | GLI3 | Sponge network | -0.222 | 0.32445 | -0.615 | 0.01482 | 0.434 |
| 15353 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | PHC3 | Sponge network | 0.826 | 1.0E-5 | 0.212 | 0.0499 | 0.434 |
| 15354 | LINC01004 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 17 | TBL1XR1 | Sponge network | 1.115 | 0 | 0.578 | 0 | 0.434 |
| 15355 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 16 | CHD2 | Sponge network | 0.75 | 0.00054 | 0.049 | 0.55371 | 0.434 |
| 15356 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p | 17 | USP6 | Sponge network | -0.031 | 0.89063 | 0.188 | 0.3871 | 0.434 |
| 15357 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 16 | CNTNAP3 | Sponge network | -1.654 | 0.00144 | -1.465 | 0.00033 | 0.434 |
| 15358 | AGAP11 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p | 11 | NGFR | Sponge network | -1.645 | 0 | -1.271 | 0.01058 | 0.434 |
| 15359 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-92a-3p | 28 | GPC6 | Sponge network | -0.563 | 0.02545 | -0.054 | 0.81465 | 0.434 |
| 15360 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 10 | DSE | Sponge network | -0.576 | 0.10121 | -0.231 | 0.05347 | 0.434 |
| 15361 | RP11-254F7.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 25 | MED12L | Sponge network | 0.176 | 0.37402 | -0.194 | 0.55299 | 0.434 |
| 15362 | FENDRR |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 47 | UNC80 | Sponge network | -1.281 | 0.00012 | -1.934 | 0 | 0.434 |
| 15363 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 21 | ST6GALNAC3 | Sponge network | -1.654 | 0.00144 | -0.987 | 0 | 0.434 |
| 15364 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-582-5p;hsa-miR-940 | 23 | QKI | Sponge network | -2.095 | 0.00112 | -0.333 | 0.04111 | 0.434 |
| 15365 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-93-5p | 55 | NTRK3 | Sponge network | -1.645 | 0 | -1.77 | 3.0E-5 | 0.434 |
| 15366 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | SOX5 | Sponge network | -0.727 | 0.04726 | -1.091 | 4.0E-5 | 0.434 |
| 15367 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-766-3p | 20 | TBL1XR1 | Sponge network | 0.873 | 0 | 0.578 | 0 | 0.434 |
| 15368 | CTD-3138B18.6 |
hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | RICTOR | Sponge network | 2.248 | 0 | 0.388 | 0.00188 | 0.434 |
| 15369 | LINC00641 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 41 | BCL2 | Sponge network | -0.222 | 0.32445 | -0.899 | 0 | 0.434 |
| 15370 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-425-5p;hsa-miR-577;hsa-miR-590-3p | 24 | ATRX | Sponge network | -0.227 | 0.31657 | 0.261 | 0.04408 | 0.434 |
| 15371 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | PDGFRA | Sponge network | -0.737 | 0.06182 | -0.372 | 0.0037 | 0.434 |
| 15372 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | SETBP1 | Sponge network | 0.16 | 0.50785 | -1.302 | 1.0E-5 | 0.434 |
| 15373 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 26 | RBMS3 | Sponge network | 1.757 | 0 | -0.519 | 0.03551 | 0.434 |
| 15374 | CTA-941F9.9 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-502-5p;hsa-miR-576-5p | 14 | TCF4 | Sponge network | -0.344 | 0.49624 | 0.016 | 0.92271 | 0.434 |
| 15375 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-940 | 43 | WDFY3 | Sponge network | 0.214 | 0.18246 | -0.201 | 0.0967 | 0.434 |
| 15376 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 53 | TMEM132B | Sponge network | -0.739 | 0.0574 | -0.83 | 0.01084 | 0.434 |
| 15377 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.083 | 0.66405 | 0.323 | 0.00792 | 0.434 |
| 15378 | RP11-822E23.8 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-877-5p | 20 | NRK | Sponge network | -0.777 | 0.16667 | 0.656 | 0.19217 | 0.434 |
| 15379 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-660-5p | 42 | AR | Sponge network | 0.214 | 0.18246 | -1.554 | 0 | 0.434 |
| 15380 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p | 52 | NTRK3 | Sponge network | -0.842 | 0.01361 | -1.77 | 3.0E-5 | 0.434 |
| 15381 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 53 | IL6ST | Sponge network | -1.203 | 0.03881 | -0.402 | 0.00676 | 0.434 |
| 15382 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-429;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 15 | LRRC4 | Sponge network | -0.563 | 0.02545 | -1.774 | 0 | 0.434 |
| 15383 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 15 | PCDH9 | Sponge network | -0.318 | 0.65847 | -1.823 | 4.0E-5 | 0.434 |
| 15384 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-590-3p | 13 | GRIN2A | Sponge network | -0.739 | 0.00044 | -2.161 | 0 | 0.434 |
| 15385 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-497-5p | 10 | NR2C2 | Sponge network | 1.178 | 0 | 0.21 | 0.03224 | 0.434 |
| 15386 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 20 | RPS6KA2 | Sponge network | -1.697 | 0.00889 | -0.57 | 0.00013 | 0.434 |
| 15387 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-628-5p | 24 | BCL6 | Sponge network | 0.249 | 0.35902 | -0.211 | 0.19489 | 0.434 |
| 15388 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-331-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 10 | MITF | Sponge network | 0.693 | 0.00076 | -0.769 | 0.00022 | 0.434 |
| 15389 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-93-3p | 34 | QKI | Sponge network | -0.693 | 0.05629 | -0.333 | 0.04111 | 0.434 |
| 15390 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 35 | BNC2 | Sponge network | 0.331 | 0.57247 | -0.491 | 0.09988 | 0.434 |
| 15391 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 56 | PRKAA2 | Sponge network | 0.997 | 0.00066 | -2.462 | 0 | 0.434 |
| 15392 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | DICER1 | Sponge network | 0.858 | 3.0E-5 | 0.042 | 0.674 | 0.434 |
| 15393 | RP11-701H24.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 14 | DTNA | Sponge network | -1.425 | 0.04204 | -1.648 | 1.0E-5 | 0.434 |
| 15394 | CTB-31O20.4 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | NF1 | Sponge network | 1.619 | 0 | 0.51 | 0 | 0.434 |
| 15395 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-421;hsa-miR-590-3p | 15 | STARD13 | Sponge network | 0.246 | 0.59912 | -0.187 | 0.27383 | 0.434 |
| 15396 | FGF14-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-96-5p | 22 | IL17RD | Sponge network | -1.582 | 8.0E-5 | -0.019 | 0.94873 | 0.434 |
| 15397 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 13 | FYN | Sponge network | -0.206 | 0.12942 | -0.131 | 0.37051 | 0.434 |
| 15398 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 43 | NTRK2 | Sponge network | 0.997 | 0.00066 | -0.736 | 0.06495 | 0.434 |
| 15399 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 10 | HIVEP1 | Sponge network | 0.537 | 0.00139 | 0.368 | 0.00064 | 0.434 |
| 15400 | LINC00476 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | NRXN3 | Sponge network | -0.615 | 0.00015 | -0.893 | 0.04186 | 0.434 |
| 15401 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-625-5p | 28 | RSPO2 | Sponge network | -0.162 | 0.47687 | -3.038 | 0 | 0.434 |
| 15402 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 59 | KIF1B | Sponge network | 0.75 | 0.00054 | -0.118 | 0.32258 | 0.434 |
| 15403 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -0.777 | 0.1134 | 0.309 | 0.17079 | 0.434 |
| 15404 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 49 | TRPS1 | Sponge network | 0.101 | 0.60919 | -0.191 | 0.44109 | 0.434 |
| 15405 | TPTEP1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | PRKCB | Sponge network | -1.216 | 0 | -1.313 | 2.0E-5 | 0.434 |
| 15406 | SNHG14 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 37 | CADM1 | Sponge network | -1.111 | 0.0003 | -0.9 | 0.00084 | 0.434 |
| 15407 | RP11-307B6.3 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421 | 16 | EPHA5 | Sponge network | -4.463 | 0.00025 | -0.957 | 0.01733 | 0.434 |
| 15408 | HOXA-AS2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-877-5p | 19 | NRK | Sponge network | 0.61 | 0.01593 | 0.656 | 0.19217 | 0.434 |
| 15409 | PCA3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | ABCB5 | Sponge network | -0.709 | 0.18168 | -0.956 | 0.04077 | 0.434 |
| 15410 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | GRIN2A | Sponge network | 0.997 | 0.00066 | -2.161 | 0 | 0.434 |
| 15411 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 18 | LHX6 | Sponge network | -1.111 | 0.0003 | -0.087 | 0.69193 | 0.434 |
| 15412 | CTC-429P9.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-3p | 13 | PTPN14 | Sponge network | 0.497 | 0.01451 | 0.144 | 0.34595 | 0.434 |
| 15413 | BOLA3-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p | 13 | PRUNE2 | Sponge network | -0.246 | 0.35034 | -1.733 | 0.00022 | 0.434 |
| 15414 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p | 18 | ABCA10 | Sponge network | -0.487 | 0.02157 | -0.571 | 0.03674 | 0.434 |
| 15415 | RP11-774O3.3 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-874-3p;hsa-miR-93-3p | 25 | CTIF | Sponge network | -0.487 | 0.02157 | -0.389 | 0.00549 | 0.434 |
| 15416 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p | 23 | PRDM2 | Sponge network | -1.161 | 0.00591 | -0.258 | 0.00721 | 0.434 |
| 15417 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | LIFR | Sponge network | -1.203 | 0.03881 | -1.794 | 0 | 0.434 |
| 15418 | ZNF667-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 17 | SEMA3C | Sponge network | -0.976 | 0.00025 | -0.272 | 0.31153 | 0.434 |
| 15419 | LINC00094 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-624-5p;hsa-miR-93-3p | 24 | WWTR1 | Sponge network | 0.253 | 0.09565 | -0.345 | 0.11997 | 0.434 |
| 15420 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-590-3p | 11 | HOOK3 | Sponge network | -1.161 | 0.00591 | 0.273 | 0.09957 | 0.434 |
| 15421 | RP11-356J5.12 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-501-3p;hsa-miR-942-5p | 12 | ZBTB20 | Sponge network | -0.899 | 6.0E-5 | -0.122 | 0.57569 | 0.434 |
| 15422 | RP11-750H9.5 |
hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p | 12 | IL6ST | Sponge network | 0.349 | 0.13152 | -0.402 | 0.00676 | 0.434 |
| 15423 | RP11-356J5.12 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | FOXO4 | Sponge network | -0.899 | 6.0E-5 | -0.516 | 2.0E-5 | 0.434 |
| 15424 | AC025171.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p | 14 | NR2C2 | Sponge network | 0.998 | 0 | 0.21 | 0.03224 | 0.434 |
| 15425 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-98-5p | 27 | LRIG1 | Sponge network | -0.355 | 0.0077 | -0.537 | 0.00588 | 0.434 |
| 15426 | RP11-21A7A.2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-324-3p;hsa-miR-589-5p | 14 | ADARB1 | Sponge network | -1.116 | 0.23686 | -0.335 | 0.06631 | 0.434 |
| 15427 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | ABI2 | Sponge network | -0.187 | 0.23787 | 0.175 | 0.14787 | 0.434 |
| 15428 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p | 10 | LHX6 | Sponge network | -0.896 | 0.00099 | -0.087 | 0.69193 | 0.434 |
| 15429 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-940 | 27 | MCC | Sponge network | -0.773 | 0.04073 | -1.005 | 0 | 0.434 |
| 15430 | FZD10-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 25 | SETBP1 | Sponge network | -0.727 | 0.04726 | -1.302 | 1.0E-5 | 0.434 |
| 15431 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p | 14 | ABCB5 | Sponge network | 0.246 | 0.59912 | -0.956 | 0.04077 | 0.434 |
| 15432 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 48 | FOXP1 | Sponge network | 0.194 | 0.64278 | 0.042 | 0.74368 | 0.434 |
| 15433 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-7-5p | 10 | CAV1 | Sponge network | -0.777 | 0.16667 | -1.013 | 6.0E-5 | 0.434 |
| 15434 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | 1.11 | 0 | 0.323 | 0.00792 | 0.434 |
| 15435 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | PLA2R1 | Sponge network | -0.576 | 0.10121 | -0.265 | 0.24676 | 0.434 |
| 15436 | BCYRN1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | SIAH1 | Sponge network | 0.311 | 0.10344 | -0.057 | 0.41415 | 0.433 |
| 15437 | U91328.19 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-501-5p;hsa-miR-589-5p | 11 | MN1 | Sponge network | -0.303 | 0.21002 | -0.358 | 0.24172 | 0.433 |
| 15438 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 11 | CITED2 | Sponge network | -0.583 | 0.14589 | -1.545 | 0 | 0.433 |
| 15439 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | 0.986 | 0.01022 | 0.323 | 0.00792 | 0.433 |
| 15440 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 34 | MITF | Sponge network | -1.216 | 0 | -0.769 | 0.00022 | 0.433 |
| 15441 | OIP5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 31 | HECA | Sponge network | 0.421 | 0.00084 | -0.217 | 0.03463 | 0.433 |
| 15442 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p | 17 | FREM2 | Sponge network | -0.842 | 0.01361 | -0.437 | 0.46353 | 0.433 |
| 15443 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | AKAP11 | Sponge network | 1.601 | 0 | 0.196 | 0.09825 | 0.433 |
| 15444 | HNRNPU-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CEP170 | Sponge network | 1.601 | 0 | 0.943 | 0 | 0.433 |
| 15445 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | PREX2 | Sponge network | 0.246 | 0.59912 | -0.272 | 0.25382 | 0.433 |
| 15446 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 33 | TOM1L2 | Sponge network | -0.737 | 0.06182 | -0.994 | 0 | 0.433 |
| 15447 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 10 | NR2C2 | Sponge network | 0.702 | 7.0E-5 | 0.21 | 0.03224 | 0.433 |
| 15448 | CTD-3064M3.3 |
hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-582-5p | 10 | KLF10 | Sponge network | -0.469 | 0.08721 | -0.783 | 0 | 0.433 |
| 15449 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 28 | USP12 | Sponge network | 0.101 | 0.60919 | -0.102 | 0.40768 | 0.433 |
| 15450 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | TIMP3 | Sponge network | 0.16 | 0.50785 | -0.546 | 0.02307 | 0.433 |
| 15451 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | CITED2 | Sponge network | -0.576 | 0.10121 | -1.545 | 0 | 0.433 |
| 15452 | OIP5-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PHF3 | Sponge network | 0.421 | 0.00084 | 0.126 | 0.19652 | 0.433 |
| 15453 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-378a-5p | 15 | ARHGAP29 | Sponge network | -0.842 | 0.01361 | 0.131 | 0.42953 | 0.433 |
| 15454 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-342-5p;hsa-miR-576-5p | 14 | SYNPO2 | Sponge network | 0.889 | 9.0E-5 | -2.342 | 0 | 0.433 |
| 15455 | CTA-363E19.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 18 | CREB1 | Sponge network | 1.055 | 0 | 0.203 | 0.00537 | 0.433 |
| 15456 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | PTPN11 | Sponge network | 0.064 | 0.72934 | 0.431 | 2.0E-5 | 0.433 |
| 15457 | DNM1P35 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | DYNC1I1 | Sponge network | 0.051 | 0.83388 | -1.12 | 0.00107 | 0.433 |
| 15458 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | NIN | Sponge network | -0.976 | 0.00025 | -0.253 | 0.13794 | 0.433 |
| 15459 | LIPE-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484 | 15 | ABL1 | Sponge network | 0.078 | 0.55803 | -0.215 | 0.07804 | 0.433 |
| 15460 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-409-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | ATRX | Sponge network | 0.659 | 3.0E-5 | 0.261 | 0.04408 | 0.433 |
| 15461 | RP11-822E23.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p | 13 | ITGB3 | Sponge network | -0.777 | 0.16667 | -0.011 | 0.95751 | 0.433 |
| 15462 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | FNBP1 | Sponge network | 0.064 | 0.72934 | -0.969 | 4.0E-5 | 0.433 |
| 15463 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 22 | PRKAA2 | Sponge network | 0.131 | 0.8436 | -2.462 | 0 | 0.433 |
| 15464 | CTC-429P9.2 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-539-5p;hsa-miR-940 | 12 | MEF2C | Sponge network | 0.043 | 0.81628 | -0.499 | 0.01448 | 0.433 |
| 15465 | AC010226.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-485-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | CADM2 | Sponge network | -0.811 | 2.0E-5 | -3.782 | 0 | 0.433 |
| 15466 | RP5-1074L1.4 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p | 10 | PHF3 | Sponge network | 1.923 | 0 | 0.126 | 0.19652 | 0.433 |
| 15467 | RP11-736K20.5 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-942-5p | 14 | GLIPR1 | Sponge network | -0.358 | 0.17255 | 0.146 | 0.3276 | 0.433 |
| 15468 | C20orf166-AS1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-582-5p | 13 | ZNF208 | Sponge network | -2.69 | 0.0001 | -0.4 | 0.22381 | 0.433 |
| 15469 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-629-5p | 19 | IGF1 | Sponge network | 0.727 | 3.0E-5 | -0.204 | 0.5898 | 0.433 |
| 15470 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | QKI | Sponge network | 1.253 | 0 | -0.333 | 0.04111 | 0.433 |
| 15471 | PWAR6 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | MSN | Sponge network | -1.575 | 6.0E-5 | -0.023 | 0.88422 | 0.433 |
| 15472 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 25 | CDH19 | Sponge network | -0.899 | 6.0E-5 | -3.434 | 0 | 0.433 |
| 15473 | JAZF1-AS1 |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | PAIP2 | Sponge network | -0.769 | 0.00035 | -0.515 | 0 | 0.433 |
| 15474 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-455-3p | 13 | DKK3 | Sponge network | -0.896 | 0.00099 | -0.052 | 0.77783 | 0.433 |
| 15475 | RP11-13K12.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-671-5p | 16 | SORCS1 | Sponge network | -0.154 | 0.78939 | -3.492 | 0 | 0.433 |
| 15476 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-508-3p | 10 | RIMS1 | Sponge network | -1.654 | 0.00144 | -3.143 | 0 | 0.433 |
| 15477 | DIO3OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-501-5p | 11 | DACT1 | Sponge network | -0.773 | 0.04073 | 0.783 | 0.00072 | 0.433 |
| 15478 | RP11-359B12.2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | BCL9 | Sponge network | 0.064 | 0.72934 | 0.321 | 0.00267 | 0.433 |
| 15479 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p | 19 | RARB | Sponge network | -1.654 | 0.00144 | -0.825 | 2.0E-5 | 0.433 |
| 15480 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 14 | CNTNAP1 | Sponge network | -2.915 | 0.00015 | 0.358 | 0.13727 | 0.433 |
| 15481 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 54 | IL6ST | Sponge network | -0.227 | 0.31657 | -0.402 | 0.00676 | 0.433 |
| 15482 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-423-5p | 10 | CBFA2T3 | Sponge network | -1.483 | 9.0E-5 | -1.221 | 1.0E-5 | 0.433 |
| 15483 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -0.769 | 0.00035 | -3.328 | 0 | 0.433 |
| 15484 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 14 | NUAK1 | Sponge network | 0.727 | 3.0E-5 | 0.904 | 0 | 0.433 |
| 15485 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ZMYND11 | Sponge network | -2.452 | 0 | -0.2 | 0.05449 | 0.433 |
| 15486 | RP11-890B15.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | PIK3CA | Sponge network | 0.101 | 0.60919 | 0.195 | 0.06908 | 0.433 |
| 15487 | RP11-13K12.1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 22 | ABL1 | Sponge network | -0.154 | 0.78939 | -0.215 | 0.07804 | 0.433 |
| 15488 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-942-5p | 16 | FBXO31 | Sponge network | -1.697 | 0.00889 | -0.085 | 0.42389 | 0.433 |
| 15489 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | FGFR1 | Sponge network | 0.082 | 0.55654 | -0.509 | 0.03957 | 0.433 |
| 15490 | HOXB-AS1 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-940 | 13 | TAL1 | Sponge network | -0.154 | 0.53954 | -0.462 | 0.00574 | 0.433 |
| 15491 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 24 | GRIN2A | Sponge network | -0.405 | 0.22013 | -2.161 | 0 | 0.433 |
| 15492 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p | 20 | MDM4 | Sponge network | 0.37 | 0.27614 | 0.345 | 0.00762 | 0.433 |
| 15493 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-93-3p | 48 | IL6ST | Sponge network | -0.465 | 0.04703 | -0.402 | 0.00676 | 0.433 |
| 15494 | RP11-2H8.2 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | RBBP6 | Sponge network | 1.089 | 0 | 0.163 | 0.05101 | 0.433 |
| 15495 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | IGF1 | Sponge network | 1.253 | 0 | -0.204 | 0.5898 | 0.433 |
| 15496 | WDFY3-AS2 |
hsa-miR-10b-3p;hsa-miR-193b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p | 10 | LRRTM1 | Sponge network | -0.942 | 0 | -3.361 | 0 | 0.433 |
| 15497 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p | 19 | CRTC3 | Sponge network | -0.154 | 0.78939 | 0.164 | 0.16786 | 0.433 |
| 15498 | CKMT2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p | 20 | SYNM | Sponge network | 0.082 | 0.55654 | -2.309 | 1.0E-5 | 0.433 |
| 15499 | CTA-14H9.5 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-7-1-3p | 10 | PLSCR4 | Sponge network | 0.145 | 0.53343 | -0.474 | 0.03833 | 0.433 |
| 15500 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 44 | MEF2C | Sponge network | -0.709 | 0.18168 | -0.499 | 0.01448 | 0.433 |
| 15501 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-98-5p | 29 | NTRK3 | Sponge network | 0.693 | 0.00076 | -1.77 | 3.0E-5 | 0.433 |
| 15502 | LINC00472 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-7-1-3p | 26 | MED12L | Sponge network | -0.842 | 0.01361 | -0.194 | 0.55299 | 0.433 |
| 15503 | DNM3OS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-935 | 16 | RELN | Sponge network | 0.572 | 0.01722 | -2.403 | 0 | 0.433 |
| 15504 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-3607-3p;hsa-miR-7-5p | 19 | KCNA6 | Sponge network | -0.693 | 0.05629 | -1.531 | 0.00013 | 0.433 |
| 15505 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 18 | SASH1 | Sponge network | -0.358 | 0.17255 | -0.502 | 0.00175 | 0.433 |
| 15506 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 43 | RUNX1T1 | Sponge network | -0.513 | 0.04703 | -0.344 | 0.2374 | 0.433 |
| 15507 | AC009948.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | IL17RD | Sponge network | 0.532 | 0.00505 | -0.019 | 0.94873 | 0.433 |
| 15508 | JAZF1-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | PDE4DIP | Sponge network | -0.769 | 0.00035 | -0.282 | 0.0257 | 0.433 |
| 15509 | BOLA3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 27 | MED12L | Sponge network | -0.246 | 0.35034 | -0.194 | 0.55299 | 0.433 |
| 15510 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-96-5p | 24 | PRKCE | Sponge network | -0.18 | 0.20343 | -0.403 | 0.00056 | 0.433 |
| 15511 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-486-5p | 16 | EPHA3 | Sponge network | 0.368 | 0.20093 | 0.185 | 0.53588 | 0.433 |
| 15512 | LINC00843 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | UBR3 | Sponge network | 1.106 | 0 | -0.021 | 0.82774 | 0.433 |
| 15513 | CTC-510F12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-335-3p | 11 | NTRK3 | Sponge network | -0.691 | 0.00146 | -1.77 | 3.0E-5 | 0.433 |
| 15514 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 15 | SCN9A | Sponge network | -4.546 | 0 | -0.525 | 0.10593 | 0.433 |
| 15515 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | AKAP11 | Sponge network | 1.205 | 0 | 0.196 | 0.09825 | 0.433 |
| 15516 | NEAT1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 18 | BAZ2B | Sponge network | 0.705 | 0.00014 | -0.276 | 0.02833 | 0.433 |
| 15517 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | PLSCR4 | Sponge network | -0.227 | 0.31657 | -0.474 | 0.03833 | 0.433 |
| 15518 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 11 | KAT6B | Sponge network | -0.663 | 0.10887 | -0.478 | 0.00018 | 0.433 |
| 15519 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-590-5p | 21 | SCN9A | Sponge network | 0.176 | 0.37402 | -0.525 | 0.10593 | 0.433 |
| 15520 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 15 | ARHGAP28 | Sponge network | -0.737 | 0.10822 | -0.911 | 0.00013 | 0.433 |
| 15521 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PHF3 | Sponge network | 0.225 | 0.30644 | 0.126 | 0.19652 | 0.433 |
| 15522 | CTC-457L16.2 |
hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-940 | 11 | SNX29 | Sponge network | 1.153 | 0.00114 | -0.34 | 0.01371 | 0.433 |
| 15523 | RP5-1112D6.4 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-497-5p;hsa-miR-582-5p | 10 | RICTOR | Sponge network | 1.379 | 0 | 0.388 | 0.00188 | 0.433 |
| 15524 | FAM66C |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 17 | ID4 | Sponge network | -0.901 | 0.00019 | -1.644 | 0 | 0.433 |
| 15525 | RP13-516M14.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-5p;hsa-miR-7-1-3p | 19 | BRD3 | Sponge network | 0.389 | 0.04293 | 0.37 | 0.00013 | 0.433 |
| 15526 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | FBXO32 | Sponge network | 0.178 | 0.29106 | -0.495 | 0.07561 | 0.433 |
| 15527 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 12 | DNAJB4 | Sponge network | 0.299 | 0.57722 | -0.7 | 1.0E-5 | 0.433 |
| 15528 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-411-5p;hsa-miR-429;hsa-miR-590-3p | 21 | USP6 | Sponge network | 0.537 | 0.00139 | 0.188 | 0.3871 | 0.433 |
| 15529 | SERTAD4-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 11 | RET | Sponge network | -0.932 | 0.00329 | -0.833 | 0.03517 | 0.433 |
| 15530 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 48 | IL6ST | Sponge network | -0.421 | 0.0114 | -0.402 | 0.00676 | 0.433 |
| 15531 | AC016747.3 |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 12 | FLI1 | Sponge network | 0.199 | 0.38015 | -0.294 | 0.10905 | 0.433 |
| 15532 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | PRKAB2 | Sponge network | 0.392 | 0.0278 | -0.367 | 0.01371 | 0.433 |
| 15533 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-671-5p | 12 | PIK3IP1 | Sponge network | -0.769 | 0.00035 | -0.187 | 0.19945 | 0.433 |
| 15534 | SNHG14 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 20 | ITGA5 | Sponge network | -1.111 | 0.0003 | 0.452 | 0.03524 | 0.433 |
| 15535 | LINC00641 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-7-1-3p | 13 | RASSF3 | Sponge network | -0.222 | 0.32445 | -0.226 | 0.11691 | 0.433 |
| 15536 | AC010226.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | LPP | Sponge network | -0.811 | 2.0E-5 | -0.459 | 0.04475 | 0.433 |
| 15537 | MYLK-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-671-5p | 23 | FOXP1 | Sponge network | -1.331 | 3.0E-5 | 0.042 | 0.74368 | 0.433 |
| 15538 | RP11-705C15.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | EPS15 | Sponge network | 0.796 | 4.0E-5 | -0.052 | 0.53998 | 0.433 |
| 15539 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-942-5p | 45 | TMEM132B | Sponge network | -0.162 | 0.47687 | -0.83 | 0.01084 | 0.433 |
| 15540 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CACNA2D1 | Sponge network | -0.899 | 6.0E-5 | -0.846 | 0.00675 | 0.433 |
| 15541 | OIP5-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 36 | ABL2 | Sponge network | 0.421 | 0.00084 | 0.82 | 0 | 0.433 |
| 15542 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-877-5p | 13 | RBMS3 | Sponge network | -1.116 | 0.23686 | -0.519 | 0.03551 | 0.433 |
| 15543 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 24 | CDH19 | Sponge network | -0.487 | 0.02157 | -3.434 | 0 | 0.433 |
| 15544 | CTA-204B4.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-454-3p;hsa-miR-590-3p | 22 | ABI2 | Sponge network | 0.765 | 7.0E-5 | 0.175 | 0.14787 | 0.433 |
| 15545 | PART1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | TPO | Sponge network | -2.925 | 0 | -1.87 | 2.0E-5 | 0.433 |
| 15546 | RP11-473I1.9 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-664a-3p | 19 | RASAL2 | Sponge network | 0.683 | 1.0E-5 | 0.869 | 0 | 0.433 |
| 15547 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | PDZRN3 | Sponge network | 0.083 | 0.66405 | -1.548 | 0 | 0.433 |
| 15548 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | LATS1 | Sponge network | 0.946 | 0 | 0.109 | 0.34208 | 0.433 |
| 15549 | SNHG16 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326 | 15 | CBFB | Sponge network | 0.961 | 0 | 1.061 | 0 | 0.433 |
| 15550 | RP11-110I1.11 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 13 | PHC3 | Sponge network | 1.034 | 0 | 0.212 | 0.0499 | 0.433 |
| 15551 | CTC-429P9.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-500a-5p | 14 | CRTC1 | Sponge network | 0.082 | 0.55397 | -0.116 | 0.28412 | 0.433 |
| 15552 | CTD-2666L21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p | 16 | ABI2 | Sponge network | 0.368 | 0.20093 | 0.175 | 0.14787 | 0.433 |
| 15553 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-96-5p | 11 | ZYX | Sponge network | 0.997 | 0.00066 | 0.019 | 0.89763 | 0.433 |
| 15554 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-590-3p | 37 | ADARB1 | Sponge network | -0.342 | 0.02288 | -0.335 | 0.06631 | 0.433 |
| 15555 | RP11-554A11.4 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-33a-3p | 12 | FMNL3 | Sponge network | -0.583 | 0.14589 | 0.555 | 4.0E-5 | 0.433 |
| 15556 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 53 | PPM1A | Sponge network | -0.739 | 0.0574 | -0.623 | 0 | 0.433 |
| 15557 | RP11-216B9.6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p | 29 | IL17RD | Sponge network | 0.174 | 0.30034 | -0.019 | 0.94873 | 0.432 |
| 15558 | RP4-584D14.7 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-362-3p | 16 | FAT3 | Sponge network | 1.294 | 0.0031 | -1.601 | 0.00051 | 0.432 |
| 15559 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-582-5p | 15 | RELN | Sponge network | -0.469 | 0.08721 | -2.403 | 0 | 0.432 |
| 15560 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-93-5p | 34 | FAT3 | Sponge network | 0.082 | 0.55397 | -1.601 | 0.00051 | 0.432 |
| 15561 | RP5-890O3.9 |
hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 10 | ABL2 | Sponge network | 0.842 | 0 | 0.82 | 0 | 0.432 |
| 15562 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-93-3p | 29 | QKI | Sponge network | -0.469 | 0.08721 | -0.333 | 0.04111 | 0.432 |
| 15563 | PART1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | CAV1 | Sponge network | -2.925 | 0 | -1.013 | 6.0E-5 | 0.432 |
| 15564 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-96-5p | 12 | FBXO32 | Sponge network | -0.187 | 0.23787 | -0.495 | 0.07561 | 0.432 |
| 15565 | SNHG14 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 23 | RPS6KA2 | Sponge network | -1.111 | 0.0003 | -0.57 | 0.00013 | 0.432 |
| 15566 | LINC00865 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 23 | LIMCH1 | Sponge network | -1.249 | 7.0E-5 | -0.915 | 1.0E-5 | 0.432 |
| 15567 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p | 10 | DYNC1I1 | Sponge network | -0.726 | 0.00132 | -1.12 | 0.00107 | 0.432 |
| 15568 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 15 | RARB | Sponge network | -4.546 | 0 | -0.825 | 2.0E-5 | 0.432 |
| 15569 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PHC3 | Sponge network | 2.094 | 0 | 0.212 | 0.0499 | 0.432 |
| 15570 | RP11-283I3.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 28 | ZFX | Sponge network | 0.614 | 1.0E-5 | 0.09 | 0.42563 | 0.432 |
| 15571 | DNM3OS |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ABCA10 | Sponge network | 0.572 | 0.01722 | -0.571 | 0.03674 | 0.432 |
| 15572 | LINC00654 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 24 | CADM1 | Sponge network | 0.374 | 0.20054 | -0.9 | 0.00084 | 0.432 |
| 15573 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ABCC9 | Sponge network | -0.342 | 0.02288 | -0.466 | 0.14121 | 0.432 |
| 15574 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 41 | NRXN1 | Sponge network | 0.997 | 0.00066 | -3.778 | 0 | 0.432 |
| 15575 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 52 | KIF1B | Sponge network | 0.95 | 0.15943 | -0.118 | 0.32258 | 0.432 |
| 15576 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | SLITRK6 | Sponge network | -0.793 | 7.0E-5 | -2.121 | 0 | 0.432 |
| 15577 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 16 | NR4A3 | Sponge network | -0.563 | 0.02545 | -1.385 | 0 | 0.432 |
| 15578 | RP11-102N12.3 |
hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | LIFR | Sponge network | -0.351 | 0.11358 | -1.794 | 0 | 0.432 |
| 15579 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 40 | DIP2C | Sponge network | 0.064 | 0.72934 | -0.436 | 0.01713 | 0.432 |
| 15580 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p | 25 | PCDH9 | Sponge network | -0.154 | 0.78939 | -1.823 | 4.0E-5 | 0.432 |
| 15581 | RP11-554A11.4 |
hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b | 10 | ITGB1 | Sponge network | -0.583 | 0.14589 | 0.524 | 0.00017 | 0.432 |
| 15582 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | CNTNAP3 | Sponge network | -1.203 | 0.03881 | -1.465 | 0.00033 | 0.432 |
| 15583 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 24 | ERC1 | Sponge network | -0.611 | 0.09607 | -0.108 | 0.38794 | 0.432 |
| 15584 | AC003090.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | TAL1 | Sponge network | -2.117 | 0.00161 | -0.462 | 0.00574 | 0.432 |
| 15585 | RP11-359B12.2 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-574-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | FHL1 | Sponge network | 0.064 | 0.72934 | -2.687 | 0 | 0.432 |
| 15586 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 30 | ESR1 | Sponge network | -0.323 | 0.01372 | -1.035 | 3.0E-5 | 0.432 |
| 15587 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | UBE4B | Sponge network | 0.72 | 0.0001 | -0.235 | 0.007 | 0.432 |
| 15588 | PCA3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 18 | CDH23 | Sponge network | -0.709 | 0.18168 | -0.974 | 0.00034 | 0.432 |
| 15589 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | CNTN1 | Sponge network | 0.395 | 0.15451 | -2.594 | 0 | 0.432 |
| 15590 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p | 20 | EBF3 | Sponge network | -1.161 | 0.00591 | -0.058 | 0.81437 | 0.432 |
| 15591 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 15 | JAZF1 | Sponge network | -1.375 | 0.08642 | -0.377 | 0.02677 | 0.432 |
| 15592 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 15 | FAM172A | Sponge network | -1.216 | 0 | -0.57 | 0 | 0.432 |
| 15593 | AC010226.4 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-590-3p | 11 | TLE4 | Sponge network | -0.811 | 2.0E-5 | -1.157 | 0 | 0.432 |
| 15594 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 30 | CTIF | Sponge network | -2.69 | 0.0001 | -0.389 | 0.00549 | 0.432 |
| 15595 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-424-5p | 14 | PTPN14 | Sponge network | 0.889 | 9.0E-5 | 0.144 | 0.34595 | 0.432 |
| 15596 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | CNTN1 | Sponge network | -0.513 | 0.04703 | -2.594 | 0 | 0.432 |
| 15597 | RP11-144G6.12 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | RBBP6 | Sponge network | 0.826 | 1.0E-5 | 0.163 | 0.05101 | 0.432 |
| 15598 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-1301-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p | 21 | CLASP2 | Sponge network | 1.254 | 0 | -0.038 | 0.71 | 0.432 |
| 15599 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p | 16 | SLC6A15 | Sponge network | -2.62 | 0 | -2.373 | 2.0E-5 | 0.432 |
| 15600 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-500a-5p | 20 | DMD | Sponge network | 0.4 | 0.14611 | -1.506 | 0 | 0.432 |
| 15601 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | MYO9A | Sponge network | 1.601 | 0 | -0.147 | 0.32373 | 0.432 |
| 15602 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | AKAP12 | Sponge network | -1.645 | 0 | -0.932 | 0.0028 | 0.432 |
| 15603 | RP11-894P9.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-940 | 13 | SIRT1 | Sponge network | 0.993 | 0 | 0.109 | 0.2593 | 0.432 |
| 15604 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | HDAC9 | Sponge network | -2.452 | 0 | -0.755 | 0.00495 | 0.432 |
| 15605 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ABCC9 | Sponge network | -1.057 | 0.00029 | -0.466 | 0.14121 | 0.432 |
| 15606 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 33 | ESR1 | Sponge network | 0.194 | 0.64278 | -1.035 | 3.0E-5 | 0.432 |
| 15607 | LINC00957 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-935;hsa-miR-940 | 12 | PHOX2B | Sponge network | -0.421 | 0.0114 | -2.68 | 0 | 0.432 |
| 15608 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-629-3p | 16 | SPTBN4 | Sponge network | -0.469 | 0.08721 | -0.786 | 0.01281 | 0.432 |
| 15609 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 36 | TACC1 | Sponge network | -0.405 | 0.22013 | -0.362 | 0.05406 | 0.432 |
| 15610 | RP11-705C15.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | NIN | Sponge network | 0.796 | 4.0E-5 | -0.253 | 0.13794 | 0.432 |
| 15611 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-590-3p | 16 | GPAM | Sponge network | 1.317 | 0 | 0.142 | 0.29732 | 0.432 |
| 15612 | AC010226.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-629-3p;hsa-miR-7-5p | 22 | IGFBP5 | Sponge network | -0.811 | 2.0E-5 | -0.806 | 0.00105 | 0.432 |
| 15613 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | DCLK1 | Sponge network | 0.214 | 0.18246 | -1.324 | 0.00014 | 0.432 |
| 15614 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | AKAP12 | Sponge network | -0.342 | 0.02288 | -0.932 | 0.0028 | 0.432 |
| 15615 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-93-5p;hsa-miR-940 | 30 | CHD5 | Sponge network | -0.612 | 0.00094 | -0.922 | 0.01521 | 0.432 |
| 15616 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | FAT4 | Sponge network | -0.421 | 0.0114 | -0.354 | 0.14694 | 0.432 |
| 15617 | DNAJC27-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 54 | CADM2 | Sponge network | -0.969 | 2.0E-5 | -3.782 | 0 | 0.432 |
| 15618 | GS1-124K5.11 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-766-3p;hsa-miR-877-5p | 30 | IL17RD | Sponge network | 0.494 | 0.00339 | -0.019 | 0.94873 | 0.432 |
| 15619 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | FOXP1 | Sponge network | 0.101 | 0.60919 | 0.042 | 0.74368 | 0.432 |
| 15620 | GNG12-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-584-5p;hsa-miR-590-3p | 12 | PCDH18 | Sponge network | -0.793 | 7.0E-5 | 0.216 | 0.24645 | 0.432 |
| 15621 | LPP-AS2 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-455-5p | 18 | ITPKB | Sponge network | -0.18 | 0.20343 | -0.704 | 0.00106 | 0.432 |
| 15622 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | PDGFRA | Sponge network | 0.997 | 0.00066 | -0.372 | 0.0037 | 0.432 |
| 15623 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 24 | PCDH10 | Sponge network | -0.563 | 0.02545 | -1.644 | 0.0078 | 0.432 |
| 15624 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-502-5p;hsa-miR-589-5p | 20 | TCF4 | Sponge network | -0.164 | 0.45106 | 0.016 | 0.92271 | 0.432 |
| 15625 | RP11-359E10.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | EPHA7 | Sponge network | 0.299 | 0.57722 | -2.197 | 3.0E-5 | 0.432 |
| 15626 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | SMARCA2 | Sponge network | -0.222 | 0.32445 | -0.529 | 0.00014 | 0.432 |
| 15627 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 23 | PTGFR | Sponge network | -2.46 | 0 | -0.233 | 0.45421 | 0.432 |
| 15628 | RAMP2-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-502-5p;hsa-miR-590-3p | 11 | PTCH1 | Sponge network | -0.513 | 0.04703 | -0.1 | 0.6179 | 0.432 |
| 15629 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-935 | 27 | RAP1A | Sponge network | -0.976 | 0.00025 | -0.929 | 0 | 0.432 |
| 15630 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 46 | ZFHX3 | Sponge network | -2.452 | 0 | 0.389 | 0.01363 | 0.432 |
| 15631 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 30 | NRXN1 | Sponge network | -0.342 | 0.02288 | -3.778 | 0 | 0.432 |
| 15632 | AC016747.3 |
hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p | 14 | MRVI1 | Sponge network | 0.199 | 0.38015 | -1.051 | 0.00022 | 0.432 |
| 15633 | RP1-151F17.2 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | TAF1 | Sponge network | 0.222 | 0.29133 | 0.39 | 3.0E-5 | 0.432 |
| 15634 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-935 | 68 | LPP | Sponge network | -0.421 | 0.0114 | -0.459 | 0.04475 | 0.432 |
| 15635 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 15 | LHX6 | Sponge network | -2.69 | 0.0001 | -0.087 | 0.69193 | 0.432 |
| 15636 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | GPC6 | Sponge network | -0.737 | 0.10822 | -0.054 | 0.81465 | 0.432 |
| 15637 | CTD-2528L19.6 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRIM13 | Sponge network | 0.953 | 0 | 0.183 | 0.06968 | 0.432 |
| 15638 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | RCAN2 | Sponge network | 0.603 | 0.00127 | -0.33 | 0.15172 | 0.432 |
| 15639 | OIP5-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | ITGB1 | Sponge network | 0.421 | 0.00084 | 0.524 | 0.00017 | 0.432 |
| 15640 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 16 | MYO9A | Sponge network | 2.163 | 0 | -0.147 | 0.32373 | 0.432 |
| 15641 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 46 | RUNX1T1 | Sponge network | 0.056 | 0.81195 | -0.344 | 0.2374 | 0.432 |
| 15642 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 43 | ZFHX4 | Sponge network | -0.612 | 0.00094 | 0.018 | 0.95574 | 0.432 |
| 15643 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p | 52 | BMPR2 | Sponge network | 0.659 | 3.0E-5 | 0.206 | 0.0635 | 0.432 |
| 15644 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PBRM1 | Sponge network | 1.063 | 0 | -0.029 | 0.78601 | 0.432 |
| 15645 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 53 | AR | Sponge network | 0.064 | 0.72934 | -1.554 | 0 | 0.432 |
| 15646 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-432-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-889-3p;hsa-miR-93-5p;hsa-miR-942-5p | 50 | UNC80 | Sponge network | -0.615 | 0.00015 | -1.934 | 0 | 0.432 |
| 15647 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-93-5p | 18 | TSHZ3 | Sponge network | -2.039 | 0.03447 | -0.556 | 0.01516 | 0.432 |
| 15648 | RP11-822E23.8 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-484;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-93-5p | 51 | DST | Sponge network | -0.777 | 0.16667 | -0.713 | 0.00096 | 0.432 |
| 15649 | RP11-464F9.1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 19 | UBR3 | Sponge network | 0.959 | 0 | -0.021 | 0.82774 | 0.432 |
| 15650 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | BNC2 | Sponge network | 0.4 | 0.14611 | -0.491 | 0.09988 | 0.432 |
| 15651 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p | 11 | ZNF208 | Sponge network | 0.4 | 0.14611 | -0.4 | 0.22381 | 0.432 |
| 15652 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-744-3p | 23 | MEF2C | Sponge network | 0.178 | 0.29106 | -0.499 | 0.01448 | 0.432 |
| 15653 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 23 | HIP1 | Sponge network | -0.576 | 0.10121 | 0.774 | 0 | 0.432 |
| 15654 | LINC00957 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | PTGFR | Sponge network | -0.421 | 0.0114 | -0.233 | 0.45421 | 0.432 |
| 15655 | H1FX-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-590-3p | 11 | MAPK10 | Sponge network | -0.206 | 0.12942 | -1.774 | 0 | 0.432 |
| 15656 | MIR143HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 33 | NCOA1 | Sponge network | -0.739 | 0.0574 | -0.432 | 0 | 0.432 |
| 15657 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | 0.082 | 0.55654 | 0.273 | 0.09957 | 0.432 |
| 15658 | RP11-540B6.6 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 18 | UBR3 | Sponge network | 0.93 | 0 | -0.021 | 0.82774 | 0.432 |
| 15659 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | RIMBP2 | Sponge network | -0.739 | 0.00044 | -1.968 | 5.0E-5 | 0.432 |
| 15660 | LINC00654 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 13 | DNMT3A | Sponge network | 0.374 | 0.20054 | 0.302 | 0.0262 | 0.432 |
| 15661 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 13 | EDA2R | Sponge network | 0.194 | 0.64278 | -0.587 | 0.01598 | 0.432 |
| 15662 | AC009948.5 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-486-5p;hsa-miR-495-3p | 17 | SYNM | Sponge network | 0.532 | 0.00505 | -2.309 | 1.0E-5 | 0.432 |
| 15663 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | RASSF2 | Sponge network | 0.249 | 0.35902 | -0.273 | 0.21961 | 0.432 |
| 15664 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 22 | RUNX1T1 | Sponge network | -1.116 | 0.23686 | -0.344 | 0.2374 | 0.432 |
| 15665 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 11 | GLI3 | Sponge network | 0.214 | 0.18246 | -0.615 | 0.01482 | 0.432 |
| 15666 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | SLITRK6 | Sponge network | -2.46 | 0 | -2.121 | 0 | 0.432 |
| 15667 | PART1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | FLNA | Sponge network | -2.925 | 0 | -0.771 | 0.01377 | 0.432 |
| 15668 | AC010226.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 14 | PDE4DIP | Sponge network | -0.811 | 2.0E-5 | -0.282 | 0.0257 | 0.432 |
| 15669 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | RCAN2 | Sponge network | 0.219 | 0.1516 | -0.33 | 0.15172 | 0.432 |
| 15670 | RP11-244O19.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-577;hsa-miR-590-3p | 14 | PIK3CA | Sponge network | -0.096 | 0.67958 | 0.195 | 0.06908 | 0.432 |
| 15671 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 23 | NTRK3 | Sponge network | -1.375 | 0.08642 | -1.77 | 3.0E-5 | 0.432 |
| 15672 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | CBL | Sponge network | 0.796 | 4.0E-5 | 0.291 | 0.01267 | 0.432 |
| 15673 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | RASSF2 | Sponge network | -0.421 | 0.0114 | -0.273 | 0.21961 | 0.432 |
| 15674 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-744-3p | 21 | HIP1 | Sponge network | -0.513 | 0.04703 | 0.774 | 0 | 0.432 |
| 15675 | RP11-166D19.1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | RASSF3 | Sponge network | -0.576 | 0.10121 | -0.226 | 0.11691 | 0.432 |
| 15676 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 22 | FBXO32 | Sponge network | 0.381 | 0.25967 | -0.495 | 0.07561 | 0.432 |
| 15677 | GNG12-AS1 |
hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | STAT5B | Sponge network | -0.793 | 7.0E-5 | -0.182 | 0.1082 | 0.432 |
| 15678 | JAZF1-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 15 | EHD3 | Sponge network | -0.769 | 0.00035 | -0.484 | 0.01747 | 0.432 |
| 15679 | AP001055.6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | FAT4 | Sponge network | 0.693 | 0.00076 | -0.354 | 0.14694 | 0.432 |
| 15680 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p | 11 | CHD1 | Sponge network | 0.946 | 0 | 0.322 | 0.00215 | 0.432 |
| 15681 | RP11-110I1.11 |
hsa-let-7d-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-590-3p | 11 | USP6 | Sponge network | 1.034 | 0 | 0.188 | 0.3871 | 0.431 |
| 15682 | GS1-124K5.11 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-590-3p | 12 | STARD13 | Sponge network | 0.494 | 0.00339 | -0.187 | 0.27383 | 0.431 |
| 15683 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | CACNA2D1 | Sponge network | -0.811 | 2.0E-5 | -0.846 | 0.00675 | 0.431 |
| 15684 | LINC00702 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-625-5p | 10 | MPL | Sponge network | -0.737 | 0.06182 | -0.624 | 0.00133 | 0.431 |
| 15685 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p | 16 | ABCA10 | Sponge network | -0.976 | 0.00025 | -0.571 | 0.03674 | 0.431 |
| 15686 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-590-3p | 18 | RAP1A | Sponge network | -0.811 | 2.0E-5 | -0.929 | 0 | 0.431 |
| 15687 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p | 24 | ZFHX3 | Sponge network | -0.469 | 0.08721 | 0.389 | 0.01363 | 0.431 |
| 15688 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 24 | LATS2 | Sponge network | -1.697 | 0.00889 | 0.159 | 0.2304 | 0.431 |
| 15689 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-577;hsa-miR-93-5p | 14 | MAP3K12 | Sponge network | -1.439 | 4.0E-5 | -0.057 | 0.59369 | 0.431 |
| 15690 | ARHGEF26-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NRG3 | Sponge network | -2.46 | 0 | -1.836 | 4.0E-5 | 0.431 |
| 15691 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-455-5p | 13 | TLL1 | Sponge network | -0.162 | 0.47687 | -1.195 | 3.0E-5 | 0.431 |
| 15692 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-744-3p | 24 | PTPN14 | Sponge network | 0.61 | 0.01593 | 0.144 | 0.34595 | 0.431 |
| 15693 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-576-5p | 27 | PBX1 | Sponge network | -0.13 | 0.48734 | -1.206 | 0 | 0.431 |
| 15694 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | PNRC1 | Sponge network | -0.896 | 0.00099 | -0.646 | 0 | 0.431 |
| 15695 | CTD-2184D3.6 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | ATM | Sponge network | 0.272 | 0.44692 | 0.178 | 0.21568 | 0.431 |
| 15696 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 42 | CHD5 | Sponge network | -0.899 | 6.0E-5 | -0.922 | 0.01521 | 0.431 |
| 15697 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 10 | TRIM33 | Sponge network | 0.917 | 0 | 0.402 | 7.0E-5 | 0.431 |
| 15698 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | PCDH10 | Sponge network | -0.899 | 6.0E-5 | -1.644 | 0.0078 | 0.431 |
| 15699 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 11 | PREX2 | Sponge network | -1.349 | 0.00017 | -0.272 | 0.25382 | 0.431 |
| 15700 | DNM3OS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-96-5p | 12 | ELMO1 | Sponge network | 0.572 | 0.01722 | 0.04 | 0.83147 | 0.431 |
| 15701 | RP11-693J15.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-671-5p | 10 | CDH23 | Sponge network | -0.72 | 0.24239 | -0.974 | 0.00034 | 0.431 |
| 15702 | LINC00657 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p | 12 | CDK6 | Sponge network | 0.91 | 0 | 0.991 | 0 | 0.431 |
| 15703 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 39 | PHC3 | Sponge network | 0.064 | 0.72934 | 0.212 | 0.0499 | 0.431 |
| 15704 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | ITGAV | Sponge network | 0.997 | 0.00066 | 0.337 | 0.01002 | 0.431 |
| 15705 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-935 | 22 | RARB | Sponge network | -0.942 | 0 | -0.825 | 2.0E-5 | 0.431 |
| 15706 | LINC00476 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | ZNF382 | Sponge network | -0.615 | 0.00015 | -0.494 | 0.03831 | 0.431 |
| 15707 | FENDRR |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | PIK3R1 | Sponge network | -1.281 | 0.00012 | -0.133 | 0.36854 | 0.431 |
| 15708 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 19 | SLC5A3 | Sponge network | 1.153 | 0 | 0.493 | 5.0E-5 | 0.431 |
| 15709 | LINC00957 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 30 | MEF2C | Sponge network | -0.421 | 0.0114 | -0.499 | 0.01448 | 0.431 |
| 15710 | CTC-429P9.3 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-940 | 12 | BAZ2B | Sponge network | 0.082 | 0.55397 | -0.276 | 0.02833 | 0.431 |
| 15711 | KB-1460A1.5 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | ITGB1 | Sponge network | 0.603 | 0.00127 | 0.524 | 0.00017 | 0.431 |
| 15712 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-576-5p | 14 | GREM1 | Sponge network | 0.176 | 0.37402 | 0.902 | 0.01691 | 0.431 |
| 15713 | CTC-296K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-940 | 19 | SNX29 | Sponge network | -2.095 | 0.00112 | -0.34 | 0.01371 | 0.431 |
| 15714 | RP11-216B9.6 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | PRUNE2 | Sponge network | 0.174 | 0.30034 | -1.733 | 0.00022 | 0.431 |
| 15715 | RP11-536O18.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-502-5p | 16 | NTRK3 | Sponge network | -0.074 | 0.90758 | -1.77 | 3.0E-5 | 0.431 |
| 15716 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 30 | PIK3R1 | Sponge network | 0.659 | 3.0E-5 | -0.133 | 0.36854 | 0.431 |
| 15717 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 22 | EBF1 | Sponge network | -0.318 | 0.65847 | -0.384 | 0.08856 | 0.431 |
| 15718 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | PRDM5 | Sponge network | -0.487 | 0.02157 | -1.09 | 0.00014 | 0.431 |
| 15719 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | NCAM2 | Sponge network | -2.117 | 0.00161 | -0.482 | 0.22565 | 0.431 |
| 15720 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-590-3p | 12 | GLIPR1 | Sponge network | -0.739 | 0.00044 | 0.146 | 0.3276 | 0.431 |
| 15721 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-424-5p | 12 | CRTC3 | Sponge network | 0.178 | 0.29106 | 0.164 | 0.16786 | 0.431 |
| 15722 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | LRRC7 | Sponge network | -0.901 | 0.00019 | -1.341 | 0.01193 | 0.431 |
| 15723 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374b-5p | 17 | ABI2 | Sponge network | 1.46 | 1.0E-5 | 0.175 | 0.14787 | 0.431 |
| 15724 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RCAN2 | Sponge network | -0.053 | 0.80685 | -0.33 | 0.15172 | 0.431 |
| 15725 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 23 | RASSF2 | Sponge network | 0.889 | 9.0E-5 | -0.273 | 0.21961 | 0.431 |
| 15726 | AC009948.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 16 | EZH1 | Sponge network | 0.532 | 0.00505 | -0.564 | 1.0E-5 | 0.431 |
| 15727 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-93-5p | 48 | CADM2 | Sponge network | -1.645 | 0 | -3.782 | 0 | 0.431 |
| 15728 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 21 | KIF1B | Sponge network | 0.537 | 0.0006 | -0.118 | 0.32258 | 0.431 |
| 15729 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 14 | CACNA2D1 | Sponge network | -0.517 | 0.02935 | -0.846 | 0.00675 | 0.431 |
| 15730 | CTD-2314G24.2 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | SFRP1 | Sponge network | 0.246 | 0.59912 | -3.566 | 0 | 0.431 |
| 15731 | TP73-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-92a-3p | 15 | ITGA5 | Sponge network | -0.563 | 0.02545 | 0.452 | 0.03524 | 0.431 |
| 15732 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SETBP1 | Sponge network | -0.206 | 0.12942 | -1.302 | 1.0E-5 | 0.431 |
| 15733 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | TACC1 | Sponge network | 0.385 | 0.10067 | -0.362 | 0.05406 | 0.431 |
| 15734 | MALAT1 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | WAC | Sponge network | 0.782 | 0.00016 | -0.054 | 0.43688 | 0.431 |
| 15735 | AC006116.24 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-539-5p | 19 | ABI2 | Sponge network | -0.473 | 0.07602 | 0.175 | 0.14787 | 0.431 |
| 15736 | TPTEP1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | ROBO1 | Sponge network | -1.216 | 0 | -0.009 | 0.96154 | 0.431 |
| 15737 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 35 | PRKAB2 | Sponge network | -1.575 | 6.0E-5 | -0.367 | 0.01371 | 0.431 |
| 15738 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 36 | CLASP2 | Sponge network | -1.575 | 6.0E-5 | -0.038 | 0.71 | 0.431 |
| 15739 | CTD-2089N3.2 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | ESR1 | Sponge network | -1.375 | 0.08642 | -1.035 | 3.0E-5 | 0.431 |
| 15740 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p | 23 | RASSF2 | Sponge network | -4.546 | 0 | -0.273 | 0.21961 | 0.431 |
| 15741 | PCA3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-96-5p | 19 | RIMBP2 | Sponge network | -0.709 | 0.18168 | -1.968 | 5.0E-5 | 0.431 |
| 15742 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 42 | PIK3R1 | Sponge network | -0.355 | 0.0077 | -0.133 | 0.36854 | 0.431 |
| 15743 | PART1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 34 | SETBP1 | Sponge network | -2.925 | 0 | -1.302 | 1.0E-5 | 0.431 |
| 15744 | GAS6-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p | 13 | EZH1 | Sponge network | 0.16 | 0.50785 | -0.564 | 1.0E-5 | 0.431 |
| 15745 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-92a-1-5p | 14 | SNX29 | Sponge network | -0.309 | 0.1145 | -0.34 | 0.01371 | 0.431 |
| 15746 | LINC00472 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-550a-5p;hsa-miR-7-1-3p | 11 | PCDH18 | Sponge network | -0.842 | 0.01361 | 0.216 | 0.24645 | 0.431 |
| 15747 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | MAF | Sponge network | -0.737 | 0.06182 | -1.257 | 0 | 0.431 |
| 15748 | RP11-506M13.3 |
hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 16 | RICTOR | Sponge network | 0.769 | 0 | 0.388 | 0.00188 | 0.431 |
| 15749 | MIR143HG |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | SYT4 | Sponge network | -0.739 | 0.0574 | -3.207 | 0 | 0.431 |
| 15750 | PART1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 13 | PRKD1 | Sponge network | -2.925 | 0 | -0.466 | 0.03583 | 0.431 |
| 15751 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-92a-3p | 15 | ZNF521 | Sponge network | 1.302 | 7.0E-5 | -0.111 | 0.58068 | 0.431 |
| 15752 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | PCDH9 | Sponge network | 0.064 | 0.72934 | -1.823 | 4.0E-5 | 0.431 |
| 15753 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | CRTC1 | Sponge network | -0.096 | 0.67958 | -0.116 | 0.28412 | 0.431 |
| 15754 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-409-3p | 22 | TGFBR3 | Sponge network | -0.309 | 0.1145 | -1.173 | 1.0E-5 | 0.431 |
| 15755 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 21 | ANK2 | Sponge network | 0.199 | 0.38015 | -1.603 | 1.0E-5 | 0.431 |
| 15756 | RP11-119F7.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-92a-3p | 51 | DST | Sponge network | 0.225 | 0.30644 | -0.713 | 0.00096 | 0.431 |
| 15757 | LINC00476 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-335-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 16 | PEG3 | Sponge network | -0.615 | 0.00015 | -1.74 | 0 | 0.431 |
| 15758 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p | 17 | RICTOR | Sponge network | 0.497 | 0.01451 | 0.388 | 0.00188 | 0.431 |
| 15759 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p | 13 | RET | Sponge network | -1.057 | 0.00029 | -0.833 | 0.03517 | 0.431 |
| 15760 | RP4-717I23.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-576-5p;hsa-miR-629-3p | 10 | RBL2 | Sponge network | 0.637 | 0.00069 | -0.282 | 0.00254 | 0.431 |
| 15761 | FENDRR |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | AFF1 | Sponge network | -1.281 | 0.00012 | -0.384 | 0.00053 | 0.431 |
| 15762 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | PDGFRA | Sponge network | -1.575 | 6.0E-5 | -0.372 | 0.0037 | 0.431 |
| 15763 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 54 | KIF1B | Sponge network | 0.537 | 0.00139 | -0.118 | 0.32258 | 0.431 |
| 15764 | AC004893.11 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | TAF1 | Sponge network | 1.317 | 0 | 0.39 | 3.0E-5 | 0.431 |
| 15765 | OIP5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | EXOC8 | Sponge network | 0.421 | 0.00084 | 0.216 | 0.00598 | 0.431 |
| 15766 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 15 | ABCC9 | Sponge network | 0.494 | 0.00339 | -0.466 | 0.14121 | 0.431 |
| 15767 | LINC00702 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 41 | ABI2 | Sponge network | -0.737 | 0.06182 | 0.175 | 0.14787 | 0.431 |
| 15768 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 20 | RASSF2 | Sponge network | -2.095 | 0.00112 | -0.273 | 0.21961 | 0.431 |
| 15769 | RP11-758N13.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-375;hsa-miR-455-3p | 11 | TSHZ3 | Sponge network | 2.939 | 3.0E-5 | -0.556 | 0.01516 | 0.431 |
| 15770 | AC083843.1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p | 12 | STAG2 | Sponge network | 1.4 | 0 | 0.252 | 0.00438 | 0.431 |
| 15771 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-93-5p | 28 | EPHA5 | Sponge network | 0.082 | 0.55397 | -0.957 | 0.01733 | 0.431 |
| 15772 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p | 31 | ESR1 | Sponge network | -2.117 | 0.00161 | -1.035 | 3.0E-5 | 0.431 |
| 15773 | RP11-344B5.2 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b | 10 | IGF1 | Sponge network | -0.055 | 0.88482 | -0.204 | 0.5898 | 0.431 |
| 15774 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-935 | 10 | DAB2 | Sponge network | -0.896 | 0.00099 | 0.431 | 0.0113 | 0.431 |
| 15775 | RP11-855A2.3 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 16 | ABL2 | Sponge network | 2.094 | 0 | 0.82 | 0 | 0.431 |
| 15776 | RP11-2B6.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | 0.539 | 0.03722 | 0.142 | 0.04938 | 0.431 |
| 15777 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-93-5p | 14 | TXNIP | Sponge network | -0.842 | 0.01361 | -1.277 | 0 | 0.431 |
| 15778 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 15 | CREBBP | Sponge network | -0.096 | 0.67958 | 0.126 | 0.15186 | 0.431 |
| 15779 | LINC00957 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-942-5p | 13 | CREBBP | Sponge network | -0.421 | 0.0114 | 0.126 | 0.15186 | 0.431 |
| 15780 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 15 | CLIP1 | Sponge network | -0.096 | 0.67958 | -0.215 | 0.08684 | 0.431 |
| 15781 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-7-5p | 10 | PDGFC | Sponge network | 0.176 | 0.37402 | -0.33 | 0.06483 | 0.431 |
| 15782 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 29 | EBF3 | Sponge network | -2.46 | 0 | -0.058 | 0.81437 | 0.431 |
| 15783 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | -2.69 | 0.0001 | -1.751 | 1.0E-5 | 0.431 |
| 15784 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3913-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 39 | HLF | Sponge network | -0.769 | 0.00035 | -2.443 | 0 | 0.431 |
| 15785 | CTC-296K1.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-331-3p | 15 | IGFBP5 | Sponge network | -2.076 | 0.00548 | -0.806 | 0.00105 | 0.431 |
| 15786 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p | 13 | FBXO32 | Sponge network | 0.959 | 0 | -0.495 | 0.07561 | 0.431 |
| 15787 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-628-5p | 18 | BCL6 | Sponge network | -2.62 | 0 | -0.211 | 0.19489 | 0.431 |
| 15788 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-5p;hsa-miR-582-5p | 29 | FBXO32 | Sponge network | 0.056 | 0.81195 | -0.495 | 0.07561 | 0.431 |
| 15789 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p | 17 | TBL1XR1 | Sponge network | 0.702 | 7.0E-5 | 0.578 | 0 | 0.431 |
| 15790 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | SYNE1 | Sponge network | -0.942 | 0 | -0.738 | 0.00316 | 0.431 |
| 15791 | AP001347.6 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | GJA1 | Sponge network | -1.161 | 0.00591 | -0.485 | 0.02179 | 0.431 |
| 15792 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | ATRX | Sponge network | 1.11 | 0 | 0.261 | 0.04408 | 0.431 |
| 15793 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | HECA | Sponge network | 0.673 | 0 | -0.217 | 0.03463 | 0.431 |
| 15794 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 23 | TGFBR2 | Sponge network | -0.576 | 0.10121 | -0.243 | 0.11408 | 0.431 |
| 15795 | CTC-457L16.2 |
hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-7-1-3p | 13 | BCL2 | Sponge network | 1.153 | 0.00114 | -0.899 | 0 | 0.431 |
| 15796 | A2M-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | PRUNE2 | Sponge network | -0.08 | 0.90092 | -1.733 | 0.00022 | 0.431 |
| 15797 | SERTAD4-AS1 |
hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-500a-5p | 13 | RYR2 | Sponge network | -0.932 | 0.00329 | -1.466 | 0.00031 | 0.431 |
| 15798 | RP11-216B9.6 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 12 | STARD13 | Sponge network | 0.174 | 0.30034 | -0.187 | 0.27383 | 0.431 |
| 15799 | LINC00641 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-362-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 41 | ARHGEF12 | Sponge network | -0.222 | 0.32445 | 0.09 | 0.35693 | 0.431 |
| 15800 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 19 | CNTNAP3 | Sponge network | -0.793 | 7.0E-5 | -1.465 | 0.00033 | 0.431 |
| 15801 | LINC00654 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 54 | AR | Sponge network | 0.374 | 0.20054 | -1.554 | 0 | 0.431 |
| 15802 | RP11-35G9.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | NAV3 | Sponge network | 0.657 | 4.0E-5 | 0.212 | 0.47729 | 0.431 |
| 15803 | ADAMTS9-AS2 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | INPP4A | Sponge network | -2.452 | 0 | 0.07 | 0.53563 | 0.431 |
| 15804 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 54 | GAS7 | Sponge network | -2.46 | 0 | -0.555 | 0.01602 | 0.431 |
| 15805 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429 | 17 | LRIG1 | Sponge network | -0.469 | 0.08721 | -0.537 | 0.00588 | 0.431 |
| 15806 | HAND2-AS1 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | RIMS2 | Sponge network | -1.697 | 0.00889 | -1.365 | 0.00208 | 0.431 |
| 15807 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | TRIP11 | Sponge network | 0.782 | 0.00016 | -0.158 | 0.06384 | 0.43 |
| 15808 | A2M-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-590-3p | 22 | DLC1 | Sponge network | -0.08 | 0.90092 | -0.382 | 0.05038 | 0.43 |
| 15809 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-532-5p | 17 | ZMYND11 | Sponge network | 0.082 | 0.55397 | -0.2 | 0.05449 | 0.43 |
| 15810 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SLC6A15 | Sponge network | -1.161 | 0.00591 | -2.373 | 2.0E-5 | 0.43 |
| 15811 | FAM66C |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 17 | CITED2 | Sponge network | -0.901 | 0.00019 | -1.545 | 0 | 0.43 |
| 15812 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p | 15 | TRIM33 | Sponge network | 0.594 | 0.00064 | 0.402 | 7.0E-5 | 0.43 |
| 15813 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 24 | PCDH10 | Sponge network | 0.056 | 0.81195 | -1.644 | 0.0078 | 0.43 |
| 15814 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 35 | EPHA3 | Sponge network | -0.899 | 6.0E-5 | 0.185 | 0.53588 | 0.43 |
| 15815 | FZD10-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935 | 13 | MAPK10 | Sponge network | -0.727 | 0.04726 | -1.774 | 0 | 0.43 |
| 15816 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 20 | NIN | Sponge network | -1.092 | 4.0E-5 | -0.253 | 0.13794 | 0.43 |
| 15817 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | FGFR1 | Sponge network | 0.225 | 0.30644 | -0.509 | 0.03957 | 0.43 |
| 15818 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-93-3p | 25 | NCOA1 | Sponge network | -1.645 | 0 | -0.432 | 0 | 0.43 |
| 15819 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p | 27 | SCN9A | Sponge network | -0.303 | 0.21002 | -0.525 | 0.10593 | 0.43 |
| 15820 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | AHNAK | Sponge network | -0.901 | 0.00019 | -1.207 | 0 | 0.43 |
| 15821 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-625-5p;hsa-miR-93-5p | 12 | LHX6 | Sponge network | -1.349 | 0.00017 | -0.087 | 0.69193 | 0.43 |
| 15822 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-424-3p;hsa-miR-577;hsa-miR-628-5p | 27 | BMPR2 | Sponge network | 0.811 | 0.03228 | 0.206 | 0.0635 | 0.43 |
| 15823 | RP11-395G23.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-93-5p | 24 | IL17RD | Sponge network | 0.381 | 0.25967 | -0.019 | 0.94873 | 0.43 |
| 15824 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-935;hsa-miR-96-5p | 29 | FBXO32 | Sponge network | -0.563 | 0.02545 | -0.495 | 0.07561 | 0.43 |
| 15825 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | JAZF1 | Sponge network | -1.216 | 0 | -0.377 | 0.02677 | 0.43 |
| 15826 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | LRRC4 | Sponge network | -0.129 | 0.57177 | -1.774 | 0 | 0.43 |
| 15827 | RP11-2H8.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | 1.089 | 0 | 0.142 | 0.04938 | 0.43 |
| 15828 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-25-3p | 10 | DKK3 | Sponge network | -0.376 | 0.00337 | -0.052 | 0.77783 | 0.43 |
| 15829 | RP1-151F17.2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 34 | IL17RD | Sponge network | 0.222 | 0.29133 | -0.019 | 0.94873 | 0.43 |
| 15830 | MAGI2-AS3 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p | 22 | IRS1 | Sponge network | -0.129 | 0.57177 | -0.026 | 0.88855 | 0.43 |
| 15831 | LINC00657 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CEP170 | Sponge network | 0.91 | 0 | 0.943 | 0 | 0.43 |
| 15832 | RP11-254F7.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-5p | 43 | DST | Sponge network | 0.176 | 0.37402 | -0.713 | 0.00096 | 0.43 |
| 15833 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CACNA2D1 | Sponge network | -1.161 | 0.00591 | -0.846 | 0.00675 | 0.43 |
| 15834 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 55 | BMPR2 | Sponge network | 0.335 | 0.20596 | 0.206 | 0.0635 | 0.43 |
| 15835 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p | 18 | IGF2 | Sponge network | 0.853 | 0.15352 | 0.848 | 0.03842 | 0.43 |
| 15836 | LINC00865 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-5p | 15 | ROBO1 | Sponge network | -1.249 | 7.0E-5 | -0.009 | 0.96154 | 0.43 |
| 15837 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ADAMTSL3 | Sponge network | -0.021 | 0.93598 | -1.559 | 0 | 0.43 |
| 15838 | RP11-326G21.1 |
hsa-let-7b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | DMD | Sponge network | -0.021 | 0.93598 | -1.506 | 0 | 0.43 |
| 15839 | RASSF8-AS1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-455-3p;hsa-miR-484 | 18 | ABL1 | Sponge network | 0.335 | 0.20596 | -0.215 | 0.07804 | 0.43 |
| 15840 | RP11-967K21.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 24 | DLC1 | Sponge network | 0.056 | 0.81195 | -0.382 | 0.05038 | 0.43 |
| 15841 | NEAT1 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 30 | ABL2 | Sponge network | 0.705 | 0.00014 | 0.82 | 0 | 0.43 |
| 15842 | EMX2OS |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | ID4 | Sponge network | -0.777 | 0.1134 | -1.644 | 0 | 0.43 |
| 15843 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-5p | 16 | FREM2 | Sponge network | 0.41 | 0.02214 | -0.437 | 0.46353 | 0.43 |
| 15844 | USP3-AS1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-589-3p;hsa-miR-889-3p | 20 | RPS6KA6 | Sponge network | -0.162 | 0.47687 | -2.989 | 0 | 0.43 |
| 15845 | RP11-532F6.3 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 14 | FMNL3 | Sponge network | -0.896 | 0.00099 | 0.555 | 4.0E-5 | 0.43 |
| 15846 | H1FX-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-625-5p | 10 | NGFR | Sponge network | -0.206 | 0.12942 | -1.271 | 0.01058 | 0.43 |
| 15847 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p | 16 | EBF1 | Sponge network | -0.932 | 0.00329 | -0.384 | 0.08856 | 0.43 |
| 15848 | LINC00672 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | PCDH9 | Sponge network | -0.227 | 0.31657 | -1.823 | 4.0E-5 | 0.43 |
| 15849 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | LRRC55 | Sponge network | -1.697 | 0.00889 | -0.026 | 0.93163 | 0.43 |
| 15850 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 24 | DDX6 | Sponge network | 0.817 | 3.0E-5 | 0.091 | 0.36428 | 0.43 |
| 15851 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 38 | TSHZ3 | Sponge network | -0.222 | 0.32445 | -0.556 | 0.01516 | 0.43 |
| 15852 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-629-5p | 20 | GRIN2A | Sponge network | -0.154 | 0.78939 | -2.161 | 0 | 0.43 |
| 15853 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-5p | 11 | LRRK2 | Sponge network | -1.645 | 0 | -1.136 | 1.0E-5 | 0.43 |
| 15854 | ADAMTS9-AS2 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-584-5p;hsa-miR-942-5p | 11 | BMPR1A | Sponge network | -2.452 | 0 | -0.366 | 0.01036 | 0.43 |
| 15855 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p | 18 | RET | Sponge network | -0.421 | 0.0114 | -0.833 | 0.03517 | 0.43 |
| 15856 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | CNTN1 | Sponge network | 0.16 | 0.50785 | -2.594 | 0 | 0.43 |
| 15857 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | TAL1 | Sponge network | -1.349 | 0.00017 | -0.462 | 0.00574 | 0.43 |
| 15858 | LINC00654 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-877-5p | 24 | NRK | Sponge network | 0.374 | 0.20054 | 0.656 | 0.19217 | 0.43 |
| 15859 | AC005562.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 14 | CEP170 | Sponge network | 0.488 | 0.00084 | 0.943 | 0 | 0.43 |
| 15860 | DIO3OS |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-582-5p | 10 | ZNF208 | Sponge network | -0.773 | 0.04073 | -0.4 | 0.22381 | 0.43 |
| 15861 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p | 19 | LRP1B | Sponge network | -0.358 | 0.17255 | -2.163 | 0 | 0.43 |
| 15862 | RP11-195F19.9 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 11 | SFRP1 | Sponge network | -0.726 | 0.00132 | -3.566 | 0 | 0.43 |
| 15863 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 62 | IL6ST | Sponge network | -0.612 | 0.00094 | -0.402 | 0.00676 | 0.43 |
| 15864 | RP11-403I13.7 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 25 | NAV3 | Sponge network | 0.395 | 0.15451 | 0.212 | 0.47729 | 0.43 |
| 15865 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | ADAMTSL3 | Sponge network | 0.4 | 0.14611 | -1.559 | 0 | 0.43 |
| 15866 | HOTAIRM1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | DYNC1I1 | Sponge network | -0.053 | 0.80685 | -1.12 | 0.00107 | 0.43 |
| 15867 | EMX2OS |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CTGF | Sponge network | -0.777 | 0.1134 | 0.321 | 0.11682 | 0.43 |
| 15868 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-7-1-3p | 15 | ADAMTSL3 | Sponge network | -0.72 | 0.24239 | -1.559 | 0 | 0.43 |
| 15869 | CTC-444N24.11 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TSC1 | Sponge network | 1.153 | 0 | 0.103 | 0.26071 | 0.43 |
| 15870 | LINC00654 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 21 | AKAP6 | Sponge network | 0.374 | 0.20054 | -1.586 | 3.0E-5 | 0.43 |
| 15871 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-146b-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-324-5p | 12 | DMD | Sponge network | -7.551 | 0 | -1.506 | 0 | 0.43 |
| 15872 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-629-3p | 14 | ITGA5 | Sponge network | -0.777 | 0.16667 | 0.452 | 0.03524 | 0.43 |
| 15873 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 26 | PGR | Sponge network | -0.513 | 0.04703 | -1.704 | 0 | 0.43 |
| 15874 | AC108488.4 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p | 10 | DNMT3A | Sponge network | 0.259 | 0.12542 | 0.302 | 0.0262 | 0.43 |
| 15875 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-93-3p | 16 | RPS6KA2 | Sponge network | -0.154 | 0.53954 | -0.57 | 0.00013 | 0.43 |
| 15876 | AC003090.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | MED12L | Sponge network | -2.117 | 0.00161 | -0.194 | 0.55299 | 0.43 |
| 15877 | LINC00920 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-486-5p | 12 | CBFB | Sponge network | 1.674 | 0 | 1.061 | 0 | 0.43 |
| 15878 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | AFF3 | Sponge network | -0.513 | 0.04703 | -2.373 | 0 | 0.43 |
| 15879 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | AHNAK | Sponge network | -0.49 | 0.04946 | -1.207 | 0 | 0.43 |
| 15880 | HCG11 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | DOCK4 | Sponge network | 0.385 | 0.10067 | 0.502 | 0.00108 | 0.43 |
| 15881 | RP11-384O8.1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | RNF180 | Sponge network | -0.738 | 0.21233 | -1.127 | 1.0E-5 | 0.43 |
| 15882 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | NF1 | Sponge network | 0.643 | 0 | 0.51 | 0 | 0.43 |
| 15883 | AP001347.6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p | 10 | THSD7A | Sponge network | -1.161 | 0.00591 | 0.242 | 0.25154 | 0.43 |
| 15884 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-940 | 22 | CHD5 | Sponge network | 0.225 | 0.30644 | -0.922 | 0.01521 | 0.43 |
| 15885 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-340-5p;hsa-miR-542-3p | 16 | DYNC1I1 | Sponge network | -0.384 | 0.40652 | -1.12 | 0.00107 | 0.43 |
| 15886 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-5p | 12 | MN1 | Sponge network | 0.246 | 0.59912 | -0.358 | 0.24172 | 0.43 |
| 15887 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-5p | 18 | NEDD4 | Sponge network | -0.384 | 0.40652 | 0.02 | 0.89834 | 0.43 |
| 15888 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | TGFBR3 | Sponge network | -1.161 | 0.00591 | -1.173 | 1.0E-5 | 0.43 |
| 15889 | RP11-589P10.7 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-18a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p | 11 | HLF | Sponge network | -2.044 | 0.00066 | -2.443 | 0 | 0.43 |
| 15890 | RP11-532F6.3 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-550a-3p | 12 | RASSF3 | Sponge network | -0.896 | 0.00099 | -0.226 | 0.11691 | 0.43 |
| 15891 | FENDRR |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 42 | BCL2 | Sponge network | -1.281 | 0.00012 | -0.899 | 0 | 0.43 |
| 15892 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | MOB1B | Sponge network | -0.323 | 0.01372 | 0.197 | 0.15266 | 0.43 |
| 15893 | CTC-444N24.11 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 32 | CCDC6 | Sponge network | 1.153 | 0 | 0.27 | 0.02555 | 0.43 |
| 15894 | LINC00957 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 15 | NCAM2 | Sponge network | -0.421 | 0.0114 | -0.482 | 0.22565 | 0.43 |
| 15895 | CTA-14H9.5 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | ZFHX4 | Sponge network | 0.145 | 0.53343 | 0.018 | 0.95574 | 0.43 |
| 15896 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 20 | LRRC4 | Sponge network | -0.576 | 0.10121 | -1.774 | 0 | 0.43 |
| 15897 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 26 | DMXL1 | Sponge network | -0.83 | 0 | -0.426 | 0.00056 | 0.43 |
| 15898 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | LRRC55 | Sponge network | -0.323 | 0.01372 | -0.026 | 0.93163 | 0.43 |
| 15899 | RASSF8-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 16 | INPP4A | Sponge network | 0.335 | 0.20596 | 0.07 | 0.53563 | 0.43 |
| 15900 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326 | 16 | HIP1 | Sponge network | -0.154 | 0.53954 | 0.774 | 0 | 0.43 |
| 15901 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | EDA2R | Sponge network | -0.376 | 0.00337 | -0.587 | 0.01598 | 0.43 |
| 15902 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p | 14 | SLITRK6 | Sponge network | -1.439 | 4.0E-5 | -2.121 | 0 | 0.43 |
| 15903 | PWAR6 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | CITED2 | Sponge network | -1.575 | 6.0E-5 | -1.545 | 0 | 0.43 |
| 15904 | RP5-994D16.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PRKAB2 | Sponge network | 0.252 | 0.08508 | -0.367 | 0.01371 | 0.43 |
| 15905 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | MCC | Sponge network | -0.72 | 0.24239 | -1.005 | 0 | 0.43 |
| 15906 | RP11-400K9.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-744-3p | 21 | LPP | Sponge network | -0.733 | 0.19469 | -0.459 | 0.04475 | 0.43 |
| 15907 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 25 | RBMS3 | Sponge network | -0.663 | 0.10887 | -0.519 | 0.03551 | 0.43 |
| 15908 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | ADAMTSL3 | Sponge network | 0.342 | 0.03129 | -1.559 | 0 | 0.43 |
| 15909 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | HOOK3 | Sponge network | 0.349 | 0.01695 | 0.273 | 0.09957 | 0.43 |
| 15910 | BOLA3-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 21 | RIMS2 | Sponge network | -0.246 | 0.35034 | -1.365 | 0.00208 | 0.43 |
| 15911 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | CUL5 | Sponge network | 1.317 | 0 | 0.026 | 0.77301 | 0.43 |
| 15912 | FAM66C |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | PLA2R1 | Sponge network | -0.901 | 0.00019 | -0.265 | 0.24676 | 0.43 |
| 15913 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 44 | PIK3R1 | Sponge network | -2.452 | 0 | -0.133 | 0.36854 | 0.43 |
| 15914 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-369-3p;hsa-miR-542-3p | 10 | PEG3 | Sponge network | -0.384 | 0.40652 | -1.74 | 0 | 0.43 |
| 15915 | CTD-2314G24.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | NAV3 | Sponge network | 0.246 | 0.59912 | 0.212 | 0.47729 | 0.43 |
| 15916 | RP11-967K21.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-93-5p | 16 | PRUNE2 | Sponge network | 0.056 | 0.81195 | -1.733 | 0.00022 | 0.43 |
| 15917 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | FAM172A | Sponge network | -0.738 | 0.21233 | -0.57 | 0 | 0.43 |
| 15918 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ERG | Sponge network | -0.323 | 0.01372 | 0.002 | 0.99011 | 0.43 |
| 15919 | RP5-1198O20.4 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | NOV | Sponge network | 0.997 | 0.00066 | -0.58 | 0.04676 | 0.43 |
| 15920 | TP73-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p | 10 | KCTD8 | Sponge network | -0.563 | 0.02545 | -3.359 | 0 | 0.43 |
| 15921 | GUSBP11 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378c;hsa-miR-424-5p | 18 | ABL2 | Sponge network | 0.856 | 0 | 0.82 | 0 | 0.43 |
| 15922 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | LRP1B | Sponge network | -1.203 | 0.03881 | -2.163 | 0 | 0.43 |
| 15923 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | EPAS1 | Sponge network | -0.769 | 0.00035 | -0.184 | 0.14418 | 0.43 |
| 15924 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-93-5p | 15 | JAZF1 | Sponge network | -1.255 | 0.00981 | -0.377 | 0.02677 | 0.43 |
| 15925 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-362-3p;hsa-miR-532-3p | 11 | CADM3 | Sponge network | -0.612 | 0.00094 | -3.663 | 0 | 0.43 |
| 15926 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | PDGFRA | Sponge network | 0.335 | 0.20596 | -0.372 | 0.0037 | 0.43 |
| 15927 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-501-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | -0.769 | 0.00035 | -2.417 | 0 | 0.43 |
| 15928 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-7-1-3p | 12 | CBL | Sponge network | 0.252 | 0.08508 | 0.291 | 0.01267 | 0.43 |
| 15929 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 22 | AKAP6 | Sponge network | 0.61 | 0.01593 | -1.586 | 3.0E-5 | 0.429 |
| 15930 | LINC00472 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-7-1-3p | 12 | LUZP2 | Sponge network | -0.842 | 0.01361 | -0.056 | 0.89416 | 0.429 |
| 15931 | RP11-968O1.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 12 | UNC5C | Sponge network | -0.829 | 0.01343 | -0.178 | 0.40214 | 0.429 |
| 15932 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 23 | ARHGAP29 | Sponge network | -0.737 | 0.06182 | 0.131 | 0.42953 | 0.429 |
| 15933 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-486-5p | 12 | DACT1 | Sponge network | -0.162 | 0.47687 | 0.783 | 0.00072 | 0.429 |
| 15934 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 30 | TRPS1 | Sponge network | -0.739 | 0.00044 | -0.191 | 0.44109 | 0.429 |
| 15935 | LINC01018 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 16 | PAK3 | Sponge network | -2.915 | 0.00015 | -0.628 | 0.07253 | 0.429 |
| 15936 | CTC-296K1.4 |
hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-3p;hsa-miR-582-3p;hsa-miR-590-5p | 10 | EBF1 | Sponge network | -2.076 | 0.00548 | -0.384 | 0.08856 | 0.429 |
| 15937 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | ZMYM2 | Sponge network | 0.946 | 0 | 0.279 | 0.01273 | 0.429 |
| 15938 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 13 | PBX1 | Sponge network | -2.039 | 0.03447 | -1.206 | 0 | 0.429 |
| 15939 | RP5-1198O20.4 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p | 15 | EHD3 | Sponge network | 0.997 | 0.00066 | -0.484 | 0.01747 | 0.429 |
| 15940 | HHIP-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 12 | UNC5C | Sponge network | -0.737 | 0.10822 | -0.178 | 0.40214 | 0.429 |
| 15941 | LINC01018 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | -2.915 | 0.00015 | 0.809 | 0.00544 | 0.429 |
| 15942 | SERTAD4-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p | 11 | NAV3 | Sponge network | -0.932 | 0.00329 | 0.212 | 0.47729 | 0.429 |
| 15943 | PCA3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | -0.709 | 0.18168 | -0.474 | 0.03833 | 0.429 |
| 15944 | LINC00702 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | RASSF3 | Sponge network | -0.737 | 0.06182 | -0.226 | 0.11691 | 0.429 |
| 15945 | CTD-2314G24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | 0.246 | 0.59912 | -1.892 | 0 | 0.429 |
| 15946 | PCA3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-5p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 45 | TRPS1 | Sponge network | -0.709 | 0.18168 | -0.191 | 0.44109 | 0.429 |
| 15947 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | NCAM2 | Sponge network | 0.61 | 0.01593 | -0.482 | 0.22565 | 0.429 |
| 15948 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p | 10 | TBRG1 | Sponge network | 0.782 | 0.00016 | -0.071 | 0.37324 | 0.429 |
| 15949 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-375;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 16 | PSIP1 | Sponge network | 0.559 | 0.00026 | -0.005 | 0.96897 | 0.429 |
| 15950 | TP73-AS1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 10 | PEA15 | Sponge network | -0.563 | 0.02545 | 0.009 | 0.93318 | 0.429 |
| 15951 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | AR | Sponge network | -0.206 | 0.12942 | -1.554 | 0 | 0.429 |
| 15952 | LINC00957 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-3p | 14 | ROBO1 | Sponge network | -0.421 | 0.0114 | -0.009 | 0.96154 | 0.429 |
| 15953 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CHD2 | Sponge network | 0.222 | 0.29133 | 0.049 | 0.55371 | 0.429 |
| 15954 | LINC00265 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-497-5p | 10 | SON | Sponge network | 1.07 | 0 | 0.168 | 0.04406 | 0.429 |
| 15955 | HHIP-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-590-3p | 22 | ASTN1 | Sponge network | -0.737 | 0.10822 | -2.389 | 2.0E-5 | 0.429 |
| 15956 | RP11-531A24.5 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | BCL9 | Sponge network | 0.75 | 0.00054 | 0.321 | 0.00267 | 0.429 |
| 15957 | ZRANB2-AS1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-935 | 12 | GRIK3 | Sponge network | -0.376 | 0.00337 | -3.208 | 0 | 0.429 |
| 15958 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-93-5p | 39 | EPHA7 | Sponge network | -0.154 | 0.78939 | -2.197 | 3.0E-5 | 0.429 |
| 15959 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | CNTN1 | Sponge network | 0.889 | 9.0E-5 | -2.594 | 0 | 0.429 |
| 15960 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | MLLT10 | Sponge network | 0.559 | 0.00026 | 0.105 | 0.33292 | 0.429 |
| 15961 | RP4-613B23.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-5p | 12 | PRKCB | Sponge network | -0.309 | 0.1145 | -1.313 | 2.0E-5 | 0.429 |
| 15962 | HHIP-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p | 12 | RELN | Sponge network | -0.737 | 0.10822 | -2.403 | 0 | 0.429 |
| 15963 | AC003090.1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | PCDH18 | Sponge network | -2.117 | 0.00161 | 0.216 | 0.24645 | 0.429 |
| 15964 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-93-3p | 14 | BNC2 | Sponge network | -1.116 | 0.23686 | -0.491 | 0.09988 | 0.429 |
| 15965 | FAM66C |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-5p | 13 | EHD3 | Sponge network | -0.901 | 0.00019 | -0.484 | 0.01747 | 0.429 |
| 15966 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | BMPR2 | Sponge network | 1.253 | 0 | 0.206 | 0.0635 | 0.429 |
| 15967 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-409-3p | 33 | EPHA5 | Sponge network | 0.331 | 0.57247 | -0.957 | 0.01733 | 0.429 |
| 15968 | RP11-159D12.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p | 32 | ARHGEF12 | Sponge network | 1.254 | 0 | 0.09 | 0.35693 | 0.429 |
| 15969 | AC006116.24 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-455-5p;hsa-miR-539-5p | 11 | PDE4DIP | Sponge network | -0.473 | 0.07602 | -0.282 | 0.0257 | 0.429 |
| 15970 | FENDRR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 33 | SNX29 | Sponge network | -1.281 | 0.00012 | -0.34 | 0.01371 | 0.429 |
| 15971 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 12 | PARK2 | Sponge network | 0.997 | 0.00066 | -1.892 | 0 | 0.429 |
| 15972 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3913-5p;hsa-miR-590-3p | 10 | RIMS1 | Sponge network | -0.769 | 0.00035 | -3.143 | 0 | 0.429 |
| 15973 | LINC00472 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | -0.842 | 0.01361 | -0.704 | 0.00106 | 0.429 |
| 15974 | MYLK-AS1 |
hsa-miR-107;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p | 10 | ELMO1 | Sponge network | -1.331 | 3.0E-5 | 0.04 | 0.83147 | 0.429 |
| 15975 | RP11-774O3.3 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | GJA1 | Sponge network | -0.487 | 0.02157 | -0.485 | 0.02179 | 0.429 |
| 15976 | LINC00641 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | PDE4DIP | Sponge network | -0.222 | 0.32445 | -0.282 | 0.0257 | 0.429 |
| 15977 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | NRXN1 | Sponge network | -0.187 | 0.23787 | -3.778 | 0 | 0.429 |
| 15978 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | KCNA6 | Sponge network | -0.777 | 0.1134 | -1.531 | 0.00013 | 0.429 |
| 15979 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 51 | QKI | Sponge network | -2.117 | 0.00161 | -0.333 | 0.04111 | 0.429 |
| 15980 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 26 | ZMYND11 | Sponge network | 0.083 | 0.66405 | -0.2 | 0.05449 | 0.429 |
| 15981 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | MOB1B | Sponge network | 0.603 | 0.00127 | 0.197 | 0.15266 | 0.429 |
| 15982 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-505-3p | 10 | CHD2 | Sponge network | 1.093 | 0 | 0.049 | 0.55371 | 0.429 |
| 15983 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 45 | IL6ST | Sponge network | 0.889 | 9.0E-5 | -0.402 | 0.00676 | 0.429 |
| 15984 | HCG11 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | DCLK1 | Sponge network | 0.385 | 0.10067 | -1.324 | 0.00014 | 0.429 |
| 15985 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-942-5p | 20 | GPC6 | Sponge network | -0.611 | 0.09607 | -0.054 | 0.81465 | 0.429 |
| 15986 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 26 | MYO9A | Sponge network | 0.219 | 0.1516 | -0.147 | 0.32373 | 0.429 |
| 15987 | RP11-758N13.1 |
hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-330-5p;hsa-miR-375;hsa-miR-424-5p | 12 | RASSF8 | Sponge network | 2.939 | 3.0E-5 | -0.122 | 0.65591 | 0.429 |
| 15988 | LINC00476 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p | 10 | HSPB7 | Sponge network | -0.615 | 0.00015 | -2.268 | 1.0E-5 | 0.429 |
| 15989 | RP5-1198O20.4 |
hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3065-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-664a-3p | 11 | LOX | Sponge network | 0.997 | 0.00066 | 1.164 | 0 | 0.429 |
| 15990 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 20 | MCC | Sponge network | -0.517 | 0.02935 | -1.005 | 0 | 0.429 |
| 15991 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 47 | SSBP2 | Sponge network | 0.572 | 0.01722 | -1.58 | 0 | 0.429 |
| 15992 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-590-3p | 10 | PRDM2 | Sponge network | -0.376 | 0.00337 | -0.258 | 0.00721 | 0.429 |
| 15993 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | GRIA3 | Sponge network | -0.739 | 0.0574 | -1.956 | 0 | 0.429 |
| 15994 | EMX2OS |
hsa-miR-185-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-542-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-940 | 10 | ILK | Sponge network | -0.777 | 0.1134 | -0.74 | 0 | 0.429 |
| 15995 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-7-1-3p | 14 | DACH2 | Sponge network | -1.249 | 7.0E-5 | -1.635 | 0.00034 | 0.429 |
| 15996 | RP11-774O3.3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | KLF10 | Sponge network | -0.487 | 0.02157 | -0.783 | 0 | 0.429 |
| 15997 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 26 | EBF1 | Sponge network | -1.057 | 0.00029 | -0.384 | 0.08856 | 0.429 |
| 15998 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 20 | LRRC55 | Sponge network | -0.513 | 0.04703 | -0.026 | 0.93163 | 0.429 |
| 15999 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-7-1-3p | 13 | PRUNE2 | Sponge network | 0.889 | 9.0E-5 | -1.733 | 0.00022 | 0.429 |
| 16000 | RP11-384O8.1 |
hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 23 | MYO9A | Sponge network | -0.738 | 0.21233 | -0.147 | 0.32373 | 0.429 |
| 16001 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 45 | MOB1B | Sponge network | -1.111 | 0.0003 | 0.197 | 0.15266 | 0.429 |
| 16002 | BMS1P20 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | MAP3K4 | Sponge network | 0.34 | 0.00273 | 0.058 | 0.54229 | 0.429 |
| 16003 | AGAP11 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-5p | 79 | LPP | Sponge network | -1.645 | 0 | -0.459 | 0.04475 | 0.429 |
| 16004 | AC003090.1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 12 | ID4 | Sponge network | -2.117 | 0.00161 | -1.644 | 0 | 0.429 |
| 16005 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-590-3p | 13 | RET | Sponge network | -1.375 | 0.08642 | -0.833 | 0.03517 | 0.429 |
| 16006 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-590-5p | 26 | TSHZ3 | Sponge network | 0.331 | 0.57247 | -0.556 | 0.01516 | 0.429 |
| 16007 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | BNC2 | Sponge network | 0.727 | 3.0E-5 | -0.491 | 0.09988 | 0.429 |
| 16008 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 17 | PRKAB2 | Sponge network | -0.517 | 0.02935 | -0.367 | 0.01371 | 0.429 |
| 16009 | RP11-305E6.4 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 27 | CREB1 | Sponge network | 1.317 | 0 | 0.203 | 0.00537 | 0.429 |
| 16010 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | SYNE1 | Sponge network | -0.726 | 0.00132 | -0.738 | 0.00316 | 0.429 |
| 16011 | USP3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 30 | SNX29 | Sponge network | -0.162 | 0.47687 | -0.34 | 0.01371 | 0.429 |
| 16012 | LINC00654 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | EYA4 | Sponge network | 0.374 | 0.20054 | 0.3 | 0.36876 | 0.429 |
| 16013 | RP1-59D14.5 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 12 | MLLT10 | Sponge network | 1.162 | 5.0E-5 | 0.105 | 0.33292 | 0.429 |
| 16014 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-542-3p | 20 | SYNPO2 | Sponge network | -0.384 | 0.40652 | -2.342 | 0 | 0.429 |
| 16015 | RP11-672A2.4 |
hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-766-3p | 17 | IL6ST | Sponge network | 1.344 | 0 | -0.402 | 0.00676 | 0.429 |
| 16016 | KB-1410C5.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 13 | BNC2 | Sponge network | 0.959 | 0 | -0.491 | 0.09988 | 0.429 |
| 16017 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | DOCK4 | Sponge network | 0.614 | 1.0E-5 | 0.502 | 0.00108 | 0.429 |
| 16018 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 49 | PPM1A | Sponge network | -2.452 | 0 | -0.623 | 0 | 0.429 |
| 16019 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 11 | ST5 | Sponge network | -0.976 | 0.00025 | -0.78 | 0 | 0.429 |
| 16020 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | NIN | Sponge network | -0.49 | 0.04946 | -0.253 | 0.13794 | 0.429 |
| 16021 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 49 | SOX5 | Sponge network | -0.222 | 0.32445 | -1.091 | 4.0E-5 | 0.429 |
| 16022 | RP11-248G5.8 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p | 20 | ABL2 | Sponge network | 1.294 | 0 | 0.82 | 0 | 0.429 |
| 16023 | MRVI1-AS1 |
hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-339-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 10 | LUZP2 | Sponge network | -1.092 | 4.0E-5 | -0.056 | 0.89416 | 0.429 |
| 16024 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p | 23 | CADM1 | Sponge network | 0.335 | 0.20596 | -0.9 | 0.00084 | 0.429 |
| 16025 | IQCH-AS1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | ERCC6 | Sponge network | 0.929 | 0 | 0.23 | 0.015 | 0.429 |
| 16026 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-671-5p | 43 | FOXP1 | Sponge network | -0.162 | 0.47687 | 0.042 | 0.74368 | 0.429 |
| 16027 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 24 | AKAP6 | Sponge network | -1.057 | 0.00029 | -1.586 | 3.0E-5 | 0.429 |
| 16028 | TP73-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | TAL1 | Sponge network | -0.563 | 0.02545 | -0.462 | 0.00574 | 0.429 |
| 16029 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -0.737 | 0.06182 | -0.252 | 0.15511 | 0.429 |
| 16030 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | ST6GAL2 | Sponge network | -0.612 | 0.00094 | -1.201 | 0.00597 | 0.429 |
| 16031 | LINC00473 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | -1.203 | 0.03881 | -0.684 | 0.00159 | 0.429 |
| 16032 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p | 18 | MAFB | Sponge network | 0.335 | 0.20596 | -0.023 | 0.89865 | 0.429 |
| 16033 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 45 | WDFY3 | Sponge network | 0.056 | 0.81195 | -0.201 | 0.0967 | 0.429 |
| 16034 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-577 | 18 | SYNPO2 | Sponge network | -0.465 | 0.04703 | -2.342 | 0 | 0.429 |
| 16035 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | FSTL5 | Sponge network | -0.323 | 0.01372 | -1.3 | 0.01619 | 0.429 |
| 16036 | RP11-6O2.3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-590-5p | 11 | INPP4A | Sponge network | 0.331 | 0.57247 | 0.07 | 0.53563 | 0.429 |
| 16037 | HCG18 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | UBR3 | Sponge network | 1.063 | 0 | -0.021 | 0.82774 | 0.429 |
| 16038 | NUTM2A-AS1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 15 | RASAL2 | Sponge network | 0.643 | 0 | 0.869 | 0 | 0.429 |
| 16039 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | MYO9A | Sponge network | -0.899 | 6.0E-5 | -0.147 | 0.32373 | 0.429 |
| 16040 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-942-5p | 26 | UNC80 | Sponge network | -4.546 | 0 | -1.934 | 0 | 0.429 |
| 16041 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRIM58 | Sponge network | -0.49 | 0.04946 | -1.692 | 0 | 0.429 |
| 16042 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-769-5p | 21 | SYNPO2 | Sponge network | -0.342 | 0.02288 | -2.342 | 0 | 0.429 |
| 16043 | LINC00092 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | OPCML | Sponge network | -1.886 | 0 | -2.498 | 0 | 0.429 |
| 16044 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 12 | NR2C2 | Sponge network | 2.368 | 2.0E-5 | 0.21 | 0.03224 | 0.429 |
| 16045 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 41 | ABI2 | Sponge network | -1.697 | 0.00889 | 0.175 | 0.14787 | 0.429 |
| 16046 | BOLA3-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-589-3p | 20 | SCN9A | Sponge network | -0.246 | 0.35034 | -0.525 | 0.10593 | 0.429 |
| 16047 | AC141928.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-96-5p | 26 | CADM1 | Sponge network | 0.853 | 0.15352 | -0.9 | 0.00084 | 0.429 |
| 16048 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-590-3p | 12 | ARHGAP29 | Sponge network | -0.376 | 0.00337 | 0.131 | 0.42953 | 0.429 |
| 16049 | RP11-403I13.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 25 | ABI2 | Sponge network | 0.619 | 0.00157 | 0.175 | 0.14787 | 0.429 |
| 16050 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | PRKAA2 | Sponge network | -0.611 | 0.09607 | -2.462 | 0 | 0.429 |
| 16051 | TP73-AS1 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 10 | EPB41L3 | Sponge network | -0.563 | 0.02545 | -1.193 | 0 | 0.429 |
| 16052 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429 | 14 | ELMO1 | Sponge network | -0.49 | 0.04946 | 0.04 | 0.83147 | 0.429 |
| 16053 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 15 | TET1 | Sponge network | 0.889 | 9.0E-5 | -0.135 | 0.52138 | 0.429 |
| 16054 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 20 | ZMYM2 | Sponge network | 1.04 | 0.00023 | 0.279 | 0.01273 | 0.429 |
| 16055 | RP11-503E24.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 15 | CREB1 | Sponge network | 2.218 | 0 | 0.203 | 0.00537 | 0.429 |
| 16056 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-409-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | MOB1B | Sponge network | 0.419 | 0.0319 | 0.197 | 0.15266 | 0.429 |
| 16057 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 21 | USP6 | Sponge network | 0.225 | 0.30644 | 0.188 | 0.3871 | 0.429 |
| 16058 | HCG11 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | ANK2 | Sponge network | 0.385 | 0.10067 | -1.603 | 1.0E-5 | 0.429 |
| 16059 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 14 | CADM1 | Sponge network | -0.517 | 0.02935 | -0.9 | 0.00084 | 0.429 |
| 16060 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-629-3p | 22 | PTPN14 | Sponge network | -0.358 | 0.17255 | 0.144 | 0.34595 | 0.429 |
| 16061 | ZNF582-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-96-5p | 24 | CADM1 | Sponge network | -1.349 | 0.00017 | -0.9 | 0.00084 | 0.429 |
| 16062 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-96-5p | 21 | TACC1 | Sponge network | -1.582 | 8.0E-5 | -0.362 | 0.05406 | 0.429 |
| 16063 | RP11-798M19.6 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 15 | GLIPR1 | Sponge network | -0.612 | 0.00094 | 0.146 | 0.3276 | 0.429 |
| 16064 | HHIP-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | PPP1R9A | Sponge network | -0.737 | 0.10822 | -0.903 | 0.04254 | 0.429 |
| 16065 | LINC00641 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | PREX2 | Sponge network | -0.222 | 0.32445 | -0.272 | 0.25382 | 0.428 |
| 16066 | TP73-AS1 |
hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-625-5p | 13 | PPP2R2C | Sponge network | -0.563 | 0.02545 | -0.959 | 0.07428 | 0.428 |
| 16067 | RP11-540B6.6 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-497-5p | 12 | CDK6 | Sponge network | 0.93 | 0 | 0.991 | 0 | 0.428 |
| 16068 | AC016747.3 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | MAPK10 | Sponge network | 0.199 | 0.38015 | -1.774 | 0 | 0.428 |
| 16069 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-93-5p | 33 | TACC1 | Sponge network | -0.777 | 0.16667 | -0.362 | 0.05406 | 0.428 |
| 16070 | HHIP-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | CDH10 | Sponge network | -0.737 | 0.10822 | -2.397 | 0 | 0.428 |
| 16071 | RP4-613B23.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-502-5p | 13 | SYT4 | Sponge network | -0.309 | 0.1145 | -3.207 | 0 | 0.428 |
| 16072 | FENDRR |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-425-3p | 12 | DUSP1 | Sponge network | -1.281 | 0.00012 | -1.754 | 0 | 0.428 |
| 16073 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | GRIN2A | Sponge network | -1.057 | 0.00029 | -2.161 | 0 | 0.428 |
| 16074 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 15 | DDX5 | Sponge network | 0.537 | 0.00139 | -0.099 | 0.17428 | 0.428 |
| 16075 | MEG3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 20 | IGF2 | Sponge network | 0.249 | 0.35902 | 0.848 | 0.03842 | 0.428 |
| 16076 | CKMT2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-96-5p | 23 | NAV3 | Sponge network | 0.082 | 0.55654 | 0.212 | 0.47729 | 0.428 |
| 16077 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-96-5p | 21 | CADM2 | Sponge network | -0.187 | 0.23787 | -3.782 | 0 | 0.428 |
| 16078 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 34 | ADARB1 | Sponge network | 0.559 | 0.00026 | -0.335 | 0.06631 | 0.428 |
| 16079 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 17 | RICTOR | Sponge network | 1.262 | 0 | 0.388 | 0.00188 | 0.428 |
| 16080 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 26 | ARHGAP29 | Sponge network | -1.111 | 0.0003 | 0.131 | 0.42953 | 0.428 |
| 16081 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | AFAP1L2 | Sponge network | -0.49 | 0.04946 | -0.284 | 0.15434 | 0.428 |
| 16082 | PART1 |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | PACRG | Sponge network | -2.925 | 0 | -2.248 | 0 | 0.428 |
| 16083 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | GRIA3 | Sponge network | -2.117 | 0.00161 | -1.956 | 0 | 0.428 |
| 16084 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 27 | ZMYM2 | Sponge network | 0.705 | 0.00014 | 0.279 | 0.01273 | 0.428 |
| 16085 | RP5-994D16.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | BMPR2 | Sponge network | 0.252 | 0.08508 | 0.206 | 0.0635 | 0.428 |
| 16086 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | SMARCA2 | Sponge network | -0.739 | 0.0574 | -0.529 | 0.00014 | 0.428 |
| 16087 | RP11-156P1.3 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 16 | MAPK10 | Sponge network | 0.214 | 0.18246 | -1.774 | 0 | 0.428 |
| 16088 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | -0.08 | 0.90092 | -1.74 | 0 | 0.428 |
| 16089 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 28 | GPC6 | Sponge network | -2.452 | 0 | -0.054 | 0.81465 | 0.428 |
| 16090 | LINC00657 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | BRD3 | Sponge network | 0.91 | 0 | 0.37 | 0.00013 | 0.428 |
| 16091 | U62631.5 |
hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-577 | 12 | RASSF2 | Sponge network | 3.115 | 0.00463 | -0.273 | 0.21961 | 0.428 |
| 16092 | ZNF667-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-484;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | RIMS2 | Sponge network | -0.976 | 0.00025 | -1.365 | 0.00208 | 0.428 |
| 16093 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 29 | SNX29 | Sponge network | -0.154 | 0.78939 | -0.34 | 0.01371 | 0.428 |
| 16094 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | CHD2 | Sponge network | 0.083 | 0.66405 | 0.049 | 0.55371 | 0.428 |
| 16095 | LINC00476 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 12 | SEPT6 | Sponge network | -0.615 | 0.00015 | -0.598 | 0.00181 | 0.428 |
| 16096 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | ADAMTSL3 | Sponge network | 0.225 | 0.30644 | -1.559 | 0 | 0.428 |
| 16097 | RP11-356J5.12 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-99b-3p | 44 | GRIK3 | Sponge network | -0.899 | 6.0E-5 | -3.208 | 0 | 0.428 |
| 16098 | RP11-458F8.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p | 13 | NF1 | Sponge network | 1.518 | 0 | 0.51 | 0 | 0.428 |
| 16099 | RP11-182J1.1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-424-5p | 10 | CSDE1 | Sponge network | -0.187 | 0.23787 | -0.493 | 0 | 0.428 |
| 16100 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-362-3p | 13 | SETBP1 | Sponge network | -0.829 | 0.01343 | -1.302 | 1.0E-5 | 0.428 |
| 16101 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 45 | TMEM132B | Sponge network | -0.709 | 0.18168 | -0.83 | 0.01084 | 0.428 |
| 16102 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NRXN3 | Sponge network | -0.342 | 0.02288 | -0.893 | 0.04186 | 0.428 |
| 16103 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-577;hsa-miR-7-1-3p | 26 | TACC1 | Sponge network | 0.389 | 0.04293 | -0.362 | 0.05406 | 0.428 |
| 16104 | AC009948.5 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-421 | 16 | PLXNC1 | Sponge network | 0.532 | 0.00505 | 0.569 | 0.00329 | 0.428 |
| 16105 | LINC00578 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-505-3p;hsa-miR-629-3p;hsa-miR-664a-3p | 17 | ASTN1 | Sponge network | 0.811 | 0.03228 | -2.389 | 2.0E-5 | 0.428 |
| 16106 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p | 15 | EZH1 | Sponge network | 0.259 | 0.12542 | -0.564 | 1.0E-5 | 0.428 |
| 16107 | CTC-296K1.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 18 | IL17RD | Sponge network | -2.095 | 0.00112 | -0.019 | 0.94873 | 0.428 |
| 16108 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | MCC | Sponge network | -2.915 | 0.00015 | -1.005 | 0 | 0.428 |
| 16109 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p | 32 | EPHA5 | Sponge network | 0.214 | 0.18246 | -0.957 | 0.01733 | 0.428 |
| 16110 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | NEDD4 | Sponge network | 0.194 | 0.64278 | 0.02 | 0.89834 | 0.428 |
| 16111 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 23 | MCC | Sponge network | 0.176 | 0.37402 | -1.005 | 0 | 0.428 |
| 16112 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | CEP170 | Sponge network | 0.72 | 0.0001 | 0.943 | 0 | 0.428 |
| 16113 | RP11-967K21.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429 | 13 | IRS1 | Sponge network | 0.056 | 0.81195 | -0.026 | 0.88855 | 0.428 |
| 16114 | LINC00957 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-671-5p | 14 | EHD3 | Sponge network | -0.421 | 0.0114 | -0.484 | 0.01747 | 0.428 |
| 16115 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 28 | MEF2C | Sponge network | 0.657 | 4.0E-5 | -0.499 | 0.01448 | 0.428 |
| 16116 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | PDGFRA | Sponge network | -0.563 | 0.02545 | -0.372 | 0.0037 | 0.428 |
| 16117 | MYLK-AS1 |
hsa-miR-1287-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-330-3p | 11 | PAIP2 | Sponge network | -1.331 | 3.0E-5 | -0.515 | 0 | 0.428 |
| 16118 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-874-3p | 18 | NR2C2 | Sponge network | 0.331 | 0.57247 | 0.21 | 0.03224 | 0.428 |
| 16119 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-532-5p | 10 | LRRC4 | Sponge network | -2.69 | 0.0001 | -1.774 | 0 | 0.428 |
| 16120 | RP11-67L2.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | BRD3 | Sponge network | 0.72 | 0.0001 | 0.37 | 0.00013 | 0.428 |
| 16121 | BVES-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-589-5p | 12 | ROBO1 | Sponge network | -2.62 | 0 | -0.009 | 0.96154 | 0.428 |
| 16122 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p | 12 | DNAJB4 | Sponge network | -0.517 | 0.02935 | -0.7 | 1.0E-5 | 0.428 |
| 16123 | LINC00863 |
hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SUFU | Sponge network | 0.189 | 0.13981 | -0.04 | 0.65489 | 0.428 |
| 16124 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-98-5p | 15 | ITGB3 | Sponge network | 0.101 | 0.60919 | -0.011 | 0.95751 | 0.428 |
| 16125 | RP3-475N16.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-495-3p;hsa-miR-625-5p | 12 | ABI2 | Sponge network | 1.056 | 0 | 0.175 | 0.14787 | 0.428 |
| 16126 | RP11-389C8.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p | 11 | MRVI1 | Sponge network | 0.727 | 3.0E-5 | -1.051 | 0.00022 | 0.428 |
| 16127 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-423-5p | 13 | TIMP3 | Sponge network | -1.654 | 0.00144 | -0.546 | 0.02307 | 0.428 |
| 16128 | HCG11 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | NAV3 | Sponge network | 0.385 | 0.10067 | 0.212 | 0.47729 | 0.428 |
| 16129 | RP11-439A17.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-576-5p | 14 | RUNX1T1 | Sponge network | 0.051 | 0.95756 | -0.344 | 0.2374 | 0.428 |
| 16130 | AC011526.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 15 | RASSF8 | Sponge network | 0.342 | 0.03129 | -0.122 | 0.65591 | 0.428 |
| 16131 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-589-3p | 34 | DIP2C | Sponge network | -0.18 | 0.20343 | -0.436 | 0.01713 | 0.428 |
| 16132 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p | 15 | SEMA3C | Sponge network | -0.777 | 0.16667 | -0.272 | 0.31153 | 0.428 |
| 16133 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-590-5p | 23 | EBF1 | Sponge network | 0.176 | 0.37402 | -0.384 | 0.08856 | 0.428 |
| 16134 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-93-5p | 27 | ZBTB4 | Sponge network | -0.227 | 0.31657 | -0.739 | 0 | 0.428 |
| 16135 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p | 16 | PKNOX1 | Sponge network | 0.064 | 0.72934 | 0.085 | 0.28917 | 0.428 |
| 16136 | MALAT1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 42 | NFIB | Sponge network | 0.782 | 0.00016 | 0.023 | 0.90795 | 0.428 |
| 16137 | RP11-57H14.4 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PHF3 | Sponge network | 0.083 | 0.66405 | 0.126 | 0.19652 | 0.428 |
| 16138 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p | 10 | PDS5B | Sponge network | 1.923 | 0 | 0.259 | 0.00962 | 0.428 |
| 16139 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-935 | 36 | PTPRT | Sponge network | -0.513 | 0.04703 | -0.907 | 0.03727 | 0.428 |
| 16140 | CTA-363E19.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 13 | TBL1XR1 | Sponge network | 1.055 | 0 | 0.578 | 0 | 0.428 |
| 16141 | AC010226.4 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-889-3p;hsa-miR-96-5p | 13 | RPS6KA6 | Sponge network | -0.811 | 2.0E-5 | -2.989 | 0 | 0.428 |
| 16142 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-421;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-652-3p;hsa-miR-940 | 38 | QKI | Sponge network | 0.214 | 0.18246 | -0.333 | 0.04111 | 0.428 |
| 16143 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-7-1-3p | 20 | RARB | Sponge network | 0.335 | 0.20596 | -0.825 | 2.0E-5 | 0.428 |
| 16144 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | PTPN12 | Sponge network | 1.254 | 0 | 0.768 | 0 | 0.428 |
| 16145 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 36 | PIK3R1 | Sponge network | 0.494 | 0.00339 | -0.133 | 0.36854 | 0.428 |
| 16146 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-577;hsa-miR-590-3p | 10 | AKAP12 | Sponge network | -1.057 | 0.00029 | -0.932 | 0.0028 | 0.428 |
| 16147 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | TIMP3 | Sponge network | -1.281 | 0.00012 | -0.546 | 0.02307 | 0.428 |
| 16148 | ACTA2-AS1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | SPOP | Sponge network | 0.194 | 0.64278 | -0.419 | 1.0E-5 | 0.428 |
| 16149 | LINC00578 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-744-3p | 36 | DST | Sponge network | 0.811 | 0.03228 | -0.713 | 0.00096 | 0.428 |
| 16150 | FZD10-AS1 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | ID4 | Sponge network | -0.727 | 0.04726 | -1.644 | 0 | 0.428 |
| 16151 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-429;hsa-miR-590-3p | 12 | RB1CC1 | Sponge network | -0.031 | 0.89063 | 0.175 | 0.12559 | 0.428 |
| 16152 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 17 | CDH19 | Sponge network | -0.513 | 0.04703 | -3.434 | 0 | 0.428 |
| 16153 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-93-5p | 21 | EPHA5 | Sponge network | -2.531 | 0 | -0.957 | 0.01733 | 0.428 |
| 16154 | CTC-510F12.2 |
hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-376c-3p;hsa-miR-766-3p | 18 | IL6ST | Sponge network | -0.691 | 0.00146 | -0.402 | 0.00676 | 0.428 |
| 16155 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p | 11 | ABL2 | Sponge network | 1.46 | 1.0E-5 | 0.82 | 0 | 0.428 |
| 16156 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-29a-5p | 13 | PBX1 | Sponge network | -0.326 | 0.28809 | -1.206 | 0 | 0.428 |
| 16157 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | TACC1 | Sponge network | -0.709 | 0.18168 | -0.362 | 0.05406 | 0.428 |
| 16158 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 13 | CHD1 | Sponge network | -0.222 | 0.32445 | 0.322 | 0.00215 | 0.428 |
| 16159 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p | 19 | RARB | Sponge network | -2.69 | 0.0001 | -0.825 | 2.0E-5 | 0.428 |
| 16160 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-32-5p;hsa-miR-326 | 10 | FAT3 | Sponge network | -0.557 | 0.11135 | -1.601 | 0.00051 | 0.428 |
| 16161 | RP11-175O19.4 |
hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 0.917 | 0 | 0.362 | 0.00066 | 0.428 |
| 16162 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-942-5p | 47 | UNC80 | Sponge network | -0.576 | 0.10121 | -1.934 | 0 | 0.428 |
| 16163 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577 | 28 | ESR1 | Sponge network | -2.69 | 0.0001 | -1.035 | 3.0E-5 | 0.428 |
| 16164 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-96-5p | 23 | RIMBP2 | Sponge network | -0.942 | 0 | -1.968 | 5.0E-5 | 0.428 |
| 16165 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 32 | GRIA3 | Sponge network | -0.737 | 0.06182 | -1.956 | 0 | 0.428 |
| 16166 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-96-5p | 32 | TCF4 | Sponge network | -0.517 | 0.02935 | 0.016 | 0.92271 | 0.428 |
| 16167 | AC138035.2 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-3614-5p;hsa-miR-539-5p | 12 | MOB1B | Sponge network | 1.073 | 0 | 0.197 | 0.15266 | 0.428 |
| 16168 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | BNC2 | Sponge network | 0.385 | 0.10067 | -0.491 | 0.09988 | 0.428 |
| 16169 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-409-5p;hsa-miR-495-3p | 11 | LIMD1 | Sponge network | 0.706 | 0 | 0.103 | 0.28978 | 0.428 |
| 16170 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-577;hsa-miR-940;hsa-miR-96-5p | 12 | LIFR | Sponge network | -0.31 | 0.33871 | -1.794 | 0 | 0.428 |
| 16171 | RP11-67L2.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-484;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-92a-3p | 58 | DST | Sponge network | 0.72 | 0.0001 | -0.713 | 0.00096 | 0.428 |
| 16172 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | BNC2 | Sponge network | 1.253 | 0 | -0.491 | 0.09988 | 0.428 |
| 16173 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | BNC2 | Sponge network | -0.08 | 0.90092 | -0.491 | 0.09988 | 0.428 |
| 16174 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 22 | ZMYM2 | Sponge network | 1.601 | 0 | 0.279 | 0.01273 | 0.428 |
| 16175 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-7-1-3p | 20 | SSBP2 | Sponge network | -0.187 | 0.23787 | -1.58 | 0 | 0.428 |
| 16176 | GNG12-AS1 |
hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 14 | CXCL12 | Sponge network | -0.793 | 7.0E-5 | -1.421 | 0 | 0.428 |
| 16177 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 50 | BNC2 | Sponge network | 0.064 | 0.72934 | -0.491 | 0.09988 | 0.428 |
| 16178 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-96-5p | 44 | KIF1B | Sponge network | 0.594 | 0.41522 | -0.118 | 0.32258 | 0.428 |
| 16179 | LINC00657 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p | 10 | ZNRF3 | Sponge network | 0.91 | 0 | 0.637 | 0.00172 | 0.428 |
| 16180 | RP11-531A24.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-7-1-3p | 20 | AKAP6 | Sponge network | 0.75 | 0.00054 | -1.586 | 3.0E-5 | 0.428 |
| 16181 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 18 | GREM1 | Sponge network | 1.302 | 7.0E-5 | 0.902 | 0.01691 | 0.428 |
| 16182 | AC025171.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-590-3p | 26 | ABL2 | Sponge network | 0.998 | 0 | 0.82 | 0 | 0.428 |
| 16183 | NDUFA6-AS1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940 | 10 | FOXO4 | Sponge network | -0.355 | 0.0077 | -0.516 | 2.0E-5 | 0.428 |
| 16184 | LINC00472 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-382-5p;hsa-miR-421;hsa-miR-450b-5p | 10 | ZNF536 | Sponge network | -0.842 | 0.01361 | -3.115 | 0 | 0.428 |
| 16185 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-98-5p | 20 | PNRC1 | Sponge network | -2.46 | 0 | -0.646 | 0 | 0.428 |
| 16186 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | SMARCA2 | Sponge network | -0.709 | 0.18168 | -0.529 | 0.00014 | 0.428 |
| 16187 | LINC00654 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 20 | MRVI1 | Sponge network | 0.374 | 0.20054 | -1.051 | 0.00022 | 0.428 |
| 16188 | RP11-57H14.4 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 46 | MDM4 | Sponge network | 0.083 | 0.66405 | 0.345 | 0.00762 | 0.428 |
| 16189 | RP11-102N12.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 13 | IGF1 | Sponge network | -0.351 | 0.11358 | -0.204 | 0.5898 | 0.428 |
| 16190 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p | 19 | CACNA2D1 | Sponge network | -1.047 | 0 | -0.846 | 0.00675 | 0.428 |
| 16191 | OIP5-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 32 | EPS15 | Sponge network | 0.421 | 0.00084 | -0.052 | 0.53998 | 0.428 |
| 16192 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-7-5p;hsa-miR-877-5p | 20 | TET1 | Sponge network | -1.654 | 0.00144 | -0.135 | 0.52138 | 0.428 |
| 16193 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | PRKAA2 | Sponge network | 0.853 | 0.15352 | -2.462 | 0 | 0.428 |
| 16194 | RP11-359B12.2 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-940 | 14 | TAF1 | Sponge network | 0.064 | 0.72934 | 0.39 | 3.0E-5 | 0.428 |
| 16195 | AC016747.3 |
hsa-miR-107;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 14 | ERG | Sponge network | 0.199 | 0.38015 | 0.002 | 0.99011 | 0.428 |
| 16196 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p | 13 | ZNF208 | Sponge network | 0.299 | 0.57722 | -0.4 | 0.22381 | 0.428 |
| 16197 | AC116366.6 |
hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-374b-3p;hsa-miR-577 | 10 | ATRX | Sponge network | 0.329 | 0.11122 | 0.261 | 0.04408 | 0.427 |
| 16198 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | TRIM33 | Sponge network | 0.702 | 7.0E-5 | 0.402 | 7.0E-5 | 0.427 |
| 16199 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 63 | TCF4 | Sponge network | 0.246 | 0.59912 | 0.016 | 0.92271 | 0.427 |
| 16200 | LINC00957 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | ST6GAL2 | Sponge network | -0.421 | 0.0114 | -1.201 | 0.00597 | 0.427 |
| 16201 | RP11-774O3.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 24 | NRK | Sponge network | -0.487 | 0.02157 | 0.656 | 0.19217 | 0.427 |
| 16202 | CTA-204B4.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-590-3p | 25 | CREB1 | Sponge network | 0.765 | 7.0E-5 | 0.203 | 0.00537 | 0.427 |
| 16203 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | DMTF1 | Sponge network | -0.222 | 0.32445 | 0.248 | 0.02726 | 0.427 |
| 16204 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-5p | 11 | GALNT13 | Sponge network | 0.176 | 0.37402 | -0.857 | 0.07439 | 0.427 |
| 16205 | BVES-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-877-5p | 16 | TET1 | Sponge network | -2.62 | 0 | -0.135 | 0.52138 | 0.427 |
| 16206 | RP1-151F17.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FBXO11 | Sponge network | 0.222 | 0.29133 | 0.142 | 0.04938 | 0.427 |
| 16207 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | DNAJB4 | Sponge network | 0.381 | 0.25967 | -0.7 | 1.0E-5 | 0.427 |
| 16208 | CTD-2314G24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 37 | DIP2C | Sponge network | 0.246 | 0.59912 | -0.436 | 0.01713 | 0.427 |
| 16209 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-582-5p | 19 | USP6 | Sponge network | 0.637 | 0.00069 | 0.188 | 0.3871 | 0.427 |
| 16210 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p | 10 | RBL2 | Sponge network | 0.178 | 0.29106 | -0.282 | 0.00254 | 0.427 |
| 16211 | RP11-73M18.8 |
hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-625-3p | 11 | ABL2 | Sponge network | 0.832 | 0 | 0.82 | 0 | 0.427 |
| 16212 | HCG11 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-369-3p;hsa-miR-629-3p;hsa-miR-744-3p | 20 | PTPN14 | Sponge network | 0.385 | 0.10067 | 0.144 | 0.34595 | 0.427 |
| 16213 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 35 | ZFHX3 | Sponge network | -0.793 | 7.0E-5 | 0.389 | 0.01363 | 0.427 |
| 16214 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 21 | TLE4 | Sponge network | -0.49 | 0.04946 | -1.157 | 0 | 0.427 |
| 16215 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | EDA2R | Sponge network | -1.203 | 0.03881 | -0.587 | 0.01598 | 0.427 |
| 16216 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-7-1-3p | 31 | RASSF2 | Sponge network | 0.194 | 0.64278 | -0.273 | 0.21961 | 0.427 |
| 16217 | AC003090.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-93-5p | 58 | DST | Sponge network | -2.117 | 0.00161 | -0.713 | 0.00096 | 0.427 |
| 16218 | CTC-444N24.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 24 | ABI2 | Sponge network | 1.153 | 0 | 0.175 | 0.14787 | 0.427 |
| 16219 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p | 27 | GPC6 | Sponge network | 0.374 | 0.20054 | -0.054 | 0.81465 | 0.427 |
| 16220 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-590-3p | 17 | RSPO2 | Sponge network | -0.737 | 0.10822 | -3.038 | 0 | 0.427 |
| 16221 | AC004540.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-423-5p;hsa-miR-484 | 15 | GRIK3 | Sponge network | -2.549 | 0.00528 | -3.208 | 0 | 0.427 |
| 16222 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | HDAC9 | Sponge network | -0.611 | 0.09607 | -0.755 | 0.00495 | 0.427 |
| 16223 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 34 | TACC1 | Sponge network | 0.199 | 0.38015 | -0.362 | 0.05406 | 0.427 |
| 16224 | FAM66C |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | GJA1 | Sponge network | -0.901 | 0.00019 | -0.485 | 0.02179 | 0.427 |
| 16225 | RP11-536O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p | 12 | MCC | Sponge network | -0.074 | 0.90758 | -1.005 | 0 | 0.427 |
| 16226 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | IGSF10 | Sponge network | -1.216 | 0 | -1.751 | 1.0E-5 | 0.427 |
| 16227 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | -0.942 | 0 | -0.615 | 0.01482 | 0.427 |
| 16228 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 64 | NTRK3 | Sponge network | 0.246 | 0.59912 | -1.77 | 3.0E-5 | 0.427 |
| 16229 | MIR497HG |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-584-5p;hsa-miR-877-5p;hsa-miR-942-5p | 14 | GLIPR1 | Sponge network | -1.439 | 4.0E-5 | 0.146 | 0.3276 | 0.427 |
| 16230 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | PLA2R1 | Sponge network | -0.487 | 0.02157 | -0.265 | 0.24676 | 0.427 |
| 16231 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p | 27 | ZBTB4 | Sponge network | 0.064 | 0.72934 | -0.739 | 0 | 0.427 |
| 16232 | RP11-307B6.3 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p | 12 | RPS6KA6 | Sponge network | -4.463 | 0.00025 | -2.989 | 0 | 0.427 |
| 16233 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-98-5p | 45 | PRKAA2 | Sponge network | -0.727 | 0.04726 | -2.462 | 0 | 0.427 |
| 16234 | RP11-326G21.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CADM2 | Sponge network | -0.021 | 0.93598 | -3.782 | 0 | 0.427 |
| 16235 | RP11-119F7.5 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-874-3p | 18 | ASTN1 | Sponge network | 0.225 | 0.30644 | -2.389 | 2.0E-5 | 0.427 |
| 16236 | HHIP-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-769-5p | 13 | SYNPO2 | Sponge network | -0.737 | 0.10822 | -2.342 | 0 | 0.427 |
| 16237 | MIR497HG |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-671-5p;hsa-miR-7-5p | 12 | EHD3 | Sponge network | -1.439 | 4.0E-5 | -0.484 | 0.01747 | 0.427 |
| 16238 | ADAMTS9-AS2 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 32 | OPCML | Sponge network | -2.452 | 0 | -2.498 | 0 | 0.427 |
| 16239 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 25 | CBL | Sponge network | 0.91 | 0 | 0.291 | 0.01267 | 0.427 |
| 16240 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | FGFR1 | Sponge network | 0.342 | 0.03129 | -0.509 | 0.03957 | 0.427 |
| 16241 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 30 | USP12 | Sponge network | -1.575 | 6.0E-5 | -0.102 | 0.40768 | 0.427 |
| 16242 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 26 | EBF1 | Sponge network | -0.513 | 0.04703 | -0.384 | 0.08856 | 0.427 |
| 16243 | FAM215B |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-7-1-3p | 15 | TSC1 | Sponge network | 0.817 | 3.0E-5 | 0.103 | 0.26071 | 0.427 |
| 16244 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | LRIG1 | Sponge network | 0.214 | 0.18246 | -0.537 | 0.00588 | 0.427 |
| 16245 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-590-3p | 15 | TMTC3 | Sponge network | 0.34 | 0.00273 | 0.123 | 0.22189 | 0.427 |
| 16246 | TP73-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p | 12 | ZNF536 | Sponge network | -0.563 | 0.02545 | -3.115 | 0 | 0.427 |
| 16247 | CTD-2554C21.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p | 25 | IGFBP5 | Sponge network | -1.654 | 0.00144 | -0.806 | 0.00105 | 0.427 |
| 16248 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374b-3p | 17 | ATRX | Sponge network | 0.946 | 0 | 0.261 | 0.04408 | 0.427 |
| 16249 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-33a-3p | 20 | PIK3C2A | Sponge network | 0.594 | 0.00064 | 0.061 | 0.53776 | 0.427 |
| 16250 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 19 | CNTNAP3 | Sponge network | -0.777 | 0.1134 | -1.465 | 0.00033 | 0.427 |
| 16251 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p | 17 | NRK | Sponge network | -0.469 | 0.08721 | 0.656 | 0.19217 | 0.427 |
| 16252 | DNM3OS |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | PRDM5 | Sponge network | 0.572 | 0.01722 | -1.09 | 0.00014 | 0.427 |
| 16253 | LINC00926 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940 | 17 | PTPRT | Sponge network | 0.756 | 0.00739 | -0.907 | 0.03727 | 0.427 |
| 16254 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 54 | QKI | Sponge network | -0.222 | 0.32445 | -0.333 | 0.04111 | 0.427 |
| 16255 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | ABL2 | Sponge network | 0.178 | 0.29106 | 0.82 | 0 | 0.427 |
| 16256 | RP11-13K12.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 72 | LPP | Sponge network | -0.154 | 0.78939 | -0.459 | 0.04475 | 0.427 |
| 16257 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 24 | CNTN1 | Sponge network | 0.214 | 0.18246 | -2.594 | 0 | 0.427 |
| 16258 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p | 12 | CITED2 | Sponge network | -0.896 | 0.00099 | -1.545 | 0 | 0.427 |
| 16259 | RP5-1074L1.4 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 1.923 | 0 | 0.362 | 0.00066 | 0.427 |
| 16260 | AC009948.5 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | PCDH18 | Sponge network | 0.532 | 0.00505 | 0.216 | 0.24645 | 0.427 |
| 16261 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 45 | EPHA7 | Sponge network | -0.777 | 0.1134 | -2.197 | 3.0E-5 | 0.427 |
| 16262 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p | 12 | ZNF292 | Sponge network | 0.614 | 1.0E-5 | 0.276 | 0.04148 | 0.427 |
| 16263 | AC103563.8 |
hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-324-5p | 11 | DTNA | Sponge network | -7.551 | 0 | -1.648 | 1.0E-5 | 0.427 |
| 16264 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-542-3p;hsa-miR-940 | 11 | HLF | Sponge network | 0.043 | 0.81628 | -2.443 | 0 | 0.427 |
| 16265 | CTD-3092A11.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 23 | ABL2 | Sponge network | 0.873 | 0 | 0.82 | 0 | 0.427 |
| 16266 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p | 15 | ZFP36L1 | Sponge network | -0.583 | 0.14589 | -0.505 | 8.0E-5 | 0.427 |
| 16267 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | DOCK4 | Sponge network | 0.214 | 0.18246 | 0.502 | 0.00108 | 0.427 |
| 16268 | PTOV1-AS1 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 12 | RASAL2 | Sponge network | 0.87 | 0 | 0.869 | 0 | 0.427 |
| 16269 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p | 21 | KIT | Sponge network | -0.323 | 0.01372 | -2.184 | 0 | 0.427 |
| 16270 | AC016747.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | ROBO1 | Sponge network | 0.199 | 0.38015 | -0.009 | 0.96154 | 0.427 |
| 16271 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 19 | PRUNE2 | Sponge network | -0.769 | 0.00035 | -1.733 | 0.00022 | 0.427 |
| 16272 | RP11-212P7.2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 20 | BCL9 | Sponge network | 0.219 | 0.1516 | 0.321 | 0.00267 | 0.427 |
| 16273 | CTC-444N24.11 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-664a-5p | 14 | CBFB | Sponge network | 1.153 | 0 | 1.061 | 0 | 0.427 |
| 16274 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 54 | THRB | Sponge network | -0.842 | 0.01361 | -1.117 | 0.0004 | 0.427 |
| 16275 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | ZMYM2 | Sponge network | 0.826 | 1.0E-5 | 0.279 | 0.01273 | 0.427 |
| 16276 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p | 23 | TACC1 | Sponge network | 0.178 | 0.29106 | -0.362 | 0.05406 | 0.427 |
| 16277 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429 | 10 | THSD7A | Sponge network | 0.056 | 0.81195 | 0.242 | 0.25154 | 0.427 |
| 16278 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-625-5p | 32 | ABI2 | Sponge network | -0.154 | 0.78939 | 0.175 | 0.14787 | 0.427 |
| 16279 | BOLA3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-629-3p | 13 | CDH23 | Sponge network | -0.246 | 0.35034 | -0.974 | 0.00034 | 0.427 |
| 16280 | LINC00672 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 11 | MAPK10 | Sponge network | -0.227 | 0.31657 | -1.774 | 0 | 0.427 |
| 16281 | SNX29P2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 25 | NAV3 | Sponge network | 0.4 | 0.14611 | 0.212 | 0.47729 | 0.427 |
| 16282 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-654-3p | 13 | NR2C2 | Sponge network | 1.157 | 0.00455 | 0.21 | 0.03224 | 0.427 |
| 16283 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 17 | LATS2 | Sponge network | -0.487 | 0.02157 | 0.159 | 0.2304 | 0.427 |
| 16284 | PCA3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ITGB3 | Sponge network | -0.709 | 0.18168 | -0.011 | 0.95751 | 0.427 |
| 16285 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 68 | TCF4 | Sponge network | 1.302 | 7.0E-5 | 0.016 | 0.92271 | 0.427 |
| 16286 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 39 | RBMS3 | Sponge network | 0.083 | 0.66405 | -0.519 | 0.03551 | 0.427 |
| 16287 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 52 | CADM2 | Sponge network | 0.335 | 0.20596 | -3.782 | 0 | 0.427 |
| 16288 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-660-5p | 47 | RUNX1T1 | Sponge network | 0.331 | 0.57247 | -0.344 | 0.2374 | 0.427 |
| 16289 | WDFY3-AS2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | KCTD8 | Sponge network | -0.942 | 0 | -3.359 | 0 | 0.427 |
| 16290 | LINC00654 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | ZDBF2 | Sponge network | 0.374 | 0.20054 | -0.998 | 0.00025 | 0.427 |
| 16291 | SH3BP5-AS1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 12 | SHPRH | Sponge network | 0.267 | 0.2937 | 0.098 | 0.40994 | 0.427 |
| 16292 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | KAT6B | Sponge network | -1.281 | 0.00012 | -0.478 | 0.00018 | 0.427 |
| 16293 | RP11-13K12.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p | 17 | HIP1 | Sponge network | -0.318 | 0.65847 | 0.774 | 0 | 0.427 |
| 16294 | PART1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | CNTNAP1 | Sponge network | -2.925 | 0 | 0.358 | 0.13727 | 0.427 |
| 16295 | MEG3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-940 | 38 | NTRK2 | Sponge network | 0.249 | 0.35902 | -0.736 | 0.06495 | 0.427 |
| 16296 | LINC00472 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-877-5p | 11 | STAT5B | Sponge network | -0.842 | 0.01361 | -0.182 | 0.1082 | 0.427 |
| 16297 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | FAM133A | Sponge network | 0.997 | 0.00066 | 0.549 | 0.29009 | 0.427 |
| 16298 | LINC00472 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-744-3p | 19 | HIP1 | Sponge network | -0.842 | 0.01361 | 0.774 | 0 | 0.427 |
| 16299 | RP11-305E6.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-628-5p | 15 | SLC5A3 | Sponge network | 1.317 | 0 | 0.493 | 5.0E-5 | 0.427 |
| 16300 | RP5-1198O20.4 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-93-3p | 30 | CTIF | Sponge network | 0.997 | 0.00066 | -0.389 | 0.00549 | 0.427 |
| 16301 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-23b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-874-3p | 12 | BCLAF1 | Sponge network | 1.122 | 0 | 0.211 | 0.00684 | 0.427 |
| 16302 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | ZNF770 | Sponge network | 0.782 | 0.00016 | 0.206 | 0.06751 | 0.427 |
| 16303 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-500b-5p | 13 | SLITRK6 | Sponge network | -0.932 | 0.00329 | -2.121 | 0 | 0.427 |
| 16304 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | NCAM2 | Sponge network | -2.46 | 0 | -0.482 | 0.22565 | 0.427 |
| 16305 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 18 | RICTOR | Sponge network | 1.4 | 0 | 0.388 | 0.00188 | 0.427 |
| 16306 | PART1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 74 | DST | Sponge network | -2.925 | 0 | -0.713 | 0.00096 | 0.427 |
| 16307 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-7-1-3p | 22 | EBF1 | Sponge network | -0.611 | 0.09607 | -0.384 | 0.08856 | 0.427 |
| 16308 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-7-5p | 39 | BCL2 | Sponge network | -0.162 | 0.47687 | -0.899 | 0 | 0.427 |
| 16309 | RP11-532F6.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-744-3p | 51 | DST | Sponge network | -0.896 | 0.00099 | -0.713 | 0.00096 | 0.427 |
| 16310 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | EPHA3 | Sponge network | 0.72 | 0.0001 | 0.185 | 0.53588 | 0.427 |
| 16311 | RP11-283I3.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 22 | BRD3 | Sponge network | 0.614 | 1.0E-5 | 0.37 | 0.00013 | 0.427 |
| 16312 | BVES-AS1 |
hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-935 | 16 | GJA1 | Sponge network | -2.62 | 0 | -0.485 | 0.02179 | 0.427 |
| 16313 | ADAMTS9-AS2 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 28 | JDP2 | Sponge network | -2.452 | 0 | -0.967 | 0 | 0.427 |
| 16314 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 46 | MEF2C | Sponge network | -0.222 | 0.32445 | -0.499 | 0.01448 | 0.427 |
| 16315 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-582-5p | 14 | ZMYM2 | Sponge network | 0.792 | 0.0049 | 0.279 | 0.01273 | 0.427 |
| 16316 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p | 27 | BTG2 | Sponge network | -0.896 | 0.00099 | -1.763 | 0 | 0.427 |
| 16317 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-3p | 25 | DST | Sponge network | 0.178 | 0.29106 | -0.713 | 0.00096 | 0.427 |
| 16318 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | ATRX | Sponge network | 0.826 | 1.0E-5 | 0.261 | 0.04408 | 0.427 |
| 16319 | LINC00702 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | NIN | Sponge network | -0.737 | 0.06182 | -0.253 | 0.13794 | 0.427 |
| 16320 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 52 | LPP | Sponge network | 0.389 | 0.04293 | -0.459 | 0.04475 | 0.427 |
| 16321 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 32 | SNX29 | Sponge network | 0.572 | 0.01722 | -0.34 | 0.01371 | 0.427 |
| 16322 | LINC00473 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-7-5p;hsa-miR-92a-3p | 14 | FLNA | Sponge network | -1.203 | 0.03881 | -0.771 | 0.01377 | 0.427 |
| 16323 | FENDRR |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p;hsa-miR-624-5p | 12 | SMARCA1 | Sponge network | -1.281 | 0.00012 | -0.684 | 0.00159 | 0.427 |
| 16324 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 31 | PRDM2 | Sponge network | 0.194 | 0.64278 | -0.258 | 0.00721 | 0.427 |
| 16325 | RP1-228H13.5 |
hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p | 15 | ABL2 | Sponge network | 1.397 | 0 | 0.82 | 0 | 0.427 |
| 16326 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 15 | SSBP2 | Sponge network | -2.039 | 0.03447 | -1.58 | 0 | 0.427 |
| 16327 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-455-3p | 13 | ABL1 | Sponge network | 0.331 | 0.57247 | -0.215 | 0.07804 | 0.427 |
| 16328 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 17 | PIK3C2A | Sponge network | 0.702 | 7.0E-5 | 0.061 | 0.53776 | 0.427 |
| 16329 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 21 | ABL1 | Sponge network | -0.355 | 0.0077 | -0.215 | 0.07804 | 0.427 |
| 16330 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-940 | 46 | GAS7 | Sponge network | -0.323 | 0.01372 | -0.555 | 0.01602 | 0.427 |
| 16331 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 52 | SSBP2 | Sponge network | 0.997 | 0.00066 | -1.58 | 0 | 0.426 |
| 16332 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 28 | RICTOR | Sponge network | 0.539 | 0.03722 | 0.388 | 0.00188 | 0.426 |
| 16333 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-940 | 13 | PEG3 | Sponge network | -0.465 | 0.04703 | -1.74 | 0 | 0.426 |
| 16334 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p | 10 | ST5 | Sponge network | -0.576 | 0.10121 | -0.78 | 0 | 0.426 |
| 16335 | LINC00957 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p | 10 | PDZD2 | Sponge network | -0.421 | 0.0114 | -0.909 | 3.0E-5 | 0.426 |
| 16336 | SNX29P2 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | TPO | Sponge network | 0.4 | 0.14611 | -1.87 | 2.0E-5 | 0.426 |
| 16337 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-501-5p | 10 | MN1 | Sponge network | 0.082 | 0.55654 | -0.358 | 0.24172 | 0.426 |
| 16338 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 56 | THRB | Sponge network | -2.915 | 0.00015 | -1.117 | 0.0004 | 0.426 |
| 16339 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | EPHA3 | Sponge network | 0.219 | 0.1516 | 0.185 | 0.53588 | 0.426 |
| 16340 | LINC00473 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p | 10 | GATA2 | Sponge network | -1.203 | 0.03881 | -0.654 | 0.0016 | 0.426 |
| 16341 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 26 | NRXN1 | Sponge network | -0.513 | 0.04703 | -3.778 | 0 | 0.426 |
| 16342 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | ZFHX3 | Sponge network | -0.901 | 0.00019 | 0.389 | 0.01363 | 0.426 |
| 16343 | HCG18 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 25 | ABI2 | Sponge network | 1.063 | 0 | 0.175 | 0.14787 | 0.426 |
| 16344 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-484;hsa-miR-590-3p | 13 | SMARCA2 | Sponge network | -0.096 | 0.67958 | -0.529 | 0.00014 | 0.426 |
| 16345 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-576-5p | 15 | CDH19 | Sponge network | 0.176 | 0.37402 | -3.434 | 0 | 0.426 |
| 16346 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p | 38 | SOX5 | Sponge network | 0.056 | 0.81195 | -1.091 | 4.0E-5 | 0.426 |
| 16347 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p | 24 | EYA4 | Sponge network | -0.777 | 0.16667 | 0.3 | 0.36876 | 0.426 |
| 16348 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-501-3p;hsa-miR-93-5p | 18 | SLITRK3 | Sponge network | -0.563 | 0.02545 | -3.328 | 0 | 0.426 |
| 16349 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 12 | LRRK2 | Sponge network | -1.057 | 0.00029 | -1.136 | 1.0E-5 | 0.426 |
| 16350 | RP11-798G7.7 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-625-5p | 10 | ZMYM2 | Sponge network | 0.858 | 3.0E-5 | 0.279 | 0.01273 | 0.426 |
| 16351 | RP11-798M19.6 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | PAIP2 | Sponge network | -0.612 | 0.00094 | -0.515 | 0 | 0.426 |
| 16352 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -0.777 | 0.16667 | -3.328 | 0 | 0.426 |
| 16353 | RP11-401P9.4 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-484 | 15 | IGF2 | Sponge network | -0.384 | 0.40652 | 0.848 | 0.03842 | 0.426 |
| 16354 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-93-5p | 37 | AR | Sponge network | 0.199 | 0.38015 | -1.554 | 0 | 0.426 |
| 16355 | CTC-429P9.1 |
hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p | 11 | SCN9A | Sponge network | 0.497 | 0.01451 | -0.525 | 0.10593 | 0.426 |
| 16356 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-214-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | LRP1B | Sponge network | -0.222 | 0.32445 | -2.163 | 0 | 0.426 |
| 16357 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-93-5p;hsa-miR-96-5p | 28 | TACC1 | Sponge network | -1.654 | 0.00144 | -0.362 | 0.05406 | 0.426 |
| 16358 | RP11-798G7.7 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-539-5p | 15 | MOB1B | Sponge network | 0.858 | 3.0E-5 | 0.197 | 0.15266 | 0.426 |
| 16359 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | ST6GALNAC3 | Sponge network | -0.737 | 0.06182 | -0.987 | 0 | 0.426 |
| 16360 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CDH19 | Sponge network | 0.246 | 0.59912 | -3.434 | 0 | 0.426 |
| 16361 | RAD51-AS1 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p | 22 | CREB1 | Sponge network | 0.768 | 7.0E-5 | 0.203 | 0.00537 | 0.426 |
| 16362 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 17 | ANK2 | Sponge network | 0.174 | 0.30034 | -1.603 | 1.0E-5 | 0.426 |
| 16363 | RP11-248G5.8 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-582-5p | 10 | PHF3 | Sponge network | 1.294 | 0 | 0.126 | 0.19652 | 0.426 |
| 16364 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p | 41 | ZFHX4 | Sponge network | -0.154 | 0.78939 | 0.018 | 0.95574 | 0.426 |
| 16365 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 28 | MOB1B | Sponge network | 0.082 | 0.55654 | 0.197 | 0.15266 | 0.426 |
| 16366 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | 0.331 | 0.57247 | 0.273 | 0.09957 | 0.426 |
| 16367 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p | 11 | PRDM5 | Sponge network | -0.096 | 0.67958 | -1.09 | 0.00014 | 0.426 |
| 16368 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | LRRK2 | Sponge network | 0.101 | 0.60919 | -1.136 | 1.0E-5 | 0.426 |
| 16369 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | NUAK1 | Sponge network | -0.737 | 0.06182 | 0.904 | 0 | 0.426 |
| 16370 | RP11-867G23.10 |
hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 11 | FGF5 | Sponge network | -2.531 | 0 | 0.549 | 0.28659 | 0.426 |
| 16371 | SNHG14 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 24 | IRS1 | Sponge network | -1.111 | 0.0003 | -0.026 | 0.88855 | 0.426 |
| 16372 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ABCC9 | Sponge network | 0.083 | 0.66405 | -0.466 | 0.14121 | 0.426 |
| 16373 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-98-5p | 11 | MPL | Sponge network | -1.697 | 0.00889 | -0.624 | 0.00133 | 0.426 |
| 16374 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-7-1-3p | 10 | RARB | Sponge network | -0.023 | 0.95109 | -0.825 | 2.0E-5 | 0.426 |
| 16375 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-378a-5p | 13 | PDS5B | Sponge network | 1.147 | 0 | 0.259 | 0.00962 | 0.426 |
| 16376 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p | 12 | CDH10 | Sponge network | -0.976 | 0.00025 | -2.397 | 0 | 0.426 |
| 16377 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 49 | ERBB4 | Sponge network | -1.575 | 6.0E-5 | -1.975 | 2.0E-5 | 0.426 |
| 16378 | HCG11 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITGB1 | Sponge network | 0.385 | 0.10067 | 0.524 | 0.00017 | 0.426 |
| 16379 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 40 | ZFHX4 | Sponge network | -0.405 | 0.22013 | 0.018 | 0.95574 | 0.426 |
| 16380 | RP11-760H22.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-342-3p;hsa-miR-374b-5p | 10 | STARD13 | Sponge network | 0.001 | 0.99753 | -0.187 | 0.27383 | 0.426 |
| 16381 | MALAT1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PHF3 | Sponge network | 0.782 | 0.00016 | 0.126 | 0.19652 | 0.426 |
| 16382 | CTD-2554C21.2 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-96-5p | 12 | RPS6KA6 | Sponge network | -1.654 | 0.00144 | -2.989 | 0 | 0.426 |
| 16383 | RP11-532F6.3 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 21 | SYT4 | Sponge network | -0.896 | 0.00099 | -3.207 | 0 | 0.426 |
| 16384 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p | 15 | PHF6 | Sponge network | 0.683 | 1.0E-5 | 0.785 | 0 | 0.426 |
| 16385 | OIP5-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | DOCK4 | Sponge network | 0.421 | 0.00084 | 0.502 | 0.00108 | 0.426 |
| 16386 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-708-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | GRIA3 | Sponge network | -0.899 | 6.0E-5 | -1.956 | 0 | 0.426 |
| 16387 | SH3RF3-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 19 | IGFBP5 | Sponge network | 0.889 | 9.0E-5 | -0.806 | 0.00105 | 0.426 |
| 16388 | CTD-2314G24.2 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 13 | RHOB | Sponge network | 0.246 | 0.59912 | -1.052 | 0 | 0.426 |
| 16389 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 41 | TGFBR3 | Sponge network | -0.899 | 6.0E-5 | -1.173 | 1.0E-5 | 0.426 |
| 16390 | HOTAIRM1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | TRPS1 | Sponge network | -0.053 | 0.80685 | -0.191 | 0.44109 | 0.426 |
| 16391 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-576-5p | 33 | PHC3 | Sponge network | -0.162 | 0.47687 | 0.212 | 0.0499 | 0.426 |
| 16392 | CALML3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-576-5p | 31 | NTRK2 | Sponge network | 0.95 | 0.15943 | -0.736 | 0.06495 | 0.426 |
| 16393 | CTD-2006H14.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ABI2 | Sponge network | 1.378 | 0 | 0.175 | 0.14787 | 0.426 |
| 16394 | FGF14-AS2 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 17 | DIP2C | Sponge network | -1.582 | 8.0E-5 | -0.436 | 0.01713 | 0.426 |
| 16395 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | LRRC4 | Sponge network | -0.49 | 0.04946 | -1.774 | 0 | 0.426 |
| 16396 | CTD-2314G24.2 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RNF180 | Sponge network | 0.246 | 0.59912 | -1.127 | 1.0E-5 | 0.426 |
| 16397 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | PDGFRA | Sponge network | -2.117 | 0.00161 | -0.372 | 0.0037 | 0.426 |
| 16398 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-628-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p | 60 | BMPR2 | Sponge network | -0.162 | 0.47687 | 0.206 | 0.0635 | 0.426 |
| 16399 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-511-5p;hsa-miR-93-3p | 16 | WWTR1 | Sponge network | -0.384 | 0.40652 | -0.345 | 0.11997 | 0.426 |
| 16400 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-340-5p | 11 | FGF5 | Sponge network | 0.331 | 0.57247 | 0.549 | 0.28659 | 0.426 |
| 16401 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | CCND2 | Sponge network | -1.281 | 0.00012 | -0.267 | 0.3112 | 0.426 |
| 16402 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-98-5p | 59 | NTRK3 | Sponge network | -0.773 | 0.04073 | -1.77 | 3.0E-5 | 0.426 |
| 16403 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-582-3p | 15 | CBL | Sponge network | 0.643 | 0 | 0.291 | 0.01267 | 0.426 |
| 16404 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | SSBP2 | Sponge network | -1.349 | 0.00017 | -1.58 | 0 | 0.426 |
| 16405 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 21 | LATS1 | Sponge network | 0.214 | 0.18246 | 0.109 | 0.34208 | 0.426 |
| 16406 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 24 | ARHGAP29 | Sponge network | -0.739 | 0.0574 | 0.131 | 0.42953 | 0.426 |
| 16407 | RASSF8-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | 0.335 | 0.20596 | 0.309 | 0.17079 | 0.426 |
| 16408 | FAM66C |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 28 | PTPN14 | Sponge network | -0.901 | 0.00019 | 0.144 | 0.34595 | 0.426 |
| 16409 | JAZF1-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-942-5p;hsa-miR-96-5p | 12 | ZBTB20 | Sponge network | -0.769 | 0.00035 | -0.122 | 0.57569 | 0.426 |
| 16410 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -0.318 | 0.65847 | -0.354 | 0.14694 | 0.426 |
| 16411 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | SETBP1 | Sponge network | 0.064 | 0.72934 | -1.302 | 1.0E-5 | 0.426 |
| 16412 | LINC00472 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421 | 14 | STARD13 | Sponge network | -0.842 | 0.01361 | -0.187 | 0.27383 | 0.426 |
| 16413 | RP11-121C2.2 |
hsa-miR-101-3p;hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 19 | UBR3 | Sponge network | 1.147 | 0 | -0.021 | 0.82774 | 0.426 |
| 16414 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-505-3p;hsa-miR-590-5p | 10 | FNBP1 | Sponge network | 0.225 | 0.30644 | -0.969 | 4.0E-5 | 0.426 |
| 16415 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-940 | 16 | IGF1 | Sponge network | 0.622 | 0.00015 | -0.204 | 0.5898 | 0.426 |
| 16416 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | ST6GAL2 | Sponge network | 0.395 | 0.15451 | -1.201 | 0.00597 | 0.426 |
| 16417 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-942-5p | 30 | NRXN1 | Sponge network | -1.645 | 0 | -3.778 | 0 | 0.426 |
| 16418 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 46 | ZFHX4 | Sponge network | 0.101 | 0.60919 | 0.018 | 0.95574 | 0.426 |
| 16419 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-7-5p;hsa-miR-93-5p | 21 | ZBTB4 | Sponge network | 0.299 | 0.57722 | -0.739 | 0 | 0.426 |
| 16420 | RP11-894P9.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-550a-5p | 19 | ARHGEF12 | Sponge network | 0.993 | 0 | 0.09 | 0.35693 | 0.426 |
| 16421 | CTD-2528L19.6 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | TAF1 | Sponge network | 0.953 | 0 | 0.39 | 3.0E-5 | 0.426 |
| 16422 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | -0.49 | 0.04946 | -0.001 | 0.99669 | 0.426 |
| 16423 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 21 | CBL | Sponge network | 0.539 | 0.03722 | 0.291 | 0.01267 | 0.426 |
| 16424 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-375 | 14 | MLLT10 | Sponge network | 1.03 | 0 | 0.105 | 0.33292 | 0.426 |
| 16425 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | SON | Sponge network | 0.178 | 0.29106 | 0.168 | 0.04406 | 0.426 |
| 16426 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SON | Sponge network | 0.539 | 0.03722 | 0.168 | 0.04406 | 0.426 |
| 16427 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-590-5p | 14 | ABCC9 | Sponge network | -0.384 | 0.40652 | -0.466 | 0.14121 | 0.426 |
| 16428 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | LRRC55 | Sponge network | 0.299 | 0.57722 | -0.026 | 0.93163 | 0.426 |
| 16429 | RP3-337H4.8 |
hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-654-3p | 11 | PHF6 | Sponge network | 1.527 | 0 | 0.785 | 0 | 0.426 |
| 16430 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-93-5p | 27 | ZBTB4 | Sponge network | -0.18 | 0.20343 | -0.739 | 0 | 0.426 |
| 16431 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-96-5p | 27 | CRTC3 | Sponge network | -0.942 | 0 | 0.164 | 0.16786 | 0.426 |
| 16432 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 40 | TSHZ3 | Sponge network | -0.899 | 6.0E-5 | -0.556 | 0.01516 | 0.426 |
| 16433 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | SCN9A | Sponge network | -0.899 | 6.0E-5 | -0.525 | 0.10593 | 0.426 |
| 16434 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 12 | MYO9A | Sponge network | -2.039 | 0.03447 | -0.147 | 0.32373 | 0.426 |
| 16435 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 27 | GPC6 | Sponge network | -1.203 | 0.03881 | -0.054 | 0.81465 | 0.426 |
| 16436 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-454-3p | 15 | MAFB | Sponge network | -0.465 | 0.04703 | -0.023 | 0.89865 | 0.426 |
| 16437 | RP11-1006G14.4 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 21 | RASSF8 | Sponge network | 0.559 | 0.00026 | -0.122 | 0.65591 | 0.426 |
| 16438 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 11 | ZMYM2 | Sponge network | 2.094 | 0 | 0.279 | 0.01273 | 0.426 |
| 16439 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 28 | PGR | Sponge network | 0.082 | 0.55654 | -1.704 | 0 | 0.426 |
| 16440 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TMEFF2 | Sponge network | 0.335 | 0.20596 | -2.426 | 0 | 0.426 |
| 16441 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | MOB1B | Sponge network | 0.064 | 0.72934 | 0.197 | 0.15266 | 0.426 |
| 16442 | PWAR6 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -1.575 | 6.0E-5 | -2.417 | 0 | 0.426 |
| 16443 | RP11-403I13.7 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-877-5p | 19 | NRK | Sponge network | 0.395 | 0.15451 | 0.656 | 0.19217 | 0.426 |
| 16444 | RP11-967K21.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-940 | 11 | MAPK10 | Sponge network | 0.056 | 0.81195 | -1.774 | 0 | 0.426 |
| 16445 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-450b-5p | 11 | DKK3 | Sponge network | 0.381 | 0.25967 | -0.052 | 0.77783 | 0.426 |
| 16446 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | UBE2QL1 | Sponge network | -2.46 | 0 | -2.417 | 0 | 0.426 |
| 16447 | RP11-25K19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p | 12 | ITGB3 | Sponge network | -1.057 | 0.00029 | -0.011 | 0.95751 | 0.426 |
| 16448 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | DCLK1 | Sponge network | 0.101 | 0.60919 | -1.324 | 0.00014 | 0.426 |
| 16449 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-497-5p | 12 | CBL | Sponge network | 0.917 | 0 | 0.291 | 0.01267 | 0.426 |
| 16450 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | ZNF770 | Sponge network | 1.11 | 0 | 0.206 | 0.06751 | 0.426 |
| 16451 | RP11-344B5.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p | 20 | RUNX1T1 | Sponge network | -0.055 | 0.88482 | -0.344 | 0.2374 | 0.426 |
| 16452 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3913-5p;hsa-miR-454-3p | 18 | ESR1 | Sponge network | -2.095 | 0.00112 | -1.035 | 3.0E-5 | 0.426 |
| 16453 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374a-3p | 16 | PIK3R1 | Sponge network | 0.542 | 0.00054 | -0.133 | 0.36854 | 0.426 |
| 16454 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 23 | TIMP3 | Sponge network | -1.249 | 7.0E-5 | -0.546 | 0.02307 | 0.426 |
| 16455 | CTC-429P9.1 |
hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-625-3p | 28 | IL6ST | Sponge network | 0.497 | 0.01451 | -0.402 | 0.00676 | 0.426 |
| 16456 | RP11-359B12.2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 20 | KLF10 | Sponge network | 0.064 | 0.72934 | -0.783 | 0 | 0.426 |
| 16457 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-582-5p;hsa-miR-590-5p | 22 | DMD | Sponge network | 0.889 | 9.0E-5 | -1.506 | 0 | 0.426 |
| 16458 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p | 13 | GREM1 | Sponge network | -0.739 | 0.00044 | 0.902 | 0.01691 | 0.426 |
| 16459 | RP11-356J5.12 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | NR3C2 | Sponge network | -0.899 | 6.0E-5 | -1.56 | 0 | 0.426 |
| 16460 | RP11-822E23.8 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 37 | GRIK3 | Sponge network | -0.777 | 0.16667 | -3.208 | 0 | 0.426 |
| 16461 | HOXB-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-629-3p | 10 | ID4 | Sponge network | -0.154 | 0.53954 | -1.644 | 0 | 0.426 |
| 16462 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p | 16 | MAFB | Sponge network | 0.727 | 3.0E-5 | -0.023 | 0.89865 | 0.426 |
| 16463 | MIR22HG |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p | 21 | JDP2 | Sponge network | -1.047 | 0 | -0.967 | 0 | 0.426 |
| 16464 | RP11-53O19.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-874-3p;hsa-miR-940 | 15 | SORCS1 | Sponge network | -0.405 | 0.22013 | -3.492 | 0 | 0.426 |
| 16465 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 31 | CHD5 | Sponge network | 0.082 | 0.55397 | -0.922 | 0.01521 | 0.426 |
| 16466 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-93-5p | 20 | RNASEL | Sponge network | -0.18 | 0.20343 | -0.272 | 0.01796 | 0.426 |
| 16467 | RP11-403I13.8 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | RASSF8 | Sponge network | 0.619 | 0.00157 | -0.122 | 0.65591 | 0.425 |
| 16468 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p | 12 | DNAJB4 | Sponge network | -0.738 | 0.21233 | -0.7 | 1.0E-5 | 0.425 |
| 16469 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p | 27 | RICTOR | Sponge network | -0.031 | 0.89063 | 0.388 | 0.00188 | 0.425 |
| 16470 | RP11-401P9.4 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-532-3p | 10 | BCL9 | Sponge network | -0.384 | 0.40652 | 0.321 | 0.00267 | 0.425 |
| 16471 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-181a-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320b;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 21 | USP12 | Sponge network | -0.031 | 0.89063 | -0.102 | 0.40768 | 0.425 |
| 16472 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429 | 14 | AHNAK | Sponge network | -2.62 | 0 | -1.207 | 0 | 0.425 |
| 16473 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PTPRB | Sponge network | -1.697 | 0.00889 | 0.105 | 0.47122 | 0.425 |
| 16474 | RP11-620J15.3 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-502-5p;hsa-miR-590-3p | 12 | FMNL3 | Sponge network | -0.739 | 0.00044 | 0.555 | 4.0E-5 | 0.425 |
| 16475 | LINC00865 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 18 | PRUNE2 | Sponge network | -1.249 | 7.0E-5 | -1.733 | 0.00022 | 0.425 |
| 16476 | BVES-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p | 12 | ABL1 | Sponge network | -2.62 | 0 | -0.215 | 0.07804 | 0.425 |
| 16477 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-671-5p | 11 | CADM3 | Sponge network | -0.206 | 0.12942 | -3.663 | 0 | 0.425 |
| 16478 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | FAT4 | Sponge network | 0.178 | 0.29106 | -0.354 | 0.14694 | 0.425 |
| 16479 | EMX2OS |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 19 | TMEFF2 | Sponge network | -0.777 | 0.1134 | -2.426 | 0 | 0.425 |
| 16480 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 21 | DST | Sponge network | -0.021 | 0.93598 | -0.713 | 0.00096 | 0.425 |
| 16481 | RP11-894P9.1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-3607-3p | 12 | RASAL2 | Sponge network | 0.993 | 0 | 0.869 | 0 | 0.425 |
| 16482 | LINC00094 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p | 17 | DYNC1I1 | Sponge network | 0.253 | 0.09565 | -1.12 | 0.00107 | 0.425 |
| 16483 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 47 | TCF4 | Sponge network | 0.657 | 4.0E-5 | 0.016 | 0.92271 | 0.425 |
| 16484 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p | 26 | THRB | Sponge network | -2.076 | 0.00548 | -1.117 | 0.0004 | 0.425 |
| 16485 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p | 12 | DCLK1 | Sponge network | 0.889 | 9.0E-5 | -1.324 | 0.00014 | 0.425 |
| 16486 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | TACC1 | Sponge network | 0.4 | 0.14611 | -0.362 | 0.05406 | 0.425 |
| 16487 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 18 | DYNC1I1 | Sponge network | 0.064 | 0.72934 | -1.12 | 0.00107 | 0.425 |
| 16488 | RP11-400K9.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-340-5p;hsa-miR-744-3p;hsa-miR-940 | 16 | HLF | Sponge network | -0.733 | 0.19469 | -2.443 | 0 | 0.425 |
| 16489 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 47 | QKI | Sponge network | -0.612 | 0.00094 | -0.333 | 0.04111 | 0.425 |
| 16490 | PGM5-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-484 | 34 | DST | Sponge network | -4.546 | 0 | -0.713 | 0.00096 | 0.425 |
| 16491 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-424-5p | 21 | ZFHX3 | Sponge network | 0.556 | 0.00298 | 0.389 | 0.01363 | 0.425 |
| 16492 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 24 | CRTC3 | Sponge network | 1.302 | 7.0E-5 | 0.164 | 0.16786 | 0.425 |
| 16493 | LINC00578 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-589-3p | 10 | FAM172A | Sponge network | 0.811 | 0.03228 | -0.57 | 0 | 0.425 |
| 16494 | RP5-1139B12.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-93-3p | 18 | QKI | Sponge network | 0.598 | 0.38481 | -0.333 | 0.04111 | 0.425 |
| 16495 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 21 | ZFHX4 | Sponge network | 0.178 | 0.29106 | 0.018 | 0.95574 | 0.425 |
| 16496 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ZMYND11 | Sponge network | 0.559 | 0.00026 | -0.2 | 0.05449 | 0.425 |
| 16497 | AP001055.6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 15 | PLSCR4 | Sponge network | 0.693 | 0.00076 | -0.474 | 0.03833 | 0.425 |
| 16498 | RP11-67L2.2 |
hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-92a-3p | 11 | FLNA | Sponge network | 0.72 | 0.0001 | -0.771 | 0.01377 | 0.425 |
| 16499 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-511-5p | 18 | EYA4 | Sponge network | 0.811 | 0.03228 | 0.3 | 0.36876 | 0.425 |
| 16500 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-940 | 28 | HLF | Sponge network | -0.246 | 0.35034 | -2.443 | 0 | 0.425 |
| 16501 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 27 | EPHA3 | Sponge network | 0.537 | 0.00139 | 0.185 | 0.53588 | 0.425 |
| 16502 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | RAP1A | Sponge network | -0.899 | 6.0E-5 | -0.929 | 0 | 0.425 |
| 16503 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-577;hsa-miR-96-5p;hsa-miR-98-5p | 11 | PLAGL1 | Sponge network | -0.769 | 0.00035 | -0.588 | 0.00912 | 0.425 |
| 16504 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-98-5p | 32 | THBS1 | Sponge network | -2.452 | 0 | 0.741 | 0.00386 | 0.425 |
| 16505 | RP11-98I9.4 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | MDM4 | Sponge network | 0.67 | 0.00048 | 0.345 | 0.00762 | 0.425 |
| 16506 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p | 23 | PGR | Sponge network | 0.4 | 0.14611 | -1.704 | 0 | 0.425 |
| 16507 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | PDGFC | Sponge network | 0.194 | 0.64278 | -0.33 | 0.06483 | 0.425 |
| 16508 | RP11-798M19.6 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-93-5p | 57 | DST | Sponge network | -0.612 | 0.00094 | -0.713 | 0.00096 | 0.425 |
| 16509 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p | 25 | AFF3 | Sponge network | 0.101 | 0.60919 | -2.373 | 0 | 0.425 |
| 16510 | LINC00473 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -1.203 | 0.03881 | 0.024 | 0.89889 | 0.425 |
| 16511 | MEG3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 19 | SORCS1 | Sponge network | 0.249 | 0.35902 | -3.492 | 0 | 0.425 |
| 16512 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-629-5p | 22 | GRIN2A | Sponge network | -1.645 | 0 | -2.161 | 0 | 0.425 |
| 16513 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-629-5p | 29 | IGF1 | Sponge network | -2.915 | 0.00015 | -0.204 | 0.5898 | 0.425 |
| 16514 | LINC00092 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 18 | ACSL6 | Sponge network | -1.886 | 0 | -1.593 | 0 | 0.425 |
| 16515 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p | 12 | ATRX | Sponge network | 1.093 | 0 | 0.261 | 0.04408 | 0.425 |
| 16516 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 36 | AKAP11 | Sponge network | -0.323 | 0.01372 | 0.196 | 0.09825 | 0.425 |
| 16517 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 18 | SCN9A | Sponge network | 0.559 | 0.00026 | -0.525 | 0.10593 | 0.425 |
| 16518 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-628-5p | 14 | BCL6 | Sponge network | -0.583 | 0.14589 | -0.211 | 0.19489 | 0.425 |
| 16519 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | LIMD1 | Sponge network | 0.401 | 0.00066 | 0.103 | 0.28978 | 0.425 |
| 16520 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-629-3p | 25 | EBF1 | Sponge network | -0.154 | 0.78939 | -0.384 | 0.08856 | 0.425 |
| 16521 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | EPAS1 | Sponge network | -0.737 | 0.06182 | -0.184 | 0.14418 | 0.425 |
| 16522 | FENDRR |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 16 | SPTBN4 | Sponge network | -1.281 | 0.00012 | -0.786 | 0.01281 | 0.425 |
| 16523 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | TGFBR2 | Sponge network | -0.125 | 0.49414 | -0.243 | 0.11408 | 0.425 |
| 16524 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p | 21 | IGF1 | Sponge network | -1.057 | 0.00029 | -0.204 | 0.5898 | 0.425 |
| 16525 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 20 | SMARCA2 | Sponge network | -0.615 | 0.00015 | -0.529 | 0.00014 | 0.425 |
| 16526 | LINC00957 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 16 | CDH19 | Sponge network | -0.421 | 0.0114 | -3.434 | 0 | 0.425 |
| 16527 | MIR497HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 44 | PTPRD | Sponge network | -1.439 | 4.0E-5 | -1.005 | 0.00064 | 0.425 |
| 16528 | GAS6-AS2 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-5p;hsa-miR-874-3p | 17 | CTIF | Sponge network | 0.16 | 0.50785 | -0.389 | 0.00549 | 0.425 |
| 16529 | LINC00957 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-93-3p | 25 | ERC1 | Sponge network | -0.421 | 0.0114 | -0.108 | 0.38794 | 0.425 |
| 16530 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | CAV1 | Sponge network | -2.117 | 0.00161 | -1.013 | 6.0E-5 | 0.425 |
| 16531 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 30 | ZBTB4 | Sponge network | 0.249 | 0.35902 | -0.739 | 0 | 0.425 |
| 16532 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | AKAP6 | Sponge network | -0.342 | 0.02288 | -1.586 | 3.0E-5 | 0.425 |
| 16533 | U91328.19 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 24 | PCDH10 | Sponge network | -0.303 | 0.21002 | -1.644 | 0.0078 | 0.425 |
| 16534 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 44 | HLF | Sponge network | -1.216 | 0 | -2.443 | 0 | 0.425 |
| 16535 | A1BG-AS1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-362-3p;hsa-miR-425-5p | 15 | MEF2C | Sponge network | -0.164 | 0.45106 | -0.499 | 0.01448 | 0.425 |
| 16536 | NR2F1-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | PDE4DIP | Sponge network | -0.49 | 0.04946 | -0.282 | 0.0257 | 0.425 |
| 16537 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 28 | DTNA | Sponge network | -0.726 | 0.00132 | -1.648 | 1.0E-5 | 0.425 |
| 16538 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-766-3p | 15 | KIAA1024 | Sponge network | 0.421 | 0.00084 | 1.083 | 0 | 0.425 |
| 16539 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | HERC2 | Sponge network | 1.254 | 0 | 0.114 | 0.30638 | 0.425 |
| 16540 | LINC00641 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | DCLK1 | Sponge network | -0.222 | 0.32445 | -1.324 | 0.00014 | 0.425 |
| 16541 | NUTM2A-AS1 |
hsa-miR-125a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | ERCC6 | Sponge network | 0.643 | 0 | 0.23 | 0.015 | 0.425 |
| 16542 | RP11-6O2.3 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-361-3p | 16 | ITPKB | Sponge network | 0.331 | 0.57247 | -0.704 | 0.00106 | 0.425 |
| 16543 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-935 | 10 | LRRN3 | Sponge network | 0.083 | 0.66405 | -0.393 | 0.16293 | 0.425 |
| 16544 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 45 | SOX5 | Sponge network | 0.194 | 0.64278 | -1.091 | 4.0E-5 | 0.425 |
| 16545 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 22 | TGFBR2 | Sponge network | -0.739 | 0.0574 | -0.243 | 0.11408 | 0.425 |
| 16546 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 19 | TMEFF2 | Sponge network | 0.997 | 0.00066 | -2.426 | 0 | 0.425 |
| 16547 | RASSF8-AS1 |
hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PCDH18 | Sponge network | 0.335 | 0.20596 | 0.216 | 0.24645 | 0.425 |
| 16548 | LINC00865 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-940 | 15 | CELF4 | Sponge network | -1.249 | 7.0E-5 | -1.135 | 0.00344 | 0.425 |
| 16549 | PWAR6 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p | 11 | ALDH1A2 | Sponge network | -1.575 | 6.0E-5 | -2.411 | 0 | 0.425 |
| 16550 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | 1.601 | 0 | 0.323 | 0.00792 | 0.425 |
| 16551 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | SSBP2 | Sponge network | -0.342 | 0.02288 | -1.58 | 0 | 0.425 |
| 16552 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-493-3p;hsa-miR-590-3p | 12 | JAKMIP2 | Sponge network | -0.899 | 6.0E-5 | -1.388 | 0 | 0.425 |
| 16553 | TPTEP1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-425-3p | 12 | DUSP1 | Sponge network | -1.216 | 0 | -1.754 | 0 | 0.425 |
| 16554 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 21 | RASSF8 | Sponge network | 0.331 | 0.57247 | -0.122 | 0.65591 | 0.425 |
| 16555 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | CNTN1 | Sponge network | -0.727 | 0.04726 | -2.594 | 0 | 0.425 |
| 16556 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-664a-3p | 21 | ITCH | Sponge network | 0.421 | 0.00084 | 0.084 | 0.34058 | 0.425 |
| 16557 | CTC-510F12.2 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-181a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p | 11 | MCC | Sponge network | -0.691 | 0.00146 | -1.005 | 0 | 0.425 |
| 16558 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p | 38 | TCF4 | Sponge network | 1.757 | 0 | 0.016 | 0.92271 | 0.425 |
| 16559 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-590-3p | 17 | DMXL1 | Sponge network | 0.178 | 0.29106 | -0.426 | 0.00056 | 0.425 |
| 16560 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | 0.572 | 0.01722 | 0.309 | 0.17079 | 0.425 |
| 16561 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 31 | FOXP1 | Sponge network | 0.559 | 0.00026 | 0.042 | 0.74368 | 0.425 |
| 16562 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 21 | NRXN3 | Sponge network | 1.302 | 7.0E-5 | -0.893 | 0.04186 | 0.425 |
| 16563 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NCAM2 | Sponge network | 0.532 | 0.00505 | -0.482 | 0.22565 | 0.425 |
| 16564 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | NEDD4 | Sponge network | 0.335 | 0.20596 | 0.02 | 0.89834 | 0.425 |
| 16565 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-877-5p | 27 | TET1 | Sponge network | -0.162 | 0.47687 | -0.135 | 0.52138 | 0.425 |
| 16566 | RP13-516M14.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-7-1-3p | 13 | AKAP6 | Sponge network | 0.389 | 0.04293 | -1.586 | 3.0E-5 | 0.425 |
| 16567 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | CBL | Sponge network | 1.294 | 0 | 0.291 | 0.01267 | 0.425 |
| 16568 | LINC00865 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 23 | PCDH10 | Sponge network | -1.249 | 7.0E-5 | -1.644 | 0.0078 | 0.425 |
| 16569 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | AKAP12 | Sponge network | -0.053 | 0.80685 | -0.932 | 0.0028 | 0.425 |
| 16570 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | USP12 | Sponge network | 0.532 | 0.00505 | -0.102 | 0.40768 | 0.425 |
| 16571 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-589-3p | 16 | TAL1 | Sponge network | -0.154 | 0.78939 | -0.462 | 0.00574 | 0.425 |
| 16572 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 32 | TACC1 | Sponge network | 0.259 | 0.12542 | -0.362 | 0.05406 | 0.425 |
| 16573 | RP11-439A17.10 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p | 14 | DTNA | Sponge network | 0.051 | 0.95756 | -1.648 | 1.0E-5 | 0.425 |
| 16574 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-93-5p;hsa-miR-935 | 17 | ARNT2 | Sponge network | -0.384 | 0.40652 | -0.332 | 0.25851 | 0.425 |
| 16575 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-935 | 45 | FAT3 | Sponge network | -0.303 | 0.21002 | -1.601 | 0.00051 | 0.425 |
| 16576 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | EPHA7 | Sponge network | 0.249 | 0.35902 | -2.197 | 3.0E-5 | 0.425 |
| 16577 | RP4-545C24.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-744-3p;hsa-miR-96-5p | 13 | HIP1 | Sponge network | 0.363 | 0.02231 | 0.774 | 0 | 0.425 |
| 16578 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | RABEP1 | Sponge network | 0.064 | 0.72934 | -0.066 | 0.43142 | 0.425 |
| 16579 | RP11-307B6.3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-589-3p | 14 | PRKCB | Sponge network | -4.463 | 0.00025 | -1.313 | 2.0E-5 | 0.425 |
| 16580 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 15 | RCAN2 | Sponge network | 0.246 | 0.59912 | -0.33 | 0.15172 | 0.425 |
| 16581 | RP11-758N13.1 |
hsa-miR-103a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-942-5p | 13 | GPC6 | Sponge network | 2.939 | 3.0E-5 | -0.054 | 0.81465 | 0.425 |
| 16582 | OIP5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 45 | ARHGEF12 | Sponge network | 0.421 | 0.00084 | 0.09 | 0.35693 | 0.425 |
| 16583 | LINC00578 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-589-3p | 12 | SORCS1 | Sponge network | 0.811 | 0.03228 | -3.492 | 0 | 0.425 |
| 16584 | TP73-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 14 | INPP4A | Sponge network | -0.563 | 0.02545 | 0.07 | 0.53563 | 0.425 |
| 16585 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-629-3p | 26 | EBF1 | Sponge network | -0.777 | 0.16667 | -0.384 | 0.08856 | 0.425 |
| 16586 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-629-3p | 14 | SPTBN4 | Sponge network | -0.976 | 0.00025 | -0.786 | 0.01281 | 0.425 |
| 16587 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-484 | 15 | RET | Sponge network | -2.69 | 0.0001 | -0.833 | 0.03517 | 0.425 |
| 16588 | GS1-124K5.11 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-877-5p | 12 | INPP4A | Sponge network | 0.494 | 0.00339 | 0.07 | 0.53563 | 0.425 |
| 16589 | AGAP11 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p | 12 | FLNA | Sponge network | -1.645 | 0 | -0.771 | 0.01377 | 0.425 |
| 16590 | LINC00476 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p | 26 | ZMYND11 | Sponge network | -0.615 | 0.00015 | -0.2 | 0.05449 | 0.425 |
| 16591 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-455-3p | 20 | MAF | Sponge network | -0.72 | 0.24239 | -1.257 | 0 | 0.425 |
| 16592 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-493-5p | 11 | PGR | Sponge network | -0.829 | 0.01343 | -1.704 | 0 | 0.425 |
| 16593 | EMX2OS |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | CDH10 | Sponge network | -0.777 | 0.1134 | -2.397 | 0 | 0.425 |
| 16594 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 21 | PRKAA2 | Sponge network | -2.039 | 0.03447 | -2.462 | 0 | 0.425 |
| 16595 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-511-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-671-5p | 35 | RBMS3 | Sponge network | 0.331 | 0.57247 | -0.519 | 0.03551 | 0.425 |
| 16596 | MYLK-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-424-5p;hsa-miR-550a-5p | 32 | CCND2 | Sponge network | -1.331 | 3.0E-5 | -0.267 | 0.3112 | 0.425 |
| 16597 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | PRKAB2 | Sponge network | 0.225 | 0.30644 | -0.367 | 0.01371 | 0.425 |
| 16598 | LINC00641 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p | 28 | ABL2 | Sponge network | -0.222 | 0.32445 | 0.82 | 0 | 0.425 |
| 16599 | SNX29P2 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | ERG | Sponge network | 0.4 | 0.14611 | 0.002 | 0.99011 | 0.425 |
| 16600 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 21 | DDX6 | Sponge network | 0.539 | 0.03722 | 0.091 | 0.36428 | 0.425 |
| 16601 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 37 | TGFBR3 | Sponge network | 0.083 | 0.66405 | -1.173 | 1.0E-5 | 0.425 |
| 16602 | TRAF3IP2-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940 | 12 | RHOB | Sponge network | -0.323 | 0.01372 | -1.052 | 0 | 0.425 |
| 16603 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 13 | CAV1 | Sponge network | -1.249 | 7.0E-5 | -1.013 | 6.0E-5 | 0.425 |
| 16604 | LINC00957 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 17 | LRRC7 | Sponge network | -0.421 | 0.0114 | -1.341 | 0.01193 | 0.425 |
| 16605 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 32 | USP12 | Sponge network | -0.129 | 0.57177 | -0.102 | 0.40768 | 0.425 |
| 16606 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-421 | 11 | RELN | Sponge network | -0.351 | 0.11358 | -2.403 | 0 | 0.425 |
| 16607 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-511-5p | 15 | ATRX | Sponge network | 0.997 | 0 | 0.261 | 0.04408 | 0.425 |
| 16608 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-629-5p | 11 | GRIN2A | Sponge network | -0.517 | 0.02935 | -2.161 | 0 | 0.425 |
| 16609 | LINC00672 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p | 39 | MDM4 | Sponge network | -0.227 | 0.31657 | 0.345 | 0.00762 | 0.425 |
| 16610 | RP11-822E23.8 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p | 11 | KLF10 | Sponge network | -0.777 | 0.16667 | -0.783 | 0 | 0.425 |
| 16611 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 40 | BNC2 | Sponge network | 0.395 | 0.15451 | -0.491 | 0.09988 | 0.425 |
| 16612 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p | 19 | AKAP12 | Sponge network | -2.925 | 0 | -0.932 | 0.0028 | 0.425 |
| 16613 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p | 14 | PLSCR4 | Sponge network | 0.342 | 0.03129 | -0.474 | 0.03833 | 0.425 |
| 16614 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-671-5p | 37 | ADARB1 | Sponge network | 0.331 | 0.57247 | -0.335 | 0.06631 | 0.425 |
| 16615 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p | 20 | SLITRK3 | Sponge network | -0.323 | 0.01372 | -3.328 | 0 | 0.425 |
| 16616 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | -0.899 | 6.0E-5 | -0.7 | 1.0E-5 | 0.425 |
| 16617 | RP11-25K19.1 |
hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 12 | CXCL12 | Sponge network | -1.057 | 0.00029 | -1.421 | 0 | 0.425 |
| 16618 | RP11-158K1.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PIK3CA | Sponge network | 0.349 | 0.01695 | 0.195 | 0.06908 | 0.425 |
| 16619 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p | 11 | PRDM5 | Sponge network | -0.738 | 0.21233 | -1.09 | 0.00014 | 0.424 |
| 16620 | PDXDC2P |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 17 | TBL1XR1 | Sponge network | 0.756 | 0 | 0.578 | 0 | 0.424 |
| 16621 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | CLASP2 | Sponge network | 0.614 | 1.0E-5 | -0.038 | 0.71 | 0.424 |
| 16622 | MEG3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429 | 18 | DYNC1I1 | Sponge network | 0.249 | 0.35902 | -1.12 | 0.00107 | 0.424 |
| 16623 | PCA3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | RELN | Sponge network | -0.709 | 0.18168 | -2.403 | 0 | 0.424 |
| 16624 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429 | 15 | CNTNAP3 | Sponge network | -0.154 | 0.53954 | -1.465 | 0.00033 | 0.424 |
| 16625 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-877-5p;hsa-miR-940 | 16 | GAS7 | Sponge network | 0.043 | 0.81628 | -0.555 | 0.01602 | 0.424 |
| 16626 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-195-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-23b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 10 | EIF1AX | Sponge network | 1.254 | 0 | 0.307 | 0.011 | 0.424 |
| 16627 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -1.349 | 0.00017 | -1.692 | 0 | 0.424 |
| 16628 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | TCF4 | Sponge network | -0.021 | 0.93598 | 0.016 | 0.92271 | 0.424 |
| 16629 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-625-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -0.096 | 0.67958 | -0.087 | 0.69193 | 0.424 |
| 16630 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 54 | TMEM132B | Sponge network | -0.129 | 0.57177 | -0.83 | 0.01084 | 0.424 |
| 16631 | PART1 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ADH1B | Sponge network | -2.925 | 0 | -3.716 | 0 | 0.424 |
| 16632 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 37 | ESR1 | Sponge network | -0.777 | 0.1134 | -1.035 | 3.0E-5 | 0.424 |
| 16633 | RP11-705C15.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-5p | 12 | CYLD | Sponge network | 0.796 | 4.0E-5 | -0.367 | 0.00171 | 0.424 |
| 16634 | AC010226.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | ASTN1 | Sponge network | -0.811 | 2.0E-5 | -2.389 | 2.0E-5 | 0.424 |
| 16635 | CTC-429P9.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-550a-3p | 22 | DIP2C | Sponge network | 0.082 | 0.55397 | -0.436 | 0.01713 | 0.424 |
| 16636 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-96-5p | 28 | EPHA3 | Sponge network | 0.082 | 0.55654 | 0.185 | 0.53588 | 0.424 |
| 16637 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | NRXN1 | Sponge network | 0.299 | 0.57722 | -3.778 | 0 | 0.424 |
| 16638 | RP11-798M19.6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-877-5p;hsa-miR-93-5p | 31 | IL17RD | Sponge network | -0.612 | 0.00094 | -0.019 | 0.94873 | 0.424 |
| 16639 | DIO3OS |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 14 | NGFR | Sponge network | -0.773 | 0.04073 | -1.271 | 0.01058 | 0.424 |
| 16640 | WDFY3-AS2 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | LIMCH1 | Sponge network | -0.942 | 0 | -0.915 | 1.0E-5 | 0.424 |
| 16641 | RP11-822E23.8 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-877-5p | 26 | ASTN1 | Sponge network | -0.777 | 0.16667 | -2.389 | 2.0E-5 | 0.424 |
| 16642 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 21 | LRRC7 | Sponge network | -0.793 | 7.0E-5 | -1.341 | 0.01193 | 0.424 |
| 16643 | AC009120.6 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | CREB1 | Sponge network | 0.919 | 0 | 0.203 | 0.00537 | 0.424 |
| 16644 | DNAJC27-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 27 | AKAP6 | Sponge network | -0.969 | 2.0E-5 | -1.586 | 3.0E-5 | 0.424 |
| 16645 | FGD5-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | PHF3 | Sponge network | 0.401 | 0.00066 | 0.126 | 0.19652 | 0.424 |
| 16646 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | EPHA5 | Sponge network | 0.246 | 0.59912 | -0.957 | 0.01733 | 0.424 |
| 16647 | RASSF8-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 15 | EZH1 | Sponge network | 0.335 | 0.20596 | -0.564 | 1.0E-5 | 0.424 |
| 16648 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-532-3p | 19 | RPS6KA2 | Sponge network | -0.096 | 0.67958 | -0.57 | 0.00013 | 0.424 |
| 16649 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p | 11 | RELN | Sponge network | -4.463 | 0.00025 | -2.403 | 0 | 0.424 |
| 16650 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 53 | CADM2 | Sponge network | -0.487 | 0.02157 | -3.782 | 0 | 0.424 |
| 16651 | AC093627.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29b-1-5p | 14 | GAS7 | Sponge network | -0.787 | 0.3928 | -0.555 | 0.01602 | 0.424 |
| 16652 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -1.216 | 0 | 0.024 | 0.89889 | 0.424 |
| 16653 | SERTAD4-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-424-5p | 22 | TGFBR3 | Sponge network | -0.932 | 0.00329 | -1.173 | 1.0E-5 | 0.424 |
| 16654 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935 | 32 | ABI2 | Sponge network | -0.896 | 0.00099 | 0.175 | 0.14787 | 0.424 |
| 16655 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 71 | AFF4 | Sponge network | 0.421 | 0.00084 | -0.266 | 0.01565 | 0.424 |
| 16656 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 35 | THRB | Sponge network | -4.546 | 0 | -1.117 | 0.0004 | 0.424 |
| 16657 | MIR22HG |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-542-3p | 17 | TLE4 | Sponge network | -1.047 | 0 | -1.157 | 0 | 0.424 |
| 16658 | EMX2OS |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | NOV | Sponge network | -0.777 | 0.1134 | -0.58 | 0.04676 | 0.424 |
| 16659 | BZRAP1-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-3p;hsa-miR-485-3p | 10 | RABEP1 | Sponge network | -0.465 | 0.04703 | -0.066 | 0.43142 | 0.424 |
| 16660 | AC018890.6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | TET1 | Sponge network | 1.61 | 0.00961 | -0.135 | 0.52138 | 0.424 |
| 16661 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 17 | UNC5C | Sponge network | -1.439 | 4.0E-5 | -0.178 | 0.40214 | 0.424 |
| 16662 | TUG1 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-654-3p | 12 | DICER1 | Sponge network | 0.972 | 0 | 0.042 | 0.674 | 0.424 |
| 16663 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-653-5p | 16 | PDS5B | Sponge network | -0.222 | 0.32445 | 0.259 | 0.00962 | 0.424 |
| 16664 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | NEDD4 | Sponge network | -0.901 | 0.00019 | 0.02 | 0.89834 | 0.424 |
| 16665 | BDNF-AS |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 15 | SLITRK6 | Sponge network | -0.668 | 3.0E-5 | -2.121 | 0 | 0.424 |
| 16666 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | THBS1 | Sponge network | -0.376 | 0.00337 | 0.741 | 0.00386 | 0.424 |
| 16667 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 20 | TCF4 | Sponge network | -1.255 | 0.00981 | 0.016 | 0.92271 | 0.424 |
| 16668 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-93-5p | 33 | AR | Sponge network | 0.381 | 0.25967 | -1.554 | 0 | 0.424 |
| 16669 | NEAT1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SON | Sponge network | 0.705 | 0.00014 | 0.168 | 0.04406 | 0.424 |
| 16670 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-532-3p;hsa-miR-671-5p | 13 | CADM3 | Sponge network | -0.513 | 0.04703 | -3.663 | 0 | 0.424 |
| 16671 | RP11-620J15.3 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | GJA1 | Sponge network | -0.739 | 0.00044 | -0.485 | 0.02179 | 0.424 |
| 16672 | PWAR6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SEMA3E | Sponge network | -1.575 | 6.0E-5 | -1.717 | 0.00137 | 0.424 |
| 16673 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-96-5p | 56 | BMPR2 | Sponge network | -0.976 | 0.00025 | 0.206 | 0.0635 | 0.424 |
| 16674 | CTA-14H9.5 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 17 | DTNA | Sponge network | 0.145 | 0.53343 | -1.648 | 1.0E-5 | 0.424 |
| 16675 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | APC | Sponge network | -0.83 | 0 | -0.255 | 0.04051 | 0.424 |
| 16676 | PCA3 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | RIMS2 | Sponge network | -0.709 | 0.18168 | -1.365 | 0.00208 | 0.424 |
| 16677 | RP11-589P10.7 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-505-3p;hsa-miR-93-3p | 13 | IL6ST | Sponge network | -2.044 | 0.00066 | -0.402 | 0.00676 | 0.424 |
| 16678 | PART1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | SPG20 | Sponge network | -2.925 | 0 | -1.16 | 0 | 0.424 |
| 16679 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PEG3 | Sponge network | -1.057 | 0.00029 | -1.74 | 0 | 0.424 |
| 16680 | PGM5-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-505-5p;hsa-miR-940 | 12 | TOM1L2 | Sponge network | -4.546 | 0 | -0.994 | 0 | 0.424 |
| 16681 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-744-3p | 27 | FAT3 | Sponge network | 0.178 | 0.29106 | -1.601 | 0.00051 | 0.424 |
| 16682 | RP11-736K20.5 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-590-3p | 11 | ATM | Sponge network | -0.358 | 0.17255 | 0.178 | 0.21568 | 0.424 |
| 16683 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p | 31 | RBMS3 | Sponge network | -0.465 | 0.04703 | -0.519 | 0.03551 | 0.424 |
| 16684 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 36 | MEF2C | Sponge network | -1.203 | 0.03881 | -0.499 | 0.01448 | 0.424 |
| 16685 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-940 | 30 | AKAP11 | Sponge network | 0.082 | 0.55654 | 0.196 | 0.09825 | 0.424 |
| 16686 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | SON | Sponge network | 0.924 | 0 | 0.168 | 0.04406 | 0.424 |
| 16687 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-7-1-3p | 16 | ADAMTSL3 | Sponge network | -4.477 | 0 | -1.559 | 0 | 0.424 |
| 16688 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | ST6GALNAC3 | Sponge network | -1.216 | 0 | -0.987 | 0 | 0.424 |
| 16689 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 25 | MOB1B | Sponge network | 0.389 | 0.04293 | 0.197 | 0.15266 | 0.424 |
| 16690 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | TRIP11 | Sponge network | -0.222 | 0.32445 | -0.158 | 0.06384 | 0.424 |
| 16691 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-590-3p | 19 | TSHZ3 | Sponge network | 0.178 | 0.29106 | -0.556 | 0.01516 | 0.424 |
| 16692 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 42 | ADARB1 | Sponge network | -1.203 | 0.03881 | -0.335 | 0.06631 | 0.424 |
| 16693 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 42 | RAP1A | Sponge network | -1.111 | 0.0003 | -0.929 | 0 | 0.424 |
| 16694 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-493-5p | 11 | PHC3 | Sponge network | 1.46 | 1.0E-5 | 0.212 | 0.0499 | 0.424 |
| 16695 | RP11-504P24.8 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 12 | PHF6 | Sponge network | 1.178 | 0 | 0.785 | 0 | 0.424 |
| 16696 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-625-5p | 29 | TGFBR3 | Sponge network | -0.469 | 0.08721 | -1.173 | 1.0E-5 | 0.424 |
| 16697 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 23 | CXXC4 | Sponge network | -1.249 | 7.0E-5 | -0.433 | 0.21055 | 0.424 |
| 16698 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-500a-3p | 11 | AFF3 | Sponge network | -4.477 | 0 | -2.373 | 0 | 0.424 |
| 16699 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-98-5p | 20 | PNRC1 | Sponge network | -1.111 | 0.0003 | -0.646 | 0 | 0.424 |
| 16700 | TRAF3IP2-AS1 |
hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | CSDE1 | Sponge network | -0.323 | 0.01372 | -0.493 | 0 | 0.424 |
| 16701 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | HOOK3 | Sponge network | 0.421 | 0.00084 | 0.273 | 0.09957 | 0.424 |
| 16702 | RP11-701H24.3 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 22 | IL6ST | Sponge network | -1.425 | 0.04204 | -0.402 | 0.00676 | 0.424 |
| 16703 | AP001347.6 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ERG | Sponge network | -1.161 | 0.00591 | 0.002 | 0.99011 | 0.424 |
| 16704 | RP11-175O19.4 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-376c-3p | 10 | DICER1 | Sponge network | 0.917 | 0 | 0.042 | 0.674 | 0.424 |
| 16705 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-629-3p | 17 | GPC6 | Sponge network | -0.154 | 0.53954 | -0.054 | 0.81465 | 0.424 |
| 16706 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 16 | CNTNAP1 | Sponge network | 0.083 | 0.66405 | 0.358 | 0.13727 | 0.424 |
| 16707 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | ARHGAP28 | Sponge network | -0.513 | 0.04703 | -0.911 | 0.00013 | 0.424 |
| 16708 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | ZFHX4 | Sponge network | -0.053 | 0.80685 | 0.018 | 0.95574 | 0.424 |
| 16709 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 31 | JAZF1 | Sponge network | -1.654 | 0.00144 | -0.377 | 0.02677 | 0.424 |
| 16710 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | SNX29 | Sponge network | 0.385 | 0.10067 | -0.34 | 0.01371 | 0.424 |
| 16711 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | 0.997 | 0.00066 | 0.126 | 0.15266 | 0.424 |
| 16712 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 16 | ITGA5 | Sponge network | -0.769 | 0.00035 | 0.452 | 0.03524 | 0.424 |
| 16713 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-5p | 16 | LATS1 | Sponge network | 0.078 | 0.55803 | 0.109 | 0.34208 | 0.424 |
| 16714 | LINC00702 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 57 | PTPRD | Sponge network | -0.737 | 0.06182 | -1.005 | 0.00064 | 0.424 |
| 16715 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ZFHX3 | Sponge network | 0.349 | 0.01695 | 0.389 | 0.01363 | 0.424 |
| 16716 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | SOX5 | Sponge network | -0.709 | 0.18168 | -1.091 | 4.0E-5 | 0.424 |
| 16717 | PCA3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-590-3p | 15 | ROBO1 | Sponge network | -0.709 | 0.18168 | -0.009 | 0.96154 | 0.424 |
| 16718 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 50 | SSBP2 | Sponge network | -0.777 | 0.1134 | -1.58 | 0 | 0.424 |
| 16719 | RP11-248G5.8 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-497-5p | 13 | CDK6 | Sponge network | 1.294 | 0 | 0.991 | 0 | 0.424 |
| 16720 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 41 | EPHA7 | Sponge network | 0.082 | 0.55654 | -2.197 | 3.0E-5 | 0.424 |
| 16721 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 67 | WDFY3 | Sponge network | -0.739 | 0.0574 | -0.201 | 0.0967 | 0.424 |
| 16722 | RP11-983P16.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-940 | 31 | NTRK2 | Sponge network | 0.41 | 0.02214 | -0.736 | 0.06495 | 0.424 |
| 16723 | LINC00473 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | KCNA6 | Sponge network | -1.203 | 0.03881 | -1.531 | 0.00013 | 0.424 |
| 16724 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-877-5p;hsa-miR-93-5p | 43 | AR | Sponge network | -0.384 | 0.40652 | -1.554 | 0 | 0.424 |
| 16725 | LINC00092 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p | 15 | CADM1 | Sponge network | -1.886 | 0 | -0.9 | 0.00084 | 0.424 |
| 16726 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-508-3p | 16 | CACNA2D1 | Sponge network | -0.154 | 0.53954 | -0.846 | 0.00675 | 0.424 |
| 16727 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-3614-5p;hsa-miR-628-5p;hsa-miR-93-3p | 12 | EPHA3 | Sponge network | -1.116 | 0.23686 | 0.185 | 0.53588 | 0.424 |
| 16728 | RP11-307B6.3 |
hsa-let-7f-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-942-5p | 27 | THRB | Sponge network | -4.463 | 0.00025 | -1.117 | 0.0004 | 0.424 |
| 16729 | MIR143HG |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | GALNT13 | Sponge network | -0.739 | 0.0574 | -0.857 | 0.07439 | 0.424 |
| 16730 | RP5-1139B12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-744-5p;hsa-miR-942-5p | 17 | NOTCH2 | Sponge network | 0.598 | 0.38481 | 0.138 | 0.35163 | 0.424 |
| 16731 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-93-5p | 27 | ST6GALNAC3 | Sponge network | -0.777 | 0.16667 | -0.987 | 0 | 0.424 |
| 16732 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | ZMYM2 | Sponge network | 1.317 | 0 | 0.279 | 0.01273 | 0.424 |
| 16733 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | KAT6B | Sponge network | 0.782 | 0.00016 | -0.478 | 0.00018 | 0.424 |
| 16734 | RP11-110I1.11 |
hsa-let-7d-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-532-3p | 10 | CBL | Sponge network | 1.034 | 0 | 0.291 | 0.01267 | 0.424 |
| 16735 | BVES-AS1 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-708-5p | 18 | FAM135B | Sponge network | -2.62 | 0 | -3.978 | 0 | 0.424 |
| 16736 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p | 15 | FGFR1 | Sponge network | -0.737 | 0.10822 | -0.509 | 0.03957 | 0.424 |
| 16737 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-33a-3p | 17 | DDX6 | Sponge network | 2.163 | 0 | 0.091 | 0.36428 | 0.424 |
| 16738 | RP11-156P1.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 39 | GRIK3 | Sponge network | 0.214 | 0.18246 | -3.208 | 0 | 0.424 |
| 16739 | CTD-3064M3.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-629-3p | 37 | DST | Sponge network | -0.469 | 0.08721 | -0.713 | 0.00096 | 0.424 |
| 16740 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p | 14 | PHC3 | Sponge network | 0.37 | 0.27614 | 0.212 | 0.0499 | 0.424 |
| 16741 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-495-3p | 10 | BCLAF1 | Sponge network | 0.972 | 0 | 0.211 | 0.00684 | 0.424 |
| 16742 | CTC-296K1.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-744-3p | 22 | DIP2C | Sponge network | -2.095 | 0.00112 | -0.436 | 0.01713 | 0.424 |
| 16743 | AC005562.1 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ITGB1 | Sponge network | 0.488 | 0.00084 | 0.524 | 0.00017 | 0.424 |
| 16744 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-877-5p | 21 | SUFU | Sponge network | -0.096 | 0.67958 | -0.04 | 0.65489 | 0.424 |
| 16745 | AC004540.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p | 12 | CXXC4 | Sponge network | -2.549 | 0.00528 | -0.433 | 0.21055 | 0.424 |
| 16746 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | PLSCR4 | Sponge network | -0.303 | 0.21002 | -0.474 | 0.03833 | 0.424 |
| 16747 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 11 | PIK3IP1 | Sponge network | -1.439 | 4.0E-5 | -0.187 | 0.19945 | 0.424 |
| 16748 | HNRNPU-AS1 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 18 | DICER1 | Sponge network | 1.601 | 0 | 0.042 | 0.674 | 0.424 |
| 16749 | RP5-887A10.1 |
hsa-miR-148b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-584-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-940 | 10 | PTPRT | Sponge network | 0.546 | 0.4892 | -0.907 | 0.03727 | 0.424 |
| 16750 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | NEDD4 | Sponge network | -1.111 | 0.0003 | 0.02 | 0.89834 | 0.424 |
| 16751 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | CHD2 | Sponge network | 1.04 | 0.00023 | 0.049 | 0.55371 | 0.424 |
| 16752 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-589-3p;hsa-miR-940 | 12 | GLI3 | Sponge network | 0.331 | 0.57247 | -0.615 | 0.01482 | 0.424 |
| 16753 | MEG3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | FYN | Sponge network | 0.249 | 0.35902 | -0.131 | 0.37051 | 0.424 |
| 16754 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-942-5p | 23 | ARHGAP28 | Sponge network | -1.249 | 7.0E-5 | -0.911 | 0.00013 | 0.423 |
| 16755 | MRVI1-AS1 |
hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-500a-3p;hsa-miR-7-1-3p | 15 | LIMCH1 | Sponge network | -1.092 | 4.0E-5 | -0.915 | 1.0E-5 | 0.423 |
| 16756 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 36 | RBMS3 | Sponge network | 0.214 | 0.18246 | -0.519 | 0.03551 | 0.423 |
| 16757 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p | 18 | RET | Sponge network | -0.842 | 0.01361 | -0.833 | 0.03517 | 0.423 |
| 16758 | SNHG14 |
hsa-miR-146b-5p;hsa-miR-24-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-92a-3p | 10 | GPX3 | Sponge network | -1.111 | 0.0003 | -2.752 | 0 | 0.423 |
| 16759 | LINC00654 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 31 | ABL2 | Sponge network | 0.374 | 0.20054 | 0.82 | 0 | 0.423 |
| 16760 | RP11-53O19.1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-493-3p | 15 | CSDE1 | Sponge network | -0.405 | 0.22013 | -0.493 | 0 | 0.423 |
| 16761 | KB-1460A1.5 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 10 | MACF1 | Sponge network | 0.603 | 0.00127 | 0.019 | 0.90335 | 0.423 |
| 16762 | CTD-2314G24.2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | 0.246 | 0.59912 | -0.704 | 0.00106 | 0.423 |
| 16763 | LINC00702 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-940 | 10 | DIRAS1 | Sponge network | -0.737 | 0.06182 | -2.518 | 0 | 0.423 |
| 16764 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940 | 25 | CRTC3 | Sponge network | -0.777 | 0.1134 | 0.164 | 0.16786 | 0.423 |
| 16765 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | LATS2 | Sponge network | -0.901 | 0.00019 | 0.159 | 0.2304 | 0.423 |
| 16766 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-877-5p | 18 | CDH19 | Sponge network | -0.777 | 0.16667 | -3.434 | 0 | 0.423 |
| 16767 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-421 | 11 | PGR | Sponge network | -4.477 | 0 | -1.704 | 0 | 0.423 |
| 16768 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-7-1-3p | 10 | CLIP1 | Sponge network | 0.299 | 0.57722 | -0.215 | 0.08684 | 0.423 |
| 16769 | RP11-536O18.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-330-3p | 12 | TSHZ3 | Sponge network | -0.074 | 0.90758 | -0.556 | 0.01516 | 0.423 |
| 16770 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-671-5p | 10 | CADM3 | Sponge network | -0.351 | 0.11358 | -3.663 | 0 | 0.423 |
| 16771 | FZD10-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 28 | CADM1 | Sponge network | -0.727 | 0.04726 | -0.9 | 0.00084 | 0.423 |
| 16772 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374a-5p | 12 | SEMA3C | Sponge network | -0.693 | 0.05629 | -0.272 | 0.31153 | 0.423 |
| 16773 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | 0.219 | 0.1516 | -0.474 | 0.03833 | 0.423 |
| 16774 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | USP12 | Sponge network | 0.673 | 0 | -0.102 | 0.40768 | 0.423 |
| 16775 | RP11-244O19.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p | 27 | ASTN1 | Sponge network | -0.096 | 0.67958 | -2.389 | 2.0E-5 | 0.423 |
| 16776 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | TACC1 | Sponge network | 0.222 | 0.29133 | -0.362 | 0.05406 | 0.423 |
| 16777 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-655-3p | 23 | PIK3C2A | Sponge network | 0.91 | 0 | 0.061 | 0.53776 | 0.423 |
| 16778 | FAM157C |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | MLLT10 | Sponge network | 0.91 | 0 | 0.105 | 0.33292 | 0.423 |
| 16779 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | PDGFRA | Sponge network | -1.886 | 0 | -0.372 | 0.0037 | 0.423 |
| 16780 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | CDHR3 | Sponge network | -0.227 | 0.31657 | -0.977 | 4.0E-5 | 0.423 |
| 16781 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-96-5p | 11 | PAK3 | Sponge network | -1.654 | 0.00144 | -0.628 | 0.07253 | 0.423 |
| 16782 | LINC01004 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | ZMYM2 | Sponge network | 1.115 | 0 | 0.279 | 0.01273 | 0.423 |
| 16783 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | PHC3 | Sponge network | 0.559 | 0.00026 | 0.212 | 0.0499 | 0.423 |
| 16784 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-93-3p | 13 | CNTNAP1 | Sponge network | -0.18 | 0.20343 | 0.358 | 0.13727 | 0.423 |
| 16785 | GNG12-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-874-3p | 25 | MXI1 | Sponge network | -0.793 | 7.0E-5 | -0.987 | 0 | 0.423 |
| 16786 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | CAV1 | Sponge network | -0.563 | 0.02545 | -1.013 | 6.0E-5 | 0.423 |
| 16787 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | CITED2 | Sponge network | -1.216 | 0 | -1.545 | 0 | 0.423 |
| 16788 | RP11-400K9.4 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-550a-5p;hsa-miR-7-1-3p | 11 | PCDH18 | Sponge network | -0.611 | 0.09607 | 0.216 | 0.24645 | 0.423 |
| 16789 | PSMD5-AS1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p | 14 | ZNF638 | Sponge network | 0.569 | 0.00067 | 0.22 | 0.01214 | 0.423 |
| 16790 | MRVI1-AS1 |
hsa-let-7f-1-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 25 | PPM1L | Sponge network | -1.092 | 4.0E-5 | -0.537 | 0.00616 | 0.423 |
| 16791 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | ATRX | Sponge network | 0.194 | 0.64278 | 0.261 | 0.04408 | 0.423 |
| 16792 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | EPHA7 | Sponge network | 0.532 | 0.00505 | -2.197 | 3.0E-5 | 0.423 |
| 16793 | PSMG3-AS1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | PDE4DIP | Sponge network | -0.125 | 0.49414 | -0.282 | 0.0257 | 0.423 |
| 16794 | LINC00472 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 10 | TPO | Sponge network | -0.842 | 0.01361 | -1.87 | 2.0E-5 | 0.423 |
| 16795 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-539-5p | 10 | PDZD2 | Sponge network | 0.532 | 0.00505 | -0.909 | 3.0E-5 | 0.423 |
| 16796 | RP11-156P1.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | ROBO1 | Sponge network | 0.214 | 0.18246 | -0.009 | 0.96154 | 0.423 |
| 16797 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-93-5p | 43 | PPM1A | Sponge network | -0.162 | 0.47687 | -0.623 | 0 | 0.423 |
| 16798 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-625-5p | 30 | JAZF1 | Sponge network | -0.342 | 0.02288 | -0.377 | 0.02677 | 0.423 |
| 16799 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 15 | PRDM5 | Sponge network | 0.997 | 0.00066 | -1.09 | 0.00014 | 0.423 |
| 16800 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 33 | SNX29 | Sponge network | -0.323 | 0.01372 | -0.34 | 0.01371 | 0.423 |
| 16801 | HCG18 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | DICER1 | Sponge network | 1.063 | 0 | 0.042 | 0.674 | 0.423 |
| 16802 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 27 | SEMA3C | Sponge network | -0.942 | 0 | -0.272 | 0.31153 | 0.423 |
| 16803 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | SASH1 | Sponge network | 0.101 | 0.60919 | -0.502 | 0.00175 | 0.423 |
| 16804 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 54 | RUNX1T1 | Sponge network | -0.738 | 0.21233 | -0.344 | 0.2374 | 0.423 |
| 16805 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p | 10 | DLC1 | Sponge network | -1.483 | 9.0E-5 | -0.382 | 0.05038 | 0.423 |
| 16806 | RP11-678G14.3 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-511-5p;hsa-miR-532-3p | 10 | MRVI1 | Sponge network | -4.477 | 0 | -1.051 | 0.00022 | 0.423 |
| 16807 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p | 23 | HIP1 | Sponge network | -0.49 | 0.04946 | 0.774 | 0 | 0.423 |
| 16808 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 16 | RELN | Sponge network | -0.096 | 0.67958 | -2.403 | 0 | 0.423 |
| 16809 | ARHGEF26-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 37 | IGFBP5 | Sponge network | -2.46 | 0 | -0.806 | 0.00105 | 0.423 |
| 16810 | RP11-400K9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-340-5p;hsa-miR-423-5p;hsa-miR-590-5p | 12 | TGFBR3 | Sponge network | -0.733 | 0.19469 | -1.173 | 1.0E-5 | 0.423 |
| 16811 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-20a-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | RICTOR | Sponge network | 1.2 | 0.01813 | 0.388 | 0.00188 | 0.423 |
| 16812 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-766-3p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -0.318 | 0.65847 | -0.564 | 1.0E-5 | 0.423 |
| 16813 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-590-3p | 12 | ZNF292 | Sponge network | -0.323 | 0.01372 | 0.276 | 0.04148 | 0.423 |
| 16814 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-505-3p | 16 | ST6GALNAC3 | Sponge network | -0.583 | 0.14589 | -0.987 | 0 | 0.423 |
| 16815 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-493-5p | 24 | PGR | Sponge network | 0.16 | 0.50785 | -1.704 | 0 | 0.423 |
| 16816 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | CDKN1C | Sponge network | -0.323 | 0.01372 | -0.929 | 0.00033 | 0.423 |
| 16817 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-625-5p | 14 | ABI2 | Sponge network | 0.858 | 3.0E-5 | 0.175 | 0.14787 | 0.423 |
| 16818 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p | 16 | KAT6B | Sponge network | -0.227 | 0.31657 | -0.478 | 0.00018 | 0.423 |
| 16819 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-33a-3p | 17 | RSPO2 | Sponge network | -0.693 | 0.05629 | -3.038 | 0 | 0.423 |
| 16820 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-576-5p | 22 | TMEM132B | Sponge network | -0.932 | 0.00329 | -0.83 | 0.01084 | 0.423 |
| 16821 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 37 | ABI2 | Sponge network | -0.487 | 0.02157 | 0.175 | 0.14787 | 0.423 |
| 16822 | RP11-212P7.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-766-3p;hsa-miR-93-5p | 18 | EZH1 | Sponge network | 0.219 | 0.1516 | -0.564 | 1.0E-5 | 0.423 |
| 16823 | MIR497HG |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 20 | PCDH17 | Sponge network | -1.439 | 4.0E-5 | 0.731 | 2.0E-5 | 0.423 |
| 16824 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940;hsa-miR-98-5p | 32 | PBX1 | Sponge network | 0.249 | 0.35902 | -1.206 | 0 | 0.423 |
| 16825 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-424-3p;hsa-miR-550a-5p;hsa-miR-590-3p | 14 | HOOK3 | Sponge network | -0.227 | 0.31657 | 0.273 | 0.09957 | 0.423 |
| 16826 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-93-5p | 12 | ANK2 | Sponge network | -0.295 | 0.02604 | -1.603 | 1.0E-5 | 0.423 |
| 16827 | RP11-2H8.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | BRD3 | Sponge network | 1.089 | 0 | 0.37 | 0.00013 | 0.423 |
| 16828 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | PCDH10 | Sponge network | 0.101 | 0.60919 | -1.644 | 0.0078 | 0.423 |
| 16829 | NDUFA6-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 23 | MRVI1 | Sponge network | -0.355 | 0.0077 | -1.051 | 0.00022 | 0.423 |
| 16830 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 27 | MYO9A | Sponge network | 0.603 | 0.00127 | -0.147 | 0.32373 | 0.423 |
| 16831 | ACTA2-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | PIK3CA | Sponge network | 0.194 | 0.64278 | 0.195 | 0.06908 | 0.423 |
| 16832 | RP11-798M19.6 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 18 | RPS6KA6 | Sponge network | -0.612 | 0.00094 | -2.989 | 0 | 0.423 |
| 16833 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 36 | RBMS3 | Sponge network | -0.342 | 0.02288 | -0.519 | 0.03551 | 0.423 |
| 16834 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p | 34 | TACC1 | Sponge network | 0.056 | 0.81195 | -0.362 | 0.05406 | 0.423 |
| 16835 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | DICER1 | Sponge network | 0.537 | 0.00139 | 0.042 | 0.674 | 0.423 |
| 16836 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-3p | 16 | DKK3 | Sponge network | -0.154 | 0.78939 | -0.052 | 0.77783 | 0.423 |
| 16837 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-484;hsa-miR-576-5p | 10 | EDA2R | Sponge network | 0.176 | 0.37402 | -0.587 | 0.01598 | 0.423 |
| 16838 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p | 13 | BCL6 | Sponge network | -4.546 | 0 | -0.211 | 0.19489 | 0.423 |
| 16839 | HNRNPU-AS1 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-376c-3p | 18 | RASAL2 | Sponge network | 1.601 | 0 | 0.869 | 0 | 0.423 |
| 16840 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | FSTL5 | Sponge network | -0.899 | 6.0E-5 | -1.3 | 0.01619 | 0.423 |
| 16841 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 17 | EZH1 | Sponge network | -0.769 | 0.00035 | -0.564 | 1.0E-5 | 0.423 |
| 16842 | MIR22HG |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 12 | PDZD2 | Sponge network | -1.047 | 0 | -0.909 | 3.0E-5 | 0.423 |
| 16843 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-550a-5p | 11 | HOOK3 | Sponge network | -0.154 | 0.78939 | 0.273 | 0.09957 | 0.423 |
| 16844 | TP73-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | IL17RD | Sponge network | -0.563 | 0.02545 | -0.019 | 0.94873 | 0.423 |
| 16845 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-590-3p | 15 | HECA | Sponge network | 1.089 | 0 | -0.217 | 0.03463 | 0.423 |
| 16846 | HAND2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 49 | NTRK2 | Sponge network | -1.697 | 0.00889 | -0.736 | 0.06495 | 0.423 |
| 16847 | LINC00641 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TPO | Sponge network | -0.222 | 0.32445 | -1.87 | 2.0E-5 | 0.423 |
| 16848 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | CUL5 | Sponge network | 0.826 | 1.0E-5 | 0.026 | 0.77301 | 0.423 |
| 16849 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-96-5p | 29 | TCF4 | Sponge network | 2.306 | 9.0E-5 | 0.016 | 0.92271 | 0.423 |
| 16850 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 58 | KIF1B | Sponge network | -0.487 | 0.02157 | -0.118 | 0.32258 | 0.423 |
| 16851 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p | 24 | MYBL1 | Sponge network | 0.537 | 0.00139 | 0.494 | 0.02152 | 0.423 |
| 16852 | OSER1-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-424-5p;hsa-miR-486-5p | 10 | DYNC1I1 | Sponge network | -0.13 | 0.48734 | -1.12 | 0.00107 | 0.423 |
| 16853 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 32 | FAT3 | Sponge network | -1.057 | 0.00029 | -1.601 | 0.00051 | 0.423 |
| 16854 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | MYO9A | Sponge network | 0.539 | 0.03722 | -0.147 | 0.32373 | 0.423 |
| 16855 | RP11-212P7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 33 | DMD | Sponge network | 0.219 | 0.1516 | -1.506 | 0 | 0.423 |
| 16856 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | PCDH10 | Sponge network | 0.532 | 0.00505 | -1.644 | 0.0078 | 0.423 |
| 16857 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 20 | NR2C2 | Sponge network | 0.222 | 0.29133 | 0.21 | 0.03224 | 0.423 |
| 16858 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 33 | RUNX1T1 | Sponge network | 0.174 | 0.30034 | -0.344 | 0.2374 | 0.423 |
| 16859 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 15 | TLL1 | Sponge network | -0.096 | 0.67958 | -1.195 | 3.0E-5 | 0.423 |
| 16860 | HOTAIRM1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NRXN3 | Sponge network | -0.053 | 0.80685 | -0.893 | 0.04186 | 0.423 |
| 16861 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 42 | RASSF2 | Sponge network | -1.216 | 0 | -0.273 | 0.21961 | 0.423 |
| 16862 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 50 | TMEM132B | Sponge network | -1.575 | 6.0E-5 | -0.83 | 0.01084 | 0.423 |
| 16863 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CLIP1 | Sponge network | -0.612 | 0.00094 | -0.215 | 0.08684 | 0.423 |
| 16864 | RP4-714D9.5 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 10 | PHF6 | Sponge network | 1.015 | 0 | 0.785 | 0 | 0.423 |
| 16865 | RP11-404E16.1 |
hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p | 10 | ZFHX3 | Sponge network | 0.097 | 0.91311 | 0.389 | 0.01363 | 0.423 |
| 16866 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | RB1CC1 | Sponge network | 0.72 | 0.0001 | 0.175 | 0.12559 | 0.423 |
| 16867 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-93-5p | 26 | ZBTB4 | Sponge network | 0.082 | 0.55397 | -0.739 | 0 | 0.423 |
| 16868 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ZFHX3 | Sponge network | 2.094 | 0 | 0.389 | 0.01363 | 0.423 |
| 16869 | RP11-182J1.1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 13 | AFF1 | Sponge network | -0.187 | 0.23787 | -0.384 | 0.00053 | 0.423 |
| 16870 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.374 | 0.20054 | 0.323 | 0.00792 | 0.423 |
| 16871 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 58 | WDFY3 | Sponge network | -0.49 | 0.04946 | -0.201 | 0.0967 | 0.423 |
| 16872 | TRAF3IP2-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NRG3 | Sponge network | -0.323 | 0.01372 | -1.836 | 4.0E-5 | 0.423 |
| 16873 | RP11-458F8.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p | 19 | MDM4 | Sponge network | 1.518 | 0 | 0.345 | 0.00762 | 0.423 |
| 16874 | MAGI2-AS3 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 25 | ATM | Sponge network | -0.129 | 0.57177 | 0.178 | 0.21568 | 0.423 |
| 16875 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-455-3p | 28 | NTRK2 | Sponge network | -1.092 | 4.0E-5 | -0.736 | 0.06495 | 0.423 |
| 16876 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-96-5p | 29 | ZFHX4 | Sponge network | -0.811 | 2.0E-5 | 0.018 | 0.95574 | 0.423 |
| 16877 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-7-5p | 14 | ESR1 | Sponge network | -0.187 | 0.23787 | -1.035 | 3.0E-5 | 0.423 |
| 16878 | ZNF32-AS2 |
hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-378c | 15 | ABL2 | Sponge network | 1.552 | 7.0E-5 | 0.82 | 0 | 0.423 |
| 16879 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-671-5p | 18 | CADM3 | Sponge network | -0.096 | 0.67958 | -3.663 | 0 | 0.423 |
| 16880 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | PIK3R1 | Sponge network | 0.673 | 0 | -0.133 | 0.36854 | 0.423 |
| 16881 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CLIP1 | Sponge network | -1.575 | 6.0E-5 | -0.215 | 0.08684 | 0.423 |
| 16882 | RP11-758N13.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-375;hsa-miR-93-3p | 16 | BNC2 | Sponge network | 2.939 | 3.0E-5 | -0.491 | 0.09988 | 0.423 |
| 16883 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 41 | PRKAB2 | Sponge network | -1.111 | 0.0003 | -0.367 | 0.01371 | 0.423 |
| 16884 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 42 | TCF4 | Sponge network | -1.057 | 0.00029 | 0.016 | 0.92271 | 0.423 |
| 16885 | RP4-791M13.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p | 13 | DYNC1I1 | Sponge network | 0.419 | 0.0319 | -1.12 | 0.00107 | 0.423 |
| 16886 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-96-5p | 38 | TCF4 | Sponge network | 0.622 | 0.00015 | 0.016 | 0.92271 | 0.423 |
| 16887 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-590-5p | 11 | AKAP12 | Sponge network | 0.331 | 0.57247 | -0.932 | 0.0028 | 0.423 |
| 16888 | MAP3K14 |
hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-26b-3p;hsa-miR-362-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 10 | GRIK3 | Sponge network | -0.295 | 0.02604 | -3.208 | 0 | 0.423 |
| 16889 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 31 | SCN9A | Sponge network | -0.942 | 0 | -0.525 | 0.10593 | 0.423 |
| 16890 | RP11-57H14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | ERCC4 | Sponge network | 0.083 | 0.66405 | 0.126 | 0.15266 | 0.423 |
| 16891 | RP11-701H24.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 11 | IGF1 | Sponge network | -1.425 | 0.04204 | -0.204 | 0.5898 | 0.423 |
| 16892 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-940 | 34 | CADM2 | Sponge network | -0.154 | 0.53954 | -3.782 | 0 | 0.423 |
| 16893 | RP11-181G12.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p | 13 | UBE4B | Sponge network | 0.556 | 0.00298 | -0.235 | 0.007 | 0.423 |
| 16894 | RP11-532F6.3 |
hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-455-5p | 10 | PLXNC1 | Sponge network | -0.896 | 0.00099 | 0.569 | 0.00329 | 0.422 |
| 16895 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 16 | CNTNAP3 | Sponge network | 0.335 | 0.20596 | -1.465 | 0.00033 | 0.422 |
| 16896 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 35 | KIF1B | Sponge network | 0.91 | 0 | -0.118 | 0.32258 | 0.422 |
| 16897 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 27 | KCNA6 | Sponge network | -0.563 | 0.02545 | -1.531 | 0.00013 | 0.422 |
| 16898 | RP11-404E16.1 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-27a-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-744-5p;hsa-miR-940 | 12 | CHD5 | Sponge network | 0.097 | 0.91311 | -0.922 | 0.01521 | 0.422 |
| 16899 | FZD10-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 18 | PRUNE2 | Sponge network | -0.727 | 0.04726 | -1.733 | 0.00022 | 0.422 |
| 16900 | RP11-254F7.2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-454-3p | 14 | ITPKB | Sponge network | 0.176 | 0.37402 | -0.704 | 0.00106 | 0.422 |
| 16901 | EMX2OS |
hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 11 | LZTS1 | Sponge network | -0.777 | 0.1134 | 1.664 | 0 | 0.422 |
| 16902 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-589-3p;hsa-miR-590-3p | 35 | ADARB1 | Sponge network | -1.161 | 0.00591 | -0.335 | 0.06631 | 0.422 |
| 16903 | RP11-6O2.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.331 | 0.57247 | -0.57 | 0 | 0.422 |
| 16904 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 22 | HIP1 | Sponge network | 1.302 | 7.0E-5 | 0.774 | 0 | 0.422 |
| 16905 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | TGFBR2 | Sponge network | -0.896 | 0.00099 | -0.243 | 0.11408 | 0.422 |
| 16906 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-493-5p;hsa-miR-940;hsa-miR-96-5p | 11 | HOOK3 | Sponge network | 0.056 | 0.8494 | 0.273 | 0.09957 | 0.422 |
| 16907 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 29 | ARHGAP28 | Sponge network | -0.129 | 0.57177 | -0.911 | 0.00013 | 0.422 |
| 16908 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-33a-5p | 19 | CADM2 | Sponge network | -2.549 | 0.00528 | -3.782 | 0 | 0.422 |
| 16909 | RP11-53O19.1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-940 | 32 | CHD5 | Sponge network | -0.405 | 0.22013 | -0.922 | 0.01521 | 0.422 |
| 16910 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | TMEFF2 | Sponge network | -0.487 | 0.02157 | -2.426 | 0 | 0.422 |
| 16911 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 35 | PRDM2 | Sponge network | -0.129 | 0.57177 | -0.258 | 0.00721 | 0.422 |
| 16912 | CTC-429P9.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p | 17 | NAV3 | Sponge network | 0.082 | 0.55397 | 0.212 | 0.47729 | 0.422 |
| 16913 | RP11-6O2.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-624-5p | 18 | WWTR1 | Sponge network | 0.331 | 0.57247 | -0.345 | 0.11997 | 0.422 |
| 16914 | PART1 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | CDKN1C | Sponge network | -2.925 | 0 | -0.929 | 0.00033 | 0.422 |
| 16915 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-495-3p | 20 | RICTOR | Sponge network | 1.117 | 0 | 0.388 | 0.00188 | 0.422 |
| 16916 | GS1-124K5.11 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | BAZ2B | Sponge network | 0.494 | 0.00339 | -0.276 | 0.02833 | 0.422 |
| 16917 | RP11-244O19.1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 16 | ATM | Sponge network | -0.096 | 0.67958 | 0.178 | 0.21568 | 0.422 |
| 16918 | RP11-758N13.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-424-5p | 17 | ZFHX4 | Sponge network | 2.939 | 3.0E-5 | 0.018 | 0.95574 | 0.422 |
| 16919 | PWAR6 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-5p | 30 | BTG2 | Sponge network | -1.575 | 6.0E-5 | -1.763 | 0 | 0.422 |
| 16920 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-940 | 14 | GLI3 | Sponge network | -0.246 | 0.35034 | -0.615 | 0.01482 | 0.422 |
| 16921 | BOLA3-AS1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b | 10 | FNBP1 | Sponge network | -0.246 | 0.35034 | -0.969 | 4.0E-5 | 0.422 |
| 16922 | RP11-999E24.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p | 15 | ETV1 | Sponge network | -0.693 | 0.05629 | -0.096 | 0.67674 | 0.422 |
| 16923 | ACTA2-AS1 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-7-5p | 10 | MLLT11 | Sponge network | 0.194 | 0.64278 | -0.661 | 0.01733 | 0.422 |
| 16924 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-7-5p | 21 | ERC1 | Sponge network | -1.654 | 0.00144 | -0.108 | 0.38794 | 0.422 |
| 16925 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | FOXP1 | Sponge network | 0.572 | 0.01722 | 0.042 | 0.74368 | 0.422 |
| 16926 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | SLITRK3 | Sponge network | -0.727 | 0.04726 | -3.328 | 0 | 0.422 |
| 16927 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-589-3p | 19 | CHD5 | Sponge network | 0.811 | 0.03228 | -0.922 | 0.01521 | 0.422 |
| 16928 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-96-5p | 16 | QKI | Sponge network | 0.348 | 0.32912 | -0.333 | 0.04111 | 0.422 |
| 16929 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | EDA2R | Sponge network | -1.281 | 0.00012 | -0.587 | 0.01598 | 0.422 |
| 16930 | BMS1P20 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p | 12 | PHF6 | Sponge network | 0.34 | 0.00273 | 0.785 | 0 | 0.422 |
| 16931 | CTC-429P9.2 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-335-5p | 10 | PAFAH1B1 | Sponge network | 0.043 | 0.81628 | -0.429 | 0 | 0.422 |
| 16932 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p | 33 | BMPR2 | Sponge network | -0.473 | 0.07602 | 0.206 | 0.0635 | 0.422 |
| 16933 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-744-3p | 19 | HLF | Sponge network | 0.811 | 0.03228 | -2.443 | 0 | 0.422 |
| 16934 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-5p | 16 | RET | Sponge network | 0.176 | 0.37402 | -0.833 | 0.03517 | 0.422 |
| 16935 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 24 | KIT | Sponge network | -0.793 | 7.0E-5 | -2.184 | 0 | 0.422 |
| 16936 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 28 | THBS1 | Sponge network | -0.563 | 0.02545 | 0.741 | 0.00386 | 0.422 |
| 16937 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MOB1B | Sponge network | 1.601 | 0 | 0.197 | 0.15266 | 0.422 |
| 16938 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 27 | HLF | Sponge network | 0.082 | 0.55397 | -2.443 | 0 | 0.422 |
| 16939 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-625-5p | 23 | ABI2 | Sponge network | -0.154 | 0.53954 | 0.175 | 0.14787 | 0.422 |
| 16940 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-5p | 22 | ATRX | Sponge network | 0.174 | 0.30034 | 0.261 | 0.04408 | 0.422 |
| 16941 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 50 | QKI | Sponge network | 0.101 | 0.60919 | -0.333 | 0.04111 | 0.422 |
| 16942 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-98-5p | 21 | PNRC1 | Sponge network | -0.49 | 0.04946 | -0.646 | 0 | 0.422 |
| 16943 | MALAT1 |
hsa-miR-130b-5p;hsa-miR-197-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-485-3p;hsa-miR-590-3p | 10 | RPL22 | Sponge network | 0.782 | 0.00016 | -0.065 | 0.5828 | 0.422 |
| 16944 | MIR22HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-503-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-708-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 29 | TOM1L2 | Sponge network | -1.047 | 0 | -0.994 | 0 | 0.422 |
| 16945 | LINC00476 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | HERC1 | Sponge network | -0.615 | 0.00015 | -0.196 | 0.08544 | 0.422 |
| 16946 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-421 | 15 | RSPO2 | Sponge network | -4.463 | 0.00025 | -3.038 | 0 | 0.422 |
| 16947 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | CHD2 | Sponge network | 0.541 | 0.00125 | 0.049 | 0.55371 | 0.422 |
| 16948 | LINC00476 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-576-5p | 16 | SFRP1 | Sponge network | -0.615 | 0.00015 | -3.566 | 0 | 0.422 |
| 16949 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-5p | 27 | PHC3 | Sponge network | 0.921 | 0.00118 | 0.212 | 0.0499 | 0.422 |
| 16950 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 27 | USP6 | Sponge network | 0.311 | 0.10344 | 0.188 | 0.3871 | 0.422 |
| 16951 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-93-5p | 34 | FAT3 | Sponge network | 0.259 | 0.12542 | -1.601 | 0.00051 | 0.422 |
| 16952 | FAM66C |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PCDH18 | Sponge network | -0.901 | 0.00019 | 0.216 | 0.24645 | 0.422 |
| 16953 | LINC00476 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-671-5p | 20 | CADM3 | Sponge network | -0.615 | 0.00015 | -3.663 | 0 | 0.422 |
| 16954 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | UBE4B | Sponge network | -0.487 | 0.02157 | -0.235 | 0.007 | 0.422 |
| 16955 | HOTAIRM1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-326;hsa-miR-539-5p;hsa-miR-590-3p | 25 | DMD | Sponge network | -0.053 | 0.80685 | -1.506 | 0 | 0.422 |
| 16956 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-455-5p;hsa-miR-93-5p | 10 | EPAS1 | Sponge network | -0.473 | 0.07602 | -0.184 | 0.14418 | 0.422 |
| 16957 | RP11-531A24.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | CREB1 | Sponge network | 0.75 | 0.00054 | 0.203 | 0.00537 | 0.422 |
| 16958 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | PDGFRA | Sponge network | 0.246 | 0.59912 | -0.372 | 0.0037 | 0.422 |
| 16959 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 48 | QKI | Sponge network | -0.405 | 0.22013 | -0.333 | 0.04111 | 0.422 |
| 16960 | CTC-471J1.2 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 21 | MDM4 | Sponge network | 1.11 | 1.0E-5 | 0.345 | 0.00762 | 0.422 |
| 16961 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-93-3p | 18 | SNX29 | Sponge network | 0.598 | 0.38481 | -0.34 | 0.01371 | 0.422 |
| 16962 | LINC00654 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 16 | MAPK10 | Sponge network | 0.374 | 0.20054 | -1.774 | 0 | 0.422 |
| 16963 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-374b-3p | 12 | PGR | Sponge network | -0.72 | 0.24239 | -1.704 | 0 | 0.422 |
| 16964 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-7-5p | 20 | ZBTB4 | Sponge network | -0.693 | 0.05629 | -0.739 | 0 | 0.422 |
| 16965 | LINC00654 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | PCDH18 | Sponge network | 0.374 | 0.20054 | 0.216 | 0.24645 | 0.422 |
| 16966 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-493-5p;hsa-miR-590-3p | 14 | MYCBP2 | Sponge network | -0.487 | 0.02157 | -0.027 | 0.82016 | 0.422 |
| 16967 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-7-1-3p | 24 | ST6GALNAC3 | Sponge network | -4.546 | 0 | -0.987 | 0 | 0.422 |
| 16968 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 43 | SASH1 | Sponge network | -0.576 | 0.10121 | -0.502 | 0.00175 | 0.422 |
| 16969 | ACTA2-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p | 17 | ATM | Sponge network | 0.194 | 0.64278 | 0.178 | 0.21568 | 0.422 |
| 16970 | LINC00672 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-98-5p | 33 | TGFBR3 | Sponge network | -0.227 | 0.31657 | -1.173 | 1.0E-5 | 0.422 |
| 16971 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-940 | 37 | CHD5 | Sponge network | -0.901 | 0.00019 | -0.922 | 0.01521 | 0.422 |
| 16972 | ADAMTS9-AS2 |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CTNNA2 | Sponge network | -2.452 | 0 | -2.547 | 0 | 0.422 |
| 16973 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 28 | ANK2 | Sponge network | 0.082 | 0.55654 | -1.603 | 1.0E-5 | 0.422 |
| 16974 | LINC00963 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-577;hsa-miR-590-3p | 11 | MACF1 | Sponge network | 0.523 | 9.0E-5 | 0.019 | 0.90335 | 0.422 |
| 16975 | RP11-890B15.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 30 | NAV3 | Sponge network | 0.101 | 0.60919 | 0.212 | 0.47729 | 0.422 |
| 16976 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-590-3p | 17 | PRKAB2 | Sponge network | 0.765 | 7.0E-5 | -0.367 | 0.01371 | 0.422 |
| 16977 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 21 | ARHGAP29 | Sponge network | -2.452 | 0 | 0.131 | 0.42953 | 0.422 |
| 16978 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-532-5p;hsa-miR-590-3p | 11 | PARK2 | Sponge network | 0.214 | 0.18246 | -1.892 | 0 | 0.422 |
| 16979 | LINC00472 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-744-3p;hsa-miR-93-5p | 54 | DST | Sponge network | -0.842 | 0.01361 | -0.713 | 0.00096 | 0.422 |
| 16980 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 28 | CUL5 | Sponge network | 0.421 | 0.00084 | 0.026 | 0.77301 | 0.422 |
| 16981 | ZNF582-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | IL17RD | Sponge network | -1.349 | 0.00017 | -0.019 | 0.94873 | 0.422 |
| 16982 | RP11-254F7.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 61 | LPP | Sponge network | 0.176 | 0.37402 | -0.459 | 0.04475 | 0.422 |
| 16983 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | KCNAB1 | Sponge network | -1.216 | 0 | -0.755 | 2.0E-5 | 0.422 |
| 16984 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-3p | 17 | PICALM | Sponge network | 0.537 | 0.00139 | 0.086 | 0.23523 | 0.422 |
| 16985 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-629-3p;hsa-miR-93-5p | 27 | ZBTB4 | Sponge network | -2.915 | 0.00015 | -0.739 | 0 | 0.422 |
| 16986 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 20 | HLF | Sponge network | -2.039 | 0.03447 | -2.443 | 0 | 0.422 |
| 16987 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | JAZF1 | Sponge network | 0.246 | 0.59912 | -0.377 | 0.02677 | 0.422 |
| 16988 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 14 | ZMYND11 | Sponge network | 0.259 | 0.12542 | -0.2 | 0.05449 | 0.422 |
| 16989 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | APC | Sponge network | -0.487 | 0.02157 | -0.255 | 0.04051 | 0.422 |
| 16990 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-93-3p | 22 | ST6GAL2 | Sponge network | -0.154 | 0.78939 | -1.201 | 0.00597 | 0.422 |
| 16991 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 25 | NIN | Sponge network | 0.214 | 0.18246 | -0.253 | 0.13794 | 0.422 |
| 16992 | AC016747.3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 11 | EGR2 | Sponge network | 0.199 | 0.38015 | 0.309 | 0.17079 | 0.422 |
| 16993 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 19 | MOB1B | Sponge network | 0.768 | 7.0E-5 | 0.197 | 0.15266 | 0.422 |
| 16994 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 35 | PGR | Sponge network | 0.064 | 0.72934 | -1.704 | 0 | 0.422 |
| 16995 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | YAP1 | Sponge network | 0.91 | 0 | 0.22 | 0.11836 | 0.422 |
| 16996 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-143-5p;hsa-miR-18a-5p;hsa-miR-210-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | CLASP2 | Sponge network | 1.2 | 0.01813 | -0.038 | 0.71 | 0.422 |
| 16997 | FZD10-AS1 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | PPP2R2C | Sponge network | -0.727 | 0.04726 | -0.959 | 0.07428 | 0.422 |
| 16998 | FGF14-AS2 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | MED12L | Sponge network | -1.582 | 8.0E-5 | -0.194 | 0.55299 | 0.422 |
| 16999 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 27 | RAP1A | Sponge network | -0.709 | 0.18168 | -0.929 | 0 | 0.422 |
| 17000 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | USP12 | Sponge network | 0.225 | 0.30644 | -0.102 | 0.40768 | 0.422 |
| 17001 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | MED12L | Sponge network | -0.358 | 0.17255 | -0.194 | 0.55299 | 0.422 |
| 17002 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-340-5p;hsa-miR-532-3p | 21 | PBX1 | Sponge network | 0.051 | 0.83388 | -1.206 | 0 | 0.422 |
| 17003 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 24 | GPC6 | Sponge network | 1.302 | 7.0E-5 | -0.054 | 0.81465 | 0.422 |
| 17004 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-874-3p;hsa-miR-940 | 20 | CRTC3 | Sponge network | -0.405 | 0.22013 | 0.164 | 0.16786 | 0.422 |
| 17005 | RP11-927P21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-576-5p | 14 | ERCC4 | Sponge network | 0.921 | 0.00118 | 0.126 | 0.15266 | 0.422 |
| 17006 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | SON | Sponge network | 0.765 | 7.0E-5 | 0.168 | 0.04406 | 0.422 |
| 17007 | RP11-927P21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-940 | 28 | CREB1 | Sponge network | 0.921 | 0.00118 | 0.203 | 0.00537 | 0.422 |
| 17008 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 42 | EPHA5 | Sponge network | -0.405 | 0.22013 | -0.957 | 0.01733 | 0.422 |
| 17009 | RP11-384O8.1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | CSDE1 | Sponge network | -0.738 | 0.21233 | -0.493 | 0 | 0.422 |
| 17010 | RP11-156P1.3 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p | 10 | HSPB7 | Sponge network | 0.214 | 0.18246 | -2.268 | 1.0E-5 | 0.422 |
| 17011 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 14 | TIMP3 | Sponge network | -0.246 | 0.35034 | -0.546 | 0.02307 | 0.422 |
| 17012 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-5p | 20 | TIMP3 | Sponge network | 0.176 | 0.37402 | -0.546 | 0.02307 | 0.421 |
| 17013 | FAM66C |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 32 | ERC1 | Sponge network | -0.901 | 0.00019 | -0.108 | 0.38794 | 0.421 |
| 17014 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199b-3p;hsa-miR-324-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | HACE1 | Sponge network | 0.75 | 0.00054 | -0.072 | 0.55239 | 0.421 |
| 17015 | SH3BP5-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-5p | 11 | FBXO11 | Sponge network | 0.267 | 0.2937 | 0.142 | 0.04938 | 0.421 |
| 17016 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 43 | PHC3 | Sponge network | 0.421 | 0.00084 | 0.212 | 0.0499 | 0.421 |
| 17017 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429 | 30 | EPHA5 | Sponge network | -0.154 | 0.53954 | -0.957 | 0.01733 | 0.421 |
| 17018 | CTD-2554C21.2 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-7-5p | 11 | EHD3 | Sponge network | -1.654 | 0.00144 | -0.484 | 0.01747 | 0.421 |
| 17019 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p | 35 | BNC2 | Sponge network | 0.199 | 0.38015 | -0.491 | 0.09988 | 0.421 |
| 17020 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | MYO9A | Sponge network | 1.2 | 0.01813 | -0.147 | 0.32373 | 0.421 |
| 17021 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.537 | 0.00139 | 0.323 | 0.00792 | 0.421 |
| 17022 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-98-5p | 32 | ZFHX3 | Sponge network | -0.227 | 0.31657 | 0.389 | 0.01363 | 0.421 |
| 17023 | RP11-597D13.9 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-582-5p | 10 | PRICKLE1 | Sponge network | 1.302 | 7.0E-5 | -0.112 | 0.67088 | 0.421 |
| 17024 | RP11-344B5.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-505-3p | 21 | NTRK3 | Sponge network | -0.055 | 0.88482 | -1.77 | 3.0E-5 | 0.421 |
| 17025 | LINC00476 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p | 22 | CDH19 | Sponge network | -0.615 | 0.00015 | -3.434 | 0 | 0.421 |
| 17026 | RAMP2-AS1 |
hsa-let-7g-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-877-5p | 16 | NRK | Sponge network | -0.513 | 0.04703 | 0.656 | 0.19217 | 0.421 |
| 17027 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-5p | 11 | LHFP | Sponge network | -0.384 | 0.40652 | -0.167 | 0.41371 | 0.421 |
| 17028 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-590-3p | 12 | KAT6B | Sponge network | 0.178 | 0.29106 | -0.478 | 0.00018 | 0.421 |
| 17029 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | TET1 | Sponge network | 0.419 | 0.0319 | -0.135 | 0.52138 | 0.421 |
| 17030 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374a-3p;hsa-miR-7-1-3p | 13 | CNTN1 | Sponge network | -0.557 | 0.11135 | -2.594 | 0 | 0.421 |
| 17031 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | HOOK3 | Sponge network | 0.873 | 0 | 0.273 | 0.09957 | 0.421 |
| 17032 | PART1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 30 | LRIG1 | Sponge network | -2.925 | 0 | -0.537 | 0.00588 | 0.421 |
| 17033 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p | 10 | PIK3CA | Sponge network | 1.46 | 1.0E-5 | 0.195 | 0.06908 | 0.421 |
| 17034 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p | 22 | CBX5 | Sponge network | 0.178 | 0.29106 | 0.165 | 0.24446 | 0.421 |
| 17035 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-629-5p;hsa-miR-940 | 28 | IGF1 | Sponge network | -1.249 | 7.0E-5 | -0.204 | 0.5898 | 0.421 |
| 17036 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 22 | GPC6 | Sponge network | 0.199 | 0.38015 | -0.054 | 0.81465 | 0.421 |
| 17037 | JAZF1-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | PCDH18 | Sponge network | -0.769 | 0.00035 | 0.216 | 0.24645 | 0.421 |
| 17038 | AC003090.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ITGB3 | Sponge network | -2.117 | 0.00161 | -0.011 | 0.95751 | 0.421 |
| 17039 | AC016747.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 14 | PTGFR | Sponge network | 0.199 | 0.38015 | -0.233 | 0.45421 | 0.421 |
| 17040 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p | 23 | CADM1 | Sponge network | -1.161 | 0.00591 | -0.9 | 0.00084 | 0.421 |
| 17041 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 22 | HIP1 | Sponge network | -0.727 | 0.04726 | 0.774 | 0 | 0.421 |
| 17042 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 55 | ADARB1 | Sponge network | -1.216 | 0 | -0.335 | 0.06631 | 0.421 |
| 17043 | RP11-121C2.2 |
hsa-miR-101-3p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 15 | KIF5B | Sponge network | 1.147 | 0 | 0.362 | 0.00066 | 0.421 |
| 17044 | SNHG14 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | LUZP2 | Sponge network | -1.111 | 0.0003 | -0.056 | 0.89416 | 0.421 |
| 17045 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-93-5p | 13 | LATS2 | Sponge network | 0.299 | 0.57722 | 0.159 | 0.2304 | 0.421 |
| 17046 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CLIP1 | Sponge network | -2.452 | 0 | -0.215 | 0.08684 | 0.421 |
| 17047 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 13 | PBRM1 | Sponge network | 0.614 | 1.0E-5 | -0.029 | 0.78601 | 0.421 |
| 17048 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 34 | ABI2 | Sponge network | -0.976 | 0.00025 | 0.175 | 0.14787 | 0.421 |
| 17049 | AC006116.24 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p | 11 | PTPN14 | Sponge network | -0.473 | 0.07602 | 0.144 | 0.34595 | 0.421 |
| 17050 | CTD-2002H8.2 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-376c-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | HECA | Sponge network | 0.931 | 1.0E-5 | -0.217 | 0.03463 | 0.421 |
| 17051 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-590-3p | 16 | SEMA3C | Sponge network | -0.358 | 0.17255 | -0.272 | 0.31153 | 0.421 |
| 17052 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | 0.246 | 0.59912 | -1.277 | 0 | 0.421 |
| 17053 | RP5-1139B12.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 10 | MCC | Sponge network | 0.598 | 0.38481 | -1.005 | 0 | 0.421 |
| 17054 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-590-3p | 16 | PTPRD | Sponge network | -1.582 | 8.0E-5 | -1.005 | 0.00064 | 0.421 |
| 17055 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -0.777 | 0.16667 | -0.087 | 0.69193 | 0.421 |
| 17056 | RP11-325K4.2 |
hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | MDM4 | Sponge network | 0.198 | 0.27624 | 0.345 | 0.00762 | 0.421 |
| 17057 | U91328.19 |
hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-940 | 10 | FOXO4 | Sponge network | -0.303 | 0.21002 | -0.516 | 2.0E-5 | 0.421 |
| 17058 | FZD10-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 61 | DST | Sponge network | -0.727 | 0.04726 | -0.713 | 0.00096 | 0.421 |
| 17059 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p | 14 | NF1 | Sponge network | 1.093 | 0 | 0.51 | 0 | 0.421 |
| 17060 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-5p | 10 | UVRAG | Sponge network | 0.335 | 0.20596 | -0.017 | 0.83865 | 0.421 |
| 17061 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-590-3p | 11 | JAKMIP2 | Sponge network | -1.203 | 0.03881 | -1.388 | 0 | 0.421 |
| 17062 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 38 | ZFHX4 | Sponge network | 0.494 | 0.00339 | 0.018 | 0.95574 | 0.421 |
| 17063 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | 0.374 | 0.20054 | -1.324 | 0.00014 | 0.421 |
| 17064 | PGM5-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-5p | 15 | ROBO1 | Sponge network | -4.546 | 0 | -0.009 | 0.96154 | 0.421 |
| 17065 | FGF14-AS2 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 10 | TLE4 | Sponge network | -1.582 | 8.0E-5 | -1.157 | 0 | 0.421 |
| 17066 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-625-5p | 26 | RSPO2 | Sponge network | -0.563 | 0.02545 | -3.038 | 0 | 0.421 |
| 17067 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-940 | 21 | IGF1 | Sponge network | -0.318 | 0.65847 | -0.204 | 0.5898 | 0.421 |
| 17068 | ATP1B3-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | CREB1 | Sponge network | 1.2 | 0.01813 | 0.203 | 0.00537 | 0.421 |
| 17069 | MIR143HG |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 12 | PLA2R1 | Sponge network | -0.739 | 0.0574 | -0.265 | 0.24676 | 0.421 |
| 17070 | GABPB1-AS1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 37 | CREB1 | Sponge network | 1.11 | 0 | 0.203 | 0.00537 | 0.421 |
| 17071 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 27 | HLF | Sponge network | -0.318 | 0.65847 | -2.443 | 0 | 0.421 |
| 17072 | FAM66C |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 27 | LRIG1 | Sponge network | -0.901 | 0.00019 | -0.537 | 0.00588 | 0.421 |
| 17073 | LINC00476 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-93-5p | 62 | CADM2 | Sponge network | -0.615 | 0.00015 | -3.782 | 0 | 0.421 |
| 17074 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-532-3p;hsa-miR-744-3p;hsa-miR-877-5p | 25 | PTPRT | Sponge network | -1.654 | 0.00144 | -0.907 | 0.03727 | 0.421 |
| 17075 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-98-5p | 20 | PNRC1 | Sponge network | -0.129 | 0.57177 | -0.646 | 0 | 0.421 |
| 17076 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-98-5p | 50 | NTRK3 | Sponge network | -0.405 | 0.22013 | -1.77 | 3.0E-5 | 0.421 |
| 17077 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-502-5p;hsa-miR-766-3p;hsa-miR-940 | 12 | CHD5 | Sponge network | 0.043 | 0.81628 | -0.922 | 0.01521 | 0.421 |
| 17078 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-589-3p | 14 | MAFB | Sponge network | -0.358 | 0.17255 | -0.023 | 0.89865 | 0.421 |
| 17079 | LINC00654 |
hsa-miR-142-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 10 | RNF180 | Sponge network | 0.374 | 0.20054 | -1.127 | 1.0E-5 | 0.421 |
| 17080 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-335-5p;hsa-miR-539-5p | 16 | PRKAA2 | Sponge network | -0.421 | 0.12556 | -2.462 | 0 | 0.421 |
| 17081 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-590-5p | 11 | SPG20 | Sponge network | 0.889 | 9.0E-5 | -1.16 | 0 | 0.421 |
| 17082 | RP13-516M14.1 |
hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429 | 10 | DYNC1I1 | Sponge network | 0.389 | 0.04293 | -1.12 | 0.00107 | 0.421 |
| 17083 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | TACC1 | Sponge network | 0.101 | 0.60919 | -0.362 | 0.05406 | 0.421 |
| 17084 | RP11-434H6.7 |
hsa-miR-106b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-382-5p;hsa-miR-664a-5p | 22 | LPP | Sponge network | 0.358 | 0.14302 | -0.459 | 0.04475 | 0.421 |
| 17085 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-532-5p | 10 | FSTL5 | Sponge network | -0.303 | 0.21002 | -1.3 | 0.01619 | 0.421 |
| 17086 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-96-5p | 33 | DMXL1 | Sponge network | -1.575 | 6.0E-5 | -0.426 | 0.00056 | 0.421 |
| 17087 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-576-5p | 16 | SEMA3E | Sponge network | -0.162 | 0.47687 | -1.717 | 0.00137 | 0.421 |
| 17088 | LINC00854 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-628-5p;hsa-miR-940 | 12 | TRIM33 | Sponge network | 1.18 | 0 | 0.402 | 7.0E-5 | 0.421 |
| 17089 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 45 | CREB3L2 | Sponge network | -0.769 | 0.00035 | -0.525 | 2.0E-5 | 0.421 |
| 17090 | AGAP11 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-532-3p | 16 | SYNM | Sponge network | -1.645 | 0 | -2.309 | 1.0E-5 | 0.421 |
| 17091 | RP11-119F7.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | FBXO11 | Sponge network | 0.225 | 0.30644 | 0.142 | 0.04938 | 0.421 |
| 17092 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p | 15 | JDP2 | Sponge network | -2.531 | 0 | -0.967 | 0 | 0.421 |
| 17093 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 13 | ZMYND11 | Sponge network | -0.583 | 0.14589 | -0.2 | 0.05449 | 0.421 |
| 17094 | AP001347.6 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 11 | KLF10 | Sponge network | -1.161 | 0.00591 | -0.783 | 0 | 0.421 |
| 17095 | RP11-244O19.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-708-5p;hsa-miR-940 | 26 | TOM1L2 | Sponge network | -0.096 | 0.67958 | -0.994 | 0 | 0.421 |
| 17096 | RP11-53O19.1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p | 19 | ABL1 | Sponge network | -0.405 | 0.22013 | -0.215 | 0.07804 | 0.421 |
| 17097 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 55 | PTPRD | Sponge network | -1.575 | 6.0E-5 | -1.005 | 0.00064 | 0.421 |
| 17098 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 33 | ADARB1 | Sponge network | 0.537 | 0.00139 | -0.335 | 0.06631 | 0.421 |
| 17099 | LINC01018 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | MLF1 | Sponge network | -2.915 | 0.00015 | -0.893 | 0.00727 | 0.421 |
| 17100 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 30 | MITF | Sponge network | 0.997 | 0.00066 | -0.769 | 0.00022 | 0.421 |
| 17101 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-940 | 30 | GAS7 | Sponge network | -0.72 | 0.24239 | -0.555 | 0.01602 | 0.421 |
| 17102 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | PHC3 | Sponge network | 0.959 | 0 | 0.212 | 0.0499 | 0.421 |
| 17103 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 27 | PTPN14 | Sponge network | -0.154 | 0.78939 | 0.144 | 0.34595 | 0.421 |
| 17104 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | NF1 | Sponge network | 0.924 | 0 | 0.51 | 0 | 0.421 |
| 17105 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | ITGB3 | Sponge network | -0.513 | 0.04703 | -0.011 | 0.95751 | 0.421 |
| 17106 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 20 | LATS2 | Sponge network | -0.096 | 0.67958 | 0.159 | 0.2304 | 0.421 |
| 17107 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | NCAM2 | Sponge network | -0.738 | 0.21233 | -0.482 | 0.22565 | 0.421 |
| 17108 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.769 | 0 | 1.083 | 0 | 0.421 |
| 17109 | RP11-672A2.4 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-424-5p | 18 | FAT3 | Sponge network | 1.344 | 0 | -1.601 | 0.00051 | 0.421 |
| 17110 | RP11-356J5.12 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | CSDE1 | Sponge network | -0.899 | 6.0E-5 | -0.493 | 0 | 0.421 |
| 17111 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-539-5p | 19 | MOB1B | Sponge network | 0.946 | 0 | 0.197 | 0.15266 | 0.421 |
| 17112 | RP4-639F20.1 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 13 | RIMS2 | Sponge network | -0.517 | 0.02935 | -1.365 | 0.00208 | 0.421 |
| 17113 | LINC00622 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 18 | ABL2 | Sponge network | 1.169 | 0.00028 | 0.82 | 0 | 0.421 |
| 17114 | RP11-473I1.9 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 20 | TMEM30A | Sponge network | 0.683 | 1.0E-5 | 0.096 | 0.31031 | 0.421 |
| 17115 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | MED12L | Sponge network | -0.976 | 0.00025 | -0.194 | 0.55299 | 0.421 |
| 17116 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CXXC4 | Sponge network | -0.323 | 0.01372 | -0.433 | 0.21055 | 0.421 |
| 17117 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | IGSF10 | Sponge network | 0.194 | 0.64278 | -1.751 | 1.0E-5 | 0.421 |
| 17118 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | ZNF208 | Sponge network | 1.302 | 7.0E-5 | -0.4 | 0.22381 | 0.421 |
| 17119 | HHIP-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PCDH18 | Sponge network | -0.737 | 0.10822 | 0.216 | 0.24645 | 0.421 |
| 17120 | GS1-124K5.11 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p | 53 | DST | Sponge network | 0.494 | 0.00339 | -0.713 | 0.00096 | 0.421 |
| 17121 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-93-5p | 33 | WDFY3 | Sponge network | -0.473 | 0.07602 | -0.201 | 0.0967 | 0.421 |
| 17122 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | AKAP11 | Sponge network | 0.532 | 0.00505 | 0.196 | 0.09825 | 0.421 |
| 17123 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | RECK | Sponge network | -0.08 | 0.90092 | -1.043 | 0 | 0.421 |
| 17124 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-331-3p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ATRX | Sponge network | 0.673 | 0 | 0.261 | 0.04408 | 0.421 |
| 17125 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 27 | PIK3C2A | Sponge network | 0.848 | 0 | 0.061 | 0.53776 | 0.421 |
| 17126 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 19 | RSPO2 | Sponge network | -0.358 | 0.17255 | -3.038 | 0 | 0.421 |
| 17127 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-628-5p | 22 | BCL6 | Sponge network | 0.082 | 0.55654 | -0.211 | 0.19489 | 0.421 |
| 17128 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 33 | CREB3L2 | Sponge network | -0.611 | 0.09607 | -0.525 | 2.0E-5 | 0.421 |
| 17129 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | -0.773 | 0.04073 | -1.047 | 0.00364 | 0.421 |
| 17130 | LINC00641 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RBBP6 | Sponge network | -0.222 | 0.32445 | 0.163 | 0.05101 | 0.421 |
| 17131 | LINC00854 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-940 | 17 | CREB1 | Sponge network | 1.18 | 0 | 0.203 | 0.00537 | 0.421 |
| 17132 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 56 | UNC80 | Sponge network | -1.111 | 0.0003 | -1.934 | 0 | 0.421 |
| 17133 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 24 | ARHGAP28 | Sponge network | -0.793 | 7.0E-5 | -0.911 | 0.00013 | 0.421 |
| 17134 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | TP53INP1 | Sponge network | -1.111 | 0.0003 | -0.496 | 0.00064 | 0.421 |
| 17135 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-501-3p;hsa-miR-93-5p | 11 | SLITRK3 | Sponge network | -1.255 | 0.00981 | -3.328 | 0 | 0.421 |
| 17136 | MIR143HG |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 34 | MXI1 | Sponge network | -0.739 | 0.0574 | -0.987 | 0 | 0.421 |
| 17137 | MRVI1-AS1 |
hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-628-5p | 12 | IRS1 | Sponge network | -1.092 | 4.0E-5 | -0.026 | 0.88855 | 0.421 |
| 17138 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-93-5p | 42 | TACC1 | Sponge network | -0.615 | 0.00015 | -0.362 | 0.05406 | 0.421 |
| 17139 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-339-5p | 10 | FNBP1 | Sponge network | -0.227 | 0.31657 | -0.969 | 4.0E-5 | 0.421 |
| 17140 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 25 | QKI | Sponge network | -0.517 | 0.02935 | -0.333 | 0.04111 | 0.421 |
| 17141 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-331-3p | 10 | ATRX | Sponge network | 1.969 | 0.00316 | 0.261 | 0.04408 | 0.421 |
| 17142 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 13 | DNAJB4 | Sponge network | -0.842 | 0.01361 | -0.7 | 1.0E-5 | 0.421 |
| 17143 | PGM5-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-589-5p | 16 | CTIF | Sponge network | -4.546 | 0 | -0.389 | 0.00549 | 0.421 |
| 17144 | RP11-1006G14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-375;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 16 | AKAP6 | Sponge network | 0.559 | 0.00026 | -1.586 | 3.0E-5 | 0.421 |
| 17145 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 54 | QKI | Sponge network | -0.942 | 0 | -0.333 | 0.04111 | 0.421 |
| 17146 | RP11-53O19.1 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-342-3p | 22 | ZBTB4 | Sponge network | -0.405 | 0.22013 | -0.739 | 0 | 0.421 |
| 17147 | RP11-21A7A.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-98-5p | 25 | NTRK3 | Sponge network | -1.116 | 0.23686 | -1.77 | 3.0E-5 | 0.421 |
| 17148 | LINC00472 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-424-5p | 15 | RPS6KA6 | Sponge network | -0.842 | 0.01361 | -2.989 | 0 | 0.421 |
| 17149 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-5p | 11 | FSTL5 | Sponge network | -0.384 | 0.40652 | -1.3 | 0.01619 | 0.421 |
| 17150 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-501-5p | 10 | DACT1 | Sponge network | 0.176 | 0.37402 | 0.783 | 0.00072 | 0.421 |
| 17151 | CTC-429P9.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p | 15 | TSC1 | Sponge network | 0.082 | 0.55397 | 0.103 | 0.26071 | 0.421 |
| 17152 | PART1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | LHFP | Sponge network | -2.925 | 0 | -0.167 | 0.41371 | 0.421 |
| 17153 | RP11-156E6.1 |
hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p;hsa-miR-497-5p | 10 | ERCC6 | Sponge network | 1.266 | 0 | 0.23 | 0.015 | 0.421 |
| 17154 | MEG3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | EGR2 | Sponge network | 0.249 | 0.35902 | 0.309 | 0.17079 | 0.421 |
| 17155 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-532-5p;hsa-miR-96-5p | 11 | LRRC4 | Sponge network | -1.349 | 0.00017 | -1.774 | 0 | 0.421 |
| 17156 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-7-1-3p | 23 | EYA4 | Sponge network | -0.405 | 0.22013 | 0.3 | 0.36876 | 0.421 |
| 17157 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 23 | ERCC4 | Sponge network | 0.064 | 0.72934 | 0.126 | 0.15266 | 0.421 |
| 17158 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | ANK2 | Sponge network | -0.08 | 0.90092 | -1.603 | 1.0E-5 | 0.421 |
| 17159 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p;hsa-miR-96-5p | 19 | LIFR | Sponge network | -1.654 | 0.00144 | -1.794 | 0 | 0.421 |
| 17160 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 28 | FOXO3 | Sponge network | 0.72 | 0.0001 | -0.04 | 0.72028 | 0.421 |
| 17161 | RP11-53O19.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | SFRP1 | Sponge network | -0.405 | 0.22013 | -3.566 | 0 | 0.421 |
| 17162 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | LRP1B | Sponge network | -0.942 | 0 | -2.163 | 0 | 0.42 |
| 17163 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-935 | 12 | ABI2 | Sponge network | -0.376 | 0.00337 | 0.175 | 0.14787 | 0.42 |
| 17164 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | ST6GAL2 | Sponge network | -2.915 | 0.00015 | -1.201 | 0.00597 | 0.42 |
| 17165 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-577;hsa-miR-664a-3p;hsa-miR-671-5p | 19 | THBS1 | Sponge network | -2.69 | 0.0001 | 0.741 | 0.00386 | 0.42 |
| 17166 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 30 | LIFR | Sponge network | -2.117 | 0.00161 | -1.794 | 0 | 0.42 |
| 17167 | RAMP2-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | SYT4 | Sponge network | -0.513 | 0.04703 | -3.207 | 0 | 0.42 |
| 17168 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | MCC | Sponge network | -0.021 | 0.93598 | -1.005 | 0 | 0.42 |
| 17169 | CTC-550B14.7 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 11 | PHF6 | Sponge network | 1.161 | 0 | 0.785 | 0 | 0.42 |
| 17170 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-628-5p | 16 | GRIA3 | Sponge network | -0.583 | 0.14589 | -1.956 | 0 | 0.42 |
| 17171 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-582-5p | 21 | ZFHX3 | Sponge network | 0.792 | 0.0049 | 0.389 | 0.01363 | 0.42 |
| 17172 | RP11-196G11.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-590-3p;hsa-miR-940 | 16 | MDM4 | Sponge network | 0.875 | 0 | 0.345 | 0.00762 | 0.42 |
| 17173 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 28 | PGR | Sponge network | -1.645 | 0 | -1.704 | 0 | 0.42 |
| 17174 | AC009948.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-590-3p | 16 | JAKMIP2 | Sponge network | 0.532 | 0.00505 | -1.388 | 0 | 0.42 |
| 17175 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | EPHA3 | Sponge network | 0.392 | 0.0278 | 0.185 | 0.53588 | 0.42 |
| 17176 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | IL6ST | Sponge network | 0.342 | 0.03129 | -0.402 | 0.00676 | 0.42 |
| 17177 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p | 11 | DCN | Sponge network | -0.842 | 0.01361 | -1.288 | 0 | 0.42 |
| 17178 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-505-3p;hsa-miR-590-3p | 15 | KIT | Sponge network | -0.737 | 0.10822 | -2.184 | 0 | 0.42 |
| 17179 | AC083843.1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p | 13 | UBR3 | Sponge network | 1.4 | 0 | -0.021 | 0.82774 | 0.42 |
| 17180 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | TRPS1 | Sponge network | -2.117 | 0.00161 | -0.191 | 0.44109 | 0.42 |
| 17181 | LINC00473 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935 | 20 | PCDH10 | Sponge network | -1.203 | 0.03881 | -1.644 | 0.0078 | 0.42 |
| 17182 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 21 | ABI2 | Sponge network | 0.889 | 9.0E-5 | 0.175 | 0.14787 | 0.42 |
| 17183 | RP11-57H14.4 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | UBR3 | Sponge network | 0.083 | 0.66405 | -0.021 | 0.82774 | 0.42 |
| 17184 | BOLA3-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | BCL9 | Sponge network | -0.246 | 0.35034 | 0.321 | 0.00267 | 0.42 |
| 17185 | LINC01018 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-96-5p | 22 | CBFA2T3 | Sponge network | -2.915 | 0.00015 | -1.221 | 1.0E-5 | 0.42 |
| 17186 | RP11-707P17.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-654-3p | 14 | RICTOR | Sponge network | 1.877 | 0 | 0.388 | 0.00188 | 0.42 |
| 17187 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 12 | LRRK2 | Sponge network | -0.777 | 0.1134 | -1.136 | 1.0E-5 | 0.42 |
| 17188 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p | 14 | THBS1 | Sponge network | -0.693 | 0.05629 | 0.741 | 0.00386 | 0.42 |
| 17189 | MIR143HG |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | CITED2 | Sponge network | -0.739 | 0.0574 | -1.545 | 0 | 0.42 |
| 17190 | AC009120.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | ERCC4 | Sponge network | 0.919 | 0 | 0.126 | 0.15266 | 0.42 |
| 17191 | CTC-429P9.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-454-3p;hsa-miR-590-3p | 16 | DLC1 | Sponge network | 0.178 | 0.29106 | -0.382 | 0.05038 | 0.42 |
| 17192 | GS1-124K5.11 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-590-3p | 27 | BRD3 | Sponge network | 0.494 | 0.00339 | 0.37 | 0.00013 | 0.42 |
| 17193 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-590-3p | 17 | MCC | Sponge network | 0.178 | 0.29106 | -1.005 | 0 | 0.42 |
| 17194 | RP11-693J15.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-671-5p | 18 | CYLD | Sponge network | -0.72 | 0.24239 | -0.367 | 0.00171 | 0.42 |
| 17195 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p | 26 | BCL2 | Sponge network | 0.082 | 0.55397 | -0.899 | 0 | 0.42 |
| 17196 | RP11-701H24.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 18 | NTRK3 | Sponge network | -1.425 | 0.04204 | -1.77 | 3.0E-5 | 0.42 |
| 17197 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p | 13 | AHNAK | Sponge network | -4.546 | 0 | -1.207 | 0 | 0.42 |
| 17198 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | CHD2 | Sponge network | 0.539 | 0.03722 | 0.049 | 0.55371 | 0.42 |
| 17199 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-93-5p | 19 | ADARB1 | Sponge network | 0.497 | 0.01451 | -0.335 | 0.06631 | 0.42 |
| 17200 | RP11-434B12.1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ATM | Sponge network | -0.031 | 0.89063 | 0.178 | 0.21568 | 0.42 |
| 17201 | AC007879.7 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326 | 11 | CBFB | Sponge network | 4.3 | 0 | 1.061 | 0 | 0.42 |
| 17202 | WDFY3-AS2 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 17 | ID4 | Sponge network | -0.942 | 0 | -1.644 | 0 | 0.42 |
| 17203 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | SEMA3C | Sponge network | -0.611 | 0.09607 | -0.272 | 0.31153 | 0.42 |
| 17204 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | ZNF770 | Sponge network | 1.601 | 0 | 0.206 | 0.06751 | 0.42 |
| 17205 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-940 | 41 | NTRK2 | Sponge network | -0.405 | 0.22013 | -0.736 | 0.06495 | 0.42 |
| 17206 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 35 | HBP1 | Sponge network | -2.46 | 0 | -0.454 | 0 | 0.42 |
| 17207 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-369-3p | 10 | AFF4 | Sponge network | 0.062 | 0.55274 | -0.266 | 0.01565 | 0.42 |
| 17208 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 15 | ZNF770 | Sponge network | 0.959 | 0 | 0.206 | 0.06751 | 0.42 |
| 17209 | LINC00472 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p | 14 | RELN | Sponge network | -0.842 | 0.01361 | -2.403 | 0 | 0.42 |
| 17210 | RP11-401P9.4 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p | 10 | PPP2R2C | Sponge network | -0.384 | 0.40652 | -0.959 | 0.07428 | 0.42 |
| 17211 | RP11-25K21.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-589-3p | 13 | QKI | Sponge network | 0.489 | 0.25786 | -0.333 | 0.04111 | 0.42 |
| 17212 | RP11-531A24.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p | 11 | FBXO11 | Sponge network | 0.75 | 0.00054 | 0.142 | 0.04938 | 0.42 |
| 17213 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | SMARCA2 | Sponge network | -2.452 | 0 | -0.529 | 0.00014 | 0.42 |
| 17214 | RP11-890B15.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 40 | AFF1 | Sponge network | 0.101 | 0.60919 | -0.384 | 0.00053 | 0.42 |
| 17215 | RP1-151F17.2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 46 | DST | Sponge network | 0.222 | 0.29133 | -0.713 | 0.00096 | 0.42 |
| 17216 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 21 | RARB | Sponge network | -0.487 | 0.02157 | -0.825 | 2.0E-5 | 0.42 |
| 17217 | CTC-429P9.5 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 11 | ERCC4 | Sponge network | 0.178 | 0.29106 | 0.126 | 0.15266 | 0.42 |
| 17218 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 11 | ZHX2 | Sponge network | -1.575 | 6.0E-5 | -0.192 | 0.18459 | 0.42 |
| 17219 | EMX2OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 54 | PTPRD | Sponge network | -0.777 | 0.1134 | -1.005 | 0.00064 | 0.42 |
| 17220 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | SLITRK6 | Sponge network | -1.111 | 0.0003 | -2.121 | 0 | 0.42 |
| 17221 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-96-5p | 14 | DPYD | Sponge network | -0.576 | 0.10121 | -0.736 | 0.00076 | 0.42 |
| 17222 | RP11-540B6.6 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p | 12 | RASAL2 | Sponge network | 0.93 | 0 | 0.869 | 0 | 0.42 |
| 17223 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-194-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-550a-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 18 | RAP1A | Sponge network | -4.546 | 0 | -0.929 | 0 | 0.42 |
| 17224 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p | 13 | SLITRK3 | Sponge network | -0.405 | 0.22013 | -3.328 | 0 | 0.42 |
| 17225 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -0.376 | 0.00337 | -0.243 | 0.11408 | 0.42 |
| 17226 | RP11-13K12.5 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-940 | 33 | GRIK3 | Sponge network | -0.318 | 0.65847 | -3.208 | 0 | 0.42 |
| 17227 | MEG3 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p | 20 | MAFB | Sponge network | 0.249 | 0.35902 | -0.023 | 0.89865 | 0.42 |
| 17228 | LINC00865 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 37 | TRPS1 | Sponge network | -1.249 | 7.0E-5 | -0.191 | 0.44109 | 0.42 |
| 17229 | RP11-216B9.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 11 | FAM172A | Sponge network | 0.174 | 0.30034 | -0.57 | 0 | 0.42 |
| 17230 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p | 10 | PTPN13 | Sponge network | -0.777 | 0.16667 | -1.208 | 0 | 0.42 |
| 17231 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p | 13 | PLSCR4 | Sponge network | -0.214 | 0.44248 | -0.474 | 0.03833 | 0.42 |
| 17232 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p | 50 | BNC2 | Sponge network | -0.615 | 0.00015 | -0.491 | 0.09988 | 0.42 |
| 17233 | RP11-119F7.5 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ITPKB | Sponge network | 0.225 | 0.30644 | -0.704 | 0.00106 | 0.42 |
| 17234 | AGAP11 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p | 11 | SH3GLB1 | Sponge network | -1.645 | 0 | -0.115 | 0.14773 | 0.42 |
| 17235 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 17 | RCAN2 | Sponge network | -0.355 | 0.0077 | -0.33 | 0.15172 | 0.42 |
| 17236 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | TBX18 | Sponge network | -1.111 | 0.0003 | 0.843 | 0.02945 | 0.42 |
| 17237 | RP11-736K20.5 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | KLF10 | Sponge network | -0.358 | 0.17255 | -0.783 | 0 | 0.42 |
| 17238 | RP11-597D13.9 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 12 | FAM133A | Sponge network | 1.302 | 7.0E-5 | 0.549 | 0.29009 | 0.42 |
| 17239 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 20 | RB1CC1 | Sponge network | 0.421 | 0.00084 | 0.175 | 0.12559 | 0.42 |
| 17240 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577 | 19 | HIP1 | Sponge network | 0.056 | 0.81195 | 0.774 | 0 | 0.42 |
| 17241 | PWAR6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 37 | ERC1 | Sponge network | -1.575 | 6.0E-5 | -0.108 | 0.38794 | 0.42 |
| 17242 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 32 | EPHA7 | Sponge network | -0.738 | 0.21233 | -2.197 | 3.0E-5 | 0.42 |
| 17243 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-5p | 14 | PHC3 | Sponge network | 0.706 | 0 | 0.212 | 0.0499 | 0.42 |
| 17244 | CTC-429P9.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-339-5p | 11 | FNBP1 | Sponge network | 0.082 | 0.55397 | -0.969 | 4.0E-5 | 0.42 |
| 17245 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 48 | EPHA5 | Sponge network | -2.46 | 0 | -0.957 | 0.01733 | 0.42 |
| 17246 | RP11-822E23.8 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-744-3p | 28 | DIP2C | Sponge network | -0.777 | 0.16667 | -0.436 | 0.01713 | 0.42 |
| 17247 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-93-3p | 45 | BMPR2 | Sponge network | 0.401 | 0.00066 | 0.206 | 0.0635 | 0.42 |
| 17248 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 51 | PAFAH1B1 | Sponge network | -0.739 | 0.0574 | -0.429 | 0 | 0.42 |
| 17249 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | PRRX1 | Sponge network | 1.302 | 7.0E-5 | 1.316 | 0 | 0.42 |
| 17250 | LINC00265 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 17 | ARHGEF12 | Sponge network | 1.07 | 0 | 0.09 | 0.35693 | 0.42 |
| 17251 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 31 | RBMS3 | Sponge network | -0.738 | 0.21233 | -0.519 | 0.03551 | 0.42 |
| 17252 | RP11-1006G14.4 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PIK3CA | Sponge network | 0.559 | 0.00026 | 0.195 | 0.06908 | 0.42 |
| 17253 | RP11-216B9.6 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b | 14 | SYNM | Sponge network | 0.174 | 0.30034 | -2.309 | 1.0E-5 | 0.42 |
| 17254 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p | 14 | RET | Sponge network | -1.886 | 0 | -0.833 | 0.03517 | 0.42 |
| 17255 | CKMT2-AS1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 20 | ATM | Sponge network | 0.082 | 0.55654 | 0.178 | 0.21568 | 0.42 |
| 17256 | RP11-144G6.12 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | DDX3X | Sponge network | 0.826 | 1.0E-5 | -0.152 | 0.07686 | 0.42 |
| 17257 | USP3-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-651-5p | 26 | PRDM2 | Sponge network | -0.162 | 0.47687 | -0.258 | 0.00721 | 0.42 |
| 17258 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-452-5p;hsa-miR-590-3p | 10 | SEPT6 | Sponge network | -0.358 | 0.17255 | -0.598 | 0.00181 | 0.42 |
| 17259 | LINC01018 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | TET1 | Sponge network | -2.915 | 0.00015 | -0.135 | 0.52138 | 0.42 |
| 17260 | RP11-400K9.4 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-942-5p | 12 | GLIPR1 | Sponge network | -0.611 | 0.09607 | 0.146 | 0.3276 | 0.42 |
| 17261 | RP11-822E23.8 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-629-3p | 10 | CXCL12 | Sponge network | -0.777 | 0.16667 | -1.421 | 0 | 0.42 |
| 17262 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 29 | CYLD | Sponge network | -0.129 | 0.57177 | -0.367 | 0.00171 | 0.42 |
| 17263 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 39 | TACC1 | Sponge network | 0.083 | 0.66405 | -0.362 | 0.05406 | 0.42 |
| 17264 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | CACNA2D1 | Sponge network | 0.064 | 0.72934 | -0.846 | 0.00675 | 0.42 |
| 17265 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | GRIA3 | Sponge network | -0.129 | 0.57177 | -1.956 | 0 | 0.42 |
| 17266 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 13 | UVRAG | Sponge network | 0.194 | 0.64278 | -0.017 | 0.83865 | 0.42 |
| 17267 | RP4-613B23.1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-502-5p | 11 | PTPN14 | Sponge network | -0.309 | 0.1145 | 0.144 | 0.34595 | 0.42 |
| 17268 | TUG1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-495-3p | 12 | STAG2 | Sponge network | 0.972 | 0 | 0.252 | 0.00438 | 0.42 |
| 17269 | RP11-46C24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | AKAP6 | Sponge network | 0.392 | 0.0278 | -1.586 | 3.0E-5 | 0.42 |
| 17270 | RP11-531A24.5 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 15 | KIF5B | Sponge network | 0.75 | 0.00054 | 0.362 | 0.00066 | 0.42 |
| 17271 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p | 15 | RBMS3 | Sponge network | -0.829 | 0.01343 | -0.519 | 0.03551 | 0.42 |
| 17272 | HCG18 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | EXOC8 | Sponge network | 1.063 | 0 | 0.216 | 0.00598 | 0.42 |
| 17273 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-28-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | HACE1 | Sponge network | 0.873 | 0 | -0.072 | 0.55239 | 0.42 |
| 17274 | THAP9-AS1 |
hsa-miR-126-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-628-5p | 10 | TRIM33 | Sponge network | 1.079 | 0 | 0.402 | 7.0E-5 | 0.42 |
| 17275 | ZSCAN16-AS1 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-493-5p;hsa-miR-532-3p | 13 | SYNM | Sponge network | -0.342 | 0.02288 | -2.309 | 1.0E-5 | 0.42 |
| 17276 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p | 18 | PHC3 | Sponge network | -1.331 | 3.0E-5 | 0.212 | 0.0499 | 0.42 |
| 17277 | ACTA2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | MSN | Sponge network | 0.194 | 0.64278 | -0.023 | 0.88422 | 0.42 |
| 17278 | RP11-182J1.1 |
hsa-miR-1271-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-24-2-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | PPM1L | Sponge network | -0.187 | 0.23787 | -0.537 | 0.00616 | 0.42 |
| 17279 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p | 15 | PRRX1 | Sponge network | -1.331 | 3.0E-5 | 1.316 | 0 | 0.42 |
| 17280 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-98-5p | 45 | PRKAA2 | Sponge network | -0.227 | 0.31657 | -2.462 | 0 | 0.42 |
| 17281 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-577 | 13 | HIP1 | Sponge network | -0.473 | 0.07602 | 0.774 | 0 | 0.42 |
| 17282 | ANKRD10-IT1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 11 | RASAL2 | Sponge network | 1.739 | 0 | 0.869 | 0 | 0.42 |
| 17283 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | NF1 | Sponge network | 1.597 | 0 | 0.51 | 0 | 0.42 |
| 17284 | MEG3 |
hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-874-3p;hsa-miR-96-5p | 10 | AJAP1 | Sponge network | 0.249 | 0.35902 | -0.561 | 0.04832 | 0.42 |
| 17285 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 15 | ZMYM2 | Sponge network | 1.262 | 0 | 0.279 | 0.01273 | 0.42 |
| 17286 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | ROBO1 | Sponge network | 0.225 | 0.30644 | -0.009 | 0.96154 | 0.42 |
| 17287 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 15 | SPTBN4 | Sponge network | -0.583 | 0.14589 | -0.786 | 0.01281 | 0.42 |
| 17288 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-199b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | TSC1 | Sponge network | 0.72 | 0.0001 | 0.103 | 0.26071 | 0.42 |
| 17289 | RP11-968O1.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p | 19 | GNAQ | Sponge network | -0.829 | 0.01343 | -0.367 | 0.00102 | 0.42 |
| 17290 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 32 | DIP2C | Sponge network | 0.083 | 0.66405 | -0.436 | 0.01713 | 0.42 |
| 17291 | FAM66C |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | RIMS1 | Sponge network | -0.901 | 0.00019 | -3.143 | 0 | 0.42 |
| 17292 | LINC00863 |
hsa-let-7a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 13 | GPAM | Sponge network | 0.189 | 0.13981 | 0.142 | 0.29732 | 0.42 |
| 17293 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | MAF | Sponge network | -0.563 | 0.02545 | -1.257 | 0 | 0.42 |
| 17294 | RP11-225B17.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 16 | CREB1 | Sponge network | 1.055 | 0 | 0.203 | 0.00537 | 0.419 |
| 17295 | RP11-356J5.12 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-940 | 11 | PHOX2B | Sponge network | -0.899 | 6.0E-5 | -2.68 | 0 | 0.419 |
| 17296 | LIPE-AS1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p | 16 | DDX3X | Sponge network | 0.078 | 0.55803 | -0.152 | 0.07686 | 0.419 |
| 17297 | WDFY3-AS2 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | RPS6KA6 | Sponge network | -0.942 | 0 | -2.989 | 0 | 0.419 |
| 17298 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CLIP1 | Sponge network | -1.697 | 0.00889 | -0.215 | 0.08684 | 0.419 |
| 17299 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | PSIP1 | Sponge network | -0.615 | 0.00015 | -0.005 | 0.96897 | 0.419 |
| 17300 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 69 | IL6ST | Sponge network | -0.615 | 0.00015 | -0.402 | 0.00676 | 0.419 |
| 17301 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-590-3p | 15 | NIN | Sponge network | -0.358 | 0.17255 | -0.253 | 0.13794 | 0.419 |
| 17302 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 18 | TIMP3 | Sponge network | 0.199 | 0.38015 | -0.546 | 0.02307 | 0.419 |
| 17303 | PWAR6 |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 11 | GLIPR2 | Sponge network | -1.575 | 6.0E-5 | -0.996 | 0 | 0.419 |
| 17304 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-93-3p | 22 | ST6GAL2 | Sponge network | -0.773 | 0.04073 | -1.201 | 0.00597 | 0.419 |
| 17305 | LINC00641 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 18 | SPG20 | Sponge network | -0.222 | 0.32445 | -1.16 | 0 | 0.419 |
| 17306 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | RET | Sponge network | -0.737 | 0.06182 | -0.833 | 0.03517 | 0.419 |
| 17307 | DIO3OS |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 21 | CDH23 | Sponge network | -0.773 | 0.04073 | -0.974 | 0.00034 | 0.419 |
| 17308 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 12 | THSD7A | Sponge network | 1.302 | 7.0E-5 | 0.242 | 0.25154 | 0.419 |
| 17309 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 47 | CHD5 | Sponge network | 0.997 | 0.00066 | -0.922 | 0.01521 | 0.419 |
| 17310 | RP11-59H7.3 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-3p | 18 | ABL2 | Sponge network | 2.163 | 0 | 0.82 | 0 | 0.419 |
| 17311 | MIR143HG |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 12 | ALDH1A2 | Sponge network | -0.739 | 0.0574 | -2.411 | 0 | 0.419 |
| 17312 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 45 | CBX5 | Sponge network | 0.494 | 0.00339 | 0.165 | 0.24446 | 0.419 |
| 17313 | GS1-124K5.11 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | PRUNE2 | Sponge network | 0.494 | 0.00339 | -1.733 | 0.00022 | 0.419 |
| 17314 | LINC00926 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-629-3p;hsa-miR-92a-3p | 12 | RASSF2 | Sponge network | 0.756 | 0.00739 | -0.273 | 0.21961 | 0.419 |
| 17315 | PGM5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 24 | IL17RD | Sponge network | -4.546 | 0 | -0.019 | 0.94873 | 0.419 |
| 17316 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-93-5p | 11 | FGFR1 | Sponge network | 0.056 | 0.81195 | -0.509 | 0.03957 | 0.419 |
| 17317 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p | 14 | SON | Sponge network | 0.594 | 0.00064 | 0.168 | 0.04406 | 0.419 |
| 17318 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ADAMTSL3 | Sponge network | -0.206 | 0.12942 | -1.559 | 0 | 0.419 |
| 17319 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 31 | EPHA3 | Sponge network | -0.942 | 0 | 0.185 | 0.53588 | 0.419 |
| 17320 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | AFF3 | Sponge network | -0.663 | 0.10887 | -2.373 | 0 | 0.419 |
| 17321 | SERTAD4-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-505-5p;hsa-miR-93-3p | 13 | CTIF | Sponge network | -0.932 | 0.00329 | -0.389 | 0.00549 | 0.419 |
| 17322 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | ETV1 | Sponge network | -0.405 | 0.22013 | -0.096 | 0.67674 | 0.419 |
| 17323 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | YAP1 | Sponge network | 0.61 | 0.01593 | 0.22 | 0.11836 | 0.419 |
| 17324 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-370-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | PCDH10 | Sponge network | -0.187 | 0.23787 | -1.644 | 0.0078 | 0.419 |
| 17325 | RP11-473I1.9 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 20 | PICALM | Sponge network | 0.683 | 1.0E-5 | 0.086 | 0.23523 | 0.419 |
| 17326 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | LRRC55 | Sponge network | 0.889 | 9.0E-5 | -0.026 | 0.93163 | 0.419 |
| 17327 | LINC00969 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 18 | RASAL2 | Sponge network | 0.683 | 1.0E-5 | 0.869 | 0 | 0.419 |
| 17328 | RP11-359B12.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-96-5p | 39 | IL17RD | Sponge network | 0.064 | 0.72934 | -0.019 | 0.94873 | 0.419 |
| 17329 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | NEDD4 | Sponge network | 0.064 | 0.72934 | 0.02 | 0.89834 | 0.419 |
| 17330 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | MOB1B | Sponge network | 1.2 | 0.01813 | 0.197 | 0.15266 | 0.419 |
| 17331 | EMX2OS |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 15 | PRDM5 | Sponge network | -0.777 | 0.1134 | -1.09 | 0.00014 | 0.419 |
| 17332 | RP11-404E16.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-342-5p | 10 | DTNA | Sponge network | 0.097 | 0.91311 | -1.648 | 1.0E-5 | 0.419 |
| 17333 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-629-3p | 18 | RASSF2 | Sponge network | -1.375 | 0.08642 | -0.273 | 0.21961 | 0.419 |
| 17334 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 60 | TCF4 | Sponge network | -0.709 | 0.18168 | 0.016 | 0.92271 | 0.419 |
| 17335 | MALAT1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TRIM13 | Sponge network | 0.782 | 0.00016 | 0.183 | 0.06968 | 0.419 |
| 17336 | RP4-639F20.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-624-5p | 12 | GJA1 | Sponge network | -0.517 | 0.02935 | -0.485 | 0.02179 | 0.419 |
| 17337 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-96-5p | 40 | JAZF1 | Sponge network | -0.942 | 0 | -0.377 | 0.02677 | 0.419 |
| 17338 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 17 | AKAP6 | Sponge network | 0.889 | 9.0E-5 | -1.586 | 3.0E-5 | 0.419 |
| 17339 | RP11-774O3.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-629-3p | 12 | SPTBN4 | Sponge network | -0.487 | 0.02157 | -0.786 | 0.01281 | 0.419 |
| 17340 | RP11-540B6.6 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-497-5p | 20 | ARHGEF12 | Sponge network | 0.93 | 0 | 0.09 | 0.35693 | 0.419 |
| 17341 | AC159540.1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p;hsa-miR-495-3p | 11 | PHF6 | Sponge network | 1.122 | 0 | 0.785 | 0 | 0.419 |
| 17342 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-769-5p | 11 | RCAN2 | Sponge network | -0.811 | 2.0E-5 | -0.33 | 0.15172 | 0.419 |
| 17343 | AC009120.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p | 20 | ABI2 | Sponge network | 0.919 | 0 | 0.175 | 0.14787 | 0.419 |
| 17344 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-93-5p | 18 | LATS2 | Sponge network | -0.162 | 0.47687 | 0.159 | 0.2304 | 0.419 |
| 17345 | BVES-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-628-5p;hsa-miR-708-5p;hsa-miR-744-3p | 18 | TOM1L2 | Sponge network | -2.62 | 0 | -0.994 | 0 | 0.419 |
| 17346 | PWAR6 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 44 | AFF1 | Sponge network | -1.575 | 6.0E-5 | -0.384 | 0.00053 | 0.419 |
| 17347 | LINC00865 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-7-1-3p | 11 | PCDH18 | Sponge network | -1.249 | 7.0E-5 | 0.216 | 0.24645 | 0.419 |
| 17348 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 11 | ERG | Sponge network | 0.299 | 0.57722 | 0.002 | 0.99011 | 0.419 |
| 17349 | DIO3OS |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 19 | CADM3 | Sponge network | -0.773 | 0.04073 | -3.663 | 0 | 0.419 |
| 17350 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 37 | PRKAB2 | Sponge network | 0.064 | 0.72934 | -0.367 | 0.01371 | 0.419 |
| 17351 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 21 | ERCC4 | Sponge network | 0.537 | 0.00139 | 0.126 | 0.15266 | 0.419 |
| 17352 | DIO3OS |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-7-5p;hsa-miR-769-5p | 24 | SETBP1 | Sponge network | -0.773 | 0.04073 | -1.302 | 1.0E-5 | 0.419 |
| 17353 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | ANK2 | Sponge network | -0.206 | 0.12942 | -1.603 | 1.0E-5 | 0.419 |
| 17354 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 52 | IL6ST | Sponge network | -1.161 | 0.00591 | -0.402 | 0.00676 | 0.419 |
| 17355 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-505-5p | 18 | HBP1 | Sponge network | -1.092 | 4.0E-5 | -0.454 | 0 | 0.419 |
| 17356 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p | 14 | TXNIP | Sponge network | -0.899 | 6.0E-5 | -1.277 | 0 | 0.419 |
| 17357 | RP11-384O8.1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 22 | RNF144A | Sponge network | -0.738 | 0.21233 | 0.594 | 0.0009 | 0.419 |
| 17358 | PCA3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | ERG | Sponge network | -0.709 | 0.18168 | 0.002 | 0.99011 | 0.419 |
| 17359 | CTA-363E19.2 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 1.055 | 0 | 0.362 | 0.00066 | 0.419 |
| 17360 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | EPHA3 | Sponge network | 0.419 | 0.0319 | 0.185 | 0.53588 | 0.419 |
| 17361 | CTD-3157E16.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 10 | GAS7 | Sponge network | -0.326 | 0.28809 | -0.555 | 0.01602 | 0.419 |
| 17362 | NDUFA6-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p | 21 | SYNM | Sponge network | -0.355 | 0.0077 | -2.309 | 1.0E-5 | 0.419 |
| 17363 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | RBMS3 | Sponge network | 0.056 | 0.8494 | -0.519 | 0.03551 | 0.419 |
| 17364 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | AKAP11 | Sponge network | 0.349 | 0.01695 | 0.196 | 0.09825 | 0.419 |
| 17365 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | APC | Sponge network | 0.101 | 0.60919 | -0.255 | 0.04051 | 0.419 |
| 17366 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 58 | PTPRD | Sponge network | -1.697 | 0.00889 | -1.005 | 0.00064 | 0.419 |
| 17367 | RP11-53O19.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p | 10 | KCTD8 | Sponge network | -0.405 | 0.22013 | -3.359 | 0 | 0.419 |
| 17368 | RP11-244H3.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PIK3CA | Sponge network | 0.659 | 3.0E-5 | 0.195 | 0.06908 | 0.419 |
| 17369 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p | 22 | BCL2 | Sponge network | -2.531 | 0 | -0.899 | 0 | 0.419 |
| 17370 | RP11-439A17.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-223-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-98-5p | 12 | DMD | Sponge network | 0.051 | 0.95756 | -1.506 | 0 | 0.419 |
| 17371 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 13 | CACNA2D1 | Sponge network | -0.318 | 0.65847 | -0.846 | 0.00675 | 0.419 |
| 17372 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-744-3p;hsa-miR-942-5p | 33 | HLF | Sponge network | -0.693 | 0.05629 | -2.443 | 0 | 0.419 |
| 17373 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-501-5p | 22 | JAZF1 | Sponge network | -0.72 | 0.24239 | -0.377 | 0.02677 | 0.419 |
| 17374 | GNG12-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-590-3p | 15 | ITGB3 | Sponge network | -0.793 | 7.0E-5 | -0.011 | 0.95751 | 0.419 |
| 17375 | ACTA2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | AFF1 | Sponge network | 0.194 | 0.64278 | -0.384 | 0.00053 | 0.419 |
| 17376 | CTC-429P9.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-532-5p | 22 | IL17RD | Sponge network | 0.178 | 0.29106 | -0.019 | 0.94873 | 0.419 |
| 17377 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | RASSF8 | Sponge network | 0.374 | 0.20054 | -0.122 | 0.65591 | 0.419 |
| 17378 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3913-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 32 | HLF | Sponge network | -0.421 | 0.0114 | -2.443 | 0 | 0.419 |
| 17379 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 23 | PDS5B | Sponge network | 0.421 | 0.00084 | 0.259 | 0.00962 | 0.419 |
| 17380 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 40 | NOTCH2 | Sponge network | -0.769 | 0.00035 | 0.138 | 0.35163 | 0.419 |
| 17381 | RP5-894A10.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-365a-3p;hsa-miR-495-3p | 12 | RICTOR | Sponge network | 0.697 | 9.0E-5 | 0.388 | 0.00188 | 0.419 |
| 17382 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | LRRC7 | Sponge network | 0.299 | 0.57722 | -1.341 | 0.01193 | 0.419 |
| 17383 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 26 | ZBTB4 | Sponge network | 0.335 | 0.20596 | -0.739 | 0 | 0.419 |
| 17384 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-429;hsa-miR-629-3p;hsa-miR-96-5p | 21 | EBF1 | Sponge network | -1.654 | 0.00144 | -0.384 | 0.08856 | 0.419 |
| 17385 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 52 | TMEM132B | Sponge network | -0.576 | 0.10121 | -0.83 | 0.01084 | 0.419 |
| 17386 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 22 | CDC73 | Sponge network | 1.254 | 0 | 0.094 | 0.20463 | 0.419 |
| 17387 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-7-1-3p | 13 | DKK3 | Sponge network | 0.61 | 0.01593 | -0.052 | 0.77783 | 0.419 |
| 17388 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | MITF | Sponge network | -0.421 | 0.0114 | -0.769 | 0.00022 | 0.419 |
| 17389 | PART1 |
hsa-let-7f-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 10 | ROR2 | Sponge network | -2.925 | 0 | -1.091 | 0.0008 | 0.419 |
| 17390 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 34 | SASH1 | Sponge network | 0.72 | 0.0001 | -0.502 | 0.00175 | 0.419 |
| 17391 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 32 | RASSF2 | Sponge network | -0.777 | 0.16667 | -0.273 | 0.21961 | 0.419 |
| 17392 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | NEDD4 | Sponge network | 0.385 | 0.10067 | 0.02 | 0.89834 | 0.419 |
| 17393 | AC106786.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-629-3p;hsa-miR-629-5p | 10 | RASSF8 | Sponge network | 1.122 | 0.02989 | -0.122 | 0.65591 | 0.419 |
| 17394 | ACTA2-AS1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | RPS6KA6 | Sponge network | 0.194 | 0.64278 | -2.989 | 0 | 0.419 |
| 17395 | CTA-204B4.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-671-5p | 11 | SHPRH | Sponge network | 0.765 | 7.0E-5 | 0.098 | 0.40994 | 0.419 |
| 17396 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-7-1-3p | 29 | PRKAB2 | Sponge network | -0.405 | 0.22013 | -0.367 | 0.01371 | 0.419 |
| 17397 | A2M-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-511-5p;hsa-miR-590-3p | 16 | MRVI1 | Sponge network | -0.08 | 0.90092 | -1.051 | 0.00022 | 0.419 |
| 17398 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-375 | 14 | BNC2 | Sponge network | 1.969 | 0.00316 | -0.491 | 0.09988 | 0.419 |
| 17399 | GAS5 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 11 | PHF6 | Sponge network | 0.728 | 0 | 0.785 | 0 | 0.419 |
| 17400 | TP73-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-940 | 13 | CELF4 | Sponge network | -0.563 | 0.02545 | -1.135 | 0.00344 | 0.419 |
| 17401 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-5p | 36 | BCL2 | Sponge network | 0.214 | 0.18246 | -0.899 | 0 | 0.419 |
| 17402 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-33a-5p | 10 | CHD1 | Sponge network | 0.706 | 0 | 0.322 | 0.00215 | 0.419 |
| 17403 | RP11-6O2.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-96-5p | 19 | PIK3CA | Sponge network | 0.331 | 0.57247 | 0.195 | 0.06908 | 0.419 |
| 17404 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 37 | CBX5 | Sponge network | 0.225 | 0.30644 | 0.165 | 0.24446 | 0.419 |
| 17405 | RP5-1198O20.4 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | ZDBF2 | Sponge network | 0.997 | 0.00066 | -0.998 | 0.00025 | 0.419 |
| 17406 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 14 | RB1CC1 | Sponge network | 0.541 | 0.00125 | 0.175 | 0.12559 | 0.419 |
| 17407 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | LRP1B | Sponge network | -1.886 | 0 | -2.163 | 0 | 0.419 |
| 17408 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-96-5p | 13 | RIMBP2 | Sponge network | -0.517 | 0.02935 | -1.968 | 5.0E-5 | 0.419 |
| 17409 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | CLASP2 | Sponge network | 0.252 | 0.08508 | -0.038 | 0.71 | 0.419 |
| 17410 | CTD-2314G24.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 15 | ZBTB20 | Sponge network | 0.246 | 0.59912 | -0.122 | 0.57569 | 0.419 |
| 17411 | RP11-156E6.1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p | 13 | RASAL2 | Sponge network | 1.266 | 0 | 0.869 | 0 | 0.418 |
| 17412 | RP11-253E3.3 |
hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-27b-3p;hsa-miR-3065-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-497-5p;hsa-miR-664a-5p | 13 | SOX11 | Sponge network | 1.764 | 0 | 2.68 | 0 | 0.418 |
| 17413 | EMX2OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940 | 36 | SNX29 | Sponge network | -0.777 | 0.1134 | -0.34 | 0.01371 | 0.418 |
| 17414 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | ST6GALNAC3 | Sponge network | -0.576 | 0.10121 | -0.987 | 0 | 0.418 |
| 17415 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | LRIG1 | Sponge network | 0.064 | 0.72934 | -0.537 | 0.00588 | 0.418 |
| 17416 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | RASSF8 | Sponge network | 0.395 | 0.15451 | -0.122 | 0.65591 | 0.418 |
| 17417 | OIP5-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 19 | MACF1 | Sponge network | 0.421 | 0.00084 | 0.019 | 0.90335 | 0.418 |
| 17418 | FAM66C |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ZNF208 | Sponge network | -0.901 | 0.00019 | -0.4 | 0.22381 | 0.418 |
| 17419 | RP11-774O3.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p | 28 | ASTN1 | Sponge network | -0.487 | 0.02157 | -2.389 | 2.0E-5 | 0.418 |
| 17420 | RP11-156P1.3 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | 0.214 | 0.18246 | -2.633 | 0 | 0.418 |
| 17421 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-93-5p | 12 | LRRC55 | Sponge network | -1.654 | 0.00144 | -0.026 | 0.93163 | 0.418 |
| 17422 | LINC00654 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 30 | NAV3 | Sponge network | 0.374 | 0.20054 | 0.212 | 0.47729 | 0.418 |
| 17423 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | RASSF8 | Sponge network | -1.886 | 0 | -0.122 | 0.65591 | 0.418 |
| 17424 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | EPHA5 | Sponge network | -0.222 | 0.32445 | -0.957 | 0.01733 | 0.418 |
| 17425 | CTD-2020K17.1 | hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p | 12 | MDM4 | Sponge network | 1.449 | 0 | 0.345 | 0.00762 | 0.418 |
| 17426 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-590-3p | 17 | IGF1 | Sponge network | -0.811 | 2.0E-5 | -0.204 | 0.5898 | 0.418 |
| 17427 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 35 | EBF1 | Sponge network | -1.216 | 0 | -0.384 | 0.08856 | 0.418 |
| 17428 | RP11-822E23.8 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 11 | RHOB | Sponge network | -0.777 | 0.16667 | -1.052 | 0 | 0.418 |
| 17429 | RP11-344B5.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b | 14 | MCC | Sponge network | -0.055 | 0.88482 | -1.005 | 0 | 0.418 |
| 17430 | MIR143HG |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 33 | RNF144A | Sponge network | -0.739 | 0.0574 | 0.594 | 0.0009 | 0.418 |
| 17431 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-369-3p;hsa-miR-374b-5p | 15 | ABI2 | Sponge network | 0.542 | 0.00054 | 0.175 | 0.14787 | 0.418 |
| 17432 | TPTEP1 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | DIRAS1 | Sponge network | -1.216 | 0 | -2.518 | 0 | 0.418 |
| 17433 | RP11-25K19.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | UNC5C | Sponge network | -1.057 | 0.00029 | -0.178 | 0.40214 | 0.418 |
| 17434 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | CREBBP | Sponge network | 0.083 | 0.66405 | 0.126 | 0.15186 | 0.418 |
| 17435 | GNG12-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-590-3p | 10 | PTPN13 | Sponge network | -0.793 | 7.0E-5 | -1.208 | 0 | 0.418 |
| 17436 | LINC00472 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-7-1-3p | 18 | GJA1 | Sponge network | -0.842 | 0.01361 | -0.485 | 0.02179 | 0.418 |
| 17437 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-877-5p;hsa-miR-93-5p | 25 | PGR | Sponge network | -1.047 | 0 | -1.704 | 0 | 0.418 |
| 17438 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p | 12 | LRRK2 | Sponge network | -0.777 | 0.16667 | -1.136 | 1.0E-5 | 0.418 |
| 17439 | RP11-597D13.9 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | MACF1 | Sponge network | 1.302 | 7.0E-5 | 0.019 | 0.90335 | 0.418 |
| 17440 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-93-5p | 14 | FAT4 | Sponge network | 0.056 | 0.81195 | -0.354 | 0.14694 | 0.418 |
| 17441 | LINC00957 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | GALNT13 | Sponge network | -0.421 | 0.0114 | -0.857 | 0.07439 | 0.418 |
| 17442 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 31 | CLASP2 | Sponge network | 0.082 | 0.55654 | -0.038 | 0.71 | 0.418 |
| 17443 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | ANK2 | Sponge network | -0.351 | 0.11358 | -1.603 | 1.0E-5 | 0.418 |
| 17444 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 31 | MYO9A | Sponge network | 0.222 | 0.29133 | -0.147 | 0.32373 | 0.418 |
| 17445 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | PHC3 | Sponge network | 1.148 | 2.0E-5 | 0.212 | 0.0499 | 0.418 |
| 17446 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-324-3p;hsa-miR-455-3p | 14 | EPHA7 | Sponge network | -1.116 | 0.23686 | -2.197 | 3.0E-5 | 0.418 |
| 17447 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p | 11 | LATS1 | Sponge network | 1.106 | 0 | 0.109 | 0.34208 | 0.418 |
| 17448 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p | 17 | RET | Sponge network | -1.654 | 0.00144 | -0.833 | 0.03517 | 0.418 |
| 17449 | LINC00865 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | GALNT13 | Sponge network | -1.249 | 7.0E-5 | -0.857 | 0.07439 | 0.418 |
| 17450 | RP11-867G23.10 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-589-3p | 14 | SCN9A | Sponge network | -2.531 | 0 | -0.525 | 0.10593 | 0.418 |
| 17451 | RP11-46C24.7 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | BRD3 | Sponge network | 0.392 | 0.0278 | 0.37 | 0.00013 | 0.418 |
| 17452 | PART1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 46 | TGFBR3 | Sponge network | -2.925 | 0 | -1.173 | 1.0E-5 | 0.418 |
| 17453 | GHRLOS |
hsa-miR-128-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-940;hsa-miR-98-5p | 13 | LIFR | Sponge network | -1.942 | 0 | -1.794 | 0 | 0.418 |
| 17454 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-629-3p;hsa-miR-942-5p | 28 | DDX6 | Sponge network | 0.594 | 0.41522 | 0.091 | 0.36428 | 0.418 |
| 17455 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 18 | AHNAK | Sponge network | -0.793 | 7.0E-5 | -1.207 | 0 | 0.418 |
| 17456 | LINC00654 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | 0.374 | 0.20054 | -1.74 | 0 | 0.418 |
| 17457 | AGAP11 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-769-5p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | -1.645 | 0 | -1.733 | 0.00022 | 0.418 |
| 17458 | CTC-429P9.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-550a-3p | 16 | BRD3 | Sponge network | 0.082 | 0.55397 | 0.37 | 0.00013 | 0.418 |
| 17459 | RP11-574K11.29 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-9-5p | 26 | CCDC6 | Sponge network | 0.997 | 0 | 0.27 | 0.02555 | 0.418 |
| 17460 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 18 | CREBBP | Sponge network | 0.997 | 0.00066 | 0.126 | 0.15186 | 0.418 |
| 17461 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 15 | RCAN2 | Sponge network | -1.349 | 0.00017 | -0.33 | 0.15172 | 0.418 |
| 17462 | HHIP-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | PTPRD | Sponge network | -0.737 | 0.10822 | -1.005 | 0.00064 | 0.418 |
| 17463 | LINC00473 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | PDE4DIP | Sponge network | -1.203 | 0.03881 | -0.282 | 0.0257 | 0.418 |
| 17464 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TAL1 | Sponge network | -0.487 | 0.02157 | -0.462 | 0.00574 | 0.418 |
| 17465 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | NIN | Sponge network | -2.452 | 0 | -0.253 | 0.13794 | 0.418 |
| 17466 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-3127-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-589-5p;hsa-miR-93-3p | 17 | RPS6KA2 | Sponge network | -0.162 | 0.47687 | -0.57 | 0.00013 | 0.418 |
| 17467 | LINC00702 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 29 | PCDH17 | Sponge network | -0.737 | 0.06182 | 0.731 | 2.0E-5 | 0.418 |
| 17468 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 37 | NRXN1 | Sponge network | 0.101 | 0.60919 | -3.778 | 0 | 0.418 |
| 17469 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 46 | BMPR2 | Sponge network | 0.331 | 0.57247 | 0.206 | 0.0635 | 0.418 |
| 17470 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-532-3p | 28 | CBL | Sponge network | -0.096 | 0.67958 | 0.291 | 0.01267 | 0.418 |
| 17471 | JAZF1-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | INPP4A | Sponge network | -0.769 | 0.00035 | 0.07 | 0.53563 | 0.418 |
| 17472 | RP11-360F5.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-590-3p | 15 | ADARB1 | Sponge network | 1.867 | 0 | -0.335 | 0.06631 | 0.418 |
| 17473 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p | 14 | AFF3 | Sponge network | -0.693 | 0.05629 | -2.373 | 0 | 0.418 |
| 17474 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940 | 31 | LIFR | Sponge network | -1.249 | 7.0E-5 | -1.794 | 0 | 0.418 |
| 17475 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-493-5p | 13 | AKAP11 | Sponge network | 0.706 | 0 | 0.196 | 0.09825 | 0.418 |
| 17476 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DOCK4 | Sponge network | 0.683 | 1.0E-5 | 0.502 | 0.00108 | 0.418 |
| 17477 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p | 10 | TGFBR2 | Sponge network | 0.693 | 0.00076 | -0.243 | 0.11408 | 0.418 |
| 17478 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p | 33 | RICTOR | Sponge network | 0.083 | 0.66405 | 0.388 | 0.00188 | 0.418 |
| 17479 | TPTEP1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | ERG | Sponge network | -1.216 | 0 | 0.002 | 0.99011 | 0.418 |
| 17480 | CTC-296K1.3 |
hsa-let-7g-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-550a-3p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-940 | 18 | PTPRT | Sponge network | -2.095 | 0.00112 | -0.907 | 0.03727 | 0.418 |
| 17481 | RP11-822E23.8 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-940 | 43 | CADM2 | Sponge network | -0.777 | 0.16667 | -3.782 | 0 | 0.418 |
| 17482 | LINC00472 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | TET1 | Sponge network | -0.842 | 0.01361 | -0.135 | 0.52138 | 0.418 |
| 17483 | RP11-144G6.12 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | UBR3 | Sponge network | 0.826 | 1.0E-5 | -0.021 | 0.82774 | 0.418 |
| 17484 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-335-3p | 10 | NTRK2 | Sponge network | -0.691 | 0.00146 | -0.736 | 0.06495 | 0.418 |
| 17485 | AC127904.2 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | SIRT1 | Sponge network | 1.308 | 0 | 0.109 | 0.2593 | 0.418 |
| 17486 | ANKRD10-IT1 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 12 | ARHGEF12 | Sponge network | 1.739 | 0 | 0.09 | 0.35693 | 0.418 |
| 17487 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 12 | PDE4DIP | Sponge network | 0.542 | 0.00054 | -0.282 | 0.0257 | 0.418 |
| 17488 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 28 | SCN9A | Sponge network | -1.281 | 0.00012 | -0.525 | 0.10593 | 0.418 |
| 17489 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | DKK3 | Sponge network | -0.096 | 0.67958 | -0.052 | 0.77783 | 0.418 |
| 17490 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-532-5p | 12 | FSTL5 | Sponge network | -0.154 | 0.78939 | -1.3 | 0.01619 | 0.418 |
| 17491 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-582-3p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | TCF4 | Sponge network | -0.31 | 0.33871 | 0.016 | 0.92271 | 0.418 |
| 17492 | CTD-2089N3.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p | 11 | CADM2 | Sponge network | -0.314 | 0.62033 | -3.782 | 0 | 0.418 |
| 17493 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | 1.317 | 0 | 0.323 | 0.00792 | 0.418 |
| 17494 | AGAP11 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-769-5p | 24 | SETBP1 | Sponge network | -1.645 | 0 | -1.302 | 1.0E-5 | 0.418 |
| 17495 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p | 12 | ZNF770 | Sponge network | 0.917 | 0 | 0.206 | 0.06751 | 0.418 |
| 17496 | TPTEP1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | TPO | Sponge network | -1.216 | 0 | -1.87 | 2.0E-5 | 0.418 |
| 17497 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p | 13 | CHD1 | Sponge network | 0.614 | 1.0E-5 | 0.322 | 0.00215 | 0.418 |
| 17498 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | IGSF10 | Sponge network | -0.896 | 0.00099 | -1.751 | 1.0E-5 | 0.418 |
| 17499 | AC005154.6 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-5p;hsa-miR-495-3p | 13 | STAG2 | Sponge network | 0.848 | 0 | 0.252 | 0.00438 | 0.418 |
| 17500 | NEAT1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | DLG1 | Sponge network | 0.705 | 0.00014 | -0.014 | 0.8926 | 0.418 |
| 17501 | LINC01018 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | TLL1 | Sponge network | -2.915 | 0.00015 | -1.195 | 3.0E-5 | 0.418 |
| 17502 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 50 | ZFHX4 | Sponge network | -0.942 | 0 | 0.018 | 0.95574 | 0.418 |
| 17503 | CTA-14H9.5 |
hsa-let-7g-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 10 | RBMS3 | Sponge network | 0.145 | 0.53343 | -0.519 | 0.03551 | 0.418 |
| 17504 | MAGI2-AS3 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | RASSF3 | Sponge network | -0.129 | 0.57177 | -0.226 | 0.11691 | 0.418 |
| 17505 | LINC00657 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ABI2 | Sponge network | 0.91 | 0 | 0.175 | 0.14787 | 0.418 |
| 17506 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-940 | 34 | PBX1 | Sponge network | -1.349 | 0.00017 | -1.206 | 0 | 0.418 |
| 17507 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 27 | ZMYM2 | Sponge network | 0.311 | 0.10344 | 0.279 | 0.01273 | 0.418 |
| 17508 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | LRRC55 | Sponge network | -1.349 | 0.00017 | -0.026 | 0.93163 | 0.418 |
| 17509 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 14 | CLIP1 | Sponge network | -0.611 | 0.09607 | -0.215 | 0.08684 | 0.418 |
| 17510 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-98-5p | 13 | SCN11A | Sponge network | -1.216 | 0 | -1.15 | 0.00299 | 0.418 |
| 17511 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 38 | ABI2 | Sponge network | 0.421 | 0.00084 | 0.175 | 0.14787 | 0.418 |
| 17512 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 17 | ERRFI1 | Sponge network | -0.49 | 0.04946 | -0.798 | 1.0E-5 | 0.418 |
| 17513 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-590-5p | 11 | THSD7A | Sponge network | -0.769 | 0.00035 | 0.242 | 0.25154 | 0.418 |
| 17514 | LINC00476 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-493-5p | 13 | KCNAB1 | Sponge network | -0.615 | 0.00015 | -0.755 | 2.0E-5 | 0.418 |
| 17515 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 18 | DAB2 | Sponge network | -1.111 | 0.0003 | 0.431 | 0.0113 | 0.418 |
| 17516 | CTD-2089N3.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p | 10 | PLSCR4 | Sponge network | -0.314 | 0.62033 | -0.474 | 0.03833 | 0.418 |
| 17517 | PART1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | THSD7B | Sponge network | -2.925 | 0 | 0.154 | 0.74723 | 0.418 |
| 17518 | LINC00473 |
hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PCDH18 | Sponge network | -1.203 | 0.03881 | 0.216 | 0.24645 | 0.418 |
| 17519 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-589-3p | 24 | TIMP3 | Sponge network | -0.154 | 0.78939 | -0.546 | 0.02307 | 0.418 |
| 17520 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-98-5p | 36 | LIFR | Sponge network | -0.129 | 0.57177 | -1.794 | 0 | 0.418 |
| 17521 | LINC00174 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-582-3p | 22 | ABL2 | Sponge network | 1.253 | 0 | 0.82 | 0 | 0.418 |
| 17522 | CALML3-AS1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-3p | 48 | DST | Sponge network | 0.95 | 0.15943 | -0.713 | 0.00096 | 0.418 |
| 17523 | RP11-248G5.8 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 16 | SLC5A3 | Sponge network | 1.294 | 0 | 0.493 | 5.0E-5 | 0.418 |
| 17524 | FZD10-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | PTCH1 | Sponge network | -0.727 | 0.04726 | -0.1 | 0.6179 | 0.418 |
| 17525 | RP11-819C21.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-500a-5p;hsa-miR-96-5p | 10 | ZBTB20 | Sponge network | 0.537 | 0.00139 | -0.122 | 0.57569 | 0.418 |
| 17526 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-664a-3p;hsa-miR-664a-5p | 24 | SOX11 | Sponge network | 1.03 | 0 | 2.68 | 0 | 0.418 |
| 17527 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | PICALM | Sponge network | 0.72 | 0.0001 | 0.086 | 0.23523 | 0.418 |
| 17528 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | ZNF208 | Sponge network | 0.61 | 0.01593 | -0.4 | 0.22381 | 0.418 |
| 17529 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429 | 10 | ZNF536 | Sponge network | -0.976 | 0.00025 | -3.115 | 0 | 0.418 |
| 17530 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RABEP1 | Sponge network | 0.537 | 0.00139 | -0.066 | 0.43142 | 0.418 |
| 17531 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-660-5p | 17 | PCDH9 | Sponge network | 0.331 | 0.57247 | -1.823 | 4.0E-5 | 0.418 |
| 17532 | HOXA-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p | 72 | LPP | Sponge network | 0.61 | 0.01593 | -0.459 | 0.04475 | 0.418 |
| 17533 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-378a-5p | 13 | PDS5B | Sponge network | 0.93 | 0 | 0.259 | 0.00962 | 0.418 |
| 17534 | RP11-434H6.7 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-132-3p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-382-5p | 12 | CAMTA1 | Sponge network | 0.358 | 0.14302 | -0.436 | 1.0E-5 | 0.418 |
| 17535 | AC016747.3 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p | 10 | CTGF | Sponge network | 0.199 | 0.38015 | 0.321 | 0.11682 | 0.418 |
| 17536 | FGF14-AS2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-660-5p | 14 | EYA4 | Sponge network | -1.582 | 8.0E-5 | 0.3 | 0.36876 | 0.418 |
| 17537 | USP3-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-486-5p;hsa-miR-877-5p;hsa-miR-942-5p | 22 | GLIPR1 | Sponge network | -0.162 | 0.47687 | 0.146 | 0.3276 | 0.418 |
| 17538 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 39 | PPM1A | Sponge network | -0.096 | 0.67958 | -0.623 | 0 | 0.418 |
| 17539 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | 0.199 | 0.38015 | -0.354 | 0.14694 | 0.417 |
| 17540 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-625-3p | 13 | ZFHX3 | Sponge network | 1.352 | 0 | 0.389 | 0.01363 | 0.417 |
| 17541 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 34 | TACC1 | Sponge network | 0.41 | 0.02214 | -0.362 | 0.05406 | 0.417 |
| 17542 | LINC00654 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | MACF1 | Sponge network | 0.374 | 0.20054 | 0.019 | 0.90335 | 0.417 |
| 17543 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-7-1-3p | 28 | PTPRD | Sponge network | -4.546 | 0 | -1.005 | 0.00064 | 0.417 |
| 17544 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-940 | 17 | MYO9A | Sponge network | 0.389 | 0.04293 | -0.147 | 0.32373 | 0.417 |
| 17545 | RP11-344B5.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-3p | 13 | TCF4 | Sponge network | -0.055 | 0.88482 | 0.016 | 0.92271 | 0.417 |
| 17546 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 35 | EBF1 | Sponge network | -1.281 | 0.00012 | -0.384 | 0.08856 | 0.417 |
| 17547 | LINC01018 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-877-5p | 10 | PHOX2B | Sponge network | -2.915 | 0.00015 | -2.68 | 0 | 0.417 |
| 17548 | HOTAIRM1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | -0.053 | 0.80685 | 0.809 | 0.00544 | 0.417 |
| 17549 | ZSCAN16-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-625-5p | 10 | NGFR | Sponge network | -0.342 | 0.02288 | -1.271 | 0.01058 | 0.417 |
| 17550 | BDNF-AS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 28 | PBX1 | Sponge network | -0.668 | 3.0E-5 | -1.206 | 0 | 0.417 |
| 17551 | RP5-1139B12.3 |
hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 11 | JAZF1 | Sponge network | 0.598 | 0.38481 | -0.377 | 0.02677 | 0.417 |
| 17552 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | FAT4 | Sponge network | 0.214 | 0.18246 | -0.354 | 0.14694 | 0.417 |
| 17553 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-450b-5p | 11 | PNRC1 | Sponge network | -0.583 | 0.14589 | -0.646 | 0 | 0.417 |
| 17554 | FENDRR |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 18 | ABCA10 | Sponge network | -1.281 | 0.00012 | -0.571 | 0.03674 | 0.417 |
| 17555 | RP4-639F20.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-942-5p;hsa-miR-96-5p | 10 | ZBTB20 | Sponge network | -0.517 | 0.02935 | -0.122 | 0.57569 | 0.417 |
| 17556 | LINC00472 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p | 29 | IL17RD | Sponge network | -0.842 | 0.01361 | -0.019 | 0.94873 | 0.417 |
| 17557 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | BCL2 | Sponge network | -2.117 | 0.00161 | -0.899 | 0 | 0.417 |
| 17558 | U91328.19 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 53 | AR | Sponge network | -0.303 | 0.21002 | -1.554 | 0 | 0.417 |
| 17559 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 21 | SETBP1 | Sponge network | -0.18 | 0.20343 | -1.302 | 1.0E-5 | 0.417 |
| 17560 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-21-5p;hsa-miR-940 | 15 | MYO9A | Sponge network | -2.095 | 0.00112 | -0.147 | 0.32373 | 0.417 |
| 17561 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | PCDH10 | Sponge network | -0.769 | 0.00035 | -1.644 | 0.0078 | 0.417 |
| 17562 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | KCNAB1 | Sponge network | -0.612 | 0.00094 | -0.755 | 2.0E-5 | 0.417 |
| 17563 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-577;hsa-miR-589-3p | 26 | TGFBR3 | Sponge network | 0.811 | 0.03228 | -1.173 | 1.0E-5 | 0.417 |
| 17564 | RP11-158K1.3 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p | 17 | ATM | Sponge network | 0.349 | 0.01695 | 0.178 | 0.21568 | 0.417 |
| 17565 | LINC00473 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | KLF10 | Sponge network | -1.203 | 0.03881 | -0.783 | 0 | 0.417 |
| 17566 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | EPHA5 | Sponge network | -0.777 | 0.1134 | -0.957 | 0.01733 | 0.417 |
| 17567 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 52 | TMEM132B | Sponge network | -0.49 | 0.04946 | -0.83 | 0.01084 | 0.417 |
| 17568 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-455-3p;hsa-miR-503-5p | 19 | RICTOR | Sponge network | 0.858 | 3.0E-5 | 0.388 | 0.00188 | 0.417 |
| 17569 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p | 10 | ABCB5 | Sponge network | 0.811 | 0.03228 | -0.956 | 0.04077 | 0.417 |
| 17570 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p | 12 | GLIPR1 | Sponge network | 0.299 | 0.57722 | 0.146 | 0.3276 | 0.417 |
| 17571 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 40 | CHD5 | Sponge network | -0.563 | 0.02545 | -0.922 | 0.01521 | 0.417 |
| 17572 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-429;hsa-miR-495-3p | 12 | CLASP2 | Sponge network | 1.106 | 0 | -0.038 | 0.71 | 0.417 |
| 17573 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | RECK | Sponge network | 0.101 | 0.60919 | -1.043 | 0 | 0.417 |
| 17574 | LINC00648 |
hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 10 | CSMD1 | Sponge network | 0.633 | 0.51678 | -0.63 | 0.18881 | 0.417 |
| 17575 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-590-3p | 15 | CNTN1 | Sponge network | 0.381 | 0.25967 | -2.594 | 0 | 0.417 |
| 17576 | RP11-620J15.3 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 19 | HIP1 | Sponge network | -0.739 | 0.00044 | 0.774 | 0 | 0.417 |
| 17577 | TPTEP1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 31 | NAV3 | Sponge network | -1.216 | 0 | 0.212 | 0.47729 | 0.417 |
| 17578 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-940;hsa-miR-96-5p | 24 | MEF2C | Sponge network | -0.326 | 0.14314 | -0.499 | 0.01448 | 0.417 |
| 17579 | RP11-98I9.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ABI2 | Sponge network | 0.67 | 0.00048 | 0.175 | 0.14787 | 0.417 |
| 17580 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 32 | MYO9A | Sponge network | -2.117 | 0.00161 | -0.147 | 0.32373 | 0.417 |
| 17581 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 12 | AKAP6 | Sponge network | -1.375 | 0.08642 | -1.586 | 3.0E-5 | 0.417 |
| 17582 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p | 13 | ITGA5 | Sponge network | -0.896 | 0.00099 | 0.452 | 0.03524 | 0.417 |
| 17583 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-539-5p | 20 | DMD | Sponge network | -0.726 | 0.00132 | -1.506 | 0 | 0.417 |
| 17584 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p | 19 | KIT | Sponge network | -0.487 | 0.02157 | -2.184 | 0 | 0.417 |
| 17585 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CLIP1 | Sponge network | 0.559 | 0.00026 | -0.215 | 0.08684 | 0.417 |
| 17586 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p | 29 | DIP2C | Sponge network | 0.253 | 0.09565 | -0.436 | 0.01713 | 0.417 |
| 17587 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 48 | UNC80 | Sponge network | -0.49 | 0.04946 | -1.934 | 0 | 0.417 |
| 17588 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 16 | UVRAG | Sponge network | -0.129 | 0.57177 | -0.017 | 0.83865 | 0.417 |
| 17589 | C20orf166-AS1 |
hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577 | 11 | FGF5 | Sponge network | -2.69 | 0.0001 | 0.549 | 0.28659 | 0.417 |
| 17590 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | -0.942 | 0 | 0.809 | 0.00544 | 0.417 |
| 17591 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3127-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p | 14 | RPS6KA2 | Sponge network | -1.654 | 0.00144 | -0.57 | 0.00013 | 0.417 |
| 17592 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-654-3p;hsa-miR-664a-3p | 13 | ITCH | Sponge network | 1.147 | 0 | 0.084 | 0.34058 | 0.417 |
| 17593 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p | 24 | SOX5 | Sponge network | -0.693 | 0.05629 | -1.091 | 4.0E-5 | 0.417 |
| 17594 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-874-3p | 28 | MXI1 | Sponge network | -0.899 | 6.0E-5 | -0.987 | 0 | 0.417 |
| 17595 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-98-5p | 13 | PNRC1 | Sponge network | -1.092 | 4.0E-5 | -0.646 | 0 | 0.417 |
| 17596 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-454-3p | 11 | THSD7A | Sponge network | 0.246 | 0.59912 | 0.242 | 0.25154 | 0.417 |
| 17597 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | MCC | Sponge network | -0.811 | 2.0E-5 | -1.005 | 0 | 0.417 |
| 17598 | CTD-2583A14.8 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-374a-3p | 13 | MDM4 | Sponge network | 1.134 | 0 | 0.345 | 0.00762 | 0.417 |
| 17599 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 36 | ESR1 | Sponge network | -1.281 | 0.00012 | -1.035 | 3.0E-5 | 0.417 |
| 17600 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 33 | LIFR | Sponge network | -0.49 | 0.04946 | -1.794 | 0 | 0.417 |
| 17601 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 44 | ADARB1 | Sponge network | -2.915 | 0.00015 | -0.335 | 0.06631 | 0.417 |
| 17602 | SNHG14 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 11 | EPB41L3 | Sponge network | -1.111 | 0.0003 | -1.193 | 0 | 0.417 |
| 17603 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | MITF | Sponge network | -0.709 | 0.18168 | -0.769 | 0.00022 | 0.417 |
| 17604 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 55 | CADM2 | Sponge network | -0.096 | 0.67958 | -3.782 | 0 | 0.417 |
| 17605 | RP11-53O19.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p | 26 | DLC1 | Sponge network | -0.405 | 0.22013 | -0.382 | 0.05038 | 0.417 |
| 17606 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-501-5p | 12 | UBE4B | Sponge network | 0.178 | 0.29106 | -0.235 | 0.007 | 0.417 |
| 17607 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-301a-3p;hsa-miR-324-3p | 13 | PTPN14 | Sponge network | 0.37 | 0.27614 | 0.144 | 0.34595 | 0.417 |
| 17608 | RP11-517B11.7 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p | 12 | USP9X | Sponge network | 0.706 | 0 | 0.222 | 0.01689 | 0.417 |
| 17609 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-590-3p | 13 | RBM5 | Sponge network | -1.575 | 6.0E-5 | -0.205 | 0.0129 | 0.417 |
| 17610 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 20 | RBL2 | Sponge network | -1.575 | 6.0E-5 | -0.282 | 0.00254 | 0.417 |
| 17611 | TP73-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-940 | 37 | NTRK2 | Sponge network | -0.563 | 0.02545 | -0.736 | 0.06495 | 0.417 |
| 17612 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-940;hsa-miR-942-5p | 31 | HLF | Sponge network | 0.16 | 0.50785 | -2.443 | 0 | 0.417 |
| 17613 | RP5-994D16.9 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-409-3p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | PIK3R1 | Sponge network | 0.252 | 0.08508 | -0.133 | 0.36854 | 0.417 |
| 17614 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | 0.101 | 0.60919 | -1.465 | 0.00033 | 0.417 |
| 17615 | LINC00473 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 23 | CBFA2T3 | Sponge network | -1.203 | 0.03881 | -1.221 | 1.0E-5 | 0.417 |
| 17616 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p | 12 | RET | Sponge network | -0.517 | 0.02935 | -0.833 | 0.03517 | 0.417 |
| 17617 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | EPHA3 | Sponge network | 0.657 | 4.0E-5 | 0.185 | 0.53588 | 0.417 |
| 17618 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 25 | MYBL1 | Sponge network | -0.323 | 0.01372 | 0.494 | 0.02152 | 0.417 |
| 17619 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | SMARCA2 | Sponge network | -1.111 | 0.0003 | -0.529 | 0.00014 | 0.417 |
| 17620 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | GAS7 | Sponge network | -0.351 | 0.11358 | -0.555 | 0.01602 | 0.417 |
| 17621 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p | 21 | RICTOR | Sponge network | 1.122 | 0 | 0.388 | 0.00188 | 0.417 |
| 17622 | PWAR6 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | LIMCH1 | Sponge network | -1.575 | 6.0E-5 | -0.915 | 1.0E-5 | 0.417 |
| 17623 | RP11-400K9.4 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | PIK3CA | Sponge network | -0.611 | 0.09607 | 0.195 | 0.06908 | 0.417 |
| 17624 | RP11-384K6.6 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-539-5p | 14 | TSC1 | Sponge network | 0.946 | 0 | 0.103 | 0.26071 | 0.417 |
| 17625 | RP11-25K19.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p | 28 | IGFBP5 | Sponge network | -1.057 | 0.00029 | -0.806 | 0.00105 | 0.417 |
| 17626 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-501-3p | 11 | LRRC7 | Sponge network | -0.154 | 0.53954 | -1.341 | 0.01193 | 0.417 |
| 17627 | LINC00473 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 23 | LIMCH1 | Sponge network | -1.203 | 0.03881 | -0.915 | 1.0E-5 | 0.417 |
| 17628 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p | 14 | SCN9A | Sponge network | -0.726 | 0.00132 | -0.525 | 0.10593 | 0.417 |
| 17629 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p | 20 | ADAMTSL3 | Sponge network | 0.082 | 0.55397 | -1.559 | 0 | 0.417 |
| 17630 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-497-5p | 12 | SON | Sponge network | 0.917 | 0 | 0.168 | 0.04406 | 0.417 |
| 17631 | BOLA3-AS1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-935;hsa-miR-940 | 31 | GRIK3 | Sponge network | -0.246 | 0.35034 | -3.208 | 0 | 0.417 |
| 17632 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | IGSF10 | Sponge network | -0.811 | 2.0E-5 | -1.751 | 1.0E-5 | 0.417 |
| 17633 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 70 | IL6ST | Sponge network | 0.064 | 0.72934 | -0.402 | 0.00676 | 0.417 |
| 17634 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 35 | IGFBP5 | Sponge network | -0.513 | 0.04703 | -0.806 | 0.00105 | 0.417 |
| 17635 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 11 | RCAN2 | Sponge network | -0.318 | 0.65847 | -0.33 | 0.15172 | 0.417 |
| 17636 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p | 14 | TLE4 | Sponge network | -0.583 | 0.14589 | -1.157 | 0 | 0.417 |
| 17637 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-589-3p | 23 | CHD5 | Sponge network | -0.358 | 0.17255 | -0.922 | 0.01521 | 0.417 |
| 17638 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | JAKMIP2 | Sponge network | 0.101 | 0.60919 | -1.388 | 0 | 0.417 |
| 17639 | RP11-67L2.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 13 | SHPRH | Sponge network | 0.72 | 0.0001 | 0.098 | 0.40994 | 0.417 |
| 17640 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-590-5p | 10 | ERG | Sponge network | 0.176 | 0.37402 | 0.002 | 0.99011 | 0.417 |
| 17641 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | SLITRK3 | Sponge network | -0.811 | 2.0E-5 | -3.328 | 0 | 0.417 |
| 17642 | RP11-66B24.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-766-3p | 14 | IL17RD | Sponge network | 0.57 | 0.1866 | -0.019 | 0.94873 | 0.417 |
| 17643 | FENDRR |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 15 | KCNAB1 | Sponge network | -1.281 | 0.00012 | -0.755 | 2.0E-5 | 0.417 |
| 17644 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-421 | 22 | TCF4 | Sponge network | -4.463 | 0.00025 | 0.016 | 0.92271 | 0.417 |
| 17645 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 28 | PBX1 | Sponge network | 0.572 | 0.01722 | -1.206 | 0 | 0.417 |
| 17646 | GS1-124K5.11 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 15 | BCL9 | Sponge network | 0.494 | 0.00339 | 0.321 | 0.00267 | 0.417 |
| 17647 | RP11-13K12.1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 16 | CXCL12 | Sponge network | -0.154 | 0.78939 | -1.421 | 0 | 0.417 |
| 17648 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 13 | ZNF770 | Sponge network | 1.266 | 0 | 0.206 | 0.06751 | 0.417 |
| 17649 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | CBFA2T3 | Sponge network | -0.727 | 0.04726 | -1.221 | 1.0E-5 | 0.417 |
| 17650 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | MCC | Sponge network | -0.726 | 0.00132 | -1.005 | 0 | 0.417 |
| 17651 | ACTA2-AS1 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ADH1B | Sponge network | 0.194 | 0.64278 | -3.716 | 0 | 0.417 |
| 17652 | PCA3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | IL17RD | Sponge network | -0.709 | 0.18168 | -0.019 | 0.94873 | 0.417 |
| 17653 | CTC-559E9.6 |
hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-7-5p | 10 | ATRX | Sponge network | 0.601 | 1.0E-5 | 0.261 | 0.04408 | 0.417 |
| 17654 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-29a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 11 | EPHA3 | Sponge network | -0.72 | 0.24239 | 0.185 | 0.53588 | 0.417 |
| 17655 | SNHG16 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-664a-3p | 12 | RASAL2 | Sponge network | 0.961 | 0 | 0.869 | 0 | 0.417 |
| 17656 | LINC00854 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-654-3p | 21 | RICTOR | Sponge network | 1.18 | 0 | 0.388 | 0.00188 | 0.417 |
| 17657 | LINC00621 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 12 | SCN9A | Sponge network | 0.131 | 0.8436 | -0.525 | 0.10593 | 0.417 |
| 17658 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | RECK | Sponge network | 0.214 | 0.18246 | -1.043 | 0 | 0.417 |
| 17659 | LINC00476 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | TPO | Sponge network | -0.615 | 0.00015 | -1.87 | 2.0E-5 | 0.417 |
| 17660 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-5p | 15 | PTPN14 | Sponge network | -0.517 | 0.02935 | 0.144 | 0.34595 | 0.417 |
| 17661 | HHIP-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | NRK | Sponge network | -0.737 | 0.10822 | 0.656 | 0.19217 | 0.417 |
| 17662 | ARHGEF26-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | LIMCH1 | Sponge network | -2.46 | 0 | -0.915 | 1.0E-5 | 0.417 |
| 17663 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | PDGFRA | Sponge network | -0.513 | 0.04703 | -0.372 | 0.0037 | 0.417 |
| 17664 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | CSMD1 | Sponge network | -0.513 | 0.04703 | -0.63 | 0.18881 | 0.417 |
| 17665 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | WWTR1 | Sponge network | 1.302 | 7.0E-5 | -0.345 | 0.11997 | 0.417 |
| 17666 | USP3-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p | 19 | ERG | Sponge network | -0.162 | 0.47687 | 0.002 | 0.99011 | 0.417 |
| 17667 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-576-5p | 13 | GREM1 | Sponge network | 0.619 | 0.00157 | 0.902 | 0.01691 | 0.417 |
| 17668 | RP11-798M19.6 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | GJA1 | Sponge network | -0.612 | 0.00094 | -0.485 | 0.02179 | 0.417 |
| 17669 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-877-5p | 19 | GRIN2A | Sponge network | -0.777 | 0.16667 | -2.161 | 0 | 0.417 |
| 17670 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | LRFN5 | Sponge network | -0.612 | 0.00094 | -1.483 | 0 | 0.417 |
| 17671 | AC006116.24 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-93-5p | 36 | DST | Sponge network | -0.473 | 0.07602 | -0.713 | 0.00096 | 0.417 |
| 17672 | RP11-589P10.7 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-93-3p | 12 | CTIF | Sponge network | -2.044 | 0.00066 | -0.389 | 0.00549 | 0.417 |
| 17673 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-34a-5p | 11 | RET | Sponge network | -0.309 | 0.1145 | -0.833 | 0.03517 | 0.416 |
| 17674 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-96-5p | 20 | CRTC3 | Sponge network | 0.331 | 0.57247 | 0.164 | 0.16786 | 0.416 |
| 17675 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421 | 11 | CAV1 | Sponge network | -1.645 | 0 | -1.013 | 6.0E-5 | 0.416 |
| 17676 | RP11-67L2.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | RASSF8 | Sponge network | 0.72 | 0.0001 | -0.122 | 0.65591 | 0.416 |
| 17677 | LINC00654 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | PDE4DIP | Sponge network | 0.374 | 0.20054 | -0.282 | 0.0257 | 0.416 |
| 17678 | RP11-418J17.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 17 | TBL1XR1 | Sponge network | 0.998 | 0 | 0.578 | 0 | 0.416 |
| 17679 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 32 | MXI1 | Sponge network | -0.576 | 0.10121 | -0.987 | 0 | 0.416 |
| 17680 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 17 | MAPK10 | Sponge network | 0.225 | 0.30644 | -1.774 | 0 | 0.416 |
| 17681 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 12 | SLC6A15 | Sponge network | -1.582 | 8.0E-5 | -2.373 | 2.0E-5 | 0.416 |
| 17682 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ADAMTSL3 | Sponge network | -0.222 | 0.32445 | -1.559 | 0 | 0.416 |
| 17683 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 13 | TXNIP | Sponge network | -0.83 | 0 | -1.277 | 0 | 0.416 |
| 17684 | RP11-389C8.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p | 18 | ABL2 | Sponge network | 0.727 | 3.0E-5 | 0.82 | 0 | 0.416 |
| 17685 | RP11-981G7.6 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 12 | SHPRH | Sponge network | 0.986 | 0.01022 | 0.098 | 0.40994 | 0.416 |
| 17686 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p | 12 | ZNF208 | Sponge network | -0.611 | 0.09607 | -0.4 | 0.22381 | 0.416 |
| 17687 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | LRRC4 | Sponge network | -0.323 | 0.01372 | -1.774 | 0 | 0.416 |
| 17688 | RP5-1120P11.1 |
hsa-miR-101-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-486-5p | 12 | CBFB | Sponge network | 2.957 | 0 | 1.061 | 0 | 0.416 |
| 17689 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-940 | 21 | MYO9A | Sponge network | -4.546 | 0 | -0.147 | 0.32373 | 0.416 |
| 17690 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 19 | NEDD4 | Sponge network | -0.842 | 0.01361 | 0.02 | 0.89834 | 0.416 |
| 17691 | NEAT1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | DMTF1 | Sponge network | 0.705 | 0.00014 | 0.248 | 0.02726 | 0.416 |
| 17692 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-5p | 14 | THSD7A | Sponge network | -0.942 | 0 | 0.242 | 0.25154 | 0.416 |
| 17693 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 27 | PGR | Sponge network | 0.374 | 0.20054 | -1.704 | 0 | 0.416 |
| 17694 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p | 20 | PTPN14 | Sponge network | 0.082 | 0.55397 | 0.144 | 0.34595 | 0.416 |
| 17695 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | SLITRK3 | Sponge network | 0.572 | 0.01722 | -3.328 | 0 | 0.416 |
| 17696 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p | 26 | RASSF2 | Sponge network | -2.62 | 0 | -0.273 | 0.21961 | 0.416 |
| 17697 | BVES-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-98-5p | 15 | ITGA5 | Sponge network | -2.62 | 0 | 0.452 | 0.03524 | 0.416 |
| 17698 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 40 | MEF2C | Sponge network | -0.612 | 0.00094 | -0.499 | 0.01448 | 0.416 |
| 17699 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 15 | KCNAB1 | Sponge network | -0.405 | 0.22013 | -0.755 | 2.0E-5 | 0.416 |
| 17700 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-590-3p | 33 | AR | Sponge network | 0.178 | 0.29106 | -1.554 | 0 | 0.416 |
| 17701 | LINC00672 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 22 | PCDH10 | Sponge network | -0.227 | 0.31657 | -1.644 | 0.0078 | 0.416 |
| 17702 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-93-3p | 44 | QKI | Sponge network | -0.154 | 0.78939 | -0.333 | 0.04111 | 0.416 |
| 17703 | CTD-2002H8.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | EPS15 | Sponge network | 0.931 | 1.0E-5 | -0.052 | 0.53998 | 0.416 |
| 17704 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 54 | AFF4 | Sponge network | 0.537 | 0.00139 | -0.266 | 0.01565 | 0.416 |
| 17705 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | MOB1B | Sponge network | 0.083 | 0.66405 | 0.197 | 0.15266 | 0.416 |
| 17706 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-625-5p | 25 | ZMYM2 | Sponge network | 1.254 | 0 | 0.279 | 0.01273 | 0.416 |
| 17707 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ZFHX3 | Sponge network | 0.953 | 0 | 0.389 | 0.01363 | 0.416 |
| 17708 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 37 | GRIA3 | Sponge network | -1.111 | 0.0003 | -1.956 | 0 | 0.416 |
| 17709 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 41 | TRPS1 | Sponge network | 0.056 | 0.81195 | -0.191 | 0.44109 | 0.416 |
| 17710 | FENDRR |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 15 | RASSF3 | Sponge network | -1.281 | 0.00012 | -0.226 | 0.11691 | 0.416 |
| 17711 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 29 | DOCK4 | Sponge network | -1.111 | 0.0003 | 0.502 | 0.00108 | 0.416 |
| 17712 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 14 | DCLK1 | Sponge network | 0.395 | 0.15451 | -1.324 | 0.00014 | 0.416 |
| 17713 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-2-5p;hsa-miR-425-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | QKI | Sponge network | -0.187 | 0.23787 | -0.333 | 0.04111 | 0.416 |
| 17714 | RP11-571M6.17 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | CREB1 | Sponge network | 1.013 | 0 | 0.203 | 0.00537 | 0.416 |
| 17715 | LINC00654 |
hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH11 | Sponge network | 0.374 | 0.20054 | 1.427 | 0 | 0.416 |
| 17716 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TIMP3 | Sponge network | -0.021 | 0.93598 | -0.546 | 0.02307 | 0.416 |
| 17717 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-590-3p | 31 | CAMTA1 | Sponge network | -0.342 | 0.02288 | -0.436 | 1.0E-5 | 0.416 |
| 17718 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-96-5p | 26 | CRTC3 | Sponge network | -1.281 | 0.00012 | 0.164 | 0.16786 | 0.416 |
| 17719 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 44 | EPHA5 | Sponge network | 0.101 | 0.60919 | -0.957 | 0.01733 | 0.416 |
| 17720 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p | 28 | SNX29 | Sponge network | 0.335 | 0.20596 | -0.34 | 0.01371 | 0.416 |
| 17721 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 31 | CYLD | Sponge network | -1.111 | 0.0003 | -0.367 | 0.00171 | 0.416 |
| 17722 | RP1-59D14.5 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-940 | 10 | BAZ2B | Sponge network | 1.162 | 5.0E-5 | -0.276 | 0.02833 | 0.416 |
| 17723 | EMX2OS |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 23 | PTGFR | Sponge network | -0.777 | 0.1134 | -0.233 | 0.45421 | 0.416 |
| 17724 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 14 | PRDM5 | Sponge network | 0.194 | 0.64278 | -1.09 | 0.00014 | 0.416 |
| 17725 | RP11-867G23.10 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-942-5p | 13 | OPCML | Sponge network | -2.531 | 0 | -2.498 | 0 | 0.416 |
| 17726 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 16 | CNTNAP3 | Sponge network | 0.056 | 0.81195 | -1.465 | 0.00033 | 0.416 |
| 17727 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 21 | TBL1XR1 | Sponge network | 0.72 | 0.0001 | 0.578 | 0 | 0.416 |
| 17728 | TPTEP1 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | NOV | Sponge network | -1.216 | 0 | -0.58 | 0.04676 | 0.416 |
| 17729 | AC016747.3 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p | 13 | MAFB | Sponge network | 0.199 | 0.38015 | -0.023 | 0.89865 | 0.416 |
| 17730 | ASH1L-AS1 |
hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | MDM4 | Sponge network | 0.948 | 0 | 0.345 | 0.00762 | 0.416 |
| 17731 | CTA-14H9.5 |
hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-508-3p;hsa-miR-7-1-3p | 10 | TACC1 | Sponge network | 0.145 | 0.53343 | -0.362 | 0.05406 | 0.416 |
| 17732 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p | 12 | RARB | Sponge network | -2.531 | 0 | -0.825 | 2.0E-5 | 0.416 |
| 17733 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | EYA4 | Sponge network | 0.619 | 0.00157 | 0.3 | 0.36876 | 0.416 |
| 17734 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | ZNF208 | Sponge network | 0.176 | 0.37402 | -0.4 | 0.22381 | 0.416 |
| 17735 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-577 | 10 | RNF144A | Sponge network | -0.473 | 0.07602 | 0.594 | 0.0009 | 0.416 |
| 17736 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-671-5p;hsa-miR-7-5p | 10 | EHD3 | Sponge network | -1.331 | 3.0E-5 | -0.484 | 0.01747 | 0.416 |
| 17737 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | CACNA2D1 | Sponge network | -0.615 | 0.00015 | -0.846 | 0.00675 | 0.416 |
| 17738 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PHC3 | Sponge network | 1.153 | 0 | 0.212 | 0.0499 | 0.416 |
| 17739 | FAM66C |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 39 | PTPRT | Sponge network | -0.901 | 0.00019 | -0.907 | 0.03727 | 0.416 |
| 17740 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 31 | RASSF8 | Sponge network | -0.899 | 6.0E-5 | -0.122 | 0.65591 | 0.416 |
| 17741 | MAP3K14 |
hsa-miR-125a-3p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 14 | IL6ST | Sponge network | -0.295 | 0.02604 | -0.402 | 0.00676 | 0.416 |
| 17742 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 24 | TGFBR2 | Sponge network | -0.49 | 0.04946 | -0.243 | 0.11408 | 0.416 |
| 17743 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-96-5p | 61 | AFF4 | Sponge network | 0.083 | 0.66405 | -0.266 | 0.01565 | 0.416 |
| 17744 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | RICTOR | Sponge network | 0.643 | 0 | 0.388 | 0.00188 | 0.416 |
| 17745 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLIP1 | Sponge network | -0.323 | 0.01372 | -0.215 | 0.08684 | 0.416 |
| 17746 | AC003090.1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 12 | RHOB | Sponge network | -2.117 | 0.00161 | -1.052 | 0 | 0.416 |
| 17747 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 32 | ANK2 | Sponge network | -0.355 | 0.0077 | -1.603 | 1.0E-5 | 0.416 |
| 17748 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 46 | SASH1 | Sponge network | -2.46 | 0 | -0.502 | 0.00175 | 0.416 |
| 17749 | CTD-2017D11.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-378c | 17 | ABL2 | Sponge network | 0.792 | 0.0049 | 0.82 | 0 | 0.416 |
| 17750 | MEG3 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | PCDH18 | Sponge network | 0.249 | 0.35902 | 0.216 | 0.24645 | 0.416 |
| 17751 | AC003090.1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | KLF10 | Sponge network | -2.117 | 0.00161 | -0.783 | 0 | 0.416 |
| 17752 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-501-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 45 | CCND2 | Sponge network | 0.194 | 0.64278 | -0.267 | 0.3112 | 0.416 |
| 17753 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CLIP1 | Sponge network | 0.064 | 0.72934 | -0.215 | 0.08684 | 0.416 |
| 17754 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-940 | 30 | NTRK2 | Sponge network | -0.318 | 0.65847 | -0.736 | 0.06495 | 0.416 |
| 17755 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | PRKAB2 | Sponge network | -0.187 | 0.23787 | -0.367 | 0.01371 | 0.416 |
| 17756 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 21 | STARD13 | Sponge network | -2.46 | 0 | -0.187 | 0.27383 | 0.416 |
| 17757 | RP11-526I2.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-365a-3p;hsa-miR-495-3p | 11 | RICTOR | Sponge network | 1.054 | 1.0E-5 | 0.388 | 0.00188 | 0.416 |
| 17758 | ZSCAN16-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 14 | PRUNE2 | Sponge network | -0.342 | 0.02288 | -1.733 | 0.00022 | 0.416 |
| 17759 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-421;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | BNC2 | Sponge network | 0.082 | 0.55654 | -0.491 | 0.09988 | 0.416 |
| 17760 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 19 | TIMP3 | Sponge network | -0.318 | 0.65847 | -0.546 | 0.02307 | 0.416 |
| 17761 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-877-5p | 29 | ANK2 | Sponge network | -0.773 | 0.04073 | -1.603 | 1.0E-5 | 0.416 |
| 17762 | RP11-384O8.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-940 | 19 | SFRP1 | Sponge network | -0.738 | 0.21233 | -3.566 | 0 | 0.416 |
| 17763 | PCA3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 47 | GAS7 | Sponge network | -0.709 | 0.18168 | -0.555 | 0.01602 | 0.416 |
| 17764 | MYLK-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p | 14 | PCDH17 | Sponge network | -1.331 | 3.0E-5 | 0.731 | 2.0E-5 | 0.416 |
| 17765 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-7-1-3p | 11 | PLSCR4 | Sponge network | 0.419 | 0.0319 | -0.474 | 0.03833 | 0.416 |
| 17766 | CTD-3064M3.3 |
hsa-miR-148b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 11 | NGFR | Sponge network | -0.469 | 0.08721 | -1.271 | 0.01058 | 0.416 |
| 17767 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 14 | GPAM | Sponge network | 1.205 | 0 | 0.142 | 0.29732 | 0.416 |
| 17768 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 29 | ZMYND11 | Sponge network | -2.46 | 0 | -0.2 | 0.05449 | 0.416 |
| 17769 | RP11-326G21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NAV3 | Sponge network | -0.021 | 0.93598 | 0.212 | 0.47729 | 0.416 |
| 17770 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-935 | 18 | RARB | Sponge network | -0.421 | 0.0114 | -0.825 | 2.0E-5 | 0.416 |
| 17771 | LINC00702 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 12 | EHD3 | Sponge network | -0.737 | 0.06182 | -0.484 | 0.01747 | 0.416 |
| 17772 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-577 | 36 | NTRK2 | Sponge network | -2.69 | 0.0001 | -0.736 | 0.06495 | 0.416 |
| 17773 | RP11-53O19.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-664a-3p;hsa-miR-874-3p | 23 | ASTN1 | Sponge network | -0.405 | 0.22013 | -2.389 | 2.0E-5 | 0.416 |
| 17774 | RP11-890B15.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-93-5p | 64 | DST | Sponge network | 0.101 | 0.60919 | -0.713 | 0.00096 | 0.416 |
| 17775 | DNM3OS |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p | 31 | ABL2 | Sponge network | 0.572 | 0.01722 | 0.82 | 0 | 0.416 |
| 17776 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p | 12 | KCNA6 | Sponge network | -0.376 | 0.00337 | -1.531 | 0.00013 | 0.416 |
| 17777 | MIR7-3HG |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-532-3p;hsa-miR-625-5p | 14 | PTPRT | Sponge network | -2.688 | 0.00397 | -0.907 | 0.03727 | 0.416 |
| 17778 | GNG12-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 18 | RELN | Sponge network | -0.793 | 7.0E-5 | -2.403 | 0 | 0.416 |
| 17779 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p | 21 | USP6 | Sponge network | 0.267 | 0.2937 | 0.188 | 0.3871 | 0.416 |
| 17780 | MIR22HG |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 16 | SIK1 | Sponge network | -1.047 | 0 | -0.98 | 0 | 0.416 |
| 17781 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 52 | CBX5 | Sponge network | -0.222 | 0.32445 | 0.165 | 0.24446 | 0.416 |
| 17782 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | EPHA5 | Sponge network | -0.421 | 0.0114 | -0.957 | 0.01733 | 0.416 |
| 17783 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 35 | CBL | Sponge network | 0.349 | 0.01695 | 0.291 | 0.01267 | 0.416 |
| 17784 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p | 29 | CBL | Sponge network | 0.082 | 0.55654 | 0.291 | 0.01267 | 0.416 |
| 17785 | LINC00957 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 10 | DIRAS1 | Sponge network | -0.421 | 0.0114 | -2.518 | 0 | 0.416 |
| 17786 | AC127904.2 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | KIF5B | Sponge network | 1.308 | 0 | 0.362 | 0.00066 | 0.416 |
| 17787 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 25 | DOCK4 | Sponge network | 0.083 | 0.66405 | 0.502 | 0.00108 | 0.416 |
| 17788 | OIP5-AS1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | SH3GLB1 | Sponge network | 0.421 | 0.00084 | -0.115 | 0.14773 | 0.416 |
| 17789 | MEG3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-744-3p;hsa-miR-96-5p | 24 | HIP1 | Sponge network | 0.249 | 0.35902 | 0.774 | 0 | 0.416 |
| 17790 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-624-5p | 25 | ANK2 | Sponge network | 0.331 | 0.57247 | -1.603 | 1.0E-5 | 0.416 |
| 17791 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-590-3p | 12 | PAK3 | Sponge network | -0.096 | 0.67958 | -0.628 | 0.07253 | 0.416 |
| 17792 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 68 | IL6ST | Sponge network | -0.942 | 0 | -0.402 | 0.00676 | 0.416 |
| 17793 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-935;hsa-miR-96-5p | 15 | ABI2 | Sponge network | 0.37 | 0.27614 | 0.175 | 0.14787 | 0.416 |
| 17794 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-935 | 20 | ARNT2 | Sponge network | -0.405 | 0.22013 | -0.332 | 0.25851 | 0.416 |
| 17795 | H19 |
hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-486-5p | 10 | DNMT3A | Sponge network | 2.439 | 0 | 0.302 | 0.0262 | 0.416 |
| 17796 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-93-5p | 37 | TACC1 | Sponge network | -0.154 | 0.78939 | -0.362 | 0.05406 | 0.416 |
| 17797 | RP11-574K11.29 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p | 23 | ARHGEF12 | Sponge network | 0.997 | 0 | 0.09 | 0.35693 | 0.416 |
| 17798 | AC005562.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 18 | NF1 | Sponge network | 0.488 | 0.00084 | 0.51 | 0 | 0.416 |
| 17799 | LINC00205 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-3p | 12 | ABL2 | Sponge network | 1.271 | 0 | 0.82 | 0 | 0.415 |
| 17800 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 25 | ST6GAL2 | Sponge network | -2.117 | 0.00161 | -1.201 | 0.00597 | 0.415 |
| 17801 | LINC00476 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | PTGFR | Sponge network | -0.615 | 0.00015 | -0.233 | 0.45421 | 0.415 |
| 17802 | RP4-717I23.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-940 | 20 | EPS15 | Sponge network | 0.637 | 0.00069 | -0.052 | 0.53998 | 0.415 |
| 17803 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-5p | 19 | SETBP1 | Sponge network | -0.384 | 0.40652 | -1.302 | 1.0E-5 | 0.415 |
| 17804 | AC007879.7 |
hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-5p | 14 | SOX11 | Sponge network | 4.3 | 0 | 2.68 | 0 | 0.415 |
| 17805 | AC009948.5 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | TAL1 | Sponge network | 0.532 | 0.00505 | -0.462 | 0.00574 | 0.415 |
| 17806 | AC016747.3 |
hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 15 | SPG20 | Sponge network | 0.199 | 0.38015 | -1.16 | 0 | 0.415 |
| 17807 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | 0.603 | 0.00127 | 0.323 | 0.00792 | 0.415 |
| 17808 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | GRIN2A | Sponge network | -0.421 | 0.0114 | -2.161 | 0 | 0.415 |
| 17809 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 30 | DIP2C | Sponge network | -0.421 | 0.0114 | -0.436 | 0.01713 | 0.415 |
| 17810 | BCYRN1 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | WAC | Sponge network | 0.311 | 0.10344 | -0.054 | 0.43688 | 0.415 |
| 17811 | BOLA3-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | MYO9A | Sponge network | -0.246 | 0.35034 | -0.147 | 0.32373 | 0.415 |
| 17812 | LINC00702 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -0.737 | 0.06182 | -1.692 | 0 | 0.415 |
| 17813 | HCG18 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FBXO11 | Sponge network | 1.063 | 0 | 0.142 | 0.04938 | 0.415 |
| 17814 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | DAB2 | Sponge network | 0.194 | 0.64278 | 0.431 | 0.0113 | 0.415 |
| 17815 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | CAV1 | Sponge network | -1.161 | 0.00591 | -1.013 | 6.0E-5 | 0.415 |
| 17816 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | AFF3 | Sponge network | -0.83 | 0 | -2.373 | 0 | 0.415 |
| 17817 | RP11-554A11.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-628-5p | 25 | TMEM132B | Sponge network | -0.583 | 0.14589 | -0.83 | 0.01084 | 0.415 |
| 17818 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | -2.915 | 0.00015 | -1.465 | 0.00033 | 0.415 |
| 17819 | MEG3 |
hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 13 | RHOB | Sponge network | 0.249 | 0.35902 | -1.052 | 0 | 0.415 |
| 17820 | RP11-67L2.2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 35 | AFF1 | Sponge network | 0.72 | 0.0001 | -0.384 | 0.00053 | 0.415 |
| 17821 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 10 | INPP4A | Sponge network | 0.889 | 9.0E-5 | 0.07 | 0.53563 | 0.415 |
| 17822 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 21 | DICER1 | Sponge network | 0.72 | 0.0001 | 0.042 | 0.674 | 0.415 |
| 17823 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p | 14 | AR | Sponge network | -7.551 | 0 | -1.554 | 0 | 0.415 |
| 17824 | LINC01018 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | FAM172A | Sponge network | -2.915 | 0.00015 | -0.57 | 0 | 0.415 |
| 17825 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-93-5p | 24 | SYNE1 | Sponge network | -0.18 | 0.20343 | -0.738 | 0.00316 | 0.415 |
| 17826 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 41 | PPM1A | Sponge network | -0.323 | 0.01372 | -0.623 | 0 | 0.415 |
| 17827 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-940 | 29 | CRTC3 | Sponge network | -0.222 | 0.32445 | 0.164 | 0.16786 | 0.415 |
| 17828 | RP11-254F19.2 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 11 | IL6ST | Sponge network | -1.428 | 0.07715 | -0.402 | 0.00676 | 0.415 |
| 17829 | DNM3OS |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | PIK3CA | Sponge network | 0.572 | 0.01722 | 0.195 | 0.06908 | 0.415 |
| 17830 | CTD-2554C21.2 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-429;hsa-miR-502-5p;hsa-miR-7-5p | 16 | PRKCB | Sponge network | -1.654 | 0.00144 | -1.313 | 2.0E-5 | 0.415 |
| 17831 | GS1-124K5.11 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 10 | FLNA | Sponge network | 0.494 | 0.00339 | -0.771 | 0.01377 | 0.415 |
| 17832 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-508-3p;hsa-miR-590-3p | 10 | NR4A3 | Sponge network | -0.358 | 0.17255 | -1.385 | 0 | 0.415 |
| 17833 | TPTEP1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 17 | ITGA5 | Sponge network | -1.216 | 0 | 0.452 | 0.03524 | 0.415 |
| 17834 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | ZFHX3 | Sponge network | 0.61 | 0.01593 | 0.389 | 0.01363 | 0.415 |
| 17835 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 22 | CREB3L2 | Sponge network | -0.376 | 0.00337 | -0.525 | 2.0E-5 | 0.415 |
| 17836 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | GALNT13 | Sponge network | -0.405 | 0.22013 | -0.857 | 0.07439 | 0.415 |
| 17837 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 24 | AKAP6 | Sponge network | 0.72 | 0.0001 | -1.586 | 3.0E-5 | 0.415 |
| 17838 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 10 | PKNOX1 | Sponge network | 0.559 | 0.00026 | 0.085 | 0.28917 | 0.415 |
| 17839 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p | 13 | DCLK1 | Sponge network | 0.082 | 0.55397 | -1.324 | 0.00014 | 0.415 |
| 17840 | LINC00641 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | BCL9 | Sponge network | -0.222 | 0.32445 | 0.321 | 0.00267 | 0.415 |
| 17841 | CTB-133G6.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-940 | 13 | MDM4 | Sponge network | 0.4 | 0.39299 | 0.345 | 0.00762 | 0.415 |
| 17842 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-5p | 34 | ERBB4 | Sponge network | -2.62 | 0 | -1.975 | 2.0E-5 | 0.415 |
| 17843 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 13 | RARB | Sponge network | -0.351 | 0.11358 | -0.825 | 2.0E-5 | 0.415 |
| 17844 | GS1-124K5.11 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 27 | CREB1 | Sponge network | 0.494 | 0.00339 | 0.203 | 0.00537 | 0.415 |
| 17845 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 49 | RUNX1T1 | Sponge network | 0.494 | 0.00339 | -0.344 | 0.2374 | 0.415 |
| 17846 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-500b-5p | 16 | GRIA3 | Sponge network | -0.932 | 0.00329 | -1.956 | 0 | 0.415 |
| 17847 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 12 | CNTNAP1 | Sponge network | -0.737 | 0.10822 | 0.358 | 0.13727 | 0.415 |
| 17848 | EMX2OS |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p | 12 | ALDH1A2 | Sponge network | -0.777 | 0.1134 | -2.411 | 0 | 0.415 |
| 17849 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-590-3p | 18 | DACT1 | Sponge network | 0.199 | 0.38015 | 0.783 | 0.00072 | 0.415 |
| 17850 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 21 | LATS2 | Sponge network | -0.777 | 0.1134 | 0.159 | 0.2304 | 0.415 |
| 17851 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-501-5p;hsa-miR-590-3p | 30 | ZFHX3 | Sponge network | 0.082 | 0.55654 | 0.389 | 0.01363 | 0.415 |
| 17852 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421 | 12 | DACT1 | Sponge network | -0.154 | 0.78939 | 0.783 | 0.00072 | 0.415 |
| 17853 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 28 | PIK3C2A | Sponge network | 0.311 | 0.10344 | 0.061 | 0.53776 | 0.415 |
| 17854 | LINC00403 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-452-5p | 11 | ANK2 | Sponge network | -3.586 | 0.00032 | -1.603 | 1.0E-5 | 0.415 |
| 17855 | LINC00865 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-935 | 41 | PTPRD | Sponge network | -1.249 | 7.0E-5 | -1.005 | 0.00064 | 0.415 |
| 17856 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 21 | USP12 | Sponge network | 0.683 | 1.0E-5 | -0.102 | 0.40768 | 0.415 |
| 17857 | LINC00578 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p | 24 | SNX29 | Sponge network | 0.811 | 0.03228 | -0.34 | 0.01371 | 0.415 |
| 17858 | AC108488.4 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-577;hsa-miR-7-1-3p | 10 | INPP4A | Sponge network | 0.259 | 0.12542 | 0.07 | 0.53563 | 0.415 |
| 17859 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-497-5p | 13 | CBL | Sponge network | 0.997 | 0 | 0.291 | 0.01267 | 0.415 |
| 17860 | RP4-668J24.2 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-511-5p;hsa-miR-532-3p | 10 | MRVI1 | Sponge network | -1.054 | 0.0706 | -1.051 | 0.00022 | 0.415 |
| 17861 | RP11-774O3.3 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p | 16 | ATM | Sponge network | -0.487 | 0.02157 | 0.178 | 0.21568 | 0.415 |
| 17862 | SNHG14 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | PIK3CA | Sponge network | -1.111 | 0.0003 | 0.195 | 0.06908 | 0.415 |
| 17863 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-92a-3p | 38 | FOXP1 | Sponge network | -0.096 | 0.67958 | 0.042 | 0.74368 | 0.415 |
| 17864 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p | 13 | DKK3 | Sponge network | -1.654 | 0.00144 | -0.052 | 0.77783 | 0.415 |
| 17865 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 14 | FGFR1 | Sponge network | 0.381 | 0.25967 | -0.509 | 0.03957 | 0.415 |
| 17866 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | BNC2 | Sponge network | -0.726 | 0.00132 | -0.491 | 0.09988 | 0.415 |
| 17867 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-625-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -0.842 | 0.01361 | -0.087 | 0.69193 | 0.415 |
| 17868 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-93-5p | 26 | JAZF1 | Sponge network | 0.299 | 0.57722 | -0.377 | 0.02677 | 0.415 |
| 17869 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | PRKAB2 | Sponge network | 0.385 | 0.10067 | -0.367 | 0.01371 | 0.415 |
| 17870 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CLIP1 | Sponge network | -1.111 | 0.0003 | -0.215 | 0.08684 | 0.415 |
| 17871 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 52 | FOXP1 | Sponge network | -0.737 | 0.06182 | 0.042 | 0.74368 | 0.415 |
| 17872 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | NIN | Sponge network | 0.083 | 0.66405 | -0.253 | 0.13794 | 0.415 |
| 17873 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 28 | ETV1 | Sponge network | -0.612 | 0.00094 | -0.096 | 0.67674 | 0.415 |
| 17874 | AC108488.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | RASSF8 | Sponge network | 0.259 | 0.12542 | -0.122 | 0.65591 | 0.415 |
| 17875 | RP11-999E24.3 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-361-3p | 13 | ITPKB | Sponge network | -0.693 | 0.05629 | -0.704 | 0.00106 | 0.415 |
| 17876 | RP11-57H14.4 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITGB1 | Sponge network | 0.083 | 0.66405 | 0.524 | 0.00017 | 0.415 |
| 17877 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | QKI | Sponge network | -0.246 | 0.35034 | -0.333 | 0.04111 | 0.415 |
| 17878 | RP11-48B3.4 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-576-5p | 17 | NUAK1 | Sponge network | 1.757 | 0 | 0.904 | 0 | 0.415 |
| 17879 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-942-5p | 22 | DDX6 | Sponge network | 0.082 | 0.55397 | 0.091 | 0.36428 | 0.415 |
| 17880 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 14 | PIK3IP1 | Sponge network | -0.49 | 0.04946 | -0.187 | 0.19945 | 0.415 |
| 17881 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | PRKAB2 | Sponge network | 0.174 | 0.30034 | -0.367 | 0.01371 | 0.415 |
| 17882 | LINC00654 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | BCL9 | Sponge network | 0.374 | 0.20054 | 0.321 | 0.00267 | 0.415 |
| 17883 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-508-3p;hsa-miR-590-3p | 12 | RIMS1 | Sponge network | -0.129 | 0.57177 | -3.143 | 0 | 0.415 |
| 17884 | HAND2-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | TLE4 | Sponge network | -1.697 | 0.00889 | -1.157 | 0 | 0.415 |
| 17885 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | HIVEP1 | Sponge network | 0.782 | 0.00016 | 0.368 | 0.00064 | 0.415 |
| 17886 | CTC-429P9.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 10 | ROBO1 | Sponge network | 0.178 | 0.29106 | -0.009 | 0.96154 | 0.415 |
| 17887 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 14 | FBXO32 | Sponge network | -1.375 | 0.08642 | -0.495 | 0.07561 | 0.415 |
| 17888 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429 | 23 | TMEM132B | Sponge network | -1.654 | 0.00144 | -0.83 | 0.01084 | 0.415 |
| 17889 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p | 14 | SON | Sponge network | 0.921 | 0.00118 | 0.168 | 0.04406 | 0.415 |
| 17890 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | NF1 | Sponge network | 1.153 | 0 | 0.51 | 0 | 0.415 |
| 17891 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | PCDH9 | Sponge network | -0.969 | 2.0E-5 | -1.823 | 4.0E-5 | 0.415 |
| 17892 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | RET | Sponge network | -0.901 | 0.00019 | -0.833 | 0.03517 | 0.415 |
| 17893 | RP11-196G18.22 |
hsa-miR-101-3p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-539-5p | 11 | KIF5B | Sponge network | 1.206 | 0 | 0.362 | 0.00066 | 0.415 |
| 17894 | MIR143HG |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 60 | PPM1L | Sponge network | -0.739 | 0.0574 | -0.537 | 0.00616 | 0.415 |
| 17895 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-940 | 13 | PBX1 | Sponge network | -1.255 | 0.00981 | -1.206 | 0 | 0.415 |
| 17896 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 59 | IL6ST | Sponge network | 0.72 | 0.0001 | -0.402 | 0.00676 | 0.415 |
| 17897 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 11 | ZFHX3 | Sponge network | 1.055 | 0 | 0.389 | 0.01363 | 0.415 |
| 17898 | FAM13A-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-93-5p | 20 | RNASEL | Sponge network | -0.83 | 0 | -0.272 | 0.01796 | 0.415 |
| 17899 | ACTA2-AS1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | CSDE1 | Sponge network | 0.194 | 0.64278 | -0.493 | 0 | 0.415 |
| 17900 | RP11-597D13.9 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | 1.302 | 7.0E-5 | -1.051 | 0.00022 | 0.415 |
| 17901 | RP11-57H14.4 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-340-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 20 | CSDE1 | Sponge network | 0.083 | 0.66405 | -0.493 | 0 | 0.415 |
| 17902 | RP11-67L2.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 18 | LATS2 | Sponge network | 0.72 | 0.0001 | 0.159 | 0.2304 | 0.415 |
| 17903 | USP3-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-935 | 17 | RELN | Sponge network | -0.162 | 0.47687 | -2.403 | 0 | 0.415 |
| 17904 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 32 | ZFHX4 | Sponge network | -0.513 | 0.04703 | 0.018 | 0.95574 | 0.415 |
| 17905 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p | 26 | PRDM2 | Sponge network | -0.976 | 0.00025 | -0.258 | 0.00721 | 0.415 |
| 17906 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 27 | CUL5 | Sponge network | 0.782 | 0.00016 | 0.026 | 0.77301 | 0.415 |
| 17907 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-7-1-3p | 10 | CAV1 | Sponge network | 0.16 | 0.50785 | -1.013 | 6.0E-5 | 0.415 |
| 17908 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | CREBBP | Sponge network | 0.194 | 0.64278 | 0.126 | 0.15186 | 0.415 |
| 17909 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p | 19 | GREM1 | Sponge network | -0.612 | 0.00094 | 0.902 | 0.01691 | 0.415 |
| 17910 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | DCLK1 | Sponge network | -0.899 | 6.0E-5 | -1.324 | 0.00014 | 0.415 |
| 17911 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 18 | SPTBN4 | Sponge network | -0.49 | 0.04946 | -0.786 | 0.01281 | 0.415 |
| 17912 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 12 | FGFR1 | Sponge network | -0.125 | 0.49414 | -0.509 | 0.03957 | 0.415 |
| 17913 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-532-5p;hsa-miR-766-3p | 46 | IL6ST | Sponge network | 0.082 | 0.55397 | -0.402 | 0.00676 | 0.415 |
| 17914 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-5p | 26 | MOB1B | Sponge network | 0.921 | 0.00118 | 0.197 | 0.15266 | 0.415 |
| 17915 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | LRRC55 | Sponge network | -1.575 | 6.0E-5 | -0.026 | 0.93163 | 0.415 |
| 17916 | GNG12-AS1 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | NR3C2 | Sponge network | -0.793 | 7.0E-5 | -1.56 | 0 | 0.415 |
| 17917 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 22 | TET1 | Sponge network | 0.222 | 0.29133 | -0.135 | 0.52138 | 0.415 |
| 17918 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 21 | ZMYM2 | Sponge network | 0.91 | 0 | 0.279 | 0.01273 | 0.415 |
| 17919 | GAS6-AS2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 20 | NAV3 | Sponge network | 0.16 | 0.50785 | 0.212 | 0.47729 | 0.415 |
| 17920 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p | 11 | THSD7A | Sponge network | 0.101 | 0.60919 | 0.242 | 0.25154 | 0.415 |
| 17921 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p | 34 | NTRK2 | Sponge network | -0.154 | 0.78939 | -0.736 | 0.06495 | 0.415 |
| 17922 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | MEF2C | Sponge network | -0.342 | 0.02288 | -0.499 | 0.01448 | 0.415 |
| 17923 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 19 | ACVR1 | Sponge network | 0.572 | 0.01722 | 0.279 | 0.00234 | 0.415 |
| 17924 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p | 12 | PDZD2 | Sponge network | -0.323 | 0.01372 | -0.909 | 3.0E-5 | 0.415 |
| 17925 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 28 | DIP2C | Sponge network | -0.513 | 0.04703 | -0.436 | 0.01713 | 0.415 |
| 17926 | PCA3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p | 33 | IGFBP5 | Sponge network | -0.709 | 0.18168 | -0.806 | 0.00105 | 0.415 |
| 17927 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 12 | RCAN2 | Sponge network | 0.765 | 7.0E-5 | -0.33 | 0.15172 | 0.415 |
| 17928 | EMX2OS |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | KLF10 | Sponge network | -0.777 | 0.1134 | -0.783 | 0 | 0.415 |
| 17929 | RP11-182J1.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-7-1-3p | 11 | MCC | Sponge network | -0.187 | 0.23787 | -1.005 | 0 | 0.415 |
| 17930 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p | 20 | MOB1B | Sponge network | 0.542 | 0.00054 | 0.197 | 0.15266 | 0.415 |
| 17931 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-5p | 28 | CNTN1 | Sponge network | -1.645 | 0 | -2.594 | 0 | 0.415 |
| 17932 | LINC00680 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c | 17 | ABL2 | Sponge network | 0.884 | 0 | 0.82 | 0 | 0.415 |
| 17933 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-940 | 59 | NFIB | Sponge network | 0.194 | 0.64278 | 0.023 | 0.90795 | 0.415 |
| 17934 | FGD5-AS1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 12 | GSK3B | Sponge network | 0.401 | 0.00066 | 0.266 | 0.00045 | 0.415 |
| 17935 | GS1-124K5.11 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-877-5p | 28 | NAV3 | Sponge network | 0.494 | 0.00339 | 0.212 | 0.47729 | 0.415 |
| 17936 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 68 | TCF4 | Sponge network | 0.374 | 0.20054 | 0.016 | 0.92271 | 0.414 |
| 17937 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ZMYND11 | Sponge network | 0.225 | 0.30644 | -0.2 | 0.05449 | 0.414 |
| 17938 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 26 | PHC3 | Sponge network | -0.227 | 0.31657 | 0.212 | 0.0499 | 0.414 |
| 17939 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-7-5p;hsa-miR-96-5p | 37 | TCF4 | Sponge network | -0.326 | 0.14314 | 0.016 | 0.92271 | 0.414 |
| 17940 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p | 17 | RET | Sponge network | -0.323 | 0.01372 | -0.833 | 0.03517 | 0.414 |
| 17941 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | PGR | Sponge network | 0.174 | 0.30034 | -1.704 | 0 | 0.414 |
| 17942 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 63 | TCF4 | Sponge network | -0.222 | 0.32445 | 0.016 | 0.92271 | 0.414 |
| 17943 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-455-3p;hsa-miR-7-1-3p | 16 | MCC | Sponge network | 0.693 | 0.00076 | -1.005 | 0 | 0.414 |
| 17944 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SLC6A15 | Sponge network | -0.901 | 0.00019 | -2.373 | 2.0E-5 | 0.414 |
| 17945 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-421 | 24 | BTG2 | Sponge network | -0.583 | 0.14589 | -1.763 | 0 | 0.414 |
| 17946 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 58 | TCF4 | Sponge network | -1.249 | 7.0E-5 | 0.016 | 0.92271 | 0.414 |
| 17947 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 34 | PRKAB2 | Sponge network | 0.311 | 0.10344 | -0.367 | 0.01371 | 0.414 |
| 17948 | FGF14-AS2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-590-3p | 10 | CELF4 | Sponge network | -1.582 | 8.0E-5 | -1.135 | 0.00344 | 0.414 |
| 17949 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DACH2 | Sponge network | -1.281 | 0.00012 | -1.635 | 0.00034 | 0.414 |
| 17950 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-942-5p | 48 | THRB | Sponge network | -0.777 | 0.16667 | -1.117 | 0.0004 | 0.414 |
| 17951 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 21 | CLASP2 | Sponge network | 0.401 | 0.00066 | -0.038 | 0.71 | 0.414 |
| 17952 | LINC00969 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p | 29 | RICTOR | Sponge network | 0.683 | 1.0E-5 | 0.388 | 0.00188 | 0.414 |
| 17953 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | TBX18 | Sponge network | -0.358 | 0.17255 | 0.843 | 0.02945 | 0.414 |
| 17954 | RP11-359E10.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | MEF2C | Sponge network | 0.299 | 0.57722 | -0.499 | 0.01448 | 0.414 |
| 17955 | LINC00702 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-590-3p | 11 | PLA2R1 | Sponge network | -0.737 | 0.06182 | -0.265 | 0.24676 | 0.414 |
| 17956 | AC005562.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | PIK3CA | Sponge network | 0.488 | 0.00084 | 0.195 | 0.06908 | 0.414 |
| 17957 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 24 | CBL | Sponge network | 0.873 | 0 | 0.291 | 0.01267 | 0.414 |
| 17958 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 34 | TACC1 | Sponge network | 0.16 | 0.50785 | -0.362 | 0.05406 | 0.414 |
| 17959 | LINC00476 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-3614-5p | 12 | NGFR | Sponge network | -0.615 | 0.00015 | -1.271 | 0.01058 | 0.414 |
| 17960 | HCG18 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 27 | BRD3 | Sponge network | 1.063 | 0 | 0.37 | 0.00013 | 0.414 |
| 17961 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 13 | BCLAF1 | Sponge network | 1.11 | 0 | 0.211 | 0.00684 | 0.414 |
| 17962 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SEMA3C | Sponge network | 0.61 | 0.01593 | -0.272 | 0.31153 | 0.414 |
| 17963 | HCG11 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | CREB1 | Sponge network | 0.385 | 0.10067 | 0.203 | 0.00537 | 0.414 |
| 17964 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ABCC9 | Sponge network | 0.61 | 0.01593 | -0.466 | 0.14121 | 0.414 |
| 17965 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 62 | THRB | Sponge network | -1.575 | 6.0E-5 | -1.117 | 0.0004 | 0.414 |
| 17966 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p | 21 | PCDH9 | Sponge network | -0.384 | 0.40652 | -1.823 | 4.0E-5 | 0.414 |
| 17967 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-511-5p | 14 | EYA4 | Sponge network | -2.531 | 0 | 0.3 | 0.36876 | 0.414 |
| 17968 | PAN3-AS1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 12 | CREB1 | Sponge network | 0.062 | 0.55274 | 0.203 | 0.00537 | 0.414 |
| 17969 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-93-3p | 46 | IL6ST | Sponge network | -0.154 | 0.53954 | -0.402 | 0.00676 | 0.414 |
| 17970 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | PHC3 | Sponge network | 0.225 | 0.30644 | 0.212 | 0.0499 | 0.414 |
| 17971 | RP11-119F7.5 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-935 | 11 | STAT5B | Sponge network | 0.225 | 0.30644 | -0.182 | 0.1082 | 0.414 |
| 17972 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p | 11 | MGA | Sponge network | 0.946 | 0 | 0.323 | 0.00792 | 0.414 |
| 17973 | AC005154.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-766-3p | 14 | KIAA1024 | Sponge network | 0.848 | 0 | 1.083 | 0 | 0.414 |
| 17974 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 33 | NFIB | Sponge network | -0.583 | 0.14589 | 0.023 | 0.90795 | 0.414 |
| 17975 | NR2F1-AS1 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 13 | LZTS1 | Sponge network | -0.49 | 0.04946 | 1.664 | 0 | 0.414 |
| 17976 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | EPG5 | Sponge network | 1.601 | 0 | 0.101 | 0.3563 | 0.414 |
| 17977 | AC108488.4 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-511-5p;hsa-miR-532-3p | 14 | MRVI1 | Sponge network | 0.259 | 0.12542 | -1.051 | 0.00022 | 0.414 |
| 17978 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FSTL5 | Sponge network | -2.915 | 0.00015 | -1.3 | 0.01619 | 0.414 |
| 17979 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | SYNE1 | Sponge network | 0.16 | 0.50785 | -0.738 | 0.00316 | 0.414 |
| 17980 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-5p | 13 | FSTL5 | Sponge network | -1.349 | 0.00017 | -1.3 | 0.01619 | 0.414 |
| 17981 | KB-1410C5.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 10 | ABI2 | Sponge network | 0.959 | 0 | 0.175 | 0.14787 | 0.414 |
| 17982 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-5p | 15 | TACC1 | Sponge network | -1.255 | 0.00981 | -0.362 | 0.05406 | 0.414 |
| 17983 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 41 | ZFHX4 | Sponge network | 1.302 | 7.0E-5 | 0.018 | 0.95574 | 0.414 |
| 17984 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-5p | 34 | GAS7 | Sponge network | 0.176 | 0.37402 | -0.555 | 0.01602 | 0.414 |
| 17985 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 59 | FOXP1 | Sponge network | -1.111 | 0.0003 | 0.042 | 0.74368 | 0.414 |
| 17986 | RP11-65J21.3 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-505-5p;hsa-miR-7-1-3p | 15 | AR | Sponge network | -0.557 | 0.11135 | -1.554 | 0 | 0.414 |
| 17987 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-590-3p | 16 | CREBBP | Sponge network | 0.349 | 0.01695 | 0.126 | 0.15186 | 0.414 |
| 17988 | ACTA2-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-550a-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | PCDH18 | Sponge network | 0.194 | 0.64278 | 0.216 | 0.24645 | 0.414 |
| 17989 | RP11-175O19.4 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 15 | CDK6 | Sponge network | 0.917 | 0 | 0.991 | 0 | 0.414 |
| 17990 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | MITF | Sponge network | -0.021 | 0.93598 | -0.769 | 0.00022 | 0.414 |
| 17991 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 28 | USP12 | Sponge network | 0.083 | 0.66405 | -0.102 | 0.40768 | 0.414 |
| 17992 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 32 | ADARB1 | Sponge network | 0.727 | 3.0E-5 | -0.335 | 0.06631 | 0.414 |
| 17993 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 32 | ABI2 | Sponge network | 0.41 | 0.02214 | 0.175 | 0.14787 | 0.414 |
| 17994 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-5p;hsa-miR-940 | 12 | MYO9A | Sponge network | 1.134 | 0 | -0.147 | 0.32373 | 0.414 |
| 17995 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-511-5p | 11 | LRRC4 | Sponge network | -1.645 | 0 | -1.774 | 0 | 0.414 |
| 17996 | RP1-151F17.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935 | 31 | DMD | Sponge network | 0.222 | 0.29133 | -1.506 | 0 | 0.414 |
| 17997 | PART1 |
hsa-let-7a-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | -2.925 | 0 | -1.628 | 0 | 0.414 |
| 17998 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | NIN | Sponge network | 0.194 | 0.64278 | -0.253 | 0.13794 | 0.414 |
| 17999 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | LRRC55 | Sponge network | 0.997 | 0.00066 | -0.026 | 0.93163 | 0.414 |
| 18000 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 32 | FBXO32 | Sponge network | -0.769 | 0.00035 | -0.495 | 0.07561 | 0.414 |
| 18001 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 11 | PSIP1 | Sponge network | 0.541 | 0.00125 | -0.005 | 0.96897 | 0.414 |
| 18002 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-576-5p | 28 | PBX1 | Sponge network | -0.611 | 0.09607 | -1.206 | 0 | 0.414 |
| 18003 | LINC00173 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 21 | SETBP1 | Sponge network | 0.056 | 0.8494 | -1.302 | 1.0E-5 | 0.414 |
| 18004 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-940 | 19 | NOTCH2 | Sponge network | -0.154 | 0.53954 | 0.138 | 0.35163 | 0.414 |
| 18005 | CTA-363E19.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 10 | PTPN11 | Sponge network | 1.055 | 0 | 0.431 | 2.0E-5 | 0.414 |
| 18006 | CTC-296K1.4 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-331-3p;hsa-miR-424-5p | 11 | CBFA2T3 | Sponge network | -2.076 | 0.00548 | -1.221 | 1.0E-5 | 0.414 |
| 18007 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p | 16 | EBF3 | Sponge network | 0.082 | 0.55397 | -0.058 | 0.81437 | 0.414 |
| 18008 | AC004540.4 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p | 14 | IL17RD | Sponge network | -2.549 | 0.00528 | -0.019 | 0.94873 | 0.414 |
| 18009 | PCA3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-96-5p | 19 | CBFA2T3 | Sponge network | -0.709 | 0.18168 | -1.221 | 1.0E-5 | 0.414 |
| 18010 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | RASSF8 | Sponge network | -0.942 | 0 | -0.122 | 0.65591 | 0.414 |
| 18011 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-5p | 10 | SEMA3E | Sponge network | -0.777 | 0.16667 | -1.717 | 0.00137 | 0.414 |
| 18012 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | -0.487 | 0.02157 | 0.126 | 0.15266 | 0.414 |
| 18013 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-98-5p | 13 | SCN11A | Sponge network | -0.942 | 0 | -1.15 | 0.00299 | 0.414 |
| 18014 | MYLK-AS1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-940 | 12 | CSRNP1 | Sponge network | -1.331 | 3.0E-5 | -1.448 | 0 | 0.414 |
| 18015 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 16 | ITCH | Sponge network | 1.317 | 0 | 0.084 | 0.34058 | 0.414 |
| 18016 | RP11-156P1.3 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 13 | NGFR | Sponge network | 0.214 | 0.18246 | -1.271 | 0.01058 | 0.414 |
| 18017 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-576-5p | 14 | ARHGAP28 | Sponge network | -0.932 | 0.00329 | -0.911 | 0.00013 | 0.414 |
| 18018 | PCA3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 17 | PNRC1 | Sponge network | -0.709 | 0.18168 | -0.646 | 0 | 0.414 |
| 18019 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | CNTNAP3 | Sponge network | -0.096 | 0.67958 | -1.465 | 0.00033 | 0.414 |
| 18020 | RP11-102N12.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-96-5p | 20 | ASTN1 | Sponge network | -0.351 | 0.11358 | -2.389 | 2.0E-5 | 0.414 |
| 18021 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | CUL5 | Sponge network | 0.222 | 0.29133 | 0.026 | 0.77301 | 0.414 |
| 18022 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-92a-3p | 25 | CRTC1 | Sponge network | 0.214 | 0.18246 | -0.116 | 0.28412 | 0.414 |
| 18023 | RP11-65J21.3 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p | 10 | ZBTB4 | Sponge network | -0.557 | 0.11135 | -0.739 | 0 | 0.414 |
| 18024 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-93-5p | 17 | TGFBR2 | Sponge network | -0.976 | 0.00025 | -0.243 | 0.11408 | 0.414 |
| 18025 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 19 | ZMYND11 | Sponge network | -0.976 | 0.00025 | -0.2 | 0.05449 | 0.414 |
| 18026 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-576-5p | 18 | GREM1 | Sponge network | 0.61 | 0.01593 | 0.902 | 0.01691 | 0.414 |
| 18027 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | DCLK1 | Sponge network | -1.057 | 0.00029 | -1.324 | 0.00014 | 0.414 |
| 18028 | RP11-571M6.17 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-625-3p | 16 | ABL2 | Sponge network | 1.013 | 0 | 0.82 | 0 | 0.414 |
| 18029 | DNM3OS |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | KCNAB1 | Sponge network | 0.572 | 0.01722 | -0.755 | 2.0E-5 | 0.414 |
| 18030 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-425-5p | 17 | GRIA3 | Sponge network | -0.693 | 0.05629 | -1.956 | 0 | 0.414 |
| 18031 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 26 | PIK3C2A | Sponge network | 1.063 | 0 | 0.061 | 0.53776 | 0.414 |
| 18032 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 49 | CHD5 | Sponge network | -2.925 | 0 | -0.922 | 0.01521 | 0.414 |
| 18033 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 10 | NR2C2 | Sponge network | 1.055 | 0 | 0.21 | 0.03224 | 0.414 |
| 18034 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | CACNA2D1 | Sponge network | 0.342 | 0.03129 | -0.846 | 0.00675 | 0.414 |
| 18035 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -0.465 | 0.04703 | 0.024 | 0.89889 | 0.414 |
| 18036 | LINC00641 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -0.222 | 0.32445 | -2.633 | 0 | 0.414 |
| 18037 | AC007879.7 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-5p | 16 | ABL2 | Sponge network | 4.3 | 0 | 0.82 | 0 | 0.414 |
| 18038 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | ZNF770 | Sponge network | 1.063 | 0 | 0.206 | 0.06751 | 0.414 |
| 18039 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 15 | AKAP6 | Sponge network | 0.419 | 0.0319 | -1.586 | 3.0E-5 | 0.414 |
| 18040 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | RET | Sponge network | -0.737 | 0.10822 | -0.833 | 0.03517 | 0.414 |
| 18041 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | FAT4 | Sponge network | 0.225 | 0.30644 | -0.354 | 0.14694 | 0.414 |
| 18042 | RP11-672A2.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-671-5p | 10 | CDH23 | Sponge network | 1.344 | 0 | -0.974 | 0.00034 | 0.414 |
| 18043 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 32 | CADM1 | Sponge network | -0.49 | 0.04946 | -0.9 | 0.00084 | 0.414 |
| 18044 | LINC00654 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | ROBO1 | Sponge network | 0.374 | 0.20054 | -0.009 | 0.96154 | 0.414 |
| 18045 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | MLLT10 | Sponge network | 1.093 | 0 | 0.105 | 0.33292 | 0.414 |
| 18046 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-200c-5p;hsa-miR-323a-3p;hsa-miR-33a-3p | 11 | DIP2C | Sponge network | 0.497 | 0.01451 | -0.436 | 0.01713 | 0.414 |
| 18047 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-5p | 21 | ESR1 | Sponge network | -1.654 | 0.00144 | -1.035 | 3.0E-5 | 0.414 |
| 18048 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-590-3p | 14 | GRIA3 | Sponge network | -1.582 | 8.0E-5 | -1.956 | 0 | 0.414 |
| 18049 | GNG12-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 19 | TAL1 | Sponge network | -0.793 | 7.0E-5 | -0.462 | 0.00574 | 0.414 |
| 18050 | RP11-672A2.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-98-5p | 10 | ZFHX3 | Sponge network | 1.344 | 0 | 0.389 | 0.01363 | 0.414 |
| 18051 | LINC00578 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-744-3p | 14 | HIP1 | Sponge network | 0.811 | 0.03228 | 0.774 | 0 | 0.414 |
| 18052 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 17 | GRIN2A | Sponge network | -0.726 | 0.00132 | -2.161 | 0 | 0.414 |
| 18053 | MIR22HG |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-940 | 17 | SFRP1 | Sponge network | -1.047 | 0 | -3.566 | 0 | 0.414 |
| 18054 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 15 | BNC2 | Sponge network | -0.557 | 0.11135 | -0.491 | 0.09988 | 0.414 |
| 18055 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | CXXC4 | Sponge network | 0.41 | 0.02214 | -0.433 | 0.21055 | 0.414 |
| 18056 | CKMT2-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | PRUNE2 | Sponge network | 0.082 | 0.55654 | -1.733 | 0.00022 | 0.414 |
| 18057 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-628-5p | 12 | GRIA3 | Sponge network | -0.376 | 0.00337 | -1.956 | 0 | 0.414 |
| 18058 | RP11-767N6.7 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-5p | 19 | MDM4 | Sponge network | 0.466 | 0.00155 | 0.345 | 0.00762 | 0.414 |
| 18059 | TP73-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | GALNT13 | Sponge network | -0.563 | 0.02545 | -0.857 | 0.07439 | 0.414 |
| 18060 | RP11-1006G14.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | SIRT1 | Sponge network | 0.559 | 0.00026 | 0.109 | 0.2593 | 0.414 |
| 18061 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 24 | PCDH10 | Sponge network | 0.16 | 0.50785 | -1.644 | 0.0078 | 0.414 |
| 18062 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-542-3p;hsa-miR-942-5p | 26 | UNC80 | Sponge network | -1.092 | 4.0E-5 | -1.934 | 0 | 0.414 |
| 18063 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 18 | PAK3 | Sponge network | -2.46 | 0 | -0.628 | 0.07253 | 0.414 |
| 18064 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 22 | SEMA3C | Sponge network | 0.246 | 0.59912 | -0.272 | 0.31153 | 0.414 |
| 18065 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-590-3p | 15 | ABCB5 | Sponge network | -0.896 | 0.00099 | -0.956 | 0.04077 | 0.414 |
| 18066 | USP3-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-940 | 17 | ZFP36L1 | Sponge network | -0.162 | 0.47687 | -0.505 | 8.0E-5 | 0.414 |
| 18067 | PCA3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | -0.709 | 0.18168 | 0.809 | 0.00544 | 0.414 |
| 18068 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 48 | PIK3R1 | Sponge network | -0.49 | 0.04946 | -0.133 | 0.36854 | 0.414 |
| 18069 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | AKAP12 | Sponge network | -0.615 | 0.00015 | -0.932 | 0.0028 | 0.414 |
| 18070 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-539-5p | 11 | RSPO2 | Sponge network | -0.309 | 0.1145 | -3.038 | 0 | 0.414 |
| 18071 | RAD51-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-495-3p | 12 | PHF6 | Sponge network | 0.768 | 7.0E-5 | 0.785 | 0 | 0.414 |
| 18072 | RP11-545G3.1 |
hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 15 | ABL2 | Sponge network | 2.622 | 0.00025 | 0.82 | 0 | 0.414 |
| 18073 | TPTEP1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 12 | ALDH1A2 | Sponge network | -1.216 | 0 | -2.411 | 0 | 0.414 |
| 18074 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-98-5p | 26 | DMD | Sponge network | -0.773 | 0.04073 | -1.506 | 0 | 0.414 |
| 18075 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 20 | LIFR | Sponge network | -1.092 | 4.0E-5 | -1.794 | 0 | 0.414 |
| 18076 | CTA-941F9.9 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-502-5p;hsa-miR-576-5p | 12 | DTNA | Sponge network | -0.344 | 0.49624 | -1.648 | 1.0E-5 | 0.414 |
| 18077 | MEG3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | PIK3CA | Sponge network | 0.249 | 0.35902 | 0.195 | 0.06908 | 0.414 |
| 18078 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | LRRC7 | Sponge network | -1.161 | 0.00591 | -1.341 | 0.01193 | 0.414 |
| 18079 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 30 | DDX6 | Sponge network | 0.532 | 0.00505 | 0.091 | 0.36428 | 0.414 |
| 18080 | LINC00957 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 19 | RNF144A | Sponge network | -0.421 | 0.0114 | 0.594 | 0.0009 | 0.414 |
| 18081 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | PIK3C2A | Sponge network | 1.2 | 0.01813 | 0.061 | 0.53776 | 0.414 |
| 18082 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 16 | RSPO2 | Sponge network | -0.376 | 0.00337 | -3.038 | 0 | 0.414 |
| 18083 | DNAJC27-AS1 |
hsa-miR-146b-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-96-5p | 12 | FHL1 | Sponge network | -0.969 | 2.0E-5 | -2.687 | 0 | 0.414 |
| 18084 | RP5-994D16.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ARHGEF12 | Sponge network | 0.252 | 0.08508 | 0.09 | 0.35693 | 0.414 |
| 18085 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p | 49 | ADARB1 | Sponge network | -0.899 | 6.0E-5 | -0.335 | 0.06631 | 0.414 |
| 18086 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CLIP1 | Sponge network | -0.49 | 0.04946 | -0.215 | 0.08684 | 0.414 |
| 18087 | HOXB-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-940 | 15 | SORCS1 | Sponge network | -0.154 | 0.53954 | -3.492 | 0 | 0.414 |
| 18088 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 29 | LRRC7 | Sponge network | -0.942 | 0 | -1.341 | 0.01193 | 0.414 |
| 18089 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 20 | SLITRK3 | Sponge network | 0.335 | 0.20596 | -3.328 | 0 | 0.414 |
| 18090 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 35 | RASSF2 | Sponge network | -0.487 | 0.02157 | -0.273 | 0.21961 | 0.414 |
| 18091 | LINC00957 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | HERC1 | Sponge network | -0.421 | 0.0114 | -0.196 | 0.08544 | 0.414 |
| 18092 | C20orf166-AS1 |
hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577 | 19 | LIFR | Sponge network | -2.69 | 0.0001 | -1.794 | 0 | 0.414 |
| 18093 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-5p | 31 | MEF2C | Sponge network | 0.176 | 0.37402 | -0.499 | 0.01448 | 0.414 |
| 18094 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 58 | RUNX1T1 | Sponge network | 0.082 | 0.55654 | -0.344 | 0.2374 | 0.414 |
| 18095 | PCA3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | NAV3 | Sponge network | -0.709 | 0.18168 | 0.212 | 0.47729 | 0.414 |
| 18096 | USP3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-455-5p | 11 | PTPN13 | Sponge network | -0.162 | 0.47687 | -1.208 | 0 | 0.414 |
| 18097 | RP11-473I1.9 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | MAP3K4 | Sponge network | 0.683 | 1.0E-5 | 0.058 | 0.54229 | 0.414 |
| 18098 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-539-5p;hsa-miR-7-1-3p | 13 | TAL1 | Sponge network | -0.611 | 0.09607 | -0.462 | 0.00574 | 0.413 |
| 18099 | TTC25 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-150-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-940 | 11 | PBX1 | Sponge network | -0.208 | 0.38261 | -1.206 | 0 | 0.413 |
| 18100 | RP11-458F8.4 |
hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-5p | 12 | ABL2 | Sponge network | 1.518 | 0 | 0.82 | 0 | 0.413 |
| 18101 | ZNF582-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 43 | DST | Sponge network | -1.349 | 0.00017 | -0.713 | 0.00096 | 0.413 |
| 18102 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p;hsa-miR-940 | 23 | AKAP11 | Sponge network | 0.946 | 0 | 0.196 | 0.09825 | 0.413 |
| 18103 | HHIP-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p | 21 | DMD | Sponge network | -0.737 | 0.10822 | -1.506 | 0 | 0.413 |
| 18104 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-93-5p | 12 | EDA2R | Sponge network | -0.154 | 0.78939 | -0.587 | 0.01598 | 0.413 |
| 18105 | LINC00092 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | UNC5C | Sponge network | -1.886 | 0 | -0.178 | 0.40214 | 0.413 |
| 18106 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-629-3p | 23 | ESR1 | Sponge network | -0.154 | 0.53954 | -1.035 | 3.0E-5 | 0.413 |
| 18107 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | RICTOR | Sponge network | 0.8 | 0 | 0.388 | 0.00188 | 0.413 |
| 18108 | CTD-2245F17.3 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-940 | 11 | GRIK3 | Sponge network | -0.421 | 0.12556 | -3.208 | 0 | 0.413 |
| 18109 | A2M-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-331-3p;hsa-miR-484 | 10 | FLNA | Sponge network | -0.08 | 0.90092 | -0.771 | 0.01377 | 0.413 |
| 18110 | RP11-890B15.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 13 | STARD13 | Sponge network | 0.101 | 0.60919 | -0.187 | 0.27383 | 0.413 |
| 18111 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-582-3p | 12 | CLIP1 | Sponge network | -0.154 | 0.53954 | -0.215 | 0.08684 | 0.413 |
| 18112 | HCG18 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p | 18 | KIF5B | Sponge network | 1.063 | 0 | 0.362 | 0.00066 | 0.413 |
| 18113 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-629-3p;hsa-miR-93-3p | 23 | EBF3 | Sponge network | -0.777 | 0.16667 | -0.058 | 0.81437 | 0.413 |
| 18114 | LINC00473 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -1.203 | 0.03881 | -0.704 | 0.00106 | 0.413 |
| 18115 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-324-3p;hsa-miR-576-5p;hsa-miR-98-5p | 15 | PBX1 | Sponge network | -1.116 | 0.23686 | -1.206 | 0 | 0.413 |
| 18116 | AP001347.6 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TAL1 | Sponge network | -1.161 | 0.00591 | -0.462 | 0.00574 | 0.413 |
| 18117 | MIR497HG |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-96-5p | 41 | CREB3L2 | Sponge network | -1.439 | 4.0E-5 | -0.525 | 2.0E-5 | 0.413 |
| 18118 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 44 | PRKAA2 | Sponge network | -0.342 | 0.02288 | -2.462 | 0 | 0.413 |
| 18119 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 27 | PGR | Sponge network | 0.199 | 0.38015 | -1.704 | 0 | 0.413 |
| 18120 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-511-5p | 20 | RBMS3 | Sponge network | 0.622 | 0.00015 | -0.519 | 0.03551 | 0.413 |
| 18121 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p | 21 | NR2C2 | Sponge network | -1.575 | 6.0E-5 | 0.21 | 0.03224 | 0.413 |
| 18122 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p | 11 | TSC22D1 | Sponge network | -0.793 | 7.0E-5 | -0.529 | 2.0E-5 | 0.413 |
| 18123 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 35 | EPHA7 | Sponge network | 0.214 | 0.18246 | -2.197 | 3.0E-5 | 0.413 |
| 18124 | CTC-429P9.5 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-93-3p | 15 | SNX29 | Sponge network | 0.178 | 0.29106 | -0.34 | 0.01371 | 0.413 |
| 18125 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-331-3p | 16 | ZBTB4 | Sponge network | 0.693 | 0.00076 | -0.739 | 0 | 0.413 |
| 18126 | CTD-2089N3.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p | 12 | FAT3 | Sponge network | -0.314 | 0.62033 | -1.601 | 0.00051 | 0.413 |
| 18127 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p | 12 | PLSCR4 | Sponge network | 2.306 | 9.0E-5 | -0.474 | 0.03833 | 0.413 |
| 18128 | RP11-35G9.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-96-5p | 22 | BCL2 | Sponge network | 0.622 | 0.00015 | -0.899 | 0 | 0.413 |
| 18129 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | CREBBP | Sponge network | -0.709 | 0.18168 | 0.126 | 0.15186 | 0.413 |
| 18130 | LINC00472 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 13 | CXCL12 | Sponge network | -0.842 | 0.01361 | -1.421 | 0 | 0.413 |
| 18131 | LINC00641 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 18 | MAPK10 | Sponge network | -0.222 | 0.32445 | -1.774 | 0 | 0.413 |
| 18132 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 18 | SCN9A | Sponge network | 0.381 | 0.25967 | -0.525 | 0.10593 | 0.413 |
| 18133 | RP11-244O19.1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 47 | PPM1L | Sponge network | -0.096 | 0.67958 | -0.537 | 0.00616 | 0.413 |
| 18134 | LINC00641 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-379-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | SH3GLB1 | Sponge network | -0.222 | 0.32445 | -0.115 | 0.14773 | 0.413 |
| 18135 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-148b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 11 | GLI3 | Sponge network | -0.465 | 0.04703 | -0.615 | 0.01482 | 0.413 |
| 18136 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p | 28 | RICTOR | Sponge network | 1.044 | 2.0E-5 | 0.388 | 0.00188 | 0.413 |
| 18137 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-7-5p | 18 | CRTC1 | Sponge network | -0.469 | 0.08721 | -0.116 | 0.28412 | 0.413 |
| 18138 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 24 | NR2C2 | Sponge network | 0.603 | 0.00127 | 0.21 | 0.03224 | 0.413 |
| 18139 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 51 | WDFY3 | Sponge network | -0.487 | 0.02157 | -0.201 | 0.0967 | 0.413 |
| 18140 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-429 | 10 | ROBO1 | Sponge network | -0.469 | 0.08721 | -0.009 | 0.96154 | 0.413 |
| 18141 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p | 53 | DTNA | Sponge network | 0.082 | 0.55654 | -1.648 | 1.0E-5 | 0.413 |
| 18142 | RP11-25K19.1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PCDH18 | Sponge network | -1.057 | 0.00029 | 0.216 | 0.24645 | 0.413 |
| 18143 | CKMT2-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p | 18 | WWTR1 | Sponge network | 0.082 | 0.55654 | -0.345 | 0.11997 | 0.413 |
| 18144 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-5p | 15 | SSBP2 | Sponge network | -1.255 | 0.00981 | -1.58 | 0 | 0.413 |
| 18145 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | EPAS1 | Sponge network | -1.203 | 0.03881 | -0.184 | 0.14418 | 0.413 |
| 18146 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-539-5p | 19 | KIF1B | Sponge network | 0.043 | 0.81628 | -0.118 | 0.32258 | 0.413 |
| 18147 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | TLE4 | Sponge network | -0.129 | 0.57177 | -1.157 | 0 | 0.413 |
| 18148 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p | 12 | ITGA5 | Sponge network | -0.739 | 0.00044 | 0.452 | 0.03524 | 0.413 |
| 18149 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | CAV1 | Sponge network | -1.203 | 0.03881 | -1.013 | 6.0E-5 | 0.413 |
| 18150 | RP11-25K19.1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 33 | AR | Sponge network | -1.057 | 0.00029 | -1.554 | 0 | 0.413 |
| 18151 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 26 | RICTOR | Sponge network | 0.986 | 0.01022 | 0.388 | 0.00188 | 0.413 |
| 18152 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 20 | EPHA3 | Sponge network | 0.659 | 3.0E-5 | 0.185 | 0.53588 | 0.413 |
| 18153 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-96-5p | 32 | FBXO32 | Sponge network | 0.72 | 0.0001 | -0.495 | 0.07561 | 0.413 |
| 18154 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 45 | TSHZ3 | Sponge network | -2.925 | 0 | -0.556 | 0.01516 | 0.413 |
| 18155 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p | 29 | FBXO32 | Sponge network | -0.777 | 0.16667 | -0.495 | 0.07561 | 0.413 |
| 18156 | RP11-121C2.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 16 | SLC5A3 | Sponge network | 1.147 | 0 | 0.493 | 5.0E-5 | 0.413 |
| 18157 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-362-3p | 12 | EPHA3 | Sponge network | -0.829 | 0.01343 | 0.185 | 0.53588 | 0.413 |
| 18158 | ADAMTS9-AS2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 30 | PDE4DIP | Sponge network | -2.452 | 0 | -0.282 | 0.0257 | 0.413 |
| 18159 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | LRP1B | Sponge network | 0.194 | 0.64278 | -2.163 | 0 | 0.413 |
| 18160 | GAS6-AS2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 19 | WWTR1 | Sponge network | 0.16 | 0.50785 | -0.345 | 0.11997 | 0.413 |
| 18161 | RP11-119F7.5 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | HERC1 | Sponge network | 0.225 | 0.30644 | -0.196 | 0.08544 | 0.413 |
| 18162 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-450b-5p | 13 | PNRC1 | Sponge network | -1.349 | 0.00017 | -0.646 | 0 | 0.413 |
| 18163 | PART1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 100 | LPP | Sponge network | -2.925 | 0 | -0.459 | 0.04475 | 0.413 |
| 18164 | CTD-3092A11.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | NIN | Sponge network | 0.873 | 0 | -0.253 | 0.13794 | 0.413 |
| 18165 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 18 | SLITRK6 | Sponge network | -1.575 | 6.0E-5 | -2.121 | 0 | 0.413 |
| 18166 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | ARHGAP29 | Sponge network | -0.096 | 0.67958 | 0.131 | 0.42953 | 0.413 |
| 18167 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-421;hsa-miR-590-3p | 18 | BCL6 | Sponge network | 0.214 | 0.18246 | -0.211 | 0.19489 | 0.413 |
| 18168 | CTD-2314G24.2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p | 12 | MACF1 | Sponge network | 0.246 | 0.59912 | 0.019 | 0.90335 | 0.413 |
| 18169 | CTD-2089N3.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 34 | LPP | Sponge network | -1.375 | 0.08642 | -0.459 | 0.04475 | 0.413 |
| 18170 | LINC00865 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-429 | 10 | THSD7A | Sponge network | -1.249 | 7.0E-5 | 0.242 | 0.25154 | 0.413 |
| 18171 | CTD-2089N3.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-3065-3p;hsa-miR-455-5p | 13 | BNC2 | Sponge network | -0.314 | 0.62033 | -0.491 | 0.09988 | 0.413 |
| 18172 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 25 | TACC1 | Sponge network | -1.886 | 0 | -0.362 | 0.05406 | 0.413 |
| 18173 | TPT1-AS1 |
hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-497-5p;hsa-miR-582-3p;hsa-miR-582-5p | 15 | BRD3 | Sponge network | 0.903 | 0 | 0.37 | 0.00013 | 0.413 |
| 18174 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TACC1 | Sponge network | -0.021 | 0.93598 | -0.362 | 0.05406 | 0.413 |
| 18175 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | NRXN3 | Sponge network | 0.219 | 0.1516 | -0.893 | 0.04186 | 0.413 |
| 18176 | RP5-1198O20.4 |
hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | GUCY1A2 | Sponge network | 0.997 | 0.00066 | 1.213 | 0 | 0.413 |
| 18177 | LINC00094 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p | 11 | INPP4A | Sponge network | 0.253 | 0.09565 | 0.07 | 0.53563 | 0.413 |
| 18178 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-940 | 33 | GAS7 | Sponge network | 0.16 | 0.50785 | -0.555 | 0.01602 | 0.413 |
| 18179 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-660-5p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | ZHX2 | Sponge network | 0.374 | 0.20054 | -0.192 | 0.18459 | 0.413 |
| 18180 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 15 | GPAM | Sponge network | 1.294 | 0 | 0.142 | 0.29732 | 0.413 |
| 18181 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | TLL1 | Sponge network | 0.532 | 0.00505 | -1.195 | 3.0E-5 | 0.413 |
| 18182 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p | 17 | PBX1 | Sponge network | -0.663 | 0.10887 | -1.206 | 0 | 0.413 |
| 18183 | AC127904.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | CREB1 | Sponge network | 1.308 | 0 | 0.203 | 0.00537 | 0.413 |
| 18184 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p | 20 | BCL6 | Sponge network | -0.563 | 0.02545 | -0.211 | 0.19489 | 0.413 |
| 18185 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 17 | CNTNAP3 | Sponge network | -0.738 | 0.21233 | -1.465 | 0.00033 | 0.413 |
| 18186 | DIO3OS |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-93-3p | 16 | WWTR1 | Sponge network | -0.773 | 0.04073 | -0.345 | 0.11997 | 0.413 |
| 18187 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-5p | 26 | TET1 | Sponge network | 0.253 | 0.09565 | -0.135 | 0.52138 | 0.413 |
| 18188 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CEP170 | Sponge network | 0.225 | 0.30644 | 0.943 | 0 | 0.413 |
| 18189 | RP11-620J15.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | CELF4 | Sponge network | -0.739 | 0.00044 | -1.135 | 0.00344 | 0.413 |
| 18190 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 12 | PTGFR | Sponge network | -1.057 | 0.00029 | -0.233 | 0.45421 | 0.413 |
| 18191 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 43 | EPHA7 | Sponge network | 0.374 | 0.20054 | -2.197 | 3.0E-5 | 0.413 |
| 18192 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | PRKAB2 | Sponge network | 0.439 | 0.00313 | -0.367 | 0.01371 | 0.413 |
| 18193 | CTA-204B4.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-590-3p | 17 | SIRT1 | Sponge network | 0.765 | 7.0E-5 | 0.109 | 0.2593 | 0.413 |
| 18194 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3613-5p | 26 | DMD | Sponge network | 0.259 | 0.12542 | -1.506 | 0 | 0.413 |
| 18195 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 15 | FYN | Sponge network | -1.697 | 0.00889 | -0.131 | 0.37051 | 0.413 |
| 18196 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SON | Sponge network | 0.559 | 0.00026 | 0.168 | 0.04406 | 0.413 |
| 18197 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | 1.205 | 0 | 0.206 | 0.06751 | 0.413 |
| 18198 | ZNF582-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 13 | CDH23 | Sponge network | -1.349 | 0.00017 | -0.974 | 0.00034 | 0.413 |
| 18199 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-26b-5p | 11 | DNAJB4 | Sponge network | -0.777 | 0.16667 | -0.7 | 1.0E-5 | 0.413 |
| 18200 | RP11-53O19.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 20 | NRK | Sponge network | -0.405 | 0.22013 | 0.656 | 0.19217 | 0.413 |
| 18201 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p;hsa-miR-96-5p | 11 | ZYX | Sponge network | -0.976 | 0.00025 | 0.019 | 0.89763 | 0.413 |
| 18202 | RP4-605O3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-411-5p;hsa-miR-590-3p | 12 | USP6 | Sponge network | 0.262 | 0.0475 | 0.188 | 0.3871 | 0.413 |
| 18203 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 16 | IGSF10 | Sponge network | -0.246 | 0.35034 | -1.751 | 1.0E-5 | 0.413 |
| 18204 | RP1-122P22.2 |
hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 10 | RNF144A | Sponge network | 0.065 | 0.73468 | 0.594 | 0.0009 | 0.413 |
| 18205 | SNHG12 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 16 | TBL1XR1 | Sponge network | 1.103 | 0 | 0.578 | 0 | 0.413 |
| 18206 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 74 | KIF1B | Sponge network | 0.421 | 0.00084 | -0.118 | 0.32258 | 0.413 |
| 18207 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 17 | MYO9A | Sponge network | 0.826 | 1.0E-5 | -0.147 | 0.32373 | 0.413 |
| 18208 | MIR497HG |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 18 | ZFP36L1 | Sponge network | -1.439 | 4.0E-5 | -0.505 | 8.0E-5 | 0.413 |
| 18209 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-655-3p | 14 | NF1 | Sponge network | 0.614 | 1.0E-5 | 0.51 | 0 | 0.413 |
| 18210 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p | 16 | EGLN1 | Sponge network | 0.542 | 0.00054 | -0.127 | 0.1536 | 0.413 |
| 18211 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-935 | 16 | ARNT2 | Sponge network | -0.246 | 0.35034 | -0.332 | 0.25851 | 0.413 |
| 18212 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | CAV1 | Sponge network | -0.08 | 0.90092 | -1.013 | 6.0E-5 | 0.413 |
| 18213 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | CNTNAP3 | Sponge network | 0.381 | 0.25967 | -1.465 | 0.00033 | 0.413 |
| 18214 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-766-3p | 41 | MAPK1 | Sponge network | 0.421 | 0.00084 | -0.017 | 0.81465 | 0.413 |
| 18215 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-590-5p | 20 | YAP1 | Sponge network | -0.384 | 0.40652 | 0.22 | 0.11836 | 0.413 |
| 18216 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3614-5p | 10 | NUAK1 | Sponge network | -0.693 | 0.05629 | 0.904 | 0 | 0.413 |
| 18217 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-93-5p | 29 | TSHZ3 | Sponge network | -0.18 | 0.20343 | -0.556 | 0.01516 | 0.413 |
| 18218 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 18 | CDH23 | Sponge network | 0.572 | 0.01722 | -0.974 | 0.00034 | 0.413 |
| 18219 | TP73-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-660-5p;hsa-miR-7-1-3p | 26 | EYA4 | Sponge network | -0.563 | 0.02545 | 0.3 | 0.36876 | 0.413 |
| 18220 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-370-3p;hsa-miR-93-3p | 15 | RPS6KA2 | Sponge network | -0.896 | 0.00099 | -0.57 | 0.00013 | 0.413 |
| 18221 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | IGSF10 | Sponge network | -1.203 | 0.03881 | -1.751 | 1.0E-5 | 0.413 |
| 18222 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 20 | NF1 | Sponge network | 1.03 | 0 | 0.51 | 0 | 0.413 |
| 18223 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 44 | DTNA | Sponge network | 0.214 | 0.18246 | -1.648 | 1.0E-5 | 0.413 |
| 18224 | DIO3OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-7-5p | 18 | RNF144A | Sponge network | -0.773 | 0.04073 | 0.594 | 0.0009 | 0.413 |
| 18225 | PWAR6 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RBM6 | Sponge network | -1.575 | 6.0E-5 | -0.137 | 0.15663 | 0.413 |
| 18226 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-940 | 15 | THBS1 | Sponge network | -1.255 | 0.00981 | 0.741 | 0.00386 | 0.413 |
| 18227 | NPSR1-AS1 |
hsa-miR-125a-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-664a-5p | 10 | CBFB | Sponge network | 3.95 | 0 | 1.061 | 0 | 0.413 |
| 18228 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | HDAC9 | Sponge network | -0.49 | 0.04946 | -0.755 | 0.00495 | 0.413 |
| 18229 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | TMEFF2 | Sponge network | -0.612 | 0.00094 | -2.426 | 0 | 0.413 |
| 18230 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p | 12 | DNAJB4 | Sponge network | -0.154 | 0.78939 | -0.7 | 1.0E-5 | 0.413 |
| 18231 | RP11-1277A3.1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-940;hsa-miR-96-5p | 12 | MYO9A | Sponge network | 0.453 | 0.01839 | -0.147 | 0.32373 | 0.413 |
| 18232 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p | 13 | MLLT10 | Sponge network | 0.93 | 0 | 0.105 | 0.33292 | 0.413 |
| 18233 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 16 | TCF4 | Sponge network | -0.733 | 0.19469 | 0.016 | 0.92271 | 0.413 |
| 18234 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-7-1-3p | 12 | DKK3 | Sponge network | -0.08 | 0.90092 | -0.052 | 0.77783 | 0.413 |
| 18235 | CTD-2647L4.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-590-5p | 11 | MYO9A | Sponge network | 0.555 | 0.00197 | -0.147 | 0.32373 | 0.413 |
| 18236 | RP11-384O8.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-671-5p | 12 | CDH23 | Sponge network | -0.738 | 0.21233 | -0.974 | 0.00034 | 0.413 |
| 18237 | RP5-890O3.9 |
hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p | 14 | MDM4 | Sponge network | 0.842 | 0 | 0.345 | 0.00762 | 0.413 |
| 18238 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-92a-3p | 17 | RUNX1T1 | Sponge network | 0.959 | 0 | -0.344 | 0.2374 | 0.413 |
| 18239 | CTA-363E19.2 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-495-3p | 11 | MYO9A | Sponge network | 1.055 | 0 | -0.147 | 0.32373 | 0.413 |
| 18240 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-582-5p | 15 | MOB1B | Sponge network | 0.972 | 0 | 0.197 | 0.15266 | 0.413 |
| 18241 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 37 | EPHA5 | Sponge network | -0.896 | 0.00099 | -0.957 | 0.01733 | 0.413 |
| 18242 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-370-3p;hsa-miR-455-5p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-93-5p | 29 | JAZF1 | Sponge network | -0.465 | 0.04703 | -0.377 | 0.02677 | 0.413 |
| 18243 | RP11-244O19.1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 10 | TAF1 | Sponge network | -0.096 | 0.67958 | 0.39 | 3.0E-5 | 0.412 |
| 18244 | RASSF8-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-92a-1-5p | 38 | GRIK3 | Sponge network | 0.335 | 0.20596 | -3.208 | 0 | 0.412 |
| 18245 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 30 | MITF | Sponge network | -0.222 | 0.32445 | -0.769 | 0.00022 | 0.412 |
| 18246 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 28 | LIFR | Sponge network | 0.335 | 0.20596 | -1.794 | 0 | 0.412 |
| 18247 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 11 | NRXN3 | Sponge network | -0.737 | 0.10822 | -0.893 | 0.04186 | 0.412 |
| 18248 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p | 10 | PRKAB2 | Sponge network | 0.497 | 0.01451 | -0.367 | 0.01371 | 0.412 |
| 18249 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 23 | MOB1B | Sponge network | 0.817 | 3.0E-5 | 0.197 | 0.15266 | 0.412 |
| 18250 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-375;hsa-miR-452-5p;hsa-miR-590-3p | 22 | ABI2 | Sponge network | -0.358 | 0.17255 | 0.175 | 0.14787 | 0.412 |
| 18251 | RAMP2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | NAV3 | Sponge network | -0.513 | 0.04703 | 0.212 | 0.47729 | 0.412 |
| 18252 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 30 | DIP2C | Sponge network | 0.41 | 0.02214 | -0.436 | 0.01713 | 0.412 |
| 18253 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 54 | TCF4 | Sponge network | -1.203 | 0.03881 | 0.016 | 0.92271 | 0.412 |
| 18254 | RP11-159D12.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p | 29 | ZFX | Sponge network | 1.254 | 0 | 0.09 | 0.42563 | 0.412 |
| 18255 | LINC00473 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 25 | LRIG1 | Sponge network | -1.203 | 0.03881 | -0.537 | 0.00588 | 0.412 |
| 18256 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-576-5p | 29 | ERC1 | Sponge network | -0.738 | 0.21233 | -0.108 | 0.38794 | 0.412 |
| 18257 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-34a-5p | 10 | NAV3 | Sponge network | 0.598 | 0.38481 | 0.212 | 0.47729 | 0.412 |
| 18258 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-5p | 17 | IGF2 | Sponge network | -0.576 | 0.10121 | 0.848 | 0.03842 | 0.412 |
| 18259 | RP11-736K20.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | TRPS1 | Sponge network | -0.358 | 0.17255 | -0.191 | 0.44109 | 0.412 |
| 18260 | RP11-1006G14.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | CREB1 | Sponge network | 0.559 | 0.00026 | 0.203 | 0.00537 | 0.412 |
| 18261 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 24 | CRTC3 | Sponge network | 0.219 | 0.1516 | 0.164 | 0.16786 | 0.412 |
| 18262 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-589-5p | 19 | GRIA3 | Sponge network | -4.546 | 0 | -1.956 | 0 | 0.412 |
| 18263 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | SLITRK6 | Sponge network | -1.886 | 0 | -2.121 | 0 | 0.412 |
| 18264 | C20orf166-AS1 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-582-5p | 50 | THRB | Sponge network | -2.69 | 0.0001 | -1.117 | 0.0004 | 0.412 |
| 18265 | DNAJC27-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-93-5p | 19 | EZH1 | Sponge network | -0.969 | 2.0E-5 | -0.564 | 1.0E-5 | 0.412 |
| 18266 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CDH10 | Sponge network | -0.942 | 0 | -2.397 | 0 | 0.412 |
| 18267 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | NF1 | Sponge network | 0.401 | 0.00066 | 0.51 | 0 | 0.412 |
| 18268 | CTD-2647L4.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-337-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-5p | 16 | BMPR2 | Sponge network | 0.555 | 0.00197 | 0.206 | 0.0635 | 0.412 |
| 18269 | AGAP11 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-576-5p | 11 | EHD3 | Sponge network | -1.645 | 0 | -0.484 | 0.01747 | 0.412 |
| 18270 | RP11-686D22.8 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-96-5p | 30 | BMPR2 | Sponge network | 0.898 | 1.0E-5 | 0.206 | 0.0635 | 0.412 |
| 18271 | RP11-379F4.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | PHC3 | Sponge network | 1.316 | 0 | 0.212 | 0.0499 | 0.412 |
| 18272 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 19 | TGFBR2 | Sponge network | -0.901 | 0.00019 | -0.243 | 0.11408 | 0.412 |
| 18273 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | GRIN2A | Sponge network | 0.335 | 0.20596 | -2.161 | 0 | 0.412 |
| 18274 | RP11-123M6.2 |
hsa-miR-103a-2-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-378c;hsa-miR-7-1-3p;hsa-miR-744-3p | 10 | LPP | Sponge network | 0.543 | 0.12356 | -0.459 | 0.04475 | 0.412 |
| 18275 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-589-5p;hsa-miR-96-5p | 19 | ARHGAP29 | Sponge network | -0.976 | 0.00025 | 0.131 | 0.42953 | 0.412 |
| 18276 | HCG11 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 26 | DLC1 | Sponge network | 0.385 | 0.10067 | -0.382 | 0.05038 | 0.412 |
| 18277 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -0.342 | 0.02288 | -0.7 | 1.0E-5 | 0.412 |
| 18278 | LINC00957 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-338-3p | 12 | ELMO1 | Sponge network | -0.421 | 0.0114 | 0.04 | 0.83147 | 0.412 |
| 18279 | C1RL-AS1 |
hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-378a-5p | 10 | CREB1 | Sponge network | 1.26 | 0 | 0.203 | 0.00537 | 0.412 |
| 18280 | LINC00476 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 15 | STAT5B | Sponge network | -0.615 | 0.00015 | -0.182 | 0.1082 | 0.412 |
| 18281 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | MAF | Sponge network | -1.575 | 6.0E-5 | -1.257 | 0 | 0.412 |
| 18282 | CTD-3092A11.2 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | UBR3 | Sponge network | 0.873 | 0 | -0.021 | 0.82774 | 0.412 |
| 18283 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 40 | BNC2 | Sponge network | -0.899 | 6.0E-5 | -0.491 | 0.09988 | 0.412 |
| 18284 | CTC-429P9.3 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-940 | 10 | MAPK10 | Sponge network | 0.082 | 0.55397 | -1.774 | 0 | 0.412 |
| 18285 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-671-5p | 14 | CYLD | Sponge network | -1.331 | 3.0E-5 | -0.367 | 0.00171 | 0.412 |
| 18286 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 32 | TRPS1 | Sponge network | -0.318 | 0.65847 | -0.191 | 0.44109 | 0.412 |
| 18287 | CTC-559E9.5 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-539-5p | 11 | PHF3 | Sponge network | 0.594 | 0.00064 | 0.126 | 0.19652 | 0.412 |
| 18288 | RP5-1074L1.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-486-5p | 13 | CBFB | Sponge network | 1.923 | 0 | 1.061 | 0 | 0.412 |
| 18289 | TPTEP1 |
hsa-miR-185-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-542-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-940 | 10 | ILK | Sponge network | -1.216 | 0 | -0.74 | 0 | 0.412 |
| 18290 | RP11-705C15.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | MDM4 | Sponge network | 0.796 | 4.0E-5 | 0.345 | 0.00762 | 0.412 |
| 18291 | HCG11 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | ATM | Sponge network | 0.385 | 0.10067 | 0.178 | 0.21568 | 0.412 |
| 18292 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p | 18 | CHD5 | Sponge network | -0.693 | 0.05629 | -0.922 | 0.01521 | 0.412 |
| 18293 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | EYA4 | Sponge network | 0.395 | 0.15451 | 0.3 | 0.36876 | 0.412 |
| 18294 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p | 14 | ABCC9 | Sponge network | -0.303 | 0.21002 | -0.466 | 0.14121 | 0.412 |
| 18295 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | NCOA1 | Sponge network | 0.194 | 0.64278 | -0.432 | 0 | 0.412 |
| 18296 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | EBF1 | Sponge network | 0.174 | 0.30034 | -0.384 | 0.08856 | 0.412 |
| 18297 | RP11-401P9.4 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-361-3p;hsa-miR-454-3p | 16 | ITPKB | Sponge network | -0.384 | 0.40652 | -0.704 | 0.00106 | 0.412 |
| 18298 | LINC00578 |
hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-577 | 12 | DOCK4 | Sponge network | 0.811 | 0.03228 | 0.502 | 0.00108 | 0.412 |
| 18299 | DNM1P35 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | GALNT13 | Sponge network | 0.051 | 0.83388 | -0.857 | 0.07439 | 0.412 |
| 18300 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-429 | 14 | ARHGAP28 | Sponge network | -0.469 | 0.08721 | -0.911 | 0.00013 | 0.412 |
| 18301 | AC003090.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-940;hsa-miR-96-5p | 32 | SNX29 | Sponge network | -2.117 | 0.00161 | -0.34 | 0.01371 | 0.412 |
| 18302 | GAS5 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p | 11 | PAFAH1B2 | Sponge network | 0.728 | 0 | 0.318 | 0.0001 | 0.412 |
| 18303 | AC016747.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-769-5p | 17 | SETBP1 | Sponge network | 0.199 | 0.38015 | -1.302 | 1.0E-5 | 0.412 |
| 18304 | ZNF582-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-5p | 12 | FMNL3 | Sponge network | -1.349 | 0.00017 | 0.555 | 4.0E-5 | 0.412 |
| 18305 | BVES-AS1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-576-5p | 22 | MED12L | Sponge network | -2.62 | 0 | -0.194 | 0.55299 | 0.412 |
| 18306 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 30 | JAZF1 | Sponge network | 0.4 | 0.14611 | -0.377 | 0.02677 | 0.412 |
| 18307 | RP11-67L2.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | PDE4DIP | Sponge network | 0.72 | 0.0001 | -0.282 | 0.0257 | 0.412 |
| 18308 | SERTAD4-AS1 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 16 | PPP1R9A | Sponge network | -0.932 | 0.00329 | -0.903 | 0.04254 | 0.412 |
| 18309 | GNG12-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p | 30 | BTG2 | Sponge network | -0.793 | 7.0E-5 | -1.763 | 0 | 0.412 |
| 18310 | LINC00969 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-625-5p | 24 | ZMYM2 | Sponge network | 0.683 | 1.0E-5 | 0.279 | 0.01273 | 0.412 |
| 18311 | RAMP2-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 15 | MRVI1 | Sponge network | -0.513 | 0.04703 | -1.051 | 0.00022 | 0.412 |
| 18312 | HOXA-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 28 | RASSF8 | Sponge network | 0.61 | 0.01593 | -0.122 | 0.65591 | 0.412 |
| 18313 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-582-5p | 24 | FBXO32 | Sponge network | 0.389 | 0.04293 | -0.495 | 0.07561 | 0.412 |
| 18314 | ZRANB2-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-942-5p | 16 | OPCML | Sponge network | -0.376 | 0.00337 | -2.498 | 0 | 0.412 |
| 18315 | RP11-890B15.3 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 14 | SHPRH | Sponge network | 0.101 | 0.60919 | 0.098 | 0.40994 | 0.412 |
| 18316 | MIR497HG |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 11 | KCNAB1 | Sponge network | -1.439 | 4.0E-5 | -0.755 | 2.0E-5 | 0.412 |
| 18317 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-576-5p | 11 | PHC3 | Sponge network | -2.039 | 0.03447 | 0.212 | 0.0499 | 0.412 |
| 18318 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 19 | RASSF2 | Sponge network | -2.531 | 0 | -0.273 | 0.21961 | 0.412 |
| 18319 | LINC00957 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | NRK | Sponge network | -0.421 | 0.0114 | 0.656 | 0.19217 | 0.412 |
| 18320 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-542-3p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | 0.194 | 0.64278 | -0.085 | 0.42389 | 0.412 |
| 18321 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-708-3p;hsa-miR-92a-1-5p | 18 | PRKAB2 | Sponge network | 0.001 | 0.99753 | -0.367 | 0.01371 | 0.412 |
| 18322 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | MDM4 | Sponge network | 0.673 | 0 | 0.345 | 0.00762 | 0.412 |
| 18323 | RP1-102E24.8 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p | 10 | ABL2 | Sponge network | 1.737 | 0 | 0.82 | 0 | 0.412 |
| 18324 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | DYNC1I1 | Sponge network | 0.219 | 0.1516 | -1.12 | 0.00107 | 0.412 |
| 18325 | LINC00476 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 30 | DLC1 | Sponge network | -0.615 | 0.00015 | -0.382 | 0.05038 | 0.412 |
| 18326 | RP11-254F19.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 11 | CADM2 | Sponge network | -1.428 | 0.07715 | -3.782 | 0 | 0.412 |
| 18327 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 42 | CREB3L2 | Sponge network | -0.563 | 0.02545 | -0.525 | 2.0E-5 | 0.412 |
| 18328 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | MLLT10 | Sponge network | 1.11 | 0 | 0.105 | 0.33292 | 0.412 |
| 18329 | CTD-2314G24.2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | RYR2 | Sponge network | 0.246 | 0.59912 | -1.466 | 0.00031 | 0.412 |
| 18330 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | NEDD4 | Sponge network | -1.249 | 7.0E-5 | 0.02 | 0.89834 | 0.412 |
| 18331 | RP11-13K12.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 24 | DIP2C | Sponge network | -0.318 | 0.65847 | -0.436 | 0.01713 | 0.412 |
| 18332 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-7-1-3p | 18 | KAT6B | Sponge network | -1.092 | 4.0E-5 | -0.478 | 0.00018 | 0.412 |
| 18333 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-493-5p | 12 | CACNA2D1 | Sponge network | -2.095 | 0.00112 | -0.846 | 0.00675 | 0.412 |
| 18334 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 21 | FGFR1 | Sponge network | -2.925 | 0 | -0.509 | 0.03957 | 0.412 |
| 18335 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FSTL5 | Sponge network | 0.572 | 0.01722 | -1.3 | 0.01619 | 0.412 |
| 18336 | PCA3 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | ZDBF2 | Sponge network | -0.709 | 0.18168 | -0.998 | 0.00025 | 0.412 |
| 18337 | NR2F1-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | LUZP2 | Sponge network | -0.49 | 0.04946 | -0.056 | 0.89416 | 0.412 |
| 18338 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -0.096 | 0.67958 | -0.243 | 0.11408 | 0.412 |
| 18339 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-582-5p | 21 | RICTOR | Sponge network | 1.206 | 0 | 0.388 | 0.00188 | 0.412 |
| 18340 | RP11-774O3.3 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p | 33 | IGFBP5 | Sponge network | -0.487 | 0.02157 | -0.806 | 0.00105 | 0.412 |
| 18341 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378c;hsa-miR-501-5p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | 0.395 | 0.15451 | 0.783 | 0.00072 | 0.412 |
| 18342 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 14 | PDS5B | Sponge network | 1.308 | 0 | 0.259 | 0.00962 | 0.412 |
| 18343 | CTA-14H9.5 |
hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-582-5p | 18 | RUNX1T1 | Sponge network | 0.145 | 0.53343 | -0.344 | 0.2374 | 0.412 |
| 18344 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-324-3p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | USP12 | Sponge network | 0.194 | 0.64278 | -0.102 | 0.40768 | 0.412 |
| 18345 | SERTAD4-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-30d-3p;hsa-miR-335-3p | 12 | STARD13 | Sponge network | -0.932 | 0.00329 | -0.187 | 0.27383 | 0.412 |
| 18346 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | MCC | Sponge network | -0.513 | 0.04703 | -1.005 | 0 | 0.412 |
| 18347 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 20 | RET | Sponge network | 0.246 | 0.59912 | -0.833 | 0.03517 | 0.412 |
| 18348 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | PBRM1 | Sponge network | 0.401 | 0.00066 | -0.029 | 0.78601 | 0.412 |
| 18349 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p | 13 | EDA2R | Sponge network | -2.62 | 0 | -0.587 | 0.01598 | 0.412 |
| 18350 | RP11-536O18.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-502-5p | 14 | TCF4 | Sponge network | -0.074 | 0.90758 | 0.016 | 0.92271 | 0.412 |
| 18351 | MEG3 |
hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-455-5p | 12 | PRDM5 | Sponge network | 0.249 | 0.35902 | -1.09 | 0.00014 | 0.412 |
| 18352 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | IGSF10 | Sponge network | 0.249 | 0.35902 | -1.751 | 1.0E-5 | 0.412 |
| 18353 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-577 | 12 | PRKAA2 | Sponge network | 0.357 | 0.72407 | -2.462 | 0 | 0.412 |
| 18354 | CKMT2-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | MRVI1 | Sponge network | 0.082 | 0.55654 | -1.051 | 0.00022 | 0.412 |
| 18355 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p | 30 | RICTOR | Sponge network | 1.317 | 0 | 0.388 | 0.00188 | 0.412 |
| 18356 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-942-5p | 40 | UNC80 | Sponge network | 0.246 | 0.59912 | -1.934 | 0 | 0.412 |
| 18357 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-505-5p | 16 | TBL1XR1 | Sponge network | 0.813 | 1.0E-5 | 0.578 | 0 | 0.412 |
| 18358 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 41 | BCL2 | Sponge network | -0.769 | 0.00035 | -0.899 | 0 | 0.412 |
| 18359 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 1.597 | 0 | 0.578 | 0 | 0.412 |
| 18360 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-577;hsa-miR-589-5p | 31 | SOX5 | Sponge network | -0.465 | 0.04703 | -1.091 | 4.0E-5 | 0.412 |
| 18361 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 18 | AKAP11 | Sponge network | 0.959 | 0 | 0.196 | 0.09825 | 0.412 |
| 18362 | RP11-244H3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CEP170 | Sponge network | 0.659 | 3.0E-5 | 0.943 | 0 | 0.412 |
| 18363 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 54 | FOXP1 | Sponge network | -1.697 | 0.00889 | 0.042 | 0.74368 | 0.412 |
| 18364 | KB-1410C5.5 |
hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 10 | MOB1B | Sponge network | 0.959 | 0 | 0.197 | 0.15266 | 0.412 |
| 18365 | FZD10-AS1 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | CDKN1C | Sponge network | -0.727 | 0.04726 | -0.929 | 0.00033 | 0.412 |
| 18366 | LINC00473 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -1.203 | 0.03881 | -1.692 | 0 | 0.412 |
| 18367 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 39 | ERC1 | Sponge network | -0.49 | 0.04946 | -0.108 | 0.38794 | 0.412 |
| 18368 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 12 | ACTB | Sponge network | -0.612 | 0.00094 | -0.247 | 0.01736 | 0.412 |
| 18369 | SH3RF3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 27 | TRPS1 | Sponge network | 0.889 | 9.0E-5 | -0.191 | 0.44109 | 0.412 |
| 18370 | CTC-429P9.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-335-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-766-3p;hsa-miR-940 | 18 | AFF4 | Sponge network | 0.043 | 0.81628 | -0.266 | 0.01565 | 0.412 |
| 18371 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | AKAP11 | Sponge network | 0.064 | 0.72934 | 0.196 | 0.09825 | 0.412 |
| 18372 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-374b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | NUAK1 | Sponge network | 0.532 | 0.00505 | 0.904 | 0 | 0.412 |
| 18373 | FGF14-AS2 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | TAL1 | Sponge network | -1.582 | 8.0E-5 | -0.462 | 0.00574 | 0.412 |
| 18374 | RP11-21A7A.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-576-5p | 12 | ZFHX4 | Sponge network | -1.116 | 0.23686 | 0.018 | 0.95574 | 0.412 |
| 18375 | RP11-384K6.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-539-5p | 16 | ABI2 | Sponge network | 0.946 | 0 | 0.175 | 0.14787 | 0.412 |
| 18376 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | EDA2R | Sponge network | -2.46 | 0 | -0.587 | 0.01598 | 0.412 |
| 18377 | LINC01018 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DCN | Sponge network | -2.915 | 0.00015 | -1.288 | 0 | 0.412 |
| 18378 | RP5-1139B12.3 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-3p | 25 | IL6ST | Sponge network | 0.598 | 0.38481 | -0.402 | 0.00676 | 0.412 |
| 18379 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-3614-5p;hsa-miR-539-5p | 14 | MOB1B | Sponge network | 0.793 | 0 | 0.197 | 0.15266 | 0.412 |
| 18380 | MIR143HG |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 34 | EYA4 | Sponge network | -0.739 | 0.0574 | 0.3 | 0.36876 | 0.412 |
| 18381 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-874-3p;hsa-miR-877-5p | 20 | CRTC3 | Sponge network | -0.842 | 0.01361 | 0.164 | 0.16786 | 0.412 |
| 18382 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | MCC | Sponge network | -0.222 | 0.32445 | -1.005 | 0 | 0.412 |
| 18383 | RP11-46C24.7 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 20 | ZMYND11 | Sponge network | 0.392 | 0.0278 | -0.2 | 0.05449 | 0.412 |
| 18384 | BOLA3-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-1-3p | 15 | ITPKB | Sponge network | -0.246 | 0.35034 | -0.704 | 0.00106 | 0.412 |
| 18385 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p | 11 | CHD2 | Sponge network | 0.856 | 0 | 0.049 | 0.55371 | 0.412 |
| 18386 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 19 | RBL2 | Sponge network | -0.739 | 0.0574 | -0.282 | 0.00254 | 0.412 |
| 18387 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | NRXN1 | Sponge network | -0.737 | 0.10822 | -3.778 | 0 | 0.412 |
| 18388 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-455-3p | 13 | DKK3 | Sponge network | -0.773 | 0.04073 | -0.052 | 0.77783 | 0.412 |
| 18389 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-940 | 23 | CRTC3 | Sponge network | -0.465 | 0.04703 | 0.164 | 0.16786 | 0.412 |
| 18390 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 28 | MCC | Sponge network | -1.057 | 0.00029 | -1.005 | 0 | 0.412 |
| 18391 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940 | 22 | CRTC3 | Sponge network | -0.323 | 0.01372 | 0.164 | 0.16786 | 0.412 |
| 18392 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | 0.494 | 0.00339 | 0.323 | 0.00792 | 0.412 |
| 18393 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | TET1 | Sponge network | 0.194 | 0.64278 | -0.135 | 0.52138 | 0.412 |
| 18394 | RP11-890B15.3 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-629-5p;hsa-miR-96-5p | 13 | WNK2 | Sponge network | 0.101 | 0.60919 | -0.352 | 0.31066 | 0.412 |
| 18395 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p | 11 | ITGA5 | Sponge network | 0.381 | 0.25967 | 0.452 | 0.03524 | 0.412 |
| 18396 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 30 | NCOA1 | Sponge network | -0.323 | 0.01372 | -0.432 | 0 | 0.412 |
| 18397 | FAM157C |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326 | 10 | NF1 | Sponge network | 0.91 | 0 | 0.51 | 0 | 0.412 |
| 18398 | LINC00702 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ZMYND11 | Sponge network | -0.737 | 0.06182 | -0.2 | 0.05449 | 0.412 |
| 18399 | RP11-750H9.5 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-200c-5p | 10 | MEF2C | Sponge network | 0.349 | 0.13152 | -0.499 | 0.01448 | 0.412 |
| 18400 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 11 | PDGFC | Sponge network | -0.563 | 0.02545 | -0.33 | 0.06483 | 0.412 |
| 18401 | EMX2OS |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | ROBO1 | Sponge network | -0.777 | 0.1134 | -0.009 | 0.96154 | 0.412 |
| 18402 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-940 | 24 | HLF | Sponge network | 0.225 | 0.30644 | -2.443 | 0 | 0.412 |
| 18403 | HAND2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p | 12 | ALDH1A2 | Sponge network | -1.697 | 0.00889 | -2.411 | 0 | 0.412 |
| 18404 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-500a-5p;hsa-miR-589-5p;hsa-miR-98-5p | 11 | ZFHX3 | Sponge network | -1.116 | 0.23686 | 0.389 | 0.01363 | 0.412 |
| 18405 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | GRIN2A | Sponge network | -0.318 | 0.65847 | -2.161 | 0 | 0.411 |
| 18406 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-550a-5p;hsa-miR-590-3p | 14 | HOOK3 | Sponge network | -0.053 | 0.80685 | 0.273 | 0.09957 | 0.411 |
| 18407 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-484 | 11 | PPM1A | Sponge network | -0.691 | 0.00146 | -0.623 | 0 | 0.411 |
| 18408 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 26 | FBXO32 | Sponge network | 0.214 | 0.18246 | -0.495 | 0.07561 | 0.411 |
| 18409 | RP11-981G7.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378c;hsa-miR-590-3p | 25 | ABL2 | Sponge network | 0.986 | 0.01022 | 0.82 | 0 | 0.411 |
| 18410 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p | 29 | CHD5 | Sponge network | 0.176 | 0.37402 | -0.922 | 0.01521 | 0.411 |
| 18411 | PSMG3-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-3p | 14 | FMNL3 | Sponge network | -0.125 | 0.49414 | 0.555 | 4.0E-5 | 0.411 |
| 18412 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | -0.738 | 0.21233 | 0.809 | 0.00544 | 0.411 |
| 18413 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 36 | CCDC6 | Sponge network | 0.683 | 1.0E-5 | 0.27 | 0.02555 | 0.411 |
| 18414 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-493-5p;hsa-miR-590-3p | 13 | MYCBP2 | Sponge network | 0.873 | 0 | -0.027 | 0.82016 | 0.411 |
| 18415 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | PDGFC | Sponge network | -0.487 | 0.02157 | -0.33 | 0.06483 | 0.411 |
| 18416 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-935 | 10 | DMD | Sponge network | -1.483 | 9.0E-5 | -1.506 | 0 | 0.411 |
| 18417 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | PBRM1 | Sponge network | 0.75 | 0.00054 | -0.029 | 0.78601 | 0.411 |
| 18418 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 51 | SSBP2 | Sponge network | -1.047 | 0 | -1.58 | 0 | 0.411 |
| 18419 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | PPM1A | Sponge network | -0.487 | 0.02157 | -0.623 | 0 | 0.411 |
| 18420 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p | 27 | FBXO32 | Sponge network | 0.174 | 0.30034 | -0.495 | 0.07561 | 0.411 |
| 18421 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 16 | FAT4 | Sponge network | 1.302 | 7.0E-5 | -0.354 | 0.14694 | 0.411 |
| 18422 | AC006116.24 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-577 | 26 | BCL2 | Sponge network | -0.473 | 0.07602 | -0.899 | 0 | 0.411 |
| 18423 | RP5-994D16.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CREB1 | Sponge network | 0.252 | 0.08508 | 0.203 | 0.00537 | 0.411 |
| 18424 | MEG3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 12 | KCNAB1 | Sponge network | 0.249 | 0.35902 | -0.755 | 2.0E-5 | 0.411 |
| 18425 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 10 | DACT1 | Sponge network | -0.896 | 0.00099 | 0.783 | 0.00072 | 0.411 |
| 18426 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p | 22 | ZMYND11 | Sponge network | 0.082 | 0.55654 | -0.2 | 0.05449 | 0.411 |
| 18427 | RP11-158K1.3 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 12 | TAF1 | Sponge network | 0.349 | 0.01695 | 0.39 | 3.0E-5 | 0.411 |
| 18428 | OIP5-AS1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-5p;hsa-miR-495-3p | 16 | STAG2 | Sponge network | 0.421 | 0.00084 | 0.252 | 0.00438 | 0.411 |
| 18429 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 29 | DIP2C | Sponge network | 0.225 | 0.30644 | -0.436 | 0.01713 | 0.411 |
| 18430 | RP11-822E23.8 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-7-5p | 22 | PRKCB | Sponge network | -0.777 | 0.16667 | -1.313 | 2.0E-5 | 0.411 |
| 18431 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-942-5p | 28 | TMEM132B | Sponge network | -0.693 | 0.05629 | -0.83 | 0.01084 | 0.411 |
| 18432 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 26 | ADAMTSL3 | Sponge network | 0.064 | 0.72934 | -1.559 | 0 | 0.411 |
| 18433 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p | 11 | FSTL5 | Sponge network | -0.405 | 0.22013 | -1.3 | 0.01619 | 0.411 |
| 18434 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TIMP3 | Sponge network | -0.323 | 0.01372 | -0.546 | 0.02307 | 0.411 |
| 18435 | MIR22HG |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | -1.047 | 0 | -0.382 | 0.05038 | 0.411 |
| 18436 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 18 | NRXN3 | Sponge network | 0.75 | 0.00054 | -0.893 | 0.04186 | 0.411 |
| 18437 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 36 | ESR1 | Sponge network | 0.997 | 0.00066 | -1.035 | 3.0E-5 | 0.411 |
| 18438 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-96-5p | 16 | CRTC3 | Sponge network | -1.654 | 0.00144 | 0.164 | 0.16786 | 0.411 |
| 18439 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-935 | 11 | RELN | Sponge network | -0.246 | 0.35034 | -2.403 | 0 | 0.411 |
| 18440 | RP11-4O1.2 |
hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-9-5p | 10 | CDK6 | Sponge network | 1.302 | 0 | 0.991 | 0 | 0.411 |
| 18441 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 23 | PRKAB2 | Sponge network | 0.331 | 0.57247 | -0.367 | 0.01371 | 0.411 |
| 18442 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-500a-3p | 10 | TSHZ3 | Sponge network | 0.598 | 0.38481 | -0.556 | 0.01516 | 0.411 |
| 18443 | RP11-532F6.3 |
hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-889-3p | 15 | RPS6KA6 | Sponge network | -0.896 | 0.00099 | -2.989 | 0 | 0.411 |
| 18444 | HAND2-AS1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | CSDE1 | Sponge network | -1.697 | 0.00889 | -0.493 | 0 | 0.411 |
| 18445 | CTC-429P9.5 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | RYR2 | Sponge network | 0.178 | 0.29106 | -1.466 | 0.00031 | 0.411 |
| 18446 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-576-5p | 33 | NTRK2 | Sponge network | -0.611 | 0.09607 | -0.736 | 0.06495 | 0.411 |
| 18447 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p | 41 | ZFHX3 | Sponge network | -0.615 | 0.00015 | 0.389 | 0.01363 | 0.411 |
| 18448 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | HIP1 | Sponge network | 0.197 | 0.25618 | 0.774 | 0 | 0.411 |
| 18449 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CDH10 | Sponge network | 0.246 | 0.59912 | -2.397 | 0 | 0.411 |
| 18450 | RP11-242D8.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p | 13 | TBL1XR1 | Sponge network | 1.404 | 0 | 0.578 | 0 | 0.411 |
| 18451 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 14 | ABCC9 | Sponge network | 1.757 | 0 | -0.466 | 0.14121 | 0.411 |
| 18452 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p | 28 | ESR1 | Sponge network | -0.842 | 0.01361 | -1.035 | 3.0E-5 | 0.411 |
| 18453 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p | 19 | FGFR1 | Sponge network | 0.064 | 0.72934 | -0.509 | 0.03957 | 0.411 |
| 18454 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-450b-5p | 14 | PKNOX1 | Sponge network | -0.162 | 0.47687 | 0.085 | 0.28917 | 0.411 |
| 18455 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | NRXN1 | Sponge network | -0.738 | 0.21233 | -3.778 | 0 | 0.411 |
| 18456 | SH3RF3-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-5p;hsa-miR-7-1-3p | 11 | ITPKB | Sponge network | 0.889 | 9.0E-5 | -0.704 | 0.00106 | 0.411 |
| 18457 | RP11-244H3.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p | 13 | EZH1 | Sponge network | 0.659 | 3.0E-5 | -0.564 | 1.0E-5 | 0.411 |
| 18458 | RP11-102N12.3 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-421 | 10 | PLXNC1 | Sponge network | -0.351 | 0.11358 | 0.569 | 0.00329 | 0.411 |
| 18459 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 21 | BCL6 | Sponge network | -0.096 | 0.67958 | -0.211 | 0.19489 | 0.411 |
| 18460 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 10 | MXI1 | Sponge network | -0.726 | 0.00132 | -0.987 | 0 | 0.411 |
| 18461 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-339-5p | 10 | SPTBN4 | Sponge network | -4.463 | 0.00025 | -0.786 | 0.01281 | 0.411 |
| 18462 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | HDAC9 | Sponge network | -1.439 | 4.0E-5 | -0.755 | 0.00495 | 0.411 |
| 18463 | FAM66C |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-93-3p | 15 | RPS6KA2 | Sponge network | -0.901 | 0.00019 | -0.57 | 0.00013 | 0.411 |
| 18464 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-660-5p;hsa-miR-93-5p | 10 | ZHX2 | Sponge network | -0.739 | 0.0574 | -0.192 | 0.18459 | 0.411 |
| 18465 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | ZFHX4 | Sponge network | -0.342 | 0.02288 | 0.018 | 0.95574 | 0.411 |
| 18466 | FENDRR |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | INPP4A | Sponge network | -1.281 | 0.00012 | 0.07 | 0.53563 | 0.411 |
| 18467 | RP11-574K11.29 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-511-5p | 19 | CREB1 | Sponge network | 0.997 | 0 | 0.203 | 0.00537 | 0.411 |
| 18468 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 14 | DYNC1I1 | Sponge network | -1.057 | 0.00029 | -1.12 | 0.00107 | 0.411 |
| 18469 | MIR143HG |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 31 | PDE4DIP | Sponge network | -0.739 | 0.0574 | -0.282 | 0.0257 | 0.411 |
| 18470 | RP11-175O19.4 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p | 11 | PHF3 | Sponge network | 0.917 | 0 | 0.126 | 0.19652 | 0.411 |
| 18471 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-7-1-3p | 19 | EBF1 | Sponge network | 0.299 | 0.57722 | -0.384 | 0.08856 | 0.411 |
| 18472 | AC127904.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p | 16 | TBL1XR1 | Sponge network | 1.308 | 0 | 0.578 | 0 | 0.411 |
| 18473 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 51 | CCND2 | Sponge network | 0.572 | 0.01722 | -0.267 | 0.3112 | 0.411 |
| 18474 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-421;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 50 | QKI | Sponge network | 0.082 | 0.55654 | -0.333 | 0.04111 | 0.411 |
| 18475 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 18 | TACC1 | Sponge network | -2.076 | 0.00548 | -0.362 | 0.05406 | 0.411 |
| 18476 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p | 16 | HIP1 | Sponge network | 0.16 | 0.50785 | 0.774 | 0 | 0.411 |
| 18477 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 39 | TRPS1 | Sponge network | 0.214 | 0.18246 | -0.191 | 0.44109 | 0.411 |
| 18478 | EMX2OS |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 15 | RHOB | Sponge network | -0.777 | 0.1134 | -1.052 | 0 | 0.411 |
| 18479 | RP11-822E23.8 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p | 70 | LPP | Sponge network | -0.777 | 0.16667 | -0.459 | 0.04475 | 0.411 |
| 18480 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 39 | ATRX | Sponge network | -0.739 | 0.0574 | 0.261 | 0.04408 | 0.411 |
| 18481 | LINC01018 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | WWTR1 | Sponge network | -2.915 | 0.00015 | -0.345 | 0.11997 | 0.411 |
| 18482 | AC109309.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b | 12 | AKAP6 | Sponge network | 0.163 | 0.87219 | -1.586 | 3.0E-5 | 0.411 |
| 18483 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p | 16 | LHX6 | Sponge network | -0.901 | 0.00019 | -0.087 | 0.69193 | 0.411 |
| 18484 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | RECK | Sponge network | 0.374 | 0.20054 | -1.043 | 0 | 0.411 |
| 18485 | LINC00702 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 16 | CITED2 | Sponge network | -0.737 | 0.06182 | -1.545 | 0 | 0.411 |
| 18486 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-629-3p | 17 | CBX5 | Sponge network | 1.056 | 0 | 0.165 | 0.24446 | 0.411 |
| 18487 | ANKRD10-IT1 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p | 13 | PIK3C2A | Sponge network | 1.739 | 0 | 0.061 | 0.53776 | 0.411 |
| 18488 | CTD-2314G24.2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-7-5p;hsa-miR-96-5p | 16 | FNBP1 | Sponge network | 0.246 | 0.59912 | -0.969 | 4.0E-5 | 0.411 |
| 18489 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421 | 23 | NTRK2 | Sponge network | -0.583 | 0.14589 | -0.736 | 0.06495 | 0.411 |
| 18490 | LINC00094 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 27 | PTPN14 | Sponge network | 0.253 | 0.09565 | 0.144 | 0.34595 | 0.411 |
| 18491 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-369-3p;hsa-miR-493-5p | 12 | PIK3R1 | Sponge network | 1.46 | 1.0E-5 | -0.133 | 0.36854 | 0.411 |
| 18492 | RP11-66B24.2 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3614-5p | 11 | HIP1 | Sponge network | 0.57 | 0.1866 | 0.774 | 0 | 0.411 |
| 18493 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 26 | PRKAB2 | Sponge network | 0.056 | 0.81195 | -0.367 | 0.01371 | 0.411 |
| 18494 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-450b-5p | 13 | RYR2 | Sponge network | -0.517 | 0.02935 | -1.466 | 0.00031 | 0.411 |
| 18495 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | PRKAA2 | Sponge network | -0.737 | 0.10822 | -2.462 | 0 | 0.411 |
| 18496 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 25 | RAP1A | Sponge network | -0.83 | 0 | -0.929 | 0 | 0.411 |
| 18497 | C20orf166-AS1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-542-3p;hsa-miR-577 | 14 | CSDE1 | Sponge network | -2.69 | 0.0001 | -0.493 | 0 | 0.411 |
| 18498 | LINC00861 |
hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-582-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 12 | TCF4 | Sponge network | 0.911 | 0.00854 | 0.016 | 0.92271 | 0.411 |
| 18499 | HHIP-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 11 | PRUNE2 | Sponge network | -0.737 | 0.10822 | -1.733 | 0.00022 | 0.411 |
| 18500 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 21 | ZFHX3 | Sponge network | -0.318 | 0.65847 | 0.389 | 0.01363 | 0.411 |
| 18501 | MEG3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-98-5p | 17 | KLF10 | Sponge network | 0.249 | 0.35902 | -0.783 | 0 | 0.411 |
| 18502 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 19 | PCDH10 | Sponge network | 0.214 | 0.18246 | -1.644 | 0.0078 | 0.411 |
| 18503 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | RBMS3 | Sponge network | 0.657 | 4.0E-5 | -0.519 | 0.03551 | 0.411 |
| 18504 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | NIN | Sponge network | 0.537 | 0.00139 | -0.253 | 0.13794 | 0.411 |
| 18505 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 27 | TACC1 | Sponge network | 1.757 | 0 | -0.362 | 0.05406 | 0.411 |
| 18506 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p | 11 | USP6 | Sponge network | 1.134 | 0 | 0.188 | 0.3871 | 0.411 |
| 18507 | CTD-2089N3.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 10 | ASTN1 | Sponge network | -0.314 | 0.62033 | -2.389 | 2.0E-5 | 0.411 |
| 18508 | AC108488.4 |
hsa-let-7g-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p | 17 | EPHA3 | Sponge network | 0.259 | 0.12542 | 0.185 | 0.53588 | 0.411 |
| 18509 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p | 10 | TRIM33 | Sponge network | 0.769 | 0 | 0.402 | 7.0E-5 | 0.411 |
| 18510 | LINC00865 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429 | 13 | NGFR | Sponge network | -1.249 | 7.0E-5 | -1.271 | 0.01058 | 0.411 |
| 18511 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-93-5p | 28 | ST6GALNAC3 | Sponge network | -2.69 | 0.0001 | -0.987 | 0 | 0.411 |
| 18512 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-7-1-3p | 28 | ANK2 | Sponge network | 0.16 | 0.50785 | -1.603 | 1.0E-5 | 0.411 |
| 18513 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 14 | CDH23 | Sponge network | -0.896 | 0.00099 | -0.974 | 0.00034 | 0.411 |
| 18514 | LINC00641 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | NAV3 | Sponge network | -0.222 | 0.32445 | 0.212 | 0.47729 | 0.411 |
| 18515 | RP11-248G5.8 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-664a-3p | 15 | RASAL2 | Sponge network | 1.294 | 0 | 0.869 | 0 | 0.411 |
| 18516 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-93-5p | 17 | CNTNAP3 | Sponge network | -0.154 | 0.78939 | -1.465 | 0.00033 | 0.411 |
| 18517 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 22 | CNTNAP3 | Sponge network | -1.697 | 0.00889 | -1.465 | 0.00033 | 0.411 |
| 18518 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 26 | SCN9A | Sponge network | 0.385 | 0.10067 | -0.525 | 0.10593 | 0.411 |
| 18519 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-93-5p | 40 | ADARB1 | Sponge network | -0.842 | 0.01361 | -0.335 | 0.06631 | 0.411 |
| 18520 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 15 | AKAP12 | Sponge network | 0.064 | 0.72934 | -0.932 | 0.0028 | 0.411 |
| 18521 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p | 26 | NTRK2 | Sponge network | -0.517 | 0.02935 | -0.736 | 0.06495 | 0.411 |
| 18522 | RP11-175O19.4 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-409-5p | 12 | STAG2 | Sponge network | 0.917 | 0 | 0.252 | 0.00438 | 0.411 |
| 18523 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 32 | PRKAB2 | Sponge network | 0.194 | 0.64278 | -0.367 | 0.01371 | 0.411 |
| 18524 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 21 | KAT6B | Sponge network | -0.793 | 7.0E-5 | -0.478 | 0.00018 | 0.411 |
| 18525 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 38 | CHD5 | Sponge network | -1.439 | 4.0E-5 | -0.922 | 0.01521 | 0.411 |
| 18526 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | WWTR1 | Sponge network | -0.08 | 0.90092 | -0.345 | 0.11997 | 0.411 |
| 18527 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-375 | 20 | ERC1 | Sponge network | -0.358 | 0.17255 | -0.108 | 0.38794 | 0.411 |
| 18528 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-935 | 31 | PTPRD | Sponge network | -1.331 | 3.0E-5 | -1.005 | 0.00064 | 0.411 |
| 18529 | CTB-31O20.4 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 15 | TBL1XR1 | Sponge network | 1.619 | 0 | 0.578 | 0 | 0.411 |
| 18530 | RP1-59D14.5 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 12 | DICER1 | Sponge network | 1.162 | 5.0E-5 | 0.042 | 0.674 | 0.411 |
| 18531 | SNX29P2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | 0.4 | 0.14611 | -0.564 | 1.0E-5 | 0.411 |
| 18532 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p | 26 | RASSF2 | Sponge network | -0.469 | 0.08721 | -0.273 | 0.21961 | 0.411 |
| 18533 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-93-5p | 35 | CHD5 | Sponge network | 0.253 | 0.09565 | -0.922 | 0.01521 | 0.411 |
| 18534 | RP11-37B2.1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-5p | 30 | CREB1 | Sponge network | 1.03 | 0 | 0.203 | 0.00537 | 0.411 |
| 18535 | LINC00672 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 46 | DTNA | Sponge network | -0.227 | 0.31657 | -1.648 | 1.0E-5 | 0.411 |
| 18536 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 26 | ARHGAP28 | Sponge network | -1.575 | 6.0E-5 | -0.911 | 0.00013 | 0.411 |
| 18537 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 19 | TGFBR2 | Sponge network | -0.323 | 0.01372 | -0.243 | 0.11408 | 0.41 |
| 18538 | RP11-57H14.4 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 21 | DICER1 | Sponge network | 0.083 | 0.66405 | 0.042 | 0.674 | 0.41 |
| 18539 | RP11-517B11.7 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-3p | 10 | PHF3 | Sponge network | 0.706 | 0 | 0.126 | 0.19652 | 0.41 |
| 18540 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 39 | CHD5 | Sponge network | -0.421 | 0.0114 | -0.922 | 0.01521 | 0.41 |
| 18541 | WAC-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 18 | TBL1XR1 | Sponge network | 0.821 | 0 | 0.578 | 0 | 0.41 |
| 18542 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | SYNE1 | Sponge network | 0.374 | 0.20054 | -0.738 | 0.00316 | 0.41 |
| 18543 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p | 14 | USP12 | Sponge network | 0.178 | 0.29106 | -0.102 | 0.40768 | 0.41 |
| 18544 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-744-5p | 11 | FGFR1 | Sponge network | 2.306 | 9.0E-5 | -0.509 | 0.03957 | 0.41 |
| 18545 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 20 | EBF1 | Sponge network | -0.206 | 0.12942 | -0.384 | 0.08856 | 0.41 |
| 18546 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | RB1CC1 | Sponge network | 0.799 | 1.0E-5 | 0.175 | 0.12559 | 0.41 |
| 18547 | FAM66C |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NRG3 | Sponge network | -0.901 | 0.00019 | -1.836 | 4.0E-5 | 0.41 |
| 18548 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-374b-5p | 15 | ABI2 | Sponge network | 0.912 | 0.00014 | 0.175 | 0.14787 | 0.41 |
| 18549 | AP001347.6 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-942-5p | 24 | IGFBP5 | Sponge network | -1.161 | 0.00591 | -0.806 | 0.00105 | 0.41 |
| 18550 | MIR143HG |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TRIM58 | Sponge network | -0.739 | 0.0574 | -1.692 | 0 | 0.41 |
| 18551 | RP11-156E6.1 |
hsa-miR-101-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-497-5p | 10 | KIF5B | Sponge network | 1.266 | 0 | 0.362 | 0.00066 | 0.41 |
| 18552 | RP5-1074L1.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 11 | KIAA1024 | Sponge network | 1.923 | 0 | 1.083 | 0 | 0.41 |
| 18553 | LINC00865 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-93-3p | 16 | RPS6KA2 | Sponge network | -1.249 | 7.0E-5 | -0.57 | 0.00013 | 0.41 |
| 18554 | RP11-166D19.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p | 14 | EHD3 | Sponge network | -0.576 | 0.10121 | -0.484 | 0.01747 | 0.41 |
| 18555 | RP4-717I23.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p | 16 | PIK3CA | Sponge network | 0.637 | 0.00069 | 0.195 | 0.06908 | 0.41 |
| 18556 | BVES-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-874-3p | 22 | MXI1 | Sponge network | -2.62 | 0 | -0.987 | 0 | 0.41 |
| 18557 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RABEP1 | Sponge network | 0.532 | 0.00505 | -0.066 | 0.43142 | 0.41 |
| 18558 | AP001347.6 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TPO | Sponge network | -1.161 | 0.00591 | -1.87 | 2.0E-5 | 0.41 |
| 18559 | LINC00865 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-5p | 14 | KLF10 | Sponge network | -1.249 | 7.0E-5 | -0.783 | 0 | 0.41 |
| 18560 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | DIP2C | Sponge network | 0.131 | 0.8436 | -0.436 | 0.01713 | 0.41 |
| 18561 | RP11-819C21.1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-96-5p | 27 | IL17RD | Sponge network | 0.537 | 0.00139 | -0.019 | 0.94873 | 0.41 |
| 18562 | CTC-429P9.2 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-26b-3p;hsa-miR-362-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-940 | 11 | GRIK3 | Sponge network | 0.043 | 0.81628 | -3.208 | 0 | 0.41 |
| 18563 | TPTEP1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | GJA1 | Sponge network | -1.216 | 0 | -0.485 | 0.02179 | 0.41 |
| 18564 | LINC00672 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-584-5p;hsa-miR-590-3p | 12 | ZNF208 | Sponge network | -0.227 | 0.31657 | -0.4 | 0.22381 | 0.41 |
| 18565 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -0.465 | 0.04703 | -0.243 | 0.11408 | 0.41 |
| 18566 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-93-5p | 15 | SLITRK3 | Sponge network | 0.056 | 0.81195 | -3.328 | 0 | 0.41 |
| 18567 | JAZF1-AS1 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | SPOP | Sponge network | -0.769 | 0.00035 | -0.419 | 1.0E-5 | 0.41 |
| 18568 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-5p | 33 | DMD | Sponge network | -1.645 | 0 | -1.506 | 0 | 0.41 |
| 18569 | DIO3OS |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-940 | 17 | SFRP1 | Sponge network | -0.773 | 0.04073 | -3.566 | 0 | 0.41 |
| 18570 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-769-5p | 25 | SYNPO2 | Sponge network | -0.355 | 0.0077 | -2.342 | 0 | 0.41 |
| 18571 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 37 | PRKAB2 | Sponge network | -1.697 | 0.00889 | -0.367 | 0.01371 | 0.41 |
| 18572 | RP11-693J15.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-93-3p;hsa-miR-940 | 22 | GRIK3 | Sponge network | -0.72 | 0.24239 | -3.208 | 0 | 0.41 |
| 18573 | CTC-444N24.11 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | 1.153 | 0 | 0.524 | 0.00017 | 0.41 |
| 18574 | RP11-686D22.8 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | QKI | Sponge network | 0.898 | 1.0E-5 | -0.333 | 0.04111 | 0.41 |
| 18575 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-5p | 23 | KCNA6 | Sponge network | -0.469 | 0.08721 | -1.531 | 0.00013 | 0.41 |
| 18576 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 13 | SEMA3E | Sponge network | -0.246 | 0.35034 | -1.717 | 0.00137 | 0.41 |
| 18577 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-5p | 14 | MAF | Sponge network | -1.255 | 0.00981 | -1.257 | 0 | 0.41 |
| 18578 | RP11-504P24.8 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c | 15 | ABL2 | Sponge network | 1.178 | 0 | 0.82 | 0 | 0.41 |
| 18579 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-93-5p | 15 | ANK2 | Sponge network | 0.497 | 0.01451 | -1.603 | 1.0E-5 | 0.41 |
| 18580 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | FAT3 | Sponge network | -0.737 | 0.10822 | -1.601 | 0.00051 | 0.41 |
| 18581 | AC006116.24 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-5p;hsa-miR-98-5p | 18 | LRIG1 | Sponge network | -0.473 | 0.07602 | -0.537 | 0.00588 | 0.41 |
| 18582 | IQCH-AS1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p | 18 | PHF6 | Sponge network | 0.929 | 0 | 0.785 | 0 | 0.41 |
| 18583 | SNHG16 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-495-3p | 13 | GSK3B | Sponge network | 0.961 | 0 | 0.266 | 0.00045 | 0.41 |
| 18584 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | CHD2 | Sponge network | 0.924 | 0 | 0.049 | 0.55371 | 0.41 |
| 18585 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-452-5p | 40 | ADARB1 | Sponge network | -0.405 | 0.22013 | -0.335 | 0.06631 | 0.41 |
| 18586 | RP11-216B9.6 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 17 | WWTR1 | Sponge network | 0.174 | 0.30034 | -0.345 | 0.11997 | 0.41 |
| 18587 | RP11-531A24.5 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | 0.75 | 0.00054 | 0.524 | 0.00017 | 0.41 |
| 18588 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-92a-3p | 12 | ZNF521 | Sponge network | 0.374 | 0.20054 | -0.111 | 0.58068 | 0.41 |
| 18589 | RP11-707P17.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 17 | TBL1XR1 | Sponge network | 1.877 | 0 | 0.578 | 0 | 0.41 |
| 18590 | AC010761.8 |
hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p | 11 | ABL2 | Sponge network | 1.638 | 0 | 0.82 | 0 | 0.41 |
| 18591 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-3p | 24 | MAF | Sponge network | -0.739 | 0.00044 | -1.257 | 0 | 0.41 |
| 18592 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | SLITRK6 | Sponge network | -0.901 | 0.00019 | -2.121 | 0 | 0.41 |
| 18593 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 19 | RPS6KA3 | Sponge network | 0.683 | 1.0E-5 | 0.256 | 0.02419 | 0.41 |
| 18594 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TRIM13 | Sponge network | 0.537 | 0.00139 | 0.183 | 0.06968 | 0.41 |
| 18595 | NDUFA6-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 44 | GRIK3 | Sponge network | -0.355 | 0.0077 | -3.208 | 0 | 0.41 |
| 18596 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-590-3p | 27 | BMPR2 | Sponge network | 0.919 | 0 | 0.206 | 0.0635 | 0.41 |
| 18597 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | MCC | Sponge network | -0.246 | 0.35034 | -1.005 | 0 | 0.41 |
| 18598 | GNG12-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p | 24 | CTIF | Sponge network | -0.793 | 7.0E-5 | -0.389 | 0.00549 | 0.41 |
| 18599 | HCG11 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | 0.385 | 0.10067 | -0.998 | 0.00025 | 0.41 |
| 18600 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-539-5p | 22 | DMD | Sponge network | -0.465 | 0.04703 | -1.506 | 0 | 0.41 |
| 18601 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 27 | BMPR2 | Sponge network | 0.959 | 0 | 0.206 | 0.0635 | 0.41 |
| 18602 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p | 26 | PTPRD | Sponge network | -0.517 | 0.02935 | -1.005 | 0.00064 | 0.41 |
| 18603 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-450b-5p | 12 | SPTBN4 | Sponge network | 0.194 | 0.64278 | -0.786 | 0.01281 | 0.41 |
| 18604 | RP11-774O3.3 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | NOV | Sponge network | -0.487 | 0.02157 | -0.58 | 0.04676 | 0.41 |
| 18605 | USP3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p | 21 | ERCC4 | Sponge network | -0.162 | 0.47687 | 0.126 | 0.15266 | 0.41 |
| 18606 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-671-5p | 12 | PIK3IP1 | Sponge network | -1.697 | 0.00889 | -0.187 | 0.19945 | 0.41 |
| 18607 | RP11-13J8.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-628-5p;hsa-miR-940 | 11 | TRIM33 | Sponge network | 3.03 | 0 | 0.402 | 7.0E-5 | 0.41 |
| 18608 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p | 15 | ERC1 | Sponge network | 0.497 | 0.01451 | -0.108 | 0.38794 | 0.41 |
| 18609 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p | 11 | SLITRK3 | Sponge network | -4.477 | 0 | -3.328 | 0 | 0.41 |
| 18610 | RP11-983P16.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | NRXN3 | Sponge network | 0.41 | 0.02214 | -0.893 | 0.04186 | 0.41 |
| 18611 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | RB1CC1 | Sponge network | 0.986 | 0.01022 | 0.175 | 0.12559 | 0.41 |
| 18612 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 21 | PTPN11 | Sponge network | 0.782 | 0.00016 | 0.431 | 2.0E-5 | 0.41 |
| 18613 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -0.384 | 0.40652 | -3.328 | 0 | 0.41 |
| 18614 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-501-3p;hsa-miR-502-3p | 33 | ZFHX4 | Sponge network | -0.384 | 0.40652 | 0.018 | 0.95574 | 0.41 |
| 18615 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-590-3p | 10 | ABCA10 | Sponge network | -0.421 | 0.0114 | -0.571 | 0.03674 | 0.41 |
| 18616 | TUG1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | UBR3 | Sponge network | 0.972 | 0 | -0.021 | 0.82774 | 0.41 |
| 18617 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | FYN | Sponge network | 0.532 | 0.00505 | -0.131 | 0.37051 | 0.41 |
| 18618 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PIK3CA | Sponge network | 1.601 | 0 | 0.195 | 0.06908 | 0.41 |
| 18619 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-93-3p | 40 | RUNX1T1 | Sponge network | 0.259 | 0.12542 | -0.344 | 0.2374 | 0.41 |
| 18620 | HAND2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 11 | FAM133A | Sponge network | -1.697 | 0.00889 | 0.549 | 0.29009 | 0.41 |
| 18621 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429 | 11 | PDZD2 | Sponge network | 0.335 | 0.20596 | -0.909 | 3.0E-5 | 0.41 |
| 18622 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | CADM2 | Sponge network | -4.477 | 0 | -3.782 | 0 | 0.41 |
| 18623 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 30 | TACC1 | Sponge network | -0.421 | 0.0114 | -0.362 | 0.05406 | 0.41 |
| 18624 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 20 | IGF1 | Sponge network | 0.657 | 4.0E-5 | -0.204 | 0.5898 | 0.41 |
| 18625 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 30 | NR2C2 | Sponge network | 0.311 | 0.10344 | 0.21 | 0.03224 | 0.41 |
| 18626 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 50 | MDM4 | Sponge network | 0.349 | 0.01695 | 0.345 | 0.00762 | 0.41 |
| 18627 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | MTSS1 | Sponge network | -0.513 | 0.04703 | -0.81 | 0.00035 | 0.41 |
| 18628 | TRAF3IP2-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | ROBO1 | Sponge network | -0.323 | 0.01372 | -0.009 | 0.96154 | 0.41 |
| 18629 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p | 10 | PDZD2 | Sponge network | -1.057 | 0.00029 | -0.909 | 3.0E-5 | 0.41 |
| 18630 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-98-5p | 27 | PBX1 | Sponge network | -0.18 | 0.20343 | -1.206 | 0 | 0.41 |
| 18631 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | SYNE1 | Sponge network | 1.302 | 7.0E-5 | -0.738 | 0.00316 | 0.41 |
| 18632 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 26 | SCN9A | Sponge network | -0.793 | 7.0E-5 | -0.525 | 0.10593 | 0.41 |
| 18633 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p | 11 | PARK2 | Sponge network | 0.572 | 0.01722 | -1.892 | 0 | 0.41 |
| 18634 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | RBMS3 | Sponge network | -0.726 | 0.00132 | -0.519 | 0.03551 | 0.41 |
| 18635 | ACTA2-AS1 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-7-5p | 10 | LZTS1 | Sponge network | 0.194 | 0.64278 | 1.664 | 0 | 0.41 |
| 18636 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | 0.572 | 0.01722 | -1.277 | 0 | 0.41 |
| 18637 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | PHC3 | Sponge network | 0.349 | 0.01695 | 0.212 | 0.0499 | 0.41 |
| 18638 | TPTEP1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | TLE4 | Sponge network | -1.216 | 0 | -1.157 | 0 | 0.41 |
| 18639 | RP11-822E23.8 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p | 18 | CDH23 | Sponge network | -0.777 | 0.16667 | -0.974 | 0.00034 | 0.41 |
| 18640 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-382-5p;hsa-miR-590-3p | 13 | TRIP11 | Sponge network | 1.254 | 0 | -0.158 | 0.06384 | 0.41 |
| 18641 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | TLE4 | Sponge network | -0.576 | 0.10121 | -1.157 | 0 | 0.41 |
| 18642 | RP11-359K18.4 | hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-577 | 10 | MDM4 | Sponge network | 0.014 | 0.96323 | 0.345 | 0.00762 | 0.41 |
| 18643 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | AKAP12 | Sponge network | 0.225 | 0.30644 | -0.932 | 0.0028 | 0.41 |
| 18644 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 27 | CRTC3 | Sponge network | -0.769 | 0.00035 | 0.164 | 0.16786 | 0.41 |
| 18645 | RP11-400K9.4 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 13 | ITGB1 | Sponge network | -0.611 | 0.09607 | 0.524 | 0.00017 | 0.41 |
| 18646 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 30 | CXXC4 | Sponge network | -1.111 | 0.0003 | -0.433 | 0.21055 | 0.41 |
| 18647 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 39 | TMEM132B | Sponge network | -0.096 | 0.67958 | -0.83 | 0.01084 | 0.41 |
| 18648 | RASSF8-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | RYR2 | Sponge network | 0.335 | 0.20596 | -1.466 | 0.00031 | 0.41 |
| 18649 | RP11-216B9.6 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-766-3p | 13 | ATM | Sponge network | 0.174 | 0.30034 | 0.178 | 0.21568 | 0.41 |
| 18650 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | CUL5 | Sponge network | 1.205 | 0 | 0.026 | 0.77301 | 0.41 |
| 18651 | HOXB-AS2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-629-3p | 25 | KIF1B | Sponge network | 1.316 | 0.01106 | -0.118 | 0.32258 | 0.41 |
| 18652 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 32 | NCOA1 | Sponge network | -1.575 | 6.0E-5 | -0.432 | 0 | 0.41 |
| 18653 | RP11-127B20.2 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 26 | MDM4 | Sponge network | 0.411 | 0.1335 | 0.345 | 0.00762 | 0.41 |
| 18654 | LINC00654 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SEMA3E | Sponge network | 0.374 | 0.20054 | -1.717 | 0.00137 | 0.41 |
| 18655 | KB-1460A1.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-766-3p | 11 | EZH1 | Sponge network | 0.603 | 0.00127 | -0.564 | 1.0E-5 | 0.41 |
| 18656 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-7-5p | 23 | BTG2 | Sponge network | -0.465 | 0.04703 | -1.763 | 0 | 0.41 |
| 18657 | RP11-195B17.1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-539-5p | 16 | DDX6 | Sponge network | 0.37 | 0.27614 | 0.091 | 0.36428 | 0.41 |
| 18658 | DIO3OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -0.773 | 0.04073 | 0.024 | 0.89889 | 0.41 |
| 18659 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-93-5p | 28 | TSHZ3 | Sponge network | -0.465 | 0.04703 | -0.556 | 0.01516 | 0.41 |
| 18660 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 34 | CHD5 | Sponge network | -1.249 | 7.0E-5 | -0.922 | 0.01521 | 0.41 |
| 18661 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | SASH1 | Sponge network | 0.083 | 0.66405 | -0.502 | 0.00175 | 0.41 |
| 18662 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | DYNC1I1 | Sponge network | -1.216 | 0 | -1.12 | 0.00107 | 0.41 |
| 18663 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 21 | SNX29 | Sponge network | -0.125 | 0.49414 | -0.34 | 0.01371 | 0.41 |
| 18664 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 14 | TIMP3 | Sponge network | -0.726 | 0.00132 | -0.546 | 0.02307 | 0.41 |
| 18665 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 18 | RBMS3 | Sponge network | 1.153 | 0.00114 | -0.519 | 0.03551 | 0.41 |
| 18666 | RP11-359E10.1 |
hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p | 10 | RPS6KA2 | Sponge network | 0.299 | 0.57722 | -0.57 | 0.00013 | 0.41 |
| 18667 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | SEPT6 | Sponge network | -0.487 | 0.02157 | -0.598 | 0.00181 | 0.41 |
| 18668 | RP11-967K21.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 71 | LPP | Sponge network | 0.056 | 0.81195 | -0.459 | 0.04475 | 0.41 |
| 18669 | USP3-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-7-5p | 30 | BTG2 | Sponge network | -0.162 | 0.47687 | -1.763 | 0 | 0.41 |
| 18670 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-625-5p | 16 | ZMYM2 | Sponge network | 1.206 | 0 | 0.279 | 0.01273 | 0.41 |
| 18671 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-576-5p | 10 | GREM1 | Sponge network | -0.318 | 0.65847 | 0.902 | 0.01691 | 0.41 |
| 18672 | ZRANB2-AS1 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-628-5p | 16 | RIMS2 | Sponge network | -0.376 | 0.00337 | -1.365 | 0.00208 | 0.41 |
| 18673 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | BCLAF1 | Sponge network | 0.75 | 0.00054 | 0.211 | 0.00684 | 0.41 |
| 18674 | A2M-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | LHFP | Sponge network | -0.08 | 0.90092 | -0.167 | 0.41371 | 0.41 |
| 18675 | ZSCAN16-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-874-3p | 28 | GRIK3 | Sponge network | -0.342 | 0.02288 | -3.208 | 0 | 0.41 |
| 18676 | CTC-490E21.10 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-93-3p | 10 | PTPN11 | Sponge network | 1.46 | 1.0E-5 | 0.431 | 2.0E-5 | 0.41 |
| 18677 | DNM3OS |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-624-5p | 12 | SMARCA1 | Sponge network | 0.572 | 0.01722 | -0.684 | 0.00159 | 0.41 |
| 18678 | AC009948.5 |
hsa-miR-107;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-96-5p | 11 | ELMO1 | Sponge network | 0.532 | 0.00505 | 0.04 | 0.83147 | 0.41 |
| 18679 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 29 | LIFR | Sponge network | 0.532 | 0.00505 | -1.794 | 0 | 0.41 |
| 18680 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-93-5p | 13 | LHX6 | Sponge network | -1.654 | 0.00144 | -0.087 | 0.69193 | 0.41 |
| 18681 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | HOOK3 | Sponge network | -0.793 | 7.0E-5 | 0.273 | 0.09957 | 0.41 |
| 18682 | PGM5-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-450b-5p | 10 | SPTBN4 | Sponge network | -4.546 | 0 | -0.786 | 0.01281 | 0.41 |
| 18683 | RP11-428J1.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-766-3p;hsa-miR-96-5p | 25 | IL17RD | Sponge network | 0.538 | 0.00076 | -0.019 | 0.94873 | 0.41 |
| 18684 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 40 | RUNX1T1 | Sponge network | 0.199 | 0.38015 | -0.344 | 0.2374 | 0.41 |
| 18685 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 11 | CNTNAP1 | Sponge network | -0.206 | 0.12942 | 0.358 | 0.13727 | 0.41 |
| 18686 | TP73-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-92a-3p | 14 | HERC1 | Sponge network | -0.563 | 0.02545 | -0.196 | 0.08544 | 0.41 |
| 18687 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | ZMYM2 | Sponge network | 0.673 | 0 | 0.279 | 0.01273 | 0.41 |
| 18688 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p;hsa-miR-98-5p | 37 | CLASP2 | Sponge network | -1.111 | 0.0003 | -0.038 | 0.71 | 0.41 |
| 18689 | AP001347.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 24 | SNX29 | Sponge network | -1.161 | 0.00591 | -0.34 | 0.01371 | 0.41 |
| 18690 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PRKAA2 | Sponge network | -0.021 | 0.93598 | -2.462 | 0 | 0.41 |
| 18691 | LINC00957 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 13 | CXCL12 | Sponge network | -0.421 | 0.0114 | -1.421 | 0 | 0.41 |
| 18692 | ZSCAN16-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | PRKCB | Sponge network | -0.342 | 0.02288 | -1.313 | 2.0E-5 | 0.41 |
| 18693 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p | 20 | ZMYND11 | Sponge network | -0.896 | 0.00099 | -0.2 | 0.05449 | 0.41 |
| 18694 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 21 | BCL6 | Sponge network | -0.901 | 0.00019 | -0.211 | 0.19489 | 0.409 |
| 18695 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-539-5p | 17 | NIN | Sponge network | -0.154 | 0.53954 | -0.253 | 0.13794 | 0.409 |
| 18696 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-942-5p | 23 | UNC80 | Sponge network | -0.72 | 0.24239 | -1.934 | 0 | 0.409 |
| 18697 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p | 25 | GAS7 | Sponge network | -2.076 | 0.00548 | -0.555 | 0.01602 | 0.409 |
| 18698 | RP11-196G18.22 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-495-3p | 14 | PHF6 | Sponge network | 1.206 | 0 | 0.785 | 0 | 0.409 |
| 18699 | FENDRR |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ZMYND11 | Sponge network | -1.281 | 0.00012 | -0.2 | 0.05449 | 0.409 |
| 18700 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 22 | BCL6 | Sponge network | -0.612 | 0.00094 | -0.211 | 0.19489 | 0.409 |
| 18701 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | LRRC7 | Sponge network | -1.582 | 8.0E-5 | -1.341 | 0.01193 | 0.409 |
| 18702 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 28 | CYLD | Sponge network | -0.739 | 0.0574 | -0.367 | 0.00171 | 0.409 |
| 18703 | HOXB-AS1 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-940 | 10 | FAM172A | Sponge network | -0.154 | 0.53954 | -0.57 | 0 | 0.409 |
| 18704 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-93-5p | 13 | SLITRK3 | Sponge network | 0.299 | 0.57722 | -3.328 | 0 | 0.409 |
| 18705 | RP11-464F9.1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-339-5p;hsa-miR-590-3p | 10 | TSC1 | Sponge network | 0.959 | 0 | 0.103 | 0.26071 | 0.409 |
| 18706 | PWAR6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 31 | CYLD | Sponge network | -1.575 | 6.0E-5 | -0.367 | 0.00171 | 0.409 |
| 18707 | AC004540.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p | 18 | DTNA | Sponge network | -2.549 | 0.00528 | -1.648 | 1.0E-5 | 0.409 |
| 18708 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 18 | ZBTB4 | Sponge network | -0.206 | 0.12942 | -0.739 | 0 | 0.409 |
| 18709 | CTD-3092A11.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-326 | 11 | SLC5A3 | Sponge network | 0.873 | 0 | 0.493 | 5.0E-5 | 0.409 |
| 18710 | LINC00702 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 14 | SEPT6 | Sponge network | -0.737 | 0.06182 | -0.598 | 0.00181 | 0.409 |
| 18711 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | TRIM33 | Sponge network | 0.539 | 0.03722 | 0.402 | 7.0E-5 | 0.409 |
| 18712 | CTC-471J1.2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-625-3p | 19 | ABL2 | Sponge network | 1.11 | 1.0E-5 | 0.82 | 0 | 0.409 |
| 18713 | RP1-59D14.5 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 19 | UBR3 | Sponge network | 1.162 | 5.0E-5 | -0.021 | 0.82774 | 0.409 |
| 18714 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 21 | SYNPO2 | Sponge network | -0.513 | 0.04703 | -2.342 | 0 | 0.409 |
| 18715 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p | 10 | THSD7A | Sponge network | -0.246 | 0.35034 | 0.242 | 0.25154 | 0.409 |
| 18716 | CTC-457L16.2 |
hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | TAL1 | Sponge network | 1.153 | 0.00114 | -0.462 | 0.00574 | 0.409 |
| 18717 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577 | 19 | AFF3 | Sponge network | -0.18 | 0.20343 | -2.373 | 0 | 0.409 |
| 18718 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | HDAC9 | Sponge network | -1.111 | 0.0003 | -0.755 | 0.00495 | 0.409 |
| 18719 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | NF1 | Sponge network | 1.317 | 0 | 0.51 | 0 | 0.409 |
| 18720 | RP11-67L2.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | CREB1 | Sponge network | 0.72 | 0.0001 | 0.203 | 0.00537 | 0.409 |
| 18721 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SLC6A15 | Sponge network | -2.117 | 0.00161 | -2.373 | 2.0E-5 | 0.409 |
| 18722 | RP11-48B3.4 |
hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p | 11 | DYNC1I1 | Sponge network | 1.757 | 0 | -1.12 | 0.00107 | 0.409 |
| 18723 | LINC00641 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 31 | KCNA6 | Sponge network | -0.222 | 0.32445 | -1.531 | 0.00013 | 0.409 |
| 18724 | SNHG14 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PCLO | Sponge network | -1.111 | 0.0003 | -1.843 | 0.00064 | 0.409 |
| 18725 | CTA-941F9.9 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-576-5p;hsa-miR-935 | 13 | PTPRD | Sponge network | -0.344 | 0.49624 | -1.005 | 0.00064 | 0.409 |
| 18726 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-93-5p | 38 | PDGFRA | Sponge network | -2.69 | 0.0001 | -0.372 | 0.0037 | 0.409 |
| 18727 | RP1-151F17.2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-96-5p | 70 | LPP | Sponge network | 0.222 | 0.29133 | -0.459 | 0.04475 | 0.409 |
| 18728 | RP11-401P9.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-93-5p | 16 | PRUNE2 | Sponge network | -0.384 | 0.40652 | -1.733 | 0.00022 | 0.409 |
| 18729 | CTC-429P9.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 10 | TSC1 | Sponge network | 0.178 | 0.29106 | 0.103 | 0.26071 | 0.409 |
| 18730 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | ADAMTSL3 | Sponge network | 0.16 | 0.50785 | -1.559 | 0 | 0.409 |
| 18731 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | MLLT10 | Sponge network | 0.421 | 0.00084 | 0.105 | 0.33292 | 0.409 |
| 18732 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-935 | 67 | LPP | Sponge network | -0.738 | 0.21233 | -0.459 | 0.04475 | 0.409 |
| 18733 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 10 | CAV1 | Sponge network | -0.726 | 0.00132 | -1.013 | 6.0E-5 | 0.409 |
| 18734 | CTC-297N7.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 12 | ARHGAP28 | Sponge network | -2.069 | 0 | -0.911 | 0.00013 | 0.409 |
| 18735 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-874-3p | 10 | GIGYF2 | Sponge network | -0.222 | 0.32445 | 0.136 | 0.04403 | 0.409 |
| 18736 | HCG18 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 14 | BCL9 | Sponge network | 1.063 | 0 | 0.321 | 0.00267 | 0.409 |
| 18737 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 18 | FAT3 | Sponge network | -2.039 | 0.03447 | -1.601 | 0.00051 | 0.409 |
| 18738 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-940 | 31 | PBX1 | Sponge network | 0.219 | 0.1516 | -1.206 | 0 | 0.409 |
| 18739 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-508-3p;hsa-miR-590-3p | 11 | RIMS1 | Sponge network | -0.358 | 0.17255 | -3.143 | 0 | 0.409 |
| 18740 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-503-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 45 | RICTOR | Sponge network | 0.311 | 0.10344 | 0.388 | 0.00188 | 0.409 |
| 18741 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | PHC3 | Sponge network | 0.997 | 0 | 0.212 | 0.0499 | 0.409 |
| 18742 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-766-3p | 21 | TBL1XR1 | Sponge network | 0.909 | 0 | 0.578 | 0 | 0.409 |
| 18743 | NNT-AS1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 13 | ZNF638 | Sponge network | 0.799 | 1.0E-5 | 0.22 | 0.01214 | 0.409 |
| 18744 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p | 38 | SOX5 | Sponge network | 0.253 | 0.09565 | -1.091 | 4.0E-5 | 0.409 |
| 18745 | AP001347.6 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | LUZP2 | Sponge network | -1.161 | 0.00591 | -0.056 | 0.89416 | 0.409 |
| 18746 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | TMEFF2 | Sponge network | -0.899 | 6.0E-5 | -2.426 | 0 | 0.409 |
| 18747 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-98-5p | 15 | PNRC1 | Sponge network | -2.62 | 0 | -0.646 | 0 | 0.409 |
| 18748 | RP11-890B15.3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-744-3p | 25 | PTPN14 | Sponge network | 0.101 | 0.60919 | 0.144 | 0.34595 | 0.409 |
| 18749 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | LATS1 | Sponge network | 0.93 | 0 | 0.109 | 0.34208 | 0.409 |
| 18750 | HAND2-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 21 | FMNL3 | Sponge network | -1.697 | 0.00889 | 0.555 | 4.0E-5 | 0.409 |
| 18751 | RP11-359B12.2 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 51 | PPM1L | Sponge network | 0.064 | 0.72934 | -0.537 | 0.00616 | 0.409 |
| 18752 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | DCLK1 | Sponge network | -0.737 | 0.10822 | -1.324 | 0.00014 | 0.409 |
| 18753 | RP11-890B15.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-671-5p | 16 | CDH23 | Sponge network | 0.101 | 0.60919 | -0.974 | 0.00034 | 0.409 |
| 18754 | PART1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 21 | CADM3 | Sponge network | -2.925 | 0 | -3.663 | 0 | 0.409 |
| 18755 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | ZNF770 | Sponge network | 0.537 | 0.00139 | 0.206 | 0.06751 | 0.409 |
| 18756 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 16 | UVRAG | Sponge network | -0.576 | 0.10121 | -0.017 | 0.83865 | 0.409 |
| 18757 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 42 | BCL2 | Sponge network | 0.572 | 0.01722 | -0.899 | 0 | 0.409 |
| 18758 | RP11-360F5.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-877-5p | 11 | RBMS3 | Sponge network | 1.867 | 0 | -0.519 | 0.03551 | 0.409 |
| 18759 | RAMP2-AS1 |
hsa-miR-146a-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 12 | ADCY1 | Sponge network | -0.513 | 0.04703 | 0.362 | 0.16982 | 0.409 |
| 18760 | PSMD5-AS1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-590-3p | 12 | TPR | Sponge network | 0.569 | 0.00067 | 0.582 | 0 | 0.409 |
| 18761 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 26 | PGR | Sponge network | -0.342 | 0.02288 | -1.704 | 0 | 0.409 |
| 18762 | RP11-6O2.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-96-5p | 18 | NCAM2 | Sponge network | 0.331 | 0.57247 | -0.482 | 0.22565 | 0.409 |
| 18763 | LINC00702 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 45 | CREB3L2 | Sponge network | -0.737 | 0.06182 | -0.525 | 2.0E-5 | 0.409 |
| 18764 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-629-3p | 13 | CHD2 | Sponge network | 0.921 | 0.00118 | 0.049 | 0.55371 | 0.409 |
| 18765 | PART1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-708-5p | 10 | MPL | Sponge network | -2.925 | 0 | -0.624 | 0.00133 | 0.409 |
| 18766 | LINC00092 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-590-3p;hsa-miR-942-5p | 14 | ARHGAP28 | Sponge network | -1.886 | 0 | -0.911 | 0.00013 | 0.409 |
| 18767 | RP11-216B9.6 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | DIP2C | Sponge network | 0.174 | 0.30034 | -0.436 | 0.01713 | 0.409 |
| 18768 | RP11-159D12.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | PHF3 | Sponge network | 1.254 | 0 | 0.126 | 0.19652 | 0.409 |
| 18769 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -0.49 | 0.04946 | 0.309 | 0.17079 | 0.409 |
| 18770 | LINC01001 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-539-5p | 11 | MOB1B | Sponge network | 1.055 | 0 | 0.197 | 0.15266 | 0.409 |
| 18771 | MIR143HG |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 13 | LZTS1 | Sponge network | -0.739 | 0.0574 | 1.664 | 0 | 0.409 |
| 18772 | RP11-195F19.9 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 47 | LPP | Sponge network | -0.726 | 0.00132 | -0.459 | 0.04475 | 0.409 |
| 18773 | RP11-206L10.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-539-5p | 11 | ABI2 | Sponge network | 0.793 | 0 | 0.175 | 0.14787 | 0.409 |
| 18774 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-340-5p | 10 | DNAJB4 | Sponge network | 0.331 | 0.57247 | -0.7 | 1.0E-5 | 0.409 |
| 18775 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 22 | YAP1 | Sponge network | 0.537 | 0.00139 | 0.22 | 0.11836 | 0.409 |
| 18776 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | EPHA3 | Sponge network | 0.614 | 1.0E-5 | 0.185 | 0.53588 | 0.409 |
| 18777 | ARHGEF26-AS1 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | RTN4 | Sponge network | -2.46 | 0 | -0.412 | 0.00014 | 0.409 |
| 18778 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-375;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | CREM | Sponge network | -0.49 | 0.04946 | -0.345 | 0.00258 | 0.409 |
| 18779 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 21 | KIT | Sponge network | -0.709 | 0.18168 | -2.184 | 0 | 0.409 |
| 18780 | FAM66C |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ITGB3 | Sponge network | -0.901 | 0.00019 | -0.011 | 0.95751 | 0.409 |
| 18781 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 10 | GIGYF2 | Sponge network | 0.959 | 0 | 0.136 | 0.04403 | 0.409 |
| 18782 | PWAR6 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | GALNT13 | Sponge network | -1.575 | 6.0E-5 | -0.857 | 0.07439 | 0.409 |
| 18783 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-628-5p | 13 | KIAA1024 | Sponge network | 0.537 | 0.00139 | 1.083 | 0 | 0.409 |
| 18784 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 38 | MAF | Sponge network | -0.739 | 0.0574 | -1.257 | 0 | 0.409 |
| 18785 | MIR143HG |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 16 | UVRAG | Sponge network | -0.739 | 0.0574 | -0.017 | 0.83865 | 0.409 |
| 18786 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-539-5p | 25 | PBX1 | Sponge network | -0.726 | 0.00132 | -1.206 | 0 | 0.409 |
| 18787 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-654-3p | 19 | NR2C2 | Sponge network | -0.227 | 0.31657 | 0.21 | 0.03224 | 0.409 |
| 18788 | FAM66C |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PCLO | Sponge network | -0.901 | 0.00019 | -1.843 | 0.00064 | 0.409 |
| 18789 | RP11-144G6.12 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-590-3p | 12 | TSC1 | Sponge network | 0.826 | 1.0E-5 | 0.103 | 0.26071 | 0.409 |
| 18790 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-361-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | CRTC1 | Sponge network | -0.323 | 0.01372 | -0.116 | 0.28412 | 0.409 |
| 18791 | LINC00702 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | FLI1 | Sponge network | -0.737 | 0.06182 | -0.294 | 0.10905 | 0.409 |
| 18792 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-655-3p | 24 | SON | Sponge network | 1.11 | 0 | 0.168 | 0.04406 | 0.409 |
| 18793 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p | 14 | PIK3C2A | Sponge network | 1.044 | 2.0E-5 | 0.061 | 0.53776 | 0.409 |
| 18794 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p | 12 | LHX6 | Sponge network | -0.739 | 0.00044 | -0.087 | 0.69193 | 0.409 |
| 18795 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | SLITRK6 | Sponge network | -0.727 | 0.04726 | -2.121 | 0 | 0.409 |
| 18796 | RP11-216B9.6 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p | 21 | RASSF8 | Sponge network | 0.174 | 0.30034 | -0.122 | 0.65591 | 0.409 |
| 18797 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-629-3p | 14 | RYR2 | Sponge network | -1.654 | 0.00144 | -1.466 | 0.00031 | 0.409 |
| 18798 | LINC00672 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 42 | AR | Sponge network | -0.227 | 0.31657 | -1.554 | 0 | 0.409 |
| 18799 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | FSTL5 | Sponge network | -0.901 | 0.00019 | -1.3 | 0.01619 | 0.409 |
| 18800 | ADAMTS9-AS2 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-708-5p | 20 | FAM135B | Sponge network | -2.452 | 0 | -3.978 | 0 | 0.409 |
| 18801 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | MYO9A | Sponge network | 0.419 | 0.0319 | -0.147 | 0.32373 | 0.409 |
| 18802 | AP001055.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-331-3p;hsa-miR-424-5p | 13 | CBFA2T3 | Sponge network | 0.693 | 0.00076 | -1.221 | 1.0E-5 | 0.409 |
| 18803 | MALAT1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-940 | 14 | DLG1 | Sponge network | 0.782 | 0.00016 | -0.014 | 0.8926 | 0.409 |
| 18804 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-652-3p;hsa-miR-93-5p | 28 | ZBTB4 | Sponge network | 0.082 | 0.55654 | -0.739 | 0 | 0.409 |
| 18805 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | CLASP2 | Sponge network | 0.559 | 0.00026 | -0.038 | 0.71 | 0.409 |
| 18806 | AC006116.24 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-577 | 22 | NTRK2 | Sponge network | -0.473 | 0.07602 | -0.736 | 0.06495 | 0.409 |
| 18807 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p | 15 | ABCA10 | Sponge network | -0.405 | 0.22013 | -0.571 | 0.03674 | 0.409 |
| 18808 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-331-5p;hsa-miR-7-1-3p | 12 | PCDH9 | Sponge network | -0.726 | 0.00132 | -1.823 | 4.0E-5 | 0.409 |
| 18809 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-93-3p | 15 | EBF3 | Sponge network | -0.72 | 0.24239 | -0.058 | 0.81437 | 0.409 |
| 18810 | GAS5 |
hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p | 10 | CDC73 | Sponge network | 0.728 | 0 | 0.094 | 0.20463 | 0.409 |
| 18811 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | PDGFRA | Sponge network | -1.057 | 0.00029 | -0.372 | 0.0037 | 0.409 |
| 18812 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | AKAP6 | Sponge network | 0.537 | 0.00139 | -1.586 | 3.0E-5 | 0.409 |
| 18813 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-508-3p;hsa-miR-590-3p | 21 | MAF | Sponge network | -0.358 | 0.17255 | -1.257 | 0 | 0.409 |
| 18814 | RP11-672A2.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-424-5p | 13 | PIK3R1 | Sponge network | 1.344 | 0 | -0.133 | 0.36854 | 0.409 |
| 18815 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | TACC1 | Sponge network | 0.374 | 0.20054 | -0.362 | 0.05406 | 0.409 |
| 18816 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-7-1-3p | 14 | DACT1 | Sponge network | -0.611 | 0.09607 | 0.783 | 0.00072 | 0.409 |
| 18817 | RP11-488L18.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p | 11 | PIK3C2A | Sponge network | 1.392 | 0 | 0.061 | 0.53776 | 0.409 |
| 18818 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-96-5p | 17 | MOB1B | Sponge network | 0.37 | 0.27614 | 0.197 | 0.15266 | 0.409 |
| 18819 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 42 | CHD5 | Sponge network | -0.303 | 0.21002 | -0.922 | 0.01521 | 0.409 |
| 18820 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | FAT4 | Sponge network | -2.46 | 0 | -0.354 | 0.14694 | 0.409 |
| 18821 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-590-3p | 20 | MLLT10 | Sponge network | 0.494 | 0.00339 | 0.105 | 0.33292 | 0.409 |
| 18822 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-590-5p | 21 | SCN9A | Sponge network | 0.889 | 9.0E-5 | -0.525 | 0.10593 | 0.409 |
| 18823 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | DYNC1I1 | Sponge network | -0.355 | 0.0077 | -1.12 | 0.00107 | 0.409 |
| 18824 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | AHNAK | Sponge network | -0.842 | 0.01361 | -1.207 | 0 | 0.409 |
| 18825 | RP11-119F7.5 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | PIK3CA | Sponge network | 0.225 | 0.30644 | 0.195 | 0.06908 | 0.409 |
| 18826 | RP11-282O18.3 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p | 10 | RASAL2 | Sponge network | 0.757 | 0 | 0.869 | 0 | 0.409 |
| 18827 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | TAL1 | Sponge network | -0.318 | 0.65847 | -0.462 | 0.00574 | 0.409 |
| 18828 | HCG11 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | ROBO1 | Sponge network | 0.385 | 0.10067 | -0.009 | 0.96154 | 0.409 |
| 18829 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | LIFR | Sponge network | -0.72 | 0.24239 | -1.794 | 0 | 0.409 |
| 18830 | RP11-758N13.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-455-3p | 11 | FBXO32 | Sponge network | 2.939 | 3.0E-5 | -0.495 | 0.07561 | 0.409 |
| 18831 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p | 18 | MYO9A | Sponge network | 0.176 | 0.37402 | -0.147 | 0.32373 | 0.409 |
| 18832 | CTC-550B14.6 |
hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | MDM4 | Sponge network | 3.885 | 0 | 0.345 | 0.00762 | 0.409 |
| 18833 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | IGSF10 | Sponge network | 0.532 | 0.00505 | -1.751 | 1.0E-5 | 0.409 |
| 18834 | RP11-53O19.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | SLC5A3 | Sponge network | 1.205 | 0 | 0.493 | 5.0E-5 | 0.409 |
| 18835 | LINC00578 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577 | 12 | FMNL3 | Sponge network | 0.811 | 0.03228 | 0.555 | 4.0E-5 | 0.409 |
| 18836 | RP11-532F6.3 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 10 | PCDH18 | Sponge network | -0.896 | 0.00099 | 0.216 | 0.24645 | 0.409 |
| 18837 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p | 13 | KCNAB1 | Sponge network | -0.162 | 0.47687 | -0.755 | 2.0E-5 | 0.409 |
| 18838 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-590-3p | 10 | RB1CC1 | Sponge network | 1.601 | 0 | 0.175 | 0.12559 | 0.409 |
| 18839 | GS1-124K5.11 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p | 26 | RASSF8 | Sponge network | 0.494 | 0.00339 | -0.122 | 0.65591 | 0.409 |
| 18840 | RP11-532F6.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 32 | AFF1 | Sponge network | -0.896 | 0.00099 | -0.384 | 0.00053 | 0.409 |
| 18841 | FENDRR |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 36 | ERC1 | Sponge network | -1.281 | 0.00012 | -0.108 | 0.38794 | 0.409 |
| 18842 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | EBF3 | Sponge network | 0.246 | 0.59912 | -0.058 | 0.81437 | 0.409 |
| 18843 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | ADARB1 | Sponge network | 0.342 | 0.03129 | -0.335 | 0.06631 | 0.409 |
| 18844 | USP3-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-452-5p;hsa-miR-625-5p | 12 | TES | Sponge network | -0.162 | 0.47687 | -0.348 | 0.00639 | 0.409 |
| 18845 | RP11-983P16.4 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | 0.41 | 0.02214 | 0.358 | 0.13727 | 0.409 |
| 18846 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-5p | 35 | TGFBR3 | Sponge network | -1.645 | 0 | -1.173 | 1.0E-5 | 0.409 |
| 18847 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 17 | GRIN2A | Sponge network | 0.299 | 0.57722 | -2.161 | 0 | 0.409 |
| 18848 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p | 19 | CRTC1 | Sponge network | -0.206 | 0.12942 | -0.116 | 0.28412 | 0.409 |
| 18849 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p | 16 | ABL1 | Sponge network | -0.739 | 0.00044 | -0.215 | 0.07804 | 0.409 |
| 18850 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 19 | CUL5 | Sponge network | 0.559 | 0.00026 | 0.026 | 0.77301 | 0.409 |
| 18851 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | -0.487 | 0.02157 | -1.047 | 0.00364 | 0.409 |
| 18852 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 22 | NF1 | Sponge network | 1.254 | 0 | 0.51 | 0 | 0.409 |
| 18853 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p | 10 | MYO9A | Sponge network | 0.062 | 0.55274 | -0.147 | 0.32373 | 0.409 |
| 18854 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 29 | DMXL1 | Sponge network | 0.101 | 0.60919 | -0.426 | 0.00056 | 0.409 |
| 18855 | EMX2OS |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 17 | FAM172A | Sponge network | -0.777 | 0.1134 | -0.57 | 0 | 0.409 |
| 18856 | AC156455.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p | 10 | NRK | Sponge network | 1.154 | 0.00041 | 0.656 | 0.19217 | 0.409 |
| 18857 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-625-3p | 26 | ZFHX3 | Sponge network | -0.358 | 0.17255 | 0.389 | 0.01363 | 0.408 |
| 18858 | RP11-819C21.1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | ERCC6 | Sponge network | 0.537 | 0.00139 | 0.23 | 0.015 | 0.408 |
| 18859 | RP11-13K12.1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p | 10 | FNBP1 | Sponge network | -0.154 | 0.78939 | -0.969 | 4.0E-5 | 0.408 |
| 18860 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | AR | Sponge network | -0.053 | 0.80685 | -1.554 | 0 | 0.408 |
| 18861 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p | 13 | CBL | Sponge network | 1.157 | 0.00455 | 0.291 | 0.01267 | 0.408 |
| 18862 | RP11-356J5.12 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-940 | 13 | RHOB | Sponge network | -0.899 | 6.0E-5 | -1.052 | 0 | 0.408 |
| 18863 | MIR497HG |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-708-5p | 10 | MPL | Sponge network | -1.439 | 4.0E-5 | -0.624 | 0.00133 | 0.408 |
| 18864 | HAND2-AS1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p | 22 | IRS1 | Sponge network | -1.697 | 0.00889 | -0.026 | 0.88855 | 0.408 |
| 18865 | AC005154.6 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PHF3 | Sponge network | 0.848 | 0 | 0.126 | 0.19652 | 0.408 |
| 18866 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | NALCN | Sponge network | -1.216 | 0 | 1.068 | 0.00237 | 0.408 |
| 18867 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | LRP1B | Sponge network | -0.612 | 0.00094 | -2.163 | 0 | 0.408 |
| 18868 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 14 | HECA | Sponge network | 1.601 | 0 | -0.217 | 0.03463 | 0.408 |
| 18869 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-339-5p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | MLLT10 | Sponge network | 0.959 | 0 | 0.105 | 0.33292 | 0.408 |
| 18870 | AC009948.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | PRUNE2 | Sponge network | 0.532 | 0.00505 | -1.733 | 0.00022 | 0.408 |
| 18871 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 28 | FOXO3 | Sponge network | 0.385 | 0.10067 | -0.04 | 0.72028 | 0.408 |
| 18872 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | EPHA5 | Sponge network | -2.117 | 0.00161 | -0.957 | 0.01733 | 0.408 |
| 18873 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-942-5p | 49 | UNC80 | Sponge network | -0.942 | 0 | -1.934 | 0 | 0.408 |
| 18874 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-542-3p | 14 | SLITRK6 | Sponge network | -2.69 | 0.0001 | -2.121 | 0 | 0.408 |
| 18875 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 38 | MOB1B | Sponge network | -0.487 | 0.02157 | 0.197 | 0.15266 | 0.408 |
| 18876 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 28 | AFF3 | Sponge network | -0.222 | 0.32445 | -2.373 | 0 | 0.408 |
| 18877 | RP11-359E10.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-330-5p;hsa-miR-671-5p;hsa-miR-940 | 10 | SORCS1 | Sponge network | 0.299 | 0.57722 | -3.492 | 0 | 0.408 |
| 18878 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | -0.206 | 0.12942 | -1.751 | 1.0E-5 | 0.408 |
| 18879 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-877-5p;hsa-miR-940 | 19 | NTRK3 | Sponge network | 0.043 | 0.81628 | -1.77 | 3.0E-5 | 0.408 |
| 18880 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | EPAS1 | Sponge network | -0.739 | 0.0574 | -0.184 | 0.14418 | 0.408 |
| 18881 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 42 | PHC3 | Sponge network | -1.111 | 0.0003 | 0.212 | 0.0499 | 0.408 |
| 18882 | ZRANB2-AS1 |
hsa-miR-1266-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | AFF1 | Sponge network | -0.376 | 0.00337 | -0.384 | 0.00053 | 0.408 |
| 18883 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 33 | PBX1 | Sponge network | 0.083 | 0.66405 | -1.206 | 0 | 0.408 |
| 18884 | RP11-967K21.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-32-5p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | LUZP2 | Sponge network | 0.056 | 0.81195 | -0.056 | 0.89416 | 0.408 |
| 18885 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-940 | 27 | AKAP11 | Sponge network | 0.594 | 0.41522 | 0.196 | 0.09825 | 0.408 |
| 18886 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-671-5p | 22 | KIT | Sponge network | -0.727 | 0.04726 | -2.184 | 0 | 0.408 |
| 18887 | RP11-678G14.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | TLL1 | Sponge network | -4.477 | 0 | -1.195 | 3.0E-5 | 0.408 |
| 18888 | RP4-639F20.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-96-5p | 12 | HIP1 | Sponge network | -0.517 | 0.02935 | 0.774 | 0 | 0.408 |
| 18889 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | MLF1 | Sponge network | -0.49 | 0.04946 | -0.893 | 0.00727 | 0.408 |
| 18890 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 11 | DAB2 | Sponge network | 0.898 | 1.0E-5 | 0.431 | 0.0113 | 0.408 |
| 18891 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | CDHR3 | Sponge network | -0.896 | 0.00099 | -0.977 | 4.0E-5 | 0.408 |
| 18892 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | SSBP2 | Sponge network | 0.299 | 0.57722 | -1.58 | 0 | 0.408 |
| 18893 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-590-3p | 13 | LRP1B | Sponge network | -1.582 | 8.0E-5 | -2.163 | 0 | 0.408 |
| 18894 | FAM66C |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-590-3p | 15 | STARD13 | Sponge network | -0.901 | 0.00019 | -0.187 | 0.27383 | 0.408 |
| 18895 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-486-5p;hsa-miR-576-5p | 19 | HDAC9 | Sponge network | -2.62 | 0 | -0.755 | 0.00495 | 0.408 |
| 18896 | PART1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | SYT4 | Sponge network | -2.925 | 0 | -3.207 | 0 | 0.408 |
| 18897 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 28 | ESR1 | Sponge network | 0.199 | 0.38015 | -1.035 | 3.0E-5 | 0.408 |
| 18898 | CTC-296K1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-5p | 13 | NAV3 | Sponge network | -2.095 | 0.00112 | 0.212 | 0.47729 | 0.408 |
| 18899 | MAGI2-AS3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-7-5p | 25 | KCNJ5 | Sponge network | -0.129 | 0.57177 | -0.281 | 0.3339 | 0.408 |
| 18900 | RP11-5C23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 27 | PIK3R1 | Sponge network | 0.274 | 0.07681 | -0.133 | 0.36854 | 0.408 |
| 18901 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-744-3p | 14 | CUL5 | Sponge network | 0.959 | 0 | 0.026 | 0.77301 | 0.408 |
| 18902 | LINC00926 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 16 | MEF2C | Sponge network | 0.756 | 0.00739 | -0.499 | 0.01448 | 0.408 |
| 18903 | RP11-283I3.6 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-576-5p | 18 | SIRT1 | Sponge network | 0.614 | 1.0E-5 | 0.109 | 0.2593 | 0.408 |
| 18904 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 62 | TCF4 | Sponge network | -0.727 | 0.04726 | 0.016 | 0.92271 | 0.408 |
| 18905 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 46 | PTPRD | Sponge network | 0.572 | 0.01722 | -1.005 | 0.00064 | 0.408 |
| 18906 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-576-5p;hsa-miR-93-5p | 16 | PDGFRA | Sponge network | -2.039 | 0.03447 | -0.372 | 0.0037 | 0.408 |
| 18907 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | PRRX1 | Sponge network | 0.299 | 0.57722 | 1.316 | 0 | 0.408 |
| 18908 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-539-5p;hsa-miR-577 | 31 | THRB | Sponge network | -0.473 | 0.07602 | -1.117 | 0.0004 | 0.408 |
| 18909 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-628-5p;hsa-miR-940 | 19 | TRIM33 | Sponge network | 0.267 | 0.2937 | 0.402 | 7.0E-5 | 0.408 |
| 18910 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-1-3p | 23 | RBMS3 | Sponge network | 0.259 | 0.12542 | -0.519 | 0.03551 | 0.408 |
| 18911 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | THBS1 | Sponge network | -1.203 | 0.03881 | 0.741 | 0.00386 | 0.408 |
| 18912 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p | 17 | CREBBP | Sponge network | -0.487 | 0.02157 | 0.126 | 0.15186 | 0.408 |
| 18913 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 28 | UNC80 | Sponge network | -0.358 | 0.17255 | -1.934 | 0 | 0.408 |
| 18914 | RP1-151F17.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 49 | MDM4 | Sponge network | 0.222 | 0.29133 | 0.345 | 0.00762 | 0.408 |
| 18915 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 51 | TCF4 | Sponge network | 0.385 | 0.10067 | 0.016 | 0.92271 | 0.408 |
| 18916 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 35 | DTNA | Sponge network | 0.889 | 9.0E-5 | -1.648 | 1.0E-5 | 0.408 |
| 18917 | CALML3-AS1 |
hsa-miR-141-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 11 | RANBP9 | Sponge network | 0.95 | 0.15943 | -0.191 | 0.04628 | 0.408 |
| 18918 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-877-5p | 20 | NAV3 | Sponge network | -0.246 | 0.35034 | 0.212 | 0.47729 | 0.408 |
| 18919 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-501-5p;hsa-miR-93-3p;hsa-miR-940 | 13 | POU2F2 | Sponge network | -0.323 | 0.01372 | -0.233 | 0.32214 | 0.408 |
| 18920 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | DMTF1 | Sponge network | 1.063 | 0 | 0.248 | 0.02726 | 0.408 |
| 18921 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ZNF536 | Sponge network | -0.769 | 0.00035 | -3.115 | 0 | 0.408 |
| 18922 | TPTEP1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 21 | RELN | Sponge network | -1.216 | 0 | -2.403 | 0 | 0.408 |
| 18923 | GS1-124K5.11 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-590-3p | 14 | ITGB1 | Sponge network | 0.494 | 0.00339 | 0.524 | 0.00017 | 0.408 |
| 18924 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ABCC9 | Sponge network | -0.737 | 0.10822 | -0.466 | 0.14121 | 0.408 |
| 18925 | MIR497HG |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | -1.439 | 4.0E-5 | 0.309 | 0.17079 | 0.408 |
| 18926 | RP11-386G11.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p | 15 | MDM4 | Sponge network | 2.031 | 0.00154 | 0.345 | 0.00762 | 0.408 |
| 18927 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-484;hsa-miR-532-3p | 16 | CBL | Sponge network | 0.453 | 0.01839 | 0.291 | 0.01267 | 0.408 |
| 18928 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PTPRB | Sponge network | -1.203 | 0.03881 | 0.105 | 0.47122 | 0.408 |
| 18929 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-942-5p | 27 | DDX6 | Sponge network | 0.082 | 0.55654 | 0.091 | 0.36428 | 0.408 |
| 18930 | CTC-296K1.4 |
hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-629-5p | 22 | DST | Sponge network | -2.076 | 0.00548 | -0.713 | 0.00096 | 0.408 |
| 18931 | RP11-46C24.7 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p | 14 | ATM | Sponge network | 0.392 | 0.0278 | 0.178 | 0.21568 | 0.408 |
| 18932 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 43 | CREB3L2 | Sponge network | -0.576 | 0.10121 | -0.525 | 2.0E-5 | 0.408 |
| 18933 | LINC00472 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-874-3p;hsa-miR-877-5p | 28 | ASTN1 | Sponge network | -0.842 | 0.01361 | -2.389 | 2.0E-5 | 0.408 |
| 18934 | LINC00641 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | SPOP | Sponge network | -0.222 | 0.32445 | -0.419 | 1.0E-5 | 0.408 |
| 18935 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p | 21 | GPC6 | Sponge network | -2.69 | 0.0001 | -0.054 | 0.81465 | 0.408 |
| 18936 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 29 | EBF1 | Sponge network | -1.203 | 0.03881 | -0.384 | 0.08856 | 0.408 |
| 18937 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 36 | ABI2 | Sponge network | 0.311 | 0.10344 | 0.175 | 0.14787 | 0.408 |
| 18938 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | CDHR3 | Sponge network | -2.452 | 0 | -0.977 | 4.0E-5 | 0.408 |
| 18939 | RP11-890B15.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | CREB1 | Sponge network | 0.101 | 0.60919 | 0.203 | 0.00537 | 0.408 |
| 18940 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | FNBP1 | Sponge network | -0.18 | 0.20343 | -0.969 | 4.0E-5 | 0.408 |
| 18941 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-577;hsa-miR-625-5p | 30 | TGFBR3 | Sponge network | -0.465 | 0.04703 | -1.173 | 1.0E-5 | 0.408 |
| 18942 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 17 | RBL2 | Sponge network | -2.452 | 0 | -0.282 | 0.00254 | 0.408 |
| 18943 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-484;hsa-miR-93-5p | 12 | EDA2R | Sponge network | -0.777 | 0.16667 | -0.587 | 0.01598 | 0.408 |
| 18944 | FAM157B |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 12 | ZMYM2 | Sponge network | 0.92 | 0 | 0.279 | 0.01273 | 0.408 |
| 18945 | LINC00174 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-374b-3p;hsa-miR-582-5p | 17 | CREB1 | Sponge network | 1.253 | 0 | 0.203 | 0.00537 | 0.408 |
| 18946 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-363-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TRIM13 | Sponge network | 0.421 | 0.00084 | 0.183 | 0.06968 | 0.408 |
| 18947 | RP4-605O3.4 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p | 11 | ATRX | Sponge network | 0.262 | 0.0475 | 0.261 | 0.04408 | 0.408 |
| 18948 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 13 | PSIP1 | Sponge network | -0.031 | 0.89063 | -0.005 | 0.96897 | 0.408 |
| 18949 | C4B-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-96-5p | 14 | RASSF8 | Sponge network | 2.306 | 9.0E-5 | -0.122 | 0.65591 | 0.408 |
| 18950 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p | 26 | SASH1 | Sponge network | -2.62 | 0 | -0.502 | 0.00175 | 0.408 |
| 18951 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | PLA2R1 | Sponge network | 0.194 | 0.64278 | -0.265 | 0.24676 | 0.408 |
| 18952 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 12 | BCL6 | Sponge network | -1.582 | 8.0E-5 | -0.211 | 0.19489 | 0.408 |
| 18953 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-let-7i-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-5p | 15 | TGFBR3 | Sponge network | -2.076 | 0.00548 | -1.173 | 1.0E-5 | 0.408 |
| 18954 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-370-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | JAZF1 | Sponge network | 0.082 | 0.55654 | -0.377 | 0.02677 | 0.408 |
| 18955 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-337-3p;hsa-miR-424-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | HOOK3 | Sponge network | 0.848 | 0 | 0.273 | 0.09957 | 0.408 |
| 18956 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | DAB2 | Sponge network | 0.532 | 0.00505 | 0.431 | 0.0113 | 0.408 |
| 18957 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-542-3p | 14 | HLF | Sponge network | -0.301 | 0.06597 | -2.443 | 0 | 0.408 |
| 18958 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p | 10 | TBX18 | Sponge network | -0.473 | 0.07602 | 0.843 | 0.02945 | 0.408 |
| 18959 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | 0.246 | 0.59912 | -0.615 | 0.01482 | 0.408 |
| 18960 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-660-5p | 25 | TIMP3 | Sponge network | -0.773 | 0.04073 | -0.546 | 0.02307 | 0.408 |
| 18961 | SNX29P2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | RASSF8 | Sponge network | 0.4 | 0.14611 | -0.122 | 0.65591 | 0.408 |
| 18962 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-5p | 18 | RASSF2 | Sponge network | -1.255 | 0.00981 | -0.273 | 0.21961 | 0.408 |
| 18963 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | PDS5B | Sponge network | 0.799 | 1.0E-5 | 0.259 | 0.00962 | 0.408 |
| 18964 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-940 | 21 | NEDD4 | Sponge network | 0.056 | 0.81195 | 0.02 | 0.89834 | 0.408 |
| 18965 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-93-5p | 16 | TXNIP | Sponge network | -0.615 | 0.00015 | -1.277 | 0 | 0.408 |
| 18966 | WDFY3-AS2 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 32 | RIMS2 | Sponge network | -0.942 | 0 | -1.365 | 0.00208 | 0.408 |
| 18967 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | RET | Sponge network | 0.572 | 0.01722 | -0.833 | 0.03517 | 0.408 |
| 18968 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-92a-3p | 34 | ZFHX4 | Sponge network | 0.199 | 0.38015 | 0.018 | 0.95574 | 0.408 |
| 18969 | PWAR6 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 13 | SPTBN4 | Sponge network | -1.575 | 6.0E-5 | -0.786 | 0.01281 | 0.408 |
| 18970 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p | 18 | TBX18 | Sponge network | -0.154 | 0.78939 | 0.843 | 0.02945 | 0.408 |
| 18971 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 31 | ABI2 | Sponge network | -0.342 | 0.02288 | 0.175 | 0.14787 | 0.408 |
| 18972 | FAM66C |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 32 | RIMS2 | Sponge network | -0.901 | 0.00019 | -1.365 | 0.00208 | 0.408 |
| 18973 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 19 | LRRC7 | Sponge network | -1.645 | 0 | -1.341 | 0.01193 | 0.408 |
| 18974 | LINC00641 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 27 | LRIG1 | Sponge network | -0.222 | 0.32445 | -0.537 | 0.00588 | 0.408 |
| 18975 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-98-5p | 13 | ITGB3 | Sponge network | -0.154 | 0.78939 | -0.011 | 0.95751 | 0.408 |
| 18976 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | HACE1 | Sponge network | 0.083 | 0.66405 | -0.072 | 0.55239 | 0.408 |
| 18977 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | TGFBR2 | Sponge network | -0.942 | 0 | -0.243 | 0.11408 | 0.408 |
| 18978 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-5p | 36 | RBMS3 | Sponge network | -1.645 | 0 | -0.519 | 0.03551 | 0.408 |
| 18979 | CTD-2314G24.2 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TPO | Sponge network | 0.246 | 0.59912 | -1.87 | 2.0E-5 | 0.408 |
| 18980 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p | 15 | ERG | Sponge network | -0.154 | 0.78939 | 0.002 | 0.99011 | 0.408 |
| 18981 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p | 10 | SYNM | Sponge network | -1.14 | 0.03513 | -2.309 | 1.0E-5 | 0.408 |
| 18982 | RP11-597D13.9 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 12 | DNMT3A | Sponge network | 1.302 | 7.0E-5 | 0.302 | 0.0262 | 0.408 |
| 18983 | RASSF8-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | 0.335 | 0.20596 | 0.524 | 0.00017 | 0.408 |
| 18984 | RP5-1198O20.4 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | HERC1 | Sponge network | 0.997 | 0.00066 | -0.196 | 0.08544 | 0.408 |
| 18985 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | HIVEP1 | Sponge network | 0.421 | 0.00084 | 0.368 | 0.00064 | 0.408 |
| 18986 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | TSHZ3 | Sponge network | -2.915 | 0.00015 | -0.556 | 0.01516 | 0.408 |
| 18987 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | LRIG1 | Sponge network | -0.611 | 0.09607 | -0.537 | 0.00588 | 0.408 |
| 18988 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p | 14 | RET | Sponge network | -0.303 | 0.21002 | -0.833 | 0.03517 | 0.408 |
| 18989 | RP11-359B12.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 23 | NRK | Sponge network | 0.064 | 0.72934 | 0.656 | 0.19217 | 0.408 |
| 18990 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | CREB3L2 | Sponge network | -1.575 | 6.0E-5 | -0.525 | 2.0E-5 | 0.408 |
| 18991 | FAM66C |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | PDE4DIP | Sponge network | -0.901 | 0.00019 | -0.282 | 0.0257 | 0.408 |
| 18992 | RP11-678G14.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p | 12 | RELN | Sponge network | -4.477 | 0 | -2.403 | 0 | 0.408 |
| 18993 | RP11-867G23.10 |
hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-423-5p;hsa-miR-424-5p | 10 | CSDE1 | Sponge network | -2.531 | 0 | -0.493 | 0 | 0.408 |
| 18994 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-485-3p | 13 | TMTC2 | Sponge network | -0.611 | 0.09607 | -0.215 | 0.18248 | 0.408 |
| 18995 | AC009948.5 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PRKCB | Sponge network | 0.532 | 0.00505 | -1.313 | 2.0E-5 | 0.408 |
| 18996 | LINC00672 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 50 | DST | Sponge network | -0.227 | 0.31657 | -0.713 | 0.00096 | 0.408 |
| 18997 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 21 | FAT4 | Sponge network | -0.942 | 0 | -0.354 | 0.14694 | 0.408 |
| 18998 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | RAP1A | Sponge network | -0.769 | 0.00035 | -0.929 | 0 | 0.408 |
| 18999 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 33 | RICTOR | Sponge network | 0.417 | 0.00445 | 0.388 | 0.00188 | 0.408 |
| 19000 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 25 | TIMP3 | Sponge network | -0.793 | 7.0E-5 | -0.546 | 0.02307 | 0.408 |
| 19001 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p | 22 | RICTOR | Sponge network | 0.419 | 0.0319 | 0.388 | 0.00188 | 0.408 |
| 19002 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | IGSF10 | Sponge network | -0.376 | 0.00337 | -1.751 | 1.0E-5 | 0.408 |
| 19003 | RP11-574K11.29 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-337-3p | 10 | RASAL2 | Sponge network | 0.997 | 0 | 0.869 | 0 | 0.408 |
| 19004 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TMEFF2 | Sponge network | 0.246 | 0.59912 | -2.426 | 0 | 0.408 |
| 19005 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-590-3p | 20 | HIP1 | Sponge network | 0.392 | 0.0278 | 0.774 | 0 | 0.408 |
| 19006 | FENDRR |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-708-5p | 19 | FAM135B | Sponge network | -1.281 | 0.00012 | -3.978 | 0 | 0.408 |
| 19007 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 14 | CHD1 | Sponge network | 0.705 | 0.00014 | 0.322 | 0.00215 | 0.408 |
| 19008 | RP5-1074L1.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 16 | SLC5A3 | Sponge network | 1.923 | 0 | 0.493 | 5.0E-5 | 0.408 |
| 19009 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-5p | 31 | TGFBR3 | Sponge network | 0.331 | 0.57247 | -1.173 | 1.0E-5 | 0.408 |
| 19010 | LPP-AS2 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-577;hsa-miR-589-3p;hsa-miR-769-5p | 22 | SYNPO2 | Sponge network | -0.18 | 0.20343 | -2.342 | 0 | 0.408 |
| 19011 | MIR22HG |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-590-5p | 11 | TLL1 | Sponge network | -1.047 | 0 | -1.195 | 3.0E-5 | 0.408 |
| 19012 | FENDRR |
hsa-miR-130b-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | ROBO2 | Sponge network | -1.281 | 0.00012 | 0.092 | 0.78768 | 0.408 |
| 19013 | ZRANB2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | ZMYND11 | Sponge network | -0.376 | 0.00337 | -0.2 | 0.05449 | 0.408 |
| 19014 | RP11-774O3.3 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 49 | PPM1L | Sponge network | -0.487 | 0.02157 | -0.537 | 0.00616 | 0.408 |
| 19015 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | SPG20 | Sponge network | -0.465 | 0.04703 | -1.16 | 0 | 0.408 |
| 19016 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-500a-5p | 10 | TRIM13 | Sponge network | 0.082 | 0.55397 | 0.183 | 0.06968 | 0.408 |
| 19017 | RP11-254F7.2 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p | 13 | SFRP1 | Sponge network | 0.176 | 0.37402 | -3.566 | 0 | 0.408 |
| 19018 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 18 | PTGFR | Sponge network | 0.064 | 0.72934 | -0.233 | 0.45421 | 0.408 |
| 19019 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | PRKAB2 | Sponge network | -0.487 | 0.02157 | -0.367 | 0.01371 | 0.408 |
| 19020 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940;hsa-miR-98-5p | 37 | PBX1 | Sponge network | 0.101 | 0.60919 | -1.206 | 0 | 0.408 |
| 19021 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-625-5p | 34 | ABI2 | Sponge network | 0.056 | 0.81195 | 0.175 | 0.14787 | 0.408 |
| 19022 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 46 | FAT3 | Sponge network | 0.16 | 0.50785 | -1.601 | 0.00051 | 0.408 |
| 19023 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-539-5p;hsa-miR-582-5p | 17 | MOB1B | Sponge network | 1.093 | 0 | 0.197 | 0.15266 | 0.408 |
| 19024 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | MOB1B | Sponge network | 0.848 | 0 | 0.197 | 0.15266 | 0.408 |
| 19025 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 14 | ITCH | Sponge network | 0.537 | 0.00139 | 0.084 | 0.34058 | 0.408 |
| 19026 | RP11-53O19.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | -0.405 | 0.22013 | 0.524 | 0.00017 | 0.408 |
| 19027 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | BCLAF1 | Sponge network | 0.91 | 0 | 0.211 | 0.00684 | 0.408 |
| 19028 | MIR22HG |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-5p | 16 | CITED2 | Sponge network | -1.047 | 0 | -1.545 | 0 | 0.407 |
| 19029 | LINC01018 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | GLI3 | Sponge network | -2.915 | 0.00015 | -0.615 | 0.01482 | 0.407 |
| 19030 | ARHGEF26-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 22 | MACF1 | Sponge network | -2.46 | 0 | 0.019 | 0.90335 | 0.407 |
| 19031 | FENDRR |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-98-5p | 16 | ITGA5 | Sponge network | -1.281 | 0.00012 | 0.452 | 0.03524 | 0.407 |
| 19032 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 40 | EPHA7 | Sponge network | 0.083 | 0.66405 | -2.197 | 3.0E-5 | 0.407 |
| 19033 | MALAT1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 14 | CEP170 | Sponge network | 0.782 | 0.00016 | 0.943 | 0 | 0.407 |
| 19034 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 25 | LRRC7 | Sponge network | -0.222 | 0.32445 | -1.341 | 0.01193 | 0.407 |
| 19035 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 32 | CBL | Sponge network | 0.532 | 0.00505 | 0.291 | 0.01267 | 0.407 |
| 19036 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-93-5p | 11 | ACSL6 | Sponge network | -0.31 | 0.33871 | -1.593 | 0 | 0.407 |
| 19037 | RP11-203M5.7 |
hsa-let-7d-5p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-495-3p | 13 | RICTOR | Sponge network | 1.684 | 1.0E-5 | 0.388 | 0.00188 | 0.407 |
| 19038 | GAS6-AS2 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-92a-3p | 13 | ITGA5 | Sponge network | 0.16 | 0.50785 | 0.452 | 0.03524 | 0.407 |
| 19039 | HOXB-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-501-3p | 12 | SEMA3C | Sponge network | -0.154 | 0.53954 | -0.272 | 0.31153 | 0.407 |
| 19040 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 25 | EPHA3 | Sponge network | -0.125 | 0.49414 | 0.185 | 0.53588 | 0.407 |
| 19041 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-3p | 25 | FZD3 | Sponge network | 0.799 | 1.0E-5 | 0.104 | 0.60316 | 0.407 |
| 19042 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-590-3p | 12 | RB1CC1 | Sponge network | 0.349 | 0.01695 | 0.175 | 0.12559 | 0.407 |
| 19043 | LINC00654 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | GALNT13 | Sponge network | 0.374 | 0.20054 | -0.857 | 0.07439 | 0.407 |
| 19044 | LINC00865 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 18 | NCAM2 | Sponge network | -1.249 | 7.0E-5 | -0.482 | 0.22565 | 0.407 |
| 19045 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | NAV3 | Sponge network | 0.61 | 0.01593 | 0.212 | 0.47729 | 0.407 |
| 19046 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | CEP170 | Sponge network | 1.089 | 0 | 0.943 | 0 | 0.407 |
| 19047 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 25 | EPHA3 | Sponge network | -0.773 | 0.04073 | 0.185 | 0.53588 | 0.407 |
| 19048 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 18 | MYBL1 | Sponge network | 0.873 | 0 | 0.494 | 0.02152 | 0.407 |
| 19049 | RP11-360F5.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-877-5p;hsa-miR-93-3p | 18 | BMPR2 | Sponge network | 1.867 | 0 | 0.206 | 0.0635 | 0.407 |
| 19050 | LINC00865 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 10 | ADH1B | Sponge network | -1.249 | 7.0E-5 | -3.716 | 0 | 0.407 |
| 19051 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-654-3p | 24 | FBXO32 | Sponge network | -0.031 | 0.89063 | -0.495 | 0.07561 | 0.407 |
| 19052 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 52 | FOXP1 | Sponge network | -2.452 | 0 | 0.042 | 0.74368 | 0.407 |
| 19053 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-369-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PEG3 | Sponge network | -0.342 | 0.02288 | -1.74 | 0 | 0.407 |
| 19054 | AC005154.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 26 | TBL1XR1 | Sponge network | 0.848 | 0 | 0.578 | 0 | 0.407 |
| 19055 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-340-5p | 25 | NTRK2 | Sponge network | -0.693 | 0.05629 | -0.736 | 0.06495 | 0.407 |
| 19056 | RP11-53O19.1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-93-3p;hsa-miR-935 | 12 | STAT5B | Sponge network | -0.405 | 0.22013 | -0.182 | 0.1082 | 0.407 |
| 19057 | GABPB1-AS1 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p | 18 | TSC1 | Sponge network | 1.11 | 0 | 0.103 | 0.26071 | 0.407 |
| 19058 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p | 21 | RNF144A | Sponge network | -0.154 | 0.78939 | 0.594 | 0.0009 | 0.407 |
| 19059 | RP11-35G9.3 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 10 | NGFR | Sponge network | 0.657 | 4.0E-5 | -1.271 | 0.01058 | 0.407 |
| 19060 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-582-5p | 21 | MOB1B | Sponge network | 1.03 | 0 | 0.197 | 0.15266 | 0.407 |
| 19061 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | SOX5 | Sponge network | -0.487 | 0.02157 | -1.091 | 4.0E-5 | 0.407 |
| 19062 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -0.351 | 0.11358 | -0.243 | 0.11408 | 0.407 |
| 19063 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p | 10 | ELMO1 | Sponge network | -0.611 | 0.09607 | 0.04 | 0.83147 | 0.407 |
| 19064 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | DYNC1I1 | Sponge network | 0.381 | 0.25967 | -1.12 | 0.00107 | 0.407 |
| 19065 | RP11-819C21.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | RASSF8 | Sponge network | 0.537 | 0.00139 | -0.122 | 0.65591 | 0.407 |
| 19066 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-3p | 52 | IL6ST | Sponge network | -0.777 | 0.16667 | -0.402 | 0.00676 | 0.407 |
| 19067 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 23 | CHD2 | Sponge network | 0.311 | 0.10344 | 0.049 | 0.55371 | 0.407 |
| 19068 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 26 | SCN9A | Sponge network | -0.769 | 0.00035 | -0.525 | 0.10593 | 0.407 |
| 19069 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p | 11 | PRKAA2 | Sponge network | -1.483 | 9.0E-5 | -2.462 | 0 | 0.407 |
| 19070 | KB-1410C5.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-744-3p;hsa-miR-92a-3p | 18 | DST | Sponge network | 0.959 | 0 | -0.713 | 0.00096 | 0.407 |
| 19071 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 21 | TCF4 | Sponge network | -0.187 | 0.23787 | 0.016 | 0.92271 | 0.407 |
| 19072 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | RASSF2 | Sponge network | -2.117 | 0.00161 | -0.273 | 0.21961 | 0.407 |
| 19073 | HOXB-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429 | 11 | FMNL3 | Sponge network | -0.154 | 0.53954 | 0.555 | 4.0E-5 | 0.407 |
| 19074 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-96-5p | 30 | FBXO32 | Sponge network | -1.349 | 0.00017 | -0.495 | 0.07561 | 0.407 |
| 19075 | RP11-458F8.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30c-5p;hsa-miR-326 | 13 | TBL1XR1 | Sponge network | 1.518 | 0 | 0.578 | 0 | 0.407 |
| 19076 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 28 | ST6GALNAC3 | Sponge network | -0.896 | 0.00099 | -0.987 | 0 | 0.407 |
| 19077 | RP11-13K12.1 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p | 11 | LZTS1 | Sponge network | -0.154 | 0.78939 | 1.664 | 0 | 0.407 |
| 19078 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | CBFA2T3 | Sponge network | -1.697 | 0.00889 | -1.221 | 1.0E-5 | 0.407 |
| 19079 | PART1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p | 17 | FNBP1 | Sponge network | -2.925 | 0 | -0.969 | 4.0E-5 | 0.407 |
| 19080 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | TACC1 | Sponge network | -0.187 | 0.23787 | -0.362 | 0.05406 | 0.407 |
| 19081 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-7-1-3p | 22 | TGFBR3 | Sponge network | -0.187 | 0.23787 | -1.173 | 1.0E-5 | 0.407 |
| 19082 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-505-5p;hsa-miR-766-3p | 18 | TBL1XR1 | Sponge network | 0.793 | 0 | 0.578 | 0 | 0.407 |
| 19083 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 17 | PTPN11 | Sponge network | 0.488 | 0.00084 | 0.431 | 2.0E-5 | 0.407 |
| 19084 | AC008746.12 |
hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374a-3p | 11 | MDM4 | Sponge network | 2.279 | 0 | 0.345 | 0.00762 | 0.407 |
| 19085 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CACNA2D1 | Sponge network | -0.737 | 0.10822 | -0.846 | 0.00675 | 0.407 |
| 19086 | TPTEP1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 14 | EHD3 | Sponge network | -1.216 | 0 | -0.484 | 0.01747 | 0.407 |
| 19087 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PTPRB | Sponge network | -0.49 | 0.04946 | 0.105 | 0.47122 | 0.407 |
| 19088 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-935 | 27 | EBF1 | Sponge network | -0.096 | 0.67958 | -0.384 | 0.08856 | 0.407 |
| 19089 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | NAV3 | Sponge network | 0.898 | 1.0E-5 | 0.212 | 0.47729 | 0.407 |
| 19090 | AC010226.4 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | ZFP36L1 | Sponge network | -0.811 | 2.0E-5 | -0.505 | 8.0E-5 | 0.407 |
| 19091 | RAMP2-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -0.513 | 0.04703 | -0.966 | 0.00035 | 0.407 |
| 19092 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-625-5p;hsa-miR-93-5p | 27 | ANK2 | Sponge network | -0.18 | 0.20343 | -1.603 | 1.0E-5 | 0.407 |
| 19093 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 29 | DMD | Sponge network | 0.61 | 0.01593 | -1.506 | 0 | 0.407 |
| 19094 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-93-3p | 14 | CNTNAP1 | Sponge network | -0.615 | 0.00015 | 0.358 | 0.13727 | 0.407 |
| 19095 | RP11-21A7A.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-98-5p | 19 | CADM2 | Sponge network | -1.116 | 0.23686 | -3.782 | 0 | 0.407 |
| 19096 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ZNF536 | Sponge network | -0.323 | 0.01372 | -3.115 | 0 | 0.407 |
| 19097 | PCA3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-629-3p | 11 | ITGA5 | Sponge network | -0.709 | 0.18168 | 0.452 | 0.03524 | 0.407 |
| 19098 | AGAP11 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p | 18 | MRVI1 | Sponge network | -1.645 | 0 | -1.051 | 0.00022 | 0.407 |
| 19099 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-493-5p | 15 | CREBBP | Sponge network | 0.614 | 1.0E-5 | 0.126 | 0.15186 | 0.407 |
| 19100 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | RPS6KA3 | Sponge network | 0.673 | 0 | 0.256 | 0.02419 | 0.407 |
| 19101 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-940 | 33 | NTRK2 | Sponge network | 0.494 | 0.00339 | -0.736 | 0.06495 | 0.407 |
| 19102 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PBRM1 | Sponge network | 1.153 | 0 | -0.029 | 0.78601 | 0.407 |
| 19103 | RP11-359B12.2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 44 | AFF1 | Sponge network | 0.064 | 0.72934 | -0.384 | 0.00053 | 0.407 |
| 19104 | RP11-2H8.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 13 | SHPRH | Sponge network | 1.089 | 0 | 0.098 | 0.40994 | 0.407 |
| 19105 | TP73-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-589-3p | 19 | KCNJ5 | Sponge network | -0.563 | 0.02545 | -0.281 | 0.3339 | 0.407 |
| 19106 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-766-3p;hsa-miR-96-5p | 30 | CADM2 | Sponge network | -0.517 | 0.02935 | -3.782 | 0 | 0.407 |
| 19107 | MEG3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-940 | 16 | FAM172A | Sponge network | 0.249 | 0.35902 | -0.57 | 0 | 0.407 |
| 19108 | RP1-151F17.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | SIRT1 | Sponge network | 0.222 | 0.29133 | 0.109 | 0.2593 | 0.407 |
| 19109 | RP11-554A11.4 |
hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3913-5p;hsa-miR-421 | 16 | RAP1A | Sponge network | -0.583 | 0.14589 | -0.929 | 0 | 0.407 |
| 19110 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 74 | KIF1B | Sponge network | -0.739 | 0.0574 | -0.118 | 0.32258 | 0.407 |
| 19111 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-93-5p | 40 | PPM1A | Sponge network | -0.793 | 7.0E-5 | -0.623 | 0 | 0.407 |
| 19112 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | NEDD4 | Sponge network | -0.222 | 0.32445 | 0.02 | 0.89834 | 0.407 |
| 19113 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-96-5p | 20 | RECK | Sponge network | -0.18 | 0.20343 | -1.043 | 0 | 0.407 |
| 19114 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | CNTN1 | Sponge network | -0.021 | 0.93598 | -2.594 | 0 | 0.407 |
| 19115 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-671-5p | 15 | ADARB1 | Sponge network | 1.344 | 0 | -0.335 | 0.06631 | 0.407 |
| 19116 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | MITF | Sponge network | 0.335 | 0.20596 | -0.769 | 0.00022 | 0.407 |
| 19117 | RP1-151F17.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | AKAP6 | Sponge network | 0.222 | 0.29133 | -1.586 | 3.0E-5 | 0.407 |
| 19118 | LINC00265 |
hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | AKAP11 | Sponge network | 1.07 | 0 | 0.196 | 0.09825 | 0.407 |
| 19119 | TUG1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 13 | AKAP11 | Sponge network | 0.972 | 0 | 0.196 | 0.09825 | 0.407 |
| 19120 | RP11-212P7.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 60 | DST | Sponge network | 0.219 | 0.1516 | -0.713 | 0.00096 | 0.407 |
| 19121 | RP11-774O3.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PIK3CA | Sponge network | -0.487 | 0.02157 | 0.195 | 0.06908 | 0.407 |
| 19122 | RP11-67L2.2 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p | 10 | MN1 | Sponge network | 0.72 | 0.0001 | -0.358 | 0.24172 | 0.407 |
| 19123 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p | 21 | CBFA2T3 | Sponge network | -0.842 | 0.01361 | -1.221 | 1.0E-5 | 0.407 |
| 19124 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-542-3p | 20 | ADAMTSL3 | Sponge network | -0.773 | 0.04073 | -1.559 | 0 | 0.407 |
| 19125 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p | 41 | ADARB1 | Sponge network | -0.738 | 0.21233 | -0.335 | 0.06631 | 0.407 |
| 19126 | DLGAP1-AS2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 12 | PHF6 | Sponge network | 2.406 | 0 | 0.785 | 0 | 0.407 |
| 19127 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 25 | PCDH9 | Sponge network | 0.082 | 0.55654 | -1.823 | 4.0E-5 | 0.407 |
| 19128 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 14 | HERC2 | Sponge network | 0.064 | 0.72934 | 0.114 | 0.30638 | 0.407 |
| 19129 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 28 | GRIN2A | Sponge network | 0.101 | 0.60919 | -2.161 | 0 | 0.407 |
| 19130 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-654-3p | 20 | RICTOR | Sponge network | 0.856 | 0 | 0.388 | 0.00188 | 0.407 |
| 19131 | RP11-13K12.5 |
hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 17 | WWTR1 | Sponge network | -0.318 | 0.65847 | -0.345 | 0.11997 | 0.407 |
| 19132 | RP11-983P16.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-935 | 14 | ARNT2 | Sponge network | 0.41 | 0.02214 | -0.332 | 0.25851 | 0.407 |
| 19133 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 20 | SLC5A3 | Sponge network | 0.421 | 0.00084 | 0.493 | 5.0E-5 | 0.407 |
| 19134 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 26 | TSHZ3 | Sponge network | -0.227 | 0.31657 | -0.556 | 0.01516 | 0.407 |
| 19135 | AC090587.5 |
hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p | 13 | GAS7 | Sponge network | -0.088 | 0.65011 | -0.555 | 0.01602 | 0.407 |
| 19136 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-424-5p | 10 | GALNT13 | Sponge network | -1.654 | 0.00144 | -0.857 | 0.07439 | 0.407 |
| 19137 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | ST6GALNAC3 | Sponge network | -0.942 | 0 | -0.987 | 0 | 0.407 |
| 19138 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ATRX | Sponge network | 1.205 | 0 | 0.261 | 0.04408 | 0.407 |
| 19139 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 32 | PDGFRA | Sponge network | -0.896 | 0.00099 | -0.372 | 0.0037 | 0.407 |
| 19140 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 19 | IGF1 | Sponge network | 0.178 | 0.29106 | -0.204 | 0.5898 | 0.407 |
| 19141 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | ERCC4 | Sponge network | 0.385 | 0.10067 | 0.126 | 0.15266 | 0.407 |
| 19142 | RP11-531A24.5 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p | 12 | CSDE1 | Sponge network | 0.75 | 0.00054 | -0.493 | 0 | 0.407 |
| 19143 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p | 17 | TRIM33 | Sponge network | 0.614 | 1.0E-5 | 0.402 | 7.0E-5 | 0.407 |
| 19144 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 22 | ZMYND11 | Sponge network | -0.405 | 0.22013 | -0.2 | 0.05449 | 0.407 |
| 19145 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-7-1-3p | 23 | PRKAB2 | Sponge network | -0.246 | 0.35034 | -0.367 | 0.01371 | 0.407 |
| 19146 | RP11-401P9.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-93-5p | 16 | EZH1 | Sponge network | -0.384 | 0.40652 | -0.564 | 1.0E-5 | 0.407 |
| 19147 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p | 32 | MAF | Sponge network | -0.901 | 0.00019 | -1.257 | 0 | 0.407 |
| 19148 | LINC01001 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326 | 10 | NF1 | Sponge network | 1.055 | 0 | 0.51 | 0 | 0.407 |
| 19149 | AC004893.11 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | NF1 | Sponge network | 1.317 | 0 | 0.51 | 0 | 0.407 |
| 19150 | LINC00654 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-940 | 36 | PTPRT | Sponge network | 0.374 | 0.20054 | -0.907 | 0.03727 | 0.407 |
| 19151 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | PLSCR4 | Sponge network | -0.726 | 0.00132 | -0.474 | 0.03833 | 0.407 |
| 19152 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DACT1 | Sponge network | 0.61 | 0.01593 | 0.783 | 0.00072 | 0.407 |
| 19153 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 11 | KIT | Sponge network | -0.693 | 0.05629 | -2.184 | 0 | 0.407 |
| 19154 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p | 18 | SPG20 | Sponge network | -0.154 | 0.78939 | -1.16 | 0 | 0.407 |
| 19155 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 37 | NCOA1 | Sponge network | -1.111 | 0.0003 | -0.432 | 0 | 0.407 |
| 19156 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 43 | PAFAH1B1 | Sponge network | 0.083 | 0.66405 | -0.429 | 0 | 0.407 |
| 19157 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-493-5p | 10 | NR2C2 | Sponge network | 0.706 | 0 | 0.21 | 0.03224 | 0.407 |
| 19158 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-942-5p | 13 | GPC6 | Sponge network | -0.517 | 0.02935 | -0.054 | 0.81465 | 0.407 |
| 19159 | RP11-10N23.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-625-5p | 11 | ABI2 | Sponge network | 1.134 | 0 | 0.175 | 0.14787 | 0.407 |
| 19160 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-93-5p | 26 | MAF | Sponge network | -0.465 | 0.04703 | -1.257 | 0 | 0.407 |
| 19161 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 13 | FSTL5 | Sponge network | -0.777 | 0.1134 | -1.3 | 0.01619 | 0.407 |
| 19162 | ZNF667-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | -0.976 | 0.00025 | -0.366 | 0.01036 | 0.407 |
| 19163 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 25 | TP53INP1 | Sponge network | -1.575 | 6.0E-5 | -0.496 | 0.00064 | 0.407 |
| 19164 | LINC00969 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-486-5p;hsa-miR-495-3p | 15 | PHF6 | Sponge network | 0.683 | 1.0E-5 | 0.785 | 0 | 0.407 |
| 19165 | RASSF8-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | ETV1 | Sponge network | 0.335 | 0.20596 | -0.096 | 0.67674 | 0.407 |
| 19166 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 27 | SCN9A | Sponge network | -2.117 | 0.00161 | -0.525 | 0.10593 | 0.407 |
| 19167 | RP11-359B12.2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 29 | DLC1 | Sponge network | 0.064 | 0.72934 | -0.382 | 0.05038 | 0.407 |
| 19168 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 44 | MCC | Sponge network | -2.925 | 0 | -1.005 | 0 | 0.407 |
| 19169 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-493-5p | 27 | RUNX1T1 | Sponge network | -0.829 | 0.01343 | -0.344 | 0.2374 | 0.407 |
| 19170 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | NEDD4 | Sponge network | 0.532 | 0.00505 | 0.02 | 0.89834 | 0.407 |
| 19171 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-935 | 43 | DIP2C | Sponge network | -0.615 | 0.00015 | -0.436 | 0.01713 | 0.407 |
| 19172 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 36 | ZFX | Sponge network | 0.75 | 0.00054 | 0.09 | 0.42563 | 0.407 |
| 19173 | CTD-2089N3.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-455-5p | 14 | BCL2 | Sponge network | -0.314 | 0.62033 | -0.899 | 0 | 0.407 |
| 19174 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p | 13 | ELMO1 | Sponge network | -0.323 | 0.01372 | 0.04 | 0.83147 | 0.407 |
| 19175 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p | 15 | RELN | Sponge network | -0.154 | 0.78939 | -2.403 | 0 | 0.406 |
| 19176 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 34 | HLF | Sponge network | 0.083 | 0.66405 | -2.443 | 0 | 0.406 |
| 19177 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | DMD | Sponge network | -0.663 | 0.10887 | -1.506 | 0 | 0.406 |
| 19178 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 21 | LRRC7 | Sponge network | -0.405 | 0.22013 | -1.341 | 0.01193 | 0.406 |
| 19179 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | ZNF208 | Sponge network | -0.612 | 0.00094 | -0.4 | 0.22381 | 0.406 |
| 19180 | HCG22 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-550a-5p | 10 | FAT2 | Sponge network | 0.816 | 0.32368 | -0.278 | 0.44877 | 0.406 |
| 19181 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-629-5p;hsa-miR-93-5p | 30 | MAF | Sponge network | -2.69 | 0.0001 | -1.257 | 0 | 0.406 |
| 19182 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p | 16 | CREBBP | Sponge network | -0.222 | 0.32445 | 0.126 | 0.15186 | 0.406 |
| 19183 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 30 | MXI1 | Sponge network | -0.487 | 0.02157 | -0.987 | 0 | 0.406 |
| 19184 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 14 | NF1 | Sponge network | 1.923 | 0 | 0.51 | 0 | 0.406 |
| 19185 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 16 | MOB1B | Sponge network | 1.205 | 0 | 0.197 | 0.15266 | 0.406 |
| 19186 | CTD-3064M3.3 |
hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-629-3p | 16 | LIFR | Sponge network | -0.469 | 0.08721 | -1.794 | 0 | 0.406 |
| 19187 | SNHG14 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 19 | POU2F2 | Sponge network | -1.111 | 0.0003 | -0.233 | 0.32214 | 0.406 |
| 19188 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 23 | SETBP1 | Sponge network | -0.355 | 0.0077 | -1.302 | 1.0E-5 | 0.406 |
| 19189 | GS1-124K5.11 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-486-5p;hsa-miR-532-3p | 16 | SYNM | Sponge network | 0.494 | 0.00339 | -2.309 | 1.0E-5 | 0.406 |
| 19190 | RP11-360F5.1 |
hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429 | 10 | ABL2 | Sponge network | 2.364 | 0.00134 | 0.82 | 0 | 0.406 |
| 19191 | LINC00702 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 45 | AFF1 | Sponge network | -0.737 | 0.06182 | -0.384 | 0.00053 | 0.406 |
| 19192 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-98-5p | 11 | ST5 | Sponge network | -0.129 | 0.57177 | -0.78 | 0 | 0.406 |
| 19193 | FENDRR |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -1.281 | 0.00012 | -0.252 | 0.15511 | 0.406 |
| 19194 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 31 | DIP2C | Sponge network | 0.082 | 0.55654 | -0.436 | 0.01713 | 0.406 |
| 19195 | RP11-400K9.4 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | GJA1 | Sponge network | -0.611 | 0.09607 | -0.485 | 0.02179 | 0.406 |
| 19196 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p | 29 | SOX5 | Sponge network | 0.199 | 0.38015 | -1.091 | 4.0E-5 | 0.406 |
| 19197 | LINC00578 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p | 15 | PRKCB | Sponge network | 0.811 | 0.03228 | -1.313 | 2.0E-5 | 0.406 |
| 19198 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p | 13 | TXNIP | Sponge network | -0.72 | 0.24239 | -1.277 | 0 | 0.406 |
| 19199 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-93-3p;hsa-miR-940 | 38 | TRPS1 | Sponge network | -0.896 | 0.00099 | -0.191 | 0.44109 | 0.406 |
| 19200 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 15 | CHD1 | Sponge network | 0.959 | 0 | 0.322 | 0.00215 | 0.406 |
| 19201 | AC074289.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-940;hsa-miR-96-5p | 14 | POU2F2 | Sponge network | -0.326 | 0.14314 | -0.233 | 0.32214 | 0.406 |
| 19202 | FZD10-AS1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 10 | SORCS2 | Sponge network | -0.727 | 0.04726 | -0.223 | 0.4253 | 0.406 |
| 19203 | RP11-389C8.2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-589-3p | 19 | KCNJ5 | Sponge network | 0.727 | 3.0E-5 | -0.281 | 0.3339 | 0.406 |
| 19204 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-409-3p;hsa-miR-590-3p | 15 | ATRX | Sponge network | 1.073 | 0.00216 | 0.261 | 0.04408 | 0.406 |
| 19205 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 17 | MYO9A | Sponge network | -0.726 | 0.00132 | -0.147 | 0.32373 | 0.406 |
| 19206 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | MLLT10 | Sponge network | 1.073 | 0.00216 | 0.105 | 0.33292 | 0.406 |
| 19207 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-3p | 29 | DTNA | Sponge network | 0.178 | 0.29106 | -1.648 | 1.0E-5 | 0.406 |
| 19208 | AC011526.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-3127-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-96-5p | 14 | SETBP1 | Sponge network | 0.342 | 0.03129 | -1.302 | 1.0E-5 | 0.406 |
| 19209 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-628-5p;hsa-miR-935 | 32 | FBXO32 | Sponge network | 0.75 | 0.00054 | -0.495 | 0.07561 | 0.406 |
| 19210 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 43 | RBMS3 | Sponge network | -0.727 | 0.04726 | -0.519 | 0.03551 | 0.406 |
| 19211 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-660-5p;hsa-miR-7-1-3p | 21 | EYA4 | Sponge network | -0.246 | 0.35034 | 0.3 | 0.36876 | 0.406 |
| 19212 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429 | 19 | GREM1 | Sponge network | -0.563 | 0.02545 | 0.902 | 0.01691 | 0.406 |
| 19213 | PSMD5-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 21 | BRD3 | Sponge network | 0.569 | 0.00067 | 0.37 | 0.00013 | 0.406 |
| 19214 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p | 16 | DYNC1I1 | Sponge network | 0.889 | 9.0E-5 | -1.12 | 0.00107 | 0.406 |
| 19215 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | NIN | Sponge network | -0.342 | 0.02288 | -0.253 | 0.13794 | 0.406 |
| 19216 | SNHG16 |
hsa-miR-100-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-363-3p | 10 | ATP2A2 | Sponge network | 0.961 | 0 | 0.604 | 0 | 0.406 |
| 19217 | LINC00092 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | RYR2 | Sponge network | -1.886 | 0 | -1.466 | 0.00031 | 0.406 |
| 19218 | PVT1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p | 10 | PHF6 | Sponge network | 1.985 | 0 | 0.785 | 0 | 0.406 |
| 19219 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 51 | AR | Sponge network | 0.61 | 0.01593 | -1.554 | 0 | 0.406 |
| 19220 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | AKAP11 | Sponge network | 1.378 | 0 | 0.196 | 0.09825 | 0.406 |
| 19221 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-577;hsa-miR-590-3p | 12 | NALCN | Sponge network | -0.096 | 0.67958 | 1.068 | 0.00237 | 0.406 |
| 19222 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 21 | FGFR1 | Sponge network | -0.615 | 0.00015 | -0.509 | 0.03957 | 0.406 |
| 19223 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 17 | SSBP2 | Sponge network | -1.375 | 0.08642 | -1.58 | 0 | 0.406 |
| 19224 | GABPB1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 25 | BRD3 | Sponge network | 1.11 | 0 | 0.37 | 0.00013 | 0.406 |
| 19225 | TUG1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p | 20 | CREB1 | Sponge network | 0.972 | 0 | 0.203 | 0.00537 | 0.406 |
| 19226 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 30 | ST6GALNAC3 | Sponge network | -0.096 | 0.67958 | -0.987 | 0 | 0.406 |
| 19227 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 18 | GAS7 | Sponge network | -1.425 | 0.04204 | -0.555 | 0.01602 | 0.406 |
| 19228 | PART1 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29b-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 11 | MLLT11 | Sponge network | -2.925 | 0 | -0.661 | 0.01733 | 0.406 |
| 19229 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 16 | CDHR3 | Sponge network | -0.031 | 0.89063 | -0.977 | 4.0E-5 | 0.406 |
| 19230 | RP4-669L17.10 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-3607-3p | 12 | RASAL2 | Sponge network | 1.093 | 0 | 0.869 | 0 | 0.406 |
| 19231 | SNHG14 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 34 | EYA4 | Sponge network | -1.111 | 0.0003 | 0.3 | 0.36876 | 0.406 |
| 19232 | LINC00957 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p | 10 | AMPH | Sponge network | -0.421 | 0.0114 | -0.916 | 5.0E-5 | 0.406 |
| 19233 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-485-3p;hsa-miR-940 | 18 | MYO9A | Sponge network | -0.465 | 0.04703 | -0.147 | 0.32373 | 0.406 |
| 19234 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 29 | MITF | Sponge network | -0.899 | 6.0E-5 | -0.769 | 0.00022 | 0.406 |
| 19235 | USP3-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-940 | 17 | TAL1 | Sponge network | -0.162 | 0.47687 | -0.462 | 0.00574 | 0.406 |
| 19236 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-874-3p | 32 | MXI1 | Sponge network | -2.452 | 0 | -0.987 | 0 | 0.406 |
| 19237 | CTC-510F12.2 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-335-3p | 11 | MEF2C | Sponge network | -0.691 | 0.00146 | -0.499 | 0.01448 | 0.406 |
| 19238 | HCG11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 14 | NRXN3 | Sponge network | 0.385 | 0.10067 | -0.893 | 0.04186 | 0.406 |
| 19239 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 60 | RUNX1T1 | Sponge network | -0.899 | 6.0E-5 | -0.344 | 0.2374 | 0.406 |
| 19240 | GNG12-AS1 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-550a-3p | 14 | RASSF3 | Sponge network | -0.793 | 7.0E-5 | -0.226 | 0.11691 | 0.406 |
| 19241 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | RASSF8 | Sponge network | -2.915 | 0.00015 | -0.122 | 0.65591 | 0.406 |
| 19242 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-550a-3p;hsa-miR-940 | 31 | MEF2C | Sponge network | 0.082 | 0.55397 | -0.499 | 0.01448 | 0.406 |
| 19243 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-590-3p;hsa-miR-940 | 19 | MYO9A | Sponge network | 0.959 | 0 | -0.147 | 0.32373 | 0.406 |
| 19244 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | PRDM5 | Sponge network | -0.323 | 0.01372 | -1.09 | 0.00014 | 0.406 |
| 19245 | RP11-159D12.2 |
hsa-miR-142-5p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | TMEM30A | Sponge network | 1.254 | 0 | 0.096 | 0.31031 | 0.406 |
| 19246 | C4B-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-5p | 13 | RBMS3 | Sponge network | 2.306 | 9.0E-5 | -0.519 | 0.03551 | 0.406 |
| 19247 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | TGFBR3 | Sponge network | 0.335 | 0.20596 | -1.173 | 1.0E-5 | 0.406 |
| 19248 | LINC00641 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 52 | GRIK3 | Sponge network | -0.222 | 0.32445 | -3.208 | 0 | 0.406 |
| 19249 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | FOXO3 | Sponge network | 1.063 | 0 | -0.04 | 0.72028 | 0.406 |
| 19250 | AC005154.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 23 | NF1 | Sponge network | 0.848 | 0 | 0.51 | 0 | 0.406 |
| 19251 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 14 | NR2C2 | Sponge network | 0.622 | 0.00015 | 0.21 | 0.03224 | 0.406 |
| 19252 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-98-5p | 23 | MITF | Sponge network | -0.227 | 0.31657 | -0.769 | 0.00022 | 0.406 |
| 19253 | GAS5-AS1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p | 12 | MDM4 | Sponge network | 0.598 | 0 | 0.345 | 0.00762 | 0.406 |
| 19254 | CTC-559E9.5 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-576-5p | 17 | ABI2 | Sponge network | 0.594 | 0.00064 | 0.175 | 0.14787 | 0.406 |
| 19255 | RP11-701H24.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 12 | TCF4 | Sponge network | -1.425 | 0.04204 | 0.016 | 0.92271 | 0.406 |
| 19256 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-92a-3p | 22 | GRIN2A | Sponge network | 0.056 | 0.81195 | -2.161 | 0 | 0.406 |
| 19257 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 35 | BNC2 | Sponge network | -0.227 | 0.31657 | -0.491 | 0.09988 | 0.406 |
| 19258 | RP11-360F5.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-3p | 14 | QKI | Sponge network | 1.867 | 0 | -0.333 | 0.04111 | 0.406 |
| 19259 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 25 | TIMP3 | Sponge network | -0.096 | 0.67958 | -0.546 | 0.02307 | 0.406 |
| 19260 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 43 | NTRK2 | Sponge network | -0.323 | 0.01372 | -0.736 | 0.06495 | 0.406 |
| 19261 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 15 | ERCC4 | Sponge network | -1.331 | 3.0E-5 | 0.126 | 0.15266 | 0.406 |
| 19262 | RAMP2-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-502-5p;hsa-miR-590-3p | 13 | FMNL3 | Sponge network | -0.513 | 0.04703 | 0.555 | 4.0E-5 | 0.406 |
| 19263 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p | 27 | SCN9A | Sponge network | -0.513 | 0.04703 | -0.525 | 0.10593 | 0.406 |
| 19264 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | ATRX | Sponge network | 0.919 | 0 | 0.261 | 0.04408 | 0.406 |
| 19265 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 27 | PIK3C2A | Sponge network | 0.683 | 1.0E-5 | 0.061 | 0.53776 | 0.406 |
| 19266 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 31 | HBP1 | Sponge network | -2.452 | 0 | -0.454 | 0 | 0.406 |
| 19267 | TPTEP1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 96 | LPP | Sponge network | -1.216 | 0 | -0.459 | 0.04475 | 0.406 |
| 19268 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-758-3p | 26 | HLF | Sponge network | -0.031 | 0.89063 | -2.443 | 0 | 0.406 |
| 19269 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-3p | 31 | PAFAH1B1 | Sponge network | 0.178 | 0.29106 | -0.429 | 0 | 0.406 |
| 19270 | RP11-196G18.24 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p | 14 | PHF6 | Sponge network | 1.262 | 0 | 0.785 | 0 | 0.406 |
| 19271 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 27 | AR | Sponge network | -4.477 | 0 | -1.554 | 0 | 0.406 |
| 19272 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | ABI2 | Sponge network | -0.031 | 0.89063 | 0.175 | 0.14787 | 0.406 |
| 19273 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 20 | CXXC4 | Sponge network | -0.13 | 0.48734 | -0.433 | 0.21055 | 0.406 |
| 19274 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 32 | GRIA3 | Sponge network | -0.323 | 0.01372 | -1.956 | 0 | 0.406 |
| 19275 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-503-5p;hsa-miR-576-5p;hsa-miR-654-3p | 36 | RICTOR | Sponge network | 0.267 | 0.2937 | 0.388 | 0.00188 | 0.406 |
| 19276 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-495-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 58 | BMPR2 | Sponge network | -0.405 | 0.22013 | 0.206 | 0.0635 | 0.406 |
| 19277 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 19 | SLC5A3 | Sponge network | 1.254 | 0 | 0.493 | 5.0E-5 | 0.406 |
| 19278 | CTC-457L16.2 |
hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-7-1-3p | 12 | PRKCB | Sponge network | 1.153 | 0.00114 | -1.313 | 2.0E-5 | 0.406 |
| 19279 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 19 | ZMYND11 | Sponge network | 0.311 | 0.10344 | -0.2 | 0.05449 | 0.406 |
| 19280 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 27 | GAS7 | Sponge network | -1.375 | 0.08642 | -0.555 | 0.01602 | 0.406 |
| 19281 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | FAT4 | Sponge network | -0.739 | 0.00044 | -0.354 | 0.14694 | 0.406 |
| 19282 | LINC00449 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | TBL1XR1 | Sponge network | 1.525 | 0 | 0.578 | 0 | 0.406 |
| 19283 | NDUFA6-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | MACF1 | Sponge network | -0.355 | 0.0077 | 0.019 | 0.90335 | 0.406 |
| 19284 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-374b-5p | 19 | CDC73 | Sponge network | 1.147 | 0 | 0.094 | 0.20463 | 0.406 |
| 19285 | LINC00641 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | RECK | Sponge network | -0.222 | 0.32445 | -1.043 | 0 | 0.406 |
| 19286 | RP11-401P9.4 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-342-3p;hsa-miR-484 | 15 | ABL1 | Sponge network | -0.384 | 0.40652 | -0.215 | 0.07804 | 0.406 |
| 19287 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | TET1 | Sponge network | -2.452 | 0 | -0.135 | 0.52138 | 0.406 |
| 19288 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 20 | ABCB5 | Sponge network | 0.997 | 0.00066 | -0.956 | 0.04077 | 0.406 |
| 19289 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | CUL5 | Sponge network | 1.055 | 0 | 0.026 | 0.77301 | 0.406 |
| 19290 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 10 | GIGYF2 | Sponge network | 0.91 | 0 | 0.136 | 0.04403 | 0.406 |
| 19291 | LINC00473 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | PRKCB | Sponge network | -1.203 | 0.03881 | -1.313 | 2.0E-5 | 0.406 |
| 19292 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 25 | ZBTB4 | Sponge network | -0.318 | 0.65847 | -0.739 | 0 | 0.406 |
| 19293 | SNX29P2 |
hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-589-3p | 14 | SYNPO2 | Sponge network | 0.4 | 0.14611 | -2.342 | 0 | 0.406 |
| 19294 | RP1-228H13.5 |
hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p | 15 | TBL1XR1 | Sponge network | 1.397 | 0 | 0.578 | 0 | 0.406 |
| 19295 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 0.959 | 0 | 0.114 | 0.30638 | 0.406 |
| 19296 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 50 | PTPRD | Sponge network | 0.335 | 0.20596 | -1.005 | 0.00064 | 0.406 |
| 19297 | PART1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | -2.925 | 0 | -0.474 | 0.03833 | 0.406 |
| 19298 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | SNX29 | Sponge network | 0.727 | 3.0E-5 | -0.34 | 0.01371 | 0.406 |
| 19299 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p | 37 | FOXP1 | Sponge network | -1.645 | 0 | 0.042 | 0.74368 | 0.406 |
| 19300 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p | 11 | EDA2R | Sponge network | -0.096 | 0.67958 | -0.587 | 0.01598 | 0.406 |
| 19301 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 10 | ZHX2 | Sponge network | -1.654 | 0.00144 | -0.192 | 0.18459 | 0.406 |
| 19302 | DNM3OS |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 18 | ATM | Sponge network | 0.572 | 0.01722 | 0.178 | 0.21568 | 0.406 |
| 19303 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-93-5p | 14 | EDA2R | Sponge network | -0.162 | 0.47687 | -0.587 | 0.01598 | 0.406 |
| 19304 | NR2F1-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | -0.49 | 0.04946 | -1.379 | 0 | 0.406 |
| 19305 | RAD51-AS1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-495-3p | 10 | STAG2 | Sponge network | 0.768 | 7.0E-5 | 0.252 | 0.00438 | 0.406 |
| 19306 | HHIP-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SYT4 | Sponge network | -0.737 | 0.10822 | -3.207 | 0 | 0.406 |
| 19307 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-26a-2-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484 | 10 | ABL1 | Sponge network | -1.116 | 0.23686 | -0.215 | 0.07804 | 0.406 |
| 19308 | RP11-967K21.1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-98-5p | 18 | KLF10 | Sponge network | 0.056 | 0.81195 | -0.783 | 0 | 0.406 |
| 19309 | LINC00702 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | TLE4 | Sponge network | -0.737 | 0.06182 | -1.157 | 0 | 0.406 |
| 19310 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-96-5p | 12 | NUAK1 | Sponge network | 2.306 | 9.0E-5 | 0.904 | 0 | 0.406 |
| 19311 | MIR600HG |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p | 16 | TSC1 | Sponge network | 0.594 | 0.41522 | 0.103 | 0.26071 | 0.406 |
| 19312 | RP11-144G6.12 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-590-3p | 16 | ZFX | Sponge network | 0.826 | 1.0E-5 | 0.09 | 0.42563 | 0.406 |
| 19313 | HOXA-AS2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-590-3p | 11 | STARD13 | Sponge network | 0.61 | 0.01593 | -0.187 | 0.27383 | 0.406 |
| 19314 | HOXA-AS2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-942-5p | 11 | ZBTB20 | Sponge network | 0.61 | 0.01593 | -0.122 | 0.57569 | 0.406 |
| 19315 | RP11-360F5.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | BNC2 | Sponge network | 1.867 | 0 | -0.491 | 0.09988 | 0.406 |
| 19316 | CD27-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 23 | ZMYND11 | Sponge network | 0.177 | 0.16681 | -0.2 | 0.05449 | 0.406 |
| 19317 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ABCC9 | Sponge network | -0.08 | 0.90092 | -0.466 | 0.14121 | 0.406 |
| 19318 | C20orf166-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-542-3p | 16 | SEMA3C | Sponge network | -2.69 | 0.0001 | -0.272 | 0.31153 | 0.406 |
| 19319 | HHIP-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-590-3p | 24 | DLC1 | Sponge network | -0.737 | 0.10822 | -0.382 | 0.05038 | 0.406 |
| 19320 | RP11-57H14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 31 | SNX29 | Sponge network | 0.083 | 0.66405 | -0.34 | 0.01371 | 0.406 |
| 19321 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | BNC2 | Sponge network | 0.374 | 0.20054 | -0.491 | 0.09988 | 0.406 |
| 19322 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-577 | 10 | KIT | Sponge network | -0.31 | 0.33871 | -2.184 | 0 | 0.406 |
| 19323 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-93-5p | 15 | LHX6 | Sponge network | -0.769 | 0.00035 | -0.087 | 0.69193 | 0.406 |
| 19324 | RP11-216B9.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 19 | NAV3 | Sponge network | 0.174 | 0.30034 | 0.212 | 0.47729 | 0.406 |
| 19325 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | MCC | Sponge network | 0.16 | 0.50785 | -1.005 | 0 | 0.406 |
| 19326 | RAMP2-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | -0.513 | 0.04703 | -1.733 | 0.00022 | 0.406 |
| 19327 | RP5-1074L1.4 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-654-3p | 11 | DICER1 | Sponge network | 1.923 | 0 | 0.042 | 0.674 | 0.406 |
| 19328 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 21 | UNC80 | Sponge network | -1.375 | 0.08642 | -1.934 | 0 | 0.406 |
| 19329 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 11 | DACT1 | Sponge network | -0.739 | 0.00044 | 0.783 | 0.00072 | 0.406 |
| 19330 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p | 22 | CUL5 | Sponge network | 0.75 | 0.00054 | 0.026 | 0.77301 | 0.405 |
| 19331 | LINC00094 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p | 11 | PRDM5 | Sponge network | 0.253 | 0.09565 | -1.09 | 0.00014 | 0.405 |
| 19332 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-93-5p | 29 | JAZF1 | Sponge network | -0.777 | 0.16667 | -0.377 | 0.02677 | 0.405 |
| 19333 | MIR497HG |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-7-1-3p | 16 | SEMA3C | Sponge network | -1.439 | 4.0E-5 | -0.272 | 0.31153 | 0.405 |
| 19334 | LINC00909 |
hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-654-3p | 15 | PHF6 | Sponge network | 0.363 | 0.00601 | 0.785 | 0 | 0.405 |
| 19335 | RP11-67L2.2 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | PREX2 | Sponge network | 0.72 | 0.0001 | -0.272 | 0.25382 | 0.405 |
| 19336 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-93-5p | 35 | CHD5 | Sponge network | 0.335 | 0.20596 | -0.922 | 0.01521 | 0.405 |
| 19337 | MIR143HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | ERCC4 | Sponge network | -0.739 | 0.0574 | 0.126 | 0.15266 | 0.405 |
| 19338 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | PIK3R1 | Sponge network | -0.976 | 0.00025 | -0.133 | 0.36854 | 0.405 |
| 19339 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 26 | HBP1 | Sponge network | -0.793 | 7.0E-5 | -0.454 | 0 | 0.405 |
| 19340 | MAGI2-AS3 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | SPOP | Sponge network | -0.129 | 0.57177 | -0.419 | 1.0E-5 | 0.405 |
| 19341 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429 | 22 | PTPN14 | Sponge network | 0.16 | 0.50785 | 0.144 | 0.34595 | 0.405 |
| 19342 | RP11-57H14.4 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | INPP4A | Sponge network | 0.083 | 0.66405 | 0.07 | 0.53563 | 0.405 |
| 19343 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | GRIA3 | Sponge network | -0.737 | 0.10822 | -1.956 | 0 | 0.405 |
| 19344 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 41 | CHD5 | Sponge network | 0.374 | 0.20054 | -0.922 | 0.01521 | 0.405 |
| 19345 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | ZMYM2 | Sponge network | 0.683 | 1.0E-5 | 0.279 | 0.01273 | 0.405 |
| 19346 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -1.654 | 0.00144 | -0.252 | 0.15511 | 0.405 |
| 19347 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p | 27 | PBX1 | Sponge network | 0.385 | 0.10067 | -1.206 | 0 | 0.405 |
| 19348 | JAZF1-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | SYT4 | Sponge network | -0.769 | 0.00035 | -3.207 | 0 | 0.405 |
| 19349 | SH3RF3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-877-5p | 19 | IL17RD | Sponge network | 0.889 | 9.0E-5 | -0.019 | 0.94873 | 0.405 |
| 19350 | AC016747.3 |
hsa-miR-107;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | PREX2 | Sponge network | 0.199 | 0.38015 | -0.272 | 0.25382 | 0.405 |
| 19351 | LINC00957 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 34 | TRPS1 | Sponge network | -0.421 | 0.0114 | -0.191 | 0.44109 | 0.405 |
| 19352 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p | 14 | ABL2 | Sponge network | 0.912 | 0.00014 | 0.82 | 0 | 0.405 |
| 19353 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | AFF3 | Sponge network | 0.997 | 0.00066 | -2.373 | 0 | 0.405 |
| 19354 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 29 | GPC6 | Sponge network | -0.769 | 0.00035 | -0.054 | 0.81465 | 0.405 |
| 19355 | AC006547.13 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-628-5p;hsa-miR-940 | 10 | TRIM33 | Sponge network | 1.159 | 0 | 0.402 | 7.0E-5 | 0.405 |
| 19356 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-330-3p;hsa-miR-590-3p | 10 | NRXN3 | Sponge network | 0.174 | 0.30034 | -0.893 | 0.04186 | 0.405 |
| 19357 | RP11-401P9.4 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-935 | 13 | MAPK10 | Sponge network | -0.384 | 0.40652 | -1.774 | 0 | 0.405 |
| 19358 | CTD-2089N3.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-3p | 15 | GAS7 | Sponge network | -0.314 | 0.62033 | -0.555 | 0.01602 | 0.405 |
| 19359 | RP11-400K9.4 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 17 | DOCK4 | Sponge network | -0.611 | 0.09607 | 0.502 | 0.00108 | 0.405 |
| 19360 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MLLT10 | Sponge network | 0.083 | 0.66405 | 0.105 | 0.33292 | 0.405 |
| 19361 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-577;hsa-miR-874-3p;hsa-miR-93-5p | 29 | TACC1 | Sponge network | -0.465 | 0.04703 | -0.362 | 0.05406 | 0.405 |
| 19362 | RP11-182J1.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-484 | 16 | DST | Sponge network | -0.187 | 0.23787 | -0.713 | 0.00096 | 0.405 |
| 19363 | RP4-668J24.2 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-424-5p;hsa-miR-511-5p | 11 | ADAMTSL3 | Sponge network | -1.054 | 0.0706 | -1.559 | 0 | 0.405 |
| 19364 | RP11-2B6.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | BRD3 | Sponge network | 0.539 | 0.03722 | 0.37 | 0.00013 | 0.405 |
| 19365 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | NR2C2 | Sponge network | 0.8 | 0 | 0.21 | 0.03224 | 0.405 |
| 19366 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 19 | IGF2 | Sponge network | -0.49 | 0.04946 | 0.848 | 0.03842 | 0.405 |
| 19367 | FZD10-AS1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | RNF180 | Sponge network | -0.727 | 0.04726 | -1.127 | 1.0E-5 | 0.405 |
| 19368 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | PAFAH1B1 | Sponge network | -0.83 | 0 | -0.429 | 0 | 0.405 |
| 19369 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 30 | MYO9A | Sponge network | -0.777 | 0.1134 | -0.147 | 0.32373 | 0.405 |
| 19370 | RP11-531A24.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-940 | 13 | FAM172A | Sponge network | 0.75 | 0.00054 | -0.57 | 0 | 0.405 |
| 19371 | ASH1L-AS1 |
hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | TBL1XR1 | Sponge network | 0.948 | 0 | 0.578 | 0 | 0.405 |
| 19372 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-877-5p | 27 | TET1 | Sponge network | 0.082 | 0.55654 | -0.135 | 0.52138 | 0.405 |
| 19373 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-340-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 57 | NFIB | Sponge network | -0.323 | 0.01372 | 0.023 | 0.90795 | 0.405 |
| 19374 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 14 | KCNAB1 | Sponge network | -0.942 | 0 | -0.755 | 2.0E-5 | 0.405 |
| 19375 | HOXA-AS2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-542-3p | 17 | SYNM | Sponge network | 0.61 | 0.01593 | -2.309 | 1.0E-5 | 0.405 |
| 19376 | RP11-159D12.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CEP170 | Sponge network | 1.254 | 0 | 0.943 | 0 | 0.405 |
| 19377 | MIR143HG |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | CDH10 | Sponge network | -0.739 | 0.0574 | -2.397 | 0 | 0.405 |
| 19378 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-654-3p | 13 | ITCH | Sponge network | 1.601 | 0 | 0.084 | 0.34058 | 0.405 |
| 19379 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | FOXP1 | Sponge network | 0.064 | 0.72934 | 0.042 | 0.74368 | 0.405 |
| 19380 | LINC00476 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LHFP | Sponge network | -0.615 | 0.00015 | -0.167 | 0.41371 | 0.405 |
| 19381 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 21 | TBL1XR1 | Sponge network | 0.537 | 0.00139 | 0.578 | 0 | 0.405 |
| 19382 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 25 | KCNA6 | Sponge network | -0.612 | 0.00094 | -1.531 | 0.00013 | 0.405 |
| 19383 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-96-5p | 23 | CRTC3 | Sponge network | 0.41 | 0.02214 | 0.164 | 0.16786 | 0.405 |
| 19384 | HOXA-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-421;hsa-miR-590-3p;hsa-miR-940 | 10 | CTNNA3 | Sponge network | 0.61 | 0.01593 | -2.046 | 0.00019 | 0.405 |
| 19385 | MYLK-AS1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-484 | 33 | KIF1B | Sponge network | -1.331 | 3.0E-5 | -0.118 | 0.32258 | 0.405 |
| 19386 | CKMT2-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-877-5p;hsa-miR-940 | 11 | PHOX2B | Sponge network | 0.082 | 0.55654 | -2.68 | 0 | 0.405 |
| 19387 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p | 32 | TRPS1 | Sponge network | 0.176 | 0.37402 | -0.191 | 0.44109 | 0.405 |
| 19388 | RP11-798M19.6 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p | 23 | ASTN1 | Sponge network | -0.612 | 0.00094 | -2.389 | 2.0E-5 | 0.405 |
| 19389 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | NR4A3 | Sponge network | -0.739 | 0.0574 | -1.385 | 0 | 0.405 |
| 19390 | U91328.19 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-624-5p | 10 | SMARCA1 | Sponge network | -0.303 | 0.21002 | -0.684 | 0.00159 | 0.405 |
| 19391 | GNG12-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 17 | ERG | Sponge network | -0.793 | 7.0E-5 | 0.002 | 0.99011 | 0.405 |
| 19392 | RP1-59D14.5 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-511-5p | 15 | ZFHX3 | Sponge network | 1.162 | 5.0E-5 | 0.389 | 0.01363 | 0.405 |
| 19393 | RP11-2B6.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CREB1 | Sponge network | 0.539 | 0.03722 | 0.203 | 0.00537 | 0.405 |
| 19394 | RP11-159D12.2 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | ERCC6 | Sponge network | 1.254 | 0 | 0.23 | 0.015 | 0.405 |
| 19395 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | ARHGAP29 | Sponge network | 0.194 | 0.64278 | 0.131 | 0.42953 | 0.405 |
| 19396 | RP11-298J20.4 |
hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p | 17 | WDFY3 | Sponge network | 0.002 | 0.99057 | -0.201 | 0.0967 | 0.405 |
| 19397 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | ZFHX4 | Sponge network | -0.187 | 0.23787 | 0.018 | 0.95574 | 0.405 |
| 19398 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 16 | PDS5B | Sponge network | 0.494 | 0.00339 | 0.259 | 0.00962 | 0.405 |
| 19399 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NOTCH2NL | Sponge network | 0.72 | 0.0001 | 0.024 | 0.84307 | 0.405 |
| 19400 | PCA3 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | MXI1 | Sponge network | -0.709 | 0.18168 | -0.987 | 0 | 0.405 |
| 19401 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940 | 17 | CRTC3 | Sponge network | 0.16 | 0.50785 | 0.164 | 0.16786 | 0.405 |
| 19402 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-744-5p | 11 | FGFR1 | Sponge network | -0.141 | 0.26434 | -0.509 | 0.03957 | 0.405 |
| 19403 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | KCNAB1 | Sponge network | -0.899 | 6.0E-5 | -0.755 | 2.0E-5 | 0.405 |
| 19404 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | MDM4 | Sponge network | 0.959 | 0 | 0.345 | 0.00762 | 0.405 |
| 19405 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 12 | HACE1 | Sponge network | 0.614 | 1.0E-5 | -0.072 | 0.55239 | 0.405 |
| 19406 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | MYO9A | Sponge network | 0.862 | 0.06213 | -0.147 | 0.32373 | 0.405 |
| 19407 | AC003090.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | FAM133A | Sponge network | -2.117 | 0.00161 | 0.549 | 0.29009 | 0.405 |
| 19408 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | FYN | Sponge network | 0.727 | 3.0E-5 | -0.131 | 0.37051 | 0.405 |
| 19409 | AC106786.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-877-5p | 19 | FAT3 | Sponge network | 1.122 | 0.02989 | -1.601 | 0.00051 | 0.405 |
| 19410 | MALAT1 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | DICER1 | Sponge network | 0.782 | 0.00016 | 0.042 | 0.674 | 0.405 |
| 19411 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 37 | CBX5 | Sponge network | 0.559 | 0.00026 | 0.165 | 0.24446 | 0.405 |
| 19412 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940;hsa-miR-98-5p | 65 | NTRK3 | Sponge network | -0.899 | 6.0E-5 | -1.77 | 3.0E-5 | 0.405 |
| 19413 | GABPB1-AS1 |
hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | TTL | Sponge network | 1.11 | 0 | 0.494 | 1.0E-5 | 0.405 |
| 19414 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-940 | 27 | CHD5 | Sponge network | -1.886 | 0 | -0.922 | 0.01521 | 0.405 |
| 19415 | RP11-434B12.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | NRXN3 | Sponge network | -0.031 | 0.89063 | -0.893 | 0.04186 | 0.405 |
| 19416 | RP11-13K12.5 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-330-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-940 | 11 | SORCS1 | Sponge network | -0.318 | 0.65847 | -3.492 | 0 | 0.405 |
| 19417 | RP11-531A24.5 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 19 | PIK3CA | Sponge network | 0.75 | 0.00054 | 0.195 | 0.06908 | 0.405 |
| 19418 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 22 | AFF3 | Sponge network | -2.915 | 0.00015 | -2.373 | 0 | 0.405 |
| 19419 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-484 | 13 | CBL | Sponge network | 1.148 | 2.0E-5 | 0.291 | 0.01267 | 0.405 |
| 19420 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 18 | TBL1XR1 | Sponge network | 0.643 | 0 | 0.578 | 0 | 0.405 |
| 19421 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-493-5p | 23 | MYBL1 | Sponge network | 0.75 | 0.00054 | 0.494 | 0.02152 | 0.405 |
| 19422 | MIR143HG |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 48 | CREB3L2 | Sponge network | -0.739 | 0.0574 | -0.525 | 2.0E-5 | 0.405 |
| 19423 | CTC-429P9.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-539-5p | 18 | DST | Sponge network | 0.043 | 0.81628 | -0.713 | 0.00096 | 0.405 |
| 19424 | RP11-33B1.4 |
hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-582-5p | 10 | ATRX | Sponge network | 0.826 | 0 | 0.261 | 0.04408 | 0.405 |
| 19425 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-424-5p;hsa-miR-576-5p | 17 | ZFHX4 | Sponge network | -2.076 | 0.00548 | 0.018 | 0.95574 | 0.405 |
| 19426 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-93-5p | 42 | AR | Sponge network | -0.18 | 0.20343 | -1.554 | 0 | 0.405 |
| 19427 | RP11-535M15.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p | 15 | RUNX1T1 | Sponge network | -1.14 | 0.03513 | -0.344 | 0.2374 | 0.405 |
| 19428 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p | 11 | LRRC55 | Sponge network | -0.517 | 0.02935 | -0.026 | 0.93163 | 0.405 |
| 19429 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-877-5p | 43 | RBMS3 | Sponge network | 0.082 | 0.55654 | -0.519 | 0.03551 | 0.405 |
| 19430 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-590-3p | 24 | EPHA5 | Sponge network | 0.178 | 0.29106 | -0.957 | 0.01733 | 0.405 |
| 19431 | RP11-13K12.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-5p | 19 | ITPKB | Sponge network | -0.154 | 0.78939 | -0.704 | 0.00106 | 0.405 |
| 19432 | RP11-102N12.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-3p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | FNBP1 | Sponge network | -0.351 | 0.11358 | -0.969 | 4.0E-5 | 0.405 |
| 19433 | LINC00672 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 16 | CDH19 | Sponge network | -0.227 | 0.31657 | -3.434 | 0 | 0.405 |
| 19434 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 20 | SLITRK6 | Sponge network | -0.49 | 0.04946 | -2.121 | 0 | 0.405 |
| 19435 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-940 | 11 | LIFR | Sponge network | -2.095 | 0.00112 | -1.794 | 0 | 0.405 |
| 19436 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 26 | NIN | Sponge network | 0.082 | 0.55654 | -0.253 | 0.13794 | 0.405 |
| 19437 | TMEM9B-AS1 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | LATS1 | Sponge network | 0.529 | 0.00552 | 0.109 | 0.34208 | 0.405 |
| 19438 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-744-3p | 15 | HIP1 | Sponge network | 0.663 | 0.00073 | 0.774 | 0 | 0.405 |
| 19439 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | RBMS3 | Sponge network | 0.537 | 0.00139 | -0.519 | 0.03551 | 0.405 |
| 19440 | LINC00641 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-98-5p | 13 | SCN11A | Sponge network | -0.222 | 0.32445 | -1.15 | 0.00299 | 0.405 |
| 19441 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p | 25 | CUX1 | Sponge network | -1.331 | 3.0E-5 | -0.039 | 0.6705 | 0.405 |
| 19442 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p | 19 | FAT4 | Sponge network | -0.738 | 0.21233 | -0.354 | 0.14694 | 0.405 |
| 19443 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-625-5p;hsa-miR-940 | 14 | SNX29 | Sponge network | -1.255 | 0.00981 | -0.34 | 0.01371 | 0.405 |
| 19444 | AC016747.3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 23 | PCDH10 | Sponge network | 0.199 | 0.38015 | -1.644 | 0.0078 | 0.405 |
| 19445 | TP73-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p | 13 | RIMS1 | Sponge network | -0.563 | 0.02545 | -3.143 | 0 | 0.405 |
| 19446 | RP11-400K9.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-671-5p | 11 | CDH23 | Sponge network | -0.611 | 0.09607 | -0.974 | 0.00034 | 0.405 |
| 19447 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 13 | DCLK1 | Sponge network | -0.342 | 0.02288 | -1.324 | 0.00014 | 0.405 |
| 19448 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 39 | TACC1 | Sponge network | 0.61 | 0.01593 | -0.362 | 0.05406 | 0.405 |
| 19449 | RP11-67L2.2 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-484 | 16 | ABL1 | Sponge network | 0.72 | 0.0001 | -0.215 | 0.07804 | 0.405 |
| 19450 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 24 | JAZF1 | Sponge network | -0.206 | 0.12942 | -0.377 | 0.02677 | 0.405 |
| 19451 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | DIP2C | Sponge network | 0.72 | 0.0001 | -0.436 | 0.01713 | 0.405 |
| 19452 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 28 | DMD | Sponge network | -0.342 | 0.02288 | -1.506 | 0 | 0.405 |
| 19453 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 28 | ANK2 | Sponge network | -0.303 | 0.21002 | -1.603 | 1.0E-5 | 0.404 |
| 19454 | FENDRR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | NCOA1 | Sponge network | -1.281 | 0.00012 | -0.432 | 0 | 0.404 |
| 19455 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-93-5p | 65 | TCF4 | Sponge network | -0.615 | 0.00015 | 0.016 | 0.92271 | 0.404 |
| 19456 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 26 | LIFR | Sponge network | -1.057 | 0.00029 | -1.794 | 0 | 0.404 |
| 19457 | RP11-48B3.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 32 | DTNA | Sponge network | 1.757 | 0 | -1.648 | 1.0E-5 | 0.404 |
| 19458 | MYLK-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-424-5p | 15 | USP12 | Sponge network | -1.331 | 3.0E-5 | -0.102 | 0.40768 | 0.404 |
| 19459 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-93-5p | 19 | EPHA5 | Sponge network | 0.497 | 0.01451 | -0.957 | 0.01733 | 0.404 |
| 19460 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429 | 10 | PIK3C2A | Sponge network | 2.364 | 0.00134 | 0.061 | 0.53776 | 0.404 |
| 19461 | RP11-536O18.1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p | 13 | FAT3 | Sponge network | -0.074 | 0.90758 | -1.601 | 0.00051 | 0.404 |
| 19462 | RP11-890B15.3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZNF844 | Sponge network | 0.101 | 0.60919 | -0.966 | 0.00035 | 0.404 |
| 19463 | H1FX-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | DTNA | Sponge network | -0.206 | 0.12942 | -1.648 | 1.0E-5 | 0.404 |
| 19464 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 24 | LRIG1 | Sponge network | -0.18 | 0.20343 | -0.537 | 0.00588 | 0.404 |
| 19465 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | ANK2 | Sponge network | -0.663 | 0.10887 | -1.603 | 1.0E-5 | 0.404 |
| 19466 | SNX29P2 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 13 | CADM3 | Sponge network | 0.4 | 0.14611 | -3.663 | 0 | 0.404 |
| 19467 | SERTAD4-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p | 10 | GLIPR1 | Sponge network | -0.932 | 0.00329 | 0.146 | 0.3276 | 0.404 |
| 19468 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 32 | RASSF2 | Sponge network | -0.513 | 0.04703 | -0.273 | 0.21961 | 0.404 |
| 19469 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 14 | PTPN11 | Sponge network | 0.953 | 0 | 0.431 | 2.0E-5 | 0.404 |
| 19470 | RP11-1006G14.4 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 15 | SHPRH | Sponge network | 0.559 | 0.00026 | 0.098 | 0.40994 | 0.404 |
| 19471 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-590-3p | 12 | PLSCR4 | Sponge network | 1.757 | 0 | -0.474 | 0.03833 | 0.404 |
| 19472 | C20orf166-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-486-5p | 14 | GLIPR1 | Sponge network | -2.69 | 0.0001 | 0.146 | 0.3276 | 0.404 |
| 19473 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | SASH1 | Sponge network | 0.532 | 0.00505 | -0.502 | 0.00175 | 0.404 |
| 19474 | RP11-25K19.1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZNF844 | Sponge network | -1.057 | 0.00029 | -0.966 | 0.00035 | 0.404 |
| 19475 | DNM3OS |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 33 | ERC1 | Sponge network | 0.572 | 0.01722 | -0.108 | 0.38794 | 0.404 |
| 19476 | RP11-35G9.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | BCL2 | Sponge network | 0.657 | 4.0E-5 | -0.899 | 0 | 0.404 |
| 19477 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-942-5p | 21 | GPC6 | Sponge network | -0.421 | 0.0114 | -0.054 | 0.81465 | 0.404 |
| 19478 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | KCNAB1 | Sponge network | 0.997 | 0.00066 | -0.755 | 2.0E-5 | 0.404 |
| 19479 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 55 | BMPR2 | Sponge network | 0.75 | 0.00054 | 0.206 | 0.0635 | 0.404 |
| 19480 | RP11-121C2.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p | 25 | ARHGEF12 | Sponge network | 1.147 | 0 | 0.09 | 0.35693 | 0.404 |
| 19481 | ADAMTS9-AS2 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-7-5p | 12 | EHD3 | Sponge network | -2.452 | 0 | -0.484 | 0.01747 | 0.404 |
| 19482 | RP11-356J5.12 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 34 | IL17RD | Sponge network | -0.899 | 6.0E-5 | -0.019 | 0.94873 | 0.404 |
| 19483 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | HECA | Sponge network | 0.194 | 0.64278 | -0.217 | 0.03463 | 0.404 |
| 19484 | RP11-35G9.5 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-500a-5p;hsa-miR-942-5p;hsa-miR-96-5p | 11 | ZBTB20 | Sponge network | 0.622 | 0.00015 | -0.122 | 0.57569 | 0.404 |
| 19485 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | EGLN1 | Sponge network | 0.683 | 1.0E-5 | -0.127 | 0.1536 | 0.404 |
| 19486 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-96-5p | 23 | CACNA2D1 | Sponge network | -0.18 | 0.20343 | -0.846 | 0.00675 | 0.404 |
| 19487 | U91328.19 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-769-5p | 19 | SYNPO2 | Sponge network | -0.303 | 0.21002 | -2.342 | 0 | 0.404 |
| 19488 | RP11-401P9.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-454-3p | 23 | DLC1 | Sponge network | -0.384 | 0.40652 | -0.382 | 0.05038 | 0.404 |
| 19489 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | SASH1 | Sponge network | -0.709 | 0.18168 | -0.502 | 0.00175 | 0.404 |
| 19490 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 11 | ATRX | Sponge network | 0.924 | 0 | 0.261 | 0.04408 | 0.404 |
| 19491 | CTC-510F12.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-34a-5p | 10 | ASTN1 | Sponge network | -0.691 | 0.00146 | -2.389 | 2.0E-5 | 0.404 |
| 19492 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 20 | RBL2 | Sponge network | -0.129 | 0.57177 | -0.282 | 0.00254 | 0.404 |
| 19493 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | MYBL1 | Sponge network | -0.739 | 0.00044 | 0.494 | 0.02152 | 0.404 |
| 19494 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p | 13 | ARHGAP28 | Sponge network | -0.583 | 0.14589 | -0.911 | 0.00013 | 0.404 |
| 19495 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 24 | PBX1 | Sponge network | 0.299 | 0.57722 | -1.206 | 0 | 0.404 |
| 19496 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-582-5p | 12 | MITF | Sponge network | -2.095 | 0.00112 | -0.769 | 0.00022 | 0.404 |
| 19497 | HOTAIRM1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | ZNF208 | Sponge network | -0.053 | 0.80685 | -0.4 | 0.22381 | 0.404 |
| 19498 | LINC00886 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p | 13 | SEMA3E | Sponge network | -0.371 | 0.24604 | -1.717 | 0.00137 | 0.404 |
| 19499 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-3613-5p;hsa-miR-493-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | ANK2 | Sponge network | 0.056 | 0.8494 | -1.603 | 1.0E-5 | 0.404 |
| 19500 | RP11-156P1.3 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | PTCH1 | Sponge network | 0.214 | 0.18246 | -0.1 | 0.6179 | 0.404 |
| 19501 | AC005618.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 29 | ABI2 | Sponge network | 0.862 | 0.06213 | 0.175 | 0.14787 | 0.404 |
| 19502 | AC016747.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p | 19 | DLC1 | Sponge network | 0.199 | 0.38015 | -0.382 | 0.05038 | 0.404 |
| 19503 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p | 10 | RB1CC1 | Sponge network | 0.946 | 0 | 0.175 | 0.12559 | 0.404 |
| 19504 | ZNF582-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-3p | 10 | ID4 | Sponge network | -1.349 | 0.00017 | -1.644 | 0 | 0.404 |
| 19505 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | -0.18 | 0.20343 | -1.751 | 1.0E-5 | 0.404 |
| 19506 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 24 | DDX6 | Sponge network | 0.67 | 0.00048 | 0.091 | 0.36428 | 0.404 |
| 19507 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 42 | NOTCH2 | Sponge network | -0.777 | 0.1134 | 0.138 | 0.35163 | 0.404 |
| 19508 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 23 | SLITRK6 | Sponge network | -0.739 | 0.0574 | -2.121 | 0 | 0.404 |
| 19509 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p | 28 | JAZF1 | Sponge network | -0.405 | 0.22013 | -0.377 | 0.02677 | 0.404 |
| 19510 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-92a-3p | 14 | DKK3 | Sponge network | 0.214 | 0.18246 | -0.052 | 0.77783 | 0.404 |
| 19511 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | ZFX | Sponge network | 0.101 | 0.60919 | 0.09 | 0.42563 | 0.404 |
| 19512 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p | 11 | ARHGEF12 | Sponge network | 0.497 | 0.01451 | 0.09 | 0.35693 | 0.404 |
| 19513 | RP11-693J15.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-335-3p | 11 | SYNPO2 | Sponge network | -0.72 | 0.24239 | -2.342 | 0 | 0.404 |
| 19514 | RP11-13K12.5 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 12 | EHD3 | Sponge network | -0.318 | 0.65847 | -0.484 | 0.01747 | 0.404 |
| 19515 | RP5-1092A3.4 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 11 | PHF6 | Sponge network | 1.495 | 0 | 0.785 | 0 | 0.404 |
| 19516 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 26 | PRKAB2 | Sponge network | 0.61 | 0.01593 | -0.367 | 0.01371 | 0.404 |
| 19517 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p | 11 | CUL5 | Sponge network | 1.294 | 0 | 0.026 | 0.77301 | 0.404 |
| 19518 | RP11-53O19.1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 12 | ZDBF2 | Sponge network | -0.405 | 0.22013 | -0.998 | 0.00025 | 0.404 |
| 19519 | HCG11 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | EZH1 | Sponge network | 0.385 | 0.10067 | -0.564 | 1.0E-5 | 0.404 |
| 19520 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MDM4 | Sponge network | -0.021 | 0.93598 | 0.345 | 0.00762 | 0.404 |
| 19521 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 1.317 | 0 | 0.322 | 0.00215 | 0.404 |
| 19522 | CROCCP2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 26 | MDM4 | Sponge network | 0.79 | 0 | 0.345 | 0.00762 | 0.404 |
| 19523 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-590-3p | 12 | AFF3 | Sponge network | 0.178 | 0.29106 | -2.373 | 0 | 0.404 |
| 19524 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-340-5p | 16 | MITF | Sponge network | -0.693 | 0.05629 | -0.769 | 0.00022 | 0.404 |
| 19525 | GNG12-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 16 | CDH23 | Sponge network | -0.793 | 7.0E-5 | -0.974 | 0.00034 | 0.404 |
| 19526 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 33 | ZFHX3 | Sponge network | -1.161 | 0.00591 | 0.389 | 0.01363 | 0.404 |
| 19527 | RP11-33B1.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 12 | TBL1XR1 | Sponge network | 0.826 | 0 | 0.578 | 0 | 0.404 |
| 19528 | LINC00957 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940 | 11 | RHOB | Sponge network | -0.421 | 0.0114 | -1.052 | 0 | 0.404 |
| 19529 | HOXA-AS2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 13 | MACF1 | Sponge network | 0.61 | 0.01593 | 0.019 | 0.90335 | 0.404 |
| 19530 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p | 20 | NAV3 | Sponge network | -0.469 | 0.08721 | 0.212 | 0.47729 | 0.404 |
| 19531 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-576-5p;hsa-miR-629-5p | 10 | IGF1 | Sponge network | -2.076 | 0.00548 | -0.204 | 0.5898 | 0.404 |
| 19532 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | NR2C2 | Sponge network | -0.096 | 0.67958 | 0.21 | 0.03224 | 0.404 |
| 19533 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 43 | NTRK3 | Sponge network | -1.057 | 0.00029 | -1.77 | 3.0E-5 | 0.404 |
| 19534 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 14 | MLLT10 | Sponge network | 0.765 | 7.0E-5 | 0.105 | 0.33292 | 0.404 |
| 19535 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 15 | LHX6 | Sponge network | -0.727 | 0.04726 | -0.087 | 0.69193 | 0.404 |
| 19536 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-98-5p | 32 | LRIG1 | Sponge network | -0.49 | 0.04946 | -0.537 | 0.00588 | 0.404 |
| 19537 | RP11-620J15.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-590-3p | 12 | CRTC1 | Sponge network | -0.739 | 0.00044 | -0.116 | 0.28412 | 0.404 |
| 19538 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | RET | Sponge network | -1.575 | 6.0E-5 | -0.833 | 0.03517 | 0.404 |
| 19539 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-877-5p | 27 | ANK2 | Sponge network | 0.494 | 0.00339 | -1.603 | 1.0E-5 | 0.404 |
| 19540 | SH3RF3-AS1 |
hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a | 13 | PLXNC1 | Sponge network | 0.889 | 9.0E-5 | 0.569 | 0.00329 | 0.404 |
| 19541 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-96-5p | 24 | BCL2 | Sponge network | -0.517 | 0.02935 | -0.899 | 0 | 0.404 |
| 19542 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 13 | CNTNAP3 | Sponge network | -0.739 | 0.00044 | -1.465 | 0.00033 | 0.404 |
| 19543 | AP001055.6 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-744-3p | 14 | IGFBP5 | Sponge network | 0.693 | 0.00076 | -0.806 | 0.00105 | 0.404 |
| 19544 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-98-5p | 10 | MPL | Sponge network | -0.129 | 0.57177 | -0.624 | 0.00133 | 0.404 |
| 19545 | HAND2-AS1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 10 | PEA15 | Sponge network | -1.697 | 0.00889 | 0.009 | 0.93318 | 0.404 |
| 19546 | RP11-158K1.3 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | DIP2C | Sponge network | 0.349 | 0.01695 | -0.436 | 0.01713 | 0.404 |
| 19547 | LINC00473 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 18 | ARNT2 | Sponge network | -1.203 | 0.03881 | -0.332 | 0.25851 | 0.404 |
| 19548 | CTD-2336O2.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-450b-5p | 20 | IL17RD | Sponge network | -0.141 | 0.26434 | -0.019 | 0.94873 | 0.404 |
| 19549 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | FGF5 | Sponge network | -0.709 | 0.18168 | 0.549 | 0.28659 | 0.404 |
| 19550 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | LRRK2 | Sponge network | -0.222 | 0.32445 | -1.136 | 1.0E-5 | 0.404 |
| 19551 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-3p | 28 | NTRK3 | Sponge network | 0.178 | 0.29106 | -1.77 | 3.0E-5 | 0.404 |
| 19552 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-935 | 41 | SSBP2 | Sponge network | -0.738 | 0.21233 | -1.58 | 0 | 0.404 |
| 19553 | LINC00957 |
hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-671-5p | 17 | KIT | Sponge network | -0.421 | 0.0114 | -2.184 | 0 | 0.404 |
| 19554 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 44 | QKI | Sponge network | -0.513 | 0.04703 | -0.333 | 0.04111 | 0.404 |
| 19555 | RP11-404E16.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-532-5p | 11 | AR | Sponge network | 0.097 | 0.91311 | -1.554 | 0 | 0.404 |
| 19556 | H19 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-5p;hsa-miR-629-3p | 10 | RASSF8 | Sponge network | 2.439 | 0 | -0.122 | 0.65591 | 0.404 |
| 19557 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 24 | EPHA3 | Sponge network | 0.349 | 0.01695 | 0.185 | 0.53588 | 0.404 |
| 19558 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-93-5p | 16 | SLITRK3 | Sponge network | -0.473 | 0.07602 | -3.328 | 0 | 0.404 |
| 19559 | DNM3OS |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | FLI1 | Sponge network | 0.572 | 0.01722 | -0.294 | 0.10905 | 0.404 |
| 19560 | EMX2OS |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SEMA3E | Sponge network | -0.777 | 0.1134 | -1.717 | 0.00137 | 0.404 |
| 19561 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | SYNE1 | Sponge network | -0.08 | 0.90092 | -0.738 | 0.00316 | 0.404 |
| 19562 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 15 | JAKMIP2 | Sponge network | -0.615 | 0.00015 | -1.388 | 0 | 0.404 |
| 19563 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p | 19 | PCDH9 | Sponge network | 0.082 | 0.55397 | -1.823 | 4.0E-5 | 0.404 |
| 19564 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 28 | PRKAB2 | Sponge network | -0.355 | 0.0077 | -0.367 | 0.01371 | 0.404 |
| 19565 | RP11-216B9.6 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 20 | DLC1 | Sponge network | 0.174 | 0.30034 | -0.382 | 0.05038 | 0.404 |
| 19566 | AC138035.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-505-5p;hsa-miR-766-3p | 15 | TBL1XR1 | Sponge network | 1.073 | 0 | 0.578 | 0 | 0.404 |
| 19567 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-143-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-98-5p | 22 | CLASP2 | Sponge network | 0.082 | 0.55397 | -0.038 | 0.71 | 0.404 |
| 19568 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-93-3p | 26 | ERC1 | Sponge network | 0.056 | 0.81195 | -0.108 | 0.38794 | 0.404 |
| 19569 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 49 | BMPR2 | Sponge network | 0.559 | 0.00026 | 0.206 | 0.0635 | 0.404 |
| 19570 | PAN3-AS1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-33a-3p;hsa-miR-369-3p | 12 | PPM1L | Sponge network | 0.062 | 0.55274 | -0.537 | 0.00616 | 0.404 |
| 19571 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CLIP1 | Sponge network | 0.083 | 0.66405 | -0.215 | 0.08684 | 0.404 |
| 19572 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-500b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | SLITRK6 | Sponge network | -0.323 | 0.01372 | -2.121 | 0 | 0.404 |
| 19573 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 36 | USP12 | Sponge network | -0.739 | 0.0574 | -0.102 | 0.40768 | 0.404 |
| 19574 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p | 17 | CADM3 | Sponge network | -0.487 | 0.02157 | -3.663 | 0 | 0.404 |
| 19575 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 31 | KCNA6 | Sponge network | 0.997 | 0.00066 | -1.531 | 0.00013 | 0.404 |
| 19576 | RP11-968O1.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-3p | 11 | GRIN2A | Sponge network | -0.829 | 0.01343 | -2.161 | 0 | 0.404 |
| 19577 | RP11-701H24.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-335-3p | 10 | CBFA2T3 | Sponge network | -1.425 | 0.04204 | -1.221 | 1.0E-5 | 0.404 |
| 19578 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-7-1-3p | 10 | TSHZ3 | Sponge network | -0.557 | 0.11135 | -0.556 | 0.01516 | 0.404 |
| 19579 | SNX29P2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-671-5p | 10 | PIK3IP1 | Sponge network | 0.4 | 0.14611 | -0.187 | 0.19945 | 0.404 |
| 19580 | MIR143HG |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p | 22 | IRS1 | Sponge network | -0.739 | 0.0574 | -0.026 | 0.88855 | 0.404 |
| 19581 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 27 | ZFHX4 | Sponge network | 0.381 | 0.25967 | 0.018 | 0.95574 | 0.404 |
| 19582 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-93-3p;hsa-miR-96-5p | 14 | EPHA3 | Sponge network | 0.622 | 0.00015 | 0.185 | 0.53588 | 0.404 |
| 19583 | CTA-204B4.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 14 | TET1 | Sponge network | 0.765 | 7.0E-5 | -0.135 | 0.52138 | 0.404 |
| 19584 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 37 | AR | Sponge network | 0.051 | 0.83388 | -1.554 | 0 | 0.404 |
| 19585 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p | 10 | AFF3 | Sponge network | 0.693 | 0.00076 | -2.373 | 0 | 0.404 |
| 19586 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-3613-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-93-5p | 17 | ANK2 | Sponge network | 0.342 | 0.03129 | -1.603 | 1.0E-5 | 0.404 |
| 19587 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | GIGYF2 | Sponge network | 0.75 | 0.00054 | 0.136 | 0.04403 | 0.404 |
| 19588 | RP11-440L14.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-424-5p | 10 | RICTOR | Sponge network | 0.599 | 0.0001 | 0.388 | 0.00188 | 0.404 |
| 19589 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CLASP2 | Sponge network | 0.953 | 0 | -0.038 | 0.71 | 0.404 |
| 19590 | RP11-678G14.3 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-7-1-3p | 24 | DTNA | Sponge network | -4.477 | 0 | -1.648 | 1.0E-5 | 0.404 |
| 19591 | LINC00641 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | PRKCB | Sponge network | -0.222 | 0.32445 | -1.313 | 2.0E-5 | 0.404 |
| 19592 | FAM66C |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 43 | BCL2 | Sponge network | -0.901 | 0.00019 | -0.899 | 0 | 0.404 |
| 19593 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-935 | 29 | DMD | Sponge network | 0.16 | 0.50785 | -1.506 | 0 | 0.404 |
| 19594 | TRIM52-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-577 | 10 | FGFR1 | Sponge network | -0.418 | 0.00068 | -0.509 | 0.03957 | 0.404 |
| 19595 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-454-3p | 27 | DLC1 | Sponge network | -0.18 | 0.20343 | -0.382 | 0.05038 | 0.404 |
| 19596 | ADAMTS9-AS2 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CTGF | Sponge network | -2.452 | 0 | 0.321 | 0.11682 | 0.403 |
| 19597 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-940 | 18 | MYO9A | Sponge network | 0.946 | 0 | -0.147 | 0.32373 | 0.403 |
| 19598 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | SOX5 | Sponge network | -1.349 | 0.00017 | -1.091 | 4.0E-5 | 0.403 |
| 19599 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 11 | GRIN2A | Sponge network | 0.178 | 0.29106 | -2.161 | 0 | 0.403 |
| 19600 | LINC00969 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p | 13 | MLLT10 | Sponge network | 0.683 | 1.0E-5 | 0.105 | 0.33292 | 0.403 |
| 19601 | RP11-554A11.4 |
hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-421 | 11 | MAFB | Sponge network | -0.583 | 0.14589 | -0.023 | 0.89865 | 0.403 |
| 19602 | RP11-981G7.6 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-376c-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | HECA | Sponge network | 0.986 | 0.01022 | -0.217 | 0.03463 | 0.403 |
| 19603 | RP11-774O3.3 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | RPS6KA6 | Sponge network | -0.487 | 0.02157 | -2.989 | 0 | 0.403 |
| 19604 | LINC00476 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-369-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | MACF1 | Sponge network | -0.615 | 0.00015 | 0.019 | 0.90335 | 0.403 |
| 19605 | RP11-540B6.6 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | KIF5B | Sponge network | 0.93 | 0 | 0.362 | 0.00066 | 0.403 |
| 19606 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p | 20 | GREM1 | Sponge network | -0.901 | 0.00019 | 0.902 | 0.01691 | 0.403 |
| 19607 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | IL6ST | Sponge network | 1.253 | 0 | -0.402 | 0.00676 | 0.403 |
| 19608 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-940 | 25 | CHD5 | Sponge network | -0.896 | 0.00099 | -0.922 | 0.01521 | 0.403 |
| 19609 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | NR2C2 | Sponge network | 1.11 | 1.0E-5 | 0.21 | 0.03224 | 0.403 |
| 19610 | HOXA-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | EYA4 | Sponge network | 0.61 | 0.01593 | 0.3 | 0.36876 | 0.403 |
| 19611 | LINC00476 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PTCH1 | Sponge network | -0.615 | 0.00015 | -0.1 | 0.6179 | 0.403 |
| 19612 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-532-5p;hsa-miR-590-3p | 17 | CXXC4 | Sponge network | 0.174 | 0.30034 | -0.433 | 0.21055 | 0.403 |
| 19613 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-589-5p;hsa-miR-93-5p | 43 | TCF4 | Sponge network | 0.082 | 0.55397 | 0.016 | 0.92271 | 0.403 |
| 19614 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | PEG3 | Sponge network | -0.355 | 0.0077 | -1.74 | 0 | 0.403 |
| 19615 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-625-5p | 22 | ZMYM2 | Sponge network | 0.75 | 0.00054 | 0.279 | 0.01273 | 0.403 |
| 19616 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p | 35 | CHD5 | Sponge network | 0.95 | 0.15943 | -0.922 | 0.01521 | 0.403 |
| 19617 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-92a-1-5p | 14 | PRKAB2 | Sponge network | -0.309 | 0.1145 | -0.367 | 0.01371 | 0.403 |
| 19618 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-589-3p;hsa-miR-93-5p | 21 | PDGFRA | Sponge network | -2.531 | 0 | -0.372 | 0.0037 | 0.403 |
| 19619 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p | 18 | NEDD4 | Sponge network | -0.154 | 0.78939 | 0.02 | 0.89834 | 0.403 |
| 19620 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 40 | ZFHX4 | Sponge network | 0.056 | 0.81195 | 0.018 | 0.95574 | 0.403 |
| 19621 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 17 | NR2C2 | Sponge network | 1.317 | 0 | 0.21 | 0.03224 | 0.403 |
| 19622 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 11 | ZNF208 | Sponge network | 0.494 | 0.00339 | -0.4 | 0.22381 | 0.403 |
| 19623 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 21 | CBL | Sponge network | 0.529 | 0.00552 | 0.291 | 0.01267 | 0.403 |
| 19624 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 46 | PDGFRA | Sponge network | -1.697 | 0.00889 | -0.372 | 0.0037 | 0.403 |
| 19625 | RP11-535M15.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p | 14 | MEF2C | Sponge network | -1.14 | 0.03513 | -0.499 | 0.01448 | 0.403 |
| 19626 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 30 | GPC6 | Sponge network | -0.777 | 0.1134 | -0.054 | 0.81465 | 0.403 |
| 19627 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-143-3p;hsa-miR-154-5p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p;hsa-miR-539-5p | 13 | LIMD1 | Sponge network | 1.147 | 0 | 0.103 | 0.28978 | 0.403 |
| 19628 | HOXB-AS1 |
hsa-let-7a-3p;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-589-5p | 12 | EYA4 | Sponge network | -0.154 | 0.53954 | 0.3 | 0.36876 | 0.403 |
| 19629 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-940 | 24 | AKAP11 | Sponge network | 0.529 | 0.00552 | 0.196 | 0.09825 | 0.403 |
| 19630 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-582-3p | 23 | SASH1 | Sponge network | -0.154 | 0.53954 | -0.502 | 0.00175 | 0.403 |
| 19631 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 29 | ZFHX4 | Sponge network | 0.051 | 0.83388 | 0.018 | 0.95574 | 0.403 |
| 19632 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 26 | BHLHE41 | Sponge network | -0.421 | 0.0114 | 0.353 | 0.12753 | 0.403 |
| 19633 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-590-3p | 11 | RBM5 | Sponge network | 0.873 | 0 | -0.205 | 0.0129 | 0.403 |
| 19634 | MIR22HG |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-629-3p | 11 | ID4 | Sponge network | -1.047 | 0 | -1.644 | 0 | 0.403 |
| 19635 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 11 | NEDD4 | Sponge network | -0.583 | 0.14589 | 0.02 | 0.89834 | 0.403 |
| 19636 | THAP9-AS1 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-654-3p | 15 | RICTOR | Sponge network | 1.079 | 0 | 0.388 | 0.00188 | 0.403 |
| 19637 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 57 | SSBP2 | Sponge network | -0.615 | 0.00015 | -1.58 | 0 | 0.403 |
| 19638 | RP11-13K12.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 16 | TET1 | Sponge network | -0.318 | 0.65847 | -0.135 | 0.52138 | 0.403 |
| 19639 | GAS6-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-940 | 34 | NTRK2 | Sponge network | 0.16 | 0.50785 | -0.736 | 0.06495 | 0.403 |
| 19640 | AC141928.1 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 11 | PPP2R2C | Sponge network | 0.853 | 0.15352 | -0.959 | 0.07428 | 0.403 |
| 19641 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 10 | ST5 | Sponge network | -2.46 | 0 | -0.78 | 0 | 0.403 |
| 19642 | AC004893.11 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p | 13 | ATM | Sponge network | 1.317 | 0 | 0.178 | 0.21568 | 0.403 |
| 19643 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 15 | CNTNAP3 | Sponge network | -1.161 | 0.00591 | -1.465 | 0.00033 | 0.403 |
| 19644 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 22 | FGF5 | Sponge network | -0.793 | 7.0E-5 | 0.549 | 0.28659 | 0.403 |
| 19645 | WDFY3-AS2 |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | CTNNA2 | Sponge network | -0.942 | 0 | -2.547 | 0 | 0.403 |
| 19646 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 45 | ERBB4 | Sponge network | -0.777 | 0.1134 | -1.975 | 2.0E-5 | 0.403 |
| 19647 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 32 | ST6GALNAC3 | Sponge network | -0.793 | 7.0E-5 | -0.987 | 0 | 0.403 |
| 19648 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-942-5p | 16 | CREBBP | Sponge network | -0.162 | 0.47687 | 0.126 | 0.15186 | 0.403 |
| 19649 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 23 | BMPR2 | Sponge network | 0.693 | 0.00076 | 0.206 | 0.0635 | 0.403 |
| 19650 | RP11-373D23.3 |
hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-493-3p | 29 | DST | Sponge network | -0.285 | 0.2172 | -0.713 | 0.00096 | 0.403 |
| 19651 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 15 | BMPR2 | Sponge network | 0.062 | 0.55274 | 0.206 | 0.0635 | 0.403 |
| 19652 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 17 | GAS7 | Sponge network | -0.295 | 0.02604 | -0.555 | 0.01602 | 0.403 |
| 19653 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | ATRX | Sponge network | 0.064 | 0.72934 | 0.261 | 0.04408 | 0.403 |
| 19654 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | ERRFI1 | Sponge network | -2.452 | 0 | -0.798 | 1.0E-5 | 0.403 |
| 19655 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-940 | 12 | FAM172A | Sponge network | 0.056 | 0.81195 | -0.57 | 0 | 0.403 |
| 19656 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p | 11 | PHF6 | Sponge network | 1.089 | 0 | 0.785 | 0 | 0.403 |
| 19657 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | NALCN | Sponge network | -0.487 | 0.02157 | 1.068 | 0.00237 | 0.403 |
| 19658 | RP11-693J15.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 12 | PCDH9 | Sponge network | -0.72 | 0.24239 | -1.823 | 4.0E-5 | 0.403 |
| 19659 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p | 46 | CCND2 | Sponge network | -0.162 | 0.47687 | -0.267 | 0.3112 | 0.403 |
| 19660 | RP5-890O3.9 |
hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p | 12 | RICTOR | Sponge network | 0.842 | 0 | 0.388 | 0.00188 | 0.403 |
| 19661 | RP11-159D12.2 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | MAP3K4 | Sponge network | 1.254 | 0 | 0.058 | 0.54229 | 0.403 |
| 19662 | RAMP2-AS1 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | -0.513 | 0.04703 | 0.321 | 0.00267 | 0.403 |
| 19663 | NEAT1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-766-3p | 16 | ATM | Sponge network | 0.705 | 0.00014 | 0.178 | 0.21568 | 0.403 |
| 19664 | TRAF3IP2-AS1 |
hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | PAIP2 | Sponge network | -0.323 | 0.01372 | -0.515 | 0 | 0.403 |
| 19665 | C4B-AS1 |
hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 10 | EBF3 | Sponge network | 2.306 | 9.0E-5 | -0.058 | 0.81437 | 0.403 |
| 19666 | TPTEP1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | NIN | Sponge network | -1.216 | 0 | -0.253 | 0.13794 | 0.403 |
| 19667 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 14 | CHD2 | Sponge network | 0.614 | 1.0E-5 | 0.049 | 0.55371 | 0.403 |
| 19668 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-93-5p | 12 | SLITRK3 | Sponge network | -0.785 | 0.00035 | -3.328 | 0 | 0.403 |
| 19669 | RP11-57H14.4 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ERCC6 | Sponge network | 0.083 | 0.66405 | 0.23 | 0.015 | 0.403 |
| 19670 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-744-3p | 22 | ADAMTSL3 | Sponge network | -0.663 | 0.10887 | -1.559 | 0 | 0.403 |
| 19671 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | LRP1B | Sponge network | -0.376 | 0.00337 | -2.163 | 0 | 0.403 |
| 19672 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p | 15 | RET | Sponge network | -0.318 | 0.65847 | -0.833 | 0.03517 | 0.403 |
| 19673 | U91328.19 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | WWTR1 | Sponge network | -0.303 | 0.21002 | -0.345 | 0.11997 | 0.403 |
| 19674 | RASSF8-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p | 12 | PARK2 | Sponge network | 0.335 | 0.20596 | -1.892 | 0 | 0.403 |
| 19675 | RP11-359B12.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | WWTR1 | Sponge network | 0.064 | 0.72934 | -0.345 | 0.11997 | 0.403 |
| 19676 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | PDS5B | Sponge network | 0.91 | 0 | 0.259 | 0.00962 | 0.403 |
| 19677 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 32 | ZFHX3 | Sponge network | -0.842 | 0.01361 | 0.389 | 0.01363 | 0.403 |
| 19678 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | KCNA6 | Sponge network | -1.161 | 0.00591 | -1.531 | 0.00013 | 0.403 |
| 19679 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 12 | PLA2R1 | Sponge network | -0.49 | 0.04946 | -0.265 | 0.24676 | 0.403 |
| 19680 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-940 | 15 | SNX29 | Sponge network | -0.72 | 0.24239 | -0.34 | 0.01371 | 0.403 |
| 19681 | ADAMTS9-AS2 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ZNF844 | Sponge network | -2.452 | 0 | -0.966 | 0.00035 | 0.403 |
| 19682 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-590-5p;hsa-miR-96-5p | 22 | EPHA3 | Sponge network | 0.331 | 0.57247 | 0.185 | 0.53588 | 0.403 |
| 19683 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-590-3p | 16 | PRDM2 | Sponge network | 0.225 | 0.30644 | -0.258 | 0.00721 | 0.403 |
| 19684 | SNHG16 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p | 16 | PHF6 | Sponge network | 0.961 | 0 | 0.785 | 0 | 0.403 |
| 19685 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 19 | PTGFR | Sponge network | 1.302 | 7.0E-5 | -0.233 | 0.45421 | 0.403 |
| 19686 | AP001055.6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 36 | LPP | Sponge network | 0.693 | 0.00076 | -0.459 | 0.04475 | 0.403 |
| 19687 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-96-5p | 13 | CRTC3 | Sponge network | 0.538 | 0.00076 | 0.164 | 0.16786 | 0.403 |
| 19688 | RP11-535M15.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p | 15 | DST | Sponge network | -1.14 | 0.03513 | -0.713 | 0.00096 | 0.403 |
| 19689 | PWAR6 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 24 | SIRT1 | Sponge network | -1.575 | 6.0E-5 | 0.109 | 0.2593 | 0.403 |
| 19690 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | KANK1 | Sponge network | -0.487 | 0.02157 | -0.55 | 0.00134 | 0.403 |
| 19691 | RP11-13K12.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p | 14 | SCN9A | Sponge network | -0.318 | 0.65847 | -0.525 | 0.10593 | 0.403 |
| 19692 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-589-3p | 14 | FAM172A | Sponge network | -0.154 | 0.78939 | -0.57 | 0 | 0.403 |
| 19693 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-7-1-3p | 13 | DNAJB4 | Sponge network | -1.349 | 0.00017 | -0.7 | 1.0E-5 | 0.403 |
| 19694 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | PHC3 | Sponge network | -0.031 | 0.89063 | 0.212 | 0.0499 | 0.403 |
| 19695 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-93-3p | 34 | AKAP11 | Sponge network | -0.615 | 0.00015 | 0.196 | 0.09825 | 0.403 |
| 19696 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-584-5p;hsa-miR-590-3p | 13 | ZNF208 | Sponge network | -0.513 | 0.04703 | -0.4 | 0.22381 | 0.403 |
| 19697 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 21 | ERCC4 | Sponge network | 0.494 | 0.00339 | 0.126 | 0.15266 | 0.403 |
| 19698 | AP001347.6 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | PREX2 | Sponge network | -1.161 | 0.00591 | -0.272 | 0.25382 | 0.403 |
| 19699 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-5p | 22 | SCN9A | Sponge network | 0.16 | 0.50785 | -0.525 | 0.10593 | 0.403 |
| 19700 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | IL17RD | Sponge network | 0.131 | 0.8436 | -0.019 | 0.94873 | 0.403 |
| 19701 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-942-5p | 20 | GPC6 | Sponge network | 0.395 | 0.15451 | -0.054 | 0.81465 | 0.403 |
| 19702 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-576-5p;hsa-miR-625-5p | 14 | ABI2 | Sponge network | -1.116 | 0.23686 | 0.175 | 0.14787 | 0.403 |
| 19703 | GNG12-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-455-5p | 10 | THSD7A | Sponge network | -0.793 | 7.0E-5 | 0.242 | 0.25154 | 0.403 |
| 19704 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p | 17 | KAT6B | Sponge network | -1.645 | 0 | -0.478 | 0.00018 | 0.403 |
| 19705 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 32 | CREB3L2 | Sponge network | -0.842 | 0.01361 | -0.525 | 2.0E-5 | 0.403 |
| 19706 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | MLLT10 | Sponge network | 1.308 | 0 | 0.105 | 0.33292 | 0.403 |
| 19707 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | HECA | Sponge network | -0.709 | 0.18168 | -0.217 | 0.03463 | 0.403 |
| 19708 | TPTEP1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | GALNT13 | Sponge network | -1.216 | 0 | -0.857 | 0.07439 | 0.403 |
| 19709 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 21 | PTPN11 | Sponge network | 0.222 | 0.29133 | 0.431 | 2.0E-5 | 0.403 |
| 19710 | TP73-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p;hsa-miR-940 | 12 | RHOB | Sponge network | -0.563 | 0.02545 | -1.052 | 0 | 0.403 |
| 19711 | RP4-639F20.1 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-942-5p | 36 | THRB | Sponge network | -0.517 | 0.02935 | -1.117 | 0.0004 | 0.403 |
| 19712 | WAC-AS1 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-590-3p | 21 | ABL2 | Sponge network | 0.821 | 0 | 0.82 | 0 | 0.403 |
| 19713 | THAP9-AS1 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-497-5p | 15 | PAFAH1B2 | Sponge network | 1.079 | 0 | 0.318 | 0.0001 | 0.403 |
| 19714 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 57 | AFF4 | Sponge network | 0.349 | 0.01695 | -0.266 | 0.01565 | 0.403 |
| 19715 | RP11-356J5.12 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | STAT5B | Sponge network | -0.899 | 6.0E-5 | -0.182 | 0.1082 | 0.403 |
| 19716 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | AHNAK | Sponge network | -0.726 | 0.00132 | -1.207 | 0 | 0.403 |
| 19717 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p | 25 | RASSF8 | Sponge network | -0.773 | 0.04073 | -0.122 | 0.65591 | 0.403 |
| 19718 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 15 | ZNF292 | Sponge network | 0.705 | 0.00014 | 0.276 | 0.04148 | 0.403 |
| 19719 | CTC-429P9.3 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-940 | 10 | PHOX2B | Sponge network | 0.082 | 0.55397 | -2.68 | 0 | 0.403 |
| 19720 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 23 | NEDD4 | Sponge network | -0.793 | 7.0E-5 | 0.02 | 0.89834 | 0.403 |
| 19721 | RP5-1198O20.4 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | KLF10 | Sponge network | 0.997 | 0.00066 | -0.783 | 0 | 0.403 |
| 19722 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | FSTL5 | Sponge network | 0.082 | 0.55654 | -1.3 | 0.01619 | 0.403 |
| 19723 | CTD-2314G24.2 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 20 | CXCL12 | Sponge network | 0.246 | 0.59912 | -1.421 | 0 | 0.403 |
| 19724 | LINC00476 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 26 | RYR2 | Sponge network | -0.615 | 0.00015 | -1.466 | 0.00031 | 0.403 |
| 19725 | LINC00476 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | KLF10 | Sponge network | -0.615 | 0.00015 | -0.783 | 0 | 0.403 |
| 19726 | MRVI1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-671-5p | 14 | CYLD | Sponge network | -1.092 | 4.0E-5 | -0.367 | 0.00171 | 0.403 |
| 19727 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 13 | PTPN11 | Sponge network | 1.308 | 0 | 0.431 | 2.0E-5 | 0.403 |
| 19728 | RP11-38L15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-423-5p;hsa-miR-590-3p | 10 | SUFU | Sponge network | 0.344 | 0.3127 | -0.04 | 0.65489 | 0.403 |
| 19729 | HOXB-AS2 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-874-3p | 10 | NR2C2 | Sponge network | 1.316 | 0.01106 | 0.21 | 0.03224 | 0.403 |
| 19730 | AC006547.13 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p | 14 | ABL2 | Sponge network | 1.159 | 0 | 0.82 | 0 | 0.403 |
| 19731 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 16 | UVRAG | Sponge network | -0.49 | 0.04946 | -0.017 | 0.83865 | 0.403 |
| 19732 | HOTAIRM1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | AKAP6 | Sponge network | -0.053 | 0.80685 | -1.586 | 3.0E-5 | 0.403 |
| 19733 | MYLK-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p | 15 | LIMCH1 | Sponge network | -1.331 | 3.0E-5 | -0.915 | 1.0E-5 | 0.403 |
| 19734 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p | 16 | TXNIP | Sponge network | -0.942 | 0 | -1.277 | 0 | 0.403 |
| 19735 | RP11-67L2.2 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 31 | UBR3 | Sponge network | 0.72 | 0.0001 | -0.021 | 0.82774 | 0.403 |
| 19736 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 29 | SNX29 | Sponge network | -1.645 | 0 | -0.34 | 0.01371 | 0.403 |
| 19737 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-497-5p;hsa-miR-582-5p | 19 | RICTOR | Sponge network | 2.368 | 2.0E-5 | 0.388 | 0.00188 | 0.403 |
| 19738 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p | 23 | ADARB1 | Sponge network | 0.37 | 0.27614 | -0.335 | 0.06631 | 0.403 |
| 19739 | KB-1410C5.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-7-1-3p;hsa-miR-940 | 10 | AKAP11 | Sponge network | 0.959 | 0 | 0.196 | 0.09825 | 0.403 |
| 19740 | RP3-402G11.28 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p | 11 | ABL2 | Sponge network | 1.119 | 5.0E-5 | 0.82 | 0 | 0.403 |
| 19741 | RP11-326G21.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-26b-3p | 13 | IGFBP5 | Sponge network | -0.021 | 0.93598 | -0.806 | 0.00105 | 0.403 |
| 19742 | AC106786.1 |
hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-98-5p | 10 | THBS1 | Sponge network | 1.122 | 0.02989 | 0.741 | 0.00386 | 0.403 |
| 19743 | CTD-2554C21.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-96-5p | 11 | SCN9A | Sponge network | -1.654 | 0.00144 | -0.525 | 0.10593 | 0.403 |
| 19744 | ZSCAN16-AS1 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 15 | SFRP1 | Sponge network | -0.342 | 0.02288 | -3.566 | 0 | 0.403 |
| 19745 | RP11-13K12.1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p | 13 | HERC1 | Sponge network | -0.154 | 0.78939 | -0.196 | 0.08544 | 0.403 |
| 19746 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p | 17 | MLLT10 | Sponge network | 0.946 | 0 | 0.105 | 0.33292 | 0.403 |
| 19747 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 43 | ADARB1 | Sponge network | 0.494 | 0.00339 | -0.335 | 0.06631 | 0.403 |
| 19748 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | PHC3 | Sponge network | 0.946 | 0 | 0.212 | 0.0499 | 0.403 |
| 19749 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | TIMP3 | Sponge network | 0.395 | 0.15451 | -0.546 | 0.02307 | 0.403 |
| 19750 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p | 17 | SOX5 | Sponge network | -2.076 | 0.00548 | -1.091 | 4.0E-5 | 0.403 |
| 19751 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 48 | AFF4 | Sponge network | -0.611 | 0.09607 | -0.266 | 0.01565 | 0.403 |
| 19752 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 18 | PTPN11 | Sponge network | 0.385 | 0.10067 | 0.431 | 2.0E-5 | 0.403 |
| 19753 | RAB30-AS1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p;hsa-miR-495-3p | 11 | PHF6 | Sponge network | 0.752 | 0 | 0.785 | 0 | 0.403 |
| 19754 | RP11-973H7.3 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p | 10 | TBL1XR1 | Sponge network | 0.901 | 0 | 0.578 | 0 | 0.402 |
| 19755 | BZRAP1-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-671-5p | 24 | DLC1 | Sponge network | -0.465 | 0.04703 | -0.382 | 0.05038 | 0.402 |
| 19756 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-671-5p | 13 | CADM3 | Sponge network | -0.318 | 0.65847 | -3.663 | 0 | 0.402 |
| 19757 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-374a-3p | 17 | MAPK1 | Sponge network | 0.542 | 0.00054 | -0.017 | 0.81465 | 0.402 |
| 19758 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-651-5p | 27 | PRDM2 | Sponge network | 0.101 | 0.60919 | -0.258 | 0.00721 | 0.402 |
| 19759 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | PTPRB | Sponge network | -0.376 | 0.00337 | 0.105 | 0.47122 | 0.402 |
| 19760 | HAND2-AS1 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | EPB41L3 | Sponge network | -1.697 | 0.00889 | -1.193 | 0 | 0.402 |
| 19761 | AC009120.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | CEP170 | Sponge network | 0.919 | 0 | 0.943 | 0 | 0.402 |
| 19762 | CTC-429P9.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-500a-5p | 10 | PKNOX1 | Sponge network | 0.082 | 0.55397 | 0.085 | 0.28917 | 0.402 |
| 19763 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-335-5p | 16 | PIK3C2A | Sponge network | 0.946 | 0 | 0.061 | 0.53776 | 0.402 |
| 19764 | RP11-244O19.1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 12 | ZNF844 | Sponge network | -0.096 | 0.67958 | -0.966 | 0.00035 | 0.402 |
| 19765 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-625-5p;hsa-miR-93-5p | 16 | LHX6 | Sponge network | -0.154 | 0.78939 | -0.087 | 0.69193 | 0.402 |
| 19766 | RP11-158K1.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | BRD3 | Sponge network | 0.349 | 0.01695 | 0.37 | 0.00013 | 0.402 |
| 19767 | GHRLOS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-7-5p | 16 | ESR1 | Sponge network | -1.942 | 0 | -1.035 | 3.0E-5 | 0.402 |
| 19768 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | EPHA7 | Sponge network | -0.342 | 0.02288 | -2.197 | 3.0E-5 | 0.402 |
| 19769 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | LRRC55 | Sponge network | 0.693 | 0.00076 | -0.026 | 0.93163 | 0.402 |
| 19770 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | JAZF1 | Sponge network | 0.101 | 0.60919 | -0.377 | 0.02677 | 0.402 |
| 19771 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 20 | DYNC1I1 | Sponge network | 0.374 | 0.20054 | -1.12 | 0.00107 | 0.402 |
| 19772 | MEG3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.249 | 0.35902 | 0.943 | 0 | 0.402 |
| 19773 | RP11-182J1.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484 | 11 | CDH23 | Sponge network | -0.187 | 0.23787 | -0.974 | 0.00034 | 0.402 |
| 19774 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-3p;hsa-miR-93-5p | 17 | MAP3K12 | Sponge network | -0.49 | 0.04946 | -0.057 | 0.59369 | 0.402 |
| 19775 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | RELN | Sponge network | 0.997 | 0.00066 | -2.403 | 0 | 0.402 |
| 19776 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 18 | PTGFR | Sponge network | 0.101 | 0.60919 | -0.233 | 0.45421 | 0.402 |
| 19777 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 49 | CREB3L2 | Sponge network | -0.49 | 0.04946 | -0.525 | 2.0E-5 | 0.402 |
| 19778 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 17 | TGFBR3 | Sponge network | 0.497 | 0.01451 | -1.173 | 1.0E-5 | 0.402 |
| 19779 | RP11-693J15.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-874-3p | 16 | ASTN1 | Sponge network | -0.72 | 0.24239 | -2.389 | 2.0E-5 | 0.402 |
| 19780 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-590-3p | 23 | ADAMTSL3 | Sponge network | 0.082 | 0.55654 | -1.559 | 0 | 0.402 |
| 19781 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 14 | MITF | Sponge network | -0.469 | 0.08721 | -0.769 | 0.00022 | 0.402 |
| 19782 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | EDA2R | Sponge network | -2.117 | 0.00161 | -0.587 | 0.01598 | 0.402 |
| 19783 | RP11-356J5.12 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 14 | NGFR | Sponge network | -0.899 | 6.0E-5 | -1.271 | 0.01058 | 0.402 |
| 19784 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | SLC5A3 | Sponge network | 1.316 | 0 | 0.493 | 5.0E-5 | 0.402 |
| 19785 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-5p | 15 | FSTL5 | Sponge network | -0.969 | 2.0E-5 | -1.3 | 0.01619 | 0.402 |
| 19786 | LINC00865 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 12 | HOMER2 | Sponge network | -1.249 | 7.0E-5 | -2.698 | 0 | 0.402 |
| 19787 | RP11-119F7.5 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-590-3p | 15 | GLIPR1 | Sponge network | 0.225 | 0.30644 | 0.146 | 0.3276 | 0.402 |
| 19788 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | 0.101 | 0.60919 | 0.126 | 0.15266 | 0.402 |
| 19789 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-7-5p;hsa-miR-93-3p | 24 | EPHA3 | Sponge network | 0.253 | 0.09565 | 0.185 | 0.53588 | 0.402 |
| 19790 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-940 | 19 | ZFP36L1 | Sponge network | -0.777 | 0.1134 | -0.505 | 8.0E-5 | 0.402 |
| 19791 | SNX29P2 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 39 | DTNA | Sponge network | 0.4 | 0.14611 | -1.648 | 1.0E-5 | 0.402 |
| 19792 | RP11-2H8.2 |
hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 15 | CHD2 | Sponge network | 1.089 | 0 | 0.049 | 0.55371 | 0.402 |
| 19793 | CTD-2336O2.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 11 | WWTR1 | Sponge network | -0.141 | 0.26434 | -0.345 | 0.11997 | 0.402 |
| 19794 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-629-3p | 28 | ZBTB4 | Sponge network | -1.249 | 7.0E-5 | -0.739 | 0 | 0.402 |
| 19795 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-577;hsa-miR-590-3p | 35 | CCND2 | Sponge network | -0.896 | 0.00099 | -0.267 | 0.3112 | 0.402 |
| 19796 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 36 | PRKAB2 | Sponge network | -0.737 | 0.06182 | -0.367 | 0.01371 | 0.402 |
| 19797 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | CACNA2D1 | Sponge network | -0.777 | 0.1134 | -0.846 | 0.00675 | 0.402 |
| 19798 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-505-5p;hsa-miR-625-5p | 19 | CTIF | Sponge network | -1.349 | 0.00017 | -0.389 | 0.00549 | 0.402 |
| 19799 | CKMT2-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | HERC1 | Sponge network | 0.082 | 0.55654 | -0.196 | 0.08544 | 0.402 |
| 19800 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | LRRC55 | Sponge network | 0.532 | 0.00505 | -0.026 | 0.93163 | 0.402 |
| 19801 | LINC00641 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | KCTD8 | Sponge network | -0.222 | 0.32445 | -3.359 | 0 | 0.402 |
| 19802 | ZRANB2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-93-5p | 14 | IL17RD | Sponge network | -0.376 | 0.00337 | -0.019 | 0.94873 | 0.402 |
| 19803 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935 | 43 | TSHZ3 | Sponge network | -0.615 | 0.00015 | -0.556 | 0.01516 | 0.402 |
| 19804 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-5p | 21 | NTRK2 | Sponge network | -4.463 | 0.00025 | -0.736 | 0.06495 | 0.402 |
| 19805 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MLLT10 | Sponge network | 1.601 | 0 | 0.105 | 0.33292 | 0.402 |
| 19806 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | EPHA3 | Sponge network | -0.737 | 0.10822 | 0.185 | 0.53588 | 0.402 |
| 19807 | U91328.21 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p | 15 | IL17RD | Sponge network | -0.735 | 0.15983 | -0.019 | 0.94873 | 0.402 |
| 19808 | AC006116.24 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-93-5p | 18 | PIK3R1 | Sponge network | -0.473 | 0.07602 | -0.133 | 0.36854 | 0.402 |
| 19809 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940 | 22 | SNX29 | Sponge network | 0.199 | 0.38015 | -0.34 | 0.01371 | 0.402 |
| 19810 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-1-5p | 26 | PRKAB2 | Sponge network | -0.227 | 0.31657 | -0.367 | 0.01371 | 0.402 |
| 19811 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 11 | FGFR1 | Sponge network | 0.693 | 0.00076 | -0.509 | 0.03957 | 0.402 |
| 19812 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-590-3p | 16 | RICTOR | Sponge network | 0.178 | 0.29106 | 0.388 | 0.00188 | 0.402 |
| 19813 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 24 | SETBP1 | Sponge network | 0.082 | 0.55654 | -1.302 | 1.0E-5 | 0.402 |
| 19814 | DIO3OS |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-942-5p | 33 | IGFBP5 | Sponge network | -0.773 | 0.04073 | -0.806 | 0.00105 | 0.402 |
| 19815 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-378a-3p | 13 | CDH23 | Sponge network | 0.889 | 9.0E-5 | -0.974 | 0.00034 | 0.402 |
| 19816 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | NF1 | Sponge network | 1.163 | 0.00018 | 0.51 | 0 | 0.402 |
| 19817 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p | 15 | MYCBP2 | Sponge network | -0.096 | 0.67958 | -0.027 | 0.82016 | 0.402 |
| 19818 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-651-5p | 21 | PRDM2 | Sponge network | -0.318 | 0.65847 | -0.258 | 0.00721 | 0.402 |
| 19819 | GNG12-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p | 12 | PREX2 | Sponge network | -0.793 | 7.0E-5 | -0.272 | 0.25382 | 0.402 |
| 19820 | SNHG10 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 12 | PHF6 | Sponge network | 0.931 | 0 | 0.785 | 0 | 0.402 |
| 19821 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 18 | ITCH | Sponge network | 0.683 | 1.0E-5 | 0.084 | 0.34058 | 0.402 |
| 19822 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 43 | MOB1B | Sponge network | -0.129 | 0.57177 | 0.197 | 0.15266 | 0.402 |
| 19823 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-7-1-3p | 16 | APC | Sponge network | -1.092 | 4.0E-5 | -0.255 | 0.04051 | 0.402 |
| 19824 | SNHG14 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-425-3p;hsa-miR-429 | 12 | DUSP1 | Sponge network | -1.111 | 0.0003 | -1.754 | 0 | 0.402 |
| 19825 | LINC00473 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-3913-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p | 18 | WWTR1 | Sponge network | -1.203 | 0.03881 | -0.345 | 0.11997 | 0.402 |
| 19826 | GABPB1-AS1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | ERCC6 | Sponge network | 1.11 | 0 | 0.23 | 0.015 | 0.402 |
| 19827 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 57 | KIF1B | Sponge network | 0.194 | 0.64278 | -0.118 | 0.32258 | 0.402 |
| 19828 | ZSCAN16-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-577 | 26 | IL17RD | Sponge network | -0.342 | 0.02288 | -0.019 | 0.94873 | 0.402 |
| 19829 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | DOCK4 | Sponge network | -0.739 | 0.0574 | 0.502 | 0.00108 | 0.402 |
| 19830 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-576-5p | 13 | HACE1 | Sponge network | -1.645 | 0 | -0.072 | 0.55239 | 0.402 |
| 19831 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 65 | NTRK3 | Sponge network | 0.532 | 0.00505 | -1.77 | 3.0E-5 | 0.402 |
| 19832 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-653-5p | 27 | DMXL1 | Sponge network | -1.645 | 0 | -0.426 | 0.00056 | 0.402 |
| 19833 | RP11-434B12.1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PHF3 | Sponge network | -0.031 | 0.89063 | 0.126 | 0.19652 | 0.402 |
| 19834 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p | 12 | RNASEL | Sponge network | -1.092 | 4.0E-5 | -0.272 | 0.01796 | 0.402 |
| 19835 | ATP1B3-AS1 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-22-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | ABL2 | Sponge network | 1.2 | 0.01813 | 0.82 | 0 | 0.402 |
| 19836 | RP11-894P9.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-5p;hsa-miR-369-3p | 10 | PTPN14 | Sponge network | 0.993 | 0 | 0.144 | 0.34595 | 0.402 |
| 19837 | RP11-130L8.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-93-5p | 52 | LPP | Sponge network | -0.663 | 0.10887 | -0.459 | 0.04475 | 0.402 |
| 19838 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-503-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 38 | RICTOR | Sponge network | 0.415 | 0.14657 | 0.388 | 0.00188 | 0.402 |
| 19839 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | FSTL5 | Sponge network | -1.203 | 0.03881 | -1.3 | 0.01619 | 0.402 |
| 19840 | KB-1460A1.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ZMYND11 | Sponge network | 0.603 | 0.00127 | -0.2 | 0.05449 | 0.402 |
| 19841 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p | 12 | LHX6 | Sponge network | -0.517 | 0.02935 | -0.087 | 0.69193 | 0.402 |
| 19842 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-5p;hsa-miR-98-5p | 26 | USP12 | Sponge network | 0.494 | 0.00339 | -0.102 | 0.40768 | 0.402 |
| 19843 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 46 | CBX5 | Sponge network | 0.537 | 0.00139 | 0.165 | 0.24446 | 0.402 |
| 19844 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p | 10 | ELMO1 | Sponge network | -0.896 | 0.00099 | 0.04 | 0.83147 | 0.402 |
| 19845 | RP11-384O8.1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-484 | 17 | ABL1 | Sponge network | -0.738 | 0.21233 | -0.215 | 0.07804 | 0.402 |
| 19846 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | 0.848 | 0 | 0.323 | 0.00792 | 0.402 |
| 19847 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | TACC1 | Sponge network | 0.693 | 0.00076 | -0.362 | 0.05406 | 0.402 |
| 19848 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-942-5p | 37 | HLF | Sponge network | -1.249 | 7.0E-5 | -2.443 | 0 | 0.402 |
| 19849 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | 0.219 | 0.1516 | -1.548 | 0 | 0.402 |
| 19850 | NR2F1-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NRG3 | Sponge network | -0.49 | 0.04946 | -1.836 | 4.0E-5 | 0.402 |
| 19851 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | PGR | Sponge network | -0.08 | 0.90092 | -1.704 | 0 | 0.402 |
| 19852 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | FLI1 | Sponge network | -0.576 | 0.10121 | -0.294 | 0.10905 | 0.402 |
| 19853 | FGF14-AS2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-582-3p;hsa-miR-590-3p | 18 | PPP1R9A | Sponge network | -1.582 | 8.0E-5 | -0.903 | 0.04254 | 0.402 |
| 19854 | RP11-345P4.7 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-495-3p | 12 | PHF6 | Sponge network | 1.597 | 0 | 0.785 | 0 | 0.402 |
| 19855 | RP11-473I1.10 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | BRD3 | Sponge network | 0.673 | 0 | 0.37 | 0.00013 | 0.402 |
| 19856 | RP11-244O19.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-940 | 20 | SIRT1 | Sponge network | -0.096 | 0.67958 | 0.109 | 0.2593 | 0.402 |
| 19857 | RP11-83A24.2 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 18 | KIF5B | Sponge network | 0.417 | 0.00445 | 0.362 | 0.00066 | 0.402 |
| 19858 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-3613-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 16 | EPHA7 | Sponge network | -1.425 | 0.04204 | -2.197 | 3.0E-5 | 0.402 |
| 19859 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 22 | CXXC4 | Sponge network | -0.405 | 0.22013 | -0.433 | 0.21055 | 0.402 |
| 19860 | OIP5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 24 | TRIM33 | Sponge network | 0.421 | 0.00084 | 0.402 | 7.0E-5 | 0.402 |
| 19861 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | RECK | Sponge network | -0.899 | 6.0E-5 | -1.043 | 0 | 0.402 |
| 19862 | PCA3 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 12 | PRDM5 | Sponge network | -0.709 | 0.18168 | -1.09 | 0.00014 | 0.402 |
| 19863 | DNM1P35 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-93-3p | 11 | CNTNAP1 | Sponge network | 0.051 | 0.83388 | 0.358 | 0.13727 | 0.402 |
| 19864 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b | 19 | ABI2 | Sponge network | -0.583 | 0.14589 | 0.175 | 0.14787 | 0.402 |
| 19865 | LINC00641 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 49 | TRPS1 | Sponge network | -0.222 | 0.32445 | -0.191 | 0.44109 | 0.402 |
| 19866 | RAMP2-AS1 |
hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PCDH18 | Sponge network | -0.513 | 0.04703 | 0.216 | 0.24645 | 0.402 |
| 19867 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | PAK3 | Sponge network | 0.997 | 0.00066 | -0.628 | 0.07253 | 0.402 |
| 19868 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 33 | TET1 | Sponge network | -0.777 | 0.1134 | -0.135 | 0.52138 | 0.402 |
| 19869 | AC156455.1 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p | 10 | COL1A2 | Sponge network | 1.154 | 0.00041 | 1.991 | 0 | 0.402 |
| 19870 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-93-3p | 21 | ST6GAL2 | Sponge network | -0.896 | 0.00099 | -1.201 | 0.00597 | 0.402 |
| 19871 | DNM3OS |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 11 | KCTD8 | Sponge network | 0.572 | 0.01722 | -3.359 | 0 | 0.402 |
| 19872 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-654-3p;hsa-miR-93-3p;hsa-miR-940 | 29 | AKAP11 | Sponge network | 0.637 | 0.00069 | 0.196 | 0.09825 | 0.402 |
| 19873 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-502-5p | 20 | PTPN14 | Sponge network | -1.161 | 0.00591 | 0.144 | 0.34595 | 0.402 |
| 19874 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | ZYX | Sponge network | -0.769 | 0.00035 | 0.019 | 0.89763 | 0.402 |
| 19875 | PCA3 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | GJA1 | Sponge network | -0.709 | 0.18168 | -0.485 | 0.02179 | 0.402 |
| 19876 | LINC00622 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | BRD3 | Sponge network | 1.169 | 0.00028 | 0.37 | 0.00013 | 0.402 |
| 19877 | CTD-3064M3.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p | 18 | EPHA5 | Sponge network | -0.469 | 0.08721 | -0.957 | 0.01733 | 0.402 |
| 19878 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | RAP1A | Sponge network | -0.49 | 0.04946 | -0.929 | 0 | 0.402 |
| 19879 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 12 | AKAP12 | Sponge network | 0.385 | 0.10067 | -0.932 | 0.0028 | 0.402 |
| 19880 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 41 | SASH1 | Sponge network | -1.216 | 0 | -0.502 | 0.00175 | 0.402 |
| 19881 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | AKAP11 | Sponge network | 0.873 | 0 | 0.196 | 0.09825 | 0.402 |
| 19882 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-940 | 21 | CRTC3 | Sponge network | 0.61 | 0.01593 | 0.164 | 0.16786 | 0.402 |
| 19883 | LINC00702 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 33 | PRDM2 | Sponge network | -0.737 | 0.06182 | -0.258 | 0.00721 | 0.402 |
| 19884 | RP11-736K20.5 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-590-3p | 11 | PIK3CA | Sponge network | -0.358 | 0.17255 | 0.195 | 0.06908 | 0.402 |
| 19885 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 33 | BHLHE41 | Sponge network | 0.572 | 0.01722 | 0.353 | 0.12753 | 0.402 |
| 19886 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429 | 19 | PRKAB2 | Sponge network | -2.62 | 0 | -0.367 | 0.01371 | 0.402 |
| 19887 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 39 | QKI | Sponge network | -0.738 | 0.21233 | -0.333 | 0.04111 | 0.402 |
| 19888 | LPP-AS2 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-769-5p;hsa-miR-93-3p | 31 | GRIK3 | Sponge network | -0.18 | 0.20343 | -3.208 | 0 | 0.402 |
| 19889 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | IGSF10 | Sponge network | 0.101 | 0.60919 | -1.751 | 1.0E-5 | 0.402 |
| 19890 | RP11-819C21.1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 13 | STAT5B | Sponge network | 0.537 | 0.00139 | -0.182 | 0.1082 | 0.402 |
| 19891 | PART1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p | 15 | ZBTB20 | Sponge network | -2.925 | 0 | -0.122 | 0.57569 | 0.402 |
| 19892 | AC006116.24 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p | 11 | STARD13 | Sponge network | -0.473 | 0.07602 | -0.187 | 0.27383 | 0.402 |
| 19893 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-589-5p | 14 | RARB | Sponge network | 0.199 | 0.38015 | -0.825 | 2.0E-5 | 0.402 |
| 19894 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | KIF1B | Sponge network | 1.163 | 0.00018 | -0.118 | 0.32258 | 0.402 |
| 19895 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 40 | RASSF2 | Sponge network | -0.901 | 0.00019 | -0.273 | 0.21961 | 0.402 |
| 19896 | PWAR6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -1.575 | 6.0E-5 | 0.024 | 0.89889 | 0.402 |
| 19897 | BZRAP1-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-493-3p;hsa-miR-505-5p | 14 | MRVI1 | Sponge network | -0.465 | 0.04703 | -1.051 | 0.00022 | 0.402 |
| 19898 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | NRXN3 | Sponge network | 0.619 | 0.00157 | -0.893 | 0.04186 | 0.402 |
| 19899 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429 | 12 | LRRK2 | Sponge network | -1.249 | 7.0E-5 | -1.136 | 1.0E-5 | 0.402 |
| 19900 | LINC00092 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-940 | 22 | NTRK2 | Sponge network | -1.886 | 0 | -0.736 | 0.06495 | 0.402 |
| 19901 | LINC01018 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-590-3p | 10 | SCN11A | Sponge network | -2.915 | 0.00015 | -1.15 | 0.00299 | 0.402 |
| 19902 | RP13-516M14.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-7-1-3p | 11 | BCL9 | Sponge network | 0.389 | 0.04293 | 0.321 | 0.00267 | 0.402 |
| 19903 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-93-3p;hsa-miR-96-5p | 18 | EBF3 | Sponge network | 0.4 | 0.14611 | -0.058 | 0.81437 | 0.402 |
| 19904 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-654-3p | 11 | ITCH | Sponge network | 0.946 | 0 | 0.084 | 0.34058 | 0.402 |
| 19905 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 30 | NTRK2 | Sponge network | 0.225 | 0.30644 | -0.736 | 0.06495 | 0.401 |
| 19906 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | MYO9A | Sponge network | 1.153 | 0.00114 | -0.147 | 0.32373 | 0.401 |
| 19907 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 12 | ZMYM2 | Sponge network | 1.148 | 2.0E-5 | 0.279 | 0.01273 | 0.401 |
| 19908 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 50 | UNC80 | Sponge network | -0.129 | 0.57177 | -1.934 | 0 | 0.401 |
| 19909 | LINC00403 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200c-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-342-5p | 16 | DTNA | Sponge network | -3.586 | 0.00032 | -1.648 | 1.0E-5 | 0.401 |
| 19910 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 17 | TBL1XR1 | Sponge network | 0.34 | 0.00273 | 0.578 | 0 | 0.401 |
| 19911 | AP001258.4 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p;hsa-miR-495-3p | 14 | PHF6 | Sponge network | 0.931 | 0 | 0.785 | 0 | 0.401 |
| 19912 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 39 | SASH1 | Sponge network | -0.777 | 0.1134 | -0.502 | 0.00175 | 0.401 |
| 19913 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 11 | PTPN11 | Sponge network | 1.03 | 0 | 0.431 | 2.0E-5 | 0.401 |
| 19914 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | SOX5 | Sponge network | -0.811 | 2.0E-5 | -1.091 | 4.0E-5 | 0.401 |
| 19915 | HCG11 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 45 | DTNA | Sponge network | 0.385 | 0.10067 | -1.648 | 1.0E-5 | 0.401 |
| 19916 | RP11-999E24.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p | 32 | CADM2 | Sponge network | -0.693 | 0.05629 | -3.782 | 0 | 0.401 |
| 19917 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 27 | ZFHX4 | Sponge network | -0.206 | 0.12942 | 0.018 | 0.95574 | 0.401 |
| 19918 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 17 | MOB1B | Sponge network | 0.178 | 0.29106 | 0.197 | 0.15266 | 0.401 |
| 19919 | LENG8-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-511-5p;hsa-miR-625-3p | 10 | ZFHX3 | Sponge network | 1.471 | 0 | 0.389 | 0.01363 | 0.401 |
| 19920 | MEG3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | PRKCB | Sponge network | 0.249 | 0.35902 | -1.313 | 2.0E-5 | 0.401 |
| 19921 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | FYN | Sponge network | 0.225 | 0.30644 | -0.131 | 0.37051 | 0.401 |
| 19922 | TPTEP1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | IL17RD | Sponge network | -1.216 | 0 | -0.019 | 0.94873 | 0.401 |
| 19923 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-7-5p;hsa-miR-93-5p | 28 | ZBTB4 | Sponge network | -0.355 | 0.0077 | -0.739 | 0 | 0.401 |
| 19924 | LINC00476 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-935 | 30 | PCDH10 | Sponge network | -0.615 | 0.00015 | -1.644 | 0.0078 | 0.401 |
| 19925 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p | 31 | PRDM2 | Sponge network | -0.901 | 0.00019 | -0.258 | 0.00721 | 0.401 |
| 19926 | RP11-473I1.10 |
hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | PIK3C2A | Sponge network | 0.673 | 0 | 0.061 | 0.53776 | 0.401 |
| 19927 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-590-3p | 14 | MITF | Sponge network | -0.739 | 0.00044 | -0.769 | 0.00022 | 0.401 |
| 19928 | RP11-379F4.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p | 15 | CREB1 | Sponge network | 1.316 | 0 | 0.203 | 0.00537 | 0.401 |
| 19929 | CTB-31O20.2 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 11 | PHF6 | Sponge network | 0.608 | 3.0E-5 | 0.785 | 0 | 0.401 |
| 19930 | RP11-53O19.1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 31 | RIMS2 | Sponge network | -0.405 | 0.22013 | -1.365 | 0.00208 | 0.401 |
| 19931 | TPTEP1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | RYR2 | Sponge network | -1.216 | 0 | -1.466 | 0.00031 | 0.401 |
| 19932 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 31 | PGR | Sponge network | -0.355 | 0.0077 | -1.704 | 0 | 0.401 |
| 19933 | RAMP2-AS1 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-5p | 16 | PTGFR | Sponge network | -0.513 | 0.04703 | -0.233 | 0.45421 | 0.401 |
| 19934 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | RB1CC1 | Sponge network | -0.222 | 0.32445 | 0.175 | 0.12559 | 0.401 |
| 19935 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | -0.976 | 0.00025 | 0.309 | 0.17079 | 0.401 |
| 19936 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-1301-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | CLASP2 | Sponge network | 0.782 | 0.00016 | -0.038 | 0.71 | 0.401 |
| 19937 | RP11-98I9.4 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | SIRT1 | Sponge network | 0.67 | 0.00048 | 0.109 | 0.2593 | 0.401 |
| 19938 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p | 12 | SPG20 | Sponge network | 0.299 | 0.57722 | -1.16 | 0 | 0.401 |
| 19939 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 14 | SASH1 | Sponge network | -2.039 | 0.03447 | -0.502 | 0.00175 | 0.401 |
| 19940 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-625-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -0.162 | 0.47687 | 0.024 | 0.89889 | 0.401 |
| 19941 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CLIP1 | Sponge network | -0.777 | 0.1134 | -0.215 | 0.08684 | 0.401 |
| 19942 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 31 | ZFHX3 | Sponge network | 0.603 | 0.00127 | 0.389 | 0.01363 | 0.401 |
| 19943 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | CDH19 | Sponge network | -0.342 | 0.02288 | -3.434 | 0 | 0.401 |
| 19944 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-550a-5p;hsa-miR-7-5p | 19 | CRTC1 | Sponge network | -1.331 | 3.0E-5 | -0.116 | 0.28412 | 0.401 |
| 19945 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-539-5p;hsa-miR-590-3p | 20 | TSC1 | Sponge network | -0.096 | 0.67958 | 0.103 | 0.26071 | 0.401 |
| 19946 | PCA3 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p | 17 | TLE4 | Sponge network | -0.709 | 0.18168 | -1.157 | 0 | 0.401 |
| 19947 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 16 | TRIM33 | Sponge network | 1.089 | 0 | 0.402 | 7.0E-5 | 0.401 |
| 19948 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 18 | HIP1 | Sponge network | 0.225 | 0.30644 | 0.774 | 0 | 0.401 |
| 19949 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-450b-5p | 22 | PPM1A | Sponge network | -0.583 | 0.14589 | -0.623 | 0 | 0.401 |
| 19950 | TP73-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 58 | DST | Sponge network | -0.563 | 0.02545 | -0.713 | 0.00096 | 0.401 |
| 19951 | ZSCAN16-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484 | 12 | EZH1 | Sponge network | -0.342 | 0.02288 | -0.564 | 1.0E-5 | 0.401 |
| 19952 | RP11-620J15.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | ETV1 | Sponge network | -0.739 | 0.00044 | -0.096 | 0.67674 | 0.401 |
| 19953 | CTD-3138B18.6 |
hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 12 | TBL1XR1 | Sponge network | 2.248 | 0 | 0.578 | 0 | 0.401 |
| 19954 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-664a-3p;hsa-miR-98-5p | 12 | SCN11A | Sponge network | -0.405 | 0.22013 | -1.15 | 0.00299 | 0.401 |
| 19955 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-7-1-3p | 17 | CLIP1 | Sponge network | -0.976 | 0.00025 | -0.215 | 0.08684 | 0.401 |
| 19956 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-671-5p | 14 | CADM3 | Sponge network | -0.738 | 0.21233 | -3.663 | 0 | 0.401 |
| 19957 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-93-5p | 32 | PIK3R1 | Sponge network | -0.611 | 0.09607 | -0.133 | 0.36854 | 0.401 |
| 19958 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-582-5p | 30 | TSHZ3 | Sponge network | -0.773 | 0.04073 | -0.556 | 0.01516 | 0.401 |
| 19959 | RP11-251G23.5 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-495-3p | 10 | STAG2 | Sponge network | 1.344 | 0 | 0.252 | 0.00438 | 0.401 |
| 19960 | MAP3K14 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | QKI | Sponge network | -0.295 | 0.02604 | -0.333 | 0.04111 | 0.401 |
| 19961 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | PDGFC | Sponge network | -2.46 | 0 | -0.33 | 0.06483 | 0.401 |
| 19962 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-93-5p | 11 | AHNAK | Sponge network | -2.531 | 0 | -1.207 | 0 | 0.401 |
| 19963 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 28 | MYO9A | Sponge network | -1.439 | 4.0E-5 | -0.147 | 0.32373 | 0.401 |
| 19964 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 31 | ANK2 | Sponge network | -0.969 | 2.0E-5 | -1.603 | 1.0E-5 | 0.401 |
| 19965 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 33 | TGFBR3 | Sponge network | -0.405 | 0.22013 | -1.173 | 1.0E-5 | 0.401 |
| 19966 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 35 | PPM1L | Sponge network | 0.683 | 1.0E-5 | -0.537 | 0.00616 | 0.401 |
| 19967 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | SMARCA2 | Sponge network | -1.575 | 6.0E-5 | -0.529 | 0.00014 | 0.401 |
| 19968 | CTA-204B4.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-766-3p | 18 | IL17RD | Sponge network | 0.765 | 7.0E-5 | -0.019 | 0.94873 | 0.401 |
| 19969 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p | 10 | ERCC4 | Sponge network | -0.187 | 0.23787 | 0.126 | 0.15266 | 0.401 |
| 19970 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 21 | MYO9A | Sponge network | 1.757 | 0 | -0.147 | 0.32373 | 0.401 |
| 19971 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-411-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 46 | SMAD2 | Sponge network | 0.101 | 0.60919 | -0.128 | 0.12732 | 0.401 |
| 19972 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-653-5p | 18 | PDS5B | Sponge network | 1.254 | 0 | 0.259 | 0.00962 | 0.401 |
| 19973 | RP11-13K12.5 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-5p | 14 | PTGFR | Sponge network | -0.318 | 0.65847 | -0.233 | 0.45421 | 0.401 |
| 19974 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 61 | LPP | Sponge network | -0.318 | 0.65847 | -0.459 | 0.04475 | 0.401 |
| 19975 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | FSTL5 | Sponge network | -2.925 | 0 | -1.3 | 0.01619 | 0.401 |
| 19976 | RP11-774O3.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | RYR2 | Sponge network | -0.487 | 0.02157 | -1.466 | 0.00031 | 0.401 |
| 19977 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-590-3p | 13 | PDS5B | Sponge network | 1.601 | 0 | 0.259 | 0.00962 | 0.401 |
| 19978 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | TSHZ3 | Sponge network | 0.727 | 3.0E-5 | -0.556 | 0.01516 | 0.401 |
| 19979 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | LRRC4 | Sponge network | -2.925 | 0 | -1.774 | 0 | 0.401 |
| 19980 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-664a-3p | 15 | GREM1 | Sponge network | 0.214 | 0.18246 | 0.902 | 0.01691 | 0.401 |
| 19981 | RP11-2H8.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p | 31 | CBX5 | Sponge network | 1.089 | 0 | 0.165 | 0.24446 | 0.401 |
| 19982 | RP11-819C21.1 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | INPP4A | Sponge network | 0.537 | 0.00139 | 0.07 | 0.53563 | 0.401 |
| 19983 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-582-5p;hsa-miR-98-5p | 37 | PRKAA2 | Sponge network | -0.773 | 0.04073 | -2.462 | 0 | 0.401 |
| 19984 | FGD5-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-200b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p | 27 | UBR3 | Sponge network | 0.401 | 0.00066 | -0.021 | 0.82774 | 0.401 |
| 19985 | LINC00578 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577 | 12 | PIK3CA | Sponge network | 0.811 | 0.03228 | 0.195 | 0.06908 | 0.401 |
| 19986 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 0.8 | 0 | 0.259 | 0.00962 | 0.401 |
| 19987 | RP11-527J8.1 |
hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p | 13 | RICTOR | Sponge network | 1.324 | 0 | 0.388 | 0.00188 | 0.401 |
| 19988 | C20orf166-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p | 12 | HERC1 | Sponge network | -2.69 | 0.0001 | -0.196 | 0.08544 | 0.401 |
| 19989 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 20 | ACVR1 | Sponge network | -0.49 | 0.04946 | 0.279 | 0.00234 | 0.401 |
| 19990 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | SASH1 | Sponge network | -0.842 | 0.01361 | -0.502 | 0.00175 | 0.401 |
| 19991 | RP11-307B6.3 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-424-5p | 11 | CSDE1 | Sponge network | -4.463 | 0.00025 | -0.493 | 0 | 0.401 |
| 19992 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 14 | ABCB5 | Sponge network | -0.612 | 0.00094 | -0.956 | 0.04077 | 0.401 |
| 19993 | RP11-53O19.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p | 18 | RYR2 | Sponge network | -0.405 | 0.22013 | -1.466 | 0.00031 | 0.401 |
| 19994 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 38 | ZFHX3 | Sponge network | -0.405 | 0.22013 | 0.389 | 0.01363 | 0.401 |
| 19995 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 40 | CREB3L2 | Sponge network | -0.096 | 0.67958 | -0.525 | 2.0E-5 | 0.401 |
| 19996 | RP11-473I1.10 |
hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TMEM30A | Sponge network | 0.673 | 0 | 0.096 | 0.31031 | 0.401 |
| 19997 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-766-3p | 10 | PHACTR4 | Sponge network | 0.311 | 0.10344 | 0.069 | 0.45203 | 0.401 |
| 19998 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-655-3p | 13 | LATS1 | Sponge network | 1.147 | 0 | 0.109 | 0.34208 | 0.401 |
| 19999 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p | 12 | TAL1 | Sponge network | -0.517 | 0.02935 | -0.462 | 0.00574 | 0.401 |
| 20000 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 43 | TACC1 | Sponge network | -0.942 | 0 | -0.362 | 0.05406 | 0.401 |
| 20001 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 36 | BCL2 | Sponge network | 0.335 | 0.20596 | -0.899 | 0 | 0.401 |
| 20002 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 15 | EBF3 | Sponge network | -0.811 | 2.0E-5 | -0.058 | 0.81437 | 0.401 |
| 20003 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-576-5p | 16 | PDE4DIP | Sponge network | 0.176 | 0.37402 | -0.282 | 0.0257 | 0.401 |
| 20004 | LINC00476 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-590-3p | 27 | SYT4 | Sponge network | -0.615 | 0.00015 | -3.207 | 0 | 0.401 |
| 20005 | RP11-67L2.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | WWTR1 | Sponge network | 0.72 | 0.0001 | -0.345 | 0.11997 | 0.401 |
| 20006 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 36 | TSHZ3 | Sponge network | 0.082 | 0.55654 | -0.556 | 0.01516 | 0.401 |
| 20007 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TSC1 | Sponge network | -0.031 | 0.89063 | 0.103 | 0.26071 | 0.401 |
| 20008 | PCA3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ZNF208 | Sponge network | -0.709 | 0.18168 | -0.4 | 0.22381 | 0.401 |
| 20009 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 40 | TMEM132B | Sponge network | -1.249 | 7.0E-5 | -0.83 | 0.01084 | 0.401 |
| 20010 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TMEFF2 | Sponge network | -1.161 | 0.00591 | -2.426 | 0 | 0.401 |
| 20011 | PCA3 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 14 | CXCL12 | Sponge network | -0.709 | 0.18168 | -1.421 | 0 | 0.401 |
| 20012 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-7-1-3p | 12 | TAL1 | Sponge network | 0.889 | 9.0E-5 | -0.462 | 0.00574 | 0.401 |
| 20013 | AC009120.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-486-5p;hsa-miR-590-3p | 16 | EPHA3 | Sponge network | 0.919 | 0 | 0.185 | 0.53588 | 0.401 |
| 20014 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-582-5p | 21 | DMD | Sponge network | 0.419 | 0.0319 | -1.506 | 0 | 0.401 |
| 20015 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 48 | NOTCH2 | Sponge network | -1.111 | 0.0003 | 0.138 | 0.35163 | 0.401 |
| 20016 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p | 10 | MAFB | Sponge network | -1.331 | 3.0E-5 | -0.023 | 0.89865 | 0.401 |
| 20017 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-93-5p | 26 | ANK2 | Sponge network | -1.645 | 0 | -1.603 | 1.0E-5 | 0.401 |
| 20018 | PWAR6 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | GJA1 | Sponge network | -1.575 | 6.0E-5 | -0.485 | 0.02179 | 0.401 |
| 20019 | GABPB1-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 15 | PHF6 | Sponge network | 1.11 | 0 | 0.785 | 0 | 0.401 |
| 20020 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | ABCC9 | Sponge network | 0.4 | 0.14611 | -0.466 | 0.14121 | 0.401 |
| 20021 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p | 22 | SOX5 | Sponge network | 0.178 | 0.29106 | -1.091 | 4.0E-5 | 0.401 |
| 20022 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577 | 25 | SSBP2 | Sponge network | 0.811 | 0.03228 | -1.58 | 0 | 0.401 |
| 20023 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 16 | NIN | Sponge network | 0.178 | 0.29106 | -0.253 | 0.13794 | 0.401 |
| 20024 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-93-5p | 12 | FBXO31 | Sponge network | -2.69 | 0.0001 | -0.085 | 0.42389 | 0.401 |
| 20025 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | PHC3 | Sponge network | 0.222 | 0.29133 | 0.212 | 0.0499 | 0.401 |
| 20026 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-940 | 16 | CRTC3 | Sponge network | 0.559 | 0.00026 | 0.164 | 0.16786 | 0.401 |
| 20027 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-940 | 24 | MYO9A | Sponge network | 1.254 | 0 | -0.147 | 0.32373 | 0.401 |
| 20028 | RP11-182J1.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | MEF2C | Sponge network | -0.187 | 0.23787 | -0.499 | 0.01448 | 0.401 |
| 20029 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | NAV3 | Sponge network | -1.375 | 0.08642 | 0.212 | 0.47729 | 0.401 |
| 20030 | RP5-1139B12.3 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-502-5p | 12 | ANK2 | Sponge network | 0.598 | 0.38481 | -1.603 | 1.0E-5 | 0.401 |
| 20031 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | 0.349 | 0.01695 | 0.809 | 0.00544 | 0.401 |
| 20032 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | RET | Sponge network | -0.769 | 0.00035 | -0.833 | 0.03517 | 0.401 |
| 20033 | RASSF8-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | DOCK4 | Sponge network | 0.335 | 0.20596 | 0.502 | 0.00108 | 0.401 |
| 20034 | RP11-102N12.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-505-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | ST6GALNAC3 | Sponge network | -0.351 | 0.11358 | -0.987 | 0 | 0.401 |
| 20035 | RP11-216B9.6 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-590-3p | 11 | ITGB1 | Sponge network | 0.174 | 0.30034 | 0.524 | 0.00017 | 0.401 |
| 20036 | RP11-798M19.6 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 12 | ID4 | Sponge network | -0.612 | 0.00094 | -1.644 | 0 | 0.401 |
| 20037 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | SCN9A | Sponge network | 0.174 | 0.30034 | -0.525 | 0.10593 | 0.401 |
| 20038 | LINC00641 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 16 | LATS2 | Sponge network | -0.222 | 0.32445 | 0.159 | 0.2304 | 0.401 |
| 20039 | LINC00092 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-345-5p;hsa-miR-3913-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 50 | LPP | Sponge network | -1.886 | 0 | -0.459 | 0.04475 | 0.401 |
| 20040 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-93-5p | 35 | CHD5 | Sponge network | -0.842 | 0.01361 | -0.922 | 0.01521 | 0.401 |
| 20041 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | TET1 | Sponge network | 0.614 | 1.0E-5 | -0.135 | 0.52138 | 0.401 |
| 20042 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-5p | 15 | USP6 | Sponge network | 0.419 | 0.0319 | 0.188 | 0.3871 | 0.401 |
| 20043 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-625-3p | 21 | CBX5 | Sponge network | 0.497 | 0.01451 | 0.165 | 0.24446 | 0.401 |
| 20044 | H1FX-AS1 |
hsa-miR-107;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-429 | 11 | ELMO1 | Sponge network | -0.206 | 0.12942 | 0.04 | 0.83147 | 0.401 |
| 20045 | HOXA-AS2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | 0.61 | 0.01593 | -1.051 | 0.00022 | 0.401 |
| 20046 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-590-3p | 13 | AKAP11 | Sponge network | 1.055 | 0 | 0.196 | 0.09825 | 0.4 |
| 20047 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-34c-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-96-5p | 52 | KIF1B | Sponge network | 0.078 | 0.55803 | -0.118 | 0.32258 | 0.4 |
| 20048 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p | 10 | PHF6 | Sponge network | 0.537 | 0.00139 | 0.785 | 0 | 0.4 |
| 20049 | LINC00094 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-93-3p | 18 | SUFU | Sponge network | 0.253 | 0.09565 | -0.04 | 0.65489 | 0.4 |
| 20050 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-582-5p | 17 | USP6 | Sponge network | 0.921 | 0.00118 | 0.188 | 0.3871 | 0.4 |
| 20051 | GABPB1-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 12 | FBXO11 | Sponge network | 1.11 | 0 | 0.142 | 0.04938 | 0.4 |
| 20052 | FENDRR |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | ZNF208 | Sponge network | -1.281 | 0.00012 | -0.4 | 0.22381 | 0.4 |
| 20053 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 54 | CADM2 | Sponge network | 0.572 | 0.01722 | -3.782 | 0 | 0.4 |
| 20054 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-505-3p | 17 | LRP1B | Sponge network | -0.583 | 0.14589 | -2.163 | 0 | 0.4 |
| 20055 | FGD5-AS1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 12 | SHPRH | Sponge network | 0.401 | 0.00066 | 0.098 | 0.40994 | 0.4 |
| 20056 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 11 | DCLK1 | Sponge network | -0.08 | 0.90092 | -1.324 | 0.00014 | 0.4 |
| 20057 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | TSC1 | Sponge network | 0.782 | 0.00016 | 0.103 | 0.26071 | 0.4 |
| 20058 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 15 | PIK3IP1 | Sponge network | -1.111 | 0.0003 | -0.187 | 0.19945 | 0.4 |
| 20059 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p | 13 | RIMBP2 | Sponge network | -0.693 | 0.05629 | -1.968 | 5.0E-5 | 0.4 |
| 20060 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 23 | SOX5 | Sponge network | -0.726 | 0.00132 | -1.091 | 4.0E-5 | 0.4 |
| 20061 | KB-1460A1.5 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | ABI2 | Sponge network | 0.603 | 0.00127 | 0.175 | 0.14787 | 0.4 |
| 20062 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p | 15 | RBMS3 | Sponge network | 0.37 | 0.27614 | -0.519 | 0.03551 | 0.4 |
| 20063 | RP11-206L10.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-539-5p | 11 | TSC1 | Sponge network | 0.793 | 0 | 0.103 | 0.26071 | 0.4 |
| 20064 | DIO3OS |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-582-5p | 13 | NRXN3 | Sponge network | -0.773 | 0.04073 | -0.893 | 0.04186 | 0.4 |
| 20065 | SNX29P2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | MED12L | Sponge network | 0.4 | 0.14611 | -0.194 | 0.55299 | 0.4 |
| 20066 | RP11-121C2.2 |
hsa-miR-101-3p;hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 15 | GSK3B | Sponge network | 1.147 | 0 | 0.266 | 0.00045 | 0.4 |
| 20067 | RP1-151F17.2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | PIK3CA | Sponge network | 0.222 | 0.29133 | 0.195 | 0.06908 | 0.4 |
| 20068 | RP11-182J1.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-484;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | PRKCB | Sponge network | -0.187 | 0.23787 | -1.313 | 2.0E-5 | 0.4 |
| 20069 | GNG12-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 13 | PAK3 | Sponge network | -0.793 | 7.0E-5 | -0.628 | 0.07253 | 0.4 |
| 20070 | RP4-791M13.3 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | MED12L | Sponge network | 0.419 | 0.0319 | -0.194 | 0.55299 | 0.4 |
| 20071 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | ARHGAP29 | Sponge network | 0.225 | 0.30644 | 0.131 | 0.42953 | 0.4 |
| 20072 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 15 | HECA | Sponge network | -1.092 | 4.0E-5 | -0.217 | 0.03463 | 0.4 |
| 20073 | CTD-3064M3.3 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-429 | 13 | PCDH17 | Sponge network | -0.469 | 0.08721 | 0.731 | 2.0E-5 | 0.4 |
| 20074 | GNG12-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-940 | 20 | ZFP36L1 | Sponge network | -0.793 | 7.0E-5 | -0.505 | 8.0E-5 | 0.4 |
| 20075 | AC093110.3 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 19 | NF1 | Sponge network | 1.044 | 2.0E-5 | 0.51 | 0 | 0.4 |
| 20076 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-5p | 35 | MOB1B | Sponge network | 0.494 | 0.00339 | 0.197 | 0.15266 | 0.4 |
| 20077 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 41 | BMPR2 | Sponge network | 1.153 | 0 | 0.206 | 0.0635 | 0.4 |
| 20078 | FAM66C |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | CDH10 | Sponge network | -0.901 | 0.00019 | -2.397 | 0 | 0.4 |
| 20079 | RP11-389C8.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | RECK | Sponge network | 0.727 | 3.0E-5 | -1.043 | 0 | 0.4 |
| 20080 | AP001347.6 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 22 | RIMS2 | Sponge network | -1.161 | 0.00591 | -1.365 | 0.00208 | 0.4 |
| 20081 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 18 | CACNA2D1 | Sponge network | -0.227 | 0.31657 | -0.846 | 0.00675 | 0.4 |
| 20082 | RP11-35G9.3 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p | 18 | ABL2 | Sponge network | 0.657 | 4.0E-5 | 0.82 | 0 | 0.4 |
| 20083 | PWAR6 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 20 | BAZ2B | Sponge network | -1.575 | 6.0E-5 | -0.276 | 0.02833 | 0.4 |
| 20084 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 14 | GPAM | Sponge network | 0.972 | 0 | 0.142 | 0.29732 | 0.4 |
| 20085 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 25 | HBP1 | Sponge network | -0.612 | 0.00094 | -0.454 | 0 | 0.4 |
| 20086 | RP11-597D13.9 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 27 | DLC1 | Sponge network | 1.302 | 7.0E-5 | -0.382 | 0.05038 | 0.4 |
| 20087 | NEAT1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 13 | GIGYF2 | Sponge network | 0.705 | 0.00014 | 0.136 | 0.04403 | 0.4 |
| 20088 | HOXA-AS2 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITGB1 | Sponge network | 0.61 | 0.01593 | 0.524 | 0.00017 | 0.4 |
| 20089 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 28 | LRP1B | Sponge network | -1.249 | 7.0E-5 | -2.163 | 0 | 0.4 |
| 20090 | TINCR |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p | 11 | PDZD2 | Sponge network | -0.664 | 0.14543 | -0.909 | 3.0E-5 | 0.4 |
| 20091 | RP11-384O8.1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 13 | KLF10 | Sponge network | -0.738 | 0.21233 | -0.783 | 0 | 0.4 |
| 20092 | RP11-531A24.5 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-625-5p | 21 | PTPN14 | Sponge network | 0.75 | 0.00054 | 0.144 | 0.34595 | 0.4 |
| 20093 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p | 13 | EDA2R | Sponge network | 0.214 | 0.18246 | -0.587 | 0.01598 | 0.4 |
| 20094 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p | 20 | HLF | Sponge network | 0.176 | 0.37402 | -2.443 | 0 | 0.4 |
| 20095 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p | 17 | ABCA10 | Sponge network | 0.335 | 0.20596 | -0.571 | 0.03674 | 0.4 |
| 20096 | RP11-403I13.7 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-532-5p | 10 | PRICKLE1 | Sponge network | 0.395 | 0.15451 | -0.112 | 0.67088 | 0.4 |
| 20097 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-500a-5p | 23 | PRKAB2 | Sponge network | 0.082 | 0.55397 | -0.367 | 0.01371 | 0.4 |
| 20098 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 48 | GAS7 | Sponge network | 0.101 | 0.60919 | -0.555 | 0.01602 | 0.4 |
| 20099 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-940 | 23 | CRTC3 | Sponge network | 0.494 | 0.00339 | 0.164 | 0.16786 | 0.4 |
| 20100 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 52 | PPM1A | Sponge network | -1.697 | 0.00889 | -0.623 | 0 | 0.4 |
| 20101 | RP11-67L2.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 23 | SIRT1 | Sponge network | 0.72 | 0.0001 | 0.109 | 0.2593 | 0.4 |
| 20102 | GNG12-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 28 | NRK | Sponge network | -0.793 | 7.0E-5 | 0.656 | 0.19217 | 0.4 |
| 20103 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 17 | NR2C2 | Sponge network | 0.417 | 0.00445 | 0.21 | 0.03224 | 0.4 |
| 20104 | RP11-13K12.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 40 | GRIK3 | Sponge network | -0.154 | 0.78939 | -3.208 | 0 | 0.4 |
| 20105 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-654-3p;hsa-miR-93-3p | 52 | BMPR2 | Sponge network | -0.227 | 0.31657 | 0.206 | 0.0635 | 0.4 |
| 20106 | AC005154.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | EXOC8 | Sponge network | 0.848 | 0 | 0.216 | 0.00598 | 0.4 |
| 20107 | DLGAP1-AS2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326 | 14 | CBFB | Sponge network | 2.406 | 0 | 1.061 | 0 | 0.4 |
| 20108 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-339-5p;hsa-miR-33a-3p | 13 | SON | Sponge network | 0.706 | 0 | 0.168 | 0.04406 | 0.4 |
| 20109 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-940 | 11 | IGF1 | Sponge network | -0.31 | 0.33871 | -0.204 | 0.5898 | 0.4 |
| 20110 | CTC-559E9.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-337-3p;hsa-miR-576-5p | 18 | ZFX | Sponge network | 0.594 | 0.00064 | 0.09 | 0.42563 | 0.4 |
| 20111 | HAND2-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-3p | 17 | PAK3 | Sponge network | -1.697 | 0.00889 | -0.628 | 0.07253 | 0.4 |
| 20112 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 42 | CREB3L2 | Sponge network | -0.901 | 0.00019 | -0.525 | 2.0E-5 | 0.4 |
| 20113 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-5p | 15 | RARB | Sponge network | -0.811 | 2.0E-5 | -0.825 | 2.0E-5 | 0.4 |
| 20114 | AC010226.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | NCAM2 | Sponge network | -0.811 | 2.0E-5 | -0.482 | 0.22565 | 0.4 |
| 20115 | LINC00957 |
hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SMARCA2 | Sponge network | -0.421 | 0.0114 | -0.529 | 0.00014 | 0.4 |
| 20116 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p | 15 | CBFA2T3 | Sponge network | -1.375 | 0.08642 | -1.221 | 1.0E-5 | 0.4 |
| 20117 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-93-5p | 17 | LHX6 | Sponge network | 0.997 | 0.00066 | -0.087 | 0.69193 | 0.4 |
| 20118 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-421 | 13 | CNTNAP3 | Sponge network | -4.477 | 0 | -1.465 | 0.00033 | 0.4 |
| 20119 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 11 | ZMYM2 | Sponge network | 0.702 | 7.0E-5 | 0.279 | 0.01273 | 0.4 |
| 20120 | HAND2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429 | 14 | ELMO1 | Sponge network | -1.697 | 0.00889 | 0.04 | 0.83147 | 0.4 |
| 20121 | CTC-296K1.4 |
hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-5p | 20 | IL6ST | Sponge network | -2.076 | 0.00548 | -0.402 | 0.00676 | 0.4 |
| 20122 | TUG1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p | 19 | ZMYM2 | Sponge network | 0.972 | 0 | 0.279 | 0.01273 | 0.4 |
| 20123 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-940 | 26 | NTRK2 | Sponge network | -0.154 | 0.53954 | -0.736 | 0.06495 | 0.4 |
| 20124 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 22 | PCDH17 | Sponge network | -0.976 | 0.00025 | 0.731 | 2.0E-5 | 0.4 |
| 20125 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FSTL5 | Sponge network | 0.064 | 0.72934 | -1.3 | 0.01619 | 0.4 |
| 20126 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | RCAN2 | Sponge network | 0.889 | 9.0E-5 | -0.33 | 0.15172 | 0.4 |
| 20127 | RP11-182J1.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 17 | WDFY3 | Sponge network | -0.187 | 0.23787 | -0.201 | 0.0967 | 0.4 |
| 20128 | HNRNPU-AS1 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TSC1 | Sponge network | 1.601 | 0 | 0.103 | 0.26071 | 0.4 |
| 20129 | LINC00476 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 23 | CDH23 | Sponge network | -0.615 | 0.00015 | -0.974 | 0.00034 | 0.4 |
| 20130 | RP11-326G21.1 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 11 | PAIP2 | Sponge network | -0.021 | 0.93598 | -0.515 | 0 | 0.4 |
| 20131 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | -0.096 | 0.67958 | -1.047 | 0.00364 | 0.4 |
| 20132 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 30 | CADM1 | Sponge network | -2.46 | 0 | -0.9 | 0.00084 | 0.4 |
| 20133 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p | 13 | TLL1 | Sponge network | -0.738 | 0.21233 | -1.195 | 3.0E-5 | 0.4 |
| 20134 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-3615;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p | 12 | DMTF1 | Sponge network | 0.415 | 0.14657 | 0.248 | 0.02726 | 0.4 |
| 20135 | RP11-473I1.9 |
hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p | 11 | RNF111 | Sponge network | 0.683 | 1.0E-5 | 0.098 | 0.21093 | 0.4 |
| 20136 | LINC00865 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-766-3p | 14 | EZH1 | Sponge network | -1.249 | 7.0E-5 | -0.564 | 1.0E-5 | 0.4 |
| 20137 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | MYO9A | Sponge network | 0.538 | 0.00076 | -0.147 | 0.32373 | 0.4 |
| 20138 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 13 | ZNF770 | Sponge network | 0.706 | 0 | 0.206 | 0.06751 | 0.4 |
| 20139 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-629-5p | 11 | UVRAG | Sponge network | -0.769 | 0.00035 | -0.017 | 0.83865 | 0.4 |
| 20140 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-577;hsa-miR-590-3p | 14 | MAP3K12 | Sponge network | -0.323 | 0.01372 | -0.057 | 0.59369 | 0.4 |
| 20141 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | SEMA3C | Sponge network | 0.395 | 0.15451 | -0.272 | 0.31153 | 0.4 |
| 20142 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | SYNE1 | Sponge network | 0.395 | 0.15451 | -0.738 | 0.00316 | 0.4 |
| 20143 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 33 | PIK3R1 | Sponge network | -0.896 | 0.00099 | -0.133 | 0.36854 | 0.4 |
| 20144 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 28 | BMPR2 | Sponge network | 0.817 | 3.0E-5 | 0.206 | 0.0635 | 0.4 |
| 20145 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 10 | SETD2 | Sponge network | 0.782 | 0.00016 | 0.031 | 0.73068 | 0.4 |
| 20146 | RP11-53O19.1 |
hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-452-5p | 13 | FNBP1 | Sponge network | -0.405 | 0.22013 | -0.969 | 4.0E-5 | 0.4 |
| 20147 | GS1-124K5.11 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p | 21 | PIK3CA | Sponge network | 0.494 | 0.00339 | 0.195 | 0.06908 | 0.4 |
| 20148 | FAM13A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 77 | LPP | Sponge network | -0.83 | 0 | -0.459 | 0.04475 | 0.4 |
| 20149 | CTD-2017D11.1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-484 | 13 | MED12L | Sponge network | 0.792 | 0.0049 | -0.194 | 0.55299 | 0.4 |
| 20150 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 12 | MLLT10 | Sponge network | 1.147 | 0 | 0.105 | 0.33292 | 0.4 |
| 20151 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | MOB1B | Sponge network | 0.415 | 0.14657 | 0.197 | 0.15266 | 0.4 |
| 20152 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | EYA4 | Sponge network | -0.421 | 0.0114 | 0.3 | 0.36876 | 0.4 |
| 20153 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | RECK | Sponge network | 0.064 | 0.72934 | -1.043 | 0 | 0.4 |
| 20154 | RP11-212P7.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | ABI2 | Sponge network | 0.219 | 0.1516 | 0.175 | 0.14787 | 0.4 |
| 20155 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-940 | 30 | IGF1 | Sponge network | -0.405 | 0.22013 | -0.204 | 0.5898 | 0.4 |
| 20156 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p | 13 | HECA | Sponge network | -0.285 | 0.2172 | -0.217 | 0.03463 | 0.4 |
| 20157 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 18 | PLSCR4 | Sponge network | -4.477 | 0 | -0.474 | 0.03833 | 0.4 |
| 20158 | FAM157A |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p | 11 | MLLT10 | Sponge network | 0.813 | 1.0E-5 | 0.105 | 0.33292 | 0.4 |
| 20159 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 23 | DOCK4 | Sponge network | 0.194 | 0.64278 | 0.502 | 0.00108 | 0.4 |
| 20160 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | MLLT10 | Sponge network | 0.225 | 0.30644 | 0.105 | 0.33292 | 0.4 |
| 20161 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 45 | NTRK2 | Sponge network | -0.942 | 0 | -0.736 | 0.06495 | 0.4 |
| 20162 | HHIP-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-532-3p | 10 | CDH23 | Sponge network | -0.737 | 0.10822 | -0.974 | 0.00034 | 0.4 |
| 20163 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p | 23 | PRDM2 | Sponge network | 0.082 | 0.55397 | -0.258 | 0.00721 | 0.4 |
| 20164 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLIP1 | Sponge network | 0.311 | 0.10344 | -0.215 | 0.08684 | 0.4 |
| 20165 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p | 17 | FGFR1 | Sponge network | -0.222 | 0.32445 | -0.509 | 0.03957 | 0.4 |
| 20166 | LINC00467 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 10 | CBFB | Sponge network | 0.884 | 0 | 1.061 | 0 | 0.4 |
| 20167 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | HOOK3 | Sponge network | -0.901 | 0.00019 | 0.273 | 0.09957 | 0.4 |
| 20168 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-221-3p | 10 | PRDM2 | Sponge network | 0.497 | 0.01451 | -0.258 | 0.00721 | 0.4 |
| 20169 | LINC00641 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-671-5p | 23 | PPP2R5C | Sponge network | -0.222 | 0.32445 | -0.314 | 3.0E-5 | 0.4 |
| 20170 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-590-3p | 18 | TGFBR3 | Sponge network | -1.582 | 8.0E-5 | -1.173 | 1.0E-5 | 0.4 |
| 20171 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 11 | ITGB3 | Sponge network | -1.582 | 8.0E-5 | -0.011 | 0.95751 | 0.4 |
| 20172 | RP11-13K12.1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-98-5p | 15 | KLF10 | Sponge network | -0.154 | 0.78939 | -0.783 | 0 | 0.4 |
| 20173 | RP11-53O19.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | KDSR | Sponge network | 1.205 | 0 | 0.239 | 0.02216 | 0.4 |
| 20174 | BVES-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-935 | 28 | PTPRD | Sponge network | -2.62 | 0 | -1.005 | 0.00064 | 0.4 |
| 20175 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | TRIM33 | Sponge network | -0.031 | 0.89063 | 0.402 | 7.0E-5 | 0.4 |
| 20176 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | FGF5 | Sponge network | -0.612 | 0.00094 | 0.549 | 0.28659 | 0.4 |
| 20177 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | HACE1 | Sponge network | 1.063 | 0 | -0.072 | 0.55239 | 0.4 |
| 20178 | HNRNPU-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | BCL9 | Sponge network | 1.601 | 0 | 0.321 | 0.00267 | 0.4 |
| 20179 | RP11-499P20.2 |
hsa-let-7d-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-493-5p;hsa-miR-505-3p | 11 | USP6 | Sponge network | 0.187 | 0.44121 | 0.188 | 0.3871 | 0.4 |
| 20180 | AC103563.9 |
hsa-miR-134-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-577 | 10 | SSBP2 | Sponge network | -6.057 | 0 | -1.58 | 0 | 0.4 |
| 20181 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-324-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | ABCC9 | Sponge network | 0.381 | 0.25967 | -0.466 | 0.14121 | 0.4 |
| 20182 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-629-5p | 11 | UVRAG | Sponge network | -0.976 | 0.00025 | -0.017 | 0.83865 | 0.4 |
| 20183 | AC010226.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-96-5p | 21 | SNX29 | Sponge network | -0.811 | 2.0E-5 | -0.34 | 0.01371 | 0.4 |
| 20184 | RP11-798M19.6 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | PRKCB | Sponge network | -0.612 | 0.00094 | -1.313 | 2.0E-5 | 0.4 |
| 20185 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-5p | 39 | JAZF1 | Sponge network | -0.615 | 0.00015 | -0.377 | 0.02677 | 0.4 |
| 20186 | AGAP11 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-5p | 13 | TLL1 | Sponge network | -1.645 | 0 | -1.195 | 3.0E-5 | 0.4 |
| 20187 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 34 | EPHA3 | Sponge network | -0.777 | 0.1134 | 0.185 | 0.53588 | 0.4 |
| 20188 | RP11-597D13.9 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-7-5p | 12 | COL1A2 | Sponge network | 1.302 | 7.0E-5 | 1.991 | 0 | 0.4 |
| 20189 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | PGR | Sponge network | 0.385 | 0.10067 | -1.704 | 0 | 0.4 |
| 20190 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p | 14 | EZH1 | Sponge network | -0.602 | 0.00331 | -0.564 | 1.0E-5 | 0.4 |
| 20191 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940 | 25 | SNX29 | Sponge network | -0.318 | 0.65847 | -0.34 | 0.01371 | 0.4 |
| 20192 | RP11-535M15.1 |
hsa-miR-146a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-589-5p | 10 | TCF4 | Sponge network | -1.14 | 0.03513 | 0.016 | 0.92271 | 0.4 |
| 20193 | RP11-473I1.9 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 15 | UBE4B | Sponge network | 0.683 | 1.0E-5 | -0.235 | 0.007 | 0.4 |
| 20194 | PART1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DNAJB4 | Sponge network | -2.925 | 0 | -0.7 | 1.0E-5 | 0.4 |
| 20195 | PCA3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 40 | NTRK2 | Sponge network | -0.709 | 0.18168 | -0.736 | 0.06495 | 0.4 |
| 20196 | LINC00702 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-96-5p | 13 | ELMO1 | Sponge network | -0.737 | 0.06182 | 0.04 | 0.83147 | 0.4 |
| 20197 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-323a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 24 | EPHA5 | Sponge network | -0.473 | 0.07602 | -0.957 | 0.01733 | 0.4 |
| 20198 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-7-1-3p | 11 | ATRX | Sponge network | 1.11 | 1.0E-5 | 0.261 | 0.04408 | 0.4 |
| 20199 | RP11-403I13.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | PRRX1 | Sponge network | 0.395 | 0.15451 | 1.316 | 0 | 0.4 |
| 20200 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-93-3p | 18 | RPS6KA2 | Sponge network | 0.194 | 0.64278 | -0.57 | 0.00013 | 0.4 |
| 20201 | RP11-428J1.5 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | BCL9 | Sponge network | 0.538 | 0.00076 | 0.321 | 0.00267 | 0.4 |
| 20202 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-3607-3p;hsa-miR-590-5p | 10 | PBRM1 | Sponge network | 0.706 | 0 | -0.029 | 0.78601 | 0.4 |
| 20203 | LINC01018 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 15 | SPTBN4 | Sponge network | -2.915 | 0.00015 | -0.786 | 0.01281 | 0.4 |
| 20204 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-576-5p | 11 | EDA2R | Sponge network | -0.246 | 0.35034 | -0.587 | 0.01598 | 0.4 |
| 20205 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-424-5p | 10 | PRKAB2 | Sponge network | -4.463 | 0.00025 | -0.367 | 0.01371 | 0.4 |
| 20206 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-5p | 25 | ERC1 | Sponge network | 0.299 | 0.57722 | -0.108 | 0.38794 | 0.4 |
| 20207 | RP11-156P1.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | FAM172A | Sponge network | 0.214 | 0.18246 | -0.57 | 0 | 0.399 |
| 20208 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PTPRB | Sponge network | -1.057 | 0.00029 | 0.105 | 0.47122 | 0.399 |
| 20209 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | RBL2 | Sponge network | 0.194 | 0.64278 | -0.282 | 0.00254 | 0.399 |
| 20210 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 68 | IL6ST | Sponge network | -1.216 | 0 | -0.402 | 0.00676 | 0.399 |
| 20211 | RP1-68D18.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-429 | 10 | ABL2 | Sponge network | 1.987 | 0 | 0.82 | 0 | 0.399 |
| 20212 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-532-3p | 14 | CADM3 | Sponge network | -0.405 | 0.22013 | -3.663 | 0 | 0.399 |
| 20213 | FGD5-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 19 | SLC5A3 | Sponge network | 0.401 | 0.00066 | 0.493 | 5.0E-5 | 0.399 |
| 20214 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 19 | RARB | Sponge network | -2.915 | 0.00015 | -0.825 | 2.0E-5 | 0.399 |
| 20215 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 31 | IGF1 | Sponge network | 0.101 | 0.60919 | -0.204 | 0.5898 | 0.399 |
| 20216 | LINC00174 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 1.253 | 0 | 0.51 | 0 | 0.399 |
| 20217 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | NEDD4 | Sponge network | 0.249 | 0.35902 | 0.02 | 0.89834 | 0.399 |
| 20218 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-421 | 17 | RSPO2 | Sponge network | -1.654 | 0.00144 | -3.038 | 0 | 0.399 |
| 20219 | HAND2-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 33 | EYA4 | Sponge network | -1.697 | 0.00889 | 0.3 | 0.36876 | 0.399 |
| 20220 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 31 | RICTOR | Sponge network | 0.603 | 0.00127 | 0.388 | 0.00188 | 0.399 |
| 20221 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-335-5p | 14 | PIK3C2A | Sponge network | 0.997 | 0 | 0.061 | 0.53776 | 0.399 |
| 20222 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 26 | CRTC3 | Sponge network | -2.46 | 0 | 0.164 | 0.16786 | 0.399 |
| 20223 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 10 | ATRX | Sponge network | 0.856 | 0 | 0.261 | 0.04408 | 0.399 |
| 20224 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | BMPR2 | Sponge network | 0.826 | 1.0E-5 | 0.206 | 0.0635 | 0.399 |
| 20225 | CTD-2554C21.2 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p | 14 | CSDE1 | Sponge network | -1.654 | 0.00144 | -0.493 | 0 | 0.399 |
| 20226 | AC003090.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 40 | NTRK2 | Sponge network | -2.117 | 0.00161 | -0.736 | 0.06495 | 0.399 |
| 20227 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | BMPR2 | Sponge network | 0.998 | 0 | 0.206 | 0.0635 | 0.399 |
| 20228 | CTD-2314G24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-98-5p | 14 | ITGB3 | Sponge network | 0.246 | 0.59912 | -0.011 | 0.95751 | 0.399 |
| 20229 | GABPB1-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 14 | KIAA1024 | Sponge network | 1.11 | 0 | 1.083 | 0 | 0.399 |
| 20230 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 25 | AR | Sponge network | -0.285 | 0.2172 | -1.554 | 0 | 0.399 |
| 20231 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 33 | ATRX | Sponge network | -0.615 | 0.00015 | 0.261 | 0.04408 | 0.399 |
| 20232 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-501-3p | 13 | SLITRK3 | Sponge network | 0.176 | 0.37402 | -3.328 | 0 | 0.399 |
| 20233 | RP11-195F19.9 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 10 | SPG20 | Sponge network | -0.726 | 0.00132 | -1.16 | 0 | 0.399 |
| 20234 | RP11-326G21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | DLC1 | Sponge network | -0.021 | 0.93598 | -0.382 | 0.05038 | 0.399 |
| 20235 | AC011526.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 17 | DMD | Sponge network | 0.342 | 0.03129 | -1.506 | 0 | 0.399 |
| 20236 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 36 | FAT3 | Sponge network | 0.174 | 0.30034 | -1.601 | 0.00051 | 0.399 |
| 20237 | RP1-59D14.5 |
hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-511-5p;hsa-miR-940 | 17 | CREB1 | Sponge network | 1.162 | 5.0E-5 | 0.203 | 0.00537 | 0.399 |
| 20238 | LINC00476 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 21 | ATM | Sponge network | -0.615 | 0.00015 | 0.178 | 0.21568 | 0.399 |
| 20239 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-664a-3p | 11 | MN1 | Sponge network | -0.355 | 0.0077 | -0.358 | 0.24172 | 0.399 |
| 20240 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p | 19 | RNF144A | Sponge network | -1.092 | 4.0E-5 | 0.594 | 0.0009 | 0.399 |
| 20241 | LINC00265 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-378a-5p | 14 | CREB1 | Sponge network | 1.07 | 0 | 0.203 | 0.00537 | 0.399 |
| 20242 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | CLASP2 | Sponge network | 0.532 | 0.00505 | -0.038 | 0.71 | 0.399 |
| 20243 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | SYNE1 | Sponge network | 0.056 | 0.8494 | -0.738 | 0.00316 | 0.399 |
| 20244 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | CBFA2T3 | Sponge network | -0.323 | 0.01372 | -1.221 | 1.0E-5 | 0.399 |
| 20245 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | NUAK1 | Sponge network | -0.318 | 0.65847 | 0.904 | 0 | 0.399 |
| 20246 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | CACNA2D1 | Sponge network | -0.246 | 0.35034 | -0.846 | 0.00675 | 0.399 |
| 20247 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 13 | ACVR1 | Sponge network | 0.898 | 1.0E-5 | 0.279 | 0.00234 | 0.399 |
| 20248 | RP1-39G22.7 |
hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | SHPRH | Sponge network | 0.439 | 0.00313 | 0.098 | 0.40994 | 0.399 |
| 20249 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p | 46 | FAT3 | Sponge network | 0.385 | 0.10067 | -1.601 | 0.00051 | 0.399 |
| 20250 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | TIMP3 | Sponge network | -2.117 | 0.00161 | -0.546 | 0.02307 | 0.399 |
| 20251 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p | 25 | PTPN14 | Sponge network | -0.769 | 0.00035 | 0.144 | 0.34595 | 0.399 |
| 20252 | LINC00641 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p | 19 | DDX3X | Sponge network | -0.222 | 0.32445 | -0.152 | 0.07686 | 0.399 |
| 20253 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 30 | PBX1 | Sponge network | 0.61 | 0.01593 | -1.206 | 0 | 0.399 |
| 20254 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 41 | ZFHX4 | Sponge network | 0.16 | 0.50785 | 0.018 | 0.95574 | 0.399 |
| 20255 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 39 | MEF2C | Sponge network | 0.225 | 0.30644 | -0.499 | 0.01448 | 0.399 |
| 20256 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | MITF | Sponge network | -1.057 | 0.00029 | -0.769 | 0.00022 | 0.399 |
| 20257 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p | 20 | KIT | Sponge network | -1.161 | 0.00591 | -2.184 | 0 | 0.399 |
| 20258 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | PIK3C2A | Sponge network | 0.858 | 3.0E-5 | 0.061 | 0.53776 | 0.399 |
| 20259 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | LRRC7 | Sponge network | -0.021 | 0.93598 | -1.341 | 0.01193 | 0.399 |
| 20260 | AP001347.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | NCOA1 | Sponge network | -1.161 | 0.00591 | -0.432 | 0 | 0.399 |
| 20261 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 15 | ST6GAL2 | Sponge network | -0.021 | 0.93598 | -1.201 | 0.00597 | 0.399 |
| 20262 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-503-5p;hsa-miR-590-3p | 11 | SMARCA1 | Sponge network | 0.214 | 0.18246 | -0.684 | 0.00159 | 0.399 |
| 20263 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p | 14 | FSTL5 | Sponge network | 0.374 | 0.20054 | -1.3 | 0.01619 | 0.399 |
| 20264 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-424-3p;hsa-miR-493-5p | 10 | HOOK3 | Sponge network | -2.62 | 0 | 0.273 | 0.09957 | 0.399 |
| 20265 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | PPM1A | Sponge network | -1.575 | 6.0E-5 | -0.623 | 0 | 0.399 |
| 20266 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 24 | AFF3 | Sponge network | 0.246 | 0.59912 | -2.373 | 0 | 0.399 |
| 20267 | BZRAP1-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-3p | 16 | DOCK4 | Sponge network | -0.465 | 0.04703 | 0.502 | 0.00108 | 0.399 |
| 20268 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 15 | FAT4 | Sponge network | -0.72 | 0.24239 | -0.354 | 0.14694 | 0.399 |
| 20269 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | UBE4B | Sponge network | 0.421 | 0.00084 | -0.235 | 0.007 | 0.399 |
| 20270 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | PHC3 | Sponge network | -0.129 | 0.57177 | 0.212 | 0.0499 | 0.399 |
| 20271 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 23 | TGFBR2 | Sponge network | 0.997 | 0.00066 | -0.243 | 0.11408 | 0.399 |
| 20272 | RP11-283I3.6 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | RASSF8 | Sponge network | 0.614 | 1.0E-5 | -0.122 | 0.65591 | 0.399 |
| 20273 | MIR143HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 58 | PTPRD | Sponge network | -0.739 | 0.0574 | -1.005 | 0.00064 | 0.399 |
| 20274 | MEG3 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-766-3p | 21 | ATM | Sponge network | 0.249 | 0.35902 | 0.178 | 0.21568 | 0.399 |
| 20275 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p | 22 | DDX6 | Sponge network | 1.317 | 0 | 0.091 | 0.36428 | 0.399 |
| 20276 | RP11-6O2.3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p | 14 | SEMA3C | Sponge network | 0.331 | 0.57247 | -0.272 | 0.31153 | 0.399 |
| 20277 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 41 | TRPS1 | Sponge network | -0.154 | 0.78939 | -0.191 | 0.44109 | 0.399 |
| 20278 | RP11-867G23.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-874-3p | 14 | MXI1 | Sponge network | -2.531 | 0 | -0.987 | 0 | 0.399 |
| 20279 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-589-5p | 29 | SOX5 | Sponge network | -0.154 | 0.53954 | -1.091 | 4.0E-5 | 0.399 |
| 20280 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -1.047 | 0 | -0.354 | 0.14694 | 0.399 |
| 20281 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 32 | PRKAB2 | Sponge network | 0.374 | 0.20054 | -0.367 | 0.01371 | 0.399 |
| 20282 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PRUNE2 | Sponge network | 0.225 | 0.30644 | -1.733 | 0.00022 | 0.399 |
| 20283 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SON | Sponge network | 1.378 | 0 | 0.168 | 0.04406 | 0.399 |
| 20284 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 21 | NEDD4 | Sponge network | -0.096 | 0.67958 | 0.02 | 0.89834 | 0.399 |
| 20285 | TPTEP1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 45 | DIP2C | Sponge network | -1.216 | 0 | -0.436 | 0.01713 | 0.399 |
| 20286 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-26b-5p | 18 | DMD | Sponge network | -0.285 | 0.2172 | -1.506 | 0 | 0.399 |
| 20287 | RP11-283I3.6 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 11 | BCL9 | Sponge network | 0.614 | 1.0E-5 | 0.321 | 0.00267 | 0.399 |
| 20288 | RP11-597D13.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 14 | STARD13 | Sponge network | 1.302 | 7.0E-5 | -0.187 | 0.27383 | 0.399 |
| 20289 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-486-5p | 11 | NR2C2 | Sponge network | 1.117 | 0 | 0.21 | 0.03224 | 0.399 |
| 20290 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PTPN11 | Sponge network | 0.34 | 0.00273 | 0.431 | 2.0E-5 | 0.399 |
| 20291 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | EDA2R | Sponge network | -0.421 | 0.0114 | -0.587 | 0.01598 | 0.399 |
| 20292 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 18 | SPG20 | Sponge network | 0.246 | 0.59912 | -1.16 | 0 | 0.399 |
| 20293 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-93-5p | 11 | RBL2 | Sponge network | 0.4 | 0.14611 | -0.282 | 0.00254 | 0.399 |
| 20294 | NEAT1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | PIK3C2A | Sponge network | 0.705 | 0.00014 | 0.061 | 0.53776 | 0.399 |
| 20295 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-589-5p | 10 | ITGB3 | Sponge network | -0.318 | 0.65847 | -0.011 | 0.95751 | 0.399 |
| 20296 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p | 39 | RICTOR | Sponge network | 0.349 | 0.01695 | 0.388 | 0.00188 | 0.399 |
| 20297 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 41 | AKAP11 | Sponge network | -1.111 | 0.0003 | 0.196 | 0.09825 | 0.399 |
| 20298 | TPTEP1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 16 | STAT5B | Sponge network | -1.216 | 0 | -0.182 | 0.1082 | 0.399 |
| 20299 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLASP2 | Sponge network | 1.153 | 0 | -0.038 | 0.71 | 0.399 |
| 20300 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 40 | CHD5 | Sponge network | 0.101 | 0.60919 | -0.922 | 0.01521 | 0.399 |
| 20301 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | LRP1B | Sponge network | -0.405 | 0.22013 | -2.163 | 0 | 0.399 |
| 20302 | LINC00641 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 18 | TRIM33 | Sponge network | -0.222 | 0.32445 | 0.402 | 7.0E-5 | 0.399 |
| 20303 | RP11-384K6.6 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-654-3p | 14 | DICER1 | Sponge network | 0.946 | 0 | 0.042 | 0.674 | 0.399 |
| 20304 | RP11-464F9.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 10 | DLG1 | Sponge network | 0.959 | 0 | -0.014 | 0.8926 | 0.399 |
| 20305 | TMEM9B-AS1 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-542-3p;hsa-miR-940 | 14 | MYO9A | Sponge network | 0.529 | 0.00552 | -0.147 | 0.32373 | 0.399 |
| 20306 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | BNC2 | Sponge network | -0.342 | 0.02288 | -0.491 | 0.09988 | 0.399 |
| 20307 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-590-3p | 12 | RICTOR | Sponge network | 0.545 | 0.05789 | 0.388 | 0.00188 | 0.399 |
| 20308 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 11 | AKAP11 | Sponge network | 1.316 | 0 | 0.196 | 0.09825 | 0.399 |
| 20309 | RP11-182J1.1 |
hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-219a-1-3p;hsa-miR-370-3p;hsa-miR-96-5p | 12 | JAZF1 | Sponge network | -0.187 | 0.23787 | -0.377 | 0.02677 | 0.399 |
| 20310 | HNRNPU-AS1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-495-3p | 13 | STAG2 | Sponge network | 1.601 | 0 | 0.252 | 0.00438 | 0.399 |
| 20311 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | -2.452 | 0 | -0.001 | 0.99669 | 0.399 |
| 20312 | RP11-158K1.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-766-3p | 13 | EZH1 | Sponge network | 0.349 | 0.01695 | -0.564 | 1.0E-5 | 0.399 |
| 20313 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-590-3p | 11 | JAKMIP2 | Sponge network | -0.513 | 0.04703 | -1.388 | 0 | 0.399 |
| 20314 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | -1.697 | 0.00889 | -0.001 | 0.99669 | 0.399 |
| 20315 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 27 | CNTN1 | Sponge network | 0.082 | 0.55654 | -2.594 | 0 | 0.399 |
| 20316 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | PTPRD | Sponge network | -0.318 | 0.65847 | -1.005 | 0.00064 | 0.399 |
| 20317 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | PDGFC | Sponge network | -0.942 | 0 | -0.33 | 0.06483 | 0.399 |
| 20318 | LIPE-AS1 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 13 | MAP3K4 | Sponge network | 0.078 | 0.55803 | 0.058 | 0.54229 | 0.399 |
| 20319 | MALAT1 |
hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | FBXW7 | Sponge network | 0.782 | 0.00016 | -0.418 | 2.0E-5 | 0.399 |
| 20320 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b | 10 | IGF1 | Sponge network | -1.14 | 0.03513 | -0.204 | 0.5898 | 0.399 |
| 20321 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 28 | TIMP3 | Sponge network | -2.46 | 0 | -0.546 | 0.02307 | 0.399 |
| 20322 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | ATRX | Sponge network | -0.487 | 0.02157 | 0.261 | 0.04408 | 0.399 |
| 20323 | SNX29P2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 10 | DCLK1 | Sponge network | 0.4 | 0.14611 | -1.324 | 0.00014 | 0.399 |
| 20324 | RP11-225B17.2 |
hsa-miR-101-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 16 | UBR3 | Sponge network | 1.055 | 0 | -0.021 | 0.82774 | 0.399 |
| 20325 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p | 14 | PTGFR | Sponge network | 0.16 | 0.50785 | -0.233 | 0.45421 | 0.399 |
| 20326 | U91328.19 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-7-1-3p | 19 | ZMYND11 | Sponge network | -0.303 | 0.21002 | -0.2 | 0.05449 | 0.399 |
| 20327 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-93-5p | 23 | TCF4 | Sponge network | 0.497 | 0.01451 | 0.016 | 0.92271 | 0.399 |
| 20328 | RP11-57H14.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | NAV3 | Sponge network | 0.083 | 0.66405 | 0.212 | 0.47729 | 0.399 |
| 20329 | RP11-57H14.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-671-5p | 27 | DLC1 | Sponge network | 0.083 | 0.66405 | -0.382 | 0.05038 | 0.399 |
| 20330 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | PTPN11 | Sponge network | 1.2 | 0.01813 | 0.431 | 2.0E-5 | 0.399 |
| 20331 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p | 15 | RYR2 | Sponge network | -0.469 | 0.08721 | -1.466 | 0.00031 | 0.399 |
| 20332 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | PAFAH1B1 | Sponge network | -1.057 | 0.00029 | -0.429 | 0 | 0.399 |
| 20333 | AGAP11 |
hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-942-5p | 19 | EMP2 | Sponge network | -1.645 | 0 | -0.423 | 0.00985 | 0.399 |
| 20334 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 38 | TSHZ3 | Sponge network | 1.302 | 7.0E-5 | -0.556 | 0.01516 | 0.399 |
| 20335 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-940 | 28 | PBX1 | Sponge network | 0.214 | 0.18246 | -1.206 | 0 | 0.399 |
| 20336 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-409-3p | 11 | PHC3 | Sponge network | 0.453 | 0.01839 | 0.212 | 0.0499 | 0.399 |
| 20337 | RP11-254F7.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-484 | 12 | CDH23 | Sponge network | 0.176 | 0.37402 | -0.974 | 0.00034 | 0.399 |
| 20338 | RAMP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-532-3p | 12 | SYNM | Sponge network | -0.513 | 0.04703 | -2.309 | 1.0E-5 | 0.399 |
| 20339 | EMX2OS |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CDH11 | Sponge network | -0.777 | 0.1134 | 1.427 | 0 | 0.399 |
| 20340 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SON | Sponge network | 0.953 | 0 | 0.168 | 0.04406 | 0.399 |
| 20341 | LINC00476 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-758-3p | 24 | CSDE1 | Sponge network | -0.615 | 0.00015 | -0.493 | 0 | 0.399 |
| 20342 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | AFF3 | Sponge network | -0.727 | 0.04726 | -2.373 | 0 | 0.399 |
| 20343 | DIO3OS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-628-5p | 43 | DTNA | Sponge network | -0.773 | 0.04073 | -1.648 | 1.0E-5 | 0.399 |
| 20344 | LINC00854 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-5p | 21 | BMPR2 | Sponge network | 1.18 | 0 | 0.206 | 0.0635 | 0.399 |
| 20345 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | PIK3C2A | Sponge network | 0.225 | 0.30644 | 0.061 | 0.53776 | 0.399 |
| 20346 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | FGF5 | Sponge network | -2.46 | 0 | 0.549 | 0.28659 | 0.399 |
| 20347 | RP4-717I23.3 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-582-3p;hsa-miR-625-3p | 25 | ABL2 | Sponge network | 0.637 | 0.00069 | 0.82 | 0 | 0.399 |
| 20348 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 14 | AKAP12 | Sponge network | 0.083 | 0.66405 | -0.932 | 0.0028 | 0.399 |
| 20349 | FZD10-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | PREX2 | Sponge network | -0.727 | 0.04726 | -0.272 | 0.25382 | 0.399 |
| 20350 | RP5-1139B12.3 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-27a-3p | 17 | MEF2C | Sponge network | 0.598 | 0.38481 | -0.499 | 0.01448 | 0.399 |
| 20351 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-942-5p | 11 | FBXO31 | Sponge network | 0.064 | 0.72934 | -0.085 | 0.42389 | 0.399 |
| 20352 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-7-1-3p | 25 | PCDH9 | Sponge network | 0.395 | 0.15451 | -1.823 | 4.0E-5 | 0.399 |
| 20353 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | PDS5B | Sponge network | 0.683 | 1.0E-5 | 0.259 | 0.00962 | 0.399 |
| 20354 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 27 | CBL | Sponge network | 0.559 | 0.00026 | 0.291 | 0.01267 | 0.399 |
| 20355 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 22 | ZFHX3 | Sponge network | 1.153 | 0 | 0.389 | 0.01363 | 0.399 |
| 20356 | TPTEP1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | THSD7B | Sponge network | -1.216 | 0 | 0.154 | 0.74723 | 0.399 |
| 20357 | LINC00403 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-339-5p | 12 | NRXN1 | Sponge network | -3.586 | 0.00032 | -3.778 | 0 | 0.399 |
| 20358 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-93-3p | 15 | PTPN11 | Sponge network | 0.078 | 0.55803 | 0.431 | 2.0E-5 | 0.399 |
| 20359 | RP11-798M19.6 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | -0.612 | 0.00094 | 0.524 | 0.00017 | 0.399 |
| 20360 | ZNF667-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 16 | CELF4 | Sponge network | -0.976 | 0.00025 | -1.135 | 0.00344 | 0.399 |
| 20361 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | RAP1A | Sponge network | -0.222 | 0.32445 | -0.929 | 0 | 0.399 |
| 20362 | RP1-68D18.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-429 | 12 | MDM4 | Sponge network | 1.987 | 0 | 0.345 | 0.00762 | 0.399 |
| 20363 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CLIP1 | Sponge network | 0.782 | 0.00016 | -0.215 | 0.08684 | 0.399 |
| 20364 | MEG3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p | 12 | JAKMIP2 | Sponge network | 0.249 | 0.35902 | -1.388 | 0 | 0.399 |
| 20365 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 56 | IL6ST | Sponge network | -1.047 | 0 | -0.402 | 0.00676 | 0.399 |
| 20366 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-582-3p | 18 | PRDM2 | Sponge network | -0.465 | 0.04703 | -0.258 | 0.00721 | 0.399 |
| 20367 | HOXA-AS2 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | 0.61 | 0.01593 | 0.358 | 0.13727 | 0.399 |
| 20368 | HOXD-AS2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-502-5p | 20 | DTNA | Sponge network | -0.208 | 0.65592 | -1.648 | 1.0E-5 | 0.399 |
| 20369 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 28 | ETV1 | Sponge network | 0.572 | 0.01722 | -0.096 | 0.67674 | 0.399 |
| 20370 | RP11-307B6.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-378a-3p;hsa-miR-378c | 21 | WDFY3 | Sponge network | -4.463 | 0.00025 | -0.201 | 0.0967 | 0.399 |
| 20371 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-940 | 27 | PBX1 | Sponge network | -0.154 | 0.53954 | -1.206 | 0 | 0.399 |
| 20372 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-652-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 32 | QKI | Sponge network | 0.622 | 0.00015 | -0.333 | 0.04111 | 0.399 |
| 20373 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-93-5p | 18 | LATS2 | Sponge network | -0.154 | 0.78939 | 0.159 | 0.2304 | 0.399 |
| 20374 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p | 10 | SON | Sponge network | 1.122 | 0 | 0.168 | 0.04406 | 0.399 |
| 20375 | CTB-179K24.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p;hsa-miR-940 | 24 | MDM4 | Sponge network | 0.841 | 0.04055 | 0.345 | 0.00762 | 0.399 |
| 20376 | NNT-AS1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-486-5p;hsa-miR-590-3p | 10 | TPR | Sponge network | 0.799 | 1.0E-5 | 0.582 | 0 | 0.399 |
| 20377 | HOTAIRM1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-424-3p;hsa-miR-532-3p | 12 | SYNM | Sponge network | -0.053 | 0.80685 | -2.309 | 1.0E-5 | 0.399 |
| 20378 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 0.385 | 0.10067 | 0.114 | 0.30638 | 0.399 |
| 20379 | FAM13A-AS1 |
hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 47 | PPARA | Sponge network | -0.83 | 0 | -0.379 | 0.00548 | 0.399 |
| 20380 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | RECK | Sponge network | -0.342 | 0.02288 | -1.043 | 0 | 0.399 |
| 20381 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | YAP1 | Sponge network | 0.997 | 0.00066 | 0.22 | 0.11836 | 0.399 |
| 20382 | RP11-10C24.1 | hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p | 10 | PHF6 | Sponge network | 0.874 | 1.0E-5 | 0.785 | 0 | 0.399 |
| 20383 | MALAT1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 26 | EPS15 | Sponge network | 0.782 | 0.00016 | -0.052 | 0.53998 | 0.399 |
| 20384 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 17 | DKK3 | Sponge network | -0.942 | 0 | -0.052 | 0.77783 | 0.398 |
| 20385 | MALAT1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | ZFX | Sponge network | 0.782 | 0.00016 | 0.09 | 0.42563 | 0.398 |
| 20386 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 19 | NIN | Sponge network | 0.614 | 1.0E-5 | -0.253 | 0.13794 | 0.398 |
| 20387 | LINC00702 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 32 | NCOA1 | Sponge network | -0.737 | 0.06182 | -0.432 | 0 | 0.398 |
| 20388 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 27 | DDX6 | Sponge network | 0.91 | 0 | 0.091 | 0.36428 | 0.398 |
| 20389 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-654-3p | 19 | RICTOR | Sponge network | 1.157 | 0.00455 | 0.388 | 0.00188 | 0.398 |
| 20390 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 12 | ZFHX4 | Sponge network | -2.039 | 0.03447 | 0.018 | 0.95574 | 0.398 |
| 20391 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | PRKAB2 | Sponge network | 0.222 | 0.29133 | -0.367 | 0.01371 | 0.398 |
| 20392 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | RABEP1 | Sponge network | 0.083 | 0.66405 | -0.066 | 0.43142 | 0.398 |
| 20393 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 11 | LHFP | Sponge network | 0.101 | 0.60919 | -0.167 | 0.41371 | 0.398 |
| 20394 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-34a-5p | 11 | TXNIP | Sponge network | -0.154 | 0.53954 | -1.277 | 0 | 0.398 |
| 20395 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | RBMS3 | Sponge network | -0.08 | 0.90092 | -0.519 | 0.03551 | 0.398 |
| 20396 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p | 28 | FBXO32 | Sponge network | 0.603 | 0.00127 | -0.495 | 0.07561 | 0.398 |
| 20397 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p | 21 | HECA | Sponge network | -0.096 | 0.67958 | -0.217 | 0.03463 | 0.398 |
| 20398 | RAD51-AS1 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p | 12 | TSC1 | Sponge network | 0.768 | 7.0E-5 | 0.103 | 0.26071 | 0.398 |
| 20399 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 21 | USP6 | Sponge network | -0.323 | 0.01372 | 0.188 | 0.3871 | 0.398 |
| 20400 | RP5-1198O20.4 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-589-3p | 22 | KCNJ5 | Sponge network | 0.997 | 0.00066 | -0.281 | 0.3339 | 0.398 |
| 20401 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 55 | THRB | Sponge network | -2.117 | 0.00161 | -1.117 | 0.0004 | 0.398 |
| 20402 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p | 28 | TACC1 | Sponge network | -0.738 | 0.21233 | -0.362 | 0.05406 | 0.398 |
| 20403 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p | 29 | GRIA3 | Sponge network | -1.575 | 6.0E-5 | -1.956 | 0 | 0.398 |
| 20404 | AC004893.11 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | CREB1 | Sponge network | 1.317 | 0 | 0.203 | 0.00537 | 0.398 |
| 20405 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-421;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p;hsa-miR-98-5p | 27 | MITF | Sponge network | 0.246 | 0.59912 | -0.769 | 0.00022 | 0.398 |
| 20406 | LINC00621 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 12 | RASSF8 | Sponge network | 0.131 | 0.8436 | -0.122 | 0.65591 | 0.398 |
| 20407 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p | 13 | DOCK4 | Sponge network | -1.331 | 3.0E-5 | 0.502 | 0.00108 | 0.398 |
| 20408 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 16 | JAKMIP2 | Sponge network | 0.064 | 0.72934 | -1.388 | 0 | 0.398 |
| 20409 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ADAMTSL3 | Sponge network | -0.08 | 0.90092 | -1.559 | 0 | 0.398 |
| 20410 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-654-3p;hsa-miR-7-1-3p | 15 | DDX6 | Sponge network | 2.094 | 0 | 0.091 | 0.36428 | 0.398 |
| 20411 | TTC25 |
hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-424-5p | 10 | EPHA7 | Sponge network | -0.208 | 0.38261 | -2.197 | 3.0E-5 | 0.398 |
| 20412 | RP11-983P16.4 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-629-5p;hsa-miR-96-5p | 13 | WNK2 | Sponge network | 0.41 | 0.02214 | -0.352 | 0.31066 | 0.398 |
| 20413 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 21 | AKAP6 | Sponge network | 1.302 | 7.0E-5 | -1.586 | 3.0E-5 | 0.398 |
| 20414 | DIO3OS |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | -0.773 | 0.04073 | -2.633 | 0 | 0.398 |
| 20415 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-93-5p | 14 | CNTNAP3 | Sponge network | -0.318 | 0.65847 | -1.465 | 0.00033 | 0.398 |
| 20416 | RP11-67L2.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p | 13 | FBXO11 | Sponge network | 0.72 | 0.0001 | 0.142 | 0.04938 | 0.398 |
| 20417 | RP11-1348G14.4 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 11 | ATRX | Sponge network | 0.597 | 0.00037 | 0.261 | 0.04408 | 0.398 |
| 20418 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-940 | 24 | CHD5 | Sponge network | -2.095 | 0.00112 | -0.922 | 0.01521 | 0.398 |
| 20419 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CXXC4 | Sponge network | 0.219 | 0.1516 | -0.433 | 0.21055 | 0.398 |
| 20420 | ANKRD10-IT1 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-7-1-3p | 15 | BMPR2 | Sponge network | 1.739 | 0 | 0.206 | 0.0635 | 0.398 |
| 20421 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-96-5p | 39 | PRKAA2 | Sponge network | 0.331 | 0.57247 | -2.462 | 0 | 0.398 |
| 20422 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | CDC73 | Sponge network | 1.055 | 0 | 0.094 | 0.20463 | 0.398 |
| 20423 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 15 | FGF5 | Sponge network | -0.738 | 0.21233 | 0.549 | 0.28659 | 0.398 |
| 20424 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | MLLT10 | Sponge network | 1.2 | 0.01813 | 0.105 | 0.33292 | 0.398 |
| 20425 | LINC00641 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-340-3p;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-940 | 61 | NFIB | Sponge network | -0.222 | 0.32445 | 0.023 | 0.90795 | 0.398 |
| 20426 | LINC00265 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 1.07 | 0 | 0.578 | 0 | 0.398 |
| 20427 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-651-5p | 26 | PRDM2 | Sponge network | -1.645 | 0 | -0.258 | 0.00721 | 0.398 |
| 20428 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PRKAB2 | Sponge network | 0.91 | 0 | -0.367 | 0.01371 | 0.398 |
| 20429 | RP5-1092A3.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-766-3p | 14 | TBL1XR1 | Sponge network | 1.495 | 0 | 0.578 | 0 | 0.398 |
| 20430 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 38 | PDGFRA | Sponge network | -0.793 | 7.0E-5 | -0.372 | 0.0037 | 0.398 |
| 20431 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | TLE4 | Sponge network | -0.358 | 0.17255 | -1.157 | 0 | 0.398 |
| 20432 | FAM157B |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-338-3p;hsa-miR-338-5p | 11 | CBL | Sponge network | 0.92 | 0 | 0.291 | 0.01267 | 0.398 |
| 20433 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CNTN1 | Sponge network | -0.206 | 0.12942 | -2.594 | 0 | 0.398 |
| 20434 | LINC00473 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-3p | 22 | RSPO2 | Sponge network | -1.203 | 0.03881 | -3.038 | 0 | 0.398 |
| 20435 | RP11-110I1.11 |
hsa-miR-10b-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | CREB1 | Sponge network | 1.034 | 0 | 0.203 | 0.00537 | 0.398 |
| 20436 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | AKAP11 | Sponge network | 0.93 | 0 | 0.196 | 0.09825 | 0.398 |
| 20437 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 39 | BCL2 | Sponge network | -0.612 | 0.00094 | -0.899 | 0 | 0.398 |
| 20438 | RP11-127B20.2 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-654-3p | 11 | BAZ2B | Sponge network | 0.411 | 0.1335 | -0.276 | 0.02833 | 0.398 |
| 20439 | RP11-890B15.3 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PHF3 | Sponge network | 0.101 | 0.60919 | 0.126 | 0.19652 | 0.398 |
| 20440 | ADAMTS9-AS2 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | PCLO | Sponge network | -2.452 | 0 | -1.843 | 0.00064 | 0.398 |
| 20441 | AC005154.6 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 14 | PHF6 | Sponge network | 0.848 | 0 | 0.785 | 0 | 0.398 |
| 20442 | PCA3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 27 | TOM1L2 | Sponge network | -0.709 | 0.18168 | -0.994 | 0 | 0.398 |
| 20443 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p | 14 | FBXO32 | Sponge network | 0.497 | 0.01451 | -0.495 | 0.07561 | 0.398 |
| 20444 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 14 | GPAM | Sponge network | 0.757 | 0 | 0.142 | 0.29732 | 0.398 |
| 20445 | LINC00094 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-361-3p;hsa-miR-625-5p | 10 | BCORL1 | Sponge network | 0.253 | 0.09565 | 0.354 | 0.01214 | 0.398 |
| 20446 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p | 13 | ABCB5 | Sponge network | -0.842 | 0.01361 | -0.956 | 0.04077 | 0.398 |
| 20447 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-655-3p | 12 | ARID2 | Sponge network | 0.683 | 1.0E-5 | 0.235 | 0.02697 | 0.398 |
| 20448 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-625-5p | 20 | ZMYM2 | Sponge network | 0.637 | 0.00069 | 0.279 | 0.01273 | 0.398 |
| 20449 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-874-3p | 34 | TACC1 | Sponge network | 0.494 | 0.00339 | -0.362 | 0.05406 | 0.398 |
| 20450 | RP11-535M15.1 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p | 10 | DIP2C | Sponge network | -1.14 | 0.03513 | -0.436 | 0.01713 | 0.398 |
| 20451 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p | 11 | BRD3 | Sponge network | 0.769 | 0 | 0.37 | 0.00013 | 0.398 |
| 20452 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-625-5p | 22 | PTPRT | Sponge network | -2.531 | 0 | -0.907 | 0.03727 | 0.398 |
| 20453 | RP11-282O18.3 |
hsa-miR-101-3p;hsa-miR-139-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 12 | GSK3B | Sponge network | 0.757 | 0 | 0.266 | 0.00045 | 0.398 |
| 20454 | RP11-389C8.2 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 11 | LATS2 | Sponge network | 0.727 | 3.0E-5 | 0.159 | 0.2304 | 0.398 |
| 20455 | RP11-819C21.1 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 10 | RANBP6 | Sponge network | 0.537 | 0.00139 | 0.175 | 0.19756 | 0.398 |
| 20456 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 19 | RBL2 | Sponge network | -0.615 | 0.00015 | -0.282 | 0.00254 | 0.398 |
| 20457 | RP11-798M19.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-744-3p | 28 | PTPN14 | Sponge network | -0.612 | 0.00094 | 0.144 | 0.34595 | 0.398 |
| 20458 | H1FX-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p | 13 | MRVI1 | Sponge network | -0.206 | 0.12942 | -1.051 | 0.00022 | 0.398 |
| 20459 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | CUL5 | Sponge network | 0.683 | 1.0E-5 | 0.026 | 0.77301 | 0.398 |
| 20460 | LINC00641 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | USP25 | Sponge network | -0.222 | 0.32445 | -0.242 | 0.01397 | 0.398 |
| 20461 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 30 | MYO9A | Sponge network | 0.374 | 0.20054 | -0.147 | 0.32373 | 0.398 |
| 20462 | LINC01018 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 24 | NAV3 | Sponge network | -2.915 | 0.00015 | 0.212 | 0.47729 | 0.398 |
| 20463 | RP4-791M13.3 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-502-5p;hsa-miR-577 | 17 | IL17RD | Sponge network | 0.419 | 0.0319 | -0.019 | 0.94873 | 0.398 |
| 20464 | RP11-244O19.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-935 | 22 | GJA1 | Sponge network | -0.096 | 0.67958 | -0.485 | 0.02179 | 0.398 |
| 20465 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-589-3p | 27 | CHD5 | Sponge network | -1.161 | 0.00591 | -0.922 | 0.01521 | 0.398 |
| 20466 | LINC00578 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-577 | 13 | PCDH17 | Sponge network | 0.811 | 0.03228 | 0.731 | 2.0E-5 | 0.398 |
| 20467 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 11 | MLLT10 | Sponge network | 0.997 | 0 | 0.105 | 0.33292 | 0.398 |
| 20468 | RP11-356J5.12 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-584-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | -0.899 | 6.0E-5 | -0.366 | 0.01036 | 0.398 |
| 20469 | RP11-119F7.5 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | PREX2 | Sponge network | 0.225 | 0.30644 | -0.272 | 0.25382 | 0.398 |
| 20470 | RP11-822E23.8 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p | 11 | ID4 | Sponge network | -0.777 | 0.16667 | -1.644 | 0 | 0.398 |
| 20471 | ARHGEF26-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-584-5p;hsa-miR-589-5p | 11 | BMPR1A | Sponge network | -2.46 | 0 | -0.366 | 0.01036 | 0.398 |
| 20472 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 37 | RAP1A | Sponge network | -0.576 | 0.10121 | -0.929 | 0 | 0.398 |
| 20473 | RP11-166D19.1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | FAM117A | Sponge network | -0.576 | 0.10121 | -1.379 | 0 | 0.398 |
| 20474 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-590-3p | 14 | MAP3K12 | Sponge network | -0.576 | 0.10121 | -0.057 | 0.59369 | 0.398 |
| 20475 | RP11-360F5.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MEF2C | Sponge network | 1.867 | 0 | -0.499 | 0.01448 | 0.398 |
| 20476 | RASSF8-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | CELF4 | Sponge network | 0.335 | 0.20596 | -1.135 | 0.00344 | 0.398 |
| 20477 | HAS2-AS1 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-7-1-3p | 10 | ZFHX4 | Sponge network | -0.011 | 0.967 | 0.018 | 0.95574 | 0.398 |
| 20478 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p | 20 | SYNE1 | Sponge network | -0.773 | 0.04073 | -0.738 | 0.00316 | 0.398 |
| 20479 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-98-5p | 19 | SEMA3C | Sponge network | -2.62 | 0 | -0.272 | 0.31153 | 0.398 |
| 20480 | FAM13A-AS1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | RPS6KA6 | Sponge network | -0.83 | 0 | -2.989 | 0 | 0.398 |
| 20481 | AC005154.6 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 20 | KIF5B | Sponge network | 0.848 | 0 | 0.362 | 0.00066 | 0.398 |
| 20482 | FAM66C |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-429 | 12 | ELMO1 | Sponge network | -0.901 | 0.00019 | 0.04 | 0.83147 | 0.398 |
| 20483 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p | 24 | ABL2 | Sponge network | -0.513 | 0.04703 | 0.82 | 0 | 0.398 |
| 20484 | RP11-216B9.6 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-375;hsa-miR-590-3p | 11 | ITPKB | Sponge network | 0.174 | 0.30034 | -0.704 | 0.00106 | 0.398 |
| 20485 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 10 | PDS5B | Sponge network | 0.594 | 0.00064 | 0.259 | 0.00962 | 0.398 |
| 20486 | ARHGEF26-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | LUZP2 | Sponge network | -2.46 | 0 | -0.056 | 0.89416 | 0.398 |
| 20487 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 19 | PCDH9 | Sponge network | 0.889 | 9.0E-5 | -1.823 | 4.0E-5 | 0.398 |
| 20488 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TIMP3 | Sponge network | 0.532 | 0.00505 | -0.546 | 0.02307 | 0.398 |
| 20489 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-877-5p | 21 | AR | Sponge network | 1.122 | 0.02989 | -1.554 | 0 | 0.398 |
| 20490 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-484 | 10 | RET | Sponge network | 0.238 | 0.70544 | -0.833 | 0.03517 | 0.398 |
| 20491 | RP4-717I23.3 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-582-5p | 10 | PHF3 | Sponge network | 0.637 | 0.00069 | 0.126 | 0.19652 | 0.398 |
| 20492 | GAS6-AS2 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-450b-5p | 12 | CXCL12 | Sponge network | 0.16 | 0.50785 | -1.421 | 0 | 0.398 |
| 20493 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 46 | RUNX1T1 | Sponge network | -0.342 | 0.02288 | -0.344 | 0.2374 | 0.398 |
| 20494 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-5p | 30 | CBL | Sponge network | 0.078 | 0.55803 | 0.291 | 0.01267 | 0.398 |
| 20495 | LPP-AS2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p | 11 | MAPK10 | Sponge network | -0.18 | 0.20343 | -1.774 | 0 | 0.398 |
| 20496 | PWAR6 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | MLF1 | Sponge network | -1.575 | 6.0E-5 | -0.893 | 0.00727 | 0.398 |
| 20497 | HCG18 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 12 | DLG1 | Sponge network | 1.063 | 0 | -0.014 | 0.8926 | 0.398 |
| 20498 | CTD-2002H8.2 |
hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ABI2 | Sponge network | 0.931 | 1.0E-5 | 0.175 | 0.14787 | 0.398 |
| 20499 | RP11-894P9.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-582-3p;hsa-miR-582-5p | 15 | BRD3 | Sponge network | 0.993 | 0 | 0.37 | 0.00013 | 0.398 |
| 20500 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ABCC9 | Sponge network | 0.72 | 0.0001 | -0.466 | 0.14121 | 0.398 |
| 20501 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p | 13 | ZNF292 | Sponge network | 0.267 | 0.2937 | 0.276 | 0.04148 | 0.398 |
| 20502 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 29 | ARID5B | Sponge network | 0.194 | 0.64278 | 0.342 | 0.01676 | 0.398 |
| 20503 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-539-5p | 15 | GRIN2A | Sponge network | -0.154 | 0.53954 | -2.161 | 0 | 0.398 |
| 20504 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-935 | 10 | LRRN3 | Sponge network | -2.46 | 0 | -0.393 | 0.16293 | 0.398 |
| 20505 | RP11-968O1.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-301a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p | 10 | CDH23 | Sponge network | -0.829 | 0.01343 | -0.974 | 0.00034 | 0.398 |
| 20506 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429 | 12 | AHNAK | Sponge network | -0.154 | 0.53954 | -1.207 | 0 | 0.398 |
| 20507 | PART1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 33 | DLC1 | Sponge network | -2.925 | 0 | -0.382 | 0.05038 | 0.398 |
| 20508 | TPT1-AS1 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-582-5p | 12 | ABI2 | Sponge network | 0.903 | 0 | 0.175 | 0.14787 | 0.398 |
| 20509 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 17 | GPAM | Sponge network | 1.147 | 0 | 0.142 | 0.29732 | 0.398 |
| 20510 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p | 19 | SYNE1 | Sponge network | 0.331 | 0.57247 | -0.738 | 0.00316 | 0.398 |
| 20511 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 40 | KIF1B | Sponge network | -1.092 | 4.0E-5 | -0.118 | 0.32258 | 0.398 |
| 20512 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | EBF1 | Sponge network | 0.246 | 0.59912 | -0.384 | 0.08856 | 0.398 |
| 20513 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 41 | TACC1 | Sponge network | -0.899 | 6.0E-5 | -0.362 | 0.05406 | 0.398 |
| 20514 | KB-1460A1.5 |
hsa-miR-101-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ZNF638 | Sponge network | 0.603 | 0.00127 | 0.22 | 0.01214 | 0.398 |
| 20515 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | PIK3C2A | Sponge network | 1.148 | 2.0E-5 | 0.061 | 0.53776 | 0.398 |
| 20516 | AC016747.3 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 31 | IGFBP5 | Sponge network | 0.199 | 0.38015 | -0.806 | 0.00105 | 0.398 |
| 20517 | CTC-444N24.11 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | UBE2D1 | Sponge network | 1.153 | 0 | 0.468 | 0 | 0.398 |
| 20518 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | TIMP3 | Sponge network | -0.942 | 0 | -0.546 | 0.02307 | 0.398 |
| 20519 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | RECK | Sponge network | -0.206 | 0.12942 | -1.043 | 0 | 0.398 |
| 20520 | RP1-151F17.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p | 18 | PTPN14 | Sponge network | 0.222 | 0.29133 | 0.144 | 0.34595 | 0.398 |
| 20521 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-576-5p | 12 | GREM1 | Sponge network | -0.246 | 0.35034 | 0.902 | 0.01691 | 0.398 |
| 20522 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 14 | RBL2 | Sponge network | 0.72 | 0.0001 | -0.282 | 0.00254 | 0.398 |
| 20523 | HCG11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SETBP1 | Sponge network | 0.385 | 0.10067 | -1.302 | 1.0E-5 | 0.398 |
| 20524 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CLIP1 | Sponge network | -0.021 | 0.93598 | -0.215 | 0.08684 | 0.398 |
| 20525 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 16 | MAP3K12 | Sponge network | -0.793 | 7.0E-5 | -0.057 | 0.59369 | 0.398 |
| 20526 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | ARHGAP29 | Sponge network | -0.323 | 0.01372 | 0.131 | 0.42953 | 0.398 |
| 20527 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-5p;hsa-miR-92b-3p | 13 | AKAP12 | Sponge network | -1.047 | 0 | -0.932 | 0.0028 | 0.398 |
| 20528 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-935 | 32 | FBXO32 | Sponge network | -1.249 | 7.0E-5 | -0.495 | 0.07561 | 0.398 |
| 20529 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | -0.355 | 0.0077 | -1.751 | 1.0E-5 | 0.398 |
| 20530 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-550a-3p | 12 | PCDH8 | Sponge network | -1.249 | 7.0E-5 | -0.997 | 0.05366 | 0.398 |
| 20531 | CTC-429P9.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 17 | CREB1 | Sponge network | 0.178 | 0.29106 | 0.203 | 0.00537 | 0.398 |
| 20532 | HOXB-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-628-5p;hsa-miR-93-3p | 22 | ERC1 | Sponge network | -0.154 | 0.53954 | -0.108 | 0.38794 | 0.398 |
| 20533 | AC009948.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.532 | 0.00505 | -0.57 | 0 | 0.398 |
| 20534 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | SETBP1 | Sponge network | 0.083 | 0.66405 | -1.302 | 1.0E-5 | 0.398 |
| 20535 | TP73-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-429 | 10 | DUSP1 | Sponge network | -0.563 | 0.02545 | -1.754 | 0 | 0.398 |
| 20536 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-877-5p;hsa-miR-940 | 39 | GAS7 | Sponge network | -0.246 | 0.35034 | -0.555 | 0.01602 | 0.398 |
| 20537 | RP11-226L15.5 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p | 13 | PHF6 | Sponge network | 0.8 | 0 | 0.785 | 0 | 0.398 |
| 20538 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p | 23 | PIK3C2A | Sponge network | -0.031 | 0.89063 | 0.061 | 0.53776 | 0.398 |
| 20539 | MIR497HG |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-96-5p | 17 | PAIP2 | Sponge network | -1.439 | 4.0E-5 | -0.515 | 0 | 0.398 |
| 20540 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | ETV1 | Sponge network | 0.051 | 0.83388 | -0.096 | 0.67674 | 0.398 |
| 20541 | RP11-159D12.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | KDSR | Sponge network | 1.254 | 0 | 0.239 | 0.02216 | 0.398 |
| 20542 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | RCAN2 | Sponge network | 0.72 | 0.0001 | -0.33 | 0.15172 | 0.398 |
| 20543 | ACTA2-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-3p | 17 | FMNL3 | Sponge network | 0.194 | 0.64278 | 0.555 | 4.0E-5 | 0.398 |
| 20544 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-501-3p;hsa-miR-625-5p | 13 | NUAK1 | Sponge network | -0.154 | 0.53954 | 0.904 | 0 | 0.398 |
| 20545 | PART1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ITPKB | Sponge network | -2.925 | 0 | -0.704 | 0.00106 | 0.398 |
| 20546 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 28 | PCDH17 | Sponge network | 0.335 | 0.20596 | 0.731 | 2.0E-5 | 0.398 |
| 20547 | LIPE-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-5p | 12 | ITGB1 | Sponge network | 0.078 | 0.55803 | 0.524 | 0.00017 | 0.398 |
| 20548 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DNAJB4 | Sponge network | -1.203 | 0.03881 | -0.7 | 1.0E-5 | 0.398 |
| 20549 | MIR143HG |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | BTG2 | Sponge network | -0.739 | 0.0574 | -1.763 | 0 | 0.398 |
| 20550 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | RNF144A | Sponge network | -1.697 | 0.00889 | 0.594 | 0.0009 | 0.398 |
| 20551 | THAP9-AS1 |
hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-497-5p | 10 | ZMYM2 | Sponge network | 1.079 | 0 | 0.279 | 0.01273 | 0.398 |
| 20552 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-940 | 10 | FAM172A | Sponge network | 0.299 | 0.57722 | -0.57 | 0 | 0.398 |
| 20553 | H19 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-589-3p | 10 | RNF144A | Sponge network | 2.439 | 0 | 0.594 | 0.0009 | 0.398 |
| 20554 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-744-5p;hsa-miR-93-5p | 19 | FGFR1 | Sponge network | -0.18 | 0.20343 | -0.509 | 0.03957 | 0.398 |
| 20555 | CTD-2002H8.2 |
hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ZFX | Sponge network | 0.931 | 1.0E-5 | 0.09 | 0.42563 | 0.398 |
| 20556 | RP11-212P7.2 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 10 | RNF180 | Sponge network | 0.219 | 0.1516 | -1.127 | 1.0E-5 | 0.397 |
| 20557 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | HDAC9 | Sponge network | -0.896 | 0.00099 | -0.755 | 0.00495 | 0.397 |
| 20558 | BZRAP1-AS1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 18 | ABL1 | Sponge network | -0.465 | 0.04703 | -0.215 | 0.07804 | 0.397 |
| 20559 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 47 | TGFBR3 | Sponge network | -1.216 | 0 | -1.173 | 1.0E-5 | 0.397 |
| 20560 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 46 | EPHA7 | Sponge network | 0.064 | 0.72934 | -2.197 | 3.0E-5 | 0.397 |
| 20561 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 26 | SASH1 | Sponge network | 0.95 | 0.15943 | -0.502 | 0.00175 | 0.397 |
| 20562 | PCA3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SEMA3C | Sponge network | -0.709 | 0.18168 | -0.272 | 0.31153 | 0.397 |
| 20563 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | SUFU | Sponge network | -0.727 | 0.04726 | -0.04 | 0.65489 | 0.397 |
| 20564 | RP11-473I1.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | ABI2 | Sponge network | 0.683 | 1.0E-5 | 0.175 | 0.14787 | 0.397 |
| 20565 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -0.563 | 0.02545 | -0.7 | 1.0E-5 | 0.397 |
| 20566 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-96-5p | 14 | ERG | Sponge network | -0.517 | 0.02935 | 0.002 | 0.99011 | 0.397 |
| 20567 | A2M-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | GLI3 | Sponge network | -0.08 | 0.90092 | -0.615 | 0.01482 | 0.397 |
| 20568 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 54 | FOXP1 | Sponge network | -0.129 | 0.57177 | 0.042 | 0.74368 | 0.397 |
| 20569 | SH3RF3-AS1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-590-5p | 30 | DST | Sponge network | 0.889 | 9.0E-5 | -0.713 | 0.00096 | 0.397 |
| 20570 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-7-1-3p | 26 | ZFHX4 | Sponge network | -0.726 | 0.00132 | 0.018 | 0.95574 | 0.397 |
| 20571 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-503-5p | 23 | RICTOR | Sponge network | 0.556 | 0.00298 | 0.388 | 0.00188 | 0.397 |
| 20572 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 15 | NF1 | Sponge network | 1.266 | 0 | 0.51 | 0 | 0.397 |
| 20573 | RP1-122P22.2 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-629-5p | 20 | DST | Sponge network | 0.065 | 0.73468 | -0.713 | 0.00096 | 0.397 |
| 20574 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | RNF144A | Sponge network | -0.487 | 0.02157 | 0.594 | 0.0009 | 0.397 |
| 20575 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-361-5p;hsa-miR-424-5p | 11 | ABCB5 | Sponge network | -1.349 | 0.00017 | -0.956 | 0.04077 | 0.397 |
| 20576 | C1RL-AS1 |
hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-365a-3p | 10 | RICTOR | Sponge network | 1.26 | 0 | 0.388 | 0.00188 | 0.397 |
| 20577 | TP73-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -0.563 | 0.02545 | -0.966 | 0.00035 | 0.397 |
| 20578 | RP11-983P16.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 22 | AKAP6 | Sponge network | 0.41 | 0.02214 | -1.586 | 3.0E-5 | 0.397 |
| 20579 | RP11-13K12.5 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-7-5p | 10 | LZTS1 | Sponge network | -0.318 | 0.65847 | 1.664 | 0 | 0.397 |
| 20580 | AC010226.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | NAV3 | Sponge network | -0.811 | 2.0E-5 | 0.212 | 0.47729 | 0.397 |
| 20581 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-655-3p;hsa-miR-92a-3p | 20 | UBE4B | Sponge network | -0.096 | 0.67958 | -0.235 | 0.007 | 0.397 |
| 20582 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-590-5p | 18 | FREM2 | Sponge network | -0.384 | 0.40652 | -0.437 | 0.46353 | 0.397 |
| 20583 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p | 19 | NRXN1 | Sponge network | 0.176 | 0.37402 | -3.778 | 0 | 0.397 |
| 20584 | NR2F1-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-425-3p;hsa-miR-429 | 12 | DUSP1 | Sponge network | -0.49 | 0.04946 | -1.754 | 0 | 0.397 |
| 20585 | RP11-439A17.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-576-5p | 13 | FAT3 | Sponge network | 0.051 | 0.95756 | -1.601 | 0.00051 | 0.397 |
| 20586 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | ATRX | Sponge network | 1.317 | 0 | 0.261 | 0.04408 | 0.397 |
| 20587 | ZNF582-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-629-3p | 21 | RYR2 | Sponge network | -1.349 | 0.00017 | -1.466 | 0.00031 | 0.397 |
| 20588 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 19 | RICTOR | Sponge network | 0.545 | 0.00064 | 0.388 | 0.00188 | 0.397 |
| 20589 | PCA3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 14 | PDZD2 | Sponge network | -0.709 | 0.18168 | -0.909 | 3.0E-5 | 0.397 |
| 20590 | HAND2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 17 | UVRAG | Sponge network | -1.697 | 0.00889 | -0.017 | 0.83865 | 0.397 |
| 20591 | RP11-6N17.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-455-5p | 12 | AKAP6 | Sponge network | 0.108 | 0.69556 | -1.586 | 3.0E-5 | 0.397 |
| 20592 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p | 13 | JDP2 | Sponge network | -1.886 | 0 | -0.967 | 0 | 0.397 |
| 20593 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 16 | SLITRK3 | Sponge network | -1.057 | 0.00029 | -3.328 | 0 | 0.397 |
| 20594 | RP11-121C2.2 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-654-3p | 13 | DICER1 | Sponge network | 1.147 | 0 | 0.042 | 0.674 | 0.397 |
| 20595 | RP11-244H3.1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | ABI2 | Sponge network | 0.659 | 3.0E-5 | 0.175 | 0.14787 | 0.397 |
| 20596 | FAM66C |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | -0.901 | 0.00019 | 0.783 | 0.00072 | 0.397 |
| 20597 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | PTPRB | Sponge network | -0.709 | 0.18168 | 0.105 | 0.47122 | 0.397 |
| 20598 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 28 | NRXN1 | Sponge network | -0.227 | 0.31657 | -3.778 | 0 | 0.397 |
| 20599 | AC011526.1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | LATS2 | Sponge network | 0.342 | 0.03129 | 0.159 | 0.2304 | 0.397 |
| 20600 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 62 | CADM2 | Sponge network | 0.997 | 0.00066 | -3.782 | 0 | 0.397 |
| 20601 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p | 24 | RICTOR | Sponge network | 0.439 | 0.00313 | 0.388 | 0.00188 | 0.397 |
| 20602 | RP11-305E6.4 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | PHF3 | Sponge network | 1.317 | 0 | 0.126 | 0.19652 | 0.397 |
| 20603 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-590-3p | 14 | PAK3 | Sponge network | -0.487 | 0.02157 | -0.628 | 0.07253 | 0.397 |
| 20604 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | CUL5 | Sponge network | 1.063 | 0 | 0.026 | 0.77301 | 0.397 |
| 20605 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SYNE1 | Sponge network | -0.125 | 0.49414 | -0.738 | 0.00316 | 0.397 |
| 20606 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 19 | NR2C2 | Sponge network | 0.194 | 0.64278 | 0.21 | 0.03224 | 0.397 |
| 20607 | RP11-216B9.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-375;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 16 | TET1 | Sponge network | 0.174 | 0.30034 | -0.135 | 0.52138 | 0.397 |
| 20608 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-539-5p | 21 | BTG2 | Sponge network | -1.092 | 4.0E-5 | -1.763 | 0 | 0.397 |
| 20609 | CTC-296K1.3 |
hsa-miR-103a-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-452-5p;hsa-miR-582-5p | 20 | BCL2 | Sponge network | -2.095 | 0.00112 | -0.899 | 0 | 0.397 |
| 20610 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-3p | 14 | UBE4B | Sponge network | -0.162 | 0.47687 | -0.235 | 0.007 | 0.397 |
| 20611 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | TGFBR3 | Sponge network | 0.16 | 0.50785 | -1.173 | 1.0E-5 | 0.397 |
| 20612 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 20 | NEDD4 | Sponge network | 0.494 | 0.00339 | 0.02 | 0.89834 | 0.397 |
| 20613 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-96-5p | 13 | EPHA3 | Sponge network | 0.898 | 1.0E-5 | 0.185 | 0.53588 | 0.397 |
| 20614 | RP11-701H24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-501-3p | 10 | TGFBR3 | Sponge network | -2.039 | 0.03447 | -1.173 | 1.0E-5 | 0.397 |
| 20615 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-362-5p;hsa-miR-450b-5p | 10 | TLE4 | Sponge network | -2.531 | 0 | -1.157 | 0 | 0.397 |
| 20616 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 26 | RSPO2 | Sponge network | -0.323 | 0.01372 | -3.038 | 0 | 0.397 |
| 20617 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-744-3p;hsa-miR-96-5p | 21 | HIP1 | Sponge network | -0.976 | 0.00025 | 0.774 | 0 | 0.397 |
| 20618 | CTC-457L16.2 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 12 | SETBP1 | Sponge network | 1.153 | 0.00114 | -1.302 | 1.0E-5 | 0.397 |
| 20619 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-590-3p | 11 | PTPN11 | Sponge network | 0.545 | 0.00064 | 0.431 | 2.0E-5 | 0.397 |
| 20620 | RP11-251G23.5 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326 | 16 | CBFB | Sponge network | 1.344 | 0 | 1.061 | 0 | 0.397 |
| 20621 | LINC00092 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-671-5p | 13 | CYLD | Sponge network | -1.886 | 0 | -0.367 | 0.00171 | 0.397 |
| 20622 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p | 29 | PPM1A | Sponge network | -2.62 | 0 | -0.623 | 0 | 0.397 |
| 20623 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p | 10 | AFAP1L2 | Sponge network | -0.405 | 0.22013 | -0.284 | 0.15434 | 0.397 |
| 20624 | RP11-212P7.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 77 | LPP | Sponge network | 0.219 | 0.1516 | -0.459 | 0.04475 | 0.397 |
| 20625 | C20orf166-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577 | 11 | SLC6A15 | Sponge network | -2.69 | 0.0001 | -2.373 | 2.0E-5 | 0.397 |
| 20626 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | NEDD4 | Sponge network | -0.976 | 0.00025 | 0.02 | 0.89834 | 0.397 |
| 20627 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TRIM13 | Sponge network | 0.559 | 0.00026 | 0.183 | 0.06968 | 0.397 |
| 20628 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 58 | WDFY3 | Sponge network | -0.576 | 0.10121 | -0.201 | 0.0967 | 0.397 |
| 20629 | RP11-701H24.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-501-3p;hsa-miR-576-5p | 10 | TET1 | Sponge network | -2.039 | 0.03447 | -0.135 | 0.52138 | 0.397 |
| 20630 | MIR143HG |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FAM133A | Sponge network | -0.739 | 0.0574 | 0.549 | 0.29009 | 0.397 |
| 20631 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 24 | AKAP11 | Sponge network | 1.11 | 0 | 0.196 | 0.09825 | 0.397 |
| 20632 | RP11-13K12.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | CDH19 | Sponge network | -0.318 | 0.65847 | -3.434 | 0 | 0.397 |
| 20633 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-190a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | CXXC4 | Sponge network | 0.419 | 0.0319 | -0.433 | 0.21055 | 0.397 |
| 20634 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 33 | ARID5B | Sponge network | -0.487 | 0.02157 | 0.342 | 0.01676 | 0.397 |
| 20635 | LINC00403 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 18 | SSBP2 | Sponge network | -3.586 | 0.00032 | -1.58 | 0 | 0.397 |
| 20636 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-7-1-3p | 13 | AKAP6 | Sponge network | -4.477 | 0 | -1.586 | 3.0E-5 | 0.397 |
| 20637 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p | 12 | CNTN1 | Sponge network | -2.039 | 0.03447 | -2.594 | 0 | 0.397 |
| 20638 | HNRNPU-AS1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | GSK3B | Sponge network | 1.601 | 0 | 0.266 | 0.00045 | 0.397 |
| 20639 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-493-5p;hsa-miR-500b-5p | 15 | SLITRK6 | Sponge network | -0.405 | 0.22013 | -2.121 | 0 | 0.397 |
| 20640 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p | 25 | PBX1 | Sponge network | 0.811 | 0.03228 | -1.206 | 0 | 0.397 |
| 20641 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 11 | ALDH1A2 | Sponge network | -2.69 | 0.0001 | -2.411 | 0 | 0.397 |
| 20642 | RASSF8-AS1 |
hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | GUCY1A2 | Sponge network | 0.335 | 0.20596 | 1.213 | 0 | 0.397 |
| 20643 | AC006116.24 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-582-3p | 18 | PRDM2 | Sponge network | -0.473 | 0.07602 | -0.258 | 0.00721 | 0.397 |
| 20644 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 18 | SCN9A | Sponge network | -0.053 | 0.80685 | -0.525 | 0.10593 | 0.397 |
| 20645 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -0.18 | 0.20343 | -0.243 | 0.11408 | 0.397 |
| 20646 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p | 35 | NOTCH2 | Sponge network | -0.154 | 0.78939 | 0.138 | 0.35163 | 0.397 |
| 20647 | C20orf166-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p | 19 | LIMCH1 | Sponge network | -2.69 | 0.0001 | -0.915 | 1.0E-5 | 0.397 |
| 20648 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | NEDD4 | Sponge network | 0.222 | 0.29133 | 0.02 | 0.89834 | 0.397 |
| 20649 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-93-5p | 37 | AR | Sponge network | 0.082 | 0.55397 | -1.554 | 0 | 0.397 |
| 20650 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 29 | JAZF1 | Sponge network | -1.057 | 0.00029 | -0.377 | 0.02677 | 0.397 |
| 20651 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | PIK3CA | Sponge network | 1.089 | 0 | 0.195 | 0.06908 | 0.397 |
| 20652 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 41 | ADARB1 | Sponge network | -0.513 | 0.04703 | -0.335 | 0.06631 | 0.397 |
| 20653 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p | 36 | GAS7 | Sponge network | -0.513 | 0.04703 | -0.555 | 0.01602 | 0.397 |
| 20654 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | PTPRB | Sponge network | 1.302 | 7.0E-5 | 0.105 | 0.47122 | 0.397 |
| 20655 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-3614-5p;hsa-miR-542-3p | 15 | TSC1 | Sponge network | 0.529 | 0.00552 | 0.103 | 0.26071 | 0.397 |
| 20656 | AC003090.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 43 | PTPRD | Sponge network | -2.117 | 0.00161 | -1.005 | 0.00064 | 0.397 |
| 20657 | AC004540.4 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-362-3p | 10 | GRIN2A | Sponge network | -2.549 | 0.00528 | -2.161 | 0 | 0.397 |
| 20658 | CTD-2245F17.3 |
hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-940 | 11 | LIFR | Sponge network | -0.421 | 0.12556 | -1.794 | 0 | 0.397 |
| 20659 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | PTPRB | Sponge network | -0.976 | 0.00025 | 0.105 | 0.47122 | 0.397 |
| 20660 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 26 | FOXO3 | Sponge network | 0.537 | 0.00139 | -0.04 | 0.72028 | 0.397 |
| 20661 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p | 13 | SYT4 | Sponge network | -4.463 | 0.00025 | -3.207 | 0 | 0.397 |
| 20662 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 11 | CHD1 | Sponge network | 0.768 | 7.0E-5 | 0.322 | 0.00215 | 0.397 |
| 20663 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 27 | EPHA7 | Sponge network | 0.174 | 0.30034 | -2.197 | 3.0E-5 | 0.397 |
| 20664 | DIO3OS |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-98-5p | 10 | HSPB7 | Sponge network | -0.773 | 0.04073 | -2.268 | 1.0E-5 | 0.397 |
| 20665 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 29 | SNX29 | Sponge network | 0.056 | 0.81195 | -0.34 | 0.01371 | 0.397 |
| 20666 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 32 | ST6GAL2 | Sponge network | -0.942 | 0 | -1.201 | 0.00597 | 0.397 |
| 20667 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | BCL2 | Sponge network | 0.064 | 0.72934 | -0.899 | 0 | 0.397 |
| 20668 | BVES-AS1 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p | 17 | LIMCH1 | Sponge network | -2.62 | 0 | -0.915 | 1.0E-5 | 0.397 |
| 20669 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-744-5p | 16 | IGF2 | Sponge network | -0.129 | 0.57177 | 0.848 | 0.03842 | 0.397 |
| 20670 | CTC-429P9.3 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-940 | 13 | SORCS1 | Sponge network | 0.082 | 0.55397 | -3.492 | 0 | 0.397 |
| 20671 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 21 | AFF3 | Sponge network | 0.572 | 0.01722 | -2.373 | 0 | 0.397 |
| 20672 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 17 | SPG20 | Sponge network | -0.738 | 0.21233 | -1.16 | 0 | 0.397 |
| 20673 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p | 13 | TXNIP | Sponge network | -0.773 | 0.04073 | -1.277 | 0 | 0.397 |
| 20674 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 20 | CRTC3 | Sponge network | -0.125 | 0.49414 | 0.164 | 0.16786 | 0.397 |
| 20675 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | ZNF521 | Sponge network | -0.727 | 0.04726 | -0.111 | 0.58068 | 0.397 |
| 20676 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-93-3p | 25 | EPHA3 | Sponge network | -0.384 | 0.40652 | 0.185 | 0.53588 | 0.397 |
| 20677 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 38 | PTPRT | Sponge network | -0.323 | 0.01372 | -0.907 | 0.03727 | 0.397 |
| 20678 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-877-5p | 19 | CRTC1 | Sponge network | -0.162 | 0.47687 | -0.116 | 0.28412 | 0.397 |
| 20679 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 30 | MCC | Sponge network | -1.047 | 0 | -1.005 | 0 | 0.397 |
| 20680 | RP11-57H14.4 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 18 | BAZ2B | Sponge network | 0.083 | 0.66405 | -0.276 | 0.02833 | 0.397 |
| 20681 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-590-3p | 15 | TRIM33 | Sponge network | 0.919 | 0 | 0.402 | 7.0E-5 | 0.397 |
| 20682 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | SYNE1 | Sponge network | -0.206 | 0.12942 | -0.738 | 0.00316 | 0.397 |
| 20683 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 52 | QKI | Sponge network | -0.709 | 0.18168 | -0.333 | 0.04111 | 0.397 |
| 20684 | RP11-535M15.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p | 13 | CADM2 | Sponge network | -1.14 | 0.03513 | -3.782 | 0 | 0.397 |
| 20685 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 13 | CDH23 | Sponge network | -0.154 | 0.53954 | -0.974 | 0.00034 | 0.397 |
| 20686 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -0.246 | 0.35034 | -0.7 | 1.0E-5 | 0.397 |
| 20687 | BOLA3-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-629-3p | 27 | IGFBP5 | Sponge network | -0.246 | 0.35034 | -0.806 | 0.00105 | 0.397 |
| 20688 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 22 | EYA4 | Sponge network | -0.318 | 0.65847 | 0.3 | 0.36876 | 0.397 |
| 20689 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-361-5p | 14 | ERG | Sponge network | -0.465 | 0.04703 | 0.002 | 0.99011 | 0.397 |
| 20690 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-429 | 13 | ST6GAL2 | Sponge network | -1.654 | 0.00144 | -1.201 | 0.00597 | 0.397 |
| 20691 | RP1-151F17.2 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | ARHGEF12 | Sponge network | 0.222 | 0.29133 | 0.09 | 0.35693 | 0.397 |
| 20692 | LINC00865 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 27 | SYT4 | Sponge network | -1.249 | 7.0E-5 | -3.207 | 0 | 0.397 |
| 20693 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-92a-3p | 17 | ITGA5 | Sponge network | 0.214 | 0.18246 | 0.452 | 0.03524 | 0.397 |
| 20694 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-3p | 16 | CITED2 | Sponge network | -0.487 | 0.02157 | -1.545 | 0 | 0.397 |
| 20695 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | LRRC55 | Sponge network | -0.842 | 0.01361 | -0.026 | 0.93163 | 0.397 |
| 20696 | MIR497HG |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 16 | INPP4A | Sponge network | -1.439 | 4.0E-5 | 0.07 | 0.53563 | 0.397 |
| 20697 | FZD10-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 39 | PTPRT | Sponge network | -0.727 | 0.04726 | -0.907 | 0.03727 | 0.397 |
| 20698 | CKMT2-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-542-3p | 10 | PRKD1 | Sponge network | 0.082 | 0.55654 | -0.466 | 0.03583 | 0.397 |
| 20699 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 11 | PDS5B | Sponge network | 0.768 | 7.0E-5 | 0.259 | 0.00962 | 0.397 |
| 20700 | USP3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 26 | TOM1L2 | Sponge network | -0.162 | 0.47687 | -0.994 | 0 | 0.397 |
| 20701 | GAS6-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 30 | DIP2C | Sponge network | 0.16 | 0.50785 | -0.436 | 0.01713 | 0.397 |
| 20702 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 22 | SETBP1 | Sponge network | -0.342 | 0.02288 | -1.302 | 1.0E-5 | 0.397 |
| 20703 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | MLF1 | Sponge network | -0.576 | 0.10121 | -0.893 | 0.00727 | 0.397 |
| 20704 | CALML3-AS1 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-625-5p | 15 | BCL9 | Sponge network | 0.95 | 0.15943 | 0.321 | 0.00267 | 0.397 |
| 20705 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 37 | RBMS3 | Sponge network | 0.61 | 0.01593 | -0.519 | 0.03551 | 0.396 |
| 20706 | LINC00641 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TMEM30A | Sponge network | -0.222 | 0.32445 | 0.096 | 0.31031 | 0.396 |
| 20707 | AC108676.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 13 | ABL2 | Sponge network | 4.346 | 2.0E-5 | 0.82 | 0 | 0.396 |
| 20708 | RP11-21A7A.2 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-30b-3p;hsa-miR-425-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-5p | 13 | SOX5 | Sponge network | -1.116 | 0.23686 | -1.091 | 4.0E-5 | 0.396 |
| 20709 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-629-3p;hsa-miR-671-5p | 12 | THBS1 | Sponge network | -0.469 | 0.08721 | 0.741 | 0.00386 | 0.396 |
| 20710 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 41 | TMEM132B | Sponge network | -1.203 | 0.03881 | -0.83 | 0.01084 | 0.396 |
| 20711 | RP11-554A11.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-505-3p | 16 | PDE4DIP | Sponge network | -0.583 | 0.14589 | -0.282 | 0.0257 | 0.396 |
| 20712 | RP11-620J15.3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | KLF10 | Sponge network | -0.739 | 0.00044 | -0.783 | 0 | 0.396 |
| 20713 | LINC00672 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 13 | INPP4A | Sponge network | -0.227 | 0.31657 | 0.07 | 0.53563 | 0.396 |
| 20714 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | -0.612 | 0.00094 | -1.465 | 0.00033 | 0.396 |
| 20715 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 23 | CDHR3 | Sponge network | -0.739 | 0.0574 | -0.977 | 4.0E-5 | 0.396 |
| 20716 | LINC00094 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-3p | 29 | ABL2 | Sponge network | 0.253 | 0.09565 | 0.82 | 0 | 0.396 |
| 20717 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | GRIA3 | Sponge network | -1.697 | 0.00889 | -1.956 | 0 | 0.396 |
| 20718 | TPTEP1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 14 | THSD7A | Sponge network | -1.216 | 0 | 0.242 | 0.25154 | 0.396 |
| 20719 | RP11-119F7.5 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-532-3p;hsa-miR-590-3p | 13 | MRVI1 | Sponge network | 0.225 | 0.30644 | -1.051 | 0.00022 | 0.396 |
| 20720 | PWAR6 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | FLI1 | Sponge network | -1.575 | 6.0E-5 | -0.294 | 0.10905 | 0.396 |
| 20721 | CTD-3092A11.2 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TSC1 | Sponge network | 0.873 | 0 | 0.103 | 0.26071 | 0.396 |
| 20722 | LINC00641 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | ZNF208 | Sponge network | -0.222 | 0.32445 | -0.4 | 0.22381 | 0.396 |
| 20723 | HHIP-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 37 | CADM2 | Sponge network | -0.737 | 0.10822 | -3.782 | 0 | 0.396 |
| 20724 | PART1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | RECK | Sponge network | -2.925 | 0 | -1.043 | 0 | 0.396 |
| 20725 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-3p | 19 | CTIF | Sponge network | -0.896 | 0.00099 | -0.389 | 0.00549 | 0.396 |
| 20726 | LINC00473 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p | 45 | GAS7 | Sponge network | -1.203 | 0.03881 | -0.555 | 0.01602 | 0.396 |
| 20727 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-7-5p | 16 | PTGFR | Sponge network | -1.161 | 0.00591 | -0.233 | 0.45421 | 0.396 |
| 20728 | CTA-363E19.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-485-3p;hsa-miR-495-3p | 16 | ZFX | Sponge network | 1.055 | 0 | 0.09 | 0.42563 | 0.396 |
| 20729 | CKMT2-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 29 | DLC1 | Sponge network | 0.082 | 0.55654 | -0.382 | 0.05038 | 0.396 |
| 20730 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | ZNF521 | Sponge network | -2.46 | 0 | -0.111 | 0.58068 | 0.396 |
| 20731 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p | 41 | QKI | Sponge network | -1.203 | 0.03881 | -0.333 | 0.04111 | 0.396 |
| 20732 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-625-5p | 13 | ZMYM2 | Sponge network | 1.162 | 5.0E-5 | 0.279 | 0.01273 | 0.396 |
| 20733 | BOLA3-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p | 14 | KLF10 | Sponge network | -0.246 | 0.35034 | -0.783 | 0 | 0.396 |
| 20734 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-590-3p | 11 | ZNF292 | Sponge network | 0.225 | 0.30644 | 0.276 | 0.04148 | 0.396 |
| 20735 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | PIK3C2A | Sponge network | 0.793 | 0 | 0.061 | 0.53776 | 0.396 |
| 20736 | DLGAP1-AS2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-33a-3p | 13 | SOX11 | Sponge network | 2.406 | 0 | 2.68 | 0 | 0.396 |
| 20737 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-590-3p | 17 | PRKAA2 | Sponge network | -1.375 | 0.08642 | -2.462 | 0 | 0.396 |
| 20738 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-629-5p;hsa-miR-940 | 25 | IGF1 | Sponge network | -0.773 | 0.04073 | -0.204 | 0.5898 | 0.396 |
| 20739 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-590-3p | 13 | GPAM | Sponge network | 0.643 | 0 | 0.142 | 0.29732 | 0.396 |
| 20740 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-342-3p | 20 | TACC1 | Sponge network | -2.095 | 0.00112 | -0.362 | 0.05406 | 0.396 |
| 20741 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 18 | GPAM | Sponge network | 1.063 | 0 | 0.142 | 0.29732 | 0.396 |
| 20742 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 16 | RECK | Sponge network | -0.405 | 0.22013 | -1.043 | 0 | 0.396 |
| 20743 | GHRLOS |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-940;hsa-miR-98-5p | 24 | CADM2 | Sponge network | -1.942 | 0 | -3.782 | 0 | 0.396 |
| 20744 | ZNF252P-AS1 |
hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 12 | TBL1XR1 | Sponge network | 1.076 | 0 | 0.578 | 0 | 0.396 |
| 20745 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MOB1B | Sponge network | 0.91 | 0 | 0.197 | 0.15266 | 0.396 |
| 20746 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 28 | PGR | Sponge network | 0.61 | 0.01593 | -1.704 | 0 | 0.396 |
| 20747 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p | 14 | TRIM33 | Sponge network | 0.8 | 0 | 0.402 | 7.0E-5 | 0.396 |
| 20748 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-7-1-3p | 22 | ZFHX3 | Sponge network | 1.61 | 0.00961 | 0.389 | 0.01363 | 0.396 |
| 20749 | PGM5-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-421 | 11 | STARD13 | Sponge network | -4.546 | 0 | -0.187 | 0.27383 | 0.396 |
| 20750 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | MED12L | Sponge network | -0.129 | 0.57177 | -0.194 | 0.55299 | 0.396 |
| 20751 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 38 | TGFBR3 | Sponge network | -0.563 | 0.02545 | -1.173 | 1.0E-5 | 0.396 |
| 20752 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 18 | PCDH17 | Sponge network | -0.513 | 0.04703 | 0.731 | 2.0E-5 | 0.396 |
| 20753 | DIO3OS |
hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-877-5p | 16 | NRK | Sponge network | -0.773 | 0.04073 | 0.656 | 0.19217 | 0.396 |
| 20754 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | CREBBP | Sponge network | 0.385 | 0.10067 | 0.126 | 0.15186 | 0.396 |
| 20755 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 25 | PIK3C2A | Sponge network | 0.222 | 0.29133 | 0.061 | 0.53776 | 0.396 |
| 20756 | RP11-25K19.1 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | LIMCH1 | Sponge network | -1.057 | 0.00029 | -0.915 | 1.0E-5 | 0.396 |
| 20757 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 12 | PCDH17 | Sponge network | 1.153 | 0.00114 | 0.731 | 2.0E-5 | 0.396 |
| 20758 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p | 17 | PTPRB | Sponge network | -0.793 | 7.0E-5 | 0.105 | 0.47122 | 0.396 |
| 20759 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-370-3p;hsa-miR-532-3p | 12 | RPS6KA2 | Sponge network | -2.117 | 0.00161 | -0.57 | 0.00013 | 0.396 |
| 20760 | RP11-867G23.10 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p | 14 | ST6GALNAC3 | Sponge network | -2.531 | 0 | -0.987 | 0 | 0.396 |
| 20761 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p | 22 | PIK3C2A | Sponge network | 1.601 | 0 | 0.061 | 0.53776 | 0.396 |
| 20762 | LINC00843 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 22 | CCDC6 | Sponge network | 1.106 | 0 | 0.27 | 0.02555 | 0.396 |
| 20763 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 47 | EPHA5 | Sponge network | -1.216 | 0 | -0.957 | 0.01733 | 0.396 |
| 20764 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NRXN3 | Sponge network | 0.537 | 0.00139 | -0.893 | 0.04186 | 0.396 |
| 20765 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ITGB3 | Sponge network | 0.064 | 0.72934 | -0.011 | 0.95751 | 0.396 |
| 20766 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | 0.225 | 0.30644 | 0.323 | 0.00792 | 0.396 |
| 20767 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-589-3p | 21 | HECA | Sponge network | -0.154 | 0.78939 | -0.217 | 0.03463 | 0.396 |
| 20768 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 47 | CCND2 | Sponge network | -0.487 | 0.02157 | -0.267 | 0.3112 | 0.396 |
| 20769 | FAM157A |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-339-5p | 19 | MDM4 | Sponge network | 0.813 | 1.0E-5 | 0.345 | 0.00762 | 0.396 |
| 20770 | CTD-2089N3.2 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p | 23 | THRB | Sponge network | -1.375 | 0.08642 | -1.117 | 0.0004 | 0.396 |
| 20771 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 55 | DTNA | Sponge network | 0.064 | 0.72934 | -1.648 | 1.0E-5 | 0.396 |
| 20772 | AC010226.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | ERG | Sponge network | -0.811 | 2.0E-5 | 0.002 | 0.99011 | 0.396 |
| 20773 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 27 | AKAP11 | Sponge network | 0.401 | 0.00066 | 0.196 | 0.09825 | 0.396 |
| 20774 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-590-3p | 19 | MYBL1 | Sponge network | 0.559 | 0.00026 | 0.494 | 0.02152 | 0.396 |
| 20775 | BCYRN1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ZNF208 | Sponge network | 0.311 | 0.10344 | -0.4 | 0.22381 | 0.396 |
| 20776 | LINC00472 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-628-5p | 13 | IRS1 | Sponge network | -0.842 | 0.01361 | -0.026 | 0.88855 | 0.396 |
| 20777 | NDUFA6-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 37 | DIP2C | Sponge network | -0.355 | 0.0077 | -0.436 | 0.01713 | 0.396 |
| 20778 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-671-5p | 32 | CHD5 | Sponge network | 0.853 | 0.15352 | -0.922 | 0.01521 | 0.396 |
| 20779 | RP11-6O2.3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-340-5p | 11 | KLF10 | Sponge network | 0.331 | 0.57247 | -0.783 | 0 | 0.396 |
| 20780 | LINC00641 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | SFRP1 | Sponge network | -0.222 | 0.32445 | -3.566 | 0 | 0.396 |
| 20781 | MEG3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | GALNT13 | Sponge network | 0.249 | 0.35902 | -0.857 | 0.07439 | 0.396 |
| 20782 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-7-1-3p | 12 | CACNA2D1 | Sponge network | -0.72 | 0.24239 | -0.846 | 0.00675 | 0.396 |
| 20783 | TPTEP1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-96-5p;hsa-miR-98-5p | 10 | PLAGL1 | Sponge network | -1.216 | 0 | -0.588 | 0.00912 | 0.396 |
| 20784 | CTC-296K1.3 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-493-5p | 15 | LIMCH1 | Sponge network | -2.095 | 0.00112 | -0.915 | 1.0E-5 | 0.396 |
| 20785 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-940 | 13 | MYO9A | Sponge network | 0.556 | 0.00298 | -0.147 | 0.32373 | 0.396 |
| 20786 | ADAMTS9-AS2 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | PAIP2 | Sponge network | -2.452 | 0 | -0.515 | 0 | 0.396 |
| 20787 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | MCC | Sponge network | 0.225 | 0.30644 | -1.005 | 0 | 0.396 |
| 20788 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | AKAP11 | Sponge network | 0.826 | 1.0E-5 | 0.196 | 0.09825 | 0.396 |
| 20789 | RP11-182J1.1 |
hsa-miR-125a-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | PHC3 | Sponge network | -0.187 | 0.23787 | 0.212 | 0.0499 | 0.396 |
| 20790 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NRXN3 | Sponge network | -0.08 | 0.90092 | -0.893 | 0.04186 | 0.396 |
| 20791 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 23 | LATS2 | Sponge network | -1.575 | 6.0E-5 | 0.159 | 0.2304 | 0.396 |
| 20792 | RP11-119F7.5 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 19 | BAZ2B | Sponge network | 0.225 | 0.30644 | -0.276 | 0.02833 | 0.396 |
| 20793 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -0.738 | 0.21233 | -2.417 | 0 | 0.396 |
| 20794 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-425-5p | 17 | FGF5 | Sponge network | -0.693 | 0.05629 | 0.549 | 0.28659 | 0.396 |
| 20795 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 25 | USP12 | Sponge network | 0.385 | 0.10067 | -0.102 | 0.40768 | 0.396 |
| 20796 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-664a-3p | 12 | ITCH | Sponge network | 1.294 | 0 | 0.084 | 0.34058 | 0.396 |
| 20797 | FENDRR |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | ID4 | Sponge network | -1.281 | 0.00012 | -1.644 | 0 | 0.396 |
| 20798 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-582-5p | 20 | USP6 | Sponge network | 0.389 | 0.04293 | 0.188 | 0.3871 | 0.396 |
| 20799 | ERVK3-1 |
hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p | 11 | CDK6 | Sponge network | 0.58 | 0 | 0.991 | 0 | 0.396 |
| 20800 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-340-5p;hsa-miR-423-5p;hsa-miR-590-5p;hsa-miR-940 | 13 | NOTCH2 | Sponge network | -0.733 | 0.19469 | 0.138 | 0.35163 | 0.396 |
| 20801 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | PDS5B | Sponge network | 1.205 | 0 | 0.259 | 0.00962 | 0.396 |
| 20802 | FZD10-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | ROBO1 | Sponge network | -0.727 | 0.04726 | -0.009 | 0.96154 | 0.396 |
| 20803 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | SON | Sponge network | 0.349 | 0.01695 | 0.168 | 0.04406 | 0.396 |
| 20804 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | AHNAK | Sponge network | -1.216 | 0 | -1.207 | 0 | 0.396 |
| 20805 | RP11-119F7.5 |
hsa-miR-10a-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | GLI3 | Sponge network | 0.225 | 0.30644 | -0.615 | 0.01482 | 0.396 |
| 20806 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-935 | 31 | ABI2 | Sponge network | -0.384 | 0.40652 | 0.175 | 0.14787 | 0.396 |
| 20807 | LENG8-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-766-3p | 15 | TBL1XR1 | Sponge network | 1.471 | 0 | 0.578 | 0 | 0.396 |
| 20808 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | RBL2 | Sponge network | -0.376 | 0.00337 | -0.282 | 0.00254 | 0.396 |
| 20809 | HCG18 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | HIP1 | Sponge network | 1.063 | 0 | 0.774 | 0 | 0.396 |
| 20810 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-940;hsa-miR-96-5p | 29 | QKI | Sponge network | -0.326 | 0.14314 | -0.333 | 0.04111 | 0.396 |
| 20811 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 17 | CACNA2D1 | Sponge network | 0.331 | 0.57247 | -0.846 | 0.00675 | 0.396 |
| 20812 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 13 | SORCS1 | Sponge network | -0.513 | 0.04703 | -3.492 | 0 | 0.396 |
| 20813 | ZNF582-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p | 18 | KCNJ5 | Sponge network | -1.349 | 0.00017 | -0.281 | 0.3339 | 0.396 |
| 20814 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-940 | 16 | PBX1 | Sponge network | -0.421 | 0.12556 | -1.206 | 0 | 0.396 |
| 20815 | BZRAP1-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-335-5p;hsa-miR-455-5p | 14 | PLXNC1 | Sponge network | -0.465 | 0.04703 | 0.569 | 0.00329 | 0.396 |
| 20816 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 41 | RUNX1T1 | Sponge network | -0.08 | 0.90092 | -0.344 | 0.2374 | 0.396 |
| 20817 | HAND2-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 35 | MXI1 | Sponge network | -1.697 | 0.00889 | -0.987 | 0 | 0.396 |
| 20818 | RP11-1277A3.1 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-96-5p | 19 | PIK3R1 | Sponge network | 0.453 | 0.01839 | -0.133 | 0.36854 | 0.396 |
| 20819 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-7-1-3p | 14 | DKK3 | Sponge network | 0.532 | 0.00505 | -0.052 | 0.77783 | 0.396 |
| 20820 | CKMT2-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | PDE4DIP | Sponge network | 0.082 | 0.55654 | -0.282 | 0.0257 | 0.396 |
| 20821 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-590-3p | 32 | MEF2C | Sponge network | -0.227 | 0.31657 | -0.499 | 0.01448 | 0.396 |
| 20822 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-5p | 37 | PRKAA2 | Sponge network | -0.154 | 0.53954 | -2.462 | 0 | 0.396 |
| 20823 | ZNF582-AS1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-450b-5p | 10 | ROBO1 | Sponge network | -1.349 | 0.00017 | -0.009 | 0.96154 | 0.396 |
| 20824 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ABCC9 | Sponge network | 0.537 | 0.00139 | -0.466 | 0.14121 | 0.396 |
| 20825 | A2M-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 26 | DIP2C | Sponge network | -0.08 | 0.90092 | -0.436 | 0.01713 | 0.396 |
| 20826 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 10 | CHD3 | Sponge network | -0.295 | 0.02604 | -0.001 | 0.99669 | 0.396 |
| 20827 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-98-5p | 23 | NTRK3 | Sponge network | 1.122 | 0.02989 | -1.77 | 3.0E-5 | 0.396 |
| 20828 | PWAR6 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | EDNRB | Sponge network | -1.575 | 6.0E-5 | -0.682 | 0.00138 | 0.396 |
| 20829 | PCA3 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | NR3C2 | Sponge network | -0.709 | 0.18168 | -1.56 | 0 | 0.396 |
| 20830 | RP4-758J18.13 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 10 | CBFB | Sponge network | 1.591 | 0 | 1.061 | 0 | 0.396 |
| 20831 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 45 | SSBP2 | Sponge network | 0.249 | 0.35902 | -1.58 | 0 | 0.396 |
| 20832 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 36 | NTRK2 | Sponge network | 0.083 | 0.66405 | -0.736 | 0.06495 | 0.396 |
| 20833 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-942-5p | 40 | UNC80 | Sponge network | -2.915 | 0.00015 | -1.934 | 0 | 0.396 |
| 20834 | KB-1572G7.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-486-5p;hsa-miR-495-3p | 13 | PHF6 | Sponge network | 0.924 | 0 | 0.785 | 0 | 0.396 |
| 20835 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | RASSF2 | Sponge network | -0.125 | 0.49414 | -0.273 | 0.21961 | 0.396 |
| 20836 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-3p;hsa-miR-590-3p | 35 | TGFBR3 | Sponge network | -0.738 | 0.21233 | -1.173 | 1.0E-5 | 0.396 |
| 20837 | CTD-2528L19.6 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZMYND11 | Sponge network | 0.953 | 0 | -0.2 | 0.05449 | 0.396 |
| 20838 | PWAR6 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | SEMA3C | Sponge network | -1.575 | 6.0E-5 | -0.272 | 0.31153 | 0.396 |
| 20839 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | FLI1 | Sponge network | -1.697 | 0.00889 | -0.294 | 0.10905 | 0.396 |
| 20840 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | CRTC1 | Sponge network | -2.452 | 0 | -0.116 | 0.28412 | 0.396 |
| 20841 | RP11-401P9.4 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-505-5p | 13 | WNK2 | Sponge network | -0.384 | 0.40652 | -0.352 | 0.31066 | 0.396 |
| 20842 | FAM215B |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 21 | EPS15 | Sponge network | 0.817 | 3.0E-5 | -0.052 | 0.53998 | 0.396 |
| 20843 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-145-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | MOB1B | Sponge network | 0.545 | 0.00064 | 0.197 | 0.15266 | 0.396 |
| 20844 | AGAP11 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p | 18 | CADM3 | Sponge network | -1.645 | 0 | -3.663 | 0 | 0.396 |
| 20845 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-576-5p | 18 | PBX1 | Sponge network | 0.889 | 9.0E-5 | -1.206 | 0 | 0.396 |
| 20846 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 69 | BMPR2 | Sponge network | -1.697 | 0.00889 | 0.206 | 0.0635 | 0.396 |
| 20847 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 53 | TRPS1 | Sponge network | 0.064 | 0.72934 | -0.191 | 0.44109 | 0.396 |
| 20848 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429 | 11 | ZFHX3 | Sponge network | 2.364 | 0.00134 | 0.389 | 0.01363 | 0.396 |
| 20849 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 32 | MAF | Sponge network | -0.777 | 0.1134 | -1.257 | 0 | 0.396 |
| 20850 | RP11-53O19.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | -0.405 | 0.22013 | 0.321 | 0.00267 | 0.396 |
| 20851 | CTD-2666L21.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-424-5p | 17 | ABL2 | Sponge network | 0.368 | 0.20093 | 0.82 | 0 | 0.396 |
| 20852 | RP11-53O19.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 29 | MDM4 | Sponge network | 1.205 | 0 | 0.345 | 0.00762 | 0.395 |
| 20853 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-877-5p | 28 | GRIN2A | Sponge network | -0.615 | 0.00015 | -2.161 | 0 | 0.395 |
| 20854 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 17 | ITGB3 | Sponge network | -1.216 | 0 | -0.011 | 0.95751 | 0.395 |
| 20855 | PWAR6 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | PIK3CA | Sponge network | -1.575 | 6.0E-5 | 0.195 | 0.06908 | 0.395 |
| 20856 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-93-5p | 10 | TXNIP | Sponge network | -1.654 | 0.00144 | -1.277 | 0 | 0.395 |
| 20857 | SERTAD4-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-502-5p | 15 | PTPN14 | Sponge network | -0.932 | 0.00329 | 0.144 | 0.34595 | 0.395 |
| 20858 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 11 | ITGAV | Sponge network | -0.376 | 0.00337 | 0.337 | 0.01002 | 0.395 |
| 20859 | JAZF1-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-942-5p | 16 | GLIPR1 | Sponge network | -0.769 | 0.00035 | 0.146 | 0.3276 | 0.395 |
| 20860 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 44 | IL6ST | Sponge network | -0.739 | 0.00044 | -0.402 | 0.00676 | 0.395 |
| 20861 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 13 | GIGYF2 | Sponge network | 0.421 | 0.00084 | 0.136 | 0.04403 | 0.395 |
| 20862 | RP11-967K21.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p | 21 | MED12L | Sponge network | 0.056 | 0.81195 | -0.194 | 0.55299 | 0.395 |
| 20863 | EMX2OS |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 18 | RPS6KA6 | Sponge network | -0.777 | 0.1134 | -2.989 | 0 | 0.395 |
| 20864 | HCG11 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | DNMT3A | Sponge network | 0.385 | 0.10067 | 0.302 | 0.0262 | 0.395 |
| 20865 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | 0.214 | 0.18246 | -0.085 | 0.42389 | 0.395 |
| 20866 | AC009948.5 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 30 | RIMS2 | Sponge network | 0.532 | 0.00505 | -1.365 | 0.00208 | 0.395 |
| 20867 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-589-3p | 11 | EPAS1 | Sponge network | -0.154 | 0.53954 | -0.184 | 0.14418 | 0.395 |
| 20868 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 13 | ERG | Sponge network | -0.318 | 0.65847 | 0.002 | 0.99011 | 0.395 |
| 20869 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-708-5p;hsa-miR-96-5p | 36 | SPRY3 | Sponge network | 0.083 | 0.66405 | 0.016 | 0.91286 | 0.395 |
| 20870 | RP11-283I3.6 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 11 | ERCC6 | Sponge network | 0.614 | 1.0E-5 | 0.23 | 0.015 | 0.395 |
| 20871 | LPP-AS2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-93-3p | 18 | WWTR1 | Sponge network | -0.18 | 0.20343 | -0.345 | 0.11997 | 0.395 |
| 20872 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-93-3p | 33 | IL6ST | Sponge network | 0.898 | 1.0E-5 | -0.402 | 0.00676 | 0.395 |
| 20873 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | LRRC7 | Sponge network | -0.899 | 6.0E-5 | -1.341 | 0.01193 | 0.395 |
| 20874 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577 | 15 | GREM1 | Sponge network | 0.199 | 0.38015 | 0.902 | 0.01691 | 0.395 |
| 20875 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-7-5p;hsa-miR-98-5p | 10 | CHD3 | Sponge network | 0.249 | 0.35902 | -0.001 | 0.99669 | 0.395 |
| 20876 | MIR600HG |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-342-3p | 12 | PHF3 | Sponge network | 0.594 | 0.41522 | 0.126 | 0.19652 | 0.395 |
| 20877 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | BTG2 | Sponge network | -2.46 | 0 | -1.763 | 0 | 0.395 |
| 20878 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 51 | UNC80 | Sponge network | -0.739 | 0.0574 | -1.934 | 0 | 0.395 |
| 20879 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 23 | IGF1 | Sponge network | -0.421 | 0.0114 | -0.204 | 0.5898 | 0.395 |
| 20880 | RP11-212P7.2 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-532-3p | 18 | SYNM | Sponge network | 0.219 | 0.1516 | -2.309 | 1.0E-5 | 0.395 |
| 20881 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 25 | APC | Sponge network | 0.537 | 0.00139 | -0.255 | 0.04051 | 0.395 |
| 20882 | DNAJC27-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 40 | DIP2C | Sponge network | -0.969 | 2.0E-5 | -0.436 | 0.01713 | 0.395 |
| 20883 | RP11-13K12.5 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | TLL1 | Sponge network | -0.318 | 0.65847 | -1.195 | 3.0E-5 | 0.395 |
| 20884 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 15 | ARHGAP28 | Sponge network | -2.039 | 0.03447 | -0.911 | 0.00013 | 0.395 |
| 20885 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p | 25 | USP12 | Sponge network | 0.082 | 0.55654 | -0.102 | 0.40768 | 0.395 |
| 20886 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | HOOK3 | Sponge network | 0.657 | 4.0E-5 | 0.273 | 0.09957 | 0.395 |
| 20887 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-7-1-3p | 10 | DNAJB4 | Sponge network | -0.318 | 0.65847 | -0.7 | 1.0E-5 | 0.395 |
| 20888 | CTC-559E9.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p | 16 | HECA | Sponge network | 0.594 | 0.00064 | -0.217 | 0.03463 | 0.395 |
| 20889 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-589-3p | 11 | ST6GALNAC3 | Sponge network | -4.463 | 0.00025 | -0.987 | 0 | 0.395 |
| 20890 | RP11-890B15.3 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 15 | PHOX2B | Sponge network | 0.101 | 0.60919 | -2.68 | 0 | 0.395 |
| 20891 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429 | 18 | GREM1 | Sponge network | -1.249 | 7.0E-5 | 0.902 | 0.01691 | 0.395 |
| 20892 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | MCC | Sponge network | -0.351 | 0.11358 | -1.005 | 0 | 0.395 |
| 20893 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 54 | BNC2 | Sponge network | -2.925 | 0 | -0.491 | 0.09988 | 0.395 |
| 20894 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-660-5p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | ZHX2 | Sponge network | -2.46 | 0 | -0.192 | 0.18459 | 0.395 |
| 20895 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 21 | TGFBR2 | Sponge network | -1.575 | 6.0E-5 | -0.243 | 0.11408 | 0.395 |
| 20896 | FAM66C |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | RNF144A | Sponge network | -0.901 | 0.00019 | 0.594 | 0.0009 | 0.395 |
| 20897 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-590-3p | 24 | KAT6B | Sponge network | -0.615 | 0.00015 | -0.478 | 0.00018 | 0.395 |
| 20898 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 56 | PPM1A | Sponge network | -1.111 | 0.0003 | -0.623 | 0 | 0.395 |
| 20899 | LINC00473 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SEMA3E | Sponge network | -1.203 | 0.03881 | -1.717 | 0.00137 | 0.395 |
| 20900 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 15 | NR2C2 | Sponge network | 0.529 | 0.00552 | 0.21 | 0.03224 | 0.395 |
| 20901 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-942-5p | 29 | UNC80 | Sponge network | -2.531 | 0 | -1.934 | 0 | 0.395 |
| 20902 | MYLK-AS1 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p | 10 | CREBBP | Sponge network | -1.331 | 3.0E-5 | 0.126 | 0.15186 | 0.395 |
| 20903 | AGAP11 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-93-5p | 12 | GLI3 | Sponge network | -1.645 | 0 | -0.615 | 0.01482 | 0.395 |
| 20904 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-493-5p | 19 | EPHA7 | Sponge network | -0.829 | 0.01343 | -2.197 | 3.0E-5 | 0.395 |
| 20905 | MIR143HG |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | PIK3CA | Sponge network | -0.739 | 0.0574 | 0.195 | 0.06908 | 0.395 |
| 20906 | RP11-574K11.29 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-497-5p;hsa-miR-9-5p | 11 | CDK6 | Sponge network | 0.997 | 0 | 0.991 | 0 | 0.395 |
| 20907 | ZNF667-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-493-5p | 12 | HERC1 | Sponge network | -0.976 | 0.00025 | -0.196 | 0.08544 | 0.395 |
| 20908 | BMS1P20 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 12 | CUL5 | Sponge network | 0.34 | 0.00273 | 0.026 | 0.77301 | 0.395 |
| 20909 | RP4-669P10.16 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-542-3p | 12 | PIK3R1 | Sponge network | -0.301 | 0.06597 | -0.133 | 0.36854 | 0.395 |
| 20910 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-361-3p | 18 | FOXO3 | Sponge network | 0.556 | 0.00298 | -0.04 | 0.72028 | 0.395 |
| 20911 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | EPHA7 | Sponge network | -0.08 | 0.90092 | -2.197 | 3.0E-5 | 0.395 |
| 20912 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 14 | RIMS1 | Sponge network | -0.899 | 6.0E-5 | -3.143 | 0 | 0.395 |
| 20913 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p | 32 | ADARB1 | Sponge network | 0.174 | 0.30034 | -0.335 | 0.06631 | 0.395 |
| 20914 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-651-5p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 57 | AFF4 | Sponge network | -1.645 | 0 | -0.266 | 0.01565 | 0.395 |
| 20915 | SH3BP5-AS1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | TAF1 | Sponge network | 0.267 | 0.2937 | 0.39 | 3.0E-5 | 0.395 |
| 20916 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 50 | PRKAA2 | Sponge network | -0.612 | 0.00094 | -2.462 | 0 | 0.395 |
| 20917 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 33 | TSHZ3 | Sponge network | 0.374 | 0.20054 | -0.556 | 0.01516 | 0.395 |
| 20918 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p | 25 | ESR1 | Sponge network | -1.057 | 0.00029 | -1.035 | 3.0E-5 | 0.395 |
| 20919 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | TIMP3 | Sponge network | -1.161 | 0.00591 | -0.546 | 0.02307 | 0.395 |
| 20920 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ZFHX3 | Sponge network | -0.021 | 0.93598 | 0.389 | 0.01363 | 0.395 |
| 20921 | LINC00957 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | AFF1 | Sponge network | -0.421 | 0.0114 | -0.384 | 0.00053 | 0.395 |
| 20922 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ABI2 | Sponge network | -1.161 | 0.00591 | 0.175 | 0.14787 | 0.395 |
| 20923 | RP11-2H8.2 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-590-3p | 18 | MYO9A | Sponge network | 1.089 | 0 | -0.147 | 0.32373 | 0.395 |
| 20924 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | TAL1 | Sponge network | -0.739 | 0.00044 | -0.462 | 0.00574 | 0.395 |
| 20925 | GNG12-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | BAZ2B | Sponge network | -0.793 | 7.0E-5 | -0.276 | 0.02833 | 0.395 |
| 20926 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-590-3p | 11 | HERC2 | Sponge network | -0.487 | 0.02157 | 0.114 | 0.30638 | 0.395 |
| 20927 | FZD10-AS1 |
hsa-miR-130b-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92b-3p | 11 | ROBO2 | Sponge network | -0.727 | 0.04726 | 0.092 | 0.78768 | 0.395 |
| 20928 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-539-5p | 13 | TSC1 | Sponge network | 0.858 | 3.0E-5 | 0.103 | 0.26071 | 0.395 |
| 20929 | ADAMTS9-AS2 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 14 | FOXO4 | Sponge network | -2.452 | 0 | -0.516 | 2.0E-5 | 0.395 |
| 20930 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 21 | EPS15 | Sponge network | 0.4 | 0.14611 | -0.052 | 0.53998 | 0.395 |
| 20931 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 60 | IL6ST | Sponge network | -1.645 | 0 | -0.402 | 0.00676 | 0.395 |
| 20932 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-889-3p;hsa-miR-93-5p;hsa-miR-942-5p | 43 | UNC80 | Sponge network | -0.162 | 0.47687 | -1.934 | 0 | 0.395 |
| 20933 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-628-5p | 29 | BMPR2 | Sponge network | -0.583 | 0.14589 | 0.206 | 0.0635 | 0.395 |
| 20934 | RP5-1085F17.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 31 | ABL2 | Sponge network | 0.541 | 0.00125 | 0.82 | 0 | 0.395 |
| 20935 | LINC00173 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | MDM4 | Sponge network | 0.056 | 0.8494 | 0.345 | 0.00762 | 0.395 |
| 20936 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p | 18 | DMD | Sponge network | 0.727 | 3.0E-5 | -1.506 | 0 | 0.395 |
| 20937 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | RELN | Sponge network | 0.214 | 0.18246 | -2.403 | 0 | 0.395 |
| 20938 | MIR497HG |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 18 | KCNJ5 | Sponge network | -1.439 | 4.0E-5 | -0.281 | 0.3339 | 0.395 |
| 20939 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PCDH10 | Sponge network | -0.342 | 0.02288 | -1.644 | 0.0078 | 0.395 |
| 20940 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p | 33 | ZFHX3 | Sponge network | 0.95 | 0.15943 | 0.389 | 0.01363 | 0.395 |
| 20941 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | CDH10 | Sponge network | -0.129 | 0.57177 | -2.397 | 0 | 0.395 |
| 20942 | PWAR6 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 30 | CADM1 | Sponge network | -1.575 | 6.0E-5 | -0.9 | 0.00084 | 0.395 |
| 20943 | RP11-822E23.8 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-5p | 26 | KCNA6 | Sponge network | -0.777 | 0.16667 | -1.531 | 0.00013 | 0.395 |
| 20944 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 33 | PRDM2 | Sponge network | -0.576 | 0.10121 | -0.258 | 0.00721 | 0.395 |
| 20945 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CAV1 | Sponge network | -0.421 | 0.0114 | -1.013 | 6.0E-5 | 0.395 |
| 20946 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | CUL5 | Sponge network | 0.91 | 0 | 0.026 | 0.77301 | 0.395 |
| 20947 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 38 | ATRX | Sponge network | -1.111 | 0.0003 | 0.261 | 0.04408 | 0.395 |
| 20948 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p | 20 | FBXO32 | Sponge network | 0.889 | 9.0E-5 | -0.495 | 0.07561 | 0.395 |
| 20949 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p | 13 | HOOK3 | Sponge network | -0.842 | 0.01361 | 0.273 | 0.09957 | 0.395 |
| 20950 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 25 | RSPO2 | Sponge network | -0.612 | 0.00094 | -3.038 | 0 | 0.395 |
| 20951 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 38 | PRKAB2 | Sponge network | 0.421 | 0.00084 | -0.367 | 0.01371 | 0.395 |
| 20952 | RP11-927P21.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-582-3p | 19 | ABL2 | Sponge network | 0.921 | 0.00118 | 0.82 | 0 | 0.395 |
| 20953 | MYLK-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-935 | 14 | OPCML | Sponge network | -1.331 | 3.0E-5 | -2.498 | 0 | 0.395 |
| 20954 | DNM3OS |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ZMYND11 | Sponge network | 0.572 | 0.01722 | -0.2 | 0.05449 | 0.395 |
| 20955 | RP11-57H14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-590-3p | 13 | ITGB3 | Sponge network | 0.083 | 0.66405 | -0.011 | 0.95751 | 0.395 |
| 20956 | ADAMTS9-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 28 | CYLD | Sponge network | -2.452 | 0 | -0.367 | 0.00171 | 0.395 |
| 20957 | RP11-83A24.2 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p;hsa-miR-654-3p | 17 | PHF6 | Sponge network | 0.417 | 0.00445 | 0.785 | 0 | 0.395 |
| 20958 | AC025171.1 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | MYO9A | Sponge network | 0.998 | 0 | -0.147 | 0.32373 | 0.395 |
| 20959 | RP11-693J15.4 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-590-5p | 16 | KCNJ5 | Sponge network | -0.72 | 0.24239 | -0.281 | 0.3339 | 0.395 |
| 20960 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 20 | CXXC4 | Sponge network | -0.246 | 0.35034 | -0.433 | 0.21055 | 0.395 |
| 20961 | HCG11 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-576-5p | 28 | ERC1 | Sponge network | 0.385 | 0.10067 | -0.108 | 0.38794 | 0.395 |
| 20962 | HAND2-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | HERC1 | Sponge network | -1.697 | 0.00889 | -0.196 | 0.08544 | 0.395 |
| 20963 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 21 | GAS7 | Sponge network | 0.348 | 0.32912 | -0.555 | 0.01602 | 0.395 |
| 20964 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | CNTN1 | Sponge network | -0.08 | 0.90092 | -2.594 | 0 | 0.395 |
| 20965 | GHRLOS |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-5p;hsa-miR-589-5p;hsa-miR-7-5p | 11 | PRKCB | Sponge network | -1.942 | 0 | -1.313 | 2.0E-5 | 0.395 |
| 20966 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 17 | GJA1 | Sponge network | 0.889 | 9.0E-5 | -0.485 | 0.02179 | 0.395 |
| 20967 | RP4-613B23.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p | 11 | ST6GALNAC3 | Sponge network | -0.309 | 0.1145 | -0.987 | 0 | 0.395 |
| 20968 | AC009948.5 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | PCDH17 | Sponge network | 0.532 | 0.00505 | 0.731 | 2.0E-5 | 0.395 |
| 20969 | LINC00957 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 15 | RPS6KA6 | Sponge network | -0.421 | 0.0114 | -2.989 | 0 | 0.395 |
| 20970 | PCA3 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 17 | GLIPR1 | Sponge network | -0.709 | 0.18168 | 0.146 | 0.3276 | 0.395 |
| 20971 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p | 16 | DYNC1I1 | Sponge network | -0.303 | 0.21002 | -1.12 | 0.00107 | 0.395 |
| 20972 | THAP9-AS1 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-9-5p | 13 | PAWR | Sponge network | 1.079 | 0 | 0.636 | 0 | 0.395 |
| 20973 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | CREB1 | Sponge network | 0.959 | 0 | 0.203 | 0.00537 | 0.395 |
| 20974 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-625-5p | 23 | RSPO2 | Sponge network | -0.842 | 0.01361 | -3.038 | 0 | 0.395 |
| 20975 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | ZMYM2 | Sponge network | 0.67 | 0.00048 | 0.279 | 0.01273 | 0.395 |
| 20976 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | NEDD4 | Sponge network | 0.61 | 0.01593 | 0.02 | 0.89834 | 0.395 |
| 20977 | AC009948.5 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | RYR2 | Sponge network | 0.532 | 0.00505 | -1.466 | 0.00031 | 0.395 |
| 20978 | LINC00648 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SLC6A15 | Sponge network | 0.633 | 0.51678 | -2.373 | 2.0E-5 | 0.395 |
| 20979 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-629-5p | 12 | UVRAG | Sponge network | -0.405 | 0.22013 | -0.017 | 0.83865 | 0.395 |
| 20980 | U91328.19 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-493-3p;hsa-miR-532-3p | 19 | MRVI1 | Sponge network | -0.303 | 0.21002 | -1.051 | 0.00022 | 0.395 |
| 20981 | RP11-535M15.1 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p | 10 | SSBP2 | Sponge network | -1.14 | 0.03513 | -1.58 | 0 | 0.395 |
| 20982 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-590-3p | 17 | PKNOX1 | Sponge network | 0.101 | 0.60919 | 0.085 | 0.28917 | 0.395 |
| 20983 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | PAFAH1B1 | Sponge network | -1.281 | 0.00012 | -0.429 | 0 | 0.395 |
| 20984 | FGD5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 27 | ABI2 | Sponge network | 0.401 | 0.00066 | 0.175 | 0.14787 | 0.395 |
| 20985 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p | 20 | SYNE1 | Sponge network | 0.494 | 0.00339 | -0.738 | 0.00316 | 0.395 |
| 20986 | C20orf166-AS1 |
hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-577 | 34 | TMEM132B | Sponge network | -2.69 | 0.0001 | -0.83 | 0.01084 | 0.395 |
| 20987 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p | 27 | FBXO32 | Sponge network | 0.559 | 0.00026 | -0.495 | 0.07561 | 0.395 |
| 20988 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 22 | FBXO32 | Sponge network | 0.538 | 0.00076 | -0.495 | 0.07561 | 0.395 |
| 20989 | ANKRD10-IT1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 13 | ABI2 | Sponge network | 1.739 | 0 | 0.175 | 0.14787 | 0.395 |
| 20990 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-96-5p | 12 | LATS2 | Sponge network | 0.331 | 0.57247 | 0.159 | 0.2304 | 0.395 |
| 20991 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | FGF5 | Sponge network | -2.452 | 0 | 0.549 | 0.28659 | 0.395 |
| 20992 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 20 | CLASP2 | Sponge network | 0.817 | 3.0E-5 | -0.038 | 0.71 | 0.395 |
| 20993 | RP11-401P9.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-935 | 71 | LPP | Sponge network | -0.384 | 0.40652 | -0.459 | 0.04475 | 0.395 |
| 20994 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 30 | GRIN2A | Sponge network | -1.216 | 0 | -2.161 | 0 | 0.395 |
| 20995 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 28 | LRIG1 | Sponge network | 0.101 | 0.60919 | -0.537 | 0.00588 | 0.395 |
| 20996 | RP11-400K9.3 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-590-5p;hsa-miR-744-3p | 10 | DIP2C | Sponge network | -0.733 | 0.19469 | -0.436 | 0.01713 | 0.395 |
| 20997 | CTD-2554C21.2 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-7-5p | 12 | RNF144A | Sponge network | -1.654 | 0.00144 | 0.594 | 0.0009 | 0.394 |
| 20998 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | LRRC4 | Sponge network | 0.194 | 0.64278 | -1.774 | 0 | 0.394 |
| 20999 | RP11-13K12.5 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-7-1-3p | 13 | ITPKB | Sponge network | -0.318 | 0.65847 | -0.704 | 0.00106 | 0.394 |
| 21000 | MIR22HG |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-493-3p;hsa-miR-532-3p;hsa-miR-542-3p | 16 | MRVI1 | Sponge network | -1.047 | 0 | -1.051 | 0.00022 | 0.394 |
| 21001 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 47 | TMEM132B | Sponge network | 0.101 | 0.60919 | -0.83 | 0.01084 | 0.394 |
| 21002 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-940 | 24 | THBS1 | Sponge network | 0.199 | 0.38015 | 0.741 | 0.00386 | 0.394 |
| 21003 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429 | 12 | RCAN2 | Sponge network | 0.056 | 0.81195 | -0.33 | 0.15172 | 0.394 |
| 21004 | RP11-890B15.3 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | 0.101 | 0.60919 | 0.524 | 0.00017 | 0.394 |
| 21005 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-29a-5p;hsa-miR-532-3p | 10 | SYNM | Sponge network | 0.497 | 0.01451 | -2.309 | 1.0E-5 | 0.394 |
| 21006 | LINC00957 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | INPP4A | Sponge network | -0.421 | 0.0114 | 0.07 | 0.53563 | 0.394 |
| 21007 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 20 | RBL2 | Sponge network | -1.111 | 0.0003 | -0.282 | 0.00254 | 0.394 |
| 21008 | LINC00094 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-92a-3p | 18 | PDE4DIP | Sponge network | 0.253 | 0.09565 | -0.282 | 0.0257 | 0.394 |
| 21009 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 22 | TRIM33 | Sponge network | 0.72 | 0.0001 | 0.402 | 7.0E-5 | 0.394 |
| 21010 | HOXA-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | DYNC1I1 | Sponge network | 0.61 | 0.01593 | -1.12 | 0.00107 | 0.394 |
| 21011 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-485-3p;hsa-miR-590-3p | 22 | PIK3C2A | Sponge network | 1.254 | 0 | 0.061 | 0.53776 | 0.394 |
| 21012 | PSMD5-AS1 |
hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p | 12 | SHPRH | Sponge network | 0.569 | 0.00067 | 0.098 | 0.40994 | 0.394 |
| 21013 | RP11-67L2.2 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | MAP3K4 | Sponge network | 0.72 | 0.0001 | 0.058 | 0.54229 | 0.394 |
| 21014 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 46 | ERBB4 | Sponge network | -1.216 | 0 | -1.975 | 2.0E-5 | 0.394 |
| 21015 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-196b-5p;hsa-miR-2355-3p;hsa-miR-26b-3p | 10 | EPHA7 | Sponge network | -0.326 | 0.28809 | -2.197 | 3.0E-5 | 0.394 |
| 21016 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PTPRB | Sponge network | 0.374 | 0.20054 | 0.105 | 0.47122 | 0.394 |
| 21017 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 54 | BMPR2 | Sponge network | 0.374 | 0.20054 | 0.206 | 0.0635 | 0.394 |
| 21018 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421 | 14 | PDE4DIP | Sponge network | -1.654 | 0.00144 | -0.282 | 0.0257 | 0.394 |
| 21019 | RP11-701H24.3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | SSBP2 | Sponge network | -1.425 | 0.04204 | -1.58 | 0 | 0.394 |
| 21020 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | BNC2 | Sponge network | -0.72 | 0.24239 | -0.491 | 0.09988 | 0.394 |
| 21021 | JAZF1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | RYR2 | Sponge network | -0.769 | 0.00035 | -1.466 | 0.00031 | 0.394 |
| 21022 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | RCAN2 | Sponge network | -2.117 | 0.00161 | -0.33 | 0.15172 | 0.394 |
| 21023 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | PHC3 | Sponge network | 0.953 | 0 | 0.212 | 0.0499 | 0.394 |
| 21024 | CTC-429P9.5 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-3p | 13 | DOCK4 | Sponge network | 0.178 | 0.29106 | 0.502 | 0.00108 | 0.394 |
| 21025 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 32 | MCC | Sponge network | 0.056 | 0.81195 | -1.005 | 0 | 0.394 |
| 21026 | MIR497HG |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 16 | POU2F2 | Sponge network | -1.439 | 4.0E-5 | -0.233 | 0.32214 | 0.394 |
| 21027 | BOLA3-AS1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p | 13 | ABL1 | Sponge network | -0.246 | 0.35034 | -0.215 | 0.07804 | 0.394 |
| 21028 | FAM66C |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 33 | ABI2 | Sponge network | -0.901 | 0.00019 | 0.175 | 0.14787 | 0.394 |
| 21029 | RP11-130L8.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 24 | DTNA | Sponge network | -0.663 | 0.10887 | -1.648 | 1.0E-5 | 0.394 |
| 21030 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-590-5p | 29 | TGFBR3 | Sponge network | 0.176 | 0.37402 | -1.173 | 1.0E-5 | 0.394 |
| 21031 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 24 | GRIN2A | Sponge network | 0.246 | 0.59912 | -2.161 | 0 | 0.394 |
| 21032 | CTC-429P9.2 |
hsa-miR-136-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-542-3p | 15 | DTNA | Sponge network | 0.043 | 0.81628 | -1.648 | 1.0E-5 | 0.394 |
| 21033 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | AFF3 | Sponge network | -1.047 | 0 | -2.373 | 0 | 0.394 |
| 21034 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | EPHA7 | Sponge network | -0.303 | 0.21002 | -2.197 | 3.0E-5 | 0.394 |
| 21035 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 32 | IL6ST | Sponge network | -0.72 | 0.24239 | -0.402 | 0.00676 | 0.394 |
| 21036 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | TACC1 | Sponge network | 0.219 | 0.1516 | -0.362 | 0.05406 | 0.394 |
| 21037 | AC006116.24 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-550a-3p;hsa-miR-577 | 10 | SYT4 | Sponge network | -0.473 | 0.07602 | -3.207 | 0 | 0.394 |
| 21038 | AC025335.1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 11 | PHF6 | Sponge network | 0.783 | 0 | 0.785 | 0 | 0.394 |
| 21039 | SH3BP5-AS1 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-7-1-3p | 17 | ZMYND11 | Sponge network | 0.267 | 0.2937 | -0.2 | 0.05449 | 0.394 |
| 21040 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | SASH1 | Sponge network | -0.733 | 0.19469 | -0.502 | 0.00175 | 0.394 |
| 21041 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CLASP2 | Sponge network | 0.67 | 0.00048 | -0.038 | 0.71 | 0.394 |
| 21042 | PART1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DCN | Sponge network | -2.925 | 0 | -1.288 | 0 | 0.394 |
| 21043 | RP11-464F9.1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p | 11 | MAT2A | Sponge network | 0.959 | 0 | -0.073 | 0.36195 | 0.394 |
| 21044 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 25 | PIK3R1 | Sponge network | -0.031 | 0.89063 | -0.133 | 0.36854 | 0.394 |
| 21045 | FAM157A |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-378a-5p | 14 | CREB1 | Sponge network | 0.813 | 1.0E-5 | 0.203 | 0.00537 | 0.394 |
| 21046 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p | 10 | ZYX | Sponge network | -0.49 | 0.04946 | 0.019 | 0.89763 | 0.394 |
| 21047 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 18 | IGF2 | Sponge network | -0.739 | 0.0574 | 0.848 | 0.03842 | 0.394 |
| 21048 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-7-1-3p | 13 | EPAS1 | Sponge network | -0.901 | 0.00019 | -0.184 | 0.14418 | 0.394 |
| 21049 | PWAR6 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | PCDH18 | Sponge network | -1.575 | 6.0E-5 | 0.216 | 0.24645 | 0.394 |
| 21050 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-940 | 14 | NEDD4 | Sponge network | -0.154 | 0.53954 | 0.02 | 0.89834 | 0.394 |
| 21051 | RP11-890B15.3 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 25 | UBR3 | Sponge network | 0.101 | 0.60919 | -0.021 | 0.82774 | 0.394 |
| 21052 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 11 | NF1 | Sponge network | 1.206 | 0 | 0.51 | 0 | 0.394 |
| 21053 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 20 | KIT | Sponge network | -0.096 | 0.67958 | -2.184 | 0 | 0.394 |
| 21054 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 24 | ZFHX3 | Sponge network | 0.91 | 0 | 0.389 | 0.01363 | 0.394 |
| 21055 | NEAT1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 14 | SHPRH | Sponge network | 0.705 | 0.00014 | 0.098 | 0.40994 | 0.394 |
| 21056 | LINC00957 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | -0.421 | 0.0114 | -2.417 | 0 | 0.394 |
| 21057 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 16 | UVRAG | Sponge network | -2.46 | 0 | -0.017 | 0.83865 | 0.394 |
| 21058 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-584-5p;hsa-miR-877-5p;hsa-miR-942-5p | 14 | GLIPR1 | Sponge network | -0.465 | 0.04703 | 0.146 | 0.3276 | 0.394 |
| 21059 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-98-5p | 70 | NTRK3 | Sponge network | -0.727 | 0.04726 | -1.77 | 3.0E-5 | 0.394 |
| 21060 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 28 | FBXO32 | Sponge network | -0.342 | 0.02288 | -0.495 | 0.07561 | 0.394 |
| 21061 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 33 | NRXN1 | Sponge network | 0.249 | 0.35902 | -3.778 | 0 | 0.394 |
| 21062 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 39 | FOXP1 | Sponge network | 0.75 | 0.00054 | 0.042 | 0.74368 | 0.394 |
| 21063 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 40 | CUX1 | Sponge network | -0.162 | 0.47687 | -0.039 | 0.6705 | 0.394 |
| 21064 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 23 | GREM1 | Sponge network | -2.46 | 0 | 0.902 | 0.01691 | 0.394 |
| 21065 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-589-5p;hsa-miR-93-5p | 11 | SLITRK3 | Sponge network | -0.726 | 0.00132 | -3.328 | 0 | 0.394 |
| 21066 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-655-3p | 18 | LATS1 | Sponge network | 0.91 | 0 | 0.109 | 0.34208 | 0.394 |
| 21067 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | -0.793 | 7.0E-5 | -1.047 | 0.00364 | 0.394 |
| 21068 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 23 | ACVR1 | Sponge network | -0.129 | 0.57177 | 0.279 | 0.00234 | 0.394 |
| 21069 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 16 | QKI | Sponge network | -0.557 | 0.11135 | -0.333 | 0.04111 | 0.394 |
| 21070 | RP11-155G14.6 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 11 | RASAL2 | Sponge network | 2.483 | 0 | 0.869 | 0 | 0.394 |
| 21071 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 22 | DDX6 | Sponge network | 0.796 | 4.0E-5 | 0.091 | 0.36428 | 0.394 |
| 21072 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-935 | 20 | DMD | Sponge network | 0.37 | 0.27614 | -1.506 | 0 | 0.394 |
| 21073 | LINC00957 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ID4 | Sponge network | -0.421 | 0.0114 | -1.644 | 0 | 0.394 |
| 21074 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-423-5p;hsa-miR-93-3p;hsa-miR-940 | 20 | NTRK3 | Sponge network | -1.483 | 9.0E-5 | -1.77 | 3.0E-5 | 0.394 |
| 21075 | LINC00472 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p | 12 | ABCA10 | Sponge network | -0.842 | 0.01361 | -0.571 | 0.03674 | 0.394 |
| 21076 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-374b-3p;hsa-miR-493-5p | 14 | GRIA3 | Sponge network | -2.095 | 0.00112 | -1.956 | 0 | 0.394 |
| 21077 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577 | 16 | TMEFF2 | Sponge network | 0.056 | 0.81195 | -2.426 | 0 | 0.394 |
| 21078 | RP11-25K19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 38 | GAS7 | Sponge network | -1.057 | 0.00029 | -0.555 | 0.01602 | 0.394 |
| 21079 | GNG12-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-940 | 22 | TOM1L2 | Sponge network | -0.793 | 7.0E-5 | -0.994 | 0 | 0.394 |
| 21080 | RP11-83A24.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 46 | MDM4 | Sponge network | 0.417 | 0.00445 | 0.345 | 0.00762 | 0.394 |
| 21081 | HOTAIRM1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p | 27 | NTRK2 | Sponge network | -0.053 | 0.80685 | -0.736 | 0.06495 | 0.394 |
| 21082 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p | 11 | FBXO32 | Sponge network | -1.255 | 0.00981 | -0.495 | 0.07561 | 0.394 |
| 21083 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | MYO9A | Sponge network | -0.053 | 0.80685 | -0.147 | 0.32373 | 0.394 |
| 21084 | CTA-204B4.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | ZMYND11 | Sponge network | 0.765 | 7.0E-5 | -0.2 | 0.05449 | 0.394 |
| 21085 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-5p | 20 | RECK | Sponge network | -1.047 | 0 | -1.043 | 0 | 0.394 |
| 21086 | LINC00641 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 19 | TLE4 | Sponge network | -0.222 | 0.32445 | -1.157 | 0 | 0.394 |
| 21087 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-671-5p;hsa-miR-877-5p | 18 | BMPR2 | Sponge network | -0.309 | 0.1145 | 0.206 | 0.0635 | 0.394 |
| 21088 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 18 | AKAP6 | Sponge network | -0.08 | 0.90092 | -1.586 | 3.0E-5 | 0.394 |
| 21089 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 20 | SCN9A | Sponge network | -0.227 | 0.31657 | -0.525 | 0.10593 | 0.394 |
| 21090 | RP11-13K12.5 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 11 | ZNF208 | Sponge network | -0.318 | 0.65847 | -0.4 | 0.22381 | 0.394 |
| 21091 | MIR22HG |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3127-5p;hsa-miR-370-3p;hsa-miR-532-3p;hsa-miR-93-3p | 15 | RPS6KA2 | Sponge network | -1.047 | 0 | -0.57 | 0.00013 | 0.394 |
| 21092 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 18 | UNC5C | Sponge network | -0.769 | 0.00035 | -0.178 | 0.40214 | 0.394 |
| 21093 | LINC00476 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-942-5p | 12 | BMPR1A | Sponge network | -0.615 | 0.00015 | -0.366 | 0.01036 | 0.394 |
| 21094 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MOB1B | Sponge network | 0.986 | 0.01022 | 0.197 | 0.15266 | 0.394 |
| 21095 | AC005562.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | ABI2 | Sponge network | 0.488 | 0.00084 | 0.175 | 0.14787 | 0.394 |
| 21096 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-92a-3p | 21 | PDE4DIP | Sponge network | 0.056 | 0.81195 | -0.282 | 0.0257 | 0.394 |
| 21097 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 32 | CNTN1 | Sponge network | 0.374 | 0.20054 | -2.594 | 0 | 0.394 |
| 21098 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p | 18 | ABL1 | Sponge network | -2.46 | 0 | -0.215 | 0.07804 | 0.394 |
| 21099 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | TACC1 | Sponge network | -1.216 | 0 | -0.362 | 0.05406 | 0.394 |
| 21100 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 34 | FBXO32 | Sponge network | 0.246 | 0.59912 | -0.495 | 0.07561 | 0.394 |
| 21101 | CTD-2314G24.2 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-590-3p | 14 | FMNL3 | Sponge network | 0.246 | 0.59912 | 0.555 | 4.0E-5 | 0.394 |
| 21102 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-940 | 29 | NOTCH2 | Sponge network | 0.056 | 0.81195 | 0.138 | 0.35163 | 0.394 |
| 21103 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 38 | TRPS1 | Sponge network | -0.563 | 0.02545 | -0.191 | 0.44109 | 0.394 |
| 21104 | LINC00654 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p | 18 | ITGB3 | Sponge network | 0.374 | 0.20054 | -0.011 | 0.95751 | 0.394 |
| 21105 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | AFF3 | Sponge network | -0.899 | 6.0E-5 | -2.373 | 0 | 0.394 |
| 21106 | RP11-13K12.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-3607-3p | 14 | PIK3CA | Sponge network | -0.154 | 0.78939 | 0.195 | 0.06908 | 0.394 |
| 21107 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p | 21 | KIF1B | Sponge network | 0.497 | 0.01451 | -0.118 | 0.32258 | 0.394 |
| 21108 | CTD-2619J13.9 |
hsa-let-7g-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-625-5p | 10 | CBX5 | Sponge network | 0.576 | 0.0022 | 0.165 | 0.24446 | 0.394 |
| 21109 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PTPRB | Sponge network | -0.513 | 0.04703 | 0.105 | 0.47122 | 0.394 |
| 21110 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-654-3p | 14 | ITCH | Sponge network | 0.222 | 0.29133 | 0.084 | 0.34058 | 0.394 |
| 21111 | RP11-46C24.7 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-766-3p | 13 | EZH1 | Sponge network | 0.392 | 0.0278 | -0.564 | 1.0E-5 | 0.394 |
| 21112 | RP11-25K19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 20 | ACSL6 | Sponge network | -1.057 | 0.00029 | -1.593 | 0 | 0.394 |
| 21113 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-7-1-3p | 10 | DCN | Sponge network | -0.246 | 0.35034 | -1.288 | 0 | 0.394 |
| 21114 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | IGSF10 | Sponge network | -0.611 | 0.09607 | -1.751 | 1.0E-5 | 0.394 |
| 21115 | SNHG14 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | PAIP2 | Sponge network | -1.111 | 0.0003 | -0.515 | 0 | 0.394 |
| 21116 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | LRIG1 | Sponge network | -0.612 | 0.00094 | -0.537 | 0.00588 | 0.394 |
| 21117 | AC016747.3 |
hsa-miR-107;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | PTPN13 | Sponge network | 0.199 | 0.38015 | -1.208 | 0 | 0.394 |
| 21118 | LINC00265 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 1.07 | 0 | 0.362 | 0.00066 | 0.394 |
| 21119 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p | 24 | LRP1B | Sponge network | -0.739 | 0.00044 | -2.163 | 0 | 0.394 |
| 21120 | PSMG3-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-505-3p;hsa-miR-7-5p | 14 | FNBP1 | Sponge network | -0.125 | 0.49414 | -0.969 | 4.0E-5 | 0.394 |
| 21121 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | TP53INP1 | Sponge network | 0.4 | 0.14611 | -0.496 | 0.00064 | 0.394 |
| 21122 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-590-3p | 29 | DTNA | Sponge network | 0.381 | 0.25967 | -1.648 | 1.0E-5 | 0.394 |
| 21123 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 61 | BMPR2 | Sponge network | -1.281 | 0.00012 | 0.206 | 0.0635 | 0.394 |
| 21124 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-532-5p;hsa-miR-590-3p | 14 | ZMYND11 | Sponge network | 0.174 | 0.30034 | -0.2 | 0.05449 | 0.394 |
| 21125 | HAND2-AS1 |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | CTNNA2 | Sponge network | -1.697 | 0.00889 | -2.547 | 0 | 0.394 |
| 21126 | CTC-429P9.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-24-3p | 19 | DLC1 | Sponge network | 0.082 | 0.55397 | -0.382 | 0.05038 | 0.394 |
| 21127 | FZD10-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 15 | EHD3 | Sponge network | -0.727 | 0.04726 | -0.484 | 0.01747 | 0.394 |
| 21128 | RP11-13K12.1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-589-3p | 10 | ZNF844 | Sponge network | -0.154 | 0.78939 | -0.966 | 0.00035 | 0.394 |
| 21129 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 12 | MAFB | Sponge network | -0.896 | 0.00099 | -0.023 | 0.89865 | 0.394 |
| 21130 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | ARHGAP28 | Sponge network | -0.376 | 0.00337 | -0.911 | 0.00013 | 0.394 |
| 21131 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p | 12 | AKAP6 | Sponge network | -2.549 | 0.00528 | -1.586 | 3.0E-5 | 0.394 |
| 21132 | SNX29P2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-671-5p | 25 | DLC1 | Sponge network | 0.4 | 0.14611 | -0.382 | 0.05038 | 0.394 |
| 21133 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p | 17 | CHD5 | Sponge network | -1.582 | 8.0E-5 | -0.922 | 0.01521 | 0.394 |
| 21134 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | CXXC4 | Sponge network | -0.901 | 0.00019 | -0.433 | 0.21055 | 0.394 |
| 21135 | RP11-400K9.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-744-3p;hsa-miR-93-3p | 20 | TOM1L2 | Sponge network | -0.611 | 0.09607 | -0.994 | 0 | 0.394 |
| 21136 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | SOX5 | Sponge network | 0.4 | 0.14611 | -1.091 | 4.0E-5 | 0.394 |
| 21137 | CTD-3092A11.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PHF3 | Sponge network | 0.873 | 0 | 0.126 | 0.19652 | 0.394 |
| 21138 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FSTL5 | Sponge network | -0.769 | 0.00035 | -1.3 | 0.01619 | 0.394 |
| 21139 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-3913-5p | 11 | FNBP1 | Sponge network | -0.738 | 0.21233 | -0.969 | 4.0E-5 | 0.394 |
| 21140 | RP11-428J1.5 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-655-3p;hsa-miR-7-1-3p | 15 | UBE4B | Sponge network | 0.538 | 0.00076 | -0.235 | 0.007 | 0.394 |
| 21141 | LINC00641 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 26 | CYLD | Sponge network | -0.222 | 0.32445 | -0.367 | 0.00171 | 0.394 |
| 21142 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 23 | PTGFR | Sponge network | -1.216 | 0 | -0.233 | 0.45421 | 0.394 |
| 21143 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 39 | CBX5 | Sponge network | 0.663 | 0.00073 | 0.165 | 0.24446 | 0.394 |
| 21144 | PGM5-AS1 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-335-3p | 15 | ACSL6 | Sponge network | -4.546 | 0 | -1.593 | 0 | 0.394 |
| 21145 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-93-3p | 45 | BMPR2 | Sponge network | 0.637 | 0.00069 | 0.206 | 0.0635 | 0.394 |
| 21146 | RP11-672A2.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-766-3p | 14 | IL17RD | Sponge network | 1.344 | 0 | -0.019 | 0.94873 | 0.394 |
| 21147 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | MOB1B | Sponge network | 1.308 | 0 | 0.197 | 0.15266 | 0.394 |
| 21148 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | 0.214 | 0.18246 | -0.001 | 0.99669 | 0.394 |
| 21149 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 21 | NRXN1 | Sponge network | -0.318 | 0.65847 | -3.778 | 0 | 0.394 |
| 21150 | RP1-102E24.8 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p | 10 | RASAL2 | Sponge network | 1.737 | 0 | 0.869 | 0 | 0.394 |
| 21151 | CTD-3064M3.3 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-582-5p | 10 | ZNF208 | Sponge network | -0.469 | 0.08721 | -0.4 | 0.22381 | 0.394 |
| 21152 | LINC00648 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | TET1 | Sponge network | 0.633 | 0.51678 | -0.135 | 0.52138 | 0.394 |
| 21153 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ABCC9 | Sponge network | 0.559 | 0.00026 | -0.466 | 0.14121 | 0.394 |
| 21154 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-532-3p | 13 | RPS6KA2 | Sponge network | -0.739 | 0.00044 | -0.57 | 0.00013 | 0.394 |
| 21155 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | CHD2 | Sponge network | 0.659 | 3.0E-5 | 0.049 | 0.55371 | 0.394 |
| 21156 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | RASSF8 | Sponge network | -0.737 | 0.10822 | -0.122 | 0.65591 | 0.394 |
| 21157 | CTC-297N7.9 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-590-5p;hsa-miR-877-5p | 10 | GRIN2A | Sponge network | -2.069 | 0 | -2.161 | 0 | 0.394 |
| 21158 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | LRRK2 | Sponge network | -0.942 | 0 | -1.136 | 1.0E-5 | 0.394 |
| 21159 | PWAR6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | CREB1 | Sponge network | -1.575 | 6.0E-5 | 0.203 | 0.00537 | 0.394 |
| 21160 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | ST6GAL2 | Sponge network | 0.61 | 0.01593 | -1.201 | 0.00597 | 0.394 |
| 21161 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-769-5p | 15 | RCAN2 | Sponge network | -0.777 | 0.16667 | -0.33 | 0.15172 | 0.393 |
| 21162 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | AR | Sponge network | 0.342 | 0.03129 | -1.554 | 0 | 0.393 |
| 21163 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-145-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-539-5p | 11 | MOB1B | Sponge network | 1.055 | 0 | 0.197 | 0.15266 | 0.393 |
| 21164 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-409-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | ST6GALNAC3 | Sponge network | -2.915 | 0.00015 | -0.987 | 0 | 0.393 |
| 21165 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 15 | DPYD | Sponge network | -1.697 | 0.00889 | -0.736 | 0.00076 | 0.393 |
| 21166 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-942-5p | 13 | RB1CC1 | Sponge network | 0.392 | 0.0278 | 0.175 | 0.12559 | 0.393 |
| 21167 | LINC00657 |
hsa-let-7g-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p | 11 | KDM3A | Sponge network | 0.91 | 0 | 0.132 | 0.22711 | 0.393 |
| 21168 | LINC00654 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 17 | SPG20 | Sponge network | 0.374 | 0.20054 | -1.16 | 0 | 0.393 |
| 21169 | RP11-121C2.2 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 14 | BRD3 | Sponge network | 1.147 | 0 | 0.37 | 0.00013 | 0.393 |
| 21170 | AC127904.2 |
hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | 1.308 | 0 | 0.098 | 0.40994 | 0.393 |
| 21171 | HAND2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | NIN | Sponge network | -1.697 | 0.00889 | -0.253 | 0.13794 | 0.393 |
| 21172 | RP11-344B5.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-664a-3p | 15 | PRRX1 | Sponge network | -0.055 | 0.88482 | 1.316 | 0 | 0.393 |
| 21173 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 12 | DAB2 | Sponge network | -0.376 | 0.00337 | 0.431 | 0.0113 | 0.393 |
| 21174 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p | 24 | RSPO2 | Sponge network | -0.777 | 0.16667 | -3.038 | 0 | 0.393 |
| 21175 | RP11-798M19.6 |
hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | CXCL12 | Sponge network | -0.612 | 0.00094 | -1.421 | 0 | 0.393 |
| 21176 | LINC00641 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 15 | JAKMIP2 | Sponge network | -0.222 | 0.32445 | -1.388 | 0 | 0.393 |
| 21177 | RP11-439A17.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-22-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-98-5p | 11 | PRKAA2 | Sponge network | 0.051 | 0.95756 | -2.462 | 0 | 0.393 |
| 21178 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 37 | DMXL1 | Sponge network | -1.111 | 0.0003 | -0.426 | 0.00056 | 0.393 |
| 21179 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 39 | AKAP11 | Sponge network | -1.575 | 6.0E-5 | 0.196 | 0.09825 | 0.393 |
| 21180 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-590-3p | 24 | FOXO3 | Sponge network | 0.765 | 7.0E-5 | -0.04 | 0.72028 | 0.393 |
| 21181 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-940 | 34 | NTRK2 | Sponge network | -0.246 | 0.35034 | -0.736 | 0.06495 | 0.393 |
| 21182 | DIO3OS |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-940 | 13 | SORCS1 | Sponge network | -0.773 | 0.04073 | -3.492 | 0 | 0.393 |
| 21183 | DNM1P35 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-423-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CELF4 | Sponge network | 0.051 | 0.83388 | -1.135 | 0.00344 | 0.393 |
| 21184 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | CSMD1 | Sponge network | -0.976 | 0.00025 | -0.63 | 0.18881 | 0.393 |
| 21185 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ABI2 | Sponge network | 0.349 | 0.01695 | 0.175 | 0.14787 | 0.393 |
| 21186 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | BCL11A | Sponge network | -1.349 | 0.00017 | -0.932 | 0.0063 | 0.393 |
| 21187 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 19 | THBS1 | Sponge network | 0.381 | 0.25967 | 0.741 | 0.00386 | 0.393 |
| 21188 | MRVI1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 17 | PRRX1 | Sponge network | -1.092 | 4.0E-5 | 1.316 | 0 | 0.393 |
| 21189 | PSMD5-AS1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | ERCC6 | Sponge network | 0.569 | 0.00067 | 0.23 | 0.015 | 0.393 |
| 21190 | RP11-33B1.4 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-582-5p | 10 | ZFHX3 | Sponge network | 0.826 | 0 | 0.389 | 0.01363 | 0.393 |
| 21191 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 20 | GRIN2A | Sponge network | -0.513 | 0.04703 | -2.161 | 0 | 0.393 |
| 21192 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 22 | TSC1 | Sponge network | 0.494 | 0.00339 | 0.103 | 0.26071 | 0.393 |
| 21193 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-375;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-590-5p | 19 | MLLT10 | Sponge network | 0.921 | 0.00118 | 0.105 | 0.33292 | 0.393 |
| 21194 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | MYO9A | Sponge network | 0.657 | 4.0E-5 | -0.147 | 0.32373 | 0.393 |
| 21195 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 18 | FGFR1 | Sponge network | -0.899 | 6.0E-5 | -0.509 | 0.03957 | 0.393 |
| 21196 | RP11-822E23.8 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-93-3p | 15 | UNC5C | Sponge network | -0.777 | 0.16667 | -0.178 | 0.40214 | 0.393 |
| 21197 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | LATS1 | Sponge network | 0.919 | 0 | 0.109 | 0.34208 | 0.393 |
| 21198 | RP11-464F9.1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | RBBP6 | Sponge network | 0.959 | 0 | 0.163 | 0.05101 | 0.393 |
| 21199 | LINC00672 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | RECK | Sponge network | -0.227 | 0.31657 | -1.043 | 0 | 0.393 |
| 21200 | RP11-798G7.7 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-455-3p | 10 | SIRT1 | Sponge network | 0.858 | 3.0E-5 | 0.109 | 0.2593 | 0.393 |
| 21201 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 33 | TACC1 | Sponge network | -1.203 | 0.03881 | -0.362 | 0.05406 | 0.393 |
| 21202 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-497-5p | 19 | RICTOR | Sponge network | 0.917 | 0 | 0.388 | 0.00188 | 0.393 |
| 21203 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-624-5p | 10 | SMARCA1 | Sponge network | -0.487 | 0.02157 | -0.684 | 0.00159 | 0.393 |
| 21204 | RP11-5C23.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 14 | CYLD | Sponge network | 0.274 | 0.07681 | -0.367 | 0.00171 | 0.393 |
| 21205 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p | 10 | ST5 | Sponge network | -2.925 | 0 | -0.78 | 0 | 0.393 |
| 21206 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 15 | CTIF | Sponge network | -0.739 | 0.00044 | -0.389 | 0.00549 | 0.393 |
| 21207 | CTC-550B14.7 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-664a-5p | 10 | CBFB | Sponge network | 1.161 | 0 | 1.061 | 0 | 0.393 |
| 21208 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-874-3p | 20 | NR2C2 | Sponge network | 0.541 | 0.00125 | 0.21 | 0.03224 | 0.393 |
| 21209 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-940 | 27 | THBS1 | Sponge network | -0.162 | 0.47687 | 0.741 | 0.00386 | 0.393 |
| 21210 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-940 | 41 | CHD5 | Sponge network | -0.942 | 0 | -0.922 | 0.01521 | 0.393 |
| 21211 | SH3RF3-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 20 | NCAM2 | Sponge network | 0.889 | 9.0E-5 | -0.482 | 0.22565 | 0.393 |
| 21212 | WDFY3-AS2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 37 | IGFBP5 | Sponge network | -0.942 | 0 | -0.806 | 0.00105 | 0.393 |
| 21213 | ARHGEF26-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-331-5p;hsa-miR-429 | 11 | DUSP1 | Sponge network | -2.46 | 0 | -1.754 | 0 | 0.393 |
| 21214 | RP11-452L6.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-425-5p | 11 | ATRX | Sponge network | 0.665 | 0.00061 | 0.261 | 0.04408 | 0.393 |
| 21215 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 37 | DIP2C | Sponge network | 0.532 | 0.00505 | -0.436 | 0.01713 | 0.393 |
| 21216 | MRVI1-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-500a-3p | 11 | FMNL3 | Sponge network | -1.092 | 4.0E-5 | 0.555 | 4.0E-5 | 0.393 |
| 21217 | LINC00476 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | KCTD8 | Sponge network | -0.615 | 0.00015 | -3.359 | 0 | 0.393 |
| 21218 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 29 | MCC | Sponge network | 0.4 | 0.14611 | -1.005 | 0 | 0.393 |
| 21219 | CTA-363E19.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p | 13 | DDX6 | Sponge network | 1.055 | 0 | 0.091 | 0.36428 | 0.393 |
| 21220 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 67 | WDFY3 | Sponge network | -1.111 | 0.0003 | -0.201 | 0.0967 | 0.393 |
| 21221 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | ST5 | Sponge network | -1.111 | 0.0003 | -0.78 | 0 | 0.393 |
| 21222 | CTC-444N24.11 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 13 | GSK3B | Sponge network | 1.153 | 0 | 0.266 | 0.00045 | 0.393 |
| 21223 | HCG18 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 13 | GSK3B | Sponge network | 1.063 | 0 | 0.266 | 0.00045 | 0.393 |
| 21224 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-134-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 28 | SSBP2 | Sponge network | -4.477 | 0 | -1.58 | 0 | 0.393 |
| 21225 | LINC00094 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-624-5p | 12 | HMCN1 | Sponge network | 0.253 | 0.09565 | 0.809 | 0.00544 | 0.393 |
| 21226 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 44 | CBX5 | Sponge network | -0.096 | 0.67958 | 0.165 | 0.24446 | 0.393 |
| 21227 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-93-5p | 12 | CNTNAP3 | Sponge network | -0.726 | 0.00132 | -1.465 | 0.00033 | 0.393 |
| 21228 | HAND2-AS1 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 10 | DSE | Sponge network | -1.697 | 0.00889 | -0.231 | 0.05347 | 0.393 |
| 21229 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | TIMP3 | Sponge network | -0.709 | 0.18168 | -0.546 | 0.02307 | 0.393 |
| 21230 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-582-5p | 10 | RB1CC1 | Sponge network | 1.294 | 0 | 0.175 | 0.12559 | 0.393 |
| 21231 | LINC00472 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-493-5p | 11 | HERC1 | Sponge network | -0.842 | 0.01361 | -0.196 | 0.08544 | 0.393 |
| 21232 | RP11-894P9.1 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p | 10 | ITGB1 | Sponge network | 0.993 | 0 | 0.524 | 0.00017 | 0.393 |
| 21233 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | TET1 | Sponge network | -1.697 | 0.00889 | -0.135 | 0.52138 | 0.393 |
| 21234 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 50 | THRB | Sponge network | -0.896 | 0.00099 | -1.117 | 0.0004 | 0.393 |
| 21235 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 50 | PRKAA2 | Sponge network | 0.249 | 0.35902 | -2.462 | 0 | 0.393 |
| 21236 | RP11-156P1.3 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-486-5p;hsa-miR-590-3p | 10 | DNMT3A | Sponge network | 0.214 | 0.18246 | 0.302 | 0.0262 | 0.393 |
| 21237 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 17 | MYCBP2 | Sponge network | 0.72 | 0.0001 | -0.027 | 0.82016 | 0.393 |
| 21238 | PCA3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DCN | Sponge network | -0.709 | 0.18168 | -1.288 | 0 | 0.393 |
| 21239 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 41 | SASH1 | Sponge network | -0.942 | 0 | -0.502 | 0.00175 | 0.393 |
| 21240 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-450b-5p | 14 | PKNOX1 | Sponge network | 0.75 | 0.00054 | 0.085 | 0.28917 | 0.393 |
| 21241 | CTD-3064M3.3 |
hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-582-5p | 10 | LUZP2 | Sponge network | -0.469 | 0.08721 | -0.056 | 0.89416 | 0.393 |
| 21242 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p | 12 | ABCB5 | Sponge network | -0.318 | 0.65847 | -0.956 | 0.04077 | 0.393 |
| 21243 | LINC00672 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-590-3p | 12 | SORCS1 | Sponge network | -0.227 | 0.31657 | -3.492 | 0 | 0.393 |
| 21244 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | FAM172A | Sponge network | -0.769 | 0.00035 | -0.57 | 0 | 0.393 |
| 21245 | MIR143HG |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | CDKN1C | Sponge network | -0.739 | 0.0574 | -0.929 | 0.00033 | 0.393 |
| 21246 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 51 | TCF4 | Sponge network | -0.738 | 0.21233 | 0.016 | 0.92271 | 0.393 |
| 21247 | TUG1 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-497-5p | 12 | CDK6 | Sponge network | 0.972 | 0 | 0.991 | 0 | 0.393 |
| 21248 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p | 19 | EBF3 | Sponge network | -1.057 | 0.00029 | -0.058 | 0.81437 | 0.393 |
| 21249 | LINC00641 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | MAP3K4 | Sponge network | -0.222 | 0.32445 | 0.058 | 0.54229 | 0.393 |
| 21250 | GS1-124K5.11 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-590-3p | 20 | ITPKB | Sponge network | 0.494 | 0.00339 | -0.704 | 0.00106 | 0.393 |
| 21251 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | DYNC1I1 | Sponge network | -0.737 | 0.10822 | -1.12 | 0.00107 | 0.393 |
| 21252 | OIP5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 18 | PKNOX1 | Sponge network | 0.421 | 0.00084 | 0.085 | 0.28917 | 0.393 |
| 21253 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | ARNT2 | Sponge network | 0.199 | 0.38015 | -0.332 | 0.25851 | 0.393 |
| 21254 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-550a-3p | 19 | NRK | Sponge network | 0.331 | 0.57247 | 0.656 | 0.19217 | 0.393 |
| 21255 | FZD10-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 23 | CADM3 | Sponge network | -0.727 | 0.04726 | -3.663 | 0 | 0.393 |
| 21256 | RP11-344B5.2 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-3p | 13 | RBMS3 | Sponge network | -0.055 | 0.88482 | -0.519 | 0.03551 | 0.393 |
| 21257 | CTD-2528L19.6 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | UBR3 | Sponge network | 0.953 | 0 | -0.021 | 0.82774 | 0.393 |
| 21258 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | RET | Sponge network | -1.216 | 0 | -0.833 | 0.03517 | 0.393 |
| 21259 | RAMP2-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-423-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | ROBO1 | Sponge network | -0.513 | 0.04703 | -0.009 | 0.96154 | 0.393 |
| 21260 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 23 | PCDH17 | Sponge network | 0.194 | 0.64278 | 0.731 | 2.0E-5 | 0.393 |
| 21261 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-576-5p;hsa-miR-93-5p | 14 | RBL2 | Sponge network | -0.611 | 0.09607 | -0.282 | 0.00254 | 0.393 |
| 21262 | DNM3OS |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | LRIG1 | Sponge network | 0.572 | 0.01722 | -0.537 | 0.00588 | 0.393 |
| 21263 | BOLA3-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-429 | 14 | KANK1 | Sponge network | -0.246 | 0.35034 | -0.55 | 0.00134 | 0.393 |
| 21264 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-500a-5p | 12 | ZNF208 | Sponge network | -0.563 | 0.02545 | -0.4 | 0.22381 | 0.393 |
| 21265 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | LRIG1 | Sponge network | 0.997 | 0.00066 | -0.537 | 0.00588 | 0.393 |
| 21266 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | PGR | Sponge network | 0.342 | 0.03129 | -1.704 | 0 | 0.393 |
| 21267 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 19 | USP6 | Sponge network | 1.04 | 0.00023 | 0.188 | 0.3871 | 0.393 |
| 21268 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-582-5p | 14 | USP6 | Sponge network | 0.993 | 0 | 0.188 | 0.3871 | 0.393 |
| 21269 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-942-5p | 41 | THRB | Sponge network | -0.693 | 0.05629 | -1.117 | 0.0004 | 0.393 |
| 21270 | DNAJC3-AS1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 10 | PHF6 | Sponge network | 0.753 | 0.00014 | 0.785 | 0 | 0.393 |
| 21271 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | CXXC4 | Sponge network | -0.563 | 0.02545 | -0.433 | 0.21055 | 0.393 |
| 21272 | RP11-130L8.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-493-3p;hsa-miR-511-5p;hsa-miR-590-3p | 14 | MRVI1 | Sponge network | -0.663 | 0.10887 | -1.051 | 0.00022 | 0.393 |
| 21273 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 63 | THRB | Sponge network | -0.899 | 6.0E-5 | -1.117 | 0.0004 | 0.393 |
| 21274 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | NCAM2 | Sponge network | 1.302 | 7.0E-5 | -0.482 | 0.22565 | 0.393 |
| 21275 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | LRRC4 | Sponge network | -1.697 | 0.00889 | -1.774 | 0 | 0.393 |
| 21276 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-93-5p | 35 | PDGFRA | Sponge network | -1.047 | 0 | -0.372 | 0.0037 | 0.393 |
| 21277 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 33 | CHD5 | Sponge network | -2.117 | 0.00161 | -0.922 | 0.01521 | 0.393 |
| 21278 | RP11-182J1.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-96-5p | 15 | ASTN1 | Sponge network | -0.187 | 0.23787 | -2.389 | 2.0E-5 | 0.393 |
| 21279 | CKMT2-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 13 | MAPK10 | Sponge network | 0.082 | 0.55654 | -1.774 | 0 | 0.393 |
| 21280 | BVES-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-486-5p;hsa-miR-877-5p;hsa-miR-942-5p | 14 | GLIPR1 | Sponge network | -2.62 | 0 | 0.146 | 0.3276 | 0.393 |
| 21281 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TRIM13 | Sponge network | 0.311 | 0.10344 | 0.183 | 0.06968 | 0.393 |
| 21282 | RP11-707P17.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 1.877 | 0 | 0.51 | 0 | 0.393 |
| 21283 | HCG11 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 13 | ABL1 | Sponge network | 0.385 | 0.10067 | -0.215 | 0.07804 | 0.393 |
| 21284 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 53 | FAT3 | Sponge network | -0.969 | 2.0E-5 | -1.601 | 0.00051 | 0.393 |
| 21285 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | DKK3 | Sponge network | -0.727 | 0.04726 | -0.052 | 0.77783 | 0.393 |
| 21286 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 29 | RSPO2 | Sponge network | -1.216 | 0 | -3.038 | 0 | 0.393 |
| 21287 | ZNF32-AS1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-576-5p | 15 | MDM4 | Sponge network | 2.413 | 1.0E-5 | 0.345 | 0.00762 | 0.393 |
| 21288 | HHIP-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | KCNA6 | Sponge network | -0.737 | 0.10822 | -1.531 | 0.00013 | 0.393 |
| 21289 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p | 10 | ZNF770 | Sponge network | 0.946 | 0 | 0.206 | 0.06751 | 0.393 |
| 21290 | GAS6-AS2 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | PCDH9 | Sponge network | 0.16 | 0.50785 | -1.823 | 4.0E-5 | 0.393 |
| 21291 | WDFY3-AS2 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 16 | RHOB | Sponge network | -0.942 | 0 | -1.052 | 0 | 0.393 |
| 21292 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 36 | RICTOR | Sponge network | 0.401 | 0.00066 | 0.388 | 0.00188 | 0.393 |
| 21293 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 13 | FGF5 | Sponge network | -0.896 | 0.00099 | 0.549 | 0.28659 | 0.393 |
| 21294 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-196b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-7-1-3p | 12 | SLC6A15 | Sponge network | -1.092 | 4.0E-5 | -2.373 | 2.0E-5 | 0.393 |
| 21295 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 19 | ZMYM2 | Sponge network | 0.752 | 0 | 0.279 | 0.01273 | 0.393 |
| 21296 | HOTAIRM1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | RASSF8 | Sponge network | -0.053 | 0.80685 | -0.122 | 0.65591 | 0.393 |
| 21297 | LINC00654 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 27 | PCDH9 | Sponge network | 0.374 | 0.20054 | -1.823 | 4.0E-5 | 0.393 |
| 21298 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 53 | UNC80 | Sponge network | -1.697 | 0.00889 | -1.934 | 0 | 0.393 |
| 21299 | RP11-390K5.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 15 | CREB1 | Sponge network | 1.148 | 2.0E-5 | 0.203 | 0.00537 | 0.393 |
| 21300 | RP11-685N10.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-3607-3p | 11 | RICTOR | Sponge network | 2.743 | 0 | 0.388 | 0.00188 | 0.393 |
| 21301 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-484 | 17 | GREM1 | Sponge network | -0.154 | 0.78939 | 0.902 | 0.01691 | 0.393 |
| 21302 | CTD-2089N3.1 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-455-5p | 10 | IL6ST | Sponge network | -0.314 | 0.62033 | -0.402 | 0.00676 | 0.393 |
| 21303 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | TACC1 | Sponge network | -0.376 | 0.00337 | -0.362 | 0.05406 | 0.393 |
| 21304 | RP11-6O2.3 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-455-3p | 10 | HERC1 | Sponge network | 0.331 | 0.57247 | -0.196 | 0.08544 | 0.393 |
| 21305 | RP11-927P21.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-3607-3p | 12 | ZMYND11 | Sponge network | 0.921 | 0.00118 | -0.2 | 0.05449 | 0.393 |
| 21306 | FAM66C |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | INPP4A | Sponge network | -0.901 | 0.00019 | 0.07 | 0.53563 | 0.393 |
| 21307 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 18 | PNRC1 | Sponge network | -0.793 | 7.0E-5 | -0.646 | 0 | 0.393 |
| 21308 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 51 | TACC1 | Sponge network | -2.925 | 0 | -0.362 | 0.05406 | 0.393 |
| 21309 | LINC00886 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-93-5p | 53 | DST | Sponge network | -0.371 | 0.24604 | -0.713 | 0.00096 | 0.393 |
| 21310 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 18 | SCN9A | Sponge network | -1.057 | 0.00029 | -0.525 | 0.10593 | 0.393 |
| 21311 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | TRIP11 | Sponge network | 0.421 | 0.00084 | -0.158 | 0.06384 | 0.393 |
| 21312 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | MYO9A | Sponge network | 0.16 | 0.50785 | -0.147 | 0.32373 | 0.393 |
| 21313 | RP1-151F17.2 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | RASSF8 | Sponge network | 0.222 | 0.29133 | -0.122 | 0.65591 | 0.393 |
| 21314 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 33 | NTRK2 | Sponge network | 0.222 | 0.29133 | -0.736 | 0.06495 | 0.393 |
| 21315 | CALML3-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p | 17 | UBE4B | Sponge network | 0.95 | 0.15943 | -0.235 | 0.007 | 0.393 |
| 21316 | RP4-613B23.1 |
hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-539-5p | 12 | RIMS2 | Sponge network | -0.309 | 0.1145 | -1.365 | 0.00208 | 0.393 |
| 21317 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-942-5p | 17 | THRB | Sponge network | -1.14 | 0.03513 | -1.117 | 0.0004 | 0.393 |
| 21318 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-935;hsa-miR-96-5p | 20 | FAT3 | Sponge network | 0.37 | 0.27614 | -1.601 | 0.00051 | 0.393 |
| 21319 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-539-5p | 12 | MGA | Sponge network | -0.303 | 0.21002 | 0.323 | 0.00792 | 0.393 |
| 21320 | MIR143HG |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 20 | TES | Sponge network | -0.739 | 0.0574 | -0.348 | 0.00639 | 0.393 |
| 21321 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-582-5p | 10 | NR2C2 | Sponge network | 0.419 | 0.0319 | 0.21 | 0.03224 | 0.393 |
| 21322 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | MED12L | Sponge network | -0.49 | 0.04946 | -0.194 | 0.55299 | 0.393 |
| 21323 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | MOB1B | Sponge network | 0.488 | 0.00084 | 0.197 | 0.15266 | 0.393 |
| 21324 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | ATRX | Sponge network | -0.709 | 0.18168 | 0.261 | 0.04408 | 0.393 |
| 21325 | RP5-1085F17.3 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | TRIM13 | Sponge network | 0.541 | 0.00125 | 0.183 | 0.06968 | 0.393 |
| 21326 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 17 | MCC | Sponge network | 0.381 | 0.25967 | -1.005 | 0 | 0.393 |
| 21327 | RP11-384O8.1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | CXCL12 | Sponge network | -0.738 | 0.21233 | -1.421 | 0 | 0.393 |
| 21328 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | PLSCR4 | Sponge network | 0.178 | 0.29106 | -0.474 | 0.03833 | 0.393 |
| 21329 | GNG12-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 15 | LUZP2 | Sponge network | -0.793 | 7.0E-5 | -0.056 | 0.89416 | 0.393 |
| 21330 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-7-1-3p | 16 | FAT4 | Sponge network | -0.246 | 0.35034 | -0.354 | 0.14694 | 0.393 |
| 21331 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-940 | 22 | CHD5 | Sponge network | 0.174 | 0.30034 | -0.922 | 0.01521 | 0.393 |
| 21332 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429 | 11 | ITGB3 | Sponge network | -0.246 | 0.35034 | -0.011 | 0.95751 | 0.393 |
| 21333 | LINC00865 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-93-3p;hsa-miR-935 | 11 | STAT5B | Sponge network | -1.249 | 7.0E-5 | -0.182 | 0.1082 | 0.393 |
| 21334 | RP11-6N17.3 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-324-3p | 12 | ADARB1 | Sponge network | 0.108 | 0.69556 | -0.335 | 0.06631 | 0.393 |
| 21335 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-369-3p;hsa-miR-495-3p | 10 | BCLAF1 | Sponge network | 1.044 | 2.0E-5 | 0.211 | 0.00684 | 0.393 |
| 21336 | CTC-550B14.7 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-374a-3p;hsa-miR-766-3p | 13 | TBL1XR1 | Sponge network | 1.161 | 0 | 0.578 | 0 | 0.393 |
| 21337 | RP11-344B5.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b | 15 | ZFHX4 | Sponge network | -0.055 | 0.88482 | 0.018 | 0.95574 | 0.393 |
| 21338 | RP11-37B2.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-582-3p | 10 | PIK3CA | Sponge network | 1.03 | 0 | 0.195 | 0.06908 | 0.393 |
| 21339 | GAS6-AS2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 46 | DST | Sponge network | 0.16 | 0.50785 | -0.713 | 0.00096 | 0.393 |
| 21340 | RP11-182J1.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | SFRP1 | Sponge network | -0.187 | 0.23787 | -3.566 | 0 | 0.393 |
| 21341 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 47 | TGFBR3 | Sponge network | -0.942 | 0 | -1.173 | 1.0E-5 | 0.393 |
| 21342 | CTB-31O20.4 |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-629-3p | 11 | DDX6 | Sponge network | 1.619 | 0 | 0.091 | 0.36428 | 0.393 |
| 21343 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 43 | BCL2 | Sponge network | -1.216 | 0 | -0.899 | 0 | 0.393 |
| 21344 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | CREBBP | Sponge network | 0.101 | 0.60919 | 0.126 | 0.15186 | 0.393 |
| 21345 | RP11-384K6.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-3p | 18 | ZFX | Sponge network | 0.946 | 0 | 0.09 | 0.42563 | 0.392 |
| 21346 | HCG11 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | 0.385 | 0.10067 | -1.051 | 0.00022 | 0.392 |
| 21347 | FAM215B |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-7-1-3p | 15 | DICER1 | Sponge network | 0.817 | 3.0E-5 | 0.042 | 0.674 | 0.392 |
| 21348 | A2M-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PCDH9 | Sponge network | -0.08 | 0.90092 | -1.823 | 4.0E-5 | 0.392 |
| 21349 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p | 17 | DLC1 | Sponge network | -0.829 | 0.01343 | -0.382 | 0.05038 | 0.392 |
| 21350 | LINC00865 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 12 | TPO | Sponge network | -1.249 | 7.0E-5 | -1.87 | 2.0E-5 | 0.392 |
| 21351 | WDFY3-AS2 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TPO | Sponge network | -0.942 | 0 | -1.87 | 2.0E-5 | 0.392 |
| 21352 | OIP5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p | 13 | LIMD1 | Sponge network | 0.421 | 0.00084 | 0.103 | 0.28978 | 0.392 |
| 21353 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | SPG20 | Sponge network | 0.082 | 0.55654 | -1.16 | 0 | 0.392 |
| 21354 | AC004893.11 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-766-3p | 35 | MDM4 | Sponge network | 1.317 | 0 | 0.345 | 0.00762 | 0.392 |
| 21355 | DNAJC27-AS1 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-7-1-3p | 31 | RIMS2 | Sponge network | -0.969 | 2.0E-5 | -1.365 | 0.00208 | 0.392 |
| 21356 | RP11-403I13.7 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-96-5p | 38 | GAS7 | Sponge network | 0.395 | 0.15451 | -0.555 | 0.01602 | 0.392 |
| 21357 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | MCC | Sponge network | -0.899 | 6.0E-5 | -1.005 | 0 | 0.392 |
| 21358 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-145-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | MOB1B | Sponge network | 2.094 | 0 | 0.197 | 0.15266 | 0.392 |
| 21359 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-940;hsa-miR-96-5p | 23 | CRTC3 | Sponge network | 0.082 | 0.55654 | 0.164 | 0.16786 | 0.392 |
| 21360 | RP11-532F6.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-590-3p | 12 | SAMHD1 | Sponge network | -0.896 | 0.00099 | 0.072 | 0.62895 | 0.392 |
| 21361 | FAM66C |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p | 14 | UVRAG | Sponge network | -0.901 | 0.00019 | -0.017 | 0.83865 | 0.392 |
| 21362 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p | 40 | LPP | Sponge network | 1.757 | 0 | -0.459 | 0.04475 | 0.392 |
| 21363 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-654-3p | 15 | ITCH | Sponge network | 0.848 | 0 | 0.084 | 0.34058 | 0.392 |
| 21364 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-423-5p;hsa-miR-629-3p;hsa-miR-93-3p | 12 | NTRK3 | Sponge network | -1.717 | 0.01522 | -1.77 | 3.0E-5 | 0.392 |
| 21365 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-590-3p | 11 | IGSF10 | Sponge network | 0.214 | 0.18246 | -1.751 | 1.0E-5 | 0.392 |
| 21366 | LINC00173 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-7-5p | 11 | PTGFR | Sponge network | 0.056 | 0.8494 | -0.233 | 0.45421 | 0.392 |
| 21367 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-744-3p | 12 | CUL5 | Sponge network | 0.768 | 7.0E-5 | 0.026 | 0.77301 | 0.392 |
| 21368 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-590-3p | 16 | STARD13 | Sponge network | -0.769 | 0.00035 | -0.187 | 0.27383 | 0.392 |
| 21369 | RP11-1006G14.4 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 12 | CSDE1 | Sponge network | 0.559 | 0.00026 | -0.493 | 0 | 0.392 |
| 21370 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | THBS1 | Sponge network | -0.246 | 0.35034 | 0.741 | 0.00386 | 0.392 |
| 21371 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 27 | USP12 | Sponge network | -0.323 | 0.01372 | -0.102 | 0.40768 | 0.392 |
| 21372 | LINC00854 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-532-3p | 13 | CBL | Sponge network | 1.18 | 0 | 0.291 | 0.01267 | 0.392 |
| 21373 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484 | 10 | SALL2 | Sponge network | 0.083 | 0.66405 | -1.398 | 2.0E-5 | 0.392 |
| 21374 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CLASP2 | Sponge network | 1.601 | 0 | -0.038 | 0.71 | 0.392 |
| 21375 | RP11-225B17.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 10 | ARHGEF12 | Sponge network | 1.055 | 0 | 0.09 | 0.35693 | 0.392 |
| 21376 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 0.919 | 0 | 0.259 | 0.00962 | 0.392 |
| 21377 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | TRIM33 | Sponge network | 0.225 | 0.30644 | 0.402 | 7.0E-5 | 0.392 |
| 21378 | LINC00473 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 24 | CADM1 | Sponge network | -1.203 | 0.03881 | -0.9 | 0.00084 | 0.392 |
| 21379 | ATP1B3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-590-3p | 13 | ARHGEF12 | Sponge network | 1.2 | 0.01813 | 0.09 | 0.35693 | 0.392 |
| 21380 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 47 | THRB | Sponge network | -1.349 | 0.00017 | -1.117 | 0.0004 | 0.392 |
| 21381 | LINC00672 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 32 | ABI2 | Sponge network | -0.227 | 0.31657 | 0.175 | 0.14787 | 0.392 |
| 21382 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 20 | ZFHX3 | Sponge network | 0.817 | 3.0E-5 | 0.389 | 0.01363 | 0.392 |
| 21383 | MEG3 |
hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 14 | HERC1 | Sponge network | 0.249 | 0.35902 | -0.196 | 0.08544 | 0.392 |
| 21384 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 19 | RARB | Sponge network | -0.899 | 6.0E-5 | -0.825 | 2.0E-5 | 0.392 |
| 21385 | LINC00641 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 23 | SORCS1 | Sponge network | -0.222 | 0.32445 | -3.492 | 0 | 0.392 |
| 21386 | CTD-3092A11.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ERCC4 | Sponge network | 0.873 | 0 | 0.126 | 0.15266 | 0.392 |
| 21387 | RP11-473I1.5 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p | 10 | PREX2 | Sponge network | 0.542 | 0.00054 | -0.272 | 0.25382 | 0.392 |
| 21388 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-940;hsa-miR-96-5p | 25 | CRTC3 | Sponge network | -1.575 | 6.0E-5 | 0.164 | 0.16786 | 0.392 |
| 21389 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PDGFC | Sponge network | 0.532 | 0.00505 | -0.33 | 0.06483 | 0.392 |
| 21390 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | ST6GALNAC3 | Sponge network | -0.739 | 0.0574 | -0.987 | 0 | 0.392 |
| 21391 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-93-5p | 25 | MCC | Sponge network | -0.384 | 0.40652 | -1.005 | 0 | 0.392 |
| 21392 | LINC00473 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 15 | SIK1 | Sponge network | -1.203 | 0.03881 | -0.98 | 0 | 0.392 |
| 21393 | PCA3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 36 | SNX29 | Sponge network | -0.709 | 0.18168 | -0.34 | 0.01371 | 0.392 |
| 21394 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | KIF1B | Sponge network | 0.252 | 0.08508 | -0.118 | 0.32258 | 0.392 |
| 21395 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-590-3p | 10 | PLA2R1 | Sponge network | -2.452 | 0 | -0.265 | 0.24676 | 0.392 |
| 21396 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p | 15 | CLASP2 | Sponge network | 1.147 | 0 | -0.038 | 0.71 | 0.392 |
| 21397 | MALAT1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p;hsa-miR-655-3p | 14 | ARID2 | Sponge network | 0.782 | 0.00016 | 0.235 | 0.02697 | 0.392 |
| 21398 | RP11-678G14.3 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | SFRP1 | Sponge network | -4.477 | 0 | -3.566 | 0 | 0.392 |
| 21399 | MEG3 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-5p | 10 | RTN4 | Sponge network | 0.249 | 0.35902 | -0.412 | 0.00014 | 0.392 |
| 21400 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | MED12L | Sponge network | -2.452 | 0 | -0.194 | 0.55299 | 0.392 |
| 21401 | CTC-429P9.2 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-940 | 10 | QKI | Sponge network | 0.043 | 0.81628 | -0.333 | 0.04111 | 0.392 |
| 21402 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484 | 10 | ZFP36L1 | Sponge network | -2.531 | 0 | -0.505 | 8.0E-5 | 0.392 |
| 21403 | DIO3OS |
hsa-miR-126-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 13 | CXCL12 | Sponge network | -0.773 | 0.04073 | -1.421 | 0 | 0.392 |
| 21404 | RP5-994D16.9 |
hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-26a-2-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | AFF1 | Sponge network | 0.252 | 0.08508 | -0.384 | 0.00053 | 0.392 |
| 21405 | TRIM52-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-421;hsa-miR-577;hsa-miR-7-1-3p | 26 | SSBP2 | Sponge network | -0.418 | 0.00068 | -1.58 | 0 | 0.392 |
| 21406 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 17 | RARB | Sponge network | -1.161 | 0.00591 | -0.825 | 2.0E-5 | 0.392 |
| 21407 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 0.873 | 0 | 0.114 | 0.30638 | 0.392 |
| 21408 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PDS5B | Sponge network | -0.323 | 0.01372 | 0.259 | 0.00962 | 0.392 |
| 21409 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-93-3p | 13 | RPS6KA2 | Sponge network | -0.611 | 0.09607 | -0.57 | 0.00013 | 0.392 |
| 21410 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-590-3p | 15 | TRIM33 | Sponge network | 1.04 | 0.00023 | 0.402 | 7.0E-5 | 0.392 |
| 21411 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p | 19 | AFF3 | Sponge network | -0.777 | 0.16667 | -2.373 | 0 | 0.392 |
| 21412 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429 | 15 | LATS2 | Sponge network | -0.469 | 0.08721 | 0.159 | 0.2304 | 0.392 |
| 21413 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | ARHGAP29 | Sponge network | -0.49 | 0.04946 | 0.131 | 0.42953 | 0.392 |
| 21414 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | MOB1B | Sponge network | 0.539 | 0.03722 | 0.197 | 0.15266 | 0.392 |
| 21415 | MIR143HG |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429 | 14 | ELMO1 | Sponge network | -0.739 | 0.0574 | 0.04 | 0.83147 | 0.392 |
| 21416 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | CNTNAP3 | Sponge network | -0.08 | 0.90092 | -1.465 | 0.00033 | 0.392 |
| 21417 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | IGSF10 | Sponge network | 0.997 | 0.00066 | -1.751 | 1.0E-5 | 0.392 |
| 21418 | FAM157B |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-766-3p | 20 | MDM4 | Sponge network | 0.92 | 0 | 0.345 | 0.00762 | 0.392 |
| 21419 | LINC00472 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 32 | ABI2 | Sponge network | -0.842 | 0.01361 | 0.175 | 0.14787 | 0.392 |
| 21420 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p | 31 | ZFHX4 | Sponge network | 0.082 | 0.55397 | 0.018 | 0.95574 | 0.392 |
| 21421 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 23 | AFF3 | Sponge network | 0.532 | 0.00505 | -2.373 | 0 | 0.392 |
| 21422 | RP11-57H14.4 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-493-5p | 10 | BMPR1A | Sponge network | 0.083 | 0.66405 | -0.366 | 0.01036 | 0.392 |
| 21423 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DNAJB4 | Sponge network | 0.064 | 0.72934 | -0.7 | 1.0E-5 | 0.392 |
| 21424 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | TACC1 | Sponge network | 0.559 | 0.00026 | -0.362 | 0.05406 | 0.392 |
| 21425 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 27 | PRDM2 | Sponge network | -0.727 | 0.04726 | -0.258 | 0.00721 | 0.392 |
| 21426 | NDUFA6-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 14 | MAPK10 | Sponge network | -0.355 | 0.0077 | -1.774 | 0 | 0.392 |
| 21427 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 10 | PHF6 | Sponge network | 1.055 | 0 | 0.785 | 0 | 0.392 |
| 21428 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 20 | HECA | Sponge network | -0.031 | 0.89063 | -0.217 | 0.03463 | 0.392 |
| 21429 | LINC00865 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 20 | PDE4DIP | Sponge network | -1.249 | 7.0E-5 | -0.282 | 0.0257 | 0.392 |
| 21430 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-93-5p;hsa-miR-940 | 21 | CADM2 | Sponge network | -1.255 | 0.00981 | -3.782 | 0 | 0.392 |
| 21431 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429 | 14 | SLITRK3 | Sponge network | -0.246 | 0.35034 | -3.328 | 0 | 0.392 |
| 21432 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 29 | EPHA3 | Sponge network | -2.915 | 0.00015 | 0.185 | 0.53588 | 0.392 |
| 21433 | RP11-66B24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p | 14 | PRKAA2 | Sponge network | 0.57 | 0.1866 | -2.462 | 0 | 0.392 |
| 21434 | RP11-2H8.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | 1.089 | 0 | 0.323 | 0.00792 | 0.392 |
| 21435 | CTC-296K1.3 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-940 | 14 | TOM1L2 | Sponge network | -2.095 | 0.00112 | -0.994 | 0 | 0.392 |
| 21436 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p | 20 | RICTOR | Sponge network | 0.983 | 0.00048 | 0.388 | 0.00188 | 0.392 |
| 21437 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 26 | ABI2 | Sponge network | -1.092 | 4.0E-5 | 0.175 | 0.14787 | 0.392 |
| 21438 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-93-5p | 27 | LRP1B | Sponge network | -0.096 | 0.67958 | -2.163 | 0 | 0.392 |
| 21439 | HAS2-AS1 |
hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p;hsa-miR-942-5p | 10 | GPC6 | Sponge network | -0.011 | 0.967 | -0.054 | 0.81465 | 0.392 |
| 21440 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-744-5p | 15 | USP12 | Sponge network | -0.376 | 0.00337 | -0.102 | 0.40768 | 0.392 |
| 21441 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 54 | THRB | Sponge network | 0.219 | 0.1516 | -1.117 | 0.0004 | 0.392 |
| 21442 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-96-5p | 14 | TACC1 | Sponge network | 0.37 | 0.27614 | -0.362 | 0.05406 | 0.392 |
| 21443 | RP11-77H9.2 |
hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-424-5p | 14 | ABL2 | Sponge network | 1.677 | 0 | 0.82 | 0 | 0.392 |
| 21444 | LINC00969 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-495-3p | 12 | STAG2 | Sponge network | 0.683 | 1.0E-5 | 0.252 | 0.00438 | 0.392 |
| 21445 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 23 | THBS1 | Sponge network | 0.727 | 3.0E-5 | 0.741 | 0.00386 | 0.392 |
| 21446 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-577;hsa-miR-590-3p | 14 | FGF5 | Sponge network | -0.096 | 0.67958 | 0.549 | 0.28659 | 0.392 |
| 21447 | FAM215B |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-582-5p;hsa-miR-940 | 12 | BAZ2B | Sponge network | 0.817 | 3.0E-5 | -0.276 | 0.02833 | 0.392 |
| 21448 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 24 | AFF4 | Sponge network | 0.826 | 1.0E-5 | -0.266 | 0.01565 | 0.392 |
| 21449 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | USP12 | Sponge network | 1.757 | 0 | -0.102 | 0.40768 | 0.392 |
| 21450 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 66 | BMPR2 | Sponge network | -2.452 | 0 | 0.206 | 0.0635 | 0.392 |
| 21451 | AGAP11 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-874-3p | 15 | SORCS1 | Sponge network | -1.645 | 0 | -3.492 | 0 | 0.392 |
| 21452 | FZD10-AS1 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PEG3 | Sponge network | -0.727 | 0.04726 | -1.74 | 0 | 0.392 |
| 21453 | RP11-13K12.5 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 19 | ST6GAL2 | Sponge network | -0.318 | 0.65847 | -1.201 | 0.00597 | 0.392 |
| 21454 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 45 | ADARB1 | Sponge network | 0.72 | 0.0001 | -0.335 | 0.06631 | 0.392 |
| 21455 | BVES-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p | 13 | RIMBP2 | Sponge network | -2.62 | 0 | -1.968 | 5.0E-5 | 0.392 |
| 21456 | DIO3OS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p | 17 | EZH1 | Sponge network | -0.773 | 0.04073 | -0.564 | 1.0E-5 | 0.392 |
| 21457 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-3p | 35 | PRKAB2 | Sponge network | -2.452 | 0 | -0.367 | 0.01371 | 0.392 |
| 21458 | RP11-620J15.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-744-3p | 65 | LPP | Sponge network | -0.739 | 0.00044 | -0.459 | 0.04475 | 0.392 |
| 21459 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 16 | PDS5B | Sponge network | 0.541 | 0.00125 | 0.259 | 0.00962 | 0.392 |
| 21460 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SYT4 | Sponge network | -0.421 | 0.0114 | -3.207 | 0 | 0.392 |
| 21461 | HCG22 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 14 | SASH1 | Sponge network | 0.816 | 0.32368 | -0.502 | 0.00175 | 0.392 |
| 21462 | MRVI1-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 16 | JDP2 | Sponge network | -1.092 | 4.0E-5 | -0.967 | 0 | 0.392 |
| 21463 | CTD-3092A11.2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 25 | ABI2 | Sponge network | 0.873 | 0 | 0.175 | 0.14787 | 0.392 |
| 21464 | RP11-798M19.6 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | TLE4 | Sponge network | -0.612 | 0.00094 | -1.157 | 0 | 0.392 |
| 21465 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-940 | 15 | ETV1 | Sponge network | -1.331 | 3.0E-5 | -0.096 | 0.67674 | 0.392 |
| 21466 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-455-3p;hsa-miR-576-5p | 10 | TSHZ3 | Sponge network | -1.116 | 0.23686 | -0.556 | 0.01516 | 0.392 |
| 21467 | CTC-429P9.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-511-5p;hsa-miR-532-3p | 12 | MRVI1 | Sponge network | 0.497 | 0.01451 | -1.051 | 0.00022 | 0.392 |
| 21468 | RP11-254F7.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p | 11 | ROBO1 | Sponge network | 0.176 | 0.37402 | -0.009 | 0.96154 | 0.392 |
| 21469 | AGAP11 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-590-5p | 14 | NOTCH2NL | Sponge network | -1.645 | 0 | 0.024 | 0.84307 | 0.392 |
| 21470 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p | 11 | LHX6 | Sponge network | -4.546 | 0 | -0.087 | 0.69193 | 0.392 |
| 21471 | RP11-418J17.1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-495-3p | 12 | PHF6 | Sponge network | 0.998 | 0 | 0.785 | 0 | 0.392 |
| 21472 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p | 20 | ZMYND11 | Sponge network | -2.69 | 0.0001 | -0.2 | 0.05449 | 0.392 |
| 21473 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ZFHX3 | Sponge network | 0.476 | 0.19203 | 0.389 | 0.01363 | 0.392 |
| 21474 | RP11-373D23.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-7-1-3p | 19 | TMEM132B | Sponge network | -0.285 | 0.2172 | -0.83 | 0.01084 | 0.392 |
| 21475 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 17 | EBF3 | Sponge network | -0.021 | 0.93598 | -0.058 | 0.81437 | 0.392 |
| 21476 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | BRD3 | Sponge network | 1.601 | 0 | 0.37 | 0.00013 | 0.392 |
| 21477 | PCA3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p | 26 | PRDM2 | Sponge network | -0.709 | 0.18168 | -0.258 | 0.00721 | 0.392 |
| 21478 | CALML3-AS1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-664a-3p | 11 | MN1 | Sponge network | 0.95 | 0.15943 | -0.358 | 0.24172 | 0.392 |
| 21479 | CTD-2245F17.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-3614-5p;hsa-miR-7-5p | 10 | EPHA3 | Sponge network | -0.421 | 0.12556 | 0.185 | 0.53588 | 0.392 |
| 21480 | HOXB-AS3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-2-3p;hsa-miR-34a-5p;hsa-miR-744-3p;hsa-miR-877-5p | 21 | FAT3 | Sponge network | 0.343 | 0.28887 | -1.601 | 0.00051 | 0.392 |
| 21481 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 46 | ZFHX3 | Sponge network | -2.46 | 0 | 0.389 | 0.01363 | 0.392 |
| 21482 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-625-3p;hsa-miR-654-3p | 37 | BMPR2 | Sponge network | 1.03 | 0 | 0.206 | 0.0635 | 0.392 |
| 21483 | AC141928.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577 | 12 | FGFR1 | Sponge network | 0.853 | 0.15352 | -0.509 | 0.03957 | 0.392 |
| 21484 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 21 | HECA | Sponge network | -0.176 | 0.59692 | -0.217 | 0.03463 | 0.392 |
| 21485 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 20 | BCL11A | Sponge network | -0.323 | 0.01372 | -0.932 | 0.0063 | 0.392 |
| 21486 | BZRAP1-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-93-5p | 11 | EGR2 | Sponge network | -0.465 | 0.04703 | 0.309 | 0.17079 | 0.392 |
| 21487 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-576-5p | 24 | FOXO3 | Sponge network | 0.95 | 0.15943 | -0.04 | 0.72028 | 0.392 |
| 21488 | LINC00672 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 15 | STARD13 | Sponge network | -0.227 | 0.31657 | -0.187 | 0.27383 | 0.392 |
| 21489 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | PDGFRA | Sponge network | -0.611 | 0.09607 | -0.372 | 0.0037 | 0.392 |
| 21490 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-940 | 24 | CHD5 | Sponge network | -0.154 | 0.53954 | -0.922 | 0.01521 | 0.392 |
| 21491 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 42 | QKI | Sponge network | 0.395 | 0.15451 | -0.333 | 0.04111 | 0.392 |
| 21492 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 36 | PIK3R1 | Sponge network | 1.302 | 7.0E-5 | -0.133 | 0.36854 | 0.392 |
| 21493 | LINC00578 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p | 20 | GRIK3 | Sponge network | 0.811 | 0.03228 | -3.208 | 0 | 0.392 |
| 21494 | AC093110.3 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 17 | TBL1XR1 | Sponge network | 1.044 | 2.0E-5 | 0.578 | 0 | 0.392 |
| 21495 | RP11-531A24.5 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-3p | 19 | ABL1 | Sponge network | 0.75 | 0.00054 | -0.215 | 0.07804 | 0.392 |
| 21496 | HOXB-AS3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-452-5p;hsa-miR-654-3p | 15 | FBXO32 | Sponge network | 0.343 | 0.28887 | -0.495 | 0.07561 | 0.392 |
| 21497 | RP11-35G9.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-96-5p | 15 | CYLD | Sponge network | 0.622 | 0.00015 | -0.367 | 0.00171 | 0.392 |
| 21498 | LINC00680 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 12 | GPAM | Sponge network | 0.884 | 0 | 0.142 | 0.29732 | 0.392 |
| 21499 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-452-5p | 14 | TIMP3 | Sponge network | -2.095 | 0.00112 | -0.546 | 0.02307 | 0.392 |
| 21500 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | DOCK4 | Sponge network | 0.064 | 0.72934 | 0.502 | 0.00108 | 0.392 |
| 21501 | RP11-25K21.6 |
hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-589-3p | 19 | IL6ST | Sponge network | 0.489 | 0.25786 | -0.402 | 0.00676 | 0.392 |
| 21502 | PWAR6 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | PDE4DIP | Sponge network | -1.575 | 6.0E-5 | -0.282 | 0.0257 | 0.392 |
| 21503 | HCG11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 30 | NOTCH2 | Sponge network | 0.385 | 0.10067 | 0.138 | 0.35163 | 0.392 |
| 21504 | LINC00476 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-877-5p | 30 | ASTN1 | Sponge network | -0.615 | 0.00015 | -2.389 | 2.0E-5 | 0.392 |
| 21505 | RP11-384O8.1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429 | 11 | NGFR | Sponge network | -0.738 | 0.21233 | -1.271 | 0.01058 | 0.392 |
| 21506 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-877-5p | 20 | NRK | Sponge network | 0.082 | 0.55654 | 0.656 | 0.19217 | 0.392 |
| 21507 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -1.697 | 0.00889 | 0.309 | 0.17079 | 0.392 |
| 21508 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-455-3p | 10 | NTRK2 | Sponge network | -0.309 | 0.1145 | -0.736 | 0.06495 | 0.392 |
| 21509 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-93-5p | 30 | TACC1 | Sponge network | 0.082 | 0.55397 | -0.362 | 0.05406 | 0.391 |
| 21510 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-576-5p | 13 | PHC3 | Sponge network | 2.163 | 0 | 0.212 | 0.0499 | 0.391 |
| 21511 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 13 | GAS7 | Sponge network | -1.717 | 0.01522 | -0.555 | 0.01602 | 0.391 |
| 21512 | U91328.19 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935 | 34 | ABI2 | Sponge network | -0.303 | 0.21002 | 0.175 | 0.14787 | 0.391 |
| 21513 | LINC00957 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-942-5p | 12 | ZBTB20 | Sponge network | -0.421 | 0.0114 | -0.122 | 0.57569 | 0.391 |
| 21514 | RP11-531A24.5 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p | 10 | WNK2 | Sponge network | 0.75 | 0.00054 | -0.352 | 0.31066 | 0.391 |
| 21515 | RP11-373D23.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p | 16 | MYO9A | Sponge network | -0.285 | 0.2172 | -0.147 | 0.32373 | 0.391 |
| 21516 | RP4-669L17.10 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-5p | 18 | BRD3 | Sponge network | 1.093 | 0 | 0.37 | 0.00013 | 0.391 |
| 21517 | FZD10-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 17 | MRVI1 | Sponge network | -0.727 | 0.04726 | -1.051 | 0.00022 | 0.391 |
| 21518 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 42 | CBX5 | Sponge network | 0.385 | 0.10067 | 0.165 | 0.24446 | 0.391 |
| 21519 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-370-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | JAZF1 | Sponge network | -0.709 | 0.18168 | -0.377 | 0.02677 | 0.391 |
| 21520 | CALML3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-542-3p;hsa-miR-664a-3p;hsa-miR-92a-1-5p | 20 | AKAP6 | Sponge network | 0.95 | 0.15943 | -1.586 | 3.0E-5 | 0.391 |
| 21521 | RP11-589P10.7 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p | 10 | LPP | Sponge network | -2.044 | 0.00066 | -0.459 | 0.04475 | 0.391 |
| 21522 | OIP5-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | FAM172A | Sponge network | 0.421 | 0.00084 | -0.57 | 0 | 0.391 |
| 21523 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SMARCA2 | Sponge network | -1.057 | 0.00029 | -0.529 | 0.00014 | 0.391 |
| 21524 | CTD-2554C21.2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-421;hsa-miR-429;hsa-miR-877-5p | 24 | AFF1 | Sponge network | -1.654 | 0.00144 | -0.384 | 0.00053 | 0.391 |
| 21525 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-96-5p | 16 | PCDH8 | Sponge network | -1.111 | 0.0003 | -0.997 | 0.05366 | 0.391 |
| 21526 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZNF844 | Sponge network | -0.739 | 0.0574 | -0.966 | 0.00035 | 0.391 |
| 21527 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 65 | IL6ST | Sponge network | 0.246 | 0.59912 | -0.402 | 0.00676 | 0.391 |
| 21528 | BZRAP1-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-940 | 14 | SFRP1 | Sponge network | -0.465 | 0.04703 | -3.566 | 0 | 0.391 |
| 21529 | WDFY3-AS2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 24 | NRK | Sponge network | -0.942 | 0 | 0.656 | 0.19217 | 0.391 |
| 21530 | PART1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 40 | IL17RD | Sponge network | -2.925 | 0 | -0.019 | 0.94873 | 0.391 |
| 21531 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 52 | SOX5 | Sponge network | -2.925 | 0 | -1.091 | 4.0E-5 | 0.391 |
| 21532 | LINC00654 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 11 | NGFR | Sponge network | 0.374 | 0.20054 | -1.271 | 0.01058 | 0.391 |
| 21533 | RP11-158K1.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 65 | LPP | Sponge network | 0.349 | 0.01695 | -0.459 | 0.04475 | 0.391 |
| 21534 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3C | Sponge network | 0.619 | 0.00157 | -0.272 | 0.31153 | 0.391 |
| 21535 | RAMP2-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SEMA3E | Sponge network | -0.513 | 0.04703 | -1.717 | 0.00137 | 0.391 |
| 21536 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-671-5p | 18 | PTPRD | Sponge network | 1.344 | 0 | -1.005 | 0.00064 | 0.391 |
| 21537 | AC016747.3 |
hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-584-5p;hsa-miR-590-3p | 10 | PCDH18 | Sponge network | 0.199 | 0.38015 | 0.216 | 0.24645 | 0.391 |
| 21538 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | ST6GAL2 | Sponge network | 0.299 | 0.57722 | -1.201 | 0.00597 | 0.391 |
| 21539 | AC141928.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484 | 11 | RET | Sponge network | 0.853 | 0.15352 | -0.833 | 0.03517 | 0.391 |
| 21540 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-940 | 41 | PTPRT | Sponge network | -2.46 | 0 | -0.907 | 0.03727 | 0.391 |
| 21541 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 52 | FAT3 | Sponge network | 0.374 | 0.20054 | -1.601 | 0.00051 | 0.391 |
| 21542 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p | 22 | PRDM2 | Sponge network | 0.537 | 0.00139 | -0.258 | 0.00721 | 0.391 |
| 21543 | DNAJC27-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | IL17RD | Sponge network | -0.969 | 2.0E-5 | -0.019 | 0.94873 | 0.391 |
| 21544 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | MAF | Sponge network | -0.769 | 0.00035 | -1.257 | 0 | 0.391 |
| 21545 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-337-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PBRM1 | Sponge network | 1.601 | 0 | -0.029 | 0.78601 | 0.391 |
| 21546 | FGD5-AS1 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-432-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 37 | PPM1L | Sponge network | 0.401 | 0.00066 | -0.537 | 0.00616 | 0.391 |
| 21547 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 22 | LATS2 | Sponge network | -1.281 | 0.00012 | 0.159 | 0.2304 | 0.391 |
| 21548 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p | 38 | UNC80 | Sponge network | -2.69 | 0.0001 | -1.934 | 0 | 0.391 |
| 21549 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | 0.299 | 0.57722 | -0.354 | 0.14694 | 0.391 |
| 21550 | MIR497HG |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 26 | OPCML | Sponge network | -1.439 | 4.0E-5 | -2.498 | 0 | 0.391 |
| 21551 | LINC01018 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | GAS7 | Sponge network | -2.915 | 0.00015 | -0.555 | 0.01602 | 0.391 |
| 21552 | CTC-444N24.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ZFX | Sponge network | 1.153 | 0 | 0.09 | 0.42563 | 0.391 |
| 21553 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 33 | MEF2C | Sponge network | -0.513 | 0.04703 | -0.499 | 0.01448 | 0.391 |
| 21554 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | EPAS1 | Sponge network | 0.997 | 0.00066 | -0.184 | 0.14418 | 0.391 |
| 21555 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 31 | APC | Sponge network | -1.111 | 0.0003 | -0.255 | 0.04051 | 0.391 |
| 21556 | LINC00476 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-93-5p | 26 | LRIG1 | Sponge network | -0.615 | 0.00015 | -0.537 | 0.00588 | 0.391 |
| 21557 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ERG | Sponge network | -1.057 | 0.00029 | 0.002 | 0.99011 | 0.391 |
| 21558 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | KAT6B | Sponge network | 0.194 | 0.64278 | -0.478 | 0.00018 | 0.391 |
| 21559 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-628-5p;hsa-miR-93-3p | 13 | ERC1 | Sponge network | -0.376 | 0.00337 | -0.108 | 0.38794 | 0.391 |
| 21560 | CTD-3092A11.2 |
hsa-let-7d-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 14 | DICER1 | Sponge network | 0.873 | 0 | 0.042 | 0.674 | 0.391 |
| 21561 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-874-3p;hsa-miR-98-5p | 11 | ST5 | Sponge network | -0.899 | 6.0E-5 | -0.78 | 0 | 0.391 |
| 21562 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DOCK4 | Sponge network | 0.673 | 0 | 0.502 | 0.00108 | 0.391 |
| 21563 | RP5-1139B12.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-671-5p;hsa-miR-93-3p | 22 | BMPR2 | Sponge network | 0.598 | 0.38481 | 0.206 | 0.0635 | 0.391 |
| 21564 | CTD-3138B18.6 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p | 12 | CREB1 | Sponge network | 2.248 | 0 | 0.203 | 0.00537 | 0.391 |
| 21565 | RASSF8-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p | 13 | RHOB | Sponge network | 0.335 | 0.20596 | -1.052 | 0 | 0.391 |
| 21566 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 34 | ERC1 | Sponge network | -0.323 | 0.01372 | -0.108 | 0.38794 | 0.391 |
| 21567 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-190a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-577;hsa-miR-940 | 14 | TRPS1 | Sponge network | 0.357 | 0.72407 | -0.191 | 0.44109 | 0.391 |
| 21568 | LINC00472 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-744-3p | 24 | PTPN14 | Sponge network | -0.842 | 0.01361 | 0.144 | 0.34595 | 0.391 |
| 21569 | RP11-473I1.9 |
hsa-let-7g-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p | 11 | KDM3A | Sponge network | 0.683 | 1.0E-5 | 0.132 | 0.22711 | 0.391 |
| 21570 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 49 | GAS7 | Sponge network | 0.082 | 0.55654 | -0.555 | 0.01602 | 0.391 |
| 21571 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | EYA4 | Sponge network | -0.021 | 0.93598 | 0.3 | 0.36876 | 0.391 |
| 21572 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | CHD2 | Sponge network | 0.919 | 0 | 0.049 | 0.55371 | 0.391 |
| 21573 | RP11-6O2.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-671-5p | 25 | DLC1 | Sponge network | 0.331 | 0.57247 | -0.382 | 0.05038 | 0.391 |
| 21574 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p | 17 | ZFHX3 | Sponge network | 0.946 | 0 | 0.389 | 0.01363 | 0.391 |
| 21575 | TUG1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 12 | ZNF770 | Sponge network | 0.972 | 0 | 0.206 | 0.06751 | 0.391 |
| 21576 | AC006116.24 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-93-5p;hsa-miR-98-5p | 16 | PRRX1 | Sponge network | -0.473 | 0.07602 | 1.316 | 0 | 0.391 |
| 21577 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 17 | PNRC1 | Sponge network | -1.575 | 6.0E-5 | -0.646 | 0 | 0.391 |
| 21578 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TET1 | Sponge network | -0.206 | 0.12942 | -0.135 | 0.52138 | 0.391 |
| 21579 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | -0.303 | 0.21002 | -0.509 | 0.03957 | 0.391 |
| 21580 | AC003090.1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 13 | MACF1 | Sponge network | -2.117 | 0.00161 | 0.019 | 0.90335 | 0.391 |
| 21581 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-874-3p | 13 | GIGYF2 | Sponge network | 0.683 | 1.0E-5 | 0.136 | 0.04403 | 0.391 |
| 21582 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | HACE1 | Sponge network | 0.683 | 1.0E-5 | -0.072 | 0.55239 | 0.391 |
| 21583 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ADAMTSL3 | Sponge network | -0.342 | 0.02288 | -1.559 | 0 | 0.391 |
| 21584 | LPP-AS2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-493-5p;hsa-miR-532-3p | 14 | SYNM | Sponge network | -0.18 | 0.20343 | -2.309 | 1.0E-5 | 0.391 |
| 21585 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | TIMP3 | Sponge network | -1.886 | 0 | -0.546 | 0.02307 | 0.391 |
| 21586 | RP11-121C2.2 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 15 | RASAL2 | Sponge network | 1.147 | 0 | 0.869 | 0 | 0.391 |
| 21587 | LINC01018 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | LSAMP | Sponge network | -2.915 | 0.00015 | -0.066 | 0.81708 | 0.391 |
| 21588 | LINC00641 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-589-5p;hsa-miR-590-3p | 18 | ROBO1 | Sponge network | -0.222 | 0.32445 | -0.009 | 0.96154 | 0.391 |
| 21589 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p | 10 | PDS5B | Sponge network | 1.134 | 0 | 0.259 | 0.00962 | 0.391 |
| 21590 | RP11-760H22.2 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p | 17 | PIK3R1 | Sponge network | 0.001 | 0.99753 | -0.133 | 0.36854 | 0.391 |
| 21591 | RP5-1198O20.4 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 10 | PXDN | Sponge network | 0.997 | 0.00066 | 1.038 | 0 | 0.391 |
| 21592 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-429;hsa-miR-590-3p | 10 | RB1CC1 | Sponge network | 0.422 | 0.27021 | 0.175 | 0.12559 | 0.391 |
| 21593 | PGM5-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 10 | ABCA10 | Sponge network | -4.546 | 0 | -0.571 | 0.03674 | 0.391 |
| 21594 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 11 | FYN | Sponge network | -0.777 | 0.16667 | -0.131 | 0.37051 | 0.391 |
| 21595 | SERTAD4-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-335-3p | 11 | EFNA5 | Sponge network | -0.932 | 0.00329 | -0.421 | 0.17868 | 0.391 |
| 21596 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 14 | DAB2 | Sponge network | -0.563 | 0.02545 | 0.431 | 0.0113 | 0.391 |
| 21597 | RP11-400K9.4 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | -0.611 | 0.09607 | 0.309 | 0.17079 | 0.391 |
| 21598 | RP11-678G14.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-339-5p;hsa-miR-505-3p | 10 | FNBP1 | Sponge network | -4.477 | 0 | -0.969 | 4.0E-5 | 0.391 |
| 21599 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-424-5p | 10 | RARB | Sponge network | -4.463 | 0.00025 | -0.825 | 2.0E-5 | 0.391 |
| 21600 | A2M-AS1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 10 | FHL1 | Sponge network | -0.08 | 0.90092 | -2.687 | 0 | 0.391 |
| 21601 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-337-3p | 11 | ZFX | Sponge network | 1.316 | 0 | 0.09 | 0.42563 | 0.391 |
| 21602 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 23 | RICTOR | Sponge network | 2.163 | 0 | 0.388 | 0.00188 | 0.391 |
| 21603 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DACT1 | Sponge network | -1.216 | 0 | 0.783 | 0.00072 | 0.391 |
| 21604 | AC009948.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-96-5p | 24 | CYLD | Sponge network | 0.532 | 0.00505 | -0.367 | 0.00171 | 0.391 |
| 21605 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 60 | THRB | Sponge network | -0.769 | 0.00035 | -1.117 | 0.0004 | 0.391 |
| 21606 | FGD5-AS1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | USP9X | Sponge network | 0.401 | 0.00066 | 0.222 | 0.01689 | 0.391 |
| 21607 | RP11-98I9.4 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PHF3 | Sponge network | 0.67 | 0.00048 | 0.126 | 0.19652 | 0.391 |
| 21608 | RP11-571M6.17 |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-590-3p | 11 | AKAP11 | Sponge network | 1.013 | 0 | 0.196 | 0.09825 | 0.391 |
| 21609 | PART1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 21 | STARD13 | Sponge network | -2.925 | 0 | -0.187 | 0.27383 | 0.391 |
| 21610 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p | 10 | DCLK1 | Sponge network | -0.465 | 0.04703 | -1.324 | 0.00014 | 0.391 |
| 21611 | CTD-2002H8.2 |
hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | LPP | Sponge network | 0.931 | 1.0E-5 | -0.459 | 0.04475 | 0.391 |
| 21612 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | DAB2 | Sponge network | -2.452 | 0 | 0.431 | 0.0113 | 0.391 |
| 21613 | RP11-400K9.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-590-5p;hsa-miR-744-3p | 17 | DST | Sponge network | -0.733 | 0.19469 | -0.713 | 0.00096 | 0.391 |
| 21614 | RP4-669P10.16 |
hsa-miR-1271-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-24-2-5p;hsa-miR-324-5p;hsa-miR-409-3p;hsa-miR-424-5p | 16 | PPM1L | Sponge network | -0.301 | 0.06597 | -0.537 | 0.00616 | 0.391 |
| 21615 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 30 | RICTOR | Sponge network | 1.757 | 0 | 0.388 | 0.00188 | 0.391 |
| 21616 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 49 | EPHA5 | Sponge network | 0.064 | 0.72934 | -0.957 | 0.01733 | 0.391 |
| 21617 | RP11-890B15.3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 19 | SFRP1 | Sponge network | 0.101 | 0.60919 | -3.566 | 0 | 0.391 |
| 21618 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SLC6A15 | Sponge network | -1.216 | 0 | -2.373 | 2.0E-5 | 0.391 |
| 21619 | GNG12-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-629-3p | 14 | ITGA5 | Sponge network | -0.793 | 7.0E-5 | 0.452 | 0.03524 | 0.391 |
| 21620 | LINC00702 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 30 | EYA4 | Sponge network | -0.737 | 0.06182 | 0.3 | 0.36876 | 0.391 |
| 21621 | BOLA3-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p | 13 | ROBO1 | Sponge network | -0.246 | 0.35034 | -0.009 | 0.96154 | 0.391 |
| 21622 | RP5-994D16.9 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 11 | ABL2 | Sponge network | 0.252 | 0.08508 | 0.82 | 0 | 0.391 |
| 21623 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-34a-5p | 12 | ERC1 | Sponge network | -0.309 | 0.1145 | -0.108 | 0.38794 | 0.391 |
| 21624 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | AKAP11 | Sponge network | 0.545 | 0.00064 | 0.196 | 0.09825 | 0.391 |
| 21625 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-874-3p | 12 | CRTC3 | Sponge network | -2.531 | 0 | 0.164 | 0.16786 | 0.391 |
| 21626 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | PSIP1 | Sponge network | 0.225 | 0.30644 | -0.005 | 0.96897 | 0.391 |
| 21627 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 14 | TET1 | Sponge network | 1.04 | 0.00023 | -0.135 | 0.52138 | 0.391 |
| 21628 | RP11-244H3.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-362-5p | 10 | ZBTB20 | Sponge network | 0.659 | 3.0E-5 | -0.122 | 0.57569 | 0.391 |
| 21629 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421 | 26 | PTPRD | Sponge network | -0.583 | 0.14589 | -1.005 | 0.00064 | 0.391 |
| 21630 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p | 10 | NALCN | Sponge network | -0.405 | 0.22013 | 1.068 | 0.00237 | 0.391 |
| 21631 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-671-5p | 19 | KCNA6 | Sponge network | -0.309 | 0.1145 | -1.531 | 0.00013 | 0.391 |
| 21632 | CTD-2089N3.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-590-3p | 15 | DIP2C | Sponge network | -1.375 | 0.08642 | -0.436 | 0.01713 | 0.391 |
| 21633 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-3p | 33 | IL6ST | Sponge network | -2.531 | 0 | -0.402 | 0.00676 | 0.391 |
| 21634 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-500a-5p | 11 | ZNF208 | Sponge network | 0.082 | 0.55397 | -0.4 | 0.22381 | 0.391 |
| 21635 | WDFY3-AS2 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PCLO | Sponge network | -0.942 | 0 | -1.843 | 0.00064 | 0.391 |
| 21636 | RP11-248G5.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-625-5p | 18 | ABI2 | Sponge network | 1.294 | 0 | 0.175 | 0.14787 | 0.391 |
| 21637 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 24 | SYNPO2 | Sponge network | 0.083 | 0.66405 | -2.342 | 0 | 0.391 |
| 21638 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-940 | 13 | TAL1 | Sponge network | -0.465 | 0.04703 | -0.462 | 0.00574 | 0.391 |
| 21639 | CTD-2245F17.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-505-3p;hsa-miR-7-5p | 13 | RBMS3 | Sponge network | -0.421 | 0.12556 | -0.519 | 0.03551 | 0.391 |
| 21640 | ARHGEF26-AS1 |
hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 23 | PAIP2 | Sponge network | -2.46 | 0 | -0.515 | 0 | 0.391 |
| 21641 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 22 | ERC1 | Sponge network | -0.896 | 0.00099 | -0.108 | 0.38794 | 0.391 |
| 21642 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | ST6GALNAC3 | Sponge network | -2.46 | 0 | -0.987 | 0 | 0.391 |
| 21643 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-362-5p | 18 | CBFA2T3 | Sponge network | -0.611 | 0.09607 | -1.221 | 1.0E-5 | 0.391 |
| 21644 | LA16c-321D4.2 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-424-5p | 12 | ABL2 | Sponge network | 2.459 | 5.0E-5 | 0.82 | 0 | 0.391 |
| 21645 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 19 | RARB | Sponge network | -0.737 | 0.10822 | -0.825 | 2.0E-5 | 0.391 |
| 21646 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-590-3p | 20 | TACC1 | Sponge network | 0.765 | 7.0E-5 | -0.362 | 0.05406 | 0.391 |
| 21647 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 27 | DDX6 | Sponge network | 0.225 | 0.30644 | 0.091 | 0.36428 | 0.391 |
| 21648 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-7-1-3p | 14 | NRXN3 | Sponge network | 0.853 | 0.15352 | -0.893 | 0.04186 | 0.391 |
| 21649 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-590-3p;hsa-miR-93-5p | 26 | PRKAA2 | Sponge network | 0.381 | 0.25967 | -2.462 | 0 | 0.391 |
| 21650 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | EDA2R | Sponge network | -0.323 | 0.01372 | -0.587 | 0.01598 | 0.391 |
| 21651 | SNHG14 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | RASSF3 | Sponge network | -1.111 | 0.0003 | -0.226 | 0.11691 | 0.391 |
| 21652 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | PDS5B | Sponge network | 0.782 | 0.00016 | 0.259 | 0.00962 | 0.391 |
| 21653 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-5p | 13 | PTPRB | Sponge network | -2.69 | 0.0001 | 0.105 | 0.47122 | 0.391 |
| 21654 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429 | 20 | PRDM2 | Sponge network | -1.654 | 0.00144 | -0.258 | 0.00721 | 0.391 |
| 21655 | OIP5-AS1 |
hsa-let-7g-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p | 11 | KDM3A | Sponge network | 0.421 | 0.00084 | 0.132 | 0.22711 | 0.391 |
| 21656 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-654-3p | 17 | NR2C2 | Sponge network | -0.176 | 0.59692 | 0.21 | 0.03224 | 0.391 |
| 21657 | RP11-35G9.5 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-96-5p | 16 | PRRX1 | Sponge network | 0.622 | 0.00015 | 1.316 | 0 | 0.391 |
| 21658 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | TLL1 | Sponge network | 0.101 | 0.60919 | -1.195 | 3.0E-5 | 0.391 |
| 21659 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -1.249 | 7.0E-5 | -0.7 | 1.0E-5 | 0.391 |
| 21660 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 30 | ZFHX4 | Sponge network | 0.389 | 0.04293 | 0.018 | 0.95574 | 0.391 |
| 21661 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 28 | BHLHE41 | Sponge network | 1.302 | 7.0E-5 | 0.353 | 0.12753 | 0.391 |
| 21662 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-96-5p | 25 | CRTC3 | Sponge network | 0.083 | 0.66405 | 0.164 | 0.16786 | 0.391 |
| 21663 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p | 15 | PTPRB | Sponge network | -0.777 | 0.16667 | 0.105 | 0.47122 | 0.391 |
| 21664 | RP11-159D12.2 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p | 14 | TRIM13 | Sponge network | 1.254 | 0 | 0.183 | 0.06968 | 0.391 |
| 21665 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | ATRX | Sponge network | 1.063 | 0 | 0.261 | 0.04408 | 0.391 |
| 21666 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-940 | 25 | PBX1 | Sponge network | 0.174 | 0.30034 | -1.206 | 0 | 0.391 |
| 21667 | LINC00957 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TBX18 | Sponge network | -0.421 | 0.0114 | 0.843 | 0.02945 | 0.391 |
| 21668 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 20 | KANK1 | Sponge network | 0.064 | 0.72934 | -0.55 | 0.00134 | 0.391 |
| 21669 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-7-1-3p | 12 | SLC6A15 | Sponge network | -4.546 | 0 | -2.373 | 2.0E-5 | 0.391 |
| 21670 | RP11-244H3.1 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 12 | INPP4A | Sponge network | 0.659 | 3.0E-5 | 0.07 | 0.53563 | 0.391 |
| 21671 | MIR22HG |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-542-3p | 21 | SYNPO2 | Sponge network | -1.047 | 0 | -2.342 | 0 | 0.391 |
| 21672 | LINC00854 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-942-5p | 19 | DDX6 | Sponge network | 1.18 | 0 | 0.091 | 0.36428 | 0.391 |
| 21673 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 24 | NCAM2 | Sponge network | -0.487 | 0.02157 | -0.482 | 0.22565 | 0.391 |
| 21674 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | RBMS3 | Sponge network | 0.419 | 0.0319 | -0.519 | 0.03551 | 0.391 |
| 21675 | FENDRR |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p | 13 | FLI1 | Sponge network | -1.281 | 0.00012 | -0.294 | 0.10905 | 0.391 |
| 21676 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | 0.569 | 0.00067 | 0.323 | 0.00792 | 0.391 |
| 21677 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p | 10 | PBRM1 | Sponge network | 0.594 | 0.41522 | -0.029 | 0.78601 | 0.391 |
| 21678 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-301a-3p | 13 | RASSF2 | Sponge network | -0.733 | 0.19469 | -0.273 | 0.21961 | 0.391 |
| 21679 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 37 | CADM2 | Sponge network | 0.176 | 0.37402 | -3.782 | 0 | 0.391 |
| 21680 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | CRTC1 | Sponge network | -0.769 | 0.00035 | -0.116 | 0.28412 | 0.39 |
| 21681 | RP11-452L6.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p | 10 | MYO9A | Sponge network | 0.665 | 0.00061 | -0.147 | 0.32373 | 0.39 |
| 21682 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-542-3p | 11 | ALDH1A2 | Sponge network | -0.769 | 0.00035 | -2.411 | 0 | 0.39 |
| 21683 | CTD-2089N3.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p | 11 | DMD | Sponge network | -0.314 | 0.62033 | -1.506 | 0 | 0.39 |
| 21684 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-339-5p;hsa-miR-421;hsa-miR-93-3p | 13 | CTIF | Sponge network | -4.463 | 0.00025 | -0.389 | 0.00549 | 0.39 |
| 21685 | RP11-356J5.12 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | MACF1 | Sponge network | -0.899 | 6.0E-5 | 0.019 | 0.90335 | 0.39 |
| 21686 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | GALNT13 | Sponge network | -0.323 | 0.01372 | -0.857 | 0.07439 | 0.39 |
| 21687 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | MOB1B | Sponge network | 0.862 | 0.06213 | 0.197 | 0.15266 | 0.39 |
| 21688 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 16 | GPAM | Sponge network | 0.401 | 0.00066 | 0.142 | 0.29732 | 0.39 |
| 21689 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 16 | PTPRD | Sponge network | -1.375 | 0.08642 | -1.005 | 0.00064 | 0.39 |
| 21690 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-450b-5p | 17 | PRDM2 | Sponge network | -4.546 | 0 | -0.258 | 0.00721 | 0.39 |
| 21691 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 36 | AR | Sponge network | -0.141 | 0.26434 | -1.554 | 0 | 0.39 |
| 21692 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | PTPN11 | Sponge network | 0.225 | 0.30644 | 0.431 | 2.0E-5 | 0.39 |
| 21693 | BOLA3-AS1 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 42 | THRB | Sponge network | -0.246 | 0.35034 | -1.117 | 0.0004 | 0.39 |
| 21694 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | RABEP1 | Sponge network | 0.194 | 0.64278 | -0.066 | 0.43142 | 0.39 |
| 21695 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 23 | EPHA3 | Sponge network | 0.61 | 0.01593 | 0.185 | 0.53588 | 0.39 |
| 21696 | ZSCAN16-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-874-3p | 12 | SORCS1 | Sponge network | -0.342 | 0.02288 | -3.492 | 0 | 0.39 |
| 21697 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | SOX5 | Sponge network | 0.853 | 0.15352 | -1.091 | 4.0E-5 | 0.39 |
| 21698 | RP11-575A19.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 12 | RUNX1T1 | Sponge network | 1.969 | 0.00316 | -0.344 | 0.2374 | 0.39 |
| 21699 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-7-5p | 22 | ESR1 | Sponge network | 0.056 | 0.8494 | -1.035 | 3.0E-5 | 0.39 |
| 21700 | ZNF32-AS2 |
hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-654-3p | 15 | RICTOR | Sponge network | 1.552 | 7.0E-5 | 0.388 | 0.00188 | 0.39 |
| 21701 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p | 22 | MYO9A | Sponge network | 0.95 | 0.15943 | -0.147 | 0.32373 | 0.39 |
| 21702 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ZFHX3 | Sponge network | 0.862 | 0.06213 | 0.389 | 0.01363 | 0.39 |
| 21703 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | DOCK4 | Sponge network | 0.537 | 0.00139 | 0.502 | 0.00108 | 0.39 |
| 21704 | CTD-2002H8.2 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ARHGEF12 | Sponge network | 0.931 | 1.0E-5 | 0.09 | 0.35693 | 0.39 |
| 21705 | KB-1410C5.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | TRPS1 | Sponge network | 0.959 | 0 | -0.191 | 0.44109 | 0.39 |
| 21706 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | SOX5 | Sponge network | 0.249 | 0.35902 | -1.091 | 4.0E-5 | 0.39 |
| 21707 | RP4-717I23.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-940 | 15 | SIRT1 | Sponge network | 0.637 | 0.00069 | 0.109 | 0.2593 | 0.39 |
| 21708 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | -2.117 | 0.00161 | -1.047 | 0.00364 | 0.39 |
| 21709 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 30 | CLASP2 | Sponge network | 0.194 | 0.64278 | -0.038 | 0.71 | 0.39 |
| 21710 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | PTPRB | Sponge network | -1.575 | 6.0E-5 | 0.105 | 0.47122 | 0.39 |
| 21711 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 17 | TXNIP | Sponge network | -0.563 | 0.02545 | -1.277 | 0 | 0.39 |
| 21712 | RP11-156E6.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p | 18 | ABL2 | Sponge network | 1.266 | 0 | 0.82 | 0 | 0.39 |
| 21713 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-511-5p;hsa-miR-625-5p | 11 | ST6GALNAC3 | Sponge network | -1.054 | 0.0706 | -0.987 | 0 | 0.39 |
| 21714 | RP11-6N17.3 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-744-3p | 16 | LPP | Sponge network | 0.108 | 0.69556 | -0.459 | 0.04475 | 0.39 |
| 21715 | RP11-283I3.6 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 14 | TRIM13 | Sponge network | 0.614 | 1.0E-5 | 0.183 | 0.06968 | 0.39 |
| 21716 | MIR600HG |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-424-5p | 20 | BRD3 | Sponge network | 0.594 | 0.41522 | 0.37 | 0.00013 | 0.39 |
| 21717 | AC009948.5 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | PRKAR1A | Sponge network | 0.532 | 0.00505 | -0.063 | 0.50314 | 0.39 |
| 21718 | LENG8-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326 | 12 | NF1 | Sponge network | 1.471 | 0 | 0.51 | 0 | 0.39 |
| 21719 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | PREX2 | Sponge network | -0.739 | 0.00044 | -0.272 | 0.25382 | 0.39 |
| 21720 | PART1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -2.925 | 0 | -2.417 | 0 | 0.39 |
| 21721 | RP11-67L2.2 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-625-5p | 11 | LZTS1 | Sponge network | 0.72 | 0.0001 | 1.664 | 0 | 0.39 |
| 21722 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p | 32 | SCN9A | Sponge network | -0.615 | 0.00015 | -0.525 | 0.10593 | 0.39 |
| 21723 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | CLASP2 | Sponge network | 0.72 | 0.0001 | -0.038 | 0.71 | 0.39 |
| 21724 | LINC00472 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 13 | KLF10 | Sponge network | -0.842 | 0.01361 | -0.783 | 0 | 0.39 |
| 21725 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 16 | ITCH | Sponge network | 0.401 | 0.00066 | 0.084 | 0.34058 | 0.39 |
| 21726 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 15 | SLITRK6 | Sponge network | -1.161 | 0.00591 | -2.121 | 0 | 0.39 |
| 21727 | CTD-2528L19.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ABI2 | Sponge network | 0.953 | 0 | 0.175 | 0.14787 | 0.39 |
| 21728 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 26 | GRIN2A | Sponge network | -0.969 | 2.0E-5 | -2.161 | 0 | 0.39 |
| 21729 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p | 13 | SPG20 | Sponge network | 0.082 | 0.55397 | -1.16 | 0 | 0.39 |
| 21730 | USP3-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-935 | 10 | SEPT6 | Sponge network | -0.162 | 0.47687 | -0.598 | 0.00181 | 0.39 |
| 21731 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-935 | 12 | LIFR | Sponge network | -0.376 | 0.00337 | -1.794 | 0 | 0.39 |
| 21732 | LINC00654 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | WWTR1 | Sponge network | 0.374 | 0.20054 | -0.345 | 0.11997 | 0.39 |
| 21733 | RP11-13K12.5 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-423-5p | 14 | ABL1 | Sponge network | -0.318 | 0.65847 | -0.215 | 0.07804 | 0.39 |
| 21734 | BZRAP1-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-582-3p;hsa-miR-93-3p | 17 | PCDH10 | Sponge network | -0.465 | 0.04703 | -1.644 | 0.0078 | 0.39 |
| 21735 | LINC00957 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ERG | Sponge network | -0.421 | 0.0114 | 0.002 | 0.99011 | 0.39 |
| 21736 | RP11-283I3.6 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | INPP4A | Sponge network | 0.614 | 1.0E-5 | 0.07 | 0.53563 | 0.39 |
| 21737 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-93-5p | 48 | UNC80 | Sponge network | -1.575 | 6.0E-5 | -1.934 | 0 | 0.39 |
| 21738 | LINC00863 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | BCL9 | Sponge network | 0.189 | 0.13981 | 0.321 | 0.00267 | 0.39 |
| 21739 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-629-5p;hsa-miR-96-5p | 20 | MAF | Sponge network | -0.517 | 0.02935 | -1.257 | 0 | 0.39 |
| 21740 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-625-5p | 14 | TES | Sponge network | -2.69 | 0.0001 | -0.348 | 0.00639 | 0.39 |
| 21741 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-96-5p | 55 | BMPR2 | Sponge network | 0.082 | 0.55654 | 0.206 | 0.0635 | 0.39 |
| 21742 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-940 | 61 | NFIB | Sponge network | -2.452 | 0 | 0.023 | 0.90795 | 0.39 |
| 21743 | BVES-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p | 19 | EYA4 | Sponge network | -2.62 | 0 | 0.3 | 0.36876 | 0.39 |
| 21744 | LINC00865 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-628-5p | 15 | IRS1 | Sponge network | -1.249 | 7.0E-5 | -0.026 | 0.88855 | 0.39 |
| 21745 | LINC01018 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-5p | 10 | PDZD2 | Sponge network | -2.915 | 0.00015 | -0.909 | 3.0E-5 | 0.39 |
| 21746 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-96-5p | 41 | SOX5 | Sponge network | 0.082 | 0.55654 | -1.091 | 4.0E-5 | 0.39 |
| 21747 | RP11-458F8.4 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30c-5p | 10 | PHF6 | Sponge network | 1.518 | 0 | 0.785 | 0 | 0.39 |
| 21748 | RP11-384O8.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | ROBO1 | Sponge network | -0.738 | 0.21233 | -0.009 | 0.96154 | 0.39 |
| 21749 | CKMT2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p | 23 | RYR2 | Sponge network | 0.082 | 0.55654 | -1.466 | 0.00031 | 0.39 |
| 21750 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 30 | DDX6 | Sponge network | 0.349 | 0.01695 | 0.091 | 0.36428 | 0.39 |
| 21751 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | HDAC9 | Sponge network | -0.129 | 0.57177 | -0.755 | 0.00495 | 0.39 |
| 21752 | TP73-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p | 20 | NRK | Sponge network | -0.563 | 0.02545 | 0.656 | 0.19217 | 0.39 |
| 21753 | TPTEP1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 22 | CDH23 | Sponge network | -1.216 | 0 | -0.974 | 0.00034 | 0.39 |
| 21754 | USP3-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-576-5p | 23 | PDE4DIP | Sponge network | -0.162 | 0.47687 | -0.282 | 0.0257 | 0.39 |
| 21755 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-93-5p | 16 | LRRC55 | Sponge network | -2.531 | 0 | -0.026 | 0.93163 | 0.39 |
| 21756 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p | 11 | TMEFF2 | Sponge network | -0.246 | 0.35034 | -2.426 | 0 | 0.39 |
| 21757 | NDUFA6-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | -0.355 | 0.0077 | -0.57 | 0 | 0.39 |
| 21758 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MYO9A | Sponge network | 0.986 | 0.01022 | -0.147 | 0.32373 | 0.39 |
| 21759 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 23 | AKAP6 | Sponge network | -0.727 | 0.04726 | -1.586 | 3.0E-5 | 0.39 |
| 21760 | AC007566.10 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 13 | TBL1XR1 | Sponge network | 2.368 | 2.0E-5 | 0.578 | 0 | 0.39 |
| 21761 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p | 12 | CHD1 | Sponge network | 1.147 | 0 | 0.322 | 0.00215 | 0.39 |
| 21762 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-940 | 13 | GALNT13 | Sponge network | -0.777 | 0.16667 | -0.857 | 0.07439 | 0.39 |
| 21763 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | TSHZ3 | Sponge network | 0.381 | 0.25967 | -0.556 | 0.01516 | 0.39 |
| 21764 | BZRAP1-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-409-5p;hsa-miR-455-5p;hsa-miR-577 | 10 | MACF1 | Sponge network | -0.465 | 0.04703 | 0.019 | 0.90335 | 0.39 |
| 21765 | U91328.21 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-589-5p | 19 | PRKAA2 | Sponge network | -0.735 | 0.15983 | -2.462 | 0 | 0.39 |
| 21766 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | ATRX | Sponge network | 0.415 | 0.14657 | 0.261 | 0.04408 | 0.39 |
| 21767 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | 0.572 | 0.01722 | -1.751 | 1.0E-5 | 0.39 |
| 21768 | RP11-73M18.8 |
hsa-let-7b-3p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-582-5p | 11 | CREB1 | Sponge network | 0.832 | 0 | 0.203 | 0.00537 | 0.39 |
| 21769 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 29 | TSHZ3 | Sponge network | -0.08 | 0.90092 | -0.556 | 0.01516 | 0.39 |
| 21770 | RP4-605O3.4 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-940 | 27 | MDM4 | Sponge network | 0.262 | 0.0475 | 0.345 | 0.00762 | 0.39 |
| 21771 | TP73-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | TPO | Sponge network | -0.563 | 0.02545 | -1.87 | 2.0E-5 | 0.39 |
| 21772 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 14 | RB1CC1 | Sponge network | 0.873 | 0 | 0.175 | 0.12559 | 0.39 |
| 21773 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 11 | CLASP2 | Sponge network | 0.706 | 0 | -0.038 | 0.71 | 0.39 |
| 21774 | LINC00621 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | LUZP2 | Sponge network | 0.131 | 0.8436 | -0.056 | 0.89416 | 0.39 |
| 21775 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-374b-5p | 14 | PIK3C2A | Sponge network | 1.923 | 0 | 0.061 | 0.53776 | 0.39 |
| 21776 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-577;hsa-miR-98-5p | 19 | ZFHX3 | Sponge network | -0.473 | 0.07602 | 0.389 | 0.01363 | 0.39 |
| 21777 | RP1-302G2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-30b-3p | 16 | AR | Sponge network | -1.483 | 9.0E-5 | -1.554 | 0 | 0.39 |
| 21778 | LINC00578 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-628-5p | 11 | BCL6 | Sponge network | 0.811 | 0.03228 | -0.211 | 0.19489 | 0.39 |
| 21779 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | 0.532 | 0.00505 | -0.243 | 0.11408 | 0.39 |
| 21780 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 28 | RSPO2 | Sponge network | 0.246 | 0.59912 | -3.038 | 0 | 0.39 |
| 21781 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | MYO9A | Sponge network | -0.021 | 0.93598 | -0.147 | 0.32373 | 0.39 |
| 21782 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 31 | FBXO32 | Sponge network | 0.532 | 0.00505 | -0.495 | 0.07561 | 0.39 |
| 21783 | RP11-999E24.3 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-93-3p | 17 | GRIK3 | Sponge network | -0.693 | 0.05629 | -3.208 | 0 | 0.39 |
| 21784 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | TLL1 | Sponge network | 0.246 | 0.59912 | -1.195 | 3.0E-5 | 0.39 |
| 21785 | RP11-102N12.3 |
hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-502-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | PRKCB | Sponge network | -0.351 | 0.11358 | -1.313 | 2.0E-5 | 0.39 |
| 21786 | DNM3OS |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-5p | 14 | PDZD2 | Sponge network | 0.572 | 0.01722 | -0.909 | 3.0E-5 | 0.39 |
| 21787 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 28 | MXI1 | Sponge network | -0.976 | 0.00025 | -0.987 | 0 | 0.39 |
| 21788 | DIO3OS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p | 33 | IL17RD | Sponge network | -0.773 | 0.04073 | -0.019 | 0.94873 | 0.39 |
| 21789 | LINC00094 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429 | 12 | ITGB1 | Sponge network | 0.253 | 0.09565 | 0.524 | 0.00017 | 0.39 |
| 21790 | RP11-1006G14.4 |
hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | HERC1 | Sponge network | 0.559 | 0.00026 | -0.196 | 0.08544 | 0.39 |
| 21791 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MCC | Sponge network | -0.737 | 0.10822 | -1.005 | 0 | 0.39 |
| 21792 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | CHD2 | Sponge network | 0.826 | 1.0E-5 | 0.049 | 0.55371 | 0.39 |
| 21793 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 30 | TGFBR3 | Sponge network | 0.299 | 0.57722 | -1.173 | 1.0E-5 | 0.39 |
| 21794 | LINC00641 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ZNF844 | Sponge network | -0.222 | 0.32445 | -0.966 | 0.00035 | 0.39 |
| 21795 | HCG11 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | 0.385 | 0.10067 | -0.704 | 0.00106 | 0.39 |
| 21796 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | INPP4A | Sponge network | -0.739 | 0.00044 | 0.07 | 0.53563 | 0.39 |
| 21797 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | MOB1B | Sponge network | 1.148 | 2.0E-5 | 0.197 | 0.15266 | 0.39 |
| 21798 | HCG18 |
hsa-let-7g-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p | 11 | KDM3A | Sponge network | 1.063 | 0 | 0.132 | 0.22711 | 0.39 |
| 21799 | RP11-53O19.1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-654-3p;hsa-miR-940 | 17 | BAZ2B | Sponge network | -0.405 | 0.22013 | -0.276 | 0.02833 | 0.39 |
| 21800 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-93-3p | 47 | QKI | Sponge network | -1.645 | 0 | -0.333 | 0.04111 | 0.39 |
| 21801 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-486-5p;hsa-miR-7-1-3p | 11 | FGF5 | Sponge network | 0.889 | 9.0E-5 | 0.549 | 0.28659 | 0.39 |
| 21802 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p | 17 | CITED2 | Sponge network | -0.129 | 0.57177 | -1.545 | 0 | 0.39 |
| 21803 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | NCAM2 | Sponge network | -0.323 | 0.01372 | -0.482 | 0.22565 | 0.39 |
| 21804 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-654-3p | 23 | RICTOR | Sponge network | 1.073 | 0.00216 | 0.388 | 0.00188 | 0.39 |
| 21805 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-5p;hsa-miR-96-5p | 12 | PAK3 | Sponge network | -1.349 | 0.00017 | -0.628 | 0.07253 | 0.39 |
| 21806 | RP11-159D12.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | BAZ2B | Sponge network | 1.254 | 0 | -0.276 | 0.02833 | 0.39 |
| 21807 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-93-5p | 33 | EPHA5 | Sponge network | -0.777 | 0.16667 | -0.957 | 0.01733 | 0.39 |
| 21808 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 31 | ABI2 | Sponge network | -0.563 | 0.02545 | 0.175 | 0.14787 | 0.39 |
| 21809 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZNF844 | Sponge network | -0.129 | 0.57177 | -0.966 | 0.00035 | 0.39 |
| 21810 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-421;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | CNTN1 | Sponge network | -0.418 | 0.00068 | -2.594 | 0 | 0.39 |
| 21811 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p | 29 | TMEM132B | Sponge network | 0.051 | 0.83388 | -0.83 | 0.01084 | 0.39 |
| 21812 | PWAR6 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | PAIP2 | Sponge network | -1.575 | 6.0E-5 | -0.515 | 0 | 0.39 |
| 21813 | AC093110.3 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p | 19 | UBR3 | Sponge network | 1.044 | 2.0E-5 | -0.021 | 0.82774 | 0.39 |
| 21814 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | EBF3 | Sponge network | -1.216 | 0 | -0.058 | 0.81437 | 0.39 |
| 21815 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-654-3p | 14 | AKAP11 | Sponge network | 0.909 | 0 | 0.196 | 0.09825 | 0.39 |
| 21816 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-590-3p;hsa-miR-671-3p | 13 | MAP3K12 | Sponge network | -0.739 | 0.00044 | -0.057 | 0.59369 | 0.39 |
| 21817 | RP5-1074L1.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | UBR3 | Sponge network | 1.923 | 0 | -0.021 | 0.82774 | 0.39 |
| 21818 | LINC00702 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ZNF536 | Sponge network | -0.737 | 0.06182 | -3.115 | 0 | 0.39 |
| 21819 | AC093110.3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-576-5p | 22 | ARHGEF12 | Sponge network | 1.044 | 2.0E-5 | 0.09 | 0.35693 | 0.39 |
| 21820 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 24 | PIK3R1 | Sponge network | 0.727 | 3.0E-5 | -0.133 | 0.36854 | 0.39 |
| 21821 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 27 | KIT | Sponge network | -2.925 | 0 | -2.184 | 0 | 0.39 |
| 21822 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | TACC1 | Sponge network | -0.318 | 0.65847 | -0.362 | 0.05406 | 0.39 |
| 21823 | WAC-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 10 | PHF6 | Sponge network | 0.821 | 0 | 0.785 | 0 | 0.39 |
| 21824 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-21-5p | 11 | BNC2 | Sponge network | -0.326 | 0.28809 | -0.491 | 0.09988 | 0.39 |
| 21825 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 37 | HLF | Sponge network | -0.969 | 2.0E-5 | -2.443 | 0 | 0.39 |
| 21826 | CTD-2528L19.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 21 | ABL2 | Sponge network | 0.953 | 0 | 0.82 | 0 | 0.39 |
| 21827 | DIO3OS |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p | 11 | NCAM2 | Sponge network | -0.773 | 0.04073 | -0.482 | 0.22565 | 0.39 |
| 21828 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-93-3p | 11 | ST6GAL2 | Sponge network | -0.932 | 0.00329 | -1.201 | 0.00597 | 0.39 |
| 21829 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-93-5p | 12 | TXNIP | Sponge network | -0.785 | 0.00035 | -1.277 | 0 | 0.39 |
| 21830 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 47 | ERBB4 | Sponge network | -2.452 | 0 | -1.975 | 2.0E-5 | 0.39 |
| 21831 | KB-1460A1.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p | 50 | DST | Sponge network | 0.603 | 0.00127 | -0.713 | 0.00096 | 0.39 |
| 21832 | RP13-516M14.1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429 | 10 | DNMT3A | Sponge network | 0.389 | 0.04293 | 0.302 | 0.0262 | 0.39 |
| 21833 | RP11-536O18.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-502-5p | 12 | DTNA | Sponge network | -0.074 | 0.90758 | -1.648 | 1.0E-5 | 0.39 |
| 21834 | CALML3-AS1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 14 | RASAL2 | Sponge network | 0.95 | 0.15943 | 0.869 | 0 | 0.39 |
| 21835 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-744-3p | 21 | HIP1 | Sponge network | 0.494 | 0.00339 | 0.774 | 0 | 0.39 |
| 21836 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p | 18 | CNTNAP3 | Sponge network | -0.222 | 0.32445 | -1.465 | 0.00033 | 0.39 |
| 21837 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | ANK2 | Sponge network | -0.125 | 0.49414 | -1.603 | 1.0E-5 | 0.39 |
| 21838 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-624-5p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-940 | 46 | NTRK3 | Sponge network | -0.246 | 0.35034 | -1.77 | 3.0E-5 | 0.39 |
| 21839 | LINC00657 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | EPG5 | Sponge network | 0.91 | 0 | 0.101 | 0.3563 | 0.39 |
| 21840 | RP11-53O19.3 |
hsa-miR-101-3p;hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | UBR3 | Sponge network | 1.205 | 0 | -0.021 | 0.82774 | 0.39 |
| 21841 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | CAV1 | Sponge network | -0.899 | 6.0E-5 | -1.013 | 6.0E-5 | 0.39 |
| 21842 | NNT-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | BRD3 | Sponge network | 0.799 | 1.0E-5 | 0.37 | 0.00013 | 0.39 |
| 21843 | LINC00702 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 33 | MXI1 | Sponge network | -0.737 | 0.06182 | -0.987 | 0 | 0.39 |
| 21844 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | CLASP2 | Sponge network | 1.063 | 0 | -0.038 | 0.71 | 0.39 |
| 21845 | RP11-2H8.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 21 | PHC3 | Sponge network | 1.089 | 0 | 0.212 | 0.0499 | 0.39 |
| 21846 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-92a-3p | 57 | TCF4 | Sponge network | 0.214 | 0.18246 | 0.016 | 0.92271 | 0.39 |
| 21847 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-93-5p | 17 | PGR | Sponge network | 0.497 | 0.01451 | -1.704 | 0 | 0.39 |
| 21848 | RP11-2B6.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PIK3CA | Sponge network | 0.539 | 0.03722 | 0.195 | 0.06908 | 0.39 |
| 21849 | CTC-510F12.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-185-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-361-3p | 10 | BTG2 | Sponge network | -0.691 | 0.00146 | -1.763 | 0 | 0.39 |
| 21850 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 22 | ZFP36L1 | Sponge network | -0.769 | 0.00035 | -0.505 | 8.0E-5 | 0.39 |
| 21851 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLI1 | Sponge network | -0.49 | 0.04946 | -0.294 | 0.10905 | 0.39 |
| 21852 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p | 11 | EDA2R | Sponge network | -0.517 | 0.02935 | -0.587 | 0.01598 | 0.39 |
| 21853 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 37 | TSHZ3 | Sponge network | 0.064 | 0.72934 | -0.556 | 0.01516 | 0.389 |
| 21854 | LINC00654 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | FYN | Sponge network | 0.374 | 0.20054 | -0.131 | 0.37051 | 0.389 |
| 21855 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p | 14 | ITGB3 | Sponge network | -0.738 | 0.21233 | -0.011 | 0.95751 | 0.389 |
| 21856 | AGAP11 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 15 | RPS6KA6 | Sponge network | -1.645 | 0 | -2.989 | 0 | 0.389 |
| 21857 | FGF14-AS2 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-590-3p | 13 | GLIPR1 | Sponge network | -1.582 | 8.0E-5 | 0.146 | 0.3276 | 0.389 |
| 21858 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | EPAS1 | Sponge network | -0.351 | 0.11358 | -0.184 | 0.14418 | 0.389 |
| 21859 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | PLSCR4 | Sponge network | -0.08 | 0.90092 | -0.474 | 0.03833 | 0.389 |
| 21860 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | GNA13 | Sponge network | 0.683 | 1.0E-5 | 0.007 | 0.93884 | 0.389 |
| 21861 | LINC00473 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-7-5p;hsa-miR-92a-3p | 27 | BTG2 | Sponge network | -1.203 | 0.03881 | -1.763 | 0 | 0.389 |
| 21862 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-5p | 24 | BTG2 | Sponge network | -0.469 | 0.08721 | -1.763 | 0 | 0.389 |
| 21863 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | ERG | Sponge network | 0.246 | 0.59912 | 0.002 | 0.99011 | 0.389 |
| 21864 | OIP5-AS1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-874-3p | 11 | ARID1B | Sponge network | 0.421 | 0.00084 | 0.003 | 0.97503 | 0.389 |
| 21865 | SNHG12 |
hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 15 | MDM4 | Sponge network | 1.103 | 0 | 0.345 | 0.00762 | 0.389 |
| 21866 | AC103563.8 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p | 12 | IL6ST | Sponge network | -7.551 | 0 | -0.402 | 0.00676 | 0.389 |
| 21867 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 17 | DYNC1I1 | Sponge network | -0.08 | 0.90092 | -1.12 | 0.00107 | 0.389 |
| 21868 | AP001372.2 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-766-3p | 16 | TBL1XR1 | Sponge network | 0.861 | 0 | 0.578 | 0 | 0.389 |
| 21869 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-625-5p | 15 | ZMYM2 | Sponge network | 0.817 | 3.0E-5 | 0.279 | 0.01273 | 0.389 |
| 21870 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 51 | RUNX1T1 | Sponge network | 0.61 | 0.01593 | -0.344 | 0.2374 | 0.389 |
| 21871 | MEG3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | INPP4A | Sponge network | 0.249 | 0.35902 | 0.07 | 0.53563 | 0.389 |
| 21872 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 11 | GPAM | Sponge network | 0.537 | 0.0006 | 0.142 | 0.29732 | 0.389 |
| 21873 | MEG3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-5p | 14 | PDZD2 | Sponge network | 0.249 | 0.35902 | -0.909 | 3.0E-5 | 0.389 |
| 21874 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940 | 36 | HLF | Sponge network | 0.572 | 0.01722 | -2.443 | 0 | 0.389 |
| 21875 | CTD-2314G24.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-7-5p | 26 | CRTC1 | Sponge network | 0.246 | 0.59912 | -0.116 | 0.28412 | 0.389 |
| 21876 | CTA-445C9.14 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 13 | PHF6 | Sponge network | 0.718 | 4.0E-5 | 0.785 | 0 | 0.389 |
| 21877 | LINC00957 |
hsa-miR-107;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | ZNF536 | Sponge network | -0.421 | 0.0114 | -3.115 | 0 | 0.389 |
| 21878 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | NF1 | Sponge network | 0.539 | 0.03722 | 0.51 | 0 | 0.389 |
| 21879 | RP11-517B11.7 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-495-3p | 12 | ERCC6 | Sponge network | 0.706 | 0 | 0.23 | 0.015 | 0.389 |
| 21880 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-93-5p | 21 | PGR | Sponge network | 0.259 | 0.12542 | -1.704 | 0 | 0.389 |
| 21881 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 19 | TES | Sponge network | -0.576 | 0.10121 | -0.348 | 0.00639 | 0.389 |
| 21882 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-940 | 28 | PBX1 | Sponge network | 0.16 | 0.50785 | -1.206 | 0 | 0.389 |
| 21883 | AP001347.6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CSMD1 | Sponge network | -1.161 | 0.00591 | -0.63 | 0.18881 | 0.389 |
| 21884 | RP11-25K19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | CRTC1 | Sponge network | -1.057 | 0.00029 | -0.116 | 0.28412 | 0.389 |
| 21885 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-29a-5p | 12 | CNTNAP3 | Sponge network | 0.176 | 0.37402 | -1.465 | 0.00033 | 0.389 |
| 21886 | CTC-429P9.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-93-5p | 24 | IL17RD | Sponge network | 0.082 | 0.55397 | -0.019 | 0.94873 | 0.389 |
| 21887 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 22 | TGFBR3 | Sponge network | -0.726 | 0.00132 | -1.173 | 1.0E-5 | 0.389 |
| 21888 | RAMP2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 14 | UNC5C | Sponge network | -0.513 | 0.04703 | -0.178 | 0.40214 | 0.389 |
| 21889 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 10 | RBMS3 | Sponge network | 1.969 | 0.00316 | -0.519 | 0.03551 | 0.389 |
| 21890 | WAC-AS1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | BRD3 | Sponge network | 0.821 | 0 | 0.37 | 0.00013 | 0.389 |
| 21891 | LINC00476 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p | 14 | RHOB | Sponge network | -0.615 | 0.00015 | -1.052 | 0 | 0.389 |
| 21892 | GNG12-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | NAV3 | Sponge network | -0.793 | 7.0E-5 | 0.212 | 0.47729 | 0.389 |
| 21893 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-455-5p | 10 | AKAP12 | Sponge network | -0.773 | 0.04073 | -0.932 | 0.0028 | 0.389 |
| 21894 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 16 | RICTOR | Sponge network | 0.252 | 0.08508 | 0.388 | 0.00188 | 0.389 |
| 21895 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | LATS2 | Sponge network | 0.381 | 0.25967 | 0.159 | 0.2304 | 0.389 |
| 21896 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-452-5p;hsa-miR-935 | 13 | RELN | Sponge network | -0.405 | 0.22013 | -2.403 | 0 | 0.389 |
| 21897 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-93-5p | 46 | PRKAA2 | Sponge network | -1.645 | 0 | -2.462 | 0 | 0.389 |
| 21898 | PCA3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PREX2 | Sponge network | -0.709 | 0.18168 | -0.272 | 0.25382 | 0.389 |
| 21899 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 43 | BCL2 | Sponge network | 0.246 | 0.59912 | -0.899 | 0 | 0.389 |
| 21900 | RP11-359E10.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-500a-3p | 14 | FMNL3 | Sponge network | 0.299 | 0.57722 | 0.555 | 4.0E-5 | 0.389 |
| 21901 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 46 | IL6ST | Sponge network | 0.4 | 0.14611 | -0.402 | 0.00676 | 0.389 |
| 21902 | AC003090.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | IL17RD | Sponge network | -2.117 | 0.00161 | -0.019 | 0.94873 | 0.389 |
| 21903 | RP5-1139B12.3 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-500a-3p | 17 | ZFHX4 | Sponge network | 0.598 | 0.38481 | 0.018 | 0.95574 | 0.389 |
| 21904 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 36 | GAS7 | Sponge network | 0.4 | 0.14611 | -0.555 | 0.01602 | 0.389 |
| 21905 | HOXA-AS2 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p | 11 | FLNA | Sponge network | 0.61 | 0.01593 | -0.771 | 0.01377 | 0.389 |
| 21906 | RP11-720L2.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p | 12 | BCL2 | Sponge network | -1.255 | 0.00981 | -0.899 | 0 | 0.389 |
| 21907 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-7-1-3p | 16 | SLC6A15 | Sponge network | -1.249 | 7.0E-5 | -2.373 | 2.0E-5 | 0.389 |
| 21908 | CTA-941F9.9 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p | 12 | EYA4 | Sponge network | -0.344 | 0.49624 | 0.3 | 0.36876 | 0.389 |
| 21909 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 32 | PHC3 | Sponge network | 0.082 | 0.55654 | 0.212 | 0.0499 | 0.389 |
| 21910 | RP11-13K12.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-501-3p | 11 | ZBTB20 | Sponge network | -0.154 | 0.78939 | -0.122 | 0.57569 | 0.389 |
| 21911 | AGAP11 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-5p | 32 | DIP2C | Sponge network | -1.645 | 0 | -0.436 | 0.01713 | 0.389 |
| 21912 | RP11-1006G14.4 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 13 | KIF5B | Sponge network | 0.559 | 0.00026 | 0.362 | 0.00066 | 0.389 |
| 21913 | FAM13A-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 16 | TLE4 | Sponge network | -0.83 | 0 | -1.157 | 0 | 0.389 |
| 21914 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 10 | GIGYF2 | Sponge network | 0.083 | 0.66405 | 0.136 | 0.04403 | 0.389 |
| 21915 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-582-5p | 15 | NR2C2 | Sponge network | 0.389 | 0.04293 | 0.21 | 0.03224 | 0.389 |
| 21916 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | FSTL5 | Sponge network | -0.615 | 0.00015 | -1.3 | 0.01619 | 0.389 |
| 21917 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 22 | JAZF1 | Sponge network | 0.174 | 0.30034 | -0.377 | 0.02677 | 0.389 |
| 21918 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | IGSF10 | Sponge network | -0.709 | 0.18168 | -1.751 | 1.0E-5 | 0.389 |
| 21919 | RP5-1198O20.4 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 11 | EPB41L3 | Sponge network | 0.997 | 0.00066 | -1.193 | 0 | 0.389 |
| 21920 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 15 | FOXP1 | Sponge network | 2.163 | 0 | 0.042 | 0.74368 | 0.389 |
| 21921 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-495-3p | 19 | RICTOR | Sponge network | 1.056 | 0 | 0.388 | 0.00188 | 0.389 |
| 21922 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 34 | AKAP11 | Sponge network | -0.405 | 0.22013 | 0.196 | 0.09825 | 0.389 |
| 21923 | CTC-296K1.3 |
hsa-miR-125a-3p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-342-3p | 11 | RIMS2 | Sponge network | -2.095 | 0.00112 | -1.365 | 0.00208 | 0.389 |
| 21924 | NNT-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 24 | ABL2 | Sponge network | 0.799 | 1.0E-5 | 0.82 | 0 | 0.389 |
| 21925 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 19 | USP6 | Sponge network | 0.539 | 0.03722 | 0.188 | 0.3871 | 0.389 |
| 21926 | AD000090.2 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-497-5p;hsa-miR-766-3p | 11 | MDM4 | Sponge network | 0.943 | 0 | 0.345 | 0.00762 | 0.389 |
| 21927 | FAM66C |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 24 | ARNT2 | Sponge network | -0.901 | 0.00019 | -0.332 | 0.25851 | 0.389 |
| 21928 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | PLSCR4 | Sponge network | 0.259 | 0.12542 | -0.474 | 0.03833 | 0.389 |
| 21929 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-96-5p | 32 | MEF2C | Sponge network | -0.18 | 0.20343 | -0.499 | 0.01448 | 0.389 |
| 21930 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 19 | TBL1XR1 | Sponge network | 1.178 | 0 | 0.578 | 0 | 0.389 |
| 21931 | LINC00641 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 25 | NRK | Sponge network | -0.222 | 0.32445 | 0.656 | 0.19217 | 0.389 |
| 21932 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | AHNAK | Sponge network | -2.117 | 0.00161 | -1.207 | 0 | 0.389 |
| 21933 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p | 16 | LHX6 | Sponge network | -0.154 | 0.53954 | -0.087 | 0.69193 | 0.389 |
| 21934 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | HDAC9 | Sponge network | -0.358 | 0.17255 | -0.755 | 0.00495 | 0.389 |
| 21935 | RP11-359B12.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 16 | ROBO1 | Sponge network | 0.064 | 0.72934 | -0.009 | 0.96154 | 0.389 |
| 21936 | RP11-400K9.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-93-5p | 27 | IL17RD | Sponge network | -0.611 | 0.09607 | -0.019 | 0.94873 | 0.389 |
| 21937 | AC074289.1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-96-5p | 15 | DOCK4 | Sponge network | -0.326 | 0.14314 | 0.502 | 0.00108 | 0.389 |
| 21938 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p | 13 | GREM1 | Sponge network | -0.811 | 2.0E-5 | 0.902 | 0.01691 | 0.389 |
| 21939 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-423-5p;hsa-miR-671-5p | 10 | CADM3 | Sponge network | 0.299 | 0.57722 | -3.663 | 0 | 0.389 |
| 21940 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 24 | TIMP3 | Sponge network | -1.203 | 0.03881 | -0.546 | 0.02307 | 0.389 |
| 21941 | LINC00641 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p | 12 | SMARCA1 | Sponge network | -0.222 | 0.32445 | -0.684 | 0.00159 | 0.389 |
| 21942 | MALAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 23 | TBL1XR1 | Sponge network | 0.782 | 0.00016 | 0.578 | 0 | 0.389 |
| 21943 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-766-3p | 17 | TBL1XR1 | Sponge network | 0.768 | 7.0E-5 | 0.578 | 0 | 0.389 |
| 21944 | RP1-302G2.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-7-5p;hsa-miR-93-3p | 10 | EPHA3 | Sponge network | -1.483 | 9.0E-5 | 0.185 | 0.53588 | 0.389 |
| 21945 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | TIMP3 | Sponge network | 0.381 | 0.25967 | -0.546 | 0.02307 | 0.389 |
| 21946 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-629-3p | 15 | EBF3 | Sponge network | -0.246 | 0.35034 | -0.058 | 0.81437 | 0.389 |
| 21947 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 11 | LATS2 | Sponge network | -4.463 | 0.00025 | 0.159 | 0.2304 | 0.389 |
| 21948 | CTA-363E19.2 |
hsa-let-7d-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 12 | DICER1 | Sponge network | 1.055 | 0 | 0.042 | 0.674 | 0.389 |
| 21949 | RP11-13K12.5 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-7-5p | 12 | FNBP1 | Sponge network | -0.318 | 0.65847 | -0.969 | 4.0E-5 | 0.389 |
| 21950 | RP11-999E24.3 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p | 11 | GJA1 | Sponge network | -0.693 | 0.05629 | -0.485 | 0.02179 | 0.389 |
| 21951 | TP73-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-935;hsa-miR-940 | 12 | PHOX2B | Sponge network | -0.563 | 0.02545 | -2.68 | 0 | 0.389 |
| 21952 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 25 | DIP2C | Sponge network | 0.559 | 0.00026 | -0.436 | 0.01713 | 0.389 |
| 21953 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | SLITRK6 | Sponge network | -0.615 | 0.00015 | -2.121 | 0 | 0.389 |
| 21954 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 23 | GRIN2A | Sponge network | 0.572 | 0.01722 | -2.161 | 0 | 0.389 |
| 21955 | RP11-822E23.8 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p | 10 | PARK2 | Sponge network | -0.777 | 0.16667 | -1.892 | 0 | 0.389 |
| 21956 | RP11-356J5.12 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 16 | KLF10 | Sponge network | -0.899 | 6.0E-5 | -0.783 | 0 | 0.389 |
| 21957 | DIO3OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-769-5p | 17 | SYNPO2 | Sponge network | -0.773 | 0.04073 | -2.342 | 0 | 0.389 |
| 21958 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 17 | NR4A3 | Sponge network | 0.249 | 0.35902 | -1.385 | 0 | 0.389 |
| 21959 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 19 | NRXN1 | Sponge network | -4.477 | 0 | -3.778 | 0 | 0.389 |
| 21960 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 52 | AR | Sponge network | 0.082 | 0.55654 | -1.554 | 0 | 0.389 |
| 21961 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 18 | PRKAR1A | Sponge network | 0.101 | 0.60919 | -0.063 | 0.50314 | 0.389 |
| 21962 | WDFY3-AS2 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | SYT4 | Sponge network | -0.942 | 0 | -3.207 | 0 | 0.389 |
| 21963 | SNX29P2 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-625-5p | 12 | NGFR | Sponge network | 0.4 | 0.14611 | -1.271 | 0.01058 | 0.389 |
| 21964 | RP11-434B12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ACVR2A | Sponge network | -0.031 | 0.89063 | -0.409 | 0.00039 | 0.389 |
| 21965 | RP11-389C8.2 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-629-3p | 10 | ITGA5 | Sponge network | 0.727 | 3.0E-5 | 0.452 | 0.03524 | 0.389 |
| 21966 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 10 | UVRAG | Sponge network | -0.612 | 0.00094 | -0.017 | 0.83865 | 0.389 |
| 21967 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 17 | SOX5 | Sponge network | 0.131 | 0.8436 | -1.091 | 4.0E-5 | 0.389 |
| 21968 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | MXI1 | Sponge network | 0.064 | 0.72934 | -0.987 | 0 | 0.389 |
| 21969 | CD27-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 30 | ABI2 | Sponge network | 0.177 | 0.16681 | 0.175 | 0.14787 | 0.389 |
| 21970 | MIR22HG |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 57 | DST | Sponge network | -1.047 | 0 | -0.713 | 0.00096 | 0.389 |
| 21971 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-577;hsa-miR-590-3p | 14 | TMEFF2 | Sponge network | -0.096 | 0.67958 | -2.426 | 0 | 0.389 |
| 21972 | RP11-890B15.3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 17 | INPP4A | Sponge network | 0.101 | 0.60919 | 0.07 | 0.53563 | 0.389 |
| 21973 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 36 | RASSF2 | Sponge network | -0.096 | 0.67958 | -0.273 | 0.21961 | 0.389 |
| 21974 | RP11-73M18.8 |
hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | MDM4 | Sponge network | 0.832 | 0 | 0.345 | 0.00762 | 0.389 |
| 21975 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-93-5p | 25 | ZBTB4 | Sponge network | 0.532 | 0.00505 | -0.739 | 0 | 0.389 |
| 21976 | KB-1460A1.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 26 | ABL2 | Sponge network | 0.603 | 0.00127 | 0.82 | 0 | 0.389 |
| 21977 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | GNA13 | Sponge network | 0.537 | 0.00139 | 0.007 | 0.93884 | 0.389 |
| 21978 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484 | 10 | PNRC1 | Sponge network | -2.531 | 0 | -0.646 | 0 | 0.389 |
| 21979 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 16 | MYBL1 | Sponge network | 1.03 | 0 | 0.494 | 0.02152 | 0.389 |
| 21980 | AC103563.8 |
hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-29a-5p | 10 | PTPRT | Sponge network | -7.551 | 0 | -0.907 | 0.03727 | 0.389 |
| 21981 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-323a-3p;hsa-miR-589-5p | 18 | PRKAA2 | Sponge network | -7.551 | 0 | -2.462 | 0 | 0.389 |
| 21982 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 17 | EBF1 | Sponge network | -0.517 | 0.02935 | -0.384 | 0.08856 | 0.389 |
| 21983 | RP11-597D13.9 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 16 | INPP4A | Sponge network | 1.302 | 7.0E-5 | 0.07 | 0.53563 | 0.389 |
| 21984 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 16 | RET | Sponge network | -0.162 | 0.47687 | -0.833 | 0.03517 | 0.389 |
| 21985 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-935 | 15 | LRRC55 | Sponge network | -0.162 | 0.47687 | -0.026 | 0.93163 | 0.389 |
| 21986 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | ZNF292 | Sponge network | 0.494 | 0.00339 | 0.276 | 0.04148 | 0.389 |
| 21987 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 12 | ERCC4 | Sponge network | 0.529 | 0.00552 | 0.126 | 0.15266 | 0.389 |
| 21988 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 46 | NTRK3 | Sponge network | 0.16 | 0.50785 | -1.77 | 3.0E-5 | 0.389 |
| 21989 | LINC00672 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p | 18 | ATM | Sponge network | -0.227 | 0.31657 | 0.178 | 0.21568 | 0.389 |
| 21990 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 15 | BHLHE41 | Sponge network | 0.693 | 0.00076 | 0.353 | 0.12753 | 0.389 |
| 21991 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | ANK2 | Sponge network | 0.727 | 3.0E-5 | -1.603 | 1.0E-5 | 0.389 |
| 21992 | LINC00843 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-495-3p | 14 | ZFX | Sponge network | 1.106 | 0 | 0.09 | 0.42563 | 0.389 |
| 21993 | LINC00969 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p | 11 | ITCH | Sponge network | 0.683 | 1.0E-5 | 0.084 | 0.34058 | 0.389 |
| 21994 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | SLITRK6 | Sponge network | -0.709 | 0.18168 | -2.121 | 0 | 0.389 |
| 21995 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-93-5p | 20 | PLSCR4 | Sponge network | -0.384 | 0.40652 | -0.474 | 0.03833 | 0.389 |
| 21996 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | DCLK1 | Sponge network | 0.082 | 0.55654 | -1.324 | 0.00014 | 0.389 |
| 21997 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-429 | 10 | LHX6 | Sponge network | -2.62 | 0 | -0.087 | 0.69193 | 0.389 |
| 21998 | RP11-798M19.6 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | -0.612 | 0.00094 | -0.226 | 0.11691 | 0.389 |
| 21999 | MEG3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p | 10 | ALDH1A2 | Sponge network | 0.249 | 0.35902 | -2.411 | 0 | 0.389 |
| 22000 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | GALNT13 | Sponge network | -0.513 | 0.04703 | -0.857 | 0.07439 | 0.389 |
| 22001 | LINC00472 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-320b | 13 | PRKAR1A | Sponge network | -0.842 | 0.01361 | -0.063 | 0.50314 | 0.389 |
| 22002 | RP11-967K21.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-532-5p | 11 | CDH23 | Sponge network | 0.056 | 0.81195 | -0.974 | 0.00034 | 0.389 |
| 22003 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 17 | CHD1 | Sponge network | 0.421 | 0.00084 | 0.322 | 0.00215 | 0.389 |
| 22004 | H1FX-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITPKB | Sponge network | -0.206 | 0.12942 | -0.704 | 0.00106 | 0.389 |
| 22005 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-5p | 22 | KCNA6 | Sponge network | -0.465 | 0.04703 | -1.531 | 0.00013 | 0.389 |
| 22006 | AC034220.3 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | NR2C2 | Sponge network | 0.309 | 0.07304 | 0.21 | 0.03224 | 0.389 |
| 22007 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 53 | IL6ST | Sponge network | 0.537 | 0.00139 | -0.402 | 0.00676 | 0.389 |
| 22008 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FAM133A | Sponge network | -2.46 | 0 | 0.549 | 0.29009 | 0.389 |
| 22009 | RP11-1006G14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | ERCC4 | Sponge network | 0.559 | 0.00026 | 0.126 | 0.15266 | 0.389 |
| 22010 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-493-5p | 10 | SYT4 | Sponge network | -1.942 | 0 | -3.207 | 0 | 0.389 |
| 22011 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-425-3p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p | 33 | SOX5 | Sponge network | -0.227 | 0.31657 | -1.091 | 4.0E-5 | 0.389 |
| 22012 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 31 | ZFHX3 | Sponge network | 0.16 | 0.50785 | 0.389 | 0.01363 | 0.389 |
| 22013 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FSTL5 | Sponge network | -0.612 | 0.00094 | -1.3 | 0.01619 | 0.389 |
| 22014 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-96-5p | 31 | PIK3R1 | Sponge network | 0.222 | 0.29133 | -0.133 | 0.36854 | 0.389 |
| 22015 | RP11-395G23.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 26 | TRPS1 | Sponge network | 0.381 | 0.25967 | -0.191 | 0.44109 | 0.389 |
| 22016 | RP11-283I3.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 24 | EPS15 | Sponge network | 0.614 | 1.0E-5 | -0.052 | 0.53998 | 0.389 |
| 22017 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-654-3p | 17 | DDX6 | Sponge network | 0.997 | 0 | 0.091 | 0.36428 | 0.389 |
| 22018 | NUTM2A-AS1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | CREB1 | Sponge network | 0.643 | 0 | 0.203 | 0.00537 | 0.389 |
| 22019 | RP11-532F6.3 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-628-5p | 13 | IRS1 | Sponge network | -0.896 | 0.00099 | -0.026 | 0.88855 | 0.389 |
| 22020 | ACTA2-AS1 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 15 | RTN4 | Sponge network | 0.194 | 0.64278 | -0.412 | 0.00014 | 0.389 |
| 22021 | RP11-359E10.1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-940 | 31 | GRIK3 | Sponge network | 0.299 | 0.57722 | -3.208 | 0 | 0.389 |
| 22022 | A2M-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p | 15 | SYNM | Sponge network | -0.08 | 0.90092 | -2.309 | 1.0E-5 | 0.389 |
| 22023 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | 0.064 | 0.72934 | 0.809 | 0.00544 | 0.389 |
| 22024 | FAM215B |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-7-1-3p | 12 | PIK3CA | Sponge network | 0.817 | 3.0E-5 | 0.195 | 0.06908 | 0.389 |
| 22025 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | NF1 | Sponge network | 1.262 | 0 | 0.51 | 0 | 0.389 |
| 22026 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-577;hsa-miR-590-3p | 20 | AFF3 | Sponge network | -0.227 | 0.31657 | -2.373 | 0 | 0.389 |
| 22027 | BZRAP1-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-93-5p | 20 | LRIG1 | Sponge network | -0.465 | 0.04703 | -0.537 | 0.00588 | 0.389 |
| 22028 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-331-5p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | HLF | Sponge network | -0.295 | 0.02604 | -2.443 | 0 | 0.389 |
| 22029 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | SUFU | Sponge network | 0.997 | 0.00066 | -0.04 | 0.65489 | 0.389 |
| 22030 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-335-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | RBL2 | Sponge network | 0.222 | 0.29133 | -0.282 | 0.00254 | 0.389 |
| 22031 | LINC00641 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-590-5p | 12 | THSD7A | Sponge network | -0.222 | 0.32445 | 0.242 | 0.25154 | 0.389 |
| 22032 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-5p | 17 | YAP1 | Sponge network | 0.331 | 0.57247 | 0.22 | 0.11836 | 0.389 |
| 22033 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-940;hsa-miR-96-5p | 34 | SNX29 | Sponge network | -0.942 | 0 | -0.34 | 0.01371 | 0.389 |
| 22034 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-93-5p | 33 | EPHA7 | Sponge network | -0.18 | 0.20343 | -2.197 | 3.0E-5 | 0.389 |
| 22035 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 30 | TSHZ3 | Sponge network | -0.405 | 0.22013 | -0.556 | 0.01516 | 0.389 |
| 22036 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | AKAP11 | Sponge network | 0.67 | 0.00048 | 0.196 | 0.09825 | 0.388 |
| 22037 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | PLSCR4 | Sponge network | -0.663 | 0.10887 | -0.474 | 0.03833 | 0.388 |
| 22038 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -0.563 | 0.02545 | 0.309 | 0.17079 | 0.388 |
| 22039 | PCA3 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ZNF536 | Sponge network | -0.709 | 0.18168 | -3.115 | 0 | 0.388 |
| 22040 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | NIN | Sponge network | 0.72 | 0.0001 | -0.253 | 0.13794 | 0.388 |
| 22041 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 10 | LHFP | Sponge network | -0.738 | 0.21233 | -0.167 | 0.41371 | 0.388 |
| 22042 | LINC00476 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-93-5p | 67 | DST | Sponge network | -0.615 | 0.00015 | -0.713 | 0.00096 | 0.388 |
| 22043 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 32 | MYO9A | Sponge network | -0.563 | 0.02545 | -0.147 | 0.32373 | 0.388 |
| 22044 | PROSER2-AS1 |
hsa-miR-129-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p | 10 | RASAL2 | Sponge network | 1.825 | 0 | 0.869 | 0 | 0.388 |
| 22045 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 34 | MCC | Sponge network | -0.612 | 0.00094 | -1.005 | 0 | 0.388 |
| 22046 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p | 17 | RICTOR | Sponge network | 0.542 | 0.00054 | 0.388 | 0.00188 | 0.388 |
| 22047 | RP4-758J18.10 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-450b-5p | 15 | DIP2C | Sponge network | -0.091 | 0.71468 | -0.436 | 0.01713 | 0.388 |
| 22048 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-744-5p | 18 | FGFR1 | Sponge network | -0.773 | 0.04073 | -0.509 | 0.03957 | 0.388 |
| 22049 | RP11-67L2.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.72 | 0.0001 | -0.57 | 0 | 0.388 |
| 22050 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 18 | WWTR1 | Sponge network | 0.225 | 0.30644 | -0.345 | 0.11997 | 0.388 |
| 22051 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 24 | LRRC7 | Sponge network | 0.249 | 0.35902 | -1.341 | 0.01193 | 0.388 |
| 22052 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | KCNA6 | Sponge network | -0.513 | 0.04703 | -1.531 | 0.00013 | 0.388 |
| 22053 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | TBL1XR1 | Sponge network | 0.832 | 0 | 0.578 | 0 | 0.388 |
| 22054 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | LRIG1 | Sponge network | -1.161 | 0.00591 | -0.537 | 0.00588 | 0.388 |
| 22055 | DIO3OS |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-7-5p | 15 | PTGFR | Sponge network | -0.773 | 0.04073 | -0.233 | 0.45421 | 0.388 |
| 22056 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-96-5p | 11 | PIK3CA | Sponge network | 0.453 | 0.01839 | 0.195 | 0.06908 | 0.388 |
| 22057 | SNX29P2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-511-5p;hsa-miR-532-3p | 14 | MRVI1 | Sponge network | 0.4 | 0.14611 | -1.051 | 0.00022 | 0.388 |
| 22058 | RP11-597D13.9 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 37 | IL17RD | Sponge network | 1.302 | 7.0E-5 | -0.019 | 0.94873 | 0.388 |
| 22059 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | KAT6B | Sponge network | -0.901 | 0.00019 | -0.478 | 0.00018 | 0.388 |
| 22060 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 19 | PTGFR | Sponge network | -0.738 | 0.21233 | -0.233 | 0.45421 | 0.388 |
| 22061 | RP11-418J17.1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-497-5p | 12 | CDK6 | Sponge network | 0.998 | 0 | 0.991 | 0 | 0.388 |
| 22062 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | NEDD4 | Sponge network | 0.889 | 9.0E-5 | 0.02 | 0.89834 | 0.388 |
| 22063 | LINC00865 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-942-5p | 13 | ZBTB20 | Sponge network | -1.249 | 7.0E-5 | -0.122 | 0.57569 | 0.388 |
| 22064 | GNG12-AS1 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 31 | RIMS2 | Sponge network | -0.793 | 7.0E-5 | -1.365 | 0.00208 | 0.388 |
| 22065 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | DOCK4 | Sponge network | -0.222 | 0.32445 | 0.502 | 0.00108 | 0.388 |
| 22066 | RP11-119F7.5 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | INPP4A | Sponge network | 0.225 | 0.30644 | 0.07 | 0.53563 | 0.388 |
| 22067 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | ATRX | Sponge network | 0.056 | 0.8494 | 0.261 | 0.04408 | 0.388 |
| 22068 | C4A-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-96-5p | 14 | RASSF8 | Sponge network | 3.345 | 0 | -0.122 | 0.65591 | 0.388 |
| 22069 | HOXA-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p | 46 | FAT3 | Sponge network | 0.61 | 0.01593 | -1.601 | 0.00051 | 0.388 |
| 22070 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 27 | SCN9A | Sponge network | -2.915 | 0.00015 | -0.525 | 0.10593 | 0.388 |
| 22071 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 27 | ERCC4 | Sponge network | -0.129 | 0.57177 | 0.126 | 0.15266 | 0.388 |
| 22072 | TUG1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 11 | CUL5 | Sponge network | 0.972 | 0 | 0.026 | 0.77301 | 0.388 |
| 22073 | RP11-890B15.3 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 17 | SORCS1 | Sponge network | 0.101 | 0.60919 | -3.492 | 0 | 0.388 |
| 22074 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-942-5p | 10 | CREBBP | Sponge network | -1.092 | 4.0E-5 | 0.126 | 0.15186 | 0.388 |
| 22075 | ACTA2-AS1 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CTGF | Sponge network | 0.194 | 0.64278 | 0.321 | 0.11682 | 0.388 |
| 22076 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-766-3p;hsa-miR-93-3p | 59 | IL6ST | Sponge network | 0.082 | 0.55654 | -0.402 | 0.00676 | 0.388 |
| 22077 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-942-5p | 40 | UNC80 | Sponge network | -1.439 | 4.0E-5 | -1.934 | 0 | 0.388 |
| 22078 | LPP-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-577 | 12 | TMEFF2 | Sponge network | -0.18 | 0.20343 | -2.426 | 0 | 0.388 |
| 22079 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-942-5p | 34 | TMEM132B | Sponge network | -2.62 | 0 | -0.83 | 0.01084 | 0.388 |
| 22080 | MEG3 |
hsa-miR-185-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-5p;hsa-miR-93-3p | 10 | DPP6 | Sponge network | 0.249 | 0.35902 | -3.777 | 0 | 0.388 |
| 22081 | EMX2OS |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 15 | SPTBN4 | Sponge network | -0.777 | 0.1134 | -0.786 | 0.01281 | 0.388 |
| 22082 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | MOB1B | Sponge network | 0.826 | 1.0E-5 | 0.197 | 0.15266 | 0.388 |
| 22083 | AP001347.6 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | RIMS1 | Sponge network | -1.161 | 0.00591 | -3.143 | 0 | 0.388 |
| 22084 | HAND2-AS1 |
hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-7-5p | 10 | CMKLR1 | Sponge network | -1.697 | 0.00889 | 0.288 | 0.17621 | 0.388 |
| 22085 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 20 | ACVR1 | Sponge network | -0.576 | 0.10121 | 0.279 | 0.00234 | 0.388 |
| 22086 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TBX18 | Sponge network | 0.61 | 0.01593 | 0.843 | 0.02945 | 0.388 |
| 22087 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | 0.559 | 0.00026 | 0.206 | 0.06751 | 0.388 |
| 22088 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 21 | HIP1 | Sponge network | 0.219 | 0.1516 | 0.774 | 0 | 0.388 |
| 22089 | TUG1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p | 10 | PDS5B | Sponge network | 0.972 | 0 | 0.259 | 0.00962 | 0.388 |
| 22090 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-93-5p | 35 | PIK3R1 | Sponge network | -0.227 | 0.31657 | -0.133 | 0.36854 | 0.388 |
| 22091 | TPTEP1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | KCTD8 | Sponge network | -1.216 | 0 | -3.359 | 0 | 0.388 |
| 22092 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-93-5p;hsa-miR-940 | 10 | GLI3 | Sponge network | 0.082 | 0.55397 | -0.615 | 0.01482 | 0.388 |
| 22093 | BVES-AS1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-132-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p | 17 | PAIP2 | Sponge network | -2.62 | 0 | -0.515 | 0 | 0.388 |
| 22094 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-98-5p | 37 | LIFR | Sponge network | -0.739 | 0.0574 | -1.794 | 0 | 0.388 |
| 22095 | H1FX-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-3p | 30 | GRIK3 | Sponge network | -0.206 | 0.12942 | -3.208 | 0 | 0.388 |
| 22096 | HAS2-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-222-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 11 | QKI | Sponge network | -0.011 | 0.967 | -0.333 | 0.04111 | 0.388 |
| 22097 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 31 | CADM1 | Sponge network | -0.576 | 0.10121 | -0.9 | 0.00084 | 0.388 |
| 22098 | U91328.19 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 28 | PCDH9 | Sponge network | -0.303 | 0.21002 | -1.823 | 4.0E-5 | 0.388 |
| 22099 | GS1-124K5.11 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p | 19 | DYNC1I1 | Sponge network | 0.494 | 0.00339 | -1.12 | 0.00107 | 0.388 |
| 22100 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 19 | CREBBP | Sponge network | 0.064 | 0.72934 | 0.126 | 0.15186 | 0.388 |
| 22101 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 19 | PRKAB2 | Sponge network | -0.583 | 0.14589 | -0.367 | 0.01371 | 0.388 |
| 22102 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-424-5p;hsa-miR-576-5p | 12 | LRRC7 | Sponge network | -0.932 | 0.00329 | -1.341 | 0.01193 | 0.388 |
| 22103 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-424-5p | 14 | EYA4 | Sponge network | -0.932 | 0.00329 | 0.3 | 0.36876 | 0.388 |
| 22104 | RP11-6O2.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 34 | TRPS1 | Sponge network | 0.331 | 0.57247 | -0.191 | 0.44109 | 0.388 |
| 22105 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-7-1-3p | 16 | TBX18 | Sponge network | -1.092 | 4.0E-5 | 0.843 | 0.02945 | 0.388 |
| 22106 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | LRP1B | Sponge network | -2.915 | 0.00015 | -2.163 | 0 | 0.388 |
| 22107 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 11 | SEPT6 | Sponge network | 0.335 | 0.20596 | -0.598 | 0.00181 | 0.388 |
| 22108 | AC011526.1 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-590-3p | 11 | PRKCB | Sponge network | 0.342 | 0.03129 | -1.313 | 2.0E-5 | 0.388 |
| 22109 | C4B-AS1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-629-5p;hsa-miR-940 | 12 | IGF1 | Sponge network | 2.306 | 9.0E-5 | -0.204 | 0.5898 | 0.388 |
| 22110 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-577 | 21 | AFF3 | Sponge network | 0.249 | 0.35902 | -2.373 | 0 | 0.388 |
| 22111 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-484;hsa-miR-628-5p;hsa-miR-629-3p | 12 | CDH23 | Sponge network | 0.811 | 0.03228 | -0.974 | 0.00034 | 0.388 |
| 22112 | RP4-798A10.7 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 13 | TBL1XR1 | Sponge network | 1.757 | 0 | 0.578 | 0 | 0.388 |
| 22113 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | LRP1B | Sponge network | -0.187 | 0.23787 | -2.163 | 0 | 0.388 |
| 22114 | LINC00473 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p | 10 | PDZD2 | Sponge network | -1.203 | 0.03881 | -0.909 | 3.0E-5 | 0.388 |
| 22115 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 17 | RICTOR | Sponge network | 0.832 | 0 | 0.388 | 0.00188 | 0.388 |
| 22116 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-577;hsa-miR-7-1-3p | 15 | TIMP3 | Sponge network | -0.418 | 0.00068 | -0.546 | 0.02307 | 0.388 |
| 22117 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | -0.342 | 0.02288 | -1.892 | 0 | 0.388 |
| 22118 | RP11-479G22.8 |
hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-374b-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 11 | NUAK1 | Sponge network | 1.308 | 0 | 0.904 | 0 | 0.388 |
| 22119 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-429 | 14 | PTPN14 | Sponge network | 1.757 | 0 | 0.144 | 0.34595 | 0.388 |
| 22120 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-660-5p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-96-5p | 49 | AFF4 | Sponge network | 0.078 | 0.55803 | -0.266 | 0.01565 | 0.388 |
| 22121 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-378a-5p | 17 | PBX1 | Sponge network | -1.375 | 0.08642 | -1.206 | 0 | 0.388 |
| 22122 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 53 | PTPRD | Sponge network | -1.216 | 0 | -1.005 | 0.00064 | 0.388 |
| 22123 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | EPHA5 | Sponge network | 0.225 | 0.30644 | -0.957 | 0.01733 | 0.388 |
| 22124 | CTC-429P9.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-769-5p;hsa-miR-940 | 27 | GRIK3 | Sponge network | 0.082 | 0.55397 | -3.208 | 0 | 0.388 |
| 22125 | LINC00654 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | PAK3 | Sponge network | 0.374 | 0.20054 | -0.628 | 0.07253 | 0.388 |
| 22126 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 18 | GRIN2A | Sponge network | -0.246 | 0.35034 | -2.161 | 0 | 0.388 |
| 22127 | HCG11 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-590-3p | 10 | IRS1 | Sponge network | 0.385 | 0.10067 | -0.026 | 0.88855 | 0.388 |
| 22128 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | FAT4 | Sponge network | -0.899 | 6.0E-5 | -0.354 | 0.14694 | 0.388 |
| 22129 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | NEDD4 | Sponge network | 0.488 | 0.00084 | 0.02 | 0.89834 | 0.388 |
| 22130 | AC141928.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-744-3p | 42 | DST | Sponge network | 0.853 | 0.15352 | -0.713 | 0.00096 | 0.388 |
| 22131 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 12 | DCLK1 | Sponge network | 0.199 | 0.38015 | -1.324 | 0.00014 | 0.388 |
| 22132 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-550a-3p | 44 | DTNA | Sponge network | 0.331 | 0.57247 | -1.648 | 1.0E-5 | 0.388 |
| 22133 | AC009948.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 25 | EPS15 | Sponge network | 0.532 | 0.00505 | -0.052 | 0.53998 | 0.388 |
| 22134 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 11 | FAM188A | Sponge network | -0.793 | 7.0E-5 | -0.44 | 0.00018 | 0.388 |
| 22135 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 32 | ARID5B | Sponge network | 1.302 | 7.0E-5 | 0.342 | 0.01676 | 0.388 |
| 22136 | C4B-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p | 12 | DLC1 | Sponge network | 2.306 | 9.0E-5 | -0.382 | 0.05038 | 0.388 |
| 22137 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 20 | PPP2R5C | Sponge network | -0.612 | 0.00094 | -0.314 | 3.0E-5 | 0.388 |
| 22138 | RP1-302G2.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p | 12 | UNC80 | Sponge network | -1.483 | 9.0E-5 | -1.934 | 0 | 0.388 |
| 22139 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | RELN | Sponge network | -0.487 | 0.02157 | -2.403 | 0 | 0.388 |
| 22140 | AP001347.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MED12L | Sponge network | -1.161 | 0.00591 | -0.194 | 0.55299 | 0.388 |
| 22141 | RP4-639F20.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 21 | MEF2C | Sponge network | -0.517 | 0.02935 | -0.499 | 0.01448 | 0.388 |
| 22142 | LINC01018 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TRIM58 | Sponge network | -2.915 | 0.00015 | -1.692 | 0 | 0.388 |
| 22143 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 47 | ZFHX4 | Sponge network | -0.899 | 6.0E-5 | 0.018 | 0.95574 | 0.388 |
| 22144 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | ERG | Sponge network | -0.942 | 0 | 0.002 | 0.99011 | 0.388 |
| 22145 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p | 37 | NTRK2 | Sponge network | 0.335 | 0.20596 | -0.736 | 0.06495 | 0.388 |
| 22146 | FAM66C |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 12 | FYN | Sponge network | -0.901 | 0.00019 | -0.131 | 0.37051 | 0.388 |
| 22147 | RP11-359B12.2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 18 | MAPK10 | Sponge network | 0.064 | 0.72934 | -1.774 | 0 | 0.388 |
| 22148 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 45 | AR | Sponge network | -0.342 | 0.02288 | -1.554 | 0 | 0.388 |
| 22149 | RP11-473I1.10 |
hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3613-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | CCDC6 | Sponge network | 0.673 | 0 | 0.27 | 0.02555 | 0.388 |
| 22150 | AC018890.6 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | MED12L | Sponge network | 1.61 | 0.00961 | -0.194 | 0.55299 | 0.388 |
| 22151 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p | 11 | KANK1 | Sponge network | -1.331 | 3.0E-5 | -0.55 | 0.00134 | 0.388 |
| 22152 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-589-5p;hsa-miR-874-3p | 10 | ST5 | Sponge network | 0.16 | 0.50785 | -0.78 | 0 | 0.388 |
| 22153 | H1FX-AS1 |
hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p | 10 | DNMT3A | Sponge network | -0.206 | 0.12942 | 0.302 | 0.0262 | 0.388 |
| 22154 | AC074289.1 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-7-5p | 17 | PRKCB | Sponge network | -0.326 | 0.14314 | -1.313 | 2.0E-5 | 0.388 |
| 22155 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FSTL5 | Sponge network | -0.222 | 0.32445 | -1.3 | 0.01619 | 0.388 |
| 22156 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 27 | MOB1B | Sponge network | 1.11 | 0 | 0.197 | 0.15266 | 0.388 |
| 22157 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p | 13 | BCL6 | Sponge network | 0.389 | 0.04293 | -0.211 | 0.19489 | 0.388 |
| 22158 | FENDRR |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | NAV3 | Sponge network | -1.281 | 0.00012 | 0.212 | 0.47729 | 0.388 |
| 22159 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 15 | CEP170 | Sponge network | -0.129 | 0.57177 | 0.943 | 0 | 0.388 |
| 22160 | FENDRR |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 43 | NOTCH2 | Sponge network | -1.281 | 0.00012 | 0.138 | 0.35163 | 0.388 |
| 22161 | LINC00641 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-98-5p | 64 | CADM2 | Sponge network | -0.222 | 0.32445 | -3.782 | 0 | 0.388 |
| 22162 | GAS5-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-374b-5p | 10 | UBE2D1 | Sponge network | 0.598 | 0 | 0.468 | 0 | 0.388 |
| 22163 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | RB1CC1 | Sponge network | 1.11 | 0 | 0.175 | 0.12559 | 0.388 |
| 22164 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | ADAMTSL3 | Sponge network | -0.727 | 0.04726 | -1.559 | 0 | 0.388 |
| 22165 | RP11-130L8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | SETBP1 | Sponge network | -0.663 | 0.10887 | -1.302 | 1.0E-5 | 0.388 |
| 22166 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | RCAN2 | Sponge network | 1.302 | 7.0E-5 | -0.33 | 0.15172 | 0.388 |
| 22167 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p | 11 | TMEFF2 | Sponge network | -0.693 | 0.05629 | -2.426 | 0 | 0.388 |
| 22168 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-940 | 42 | QKI | Sponge network | -0.773 | 0.04073 | -0.333 | 0.04111 | 0.388 |
| 22169 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-766-3p | 12 | EZH1 | Sponge network | -0.246 | 0.35034 | -0.564 | 1.0E-5 | 0.388 |
| 22170 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-577 | 17 | SCN9A | Sponge network | 0.419 | 0.0319 | -0.525 | 0.10593 | 0.388 |
| 22171 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | 0.768 | 7.0E-5 | 0.206 | 0.06751 | 0.388 |
| 22172 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TMEFF2 | Sponge network | -0.323 | 0.01372 | -2.426 | 0 | 0.388 |
| 22173 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 30 | MTSS1 | Sponge network | -0.563 | 0.02545 | -0.81 | 0.00035 | 0.388 |
| 22174 | RP11-305E6.4 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-590-3p | 12 | KDSR | Sponge network | 1.317 | 0 | 0.239 | 0.02216 | 0.388 |
| 22175 | RP4-639F20.1 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-542-3p | 10 | CSDE1 | Sponge network | -0.517 | 0.02935 | -0.493 | 0 | 0.388 |
| 22176 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | LRP1B | Sponge network | -0.487 | 0.02157 | -2.163 | 0 | 0.388 |
| 22177 | RP11-373D23.3 |
hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-503-5p | 11 | TOM1L2 | Sponge network | -0.285 | 0.2172 | -0.994 | 0 | 0.388 |
| 22178 | RP11-2H8.2 |
hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 11 | CHD1 | Sponge network | 1.089 | 0 | 0.322 | 0.00215 | 0.388 |
| 22179 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 32 | USP12 | Sponge network | -1.281 | 0.00012 | -0.102 | 0.40768 | 0.388 |
| 22180 | CTD-2245F17.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-629-3p | 14 | AR | Sponge network | -0.421 | 0.12556 | -1.554 | 0 | 0.388 |
| 22181 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | RASSF8 | Sponge network | -0.342 | 0.02288 | -0.122 | 0.65591 | 0.388 |
| 22182 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | CHD2 | Sponge network | 0.556 | 0.00298 | 0.049 | 0.55371 | 0.388 |
| 22183 | PSMG3-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-877-5p | 40 | GAS7 | Sponge network | -0.125 | 0.49414 | -0.555 | 0.01602 | 0.387 |
| 22184 | RP11-760H22.2 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | LRIG1 | Sponge network | 0.001 | 0.99753 | -0.537 | 0.00588 | 0.387 |
| 22185 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 79 | NTRK3 | Sponge network | -2.925 | 0 | -1.77 | 3.0E-5 | 0.387 |
| 22186 | SNX29P2 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | PCDH9 | Sponge network | 0.4 | 0.14611 | -1.823 | 4.0E-5 | 0.387 |
| 22187 | CTA-204B4.2 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-505-5p | 12 | WNK2 | Sponge network | 0.765 | 7.0E-5 | -0.352 | 0.31066 | 0.387 |
| 22188 | CTD-2314G24.2 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 45 | GRIK3 | Sponge network | 0.246 | 0.59912 | -3.208 | 0 | 0.387 |
| 22189 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-93-5p | 35 | SASH1 | Sponge network | -0.162 | 0.47687 | -0.502 | 0.00175 | 0.387 |
| 22190 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p | 25 | DDX6 | Sponge network | 1.063 | 0 | 0.091 | 0.36428 | 0.387 |
| 22191 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-96-5p;hsa-miR-98-5p | 20 | MITF | Sponge network | -0.18 | 0.20343 | -0.769 | 0.00022 | 0.387 |
| 22192 | RP4-798A10.7 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 10 | PHF6 | Sponge network | 1.757 | 0 | 0.785 | 0 | 0.387 |
| 22193 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p | 10 | PDS5B | Sponge network | 0.752 | 0 | 0.259 | 0.00962 | 0.387 |
| 22194 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 11 | PDGFC | Sponge network | -0.842 | 0.01361 | -0.33 | 0.06483 | 0.387 |
| 22195 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | ERBB4 | Sponge network | -2.117 | 0.00161 | -1.975 | 2.0E-5 | 0.387 |
| 22196 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TBX18 | Sponge network | -1.697 | 0.00889 | 0.843 | 0.02945 | 0.387 |
| 22197 | RP11-53O19.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p | 13 | STARD13 | Sponge network | -0.405 | 0.22013 | -0.187 | 0.27383 | 0.387 |
| 22198 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p | 17 | IGF2 | Sponge network | -0.737 | 0.06182 | 0.848 | 0.03842 | 0.387 |
| 22199 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 32 | CHD5 | Sponge network | 0.559 | 0.00026 | -0.922 | 0.01521 | 0.387 |
| 22200 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | RAP1A | Sponge network | -0.737 | 0.06182 | -0.929 | 0 | 0.387 |
| 22201 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-940 | 42 | CHD5 | Sponge network | 0.494 | 0.00339 | -0.922 | 0.01521 | 0.387 |
| 22202 | IGF2-AS |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-625-5p | 10 | NUAK1 | Sponge network | 3.339 | 0.00237 | 0.904 | 0 | 0.387 |
| 22203 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 39 | PAFAH1B1 | Sponge network | 0.082 | 0.55397 | -0.429 | 0 | 0.387 |
| 22204 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | USP12 | Sponge network | 0.349 | 0.01695 | -0.102 | 0.40768 | 0.387 |
| 22205 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SSBP2 | Sponge network | -0.021 | 0.93598 | -1.58 | 0 | 0.387 |
| 22206 | ARHGEF26-AS1 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-708-5p | 21 | FAM135B | Sponge network | -2.46 | 0 | -3.978 | 0 | 0.387 |
| 22207 | RP11-307B6.3 |
hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-194-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-421 | 12 | RAP1A | Sponge network | -4.463 | 0.00025 | -0.929 | 0 | 0.387 |
| 22208 | AC093110.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 17 | ABL2 | Sponge network | 1.044 | 2.0E-5 | 0.82 | 0 | 0.387 |
| 22209 | RP11-678G14.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p | 13 | CADM3 | Sponge network | -4.477 | 0 | -3.663 | 0 | 0.387 |
| 22210 | RP11-888D10.4 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 21 | CREB1 | Sponge network | 0.702 | 7.0E-5 | 0.203 | 0.00537 | 0.387 |
| 22211 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | PDS5B | Sponge network | 0.559 | 0.00026 | 0.259 | 0.00962 | 0.387 |
| 22212 | FZD10-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | THSD7B | Sponge network | -0.727 | 0.04726 | 0.154 | 0.74723 | 0.387 |
| 22213 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | IL6ST | Sponge network | -0.021 | 0.93598 | -0.402 | 0.00676 | 0.387 |
| 22214 | GNG12-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | PRKCB | Sponge network | -0.793 | 7.0E-5 | -1.313 | 2.0E-5 | 0.387 |
| 22215 | LINC00641 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ARNT | Sponge network | -0.222 | 0.32445 | -0.08 | 0.26073 | 0.387 |
| 22216 | RP11-123M6.2 |
hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-7-1-3p | 13 | IL6ST | Sponge network | 0.543 | 0.12356 | -0.402 | 0.00676 | 0.387 |
| 22217 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-335-5p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | TP53INP1 | Sponge network | 0.796 | 4.0E-5 | -0.496 | 0.00064 | 0.387 |
| 22218 | RP11-13K12.5 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | RNF144A | Sponge network | -0.318 | 0.65847 | 0.594 | 0.0009 | 0.387 |
| 22219 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p | 14 | PTGFR | Sponge network | -0.246 | 0.35034 | -0.233 | 0.45421 | 0.387 |
| 22220 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-942-5p | 15 | ARHGAP28 | Sponge network | -0.72 | 0.24239 | -0.911 | 0.00013 | 0.387 |
| 22221 | WDFY3-AS2 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | ZDBF2 | Sponge network | -0.942 | 0 | -0.998 | 0.00025 | 0.387 |
| 22222 | LINC00473 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3913-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-942-5p | 37 | HLF | Sponge network | -1.203 | 0.03881 | -2.443 | 0 | 0.387 |
| 22223 | RP11-195F19.9 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 18 | TRPS1 | Sponge network | -0.726 | 0.00132 | -0.191 | 0.44109 | 0.387 |
| 22224 | BVES-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-576-5p | 12 | HERC1 | Sponge network | -2.62 | 0 | -0.196 | 0.08544 | 0.387 |
| 22225 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | GNA13 | Sponge network | -0.222 | 0.32445 | 0.007 | 0.93884 | 0.387 |
| 22226 | LPP-AS2 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-96-5p | 11 | PAK3 | Sponge network | -0.18 | 0.20343 | -0.628 | 0.07253 | 0.387 |
| 22227 | RP11-758N13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-744-5p | 13 | FGFR1 | Sponge network | 2.939 | 3.0E-5 | -0.509 | 0.03957 | 0.387 |
| 22228 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 43 | CHD5 | Sponge network | -0.355 | 0.0077 | -0.922 | 0.01521 | 0.387 |
| 22229 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 31 | RICTOR | Sponge network | -0.323 | 0.01372 | 0.388 | 0.00188 | 0.387 |
| 22230 | LINC00654 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 17 | RPS6KA2 | Sponge network | 0.374 | 0.20054 | -0.57 | 0.00013 | 0.387 |
| 22231 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SON | Sponge network | 0.225 | 0.30644 | 0.168 | 0.04406 | 0.387 |
| 22232 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | DPYD | Sponge network | -0.769 | 0.00035 | -0.736 | 0.00076 | 0.387 |
| 22233 | RP11-359B12.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-940 | 24 | SIRT1 | Sponge network | 0.064 | 0.72934 | 0.109 | 0.2593 | 0.387 |
| 22234 | LINC00969 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-3p | 17 | ATRX | Sponge network | 0.683 | 1.0E-5 | 0.261 | 0.04408 | 0.387 |
| 22235 | SH3RF3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 47 | LPP | Sponge network | 0.889 | 9.0E-5 | -0.459 | 0.04475 | 0.387 |
| 22236 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | GALNT13 | Sponge network | -2.452 | 0 | -0.857 | 0.07439 | 0.387 |
| 22237 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 48 | TMEM132B | Sponge network | -0.737 | 0.06182 | -0.83 | 0.01084 | 0.387 |
| 22238 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | ANK2 | Sponge network | 0.131 | 0.8436 | -1.603 | 1.0E-5 | 0.387 |
| 22239 | DNM3OS |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 12 | SEPT6 | Sponge network | 0.572 | 0.01722 | -0.598 | 0.00181 | 0.387 |
| 22240 | RP5-1198O20.4 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-940 | 11 | PHOX2B | Sponge network | 0.997 | 0.00066 | -2.68 | 0 | 0.387 |
| 22241 | RP11-927P21.1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p | 23 | ABI2 | Sponge network | 0.921 | 0.00118 | 0.175 | 0.14787 | 0.387 |
| 22242 | RP1-122P22.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 27 | RUNX1T1 | Sponge network | 0.065 | 0.73468 | -0.344 | 0.2374 | 0.387 |
| 22243 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 14 | PKNOX1 | Sponge network | 0.537 | 0.00139 | 0.085 | 0.28917 | 0.387 |
| 22244 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | RECK | Sponge network | -0.727 | 0.04726 | -1.043 | 0 | 0.387 |
| 22245 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | MOB1B | Sponge network | 0.392 | 0.0278 | 0.197 | 0.15266 | 0.387 |
| 22246 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p | 11 | ATRX | Sponge network | 1.46 | 1.0E-5 | 0.261 | 0.04408 | 0.387 |
| 22247 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p | 12 | IGF2 | Sponge network | 0.225 | 0.30644 | 0.848 | 0.03842 | 0.387 |
| 22248 | FAM157B |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-505-5p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 0.92 | 0 | 0.578 | 0 | 0.387 |
| 22249 | PTOV1-AS1 |
hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-378a-3p;hsa-miR-378c | 10 | BRD3 | Sponge network | 0.87 | 0 | 0.37 | 0.00013 | 0.387 |
| 22250 | MIR143HG |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 10 | DIRAS1 | Sponge network | -0.739 | 0.0574 | -2.518 | 0 | 0.387 |
| 22251 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 47 | ADARB1 | Sponge network | -0.355 | 0.0077 | -0.335 | 0.06631 | 0.387 |
| 22252 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | EDA2R | Sponge network | -2.915 | 0.00015 | -0.587 | 0.01598 | 0.387 |
| 22253 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | CXXC4 | Sponge network | -0.942 | 0 | -0.433 | 0.21055 | 0.387 |
| 22254 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 15 | KCNAB1 | Sponge network | 0.083 | 0.66405 | -0.755 | 2.0E-5 | 0.387 |
| 22255 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | LRRC7 | Sponge network | -0.187 | 0.23787 | -1.341 | 0.01193 | 0.387 |
| 22256 | LINC00173 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-96-5p | 12 | ELMO1 | Sponge network | 0.056 | 0.8494 | 0.04 | 0.83147 | 0.387 |
| 22257 | KB-1460A1.5 |
hsa-miR-101-3p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 15 | KIF5B | Sponge network | 0.603 | 0.00127 | 0.362 | 0.00066 | 0.387 |
| 22258 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-629-5p;hsa-miR-942-5p | 28 | UNC80 | Sponge network | -0.517 | 0.02935 | -1.934 | 0 | 0.387 |
| 22259 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 13 | MAF | Sponge network | -0.583 | 0.14589 | -1.257 | 0 | 0.387 |
| 22260 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p | 18 | DYNC1I1 | Sponge network | 0.101 | 0.60919 | -1.12 | 0.00107 | 0.387 |
| 22261 | CTA-228A9.3 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-376c-3p;hsa-miR-766-3p | 13 | TBL1XR1 | Sponge network | 2.466 | 0 | 0.578 | 0 | 0.387 |
| 22262 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | AHNAK | Sponge network | 0.101 | 0.60919 | -1.207 | 0 | 0.387 |
| 22263 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-539-5p | 27 | NRXN1 | Sponge network | -0.154 | 0.78939 | -3.778 | 0 | 0.387 |
| 22264 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-874-3p;hsa-miR-940 | 16 | CRTC3 | Sponge network | 0.349 | 0.01695 | 0.164 | 0.16786 | 0.387 |
| 22265 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | MOB1B | Sponge network | -0.612 | 0.00094 | 0.197 | 0.15266 | 0.387 |
| 22266 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | MCC | Sponge network | -0.053 | 0.80685 | -1.005 | 0 | 0.387 |
| 22267 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-5p | 21 | CLASP2 | Sponge network | 0.214 | 0.18246 | -0.038 | 0.71 | 0.387 |
| 22268 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-940;hsa-miR-96-5p | 16 | POU2F2 | Sponge network | -2.452 | 0 | -0.233 | 0.32214 | 0.387 |
| 22269 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | ST5 | Sponge network | -0.49 | 0.04946 | -0.78 | 0 | 0.387 |
| 22270 | GNG12-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 17 | RIMBP2 | Sponge network | -0.793 | 7.0E-5 | -1.968 | 5.0E-5 | 0.387 |
| 22271 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 42 | IL6ST | Sponge network | 0.614 | 1.0E-5 | -0.402 | 0.00676 | 0.387 |
| 22272 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 19 | ZNF770 | Sponge network | 0.91 | 0 | 0.206 | 0.06751 | 0.387 |
| 22273 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 15 | SSBP2 | Sponge network | -0.295 | 0.02604 | -1.58 | 0 | 0.387 |
| 22274 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-590-3p | 18 | GAS7 | Sponge network | 0.178 | 0.29106 | -0.555 | 0.01602 | 0.387 |
| 22275 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-96-5p | 38 | ZFHX4 | Sponge network | 0.331 | 0.57247 | 0.018 | 0.95574 | 0.387 |
| 22276 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | SYNE1 | Sponge network | 0.083 | 0.66405 | -0.738 | 0.00316 | 0.387 |
| 22277 | FENDRR |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | TET1 | Sponge network | -1.281 | 0.00012 | -0.135 | 0.52138 | 0.387 |
| 22278 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | GPC6 | Sponge network | 0.342 | 0.03129 | -0.054 | 0.81465 | 0.387 |
| 22279 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-429 | 16 | CLIP1 | Sponge network | -2.62 | 0 | -0.215 | 0.08684 | 0.387 |
| 22280 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | TRIM33 | Sponge network | 0.439 | 0.00313 | 0.402 | 7.0E-5 | 0.387 |
| 22281 | RP11-750H9.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-224-5p | 10 | RUNX1T1 | Sponge network | 0.349 | 0.13152 | -0.344 | 0.2374 | 0.387 |
| 22282 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 35 | MOB1B | Sponge network | -0.793 | 7.0E-5 | 0.197 | 0.15266 | 0.387 |
| 22283 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-940 | 24 | AKAP11 | Sponge network | 0.331 | 0.57247 | 0.196 | 0.09825 | 0.387 |
| 22284 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | UBE4B | Sponge network | -0.355 | 0.0077 | -0.235 | 0.007 | 0.387 |
| 22285 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | PDGFRA | Sponge network | 0.194 | 0.64278 | -0.372 | 0.0037 | 0.387 |
| 22286 | ZNF582-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-940;hsa-miR-96-5p | 13 | POU2F2 | Sponge network | -1.349 | 0.00017 | -0.233 | 0.32214 | 0.387 |
| 22287 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 16 | MOB1B | Sponge network | 1.923 | 0 | 0.197 | 0.15266 | 0.387 |
| 22288 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | RARB | Sponge network | -1.349 | 0.00017 | -0.825 | 2.0E-5 | 0.387 |
| 22289 | AC004540.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-5p | 15 | PTPRT | Sponge network | -2.549 | 0.00528 | -0.907 | 0.03727 | 0.387 |
| 22290 | RP11-571M6.17 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | MDM4 | Sponge network | 1.013 | 0 | 0.345 | 0.00762 | 0.387 |
| 22291 | MIR143HG |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 24 | ERRFI1 | Sponge network | -0.739 | 0.0574 | -0.798 | 1.0E-5 | 0.387 |
| 22292 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-5p | 14 | SYNE1 | Sponge network | 0.622 | 0.00015 | -0.738 | 0.00316 | 0.387 |
| 22293 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 20 | RBMS3 | Sponge network | -0.206 | 0.12942 | -0.519 | 0.03551 | 0.387 |
| 22294 | RP11-175O19.4 |
hsa-miR-10b-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p | 20 | ARHGEF12 | Sponge network | 0.917 | 0 | 0.09 | 0.35693 | 0.387 |
| 22295 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-455-5p;hsa-miR-577 | 12 | SLC6A15 | Sponge network | -0.473 | 0.07602 | -2.373 | 2.0E-5 | 0.387 |
| 22296 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p | 10 | PTPRB | Sponge network | 0.811 | 0.03228 | 0.105 | 0.47122 | 0.387 |
| 22297 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | CXXC4 | Sponge network | -1.203 | 0.03881 | -0.433 | 0.21055 | 0.387 |
| 22298 | RP11-119F7.5 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p | 14 | ABL1 | Sponge network | 0.225 | 0.30644 | -0.215 | 0.07804 | 0.387 |
| 22299 | JAZF1-AS1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484 | 16 | ABL1 | Sponge network | -0.769 | 0.00035 | -0.215 | 0.07804 | 0.387 |
| 22300 | CTA-363E19.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p | 13 | CBL | Sponge network | 1.055 | 0 | 0.291 | 0.01267 | 0.387 |
| 22301 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-3p | 15 | ITGB3 | Sponge network | -0.323 | 0.01372 | -0.011 | 0.95751 | 0.387 |
| 22302 | BZRAP1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-424-3p | 13 | SYNM | Sponge network | -0.465 | 0.04703 | -2.309 | 1.0E-5 | 0.387 |
| 22303 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p | 23 | RASSF2 | Sponge network | 0.622 | 0.00015 | -0.273 | 0.21961 | 0.387 |
| 22304 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | EYA4 | Sponge network | 1.302 | 7.0E-5 | 0.3 | 0.36876 | 0.387 |
| 22305 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 19 | LRRC55 | Sponge network | -0.487 | 0.02157 | -0.026 | 0.93163 | 0.387 |
| 22306 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-769-5p | 10 | RCAN2 | Sponge network | 0.381 | 0.25967 | -0.33 | 0.15172 | 0.387 |
| 22307 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | SUFU | Sponge network | -0.421 | 0.0114 | -0.04 | 0.65489 | 0.387 |
| 22308 | ADAMTS9-AS2 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | RASSF3 | Sponge network | -2.452 | 0 | -0.226 | 0.11691 | 0.387 |
| 22309 | RP11-48B3.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 14 | SETBP1 | Sponge network | 1.757 | 0 | -1.302 | 1.0E-5 | 0.387 |
| 22310 | AC005618.6 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 23 | RASSF8 | Sponge network | 0.862 | 0.06213 | -0.122 | 0.65591 | 0.387 |
| 22311 | AGAP11 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p | 17 | GJA1 | Sponge network | -1.645 | 0 | -0.485 | 0.02179 | 0.387 |
| 22312 | RP11-672A2.4 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p | 11 | PTPN14 | Sponge network | 1.344 | 0 | 0.144 | 0.34595 | 0.387 |
| 22313 | BVES-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-629-3p | 10 | SPTBN4 | Sponge network | -2.62 | 0 | -0.786 | 0.01281 | 0.387 |
| 22314 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 11 | FGF5 | Sponge network | -0.246 | 0.35034 | 0.549 | 0.28659 | 0.387 |
| 22315 | RP11-244O19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 28 | PRDM2 | Sponge network | -0.096 | 0.67958 | -0.258 | 0.00721 | 0.387 |
| 22316 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 18 | LHX6 | Sponge network | -1.697 | 0.00889 | -0.087 | 0.69193 | 0.387 |
| 22317 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 57 | CCND2 | Sponge network | -0.737 | 0.06182 | -0.267 | 0.3112 | 0.387 |
| 22318 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | PTPRB | Sponge network | 0.249 | 0.35902 | 0.105 | 0.47122 | 0.387 |
| 22319 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | -0.465 | 0.04703 | -1.047 | 0.00364 | 0.387 |
| 22320 | NEAT1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-942-5p | 13 | RB1CC1 | Sponge network | 0.705 | 0.00014 | 0.175 | 0.12559 | 0.387 |
| 22321 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-671-5p | 21 | KIT | Sponge network | -0.162 | 0.47687 | -2.184 | 0 | 0.387 |
| 22322 | RASSF8-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p | 12 | KCTD8 | Sponge network | 0.335 | 0.20596 | -3.359 | 0 | 0.387 |
| 22323 | RP11-6O2.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-624-5p | 17 | CDH19 | Sponge network | 0.331 | 0.57247 | -3.434 | 0 | 0.387 |
| 22324 | PSMD5-AS1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | TSC1 | Sponge network | 0.569 | 0.00067 | 0.103 | 0.26071 | 0.387 |
| 22325 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 12 | PLA2R1 | Sponge network | -2.46 | 0 | -0.265 | 0.24676 | 0.387 |
| 22326 | RP5-1198O20.4 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 14 | RHOB | Sponge network | 0.997 | 0.00066 | -1.052 | 0 | 0.387 |
| 22327 | RP11-927P21.1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-940 | 12 | BAZ2B | Sponge network | 0.921 | 0.00118 | -0.276 | 0.02833 | 0.387 |
| 22328 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429 | 10 | CDH10 | Sponge network | -2.69 | 0.0001 | -2.397 | 0 | 0.387 |
| 22329 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p | 26 | KCNA6 | Sponge network | -0.096 | 0.67958 | -1.531 | 0.00013 | 0.387 |
| 22330 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-625-5p | 35 | CBX5 | Sponge network | 0.214 | 0.18246 | 0.165 | 0.24446 | 0.387 |
| 22331 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | SEMA3C | Sponge network | -0.738 | 0.21233 | -0.272 | 0.31153 | 0.387 |
| 22332 | RP11-156P1.3 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p | 27 | CTIF | Sponge network | 0.214 | 0.18246 | -0.389 | 0.00549 | 0.387 |
| 22333 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 44 | THRB | Sponge network | -0.739 | 0.00044 | -1.117 | 0.0004 | 0.387 |
| 22334 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 48 | SOX5 | Sponge network | -1.216 | 0 | -1.091 | 4.0E-5 | 0.387 |
| 22335 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-940 | 22 | SNX29 | Sponge network | 0.16 | 0.50785 | -0.34 | 0.01371 | 0.387 |
| 22336 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-942-5p | 30 | TMEM132B | Sponge network | -0.358 | 0.17255 | -0.83 | 0.01084 | 0.387 |
| 22337 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | ADAMTSL3 | Sponge network | -0.738 | 0.21233 | -1.559 | 0 | 0.387 |
| 22338 | LINC00865 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 12 | RHOB | Sponge network | -1.249 | 7.0E-5 | -1.052 | 0 | 0.387 |
| 22339 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-625-5p;hsa-miR-655-3p | 13 | ARID2 | Sponge network | 0.537 | 0.00139 | 0.235 | 0.02697 | 0.387 |
| 22340 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | PGR | Sponge network | -0.053 | 0.80685 | -1.704 | 0 | 0.387 |
| 22341 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-424-5p | 10 | LRRC55 | Sponge network | 1.344 | 0 | -0.026 | 0.93163 | 0.387 |
| 22342 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 33 | LRIG1 | Sponge network | -2.46 | 0 | -0.537 | 0.00588 | 0.387 |
| 22343 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 10 | HACE1 | Sponge network | 0.706 | 0 | -0.072 | 0.55239 | 0.387 |
| 22344 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-3p | 36 | ZFHX4 | Sponge network | -0.227 | 0.31657 | 0.018 | 0.95574 | 0.387 |
| 22345 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 29 | CREB3L2 | Sponge network | -4.546 | 0 | -0.525 | 2.0E-5 | 0.387 |
| 22346 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | TSC1 | Sponge network | 0.614 | 1.0E-5 | 0.103 | 0.26071 | 0.387 |
| 22347 | LINC00957 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | BCL6 | Sponge network | -0.421 | 0.0114 | -0.211 | 0.19489 | 0.387 |
| 22348 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | NCOA1 | Sponge network | 0.101 | 0.60919 | -0.432 | 0 | 0.387 |
| 22349 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-495-3p | 22 | RICTOR | Sponge network | 0.752 | 0 | 0.388 | 0.00188 | 0.387 |
| 22350 | BDNF-AS |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-340-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 11 | KCNAB1 | Sponge network | -0.668 | 3.0E-5 | -0.755 | 2.0E-5 | 0.387 |
| 22351 | LINC00094 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-486-5p | 12 | DNMT3A | Sponge network | 0.253 | 0.09565 | 0.302 | 0.0262 | 0.387 |
| 22352 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-660-5p;hsa-miR-93-3p | 32 | RUNX1T1 | Sponge network | -0.72 | 0.24239 | -0.344 | 0.2374 | 0.387 |
| 22353 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | NIN | Sponge network | 0.299 | 0.57722 | -0.253 | 0.13794 | 0.387 |
| 22354 | MALAT1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 35 | ARHGEF12 | Sponge network | 0.782 | 0.00016 | 0.09 | 0.35693 | 0.387 |
| 22355 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-590-5p | 10 | AKAP12 | Sponge network | 0.395 | 0.15451 | -0.932 | 0.0028 | 0.387 |
| 22356 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-493-5p;hsa-miR-577;hsa-miR-654-3p | 22 | MYBL1 | Sponge network | 0.637 | 0.00069 | 0.494 | 0.02152 | 0.387 |
| 22357 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 72 | TCF4 | Sponge network | -2.46 | 0 | 0.016 | 0.92271 | 0.387 |
| 22358 | RP11-968O1.5 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-532-5p | 29 | IL6ST | Sponge network | -0.829 | 0.01343 | -0.402 | 0.00676 | 0.387 |
| 22359 | LINC00472 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p | 12 | FAM172A | Sponge network | -0.842 | 0.01361 | -0.57 | 0 | 0.387 |
| 22360 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 14 | LRRC55 | Sponge network | -1.161 | 0.00591 | -0.026 | 0.93163 | 0.387 |
| 22361 | RP11-395G23.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | ABI2 | Sponge network | 0.381 | 0.25967 | 0.175 | 0.14787 | 0.387 |
| 22362 | NDUFA6-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 83 | LPP | Sponge network | -0.355 | 0.0077 | -0.459 | 0.04475 | 0.387 |
| 22363 | RP3-467N11.1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p;hsa-miR-495-3p | 13 | PHF6 | Sponge network | 0.953 | 0 | 0.785 | 0 | 0.387 |
| 22364 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 11 | NR2C2 | Sponge network | 1.262 | 0 | 0.21 | 0.03224 | 0.387 |
| 22365 | RP11-83A24.2 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-409-5p;hsa-miR-590-3p | 12 | TPR | Sponge network | 0.417 | 0.00445 | 0.582 | 0 | 0.387 |
| 22366 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 11 | LATS2 | Sponge network | 0.176 | 0.37402 | 0.159 | 0.2304 | 0.387 |
| 22367 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p | 11 | ABCA10 | Sponge network | -2.69 | 0.0001 | -0.571 | 0.03674 | 0.387 |
| 22368 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 30 | BNC2 | Sponge network | 0.174 | 0.30034 | -0.491 | 0.09988 | 0.387 |
| 22369 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 18 | RICTOR | Sponge network | 1.266 | 0 | 0.388 | 0.00188 | 0.387 |
| 22370 | RP11-526I2.5 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 10 | PHF6 | Sponge network | 1.054 | 1.0E-5 | 0.785 | 0 | 0.386 |
| 22371 | LINC00654 |
hsa-miR-126-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | IRS1 | Sponge network | 0.374 | 0.20054 | -0.026 | 0.88855 | 0.386 |
| 22372 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 13 | ABCB5 | Sponge network | 0.335 | 0.20596 | -0.956 | 0.04077 | 0.386 |
| 22373 | MEG3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-664a-5p | 18 | SOX11 | Sponge network | 0.249 | 0.35902 | 2.68 | 0 | 0.386 |
| 22374 | ZNF667-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 21 | KCNJ5 | Sponge network | -0.976 | 0.00025 | -0.281 | 0.3339 | 0.386 |
| 22375 | MALAT1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | PIK3CA | Sponge network | 0.782 | 0.00016 | 0.195 | 0.06908 | 0.386 |
| 22376 | LINC00641 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-5p | 11 | MLLT11 | Sponge network | -0.222 | 0.32445 | -0.661 | 0.01733 | 0.386 |
| 22377 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | CAV1 | Sponge network | -0.222 | 0.32445 | -1.013 | 6.0E-5 | 0.386 |
| 22378 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | DKK3 | Sponge network | -0.737 | 0.10822 | -0.052 | 0.77783 | 0.386 |
| 22379 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | ST6GALNAC3 | Sponge network | 0.335 | 0.20596 | -0.987 | 0 | 0.386 |
| 22380 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 0.222 | 0.29133 | 0.943 | 0 | 0.386 |
| 22381 | RP11-571M6.17 |
hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | 1.013 | 0 | 0.206 | 0.06751 | 0.386 |
| 22382 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 18 | GRIA3 | Sponge network | -0.358 | 0.17255 | -1.956 | 0 | 0.386 |
| 22383 | USP3-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-940 | 15 | GALNT13 | Sponge network | -0.162 | 0.47687 | -0.857 | 0.07439 | 0.386 |
| 22384 | RP11-53O19.3 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | USP9X | Sponge network | 1.205 | 0 | 0.222 | 0.01689 | 0.386 |
| 22385 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p | 11 | DCLK1 | Sponge network | -0.351 | 0.11358 | -1.324 | 0.00014 | 0.386 |
| 22386 | LINC00861 |
hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 13 | QKI | Sponge network | 0.911 | 0.00854 | -0.333 | 0.04111 | 0.386 |
| 22387 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-330-5p | 15 | CNTN1 | Sponge network | -0.829 | 0.01343 | -2.594 | 0 | 0.386 |
| 22388 | HOXB-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p | 21 | PRKCB | Sponge network | -0.154 | 0.53954 | -1.313 | 2.0E-5 | 0.386 |
| 22389 | HHIP-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | GALNT13 | Sponge network | -0.737 | 0.10822 | -0.857 | 0.07439 | 0.386 |
| 22390 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p | 33 | CBX5 | Sponge network | 1.757 | 0 | 0.165 | 0.24446 | 0.386 |
| 22391 | LINC00865 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-935 | 21 | ARNT2 | Sponge network | -1.249 | 7.0E-5 | -0.332 | 0.25851 | 0.386 |
| 22392 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | NEDD4 | Sponge network | -0.942 | 0 | 0.02 | 0.89834 | 0.386 |
| 22393 | ANKRD10-IT1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p | 11 | SLC5A3 | Sponge network | 1.739 | 0 | 0.493 | 5.0E-5 | 0.386 |
| 22394 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p | 10 | PSIP1 | Sponge network | 1.153 | 0 | -0.005 | 0.96897 | 0.386 |
| 22395 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | RECK | Sponge network | 0.16 | 0.50785 | -1.043 | 0 | 0.386 |
| 22396 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-375 | 10 | DOCK4 | Sponge network | 0.858 | 3.0E-5 | 0.502 | 0.00108 | 0.386 |
| 22397 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 38 | RBMS3 | Sponge network | 0.219 | 0.1516 | -0.519 | 0.03551 | 0.386 |
| 22398 | PWAR6 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 34 | PTPRT | Sponge network | -1.575 | 6.0E-5 | -0.907 | 0.03727 | 0.386 |
| 22399 | LINC00578 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-505-3p;hsa-miR-589-3p | 15 | KCNJ5 | Sponge network | 0.811 | 0.03228 | -0.281 | 0.3339 | 0.386 |
| 22400 | RP11-48B3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 29 | RUNX1T1 | Sponge network | 1.757 | 0 | -0.344 | 0.2374 | 0.386 |
| 22401 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 15 | ZNF770 | Sponge network | 1.294 | 0 | 0.206 | 0.06751 | 0.386 |
| 22402 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 13 | GPAM | Sponge network | 0.539 | 0.03722 | 0.142 | 0.29732 | 0.386 |
| 22403 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | NCAM2 | Sponge network | -0.769 | 0.00035 | -0.482 | 0.22565 | 0.386 |
| 22404 | RP11-13K12.5 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p | 12 | SPG20 | Sponge network | -0.318 | 0.65847 | -1.16 | 0 | 0.386 |
| 22405 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-29a-5p | 11 | CNTNAP3 | Sponge network | -0.829 | 0.01343 | -1.465 | 0.00033 | 0.386 |
| 22406 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p | 24 | PBX1 | Sponge network | -0.053 | 0.80685 | -1.206 | 0 | 0.386 |
| 22407 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | LRRC55 | Sponge network | -0.72 | 0.24239 | -0.026 | 0.93163 | 0.386 |
| 22408 | RP11-403I13.8 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | NAV3 | Sponge network | 0.619 | 0.00157 | 0.212 | 0.47729 | 0.386 |
| 22409 | LINC00473 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 19 | PCDH17 | Sponge network | -1.203 | 0.03881 | 0.731 | 2.0E-5 | 0.386 |
| 22410 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p | 42 | PAFAH1B1 | Sponge network | -1.645 | 0 | -0.429 | 0 | 0.386 |
| 22411 | RP11-67L2.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 13 | DLG1 | Sponge network | 0.72 | 0.0001 | -0.014 | 0.8926 | 0.386 |
| 22412 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 25 | RARB | Sponge network | -1.216 | 0 | -0.825 | 2.0E-5 | 0.386 |
| 22413 | LINC01018 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 74 | LPP | Sponge network | -2.915 | 0.00015 | -0.459 | 0.04475 | 0.386 |
| 22414 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 29 | CLASP2 | Sponge network | 0.267 | 0.2937 | -0.038 | 0.71 | 0.386 |
| 22415 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 20 | CBFA2T3 | Sponge network | -0.513 | 0.04703 | -1.221 | 1.0E-5 | 0.386 |
| 22416 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-655-3p | 18 | LATS1 | Sponge network | 1.601 | 0 | 0.109 | 0.34208 | 0.386 |
| 22417 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-495-3p | 14 | DDX6 | Sponge network | 1.205 | 0 | 0.091 | 0.36428 | 0.386 |
| 22418 | LINC00702 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | -0.737 | 0.06182 | -0.001 | 0.99669 | 0.386 |
| 22419 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-539-5p | 15 | CLASP2 | Sponge network | 0.946 | 0 | -0.038 | 0.71 | 0.386 |
| 22420 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 16 | DKK3 | Sponge network | 0.246 | 0.59912 | -0.052 | 0.77783 | 0.386 |
| 22421 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 33 | BHLHE41 | Sponge network | -0.49 | 0.04946 | 0.353 | 0.12753 | 0.386 |
| 22422 | RP4-717I23.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 13 | ERCC4 | Sponge network | 0.637 | 0.00069 | 0.126 | 0.15266 | 0.386 |
| 22423 | PART1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 69 | RUNX1T1 | Sponge network | -2.925 | 0 | -0.344 | 0.2374 | 0.386 |
| 22424 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 19 | LRRC55 | Sponge network | -1.203 | 0.03881 | -0.026 | 0.93163 | 0.386 |
| 22425 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-98-5p | 20 | USP12 | Sponge network | 0.056 | 0.81195 | -0.102 | 0.40768 | 0.386 |
| 22426 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | ST6GALNAC3 | Sponge network | -1.349 | 0.00017 | -0.987 | 0 | 0.386 |
| 22427 | A1BG-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-93-3p | 11 | SUFU | Sponge network | -0.164 | 0.45106 | -0.04 | 0.65489 | 0.386 |
| 22428 | LINC00476 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 22 | GLIPR1 | Sponge network | -0.615 | 0.00015 | 0.146 | 0.3276 | 0.386 |
| 22429 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -2.46 | 0 | -1.692 | 0 | 0.386 |
| 22430 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 21 | NR2C2 | Sponge network | 0.569 | 0.00067 | 0.21 | 0.03224 | 0.386 |
| 22431 | RP11-119F7.5 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | 0.225 | 0.30644 | 0.098 | 0.40994 | 0.386 |
| 22432 | LINC00702 |
hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-660-5p | 10 | DNER | Sponge network | -0.737 | 0.06182 | -3.787 | 0 | 0.386 |
| 22433 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p | 15 | RASSF2 | Sponge network | -0.309 | 0.1145 | -0.273 | 0.21961 | 0.386 |
| 22434 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p | 20 | KCNA6 | Sponge network | -0.154 | 0.53954 | -1.531 | 0.00013 | 0.386 |
| 22435 | CTD-2089N3.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-455-5p | 10 | TSHZ3 | Sponge network | -0.314 | 0.62033 | -0.556 | 0.01516 | 0.386 |
| 22436 | RP11-67L2.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 38 | IL17RD | Sponge network | 0.72 | 0.0001 | -0.019 | 0.94873 | 0.386 |
| 22437 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 25 | CBFA2T3 | Sponge network | -2.46 | 0 | -1.221 | 1.0E-5 | 0.386 |
| 22438 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-223-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | EPHA3 | Sponge network | -0.021 | 0.93598 | 0.185 | 0.53588 | 0.386 |
| 22439 | HOXA-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 33 | ABI2 | Sponge network | 0.61 | 0.01593 | 0.175 | 0.14787 | 0.386 |
| 22440 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 10 | PHC3 | Sponge network | 1.597 | 0 | 0.212 | 0.0499 | 0.386 |
| 22441 | LINC00969 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 11 | BCLAF1 | Sponge network | 0.683 | 1.0E-5 | 0.211 | 0.00684 | 0.386 |
| 22442 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | RCAN2 | Sponge network | 0.862 | 0.06213 | -0.33 | 0.15172 | 0.386 |
| 22443 | RP11-156P1.3 |
hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | 0.214 | 0.18246 | 0.154 | 0.74723 | 0.386 |
| 22444 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | EPHA5 | Sponge network | -0.611 | 0.09607 | -0.957 | 0.01733 | 0.386 |
| 22445 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-652-3p | 27 | ZBTB4 | Sponge network | 0.16 | 0.50785 | -0.739 | 0 | 0.386 |
| 22446 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-655-3p | 26 | PIK3C2A | Sponge network | 0.72 | 0.0001 | 0.061 | 0.53776 | 0.386 |
| 22447 | RP13-516M14.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p | 26 | DMD | Sponge network | 0.389 | 0.04293 | -1.506 | 0 | 0.386 |
| 22448 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 14 | FSTL5 | Sponge network | 0.997 | 0.00066 | -1.3 | 0.01619 | 0.386 |
| 22449 | LINC00886 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-5p | 32 | DMD | Sponge network | -0.371 | 0.24604 | -1.506 | 0 | 0.386 |
| 22450 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-324-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 32 | EPHA7 | Sponge network | 0.051 | 0.83388 | -2.197 | 3.0E-5 | 0.386 |
| 22451 | C4B-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-93-3p | 24 | RUNX1T1 | Sponge network | 2.306 | 9.0E-5 | -0.344 | 0.2374 | 0.386 |
| 22452 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PRRX1 | Sponge network | -0.021 | 0.93598 | 1.316 | 0 | 0.386 |
| 22453 | MIR143HG |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | FAM117A | Sponge network | -0.739 | 0.0574 | -1.379 | 0 | 0.386 |
| 22454 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 23 | USP12 | Sponge network | -0.513 | 0.04703 | -0.102 | 0.40768 | 0.386 |
| 22455 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 39 | ETV1 | Sponge network | -0.739 | 0.0574 | -0.096 | 0.67674 | 0.386 |
| 22456 | LINC00641 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | KIF5B | Sponge network | -0.222 | 0.32445 | 0.362 | 0.00066 | 0.386 |
| 22457 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-532-3p | 15 | PBX1 | Sponge network | -0.206 | 0.12942 | -1.206 | 0 | 0.386 |
| 22458 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 29 | EPHA3 | Sponge network | -0.355 | 0.0077 | 0.185 | 0.53588 | 0.386 |
| 22459 | RASSF8-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | KLF10 | Sponge network | 0.335 | 0.20596 | -0.783 | 0 | 0.386 |
| 22460 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 22 | USP12 | Sponge network | -1.092 | 4.0E-5 | -0.102 | 0.40768 | 0.386 |
| 22461 | CTC-550B14.7 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-374a-3p;hsa-miR-409-5p | 14 | CCDC6 | Sponge network | 1.161 | 0 | 0.27 | 0.02555 | 0.386 |
| 22462 | GAS6-AS2 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-342-3p | 13 | ABL1 | Sponge network | 0.16 | 0.50785 | -0.215 | 0.07804 | 0.386 |
| 22463 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 20 | USP6 | Sponge network | 0.541 | 0.00125 | 0.188 | 0.3871 | 0.386 |
| 22464 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-502-5p | 19 | ANK2 | Sponge network | 0.622 | 0.00015 | -1.603 | 1.0E-5 | 0.386 |
| 22465 | PART1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | -2.925 | 0 | -1.324 | 0.00014 | 0.386 |
| 22466 | RP5-1139B12.3 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 16 | RASSF2 | Sponge network | 0.598 | 0.38481 | -0.273 | 0.21961 | 0.386 |
| 22467 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 15 | PBX1 | Sponge network | 0.419 | 0.0319 | -1.206 | 0 | 0.386 |
| 22468 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | HACE1 | Sponge network | 1.205 | 0 | -0.072 | 0.55239 | 0.386 |
| 22469 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-629-3p;hsa-miR-654-3p | 12 | PSIP1 | Sponge network | 0.637 | 0.00069 | -0.005 | 0.96897 | 0.386 |
| 22470 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 0.91 | 0 | 0.114 | 0.30638 | 0.386 |
| 22471 | RP11-401P9.4 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-590-5p | 11 | PTCH1 | Sponge network | -0.384 | 0.40652 | -0.1 | 0.6179 | 0.386 |
| 22472 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | MAF | Sponge network | -2.46 | 0 | -1.257 | 0 | 0.386 |
| 22473 | FAM66C |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | HERC1 | Sponge network | -0.901 | 0.00019 | -0.196 | 0.08544 | 0.386 |
| 22474 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | APC | Sponge network | 0.194 | 0.64278 | -0.255 | 0.04051 | 0.386 |
| 22475 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-374a-3p;hsa-miR-495-3p | 13 | MYO9A | Sponge network | 0.997 | 0 | -0.147 | 0.32373 | 0.386 |
| 22476 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 27 | PIK3R1 | Sponge network | 0.683 | 1.0E-5 | -0.133 | 0.36854 | 0.386 |
| 22477 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-450b-5p | 15 | PBX1 | Sponge network | -0.021 | 0.93598 | -1.206 | 0 | 0.386 |
| 22478 | RP11-67L2.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | NAV3 | Sponge network | 0.72 | 0.0001 | 0.212 | 0.47729 | 0.386 |
| 22479 | LINC00472 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-550a-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p | 31 | PTPRT | Sponge network | -0.842 | 0.01361 | -0.907 | 0.03727 | 0.386 |
| 22480 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 28 | MCC | Sponge network | -0.465 | 0.04703 | -1.005 | 0 | 0.386 |
| 22481 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 38 | PRKAB2 | Sponge network | -0.129 | 0.57177 | -0.367 | 0.01371 | 0.386 |
| 22482 | BCYRN1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | ITGB1 | Sponge network | 0.311 | 0.10344 | 0.524 | 0.00017 | 0.386 |
| 22483 | LINC00641 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p | 12 | LIMD1 | Sponge network | -0.222 | 0.32445 | 0.103 | 0.28978 | 0.386 |
| 22484 | LINC00672 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | RYR2 | Sponge network | -0.227 | 0.31657 | -1.466 | 0.00031 | 0.386 |
| 22485 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-766-3p;hsa-miR-940 | 17 | CADM2 | Sponge network | -1.483 | 9.0E-5 | -3.782 | 0 | 0.386 |
| 22486 | LINC00092 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-671-5p | 24 | CREB3L2 | Sponge network | -1.886 | 0 | -0.525 | 2.0E-5 | 0.386 |
| 22487 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 27 | ARHGAP28 | Sponge network | -0.49 | 0.04946 | -0.911 | 0.00013 | 0.386 |
| 22488 | CTC-429P9.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-877-5p | 13 | CCND2 | Sponge network | 0.043 | 0.81628 | -0.267 | 0.3112 | 0.386 |
| 22489 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 36 | ADARB1 | Sponge network | 0.259 | 0.12542 | -0.335 | 0.06631 | 0.386 |
| 22490 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SON | Sponge network | 0.545 | 0.00064 | 0.168 | 0.04406 | 0.386 |
| 22491 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p | 11 | GPAM | Sponge network | 0.93 | 0 | 0.142 | 0.29732 | 0.386 |
| 22492 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 41 | QKI | Sponge network | -0.08 | 0.90092 | -0.333 | 0.04111 | 0.386 |
| 22493 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | SMARCA2 | Sponge network | -0.612 | 0.00094 | -0.529 | 0.00014 | 0.386 |
| 22494 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-590-3p | 12 | RB1CC1 | Sponge network | 1.254 | 0 | 0.175 | 0.12559 | 0.386 |
| 22495 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 15 | CNTNAP3 | Sponge network | -0.141 | 0.26434 | -1.465 | 0.00033 | 0.386 |
| 22496 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 46 | TMEM132B | Sponge network | -1.281 | 0.00012 | -0.83 | 0.01084 | 0.386 |
| 22497 | AC010226.4 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | RYR2 | Sponge network | -0.811 | 2.0E-5 | -1.466 | 0.00031 | 0.386 |
| 22498 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | GRIN2A | Sponge network | -0.738 | 0.21233 | -2.161 | 0 | 0.386 |
| 22499 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p | 12 | CUL5 | Sponge network | 0.993 | 0 | 0.026 | 0.77301 | 0.386 |
| 22500 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TSC1 | Sponge network | 0.673 | 0 | 0.103 | 0.26071 | 0.386 |
| 22501 | LINC00578 |
hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577 | 10 | ITGAV | Sponge network | 0.811 | 0.03228 | 0.337 | 0.01002 | 0.386 |
| 22502 | RP11-968O1.5 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-493-5p;hsa-miR-532-3p | 10 | SYNM | Sponge network | -0.829 | 0.01343 | -2.309 | 1.0E-5 | 0.386 |
| 22503 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 18 | SASH1 | Sponge network | 0.178 | 0.29106 | -0.502 | 0.00175 | 0.386 |
| 22504 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27b-5p | 10 | KCNA6 | Sponge network | -7.551 | 0 | -1.531 | 0.00013 | 0.386 |
| 22505 | RP11-379F4.7 |
hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 11 | RASAL2 | Sponge network | 1.316 | 0 | 0.869 | 0 | 0.386 |
| 22506 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-500a-3p;hsa-miR-940 | 23 | HLF | Sponge network | -4.477 | 0 | -2.443 | 0 | 0.386 |
| 22507 | LINC00672 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-590-3p | 12 | ABCA10 | Sponge network | -0.227 | 0.31657 | -0.571 | 0.03674 | 0.386 |
| 22508 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | MGA | Sponge network | -0.222 | 0.32445 | 0.323 | 0.00792 | 0.386 |
| 22509 | GUSBP11 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-628-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.856 | 0 | 1.083 | 0 | 0.386 |
| 22510 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p | 31 | MOB1B | Sponge network | -0.096 | 0.67958 | 0.197 | 0.15266 | 0.386 |
| 22511 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-877-5p | 39 | FAT3 | Sponge network | -0.773 | 0.04073 | -1.601 | 0.00051 | 0.386 |
| 22512 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-590-3p;hsa-miR-654-3p | 11 | NR2C2 | Sponge network | 0.439 | 0.00313 | 0.21 | 0.03224 | 0.386 |
| 22513 | RP11-434H6.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p | 11 | RICTOR | Sponge network | 0.358 | 0.14302 | 0.388 | 0.00188 | 0.386 |
| 22514 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p | 16 | BCL6 | Sponge network | -1.349 | 0.00017 | -0.211 | 0.19489 | 0.386 |
| 22515 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-222-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | ESR1 | Sponge network | 0.342 | 0.03129 | -1.035 | 3.0E-5 | 0.386 |
| 22516 | RP11-360F5.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | PHC3 | Sponge network | 1.867 | 0 | 0.212 | 0.0499 | 0.386 |
| 22517 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 31 | ST6GAL2 | Sponge network | -0.323 | 0.01372 | -1.201 | 0.00597 | 0.386 |
| 22518 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-495-3p;hsa-miR-576-5p | 16 | ATRX | Sponge network | 1.044 | 2.0E-5 | 0.261 | 0.04408 | 0.386 |
| 22519 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374a-3p;hsa-miR-532-3p | 25 | CBL | Sponge network | 0.765 | 7.0E-5 | 0.291 | 0.01267 | 0.386 |
| 22520 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-96-5p | 32 | HBP1 | Sponge network | -0.942 | 0 | -0.454 | 0 | 0.386 |
| 22521 | RASSF8-AS1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p | 19 | IRS1 | Sponge network | 0.335 | 0.20596 | -0.026 | 0.88855 | 0.386 |
| 22522 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-655-3p | 27 | LATS1 | Sponge network | 0.349 | 0.01695 | 0.109 | 0.34208 | 0.386 |
| 22523 | ZNF582-AS1 |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-96-5p | 12 | PAIP2 | Sponge network | -1.349 | 0.00017 | -0.515 | 0 | 0.386 |
| 22524 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | 0.4 | 0.14611 | -1.751 | 1.0E-5 | 0.386 |
| 22525 | MIR497HG |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | GALNT13 | Sponge network | -1.439 | 4.0E-5 | -0.857 | 0.07439 | 0.386 |
| 22526 | RP11-798M19.6 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | SIAH1 | Sponge network | -0.612 | 0.00094 | -0.057 | 0.41415 | 0.386 |
| 22527 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-423-5p;hsa-miR-484 | 15 | TGFBR3 | Sponge network | -2.549 | 0.00528 | -1.173 | 1.0E-5 | 0.386 |
| 22528 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 31 | USP12 | Sponge network | 0.421 | 0.00084 | -0.102 | 0.40768 | 0.386 |
| 22529 | BOLA3-AS1 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | SFRP1 | Sponge network | -0.246 | 0.35034 | -3.566 | 0 | 0.386 |
| 22530 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 24 | NUAK1 | Sponge network | -0.777 | 0.1134 | 0.904 | 0 | 0.386 |
| 22531 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | UBE4B | Sponge network | 0.349 | 0.01695 | -0.235 | 0.007 | 0.386 |
| 22532 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-454-3p;hsa-miR-671-5p | 11 | DLC1 | Sponge network | 0.598 | 0.38481 | -0.382 | 0.05038 | 0.386 |
| 22533 | DNM1P35 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | RCAN2 | Sponge network | 0.051 | 0.83388 | -0.33 | 0.15172 | 0.386 |
| 22534 | LINC00654 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ZNF208 | Sponge network | 0.374 | 0.20054 | -0.4 | 0.22381 | 0.386 |
| 22535 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-93-5p | 20 | MAF | Sponge network | -0.473 | 0.07602 | -1.257 | 0 | 0.386 |
| 22536 | HCG11 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | INPP4A | Sponge network | 0.385 | 0.10067 | 0.07 | 0.53563 | 0.386 |
| 22537 | LINC00702 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 34 | OPCML | Sponge network | -0.737 | 0.06182 | -2.498 | 0 | 0.386 |
| 22538 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-96-5p | 17 | JAZF1 | Sponge network | -0.517 | 0.02935 | -0.377 | 0.02677 | 0.386 |
| 22539 | MIR143HG |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | AFAP1L2 | Sponge network | -0.739 | 0.0574 | -0.284 | 0.15434 | 0.386 |
| 22540 | LINC00957 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 19 | ABL1 | Sponge network | -0.421 | 0.0114 | -0.215 | 0.07804 | 0.386 |
| 22541 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | THRB | Sponge network | -0.021 | 0.93598 | -1.117 | 0.0004 | 0.385 |
| 22542 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 27 | YAP1 | Sponge network | 0.064 | 0.72934 | 0.22 | 0.11836 | 0.385 |
| 22543 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | FLNA | Sponge network | -0.355 | 0.0077 | -0.771 | 0.01377 | 0.385 |
| 22544 | AC009948.5 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-877-5p;hsa-miR-96-5p | 22 | ASTN1 | Sponge network | 0.532 | 0.00505 | -2.389 | 2.0E-5 | 0.385 |
| 22545 | CTC-429P9.5 |
hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 10 | SPG20 | Sponge network | 0.178 | 0.29106 | -1.16 | 0 | 0.385 |
| 22546 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-582-5p | 16 | PIK3C2A | Sponge network | 0.817 | 3.0E-5 | 0.061 | 0.53776 | 0.385 |
| 22547 | FZD10-AS1 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | SFRP1 | Sponge network | -0.727 | 0.04726 | -3.566 | 0 | 0.385 |
| 22548 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-484;hsa-miR-577;hsa-miR-93-3p | 19 | EBF3 | Sponge network | -0.465 | 0.04703 | -0.058 | 0.81437 | 0.385 |
| 22549 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 25 | RNF144A | Sponge network | 0.395 | 0.15451 | 0.594 | 0.0009 | 0.385 |
| 22550 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 15 | RBL2 | Sponge network | 0.064 | 0.72934 | -0.282 | 0.00254 | 0.385 |
| 22551 | HAND2-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 19 | TES | Sponge network | -1.697 | 0.00889 | -0.348 | 0.00639 | 0.385 |
| 22552 | LINC01018 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ITPKB | Sponge network | -2.915 | 0.00015 | -0.704 | 0.00106 | 0.385 |
| 22553 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-33a-3p | 17 | MYO9A | Sponge network | -0.693 | 0.05629 | -0.147 | 0.32373 | 0.385 |
| 22554 | PCA3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | TAL1 | Sponge network | -0.709 | 0.18168 | -0.462 | 0.00574 | 0.385 |
| 22555 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-425-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 52 | DTNA | Sponge network | -0.355 | 0.0077 | -1.648 | 1.0E-5 | 0.385 |
| 22556 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-532-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | LRRC4 | Sponge network | -1.047 | 0 | -1.774 | 0 | 0.385 |
| 22557 | RP11-774O3.3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | INPP4A | Sponge network | -0.487 | 0.02157 | 0.07 | 0.53563 | 0.385 |
| 22558 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -2.46 | 0 | -0.252 | 0.15511 | 0.385 |
| 22559 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 12 | PCDH17 | Sponge network | -1.092 | 4.0E-5 | 0.731 | 2.0E-5 | 0.385 |
| 22560 | RP11-597D13.9 |
hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p | 11 | FLNA | Sponge network | 1.302 | 7.0E-5 | -0.771 | 0.01377 | 0.385 |
| 22561 | AC005154.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 14 | CEP170 | Sponge network | 0.848 | 0 | 0.943 | 0 | 0.385 |
| 22562 | AC003090.1 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | SPOP | Sponge network | -2.117 | 0.00161 | -0.419 | 1.0E-5 | 0.385 |
| 22563 | LINC00641 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 30 | PCDH10 | Sponge network | -0.222 | 0.32445 | -1.644 | 0.0078 | 0.385 |
| 22564 | LINC00654 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 45 | GAS7 | Sponge network | 0.374 | 0.20054 | -0.555 | 0.01602 | 0.385 |
| 22565 | RP11-983P16.4 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-484 | 18 | ABL1 | Sponge network | 0.41 | 0.02214 | -0.215 | 0.07804 | 0.385 |
| 22566 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | RNF144A | Sponge network | 0.381 | 0.25967 | 0.594 | 0.0009 | 0.385 |
| 22567 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 19 | PRDM2 | Sponge network | 0.385 | 0.10067 | -0.258 | 0.00721 | 0.385 |
| 22568 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-542-3p;hsa-miR-942-5p | 12 | ERRFI1 | Sponge network | -1.092 | 4.0E-5 | -0.798 | 1.0E-5 | 0.385 |
| 22569 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZFHX3 | Sponge network | 1.308 | 0 | 0.389 | 0.01363 | 0.385 |
| 22570 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 30 | MCC | Sponge network | -0.668 | 3.0E-5 | -1.005 | 0 | 0.385 |
| 22571 | FENDRR |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-7-1-3p | 11 | PACRG | Sponge network | -1.281 | 0.00012 | -2.248 | 0 | 0.385 |
| 22572 | RP11-497H16.9 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TMEM30A | Sponge network | 0.545 | 0.00064 | 0.096 | 0.31031 | 0.385 |
| 22573 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-5p | 24 | RAP1A | Sponge network | -0.031 | 0.89063 | -0.929 | 0 | 0.385 |
| 22574 | GNG12-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-455-3p | 18 | ABL1 | Sponge network | -0.793 | 7.0E-5 | -0.215 | 0.07804 | 0.385 |
| 22575 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p | 18 | TCF12 | Sponge network | 0.614 | 1.0E-5 | 0.174 | 0.13271 | 0.385 |
| 22576 | RP11-819C21.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-744-3p | 48 | DST | Sponge network | 0.537 | 0.00139 | -0.713 | 0.00096 | 0.385 |
| 22577 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 22 | EPHA5 | Sponge network | -1.582 | 8.0E-5 | -0.957 | 0.01733 | 0.385 |
| 22578 | DNAJC27-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 16 | CELF4 | Sponge network | -0.969 | 2.0E-5 | -1.135 | 0.00344 | 0.385 |
| 22579 | AC009948.5 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 12 | TAF1 | Sponge network | 0.532 | 0.00505 | 0.39 | 3.0E-5 | 0.385 |
| 22580 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-5p | 29 | AR | Sponge network | -0.829 | 0.01343 | -1.554 | 0 | 0.385 |
| 22581 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-7-1-3p | 13 | SASH1 | Sponge network | -0.285 | 0.2172 | -0.502 | 0.00175 | 0.385 |
| 22582 | RP11-119F7.5 |
hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PAK3 | Sponge network | 0.225 | 0.30644 | -0.628 | 0.07253 | 0.385 |
| 22583 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | LRRC55 | Sponge network | -0.901 | 0.00019 | -0.026 | 0.93163 | 0.385 |
| 22584 | RP11-13K12.1 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p | 10 | DIRAS1 | Sponge network | -0.154 | 0.78939 | -2.518 | 0 | 0.385 |
| 22585 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 50 | SSBP2 | Sponge network | 0.101 | 0.60919 | -1.58 | 0 | 0.385 |
| 22586 | RP11-216B9.6 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | MRVI1 | Sponge network | 0.174 | 0.30034 | -1.051 | 0.00022 | 0.385 |
| 22587 | LINC00472 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-877-5p | 12 | INPP4A | Sponge network | -0.842 | 0.01361 | 0.07 | 0.53563 | 0.385 |
| 22588 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 17 | NEDD4 | Sponge network | 0.214 | 0.18246 | 0.02 | 0.89834 | 0.385 |
| 22589 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 32 | BHLHE41 | Sponge network | 0.997 | 0.00066 | 0.353 | 0.12753 | 0.385 |
| 22590 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | RBL2 | Sponge network | 0.083 | 0.66405 | -0.282 | 0.00254 | 0.385 |
| 22591 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 33 | TGFBR3 | Sponge network | 0.214 | 0.18246 | -1.173 | 1.0E-5 | 0.385 |
| 22592 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | RASSF8 | Sponge network | 0.219 | 0.1516 | -0.122 | 0.65591 | 0.385 |
| 22593 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-96-5p | 31 | CREB3L2 | Sponge network | -1.654 | 0.00144 | -0.525 | 2.0E-5 | 0.385 |
| 22594 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-5p | 16 | SYT4 | Sponge network | -0.469 | 0.08721 | -3.207 | 0 | 0.385 |
| 22595 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-629-5p | 10 | UVRAG | Sponge network | -0.517 | 0.02935 | -0.017 | 0.83865 | 0.385 |
| 22596 | RP11-119F7.5 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-424-3p;hsa-miR-493-5p;hsa-miR-532-3p | 15 | SYNM | Sponge network | 0.225 | 0.30644 | -2.309 | 1.0E-5 | 0.385 |
| 22597 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 26 | MXI1 | Sponge network | -0.405 | 0.22013 | -0.987 | 0 | 0.385 |
| 22598 | RP11-359B12.2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | PIK3CA | Sponge network | 0.064 | 0.72934 | 0.195 | 0.06908 | 0.385 |
| 22599 | ZNF582-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 11 | EHD3 | Sponge network | -1.349 | 0.00017 | -0.484 | 0.01747 | 0.385 |
| 22600 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-940;hsa-miR-96-5p | 30 | SNX29 | Sponge network | 0.101 | 0.60919 | -0.34 | 0.01371 | 0.385 |
| 22601 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DACT1 | Sponge network | 0.727 | 3.0E-5 | 0.783 | 0.00072 | 0.385 |
| 22602 | RP11-999E24.3 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-425-5p | 14 | RYR2 | Sponge network | -0.693 | 0.05629 | -1.466 | 0.00031 | 0.385 |
| 22603 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-769-5p | 18 | RCAN2 | Sponge network | -0.563 | 0.02545 | -0.33 | 0.15172 | 0.385 |
| 22604 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-590-3p | 19 | AKAP11 | Sponge network | 0.919 | 0 | 0.196 | 0.09825 | 0.385 |
| 22605 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | NEDD4 | Sponge network | 1.302 | 7.0E-5 | 0.02 | 0.89834 | 0.385 |
| 22606 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 24 | NOTCH2 | Sponge network | 0.683 | 1.0E-5 | 0.138 | 0.35163 | 0.385 |
| 22607 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-421;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | ZNF521 | Sponge network | 0.16 | 0.50785 | -0.111 | 0.58068 | 0.385 |
| 22608 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CDH11 | Sponge network | -1.111 | 0.0003 | 1.427 | 0 | 0.385 |
| 22609 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 10 | PDGFC | Sponge network | -4.546 | 0 | -0.33 | 0.06483 | 0.385 |
| 22610 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-92a-1-5p | 20 | PRKAB2 | Sponge network | 0.368 | 0.20093 | -0.367 | 0.01371 | 0.385 |
| 22611 | RP11-758N13.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-375;hsa-miR-532-3p | 22 | GAS7 | Sponge network | 2.939 | 3.0E-5 | -0.555 | 0.01602 | 0.385 |
| 22612 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-628-5p | 21 | FOXP1 | Sponge network | -0.583 | 0.14589 | 0.042 | 0.74368 | 0.385 |
| 22613 | LINC00473 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-629-3p;hsa-miR-92a-3p | 12 | ITGA5 | Sponge network | -1.203 | 0.03881 | 0.452 | 0.03524 | 0.385 |
| 22614 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | DKK3 | Sponge network | 0.225 | 0.30644 | -0.052 | 0.77783 | 0.385 |
| 22615 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-590-3p | 11 | FOXO3 | Sponge network | 1.163 | 0.00018 | -0.04 | 0.72028 | 0.385 |
| 22616 | GS1-124K5.11 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | PHF3 | Sponge network | 0.494 | 0.00339 | 0.126 | 0.19652 | 0.385 |
| 22617 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 57 | DTNA | Sponge network | -0.969 | 2.0E-5 | -1.648 | 1.0E-5 | 0.385 |
| 22618 | RP11-141C7.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-497-5p | 12 | TBL1XR1 | Sponge network | 2.412 | 0 | 0.578 | 0 | 0.385 |
| 22619 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | LRRC55 | Sponge network | -0.769 | 0.00035 | -0.026 | 0.93163 | 0.385 |
| 22620 | DNM3OS |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | HERC1 | Sponge network | 0.572 | 0.01722 | -0.196 | 0.08544 | 0.385 |
| 22621 | RP5-1074L1.4 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | ERCC6 | Sponge network | 1.923 | 0 | 0.23 | 0.015 | 0.385 |
| 22622 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-429;hsa-miR-484;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-940 | 13 | CRTC3 | Sponge network | -0.738 | 0.21233 | 0.164 | 0.16786 | 0.385 |
| 22623 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | HDAC9 | Sponge network | -0.576 | 0.10121 | -0.755 | 0.00495 | 0.385 |
| 22624 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | FBXO32 | Sponge network | 0.342 | 0.03129 | -0.495 | 0.07561 | 0.385 |
| 22625 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-93-3p | 53 | BMPR2 | Sponge network | -0.154 | 0.78939 | 0.206 | 0.0635 | 0.385 |
| 22626 | RP11-254F7.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-501-5p;hsa-miR-7-5p | 24 | IGFBP5 | Sponge network | 0.176 | 0.37402 | -0.806 | 0.00105 | 0.385 |
| 22627 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | PEG3 | Sponge network | 0.61 | 0.01593 | -1.74 | 0 | 0.385 |
| 22628 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-532-3p;hsa-miR-93-5p | 27 | DST | Sponge network | 0.497 | 0.01451 | -0.713 | 0.00096 | 0.385 |
| 22629 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | FSTL5 | Sponge network | -1.057 | 0.00029 | -1.3 | 0.01619 | 0.385 |
| 22630 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | NIN | Sponge network | 0.683 | 1.0E-5 | -0.253 | 0.13794 | 0.385 |
| 22631 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | MLLT10 | Sponge network | 0.064 | 0.72934 | 0.105 | 0.33292 | 0.385 |
| 22632 | PCA3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PTPN13 | Sponge network | -0.709 | 0.18168 | -1.208 | 0 | 0.385 |
| 22633 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-7-5p | 23 | ZBTB4 | Sponge network | 0.176 | 0.37402 | -0.739 | 0 | 0.385 |
| 22634 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-93-5p | 13 | TRIM3 | Sponge network | -0.976 | 0.00025 | -0.577 | 0 | 0.385 |
| 22635 | FGD5-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 33 | CREB1 | Sponge network | 0.401 | 0.00066 | 0.203 | 0.00537 | 0.385 |
| 22636 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-93-5p | 28 | LRP1B | Sponge network | -1.645 | 0 | -2.163 | 0 | 0.385 |
| 22637 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 32 | CBL | Sponge network | 0.101 | 0.60919 | 0.291 | 0.01267 | 0.385 |
| 22638 | RP11-693J15.4 |
hsa-miR-1301-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | SFRP1 | Sponge network | -0.72 | 0.24239 | -3.566 | 0 | 0.385 |
| 22639 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | SOX5 | Sponge network | 0.083 | 0.66405 | -1.091 | 4.0E-5 | 0.385 |
| 22640 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | NRXN1 | Sponge network | -0.222 | 0.32445 | -3.778 | 0 | 0.385 |
| 22641 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 51 | CADM2 | Sponge network | 0.249 | 0.35902 | -3.782 | 0 | 0.385 |
| 22642 | PWAR6 |
hsa-miR-140-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | CHD6 | Sponge network | -1.575 | 6.0E-5 | 0.32 | 0.01326 | 0.385 |
| 22643 | CTD-2554C21.2 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-508-3p | 43 | THRB | Sponge network | -1.654 | 0.00144 | -1.117 | 0.0004 | 0.385 |
| 22644 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 52 | PRKAA2 | Sponge network | -0.969 | 2.0E-5 | -2.462 | 0 | 0.385 |
| 22645 | RP11-822E23.8 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-942-5p | 13 | ZBTB20 | Sponge network | -0.777 | 0.16667 | -0.122 | 0.57569 | 0.385 |
| 22646 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | THBS1 | Sponge network | -0.318 | 0.65847 | 0.741 | 0.00386 | 0.385 |
| 22647 | RP11-554A11.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-628-5p | 14 | TOM1L2 | Sponge network | -0.583 | 0.14589 | -0.994 | 0 | 0.385 |
| 22648 | RP11-57H14.4 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | PIK3CA | Sponge network | 0.083 | 0.66405 | 0.195 | 0.06908 | 0.385 |
| 22649 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | ADAMTSL3 | Sponge network | -0.355 | 0.0077 | -1.559 | 0 | 0.385 |
| 22650 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-625-5p | 29 | PTPRT | Sponge network | -2.69 | 0.0001 | -0.907 | 0.03727 | 0.385 |
| 22651 | RP11-67L2.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p | 12 | THSD7A | Sponge network | 0.72 | 0.0001 | 0.242 | 0.25154 | 0.385 |
| 22652 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-378c;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 30 | ERC1 | Sponge network | 0.082 | 0.55654 | -0.108 | 0.38794 | 0.385 |
| 22653 | LINC00702 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 23 | HIP1 | Sponge network | -0.737 | 0.06182 | 0.774 | 0 | 0.385 |
| 22654 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-942-5p | 40 | TMEM132B | Sponge network | -1.439 | 4.0E-5 | -0.83 | 0.01084 | 0.385 |
| 22655 | RP1-59D14.5 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 12 | NF1 | Sponge network | 1.162 | 5.0E-5 | 0.51 | 0 | 0.385 |
| 22656 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -0.08 | 0.90092 | -0.087 | 0.69193 | 0.385 |
| 22657 | RP11-48B3.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-577;hsa-miR-590-3p | 15 | NRK | Sponge network | 1.757 | 0 | 0.656 | 0.19217 | 0.385 |
| 22658 | DNAJC27-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p | 21 | SYNM | Sponge network | -0.969 | 2.0E-5 | -2.309 | 1.0E-5 | 0.385 |
| 22659 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 51 | CBX5 | Sponge network | 0.421 | 0.00084 | 0.165 | 0.24446 | 0.385 |
| 22660 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | ARHGAP29 | Sponge network | -1.575 | 6.0E-5 | 0.131 | 0.42953 | 0.385 |
| 22661 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-590-5p;hsa-miR-874-3p | 10 | DCUN1D3 | Sponge network | 0.078 | 0.55803 | 0.152 | 0.1769 | 0.385 |
| 22662 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p | 33 | FBXO32 | Sponge network | 0.349 | 0.01695 | -0.495 | 0.07561 | 0.385 |
| 22663 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-409-3p | 17 | BCL2 | Sponge network | -0.309 | 0.1145 | -0.899 | 0 | 0.385 |
| 22664 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | TSHZ3 | Sponge network | 0.61 | 0.01593 | -0.556 | 0.01516 | 0.385 |
| 22665 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | 0.083 | 0.66405 | 0.809 | 0.00544 | 0.385 |
| 22666 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | DKK3 | Sponge network | -1.203 | 0.03881 | -0.052 | 0.77783 | 0.385 |
| 22667 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p | 16 | MYCBP2 | Sponge network | 0.101 | 0.60919 | -0.027 | 0.82016 | 0.385 |
| 22668 | AP001055.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-744-3p | 15 | PTPN14 | Sponge network | 0.693 | 0.00076 | 0.144 | 0.34595 | 0.385 |
| 22669 | ZSCAN16-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | WWTR1 | Sponge network | -0.342 | 0.02288 | -0.345 | 0.11997 | 0.385 |
| 22670 | FAM157B |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3614-5p | 10 | MOB1B | Sponge network | 0.92 | 0 | 0.197 | 0.15266 | 0.385 |
| 22671 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p | 28 | RICTOR | Sponge network | 0.657 | 4.0E-5 | 0.388 | 0.00188 | 0.385 |
| 22672 | CTC-429P9.5 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-3p | 16 | GRIK3 | Sponge network | 0.178 | 0.29106 | -3.208 | 0 | 0.385 |
| 22673 | RP11-13K12.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-629-3p | 12 | SPTBN4 | Sponge network | -0.154 | 0.78939 | -0.786 | 0.01281 | 0.385 |
| 22674 | LINC00403 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-30d-3p;hsa-miR-424-5p | 14 | FAT3 | Sponge network | -3.586 | 0.00032 | -1.601 | 0.00051 | 0.385 |
| 22675 | LINC00174 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 1.253 | 0 | 0.362 | 0.00066 | 0.385 |
| 22676 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZNF844 | Sponge network | 0.225 | 0.30644 | -0.966 | 0.00035 | 0.385 |
| 22677 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-624-5p | 11 | PCDH8 | Sponge network | -0.358 | 0.17255 | -0.997 | 0.05366 | 0.385 |
| 22678 | ZNF582-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-590-5p | 14 | ITGB3 | Sponge network | -1.349 | 0.00017 | -0.011 | 0.95751 | 0.385 |
| 22679 | H1FX-AS1 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PEG3 | Sponge network | -0.206 | 0.12942 | -1.74 | 0 | 0.385 |
| 22680 | RP11-25K19.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | DIP2C | Sponge network | -1.057 | 0.00029 | -0.436 | 0.01713 | 0.385 |
| 22681 | FENDRR |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | RTN4 | Sponge network | -1.281 | 0.00012 | -0.412 | 0.00014 | 0.385 |
| 22682 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-7-1-3p | 11 | CDH10 | Sponge network | -0.969 | 2.0E-5 | -2.397 | 0 | 0.385 |
| 22683 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | ATRX | Sponge network | 0.419 | 0.0319 | 0.261 | 0.04408 | 0.385 |
| 22684 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | KCNA6 | Sponge network | 0.299 | 0.57722 | -1.531 | 0.00013 | 0.385 |
| 22685 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b | 10 | RBM5 | Sponge network | 0.75 | 0.00054 | -0.205 | 0.0129 | 0.385 |
| 22686 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | RCAN2 | Sponge network | -2.915 | 0.00015 | -0.33 | 0.15172 | 0.385 |
| 22687 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | SON | Sponge network | 0.856 | 0 | 0.168 | 0.04406 | 0.385 |
| 22688 | LINC00672 |
hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | ITGB1 | Sponge network | -0.227 | 0.31657 | 0.524 | 0.00017 | 0.385 |
| 22689 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 31 | BHLHE41 | Sponge network | -0.576 | 0.10121 | 0.353 | 0.12753 | 0.385 |
| 22690 | AC141928.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 20 | DIP2C | Sponge network | 0.853 | 0.15352 | -0.436 | 0.01713 | 0.385 |
| 22691 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-590-3p | 13 | NIN | Sponge network | -0.739 | 0.00044 | -0.253 | 0.13794 | 0.385 |
| 22692 | SNHG1 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 12 | PAFAH1B2 | Sponge network | 1.265 | 0 | 0.318 | 0.0001 | 0.385 |
| 22693 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-96-5p | 31 | BMPR2 | Sponge network | -0.517 | 0.02935 | 0.206 | 0.0635 | 0.385 |
| 22694 | LINC00473 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 14 | CSRNP1 | Sponge network | -1.203 | 0.03881 | -1.448 | 0 | 0.385 |
| 22695 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-93-5p | 16 | EZH1 | Sponge network | 0.246 | 0.59912 | -0.564 | 1.0E-5 | 0.385 |
| 22696 | HAND2-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NRG3 | Sponge network | -1.697 | 0.00889 | -1.836 | 4.0E-5 | 0.385 |
| 22697 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TIMP3 | Sponge network | -0.08 | 0.90092 | -0.546 | 0.02307 | 0.385 |
| 22698 | LINC00174 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 20 | TBL1XR1 | Sponge network | 1.253 | 0 | 0.578 | 0 | 0.385 |
| 22699 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-940 | 15 | GLI3 | Sponge network | -0.405 | 0.22013 | -0.615 | 0.01482 | 0.385 |
| 22700 | A2M-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 12 | MAPK10 | Sponge network | -0.08 | 0.90092 | -1.774 | 0 | 0.385 |
| 22701 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p | 10 | EDA2R | Sponge network | -0.513 | 0.04703 | -0.587 | 0.01598 | 0.385 |
| 22702 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 29 | GRIA3 | Sponge network | -0.942 | 0 | -1.956 | 0 | 0.385 |
| 22703 | WDFY3-AS2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | PDE4DIP | Sponge network | -0.942 | 0 | -0.282 | 0.0257 | 0.385 |
| 22704 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -2.039 | 0.03447 | -0.133 | 0.36854 | 0.385 |
| 22705 | RP5-1198O20.4 |
hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 11 | CDH13 | Sponge network | 0.997 | 0.00066 | 0.634 | 0.00094 | 0.385 |
| 22706 | LINC00672 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-32-5p | 11 | RASSF3 | Sponge network | -0.227 | 0.31657 | -0.226 | 0.11691 | 0.385 |
| 22707 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | BCL11A | Sponge network | -1.575 | 6.0E-5 | -0.932 | 0.0063 | 0.385 |
| 22708 | RP1-302G2.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p | 17 | EPHA7 | Sponge network | -1.483 | 9.0E-5 | -2.197 | 3.0E-5 | 0.385 |
| 22709 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-550a-5p;hsa-miR-766-3p;hsa-miR-93-5p | 33 | CHD5 | Sponge network | -0.227 | 0.31657 | -0.922 | 0.01521 | 0.385 |
| 22710 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | EPHA5 | Sponge network | -0.318 | 0.65847 | -0.957 | 0.01733 | 0.385 |
| 22711 | NNT-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | BCL9 | Sponge network | 0.799 | 1.0E-5 | 0.321 | 0.00267 | 0.385 |
| 22712 | CTC-559E9.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p | 19 | ABL2 | Sponge network | 0.594 | 0.00064 | 0.82 | 0 | 0.385 |
| 22713 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-29a-5p | 13 | EPHA3 | Sponge network | 0.594 | 0.00064 | 0.185 | 0.53588 | 0.385 |
| 22714 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 53 | FOXP1 | Sponge network | -0.576 | 0.10121 | 0.042 | 0.74368 | 0.385 |
| 22715 | RP11-750H9.5 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-224-5p | 10 | IGFBP5 | Sponge network | 0.349 | 0.13152 | -0.806 | 0.00105 | 0.385 |
| 22716 | ZSCAN16-AS1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 14 | CSDE1 | Sponge network | -0.342 | 0.02288 | -0.493 | 0 | 0.385 |
| 22717 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-629-3p | 13 | ITGA5 | Sponge network | -0.246 | 0.35034 | 0.452 | 0.03524 | 0.385 |
| 22718 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-429 | 14 | GRIA3 | Sponge network | -1.654 | 0.00144 | -1.956 | 0 | 0.384 |
| 22719 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 33 | TET1 | Sponge network | -0.737 | 0.06182 | -0.135 | 0.52138 | 0.384 |
| 22720 | HHIP-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | NAV3 | Sponge network | -0.737 | 0.10822 | 0.212 | 0.47729 | 0.384 |
| 22721 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-942-5p | 44 | THRB | Sponge network | -0.358 | 0.17255 | -1.117 | 0.0004 | 0.384 |
| 22722 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-495-3p;hsa-miR-7-1-3p | 20 | CLIP1 | Sponge network | -0.405 | 0.22013 | -0.215 | 0.08684 | 0.384 |
| 22723 | LINC00578 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 31 | CADM2 | Sponge network | 0.811 | 0.03228 | -3.782 | 0 | 0.384 |
| 22724 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | SLC6A15 | Sponge network | -0.576 | 0.10121 | -2.373 | 2.0E-5 | 0.384 |
| 22725 | RP5-1198O20.4 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | RIMS2 | Sponge network | 0.997 | 0.00066 | -1.365 | 0.00208 | 0.384 |
| 22726 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-495-3p | 14 | NF1 | Sponge network | 1.344 | 0 | 0.51 | 0 | 0.384 |
| 22727 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 27 | ERCC4 | Sponge network | -1.111 | 0.0003 | 0.126 | 0.15266 | 0.384 |
| 22728 | RP11-244H3.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p | 62 | LPP | Sponge network | 0.659 | 3.0E-5 | -0.459 | 0.04475 | 0.384 |
| 22729 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 60 | LPP | Sponge network | 0.259 | 0.12542 | -0.459 | 0.04475 | 0.384 |
| 22730 | RP11-46C24.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | 0.392 | 0.0278 | 0.809 | 0.00544 | 0.384 |
| 22731 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p | 11 | BMPR2 | Sponge network | 0.793 | 0 | 0.206 | 0.0635 | 0.384 |
| 22732 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | CREBBP | Sponge network | -1.111 | 0.0003 | 0.126 | 0.15186 | 0.384 |
| 22733 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | RECK | Sponge network | -0.351 | 0.11358 | -1.043 | 0 | 0.384 |
| 22734 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-331-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p | 14 | SLITRK6 | Sponge network | -0.384 | 0.40652 | -2.121 | 0 | 0.384 |
| 22735 | CTC-429P9.2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-335-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p | 11 | RYR2 | Sponge network | 0.043 | 0.81628 | -1.466 | 0.00031 | 0.384 |
| 22736 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-96-5p | 24 | PIK3R1 | Sponge network | -0.517 | 0.02935 | -0.133 | 0.36854 | 0.384 |
| 22737 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | ZFHX4 | Sponge network | 1.253 | 0 | 0.018 | 0.95574 | 0.384 |
| 22738 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | NALCN | Sponge network | 1.757 | 0 | 1.068 | 0.00237 | 0.384 |
| 22739 | GNG12-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 35 | ACVR2A | Sponge network | -0.793 | 7.0E-5 | -0.409 | 0.00039 | 0.384 |
| 22740 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-409-3p;hsa-miR-590-3p | 19 | PHC3 | Sponge network | 0.765 | 7.0E-5 | 0.212 | 0.0499 | 0.384 |
| 22741 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | -0.162 | 0.47687 | -1.751 | 1.0E-5 | 0.384 |
| 22742 | RP4-791M13.3 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-940 | 22 | CHD5 | Sponge network | 0.419 | 0.0319 | -0.922 | 0.01521 | 0.384 |
| 22743 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SMARCA2 | Sponge network | 0.194 | 0.64278 | -0.529 | 0.00014 | 0.384 |
| 22744 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p | 13 | EDA2R | Sponge network | -0.08 | 0.90092 | -0.587 | 0.01598 | 0.384 |
| 22745 | NUTM2A-AS1 |
hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-497-5p | 10 | CDK6 | Sponge network | 0.643 | 0 | 0.991 | 0 | 0.384 |
| 22746 | BCYRN1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 25 | SIRT1 | Sponge network | 0.311 | 0.10344 | 0.109 | 0.2593 | 0.384 |
| 22747 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | MLLT10 | Sponge network | 0.924 | 0 | 0.105 | 0.33292 | 0.384 |
| 22748 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | AKAP11 | Sponge network | 0.643 | 0 | 0.196 | 0.09825 | 0.384 |
| 22749 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-7-1-3p | 14 | MED12L | Sponge network | 0.238 | 0.70544 | -0.194 | 0.55299 | 0.384 |
| 22750 | RP5-1139B12.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-423-3p;hsa-miR-454-3p | 10 | ROBO1 | Sponge network | 0.598 | 0.38481 | -0.009 | 0.96154 | 0.384 |
| 22751 | RP11-307B6.3 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-532-5p | 10 | RIMS2 | Sponge network | -4.463 | 0.00025 | -1.365 | 0.00208 | 0.384 |
| 22752 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-590-3p | 29 | AFF4 | Sponge network | 0.178 | 0.29106 | -0.266 | 0.01565 | 0.384 |
| 22753 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 13 | DLG1 | Sponge network | 0.683 | 1.0E-5 | -0.014 | 0.8926 | 0.384 |
| 22754 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 25 | AFF3 | Sponge network | -0.612 | 0.00094 | -2.373 | 0 | 0.384 |
| 22755 | PART1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 16 | PHOX2B | Sponge network | -2.925 | 0 | -2.68 | 0 | 0.384 |
| 22756 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 45 | THRB | Sponge network | 0.176 | 0.37402 | -1.117 | 0.0004 | 0.384 |
| 22757 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p | 21 | PLSCR4 | Sponge network | 0.214 | 0.18246 | -0.474 | 0.03833 | 0.384 |
| 22758 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | PHC3 | Sponge network | 1.063 | 0 | 0.212 | 0.0499 | 0.384 |
| 22759 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 33 | TGFBR3 | Sponge network | -0.513 | 0.04703 | -1.173 | 1.0E-5 | 0.384 |
| 22760 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-935 | 29 | DIP2C | Sponge network | 0.75 | 0.00054 | -0.436 | 0.01713 | 0.384 |
| 22761 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-940 | 24 | AKAP11 | Sponge network | 0.174 | 0.30034 | 0.196 | 0.09825 | 0.384 |
| 22762 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p | 10 | ELMO1 | Sponge network | -0.583 | 0.14589 | 0.04 | 0.83147 | 0.384 |
| 22763 | RP11-182J1.1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-766-3p | 18 | CHD5 | Sponge network | -0.187 | 0.23787 | -0.922 | 0.01521 | 0.384 |
| 22764 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-589-3p | 10 | ABCA10 | Sponge network | 0.4 | 0.14611 | -0.571 | 0.03674 | 0.384 |
| 22765 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-582-3p;hsa-miR-590-3p | 26 | ABL2 | Sponge network | 0.101 | 0.60919 | 0.82 | 0 | 0.384 |
| 22766 | HCG11 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | BCL9 | Sponge network | 0.385 | 0.10067 | 0.321 | 0.00267 | 0.384 |
| 22767 | FZD10-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | -0.727 | 0.04726 | -0.998 | 0.00025 | 0.384 |
| 22768 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | EYA4 | Sponge network | 0.889 | 9.0E-5 | 0.3 | 0.36876 | 0.384 |
| 22769 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 30 | ZFHX3 | Sponge network | 0.848 | 0 | 0.389 | 0.01363 | 0.384 |
| 22770 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p | 24 | ABL2 | Sponge network | -0.154 | 0.78939 | 0.82 | 0 | 0.384 |
| 22771 | RP1-302G2.5 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-7-5p | 15 | IGFBP5 | Sponge network | -1.483 | 9.0E-5 | -0.806 | 0.00105 | 0.384 |
| 22772 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-493-5p;hsa-miR-654-3p | 12 | NR2C2 | Sponge network | 0.909 | 0 | 0.21 | 0.03224 | 0.384 |
| 22773 | WDFY3-AS2 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 10 | DIRAS1 | Sponge network | -0.942 | 0 | -2.518 | 0 | 0.384 |
| 22774 | BZRAP1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-874-3p;hsa-miR-877-5p | 27 | ASTN1 | Sponge network | -0.465 | 0.04703 | -2.389 | 2.0E-5 | 0.384 |
| 22775 | RP11-473I1.9 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | AFF1 | Sponge network | 0.683 | 1.0E-5 | -0.384 | 0.00053 | 0.384 |
| 22776 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 11 | RCAN2 | Sponge network | 0.541 | 0.00125 | -0.33 | 0.15172 | 0.384 |
| 22777 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 27 | SCN9A | Sponge network | 0.374 | 0.20054 | -0.525 | 0.10593 | 0.384 |
| 22778 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | INPP4A | Sponge network | 0.299 | 0.57722 | 0.07 | 0.53563 | 0.384 |
| 22779 | PGM5-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | PTPRT | Sponge network | -4.546 | 0 | -0.907 | 0.03727 | 0.384 |
| 22780 | OIP5-AS1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 19 | CSDE1 | Sponge network | 0.421 | 0.00084 | -0.493 | 0 | 0.384 |
| 22781 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-590-3p | 11 | PNRC1 | Sponge network | -0.739 | 0.00044 | -0.646 | 0 | 0.384 |
| 22782 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p;hsa-miR-629-3p | 14 | CHD2 | Sponge network | 0.946 | 0 | 0.049 | 0.55371 | 0.384 |
| 22783 | AC005154.6 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 20 | SIRT1 | Sponge network | 0.848 | 0 | 0.109 | 0.2593 | 0.384 |
| 22784 | SH3BP5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-539-5p | 13 | DDX5 | Sponge network | 0.267 | 0.2937 | -0.099 | 0.17428 | 0.384 |
| 22785 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-940 | 21 | CRTC3 | Sponge network | -1.249 | 7.0E-5 | 0.164 | 0.16786 | 0.384 |
| 22786 | AC009120.6 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-590-3p | 11 | DOCK4 | Sponge network | 0.919 | 0 | 0.502 | 0.00108 | 0.384 |
| 22787 | RP11-195F19.9 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 12 | PDE4DIP | Sponge network | -0.726 | 0.00132 | -0.282 | 0.0257 | 0.384 |
| 22788 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | AFAP1L2 | Sponge network | -0.942 | 0 | -0.284 | 0.15434 | 0.384 |
| 22789 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | PRRX1 | Sponge network | 0.199 | 0.38015 | 1.316 | 0 | 0.384 |
| 22790 | RP11-46C24.7 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 32 | DIP2C | Sponge network | 0.392 | 0.0278 | -0.436 | 0.01713 | 0.384 |
| 22791 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-92a-3p | 11 | DKK3 | Sponge network | -0.738 | 0.21233 | -0.052 | 0.77783 | 0.384 |
| 22792 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-425-5p;hsa-miR-550a-3p | 40 | DTNA | Sponge network | 0.082 | 0.55397 | -1.648 | 1.0E-5 | 0.384 |
| 22793 | BCYRN1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RBM6 | Sponge network | 0.311 | 0.10344 | -0.137 | 0.15663 | 0.384 |
| 22794 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-93-3p | 49 | BMPR2 | Sponge network | -0.896 | 0.00099 | 0.206 | 0.0635 | 0.384 |
| 22795 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429 | 17 | DYNC1I1 | Sponge network | 0.16 | 0.50785 | -1.12 | 0.00107 | 0.384 |
| 22796 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | FREM2 | Sponge network | 0.225 | 0.30644 | -0.437 | 0.46353 | 0.384 |
| 22797 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 23 | TP53INP1 | Sponge network | 0.532 | 0.00505 | -0.496 | 0.00064 | 0.384 |
| 22798 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-96-5p | 47 | AFF4 | Sponge network | 0.614 | 1.0E-5 | -0.266 | 0.01565 | 0.384 |
| 22799 | RP11-182J1.1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 12 | EPHA5 | Sponge network | -0.187 | 0.23787 | -0.957 | 0.01733 | 0.384 |
| 22800 | RP11-822E23.8 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p | 35 | CREB3L2 | Sponge network | -0.777 | 0.16667 | -0.525 | 2.0E-5 | 0.384 |
| 22801 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-590-3p | 28 | ABL2 | Sponge network | 0.335 | 0.20596 | 0.82 | 0 | 0.384 |
| 22802 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 12 | PIK3IP1 | Sponge network | -0.737 | 0.06182 | -0.187 | 0.19945 | 0.384 |
| 22803 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TRIP11 | Sponge network | 0.537 | 0.00139 | -0.158 | 0.06384 | 0.384 |
| 22804 | USP3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-671-5p | 17 | CDH23 | Sponge network | -0.162 | 0.47687 | -0.974 | 0.00034 | 0.384 |
| 22805 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | ERG | Sponge network | -0.351 | 0.11358 | 0.002 | 0.99011 | 0.384 |
| 22806 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-450b-5p | 15 | MOB1B | Sponge network | 0.912 | 0.00014 | 0.197 | 0.15266 | 0.384 |
| 22807 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-330-3p;hsa-miR-33a-3p | 17 | MOB1B | Sponge network | 2.163 | 0 | 0.197 | 0.15266 | 0.384 |
| 22808 | LINC00476 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p | 39 | IL17RD | Sponge network | -0.615 | 0.00015 | -0.019 | 0.94873 | 0.384 |
| 22809 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 47 | CADM2 | Sponge network | -0.513 | 0.04703 | -3.782 | 0 | 0.384 |
| 22810 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-589-3p | 21 | PDGFRA | Sponge network | 0.811 | 0.03228 | -0.372 | 0.0037 | 0.384 |
| 22811 | LINC00657 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-337-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 18 | PICALM | Sponge network | 0.91 | 0 | 0.086 | 0.23523 | 0.384 |
| 22812 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-29b-1-5p | 10 | TCF4 | Sponge network | -0.326 | 0.28809 | 0.016 | 0.92271 | 0.384 |
| 22813 | RP11-244H3.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | FMNL3 | Sponge network | 0.659 | 3.0E-5 | 0.555 | 4.0E-5 | 0.384 |
| 22814 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 12 | RCAN2 | Sponge network | 1.757 | 0 | -0.33 | 0.15172 | 0.384 |
| 22815 | DNAJC27-AS1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | -0.969 | 2.0E-5 | -2.633 | 0 | 0.384 |
| 22816 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | SOX5 | Sponge network | 0.385 | 0.10067 | -1.091 | 4.0E-5 | 0.384 |
| 22817 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TLL1 | Sponge network | -0.737 | 0.10822 | -1.195 | 3.0E-5 | 0.384 |
| 22818 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-590-3p | 12 | UVRAG | Sponge network | -0.899 | 6.0E-5 | -0.017 | 0.83865 | 0.384 |
| 22819 | U91328.19 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 39 | TGFBR3 | Sponge network | -0.303 | 0.21002 | -1.173 | 1.0E-5 | 0.384 |
| 22820 | CTA-941F9.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-935 | 23 | LPP | Sponge network | -0.344 | 0.49624 | -0.459 | 0.04475 | 0.384 |
| 22821 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | IL17RD | Sponge network | -1.654 | 0.00144 | -0.019 | 0.94873 | 0.384 |
| 22822 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 84 | AFF4 | Sponge network | -1.111 | 0.0003 | -0.266 | 0.01565 | 0.384 |
| 22823 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | AKAP11 | Sponge network | 1.2 | 0.01813 | 0.196 | 0.09825 | 0.384 |
| 22824 | RP11-53O19.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-7-1-3p | 26 | SYT4 | Sponge network | -0.405 | 0.22013 | -3.207 | 0 | 0.384 |
| 22825 | LINC00265 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 21 | MDM4 | Sponge network | 1.07 | 0 | 0.345 | 0.00762 | 0.384 |
| 22826 | RP11-122K13.12 |
hsa-let-7a-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p | 10 | RICTOR | Sponge network | 1.219 | 0 | 0.388 | 0.00188 | 0.384 |
| 22827 | GS1-124K5.11 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-590-3p | 21 | SYNPO2 | Sponge network | 0.494 | 0.00339 | -2.342 | 0 | 0.384 |
| 22828 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-3p | 10 | BCL11A | Sponge network | -1.886 | 0 | -0.932 | 0.0063 | 0.384 |
| 22829 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | TLL1 | Sponge network | -0.899 | 6.0E-5 | -1.195 | 3.0E-5 | 0.384 |
| 22830 | RP11-535M15.1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 13 | QKI | Sponge network | -1.14 | 0.03513 | -0.333 | 0.04111 | 0.384 |
| 22831 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-539-5p | 13 | NEDD4 | Sponge network | -0.473 | 0.07602 | 0.02 | 0.89834 | 0.384 |
| 22832 | RP11-532F6.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 16 | TET1 | Sponge network | -0.896 | 0.00099 | -0.135 | 0.52138 | 0.384 |
| 22833 | AC074289.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-7-5p;hsa-miR-96-5p | 29 | BCL2 | Sponge network | -0.326 | 0.14314 | -0.899 | 0 | 0.384 |
| 22834 | HCG11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-93-5p | 30 | CHD5 | Sponge network | 0.385 | 0.10067 | -0.922 | 0.01521 | 0.384 |
| 22835 | LINC00963 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 34 | ABI2 | Sponge network | 0.523 | 9.0E-5 | 0.175 | 0.14787 | 0.384 |
| 22836 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LHFP | Sponge network | 0.064 | 0.72934 | -0.167 | 0.41371 | 0.384 |
| 22837 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 48 | BMPR2 | Sponge network | -0.842 | 0.01361 | 0.206 | 0.0635 | 0.384 |
| 22838 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 11 | CLIP1 | Sponge network | 0.178 | 0.29106 | -0.215 | 0.08684 | 0.384 |
| 22839 | RP11-428J1.5 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 28 | NOTCH2 | Sponge network | 0.538 | 0.00076 | 0.138 | 0.35163 | 0.384 |
| 22840 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PLSCR4 | Sponge network | -0.222 | 0.32445 | -0.474 | 0.03833 | 0.384 |
| 22841 | RP11-48B3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 21 | DMD | Sponge network | 1.757 | 0 | -1.506 | 0 | 0.384 |
| 22842 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 22 | DOCK4 | Sponge network | -0.576 | 0.10121 | 0.502 | 0.00108 | 0.384 |
| 22843 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 24 | EPHA7 | Sponge network | 0.889 | 9.0E-5 | -2.197 | 3.0E-5 | 0.384 |
| 22844 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-582-5p | 14 | RICTOR | Sponge network | 0.757 | 0 | 0.388 | 0.00188 | 0.384 |
| 22845 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 10 | CHD2 | Sponge network | 0.419 | 0.0319 | 0.049 | 0.55371 | 0.384 |
| 22846 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p | 26 | CBL | Sponge network | 1.063 | 0 | 0.291 | 0.01267 | 0.384 |
| 22847 | RP11-152F13.8 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-766-3p | 12 | MDM4 | Sponge network | 0.843 | 0.01775 | 0.345 | 0.00762 | 0.384 |
| 22848 | AC004893.11 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | PIK3CA | Sponge network | 1.317 | 0 | 0.195 | 0.06908 | 0.384 |
| 22849 | CTD-3025N20.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-590-3p;hsa-miR-654-3p | 15 | RICTOR | Sponge network | 1.046 | 0 | 0.388 | 0.00188 | 0.384 |
| 22850 | RP11-360F5.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-877-5p | 12 | NAV3 | Sponge network | 1.867 | 0 | 0.212 | 0.47729 | 0.384 |
| 22851 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 17 | PTPN11 | Sponge network | 0.532 | 0.00505 | 0.431 | 2.0E-5 | 0.384 |
| 22852 | HNRNPU-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | SIRT1 | Sponge network | 1.601 | 0 | 0.109 | 0.2593 | 0.384 |
| 22853 | RP11-159D12.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-940 | 20 | EPS15 | Sponge network | 1.254 | 0 | -0.052 | 0.53998 | 0.384 |
| 22854 | ZSCAN16-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | CELF4 | Sponge network | -0.342 | 0.02288 | -1.135 | 0.00344 | 0.384 |
| 22855 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 36 | MYO9A | Sponge network | -0.942 | 0 | -0.147 | 0.32373 | 0.384 |
| 22856 | CTD-2666L21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-33a-3p;hsa-miR-493-5p | 11 | DMD | Sponge network | 0.368 | 0.20093 | -1.506 | 0 | 0.384 |
| 22857 | LINC00667 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PDE4DIP | Sponge network | -0.235 | 0.15142 | -0.282 | 0.0257 | 0.384 |
| 22858 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | ITGB3 | Sponge network | 0.385 | 0.10067 | -0.011 | 0.95751 | 0.384 |
| 22859 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 13 | IL6ST | Sponge network | -0.326 | 0.28809 | -0.402 | 0.00676 | 0.384 |
| 22860 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 30 | ABI2 | Sponge network | -0.611 | 0.09607 | 0.175 | 0.14787 | 0.384 |
| 22861 | LINC00092 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 21 | PTPRD | Sponge network | -1.886 | 0 | -1.005 | 0.00064 | 0.384 |
| 22862 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 31 | MOB1B | Sponge network | 0.267 | 0.2937 | 0.197 | 0.15266 | 0.384 |
| 22863 | RP11-802E16.3 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 14 | PHF6 | Sponge network | 2.048 | 0 | 0.785 | 0 | 0.384 |
| 22864 | LINC00865 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 36 | GRIK3 | Sponge network | -1.249 | 7.0E-5 | -3.208 | 0 | 0.384 |
| 22865 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-5p | 30 | CLASP2 | Sponge network | -0.162 | 0.47687 | -0.038 | 0.71 | 0.384 |
| 22866 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-590-3p | 12 | ZFHX3 | Sponge network | 1.148 | 2.0E-5 | 0.389 | 0.01363 | 0.384 |
| 22867 | TP73-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 15 | SPTBN4 | Sponge network | -0.563 | 0.02545 | -0.786 | 0.01281 | 0.384 |
| 22868 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-582-5p | 16 | ATRX | Sponge network | 0.792 | 0.0049 | 0.261 | 0.04408 | 0.384 |
| 22869 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-590-3p | 16 | CREBBP | Sponge network | 0.537 | 0.00139 | 0.126 | 0.15186 | 0.384 |
| 22870 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PDGFC | Sponge network | -2.117 | 0.00161 | -0.33 | 0.06483 | 0.384 |
| 22871 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 30 | UNC80 | Sponge network | -0.739 | 0.00044 | -1.934 | 0 | 0.384 |
| 22872 | HHIP-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-590-3p | 12 | PTCH1 | Sponge network | -0.737 | 0.10822 | -0.1 | 0.6179 | 0.384 |
| 22873 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-484;hsa-miR-577 | 10 | MYCBP2 | Sponge network | -0.465 | 0.04703 | -0.027 | 0.82016 | 0.384 |
| 22874 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-7-1-3p | 22 | CNTN1 | Sponge network | -0.303 | 0.21002 | -2.594 | 0 | 0.384 |
| 22875 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 19 | RNF144A | Sponge network | -0.08 | 0.90092 | 0.594 | 0.0009 | 0.384 |
| 22876 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 33 | DDX6 | Sponge network | 0.101 | 0.60919 | 0.091 | 0.36428 | 0.384 |
| 22877 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 30 | BNC2 | Sponge network | 0.259 | 0.12542 | -0.491 | 0.09988 | 0.384 |
| 22878 | CD27-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-766-3p | 14 | EZH1 | Sponge network | 0.177 | 0.16681 | -0.564 | 1.0E-5 | 0.384 |
| 22879 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-655-3p | 10 | ARID2 | Sponge network | 0.542 | 0.00054 | 0.235 | 0.02697 | 0.384 |
| 22880 | RP11-251G23.5 |
hsa-miR-101-3p;hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p | 14 | UBR3 | Sponge network | 1.344 | 0 | -0.021 | 0.82774 | 0.384 |
| 22881 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-624-5p | 11 | SMARCA1 | Sponge network | -1.216 | 0 | -0.684 | 0.00159 | 0.384 |
| 22882 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-589-3p | 12 | TAL1 | Sponge network | 0.176 | 0.37402 | -0.462 | 0.00574 | 0.384 |
| 22883 | USP3-AS1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-628-5p | 32 | RIMS2 | Sponge network | -0.162 | 0.47687 | -1.365 | 0.00208 | 0.384 |
| 22884 | NEAT1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | NF1 | Sponge network | 0.705 | 0.00014 | 0.51 | 0 | 0.384 |
| 22885 | RAMP2-AS1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p | 11 | CACNA1E | Sponge network | -0.513 | 0.04703 | 1.752 | 0 | 0.384 |
| 22886 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 11 | ARHGAP28 | Sponge network | -1.331 | 3.0E-5 | -0.911 | 0.00013 | 0.384 |
| 22887 | LINC00954 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-582-5p | 13 | CREB1 | Sponge network | 1.157 | 0.00455 | 0.203 | 0.00537 | 0.384 |
| 22888 | RP11-46C24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-935 | 31 | DMD | Sponge network | 0.392 | 0.0278 | -1.506 | 0 | 0.384 |
| 22889 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | FYN | Sponge network | -1.349 | 0.00017 | -0.131 | 0.37051 | 0.384 |
| 22890 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-196b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-942-5p | 10 | ARHGAP28 | Sponge network | -0.326 | 0.28809 | -0.911 | 0.00013 | 0.384 |
| 22891 | FAM66C |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 12 | PEA15 | Sponge network | -0.901 | 0.00019 | 0.009 | 0.93318 | 0.384 |
| 22892 | RP11-356J5.12 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-452-5p;hsa-miR-590-5p;hsa-miR-7-5p | 17 | FNBP1 | Sponge network | -0.899 | 6.0E-5 | -0.969 | 4.0E-5 | 0.384 |
| 22893 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 16 | CHD1 | Sponge network | 0.683 | 1.0E-5 | 0.322 | 0.00215 | 0.384 |
| 22894 | RP11-819C21.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 28 | EPS15 | Sponge network | 0.537 | 0.00139 | -0.052 | 0.53998 | 0.384 |
| 22895 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 40 | NTRK3 | Sponge network | -0.206 | 0.12942 | -1.77 | 3.0E-5 | 0.384 |
| 22896 | ADAMTS9-AS2 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 29 | EFNA5 | Sponge network | -2.452 | 0 | -0.421 | 0.17868 | 0.384 |
| 22897 | EMX2OS |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | SPOP | Sponge network | -0.777 | 0.1134 | -0.419 | 1.0E-5 | 0.384 |
| 22898 | SNHG14 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 23 | PRKAR1A | Sponge network | -1.111 | 0.0003 | -0.063 | 0.50314 | 0.384 |
| 22899 | CTD-2002H8.2 |
hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | PIK3R1 | Sponge network | 0.931 | 1.0E-5 | -0.133 | 0.36854 | 0.384 |
| 22900 | MIR143HG |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 16 | EHD3 | Sponge network | -0.739 | 0.0574 | -0.484 | 0.01747 | 0.384 |
| 22901 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-505-3p | 14 | JAKMIP2 | Sponge network | 0.395 | 0.15451 | -1.388 | 0 | 0.384 |
| 22902 | RP11-401P9.4 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-5p | 12 | ID4 | Sponge network | -0.384 | 0.40652 | -1.644 | 0 | 0.384 |
| 22903 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | TBL1XR1 | Sponge network | 1.378 | 0 | 0.578 | 0 | 0.384 |
| 22904 | FAM66C |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SEMA3C | Sponge network | -0.901 | 0.00019 | -0.272 | 0.31153 | 0.384 |
| 22905 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 16 | AKAP11 | Sponge network | 0.997 | 0 | 0.196 | 0.09825 | 0.384 |
| 22906 | CTD-2314G24.2 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ADH1B | Sponge network | 0.246 | 0.59912 | -3.716 | 0 | 0.384 |
| 22907 | JAZF1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 33 | AFF1 | Sponge network | -0.769 | 0.00035 | -0.384 | 0.00053 | 0.384 |
| 22908 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-576-5p | 17 | PDS5B | Sponge network | 0.75 | 0.00054 | 0.259 | 0.00962 | 0.384 |
| 22909 | RP11-2B6.2 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DICER1 | Sponge network | 0.539 | 0.03722 | 0.042 | 0.674 | 0.384 |
| 22910 | RP11-121C2.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-495-3p | 20 | ZFX | Sponge network | 1.147 | 0 | 0.09 | 0.42563 | 0.384 |
| 22911 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -0.154 | 0.78939 | 0.024 | 0.89889 | 0.384 |
| 22912 | LINC00094 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-671-5p | 27 | DLC1 | Sponge network | 0.253 | 0.09565 | -0.382 | 0.05038 | 0.384 |
| 22913 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-493-5p | 14 | SYNE1 | Sponge network | -0.829 | 0.01343 | -0.738 | 0.00316 | 0.383 |
| 22914 | RP5-1139B12.3 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-454-3p | 15 | ZBTB4 | Sponge network | 0.598 | 0.38481 | -0.739 | 0 | 0.383 |
| 22915 | CKMT2-AS1 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | 0.082 | 0.55654 | -1.628 | 0 | 0.383 |
| 22916 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | PDGFRA | Sponge network | -0.487 | 0.02157 | -0.372 | 0.0037 | 0.383 |
| 22917 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-671-5p | 38 | PTPRD | Sponge network | -0.773 | 0.04073 | -1.005 | 0.00064 | 0.383 |
| 22918 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -0.611 | 0.09607 | -0.087 | 0.69193 | 0.383 |
| 22919 | RP11-473I1.9 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-628-5p | 23 | ERC1 | Sponge network | 0.683 | 1.0E-5 | -0.108 | 0.38794 | 0.383 |
| 22920 | TINCR |
hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-96-5p | 22 | MTSS1 | Sponge network | -0.664 | 0.14543 | -0.81 | 0.00035 | 0.383 |
| 22921 | RP11-597D13.9 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | RNF180 | Sponge network | 1.302 | 7.0E-5 | -1.127 | 1.0E-5 | 0.383 |
| 22922 | ACTA2-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 16 | LUZP2 | Sponge network | 0.194 | 0.64278 | -0.056 | 0.89416 | 0.383 |
| 22923 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p | 25 | MYBL1 | Sponge network | -0.222 | 0.32445 | 0.494 | 0.02152 | 0.383 |
| 22924 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-93-3p | 35 | IL6ST | Sponge network | 0.622 | 0.00015 | -0.402 | 0.00676 | 0.383 |
| 22925 | LINC00702 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FAM133A | Sponge network | -0.737 | 0.06182 | 0.549 | 0.29009 | 0.383 |
| 22926 | CTC-559E9.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p | 10 | USP6 | Sponge network | 0.601 | 1.0E-5 | 0.188 | 0.3871 | 0.383 |
| 22927 | LINC00472 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p | 18 | RYR2 | Sponge network | -0.842 | 0.01361 | -1.466 | 0.00031 | 0.383 |
| 22928 | RP11-473I1.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-369-3p | 18 | WDFY3 | Sponge network | 0.542 | 0.00054 | -0.201 | 0.0967 | 0.383 |
| 22929 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 22 | ERRFI1 | Sponge network | -0.129 | 0.57177 | -0.798 | 1.0E-5 | 0.383 |
| 22930 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p | 10 | JAKMIP2 | Sponge network | 0.082 | 0.55654 | -1.388 | 0 | 0.383 |
| 22931 | RP11-161M6.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-339-5p | 11 | NTRK2 | Sponge network | 0.61 | 0.06611 | -0.736 | 0.06495 | 0.383 |
| 22932 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 35 | SCN9A | Sponge network | 0.064 | 0.72934 | -0.525 | 0.10593 | 0.383 |
| 22933 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | CXXC4 | Sponge network | -0.793 | 7.0E-5 | -0.433 | 0.21055 | 0.383 |
| 22934 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 27 | DMXL1 | Sponge network | 0.083 | 0.66405 | -0.426 | 0.00056 | 0.383 |
| 22935 | RP11-38L15.3 |
hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 17 | GAS7 | Sponge network | 0.344 | 0.3127 | -0.555 | 0.01602 | 0.383 |
| 22936 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 19 | SCN9A | Sponge network | -1.203 | 0.03881 | -0.525 | 0.10593 | 0.383 |
| 22937 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 28 | SSBP2 | Sponge network | -0.517 | 0.02935 | -1.58 | 0 | 0.383 |
| 22938 | RP11-672A2.4 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p | 12 | KCNJ5 | Sponge network | 1.344 | 0 | -0.281 | 0.3339 | 0.383 |
| 22939 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 51 | NFIB | Sponge network | -0.487 | 0.02157 | 0.023 | 0.90795 | 0.383 |
| 22940 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-590-3p | 10 | PTPN13 | Sponge network | 0.101 | 0.60919 | -1.208 | 0 | 0.383 |
| 22941 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 27 | IGF1 | Sponge network | -0.727 | 0.04726 | -0.204 | 0.5898 | 0.383 |
| 22942 | RP11-819C21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NAV3 | Sponge network | 0.537 | 0.00139 | 0.212 | 0.47729 | 0.383 |
| 22943 | RP11-693J15.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-455-3p | 11 | RELN | Sponge network | -0.72 | 0.24239 | -2.403 | 0 | 0.383 |
| 22944 | LINC00969 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450a-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-628-5p;hsa-miR-629-3p | 15 | CHD2 | Sponge network | 0.683 | 1.0E-5 | 0.049 | 0.55371 | 0.383 |
| 22945 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-500a-3p;hsa-miR-501-3p | 12 | TET1 | Sponge network | 0.453 | 0.01839 | -0.135 | 0.52138 | 0.383 |
| 22946 | RP11-452L6.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p | 11 | ABL2 | Sponge network | 0.665 | 0.00061 | 0.82 | 0 | 0.383 |
| 22947 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | CREBBP | Sponge network | -0.129 | 0.57177 | 0.126 | 0.15186 | 0.383 |
| 22948 | DIO3OS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p | 26 | DIP2C | Sponge network | -0.773 | 0.04073 | -0.436 | 0.01713 | 0.383 |
| 22949 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | BCL2 | Sponge network | -0.563 | 0.02545 | -0.899 | 0 | 0.383 |
| 22950 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-502-5p | 12 | IL17RD | Sponge network | 0.272 | 0.44692 | -0.019 | 0.94873 | 0.383 |
| 22951 | RP11-434B12.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p | 41 | DST | Sponge network | -0.031 | 0.89063 | -0.713 | 0.00096 | 0.383 |
| 22952 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | LRIG1 | Sponge network | -0.187 | 0.23787 | -0.537 | 0.00588 | 0.383 |
| 22953 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-370-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 23 | PCDH10 | Sponge network | 0.082 | 0.55654 | -1.644 | 0.0078 | 0.383 |
| 22954 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 47 | PRKAA2 | Sponge network | -0.769 | 0.00035 | -2.462 | 0 | 0.383 |
| 22955 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 20 | BCL6 | Sponge network | 0.572 | 0.01722 | -0.211 | 0.19489 | 0.383 |
| 22956 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | THBS1 | Sponge network | -1.349 | 0.00017 | 0.741 | 0.00386 | 0.383 |
| 22957 | DNM1P35 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p | 32 | FAT3 | Sponge network | 0.051 | 0.83388 | -1.601 | 0.00051 | 0.383 |
| 22958 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | EDA2R | Sponge network | -0.942 | 0 | -0.587 | 0.01598 | 0.383 |
| 22959 | AC138035.2 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p | 11 | PHF6 | Sponge network | 1.073 | 0 | 0.785 | 0 | 0.383 |
| 22960 | AC016747.3 |
hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p | 11 | FLNA | Sponge network | 0.199 | 0.38015 | -0.771 | 0.01377 | 0.383 |
| 22961 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-429 | 10 | ELMO1 | Sponge network | -0.469 | 0.08721 | 0.04 | 0.83147 | 0.383 |
| 22962 | FLNB-AS1 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | ABL2 | Sponge network | 1.163 | 0.00018 | 0.82 | 0 | 0.383 |
| 22963 | RP1-228H13.5 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-664a-5p | 17 | CBFB | Sponge network | 1.397 | 0 | 1.061 | 0 | 0.383 |
| 22964 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 33 | NFIB | Sponge network | -1.331 | 3.0E-5 | 0.023 | 0.90795 | 0.383 |
| 22965 | RP11-554A11.4 |
hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p | 11 | KAT6B | Sponge network | -0.583 | 0.14589 | -0.478 | 0.00018 | 0.383 |
| 22966 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | FGF5 | Sponge network | -1.111 | 0.0003 | 0.549 | 0.28659 | 0.383 |
| 22967 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | TRPS1 | Sponge network | -0.206 | 0.12942 | -0.191 | 0.44109 | 0.383 |
| 22968 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p | 15 | HOOK3 | Sponge network | 0.603 | 0.00127 | 0.273 | 0.09957 | 0.383 |
| 22969 | TRIM52-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-532-3p | 10 | CADM3 | Sponge network | -0.418 | 0.00068 | -3.663 | 0 | 0.383 |
| 22970 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p | 24 | MAF | Sponge network | -0.896 | 0.00099 | -1.257 | 0 | 0.383 |
| 22971 | NUTM2A-AS1 |
hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | BRD3 | Sponge network | 0.643 | 0 | 0.37 | 0.00013 | 0.383 |
| 22972 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 17 | ZFP36L1 | Sponge network | -1.349 | 0.00017 | -0.505 | 8.0E-5 | 0.383 |
| 22973 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | DYNC1I1 | Sponge network | 0.082 | 0.55654 | -1.12 | 0.00107 | 0.383 |
| 22974 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | NCAM2 | Sponge network | -0.563 | 0.02545 | -0.482 | 0.22565 | 0.383 |
| 22975 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | SYNE1 | Sponge network | 0.174 | 0.30034 | -0.738 | 0.00316 | 0.383 |
| 22976 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 16 | PTPN11 | Sponge network | 0.082 | 0.55654 | 0.431 | 2.0E-5 | 0.383 |
| 22977 | RP11-693J15.4 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-7-1-3p | 10 | CDH19 | Sponge network | -0.72 | 0.24239 | -3.434 | 0 | 0.383 |
| 22978 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 28 | PRKAA2 | Sponge network | 0.889 | 9.0E-5 | -2.462 | 0 | 0.383 |
| 22979 | CTD-2528L19.6 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | KIF5B | Sponge network | 0.953 | 0 | 0.362 | 0.00066 | 0.383 |
| 22980 | DNAJC3-AS1 |
hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-582-5p | 15 | RICTOR | Sponge network | 0.753 | 0.00014 | 0.388 | 0.00188 | 0.383 |
| 22981 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRIP11 | Sponge network | 1.153 | 0 | -0.158 | 0.06384 | 0.383 |
| 22982 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-625-5p | 22 | PTPRT | Sponge network | -0.469 | 0.08721 | -0.907 | 0.03727 | 0.383 |
| 22983 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ETV1 | Sponge network | 0.619 | 0.00157 | -0.096 | 0.67674 | 0.383 |
| 22984 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | PIK3CA | Sponge network | 0.998 | 0 | 0.195 | 0.06908 | 0.383 |
| 22985 | MIR497HG |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 21 | PDE4DIP | Sponge network | -1.439 | 4.0E-5 | -0.282 | 0.0257 | 0.383 |
| 22986 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p | 12 | KCNA6 | Sponge network | -1.425 | 0.04204 | -1.531 | 0.00013 | 0.383 |
| 22987 | TTC28-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 15 | PHF6 | Sponge network | 0.373 | 0.00068 | 0.785 | 0 | 0.383 |
| 22988 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | NEDD4 | Sponge network | -2.452 | 0 | 0.02 | 0.89834 | 0.383 |
| 22989 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-96-5p | 19 | PRRX1 | Sponge network | -0.517 | 0.02935 | 1.316 | 0 | 0.383 |
| 22990 | MRVI1-AS1 |
hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 14 | OPCML | Sponge network | -1.092 | 4.0E-5 | -2.498 | 0 | 0.383 |
| 22991 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 29 | ESR1 | Sponge network | -1.161 | 0.00591 | -1.035 | 3.0E-5 | 0.383 |
| 22992 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-500a-5p | 17 | LIFR | Sponge network | -0.932 | 0.00329 | -1.794 | 0 | 0.383 |
| 22993 | LINC00863 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PDE4DIP | Sponge network | 0.189 | 0.13981 | -0.282 | 0.0257 | 0.383 |
| 22994 | MIR22HG |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 17 | CADM3 | Sponge network | -1.047 | 0 | -3.663 | 0 | 0.383 |
| 22995 | SNHG14 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FAM133A | Sponge network | -1.111 | 0.0003 | 0.549 | 0.29009 | 0.383 |
| 22996 | AGAP11 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-629-3p | 25 | PTPN14 | Sponge network | -1.645 | 0 | 0.144 | 0.34595 | 0.383 |
| 22997 | AC018890.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-5p;hsa-miR-96-5p | 17 | CADM1 | Sponge network | 1.61 | 0.00961 | -0.9 | 0.00084 | 0.383 |
| 22998 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 23 | RICTOR | Sponge network | 0.174 | 0.30034 | 0.388 | 0.00188 | 0.383 |
| 22999 | CTB-31O20.2 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 11 | CBFB | Sponge network | 0.608 | 3.0E-5 | 1.061 | 0 | 0.383 |
| 23000 | AC010226.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-590-3p | 25 | TGFBR3 | Sponge network | -0.811 | 2.0E-5 | -1.173 | 1.0E-5 | 0.383 |
| 23001 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | 0.395 | 0.15451 | -1.047 | 0.00364 | 0.383 |
| 23002 | AGAP11 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-629-3p | 16 | RYR2 | Sponge network | -1.645 | 0 | -1.466 | 0.00031 | 0.383 |
| 23003 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 58 | AR | Sponge network | -0.969 | 2.0E-5 | -1.554 | 0 | 0.383 |
| 23004 | FENDRR |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZNF844 | Sponge network | -1.281 | 0.00012 | -0.966 | 0.00035 | 0.383 |
| 23005 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | ATRX | Sponge network | 0.349 | 0.01695 | 0.261 | 0.04408 | 0.383 |
| 23006 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 14 | TBX18 | Sponge network | 1.757 | 0 | 0.843 | 0.02945 | 0.383 |
| 23007 | RP11-305E6.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 13 | KIAA1024 | Sponge network | 1.317 | 0 | 1.083 | 0 | 0.383 |
| 23008 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-590-3p | 10 | MLF1 | Sponge network | -0.793 | 7.0E-5 | -0.893 | 0.00727 | 0.383 |
| 23009 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-7-5p | 15 | CRTC3 | Sponge network | -1.161 | 0.00591 | 0.164 | 0.16786 | 0.383 |
| 23010 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 25 | TSHZ3 | Sponge network | 0.082 | 0.55397 | -0.556 | 0.01516 | 0.383 |
| 23011 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-629-3p | 27 | PIK3R1 | Sponge network | -0.469 | 0.08721 | -0.133 | 0.36854 | 0.383 |
| 23012 | RP11-798M19.6 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 28 | RIMS2 | Sponge network | -0.612 | 0.00094 | -1.365 | 0.00208 | 0.383 |
| 23013 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | -0.154 | 0.78939 | -1.047 | 0.00364 | 0.383 |
| 23014 | FAM215B |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-7-1-3p | 11 | PHF3 | Sponge network | 0.817 | 3.0E-5 | 0.126 | 0.19652 | 0.383 |
| 23015 | RP11-35G9.5 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-93-3p;hsa-miR-96-5p | 17 | DOCK4 | Sponge network | 0.622 | 0.00015 | 0.502 | 0.00108 | 0.383 |
| 23016 | RP11-356J5.12 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NRG3 | Sponge network | -0.899 | 6.0E-5 | -1.836 | 4.0E-5 | 0.383 |
| 23017 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 37 | RASSF2 | Sponge network | -1.203 | 0.03881 | -0.273 | 0.21961 | 0.383 |
| 23018 | RP11-867G23.10 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-93-3p | 18 | TOM1L2 | Sponge network | -2.531 | 0 | -0.994 | 0 | 0.383 |
| 23019 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940;hsa-miR-96-5p | 20 | CRTC3 | Sponge network | -1.349 | 0.00017 | 0.164 | 0.16786 | 0.383 |
| 23020 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 28 | RBMS3 | Sponge network | -0.053 | 0.80685 | -0.519 | 0.03551 | 0.383 |
| 23021 | LINC00654 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-96-5p | 13 | WNK2 | Sponge network | 0.374 | 0.20054 | -0.352 | 0.31066 | 0.383 |
| 23022 | RP11-434B12.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | ZNF208 | Sponge network | -0.031 | 0.89063 | -0.4 | 0.22381 | 0.383 |
| 23023 | RP5-1198O20.4 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | PRKCB | Sponge network | 0.997 | 0.00066 | -1.313 | 2.0E-5 | 0.383 |
| 23024 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | PPM1A | Sponge network | -0.726 | 0.00132 | -0.623 | 0 | 0.383 |
| 23025 | CTD-3138B18.6 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-331-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | PHC3 | Sponge network | 2.248 | 0 | 0.212 | 0.0499 | 0.383 |
| 23026 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 50 | AR | Sponge network | 1.302 | 7.0E-5 | -1.554 | 0 | 0.383 |
| 23027 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-93-5p | 37 | PPM1A | Sponge network | -1.645 | 0 | -0.623 | 0 | 0.383 |
| 23028 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 21 | CTIF | Sponge network | -0.612 | 0.00094 | -0.389 | 0.00549 | 0.383 |
| 23029 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 49 | FOXP1 | Sponge network | 0.997 | 0.00066 | 0.042 | 0.74368 | 0.383 |
| 23030 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-655-3p | 14 | LATS1 | Sponge network | 0.594 | 0.41522 | 0.109 | 0.34208 | 0.383 |
| 23031 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | DIP2C | Sponge network | 0.381 | 0.25967 | -0.436 | 0.01713 | 0.383 |
| 23032 | LINC00865 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429 | 16 | PTGFR | Sponge network | -1.249 | 7.0E-5 | -0.233 | 0.45421 | 0.383 |
| 23033 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p | 11 | USP6 | Sponge network | 0.37 | 0.27614 | 0.188 | 0.3871 | 0.383 |
| 23034 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 17 | EBF3 | Sponge network | 1.757 | 0 | -0.058 | 0.81437 | 0.383 |
| 23035 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CLIP1 | Sponge network | 0.101 | 0.60919 | -0.215 | 0.08684 | 0.383 |
| 23036 | TMEM9B-AS1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-582-5p | 19 | DDX6 | Sponge network | 0.529 | 0.00552 | 0.091 | 0.36428 | 0.383 |
| 23037 | ZNF667-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 18 | LUZP2 | Sponge network | -0.976 | 0.00025 | -0.056 | 0.89416 | 0.383 |
| 23038 | TTC25 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | CADM2 | Sponge network | -0.208 | 0.38261 | -3.782 | 0 | 0.383 |
| 23039 | AC009120.5 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-93-3p | 10 | BMPR2 | Sponge network | 1.383 | 0 | 0.206 | 0.0635 | 0.383 |
| 23040 | RP11-384K6.6 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | UBR3 | Sponge network | 0.946 | 0 | -0.021 | 0.82774 | 0.383 |
| 23041 | U91328.19 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-342-3p | 15 | STARD13 | Sponge network | -0.303 | 0.21002 | -0.187 | 0.27383 | 0.383 |
| 23042 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 11 | ZNF770 | Sponge network | 0.614 | 1.0E-5 | 0.206 | 0.06751 | 0.383 |
| 23043 | FZD10-AS1 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-98-5p | 10 | HSPB7 | Sponge network | -0.727 | 0.04726 | -2.268 | 1.0E-5 | 0.383 |
| 23044 | H1FX-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-629-5p | 16 | IGF1 | Sponge network | -0.206 | 0.12942 | -0.204 | 0.5898 | 0.383 |
| 23045 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p | 37 | BMPR2 | Sponge network | 1.063 | 0 | 0.206 | 0.0635 | 0.383 |
| 23046 | AC009948.5 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-589-3p | 16 | MAFB | Sponge network | 0.532 | 0.00505 | -0.023 | 0.89865 | 0.383 |
| 23047 | TPTEP1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 15 | RIMS1 | Sponge network | -1.216 | 0 | -3.143 | 0 | 0.383 |
| 23048 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-92a-3p | 14 | IL17RD | Sponge network | 0.959 | 0 | -0.019 | 0.94873 | 0.383 |
| 23049 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-3p | 17 | ABL2 | Sponge network | 0.174 | 0.30034 | 0.82 | 0 | 0.383 |
| 23050 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | NTRK3 | Sponge network | -0.351 | 0.11358 | -1.77 | 3.0E-5 | 0.383 |
| 23051 | RP11-53O19.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 24 | BRD3 | Sponge network | -0.405 | 0.22013 | 0.37 | 0.00013 | 0.383 |
| 23052 | LINC00954 |
hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-365a-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-550a-3p | 10 | CACNA1E | Sponge network | 1.157 | 0.00455 | 1.752 | 0 | 0.383 |
| 23053 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SAMHD1 | Sponge network | 0.532 | 0.00505 | 0.072 | 0.62895 | 0.383 |
| 23054 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-625-3p | 15 | BMPR2 | Sponge network | 1.055 | 0 | 0.206 | 0.0635 | 0.383 |
| 23055 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 34 | PRDM2 | Sponge network | 0.997 | 0.00066 | -0.258 | 0.00721 | 0.382 |
| 23056 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-589-5p | 15 | SLITRK3 | Sponge network | 0.16 | 0.50785 | -3.328 | 0 | 0.382 |
| 23057 | C20orf166-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-429 | 10 | DUSP1 | Sponge network | -2.69 | 0.0001 | -1.754 | 0 | 0.382 |
| 23058 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | GPC6 | Sponge network | -0.811 | 2.0E-5 | -0.054 | 0.81465 | 0.382 |
| 23059 | CTC-444N24.11 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-497-5p | 11 | CDK6 | Sponge network | 1.153 | 0 | 0.991 | 0 | 0.382 |
| 23060 | GAS6-AS2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p | 26 | DLC1 | Sponge network | 0.16 | 0.50785 | -0.382 | 0.05038 | 0.382 |
| 23061 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-577 | 20 | GREM1 | Sponge network | -0.096 | 0.67958 | 0.902 | 0.01691 | 0.382 |
| 23062 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p | 13 | AKAP12 | Sponge network | -0.727 | 0.04726 | -0.932 | 0.0028 | 0.382 |
| 23063 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 21 | MYBL1 | Sponge network | -0.342 | 0.02288 | 0.494 | 0.02152 | 0.382 |
| 23064 | A2M-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p | 10 | AMPH | Sponge network | -0.08 | 0.90092 | -0.916 | 5.0E-5 | 0.382 |
| 23065 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-93-3p | 30 | ERC1 | Sponge network | -2.69 | 0.0001 | -0.108 | 0.38794 | 0.382 |
| 23066 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p | 30 | DDX6 | Sponge network | 1.11 | 0 | 0.091 | 0.36428 | 0.382 |
| 23067 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | YAP1 | Sponge network | 0.826 | 1.0E-5 | 0.22 | 0.11836 | 0.382 |
| 23068 | AC138035.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-378a-5p | 15 | CREB1 | Sponge network | 1.073 | 0 | 0.203 | 0.00537 | 0.382 |
| 23069 | RP4-791M13.3 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p | 10 | DNMT3A | Sponge network | 0.419 | 0.0319 | 0.302 | 0.0262 | 0.382 |
| 23070 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-935;hsa-miR-940 | 20 | PTPRT | Sponge network | -1.331 | 3.0E-5 | -0.907 | 0.03727 | 0.382 |
| 23071 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-93-5p | 15 | LATS2 | Sponge network | 0.056 | 0.81195 | 0.159 | 0.2304 | 0.382 |
| 23072 | RP11-13K12.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 30 | IGFBP5 | Sponge network | -0.154 | 0.78939 | -0.806 | 0.00105 | 0.382 |
| 23073 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 41 | PPM1A | Sponge network | -0.899 | 6.0E-5 | -0.623 | 0 | 0.382 |
| 23074 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-93-3p | 31 | ERC1 | Sponge network | 0.083 | 0.66405 | -0.108 | 0.38794 | 0.382 |
| 23075 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | PBRM1 | Sponge network | 0.064 | 0.72934 | -0.029 | 0.78601 | 0.382 |
| 23076 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | ZMYND11 | Sponge network | 0.421 | 0.00084 | -0.2 | 0.05449 | 0.382 |
| 23077 | JAZF1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 20 | KCNJ5 | Sponge network | -0.769 | 0.00035 | -0.281 | 0.3339 | 0.382 |
| 23078 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-708-5p;hsa-miR-96-5p | 40 | SPRY3 | Sponge network | 0.72 | 0.0001 | 0.016 | 0.91286 | 0.382 |
| 23079 | RP11-360F5.3 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-590-3p | 11 | PRRX1 | Sponge network | 1.867 | 0 | 1.316 | 0 | 0.382 |
| 23080 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-30a-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 14 | SOX11 | Sponge network | 1.055 | 0 | 2.68 | 0 | 0.382 |
| 23081 | AGAP11 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-369-3p;hsa-miR-576-5p | 15 | KLF10 | Sponge network | -1.645 | 0 | -0.783 | 0 | 0.382 |
| 23082 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-5p | 27 | PIK3R1 | Sponge network | 0.889 | 9.0E-5 | -0.133 | 0.36854 | 0.382 |
| 23083 | MIR143HG |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLI1 | Sponge network | -0.739 | 0.0574 | -0.294 | 0.10905 | 0.382 |
| 23084 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | PDGFRA | Sponge network | -1.216 | 0 | -0.372 | 0.0037 | 0.382 |
| 23085 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | MITF | Sponge network | -1.047 | 0 | -0.769 | 0.00022 | 0.382 |
| 23086 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 18 | TRIM33 | Sponge network | 0.683 | 1.0E-5 | 0.402 | 7.0E-5 | 0.382 |
| 23087 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 13 | UVRAG | Sponge network | 0.214 | 0.18246 | -0.017 | 0.83865 | 0.382 |
| 23088 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | LRIG1 | Sponge network | -2.117 | 0.00161 | -0.537 | 0.00588 | 0.382 |
| 23089 | CTA-941F9.9 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-29b-1-5p;hsa-miR-576-5p | 15 | IL6ST | Sponge network | -0.344 | 0.49624 | -0.402 | 0.00676 | 0.382 |
| 23090 | MIR497HG |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-5p | 11 | RTN4 | Sponge network | -1.439 | 4.0E-5 | -0.412 | 0.00014 | 0.382 |
| 23091 | RP11-400K9.4 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-942-5p | 11 | ZBTB20 | Sponge network | -0.611 | 0.09607 | -0.122 | 0.57569 | 0.382 |
| 23092 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.862 | 0.06213 | 0.323 | 0.00792 | 0.382 |
| 23093 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-590-3p | 12 | DDX5 | Sponge network | 0.082 | 0.55654 | -0.099 | 0.17428 | 0.382 |
| 23094 | RP5-1139B12.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p | 11 | SYNM | Sponge network | 0.598 | 0.38481 | -2.309 | 1.0E-5 | 0.382 |
| 23095 | RP11-2B6.2 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PHF3 | Sponge network | 0.539 | 0.03722 | 0.126 | 0.19652 | 0.382 |
| 23096 | HAND2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 44 | AFF1 | Sponge network | -1.697 | 0.00889 | -0.384 | 0.00053 | 0.382 |
| 23097 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 21 | CADM1 | Sponge network | -0.206 | 0.12942 | -0.9 | 0.00084 | 0.382 |
| 23098 | RP5-994D16.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PHC3 | Sponge network | 0.252 | 0.08508 | 0.212 | 0.0499 | 0.382 |
| 23099 | AC010226.4 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | PAIP2 | Sponge network | -0.811 | 2.0E-5 | -0.515 | 0 | 0.382 |
| 23100 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 30 | MOB1B | Sponge network | 0.174 | 0.30034 | 0.197 | 0.15266 | 0.382 |
| 23101 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-182-5p | 10 | STARD13 | Sponge network | -2.531 | 0 | -0.187 | 0.27383 | 0.382 |
| 23102 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | PAFAH1B1 | Sponge network | -0.709 | 0.18168 | -0.429 | 0 | 0.382 |
| 23103 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 48 | FAT3 | Sponge network | 0.253 | 0.09565 | -1.601 | 0.00051 | 0.382 |
| 23104 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | NALCN | Sponge network | -0.323 | 0.01372 | 1.068 | 0.00237 | 0.382 |
| 23105 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | GRIN2A | Sponge network | -0.222 | 0.32445 | -2.161 | 0 | 0.382 |
| 23106 | LINC00662 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p | 20 | CADM1 | Sponge network | 0.213 | 0.32297 | -0.9 | 0.00084 | 0.382 |
| 23107 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 15 | TRIM3 | Sponge network | -2.69 | 0.0001 | -0.577 | 0 | 0.382 |
| 23108 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-3p | 16 | LATS1 | Sponge network | 0.817 | 3.0E-5 | 0.109 | 0.34208 | 0.382 |
| 23109 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | PREX2 | Sponge network | -2.46 | 0 | -0.272 | 0.25382 | 0.382 |
| 23110 | CKMT2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | IL17RD | Sponge network | 0.082 | 0.55654 | -0.019 | 0.94873 | 0.382 |
| 23111 | USP3-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-5p | 26 | KCNA6 | Sponge network | -0.162 | 0.47687 | -1.531 | 0.00013 | 0.382 |
| 23112 | LINC00476 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-5p | 31 | BTG2 | Sponge network | -0.615 | 0.00015 | -1.763 | 0 | 0.382 |
| 23113 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 13 | INPP4A | Sponge network | -0.246 | 0.35034 | 0.07 | 0.53563 | 0.382 |
| 23114 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-324-5p;hsa-miR-542-3p;hsa-miR-98-5p | 12 | DMD | Sponge network | -0.301 | 0.06597 | -1.506 | 0 | 0.382 |
| 23115 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-93-5p | 11 | PLSCR4 | Sponge network | 0.497 | 0.01451 | -0.474 | 0.03833 | 0.382 |
| 23116 | RP4-668J24.2 |
hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-532-3p | 11 | PBX1 | Sponge network | -1.054 | 0.0706 | -1.206 | 0 | 0.382 |
| 23117 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p | 17 | CREB3L2 | Sponge network | 0.889 | 9.0E-5 | -0.525 | 2.0E-5 | 0.382 |
| 23118 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 30 | HLF | Sponge network | 0.222 | 0.29133 | -2.443 | 0 | 0.382 |
| 23119 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | TCF4 | Sponge network | -0.351 | 0.11358 | 0.016 | 0.92271 | 0.382 |
| 23120 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p | 11 | ABCA10 | Sponge network | -0.896 | 0.00099 | -0.571 | 0.03674 | 0.382 |
| 23121 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | -0.777 | 0.16667 | -1.277 | 0 | 0.382 |
| 23122 | RP11-535M15.1 |
hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c | 10 | TACC1 | Sponge network | -1.14 | 0.03513 | -0.362 | 0.05406 | 0.382 |
| 23123 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-362-5p;hsa-miR-590-3p | 13 | TLE4 | Sponge network | -0.096 | 0.67958 | -1.157 | 0 | 0.382 |
| 23124 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p | 10 | HOOK3 | Sponge network | 0.95 | 0.15943 | 0.273 | 0.09957 | 0.382 |
| 23125 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 54 | AR | Sponge network | 0.219 | 0.1516 | -1.554 | 0 | 0.382 |
| 23126 | RP11-439A17.10 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p | 12 | AR | Sponge network | 0.051 | 0.95756 | -1.554 | 0 | 0.382 |
| 23127 | GAS6-AS2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 13 | MAPK10 | Sponge network | 0.16 | 0.50785 | -1.774 | 0 | 0.382 |
| 23128 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | ANK2 | Sponge network | -4.477 | 0 | -1.603 | 1.0E-5 | 0.382 |
| 23129 | HCG11 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 20 | NUAK1 | Sponge network | 0.385 | 0.10067 | 0.904 | 0 | 0.382 |
| 23130 | SNHG14 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 48 | PPP1R9A | Sponge network | -1.111 | 0.0003 | -0.903 | 0.04254 | 0.382 |
| 23131 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-940 | 53 | WDFY3 | Sponge network | -0.901 | 0.00019 | -0.201 | 0.0967 | 0.382 |
| 23132 | RP11-774O3.3 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | SH3GLB1 | Sponge network | -0.487 | 0.02157 | -0.115 | 0.14773 | 0.382 |
| 23133 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-96-5p | 13 | DAB2 | Sponge network | 0.395 | 0.15451 | 0.431 | 0.0113 | 0.382 |
| 23134 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | DAB2 | Sponge network | -0.769 | 0.00035 | 0.431 | 0.0113 | 0.382 |
| 23135 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | CDC73 | Sponge network | 1.205 | 0 | 0.094 | 0.20463 | 0.382 |
| 23136 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | CNTN1 | Sponge network | 0.385 | 0.10067 | -2.594 | 0 | 0.382 |
| 23137 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-590-3p | 21 | LATS1 | Sponge network | 0.101 | 0.60919 | 0.109 | 0.34208 | 0.382 |
| 23138 | RP11-156P1.3 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-92a-3p | 12 | SPTBN4 | Sponge network | 0.214 | 0.18246 | -0.786 | 0.01281 | 0.382 |
| 23139 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | EXOC8 | Sponge network | 0.537 | 0.00139 | 0.216 | 0.00598 | 0.382 |
| 23140 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 23 | EBF3 | Sponge network | -0.793 | 7.0E-5 | -0.058 | 0.81437 | 0.382 |
| 23141 | AC010226.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-590-3p | 13 | STARD13 | Sponge network | -0.811 | 2.0E-5 | -0.187 | 0.27383 | 0.382 |
| 23142 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 22 | ACVR1 | Sponge network | 1.302 | 7.0E-5 | 0.279 | 0.00234 | 0.382 |
| 23143 | ZNF582-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | DIP2C | Sponge network | -1.349 | 0.00017 | -0.436 | 0.01713 | 0.382 |
| 23144 | SNX29P2 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-96-5p | 14 | CXCL12 | Sponge network | 0.4 | 0.14611 | -1.421 | 0 | 0.382 |
| 23145 | RP11-1006G14.4 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-744-3p | 41 | DST | Sponge network | 0.559 | 0.00026 | -0.713 | 0.00096 | 0.382 |
| 23146 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 40 | SOX5 | Sponge network | -0.738 | 0.21233 | -1.091 | 4.0E-5 | 0.382 |
| 23147 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 42 | ZFHX4 | Sponge network | 0.082 | 0.55654 | 0.018 | 0.95574 | 0.382 |
| 23148 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p | 65 | NTRK3 | Sponge network | -0.615 | 0.00015 | -1.77 | 3.0E-5 | 0.382 |
| 23149 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ZFX | Sponge network | 0.559 | 0.00026 | 0.09 | 0.42563 | 0.382 |
| 23150 | RP11-531A24.5 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 32 | ARHGEF12 | Sponge network | 0.75 | 0.00054 | 0.09 | 0.35693 | 0.382 |
| 23151 | LINC00854 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p | 11 | USP6 | Sponge network | 1.18 | 0 | 0.188 | 0.3871 | 0.382 |
| 23152 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 20 | SCN9A | Sponge network | 0.178 | 0.29106 | -0.525 | 0.10593 | 0.382 |
| 23153 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p | 14 | PSIP1 | Sponge network | 0.75 | 0.00054 | -0.005 | 0.96897 | 0.382 |
| 23154 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p | 34 | LIFR | Sponge network | -0.576 | 0.10121 | -1.794 | 0 | 0.382 |
| 23155 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | PTPRT | Sponge network | -1.161 | 0.00591 | -0.907 | 0.03727 | 0.382 |
| 23156 | U91328.19 |
hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | BCL9 | Sponge network | -0.303 | 0.21002 | 0.321 | 0.00267 | 0.382 |
| 23157 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 18 | SYNE1 | Sponge network | -0.663 | 0.10887 | -0.738 | 0.00316 | 0.382 |
| 23158 | AGAP11 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p | 25 | ZMYND11 | Sponge network | -1.645 | 0 | -0.2 | 0.05449 | 0.382 |
| 23159 | CTD-3064M3.3 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p | 11 | FMNL3 | Sponge network | -0.469 | 0.08721 | 0.555 | 4.0E-5 | 0.382 |
| 23160 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-486-5p | 11 | MYO9A | Sponge network | 0.368 | 0.20093 | -0.147 | 0.32373 | 0.382 |
| 23161 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-940 | 50 | WDFY3 | Sponge network | -1.249 | 7.0E-5 | -0.201 | 0.0967 | 0.382 |
| 23162 | RP11-47A8.5 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-625-5p;hsa-miR-92a-1-5p | 10 | SNX29 | Sponge network | 0.103 | 0.43169 | -0.34 | 0.01371 | 0.382 |
| 23163 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 10 | UVRAG | Sponge network | -0.513 | 0.04703 | -0.017 | 0.83865 | 0.382 |
| 23164 | SNX29P2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-96-5p | 24 | ASTN1 | Sponge network | 0.4 | 0.14611 | -2.389 | 2.0E-5 | 0.382 |
| 23165 | CTD-2139B15.2 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-590-5p | 11 | UBE2D1 | Sponge network | 0.286 | 0.00612 | 0.468 | 0 | 0.382 |
| 23166 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-625-5p | 14 | ANK2 | Sponge network | -1.054 | 0.0706 | -1.603 | 1.0E-5 | 0.382 |
| 23167 | PGM5-AS1 |
hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-378a-3p;hsa-miR-589-5p | 10 | RPS6KA2 | Sponge network | -4.546 | 0 | -0.57 | 0.00013 | 0.382 |
| 23168 | TPTEP1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 34 | CTIF | Sponge network | -1.216 | 0 | -0.389 | 0.00549 | 0.382 |
| 23169 | AGAP11 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-590-5p | 10 | MAPK10 | Sponge network | -1.645 | 0 | -1.774 | 0 | 0.382 |
| 23170 | PCA3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 34 | PTPRT | Sponge network | -0.709 | 0.18168 | -0.907 | 0.03727 | 0.382 |
| 23171 | HCG11 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | LATS2 | Sponge network | 0.385 | 0.10067 | 0.159 | 0.2304 | 0.382 |
| 23172 | AC004893.11 |
hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 10 | CDK6 | Sponge network | 1.317 | 0 | 0.991 | 0 | 0.382 |
| 23173 | RP11-760H22.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-92a-1-5p | 28 | PPARA | Sponge network | 0.001 | 0.99753 | -0.379 | 0.00548 | 0.382 |
| 23174 | RP11-554A11.4 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-628-5p | 10 | IRS1 | Sponge network | -0.583 | 0.14589 | -0.026 | 0.88855 | 0.382 |
| 23175 | PART1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-877-5p | 32 | ASTN1 | Sponge network | -2.925 | 0 | -2.389 | 2.0E-5 | 0.382 |
| 23176 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | ST6GAL2 | Sponge network | 0.619 | 0.00157 | -1.201 | 0.00597 | 0.382 |
| 23177 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 21 | TGFBR3 | Sponge network | -0.285 | 0.2172 | -1.173 | 1.0E-5 | 0.382 |
| 23178 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p | 20 | CRTC1 | Sponge network | -0.421 | 0.0114 | -0.116 | 0.28412 | 0.382 |
| 23179 | RP11-967K21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577 | 14 | NRK | Sponge network | 0.056 | 0.81195 | 0.656 | 0.19217 | 0.382 |
| 23180 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | EBF1 | Sponge network | -1.375 | 0.08642 | -0.384 | 0.08856 | 0.382 |
| 23181 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 33 | BHLHE41 | Sponge network | -0.739 | 0.0574 | 0.353 | 0.12753 | 0.382 |
| 23182 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | PDGFRA | Sponge network | 0.342 | 0.03129 | -0.372 | 0.0037 | 0.382 |
| 23183 | LINC00865 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 27 | NAV3 | Sponge network | -1.249 | 7.0E-5 | 0.212 | 0.47729 | 0.382 |
| 23184 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-655-3p | 18 | NF1 | Sponge network | 1.147 | 0 | 0.51 | 0 | 0.382 |
| 23185 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 53 | KIF1B | Sponge network | 0.222 | 0.29133 | -0.118 | 0.32258 | 0.382 |
| 23186 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | SASH1 | Sponge network | -0.318 | 0.65847 | -0.502 | 0.00175 | 0.382 |
| 23187 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p | 10 | NEDD4 | Sponge network | -0.932 | 0.00329 | 0.02 | 0.89834 | 0.382 |
| 23188 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 11 | ZNF208 | Sponge network | 0.537 | 0.00139 | -0.4 | 0.22381 | 0.382 |
| 23189 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-7-5p | 31 | ATRX | Sponge network | -0.162 | 0.47687 | 0.261 | 0.04408 | 0.382 |
| 23190 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-93-5p | 15 | CNTNAP3 | Sponge network | 0.299 | 0.57722 | -1.465 | 0.00033 | 0.382 |
| 23191 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | ST6GAL2 | Sponge network | 0.16 | 0.50785 | -1.201 | 0.00597 | 0.382 |
| 23192 | RP11-597D13.9 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p | 11 | DCLK1 | Sponge network | 1.302 | 7.0E-5 | -1.324 | 0.00014 | 0.382 |
| 23193 | LINC00092 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p | 15 | ARNT2 | Sponge network | -1.886 | 0 | -0.332 | 0.25851 | 0.382 |
| 23194 | PWAR6 |
hsa-miR-10b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 10 | EXTL2 | Sponge network | -1.575 | 6.0E-5 | 0.109 | 0.37375 | 0.382 |
| 23195 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | HACE1 | Sponge network | 0.67 | 0.00048 | -0.072 | 0.55239 | 0.382 |
| 23196 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PLSCR4 | Sponge network | -0.342 | 0.02288 | -0.474 | 0.03833 | 0.382 |
| 23197 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | 0.194 | 0.64278 | 0.126 | 0.15266 | 0.382 |
| 23198 | CTD-2314G24.2 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 11 | THSD7B | Sponge network | 0.246 | 0.59912 | 0.154 | 0.74723 | 0.382 |
| 23199 | RP11-379F4.7 |
hsa-miR-101-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 14 | RICTOR | Sponge network | 1.316 | 0 | 0.388 | 0.00188 | 0.382 |
| 23200 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 49 | QKI | Sponge network | -2.915 | 0.00015 | -0.333 | 0.04111 | 0.382 |
| 23201 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 34 | SCN9A | Sponge network | 0.421 | 0.00084 | -0.525 | 0.10593 | 0.382 |
| 23202 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 28 | EPHA3 | Sponge network | -0.18 | 0.20343 | 0.185 | 0.53588 | 0.382 |
| 23203 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SEMA3C | Sponge network | -0.323 | 0.01372 | -0.272 | 0.31153 | 0.382 |
| 23204 | TP73-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | NIN | Sponge network | -0.563 | 0.02545 | -0.253 | 0.13794 | 0.382 |
| 23205 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | LRRK2 | Sponge network | -0.227 | 0.31657 | -1.136 | 1.0E-5 | 0.382 |
| 23206 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PLSCR4 | Sponge network | -0.141 | 0.26434 | -0.474 | 0.03833 | 0.382 |
| 23207 | LINC00641 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 16 | RHOB | Sponge network | -0.222 | 0.32445 | -1.052 | 0 | 0.382 |
| 23208 | OIP5-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | KDSR | Sponge network | 0.421 | 0.00084 | 0.239 | 0.02216 | 0.382 |
| 23209 | CTD-2528L19.6 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-5p | 10 | RANBP6 | Sponge network | 0.953 | 0 | 0.175 | 0.19756 | 0.382 |
| 23210 | RP11-6O2.3 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-340-5p | 16 | CSDE1 | Sponge network | 0.331 | 0.57247 | -0.493 | 0 | 0.382 |
| 23211 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 28 | TACC1 | Sponge network | -0.811 | 2.0E-5 | -0.362 | 0.05406 | 0.382 |
| 23212 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-93-5p | 40 | ADARB1 | Sponge network | -0.384 | 0.40652 | -0.335 | 0.06631 | 0.382 |
| 23213 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SYNE1 | Sponge network | -0.342 | 0.02288 | -0.738 | 0.00316 | 0.382 |
| 23214 | HCG11 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-629-3p | 15 | CDH23 | Sponge network | 0.385 | 0.10067 | -0.974 | 0.00034 | 0.382 |
| 23215 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-655-3p | 17 | KANK1 | Sponge network | 0.083 | 0.66405 | -0.55 | 0.00134 | 0.382 |
| 23216 | KB-1460A1.5 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-629-5p | 10 | WNK2 | Sponge network | 0.603 | 0.00127 | -0.352 | 0.31066 | 0.382 |
| 23217 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 19 | PTPRD | Sponge network | -0.376 | 0.00337 | -1.005 | 0.00064 | 0.382 |
| 23218 | ZNF582-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | MED12L | Sponge network | -1.349 | 0.00017 | -0.194 | 0.55299 | 0.382 |
| 23219 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TGFBR3 | Sponge network | -0.031 | 0.89063 | -1.173 | 1.0E-5 | 0.382 |
| 23220 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p | 12 | ABL2 | Sponge network | 0.37 | 0.27614 | 0.82 | 0 | 0.382 |
| 23221 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 22 | NR2C2 | Sponge network | 0.064 | 0.72934 | 0.21 | 0.03224 | 0.382 |
| 23222 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p | 14 | DCLK1 | Sponge network | -0.773 | 0.04073 | -1.324 | 0.00014 | 0.382 |
| 23223 | RP11-359B12.2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | RYR2 | Sponge network | 0.064 | 0.72934 | -1.466 | 0.00031 | 0.382 |
| 23224 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-484 | 12 | RET | Sponge network | -4.546 | 0 | -0.833 | 0.03517 | 0.382 |
| 23225 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 18 | FNBP1 | Sponge network | -0.942 | 0 | -0.969 | 4.0E-5 | 0.382 |
| 23226 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-98-5p | 34 | TGFBR3 | Sponge network | -0.773 | 0.04073 | -1.173 | 1.0E-5 | 0.382 |
| 23227 | RP11-48B3.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 13 | LATS2 | Sponge network | 1.757 | 0 | 0.159 | 0.2304 | 0.382 |
| 23228 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PTPRB | Sponge network | -0.421 | 0.0114 | 0.105 | 0.47122 | 0.382 |
| 23229 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 25 | ARID5B | Sponge network | 0.225 | 0.30644 | 0.342 | 0.01676 | 0.382 |
| 23230 | ATP1B3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 12 | ABI2 | Sponge network | 1.2 | 0.01813 | 0.175 | 0.14787 | 0.382 |
| 23231 | LINC00473 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-877-5p | 21 | NRK | Sponge network | -1.203 | 0.03881 | 0.656 | 0.19217 | 0.382 |
| 23232 | LINC00680 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p | 18 | RICTOR | Sponge network | 0.884 | 0 | 0.388 | 0.00188 | 0.382 |
| 23233 | CTC-429P9.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 10 | FOXO3 | Sponge network | 0.497 | 0.01451 | -0.04 | 0.72028 | 0.382 |
| 23234 | LINC00472 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-942-5p | 27 | IGFBP5 | Sponge network | -0.842 | 0.01361 | -0.806 | 0.00105 | 0.382 |
| 23235 | TPTEP1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 67 | DST | Sponge network | -1.216 | 0 | -0.713 | 0.00096 | 0.382 |
| 23236 | C20orf166-AS1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 11 | PEA15 | Sponge network | -2.69 | 0.0001 | 0.009 | 0.93318 | 0.382 |
| 23237 | PTOV1-AS1 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-378a-3p;hsa-miR-378c | 15 | ABL2 | Sponge network | 0.87 | 0 | 0.82 | 0 | 0.382 |
| 23238 | CTC-559E9.5 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p | 13 | PIK3CA | Sponge network | 0.594 | 0.00064 | 0.195 | 0.06908 | 0.382 |
| 23239 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-424-5p | 12 | CRTC3 | Sponge network | -0.932 | 0.00329 | 0.164 | 0.16786 | 0.382 |
| 23240 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-590-3p | 16 | SLITRK3 | Sponge network | -0.899 | 6.0E-5 | -3.328 | 0 | 0.382 |
| 23241 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p;hsa-miR-940 | 14 | BHLHE41 | Sponge network | -1.255 | 0.00981 | 0.353 | 0.12753 | 0.382 |
| 23242 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 20 | ARNT2 | Sponge network | -0.096 | 0.67958 | -0.332 | 0.25851 | 0.382 |
| 23243 | RP11-720L2.4 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 20 | DST | Sponge network | -1.255 | 0.00981 | -0.713 | 0.00096 | 0.382 |
| 23244 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-411-5p;hsa-miR-590-3p | 17 | USP6 | Sponge network | 0.765 | 7.0E-5 | 0.188 | 0.3871 | 0.382 |
| 23245 | CTC-296K1.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-671-5p | 18 | KCNA6 | Sponge network | -2.095 | 0.00112 | -1.531 | 0.00013 | 0.382 |
| 23246 | HAND2-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PCDH18 | Sponge network | -1.697 | 0.00889 | 0.216 | 0.24645 | 0.382 |
| 23247 | AC159540.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | RICTOR | Sponge network | 1.634 | 0 | 0.388 | 0.00188 | 0.381 |
| 23248 | LINC00476 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | -0.615 | 0.00015 | -1.628 | 0 | 0.381 |
| 23249 | AC005618.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | 0.862 | 0.06213 | 0.809 | 0.00544 | 0.381 |
| 23250 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 52 | QKI | Sponge network | -0.793 | 7.0E-5 | -0.333 | 0.04111 | 0.381 |
| 23251 | AC127904.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PHF3 | Sponge network | 1.308 | 0 | 0.126 | 0.19652 | 0.381 |
| 23252 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-93-5p | 43 | BNC2 | Sponge network | 0.056 | 0.81195 | -0.491 | 0.09988 | 0.381 |
| 23253 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-96-5p | 22 | CBFA2T3 | Sponge network | -1.654 | 0.00144 | -1.221 | 1.0E-5 | 0.381 |
| 23254 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | NR2C2 | Sponge network | 1.073 | 0.00216 | 0.21 | 0.03224 | 0.381 |
| 23255 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 12 | MCC | Sponge network | 0.342 | 0.03129 | -1.005 | 0 | 0.381 |
| 23256 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 21 | PRKAR1A | Sponge network | -0.129 | 0.57177 | -0.063 | 0.50314 | 0.381 |
| 23257 | HCG11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | MED12L | Sponge network | 0.385 | 0.10067 | -0.194 | 0.55299 | 0.381 |
| 23258 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | DDX6 | Sponge network | -0.323 | 0.01372 | 0.091 | 0.36428 | 0.381 |
| 23259 | LINC00969 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p | 10 | CHD1 | Sponge network | 0.683 | 1.0E-5 | 0.322 | 0.00215 | 0.381 |
| 23260 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 32 | DMXL1 | Sponge network | 0.421 | 0.00084 | -0.426 | 0.00056 | 0.381 |
| 23261 | LINC00957 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-450b-5p | 11 | SPTBN4 | Sponge network | -0.421 | 0.0114 | -0.786 | 0.01281 | 0.381 |
| 23262 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ZNF208 | Sponge network | 0.246 | 0.59912 | -0.4 | 0.22381 | 0.381 |
| 23263 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p | 15 | TLL1 | Sponge network | -0.154 | 0.78939 | -1.195 | 3.0E-5 | 0.381 |
| 23264 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p | 11 | TXNIP | Sponge network | 0.16 | 0.50785 | -1.277 | 0 | 0.381 |
| 23265 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 33 | PRKAB2 | Sponge network | -0.901 | 0.00019 | -0.367 | 0.01371 | 0.381 |
| 23266 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 51 | TCF4 | Sponge network | -0.08 | 0.90092 | 0.016 | 0.92271 | 0.381 |
| 23267 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-96-5p | 10 | TLL1 | Sponge network | -0.517 | 0.02935 | -1.195 | 3.0E-5 | 0.381 |
| 23268 | RP11-65J21.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 11 | AKAP6 | Sponge network | -0.557 | 0.11135 | -1.586 | 3.0E-5 | 0.381 |
| 23269 | MIR600HG |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-655-3p;hsa-miR-940 | 10 | TAF1 | Sponge network | 0.594 | 0.41522 | 0.39 | 3.0E-5 | 0.381 |
| 23270 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | RBMS3 | Sponge network | -0.031 | 0.89063 | -0.519 | 0.03551 | 0.381 |
| 23271 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 22 | CDHR3 | Sponge network | -0.323 | 0.01372 | -0.977 | 4.0E-5 | 0.381 |
| 23272 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-425-5p;hsa-miR-628-5p;hsa-miR-7-5p | 15 | ERC1 | Sponge network | -0.726 | 0.00132 | -0.108 | 0.38794 | 0.381 |
| 23273 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | 0.083 | 0.66405 | -1.74 | 0 | 0.381 |
| 23274 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | ANK2 | Sponge network | 1.302 | 7.0E-5 | -1.603 | 1.0E-5 | 0.381 |
| 23275 | NDUFA6-AS1 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-98-5p | 10 | HSPB7 | Sponge network | -0.355 | 0.0077 | -2.268 | 1.0E-5 | 0.381 |
| 23276 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | 0.997 | 0.00066 | -0.252 | 0.15511 | 0.381 |
| 23277 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 23 | ITGAV | Sponge network | 0.249 | 0.35902 | 0.337 | 0.01002 | 0.381 |
| 23278 | HHIP-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | PTPRT | Sponge network | -0.737 | 0.10822 | -0.907 | 0.03727 | 0.381 |
| 23279 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-369-3p;hsa-miR-576-5p | 10 | CDHR3 | Sponge network | -0.932 | 0.00329 | -0.977 | 4.0E-5 | 0.381 |
| 23280 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-589-5p;hsa-miR-935 | 19 | RARB | Sponge network | -0.162 | 0.47687 | -0.825 | 2.0E-5 | 0.381 |
| 23281 | RP11-305E6.4 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p | 17 | TSC1 | Sponge network | 1.317 | 0 | 0.103 | 0.26071 | 0.381 |
| 23282 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-589-3p | 13 | ABCC9 | Sponge network | -0.773 | 0.04073 | -0.466 | 0.14121 | 0.381 |
| 23283 | FENDRR |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p | 12 | NGFR | Sponge network | -1.281 | 0.00012 | -1.271 | 0.01058 | 0.381 |
| 23284 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-629-3p | 22 | RASSF2 | Sponge network | 0.811 | 0.03228 | -0.273 | 0.21961 | 0.381 |
| 23285 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | AFF3 | Sponge network | -0.777 | 0.1134 | -2.373 | 0 | 0.381 |
| 23286 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-93-5p | 31 | PPM1A | Sponge network | -2.69 | 0.0001 | -0.623 | 0 | 0.381 |
| 23287 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PRKAB2 | Sponge network | 0.417 | 0.00445 | -0.367 | 0.01371 | 0.381 |
| 23288 | DIO3OS |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-339-5p;hsa-miR-7-5p | 11 | FNBP1 | Sponge network | -0.773 | 0.04073 | -0.969 | 4.0E-5 | 0.381 |
| 23289 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-940 | 37 | GAS7 | Sponge network | 0.056 | 0.81195 | -0.555 | 0.01602 | 0.381 |
| 23290 | RP11-102N12.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-671-5p | 19 | GRIK3 | Sponge network | -0.351 | 0.11358 | -3.208 | 0 | 0.381 |
| 23291 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 31 | ERBB4 | Sponge network | -4.546 | 0 | -1.975 | 2.0E-5 | 0.381 |
| 23292 | RP11-251G23.5 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p | 14 | ABL2 | Sponge network | 1.344 | 0 | 0.82 | 0 | 0.381 |
| 23293 | LINC00265 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 11 | RPS6KA3 | Sponge network | 1.07 | 0 | 0.256 | 0.02419 | 0.381 |
| 23294 | CTD-3092A11.2 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | UBE2D1 | Sponge network | 0.873 | 0 | 0.468 | 0 | 0.381 |
| 23295 | ARHGEF26-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | ROBO1 | Sponge network | -2.46 | 0 | -0.009 | 0.96154 | 0.381 |
| 23296 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 64 | NTRK3 | Sponge network | -0.096 | 0.67958 | -1.77 | 3.0E-5 | 0.381 |
| 23297 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | ZNF292 | Sponge network | 0.222 | 0.29133 | 0.276 | 0.04148 | 0.381 |
| 23298 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-655-3p | 21 | PIK3C2A | Sponge network | 0.614 | 1.0E-5 | 0.061 | 0.53776 | 0.381 |
| 23299 | LINC00473 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DACT1 | Sponge network | -1.203 | 0.03881 | 0.783 | 0.00072 | 0.381 |
| 23300 | RP11-83A24.2 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-409-5p;hsa-miR-495-3p | 13 | STAG2 | Sponge network | 0.417 | 0.00445 | 0.252 | 0.00438 | 0.381 |
| 23301 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 22 | ACVR1 | Sponge network | 0.997 | 0.00066 | 0.279 | 0.00234 | 0.381 |
| 23302 | RP11-535M15.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p | 13 | GAS7 | Sponge network | -1.14 | 0.03513 | -0.555 | 0.01602 | 0.381 |
| 23303 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | EYA4 | Sponge network | 0.194 | 0.64278 | 0.3 | 0.36876 | 0.381 |
| 23304 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-874-3p | 31 | CTIF | Sponge network | -0.942 | 0 | -0.389 | 0.00549 | 0.381 |
| 23305 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-940;hsa-miR-96-5p | 40 | GAS7 | Sponge network | 1.302 | 7.0E-5 | -0.555 | 0.01602 | 0.381 |
| 23306 | RP5-1074L1.4 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-497-5p | 12 | CDK6 | Sponge network | 1.923 | 0 | 0.991 | 0 | 0.381 |
| 23307 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-744-3p | 23 | ADAMTSL3 | Sponge network | 0.494 | 0.00339 | -1.559 | 0 | 0.381 |
| 23308 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | NCOA1 | Sponge network | -0.421 | 0.0114 | -0.432 | 0 | 0.381 |
| 23309 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-375;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p | 17 | PSIP1 | Sponge network | 0.082 | 0.55654 | -0.005 | 0.96897 | 0.381 |
| 23310 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 23 | AR | Sponge network | 0.693 | 0.00076 | -1.554 | 0 | 0.381 |
| 23311 | RP11-384K6.6 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-940 | 11 | SIRT1 | Sponge network | 0.946 | 0 | 0.109 | 0.2593 | 0.381 |
| 23312 | BZRAP1-AS1 |
hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-93-5p | 12 | PRUNE2 | Sponge network | -0.465 | 0.04703 | -1.733 | 0.00022 | 0.381 |
| 23313 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-493-5p;hsa-miR-590-3p | 18 | MYBL1 | Sponge network | 0.225 | 0.30644 | 0.494 | 0.02152 | 0.381 |
| 23314 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | NIN | Sponge network | 0.997 | 0.00066 | -0.253 | 0.13794 | 0.381 |
| 23315 | RP11-705C15.3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | RASSF2 | Sponge network | 0.796 | 4.0E-5 | -0.273 | 0.21961 | 0.381 |
| 23316 | CTD-3138B18.6 |
hsa-miR-101-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | NF1 | Sponge network | 2.248 | 0 | 0.51 | 0 | 0.381 |
| 23317 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-542-3p;hsa-miR-582-5p | 22 | PHC3 | Sponge network | 0.529 | 0.00552 | 0.212 | 0.0499 | 0.381 |
| 23318 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 12 | CNTNAP1 | Sponge network | 0.395 | 0.15451 | 0.358 | 0.13727 | 0.381 |
| 23319 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | SMARCA2 | Sponge network | -0.769 | 0.00035 | -0.529 | 0.00014 | 0.381 |
| 23320 | RP11-48B3.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | ADARB1 | Sponge network | 1.757 | 0 | -0.335 | 0.06631 | 0.381 |
| 23321 | NDUFA6-AS1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | RNF180 | Sponge network | -0.355 | 0.0077 | -1.127 | 1.0E-5 | 0.381 |
| 23322 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p | 13 | LHX6 | Sponge network | 0.194 | 0.64278 | -0.087 | 0.69193 | 0.381 |
| 23323 | CTD-2666L21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p | 19 | CREB1 | Sponge network | 0.368 | 0.20093 | 0.203 | 0.00537 | 0.381 |
| 23324 | RP11-890B15.3 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-590-3p | 10 | ARID1B | Sponge network | 0.101 | 0.60919 | 0.003 | 0.97503 | 0.381 |
| 23325 | CTD-2089N3.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-146a-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-378a-3p;hsa-miR-455-5p | 14 | NTRK3 | Sponge network | -0.314 | 0.62033 | -1.77 | 3.0E-5 | 0.381 |
| 23326 | RP11-506M13.3 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p | 12 | RASAL2 | Sponge network | 0.769 | 0 | 0.869 | 0 | 0.381 |
| 23327 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577 | 16 | GRIN2A | Sponge network | 0.811 | 0.03228 | -2.161 | 0 | 0.381 |
| 23328 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | DKK3 | Sponge network | -2.925 | 0 | -0.052 | 0.77783 | 0.381 |
| 23329 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-582-5p | 22 | PRKAA2 | Sponge network | 0.792 | 0.0049 | -2.462 | 0 | 0.381 |
| 23330 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 12 | CNTNAP3 | Sponge network | -1.886 | 0 | -1.465 | 0.00033 | 0.381 |
| 23331 | RP11-98I9.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | 0.67 | 0.00048 | 0.142 | 0.04938 | 0.381 |
| 23332 | RP11-158K1.3 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | ARHGEF12 | Sponge network | 0.349 | 0.01695 | 0.09 | 0.35693 | 0.381 |
| 23333 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-7-1-3p | 15 | HDAC9 | Sponge network | -1.092 | 4.0E-5 | -0.755 | 0.00495 | 0.381 |
| 23334 | BOLA3-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | LUZP2 | Sponge network | -0.246 | 0.35034 | -0.056 | 0.89416 | 0.381 |
| 23335 | TPTEP1 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | RIMS2 | Sponge network | -1.216 | 0 | -1.365 | 0.00208 | 0.381 |
| 23336 | MIR143HG |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NRG3 | Sponge network | -0.739 | 0.0574 | -1.836 | 4.0E-5 | 0.381 |
| 23337 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | HECA | Sponge network | 0.272 | 0.44692 | -0.217 | 0.03463 | 0.381 |
| 23338 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p | 23 | BCL6 | Sponge network | -0.222 | 0.32445 | -0.211 | 0.19489 | 0.381 |
| 23339 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p | 15 | FREM2 | Sponge network | -0.18 | 0.20343 | -0.437 | 0.46353 | 0.381 |
| 23340 | SH3BP5-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | CREB1 | Sponge network | 0.267 | 0.2937 | 0.203 | 0.00537 | 0.381 |
| 23341 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-425-5p;hsa-miR-590-3p | 11 | ITGB3 | Sponge network | -0.421 | 0.0114 | -0.011 | 0.95751 | 0.381 |
| 23342 | CTA-204B4.2 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 13 | UBR3 | Sponge network | 0.765 | 7.0E-5 | -0.021 | 0.82774 | 0.381 |
| 23343 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p | 33 | QKI | Sponge network | 1.757 | 0 | -0.333 | 0.04111 | 0.381 |
| 23344 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-96-5p | 26 | MITF | Sponge network | 0.082 | 0.55654 | -0.769 | 0.00022 | 0.381 |
| 23345 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | PDGFRA | Sponge network | 0.727 | 3.0E-5 | -0.372 | 0.0037 | 0.381 |
| 23346 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 27 | FOXO3 | Sponge network | 0.349 | 0.01695 | -0.04 | 0.72028 | 0.381 |
| 23347 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 42 | MOB1B | Sponge network | 0.311 | 0.10344 | 0.197 | 0.15266 | 0.381 |
| 23348 | RP11-819C21.1 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | TMEM30A | Sponge network | 0.537 | 0.00139 | 0.096 | 0.31031 | 0.381 |
| 23349 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-589-5p;hsa-miR-93-3p | 15 | RPS6KA2 | Sponge network | -0.323 | 0.01372 | -0.57 | 0.00013 | 0.381 |
| 23350 | EMX2OS |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 18 | IGF2 | Sponge network | -0.777 | 0.1134 | 0.848 | 0.03842 | 0.381 |
| 23351 | ZNF32-AS2 |
hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-539-5p | 10 | MOB1B | Sponge network | 1.552 | 7.0E-5 | 0.197 | 0.15266 | 0.381 |
| 23352 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-429 | 17 | MED12L | Sponge network | -0.469 | 0.08721 | -0.194 | 0.55299 | 0.381 |
| 23353 | AC018890.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429 | 24 | ABL2 | Sponge network | 1.61 | 0.00961 | 0.82 | 0 | 0.381 |
| 23354 | AC108488.4 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | 0.259 | 0.12542 | -0.704 | 0.00106 | 0.381 |
| 23355 | EIF3J-AS1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | MOB1B | Sponge network | 0.331 | 0.00509 | 0.197 | 0.15266 | 0.381 |
| 23356 | NNT-AS1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | KIF5B | Sponge network | 0.799 | 1.0E-5 | 0.362 | 0.00066 | 0.381 |
| 23357 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-577;hsa-miR-590-3p | 18 | PRKAR1A | Sponge network | -0.096 | 0.67958 | -0.063 | 0.50314 | 0.381 |
| 23358 | RP11-212P7.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 26 | NAV3 | Sponge network | 0.219 | 0.1516 | 0.212 | 0.47729 | 0.381 |
| 23359 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 28 | ZMYM2 | Sponge network | 1.11 | 0 | 0.279 | 0.01273 | 0.381 |
| 23360 | RP11-21A7A.2 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-625-5p | 12 | SFRP1 | Sponge network | -1.116 | 0.23686 | -3.566 | 0 | 0.381 |
| 23361 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 31 | MOB1B | Sponge network | 0.249 | 0.35902 | 0.197 | 0.15266 | 0.381 |
| 23362 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 25 | THBS1 | Sponge network | -0.896 | 0.00099 | 0.741 | 0.00386 | 0.381 |
| 23363 | RP4-584D14.7 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-502-5p;hsa-miR-625-5p | 11 | PTPN14 | Sponge network | 1.294 | 0.0031 | 0.144 | 0.34595 | 0.381 |
| 23364 | EIF3J-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | BRD3 | Sponge network | 0.331 | 0.00509 | 0.37 | 0.00013 | 0.381 |
| 23365 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 35 | MOB1B | Sponge network | 0.572 | 0.01722 | 0.197 | 0.15266 | 0.381 |
| 23366 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PAK3 | Sponge network | 0.214 | 0.18246 | -0.628 | 0.07253 | 0.381 |
| 23367 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-874-3p | 19 | CRTC3 | Sponge network | -2.69 | 0.0001 | 0.164 | 0.16786 | 0.381 |
| 23368 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p | 11 | ZNF292 | Sponge network | 0.946 | 0 | 0.276 | 0.04148 | 0.381 |
| 23369 | RP11-196G18.23 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | BRD3 | Sponge network | 0.724 | 0.00039 | 0.37 | 0.00013 | 0.381 |
| 23370 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-98-5p | 33 | THBS1 | Sponge network | -0.769 | 0.00035 | 0.741 | 0.00386 | 0.381 |
| 23371 | AC025171.1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | HIP1 | Sponge network | 0.998 | 0 | 0.774 | 0 | 0.381 |
| 23372 | AP001347.6 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZNF844 | Sponge network | -1.161 | 0.00591 | -0.966 | 0.00035 | 0.381 |
| 23373 | RP11-283I3.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 10 | FAM172A | Sponge network | 0.614 | 1.0E-5 | -0.57 | 0 | 0.381 |
| 23374 | RP11-67L2.2 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | INPP4A | Sponge network | 0.72 | 0.0001 | 0.07 | 0.53563 | 0.381 |
| 23375 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -1.575 | 6.0E-5 | -0.252 | 0.15511 | 0.381 |
| 23376 | SH3BP5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | TSC1 | Sponge network | 0.267 | 0.2937 | 0.103 | 0.26071 | 0.381 |
| 23377 | AC005618.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 28 | ABL2 | Sponge network | 0.862 | 0.06213 | 0.82 | 0 | 0.381 |
| 23378 | NDUFA6-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 27 | PDE4DIP | Sponge network | -0.355 | 0.0077 | -0.282 | 0.0257 | 0.381 |
| 23379 | RP4-668J24.2 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-628-5p | 19 | DTNA | Sponge network | -1.054 | 0.0706 | -1.648 | 1.0E-5 | 0.381 |
| 23380 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-940;hsa-miR-96-5p | 21 | CRTC3 | Sponge network | -0.709 | 0.18168 | 0.164 | 0.16786 | 0.381 |
| 23381 | MALAT1 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-550a-5p;hsa-miR-590-3p | 38 | CCDC6 | Sponge network | 0.782 | 0.00016 | 0.27 | 0.02555 | 0.381 |
| 23382 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-766-3p | 23 | TBL1XR1 | Sponge network | 0.417 | 0.00445 | 0.578 | 0 | 0.381 |
| 23383 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 28 | BNC2 | Sponge network | 0.056 | 0.8494 | -0.491 | 0.09988 | 0.381 |
| 23384 | RP11-440L14.4 |
hsa-let-7g-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-5p;hsa-miR-590-3p | 11 | ABL2 | Sponge network | 0.545 | 0.05789 | 0.82 | 0 | 0.381 |
| 23385 | RP4-717I23.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p | 10 | ZNF208 | Sponge network | 0.637 | 0.00069 | -0.4 | 0.22381 | 0.381 |
| 23386 | TP73-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-96-5p | 14 | ZBTB20 | Sponge network | -0.563 | 0.02545 | -0.122 | 0.57569 | 0.381 |
| 23387 | RP4-614O4.11 |
hsa-let-7d-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 10 | RICTOR | Sponge network | 1.042 | 0.00177 | 0.388 | 0.00188 | 0.381 |
| 23388 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-485-3p | 10 | SASH1 | Sponge network | 0.043 | 0.81628 | -0.502 | 0.00175 | 0.381 |
| 23389 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p | 13 | RARB | Sponge network | -0.583 | 0.14589 | -0.825 | 2.0E-5 | 0.381 |
| 23390 | C4B-AS1 |
hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 18 | MEF2C | Sponge network | 2.306 | 9.0E-5 | -0.499 | 0.01448 | 0.381 |
| 23391 | LINC00865 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p | 15 | RPS6KA6 | Sponge network | -1.249 | 7.0E-5 | -2.989 | 0 | 0.381 |
| 23392 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | ABCA10 | Sponge network | -0.942 | 0 | -0.571 | 0.03674 | 0.381 |
| 23393 | RP11-344B5.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-320a;hsa-miR-320b | 15 | WDFY3 | Sponge network | -0.055 | 0.88482 | -0.201 | 0.0967 | 0.381 |
| 23394 | FZD10-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 14 | SORCS1 | Sponge network | -0.727 | 0.04726 | -3.492 | 0 | 0.381 |
| 23395 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | PDGFRA | Sponge network | 0.374 | 0.20054 | -0.372 | 0.0037 | 0.381 |
| 23396 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-5p | 43 | TCF4 | Sponge network | 0.174 | 0.30034 | 0.016 | 0.92271 | 0.381 |
| 23397 | RP11-360F5.3 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-93-3p | 20 | IL6ST | Sponge network | 1.867 | 0 | -0.402 | 0.00676 | 0.381 |
| 23398 | RP11-35G9.5 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-589-3p | 10 | PCDH9 | Sponge network | 0.622 | 0.00015 | -1.823 | 4.0E-5 | 0.381 |
| 23399 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | BNC2 | Sponge network | 1.153 | 0.00114 | -0.491 | 0.09988 | 0.381 |
| 23400 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p | 11 | TXNIP | Sponge network | -0.737 | 0.10822 | -1.277 | 0 | 0.381 |
| 23401 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 46 | TRPS1 | Sponge network | 0.222 | 0.29133 | -0.191 | 0.44109 | 0.381 |
| 23402 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | TAL1 | Sponge network | 0.083 | 0.66405 | -0.462 | 0.00574 | 0.381 |
| 23403 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 36 | SASH1 | Sponge network | 0.421 | 0.00084 | -0.502 | 0.00175 | 0.381 |
| 23404 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p | 14 | ITGA5 | Sponge network | -0.612 | 0.00094 | 0.452 | 0.03524 | 0.381 |
| 23405 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ZFX | Sponge network | 0.532 | 0.00505 | 0.09 | 0.42563 | 0.381 |
| 23406 | RP11-254F7.2 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p | 28 | GRIK3 | Sponge network | 0.176 | 0.37402 | -3.208 | 0 | 0.381 |
| 23407 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | KAT6B | Sponge network | 0.537 | 0.00139 | -0.478 | 0.00018 | 0.381 |
| 23408 | RP11-13K12.5 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | MEF2C | Sponge network | -0.318 | 0.65847 | -0.499 | 0.01448 | 0.381 |
| 23409 | RP11-410L14.2 |
hsa-let-7d-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-497-5p | 18 | MDM4 | Sponge network | -0.052 | 0.76839 | 0.345 | 0.00762 | 0.381 |
| 23410 | RP11-48B3.4 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 16 | ATM | Sponge network | 1.757 | 0 | 0.178 | 0.21568 | 0.381 |
| 23411 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-628-5p | 17 | FOXP1 | Sponge network | -0.376 | 0.00337 | 0.042 | 0.74368 | 0.381 |
| 23412 | LINC00863 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | CBX5 | Sponge network | 0.189 | 0.13981 | 0.165 | 0.24446 | 0.381 |
| 23413 | SH3BP5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p | 10 | EZH1 | Sponge network | 0.267 | 0.2937 | -0.564 | 1.0E-5 | 0.381 |
| 23414 | RP1-59D14.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-625-5p | 15 | ABI2 | Sponge network | 1.162 | 5.0E-5 | 0.175 | 0.14787 | 0.381 |
| 23415 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SMARCA2 | Sponge network | -0.83 | 0 | -0.529 | 0.00014 | 0.381 |
| 23416 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-93-5p | 18 | PDGFRA | Sponge network | -1.255 | 0.00981 | -0.372 | 0.0037 | 0.381 |
| 23417 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | CACNA2D1 | Sponge network | -0.08 | 0.90092 | -0.846 | 0.00675 | 0.381 |
| 23418 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p | 11 | MAP3K12 | Sponge network | -1.331 | 3.0E-5 | -0.057 | 0.59369 | 0.381 |
| 23419 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | RELN | Sponge network | 0.194 | 0.64278 | -2.403 | 0 | 0.381 |
| 23420 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | EPAS1 | Sponge network | -1.281 | 0.00012 | -0.184 | 0.14418 | 0.381 |
| 23421 | RP11-53O19.3 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 11 | RASAL2 | Sponge network | 1.205 | 0 | 0.869 | 0 | 0.38 |
| 23422 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | QKI | Sponge network | 0.385 | 0.10067 | -0.333 | 0.04111 | 0.38 |
| 23423 | TPTEP1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 19 | PAK3 | Sponge network | -1.216 | 0 | -0.628 | 0.07253 | 0.38 |
| 23424 | RP11-344B5.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-3p | 10 | TSHZ3 | Sponge network | -0.055 | 0.88482 | -0.556 | 0.01516 | 0.38 |
| 23425 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-655-3p | 20 | LATS1 | Sponge network | -0.096 | 0.67958 | 0.109 | 0.34208 | 0.38 |
| 23426 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30e-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 54 | CCND2 | Sponge network | 0.997 | 0.00066 | -0.267 | 0.3112 | 0.38 |
| 23427 | PART1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-92a-3p | 26 | RIMBP2 | Sponge network | -2.925 | 0 | -1.968 | 5.0E-5 | 0.38 |
| 23428 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 14 | NCAM2 | Sponge network | -0.096 | 0.67958 | -0.482 | 0.22565 | 0.38 |
| 23429 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-590-5p | 17 | PIK3C2A | Sponge network | 0.706 | 0 | 0.061 | 0.53776 | 0.38 |
| 23430 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-425-5p;hsa-miR-7-5p | 10 | ERC1 | Sponge network | -0.187 | 0.23787 | -0.108 | 0.38794 | 0.38 |
| 23431 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | ANK2 | Sponge network | -0.053 | 0.80685 | -1.603 | 1.0E-5 | 0.38 |
| 23432 | RP11-158K1.3 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 46 | DST | Sponge network | 0.349 | 0.01695 | -0.713 | 0.00096 | 0.38 |
| 23433 | MAPKAPK5-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-486-5p | 16 | CBFB | Sponge network | 0.765 | 0 | 1.061 | 0 | 0.38 |
| 23434 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-590-5p | 23 | GRIA3 | Sponge network | -1.349 | 0.00017 | -1.956 | 0 | 0.38 |
| 23435 | H1FX-AS1 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | SFRP1 | Sponge network | -0.206 | 0.12942 | -3.566 | 0 | 0.38 |
| 23436 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | HECA | Sponge network | -0.323 | 0.01372 | -0.217 | 0.03463 | 0.38 |
| 23437 | RP11-6O2.3 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-590-5p;hsa-miR-653-5p | 21 | UBR3 | Sponge network | 0.331 | 0.57247 | -0.021 | 0.82774 | 0.38 |
| 23438 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p | 15 | HIP1 | Sponge network | 0.75 | 0.00054 | 0.774 | 0 | 0.38 |
| 23439 | HOXB-AS2 |
hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429 | 12 | ABL2 | Sponge network | 1.316 | 0.01106 | 0.82 | 0 | 0.38 |
| 23440 | ADAMTS9-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-708-3p;hsa-miR-96-5p | 17 | UNC5C | Sponge network | -2.452 | 0 | -0.178 | 0.40214 | 0.38 |
| 23441 | LINC00654 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-590-3p | 10 | NOV | Sponge network | 0.374 | 0.20054 | -0.58 | 0.04676 | 0.38 |
| 23442 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 17 | ZMYM2 | Sponge network | 0.924 | 0 | 0.279 | 0.01273 | 0.38 |
| 23443 | LINC00173 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p | 31 | RUNX1T1 | Sponge network | 0.056 | 0.8494 | -0.344 | 0.2374 | 0.38 |
| 23444 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 11 | PDZD2 | Sponge network | -0.384 | 0.40652 | -0.909 | 3.0E-5 | 0.38 |
| 23445 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 17 | NF1 | Sponge network | 1.205 | 0 | 0.51 | 0 | 0.38 |
| 23446 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p | 15 | AFF3 | Sponge network | -0.726 | 0.00132 | -2.373 | 0 | 0.38 |
| 23447 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-5p | 14 | RET | Sponge network | -0.384 | 0.40652 | -0.833 | 0.03517 | 0.38 |
| 23448 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | AKAP12 | Sponge network | 0.219 | 0.1516 | -0.932 | 0.0028 | 0.38 |
| 23449 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-511-5p | 17 | EYA4 | Sponge network | 0.056 | 0.81195 | 0.3 | 0.36876 | 0.38 |
| 23450 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | LRIG1 | Sponge network | 0.41 | 0.02214 | -0.537 | 0.00588 | 0.38 |
| 23451 | RP11-156P1.3 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 12 | LZTS1 | Sponge network | 0.214 | 0.18246 | 1.664 | 0 | 0.38 |
| 23452 | AC007566.10 |
hsa-let-7g-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-582-3p | 13 | ABL2 | Sponge network | 2.368 | 2.0E-5 | 0.82 | 0 | 0.38 |
| 23453 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p | 20 | CLASP2 | Sponge network | 0.594 | 0.00064 | -0.038 | 0.71 | 0.38 |
| 23454 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 29 | ESR1 | Sponge network | -2.915 | 0.00015 | -1.035 | 3.0E-5 | 0.38 |
| 23455 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-429 | 10 | DNAJB4 | Sponge network | -0.469 | 0.08721 | -0.7 | 1.0E-5 | 0.38 |
| 23456 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-582-3p | 23 | CBL | Sponge network | 1.254 | 0 | 0.291 | 0.01267 | 0.38 |
| 23457 | CTA-14H9.5 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | ZFHX3 | Sponge network | 0.145 | 0.53343 | 0.389 | 0.01363 | 0.38 |
| 23458 | AC006547.13 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-654-3p;hsa-miR-874-3p | 13 | NR2C2 | Sponge network | 1.159 | 0 | 0.21 | 0.03224 | 0.38 |
| 23459 | HOXA-AS2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 21 | HIP1 | Sponge network | 0.61 | 0.01593 | 0.774 | 0 | 0.38 |
| 23460 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-940;hsa-miR-96-5p | 22 | ZFP36L1 | Sponge network | -0.737 | 0.06182 | -0.505 | 8.0E-5 | 0.38 |
| 23461 | RP11-867G23.10 |
hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-550a-3p | 12 | RAP1A | Sponge network | -2.531 | 0 | -0.929 | 0 | 0.38 |
| 23462 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 61 | TCF4 | Sponge network | 0.064 | 0.72934 | 0.016 | 0.92271 | 0.38 |
| 23463 | AC005618.6 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-628-5p | 14 | IRS1 | Sponge network | 0.862 | 0.06213 | -0.026 | 0.88855 | 0.38 |
| 23464 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-582-3p | 23 | PBX1 | Sponge network | -0.031 | 0.89063 | -1.206 | 0 | 0.38 |
| 23465 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-93-5p;hsa-miR-940 | 30 | CHD5 | Sponge network | -1.349 | 0.00017 | -0.922 | 0.01521 | 0.38 |
| 23466 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 18 | NEDD4 | Sponge network | 1.757 | 0 | 0.02 | 0.89834 | 0.38 |
| 23467 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 59 | WDFY3 | Sponge network | 0.997 | 0.00066 | -0.201 | 0.0967 | 0.38 |
| 23468 | HOXA-AS2 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-542-3p | 11 | PRKD1 | Sponge network | 0.61 | 0.01593 | -0.466 | 0.03583 | 0.38 |
| 23469 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-940 | 20 | MCC | Sponge network | -0.829 | 0.01343 | -1.005 | 0 | 0.38 |
| 23470 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | GALNT13 | Sponge network | 0.997 | 0.00066 | -0.857 | 0.07439 | 0.38 |
| 23471 | AC108488.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-7-1-3p | 17 | TET1 | Sponge network | 0.259 | 0.12542 | -0.135 | 0.52138 | 0.38 |
| 23472 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-93-3p;hsa-miR-93-5p | 41 | SSBP2 | Sponge network | -0.18 | 0.20343 | -1.58 | 0 | 0.38 |
| 23473 | RP11-403I13.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 0.619 | 0.00157 | 0.943 | 0 | 0.38 |
| 23474 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | PLSCR4 | Sponge network | -0.206 | 0.12942 | -0.474 | 0.03833 | 0.38 |
| 23475 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SLC6A15 | Sponge network | -0.323 | 0.01372 | -2.373 | 2.0E-5 | 0.38 |
| 23476 | RP11-359B12.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | NAV3 | Sponge network | 0.064 | 0.72934 | 0.212 | 0.47729 | 0.38 |
| 23477 | GAS6-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | ERG | Sponge network | 0.16 | 0.50785 | 0.002 | 0.99011 | 0.38 |
| 23478 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | MYO9A | Sponge network | 0.267 | 0.2937 | -0.147 | 0.32373 | 0.38 |
| 23479 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | RASSF2 | Sponge network | 0.246 | 0.59912 | -0.273 | 0.21961 | 0.38 |
| 23480 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-455-5p | 16 | AKAP6 | Sponge network | -0.465 | 0.04703 | -1.586 | 3.0E-5 | 0.38 |
| 23481 | AC009948.5 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-577 | 10 | AMPH | Sponge network | 0.532 | 0.00505 | -0.916 | 5.0E-5 | 0.38 |
| 23482 | LINC00473 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-3p | 37 | CREB3L2 | Sponge network | -1.203 | 0.03881 | -0.525 | 2.0E-5 | 0.38 |
| 23483 | ZSCAN16-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 70 | LPP | Sponge network | -0.342 | 0.02288 | -0.459 | 0.04475 | 0.38 |
| 23484 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 28 | ZFHX4 | Sponge network | 0.259 | 0.12542 | 0.018 | 0.95574 | 0.38 |
| 23485 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-874-3p | 10 | NR2C2 | Sponge network | 1.352 | 0 | 0.21 | 0.03224 | 0.38 |
| 23486 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p | 15 | AFF3 | Sponge network | 0.176 | 0.37402 | -2.373 | 0 | 0.38 |
| 23487 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | UVRAG | Sponge network | 0.064 | 0.72934 | -0.017 | 0.83865 | 0.38 |
| 23488 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | NALCN | Sponge network | -0.769 | 0.00035 | 1.068 | 0.00237 | 0.38 |
| 23489 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-93-5p | 16 | RBL2 | Sponge network | -0.162 | 0.47687 | -0.282 | 0.00254 | 0.38 |
| 23490 | CTD-3092A11.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-365a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ARHGEF12 | Sponge network | 0.873 | 0 | 0.09 | 0.35693 | 0.38 |
| 23491 | AC159540.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 11 | PHF6 | Sponge network | 1.634 | 0 | 0.785 | 0 | 0.38 |
| 23492 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p | 18 | MAFB | Sponge network | -1.216 | 0 | -0.023 | 0.89865 | 0.38 |
| 23493 | RP11-206L10.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 10 | PHF6 | Sponge network | 0.793 | 0 | 0.785 | 0 | 0.38 |
| 23494 | CTC-429P9.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-539-5p;hsa-miR-940 | 12 | TRPS1 | Sponge network | 0.043 | 0.81628 | -0.191 | 0.44109 | 0.38 |
| 23495 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CACNA2D1 | Sponge network | 0.225 | 0.30644 | -0.846 | 0.00675 | 0.38 |
| 23496 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | SYNE1 | Sponge network | 0.259 | 0.12542 | -0.738 | 0.00316 | 0.38 |
| 23497 | TRAF3IP2-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 10 | PKD1L2 | Sponge network | -0.323 | 0.01372 | -0.971 | 0.00438 | 0.38 |
| 23498 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | AKAP11 | Sponge network | 0.539 | 0.03722 | 0.196 | 0.09825 | 0.38 |
| 23499 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | -0.303 | 0.21002 | 0.358 | 0.13727 | 0.38 |
| 23500 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-33a-5p | 12 | CHD1 | Sponge network | 1.044 | 2.0E-5 | 0.322 | 0.00215 | 0.38 |
| 23501 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | LRRC7 | Sponge network | -1.349 | 0.00017 | -1.341 | 0.01193 | 0.38 |
| 23502 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-7-5p | 11 | PTGFR | Sponge network | 0.174 | 0.30034 | -0.233 | 0.45421 | 0.38 |
| 23503 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p | 14 | ZFHX4 | Sponge network | -1.14 | 0.03513 | 0.018 | 0.95574 | 0.38 |
| 23504 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-7-5p | 21 | RNF144A | Sponge network | -0.777 | 0.16667 | 0.594 | 0.0009 | 0.38 |
| 23505 | RP5-1085F17.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 25 | ABI2 | Sponge network | 0.541 | 0.00125 | 0.175 | 0.14787 | 0.38 |
| 23506 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | CREB1 | Sponge network | 0.998 | 0 | 0.203 | 0.00537 | 0.38 |
| 23507 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 28 | NUAK1 | Sponge network | -0.739 | 0.0574 | 0.904 | 0 | 0.38 |
| 23508 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | STARD13 | Sponge network | -2.117 | 0.00161 | -0.187 | 0.27383 | 0.38 |
| 23509 | AC108488.4 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | LRIG1 | Sponge network | 0.259 | 0.12542 | -0.537 | 0.00588 | 0.38 |
| 23510 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ARID5B | Sponge network | 0.659 | 3.0E-5 | 0.342 | 0.01676 | 0.38 |
| 23511 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | PRKAR1A | Sponge network | 0.214 | 0.18246 | -0.063 | 0.50314 | 0.38 |
| 23512 | IQCH-AS1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 20 | TBL1XR1 | Sponge network | 0.929 | 0 | 0.578 | 0 | 0.38 |
| 23513 | RP11-46C24.7 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | 0.392 | 0.0278 | -1.12 | 0.00107 | 0.38 |
| 23514 | RP11-53O19.1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-576-5p | 14 | ID4 | Sponge network | -0.405 | 0.22013 | -1.644 | 0 | 0.38 |
| 23515 | RP11-434H6.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p | 10 | ACVR2A | Sponge network | 0.358 | 0.14302 | -0.409 | 0.00039 | 0.38 |
| 23516 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429 | 11 | MYO9A | Sponge network | 2.364 | 0.00134 | -0.147 | 0.32373 | 0.38 |
| 23517 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 15 | CNTNAP1 | Sponge network | 0.219 | 0.1516 | 0.358 | 0.13727 | 0.38 |
| 23518 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 20 | SASH1 | Sponge network | -0.583 | 0.14589 | -0.502 | 0.00175 | 0.38 |
| 23519 | TPTEP1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ZNF844 | Sponge network | -1.216 | 0 | -0.966 | 0.00035 | 0.38 |
| 23520 | LINC00957 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | LIMCH1 | Sponge network | -0.421 | 0.0114 | -0.915 | 1.0E-5 | 0.38 |
| 23521 | FGD5-AS1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-493-5p;hsa-miR-590-3p | 10 | ZNF638 | Sponge network | 0.401 | 0.00066 | 0.22 | 0.01214 | 0.38 |
| 23522 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p | 11 | PRDM5 | Sponge network | -0.384 | 0.40652 | -1.09 | 0.00014 | 0.38 |
| 23523 | RP11-531A24.5 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | 0.75 | 0.00054 | 0.358 | 0.13727 | 0.38 |
| 23524 | CTC-429P9.3 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p | 16 | CSDE1 | Sponge network | 0.082 | 0.55397 | -0.493 | 0 | 0.38 |
| 23525 | RP11-175O19.4 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 13 | PAWR | Sponge network | 0.917 | 0 | 0.636 | 0 | 0.38 |
| 23526 | AC016747.3 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 22 | PCDH9 | Sponge network | 0.199 | 0.38015 | -1.823 | 4.0E-5 | 0.38 |
| 23527 | RP11-212P7.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 24 | TET1 | Sponge network | 0.219 | 0.1516 | -0.135 | 0.52138 | 0.38 |
| 23528 | CTC-429P9.5 |
hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p | 20 | ARHGEF12 | Sponge network | 0.178 | 0.29106 | 0.09 | 0.35693 | 0.38 |
| 23529 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | CBL | Sponge network | 1.147 | 0 | 0.291 | 0.01267 | 0.38 |
| 23530 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-744-3p | 25 | FAT3 | Sponge network | 0.693 | 0.00076 | -1.601 | 0.00051 | 0.38 |
| 23531 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-98-5p | 26 | THBS1 | Sponge network | -0.154 | 0.78939 | 0.741 | 0.00386 | 0.38 |
| 23532 | SNX29P2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 70 | LPP | Sponge network | 0.4 | 0.14611 | -0.459 | 0.04475 | 0.38 |
| 23533 | CTA-204B4.2 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-454-3p;hsa-miR-671-5p;hsa-miR-766-3p | 17 | CHD5 | Sponge network | 0.765 | 7.0E-5 | -0.922 | 0.01521 | 0.38 |
| 23534 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p | 28 | ADARB1 | Sponge network | 0.389 | 0.04293 | -0.335 | 0.06631 | 0.38 |
| 23535 | RP11-46C24.7 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-369-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 12 | MACF1 | Sponge network | 0.392 | 0.0278 | 0.019 | 0.90335 | 0.38 |
| 23536 | RP11-517B11.7 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-409-5p;hsa-miR-432-5p;hsa-miR-495-3p | 10 | STAG2 | Sponge network | 0.706 | 0 | 0.252 | 0.00438 | 0.38 |
| 23537 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577 | 12 | INPP4A | Sponge network | -2.69 | 0.0001 | 0.07 | 0.53563 | 0.38 |
| 23538 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p | 10 | ST5 | Sponge network | -0.323 | 0.01372 | -0.78 | 0 | 0.38 |
| 23539 | LINC00641 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NRG3 | Sponge network | -0.222 | 0.32445 | -1.836 | 4.0E-5 | 0.38 |
| 23540 | LINC00843 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-429 | 10 | HECA | Sponge network | 1.106 | 0 | -0.217 | 0.03463 | 0.38 |
| 23541 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7b-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 43 | EPHA7 | Sponge network | -0.355 | 0.0077 | -2.197 | 3.0E-5 | 0.38 |
| 23542 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-590-3p | 12 | PRDM5 | Sponge network | -0.899 | 6.0E-5 | -1.09 | 0.00014 | 0.38 |
| 23543 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-628-5p | 17 | BCL6 | Sponge network | -1.161 | 0.00591 | -0.211 | 0.19489 | 0.38 |
| 23544 | FAM157A |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 10 | PHF6 | Sponge network | 0.813 | 1.0E-5 | 0.785 | 0 | 0.38 |
| 23545 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | CRTC3 | Sponge network | -0.811 | 2.0E-5 | 0.164 | 0.16786 | 0.38 |
| 23546 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | PIK3R1 | Sponge network | -0.811 | 2.0E-5 | -0.133 | 0.36854 | 0.38 |
| 23547 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 19 | PTGFR | Sponge network | 0.214 | 0.18246 | -0.233 | 0.45421 | 0.38 |
| 23548 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-940 | 26 | CADM2 | Sponge network | -0.829 | 0.01343 | -3.782 | 0 | 0.38 |
| 23549 | RP11-967K21.1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 37 | PPM1L | Sponge network | 0.056 | 0.81195 | -0.537 | 0.00616 | 0.38 |
| 23550 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-335-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 14 | PNRC1 | Sponge network | -0.777 | 0.1134 | -0.646 | 0 | 0.38 |
| 23551 | RP11-574K11.29 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | UBR3 | Sponge network | 0.997 | 0 | -0.021 | 0.82774 | 0.38 |
| 23552 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p | 23 | AKAP11 | Sponge network | 1.757 | 0 | 0.196 | 0.09825 | 0.38 |
| 23553 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-629-3p;hsa-miR-942-5p | 25 | GPC6 | Sponge network | -0.773 | 0.04073 | -0.054 | 0.81465 | 0.38 |
| 23554 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p | 25 | EBF1 | Sponge network | -0.227 | 0.31657 | -0.384 | 0.08856 | 0.38 |
| 23555 | RP1-59D14.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p | 18 | CBL | Sponge network | 1.162 | 5.0E-5 | 0.291 | 0.01267 | 0.38 |
| 23556 | MIR22HG |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-93-3p;hsa-miR-940 | 42 | GRIK3 | Sponge network | -1.047 | 0 | -3.208 | 0 | 0.38 |
| 23557 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p | 41 | IL6ST | Sponge network | 0.657 | 4.0E-5 | -0.402 | 0.00676 | 0.38 |
| 23558 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-582-5p | 13 | DOCK4 | Sponge network | 0.529 | 0.00552 | 0.502 | 0.00108 | 0.38 |
| 23559 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-664a-3p | 13 | RCAN2 | Sponge network | 0.95 | 0.15943 | -0.33 | 0.15172 | 0.38 |
| 23560 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 12 | PLSCR4 | Sponge network | 0.174 | 0.30034 | -0.474 | 0.03833 | 0.38 |
| 23561 | RP11-701H24.3 |
hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 10 | TACC1 | Sponge network | -1.425 | 0.04204 | -0.362 | 0.05406 | 0.38 |
| 23562 | RP11-473I1.5 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-655-3p | 14 | TMEM30A | Sponge network | 0.542 | 0.00054 | 0.096 | 0.31031 | 0.38 |
| 23563 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | PHC3 | Sponge network | 0.693 | 0.00076 | 0.212 | 0.0499 | 0.38 |
| 23564 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940 | 41 | MEF2C | Sponge network | -0.777 | 0.1134 | -0.499 | 0.01448 | 0.38 |
| 23565 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | BCL2 | Sponge network | 0.532 | 0.00505 | -0.899 | 0 | 0.38 |
| 23566 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | HACE1 | Sponge network | -0.323 | 0.01372 | -0.072 | 0.55239 | 0.38 |
| 23567 | ZNF582-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-877-5p | 13 | GLIPR1 | Sponge network | -1.349 | 0.00017 | 0.146 | 0.3276 | 0.38 |
| 23568 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 57 | SMAD4 | Sponge network | 0.101 | 0.60919 | -0.313 | 0.00413 | 0.38 |
| 23569 | RP11-53O19.3 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p | 11 | CBFB | Sponge network | 1.205 | 0 | 1.061 | 0 | 0.38 |
| 23570 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -0.513 | 0.04703 | 0.309 | 0.17079 | 0.38 |
| 23571 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p | 13 | ARID2 | Sponge network | 1.153 | 0 | 0.235 | 0.02697 | 0.38 |
| 23572 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-542-3p | 16 | PDE4DIP | Sponge network | -0.517 | 0.02935 | -0.282 | 0.0257 | 0.38 |
| 23573 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-5p | 19 | CXXC4 | Sponge network | -2.62 | 0 | -0.433 | 0.21055 | 0.38 |
| 23574 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 30 | TACC1 | Sponge network | 0.75 | 0.00054 | -0.362 | 0.05406 | 0.38 |
| 23575 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | SASH1 | Sponge network | -0.376 | 0.00337 | -0.502 | 0.00175 | 0.38 |
| 23576 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-96-5p | 23 | AFF4 | Sponge network | -0.187 | 0.23787 | -0.266 | 0.01565 | 0.38 |
| 23577 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | RBMS3 | Sponge network | -0.285 | 0.2172 | -0.519 | 0.03551 | 0.38 |
| 23578 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-7-1-3p | 19 | CXXC4 | Sponge network | -0.842 | 0.01361 | -0.433 | 0.21055 | 0.38 |
| 23579 | ZRANB2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-3p | 15 | TRPS1 | Sponge network | -0.376 | 0.00337 | -0.191 | 0.44109 | 0.38 |
| 23580 | LINC00473 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 12 | KCNAB1 | Sponge network | -1.203 | 0.03881 | -0.755 | 2.0E-5 | 0.38 |
| 23581 | RP11-244O19.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 15 | FMNL3 | Sponge network | -0.096 | 0.67958 | 0.555 | 4.0E-5 | 0.38 |
| 23582 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 17 | HIP1 | Sponge network | 0.559 | 0.00026 | 0.774 | 0 | 0.38 |
| 23583 | AC138035.2 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | PIK3C2A | Sponge network | 1.073 | 0 | 0.061 | 0.53776 | 0.38 |
| 23584 | RP11-504P24.4 |
hsa-let-7g-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 10 | BRD3 | Sponge network | 0.624 | 0.0004 | 0.37 | 0.00013 | 0.38 |
| 23585 | LINC00654 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-590-3p | 15 | STARD13 | Sponge network | 0.374 | 0.20054 | -0.187 | 0.27383 | 0.38 |
| 23586 | RP11-815I9.4 |
hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-365a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 14 | ARHGEF12 | Sponge network | 0.537 | 0.0006 | 0.09 | 0.35693 | 0.38 |
| 23587 | RP11-35G9.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | EPS15 | Sponge network | 0.657 | 4.0E-5 | -0.052 | 0.53998 | 0.38 |
| 23588 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-942-5p | 39 | TMEM132B | Sponge network | -1.645 | 0 | -0.83 | 0.01084 | 0.38 |
| 23589 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 41 | NOTCH2 | Sponge network | 0.064 | 0.72934 | 0.138 | 0.35163 | 0.38 |
| 23590 | KB-431C1.4 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p | 10 | ERCC6 | Sponge network | 0.909 | 0 | 0.23 | 0.015 | 0.38 |
| 23591 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | MYO9A | Sponge network | 0.61 | 0.01593 | -0.147 | 0.32373 | 0.38 |
| 23592 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-590-3p | 10 | ZNF844 | Sponge network | -0.896 | 0.00099 | -0.966 | 0.00035 | 0.38 |
| 23593 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | EPAS1 | Sponge network | -0.611 | 0.09607 | -0.184 | 0.14418 | 0.38 |
| 23594 | LINC00648 |
hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | MED12L | Sponge network | 0.633 | 0.51678 | -0.194 | 0.55299 | 0.38 |
| 23595 | FAM157B |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-505-5p | 15 | ARHGEF12 | Sponge network | 0.92 | 0 | 0.09 | 0.35693 | 0.38 |
| 23596 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-628-5p | 27 | GRIA3 | Sponge network | -1.249 | 7.0E-5 | -1.956 | 0 | 0.38 |
| 23597 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 33 | ZBTB4 | Sponge network | -2.925 | 0 | -0.739 | 0 | 0.38 |
| 23598 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PLSCR4 | Sponge network | 0.72 | 0.0001 | -0.474 | 0.03833 | 0.38 |
| 23599 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CACNA2D1 | Sponge network | 0.727 | 3.0E-5 | -0.846 | 0.00675 | 0.38 |
| 23600 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | HIP1 | Sponge network | 0.538 | 0.00076 | 0.774 | 0 | 0.38 |
| 23601 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-576-5p | 12 | NCAM2 | Sponge network | 0.176 | 0.37402 | -0.482 | 0.22565 | 0.38 |
| 23602 | CTC-296K1.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-342-3p;hsa-miR-582-5p | 10 | STARD13 | Sponge network | -2.095 | 0.00112 | -0.187 | 0.27383 | 0.38 |
| 23603 | RP11-284N8.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-940 | 14 | PTPRT | Sponge network | -0.31 | 0.33871 | -0.907 | 0.03727 | 0.38 |
| 23604 | DNM3OS |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-501-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | 0.572 | 0.01722 | -2.417 | 0 | 0.38 |
| 23605 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 56 | AR | Sponge network | -0.355 | 0.0077 | -1.554 | 0 | 0.38 |
| 23606 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p | 11 | ARHGAP29 | Sponge network | 0.889 | 9.0E-5 | 0.131 | 0.42953 | 0.38 |
| 23607 | RP11-344B5.2 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b | 12 | EYA4 | Sponge network | -0.055 | 0.88482 | 0.3 | 0.36876 | 0.38 |
| 23608 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-5p;hsa-miR-940 | 12 | CRTC3 | Sponge network | 0.174 | 0.30034 | 0.164 | 0.16786 | 0.38 |
| 23609 | CTA-363E19.2 |
hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 10 | RASAL2 | Sponge network | 1.055 | 0 | 0.869 | 0 | 0.38 |
| 23610 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 17 | POU2F2 | Sponge network | -0.769 | 0.00035 | -0.233 | 0.32214 | 0.38 |
| 23611 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 17 | QKI | Sponge network | -0.31 | 0.33871 | -0.333 | 0.04111 | 0.38 |
| 23612 | GS1-124K5.11 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | DCLK1 | Sponge network | 0.494 | 0.00339 | -1.324 | 0.00014 | 0.38 |
| 23613 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-7-5p | 19 | KCNA6 | Sponge network | 0.176 | 0.37402 | -1.531 | 0.00013 | 0.38 |
| 23614 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 18 | LHX6 | Sponge network | 0.249 | 0.35902 | -0.087 | 0.69193 | 0.38 |
| 23615 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-7-1-3p | 20 | CBL | Sponge network | 1.153 | 0.00114 | 0.291 | 0.01267 | 0.38 |
| 23616 | LINC00094 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-629-3p | 26 | RASSF8 | Sponge network | 0.253 | 0.09565 | -0.122 | 0.65591 | 0.38 |
| 23617 | RP4-791M13.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-7-1-3p | 20 | ANK2 | Sponge network | 0.419 | 0.0319 | -1.603 | 1.0E-5 | 0.38 |
| 23618 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-378a-5p | 10 | PDS5B | Sponge network | 0.903 | 0 | 0.259 | 0.00962 | 0.379 |
| 23619 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 25 | CYLD | Sponge network | -0.769 | 0.00035 | -0.367 | 0.00171 | 0.379 |
| 23620 | LINC00578 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p | 27 | CCND2 | Sponge network | 0.811 | 0.03228 | -0.267 | 0.3112 | 0.379 |
| 23621 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-93-5p | 19 | LRRC55 | Sponge network | -0.154 | 0.78939 | -0.026 | 0.93163 | 0.379 |
| 23622 | A1BG-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-93-3p | 10 | EPHA3 | Sponge network | -0.164 | 0.45106 | 0.185 | 0.53588 | 0.379 |
| 23623 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 21 | KIT | Sponge network | 0.335 | 0.20596 | -2.184 | 0 | 0.379 |
| 23624 | RP11-158K1.3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | BCL9 | Sponge network | 0.349 | 0.01695 | 0.321 | 0.00267 | 0.379 |
| 23625 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | -0.969 | 2.0E-5 | -0.474 | 0.03833 | 0.379 |
| 23626 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 23 | HECA | Sponge network | 0.385 | 0.10067 | -0.217 | 0.03463 | 0.379 |
| 23627 | RP11-867G23.10 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-629-3p | 11 | LIFR | Sponge network | -2.531 | 0 | -1.794 | 0 | 0.379 |
| 23628 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 16 | FBXO31 | Sponge network | -0.576 | 0.10121 | -0.085 | 0.42389 | 0.379 |
| 23629 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 26 | RIMBP2 | Sponge network | -2.452 | 0 | -1.968 | 5.0E-5 | 0.379 |
| 23630 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 22 | MYO9A | Sponge network | 1.11 | 0 | -0.147 | 0.32373 | 0.379 |
| 23631 | LPP-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p | 32 | AFF1 | Sponge network | -0.18 | 0.20343 | -0.384 | 0.00053 | 0.379 |
| 23632 | LPP-AS2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-5p | 26 | BTG2 | Sponge network | -0.18 | 0.20343 | -1.763 | 0 | 0.379 |
| 23633 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p | 28 | ESR1 | Sponge network | -0.125 | 0.49414 | -1.035 | 3.0E-5 | 0.379 |
| 23634 | RP11-384K6.6 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | ERCC6 | Sponge network | 0.946 | 0 | 0.23 | 0.015 | 0.379 |
| 23635 | BOLA3-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-454-3p;hsa-miR-744-5p | 11 | IGF2 | Sponge network | -0.246 | 0.35034 | 0.848 | 0.03842 | 0.379 |
| 23636 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | RSPO2 | Sponge network | -0.021 | 0.93598 | -3.038 | 0 | 0.379 |
| 23637 | LINC00473 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 34 | IL17RD | Sponge network | -1.203 | 0.03881 | -0.019 | 0.94873 | 0.379 |
| 23638 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 54 | WDFY3 | Sponge network | 0.532 | 0.00505 | -0.201 | 0.0967 | 0.379 |
| 23639 | DIO3OS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-940 | 12 | GALNT13 | Sponge network | -0.773 | 0.04073 | -0.857 | 0.07439 | 0.379 |
| 23640 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 13 | HMCN1 | Sponge network | 0.421 | 0.00084 | 0.809 | 0.00544 | 0.379 |
| 23641 | MIR600HG |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-502-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 14 | SHPRH | Sponge network | 0.594 | 0.41522 | 0.098 | 0.40994 | 0.379 |
| 23642 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-766-3p;hsa-miR-940 | 29 | AFF4 | Sponge network | 0.946 | 0 | -0.266 | 0.01565 | 0.379 |
| 23643 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-7-1-3p | 13 | DKK3 | Sponge network | -0.053 | 0.80685 | -0.052 | 0.77783 | 0.379 |
| 23644 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | RNF144A | Sponge network | 0.532 | 0.00505 | 0.594 | 0.0009 | 0.379 |
| 23645 | SNHG16 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-497-5p;hsa-miR-664a-3p | 17 | SOX11 | Sponge network | 0.961 | 0 | 2.68 | 0 | 0.379 |
| 23646 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 38 | DTNA | Sponge network | 0.395 | 0.15451 | -1.648 | 1.0E-5 | 0.379 |
| 23647 | KCNJ2-AS1 |
hsa-let-7i-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | TGFBR3 | Sponge network | -0.436 | 0.05325 | -1.173 | 1.0E-5 | 0.379 |
| 23648 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-93-5p | 11 | EDA2R | Sponge network | 0.299 | 0.57722 | -0.587 | 0.01598 | 0.379 |
| 23649 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRIM58 | Sponge network | -1.697 | 0.00889 | -1.692 | 0 | 0.379 |
| 23650 | RP11-248G5.8 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-766-3p | 10 | ATM | Sponge network | 1.294 | 0 | 0.178 | 0.21568 | 0.379 |
| 23651 | DNM3OS |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 41 | GRIK3 | Sponge network | 0.572 | 0.01722 | -3.208 | 0 | 0.379 |
| 23652 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-484;hsa-miR-7-5p | 13 | FLNA | Sponge network | -0.342 | 0.02288 | -0.771 | 0.01377 | 0.379 |
| 23653 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 39 | ATRX | Sponge network | 0.421 | 0.00084 | 0.261 | 0.04408 | 0.379 |
| 23654 | RP1-59D14.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 18 | BRD3 | Sponge network | 1.162 | 5.0E-5 | 0.37 | 0.00013 | 0.379 |
| 23655 | RP11-395G23.3 |
hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | PIK3CA | Sponge network | 0.381 | 0.25967 | 0.195 | 0.06908 | 0.379 |
| 23656 | RP11-244O19.1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 26 | SYT4 | Sponge network | -0.096 | 0.67958 | -3.207 | 0 | 0.379 |
| 23657 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-582-5p;hsa-miR-942-5p | 13 | RB1CC1 | Sponge network | 0.637 | 0.00069 | 0.175 | 0.12559 | 0.379 |
| 23658 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 33 | NCOA1 | Sponge network | -0.129 | 0.57177 | -0.432 | 0 | 0.379 |
| 23659 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-942-5p | 32 | DDX6 | Sponge network | 0.637 | 0.00069 | 0.091 | 0.36428 | 0.379 |
| 23660 | TP73-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | PCDH18 | Sponge network | -0.563 | 0.02545 | 0.216 | 0.24645 | 0.379 |
| 23661 | LINC00672 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-98-5p | 48 | CADM2 | Sponge network | -0.227 | 0.31657 | -3.782 | 0 | 0.379 |
| 23662 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | SASH1 | Sponge network | 0.249 | 0.35902 | -0.502 | 0.00175 | 0.379 |
| 23663 | RP11-389C8.2 |
hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | SYNPO2 | Sponge network | 0.727 | 3.0E-5 | -2.342 | 0 | 0.379 |
| 23664 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 24 | BCL11A | Sponge network | -2.46 | 0 | -0.932 | 0.0063 | 0.379 |
| 23665 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | SEMA3C | Sponge network | 0.299 | 0.57722 | -0.272 | 0.31153 | 0.379 |
| 23666 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 32 | APC | Sponge network | -1.575 | 6.0E-5 | -0.255 | 0.04051 | 0.379 |
| 23667 | ZNF582-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p | 17 | SYT4 | Sponge network | -1.349 | 0.00017 | -3.207 | 0 | 0.379 |
| 23668 | RP11-597D13.9 |
hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | 1.302 | 7.0E-5 | -0.615 | 0.01482 | 0.379 |
| 23669 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | HACE1 | Sponge network | -0.031 | 0.89063 | -0.072 | 0.55239 | 0.379 |
| 23670 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-940 | 36 | NTRK2 | Sponge network | 0.056 | 0.81195 | -0.736 | 0.06495 | 0.379 |
| 23671 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p | 12 | FGFR1 | Sponge network | 0.41 | 0.02214 | -0.509 | 0.03957 | 0.379 |
| 23672 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | DDX6 | Sponge network | 0.194 | 0.64278 | 0.091 | 0.36428 | 0.379 |
| 23673 | RP11-822E23.8 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-7-5p | 13 | IRS1 | Sponge network | -0.777 | 0.16667 | -0.026 | 0.88855 | 0.379 |
| 23674 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p | 10 | JAKMIP2 | Sponge network | 0.889 | 9.0E-5 | -1.388 | 0 | 0.379 |
| 23675 | BVES-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-375;hsa-miR-429 | 15 | NIN | Sponge network | -2.62 | 0 | -0.253 | 0.13794 | 0.379 |
| 23676 | ARHGEF26-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZNF844 | Sponge network | -2.46 | 0 | -0.966 | 0.00035 | 0.379 |
| 23677 | BVES-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p | 32 | NTRK2 | Sponge network | -2.62 | 0 | -0.736 | 0.06495 | 0.379 |
| 23678 | RP5-994D16.9 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TSC1 | Sponge network | 0.252 | 0.08508 | 0.103 | 0.26071 | 0.379 |
| 23679 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p | 10 | FSTL5 | Sponge network | 0.253 | 0.09565 | -1.3 | 0.01619 | 0.379 |
| 23680 | CTD-2336O2.1 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 21 | DIP2C | Sponge network | -0.141 | 0.26434 | -0.436 | 0.01713 | 0.379 |
| 23681 | FAM66C |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484 | 16 | ABL1 | Sponge network | -0.901 | 0.00019 | -0.215 | 0.07804 | 0.379 |
| 23682 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-7-5p;hsa-miR-96-5p | 11 | ZYX | Sponge network | 0.194 | 0.64278 | 0.019 | 0.89763 | 0.379 |
| 23683 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p | 14 | DDX6 | Sponge network | 1.597 | 0 | 0.091 | 0.36428 | 0.379 |
| 23684 | H1FX-AS1 |
hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | MEF2C | Sponge network | -0.206 | 0.12942 | -0.499 | 0.01448 | 0.379 |
| 23685 | MALAT1 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 21 | TMEM30A | Sponge network | 0.782 | 0.00016 | 0.096 | 0.31031 | 0.379 |
| 23686 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | ABCB5 | Sponge network | -0.899 | 6.0E-5 | -0.956 | 0.04077 | 0.379 |
| 23687 | LINC00173 |
hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 14 | ITPKB | Sponge network | 0.056 | 0.8494 | -0.704 | 0.00106 | 0.379 |
| 23688 | RP1-257A7.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p | 13 | MDM4 | Sponge network | 0.849 | 0.00178 | 0.345 | 0.00762 | 0.379 |
| 23689 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RB1CC1 | Sponge network | 0.603 | 0.00127 | 0.175 | 0.12559 | 0.379 |
| 23690 | RP11-390K5.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-365a-3p;hsa-miR-590-3p | 17 | ARHGEF12 | Sponge network | 1.148 | 2.0E-5 | 0.09 | 0.35693 | 0.379 |
| 23691 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-935;hsa-miR-96-5p | 33 | FBXO32 | Sponge network | 0.083 | 0.66405 | -0.495 | 0.07561 | 0.379 |
| 23692 | RP1-122P22.2 |
hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-532-3p | 15 | PBX1 | Sponge network | 0.065 | 0.73468 | -1.206 | 0 | 0.379 |
| 23693 | U91328.19 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -0.303 | 0.21002 | -0.252 | 0.15511 | 0.379 |
| 23694 | RP11-758N13.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-93-3p | 26 | RUNX1T1 | Sponge network | 2.939 | 3.0E-5 | -0.344 | 0.2374 | 0.379 |
| 23695 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | WWTR1 | Sponge network | 0.395 | 0.15451 | -0.345 | 0.11997 | 0.379 |
| 23696 | RP11-67L2.2 |
hsa-miR-125a-3p;hsa-miR-149-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | CDC27 | Sponge network | 0.72 | 0.0001 | 0.384 | 0 | 0.379 |
| 23697 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 11 | AKAP12 | Sponge network | 0.222 | 0.29133 | -0.932 | 0.0028 | 0.379 |
| 23698 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | PIK3R1 | Sponge network | 0.225 | 0.30644 | -0.133 | 0.36854 | 0.379 |
| 23699 | U91328.19 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-624-5p | 13 | HMCN1 | Sponge network | -0.303 | 0.21002 | 0.809 | 0.00544 | 0.379 |
| 23700 | RP11-848P1.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | GAS7 | Sponge network | 1.253 | 0 | -0.555 | 0.01602 | 0.379 |
| 23701 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-7-5p | 15 | TET1 | Sponge network | -0.469 | 0.08721 | -0.135 | 0.52138 | 0.379 |
| 23702 | FAM13A-AS1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-181d-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-664a-5p | 11 | TET2 | Sponge network | -0.83 | 0 | -0.222 | 0.04665 | 0.379 |
| 23703 | PART1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | LRP1B | Sponge network | -2.925 | 0 | -2.163 | 0 | 0.379 |
| 23704 | LINC00173 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 24 | BCL2 | Sponge network | 0.056 | 0.8494 | -0.899 | 0 | 0.379 |
| 23705 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b | 12 | MYCBP2 | Sponge network | 0.082 | 0.55397 | -0.027 | 0.82016 | 0.379 |
| 23706 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p | 24 | PRDM2 | Sponge network | -0.842 | 0.01361 | -0.258 | 0.00721 | 0.379 |
| 23707 | AC141928.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 21 | BRD3 | Sponge network | 0.853 | 0.15352 | 0.37 | 0.00013 | 0.379 |
| 23708 | ZSCAN16-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 14 | MRVI1 | Sponge network | -0.342 | 0.02288 | -1.051 | 0.00022 | 0.379 |
| 23709 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | EPAS1 | Sponge network | -0.842 | 0.01361 | -0.184 | 0.14418 | 0.379 |
| 23710 | C20orf166-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p | 10 | NRG3 | Sponge network | -2.69 | 0.0001 | -1.836 | 4.0E-5 | 0.379 |
| 23711 | AP001347.6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | ABCA10 | Sponge network | -1.161 | 0.00591 | -0.571 | 0.03674 | 0.379 |
| 23712 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 27 | NRXN1 | Sponge network | -0.727 | 0.04726 | -3.778 | 0 | 0.379 |
| 23713 | RP11-123M6.2 |
hsa-miR-130b-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-629-5p;hsa-miR-744-3p | 13 | DST | Sponge network | 0.543 | 0.12356 | -0.713 | 0.00096 | 0.379 |
| 23714 | RAB30-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 12 | TRIM33 | Sponge network | 0.752 | 0 | 0.402 | 7.0E-5 | 0.379 |
| 23715 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 18 | TBL1XR1 | Sponge network | 0.769 | 0 | 0.578 | 0 | 0.379 |
| 23716 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-653-5p | 13 | DMXL1 | Sponge network | -0.376 | 0.00337 | -0.426 | 0.00056 | 0.379 |
| 23717 | GS1-124K5.11 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-486-5p;hsa-miR-590-3p | 12 | DNMT3A | Sponge network | 0.494 | 0.00339 | 0.302 | 0.0262 | 0.379 |
| 23718 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p | 30 | PDGFRA | Sponge network | -2.62 | 0 | -0.372 | 0.0037 | 0.379 |
| 23719 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p | 14 | PRKCE | Sponge network | -1.331 | 3.0E-5 | -0.403 | 0.00056 | 0.379 |
| 23720 | SERTAD4-AS1 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p | 11 | RIMS2 | Sponge network | -0.932 | 0.00329 | -1.365 | 0.00208 | 0.379 |
| 23721 | TRIM52-AS1 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | SFRP1 | Sponge network | -0.418 | 0.00068 | -3.566 | 0 | 0.379 |
| 23722 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 18 | EBF3 | Sponge network | -0.738 | 0.21233 | -0.058 | 0.81437 | 0.379 |
| 23723 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 14 | FBXO31 | Sponge network | -0.355 | 0.0077 | -0.085 | 0.42389 | 0.379 |
| 23724 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | NRXN1 | Sponge network | -0.246 | 0.35034 | -3.778 | 0 | 0.379 |
| 23725 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-484 | 11 | SMARCA2 | Sponge network | -0.465 | 0.04703 | -0.529 | 0.00014 | 0.379 |
| 23726 | LINC00174 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-539-5p;hsa-miR-582-5p | 14 | MOB1B | Sponge network | 1.253 | 0 | 0.197 | 0.15266 | 0.379 |
| 23727 | AP001347.6 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | PRKCB | Sponge network | -1.161 | 0.00591 | -1.313 | 2.0E-5 | 0.379 |
| 23728 | RP11-48B3.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 11 | CEP170 | Sponge network | 1.757 | 0 | 0.943 | 0 | 0.379 |
| 23729 | AC004540.4 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-362-3p | 17 | FAT3 | Sponge network | -2.549 | 0.00528 | -1.601 | 0.00051 | 0.379 |
| 23730 | ANKRD10-IT1 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | UBR3 | Sponge network | 1.739 | 0 | -0.021 | 0.82774 | 0.379 |
| 23731 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 17 | SON | Sponge network | 1.205 | 0 | 0.168 | 0.04406 | 0.379 |
| 23732 | LINC00654 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-96-5p | 11 | ZBTB20 | Sponge network | 0.374 | 0.20054 | -0.122 | 0.57569 | 0.379 |
| 23733 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-940 | 34 | PBX1 | Sponge network | 0.331 | 0.57247 | -1.206 | 0 | 0.379 |
| 23734 | LINC00092 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-590-3p | 18 | NCOA1 | Sponge network | -1.886 | 0 | -0.432 | 0 | 0.379 |
| 23735 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 21 | KIT | Sponge network | 0.572 | 0.01722 | -2.184 | 0 | 0.379 |
| 23736 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935 | 49 | PTPRD | Sponge network | -0.563 | 0.02545 | -1.005 | 0.00064 | 0.379 |
| 23737 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p | 20 | DDX6 | Sponge network | 0.768 | 7.0E-5 | 0.091 | 0.36428 | 0.379 |
| 23738 | LINC00476 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-671-5p | 24 | PPP2R5C | Sponge network | -0.615 | 0.00015 | -0.314 | 3.0E-5 | 0.379 |
| 23739 | HNRNPU-AS1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | ERCC6 | Sponge network | 1.601 | 0 | 0.23 | 0.015 | 0.379 |
| 23740 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 20 | SLC6A15 | Sponge network | -0.793 | 7.0E-5 | -2.373 | 2.0E-5 | 0.379 |
| 23741 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-542-3p | 19 | SYT4 | Sponge network | -0.517 | 0.02935 | -3.207 | 0 | 0.379 |
| 23742 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-589-3p | 21 | KCNA6 | Sponge network | 0.811 | 0.03228 | -1.531 | 0.00013 | 0.379 |
| 23743 | RP11-384K6.6 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-424-5p | 10 | DOCK4 | Sponge network | 0.946 | 0 | 0.502 | 0.00108 | 0.379 |
| 23744 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p | 22 | CBFA2T3 | Sponge network | -0.487 | 0.02157 | -1.221 | 1.0E-5 | 0.379 |
| 23745 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-34a-5p;hsa-miR-370-3p;hsa-miR-7-1-3p | 10 | ZMYND11 | Sponge network | -0.187 | 0.23787 | -0.2 | 0.05449 | 0.379 |
| 23746 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | JAZF1 | Sponge network | 0.342 | 0.03129 | -0.377 | 0.02677 | 0.379 |
| 23747 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | -0.303 | 0.21002 | -1.751 | 1.0E-5 | 0.379 |
| 23748 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p | 24 | MTSS1 | Sponge network | -0.154 | 0.78939 | -0.81 | 0.00035 | 0.379 |
| 23749 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 54 | CADM2 | Sponge network | 0.056 | 0.81195 | -3.782 | 0 | 0.379 |
| 23750 | SH3RF3-AS1 |
hsa-miR-126-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p | 13 | IRS1 | Sponge network | 0.889 | 9.0E-5 | -0.026 | 0.88855 | 0.379 |
| 23751 | RP4-669L17.10 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 1.093 | 0 | 0.362 | 0.00066 | 0.379 |
| 23752 | CTC-429P9.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-532-5p | 19 | RYR2 | Sponge network | 0.082 | 0.55397 | -1.466 | 0.00031 | 0.379 |
| 23753 | FAM13A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | GNAQ | Sponge network | -0.83 | 0 | -0.367 | 0.00102 | 0.379 |
| 23754 | RP11-968O1.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-493-5p | 14 | DIP2C | Sponge network | -0.829 | 0.01343 | -0.436 | 0.01713 | 0.379 |
| 23755 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | DKK3 | Sponge network | 1.302 | 7.0E-5 | -0.052 | 0.77783 | 0.379 |
| 23756 | PSMD5-AS1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 19 | PHF6 | Sponge network | 0.569 | 0.00067 | 0.785 | 0 | 0.379 |
| 23757 | AC016747.3 |
hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-455-5p | 14 | PLXNC1 | Sponge network | 0.199 | 0.38015 | 0.569 | 0.00329 | 0.379 |
| 23758 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p | 23 | EBF1 | Sponge network | 0.199 | 0.38015 | -0.384 | 0.08856 | 0.379 |
| 23759 | RP11-13K12.5 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | NEDD4 | Sponge network | -0.318 | 0.65847 | 0.02 | 0.89834 | 0.379 |
| 23760 | RP3-523E19.2 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p | 12 | CCDC6 | Sponge network | 2.887 | 0 | 0.27 | 0.02555 | 0.379 |
| 23761 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 35 | CBL | Sponge network | 0.488 | 0.00084 | 0.291 | 0.01267 | 0.379 |
| 23762 | PCA3 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-5p | 28 | BTG2 | Sponge network | -0.709 | 0.18168 | -1.763 | 0 | 0.379 |
| 23763 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p | 45 | GAS7 | Sponge network | -0.727 | 0.04726 | -0.555 | 0.01602 | 0.379 |
| 23764 | RP11-53O19.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-942-5p | 11 | ZBTB20 | Sponge network | -0.405 | 0.22013 | -0.122 | 0.57569 | 0.379 |
| 23765 | PWAR6 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 14 | EHD3 | Sponge network | -1.575 | 6.0E-5 | -0.484 | 0.01747 | 0.379 |
| 23766 | FENDRR |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-942-5p | 26 | OPCML | Sponge network | -1.281 | 0.00012 | -2.498 | 0 | 0.379 |
| 23767 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 11 | ABCA10 | Sponge network | -0.693 | 0.05629 | -0.571 | 0.03674 | 0.379 |
| 23768 | CTC-559E9.5 |
hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-629-5p | 17 | UBR3 | Sponge network | 0.594 | 0.00064 | -0.021 | 0.82774 | 0.379 |
| 23769 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | SLC6A15 | Sponge network | -0.969 | 2.0E-5 | -2.373 | 2.0E-5 | 0.379 |
| 23770 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | 0.083 | 0.66405 | -1.324 | 0.00014 | 0.379 |
| 23771 | U62631.5 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-940 | 13 | MDM4 | Sponge network | 3.115 | 0.00463 | 0.345 | 0.00762 | 0.379 |
| 23772 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 14 | GLI3 | Sponge network | 0.082 | 0.55654 | -0.615 | 0.01482 | 0.379 |
| 23773 | AC004893.11 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | PHF3 | Sponge network | 1.317 | 0 | 0.126 | 0.19652 | 0.379 |
| 23774 | RP11-48B3.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 17 | DLC1 | Sponge network | 1.757 | 0 | -0.382 | 0.05038 | 0.379 |
| 23775 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | ZMYM2 | Sponge network | 0.222 | 0.29133 | 0.279 | 0.01273 | 0.379 |
| 23776 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | SASH1 | Sponge network | -0.421 | 0.0114 | -0.502 | 0.00175 | 0.379 |
| 23777 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 19 | TGFBR2 | Sponge network | -1.203 | 0.03881 | -0.243 | 0.11408 | 0.379 |
| 23778 | RP11-597D13.9 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | DOCK4 | Sponge network | 1.302 | 7.0E-5 | 0.502 | 0.00108 | 0.379 |
| 23779 | FAM13A-AS1 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ADH1B | Sponge network | -0.83 | 0 | -3.716 | 0 | 0.379 |
| 23780 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 22 | PRKAA2 | Sponge network | 0.178 | 0.29106 | -2.462 | 0 | 0.379 |
| 23781 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-539-5p | 20 | KIF1B | Sponge network | 0.793 | 0 | -0.118 | 0.32258 | 0.379 |
| 23782 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-98-5p | 34 | THBS1 | Sponge network | -1.216 | 0 | 0.741 | 0.00386 | 0.379 |
| 23783 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p | 13 | PRKAB2 | Sponge network | 1.106 | 0 | -0.367 | 0.01371 | 0.379 |
| 23784 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-3p | 10 | LRRC55 | Sponge network | -2.095 | 0.00112 | -0.026 | 0.93163 | 0.379 |
| 23785 | A2M-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | DTNA | Sponge network | -0.08 | 0.90092 | -1.648 | 1.0E-5 | 0.379 |
| 23786 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 28 | IGF1 | Sponge network | 0.214 | 0.18246 | -0.204 | 0.5898 | 0.379 |
| 23787 | LINC00672 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-98-5p | 15 | KLF10 | Sponge network | -0.227 | 0.31657 | -0.783 | 0 | 0.379 |
| 23788 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 25 | RNF144A | Sponge network | -0.727 | 0.04726 | 0.594 | 0.0009 | 0.379 |
| 23789 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-940;hsa-miR-942-5p | 36 | NOTCH2 | Sponge network | -0.162 | 0.47687 | 0.138 | 0.35163 | 0.379 |
| 23790 | RP11-119F7.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 19 | SYNPO2 | Sponge network | 0.225 | 0.30644 | -2.342 | 0 | 0.378 |
| 23791 | SNHG14 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 11 | DSE | Sponge network | -1.111 | 0.0003 | -0.231 | 0.05347 | 0.378 |
| 23792 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 17 | CRTC3 | Sponge network | -0.318 | 0.65847 | 0.164 | 0.16786 | 0.378 |
| 23793 | NDUFA6-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | CRTC1 | Sponge network | -0.355 | 0.0077 | -0.116 | 0.28412 | 0.378 |
| 23794 | RP11-798G7.7 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-375 | 12 | MLLT10 | Sponge network | 0.858 | 3.0E-5 | 0.105 | 0.33292 | 0.378 |
| 23795 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-542-3p | 10 | RARB | Sponge network | 0.056 | 0.81195 | -0.825 | 2.0E-5 | 0.378 |
| 23796 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 25 | PHC3 | Sponge network | 0.214 | 0.18246 | 0.212 | 0.0499 | 0.378 |
| 23797 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p | 19 | RET | Sponge network | -0.777 | 0.16667 | -0.833 | 0.03517 | 0.378 |
| 23798 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | PBRM1 | Sponge network | 0.826 | 1.0E-5 | -0.029 | 0.78601 | 0.378 |
| 23799 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 26 | SYNPO2 | Sponge network | 0.082 | 0.55654 | -2.342 | 0 | 0.378 |
| 23800 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | CACNA2D1 | Sponge network | 0.299 | 0.57722 | -0.846 | 0.00675 | 0.378 |
| 23801 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | NUAK1 | Sponge network | -0.976 | 0.00025 | 0.904 | 0 | 0.378 |
| 23802 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-93-5p | 16 | RBL2 | Sponge network | -1.645 | 0 | -0.282 | 0.00254 | 0.378 |
| 23803 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3913-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-942-5p | 29 | HLF | Sponge network | -0.342 | 0.02288 | -2.443 | 0 | 0.378 |
| 23804 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 38 | PIK3R1 | Sponge network | -0.612 | 0.00094 | -0.133 | 0.36854 | 0.378 |
| 23805 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 16 | CDH19 | Sponge network | -0.154 | 0.78939 | -3.434 | 0 | 0.378 |
| 23806 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-378a-5p | 10 | PDS5B | Sponge network | 0.924 | 0 | 0.259 | 0.00962 | 0.378 |
| 23807 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429 | 12 | RCAN2 | Sponge network | 0.253 | 0.09565 | -0.33 | 0.15172 | 0.378 |
| 23808 | TRAF3IP2-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 27 | PDE4DIP | Sponge network | -0.323 | 0.01372 | -0.282 | 0.0257 | 0.378 |
| 23809 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-93-5p | 30 | JAZF1 | Sponge network | -1.203 | 0.03881 | -0.377 | 0.02677 | 0.378 |
| 23810 | CTA-941F9.9 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-505-5p | 10 | ASTN1 | Sponge network | -0.344 | 0.49624 | -2.389 | 2.0E-5 | 0.378 |
| 23811 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 21 | ARHGAP29 | Sponge network | 0.997 | 0.00066 | 0.131 | 0.42953 | 0.378 |
| 23812 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-93-5p | 31 | AFF4 | Sponge network | 0.497 | 0.01451 | -0.266 | 0.01565 | 0.378 |
| 23813 | RP11-890B15.3 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TPO | Sponge network | 0.101 | 0.60919 | -1.87 | 2.0E-5 | 0.378 |
| 23814 | RP11-389C8.2 |
hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | GLI3 | Sponge network | 0.727 | 3.0E-5 | -0.615 | 0.01482 | 0.378 |
| 23815 | RP11-35G9.3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 10 | FLI1 | Sponge network | 0.657 | 4.0E-5 | -0.294 | 0.10905 | 0.378 |
| 23816 | CTB-113P19.4 |
hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | TP63 | Sponge network | 0.906 | 0.31715 | -0.363 | 0.38191 | 0.378 |
| 23817 | RP11-244O19.1 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-493-3p;hsa-miR-590-3p | 11 | ARID1B | Sponge network | -0.096 | 0.67958 | 0.003 | 0.97503 | 0.378 |
| 23818 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | ERCC4 | Sponge network | -0.896 | 0.00099 | 0.126 | 0.15266 | 0.378 |
| 23819 | GNG12-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-5p | 39 | GNAQ | Sponge network | -0.793 | 7.0E-5 | -0.367 | 0.00102 | 0.378 |
| 23820 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-96-5p | 20 | CRTC3 | Sponge network | 0.395 | 0.15451 | 0.164 | 0.16786 | 0.378 |
| 23821 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-375 | 11 | MLLT10 | Sponge network | 0.856 | 0 | 0.105 | 0.33292 | 0.378 |
| 23822 | RP11-159D12.2 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p | 28 | ABL2 | Sponge network | 1.254 | 0 | 0.82 | 0 | 0.378 |
| 23823 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p | 15 | RB1CC1 | Sponge network | 0.95 | 0.15943 | 0.175 | 0.12559 | 0.378 |
| 23824 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | TACC1 | Sponge network | 0.727 | 3.0E-5 | -0.362 | 0.05406 | 0.378 |
| 23825 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p | 16 | CITED2 | Sponge network | -0.793 | 7.0E-5 | -1.545 | 0 | 0.378 |
| 23826 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-450b-5p;hsa-miR-93-3p | 11 | AKAP11 | Sponge network | 0.912 | 0.00014 | 0.196 | 0.09825 | 0.378 |
| 23827 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | 0.559 | 0.00026 | 0.809 | 0.00544 | 0.378 |
| 23828 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-671-5p | 24 | RBMS3 | Sponge network | 0.381 | 0.25967 | -0.519 | 0.03551 | 0.378 |
| 23829 | LINC00641 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 14 | CEP170 | Sponge network | -0.222 | 0.32445 | 0.943 | 0 | 0.378 |
| 23830 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 38 | FOXP1 | Sponge network | 0.349 | 0.01695 | 0.042 | 0.74368 | 0.378 |
| 23831 | AC010226.4 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | KLF10 | Sponge network | -0.811 | 2.0E-5 | -0.783 | 0 | 0.378 |
| 23832 | CTC-510F12.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-766-3p | 10 | MDM4 | Sponge network | -0.691 | 0.00146 | 0.345 | 0.00762 | 0.378 |
| 23833 | LINC00092 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | DIP2C | Sponge network | -1.886 | 0 | -0.436 | 0.01713 | 0.378 |
| 23834 | AC127904.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TSC1 | Sponge network | 1.308 | 0 | 0.103 | 0.26071 | 0.378 |
| 23835 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 19 | CDHR3 | Sponge network | -0.83 | 0 | -0.977 | 4.0E-5 | 0.378 |
| 23836 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 28 | GRIN2A | Sponge network | -0.355 | 0.0077 | -2.161 | 0 | 0.378 |
| 23837 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 19 | HIP1 | Sponge network | -0.125 | 0.49414 | 0.774 | 0 | 0.378 |
| 23838 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-629-3p | 10 | EBF3 | Sponge network | 1.153 | 0.00114 | -0.058 | 0.81437 | 0.378 |
| 23839 | MYLK-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p | 11 | PRKAR1A | Sponge network | -1.331 | 3.0E-5 | -0.063 | 0.50314 | 0.378 |
| 23840 | FAM13A-AS1 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 24 | NCOA1 | Sponge network | -0.83 | 0 | -0.432 | 0 | 0.378 |
| 23841 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | -2.915 | 0.00015 | -0.7 | 1.0E-5 | 0.378 |
| 23842 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-93-5p | 32 | PRKAA2 | Sponge network | 0.051 | 0.83388 | -2.462 | 0 | 0.378 |
| 23843 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-625-5p | 16 | CADM1 | Sponge network | -2.531 | 0 | -0.9 | 0.00084 | 0.378 |
| 23844 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-429 | 10 | TSC22D1 | Sponge network | -0.612 | 0.00094 | -0.529 | 2.0E-5 | 0.378 |
| 23845 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-3p;hsa-miR-589-3p;hsa-miR-874-3p;hsa-miR-96-5p | 32 | TACC1 | Sponge network | 0.331 | 0.57247 | -0.362 | 0.05406 | 0.378 |
| 23846 | AC016747.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 37 | TRPS1 | Sponge network | 0.199 | 0.38015 | -0.191 | 0.44109 | 0.378 |
| 23847 | HCG11 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-590-3p | 17 | DYNC1I1 | Sponge network | 0.385 | 0.10067 | -1.12 | 0.00107 | 0.378 |
| 23848 | OIP5-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 35 | ERC1 | Sponge network | 0.421 | 0.00084 | -0.108 | 0.38794 | 0.378 |
| 23849 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-96-5p | 13 | DPYD | Sponge network | -2.452 | 0 | -0.736 | 0.00076 | 0.378 |
| 23850 | RP11-305E6.4 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-363-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | TRIM13 | Sponge network | 1.317 | 0 | 0.183 | 0.06968 | 0.378 |
| 23851 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-629-5p | 16 | TCF4 | Sponge network | -2.076 | 0.00548 | 0.016 | 0.92271 | 0.378 |
| 23852 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429 | 13 | ARID5B | Sponge network | 1.106 | 0 | 0.342 | 0.01676 | 0.378 |
| 23853 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 32 | MXI1 | Sponge network | -1.216 | 0 | -0.987 | 0 | 0.378 |
| 23854 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | SPG20 | Sponge network | 0.214 | 0.18246 | -1.16 | 0 | 0.378 |
| 23855 | HOXA-AS2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | 0.61 | 0.01593 | -0.615 | 0.01482 | 0.378 |
| 23856 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-339-5p | 20 | KIF1B | Sponge network | 0.813 | 1.0E-5 | -0.118 | 0.32258 | 0.378 |
| 23857 | RP11-158K1.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.349 | 0.01695 | -0.57 | 0 | 0.378 |
| 23858 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-582-3p;hsa-miR-590-3p | 21 | SASH1 | Sponge network | -1.582 | 8.0E-5 | -0.502 | 0.00175 | 0.378 |
| 23859 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-93-5p | 14 | TXNIP | Sponge network | -0.777 | 0.1134 | -1.277 | 0 | 0.378 |
| 23860 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-532-5p | 13 | PTPN14 | Sponge network | -4.463 | 0.00025 | 0.144 | 0.34595 | 0.378 |
| 23861 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | CXXC4 | Sponge network | -1.575 | 6.0E-5 | -0.433 | 0.21055 | 0.378 |
| 23862 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 14 | FREM2 | Sponge network | -0.513 | 0.04703 | -0.437 | 0.46353 | 0.378 |
| 23863 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-502-5p | 13 | CHD5 | Sponge network | -0.932 | 0.00329 | -0.922 | 0.01521 | 0.378 |
| 23864 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | SASH1 | Sponge network | 0.889 | 9.0E-5 | -0.502 | 0.00175 | 0.378 |
| 23865 | A2M-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-484 | 15 | CDH23 | Sponge network | -0.08 | 0.90092 | -0.974 | 0.00034 | 0.378 |
| 23866 | ADAMTS9-AS2 |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 20 | JUN | Sponge network | -2.452 | 0 | -0.932 | 0 | 0.378 |
| 23867 | CTC-444N24.11 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-429;hsa-miR-590-3p | 11 | PICALM | Sponge network | 1.153 | 0 | 0.086 | 0.23523 | 0.378 |
| 23868 | MIR143HG |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | LUZP2 | Sponge network | -0.739 | 0.0574 | -0.056 | 0.89416 | 0.378 |
| 23869 | PSMG3-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | MACF1 | Sponge network | -0.125 | 0.49414 | 0.019 | 0.90335 | 0.378 |
| 23870 | RP5-994D16.9 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 10 | TET1 | Sponge network | 0.252 | 0.08508 | -0.135 | 0.52138 | 0.378 |
| 23871 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-590-3p | 18 | TLE4 | Sponge network | -0.899 | 6.0E-5 | -1.157 | 0 | 0.378 |
| 23872 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | MYBL1 | Sponge network | 0.796 | 4.0E-5 | 0.494 | 0.02152 | 0.378 |
| 23873 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 39 | BCL2 | Sponge network | -0.355 | 0.0077 | -0.899 | 0 | 0.378 |
| 23874 | CTD-2002H8.2 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | SPRY3 | Sponge network | 0.931 | 1.0E-5 | 0.016 | 0.91286 | 0.378 |
| 23875 | PGM5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-484;hsa-miR-7-1-3p | 21 | MED12L | Sponge network | -4.546 | 0 | -0.194 | 0.55299 | 0.378 |
| 23876 | LINC00473 |
hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | RPS6KA6 | Sponge network | -1.203 | 0.03881 | -2.989 | 0 | 0.378 |
| 23877 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 31 | ZFHX3 | Sponge network | -0.612 | 0.00094 | 0.389 | 0.01363 | 0.378 |
| 23878 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 18 | CHD2 | Sponge network | 0.349 | 0.01695 | 0.049 | 0.55371 | 0.378 |
| 23879 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 32 | EYA4 | Sponge network | -2.46 | 0 | 0.3 | 0.36876 | 0.378 |
| 23880 | RP11-48B3.4 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 36 | MDM4 | Sponge network | 1.757 | 0 | 0.345 | 0.00762 | 0.378 |
| 23881 | CTC-444N24.11 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 1.153 | 0 | 0.321 | 0.00267 | 0.378 |
| 23882 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ETV1 | Sponge network | -0.342 | 0.02288 | -0.096 | 0.67674 | 0.378 |
| 23883 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | PLA2R1 | Sponge network | -0.942 | 0 | -0.265 | 0.24676 | 0.378 |
| 23884 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 25 | EPHA3 | Sponge network | 0.41 | 0.02214 | 0.185 | 0.53588 | 0.378 |
| 23885 | AP001347.6 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 12 | CELF4 | Sponge network | -1.161 | 0.00591 | -1.135 | 0.00344 | 0.378 |
| 23886 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p | 32 | RICTOR | Sponge network | 0.392 | 0.0278 | 0.388 | 0.00188 | 0.378 |
| 23887 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | CDH10 | Sponge network | -0.769 | 0.00035 | -2.397 | 0 | 0.378 |
| 23888 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 35 | BMPR2 | Sponge network | 0.765 | 7.0E-5 | 0.206 | 0.0635 | 0.378 |
| 23889 | PWAR6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | DACT1 | Sponge network | -1.575 | 6.0E-5 | 0.783 | 0.00072 | 0.378 |
| 23890 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 15 | PNRC1 | Sponge network | -0.737 | 0.06182 | -0.646 | 0 | 0.378 |
| 23891 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | PIK3CA | Sponge network | 0.919 | 0 | 0.195 | 0.06908 | 0.378 |
| 23892 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 21 | NR2C2 | Sponge network | 0.392 | 0.0278 | 0.21 | 0.03224 | 0.378 |
| 23893 | FAM66C |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | CRTC1 | Sponge network | -0.901 | 0.00019 | -0.116 | 0.28412 | 0.378 |
| 23894 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-589-5p;hsa-miR-96-5p | 16 | ARHGAP29 | Sponge network | -1.439 | 4.0E-5 | 0.131 | 0.42953 | 0.378 |
| 23895 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577 | 15 | DNAJB4 | Sponge network | 0.056 | 0.81195 | -0.7 | 1.0E-5 | 0.378 |
| 23896 | MALAT1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 22 | NF1 | Sponge network | 0.782 | 0.00016 | 0.51 | 0 | 0.378 |
| 23897 | RP11-25K19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p | 25 | SNX29 | Sponge network | -1.057 | 0.00029 | -0.34 | 0.01371 | 0.378 |
| 23898 | C4B-AS1 |
hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 12 | EPHA3 | Sponge network | 2.306 | 9.0E-5 | 0.185 | 0.53588 | 0.378 |
| 23899 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p | 15 | ITGA5 | Sponge network | -0.487 | 0.02157 | 0.452 | 0.03524 | 0.378 |
| 23900 | DNAJC27-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-671-5p;hsa-miR-940 | 18 | SORCS1 | Sponge network | -0.969 | 2.0E-5 | -3.492 | 0 | 0.378 |
| 23901 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p | 10 | CRTC1 | Sponge network | -0.583 | 0.14589 | -0.116 | 0.28412 | 0.378 |
| 23902 | RP11-458D21.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-429 | 24 | ABL2 | Sponge network | 0.663 | 0.00073 | 0.82 | 0 | 0.378 |
| 23903 | PART1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 41 | IGFBP5 | Sponge network | -2.925 | 0 | -0.806 | 0.00105 | 0.378 |
| 23904 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 22 | ZMYM2 | Sponge network | -0.222 | 0.32445 | 0.279 | 0.01273 | 0.378 |
| 23905 | BOLA3-AS1 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-7-1-3p | 16 | DOCK4 | Sponge network | -0.246 | 0.35034 | 0.502 | 0.00108 | 0.378 |
| 23906 | LINC01018 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 31 | SNX29 | Sponge network | -2.915 | 0.00015 | -0.34 | 0.01371 | 0.378 |
| 23907 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-495-3p | 10 | BCLAF1 | Sponge network | 0.559 | 0.00026 | 0.211 | 0.00684 | 0.378 |
| 23908 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-940 | 30 | MDM4 | Sponge network | 0.174 | 0.30034 | 0.345 | 0.00762 | 0.378 |
| 23909 | RP11-65J21.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 16 | TRPS1 | Sponge network | -0.557 | 0.11135 | -0.191 | 0.44109 | 0.378 |
| 23910 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-93-5p | 20 | TACC1 | Sponge network | 0.497 | 0.01451 | -0.362 | 0.05406 | 0.378 |
| 23911 | SH3RF3-AS1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | PCDH10 | Sponge network | 0.889 | 9.0E-5 | -1.644 | 0.0078 | 0.378 |
| 23912 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | PRKAA2 | Sponge network | -0.663 | 0.10887 | -2.462 | 0 | 0.378 |
| 23913 | RP11-473I1.9 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-5p;hsa-miR-495-3p | 14 | STAG2 | Sponge network | 0.683 | 1.0E-5 | 0.252 | 0.00438 | 0.378 |
| 23914 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-654-3p | 31 | RICTOR | Sponge network | -0.405 | 0.22013 | 0.388 | 0.00188 | 0.378 |
| 23915 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p | 24 | ZFHX3 | Sponge network | 0.921 | 0.00118 | 0.389 | 0.01363 | 0.378 |
| 23916 | CKMT2-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 19 | SIRT1 | Sponge network | 0.082 | 0.55654 | 0.109 | 0.2593 | 0.378 |
| 23917 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p | 35 | NTRK2 | Sponge network | -0.513 | 0.04703 | -0.736 | 0.06495 | 0.378 |
| 23918 | SNHG16 |
hsa-miR-125a-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | ERCC6 | Sponge network | 0.961 | 0 | 0.23 | 0.015 | 0.378 |
| 23919 | RP11-166D19.1 |
hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-590-3p | 11 | EGR1 | Sponge network | -0.576 | 0.10121 | -1.195 | 0 | 0.378 |
| 23920 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-7-1-3p | 12 | DIP2C | Sponge network | -0.285 | 0.2172 | -0.436 | 0.01713 | 0.378 |
| 23921 | CTD-2314G24.2 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-508-3p;hsa-miR-7-1-3p | 11 | RASSF3 | Sponge network | 0.246 | 0.59912 | -0.226 | 0.11691 | 0.378 |
| 23922 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-3p;hsa-miR-93-5p | 18 | MAP3K12 | Sponge network | 0.335 | 0.20596 | -0.057 | 0.59369 | 0.378 |
| 23923 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | GPC6 | Sponge network | -2.117 | 0.00161 | -0.054 | 0.81465 | 0.378 |
| 23924 | C20orf166-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-5p | 12 | PCDH18 | Sponge network | -2.69 | 0.0001 | 0.216 | 0.24645 | 0.378 |
| 23925 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-532-3p | 18 | CBL | Sponge network | 1.317 | 0 | 0.291 | 0.01267 | 0.378 |
| 23926 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 22 | THBS1 | Sponge network | 0.395 | 0.15451 | 0.741 | 0.00386 | 0.378 |
| 23927 | RP11-890B15.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 23 | NRK | Sponge network | 0.101 | 0.60919 | 0.656 | 0.19217 | 0.378 |
| 23928 | NNT-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 22 | ABI2 | Sponge network | 0.799 | 1.0E-5 | 0.175 | 0.14787 | 0.378 |
| 23929 | USP3-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-671-5p | 10 | PIK3IP1 | Sponge network | -0.162 | 0.47687 | -0.187 | 0.19945 | 0.378 |
| 23930 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-577;hsa-miR-590-3p | 20 | KIT | Sponge network | -0.899 | 6.0E-5 | -2.184 | 0 | 0.378 |
| 23931 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 16 | HOOK3 | Sponge network | 0.219 | 0.1516 | 0.273 | 0.09957 | 0.378 |
| 23932 | RP11-166D19.1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | HERC1 | Sponge network | -0.576 | 0.10121 | -0.196 | 0.08544 | 0.378 |
| 23933 | RP11-66B24.2 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-27a-3p | 10 | CADM1 | Sponge network | 0.57 | 0.1866 | -0.9 | 0.00084 | 0.378 |
| 23934 | RP11-282O18.3 |
hsa-miR-101-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 12 | UBR3 | Sponge network | 0.757 | 0 | -0.021 | 0.82774 | 0.378 |
| 23935 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 26 | TMTC3 | Sponge network | 0.91 | 0 | 0.123 | 0.22189 | 0.378 |
| 23936 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-362-3p | 26 | PDGFRA | Sponge network | -0.693 | 0.05629 | -0.372 | 0.0037 | 0.378 |
| 23937 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-576-5p | 23 | CBL | Sponge network | 0.401 | 0.00066 | 0.291 | 0.01267 | 0.378 |
| 23938 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | -0.777 | 0.16667 | -1.751 | 1.0E-5 | 0.378 |
| 23939 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-877-5p;hsa-miR-93-5p | 29 | PGR | Sponge network | -0.969 | 2.0E-5 | -1.704 | 0 | 0.378 |
| 23940 | AC108488.4 |
hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-874-3p;hsa-miR-940 | 15 | CRTC3 | Sponge network | 0.259 | 0.12542 | 0.164 | 0.16786 | 0.378 |
| 23941 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-93-5p | 11 | LHX6 | Sponge network | -0.351 | 0.11358 | -0.087 | 0.69193 | 0.378 |
| 23942 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-7-1-3p | 16 | TCF4 | Sponge network | -0.011 | 0.967 | 0.016 | 0.92271 | 0.378 |
| 23943 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ZFHX3 | Sponge network | 0.539 | 0.03722 | 0.389 | 0.01363 | 0.378 |
| 23944 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | NR4A3 | Sponge network | -0.777 | 0.16667 | -1.385 | 0 | 0.378 |
| 23945 | CTD-2245F17.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-625-5p;hsa-miR-940 | 15 | PTPRT | Sponge network | -0.421 | 0.12556 | -0.907 | 0.03727 | 0.378 |
| 23946 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | PDGFRA | Sponge network | -1.349 | 0.00017 | -0.372 | 0.0037 | 0.378 |
| 23947 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | CXXC4 | Sponge network | -2.46 | 0 | -0.433 | 0.21055 | 0.378 |
| 23948 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-671-5p | 17 | FAT4 | Sponge network | -0.773 | 0.04073 | -0.354 | 0.14694 | 0.378 |
| 23949 | PCA3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | RET | Sponge network | -0.709 | 0.18168 | -0.833 | 0.03517 | 0.378 |
| 23950 | RP11-33B1.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | ARHGEF12 | Sponge network | 0.826 | 0 | 0.09 | 0.35693 | 0.378 |
| 23951 | MAGI2-AS3 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NRG3 | Sponge network | -0.129 | 0.57177 | -1.836 | 4.0E-5 | 0.378 |
| 23952 | RP11-379F4.7 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | NF1 | Sponge network | 1.316 | 0 | 0.51 | 0 | 0.378 |
| 23953 | FENDRR |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | SPOP | Sponge network | -1.281 | 0.00012 | -0.419 | 1.0E-5 | 0.378 |
| 23954 | ZNF667-AS1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-429;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | IRS1 | Sponge network | -0.976 | 0.00025 | -0.026 | 0.88855 | 0.378 |
| 23955 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | TCF4 | Sponge network | -0.18 | 0.20343 | 0.016 | 0.92271 | 0.378 |
| 23956 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | 0.572 | 0.01722 | -0.001 | 0.99669 | 0.378 |
| 23957 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-3p | 36 | BNC2 | Sponge network | 0.494 | 0.00339 | -0.491 | 0.09988 | 0.378 |
| 23958 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p | 14 | SON | Sponge network | 1.04 | 0.00023 | 0.168 | 0.04406 | 0.378 |
| 23959 | CTD-3092A11.2 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-9-5p | 27 | CCDC6 | Sponge network | 0.873 | 0 | 0.27 | 0.02555 | 0.378 |
| 23960 | PTOV1-AS1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-654-3p | 13 | PHF6 | Sponge network | 0.87 | 0 | 0.785 | 0 | 0.378 |
| 23961 | MIR143HG |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 25 | ATM | Sponge network | -0.739 | 0.0574 | 0.178 | 0.21568 | 0.378 |
| 23962 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p | 12 | PGR | Sponge network | -2.549 | 0.00528 | -1.704 | 0 | 0.378 |
| 23963 | AC009120.5 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-625-5p | 16 | LPP | Sponge network | 1.383 | 0 | -0.459 | 0.04475 | 0.378 |
| 23964 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-590-3p | 15 | RELN | Sponge network | 0.082 | 0.55654 | -2.403 | 0 | 0.378 |
| 23965 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p | 18 | PNRC1 | Sponge network | -0.901 | 0.00019 | -0.646 | 0 | 0.378 |
| 23966 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 46 | BMPR2 | Sponge network | 0.848 | 0 | 0.206 | 0.0635 | 0.378 |
| 23967 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p | 10 | USP25 | Sponge network | -0.487 | 0.02157 | -0.242 | 0.01397 | 0.378 |
| 23968 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 19 | GREM1 | Sponge network | -0.487 | 0.02157 | 0.902 | 0.01691 | 0.378 |
| 23969 | BCYRN1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | CREB1 | Sponge network | 0.311 | 0.10344 | 0.203 | 0.00537 | 0.378 |
| 23970 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-590-3p | 11 | RB1CC1 | Sponge network | 1.073 | 0.00216 | 0.175 | 0.12559 | 0.378 |
| 23971 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PRDM11 | Sponge network | -1.697 | 0.00889 | -0.252 | 0.15511 | 0.378 |
| 23972 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | TMTC2 | Sponge network | -0.096 | 0.67958 | -0.215 | 0.18248 | 0.378 |
| 23973 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 17 | DOCK4 | Sponge network | 0.299 | 0.57722 | 0.502 | 0.00108 | 0.378 |
| 23974 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SAMHD1 | Sponge network | -0.129 | 0.57177 | 0.072 | 0.62895 | 0.378 |
| 23975 | DIO3OS |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p | 13 | SPG20 | Sponge network | -0.773 | 0.04073 | -1.16 | 0 | 0.378 |
| 23976 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | MOB1B | Sponge network | 1.317 | 0 | 0.197 | 0.15266 | 0.378 |
| 23977 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | SYNE1 | Sponge network | -2.925 | 0 | -0.738 | 0.00316 | 0.378 |
| 23978 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-654-3p | 18 | TP53INP1 | Sponge network | -0.176 | 0.59692 | -0.496 | 0.00064 | 0.378 |
| 23979 | GHRLOS |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-942-5p | 14 | NRXN1 | Sponge network | -1.942 | 0 | -3.778 | 0 | 0.378 |
| 23980 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | PDE4DIP | Sponge network | 1.302 | 7.0E-5 | -0.282 | 0.0257 | 0.378 |
| 23981 | CTD-2554C21.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-532-3p;hsa-miR-769-5p;hsa-miR-93-5p | 38 | WDFY3 | Sponge network | -1.654 | 0.00144 | -0.201 | 0.0967 | 0.378 |
| 23982 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-93-5p | 16 | LATS2 | Sponge network | -2.69 | 0.0001 | 0.159 | 0.2304 | 0.378 |
| 23983 | C20orf166-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-92a-1-5p | 15 | LUZP2 | Sponge network | -2.69 | 0.0001 | -0.056 | 0.89416 | 0.378 |
| 23984 | RP11-66B24.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p | 16 | NTRK2 | Sponge network | 0.57 | 0.1866 | -0.736 | 0.06495 | 0.378 |
| 23985 | RP11-535M15.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p | 11 | EPHA7 | Sponge network | -1.14 | 0.03513 | -2.197 | 3.0E-5 | 0.378 |
| 23986 | RP11-2H8.2 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-629-3p | 23 | DDX6 | Sponge network | 1.089 | 0 | 0.091 | 0.36428 | 0.378 |
| 23987 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | MOB1B | Sponge network | 0.417 | 0.00445 | 0.197 | 0.15266 | 0.378 |
| 23988 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 18 | MAP3K12 | Sponge network | -0.739 | 0.0574 | -0.057 | 0.59369 | 0.378 |
| 23989 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-495-3p | 13 | TRIM33 | Sponge network | 0.706 | 0 | 0.402 | 7.0E-5 | 0.378 |
| 23990 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 28 | PBX1 | Sponge network | -0.738 | 0.21233 | -1.206 | 0 | 0.378 |
| 23991 | LINC00702 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | CDKN1C | Sponge network | -0.737 | 0.06182 | -0.929 | 0.00033 | 0.378 |
| 23992 | RP11-540B6.6 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p | 15 | SLC5A3 | Sponge network | 0.93 | 0 | 0.493 | 5.0E-5 | 0.378 |
| 23993 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-369-3p | 29 | PPP1R9A | Sponge network | -0.384 | 0.40652 | -0.903 | 0.04254 | 0.377 |
| 23994 | HCG11 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DACT1 | Sponge network | 0.385 | 0.10067 | 0.783 | 0.00072 | 0.377 |
| 23995 | RP4-639F20.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 30 | WDFY3 | Sponge network | -0.517 | 0.02935 | -0.201 | 0.0967 | 0.377 |
| 23996 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 50 | MDM4 | Sponge network | 0.194 | 0.64278 | 0.345 | 0.00762 | 0.377 |
| 23997 | RP11-531A24.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-655-3p;hsa-miR-935 | 32 | DMD | Sponge network | 0.75 | 0.00054 | -1.506 | 0 | 0.377 |
| 23998 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-590-5p;hsa-miR-96-5p | 27 | SCN9A | Sponge network | -0.969 | 2.0E-5 | -0.525 | 0.10593 | 0.377 |
| 23999 | TRAF3IP2-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | FAM117A | Sponge network | -0.323 | 0.01372 | -1.379 | 0 | 0.377 |
| 24000 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | -0.727 | 0.04726 | -1.751 | 1.0E-5 | 0.377 |
| 24001 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | PHC3 | Sponge network | 0.997 | 0.00066 | 0.212 | 0.0499 | 0.377 |
| 24002 | ZSCAN16-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-874-3p | 25 | ASTN1 | Sponge network | -0.342 | 0.02288 | -2.389 | 2.0E-5 | 0.377 |
| 24003 | RP11-531A24.5 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-495-3p | 13 | ERCC6 | Sponge network | 0.75 | 0.00054 | 0.23 | 0.015 | 0.377 |
| 24004 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 31 | MOB1B | Sponge network | 0.541 | 0.00125 | 0.197 | 0.15266 | 0.377 |
| 24005 | LINC01018 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TPO | Sponge network | -2.915 | 0.00015 | -1.87 | 2.0E-5 | 0.377 |
| 24006 | RP1-257A7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-629-3p | 11 | EBF1 | Sponge network | 0.849 | 0.00178 | -0.384 | 0.08856 | 0.377 |
| 24007 | AP001055.6 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-424-5p | 12 | ASTN1 | Sponge network | 0.693 | 0.00076 | -2.389 | 2.0E-5 | 0.377 |
| 24008 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-654-3p | 16 | NR2C2 | Sponge network | 1.03 | 0 | 0.21 | 0.03224 | 0.377 |
| 24009 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-455-5p | 10 | PRDM5 | Sponge network | -0.154 | 0.78939 | -1.09 | 0.00014 | 0.377 |
| 24010 | RP11-225B17.2 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-590-3p | 11 | BRD3 | Sponge network | 1.055 | 0 | 0.37 | 0.00013 | 0.377 |
| 24011 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | CCND2 | Sponge network | -0.83 | 0 | -0.267 | 0.3112 | 0.377 |
| 24012 | RP11-574K11.29 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p;hsa-miR-654-3p | 15 | PHF6 | Sponge network | 0.997 | 0 | 0.785 | 0 | 0.377 |
| 24013 | RP11-473I1.5 |
hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p | 11 | GNA13 | Sponge network | 0.542 | 0.00054 | 0.007 | 0.93884 | 0.377 |
| 24014 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p | 15 | RECK | Sponge network | 0.056 | 0.81195 | -1.043 | 0 | 0.377 |
| 24015 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | FGFR1 | Sponge network | -0.355 | 0.0077 | -0.509 | 0.03957 | 0.377 |
| 24016 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 27 | MAF | Sponge network | -0.421 | 0.0114 | -1.257 | 0 | 0.377 |
| 24017 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 24 | TBL1XR1 | Sponge network | 0.924 | 0 | 0.578 | 0 | 0.377 |
| 24018 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-493-5p | 15 | KAT6B | Sponge network | -2.62 | 0 | -0.478 | 0.00018 | 0.377 |
| 24019 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-766-3p | 37 | MAPK1 | Sponge network | 0.683 | 1.0E-5 | -0.017 | 0.81465 | 0.377 |
| 24020 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | ABCB5 | Sponge network | -1.203 | 0.03881 | -0.956 | 0.04077 | 0.377 |
| 24021 | RP11-35G9.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-744-3p;hsa-miR-940 | 16 | EPS15 | Sponge network | 0.622 | 0.00015 | -0.052 | 0.53998 | 0.377 |
| 24022 | RP11-983P16.4 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-542-3p | 10 | MACF1 | Sponge network | 0.41 | 0.02214 | 0.019 | 0.90335 | 0.377 |
| 24023 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p | 30 | PGR | Sponge network | -0.303 | 0.21002 | -1.704 | 0 | 0.377 |
| 24024 | RASSF8-AS1 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ADH1B | Sponge network | 0.335 | 0.20596 | -3.716 | 0 | 0.377 |
| 24025 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | MOB1B | Sponge network | 0.959 | 0 | 0.197 | 0.15266 | 0.377 |
| 24026 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | MOB1B | Sponge network | 0.998 | 0 | 0.197 | 0.15266 | 0.377 |
| 24027 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 53 | CCND2 | Sponge network | -0.576 | 0.10121 | -0.267 | 0.3112 | 0.377 |
| 24028 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | SLC6A15 | Sponge network | -0.739 | 0.0574 | -2.373 | 2.0E-5 | 0.377 |
| 24029 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | DACH2 | Sponge network | -1.111 | 0.0003 | -1.635 | 0.00034 | 0.377 |
| 24030 | LINC00969 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 18 | PHC3 | Sponge network | 0.683 | 1.0E-5 | 0.212 | 0.0499 | 0.377 |
| 24031 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-7-5p | 10 | CAV1 | Sponge network | -1.047 | 0 | -1.013 | 6.0E-5 | 0.377 |
| 24032 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SMARCA2 | Sponge network | 0.101 | 0.60919 | -0.529 | 0.00014 | 0.377 |
| 24033 | AC108488.4 |
hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-342-3p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 18 | PCDH10 | Sponge network | 0.259 | 0.12542 | -1.644 | 0.0078 | 0.377 |
| 24034 | RP11-597D13.9 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-942-5p;hsa-miR-96-5p | 10 | ZBTB20 | Sponge network | 1.302 | 7.0E-5 | -0.122 | 0.57569 | 0.377 |
| 24035 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-93-5p | 16 | CDHR3 | Sponge network | -1.439 | 4.0E-5 | -0.977 | 4.0E-5 | 0.377 |
| 24036 | U91328.19 |
hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 11 | STAT5B | Sponge network | -0.303 | 0.21002 | -0.182 | 0.1082 | 0.377 |
| 24037 | RP11-21A7A.2 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-628-5p | 31 | LPP | Sponge network | -1.116 | 0.23686 | -0.459 | 0.04475 | 0.377 |
| 24038 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-766-3p;hsa-miR-96-5p | 16 | IL17RD | Sponge network | -0.187 | 0.23787 | -0.019 | 0.94873 | 0.377 |
| 24039 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429 | 18 | EBF3 | Sponge network | 0.16 | 0.50785 | -0.058 | 0.81437 | 0.377 |
| 24040 | RP11-216B9.6 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | PCDH9 | Sponge network | 0.174 | 0.30034 | -1.823 | 4.0E-5 | 0.377 |
| 24041 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | LRIG1 | Sponge network | -0.563 | 0.02545 | -0.537 | 0.00588 | 0.377 |
| 24042 | AC003090.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ABCA10 | Sponge network | -2.117 | 0.00161 | -0.571 | 0.03674 | 0.377 |
| 24043 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PCDH10 | Sponge network | -1.057 | 0.00029 | -1.644 | 0.0078 | 0.377 |
| 24044 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 24 | MYO9A | Sponge network | -0.513 | 0.04703 | -0.147 | 0.32373 | 0.377 |
| 24045 | PGM5-AS1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | PPP1R9A | Sponge network | -4.546 | 0 | -0.903 | 0.04254 | 0.377 |
| 24046 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-195-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 10 | ZNF770 | Sponge network | 0.832 | 0 | 0.206 | 0.06751 | 0.377 |
| 24047 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 10 | RB1CC1 | Sponge network | 1.04 | 0.00023 | 0.175 | 0.12559 | 0.377 |
| 24048 | RP11-720L2.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-625-5p;hsa-miR-93-5p | 29 | LPP | Sponge network | -1.255 | 0.00981 | -0.459 | 0.04475 | 0.377 |
| 24049 | RP11-325K4.3 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | MDM4 | Sponge network | 0.178 | 0.31659 | 0.345 | 0.00762 | 0.377 |
| 24050 | RP11-693J15.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-942-5p | 21 | IGFBP5 | Sponge network | -0.72 | 0.24239 | -0.806 | 0.00105 | 0.377 |
| 24051 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-942-5p | 31 | UNC80 | Sponge network | -0.342 | 0.02288 | -1.934 | 0 | 0.377 |
| 24052 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577 | 16 | NRK | Sponge network | 0.259 | 0.12542 | 0.656 | 0.19217 | 0.377 |
| 24053 | TP73-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-93-3p | 31 | ERC1 | Sponge network | -0.563 | 0.02545 | -0.108 | 0.38794 | 0.377 |
| 24054 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577 | 22 | GREM1 | Sponge network | -2.925 | 0 | 0.902 | 0.01691 | 0.377 |
| 24055 | RP11-400K9.4 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p | 12 | PLXNC1 | Sponge network | -0.611 | 0.09607 | 0.569 | 0.00329 | 0.377 |
| 24056 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | FAT4 | Sponge network | 0.064 | 0.72934 | -0.354 | 0.14694 | 0.377 |
| 24057 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 11 | CNTNAP1 | Sponge network | -0.342 | 0.02288 | 0.358 | 0.13727 | 0.377 |
| 24058 | RP11-57H14.4 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-501-3p;hsa-miR-96-5p | 10 | ZBTB20 | Sponge network | 0.083 | 0.66405 | -0.122 | 0.57569 | 0.377 |
| 24059 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 30 | RBMS3 | Sponge network | 0.389 | 0.04293 | -0.519 | 0.03551 | 0.377 |
| 24060 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p | 12 | AKAP6 | Sponge network | -0.285 | 0.2172 | -1.586 | 3.0E-5 | 0.377 |
| 24061 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | PDGFRA | Sponge network | -0.842 | 0.01361 | -0.372 | 0.0037 | 0.377 |
| 24062 | LINC01018 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 12 | RHOB | Sponge network | -2.915 | 0.00015 | -1.052 | 0 | 0.377 |
| 24063 | RP11-999E24.3 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-93-3p | 12 | CTIF | Sponge network | -0.693 | 0.05629 | -0.389 | 0.00549 | 0.377 |
| 24064 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | ST6GAL2 | Sponge network | -0.246 | 0.35034 | -1.201 | 0.00597 | 0.377 |
| 24065 | MIR497HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | NCOA1 | Sponge network | -1.439 | 4.0E-5 | -0.432 | 0 | 0.377 |
| 24066 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p | 14 | ABL2 | Sponge network | 0.453 | 0.01839 | 0.82 | 0 | 0.377 |
| 24067 | LINC00654 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p | 36 | IGFBP5 | Sponge network | 0.374 | 0.20054 | -0.806 | 0.00105 | 0.377 |
| 24068 | RP4-613B23.1 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-539-5p | 29 | THRB | Sponge network | -0.309 | 0.1145 | -1.117 | 0.0004 | 0.377 |
| 24069 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | SCN9A | Sponge network | -0.811 | 2.0E-5 | -0.525 | 0.10593 | 0.377 |
| 24070 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 20 | RASSF8 | Sponge network | -1.057 | 0.00029 | -0.122 | 0.65591 | 0.377 |
| 24071 | RP11-464F9.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 14 | SLC5A3 | Sponge network | 0.959 | 0 | 0.493 | 5.0E-5 | 0.377 |
| 24072 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p | 11 | LRRC55 | Sponge network | 0.622 | 0.00015 | -0.026 | 0.93163 | 0.377 |
| 24073 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 33 | AR | Sponge network | -0.663 | 0.10887 | -1.554 | 0 | 0.377 |
| 24074 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 13 | PTGFR | Sponge network | -0.08 | 0.90092 | -0.233 | 0.45421 | 0.377 |
| 24075 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p | 12 | FSTL5 | Sponge network | 0.056 | 0.81195 | -1.3 | 0.01619 | 0.377 |
| 24076 | RP11-166D19.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 29 | PDE4DIP | Sponge network | -0.576 | 0.10121 | -0.282 | 0.0257 | 0.377 |
| 24077 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 22 | NUAK1 | Sponge network | -0.513 | 0.04703 | 0.904 | 0 | 0.377 |
| 24078 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | 0.219 | 0.1516 | -0.509 | 0.03957 | 0.377 |
| 24079 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-93-5p;hsa-miR-942-5p | 12 | FBXO31 | Sponge network | -0.612 | 0.00094 | -0.085 | 0.42389 | 0.377 |
| 24080 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | AHNAK | Sponge network | -0.612 | 0.00094 | -1.207 | 0 | 0.377 |
| 24081 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | EGFR | Sponge network | -0.421 | 0.0114 | -0.175 | 0.48451 | 0.377 |
| 24082 | RP11-248G5.8 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-625-5p;hsa-miR-766-3p | 29 | MDM4 | Sponge network | 1.294 | 0 | 0.345 | 0.00762 | 0.377 |
| 24083 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 29 | CNTN1 | Sponge network | 0.225 | 0.30644 | -2.594 | 0 | 0.377 |
| 24084 | LINC00672 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | TAL1 | Sponge network | -0.227 | 0.31657 | -0.462 | 0.00574 | 0.377 |
| 24085 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-330-3p | 12 | JDP2 | Sponge network | -1.331 | 3.0E-5 | -0.967 | 0 | 0.377 |
| 24086 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-590-3p | 11 | LATS1 | Sponge network | 0.67 | 0.00048 | 0.109 | 0.34208 | 0.377 |
| 24087 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 12 | ZNF208 | Sponge network | -0.096 | 0.67958 | -0.4 | 0.22381 | 0.377 |
| 24088 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 19 | TMEFF2 | Sponge network | -0.405 | 0.22013 | -2.426 | 0 | 0.377 |
| 24089 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-455-3p | 16 | PPM1L | Sponge network | -0.309 | 0.1145 | -0.537 | 0.00616 | 0.377 |
| 24090 | RP11-728F11.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-7-1-3p | 17 | SOX5 | Sponge network | 0.238 | 0.70544 | -1.091 | 4.0E-5 | 0.377 |
| 24091 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 12 | UVRAG | Sponge network | 0.246 | 0.59912 | -0.017 | 0.83865 | 0.377 |
| 24092 | RP11-531A24.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-429 | 14 | STARD13 | Sponge network | 0.75 | 0.00054 | -0.187 | 0.27383 | 0.377 |
| 24093 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SON | Sponge network | 0.986 | 0.01022 | 0.168 | 0.04406 | 0.377 |
| 24094 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p | 11 | EDA2R | Sponge network | -0.737 | 0.10822 | -0.587 | 0.01598 | 0.377 |
| 24095 | RAMP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 16 | EZH1 | Sponge network | -0.513 | 0.04703 | -0.564 | 1.0E-5 | 0.377 |
| 24096 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | TCF4 | Sponge network | 0.056 | 0.8494 | 0.016 | 0.92271 | 0.377 |
| 24097 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429 | 12 | CITED2 | Sponge network | -1.654 | 0.00144 | -1.545 | 0 | 0.377 |
| 24098 | LINC00963 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p | 29 | ABL2 | Sponge network | 0.523 | 9.0E-5 | 0.82 | 0 | 0.377 |
| 24099 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-629-3p | 13 | ITGA5 | Sponge network | 0.331 | 0.57247 | 0.452 | 0.03524 | 0.377 |
| 24100 | RP1-59D14.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-940 | 11 | SIRT1 | Sponge network | 1.162 | 5.0E-5 | 0.109 | 0.2593 | 0.377 |
| 24101 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 35 | TSHZ3 | Sponge network | 0.395 | 0.15451 | -0.556 | 0.01516 | 0.377 |
| 24102 | LINC00472 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p | 11 | UVRAG | Sponge network | -0.842 | 0.01361 | -0.017 | 0.83865 | 0.377 |
| 24103 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-660-5p | 18 | RAP1A | Sponge network | -1.582 | 8.0E-5 | -0.929 | 0 | 0.377 |
| 24104 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-502-3p | 16 | CHD5 | Sponge network | 0.37 | 0.27614 | -0.922 | 0.01521 | 0.377 |
| 24105 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p | 20 | TET1 | Sponge network | -0.227 | 0.31657 | -0.135 | 0.52138 | 0.377 |
| 24106 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | CACNA2D1 | Sponge network | -2.925 | 0 | -0.846 | 0.00675 | 0.377 |
| 24107 | TPTEP1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | LSAMP | Sponge network | -1.216 | 0 | -0.066 | 0.81708 | 0.377 |
| 24108 | RP11-390K5.6 |
hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | DICER1 | Sponge network | 1.148 | 2.0E-5 | 0.042 | 0.674 | 0.377 |
| 24109 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p | 14 | TET1 | Sponge network | -0.583 | 0.14589 | -0.135 | 0.52138 | 0.377 |
| 24110 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | HDAC9 | Sponge network | -0.739 | 0.0574 | -0.755 | 0.00495 | 0.377 |
| 24111 | HCG11 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | FGFR1 | Sponge network | 0.385 | 0.10067 | -0.509 | 0.03957 | 0.377 |
| 24112 | LPP-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577 | 45 | DTNA | Sponge network | -0.18 | 0.20343 | -1.648 | 1.0E-5 | 0.377 |
| 24113 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | PNRC1 | Sponge network | -0.021 | 0.93598 | -0.646 | 0 | 0.377 |
| 24114 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 43 | ZFHX4 | Sponge network | 0.219 | 0.1516 | 0.018 | 0.95574 | 0.377 |
| 24115 | RP11-148O21.2 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-361-5p;hsa-miR-500a-5p;hsa-miR-589-3p | 15 | NTRK3 | Sponge network | 0.233 | 0.81029 | -1.77 | 3.0E-5 | 0.377 |
| 24116 | RP11-401P9.4 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-874-3p | 11 | SORCS1 | Sponge network | -0.384 | 0.40652 | -3.492 | 0 | 0.377 |
| 24117 | RP11-53O19.3 |
hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | MAP3K4 | Sponge network | 1.205 | 0 | 0.058 | 0.54229 | 0.377 |
| 24118 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-625-5p | 19 | ZMYM2 | Sponge network | 0.614 | 1.0E-5 | 0.279 | 0.01273 | 0.377 |
| 24119 | RP11-554A11.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-425-5p | 23 | PTPRT | Sponge network | -0.583 | 0.14589 | -0.907 | 0.03727 | 0.377 |
| 24120 | RP11-113K21.4 |
hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-582-5p | 12 | RICTOR | Sponge network | 0.676 | 0 | 0.388 | 0.00188 | 0.377 |
| 24121 | RP11-403I13.8 |
hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-96-5p | 10 | DAB2 | Sponge network | 0.619 | 0.00157 | 0.431 | 0.0113 | 0.377 |
| 24122 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 16 | GLI3 | Sponge network | 0.064 | 0.72934 | -0.615 | 0.01482 | 0.377 |
| 24123 | RP11-597D13.9 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 17 | FMNL3 | Sponge network | 1.302 | 7.0E-5 | 0.555 | 4.0E-5 | 0.377 |
| 24124 | RP11-345P4.7 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | UBR3 | Sponge network | 1.597 | 0 | -0.021 | 0.82774 | 0.377 |
| 24125 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 27 | THBS1 | Sponge network | 0.374 | 0.20054 | 0.741 | 0.00386 | 0.377 |
| 24126 | RP11-774O3.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | PDE4DIP | Sponge network | -0.487 | 0.02157 | -0.282 | 0.0257 | 0.377 |
| 24127 | RP11-554A11.4 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-3065-3p;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p | 25 | AFF1 | Sponge network | -0.583 | 0.14589 | -0.384 | 0.00053 | 0.377 |
| 24128 | LINC00472 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 13 | SPOP | Sponge network | -0.842 | 0.01361 | -0.419 | 1.0E-5 | 0.377 |
| 24129 | RP11-867G23.10 |
hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-378c;hsa-miR-484;hsa-miR-589-3p | 14 | MED12L | Sponge network | -2.531 | 0 | -0.194 | 0.55299 | 0.377 |
| 24130 | RP11-701H24.7 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p | 12 | BCL2 | Sponge network | -2.039 | 0.03447 | -0.899 | 0 | 0.377 |
| 24131 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PRKAA2 | Sponge network | -0.206 | 0.12942 | -2.462 | 0 | 0.377 |
| 24132 | PART1 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | RTN4 | Sponge network | -2.925 | 0 | -0.412 | 0.00014 | 0.377 |
| 24133 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | EBF1 | Sponge network | -0.351 | 0.11358 | -0.384 | 0.08856 | 0.377 |
| 24134 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | CCND2 | Sponge network | 0.101 | 0.60919 | -0.267 | 0.3112 | 0.377 |
| 24135 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-577 | 18 | GREM1 | Sponge network | -0.793 | 7.0E-5 | 0.902 | 0.01691 | 0.377 |
| 24136 | NUTM2A-AS1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | PHF3 | Sponge network | 0.643 | 0 | 0.126 | 0.19652 | 0.377 |
| 24137 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | 0.101 | 0.60919 | 0.323 | 0.00792 | 0.377 |
| 24138 | LINC00472 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 28 | SYT4 | Sponge network | -0.842 | 0.01361 | -3.207 | 0 | 0.377 |
| 24139 | WDFY3-AS2 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 13 | PHOX2B | Sponge network | -0.942 | 0 | -2.68 | 0 | 0.377 |
| 24140 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | CRTC3 | Sponge network | 0.619 | 0.00157 | 0.164 | 0.16786 | 0.377 |
| 24141 | RP11-13K12.5 |
hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 11 | KLF10 | Sponge network | -0.318 | 0.65847 | -0.783 | 0 | 0.377 |
| 24142 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 20 | KIT | Sponge network | 0.374 | 0.20054 | -2.184 | 0 | 0.377 |
| 24143 | PGM5-AS1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-660-5p | 13 | IRS1 | Sponge network | -4.546 | 0 | -0.026 | 0.88855 | 0.377 |
| 24144 | RP5-855D21.3 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 14 | MDM4 | Sponge network | 1.728 | 0 | 0.345 | 0.00762 | 0.377 |
| 24145 | RP11-526I2.5 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-495-3p | 15 | CREB1 | Sponge network | 1.054 | 1.0E-5 | 0.203 | 0.00537 | 0.377 |
| 24146 | PCA3 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 16 | ZFP36L1 | Sponge network | -0.709 | 0.18168 | -0.505 | 8.0E-5 | 0.377 |
| 24147 | LINC00174 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-539-5p;hsa-miR-582-5p | 10 | PHF3 | Sponge network | 1.253 | 0 | 0.126 | 0.19652 | 0.377 |
| 24148 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 24 | ETV1 | Sponge network | -0.421 | 0.0114 | -0.096 | 0.67674 | 0.377 |
| 24149 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-98-5p | 30 | DMD | Sponge network | -0.727 | 0.04726 | -1.506 | 0 | 0.377 |
| 24150 | RP11-855A2.5 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-629-3p | 11 | ASTN1 | Sponge network | -1.717 | 0.01522 | -2.389 | 2.0E-5 | 0.377 |
| 24151 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p | 22 | EYA4 | Sponge network | -0.738 | 0.21233 | 0.3 | 0.36876 | 0.377 |
| 24152 | RP11-458D21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-766-3p | 19 | IL17RD | Sponge network | 0.663 | 0.00073 | -0.019 | 0.94873 | 0.377 |
| 24153 | RP11-159D12.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | RPS6KA3 | Sponge network | 1.254 | 0 | 0.256 | 0.02419 | 0.377 |
| 24154 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 14 | CNTNAP1 | Sponge network | -0.355 | 0.0077 | 0.358 | 0.13727 | 0.377 |
| 24155 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | ZMYM2 | Sponge network | 0.8 | 0 | 0.279 | 0.01273 | 0.377 |
| 24156 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 21 | EYA4 | Sponge network | -0.513 | 0.04703 | 0.3 | 0.36876 | 0.377 |
| 24157 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-214-5p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 62 | SMAD4 | Sponge network | 0.421 | 0.00084 | -0.313 | 0.00413 | 0.377 |
| 24158 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | LRP1B | Sponge network | -0.513 | 0.04703 | -2.163 | 0 | 0.377 |
| 24159 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-374b-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | ATRX | Sponge network | 0.199 | 0.52763 | 0.261 | 0.04408 | 0.377 |
| 24160 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | SMARCA2 | Sponge network | -0.811 | 2.0E-5 | -0.529 | 0.00014 | 0.377 |
| 24161 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | ZMYM2 | Sponge network | 0.873 | 0 | 0.279 | 0.01273 | 0.377 |
| 24162 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-98-5p | 38 | TGFBR3 | Sponge network | -0.154 | 0.78939 | -1.173 | 1.0E-5 | 0.377 |
| 24163 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-769-5p | 17 | RCAN2 | Sponge network | -1.249 | 7.0E-5 | -0.33 | 0.15172 | 0.377 |
| 24164 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p | 14 | SPTBN4 | Sponge network | -2.69 | 0.0001 | -0.786 | 0.01281 | 0.376 |
| 24165 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NR4A3 | Sponge network | -0.513 | 0.04703 | -1.385 | 0 | 0.376 |
| 24166 | RP11-649A18.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-664a-5p | 11 | CBFB | Sponge network | 2.218 | 0 | 1.061 | 0 | 0.376 |
| 24167 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-301a-3p | 12 | RASSF2 | Sponge network | 0.043 | 0.81628 | -0.273 | 0.21961 | 0.376 |
| 24168 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-93-3p | 16 | ERC1 | Sponge network | -2.531 | 0 | -0.108 | 0.38794 | 0.376 |
| 24169 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | BNC2 | Sponge network | -0.053 | 0.80685 | -0.491 | 0.09988 | 0.376 |
| 24170 | RP11-597D13.9 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 19 | LATS2 | Sponge network | 1.302 | 7.0E-5 | 0.159 | 0.2304 | 0.376 |
| 24171 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 25 | MITF | Sponge network | -0.83 | 0 | -0.769 | 0.00022 | 0.376 |
| 24172 | RP4-717I23.3 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-370-3p | 11 | ZMYND11 | Sponge network | 0.637 | 0.00069 | -0.2 | 0.05449 | 0.376 |
| 24173 | RP11-148O21.2 |
hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-589-3p | 13 | IL6ST | Sponge network | 0.233 | 0.81029 | -0.402 | 0.00676 | 0.376 |
| 24174 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ABCA10 | Sponge network | -0.899 | 6.0E-5 | -0.571 | 0.03674 | 0.376 |
| 24175 | RP11-3K16.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p | 11 | MDM4 | Sponge network | 0.749 | 0.17788 | 0.345 | 0.00762 | 0.376 |
| 24176 | RP11-819C21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 15 | STARD13 | Sponge network | 0.537 | 0.00139 | -0.187 | 0.27383 | 0.376 |
| 24177 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-590-5p | 24 | PHC3 | Sponge network | 0.331 | 0.57247 | 0.212 | 0.0499 | 0.376 |
| 24178 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | TIMP3 | Sponge network | 0.082 | 0.55654 | -0.546 | 0.02307 | 0.376 |
| 24179 | RP11-894P9.1 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-582-3p | 16 | FOXO3 | Sponge network | 0.993 | 0 | -0.04 | 0.72028 | 0.376 |
| 24180 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 17 | PTPRD | Sponge network | -1.255 | 0.00981 | -1.005 | 0.00064 | 0.376 |
| 24181 | SNHG14 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p | 22 | MAFB | Sponge network | -1.111 | 0.0003 | -0.023 | 0.89865 | 0.376 |
| 24182 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | TACC1 | Sponge network | -0.031 | 0.89063 | -0.362 | 0.05406 | 0.376 |
| 24183 | RP11-967K21.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-940 | 18 | SFRP1 | Sponge network | 0.056 | 0.81195 | -3.566 | 0 | 0.376 |
| 24184 | MAGI2-AS3 |
hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30c-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 12 | CDH13 | Sponge network | -0.129 | 0.57177 | 0.634 | 0.00094 | 0.376 |
| 24185 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 16 | SPTBN4 | Sponge network | -2.46 | 0 | -0.786 | 0.01281 | 0.376 |
| 24186 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 16 | CXXC4 | Sponge network | 1.757 | 0 | -0.433 | 0.21055 | 0.376 |
| 24187 | RP11-400K9.4 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 11 | EHD3 | Sponge network | -0.611 | 0.09607 | -0.484 | 0.01747 | 0.376 |
| 24188 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-550a-3p | 18 | HECA | Sponge network | 0.082 | 0.55397 | -0.217 | 0.03463 | 0.376 |
| 24189 | DNAJC27-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 17 | MAPK10 | Sponge network | -0.969 | 2.0E-5 | -1.774 | 0 | 0.376 |
| 24190 | TRIM52-AS1 |
hsa-miR-1266-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-96-5p | 14 | ASTN1 | Sponge network | -0.418 | 0.00068 | -2.389 | 2.0E-5 | 0.376 |
| 24191 | CALML3-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p | 24 | BRD3 | Sponge network | 0.95 | 0.15943 | 0.37 | 0.00013 | 0.376 |
| 24192 | GS1-124K5.11 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-940 | 22 | SIRT1 | Sponge network | 0.494 | 0.00339 | 0.109 | 0.2593 | 0.376 |
| 24193 | FENDRR |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | NCAM2 | Sponge network | -1.281 | 0.00012 | -0.482 | 0.22565 | 0.376 |
| 24194 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 10 | RECK | Sponge network | 0.178 | 0.29106 | -1.043 | 0 | 0.376 |
| 24195 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | MAF | Sponge network | -1.281 | 0.00012 | -1.257 | 0 | 0.376 |
| 24196 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 19 | TGFBR2 | Sponge network | 0.374 | 0.20054 | -0.243 | 0.11408 | 0.376 |
| 24197 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 37 | RBMS3 | Sponge network | 0.222 | 0.29133 | -0.519 | 0.03551 | 0.376 |
| 24198 | RP11-254F7.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p | 15 | BRD3 | Sponge network | 0.176 | 0.37402 | 0.37 | 0.00013 | 0.376 |
| 24199 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 12 | SPTBN4 | Sponge network | -2.117 | 0.00161 | -0.786 | 0.01281 | 0.376 |
| 24200 | RP11-67L2.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | 0.72 | 0.0001 | 0.358 | 0.13727 | 0.376 |
| 24201 | RP11-35G9.3 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | DOCK4 | Sponge network | 0.657 | 4.0E-5 | 0.502 | 0.00108 | 0.376 |
| 24202 | RP11-760H22.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-655-3p | 19 | DMD | Sponge network | 0.001 | 0.99753 | -1.506 | 0 | 0.376 |
| 24203 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-577 | 10 | LRRC7 | Sponge network | -0.473 | 0.07602 | -1.341 | 0.01193 | 0.376 |
| 24204 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-576-5p | 10 | ATRX | Sponge network | -2.039 | 0.03447 | 0.261 | 0.04408 | 0.376 |
| 24205 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-96-5p | 11 | DAB2 | Sponge network | -0.517 | 0.02935 | 0.431 | 0.0113 | 0.376 |
| 24206 | U91328.19 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-503-5p | 18 | ABL1 | Sponge network | -0.303 | 0.21002 | -0.215 | 0.07804 | 0.376 |
| 24207 | RP11-48B3.4 |
hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p | 10 | LRRC55 | Sponge network | 1.757 | 0 | -0.026 | 0.93163 | 0.376 |
| 24208 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 25 | RSPO2 | Sponge network | -1.161 | 0.00591 | -3.038 | 0 | 0.376 |
| 24209 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p | 12 | FAT4 | Sponge network | -0.517 | 0.02935 | -0.354 | 0.14694 | 0.376 |
| 24210 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-942-5p;hsa-miR-98-5p | 24 | ARHGAP28 | Sponge network | 0.249 | 0.35902 | -0.911 | 0.00013 | 0.376 |
| 24211 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p | 12 | PLSCR4 | Sponge network | 3.345 | 0 | -0.474 | 0.03833 | 0.376 |
| 24212 | LINC01018 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | RYR2 | Sponge network | -2.915 | 0.00015 | -1.466 | 0.00031 | 0.376 |
| 24213 | RP11-403I13.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-93-3p | 42 | RUNX1T1 | Sponge network | 0.395 | 0.15451 | -0.344 | 0.2374 | 0.376 |
| 24214 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 59 | WDFY3 | Sponge network | -2.452 | 0 | -0.201 | 0.0967 | 0.376 |
| 24215 | AC009948.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | SIRT1 | Sponge network | 0.532 | 0.00505 | 0.109 | 0.2593 | 0.376 |
| 24216 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-877-5p | 12 | ANK2 | Sponge network | 1.122 | 0.02989 | -1.603 | 1.0E-5 | 0.376 |
| 24217 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 26 | SOX5 | Sponge network | 1.757 | 0 | -1.091 | 4.0E-5 | 0.376 |
| 24218 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-628-5p | 24 | CDC73 | Sponge network | 0.421 | 0.00084 | 0.094 | 0.20463 | 0.376 |
| 24219 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-5p | 28 | BTG2 | Sponge network | -0.612 | 0.00094 | -1.763 | 0 | 0.376 |
| 24220 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | DIP2C | Sponge network | -0.342 | 0.02288 | -0.436 | 0.01713 | 0.376 |
| 24221 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | PREX2 | Sponge network | 0.083 | 0.66405 | -0.272 | 0.25382 | 0.376 |
| 24222 | PWAR6 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | LUZP2 | Sponge network | -1.575 | 6.0E-5 | -0.056 | 0.89416 | 0.376 |
| 24223 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-3p | 18 | UNC5C | Sponge network | -0.49 | 0.04946 | -0.178 | 0.40214 | 0.376 |
| 24224 | RP11-473I1.10 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZFHX3 | Sponge network | 0.673 | 0 | 0.389 | 0.01363 | 0.376 |
| 24225 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | PBRM1 | Sponge network | 0.225 | 0.30644 | -0.029 | 0.78601 | 0.376 |
| 24226 | AC010761.8 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-582-5p | 11 | RICTOR | Sponge network | 1.638 | 0 | 0.388 | 0.00188 | 0.376 |
| 24227 | RP11-296I10.3 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-3614-5p | 16 | ABL2 | Sponge network | 0.592 | 0.00014 | 0.82 | 0 | 0.376 |
| 24228 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b | 10 | CLIP1 | Sponge network | 0.37 | 0.27614 | -0.215 | 0.08684 | 0.376 |
| 24229 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 51 | IL6ST | Sponge network | -1.349 | 0.00017 | -0.402 | 0.00676 | 0.376 |
| 24230 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-940 | 33 | ETV1 | Sponge network | -0.162 | 0.47687 | -0.096 | 0.67674 | 0.376 |
| 24231 | RP11-434B12.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | CREB1 | Sponge network | -0.031 | 0.89063 | 0.203 | 0.00537 | 0.376 |
| 24232 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | DACT1 | Sponge network | 0.532 | 0.00505 | 0.783 | 0.00072 | 0.376 |
| 24233 | RP11-326G21.1 |
hsa-let-7b-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TGFBR3 | Sponge network | -0.021 | 0.93598 | -1.173 | 1.0E-5 | 0.376 |
| 24234 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | ST6GAL2 | Sponge network | -0.405 | 0.22013 | -1.201 | 0.00597 | 0.376 |
| 24235 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-708-5p;hsa-miR-766-3p | 16 | EZH1 | Sponge network | 0.41 | 0.02214 | -0.564 | 1.0E-5 | 0.376 |
| 24236 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-550a-5p;hsa-miR-940;hsa-miR-96-5p | 11 | HOOK3 | Sponge network | 0.622 | 0.00015 | 0.273 | 0.09957 | 0.376 |
| 24237 | AC006547.13 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-654-3p;hsa-miR-940 | 10 | BAZ2B | Sponge network | 1.159 | 0 | -0.276 | 0.02833 | 0.376 |
| 24238 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 21 | CSMD1 | Sponge network | 0.225 | 0.30644 | -0.63 | 0.18881 | 0.376 |
| 24239 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-629-5p | 30 | MAF | Sponge network | -1.249 | 7.0E-5 | -1.257 | 0 | 0.376 |
| 24240 | EMX2OS |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | PREX2 | Sponge network | -0.777 | 0.1134 | -0.272 | 0.25382 | 0.376 |
| 24241 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-590-3p | 24 | MYBL1 | Sponge network | 0.082 | 0.55654 | 0.494 | 0.02152 | 0.376 |
| 24242 | LINC00969 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 13 | GSK3B | Sponge network | 0.683 | 1.0E-5 | 0.266 | 0.00045 | 0.376 |
| 24243 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-375;hsa-miR-590-3p | 14 | MYBL1 | Sponge network | 0.174 | 0.30034 | 0.494 | 0.02152 | 0.376 |
| 24244 | LINC00843 |
hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-424-5p;hsa-miR-429 | 10 | DOCK4 | Sponge network | 1.106 | 0 | 0.502 | 0.00108 | 0.376 |
| 24245 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 30 | NTRK2 | Sponge network | -0.739 | 0.00044 | -0.736 | 0.06495 | 0.376 |
| 24246 | LINC00641 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-584-5p;hsa-miR-590-3p | 19 | GLIPR1 | Sponge network | -0.222 | 0.32445 | 0.146 | 0.3276 | 0.376 |
| 24247 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | ANK2 | Sponge network | 0.259 | 0.12542 | -1.603 | 1.0E-5 | 0.376 |
| 24248 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | ST6GALNAC3 | Sponge network | 0.246 | 0.59912 | -0.987 | 0 | 0.376 |
| 24249 | MIR143HG |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p | 24 | HIP1 | Sponge network | -0.739 | 0.0574 | 0.774 | 0 | 0.376 |
| 24250 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 12 | PCDH10 | Sponge network | 0.178 | 0.29106 | -1.644 | 0.0078 | 0.376 |
| 24251 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 30 | ARID5B | Sponge network | 0.249 | 0.35902 | 0.342 | 0.01676 | 0.376 |
| 24252 | CTA-204B4.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-590-3p | 14 | AKAP6 | Sponge network | 0.765 | 7.0E-5 | -1.586 | 3.0E-5 | 0.376 |
| 24253 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-576-5p | 14 | CACNA2D1 | Sponge network | 0.176 | 0.37402 | -0.846 | 0.00675 | 0.376 |
| 24254 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 13 | LRRK2 | Sponge network | -0.793 | 7.0E-5 | -1.136 | 1.0E-5 | 0.376 |
| 24255 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 45 | ZFHX4 | Sponge network | -0.727 | 0.04726 | 0.018 | 0.95574 | 0.376 |
| 24256 | AC083843.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p | 20 | CREB1 | Sponge network | 1.4 | 0 | 0.203 | 0.00537 | 0.376 |
| 24257 | LINC00957 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | -0.421 | 0.0114 | -0.684 | 0.00159 | 0.376 |
| 24258 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-30b-3p | 12 | KCNA6 | Sponge network | -0.691 | 0.00146 | -1.531 | 0.00013 | 0.376 |
| 24259 | RP11-212P7.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-96-5p | 11 | ZBTB20 | Sponge network | 0.219 | 0.1516 | -0.122 | 0.57569 | 0.376 |
| 24260 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p | 36 | SSBP2 | Sponge network | 0.176 | 0.37402 | -1.58 | 0 | 0.376 |
| 24261 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 45 | SOX5 | Sponge network | 0.064 | 0.72934 | -1.091 | 4.0E-5 | 0.376 |
| 24262 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 21 | CADM3 | Sponge network | -0.154 | 0.78939 | -3.663 | 0 | 0.376 |
| 24263 | PGM5-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-660-5p;hsa-miR-7-1-3p | 17 | LIMCH1 | Sponge network | -4.546 | 0 | -0.915 | 1.0E-5 | 0.376 |
| 24264 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-3p | 11 | SON | Sponge network | 1.361 | 0 | 0.168 | 0.04406 | 0.376 |
| 24265 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 18 | GPAM | Sponge network | 0.569 | 0.00067 | 0.142 | 0.29732 | 0.376 |
| 24266 | RP11-890B15.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | PDE4DIP | Sponge network | 0.101 | 0.60919 | -0.282 | 0.0257 | 0.376 |
| 24267 | GNG12-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-942-5p | 25 | OPCML | Sponge network | -0.793 | 7.0E-5 | -2.498 | 0 | 0.376 |
| 24268 | CTA-14H9.5 |
hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-582-5p | 13 | FAT3 | Sponge network | 0.145 | 0.53343 | -1.601 | 0.00051 | 0.376 |
| 24269 | MRPL23-AS1 |
hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-502-5p;hsa-miR-766-3p | 15 | IL17RD | Sponge network | -0.278 | 0.76559 | -0.019 | 0.94873 | 0.376 |
| 24270 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p | 25 | MYO9A | Sponge network | 0.253 | 0.09565 | -0.147 | 0.32373 | 0.376 |
| 24271 | PART1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 21 | GLI3 | Sponge network | -2.925 | 0 | -0.615 | 0.01482 | 0.376 |
| 24272 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | EPHA7 | Sponge network | -0.726 | 0.00132 | -2.197 | 3.0E-5 | 0.376 |
| 24273 | LINC00403 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-452-5p | 16 | TMEM132B | Sponge network | -3.586 | 0.00032 | -0.83 | 0.01084 | 0.376 |
| 24274 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 14 | RIMS1 | Sponge network | 0.194 | 0.64278 | -3.143 | 0 | 0.376 |
| 24275 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 20 | KANK1 | Sponge network | -0.222 | 0.32445 | -0.55 | 0.00134 | 0.376 |
| 24276 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-590-3p | 18 | ZMYND11 | Sponge network | 0.494 | 0.00339 | -0.2 | 0.05449 | 0.376 |
| 24277 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | CREM | Sponge network | -0.129 | 0.57177 | -0.345 | 0.00258 | 0.376 |
| 24278 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-93-5p | 25 | RNASEL | Sponge network | -0.793 | 7.0E-5 | -0.272 | 0.01796 | 0.376 |
| 24279 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-942-5p | 26 | HLF | Sponge network | 0.214 | 0.18246 | -2.443 | 0 | 0.376 |
| 24280 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 56 | TCF4 | Sponge network | 0.082 | 0.55654 | 0.016 | 0.92271 | 0.376 |
| 24281 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 17 | RPS6KA2 | Sponge network | -1.216 | 0 | -0.57 | 0.00013 | 0.376 |
| 24282 | DIO3OS |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-877-5p | 15 | CDH19 | Sponge network | -0.773 | 0.04073 | -3.434 | 0 | 0.376 |
| 24283 | RP11-251G23.5 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 14 | ZFX | Sponge network | 1.344 | 0 | 0.09 | 0.42563 | 0.376 |
| 24284 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-629-3p;hsa-miR-935;hsa-miR-96-5p | 18 | EBF1 | Sponge network | -0.326 | 0.14314 | -0.384 | 0.08856 | 0.376 |
| 24285 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 12 | GRIN2A | Sponge network | 0.051 | 0.83388 | -2.161 | 0 | 0.376 |
| 24286 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-590-5p | 10 | FSTL5 | Sponge network | 0.331 | 0.57247 | -1.3 | 0.01619 | 0.376 |
| 24287 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-330-5p | 11 | RNASEL | Sponge network | -0.829 | 0.01343 | -0.272 | 0.01796 | 0.376 |
| 24288 | CTA-363E19.2 |
hsa-miR-150-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | GSK3B | Sponge network | 1.055 | 0 | 0.266 | 0.00045 | 0.376 |
| 24289 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | SCN9A | Sponge network | 0.051 | 0.83388 | -0.525 | 0.10593 | 0.376 |
| 24290 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-625-5p | 23 | RSPO2 | Sponge network | -1.047 | 0 | -3.038 | 0 | 0.376 |
| 24291 | AC009948.5 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-628-5p | 28 | ERC1 | Sponge network | 0.532 | 0.00505 | -0.108 | 0.38794 | 0.376 |
| 24292 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-5p | 25 | CLASP2 | Sponge network | 0.637 | 0.00069 | -0.038 | 0.71 | 0.376 |
| 24293 | RP11-182L21.6 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p | 10 | BRD3 | Sponge network | 0.353 | 0.00175 | 0.37 | 0.00013 | 0.376 |
| 24294 | RP11-473I1.5 |
hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-942-5p | 11 | PICALM | Sponge network | 0.542 | 0.00054 | 0.086 | 0.23523 | 0.376 |
| 24295 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 20 | TCF12 | Sponge network | 0.537 | 0.00139 | 0.174 | 0.13271 | 0.376 |
| 24296 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-539-5p;hsa-miR-98-5p | 17 | MITF | Sponge network | -0.473 | 0.07602 | -0.769 | 0.00022 | 0.376 |
| 24297 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | HOOK3 | Sponge network | 0.392 | 0.0278 | 0.273 | 0.09957 | 0.376 |
| 24298 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-495-3p;hsa-miR-589-3p;hsa-miR-629-3p | 24 | USP12 | Sponge network | 0.331 | 0.57247 | -0.102 | 0.40768 | 0.376 |
| 24299 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-337-3p;hsa-miR-424-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | HOOK3 | Sponge network | 0.659 | 3.0E-5 | 0.273 | 0.09957 | 0.376 |
| 24300 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | LHFP | Sponge network | -0.125 | 0.49414 | -0.167 | 0.41371 | 0.376 |
| 24301 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | MLLT10 | Sponge network | 1.205 | 0 | 0.105 | 0.33292 | 0.376 |
| 24302 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577 | 24 | PRKAA2 | Sponge network | 0.811 | 0.03228 | -2.462 | 0 | 0.376 |
| 24303 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | ST5 | Sponge network | -1.697 | 0.00889 | -0.78 | 0 | 0.376 |
| 24304 | DNM1P35 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 17 | AKAP6 | Sponge network | 0.051 | 0.83388 | -1.586 | 3.0E-5 | 0.376 |
| 24305 | AC016747.3 |
hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 12 | TMEFF2 | Sponge network | 0.199 | 0.38015 | -2.426 | 0 | 0.376 |
| 24306 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-7-1-3p | 23 | TET1 | Sponge network | 0.853 | 0.15352 | -0.135 | 0.52138 | 0.376 |
| 24307 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p | 17 | CBL | Sponge network | 1.361 | 0 | 0.291 | 0.01267 | 0.376 |
| 24308 | SNHG16 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-154-5p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-452-5p;hsa-miR-495-3p | 10 | LIMD1 | Sponge network | 0.961 | 0 | 0.103 | 0.28978 | 0.376 |
| 24309 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 78 | AFF4 | Sponge network | -0.129 | 0.57177 | -0.266 | 0.01565 | 0.376 |
| 24310 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | KAT6B | Sponge network | -0.899 | 6.0E-5 | -0.478 | 0.00018 | 0.376 |
| 24311 | RP11-46C24.7 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 11 | TAF1 | Sponge network | 0.392 | 0.0278 | 0.39 | 3.0E-5 | 0.376 |
| 24312 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -0.473 | 0.07602 | -0.243 | 0.11408 | 0.376 |
| 24313 | SERTAD4-AS1 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-362-5p | 13 | OPCML | Sponge network | -0.932 | 0.00329 | -2.498 | 0 | 0.376 |
| 24314 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | USP12 | Sponge network | 0.559 | 0.00026 | -0.102 | 0.40768 | 0.376 |
| 24315 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-589-5p | 10 | EYA4 | Sponge network | -0.309 | 0.1145 | 0.3 | 0.36876 | 0.376 |
| 24316 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 34 | NFIB | Sponge network | -1.092 | 4.0E-5 | 0.023 | 0.90795 | 0.376 |
| 24317 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-93-5p | 16 | FGFR1 | Sponge network | 0.259 | 0.12542 | -0.509 | 0.03957 | 0.376 |
| 24318 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 26 | NIN | Sponge network | -0.405 | 0.22013 | -0.253 | 0.13794 | 0.376 |
| 24319 | LPP-AS2 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p | 10 | PRKD1 | Sponge network | -0.18 | 0.20343 | -0.466 | 0.03583 | 0.376 |
| 24320 | H1FX-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 14 | WWTR1 | Sponge network | -0.206 | 0.12942 | -0.345 | 0.11997 | 0.375 |
| 24321 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 39 | AR | Sponge network | 0.853 | 0.15352 | -1.554 | 0 | 0.375 |
| 24322 | HNRNPU-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ZFX | Sponge network | 1.601 | 0 | 0.09 | 0.42563 | 0.375 |
| 24323 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | NTRK3 | Sponge network | -0.418 | 0.00068 | -1.77 | 3.0E-5 | 0.375 |
| 24324 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p | 45 | WDFY3 | Sponge network | 0.95 | 0.15943 | -0.201 | 0.0967 | 0.375 |
| 24325 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | PEG3 | Sponge network | 0.4 | 0.14611 | -1.74 | 0 | 0.375 |
| 24326 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-501-3p | 14 | KAT6B | Sponge network | 0.082 | 0.55397 | -0.478 | 0.00018 | 0.375 |
| 24327 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-655-3p | 19 | PIK3C2A | Sponge network | 1.147 | 0 | 0.061 | 0.53776 | 0.375 |
| 24328 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | GRIA3 | Sponge network | -0.769 | 0.00035 | -1.956 | 0 | 0.375 |
| 24329 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 16 | CNTNAP3 | Sponge network | 0.249 | 0.35902 | -1.465 | 0.00033 | 0.375 |
| 24330 | FZD10-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TMEFF2 | Sponge network | -0.727 | 0.04726 | -2.426 | 0 | 0.375 |
| 24331 | RP11-597D13.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 31 | ABI2 | Sponge network | 1.302 | 7.0E-5 | 0.175 | 0.14787 | 0.375 |
| 24332 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | PEG3 | Sponge network | 1.302 | 7.0E-5 | -1.74 | 0 | 0.375 |
| 24333 | PART1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ZNF536 | Sponge network | -2.925 | 0 | -3.115 | 0 | 0.375 |
| 24334 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-7-1-3p | 20 | CACNA2D1 | Sponge network | -0.303 | 0.21002 | -0.846 | 0.00675 | 0.375 |
| 24335 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | EBF3 | Sponge network | -0.08 | 0.90092 | -0.058 | 0.81437 | 0.375 |
| 24336 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 1.063 | 0 | 0.114 | 0.30638 | 0.375 |
| 24337 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | GRIN2A | Sponge network | 0.225 | 0.30644 | -2.161 | 0 | 0.375 |
| 24338 | AC108488.4 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | 0.259 | 0.12542 | -1.74 | 0 | 0.375 |
| 24339 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | ATRX | Sponge network | 0.657 | 4.0E-5 | 0.261 | 0.04408 | 0.375 |
| 24340 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500b-5p;hsa-miR-590-3p | 20 | RSPO2 | Sponge network | 0.335 | 0.20596 | -3.038 | 0 | 0.375 |
| 24341 | LINC00672 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-98-5p | 24 | LRIG1 | Sponge network | -0.227 | 0.31657 | -0.537 | 0.00588 | 0.375 |
| 24342 | RP11-410L14.2 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-127-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-450b-5p | 12 | CAMTA1 | Sponge network | -0.052 | 0.76839 | -0.436 | 1.0E-5 | 0.375 |
| 24343 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 31 | PRKCE | Sponge network | -2.452 | 0 | -0.403 | 0.00056 | 0.375 |
| 24344 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-940;hsa-miR-96-5p | 17 | MYO9A | Sponge network | 0.622 | 0.00015 | -0.147 | 0.32373 | 0.375 |
| 24345 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-500a-3p | 16 | PRKCE | Sponge network | -1.092 | 4.0E-5 | -0.403 | 0.00056 | 0.375 |
| 24346 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 33 | SSBP2 | Sponge network | -0.318 | 0.65847 | -1.58 | 0 | 0.375 |
| 24347 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 19 | KIT | Sponge network | 0.532 | 0.00505 | -2.184 | 0 | 0.375 |
| 24348 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | AHNAK | Sponge network | -0.777 | 0.1134 | -1.207 | 0 | 0.375 |
| 24349 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p | 12 | RICTOR | Sponge network | 1.46 | 1.0E-5 | 0.388 | 0.00188 | 0.375 |
| 24350 | RP11-274B21.10 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 26 | MDM4 | Sponge network | 1.073 | 0.00216 | 0.345 | 0.00762 | 0.375 |
| 24351 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p | 16 | KANK1 | Sponge network | -0.162 | 0.47687 | -0.55 | 0.00134 | 0.375 |
| 24352 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-7-1-3p | 17 | SOX5 | Sponge network | 1.153 | 0.00114 | -1.091 | 4.0E-5 | 0.375 |
| 24353 | LINC00657 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 31 | CCDC6 | Sponge network | 0.91 | 0 | 0.27 | 0.02555 | 0.375 |
| 24354 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-93-5p | 21 | PLSCR4 | Sponge network | 0.082 | 0.55397 | -0.474 | 0.03833 | 0.375 |
| 24355 | ERVH48-1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FBXO11 | Sponge network | 1.04 | 0.00023 | 0.142 | 0.04938 | 0.375 |
| 24356 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-34a-5p;hsa-miR-96-5p | 26 | FAT3 | Sponge network | -0.418 | 0.00068 | -1.601 | 0.00051 | 0.375 |
| 24357 | FZD10-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p | 16 | GLI3 | Sponge network | -0.727 | 0.04726 | -0.615 | 0.01482 | 0.375 |
| 24358 | RP11-400K9.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-576-5p | 18 | PRKAA2 | Sponge network | -0.733 | 0.19469 | -2.462 | 0 | 0.375 |
| 24359 | RP11-283I3.6 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 29 | ARHGEF12 | Sponge network | 0.614 | 1.0E-5 | 0.09 | 0.35693 | 0.375 |
| 24360 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PHC3 | Sponge network | 1.378 | 0 | 0.212 | 0.0499 | 0.375 |
| 24361 | RP4-613B23.1 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-589-5p | 11 | ZFHX3 | Sponge network | -0.309 | 0.1145 | 0.389 | 0.01363 | 0.375 |
| 24362 | RP11-6O2.3 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-589-3p;hsa-miR-653-5p;hsa-miR-660-5p;hsa-miR-96-5p | 36 | PPM1L | Sponge network | 0.331 | 0.57247 | -0.537 | 0.00616 | 0.375 |
| 24363 | ANKRD10-IT1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-32-5p | 10 | SOX11 | Sponge network | 1.739 | 0 | 2.68 | 0 | 0.375 |
| 24364 | MIR22HG |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-940 | 13 | MAPK10 | Sponge network | -1.047 | 0 | -1.774 | 0 | 0.375 |
| 24365 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 57 | IL6ST | Sponge network | -0.83 | 0 | -0.402 | 0.00676 | 0.375 |
| 24366 | LINC00621 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | DTNA | Sponge network | 0.131 | 0.8436 | -1.648 | 1.0E-5 | 0.375 |
| 24367 | RP11-244O19.1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-590-3p | 14 | PTCH1 | Sponge network | -0.096 | 0.67958 | -0.1 | 0.6179 | 0.375 |
| 24368 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NR4A3 | Sponge network | -1.057 | 0.00029 | -1.385 | 0 | 0.375 |
| 24369 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-940;hsa-miR-96-5p | 17 | MYO9A | Sponge network | 0.594 | 0.41522 | -0.147 | 0.32373 | 0.375 |
| 24370 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-93-3p | 23 | ST6GAL2 | Sponge network | -0.777 | 0.16667 | -1.201 | 0.00597 | 0.375 |
| 24371 | DNAJC27-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-98-5p | 37 | DMD | Sponge network | -0.969 | 2.0E-5 | -1.506 | 0 | 0.375 |
| 24372 | AC083843.1 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 11 | DICER1 | Sponge network | 1.4 | 0 | 0.042 | 0.674 | 0.375 |
| 24373 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p | 12 | SMARCA1 | Sponge network | -1.697 | 0.00889 | -0.684 | 0.00159 | 0.375 |
| 24374 | SERHL |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-625-5p | 26 | TGFBR3 | Sponge network | -0.602 | 0.00331 | -1.173 | 1.0E-5 | 0.375 |
| 24375 | GNG12-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-625-5p | 13 | TES | Sponge network | -0.793 | 7.0E-5 | -0.348 | 0.00639 | 0.375 |
| 24376 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | MLF1 | Sponge network | -2.452 | 0 | -0.893 | 0.00727 | 0.375 |
| 24377 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-425-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | ATRX | Sponge network | 0.959 | 0 | 0.261 | 0.04408 | 0.375 |
| 24378 | RP5-1198O20.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | LUZP2 | Sponge network | 0.997 | 0.00066 | -0.056 | 0.89416 | 0.375 |
| 24379 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-93-3p | 10 | EPHA3 | Sponge network | 0.912 | 0.00014 | 0.185 | 0.53588 | 0.375 |
| 24380 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 12 | KAT6B | Sponge network | -0.376 | 0.00337 | -0.478 | 0.00018 | 0.375 |
| 24381 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 30 | RNASEL | Sponge network | -1.111 | 0.0003 | -0.272 | 0.01796 | 0.375 |
| 24382 | LIPE-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-409-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p | 13 | MACF1 | Sponge network | 0.078 | 0.55803 | 0.019 | 0.90335 | 0.375 |
| 24383 | HOXB-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-589-3p | 13 | MAFB | Sponge network | -0.154 | 0.53954 | -0.023 | 0.89865 | 0.375 |
| 24384 | PCA3 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-505-5p;hsa-miR-629-5p;hsa-miR-96-5p | 14 | WNK2 | Sponge network | -0.709 | 0.18168 | -0.352 | 0.31066 | 0.375 |
| 24385 | RP1-39G22.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | ABI2 | Sponge network | 0.439 | 0.00313 | 0.175 | 0.14787 | 0.375 |
| 24386 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-940 | 31 | CHD5 | Sponge network | 0.537 | 0.00139 | -0.922 | 0.01521 | 0.375 |
| 24387 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-629-3p | 31 | DST | Sponge network | -1.582 | 8.0E-5 | -0.713 | 0.00096 | 0.375 |
| 24388 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p | 12 | CDHR3 | Sponge network | 0.001 | 0.99753 | -0.977 | 4.0E-5 | 0.375 |
| 24389 | OIP5-AS1 |
hsa-miR-140-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | CHD6 | Sponge network | 0.421 | 0.00084 | 0.32 | 0.01326 | 0.375 |
| 24390 | RP11-119F7.5 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-550a-3p | 10 | CACNA1E | Sponge network | 0.225 | 0.30644 | 1.752 | 0 | 0.375 |
| 24391 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-766-3p;hsa-miR-940 | 33 | CHD5 | Sponge network | 0.392 | 0.0278 | -0.922 | 0.01521 | 0.375 |
| 24392 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | NUAK1 | Sponge network | -1.697 | 0.00889 | 0.904 | 0 | 0.375 |
| 24393 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-5p | 30 | MAF | Sponge network | -0.323 | 0.01372 | -1.257 | 0 | 0.375 |
| 24394 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p | 10 | DLC1 | Sponge network | -0.421 | 0.12556 | -0.382 | 0.05038 | 0.375 |
| 24395 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | CREBBP | Sponge network | -0.737 | 0.06182 | 0.126 | 0.15186 | 0.375 |
| 24396 | AC006547.13 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p | 11 | USP6 | Sponge network | 1.159 | 0 | 0.188 | 0.3871 | 0.375 |
| 24397 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-5p | 34 | MTSS1 | Sponge network | -0.487 | 0.02157 | -0.81 | 0.00035 | 0.375 |
| 24398 | LINC00654 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 25 | RNF144A | Sponge network | 0.374 | 0.20054 | 0.594 | 0.0009 | 0.375 |
| 24399 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 21 | RCAN2 | Sponge network | -1.216 | 0 | -0.33 | 0.15172 | 0.375 |
| 24400 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-98-5p | 21 | TGFBR3 | Sponge network | -0.301 | 0.06597 | -1.173 | 1.0E-5 | 0.375 |
| 24401 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.385 | 0.10067 | 0.323 | 0.00792 | 0.375 |
| 24402 | MIR143HG |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | LIMCH1 | Sponge network | -0.739 | 0.0574 | -0.915 | 1.0E-5 | 0.375 |
| 24403 | RP11-672A2.4 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-301a-3p;hsa-miR-30b-3p | 10 | SOX5 | Sponge network | 1.344 | 0 | -1.091 | 4.0E-5 | 0.375 |
| 24404 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-590-3p | 32 | ZFX | Sponge network | -0.096 | 0.67958 | 0.09 | 0.42563 | 0.375 |
| 24405 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ABCC9 | Sponge network | -0.053 | 0.80685 | -0.466 | 0.14121 | 0.375 |
| 24406 | RP11-57H14.4 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PTCH1 | Sponge network | 0.083 | 0.66405 | -0.1 | 0.6179 | 0.375 |
| 24407 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 29 | TSHZ3 | Sponge network | 0.225 | 0.30644 | -0.556 | 0.01516 | 0.375 |
| 24408 | RP11-98I9.4 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | ATM | Sponge network | 0.67 | 0.00048 | 0.178 | 0.21568 | 0.375 |
| 24409 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-940 | 37 | HLF | Sponge network | -0.303 | 0.21002 | -2.443 | 0 | 0.375 |
| 24410 | FENDRR |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 16 | CSRNP1 | Sponge network | -1.281 | 0.00012 | -1.448 | 0 | 0.375 |
| 24411 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-5p | 38 | MOB1B | Sponge network | -0.162 | 0.47687 | 0.197 | 0.15266 | 0.375 |
| 24412 | BOLA3-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p | 13 | FREM2 | Sponge network | -0.246 | 0.35034 | -0.437 | 0.46353 | 0.375 |
| 24413 | RP11-531A24.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p | 42 | DST | Sponge network | 0.75 | 0.00054 | -0.713 | 0.00096 | 0.375 |
| 24414 | CTC-457L16.2 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 14 | PCDH9 | Sponge network | 1.153 | 0.00114 | -1.823 | 4.0E-5 | 0.375 |
| 24415 | RP11-1246C19.1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-539-5p | 10 | TSC1 | Sponge network | 1.352 | 0 | 0.103 | 0.26071 | 0.375 |
| 24416 | SH3RF3-AS1 |
hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b | 12 | JAZF1 | Sponge network | 0.889 | 9.0E-5 | -0.377 | 0.02677 | 0.375 |
| 24417 | CTC-429P9.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p | 10 | SEMA3C | Sponge network | 0.497 | 0.01451 | -0.272 | 0.31153 | 0.375 |
| 24418 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p | 29 | ZFHX3 | Sponge network | -0.896 | 0.00099 | 0.389 | 0.01363 | 0.375 |
| 24419 | RP5-894A10.6 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-365a-3p;hsa-miR-495-3p | 14 | ARHGEF12 | Sponge network | 0.697 | 9.0E-5 | 0.09 | 0.35693 | 0.375 |
| 24420 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 12 | CSRNP1 | Sponge network | -1.092 | 4.0E-5 | -1.448 | 0 | 0.375 |
| 24421 | PCA3 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 17 | CSDE1 | Sponge network | -0.709 | 0.18168 | -0.493 | 0 | 0.375 |
| 24422 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | EPS15 | Sponge network | -0.376 | 0.00337 | -0.052 | 0.53998 | 0.375 |
| 24423 | RP11-53O19.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-502-5p | 26 | PTPN14 | Sponge network | -0.405 | 0.22013 | 0.144 | 0.34595 | 0.375 |
| 24424 | LINC00174 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | ERCC6 | Sponge network | 1.253 | 0 | 0.23 | 0.015 | 0.375 |
| 24425 | MAP3K14 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-25-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-624-5p | 12 | NOTCH2 | Sponge network | -0.295 | 0.02604 | 0.138 | 0.35163 | 0.375 |
| 24426 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | TSHZ3 | Sponge network | 0.385 | 0.10067 | -0.556 | 0.01516 | 0.375 |
| 24427 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-93-3p | 48 | BMPR2 | Sponge network | 0.174 | 0.30034 | 0.206 | 0.0635 | 0.375 |
| 24428 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | LRRK2 | Sponge network | -0.615 | 0.00015 | -1.136 | 1.0E-5 | 0.375 |
| 24429 | RP11-967K21.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-375;hsa-miR-429 | 16 | ITPKB | Sponge network | 0.056 | 0.81195 | -0.704 | 0.00106 | 0.375 |
| 24430 | RP11-848P1.3 |
hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | PIK3R1 | Sponge network | 1.253 | 0 | -0.133 | 0.36854 | 0.375 |
| 24431 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-98-5p | 20 | AKAP6 | Sponge network | -0.18 | 0.20343 | -1.586 | 3.0E-5 | 0.375 |
| 24432 | MIR143HG |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 15 | FYN | Sponge network | -0.739 | 0.0574 | -0.131 | 0.37051 | 0.375 |
| 24433 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p | 13 | FAT4 | Sponge network | 2.306 | 9.0E-5 | -0.354 | 0.14694 | 0.375 |
| 24434 | RP11-517B11.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 13 | RPS6KA3 | Sponge network | 0.706 | 0 | 0.256 | 0.02419 | 0.375 |
| 24435 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | RBL2 | Sponge network | -0.896 | 0.00099 | -0.282 | 0.00254 | 0.375 |
| 24436 | RP11-867G23.10 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-625-5p | 15 | CRTC1 | Sponge network | -2.531 | 0 | -0.116 | 0.28412 | 0.375 |
| 24437 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 36 | RBMS3 | Sponge network | 0.72 | 0.0001 | -0.519 | 0.03551 | 0.375 |
| 24438 | RP11-973H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-342-3p | 13 | RICTOR | Sponge network | 0.901 | 0 | 0.388 | 0.00188 | 0.375 |
| 24439 | FENDRR |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 11 | ALDH1A2 | Sponge network | -1.281 | 0.00012 | -2.411 | 0 | 0.375 |
| 24440 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 33 | TMTC3 | Sponge network | 0.421 | 0.00084 | 0.123 | 0.22189 | 0.375 |
| 24441 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181a-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-590-3p | 17 | PIK3C2A | Sponge network | 0.545 | 0.00064 | 0.061 | 0.53776 | 0.375 |
| 24442 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 45 | KIF1B | Sponge network | 0.614 | 1.0E-5 | -0.118 | 0.32258 | 0.375 |
| 24443 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p | 17 | SLITRK3 | Sponge network | -0.738 | 0.21233 | -3.328 | 0 | 0.375 |
| 24444 | RP11-21A7A.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-18a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-942-5p | 14 | UNC80 | Sponge network | -1.116 | 0.23686 | -1.934 | 0 | 0.375 |
| 24445 | RP11-244O19.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3913-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | IGFBP5 | Sponge network | -0.096 | 0.67958 | -0.806 | 0.00105 | 0.375 |
| 24446 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | NEDD4 | Sponge network | -2.46 | 0 | 0.02 | 0.89834 | 0.375 |
| 24447 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-93-3p;hsa-miR-940 | 26 | SNX29 | Sponge network | -0.405 | 0.22013 | -0.34 | 0.01371 | 0.375 |
| 24448 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-940 | 26 | CHD5 | Sponge network | 0.16 | 0.50785 | -0.922 | 0.01521 | 0.375 |
| 24449 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | FAT4 | Sponge network | -0.777 | 0.1134 | -0.354 | 0.14694 | 0.375 |
| 24450 | LINC00865 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-93-3p | 15 | UNC5C | Sponge network | -1.249 | 7.0E-5 | -0.178 | 0.40214 | 0.375 |
| 24451 | RP11-25K19.1 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p | 14 | FAM135B | Sponge network | -1.057 | 0.00029 | -3.978 | 0 | 0.375 |
| 24452 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ZFHX3 | Sponge network | -0.513 | 0.04703 | 0.389 | 0.01363 | 0.375 |
| 24453 | MIR17HG |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-664a-5p | 13 | CBFB | Sponge network | 1.699 | 0 | 1.061 | 0 | 0.375 |
| 24454 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 20 | RASSF8 | Sponge network | 0.727 | 3.0E-5 | -0.122 | 0.65591 | 0.375 |
| 24455 | RP11-166D19.1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | PAIP2 | Sponge network | -0.576 | 0.10121 | -0.515 | 0 | 0.375 |
| 24456 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-590-3p | 27 | ABL2 | Sponge network | -0.096 | 0.67958 | 0.82 | 0 | 0.375 |
| 24457 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-369-3p;hsa-miR-874-3p | 10 | BCLAF1 | Sponge network | 0.541 | 0.00125 | 0.211 | 0.00684 | 0.375 |
| 24458 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-940 | 11 | HOOK3 | Sponge network | -4.546 | 0 | 0.273 | 0.09957 | 0.375 |
| 24459 | ZNF582-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | PTPRD | Sponge network | -1.349 | 0.00017 | -1.005 | 0.00064 | 0.375 |
| 24460 | RP11-21A7A.2 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-625-5p | 11 | NGFR | Sponge network | -1.116 | 0.23686 | -1.271 | 0.01058 | 0.375 |
| 24461 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CLASP2 | Sponge network | 0.873 | 0 | -0.038 | 0.71 | 0.375 |
| 24462 | GS1-124K5.11 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 19 | MRVI1 | Sponge network | 0.494 | 0.00339 | -1.051 | 0.00022 | 0.375 |
| 24463 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | FOXP1 | Sponge network | 0.614 | 1.0E-5 | 0.042 | 0.74368 | 0.375 |
| 24464 | FAM157A |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-505-5p | 14 | ARHGEF12 | Sponge network | 0.813 | 1.0E-5 | 0.09 | 0.35693 | 0.375 |
| 24465 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 36 | GAS7 | Sponge network | -0.738 | 0.21233 | -0.555 | 0.01602 | 0.375 |
| 24466 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p | 11 | CNTNAP1 | Sponge network | 0.381 | 0.25967 | 0.358 | 0.13727 | 0.375 |
| 24467 | RP11-47A8.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-423-5p;hsa-miR-744-5p;hsa-miR-942-5p | 20 | NOTCH2 | Sponge network | 0.103 | 0.43169 | 0.138 | 0.35163 | 0.375 |
| 24468 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 25 | LRRC7 | Sponge network | -0.615 | 0.00015 | -1.341 | 0.01193 | 0.375 |
| 24469 | AC009120.5 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-590-5p | 11 | FOXP1 | Sponge network | 1.383 | 0 | 0.042 | 0.74368 | 0.375 |
| 24470 | AC005618.6 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-92a-3p | 25 | IL17RD | Sponge network | 0.862 | 0.06213 | -0.019 | 0.94873 | 0.375 |
| 24471 | CTC-550B14.6 |
hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p | 11 | ABL2 | Sponge network | 3.885 | 0 | 0.82 | 0 | 0.375 |
| 24472 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p | 15 | PBX1 | Sponge network | -2.549 | 0.00528 | -1.206 | 0 | 0.375 |
| 24473 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-625-3p | 21 | ZFHX3 | Sponge network | 1.03 | 0 | 0.389 | 0.01363 | 0.375 |
| 24474 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | AFF3 | Sponge network | -2.925 | 0 | -2.373 | 0 | 0.375 |
| 24475 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-590-3p | 12 | RBL2 | Sponge network | 0.392 | 0.0278 | -0.282 | 0.00254 | 0.375 |
| 24476 | HOXA-AS2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | BCL9 | Sponge network | 0.61 | 0.01593 | 0.321 | 0.00267 | 0.375 |
| 24477 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 42 | UNC80 | Sponge network | -0.976 | 0.00025 | -1.934 | 0 | 0.375 |
| 24478 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | NF1 | Sponge network | 1.378 | 0 | 0.51 | 0 | 0.375 |
| 24479 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-92a-3p | 11 | UBE4B | Sponge network | 0.056 | 0.81195 | -0.235 | 0.007 | 0.375 |
| 24480 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | EPAS1 | Sponge network | -0.576 | 0.10121 | -0.184 | 0.14418 | 0.375 |
| 24481 | RP11-395G23.3 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p | 18 | PTPN14 | Sponge network | 0.381 | 0.25967 | 0.144 | 0.34595 | 0.375 |
| 24482 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-374a-3p | 14 | MOB1B | Sponge network | 0.997 | 0 | 0.197 | 0.15266 | 0.375 |
| 24483 | GNG12-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | ZDBF2 | Sponge network | -0.793 | 7.0E-5 | -0.998 | 0.00025 | 0.375 |
| 24484 | RP4-791M13.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 16 | ABL2 | Sponge network | 0.419 | 0.0319 | 0.82 | 0 | 0.375 |
| 24485 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 12 | DYNC1I1 | Sponge network | 0.178 | 0.29106 | -1.12 | 0.00107 | 0.375 |
| 24486 | RP11-73M18.8 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p | 12 | CBFB | Sponge network | 0.832 | 0 | 1.061 | 0 | 0.375 |
| 24487 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 45 | ERBB4 | Sponge network | -0.969 | 2.0E-5 | -1.975 | 2.0E-5 | 0.375 |
| 24488 | U91328.19 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-940 | 14 | CELF4 | Sponge network | -0.303 | 0.21002 | -1.135 | 0.00344 | 0.375 |
| 24489 | RP11-585P4.5 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-664a-5p | 14 | CBFB | Sponge network | 1.046 | 1.0E-5 | 1.061 | 0 | 0.375 |
| 24490 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 47 | AR | Sponge network | 0.395 | 0.15451 | -1.554 | 0 | 0.374 |
| 24491 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 41 | TACC1 | Sponge network | 0.72 | 0.0001 | -0.362 | 0.05406 | 0.374 |
| 24492 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-577;hsa-miR-590-3p | 17 | KAT6B | Sponge network | -0.096 | 0.67958 | -0.478 | 0.00018 | 0.374 |
| 24493 | LINC00472 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 23 | PDE4DIP | Sponge network | -0.842 | 0.01361 | -0.282 | 0.0257 | 0.374 |
| 24494 | RP11-678G14.3 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-484;hsa-miR-7-1-3p | 11 | NRXN3 | Sponge network | -4.477 | 0 | -0.893 | 0.04186 | 0.374 |
| 24495 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-940 | 17 | TAL1 | Sponge network | -0.777 | 0.16667 | -0.462 | 0.00574 | 0.374 |
| 24496 | RP4-717I23.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-766-3p | 11 | KIAA1024 | Sponge network | 0.637 | 0.00069 | 1.083 | 0 | 0.374 |
| 24497 | HOXA-AS2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 43 | TRPS1 | Sponge network | 0.61 | 0.01593 | -0.191 | 0.44109 | 0.374 |
| 24498 | PART1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 20 | CXCL12 | Sponge network | -2.925 | 0 | -1.421 | 0 | 0.374 |
| 24499 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | PHC3 | Sponge network | 0.572 | 0.01722 | 0.212 | 0.0499 | 0.374 |
| 24500 | RP5-1139B12.3 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-454-3p | 18 | FAT3 | Sponge network | 0.598 | 0.38481 | -1.601 | 0.00051 | 0.374 |
| 24501 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 23 | MYBL1 | Sponge network | 0.532 | 0.00505 | 0.494 | 0.02152 | 0.374 |
| 24502 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-625-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -2.117 | 0.00161 | -0.087 | 0.69193 | 0.374 |
| 24503 | AC109309.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-330-3p | 12 | PRKAA2 | Sponge network | 0.163 | 0.87219 | -2.462 | 0 | 0.374 |
| 24504 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 14 | NF1 | Sponge network | 0.856 | 0 | 0.51 | 0 | 0.374 |
| 24505 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p | 32 | PPM1A | Sponge network | -0.896 | 0.00099 | -0.623 | 0 | 0.374 |
| 24506 | RP4-791M13.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | RASSF8 | Sponge network | 0.419 | 0.0319 | -0.122 | 0.65591 | 0.374 |
| 24507 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-655-3p | 24 | LATS1 | Sponge network | 0.75 | 0.00054 | 0.109 | 0.34208 | 0.374 |
| 24508 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p | 10 | HOOK3 | Sponge network | 0.176 | 0.37402 | 0.273 | 0.09957 | 0.374 |
| 24509 | RP11-890B15.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-96-5p | 25 | CYLD | Sponge network | 0.101 | 0.60919 | -0.367 | 0.00171 | 0.374 |
| 24510 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p | 17 | RIMBP2 | Sponge network | -2.69 | 0.0001 | -1.968 | 5.0E-5 | 0.374 |
| 24511 | RP11-798G7.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-375;hsa-miR-625-5p | 12 | PTPN14 | Sponge network | 0.858 | 3.0E-5 | 0.144 | 0.34595 | 0.374 |
| 24512 | ZNF582-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 68 | LPP | Sponge network | -1.349 | 0.00017 | -0.459 | 0.04475 | 0.374 |
| 24513 | CTC-471J1.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-7-1-3p | 19 | CREB1 | Sponge network | 1.11 | 1.0E-5 | 0.203 | 0.00537 | 0.374 |
| 24514 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-511-5p;hsa-miR-542-3p | 19 | ADAMTSL3 | Sponge network | -0.384 | 0.40652 | -1.559 | 0 | 0.374 |
| 24515 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 24 | EBF3 | Sponge network | -0.615 | 0.00015 | -0.058 | 0.81437 | 0.374 |
| 24516 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-766-3p | 36 | AFF4 | Sponge network | -1.092 | 4.0E-5 | -0.266 | 0.01565 | 0.374 |
| 24517 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-33a-5p;hsa-miR-455-5p | 14 | SLITRK3 | Sponge network | -0.773 | 0.04073 | -3.328 | 0 | 0.374 |
| 24518 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-655-3p | 18 | NF1 | Sponge network | 0.75 | 0.00054 | 0.51 | 0 | 0.374 |
| 24519 | RP11-506M13.3 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-582-3p | 16 | ABL2 | Sponge network | 0.769 | 0 | 0.82 | 0 | 0.374 |
| 24520 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | LRP1B | Sponge network | -1.161 | 0.00591 | -2.163 | 0 | 0.374 |
| 24521 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-940 | 42 | NTRK2 | Sponge network | -0.969 | 2.0E-5 | -0.736 | 0.06495 | 0.374 |
| 24522 | PWAR6 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | -1.575 | 6.0E-5 | -0.226 | 0.11691 | 0.374 |
| 24523 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 25 | KIT | Sponge network | 0.246 | 0.59912 | -2.184 | 0 | 0.374 |
| 24524 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | HIVEP1 | Sponge network | 1.063 | 0 | 0.368 | 0.00064 | 0.374 |
| 24525 | LINC00476 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p | 13 | AMPH | Sponge network | -0.615 | 0.00015 | -0.916 | 5.0E-5 | 0.374 |
| 24526 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CACNA2D1 | Sponge network | 0.421 | 0.00084 | -0.846 | 0.00675 | 0.374 |
| 24527 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | AKAP12 | Sponge network | 0.72 | 0.0001 | -0.932 | 0.0028 | 0.374 |
| 24528 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 18 | ST6GALNAC3 | Sponge network | -0.72 | 0.24239 | -0.987 | 0 | 0.374 |
| 24529 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 34 | MOB1B | Sponge network | 0.335 | 0.20596 | 0.197 | 0.15266 | 0.374 |
| 24530 | LINC00957 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | TMEM132B | Sponge network | -0.421 | 0.0114 | -0.83 | 0.01084 | 0.374 |
| 24531 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 16 | PSIP1 | Sponge network | 0.064 | 0.72934 | -0.005 | 0.96897 | 0.374 |
| 24532 | AC116366.6 |
hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940 | 15 | MEF2C | Sponge network | 0.329 | 0.11122 | -0.499 | 0.01448 | 0.374 |
| 24533 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-337-3p;hsa-miR-33a-3p | 11 | RPS6KA3 | Sponge network | 0.542 | 0.00054 | 0.256 | 0.02419 | 0.374 |
| 24534 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 40 | ESR1 | Sponge network | -2.46 | 0 | -1.035 | 3.0E-5 | 0.374 |
| 24535 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 45 | IL6ST | Sponge network | 0.176 | 0.37402 | -0.402 | 0.00676 | 0.374 |
| 24536 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 37 | QKI | Sponge network | 0.4 | 0.14611 | -0.333 | 0.04111 | 0.374 |
| 24537 | PART1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | TLL1 | Sponge network | -2.925 | 0 | -1.195 | 3.0E-5 | 0.374 |
| 24538 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | IGSF10 | Sponge network | -2.925 | 0 | -1.751 | 1.0E-5 | 0.374 |
| 24539 | RP11-1006G14.4 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3127-5p;hsa-miR-455-3p | 13 | ABL1 | Sponge network | 0.559 | 0.00026 | -0.215 | 0.07804 | 0.374 |
| 24540 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 34 | ST6GALNAC3 | Sponge network | -0.323 | 0.01372 | -0.987 | 0 | 0.374 |
| 24541 | TRAF3IP2-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | -0.323 | 0.01372 | 0.524 | 0.00017 | 0.374 |
| 24542 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 30 | ST6GAL2 | Sponge network | -0.563 | 0.02545 | -1.201 | 0.00597 | 0.374 |
| 24543 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | CXXC4 | Sponge network | 0.225 | 0.30644 | -0.433 | 0.21055 | 0.374 |
| 24544 | MAGI2-AS3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | LUZP2 | Sponge network | -0.129 | 0.57177 | -0.056 | 0.89416 | 0.374 |
| 24545 | HCG11 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 27 | DIP2C | Sponge network | 0.385 | 0.10067 | -0.436 | 0.01713 | 0.374 |
| 24546 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-493-5p | 17 | USP6 | Sponge network | 0.056 | 0.8494 | 0.188 | 0.3871 | 0.374 |
| 24547 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | ZFX | Sponge network | 0.72 | 0.0001 | 0.09 | 0.42563 | 0.374 |
| 24548 | RP11-83A24.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PHF3 | Sponge network | 0.417 | 0.00445 | 0.126 | 0.19652 | 0.374 |
| 24549 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 16 | ARNT2 | Sponge network | -0.358 | 0.17255 | -0.332 | 0.25851 | 0.374 |
| 24550 | LINC01018 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 11 | CELF4 | Sponge network | -2.915 | 0.00015 | -1.135 | 0.00344 | 0.374 |
| 24551 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-628-5p | 27 | RUNX1T1 | Sponge network | -0.726 | 0.00132 | -0.344 | 0.2374 | 0.374 |
| 24552 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p | 15 | GREM1 | Sponge network | -0.738 | 0.21233 | 0.902 | 0.01691 | 0.374 |
| 24553 | DNM1P35 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-628-5p | 14 | GRIA3 | Sponge network | 0.051 | 0.83388 | -1.956 | 0 | 0.374 |
| 24554 | PART1 |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CTNNA2 | Sponge network | -2.925 | 0 | -2.547 | 0 | 0.374 |
| 24555 | AP001056.1 |
hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-96-5p | 10 | GAS7 | Sponge network | 1.062 | 0.00912 | -0.555 | 0.01602 | 0.374 |
| 24556 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 28 | ST6GALNAC3 | Sponge network | -0.18 | 0.20343 | -0.987 | 0 | 0.374 |
| 24557 | CTC-559E9.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-877-5p | 22 | EPS15 | Sponge network | 0.594 | 0.00064 | -0.052 | 0.53998 | 0.374 |
| 24558 | LINC00843 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429 | 15 | EPS15 | Sponge network | 1.106 | 0 | -0.052 | 0.53998 | 0.374 |
| 24559 | TP73-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 27 | RIMS2 | Sponge network | -0.563 | 0.02545 | -1.365 | 0.00208 | 0.374 |
| 24560 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-940 | 14 | HOOK3 | Sponge network | 0.056 | 0.81195 | 0.273 | 0.09957 | 0.374 |
| 24561 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | PEG3 | Sponge network | 0.174 | 0.30034 | -1.74 | 0 | 0.374 |
| 24562 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 24 | ANK2 | Sponge network | 0.395 | 0.15451 | -1.603 | 1.0E-5 | 0.374 |
| 24563 | ATP1B3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-96-5p | 22 | LPP | Sponge network | 1.2 | 0.01813 | -0.459 | 0.04475 | 0.374 |
| 24564 | RP11-403I13.8 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p | 10 | CACNA1E | Sponge network | 0.619 | 0.00157 | 1.752 | 0 | 0.374 |
| 24565 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | CNTN1 | Sponge network | -0.969 | 2.0E-5 | -2.594 | 0 | 0.374 |
| 24566 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | NRXN3 | Sponge network | 0.72 | 0.0001 | -0.893 | 0.04186 | 0.374 |
| 24567 | LINC00472 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-766-3p | 11 | ATM | Sponge network | -0.842 | 0.01361 | 0.178 | 0.21568 | 0.374 |
| 24568 | CKMT2-AS1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | CSDE1 | Sponge network | 0.082 | 0.55654 | -0.493 | 0 | 0.374 |
| 24569 | LOXL1-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | HIP1 | Sponge network | 0.487 | 0.04053 | 0.774 | 0 | 0.374 |
| 24570 | A1BG-AS1 |
hsa-miR-1266-5p;hsa-miR-15a-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-378a-5p | 11 | ASTN1 | Sponge network | -0.164 | 0.45106 | -2.389 | 2.0E-5 | 0.374 |
| 24571 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 36 | NRXN1 | Sponge network | -0.969 | 2.0E-5 | -3.778 | 0 | 0.374 |
| 24572 | BDNF-AS |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | PCDH9 | Sponge network | -0.668 | 3.0E-5 | -1.823 | 4.0E-5 | 0.374 |
| 24573 | RP11-326G21.1 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RIMS2 | Sponge network | -0.021 | 0.93598 | -1.365 | 0.00208 | 0.374 |
| 24574 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | DPYD | Sponge network | -0.83 | 0 | -0.736 | 0.00076 | 0.374 |
| 24575 | AP001347.6 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | GALNT13 | Sponge network | -1.161 | 0.00591 | -0.857 | 0.07439 | 0.374 |
| 24576 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-664a-3p;hsa-miR-874-3p | 19 | MXI1 | Sponge network | -2.69 | 0.0001 | -0.987 | 0 | 0.374 |
| 24577 | MIR181A2HG |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | MDM4 | Sponge network | 1.822 | 1.0E-5 | 0.345 | 0.00762 | 0.374 |
| 24578 | A2M-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 19 | SYNPO2 | Sponge network | -0.08 | 0.90092 | -2.342 | 0 | 0.374 |
| 24579 | LINC00657 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DICER1 | Sponge network | 0.91 | 0 | 0.042 | 0.674 | 0.374 |
| 24580 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-935 | 17 | HBP1 | Sponge network | -1.331 | 3.0E-5 | -0.454 | 0 | 0.374 |
| 24581 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 25 | CRTC3 | Sponge network | -1.203 | 0.03881 | 0.164 | 0.16786 | 0.374 |
| 24582 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-7-1-3p | 10 | CAV1 | Sponge network | -0.246 | 0.35034 | -1.013 | 6.0E-5 | 0.374 |
| 24583 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | CDH19 | Sponge network | -0.969 | 2.0E-5 | -3.434 | 0 | 0.374 |
| 24584 | LINC00578 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-501-5p | 15 | CTIF | Sponge network | 0.811 | 0.03228 | -0.389 | 0.00549 | 0.374 |
| 24585 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 10 | TPO | Sponge network | -0.513 | 0.04703 | -1.87 | 2.0E-5 | 0.374 |
| 24586 | MIR22HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-877-5p | 18 | CDH19 | Sponge network | -1.047 | 0 | -3.434 | 0 | 0.374 |
| 24587 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | NOTCH2NL | Sponge network | 0.194 | 0.64278 | 0.024 | 0.84307 | 0.374 |
| 24588 | TP73-AS1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-96-5p | 16 | RPS6KA6 | Sponge network | -0.563 | 0.02545 | -2.989 | 0 | 0.374 |
| 24589 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935 | 28 | ABI2 | Sponge network | -0.513 | 0.04703 | 0.175 | 0.14787 | 0.374 |
| 24590 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 15 | ARHGAP28 | Sponge network | -1.582 | 8.0E-5 | -0.911 | 0.00013 | 0.374 |
| 24591 | RP11-705C15.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | SNX29 | Sponge network | 0.796 | 4.0E-5 | -0.34 | 0.01371 | 0.374 |
| 24592 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 21 | UNC80 | Sponge network | -1.582 | 8.0E-5 | -1.934 | 0 | 0.374 |
| 24593 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 13 | FLI1 | Sponge network | 0.335 | 0.20596 | -0.294 | 0.10905 | 0.374 |
| 24594 | MEG3 |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p | 11 | GLIPR2 | Sponge network | 0.249 | 0.35902 | -0.996 | 0 | 0.374 |
| 24595 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-874-3p;hsa-miR-877-5p | 16 | CRTC3 | Sponge network | -2.62 | 0 | 0.164 | 0.16786 | 0.374 |
| 24596 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-625-5p | 18 | CSMD1 | Sponge network | -0.154 | 0.78939 | -0.63 | 0.18881 | 0.374 |
| 24597 | ADAMTS9-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 47 | GNAQ | Sponge network | -2.452 | 0 | -0.367 | 0.00102 | 0.374 |
| 24598 | RP11-403I13.7 |
hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 11 | FLNA | Sponge network | 0.395 | 0.15451 | -0.771 | 0.01377 | 0.374 |
| 24599 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | NUAK1 | Sponge network | 0.246 | 0.59912 | 0.904 | 0 | 0.374 |
| 24600 | RP11-686D22.8 |
hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | HIP1 | Sponge network | 0.898 | 1.0E-5 | 0.774 | 0 | 0.374 |
| 24601 | LINC00476 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-93-5p | 62 | AR | Sponge network | -0.615 | 0.00015 | -1.554 | 0 | 0.374 |
| 24602 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429 | 16 | SPG20 | Sponge network | 0.056 | 0.81195 | -1.16 | 0 | 0.374 |
| 24603 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-3065-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 13 | CBL | Sponge network | 0.693 | 0.00076 | 0.291 | 0.01267 | 0.374 |
| 24604 | SNHG14 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ARNT | Sponge network | -1.111 | 0.0003 | -0.08 | 0.26073 | 0.374 |
| 24605 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 34 | BHLHE41 | Sponge network | 0.249 | 0.35902 | 0.353 | 0.12753 | 0.374 |
| 24606 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577 | 15 | NIN | Sponge network | 0.811 | 0.03228 | -0.253 | 0.13794 | 0.374 |
| 24607 | RP11-212P7.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 18 | LUZP2 | Sponge network | 0.219 | 0.1516 | -0.056 | 0.89416 | 0.374 |
| 24608 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p | 33 | EPHA5 | Sponge network | -0.693 | 0.05629 | -0.957 | 0.01733 | 0.374 |
| 24609 | CTA-204B4.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 15 | ERCC4 | Sponge network | 0.765 | 7.0E-5 | 0.126 | 0.15266 | 0.374 |
| 24610 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 22 | CUL5 | Sponge network | 1.11 | 0 | 0.026 | 0.77301 | 0.374 |
| 24611 | LINC00963 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | STARD13 | Sponge network | 0.523 | 9.0E-5 | -0.187 | 0.27383 | 0.374 |
| 24612 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | MYO9A | Sponge network | 0.848 | 0 | -0.147 | 0.32373 | 0.374 |
| 24613 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | ST6GAL2 | Sponge network | 0.532 | 0.00505 | -1.201 | 0.00597 | 0.374 |
| 24614 | LINC00886 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-93-5p | 18 | RNASEL | Sponge network | -0.371 | 0.24604 | -0.272 | 0.01796 | 0.374 |
| 24615 | MIR22HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-590-5p | 14 | ITGB3 | Sponge network | -1.047 | 0 | -0.011 | 0.95751 | 0.374 |
| 24616 | WDFY3-AS2 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-493-5p;hsa-miR-584-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | -0.942 | 0 | -0.366 | 0.01036 | 0.374 |
| 24617 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 11 | BCL6 | Sponge network | -0.206 | 0.12942 | -0.211 | 0.19489 | 0.374 |
| 24618 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p | 14 | PCDH9 | Sponge network | -0.351 | 0.11358 | -1.823 | 4.0E-5 | 0.374 |
| 24619 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | PHC3 | Sponge network | 0.705 | 0.00014 | 0.212 | 0.0499 | 0.374 |
| 24620 | SNHG14 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | CREM | Sponge network | -1.111 | 0.0003 | -0.345 | 0.00258 | 0.374 |
| 24621 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | CDHR3 | Sponge network | -1.281 | 0.00012 | -0.977 | 4.0E-5 | 0.374 |
| 24622 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 16 | CHD1 | Sponge network | 0.848 | 0 | 0.322 | 0.00215 | 0.374 |
| 24623 | LINC00094 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p | 19 | PIK3CA | Sponge network | 0.253 | 0.09565 | 0.195 | 0.06908 | 0.374 |
| 24624 | WDFY3-AS2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 16 | ZBTB20 | Sponge network | -0.942 | 0 | -0.122 | 0.57569 | 0.374 |
| 24625 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 32 | QKI | Sponge network | 0.056 | 0.8494 | -0.333 | 0.04111 | 0.374 |
| 24626 | CTC-296K1.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-582-5p;hsa-miR-940 | 18 | PPP1R9A | Sponge network | -2.095 | 0.00112 | -0.903 | 0.04254 | 0.374 |
| 24627 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-501-5p | 20 | NTRK2 | Sponge network | -2.531 | 0 | -0.736 | 0.06495 | 0.374 |
| 24628 | RP11-196G18.24 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 24 | MDM4 | Sponge network | 1.262 | 0 | 0.345 | 0.00762 | 0.374 |
| 24629 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-98-5p | 11 | DMD | Sponge network | 1.344 | 0 | -1.506 | 0 | 0.374 |
| 24630 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | HECA | Sponge network | -2.452 | 0 | -0.217 | 0.03463 | 0.374 |
| 24631 | RP11-822E23.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 30 | SNX29 | Sponge network | -0.777 | 0.16667 | -0.34 | 0.01371 | 0.374 |
| 24632 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 34 | RICTOR | Sponge network | 0.541 | 0.00125 | 0.388 | 0.00188 | 0.374 |
| 24633 | RP11-67L2.2 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SH3GLB1 | Sponge network | 0.72 | 0.0001 | -0.115 | 0.14773 | 0.374 |
| 24634 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-7-1-3p | 19 | USP12 | Sponge network | 0.75 | 0.00054 | -0.102 | 0.40768 | 0.374 |
| 24635 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 19 | SUFU | Sponge network | -0.125 | 0.49414 | -0.04 | 0.65489 | 0.374 |
| 24636 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p | 49 | FOXP1 | Sponge network | -0.615 | 0.00015 | 0.042 | 0.74368 | 0.374 |
| 24637 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p | 18 | DYNC1I1 | Sponge network | -1.645 | 0 | -1.12 | 0.00107 | 0.374 |
| 24638 | HNRNPU-AS1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-9-5p | 33 | CCDC6 | Sponge network | 1.601 | 0 | 0.27 | 0.02555 | 0.374 |
| 24639 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p | 17 | EYA4 | Sponge network | 1.757 | 0 | 0.3 | 0.36876 | 0.374 |
| 24640 | PCA3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | KLF10 | Sponge network | -0.709 | 0.18168 | -0.783 | 0 | 0.374 |
| 24641 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-96-5p | 15 | EBF3 | Sponge network | 0.056 | 0.8494 | -0.058 | 0.81437 | 0.374 |
| 24642 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 53 | THRB | Sponge network | -0.612 | 0.00094 | -1.117 | 0.0004 | 0.374 |
| 24643 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-93-5p | 33 | LRP1B | Sponge network | -0.615 | 0.00015 | -2.163 | 0 | 0.374 |
| 24644 | RP11-488L18.10 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29c-3p;hsa-miR-30a-3p | 10 | ABL2 | Sponge network | 1.392 | 0 | 0.82 | 0 | 0.374 |
| 24645 | CKMT2-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p | 14 | BCL9 | Sponge network | 0.082 | 0.55654 | 0.321 | 0.00267 | 0.374 |
| 24646 | ZRANB2-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 31 | DST | Sponge network | -0.376 | 0.00337 | -0.713 | 0.00096 | 0.374 |
| 24647 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | RABEP1 | Sponge network | 0.225 | 0.30644 | -0.066 | 0.43142 | 0.374 |
| 24648 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | CCND2 | Sponge network | -1.057 | 0.00029 | -0.267 | 0.3112 | 0.374 |
| 24649 | RP11-822E23.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p | 24 | CBFA2T3 | Sponge network | -0.777 | 0.16667 | -1.221 | 1.0E-5 | 0.374 |
| 24650 | CTC-429P9.5 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p | 19 | NCOA1 | Sponge network | 0.178 | 0.29106 | -0.432 | 0 | 0.374 |
| 24651 | RP11-67L2.2 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p | 23 | SYNM | Sponge network | 0.72 | 0.0001 | -2.309 | 1.0E-5 | 0.374 |
| 24652 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | ZNF536 | Sponge network | -0.49 | 0.04946 | -3.115 | 0 | 0.374 |
| 24653 | MIR143HG |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-942-5p | 35 | OPCML | Sponge network | -0.739 | 0.0574 | -2.498 | 0 | 0.374 |
| 24654 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | ERG | Sponge network | 0.101 | 0.60919 | 0.002 | 0.99011 | 0.374 |
| 24655 | AP000240.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p | 10 | TBL1XR1 | Sponge network | 1.462 | 0 | 0.578 | 0 | 0.374 |
| 24656 | LINC00578 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-629-3p | 10 | SPTBN4 | Sponge network | 0.811 | 0.03228 | -0.786 | 0.01281 | 0.373 |
| 24657 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429 | 11 | CBFA2T3 | Sponge network | 0.727 | 3.0E-5 | -1.221 | 1.0E-5 | 0.373 |
| 24658 | HCG18 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | SIRT1 | Sponge network | 1.063 | 0 | 0.109 | 0.2593 | 0.373 |
| 24659 | FAM157C |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 0.91 | 0 | 0.578 | 0 | 0.373 |
| 24660 | U91328.19 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | -0.303 | 0.21002 | -1.74 | 0 | 0.373 |
| 24661 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 17 | TGFBR2 | Sponge network | 0.214 | 0.18246 | -0.243 | 0.11408 | 0.373 |
| 24662 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-5p | 42 | EPHA5 | Sponge network | 0.056 | 0.81195 | -0.957 | 0.01733 | 0.373 |
| 24663 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-576-5p | 18 | ERCC4 | Sponge network | 0.614 | 1.0E-5 | 0.126 | 0.15266 | 0.373 |
| 24664 | RP11-254F7.2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-576-5p | 11 | KLF10 | Sponge network | 0.176 | 0.37402 | -0.783 | 0 | 0.373 |
| 24665 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-625-5p | 10 | MPL | Sponge network | -0.777 | 0.1134 | -0.624 | 0.00133 | 0.373 |
| 24666 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-5p | 11 | AKAP6 | Sponge network | -0.829 | 0.01343 | -1.586 | 3.0E-5 | 0.373 |
| 24667 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-93-5p | 14 | TRIM3 | Sponge network | 0.246 | 0.59912 | -0.577 | 0 | 0.373 |
| 24668 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | RASSF2 | Sponge network | 0.657 | 4.0E-5 | -0.273 | 0.21961 | 0.373 |
| 24669 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 26 | FBXO32 | Sponge network | -1.057 | 0.00029 | -0.495 | 0.07561 | 0.373 |
| 24670 | CTD-2314G24.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 48 | GAS7 | Sponge network | 0.246 | 0.59912 | -0.555 | 0.01602 | 0.373 |
| 24671 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-93-3p | 24 | NCOA1 | Sponge network | -0.18 | 0.20343 | -0.432 | 0 | 0.373 |
| 24672 | CTC-510F12.2 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-376c-3p | 14 | BMPR2 | Sponge network | -0.691 | 0.00146 | 0.206 | 0.0635 | 0.373 |
| 24673 | RP11-822E23.8 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-589-3p | 35 | TGFBR3 | Sponge network | -0.777 | 0.16667 | -1.173 | 1.0E-5 | 0.373 |
| 24674 | RP13-516M14.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577 | 39 | DST | Sponge network | 0.389 | 0.04293 | -0.713 | 0.00096 | 0.373 |
| 24675 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | NIN | Sponge network | 0.335 | 0.20596 | -0.253 | 0.13794 | 0.373 |
| 24676 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-3065-3p;hsa-miR-424-5p;hsa-miR-455-3p | 11 | CRTC3 | Sponge network | 0.693 | 0.00076 | 0.164 | 0.16786 | 0.373 |
| 24677 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-590-3p | 11 | HLF | Sponge network | -1.425 | 0.04204 | -2.443 | 0 | 0.373 |
| 24678 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-935 | 29 | ABI2 | Sponge network | 0.16 | 0.50785 | 0.175 | 0.14787 | 0.373 |
| 24679 | RP13-516M14.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | RASSF8 | Sponge network | 0.389 | 0.04293 | -0.122 | 0.65591 | 0.373 |
| 24680 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CNTN1 | Sponge network | 0.61 | 0.01593 | -2.594 | 0 | 0.373 |
| 24681 | U91328.19 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 19 | BRD3 | Sponge network | -0.303 | 0.21002 | 0.37 | 0.00013 | 0.373 |
| 24682 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | MOB1B | Sponge network | 0.705 | 0.00014 | 0.197 | 0.15266 | 0.373 |
| 24683 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-590-3p | 11 | PLA2R1 | Sponge network | -1.161 | 0.00591 | -0.265 | 0.24676 | 0.373 |
| 24684 | AC103563.8 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p | 10 | ACSL6 | Sponge network | -7.551 | 0 | -1.593 | 0 | 0.373 |
| 24685 | LINC00957 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421 | 14 | MAFB | Sponge network | -0.421 | 0.0114 | -0.023 | 0.89865 | 0.373 |
| 24686 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | DOCK4 | Sponge network | -0.487 | 0.02157 | 0.502 | 0.00108 | 0.373 |
| 24687 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 18 | FOXO3 | Sponge network | 0.986 | 0.01022 | -0.04 | 0.72028 | 0.373 |
| 24688 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 34 | LIFR | Sponge network | -0.737 | 0.06182 | -1.794 | 0 | 0.373 |
| 24689 | ADAMTS9-AS2 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | PTCH1 | Sponge network | -2.452 | 0 | -0.1 | 0.6179 | 0.373 |
| 24690 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 51 | WDFY3 | Sponge network | -0.222 | 0.32445 | -0.201 | 0.0967 | 0.373 |
| 24691 | RP11-13K12.5 |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p | 15 | AFF3 | Sponge network | -0.318 | 0.65847 | -2.373 | 0 | 0.373 |
| 24692 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-92a-1-5p | 28 | PRKAB2 | Sponge network | -0.154 | 0.78939 | -0.367 | 0.01371 | 0.373 |
| 24693 | LINC00641 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-214-5p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-874-3p | 36 | MXI1 | Sponge network | -0.222 | 0.32445 | -0.987 | 0 | 0.373 |
| 24694 | LINC00641 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | HSPB7 | Sponge network | -0.222 | 0.32445 | -2.268 | 1.0E-5 | 0.373 |
| 24695 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p | 18 | CNTNAP3 | Sponge network | -0.899 | 6.0E-5 | -1.465 | 0.00033 | 0.373 |
| 24696 | RP11-144G6.12 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-320a;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | RPS6KA3 | Sponge network | 0.826 | 1.0E-5 | 0.256 | 0.02419 | 0.373 |
| 24697 | RP11-574K11.29 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-495-3p;hsa-miR-654-3p | 10 | DICER1 | Sponge network | 0.997 | 0 | 0.042 | 0.674 | 0.373 |
| 24698 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-96-5p | 15 | PIK3R1 | Sponge network | 0.37 | 0.27614 | -0.133 | 0.36854 | 0.373 |
| 24699 | LINC00654 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-96-5p | 25 | ASTN1 | Sponge network | 0.374 | 0.20054 | -2.389 | 2.0E-5 | 0.373 |
| 24700 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-671-5p | 18 | KIT | Sponge network | -0.773 | 0.04073 | -2.184 | 0 | 0.373 |
| 24701 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-96-5p | 15 | LRRC7 | Sponge network | 0.331 | 0.57247 | -1.341 | 0.01193 | 0.373 |
| 24702 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | CNTNAP3 | Sponge network | -0.053 | 0.80685 | -1.465 | 0.00033 | 0.373 |
| 24703 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-96-5p | 23 | MYO9A | Sponge network | -0.18 | 0.20343 | -0.147 | 0.32373 | 0.373 |
| 24704 | RP11-685N10.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p | 16 | TBL1XR1 | Sponge network | 2.743 | 0 | 0.578 | 0 | 0.373 |
| 24705 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | PIK3R1 | Sponge network | -1.439 | 4.0E-5 | -0.133 | 0.36854 | 0.373 |
| 24706 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SEMA3E | Sponge network | -0.769 | 0.00035 | -1.717 | 0.00137 | 0.373 |
| 24707 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-5p | 31 | ESR1 | Sponge network | -0.777 | 0.16667 | -1.035 | 3.0E-5 | 0.373 |
| 24708 | AC010226.4 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | GLIPR1 | Sponge network | -0.811 | 2.0E-5 | 0.146 | 0.3276 | 0.373 |
| 24709 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | SON | Sponge network | 1.044 | 2.0E-5 | 0.168 | 0.04406 | 0.373 |
| 24710 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TBX18 | Sponge network | -1.575 | 6.0E-5 | 0.843 | 0.02945 | 0.373 |
| 24711 | RP11-244O19.1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p | 20 | RPS6KA6 | Sponge network | -0.096 | 0.67958 | -2.989 | 0 | 0.373 |
| 24712 | U62631.5 |
hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-33a-5p | 11 | TCF4 | Sponge network | 3.115 | 0.00463 | 0.016 | 0.92271 | 0.373 |
| 24713 | RP11-13K12.5 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-942-5p | 26 | IGFBP5 | Sponge network | -0.318 | 0.65847 | -0.806 | 0.00105 | 0.373 |
| 24714 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 15 | RET | Sponge network | -0.738 | 0.21233 | -0.833 | 0.03517 | 0.373 |
| 24715 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 13 | SLITRK3 | Sponge network | -0.342 | 0.02288 | -3.328 | 0 | 0.373 |
| 24716 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-335-3p | 13 | QKI | Sponge network | -0.691 | 0.00146 | -0.333 | 0.04111 | 0.373 |
| 24717 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 50 | CCND2 | Sponge network | -0.222 | 0.32445 | -0.267 | 0.3112 | 0.373 |
| 24718 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940;hsa-miR-96-5p | 54 | GAS7 | Sponge network | -0.942 | 0 | -0.555 | 0.01602 | 0.373 |
| 24719 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 26 | PCDH17 | Sponge network | -0.49 | 0.04946 | 0.731 | 2.0E-5 | 0.373 |
| 24720 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DCN | Sponge network | -0.08 | 0.90092 | -1.288 | 0 | 0.373 |
| 24721 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 31 | RSPO2 | Sponge network | -0.942 | 0 | -3.038 | 0 | 0.373 |
| 24722 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-766-3p | 20 | TBL1XR1 | Sponge network | 0.614 | 1.0E-5 | 0.578 | 0 | 0.373 |
| 24723 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-940 | 32 | PBX1 | Sponge network | -0.777 | 0.16667 | -1.206 | 0 | 0.373 |
| 24724 | RP11-977G19.11 |
hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 10 | ABL2 | Sponge network | 0.957 | 0 | 0.82 | 0 | 0.373 |
| 24725 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | CITED2 | Sponge network | -0.942 | 0 | -1.545 | 0 | 0.373 |
| 24726 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | RBL2 | Sponge network | 0.683 | 1.0E-5 | -0.282 | 0.00254 | 0.373 |
| 24727 | SNHG14 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 31 | PCDH17 | Sponge network | -1.111 | 0.0003 | 0.731 | 2.0E-5 | 0.373 |
| 24728 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p | 11 | TCF4 | Sponge network | -0.691 | 0.00146 | 0.016 | 0.92271 | 0.373 |
| 24729 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PBRM1 | Sponge network | -0.323 | 0.01372 | -0.029 | 0.78601 | 0.373 |
| 24730 | RP11-156P1.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-874-3p | 24 | ASTN1 | Sponge network | 0.214 | 0.18246 | -2.389 | 2.0E-5 | 0.373 |
| 24731 | HCG11 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p | 65 | LPP | Sponge network | 0.385 | 0.10067 | -0.459 | 0.04475 | 0.373 |
| 24732 | RP11-798M19.6 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940 | 10 | FOXO4 | Sponge network | -0.612 | 0.00094 | -0.516 | 2.0E-5 | 0.373 |
| 24733 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-93-3p | 16 | RUNX1T1 | Sponge network | -1.483 | 9.0E-5 | -0.344 | 0.2374 | 0.373 |
| 24734 | FENDRR |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | GNAQ | Sponge network | -1.281 | 0.00012 | -0.367 | 0.00102 | 0.373 |
| 24735 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p | 45 | FAT3 | Sponge network | 0.494 | 0.00339 | -1.601 | 0.00051 | 0.373 |
| 24736 | RP11-589P10.7 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-185-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-338-3p | 12 | PBX1 | Sponge network | -2.044 | 0.00066 | -1.206 | 0 | 0.373 |
| 24737 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | RCAN2 | Sponge network | -0.08 | 0.90092 | -0.33 | 0.15172 | 0.373 |
| 24738 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | BNC2 | Sponge network | -1.057 | 0.00029 | -0.491 | 0.09988 | 0.373 |
| 24739 | CTD-2336O2.1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | DTNA | Sponge network | -0.141 | 0.26434 | -1.648 | 1.0E-5 | 0.373 |
| 24740 | EMX2OS |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 24 | RIMBP2 | Sponge network | -0.777 | 0.1134 | -1.968 | 5.0E-5 | 0.373 |
| 24741 | RP5-1139B12.3 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p | 12 | HIP1 | Sponge network | 0.598 | 0.38481 | 0.774 | 0 | 0.373 |
| 24742 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-362-3p | 14 | ADARB1 | Sponge network | 1.294 | 0.0031 | -0.335 | 0.06631 | 0.373 |
| 24743 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 26 | EBF3 | Sponge network | -0.942 | 0 | -0.058 | 0.81437 | 0.373 |
| 24744 | AC005154.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | PIK3CA | Sponge network | 0.848 | 0 | 0.195 | 0.06908 | 0.373 |
| 24745 | SNHG14 |
hsa-miR-10b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 10 | EXTL2 | Sponge network | -1.111 | 0.0003 | 0.109 | 0.37375 | 0.373 |
| 24746 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-5p | 43 | ADARB1 | Sponge network | -0.777 | 0.16667 | -0.335 | 0.06631 | 0.373 |
| 24747 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | AHNAK | Sponge network | -0.942 | 0 | -1.207 | 0 | 0.373 |
| 24748 | FZD10-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 23 | CDH23 | Sponge network | -0.727 | 0.04726 | -0.974 | 0.00034 | 0.373 |
| 24749 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PHC3 | Sponge network | 0.476 | 0.19203 | 0.212 | 0.0499 | 0.373 |
| 24750 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | QKI | Sponge network | -0.342 | 0.02288 | -0.333 | 0.04111 | 0.373 |
| 24751 | CTC-296K1.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-744-3p | 28 | DST | Sponge network | -2.095 | 0.00112 | -0.713 | 0.00096 | 0.373 |
| 24752 | LINC00957 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SEMA3E | Sponge network | -0.421 | 0.0114 | -1.717 | 0.00137 | 0.373 |
| 24753 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 13 | LRRK2 | Sponge network | -2.915 | 0.00015 | -1.136 | 1.0E-5 | 0.373 |
| 24754 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ZNF536 | Sponge network | -0.576 | 0.10121 | -3.115 | 0 | 0.373 |
| 24755 | RP11-130L8.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | TET1 | Sponge network | -0.663 | 0.10887 | -0.135 | 0.52138 | 0.373 |
| 24756 | RP11-46C24.7 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | BCL9 | Sponge network | 0.392 | 0.0278 | 0.321 | 0.00267 | 0.373 |
| 24757 | AC093110.3 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 24 | CCDC6 | Sponge network | 1.044 | 2.0E-5 | 0.27 | 0.02555 | 0.373 |
| 24758 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-485-3p | 16 | PDZD2 | Sponge network | -1.697 | 0.00889 | -0.909 | 3.0E-5 | 0.373 |
| 24759 | EMX2OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | RNF144A | Sponge network | -0.777 | 0.1134 | 0.594 | 0.0009 | 0.373 |
| 24760 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-339-5p;hsa-miR-582-5p | 18 | KIF1B | Sponge network | 0.757 | 0 | -0.118 | 0.32258 | 0.373 |
| 24761 | RP11-119F7.5 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PTCH1 | Sponge network | 0.225 | 0.30644 | -0.1 | 0.6179 | 0.373 |
| 24762 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-877-5p | 25 | PGR | Sponge network | 0.494 | 0.00339 | -1.704 | 0 | 0.373 |
| 24763 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | EGLN1 | Sponge network | 0.673 | 0 | -0.127 | 0.1536 | 0.373 |
| 24764 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p | 12 | CHD1 | Sponge network | 0.917 | 0 | 0.322 | 0.00215 | 0.373 |
| 24765 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-940 | 39 | WDFY3 | Sponge network | 0.331 | 0.57247 | -0.201 | 0.0967 | 0.373 |
| 24766 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | SLITRK3 | Sponge network | -1.203 | 0.03881 | -3.328 | 0 | 0.373 |
| 24767 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | ANK3 | Sponge network | 0.826 | 1.0E-5 | -0.55 | 0.00086 | 0.373 |
| 24768 | AC108488.4 |
hsa-let-7b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | NEDD4 | Sponge network | 0.259 | 0.12542 | 0.02 | 0.89834 | 0.373 |
| 24769 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | JAZF1 | Sponge network | 0.727 | 3.0E-5 | -0.377 | 0.02677 | 0.373 |
| 24770 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p | 18 | BRD3 | Sponge network | 1.04 | 0.00023 | 0.37 | 0.00013 | 0.373 |
| 24771 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-942-5p | 12 | CREBBP | Sponge network | -0.611 | 0.09607 | 0.126 | 0.15186 | 0.373 |
| 24772 | BVES-AS1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-744-3p | 19 | PTPN14 | Sponge network | -2.62 | 0 | 0.144 | 0.34595 | 0.373 |
| 24773 | PCA3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 27 | RSPO2 | Sponge network | -0.709 | 0.18168 | -3.038 | 0 | 0.373 |
| 24774 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-532-5p;hsa-miR-625-5p | 21 | PTPN14 | Sponge network | 0.056 | 0.81195 | 0.144 | 0.34595 | 0.373 |
| 24775 | RP11-359B12.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PDE4DIP | Sponge network | 0.064 | 0.72934 | -0.282 | 0.0257 | 0.373 |
| 24776 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p | 18 | CADM3 | Sponge network | -0.899 | 6.0E-5 | -3.663 | 0 | 0.373 |
| 24777 | RP11-35G9.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p | 19 | CYLD | Sponge network | 0.657 | 4.0E-5 | -0.367 | 0.00171 | 0.373 |
| 24778 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-96-5p | 35 | ZFHX4 | Sponge network | -0.18 | 0.20343 | 0.018 | 0.95574 | 0.373 |
| 24779 | BVES-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-877-5p | 13 | INPP4A | Sponge network | -2.62 | 0 | 0.07 | 0.53563 | 0.373 |
| 24780 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | PHC3 | Sponge network | 0.702 | 7.0E-5 | 0.212 | 0.0499 | 0.373 |
| 24781 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-744-3p | 14 | HIP1 | Sponge network | 0.178 | 0.29106 | 0.774 | 0 | 0.373 |
| 24782 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 44 | IL6ST | Sponge network | 0.299 | 0.57722 | -0.402 | 0.00676 | 0.373 |
| 24783 | DNM3OS |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 55 | NFIB | Sponge network | 0.572 | 0.01722 | 0.023 | 0.90795 | 0.373 |
| 24784 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | SYNE1 | Sponge network | -0.355 | 0.0077 | -0.738 | 0.00316 | 0.373 |
| 24785 | U91328.21 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320b | 14 | DTNA | Sponge network | -0.735 | 0.15983 | -1.648 | 1.0E-5 | 0.373 |
| 24786 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-330-3p | 10 | CADM3 | Sponge network | 0.176 | 0.37402 | -3.663 | 0 | 0.373 |
| 24787 | LINC00173 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-375;hsa-miR-421;hsa-miR-455-5p;hsa-miR-7-1-3p | 15 | AKAP6 | Sponge network | 0.056 | 0.8494 | -1.586 | 3.0E-5 | 0.373 |
| 24788 | RP11-216B9.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 12 | NRK | Sponge network | 0.174 | 0.30034 | 0.656 | 0.19217 | 0.373 |
| 24789 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-337-3p;hsa-miR-3614-5p;hsa-miR-582-5p | 11 | MOB1B | Sponge network | 2.368 | 2.0E-5 | 0.197 | 0.15266 | 0.373 |
| 24790 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-935 | 26 | EBF1 | Sponge network | -0.405 | 0.22013 | -0.384 | 0.08856 | 0.373 |
| 24791 | RP11-2H8.2 |
hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 1.089 | 0 | 0.114 | 0.30638 | 0.373 |
| 24792 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | CDKN1C | Sponge network | 0.219 | 0.1516 | -0.929 | 0.00033 | 0.373 |
| 24793 | ACTA2-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PDE4DIP | Sponge network | 0.194 | 0.64278 | -0.282 | 0.0257 | 0.373 |
| 24794 | RP11-403I13.8 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 10 | CNTNAP1 | Sponge network | 0.619 | 0.00157 | 0.358 | 0.13727 | 0.373 |
| 24795 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 13 | LHX6 | Sponge network | -0.318 | 0.65847 | -0.087 | 0.69193 | 0.373 |
| 24796 | RP11-57H14.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 12 | ROBO1 | Sponge network | 0.083 | 0.66405 | -0.009 | 0.96154 | 0.373 |
| 24797 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | TSC1 | Sponge network | 0.523 | 9.0E-5 | 0.103 | 0.26071 | 0.373 |
| 24798 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 25 | USP12 | Sponge network | 0.335 | 0.20596 | -0.102 | 0.40768 | 0.373 |
| 24799 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 28 | BNC2 | Sponge network | 0.381 | 0.25967 | -0.491 | 0.09988 | 0.373 |
| 24800 | LINC00173 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-539-5p | 13 | RYR2 | Sponge network | 0.056 | 0.8494 | -1.466 | 0.00031 | 0.373 |
| 24801 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MTSS1 | Sponge network | -1.161 | 0.00591 | -0.81 | 0.00035 | 0.373 |
| 24802 | RP11-1006G14.4 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | 0.559 | 0.00026 | 0.358 | 0.13727 | 0.373 |
| 24803 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 10 | ARHGAP28 | Sponge network | -1.375 | 0.08642 | -0.911 | 0.00013 | 0.373 |
| 24804 | NEAT1 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TSC1 | Sponge network | 0.705 | 0.00014 | 0.103 | 0.26071 | 0.373 |
| 24805 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p | 14 | NR2C2 | Sponge network | 0.368 | 0.20093 | 0.21 | 0.03224 | 0.373 |
| 24806 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 48 | BCL2 | Sponge network | -2.46 | 0 | -0.899 | 0 | 0.373 |
| 24807 | ZNF667-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | PIK3CA | Sponge network | -0.976 | 0.00025 | 0.195 | 0.06908 | 0.373 |
| 24808 | RP4-669P10.16 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-424-5p | 10 | PRKAB2 | Sponge network | -0.301 | 0.06597 | -0.367 | 0.01371 | 0.373 |
| 24809 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 24 | BMPR2 | Sponge network | 1.2 | 0.01813 | 0.206 | 0.0635 | 0.373 |
| 24810 | MIR497HG |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 25 | PTPN14 | Sponge network | -1.439 | 4.0E-5 | 0.144 | 0.34595 | 0.373 |
| 24811 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | MLLT10 | Sponge network | 0.539 | 0.03722 | 0.105 | 0.33292 | 0.373 |
| 24812 | RP11-736K20.5 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 26 | AFF1 | Sponge network | -0.358 | 0.17255 | -0.384 | 0.00053 | 0.373 |
| 24813 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 22 | CDH19 | Sponge network | 0.532 | 0.00505 | -3.434 | 0 | 0.373 |
| 24814 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 32 | DDX6 | Sponge network | -1.575 | 6.0E-5 | 0.091 | 0.36428 | 0.373 |
| 24815 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-590-3p | 14 | SLITRK3 | Sponge network | -0.737 | 0.10822 | -3.328 | 0 | 0.373 |
| 24816 | TUG1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-582-3p | 13 | FOXO3 | Sponge network | 0.972 | 0 | -0.04 | 0.72028 | 0.373 |
| 24817 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p | 17 | ZBTB4 | Sponge network | -0.829 | 0.01343 | -0.739 | 0 | 0.373 |
| 24818 | CTC-297N7.9 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-93-3p | 24 | SSBP2 | Sponge network | -2.069 | 0 | -1.58 | 0 | 0.373 |
| 24819 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 27 | TMEM132B | Sponge network | -0.726 | 0.00132 | -0.83 | 0.01084 | 0.373 |
| 24820 | KB-1460A1.5 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 0.603 | 0.00127 | 0.321 | 0.00267 | 0.373 |
| 24821 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | -0.769 | 0.00035 | -0.001 | 0.99669 | 0.373 |
| 24822 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | ARHGAP29 | Sponge network | -0.901 | 0.00019 | 0.131 | 0.42953 | 0.373 |
| 24823 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | NF1 | Sponge network | 1.352 | 0 | 0.51 | 0 | 0.373 |
| 24824 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 38 | DDX6 | Sponge network | 0.415 | 0.14657 | 0.091 | 0.36428 | 0.373 |
| 24825 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | ZNF208 | Sponge network | 0.862 | 0.06213 | -0.4 | 0.22381 | 0.373 |
| 24826 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-493-5p;hsa-miR-590-3p | 12 | CDC73 | Sponge network | 0.873 | 0 | 0.094 | 0.20463 | 0.373 |
| 24827 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 41 | FOXP1 | Sponge network | -0.405 | 0.22013 | 0.042 | 0.74368 | 0.373 |
| 24828 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 12 | ITGB3 | Sponge network | -0.739 | 0.00044 | -0.011 | 0.95751 | 0.373 |
| 24829 | CTA-363E19.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-539-5p | 12 | CLASP2 | Sponge network | 1.055 | 0 | -0.038 | 0.71 | 0.373 |
| 24830 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-93-5p;hsa-miR-940 | 37 | CHD5 | Sponge network | 0.246 | 0.59912 | -0.922 | 0.01521 | 0.373 |
| 24831 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | NALCN | Sponge network | -1.281 | 0.00012 | 1.068 | 0.00237 | 0.373 |
| 24832 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 48 | THRB | Sponge network | 0.056 | 0.81195 | -1.117 | 0.0004 | 0.373 |
| 24833 | RP11-2H8.2 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-766-3p | 12 | ATM | Sponge network | 1.089 | 0 | 0.178 | 0.21568 | 0.373 |
| 24834 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429 | 16 | AFF3 | Sponge network | -0.154 | 0.53954 | -2.373 | 0 | 0.373 |
| 24835 | RP11-119F7.5 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PCDH18 | Sponge network | 0.225 | 0.30644 | 0.216 | 0.24645 | 0.373 |
| 24836 | RP11-121C2.2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p | 15 | CBFB | Sponge network | 1.147 | 0 | 1.061 | 0 | 0.373 |
| 24837 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p | 15 | CLIP1 | Sponge network | 0.95 | 0.15943 | -0.215 | 0.08684 | 0.373 |
| 24838 | LINC00654 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-532-3p | 16 | SYNM | Sponge network | 0.374 | 0.20054 | -2.309 | 1.0E-5 | 0.373 |
| 24839 | RP11-359E10.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-625-5p;hsa-miR-7-5p | 23 | IGFBP5 | Sponge network | 0.299 | 0.57722 | -0.806 | 0.00105 | 0.373 |
| 24840 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | SLITRK6 | Sponge network | -0.737 | 0.06182 | -2.121 | 0 | 0.373 |
| 24841 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | PRKAA2 | Sponge network | -0.351 | 0.11358 | -2.462 | 0 | 0.373 |
| 24842 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 21 | PRKAB2 | Sponge network | -0.318 | 0.65847 | -0.367 | 0.01371 | 0.373 |
| 24843 | RP11-196G18.23 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-200b-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | BCL9 | Sponge network | 0.724 | 0.00039 | 0.321 | 0.00267 | 0.373 |
| 24844 | RP1-151F17.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | STARD13 | Sponge network | 0.222 | 0.29133 | -0.187 | 0.27383 | 0.373 |
| 24845 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-93-3p | 46 | RUNX1T1 | Sponge network | -0.384 | 0.40652 | -0.344 | 0.2374 | 0.373 |
| 24846 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-532-3p | 14 | SYNM | Sponge network | 0.259 | 0.12542 | -2.309 | 1.0E-5 | 0.373 |
| 24847 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-93-3p;hsa-miR-93-5p | 35 | PAFAH1B1 | Sponge network | -2.69 | 0.0001 | -0.429 | 0 | 0.373 |
| 24848 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 25 | MYBL1 | Sponge network | 0.392 | 0.0278 | 0.494 | 0.02152 | 0.373 |
| 24849 | HCG11 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | IGSF10 | Sponge network | 0.385 | 0.10067 | -1.751 | 1.0E-5 | 0.373 |
| 24850 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 19 | PRRX1 | Sponge network | 0.16 | 0.50785 | 1.316 | 0 | 0.373 |
| 24851 | JAZF1-AS1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | RASSF3 | Sponge network | -0.769 | 0.00035 | -0.226 | 0.11691 | 0.373 |
| 24852 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | AKAP11 | Sponge network | 0.848 | 0 | 0.196 | 0.09825 | 0.373 |
| 24853 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-942-5p | 20 | TMEM132B | Sponge network | -4.463 | 0.00025 | -0.83 | 0.01084 | 0.373 |
| 24854 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-628-5p | 27 | ERC1 | Sponge network | 0.349 | 0.01695 | -0.108 | 0.38794 | 0.373 |
| 24855 | LINC00476 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 19 | BAZ2B | Sponge network | -0.615 | 0.00015 | -0.276 | 0.02833 | 0.373 |
| 24856 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p | 11 | PAK3 | Sponge network | -0.405 | 0.22013 | -0.628 | 0.07253 | 0.373 |
| 24857 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 52 | KIF1B | Sponge network | 0.385 | 0.10067 | -0.118 | 0.32258 | 0.373 |
| 24858 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 10 | DMD | Sponge network | -1.255 | 0.00981 | -1.506 | 0 | 0.373 |
| 24859 | EMX2OS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | RELN | Sponge network | -0.777 | 0.1134 | -2.403 | 0 | 0.373 |
| 24860 | LINC00865 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p | 16 | RIMBP2 | Sponge network | -1.249 | 7.0E-5 | -1.968 | 5.0E-5 | 0.373 |
| 24861 | GNG12-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 46 | GAS7 | Sponge network | -0.793 | 7.0E-5 | -0.555 | 0.01602 | 0.373 |
| 24862 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 13 | PTPN11 | Sponge network | 1.757 | 0 | 0.431 | 2.0E-5 | 0.373 |
| 24863 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | FGFR1 | Sponge network | 0.174 | 0.30034 | -0.509 | 0.03957 | 0.373 |
| 24864 | RP11-98I9.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | UBR3 | Sponge network | 0.67 | 0.00048 | -0.021 | 0.82774 | 0.372 |
| 24865 | AP001347.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-5p | 16 | CRTC1 | Sponge network | -1.161 | 0.00591 | -0.116 | 0.28412 | 0.372 |
| 24866 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 11 | PARK2 | Sponge network | -0.738 | 0.21233 | -1.892 | 0 | 0.372 |
| 24867 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p | 13 | BCL6 | Sponge network | -2.531 | 0 | -0.211 | 0.19489 | 0.372 |
| 24868 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-455-5p | 10 | AMPH | Sponge network | -0.154 | 0.78939 | -0.916 | 5.0E-5 | 0.372 |
| 24869 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-93-5p | 33 | RASSF2 | Sponge network | -0.18 | 0.20343 | -0.273 | 0.21961 | 0.372 |
| 24870 | MIR497HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-93-5p | 26 | ACSL6 | Sponge network | -1.439 | 4.0E-5 | -1.593 | 0 | 0.372 |
| 24871 | FZD10-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 38 | NTRK2 | Sponge network | -0.727 | 0.04726 | -0.736 | 0.06495 | 0.372 |
| 24872 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-532-3p | 17 | PBX1 | Sponge network | 0.497 | 0.01451 | -1.206 | 0 | 0.372 |
| 24873 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 50 | THRB | Sponge network | 0.335 | 0.20596 | -1.117 | 0.0004 | 0.372 |
| 24874 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-625-3p | 33 | ZFHX3 | Sponge network | 0.253 | 0.09565 | 0.389 | 0.01363 | 0.372 |
| 24875 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | QKI | Sponge network | 1.153 | 0.00114 | -0.333 | 0.04111 | 0.372 |
| 24876 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p | 34 | RICTOR | Sponge network | 0.101 | 0.60919 | 0.388 | 0.00188 | 0.372 |
| 24877 | RP11-379F4.7 |
hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-654-3p | 13 | DDX6 | Sponge network | 1.316 | 0 | 0.091 | 0.36428 | 0.372 |
| 24878 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 51 | IL6ST | Sponge network | 0.659 | 3.0E-5 | -0.402 | 0.00676 | 0.372 |
| 24879 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | NRXN3 | Sponge network | -0.969 | 2.0E-5 | -0.893 | 0.04186 | 0.372 |
| 24880 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-590-3p | 13 | APC | Sponge network | 0.178 | 0.29106 | -0.255 | 0.04051 | 0.372 |
| 24881 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | PRRX1 | Sponge network | -0.611 | 0.09607 | 1.316 | 0 | 0.372 |
| 24882 | RP11-48B3.4 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p | 16 | CHD5 | Sponge network | 1.757 | 0 | -0.922 | 0.01521 | 0.372 |
| 24883 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 47 | CBX5 | Sponge network | -0.323 | 0.01372 | 0.165 | 0.24446 | 0.372 |
| 24884 | RP11-158K1.3 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | UBR3 | Sponge network | 0.349 | 0.01695 | -0.021 | 0.82774 | 0.372 |
| 24885 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 27 | RNF144A | Sponge network | 1.302 | 7.0E-5 | 0.594 | 0.0009 | 0.372 |
| 24886 | MYLK-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 13 | EYA4 | Sponge network | -1.331 | 3.0E-5 | 0.3 | 0.36876 | 0.372 |
| 24887 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | EPHA5 | Sponge network | -0.811 | 2.0E-5 | -0.957 | 0.01733 | 0.372 |
| 24888 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | TET1 | Sponge network | 0.41 | 0.02214 | -0.135 | 0.52138 | 0.372 |
| 24889 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | WWTR1 | Sponge network | 1.757 | 0 | -0.345 | 0.11997 | 0.372 |
| 24890 | RP11-65J21.3 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-629-5p | 15 | CTIF | Sponge network | -0.557 | 0.11135 | -0.389 | 0.00549 | 0.372 |
| 24891 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | MYO9A | Sponge network | 2.094 | 0 | -0.147 | 0.32373 | 0.372 |
| 24892 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | EPHA7 | Sponge network | 0.259 | 0.12542 | -2.197 | 3.0E-5 | 0.372 |
| 24893 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | MTSS1 | Sponge network | -0.901 | 0.00019 | -0.81 | 0.00035 | 0.372 |
| 24894 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p | 28 | RICTOR | Sponge network | 0.056 | 0.81195 | 0.388 | 0.00188 | 0.372 |
| 24895 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 33 | PRRX1 | Sponge network | -1.111 | 0.0003 | 1.316 | 0 | 0.372 |
| 24896 | PART1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-590-5p | 20 | PDZD2 | Sponge network | -2.925 | 0 | -0.909 | 3.0E-5 | 0.372 |
| 24897 | RP11-38L15.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 16 | AR | Sponge network | 0.344 | 0.3127 | -1.554 | 0 | 0.372 |
| 24898 | CTD-2245F17.3 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-378a-5p;hsa-miR-502-5p | 14 | DTNA | Sponge network | -0.421 | 0.12556 | -1.648 | 1.0E-5 | 0.372 |
| 24899 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 44 | TMEM132B | Sponge network | -1.216 | 0 | -0.83 | 0.01084 | 0.372 |
| 24900 | CTD-2089N3.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-455-5p | 14 | UNC80 | Sponge network | -0.314 | 0.62033 | -1.934 | 0 | 0.372 |
| 24901 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | EDA2R | Sponge network | -0.793 | 7.0E-5 | -0.587 | 0.01598 | 0.372 |
| 24902 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-629-3p | 15 | USP12 | Sponge network | -0.583 | 0.14589 | -0.102 | 0.40768 | 0.372 |
| 24903 | AC093110.3 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p | 12 | TSC1 | Sponge network | 1.044 | 2.0E-5 | 0.103 | 0.26071 | 0.372 |
| 24904 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 20 | ARHGAP28 | Sponge network | -1.161 | 0.00591 | -0.911 | 0.00013 | 0.372 |
| 24905 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-450a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940;hsa-miR-98-5p | 37 | PBX1 | Sponge network | -0.969 | 2.0E-5 | -1.206 | 0 | 0.372 |
| 24906 | TPTEP1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | BTG2 | Sponge network | -1.216 | 0 | -1.763 | 0 | 0.372 |
| 24907 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-342-3p | 10 | ZNRF3 | Sponge network | 1.147 | 0 | 0.637 | 0.00172 | 0.372 |
| 24908 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | 0.335 | 0.20596 | -2.417 | 0 | 0.372 |
| 24909 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-214-5p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 58 | SMAD4 | Sponge network | -0.222 | 0.32445 | -0.313 | 0.00413 | 0.372 |
| 24910 | MALAT1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-495-3p | 11 | PHF6 | Sponge network | 0.782 | 0.00016 | 0.785 | 0 | 0.372 |
| 24911 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 14 | FGFR1 | Sponge network | 0.178 | 0.29106 | -0.509 | 0.03957 | 0.372 |
| 24912 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-671-5p;hsa-miR-940 | 32 | MDM4 | Sponge network | 0.453 | 0.01839 | 0.345 | 0.00762 | 0.372 |
| 24913 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-671-5p | 21 | KIT | Sponge network | -1.047 | 0 | -2.184 | 0 | 0.372 |
| 24914 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p | 17 | ZMYND11 | Sponge network | 0.056 | 0.81195 | -0.2 | 0.05449 | 0.372 |
| 24915 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-629-5p | 52 | KIF1B | Sponge network | -1.645 | 0 | -0.118 | 0.32258 | 0.372 |
| 24916 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-484;hsa-miR-7-1-3p | 15 | TRPS1 | Sponge network | -0.187 | 0.23787 | -0.191 | 0.44109 | 0.372 |
| 24917 | BOLA3-AS1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p | 12 | KIT | Sponge network | -0.246 | 0.35034 | -2.184 | 0 | 0.372 |
| 24918 | RP11-736K20.5 |
hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p | 15 | ZMYND11 | Sponge network | -0.358 | 0.17255 | -0.2 | 0.05449 | 0.372 |
| 24919 | CTC-429P9.5 |
hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | UBR3 | Sponge network | 0.178 | 0.29106 | -0.021 | 0.82774 | 0.372 |
| 24920 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 15 | CNTNAP3 | Sponge network | 0.532 | 0.00505 | -1.465 | 0.00033 | 0.372 |
| 24921 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CLIP1 | Sponge network | -0.709 | 0.18168 | -0.215 | 0.08684 | 0.372 |
| 24922 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | PEG3 | Sponge network | -0.969 | 2.0E-5 | -1.74 | 0 | 0.372 |
| 24923 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p | 12 | HIVEP1 | Sponge network | 0.532 | 0.00505 | 0.368 | 0.00064 | 0.372 |
| 24924 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 52 | ZFHX4 | Sponge network | -2.925 | 0 | 0.018 | 0.95574 | 0.372 |
| 24925 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-629-3p | 10 | ITGA5 | Sponge network | -1.349 | 0.00017 | 0.452 | 0.03524 | 0.372 |
| 24926 | MAP3K14 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 12 | BNC2 | Sponge network | -0.295 | 0.02604 | -0.491 | 0.09988 | 0.372 |
| 24927 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 43 | EPHA7 | Sponge network | -0.969 | 2.0E-5 | -2.197 | 3.0E-5 | 0.372 |
| 24928 | MRVI1-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-452-5p;hsa-miR-98-5p | 12 | TES | Sponge network | -1.092 | 4.0E-5 | -0.348 | 0.00639 | 0.372 |
| 24929 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 28 | ZBTB4 | Sponge network | -0.777 | 0.16667 | -0.739 | 0 | 0.372 |
| 24930 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-590-3p | 10 | ARID5B | Sponge network | 0.178 | 0.29106 | 0.342 | 0.01676 | 0.372 |
| 24931 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 17 | ARID5B | Sponge network | 0.727 | 3.0E-5 | 0.342 | 0.01676 | 0.372 |
| 24932 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-495-3p | 11 | ZNF292 | Sponge network | 1.044 | 2.0E-5 | 0.276 | 0.04148 | 0.372 |
| 24933 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 21 | DDX6 | Sponge network | 1.308 | 0 | 0.091 | 0.36428 | 0.372 |
| 24934 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 22 | HECA | Sponge network | 0.494 | 0.00339 | -0.217 | 0.03463 | 0.372 |
| 24935 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 35 | EPHA3 | Sponge network | -2.925 | 0 | 0.185 | 0.53588 | 0.372 |
| 24936 | U91328.19 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-625-3p;hsa-miR-660-5p | 15 | RIMBP2 | Sponge network | -0.303 | 0.21002 | -1.968 | 5.0E-5 | 0.372 |
| 24937 | CTD-2245F17.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-378a-3p;hsa-miR-940 | 13 | WDFY3 | Sponge network | -0.421 | 0.12556 | -0.201 | 0.0967 | 0.372 |
| 24938 | CTC-301O7.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 12 | ADARB1 | Sponge network | 0.292 | 0.43384 | -0.335 | 0.06631 | 0.372 |
| 24939 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 10 | TBX18 | Sponge network | -0.739 | 0.00044 | 0.843 | 0.02945 | 0.372 |
| 24940 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 18 | NOTCH2 | Sponge network | 0.673 | 0 | 0.138 | 0.35163 | 0.372 |
| 24941 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | CDKN1C | Sponge network | -0.129 | 0.57177 | -0.929 | 0.00033 | 0.372 |
| 24942 | RP11-67L2.2 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | CSDE1 | Sponge network | 0.72 | 0.0001 | -0.493 | 0 | 0.372 |
| 24943 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 24 | HLF | Sponge network | -0.726 | 0.00132 | -2.443 | 0 | 0.372 |
| 24944 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-93-3p | 46 | RUNX1T1 | Sponge network | -0.465 | 0.04703 | -0.344 | 0.2374 | 0.372 |
| 24945 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 15 | FGFR1 | Sponge network | -0.227 | 0.31657 | -0.509 | 0.03957 | 0.372 |
| 24946 | RP11-999E24.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-93-3p | 12 | RPS6KA2 | Sponge network | -0.693 | 0.05629 | -0.57 | 0.00013 | 0.372 |
| 24947 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | 0.572 | 0.01722 | 0.126 | 0.15266 | 0.372 |
| 24948 | RP1-39G22.7 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p;hsa-miR-654-3p | 12 | PHF6 | Sponge network | 0.439 | 0.00313 | 0.785 | 0 | 0.372 |
| 24949 | LINC00641 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 29 | NCOA1 | Sponge network | -0.222 | 0.32445 | -0.432 | 0 | 0.372 |
| 24950 | FAM66C |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 31 | NCOA1 | Sponge network | -0.901 | 0.00019 | -0.432 | 0 | 0.372 |
| 24951 | RP11-35G9.5 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-589-3p | 15 | PRKCB | Sponge network | 0.622 | 0.00015 | -1.313 | 2.0E-5 | 0.372 |
| 24952 | LINC00672 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-3p | 19 | PRKCB | Sponge network | -0.227 | 0.31657 | -1.313 | 2.0E-5 | 0.372 |
| 24953 | ZNF667-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-5p | 10 | PCLO | Sponge network | -0.976 | 0.00025 | -1.843 | 0.00064 | 0.372 |
| 24954 | AC005618.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | BRD3 | Sponge network | 0.862 | 0.06213 | 0.37 | 0.00013 | 0.372 |
| 24955 | RP11-102N12.3 |
hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | CNTN1 | Sponge network | -0.351 | 0.11358 | -2.594 | 0 | 0.372 |
| 24956 | LINC00957 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940 | 20 | LIFR | Sponge network | -0.421 | 0.0114 | -1.794 | 0 | 0.372 |
| 24957 | FAM66C |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -0.901 | 0.00019 | -1.692 | 0 | 0.372 |
| 24958 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | TACC1 | Sponge network | -0.513 | 0.04703 | -0.362 | 0.05406 | 0.372 |
| 24959 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 16 | NF1 | Sponge network | 0.873 | 0 | 0.51 | 0 | 0.372 |
| 24960 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-421 | 10 | BCL6 | Sponge network | 0.056 | 0.8494 | -0.211 | 0.19489 | 0.372 |
| 24961 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-34a-5p;hsa-miR-486-5p;hsa-miR-590-3p | 12 | CEP170 | Sponge network | 0.082 | 0.55654 | 0.943 | 0 | 0.372 |
| 24962 | CTC-559E9.6 |
hsa-miR-125a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p | 10 | PHC3 | Sponge network | 0.601 | 1.0E-5 | 0.212 | 0.0499 | 0.372 |
| 24963 | RP11-473I1.5 |
hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-374b-5p | 10 | DMXL1 | Sponge network | 0.542 | 0.00054 | -0.426 | 0.00056 | 0.372 |
| 24964 | RP11-53O19.3 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | ERCC6 | Sponge network | 1.205 | 0 | 0.23 | 0.015 | 0.372 |
| 24965 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 40 | RAP1A | Sponge network | -2.925 | 0 | -0.929 | 0 | 0.372 |
| 24966 | RP11-1006G14.4 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PHF3 | Sponge network | 0.559 | 0.00026 | 0.126 | 0.19652 | 0.372 |
| 24967 | SNHG14 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CTGF | Sponge network | -1.111 | 0.0003 | 0.321 | 0.11682 | 0.372 |
| 24968 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-542-3p | 10 | HLF | Sponge network | -0.557 | 0.11135 | -2.443 | 0 | 0.372 |
| 24969 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-589-3p | 16 | MAFB | Sponge network | -0.487 | 0.02157 | -0.023 | 0.89865 | 0.372 |
| 24970 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | RNF144A | Sponge network | -0.563 | 0.02545 | 0.594 | 0.0009 | 0.372 |
| 24971 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 29 | ABL2 | Sponge network | 0.194 | 0.64278 | 0.82 | 0 | 0.372 |
| 24972 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | HOOK3 | Sponge network | -0.358 | 0.17255 | 0.273 | 0.09957 | 0.372 |
| 24973 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-940 | 18 | NEDD4 | Sponge network | 0.331 | 0.57247 | 0.02 | 0.89834 | 0.372 |
| 24974 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-654-3p | 18 | BMPR2 | Sponge network | 0.997 | 0 | 0.206 | 0.0635 | 0.372 |
| 24975 | GS1-124K5.11 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | RBBP6 | Sponge network | 0.494 | 0.00339 | 0.163 | 0.05101 | 0.372 |
| 24976 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-98-5p | 25 | ZFHX3 | Sponge network | -2.62 | 0 | 0.389 | 0.01363 | 0.372 |
| 24977 | LINC00865 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | TAL1 | Sponge network | -1.249 | 7.0E-5 | -0.462 | 0.00574 | 0.372 |
| 24978 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-3p | 20 | MOB1B | Sponge network | 0.439 | 0.00313 | 0.197 | 0.15266 | 0.372 |
| 24979 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NRXN3 | Sponge network | 0.559 | 0.00026 | -0.893 | 0.04186 | 0.372 |
| 24980 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 55 | QKI | Sponge network | 0.064 | 0.72934 | -0.333 | 0.04111 | 0.372 |
| 24981 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 35 | PTPRT | Sponge network | 0.246 | 0.59912 | -0.907 | 0.03727 | 0.372 |
| 24982 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | GALNT13 | Sponge network | 0.246 | 0.59912 | -0.857 | 0.07439 | 0.372 |
| 24983 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 20 | TGFBR2 | Sponge network | 0.572 | 0.01722 | -0.243 | 0.11408 | 0.372 |
| 24984 | CTC-296K1.3 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-550a-3p | 13 | NRK | Sponge network | -2.095 | 0.00112 | 0.656 | 0.19217 | 0.372 |
| 24985 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-590-3p | 19 | JAKMIP2 | Sponge network | -0.942 | 0 | -1.388 | 0 | 0.372 |
| 24986 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | TET1 | Sponge network | 0.997 | 0.00066 | -0.135 | 0.52138 | 0.372 |
| 24987 | SNHG14 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 36 | CTIF | Sponge network | -1.111 | 0.0003 | -0.389 | 0.00549 | 0.372 |
| 24988 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-93-3p | 10 | FYN | Sponge network | -0.154 | 0.53954 | -0.131 | 0.37051 | 0.372 |
| 24989 | RP4-791M13.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 31 | CBX5 | Sponge network | 0.419 | 0.0319 | 0.165 | 0.24446 | 0.372 |
| 24990 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p | 24 | ST6GALNAC3 | Sponge network | 0.176 | 0.37402 | -0.987 | 0 | 0.372 |
| 24991 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 34 | HLF | Sponge network | 0.082 | 0.55654 | -2.443 | 0 | 0.372 |
| 24992 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | HOOK3 | Sponge network | 0.214 | 0.18246 | 0.273 | 0.09957 | 0.372 |
| 24993 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | KAT6B | Sponge network | -2.46 | 0 | -0.478 | 0.00018 | 0.372 |
| 24994 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-429 | 11 | GREM1 | Sponge network | -1.654 | 0.00144 | 0.902 | 0.01691 | 0.372 |
| 24995 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-3p | 52 | IL6ST | Sponge network | -0.18 | 0.20343 | -0.402 | 0.00676 | 0.372 |
| 24996 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-576-5p | 10 | EDA2R | Sponge network | -0.405 | 0.22013 | -0.587 | 0.01598 | 0.372 |
| 24997 | TPTEP1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 31 | TOM1L2 | Sponge network | -1.216 | 0 | -0.994 | 0 | 0.372 |
| 24998 | RP11-158K1.3 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | PREX2 | Sponge network | 0.349 | 0.01695 | -0.272 | 0.25382 | 0.372 |
| 24999 | MEG3 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 47 | MDM4 | Sponge network | 0.249 | 0.35902 | 0.345 | 0.00762 | 0.372 |
| 25000 | PSMG3-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-590-3p | 16 | PTCH1 | Sponge network | -0.125 | 0.49414 | -0.1 | 0.6179 | 0.372 |
| 25001 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 19 | ADARB1 | Sponge network | -1.375 | 0.08642 | -0.335 | 0.06631 | 0.372 |
| 25002 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-940 | 13 | TAL1 | Sponge network | 0.082 | 0.55397 | -0.462 | 0.00574 | 0.372 |
| 25003 | RP1-59D14.5 |
hsa-miR-130a-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p | 12 | RASAL2 | Sponge network | 1.162 | 5.0E-5 | 0.869 | 0 | 0.372 |
| 25004 | FGF14-AS2 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-874-3p | 17 | CTIF | Sponge network | -1.582 | 8.0E-5 | -0.389 | 0.00549 | 0.372 |
| 25005 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 16 | PDS5B | Sponge network | 0.225 | 0.30644 | 0.259 | 0.00962 | 0.372 |
| 25006 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-655-3p | 24 | PIK3C2A | Sponge network | 0.75 | 0.00054 | 0.061 | 0.53776 | 0.372 |
| 25007 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p | 15 | CREBBP | Sponge network | -0.154 | 0.78939 | 0.126 | 0.15186 | 0.372 |
| 25008 | RP11-6O2.3 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-550a-3p | 12 | RASSF3 | Sponge network | 0.331 | 0.57247 | -0.226 | 0.11691 | 0.372 |
| 25009 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 25 | GPC6 | Sponge network | -0.513 | 0.04703 | -0.054 | 0.81465 | 0.372 |
| 25010 | RP11-360F5.3 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 12 | MDM4 | Sponge network | 1.867 | 0 | 0.345 | 0.00762 | 0.372 |
| 25011 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-671-5p | 48 | ADARB1 | Sponge network | -0.942 | 0 | -0.335 | 0.06631 | 0.372 |
| 25012 | AC003090.1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | PTCH1 | Sponge network | -2.117 | 0.00161 | -0.1 | 0.6179 | 0.372 |
| 25013 | PVT1 |
hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-497-5p | 10 | TBL1XR1 | Sponge network | 1.985 | 0 | 0.578 | 0 | 0.372 |
| 25014 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p | 10 | BCLAF1 | Sponge network | 0.702 | 7.0E-5 | 0.211 | 0.00684 | 0.372 |
| 25015 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-5p;hsa-miR-330-3p | 13 | ZBTB4 | Sponge network | 0.889 | 9.0E-5 | -0.739 | 0 | 0.372 |
| 25016 | LINC00963 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-96-5p | 19 | CRTC3 | Sponge network | 0.523 | 9.0E-5 | 0.164 | 0.16786 | 0.372 |
| 25017 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-654-3p | 28 | FBXO32 | Sponge network | 0.385 | 0.10067 | -0.495 | 0.07561 | 0.372 |
| 25018 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-590-5p | 10 | PCDH8 | Sponge network | -0.147 | 0.40452 | -0.997 | 0.05366 | 0.372 |
| 25019 | RP11-158K1.3 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PHF3 | Sponge network | 0.349 | 0.01695 | 0.126 | 0.19652 | 0.372 |
| 25020 | AC108676.1 |
hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-654-3p | 18 | RICTOR | Sponge network | 4.346 | 2.0E-5 | 0.388 | 0.00188 | 0.372 |
| 25021 | U91328.19 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-484;hsa-miR-7-5p | 12 | FLNA | Sponge network | -0.303 | 0.21002 | -0.771 | 0.01377 | 0.372 |
| 25022 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-369-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | PRDM2 | Sponge network | 0.931 | 1.0E-5 | -0.258 | 0.00721 | 0.372 |
| 25023 | DNAJC27-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 13 | PHOX2B | Sponge network | -0.969 | 2.0E-5 | -2.68 | 0 | 0.372 |
| 25024 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-582-5p | 23 | KIF1B | Sponge network | 0.993 | 0 | -0.118 | 0.32258 | 0.372 |
| 25025 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 72 | AFF4 | Sponge network | -1.575 | 6.0E-5 | -0.266 | 0.01565 | 0.372 |
| 25026 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 13 | PCDH10 | Sponge network | -0.206 | 0.12942 | -1.644 | 0.0078 | 0.372 |
| 25027 | TRAF3IP2-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | GJA1 | Sponge network | -0.323 | 0.01372 | -0.485 | 0.02179 | 0.372 |
| 25028 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p | 25 | PBX1 | Sponge network | 0.95 | 0.15943 | -1.206 | 0 | 0.372 |
| 25029 | ARHGEF26-AS1 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | CSDE1 | Sponge network | -2.46 | 0 | -0.493 | 0 | 0.372 |
| 25030 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | TSC1 | Sponge network | 0.064 | 0.72934 | 0.103 | 0.26071 | 0.372 |
| 25031 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-429;hsa-miR-96-5p | 10 | DOCK4 | Sponge network | 0.348 | 0.32912 | 0.502 | 0.00108 | 0.372 |
| 25032 | OIP5-AS1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-664a-3p | 19 | RASAL2 | Sponge network | 0.421 | 0.00084 | 0.869 | 0 | 0.372 |
| 25033 | FLNB-AS1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | KIF5B | Sponge network | 1.163 | 0.00018 | 0.362 | 0.00066 | 0.372 |
| 25034 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-942-5p | 24 | DDX6 | Sponge network | 0.765 | 7.0E-5 | 0.091 | 0.36428 | 0.372 |
| 25035 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 10 | UBE2QL1 | Sponge network | -0.246 | 0.35034 | -2.417 | 0 | 0.372 |
| 25036 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | MYO9A | Sponge network | 1.04 | 0.00023 | -0.147 | 0.32373 | 0.372 |
| 25037 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-935 | 29 | DMD | Sponge network | -0.08 | 0.90092 | -1.506 | 0 | 0.372 |
| 25038 | RP11-57H14.4 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 22 | SIRT1 | Sponge network | 0.083 | 0.66405 | 0.109 | 0.2593 | 0.372 |
| 25039 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | KAT6B | Sponge network | 0.064 | 0.72934 | -0.478 | 0.00018 | 0.372 |
| 25040 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 26 | PLSCR4 | Sponge network | -0.615 | 0.00015 | -0.474 | 0.03833 | 0.372 |
| 25041 | ZNF32-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-342-3p;hsa-miR-378c | 10 | BRD3 | Sponge network | 1.552 | 7.0E-5 | 0.37 | 0.00013 | 0.372 |
| 25042 | GS1-124K5.11 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 20 | ABL1 | Sponge network | 0.494 | 0.00339 | -0.215 | 0.07804 | 0.372 |
| 25043 | LINC00403 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-424-5p | 13 | ASTN1 | Sponge network | -3.586 | 0.00032 | -2.389 | 2.0E-5 | 0.372 |
| 25044 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 11 | EPHA5 | Sponge network | 0.959 | 0 | -0.957 | 0.01733 | 0.372 |
| 25045 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | HOOK3 | Sponge network | 0.619 | 0.00157 | 0.273 | 0.09957 | 0.372 |
| 25046 | RP11-597D13.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 23 | SETBP1 | Sponge network | 1.302 | 7.0E-5 | -1.302 | 1.0E-5 | 0.372 |
| 25047 | RP11-166D19.1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NRG3 | Sponge network | -0.576 | 0.10121 | -1.836 | 4.0E-5 | 0.371 |
| 25048 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | ZFHX3 | Sponge network | 0.8 | 0 | 0.389 | 0.01363 | 0.371 |
| 25049 | H19 |
hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p | 28 | CBX5 | Sponge network | 2.439 | 0 | 0.165 | 0.24446 | 0.371 |
| 25050 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 28 | ARHGAP28 | Sponge network | 0.335 | 0.20596 | -0.911 | 0.00013 | 0.371 |
| 25051 | RP11-2H8.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-628-5p | 20 | FOXP1 | Sponge network | 1.089 | 0 | 0.042 | 0.74368 | 0.371 |
| 25052 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DNAJB4 | Sponge network | 0.225 | 0.30644 | -0.7 | 1.0E-5 | 0.371 |
| 25053 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 26 | LRIG1 | Sponge network | -0.769 | 0.00035 | -0.537 | 0.00588 | 0.371 |
| 25054 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | KCNA6 | Sponge network | -0.318 | 0.65847 | -1.531 | 0.00013 | 0.371 |
| 25055 | RP11-531A24.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p | 23 | ERCC4 | Sponge network | 0.75 | 0.00054 | 0.126 | 0.15266 | 0.371 |
| 25056 | LINC00622 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | CBX5 | Sponge network | 1.169 | 0.00028 | 0.165 | 0.24446 | 0.371 |
| 25057 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 33 | PRKAB2 | Sponge network | -0.942 | 0 | -0.367 | 0.01371 | 0.371 |
| 25058 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p | 10 | ST5 | Sponge network | -1.575 | 6.0E-5 | -0.78 | 0 | 0.371 |
| 25059 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 24 | PGR | Sponge network | -0.727 | 0.04726 | -1.704 | 0 | 0.371 |
| 25060 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 14 | DYNC1I1 | Sponge network | 0.199 | 0.38015 | -1.12 | 0.00107 | 0.371 |
| 25061 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-93-5p | 27 | MAF | Sponge network | -0.611 | 0.09607 | -1.257 | 0 | 0.371 |
| 25062 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | LIFR | Sponge network | -2.915 | 0.00015 | -1.794 | 0 | 0.371 |
| 25063 | RP11-248G5.8 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-664a-5p | 16 | CBFB | Sponge network | 1.294 | 0 | 1.061 | 0 | 0.371 |
| 25064 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | ANK2 | Sponge network | 0.657 | 4.0E-5 | -1.603 | 1.0E-5 | 0.371 |
| 25065 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 31 | HLF | Sponge network | -0.465 | 0.04703 | -2.443 | 0 | 0.371 |
| 25066 | MIR143HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 57 | CUX1 | Sponge network | -0.739 | 0.0574 | -0.039 | 0.6705 | 0.371 |
| 25067 | LINC00641 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p | 29 | ASTN1 | Sponge network | -0.222 | 0.32445 | -2.389 | 2.0E-5 | 0.371 |
| 25068 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-940 | 17 | NEDD4 | Sponge network | -0.773 | 0.04073 | 0.02 | 0.89834 | 0.371 |
| 25069 | RP13-516M14.1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-940 | 15 | BAZ2B | Sponge network | 0.389 | 0.04293 | -0.276 | 0.02833 | 0.371 |
| 25070 | RP11-977G19.11 |
hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-940 | 13 | CREB1 | Sponge network | 0.957 | 0 | 0.203 | 0.00537 | 0.371 |
| 25071 | AC138035.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326 | 10 | BRD3 | Sponge network | 1.073 | 0 | 0.37 | 0.00013 | 0.371 |
| 25072 | H19 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p | 16 | TET1 | Sponge network | 2.439 | 0 | -0.135 | 0.52138 | 0.371 |
| 25073 | RP11-216B9.6 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | INPP4A | Sponge network | 0.174 | 0.30034 | 0.07 | 0.53563 | 0.371 |
| 25074 | HCG11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-93-5p | 40 | ADARB1 | Sponge network | 0.385 | 0.10067 | -0.335 | 0.06631 | 0.371 |
| 25075 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p | 20 | SETBP1 | Sponge network | -0.738 | 0.21233 | -1.302 | 1.0E-5 | 0.371 |
| 25076 | RP11-464F9.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-590-3p | 21 | ZFX | Sponge network | 0.959 | 0 | 0.09 | 0.42563 | 0.371 |
| 25077 | DNM3OS |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-7-1-3p | 13 | RASSF3 | Sponge network | 0.572 | 0.01722 | -0.226 | 0.11691 | 0.371 |
| 25078 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | ADAMTSL3 | Sponge network | -1.047 | 0 | -1.559 | 0 | 0.371 |
| 25079 | RP11-226L15.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | ABI2 | Sponge network | 0.8 | 0 | 0.175 | 0.14787 | 0.371 |
| 25080 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-624-5p;hsa-miR-7-1-3p | 15 | BCL11A | Sponge network | -4.546 | 0 | -0.932 | 0.0063 | 0.371 |
| 25081 | FAM157A |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p | 12 | PIK3C2A | Sponge network | 0.813 | 1.0E-5 | 0.061 | 0.53776 | 0.371 |
| 25082 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | KAT6B | Sponge network | -0.49 | 0.04946 | -0.478 | 0.00018 | 0.371 |
| 25083 | RP13-516M14.1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | 0.389 | 0.04293 | -0.704 | 0.00106 | 0.371 |
| 25084 | RP11-498C9.15 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 13 | CBFB | Sponge network | 1.087 | 0 | 1.061 | 0 | 0.371 |
| 25085 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | ABCA10 | Sponge network | -0.096 | 0.67958 | -0.571 | 0.03674 | 0.371 |
| 25086 | C4B-AS1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 20 | BNC2 | Sponge network | 2.306 | 9.0E-5 | -0.491 | 0.09988 | 0.371 |
| 25087 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CACNA2D1 | Sponge network | -0.612 | 0.00094 | -0.846 | 0.00675 | 0.371 |
| 25088 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 32 | IGF1 | Sponge network | 1.302 | 7.0E-5 | -0.204 | 0.5898 | 0.371 |
| 25089 | CTC-296K1.3 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-550a-3p | 13 | SYT4 | Sponge network | -2.095 | 0.00112 | -3.207 | 0 | 0.371 |
| 25090 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | PRKCE | Sponge network | -0.901 | 0.00019 | -0.403 | 0.00056 | 0.371 |
| 25091 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-421 | 18 | NCOA1 | Sponge network | -0.583 | 0.14589 | -0.432 | 0 | 0.371 |
| 25092 | FAM215B |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-7-1-3p | 13 | TET1 | Sponge network | 0.817 | 3.0E-5 | -0.135 | 0.52138 | 0.371 |
| 25093 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 40 | TSHZ3 | Sponge network | -0.727 | 0.04726 | -0.556 | 0.01516 | 0.371 |
| 25094 | LINC01018 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | NCAM2 | Sponge network | -2.915 | 0.00015 | -0.482 | 0.22565 | 0.371 |
| 25095 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-335-3p | 11 | PTPN14 | Sponge network | -0.021 | 0.93598 | 0.144 | 0.34595 | 0.371 |
| 25096 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-96-5p | 14 | ZFHX4 | Sponge network | 0.348 | 0.32912 | 0.018 | 0.95574 | 0.371 |
| 25097 | ADAMTS9-AS2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | LUZP2 | Sponge network | -2.452 | 0 | -0.056 | 0.89416 | 0.371 |
| 25098 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 34 | BCL2 | Sponge network | -0.513 | 0.04703 | -0.899 | 0 | 0.371 |
| 25099 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-421;hsa-miR-7-1-3p | 13 | CNTN1 | Sponge network | -4.477 | 0 | -2.594 | 0 | 0.371 |
| 25100 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-590-5p;hsa-miR-93-5p | 21 | RNASEL | Sponge network | -1.047 | 0 | -0.272 | 0.01796 | 0.371 |
| 25101 | RP11-226L15.5 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | KIF5B | Sponge network | 0.8 | 0 | 0.362 | 0.00066 | 0.371 |
| 25102 | RP11-110I1.11 |
hsa-miR-10b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-590-3p | 11 | ARHGEF12 | Sponge network | 1.034 | 0 | 0.09 | 0.35693 | 0.371 |
| 25103 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-193b-3p | 11 | MYBL1 | Sponge network | 1.056 | 0 | 0.494 | 0.02152 | 0.371 |
| 25104 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 13 | HOOK3 | Sponge network | 1.302 | 7.0E-5 | 0.273 | 0.09957 | 0.371 |
| 25105 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | PCDH10 | Sponge network | 0.064 | 0.72934 | -1.644 | 0.0078 | 0.371 |
| 25106 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 24 | SCN9A | Sponge network | 1.757 | 0 | -0.525 | 0.10593 | 0.371 |
| 25107 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | LRRC55 | Sponge network | -0.611 | 0.09607 | -0.026 | 0.93163 | 0.371 |
| 25108 | AC083843.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p | 15 | PIK3C2A | Sponge network | 1.4 | 0 | 0.061 | 0.53776 | 0.371 |
| 25109 | GUSBP11 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 12 | PHF6 | Sponge network | 0.856 | 0 | 0.785 | 0 | 0.371 |
| 25110 | ZSCAN16-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | PRKACA | Sponge network | -0.342 | 0.02288 | -0.255 | 0.0012 | 0.371 |
| 25111 | RP11-182J1.1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 18 | RASSF2 | Sponge network | -0.187 | 0.23787 | -0.273 | 0.21961 | 0.371 |
| 25112 | DNAJC27-AS1 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-98-5p | 11 | HSPB7 | Sponge network | -0.969 | 2.0E-5 | -2.268 | 1.0E-5 | 0.371 |
| 25113 | MEG3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p | 16 | ABCB5 | Sponge network | 0.249 | 0.35902 | -0.956 | 0.04077 | 0.371 |
| 25114 | PCA3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 20 | BAZ2B | Sponge network | -0.709 | 0.18168 | -0.276 | 0.02833 | 0.371 |
| 25115 | OIP5-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | BRD3 | Sponge network | 0.421 | 0.00084 | 0.37 | 0.00013 | 0.371 |
| 25116 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 49 | TRPS1 | Sponge network | 0.72 | 0.0001 | -0.191 | 0.44109 | 0.371 |
| 25117 | CTA-363E19.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-625-3p | 11 | ZFHX3 | Sponge network | 1.055 | 0 | 0.389 | 0.01363 | 0.371 |
| 25118 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | -0.513 | 0.04703 | 0.783 | 0.00072 | 0.371 |
| 25119 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935 | 34 | TSHZ3 | Sponge network | -0.384 | 0.40652 | -0.556 | 0.01516 | 0.371 |
| 25120 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-96-5p | 18 | MED12L | Sponge network | -0.517 | 0.02935 | -0.194 | 0.55299 | 0.371 |
| 25121 | CTD-2303H24.2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | PIK3CA | Sponge network | 0.415 | 0.14657 | 0.195 | 0.06908 | 0.371 |
| 25122 | C20orf166-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-542-3p | 12 | ITGB1 | Sponge network | -2.69 | 0.0001 | 0.524 | 0.00017 | 0.371 |
| 25123 | TP73-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-935 | 19 | GJA1 | Sponge network | -0.563 | 0.02545 | -0.485 | 0.02179 | 0.371 |
| 25124 | HNRNPU-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 10 | DLG1 | Sponge network | 1.601 | 0 | -0.014 | 0.8926 | 0.371 |
| 25125 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 30 | CBX5 | Sponge network | 0.919 | 0 | 0.165 | 0.24446 | 0.371 |
| 25126 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p | 29 | FBXO32 | Sponge network | 0.494 | 0.00339 | -0.495 | 0.07561 | 0.371 |
| 25127 | HOXA-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 23 | SETBP1 | Sponge network | 0.61 | 0.01593 | -1.302 | 1.0E-5 | 0.371 |
| 25128 | AC025171.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | EPHA3 | Sponge network | 0.998 | 0 | 0.185 | 0.53588 | 0.371 |
| 25129 | RP11-296I10.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326 | 12 | BRD3 | Sponge network | 0.592 | 0.00014 | 0.37 | 0.00013 | 0.371 |
| 25130 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | CHD2 | Sponge network | 1.044 | 2.0E-5 | 0.049 | 0.55371 | 0.371 |
| 25131 | RP1-151F17.2 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 12 | RNF180 | Sponge network | 0.222 | 0.29133 | -1.127 | 1.0E-5 | 0.371 |
| 25132 | RP11-57H14.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-484;hsa-miR-590-3p | 12 | FBXO11 | Sponge network | 0.083 | 0.66405 | 0.142 | 0.04938 | 0.371 |
| 25133 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-493-5p | 11 | SEMA3E | Sponge network | -0.384 | 0.40652 | -1.717 | 0.00137 | 0.371 |
| 25134 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 14 | NF1 | Sponge network | 0.903 | 0 | 0.51 | 0 | 0.371 |
| 25135 | RP11-1006G14.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | NAV3 | Sponge network | 0.559 | 0.00026 | 0.212 | 0.47729 | 0.371 |
| 25136 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 30 | USP12 | Sponge network | 0.064 | 0.72934 | -0.102 | 0.40768 | 0.371 |
| 25137 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-484 | 20 | CBX5 | Sponge network | 0.592 | 0.00014 | 0.165 | 0.24446 | 0.371 |
| 25138 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PBRM1 | Sponge network | 0.72 | 0.0001 | -0.029 | 0.78601 | 0.371 |
| 25139 | A1BG-AS1 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p | 17 | BCL2 | Sponge network | -0.164 | 0.45106 | -0.899 | 0 | 0.371 |
| 25140 | RP1-59D14.5 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-628-5p | 10 | VPS53 | Sponge network | 1.162 | 5.0E-5 | 0.103 | 0.22667 | 0.371 |
| 25141 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | AR | Sponge network | -0.08 | 0.90092 | -1.554 | 0 | 0.371 |
| 25142 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 14 | LRRC4 | Sponge network | -1.203 | 0.03881 | -1.774 | 0 | 0.371 |
| 25143 | AC018890.6 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | DOCK4 | Sponge network | 1.61 | 0.00961 | 0.502 | 0.00108 | 0.371 |
| 25144 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | CREB3L2 | Sponge network | -1.349 | 0.00017 | -0.525 | 2.0E-5 | 0.371 |
| 25145 | RP11-344B5.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-664a-3p | 12 | NUAK1 | Sponge network | -0.055 | 0.88482 | 0.904 | 0 | 0.371 |
| 25146 | RP11-758N13.1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-424-5p | 21 | FAT3 | Sponge network | 2.939 | 3.0E-5 | -1.601 | 0.00051 | 0.371 |
| 25147 | TPTEP1 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-542-3p | 10 | ZNF331 | Sponge network | -1.216 | 0 | -1.251 | 0 | 0.371 |
| 25148 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p | 15 | GREM1 | Sponge network | -2.915 | 0.00015 | 0.902 | 0.01691 | 0.371 |
| 25149 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TBX18 | Sponge network | 1.302 | 7.0E-5 | 0.843 | 0.02945 | 0.371 |
| 25150 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | ANK2 | Sponge network | 1.153 | 0.00114 | -1.603 | 1.0E-5 | 0.371 |
| 25151 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-455-3p | 21 | PLSCR4 | Sponge network | 0.331 | 0.57247 | -0.474 | 0.03833 | 0.371 |
| 25152 | RP11-359B12.2 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 18 | CXCL12 | Sponge network | 0.064 | 0.72934 | -1.421 | 0 | 0.371 |
| 25153 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-181a-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-98-5p | 22 | USP12 | Sponge network | -2.62 | 0 | -0.102 | 0.40768 | 0.371 |
| 25154 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 11 | BCL6 | Sponge network | -0.517 | 0.02935 | -0.211 | 0.19489 | 0.371 |
| 25155 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ST6GALNAC3 | Sponge network | -0.737 | 0.10822 | -0.987 | 0 | 0.371 |
| 25156 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | RIMBP2 | Sponge network | -0.421 | 0.0114 | -1.968 | 5.0E-5 | 0.371 |
| 25157 | GNG12-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 10 | FAM117A | Sponge network | -0.793 | 7.0E-5 | -1.379 | 0 | 0.371 |
| 25158 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 46 | FZD3 | Sponge network | 0.537 | 0.00139 | 0.104 | 0.60316 | 0.371 |
| 25159 | RP11-156E6.1 |
hsa-miR-100-5p;hsa-miR-101-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 10 | ATP2A2 | Sponge network | 1.266 | 0 | 0.604 | 0 | 0.371 |
| 25160 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | APC | Sponge network | -0.739 | 0.0574 | -0.255 | 0.04051 | 0.371 |
| 25161 | RP11-57H14.4 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | MRVI1 | Sponge network | 0.083 | 0.66405 | -1.051 | 0.00022 | 0.371 |
| 25162 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-484 | 11 | CUL5 | Sponge network | 0.93 | 0 | 0.026 | 0.77301 | 0.371 |
| 25163 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-429 | 14 | LHX6 | Sponge network | -0.738 | 0.21233 | -0.087 | 0.69193 | 0.371 |
| 25164 | MEG3 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 17 | ID4 | Sponge network | 0.249 | 0.35902 | -1.644 | 0 | 0.371 |
| 25165 | RP11-531A24.5 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 23 | RASSF8 | Sponge network | 0.75 | 0.00054 | -0.122 | 0.65591 | 0.371 |
| 25166 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | ATRX | Sponge network | 0.4 | 0.14611 | 0.261 | 0.04408 | 0.371 |
| 25167 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | TRIM33 | Sponge network | 0.083 | 0.66405 | 0.402 | 7.0E-5 | 0.371 |
| 25168 | BOLA3-AS1 |
hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-935 | 19 | RNF144A | Sponge network | -0.246 | 0.35034 | 0.594 | 0.0009 | 0.371 |
| 25169 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | RICTOR | Sponge network | 0.959 | 0 | 0.388 | 0.00188 | 0.371 |
| 25170 | HOTAIRM1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | ABCA10 | Sponge network | -0.053 | 0.80685 | -0.571 | 0.03674 | 0.371 |
| 25171 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-93-3p | 21 | EBF3 | Sponge network | -0.773 | 0.04073 | -0.058 | 0.81437 | 0.371 |
| 25172 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 25 | KIF1B | Sponge network | -0.285 | 0.2172 | -0.118 | 0.32258 | 0.371 |
| 25173 | AC108488.4 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 18 | WWTR1 | Sponge network | 0.259 | 0.12542 | -0.345 | 0.11997 | 0.371 |
| 25174 | MIR497HG |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | LIMCH1 | Sponge network | -1.439 | 4.0E-5 | -0.915 | 1.0E-5 | 0.371 |
| 25175 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-511-5p | 17 | ZFHX3 | Sponge network | 0.592 | 0.00014 | 0.389 | 0.01363 | 0.371 |
| 25176 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-576-5p | 11 | AKAP12 | Sponge network | 0.253 | 0.09565 | -0.932 | 0.0028 | 0.371 |
| 25177 | HOXB-AS2 |
hsa-miR-101-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-576-5p | 16 | FOXO3 | Sponge network | 1.316 | 0.01106 | -0.04 | 0.72028 | 0.371 |
| 25178 | PWAR6 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | MXI1 | Sponge network | -1.575 | 6.0E-5 | -0.987 | 0 | 0.371 |
| 25179 | RP11-130L8.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-93-5p | 13 | EZH1 | Sponge network | -0.663 | 0.10887 | -0.564 | 1.0E-5 | 0.371 |
| 25180 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 42 | EPHA5 | Sponge network | -0.227 | 0.31657 | -0.957 | 0.01733 | 0.371 |
| 25181 | RP11-685N10.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-3607-3p | 16 | MDM4 | Sponge network | 2.743 | 0 | 0.345 | 0.00762 | 0.371 |
| 25182 | RP5-1139B12.3 |
hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-505-5p | 29 | LPP | Sponge network | 0.598 | 0.38481 | -0.459 | 0.04475 | 0.371 |
| 25183 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-660-5p | 16 | CXXC4 | Sponge network | -1.582 | 8.0E-5 | -0.433 | 0.21055 | 0.371 |
| 25184 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-590-3p | 15 | NR2C2 | Sponge network | 1.757 | 0 | 0.21 | 0.03224 | 0.371 |
| 25185 | RP11-13K12.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-3p | 16 | BCL9 | Sponge network | -0.154 | 0.78939 | 0.321 | 0.00267 | 0.371 |
| 25186 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 69 | THRB | Sponge network | -2.925 | 0 | -1.117 | 0.0004 | 0.371 |
| 25187 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 13 | BCLAF1 | Sponge network | 0.848 | 0 | 0.211 | 0.00684 | 0.371 |
| 25188 | RP11-968O1.5 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p | 11 | WWTR1 | Sponge network | -0.829 | 0.01343 | -0.345 | 0.11997 | 0.371 |
| 25189 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 25 | DOCK4 | Sponge network | -1.575 | 6.0E-5 | 0.502 | 0.00108 | 0.371 |
| 25190 | NEAT1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 22 | UBR3 | Sponge network | 0.705 | 0.00014 | -0.021 | 0.82774 | 0.371 |
| 25191 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLIP1 | Sponge network | -0.901 | 0.00019 | -0.215 | 0.08684 | 0.371 |
| 25192 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | FREM2 | Sponge network | -2.46 | 0 | -0.437 | 0.46353 | 0.371 |
| 25193 | HOXB-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-429 | 12 | PCDH17 | Sponge network | -0.154 | 0.53954 | 0.731 | 2.0E-5 | 0.371 |
| 25194 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | RICTOR | Sponge network | 0.272 | 0.44692 | 0.388 | 0.00188 | 0.371 |
| 25195 | PART1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | CDH10 | Sponge network | -2.925 | 0 | -2.397 | 0 | 0.371 |
| 25196 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | TIMP3 | Sponge network | -2.915 | 0.00015 | -0.546 | 0.02307 | 0.371 |
| 25197 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | BTG2 | Sponge network | -1.697 | 0.00889 | -1.763 | 0 | 0.371 |
| 25198 | RP11-983P16.4 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 15 | CELF4 | Sponge network | 0.41 | 0.02214 | -1.135 | 0.00344 | 0.371 |
| 25199 | SNX29P2 |
hsa-let-7d-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-877-5p;hsa-miR-942-5p | 12 | GLIPR1 | Sponge network | 0.4 | 0.14611 | 0.146 | 0.3276 | 0.371 |
| 25200 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | KAT6B | Sponge network | -0.83 | 0 | -0.478 | 0.00018 | 0.371 |
| 25201 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 31 | MYO9A | Sponge network | 1.302 | 7.0E-5 | -0.147 | 0.32373 | 0.371 |
| 25202 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 35 | JAZF1 | Sponge network | 0.064 | 0.72934 | -0.377 | 0.02677 | 0.371 |
| 25203 | RP11-479G22.8 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p | 11 | PICALM | Sponge network | 1.308 | 0 | 0.086 | 0.23523 | 0.371 |
| 25204 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-942-5p | 18 | NOTCH2 | Sponge network | -0.376 | 0.00337 | 0.138 | 0.35163 | 0.371 |
| 25205 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | DMXL1 | Sponge network | -0.096 | 0.67958 | -0.426 | 0.00056 | 0.371 |
| 25206 | GHRLOS |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-940 | 16 | PTPRT | Sponge network | -1.942 | 0 | -0.907 | 0.03727 | 0.371 |
| 25207 | A2M-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITPKB | Sponge network | -0.08 | 0.90092 | -0.704 | 0.00106 | 0.371 |
| 25208 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 17 | SPTBN4 | Sponge network | 0.997 | 0.00066 | -0.786 | 0.01281 | 0.371 |
| 25209 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-874-3p | 10 | BCLAF1 | Sponge network | 0.083 | 0.66405 | 0.211 | 0.00684 | 0.371 |
| 25210 | RP11-384O8.1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 10 | RHOB | Sponge network | -0.738 | 0.21233 | -1.052 | 0 | 0.371 |
| 25211 | ZNF582-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 22 | PRRX1 | Sponge network | -1.349 | 0.00017 | 1.316 | 0 | 0.371 |
| 25212 | RP11-326G21.1 |
hsa-let-7b-5p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | GAS7 | Sponge network | -0.021 | 0.93598 | -0.555 | 0.01602 | 0.371 |
| 25213 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-93-5p | 24 | ADARB1 | Sponge network | 0.056 | 0.8494 | -0.335 | 0.06631 | 0.371 |
| 25214 | RP11-535M15.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-330-3p;hsa-miR-338-3p | 10 | TGFBR3 | Sponge network | -1.14 | 0.03513 | -1.173 | 1.0E-5 | 0.371 |
| 25215 | TRAF3IP2-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p | 11 | FBXO11 | Sponge network | -0.323 | 0.01372 | 0.142 | 0.04938 | 0.371 |
| 25216 | PCBP1-AS1 |
hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-214-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-652-3p;hsa-miR-940 | 11 | SLC12A6 | Sponge network | -0.567 | 0 | -0.343 | 0.00043 | 0.371 |
| 25217 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p | 11 | AFAP1L2 | Sponge network | -2.452 | 0 | -0.284 | 0.15434 | 0.371 |
| 25218 | RP11-815I9.4 |
hsa-let-7g-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-361-3p;hsa-miR-576-5p | 12 | BRD3 | Sponge network | 0.537 | 0.0006 | 0.37 | 0.00013 | 0.371 |
| 25219 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-96-5p | 28 | NRXN1 | Sponge network | 0.331 | 0.57247 | -3.778 | 0 | 0.371 |
| 25220 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | NRXN3 | Sponge network | 0.862 | 0.06213 | -0.893 | 0.04186 | 0.371 |
| 25221 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p | 17 | CRTC3 | Sponge network | -0.384 | 0.40652 | 0.164 | 0.16786 | 0.371 |
| 25222 | HAND2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 39 | ERC1 | Sponge network | -1.697 | 0.00889 | -0.108 | 0.38794 | 0.371 |
| 25223 | FAM13A-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-92b-3p;hsa-miR-940 | 18 | BAZ2B | Sponge network | -0.83 | 0 | -0.276 | 0.02833 | 0.371 |
| 25224 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | GNA13 | Sponge network | 1.254 | 0 | 0.007 | 0.93884 | 0.371 |
| 25225 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 31 | IGF1 | Sponge network | -0.615 | 0.00015 | -0.204 | 0.5898 | 0.371 |
| 25226 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 12 | SEPT6 | Sponge network | 0.246 | 0.59912 | -0.598 | 0.00181 | 0.371 |
| 25227 | HOXA-AS2 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 11 | LZTS1 | Sponge network | 0.61 | 0.01593 | 1.664 | 0 | 0.371 |
| 25228 | SNHG14 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | PTCH1 | Sponge network | -1.111 | 0.0003 | -0.1 | 0.6179 | 0.371 |
| 25229 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-935 | 50 | PTPRD | Sponge network | -0.942 | 0 | -1.005 | 0.00064 | 0.371 |
| 25230 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | SUFU | Sponge network | -0.487 | 0.02157 | -0.04 | 0.65489 | 0.371 |
| 25231 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-29a-5p | 13 | BCL2 | Sponge network | -4.463 | 0.00025 | -0.899 | 0 | 0.371 |
| 25232 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 47 | CCND2 | Sponge network | -0.096 | 0.67958 | -0.267 | 0.3112 | 0.371 |
| 25233 | LIPE-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-34c-5p;hsa-miR-582-5p | 14 | KLF10 | Sponge network | 0.078 | 0.55803 | -0.783 | 0 | 0.371 |
| 25234 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-93-3p | 42 | IL6ST | Sponge network | -0.693 | 0.05629 | -0.402 | 0.00676 | 0.371 |
| 25235 | H1FX-AS1 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p | 15 | AFF3 | Sponge network | -0.206 | 0.12942 | -2.373 | 0 | 0.371 |
| 25236 | CTA-445C9.14 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 0.718 | 4.0E-5 | 0.578 | 0 | 0.371 |
| 25237 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | LSAMP | Sponge network | -0.615 | 0.00015 | -0.066 | 0.81708 | 0.371 |
| 25238 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 12 | NALCN | Sponge network | -0.942 | 0 | 1.068 | 0.00237 | 0.371 |
| 25239 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30b-5p | 10 | GPAM | Sponge network | 0.542 | 0.00054 | 0.142 | 0.29732 | 0.371 |
| 25240 | RP11-13K12.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-5p | 14 | KANK1 | Sponge network | -0.154 | 0.78939 | -0.55 | 0.00134 | 0.371 |
| 25241 | RP11-403I13.7 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-744-3p | 49 | DST | Sponge network | 0.395 | 0.15451 | -0.713 | 0.00096 | 0.371 |
| 25242 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-429 | 12 | ELMO1 | Sponge network | -0.125 | 0.49414 | 0.04 | 0.83147 | 0.371 |
| 25243 | HCG11 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PCDH10 | Sponge network | 0.385 | 0.10067 | -1.644 | 0.0078 | 0.371 |
| 25244 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 23 | HIP1 | Sponge network | 0.194 | 0.64278 | 0.774 | 0 | 0.371 |
| 25245 | USP3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-93-5p | 27 | NIN | Sponge network | -0.162 | 0.47687 | -0.253 | 0.13794 | 0.371 |
| 25246 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 22 | USP6 | Sponge network | 0.848 | 0 | 0.188 | 0.3871 | 0.371 |
| 25247 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 11 | KCNAB1 | Sponge network | -0.342 | 0.02288 | -0.755 | 2.0E-5 | 0.371 |
| 25248 | LINC00648 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | ASTN1 | Sponge network | 0.633 | 0.51678 | -2.389 | 2.0E-5 | 0.371 |
| 25249 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-192-3p;hsa-miR-200b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 16 | FZD3 | Sponge network | 1.055 | 0 | 0.104 | 0.60316 | 0.371 |
| 25250 | HAND2-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | -1.697 | 0.00889 | -1.379 | 0 | 0.37 |
| 25251 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | KCNA6 | Sponge network | -0.187 | 0.23787 | -1.531 | 0.00013 | 0.37 |
| 25252 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 31 | EPHA7 | Sponge network | 0.41 | 0.02214 | -2.197 | 3.0E-5 | 0.37 |
| 25253 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-7-1-3p | 13 | NUAK1 | Sponge network | 0.225 | 0.30644 | 0.904 | 0 | 0.37 |
| 25254 | FZD10-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | PCDH9 | Sponge network | -0.727 | 0.04726 | -1.823 | 4.0E-5 | 0.37 |
| 25255 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-495-3p | 11 | NF1 | Sponge network | 1.056 | 0 | 0.51 | 0 | 0.37 |
| 25256 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | ARNT2 | Sponge network | -0.899 | 6.0E-5 | -0.332 | 0.25851 | 0.37 |
| 25257 | LINC00957 |
hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DACT1 | Sponge network | -0.421 | 0.0114 | 0.783 | 0.00072 | 0.37 |
| 25258 | RP11-574K11.29 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | KIF5B | Sponge network | 0.997 | 0 | 0.362 | 0.00066 | 0.37 |
| 25259 | SH3RF3-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p | 16 | KCNJ5 | Sponge network | 0.889 | 9.0E-5 | -0.281 | 0.3339 | 0.37 |
| 25260 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | NR4A3 | Sponge network | -0.811 | 2.0E-5 | -1.385 | 0 | 0.37 |
| 25261 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 37 | ZFHX4 | Sponge network | 0.4 | 0.14611 | 0.018 | 0.95574 | 0.37 |
| 25262 | HOTAIRM1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MED12L | Sponge network | -0.053 | 0.80685 | -0.194 | 0.55299 | 0.37 |
| 25263 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PRDM11 | Sponge network | -0.563 | 0.02545 | -0.252 | 0.15511 | 0.37 |
| 25264 | CTA-363E19.2 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-485-3p | 10 | SIRT1 | Sponge network | 1.055 | 0 | 0.109 | 0.2593 | 0.37 |
| 25265 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577 | 21 | TET1 | Sponge network | -2.69 | 0.0001 | -0.135 | 0.52138 | 0.37 |
| 25266 | LINC00865 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-629-5p | 12 | UVRAG | Sponge network | -1.249 | 7.0E-5 | -0.017 | 0.83865 | 0.37 |
| 25267 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-576-5p | 18 | ERCC4 | Sponge network | -0.611 | 0.09607 | 0.126 | 0.15266 | 0.37 |
| 25268 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | CLASP2 | Sponge network | -0.487 | 0.02157 | -0.038 | 0.71 | 0.37 |
| 25269 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-589-3p | 24 | RNASEL | Sponge network | -0.487 | 0.02157 | -0.272 | 0.01796 | 0.37 |
| 25270 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p | 10 | EPHA5 | Sponge network | 1.344 | 0 | -0.957 | 0.01733 | 0.37 |
| 25271 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | PDE4DIP | Sponge network | -0.738 | 0.21233 | -0.282 | 0.0257 | 0.37 |
| 25272 | FGD5-AS1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p | 21 | DDX3X | Sponge network | 0.401 | 0.00066 | -0.152 | 0.07686 | 0.37 |
| 25273 | CKMT2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-877-5p | 35 | AFF1 | Sponge network | 0.082 | 0.55654 | -0.384 | 0.00053 | 0.37 |
| 25274 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LHFP | Sponge network | -0.899 | 6.0E-5 | -0.167 | 0.41371 | 0.37 |
| 25275 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | SYNE1 | Sponge network | 0.898 | 1.0E-5 | -0.738 | 0.00316 | 0.37 |
| 25276 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | HACE1 | Sponge network | 0.559 | 0.00026 | -0.072 | 0.55239 | 0.37 |
| 25277 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 25 | MITF | Sponge network | -0.405 | 0.22013 | -0.769 | 0.00022 | 0.37 |
| 25278 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 27 | ERCC4 | Sponge network | -1.575 | 6.0E-5 | 0.126 | 0.15266 | 0.37 |
| 25279 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-582-3p | 25 | CBL | Sponge network | -0.465 | 0.04703 | 0.291 | 0.01267 | 0.37 |
| 25280 | RP11-156P1.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p | 10 | PIK3IP1 | Sponge network | 0.214 | 0.18246 | -0.187 | 0.19945 | 0.37 |
| 25281 | SOX2-OT |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CTNNA2 | Sponge network | -1.264 | 6.0E-5 | -2.547 | 0 | 0.37 |
| 25282 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 44 | NTRK3 | Sponge network | 0.082 | 0.55397 | -1.77 | 3.0E-5 | 0.37 |
| 25283 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 41 | TRPS1 | Sponge network | 0.082 | 0.55654 | -0.191 | 0.44109 | 0.37 |
| 25284 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p | 11 | NALCN | Sponge network | -2.62 | 0 | 1.068 | 0.00237 | 0.37 |
| 25285 | RP11-244H3.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-590-3p | 22 | ABL2 | Sponge network | 0.659 | 3.0E-5 | 0.82 | 0 | 0.37 |
| 25286 | RP11-774O3.3 |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-7-1-3p | 11 | PACRG | Sponge network | -0.487 | 0.02157 | -2.248 | 0 | 0.37 |
| 25287 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | FOXP1 | Sponge network | 0.249 | 0.35902 | 0.042 | 0.74368 | 0.37 |
| 25288 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TIMP3 | Sponge network | -0.342 | 0.02288 | -0.546 | 0.02307 | 0.37 |
| 25289 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 37 | ZFHX3 | Sponge network | 0.246 | 0.59912 | 0.389 | 0.01363 | 0.37 |
| 25290 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-501-5p | 20 | MAFB | Sponge network | -2.452 | 0 | -0.023 | 0.89865 | 0.37 |
| 25291 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p | 25 | SYNPO2 | Sponge network | -0.969 | 2.0E-5 | -2.342 | 0 | 0.37 |
| 25292 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-660-5p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | ZHX2 | Sponge network | -0.737 | 0.06182 | -0.192 | 0.18459 | 0.37 |
| 25293 | RP11-307B6.3 |
hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-93-3p | 18 | BMPR2 | Sponge network | -4.463 | 0.00025 | 0.206 | 0.0635 | 0.37 |
| 25294 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-940;hsa-miR-98-5p | 26 | PBX1 | Sponge network | -0.773 | 0.04073 | -1.206 | 0 | 0.37 |
| 25295 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 29 | THBS1 | Sponge network | -1.281 | 0.00012 | 0.741 | 0.00386 | 0.37 |
| 25296 | TTC25 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-5p | 12 | AKAP6 | Sponge network | -0.208 | 0.38261 | -1.586 | 3.0E-5 | 0.37 |
| 25297 | RP11-25K21.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-589-3p | 10 | ESR1 | Sponge network | 0.489 | 0.25786 | -1.035 | 3.0E-5 | 0.37 |
| 25298 | CTC-444N24.11 |
hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DOCK4 | Sponge network | 1.153 | 0 | 0.502 | 0.00108 | 0.37 |
| 25299 | AC156455.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-664a-5p | 13 | SOX11 | Sponge network | 1.154 | 0.00041 | 2.68 | 0 | 0.37 |
| 25300 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | DCLK1 | Sponge network | 0.064 | 0.72934 | -1.324 | 0.00014 | 0.37 |
| 25301 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-940 | 48 | QKI | Sponge network | 0.056 | 0.81195 | -0.333 | 0.04111 | 0.37 |
| 25302 | FZD10-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ZNF536 | Sponge network | -0.727 | 0.04726 | -3.115 | 0 | 0.37 |
| 25303 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | KIF1B | Sponge network | 1.153 | 0 | -0.118 | 0.32258 | 0.37 |
| 25304 | LINC00672 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | DCLK1 | Sponge network | -0.227 | 0.31657 | -1.324 | 0.00014 | 0.37 |
| 25305 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PTPRB | Sponge network | -0.896 | 0.00099 | 0.105 | 0.47122 | 0.37 |
| 25306 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | TSC1 | Sponge network | -0.323 | 0.01372 | 0.103 | 0.26071 | 0.37 |
| 25307 | RP11-2H8.2 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-744-3p | 14 | KIF5B | Sponge network | 1.089 | 0 | 0.362 | 0.00066 | 0.37 |
| 25308 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 17 | ABCC9 | Sponge network | 0.75 | 0.00054 | -0.466 | 0.14121 | 0.37 |
| 25309 | PART1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | PRDM5 | Sponge network | -2.925 | 0 | -1.09 | 0.00014 | 0.37 |
| 25310 | RP11-102N12.3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | -0.351 | 0.11358 | 0.309 | 0.17079 | 0.37 |
| 25311 | CTA-941F9.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-502-5p | 11 | PTPN14 | Sponge network | -0.344 | 0.49624 | 0.144 | 0.34595 | 0.37 |
| 25312 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-93-5p | 16 | RASSF2 | Sponge network | 0.348 | 0.32912 | -0.273 | 0.21961 | 0.37 |
| 25313 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 21 | TGFBR2 | Sponge network | -1.697 | 0.00889 | -0.243 | 0.11408 | 0.37 |
| 25314 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-98-5p | 26 | ARHGAP28 | Sponge network | -2.46 | 0 | -0.911 | 0.00013 | 0.37 |
| 25315 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-450b-5p | 11 | ZFHX3 | Sponge network | 0.912 | 0.00014 | 0.389 | 0.01363 | 0.37 |
| 25316 | RP11-384O8.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935;hsa-miR-940 | 42 | GRIK3 | Sponge network | -0.738 | 0.21233 | -3.208 | 0 | 0.37 |
| 25317 | TMEM9B-AS1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-3614-5p;hsa-miR-582-5p;hsa-miR-589-3p | 17 | MOB1B | Sponge network | 0.529 | 0.00552 | 0.197 | 0.15266 | 0.37 |
| 25318 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 11 | HOOK3 | Sponge network | 0.415 | 0.14657 | 0.273 | 0.09957 | 0.37 |
| 25319 | CTB-179K24.3 |
hsa-miR-10b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | NR2C2 | Sponge network | 0.841 | 0.04055 | 0.21 | 0.03224 | 0.37 |
| 25320 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-98-5p | 26 | KCNA6 | Sponge network | -0.773 | 0.04073 | -1.531 | 0.00013 | 0.37 |
| 25321 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 23 | RARB | Sponge network | -1.697 | 0.00889 | -0.825 | 2.0E-5 | 0.37 |
| 25322 | RP11-59H7.3 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 12 | ATM | Sponge network | 2.163 | 0 | 0.178 | 0.21568 | 0.37 |
| 25323 | AP001469.9 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p | 11 | PHF6 | Sponge network | 1.096 | 0 | 0.785 | 0 | 0.37 |
| 25324 | RP11-531A24.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p | 16 | PRUNE2 | Sponge network | 0.75 | 0.00054 | -1.733 | 0.00022 | 0.37 |
| 25325 | USP3-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p | 16 | TLE4 | Sponge network | -0.162 | 0.47687 | -1.157 | 0 | 0.37 |
| 25326 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 15 | TRIM3 | Sponge network | -0.739 | 0.0574 | -0.577 | 0 | 0.37 |
| 25327 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 21 | RET | Sponge network | 0.997 | 0.00066 | -0.833 | 0.03517 | 0.37 |
| 25328 | ZNF582-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 13 | KLF10 | Sponge network | -1.349 | 0.00017 | -0.783 | 0 | 0.37 |
| 25329 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 58 | IL6ST | Sponge network | -0.405 | 0.22013 | -0.402 | 0.00676 | 0.37 |
| 25330 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | PHC3 | Sponge network | 1.055 | 0 | 0.212 | 0.0499 | 0.37 |
| 25331 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | SEMA3E | Sponge network | -0.896 | 0.00099 | -1.717 | 0.00137 | 0.37 |
| 25332 | LINC00963 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-93-3p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | 0.523 | 9.0E-5 | 0.358 | 0.13727 | 0.37 |
| 25333 | AC108488.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-7-1-3p | 21 | DIP2C | Sponge network | 0.259 | 0.12542 | -0.436 | 0.01713 | 0.37 |
| 25334 | NR2F1-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-5p | 15 | EHD3 | Sponge network | -0.49 | 0.04946 | -0.484 | 0.01747 | 0.37 |
| 25335 | FZD10-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 37 | IGFBP5 | Sponge network | -0.727 | 0.04726 | -0.806 | 0.00105 | 0.37 |
| 25336 | RP11-344B5.2 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-224-5p | 15 | BCL2 | Sponge network | -0.055 | 0.88482 | -0.899 | 0 | 0.37 |
| 25337 | MEG3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-942-5p | 16 | ERRFI1 | Sponge network | 0.249 | 0.35902 | -0.798 | 1.0E-5 | 0.37 |
| 25338 | RP5-894A10.6 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-495-3p | 10 | GSK3B | Sponge network | 0.697 | 9.0E-5 | 0.266 | 0.00045 | 0.37 |
| 25339 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3913-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 29 | ZBTB4 | Sponge network | -1.047 | 0 | -0.739 | 0 | 0.37 |
| 25340 | AC009120.6 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-3614-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | TSC1 | Sponge network | 0.919 | 0 | 0.103 | 0.26071 | 0.37 |
| 25341 | RP11-195F19.9 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-93-5p | 15 | IL17RD | Sponge network | -0.726 | 0.00132 | -0.019 | 0.94873 | 0.37 |
| 25342 | DIO3OS |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-660-5p | 23 | PCDH9 | Sponge network | -0.773 | 0.04073 | -1.823 | 4.0E-5 | 0.37 |
| 25343 | LINC00641 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 24 | CDH19 | Sponge network | -0.222 | 0.32445 | -3.434 | 0 | 0.37 |
| 25344 | TRAF3IP2-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 39 | IGFBP5 | Sponge network | -0.323 | 0.01372 | -0.806 | 0.00105 | 0.37 |
| 25345 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3065-3p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 36 | CBL | Sponge network | 0.194 | 0.64278 | 0.291 | 0.01267 | 0.37 |
| 25346 | RP11-554A11.4 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-628-5p | 16 | RIMS2 | Sponge network | -0.583 | 0.14589 | -1.365 | 0.00208 | 0.37 |
| 25347 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | HERC2 | Sponge network | 1.11 | 0 | 0.114 | 0.30638 | 0.37 |
| 25348 | TRIM52-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-361-3p | 18 | ADARB1 | Sponge network | -0.418 | 0.00068 | -0.335 | 0.06631 | 0.37 |
| 25349 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-96-5p | 20 | CRTC3 | Sponge network | -2.915 | 0.00015 | 0.164 | 0.16786 | 0.37 |
| 25350 | LINC00473 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p | 35 | NTRK2 | Sponge network | -1.203 | 0.03881 | -0.736 | 0.06495 | 0.37 |
| 25351 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-93-5p | 16 | RNASEL | Sponge network | -0.227 | 0.31657 | -0.272 | 0.01796 | 0.37 |
| 25352 | RP11-545G3.1 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-424-5p | 15 | MDM4 | Sponge network | 2.622 | 0.00025 | 0.345 | 0.00762 | 0.37 |
| 25353 | AC010226.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 16 | DIP2C | Sponge network | -0.811 | 2.0E-5 | -0.436 | 0.01713 | 0.37 |
| 25354 | RP11-244H3.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-744-3p | 21 | PTPN14 | Sponge network | 0.659 | 3.0E-5 | 0.144 | 0.34595 | 0.37 |
| 25355 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 42 | ERBB4 | Sponge network | -0.901 | 0.00019 | -1.975 | 2.0E-5 | 0.37 |
| 25356 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p | 10 | PDS5B | Sponge network | 0.817 | 3.0E-5 | 0.259 | 0.00962 | 0.37 |
| 25357 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | TAL1 | Sponge network | 0.997 | 0.00066 | -0.462 | 0.00574 | 0.37 |
| 25358 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-628-5p | 32 | MEF2C | Sponge network | -0.154 | 0.78939 | -0.499 | 0.01448 | 0.37 |
| 25359 | TPTEP1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 46 | CREB3L2 | Sponge network | -1.216 | 0 | -0.525 | 2.0E-5 | 0.37 |
| 25360 | PTOV1-AS1 |
hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-654-3p | 11 | RICTOR | Sponge network | 0.87 | 0 | 0.388 | 0.00188 | 0.37 |
| 25361 | MIR497HG |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 10 | RASSF3 | Sponge network | -1.439 | 4.0E-5 | -0.226 | 0.11691 | 0.37 |
| 25362 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 37 | GAS7 | Sponge network | 0.385 | 0.10067 | -0.555 | 0.01602 | 0.37 |
| 25363 | GNG12-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 26 | SYT4 | Sponge network | -0.793 | 7.0E-5 | -3.207 | 0 | 0.37 |
| 25364 | AC006042.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p | 13 | MDM4 | Sponge network | 1.671 | 2.0E-5 | 0.345 | 0.00762 | 0.37 |
| 25365 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | HDAC9 | Sponge network | -1.697 | 0.00889 | -0.755 | 0.00495 | 0.37 |
| 25366 | RP11-158K1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | CREB1 | Sponge network | 0.349 | 0.01695 | 0.203 | 0.00537 | 0.37 |
| 25367 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-454-3p | 11 | RARB | Sponge network | -2.095 | 0.00112 | -0.825 | 2.0E-5 | 0.37 |
| 25368 | LINC00969 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p | 17 | SON | Sponge network | 0.683 | 1.0E-5 | 0.168 | 0.04406 | 0.37 |
| 25369 | SNHG16 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-664a-3p | 10 | ITCH | Sponge network | 0.961 | 0 | 0.084 | 0.34058 | 0.37 |
| 25370 | HOXD-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-500a-5p | 14 | PRKAA2 | Sponge network | -0.208 | 0.65592 | -2.462 | 0 | 0.37 |
| 25371 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-940 | 32 | CHD5 | Sponge network | -0.738 | 0.21233 | -0.922 | 0.01521 | 0.37 |
| 25372 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-493-5p | 25 | TMEM132B | Sponge network | -2.095 | 0.00112 | -0.83 | 0.01084 | 0.37 |
| 25373 | RP11-758N13.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-3p | 15 | PLSCR4 | Sponge network | 2.939 | 3.0E-5 | -0.474 | 0.03833 | 0.37 |
| 25374 | RP11-728F11.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-7-1-3p | 11 | CXXC4 | Sponge network | 0.238 | 0.70544 | -0.433 | 0.21055 | 0.37 |
| 25375 | BVES-AS1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p | 12 | NR3C2 | Sponge network | -2.62 | 0 | -1.56 | 0 | 0.37 |
| 25376 | RP11-373D23.3 |
hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-493-3p | 10 | MRVI1 | Sponge network | -0.285 | 0.2172 | -1.051 | 0.00022 | 0.37 |
| 25377 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 25 | MAF | Sponge network | -1.161 | 0.00591 | -1.257 | 0 | 0.37 |
| 25378 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 15 | TRIM13 | Sponge network | -0.323 | 0.01372 | 0.183 | 0.06968 | 0.37 |
| 25379 | DIO3OS |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-877-5p | 24 | NAV3 | Sponge network | -0.773 | 0.04073 | 0.212 | 0.47729 | 0.37 |
| 25380 | AC006116.24 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-577 | 10 | PRKAR1A | Sponge network | -0.473 | 0.07602 | -0.063 | 0.50314 | 0.37 |
| 25381 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-664a-3p;hsa-miR-664a-5p | 21 | SOX11 | Sponge network | 0.903 | 0 | 2.68 | 0 | 0.37 |
| 25382 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-502-5p | 13 | MXI1 | Sponge network | -0.517 | 0.02935 | -0.987 | 0 | 0.37 |
| 25383 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PTGFR | Sponge network | 0.082 | 0.55654 | -0.233 | 0.45421 | 0.37 |
| 25384 | LINC00472 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p | 10 | EHD3 | Sponge network | -0.842 | 0.01361 | -0.484 | 0.01747 | 0.37 |
| 25385 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 12 | NALCN | Sponge network | -1.161 | 0.00591 | 1.068 | 0.00237 | 0.37 |
| 25386 | HCG11 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | FAM172A | Sponge network | 0.385 | 0.10067 | -0.57 | 0 | 0.37 |
| 25387 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-484;hsa-miR-590-3p | 10 | EDA2R | Sponge network | -1.886 | 0 | -0.587 | 0.01598 | 0.37 |
| 25388 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-365a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p | 30 | RICTOR | Sponge network | 0.663 | 0.00073 | 0.388 | 0.00188 | 0.37 |
| 25389 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | HIP1 | Sponge network | 0.919 | 0 | 0.774 | 0 | 0.37 |
| 25390 | HAND2-AS1 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | CDKN1C | Sponge network | -1.697 | 0.00889 | -0.929 | 0.00033 | 0.37 |
| 25391 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 25 | TACC1 | Sponge network | -0.693 | 0.05629 | -0.362 | 0.05406 | 0.37 |
| 25392 | FGD5-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p | 28 | ABL2 | Sponge network | 0.401 | 0.00066 | 0.82 | 0 | 0.37 |
| 25393 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p | 10 | PDZD2 | Sponge network | 0.853 | 0.15352 | -0.909 | 3.0E-5 | 0.37 |
| 25394 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-590-5p;hsa-miR-7-1-3p | 37 | WDFY3 | Sponge network | 0.889 | 9.0E-5 | -0.201 | 0.0967 | 0.37 |
| 25395 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-485-3p;hsa-miR-877-5p | 11 | CDH19 | Sponge network | -0.465 | 0.04703 | -3.434 | 0 | 0.37 |
| 25396 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-708-5p | 32 | RAP1A | Sponge network | -0.487 | 0.02157 | -0.929 | 0 | 0.37 |
| 25397 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 11 | MGA | Sponge network | 1.153 | 0 | 0.323 | 0.00792 | 0.37 |
| 25398 | LINC00672 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 33 | GRIK3 | Sponge network | -0.227 | 0.31657 | -3.208 | 0 | 0.37 |
| 25399 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 44 | FZD3 | Sponge network | 0.75 | 0.00054 | 0.104 | 0.60316 | 0.37 |
| 25400 | CTA-204B4.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 19 | DMD | Sponge network | 0.765 | 7.0E-5 | -1.506 | 0 | 0.37 |
| 25401 | LPP-AS2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p | 15 | PEG3 | Sponge network | -0.18 | 0.20343 | -1.74 | 0 | 0.37 |
| 25402 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-3p | 32 | CBX5 | Sponge network | 0.765 | 7.0E-5 | 0.165 | 0.24446 | 0.37 |
| 25403 | MIR600HG |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-655-3p | 15 | HECA | Sponge network | 0.594 | 0.41522 | -0.217 | 0.03463 | 0.37 |
| 25404 | TRAF3IP2-AS1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 13 | SHPRH | Sponge network | -0.323 | 0.01372 | 0.098 | 0.40994 | 0.37 |
| 25405 | RP11-46C24.7 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577 | 10 | MLLT11 | Sponge network | 0.392 | 0.0278 | -0.661 | 0.01733 | 0.37 |
| 25406 | RP1-39G22.7 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-5p;hsa-miR-590-3p | 14 | BRD3 | Sponge network | 0.439 | 0.00313 | 0.37 | 0.00013 | 0.37 |
| 25407 | PCA3 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | SYT4 | Sponge network | -0.709 | 0.18168 | -3.207 | 0 | 0.37 |
| 25408 | RP11-597D13.9 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p | 16 | DYNC1I1 | Sponge network | 1.302 | 7.0E-5 | -1.12 | 0.00107 | 0.37 |
| 25409 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 50 | FOXP1 | Sponge network | 0.421 | 0.00084 | 0.042 | 0.74368 | 0.37 |
| 25410 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-3127-5p;hsa-miR-378a-3p | 10 | RPS6KA2 | Sponge network | 0.176 | 0.37402 | -0.57 | 0.00013 | 0.37 |
| 25411 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | CACNA2D1 | Sponge network | 0.4 | 0.14611 | -0.846 | 0.00675 | 0.37 |
| 25412 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p | 15 | PDE4DIP | Sponge network | -0.777 | 0.16667 | -0.282 | 0.0257 | 0.37 |
| 25413 | CALML3-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-369-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p | 13 | MACF1 | Sponge network | 0.95 | 0.15943 | 0.019 | 0.90335 | 0.37 |
| 25414 | CTC-296K1.4 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | THBS1 | Sponge network | -2.076 | 0.00548 | 0.741 | 0.00386 | 0.37 |
| 25415 | RP11-13K12.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p | 12 | INPP4A | Sponge network | -0.154 | 0.78939 | 0.07 | 0.53563 | 0.37 |
| 25416 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 16 | HIP1 | Sponge network | 0.862 | 0.06213 | 0.774 | 0 | 0.37 |
| 25417 | CTC-296K1.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-940 | 23 | NTRK2 | Sponge network | -2.095 | 0.00112 | -0.736 | 0.06495 | 0.37 |
| 25418 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | NR4A3 | Sponge network | -1.697 | 0.00889 | -1.385 | 0 | 0.37 |
| 25419 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p | 11 | PDZD2 | Sponge network | -0.668 | 3.0E-5 | -0.909 | 3.0E-5 | 0.37 |
| 25420 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 44 | MEF2C | Sponge network | -1.249 | 7.0E-5 | -0.499 | 0.01448 | 0.37 |
| 25421 | RP11-160O5.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-455-3p;hsa-miR-539-5p | 21 | THRB | Sponge network | -0.327 | 0.07353 | -1.117 | 0.0004 | 0.37 |
| 25422 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLIP1 | Sponge network | 0.572 | 0.01722 | -0.215 | 0.08684 | 0.37 |
| 25423 | RP11-517B11.7 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-590-5p | 19 | UBR3 | Sponge network | 0.706 | 0 | -0.021 | 0.82774 | 0.37 |
| 25424 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-590-5p | 13 | PIK3C2A | Sponge network | 1.134 | 0 | 0.061 | 0.53776 | 0.37 |
| 25425 | THAP9-AS1 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 12 | RASAL2 | Sponge network | 1.079 | 0 | 0.869 | 0 | 0.37 |
| 25426 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 41 | BNC2 | Sponge network | -0.513 | 0.04703 | -0.491 | 0.09988 | 0.37 |
| 25427 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-577;hsa-miR-93-5p | 26 | ZBTB4 | Sponge network | 0.056 | 0.81195 | -0.739 | 0 | 0.37 |
| 25428 | GAS6-AS2 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | TPO | Sponge network | 0.16 | 0.50785 | -1.87 | 2.0E-5 | 0.37 |
| 25429 | HCG11 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | FAT4 | Sponge network | 0.385 | 0.10067 | -0.354 | 0.14694 | 0.37 |
| 25430 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | EBF3 | Sponge network | 0.082 | 0.55654 | -0.058 | 0.81437 | 0.37 |
| 25431 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p | 11 | GAS1 | Sponge network | -0.942 | 0 | -1.047 | 0.00364 | 0.37 |
| 25432 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-3p | 18 | UNC5C | Sponge network | -0.129 | 0.57177 | -0.178 | 0.40214 | 0.37 |
| 25433 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 53 | BMPR2 | Sponge network | 0.782 | 0.00016 | 0.206 | 0.0635 | 0.37 |
| 25434 | CTD-2089N3.1 |
hsa-let-7b-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p | 14 | AR | Sponge network | -0.314 | 0.62033 | -1.554 | 0 | 0.37 |
| 25435 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | LRRC7 | Sponge network | -0.342 | 0.02288 | -1.341 | 0.01193 | 0.37 |
| 25436 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p | 18 | GREM1 | Sponge network | 0.374 | 0.20054 | 0.902 | 0.01691 | 0.37 |
| 25437 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 31 | PGR | Sponge network | 1.302 | 7.0E-5 | -1.704 | 0 | 0.37 |
| 25438 | ZNF582-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-96-5p | 12 | RPS6KA6 | Sponge network | -1.349 | 0.00017 | -2.989 | 0 | 0.37 |
| 25439 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | 0.532 | 0.00505 | 0.309 | 0.17079 | 0.37 |
| 25440 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | GRIA3 | Sponge network | -0.901 | 0.00019 | -1.956 | 0 | 0.37 |
| 25441 | LINC00472 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-7-1-3p | 22 | LIMCH1 | Sponge network | -0.842 | 0.01361 | -0.915 | 1.0E-5 | 0.37 |
| 25442 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | WWTR1 | Sponge network | 0.219 | 0.1516 | -0.345 | 0.11997 | 0.37 |
| 25443 | PWAR6 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 21 | RPS6KA2 | Sponge network | -1.575 | 6.0E-5 | -0.57 | 0.00013 | 0.37 |
| 25444 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-505-3p | 10 | SON | Sponge network | 1.093 | 0 | 0.168 | 0.04406 | 0.37 |
| 25445 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | LRRK2 | Sponge network | -0.612 | 0.00094 | -1.136 | 1.0E-5 | 0.37 |
| 25446 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p | 18 | PHC3 | Sponge network | 1.044 | 2.0E-5 | 0.212 | 0.0499 | 0.37 |
| 25447 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 37 | PIK3R1 | Sponge network | 0.331 | 0.57247 | -0.133 | 0.36854 | 0.37 |
| 25448 | LINC00702 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 55 | PPM1L | Sponge network | -0.737 | 0.06182 | -0.537 | 0.00616 | 0.37 |
| 25449 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | NF1 | Sponge network | 0.494 | 0.00339 | 0.51 | 0 | 0.37 |
| 25450 | AC004540.4 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p | 10 | RBMS3 | Sponge network | -2.549 | 0.00528 | -0.519 | 0.03551 | 0.37 |
| 25451 | HHIP-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | LUZP2 | Sponge network | -0.737 | 0.10822 | -0.056 | 0.89416 | 0.37 |
| 25452 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 18 | THBS1 | Sponge network | 1.757 | 0 | 0.741 | 0.00386 | 0.37 |
| 25453 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | RASSF8 | Sponge network | 0.853 | 0.15352 | -0.122 | 0.65591 | 0.37 |
| 25454 | RP11-66B24.2 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-378c | 11 | MED12L | Sponge network | 0.57 | 0.1866 | -0.194 | 0.55299 | 0.37 |
| 25455 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | RABEP1 | Sponge network | -0.487 | 0.02157 | -0.066 | 0.43142 | 0.37 |
| 25456 | A2M-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 23 | PCDH10 | Sponge network | -0.08 | 0.90092 | -1.644 | 0.0078 | 0.37 |
| 25457 | CTD-2017D11.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-582-5p | 16 | DMD | Sponge network | 0.792 | 0.0049 | -1.506 | 0 | 0.37 |
| 25458 | RP11-282O18.3 |
hsa-miR-101-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 12 | PHF6 | Sponge network | 0.757 | 0 | 0.785 | 0 | 0.37 |
| 25459 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-93-5p | 12 | TRIM3 | Sponge network | -1.654 | 0.00144 | -0.577 | 0 | 0.369 |
| 25460 | RP4-669P10.16 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-629-5p | 30 | KIF1B | Sponge network | -0.301 | 0.06597 | -0.118 | 0.32258 | 0.369 |
| 25461 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-93-5p | 18 | SLITRK3 | Sponge network | 0.249 | 0.35902 | -3.328 | 0 | 0.369 |
| 25462 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 29 | SCN9A | Sponge network | 0.537 | 0.00139 | -0.525 | 0.10593 | 0.369 |
| 25463 | ZNF667-AS1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-508-3p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | -0.976 | 0.00025 | -0.226 | 0.11691 | 0.369 |
| 25464 | FENDRR |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-628-5p;hsa-miR-708-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 32 | TOM1L2 | Sponge network | -1.281 | 0.00012 | -0.994 | 0 | 0.369 |
| 25465 | PWAR6 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PCLO | Sponge network | -1.575 | 6.0E-5 | -1.843 | 0.00064 | 0.369 |
| 25466 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-93-5p;hsa-miR-935 | 11 | DAB2 | Sponge network | 1.153 | 0.00114 | 0.431 | 0.0113 | 0.369 |
| 25467 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 44 | ERBB4 | Sponge network | -1.264 | 6.0E-5 | -1.975 | 2.0E-5 | 0.369 |
| 25468 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | ATRX | Sponge network | -0.612 | 0.00094 | 0.261 | 0.04408 | 0.369 |
| 25469 | LINC00476 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | RIMBP2 | Sponge network | -0.615 | 0.00015 | -1.968 | 5.0E-5 | 0.369 |
| 25470 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-654-3p | 16 | ITCH | Sponge network | 1.11 | 0 | 0.084 | 0.34058 | 0.369 |
| 25471 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p | 23 | SCN9A | Sponge network | -0.154 | 0.78939 | -0.525 | 0.10593 | 0.369 |
| 25472 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ATRX | Sponge network | 0.272 | 0.44692 | 0.261 | 0.04408 | 0.369 |
| 25473 | RAMP2-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | MACF1 | Sponge network | -0.513 | 0.04703 | 0.019 | 0.90335 | 0.369 |
| 25474 | RP11-195F19.9 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-93-5p | 27 | DST | Sponge network | -0.726 | 0.00132 | -0.713 | 0.00096 | 0.369 |
| 25475 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | SYNE1 | Sponge network | -4.477 | 0 | -0.738 | 0.00316 | 0.369 |
| 25476 | RP11-25K19.1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 11 | HERC1 | Sponge network | -1.057 | 0.00029 | -0.196 | 0.08544 | 0.369 |
| 25477 | RP11-6O2.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-374a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-96-5p | 46 | CADM2 | Sponge network | 0.331 | 0.57247 | -3.782 | 0 | 0.369 |
| 25478 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | ATRX | Sponge network | 1.61 | 0.00961 | 0.261 | 0.04408 | 0.369 |
| 25479 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 13 | DCN | Sponge network | -0.727 | 0.04726 | -1.288 | 0 | 0.369 |
| 25480 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-940 | 23 | THBS1 | Sponge network | -0.738 | 0.21233 | 0.741 | 0.00386 | 0.369 |
| 25481 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | ABCB5 | Sponge network | -0.737 | 0.10822 | -0.956 | 0.04077 | 0.369 |
| 25482 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p | 18 | RET | Sponge network | -0.096 | 0.67958 | -0.833 | 0.03517 | 0.369 |
| 25483 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-940;hsa-miR-96-5p | 33 | SNX29 | Sponge network | 0.064 | 0.72934 | -0.34 | 0.01371 | 0.369 |
| 25484 | MEG3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 36 | ERC1 | Sponge network | 0.249 | 0.35902 | -0.108 | 0.38794 | 0.369 |
| 25485 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TIMP3 | Sponge network | 0.101 | 0.60919 | -0.546 | 0.02307 | 0.369 |
| 25486 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-337-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | KDSR | Sponge network | 1.153 | 0 | 0.239 | 0.02216 | 0.369 |
| 25487 | PWAR6 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-96-5p | 22 | IRS1 | Sponge network | -1.575 | 6.0E-5 | -0.026 | 0.88855 | 0.369 |
| 25488 | RP13-516M14.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-577;hsa-miR-940 | 26 | NTRK2 | Sponge network | 0.389 | 0.04293 | -0.736 | 0.06495 | 0.369 |
| 25489 | RP11-701H24.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 10 | MED12L | Sponge network | -1.425 | 0.04204 | -0.194 | 0.55299 | 0.369 |
| 25490 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | CREM | Sponge network | -0.323 | 0.01372 | -0.345 | 0.00258 | 0.369 |
| 25491 | LINC00173 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-421;hsa-miR-539-5p | 11 | SYNPO2 | Sponge network | 0.056 | 0.8494 | -2.342 | 0 | 0.369 |
| 25492 | NR2F1-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | JDP2 | Sponge network | -0.49 | 0.04946 | -0.967 | 0 | 0.369 |
| 25493 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | HACE1 | Sponge network | 0.953 | 0 | -0.072 | 0.55239 | 0.369 |
| 25494 | AC011526.1 |
hsa-miR-141-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-361-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-769-5p | 12 | GRIK3 | Sponge network | 0.342 | 0.03129 | -3.208 | 0 | 0.369 |
| 25495 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p | 20 | MAPK1 | Sponge network | 0.673 | 0 | -0.017 | 0.81465 | 0.369 |
| 25496 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-550a-5p;hsa-miR-590-5p | 13 | TSHZ3 | Sponge network | 2.306 | 9.0E-5 | -0.556 | 0.01516 | 0.369 |
| 25497 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-940 | 55 | WDFY3 | Sponge network | 0.101 | 0.60919 | -0.201 | 0.0967 | 0.369 |
| 25498 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 27 | ZFHX3 | Sponge network | 0.663 | 0.00073 | 0.389 | 0.01363 | 0.369 |
| 25499 | RP11-121C2.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-495-3p;hsa-miR-539-5p | 11 | TSC1 | Sponge network | 1.147 | 0 | 0.103 | 0.26071 | 0.369 |
| 25500 | LINC00672 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | PTGFR | Sponge network | -0.227 | 0.31657 | -0.233 | 0.45421 | 0.369 |
| 25501 | LINC00641 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-98-5p | 19 | PICALM | Sponge network | -0.222 | 0.32445 | 0.086 | 0.23523 | 0.369 |
| 25502 | TPTEP1 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | MSN | Sponge network | -1.216 | 0 | -0.023 | 0.88422 | 0.369 |
| 25503 | RP11-428J1.5 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | DIP2C | Sponge network | 0.538 | 0.00076 | -0.436 | 0.01713 | 0.369 |
| 25504 | ZSCAN16-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 13 | CXCL12 | Sponge network | -0.342 | 0.02288 | -1.421 | 0 | 0.369 |
| 25505 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-889-3p;hsa-miR-93-5p | 25 | UNC80 | Sponge network | -0.811 | 2.0E-5 | -1.934 | 0 | 0.369 |
| 25506 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | TSHZ3 | Sponge network | 0.259 | 0.12542 | -0.556 | 0.01516 | 0.369 |
| 25507 | RP11-686D22.8 |
hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | PIK3CA | Sponge network | 0.898 | 1.0E-5 | 0.195 | 0.06908 | 0.369 |
| 25508 | LINC00473 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 28 | PPP1R9A | Sponge network | -1.203 | 0.03881 | -0.903 | 0.04254 | 0.369 |
| 25509 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | 0.532 | 0.00505 | -1.047 | 0.00364 | 0.369 |
| 25510 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p | 18 | CBFA2T3 | Sponge network | -4.477 | 0 | -1.221 | 1.0E-5 | 0.369 |
| 25511 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-7-1-3p | 18 | APC | Sponge network | -0.611 | 0.09607 | -0.255 | 0.04051 | 0.369 |
| 25512 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-625-3p | 24 | ABL2 | Sponge network | 0.41 | 0.02214 | 0.82 | 0 | 0.369 |
| 25513 | CTD-2184D3.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | ABL2 | Sponge network | 0.272 | 0.44692 | 0.82 | 0 | 0.369 |
| 25514 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 25 | EPHA7 | Sponge network | -0.031 | 0.89063 | -2.197 | 3.0E-5 | 0.369 |
| 25515 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | TRPS1 | Sponge network | 0.131 | 0.8436 | -0.191 | 0.44109 | 0.369 |
| 25516 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-766-3p | 10 | TBL1XR1 | Sponge network | 2.094 | 0 | 0.578 | 0 | 0.369 |
| 25517 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 38 | PIK3R1 | Sponge network | 0.335 | 0.20596 | -0.133 | 0.36854 | 0.369 |
| 25518 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 10 | THBS1 | Sponge network | 1.308 | 0 | 0.741 | 0.00386 | 0.369 |
| 25519 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 10 | PDGFRA | Sponge network | -1.054 | 0.0706 | -0.372 | 0.0037 | 0.369 |
| 25520 | RP11-159D12.2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p;hsa-miR-664a-5p | 17 | CBFB | Sponge network | 1.254 | 0 | 1.061 | 0 | 0.369 |
| 25521 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | DKK3 | Sponge network | 0.056 | 0.81195 | -0.052 | 0.77783 | 0.369 |
| 25522 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | CACNA2D1 | Sponge network | 0.083 | 0.66405 | -0.846 | 0.00675 | 0.369 |
| 25523 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 18 | PTPN11 | Sponge network | 0.603 | 0.00127 | 0.431 | 2.0E-5 | 0.369 |
| 25524 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 17 | UBE4B | Sponge network | 0.537 | 0.00139 | -0.235 | 0.007 | 0.369 |
| 25525 | RP11-13K12.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-92a-1-5p | 11 | LUZP2 | Sponge network | -0.154 | 0.78939 | -0.056 | 0.89416 | 0.369 |
| 25526 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 14 | ZFHX3 | Sponge network | -0.187 | 0.23787 | 0.389 | 0.01363 | 0.369 |
| 25527 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 29 | PCDH17 | Sponge network | 1.302 | 7.0E-5 | 0.731 | 2.0E-5 | 0.369 |
| 25528 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 19 | RARB | Sponge network | -0.323 | 0.01372 | -0.825 | 2.0E-5 | 0.369 |
| 25529 | GNG12-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 32 | ETV1 | Sponge network | -0.793 | 7.0E-5 | -0.096 | 0.67674 | 0.369 |
| 25530 | RP11-822E23.8 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-335-3p;hsa-miR-335-5p | 11 | FMNL3 | Sponge network | -0.777 | 0.16667 | 0.555 | 4.0E-5 | 0.369 |
| 25531 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | HECA | Sponge network | -0.739 | 0.0574 | -0.217 | 0.03463 | 0.369 |
| 25532 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 13 | BHLHE41 | Sponge network | 1.344 | 0 | 0.353 | 0.12753 | 0.369 |
| 25533 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-335-3p;hsa-miR-338-3p | 11 | ELMO1 | Sponge network | -0.777 | 0.16667 | 0.04 | 0.83147 | 0.369 |
| 25534 | ARHGEF26-AS1 |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-542-3p | 10 | CADM4 | Sponge network | -2.46 | 0 | -0.635 | 0.00305 | 0.369 |
| 25535 | RP11-686O6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-577 | 13 | ZFHX3 | Sponge network | 0.453 | 0.01971 | 0.389 | 0.01363 | 0.369 |
| 25536 | LINC00472 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | NIN | Sponge network | -0.842 | 0.01361 | -0.253 | 0.13794 | 0.369 |
| 25537 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p | 21 | RET | Sponge network | -1.697 | 0.00889 | -0.833 | 0.03517 | 0.369 |
| 25538 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | SON | Sponge network | 0.702 | 7.0E-5 | 0.168 | 0.04406 | 0.369 |
| 25539 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-29b-3p;hsa-miR-330-3p | 11 | MAFB | Sponge network | 0.889 | 9.0E-5 | -0.023 | 0.89865 | 0.369 |
| 25540 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 31 | PTPRD | Sponge network | 0.889 | 9.0E-5 | -1.005 | 0.00064 | 0.369 |
| 25541 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-511-5p | 14 | ATRX | Sponge network | 0.93 | 0 | 0.261 | 0.04408 | 0.369 |
| 25542 | U91328.21 |
hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p | 13 | MOB1B | Sponge network | -0.735 | 0.15983 | 0.197 | 0.15266 | 0.369 |
| 25543 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p | 34 | CLASP2 | Sponge network | -0.615 | 0.00015 | -0.038 | 0.71 | 0.369 |
| 25544 | FAM66C |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ZMYND11 | Sponge network | -0.901 | 0.00019 | -0.2 | 0.05449 | 0.369 |
| 25545 | OSER1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-93-5p | 24 | IL17RD | Sponge network | -0.13 | 0.48734 | -0.019 | 0.94873 | 0.369 |
| 25546 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | EPAS1 | Sponge network | -0.49 | 0.04946 | -0.184 | 0.14418 | 0.369 |
| 25547 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 38 | PTPRD | Sponge network | -0.739 | 0.00044 | -1.005 | 0.00064 | 0.369 |
| 25548 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 23 | FOXP1 | Sponge network | -0.021 | 0.93598 | 0.042 | 0.74368 | 0.369 |
| 25549 | LINC00865 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 13 | CDH10 | Sponge network | -1.249 | 7.0E-5 | -2.397 | 0 | 0.369 |
| 25550 | HOXB-AS1 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-940 | 19 | ETV1 | Sponge network | -0.154 | 0.53954 | -0.096 | 0.67674 | 0.369 |
| 25551 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 20 | PRDM2 | Sponge network | -0.421 | 0.0114 | -0.258 | 0.00721 | 0.369 |
| 25552 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | SLC6A15 | Sponge network | -0.737 | 0.06182 | -2.373 | 2.0E-5 | 0.369 |
| 25553 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 30 | CRTC3 | Sponge network | -1.216 | 0 | 0.164 | 0.16786 | 0.369 |
| 25554 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-629-3p | 18 | BHLHE41 | Sponge network | -0.583 | 0.14589 | 0.353 | 0.12753 | 0.369 |
| 25555 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-212-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-493-5p;hsa-miR-590-3p | 14 | MYCBP2 | Sponge network | 0.782 | 0.00016 | -0.027 | 0.82016 | 0.369 |
| 25556 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-93-5p | 13 | TRIM3 | Sponge network | -0.793 | 7.0E-5 | -0.577 | 0 | 0.369 |
| 25557 | RP1-228H13.5 |
hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 12 | RASAL2 | Sponge network | 1.397 | 0 | 0.869 | 0 | 0.369 |
| 25558 | CTC-429P9.5 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | WWTR1 | Sponge network | 0.178 | 0.29106 | -0.345 | 0.11997 | 0.369 |
| 25559 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p | 23 | PTPN14 | Sponge network | -0.469 | 0.08721 | 0.144 | 0.34595 | 0.369 |
| 25560 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p | 28 | KIF1B | Sponge network | 1.106 | 0 | -0.118 | 0.32258 | 0.369 |
| 25561 | BCYRN1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | 0.311 | 0.10344 | -0.366 | 0.01036 | 0.369 |
| 25562 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | LRRC55 | Sponge network | -0.777 | 0.1134 | -0.026 | 0.93163 | 0.369 |
| 25563 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | USP12 | Sponge network | 0.401 | 0.00066 | -0.102 | 0.40768 | 0.369 |
| 25564 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-3p | 38 | PRKAB2 | Sponge network | -2.925 | 0 | -0.367 | 0.01371 | 0.369 |
| 25565 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | QKI | Sponge network | -0.351 | 0.11358 | -0.333 | 0.04111 | 0.369 |
| 25566 | MRVI1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-423-5p;hsa-miR-7-1-3p | 13 | SUFU | Sponge network | -1.092 | 4.0E-5 | -0.04 | 0.65489 | 0.369 |
| 25567 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | LRIG1 | Sponge network | 0.16 | 0.50785 | -0.537 | 0.00588 | 0.369 |
| 25568 | MIR143HG |
hsa-miR-148b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | GNAI2 | Sponge network | -0.739 | 0.0574 | -0.063 | 0.54532 | 0.369 |
| 25569 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | NF1 | Sponge network | 0.79 | 0 | 0.51 | 0 | 0.369 |
| 25570 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p | 14 | PREX2 | Sponge network | 0.064 | 0.72934 | -0.272 | 0.25382 | 0.369 |
| 25571 | MIR22HG |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-942-5p | 15 | ERRFI1 | Sponge network | -1.047 | 0 | -0.798 | 1.0E-5 | 0.369 |
| 25572 | RAMP2-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-484 | 16 | ABL1 | Sponge network | -0.513 | 0.04703 | -0.215 | 0.07804 | 0.369 |
| 25573 | WDFY3-AS2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | MACF1 | Sponge network | -0.942 | 0 | 0.019 | 0.90335 | 0.369 |
| 25574 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 27 | ERC1 | Sponge network | -0.769 | 0.00035 | -0.108 | 0.38794 | 0.369 |
| 25575 | RP4-584D14.7 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-577 | 11 | ZFHX3 | Sponge network | 1.294 | 0.0031 | 0.389 | 0.01363 | 0.369 |
| 25576 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-3p;hsa-miR-7-1-3p | 12 | UBE4B | Sponge network | -1.092 | 4.0E-5 | -0.235 | 0.007 | 0.369 |
| 25577 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -0.777 | 0.16667 | -0.243 | 0.11408 | 0.369 |
| 25578 | AGAP11 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-590-5p | 15 | ABCA10 | Sponge network | -1.645 | 0 | -0.571 | 0.03674 | 0.369 |
| 25579 | OIP5-AS1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 42 | CCDC6 | Sponge network | 0.421 | 0.00084 | 0.27 | 0.02555 | 0.369 |
| 25580 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-92a-3p | 29 | RASSF2 | Sponge network | 0.214 | 0.18246 | -0.273 | 0.21961 | 0.369 |
| 25581 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NAV3 | Sponge network | -0.206 | 0.12942 | 0.212 | 0.47729 | 0.369 |
| 25582 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | PARK2 | Sponge network | 0.082 | 0.55654 | -1.892 | 0 | 0.369 |
| 25583 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | DOCK4 | Sponge network | -0.737 | 0.06182 | 0.502 | 0.00108 | 0.369 |
| 25584 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-590-3p | 17 | NRXN1 | Sponge network | -0.663 | 0.10887 | -3.778 | 0 | 0.369 |
| 25585 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 28 | NRXN1 | Sponge network | 0.4 | 0.14611 | -3.778 | 0 | 0.369 |
| 25586 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | ZMYM2 | Sponge network | 1.205 | 0 | 0.279 | 0.01273 | 0.369 |
| 25587 | USP3-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-495-3p;hsa-miR-625-5p | 12 | BCL9 | Sponge network | -0.162 | 0.47687 | 0.321 | 0.00267 | 0.369 |
| 25588 | RP4-791M13.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-577 | 26 | DST | Sponge network | 0.419 | 0.0319 | -0.713 | 0.00096 | 0.369 |
| 25589 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-582-3p;hsa-miR-590-3p | 12 | UVRAG | Sponge network | 0.083 | 0.66405 | -0.017 | 0.83865 | 0.369 |
| 25590 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p | 14 | RET | Sponge network | -0.342 | 0.02288 | -0.833 | 0.03517 | 0.369 |
| 25591 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-532-5p | 11 | LRRC55 | Sponge network | -1.886 | 0 | -0.026 | 0.93163 | 0.369 |
| 25592 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-3p | 11 | PBRM1 | Sponge network | 1.254 | 0 | -0.029 | 0.78601 | 0.369 |
| 25593 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-21-5p;hsa-miR-424-5p | 14 | UNC80 | Sponge network | -0.187 | 0.23787 | -1.934 | 0 | 0.369 |
| 25594 | HOXA-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 31 | DIP2C | Sponge network | 0.61 | 0.01593 | -0.436 | 0.01713 | 0.369 |
| 25595 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-542-3p | 11 | KANK1 | Sponge network | -0.517 | 0.02935 | -0.55 | 0.00134 | 0.369 |
| 25596 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 18 | TXNIP | Sponge network | -0.355 | 0.0077 | -1.277 | 0 | 0.369 |
| 25597 | FAM157B |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-3p | 20 | KIF1B | Sponge network | 0.92 | 0 | -0.118 | 0.32258 | 0.369 |
| 25598 | CALML3-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-625-5p | 31 | ABI2 | Sponge network | 0.95 | 0.15943 | 0.175 | 0.14787 | 0.369 |
| 25599 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-5p | 33 | TSHZ3 | Sponge network | 0.056 | 0.81195 | -0.556 | 0.01516 | 0.369 |
| 25600 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 33 | BMPR2 | Sponge network | 1.601 | 0 | 0.206 | 0.0635 | 0.369 |
| 25601 | LINC00702 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 22 | IRS1 | Sponge network | -0.737 | 0.06182 | -0.026 | 0.88855 | 0.369 |
| 25602 | RP11-35G9.3 |
hsa-let-7d-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-486-5p;hsa-miR-590-3p | 10 | GLIPR1 | Sponge network | 0.657 | 4.0E-5 | 0.146 | 0.3276 | 0.369 |
| 25603 | C4A-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-628-5p;hsa-miR-93-3p | 26 | RUNX1T1 | Sponge network | 3.345 | 0 | -0.344 | 0.2374 | 0.369 |
| 25604 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-500a-5p;hsa-miR-942-5p | 12 | HLF | Sponge network | -1.116 | 0.23686 | -2.443 | 0 | 0.369 |
| 25605 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-889-3p;hsa-miR-93-5p;hsa-miR-942-5p | 42 | UNC80 | Sponge network | -0.793 | 7.0E-5 | -1.934 | 0 | 0.369 |
| 25606 | ZNF667-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | -0.976 | 0.00025 | 0.524 | 0.00017 | 0.369 |
| 25607 | CD27-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 21 | AKAP6 | Sponge network | 0.177 | 0.16681 | -1.586 | 3.0E-5 | 0.369 |
| 25608 | CTC-487M23.5 |
hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p | 12 | FBXO32 | Sponge network | 0.912 | 0.00014 | -0.495 | 0.07561 | 0.369 |
| 25609 | RP11-48B3.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p | 15 | DIP2C | Sponge network | 1.757 | 0 | -0.436 | 0.01713 | 0.369 |
| 25610 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 21 | MEF2C | Sponge network | -0.663 | 0.10887 | -0.499 | 0.01448 | 0.369 |
| 25611 | RP11-57H14.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PDE4DIP | Sponge network | 0.083 | 0.66405 | -0.282 | 0.0257 | 0.369 |
| 25612 | BMS1P20 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | CCDC6 | Sponge network | 0.34 | 0.00273 | 0.27 | 0.02555 | 0.369 |
| 25613 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 14 | NCAM2 | Sponge network | 0.16 | 0.50785 | -0.482 | 0.22565 | 0.369 |
| 25614 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | KAT6B | Sponge network | 0.335 | 0.20596 | -0.478 | 0.00018 | 0.369 |
| 25615 | AC010226.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | ACSL6 | Sponge network | -0.811 | 2.0E-5 | -1.593 | 0 | 0.369 |
| 25616 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | RET | Sponge network | -0.942 | 0 | -0.833 | 0.03517 | 0.369 |
| 25617 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 52 | WDFY3 | Sponge network | -0.976 | 0.00025 | -0.201 | 0.0967 | 0.369 |
| 25618 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 21 | GPC6 | Sponge network | 0.214 | 0.18246 | -0.054 | 0.81465 | 0.369 |
| 25619 | PART1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p | 21 | RPS6KA2 | Sponge network | -2.925 | 0 | -0.57 | 0.00013 | 0.369 |
| 25620 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-628-5p | 11 | ERC1 | Sponge network | 0.673 | 0 | -0.108 | 0.38794 | 0.369 |
| 25621 | RP11-620J15.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p | 42 | DST | Sponge network | -0.739 | 0.00044 | -0.713 | 0.00096 | 0.369 |
| 25622 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-331-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-5p | 16 | BCL11A | Sponge network | -0.384 | 0.40652 | -0.932 | 0.0063 | 0.369 |
| 25623 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | RICTOR | Sponge network | 1.163 | 0.00018 | 0.388 | 0.00188 | 0.369 |
| 25624 | RP11-983P16.4 |
hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p | 15 | SYNM | Sponge network | 0.41 | 0.02214 | -2.309 | 1.0E-5 | 0.369 |
| 25625 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200c-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-629-3p;hsa-miR-654-3p | 14 | PSIP1 | Sponge network | 0.267 | 0.2937 | -0.005 | 0.96897 | 0.369 |
| 25626 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SYNE1 | Sponge network | -0.737 | 0.10822 | -0.738 | 0.00316 | 0.369 |
| 25627 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 30 | ERBB4 | Sponge network | -1.582 | 8.0E-5 | -1.975 | 2.0E-5 | 0.369 |
| 25628 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 12 | EDA2R | Sponge network | -0.738 | 0.21233 | -0.587 | 0.01598 | 0.369 |
| 25629 | FZD10-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 24 | LIMCH1 | Sponge network | -0.727 | 0.04726 | -0.915 | 1.0E-5 | 0.369 |
| 25630 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-5p | 44 | WDFY3 | Sponge network | 0.078 | 0.55803 | -0.201 | 0.0967 | 0.369 |
| 25631 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p | 14 | NUAK1 | Sponge network | -0.583 | 0.14589 | 0.904 | 0 | 0.369 |
| 25632 | RP11-571M6.17 |
hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p | 11 | DDX6 | Sponge network | 1.013 | 0 | 0.091 | 0.36428 | 0.369 |
| 25633 | RP11-410L14.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-450b-5p | 11 | FBXO32 | Sponge network | -0.052 | 0.76839 | -0.495 | 0.07561 | 0.369 |
| 25634 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | MAF | Sponge network | -0.31 | 0.33871 | -1.257 | 0 | 0.369 |
| 25635 | AC005154.6 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | EPG5 | Sponge network | 0.848 | 0 | 0.101 | 0.3563 | 0.369 |
| 25636 | RP11-13K12.5 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 27 | TMEM132B | Sponge network | -0.318 | 0.65847 | -0.83 | 0.01084 | 0.369 |
| 25637 | RP11-404E16.1 |
hsa-miR-10b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-532-5p | 12 | PTPN14 | Sponge network | 0.097 | 0.91311 | 0.144 | 0.34595 | 0.369 |
| 25638 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 35 | NRXN1 | Sponge network | -0.83 | 0 | -3.778 | 0 | 0.369 |
| 25639 | A2M-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | IL17RD | Sponge network | -0.08 | 0.90092 | -0.019 | 0.94873 | 0.369 |
| 25640 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | TACC1 | Sponge network | -0.18 | 0.20343 | -0.362 | 0.05406 | 0.369 |
| 25641 | LINC00926 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-324-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 19 | NTRK3 | Sponge network | 0.756 | 0.00739 | -1.77 | 3.0E-5 | 0.369 |
| 25642 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-874-3p | 10 | BCLAF1 | Sponge network | 0.924 | 0 | 0.211 | 0.00684 | 0.369 |
| 25643 | RP11-166D19.1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-942-5p | 30 | OPCML | Sponge network | -0.576 | 0.10121 | -2.498 | 0 | 0.369 |
| 25644 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-96-5p | 10 | PIK3CA | Sponge network | -1.654 | 0.00144 | 0.195 | 0.06908 | 0.369 |
| 25645 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | KCNAB1 | Sponge network | -2.117 | 0.00161 | -0.755 | 2.0E-5 | 0.369 |
| 25646 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-577 | 13 | PRKAB2 | Sponge network | -0.473 | 0.07602 | -0.367 | 0.01371 | 0.369 |
| 25647 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 53 | PAFAH1B1 | Sponge network | -0.737 | 0.06182 | -0.429 | 0 | 0.369 |
| 25648 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 19 | LATS2 | Sponge network | 0.199 | 0.38015 | 0.159 | 0.2304 | 0.369 |
| 25649 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | LRRC4 | Sponge network | -0.942 | 0 | -1.774 | 0 | 0.369 |
| 25650 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 22 | GPC6 | Sponge network | 0.385 | 0.10067 | -0.054 | 0.81465 | 0.369 |
| 25651 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | LATS2 | Sponge network | -0.563 | 0.02545 | 0.159 | 0.2304 | 0.369 |
| 25652 | CTC-457L16.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-671-5p | 15 | CYLD | Sponge network | 1.153 | 0.00114 | -0.367 | 0.00171 | 0.369 |
| 25653 | RP11-983P16.4 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-7-5p | 10 | MLLT11 | Sponge network | 0.41 | 0.02214 | -0.661 | 0.01733 | 0.369 |
| 25654 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | DOCK4 | Sponge network | -0.49 | 0.04946 | 0.502 | 0.00108 | 0.368 |
| 25655 | LINC00578 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p | 20 | ESR1 | Sponge network | 0.811 | 0.03228 | -1.035 | 3.0E-5 | 0.368 |
| 25656 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-628-5p | 22 | BMPR2 | Sponge network | 1.361 | 0 | 0.206 | 0.0635 | 0.368 |
| 25657 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p | 11 | CHD1 | Sponge network | 0.832 | 0 | 0.322 | 0.00215 | 0.368 |
| 25658 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-940 | 25 | IGF1 | Sponge network | -0.246 | 0.35034 | -0.204 | 0.5898 | 0.368 |
| 25659 | RP11-981G7.6 |
hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | TAF1 | Sponge network | 0.986 | 0.01022 | 0.39 | 3.0E-5 | 0.368 |
| 25660 | LINC00909 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-664a-5p | 15 | SMAD4 | Sponge network | 0.363 | 0.00601 | -0.313 | 0.00413 | 0.368 |
| 25661 | AC009120.6 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 12 | ERCC6 | Sponge network | 0.919 | 0 | 0.23 | 0.015 | 0.368 |
| 25662 | RP5-1074L1.4 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 13 | GSK3B | Sponge network | 1.923 | 0 | 0.266 | 0.00045 | 0.368 |
| 25663 | MEG3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-660-5p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | ZHX2 | Sponge network | 0.249 | 0.35902 | -0.192 | 0.18459 | 0.368 |
| 25664 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 10 | ZFX | Sponge network | 0.178 | 0.29106 | 0.09 | 0.42563 | 0.368 |
| 25665 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 34 | QKI | Sponge network | -0.318 | 0.65847 | -0.333 | 0.04111 | 0.368 |
| 25666 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-5p | 10 | PRKAB2 | Sponge network | 1.134 | 0 | -0.367 | 0.01371 | 0.368 |
| 25667 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 36 | SNX29 | Sponge network | 0.249 | 0.35902 | -0.34 | 0.01371 | 0.368 |
| 25668 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p | 36 | BMPR2 | Sponge network | 1.757 | 0 | 0.206 | 0.0635 | 0.368 |
| 25669 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 32 | BHLHE41 | Sponge network | -0.737 | 0.06182 | 0.353 | 0.12753 | 0.368 |
| 25670 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 11 | CHD1 | Sponge network | 0.702 | 7.0E-5 | 0.322 | 0.00215 | 0.368 |
| 25671 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 11 | AKAP11 | Sponge network | 1.923 | 0 | 0.196 | 0.09825 | 0.368 |
| 25672 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-935 | 24 | PCDH10 | Sponge network | -0.513 | 0.04703 | -1.644 | 0.0078 | 0.368 |
| 25673 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | EPHA5 | Sponge network | 0.299 | 0.57722 | -0.957 | 0.01733 | 0.368 |
| 25674 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | SUFU | Sponge network | -0.769 | 0.00035 | -0.04 | 0.65489 | 0.368 |
| 25675 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | WWTR1 | Sponge network | -0.513 | 0.04703 | -0.345 | 0.11997 | 0.368 |
| 25676 | FAM66C |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-532-5p | 10 | PIK3IP1 | Sponge network | -0.901 | 0.00019 | -0.187 | 0.19945 | 0.368 |
| 25677 | DNM1P35 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-671-5p | 23 | DLC1 | Sponge network | 0.051 | 0.83388 | -0.382 | 0.05038 | 0.368 |
| 25678 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | -2.915 | 0.00015 | -1.277 | 0 | 0.368 |
| 25679 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p | 10 | ADARB1 | Sponge network | 0.959 | 0 | -0.335 | 0.06631 | 0.368 |
| 25680 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ZNF208 | Sponge network | -0.899 | 6.0E-5 | -0.4 | 0.22381 | 0.368 |
| 25681 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-539-5p | 15 | MOB1B | Sponge network | 1.122 | 0 | 0.197 | 0.15266 | 0.368 |
| 25682 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-3p | 15 | SSBP2 | Sponge network | -0.733 | 0.19469 | -1.58 | 0 | 0.368 |
| 25683 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | CXXC4 | Sponge network | -0.739 | 0.00044 | -0.433 | 0.21055 | 0.368 |
| 25684 | JAZF1-AS1 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | EDNRB | Sponge network | -0.769 | 0.00035 | -0.682 | 0.00138 | 0.368 |
| 25685 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | SLITRK6 | Sponge network | -0.576 | 0.10121 | -2.121 | 0 | 0.368 |
| 25686 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 39 | SPRY3 | Sponge network | -0.222 | 0.32445 | 0.016 | 0.91286 | 0.368 |
| 25687 | BMS1P20 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | ARHGEF12 | Sponge network | 0.34 | 0.00273 | 0.09 | 0.35693 | 0.368 |
| 25688 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-590-3p | 10 | CREBBP | Sponge network | 1.601 | 0 | 0.126 | 0.15186 | 0.368 |
| 25689 | CTD-2314G24.2 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-421 | 12 | PLXNC1 | Sponge network | 0.246 | 0.59912 | 0.569 | 0.00329 | 0.368 |
| 25690 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-660-5p | 15 | RIMBP2 | Sponge network | -0.246 | 0.35034 | -1.968 | 5.0E-5 | 0.368 |
| 25691 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-96-5p | 33 | BCL2 | Sponge network | -1.654 | 0.00144 | -0.899 | 0 | 0.368 |
| 25692 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | EPHA3 | Sponge network | 0.75 | 0.00054 | 0.185 | 0.53588 | 0.368 |
| 25693 | AC011526.1 |
hsa-let-7g-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 13 | PCDH10 | Sponge network | 0.342 | 0.03129 | -1.644 | 0.0078 | 0.368 |
| 25694 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | PPM1A | Sponge network | -0.303 | 0.21002 | -0.623 | 0 | 0.368 |
| 25695 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-628-5p | 25 | FBXO32 | Sponge network | -0.773 | 0.04073 | -0.495 | 0.07561 | 0.368 |
| 25696 | RP11-6N17.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p | 12 | AFF1 | Sponge network | 0.108 | 0.69556 | -0.384 | 0.00053 | 0.368 |
| 25697 | RP11-390K5.6 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-590-3p | 11 | UBR3 | Sponge network | 1.148 | 2.0E-5 | -0.021 | 0.82774 | 0.368 |
| 25698 | PART1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PCLO | Sponge network | -2.925 | 0 | -1.843 | 0.00064 | 0.368 |
| 25699 | CTD-2336O2.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 33 | TRPS1 | Sponge network | -0.141 | 0.26434 | -0.191 | 0.44109 | 0.368 |
| 25700 | LINC00621 |
hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 16 | RNF144A | Sponge network | 0.131 | 0.8436 | 0.594 | 0.0009 | 0.368 |
| 25701 | RP11-121C2.2 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p | 11 | MAT2A | Sponge network | 1.147 | 0 | -0.073 | 0.36195 | 0.368 |
| 25702 | CTC-487M23.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 28 | BMPR2 | Sponge network | 0.912 | 0.00014 | 0.206 | 0.0635 | 0.368 |
| 25703 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-590-5p | 10 | THSD7A | Sponge network | 0.214 | 0.18246 | 0.242 | 0.25154 | 0.368 |
| 25704 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 19 | PDS5B | Sponge network | 1.11 | 0 | 0.259 | 0.00962 | 0.368 |
| 25705 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-193b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PTPN11 | Sponge network | 0.214 | 0.18246 | 0.431 | 2.0E-5 | 0.368 |
| 25706 | JAZF1-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-577;hsa-miR-590-3p | 22 | NRK | Sponge network | -0.769 | 0.00035 | 0.656 | 0.19217 | 0.368 |
| 25707 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-181a-2-3p;hsa-miR-2110 | 10 | USP12 | Sponge network | -0.691 | 0.00146 | -0.102 | 0.40768 | 0.368 |
| 25708 | RP11-212P7.2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ITGB1 | Sponge network | 0.219 | 0.1516 | 0.524 | 0.00017 | 0.368 |
| 25709 | RP1-151F17.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 19 | BAZ2B | Sponge network | 0.222 | 0.29133 | -0.276 | 0.02833 | 0.368 |
| 25710 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-330-3p;hsa-miR-374b-5p;hsa-miR-655-3p | 14 | APC | Sponge network | 0.542 | 0.00054 | -0.255 | 0.04051 | 0.368 |
| 25711 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | DSE | Sponge network | -2.452 | 0 | -0.231 | 0.05347 | 0.368 |
| 25712 | RP5-1092A3.4 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-664a-5p | 13 | CBFB | Sponge network | 1.495 | 0 | 1.061 | 0 | 0.368 |
| 25713 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-98-5p | 20 | PRRX1 | Sponge network | -0.773 | 0.04073 | 1.316 | 0 | 0.368 |
| 25714 | FZD10-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 26 | NAV3 | Sponge network | -0.727 | 0.04726 | 0.212 | 0.47729 | 0.368 |
| 25715 | LINC00094 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-429;hsa-miR-582-5p | 13 | PRKAR1A | Sponge network | 0.253 | 0.09565 | -0.063 | 0.50314 | 0.368 |
| 25716 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-590-3p | 10 | PNRC1 | Sponge network | -1.582 | 8.0E-5 | -0.646 | 0 | 0.368 |
| 25717 | CALML3-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-542-3p | 15 | KANK1 | Sponge network | 0.95 | 0.15943 | -0.55 | 0.00134 | 0.368 |
| 25718 | LINC00654 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ITGB1 | Sponge network | 0.374 | 0.20054 | 0.524 | 0.00017 | 0.368 |
| 25719 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-3065-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-671-5p | 13 | CRTC3 | Sponge network | 0.727 | 3.0E-5 | 0.164 | 0.16786 | 0.368 |
| 25720 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-5p | 19 | SETBP1 | Sponge network | 0.174 | 0.30034 | -1.302 | 1.0E-5 | 0.368 |
| 25721 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-7-1-3p | 11 | NUAK1 | Sponge network | 0.693 | 0.00076 | 0.904 | 0 | 0.368 |
| 25722 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | LRRC55 | Sponge network | 0.657 | 4.0E-5 | -0.026 | 0.93163 | 0.368 |
| 25723 | RP11-158K1.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 26 | RASSF8 | Sponge network | 0.349 | 0.01695 | -0.122 | 0.65591 | 0.368 |
| 25724 | FZD10-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p | 15 | SPG20 | Sponge network | -0.727 | 0.04726 | -1.16 | 0 | 0.368 |
| 25725 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | TSHZ3 | Sponge network | -1.057 | 0.00029 | -0.556 | 0.01516 | 0.368 |
| 25726 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ETV1 | Sponge network | -0.021 | 0.93598 | -0.096 | 0.67674 | 0.368 |
| 25727 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p | 26 | GRIA3 | Sponge network | -0.612 | 0.00094 | -1.956 | 0 | 0.368 |
| 25728 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | CDH19 | Sponge network | -0.738 | 0.21233 | -3.434 | 0 | 0.368 |
| 25729 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | PRRX1 | Sponge network | 1.153 | 0.00114 | 1.316 | 0 | 0.368 |
| 25730 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-539-5p | 11 | PDZD2 | Sponge network | -0.405 | 0.22013 | -0.909 | 3.0E-5 | 0.368 |
| 25731 | RP11-620J15.3 |
hsa-miR-10a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-30d-5p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | HDAC9 | Sponge network | -0.739 | 0.00044 | -0.755 | 0.00495 | 0.368 |
| 25732 | RP11-46C24.7 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-495-3p;hsa-miR-542-3p | 15 | SYNM | Sponge network | 0.392 | 0.0278 | -2.309 | 1.0E-5 | 0.368 |
| 25733 | BVES-AS1 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-550a-3p | 13 | RASSF3 | Sponge network | -2.62 | 0 | -0.226 | 0.11691 | 0.368 |
| 25734 | RP1-302G2.5 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p | 11 | PDGFRA | Sponge network | -1.483 | 9.0E-5 | -0.372 | 0.0037 | 0.368 |
| 25735 | LINC00342 |
hsa-miR-10b-3p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p | 10 | ATRX | Sponge network | 0.297 | 0.19933 | 0.261 | 0.04408 | 0.368 |
| 25736 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-590-3p | 10 | RBM5 | Sponge network | 0.782 | 0.00016 | -0.205 | 0.0129 | 0.368 |
| 25737 | OIP5-AS1 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | TMEM30A | Sponge network | 0.421 | 0.00084 | 0.096 | 0.31031 | 0.368 |
| 25738 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | AHNAK | Sponge network | 0.72 | 0.0001 | -1.207 | 0 | 0.368 |
| 25739 | RP11-216L13.19 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-654-3p | 11 | PHF6 | Sponge network | 1.674 | 0 | 0.785 | 0 | 0.368 |
| 25740 | HOTAIRM1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 53 | LPP | Sponge network | -0.053 | 0.80685 | -0.459 | 0.04475 | 0.368 |
| 25741 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p | 26 | ATRX | Sponge network | -0.096 | 0.67958 | 0.261 | 0.04408 | 0.368 |
| 25742 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 20 | NUAK1 | Sponge network | -0.154 | 0.78939 | 0.904 | 0 | 0.368 |
| 25743 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 27 | TMTC3 | Sponge network | 0.683 | 1.0E-5 | 0.123 | 0.22189 | 0.368 |
| 25744 | PSMG3-AS1 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | NAV3 | Sponge network | -0.125 | 0.49414 | 0.212 | 0.47729 | 0.368 |
| 25745 | LINC00641 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 56 | PPM1L | Sponge network | -0.222 | 0.32445 | -0.537 | 0.00616 | 0.368 |
| 25746 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-425-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 26 | QKI | Sponge network | -0.726 | 0.00132 | -0.333 | 0.04111 | 0.368 |
| 25747 | LINC00843 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-429;hsa-miR-495-3p | 12 | ABI2 | Sponge network | 1.106 | 0 | 0.175 | 0.14787 | 0.368 |
| 25748 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 22 | BCL2 | Sponge network | -0.351 | 0.11358 | -0.899 | 0 | 0.368 |
| 25749 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p | 23 | MOB1B | Sponge network | 0.082 | 0.55397 | 0.197 | 0.15266 | 0.368 |
| 25750 | AC025171.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | PHC3 | Sponge network | 0.998 | 0 | 0.212 | 0.0499 | 0.368 |
| 25751 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | CHD2 | Sponge network | 0.986 | 0.01022 | 0.049 | 0.55371 | 0.368 |
| 25752 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-877-5p | 16 | GRIN2A | Sponge network | -0.773 | 0.04073 | -2.161 | 0 | 0.368 |
| 25753 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-505-3p;hsa-miR-582-5p | 14 | USP6 | Sponge network | 1.093 | 0 | 0.188 | 0.3871 | 0.368 |
| 25754 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | MYO9A | Sponge network | 1.153 | 0 | -0.147 | 0.32373 | 0.368 |
| 25755 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 24 | PGR | Sponge network | 0.083 | 0.66405 | -1.704 | 0 | 0.368 |
| 25756 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-629-3p | 24 | ZBTB4 | Sponge network | 0.331 | 0.57247 | -0.739 | 0 | 0.368 |
| 25757 | FAM157A |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 13 | UBR3 | Sponge network | 0.813 | 1.0E-5 | -0.021 | 0.82774 | 0.368 |
| 25758 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 20 | RECK | Sponge network | 0.082 | 0.55654 | -1.043 | 0 | 0.368 |
| 25759 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-376c-3p;hsa-miR-495-3p | 19 | TBL1XR1 | Sponge network | 0.8 | 0 | 0.578 | 0 | 0.368 |
| 25760 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | MCC | Sponge network | 0.385 | 0.10067 | -1.005 | 0 | 0.368 |
| 25761 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.335 | 0.20596 | 0.943 | 0 | 0.368 |
| 25762 | RP11-967K21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-940 | 28 | ETV1 | Sponge network | 0.056 | 0.81195 | -0.096 | 0.67674 | 0.368 |
| 25763 | RP1-151F17.2 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | SH3GLB1 | Sponge network | 0.222 | 0.29133 | -0.115 | 0.14773 | 0.368 |
| 25764 | ACTA2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-96-5p | 23 | CYLD | Sponge network | 0.194 | 0.64278 | -0.367 | 0.00171 | 0.368 |
| 25765 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PDE4DIP | Sponge network | 0.335 | 0.20596 | -0.282 | 0.0257 | 0.368 |
| 25766 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 61 | KIF1B | Sponge network | 0.101 | 0.60919 | -0.118 | 0.32258 | 0.368 |
| 25767 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 36 | PPP1R9A | Sponge network | -0.793 | 7.0E-5 | -0.903 | 0.04254 | 0.368 |
| 25768 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 20 | KCNA6 | Sponge network | -0.08 | 0.90092 | -1.531 | 0.00013 | 0.368 |
| 25769 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-331-3p;hsa-miR-342-5p | 13 | ATRX | Sponge network | 0.556 | 0.00298 | 0.261 | 0.04408 | 0.368 |
| 25770 | RP11-503E24.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-374b-5p | 13 | PIK3C2A | Sponge network | 2.218 | 0 | 0.061 | 0.53776 | 0.368 |
| 25771 | MEG3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p | 11 | FAM133A | Sponge network | 0.249 | 0.35902 | 0.549 | 0.29009 | 0.368 |
| 25772 | AC005154.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-3677-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p | 18 | PRKAA1 | Sponge network | 0.848 | 0 | 0.317 | 0.0031 | 0.368 |
| 25773 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | RB1CC1 | Sponge network | 0.67 | 0.00048 | 0.175 | 0.12559 | 0.368 |
| 25774 | PART1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MLF1 | Sponge network | -2.925 | 0 | -0.893 | 0.00727 | 0.368 |
| 25775 | RP11-305E6.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p | 11 | CBFB | Sponge network | 1.317 | 0 | 1.061 | 0 | 0.368 |
| 25776 | PSMG3-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429 | 10 | NGFR | Sponge network | -0.125 | 0.49414 | -1.271 | 0.01058 | 0.368 |
| 25777 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | MLLT10 | Sponge network | 1.063 | 0 | 0.105 | 0.33292 | 0.368 |
| 25778 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | EBF1 | Sponge network | -0.187 | 0.23787 | -0.384 | 0.08856 | 0.368 |
| 25779 | RP11-693J15.4 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | TAL1 | Sponge network | -0.72 | 0.24239 | -0.462 | 0.00574 | 0.368 |
| 25780 | LINC00641 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 18 | CXCL12 | Sponge network | -0.222 | 0.32445 | -1.421 | 0 | 0.368 |
| 25781 | JAZF1-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 20 | ID4 | Sponge network | -0.769 | 0.00035 | -1.644 | 0 | 0.368 |
| 25782 | RP3-402G11.26 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 17 | GRIK3 | Sponge network | -0.254 | 0.19303 | -3.208 | 0 | 0.368 |
| 25783 | JAZF1-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | TPO | Sponge network | -0.769 | 0.00035 | -1.87 | 2.0E-5 | 0.368 |
| 25784 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-96-5p | 12 | CRTC3 | Sponge network | 0.998 | 0 | 0.164 | 0.16786 | 0.368 |
| 25785 | FZD10-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ERG | Sponge network | -0.727 | 0.04726 | 0.002 | 0.99011 | 0.368 |
| 25786 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 53 | PPM1A | Sponge network | -2.46 | 0 | -0.623 | 0 | 0.368 |
| 25787 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-96-5p | 13 | CBFA2T3 | Sponge network | -0.187 | 0.23787 | -1.221 | 1.0E-5 | 0.368 |
| 25788 | RP11-77H9.2 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | SON | Sponge network | 1.677 | 0 | 0.168 | 0.04406 | 0.368 |
| 25789 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 32 | CHD5 | Sponge network | 0.259 | 0.12542 | -0.922 | 0.01521 | 0.368 |
| 25790 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ZNF536 | Sponge network | -0.942 | 0 | -3.115 | 0 | 0.368 |
| 25791 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p | 41 | TACC1 | Sponge network | -0.355 | 0.0077 | -0.362 | 0.05406 | 0.368 |
| 25792 | DNM3OS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 11 | UVRAG | Sponge network | 0.572 | 0.01722 | -0.017 | 0.83865 | 0.368 |
| 25793 | BOLA3-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 19 | SYT4 | Sponge network | -0.246 | 0.35034 | -3.207 | 0 | 0.368 |
| 25794 | BCYRN1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 25 | AKAP6 | Sponge network | 0.311 | 0.10344 | -1.586 | 3.0E-5 | 0.368 |
| 25795 | USP3-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-92a-1-5p | 16 | LUZP2 | Sponge network | -0.162 | 0.47687 | -0.056 | 0.89416 | 0.368 |
| 25796 | ARHGEF26-AS1 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | GABRB2 | Sponge network | -2.46 | 0 | -1.841 | 0.00029 | 0.368 |
| 25797 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p | 13 | NF1 | Sponge network | 0.663 | 0.00073 | 0.51 | 0 | 0.368 |
| 25798 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30c-5p;hsa-miR-3607-3p | 10 | TBL1XR1 | Sponge network | 0.537 | 0.0006 | 0.578 | 0 | 0.368 |
| 25799 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p | 12 | PAK3 | Sponge network | -0.246 | 0.35034 | -0.628 | 0.07253 | 0.368 |
| 25800 | RP11-119F7.5 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | 0.225 | 0.30644 | 0.524 | 0.00017 | 0.368 |
| 25801 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-935 | 14 | LRRC55 | Sponge network | -0.896 | 0.00099 | -0.026 | 0.93163 | 0.368 |
| 25802 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-93-5p | 13 | CNTNAP3 | Sponge network | -1.047 | 0 | -1.465 | 0.00033 | 0.368 |
| 25803 | RP11-927P21.1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 52 | LPP | Sponge network | 0.921 | 0.00118 | -0.459 | 0.04475 | 0.368 |
| 25804 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-337-3p;hsa-miR-576-5p | 15 | CUL5 | Sponge network | 0.594 | 0.00064 | 0.026 | 0.77301 | 0.368 |
| 25805 | LINC00265 |
hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-539-5p | 13 | MOB1B | Sponge network | 1.07 | 0 | 0.197 | 0.15266 | 0.368 |
| 25806 | NEAT1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | EPG5 | Sponge network | 0.705 | 0.00014 | 0.101 | 0.3563 | 0.368 |
| 25807 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 12 | FLI1 | Sponge network | 0.532 | 0.00505 | -0.294 | 0.10905 | 0.368 |
| 25808 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-93-3p | 38 | RUNX1T1 | Sponge network | -0.206 | 0.12942 | -0.344 | 0.2374 | 0.368 |
| 25809 | PART1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | LRFN5 | Sponge network | -2.925 | 0 | -1.483 | 0 | 0.368 |
| 25810 | LINC00173 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-874-3p;hsa-miR-940 | 11 | SORCS1 | Sponge network | 0.056 | 0.8494 | -3.492 | 0 | 0.368 |
| 25811 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | NEDD4 | Sponge network | 0.101 | 0.60919 | 0.02 | 0.89834 | 0.368 |
| 25812 | LPP-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-93-5p | 13 | EZH1 | Sponge network | -0.18 | 0.20343 | -0.564 | 1.0E-5 | 0.368 |
| 25813 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-5p | 25 | ATRX | Sponge network | 0.331 | 0.57247 | 0.261 | 0.04408 | 0.368 |
| 25814 | RP11-996F15.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 28 | MDM4 | Sponge network | 0.983 | 0.00048 | 0.345 | 0.00762 | 0.368 |
| 25815 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-29a-5p | 10 | CNTNAP3 | Sponge network | -7.551 | 0 | -1.465 | 0.00033 | 0.368 |
| 25816 | LINC00667 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | UBE4B | Sponge network | -0.235 | 0.15142 | -0.235 | 0.007 | 0.368 |
| 25817 | HOXB-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-429 | 23 | BCL2 | Sponge network | -0.154 | 0.53954 | -0.899 | 0 | 0.368 |
| 25818 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | NOTCH2NL | Sponge network | -0.487 | 0.02157 | 0.024 | 0.84307 | 0.368 |
| 25819 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p | 15 | DCLK1 | Sponge network | -1.645 | 0 | -1.324 | 0.00014 | 0.368 |
| 25820 | FENDRR |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-340-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 58 | NFIB | Sponge network | -1.281 | 0.00012 | 0.023 | 0.90795 | 0.368 |
| 25821 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p | 54 | TCF4 | Sponge network | -0.773 | 0.04073 | 0.016 | 0.92271 | 0.368 |
| 25822 | RP11-359B12.2 |
hsa-miR-10b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 10 | EXTL2 | Sponge network | 0.064 | 0.72934 | 0.109 | 0.37375 | 0.368 |
| 25823 | LINC00865 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-940 | 35 | PTPRT | Sponge network | -1.249 | 7.0E-5 | -0.907 | 0.03727 | 0.368 |
| 25824 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-96-5p | 45 | FAT3 | Sponge network | 0.395 | 0.15451 | -1.601 | 0.00051 | 0.368 |
| 25825 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | EXOC8 | Sponge network | 1.254 | 0 | 0.216 | 0.00598 | 0.368 |
| 25826 | RP11-356J5.12 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RTN4 | Sponge network | -0.899 | 6.0E-5 | -0.412 | 0.00014 | 0.368 |
| 25827 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935 | 43 | FAT3 | Sponge network | -0.513 | 0.04703 | -1.601 | 0.00051 | 0.368 |
| 25828 | RP11-182J1.1 |
hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 14 | PTPN14 | Sponge network | -0.187 | 0.23787 | 0.144 | 0.34595 | 0.368 |
| 25829 | BMS1P20 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | SLC5A3 | Sponge network | 0.34 | 0.00273 | 0.493 | 5.0E-5 | 0.368 |
| 25830 | LINC00403 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-452-5p;hsa-miR-589-5p | 15 | ADARB1 | Sponge network | -3.586 | 0.00032 | -0.335 | 0.06631 | 0.368 |
| 25831 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 28 | CBFA2T3 | Sponge network | -1.216 | 0 | -1.221 | 1.0E-5 | 0.368 |
| 25832 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 11 | KIAA1024 | Sponge network | 1.089 | 0 | 1.083 | 0 | 0.368 |
| 25833 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 25 | TET1 | Sponge network | 0.311 | 0.10344 | -0.135 | 0.52138 | 0.368 |
| 25834 | SERTAD4-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-576-5p | 18 | NTRK2 | Sponge network | -0.932 | 0.00329 | -0.736 | 0.06495 | 0.368 |
| 25835 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 51 | THRB | Sponge network | 0.374 | 0.20054 | -1.117 | 0.0004 | 0.368 |
| 25836 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | TSHZ3 | Sponge network | -0.737 | 0.10822 | -0.556 | 0.01516 | 0.368 |
| 25837 | RAMP2-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-744-3p | 16 | IGF2 | Sponge network | -0.513 | 0.04703 | 0.848 | 0.03842 | 0.368 |
| 25838 | NDUFA6-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | KANK1 | Sponge network | -0.355 | 0.0077 | -0.55 | 0.00134 | 0.368 |
| 25839 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-424-5p | 11 | LRP1B | Sponge network | -3.586 | 0.00032 | -2.163 | 0 | 0.368 |
| 25840 | RP11-182J1.1 |
hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-7-5p | 11 | KCNJ5 | Sponge network | -0.187 | 0.23787 | -0.281 | 0.3339 | 0.368 |
| 25841 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 61 | WDFY3 | Sponge network | -1.575 | 6.0E-5 | -0.201 | 0.0967 | 0.368 |
| 25842 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | DPYD | Sponge network | -0.739 | 0.0574 | -0.736 | 0.00076 | 0.368 |
| 25843 | RP11-57H14.4 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SH3GLB1 | Sponge network | 0.083 | 0.66405 | -0.115 | 0.14773 | 0.368 |
| 25844 | C4B-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-589-3p;hsa-miR-96-5p | 16 | ZFHX4 | Sponge network | 2.306 | 9.0E-5 | 0.018 | 0.95574 | 0.368 |
| 25845 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PLSCR4 | Sponge network | -1.057 | 0.00029 | -0.474 | 0.03833 | 0.368 |
| 25846 | RP11-359B12.2 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 15 | RHOB | Sponge network | 0.064 | 0.72934 | -1.052 | 0 | 0.368 |
| 25847 | MIR497HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 22 | PRRX1 | Sponge network | -1.439 | 4.0E-5 | 1.316 | 0 | 0.368 |
| 25848 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-942-5p | 11 | PCDH17 | Sponge network | -0.376 | 0.00337 | 0.731 | 2.0E-5 | 0.368 |
| 25849 | RP11-212P7.2 |
hsa-miR-141-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 11 | STAT5B | Sponge network | 0.219 | 0.1516 | -0.182 | 0.1082 | 0.368 |
| 25850 | RP11-890B15.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | KCTD8 | Sponge network | 0.101 | 0.60919 | -3.359 | 0 | 0.368 |
| 25851 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | ZMYND11 | Sponge network | -0.942 | 0 | -0.2 | 0.05449 | 0.368 |
| 25852 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-7-1-3p | 16 | MLLT10 | Sponge network | 0.389 | 0.04293 | 0.105 | 0.33292 | 0.367 |
| 25853 | LINC00672 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-590-3p | 22 | NAV3 | Sponge network | -0.227 | 0.31657 | 0.212 | 0.47729 | 0.367 |
| 25854 | MRPL23-AS1 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p | 19 | CHD5 | Sponge network | -0.278 | 0.76559 | -0.922 | 0.01521 | 0.367 |
| 25855 | RP11-195B17.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-33a-5p | 13 | AKAP11 | Sponge network | 0.37 | 0.27614 | 0.196 | 0.09825 | 0.367 |
| 25856 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | 0.082 | 0.55654 | -1.09 | 0.00014 | 0.367 |
| 25857 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 39 | MOB1B | Sponge network | 0.194 | 0.64278 | 0.197 | 0.15266 | 0.367 |
| 25858 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 25 | PCDH17 | Sponge network | 0.246 | 0.59912 | 0.731 | 2.0E-5 | 0.367 |
| 25859 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 20 | DOCK4 | Sponge network | -0.563 | 0.02545 | 0.502 | 0.00108 | 0.367 |
| 25860 | RASSF8-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 14 | EHD3 | Sponge network | 0.335 | 0.20596 | -0.484 | 0.01747 | 0.367 |
| 25861 | RP11-307B6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-93-3p | 16 | SNX29 | Sponge network | -4.463 | 0.00025 | -0.34 | 0.01371 | 0.367 |
| 25862 | RP11-73M18.10 |
hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-497-5p | 13 | DDX3X | Sponge network | 0.85 | 0 | -0.152 | 0.07686 | 0.367 |
| 25863 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429 | 15 | BCL6 | Sponge network | -2.69 | 0.0001 | -0.211 | 0.19489 | 0.367 |
| 25864 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | PRKAB2 | Sponge network | 0.16 | 0.50785 | -0.367 | 0.01371 | 0.367 |
| 25865 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 63 | THRB | Sponge network | -0.323 | 0.01372 | -1.117 | 0.0004 | 0.367 |
| 25866 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLIP1 | Sponge network | 0.385 | 0.10067 | -0.215 | 0.08684 | 0.367 |
| 25867 | CTD-2245F17.3 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-652-3p;hsa-miR-940 | 12 | QKI | Sponge network | -0.421 | 0.12556 | -0.333 | 0.04111 | 0.367 |
| 25868 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-628-5p | 15 | GRIA3 | Sponge network | -0.726 | 0.00132 | -1.956 | 0 | 0.367 |
| 25869 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484 | 16 | IGF2 | Sponge network | -2.69 | 0.0001 | 0.848 | 0.03842 | 0.367 |
| 25870 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-874-3p | 20 | MXI1 | Sponge network | -0.583 | 0.14589 | -0.987 | 0 | 0.367 |
| 25871 | LINC00473 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-942-5p | 26 | OPCML | Sponge network | -1.203 | 0.03881 | -2.498 | 0 | 0.367 |
| 25872 | HCG18 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 10 | RAPGEF5 | Sponge network | 1.063 | 0 | 0.536 | 4.0E-5 | 0.367 |
| 25873 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | EPHA5 | Sponge network | -0.769 | 0.00035 | -0.957 | 0.01733 | 0.367 |
| 25874 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | CSMD1 | Sponge network | -0.318 | 0.65847 | -0.63 | 0.18881 | 0.367 |
| 25875 | RP11-57H14.4 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 42 | GRIK3 | Sponge network | 0.083 | 0.66405 | -3.208 | 0 | 0.367 |
| 25876 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | KAT6B | Sponge network | -0.031 | 0.89063 | -0.478 | 0.00018 | 0.367 |
| 25877 | PSMG3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | CRTC1 | Sponge network | -0.125 | 0.49414 | -0.116 | 0.28412 | 0.367 |
| 25878 | LINC00092 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-708-5p;hsa-miR-940 | 19 | TOM1L2 | Sponge network | -1.886 | 0 | -0.994 | 0 | 0.367 |
| 25879 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | LRRC4 | Sponge network | -0.096 | 0.67958 | -1.774 | 0 | 0.367 |
| 25880 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 32 | ANK2 | Sponge network | 0.219 | 0.1516 | -1.603 | 1.0E-5 | 0.367 |
| 25881 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CXXC4 | Sponge network | -1.057 | 0.00029 | -0.433 | 0.21055 | 0.367 |
| 25882 | PWAR6 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 43 | PPP1R9A | Sponge network | -1.575 | 6.0E-5 | -0.903 | 0.04254 | 0.367 |
| 25883 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | TSHZ3 | Sponge network | -0.053 | 0.80685 | -0.556 | 0.01516 | 0.367 |
| 25884 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p | 43 | BNC2 | Sponge network | -1.645 | 0 | -0.491 | 0.09988 | 0.367 |
| 25885 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 21 | CDHR3 | Sponge network | -2.46 | 0 | -0.977 | 4.0E-5 | 0.367 |
| 25886 | LINC00865 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 77 | LPP | Sponge network | -1.249 | 7.0E-5 | -0.459 | 0.04475 | 0.367 |
| 25887 | PSMD5-AS1 |
hsa-miR-139-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-497-5p | 10 | SUZ12 | Sponge network | 0.569 | 0.00067 | 0.619 | 0 | 0.367 |
| 25888 | LINC00092 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p | 14 | RIMS2 | Sponge network | -1.886 | 0 | -1.365 | 0.00208 | 0.367 |
| 25889 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p | 14 | PDZD2 | Sponge network | 0.101 | 0.60919 | -0.909 | 3.0E-5 | 0.367 |
| 25890 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | RIMBP2 | Sponge network | -0.323 | 0.01372 | -1.968 | 5.0E-5 | 0.367 |
| 25891 | DNM3OS |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-93-3p | 16 | RPS6KA2 | Sponge network | 0.572 | 0.01722 | -0.57 | 0.00013 | 0.367 |
| 25892 | GABPB1-AS1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | PHF3 | Sponge network | 1.11 | 0 | 0.126 | 0.19652 | 0.367 |
| 25893 | U91328.19 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-369-3p | 16 | KLF10 | Sponge network | -0.303 | 0.21002 | -0.783 | 0 | 0.367 |
| 25894 | C20orf166-AS1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p | 11 | FOXO4 | Sponge network | -2.69 | 0.0001 | -0.516 | 2.0E-5 | 0.367 |
| 25895 | U91328.19 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-935;hsa-miR-940 | 12 | MAPK10 | Sponge network | -0.303 | 0.21002 | -1.774 | 0 | 0.367 |
| 25896 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | PDGFC | Sponge network | -1.161 | 0.00591 | -0.33 | 0.06483 | 0.367 |
| 25897 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 10 | PHC3 | Sponge network | 0.545 | 0.05789 | 0.212 | 0.0499 | 0.367 |
| 25898 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SYNE1 | Sponge network | 0.657 | 4.0E-5 | -0.738 | 0.00316 | 0.367 |
| 25899 | CTC-429P9.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-501-3p | 13 | TET1 | Sponge network | 0.082 | 0.55397 | -0.135 | 0.52138 | 0.367 |
| 25900 | LINC00472 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-511-5p;hsa-miR-625-5p | 11 | CELF4 | Sponge network | -0.842 | 0.01361 | -1.135 | 0.00344 | 0.367 |
| 25901 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | LRRC7 | Sponge network | 0.246 | 0.59912 | -1.341 | 0.01193 | 0.367 |
| 25902 | PART1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 20 | RELN | Sponge network | -2.925 | 0 | -2.403 | 0 | 0.367 |
| 25903 | RP11-35G9.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | CREB1 | Sponge network | 0.657 | 4.0E-5 | 0.203 | 0.00537 | 0.367 |
| 25904 | HOTAIRM1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | DCLK1 | Sponge network | -0.053 | 0.80685 | -1.324 | 0.00014 | 0.367 |
| 25905 | LINC00654 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 41 | NTRK2 | Sponge network | 0.374 | 0.20054 | -0.736 | 0.06495 | 0.367 |
| 25906 | PWAR6 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ZNF638 | Sponge network | -1.575 | 6.0E-5 | 0.22 | 0.01214 | 0.367 |
| 25907 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | CBL | Sponge network | 0.225 | 0.30644 | 0.291 | 0.01267 | 0.367 |
| 25908 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | ZFX | Sponge network | 0.064 | 0.72934 | 0.09 | 0.42563 | 0.367 |
| 25909 | HOXA-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 34 | NTRK2 | Sponge network | 0.61 | 0.01593 | -0.736 | 0.06495 | 0.367 |
| 25910 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | RBL2 | Sponge network | -0.901 | 0.00019 | -0.282 | 0.00254 | 0.367 |
| 25911 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 11 | STC1 | Sponge network | -0.129 | 0.57177 | 1.017 | 0 | 0.367 |
| 25912 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-874-3p;hsa-miR-98-5p | 11 | ST5 | Sponge network | -0.942 | 0 | -0.78 | 0 | 0.367 |
| 25913 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-590-3p | 14 | ITGB3 | Sponge network | -0.08 | 0.90092 | -0.011 | 0.95751 | 0.367 |
| 25914 | MIR143HG |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-940 | 44 | PTPRT | Sponge network | -0.739 | 0.0574 | -0.907 | 0.03727 | 0.367 |
| 25915 | MIR600HG |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-940 | 43 | MDM4 | Sponge network | 0.594 | 0.41522 | 0.345 | 0.00762 | 0.367 |
| 25916 | RP11-890B15.3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | PRKCB | Sponge network | 0.101 | 0.60919 | -1.313 | 2.0E-5 | 0.367 |
| 25917 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429 | 12 | LRRC4 | Sponge network | -1.249 | 7.0E-5 | -1.774 | 0 | 0.367 |
| 25918 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 34 | MAF | Sponge network | -0.942 | 0 | -1.257 | 0 | 0.367 |
| 25919 | RP5-855D21.3 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 10 | NF1 | Sponge network | 1.728 | 0 | 0.51 | 0 | 0.367 |
| 25920 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-362-3p | 23 | SSBP2 | Sponge network | -2.549 | 0.00528 | -1.58 | 0 | 0.367 |
| 25921 | FENDRR |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-660-5p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | ZHX2 | Sponge network | -1.281 | 0.00012 | -0.192 | 0.18459 | 0.367 |
| 25922 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p | 30 | UNC80 | Sponge network | -0.246 | 0.35034 | -1.934 | 0 | 0.367 |
| 25923 | RP11-428J1.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | TRPS1 | Sponge network | 0.538 | 0.00076 | -0.191 | 0.44109 | 0.367 |
| 25924 | RP11-25K21.6 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-589-3p | 11 | BNC2 | Sponge network | 0.489 | 0.25786 | -0.491 | 0.09988 | 0.367 |
| 25925 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-93-5p | 20 | GAS7 | Sponge network | 0.497 | 0.01451 | -0.555 | 0.01602 | 0.367 |
| 25926 | RP11-428J1.5 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-429 | 17 | ERC1 | Sponge network | 0.538 | 0.00076 | -0.108 | 0.38794 | 0.367 |
| 25927 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | CHD2 | Sponge network | 1.205 | 0 | 0.049 | 0.55371 | 0.367 |
| 25928 | HCG11 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 10 | LHFP | Sponge network | 0.385 | 0.10067 | -0.167 | 0.41371 | 0.367 |
| 25929 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | RCAN2 | Sponge network | -0.942 | 0 | -0.33 | 0.15172 | 0.367 |
| 25930 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-96-5p | 62 | BMPR2 | Sponge network | 0.311 | 0.10344 | 0.206 | 0.0635 | 0.367 |
| 25931 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-34a-5p | 10 | FOXP1 | Sponge network | -0.691 | 0.00146 | 0.042 | 0.74368 | 0.367 |
| 25932 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-375;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PTPN12 | Sponge network | 0.72 | 0.0001 | 0.768 | 0 | 0.367 |
| 25933 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-625-5p;hsa-miR-93-5p | 12 | LHX6 | Sponge network | 0.299 | 0.57722 | -0.087 | 0.69193 | 0.367 |
| 25934 | RP11-503E24.2 |
hsa-miR-127-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-495-3p | 14 | CCDC6 | Sponge network | 2.218 | 0 | 0.27 | 0.02555 | 0.367 |
| 25935 | RP11-141C7.4 |
hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 12 | MDM4 | Sponge network | 2.412 | 0 | 0.345 | 0.00762 | 0.367 |
| 25936 | LINC00969 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p | 27 | CREB1 | Sponge network | 0.683 | 1.0E-5 | 0.203 | 0.00537 | 0.367 |
| 25937 | RP11-166D19.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-5p | 23 | KCNJ5 | Sponge network | -0.576 | 0.10121 | -0.281 | 0.3339 | 0.367 |
| 25938 | CTD-2666L21.1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-424-5p | 27 | MDM4 | Sponge network | 0.368 | 0.20093 | 0.345 | 0.00762 | 0.367 |
| 25939 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 15 | SOX5 | Sponge network | -1.255 | 0.00981 | -1.091 | 4.0E-5 | 0.367 |
| 25940 | CTD-2314G24.2 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-484 | 16 | ABL1 | Sponge network | 0.246 | 0.59912 | -0.215 | 0.07804 | 0.367 |
| 25941 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-671-3p;hsa-miR-93-5p | 17 | MAP3K12 | Sponge network | -0.976 | 0.00025 | -0.057 | 0.59369 | 0.367 |
| 25942 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | TMEFF2 | Sponge network | -0.969 | 2.0E-5 | -2.426 | 0 | 0.367 |
| 25943 | RP11-802E16.3 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 2.048 | 0 | 0.51 | 0 | 0.367 |
| 25944 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | ANK2 | Sponge network | -0.668 | 3.0E-5 | -1.603 | 1.0E-5 | 0.367 |
| 25945 | U91328.19 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 16 | UBE4B | Sponge network | -0.303 | 0.21002 | -0.235 | 0.007 | 0.367 |
| 25946 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p | 56 | RUNX1T1 | Sponge network | 0.083 | 0.66405 | -0.344 | 0.2374 | 0.367 |
| 25947 | LPP-AS2 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | -0.18 | 0.20343 | -2.633 | 0 | 0.367 |
| 25948 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 29 | HBP1 | Sponge network | -0.976 | 0.00025 | -0.454 | 0 | 0.367 |
| 25949 | AC009120.6 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 10 | BCL9 | Sponge network | 0.919 | 0 | 0.321 | 0.00267 | 0.367 |
| 25950 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92a-3p;hsa-miR-940 | 24 | THBS1 | Sponge network | 0.214 | 0.18246 | 0.741 | 0.00386 | 0.367 |
| 25951 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-590-3p | 25 | MYBL1 | Sponge network | -0.615 | 0.00015 | 0.494 | 0.02152 | 0.367 |
| 25952 | CTD-2314G24.2 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | GLIPR1 | Sponge network | 0.246 | 0.59912 | 0.146 | 0.3276 | 0.367 |
| 25953 | CTD-3092A11.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-376c-3p;hsa-miR-625-5p | 11 | ARID2 | Sponge network | 0.873 | 0 | 0.235 | 0.02697 | 0.367 |
| 25954 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 12 | ZNF208 | Sponge network | -0.737 | 0.10822 | -0.4 | 0.22381 | 0.367 |
| 25955 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-5p | 43 | PTPRD | Sponge network | -0.154 | 0.78939 | -1.005 | 0.00064 | 0.367 |
| 25956 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-27b-3p;hsa-miR-664a-5p | 12 | SOX11 | Sponge network | 1.056 | 0 | 2.68 | 0 | 0.367 |
| 25957 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 33 | ADARB1 | Sponge network | -0.206 | 0.12942 | -0.335 | 0.06631 | 0.367 |
| 25958 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 20 | DIP2C | Sponge network | 0.389 | 0.04293 | -0.436 | 0.01713 | 0.367 |
| 25959 | GHRLOS |
hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-7-5p | 17 | BTG2 | Sponge network | -1.942 | 0 | -1.763 | 0 | 0.367 |
| 25960 | BZRAP1-AS1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | -0.465 | 0.04703 | -2.633 | 0 | 0.367 |
| 25961 | RP1-102E24.8 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p | 11 | NF1 | Sponge network | 1.737 | 0 | 0.51 | 0 | 0.367 |
| 25962 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | PRKAA2 | Sponge network | 0.61 | 0.01593 | -2.462 | 0 | 0.367 |
| 25963 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-590-3p | 12 | NRXN1 | Sponge network | 0.178 | 0.29106 | -3.778 | 0 | 0.367 |
| 25964 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 25 | EPS15 | Sponge network | -0.323 | 0.01372 | -0.052 | 0.53998 | 0.367 |
| 25965 | RP11-967K21.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | ROBO1 | Sponge network | 0.056 | 0.81195 | -0.009 | 0.96154 | 0.367 |
| 25966 | C20orf166-AS1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-542-3p | 21 | RAP1A | Sponge network | -2.69 | 0.0001 | -0.929 | 0 | 0.367 |
| 25967 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NUAK1 | Sponge network | 1.302 | 7.0E-5 | 0.904 | 0 | 0.367 |
| 25968 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | NUAK1 | Sponge network | 0.395 | 0.15451 | 0.904 | 0 | 0.367 |
| 25969 | RP11-95D17.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-625-3p | 18 | ABL2 | Sponge network | 0.75 | 0 | 0.82 | 0 | 0.367 |
| 25970 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | FOXO3 | Sponge network | 0.683 | 1.0E-5 | -0.04 | 0.72028 | 0.367 |
| 25971 | RP11-212P7.2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-369-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | MACF1 | Sponge network | 0.219 | 0.1516 | 0.019 | 0.90335 | 0.367 |
| 25972 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | NIN | Sponge network | 0.559 | 0.00026 | -0.253 | 0.13794 | 0.367 |
| 25973 | BOLA3-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | BRD3 | Sponge network | -0.246 | 0.35034 | 0.37 | 0.00013 | 0.367 |
| 25974 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 27 | AKAP11 | Sponge network | 0.862 | 0.06213 | 0.196 | 0.09825 | 0.367 |
| 25975 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | MYO9A | Sponge network | -1.249 | 7.0E-5 | -0.147 | 0.32373 | 0.367 |
| 25976 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | HACE1 | Sponge network | 0.72 | 0.0001 | -0.072 | 0.55239 | 0.367 |
| 25977 | CTC-297N7.9 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-877-5p | 20 | PTPRT | Sponge network | -2.069 | 0 | -0.907 | 0.03727 | 0.367 |
| 25978 | RASSF8-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 15 | STARD13 | Sponge network | 0.335 | 0.20596 | -0.187 | 0.27383 | 0.367 |
| 25979 | RP11-47A8.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p | 10 | HIP1 | Sponge network | 0.103 | 0.43169 | 0.774 | 0 | 0.367 |
| 25980 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 31 | PBX1 | Sponge network | 0.41 | 0.02214 | -1.206 | 0 | 0.367 |
| 25981 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | TGFBR3 | Sponge network | 0.225 | 0.30644 | -1.173 | 1.0E-5 | 0.367 |
| 25982 | C20orf166-AS1 |
hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 40 | CUX1 | Sponge network | -2.69 | 0.0001 | -0.039 | 0.6705 | 0.367 |
| 25983 | TPTEP1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | FAM117A | Sponge network | -1.216 | 0 | -1.379 | 0 | 0.367 |
| 25984 | DNM3OS |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | CREB1 | Sponge network | 0.572 | 0.01722 | 0.203 | 0.00537 | 0.367 |
| 25985 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-629-3p | 28 | DDX6 | Sponge network | -0.096 | 0.67958 | 0.091 | 0.36428 | 0.367 |
| 25986 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p | 27 | PAFAH1B1 | Sponge network | -2.62 | 0 | -0.429 | 0 | 0.367 |
| 25987 | RP11-822E23.8 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p | 12 | STARD13 | Sponge network | -0.777 | 0.16667 | -0.187 | 0.27383 | 0.367 |
| 25988 | SNHG14 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 31 | EPS15 | Sponge network | -1.111 | 0.0003 | -0.052 | 0.53998 | 0.367 |
| 25989 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-335-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-497-5p;hsa-miR-503-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 31 | RICTOR | Sponge network | 0.659 | 3.0E-5 | 0.388 | 0.00188 | 0.367 |
| 25990 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p | 15 | LATS2 | Sponge network | -0.773 | 0.04073 | 0.159 | 0.2304 | 0.367 |
| 25991 | RP11-282O18.3 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-505-5p | 14 | ARHGEF12 | Sponge network | 0.757 | 0 | 0.09 | 0.35693 | 0.367 |
| 25992 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-577 | 14 | EPHA5 | Sponge network | 1.294 | 0.0031 | -0.957 | 0.01733 | 0.367 |
| 25993 | RP11-479G22.8 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | BRD3 | Sponge network | 1.308 | 0 | 0.37 | 0.00013 | 0.367 |
| 25994 | FENDRR |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 27 | PCDH17 | Sponge network | -1.281 | 0.00012 | 0.731 | 2.0E-5 | 0.367 |
| 25995 | AC005154.6 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 19 | BAZ2B | Sponge network | 0.848 | 0 | -0.276 | 0.02833 | 0.367 |
| 25996 | AP001347.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-5p | 19 | CBFA2T3 | Sponge network | -1.161 | 0.00591 | -1.221 | 1.0E-5 | 0.367 |
| 25997 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 16 | CNTNAP3 | Sponge network | -0.896 | 0.00099 | -1.465 | 0.00033 | 0.367 |
| 25998 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 37 | UNC80 | Sponge network | -0.421 | 0.0114 | -1.934 | 0 | 0.367 |
| 25999 | MEG3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-629-3p | 24 | RYR2 | Sponge network | 0.249 | 0.35902 | -1.466 | 0.00031 | 0.367 |
| 26000 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | ADARB1 | Sponge network | -0.295 | 0.02604 | -0.335 | 0.06631 | 0.367 |
| 26001 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-3p | 31 | MAF | Sponge network | -0.487 | 0.02157 | -1.257 | 0 | 0.367 |
| 26002 | ZNF582-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p | 11 | RPS6KA2 | Sponge network | -1.349 | 0.00017 | -0.57 | 0.00013 | 0.367 |
| 26003 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-96-5p | 16 | LRRC7 | Sponge network | -1.654 | 0.00144 | -1.341 | 0.01193 | 0.367 |
| 26004 | AC003090.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | TET1 | Sponge network | -2.117 | 0.00161 | -0.135 | 0.52138 | 0.367 |
| 26005 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p | 11 | TRIM33 | Sponge network | 1.117 | 0 | 0.402 | 7.0E-5 | 0.367 |
| 26006 | FAM66C |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-429 | 10 | DUSP1 | Sponge network | -0.901 | 0.00019 | -1.754 | 0 | 0.367 |
| 26007 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-455-3p | 12 | RARB | Sponge network | -0.469 | 0.08721 | -0.825 | 2.0E-5 | 0.367 |
| 26008 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-501-5p | 16 | RET | Sponge network | -0.405 | 0.22013 | -0.833 | 0.03517 | 0.367 |
| 26009 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p | 25 | PTPN14 | Sponge network | -2.69 | 0.0001 | 0.144 | 0.34595 | 0.367 |
| 26010 | AP000240.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p | 12 | CREB1 | Sponge network | 1.462 | 0 | 0.203 | 0.00537 | 0.367 |
| 26011 | RP5-894A10.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 13 | TBL1XR1 | Sponge network | 0.697 | 9.0E-5 | 0.578 | 0 | 0.367 |
| 26012 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p | 16 | DMD | Sponge network | 0.131 | 0.8436 | -1.506 | 0 | 0.367 |
| 26013 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p | 19 | PRKAB2 | Sponge network | -1.654 | 0.00144 | -0.367 | 0.01371 | 0.367 |
| 26014 | LINC00954 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429 | 16 | ABL2 | Sponge network | 1.157 | 0.00455 | 0.82 | 0 | 0.367 |
| 26015 | RP5-855D21.3 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 12 | RICTOR | Sponge network | 1.728 | 0 | 0.388 | 0.00188 | 0.367 |
| 26016 | FAM66C |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -0.901 | 0.00019 | -2.417 | 0 | 0.367 |
| 26017 | RP11-356J5.12 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p | 28 | ASTN1 | Sponge network | -0.899 | 6.0E-5 | -2.389 | 2.0E-5 | 0.367 |
| 26018 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 36 | PRKAB2 | Sponge network | -0.576 | 0.10121 | -0.367 | 0.01371 | 0.367 |
| 26019 | RP11-344B5.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-27a-3p | 12 | ANK2 | Sponge network | -0.055 | 0.88482 | -1.603 | 1.0E-5 | 0.367 |
| 26020 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3913-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 31 | ESR1 | Sponge network | -0.096 | 0.67958 | -1.035 | 3.0E-5 | 0.367 |
| 26021 | RP11-389C8.2 |
hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 16 | NRK | Sponge network | 0.727 | 3.0E-5 | 0.656 | 0.19217 | 0.367 |
| 26022 | CTD-3092A11.2 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-411-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TMEM30A | Sponge network | 0.873 | 0 | 0.096 | 0.31031 | 0.367 |
| 26023 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p | 14 | PDS5B | Sponge network | 0.594 | 0.41522 | 0.259 | 0.00962 | 0.367 |
| 26024 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-942-5p | 17 | FBXO31 | Sponge network | -0.129 | 0.57177 | -0.085 | 0.42389 | 0.367 |
| 26025 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 48 | WDFY3 | Sponge network | -0.513 | 0.04703 | -0.201 | 0.0967 | 0.367 |
| 26026 | TTC25 |
hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-5p | 10 | THRB | Sponge network | -0.208 | 0.38261 | -1.117 | 0.0004 | 0.367 |
| 26027 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | RNF144A | Sponge network | 0.194 | 0.64278 | 0.594 | 0.0009 | 0.367 |
| 26028 | RP11-344B5.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b | 11 | PLSCR4 | Sponge network | -0.055 | 0.88482 | -0.474 | 0.03833 | 0.367 |
| 26029 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 33 | CREB3L2 | Sponge network | -0.896 | 0.00099 | -0.525 | 2.0E-5 | 0.367 |
| 26030 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-330-5p | 17 | GPC6 | Sponge network | 0.176 | 0.37402 | -0.054 | 0.81465 | 0.367 |
| 26031 | SNHG14 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935 | 35 | ACSL6 | Sponge network | -1.111 | 0.0003 | -1.593 | 0 | 0.367 |
| 26032 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PLSCR4 | Sponge network | 0.862 | 0.06213 | -0.474 | 0.03833 | 0.367 |
| 26033 | HOXD-AS2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-3607-3p | 15 | SOX5 | Sponge network | -0.208 | 0.65592 | -1.091 | 4.0E-5 | 0.367 |
| 26034 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p | 14 | SOX11 | Sponge network | 0.889 | 9.0E-5 | 2.68 | 0 | 0.367 |
| 26035 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CNTN1 | Sponge network | -0.053 | 0.80685 | -2.594 | 0 | 0.367 |
| 26036 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -0.769 | 0.00035 | -0.252 | 0.15511 | 0.367 |
| 26037 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-576-5p | 23 | FOXP1 | Sponge network | 0.594 | 0.00064 | 0.042 | 0.74368 | 0.367 |
| 26038 | PGM5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 12 | LRIG1 | Sponge network | -4.546 | 0 | -0.537 | 0.00588 | 0.366 |
| 26039 | RP11-1006G14.4 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | UBR3 | Sponge network | 0.559 | 0.00026 | -0.021 | 0.82774 | 0.366 |
| 26040 | LINC00672 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-93-5p | 30 | IL17RD | Sponge network | -0.227 | 0.31657 | -0.019 | 0.94873 | 0.366 |
| 26041 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-450a-5p;hsa-miR-497-5p;hsa-miR-628-5p | 13 | CHD2 | Sponge network | 0.983 | 0.00048 | 0.049 | 0.55371 | 0.366 |
| 26042 | RP11-6O2.3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-629-3p | 11 | BCL9 | Sponge network | 0.331 | 0.57247 | 0.321 | 0.00267 | 0.366 |
| 26043 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p | 15 | SCN9A | Sponge network | -2.095 | 0.00112 | -0.525 | 0.10593 | 0.366 |
| 26044 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-940 | 22 | IGF1 | Sponge network | 0.16 | 0.50785 | -0.204 | 0.5898 | 0.366 |
| 26045 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 47 | CADM2 | Sponge network | -0.18 | 0.20343 | -3.782 | 0 | 0.366 |
| 26046 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-455-3p | 12 | DKK3 | Sponge network | -0.811 | 2.0E-5 | -0.052 | 0.77783 | 0.366 |
| 26047 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p | 16 | TMTC3 | Sponge network | 1.266 | 0 | 0.123 | 0.22189 | 0.366 |
| 26048 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 34 | HBP1 | Sponge network | -1.216 | 0 | -0.454 | 0 | 0.366 |
| 26049 | RP11-216B9.6 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p | 43 | DST | Sponge network | 0.174 | 0.30034 | -0.713 | 0.00096 | 0.366 |
| 26050 | USP3-AS1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-505-5p | 13 | WNK2 | Sponge network | -0.162 | 0.47687 | -0.352 | 0.31066 | 0.366 |
| 26051 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-577 | 16 | IL17RD | Sponge network | 0.357 | 0.72407 | -0.019 | 0.94873 | 0.366 |
| 26052 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p | 27 | KIF1B | Sponge network | 0.93 | 0 | -0.118 | 0.32258 | 0.366 |
| 26053 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | NIN | Sponge network | 0.064 | 0.72934 | -0.253 | 0.13794 | 0.366 |
| 26054 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 28 | ZMYM2 | Sponge network | 0.421 | 0.00084 | 0.279 | 0.01273 | 0.366 |
| 26055 | AC005154.6 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | UBR3 | Sponge network | 0.848 | 0 | -0.021 | 0.82774 | 0.366 |
| 26056 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | THBS1 | Sponge network | -1.582 | 8.0E-5 | 0.741 | 0.00386 | 0.366 |
| 26057 | AGAP11 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p | 11 | MN1 | Sponge network | -1.645 | 0 | -0.358 | 0.24172 | 0.366 |
| 26058 | MAPKAPK5-AS1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-766-3p | 13 | TBL1XR1 | Sponge network | 0.765 | 0 | 0.578 | 0 | 0.366 |
| 26059 | TRAF3IP2-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | ID4 | Sponge network | -0.323 | 0.01372 | -1.644 | 0 | 0.366 |
| 26060 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | PDGFC | Sponge network | -0.899 | 6.0E-5 | -0.33 | 0.06483 | 0.366 |
| 26061 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | NEDD4 | Sponge network | -1.161 | 0.00591 | 0.02 | 0.89834 | 0.366 |
| 26062 | LINC00473 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | ID4 | Sponge network | -1.203 | 0.03881 | -1.644 | 0 | 0.366 |
| 26063 | RP11-727A23.5 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-550a-3p | 13 | BRD3 | Sponge network | 1.117 | 0 | 0.37 | 0.00013 | 0.366 |
| 26064 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 27 | FAT3 | Sponge network | 0.381 | 0.25967 | -1.601 | 0.00051 | 0.366 |
| 26065 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-7-1-3p | 10 | KCNAB1 | Sponge network | 0.559 | 0.00026 | -0.755 | 2.0E-5 | 0.366 |
| 26066 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-96-5p | 34 | BMPR2 | Sponge network | -1.654 | 0.00144 | 0.206 | 0.0635 | 0.366 |
| 26067 | AC009948.5 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ZNF844 | Sponge network | 0.532 | 0.00505 | -0.966 | 0.00035 | 0.366 |
| 26068 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | USP12 | Sponge network | 1.308 | 0 | -0.102 | 0.40768 | 0.366 |
| 26069 | RP11-156P1.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 21 | RYR2 | Sponge network | 0.214 | 0.18246 | -1.466 | 0.00031 | 0.366 |
| 26070 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | SEMA3C | Sponge network | -1.349 | 0.00017 | -0.272 | 0.31153 | 0.366 |
| 26071 | RP11-13K12.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p | 24 | BRD3 | Sponge network | -0.154 | 0.78939 | 0.37 | 0.00013 | 0.366 |
| 26072 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | PDE4DIP | Sponge network | -0.376 | 0.00337 | -0.282 | 0.0257 | 0.366 |
| 26073 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 34 | DDX6 | Sponge network | 0.848 | 0 | 0.091 | 0.36428 | 0.366 |
| 26074 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-590-3p | 17 | FOXO3 | Sponge network | 1.601 | 0 | -0.04 | 0.72028 | 0.366 |
| 26075 | TPTEP1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 11 | PEA15 | Sponge network | -1.216 | 0 | 0.009 | 0.93318 | 0.366 |
| 26076 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-582-5p | 19 | AFF3 | Sponge network | -0.773 | 0.04073 | -2.373 | 0 | 0.366 |
| 26077 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 14 | FBXO31 | Sponge network | -2.452 | 0 | -0.085 | 0.42389 | 0.366 |
| 26078 | FAM66C |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 15 | CELF4 | Sponge network | -0.901 | 0.00019 | -1.135 | 0.00344 | 0.366 |
| 26079 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 29 | LRP1B | Sponge network | -0.738 | 0.21233 | -2.163 | 0 | 0.366 |
| 26080 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 51 | PPM1A | Sponge network | -0.129 | 0.57177 | -0.623 | 0 | 0.366 |
| 26081 | ZNF667-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 19 | JDP2 | Sponge network | -0.976 | 0.00025 | -0.967 | 0 | 0.366 |
| 26082 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940 | 19 | CRTC3 | Sponge network | 0.199 | 0.38015 | 0.164 | 0.16786 | 0.366 |
| 26083 | RP11-403I13.7 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 25 | PCDH10 | Sponge network | 0.395 | 0.15451 | -1.644 | 0.0078 | 0.366 |
| 26084 | RP5-894A10.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-154-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-485-3p;hsa-miR-495-3p | 10 | LIMD1 | Sponge network | 0.697 | 9.0E-5 | 0.103 | 0.28978 | 0.366 |
| 26085 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | LRP1B | Sponge network | -0.563 | 0.02545 | -2.163 | 0 | 0.366 |
| 26086 | SNHG14 |
hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | PACRG | Sponge network | -1.111 | 0.0003 | -2.248 | 0 | 0.366 |
| 26087 | MAGI2-AS3 |
hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-660-5p | 10 | DNER | Sponge network | -0.129 | 0.57177 | -3.787 | 0 | 0.366 |
| 26088 | CTD-2583A14.8 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p | 10 | RICTOR | Sponge network | 1.134 | 0 | 0.388 | 0.00188 | 0.366 |
| 26089 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-590-5p;hsa-miR-708-3p | 26 | PRKAB2 | Sponge network | -0.371 | 0.24604 | -0.367 | 0.01371 | 0.366 |
| 26090 | PSMG3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p | 16 | CDH23 | Sponge network | -0.125 | 0.49414 | -0.974 | 0.00034 | 0.366 |
| 26091 | RP11-181G12.2 |
hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | UBR3 | Sponge network | 0.556 | 0.00298 | -0.021 | 0.82774 | 0.366 |
| 26092 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | 0.392 | 0.0278 | 0.323 | 0.00792 | 0.366 |
| 26093 | C20orf166-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-454-3p | 28 | AFF1 | Sponge network | -2.69 | 0.0001 | -0.384 | 0.00053 | 0.366 |
| 26094 | LINC00476 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 27 | NIN | Sponge network | -0.615 | 0.00015 | -0.253 | 0.13794 | 0.366 |
| 26095 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 37 | SNX29 | Sponge network | -2.46 | 0 | -0.34 | 0.01371 | 0.366 |
| 26096 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-766-3p | 28 | CHD5 | Sponge network | 0.603 | 0.00127 | -0.922 | 0.01521 | 0.366 |
| 26097 | AC108488.4 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | 0.259 | 0.12542 | -2.633 | 0 | 0.366 |
| 26098 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-542-3p | 21 | EPHA5 | Sponge network | -0.517 | 0.02935 | -0.957 | 0.01733 | 0.366 |
| 26099 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 16 | CYLD | Sponge network | 0.898 | 1.0E-5 | -0.367 | 0.00171 | 0.366 |
| 26100 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | FAM133A | Sponge network | 0.194 | 0.64278 | 0.549 | 0.29009 | 0.366 |
| 26101 | SERTAD4-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-369-3p | 17 | GNAQ | Sponge network | -0.932 | 0.00329 | -0.367 | 0.00102 | 0.366 |
| 26102 | FZD10-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | NRXN3 | Sponge network | -0.727 | 0.04726 | -0.893 | 0.04186 | 0.366 |
| 26103 | RP11-395G23.3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-324-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | PCDH10 | Sponge network | 0.381 | 0.25967 | -1.644 | 0.0078 | 0.366 |
| 26104 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-744-3p | 31 | DIP2C | Sponge network | 0.494 | 0.00339 | -0.436 | 0.01713 | 0.366 |
| 26105 | TMEM9B-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p | 10 | TCF12 | Sponge network | 0.529 | 0.00552 | 0.174 | 0.13271 | 0.366 |
| 26106 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 24 | SCN9A | Sponge network | 0.199 | 0.38015 | -0.525 | 0.10593 | 0.366 |
| 26107 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p | 27 | UNC80 | Sponge network | -0.737 | 0.10822 | -1.934 | 0 | 0.366 |
| 26108 | AC005562.1 |
hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 11 | TAF1 | Sponge network | 0.488 | 0.00084 | 0.39 | 3.0E-5 | 0.366 |
| 26109 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DACH2 | Sponge network | -0.513 | 0.04703 | -1.635 | 0.00034 | 0.366 |
| 26110 | NEAT1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | HECA | Sponge network | 0.705 | 0.00014 | -0.217 | 0.03463 | 0.366 |
| 26111 | SNHG14 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | ITGB1 | Sponge network | -1.111 | 0.0003 | 0.524 | 0.00017 | 0.366 |
| 26112 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p | 11 | SYNPO2 | Sponge network | 0.497 | 0.01451 | -2.342 | 0 | 0.366 |
| 26113 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p | 14 | CNTNAP3 | Sponge network | -0.693 | 0.05629 | -1.465 | 0.00033 | 0.366 |
| 26114 | RP11-967K21.1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 12 | NGFR | Sponge network | 0.056 | 0.81195 | -1.271 | 0.01058 | 0.366 |
| 26115 | HCG11 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | NIN | Sponge network | 0.385 | 0.10067 | -0.253 | 0.13794 | 0.366 |
| 26116 | RP11-1006G14.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-590-3p | 14 | NRK | Sponge network | 0.559 | 0.00026 | 0.656 | 0.19217 | 0.366 |
| 26117 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ABCA10 | Sponge network | 0.225 | 0.30644 | -0.571 | 0.03674 | 0.366 |
| 26118 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421 | 11 | JAKMIP2 | Sponge network | -0.154 | 0.78939 | -1.388 | 0 | 0.366 |
| 26119 | LINC00342 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-299-5p | 11 | MAT2A | Sponge network | 0.297 | 0.19933 | -0.073 | 0.36195 | 0.366 |
| 26120 | LINC01018 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-589-5p;hsa-miR-590-3p | 15 | ROBO1 | Sponge network | -2.915 | 0.00015 | -0.009 | 0.96154 | 0.366 |
| 26121 | CTC-429P9.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-940 | 13 | SIRT1 | Sponge network | 0.082 | 0.55397 | 0.109 | 0.2593 | 0.366 |
| 26122 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | FAT4 | Sponge network | -0.726 | 0.00132 | -0.354 | 0.14694 | 0.366 |
| 26123 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 27 | MCC | Sponge network | -0.08 | 0.90092 | -1.005 | 0 | 0.366 |
| 26124 | RP11-35G9.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PRRX1 | Sponge network | 0.657 | 4.0E-5 | 1.316 | 0 | 0.366 |
| 26125 | AC083843.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-539-5p | 14 | ARHGEF12 | Sponge network | 1.4 | 0 | 0.09 | 0.35693 | 0.366 |
| 26126 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3C | Sponge network | 0.222 | 0.29133 | -0.272 | 0.31153 | 0.366 |
| 26127 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935 | 21 | PCDH10 | Sponge network | 0.225 | 0.30644 | -1.644 | 0.0078 | 0.366 |
| 26128 | LINC00641 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p | 11 | PRKD1 | Sponge network | -0.222 | 0.32445 | -0.466 | 0.03583 | 0.366 |
| 26129 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 22 | DICER1 | Sponge network | 0.614 | 1.0E-5 | 0.042 | 0.674 | 0.366 |
| 26130 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | HECA | Sponge network | -0.421 | 0.0114 | -0.217 | 0.03463 | 0.366 |
| 26131 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 14 | DPYD | Sponge network | -0.49 | 0.04946 | -0.736 | 0.00076 | 0.366 |
| 26132 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | CHD2 | Sponge network | 0.959 | 0 | 0.049 | 0.55371 | 0.366 |
| 26133 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 29 | USP12 | Sponge network | 0.572 | 0.01722 | -0.102 | 0.40768 | 0.366 |
| 26134 | SNHG14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 26 | ARNT2 | Sponge network | -1.111 | 0.0003 | -0.332 | 0.25851 | 0.366 |
| 26135 | AC093110.3 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p | 22 | CREB1 | Sponge network | 1.044 | 2.0E-5 | 0.203 | 0.00537 | 0.366 |
| 26136 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-323a-3p | 14 | MTSS1 | Sponge network | -0.473 | 0.07602 | -0.81 | 0.00035 | 0.366 |
| 26137 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-576-5p | 19 | CBL | Sponge network | 1.317 | 0 | 0.291 | 0.01267 | 0.366 |
| 26138 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | BNC2 | Sponge network | -0.727 | 0.04726 | -0.491 | 0.09988 | 0.366 |
| 26139 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-940 | 54 | WDFY3 | Sponge network | 0.194 | 0.64278 | -0.201 | 0.0967 | 0.366 |
| 26140 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326 | 10 | TBL1XR1 | Sponge network | 2.163 | 0 | 0.578 | 0 | 0.366 |
| 26141 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-511-5p | 15 | ZFHX3 | Sponge network | -0.278 | 0.76559 | 0.389 | 0.01363 | 0.366 |
| 26142 | RP11-65J21.3 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p | 10 | MRVI1 | Sponge network | -0.557 | 0.11135 | -1.051 | 0.00022 | 0.366 |
| 26143 | LINC00662 |
hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | RNF144A | Sponge network | 0.213 | 0.32297 | 0.594 | 0.0009 | 0.366 |
| 26144 | LINC00476 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-877-5p | 21 | CBFA2T3 | Sponge network | -0.615 | 0.00015 | -1.221 | 1.0E-5 | 0.366 |
| 26145 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-7-1-3p | 21 | MED12L | Sponge network | 0.389 | 0.04293 | -0.194 | 0.55299 | 0.366 |
| 26146 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 10 | IGF1 | Sponge network | -0.187 | 0.23787 | -0.204 | 0.5898 | 0.366 |
| 26147 | LINC00662 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PRKAA2 | Sponge network | 0.213 | 0.32297 | -2.462 | 0 | 0.366 |
| 26148 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-539-5p | 15 | ITCH | Sponge network | 0.594 | 0.00064 | 0.084 | 0.34058 | 0.366 |
| 26149 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 19 | RET | Sponge network | 0.194 | 0.64278 | -0.833 | 0.03517 | 0.366 |
| 26150 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | ZMYM2 | Sponge network | 0.643 | 0 | 0.279 | 0.01273 | 0.366 |
| 26151 | LINC00957 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 27 | CTIF | Sponge network | -0.421 | 0.0114 | -0.389 | 0.00549 | 0.366 |
| 26152 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-5p | 26 | SASH1 | Sponge network | 0.331 | 0.57247 | -0.502 | 0.00175 | 0.366 |
| 26153 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 15 | TACC1 | Sponge network | -0.557 | 0.11135 | -0.362 | 0.05406 | 0.366 |
| 26154 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p | 10 | ZNF292 | Sponge network | 2.163 | 0 | 0.276 | 0.04148 | 0.366 |
| 26155 | RP11-166D19.1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429 | 16 | PLXNC1 | Sponge network | -0.576 | 0.10121 | 0.569 | 0.00329 | 0.366 |
| 26156 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | NOTCH2 | Sponge network | 0.299 | 0.57722 | 0.138 | 0.35163 | 0.366 |
| 26157 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-93-5p | 10 | TXNIP | Sponge network | 0.082 | 0.55397 | -1.277 | 0 | 0.366 |
| 26158 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | THBS1 | Sponge network | -1.575 | 6.0E-5 | 0.741 | 0.00386 | 0.366 |
| 26159 | HAS2-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | RASSF8 | Sponge network | -0.011 | 0.967 | -0.122 | 0.65591 | 0.366 |
| 26160 | PPP1R26-AS1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326 | 11 | TBL1XR1 | Sponge network | 1.454 | 0 | 0.578 | 0 | 0.366 |
| 26161 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-423-3p;hsa-miR-424-5p | 19 | UBE4B | Sponge network | -0.154 | 0.78939 | -0.235 | 0.007 | 0.366 |
| 26162 | AGAP11 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-93-5p | 24 | NIN | Sponge network | -1.645 | 0 | -0.253 | 0.13794 | 0.366 |
| 26163 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 35 | BNC2 | Sponge network | 0.619 | 0.00157 | -0.491 | 0.09988 | 0.366 |
| 26164 | DNM3OS |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 35 | NTRK2 | Sponge network | 0.572 | 0.01722 | -0.736 | 0.06495 | 0.366 |
| 26165 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-511-5p | 19 | BCL6 | Sponge network | -1.645 | 0 | -0.211 | 0.19489 | 0.366 |
| 26166 | RP11-693J15.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | NIN | Sponge network | -0.72 | 0.24239 | -0.253 | 0.13794 | 0.366 |
| 26167 | RP11-57H14.4 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-96-5p | 15 | WNK2 | Sponge network | 0.083 | 0.66405 | -0.352 | 0.31066 | 0.366 |
| 26168 | CTD-2002H8.2 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-1271-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | EPHA3 | Sponge network | 0.931 | 1.0E-5 | 0.185 | 0.53588 | 0.366 |
| 26169 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-532-5p | 18 | BMPR2 | Sponge network | -2.039 | 0.03447 | 0.206 | 0.0635 | 0.366 |
| 26170 | RP5-1198O20.4 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 39 | AFF1 | Sponge network | 0.997 | 0.00066 | -0.384 | 0.00053 | 0.366 |
| 26171 | KCNJ2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | NTRK2 | Sponge network | -0.436 | 0.05325 | -0.736 | 0.06495 | 0.366 |
| 26172 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 49 | THRB | Sponge network | -0.513 | 0.04703 | -1.117 | 0.0004 | 0.366 |
| 26173 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 13 | GPAM | Sponge network | 0.959 | 0 | 0.142 | 0.29732 | 0.366 |
| 26174 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 40 | FAT3 | Sponge network | -0.053 | 0.80685 | -1.601 | 0.00051 | 0.366 |
| 26175 | DNM3OS |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PDE4DIP | Sponge network | 0.572 | 0.01722 | -0.282 | 0.0257 | 0.366 |
| 26176 | MIR143HG |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 17 | ACTB | Sponge network | -0.739 | 0.0574 | -0.247 | 0.01736 | 0.366 |
| 26177 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-423-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 18 | UBE4B | Sponge network | 0.101 | 0.60919 | -0.235 | 0.007 | 0.366 |
| 26178 | C20orf166-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-93-5p | 38 | CCND2 | Sponge network | -2.69 | 0.0001 | -0.267 | 0.3112 | 0.366 |
| 26179 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-98-5p | 12 | ITGB3 | Sponge network | 0.056 | 0.81195 | -0.011 | 0.95751 | 0.366 |
| 26180 | LINC00865 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-940 | 38 | NTRK2 | Sponge network | -1.249 | 7.0E-5 | -0.736 | 0.06495 | 0.366 |
| 26181 | JAZF1-AS1 |
hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | PACRG | Sponge network | -0.769 | 0.00035 | -2.248 | 0 | 0.366 |
| 26182 | PDXDC2P |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 11 | PRKAA1 | Sponge network | 0.756 | 0 | 0.317 | 0.0031 | 0.366 |
| 26183 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | AKAP11 | Sponge network | 0.817 | 3.0E-5 | 0.196 | 0.09825 | 0.366 |
| 26184 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 43 | SASH1 | Sponge network | -0.901 | 0.00019 | -0.502 | 0.00175 | 0.366 |
| 26185 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.614 | 1.0E-5 | 1.083 | 0 | 0.366 |
| 26186 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 19 | NEDD4 | Sponge network | -0.611 | 0.09607 | 0.02 | 0.89834 | 0.366 |
| 26187 | RP11-404E16.1 |
hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-424-5p | 12 | RUNX1T1 | Sponge network | 0.097 | 0.91311 | -0.344 | 0.2374 | 0.366 |
| 26188 | RP11-403I13.7 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-589-5p | 12 | ROBO1 | Sponge network | 0.395 | 0.15451 | -0.009 | 0.96154 | 0.366 |
| 26189 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RB1CC1 | Sponge network | 0.569 | 0.00067 | 0.175 | 0.12559 | 0.366 |
| 26190 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 27 | FOXP1 | Sponge network | 0.174 | 0.30034 | 0.042 | 0.74368 | 0.366 |
| 26191 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 26 | ABI2 | Sponge network | -0.318 | 0.65847 | 0.175 | 0.14787 | 0.366 |
| 26192 | RP11-46C24.7 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITGB1 | Sponge network | 0.392 | 0.0278 | 0.524 | 0.00017 | 0.366 |
| 26193 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 18 | ARNT2 | Sponge network | -0.513 | 0.04703 | -0.332 | 0.25851 | 0.366 |
| 26194 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 23 | IGF1 | Sponge network | -0.342 | 0.02288 | -0.204 | 0.5898 | 0.366 |
| 26195 | CD27-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-542-3p | 17 | DYNC1I1 | Sponge network | 0.177 | 0.16681 | -1.12 | 0.00107 | 0.366 |
| 26196 | RP11-2B6.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-624-5p | 11 | SIRT1 | Sponge network | 0.539 | 0.03722 | 0.109 | 0.2593 | 0.366 |
| 26197 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-338-3p | 10 | ELMO1 | Sponge network | -2.531 | 0 | 0.04 | 0.83147 | 0.366 |
| 26198 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DACT1 | Sponge network | 0.219 | 0.1516 | 0.783 | 0.00072 | 0.366 |
| 26199 | RP11-672A2.4 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-424-5p | 17 | EPHA7 | Sponge network | 1.344 | 0 | -2.197 | 3.0E-5 | 0.366 |
| 26200 | LINC00863 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | HIP1 | Sponge network | 0.189 | 0.13981 | 0.774 | 0 | 0.366 |
| 26201 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | PRKAA2 | Sponge network | 0.41 | 0.02214 | -2.462 | 0 | 0.366 |
| 26202 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CLIP1 | Sponge network | 0.532 | 0.00505 | -0.215 | 0.08684 | 0.366 |
| 26203 | RP11-403I13.8 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | 0.619 | 0.00157 | 0.524 | 0.00017 | 0.366 |
| 26204 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-25-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 12 | DKK3 | Sponge network | -4.546 | 0 | -0.052 | 0.77783 | 0.366 |
| 26205 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 39 | CHD5 | Sponge network | 0.572 | 0.01722 | -0.922 | 0.01521 | 0.366 |
| 26206 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-429;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-935 | 31 | ABI2 | Sponge network | -2.62 | 0 | 0.175 | 0.14787 | 0.366 |
| 26207 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-5p;hsa-miR-625-3p | 17 | BMPR2 | Sponge network | 0.537 | 0.0006 | 0.206 | 0.0635 | 0.366 |
| 26208 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-590-3p | 19 | HIP1 | Sponge network | 0.381 | 0.25967 | 0.774 | 0 | 0.366 |
| 26209 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-429 | 13 | CXXC4 | Sponge network | -1.654 | 0.00144 | -0.433 | 0.21055 | 0.366 |
| 26210 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | PLSCR4 | Sponge network | 0.131 | 0.8436 | -0.474 | 0.03833 | 0.366 |
| 26211 | RP1-59D14.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-629-3p | 32 | KIF1B | Sponge network | 1.162 | 5.0E-5 | -0.118 | 0.32258 | 0.366 |
| 26212 | LINC00963 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 17 | BCL9 | Sponge network | 0.523 | 9.0E-5 | 0.321 | 0.00267 | 0.366 |
| 26213 | RP11-798M19.6 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | SYT4 | Sponge network | -0.612 | 0.00094 | -3.207 | 0 | 0.366 |
| 26214 | LINC00654 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 28 | PCDH10 | Sponge network | 0.374 | 0.20054 | -1.644 | 0.0078 | 0.366 |
| 26215 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 43 | FZD3 | Sponge network | -0.222 | 0.32445 | 0.104 | 0.60316 | 0.366 |
| 26216 | RP11-479G22.8 |
hsa-miR-101-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | KIF5B | Sponge network | 1.308 | 0 | 0.362 | 0.00066 | 0.366 |
| 26217 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-7-1-3p | 24 | DTNA | Sponge network | -0.351 | 0.11358 | -1.648 | 1.0E-5 | 0.366 |
| 26218 | EMX2OS |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | CTNNA2 | Sponge network | -0.777 | 0.1134 | -2.547 | 0 | 0.366 |
| 26219 | RP4-613B23.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-361-5p | 13 | PPM1A | Sponge network | -0.309 | 0.1145 | -0.623 | 0 | 0.366 |
| 26220 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-5p | 26 | RNASEL | Sponge network | -2.452 | 0 | -0.272 | 0.01796 | 0.366 |
| 26221 | RP11-67L2.2 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-708-5p | 32 | CCDC6 | Sponge network | 0.72 | 0.0001 | 0.27 | 0.02555 | 0.366 |
| 26222 | RP4-791M13.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 19 | BRD3 | Sponge network | 0.419 | 0.0319 | 0.37 | 0.00013 | 0.366 |
| 26223 | WDFY3-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-96-5p | 90 | LPP | Sponge network | -0.942 | 0 | -0.459 | 0.04475 | 0.366 |
| 26224 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 12 | ABCB5 | Sponge network | -1.161 | 0.00591 | -0.956 | 0.04077 | 0.366 |
| 26225 | CTC-559E9.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-34a-5p | 11 | ZMYND11 | Sponge network | 0.594 | 0.00064 | -0.2 | 0.05449 | 0.366 |
| 26226 | LPP-AS2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 10 | AMPH | Sponge network | -0.18 | 0.20343 | -0.916 | 5.0E-5 | 0.366 |
| 26227 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 10 | FAM188A | Sponge network | -2.62 | 0 | -0.44 | 0.00018 | 0.366 |
| 26228 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 26 | GRIN2A | Sponge network | 0.219 | 0.1516 | -2.161 | 0 | 0.366 |
| 26229 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p | 27 | RUNX1T1 | Sponge network | 0.419 | 0.0319 | -0.344 | 0.2374 | 0.366 |
| 26230 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 18 | NUAK1 | Sponge network | 0.176 | 0.37402 | 0.904 | 0 | 0.366 |
| 26231 | RP11-25K19.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 60 | LPP | Sponge network | -1.057 | 0.00029 | -0.459 | 0.04475 | 0.366 |
| 26232 | EMX2OS |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-93-3p | 28 | CTIF | Sponge network | -0.777 | 0.1134 | -0.389 | 0.00549 | 0.366 |
| 26233 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-935;hsa-miR-940 | 27 | LIFR | Sponge network | -0.096 | 0.67958 | -1.794 | 0 | 0.366 |
| 26234 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | AHNAK | Sponge network | -2.46 | 0 | -1.207 | 0 | 0.366 |
| 26235 | LINC00641 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | PRKAR1A | Sponge network | -0.222 | 0.32445 | -0.063 | 0.50314 | 0.366 |
| 26236 | RP11-1006G14.4 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | SH3GLB1 | Sponge network | 0.559 | 0.00026 | -0.115 | 0.14773 | 0.366 |
| 26237 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SMARCA2 | Sponge network | -0.323 | 0.01372 | -0.529 | 0.00014 | 0.365 |
| 26238 | RP11-6O2.3 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-335-5p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-940 | 11 | RHOB | Sponge network | 0.331 | 0.57247 | -1.052 | 0 | 0.365 |
| 26239 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-877-5p | 19 | CBFA2T3 | Sponge network | 0.335 | 0.20596 | -1.221 | 1.0E-5 | 0.365 |
| 26240 | OSER1-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p | 17 | CADM1 | Sponge network | -0.13 | 0.48734 | -0.9 | 0.00084 | 0.365 |
| 26241 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 20 | SOX11 | Sponge network | 1.317 | 0 | 2.68 | 0 | 0.365 |
| 26242 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-942-5p | 37 | THRB | Sponge network | -1.886 | 0 | -1.117 | 0.0004 | 0.365 |
| 26243 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-98-5p | 14 | PBX1 | Sponge network | -0.301 | 0.06597 | -1.206 | 0 | 0.365 |
| 26244 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 35 | TGFBR3 | Sponge network | -0.421 | 0.0114 | -1.173 | 1.0E-5 | 0.365 |
| 26245 | LINC00657 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-664a-3p | 15 | PRKAA1 | Sponge network | 0.91 | 0 | 0.317 | 0.0031 | 0.365 |
| 26246 | CTC-559E9.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p | 38 | LPP | Sponge network | 0.594 | 0.00064 | -0.459 | 0.04475 | 0.365 |
| 26247 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 30 | ADARB1 | Sponge network | -1.057 | 0.00029 | -0.335 | 0.06631 | 0.365 |
| 26248 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | PLSCR4 | Sponge network | 0.082 | 0.55654 | -0.474 | 0.03833 | 0.365 |
| 26249 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PTPRB | Sponge network | -0.737 | 0.10822 | 0.105 | 0.47122 | 0.365 |
| 26250 | GHRLOS |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-7-5p | 18 | BCL2 | Sponge network | -1.942 | 0 | -0.899 | 0 | 0.365 |
| 26251 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | ST6GALNAC3 | Sponge network | -1.161 | 0.00591 | -0.987 | 0 | 0.365 |
| 26252 | RP11-46C24.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NRXN3 | Sponge network | 0.392 | 0.0278 | -0.893 | 0.04186 | 0.365 |
| 26253 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 12 | DCN | Sponge network | -0.899 | 6.0E-5 | -1.288 | 0 | 0.365 |
| 26254 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 58 | IL6ST | Sponge network | -0.154 | 0.78939 | -0.402 | 0.00676 | 0.365 |
| 26255 | LINC00641 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-577;hsa-miR-98-5p | 10 | PLAGL1 | Sponge network | -0.222 | 0.32445 | -0.588 | 0.00912 | 0.365 |
| 26256 | PGM5-AS1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-423-5p;hsa-miR-484 | 10 | ABL1 | Sponge network | -4.546 | 0 | -0.215 | 0.07804 | 0.365 |
| 26257 | ANKRD10-IT1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-32-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 10 | TSC1 | Sponge network | 1.739 | 0 | 0.103 | 0.26071 | 0.365 |
| 26258 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-93-5p | 12 | TXNIP | Sponge network | -1.203 | 0.03881 | -1.277 | 0 | 0.365 |
| 26259 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 19 | MCC | Sponge network | -0.663 | 0.10887 | -1.005 | 0 | 0.365 |
| 26260 | FAM66C |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 22 | RIMBP2 | Sponge network | -0.901 | 0.00019 | -1.968 | 5.0E-5 | 0.365 |
| 26261 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CXXC4 | Sponge network | 0.335 | 0.20596 | -0.433 | 0.21055 | 0.365 |
| 26262 | LINC00654 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 16 | UNC5C | Sponge network | 0.374 | 0.20054 | -0.178 | 0.40214 | 0.365 |
| 26263 | EMX2OS |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p | 15 | ZNF208 | Sponge network | -0.777 | 0.1134 | -0.4 | 0.22381 | 0.365 |
| 26264 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | LRIG1 | Sponge network | -1.057 | 0.00029 | -0.537 | 0.00588 | 0.365 |
| 26265 | LINC00092 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-590-3p;hsa-miR-708-5p | 12 | FAM135B | Sponge network | -1.886 | 0 | -3.978 | 0 | 0.365 |
| 26266 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PBRM1 | Sponge network | 0.559 | 0.00026 | -0.029 | 0.78601 | 0.365 |
| 26267 | U91328.19 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-532-5p | 10 | PARK2 | Sponge network | -0.303 | 0.21002 | -1.892 | 0 | 0.365 |
| 26268 | RP11-389C8.2 |
hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ITPKB | Sponge network | 0.727 | 3.0E-5 | -0.704 | 0.00106 | 0.365 |
| 26269 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZNF844 | Sponge network | -0.737 | 0.06182 | -0.966 | 0.00035 | 0.365 |
| 26270 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 46 | DTNA | Sponge network | 0.219 | 0.1516 | -1.648 | 1.0E-5 | 0.365 |
| 26271 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 45 | TMEM132B | Sponge network | -0.899 | 6.0E-5 | -0.83 | 0.01084 | 0.365 |
| 26272 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 30 | FOXO3 | Sponge network | -0.222 | 0.32445 | -0.04 | 0.72028 | 0.365 |
| 26273 | CTD-2245F17.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-629-3p | 12 | ASTN1 | Sponge network | -0.421 | 0.12556 | -2.389 | 2.0E-5 | 0.365 |
| 26274 | RP11-35G9.5 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p | 14 | ABL2 | Sponge network | 0.622 | 0.00015 | 0.82 | 0 | 0.365 |
| 26275 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MITF | Sponge network | -0.342 | 0.02288 | -0.769 | 0.00022 | 0.365 |
| 26276 | USP3-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-576-5p | 21 | BRD3 | Sponge network | -0.162 | 0.47687 | 0.37 | 0.00013 | 0.365 |
| 26277 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 26 | EBF1 | Sponge network | 0.082 | 0.55654 | -0.384 | 0.08856 | 0.365 |
| 26278 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -0.942 | 0 | 0.024 | 0.89889 | 0.365 |
| 26279 | HOTAIRM1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p | 38 | CADM2 | Sponge network | -0.053 | 0.80685 | -3.782 | 0 | 0.365 |
| 26280 | CKMT2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | ERCC4 | Sponge network | 0.082 | 0.55654 | 0.126 | 0.15266 | 0.365 |
| 26281 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | ST6GALNAC3 | Sponge network | -0.811 | 2.0E-5 | -0.987 | 0 | 0.365 |
| 26282 | CTC-297N7.5 |
hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-339-5p | 10 | THRB | Sponge network | -1.9 | 0 | -1.117 | 0.0004 | 0.365 |
| 26283 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-451a;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 37 | BCL2 | Sponge network | -0.342 | 0.02288 | -0.899 | 0 | 0.365 |
| 26284 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-7-1-3p | 11 | CACNA2D1 | Sponge network | -4.477 | 0 | -0.846 | 0.00675 | 0.365 |
| 26285 | LINC00672 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-98-5p | 12 | ITGB3 | Sponge network | -0.227 | 0.31657 | -0.011 | 0.95751 | 0.365 |
| 26286 | RP11-439A17.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-2110;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-576-5p | 12 | ZFHX4 | Sponge network | 0.051 | 0.95756 | 0.018 | 0.95574 | 0.365 |
| 26287 | GS1-124K5.11 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p | 12 | FBXO11 | Sponge network | 0.494 | 0.00339 | 0.142 | 0.04938 | 0.365 |
| 26288 | AP001347.6 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-590-3p | 13 | TLE4 | Sponge network | -1.161 | 0.00591 | -1.157 | 0 | 0.365 |
| 26289 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-192-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | USP12 | Sponge network | 0.931 | 1.0E-5 | -0.102 | 0.40768 | 0.365 |
| 26290 | RP11-152N13.5 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30c-5p | 10 | PHF6 | Sponge network | 0.699 | 3.0E-5 | 0.785 | 0 | 0.365 |
| 26291 | DLGAP1-AS2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-326 | 10 | NF1 | Sponge network | 2.406 | 0 | 0.51 | 0 | 0.365 |
| 26292 | FENDRR |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | PPM1L | Sponge network | -1.281 | 0.00012 | -0.537 | 0.00616 | 0.365 |
| 26293 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | PGR | Sponge network | -0.663 | 0.10887 | -1.704 | 0 | 0.365 |
| 26294 | RP11-161M6.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 10 | EPHA3 | Sponge network | 0.61 | 0.06611 | 0.185 | 0.53588 | 0.365 |
| 26295 | SOX2-OT |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-889-3p;hsa-miR-93-5p;hsa-miR-942-5p | 45 | UNC80 | Sponge network | -1.264 | 6.0E-5 | -1.934 | 0 | 0.365 |
| 26296 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | CACNA2D1 | Sponge network | 0.082 | 0.55654 | -0.846 | 0.00675 | 0.365 |
| 26297 | RP11-774O3.3 |
hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-625-5p | 11 | SLC12A6 | Sponge network | -0.487 | 0.02157 | -0.343 | 0.00043 | 0.365 |
| 26298 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 42 | NTRK2 | Sponge network | 0.101 | 0.60919 | -0.736 | 0.06495 | 0.365 |
| 26299 | CTC-559E9.6 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-32-5p | 13 | ZFHX3 | Sponge network | 0.601 | 1.0E-5 | 0.389 | 0.01363 | 0.365 |
| 26300 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | -0.899 | 6.0E-5 | -0.615 | 0.01482 | 0.365 |
| 26301 | RP11-25K21.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-374a-3p | 15 | GAS7 | Sponge network | 0.489 | 0.25786 | -0.555 | 0.01602 | 0.365 |
| 26302 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-93-3p;hsa-miR-93-5p | 37 | SSBP2 | Sponge network | -0.465 | 0.04703 | -1.58 | 0 | 0.365 |
| 26303 | HAND2-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 31 | PDE4DIP | Sponge network | -1.697 | 0.00889 | -0.282 | 0.0257 | 0.365 |
| 26304 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-671-5p | 18 | ADARB1 | Sponge network | -0.285 | 0.2172 | -0.335 | 0.06631 | 0.365 |
| 26305 | AC010226.4 |
hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | CYLD | Sponge network | -0.811 | 2.0E-5 | -0.367 | 0.00171 | 0.365 |
| 26306 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-940 | 17 | MYO9A | Sponge network | 0.792 | 0.0049 | -0.147 | 0.32373 | 0.365 |
| 26307 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p | 19 | CREB3L2 | Sponge network | -4.463 | 0.00025 | -0.525 | 2.0E-5 | 0.365 |
| 26308 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ATRX | Sponge network | 0.392 | 0.0278 | 0.261 | 0.04408 | 0.365 |
| 26309 | LINC00865 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 15 | PLAG1 | Sponge network | -1.249 | 7.0E-5 | -0.315 | 0.26532 | 0.365 |
| 26310 | RP11-181G12.2 |
hsa-let-7d-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-671-5p;hsa-miR-940 | 29 | MDM4 | Sponge network | 0.556 | 0.00298 | 0.345 | 0.00762 | 0.365 |
| 26311 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p | 11 | FAT3 | Sponge network | -1.14 | 0.03513 | -1.601 | 0.00051 | 0.365 |
| 26312 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p | 23 | IGF1 | Sponge network | -0.384 | 0.40652 | -0.204 | 0.5898 | 0.365 |
| 26313 | TUG1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p | 14 | ABI2 | Sponge network | 0.972 | 0 | 0.175 | 0.14787 | 0.365 |
| 26314 | CTD-2089N3.2 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-652-3p | 20 | QKI | Sponge network | -1.375 | 0.08642 | -0.333 | 0.04111 | 0.365 |
| 26315 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 30 | MEF2C | Sponge network | 0.174 | 0.30034 | -0.499 | 0.01448 | 0.365 |
| 26316 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 15 | THBS1 | Sponge network | -0.932 | 0.00329 | 0.741 | 0.00386 | 0.365 |
| 26317 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-577;hsa-miR-93-5p | 25 | IL17RD | Sponge network | 0.259 | 0.12542 | -0.019 | 0.94873 | 0.365 |
| 26318 | RP11-403I13.8 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NUAK1 | Sponge network | 0.619 | 0.00157 | 0.904 | 0 | 0.365 |
| 26319 | RP11-360F5.3 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-93-3p | 12 | RUNX1T1 | Sponge network | 1.867 | 0 | -0.344 | 0.2374 | 0.365 |
| 26320 | AGAP11 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-874-3p | 26 | ASTN1 | Sponge network | -1.645 | 0 | -2.389 | 2.0E-5 | 0.365 |
| 26321 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 26 | TMEM132B | Sponge network | -1.582 | 8.0E-5 | -0.83 | 0.01084 | 0.365 |
| 26322 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 50 | THRB | Sponge network | 0.61 | 0.01593 | -1.117 | 0.0004 | 0.365 |
| 26323 | ZNF582-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 12 | INPP4A | Sponge network | -1.349 | 0.00017 | 0.07 | 0.53563 | 0.365 |
| 26324 | LINC00969 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | ERCC6 | Sponge network | 0.683 | 1.0E-5 | 0.23 | 0.015 | 0.365 |
| 26325 | RP11-37B2.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-625-5p | 34 | MDM4 | Sponge network | 1.03 | 0 | 0.345 | 0.00762 | 0.365 |
| 26326 | ZRANB2-AS1 |
hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | CYLD | Sponge network | -0.376 | 0.00337 | -0.367 | 0.00171 | 0.365 |
| 26327 | CKMT2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 31 | SNX29 | Sponge network | 0.082 | 0.55654 | -0.34 | 0.01371 | 0.365 |
| 26328 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | LATS1 | Sponge network | 0.178 | 0.29106 | 0.109 | 0.34208 | 0.365 |
| 26329 | FZD10-AS1 |
hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | -0.727 | 0.04726 | -0.684 | 0.00159 | 0.365 |
| 26330 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | PCDH9 | Sponge network | 0.083 | 0.66405 | -1.823 | 4.0E-5 | 0.365 |
| 26331 | AC005154.6 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 19 | RASAL2 | Sponge network | 0.848 | 0 | 0.869 | 0 | 0.365 |
| 26332 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 53 | CCND2 | Sponge network | 0.249 | 0.35902 | -0.267 | 0.3112 | 0.365 |
| 26333 | BOLA3-AS1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-505-5p | 10 | WNK2 | Sponge network | -0.246 | 0.35034 | -0.352 | 0.31066 | 0.365 |
| 26334 | RP11-434B12.1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | DTNA | Sponge network | -0.031 | 0.89063 | -1.648 | 1.0E-5 | 0.365 |
| 26335 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | ITCH | Sponge network | 0.782 | 0.00016 | 0.084 | 0.34058 | 0.365 |
| 26336 | CTD-2314G24.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | 0.246 | 0.59912 | -0.974 | 0.00034 | 0.365 |
| 26337 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-484;hsa-miR-590-3p | 14 | PKNOX1 | Sponge network | -0.096 | 0.67958 | 0.085 | 0.28917 | 0.365 |
| 26338 | RP11-158K1.3 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 16 | SHPRH | Sponge network | 0.349 | 0.01695 | 0.098 | 0.40994 | 0.365 |
| 26339 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 63 | KIF1B | Sponge network | 0.997 | 0.00066 | -0.118 | 0.32258 | 0.365 |
| 26340 | GAS6-AS2 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-940 | 14 | SFRP1 | Sponge network | 0.16 | 0.50785 | -3.566 | 0 | 0.365 |
| 26341 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 15 | IGF2 | Sponge network | 0.335 | 0.20596 | 0.848 | 0.03842 | 0.365 |
| 26342 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-590-3p | 27 | RICTOR | Sponge network | 1.04 | 0.00023 | 0.388 | 0.00188 | 0.365 |
| 26343 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 15 | RELN | Sponge network | -0.612 | 0.00094 | -2.403 | 0 | 0.365 |
| 26344 | RP1-102E24.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326 | 14 | TBL1XR1 | Sponge network | 1.737 | 0 | 0.578 | 0 | 0.365 |
| 26345 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-93-3p;hsa-miR-940 | 14 | CNTNAP1 | Sponge network | -0.773 | 0.04073 | 0.358 | 0.13727 | 0.365 |
| 26346 | H19 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-664a-3p | 10 | NRXN3 | Sponge network | 2.439 | 0 | -0.893 | 0.04186 | 0.365 |
| 26347 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PBRM1 | Sponge network | 0.91 | 0 | -0.029 | 0.78601 | 0.365 |
| 26348 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-93-5p | 19 | LATS2 | Sponge network | -1.645 | 0 | 0.159 | 0.2304 | 0.365 |
| 26349 | FZD10-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-92a-1-5p;hsa-miR-935 | 46 | GRIK3 | Sponge network | -0.727 | 0.04726 | -3.208 | 0 | 0.365 |
| 26350 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | HOOK3 | Sponge network | 0.862 | 0.06213 | 0.273 | 0.09957 | 0.365 |
| 26351 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 32 | CRTC1 | Sponge network | -0.739 | 0.0574 | -0.116 | 0.28412 | 0.365 |
| 26352 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-942-5p | 22 | GPC6 | Sponge network | -0.162 | 0.47687 | -0.054 | 0.81465 | 0.365 |
| 26353 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-342-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 12 | FBXO31 | Sponge network | 0.16 | 0.50785 | -0.085 | 0.42389 | 0.365 |
| 26354 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 32 | UNC80 | Sponge network | -0.513 | 0.04703 | -1.934 | 0 | 0.365 |
| 26355 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 20 | THBS1 | Sponge network | 0.176 | 0.37402 | 0.741 | 0.00386 | 0.365 |
| 26356 | DIO3OS |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-942-5p | 11 | ZBTB20 | Sponge network | -0.773 | 0.04073 | -0.122 | 0.57569 | 0.365 |
| 26357 | RP11-890B15.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | BRD3 | Sponge network | 0.101 | 0.60919 | 0.37 | 0.00013 | 0.365 |
| 26358 | SH3BP5-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-654-3p;hsa-miR-940 | 15 | BAZ2B | Sponge network | 0.267 | 0.2937 | -0.276 | 0.02833 | 0.365 |
| 26359 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 18 | PTPN11 | Sponge network | 0.101 | 0.60919 | 0.431 | 2.0E-5 | 0.365 |
| 26360 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 27 | BCL11A | Sponge network | -1.111 | 0.0003 | -0.932 | 0.0063 | 0.365 |
| 26361 | GAS6-AS2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-429;hsa-miR-7-1-3p | 15 | ITPKB | Sponge network | 0.16 | 0.50785 | -0.704 | 0.00106 | 0.365 |
| 26362 | RP11-701H24.3 |
hsa-miR-130b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-301a-3p | 10 | ZBTB4 | Sponge network | -1.425 | 0.04204 | -0.739 | 0 | 0.365 |
| 26363 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CHD2 | Sponge network | 0.225 | 0.30644 | 0.049 | 0.55371 | 0.365 |
| 26364 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-589-5p | 23 | EYA4 | Sponge network | -0.154 | 0.78939 | 0.3 | 0.36876 | 0.365 |
| 26365 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 16 | GPAM | Sponge network | 0.75 | 0.00054 | 0.142 | 0.29732 | 0.365 |
| 26366 | NR2F1-AS1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | RASSF3 | Sponge network | -0.49 | 0.04946 | -0.226 | 0.11691 | 0.365 |
| 26367 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RECK | Sponge network | -0.663 | 0.10887 | -1.043 | 0 | 0.365 |
| 26368 | AGAP11 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p | 12 | RIMS1 | Sponge network | -1.645 | 0 | -3.143 | 0 | 0.365 |
| 26369 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | RECK | Sponge network | -4.477 | 0 | -1.043 | 0 | 0.365 |
| 26370 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | TET1 | Sponge network | 0.919 | 0 | -0.135 | 0.52138 | 0.365 |
| 26371 | LINC00863 |
hsa-let-7a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-671-5p | 12 | CRTC3 | Sponge network | 0.189 | 0.13981 | 0.164 | 0.16786 | 0.365 |
| 26372 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 34 | EPHA5 | Sponge network | -1.057 | 0.00029 | -0.957 | 0.01733 | 0.365 |
| 26373 | RP11-65J21.3 |
hsa-let-7d-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 10 | PRRX1 | Sponge network | -0.557 | 0.11135 | 1.316 | 0 | 0.365 |
| 26374 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | RASSF2 | Sponge network | 0.101 | 0.60919 | -0.273 | 0.21961 | 0.365 |
| 26375 | LINC00578 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 13 | SASH1 | Sponge network | 0.811 | 0.03228 | -0.502 | 0.00175 | 0.365 |
| 26376 | RP11-119F7.5 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | RIMS2 | Sponge network | 0.225 | 0.30644 | -1.365 | 0.00208 | 0.365 |
| 26377 | PGM5-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-511-5p | 17 | BTG2 | Sponge network | -4.546 | 0 | -1.763 | 0 | 0.365 |
| 26378 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 45 | WDFY3 | Sponge network | 0.488 | 0.00084 | -0.201 | 0.0967 | 0.365 |
| 26379 | RP11-774O3.3 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 12 | SPOP | Sponge network | -0.487 | 0.02157 | -0.419 | 1.0E-5 | 0.365 |
| 26380 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | ZNF292 | Sponge network | 0.417 | 0.00445 | 0.276 | 0.04148 | 0.365 |
| 26381 | AGAP11 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-455-3p | 15 | TLE4 | Sponge network | -1.645 | 0 | -1.157 | 0 | 0.365 |
| 26382 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p | 21 | ZFHX3 | Sponge network | 0.799 | 1.0E-5 | 0.389 | 0.01363 | 0.365 |
| 26383 | RP11-13K12.5 |
hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-671-5p | 13 | KIT | Sponge network | -0.318 | 0.65847 | -2.184 | 0 | 0.365 |
| 26384 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | PPM1A | Sponge network | -0.737 | 0.06182 | -0.623 | 0 | 0.365 |
| 26385 | RP11-798M19.6 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 32 | GRIK3 | Sponge network | -0.612 | 0.00094 | -3.208 | 0 | 0.365 |
| 26386 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 17 | ZNF292 | Sponge network | 0.72 | 0.0001 | 0.276 | 0.04148 | 0.365 |
| 26387 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLIP1 | Sponge network | -0.031 | 0.89063 | -0.215 | 0.08684 | 0.365 |
| 26388 | RP4-669P10.16 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-542-3p | 11 | SYNM | Sponge network | -0.301 | 0.06597 | -2.309 | 1.0E-5 | 0.365 |
| 26389 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p | 11 | ANK2 | Sponge network | 0.37 | 0.27614 | -1.603 | 1.0E-5 | 0.365 |
| 26390 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | NCOA1 | Sponge network | -0.096 | 0.67958 | -0.432 | 0 | 0.365 |
| 26391 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | EPHA5 | Sponge network | -0.899 | 6.0E-5 | -0.957 | 0.01733 | 0.365 |
| 26392 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p | 21 | PLSCR4 | Sponge network | 0.494 | 0.00339 | -0.474 | 0.03833 | 0.365 |
| 26393 | ZSCAN16-AS1 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | RIMS2 | Sponge network | -0.342 | 0.02288 | -1.365 | 0.00208 | 0.365 |
| 26394 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | AFF3 | Sponge network | -0.021 | 0.93598 | -2.373 | 0 | 0.365 |
| 26395 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-5p | 36 | TSHZ3 | Sponge network | -1.645 | 0 | -0.556 | 0.01516 | 0.365 |
| 26396 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-93-5p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | -0.611 | 0.09607 | -0.085 | 0.42389 | 0.365 |
| 26397 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | EPHA7 | Sponge network | 0.385 | 0.10067 | -2.197 | 3.0E-5 | 0.365 |
| 26398 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 29 | EPHA3 | Sponge network | -0.303 | 0.21002 | 0.185 | 0.53588 | 0.365 |
| 26399 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-877-5p | 22 | CRTC1 | Sponge network | -0.615 | 0.00015 | -0.116 | 0.28412 | 0.365 |
| 26400 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p | 15 | ABCC9 | Sponge network | -0.18 | 0.20343 | -0.466 | 0.14121 | 0.365 |
| 26401 | CTD-2089N3.2 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | LIFR | Sponge network | -1.375 | 0.08642 | -1.794 | 0 | 0.365 |
| 26402 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p | 17 | GJA1 | Sponge network | -0.358 | 0.17255 | -0.485 | 0.02179 | 0.365 |
| 26403 | RP11-35G9.5 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-93-3p | 30 | RUNX1T1 | Sponge network | 0.622 | 0.00015 | -0.344 | 0.2374 | 0.365 |
| 26404 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-7-1-3p | 11 | CLIP1 | Sponge network | 0.389 | 0.04293 | -0.215 | 0.08684 | 0.365 |
| 26405 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-629-5p | 10 | UVRAG | Sponge network | -2.69 | 0.0001 | -0.017 | 0.83865 | 0.365 |
| 26406 | MIR17HG |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-497-5p;hsa-miR-654-3p | 10 | RICTOR | Sponge network | 1.699 | 0 | 0.388 | 0.00188 | 0.365 |
| 26407 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-651-5p | 26 | PRDM2 | Sponge network | -2.117 | 0.00161 | -0.258 | 0.00721 | 0.365 |
| 26408 | RP1-151F17.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 23 | NAV3 | Sponge network | 0.222 | 0.29133 | 0.212 | 0.47729 | 0.365 |
| 26409 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 18 | PSIP1 | Sponge network | 0.311 | 0.10344 | -0.005 | 0.96897 | 0.365 |
| 26410 | FAM66C |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FAM133A | Sponge network | -0.901 | 0.00019 | 0.549 | 0.29009 | 0.365 |
| 26411 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SYNE1 | Sponge network | 0.659 | 3.0E-5 | -0.738 | 0.00316 | 0.365 |
| 26412 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-7-5p;hsa-miR-940 | 12 | CRTC3 | Sponge network | 0.419 | 0.0319 | 0.164 | 0.16786 | 0.365 |
| 26413 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-342-3p;hsa-miR-93-5p | 11 | FBXO31 | Sponge network | -0.709 | 0.18168 | -0.085 | 0.42389 | 0.365 |
| 26414 | RP11-701H24.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 17 | FAT3 | Sponge network | -1.425 | 0.04204 | -1.601 | 0.00051 | 0.365 |
| 26415 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | -0.096 | 0.67958 | -1.751 | 1.0E-5 | 0.365 |
| 26416 | RP11-597D13.9 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | PCDH18 | Sponge network | 1.302 | 7.0E-5 | 0.216 | 0.24645 | 0.365 |
| 26417 | LINC01018 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-671-5p | 29 | DLC1 | Sponge network | -2.915 | 0.00015 | -0.382 | 0.05038 | 0.365 |
| 26418 | RP11-458D21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 22 | BRD3 | Sponge network | 0.663 | 0.00073 | 0.37 | 0.00013 | 0.365 |
| 26419 | RP4-669H2.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p | 10 | USP6 | Sponge network | 1.711 | 0.00019 | 0.188 | 0.3871 | 0.365 |
| 26420 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | ABCA10 | Sponge network | 0.537 | 0.00139 | -0.571 | 0.03674 | 0.365 |
| 26421 | RP11-345P4.7 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-374b-3p;hsa-miR-495-3p | 17 | CREB1 | Sponge network | 1.597 | 0 | 0.203 | 0.00537 | 0.365 |
| 26422 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 34 | ZFHX4 | Sponge network | 0.862 | 0.06213 | 0.018 | 0.95574 | 0.365 |
| 26423 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | EPHA3 | Sponge network | 0.395 | 0.15451 | 0.185 | 0.53588 | 0.365 |
| 26424 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-5p;hsa-miR-424-3p;hsa-miR-628-5p | 17 | BMPR2 | Sponge network | 1.162 | 5.0E-5 | 0.206 | 0.0635 | 0.365 |
| 26425 | RP11-356J5.12 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-505-5p | 11 | WNK2 | Sponge network | -0.899 | 6.0E-5 | -0.352 | 0.31066 | 0.365 |
| 26426 | SNHG14 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 25 | HIP1 | Sponge network | -1.111 | 0.0003 | 0.774 | 0 | 0.365 |
| 26427 | KCNJ2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 12 | BCL11A | Sponge network | -0.436 | 0.05325 | -0.932 | 0.0063 | 0.365 |
| 26428 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-93-5p | 14 | PIK3R1 | Sponge network | -1.255 | 0.00981 | -0.133 | 0.36854 | 0.365 |
| 26429 | AC009948.5 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 19 | SORCS1 | Sponge network | 0.532 | 0.00505 | -3.492 | 0 | 0.365 |
| 26430 | RP11-758N13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p | 12 | NUAK1 | Sponge network | 2.939 | 3.0E-5 | 0.904 | 0 | 0.365 |
| 26431 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-93-5p | 16 | LRRC55 | Sponge network | -2.69 | 0.0001 | -0.026 | 0.93163 | 0.365 |
| 26432 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p | 35 | CAMTA1 | Sponge network | -0.612 | 0.00094 | -0.436 | 1.0E-5 | 0.365 |
| 26433 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-7-1-3p | 10 | CNTN1 | Sponge network | 0.693 | 0.00076 | -2.594 | 0 | 0.365 |
| 26434 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-7-1-3p | 22 | EYA4 | Sponge network | -1.092 | 4.0E-5 | 0.3 | 0.36876 | 0.364 |
| 26435 | AC016747.3 |
hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | ZDBF2 | Sponge network | 0.199 | 0.38015 | -0.998 | 0.00025 | 0.364 |
| 26436 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PDE4DIP | Sponge network | 0.349 | 0.01695 | -0.282 | 0.0257 | 0.364 |
| 26437 | RP11-195F19.9 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 12 | JDP2 | Sponge network | -0.726 | 0.00132 | -0.967 | 0 | 0.364 |
| 26438 | FGD5-AS1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p;hsa-miR-495-3p | 13 | PHF6 | Sponge network | 0.401 | 0.00066 | 0.785 | 0 | 0.364 |
| 26439 | LINC00654 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 23 | RIMBP2 | Sponge network | 0.374 | 0.20054 | -1.968 | 5.0E-5 | 0.364 |
| 26440 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | RCAN2 | Sponge network | 0.349 | 0.01695 | -0.33 | 0.15172 | 0.364 |
| 26441 | KB-1572G7.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-378a-5p;hsa-miR-495-3p | 19 | CREB1 | Sponge network | 0.924 | 0 | 0.203 | 0.00537 | 0.364 |
| 26442 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-502-3p | 20 | RUNX1T1 | Sponge network | 0.37 | 0.27614 | -0.344 | 0.2374 | 0.364 |
| 26443 | GHRLOS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-3p | 10 | RELN | Sponge network | -1.942 | 0 | -2.403 | 0 | 0.364 |
| 26444 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | ST6GALNAC3 | Sponge network | 0.727 | 3.0E-5 | -0.987 | 0 | 0.364 |
| 26445 | LINC00865 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429 | 22 | CBFA2T3 | Sponge network | -1.249 | 7.0E-5 | -1.221 | 1.0E-5 | 0.364 |
| 26446 | RP11-806O11.1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ATRX | Sponge network | 0.284 | 0.52463 | 0.261 | 0.04408 | 0.364 |
| 26447 | CTC-297N7.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-3p | 21 | NTRK3 | Sponge network | -2.069 | 0 | -1.77 | 3.0E-5 | 0.364 |
| 26448 | PCA3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CDH10 | Sponge network | -0.709 | 0.18168 | -2.397 | 0 | 0.364 |
| 26449 | HCG18 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 18 | SLC5A3 | Sponge network | 1.063 | 0 | 0.493 | 5.0E-5 | 0.364 |
| 26450 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p | 23 | CXXC4 | Sponge network | 0.056 | 0.81195 | -0.433 | 0.21055 | 0.364 |
| 26451 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | ABCC9 | Sponge network | 0.395 | 0.15451 | -0.466 | 0.14121 | 0.364 |
| 26452 | RP11-48B3.4 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p | 17 | ANK2 | Sponge network | 1.757 | 0 | -1.603 | 1.0E-5 | 0.364 |
| 26453 | RAB30-AS1 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-625-3p | 18 | ABL2 | Sponge network | 0.752 | 0 | 0.82 | 0 | 0.364 |
| 26454 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-511-5p | 22 | BTG2 | Sponge network | -0.739 | 0.00044 | -1.763 | 0 | 0.364 |
| 26455 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | PLSCR4 | Sponge network | 0.853 | 0.15352 | -0.474 | 0.03833 | 0.364 |
| 26456 | BOLA3-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-629-3p | 10 | CXCL12 | Sponge network | -0.246 | 0.35034 | -1.421 | 0 | 0.364 |
| 26457 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 35 | AKAP11 | Sponge network | 0.194 | 0.64278 | 0.196 | 0.09825 | 0.364 |
| 26458 | LPP-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-577 | 12 | SEMA3E | Sponge network | -0.18 | 0.20343 | -1.717 | 0.00137 | 0.364 |
| 26459 | LINC00476 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-5p | 14 | MN1 | Sponge network | -0.615 | 0.00015 | -0.358 | 0.24172 | 0.364 |
| 26460 | LINC00476 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 18 | TAL1 | Sponge network | -0.615 | 0.00015 | -0.462 | 0.00574 | 0.364 |
| 26461 | RP11-967K21.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 14 | BCL9 | Sponge network | 0.056 | 0.81195 | 0.321 | 0.00267 | 0.364 |
| 26462 | NEAT1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | ARHGEF12 | Sponge network | 0.705 | 0.00014 | 0.09 | 0.35693 | 0.364 |
| 26463 | AC005154.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-940 | 15 | DLG1 | Sponge network | 0.848 | 0 | -0.014 | 0.8926 | 0.364 |
| 26464 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 20 | CRTC3 | Sponge network | -0.793 | 7.0E-5 | 0.164 | 0.16786 | 0.364 |
| 26465 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500b-5p | 15 | RSPO2 | Sponge network | 0.811 | 0.03228 | -3.038 | 0 | 0.364 |
| 26466 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-93-5p | 19 | SLITRK3 | Sponge network | -0.969 | 2.0E-5 | -3.328 | 0 | 0.364 |
| 26467 | RP11-216B9.6 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p | 35 | DTNA | Sponge network | 0.174 | 0.30034 | -1.648 | 1.0E-5 | 0.364 |
| 26468 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | EPHA5 | Sponge network | -1.161 | 0.00591 | -0.957 | 0.01733 | 0.364 |
| 26469 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 30 | EYA4 | Sponge network | -2.452 | 0 | 0.3 | 0.36876 | 0.364 |
| 26470 | CALML3-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 11 | FLNA | Sponge network | 0.95 | 0.15943 | -0.771 | 0.01377 | 0.364 |
| 26471 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | EBF3 | Sponge network | 1.302 | 7.0E-5 | -0.058 | 0.81437 | 0.364 |
| 26472 | RP11-506M13.3 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 11 | PAFAH1B2 | Sponge network | 0.769 | 0 | 0.318 | 0.0001 | 0.364 |
| 26473 | HOTAIRM1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PEG3 | Sponge network | -0.053 | 0.80685 | -1.74 | 0 | 0.364 |
| 26474 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | TOPORS | Sponge network | -0.222 | 0.32445 | -0.226 | 0.00524 | 0.364 |
| 26475 | RP11-37B2.1 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p | 13 | SIRT1 | Sponge network | 1.03 | 0 | 0.109 | 0.2593 | 0.364 |
| 26476 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -0.31 | 0.33871 | -0.243 | 0.11408 | 0.364 |
| 26477 | RP11-400K9.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 21 | PDE4DIP | Sponge network | -0.611 | 0.09607 | -0.282 | 0.0257 | 0.364 |
| 26478 | RP11-554A11.4 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p | 11 | OPCML | Sponge network | -0.583 | 0.14589 | -2.498 | 0 | 0.364 |
| 26479 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PIK3R1 | Sponge network | -0.295 | 0.02604 | -0.133 | 0.36854 | 0.364 |
| 26480 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 26 | CHD5 | Sponge network | 0.056 | 0.81195 | -0.922 | 0.01521 | 0.364 |
| 26481 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-432-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 51 | PPM1L | Sponge network | -0.355 | 0.0077 | -0.537 | 0.00616 | 0.364 |
| 26482 | HCG11 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | BRD3 | Sponge network | 0.385 | 0.10067 | 0.37 | 0.00013 | 0.364 |
| 26483 | LINC00886 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-542-3p | 17 | DYNC1I1 | Sponge network | -0.371 | 0.24604 | -1.12 | 0.00107 | 0.364 |
| 26484 | RP11-798G7.7 |
hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | UBR3 | Sponge network | 0.858 | 3.0E-5 | -0.021 | 0.82774 | 0.364 |
| 26485 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 13 | DCLK1 | Sponge network | -0.125 | 0.49414 | -1.324 | 0.00014 | 0.364 |
| 26486 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-654-3p | 16 | ITCH | Sponge network | 0.72 | 0.0001 | 0.084 | 0.34058 | 0.364 |
| 26487 | RP11-130L8.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-3607-3p;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | PRUNE2 | Sponge network | -0.663 | 0.10887 | -1.733 | 0.00022 | 0.364 |
| 26488 | LINC00473 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935 | 73 | LPP | Sponge network | -1.203 | 0.03881 | -0.459 | 0.04475 | 0.364 |
| 26489 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | DKK3 | Sponge network | -0.777 | 0.1134 | -0.052 | 0.77783 | 0.364 |
| 26490 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-501-5p | 11 | TXNIP | Sponge network | -1.161 | 0.00591 | -1.277 | 0 | 0.364 |
| 26491 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p | 12 | CBFA2T3 | Sponge network | -0.376 | 0.00337 | -1.221 | 1.0E-5 | 0.364 |
| 26492 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | PDS5B | Sponge network | 0.848 | 0 | 0.259 | 0.00962 | 0.364 |
| 26493 | FZD10-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 23 | SYNPO2 | Sponge network | -0.727 | 0.04726 | -2.342 | 0 | 0.364 |
| 26494 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | NCAM2 | Sponge network | 0.862 | 0.06213 | -0.482 | 0.22565 | 0.364 |
| 26495 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-93-3p | 28 | ST6GAL2 | Sponge network | -1.645 | 0 | -1.201 | 0.00597 | 0.364 |
| 26496 | FAM66C |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | CSDE1 | Sponge network | -0.901 | 0.00019 | -0.493 | 0 | 0.364 |
| 26497 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p | 17 | PTPRB | Sponge network | -0.162 | 0.47687 | 0.105 | 0.47122 | 0.364 |
| 26498 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 28 | EPHA5 | Sponge network | 0.889 | 9.0E-5 | -0.957 | 0.01733 | 0.364 |
| 26499 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 58 | IL6ST | Sponge network | 0.083 | 0.66405 | -0.402 | 0.00676 | 0.364 |
| 26500 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | 0.537 | 0.00139 | 0.809 | 0.00544 | 0.364 |
| 26501 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p | 19 | ARHGAP29 | Sponge network | -0.222 | 0.32445 | 0.131 | 0.42953 | 0.364 |
| 26502 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 45 | RBMS3 | Sponge network | -0.969 | 2.0E-5 | -0.519 | 0.03551 | 0.364 |
| 26503 | BAIAP2-AS1 |
hsa-let-7g-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c | 11 | BRD3 | Sponge network | 0.702 | 6.0E-5 | 0.37 | 0.00013 | 0.364 |
| 26504 | RP5-1139B12.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 14 | IGF1 | Sponge network | 0.598 | 0.38481 | -0.204 | 0.5898 | 0.364 |
| 26505 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 14 | AKAP6 | Sponge network | 0.4 | 0.14611 | -1.586 | 3.0E-5 | 0.364 |
| 26506 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-582-5p | 21 | PPP1R9A | Sponge network | -0.469 | 0.08721 | -0.903 | 0.04254 | 0.364 |
| 26507 | RP11-156P1.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | BRD3 | Sponge network | 0.214 | 0.18246 | 0.37 | 0.00013 | 0.364 |
| 26508 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 15 | CLIP1 | Sponge network | -0.793 | 7.0E-5 | -0.215 | 0.08684 | 0.364 |
| 26509 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 23 | ADAMTSL3 | Sponge network | 0.385 | 0.10067 | -1.559 | 0 | 0.364 |
| 26510 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-655-3p | 33 | DMD | Sponge network | 0.72 | 0.0001 | -1.506 | 0 | 0.364 |
| 26511 | RP11-175O19.4 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-497-5p | 10 | RPS6KA3 | Sponge network | 0.917 | 0 | 0.256 | 0.02419 | 0.364 |
| 26512 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 23 | KIT | Sponge network | 0.194 | 0.64278 | -2.184 | 0 | 0.364 |
| 26513 | LINC01018 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | LRIG1 | Sponge network | -2.915 | 0.00015 | -0.537 | 0.00588 | 0.364 |
| 26514 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | -0.384 | 0.40652 | -1.751 | 1.0E-5 | 0.364 |
| 26515 | ZSCAN16-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 19 | ARNT2 | Sponge network | -0.342 | 0.02288 | -0.332 | 0.25851 | 0.364 |
| 26516 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-484 | 13 | DDX6 | Sponge network | 0.93 | 0 | 0.091 | 0.36428 | 0.364 |
| 26517 | TINCR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-96-5p | 21 | HBP1 | Sponge network | -0.664 | 0.14543 | -0.454 | 0 | 0.364 |
| 26518 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 14 | PRDM5 | Sponge network | 0.101 | 0.60919 | -1.09 | 0.00014 | 0.364 |
| 26519 | RP4-669L17.10 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-539-5p | 12 | TSC1 | Sponge network | 1.093 | 0 | 0.103 | 0.26071 | 0.364 |
| 26520 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 12 | MLLT10 | Sponge network | 0.439 | 0.00313 | 0.105 | 0.33292 | 0.364 |
| 26521 | H1FX-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-93-3p | 21 | CTIF | Sponge network | -0.206 | 0.12942 | -0.389 | 0.00549 | 0.364 |
| 26522 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 44 | EPHA5 | Sponge network | -0.615 | 0.00015 | -0.957 | 0.01733 | 0.364 |
| 26523 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-877-5p | 11 | PGR | Sponge network | 1.122 | 0.02989 | -1.704 | 0 | 0.364 |
| 26524 | PSMG3-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p | 12 | MAPK10 | Sponge network | -0.125 | 0.49414 | -1.774 | 0 | 0.364 |
| 26525 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-590-5p | 21 | CNTN1 | Sponge network | -0.384 | 0.40652 | -2.594 | 0 | 0.364 |
| 26526 | LINC01018 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | HIP1 | Sponge network | -2.915 | 0.00015 | 0.774 | 0 | 0.364 |
| 26527 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 22 | HIP1 | Sponge network | 0.064 | 0.72934 | 0.774 | 0 | 0.364 |
| 26528 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p | 13 | PKNOX1 | Sponge network | -0.612 | 0.00094 | 0.085 | 0.28917 | 0.364 |
| 26529 | RP11-25K19.1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 12 | RHOB | Sponge network | -1.057 | 0.00029 | -1.052 | 0 | 0.364 |
| 26530 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 45 | CBX5 | Sponge network | -0.487 | 0.02157 | 0.165 | 0.24446 | 0.364 |
| 26531 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-542-3p | 10 | TSC22D1 | Sponge network | -2.452 | 0 | -0.529 | 2.0E-5 | 0.364 |
| 26532 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-93-5p | 35 | UNC80 | Sponge network | -1.349 | 0.00017 | -1.934 | 0 | 0.364 |
| 26533 | RP11-83A24.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 13 | KIAA1024 | Sponge network | 0.417 | 0.00445 | 1.083 | 0 | 0.364 |
| 26534 | LINC00969 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-766-3p | 14 | ATM | Sponge network | 0.683 | 1.0E-5 | 0.178 | 0.21568 | 0.364 |
| 26535 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p | 50 | THRB | Sponge network | -0.303 | 0.21002 | -1.117 | 0.0004 | 0.364 |
| 26536 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | EYA4 | Sponge network | -0.942 | 0 | 0.3 | 0.36876 | 0.364 |
| 26537 | BVES-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429 | 13 | CSMD1 | Sponge network | -2.62 | 0 | -0.63 | 0.18881 | 0.364 |
| 26538 | CTD-2245F17.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-629-3p;hsa-miR-7-5p | 12 | ESR1 | Sponge network | -0.421 | 0.12556 | -1.035 | 3.0E-5 | 0.364 |
| 26539 | C4B-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940;hsa-miR-96-5p | 21 | GAS7 | Sponge network | 2.306 | 9.0E-5 | -0.555 | 0.01602 | 0.364 |
| 26540 | AC093110.3 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-495-3p | 11 | DICER1 | Sponge network | 1.044 | 2.0E-5 | 0.042 | 0.674 | 0.364 |
| 26541 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-629-3p | 15 | THBS1 | Sponge network | -1.654 | 0.00144 | 0.741 | 0.00386 | 0.364 |
| 26542 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-93-3p | 37 | BMPR2 | Sponge network | -0.154 | 0.53954 | 0.206 | 0.0635 | 0.364 |
| 26543 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-5p;hsa-miR-744-3p | 10 | CUL5 | Sponge network | 1.134 | 0 | 0.026 | 0.77301 | 0.364 |
| 26544 | CTA-204B4.2 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-744-3p | 38 | LPP | Sponge network | 0.765 | 7.0E-5 | -0.459 | 0.04475 | 0.364 |
| 26545 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | KAT6B | Sponge network | -0.612 | 0.00094 | -0.478 | 0.00018 | 0.364 |
| 26546 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 20 | PCDH10 | Sponge network | 0.174 | 0.30034 | -1.644 | 0.0078 | 0.364 |
| 26547 | CTC-429P9.2 |
hsa-miR-1266-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-485-3p;hsa-miR-877-5p | 10 | AFF1 | Sponge network | 0.043 | 0.81628 | -0.384 | 0.00053 | 0.364 |
| 26548 | TPTEP1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 24 | ZFP36L1 | Sponge network | -1.216 | 0 | -0.505 | 8.0E-5 | 0.364 |
| 26549 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 34 | DMXL1 | Sponge network | -2.452 | 0 | -0.426 | 0.00056 | 0.364 |
| 26550 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-93-5p | 16 | LHX6 | Sponge network | -1.575 | 6.0E-5 | -0.087 | 0.69193 | 0.364 |
| 26551 | RP11-439A17.10 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-5p | 10 | TCF4 | Sponge network | 0.051 | 0.95756 | 0.016 | 0.92271 | 0.364 |
| 26552 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 19 | GPC6 | Sponge network | 0.619 | 0.00157 | -0.054 | 0.81465 | 0.364 |
| 26553 | TPTEP1 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 10 | EPB41L3 | Sponge network | -1.216 | 0 | -1.193 | 0 | 0.364 |
| 26554 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | DPYD | Sponge network | -0.737 | 0.06182 | -0.736 | 0.00076 | 0.364 |
| 26555 | LINC00969 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p | 11 | MAT2A | Sponge network | 0.683 | 1.0E-5 | -0.073 | 0.36195 | 0.364 |
| 26556 | MIR600HG |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-96-5p | 25 | PIK3R1 | Sponge network | 0.594 | 0.41522 | -0.133 | 0.36854 | 0.364 |
| 26557 | RP11-359B12.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PHF3 | Sponge network | 0.064 | 0.72934 | 0.126 | 0.19652 | 0.364 |
| 26558 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-877-5p | 10 | FLI1 | Sponge network | -0.777 | 0.16667 | -0.294 | 0.10905 | 0.364 |
| 26559 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | TAL1 | Sponge network | -2.46 | 0 | -0.462 | 0.00574 | 0.364 |
| 26560 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-429 | 11 | ELMO1 | Sponge network | -2.69 | 0.0001 | 0.04 | 0.83147 | 0.364 |
| 26561 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-98-5p | 50 | PRKAA2 | Sponge network | -0.355 | 0.0077 | -2.462 | 0 | 0.364 |
| 26562 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-539-5p;hsa-miR-766-3p | 20 | AFF4 | Sponge network | 0.858 | 3.0E-5 | -0.266 | 0.01565 | 0.364 |
| 26563 | RP11-434B12.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PRUNE2 | Sponge network | -0.031 | 0.89063 | -1.733 | 0.00022 | 0.364 |
| 26564 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-93-3p | 26 | TRPS1 | Sponge network | -0.469 | 0.08721 | -0.191 | 0.44109 | 0.364 |
| 26565 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-582-5p | 22 | PIK3C2A | Sponge network | 0.637 | 0.00069 | 0.061 | 0.53776 | 0.364 |
| 26566 | GNG12-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p | 22 | JDP2 | Sponge network | -0.793 | 7.0E-5 | -0.967 | 0 | 0.364 |
| 26567 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 47 | FOXP1 | Sponge network | -0.899 | 6.0E-5 | 0.042 | 0.74368 | 0.364 |
| 26568 | LINC00476 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ZNF208 | Sponge network | -0.615 | 0.00015 | -0.4 | 0.22381 | 0.364 |
| 26569 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-5p | 20 | CRTC1 | Sponge network | 0.051 | 0.83388 | -0.116 | 0.28412 | 0.364 |
| 26570 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-501-3p | 19 | APC | Sponge network | -1.047 | 0 | -0.255 | 0.04051 | 0.364 |
| 26571 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 15 | CXXC4 | Sponge network | -0.318 | 0.65847 | -0.433 | 0.21055 | 0.364 |
| 26572 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | DKK3 | Sponge network | -0.323 | 0.01372 | -0.052 | 0.77783 | 0.364 |
| 26573 | CTC-559E9.6 |
hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-654-3p | 14 | PIK3R1 | Sponge network | 0.601 | 1.0E-5 | -0.133 | 0.36854 | 0.364 |
| 26574 | ZRANB2-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p | 10 | BTG2 | Sponge network | -0.376 | 0.00337 | -1.763 | 0 | 0.364 |
| 26575 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | MED12L | Sponge network | -0.576 | 0.10121 | -0.194 | 0.55299 | 0.364 |
| 26576 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-628-5p | 12 | FBXO32 | Sponge network | -1.116 | 0.23686 | -0.495 | 0.07561 | 0.364 |
| 26577 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | MYBL1 | Sponge network | 1.378 | 0 | 0.494 | 0.02152 | 0.364 |
| 26578 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p | 14 | PBX1 | Sponge network | -1.425 | 0.04204 | -1.206 | 0 | 0.364 |
| 26579 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-93-5p | 10 | EZH1 | Sponge network | 0.056 | 0.8494 | -0.564 | 1.0E-5 | 0.364 |
| 26580 | NR2F1-AS1 |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p | 19 | PAIP2 | Sponge network | -0.49 | 0.04946 | -0.515 | 0 | 0.364 |
| 26581 | LINC01018 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | MED12L | Sponge network | -2.915 | 0.00015 | -0.194 | 0.55299 | 0.364 |
| 26582 | RP5-1120P11.1 |
hsa-miR-101-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-486-5p | 10 | PHF6 | Sponge network | 2.957 | 0 | 0.785 | 0 | 0.364 |
| 26583 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-942-5p | 31 | HLF | Sponge network | -0.513 | 0.04703 | -2.443 | 0 | 0.364 |
| 26584 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-532-3p;hsa-miR-550a-5p | 15 | RIMBP2 | Sponge network | -0.384 | 0.40652 | -1.968 | 5.0E-5 | 0.364 |
| 26585 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p | 10 | CXXC4 | Sponge network | -0.517 | 0.02935 | -0.433 | 0.21055 | 0.364 |
| 26586 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | AKAP6 | Sponge network | 0.131 | 0.8436 | -1.586 | 3.0E-5 | 0.364 |
| 26587 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | DMXL1 | Sponge network | -0.323 | 0.01372 | -0.426 | 0.00056 | 0.364 |
| 26588 | NNT-AS1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 16 | PHF6 | Sponge network | 0.799 | 1.0E-5 | 0.785 | 0 | 0.364 |
| 26589 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-576-5p | 13 | PTPRD | Sponge network | 1.122 | 0.02989 | -1.005 | 0.00064 | 0.364 |
| 26590 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p | 14 | CLIP1 | Sponge network | -2.69 | 0.0001 | -0.215 | 0.08684 | 0.364 |
| 26591 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p | 17 | EYA4 | Sponge network | 0.381 | 0.25967 | 0.3 | 0.36876 | 0.364 |
| 26592 | PWAR6 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | EPB41L3 | Sponge network | -1.575 | 6.0E-5 | -1.193 | 0 | 0.364 |
| 26593 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-589-5p | 14 | RARB | Sponge network | -0.309 | 0.1145 | -0.825 | 2.0E-5 | 0.364 |
| 26594 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CXXC4 | Sponge network | -0.737 | 0.10822 | -0.433 | 0.21055 | 0.364 |
| 26595 | SERTAD4-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-576-5p | 23 | NFIB | Sponge network | -0.932 | 0.00329 | 0.023 | 0.90795 | 0.364 |
| 26596 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-935 | 17 | ARNT2 | Sponge network | -1.331 | 3.0E-5 | -0.332 | 0.25851 | 0.364 |
| 26597 | LINC00472 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 12 | HOMER2 | Sponge network | -0.842 | 0.01361 | -2.698 | 0 | 0.364 |
| 26598 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 17 | PSIP1 | Sponge network | 0.083 | 0.66405 | -0.005 | 0.96897 | 0.364 |
| 26599 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | TACC1 | Sponge network | -0.053 | 0.80685 | -0.362 | 0.05406 | 0.364 |
| 26600 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-935;hsa-miR-96-5p | 38 | MTSS1 | Sponge network | -2.46 | 0 | -0.81 | 0.00035 | 0.364 |
| 26601 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 18 | PRKAR1A | Sponge network | -0.769 | 0.00035 | -0.063 | 0.50314 | 0.364 |
| 26602 | DIO3OS |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-629-3p | 14 | ID4 | Sponge network | -0.773 | 0.04073 | -1.644 | 0 | 0.364 |
| 26603 | RP11-159D12.2 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p | 13 | MAT2A | Sponge network | 1.254 | 0 | -0.073 | 0.36195 | 0.364 |
| 26604 | RP11-2B6.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p | 11 | ERCC4 | Sponge network | 0.539 | 0.03722 | 0.126 | 0.15266 | 0.364 |
| 26605 | FAM157B |
hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p | 10 | AKAP11 | Sponge network | 0.92 | 0 | 0.196 | 0.09825 | 0.364 |
| 26606 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 24 | MCC | Sponge network | 0.056 | 0.8494 | -1.005 | 0 | 0.364 |
| 26607 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 21 | CDC73 | Sponge network | 0.537 | 0.00139 | 0.094 | 0.20463 | 0.364 |
| 26608 | RP11-389C8.2 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p | 10 | STARD13 | Sponge network | 0.727 | 3.0E-5 | -0.187 | 0.27383 | 0.364 |
| 26609 | CTD-2647L4.4 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-495-3p | 10 | CREB1 | Sponge network | 0.555 | 0.00197 | 0.203 | 0.00537 | 0.364 |
| 26610 | RP11-254F7.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p | 22 | ETV1 | Sponge network | 0.176 | 0.37402 | -0.096 | 0.67674 | 0.364 |
| 26611 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 35 | ABL2 | Sponge network | -0.129 | 0.57177 | 0.82 | 0 | 0.364 |
| 26612 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | LIMD1 | Sponge network | 0.75 | 0.00054 | 0.103 | 0.28978 | 0.364 |
| 26613 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p | 15 | BCL6 | Sponge network | 0.199 | 0.38015 | -0.211 | 0.19489 | 0.364 |
| 26614 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 16 | SUFU | Sponge network | 0.16 | 0.50785 | -0.04 | 0.65489 | 0.364 |
| 26615 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 62 | AFF4 | Sponge network | 0.532 | 0.00505 | -0.266 | 0.01565 | 0.364 |
| 26616 | AC010226.4 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-330-3p;hsa-miR-590-3p | 12 | JDP2 | Sponge network | -0.811 | 2.0E-5 | -0.967 | 0 | 0.364 |
| 26617 | RP11-736K20.5 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | CSDE1 | Sponge network | -0.358 | 0.17255 | -0.493 | 0 | 0.364 |
| 26618 | RP11-434B12.1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | -0.031 | 0.89063 | 0.098 | 0.40994 | 0.364 |
| 26619 | OIP5-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 27 | SIRT1 | Sponge network | 0.421 | 0.00084 | 0.109 | 0.2593 | 0.364 |
| 26620 | RP11-401P9.4 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-342-3p | 13 | STARD13 | Sponge network | -0.384 | 0.40652 | -0.187 | 0.27383 | 0.364 |
| 26621 | LINC00473 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-590-3p | 17 | STARD13 | Sponge network | -1.203 | 0.03881 | -0.187 | 0.27383 | 0.364 |
| 26622 | AP001055.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-331-3p | 10 | CDH23 | Sponge network | 0.693 | 0.00076 | -0.974 | 0.00034 | 0.364 |
| 26623 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-221-3p;hsa-miR-330-3p;hsa-miR-7-1-3p | 15 | PLSCR4 | Sponge network | 0.238 | 0.70544 | -0.474 | 0.03833 | 0.364 |
| 26624 | RP13-516M14.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429 | 22 | ABL2 | Sponge network | 0.389 | 0.04293 | 0.82 | 0 | 0.364 |
| 26625 | HOXD-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p | 11 | ASTN1 | Sponge network | -0.208 | 0.65592 | -2.389 | 2.0E-5 | 0.364 |
| 26626 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-940 | 25 | NTRK2 | Sponge network | 0.174 | 0.30034 | -0.736 | 0.06495 | 0.364 |
| 26627 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | IGSF10 | Sponge network | -0.615 | 0.00015 | -1.751 | 1.0E-5 | 0.364 |
| 26628 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p | 11 | NALCN | Sponge network | 0.253 | 0.09565 | 1.068 | 0.00237 | 0.364 |
| 26629 | AC009948.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p | 11 | FBXO11 | Sponge network | 0.532 | 0.00505 | 0.142 | 0.04938 | 0.364 |
| 26630 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-339-5p | 11 | MLLT10 | Sponge network | 0.79 | 0 | 0.105 | 0.33292 | 0.364 |
| 26631 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 11 | RIMS1 | Sponge network | -0.487 | 0.02157 | -3.143 | 0 | 0.364 |
| 26632 | RP11-212P7.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 19 | MRVI1 | Sponge network | 0.219 | 0.1516 | -1.051 | 0.00022 | 0.364 |
| 26633 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p | 11 | ZNF292 | Sponge network | 0.082 | 0.55397 | 0.276 | 0.04148 | 0.364 |
| 26634 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | LRP1B | Sponge network | -0.421 | 0.0114 | -2.163 | 0 | 0.364 |
| 26635 | RP11-6O2.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-590-5p | 14 | SPG20 | Sponge network | 0.331 | 0.57247 | -1.16 | 0 | 0.364 |
| 26636 | AC083843.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 11 | CUL5 | Sponge network | 1.4 | 0 | 0.026 | 0.77301 | 0.364 |
| 26637 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p | 19 | NTRK2 | Sponge network | 0.178 | 0.29106 | -0.736 | 0.06495 | 0.364 |
| 26638 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | PAK3 | Sponge network | 0.101 | 0.60919 | -0.628 | 0.07253 | 0.364 |
| 26639 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-671-5p | 25 | ARID5B | Sponge network | -0.096 | 0.67958 | 0.342 | 0.01676 | 0.364 |
| 26640 | AC016747.3 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-940 | 13 | SFRP1 | Sponge network | 0.199 | 0.38015 | -3.566 | 0 | 0.363 |
| 26641 | RP11-597D13.8 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-185-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-671-5p | 10 | GAS7 | Sponge network | 0.936 | 0.03889 | -0.555 | 0.01602 | 0.363 |
| 26642 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 41 | NFIB | Sponge network | -0.031 | 0.89063 | 0.023 | 0.90795 | 0.363 |
| 26643 | AC127904.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PIK3CA | Sponge network | 1.308 | 0 | 0.195 | 0.06908 | 0.363 |
| 26644 | RP11-25K19.1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | OPCML | Sponge network | -1.057 | 0.00029 | -2.498 | 0 | 0.363 |
| 26645 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-96-5p | 49 | FAT3 | Sponge network | 0.083 | 0.66405 | -1.601 | 0.00051 | 0.363 |
| 26646 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3614-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p | 23 | JAZF1 | Sponge network | 0.214 | 0.18246 | -0.377 | 0.02677 | 0.363 |
| 26647 | RP5-855D21.3 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p | 11 | ABL2 | Sponge network | 1.728 | 0 | 0.82 | 0 | 0.363 |
| 26648 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-486-5p | 22 | LRIG1 | Sponge network | 0.176 | 0.37402 | -0.537 | 0.00588 | 0.363 |
| 26649 | LINC00476 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 15 | RIMS1 | Sponge network | -0.615 | 0.00015 | -3.143 | 0 | 0.363 |
| 26650 | PCA3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | -0.709 | 0.18168 | -0.001 | 0.99669 | 0.363 |
| 26651 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 12 | FOXP1 | Sponge network | -2.039 | 0.03447 | 0.042 | 0.74368 | 0.363 |
| 26652 | CKMT2-AS1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 11 | FHL1 | Sponge network | 0.082 | 0.55654 | -2.687 | 0 | 0.363 |
| 26653 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-374a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | ABCC9 | Sponge network | 0.051 | 0.83388 | -0.466 | 0.14121 | 0.363 |
| 26654 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-935 | 27 | FBXO32 | Sponge network | 0.392 | 0.0278 | -0.495 | 0.07561 | 0.363 |
| 26655 | RP11-2H8.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-590-3p | 11 | PHF3 | Sponge network | 1.089 | 0 | 0.126 | 0.19652 | 0.363 |
| 26656 | AC018890.6 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-550a-3p | 12 | CACNA1E | Sponge network | 1.61 | 0.00961 | 1.752 | 0 | 0.363 |
| 26657 | RP11-102N12.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-96-5p | 11 | UNC5C | Sponge network | -0.351 | 0.11358 | -0.178 | 0.40214 | 0.363 |
| 26658 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-382-5p;hsa-miR-450a-5p | 20 | PBX1 | Sponge network | -0.285 | 0.2172 | -1.206 | 0 | 0.363 |
| 26659 | LINC00476 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-877-5p | 27 | NAV3 | Sponge network | -0.615 | 0.00015 | 0.212 | 0.47729 | 0.363 |
| 26660 | PDXDC2P |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-497-5p;hsa-miR-9-5p | 11 | CDK6 | Sponge network | 0.756 | 0 | 0.991 | 0 | 0.363 |
| 26661 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 17 | SON | Sponge network | 1.317 | 0 | 0.168 | 0.04406 | 0.363 |
| 26662 | WDFY3-AS2 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | SPOP | Sponge network | -0.942 | 0 | -0.419 | 1.0E-5 | 0.363 |
| 26663 | MRVI1-AS1 |
hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-7-1-3p | 11 | DOCK4 | Sponge network | -1.092 | 4.0E-5 | 0.502 | 0.00108 | 0.363 |
| 26664 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 17 | AKAP11 | Sponge network | 0.799 | 1.0E-5 | 0.196 | 0.09825 | 0.363 |
| 26665 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 27 | PRKAB2 | Sponge network | -0.976 | 0.00025 | -0.367 | 0.01371 | 0.363 |
| 26666 | HCG18 |
hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 12 | CDK6 | Sponge network | 1.063 | 0 | 0.991 | 0 | 0.363 |
| 26667 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 34 | EPHA7 | Sponge network | 0.4 | 0.14611 | -2.197 | 3.0E-5 | 0.363 |
| 26668 | RP11-822E23.8 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p | 31 | IL17RD | Sponge network | -0.777 | 0.16667 | -0.019 | 0.94873 | 0.363 |
| 26669 | CTC-297N7.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-584-5p;hsa-miR-590-5p | 18 | CADM2 | Sponge network | -2.069 | 0 | -3.782 | 0 | 0.363 |
| 26670 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 26 | USP12 | Sponge network | -1.249 | 7.0E-5 | -0.102 | 0.40768 | 0.363 |
| 26671 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-935 | 42 | FAT3 | Sponge network | 0.75 | 0.00054 | -1.601 | 0.00051 | 0.363 |
| 26672 | LINC00621 |
hsa-miR-10a-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | SETBP1 | Sponge network | 0.131 | 0.8436 | -1.302 | 1.0E-5 | 0.363 |
| 26673 | MIR22HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-708-3p;hsa-miR-93-3p | 15 | UNC5C | Sponge network | -1.047 | 0 | -0.178 | 0.40214 | 0.363 |
| 26674 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 25 | MYBL1 | Sponge network | 0.083 | 0.66405 | 0.494 | 0.02152 | 0.363 |
| 26675 | FENDRR |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 13 | EHD3 | Sponge network | -1.281 | 0.00012 | -0.484 | 0.01747 | 0.363 |
| 26676 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 48 | PPM1A | Sponge network | -1.281 | 0.00012 | -0.623 | 0 | 0.363 |
| 26677 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | USP25 | Sponge network | -0.129 | 0.57177 | -0.242 | 0.01397 | 0.363 |
| 26678 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 29 | RUNX1T1 | Sponge network | -0.663 | 0.10887 | -0.344 | 0.2374 | 0.363 |
| 26679 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | DKK3 | Sponge network | -2.46 | 0 | -0.052 | 0.77783 | 0.363 |
| 26680 | H19 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p | 12 | NAV3 | Sponge network | 2.439 | 0 | 0.212 | 0.47729 | 0.363 |
| 26681 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 35 | ESR1 | Sponge network | 0.246 | 0.59912 | -1.035 | 3.0E-5 | 0.363 |
| 26682 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26b-3p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-940 | 11 | EPS15 | Sponge network | 0.993 | 0 | -0.052 | 0.53998 | 0.363 |
| 26683 | PCA3 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | LIMCH1 | Sponge network | -0.709 | 0.18168 | -0.915 | 1.0E-5 | 0.363 |
| 26684 | RP11-597D13.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 28 | TET1 | Sponge network | 1.302 | 7.0E-5 | -0.135 | 0.52138 | 0.363 |
| 26685 | AC093110.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-495-3p | 15 | TRIM33 | Sponge network | 1.044 | 2.0E-5 | 0.402 | 7.0E-5 | 0.363 |
| 26686 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-625-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | 0.246 | 0.59912 | -0.087 | 0.69193 | 0.363 |
| 26687 | SNHG14 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 12 | PEA15 | Sponge network | -1.111 | 0.0003 | 0.009 | 0.93318 | 0.363 |
| 26688 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | NIN | Sponge network | 0.225 | 0.30644 | -0.253 | 0.13794 | 0.363 |
| 26689 | USP3-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-455-5p | 12 | PAK3 | Sponge network | -0.162 | 0.47687 | -0.628 | 0.07253 | 0.363 |
| 26690 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | LRRC7 | Sponge network | 0.101 | 0.60919 | -1.341 | 0.01193 | 0.363 |
| 26691 | RP11-65J21.3 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-7-1-3p | 11 | NOTCH2 | Sponge network | -0.557 | 0.11135 | 0.138 | 0.35163 | 0.363 |
| 26692 | RP11-5C23.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p | 10 | EZH1 | Sponge network | 0.274 | 0.07681 | -0.564 | 1.0E-5 | 0.363 |
| 26693 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | PHC3 | Sponge network | 0.274 | 0.07681 | 0.212 | 0.0499 | 0.363 |
| 26694 | AC093627.8 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-625-5p;hsa-miR-7-5p | 15 | IGFBP5 | Sponge network | -0.787 | 0.3928 | -0.806 | 0.00105 | 0.363 |
| 26695 | RP11-121C2.2 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | USP9X | Sponge network | 1.147 | 0 | 0.222 | 0.01689 | 0.363 |
| 26696 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p | 13 | TCF12 | Sponge network | 0.542 | 0.00054 | 0.174 | 0.13271 | 0.363 |
| 26697 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | FSTL5 | Sponge network | -1.216 | 0 | -1.3 | 0.01619 | 0.363 |
| 26698 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p | 17 | PNRC1 | Sponge network | -0.162 | 0.47687 | -0.646 | 0 | 0.363 |
| 26699 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-363-3p;hsa-miR-374b-5p | 10 | KIAA1024 | Sponge network | 1.601 | 0 | 1.083 | 0 | 0.363 |
| 26700 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | TRPS1 | Sponge network | 0.225 | 0.30644 | -0.191 | 0.44109 | 0.363 |
| 26701 | FENDRR |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 25 | SEMA3C | Sponge network | -1.281 | 0.00012 | -0.272 | 0.31153 | 0.363 |
| 26702 | FAM157B |
hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | MLLT10 | Sponge network | 0.92 | 0 | 0.105 | 0.33292 | 0.363 |
| 26703 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-5p | 18 | RASSF8 | Sponge network | -0.384 | 0.40652 | -0.122 | 0.65591 | 0.363 |
| 26704 | RP11-38L15.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-345-5p;hsa-miR-590-3p | 16 | ADARB1 | Sponge network | 0.344 | 0.3127 | -0.335 | 0.06631 | 0.363 |
| 26705 | LINC00342 |
hsa-miR-126-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p | 10 | TRIM33 | Sponge network | 0.297 | 0.19933 | 0.402 | 7.0E-5 | 0.363 |
| 26706 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | APC | Sponge network | -0.709 | 0.18168 | -0.255 | 0.04051 | 0.363 |
| 26707 | RP11-35G9.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 22 | SNX29 | Sponge network | 0.622 | 0.00015 | -0.34 | 0.01371 | 0.363 |
| 26708 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 29 | AKAP11 | Sponge network | 0.488 | 0.00084 | 0.196 | 0.09825 | 0.363 |
| 26709 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p | 10 | ZMYND11 | Sponge network | -0.517 | 0.02935 | -0.2 | 0.05449 | 0.363 |
| 26710 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | PBRM1 | Sponge network | 0.782 | 0.00016 | -0.029 | 0.78601 | 0.363 |
| 26711 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | TCF4 | Sponge network | 0.348 | 0.32912 | 0.016 | 0.92271 | 0.363 |
| 26712 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p | 18 | ZMYM2 | Sponge network | 0.663 | 0.00073 | 0.279 | 0.01273 | 0.363 |
| 26713 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p | 15 | PKNOX1 | Sponge network | -0.615 | 0.00015 | 0.085 | 0.28917 | 0.363 |
| 26714 | NDUFA6-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 17 | KLF10 | Sponge network | -0.355 | 0.0077 | -0.783 | 0 | 0.363 |
| 26715 | LINC01018 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | PTPRD | Sponge network | -2.915 | 0.00015 | -1.005 | 0.00064 | 0.363 |
| 26716 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p;hsa-miR-98-5p | 28 | MITF | Sponge network | 0.101 | 0.60919 | -0.769 | 0.00022 | 0.363 |
| 26717 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | CACNA2D1 | Sponge network | 0.72 | 0.0001 | -0.846 | 0.00675 | 0.363 |
| 26718 | RP1-122P22.2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p | 12 | DIP2C | Sponge network | 0.065 | 0.73468 | -0.436 | 0.01713 | 0.363 |
| 26719 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p | 20 | LRRC7 | Sponge network | -0.154 | 0.78939 | -1.341 | 0.01193 | 0.363 |
| 26720 | HOXA-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 46 | DTNA | Sponge network | 0.61 | 0.01593 | -1.648 | 1.0E-5 | 0.363 |
| 26721 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p | 10 | CAV1 | Sponge network | -0.227 | 0.31657 | -1.013 | 6.0E-5 | 0.363 |
| 26722 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-93-5p;hsa-miR-935 | 19 | LRRC55 | Sponge network | -0.096 | 0.67958 | -0.026 | 0.93163 | 0.363 |
| 26723 | MIR600HG |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p | 18 | DICER1 | Sponge network | 0.594 | 0.41522 | 0.042 | 0.674 | 0.363 |
| 26724 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 20 | EPHA3 | Sponge network | 1.757 | 0 | 0.185 | 0.53588 | 0.363 |
| 26725 | FAM157B |
hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 11 | FOXO3 | Sponge network | 0.92 | 0 | -0.04 | 0.72028 | 0.363 |
| 26726 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-342-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | ATRX | Sponge network | 0.8 | 0 | 0.261 | 0.04408 | 0.363 |
| 26727 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | SLITRK6 | Sponge network | -0.487 | 0.02157 | -2.121 | 0 | 0.363 |
| 26728 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p | 13 | PDGFRA | Sponge network | -0.309 | 0.1145 | -0.372 | 0.0037 | 0.363 |
| 26729 | RP11-166D19.1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | JDP2 | Sponge network | -0.576 | 0.10121 | -0.967 | 0 | 0.363 |
| 26730 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 11 | RB1CC1 | Sponge network | 1.757 | 0 | 0.175 | 0.12559 | 0.363 |
| 26731 | PGM5-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-942-5p | 10 | GLIPR1 | Sponge network | -4.546 | 0 | 0.146 | 0.3276 | 0.363 |
| 26732 | RAMP2-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ITPKB | Sponge network | -0.513 | 0.04703 | -0.704 | 0.00106 | 0.363 |
| 26733 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p | 18 | SYT4 | Sponge network | -1.654 | 0.00144 | -3.207 | 0 | 0.363 |
| 26734 | NNT-AS1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | PHF3 | Sponge network | 0.799 | 1.0E-5 | 0.126 | 0.19652 | 0.363 |
| 26735 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | FREM2 | Sponge network | 0.374 | 0.20054 | -0.437 | 0.46353 | 0.363 |
| 26736 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-532-5p;hsa-miR-629-3p | 18 | PTPN14 | Sponge network | 0.174 | 0.30034 | 0.144 | 0.34595 | 0.363 |
| 26737 | RP11-98I9.4 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PIK3CA | Sponge network | 0.67 | 0.00048 | 0.195 | 0.06908 | 0.363 |
| 26738 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-142-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-532-3p;hsa-miR-940 | 21 | PBX1 | Sponge network | -4.477 | 0 | -1.206 | 0 | 0.363 |
| 26739 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | -0.668 | 3.0E-5 | -1.09 | 0.00014 | 0.363 |
| 26740 | AC009948.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 36 | NTRK2 | Sponge network | 0.532 | 0.00505 | -0.736 | 0.06495 | 0.363 |
| 26741 | EMX2OS |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-589-3p | 20 | MAFB | Sponge network | -0.777 | 0.1134 | -0.023 | 0.89865 | 0.363 |
| 26742 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | PREX2 | Sponge network | -0.738 | 0.21233 | -0.272 | 0.25382 | 0.363 |
| 26743 | CTC-301O7.4 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p | 10 | RASSF2 | Sponge network | 0.292 | 0.43384 | -0.273 | 0.21961 | 0.363 |
| 26744 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | GALNT13 | Sponge network | -0.942 | 0 | -0.857 | 0.07439 | 0.363 |
| 26745 | RP11-384K6.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p | 19 | BRD3 | Sponge network | 0.946 | 0 | 0.37 | 0.00013 | 0.363 |
| 26746 | LINC00963 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ITGB1 | Sponge network | 0.523 | 9.0E-5 | 0.524 | 0.00017 | 0.363 |
| 26747 | MALAT1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 12 | KIAA1024 | Sponge network | 0.782 | 0.00016 | 1.083 | 0 | 0.363 |
| 26748 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-93-3p | 21 | WWTR1 | Sponge network | 0.494 | 0.00339 | -0.345 | 0.11997 | 0.363 |
| 26749 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | HDAC9 | Sponge network | 0.4 | 0.14611 | -0.755 | 0.00495 | 0.363 |
| 26750 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-654-3p | 20 | RICTOR | Sponge network | 0.909 | 0 | 0.388 | 0.00188 | 0.363 |
| 26751 | RP11-597D13.9 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 31 | PCDH10 | Sponge network | 1.302 | 7.0E-5 | -1.644 | 0.0078 | 0.363 |
| 26752 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-590-3p | 22 | USP6 | Sponge network | 0.494 | 0.00339 | 0.188 | 0.3871 | 0.363 |
| 26753 | BVES-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-3p | 24 | ETV1 | Sponge network | -2.62 | 0 | -0.096 | 0.67674 | 0.363 |
| 26754 | RP11-855A2.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ABI2 | Sponge network | 2.094 | 0 | 0.175 | 0.14787 | 0.363 |
| 26755 | RP11-373D23.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-7-1-3p | 12 | WWTR1 | Sponge network | -0.285 | 0.2172 | -0.345 | 0.11997 | 0.363 |
| 26756 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 11 | YAP1 | Sponge network | 0.34 | 0.00273 | 0.22 | 0.11836 | 0.363 |
| 26757 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 20 | NOTCH2 | Sponge network | -0.187 | 0.23787 | 0.138 | 0.35163 | 0.363 |
| 26758 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CLIP1 | Sponge network | -1.161 | 0.00591 | -0.215 | 0.08684 | 0.363 |
| 26759 | RAMP2-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | ID4 | Sponge network | -0.513 | 0.04703 | -1.644 | 0 | 0.363 |
| 26760 | HOTAIRM1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SETBP1 | Sponge network | -0.053 | 0.80685 | -1.302 | 1.0E-5 | 0.363 |
| 26761 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-942-5p | 30 | THRB | Sponge network | -0.376 | 0.00337 | -1.117 | 0.0004 | 0.363 |
| 26762 | LINC00672 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-758-3p | 22 | CSDE1 | Sponge network | -0.227 | 0.31657 | -0.493 | 0 | 0.363 |
| 26763 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p | 15 | MYO9A | Sponge network | 1.361 | 0 | -0.147 | 0.32373 | 0.363 |
| 26764 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 34 | PPP1R9A | Sponge network | -0.513 | 0.04703 | -0.903 | 0.04254 | 0.363 |
| 26765 | CTC-457L16.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 20 | GAS7 | Sponge network | 1.153 | 0.00114 | -0.555 | 0.01602 | 0.363 |
| 26766 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | -0.942 | 0 | -3.143 | 0 | 0.363 |
| 26767 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 46 | TCF4 | Sponge network | -0.342 | 0.02288 | 0.016 | 0.92271 | 0.363 |
| 26768 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-93-5p | 15 | LATS2 | Sponge network | -0.318 | 0.65847 | 0.159 | 0.2304 | 0.363 |
| 26769 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-16-2-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-655-3p | 15 | PIK3C2A | Sponge network | 0.768 | 7.0E-5 | 0.061 | 0.53776 | 0.363 |
| 26770 | RP11-967K21.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p | 16 | ASTN1 | Sponge network | 0.056 | 0.81195 | -2.389 | 2.0E-5 | 0.363 |
| 26771 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-424-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | HOOK3 | Sponge network | 1.254 | 0 | 0.273 | 0.09957 | 0.363 |
| 26772 | RP11-359E10.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | NEDD4 | Sponge network | 0.299 | 0.57722 | 0.02 | 0.89834 | 0.363 |
| 26773 | PART1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-7-1-3p | 30 | WWTR1 | Sponge network | -2.925 | 0 | -0.345 | 0.11997 | 0.363 |
| 26774 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-93-5p;hsa-miR-942-5p | 10 | FBXO31 | Sponge network | -2.531 | 0 | -0.085 | 0.42389 | 0.363 |
| 26775 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | PTPRB | Sponge network | -0.096 | 0.67958 | 0.105 | 0.47122 | 0.363 |
| 26776 | EMX2OS |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | NIN | Sponge network | -0.777 | 0.1134 | -0.253 | 0.13794 | 0.363 |
| 26777 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-423-5p;hsa-miR-455-5p | 11 | PAK3 | Sponge network | -0.154 | 0.78939 | -0.628 | 0.07253 | 0.363 |
| 26778 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | CBL | Sponge network | 0.924 | 0 | 0.291 | 0.01267 | 0.363 |
| 26779 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-5p | 20 | CLASP2 | Sponge network | 0.921 | 0.00118 | -0.038 | 0.71 | 0.363 |
| 26780 | AC109309.4 |
hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-330-3p | 10 | IL17RD | Sponge network | 0.163 | 0.87219 | -0.019 | 0.94873 | 0.363 |
| 26781 | RP11-216B9.6 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | SYNPO2 | Sponge network | 0.174 | 0.30034 | -2.342 | 0 | 0.363 |
| 26782 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 16 | CITED2 | Sponge network | -0.612 | 0.00094 | -1.545 | 0 | 0.363 |
| 26783 | RP11-434B12.1 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | -0.031 | 0.89063 | -0.704 | 0.00106 | 0.363 |
| 26784 | SNX29P2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-671-5p;hsa-miR-96-5p | 21 | CYLD | Sponge network | 0.4 | 0.14611 | -0.367 | 0.00171 | 0.363 |
| 26785 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-582-3p;hsa-miR-664a-5p | 36 | ADARB1 | Sponge network | 0.41 | 0.02214 | -0.335 | 0.06631 | 0.363 |
| 26786 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | SYNE1 | Sponge network | 0.72 | 0.0001 | -0.738 | 0.00316 | 0.363 |
| 26787 | AC074289.1 |
hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p | 14 | SYNE1 | Sponge network | -0.326 | 0.14314 | -0.738 | 0.00316 | 0.363 |
| 26788 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-7-5p;hsa-miR-93-5p | 11 | GLI3 | Sponge network | -0.384 | 0.40652 | -0.615 | 0.01482 | 0.363 |
| 26789 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | 0.494 | 0.00339 | -1.548 | 0 | 0.363 |
| 26790 | AC108488.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-7-1-3p | 37 | DTNA | Sponge network | 0.259 | 0.12542 | -1.648 | 1.0E-5 | 0.363 |
| 26791 | HOXA-AS2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 18 | PRUNE2 | Sponge network | 0.61 | 0.01593 | -1.733 | 0.00022 | 0.363 |
| 26792 | PART1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | TLE4 | Sponge network | -2.925 | 0 | -1.157 | 0 | 0.363 |
| 26793 | RP11-53O19.3 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 24 | CREB1 | Sponge network | 1.205 | 0 | 0.203 | 0.00537 | 0.363 |
| 26794 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 52 | PAFAH1B1 | Sponge network | -0.129 | 0.57177 | -0.429 | 0 | 0.363 |
| 26795 | RP11-166D19.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | PIK3CA | Sponge network | -0.576 | 0.10121 | 0.195 | 0.06908 | 0.363 |
| 26796 | RP11-701H24.3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p | 13 | RASSF2 | Sponge network | -1.425 | 0.04204 | -0.273 | 0.21961 | 0.363 |
| 26797 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 10 | MCC | Sponge network | 1.122 | 0.02989 | -1.005 | 0 | 0.363 |
| 26798 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-93-5p | 17 | CDHR3 | Sponge network | -0.371 | 0.24604 | -0.977 | 4.0E-5 | 0.363 |
| 26799 | IQCH-AS1 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 12 | RASAL2 | Sponge network | 0.929 | 0 | 0.869 | 0 | 0.363 |
| 26800 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | PPM1A | Sponge network | 0.083 | 0.66405 | -0.623 | 0 | 0.363 |
| 26801 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-93-5p | 28 | MAF | Sponge network | -0.777 | 0.16667 | -1.257 | 0 | 0.363 |
| 26802 | MIR22HG |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | PREX2 | Sponge network | -1.047 | 0 | -0.272 | 0.25382 | 0.363 |
| 26803 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 13 | ERRFI1 | Sponge network | -0.612 | 0.00094 | -0.798 | 1.0E-5 | 0.363 |
| 26804 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 18 | NCAM2 | Sponge network | -0.246 | 0.35034 | -0.482 | 0.22565 | 0.363 |
| 26805 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -0.487 | 0.02157 | -1.692 | 0 | 0.363 |
| 26806 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | PLSCR4 | Sponge network | 0.051 | 0.83388 | -0.474 | 0.03833 | 0.363 |
| 26807 | PART1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | FAM117A | Sponge network | -2.925 | 0 | -1.379 | 0 | 0.363 |
| 26808 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-330-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MOB1B | Sponge network | -0.053 | 0.80685 | 0.197 | 0.15266 | 0.363 |
| 26809 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 57 | IL6ST | Sponge network | -1.249 | 7.0E-5 | -0.402 | 0.00676 | 0.363 |
| 26810 | MEG3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-5p | 20 | RET | Sponge network | 0.249 | 0.35902 | -0.833 | 0.03517 | 0.363 |
| 26811 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 12 | SEPT6 | Sponge network | 0.194 | 0.64278 | -0.598 | 0.00181 | 0.363 |
| 26812 | MIR143HG |
hsa-miR-10b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 10 | EXTL2 | Sponge network | -0.739 | 0.0574 | 0.109 | 0.37375 | 0.363 |
| 26813 | LINC00654 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p | 13 | CACNA1E | Sponge network | 0.374 | 0.20054 | 1.752 | 0 | 0.363 |
| 26814 | AC116366.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-935 | 13 | EBF1 | Sponge network | 0.329 | 0.11122 | -0.384 | 0.08856 | 0.363 |
| 26815 | AP001258.4 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 11 | CDK6 | Sponge network | 0.931 | 0 | 0.991 | 0 | 0.363 |
| 26816 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 17 | ZFHX4 | Sponge network | 0.342 | 0.03129 | 0.018 | 0.95574 | 0.363 |
| 26817 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 35 | NTRK3 | Sponge network | 0.622 | 0.00015 | -1.77 | 3.0E-5 | 0.363 |
| 26818 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 17 | TXNIP | Sponge network | -2.925 | 0 | -1.277 | 0 | 0.363 |
| 26819 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-93-5p | 20 | ZBTB4 | Sponge network | -0.785 | 0.00035 | -0.739 | 0 | 0.363 |
| 26820 | NNT-AS1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p | 12 | STAG2 | Sponge network | 0.799 | 1.0E-5 | 0.252 | 0.00438 | 0.363 |
| 26821 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 15 | RBL2 | Sponge network | -0.227 | 0.31657 | -0.282 | 0.00254 | 0.363 |
| 26822 | C4B-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-486-5p | 12 | NRK | Sponge network | 2.306 | 9.0E-5 | 0.656 | 0.19217 | 0.363 |
| 26823 | RP11-25K19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MED12L | Sponge network | -1.057 | 0.00029 | -0.194 | 0.55299 | 0.363 |
| 26824 | RP11-159D12.2 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-9-5p | 10 | SPTBN1 | Sponge network | 1.254 | 0 | 0.561 | 1.0E-5 | 0.363 |
| 26825 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940 | 38 | NOTCH2 | Sponge network | -0.563 | 0.02545 | 0.138 | 0.35163 | 0.363 |
| 26826 | CTD-3064M3.3 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-429;hsa-miR-500a-3p | 15 | LIMCH1 | Sponge network | -0.469 | 0.08721 | -0.915 | 1.0E-5 | 0.363 |
| 26827 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p | 13 | EBF1 | Sponge network | 0.497 | 0.01451 | -0.384 | 0.08856 | 0.363 |
| 26828 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | LRRC4 | Sponge network | -0.727 | 0.04726 | -1.774 | 0 | 0.363 |
| 26829 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-708-5p | 48 | AFF4 | Sponge network | 0.401 | 0.00066 | -0.266 | 0.01565 | 0.363 |
| 26830 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 38 | ESR1 | Sponge network | -0.942 | 0 | -1.035 | 3.0E-5 | 0.363 |
| 26831 | RP11-727A23.5 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c | 23 | ABL2 | Sponge network | 1.117 | 0 | 0.82 | 0 | 0.362 |
| 26832 | RP11-672A2.4 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p | 14 | WDFY3 | Sponge network | 1.344 | 0 | -0.201 | 0.0967 | 0.362 |
| 26833 | CTD-2554C21.2 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-629-5p | 23 | CTIF | Sponge network | -1.654 | 0.00144 | -0.389 | 0.00549 | 0.362 |
| 26834 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-3p | 12 | UNC5C | Sponge network | -0.896 | 0.00099 | -0.178 | 0.40214 | 0.362 |
| 26835 | CKMT2-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | FMNL3 | Sponge network | 0.082 | 0.55654 | 0.555 | 4.0E-5 | 0.362 |
| 26836 | RP11-175O19.4 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.917 | 0 | 1.083 | 0 | 0.362 |
| 26837 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p | 12 | SYNE1 | Sponge network | 0.37 | 0.27614 | -0.738 | 0.00316 | 0.362 |
| 26838 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-96-5p | 15 | EBF1 | Sponge network | -0.31 | 0.33871 | -0.384 | 0.08856 | 0.362 |
| 26839 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-199b-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 22 | SMAD4 | Sponge network | 0.542 | 0.00054 | -0.313 | 0.00413 | 0.362 |
| 26840 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 33 | PRKAA2 | Sponge network | 0.389 | 0.04293 | -2.462 | 0 | 0.362 |
| 26841 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-652-3p | 26 | ZBTB4 | Sponge network | 0.72 | 0.0001 | -0.739 | 0 | 0.362 |
| 26842 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-93-5p | 16 | TXNIP | Sponge network | 0.101 | 0.60919 | -1.277 | 0 | 0.362 |
| 26843 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-744-3p | 18 | HIP1 | Sponge network | -0.13 | 0.48734 | 0.774 | 0 | 0.362 |
| 26844 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 30 | MCC | Sponge network | -0.176 | 0.59692 | -1.005 | 0 | 0.362 |
| 26845 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429 | 13 | CITED2 | Sponge network | -0.563 | 0.02545 | -1.545 | 0 | 0.362 |
| 26846 | RP11-152F13.8 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p | 10 | CCDC6 | Sponge network | 0.843 | 0.01775 | 0.27 | 0.02555 | 0.362 |
| 26847 | HHIP-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | PCDH10 | Sponge network | -0.737 | 0.10822 | -1.644 | 0.0078 | 0.362 |
| 26848 | AC011526.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | ASTN1 | Sponge network | 0.342 | 0.03129 | -2.389 | 2.0E-5 | 0.362 |
| 26849 | FAM66C |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | TLE4 | Sponge network | -0.901 | 0.00019 | -1.157 | 0 | 0.362 |
| 26850 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | CREBBP | Sponge network | 0.421 | 0.00084 | 0.126 | 0.15186 | 0.362 |
| 26851 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | CDH10 | Sponge network | -0.563 | 0.02545 | -2.397 | 0 | 0.362 |
| 26852 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | QKI | Sponge network | 0.657 | 4.0E-5 | -0.333 | 0.04111 | 0.362 |
| 26853 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | CNTN1 | Sponge network | -0.355 | 0.0077 | -2.594 | 0 | 0.362 |
| 26854 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | TAL1 | Sponge network | -0.246 | 0.35034 | -0.462 | 0.00574 | 0.362 |
| 26855 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TRIM13 | Sponge network | 0.083 | 0.66405 | 0.183 | 0.06968 | 0.362 |
| 26856 | RP11-999E24.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-93-3p | 25 | TRPS1 | Sponge network | -0.693 | 0.05629 | -0.191 | 0.44109 | 0.362 |
| 26857 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-708-5p | 15 | RAP1A | Sponge network | -1.886 | 0 | -0.929 | 0 | 0.362 |
| 26858 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 26 | APC | Sponge network | 0.349 | 0.01695 | -0.255 | 0.04051 | 0.362 |
| 26859 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 43 | GAS7 | Sponge network | -0.355 | 0.0077 | -0.555 | 0.01602 | 0.362 |
| 26860 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-935 | 10 | LRRN3 | Sponge network | -0.942 | 0 | -0.393 | 0.16293 | 0.362 |
| 26861 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 23 | BCL2 | Sponge network | -1.582 | 8.0E-5 | -0.899 | 0 | 0.362 |
| 26862 | FAM215B |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p | 14 | HECA | Sponge network | 0.817 | 3.0E-5 | -0.217 | 0.03463 | 0.362 |
| 26863 | IQCH-AS1 |
hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-363-3p;hsa-miR-497-5p | 10 | E2F3 | Sponge network | 0.929 | 0 | 1.289 | 0 | 0.362 |
| 26864 | RP11-344B5.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-3p | 19 | DST | Sponge network | -0.055 | 0.88482 | -0.713 | 0.00096 | 0.362 |
| 26865 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 46 | TMEM132B | Sponge network | -0.222 | 0.32445 | -0.83 | 0.01084 | 0.362 |
| 26866 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | -0.053 | 0.80685 | -0.509 | 0.03957 | 0.362 |
| 26867 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PDE4DIP | Sponge network | 0.532 | 0.00505 | -0.282 | 0.0257 | 0.362 |
| 26868 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-96-5p | 39 | SOX5 | Sponge network | -0.18 | 0.20343 | -1.091 | 4.0E-5 | 0.362 |
| 26869 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p | 28 | ESR1 | Sponge network | 0.214 | 0.18246 | -1.035 | 3.0E-5 | 0.362 |
| 26870 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 35 | FOXP1 | Sponge network | 0.225 | 0.30644 | 0.042 | 0.74368 | 0.362 |
| 26871 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p | 18 | MYBL1 | Sponge network | 0.765 | 7.0E-5 | 0.494 | 0.02152 | 0.362 |
| 26872 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p | 19 | PTPN14 | Sponge network | 0.176 | 0.37402 | 0.144 | 0.34595 | 0.362 |
| 26873 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 0.637 | 0.00069 | 0.578 | 0 | 0.362 |
| 26874 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 24 | RASSF8 | Sponge network | -0.303 | 0.21002 | -0.122 | 0.65591 | 0.362 |
| 26875 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 30 | NEDD4 | Sponge network | -1.575 | 6.0E-5 | 0.02 | 0.89834 | 0.362 |
| 26876 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-7-5p | 12 | BTG2 | Sponge network | -1.582 | 8.0E-5 | -1.763 | 0 | 0.362 |
| 26877 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 26 | PGR | Sponge network | 0.219 | 0.1516 | -1.704 | 0 | 0.362 |
| 26878 | CTD-2184D3.6 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | MDM4 | Sponge network | 0.272 | 0.44692 | 0.345 | 0.00762 | 0.362 |
| 26879 | RP11-244O19.1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | ID4 | Sponge network | -0.096 | 0.67958 | -1.644 | 0 | 0.362 |
| 26880 | CTD-2336O2.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | AKAP6 | Sponge network | -0.141 | 0.26434 | -1.586 | 3.0E-5 | 0.362 |
| 26881 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 23 | ACVR1 | Sponge network | -0.739 | 0.0574 | 0.279 | 0.00234 | 0.362 |
| 26882 | MIR143HG |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-429 | 12 | DUSP1 | Sponge network | -0.739 | 0.0574 | -1.754 | 0 | 0.362 |
| 26883 | RP11-53O19.1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-940 | 10 | RHOB | Sponge network | -0.405 | 0.22013 | -1.052 | 0 | 0.362 |
| 26884 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p | 10 | NAV3 | Sponge network | 1.46 | 1.0E-5 | 0.212 | 0.47729 | 0.362 |
| 26885 | LINC00094 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p | 11 | UVRAG | Sponge network | 0.253 | 0.09565 | -0.017 | 0.83865 | 0.362 |
| 26886 | DNAJC27-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | -0.969 | 2.0E-5 | -1.379 | 0 | 0.362 |
| 26887 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-590-3p | 16 | RET | Sponge network | -1.161 | 0.00591 | -0.833 | 0.03517 | 0.362 |
| 26888 | CTC-378H22.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-501-5p | 13 | UNC80 | Sponge network | 0.001 | 0.99663 | -1.934 | 0 | 0.362 |
| 26889 | RP11-195F19.9 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p | 16 | ASTN1 | Sponge network | -0.726 | 0.00132 | -2.389 | 2.0E-5 | 0.362 |
| 26890 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 51 | CCND2 | Sponge network | 0.064 | 0.72934 | -0.267 | 0.3112 | 0.362 |
| 26891 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | LRIG1 | Sponge network | 0.083 | 0.66405 | -0.537 | 0.00588 | 0.362 |
| 26892 | RP11-326C3.7 |
hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-424-5p | 12 | AR | Sponge network | 0.295 | 0.28897 | -1.554 | 0 | 0.362 |
| 26893 | RP11-6O2.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-550a-3p | 13 | BRD3 | Sponge network | 0.331 | 0.57247 | 0.37 | 0.00013 | 0.362 |
| 26894 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p | 10 | ZNF536 | Sponge network | -1.349 | 0.00017 | -3.115 | 0 | 0.362 |
| 26895 | LINC00702 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 24 | KCNJ5 | Sponge network | -0.737 | 0.06182 | -0.281 | 0.3339 | 0.362 |
| 26896 | CTD-2245F17.3 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-629-3p | 14 | RASSF2 | Sponge network | -0.421 | 0.12556 | -0.273 | 0.21961 | 0.362 |
| 26897 | NEAT1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-940 | 19 | SIRT1 | Sponge network | 0.705 | 0.00014 | 0.109 | 0.2593 | 0.362 |
| 26898 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 29 | USP12 | Sponge network | -0.737 | 0.06182 | -0.102 | 0.40768 | 0.362 |
| 26899 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-654-3p | 29 | FBXO32 | Sponge network | 0.177 | 0.16681 | -0.495 | 0.07561 | 0.362 |
| 26900 | RP1-302G2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-940 | 13 | MCC | Sponge network | -1.483 | 9.0E-5 | -1.005 | 0 | 0.362 |
| 26901 | OIP5-AS1 |
hsa-miR-16-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-654-3p | 10 | USP25 | Sponge network | 0.421 | 0.00084 | -0.242 | 0.01397 | 0.362 |
| 26902 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | HACE1 | Sponge network | 0.959 | 0 | -0.072 | 0.55239 | 0.362 |
| 26903 | LINC00861 |
hsa-miR-103a-2-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-577;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-877-5p | 10 | RBMS3 | Sponge network | 0.911 | 0.00854 | -0.519 | 0.03551 | 0.362 |
| 26904 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-505-3p | 24 | PAFAH1B1 | Sponge network | -0.583 | 0.14589 | -0.429 | 0 | 0.362 |
| 26905 | RP11-244O19.1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-628-5p | 17 | IRS1 | Sponge network | -0.096 | 0.67958 | -0.026 | 0.88855 | 0.362 |
| 26906 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | CXXC4 | Sponge network | -0.49 | 0.04946 | -0.433 | 0.21055 | 0.362 |
| 26907 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 44 | CHD5 | Sponge network | -0.969 | 2.0E-5 | -0.922 | 0.01521 | 0.362 |
| 26908 | RP4-668J24.2 |
hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p | 12 | PPP1R9A | Sponge network | -1.054 | 0.0706 | -0.903 | 0.04254 | 0.362 |
| 26909 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-93-5p | 13 | EDA2R | Sponge network | -0.465 | 0.04703 | -0.587 | 0.01598 | 0.362 |
| 26910 | HCG18 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p;hsa-miR-655-3p | 13 | ARID2 | Sponge network | 1.063 | 0 | 0.235 | 0.02697 | 0.362 |
| 26911 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p | 13 | LATS1 | Sponge network | 1.044 | 2.0E-5 | 0.109 | 0.34208 | 0.362 |
| 26912 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-766-3p;hsa-miR-940 | 26 | CHD5 | Sponge network | 0.389 | 0.04293 | -0.922 | 0.01521 | 0.362 |
| 26913 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 28 | ITGAV | Sponge network | -0.739 | 0.0574 | 0.337 | 0.01002 | 0.362 |
| 26914 | RP11-254F7.2 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-503-5p | 13 | ABL1 | Sponge network | 0.176 | 0.37402 | -0.215 | 0.07804 | 0.362 |
| 26915 | CTD-3064M3.3 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p | 14 | HIP1 | Sponge network | -0.469 | 0.08721 | 0.774 | 0 | 0.362 |
| 26916 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-935;hsa-miR-96-5p | 40 | MTSS1 | Sponge network | -1.111 | 0.0003 | -0.81 | 0.00035 | 0.362 |
| 26917 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 23 | MLLT10 | Sponge network | 0.614 | 1.0E-5 | 0.105 | 0.33292 | 0.362 |
| 26918 | PWAR6 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-671-5p | 13 | PIK3IP1 | Sponge network | -1.575 | 6.0E-5 | -0.187 | 0.19945 | 0.362 |
| 26919 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 21 | ZFHX3 | Sponge network | 1.601 | 0 | 0.389 | 0.01363 | 0.362 |
| 26920 | SERTAD4-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 16 | BTG2 | Sponge network | -0.932 | 0.00329 | -1.763 | 0 | 0.362 |
| 26921 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | LRP1B | Sponge network | -0.777 | 0.1134 | -2.163 | 0 | 0.362 |
| 26922 | LINC00969 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 13 | DICER1 | Sponge network | 0.683 | 1.0E-5 | 0.042 | 0.674 | 0.362 |
| 26923 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITGAV | Sponge network | 0.683 | 1.0E-5 | 0.337 | 0.01002 | 0.362 |
| 26924 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | CDC73 | Sponge network | 0.953 | 0 | 0.094 | 0.20463 | 0.362 |
| 26925 | RP11-428J1.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 21 | ABL2 | Sponge network | 0.538 | 0.00076 | 0.82 | 0 | 0.362 |
| 26926 | RP11-156E6.1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-374b-5p | 13 | UBE2D1 | Sponge network | 1.266 | 0 | 0.468 | 0 | 0.362 |
| 26927 | RP11-497H16.9 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | UBE2D1 | Sponge network | 0.545 | 0.00064 | 0.468 | 0 | 0.362 |
| 26928 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429 | 13 | CNTNAP3 | Sponge network | -0.246 | 0.35034 | -1.465 | 0.00033 | 0.362 |
| 26929 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | CDH19 | Sponge network | 0.101 | 0.60919 | -3.434 | 0 | 0.362 |
| 26930 | GAS6-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | NIN | Sponge network | 0.16 | 0.50785 | -0.253 | 0.13794 | 0.362 |
| 26931 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | LRRC4 | Sponge network | -0.896 | 0.00099 | -1.774 | 0 | 0.362 |
| 26932 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 20 | SUFU | Sponge network | -0.355 | 0.0077 | -0.04 | 0.65489 | 0.362 |
| 26933 | RP11-26J3.3 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-30a-5p;hsa-miR-30c-5p | 10 | PHF6 | Sponge network | 0.97 | 0 | 0.785 | 0 | 0.362 |
| 26934 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p | 11 | FREM2 | Sponge network | 0.663 | 0.00073 | -0.437 | 0.46353 | 0.362 |
| 26935 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 12 | PGR | Sponge network | -0.206 | 0.12942 | -1.704 | 0 | 0.362 |
| 26936 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 26 | PBX1 | Sponge network | 0.532 | 0.00505 | -1.206 | 0 | 0.362 |
| 26937 | ZSCAN16-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p | 26 | DLC1 | Sponge network | -0.342 | 0.02288 | -0.382 | 0.05038 | 0.362 |
| 26938 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p | 14 | TCF4 | Sponge network | 2.364 | 0.00134 | 0.016 | 0.92271 | 0.362 |
| 26939 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-501-5p | 13 | TXNIP | Sponge network | -0.222 | 0.32445 | -1.277 | 0 | 0.362 |
| 26940 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-96-5p | 17 | CRTC3 | Sponge network | 0.853 | 0.15352 | 0.164 | 0.16786 | 0.362 |
| 26941 | RP11-2B6.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | HECA | Sponge network | 0.539 | 0.03722 | -0.217 | 0.03463 | 0.362 |
| 26942 | RP11-403I13.7 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-877-5p;hsa-miR-96-5p | 21 | ASTN1 | Sponge network | 0.395 | 0.15451 | -2.389 | 2.0E-5 | 0.362 |
| 26943 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-93-5p | 42 | ADARB1 | Sponge network | -0.303 | 0.21002 | -0.335 | 0.06631 | 0.362 |
| 26944 | AC011526.1 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | DTNA | Sponge network | 0.342 | 0.03129 | -1.648 | 1.0E-5 | 0.362 |
| 26945 | RP11-758N13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-424-5p | 17 | BHLHE41 | Sponge network | 2.939 | 3.0E-5 | 0.353 | 0.12753 | 0.362 |
| 26946 | RP11-968O1.5 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-940 | 10 | SFRP1 | Sponge network | -0.829 | 0.01343 | -3.566 | 0 | 0.362 |
| 26947 | LINC00094 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-93-5p | 18 | ARNT2 | Sponge network | 0.253 | 0.09565 | -0.332 | 0.25851 | 0.362 |
| 26948 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | HACE1 | Sponge network | 0.101 | 0.60919 | -0.072 | 0.55239 | 0.362 |
| 26949 | H1FX-AS1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CDH19 | Sponge network | -0.206 | 0.12942 | -3.434 | 0 | 0.362 |
| 26950 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p | 28 | CREB3L2 | Sponge network | -2.531 | 0 | -0.525 | 2.0E-5 | 0.362 |
| 26951 | LINC00654 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 21 | PTGFR | Sponge network | 0.374 | 0.20054 | -0.233 | 0.45421 | 0.362 |
| 26952 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 10 | ARNT | Sponge network | -0.129 | 0.57177 | -0.08 | 0.26073 | 0.362 |
| 26953 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-624-5p | 10 | SMARCA1 | Sponge network | -0.899 | 6.0E-5 | -0.684 | 0.00159 | 0.362 |
| 26954 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-7-1-3p | 16 | SLC6A15 | Sponge network | -0.842 | 0.01361 | -2.373 | 2.0E-5 | 0.362 |
| 26955 | RP11-155G14.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326 | 11 | TBL1XR1 | Sponge network | 2.483 | 0 | 0.578 | 0 | 0.362 |
| 26956 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-744-3p | 19 | PTPRT | Sponge network | 0.178 | 0.29106 | -0.907 | 0.03727 | 0.362 |
| 26957 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | SEMA3C | Sponge network | -0.739 | 0.00044 | -0.272 | 0.31153 | 0.362 |
| 26958 | LPP-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-493-5p | 10 | NRXN3 | Sponge network | -0.18 | 0.20343 | -0.893 | 0.04186 | 0.362 |
| 26959 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-874-3p | 10 | BCLAF1 | Sponge network | 0.267 | 0.2937 | 0.211 | 0.00684 | 0.362 |
| 26960 | LINC00654 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 10 | PXDN | Sponge network | 0.374 | 0.20054 | 1.038 | 0 | 0.362 |
| 26961 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p | 12 | ZFP36L1 | Sponge network | -0.611 | 0.09607 | -0.505 | 8.0E-5 | 0.362 |
| 26962 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-375;hsa-miR-93-3p;hsa-miR-96-5p | 13 | EBF3 | Sponge network | 0.622 | 0.00015 | -0.058 | 0.81437 | 0.362 |
| 26963 | CTC-297N7.9 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-590-5p | 10 | GRIA3 | Sponge network | -2.069 | 0 | -1.956 | 0 | 0.362 |
| 26964 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-7-1-3p | 12 | BNC2 | Sponge network | -0.011 | 0.967 | -0.491 | 0.09988 | 0.362 |
| 26965 | MALAT1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 19 | ZMYND11 | Sponge network | 0.782 | 0.00016 | -0.2 | 0.05449 | 0.362 |
| 26966 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-3p;hsa-miR-339-5p | 14 | MYBL1 | Sponge network | 0.993 | 0 | 0.494 | 0.02152 | 0.362 |
| 26967 | TRAM2-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | PHF3 | Sponge network | 0.336 | 0.00739 | 0.126 | 0.19652 | 0.362 |
| 26968 | LINC00472 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | PPM1L | Sponge network | -0.842 | 0.01361 | -0.537 | 0.00616 | 0.362 |
| 26969 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | NALCN | Sponge network | 0.082 | 0.55654 | 1.068 | 0.00237 | 0.362 |
| 26970 | RP13-516M14.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-429 | 18 | PTPN14 | Sponge network | 0.389 | 0.04293 | 0.144 | 0.34595 | 0.362 |
| 26971 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SMARCA2 | Sponge network | -1.281 | 0.00012 | -0.529 | 0.00014 | 0.362 |
| 26972 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-5p | 11 | LRRK2 | Sponge network | -1.047 | 0 | -1.136 | 1.0E-5 | 0.362 |
| 26973 | RP4-669P10.16 |
hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-98-5p | 12 | LRIG1 | Sponge network | -0.301 | 0.06597 | -0.537 | 0.00588 | 0.362 |
| 26974 | RP11-464F9.1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p | 18 | DDX3X | Sponge network | 0.959 | 0 | -0.152 | 0.07686 | 0.362 |
| 26975 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | -0.969 | 2.0E-5 | -0.509 | 0.03957 | 0.362 |
| 26976 | RP11-123M6.2 |
hsa-miR-103a-2-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-330-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 10 | BMPR2 | Sponge network | 0.543 | 0.12356 | 0.206 | 0.0635 | 0.362 |
| 26977 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 42 | TCF4 | Sponge network | -0.227 | 0.31657 | 0.016 | 0.92271 | 0.362 |
| 26978 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 50 | WDFY3 | Sponge network | 0.253 | 0.09565 | -0.201 | 0.0967 | 0.362 |
| 26979 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | SLITRK3 | Sponge network | -0.206 | 0.12942 | -3.328 | 0 | 0.362 |
| 26980 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MOB1B | Sponge network | -0.021 | 0.93598 | 0.197 | 0.15266 | 0.362 |
| 26981 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-940 | 18 | CREB1 | Sponge network | 0.529 | 0.00552 | 0.203 | 0.00537 | 0.362 |
| 26982 | RP11-119F7.5 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-940 | 14 | SORCS1 | Sponge network | 0.225 | 0.30644 | -3.492 | 0 | 0.362 |
| 26983 | AGAP11 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-5p | 14 | RIT1 | Sponge network | -1.645 | 0 | -0.296 | 0.00054 | 0.362 |
| 26984 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p | 28 | TCF4 | Sponge network | 0.368 | 0.20093 | 0.016 | 0.92271 | 0.362 |
| 26985 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 23 | USP12 | Sponge network | -0.246 | 0.35034 | -0.102 | 0.40768 | 0.362 |
| 26986 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 42 | NTRK2 | Sponge network | -0.355 | 0.0077 | -0.736 | 0.06495 | 0.362 |
| 26987 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 33 | PRDM2 | Sponge network | -0.49 | 0.04946 | -0.258 | 0.00721 | 0.362 |
| 26988 | LINC00473 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | PTPN13 | Sponge network | -1.203 | 0.03881 | -1.208 | 0 | 0.362 |
| 26989 | CTC-444N24.11 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-497-5p | 11 | PAFAH1B2 | Sponge network | 1.153 | 0 | 0.318 | 0.0001 | 0.362 |
| 26990 | LINC00476 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | TMEFF2 | Sponge network | -0.615 | 0.00015 | -2.426 | 0 | 0.362 |
| 26991 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 10 | RCAN2 | Sponge network | 0.368 | 0.20093 | -0.33 | 0.15172 | 0.362 |
| 26992 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p | 11 | SNX29 | Sponge network | 0.497 | 0.01451 | -0.34 | 0.01371 | 0.362 |
| 26993 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 37 | IL6ST | Sponge network | 0.056 | 0.8494 | -0.402 | 0.00676 | 0.362 |
| 26994 | BMS1P20 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DICER1 | Sponge network | 0.34 | 0.00273 | 0.042 | 0.674 | 0.362 |
| 26995 | ZNF582-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-940 | 33 | PTPRT | Sponge network | -1.349 | 0.00017 | -0.907 | 0.03727 | 0.362 |
| 26996 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p | 27 | MTSS1 | Sponge network | 0.95 | 0.15943 | -0.81 | 0.00035 | 0.362 |
| 26997 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 19 | NR4A3 | Sponge network | -1.216 | 0 | -1.385 | 0 | 0.362 |
| 26998 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-625-3p | 18 | ZFHX4 | Sponge network | 0.497 | 0.01451 | 0.018 | 0.95574 | 0.362 |
| 26999 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-708-3p | 18 | HLF | Sponge network | -0.285 | 0.2172 | -2.443 | 0 | 0.362 |
| 27000 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | RNF144A | Sponge network | -1.161 | 0.00591 | 0.594 | 0.0009 | 0.362 |
| 27001 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-590-5p | 18 | PIK3C2A | Sponge network | 0.921 | 0.00118 | 0.061 | 0.53776 | 0.362 |
| 27002 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-493-5p | 15 | SLITRK6 | Sponge network | -0.842 | 0.01361 | -2.121 | 0 | 0.362 |
| 27003 | RP11-53O19.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-940 | 12 | CELF4 | Sponge network | -0.405 | 0.22013 | -1.135 | 0.00344 | 0.362 |
| 27004 | U91328.19 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | -0.303 | 0.21002 | -0.998 | 0.00025 | 0.362 |
| 27005 | RP11-981G7.6 |
hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DICER1 | Sponge network | 0.986 | 0.01022 | 0.042 | 0.674 | 0.362 |
| 27006 | RP11-395G23.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 12 | MRVI1 | Sponge network | 0.381 | 0.25967 | -1.051 | 0.00022 | 0.362 |
| 27007 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 36 | PBX1 | Sponge network | 0.082 | 0.55654 | -1.206 | 0 | 0.362 |
| 27008 | RP11-13K12.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-421 | 12 | STARD13 | Sponge network | -0.154 | 0.78939 | -0.187 | 0.27383 | 0.362 |
| 27009 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 54 | KIF1B | Sponge network | 0.349 | 0.01695 | -0.118 | 0.32258 | 0.362 |
| 27010 | RP11-672A2.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p | 15 | DTNA | Sponge network | 1.344 | 0 | -1.648 | 1.0E-5 | 0.362 |
| 27011 | HAND2-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 16 | POU2F2 | Sponge network | -1.697 | 0.00889 | -0.233 | 0.32214 | 0.362 |
| 27012 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | DACH2 | Sponge network | -0.901 | 0.00019 | -1.635 | 0.00034 | 0.362 |
| 27013 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -0.727 | 0.04726 | -0.354 | 0.14694 | 0.362 |
| 27014 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-340-5p;hsa-miR-374a-3p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-96-5p | 52 | AFF4 | Sponge network | 0.331 | 0.57247 | -0.266 | 0.01565 | 0.362 |
| 27015 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-3607-3p;hsa-miR-539-5p | 14 | MOB1B | Sponge network | 1.178 | 0 | 0.197 | 0.15266 | 0.362 |
| 27016 | RP11-356J5.12 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-874-3p | 25 | CTIF | Sponge network | -0.899 | 6.0E-5 | -0.389 | 0.00549 | 0.362 |
| 27017 | RP13-225O21.2 |
hsa-miR-1271-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-505-5p | 11 | ARHGEF12 | Sponge network | 1.279 | 8.0E-5 | 0.09 | 0.35693 | 0.362 |
| 27018 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | ITGAV | Sponge network | -0.737 | 0.06182 | 0.337 | 0.01002 | 0.362 |
| 27019 | AGAP11 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-590-5p | 15 | TMEFF2 | Sponge network | -1.645 | 0 | -2.426 | 0 | 0.362 |
| 27020 | LINC01018 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 12 | HIC1 | Sponge network | -2.915 | 0.00015 | 0.024 | 0.89889 | 0.362 |
| 27021 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 28 | ARID5B | Sponge network | 0.782 | 0.00016 | 0.342 | 0.01676 | 0.362 |
| 27022 | RP11-216B9.6 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | TAL1 | Sponge network | 0.174 | 0.30034 | -0.462 | 0.00574 | 0.362 |
| 27023 | RP11-2H8.2 |
hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 12 | ZNF770 | Sponge network | 1.089 | 0 | 0.206 | 0.06751 | 0.362 |
| 27024 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | TGFBR3 | Sponge network | 0.174 | 0.30034 | -1.173 | 1.0E-5 | 0.361 |
| 27025 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-590-5p | 13 | THSD7A | Sponge network | 0.082 | 0.55654 | 0.242 | 0.25154 | 0.361 |
| 27026 | TRAF3IP2-AS1 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-874-3p | 14 | ARID1B | Sponge network | -0.323 | 0.01372 | 0.003 | 0.97503 | 0.361 |
| 27027 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | EYA4 | Sponge network | -1.349 | 0.00017 | 0.3 | 0.36876 | 0.361 |
| 27028 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | NEDD4 | Sponge network | -1.281 | 0.00012 | 0.02 | 0.89834 | 0.361 |
| 27029 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p | 12 | TSC22D1 | Sponge network | -0.323 | 0.01372 | -0.529 | 2.0E-5 | 0.361 |
| 27030 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | CRTC1 | Sponge network | -0.49 | 0.04946 | -0.116 | 0.28412 | 0.361 |
| 27031 | AC034220.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 49 | AFF4 | Sponge network | 0.309 | 0.07304 | -0.266 | 0.01565 | 0.361 |
| 27032 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429 | 20 | ARHGAP28 | Sponge network | -2.69 | 0.0001 | -0.911 | 0.00013 | 0.361 |
| 27033 | RP11-395G23.3 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | FAM172A | Sponge network | 0.381 | 0.25967 | -0.57 | 0 | 0.361 |
| 27034 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-34a-5p | 10 | RNASEL | Sponge network | -0.187 | 0.23787 | -0.272 | 0.01796 | 0.361 |
| 27035 | RP11-47L3.1 | hsa-miR-106a-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p | 10 | GAS7 | Sponge network | 0.166 | 0.49046 | -0.555 | 0.01602 | 0.361 |
| 27036 | RP11-439A17.10 |
hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-194-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-590-5p | 10 | DIP2C | Sponge network | 0.051 | 0.95756 | -0.436 | 0.01713 | 0.361 |
| 27037 | RP5-994D16.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 19 | AFF4 | Sponge network | 0.252 | 0.08508 | -0.266 | 0.01565 | 0.361 |
| 27038 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p | 27 | LPP | Sponge network | 0.993 | 0 | -0.459 | 0.04475 | 0.361 |
| 27039 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-582-5p | 12 | ZMYM2 | Sponge network | 0.757 | 0 | 0.279 | 0.01273 | 0.361 |
| 27040 | TPTEP1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ZNF536 | Sponge network | -1.216 | 0 | -3.115 | 0 | 0.361 |
| 27041 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-590-3p | 10 | DCN | Sponge network | -0.096 | 0.67958 | -1.288 | 0 | 0.361 |
| 27042 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 59 | CCND2 | Sponge network | -1.111 | 0.0003 | -0.267 | 0.3112 | 0.361 |
| 27043 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | EDA2R | Sponge network | -0.811 | 2.0E-5 | -0.587 | 0.01598 | 0.361 |
| 27044 | RP11-48B3.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 17 | NAV3 | Sponge network | 1.757 | 0 | 0.212 | 0.47729 | 0.361 |
| 27045 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | ARID5B | Sponge network | -0.709 | 0.18168 | 0.342 | 0.01676 | 0.361 |
| 27046 | RP11-226L15.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-628-5p | 10 | KIAA1024 | Sponge network | 0.8 | 0 | 1.083 | 0 | 0.361 |
| 27047 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-382-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 31 | NRXN1 | Sponge network | -1.264 | 6.0E-5 | -3.778 | 0 | 0.361 |
| 27048 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | TGFBR2 | Sponge network | 0.246 | 0.59912 | -0.243 | 0.11408 | 0.361 |
| 27049 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 21 | AFF3 | Sponge network | -0.342 | 0.02288 | -2.373 | 0 | 0.361 |
| 27050 | AGAP11 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 26 | CTIF | Sponge network | -1.645 | 0 | -0.389 | 0.00549 | 0.361 |
| 27051 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 65 | IL6ST | Sponge network | -0.777 | 0.1134 | -0.402 | 0.00676 | 0.361 |
| 27052 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-484;hsa-miR-550a-5p;hsa-miR-629-3p;hsa-miR-664a-5p | 28 | DST | Sponge network | 1.316 | 0.01106 | -0.713 | 0.00096 | 0.361 |
| 27053 | CTC-510F12.2 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-484 | 16 | KIF1B | Sponge network | -0.691 | 0.00146 | -0.118 | 0.32258 | 0.361 |
| 27054 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | BNC2 | Sponge network | 0.622 | 0.00015 | -0.491 | 0.09988 | 0.361 |
| 27055 | LINC00403 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-324-3p | 10 | ZBTB4 | Sponge network | -3.586 | 0.00032 | -0.739 | 0 | 0.361 |
| 27056 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 22 | ARID5B | Sponge network | 0.056 | 0.8494 | 0.342 | 0.01676 | 0.361 |
| 27057 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | RABEP1 | Sponge network | -0.612 | 0.00094 | -0.066 | 0.43142 | 0.361 |
| 27058 | LINC00957 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p | 24 | BTG2 | Sponge network | -0.421 | 0.0114 | -1.763 | 0 | 0.361 |
| 27059 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | SASH1 | Sponge network | -0.323 | 0.01372 | -0.502 | 0.00175 | 0.361 |
| 27060 | AGAP11 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-455-5p | 15 | ITPKB | Sponge network | -1.645 | 0 | -0.704 | 0.00106 | 0.361 |
| 27061 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-542-3p | 12 | ABCB5 | Sponge network | -0.517 | 0.02935 | -0.956 | 0.04077 | 0.361 |
| 27062 | CTC-444N24.11 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | USP9X | Sponge network | 1.153 | 0 | 0.222 | 0.01689 | 0.361 |
| 27063 | RP11-48B3.4 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 15 | PIK3CA | Sponge network | 1.757 | 0 | 0.195 | 0.06908 | 0.361 |
| 27064 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 30 | CADM1 | Sponge network | -2.452 | 0 | -0.9 | 0.00084 | 0.361 |
| 27065 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 18 | RCAN2 | Sponge network | -1.203 | 0.03881 | -0.33 | 0.15172 | 0.361 |
| 27066 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-33a-3p | 10 | PTPRB | Sponge network | 0.497 | 0.01451 | 0.105 | 0.47122 | 0.361 |
| 27067 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | PRDM5 | Sponge network | 0.246 | 0.59912 | -1.09 | 0.00014 | 0.361 |
| 27068 | RP13-225O21.2 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 10 | RICTOR | Sponge network | 1.279 | 8.0E-5 | 0.388 | 0.00188 | 0.361 |
| 27069 | AC103563.9 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-455-5p;hsa-miR-577 | 12 | BCL2 | Sponge network | -6.057 | 0 | -0.899 | 0 | 0.361 |
| 27070 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 13 | EYA4 | Sponge network | 0.174 | 0.30034 | 0.3 | 0.36876 | 0.361 |
| 27071 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-590-3p | 10 | LRRK2 | Sponge network | -1.203 | 0.03881 | -1.136 | 1.0E-5 | 0.361 |
| 27072 | GAS6-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-92a-3p | 28 | IL17RD | Sponge network | 0.16 | 0.50785 | -0.019 | 0.94873 | 0.361 |
| 27073 | LINC00476 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | SPG20 | Sponge network | -0.615 | 0.00015 | -1.16 | 0 | 0.361 |
| 27074 | CTD-2583A14.8 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-374a-3p | 11 | TBL1XR1 | Sponge network | 1.134 | 0 | 0.578 | 0 | 0.361 |
| 27075 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-590-3p | 16 | TMTC3 | Sponge network | 0.673 | 0 | 0.123 | 0.22189 | 0.361 |
| 27076 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | NEDD4 | Sponge network | 0.559 | 0.00026 | 0.02 | 0.89834 | 0.361 |
| 27077 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-590-3p | 24 | PRKCE | Sponge network | -1.161 | 0.00591 | -0.403 | 0.00056 | 0.361 |
| 27078 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-935 | 31 | DMD | Sponge network | 0.537 | 0.00139 | -1.506 | 0 | 0.361 |
| 27079 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 28 | ATRX | Sponge network | -0.793 | 7.0E-5 | 0.261 | 0.04408 | 0.361 |
| 27080 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 27 | RSPO2 | Sponge network | 0.997 | 0.00066 | -3.038 | 0 | 0.361 |
| 27081 | RP11-798G7.7 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-455-3p;hsa-miR-539-5p | 17 | ARHGEF12 | Sponge network | 0.858 | 3.0E-5 | 0.09 | 0.35693 | 0.361 |
| 27082 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | NCAM2 | Sponge network | -0.08 | 0.90092 | -0.482 | 0.22565 | 0.361 |
| 27083 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 25 | SEMA3C | Sponge network | -2.46 | 0 | -0.272 | 0.31153 | 0.361 |
| 27084 | LINC00641 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | RELN | Sponge network | -0.222 | 0.32445 | -2.403 | 0 | 0.361 |
| 27085 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-590-3p | 15 | DACT1 | Sponge network | -0.096 | 0.67958 | 0.783 | 0.00072 | 0.361 |
| 27086 | LINC00173 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 11 | GLI3 | Sponge network | 0.056 | 0.8494 | -0.615 | 0.01482 | 0.361 |
| 27087 | CTA-363E19.2 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-485-3p;hsa-miR-495-3p | 14 | PIK3R1 | Sponge network | 1.055 | 0 | -0.133 | 0.36854 | 0.361 |
| 27088 | HNRNPU-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ABI2 | Sponge network | 1.601 | 0 | 0.175 | 0.14787 | 0.361 |
| 27089 | RP11-597D13.9 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | RECK | Sponge network | 1.302 | 7.0E-5 | -1.043 | 0 | 0.361 |
| 27090 | RP11-890B15.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p | 11 | FBXO11 | Sponge network | 0.101 | 0.60919 | 0.142 | 0.04938 | 0.361 |
| 27091 | RAD51-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 11 | UBR3 | Sponge network | 0.768 | 7.0E-5 | -0.021 | 0.82774 | 0.361 |
| 27092 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | EYA4 | Sponge network | 0.225 | 0.30644 | 0.3 | 0.36876 | 0.361 |
| 27093 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | SOX5 | Sponge network | -0.737 | 0.10822 | -1.091 | 4.0E-5 | 0.361 |
| 27094 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | PTPRB | Sponge network | -0.611 | 0.09607 | 0.105 | 0.47122 | 0.361 |
| 27095 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p | 22 | PBX1 | Sponge network | 0.853 | 0.15352 | -1.206 | 0 | 0.361 |
| 27096 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p | 10 | TBRG1 | Sponge network | -0.612 | 0.00094 | -0.071 | 0.37324 | 0.361 |
| 27097 | LINC00654 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DCN | Sponge network | 0.374 | 0.20054 | -1.288 | 0 | 0.361 |
| 27098 | PWAR6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | EPS15 | Sponge network | -1.575 | 6.0E-5 | -0.052 | 0.53998 | 0.361 |
| 27099 | LINC01018 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 14 | CITED2 | Sponge network | -2.915 | 0.00015 | -1.545 | 0 | 0.361 |
| 27100 | RP11-554A11.4 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-769-5p | 29 | WDFY3 | Sponge network | -0.583 | 0.14589 | -0.201 | 0.0967 | 0.361 |
| 27101 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 21 | GRIN2A | Sponge network | 0.4 | 0.14611 | -2.161 | 0 | 0.361 |
| 27102 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DACH2 | Sponge network | -1.057 | 0.00029 | -1.635 | 0.00034 | 0.361 |
| 27103 | MIR143HG |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940 | 13 | FOXO4 | Sponge network | -0.739 | 0.0574 | -0.516 | 2.0E-5 | 0.361 |
| 27104 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 17 | PPM1L | Sponge network | 0.673 | 0 | -0.537 | 0.00616 | 0.361 |
| 27105 | CTD-3092A11.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZMYND11 | Sponge network | 0.873 | 0 | -0.2 | 0.05449 | 0.361 |
| 27106 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 13 | PTPN11 | Sponge network | 0.919 | 0 | 0.431 | 2.0E-5 | 0.361 |
| 27107 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-589-5p | 14 | RPS6KA2 | Sponge network | 0.16 | 0.50785 | -0.57 | 0.00013 | 0.361 |
| 27108 | CTD-3064M3.3 |
hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429 | 12 | MYO9A | Sponge network | -0.469 | 0.08721 | -0.147 | 0.32373 | 0.361 |
| 27109 | LINC01001 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326 | 11 | BRD3 | Sponge network | 1.055 | 0 | 0.37 | 0.00013 | 0.361 |
| 27110 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 13 | CNTNAP1 | Sponge network | -0.727 | 0.04726 | 0.358 | 0.13727 | 0.361 |
| 27111 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-935 | 24 | MTSS1 | Sponge network | -0.246 | 0.35034 | -0.81 | 0.00035 | 0.361 |
| 27112 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-625-5p | 13 | RSPO2 | Sponge network | -0.206 | 0.12942 | -3.038 | 0 | 0.361 |
| 27113 | BZRAP1-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-370-3p | 10 | TPO | Sponge network | -0.465 | 0.04703 | -1.87 | 2.0E-5 | 0.361 |
| 27114 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p | 15 | LATS2 | Sponge network | -0.896 | 0.00099 | 0.159 | 0.2304 | 0.361 |
| 27115 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p | 11 | CSRNP1 | Sponge network | -2.531 | 0 | -1.448 | 0 | 0.361 |
| 27116 | RP11-384K6.6 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p | 10 | ITGB1 | Sponge network | 0.946 | 0 | 0.524 | 0.00017 | 0.361 |
| 27117 | EMX2OS |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | RTN4 | Sponge network | -0.777 | 0.1134 | -0.412 | 0.00014 | 0.361 |
| 27118 | CTD-2666L21.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-378a-5p;hsa-miR-486-5p | 21 | ADARB1 | Sponge network | 0.368 | 0.20093 | -0.335 | 0.06631 | 0.361 |
| 27119 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 52 | NFIB | Sponge network | -0.162 | 0.47687 | 0.023 | 0.90795 | 0.361 |
| 27120 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-942-5p | 45 | UNC80 | Sponge network | -0.769 | 0.00035 | -1.934 | 0 | 0.361 |
| 27121 | LINC00094 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p | 16 | KLF10 | Sponge network | 0.253 | 0.09565 | -0.783 | 0 | 0.361 |
| 27122 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | 0.082 | 0.55397 | -0.509 | 0.03957 | 0.361 |
| 27123 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TBX18 | Sponge network | 0.862 | 0.06213 | 0.843 | 0.02945 | 0.361 |
| 27124 | RAMP2-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 50 | DST | Sponge network | -0.513 | 0.04703 | -0.713 | 0.00096 | 0.361 |
| 27125 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-409-3p;hsa-miR-590-5p | 11 | ITGB3 | Sponge network | 0.176 | 0.37402 | -0.011 | 0.95751 | 0.361 |
| 27126 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-590-3p | 14 | TRIM33 | Sponge network | 0.986 | 0.01022 | 0.402 | 7.0E-5 | 0.361 |
| 27127 | RP5-1139B12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-874-3p | 11 | TACC1 | Sponge network | 0.598 | 0.38481 | -0.362 | 0.05406 | 0.361 |
| 27128 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 18 | ZFHX4 | Sponge network | 0.065 | 0.73468 | 0.018 | 0.95574 | 0.361 |
| 27129 | ZNF667-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-429;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 35 | AFF1 | Sponge network | -0.976 | 0.00025 | -0.384 | 0.00053 | 0.361 |
| 27130 | SNHG16 |
hsa-let-7a-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p | 15 | CDC73 | Sponge network | 0.961 | 0 | 0.094 | 0.20463 | 0.361 |
| 27131 | RP11-597D13.9 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-96-5p | 70 | LPP | Sponge network | 1.302 | 7.0E-5 | -0.459 | 0.04475 | 0.361 |
| 27132 | RP11-48B3.4 |
hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 14 | DOCK4 | Sponge network | 1.757 | 0 | 0.502 | 0.00108 | 0.361 |
| 27133 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 41 | FOXP1 | Sponge network | 0.083 | 0.66405 | 0.042 | 0.74368 | 0.361 |
| 27134 | LINC00957 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-940 | 33 | PTPRT | Sponge network | -0.421 | 0.0114 | -0.907 | 0.03727 | 0.361 |
| 27135 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-484 | 11 | PNRC1 | Sponge network | -0.465 | 0.04703 | -0.646 | 0 | 0.361 |
| 27136 | PCBP1-AS1 |
hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2355-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-432-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-92b-3p | 16 | IKZF2 | Sponge network | -0.567 | 0 | -0.284 | 0.1942 | 0.361 |
| 27137 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 24 | ARHGAP29 | Sponge network | -1.697 | 0.00889 | 0.131 | 0.42953 | 0.361 |
| 27138 | DIO3OS |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-542-3p | 15 | SYNM | Sponge network | -0.773 | 0.04073 | -2.309 | 1.0E-5 | 0.361 |
| 27139 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 17 | AKAP11 | Sponge network | 1.317 | 0 | 0.196 | 0.09825 | 0.361 |
| 27140 | USP3-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p | 11 | ZNF536 | Sponge network | -0.162 | 0.47687 | -3.115 | 0 | 0.361 |
| 27141 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 28 | MCC | Sponge network | 0.214 | 0.18246 | -1.005 | 0 | 0.361 |
| 27142 | RP11-672A2.4 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-330-5p | 17 | DST | Sponge network | 1.344 | 0 | -0.713 | 0.00096 | 0.361 |
| 27143 | BDNF-AS |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | HOMER2 | Sponge network | -0.668 | 3.0E-5 | -2.698 | 0 | 0.361 |
| 27144 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-942-5p | 30 | DDX6 | Sponge network | 0.494 | 0.00339 | 0.091 | 0.36428 | 0.361 |
| 27145 | EIF3J-AS1 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MYO9A | Sponge network | 0.331 | 0.00509 | -0.147 | 0.32373 | 0.361 |
| 27146 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-628-5p | 17 | ACVR1 | Sponge network | 0.61 | 0.01593 | 0.279 | 0.00234 | 0.361 |
| 27147 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 22 | DICER1 | Sponge network | 0.349 | 0.01695 | 0.042 | 0.674 | 0.361 |
| 27148 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-744-3p | 27 | PTPN14 | Sponge network | -2.46 | 0 | 0.144 | 0.34595 | 0.361 |
| 27149 | FAM157C |
hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-378a-5p;hsa-miR-511-5p | 14 | CREB1 | Sponge network | 0.91 | 0 | 0.203 | 0.00537 | 0.361 |
| 27150 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | MYO9A | Sponge network | 0.705 | 0.00014 | -0.147 | 0.32373 | 0.361 |
| 27151 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-942-5p | 15 | CREBBP | Sponge network | -0.323 | 0.01372 | 0.126 | 0.15186 | 0.361 |
| 27152 | RP11-359B12.2 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577 | 11 | MLLT11 | Sponge network | 0.064 | 0.72934 | -0.661 | 0.01733 | 0.361 |
| 27153 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-625-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -0.162 | 0.47687 | -0.087 | 0.69193 | 0.361 |
| 27154 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | CLASP2 | Sponge network | 0.959 | 0 | -0.038 | 0.71 | 0.361 |
| 27155 | RP11-532F6.3 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-935 | 22 | OPCML | Sponge network | -0.896 | 0.00099 | -2.498 | 0 | 0.361 |
| 27156 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | EGLN1 | Sponge network | 0.421 | 0.00084 | -0.127 | 0.1536 | 0.361 |
| 27157 | RP1-151F17.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-29a-5p | 11 | NTRK2 | Sponge network | -0.321 | 0.22761 | -0.736 | 0.06495 | 0.361 |
| 27158 | HOTAIRM1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ABI2 | Sponge network | -0.053 | 0.80685 | 0.175 | 0.14787 | 0.361 |
| 27159 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 22 | DOCK4 | Sponge network | -0.976 | 0.00025 | 0.502 | 0.00108 | 0.361 |
| 27160 | RP11-720L2.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-940 | 13 | GRIK3 | Sponge network | -1.255 | 0.00981 | -3.208 | 0 | 0.361 |
| 27161 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | ZNF208 | Sponge network | -0.769 | 0.00035 | -0.4 | 0.22381 | 0.361 |
| 27162 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-93-5p | 19 | NIN | Sponge network | 0.082 | 0.55397 | -0.253 | 0.13794 | 0.361 |
| 27163 | SOX2-OT |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 54 | CADM2 | Sponge network | -1.264 | 6.0E-5 | -3.782 | 0 | 0.361 |
| 27164 | SNHG14 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 34 | RNF144A | Sponge network | -1.111 | 0.0003 | 0.594 | 0.0009 | 0.361 |
| 27165 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p | 15 | ABCB5 | Sponge network | -0.154 | 0.78939 | -0.956 | 0.04077 | 0.361 |
| 27166 | PSMG3-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -0.125 | 0.49414 | -0.704 | 0.00106 | 0.361 |
| 27167 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 30 | PRKAA2 | Sponge network | 0.419 | 0.0319 | -2.462 | 0 | 0.361 |
| 27168 | LINC00702 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | AFAP1L2 | Sponge network | -0.737 | 0.06182 | -0.284 | 0.15434 | 0.361 |
| 27169 | SNHG14 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 56 | MDM4 | Sponge network | -1.111 | 0.0003 | 0.345 | 0.00762 | 0.361 |
| 27170 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-629-3p | 19 | AR | Sponge network | -2.549 | 0.00528 | -1.554 | 0 | 0.361 |
| 27171 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-93-5p | 49 | TCF4 | Sponge network | -0.384 | 0.40652 | 0.016 | 0.92271 | 0.361 |
| 27172 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | ABCB5 | Sponge network | -1.216 | 0 | -0.956 | 0.04077 | 0.361 |
| 27173 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | SMARCA2 | Sponge network | -0.737 | 0.06182 | -0.529 | 0.00014 | 0.361 |
| 27174 | RP5-1198O20.4 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | RTN4 | Sponge network | 0.997 | 0.00066 | -0.412 | 0.00014 | 0.361 |
| 27175 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-429 | 12 | AKAP11 | Sponge network | 1.361 | 0 | 0.196 | 0.09825 | 0.361 |
| 27176 | RP11-867G23.10 |
hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-338-3p | 10 | LIMCH1 | Sponge network | -2.531 | 0 | -0.915 | 1.0E-5 | 0.361 |
| 27177 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | FOXO3 | Sponge network | 1.757 | 0 | -0.04 | 0.72028 | 0.361 |
| 27178 | MIR22HG |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p | 12 | RIMS1 | Sponge network | -1.047 | 0 | -3.143 | 0 | 0.361 |
| 27179 | RP11-35G9.3 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 20 | PCDH9 | Sponge network | 0.657 | 4.0E-5 | -1.823 | 4.0E-5 | 0.361 |
| 27180 | RP11-479G22.8 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | RASSF8 | Sponge network | 1.308 | 0 | -0.122 | 0.65591 | 0.361 |
| 27181 | FGF14-AS2 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 12 | CSMD1 | Sponge network | -1.582 | 8.0E-5 | -0.63 | 0.18881 | 0.361 |
| 27182 | RP11-119F7.5 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | CSDE1 | Sponge network | 0.225 | 0.30644 | -0.493 | 0 | 0.361 |
| 27183 | RP5-1198O20.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-93-3p;hsa-miR-940 | 51 | GRIK3 | Sponge network | 0.997 | 0.00066 | -3.208 | 0 | 0.361 |
| 27184 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 15 | LHX6 | Sponge network | 0.374 | 0.20054 | -0.087 | 0.69193 | 0.361 |
| 27185 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 16 | DKK3 | Sponge network | -0.612 | 0.00094 | -0.052 | 0.77783 | 0.361 |
| 27186 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-495-3p | 11 | PHC3 | Sponge network | 1.056 | 0 | 0.212 | 0.0499 | 0.361 |
| 27187 | MALAT1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 31 | ABI2 | Sponge network | 0.782 | 0.00016 | 0.175 | 0.14787 | 0.361 |
| 27188 | LINC00657 |
hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DOCK4 | Sponge network | 0.91 | 0 | 0.502 | 0.00108 | 0.361 |
| 27189 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | PRKAA2 | Sponge network | 0.385 | 0.10067 | -2.462 | 0 | 0.361 |
| 27190 | LINC00654 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 35 | ABI2 | Sponge network | 0.374 | 0.20054 | 0.175 | 0.14787 | 0.361 |
| 27191 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-96-5p | 35 | TCF4 | Sponge network | 0.898 | 1.0E-5 | 0.016 | 0.92271 | 0.361 |
| 27192 | LINC00969 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-539-5p | 11 | TSC1 | Sponge network | 0.683 | 1.0E-5 | 0.103 | 0.26071 | 0.361 |
| 27193 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-96-5p | 21 | SCN9A | Sponge network | -1.349 | 0.00017 | -0.525 | 0.10593 | 0.361 |
| 27194 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | TLL1 | Sponge network | 0.299 | 0.57722 | -1.195 | 3.0E-5 | 0.361 |
| 27195 | MALAT1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | EPG5 | Sponge network | 0.782 | 0.00016 | 0.101 | 0.3563 | 0.361 |
| 27196 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-3p | 21 | TMTC3 | Sponge network | 1.089 | 0 | 0.123 | 0.22189 | 0.361 |
| 27197 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | MAP3K12 | Sponge network | -1.203 | 0.03881 | -0.057 | 0.59369 | 0.361 |
| 27198 | FAM66C |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | GLIPR1 | Sponge network | -0.901 | 0.00019 | 0.146 | 0.3276 | 0.361 |
| 27199 | RP11-758N13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p | 14 | PGR | Sponge network | 2.939 | 3.0E-5 | -1.704 | 0 | 0.361 |
| 27200 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 31 | NTRK3 | Sponge network | 0.342 | 0.03129 | -1.77 | 3.0E-5 | 0.361 |
| 27201 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 16 | ZMYM2 | Sponge network | 1.044 | 2.0E-5 | 0.279 | 0.01273 | 0.361 |
| 27202 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-98-5p | 37 | PBX1 | Sponge network | 0.246 | 0.59912 | -1.206 | 0 | 0.361 |
| 27203 | PWAR6 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | EYA4 | Sponge network | -1.575 | 6.0E-5 | 0.3 | 0.36876 | 0.361 |
| 27204 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-625-3p | 12 | ZFHX3 | Sponge network | 0.903 | 0 | 0.389 | 0.01363 | 0.361 |
| 27205 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-375 | 15 | MYBL1 | Sponge network | -0.326 | 0.14314 | 0.494 | 0.02152 | 0.361 |
| 27206 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-654-3p | 16 | TP53INP1 | Sponge network | -0.567 | 0 | -0.496 | 0.00064 | 0.361 |
| 27207 | AC009120.6 |
hsa-let-7i-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-590-3p | 14 | DICER1 | Sponge network | 0.919 | 0 | 0.042 | 0.674 | 0.361 |
| 27208 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 44 | CBX5 | Sponge network | 0.72 | 0.0001 | 0.165 | 0.24446 | 0.361 |
| 27209 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 17 | RB1CC1 | Sponge network | 0.683 | 1.0E-5 | 0.175 | 0.12559 | 0.361 |
| 27210 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-539-5p;hsa-miR-576-5p | 17 | TSC1 | Sponge network | 0.921 | 0.00118 | 0.103 | 0.26071 | 0.361 |
| 27211 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | HBP1 | Sponge network | -1.575 | 6.0E-5 | -0.454 | 0 | 0.361 |
| 27212 | LINC00657 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ZFX | Sponge network | 0.91 | 0 | 0.09 | 0.42563 | 0.361 |
| 27213 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 40 | NTRK2 | Sponge network | 0.214 | 0.18246 | -0.736 | 0.06495 | 0.361 |
| 27214 | PSMD5-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 26 | ABL2 | Sponge network | 0.569 | 0.00067 | 0.82 | 0 | 0.361 |
| 27215 | RP11-439A17.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-590-5p | 13 | SNX29 | Sponge network | 0.051 | 0.95756 | -0.34 | 0.01371 | 0.361 |
| 27216 | DNM3OS |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-96-5p | 17 | IRS1 | Sponge network | 0.572 | 0.01722 | -0.026 | 0.88855 | 0.361 |
| 27217 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 17 | ZMYND11 | Sponge network | 0.349 | 0.01695 | -0.2 | 0.05449 | 0.361 |
| 27218 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-940 | 41 | PTPRT | Sponge network | -0.129 | 0.57177 | -0.907 | 0.03727 | 0.361 |
| 27219 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 22 | ZMYM2 | Sponge network | 0.417 | 0.00445 | 0.279 | 0.01273 | 0.361 |
| 27220 | CTC-559E9.5 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-539-5p | 13 | KIF5B | Sponge network | 0.594 | 0.00064 | 0.362 | 0.00066 | 0.361 |
| 27221 | RP11-686D22.8 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ITGB1 | Sponge network | 0.898 | 1.0E-5 | 0.524 | 0.00017 | 0.361 |
| 27222 | RP11-360F5.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p | 14 | GAS7 | Sponge network | 1.867 | 0 | -0.555 | 0.01602 | 0.361 |
| 27223 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | KCNA6 | Sponge network | -0.611 | 0.09607 | -1.531 | 0.00013 | 0.361 |
| 27224 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 33 | ARID5B | Sponge network | -0.129 | 0.57177 | 0.342 | 0.01676 | 0.361 |
| 27225 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429 | 13 | CREBBP | Sponge network | 0.056 | 0.81195 | 0.126 | 0.15186 | 0.361 |
| 27226 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-500a-3p | 13 | ARHGAP29 | Sponge network | -1.092 | 4.0E-5 | 0.131 | 0.42953 | 0.361 |
| 27227 | SNHG1 |
hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-337-3p | 11 | CDC73 | Sponge network | 1.265 | 0 | 0.094 | 0.20463 | 0.361 |
| 27228 | PTOV1-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-766-3p | 15 | TBL1XR1 | Sponge network | 0.87 | 0 | 0.578 | 0 | 0.36 |
| 27229 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | CUL5 | Sponge network | 0.083 | 0.66405 | 0.026 | 0.77301 | 0.36 |
| 27230 | SH3RF3-AS1 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p | 11 | PTCH1 | Sponge network | 0.889 | 9.0E-5 | -0.1 | 0.6179 | 0.36 |
| 27231 | TRAF3IP2-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | PTCH1 | Sponge network | -0.323 | 0.01372 | -0.1 | 0.6179 | 0.36 |
| 27232 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-340-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 18 | RNF144A | Sponge network | 0.16 | 0.50785 | 0.594 | 0.0009 | 0.36 |
| 27233 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 19 | EBF3 | Sponge network | 0.199 | 0.38015 | -0.058 | 0.81437 | 0.36 |
| 27234 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 39 | MCC | Sponge network | -0.615 | 0.00015 | -1.005 | 0 | 0.36 |
| 27235 | AGAP11 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-590-5p | 14 | FNBP1 | Sponge network | -1.645 | 0 | -0.969 | 4.0E-5 | 0.36 |
| 27236 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p | 11 | ZNF292 | Sponge network | 1.03 | 0 | 0.276 | 0.04148 | 0.36 |
| 27237 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | PTPN13 | Sponge network | -0.738 | 0.21233 | -1.208 | 0 | 0.36 |
| 27238 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-577;hsa-miR-92a-1-5p | 20 | PRKAB2 | Sponge network | -2.69 | 0.0001 | -0.367 | 0.01371 | 0.36 |
| 27239 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | EPHA5 | Sponge network | -0.021 | 0.93598 | -0.957 | 0.01733 | 0.36 |
| 27240 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | FYN | Sponge network | -1.161 | 0.00591 | -0.131 | 0.37051 | 0.36 |
| 27241 | RP11-244H3.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | 0.659 | 3.0E-5 | 0.142 | 0.04938 | 0.36 |
| 27242 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | CNTN1 | Sponge network | 0.064 | 0.72934 | -2.594 | 0 | 0.36 |
| 27243 | RP11-159D12.2 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | USP9X | Sponge network | 1.254 | 0 | 0.222 | 0.01689 | 0.36 |
| 27244 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-877-5p | 16 | CDH19 | Sponge network | -0.384 | 0.40652 | -3.434 | 0 | 0.36 |
| 27245 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 15 | NEDD4 | Sponge network | -0.739 | 0.00044 | 0.02 | 0.89834 | 0.36 |
| 27246 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 18 | EBF1 | Sponge network | 1.757 | 0 | -0.384 | 0.08856 | 0.36 |
| 27247 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p | 29 | JAZF1 | Sponge network | -0.738 | 0.21233 | -0.377 | 0.02677 | 0.36 |
| 27248 | RP11-327P2.5 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-505-5p;hsa-miR-629-5p | 11 | WNK2 | Sponge network | -0.147 | 0.40452 | -0.352 | 0.31066 | 0.36 |
| 27249 | CTC-487M23.5 |
hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-450b-5p | 16 | SCN9A | Sponge network | 0.912 | 0.00014 | -0.525 | 0.10593 | 0.36 |
| 27250 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 23 | ERC1 | Sponge network | -0.318 | 0.65847 | -0.108 | 0.38794 | 0.36 |
| 27251 | LINC01018 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-590-5p | 10 | THSD7A | Sponge network | -2.915 | 0.00015 | 0.242 | 0.25154 | 0.36 |
| 27252 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-93-5p;hsa-miR-940 | 19 | HLF | Sponge network | -1.255 | 0.00981 | -2.443 | 0 | 0.36 |
| 27253 | RP11-13K12.1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p | 15 | PTCH1 | Sponge network | -0.154 | 0.78939 | -0.1 | 0.6179 | 0.36 |
| 27254 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 28 | EPHA5 | Sponge network | -0.246 | 0.35034 | -0.957 | 0.01733 | 0.36 |
| 27255 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | LRRC55 | Sponge network | -0.318 | 0.65847 | -0.026 | 0.93163 | 0.36 |
| 27256 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | SEMA3E | Sponge network | -0.405 | 0.22013 | -1.717 | 0.00137 | 0.36 |
| 27257 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-493-5p;hsa-miR-495-3p | 20 | SMAD2 | Sponge network | 0.706 | 0 | -0.128 | 0.12732 | 0.36 |
| 27258 | RP11-156E6.1 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-497-5p | 15 | PAFAH1B2 | Sponge network | 1.266 | 0 | 0.318 | 0.0001 | 0.36 |
| 27259 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p | 41 | CBX5 | Sponge network | 0.594 | 0.41522 | 0.165 | 0.24446 | 0.36 |
| 27260 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-671-5p | 27 | PTPRD | Sponge network | -0.154 | 0.53954 | -1.005 | 0.00064 | 0.36 |
| 27261 | RP11-440L14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 15 | LPP | Sponge network | 0.545 | 0.05789 | -0.459 | 0.04475 | 0.36 |
| 27262 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 20 | RET | Sponge network | -0.615 | 0.00015 | -0.833 | 0.03517 | 0.36 |
| 27263 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | GNA13 | Sponge network | 0.421 | 0.00084 | 0.007 | 0.93884 | 0.36 |
| 27264 | GABPB1-AS1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-5p;hsa-miR-495-3p | 15 | STAG2 | Sponge network | 1.11 | 0 | 0.252 | 0.00438 | 0.36 |
| 27265 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-629-5p;hsa-miR-93-5p | 42 | UNC80 | Sponge network | -0.563 | 0.02545 | -1.934 | 0 | 0.36 |
| 27266 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3913-5p;hsa-miR-590-3p | 10 | RIMS1 | Sponge network | -0.096 | 0.67958 | -3.143 | 0 | 0.36 |
| 27267 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 31 | CADM1 | Sponge network | -0.129 | 0.57177 | -0.9 | 0.00084 | 0.36 |
| 27268 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 54 | RUNX1T1 | Sponge network | -0.727 | 0.04726 | -0.344 | 0.2374 | 0.36 |
| 27269 | RASSF8-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-421;hsa-miR-429 | 16 | PLXNC1 | Sponge network | 0.335 | 0.20596 | 0.569 | 0.00329 | 0.36 |
| 27270 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 14 | RCAN2 | Sponge network | 0.385 | 0.10067 | -0.33 | 0.15172 | 0.36 |
| 27271 | RP11-6O2.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-96-5p | 19 | LRIG1 | Sponge network | 0.331 | 0.57247 | -0.537 | 0.00588 | 0.36 |
| 27272 | RP11-195B17.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-539-5p | 22 | DST | Sponge network | 0.37 | 0.27614 | -0.713 | 0.00096 | 0.36 |
| 27273 | CTD-2666L21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-424-5p | 13 | BRD3 | Sponge network | 0.368 | 0.20093 | 0.37 | 0.00013 | 0.36 |
| 27274 | A1BG-AS1 |
hsa-miR-107;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-589-3p | 11 | ST6GALNAC3 | Sponge network | -0.164 | 0.45106 | -0.987 | 0 | 0.36 |
| 27275 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | MITF | Sponge network | 0.199 | 0.38015 | -0.769 | 0.00022 | 0.36 |
| 27276 | LINC00621 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | HIP1 | Sponge network | 0.131 | 0.8436 | 0.774 | 0 | 0.36 |
| 27277 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p | 17 | NIN | Sponge network | -0.583 | 0.14589 | -0.253 | 0.13794 | 0.36 |
| 27278 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 24 | EPHA3 | Sponge network | 0.559 | 0.00026 | 0.185 | 0.53588 | 0.36 |
| 27279 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-96-5p | 65 | AFF4 | Sponge network | 0.194 | 0.64278 | -0.266 | 0.01565 | 0.36 |
| 27280 | LINC00622 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZFHX3 | Sponge network | 1.169 | 0.00028 | 0.389 | 0.01363 | 0.36 |
| 27281 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | RPS6KA3 | Sponge network | 0.537 | 0.00139 | 0.256 | 0.02419 | 0.36 |
| 27282 | AC006116.24 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p | 16 | AFF1 | Sponge network | -0.473 | 0.07602 | -0.384 | 0.00053 | 0.36 |
| 27283 | EMX2OS |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NRG3 | Sponge network | -0.777 | 0.1134 | -1.836 | 4.0E-5 | 0.36 |
| 27284 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-7-5p | 11 | GLI3 | Sponge network | 0.395 | 0.15451 | -0.615 | 0.01482 | 0.36 |
| 27285 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 17 | SASH1 | Sponge network | -2.095 | 0.00112 | -0.502 | 0.00175 | 0.36 |
| 27286 | DNAJC27-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-7-1-3p | 11 | TPO | Sponge network | -0.969 | 2.0E-5 | -1.87 | 2.0E-5 | 0.36 |
| 27287 | LINC00476 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RBM6 | Sponge network | -0.615 | 0.00015 | -0.137 | 0.15663 | 0.36 |
| 27288 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-660-5p;hsa-miR-93-5p | 10 | ZHX2 | Sponge network | -0.777 | 0.1134 | -0.192 | 0.18459 | 0.36 |
| 27289 | LINC00473 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 13 | ZNF208 | Sponge network | -1.203 | 0.03881 | -0.4 | 0.22381 | 0.36 |
| 27290 | TP73-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-629-5p | 12 | UVRAG | Sponge network | -0.563 | 0.02545 | -0.017 | 0.83865 | 0.36 |
| 27291 | RP11-156P1.3 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-550a-3p | 10 | CACNA1E | Sponge network | 0.214 | 0.18246 | 1.752 | 0 | 0.36 |
| 27292 | LINC00094 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-486-5p | 14 | NRK | Sponge network | 0.253 | 0.09565 | 0.656 | 0.19217 | 0.36 |
| 27293 | RP11-244H3.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | NAV3 | Sponge network | 0.659 | 3.0E-5 | 0.212 | 0.47729 | 0.36 |
| 27294 | FAM66C |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429 | 10 | SCN5A | Sponge network | -0.901 | 0.00019 | -0.259 | 0.48224 | 0.36 |
| 27295 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 22 | PICALM | Sponge network | 0.421 | 0.00084 | 0.086 | 0.23523 | 0.36 |
| 27296 | RP5-855D21.1 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-376c-3p | 12 | BMPR2 | Sponge network | 1.115 | 0.00188 | 0.206 | 0.0635 | 0.36 |
| 27297 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TBX18 | Sponge network | -2.452 | 0 | 0.843 | 0.02945 | 0.36 |
| 27298 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-5p | 14 | ZMYM2 | Sponge network | 0.921 | 0.00118 | 0.279 | 0.01273 | 0.36 |
| 27299 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | MEF2C | Sponge network | 0.889 | 9.0E-5 | -0.499 | 0.01448 | 0.36 |
| 27300 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-744-5p;hsa-miR-93-5p | 13 | CHD5 | Sponge network | -0.376 | 0.00337 | -0.922 | 0.01521 | 0.36 |
| 27301 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | SSBP2 | Sponge network | -0.737 | 0.10822 | -1.58 | 0 | 0.36 |
| 27302 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-935 | 16 | LRRC55 | Sponge network | -2.62 | 0 | -0.026 | 0.93163 | 0.36 |
| 27303 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-361-5p | 17 | UNC80 | Sponge network | -0.309 | 0.1145 | -1.934 | 0 | 0.36 |
| 27304 | SOX2-OT |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-935;hsa-miR-940;hsa-miR-99b-3p | 46 | GRIK3 | Sponge network | -1.264 | 6.0E-5 | -3.208 | 0 | 0.36 |
| 27305 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 53 | BMPR2 | Sponge network | 0.488 | 0.00084 | 0.206 | 0.0635 | 0.36 |
| 27306 | RP11-760H22.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-501-3p | 13 | TGFBR3 | Sponge network | 0.001 | 0.99753 | -1.173 | 1.0E-5 | 0.36 |
| 27307 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 28 | PCDH10 | Sponge network | -0.969 | 2.0E-5 | -1.644 | 0.0078 | 0.36 |
| 27308 | MRPL23-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-92a-1-5p | 11 | AKAP6 | Sponge network | -0.278 | 0.76559 | -1.586 | 3.0E-5 | 0.36 |
| 27309 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-940 | 48 | CADM2 | Sponge network | -0.738 | 0.21233 | -3.782 | 0 | 0.36 |
| 27310 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 11 | INPP4A | Sponge network | -0.125 | 0.49414 | 0.07 | 0.53563 | 0.36 |
| 27311 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-625-5p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -0.096 | 0.67958 | 0.024 | 0.89889 | 0.36 |
| 27312 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-576-5p | 18 | DMXL1 | Sponge network | -0.611 | 0.09607 | -0.426 | 0.00056 | 0.36 |
| 27313 | FGD5-AS1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | ERCC6 | Sponge network | 0.401 | 0.00066 | 0.23 | 0.015 | 0.36 |
| 27314 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 12 | DACT1 | Sponge network | 0.4 | 0.14611 | 0.783 | 0.00072 | 0.36 |
| 27315 | USP3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-93-5p | 25 | PRRX1 | Sponge network | -0.162 | 0.47687 | 1.316 | 0 | 0.36 |
| 27316 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-940 | 26 | IGF1 | Sponge network | 0.056 | 0.81195 | -0.204 | 0.5898 | 0.36 |
| 27317 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-495-3p | 11 | MYO9A | Sponge network | 1.056 | 0 | -0.147 | 0.32373 | 0.36 |
| 27318 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | LATS1 | Sponge network | 1.163 | 0.00018 | 0.109 | 0.34208 | 0.36 |
| 27319 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-628-5p | 19 | BCL6 | Sponge network | -0.405 | 0.22013 | -0.211 | 0.19489 | 0.36 |
| 27320 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 15 | AKAP13 | Sponge network | 0.4 | 0.14611 | 0.028 | 0.7729 | 0.36 |
| 27321 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-96-5p | 60 | NTRK3 | Sponge network | 0.395 | 0.15451 | -1.77 | 3.0E-5 | 0.36 |
| 27322 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p | 22 | AKAP11 | Sponge network | 0.663 | 0.00073 | 0.196 | 0.09825 | 0.36 |
| 27323 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 15 | TSHZ3 | Sponge network | 0.497 | 0.01451 | -0.556 | 0.01516 | 0.36 |
| 27324 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-5p | 18 | ESR1 | Sponge network | -0.517 | 0.02935 | -1.035 | 3.0E-5 | 0.36 |
| 27325 | RP11-822E23.8 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-589-3p | 16 | RPS6KA6 | Sponge network | -0.777 | 0.16667 | -2.989 | 0 | 0.36 |
| 27326 | SH3BP5-AS1 |
hsa-miR-106b-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | RABEP1 | Sponge network | 0.267 | 0.2937 | -0.066 | 0.43142 | 0.36 |
| 27327 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | AKAP11 | Sponge network | -0.031 | 0.89063 | 0.196 | 0.09825 | 0.36 |
| 27328 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 24 | KANK1 | Sponge network | -1.575 | 6.0E-5 | -0.55 | 0.00134 | 0.36 |
| 27329 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | SOX5 | Sponge network | -0.08 | 0.90092 | -1.091 | 4.0E-5 | 0.36 |
| 27330 | LINC00472 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | FYN | Sponge network | -0.842 | 0.01361 | -0.131 | 0.37051 | 0.36 |
| 27331 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 22 | BCL6 | Sponge network | 0.064 | 0.72934 | -0.211 | 0.19489 | 0.36 |
| 27332 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | NR4A3 | Sponge network | -1.575 | 6.0E-5 | -1.385 | 0 | 0.36 |
| 27333 | RP11-968O1.5 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-224-5p | 10 | FAM135B | Sponge network | -0.829 | 0.01343 | -3.978 | 0 | 0.36 |
| 27334 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p | 12 | DPYD | Sponge network | -0.487 | 0.02157 | -0.736 | 0.00076 | 0.36 |
| 27335 | SERHL |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-3607-3p | 21 | HLF | Sponge network | -0.602 | 0.00331 | -2.443 | 0 | 0.36 |
| 27336 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-93-5p | 15 | FAT4 | Sponge network | 0.082 | 0.55397 | -0.354 | 0.14694 | 0.36 |
| 27337 | RP11-434B12.1 |
hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-495-3p | 15 | SYNM | Sponge network | -0.031 | 0.89063 | -2.309 | 1.0E-5 | 0.36 |
| 27338 | RP4-717I23.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-625-5p | 12 | CEP170 | Sponge network | 0.637 | 0.00069 | 0.943 | 0 | 0.36 |
| 27339 | CTC-301O7.4 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 11 | TCF4 | Sponge network | 0.292 | 0.43384 | 0.016 | 0.92271 | 0.36 |
| 27340 | RP11-98I9.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p | 14 | TRIM33 | Sponge network | 0.67 | 0.00048 | 0.402 | 7.0E-5 | 0.36 |
| 27341 | H1FX-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 16 | NRXN1 | Sponge network | -0.206 | 0.12942 | -3.778 | 0 | 0.36 |
| 27342 | AC108488.4 |
hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-629-3p | 24 | PTPN14 | Sponge network | 0.259 | 0.12542 | 0.144 | 0.34595 | 0.36 |
| 27343 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 32 | ARID5B | Sponge network | -0.222 | 0.32445 | 0.342 | 0.01676 | 0.36 |
| 27344 | RP11-439A17.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | GAS7 | Sponge network | 0.051 | 0.95756 | -0.555 | 0.01602 | 0.36 |
| 27345 | RP11-774O3.3 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | RIMS2 | Sponge network | -0.487 | 0.02157 | -1.365 | 0.00208 | 0.36 |
| 27346 | RP11-196G18.22 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-625-5p;hsa-miR-766-3p | 25 | MDM4 | Sponge network | 1.206 | 0 | 0.345 | 0.00762 | 0.36 |
| 27347 | GHRLOS |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-942-5p | 12 | OPCML | Sponge network | -1.942 | 0 | -2.498 | 0 | 0.36 |
| 27348 | HOXA-AS2 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | PCDH10 | Sponge network | 0.61 | 0.01593 | -1.644 | 0.0078 | 0.36 |
| 27349 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 20 | EPHA7 | Sponge network | 0.419 | 0.0319 | -2.197 | 3.0E-5 | 0.36 |
| 27350 | SNHG14 |
hsa-miR-140-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | CHD6 | Sponge network | -1.111 | 0.0003 | 0.32 | 0.01326 | 0.36 |
| 27351 | NEAT1 |
hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 18 | RASAL2 | Sponge network | 0.705 | 0.00014 | 0.869 | 0 | 0.36 |
| 27352 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-382-5p;hsa-miR-421 | 15 | SYNPO2 | Sponge network | -0.285 | 0.2172 | -2.342 | 0 | 0.36 |
| 27353 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-93-5p | 20 | IL17RD | Sponge network | 0.497 | 0.01451 | -0.019 | 0.94873 | 0.36 |
| 27354 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-33a-3p | 10 | ARNT2 | Sponge network | -0.309 | 0.1145 | -0.332 | 0.25851 | 0.36 |
| 27355 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | LRRC55 | Sponge network | -2.117 | 0.00161 | -0.026 | 0.93163 | 0.36 |
| 27356 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-454-3p | 26 | ZBTB4 | Sponge network | -0.738 | 0.21233 | -0.739 | 0 | 0.36 |
| 27357 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-320b;hsa-miR-424-5p | 10 | DYNC1I1 | Sponge network | 0.082 | 0.55397 | -1.12 | 0.00107 | 0.36 |
| 27358 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p | 37 | QKI | Sponge network | -0.227 | 0.31657 | -0.333 | 0.04111 | 0.36 |
| 27359 | LINC00173 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-7-1-3p | 11 | NRXN3 | Sponge network | 0.056 | 0.8494 | -0.893 | 0.04186 | 0.36 |
| 27360 | RP11-400K9.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-5p;hsa-miR-940 | 17 | WDFY3 | Sponge network | -0.733 | 0.19469 | -0.201 | 0.0967 | 0.36 |
| 27361 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 16 | BCL6 | Sponge network | -2.117 | 0.00161 | -0.211 | 0.19489 | 0.36 |
| 27362 | RP11-1246C19.1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 1.352 | 0 | 0.362 | 0.00066 | 0.36 |
| 27363 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 52 | SSBP2 | Sponge network | -0.222 | 0.32445 | -1.58 | 0 | 0.36 |
| 27364 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | HIP1 | Sponge network | 0.683 | 1.0E-5 | 0.774 | 0 | 0.36 |
| 27365 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-590-3p | 14 | PNRC1 | Sponge network | -0.487 | 0.02157 | -0.646 | 0 | 0.36 |
| 27366 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p | 11 | SLITRK3 | Sponge network | -0.418 | 0.00068 | -3.328 | 0 | 0.36 |
| 27367 | TRAF3IP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | MSN | Sponge network | -0.323 | 0.01372 | -0.023 | 0.88422 | 0.36 |
| 27368 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 64 | NTRK3 | Sponge network | 0.374 | 0.20054 | -1.77 | 3.0E-5 | 0.36 |
| 27369 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 25 | EBF3 | Sponge network | 0.374 | 0.20054 | -0.058 | 0.81437 | 0.36 |
| 27370 | RP11-497H16.9 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PHF3 | Sponge network | 0.545 | 0.00064 | 0.126 | 0.19652 | 0.36 |
| 27371 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p | 13 | AHNAK | Sponge network | 0.214 | 0.18246 | -1.207 | 0 | 0.36 |
| 27372 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 17 | RBL2 | Sponge network | -0.612 | 0.00094 | -0.282 | 0.00254 | 0.36 |
| 27373 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 20 | CRTC3 | Sponge network | 0.659 | 3.0E-5 | 0.164 | 0.16786 | 0.36 |
| 27374 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | ST6GAL2 | Sponge network | -0.709 | 0.18168 | -1.201 | 0.00597 | 0.36 |
| 27375 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940 | 50 | MEF2C | Sponge network | -2.925 | 0 | -0.499 | 0.01448 | 0.36 |
| 27376 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 33 | PHC3 | Sponge network | -0.709 | 0.18168 | 0.212 | 0.0499 | 0.36 |
| 27377 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p | 13 | CBL | Sponge network | 1.044 | 2.0E-5 | 0.291 | 0.01267 | 0.36 |
| 27378 | FENDRR |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 29 | NRK | Sponge network | -1.281 | 0.00012 | 0.656 | 0.19217 | 0.36 |
| 27379 | FLNB-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | ARHGEF12 | Sponge network | 1.163 | 0.00018 | 0.09 | 0.35693 | 0.36 |
| 27380 | AF131215.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | TET1 | Sponge network | -0.176 | 0.59692 | -0.135 | 0.52138 | 0.36 |
| 27381 | RP11-67L2.2 |
hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-874-3p | 12 | ARID1B | Sponge network | 0.72 | 0.0001 | 0.003 | 0.97503 | 0.36 |
| 27382 | FAM66C |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | HIP1 | Sponge network | -0.901 | 0.00019 | 0.774 | 0 | 0.36 |
| 27383 | GNG12-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 41 | NTRK2 | Sponge network | -0.793 | 7.0E-5 | -0.736 | 0.06495 | 0.36 |
| 27384 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | PLSCR4 | Sponge network | -0.351 | 0.11358 | -0.474 | 0.03833 | 0.36 |
| 27385 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-374b-3p | 10 | GRIA3 | Sponge network | -4.463 | 0.00025 | -1.956 | 0 | 0.36 |
| 27386 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | ZFX | Sponge network | 0.083 | 0.66405 | 0.09 | 0.42563 | 0.36 |
| 27387 | AC093110.3 |
hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-539-5p | 10 | PHF3 | Sponge network | 1.044 | 2.0E-5 | 0.126 | 0.19652 | 0.36 |
| 27388 | CTC-523E23.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-429 | 13 | KCNA6 | Sponge network | -0.235 | 0.35308 | -1.531 | 0.00013 | 0.36 |
| 27389 | SNHG12 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p | 12 | MAT2A | Sponge network | 1.103 | 0 | -0.073 | 0.36195 | 0.36 |
| 27390 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 47 | TMEM132B | Sponge network | 0.194 | 0.64278 | -0.83 | 0.01084 | 0.36 |
| 27391 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-942-5p | 21 | ARHGAP28 | Sponge network | -1.047 | 0 | -0.911 | 0.00013 | 0.36 |
| 27392 | RP11-73M18.10 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-342-3p;hsa-miR-497-5p | 11 | KIF1B | Sponge network | 0.85 | 0 | -0.118 | 0.32258 | 0.36 |
| 27393 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-93-3p | 35 | SSBP2 | Sponge network | -0.773 | 0.04073 | -1.58 | 0 | 0.36 |
| 27394 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3E | Sponge network | -1.216 | 0 | -1.717 | 0.00137 | 0.36 |
| 27395 | RP11-46C24.7 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 16 | UBE4B | Sponge network | 0.392 | 0.0278 | -0.235 | 0.007 | 0.36 |
| 27396 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-940 | 26 | AKAP11 | Sponge network | 0.056 | 0.81195 | 0.196 | 0.09825 | 0.36 |
| 27397 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-577 | 18 | TIMP3 | Sponge network | -0.465 | 0.04703 | -0.546 | 0.02307 | 0.36 |
| 27398 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-940 | 27 | BHLHE41 | Sponge network | 0.194 | 0.64278 | 0.353 | 0.12753 | 0.36 |
| 27399 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-590-3p | 30 | CCND2 | Sponge network | 0.178 | 0.29106 | -0.267 | 0.3112 | 0.36 |
| 27400 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 45 | PAFAH1B1 | Sponge network | -0.323 | 0.01372 | -0.429 | 0 | 0.36 |
| 27401 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 24 | YAP1 | Sponge network | 0.311 | 0.10344 | 0.22 | 0.11836 | 0.36 |
| 27402 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 33 | SCN9A | Sponge network | -2.925 | 0 | -0.525 | 0.10593 | 0.36 |
| 27403 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 21 | TIMP3 | Sponge network | -0.842 | 0.01361 | -0.546 | 0.02307 | 0.36 |
| 27404 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p | 34 | IL6ST | Sponge network | 0.274 | 0.07681 | -0.402 | 0.00676 | 0.36 |
| 27405 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 77 | AFF4 | Sponge network | -0.739 | 0.0574 | -0.266 | 0.01565 | 0.36 |
| 27406 | AC010226.4 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-590-3p | 16 | NRK | Sponge network | -0.811 | 2.0E-5 | 0.656 | 0.19217 | 0.36 |
| 27407 | RP11-597D13.9 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 15 | ITGA5 | Sponge network | 1.302 | 7.0E-5 | 0.452 | 0.03524 | 0.36 |
| 27408 | RP11-540B6.6 |
hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | BRD3 | Sponge network | 0.93 | 0 | 0.37 | 0.00013 | 0.36 |
| 27409 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | ARID5B | Sponge network | 0.572 | 0.01722 | 0.342 | 0.01676 | 0.36 |
| 27410 | RP11-13K12.5 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 15 | RIMS2 | Sponge network | -0.318 | 0.65847 | -1.365 | 0.00208 | 0.36 |
| 27411 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 14 | ARHGAP28 | Sponge network | -4.477 | 0 | -0.911 | 0.00013 | 0.36 |
| 27412 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 29 | AKAP11 | Sponge network | 0.222 | 0.29133 | 0.196 | 0.09825 | 0.36 |
| 27413 | AP006216.10 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-590-3p | 12 | MDM4 | Sponge network | 0.779 | 0.00016 | 0.345 | 0.00762 | 0.36 |
| 27414 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TAL1 | Sponge network | -1.057 | 0.00029 | -0.462 | 0.00574 | 0.36 |
| 27415 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | -0.727 | 0.04726 | 0.783 | 0.00072 | 0.36 |
| 27416 | AC025335.1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-376c-3p | 13 | RASAL2 | Sponge network | 0.783 | 0 | 0.869 | 0 | 0.36 |
| 27417 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-629-3p;hsa-miR-942-5p | 12 | GPC6 | Sponge network | -2.531 | 0 | -0.054 | 0.81465 | 0.36 |
| 27418 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 21 | RNASEL | Sponge network | -1.654 | 0.00144 | -0.272 | 0.01796 | 0.36 |
| 27419 | HAND2-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 29 | PCDH17 | Sponge network | -1.697 | 0.00889 | 0.731 | 2.0E-5 | 0.36 |
| 27420 | RP11-401P9.4 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 13 | STAT5B | Sponge network | -0.384 | 0.40652 | -0.182 | 0.1082 | 0.36 |
| 27421 | PCA3 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RTN4 | Sponge network | -0.709 | 0.18168 | -0.412 | 0.00014 | 0.36 |
| 27422 | RP11-798M19.6 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ADH1B | Sponge network | -0.612 | 0.00094 | -3.716 | 0 | 0.36 |
| 27423 | JAZF1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 28 | EFNA5 | Sponge network | -0.769 | 0.00035 | -0.421 | 0.17868 | 0.36 |
| 27424 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-5p | 15 | LRRC55 | Sponge network | 0.178 | 0.29106 | -0.026 | 0.93163 | 0.36 |
| 27425 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | MITF | Sponge network | -2.925 | 0 | -0.769 | 0.00022 | 0.36 |
| 27426 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-409-3p | 12 | ATRX | Sponge network | 0.368 | 0.20093 | 0.261 | 0.04408 | 0.36 |
| 27427 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-421;hsa-miR-590-3p | 13 | CNTN1 | Sponge network | 0.178 | 0.29106 | -2.594 | 0 | 0.36 |
| 27428 | RP11-701H24.3 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-511-5p;hsa-miR-590-3p | 10 | ZFHX3 | Sponge network | -1.425 | 0.04204 | 0.389 | 0.01363 | 0.36 |
| 27429 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | RCAN2 | Sponge network | 0.659 | 3.0E-5 | -0.33 | 0.15172 | 0.36 |
| 27430 | RP11-517B11.7 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-374a-3p;hsa-miR-495-3p | 13 | SLC5A3 | Sponge network | 0.706 | 0 | 0.493 | 5.0E-5 | 0.36 |
| 27431 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | EPHA3 | Sponge network | -0.727 | 0.04726 | 0.185 | 0.53588 | 0.36 |
| 27432 | DNM3OS |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ADH1B | Sponge network | 0.572 | 0.01722 | -3.716 | 0 | 0.36 |
| 27433 | AC093627.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-29b-1-5p | 18 | RUNX1T1 | Sponge network | -0.787 | 0.3928 | -0.344 | 0.2374 | 0.36 |
| 27434 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-511-5p;hsa-miR-590-3p | 14 | BCL6 | Sponge network | -0.358 | 0.17255 | -0.211 | 0.19489 | 0.36 |
| 27435 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | LRRC7 | Sponge network | 0.335 | 0.20596 | -1.341 | 0.01193 | 0.36 |
| 27436 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-577;hsa-miR-590-3p | 10 | SEMA3E | Sponge network | -0.096 | 0.67958 | -1.717 | 0.00137 | 0.36 |
| 27437 | RP11-159D12.2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 22 | ABI2 | Sponge network | 1.254 | 0 | 0.175 | 0.14787 | 0.36 |
| 27438 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-589-5p | 22 | EYA4 | Sponge network | -2.69 | 0.0001 | 0.3 | 0.36876 | 0.359 |
| 27439 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 50 | IL6ST | Sponge network | 0.214 | 0.18246 | -0.402 | 0.00676 | 0.359 |
| 27440 | RP11-497H16.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | ABI2 | Sponge network | 0.545 | 0.00064 | 0.175 | 0.14787 | 0.359 |
| 27441 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | PRKAA2 | Sponge network | -1.264 | 6.0E-5 | -2.462 | 0 | 0.359 |
| 27442 | EMX2OS |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 34 | ABI2 | Sponge network | -0.777 | 0.1134 | 0.175 | 0.14787 | 0.359 |
| 27443 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | TLE4 | Sponge network | 0.101 | 0.60919 | -1.157 | 0 | 0.359 |
| 27444 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 24 | USP12 | Sponge network | 0.782 | 0.00016 | -0.102 | 0.40768 | 0.359 |
| 27445 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | 0.997 | 0.00066 | 0.309 | 0.17079 | 0.359 |
| 27446 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-96-5p | 27 | EBF1 | Sponge network | -0.18 | 0.20343 | -0.384 | 0.08856 | 0.359 |
| 27447 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 12 | CNTNAP3 | Sponge network | -0.513 | 0.04703 | -1.465 | 0.00033 | 0.359 |
| 27448 | SH3RF3-AS1 |
hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-505-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p | 15 | CTIF | Sponge network | 0.889 | 9.0E-5 | -0.389 | 0.00549 | 0.359 |
| 27449 | LINC01018 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | -2.915 | 0.00015 | 0.783 | 0.00072 | 0.359 |
| 27450 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 12 | KCNAB1 | Sponge network | -0.563 | 0.02545 | -0.755 | 2.0E-5 | 0.359 |
| 27451 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 29 | RBMS3 | Sponge network | 0.659 | 3.0E-5 | -0.519 | 0.03551 | 0.359 |
| 27452 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 37 | LIFR | Sponge network | -0.942 | 0 | -1.794 | 0 | 0.359 |
| 27453 | ANKRD10-IT1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-7-1-3p | 12 | BRD3 | Sponge network | 1.739 | 0 | 0.37 | 0.00013 | 0.359 |
| 27454 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | SASH1 | Sponge network | -1.439 | 4.0E-5 | -0.502 | 0.00175 | 0.359 |
| 27455 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p | 10 | MLLT10 | Sponge network | 0.832 | 0 | 0.105 | 0.33292 | 0.359 |
| 27456 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-93-3p | 14 | TOM1L2 | Sponge network | -4.463 | 0.00025 | -0.994 | 0 | 0.359 |
| 27457 | CTC-559E9.5 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-539-5p;hsa-miR-576-5p | 14 | TSC1 | Sponge network | 0.594 | 0.00064 | 0.103 | 0.26071 | 0.359 |
| 27458 | RP11-672A2.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-505-5p | 15 | ASTN1 | Sponge network | 1.344 | 0 | -2.389 | 2.0E-5 | 0.359 |
| 27459 | GNG12-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 27 | PRDM2 | Sponge network | -0.793 | 7.0E-5 | -0.258 | 0.00721 | 0.359 |
| 27460 | FENDRR |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TRIM58 | Sponge network | -1.281 | 0.00012 | -1.692 | 0 | 0.359 |
| 27461 | ASH1L-AS1 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | RICTOR | Sponge network | 0.948 | 0 | 0.388 | 0.00188 | 0.359 |
| 27462 | RP11-517B11.7 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 15 | PHF6 | Sponge network | 0.706 | 0 | 0.785 | 0 | 0.359 |
| 27463 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-590-3p | 10 | RB1CC1 | Sponge network | 0.539 | 0.03722 | 0.175 | 0.12559 | 0.359 |
| 27464 | RP11-6O2.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-96-5p | 18 | SETBP1 | Sponge network | 0.331 | 0.57247 | -1.302 | 1.0E-5 | 0.359 |
| 27465 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | RET | Sponge network | -2.915 | 0.00015 | -0.833 | 0.03517 | 0.359 |
| 27466 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 35 | RBMS3 | Sponge network | -0.125 | 0.49414 | -0.519 | 0.03551 | 0.359 |
| 27467 | PART1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 14 | PTPN13 | Sponge network | -2.925 | 0 | -1.208 | 0 | 0.359 |
| 27468 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-450b-5p | 21 | PRDM2 | Sponge network | -0.583 | 0.14589 | -0.258 | 0.00721 | 0.359 |
| 27469 | PWAR6 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 16 | POU2F2 | Sponge network | -1.575 | 6.0E-5 | -0.233 | 0.32214 | 0.359 |
| 27470 | ZRANB2-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 10 | PIK3CA | Sponge network | -0.376 | 0.00337 | 0.195 | 0.06908 | 0.359 |
| 27471 | ANKRD10-IT1 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 10 | MYO9A | Sponge network | 1.739 | 0 | -0.147 | 0.32373 | 0.359 |
| 27472 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p | 19 | FGFR1 | Sponge network | 0.083 | 0.66405 | -0.509 | 0.03957 | 0.359 |
| 27473 | SERTAD4-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p | 22 | CREB3L2 | Sponge network | -0.932 | 0.00329 | -0.525 | 2.0E-5 | 0.359 |
| 27474 | RP11-359B12.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | CREB1 | Sponge network | 0.064 | 0.72934 | 0.203 | 0.00537 | 0.359 |
| 27475 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p | 17 | LATS1 | Sponge network | 1.254 | 0 | 0.109 | 0.34208 | 0.359 |
| 27476 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 13 | MAF | Sponge network | -0.376 | 0.00337 | -1.257 | 0 | 0.359 |
| 27477 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p | 15 | HOOK3 | Sponge network | -0.615 | 0.00015 | 0.273 | 0.09957 | 0.359 |
| 27478 | RP11-181G12.2 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-550a-5p;hsa-miR-671-5p;hsa-miR-940 | 23 | CHD5 | Sponge network | 0.556 | 0.00298 | -0.922 | 0.01521 | 0.359 |
| 27479 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SLC6A15 | Sponge network | -0.942 | 0 | -2.373 | 2.0E-5 | 0.359 |
| 27480 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | PDE4DIP | Sponge network | -0.246 | 0.35034 | -0.282 | 0.0257 | 0.359 |
| 27481 | RP11-458F8.4 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30c-5p;hsa-miR-30e-5p | 14 | CREB1 | Sponge network | 1.518 | 0 | 0.203 | 0.00537 | 0.359 |
| 27482 | RP11-121C2.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 1.147 | 0 | 1.083 | 0 | 0.359 |
| 27483 | AC093627.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-625-5p;hsa-miR-7-5p | 13 | SNX29 | Sponge network | -0.787 | 0.3928 | -0.34 | 0.01371 | 0.359 |
| 27484 | RP11-121C2.2 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 14 | CDK6 | Sponge network | 1.147 | 0 | 0.991 | 0 | 0.359 |
| 27485 | LINC00702 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-542-3p | 10 | ZNF331 | Sponge network | -0.737 | 0.06182 | -1.251 | 0 | 0.359 |
| 27486 | FGF14-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | LRIG1 | Sponge network | -1.582 | 8.0E-5 | -0.537 | 0.00588 | 0.359 |
| 27487 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 29 | PGR | Sponge network | 0.222 | 0.29133 | -1.704 | 0 | 0.359 |
| 27488 | RP11-575A19.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p | 16 | LPP | Sponge network | 1.969 | 0.00316 | -0.459 | 0.04475 | 0.359 |
| 27489 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | PHC3 | Sponge network | 0.249 | 0.35902 | 0.212 | 0.0499 | 0.359 |
| 27490 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-940 | 21 | IGF1 | Sponge network | 0.082 | 0.55397 | -0.204 | 0.5898 | 0.359 |
| 27491 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 28 | TGFBR3 | Sponge network | 0.537 | 0.00139 | -1.173 | 1.0E-5 | 0.359 |
| 27492 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-576-5p | 12 | RIMBP2 | Sponge network | -0.318 | 0.65847 | -1.968 | 5.0E-5 | 0.359 |
| 27493 | RP11-736K20.5 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-511-5p | 17 | BTG2 | Sponge network | -0.358 | 0.17255 | -1.763 | 0 | 0.359 |
| 27494 | RP11-531A24.5 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 28 | ERC1 | Sponge network | 0.75 | 0.00054 | -0.108 | 0.38794 | 0.359 |
| 27495 | RP13-516M14.1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 39 | DTNA | Sponge network | 0.389 | 0.04293 | -1.648 | 1.0E-5 | 0.359 |
| 27496 | CTC-510F12.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-23a-3p;hsa-miR-335-3p | 10 | UNC80 | Sponge network | -0.691 | 0.00146 | -1.934 | 0 | 0.359 |
| 27497 | HCG18 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | TSC1 | Sponge network | 1.063 | 0 | 0.103 | 0.26071 | 0.359 |
| 27498 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-370-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 34 | CHD5 | Sponge network | 0.082 | 0.55654 | -0.922 | 0.01521 | 0.359 |
| 27499 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | ST6GALNAC3 | Sponge network | -0.611 | 0.09607 | -0.987 | 0 | 0.359 |
| 27500 | LINC00641 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 48 | GAS7 | Sponge network | -0.222 | 0.32445 | -0.555 | 0.01602 | 0.359 |
| 27501 | FAM66C |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 38 | PPP1R9A | Sponge network | -0.901 | 0.00019 | -0.903 | 0.04254 | 0.359 |
| 27502 | LINC00476 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-935 | 36 | PTPRT | Sponge network | -0.615 | 0.00015 | -0.907 | 0.03727 | 0.359 |
| 27503 | AC009948.5 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | RELN | Sponge network | 0.532 | 0.00505 | -2.403 | 0 | 0.359 |
| 27504 | C4B-AS1 |
hsa-let-7d-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 14 | THBS1 | Sponge network | 2.306 | 9.0E-5 | 0.741 | 0.00386 | 0.359 |
| 27505 | AF131215.9 |
hsa-let-7d-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | TLE4 | Sponge network | -0.322 | 0.25945 | -1.157 | 0 | 0.359 |
| 27506 | USP3-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-589-3p | 24 | SYT4 | Sponge network | -0.162 | 0.47687 | -3.207 | 0 | 0.359 |
| 27507 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-589-3p | 25 | ADARB1 | Sponge network | 2.306 | 9.0E-5 | -0.335 | 0.06631 | 0.359 |
| 27508 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 34 | MYO9A | Sponge network | 0.311 | 0.10344 | -0.147 | 0.32373 | 0.359 |
| 27509 | RP11-479G22.8 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 1.308 | 0 | 0.321 | 0.00267 | 0.359 |
| 27510 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-93-3p | 50 | RUNX1T1 | Sponge network | -0.18 | 0.20343 | -0.344 | 0.2374 | 0.359 |
| 27511 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-590-5p | 16 | PDZD2 | Sponge network | -0.899 | 6.0E-5 | -0.909 | 3.0E-5 | 0.359 |
| 27512 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-93-5p | 34 | HLF | Sponge network | -0.18 | 0.20343 | -2.443 | 0 | 0.359 |
| 27513 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | USP6 | Sponge network | 1.11 | 0 | 0.188 | 0.3871 | 0.359 |
| 27514 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 25 | TMTC3 | Sponge network | 0.559 | 0.00026 | 0.123 | 0.22189 | 0.359 |
| 27515 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-590-3p | 12 | PSIP1 | Sponge network | 0.532 | 0.00505 | -0.005 | 0.96897 | 0.359 |
| 27516 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | SLITRK3 | Sponge network | -0.615 | 0.00015 | -3.328 | 0 | 0.359 |
| 27517 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p | 18 | CADM1 | Sponge network | -0.469 | 0.08721 | -0.9 | 0.00084 | 0.359 |
| 27518 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 15 | PKNOX1 | Sponge network | 0.572 | 0.01722 | 0.085 | 0.28917 | 0.359 |
| 27519 | PSMG3-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLI1 | Sponge network | -0.125 | 0.49414 | -0.294 | 0.10905 | 0.359 |
| 27520 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-96-5p | 68 | LPP | Sponge network | 0.41 | 0.02214 | -0.459 | 0.04475 | 0.359 |
| 27521 | A2M-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 17 | SFRP1 | Sponge network | -0.08 | 0.90092 | -3.566 | 0 | 0.359 |
| 27522 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-429;hsa-miR-590-3p | 10 | DDX5 | Sponge network | -0.031 | 0.89063 | -0.099 | 0.17428 | 0.359 |
| 27523 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 10 | GALNT13 | Sponge network | -0.726 | 0.00132 | -0.857 | 0.07439 | 0.359 |
| 27524 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 38 | CBX5 | Sponge network | 0.603 | 0.00127 | 0.165 | 0.24446 | 0.359 |
| 27525 | RP11-196G11.4 |
hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-365a-3p;hsa-miR-590-3p | 13 | RICTOR | Sponge network | 0.875 | 0 | 0.388 | 0.00188 | 0.359 |
| 27526 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-33a-5p;hsa-miR-539-5p | 16 | SSBP2 | Sponge network | -0.214 | 0.44248 | -1.58 | 0 | 0.359 |
| 27527 | MIR600HG |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-940 | 30 | CHD5 | Sponge network | 0.594 | 0.41522 | -0.922 | 0.01521 | 0.359 |
| 27528 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | AHNAK | Sponge network | -0.83 | 0 | -1.207 | 0 | 0.359 |
| 27529 | BDNF-AS |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | PRUNE2 | Sponge network | -0.668 | 3.0E-5 | -1.733 | 0.00022 | 0.359 |
| 27530 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | HOOK3 | Sponge network | 0.381 | 0.25967 | 0.273 | 0.09957 | 0.359 |
| 27531 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-505-5p;hsa-miR-590-3p | 11 | RBM5 | Sponge network | -0.227 | 0.31657 | -0.205 | 0.0129 | 0.359 |
| 27532 | EMX2OS |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | TAL1 | Sponge network | -0.777 | 0.1134 | -0.462 | 0.00574 | 0.359 |
| 27533 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 44 | PAFAH1B1 | Sponge network | 0.194 | 0.64278 | -0.429 | 0 | 0.359 |
| 27534 | LINC01018 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | -2.915 | 0.00015 | -0.366 | 0.01036 | 0.359 |
| 27535 | AF131215.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 43 | MDM4 | Sponge network | -0.176 | 0.59692 | 0.345 | 0.00762 | 0.359 |
| 27536 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-3p | 15 | AR | Sponge network | -0.214 | 0.44248 | -1.554 | 0 | 0.359 |
| 27537 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-374a-3p;hsa-miR-590-5p | 11 | CBL | Sponge network | 1.134 | 0 | 0.291 | 0.01267 | 0.359 |
| 27538 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-374b-3p;hsa-miR-590-3p | 18 | SPRY3 | Sponge network | 0.178 | 0.29106 | 0.016 | 0.91286 | 0.359 |
| 27539 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ATRX | Sponge network | 0.417 | 0.00445 | 0.261 | 0.04408 | 0.359 |
| 27540 | TRAF3IP2-AS1 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | RASSF3 | Sponge network | -0.323 | 0.01372 | -0.226 | 0.11691 | 0.359 |
| 27541 | MIR143HG |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | TMTC2 | Sponge network | -0.739 | 0.0574 | -0.215 | 0.18248 | 0.359 |
| 27542 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p | 17 | SCN9A | Sponge network | 0.082 | 0.55397 | -0.525 | 0.10593 | 0.359 |
| 27543 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | FGF5 | Sponge network | 0.532 | 0.00505 | 0.549 | 0.28659 | 0.359 |
| 27544 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 39 | ZFHX3 | Sponge network | -1.281 | 0.00012 | 0.389 | 0.01363 | 0.359 |
| 27545 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-33a-3p | 10 | LRRC7 | Sponge network | -0.693 | 0.05629 | -1.341 | 0.01193 | 0.359 |
| 27546 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p | 14 | CNTNAP3 | Sponge network | 0.331 | 0.57247 | -1.465 | 0.00033 | 0.359 |
| 27547 | RP11-620J15.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | RYR2 | Sponge network | -0.739 | 0.00044 | -1.466 | 0.00031 | 0.359 |
| 27548 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-590-3p | 10 | GPAM | Sponge network | 0.336 | 0.00739 | 0.142 | 0.29732 | 0.359 |
| 27549 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-539-5p | 14 | PRKAA2 | Sponge network | 0.043 | 0.81628 | -2.462 | 0 | 0.359 |
| 27550 | TRAF3IP2-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | BRD3 | Sponge network | -0.323 | 0.01372 | 0.37 | 0.00013 | 0.359 |
| 27551 | LINC00476 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ITGB3 | Sponge network | -0.615 | 0.00015 | -0.011 | 0.95751 | 0.359 |
| 27552 | RP11-25K21.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29a-5p | 12 | PRRX1 | Sponge network | 0.489 | 0.25786 | 1.316 | 0 | 0.359 |
| 27553 | PSMG3-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 34 | GRIK3 | Sponge network | -0.125 | 0.49414 | -3.208 | 0 | 0.359 |
| 27554 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PDE4DIP | Sponge network | 0.998 | 0 | -0.282 | 0.0257 | 0.359 |
| 27555 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-539-5p;hsa-miR-576-5p | 11 | TP53INP1 | Sponge network | 0.594 | 0.00064 | -0.496 | 0.00064 | 0.359 |
| 27556 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | AR | Sponge network | 0.385 | 0.10067 | -1.554 | 0 | 0.359 |
| 27557 | RP11-479G22.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-26a-2-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 14 | ABI2 | Sponge network | 1.308 | 0 | 0.175 | 0.14787 | 0.359 |
| 27558 | MRPL23-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-502-5p | 25 | DTNA | Sponge network | -0.278 | 0.76559 | -1.648 | 1.0E-5 | 0.359 |
| 27559 | AGAP11 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-3913-5p;hsa-miR-421 | 33 | AFF1 | Sponge network | -1.645 | 0 | -0.384 | 0.00053 | 0.359 |
| 27560 | RP11-175O19.4 |
hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-497-5p | 14 | SLC5A3 | Sponge network | 0.917 | 0 | 0.493 | 5.0E-5 | 0.359 |
| 27561 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p | 19 | EBF3 | Sponge network | 0.214 | 0.18246 | -0.058 | 0.81437 | 0.359 |
| 27562 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-539-5p | 11 | SSBP2 | Sponge network | 0.043 | 0.81628 | -1.58 | 0 | 0.359 |
| 27563 | CTD-3064M3.3 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-582-5p | 14 | GJA1 | Sponge network | -0.469 | 0.08721 | -0.485 | 0.02179 | 0.359 |
| 27564 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 22 | TBL1XR1 | Sponge network | 1.03 | 0 | 0.578 | 0 | 0.359 |
| 27565 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TSC1 | Sponge network | 0.559 | 0.00026 | 0.103 | 0.26071 | 0.359 |
| 27566 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-93-5p | 17 | CNTNAP3 | Sponge network | -0.303 | 0.21002 | -1.465 | 0.00033 | 0.359 |
| 27567 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-3607-3p | 11 | RICTOR | Sponge network | 0.37 | 0.27614 | 0.388 | 0.00188 | 0.359 |
| 27568 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | DDX5 | Sponge network | -1.575 | 6.0E-5 | -0.099 | 0.17428 | 0.359 |
| 27569 | ZNF32-AS2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-654-3p | 13 | DDX6 | Sponge network | 1.552 | 7.0E-5 | 0.091 | 0.36428 | 0.359 |
| 27570 | RP1-151F17.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | 0.222 | 0.29133 | 0.098 | 0.40994 | 0.359 |
| 27571 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 22 | TRIM33 | Sponge network | 0.417 | 0.00445 | 0.402 | 7.0E-5 | 0.359 |
| 27572 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p | 47 | AR | Sponge network | 0.494 | 0.00339 | -1.554 | 0 | 0.359 |
| 27573 | CTD-2314G24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-96-5p | 22 | CBFA2T3 | Sponge network | 0.246 | 0.59912 | -1.221 | 1.0E-5 | 0.359 |
| 27574 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-424-5p | 14 | MYBL1 | Sponge network | 0.299 | 0.57722 | 0.494 | 0.02152 | 0.359 |
| 27575 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-424-5p | 12 | RECK | Sponge network | 0.082 | 0.55397 | -1.043 | 0 | 0.359 |
| 27576 | LINC00578 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-628-5p | 10 | ACVR1 | Sponge network | 0.811 | 0.03228 | 0.279 | 0.00234 | 0.359 |
| 27577 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-625-5p | 14 | TGFBR3 | Sponge network | -1.255 | 0.00981 | -1.173 | 1.0E-5 | 0.359 |
| 27578 | AC016747.3 |
hsa-miR-107;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-769-5p | 15 | SYNPO2 | Sponge network | 0.199 | 0.38015 | -2.342 | 0 | 0.359 |
| 27579 | USP3-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-589-3p | 11 | ZNF844 | Sponge network | -0.162 | 0.47687 | -0.966 | 0.00035 | 0.359 |
| 27580 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 17 | MAP3K12 | Sponge network | -0.129 | 0.57177 | -0.057 | 0.59369 | 0.359 |
| 27581 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 32 | RICTOR | Sponge network | 0.569 | 0.00067 | 0.388 | 0.00188 | 0.359 |
| 27582 | CTC-378H22.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-877-5p | 12 | ASTN1 | Sponge network | 0.001 | 0.99663 | -2.389 | 2.0E-5 | 0.359 |
| 27583 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-625-5p | 14 | ABI2 | Sponge network | -0.278 | 0.76559 | 0.175 | 0.14787 | 0.359 |
| 27584 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 40 | ABI2 | Sponge network | -2.46 | 0 | 0.175 | 0.14787 | 0.359 |
| 27585 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 43 | PTPRD | Sponge network | -0.421 | 0.0114 | -1.005 | 0.00064 | 0.359 |
| 27586 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | NEDD4 | Sponge network | 0.225 | 0.30644 | 0.02 | 0.89834 | 0.359 |
| 27587 | RP11-403I13.8 |
hsa-miR-126-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | IRS1 | Sponge network | 0.619 | 0.00157 | -0.026 | 0.88855 | 0.359 |
| 27588 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-582-5p | 14 | MOB1B | Sponge network | 1.262 | 0 | 0.197 | 0.15266 | 0.359 |
| 27589 | LINC00843 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p | 11 | PIK3R1 | Sponge network | 1.106 | 0 | -0.133 | 0.36854 | 0.359 |
| 27590 | RP11-798M19.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 21 | TOM1L2 | Sponge network | -0.612 | 0.00094 | -0.994 | 0 | 0.359 |
| 27591 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 21 | EBF3 | Sponge network | -0.342 | 0.02288 | -0.058 | 0.81437 | 0.359 |
| 27592 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-429 | 12 | CBL | Sponge network | 2.364 | 0.00134 | 0.291 | 0.01267 | 0.359 |
| 27593 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 22 | HIP1 | Sponge network | 0.083 | 0.66405 | 0.774 | 0 | 0.359 |
| 27594 | CTD-3092A11.2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ITGB1 | Sponge network | 0.873 | 0 | 0.524 | 0.00017 | 0.359 |
| 27595 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-942-5p | 10 | NRXN1 | Sponge network | -1.14 | 0.03513 | -3.778 | 0 | 0.359 |
| 27596 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 15 | ZFHX3 | Sponge network | 1.073 | 0.00216 | 0.389 | 0.01363 | 0.359 |
| 27597 | ZSCAN16-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-942-5p | 15 | GLIPR1 | Sponge network | -0.342 | 0.02288 | 0.146 | 0.3276 | 0.359 |
| 27598 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-450b-5p | 13 | RBMS3 | Sponge network | 0.912 | 0.00014 | -0.519 | 0.03551 | 0.359 |
| 27599 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-629-5p;hsa-miR-96-5p | 39 | KIF1B | Sponge network | -0.517 | 0.02935 | -0.118 | 0.32258 | 0.359 |
| 27600 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 20 | PIK3C2A | Sponge network | 0.873 | 0 | 0.061 | 0.53776 | 0.359 |
| 27601 | RP11-473I1.5 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-655-3p | 10 | PIK3C2A | Sponge network | 0.542 | 0.00054 | 0.061 | 0.53776 | 0.359 |
| 27602 | HOXA-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | FAM133A | Sponge network | 0.61 | 0.01593 | 0.549 | 0.29009 | 0.359 |
| 27603 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 21 | CNTNAP3 | Sponge network | -2.925 | 0 | -1.465 | 0.00033 | 0.359 |
| 27604 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-576-5p | 10 | HECA | Sponge network | 1.316 | 0.01106 | -0.217 | 0.03463 | 0.359 |
| 27605 | FGF14-AS2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p | 19 | PTPRT | Sponge network | -1.582 | 8.0E-5 | -0.907 | 0.03727 | 0.359 |
| 27606 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 28 | MOB1B | Sponge network | -0.227 | 0.31657 | 0.197 | 0.15266 | 0.359 |
| 27607 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-375;hsa-miR-429;hsa-miR-7-1-3p | 11 | VEZT | Sponge network | 1.153 | 0 | 0.259 | 0.00296 | 0.359 |
| 27608 | RP11-130L8.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | SPG20 | Sponge network | -0.663 | 0.10887 | -1.16 | 0 | 0.359 |
| 27609 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-98-5p | 42 | PRKAA2 | Sponge network | 0.494 | 0.00339 | -2.462 | 0 | 0.359 |
| 27610 | RP11-298J20.4 |
hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-671-5p | 19 | BMPR2 | Sponge network | 0.002 | 0.99057 | 0.206 | 0.0635 | 0.359 |
| 27611 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-374a-3p | 17 | GREM1 | Sponge network | -0.842 | 0.01361 | 0.902 | 0.01691 | 0.359 |
| 27612 | RP11-727A23.5 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p | 13 | PHF6 | Sponge network | 1.117 | 0 | 0.785 | 0 | 0.359 |
| 27613 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 53 | IL6ST | Sponge network | -0.176 | 0.59692 | -0.402 | 0.00676 | 0.359 |
| 27614 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-501-3p;hsa-miR-96-5p | 20 | ABI2 | Sponge network | 0.453 | 0.01839 | 0.175 | 0.14787 | 0.359 |
| 27615 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 16 | TP53INP1 | Sponge network | 0.657 | 4.0E-5 | -0.496 | 0.00064 | 0.359 |
| 27616 | RP11-53O19.1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 25 | MED12L | Sponge network | -0.405 | 0.22013 | -0.194 | 0.55299 | 0.359 |
| 27617 | FENDRR |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 21 | IRS1 | Sponge network | -1.281 | 0.00012 | -0.026 | 0.88855 | 0.359 |
| 27618 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | NEDD4 | Sponge network | 0.537 | 0.00139 | 0.02 | 0.89834 | 0.359 |
| 27619 | LINC00672 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p | 11 | KCNAB1 | Sponge network | -0.227 | 0.31657 | -0.755 | 2.0E-5 | 0.359 |
| 27620 | LPP-AS2 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | FAT3 | Sponge network | -0.18 | 0.20343 | -1.601 | 0.00051 | 0.359 |
| 27621 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-625-5p | 18 | RSPO2 | Sponge network | -0.154 | 0.53954 | -3.038 | 0 | 0.359 |
| 27622 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 30 | CNTN1 | Sponge network | -0.615 | 0.00015 | -2.594 | 0 | 0.359 |
| 27623 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-196a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-424-5p | 12 | CBFA2T3 | Sponge network | -0.295 | 0.02604 | -1.221 | 1.0E-5 | 0.359 |
| 27624 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-5p | 15 | PTGFR | Sponge network | -0.737 | 0.10822 | -0.233 | 0.45421 | 0.359 |
| 27625 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | RCAN2 | Sponge network | -0.969 | 2.0E-5 | -0.33 | 0.15172 | 0.359 |
| 27626 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | FGF5 | Sponge network | -0.021 | 0.93598 | 0.549 | 0.28659 | 0.359 |
| 27627 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | ST6GAL2 | Sponge network | -0.053 | 0.80685 | -1.201 | 0.00597 | 0.359 |
| 27628 | RP11-531A24.5 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-495-3p | 10 | STAG2 | Sponge network | 0.75 | 0.00054 | 0.252 | 0.00438 | 0.359 |
| 27629 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 22 | ABL2 | Sponge network | 0.222 | 0.29133 | 0.82 | 0 | 0.359 |
| 27630 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p | 17 | RARB | Sponge network | -1.645 | 0 | -0.825 | 2.0E-5 | 0.359 |
| 27631 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | AHNAK | Sponge network | 0.083 | 0.66405 | -1.207 | 0 | 0.359 |
| 27632 | RP11-5C23.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 12 | ZMYND11 | Sponge network | 0.274 | 0.07681 | -0.2 | 0.05449 | 0.359 |
| 27633 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p | 11 | PTPN14 | Sponge network | -1.582 | 8.0E-5 | 0.144 | 0.34595 | 0.359 |
| 27634 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p | 12 | CRTC3 | Sponge network | 1.757 | 0 | 0.164 | 0.16786 | 0.359 |
| 27635 | RP11-356J5.12 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | BTG2 | Sponge network | -0.899 | 6.0E-5 | -1.763 | 0 | 0.359 |
| 27636 | CTA-14H9.5 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 11 | AR | Sponge network | 0.145 | 0.53343 | -1.554 | 0 | 0.359 |
| 27637 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-576-5p | 10 | RIMBP2 | Sponge network | 0.176 | 0.37402 | -1.968 | 5.0E-5 | 0.359 |
| 27638 | FAM157C |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p | 13 | PHF6 | Sponge network | 0.91 | 0 | 0.785 | 0 | 0.359 |
| 27639 | H1FX-AS1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 12 | GRIN2A | Sponge network | -0.206 | 0.12942 | -2.161 | 0 | 0.359 |
| 27640 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p | 27 | HLF | Sponge network | 0.95 | 0.15943 | -2.443 | 0 | 0.359 |
| 27641 | CKMT2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ITGB3 | Sponge network | 0.082 | 0.55654 | -0.011 | 0.95751 | 0.359 |
| 27642 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 19 | CRTC3 | Sponge network | -0.513 | 0.04703 | 0.164 | 0.16786 | 0.359 |
| 27643 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-7-1-3p | 31 | RASSF2 | Sponge network | -1.161 | 0.00591 | -0.273 | 0.21961 | 0.359 |
| 27644 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-589-3p | 10 | CDH10 | Sponge network | -0.162 | 0.47687 | -2.397 | 0 | 0.359 |
| 27645 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 51 | WDFY3 | Sponge network | 0.083 | 0.66405 | -0.201 | 0.0967 | 0.359 |
| 27646 | FAM66C |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | SYT4 | Sponge network | -0.901 | 0.00019 | -3.207 | 0 | 0.359 |
| 27647 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-369-3p;hsa-miR-495-3p | 22 | BMPR2 | Sponge network | 1.044 | 2.0E-5 | 0.206 | 0.0635 | 0.359 |
| 27648 | RP11-527J8.1 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-9-5p | 13 | CCDC6 | Sponge network | 1.324 | 0 | 0.27 | 0.02555 | 0.359 |
| 27649 | RP11-410L14.2 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | PPP2R5C | Sponge network | -0.052 | 0.76839 | -0.314 | 3.0E-5 | 0.359 |
| 27650 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-500b-5p;hsa-miR-590-5p | 30 | ACVR2A | Sponge network | -1.645 | 0 | -0.409 | 0.00039 | 0.359 |
| 27651 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 34 | CADM1 | Sponge network | -1.216 | 0 | -0.9 | 0.00084 | 0.359 |
| 27652 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | DKK3 | Sponge network | -0.899 | 6.0E-5 | -0.052 | 0.77783 | 0.359 |
| 27653 | LINC00473 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | RYR2 | Sponge network | -1.203 | 0.03881 | -1.466 | 0.00031 | 0.359 |
| 27654 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p | 17 | DDX6 | Sponge network | 1.294 | 0 | 0.091 | 0.36428 | 0.359 |
| 27655 | LINC01001 |
hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-766-3p | 15 | TBL1XR1 | Sponge network | 1.055 | 0 | 0.578 | 0 | 0.359 |
| 27656 | LINC01001 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p | 13 | CREB1 | Sponge network | 1.055 | 0 | 0.203 | 0.00537 | 0.359 |
| 27657 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-3607-3p | 13 | IGSF10 | Sponge network | -0.773 | 0.04073 | -1.751 | 1.0E-5 | 0.359 |
| 27658 | RP11-195F19.9 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-5p | 11 | PTGFR | Sponge network | -0.726 | 0.00132 | -0.233 | 0.45421 | 0.359 |
| 27659 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 36 | CHD5 | Sponge network | -0.777 | 0.16667 | -0.922 | 0.01521 | 0.359 |
| 27660 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-5p | 11 | FOXO3 | Sponge network | 1.106 | 0 | -0.04 | 0.72028 | 0.359 |
| 27661 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p | 19 | PRRX1 | Sponge network | -0.376 | 0.00337 | 1.316 | 0 | 0.358 |
| 27662 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | PRKAB2 | Sponge network | 1.089 | 0 | -0.367 | 0.01371 | 0.358 |
| 27663 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-940;hsa-miR-96-5p | 13 | HOOK3 | Sponge network | 0.4 | 0.14611 | 0.273 | 0.09957 | 0.358 |
| 27664 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | LRRC7 | Sponge network | -0.739 | 0.00044 | -1.341 | 0.01193 | 0.358 |
| 27665 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-589-3p | 26 | USP12 | Sponge network | -0.162 | 0.47687 | -0.102 | 0.40768 | 0.358 |
| 27666 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 44 | BCL2 | Sponge network | -0.777 | 0.1134 | -0.899 | 0 | 0.358 |
| 27667 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p | 10 | CDH10 | Sponge network | -0.246 | 0.35034 | -2.397 | 0 | 0.358 |
| 27668 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | AHNAK | Sponge network | -0.323 | 0.01372 | -1.207 | 0 | 0.358 |
| 27669 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | RASSF8 | Sponge network | 0.083 | 0.66405 | -0.122 | 0.65591 | 0.358 |
| 27670 | AP001055.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 17 | KCNA6 | Sponge network | 0.693 | 0.00076 | -1.531 | 0.00013 | 0.358 |
| 27671 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 15 | PRKAA1 | Sponge network | 1.923 | 0 | 0.317 | 0.0031 | 0.358 |
| 27672 | PGM5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-660-5p | 14 | CRTC1 | Sponge network | -4.546 | 0 | -0.116 | 0.28412 | 0.358 |
| 27673 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-625-3p | 19 | TMTC3 | Sponge network | 1.317 | 0 | 0.123 | 0.22189 | 0.358 |
| 27674 | RP11-6O2.3 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-450b-5p | 10 | TPO | Sponge network | 0.331 | 0.57247 | -1.87 | 2.0E-5 | 0.358 |
| 27675 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-455-3p | 10 | TLE4 | Sponge network | -0.72 | 0.24239 | -1.157 | 0 | 0.358 |
| 27676 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 17 | RBL2 | Sponge network | -0.709 | 0.18168 | -0.282 | 0.00254 | 0.358 |
| 27677 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 25 | ARNT2 | Sponge network | -0.129 | 0.57177 | -0.332 | 0.25851 | 0.358 |
| 27678 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p | 36 | BNC2 | Sponge network | -0.384 | 0.40652 | -0.491 | 0.09988 | 0.358 |
| 27679 | NR2F1-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | HERC1 | Sponge network | -0.49 | 0.04946 | -0.196 | 0.08544 | 0.358 |
| 27680 | RP4-758J18.10 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p | 17 | TACC1 | Sponge network | -0.091 | 0.71468 | -0.362 | 0.05406 | 0.358 |
| 27681 | GS1-124K5.11 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-877-5p | 21 | NRK | Sponge network | 0.494 | 0.00339 | 0.656 | 0.19217 | 0.358 |
| 27682 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-484 | 10 | BCL11A | Sponge network | 0.792 | 0.0049 | -0.932 | 0.0063 | 0.358 |
| 27683 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p | 15 | TRIM33 | Sponge network | 1.03 | 0 | 0.402 | 7.0E-5 | 0.358 |
| 27684 | EMX2OS |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | SEMA3C | Sponge network | -0.777 | 0.1134 | -0.272 | 0.31153 | 0.358 |
| 27685 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 22 | ZBTB4 | Sponge network | 0.199 | 0.38015 | -0.739 | 0 | 0.358 |
| 27686 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7b-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 47 | BNC2 | Sponge network | -0.355 | 0.0077 | -0.491 | 0.09988 | 0.358 |
| 27687 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | DMTF1 | Sponge network | 0.659 | 3.0E-5 | 0.248 | 0.02726 | 0.358 |
| 27688 | AC090587.4 |
hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-532-3p | 10 | PPM1L | Sponge network | -0.125 | 0.60166 | -0.537 | 0.00616 | 0.358 |
| 27689 | HAR1A |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-423-5p;hsa-miR-877-5p | 11 | CBFA2T3 | Sponge network | -0.672 | 0.19447 | -1.221 | 1.0E-5 | 0.358 |
| 27690 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 18 | CDH19 | Sponge network | 0.225 | 0.30644 | -3.434 | 0 | 0.358 |
| 27691 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | LATS2 | Sponge network | 0.214 | 0.18246 | 0.159 | 0.2304 | 0.358 |
| 27692 | LINC00702 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | PIK3CA | Sponge network | -0.737 | 0.06182 | 0.195 | 0.06908 | 0.358 |
| 27693 | RP4-669H2.1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p | 10 | ZFHX3 | Sponge network | 1.711 | 0.00019 | 0.389 | 0.01363 | 0.358 |
| 27694 | AC141928.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | ABI2 | Sponge network | 0.853 | 0.15352 | 0.175 | 0.14787 | 0.358 |
| 27695 | RP4-791M13.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-425-5p;hsa-miR-7-1-3p | 16 | DIP2C | Sponge network | 0.419 | 0.0319 | -0.436 | 0.01713 | 0.358 |
| 27696 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-590-3p | 11 | RBM5 | Sponge network | 0.194 | 0.64278 | -0.205 | 0.0129 | 0.358 |
| 27697 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 24 | EBF3 | Sponge network | -0.612 | 0.00094 | -0.058 | 0.81437 | 0.358 |
| 27698 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-874-3p | 11 | GIGYF2 | Sponge network | 1.063 | 0 | 0.136 | 0.04403 | 0.358 |
| 27699 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 45 | RBMS3 | Sponge network | 0.311 | 0.10344 | -0.519 | 0.03551 | 0.358 |
| 27700 | AC009948.5 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 39 | AFF1 | Sponge network | 0.532 | 0.00505 | -0.384 | 0.00053 | 0.358 |
| 27701 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | HDAC9 | Sponge network | -1.575 | 6.0E-5 | -0.755 | 0.00495 | 0.358 |
| 27702 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-93-5p | 12 | EPAS1 | Sponge network | -1.047 | 0 | -0.184 | 0.14418 | 0.358 |
| 27703 | HCG11 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-501-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 17 | UBE4B | Sponge network | 0.385 | 0.10067 | -0.235 | 0.007 | 0.358 |
| 27704 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-874-3p | 11 | NR2C2 | Sponge network | 0.592 | 0.00014 | 0.21 | 0.03224 | 0.358 |
| 27705 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 17 | KCNA6 | Sponge network | -0.72 | 0.24239 | -1.531 | 0.00013 | 0.358 |
| 27706 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 38 | PHC3 | Sponge network | -0.615 | 0.00015 | 0.212 | 0.0499 | 0.358 |
| 27707 | C4B-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-744-3p;hsa-miR-96-5p | 15 | HIP1 | Sponge network | 2.306 | 9.0E-5 | 0.774 | 0 | 0.358 |
| 27708 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 26 | SASH1 | Sponge network | -0.513 | 0.04703 | -0.502 | 0.00175 | 0.358 |
| 27709 | RP11-755F10.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-215-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-532-3p | 12 | DST | Sponge network | 0.952 | 0 | -0.713 | 0.00096 | 0.358 |
| 27710 | RP11-968O1.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-493-5p | 23 | FAT3 | Sponge network | -0.829 | 0.01343 | -1.601 | 0.00051 | 0.358 |
| 27711 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CDH10 | Sponge network | -0.612 | 0.00094 | -2.397 | 0 | 0.358 |
| 27712 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 37 | RAP1A | Sponge network | -0.129 | 0.57177 | -0.929 | 0 | 0.358 |
| 27713 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PLSCR4 | Sponge network | 0.064 | 0.72934 | -0.474 | 0.03833 | 0.358 |
| 27714 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | PGR | Sponge network | -0.83 | 0 | -1.704 | 0 | 0.358 |
| 27715 | RP11-464F9.1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-744-3p | 13 | KIF5B | Sponge network | 0.959 | 0 | 0.362 | 0.00066 | 0.358 |
| 27716 | TPTEP1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | MACF1 | Sponge network | -1.216 | 0 | 0.019 | 0.90335 | 0.358 |
| 27717 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-940;hsa-miR-96-5p | 39 | MEF2C | Sponge network | 0.082 | 0.55654 | -0.499 | 0.01448 | 0.358 |
| 27718 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 13 | FYN | Sponge network | -0.125 | 0.49414 | -0.131 | 0.37051 | 0.358 |
| 27719 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | MCC | Sponge network | 0.727 | 3.0E-5 | -1.005 | 0 | 0.358 |
| 27720 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 22 | PPM1A | Sponge network | 0.178 | 0.29106 | -0.623 | 0 | 0.358 |
| 27721 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p | 43 | BMPR2 | Sponge network | 0.921 | 0.00118 | 0.206 | 0.0635 | 0.358 |
| 27722 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-455-5p;hsa-miR-93-5p;hsa-miR-940 | 49 | WDFY3 | Sponge network | -0.162 | 0.47687 | -0.201 | 0.0967 | 0.358 |
| 27723 | HCG18 |
hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | DOCK4 | Sponge network | 1.063 | 0 | 0.502 | 0.00108 | 0.358 |
| 27724 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 38 | PPP1R9A | Sponge network | -0.899 | 6.0E-5 | -0.903 | 0.04254 | 0.358 |
| 27725 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-493-5p | 14 | MYCBP2 | Sponge network | 0.75 | 0.00054 | -0.027 | 0.82016 | 0.358 |
| 27726 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-424-5p | 11 | RET | Sponge network | -4.463 | 0.00025 | -0.833 | 0.03517 | 0.358 |
| 27727 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-454-3p | 13 | RARB | Sponge network | 0.176 | 0.37402 | -0.825 | 2.0E-5 | 0.358 |
| 27728 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-542-3p | 10 | FBXO31 | Sponge network | 0.083 | 0.66405 | -0.085 | 0.42389 | 0.358 |
| 27729 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p | 15 | DYNC1I1 | Sponge network | -0.285 | 0.2172 | -1.12 | 0.00107 | 0.358 |
| 27730 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 42 | SASH1 | Sponge network | -0.49 | 0.04946 | -0.502 | 0.00175 | 0.358 |
| 27731 | RP11-59H7.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 17 | CREB1 | Sponge network | 2.163 | 0 | 0.203 | 0.00537 | 0.358 |
| 27732 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | TLL1 | Sponge network | -0.612 | 0.00094 | -1.195 | 3.0E-5 | 0.358 |
| 27733 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-539-5p | 16 | PGR | Sponge network | 0.37 | 0.27614 | -1.704 | 0 | 0.358 |
| 27734 | CTD-2666L21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p | 29 | RUNX1T1 | Sponge network | 0.368 | 0.20093 | -0.344 | 0.2374 | 0.358 |
| 27735 | AC005618.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | AKAP6 | Sponge network | 0.862 | 0.06213 | -1.586 | 3.0E-5 | 0.358 |
| 27736 | RP11-981G7.6 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p | 12 | FBXO11 | Sponge network | 0.986 | 0.01022 | 0.142 | 0.04938 | 0.358 |
| 27737 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 20 | CLASP2 | Sponge network | 0.765 | 7.0E-5 | -0.038 | 0.71 | 0.358 |
| 27738 | HOXB-AS1 |
hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429 | 12 | ELMO1 | Sponge network | -0.154 | 0.53954 | 0.04 | 0.83147 | 0.358 |
| 27739 | RP11-774O3.3 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PCDH18 | Sponge network | -0.487 | 0.02157 | 0.216 | 0.24645 | 0.358 |
| 27740 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 28 | LIFR | Sponge network | -1.047 | 0 | -1.794 | 0 | 0.358 |
| 27741 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 15 | CEP170 | Sponge network | 0.75 | 0.00054 | 0.943 | 0 | 0.358 |
| 27742 | AC016747.3 |
hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 19 | NCAM2 | Sponge network | 0.199 | 0.38015 | -0.482 | 0.22565 | 0.358 |
| 27743 | RP11-774O3.3 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-590-3p | 15 | GLIPR1 | Sponge network | -0.487 | 0.02157 | 0.146 | 0.3276 | 0.358 |
| 27744 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-651-5p;hsa-miR-766-3p;hsa-miR-940 | 33 | AFF4 | Sponge network | -1.331 | 3.0E-5 | -0.266 | 0.01565 | 0.358 |
| 27745 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-539-5p | 17 | RSPO2 | Sponge network | -0.726 | 0.00132 | -3.038 | 0 | 0.358 |
| 27746 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 25 | MITF | Sponge network | -1.249 | 7.0E-5 | -0.769 | 0.00022 | 0.358 |
| 27747 | FGD5-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p | 16 | DOCK4 | Sponge network | 0.401 | 0.00066 | 0.502 | 0.00108 | 0.358 |
| 27748 | AC003090.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p | 25 | BTG2 | Sponge network | -2.117 | 0.00161 | -1.763 | 0 | 0.358 |
| 27749 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 31 | PTPRT | Sponge network | -0.976 | 0.00025 | -0.907 | 0.03727 | 0.358 |
| 27750 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-96-5p | 33 | MOB1B | Sponge network | 0.331 | 0.57247 | 0.197 | 0.15266 | 0.358 |
| 27751 | RP11-434B12.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 18 | RASSF8 | Sponge network | -0.031 | 0.89063 | -0.122 | 0.65591 | 0.358 |
| 27752 | FGD5-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | TCF12 | Sponge network | 0.401 | 0.00066 | 0.174 | 0.13271 | 0.358 |
| 27753 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 61 | AFF4 | Sponge network | -0.096 | 0.67958 | -0.266 | 0.01565 | 0.358 |
| 27754 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 14 | SEMA3C | Sponge network | 0.176 | 0.37402 | -0.272 | 0.31153 | 0.358 |
| 27755 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-628-5p | 20 | GRIA3 | Sponge network | -0.842 | 0.01361 | -1.956 | 0 | 0.358 |
| 27756 | HNRNPU-AS1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | USP9X | Sponge network | 1.601 | 0 | 0.222 | 0.01689 | 0.358 |
| 27757 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | DKK3 | Sponge network | 0.219 | 0.1516 | -0.052 | 0.77783 | 0.358 |
| 27758 | AC003090.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | ROBO1 | Sponge network | -2.117 | 0.00161 | -0.009 | 0.96154 | 0.358 |
| 27759 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | KDSR | Sponge network | 0.72 | 0.0001 | 0.239 | 0.02216 | 0.358 |
| 27760 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-497-5p;hsa-miR-664a-3p;hsa-miR-664a-5p | 21 | SOX11 | Sponge network | 0.972 | 0 | 2.68 | 0 | 0.358 |
| 27761 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-625-5p | 22 | YAP1 | Sponge network | -0.162 | 0.47687 | 0.22 | 0.11836 | 0.358 |
| 27762 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p | 10 | RB1CC1 | Sponge network | 0.389 | 0.04293 | 0.175 | 0.12559 | 0.358 |
| 27763 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 57 | KIF1B | Sponge network | -0.355 | 0.0077 | -0.118 | 0.32258 | 0.358 |
| 27764 | LINC00641 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 22 | SEMA3C | Sponge network | -0.222 | 0.32445 | -0.272 | 0.31153 | 0.358 |
| 27765 | RAMP2-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-590-3p | 13 | KLF10 | Sponge network | -0.513 | 0.04703 | -0.783 | 0 | 0.358 |
| 27766 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-323a-3p | 11 | JAZF1 | Sponge network | -7.551 | 0 | -0.377 | 0.02677 | 0.358 |
| 27767 | RP11-35G9.3 |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p | 10 | DCLK1 | Sponge network | 0.657 | 4.0E-5 | -1.324 | 0.00014 | 0.358 |
| 27768 | RP11-1006G14.4 |
hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-495-3p | 12 | SYNM | Sponge network | 0.559 | 0.00026 | -2.309 | 1.0E-5 | 0.358 |
| 27769 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-590-3p | 10 | LATS1 | Sponge network | 0.545 | 0.00064 | 0.109 | 0.34208 | 0.358 |
| 27770 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 27 | EBF1 | Sponge network | 0.494 | 0.00339 | -0.384 | 0.08856 | 0.358 |
| 27771 | AD000090.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-497-5p;hsa-miR-766-3p | 12 | TBL1XR1 | Sponge network | 0.943 | 0 | 0.578 | 0 | 0.358 |
| 27772 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p | 15 | KIF1B | Sponge network | 0.062 | 0.55274 | -0.118 | 0.32258 | 0.358 |
| 27773 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-590-3p | 25 | TSHZ3 | Sponge network | 0.174 | 0.30034 | -0.556 | 0.01516 | 0.358 |
| 27774 | BOLA3-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-940 | 12 | SORCS1 | Sponge network | -0.246 | 0.35034 | -3.492 | 0 | 0.358 |
| 27775 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 35 | JAZF1 | Sponge network | -0.777 | 0.1134 | -0.377 | 0.02677 | 0.358 |
| 27776 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-93-5p | 37 | ADARB1 | Sponge network | -0.08 | 0.90092 | -0.335 | 0.06631 | 0.358 |
| 27777 | AC083843.1 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-495-3p;hsa-miR-99a-5p | 23 | CCDC6 | Sponge network | 1.4 | 0 | 0.27 | 0.02555 | 0.358 |
| 27778 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | FYN | Sponge network | -2.117 | 0.00161 | -0.131 | 0.37051 | 0.358 |
| 27779 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421 | 11 | LRRK2 | Sponge network | -0.154 | 0.78939 | -1.136 | 1.0E-5 | 0.358 |
| 27780 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-369-3p;hsa-miR-532-5p | 20 | PTPN14 | Sponge network | -0.384 | 0.40652 | 0.144 | 0.34595 | 0.358 |
| 27781 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-532-5p | 15 | PTPRB | Sponge network | -0.154 | 0.78939 | 0.105 | 0.47122 | 0.358 |
| 27782 | RP11-264B14.1 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-22-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-628-5p;hsa-miR-96-5p | 10 | FOXP1 | Sponge network | 0.557 | 0.00564 | 0.042 | 0.74368 | 0.358 |
| 27783 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-942-5p | 42 | UNC80 | Sponge network | -0.612 | 0.00094 | -1.934 | 0 | 0.358 |
| 27784 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-450b-5p | 18 | DMXL1 | Sponge network | -1.092 | 4.0E-5 | -0.426 | 0.00056 | 0.358 |
| 27785 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-874-3p;hsa-miR-93-5p | 37 | TACC1 | Sponge network | 0.253 | 0.09565 | -0.362 | 0.05406 | 0.358 |
| 27786 | MEG3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 52 | GRIK3 | Sponge network | 0.249 | 0.35902 | -3.208 | 0 | 0.358 |
| 27787 | CTC-378H22.2 |
hsa-miR-106a-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-744-3p;hsa-miR-877-5p | 16 | NTRK3 | Sponge network | 0.001 | 0.99663 | -1.77 | 3.0E-5 | 0.358 |
| 27788 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SEMA3E | Sponge network | 0.246 | 0.59912 | -1.717 | 0.00137 | 0.358 |
| 27789 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | MED12L | Sponge network | 0.194 | 0.64278 | -0.194 | 0.55299 | 0.358 |
| 27790 | RP11-206L10.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-505-5p;hsa-miR-539-5p | 14 | ARHGEF12 | Sponge network | 0.793 | 0 | 0.09 | 0.35693 | 0.358 |
| 27791 | RP11-464F9.1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p | 11 | GSK3B | Sponge network | 0.959 | 0 | 0.266 | 0.00045 | 0.358 |
| 27792 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | PRKAA2 | Sponge network | -0.13 | 0.48734 | -2.462 | 0 | 0.358 |
| 27793 | SNHG1 |
hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | SLC5A3 | Sponge network | 1.265 | 0 | 0.493 | 5.0E-5 | 0.358 |
| 27794 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 11 | POU2F2 | Sponge network | -0.896 | 0.00099 | -0.233 | 0.32214 | 0.358 |
| 27795 | CTD-2002H8.2 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | FOXP1 | Sponge network | 0.931 | 1.0E-5 | 0.042 | 0.74368 | 0.358 |
| 27796 | FAM66C |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 44 | PTPRD | Sponge network | -0.901 | 0.00019 | -1.005 | 0.00064 | 0.358 |
| 27797 | MRVI1-AS1 |
hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p | 16 | MED12L | Sponge network | -1.092 | 4.0E-5 | -0.194 | 0.55299 | 0.358 |
| 27798 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-485-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SAMHD1 | Sponge network | -0.769 | 0.00035 | 0.072 | 0.62895 | 0.358 |
| 27799 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 37 | TRPS1 | Sponge network | -0.738 | 0.21233 | -0.191 | 0.44109 | 0.358 |
| 27800 | RP5-1139B12.3 |
hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-454-3p | 15 | DIP2C | Sponge network | 0.598 | 0.38481 | -0.436 | 0.01713 | 0.358 |
| 27801 | HCG18 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 29 | SOX11 | Sponge network | 1.063 | 0 | 2.68 | 0 | 0.358 |
| 27802 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-628-5p | 29 | GRIA3 | Sponge network | 0.246 | 0.59912 | -1.956 | 0 | 0.358 |
| 27803 | LINC00969 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 19 | MOB1B | Sponge network | 0.683 | 1.0E-5 | 0.197 | 0.15266 | 0.358 |
| 27804 | RP11-389C8.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-3p | 13 | RPS6KA2 | Sponge network | 0.727 | 3.0E-5 | -0.57 | 0.00013 | 0.358 |
| 27805 | AC093627.8 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p | 14 | BNC2 | Sponge network | -0.787 | 0.3928 | -0.491 | 0.09988 | 0.358 |
| 27806 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 17 | LATS2 | Sponge network | -1.654 | 0.00144 | 0.159 | 0.2304 | 0.358 |
| 27807 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | PHC3 | Sponge network | 1.04 | 0.00023 | 0.212 | 0.0499 | 0.358 |
| 27808 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-192-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | BMPR2 | Sponge network | 1.055 | 0 | 0.206 | 0.0635 | 0.358 |
| 27809 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TIMP3 | Sponge network | -0.737 | 0.10822 | -0.546 | 0.02307 | 0.358 |
| 27810 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | 0.997 | 0.00066 | -3.143 | 0 | 0.358 |
| 27811 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | EBF3 | Sponge network | 0.083 | 0.66405 | -0.058 | 0.81437 | 0.358 |
| 27812 | HOTAIRM1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | PRUNE2 | Sponge network | -0.053 | 0.80685 | -1.733 | 0.00022 | 0.358 |
| 27813 | RP11-400K9.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-590-5p;hsa-miR-93-3p | 16 | BMPR2 | Sponge network | -0.733 | 0.19469 | 0.206 | 0.0635 | 0.358 |
| 27814 | FZD10-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 37 | TRPS1 | Sponge network | -0.727 | 0.04726 | -0.191 | 0.44109 | 0.358 |
| 27815 | TP73-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-660-5p;hsa-miR-7-1-3p | 23 | LIMCH1 | Sponge network | -0.563 | 0.02545 | -0.915 | 1.0E-5 | 0.358 |
| 27816 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-96-5p | 26 | CREB3L2 | Sponge network | -1.582 | 8.0E-5 | -0.525 | 2.0E-5 | 0.358 |
| 27817 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 53 | NTRK3 | Sponge network | 0.056 | 0.81195 | -1.77 | 3.0E-5 | 0.358 |
| 27818 | CTD-3092A11.2 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p | 11 | RASAL2 | Sponge network | 0.873 | 0 | 0.869 | 0 | 0.358 |
| 27819 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-7-1-3p | 18 | ZFHX3 | Sponge network | 0.411 | 0.1335 | 0.389 | 0.01363 | 0.358 |
| 27820 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 12 | DNAJB4 | Sponge network | 0.082 | 0.55654 | -0.7 | 1.0E-5 | 0.358 |
| 27821 | ANKRD10-IT1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-7-1-3p | 11 | ZFHX3 | Sponge network | 1.739 | 0 | 0.389 | 0.01363 | 0.358 |
| 27822 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | LRP1B | Sponge network | 0.131 | 0.8436 | -2.163 | 0 | 0.358 |
| 27823 | RP11-212P7.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-877-5p | 18 | NRK | Sponge network | 0.219 | 0.1516 | 0.656 | 0.19217 | 0.358 |
| 27824 | GHRLOS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-493-5p;hsa-miR-589-5p | 12 | GRIA3 | Sponge network | -1.942 | 0 | -1.956 | 0 | 0.358 |
| 27825 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-582-5p | 11 | TBX18 | Sponge network | -0.773 | 0.04073 | 0.843 | 0.02945 | 0.358 |
| 27826 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-93-5p | 38 | PAFAH1B1 | Sponge network | -0.793 | 7.0E-5 | -0.429 | 0 | 0.358 |
| 27827 | RP11-819C21.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITPKB | Sponge network | 0.537 | 0.00139 | -0.704 | 0.00106 | 0.358 |
| 27828 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 14 | LATS2 | Sponge network | -1.161 | 0.00591 | 0.159 | 0.2304 | 0.358 |
| 27829 | BOLA3-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-429;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-940 | 10 | RHOB | Sponge network | -0.246 | 0.35034 | -1.052 | 0 | 0.358 |
| 27830 | AC009120.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ZFX | Sponge network | 0.919 | 0 | 0.09 | 0.42563 | 0.358 |
| 27831 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-93-5p | 14 | LATS2 | Sponge network | -0.777 | 0.16667 | 0.159 | 0.2304 | 0.358 |
| 27832 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p | 26 | ZFHX4 | Sponge network | 0.174 | 0.30034 | 0.018 | 0.95574 | 0.358 |
| 27833 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-589-3p | 11 | RNASEL | Sponge network | -4.463 | 0.00025 | -0.272 | 0.01796 | 0.358 |
| 27834 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | TACC1 | Sponge network | 0.246 | 0.59912 | -0.362 | 0.05406 | 0.358 |
| 27835 | HCG11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | EYA4 | Sponge network | 0.385 | 0.10067 | 0.3 | 0.36876 | 0.358 |
| 27836 | TPTEP1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 17 | KLF10 | Sponge network | -1.216 | 0 | -0.783 | 0 | 0.358 |
| 27837 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 13 | CDK6 | Sponge network | 0.683 | 1.0E-5 | 0.991 | 0 | 0.358 |
| 27838 | PWAR6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ARNT | Sponge network | -1.575 | 6.0E-5 | -0.08 | 0.26073 | 0.358 |
| 27839 | ZNF582-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 22 | LRIG1 | Sponge network | -1.349 | 0.00017 | -0.537 | 0.00588 | 0.358 |
| 27840 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 53 | NFIB | Sponge network | -0.612 | 0.00094 | 0.023 | 0.90795 | 0.358 |
| 27841 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 17 | POU2F2 | Sponge network | -0.129 | 0.57177 | -0.233 | 0.32214 | 0.358 |
| 27842 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 28 | RNASEL | Sponge network | -0.739 | 0.0574 | -0.272 | 0.01796 | 0.358 |
| 27843 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-93-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | -0.465 | 0.04703 | 0.358 | 0.13727 | 0.358 |
| 27844 | RP11-226L15.5 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 27 | MDM4 | Sponge network | 0.8 | 0 | 0.345 | 0.00762 | 0.358 |
| 27845 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-7-1-3p | 18 | SON | Sponge network | 0.267 | 0.2937 | 0.168 | 0.04406 | 0.358 |
| 27846 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p | 14 | FGF5 | Sponge network | -1.439 | 4.0E-5 | 0.549 | 0.28659 | 0.358 |
| 27847 | CTA-14H9.5 |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-508-3p | 17 | DST | Sponge network | 0.145 | 0.53343 | -0.713 | 0.00096 | 0.358 |
| 27848 | LINC00702 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | CDH10 | Sponge network | -0.737 | 0.06182 | -2.397 | 0 | 0.358 |
| 27849 | RP11-159D12.2 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 14 | GSK3B | Sponge network | 1.254 | 0 | 0.266 | 0.00045 | 0.358 |
| 27850 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-940 | 34 | NTRK2 | Sponge network | 0.219 | 0.1516 | -0.736 | 0.06495 | 0.358 |
| 27851 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-27a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | NEDD4 | Sponge network | -0.021 | 0.93598 | 0.02 | 0.89834 | 0.358 |
| 27852 | CTD-3064H18.1 |
hsa-miR-106b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 11 | TCF4 | Sponge network | -0.983 | 0.01395 | 0.016 | 0.92271 | 0.357 |
| 27853 | HOTAIRM1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-590-3p | 13 | MRVI1 | Sponge network | -0.053 | 0.80685 | -1.051 | 0.00022 | 0.357 |
| 27854 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-335-5p;hsa-miR-425-5p;hsa-miR-484 | 15 | CBL | Sponge network | 0.592 | 0.00014 | 0.291 | 0.01267 | 0.357 |
| 27855 | CTD-3138B18.6 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | BMPR2 | Sponge network | 2.248 | 0 | 0.206 | 0.0635 | 0.357 |
| 27856 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-577;hsa-miR-7-1-3p | 13 | ADAMTSL3 | Sponge network | 0.419 | 0.0319 | -1.559 | 0 | 0.357 |
| 27857 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | PRRX1 | Sponge network | -0.739 | 0.00044 | 1.316 | 0 | 0.357 |
| 27858 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 29 | GRIA3 | Sponge network | -1.281 | 0.00012 | -1.956 | 0 | 0.357 |
| 27859 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p | 19 | MITF | Sponge network | -0.777 | 0.16667 | -0.769 | 0.00022 | 0.357 |
| 27860 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-337-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | HOOK3 | Sponge network | 1.11 | 0 | 0.273 | 0.09957 | 0.357 |
| 27861 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p | 18 | LRP1B | Sponge network | 0.331 | 0.57247 | -2.163 | 0 | 0.357 |
| 27862 | LINC00702 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | FYN | Sponge network | -0.737 | 0.06182 | -0.131 | 0.37051 | 0.357 |
| 27863 | LINC00863 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 16 | UBE4B | Sponge network | 0.189 | 0.13981 | -0.235 | 0.007 | 0.357 |
| 27864 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-629-3p | 20 | PTPN14 | Sponge network | -0.246 | 0.35034 | 0.144 | 0.34595 | 0.357 |
| 27865 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-589-3p | 16 | PDGFRA | Sponge network | -0.164 | 0.45106 | -0.372 | 0.0037 | 0.357 |
| 27866 | CTC-378H22.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-744-3p | 16 | IGFBP5 | Sponge network | 0.001 | 0.99663 | -0.806 | 0.00105 | 0.357 |
| 27867 | AC009948.5 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | DNMT3A | Sponge network | 0.532 | 0.00505 | 0.302 | 0.0262 | 0.357 |
| 27868 | AC093110.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-576-5p | 11 | SIRT1 | Sponge network | 1.044 | 2.0E-5 | 0.109 | 0.2593 | 0.357 |
| 27869 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | ST6GALNAC3 | Sponge network | -2.925 | 0 | -0.987 | 0 | 0.357 |
| 27870 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 18 | LRRC55 | Sponge network | -0.246 | 0.35034 | -0.026 | 0.93163 | 0.357 |
| 27871 | RP11-822E23.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-93-5p | 23 | PRRX1 | Sponge network | -0.777 | 0.16667 | 1.316 | 0 | 0.357 |
| 27872 | BVES-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-378c;hsa-miR-429;hsa-miR-576-5p;hsa-miR-628-5p | 26 | ERC1 | Sponge network | -2.62 | 0 | -0.108 | 0.38794 | 0.357 |
| 27873 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 16 | ZBTB4 | Sponge network | 0.497 | 0.01451 | -0.739 | 0 | 0.357 |
| 27874 | TP73-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-96-5p | 17 | PRKAR1A | Sponge network | -0.563 | 0.02545 | -0.063 | 0.50314 | 0.357 |
| 27875 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | EPAS1 | Sponge network | -0.513 | 0.04703 | -0.184 | 0.14418 | 0.357 |
| 27876 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 45 | NOTCH2 | Sponge network | -1.216 | 0 | 0.138 | 0.35163 | 0.357 |
| 27877 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-590-5p | 14 | ABCC9 | Sponge network | -1.645 | 0 | -0.466 | 0.14121 | 0.357 |
| 27878 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-486-5p | 19 | PIK3R1 | Sponge network | -0.309 | 0.1145 | -0.133 | 0.36854 | 0.357 |
| 27879 | SOX2-OT |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-214-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | LRP1B | Sponge network | -1.264 | 6.0E-5 | -2.163 | 0 | 0.357 |
| 27880 | AC016747.3 |
hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-5p | 18 | PRKCB | Sponge network | 0.199 | 0.38015 | -1.313 | 2.0E-5 | 0.357 |
| 27881 | RP11-67L2.2 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | TMEM30A | Sponge network | 0.72 | 0.0001 | 0.096 | 0.31031 | 0.357 |
| 27882 | RP11-159D12.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-590-3p | 14 | ZMYND11 | Sponge network | 1.254 | 0 | -0.2 | 0.05449 | 0.357 |
| 27883 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p | 11 | AKAP6 | Sponge network | 1.46 | 1.0E-5 | -1.586 | 3.0E-5 | 0.357 |
| 27884 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | 0.41 | 0.02214 | -0.466 | 0.14121 | 0.357 |
| 27885 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 57 | WDFY3 | Sponge network | 0.374 | 0.20054 | -0.201 | 0.0967 | 0.357 |
| 27886 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 48 | SOX5 | Sponge network | 0.997 | 0.00066 | -1.091 | 4.0E-5 | 0.357 |
| 27887 | RP11-48B3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 12 | AKAP6 | Sponge network | 1.757 | 0 | -1.586 | 3.0E-5 | 0.357 |
| 27888 | GNG12-AS1 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-590-3p | 11 | ACTB | Sponge network | -0.793 | 7.0E-5 | -0.247 | 0.01736 | 0.357 |
| 27889 | RP11-693J15.4 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-362-5p | 13 | MRVI1 | Sponge network | -0.72 | 0.24239 | -1.051 | 0.00022 | 0.357 |
| 27890 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | PDE4DIP | Sponge network | -0.739 | 0.00044 | -0.282 | 0.0257 | 0.357 |
| 27891 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 18 | ABI2 | Sponge network | -0.726 | 0.00132 | 0.175 | 0.14787 | 0.357 |
| 27892 | MIR600HG |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-550a-5p;hsa-miR-942-5p | 24 | ARHGEF12 | Sponge network | 0.594 | 0.41522 | 0.09 | 0.35693 | 0.357 |
| 27893 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 18 | RSPO2 | Sponge network | -0.517 | 0.02935 | -3.038 | 0 | 0.357 |
| 27894 | SNX29P2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-92a-3p | 11 | MACF1 | Sponge network | 0.4 | 0.14611 | 0.019 | 0.90335 | 0.357 |
| 27895 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-361-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 20 | KAT6B | Sponge network | -0.976 | 0.00025 | -0.478 | 0.00018 | 0.357 |
| 27896 | RP11-65J21.3 |
hsa-let-7d-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p | 12 | SNX29 | Sponge network | -0.557 | 0.11135 | -0.34 | 0.01371 | 0.357 |
| 27897 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | ZMYM2 | Sponge network | 1.597 | 0 | 0.279 | 0.01273 | 0.357 |
| 27898 | RP1-151F17.2 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-877-5p | 16 | GLIPR1 | Sponge network | 0.222 | 0.29133 | 0.146 | 0.3276 | 0.357 |
| 27899 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p | 32 | ERBB4 | Sponge network | -0.358 | 0.17255 | -1.975 | 2.0E-5 | 0.357 |
| 27900 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | SCN9A | Sponge network | -0.141 | 0.26434 | -0.525 | 0.10593 | 0.357 |
| 27901 | CTD-2089N3.2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | RYR2 | Sponge network | -1.375 | 0.08642 | -1.466 | 0.00031 | 0.357 |
| 27902 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p | 21 | PRDM2 | Sponge network | -0.384 | 0.40652 | -0.258 | 0.00721 | 0.357 |
| 27903 | OSER1-AS1 |
hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p | 27 | CHD5 | Sponge network | -0.13 | 0.48734 | -0.922 | 0.01521 | 0.357 |
| 27904 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p | 22 | PRDM2 | Sponge network | 0.494 | 0.00339 | -0.258 | 0.00721 | 0.357 |
| 27905 | FAM215B |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-664a-5p | 10 | RAPGEF5 | Sponge network | 0.817 | 3.0E-5 | 0.536 | 4.0E-5 | 0.357 |
| 27906 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-766-3p | 18 | TBL1XR1 | Sponge network | 1.093 | 0 | 0.578 | 0 | 0.357 |
| 27907 | GS1-124K5.11 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p | 48 | DTNA | Sponge network | 0.494 | 0.00339 | -1.648 | 1.0E-5 | 0.357 |
| 27908 | RP11-701H24.3 |
hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-590-3p | 12 | ZFHX4 | Sponge network | -1.425 | 0.04204 | 0.018 | 0.95574 | 0.357 |
| 27909 | LINC00865 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-500a-5p | 12 | MACF1 | Sponge network | -1.249 | 7.0E-5 | 0.019 | 0.90335 | 0.357 |
| 27910 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-455-5p | 11 | THSD7A | Sponge network | 0.083 | 0.66405 | 0.242 | 0.25154 | 0.357 |
| 27911 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 45 | CADM2 | Sponge network | -0.246 | 0.35034 | -3.782 | 0 | 0.357 |
| 27912 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-664a-3p;hsa-miR-942-5p | 19 | ERRFI1 | Sponge network | -0.976 | 0.00025 | -0.798 | 1.0E-5 | 0.357 |
| 27913 | RP11-678G14.2 |
hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p | 11 | TCF4 | Sponge network | -1.765 | 0.0778 | 0.016 | 0.92271 | 0.357 |
| 27914 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-3p | 21 | EBF3 | Sponge network | 0.056 | 0.81195 | -0.058 | 0.81437 | 0.357 |
| 27915 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | 0.61 | 0.01593 | -1.603 | 1.0E-5 | 0.357 |
| 27916 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-708-5p | 28 | RAP1A | Sponge network | -1.645 | 0 | -0.929 | 0 | 0.357 |
| 27917 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | CUL5 | Sponge network | 0.67 | 0.00048 | 0.026 | 0.77301 | 0.357 |
| 27918 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-503-5p;hsa-miR-93-5p | 34 | CCND2 | Sponge network | 0.082 | 0.55397 | -0.267 | 0.3112 | 0.357 |
| 27919 | RP11-121C2.2 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p | 18 | DDX3X | Sponge network | 1.147 | 0 | -0.152 | 0.07686 | 0.357 |
| 27920 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-942-5p | 13 | THRB | Sponge network | -0.326 | 0.28809 | -1.117 | 0.0004 | 0.357 |
| 27921 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-93-5p | 11 | RBL2 | Sponge network | -0.465 | 0.04703 | -0.282 | 0.00254 | 0.357 |
| 27922 | RP11-216L13.19 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-664a-5p | 11 | CBFB | Sponge network | 1.674 | 0 | 1.061 | 0 | 0.357 |
| 27923 | PART1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 31 | RSPO2 | Sponge network | -2.925 | 0 | -3.038 | 0 | 0.357 |
| 27924 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-424-5p | 15 | EPHA7 | Sponge network | -3.586 | 0.00032 | -2.197 | 3.0E-5 | 0.357 |
| 27925 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | -0.235 | 0.15142 | 0.323 | 0.00792 | 0.357 |
| 27926 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-539-5p | 12 | BCL11A | Sponge network | -0.465 | 0.04703 | -0.932 | 0.0063 | 0.357 |
| 27927 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | IGSF10 | Sponge network | -0.969 | 2.0E-5 | -1.751 | 1.0E-5 | 0.357 |
| 27928 | LINC00865 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-940 | 12 | FAM172A | Sponge network | -1.249 | 7.0E-5 | -0.57 | 0 | 0.357 |
| 27929 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | BCL11A | Sponge network | -0.709 | 0.18168 | -0.932 | 0.0063 | 0.357 |
| 27930 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 11 | LRRC7 | Sponge network | -1.886 | 0 | -1.341 | 0.01193 | 0.357 |
| 27931 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | DACH2 | Sponge network | -2.117 | 0.00161 | -1.635 | 0.00034 | 0.357 |
| 27932 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 27 | EBF3 | Sponge network | 0.064 | 0.72934 | -0.058 | 0.81437 | 0.357 |
| 27933 | LINC00621 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | CNTN1 | Sponge network | 0.131 | 0.8436 | -2.594 | 0 | 0.357 |
| 27934 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 25 | GPC6 | Sponge network | -0.323 | 0.01372 | -0.054 | 0.81465 | 0.357 |
| 27935 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.194 | 0.64278 | 0.943 | 0 | 0.357 |
| 27936 | LINC01001 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p | 10 | PHF6 | Sponge network | 1.055 | 0 | 0.785 | 0 | 0.357 |
| 27937 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | ERRFI1 | Sponge network | 0.335 | 0.20596 | -0.798 | 1.0E-5 | 0.357 |
| 27938 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | JAZF1 | Sponge network | -0.053 | 0.80685 | -0.377 | 0.02677 | 0.357 |
| 27939 | RP11-158K1.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-766-3p | 32 | IL17RD | Sponge network | 0.349 | 0.01695 | -0.019 | 0.94873 | 0.357 |
| 27940 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | MITF | Sponge network | 0.274 | 0.07681 | -0.769 | 0.00022 | 0.357 |
| 27941 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | SLITRK3 | Sponge network | -0.355 | 0.0077 | -3.328 | 0 | 0.357 |
| 27942 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | LIFR | Sponge network | -1.161 | 0.00591 | -1.794 | 0 | 0.357 |
| 27943 | AC016747.3 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | CADM1 | Sponge network | 0.199 | 0.38015 | -0.9 | 0.00084 | 0.357 |
| 27944 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | EPAS1 | Sponge network | 0.374 | 0.20054 | -0.184 | 0.14418 | 0.357 |
| 27945 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | BCL2 | Sponge network | 0.083 | 0.66405 | -0.899 | 0 | 0.357 |
| 27946 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 34 | PPP1R9A | Sponge network | 0.225 | 0.30644 | -0.903 | 0.04254 | 0.357 |
| 27947 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | AHNAK | Sponge network | -0.709 | 0.18168 | -1.207 | 0 | 0.357 |
| 27948 | AC009948.5 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-342-3p | 18 | ABL1 | Sponge network | 0.532 | 0.00505 | -0.215 | 0.07804 | 0.357 |
| 27949 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 16 | SLITRK3 | Sponge network | 0.214 | 0.18246 | -3.328 | 0 | 0.357 |
| 27950 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940 | 17 | CRTC3 | Sponge network | 0.862 | 0.06213 | 0.164 | 0.16786 | 0.357 |
| 27951 | RP11-216L13.19 |
hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 11 | TBL1XR1 | Sponge network | 1.674 | 0 | 0.578 | 0 | 0.357 |
| 27952 | BDNF-AS |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | DIP2C | Sponge network | -0.668 | 3.0E-5 | -0.436 | 0.01713 | 0.357 |
| 27953 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 26 | BMPR2 | Sponge network | 0.37 | 0.27614 | 0.206 | 0.0635 | 0.357 |
| 27954 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-5p | 11 | THSD7A | Sponge network | 0.064 | 0.72934 | 0.242 | 0.25154 | 0.357 |
| 27955 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 21 | EPHA5 | Sponge network | 1.46 | 1.0E-5 | -0.957 | 0.01733 | 0.357 |
| 27956 | LINC00672 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 15 | SPG20 | Sponge network | -0.227 | 0.31657 | -1.16 | 0 | 0.357 |
| 27957 | RP11-597D13.9 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p | 14 | EZH1 | Sponge network | 1.302 | 7.0E-5 | -0.564 | 1.0E-5 | 0.357 |
| 27958 | RP11-439A17.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p | 21 | LPP | Sponge network | 0.051 | 0.95756 | -0.459 | 0.04475 | 0.357 |
| 27959 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p | 38 | CAMTA1 | Sponge network | -0.793 | 7.0E-5 | -0.436 | 1.0E-5 | 0.357 |
| 27960 | AC034220.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PIK3CA | Sponge network | 0.309 | 0.07304 | 0.195 | 0.06908 | 0.357 |
| 27961 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | DOCK4 | Sponge network | 0.374 | 0.20054 | 0.502 | 0.00108 | 0.357 |
| 27962 | AC003090.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-96-5p | 12 | ELMO1 | Sponge network | -2.117 | 0.00161 | 0.04 | 0.83147 | 0.357 |
| 27963 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-590-5p | 10 | ABCA10 | Sponge network | 0.176 | 0.37402 | -0.571 | 0.03674 | 0.357 |
| 27964 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-96-5p | 22 | FOXO3 | Sponge network | 0.853 | 0.15352 | -0.04 | 0.72028 | 0.357 |
| 27965 | RP11-532F6.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-532-5p;hsa-miR-577 | 27 | IL17RD | Sponge network | -0.896 | 0.00099 | -0.019 | 0.94873 | 0.357 |
| 27966 | CTC-378H22.2 |
hsa-miR-106a-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-338-3p;hsa-miR-532-3p;hsa-miR-877-5p | 19 | GAS7 | Sponge network | 0.001 | 0.99663 | -0.555 | 0.01602 | 0.357 |
| 27967 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 13 | TLL1 | Sponge network | -0.405 | 0.22013 | -1.195 | 3.0E-5 | 0.357 |
| 27968 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-93-5p;hsa-miR-935 | 20 | ARNT2 | Sponge network | -0.976 | 0.00025 | -0.332 | 0.25851 | 0.357 |
| 27969 | RP1-59D14.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 13 | CLASP2 | Sponge network | 1.162 | 5.0E-5 | -0.038 | 0.71 | 0.357 |
| 27970 | DIO3OS |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-877-5p | 26 | ASTN1 | Sponge network | -0.773 | 0.04073 | -2.389 | 2.0E-5 | 0.357 |
| 27971 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | PRKCE | Sponge network | -0.615 | 0.00015 | -0.403 | 0.00056 | 0.357 |
| 27972 | LINC00702 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 59 | CUX1 | Sponge network | -0.737 | 0.06182 | -0.039 | 0.6705 | 0.357 |
| 27973 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 18 | TGFBR2 | Sponge network | 0.4 | 0.14611 | -0.243 | 0.11408 | 0.357 |
| 27974 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | RNF144A | Sponge network | -0.739 | 0.00044 | 0.594 | 0.0009 | 0.357 |
| 27975 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 12 | ERCC4 | Sponge network | 0.598 | 0.38481 | 0.126 | 0.15266 | 0.357 |
| 27976 | TP73-AS1 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | ADCY1 | Sponge network | -0.563 | 0.02545 | 0.362 | 0.16982 | 0.357 |
| 27977 | AC004893.11 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 1.317 | 0 | 0.578 | 0 | 0.357 |
| 27978 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429 | 22 | ARHGAP28 | Sponge network | -0.563 | 0.02545 | -0.911 | 0.00013 | 0.357 |
| 27979 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 18 | ETV1 | Sponge network | -1.092 | 4.0E-5 | -0.096 | 0.67674 | 0.357 |
| 27980 | LINC00094 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-582-5p;hsa-miR-655-3p | 34 | DMD | Sponge network | 0.253 | 0.09565 | -1.506 | 0 | 0.357 |
| 27981 | RP11-822E23.8 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p | 15 | ITPKB | Sponge network | -0.777 | 0.16667 | -0.704 | 0.00106 | 0.357 |
| 27982 | HCG11 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | FMNL3 | Sponge network | 0.385 | 0.10067 | 0.555 | 4.0E-5 | 0.357 |
| 27983 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-93-3p | 12 | NCOA1 | Sponge network | -0.295 | 0.02604 | -0.432 | 0 | 0.357 |
| 27984 | CTC-429P9.2 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-940 | 12 | NTRK2 | Sponge network | 0.043 | 0.81628 | -0.736 | 0.06495 | 0.357 |
| 27985 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-361-3p | 11 | EBF1 | Sponge network | 0.598 | 0.38481 | -0.384 | 0.08856 | 0.357 |
| 27986 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | ACVR2A | Sponge network | -0.323 | 0.01372 | -0.409 | 0.00039 | 0.357 |
| 27987 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p | 26 | PPP1R9A | Sponge network | -2.62 | 0 | -0.903 | 0.04254 | 0.357 |
| 27988 | TPTEP1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 14 | SEPT6 | Sponge network | -1.216 | 0 | -0.598 | 0.00181 | 0.357 |
| 27989 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-93-5p | 15 | EDA2R | Sponge network | -2.69 | 0.0001 | -0.587 | 0.01598 | 0.357 |
| 27990 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421 | 10 | FAT2 | Sponge network | -0.487 | 0.02157 | -0.278 | 0.44877 | 0.357 |
| 27991 | AC004893.11 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 12 | RASAL2 | Sponge network | 1.317 | 0 | 0.869 | 0 | 0.357 |
| 27992 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FAM188A | Sponge network | -0.222 | 0.32445 | -0.44 | 0.00018 | 0.357 |
| 27993 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-624-5p | 14 | LATS1 | Sponge network | 0.539 | 0.03722 | 0.109 | 0.34208 | 0.357 |
| 27994 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-877-5p;hsa-miR-93-5p | 43 | AR | Sponge network | -0.465 | 0.04703 | -1.554 | 0 | 0.357 |
| 27995 | AC005154.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 10 | SKIL | Sponge network | 0.848 | 0 | 0.559 | 8.0E-5 | 0.357 |
| 27996 | RP11-216B9.6 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | MAPK10 | Sponge network | 0.174 | 0.30034 | -1.774 | 0 | 0.357 |
| 27997 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 25 | IGF1 | Sponge network | -0.227 | 0.31657 | -0.204 | 0.5898 | 0.357 |
| 27998 | PTOV1-AS1 |
hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p | 10 | SLC5A3 | Sponge network | 0.87 | 0 | 0.493 | 5.0E-5 | 0.357 |
| 27999 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-940 | 32 | PTPRT | Sponge network | -0.896 | 0.00099 | -0.907 | 0.03727 | 0.357 |
| 28000 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-582-5p | 14 | AFF3 | Sponge network | 0.889 | 9.0E-5 | -2.373 | 0 | 0.357 |
| 28001 | LINC00476 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 39 | ABI2 | Sponge network | -0.615 | 0.00015 | 0.175 | 0.14787 | 0.357 |
| 28002 | FENDRR |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 32 | RIMS2 | Sponge network | -1.281 | 0.00012 | -1.365 | 0.00208 | 0.357 |
| 28003 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 16 | HOOK3 | Sponge network | -2.117 | 0.00161 | 0.273 | 0.09957 | 0.357 |
| 28004 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 62 | IL6ST | Sponge network | -0.899 | 6.0E-5 | -0.402 | 0.00676 | 0.357 |
| 28005 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | EBF3 | Sponge network | 0.385 | 0.10067 | -0.058 | 0.81437 | 0.357 |
| 28006 | HCG11 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 11 | STARD13 | Sponge network | 0.385 | 0.10067 | -0.187 | 0.27383 | 0.357 |
| 28007 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577 | 20 | KAT6B | Sponge network | -0.18 | 0.20343 | -0.478 | 0.00018 | 0.357 |
| 28008 | RP11-531A24.5 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 22 | DICER1 | Sponge network | 0.75 | 0.00054 | 0.042 | 0.674 | 0.357 |
| 28009 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | TAL1 | Sponge network | 0.299 | 0.57722 | -0.462 | 0.00574 | 0.357 |
| 28010 | LINC00173 |
hsa-let-7d-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-421 | 15 | PRDM2 | Sponge network | 0.056 | 0.8494 | -0.258 | 0.00721 | 0.357 |
| 28011 | RP11-244H3.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 21 | EPS15 | Sponge network | 0.659 | 3.0E-5 | -0.052 | 0.53998 | 0.357 |
| 28012 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | AFAP1L2 | Sponge network | -0.129 | 0.57177 | -0.284 | 0.15434 | 0.357 |
| 28013 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-625-5p;hsa-miR-628-5p | 31 | LPP | Sponge network | 1.162 | 5.0E-5 | -0.459 | 0.04475 | 0.357 |
| 28014 | MYLK-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p | 12 | HIP1 | Sponge network | -1.331 | 3.0E-5 | 0.774 | 0 | 0.357 |
| 28015 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-493-5p | 14 | MLLT10 | Sponge network | 1.044 | 2.0E-5 | 0.105 | 0.33292 | 0.357 |
| 28016 | RP11-307B6.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-5p | 10 | PDE4DIP | Sponge network | -4.463 | 0.00025 | -0.282 | 0.0257 | 0.357 |
| 28017 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 21 | ARNT2 | Sponge network | -0.738 | 0.21233 | -0.332 | 0.25851 | 0.357 |
| 28018 | LINC00702 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 17 | TES | Sponge network | -0.737 | 0.06182 | -0.348 | 0.00639 | 0.357 |
| 28019 | EMX2OS |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 38 | ERC1 | Sponge network | -0.777 | 0.1134 | -0.108 | 0.38794 | 0.357 |
| 28020 | RP11-345P4.7 |
hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 10 | BRD3 | Sponge network | 1.597 | 0 | 0.37 | 0.00013 | 0.357 |
| 28021 | RP11-999E24.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p | 14 | CBFA2T3 | Sponge network | -0.693 | 0.05629 | -1.221 | 1.0E-5 | 0.357 |
| 28022 | BZRAP1-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-577 | 10 | ZNF844 | Sponge network | -0.465 | 0.04703 | -0.966 | 0.00035 | 0.357 |
| 28023 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | RICTOR | Sponge network | 0.79 | 0 | 0.388 | 0.00188 | 0.357 |
| 28024 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-935 | 15 | RELN | Sponge network | -0.738 | 0.21233 | -2.403 | 0 | 0.357 |
| 28025 | AC003090.1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | OPCML | Sponge network | -2.117 | 0.00161 | -2.498 | 0 | 0.357 |
| 28026 | LINC00672 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-629-3p | 21 | PTPN14 | Sponge network | -0.227 | 0.31657 | 0.144 | 0.34595 | 0.357 |
| 28027 | MYLK-AS1 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p | 12 | UBE4B | Sponge network | -1.331 | 3.0E-5 | -0.235 | 0.007 | 0.357 |
| 28028 | TPT1-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-5p | 19 | CREB1 | Sponge network | 0.903 | 0 | 0.203 | 0.00537 | 0.357 |
| 28029 | CTC-487M23.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-92a-3p | 14 | PRKAB2 | Sponge network | 0.912 | 0.00014 | -0.367 | 0.01371 | 0.357 |
| 28030 | AC108488.4 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | AR | Sponge network | 0.259 | 0.12542 | -1.554 | 0 | 0.357 |
| 28031 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-421 | 13 | PGR | Sponge network | -0.285 | 0.2172 | -1.704 | 0 | 0.357 |
| 28032 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-93-3p | 16 | PTPN11 | Sponge network | 0.253 | 0.09565 | 0.431 | 2.0E-5 | 0.357 |
| 28033 | RP11-999E24.3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-340-5p | 12 | KLF10 | Sponge network | -0.693 | 0.05629 | -0.783 | 0 | 0.357 |
| 28034 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | TIMP3 | Sponge network | 0.619 | 0.00157 | -0.546 | 0.02307 | 0.357 |
| 28035 | RP11-359B12.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 15 | SHPRH | Sponge network | 0.064 | 0.72934 | 0.098 | 0.40994 | 0.357 |
| 28036 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | MAF | Sponge network | 0.572 | 0.01722 | -1.257 | 0 | 0.357 |
| 28037 | PCA3 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 10 | FOXO4 | Sponge network | -0.709 | 0.18168 | -0.516 | 2.0E-5 | 0.357 |
| 28038 | HCG11 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 21 | SYNPO2 | Sponge network | 0.385 | 0.10067 | -2.342 | 0 | 0.357 |
| 28039 | CTD-2554C21.2 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429 | 20 | LIMCH1 | Sponge network | -1.654 | 0.00144 | -0.915 | 1.0E-5 | 0.357 |
| 28040 | BVES-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-629-3p | 10 | POU2F2 | Sponge network | -2.62 | 0 | -0.233 | 0.32214 | 0.357 |
| 28041 | AC003090.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | GALNT13 | Sponge network | -2.117 | 0.00161 | -0.857 | 0.07439 | 0.357 |
| 28042 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-590-5p | 13 | SYNE1 | Sponge network | 2.306 | 9.0E-5 | -0.738 | 0.00316 | 0.357 |
| 28043 | AP001347.6 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-590-3p | 19 | RYR2 | Sponge network | -1.161 | 0.00591 | -1.466 | 0.00031 | 0.357 |
| 28044 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p | 29 | HBP1 | Sponge network | -0.901 | 0.00019 | -0.454 | 0 | 0.357 |
| 28045 | RP11-389C8.2 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | ATM | Sponge network | 0.727 | 3.0E-5 | 0.178 | 0.21568 | 0.357 |
| 28046 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p | 20 | NCAM2 | Sponge network | -0.793 | 7.0E-5 | -0.482 | 0.22565 | 0.357 |
| 28047 | RP5-1139B12.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 13 | ESR1 | Sponge network | 0.598 | 0.38481 | -1.035 | 3.0E-5 | 0.357 |
| 28048 | RP11-464F9.1 |
hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | DOCK4 | Sponge network | 0.959 | 0 | 0.502 | 0.00108 | 0.357 |
| 28049 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | NR4A3 | Sponge network | 0.572 | 0.01722 | -1.385 | 0 | 0.357 |
| 28050 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 38 | LIFR | Sponge network | -2.46 | 0 | -1.794 | 0 | 0.357 |
| 28051 | LPP-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-455-5p | 10 | MN1 | Sponge network | -0.18 | 0.20343 | -0.358 | 0.24172 | 0.356 |
| 28052 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | HECA | Sponge network | -0.129 | 0.57177 | -0.217 | 0.03463 | 0.356 |
| 28053 | H1FX-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PCDH9 | Sponge network | -0.206 | 0.12942 | -1.823 | 4.0E-5 | 0.356 |
| 28054 | HAND2-AS1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | NR3C2 | Sponge network | -1.697 | 0.00889 | -1.56 | 0 | 0.356 |
| 28055 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-452-5p;hsa-miR-590-3p | 20 | PHC3 | Sponge network | -0.358 | 0.17255 | 0.212 | 0.0499 | 0.356 |
| 28056 | RAD51-AS1 |
hsa-miR-1266-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 17 | ABL2 | Sponge network | 0.768 | 7.0E-5 | 0.82 | 0 | 0.356 |
| 28057 | RP11-728F11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-7-1-3p | 21 | PRKAA2 | Sponge network | 0.238 | 0.70544 | -2.462 | 0 | 0.356 |
| 28058 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 22 | MOB1B | Sponge network | 0.919 | 0 | 0.197 | 0.15266 | 0.356 |
| 28059 | RP11-473I1.10 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | FOXO3 | Sponge network | 0.673 | 0 | -0.04 | 0.72028 | 0.356 |
| 28060 | HCG11 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | PRUNE2 | Sponge network | 0.385 | 0.10067 | -1.733 | 0.00022 | 0.356 |
| 28061 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | TCF12 | Sponge network | 0.064 | 0.72934 | 0.174 | 0.13271 | 0.356 |
| 28062 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 44 | TMEM132B | Sponge network | -0.405 | 0.22013 | -0.83 | 0.01084 | 0.356 |
| 28063 | CALML3-AS1 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-361-3p;hsa-miR-625-5p | 10 | BCORL1 | Sponge network | 0.95 | 0.15943 | 0.354 | 0.01214 | 0.356 |
| 28064 | RP11-705C15.3 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 13 | NAV3 | Sponge network | 0.796 | 4.0E-5 | 0.212 | 0.47729 | 0.356 |
| 28065 | AC109309.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b | 13 | EPHA7 | Sponge network | 0.163 | 0.87219 | -2.197 | 3.0E-5 | 0.356 |
| 28066 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p | 12 | ESR1 | Sponge network | -0.932 | 0.00329 | -1.035 | 3.0E-5 | 0.356 |
| 28067 | TTC28-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p | 17 | BRD3 | Sponge network | 0.373 | 0.00068 | 0.37 | 0.00013 | 0.356 |
| 28068 | LINC00957 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 13 | RCAN2 | Sponge network | -0.421 | 0.0114 | -0.33 | 0.15172 | 0.356 |
| 28069 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 58 | CADM2 | Sponge network | 0.101 | 0.60919 | -3.782 | 0 | 0.356 |
| 28070 | RP11-395G23.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 15 | NAV3 | Sponge network | 0.381 | 0.25967 | 0.212 | 0.47729 | 0.356 |
| 28071 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 13 | PTPRD | Sponge network | -0.011 | 0.967 | -1.005 | 0.00064 | 0.356 |
| 28072 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429 | 11 | UBE4B | Sponge network | -1.654 | 0.00144 | -0.235 | 0.007 | 0.356 |
| 28073 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 46 | QKI | Sponge network | 0.222 | 0.29133 | -0.333 | 0.04111 | 0.356 |
| 28074 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 15 | SEMA3C | Sponge network | -0.096 | 0.67958 | -0.272 | 0.31153 | 0.356 |
| 28075 | CTD-2245F17.3 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-335-5p;hsa-miR-505-3p;hsa-miR-629-3p;hsa-miR-766-3p | 17 | IL6ST | Sponge network | -0.421 | 0.12556 | -0.402 | 0.00676 | 0.356 |
| 28076 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-940 | 29 | NTRK2 | Sponge network | 0.082 | 0.55397 | -0.736 | 0.06495 | 0.356 |
| 28077 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 49 | UNC80 | Sponge network | -1.216 | 0 | -1.934 | 0 | 0.356 |
| 28078 | GNG12-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 17 | CELF4 | Sponge network | -0.793 | 7.0E-5 | -1.135 | 0.00344 | 0.356 |
| 28079 | LINC00662 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484 | 10 | WNK2 | Sponge network | 0.213 | 0.32297 | -0.352 | 0.31066 | 0.356 |
| 28080 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 20 | ATRX | Sponge network | 0.274 | 0.07681 | 0.261 | 0.04408 | 0.356 |
| 28081 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | ZFX | Sponge network | -0.323 | 0.01372 | 0.09 | 0.42563 | 0.356 |
| 28082 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940 | 37 | HLF | Sponge network | 0.532 | 0.00505 | -2.443 | 0 | 0.356 |
| 28083 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-942-5p | 44 | UNC80 | Sponge network | -0.969 | 2.0E-5 | -1.934 | 0 | 0.356 |
| 28084 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 13 | BCL11A | Sponge network | 0.056 | 0.8494 | -0.932 | 0.0063 | 0.356 |
| 28085 | RASSF8-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | SYT4 | Sponge network | 0.335 | 0.20596 | -3.207 | 0 | 0.356 |
| 28086 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | TET1 | Sponge network | 0.559 | 0.00026 | -0.135 | 0.52138 | 0.356 |
| 28087 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | TBX18 | Sponge network | -0.738 | 0.21233 | 0.843 | 0.02945 | 0.356 |
| 28088 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-93-5p | 10 | EDA2R | Sponge network | -0.318 | 0.65847 | -0.587 | 0.01598 | 0.356 |
| 28089 | RP11-401P9.4 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p | 10 | RADIL | Sponge network | -0.384 | 0.40652 | -1.628 | 0 | 0.356 |
| 28090 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p | 35 | CHD5 | Sponge network | -1.203 | 0.03881 | -0.922 | 0.01521 | 0.356 |
| 28091 | U91328.19 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-877-5p | 23 | NRK | Sponge network | -0.303 | 0.21002 | 0.656 | 0.19217 | 0.356 |
| 28092 | HCG11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | ZNF208 | Sponge network | 0.385 | 0.10067 | -0.4 | 0.22381 | 0.356 |
| 28093 | RP11-359B12.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 10 | SPTBN4 | Sponge network | 0.064 | 0.72934 | -0.786 | 0.01281 | 0.356 |
| 28094 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | TAL1 | Sponge network | 0.101 | 0.60919 | -0.462 | 0.00574 | 0.356 |
| 28095 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 28 | SSBP2 | Sponge network | 0.381 | 0.25967 | -1.58 | 0 | 0.356 |
| 28096 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 17 | ZFP36L1 | Sponge network | -0.901 | 0.00019 | -0.505 | 8.0E-5 | 0.356 |
| 28097 | RP11-2H8.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 20 | TBL1XR1 | Sponge network | 1.089 | 0 | 0.578 | 0 | 0.356 |
| 28098 | RP11-244O19.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-671-5p | 22 | CYLD | Sponge network | -0.096 | 0.67958 | -0.367 | 0.00171 | 0.356 |
| 28099 | LIPE-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-34c-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 18 | UBE4B | Sponge network | 0.078 | 0.55803 | -0.235 | 0.007 | 0.356 |
| 28100 | RP11-46C24.7 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-935 | 65 | LPP | Sponge network | 0.392 | 0.0278 | -0.459 | 0.04475 | 0.356 |
| 28101 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940 | 24 | LIFR | Sponge network | 0.225 | 0.30644 | -1.794 | 0 | 0.356 |
| 28102 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-24-2-5p;hsa-miR-331-5p | 12 | NTRK3 | Sponge network | -0.326 | 0.28809 | -1.77 | 3.0E-5 | 0.356 |
| 28103 | RP13-1039J1.4 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-582-5p;hsa-miR-589-3p | 15 | THRB | Sponge network | 0.356 | 0.33467 | -1.117 | 0.0004 | 0.356 |
| 28104 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-532-3p | 11 | CADM3 | Sponge network | -0.246 | 0.35034 | -3.663 | 0 | 0.356 |
| 28105 | CTD-2314G24.2 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PTCH1 | Sponge network | 0.246 | 0.59912 | -0.1 | 0.6179 | 0.356 |
| 28106 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | PRKAA2 | Sponge network | 0.572 | 0.01722 | -2.462 | 0 | 0.356 |
| 28107 | CTB-31O20.4 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p | 17 | CREB1 | Sponge network | 1.619 | 0 | 0.203 | 0.00537 | 0.356 |
| 28108 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 26 | RSPO2 | Sponge network | -0.487 | 0.02157 | -3.038 | 0 | 0.356 |
| 28109 | RP11-244H3.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 19 | ERCC4 | Sponge network | 0.659 | 3.0E-5 | 0.126 | 0.15266 | 0.356 |
| 28110 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 32 | TGFBR3 | Sponge network | 0.494 | 0.00339 | -1.173 | 1.0E-5 | 0.356 |
| 28111 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-542-3p;hsa-miR-93-5p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | 0.101 | 0.60919 | -0.085 | 0.42389 | 0.356 |
| 28112 | RP11-384O8.1 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p | 10 | RTN4 | Sponge network | -0.738 | 0.21233 | -0.412 | 0.00014 | 0.356 |
| 28113 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-766-3p | 16 | TBL1XR1 | Sponge network | 0.539 | 0.03722 | 0.578 | 0 | 0.356 |
| 28114 | RP1-59D14.5 |
hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 11 | PIK3C2A | Sponge network | 1.162 | 5.0E-5 | 0.061 | 0.53776 | 0.356 |
| 28115 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-577;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -0.303 | 0.21002 | -0.7 | 1.0E-5 | 0.356 |
| 28116 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p | 14 | GALNT13 | Sponge network | -0.154 | 0.78939 | -0.857 | 0.07439 | 0.356 |
| 28117 | RP11-760H22.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-342-3p;hsa-miR-374b-5p | 11 | TCF12 | Sponge network | 0.001 | 0.99753 | 0.174 | 0.13271 | 0.356 |
| 28118 | RP1-151F17.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | TGFBR3 | Sponge network | 0.222 | 0.29133 | -1.173 | 1.0E-5 | 0.356 |
| 28119 | RP5-994D16.9 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZFX | Sponge network | 0.252 | 0.08508 | 0.09 | 0.42563 | 0.356 |
| 28120 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 22 | GRIN2A | Sponge network | 0.249 | 0.35902 | -2.161 | 0 | 0.356 |
| 28121 | PART1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | ABCA10 | Sponge network | -2.925 | 0 | -0.571 | 0.03674 | 0.356 |
| 28122 | RP11-46C24.7 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | RASSF8 | Sponge network | 0.392 | 0.0278 | -0.122 | 0.65591 | 0.356 |
| 28123 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | FYN | Sponge network | -0.096 | 0.67958 | -0.131 | 0.37051 | 0.356 |
| 28124 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | DKK3 | Sponge network | 0.222 | 0.29133 | -0.052 | 0.77783 | 0.356 |
| 28125 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-3614-5p | 13 | HIP1 | Sponge network | 0.592 | 0.00014 | 0.774 | 0 | 0.356 |
| 28126 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-7-5p;hsa-miR-93-3p | 22 | ERC1 | Sponge network | -0.469 | 0.08721 | -0.108 | 0.38794 | 0.356 |
| 28127 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577 | 27 | NTRK2 | Sponge network | 0.811 | 0.03228 | -0.736 | 0.06495 | 0.356 |
| 28128 | RP11-244H3.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p | 20 | CYLD | Sponge network | 0.659 | 3.0E-5 | -0.367 | 0.00171 | 0.356 |
| 28129 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-98-5p | 11 | SCN11A | Sponge network | 0.494 | 0.00339 | -1.15 | 0.00299 | 0.356 |
| 28130 | A2M-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -0.08 | 0.90092 | -0.564 | 1.0E-5 | 0.356 |
| 28131 | NDUFA6-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-376c-3p;hsa-miR-493-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 31 | NCOA1 | Sponge network | -0.355 | 0.0077 | -0.432 | 0 | 0.356 |
| 28132 | RP11-983P16.4 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-342-3p | 11 | STARD13 | Sponge network | 0.41 | 0.02214 | -0.187 | 0.27383 | 0.356 |
| 28133 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PDS5B | Sponge network | 0.67 | 0.00048 | 0.259 | 0.00962 | 0.356 |
| 28134 | MRPL23-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-331-3p;hsa-miR-34a-5p | 14 | PRKAA2 | Sponge network | -0.278 | 0.76559 | -2.462 | 0 | 0.356 |
| 28135 | CTD-2528L19.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | BRD3 | Sponge network | 0.953 | 0 | 0.37 | 0.00013 | 0.356 |
| 28136 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 35 | CBL | Sponge network | 0.249 | 0.35902 | 0.291 | 0.01267 | 0.356 |
| 28137 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p | 15 | CBL | Sponge network | 0.37 | 0.27614 | 0.291 | 0.01267 | 0.356 |
| 28138 | CTD-2314G24.2 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | ZDBF2 | Sponge network | 0.246 | 0.59912 | -0.998 | 0.00025 | 0.356 |
| 28139 | LINC00476 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-935 | 15 | RELN | Sponge network | -0.615 | 0.00015 | -2.403 | 0 | 0.356 |
| 28140 | TINCR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 39 | CHD5 | Sponge network | -0.664 | 0.14543 | -0.922 | 0.01521 | 0.356 |
| 28141 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-935 | 63 | LPP | Sponge network | -0.246 | 0.35034 | -0.459 | 0.04475 | 0.356 |
| 28142 | AC127904.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 1.308 | 0 | 0.51 | 0 | 0.356 |
| 28143 | DNM3OS |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RTN4 | Sponge network | 0.572 | 0.01722 | -0.412 | 0.00014 | 0.356 |
| 28144 | EIF3J-AS1 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MLLT10 | Sponge network | 0.331 | 0.00509 | 0.105 | 0.33292 | 0.356 |
| 28145 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p | 12 | TSC1 | Sponge network | 0.497 | 0.01451 | 0.103 | 0.26071 | 0.356 |
| 28146 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | TCF4 | Sponge network | -0.206 | 0.12942 | 0.016 | 0.92271 | 0.356 |
| 28147 | AC009120.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | 0.919 | 0 | 0.809 | 0.00544 | 0.356 |
| 28148 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p | 11 | ALDH1A2 | Sponge network | -0.323 | 0.01372 | -2.411 | 0 | 0.356 |
| 28149 | DNM1P35 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | PCDH9 | Sponge network | 0.051 | 0.83388 | -1.823 | 4.0E-5 | 0.356 |
| 28150 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-7-1-3p | 10 | DKK3 | Sponge network | 0.727 | 3.0E-5 | -0.052 | 0.77783 | 0.356 |
| 28151 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 14 | MOB1B | Sponge network | 1.4 | 0 | 0.197 | 0.15266 | 0.356 |
| 28152 | RP11-155G14.6 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-30a-5p;hsa-miR-326 | 10 | SOX11 | Sponge network | 2.483 | 0 | 2.68 | 0 | 0.356 |
| 28153 | RP4-639F20.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 20 | TRPS1 | Sponge network | -0.517 | 0.02935 | -0.191 | 0.44109 | 0.356 |
| 28154 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-935 | 22 | LRRC7 | Sponge network | -0.303 | 0.21002 | -1.341 | 0.01193 | 0.356 |
| 28155 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | RBL2 | Sponge network | 0.782 | 0.00016 | -0.282 | 0.00254 | 0.356 |
| 28156 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3065-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 36 | CBL | Sponge network | 0.997 | 0.00066 | 0.291 | 0.01267 | 0.356 |
| 28157 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 15 | GIGYF2 | Sponge network | 0.848 | 0 | 0.136 | 0.04403 | 0.356 |
| 28158 | LINC00641 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 46 | NTRK2 | Sponge network | -0.222 | 0.32445 | -0.736 | 0.06495 | 0.356 |
| 28159 | RP11-384O8.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-940 | 33 | MEF2C | Sponge network | -0.738 | 0.21233 | -0.499 | 0.01448 | 0.356 |
| 28160 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 28 | APC | Sponge network | 0.72 | 0.0001 | -0.255 | 0.04051 | 0.356 |
| 28161 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | PPM1A | Sponge network | 0.194 | 0.64278 | -0.623 | 0 | 0.356 |
| 28162 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p | 18 | ZFHX3 | Sponge network | 1.317 | 0 | 0.389 | 0.01363 | 0.356 |
| 28163 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LHFP | Sponge network | 0.082 | 0.55654 | -0.167 | 0.41371 | 0.356 |
| 28164 | LINC00094 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-92a-1-5p | 22 | AKAP6 | Sponge network | 0.253 | 0.09565 | -1.586 | 3.0E-5 | 0.356 |
| 28165 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 17 | NCAM2 | Sponge network | -1.161 | 0.00591 | -0.482 | 0.22565 | 0.356 |
| 28166 | LINC00472 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 20 | EFNA5 | Sponge network | -0.842 | 0.01361 | -0.421 | 0.17868 | 0.356 |
| 28167 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | RABEP1 | Sponge network | 0.572 | 0.01722 | -0.066 | 0.43142 | 0.356 |
| 28168 | CTC-444N24.11 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 11 | ERCC4 | Sponge network | 1.153 | 0 | 0.126 | 0.15266 | 0.356 |
| 28169 | AC108488.4 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 19 | NAV3 | Sponge network | 0.259 | 0.12542 | 0.212 | 0.47729 | 0.356 |
| 28170 | LINC00957 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-3p | 14 | FMNL3 | Sponge network | -0.421 | 0.0114 | 0.555 | 4.0E-5 | 0.356 |
| 28171 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-93-3p | 19 | TRPS1 | Sponge network | 1.46 | 1.0E-5 | -0.191 | 0.44109 | 0.356 |
| 28172 | TP73-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-93-3p | 30 | CTIF | Sponge network | -0.563 | 0.02545 | -0.389 | 0.00549 | 0.356 |
| 28173 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 17 | CAV1 | Sponge network | 0.246 | 0.59912 | -1.013 | 6.0E-5 | 0.356 |
| 28174 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | USP12 | Sponge network | 0.252 | 0.08508 | -0.102 | 0.40768 | 0.356 |
| 28175 | AC108488.4 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 20 | PCDH9 | Sponge network | 0.259 | 0.12542 | -1.823 | 4.0E-5 | 0.356 |
| 28176 | RP4-669H2.1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-378a-3p;hsa-miR-378c | 15 | ABL2 | Sponge network | 1.711 | 0.00019 | 0.82 | 0 | 0.356 |
| 28177 | LINC00702 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 26 | LIMCH1 | Sponge network | -0.737 | 0.06182 | -0.915 | 1.0E-5 | 0.356 |
| 28178 | RP11-33B1.4 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 10 | BRD3 | Sponge network | 0.826 | 0 | 0.37 | 0.00013 | 0.356 |
| 28179 | LINC00969 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p | 13 | CUL5 | Sponge network | 0.683 | 1.0E-5 | 0.026 | 0.77301 | 0.356 |
| 28180 | RP11-819C21.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p | 19 | RYR2 | Sponge network | 0.537 | 0.00139 | -1.466 | 0.00031 | 0.356 |
| 28181 | RP4-614O4.11 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p | 10 | ZFHX3 | Sponge network | 1.042 | 0.00177 | 0.389 | 0.01363 | 0.356 |
| 28182 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-331-3p | 13 | EPHA7 | Sponge network | 0.598 | 0.38481 | -2.197 | 3.0E-5 | 0.356 |
| 28183 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p;hsa-miR-98-5p | 45 | CREB3L2 | Sponge network | -0.942 | 0 | -0.525 | 2.0E-5 | 0.356 |
| 28184 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500b-5p;hsa-miR-590-5p | 21 | RSPO2 | Sponge network | -1.645 | 0 | -3.038 | 0 | 0.356 |
| 28185 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 22 | LRRC7 | Sponge network | -1.203 | 0.03881 | -1.341 | 0.01193 | 0.356 |
| 28186 | RP11-264B17.2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 10 | CBFB | Sponge network | 1.82 | 0 | 1.061 | 0 | 0.356 |
| 28187 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | TRIP11 | Sponge network | 0.683 | 1.0E-5 | -0.158 | 0.06384 | 0.356 |
| 28188 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p | 32 | PIK3R1 | Sponge network | 0.75 | 0.00054 | -0.133 | 0.36854 | 0.356 |
| 28189 | C4B-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | SETBP1 | Sponge network | 2.306 | 9.0E-5 | -1.302 | 1.0E-5 | 0.356 |
| 28190 | AC034220.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | CREB1 | Sponge network | 0.309 | 0.07304 | 0.203 | 0.00537 | 0.356 |
| 28191 | RP11-5C23.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 49 | LPP | Sponge network | 0.274 | 0.07681 | -0.459 | 0.04475 | 0.356 |
| 28192 | LINC00173 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | NAV3 | Sponge network | 0.056 | 0.8494 | 0.212 | 0.47729 | 0.356 |
| 28193 | PCA3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-744-3p | 26 | PTPN14 | Sponge network | -0.709 | 0.18168 | 0.144 | 0.34595 | 0.356 |
| 28194 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 28 | LIFR | Sponge network | -0.487 | 0.02157 | -1.794 | 0 | 0.356 |
| 28195 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-653-5p | 14 | DMXL1 | Sponge network | 0.545 | 0.00064 | -0.426 | 0.00056 | 0.356 |
| 28196 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429 | 16 | ZMYND11 | Sponge network | -1.654 | 0.00144 | -0.2 | 0.05449 | 0.356 |
| 28197 | RP3-467N11.1 |
hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | TBL1XR1 | Sponge network | 0.953 | 0 | 0.578 | 0 | 0.356 |
| 28198 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-877-5p;hsa-miR-93-5p | 28 | ANK2 | Sponge network | -1.047 | 0 | -1.603 | 1.0E-5 | 0.356 |
| 28199 | BOLA3-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 23 | ST6GALNAC3 | Sponge network | -0.246 | 0.35034 | -0.987 | 0 | 0.356 |
| 28200 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 68 | KIF1B | Sponge network | -0.737 | 0.06182 | -0.118 | 0.32258 | 0.356 |
| 28201 | DNM1P35 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | RET | Sponge network | 0.051 | 0.83388 | -0.833 | 0.03517 | 0.356 |
| 28202 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p | 15 | ABCC9 | Sponge network | 0.253 | 0.09565 | -0.466 | 0.14121 | 0.356 |
| 28203 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | SOX5 | Sponge network | 1.61 | 0.00961 | -1.091 | 4.0E-5 | 0.356 |
| 28204 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | CACNA2D1 | Sponge network | 1.302 | 7.0E-5 | -0.846 | 0.00675 | 0.356 |
| 28205 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-654-3p | 14 | ITCH | Sponge network | 0.705 | 0.00014 | 0.084 | 0.34058 | 0.356 |
| 28206 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 24 | GRIA3 | Sponge network | -1.161 | 0.00591 | -1.956 | 0 | 0.356 |
| 28207 | HCG18 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | RBBP6 | Sponge network | 1.063 | 0 | 0.163 | 0.05101 | 0.356 |
| 28208 | RP11-2B6.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ABI2 | Sponge network | 0.539 | 0.03722 | 0.175 | 0.14787 | 0.356 |
| 28209 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | TAL1 | Sponge network | -0.738 | 0.21233 | -0.462 | 0.00574 | 0.356 |
| 28210 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-769-5p;hsa-miR-93-5p | 49 | WDFY3 | Sponge network | -0.154 | 0.78939 | -0.201 | 0.0967 | 0.356 |
| 28211 | RP11-326G21.1 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZMYND11 | Sponge network | -0.021 | 0.93598 | -0.2 | 0.05449 | 0.356 |
| 28212 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 24 | MYO9A | Sponge network | 0.41 | 0.02214 | -0.147 | 0.32373 | 0.356 |
| 28213 | RP11-130L8.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ZNF208 | Sponge network | -0.663 | 0.10887 | -0.4 | 0.22381 | 0.356 |
| 28214 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 39 | QKI | Sponge network | 0.619 | 0.00157 | -0.333 | 0.04111 | 0.356 |
| 28215 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | MOB1B | Sponge network | 0.401 | 0.00066 | 0.197 | 0.15266 | 0.356 |
| 28216 | RP11-798M19.6 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 18 | BAZ2B | Sponge network | -0.612 | 0.00094 | -0.276 | 0.02833 | 0.356 |
| 28217 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | PRKAA2 | Sponge network | 0.219 | 0.1516 | -2.462 | 0 | 0.356 |
| 28218 | RP11-983P16.4 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-486-5p;hsa-miR-96-5p | 10 | DNMT3A | Sponge network | 0.41 | 0.02214 | 0.302 | 0.0262 | 0.356 |
| 28219 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-744-3p | 28 | PTPN14 | Sponge network | -0.942 | 0 | 0.144 | 0.34595 | 0.356 |
| 28220 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 11 | PDS5B | Sponge network | 0.174 | 0.30034 | 0.259 | 0.00962 | 0.356 |
| 28221 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-7-5p | 18 | ERC1 | Sponge network | -0.517 | 0.02935 | -0.108 | 0.38794 | 0.356 |
| 28222 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | RET | Sponge network | -0.896 | 0.00099 | -0.833 | 0.03517 | 0.356 |
| 28223 | LOXL1-AS1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 15 | ARNT2 | Sponge network | 0.487 | 0.04053 | -0.332 | 0.25851 | 0.356 |
| 28224 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-628-5p | 33 | FOXP1 | Sponge network | -2.62 | 0 | 0.042 | 0.74368 | 0.356 |
| 28225 | DLGAP1-AS2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p | 14 | TBL1XR1 | Sponge network | 2.406 | 0 | 0.578 | 0 | 0.356 |
| 28226 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-940 | 37 | HLF | Sponge network | 0.056 | 0.81195 | -2.443 | 0 | 0.355 |
| 28227 | RP11-345P4.7 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | ERCC6 | Sponge network | 1.597 | 0 | 0.23 | 0.015 | 0.355 |
| 28228 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-339-5p | 12 | CHD1 | Sponge network | 0.637 | 0.00069 | 0.322 | 0.00215 | 0.355 |
| 28229 | LINC00094 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-92a-3p | 14 | NRXN3 | Sponge network | 0.253 | 0.09565 | -0.893 | 0.04186 | 0.355 |
| 28230 | RP11-212P7.2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 29 | PCDH10 | Sponge network | 0.219 | 0.1516 | -1.644 | 0.0078 | 0.355 |
| 28231 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-942-5p | 15 | RB1CC1 | Sponge network | 0.415 | 0.14657 | 0.175 | 0.12559 | 0.355 |
| 28232 | RP11-758N13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p | 12 | SYNE1 | Sponge network | 2.939 | 3.0E-5 | -0.738 | 0.00316 | 0.355 |
| 28233 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-542-3p | 13 | TLE4 | Sponge network | -0.611 | 0.09607 | -1.157 | 0 | 0.355 |
| 28234 | CTC-523E23.11 |
hsa-miR-106a-5p;hsa-miR-10a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-429 | 12 | RASSF2 | Sponge network | -0.235 | 0.35308 | -0.273 | 0.21961 | 0.355 |
| 28235 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p | 25 | SNX29 | Sponge network | -0.469 | 0.08721 | -0.34 | 0.01371 | 0.355 |
| 28236 | TRAF3IP2-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | LIMCH1 | Sponge network | -0.323 | 0.01372 | -0.915 | 1.0E-5 | 0.355 |
| 28237 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-7-1-3p | 19 | TSHZ3 | Sponge network | -4.477 | 0 | -0.556 | 0.01516 | 0.355 |
| 28238 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 24 | TBL1XR1 | Sponge network | 0.705 | 0.00014 | 0.578 | 0 | 0.355 |
| 28239 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 15 | MAP3K12 | Sponge network | 0.572 | 0.01722 | -0.057 | 0.59369 | 0.355 |
| 28240 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | MCC | Sponge network | 0.064 | 0.72934 | -1.005 | 0 | 0.355 |
| 28241 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | LRRC4 | Sponge network | -0.777 | 0.1134 | -1.774 | 0 | 0.355 |
| 28242 | RP11-159D12.2 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-379-5p;hsa-miR-590-3p | 11 | SH3GLB1 | Sponge network | 1.254 | 0 | -0.115 | 0.14773 | 0.355 |
| 28243 | RP11-46C24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | ERCC4 | Sponge network | 0.392 | 0.0278 | 0.126 | 0.15266 | 0.355 |
| 28244 | LINC00865 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-3p | 11 | FYN | Sponge network | -1.249 | 7.0E-5 | -0.131 | 0.37051 | 0.355 |
| 28245 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | CBL | Sponge network | 1.316 | 0 | 0.291 | 0.01267 | 0.355 |
| 28246 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-589-3p | 14 | PPP1R9A | Sponge network | -4.463 | 0.00025 | -0.903 | 0.04254 | 0.355 |
| 28247 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-629-3p | 13 | POU2F2 | Sponge network | 0.335 | 0.20596 | -0.233 | 0.32214 | 0.355 |
| 28248 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-590-3p | 10 | PCDH8 | Sponge network | -1.161 | 0.00591 | -0.997 | 0.05366 | 0.355 |
| 28249 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 14 | GPAM | Sponge network | 1.266 | 0 | 0.142 | 0.29732 | 0.355 |
| 28250 | LINC00476 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | PAK3 | Sponge network | -0.615 | 0.00015 | -0.628 | 0.07253 | 0.355 |
| 28251 | RP11-195F19.9 |
hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-33a-3p | 10 | CSDE1 | Sponge network | -0.726 | 0.00132 | -0.493 | 0 | 0.355 |
| 28252 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-429;hsa-miR-629-3p | 10 | ITGA5 | Sponge network | -1.654 | 0.00144 | 0.452 | 0.03524 | 0.355 |
| 28253 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 18 | CRTC3 | Sponge network | -2.117 | 0.00161 | 0.164 | 0.16786 | 0.355 |
| 28254 | AC006116.24 |
hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-539-5p;hsa-miR-582-3p | 11 | RYR2 | Sponge network | -0.473 | 0.07602 | -1.466 | 0.00031 | 0.355 |
| 28255 | CTC-490E21.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 11 | ARHGEF12 | Sponge network | 1.46 | 1.0E-5 | 0.09 | 0.35693 | 0.355 |
| 28256 | CTC-429P9.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p | 10 | TCF12 | Sponge network | 0.497 | 0.01451 | 0.174 | 0.13271 | 0.355 |
| 28257 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-940 | 34 | PPP1R9A | Sponge network | -0.162 | 0.47687 | -0.903 | 0.04254 | 0.355 |
| 28258 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SLC6A15 | Sponge network | 0.335 | 0.20596 | -2.373 | 2.0E-5 | 0.355 |
| 28259 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 14 | HERC2 | Sponge network | 0.782 | 0.00016 | 0.114 | 0.30638 | 0.355 |
| 28260 | LINC00865 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -1.249 | 7.0E-5 | -0.704 | 0.00106 | 0.355 |
| 28261 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | PRKAB2 | Sponge network | 1.04 | 0.00023 | -0.367 | 0.01371 | 0.355 |
| 28262 | RP11-439A17.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-98-5p | 14 | NTRK3 | Sponge network | 0.051 | 0.95756 | -1.77 | 3.0E-5 | 0.355 |
| 28263 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-96-5p | 14 | PCDH8 | Sponge network | 0.374 | 0.20054 | -0.997 | 0.05366 | 0.355 |
| 28264 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-629-3p | 21 | RASSF2 | Sponge network | 0.898 | 1.0E-5 | -0.273 | 0.21961 | 0.355 |
| 28265 | RP11-35G9.5 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-96-5p | 15 | ASTN1 | Sponge network | 0.622 | 0.00015 | -2.389 | 2.0E-5 | 0.355 |
| 28266 | PART1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 14 | KCNAB1 | Sponge network | -2.925 | 0 | -0.755 | 2.0E-5 | 0.355 |
| 28267 | SNHG16 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 14 | NF1 | Sponge network | 0.961 | 0 | 0.51 | 0 | 0.355 |
| 28268 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 14 | HACE1 | Sponge network | 0.267 | 0.2937 | -0.072 | 0.55239 | 0.355 |
| 28269 | MAGI2-AS3 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | BTG2 | Sponge network | -0.129 | 0.57177 | -1.763 | 0 | 0.355 |
| 28270 | RP11-283I3.6 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-550a-3p | 10 | CACNA1E | Sponge network | 0.614 | 1.0E-5 | 1.752 | 0 | 0.355 |
| 28271 | RP11-384K6.6 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-5p | 25 | CCDC6 | Sponge network | 0.946 | 0 | 0.27 | 0.02555 | 0.355 |
| 28272 | RP11-159D12.2 |
hsa-let-7e-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 19 | COPS2 | Sponge network | 1.254 | 0 | -0.178 | 0.04974 | 0.355 |
| 28273 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-339-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 28 | PRKCE | Sponge network | -1.281 | 0.00012 | -0.403 | 0.00056 | 0.355 |
| 28274 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-331-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | BCL11A | Sponge network | -0.793 | 7.0E-5 | -0.932 | 0.0063 | 0.355 |
| 28275 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 26 | DMXL1 | Sponge network | 0.194 | 0.64278 | -0.426 | 0.00056 | 0.355 |
| 28276 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-7-1-3p | 14 | KCNA6 | Sponge network | 0.225 | 0.30644 | -1.531 | 0.00013 | 0.355 |
| 28277 | NDUFA6-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | SFRP1 | Sponge network | -0.355 | 0.0077 | -3.566 | 0 | 0.355 |
| 28278 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-625-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | 0.532 | 0.00505 | -0.087 | 0.69193 | 0.355 |
| 28279 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-940 | 41 | QKI | Sponge network | 0.16 | 0.50785 | -0.333 | 0.04111 | 0.355 |
| 28280 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ZFHX3 | Sponge network | 0.659 | 3.0E-5 | 0.389 | 0.01363 | 0.355 |
| 28281 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-532-5p;hsa-miR-940 | 27 | NTRK3 | Sponge network | -0.829 | 0.01343 | -1.77 | 3.0E-5 | 0.355 |
| 28282 | RP11-156P1.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-940 | 16 | SORCS1 | Sponge network | 0.214 | 0.18246 | -3.492 | 0 | 0.355 |
| 28283 | KB-1460A1.5 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | AKAP6 | Sponge network | 0.603 | 0.00127 | -1.586 | 3.0E-5 | 0.355 |
| 28284 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SYT4 | Sponge network | -0.206 | 0.12942 | -3.207 | 0 | 0.355 |
| 28285 | ACTA2-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | BRD3 | Sponge network | 0.194 | 0.64278 | 0.37 | 0.00013 | 0.355 |
| 28286 | LINC00865 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-671-5p | 12 | EHD3 | Sponge network | -1.249 | 7.0E-5 | -0.484 | 0.01747 | 0.355 |
| 28287 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p | 14 | CNTNAP3 | Sponge network | -0.517 | 0.02935 | -1.465 | 0.00033 | 0.355 |
| 28288 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 18 | LRP1B | Sponge network | -4.477 | 0 | -2.163 | 0 | 0.355 |
| 28289 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-935 | 14 | DIP2C | Sponge network | 0.37 | 0.27614 | -0.436 | 0.01713 | 0.355 |
| 28290 | RP11-400K9.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-340-5p | 12 | IL17RD | Sponge network | -0.733 | 0.19469 | -0.019 | 0.94873 | 0.355 |
| 28291 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SLC6A15 | Sponge network | 0.225 | 0.30644 | -2.373 | 2.0E-5 | 0.355 |
| 28292 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | NIN | Sponge network | -0.612 | 0.00094 | -0.253 | 0.13794 | 0.355 |
| 28293 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 12 | GPAM | Sponge network | 0.673 | 0 | 0.142 | 0.29732 | 0.355 |
| 28294 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 17 | UNC5C | Sponge network | 0.572 | 0.01722 | -0.178 | 0.40214 | 0.355 |
| 28295 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-651-5p | 26 | PRDM2 | Sponge network | -0.154 | 0.78939 | -0.258 | 0.00721 | 0.355 |
| 28296 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-93-3p | 20 | FOXO3 | Sponge network | -0.176 | 0.59692 | -0.04 | 0.72028 | 0.355 |
| 28297 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-5p | 37 | ADARB1 | Sponge network | 0.4 | 0.14611 | -0.335 | 0.06631 | 0.355 |
| 28298 | C20orf166-AS1 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-542-3p | 18 | PDE4DIP | Sponge network | -2.69 | 0.0001 | -0.282 | 0.0257 | 0.355 |
| 28299 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-590-3p | 13 | RBM5 | Sponge network | -0.487 | 0.02157 | -0.205 | 0.0129 | 0.355 |
| 28300 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 16 | LRRC7 | Sponge network | 0.176 | 0.37402 | -1.341 | 0.01193 | 0.355 |
| 28301 | RP11-196G18.22 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-376c-3p | 12 | RASAL2 | Sponge network | 1.206 | 0 | 0.869 | 0 | 0.355 |
| 28302 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 43 | CUX1 | Sponge network | -0.096 | 0.67958 | -0.039 | 0.6705 | 0.355 |
| 28303 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 23 | BCL11A | Sponge network | -2.452 | 0 | -0.932 | 0.0063 | 0.355 |
| 28304 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p | 19 | GREM1 | Sponge network | -1.203 | 0.03881 | 0.902 | 0.01691 | 0.355 |
| 28305 | RP11-819C21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 19 | NRK | Sponge network | 0.537 | 0.00139 | 0.656 | 0.19217 | 0.355 |
| 28306 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 14 | MITF | Sponge network | -0.726 | 0.00132 | -0.769 | 0.00022 | 0.355 |
| 28307 | RP11-307B6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 12 | CBFA2T3 | Sponge network | -4.463 | 0.00025 | -1.221 | 1.0E-5 | 0.355 |
| 28308 | RP11-53O19.1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-374a-5p;hsa-miR-942-5p | 16 | GLIPR1 | Sponge network | -0.405 | 0.22013 | 0.146 | 0.3276 | 0.355 |
| 28309 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 50 | BMPR2 | Sponge network | -0.513 | 0.04703 | 0.206 | 0.0635 | 0.355 |
| 28310 | CROCCP2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p | 10 | PHF3 | Sponge network | 0.79 | 0 | 0.126 | 0.19652 | 0.355 |
| 28311 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 23 | SASH1 | Sponge network | -4.546 | 0 | -0.502 | 0.00175 | 0.355 |
| 28312 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-2355-3p;hsa-miR-3613-5p;hsa-miR-93-5p | 14 | EPHA7 | Sponge network | 0.497 | 0.01451 | -2.197 | 3.0E-5 | 0.355 |
| 28313 | RP11-758N13.1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-345-5p;hsa-miR-942-5p | 15 | IGFBP5 | Sponge network | 2.939 | 3.0E-5 | -0.806 | 0.00105 | 0.355 |
| 28314 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 58 | PRKAA2 | Sponge network | -0.615 | 0.00015 | -2.462 | 0 | 0.355 |
| 28315 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-628-5p | 11 | CAV1 | Sponge network | -0.154 | 0.78939 | -1.013 | 6.0E-5 | 0.355 |
| 28316 | CTC-510F12.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-376c-3p | 10 | PDE4DIP | Sponge network | -0.691 | 0.00146 | -0.282 | 0.0257 | 0.355 |
| 28317 | RP11-395G23.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-532-3p | 13 | SYNM | Sponge network | 0.381 | 0.25967 | -2.309 | 1.0E-5 | 0.355 |
| 28318 | EMX2OS |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p;hsa-miR-624-5p | 12 | SMARCA1 | Sponge network | -0.777 | 0.1134 | -0.684 | 0.00159 | 0.355 |
| 28319 | OIP5-AS1 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 13 | LZTS1 | Sponge network | 0.421 | 0.00084 | 1.664 | 0 | 0.355 |
| 28320 | USP3-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-361-5p | 10 | PCDH18 | Sponge network | -0.162 | 0.47687 | 0.216 | 0.24645 | 0.355 |
| 28321 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-590-3p | 11 | PLA2R1 | Sponge network | 0.101 | 0.60919 | -0.265 | 0.24676 | 0.355 |
| 28322 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-590-3p | 34 | WDFY3 | Sponge network | -0.358 | 0.17255 | -0.201 | 0.0967 | 0.355 |
| 28323 | RP11-890B15.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p | 24 | RYR2 | Sponge network | 0.101 | 0.60919 | -1.466 | 0.00031 | 0.355 |
| 28324 | OIP5-AS1 |
hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 13 | CDK6 | Sponge network | 0.421 | 0.00084 | 0.991 | 0 | 0.355 |
| 28325 | CTD-2303H24.2 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PHF3 | Sponge network | 0.415 | 0.14657 | 0.126 | 0.19652 | 0.355 |
| 28326 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-766-3p | 11 | PHACTR4 | Sponge network | 0.782 | 0.00016 | 0.069 | 0.45203 | 0.355 |
| 28327 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-429;hsa-miR-577 | 18 | MYBL1 | Sponge network | 0.389 | 0.04293 | 0.494 | 0.02152 | 0.355 |
| 28328 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3913-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 33 | ESR1 | Sponge network | -1.203 | 0.03881 | -1.035 | 3.0E-5 | 0.355 |
| 28329 | C4A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-589-3p;hsa-miR-96-5p | 18 | ZFHX4 | Sponge network | 3.345 | 0 | 0.018 | 0.95574 | 0.355 |
| 28330 | CTC-457L16.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p | 23 | RUNX1T1 | Sponge network | 1.153 | 0.00114 | -0.344 | 0.2374 | 0.355 |
| 28331 | LINC00476 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-93-3p | 29 | WWTR1 | Sponge network | -0.615 | 0.00015 | -0.345 | 0.11997 | 0.355 |
| 28332 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484 | 10 | RET | Sponge network | -0.187 | 0.23787 | -0.833 | 0.03517 | 0.355 |
| 28333 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | NUAK1 | Sponge network | 0.374 | 0.20054 | 0.904 | 0 | 0.355 |
| 28334 | RP11-156P1.3 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940 | 12 | FOXO4 | Sponge network | 0.214 | 0.18246 | -0.516 | 2.0E-5 | 0.355 |
| 28335 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p | 18 | PAFAH1B1 | Sponge network | 0.542 | 0.00054 | -0.429 | 0 | 0.355 |
| 28336 | FZD10-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-532-3p | 15 | SYNM | Sponge network | -0.727 | 0.04726 | -2.309 | 1.0E-5 | 0.355 |
| 28337 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | NIN | Sponge network | -0.421 | 0.0114 | -0.253 | 0.13794 | 0.355 |
| 28338 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-455-3p;hsa-miR-577;hsa-miR-7-1-3p | 16 | TSHZ3 | Sponge network | -0.418 | 0.00068 | -0.556 | 0.01516 | 0.355 |
| 28339 | CALML3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-664a-3p | 16 | NRXN3 | Sponge network | 0.95 | 0.15943 | -0.893 | 0.04186 | 0.355 |
| 28340 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-30a-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p | 10 | PDS5B | Sponge network | 1.4 | 0 | 0.259 | 0.00962 | 0.355 |
| 28341 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 27 | JAZF1 | Sponge network | 1.302 | 7.0E-5 | -0.377 | 0.02677 | 0.355 |
| 28342 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-330-3p | 11 | APC | Sponge network | -1.331 | 3.0E-5 | -0.255 | 0.04051 | 0.355 |
| 28343 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 32 | EPHA3 | Sponge network | 0.421 | 0.00084 | 0.185 | 0.53588 | 0.355 |
| 28344 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | PTPRB | Sponge network | -0.227 | 0.31657 | 0.105 | 0.47122 | 0.355 |
| 28345 | LINC00702 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-940 | 66 | NFIB | Sponge network | -0.737 | 0.06182 | 0.023 | 0.90795 | 0.355 |
| 28346 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 53 | QKI | Sponge network | 1.302 | 7.0E-5 | -0.333 | 0.04111 | 0.355 |
| 28347 | RP4-717I23.3 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-577 | 10 | MACF1 | Sponge network | 0.637 | 0.00069 | 0.019 | 0.90335 | 0.355 |
| 28348 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-628-5p | 36 | TMEM132B | Sponge network | -0.154 | 0.78939 | -0.83 | 0.01084 | 0.355 |
| 28349 | FENDRR |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-335-3p;hsa-miR-96-5p | 11 | ELMO1 | Sponge network | -1.281 | 0.00012 | 0.04 | 0.83147 | 0.355 |
| 28350 | MIR22HG |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-5p | 18 | SPG20 | Sponge network | -1.047 | 0 | -1.16 | 0 | 0.355 |
| 28351 | LINC00863 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 11 | DNMT3A | Sponge network | 0.189 | 0.13981 | 0.302 | 0.0262 | 0.355 |
| 28352 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3065-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p | 23 | CHD5 | Sponge network | 0.889 | 9.0E-5 | -0.922 | 0.01521 | 0.355 |
| 28353 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | BCL11A | Sponge network | -0.615 | 0.00015 | -0.932 | 0.0063 | 0.355 |
| 28354 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429 | 23 | MYBL1 | Sponge network | 0.253 | 0.09565 | 0.494 | 0.02152 | 0.355 |
| 28355 | RP11-400K9.4 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-7-5p | 15 | IRS1 | Sponge network | -0.611 | 0.09607 | -0.026 | 0.88855 | 0.355 |
| 28356 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-576-5p | 18 | CBL | Sponge network | 0.594 | 0.00064 | 0.291 | 0.01267 | 0.355 |
| 28357 | CTC-296K1.4 |
hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-500a-5p;hsa-miR-590-5p | 15 | SNX29 | Sponge network | -2.076 | 0.00548 | -0.34 | 0.01371 | 0.355 |
| 28358 | RP11-983P16.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | SEMA3E | Sponge network | 0.41 | 0.02214 | -1.717 | 0.00137 | 0.355 |
| 28359 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 18 | RICTOR | Sponge network | 0.706 | 0 | 0.388 | 0.00188 | 0.355 |
| 28360 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 50 | KIF1B | Sponge network | 0.559 | 0.00026 | -0.118 | 0.32258 | 0.355 |
| 28361 | LINC00654 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | 0.374 | 0.20054 | -0.615 | 0.01482 | 0.355 |
| 28362 | LINC00092 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-532-5p;hsa-miR-589-3p | 22 | IL17RD | Sponge network | -1.886 | 0 | -0.019 | 0.94873 | 0.355 |
| 28363 | FAM157B |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-378a-5p | 16 | CREB1 | Sponge network | 0.92 | 0 | 0.203 | 0.00537 | 0.355 |
| 28364 | LINC00472 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-7-1-3p | 21 | ZMYND11 | Sponge network | -0.842 | 0.01361 | -0.2 | 0.05449 | 0.355 |
| 28365 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-409-3p | 19 | MEF2C | Sponge network | 0.497 | 0.01451 | -0.499 | 0.01448 | 0.355 |
| 28366 | RP11-282O18.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-582-5p | 14 | CREB1 | Sponge network | 0.757 | 0 | 0.203 | 0.00537 | 0.355 |
| 28367 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ABCC9 | Sponge network | 0.219 | 0.1516 | -0.466 | 0.14121 | 0.355 |
| 28368 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | DDX5 | Sponge network | 0.222 | 0.29133 | -0.099 | 0.17428 | 0.355 |
| 28369 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 14 | RIMS1 | Sponge network | 0.246 | 0.59912 | -3.143 | 0 | 0.355 |
| 28370 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p | 14 | PSIP1 | Sponge network | 0.614 | 1.0E-5 | -0.005 | 0.96897 | 0.355 |
| 28371 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 32 | EPHA5 | Sponge network | -0.739 | 0.00044 | -0.957 | 0.01733 | 0.355 |
| 28372 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | RECK | Sponge network | -0.125 | 0.49414 | -1.043 | 0 | 0.355 |
| 28373 | RP11-428J1.5 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | BRD3 | Sponge network | 0.538 | 0.00076 | 0.37 | 0.00013 | 0.355 |
| 28374 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-589-3p | 10 | TET1 | Sponge network | -4.463 | 0.00025 | -0.135 | 0.52138 | 0.355 |
| 28375 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-223-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 11 | PDS5B | Sponge network | 0.643 | 0 | 0.259 | 0.00962 | 0.355 |
| 28376 | RP11-981G7.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.986 | 0.01022 | 1.083 | 0 | 0.355 |
| 28377 | CTC-559E9.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-877-5p | 10 | TET1 | Sponge network | 0.594 | 0.00064 | -0.135 | 0.52138 | 0.355 |
| 28378 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-744-3p;hsa-miR-96-5p | 20 | HIP1 | Sponge network | 0.395 | 0.15451 | 0.774 | 0 | 0.355 |
| 28379 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3065-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 36 | CBL | Sponge network | 0.064 | 0.72934 | 0.291 | 0.01267 | 0.355 |
| 28380 | LINC00476 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ZNF536 | Sponge network | -0.615 | 0.00015 | -3.115 | 0 | 0.355 |
| 28381 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-500a-5p | 17 | ZFHX3 | Sponge network | 2.163 | 0 | 0.389 | 0.01363 | 0.355 |
| 28382 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326 | 10 | NF1 | Sponge network | 1.361 | 0 | 0.51 | 0 | 0.355 |
| 28383 | CTD-2349P21.10 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | CREB1 | Sponge network | 1.341 | 0.00014 | 0.203 | 0.00537 | 0.355 |
| 28384 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p | 12 | HIP1 | Sponge network | 0.497 | 0.01451 | 0.774 | 0 | 0.355 |
| 28385 | RP11-418J17.1 |
hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-582-3p | 12 | ABL2 | Sponge network | 0.998 | 0 | 0.82 | 0 | 0.355 |
| 28386 | LPP-AS2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-96-5p | 14 | HDAC9 | Sponge network | -0.18 | 0.20343 | -0.755 | 0.00495 | 0.355 |
| 28387 | LINC00174 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p | 14 | CBL | Sponge network | 1.253 | 0 | 0.291 | 0.01267 | 0.355 |
| 28388 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 13 | BCLAF1 | Sponge network | 0.417 | 0.00445 | 0.211 | 0.00684 | 0.355 |
| 28389 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-24-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 19 | MXI1 | Sponge network | -0.83 | 0 | -0.987 | 0 | 0.355 |
| 28390 | RP11-973H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-145-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-539-5p | 11 | MOB1B | Sponge network | 0.901 | 0 | 0.197 | 0.15266 | 0.355 |
| 28391 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 10 | MLLT10 | Sponge network | 0.702 | 7.0E-5 | 0.105 | 0.33292 | 0.355 |
| 28392 | LINC00094 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-93-5p | 17 | LATS2 | Sponge network | 0.253 | 0.09565 | 0.159 | 0.2304 | 0.355 |
| 28393 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 10 | THBS1 | Sponge network | 0.959 | 0 | 0.741 | 0.00386 | 0.355 |
| 28394 | U91328.19 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-7-1-3p | 15 | ITPKB | Sponge network | -0.303 | 0.21002 | -0.704 | 0.00106 | 0.355 |
| 28395 | RP11-867G23.10 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-744-5p | 10 | IGF2 | Sponge network | -2.531 | 0 | 0.848 | 0.03842 | 0.355 |
| 28396 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-576-5p;hsa-miR-744-3p | 19 | HIP1 | Sponge network | -0.611 | 0.09607 | 0.774 | 0 | 0.355 |
| 28397 | ACTA2-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 13 | PIK3IP1 | Sponge network | 0.194 | 0.64278 | -0.187 | 0.19945 | 0.355 |
| 28398 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p | 10 | GAS1 | Sponge network | -1.203 | 0.03881 | -1.047 | 0.00364 | 0.355 |
| 28399 | TRAF3IP2-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | BTG2 | Sponge network | -0.323 | 0.01372 | -1.763 | 0 | 0.355 |
| 28400 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | MYO9A | Sponge network | 1.063 | 0 | -0.147 | 0.32373 | 0.355 |
| 28401 | LINC00473 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p | 14 | ABCA10 | Sponge network | -1.203 | 0.03881 | -0.571 | 0.03674 | 0.355 |
| 28402 | SNHG14 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | RTN4 | Sponge network | -1.111 | 0.0003 | -0.412 | 0.00014 | 0.355 |
| 28403 | KB-431C1.4 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-493-5p;hsa-miR-495-3p | 18 | CREB1 | Sponge network | 0.909 | 0 | 0.203 | 0.00537 | 0.355 |
| 28404 | CTD-2245F17.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-7-5p | 15 | TCF4 | Sponge network | -0.421 | 0.12556 | 0.016 | 0.92271 | 0.355 |
| 28405 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-374b-5p;hsa-miR-625-5p | 14 | NUAK1 | Sponge network | 0.381 | 0.25967 | 0.904 | 0 | 0.355 |
| 28406 | GABPB1-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | ERCC4 | Sponge network | 1.11 | 0 | 0.126 | 0.15266 | 0.355 |
| 28407 | RP11-701H24.7 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 14 | MDM4 | Sponge network | -2.039 | 0.03447 | 0.345 | 0.00762 | 0.355 |
| 28408 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | ITCH | Sponge network | 1.254 | 0 | 0.084 | 0.34058 | 0.355 |
| 28409 | RP5-1085F17.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 16 | BAZ2B | Sponge network | 0.541 | 0.00125 | -0.276 | 0.02833 | 0.355 |
| 28410 | RP11-158K1.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TET1 | Sponge network | 0.349 | 0.01695 | -0.135 | 0.52138 | 0.355 |
| 28411 | LINC00473 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-590-3p | 15 | TLE4 | Sponge network | -1.203 | 0.03881 | -1.157 | 0 | 0.355 |
| 28412 | FENDRR |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | GLIPR1 | Sponge network | -1.281 | 0.00012 | 0.146 | 0.3276 | 0.355 |
| 28413 | LINC00654 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p | 15 | FLNA | Sponge network | 0.374 | 0.20054 | -0.771 | 0.01377 | 0.355 |
| 28414 | AC009948.5 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 16 | SFRP1 | Sponge network | 0.532 | 0.00505 | -3.566 | 0 | 0.355 |
| 28415 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-664a-5p | 22 | SMAD4 | Sponge network | 0.706 | 0 | -0.313 | 0.00413 | 0.355 |
| 28416 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | DMXL1 | Sponge network | -0.739 | 0.0574 | -0.426 | 0.00056 | 0.355 |
| 28417 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | TLE4 | Sponge network | 0.194 | 0.64278 | -1.157 | 0 | 0.355 |
| 28418 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-429;hsa-miR-589-5p | 12 | ARHGAP29 | Sponge network | -0.154 | 0.53954 | 0.131 | 0.42953 | 0.355 |
| 28419 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 39 | EPHA7 | Sponge network | -0.727 | 0.04726 | -2.197 | 3.0E-5 | 0.355 |
| 28420 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | PLSCR4 | Sponge network | 0.727 | 3.0E-5 | -0.474 | 0.03833 | 0.355 |
| 28421 | CTD-2528L19.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | PIK3CA | Sponge network | 0.953 | 0 | 0.195 | 0.06908 | 0.355 |
| 28422 | RP11-819C21.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 10 | DLG1 | Sponge network | 0.537 | 0.00139 | -0.014 | 0.8926 | 0.355 |
| 28423 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | KAT6B | Sponge network | -0.129 | 0.57177 | -0.478 | 0.00018 | 0.355 |
| 28424 | RP11-400K9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-940 | 13 | PBX1 | Sponge network | -0.733 | 0.19469 | -1.206 | 0 | 0.355 |
| 28425 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-362-3p | 12 | ST6GALNAC3 | Sponge network | -2.549 | 0.00528 | -0.987 | 0 | 0.355 |
| 28426 | RP11-701H24.3 |
hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 13 | DST | Sponge network | -1.425 | 0.04204 | -0.713 | 0.00096 | 0.355 |
| 28427 | RP11-490M8.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-539-5p | 13 | DIP2C | Sponge network | -0.214 | 0.44248 | -0.436 | 0.01713 | 0.355 |
| 28428 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-629-5p | 26 | MAF | Sponge network | -0.773 | 0.04073 | -1.257 | 0 | 0.355 |
| 28429 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 34 | MOB1B | Sponge network | 0.659 | 3.0E-5 | 0.197 | 0.15266 | 0.355 |
| 28430 | CTC-297N7.9 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-29b-1-5p;hsa-miR-590-5p | 13 | GNAQ | Sponge network | -2.069 | 0 | -0.367 | 0.00102 | 0.355 |
| 28431 | HOTAIRM1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 11 | CNTNAP1 | Sponge network | -0.053 | 0.80685 | 0.358 | 0.13727 | 0.355 |
| 28432 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-7-1-3p | 22 | THRB | Sponge network | -0.187 | 0.23787 | -1.117 | 0.0004 | 0.355 |
| 28433 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 63 | BMPR2 | Sponge network | 0.219 | 0.1516 | 0.206 | 0.0635 | 0.355 |
| 28434 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 13 | TRIP11 | Sponge network | 0.817 | 3.0E-5 | -0.158 | 0.06384 | 0.355 |
| 28435 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-93-3p | 21 | EBF3 | Sponge network | -1.249 | 7.0E-5 | -0.058 | 0.81437 | 0.355 |
| 28436 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-96-5p | 24 | DMXL1 | Sponge network | 0.614 | 1.0E-5 | -0.426 | 0.00056 | 0.355 |
| 28437 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 44 | EPHA5 | Sponge network | -0.942 | 0 | -0.957 | 0.01733 | 0.354 |
| 28438 | PAXIP1-AS1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-766-3p | 18 | TBL1XR1 | Sponge network | 0.66 | 1.0E-5 | 0.578 | 0 | 0.354 |
| 28439 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-577;hsa-miR-7-1-3p | 19 | TET1 | Sponge network | 0.389 | 0.04293 | -0.135 | 0.52138 | 0.354 |
| 28440 | MEG3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p | 11 | PARK2 | Sponge network | 0.249 | 0.35902 | -1.892 | 0 | 0.354 |
| 28441 | HOXA-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | LHFP | Sponge network | 0.61 | 0.01593 | -0.167 | 0.41371 | 0.354 |
| 28442 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-301a-3p | 11 | NCOA1 | Sponge network | 0.043 | 0.81628 | -0.432 | 0 | 0.354 |
| 28443 | FAM13A-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | -0.83 | 0 | -0.57 | 0 | 0.354 |
| 28444 | MIR22HG |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 13 | SORCS1 | Sponge network | -1.047 | 0 | -3.492 | 0 | 0.354 |
| 28445 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 42 | NOTCH2 | Sponge network | -0.942 | 0 | 0.138 | 0.35163 | 0.354 |
| 28446 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p | 46 | CBX5 | Sponge network | 0.253 | 0.09565 | 0.165 | 0.24446 | 0.354 |
| 28447 | ACTA2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-628-5p;hsa-miR-708-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 28 | TOM1L2 | Sponge network | 0.194 | 0.64278 | -0.994 | 0 | 0.354 |
| 28448 | A2M-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-96-5p | 11 | CXCL12 | Sponge network | -0.08 | 0.90092 | -1.421 | 0 | 0.354 |
| 28449 | LPP-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 22 | NAV3 | Sponge network | -0.18 | 0.20343 | 0.212 | 0.47729 | 0.354 |
| 28450 | RP11-150O12.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-3614-5p | 17 | ABL2 | Sponge network | 2.605 | 2.0E-5 | 0.82 | 0 | 0.354 |
| 28451 | FAM13A-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | SLC2A13 | Sponge network | -0.83 | 0 | -0.331 | 0.03658 | 0.354 |
| 28452 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 28 | PCDH17 | Sponge network | -2.452 | 0 | 0.731 | 2.0E-5 | 0.354 |
| 28453 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | AHNAK | Sponge network | 0.299 | 0.57722 | -1.207 | 0 | 0.354 |
| 28454 | FAM157B |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-2355-3p;hsa-miR-338-5p;hsa-miR-3614-5p | 11 | TSC1 | Sponge network | 0.92 | 0 | 0.103 | 0.26071 | 0.354 |
| 28455 | LINC00205 |
hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p | 12 | TBL1XR1 | Sponge network | 1.271 | 0 | 0.578 | 0 | 0.354 |
| 28456 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-424-5p | 16 | CBX5 | Sponge network | 0.972 | 0 | 0.165 | 0.24446 | 0.354 |
| 28457 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-192-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | BMPR2 | Sponge network | 1.378 | 0 | 0.206 | 0.0635 | 0.354 |
| 28458 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-584-5p | 11 | ZNF208 | Sponge network | -0.465 | 0.04703 | -0.4 | 0.22381 | 0.354 |
| 28459 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 28 | FOXP1 | Sponge network | -1.092 | 4.0E-5 | 0.042 | 0.74368 | 0.354 |
| 28460 | RP11-25K19.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | RABEP1 | Sponge network | -1.057 | 0.00029 | -0.066 | 0.43142 | 0.354 |
| 28461 | RP5-1198O20.4 |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 11 | GLIPR2 | Sponge network | 0.997 | 0.00066 | -0.996 | 0 | 0.354 |
| 28462 | CALML3-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429 | 26 | ABL2 | Sponge network | 0.95 | 0.15943 | 0.82 | 0 | 0.354 |
| 28463 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p | 21 | TRPS1 | Sponge network | 0.178 | 0.29106 | -0.191 | 0.44109 | 0.354 |
| 28464 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | 0.083 | 0.66405 | -1.892 | 0 | 0.354 |
| 28465 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 1.117 | 0 | 0.578 | 0 | 0.354 |
| 28466 | RP11-180O5.2 |
hsa-let-7a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 11 | TBL1XR1 | Sponge network | 3.607 | 0.0001 | 0.578 | 0 | 0.354 |
| 28467 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | SLITRK6 | Sponge network | -2.925 | 0 | -2.121 | 0 | 0.354 |
| 28468 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ZFHX3 | Sponge network | 0.272 | 0.44692 | 0.389 | 0.01363 | 0.354 |
| 28469 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-629-3p | 35 | KIF1B | Sponge network | 0.946 | 0 | -0.118 | 0.32258 | 0.354 |
| 28470 | RP3-402G11.26 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-590-3p | 11 | DMD | Sponge network | -0.254 | 0.19303 | -1.506 | 0 | 0.354 |
| 28471 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-93-5p | 34 | RASSF2 | Sponge network | -2.69 | 0.0001 | -0.273 | 0.21961 | 0.354 |
| 28472 | RP11-6O2.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-584-5p;hsa-miR-590-5p | 13 | ZNF208 | Sponge network | 0.331 | 0.57247 | -0.4 | 0.22381 | 0.354 |
| 28473 | RP11-6O2.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-589-3p | 31 | AFF1 | Sponge network | 0.331 | 0.57247 | -0.384 | 0.00053 | 0.354 |
| 28474 | RP4-669P10.16 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-93-3p | 17 | NCOA1 | Sponge network | -0.301 | 0.06597 | -0.432 | 0 | 0.354 |
| 28475 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-93-5p | 14 | EPAS1 | Sponge network | -2.69 | 0.0001 | -0.184 | 0.14418 | 0.354 |
| 28476 | LPP-AS2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-577 | 10 | MACF1 | Sponge network | -0.18 | 0.20343 | 0.019 | 0.90335 | 0.354 |
| 28477 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | MYO9A | Sponge network | 0.395 | 0.15451 | -0.147 | 0.32373 | 0.354 |
| 28478 | AC016747.3 |
hsa-let-7a-3p;hsa-miR-10a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | HDAC9 | Sponge network | 0.199 | 0.38015 | -0.755 | 0.00495 | 0.354 |
| 28479 | NDUFA6-AS1 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-493-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-874-3p | 14 | ARID1B | Sponge network | -0.355 | 0.0077 | 0.003 | 0.97503 | 0.354 |
| 28480 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-93-5p | 30 | SSBP2 | Sponge network | -0.785 | 0.00035 | -1.58 | 0 | 0.354 |
| 28481 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | CNTN1 | Sponge network | -0.141 | 0.26434 | -2.594 | 0 | 0.354 |
| 28482 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 17 | MAP3K12 | Sponge network | -0.737 | 0.06182 | -0.057 | 0.59369 | 0.354 |
| 28483 | AP001347.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-425-5p;hsa-miR-590-3p | 11 | ITGB3 | Sponge network | -1.161 | 0.00591 | -0.011 | 0.95751 | 0.354 |
| 28484 | LINC00861 |
hsa-miR-130b-3p;hsa-miR-196b-5p;hsa-miR-224-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 10 | SNX29 | Sponge network | 0.911 | 0.00854 | -0.34 | 0.01371 | 0.354 |
| 28485 | RP11-403I13.7 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 12 | PRUNE2 | Sponge network | 0.395 | 0.15451 | -1.733 | 0.00022 | 0.354 |
| 28486 | AF131215.2 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | SETBP1 | Sponge network | -0.176 | 0.59692 | -1.302 | 1.0E-5 | 0.354 |
| 28487 | HOXB-AS3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-339-5p | 13 | STARD13 | Sponge network | 0.343 | 0.28887 | -0.187 | 0.27383 | 0.354 |
| 28488 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-539-5p;hsa-miR-766-3p;hsa-miR-940 | 18 | CADM2 | Sponge network | -0.421 | 0.12556 | -3.782 | 0 | 0.354 |
| 28489 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429 | 13 | ABCB5 | Sponge network | -1.654 | 0.00144 | -0.956 | 0.04077 | 0.354 |
| 28490 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-577 | 14 | MYBL1 | Sponge network | 0.419 | 0.0319 | 0.494 | 0.02152 | 0.354 |
| 28491 | RP11-401P9.4 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-590-5p;hsa-miR-7-5p | 12 | FNBP1 | Sponge network | -0.384 | 0.40652 | -0.969 | 4.0E-5 | 0.354 |
| 28492 | RP11-736K20.5 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 17 | LIMCH1 | Sponge network | -0.358 | 0.17255 | -0.915 | 1.0E-5 | 0.354 |
| 28493 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 21 | USP6 | Sponge network | 1.153 | 0 | 0.188 | 0.3871 | 0.354 |
| 28494 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-5p | 21 | MOB1B | Sponge network | 1.04 | 0.00023 | 0.197 | 0.15266 | 0.354 |
| 28495 | LINC00654 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | TAL1 | Sponge network | 0.374 | 0.20054 | -0.462 | 0.00574 | 0.354 |
| 28496 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | NOTCH2 | Sponge network | -1.161 | 0.00591 | 0.138 | 0.35163 | 0.354 |
| 28497 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-409-5p;hsa-miR-495-3p;hsa-miR-539-5p | 11 | LIMD1 | Sponge network | 0.537 | 0.00139 | 0.103 | 0.28978 | 0.354 |
| 28498 | MEG3 |
hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 12 | PHOX2B | Sponge network | 0.249 | 0.35902 | -2.68 | 0 | 0.354 |
| 28499 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | FBXO32 | Sponge network | 1.308 | 0 | -0.495 | 0.07561 | 0.354 |
| 28500 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | MED12L | Sponge network | 0.614 | 1.0E-5 | -0.194 | 0.55299 | 0.354 |
| 28501 | AC159540.2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-629-3p | 12 | DDX6 | Sponge network | 1.634 | 0 | 0.091 | 0.36428 | 0.354 |
| 28502 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | USP12 | Sponge network | 0.272 | 0.44692 | -0.102 | 0.40768 | 0.354 |
| 28503 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p | 16 | RET | Sponge network | 0.101 | 0.60919 | -0.833 | 0.03517 | 0.354 |
| 28504 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 49 | PTPRD | Sponge network | -0.727 | 0.04726 | -1.005 | 0.00064 | 0.354 |
| 28505 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | 1.153 | 0.00114 | -0.243 | 0.11408 | 0.354 |
| 28506 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 12 | ZMYM2 | Sponge network | 1.344 | 0 | 0.279 | 0.01273 | 0.354 |
| 28507 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-450b-5p;hsa-miR-532-3p | 19 | PBX1 | Sponge network | 0.381 | 0.25967 | -1.206 | 0 | 0.354 |
| 28508 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-429;hsa-miR-582-5p | 10 | USP6 | Sponge network | 1.157 | 0.00455 | 0.188 | 0.3871 | 0.354 |
| 28509 | RP11-867G23.10 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-769-5p;hsa-miR-93-5p | 28 | WDFY3 | Sponge network | -2.531 | 0 | -0.201 | 0.0967 | 0.354 |
| 28510 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-629-3p | 13 | ITGA5 | Sponge network | -0.318 | 0.65847 | 0.452 | 0.03524 | 0.354 |
| 28511 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-3p;hsa-miR-335-3p;hsa-miR-590-3p | 12 | TGFBR3 | Sponge network | -1.425 | 0.04204 | -1.173 | 1.0E-5 | 0.354 |
| 28512 | RP11-981G7.6 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PHF3 | Sponge network | 0.986 | 0.01022 | 0.126 | 0.19652 | 0.354 |
| 28513 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | APC | Sponge network | -0.129 | 0.57177 | -0.255 | 0.04051 | 0.354 |
| 28514 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p | 19 | FBXO32 | Sponge network | 0.259 | 0.12542 | -0.495 | 0.07561 | 0.354 |
| 28515 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-7-1-3p | 13 | EPHA5 | Sponge network | 0.693 | 0.00076 | -0.957 | 0.01733 | 0.354 |
| 28516 | CTC-550B14.7 |
hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30c-2-3p;hsa-miR-365a-3p;hsa-miR-374a-3p | 14 | VPS53 | Sponge network | 1.161 | 0 | 0.103 | 0.22667 | 0.354 |
| 28517 | RP11-400K9.4 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p | 13 | CSDE1 | Sponge network | -0.611 | 0.09607 | -0.493 | 0 | 0.354 |
| 28518 | AC010226.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-96-5p | 10 | ELMO1 | Sponge network | -0.811 | 2.0E-5 | 0.04 | 0.83147 | 0.354 |
| 28519 | OIP5-AS1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p | 24 | DDX3X | Sponge network | 0.421 | 0.00084 | -0.152 | 0.07686 | 0.354 |
| 28520 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | CBL | Sponge network | 0.619 | 0.00157 | 0.291 | 0.01267 | 0.354 |
| 28521 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-98-5p | 24 | CLASP2 | Sponge network | -0.227 | 0.31657 | -0.038 | 0.71 | 0.354 |
| 28522 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-375 | 11 | ZFHX4 | Sponge network | 1.969 | 0.00316 | 0.018 | 0.95574 | 0.354 |
| 28523 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TRIP11 | Sponge network | 0.72 | 0.0001 | -0.158 | 0.06384 | 0.354 |
| 28524 | MEG3 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 17 | IRS1 | Sponge network | 0.249 | 0.35902 | -0.026 | 0.88855 | 0.354 |
| 28525 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p | 16 | ATRX | Sponge network | 1.147 | 0 | 0.261 | 0.04408 | 0.354 |
| 28526 | AF131215.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p | 20 | ERCC4 | Sponge network | -0.176 | 0.59692 | 0.126 | 0.15266 | 0.354 |
| 28527 | RP4-584D14.7 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-625-5p | 29 | LPP | Sponge network | 1.294 | 0.0031 | -0.459 | 0.04475 | 0.354 |
| 28528 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 33 | EPHA7 | Sponge network | 0.494 | 0.00339 | -2.197 | 3.0E-5 | 0.354 |
| 28529 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | DPYD | Sponge network | -1.439 | 4.0E-5 | -0.736 | 0.00076 | 0.354 |
| 28530 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | PLSCR4 | Sponge network | -0.668 | 3.0E-5 | -0.474 | 0.03833 | 0.354 |
| 28531 | PART1 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | NR3C2 | Sponge network | -2.925 | 0 | -1.56 | 0 | 0.354 |
| 28532 | LINC00578 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-589-3p | 10 | YAP1 | Sponge network | 0.811 | 0.03228 | 0.22 | 0.11836 | 0.354 |
| 28533 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p | 18 | RPS6KA2 | Sponge network | -0.942 | 0 | -0.57 | 0.00013 | 0.354 |
| 28534 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-940 | 32 | AKAP11 | Sponge network | 0.541 | 0.00125 | 0.196 | 0.09825 | 0.354 |
| 28535 | RP11-67L2.2 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | RNF180 | Sponge network | 0.72 | 0.0001 | -1.127 | 1.0E-5 | 0.354 |
| 28536 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 18 | PAFAH1B1 | Sponge network | -0.376 | 0.00337 | -0.429 | 0 | 0.354 |
| 28537 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p | 37 | PRKAA2 | Sponge network | 0.214 | 0.18246 | -2.462 | 0 | 0.354 |
| 28538 | AGAP11 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-629-3p | 16 | CDH23 | Sponge network | -1.645 | 0 | -0.974 | 0.00034 | 0.354 |
| 28539 | HAND2-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 48 | CREB3L2 | Sponge network | -1.697 | 0.00889 | -0.525 | 2.0E-5 | 0.354 |
| 28540 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | MLLT10 | Sponge network | 0.659 | 3.0E-5 | 0.105 | 0.33292 | 0.354 |
| 28541 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PTPRB | Sponge network | 0.101 | 0.60919 | 0.105 | 0.47122 | 0.354 |
| 28542 | CKMT2-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | KLF10 | Sponge network | 0.082 | 0.55654 | -0.783 | 0 | 0.354 |
| 28543 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-671-5p | 21 | KIT | Sponge network | -0.154 | 0.78939 | -2.184 | 0 | 0.354 |
| 28544 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | ABCB5 | Sponge network | -0.739 | 0.00044 | -0.956 | 0.04077 | 0.354 |
| 28545 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-93-5p;hsa-miR-940 | 26 | MCC | Sponge network | 0.082 | 0.55397 | -1.005 | 0 | 0.354 |
| 28546 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374a-3p | 14 | TMTC3 | Sponge network | 0.757 | 0 | 0.123 | 0.22189 | 0.354 |
| 28547 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-590-3p | 38 | THRB | Sponge network | -0.811 | 2.0E-5 | -1.117 | 0.0004 | 0.354 |
| 28548 | AGAP11 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-421;hsa-miR-590-5p | 17 | SPG20 | Sponge network | -1.645 | 0 | -1.16 | 0 | 0.354 |
| 28549 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | 0.61 | 0.01593 | -0.684 | 0.00159 | 0.354 |
| 28550 | WDFY3-AS2 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 24 | GJA1 | Sponge network | -0.942 | 0 | -0.485 | 0.02179 | 0.354 |
| 28551 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 12 | SUFU | Sponge network | -0.351 | 0.11358 | -0.04 | 0.65489 | 0.354 |
| 28552 | ACTA2-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | ID4 | Sponge network | 0.194 | 0.64278 | -1.644 | 0 | 0.354 |
| 28553 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p | 13 | ZMYND11 | Sponge network | 0.176 | 0.37402 | -0.2 | 0.05449 | 0.354 |
| 28554 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 38 | TMEM132B | Sponge network | 0.335 | 0.20596 | -0.83 | 0.01084 | 0.354 |
| 28555 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p | 17 | MLLT10 | Sponge network | 0.082 | 0.55397 | 0.105 | 0.33292 | 0.354 |
| 28556 | CTD-2554C21.2 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429 | 15 | GJA1 | Sponge network | -1.654 | 0.00144 | -0.485 | 0.02179 | 0.354 |
| 28557 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-940 | 23 | HLF | Sponge network | 0.389 | 0.04293 | -2.443 | 0 | 0.354 |
| 28558 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | CITED2 | Sponge network | -2.117 | 0.00161 | -1.545 | 0 | 0.354 |
| 28559 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | ABI2 | Sponge network | -2.452 | 0 | 0.175 | 0.14787 | 0.354 |
| 28560 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | EGLN1 | Sponge network | 0.401 | 0.00066 | -0.127 | 0.1536 | 0.354 |
| 28561 | OIP5-AS1 |
hsa-let-7e-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-497-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-942-5p | 30 | COPS2 | Sponge network | 0.421 | 0.00084 | -0.178 | 0.04974 | 0.354 |
| 28562 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 15 | DAB2 | Sponge network | -1.281 | 0.00012 | 0.431 | 0.0113 | 0.354 |
| 28563 | RP11-57H14.4 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | KIF5B | Sponge network | 0.083 | 0.66405 | 0.362 | 0.00066 | 0.354 |
| 28564 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | CDHR3 | Sponge network | -1.575 | 6.0E-5 | -0.977 | 4.0E-5 | 0.354 |
| 28565 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-7-1-3p | 12 | TSHZ3 | Sponge network | -0.285 | 0.2172 | -0.556 | 0.01516 | 0.354 |
| 28566 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 20 | CRTC3 | Sponge network | -0.777 | 0.16667 | 0.164 | 0.16786 | 0.354 |
| 28567 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 34 | FBXO32 | Sponge network | -0.355 | 0.0077 | -0.495 | 0.07561 | 0.354 |
| 28568 | RP11-46C24.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TET1 | Sponge network | 0.392 | 0.0278 | -0.135 | 0.52138 | 0.354 |
| 28569 | A1BG-AS1 |
hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-425-5p;hsa-miR-532-3p | 11 | PTPRT | Sponge network | -0.164 | 0.45106 | -0.907 | 0.03727 | 0.354 |
| 28570 | LINC01018 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ZNF208 | Sponge network | -2.915 | 0.00015 | -0.4 | 0.22381 | 0.354 |
| 28571 | RP11-1277A3.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-409-3p;hsa-miR-511-5p | 13 | ATRX | Sponge network | 0.453 | 0.01839 | 0.261 | 0.04408 | 0.354 |
| 28572 | RP11-307B6.3 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-338-3p;hsa-miR-421 | 11 | LIMCH1 | Sponge network | -4.463 | 0.00025 | -0.915 | 1.0E-5 | 0.354 |
| 28573 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | NCOA1 | Sponge network | -0.976 | 0.00025 | -0.432 | 0 | 0.354 |
| 28574 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-423-5p;hsa-miR-629-3p | 24 | NTRK3 | Sponge network | -2.549 | 0.00528 | -1.77 | 3.0E-5 | 0.354 |
| 28575 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -1.111 | 0.0003 | 0.309 | 0.17079 | 0.354 |
| 28576 | RP11-384O8.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 13 | SORCS1 | Sponge network | -0.738 | 0.21233 | -3.492 | 0 | 0.354 |
| 28577 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577;hsa-miR-671-5p;hsa-miR-93-5p | 37 | PTPRD | Sponge network | -2.69 | 0.0001 | -1.005 | 0.00064 | 0.354 |
| 28578 | RP11-53O19.1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | SPOP | Sponge network | -0.405 | 0.22013 | -0.419 | 1.0E-5 | 0.354 |
| 28579 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-942-5p | 16 | FBXO31 | Sponge network | 0.997 | 0.00066 | -0.085 | 0.42389 | 0.354 |
| 28580 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-940 | 31 | NTRK2 | Sponge network | 0.299 | 0.57722 | -0.736 | 0.06495 | 0.354 |
| 28581 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-653-5p | 12 | PDS5B | Sponge network | 0.082 | 0.55397 | 0.259 | 0.00962 | 0.354 |
| 28582 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 58 | KIF1B | Sponge network | -0.709 | 0.18168 | -0.118 | 0.32258 | 0.354 |
| 28583 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | DCLK1 | Sponge network | -0.206 | 0.12942 | -1.324 | 0.00014 | 0.354 |
| 28584 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-3p | 25 | QKI | Sponge network | 0.178 | 0.29106 | -0.333 | 0.04111 | 0.354 |
| 28585 | RP11-244O19.1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 10 | FAM117A | Sponge network | -0.096 | 0.67958 | -1.379 | 0 | 0.354 |
| 28586 | AC006116.24 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-33a-5p | 11 | EYA4 | Sponge network | -0.473 | 0.07602 | 0.3 | 0.36876 | 0.354 |
| 28587 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | TACC1 | Sponge network | 0.342 | 0.03129 | -0.362 | 0.05406 | 0.354 |
| 28588 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 44 | BMPR2 | Sponge network | -0.031 | 0.89063 | 0.206 | 0.0635 | 0.354 |
| 28589 | PGM5-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421 | 12 | MAFB | Sponge network | -4.546 | 0 | -0.023 | 0.89865 | 0.354 |
| 28590 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-532-5p | 19 | BNC2 | Sponge network | -0.829 | 0.01343 | -0.491 | 0.09988 | 0.354 |
| 28591 | CTC-429P9.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-877-5p | 10 | ASTN1 | Sponge network | 0.043 | 0.81628 | -2.389 | 2.0E-5 | 0.354 |
| 28592 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 19 | KIF1B | Sponge network | 1.597 | 0 | -0.118 | 0.32258 | 0.354 |
| 28593 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 27 | PCDH17 | Sponge network | -0.576 | 0.10121 | 0.731 | 2.0E-5 | 0.354 |
| 28594 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-590-5p;hsa-miR-93-5p | 21 | RNASEL | Sponge network | -2.117 | 0.00161 | -0.272 | 0.01796 | 0.354 |
| 28595 | RP11-819C21.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PRUNE2 | Sponge network | 0.537 | 0.00139 | -1.733 | 0.00022 | 0.354 |
| 28596 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p | 42 | SSBP2 | Sponge network | 0.082 | 0.55654 | -1.58 | 0 | 0.354 |
| 28597 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-3p | 50 | KIF1B | Sponge network | 0.214 | 0.18246 | -0.118 | 0.32258 | 0.354 |
| 28598 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-539-5p | 13 | HECA | Sponge network | 0.858 | 3.0E-5 | -0.217 | 0.03463 | 0.354 |
| 28599 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | TSHZ3 | Sponge network | -0.206 | 0.12942 | -0.556 | 0.01516 | 0.354 |
| 28600 | LENG8-AS1 |
hsa-let-7a-3p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-664a-5p | 14 | SOX11 | Sponge network | 1.471 | 0 | 2.68 | 0 | 0.354 |
| 28601 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-629-3p | 25 | RASSF2 | Sponge network | -0.154 | 0.53954 | -0.273 | 0.21961 | 0.354 |
| 28602 | LPP-AS2 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-96-5p | 22 | EFNA5 | Sponge network | -0.18 | 0.20343 | -0.421 | 0.17868 | 0.354 |
| 28603 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | DACH2 | Sponge network | -2.452 | 0 | -1.635 | 0.00034 | 0.354 |
| 28604 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 17 | NR4A3 | Sponge network | -2.117 | 0.00161 | -1.385 | 0 | 0.354 |
| 28605 | AC090587.5 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-589-3p;hsa-miR-93-3p | 11 | TRPS1 | Sponge network | -0.088 | 0.65011 | -0.191 | 0.44109 | 0.354 |
| 28606 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | JAZF1 | Sponge network | 0.178 | 0.29106 | -0.377 | 0.02677 | 0.354 |
| 28607 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-378c;hsa-miR-429;hsa-miR-96-5p | 21 | MED12L | Sponge network | -1.654 | 0.00144 | -0.194 | 0.55299 | 0.354 |
| 28608 | SNHG14 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 30 | CSMD1 | Sponge network | -1.111 | 0.0003 | -0.63 | 0.18881 | 0.354 |
| 28609 | CTD-3126B10.4 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | ZFHX3 | Sponge network | 0.778 | 0.00101 | 0.389 | 0.01363 | 0.354 |
| 28610 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-940 | 29 | PTPRT | Sponge network | -0.318 | 0.65847 | -0.907 | 0.03727 | 0.354 |
| 28611 | CTC-487M23.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-664a-5p;hsa-miR-92a-3p | 35 | LPP | Sponge network | 0.912 | 0.00014 | -0.459 | 0.04475 | 0.354 |
| 28612 | RP11-158K1.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p | 11 | FBXO11 | Sponge network | 0.349 | 0.01695 | 0.142 | 0.04938 | 0.354 |
| 28613 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-3p | 23 | ZFHX3 | Sponge network | -0.693 | 0.05629 | 0.389 | 0.01363 | 0.354 |
| 28614 | LINC00702 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p | 10 | DUSP1 | Sponge network | -0.737 | 0.06182 | -1.754 | 0 | 0.354 |
| 28615 | AC108676.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-628-5p | 12 | TRIM33 | Sponge network | 4.346 | 2.0E-5 | 0.402 | 7.0E-5 | 0.354 |
| 28616 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-7-5p | 15 | CLASP2 | Sponge network | -1.331 | 3.0E-5 | -0.038 | 0.71 | 0.354 |
| 28617 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 17 | CLIP1 | Sponge network | 0.249 | 0.35902 | -0.215 | 0.08684 | 0.354 |
| 28618 | WDFY3-AS2 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-935 | 12 | STAT5B | Sponge network | -0.942 | 0 | -0.182 | 0.1082 | 0.354 |
| 28619 | RP11-244O19.1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-877-5p | 15 | GLIPR1 | Sponge network | -0.096 | 0.67958 | 0.146 | 0.3276 | 0.354 |
| 28620 | RP11-67L2.2 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | KIF5B | Sponge network | 0.72 | 0.0001 | 0.362 | 0.00066 | 0.354 |
| 28621 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p | 11 | THSD7A | Sponge network | -0.738 | 0.21233 | 0.242 | 0.25154 | 0.354 |
| 28622 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -0.323 | 0.01372 | -1.692 | 0 | 0.354 |
| 28623 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | ZMYM2 | Sponge network | 0.539 | 0.03722 | 0.279 | 0.01273 | 0.354 |
| 28624 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 15 | DKK3 | Sponge network | -0.405 | 0.22013 | -0.052 | 0.77783 | 0.354 |
| 28625 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-93-3p | 21 | DOCK4 | Sponge network | 0.253 | 0.09565 | 0.502 | 0.00108 | 0.354 |
| 28626 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 47 | TRPS1 | Sponge network | -0.612 | 0.00094 | -0.191 | 0.44109 | 0.354 |
| 28627 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | 0.4 | 0.14611 | 0.024 | 0.89889 | 0.354 |
| 28628 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 53 | CCND2 | Sponge network | -1.575 | 6.0E-5 | -0.267 | 0.3112 | 0.354 |
| 28629 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p | 28 | GPC6 | Sponge network | -0.096 | 0.67958 | -0.054 | 0.81465 | 0.354 |
| 28630 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-577 | 26 | PHC3 | Sponge network | -0.18 | 0.20343 | 0.212 | 0.0499 | 0.354 |
| 28631 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p | 17 | LATS1 | Sponge network | 0.873 | 0 | 0.109 | 0.34208 | 0.354 |
| 28632 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-93-3p | 27 | NCOA1 | Sponge network | -0.154 | 0.78939 | -0.432 | 0 | 0.354 |
| 28633 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374b-5p;hsa-miR-450b-5p | 14 | DIP2C | Sponge network | 0.912 | 0.00014 | -0.436 | 0.01713 | 0.354 |
| 28634 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-532-3p | 12 | RPS6KA2 | Sponge network | -0.246 | 0.35034 | -0.57 | 0.00013 | 0.354 |
| 28635 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-625-3p | 24 | RAP1A | Sponge network | -0.227 | 0.31657 | -0.929 | 0 | 0.354 |
| 28636 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p | 10 | UVRAG | Sponge network | 0.056 | 0.81195 | -0.017 | 0.83865 | 0.354 |
| 28637 | RP11-166D19.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | LUZP2 | Sponge network | -0.576 | 0.10121 | -0.056 | 0.89416 | 0.354 |
| 28638 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | MEF2C | Sponge network | -2.915 | 0.00015 | -0.499 | 0.01448 | 0.354 |
| 28639 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-92a-3p | 10 | ZNF521 | Sponge network | 0.657 | 4.0E-5 | -0.111 | 0.58068 | 0.354 |
| 28640 | AC005618.6 |
hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | ZNF382 | Sponge network | 0.862 | 0.06213 | -0.494 | 0.03831 | 0.354 |
| 28641 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-96-5p | 13 | NUAK1 | Sponge network | 3.345 | 0 | 0.904 | 0 | 0.354 |
| 28642 | CTA-941F9.9 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-576-5p;hsa-miR-935 | 13 | SSBP2 | Sponge network | -0.344 | 0.49624 | -1.58 | 0 | 0.354 |
| 28643 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 41 | RBMS3 | Sponge network | -0.355 | 0.0077 | -0.519 | 0.03551 | 0.354 |
| 28644 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-628-5p | 17 | GRIA3 | Sponge network | -0.154 | 0.53954 | -1.956 | 0 | 0.354 |
| 28645 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-942-5p | 15 | NOTCH2 | Sponge network | 1.316 | 0.01106 | 0.138 | 0.35163 | 0.354 |
| 28646 | EMX2OS |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | EPB41L3 | Sponge network | -0.777 | 0.1134 | -1.193 | 0 | 0.354 |
| 28647 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CLASP2 | Sponge network | 0.545 | 0.00064 | -0.038 | 0.71 | 0.354 |
| 28648 | HAR1A |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-744-3p | 11 | ADAMTSL3 | Sponge network | -0.672 | 0.19447 | -1.559 | 0 | 0.354 |
| 28649 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-484 | 11 | CHD5 | Sponge network | -2.549 | 0.00528 | -0.922 | 0.01521 | 0.354 |
| 28650 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-93-3p | 12 | DOCK4 | Sponge network | 1.46 | 1.0E-5 | 0.502 | 0.00108 | 0.354 |
| 28651 | RP11-53O19.1 |
hsa-miR-142-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 11 | CAV1 | Sponge network | -0.405 | 0.22013 | -1.013 | 6.0E-5 | 0.354 |
| 28652 | RP11-536O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 15 | RASSF2 | Sponge network | -0.074 | 0.90758 | -0.273 | 0.21961 | 0.354 |
| 28653 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SMARCA2 | Sponge network | -0.129 | 0.57177 | -0.529 | 0.00014 | 0.353 |
| 28654 | LINC00957 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | FLI1 | Sponge network | -0.421 | 0.0114 | -0.294 | 0.10905 | 0.353 |
| 28655 | SNHG14 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 35 | MXI1 | Sponge network | -1.111 | 0.0003 | -0.987 | 0 | 0.353 |
| 28656 | GABPB1-AS1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-744-3p | 18 | KIF5B | Sponge network | 1.11 | 0 | 0.362 | 0.00066 | 0.353 |
| 28657 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-874-3p | 11 | BCLAF1 | Sponge network | 0.637 | 0.00069 | 0.211 | 0.00684 | 0.353 |
| 28658 | DNAJC3-AS1 |
hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-5p | 14 | CREB1 | Sponge network | 0.753 | 0.00014 | 0.203 | 0.00537 | 0.353 |
| 28659 | RP11-307B6.3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p | 13 | ZFHX3 | Sponge network | -4.463 | 0.00025 | 0.389 | 0.01363 | 0.353 |
| 28660 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | BMPR2 | Sponge network | 0.539 | 0.03722 | 0.206 | 0.0635 | 0.353 |
| 28661 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | -0.769 | 0.00035 | -1.465 | 0.00033 | 0.353 |
| 28662 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-539-5p | 10 | MGA | Sponge network | -0.563 | 0.02545 | 0.323 | 0.00792 | 0.353 |
| 28663 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-625-5p;hsa-miR-93-5p | 22 | JAZF1 | Sponge network | 0.259 | 0.12542 | -0.377 | 0.02677 | 0.353 |
| 28664 | RP1-257A7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p | 14 | TCF4 | Sponge network | 0.849 | 0.00178 | 0.016 | 0.92271 | 0.353 |
| 28665 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-590-3p | 23 | MXI1 | Sponge network | -0.096 | 0.67958 | -0.987 | 0 | 0.353 |
| 28666 | LINC00173 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-96-5p | 15 | CYLD | Sponge network | 0.056 | 0.8494 | -0.367 | 0.00171 | 0.353 |
| 28667 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | LRRC7 | Sponge network | -2.915 | 0.00015 | -1.341 | 0.01193 | 0.353 |
| 28668 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | THBS1 | Sponge network | -0.709 | 0.18168 | 0.741 | 0.00386 | 0.353 |
| 28669 | RP11-59H7.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-576-5p | 14 | BRD3 | Sponge network | 2.163 | 0 | 0.37 | 0.00013 | 0.353 |
| 28670 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-7-1-3p | 10 | SLC6A15 | Sponge network | -4.477 | 0 | -2.373 | 2.0E-5 | 0.353 |
| 28671 | RP11-686D22.4 |
hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-577;hsa-miR-590-3p | 12 | RUNX1T1 | Sponge network | 0.793 | 5.0E-5 | -0.344 | 0.2374 | 0.353 |
| 28672 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | EYA4 | Sponge network | -0.323 | 0.01372 | 0.3 | 0.36876 | 0.353 |
| 28673 | MRPL23-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p | 13 | MOB1B | Sponge network | -0.278 | 0.76559 | 0.197 | 0.15266 | 0.353 |
| 28674 | RP11-774O3.3 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-874-3p | 14 | ARID1B | Sponge network | -0.487 | 0.02157 | 0.003 | 0.97503 | 0.353 |
| 28675 | AC093627.8 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-7-5p | 10 | ESR1 | Sponge network | -0.787 | 0.3928 | -1.035 | 3.0E-5 | 0.353 |
| 28676 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 47 | PPP1R9A | Sponge network | -2.46 | 0 | -0.903 | 0.04254 | 0.353 |
| 28677 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | RBMS3 | Sponge network | 0.559 | 0.00026 | -0.519 | 0.03551 | 0.353 |
| 28678 | HOXD-AS2 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-502-5p | 24 | AR | Sponge network | -0.208 | 0.65592 | -1.554 | 0 | 0.353 |
| 28679 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-582-5p | 26 | FBXO32 | Sponge network | 0.95 | 0.15943 | -0.495 | 0.07561 | 0.353 |
| 28680 | HOTAIRM1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | DIP2C | Sponge network | -0.053 | 0.80685 | -0.436 | 0.01713 | 0.353 |
| 28681 | RP11-57H14.4 |
hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-935;hsa-miR-940 | 12 | PHOX2B | Sponge network | 0.083 | 0.66405 | -2.68 | 0 | 0.353 |
| 28682 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 23 | TIMP3 | Sponge network | 0.374 | 0.20054 | -0.546 | 0.02307 | 0.353 |
| 28683 | LINC00672 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 48 | NFIB | Sponge network | -0.227 | 0.31657 | 0.023 | 0.90795 | 0.353 |
| 28684 | CTD-2528L19.6 |
hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TMEM30A | Sponge network | 0.953 | 0 | 0.096 | 0.31031 | 0.353 |
| 28685 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 25 | LRRC7 | Sponge network | -1.281 | 0.00012 | -1.341 | 0.01193 | 0.353 |
| 28686 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 20 | BCL11A | Sponge network | -0.976 | 0.00025 | -0.932 | 0.0063 | 0.353 |
| 28687 | HOTAIRM1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | SYNPO2 | Sponge network | -0.053 | 0.80685 | -2.342 | 0 | 0.353 |
| 28688 | RP11-307B6.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-589-3p | 15 | AFF1 | Sponge network | -4.463 | 0.00025 | -0.384 | 0.00053 | 0.353 |
| 28689 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-93-3p;hsa-miR-940 | 14 | AKAP11 | Sponge network | 0.453 | 0.01839 | 0.196 | 0.09825 | 0.353 |
| 28690 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 30 | USP12 | Sponge network | 0.603 | 0.00127 | -0.102 | 0.40768 | 0.353 |
| 28691 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | AKAP11 | Sponge network | 0.272 | 0.44692 | 0.196 | 0.09825 | 0.353 |
| 28692 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-93-3p | 12 | PTPN11 | Sponge network | 2.163 | 0 | 0.431 | 2.0E-5 | 0.353 |
| 28693 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-935;hsa-miR-96-5p | 16 | FBXO32 | Sponge network | 0.37 | 0.27614 | -0.495 | 0.07561 | 0.353 |
| 28694 | RP11-283I3.6 |
hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | GSK3B | Sponge network | 0.614 | 1.0E-5 | 0.266 | 0.00045 | 0.353 |
| 28695 | PAN3-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p | 13 | CBX5 | Sponge network | 0.062 | 0.55274 | 0.165 | 0.24446 | 0.353 |
| 28696 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -0.612 | 0.00094 | -0.354 | 0.14694 | 0.353 |
| 28697 | LINC00865 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | -1.249 | 7.0E-5 | 0.783 | 0.00072 | 0.353 |
| 28698 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 43 | CADM2 | Sponge network | 0.225 | 0.30644 | -3.782 | 0 | 0.353 |
| 28699 | LINC00865 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 27 | KCNA6 | Sponge network | -1.249 | 7.0E-5 | -1.531 | 0.00013 | 0.353 |
| 28700 | HOXA-AS2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-590-3p | 28 | ABL2 | Sponge network | 0.61 | 0.01593 | 0.82 | 0 | 0.353 |
| 28701 | RP1-59D14.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 16 | TBL1XR1 | Sponge network | 1.162 | 5.0E-5 | 0.578 | 0 | 0.353 |
| 28702 | SH3BP5-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-940 | 23 | SIRT1 | Sponge network | 0.267 | 0.2937 | 0.109 | 0.2593 | 0.353 |
| 28703 | RP11-46C24.7 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | 0.392 | 0.0278 | 0.358 | 0.13727 | 0.353 |
| 28704 | CTB-113P19.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | TRPS1 | Sponge network | 0.906 | 0.31715 | -0.191 | 0.44109 | 0.353 |
| 28705 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 33 | PTPRT | Sponge network | -0.777 | 0.16667 | -0.907 | 0.03727 | 0.353 |
| 28706 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-577;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p | 35 | CREB3L2 | Sponge network | -2.69 | 0.0001 | -0.525 | 2.0E-5 | 0.353 |
| 28707 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-629-3p | 43 | CBX5 | Sponge network | -0.154 | 0.78939 | 0.165 | 0.24446 | 0.353 |
| 28708 | RP11-798M19.6 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 10 | SPTBN4 | Sponge network | -0.612 | 0.00094 | -0.786 | 0.01281 | 0.353 |
| 28709 | ARHGAP5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-542-3p | 11 | KANK1 | Sponge network | -0.644 | 0.00021 | -0.55 | 0.00134 | 0.353 |
| 28710 | DNM1P35 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-590-3p | 12 | MRVI1 | Sponge network | 0.051 | 0.83388 | -1.051 | 0.00022 | 0.353 |
| 28711 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | KCNA6 | Sponge network | 0.4 | 0.14611 | -1.531 | 0.00013 | 0.353 |
| 28712 | TP73-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 81 | LPP | Sponge network | -0.563 | 0.02545 | -0.459 | 0.04475 | 0.353 |
| 28713 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 26 | TBL1XR1 | Sponge network | 1.11 | 0 | 0.578 | 0 | 0.353 |
| 28714 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | PPM1A | Sponge network | -0.376 | 0.00337 | -0.623 | 0 | 0.353 |
| 28715 | H1FX-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p | 15 | SYNPO2 | Sponge network | -0.206 | 0.12942 | -2.342 | 0 | 0.353 |
| 28716 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p | 12 | MOB1B | Sponge network | 1.316 | 0 | 0.197 | 0.15266 | 0.353 |
| 28717 | A1BG-AS1 |
hsa-miR-141-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-671-5p;hsa-miR-93-3p | 16 | GRIK3 | Sponge network | -0.164 | 0.45106 | -3.208 | 0 | 0.353 |
| 28718 | AC025171.1 |
hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | DOCK4 | Sponge network | 0.998 | 0 | 0.502 | 0.00108 | 0.353 |
| 28719 | U91328.21 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b | 13 | DMD | Sponge network | -0.735 | 0.15983 | -1.506 | 0 | 0.353 |
| 28720 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-335-3p | 11 | APC | Sponge network | 0.497 | 0.01451 | -0.255 | 0.04051 | 0.353 |
| 28721 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ZFHX4 | Sponge network | -1.057 | 0.00029 | 0.018 | 0.95574 | 0.353 |
| 28722 | LENG8-AS1 |
hsa-let-7a-3p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-374a-3p;hsa-miR-539-5p | 10 | MOB1B | Sponge network | 1.471 | 0 | 0.197 | 0.15266 | 0.353 |
| 28723 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ZMYND11 | Sponge network | 0.997 | 0.00066 | -0.2 | 0.05449 | 0.353 |
| 28724 | AC005154.6 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | ERCC6 | Sponge network | 0.848 | 0 | 0.23 | 0.015 | 0.353 |
| 28725 | H1FX-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 17 | ESR1 | Sponge network | -0.206 | 0.12942 | -1.035 | 3.0E-5 | 0.353 |
| 28726 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | ARNT2 | Sponge network | 0.532 | 0.00505 | -0.332 | 0.25851 | 0.353 |
| 28727 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-96-5p | 27 | KIF1B | Sponge network | 0.37 | 0.27614 | -0.118 | 0.32258 | 0.353 |
| 28728 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | ABCC9 | Sponge network | -0.355 | 0.0077 | -0.466 | 0.14121 | 0.353 |
| 28729 | RP11-155G14.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-539-5p | 13 | DDX6 | Sponge network | 2.483 | 0 | 0.091 | 0.36428 | 0.353 |
| 28730 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-92b-3p | 10 | AKAP12 | Sponge network | 0.4 | 0.14611 | -0.932 | 0.0028 | 0.353 |
| 28731 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 0.998 | 0 | 0.943 | 0 | 0.353 |
| 28732 | FAM13A-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | UNC5D | Sponge network | -0.83 | 0 | -3.646 | 0 | 0.353 |
| 28733 | CTD-2554C21.2 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-429 | 16 | RIMS2 | Sponge network | -1.654 | 0.00144 | -1.365 | 0.00208 | 0.353 |
| 28734 | RP5-1047A19.4 |
hsa-miR-125a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p | 11 | CCDC6 | Sponge network | 0.745 | 1.0E-5 | 0.27 | 0.02555 | 0.353 |
| 28735 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 43 | BNC2 | Sponge network | 0.083 | 0.66405 | -0.491 | 0.09988 | 0.353 |
| 28736 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | PRKAB2 | Sponge network | -0.513 | 0.04703 | -0.367 | 0.01371 | 0.353 |
| 28737 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 24 | NRXN1 | Sponge network | -0.08 | 0.90092 | -3.778 | 0 | 0.353 |
| 28738 | LINC00926 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 20 | TCF4 | Sponge network | 0.756 | 0.00739 | 0.016 | 0.92271 | 0.353 |
| 28739 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p | 21 | NUAK1 | Sponge network | 0.253 | 0.09565 | 0.904 | 0 | 0.353 |
| 28740 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 42 | CADM2 | Sponge network | 0.4 | 0.14611 | -3.782 | 0 | 0.353 |
| 28741 | RP11-212P7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 47 | RUNX1T1 | Sponge network | 0.219 | 0.1516 | -0.344 | 0.2374 | 0.353 |
| 28742 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-628-5p | 23 | GRIA3 | Sponge network | -0.162 | 0.47687 | -1.956 | 0 | 0.353 |
| 28743 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p | 43 | CHD5 | Sponge network | -0.727 | 0.04726 | -0.922 | 0.01521 | 0.353 |
| 28744 | LINC00657 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-590-3p | 16 | DDX3X | Sponge network | 0.91 | 0 | -0.152 | 0.07686 | 0.353 |
| 28745 | ZNF582-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-7-1-3p | 18 | RIMS2 | Sponge network | -1.349 | 0.00017 | -1.365 | 0.00208 | 0.353 |
| 28746 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-940 | 31 | MDM4 | Sponge network | 0.419 | 0.0319 | 0.345 | 0.00762 | 0.353 |
| 28747 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 28 | USP12 | Sponge network | 0.997 | 0.00066 | -0.102 | 0.40768 | 0.353 |
| 28748 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | SCN9A | Sponge network | -0.355 | 0.0077 | -0.525 | 0.10593 | 0.353 |
| 28749 | RP11-182J1.1 |
hsa-miR-135b-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | MOB1B | Sponge network | -0.187 | 0.23787 | 0.197 | 0.15266 | 0.353 |
| 28750 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p | 28 | ZFHX3 | Sponge network | 0.853 | 0.15352 | 0.389 | 0.01363 | 0.353 |
| 28751 | DIO3OS |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-769-5p | 13 | PRUNE2 | Sponge network | -0.773 | 0.04073 | -1.733 | 0.00022 | 0.353 |
| 28752 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 29 | ARHGAP28 | Sponge network | -0.739 | 0.0574 | -0.911 | 0.00013 | 0.353 |
| 28753 | CTC-559E9.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 14 | ERCC4 | Sponge network | 0.594 | 0.00064 | 0.126 | 0.15266 | 0.353 |
| 28754 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 27 | ARHGAP28 | Sponge network | -1.216 | 0 | -0.911 | 0.00013 | 0.353 |
| 28755 | RP1-122P22.2 |
hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 17 | NTRK3 | Sponge network | 0.065 | 0.73468 | -1.77 | 3.0E-5 | 0.353 |
| 28756 | PWAR6 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | USP25 | Sponge network | -1.575 | 6.0E-5 | -0.242 | 0.01397 | 0.353 |
| 28757 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 13 | LRRC55 | Sponge network | 0.225 | 0.30644 | -0.026 | 0.93163 | 0.353 |
| 28758 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484 | 12 | PNRC1 | Sponge network | -2.69 | 0.0001 | -0.646 | 0 | 0.353 |
| 28759 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-590-3p | 17 | CADM1 | Sponge network | -0.358 | 0.17255 | -0.9 | 0.00084 | 0.353 |
| 28760 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-421;hsa-miR-424-5p | 10 | DYNC1I1 | Sponge network | -4.477 | 0 | -1.12 | 0.00107 | 0.353 |
| 28761 | LINC00403 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-424-5p | 10 | SYT4 | Sponge network | -3.586 | 0.00032 | -3.207 | 0 | 0.353 |
| 28762 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 12 | SLITRK3 | Sponge network | -0.141 | 0.26434 | -3.328 | 0 | 0.353 |
| 28763 | LINC00578 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-664a-3p | 11 | SOX11 | Sponge network | 0.811 | 0.03228 | 2.68 | 0 | 0.353 |
| 28764 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 17 | LRRC7 | Sponge network | -0.227 | 0.31657 | -1.341 | 0.01193 | 0.353 |
| 28765 | RP11-888D10.4 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 17 | ABL2 | Sponge network | 0.702 | 7.0E-5 | 0.82 | 0 | 0.353 |
| 28766 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 20 | CHD2 | Sponge network | 0.415 | 0.14657 | 0.049 | 0.55371 | 0.353 |
| 28767 | LINC00403 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-424-5p | 12 | PPM1A | Sponge network | -3.586 | 0.00032 | -0.623 | 0 | 0.353 |
| 28768 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 20 | MITF | Sponge network | -1.203 | 0.03881 | -0.769 | 0.00022 | 0.353 |
| 28769 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 26 | PBX1 | Sponge network | 0.389 | 0.04293 | -1.206 | 0 | 0.353 |
| 28770 | THAP9-AS1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-664a-3p | 13 | PRKAA1 | Sponge network | 1.079 | 0 | 0.317 | 0.0031 | 0.353 |
| 28771 | RP11-774O3.3 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | RTN4 | Sponge network | -0.487 | 0.02157 | -0.412 | 0.00014 | 0.353 |
| 28772 | MYLK-AS1 |
hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-484 | 17 | CADM1 | Sponge network | -1.331 | 3.0E-5 | -0.9 | 0.00084 | 0.353 |
| 28773 | MIR600HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-589-3p | 19 | PRDM2 | Sponge network | 0.594 | 0.41522 | -0.258 | 0.00721 | 0.353 |
| 28774 | RP11-597D13.9 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-532-3p | 15 | SYNM | Sponge network | 1.302 | 7.0E-5 | -2.309 | 1.0E-5 | 0.353 |
| 28775 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | ZMYM2 | Sponge network | 0.769 | 0 | 0.279 | 0.01273 | 0.353 |
| 28776 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 42 | NOTCH2 | Sponge network | -2.452 | 0 | 0.138 | 0.35163 | 0.353 |
| 28777 | HCG18 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | ERCC4 | Sponge network | 1.063 | 0 | 0.126 | 0.15266 | 0.353 |
| 28778 | RP11-48B3.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p | 31 | DST | Sponge network | 1.757 | 0 | -0.713 | 0.00096 | 0.353 |
| 28779 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 39 | NTRK2 | Sponge network | 0.064 | 0.72934 | -0.736 | 0.06495 | 0.353 |
| 28780 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 11 | NEDD4 | Sponge network | -0.469 | 0.08721 | 0.02 | 0.89834 | 0.353 |
| 28781 | BVES-AS1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-589-5p | 10 | SORCS2 | Sponge network | -2.62 | 0 | -0.223 | 0.4253 | 0.353 |
| 28782 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-590-3p | 10 | MAPK10 | Sponge network | 0.727 | 3.0E-5 | -1.774 | 0 | 0.353 |
| 28783 | LINC00094 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-299-5p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-655-3p | 12 | NF1 | Sponge network | 0.253 | 0.09565 | 0.51 | 0 | 0.353 |
| 28784 | RP11-517B11.7 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-339-5p;hsa-miR-590-5p | 10 | MAP3K4 | Sponge network | 0.706 | 0 | 0.058 | 0.54229 | 0.353 |
| 28785 | BVES-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-874-3p;hsa-miR-98-5p | 31 | CREB3L2 | Sponge network | -2.62 | 0 | -0.525 | 2.0E-5 | 0.353 |
| 28786 | CTA-204B4.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p | 34 | DST | Sponge network | 0.765 | 7.0E-5 | -0.713 | 0.00096 | 0.353 |
| 28787 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | LRRC4 | Sponge network | -0.709 | 0.18168 | -1.774 | 0 | 0.353 |
| 28788 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 22 | MYBL1 | Sponge network | 0.657 | 4.0E-5 | 0.494 | 0.02152 | 0.353 |
| 28789 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 33 | ATRX | Sponge network | 0.249 | 0.35902 | 0.261 | 0.04408 | 0.353 |
| 28790 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-582-5p | 32 | FBXO32 | Sponge network | 0.253 | 0.09565 | -0.495 | 0.07561 | 0.353 |
| 28791 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 54 | IL6ST | Sponge network | -0.513 | 0.04703 | -0.402 | 0.00676 | 0.353 |
| 28792 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 18 | BCL2 | Sponge network | -1.375 | 0.08642 | -0.899 | 0 | 0.353 |
| 28793 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p | 18 | ABI2 | Sponge network | -4.463 | 0.00025 | 0.175 | 0.14787 | 0.353 |
| 28794 | RP11-148O21.2 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-500b-5p | 11 | ACSL6 | Sponge network | 0.233 | 0.81029 | -1.593 | 0 | 0.353 |
| 28795 | PSMG3-AS1 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 23 | DLC1 | Sponge network | -0.125 | 0.49414 | -0.382 | 0.05038 | 0.353 |
| 28796 | LINC00680 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 16 | TBL1XR1 | Sponge network | 0.884 | 0 | 0.578 | 0 | 0.353 |
| 28797 | RP5-890O3.9 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-495-3p | 10 | TBL1XR1 | Sponge network | 0.842 | 0 | 0.578 | 0 | 0.353 |
| 28798 | RP11-244H3.1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | DOCK4 | Sponge network | 0.659 | 3.0E-5 | 0.502 | 0.00108 | 0.353 |
| 28799 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p | 21 | PCDH17 | Sponge network | -0.096 | 0.67958 | 0.731 | 2.0E-5 | 0.353 |
| 28800 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-92a-3p;hsa-miR-940 | 35 | PPP1R9A | Sponge network | 0.056 | 0.81195 | -0.903 | 0.04254 | 0.353 |
| 28801 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 53 | ERBB4 | Sponge network | -2.925 | 0 | -1.975 | 2.0E-5 | 0.353 |
| 28802 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-590-3p | 18 | RET | Sponge network | -0.793 | 7.0E-5 | -0.833 | 0.03517 | 0.353 |
| 28803 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-590-3p | 12 | PTPN11 | Sponge network | 0.67 | 0.00048 | 0.431 | 2.0E-5 | 0.353 |
| 28804 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p | 17 | SASH1 | Sponge network | 0.594 | 0.41522 | -0.502 | 0.00175 | 0.353 |
| 28805 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-671-5p;hsa-miR-940 | 24 | CHD5 | Sponge network | 0.222 | 0.29133 | -0.922 | 0.01521 | 0.353 |
| 28806 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-582-5p | 12 | DDX6 | Sponge network | 0.832 | 0 | 0.091 | 0.36428 | 0.353 |
| 28807 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-93-5p | 14 | IGSF10 | Sponge network | -1.645 | 0 | -1.751 | 1.0E-5 | 0.353 |
| 28808 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 26 | PHC3 | Sponge network | 0.401 | 0.00066 | 0.212 | 0.0499 | 0.353 |
| 28809 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-93-5p | 11 | FBXO31 | Sponge network | 0.299 | 0.57722 | -0.085 | 0.42389 | 0.353 |
| 28810 | FAM215B |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 18 | ABI2 | Sponge network | 0.817 | 3.0E-5 | 0.175 | 0.14787 | 0.353 |
| 28811 | RP11-1006G14.4 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p | 15 | RYR2 | Sponge network | 0.559 | 0.00026 | -1.466 | 0.00031 | 0.353 |
| 28812 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | UBE2QL1 | Sponge network | -0.942 | 0 | -2.417 | 0 | 0.353 |
| 28813 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484 | 11 | ITGA5 | Sponge network | 0.176 | 0.37402 | 0.452 | 0.03524 | 0.353 |
| 28814 | MYLK-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p | 11 | DMXL1 | Sponge network | -1.331 | 3.0E-5 | -0.426 | 0.00056 | 0.353 |
| 28815 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | -0.053 | 0.80685 | -1.751 | 1.0E-5 | 0.353 |
| 28816 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-532-3p;hsa-miR-940 | 21 | PTPRT | Sponge network | -0.829 | 0.01343 | -0.907 | 0.03727 | 0.353 |
| 28817 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-508-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-5p | 27 | RBMS3 | Sponge network | 0.051 | 0.83388 | -0.519 | 0.03551 | 0.353 |
| 28818 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-877-5p | 23 | PGR | Sponge network | 0.395 | 0.15451 | -1.704 | 0 | 0.353 |
| 28819 | RP5-1139B12.3 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 10 | MAFB | Sponge network | 0.598 | 0.38481 | -0.023 | 0.89865 | 0.353 |
| 28820 | CTD-3092A11.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | EPS15 | Sponge network | 0.873 | 0 | -0.052 | 0.53998 | 0.353 |
| 28821 | RP1-151F17.2 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | KIF5B | Sponge network | 0.222 | 0.29133 | 0.362 | 0.00066 | 0.353 |
| 28822 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 23 | PHC3 | Sponge network | 0.174 | 0.30034 | 0.212 | 0.0499 | 0.353 |
| 28823 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 21 | RARB | Sponge network | 0.194 | 0.64278 | -0.825 | 2.0E-5 | 0.353 |
| 28824 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 25 | KANK1 | Sponge network | -1.111 | 0.0003 | -0.55 | 0.00134 | 0.353 |
| 28825 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-7-1-3p | 18 | CBX5 | Sponge network | 0.796 | 4.0E-5 | 0.165 | 0.24446 | 0.353 |
| 28826 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-590-3p | 14 | FGF5 | Sponge network | -0.739 | 0.00044 | 0.549 | 0.28659 | 0.353 |
| 28827 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-582-5p | 13 | NR2C2 | Sponge network | 0.663 | 0.00073 | 0.21 | 0.03224 | 0.353 |
| 28828 | CTC-296K1.4 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p | 18 | QKI | Sponge network | -2.076 | 0.00548 | -0.333 | 0.04111 | 0.353 |
| 28829 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | -0.342 | 0.02288 | -1.3 | 0.01619 | 0.353 |
| 28830 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-96-5p | 26 | PDGFRA | Sponge network | -1.582 | 8.0E-5 | -0.372 | 0.0037 | 0.353 |
| 28831 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | EPAS1 | Sponge network | -1.654 | 0.00144 | -0.184 | 0.14418 | 0.353 |
| 28832 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-96-5p | 10 | PCDH8 | Sponge network | 0.853 | 0.15352 | -0.997 | 0.05366 | 0.353 |
| 28833 | BDNF-AS |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | EPHA7 | Sponge network | -0.668 | 3.0E-5 | -2.197 | 3.0E-5 | 0.353 |
| 28834 | RP11-499P20.2 |
hsa-let-7d-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-455-3p | 17 | RICTOR | Sponge network | 0.187 | 0.44121 | 0.388 | 0.00188 | 0.353 |
| 28835 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 22 | EBF3 | Sponge network | -0.227 | 0.31657 | -0.058 | 0.81437 | 0.353 |
| 28836 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | TAL1 | Sponge network | 0.246 | 0.59912 | -0.462 | 0.00574 | 0.353 |
| 28837 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EPAS1 | Sponge network | -0.318 | 0.65847 | -0.184 | 0.14418 | 0.353 |
| 28838 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 29 | MOB1B | Sponge network | 0.619 | 0.00157 | 0.197 | 0.15266 | 0.353 |
| 28839 | BCYRN1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-5p;hsa-miR-495-3p | 12 | STAG2 | Sponge network | 0.311 | 0.10344 | 0.252 | 0.00438 | 0.353 |
| 28840 | AC106786.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-877-5p | 13 | RBMS3 | Sponge network | 1.122 | 0.02989 | -0.519 | 0.03551 | 0.353 |
| 28841 | RP11-244O19.1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 10 | PCDH18 | Sponge network | -0.096 | 0.67958 | 0.216 | 0.24645 | 0.353 |
| 28842 | RP11-35G9.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-96-5p | 15 | SETBP1 | Sponge network | 0.622 | 0.00015 | -1.302 | 1.0E-5 | 0.353 |
| 28843 | CTD-2089N3.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3065-3p | 14 | CCND2 | Sponge network | -0.314 | 0.62033 | -0.267 | 0.3112 | 0.353 |
| 28844 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 12 | CLIP1 | Sponge network | -4.546 | 0 | -0.215 | 0.08684 | 0.353 |
| 28845 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577 | 15 | GREM1 | Sponge network | 0.056 | 0.81195 | 0.902 | 0.01691 | 0.353 |
| 28846 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | ZFX | Sponge network | 0.349 | 0.01695 | 0.09 | 0.42563 | 0.353 |
| 28847 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 12 | TRIM33 | Sponge network | 1.163 | 0.00018 | 0.402 | 7.0E-5 | 0.353 |
| 28848 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 24 | PRKAB2 | Sponge network | 0.401 | 0.00066 | -0.367 | 0.01371 | 0.353 |
| 28849 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-7-1-3p | 23 | IL6ST | Sponge network | -0.351 | 0.11358 | -0.402 | 0.00676 | 0.353 |
| 28850 | MAGI2-AS3 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | -0.129 | 0.57177 | -1.379 | 0 | 0.353 |
| 28851 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-744-5p | 18 | NOTCH2 | Sponge network | -0.309 | 0.1145 | 0.138 | 0.35163 | 0.353 |
| 28852 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | LRP1B | Sponge network | 0.225 | 0.30644 | -2.163 | 0 | 0.353 |
| 28853 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 30 | CBL | Sponge network | 0.659 | 3.0E-5 | 0.291 | 0.01267 | 0.353 |
| 28854 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 26 | PRKAB2 | Sponge network | -1.249 | 7.0E-5 | -0.367 | 0.01371 | 0.353 |
| 28855 | CTD-3092A11.2 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 15 | PIK3R1 | Sponge network | 0.873 | 0 | -0.133 | 0.36854 | 0.353 |
| 28856 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 34 | TRPS1 | Sponge network | -0.777 | 0.16667 | -0.191 | 0.44109 | 0.353 |
| 28857 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 34 | PRKAB2 | Sponge network | -1.281 | 0.00012 | -0.367 | 0.01371 | 0.353 |
| 28858 | ZNF582-AS1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-3127-5p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-503-5p | 17 | ABL1 | Sponge network | -1.349 | 0.00017 | -0.215 | 0.07804 | 0.353 |
| 28859 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 27 | TMTC3 | Sponge network | 0.782 | 0.00016 | 0.123 | 0.22189 | 0.353 |
| 28860 | RP11-755F10.1 |
hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-24-1-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-589-3p | 11 | IL6ST | Sponge network | 0.952 | 0 | -0.402 | 0.00676 | 0.353 |
| 28861 | CALML3-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-92a-1-5p | 44 | TRPS1 | Sponge network | 0.95 | 0.15943 | -0.191 | 0.44109 | 0.353 |
| 28862 | RP11-589P10.7 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-671-5p | 12 | CREB3L2 | Sponge network | -2.044 | 0.00066 | -0.525 | 2.0E-5 | 0.353 |
| 28863 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | EPHA3 | Sponge network | 0.222 | 0.29133 | 0.185 | 0.53588 | 0.352 |
| 28864 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 16 | NEDD4 | Sponge network | -0.358 | 0.17255 | 0.02 | 0.89834 | 0.352 |
| 28865 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | THBS1 | Sponge network | -0.739 | 0.00044 | 0.741 | 0.00386 | 0.352 |
| 28866 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 55 | WDFY3 | Sponge network | -1.281 | 0.00012 | -0.201 | 0.0967 | 0.352 |
| 28867 | AP001258.4 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 13 | TBL1XR1 | Sponge network | 0.931 | 0 | 0.578 | 0 | 0.352 |
| 28868 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-340-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 26 | NFIB | Sponge network | 0.178 | 0.29106 | 0.023 | 0.90795 | 0.352 |
| 28869 | RP11-156P1.3 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-92a-3p | 51 | DST | Sponge network | 0.214 | 0.18246 | -0.713 | 0.00096 | 0.352 |
| 28870 | AC016747.3 |
hsa-miR-130b-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 10 | SORCS1 | Sponge network | 0.199 | 0.38015 | -3.492 | 0 | 0.352 |
| 28871 | AC005618.6 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | 0.862 | 0.06213 | 0.358 | 0.13727 | 0.352 |
| 28872 | SNHG14 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 38 | CREB1 | Sponge network | -1.111 | 0.0003 | 0.203 | 0.00537 | 0.352 |
| 28873 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-664a-3p | 14 | ERRFI1 | Sponge network | -2.69 | 0.0001 | -0.798 | 1.0E-5 | 0.352 |
| 28874 | RP11-678G14.3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-484;hsa-miR-7-1-3p | 12 | PRKCB | Sponge network | -4.477 | 0 | -1.313 | 2.0E-5 | 0.352 |
| 28875 | RP11-927P21.1 |
hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p | 11 | PHF3 | Sponge network | 0.921 | 0.00118 | 0.126 | 0.19652 | 0.352 |
| 28876 | RP11-535M15.1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-942-5p | 15 | IGFBP5 | Sponge network | -1.14 | 0.03513 | -0.806 | 0.00105 | 0.352 |
| 28877 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CXXC4 | Sponge network | -0.235 | 0.15142 | -0.433 | 0.21055 | 0.352 |
| 28878 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 20 | FOXP1 | Sponge network | 0.817 | 3.0E-5 | 0.042 | 0.74368 | 0.352 |
| 28879 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | LATS1 | Sponge network | 1.153 | 0 | 0.109 | 0.34208 | 0.352 |
| 28880 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | KCNAB1 | Sponge network | 0.532 | 0.00505 | -0.755 | 2.0E-5 | 0.352 |
| 28881 | RP11-678G14.3 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-3p | 18 | ASTN1 | Sponge network | -4.477 | 0 | -2.389 | 2.0E-5 | 0.352 |
| 28882 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 45 | TMEM132B | Sponge network | 0.997 | 0.00066 | -0.83 | 0.01084 | 0.352 |
| 28883 | RP11-53O19.1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-942-5p | 24 | IGFBP5 | Sponge network | -0.405 | 0.22013 | -0.806 | 0.00105 | 0.352 |
| 28884 | RP11-531A24.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 27 | TGFBR3 | Sponge network | 0.75 | 0.00054 | -1.173 | 1.0E-5 | 0.352 |
| 28885 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-486-5p | 23 | RBMS3 | Sponge network | 0.368 | 0.20093 | -0.519 | 0.03551 | 0.352 |
| 28886 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 36 | ZFHX3 | Sponge network | -0.899 | 6.0E-5 | 0.389 | 0.01363 | 0.352 |
| 28887 | AP001055.6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-7-1-3p | 17 | TRPS1 | Sponge network | 0.693 | 0.00076 | -0.191 | 0.44109 | 0.352 |
| 28888 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 45 | PDGFRA | Sponge network | -0.942 | 0 | -0.372 | 0.0037 | 0.352 |
| 28889 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | LRRC4 | Sponge network | -0.899 | 6.0E-5 | -1.774 | 0 | 0.352 |
| 28890 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 46 | PAFAH1B1 | Sponge network | -0.899 | 6.0E-5 | -0.429 | 0 | 0.352 |
| 28891 | RP11-774O3.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | BAZ2B | Sponge network | -0.487 | 0.02157 | -0.276 | 0.02833 | 0.352 |
| 28892 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 26 | JAZF1 | Sponge network | 0.082 | 0.55397 | -0.377 | 0.02677 | 0.352 |
| 28893 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-93-5p | 12 | RBL2 | Sponge network | 0.082 | 0.55397 | -0.282 | 0.00254 | 0.352 |
| 28894 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-940 | 27 | SNX29 | Sponge network | -0.246 | 0.35034 | -0.34 | 0.01371 | 0.352 |
| 28895 | RP11-95D17.1 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 10 | RASAL2 | Sponge network | 0.75 | 0 | 0.869 | 0 | 0.352 |
| 28896 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-577;hsa-miR-629-3p | 24 | ZBTB4 | Sponge network | 0.083 | 0.66405 | -0.739 | 0 | 0.352 |
| 28897 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | CDHR3 | Sponge network | -0.222 | 0.32445 | -0.977 | 4.0E-5 | 0.352 |
| 28898 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | DIP2C | Sponge network | -0.031 | 0.89063 | -0.436 | 0.01713 | 0.352 |
| 28899 | RP11-649A18.4 |
hsa-miR-129-5p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-497-5p;hsa-miR-9-5p | 12 | CCDC6 | Sponge network | 2.218 | 0 | 0.27 | 0.02555 | 0.352 |
| 28900 | RP5-1021I20.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-93-5p | 11 | EZH1 | Sponge network | -0.785 | 0.00035 | -0.564 | 1.0E-5 | 0.352 |
| 28901 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-550a-3p | 18 | NRK | Sponge network | 0.082 | 0.55397 | 0.656 | 0.19217 | 0.352 |
| 28902 | AC093627.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-625-5p | 11 | MCC | Sponge network | -0.787 | 0.3928 | -1.005 | 0 | 0.352 |
| 28903 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p | 15 | PDS5B | Sponge network | 0.637 | 0.00069 | 0.259 | 0.00962 | 0.352 |
| 28904 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SLC6A15 | Sponge network | -1.264 | 6.0E-5 | -2.373 | 2.0E-5 | 0.352 |
| 28905 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | FAT4 | Sponge network | -0.351 | 0.11358 | -0.354 | 0.14694 | 0.352 |
| 28906 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 18 | CYLD | Sponge network | -0.358 | 0.17255 | -0.367 | 0.00171 | 0.352 |
| 28907 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CEP170 | Sponge network | 0.997 | 0.00066 | 0.943 | 0 | 0.352 |
| 28908 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ZNF536 | Sponge network | -0.129 | 0.57177 | -3.115 | 0 | 0.352 |
| 28909 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-155-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-7-1-3p | 10 | CBL | Sponge network | 0.959 | 0 | 0.291 | 0.01267 | 0.352 |
| 28910 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p | 14 | TBX18 | Sponge network | -0.777 | 0.16667 | 0.843 | 0.02945 | 0.352 |
| 28911 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | IGSF10 | Sponge network | -0.612 | 0.00094 | -1.751 | 1.0E-5 | 0.352 |
| 28912 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-331-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 23 | PHC3 | Sponge network | 0.4 | 0.14611 | 0.212 | 0.0499 | 0.352 |
| 28913 | LINC00473 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 28 | EYA4 | Sponge network | -1.203 | 0.03881 | 0.3 | 0.36876 | 0.352 |
| 28914 | LINC00886 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-93-5p | 25 | PLSCR4 | Sponge network | -0.371 | 0.24604 | -0.474 | 0.03833 | 0.352 |
| 28915 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p | 11 | ARHGAP29 | Sponge network | 1.757 | 0 | 0.131 | 0.42953 | 0.352 |
| 28916 | RP11-248G5.8 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-495-3p | 13 | GSK3B | Sponge network | 1.294 | 0 | 0.266 | 0.00045 | 0.352 |
| 28917 | TP73-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-589-3p | 15 | ABCA10 | Sponge network | -0.563 | 0.02545 | -0.571 | 0.03674 | 0.352 |
| 28918 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-744-3p;hsa-miR-92a-3p | 16 | FAT3 | Sponge network | 0.959 | 0 | -1.601 | 0.00051 | 0.352 |
| 28919 | RP4-791M13.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | SETBP1 | Sponge network | 0.419 | 0.0319 | -1.302 | 1.0E-5 | 0.352 |
| 28920 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 18 | ZMYND11 | Sponge network | -0.611 | 0.09607 | -0.2 | 0.05449 | 0.352 |
| 28921 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 22 | PBX1 | Sponge network | -0.322 | 0.25945 | -1.206 | 0 | 0.352 |
| 28922 | LINC00578 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-589-3p | 11 | TAL1 | Sponge network | 0.811 | 0.03228 | -0.462 | 0.00574 | 0.352 |
| 28923 | AC007879.7 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326 | 12 | TBL1XR1 | Sponge network | 4.3 | 0 | 0.578 | 0 | 0.352 |
| 28924 | LPP-AS2 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-7-5p | 19 | PRKCB | Sponge network | -0.18 | 0.20343 | -1.313 | 2.0E-5 | 0.352 |
| 28925 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-7-1-3p | 13 | FGF5 | Sponge network | -0.726 | 0.00132 | 0.549 | 0.28659 | 0.352 |
| 28926 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | MAP3K12 | Sponge network | -1.281 | 0.00012 | -0.057 | 0.59369 | 0.352 |
| 28927 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | NCAM2 | Sponge network | -0.405 | 0.22013 | -0.482 | 0.22565 | 0.352 |
| 28928 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | ST6GALNAC3 | Sponge network | -1.697 | 0.00889 | -0.987 | 0 | 0.352 |
| 28929 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | NCOA1 | Sponge network | -0.611 | 0.09607 | -0.432 | 0 | 0.352 |
| 28930 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PBRM1 | Sponge network | -0.487 | 0.02157 | -0.029 | 0.78601 | 0.352 |
| 28931 | RP11-384K6.6 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-330-3p | 14 | AFF1 | Sponge network | 0.946 | 0 | -0.384 | 0.00053 | 0.352 |
| 28932 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 30 | PRRX1 | Sponge network | -1.216 | 0 | 1.316 | 0 | 0.352 |
| 28933 | RP11-212P7.2 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 12 | FLNA | Sponge network | 0.219 | 0.1516 | -0.771 | 0.01377 | 0.352 |
| 28934 | AC025171.1 |
hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | RNF144A | Sponge network | 0.998 | 0 | 0.594 | 0.0009 | 0.352 |
| 28935 | RAMP2-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p | 13 | FLNA | Sponge network | -0.513 | 0.04703 | -0.771 | 0.01377 | 0.352 |
| 28936 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-425-5p | 20 | ERC1 | Sponge network | 0.082 | 0.55397 | -0.108 | 0.38794 | 0.352 |
| 28937 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-508-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 19 | TGFBR2 | Sponge network | -0.563 | 0.02545 | -0.243 | 0.11408 | 0.352 |
| 28938 | RP11-532F6.3 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-5p | 37 | PPM1L | Sponge network | -0.896 | 0.00099 | -0.537 | 0.00616 | 0.352 |
| 28939 | RP5-1139B12.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 12 | SOX5 | Sponge network | 0.598 | 0.38481 | -1.091 | 4.0E-5 | 0.352 |
| 28940 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | -0.227 | 0.31657 | -1.3 | 0.01619 | 0.352 |
| 28941 | LINC00449 |
hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 11 | CBFB | Sponge network | 1.525 | 0 | 1.061 | 0 | 0.352 |
| 28942 | RP11-822E23.8 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p | 10 | ROBO1 | Sponge network | -0.777 | 0.16667 | -0.009 | 0.96154 | 0.352 |
| 28943 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p | 34 | BCL2 | Sponge network | -0.738 | 0.21233 | -0.899 | 0 | 0.352 |
| 28944 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | KCNA6 | Sponge network | -0.726 | 0.00132 | -1.531 | 0.00013 | 0.352 |
| 28945 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | EPHA3 | Sponge network | -0.342 | 0.02288 | 0.185 | 0.53588 | 0.352 |
| 28946 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 27 | PRKAB2 | Sponge network | 0.494 | 0.00339 | -0.367 | 0.01371 | 0.352 |
| 28947 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-629-3p | 17 | GPC6 | Sponge network | -1.654 | 0.00144 | -0.054 | 0.81465 | 0.352 |
| 28948 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p | 16 | CLIP1 | Sponge network | 0.494 | 0.00339 | -0.215 | 0.08684 | 0.352 |
| 28949 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-5p;hsa-miR-93-5p | 32 | UNC80 | Sponge network | -1.654 | 0.00144 | -1.934 | 0 | 0.352 |
| 28950 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 11 | SEPT6 | Sponge network | -2.46 | 0 | -0.598 | 0.00181 | 0.352 |
| 28951 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | SOX5 | Sponge network | -0.72 | 0.24239 | -1.091 | 4.0E-5 | 0.352 |
| 28952 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-96-5p;hsa-miR-98-5p | 19 | LIFR | Sponge network | -0.18 | 0.20343 | -1.794 | 0 | 0.352 |
| 28953 | HCG11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.385 | 0.10067 | 0.943 | 0 | 0.352 |
| 28954 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | TIMP3 | Sponge network | 1.302 | 7.0E-5 | -0.546 | 0.02307 | 0.352 |
| 28955 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-505-3p;hsa-miR-92a-3p | 13 | PDE4DIP | Sponge network | 0.453 | 0.01839 | -0.282 | 0.0257 | 0.352 |
| 28956 | ZNF667-AS1 |
hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-874-3p;hsa-miR-96-5p | 10 | AJAP1 | Sponge network | -0.976 | 0.00025 | -0.561 | 0.04832 | 0.352 |
| 28957 | NDUFA6-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 35 | IL17RD | Sponge network | -0.355 | 0.0077 | -0.019 | 0.94873 | 0.352 |
| 28958 | LINC00092 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | CSMD1 | Sponge network | -1.886 | 0 | -0.63 | 0.18881 | 0.352 |
| 28959 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | FOXO3 | Sponge network | 0.374 | 0.20054 | -0.04 | 0.72028 | 0.352 |
| 28960 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-96-5p | 33 | HBP1 | Sponge network | -0.576 | 0.10121 | -0.454 | 0 | 0.352 |
| 28961 | RP11-359B12.2 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p | 25 | UBR3 | Sponge network | 0.064 | 0.72934 | -0.021 | 0.82774 | 0.352 |
| 28962 | LINC00174 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-582-5p | 10 | ZFHX3 | Sponge network | 1.253 | 0 | 0.389 | 0.01363 | 0.352 |
| 28963 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-3127-5p | 11 | RPS6KA2 | Sponge network | -1.161 | 0.00591 | -0.57 | 0.00013 | 0.352 |
| 28964 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-455-3p | 12 | PIK3R1 | Sponge network | 0.858 | 3.0E-5 | -0.133 | 0.36854 | 0.352 |
| 28965 | LINC00094 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-532-3p | 18 | SYNM | Sponge network | 0.253 | 0.09565 | -2.309 | 1.0E-5 | 0.352 |
| 28966 | RASSF8-AS1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | ATM | Sponge network | 0.335 | 0.20596 | 0.178 | 0.21568 | 0.352 |
| 28967 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | ATRX | Sponge network | 0.41 | 0.02214 | 0.261 | 0.04408 | 0.352 |
| 28968 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 49 | THRB | Sponge network | -0.421 | 0.0114 | -1.117 | 0.0004 | 0.352 |
| 28969 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-942-5p | 25 | DDX6 | Sponge network | 0.921 | 0.00118 | 0.091 | 0.36428 | 0.352 |
| 28970 | RP11-156P1.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-7-5p | 21 | KCNJ5 | Sponge network | 0.214 | 0.18246 | -0.281 | 0.3339 | 0.352 |
| 28971 | RP11-403I13.8 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-7-5p | 10 | COL1A2 | Sponge network | 0.619 | 0.00157 | 1.991 | 0 | 0.352 |
| 28972 | AC011526.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | HIP1 | Sponge network | 0.342 | 0.03129 | 0.774 | 0 | 0.352 |
| 28973 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-629-3p;hsa-miR-7-5p | 19 | ESR1 | Sponge network | -0.326 | 0.14314 | -1.035 | 3.0E-5 | 0.352 |
| 28974 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ANK2 | Sponge network | -0.031 | 0.89063 | -1.603 | 1.0E-5 | 0.352 |
| 28975 | RP11-344B5.2 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b | 15 | MEF2C | Sponge network | -0.055 | 0.88482 | -0.499 | 0.01448 | 0.352 |
| 28976 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-342-3p | 12 | DKK3 | Sponge network | -2.095 | 0.00112 | -0.052 | 0.77783 | 0.352 |
| 28977 | LPP-AS2 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p | 17 | GJA1 | Sponge network | -0.18 | 0.20343 | -0.485 | 0.02179 | 0.352 |
| 28978 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 28 | USP12 | Sponge network | -0.303 | 0.21002 | -0.102 | 0.40768 | 0.352 |
| 28979 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p | 16 | ERCC4 | Sponge network | 0.4 | 0.14611 | 0.126 | 0.15266 | 0.352 |
| 28980 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 20 | TES | Sponge network | -0.49 | 0.04946 | -0.348 | 0.00639 | 0.352 |
| 28981 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-93-5p | 11 | TXNIP | Sponge network | -0.18 | 0.20343 | -1.277 | 0 | 0.352 |
| 28982 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-93-5p;hsa-miR-942-5p | 14 | FBXO31 | Sponge network | -1.439 | 4.0E-5 | -0.085 | 0.42389 | 0.352 |
| 28983 | BOLA3-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | -0.246 | 0.35034 | 0.524 | 0.00017 | 0.352 |
| 28984 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-503-5p | 19 | ABL1 | Sponge network | -1.654 | 0.00144 | -0.215 | 0.07804 | 0.352 |
| 28985 | EMX2OS |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | MSN | Sponge network | -0.777 | 0.1134 | -0.023 | 0.88422 | 0.352 |
| 28986 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 26 | LRP1B | Sponge network | -0.227 | 0.31657 | -2.163 | 0 | 0.352 |
| 28987 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | TIMP3 | Sponge network | -0.899 | 6.0E-5 | -0.546 | 0.02307 | 0.352 |
| 28988 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p | 24 | BMPR2 | Sponge network | 1.317 | 0 | 0.206 | 0.0635 | 0.352 |
| 28989 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | ST6GALNAC3 | Sponge network | -0.421 | 0.0114 | -0.987 | 0 | 0.352 |
| 28990 | RP11-384O8.1 |
hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 10 | DACT1 | Sponge network | -0.738 | 0.21233 | 0.783 | 0.00072 | 0.352 |
| 28991 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p | 32 | TMEM132B | Sponge network | 0.176 | 0.37402 | -0.83 | 0.01084 | 0.352 |
| 28992 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | ITGAV | Sponge network | -0.576 | 0.10121 | 0.337 | 0.01002 | 0.352 |
| 28993 | RP11-263K19.4 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429 | 19 | ABL2 | Sponge network | 0.859 | 0.00331 | 0.82 | 0 | 0.352 |
| 28994 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PHC3 | Sponge network | 0.545 | 0.00064 | 0.212 | 0.0499 | 0.352 |
| 28995 | RP1-122P22.2 |
hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-338-3p;hsa-miR-532-3p;hsa-miR-629-3p | 15 | GAS7 | Sponge network | 0.065 | 0.73468 | -0.555 | 0.01602 | 0.352 |
| 28996 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-671-5p | 19 | DLC1 | Sponge network | -0.72 | 0.24239 | -0.382 | 0.05038 | 0.352 |
| 28997 | AC018890.6 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | BCL9 | Sponge network | 1.61 | 0.00961 | 0.321 | 0.00267 | 0.352 |
| 28998 | RP11-774O3.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | LUZP2 | Sponge network | -0.487 | 0.02157 | -0.056 | 0.89416 | 0.352 |
| 28999 | PCA3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | AFF1 | Sponge network | -0.709 | 0.18168 | -0.384 | 0.00053 | 0.352 |
| 29000 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-576-5p | 29 | PBX1 | Sponge network | -0.147 | 0.40452 | -1.206 | 0 | 0.352 |
| 29001 | MRVI1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 10 | UNC5C | Sponge network | -1.092 | 4.0E-5 | -0.178 | 0.40214 | 0.352 |
| 29002 | RP11-159D12.2 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-590-3p | 21 | DDX3X | Sponge network | 1.254 | 0 | -0.152 | 0.07686 | 0.352 |
| 29003 | RP3-523E19.2 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-30a-3p;hsa-miR-339-5p | 11 | MDM4 | Sponge network | 2.887 | 0 | 0.345 | 0.00762 | 0.352 |
| 29004 | NR2F1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 29 | EFNA5 | Sponge network | -0.49 | 0.04946 | -0.421 | 0.17868 | 0.352 |
| 29005 | RP11-2H8.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p | 14 | SIRT1 | Sponge network | 1.089 | 0 | 0.109 | 0.2593 | 0.352 |
| 29006 | RP11-540B6.6 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 29 | CCDC6 | Sponge network | 0.93 | 0 | 0.27 | 0.02555 | 0.352 |
| 29007 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940;hsa-miR-96-5p | 44 | GAS7 | Sponge network | 0.064 | 0.72934 | -0.555 | 0.01602 | 0.352 |
| 29008 | RP11-434H6.7 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-149-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-409-3p | 11 | NFIB | Sponge network | 0.358 | 0.14302 | 0.023 | 0.90795 | 0.352 |
| 29009 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-342-3p | 17 | GRIN2A | Sponge network | 0.082 | 0.55397 | -2.161 | 0 | 0.352 |
| 29010 | MIR22HG |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p | 14 | RELN | Sponge network | -1.047 | 0 | -2.403 | 0 | 0.352 |
| 29011 | CTD-3138B18.6 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | TSC1 | Sponge network | 2.248 | 0 | 0.103 | 0.26071 | 0.352 |
| 29012 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p | 12 | CREB1 | Sponge network | 0.769 | 0 | 0.203 | 0.00537 | 0.352 |
| 29013 | HAND2-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 32 | OPCML | Sponge network | -1.697 | 0.00889 | -2.498 | 0 | 0.352 |
| 29014 | AF131215.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 24 | PTPRT | Sponge network | -0.176 | 0.59692 | -0.907 | 0.03727 | 0.352 |
| 29015 | AC007879.7 |
hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326 | 10 | SLC5A3 | Sponge network | 4.3 | 0 | 0.493 | 5.0E-5 | 0.352 |
| 29016 | FENDRR |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | FYN | Sponge network | -1.281 | 0.00012 | -0.131 | 0.37051 | 0.352 |
| 29017 | LINC00657 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p | 15 | ATM | Sponge network | 0.91 | 0 | 0.178 | 0.21568 | 0.352 |
| 29018 | CTC-429P9.2 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-940 | 15 | NFIB | Sponge network | 0.043 | 0.81628 | 0.023 | 0.90795 | 0.352 |
| 29019 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 21 | NF1 | Sponge network | 0.569 | 0.00067 | 0.51 | 0 | 0.352 |
| 29020 | RP11-66B24.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-375 | 15 | ZFHX4 | Sponge network | 0.57 | 0.1866 | 0.018 | 0.95574 | 0.352 |
| 29021 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 19 | CDC73 | Sponge network | 1.063 | 0 | 0.094 | 0.20463 | 0.352 |
| 29022 | RP1-151F17.2 |
hsa-miR-10b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 10 | EXTL2 | Sponge network | 0.222 | 0.29133 | 0.109 | 0.37375 | 0.352 |
| 29023 | RP11-6O2.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 18 | NAV3 | Sponge network | 0.331 | 0.57247 | 0.212 | 0.47729 | 0.352 |
| 29024 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 17 | HECA | Sponge network | -0.318 | 0.65847 | -0.217 | 0.03463 | 0.352 |
| 29025 | HCG11 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | RECK | Sponge network | 0.385 | 0.10067 | -1.043 | 0 | 0.352 |
| 29026 | OIP5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-654-3p | 41 | PIK3R1 | Sponge network | 0.421 | 0.00084 | -0.133 | 0.36854 | 0.352 |
| 29027 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 56 | THRB | Sponge network | 0.246 | 0.59912 | -1.117 | 0.0004 | 0.352 |
| 29028 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | BNC2 | Sponge network | -0.18 | 0.20343 | -0.491 | 0.09988 | 0.352 |
| 29029 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SEMA3E | Sponge network | -1.161 | 0.00591 | -1.717 | 0.00137 | 0.352 |
| 29030 | HCG11 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | PRDM5 | Sponge network | 0.385 | 0.10067 | -1.09 | 0.00014 | 0.352 |
| 29031 | RP4-668J24.2 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-424-5p;hsa-miR-628-5p;hsa-miR-629-3p | 12 | RASSF8 | Sponge network | -1.054 | 0.0706 | -0.122 | 0.65591 | 0.352 |
| 29032 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-940 | 12 | FAM172A | Sponge network | -0.969 | 2.0E-5 | -0.57 | 0 | 0.352 |
| 29033 | CTD-3064M3.3 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-424-5p | 11 | RPS6KA6 | Sponge network | -0.469 | 0.08721 | -2.989 | 0 | 0.352 |
| 29034 | RP13-516M14.1 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-766-3p | 14 | ATM | Sponge network | 0.389 | 0.04293 | 0.178 | 0.21568 | 0.352 |
| 29035 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-10a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-7-1-3p | 20 | DDX6 | Sponge network | 0.34 | 0.00273 | 0.091 | 0.36428 | 0.352 |
| 29036 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-576-5p | 17 | HIP1 | Sponge network | 0.95 | 0.15943 | 0.774 | 0 | 0.352 |
| 29037 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-629-3p | 24 | DDX6 | Sponge network | 0.331 | 0.57247 | 0.091 | 0.36428 | 0.352 |
| 29038 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p | 17 | CNTN1 | Sponge network | 0.082 | 0.55397 | -2.594 | 0 | 0.352 |
| 29039 | RP11-130L8.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-493-3p;hsa-miR-590-3p | 16 | NCOA1 | Sponge network | -0.663 | 0.10887 | -0.432 | 0 | 0.352 |
| 29040 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | PCDH8 | Sponge network | -1.575 | 6.0E-5 | -0.997 | 0.05366 | 0.352 |
| 29041 | LINC00662 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-577 | 15 | IL17RD | Sponge network | 0.213 | 0.32297 | -0.019 | 0.94873 | 0.352 |
| 29042 | AC141928.1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p | 11 | CACNA1E | Sponge network | 0.853 | 0.15352 | 1.752 | 0 | 0.352 |
| 29043 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 11 | GPAM | Sponge network | 0.903 | 0 | 0.142 | 0.29732 | 0.352 |
| 29044 | TP73-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-935 | 13 | SEPT6 | Sponge network | -0.563 | 0.02545 | -0.598 | 0.00181 | 0.352 |
| 29045 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-5p | 21 | LIFR | Sponge network | 0.176 | 0.37402 | -1.794 | 0 | 0.352 |
| 29046 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-500b-5p | 11 | SCN9A | Sponge network | -0.557 | 0.11135 | -0.525 | 0.10593 | 0.352 |
| 29047 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p | 11 | NUAK1 | Sponge network | 0.912 | 0.00014 | 0.904 | 0 | 0.352 |
| 29048 | RBPMS-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | PLSCR4 | Sponge network | 0.144 | 0.64989 | -0.474 | 0.03833 | 0.352 |
| 29049 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-96-5p | 15 | IL17RD | Sponge network | 0.37 | 0.27614 | -0.019 | 0.94873 | 0.352 |
| 29050 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CNTN1 | Sponge network | -0.342 | 0.02288 | -2.594 | 0 | 0.352 |
| 29051 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-96-5p | 42 | FAT3 | Sponge network | 0.537 | 0.00139 | -1.601 | 0.00051 | 0.352 |
| 29052 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 25 | PRKAB2 | Sponge network | 0.082 | 0.55654 | -0.367 | 0.01371 | 0.352 |
| 29053 | ADAMTS9-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-7-5p;hsa-miR-96-5p | 10 | ZYX | Sponge network | -2.452 | 0 | 0.019 | 0.89763 | 0.352 |
| 29054 | AC006116.24 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 17 | LRP1B | Sponge network | -0.473 | 0.07602 | -2.163 | 0 | 0.352 |
| 29055 | LINC00672 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-654-3p | 16 | BAZ2B | Sponge network | -0.227 | 0.31657 | -0.276 | 0.02833 | 0.352 |
| 29056 | RP11-535M15.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-3p | 15 | NTRK3 | Sponge network | -1.14 | 0.03513 | -1.77 | 3.0E-5 | 0.352 |
| 29057 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-96-5p | 26 | RASSF8 | Sponge network | -0.18 | 0.20343 | -0.122 | 0.65591 | 0.352 |
| 29058 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 12 | ZNF770 | Sponge network | 0.909 | 0 | 0.206 | 0.06751 | 0.352 |
| 29059 | HOXB-AS3 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-505-5p;hsa-miR-629-5p | 12 | WNK2 | Sponge network | 0.343 | 0.28887 | -0.352 | 0.31066 | 0.352 |
| 29060 | HAS2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p | 10 | NTRK2 | Sponge network | -0.011 | 0.967 | -0.736 | 0.06495 | 0.352 |
| 29061 | AC074289.1 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-629-3p | 23 | RASSF2 | Sponge network | -0.326 | 0.14314 | -0.273 | 0.21961 | 0.352 |
| 29062 | FZD10-AS1 |
hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-550a-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PCDH18 | Sponge network | -0.727 | 0.04726 | 0.216 | 0.24645 | 0.352 |
| 29063 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 50 | TMEM132B | Sponge network | -2.925 | 0 | -0.83 | 0.01084 | 0.352 |
| 29064 | RP11-156E6.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-497-5p | 12 | BRD3 | Sponge network | 1.266 | 0 | 0.37 | 0.00013 | 0.352 |
| 29065 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 16 | TXNIP | Sponge network | -1.645 | 0 | -1.277 | 0 | 0.352 |
| 29066 | RP11-473I1.9 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 0.683 | 1.0E-5 | 0.321 | 0.00267 | 0.352 |
| 29067 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SON | Sponge network | 0.101 | 0.60919 | 0.168 | 0.04406 | 0.352 |
| 29068 | RAB11B-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-181a-2-3p;hsa-miR-185-3p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p | 13 | CAMTA1 | Sponge network | -0.508 | 2.0E-5 | -0.436 | 1.0E-5 | 0.352 |
| 29069 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p | 11 | PRDM5 | Sponge network | -0.08 | 0.90092 | -1.09 | 0.00014 | 0.352 |
| 29070 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-539-5p | 13 | CDHR3 | Sponge network | -1.092 | 4.0E-5 | -0.977 | 4.0E-5 | 0.352 |
| 29071 | AC034220.3 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | MYO9A | Sponge network | 0.309 | 0.07304 | -0.147 | 0.32373 | 0.352 |
| 29072 | LPP-AS2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-505-3p | 20 | PDE4DIP | Sponge network | -0.18 | 0.20343 | -0.282 | 0.0257 | 0.352 |
| 29073 | RP11-999E24.3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-374a-5p | 20 | ABI2 | Sponge network | -0.693 | 0.05629 | 0.175 | 0.14787 | 0.352 |
| 29074 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | LRRC7 | Sponge network | 0.532 | 0.00505 | -1.341 | 0.01193 | 0.352 |
| 29075 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DNAJB4 | Sponge network | -1.057 | 0.00029 | -0.7 | 1.0E-5 | 0.352 |
| 29076 | FZD10-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 22 | PCDH10 | Sponge network | -0.727 | 0.04726 | -1.644 | 0.0078 | 0.352 |
| 29077 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-93-5p;hsa-miR-940 | 35 | GAS7 | Sponge network | 0.082 | 0.55397 | -0.555 | 0.01602 | 0.351 |
| 29078 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p | 39 | SSBP2 | Sponge network | 0.199 | 0.38015 | -1.58 | 0 | 0.351 |
| 29079 | CTD-2314G24.2 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | SPOP | Sponge network | 0.246 | 0.59912 | -0.419 | 1.0E-5 | 0.351 |
| 29080 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 17 | NCOA1 | Sponge network | -0.187 | 0.23787 | -0.432 | 0 | 0.351 |
| 29081 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p | 14 | ZMYM2 | Sponge network | 0.903 | 0 | 0.279 | 0.01273 | 0.351 |
| 29082 | ZRANB2-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-935 | 19 | PTPRT | Sponge network | -0.376 | 0.00337 | -0.907 | 0.03727 | 0.351 |
| 29083 | BCYRN1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-935 | 36 | DMD | Sponge network | 0.311 | 0.10344 | -1.506 | 0 | 0.351 |
| 29084 | CTD-2002H8.2 |
hsa-miR-130a-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-374b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | DMXL1 | Sponge network | 0.931 | 1.0E-5 | -0.426 | 0.00056 | 0.351 |
| 29085 | RP11-276H19.2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-338-5p;hsa-miR-660-5p | 10 | EYA4 | Sponge network | -0.472 | 0.48851 | 0.3 | 0.36876 | 0.351 |
| 29086 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | PTPRB | Sponge network | -0.901 | 0.00019 | 0.105 | 0.47122 | 0.351 |
| 29087 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 26 | RASSF8 | Sponge network | -0.727 | 0.04726 | -0.122 | 0.65591 | 0.351 |
| 29088 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-576-5p | 15 | MYO9A | Sponge network | 1.044 | 2.0E-5 | -0.147 | 0.32373 | 0.351 |
| 29089 | LINC01018 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | PPP1R9A | Sponge network | -2.915 | 0.00015 | -0.903 | 0.04254 | 0.351 |
| 29090 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429 | 14 | IGSF10 | Sponge network | -0.154 | 0.53954 | -1.751 | 1.0E-5 | 0.351 |
| 29091 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p | 14 | SLC5A3 | Sponge network | 1.089 | 0 | 0.493 | 5.0E-5 | 0.351 |
| 29092 | AC116366.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-940 | 21 | NFIB | Sponge network | 0.329 | 0.11122 | 0.023 | 0.90795 | 0.351 |
| 29093 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | KCNA6 | Sponge network | -0.487 | 0.02157 | -1.531 | 0.00013 | 0.351 |
| 29094 | RP5-894A10.6 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-495-3p | 13 | CREB1 | Sponge network | 0.697 | 9.0E-5 | 0.203 | 0.00537 | 0.351 |
| 29095 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 28 | BNC2 | Sponge network | 1.757 | 0 | -0.491 | 0.09988 | 0.351 |
| 29096 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-3p | 25 | PDGFRA | Sponge network | -0.829 | 0.01343 | -0.372 | 0.0037 | 0.351 |
| 29097 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | PEG3 | Sponge network | 0.494 | 0.00339 | -1.74 | 0 | 0.351 |
| 29098 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-589-5p | 28 | SOX5 | Sponge network | 0.082 | 0.55397 | -1.091 | 4.0E-5 | 0.351 |
| 29099 | RP3-475N16.1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-455-3p;hsa-miR-495-3p | 19 | CREB1 | Sponge network | 1.056 | 0 | 0.203 | 0.00537 | 0.351 |
| 29100 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 42 | PPP1R9A | Sponge network | -2.452 | 0 | -0.903 | 0.04254 | 0.351 |
| 29101 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | TET1 | Sponge network | 0.619 | 0.00157 | -0.135 | 0.52138 | 0.351 |
| 29102 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | LRRK2 | Sponge network | 0.214 | 0.18246 | -1.136 | 1.0E-5 | 0.351 |
| 29103 | AF131215.9 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 34 | PPP1R9A | Sponge network | -0.322 | 0.25945 | -0.903 | 0.04254 | 0.351 |
| 29104 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 58 | CADM2 | Sponge network | 0.532 | 0.00505 | -3.782 | 0 | 0.351 |
| 29105 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-708-5p | 30 | RAP1A | Sponge network | 0.194 | 0.64278 | -0.929 | 0 | 0.351 |
| 29106 | RP11-517B11.7 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p | 17 | ABL2 | Sponge network | 0.706 | 0 | 0.82 | 0 | 0.351 |
| 29107 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p | 17 | ESR1 | Sponge network | 0.622 | 0.00015 | -1.035 | 3.0E-5 | 0.351 |
| 29108 | GAS6-AS2 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-935 | 11 | STAT5B | Sponge network | 0.16 | 0.50785 | -0.182 | 0.1082 | 0.351 |
| 29109 | AC108488.4 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 12 | SEMA3C | Sponge network | 0.259 | 0.12542 | -0.272 | 0.31153 | 0.351 |
| 29110 | RP11-37B2.1 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-582-5p | 12 | DOCK4 | Sponge network | 1.03 | 0 | 0.502 | 0.00108 | 0.351 |
| 29111 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 26 | ADAMTSL3 | Sponge network | -0.969 | 2.0E-5 | -1.559 | 0 | 0.351 |
| 29112 | CTD-2314G24.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | WWTR1 | Sponge network | 0.246 | 0.59912 | -0.345 | 0.11997 | 0.351 |
| 29113 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | NRXN1 | Sponge network | 0.342 | 0.03129 | -3.778 | 0 | 0.351 |
| 29114 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 45 | MOB1B | Sponge network | -0.739 | 0.0574 | 0.197 | 0.15266 | 0.351 |
| 29115 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 23 | SLC6A15 | Sponge network | -0.405 | 0.22013 | -2.373 | 2.0E-5 | 0.351 |
| 29116 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | GALNT13 | Sponge network | 0.225 | 0.30644 | -0.857 | 0.07439 | 0.351 |
| 29117 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 33 | TGFBR3 | Sponge network | -0.342 | 0.02288 | -1.173 | 1.0E-5 | 0.351 |
| 29118 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-877-5p | 10 | NAV3 | Sponge network | -1.116 | 0.23686 | 0.212 | 0.47729 | 0.351 |
| 29119 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 23 | MXI1 | Sponge network | -0.421 | 0.0114 | -0.987 | 0 | 0.351 |
| 29120 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-590-3p | 16 | TBX18 | Sponge network | 0.082 | 0.55654 | 0.843 | 0.02945 | 0.351 |
| 29121 | RP11-356J5.12 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 18 | RPS6KA6 | Sponge network | -0.899 | 6.0E-5 | -2.989 | 0 | 0.351 |
| 29122 | RP11-504P24.8 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-495-3p | 16 | CREB1 | Sponge network | 1.178 | 0 | 0.203 | 0.00537 | 0.351 |
| 29123 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 29 | RNF144A | Sponge network | -1.216 | 0 | 0.594 | 0.0009 | 0.351 |
| 29124 | TTC28-AS1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-654-3p | 10 | ZNF638 | Sponge network | 0.373 | 0.00068 | 0.22 | 0.01214 | 0.351 |
| 29125 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 17 | PKNOX1 | Sponge network | -1.697 | 0.00889 | 0.085 | 0.28917 | 0.351 |
| 29126 | ZNF667-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 23 | PDE4DIP | Sponge network | -0.976 | 0.00025 | -0.282 | 0.0257 | 0.351 |
| 29127 | PSMG3-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p | 15 | MRVI1 | Sponge network | -0.125 | 0.49414 | -1.051 | 0.00022 | 0.351 |
| 29128 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-374b-5p | 15 | CDC73 | Sponge network | 1.4 | 0 | 0.094 | 0.20463 | 0.351 |
| 29129 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 45 | PIK3R1 | Sponge network | -0.615 | 0.00015 | -0.133 | 0.36854 | 0.351 |
| 29130 | AC108676.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-654-3p | 12 | NR2C2 | Sponge network | 4.346 | 2.0E-5 | 0.21 | 0.03224 | 0.351 |
| 29131 | RP11-356J5.12 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | SYT4 | Sponge network | -0.899 | 6.0E-5 | -3.207 | 0 | 0.351 |
| 29132 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-940;hsa-miR-96-5p | 16 | CRTC3 | Sponge network | 0.537 | 0.00139 | 0.164 | 0.16786 | 0.351 |
| 29133 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p | 15 | PIK3CA | Sponge network | 1.254 | 0 | 0.195 | 0.06908 | 0.351 |
| 29134 | AC138035.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-539-5p | 10 | ABI2 | Sponge network | 1.073 | 0 | 0.175 | 0.14787 | 0.351 |
| 29135 | LPP-AS2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p | 14 | GLI3 | Sponge network | -0.18 | 0.20343 | -0.615 | 0.01482 | 0.351 |
| 29136 | CD27-AS1 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | SPOP | Sponge network | 0.177 | 0.16681 | -0.419 | 1.0E-5 | 0.351 |
| 29137 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p | 13 | JAKMIP2 | Sponge network | -0.18 | 0.20343 | -1.388 | 0 | 0.351 |
| 29138 | RP11-815I9.4 |
hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | DICER1 | Sponge network | 0.537 | 0.0006 | 0.042 | 0.674 | 0.351 |
| 29139 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-330-5p | 15 | BTG2 | Sponge network | -1.375 | 0.08642 | -1.763 | 0 | 0.351 |
| 29140 | RP11-497H16.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ARHGEF12 | Sponge network | 0.545 | 0.00064 | 0.09 | 0.35693 | 0.351 |
| 29141 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 53 | CCND2 | Sponge network | -1.216 | 0 | -0.267 | 0.3112 | 0.351 |
| 29142 | PCA3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p | 21 | CRTC1 | Sponge network | -0.709 | 0.18168 | -0.116 | 0.28412 | 0.351 |
| 29143 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p | 16 | TBX18 | Sponge network | -0.246 | 0.35034 | 0.843 | 0.02945 | 0.351 |
| 29144 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 20 | RNF144A | Sponge network | 1.757 | 0 | 0.594 | 0.0009 | 0.351 |
| 29145 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-93-5p;hsa-miR-940 | 26 | BHLHE41 | Sponge network | 0.4 | 0.14611 | 0.353 | 0.12753 | 0.351 |
| 29146 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-7-1-3p | 33 | ZFHX3 | Sponge network | 0.177 | 0.16681 | 0.389 | 0.01363 | 0.351 |
| 29147 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 12 | ZNF208 | Sponge network | -0.246 | 0.35034 | -0.4 | 0.22381 | 0.351 |
| 29148 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | MAF | Sponge network | 0.249 | 0.35902 | -1.257 | 0 | 0.351 |
| 29149 | LINC00702 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-7-5p | 11 | COL1A2 | Sponge network | -0.737 | 0.06182 | 1.991 | 0 | 0.351 |
| 29150 | RP11-822E23.8 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-589-3p | 23 | SYT4 | Sponge network | -0.777 | 0.16667 | -3.207 | 0 | 0.351 |
| 29151 | IGF2-AS |
hsa-let-7g-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-486-5p | 10 | PRRX1 | Sponge network | 3.339 | 0.00237 | 1.316 | 0 | 0.351 |
| 29152 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | ARID5B | Sponge network | 0.537 | 0.00139 | 0.342 | 0.01676 | 0.351 |
| 29153 | SERTAD4-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-576-5p | 18 | UNC80 | Sponge network | -0.932 | 0.00329 | -1.934 | 0 | 0.351 |
| 29154 | DNAJC27-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | PRUNE2 | Sponge network | -0.969 | 2.0E-5 | -1.733 | 0.00022 | 0.351 |
| 29155 | A1BG-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-671-5p | 11 | FAT4 | Sponge network | -0.164 | 0.45106 | -0.354 | 0.14694 | 0.351 |
| 29156 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-502-5p | 15 | BCL11A | Sponge network | -2.62 | 0 | -0.932 | 0.0063 | 0.351 |
| 29157 | RP11-356J5.12 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | -0.899 | 6.0E-5 | -0.998 | 0.00025 | 0.351 |
| 29158 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PCDH8 | Sponge network | -0.901 | 0.00019 | -0.997 | 0.05366 | 0.351 |
| 29159 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-664a-5p;hsa-miR-766-3p | 23 | MAPK1 | Sponge network | 0.706 | 0 | -0.017 | 0.81465 | 0.351 |
| 29160 | NEAT1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PIK3CA | Sponge network | 0.705 | 0.00014 | 0.195 | 0.06908 | 0.351 |
| 29161 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-577;hsa-miR-7-1-3p | 15 | ADAMTSL3 | Sponge network | -0.418 | 0.00068 | -1.559 | 0 | 0.351 |
| 29162 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 30 | BHLHE41 | Sponge network | -1.697 | 0.00889 | 0.353 | 0.12753 | 0.351 |
| 29163 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-577 | 19 | CNTN1 | Sponge network | -0.465 | 0.04703 | -2.594 | 0 | 0.351 |
| 29164 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-93-5p | 39 | EPHA5 | Sponge network | -0.154 | 0.78939 | -0.957 | 0.01733 | 0.351 |
| 29165 | RP11-693J15.4 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-374b-3p | 12 | RPS6KA6 | Sponge network | -0.72 | 0.24239 | -2.989 | 0 | 0.351 |
| 29166 | RP3-402G11.26 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 16 | PRKAA2 | Sponge network | -0.254 | 0.19303 | -2.462 | 0 | 0.351 |
| 29167 | BCYRN1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 10 | SKIL | Sponge network | 0.311 | 0.10344 | 0.559 | 8.0E-5 | 0.351 |
| 29168 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 12 | PIK3C2A | Sponge network | 0.924 | 0 | 0.061 | 0.53776 | 0.351 |
| 29169 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-501-5p | 16 | MAFB | Sponge network | 0.395 | 0.15451 | -0.023 | 0.89865 | 0.351 |
| 29170 | RP11-798M19.6 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-539-5p | 10 | PIK3IP1 | Sponge network | -0.612 | 0.00094 | -0.187 | 0.19945 | 0.351 |
| 29171 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-590-3p | 18 | STARD13 | Sponge network | 0.082 | 0.55654 | -0.187 | 0.27383 | 0.351 |
| 29172 | RP4-669P10.16 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-542-3p | 11 | DIP2C | Sponge network | -0.301 | 0.06597 | -0.436 | 0.01713 | 0.351 |
| 29173 | PCA3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-409-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-940 | 49 | PPARA | Sponge network | -0.709 | 0.18168 | -0.379 | 0.00548 | 0.351 |
| 29174 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-193a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | NF1 | Sponge network | 0.222 | 0.29133 | 0.51 | 0 | 0.351 |
| 29175 | RP11-283I3.6 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-576-5p | 20 | UBR3 | Sponge network | 0.614 | 1.0E-5 | -0.021 | 0.82774 | 0.351 |
| 29176 | RP11-802E16.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 15 | TBL1XR1 | Sponge network | 2.048 | 0 | 0.578 | 0 | 0.351 |
| 29177 | RP11-212P7.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ZMYND11 | Sponge network | 0.219 | 0.1516 | -0.2 | 0.05449 | 0.351 |
| 29178 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p | 11 | KIT | Sponge network | 0.889 | 9.0E-5 | -2.184 | 0 | 0.351 |
| 29179 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-625-5p | 13 | YAP1 | Sponge network | 1.03 | 0 | 0.22 | 0.11836 | 0.351 |
| 29180 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | GAS7 | Sponge network | -0.18 | 0.20343 | -0.555 | 0.01602 | 0.351 |
| 29181 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PRKAB2 | Sponge network | 0.422 | 0.27021 | -0.367 | 0.01371 | 0.351 |
| 29182 | CTC-510F12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-484;hsa-miR-935 | 18 | NFIB | Sponge network | -0.691 | 0.00146 | 0.023 | 0.90795 | 0.351 |
| 29183 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-940 | 10 | CRTC3 | Sponge network | 0.272 | 0.44692 | 0.164 | 0.16786 | 0.351 |
| 29184 | OSER1-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-486-5p;hsa-miR-7-1-3p | 15 | AKAP6 | Sponge network | -0.13 | 0.48734 | -1.586 | 3.0E-5 | 0.351 |
| 29185 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-96-5p | 15 | PCDH8 | Sponge network | -2.46 | 0 | -0.997 | 0.05366 | 0.351 |
| 29186 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | USP6 | Sponge network | 0.659 | 3.0E-5 | 0.188 | 0.3871 | 0.351 |
| 29187 | RP11-999E24.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-7-5p | 13 | TET1 | Sponge network | -0.693 | 0.05629 | -0.135 | 0.52138 | 0.351 |
| 29188 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | MLF1 | Sponge network | -0.129 | 0.57177 | -0.893 | 0.00727 | 0.351 |
| 29189 | RP11-359B12.2 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 13 | FOXO4 | Sponge network | 0.064 | 0.72934 | -0.516 | 2.0E-5 | 0.351 |
| 29190 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p | 15 | RPS6KA2 | Sponge network | 0.335 | 0.20596 | -0.57 | 0.00013 | 0.351 |
| 29191 | AC034220.3 |
hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-590-3p | 17 | DMXL1 | Sponge network | 0.309 | 0.07304 | -0.426 | 0.00056 | 0.351 |
| 29192 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p | 18 | USP6 | Sponge network | 0.082 | 0.55397 | 0.188 | 0.3871 | 0.351 |
| 29193 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 56 | THRB | Sponge network | -0.709 | 0.18168 | -1.117 | 0.0004 | 0.351 |
| 29194 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-93-5p | 14 | RNASEL | Sponge network | -0.811 | 2.0E-5 | -0.272 | 0.01796 | 0.351 |
| 29195 | LINC01018 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | ZNF536 | Sponge network | -2.915 | 0.00015 | -3.115 | 0 | 0.351 |
| 29196 | AGAP11 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p | 40 | NTRK2 | Sponge network | -1.645 | 0 | -0.736 | 0.06495 | 0.351 |
| 29197 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-940 | 23 | PPP1R9A | Sponge network | -1.331 | 3.0E-5 | -0.903 | 0.04254 | 0.351 |
| 29198 | AC003090.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | CSMD1 | Sponge network | -2.117 | 0.00161 | -0.63 | 0.18881 | 0.351 |
| 29199 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | USP12 | Sponge network | -0.318 | 0.65847 | -0.102 | 0.40768 | 0.351 |
| 29200 | RP11-822E23.8 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-335-5p | 13 | PLXNC1 | Sponge network | -0.777 | 0.16667 | 0.569 | 0.00329 | 0.351 |
| 29201 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 12 | PAFAH1B1 | Sponge network | 0.497 | 0.01451 | -0.429 | 0 | 0.351 |
| 29202 | CTC-378H22.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-455-5p | 14 | RUNX1T1 | Sponge network | 0.001 | 0.99663 | -0.344 | 0.2374 | 0.351 |
| 29203 | HOXB-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-671-5p | 12 | EHD3 | Sponge network | -0.154 | 0.53954 | -0.484 | 0.01747 | 0.351 |
| 29204 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-362-3p | 13 | GREM1 | Sponge network | -0.737 | 0.10822 | 0.902 | 0.01691 | 0.351 |
| 29205 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p | 15 | PCDH10 | Sponge network | 0.082 | 0.55397 | -1.644 | 0.0078 | 0.351 |
| 29206 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | PTPN11 | Sponge network | 0.765 | 7.0E-5 | 0.431 | 2.0E-5 | 0.351 |
| 29207 | CTC-510F12.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-335-3p | 13 | TMEM132B | Sponge network | -0.691 | 0.00146 | -0.83 | 0.01084 | 0.351 |
| 29208 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 10 | UVRAG | Sponge network | 1.302 | 7.0E-5 | -0.017 | 0.83865 | 0.351 |
| 29209 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p | 11 | KCNAB1 | Sponge network | -0.896 | 0.00099 | -0.755 | 2.0E-5 | 0.351 |
| 29210 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | ERCC4 | Sponge network | -0.358 | 0.17255 | 0.126 | 0.15266 | 0.351 |
| 29211 | LINC00854 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-628-5p;hsa-miR-7-5p | 24 | TCF4 | Sponge network | 1.18 | 0 | 0.016 | 0.92271 | 0.351 |
| 29212 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.401 | 0.00066 | 0.323 | 0.00792 | 0.351 |
| 29213 | HHIP-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | AKAP6 | Sponge network | -0.737 | 0.10822 | -1.586 | 3.0E-5 | 0.351 |
| 29214 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-708-5p;hsa-miR-93-3p;hsa-miR-940 | 27 | TOM1L2 | Sponge network | -1.439 | 4.0E-5 | -0.994 | 0 | 0.351 |
| 29215 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 40 | BMPR2 | Sponge network | 0.299 | 0.57722 | 0.206 | 0.0635 | 0.351 |
| 29216 | CKMT2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p | 10 | SCN11A | Sponge network | 0.082 | 0.55654 | -1.15 | 0.00299 | 0.351 |
| 29217 | RP4-639F20.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p | 13 | CDH23 | Sponge network | -0.517 | 0.02935 | -0.974 | 0.00034 | 0.351 |
| 29218 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 14 | ST6GAL2 | Sponge network | -0.141 | 0.26434 | -1.201 | 0.00597 | 0.351 |
| 29219 | RP11-403I13.8 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | DNMT3A | Sponge network | 0.619 | 0.00157 | 0.302 | 0.0262 | 0.351 |
| 29220 | LINC00094 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-92a-3p | 14 | ITGA5 | Sponge network | 0.253 | 0.09565 | 0.452 | 0.03524 | 0.351 |
| 29221 | LINC01004 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | RICTOR | Sponge network | 1.115 | 0 | 0.388 | 0.00188 | 0.351 |
| 29222 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-182-5p | 10 | STARD13 | Sponge network | -1.349 | 0.00017 | -0.187 | 0.27383 | 0.351 |
| 29223 | RP6-65G23.3 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | CBFB | Sponge network | 0.272 | 0.24062 | 1.061 | 0 | 0.351 |
| 29224 | RP11-532F6.3 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935 | 21 | GJA1 | Sponge network | -0.896 | 0.00099 | -0.485 | 0.02179 | 0.351 |
| 29225 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 20 | IGF1 | Sponge network | 1.757 | 0 | -0.204 | 0.5898 | 0.351 |
| 29226 | LINC00657 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | SIRT1 | Sponge network | 0.91 | 0 | 0.109 | 0.2593 | 0.351 |
| 29227 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | LRIG1 | Sponge network | 0.082 | 0.55654 | -0.537 | 0.00588 | 0.351 |
| 29228 | RP11-428J1.5 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | WWTR1 | Sponge network | 0.538 | 0.00076 | -0.345 | 0.11997 | 0.351 |
| 29229 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 11 | DNAJB4 | Sponge network | 0.889 | 9.0E-5 | -0.7 | 1.0E-5 | 0.351 |
| 29230 | RP11-98I9.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CEP170 | Sponge network | 0.67 | 0.00048 | 0.943 | 0 | 0.351 |
| 29231 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | LRP1B | Sponge network | -0.611 | 0.09607 | -2.163 | 0 | 0.351 |
| 29232 | RP11-248G5.8 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-539-5p | 10 | TCF12 | Sponge network | 1.294 | 0 | 0.174 | 0.13271 | 0.351 |
| 29233 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p | 23 | ESR1 | Sponge network | 0.889 | 9.0E-5 | -1.035 | 3.0E-5 | 0.351 |
| 29234 | RP11-1277A3.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-629-3p;hsa-miR-96-5p | 12 | RASSF8 | Sponge network | 0.453 | 0.01839 | -0.122 | 0.65591 | 0.351 |
| 29235 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | IGSF10 | Sponge network | -0.222 | 0.32445 | -1.751 | 1.0E-5 | 0.351 |
| 29236 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p | 10 | PLA2R1 | Sponge network | -0.777 | 0.16667 | -0.265 | 0.24676 | 0.351 |
| 29237 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | PTPRB | Sponge network | 0.494 | 0.00339 | 0.105 | 0.47122 | 0.351 |
| 29238 | RP11-359B12.2 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | 0.064 | 0.72934 | -0.226 | 0.11691 | 0.351 |
| 29239 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 49 | NTRK3 | Sponge network | -0.465 | 0.04703 | -1.77 | 3.0E-5 | 0.351 |
| 29240 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-484;hsa-miR-590-3p | 27 | MED12L | Sponge network | 0.494 | 0.00339 | -0.194 | 0.55299 | 0.351 |
| 29241 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-940 | 47 | WDFY3 | Sponge network | 0.61 | 0.01593 | -0.201 | 0.0967 | 0.351 |
| 29242 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | PHC3 | Sponge network | 0.657 | 4.0E-5 | 0.212 | 0.0499 | 0.351 |
| 29243 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-429 | 20 | ABL2 | Sponge network | 0.056 | 0.81195 | 0.82 | 0 | 0.351 |
| 29244 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-654-3p | 18 | TP53INP1 | Sponge network | 0.637 | 0.00069 | -0.496 | 0.00064 | 0.351 |
| 29245 | RP11-403I13.7 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 28 | ABI2 | Sponge network | 0.395 | 0.15451 | 0.175 | 0.14787 | 0.351 |
| 29246 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 16 | ARNT2 | Sponge network | -0.517 | 0.02935 | -0.332 | 0.25851 | 0.351 |
| 29247 | RP11-707P17.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-495-3p | 11 | CREB1 | Sponge network | 1.877 | 0 | 0.203 | 0.00537 | 0.351 |
| 29248 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-590-5p;hsa-miR-96-5p | 24 | CNTN1 | Sponge network | 0.331 | 0.57247 | -2.594 | 0 | 0.351 |
| 29249 | SNHG14 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-940 | 28 | SIRT1 | Sponge network | -1.111 | 0.0003 | 0.109 | 0.2593 | 0.351 |
| 29250 | RP11-384O8.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p | 16 | NRK | Sponge network | -0.738 | 0.21233 | 0.656 | 0.19217 | 0.351 |
| 29251 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-5p | 16 | FSTL5 | Sponge network | 0.249 | 0.35902 | -1.3 | 0.01619 | 0.351 |
| 29252 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 33 | MXI1 | Sponge network | -0.49 | 0.04946 | -0.987 | 0 | 0.351 |
| 29253 | TRAF3IP2-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | BCL9 | Sponge network | -0.323 | 0.01372 | 0.321 | 0.00267 | 0.351 |
| 29254 | USP3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-93-3p | 25 | NCOA1 | Sponge network | -0.162 | 0.47687 | -0.432 | 0 | 0.351 |
| 29255 | RP11-686D22.4 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p | 13 | QKI | Sponge network | 0.793 | 5.0E-5 | -0.333 | 0.04111 | 0.351 |
| 29256 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-424-5p | 17 | RARB | Sponge network | -0.777 | 0.16667 | -0.825 | 2.0E-5 | 0.351 |
| 29257 | HCG11 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PTCH1 | Sponge network | 0.385 | 0.10067 | -0.1 | 0.6179 | 0.351 |
| 29258 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | LRP1B | Sponge network | -0.969 | 2.0E-5 | -2.163 | 0 | 0.35 |
| 29259 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-940 | 67 | NFIB | Sponge network | -0.129 | 0.57177 | 0.023 | 0.90795 | 0.35 |
| 29260 | CTC-429P9.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 11 | CRTC1 | Sponge network | 0.178 | 0.29106 | -0.116 | 0.28412 | 0.35 |
| 29261 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | TSC1 | Sponge network | 0.214 | 0.18246 | 0.103 | 0.26071 | 0.35 |
| 29262 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 22 | ETV1 | Sponge network | 0.395 | 0.15451 | -0.096 | 0.67674 | 0.35 |
| 29263 | LPP-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-96-5p | 27 | ASTN1 | Sponge network | -0.18 | 0.20343 | -2.389 | 2.0E-5 | 0.35 |
| 29264 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-7-1-3p | 19 | KAT6B | Sponge network | 0.4 | 0.14611 | -0.478 | 0.00018 | 0.35 |
| 29265 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | LRRC55 | Sponge network | -0.222 | 0.32445 | -0.026 | 0.93163 | 0.35 |
| 29266 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-98-5p | 17 | TGFBR3 | Sponge network | -1.116 | 0.23686 | -1.173 | 1.0E-5 | 0.35 |
| 29267 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | ARID5B | Sponge network | -1.575 | 6.0E-5 | 0.342 | 0.01676 | 0.35 |
| 29268 | RP5-1198O20.4 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ADH1B | Sponge network | 0.997 | 0.00066 | -3.716 | 0 | 0.35 |
| 29269 | LINC00865 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429 | 14 | CITED2 | Sponge network | -1.249 | 7.0E-5 | -1.545 | 0 | 0.35 |
| 29270 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 29 | MOB1B | Sponge network | -0.738 | 0.21233 | 0.197 | 0.15266 | 0.35 |
| 29271 | RP11-345P4.7 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 1.597 | 0 | 0.362 | 0.00066 | 0.35 |
| 29272 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TET1 | Sponge network | 0.862 | 0.06213 | -0.135 | 0.52138 | 0.35 |
| 29273 | HHIP-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-532-3p | 12 | SYNM | Sponge network | -0.737 | 0.10822 | -2.309 | 1.0E-5 | 0.35 |
| 29274 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-671-5p | 14 | KIT | Sponge network | -0.176 | 0.59692 | -2.184 | 0 | 0.35 |
| 29275 | RP11-458D21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | PDE4DIP | Sponge network | 0.663 | 0.00073 | -0.282 | 0.0257 | 0.35 |
| 29276 | RP11-360F5.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-629-3p | 10 | DDX6 | Sponge network | 1.867 | 0 | 0.091 | 0.36428 | 0.35 |
| 29277 | RP11-127B20.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 30 | LPP | Sponge network | 0.411 | 0.1335 | -0.459 | 0.04475 | 0.35 |
| 29278 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 13 | LATS2 | Sponge network | -0.726 | 0.00132 | 0.159 | 0.2304 | 0.35 |
| 29279 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-589-3p | 14 | ABCA10 | Sponge network | -0.777 | 0.16667 | -0.571 | 0.03674 | 0.35 |
| 29280 | RP11-819C21.1 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-342-3p | 17 | ABL1 | Sponge network | 0.537 | 0.00139 | -0.215 | 0.07804 | 0.35 |
| 29281 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 43 | SSBP2 | Sponge network | 0.16 | 0.50785 | -1.58 | 0 | 0.35 |
| 29282 | BOLA3-AS1 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p | 11 | CSDE1 | Sponge network | -0.246 | 0.35034 | -0.493 | 0 | 0.35 |
| 29283 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | FZD3 | Sponge network | 0.953 | 0 | 0.104 | 0.60316 | 0.35 |
| 29284 | PART1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 49 | NTRK2 | Sponge network | -2.925 | 0 | -0.736 | 0.06495 | 0.35 |
| 29285 | PCA3 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | CITED2 | Sponge network | -0.709 | 0.18168 | -1.545 | 0 | 0.35 |
| 29286 | AC108676.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 20 | MDM4 | Sponge network | 4.346 | 2.0E-5 | 0.345 | 0.00762 | 0.35 |
| 29287 | AC016747.3 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | CXCL12 | Sponge network | 0.199 | 0.38015 | -1.421 | 0 | 0.35 |
| 29288 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 10 | PTPN14 | Sponge network | 0.542 | 0.00054 | 0.144 | 0.34595 | 0.35 |
| 29289 | RP11-110I1.11 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-590-3p;hsa-miR-940 | 10 | AKAP11 | Sponge network | 1.034 | 0 | 0.196 | 0.09825 | 0.35 |
| 29290 | BDNF-AS |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 52 | DST | Sponge network | -0.668 | 3.0E-5 | -0.713 | 0.00096 | 0.35 |
| 29291 | RP11-474O21.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-93-5p | 14 | GAS7 | Sponge network | -0.023 | 0.95109 | -0.555 | 0.01602 | 0.35 |
| 29292 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-577;hsa-miR-7-1-3p | 15 | TIMP3 | Sponge network | 0.259 | 0.12542 | -0.546 | 0.02307 | 0.35 |
| 29293 | EIF3J-AS1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CREB1 | Sponge network | 0.331 | 0.00509 | 0.203 | 0.00537 | 0.35 |
| 29294 | HOXA-AS2 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | DCLK1 | Sponge network | 0.61 | 0.01593 | -1.324 | 0.00014 | 0.35 |
| 29295 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-935 | 29 | FBXO32 | Sponge network | -0.384 | 0.40652 | -0.495 | 0.07561 | 0.35 |
| 29296 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-497-5p | 25 | CBL | Sponge network | 1.089 | 0 | 0.291 | 0.01267 | 0.35 |
| 29297 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | MITF | Sponge network | -2.915 | 0.00015 | -0.769 | 0.00022 | 0.35 |
| 29298 | AP001347.6 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | EYA4 | Sponge network | -1.161 | 0.00591 | 0.3 | 0.36876 | 0.35 |
| 29299 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 12 | PRKAA1 | Sponge network | 0.826 | 1.0E-5 | 0.317 | 0.0031 | 0.35 |
| 29300 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 11 | ATRX | Sponge network | 0.702 | 7.0E-5 | 0.261 | 0.04408 | 0.35 |
| 29301 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 17 | NR2C2 | Sponge network | 0.174 | 0.30034 | 0.21 | 0.03224 | 0.35 |
| 29302 | MALAT1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | ACVR2A | Sponge network | 0.782 | 0.00016 | -0.409 | 0.00039 | 0.35 |
| 29303 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CLIP1 | Sponge network | 0.335 | 0.20596 | -0.215 | 0.08684 | 0.35 |
| 29304 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-5p | 28 | THRB | Sponge network | -2.095 | 0.00112 | -1.117 | 0.0004 | 0.35 |
| 29305 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-376c-3p | 15 | TBL1XR1 | Sponge network | 0.919 | 0 | 0.578 | 0 | 0.35 |
| 29306 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-589-3p | 15 | PIK3R1 | Sponge network | -4.463 | 0.00025 | -0.133 | 0.36854 | 0.35 |
| 29307 | LINC00865 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 31 | SNX29 | Sponge network | -1.249 | 7.0E-5 | -0.34 | 0.01371 | 0.35 |
| 29308 | A1BG-AS1 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-671-5p | 10 | KIT | Sponge network | -0.164 | 0.45106 | -2.184 | 0 | 0.35 |
| 29309 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 30 | UNC80 | Sponge network | -0.583 | 0.14589 | -1.934 | 0 | 0.35 |
| 29310 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | ETV1 | Sponge network | 0.381 | 0.25967 | -0.096 | 0.67674 | 0.35 |
| 29311 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | WDFY3 | Sponge network | -0.842 | 0.01361 | -0.201 | 0.0967 | 0.35 |
| 29312 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 34 | ST6GALNAC3 | Sponge network | -0.899 | 6.0E-5 | -0.987 | 0 | 0.35 |
| 29313 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | 0.342 | 0.03129 | -0.243 | 0.11408 | 0.35 |
| 29314 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-505-5p | 17 | TBL1XR1 | Sponge network | 0.439 | 0.00313 | 0.578 | 0 | 0.35 |
| 29315 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-26b-3p;hsa-miR-539-5p;hsa-miR-98-5p | 13 | RSPO2 | Sponge network | -0.473 | 0.07602 | -3.038 | 0 | 0.35 |
| 29316 | OSER1-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-582-5p | 22 | DMD | Sponge network | -0.13 | 0.48734 | -1.506 | 0 | 0.35 |
| 29317 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | PLSCR4 | Sponge network | -0.727 | 0.04726 | -0.474 | 0.03833 | 0.35 |
| 29318 | RP11-728F11.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 14 | CADM1 | Sponge network | 0.238 | 0.70544 | -0.9 | 0.00084 | 0.35 |
| 29319 | RP11-119F7.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-7-1-3p | 12 | PLAG1 | Sponge network | 0.225 | 0.30644 | -0.315 | 0.26532 | 0.35 |
| 29320 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 19 | TRIM33 | Sponge network | 0.494 | 0.00339 | 0.402 | 7.0E-5 | 0.35 |
| 29321 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 28 | TET1 | Sponge network | 0.101 | 0.60919 | -0.135 | 0.52138 | 0.35 |
| 29322 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 33 | BCL2 | Sponge network | -0.727 | 0.04726 | -0.899 | 0 | 0.35 |
| 29323 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 43 | PRKAA2 | Sponge network | -0.031 | 0.89063 | -2.462 | 0 | 0.35 |
| 29324 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 26 | NTRK2 | Sponge network | 1.757 | 0 | -0.736 | 0.06495 | 0.35 |
| 29325 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p | 20 | ERCC4 | Sponge network | 0.056 | 0.81195 | 0.126 | 0.15266 | 0.35 |
| 29326 | RP11-927P21.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p | 18 | PIK3CA | Sponge network | 0.921 | 0.00118 | 0.195 | 0.06908 | 0.35 |
| 29327 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 15 | ANK2 | Sponge network | 0.238 | 0.70544 | -1.603 | 1.0E-5 | 0.35 |
| 29328 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | ITGAV | Sponge network | -0.611 | 0.09607 | 0.337 | 0.01002 | 0.35 |
| 29329 | RP11-35G9.3 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ERG | Sponge network | 0.657 | 4.0E-5 | 0.002 | 0.99011 | 0.35 |
| 29330 | CTC-429P9.5 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 10 | GLIPR1 | Sponge network | 0.178 | 0.29106 | 0.146 | 0.3276 | 0.35 |
| 29331 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 24 | SOX11 | Sponge network | 1.153 | 0 | 2.68 | 0 | 0.35 |
| 29332 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-874-3p | 36 | TACC1 | Sponge network | 0.95 | 0.15943 | -0.362 | 0.05406 | 0.35 |
| 29333 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 16 | FOXO3 | Sponge network | 0.826 | 1.0E-5 | -0.04 | 0.72028 | 0.35 |
| 29334 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-374a-5p;hsa-miR-450b-5p | 14 | ST6GAL2 | Sponge network | 0.381 | 0.25967 | -1.201 | 0.00597 | 0.35 |
| 29335 | RP11-395G23.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | PRRX1 | Sponge network | 0.381 | 0.25967 | 1.316 | 0 | 0.35 |
| 29336 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | BMPR2 | Sponge network | 0.419 | 0.0319 | 0.206 | 0.0635 | 0.35 |
| 29337 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p | 10 | USP12 | Sponge network | 1.106 | 0 | -0.102 | 0.40768 | 0.35 |
| 29338 | ARHGEF26-AS1 |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 20 | JUN | Sponge network | -2.46 | 0 | -0.932 | 0 | 0.35 |
| 29339 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-628-5p;hsa-miR-7-5p | 24 | ERC1 | Sponge network | -1.161 | 0.00591 | -0.108 | 0.38794 | 0.35 |
| 29340 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | ARHGAP28 | Sponge network | -0.487 | 0.02157 | -0.911 | 0.00013 | 0.35 |
| 29341 | AF131215.9 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-590-3p | 14 | LIMCH1 | Sponge network | -0.322 | 0.25945 | -0.915 | 1.0E-5 | 0.35 |
| 29342 | MIR497HG |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-92a-1-5p;hsa-miR-96-5p | 12 | PRKACA | Sponge network | -1.439 | 4.0E-5 | -0.255 | 0.0012 | 0.35 |
| 29343 | CTC-297N7.9 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-5p;hsa-miR-93-3p | 12 | UNC5C | Sponge network | -2.069 | 0 | -0.178 | 0.40214 | 0.35 |
| 29344 | NDUFA6-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 39 | AFF1 | Sponge network | -0.355 | 0.0077 | -0.384 | 0.00053 | 0.35 |
| 29345 | RP11-212P7.2 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | 0.219 | 0.1516 | -0.252 | 0.15511 | 0.35 |
| 29346 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | MITF | Sponge network | 0.299 | 0.57722 | -0.769 | 0.00022 | 0.35 |
| 29347 | NR2F1-AS1 |
hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-590-3p | 11 | EGR1 | Sponge network | -0.49 | 0.04946 | -1.195 | 0 | 0.35 |
| 29348 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-940 | 25 | SNX29 | Sponge network | -0.738 | 0.21233 | -0.34 | 0.01371 | 0.35 |
| 29349 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-7-5p | 19 | EPHA3 | Sponge network | 0.001 | 0.99753 | 0.185 | 0.53588 | 0.35 |
| 29350 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-93-5p | 14 | CHD5 | Sponge network | 0.497 | 0.01451 | -0.922 | 0.01521 | 0.35 |
| 29351 | CTC-490E21.10 |
hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-5p | 10 | TMEM30A | Sponge network | 1.46 | 1.0E-5 | 0.096 | 0.31031 | 0.35 |
| 29352 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-326;hsa-miR-539-5p;hsa-miR-590-3p | 12 | DDX5 | Sponge network | -0.323 | 0.01372 | -0.099 | 0.17428 | 0.35 |
| 29353 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p | 13 | RASSF2 | Sponge network | -7.551 | 0 | -0.273 | 0.21961 | 0.35 |
| 29354 | LINC00641 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | TMTC2 | Sponge network | -0.222 | 0.32445 | -0.215 | 0.18248 | 0.35 |
| 29355 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-429 | 10 | GREM1 | Sponge network | 0.727 | 3.0E-5 | 0.902 | 0.01691 | 0.35 |
| 29356 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | KDSR | Sponge network | 0.537 | 0.00139 | 0.239 | 0.02216 | 0.35 |
| 29357 | LINC00476 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 32 | ZFX | Sponge network | -0.615 | 0.00015 | 0.09 | 0.42563 | 0.35 |
| 29358 | PGM5-AS1 |
hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-624-5p;hsa-miR-7-1-3p | 12 | GJA1 | Sponge network | -4.546 | 0 | -0.485 | 0.02179 | 0.35 |
| 29359 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-654-3p | 13 | BMPR2 | Sponge network | 1.316 | 0 | 0.206 | 0.0635 | 0.35 |
| 29360 | EIF3J-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p | 28 | RICTOR | Sponge network | 0.331 | 0.00509 | 0.388 | 0.00188 | 0.35 |
| 29361 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PPP1R9A | Sponge network | -0.206 | 0.12942 | -0.903 | 0.04254 | 0.35 |
| 29362 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-96-5p | 14 | PCDH8 | Sponge network | -0.976 | 0.00025 | -0.997 | 0.05366 | 0.35 |
| 29363 | RP1-59D14.5 |
hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-940 | 18 | AKAP11 | Sponge network | 1.162 | 5.0E-5 | 0.196 | 0.09825 | 0.35 |
| 29364 | HHIP-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-769-5p | 24 | GRIK3 | Sponge network | -0.737 | 0.10822 | -3.208 | 0 | 0.35 |
| 29365 | AC003090.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | RIMBP2 | Sponge network | -2.117 | 0.00161 | -1.968 | 5.0E-5 | 0.35 |
| 29366 | RP11-758N13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-505-5p | 25 | AR | Sponge network | 2.939 | 3.0E-5 | -1.554 | 0 | 0.35 |
| 29367 | CTD-3025N20.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 14 | FZD3 | Sponge network | 1.046 | 0 | 0.104 | 0.60316 | 0.35 |
| 29368 | RP11-678G14.3 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | PCDH9 | Sponge network | -4.477 | 0 | -1.823 | 4.0E-5 | 0.35 |
| 29369 | RP11-359B12.2 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | ZDBF2 | Sponge network | 0.064 | 0.72934 | -0.998 | 0.00025 | 0.35 |
| 29370 | ERVH48-1 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-590-3p | 24 | CREB1 | Sponge network | 1.04 | 0.00023 | 0.203 | 0.00537 | 0.35 |
| 29371 | KB-1410C5.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | ZFHX4 | Sponge network | 0.959 | 0 | 0.018 | 0.95574 | 0.35 |
| 29372 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-5p | 15 | TBX18 | Sponge network | 0.95 | 0.15943 | 0.843 | 0.02945 | 0.35 |
| 29373 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-7-1-3p | 15 | AR | Sponge network | 0.959 | 0 | -1.554 | 0 | 0.35 |
| 29374 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 14 | PTPN11 | Sponge network | 0.541 | 0.00125 | 0.431 | 2.0E-5 | 0.35 |
| 29375 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-589-5p | 31 | CBX5 | Sponge network | 0.082 | 0.55397 | 0.165 | 0.24446 | 0.35 |
| 29376 | LINC00473 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p | 19 | EZH1 | Sponge network | -1.203 | 0.03881 | -0.564 | 1.0E-5 | 0.35 |
| 29377 | LA16c-321D4.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 15 | RICTOR | Sponge network | 2.459 | 5.0E-5 | 0.388 | 0.00188 | 0.35 |
| 29378 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-532-5p;hsa-miR-590-3p | 15 | SLITRK6 | Sponge network | -1.203 | 0.03881 | -2.121 | 0 | 0.35 |
| 29379 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-331-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | ATRX | Sponge network | 0.799 | 1.0E-5 | 0.261 | 0.04408 | 0.35 |
| 29380 | LINC00476 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | PAIP2 | Sponge network | -0.615 | 0.00015 | -0.515 | 0 | 0.35 |
| 29381 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 18 | GPAM | Sponge network | 0.683 | 1.0E-5 | 0.142 | 0.29732 | 0.35 |
| 29382 | CTD-2528L19.6 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TSC1 | Sponge network | 0.953 | 0 | 0.103 | 0.26071 | 0.35 |
| 29383 | SH3RF3-AS1 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-582-5p | 15 | KLF10 | Sponge network | 0.889 | 9.0E-5 | -0.783 | 0 | 0.35 |
| 29384 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | AKAP12 | Sponge network | 0.349 | 0.01695 | -0.932 | 0.0028 | 0.35 |
| 29385 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p | 10 | DMTF1 | Sponge network | 0.532 | 0.00505 | 0.248 | 0.02726 | 0.35 |
| 29386 | CTD-2184D3.6 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PCDH9 | Sponge network | 0.272 | 0.44692 | -1.823 | 4.0E-5 | 0.35 |
| 29387 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | TAL1 | Sponge network | 0.064 | 0.72934 | -0.462 | 0.00574 | 0.35 |
| 29388 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-628-5p | 12 | CDC73 | Sponge network | 1.923 | 0 | 0.094 | 0.20463 | 0.35 |
| 29389 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | NRXN1 | Sponge network | 0.056 | 0.8494 | -3.778 | 0 | 0.35 |
| 29390 | LINC00092 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-589-3p | 11 | KCNJ5 | Sponge network | -1.886 | 0 | -0.281 | 0.3339 | 0.35 |
| 29391 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-93-5p | 47 | AR | Sponge network | -1.047 | 0 | -1.554 | 0 | 0.35 |
| 29392 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 43 | ADARB1 | Sponge network | -1.645 | 0 | -0.335 | 0.06631 | 0.35 |
| 29393 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-30b-3p;hsa-miR-34a-5p | 10 | GPC6 | Sponge network | -0.187 | 0.23787 | -0.054 | 0.81465 | 0.35 |
| 29394 | RP11-798M19.6 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 37 | ACVR2A | Sponge network | -0.612 | 0.00094 | -0.409 | 0.00039 | 0.35 |
| 29395 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-7-1-3p | 13 | SON | Sponge network | -0.176 | 0.59692 | 0.168 | 0.04406 | 0.35 |
| 29396 | HHIP-AS1 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-32-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | JUN | Sponge network | -0.737 | 0.10822 | -0.932 | 0 | 0.35 |
| 29397 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-330-3p;hsa-miR-590-3p | 11 | NEDD4 | Sponge network | -1.582 | 8.0E-5 | 0.02 | 0.89834 | 0.35 |
| 29398 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 11 | PIK3C2A | Sponge network | 2.094 | 0 | 0.061 | 0.53776 | 0.35 |
| 29399 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-664a-5p | 30 | ADARB1 | Sponge network | 0.614 | 1.0E-5 | -0.335 | 0.06631 | 0.35 |
| 29400 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-625-5p | 25 | ZMYM2 | Sponge network | 1.03 | 0 | 0.279 | 0.01273 | 0.35 |
| 29401 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p | 30 | PPP1R9A | Sponge network | -0.18 | 0.20343 | -0.903 | 0.04254 | 0.35 |
| 29402 | RP11-25K19.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-629-3p | 11 | POU2F2 | Sponge network | -1.057 | 0.00029 | -0.233 | 0.32214 | 0.35 |
| 29403 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | NUAK1 | Sponge network | 0.614 | 1.0E-5 | 0.904 | 0 | 0.35 |
| 29404 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429 | 17 | LRP1B | Sponge network | -0.154 | 0.53954 | -2.163 | 0 | 0.35 |
| 29405 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-335-5p;hsa-miR-7-5p | 14 | ZBTB4 | Sponge network | -0.285 | 0.2172 | -0.739 | 0 | 0.35 |
| 29406 | LINC00476 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PREX2 | Sponge network | -0.615 | 0.00015 | -0.272 | 0.25382 | 0.35 |
| 29407 | SERTAD4-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-369-3p;hsa-miR-576-5p | 10 | PDE4DIP | Sponge network | -0.932 | 0.00329 | -0.282 | 0.0257 | 0.35 |
| 29408 | H1FX-AS1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 30 | QKI | Sponge network | -0.206 | 0.12942 | -0.333 | 0.04111 | 0.35 |
| 29409 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-589-3p;hsa-miR-93-5p | 11 | EPAS1 | Sponge network | -2.531 | 0 | -0.184 | 0.14418 | 0.35 |
| 29410 | JAZF1-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | HERC1 | Sponge network | -0.769 | 0.00035 | -0.196 | 0.08544 | 0.35 |
| 29411 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p | 11 | RCAN2 | Sponge network | 0.594 | 0.00064 | -0.33 | 0.15172 | 0.35 |
| 29412 | RP11-458D21.1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 10 | PHF3 | Sponge network | 0.663 | 0.00073 | 0.126 | 0.19652 | 0.35 |
| 29413 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p | 16 | TBX18 | Sponge network | 0.056 | 0.81195 | 0.843 | 0.02945 | 0.35 |
| 29414 | LINC00641 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 15 | NGFR | Sponge network | -0.222 | 0.32445 | -1.271 | 0.01058 | 0.35 |
| 29415 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-93-3p;hsa-miR-93-5p | 11 | FYN | Sponge network | -2.69 | 0.0001 | -0.131 | 0.37051 | 0.35 |
| 29416 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p | 10 | ZNF292 | Sponge network | 0.603 | 0.00127 | 0.276 | 0.04148 | 0.35 |
| 29417 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 60 | BMPR2 | Sponge network | -0.942 | 0 | 0.206 | 0.0635 | 0.35 |
| 29418 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | TMTC3 | Sponge network | 0.873 | 0 | 0.123 | 0.22189 | 0.35 |
| 29419 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 19 | RNF144A | Sponge network | -0.513 | 0.04703 | 0.594 | 0.0009 | 0.35 |
| 29420 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-7-5p;hsa-miR-96-5p | 16 | CRTC3 | Sponge network | -0.18 | 0.20343 | 0.164 | 0.16786 | 0.35 |
| 29421 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-98-5p | 23 | THBS1 | Sponge network | -2.62 | 0 | 0.741 | 0.00386 | 0.35 |
| 29422 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | SYNE1 | Sponge network | -0.727 | 0.04726 | -0.738 | 0.00316 | 0.35 |
| 29423 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 19 | SYNE1 | Sponge network | 1.757 | 0 | -0.738 | 0.00316 | 0.35 |
| 29424 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-23a-3p | 10 | DIP2C | Sponge network | -2.549 | 0.00528 | -0.436 | 0.01713 | 0.35 |
| 29425 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | EPHA7 | Sponge network | -0.053 | 0.80685 | -2.197 | 3.0E-5 | 0.35 |
| 29426 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-590-3p | 15 | USP6 | Sponge network | 1.073 | 0.00216 | 0.188 | 0.3871 | 0.35 |
| 29427 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 20 | DYNC1I1 | Sponge network | -0.727 | 0.04726 | -1.12 | 0.00107 | 0.35 |
| 29428 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p | 14 | CACNA2D1 | Sponge network | 0.082 | 0.55397 | -0.846 | 0.00675 | 0.35 |
| 29429 | SNHG14 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 12 | PLA2R1 | Sponge network | -1.111 | 0.0003 | -0.265 | 0.24676 | 0.35 |
| 29430 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 32 | IGF1 | Sponge network | -0.222 | 0.32445 | -0.204 | 0.5898 | 0.35 |
| 29431 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | KCNAB1 | Sponge network | 0.537 | 0.00139 | -0.755 | 2.0E-5 | 0.35 |
| 29432 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | RNF144A | Sponge network | 0.101 | 0.60919 | 0.594 | 0.0009 | 0.35 |
| 29433 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p | 20 | ZMYM2 | Sponge network | 0.494 | 0.00339 | 0.279 | 0.01273 | 0.35 |
| 29434 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | SNX29 | Sponge network | -0.517 | 0.02935 | -0.34 | 0.01371 | 0.35 |
| 29435 | RP11-1277A3.1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-484;hsa-miR-96-5p | 14 | MED12L | Sponge network | 0.453 | 0.01839 | -0.194 | 0.55299 | 0.35 |
| 29436 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p | 13 | PCDH8 | Sponge network | -0.49 | 0.04946 | -0.997 | 0.05366 | 0.35 |
| 29437 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-424-5p | 21 | RUNX1T1 | Sponge network | -3.586 | 0.00032 | -0.344 | 0.2374 | 0.35 |
| 29438 | AP001055.6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p | 13 | EZH1 | Sponge network | 0.693 | 0.00076 | -0.564 | 1.0E-5 | 0.35 |
| 29439 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-744-3p | 18 | HIP1 | Sponge network | -0.162 | 0.47687 | 0.774 | 0 | 0.35 |
| 29440 | OIP5-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-199b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | TSC1 | Sponge network | 0.421 | 0.00084 | 0.103 | 0.26071 | 0.35 |
| 29441 | RP11-701H24.7 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-93-5p | 19 | DST | Sponge network | -2.039 | 0.03447 | -0.713 | 0.00096 | 0.35 |
| 29442 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-577 | 10 | FGFR1 | Sponge network | 1.294 | 0.0031 | -0.509 | 0.03957 | 0.35 |
| 29443 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 16 | TXNIP | Sponge network | -1.047 | 0 | -1.277 | 0 | 0.35 |
| 29444 | RP4-669P10.16 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-409-3p | 16 | AFF1 | Sponge network | -0.301 | 0.06597 | -0.384 | 0.00053 | 0.35 |
| 29445 | RP11-359E10.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | GJA1 | Sponge network | 0.299 | 0.57722 | -0.485 | 0.02179 | 0.35 |
| 29446 | RP4-669L17.10 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p;hsa-miR-495-3p | 10 | PHF6 | Sponge network | 1.093 | 0 | 0.785 | 0 | 0.35 |
| 29447 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | CLASP2 | Sponge network | 0.385 | 0.10067 | -0.038 | 0.71 | 0.35 |
| 29448 | RP11-806L2.6 |
hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-625-5p | 10 | MDM4 | Sponge network | 2.103 | 0.0002 | 0.345 | 0.00762 | 0.35 |
| 29449 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 17 | FOXO3 | Sponge network | 1.317 | 0 | -0.04 | 0.72028 | 0.35 |
| 29450 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 23 | SON | Sponge network | 0.532 | 0.00505 | 0.168 | 0.04406 | 0.35 |
| 29451 | HCG11 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p | 12 | PDZD2 | Sponge network | 0.385 | 0.10067 | -0.909 | 3.0E-5 | 0.35 |
| 29452 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 23 | CXXC4 | Sponge network | -1.281 | 0.00012 | -0.433 | 0.21055 | 0.35 |
| 29453 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-664a-5p | 14 | ADARB1 | Sponge network | 0.912 | 0.00014 | -0.335 | 0.06631 | 0.35 |
| 29454 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p | 21 | PRDM2 | Sponge network | 0.4 | 0.14611 | -0.258 | 0.00721 | 0.35 |
| 29455 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-495-3p;hsa-miR-532-3p | 10 | USP12 | Sponge network | 0.993 | 0 | -0.102 | 0.40768 | 0.35 |
| 29456 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-96-5p | 22 | MITF | Sponge network | 0.331 | 0.57247 | -0.769 | 0.00022 | 0.35 |
| 29457 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 19 | NRXN1 | Sponge network | -0.726 | 0.00132 | -3.778 | 0 | 0.35 |
| 29458 | RP4-758J18.10 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-21-3p | 11 | DYNC1I1 | Sponge network | -0.091 | 0.71468 | -1.12 | 0.00107 | 0.349 |
| 29459 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 15 | FZD3 | Sponge network | 0.48 | 0.05578 | 0.104 | 0.60316 | 0.349 |
| 29460 | CTD-2017D11.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-940 | 20 | CREB1 | Sponge network | 0.792 | 0.0049 | 0.203 | 0.00537 | 0.349 |
| 29461 | CTC-297N7.9 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p | 12 | NRXN1 | Sponge network | -2.069 | 0 | -3.778 | 0 | 0.349 |
| 29462 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 55 | PRKAA2 | Sponge network | 0.064 | 0.72934 | -2.462 | 0 | 0.349 |
| 29463 | RP11-758N13.1 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p | 11 | CNTN1 | Sponge network | 2.939 | 3.0E-5 | -2.594 | 0 | 0.349 |
| 29464 | RP11-728F11.4 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-7-1-3p | 17 | EPHA7 | Sponge network | 0.238 | 0.70544 | -2.197 | 3.0E-5 | 0.349 |
| 29465 | HCG11 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 15 | MAPK10 | Sponge network | 0.385 | 0.10067 | -1.774 | 0 | 0.349 |
| 29466 | RP11-575A19.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-629-3p | 17 | NFIB | Sponge network | 1.969 | 0.00316 | 0.023 | 0.90795 | 0.349 |
| 29467 | RP11-890B15.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 21 | SIRT1 | Sponge network | 0.101 | 0.60919 | 0.109 | 0.2593 | 0.349 |
| 29468 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-96-5p | 15 | NUAK1 | Sponge network | -0.517 | 0.02935 | 0.904 | 0 | 0.349 |
| 29469 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 23 | SCN9A | Sponge network | 0.349 | 0.01695 | -0.525 | 0.10593 | 0.349 |
| 29470 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-500a-3p | 15 | RUNX1T1 | Sponge network | -7.551 | 0 | -0.344 | 0.2374 | 0.349 |
| 29471 | RP11-2H8.2 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 1.089 | 0 | 0.259 | 0.00962 | 0.349 |
| 29472 | H1FX-AS1 |
hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p | 15 | PAIP2 | Sponge network | -0.206 | 0.12942 | -0.515 | 0 | 0.349 |
| 29473 | LINC00621 |
hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-331-5p;hsa-miR-590-3p | 10 | PCDH9 | Sponge network | 0.131 | 0.8436 | -1.823 | 4.0E-5 | 0.349 |
| 29474 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | TACC1 | Sponge network | 0.537 | 0.00139 | -0.362 | 0.05406 | 0.349 |
| 29475 | FGF14-AS2 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 12 | OPCML | Sponge network | -1.582 | 8.0E-5 | -2.498 | 0 | 0.349 |
| 29476 | AC009948.5 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 12 | ABCB5 | Sponge network | 0.532 | 0.00505 | -0.956 | 0.04077 | 0.349 |
| 29477 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-30c-5p;hsa-miR-93-3p | 11 | EBF3 | Sponge network | 0.912 | 0.00014 | -0.058 | 0.81437 | 0.349 |
| 29478 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 37 | KIF1B | Sponge network | 1.601 | 0 | -0.118 | 0.32258 | 0.349 |
| 29479 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 25 | MYO9A | Sponge network | 0.415 | 0.14657 | -0.147 | 0.32373 | 0.349 |
| 29480 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | RABEP1 | Sponge network | -0.739 | 0.0574 | -0.066 | 0.43142 | 0.349 |
| 29481 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-940 | 19 | CRTC3 | Sponge network | 0.082 | 0.55397 | 0.164 | 0.16786 | 0.349 |
| 29482 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | EPHA3 | Sponge network | -0.726 | 0.00132 | 0.185 | 0.53588 | 0.349 |
| 29483 | EMX2OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-590-3p | 12 | ITGB3 | Sponge network | -0.777 | 0.1134 | -0.011 | 0.95751 | 0.349 |
| 29484 | SNHG6 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p | 11 | PHF6 | Sponge network | 0.668 | 0 | 0.785 | 0 | 0.349 |
| 29485 | RP3-402G11.26 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-940 | 11 | PBX1 | Sponge network | -0.254 | 0.19303 | -1.206 | 0 | 0.349 |
| 29486 | CTC-550B14.6 |
hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | TBL1XR1 | Sponge network | 3.885 | 0 | 0.578 | 0 | 0.349 |
| 29487 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 26 | TSHZ3 | Sponge network | -0.513 | 0.04703 | -0.556 | 0.01516 | 0.349 |
| 29488 | LINC00476 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p | 13 | ELMO1 | Sponge network | -0.615 | 0.00015 | 0.04 | 0.83147 | 0.349 |
| 29489 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-361-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | CAV1 | Sponge network | 0.064 | 0.72934 | -1.013 | 6.0E-5 | 0.349 |
| 29490 | RP11-540B6.6 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p | 12 | MAT2A | Sponge network | 0.93 | 0 | -0.073 | 0.36195 | 0.349 |
| 29491 | FAM13A-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 57 | CADM2 | Sponge network | -0.83 | 0 | -3.782 | 0 | 0.349 |
| 29492 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p | 16 | PRKAA1 | Sponge network | 0.417 | 0.00445 | 0.317 | 0.0031 | 0.349 |
| 29493 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ADAMTSL3 | Sponge network | 0.083 | 0.66405 | -1.559 | 0 | 0.349 |
| 29494 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 20 | KAT6B | Sponge network | 0.75 | 0.00054 | -0.478 | 0.00018 | 0.349 |
| 29495 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | ZFHX4 | Sponge network | 0.41 | 0.02214 | 0.018 | 0.95574 | 0.349 |
| 29496 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 17 | PNRC1 | Sponge network | -0.615 | 0.00015 | -0.646 | 0 | 0.349 |
| 29497 | LINC01018 |
hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | PPP2R2C | Sponge network | -2.915 | 0.00015 | -0.959 | 0.07428 | 0.349 |
| 29498 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b | 13 | USP12 | Sponge network | -0.517 | 0.02935 | -0.102 | 0.40768 | 0.349 |
| 29499 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | AKAP13 | Sponge network | -0.487 | 0.02157 | 0.028 | 0.7729 | 0.349 |
| 29500 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | PDGFC | Sponge network | -0.777 | 0.1134 | -0.33 | 0.06483 | 0.349 |
| 29501 | AGAP11 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-766-3p;hsa-miR-93-5p | 28 | IL17RD | Sponge network | -1.645 | 0 | -0.019 | 0.94873 | 0.349 |
| 29502 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-484 | 17 | GREM1 | Sponge network | -0.773 | 0.04073 | 0.902 | 0.01691 | 0.349 |
| 29503 | PSMG3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | SETBP1 | Sponge network | -0.125 | 0.49414 | -1.302 | 1.0E-5 | 0.349 |
| 29504 | LINC00174 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p | 10 | PHF6 | Sponge network | 1.253 | 0 | 0.785 | 0 | 0.349 |
| 29505 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 46 | UNC80 | Sponge network | -0.777 | 0.1134 | -1.934 | 0 | 0.349 |
| 29506 | RP11-119F7.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | STARD13 | Sponge network | 0.225 | 0.30644 | -0.187 | 0.27383 | 0.349 |
| 29507 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | NRXN1 | Sponge network | 0.532 | 0.00505 | -3.778 | 0 | 0.349 |
| 29508 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p | 24 | ZFHX3 | Sponge network | 0.051 | 0.83388 | 0.389 | 0.01363 | 0.349 |
| 29509 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-660-5p;hsa-miR-940 | 14 | ZFP36L1 | Sponge network | -4.546 | 0 | -0.505 | 8.0E-5 | 0.349 |
| 29510 | LINC00472 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p | 10 | ITGA5 | Sponge network | -0.842 | 0.01361 | 0.452 | 0.03524 | 0.349 |
| 29511 | C4A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-590-5p | 16 | RBMS3 | Sponge network | 3.345 | 0 | -0.519 | 0.03551 | 0.349 |
| 29512 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 37 | BMPR2 | Sponge network | 0.873 | 0 | 0.206 | 0.0635 | 0.349 |
| 29513 | RP11-248G5.8 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-363-3p;hsa-miR-766-3p | 11 | KIAA1024 | Sponge network | 1.294 | 0 | 1.083 | 0 | 0.349 |
| 29514 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | PGR | Sponge network | 0.051 | 0.83388 | -1.704 | 0 | 0.349 |
| 29515 | EIF3J-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-766-3p | 22 | IL17RD | Sponge network | 0.331 | 0.00509 | -0.019 | 0.94873 | 0.349 |
| 29516 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 40 | RUNX1T1 | Sponge network | 0.537 | 0.00139 | -0.344 | 0.2374 | 0.349 |
| 29517 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-629-3p | 22 | DDX6 | Sponge network | 0.919 | 0 | 0.091 | 0.36428 | 0.349 |
| 29518 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-577 | 25 | NTRK2 | Sponge network | 0.853 | 0.15352 | -0.736 | 0.06495 | 0.349 |
| 29519 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 23 | DICER1 | Sponge network | 0.101 | 0.60919 | 0.042 | 0.674 | 0.349 |
| 29520 | KB-1460A1.5 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-7-5p | 11 | FLNA | Sponge network | 0.603 | 0.00127 | -0.771 | 0.01377 | 0.349 |
| 29521 | CTA-14H9.5 |
hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-7-1-3p | 16 | NTRK3 | Sponge network | 0.145 | 0.53343 | -1.77 | 3.0E-5 | 0.349 |
| 29522 | CTD-2528L19.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 10 | ARID2 | Sponge network | 0.953 | 0 | 0.235 | 0.02697 | 0.349 |
| 29523 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | MLLT10 | Sponge network | 0.082 | 0.55654 | 0.105 | 0.33292 | 0.349 |
| 29524 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 38 | ABI2 | Sponge network | -0.942 | 0 | 0.175 | 0.14787 | 0.349 |
| 29525 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 24 | NR2C2 | Sponge network | -1.111 | 0.0003 | 0.21 | 0.03224 | 0.349 |
| 29526 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-589-3p;hsa-miR-93-5p | 12 | EPAS1 | Sponge network | -0.777 | 0.16667 | -0.184 | 0.14418 | 0.349 |
| 29527 | RP11-195B17.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-96-5p | 10 | RASSF8 | Sponge network | 0.37 | 0.27614 | -0.122 | 0.65591 | 0.349 |
| 29528 | LINC00205 |
hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p | 11 | SOX11 | Sponge network | 1.271 | 0 | 2.68 | 0 | 0.349 |
| 29529 | RP11-517B11.7 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-212-3p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 10 | SIRT1 | Sponge network | 0.706 | 0 | 0.109 | 0.2593 | 0.349 |
| 29530 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 18 | RBL2 | Sponge network | -0.737 | 0.06182 | -0.282 | 0.00254 | 0.349 |
| 29531 | C20orf166-AS1 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-429 | 12 | IRS1 | Sponge network | -2.69 | 0.0001 | -0.026 | 0.88855 | 0.349 |
| 29532 | AC009948.5 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | LUZP2 | Sponge network | 0.532 | 0.00505 | -0.056 | 0.89416 | 0.349 |
| 29533 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p | 13 | CHD1 | Sponge network | 1.294 | 0 | 0.322 | 0.00215 | 0.349 |
| 29534 | LINC00476 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 11 | DCN | Sponge network | -0.615 | 0.00015 | -1.288 | 0 | 0.349 |
| 29535 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 30 | APC | Sponge network | 0.421 | 0.00084 | -0.255 | 0.04051 | 0.349 |
| 29536 | RP11-158K1.3 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITGB1 | Sponge network | 0.349 | 0.01695 | 0.524 | 0.00017 | 0.349 |
| 29537 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 51 | CCND2 | Sponge network | -2.452 | 0 | -0.267 | 0.3112 | 0.349 |
| 29538 | AC003090.1 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CTGF | Sponge network | -2.117 | 0.00161 | 0.321 | 0.11682 | 0.349 |
| 29539 | RP11-439L18.2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-454-3p | 10 | AFF1 | Sponge network | 0.224 | 0.48719 | -0.384 | 0.00053 | 0.349 |
| 29540 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 24 | ADAMTSL3 | Sponge network | 1.302 | 7.0E-5 | -1.559 | 0 | 0.349 |
| 29541 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-671-5p | 10 | FOXP1 | Sponge network | -0.309 | 0.1145 | 0.042 | 0.74368 | 0.349 |
| 29542 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-550a-3p;hsa-miR-653-5p | 21 | DMXL1 | Sponge network | 0.082 | 0.55397 | -0.426 | 0.00056 | 0.349 |
| 29543 | RP11-479G22.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 1.308 | 0 | 0.943 | 0 | 0.349 |
| 29544 | HCG22 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-93-3p;hsa-miR-96-5p | 18 | SNX29 | Sponge network | 0.816 | 0.32368 | -0.34 | 0.01371 | 0.349 |
| 29545 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p | 18 | NF1 | Sponge network | 0.083 | 0.66405 | 0.51 | 0 | 0.349 |
| 29546 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-30e-5p;hsa-miR-590-3p | 12 | MOB1B | Sponge network | 0.545 | 0.05789 | 0.197 | 0.15266 | 0.349 |
| 29547 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | LRRC55 | Sponge network | -0.206 | 0.12942 | -0.026 | 0.93163 | 0.349 |
| 29548 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p | 19 | CBL | Sponge network | 1.03 | 0 | 0.291 | 0.01267 | 0.349 |
| 29549 | RP11-244O19.1 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 10 | DIRAS1 | Sponge network | -0.096 | 0.67958 | -2.518 | 0 | 0.349 |
| 29550 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 18 | BMPR2 | Sponge network | 1.056 | 0 | 0.206 | 0.0635 | 0.349 |
| 29551 | HOXB-AS1 |
hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-940 | 10 | ZFP36L1 | Sponge network | -0.154 | 0.53954 | -0.505 | 8.0E-5 | 0.349 |
| 29552 | RP11-384K6.6 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | DDX3X | Sponge network | 0.946 | 0 | -0.152 | 0.07686 | 0.349 |
| 29553 | CTD-3064M3.3 |
hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p | 10 | ARHGAP29 | Sponge network | -0.469 | 0.08721 | 0.131 | 0.42953 | 0.349 |
| 29554 | CTD-2666L21.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-24-3p | 12 | DLC1 | Sponge network | 0.368 | 0.20093 | -0.382 | 0.05038 | 0.349 |
| 29555 | LINC00886 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-877-5p | 26 | EGFR | Sponge network | -0.371 | 0.24604 | -0.175 | 0.48451 | 0.349 |
| 29556 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p | 10 | PCDH8 | Sponge network | -0.154 | 0.78939 | -0.997 | 0.05366 | 0.349 |
| 29557 | LINC00578 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-664a-3p | 18 | RIMS2 | Sponge network | 0.811 | 0.03228 | -1.365 | 0.00208 | 0.349 |
| 29558 | AC018890.6 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | CSMD1 | Sponge network | 1.61 | 0.00961 | -0.63 | 0.18881 | 0.349 |
| 29559 | AC025171.1 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PHF3 | Sponge network | 0.998 | 0 | 0.126 | 0.19652 | 0.349 |
| 29560 | FAM13A-AS1 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 26 | KCNA6 | Sponge network | -0.83 | 0 | -1.531 | 0.00013 | 0.349 |
| 29561 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p | 20 | RASSF2 | Sponge network | -4.463 | 0.00025 | -0.273 | 0.21961 | 0.349 |
| 29562 | LINC00173 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | TGFBR3 | Sponge network | 0.056 | 0.8494 | -1.173 | 1.0E-5 | 0.349 |
| 29563 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-421 | 18 | AFF3 | Sponge network | -0.154 | 0.78939 | -2.373 | 0 | 0.349 |
| 29564 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | RICTOR | Sponge network | 0.619 | 0.00157 | 0.388 | 0.00188 | 0.349 |
| 29565 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-3p;hsa-miR-93-5p | 19 | MAP3K12 | Sponge network | -1.697 | 0.00889 | -0.057 | 0.59369 | 0.349 |
| 29566 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-93-5p | 28 | PPM1A | Sponge network | 0.082 | 0.55397 | -0.623 | 0 | 0.349 |
| 29567 | CKMT2-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 16 | SORCS1 | Sponge network | 0.082 | 0.55654 | -3.492 | 0 | 0.349 |
| 29568 | RP4-798A10.7 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | CBFB | Sponge network | 1.757 | 0 | 1.061 | 0 | 0.349 |
| 29569 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 18 | ARHGAP29 | Sponge network | 1.302 | 7.0E-5 | 0.131 | 0.42953 | 0.349 |
| 29570 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-3127-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | TACC1 | Sponge network | -0.295 | 0.02604 | -0.362 | 0.05406 | 0.349 |
| 29571 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-3p | 33 | TRPS1 | Sponge network | -0.18 | 0.20343 | -0.191 | 0.44109 | 0.349 |
| 29572 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 34 | NTRK2 | Sponge network | -0.612 | 0.00094 | -0.736 | 0.06495 | 0.349 |
| 29573 | HOXA-AS2 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-874-3p | 10 | HSPB7 | Sponge network | 0.61 | 0.01593 | -2.268 | 1.0E-5 | 0.349 |
| 29574 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-7-1-3p | 13 | ABCC9 | Sponge network | 0.389 | 0.04293 | -0.466 | 0.14121 | 0.349 |
| 29575 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 15 | ZMYM2 | Sponge network | 1.757 | 0 | 0.279 | 0.01273 | 0.349 |
| 29576 | RP11-536O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3065-3p | 16 | ADARB1 | Sponge network | -0.074 | 0.90758 | -0.335 | 0.06631 | 0.349 |
| 29577 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p | 12 | FSTL5 | Sponge network | -0.487 | 0.02157 | -1.3 | 0.01619 | 0.349 |
| 29578 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-502-5p | 14 | TGFBR2 | Sponge network | -2.62 | 0 | -0.243 | 0.11408 | 0.349 |
| 29579 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p | 26 | ZFHX3 | Sponge network | -0.739 | 0.00044 | 0.389 | 0.01363 | 0.349 |
| 29580 | RP11-305E6.4 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 15 | UBE2D1 | Sponge network | 1.317 | 0 | 0.468 | 0 | 0.349 |
| 29581 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-940 | 28 | AFF4 | Sponge network | 0.959 | 0 | -0.266 | 0.01565 | 0.349 |
| 29582 | PCA3 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | -0.709 | 0.18168 | -0.226 | 0.11691 | 0.349 |
| 29583 | LINC00702 |
hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-664a-3p | 10 | LOX | Sponge network | -0.737 | 0.06182 | 1.164 | 0 | 0.349 |
| 29584 | TRAF3IP2-AS1 |
hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 12 | FBXW7 | Sponge network | -0.323 | 0.01372 | -0.418 | 2.0E-5 | 0.349 |
| 29585 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 18 | NFIB | Sponge network | 0.959 | 0 | 0.023 | 0.90795 | 0.349 |
| 29586 | PWAR6 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 20 | FMNL3 | Sponge network | -1.575 | 6.0E-5 | 0.555 | 4.0E-5 | 0.349 |
| 29587 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-539-5p | 35 | TMEM132B | Sponge network | 0.331 | 0.57247 | -0.83 | 0.01084 | 0.349 |
| 29588 | LINC00641 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-505-5p | 13 | WNK2 | Sponge network | -0.222 | 0.32445 | -0.352 | 0.31066 | 0.349 |
| 29589 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | ARNT2 | Sponge network | -0.769 | 0.00035 | -0.332 | 0.25851 | 0.349 |
| 29590 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 22 | RARB | Sponge network | -0.777 | 0.1134 | -0.825 | 2.0E-5 | 0.349 |
| 29591 | LINC01018 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | ARNT2 | Sponge network | -2.915 | 0.00015 | -0.332 | 0.25851 | 0.349 |
| 29592 | MIR22HG |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-940 | 13 | PEG3 | Sponge network | -1.047 | 0 | -1.74 | 0 | 0.349 |
| 29593 | HAR1A |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b | 13 | CNTN1 | Sponge network | -0.672 | 0.19447 | -2.594 | 0 | 0.349 |
| 29594 | LINC00672 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-542-3p | 15 | SFRP1 | Sponge network | -0.227 | 0.31657 | -3.566 | 0 | 0.349 |
| 29595 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 42 | CHD5 | Sponge network | 0.61 | 0.01593 | -0.922 | 0.01521 | 0.349 |
| 29596 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 35 | ARID5B | Sponge network | -0.739 | 0.0574 | 0.342 | 0.01676 | 0.349 |
| 29597 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ATRX | Sponge network | 1.253 | 0 | 0.261 | 0.04408 | 0.349 |
| 29598 | HCG18 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 30 | ARHGEF12 | Sponge network | 1.063 | 0 | 0.09 | 0.35693 | 0.349 |
| 29599 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p | 14 | FSTL5 | Sponge network | 0.101 | 0.60919 | -1.3 | 0.01619 | 0.349 |
| 29600 | HCG18 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 19 | PRKAA1 | Sponge network | 1.063 | 0 | 0.317 | 0.0031 | 0.349 |
| 29601 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p | 23 | HECA | Sponge network | 0.056 | 0.81195 | -0.217 | 0.03463 | 0.349 |
| 29602 | RP11-440L14.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-5p;hsa-miR-590-3p | 14 | CREB1 | Sponge network | 0.545 | 0.05789 | 0.203 | 0.00537 | 0.349 |
| 29603 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-500b-5p | 17 | SLITRK6 | Sponge network | -1.249 | 7.0E-5 | -2.121 | 0 | 0.349 |
| 29604 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-451a;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | BCL2 | Sponge network | 0.997 | 0.00066 | -0.899 | 0 | 0.349 |
| 29605 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | PRRX1 | Sponge network | -0.318 | 0.65847 | 1.316 | 0 | 0.349 |
| 29606 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 16 | RBL2 | Sponge network | -0.323 | 0.01372 | -0.282 | 0.00254 | 0.349 |
| 29607 | RP5-1139B12.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-671-5p | 10 | CDH23 | Sponge network | 0.598 | 0.38481 | -0.974 | 0.00034 | 0.349 |
| 29608 | RP11-360F5.3 |
hsa-let-7d-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | RICTOR | Sponge network | 1.867 | 0 | 0.388 | 0.00188 | 0.349 |
| 29609 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-550a-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | DMXL1 | Sponge network | -1.047 | 0 | -0.426 | 0.00056 | 0.349 |
| 29610 | PRKAG2-AS1 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DTNA | Sponge network | -0.622 | 0.02807 | -1.648 | 1.0E-5 | 0.349 |
| 29611 | RP11-473I1.10 |
hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-624-5p | 11 | SIRT1 | Sponge network | 0.673 | 0 | 0.109 | 0.2593 | 0.349 |
| 29612 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MOB1B | Sponge network | 0.272 | 0.44692 | 0.197 | 0.15266 | 0.349 |
| 29613 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-450b-5p | 13 | DKK3 | Sponge network | 0.174 | 0.30034 | -0.052 | 0.77783 | 0.349 |
| 29614 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-590-3p | 13 | RB1CC1 | Sponge network | 0.083 | 0.66405 | 0.175 | 0.12559 | 0.349 |
| 29615 | RP11-379F4.7 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | UBR3 | Sponge network | 1.316 | 0 | -0.021 | 0.82774 | 0.349 |
| 29616 | LINC00843 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | RPS6KA3 | Sponge network | 1.106 | 0 | 0.256 | 0.02419 | 0.349 |
| 29617 | USP3-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-940 | 10 | FAM117A | Sponge network | -0.162 | 0.47687 | -1.379 | 0 | 0.349 |
| 29618 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-940 | 33 | HLF | Sponge network | -0.738 | 0.21233 | -2.443 | 0 | 0.349 |
| 29619 | AC010226.4 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-96-5p | 28 | CREB3L2 | Sponge network | -0.811 | 2.0E-5 | -0.525 | 2.0E-5 | 0.349 |
| 29620 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 16 | BCL6 | Sponge network | -0.896 | 0.00099 | -0.211 | 0.19489 | 0.349 |
| 29621 | HOXA-AS2 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | 0.61 | 0.01593 | -2.633 | 0 | 0.349 |
| 29622 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-532-3p | 31 | CBL | Sponge network | -0.154 | 0.78939 | 0.291 | 0.01267 | 0.349 |
| 29623 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 18 | NRK | Sponge network | -0.342 | 0.02288 | 0.656 | 0.19217 | 0.349 |
| 29624 | LINC00641 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PTPN13 | Sponge network | -0.222 | 0.32445 | -1.208 | 0 | 0.349 |
| 29625 | CTC-429P9.2 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-485-3p | 11 | SMAD4 | Sponge network | 0.043 | 0.81628 | -0.313 | 0.00413 | 0.349 |
| 29626 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RASSF8 | Sponge network | -0.663 | 0.10887 | -0.122 | 0.65591 | 0.349 |
| 29627 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | LIFR | Sponge network | -0.663 | 0.10887 | -1.794 | 0 | 0.349 |
| 29628 | ZNF833P |
hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-766-3p | 12 | MDM4 | Sponge network | 0.688 | 1.0E-5 | 0.345 | 0.00762 | 0.349 |
| 29629 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | EPHA5 | Sponge network | -0.842 | 0.01361 | -0.957 | 0.01733 | 0.349 |
| 29630 | RP11-159D12.2 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 18 | RASAL2 | Sponge network | 1.254 | 0 | 0.869 | 0 | 0.349 |
| 29631 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-93-3p | 31 | AKAP11 | Sponge network | -0.384 | 0.40652 | 0.196 | 0.09825 | 0.349 |
| 29632 | LINC00641 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 16 | PDZD2 | Sponge network | -0.222 | 0.32445 | -0.909 | 3.0E-5 | 0.349 |
| 29633 | RP11-73M18.8 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 21 | CCDC6 | Sponge network | 0.832 | 0 | 0.27 | 0.02555 | 0.349 |
| 29634 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 15 | ARHGAP29 | Sponge network | -0.769 | 0.00035 | 0.131 | 0.42953 | 0.349 |
| 29635 | LINC00654 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | SYNPO2 | Sponge network | 0.374 | 0.20054 | -2.342 | 0 | 0.349 |
| 29636 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | DCLK1 | Sponge network | -0.355 | 0.0077 | -1.324 | 0.00014 | 0.349 |
| 29637 | HCG18 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | EPG5 | Sponge network | 1.063 | 0 | 0.101 | 0.3563 | 0.349 |
| 29638 | CTC-296K1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-374b-3p | 20 | UNC80 | Sponge network | -2.095 | 0.00112 | -1.934 | 0 | 0.349 |
| 29639 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 12 | FGF5 | Sponge network | 0.16 | 0.50785 | 0.549 | 0.28659 | 0.349 |
| 29640 | RP1-122P22.2 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-532-3p | 16 | WDFY3 | Sponge network | 0.065 | 0.73468 | -0.201 | 0.0967 | 0.349 |
| 29641 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | TET1 | Sponge network | 0.72 | 0.0001 | -0.135 | 0.52138 | 0.349 |
| 29642 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 19 | PTPN11 | Sponge network | 0.349 | 0.01695 | 0.431 | 2.0E-5 | 0.349 |
| 29643 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 19 | GPC6 | Sponge network | -1.057 | 0.00029 | -0.054 | 0.81465 | 0.349 |
| 29644 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | SETBP1 | Sponge network | -0.969 | 2.0E-5 | -1.302 | 1.0E-5 | 0.349 |
| 29645 | FZD10-AS1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | RPS6KA6 | Sponge network | -0.727 | 0.04726 | -2.989 | 0 | 0.349 |
| 29646 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-370-3p | 19 | JAZF1 | Sponge network | -0.285 | 0.2172 | -0.377 | 0.02677 | 0.349 |
| 29647 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p | 12 | TMEFF2 | Sponge network | -0.154 | 0.78939 | -2.426 | 0 | 0.349 |
| 29648 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-378a-5p | 14 | BMPR2 | Sponge network | 0.813 | 1.0E-5 | 0.206 | 0.0635 | 0.349 |
| 29649 | SNX29P2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 20 | TET1 | Sponge network | 0.4 | 0.14611 | -0.135 | 0.52138 | 0.349 |
| 29650 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 10 | CUL5 | Sponge network | 0.752 | 0 | 0.026 | 0.77301 | 0.349 |
| 29651 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 11 | SEPT6 | Sponge network | -0.096 | 0.67958 | -0.598 | 0.00181 | 0.349 |
| 29652 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-940 | 15 | CRTC3 | Sponge network | 1.153 | 0.00114 | 0.164 | 0.16786 | 0.349 |
| 29653 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 14 | ZFX | Sponge network | 0.993 | 0 | 0.09 | 0.42563 | 0.349 |
| 29654 | HOXA-AS2 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | LATS2 | Sponge network | 0.61 | 0.01593 | 0.159 | 0.2304 | 0.349 |
| 29655 | RP11-251G23.5 |
hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-376c-3p | 10 | PAWR | Sponge network | 1.344 | 0 | 0.636 | 0 | 0.349 |
| 29656 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-590-5p | 17 | WDFY3 | Sponge network | 0.537 | 0.0006 | -0.201 | 0.0967 | 0.349 |
| 29657 | SH3RF3-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-7-1-3p | 10 | BCL9 | Sponge network | 0.889 | 9.0E-5 | 0.321 | 0.00267 | 0.349 |
| 29658 | CTC-429P9.3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-940 | 11 | SFRP1 | Sponge network | 0.082 | 0.55397 | -3.566 | 0 | 0.348 |
| 29659 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 16 | ZMYM2 | Sponge network | 0.856 | 0 | 0.279 | 0.01273 | 0.348 |
| 29660 | RP11-473I1.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | BRD3 | Sponge network | 0.683 | 1.0E-5 | 0.37 | 0.00013 | 0.348 |
| 29661 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 30 | SASH1 | Sponge network | -0.405 | 0.22013 | -0.502 | 0.00175 | 0.348 |
| 29662 | CTD-2666L21.1 |
hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-493-5p | 11 | SYNE1 | Sponge network | 0.368 | 0.20093 | -0.738 | 0.00316 | 0.348 |
| 29663 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-660-5p;hsa-miR-93-5p | 11 | ZHX2 | Sponge network | -1.697 | 0.00889 | -0.192 | 0.18459 | 0.348 |
| 29664 | GS1-124K5.11 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-590-3p | 11 | PTCH1 | Sponge network | 0.494 | 0.00339 | -0.1 | 0.6179 | 0.348 |
| 29665 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | LRRC7 | Sponge network | 0.056 | 0.8494 | -1.341 | 0.01193 | 0.348 |
| 29666 | RP11-428J1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-590-3p | 11 | CNTNAP3 | Sponge network | 0.538 | 0.00076 | -1.465 | 0.00033 | 0.348 |
| 29667 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-223-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | CXXC4 | Sponge network | 0.663 | 0.00073 | -0.433 | 0.21055 | 0.348 |
| 29668 | CD27-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-339-5p | 14 | STARD13 | Sponge network | 0.177 | 0.16681 | -0.187 | 0.27383 | 0.348 |
| 29669 | AC083843.1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | GSK3B | Sponge network | 1.4 | 0 | 0.266 | 0.00045 | 0.348 |
| 29670 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 20 | PDE4DIP | Sponge network | 0.225 | 0.30644 | -0.282 | 0.0257 | 0.348 |
| 29671 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 24 | HIP1 | Sponge network | 0.101 | 0.60919 | 0.774 | 0 | 0.348 |
| 29672 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-452-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-766-3p | 52 | IL6ST | Sponge network | 0.331 | 0.57247 | -0.402 | 0.00676 | 0.348 |
| 29673 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | LRRC7 | Sponge network | 0.178 | 0.29106 | -1.341 | 0.01193 | 0.348 |
| 29674 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p | 16 | IL17RD | Sponge network | -0.021 | 0.93598 | -0.019 | 0.94873 | 0.348 |
| 29675 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | RNF144A | Sponge network | 0.219 | 0.1516 | 0.594 | 0.0009 | 0.348 |
| 29676 | RP11-253E3.3 |
hsa-miR-101-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-497-5p | 10 | NF1 | Sponge network | 1.764 | 0 | 0.51 | 0 | 0.348 |
| 29677 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 25 | SOX5 | Sponge network | 0.811 | 0.03228 | -1.091 | 4.0E-5 | 0.348 |
| 29678 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-766-3p | 51 | MDM4 | Sponge network | 0.683 | 1.0E-5 | 0.345 | 0.00762 | 0.348 |
| 29679 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | UBE2QL1 | Sponge network | 0.246 | 0.59912 | -2.417 | 0 | 0.348 |
| 29680 | C4B-AS1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-629-3p;hsa-miR-744-3p | 18 | IGFBP5 | Sponge network | 2.306 | 9.0E-5 | -0.806 | 0.00105 | 0.348 |
| 29681 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 25 | SEMA3C | Sponge network | -1.216 | 0 | -0.272 | 0.31153 | 0.348 |
| 29682 | RP11-532F6.3 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | NR3C2 | Sponge network | -0.896 | 0.00099 | -1.56 | 0 | 0.348 |
| 29683 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-93-5p | 12 | LRRC55 | Sponge network | -0.811 | 2.0E-5 | -0.026 | 0.93163 | 0.348 |
| 29684 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | NEDD4 | Sponge network | -0.323 | 0.01372 | 0.02 | 0.89834 | 0.348 |
| 29685 | FAM13A-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | PDE4DIP | Sponge network | -0.83 | 0 | -0.282 | 0.0257 | 0.348 |
| 29686 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 16 | CNTNAP3 | Sponge network | 0.572 | 0.01722 | -1.465 | 0.00033 | 0.348 |
| 29687 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-940 | 25 | THBS1 | Sponge network | -0.096 | 0.67958 | 0.741 | 0.00386 | 0.348 |
| 29688 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CACNA2D1 | Sponge network | 0.683 | 1.0E-5 | -0.846 | 0.00675 | 0.348 |
| 29689 | HAR1A |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p | 11 | RECK | Sponge network | -0.672 | 0.19447 | -1.043 | 0 | 0.348 |
| 29690 | FAM66C |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p | 16 | IRS1 | Sponge network | -0.901 | 0.00019 | -0.026 | 0.88855 | 0.348 |
| 29691 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | SASH1 | Sponge network | -1.281 | 0.00012 | -0.502 | 0.00175 | 0.348 |
| 29692 | HOXA-AS2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ZMYND11 | Sponge network | 0.61 | 0.01593 | -0.2 | 0.05449 | 0.348 |
| 29693 | CTD-2017D11.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-5p | 13 | BRD3 | Sponge network | 0.792 | 0.0049 | 0.37 | 0.00013 | 0.348 |
| 29694 | RP11-400K9.3 |
hsa-let-7d-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-940 | 13 | THBS1 | Sponge network | -0.733 | 0.19469 | 0.741 | 0.00386 | 0.348 |
| 29695 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-424-5p | 10 | FGFR1 | Sponge network | -1.054 | 0.0706 | -0.509 | 0.03957 | 0.348 |
| 29696 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-33a-3p | 12 | CLASP2 | Sponge network | 0.93 | 0 | -0.038 | 0.71 | 0.348 |
| 29697 | RP11-35G9.5 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p | 10 | NRK | Sponge network | 0.622 | 0.00015 | 0.656 | 0.19217 | 0.348 |
| 29698 | LINC01018 |
hsa-miR-126-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 16 | CXCL12 | Sponge network | -2.915 | 0.00015 | -1.421 | 0 | 0.348 |
| 29699 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | GRIA3 | Sponge network | -0.777 | 0.1134 | -1.956 | 0 | 0.348 |
| 29700 | AC141928.1 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 25 | ANK2 | Sponge network | 0.853 | 0.15352 | -1.603 | 1.0E-5 | 0.348 |
| 29701 | AC034220.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 16 | TBL1XR1 | Sponge network | 0.309 | 0.07304 | 0.578 | 0 | 0.348 |
| 29702 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-7-1-3p | 13 | TBX18 | Sponge network | 0.395 | 0.15451 | 0.843 | 0.02945 | 0.348 |
| 29703 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | AKAP11 | Sponge network | 0.953 | 0 | 0.196 | 0.09825 | 0.348 |
| 29704 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 16 | CREBBP | Sponge network | -0.358 | 0.17255 | 0.126 | 0.15186 | 0.348 |
| 29705 | CTD-2666L21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-192-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-455-3p;hsa-miR-493-5p | 14 | ZFX | Sponge network | 0.368 | 0.20093 | 0.09 | 0.42563 | 0.348 |
| 29706 | CTC-457L16.2 |
hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | MDM4 | Sponge network | 1.153 | 0.00114 | 0.345 | 0.00762 | 0.348 |
| 29707 | LINC00657 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | RPS6KA3 | Sponge network | 0.91 | 0 | 0.256 | 0.02419 | 0.348 |
| 29708 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | KANK1 | Sponge network | -0.323 | 0.01372 | -0.55 | 0.00134 | 0.348 |
| 29709 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2355-3p;hsa-miR-339-5p;hsa-miR-424-5p | 10 | BTG2 | Sponge network | -3.586 | 0.00032 | -1.763 | 0 | 0.348 |
| 29710 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | 0.335 | 0.20596 | -1.692 | 0 | 0.348 |
| 29711 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 16 | CNTN1 | Sponge network | 0.342 | 0.03129 | -2.594 | 0 | 0.348 |
| 29712 | HOXA-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 25 | SYNPO2 | Sponge network | 0.61 | 0.01593 | -2.342 | 0 | 0.348 |
| 29713 | CKMT2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 28 | CREB1 | Sponge network | 0.082 | 0.55654 | 0.203 | 0.00537 | 0.348 |
| 29714 | FAM215B |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p | 10 | PRDM2 | Sponge network | 0.817 | 3.0E-5 | -0.258 | 0.00721 | 0.348 |
| 29715 | LINC00641 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 41 | ACVR2A | Sponge network | -0.222 | 0.32445 | -0.409 | 0.00039 | 0.348 |
| 29716 | RP4-717I23.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-5p;hsa-miR-935 | 56 | LPP | Sponge network | 0.637 | 0.00069 | -0.459 | 0.04475 | 0.348 |
| 29717 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | BCL11A | Sponge network | -1.161 | 0.00591 | -0.932 | 0.0063 | 0.348 |
| 29718 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 14 | ZNF770 | Sponge network | 0.702 | 7.0E-5 | 0.206 | 0.06751 | 0.348 |
| 29719 | RP11-473I1.10 |
hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | AFF1 | Sponge network | 0.673 | 0 | -0.384 | 0.00053 | 0.348 |
| 29720 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | DOCK4 | Sponge network | 0.75 | 0.00054 | 0.502 | 0.00108 | 0.348 |
| 29721 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | ABCB5 | Sponge network | -0.942 | 0 | -0.956 | 0.04077 | 0.348 |
| 29722 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 10 | ST5 | Sponge network | -0.355 | 0.0077 | -0.78 | 0 | 0.348 |
| 29723 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 27 | ST6GAL2 | Sponge network | -0.769 | 0.00035 | -1.201 | 0.00597 | 0.348 |
| 29724 | DNAJC3-AS1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-582-5p | 14 | BRD3 | Sponge network | 0.753 | 0.00014 | 0.37 | 0.00013 | 0.348 |
| 29725 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | -0.727 | 0.04726 | -0.466 | 0.14121 | 0.348 |
| 29726 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p | 19 | PIK3R1 | Sponge network | 0.529 | 0.00552 | -0.133 | 0.36854 | 0.348 |
| 29727 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-98-5p | 23 | USP12 | Sponge network | -0.154 | 0.78939 | -0.102 | 0.40768 | 0.348 |
| 29728 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | AKAP6 | Sponge network | 0.252 | 0.08508 | -1.586 | 3.0E-5 | 0.348 |
| 29729 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-493-5p | 15 | SLITRK6 | Sponge network | -0.976 | 0.00025 | -2.121 | 0 | 0.348 |
| 29730 | FZD10-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p | 10 | THSD7A | Sponge network | -0.727 | 0.04726 | 0.242 | 0.25154 | 0.348 |
| 29731 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-940 | 17 | IGF1 | Sponge network | 0.056 | 0.8494 | -0.204 | 0.5898 | 0.348 |
| 29732 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-501-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 25 | APC | Sponge network | 0.614 | 1.0E-5 | -0.255 | 0.04051 | 0.348 |
| 29733 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940;hsa-miR-96-5p | 69 | AFF4 | Sponge network | -2.452 | 0 | -0.266 | 0.01565 | 0.348 |
| 29734 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 11 | GPC6 | Sponge network | -0.021 | 0.93598 | -0.054 | 0.81465 | 0.348 |
| 29735 | RP11-384O8.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p | 14 | STARD13 | Sponge network | -0.738 | 0.21233 | -0.187 | 0.27383 | 0.348 |
| 29736 | RP11-195F19.9 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 18 | AFF1 | Sponge network | -0.726 | 0.00132 | -0.384 | 0.00053 | 0.348 |
| 29737 | CTC-559E9.6 |
hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-455-3p;hsa-miR-940 | 13 | CREB1 | Sponge network | 0.601 | 1.0E-5 | 0.203 | 0.00537 | 0.348 |
| 29738 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-935 | 30 | MTSS1 | Sponge network | -0.405 | 0.22013 | -0.81 | 0.00035 | 0.348 |
| 29739 | CTC-296K1.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 15 | MED12L | Sponge network | -2.095 | 0.00112 | -0.194 | 0.55299 | 0.348 |
| 29740 | RP4-717I23.3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-935 | 22 | ABI2 | Sponge network | 0.637 | 0.00069 | 0.175 | 0.14787 | 0.348 |
| 29741 | RP11-156P1.3 |
hsa-miR-125a-3p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 13 | ATM | Sponge network | 0.214 | 0.18246 | 0.178 | 0.21568 | 0.348 |
| 29742 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p | 24 | FBXO32 | Sponge network | 1.757 | 0 | -0.495 | 0.07561 | 0.348 |
| 29743 | RP11-244H3.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-424-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | RABEP1 | Sponge network | 0.659 | 3.0E-5 | -0.066 | 0.43142 | 0.348 |
| 29744 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | CLASP2 | Sponge network | 0.826 | 1.0E-5 | -0.038 | 0.71 | 0.348 |
| 29745 | RP11-102N12.3 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | RBMS3 | Sponge network | -0.351 | 0.11358 | -0.519 | 0.03551 | 0.348 |
| 29746 | RP11-434B12.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | BRD3 | Sponge network | -0.031 | 0.89063 | 0.37 | 0.00013 | 0.348 |
| 29747 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 22 | SMAD4 | Sponge network | -0.376 | 0.00337 | -0.313 | 0.00413 | 0.348 |
| 29748 | H19 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p | 11 | GPC6 | Sponge network | 2.439 | 0 | -0.054 | 0.81465 | 0.348 |
| 29749 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 24 | ANK2 | Sponge network | 0.051 | 0.83388 | -1.603 | 1.0E-5 | 0.348 |
| 29750 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | TAL1 | Sponge network | -0.899 | 6.0E-5 | -0.462 | 0.00574 | 0.348 |
| 29751 | U91328.21 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-330-3p | 12 | PLSCR4 | Sponge network | -0.735 | 0.15983 | -0.474 | 0.03833 | 0.348 |
| 29752 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | NEDD4 | Sponge network | -1.697 | 0.00889 | 0.02 | 0.89834 | 0.348 |
| 29753 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 29 | USP12 | Sponge network | -0.405 | 0.22013 | -0.102 | 0.40768 | 0.348 |
| 29754 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-501-3p;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | 0.101 | 0.60919 | -0.684 | 0.00159 | 0.348 |
| 29755 | WDFY3-AS2 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | RTN4 | Sponge network | -0.942 | 0 | -0.412 | 0.00014 | 0.348 |
| 29756 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | THBS1 | Sponge network | -1.375 | 0.08642 | 0.741 | 0.00386 | 0.348 |
| 29757 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 16 | MYO9A | Sponge network | -0.557 | 0.11135 | -0.147 | 0.32373 | 0.348 |
| 29758 | LINC00092 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 10 | NRK | Sponge network | -1.886 | 0 | 0.656 | 0.19217 | 0.348 |
| 29759 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 41 | BMPR2 | Sponge network | -0.246 | 0.35034 | 0.206 | 0.0635 | 0.348 |
| 29760 | MYLK-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p | 10 | ACVR1 | Sponge network | -1.331 | 3.0E-5 | 0.279 | 0.00234 | 0.348 |
| 29761 | LINC00654 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | PTPN13 | Sponge network | 0.374 | 0.20054 | -1.208 | 0 | 0.348 |
| 29762 | RP11-384O8.1 |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | AFF3 | Sponge network | -0.738 | 0.21233 | -2.373 | 0 | 0.348 |
| 29763 | MIR497HG |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-590-5p | 15 | PTCH1 | Sponge network | -1.439 | 4.0E-5 | -0.1 | 0.6179 | 0.348 |
| 29764 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DACH2 | Sponge network | -0.49 | 0.04946 | -1.635 | 0.00034 | 0.348 |
| 29765 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PDS5B | Sponge network | 1.317 | 0 | 0.259 | 0.00962 | 0.348 |
| 29766 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 14 | TRIM3 | Sponge network | -0.737 | 0.06182 | -0.577 | 0 | 0.348 |
| 29767 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-93-5p | 14 | AHNAK | Sponge network | -0.18 | 0.20343 | -1.207 | 0 | 0.348 |
| 29768 | PSMG3-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | HERC1 | Sponge network | -0.125 | 0.49414 | -0.196 | 0.08544 | 0.348 |
| 29769 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | PPM1A | Sponge network | -1.057 | 0.00029 | -0.623 | 0 | 0.348 |
| 29770 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 41 | TGFBR3 | Sponge network | -0.769 | 0.00035 | -1.173 | 1.0E-5 | 0.348 |
| 29771 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-5p | 14 | ZMYM2 | Sponge network | 1.61 | 0.00961 | 0.279 | 0.01273 | 0.348 |
| 29772 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-93-5p | 30 | IL17RD | Sponge network | -0.421 | 0.0114 | -0.019 | 0.94873 | 0.348 |
| 29773 | PSMG3-AS1 |
hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | FLNA | Sponge network | -0.125 | 0.49414 | -0.771 | 0.01377 | 0.348 |
| 29774 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 17 | DKK3 | Sponge network | -2.915 | 0.00015 | -0.052 | 0.77783 | 0.348 |
| 29775 | LINC00702 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | LUZP2 | Sponge network | -0.737 | 0.06182 | -0.056 | 0.89416 | 0.348 |
| 29776 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p | 10 | IGSF10 | Sponge network | -0.829 | 0.01343 | -1.751 | 1.0E-5 | 0.348 |
| 29777 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | ARID5B | Sponge network | 0.532 | 0.00505 | 0.342 | 0.01676 | 0.348 |
| 29778 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 29 | LRIG1 | Sponge network | -0.899 | 6.0E-5 | -0.537 | 0.00588 | 0.348 |
| 29779 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-7-1-3p | 12 | SEMA3C | Sponge network | 0.16 | 0.50785 | -0.272 | 0.31153 | 0.348 |
| 29780 | DNAJC27-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 23 | SFRP1 | Sponge network | -0.969 | 2.0E-5 | -3.566 | 0 | 0.348 |
| 29781 | RP4-669P10.16 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-766-3p | 14 | CHD5 | Sponge network | -0.301 | 0.06597 | -0.922 | 0.01521 | 0.348 |
| 29782 | SNHG10 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p | 11 | TBL1XR1 | Sponge network | 0.931 | 0 | 0.578 | 0 | 0.348 |
| 29783 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 18 | NEDD4 | Sponge network | 0.174 | 0.30034 | 0.02 | 0.89834 | 0.348 |
| 29784 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 16 | ABCB5 | Sponge network | 0.374 | 0.20054 | -0.956 | 0.04077 | 0.348 |
| 29785 | BVES-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-98-5p | 14 | ZFP36L1 | Sponge network | -2.62 | 0 | -0.505 | 8.0E-5 | 0.348 |
| 29786 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-629-3p | 22 | CBX5 | Sponge network | 0.453 | 0.01839 | 0.165 | 0.24446 | 0.348 |
| 29787 | CTC-296K1.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-576-5p;hsa-miR-590-5p | 15 | PTPRT | Sponge network | -2.076 | 0.00548 | -0.907 | 0.03727 | 0.348 |
| 29788 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-654-3p | 36 | PIK3R1 | Sponge network | 0.349 | 0.01695 | -0.133 | 0.36854 | 0.348 |
| 29789 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | FOXP1 | Sponge network | 0.545 | 0.00064 | 0.042 | 0.74368 | 0.348 |
| 29790 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-369-3p | 11 | PNRC1 | Sponge network | -0.611 | 0.09607 | -0.646 | 0 | 0.348 |
| 29791 | OIP5-AS1 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | WAC | Sponge network | 0.421 | 0.00084 | -0.054 | 0.43688 | 0.348 |
| 29792 | CTD-2666L21.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-744-3p | 38 | LPP | Sponge network | 0.368 | 0.20093 | -0.459 | 0.04475 | 0.348 |
| 29793 | DNAJC27-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-96-5p | 27 | ASTN1 | Sponge network | -0.969 | 2.0E-5 | -2.389 | 2.0E-5 | 0.348 |
| 29794 | RP5-894A10.6 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-485-3p | 12 | UBR3 | Sponge network | 0.697 | 9.0E-5 | -0.021 | 0.82774 | 0.348 |
| 29795 | C4A-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-96-5p | 12 | EPHA3 | Sponge network | 3.345 | 0 | 0.185 | 0.53588 | 0.348 |
| 29796 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | CUL5 | Sponge network | -0.031 | 0.89063 | 0.026 | 0.77301 | 0.348 |
| 29797 | RP11-274B21.10 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | TSC1 | Sponge network | 1.073 | 0.00216 | 0.103 | 0.26071 | 0.348 |
| 29798 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p | 12 | HIVEP1 | Sponge network | -0.487 | 0.02157 | 0.368 | 0.00064 | 0.348 |
| 29799 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 33 | TOM1L2 | Sponge network | -1.697 | 0.00889 | -0.994 | 0 | 0.348 |
| 29800 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p | 21 | TSC1 | Sponge network | 0.331 | 0.57247 | 0.103 | 0.26071 | 0.348 |
| 29801 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-424-3p | 11 | HOOK3 | Sponge network | -0.693 | 0.05629 | 0.273 | 0.09957 | 0.348 |
| 29802 | BVES-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-942-5p | 12 | ERRFI1 | Sponge network | -2.62 | 0 | -0.798 | 1.0E-5 | 0.348 |
| 29803 | FGD5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 28 | ZFX | Sponge network | 0.401 | 0.00066 | 0.09 | 0.42563 | 0.348 |
| 29804 | LINC00092 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | CRTC1 | Sponge network | -1.886 | 0 | -0.116 | 0.28412 | 0.348 |
| 29805 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 25 | PDGFRA | Sponge network | 0.199 | 0.38015 | -0.372 | 0.0037 | 0.348 |
| 29806 | HOTAIRM1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DNAJB4 | Sponge network | -0.053 | 0.80685 | -0.7 | 1.0E-5 | 0.348 |
| 29807 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-93-5p | 20 | JAZF1 | Sponge network | -0.726 | 0.00132 | -0.377 | 0.02677 | 0.348 |
| 29808 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | PHC3 | Sponge network | 0.768 | 7.0E-5 | 0.212 | 0.0499 | 0.348 |
| 29809 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-429 | 14 | CLIP1 | Sponge network | 0.056 | 0.81195 | -0.215 | 0.08684 | 0.348 |
| 29810 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | PHC3 | Sponge network | 0.924 | 0 | 0.212 | 0.0499 | 0.348 |
| 29811 | RP11-479G22.8 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | KDSR | Sponge network | 1.308 | 0 | 0.239 | 0.02216 | 0.348 |
| 29812 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-589-3p | 53 | THRB | Sponge network | -0.154 | 0.78939 | -1.117 | 0.0004 | 0.348 |
| 29813 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | LHFP | Sponge network | -0.355 | 0.0077 | -0.167 | 0.41371 | 0.348 |
| 29814 | WAC-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-374b-5p | 14 | RASAL2 | Sponge network | 0.821 | 0 | 0.869 | 0 | 0.348 |
| 29815 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 15 | ADAMTSL3 | Sponge network | 0.174 | 0.30034 | -1.559 | 0 | 0.348 |
| 29816 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 39 | DDX6 | Sponge network | 0.311 | 0.10344 | 0.091 | 0.36428 | 0.348 |
| 29817 | RAMP2-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 12 | TMEFF2 | Sponge network | -0.513 | 0.04703 | -2.426 | 0 | 0.348 |
| 29818 | RP11-359E10.1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 18 | ABL2 | Sponge network | 0.299 | 0.57722 | 0.82 | 0 | 0.348 |
| 29819 | PGM5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-7-1-3p | 11 | ZMYND11 | Sponge network | -4.546 | 0 | -0.2 | 0.05449 | 0.348 |
| 29820 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | MYO9A | Sponge network | 0.91 | 0 | -0.147 | 0.32373 | 0.348 |
| 29821 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | LPP | Sponge network | 0.056 | 0.8494 | -0.459 | 0.04475 | 0.348 |
| 29822 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-5p | 22 | RECK | Sponge network | -1.645 | 0 | -1.043 | 0 | 0.348 |
| 29823 | RP11-389C8.2 |
hsa-let-7d-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-942-5p | 14 | GLIPR1 | Sponge network | 0.727 | 3.0E-5 | 0.146 | 0.3276 | 0.348 |
| 29824 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-500b-5p;hsa-miR-590-3p | 26 | SCN9A | Sponge network | 0.659 | 3.0E-5 | -0.525 | 0.10593 | 0.348 |
| 29825 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 23 | SCN9A | Sponge network | -0.08 | 0.90092 | -0.525 | 0.10593 | 0.348 |
| 29826 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-539-5p;hsa-miR-582-5p | 18 | MOB1B | Sponge network | 1.206 | 0 | 0.197 | 0.15266 | 0.348 |
| 29827 | RP11-798G7.7 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p | 11 | ARID5B | Sponge network | 0.858 | 3.0E-5 | 0.342 | 0.01676 | 0.348 |
| 29828 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-511-5p;hsa-miR-940 | 13 | TRIM33 | Sponge network | 0.389 | 0.04293 | 0.402 | 7.0E-5 | 0.348 |
| 29829 | MIR497HG |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p;hsa-miR-940 | 15 | JUN | Sponge network | -1.439 | 4.0E-5 | -0.932 | 0 | 0.348 |
| 29830 | ZNF582-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 21 | TET1 | Sponge network | -1.349 | 0.00017 | -0.135 | 0.52138 | 0.348 |
| 29831 | CTA-204B4.2 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 17 | MED12L | Sponge network | 0.765 | 7.0E-5 | -0.194 | 0.55299 | 0.348 |
| 29832 | TPTEP1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484 | 20 | ABL1 | Sponge network | -1.216 | 0 | -0.215 | 0.07804 | 0.348 |
| 29833 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-424-5p | 11 | RARB | Sponge network | -1.054 | 0.0706 | -0.825 | 2.0E-5 | 0.348 |
| 29834 | GS1-124K5.11 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 23 | PCDH9 | Sponge network | 0.494 | 0.00339 | -1.823 | 4.0E-5 | 0.348 |
| 29835 | LINC00094 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-624-5p;hsa-miR-92a-3p;hsa-miR-942-5p | 34 | NOTCH2 | Sponge network | 0.253 | 0.09565 | 0.138 | 0.35163 | 0.348 |
| 29836 | CTB-31O20.4 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-328-3p;hsa-miR-374b-3p;hsa-miR-628-5p | 13 | VPS53 | Sponge network | 1.619 | 0 | 0.103 | 0.22667 | 0.348 |
| 29837 | PART1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | LIMCH1 | Sponge network | -2.925 | 0 | -0.915 | 1.0E-5 | 0.348 |
| 29838 | RP11-403I13.8 |
hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | ZDBF2 | Sponge network | 0.619 | 0.00157 | -0.998 | 0.00025 | 0.348 |
| 29839 | RP5-1085F17.3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 12 | BCL9 | Sponge network | 0.541 | 0.00125 | 0.321 | 0.00267 | 0.348 |
| 29840 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 22 | SPG20 | Sponge network | 0.064 | 0.72934 | -1.16 | 0 | 0.348 |
| 29841 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | NCOA1 | Sponge network | -0.942 | 0 | -0.432 | 0 | 0.348 |
| 29842 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-942-5p | 24 | NRXN1 | Sponge network | 0.082 | 0.55397 | -3.778 | 0 | 0.348 |
| 29843 | EMX2OS |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | EYA4 | Sponge network | -0.777 | 0.1134 | 0.3 | 0.36876 | 0.348 |
| 29844 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | MED12L | Sponge network | 0.537 | 0.00139 | -0.194 | 0.55299 | 0.348 |
| 29845 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 10 | AMPH | Sponge network | -0.405 | 0.22013 | -0.916 | 5.0E-5 | 0.348 |
| 29846 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | DOCK4 | Sponge network | 0.523 | 9.0E-5 | 0.502 | 0.00108 | 0.348 |
| 29847 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p | 20 | CXXC4 | Sponge network | 0.494 | 0.00339 | -0.433 | 0.21055 | 0.348 |
| 29848 | RP5-1139B12.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-502-5p | 11 | PTPN14 | Sponge network | 0.598 | 0.38481 | 0.144 | 0.34595 | 0.348 |
| 29849 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | SLITRK3 | Sponge network | 0.381 | 0.25967 | -3.328 | 0 | 0.348 |
| 29850 | RP11-344B5.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b | 16 | IGFBP5 | Sponge network | -0.055 | 0.88482 | -0.806 | 0.00105 | 0.348 |
| 29851 | HAND2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-651-5p | 36 | PRDM2 | Sponge network | -1.697 | 0.00889 | -0.258 | 0.00721 | 0.348 |
| 29852 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PLSCR4 | Sponge network | 0.392 | 0.0278 | -0.474 | 0.03833 | 0.348 |
| 29853 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 21 | GRIN2A | Sponge network | 0.374 | 0.20054 | -2.161 | 0 | 0.348 |
| 29854 | RP11-67L2.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | MRVI1 | Sponge network | 0.72 | 0.0001 | -1.051 | 0.00022 | 0.348 |
| 29855 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-455-3p | 26 | RICTOR | Sponge network | 0.594 | 0.41522 | 0.388 | 0.00188 | 0.348 |
| 29856 | RP11-35G9.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | ROBO1 | Sponge network | 0.657 | 4.0E-5 | -0.009 | 0.96154 | 0.348 |
| 29857 | U91328.19 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-935 | 74 | LPP | Sponge network | -0.303 | 0.21002 | -0.459 | 0.04475 | 0.348 |
| 29858 | THAP9-AS1 |
hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-628-5p | 14 | CDC73 | Sponge network | 1.079 | 0 | 0.094 | 0.20463 | 0.348 |
| 29859 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-940 | 22 | AKAP11 | Sponge network | 0.921 | 0.00118 | 0.196 | 0.09825 | 0.348 |
| 29860 | CTC-487M23.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p | 24 | MDM4 | Sponge network | 0.912 | 0.00014 | 0.345 | 0.00762 | 0.348 |
| 29861 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | CHD2 | Sponge network | 0.101 | 0.60919 | 0.049 | 0.55371 | 0.348 |
| 29862 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-629-3p | 29 | DDX6 | Sponge network | 0.541 | 0.00125 | 0.091 | 0.36428 | 0.348 |
| 29863 | ARHGEF26-AS1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 13 | FOXO4 | Sponge network | -2.46 | 0 | -0.516 | 2.0E-5 | 0.348 |
| 29864 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | GPC6 | Sponge network | 0.225 | 0.30644 | -0.054 | 0.81465 | 0.348 |
| 29865 | ADAMTS9-AS2 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 20 | FMNL3 | Sponge network | -2.452 | 0 | 0.555 | 4.0E-5 | 0.348 |
| 29866 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MLF1 | Sponge network | -0.942 | 0 | -0.893 | 0.00727 | 0.348 |
| 29867 | FAM66C |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 16 | UNC5C | Sponge network | -0.901 | 0.00019 | -0.178 | 0.40214 | 0.348 |
| 29868 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 39 | SOX5 | Sponge network | -0.303 | 0.21002 | -1.091 | 4.0E-5 | 0.348 |
| 29869 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-935 | 35 | HBP1 | Sponge network | -0.739 | 0.0574 | -0.454 | 0 | 0.348 |
| 29870 | RP11-254F7.2 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 10 | ID4 | Sponge network | 0.176 | 0.37402 | -1.644 | 0 | 0.348 |
| 29871 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 62 | RUNX1T1 | Sponge network | -0.355 | 0.0077 | -0.344 | 0.2374 | 0.348 |
| 29872 | FZD10-AS1 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 20 | PTGFR | Sponge network | -0.727 | 0.04726 | -0.233 | 0.45421 | 0.348 |
| 29873 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | ABCB5 | Sponge network | -1.281 | 0.00012 | -0.956 | 0.04077 | 0.348 |
| 29874 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | EPHA5 | Sponge network | 0.222 | 0.29133 | -0.957 | 0.01733 | 0.348 |
| 29875 | RP11-967K21.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577 | 16 | INPP4A | Sponge network | 0.056 | 0.81195 | 0.07 | 0.53563 | 0.348 |
| 29876 | PDXDC2P |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-495-3p;hsa-miR-654-3p | 13 | PHF6 | Sponge network | 0.756 | 0 | 0.785 | 0 | 0.348 |
| 29877 | U91328.19 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-500a-5p;hsa-miR-577 | 13 | MACF1 | Sponge network | -0.303 | 0.21002 | 0.019 | 0.90335 | 0.348 |
| 29878 | RP11-254F7.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-7-5p | 17 | CRTC1 | Sponge network | 0.176 | 0.37402 | -0.116 | 0.28412 | 0.348 |
| 29879 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | DYNC1I1 | Sponge network | -0.663 | 0.10887 | -1.12 | 0.00107 | 0.348 |
| 29880 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-766-3p | 22 | CHD5 | Sponge network | 0.238 | 0.70544 | -0.922 | 0.01521 | 0.348 |
| 29881 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 51 | BMPR2 | Sponge network | -0.769 | 0.00035 | 0.206 | 0.0635 | 0.348 |
| 29882 | RP11-517B11.7 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-409-5p;hsa-miR-495-3p;hsa-miR-664a-3p;hsa-miR-9-5p | 27 | CCDC6 | Sponge network | 0.706 | 0 | 0.27 | 0.02555 | 0.348 |
| 29883 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 36 | EPHA5 | Sponge network | 0.537 | 0.00139 | -0.957 | 0.01733 | 0.348 |
| 29884 | CTD-2089N3.1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p | 13 | BTG2 | Sponge network | -0.314 | 0.62033 | -1.763 | 0 | 0.348 |
| 29885 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-7-1-3p | 23 | NOTCH2 | Sponge network | 0.614 | 1.0E-5 | 0.138 | 0.35163 | 0.348 |
| 29886 | AC009120.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484 | 10 | CRTC3 | Sponge network | 0.919 | 0 | 0.164 | 0.16786 | 0.348 |
| 29887 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 13 | WWTR1 | Sponge network | 0.853 | 0.15352 | -0.345 | 0.11997 | 0.348 |
| 29888 | PCA3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | GALNT13 | Sponge network | -0.709 | 0.18168 | -0.857 | 0.07439 | 0.348 |
| 29889 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | AKAP11 | Sponge network | 2.368 | 2.0E-5 | 0.196 | 0.09825 | 0.348 |
| 29890 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p | 16 | AFF3 | Sponge network | 0.727 | 3.0E-5 | -2.373 | 0 | 0.348 |
| 29891 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 39 | FZD3 | Sponge network | 0.559 | 0.00026 | 0.104 | 0.60316 | 0.348 |
| 29892 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | MOB1B | Sponge network | 0.702 | 7.0E-5 | 0.197 | 0.15266 | 0.348 |
| 29893 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-654-3p | 11 | USP25 | Sponge network | 0.194 | 0.64278 | -0.242 | 0.01397 | 0.348 |
| 29894 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | 0.056 | 0.81195 | -0.615 | 0.01482 | 0.347 |
| 29895 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | EPHA7 | Sponge network | -0.663 | 0.10887 | -2.197 | 3.0E-5 | 0.347 |
| 29896 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | MITF | Sponge network | -0.737 | 0.10822 | -0.769 | 0.00022 | 0.347 |
| 29897 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 13 | FOXO3 | Sponge network | 1.148 | 2.0E-5 | -0.04 | 0.72028 | 0.347 |
| 29898 | LINC00854 |
hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-5p | 13 | MOB1B | Sponge network | 1.18 | 0 | 0.197 | 0.15266 | 0.347 |
| 29899 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-96-5p | 16 | ACVR1 | Sponge network | -0.18 | 0.20343 | 0.279 | 0.00234 | 0.347 |
| 29900 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 15 | PKNOX1 | Sponge network | -0.323 | 0.01372 | 0.085 | 0.28917 | 0.347 |
| 29901 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p;hsa-miR-942-5p | 51 | UNC80 | Sponge network | -0.737 | 0.06182 | -1.934 | 0 | 0.347 |
| 29902 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-93-5p | 18 | PLSCR4 | Sponge network | -0.18 | 0.20343 | -0.474 | 0.03833 | 0.347 |
| 29903 | RP11-434H6.7 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-29b-1-5p;hsa-miR-409-3p | 12 | PHC3 | Sponge network | 0.358 | 0.14302 | 0.212 | 0.0499 | 0.347 |
| 29904 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-942-5p | 18 | ARHGEF12 | Sponge network | 1.162 | 5.0E-5 | 0.09 | 0.35693 | 0.347 |
| 29905 | RP11-428J1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | AKAP6 | Sponge network | 0.538 | 0.00076 | -1.586 | 3.0E-5 | 0.347 |
| 29906 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-331-5p;hsa-miR-98-5p | 16 | USP12 | Sponge network | -0.473 | 0.07602 | -0.102 | 0.40768 | 0.347 |
| 29907 | RP4-791M13.3 |
hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-577 | 14 | HIP1 | Sponge network | 0.419 | 0.0319 | 0.774 | 0 | 0.347 |
| 29908 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 17 | NR2C2 | Sponge network | 0.799 | 1.0E-5 | 0.21 | 0.03224 | 0.347 |
| 29909 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-92a-3p | 21 | AFF4 | Sponge network | 0.912 | 0.00014 | -0.266 | 0.01565 | 0.347 |
| 29910 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p | 16 | ERG | Sponge network | -0.738 | 0.21233 | 0.002 | 0.99011 | 0.347 |
| 29911 | RP11-571M6.17 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ZMYM2 | Sponge network | 1.013 | 0 | 0.279 | 0.01273 | 0.347 |
| 29912 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-744-3p | 14 | CUL5 | Sponge network | 0.946 | 0 | 0.026 | 0.77301 | 0.347 |
| 29913 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | ITGAV | Sponge network | -0.49 | 0.04946 | 0.337 | 0.01002 | 0.347 |
| 29914 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | EPAS1 | Sponge network | 0.249 | 0.35902 | -0.184 | 0.14418 | 0.347 |
| 29915 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p | 26 | NRXN1 | Sponge network | -0.303 | 0.21002 | -3.778 | 0 | 0.347 |
| 29916 | EIF3J-AS1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-370-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ZMYND11 | Sponge network | 0.331 | 0.00509 | -0.2 | 0.05449 | 0.347 |
| 29917 | CTC-490E21.10 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p | 10 | NRK | Sponge network | 1.46 | 1.0E-5 | 0.656 | 0.19217 | 0.347 |
| 29918 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | DAB2 | Sponge network | -0.777 | 0.1134 | 0.431 | 0.0113 | 0.347 |
| 29919 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940 | 47 | QKI | Sponge network | -0.899 | 6.0E-5 | -0.333 | 0.04111 | 0.347 |
| 29920 | RP11-597D13.9 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | PIK3CA | Sponge network | 1.302 | 7.0E-5 | 0.195 | 0.06908 | 0.347 |
| 29921 | RP11-620J15.3 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-744-3p | 23 | PTPN14 | Sponge network | -0.739 | 0.00044 | 0.144 | 0.34595 | 0.347 |
| 29922 | RP11-531A24.5 |
hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p | 17 | SYNM | Sponge network | 0.75 | 0.00054 | -2.309 | 1.0E-5 | 0.347 |
| 29923 | LIPE-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 22 | UBR3 | Sponge network | 0.078 | 0.55803 | -0.021 | 0.82774 | 0.347 |
| 29924 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | ARNT2 | Sponge network | -1.575 | 6.0E-5 | -0.332 | 0.25851 | 0.347 |
| 29925 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 12 | CNTNAP3 | Sponge network | 0.385 | 0.10067 | -1.465 | 0.00033 | 0.347 |
| 29926 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p | 21 | CRTC3 | Sponge network | 0.95 | 0.15943 | 0.164 | 0.16786 | 0.347 |
| 29927 | DNAJC27-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | ZDBF2 | Sponge network | -0.969 | 2.0E-5 | -0.998 | 0.00025 | 0.347 |
| 29928 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 29 | PBX1 | Sponge network | 0.349 | 0.01695 | -1.206 | 0 | 0.347 |
| 29929 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p | 15 | HLF | Sponge network | -0.021 | 0.93598 | -2.443 | 0 | 0.347 |
| 29930 | RP11-212P7.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | PRUNE2 | Sponge network | 0.219 | 0.1516 | -1.733 | 0.00022 | 0.347 |
| 29931 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 42 | EPHA7 | Sponge network | 1.302 | 7.0E-5 | -2.197 | 3.0E-5 | 0.347 |
| 29932 | LINC00865 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-550a-3p | 22 | NRK | Sponge network | -1.249 | 7.0E-5 | 0.656 | 0.19217 | 0.347 |
| 29933 | FAM66C |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 10 | MSN | Sponge network | -0.901 | 0.00019 | -0.023 | 0.88422 | 0.347 |
| 29934 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | BHLHE41 | Sponge network | -0.358 | 0.17255 | 0.353 | 0.12753 | 0.347 |
| 29935 | LINC00865 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-629-3p | 26 | RYR2 | Sponge network | -1.249 | 7.0E-5 | -1.466 | 0.00031 | 0.347 |
| 29936 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p | 11 | GAS1 | Sponge network | -1.249 | 7.0E-5 | -1.047 | 0.00364 | 0.347 |
| 29937 | FAM66C |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 43 | UNC80 | Sponge network | -0.901 | 0.00019 | -1.934 | 0 | 0.347 |
| 29938 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 15 | PLSCR4 | Sponge network | -0.285 | 0.2172 | -0.474 | 0.03833 | 0.347 |
| 29939 | ADAMTS9-AS2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-708-3p;hsa-miR-96-5p | 17 | UNC5D | Sponge network | -2.452 | 0 | -3.646 | 0 | 0.347 |
| 29940 | RP11-395A13.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p | 15 | NTRK2 | Sponge network | 0.057 | 0.61225 | -0.736 | 0.06495 | 0.347 |
| 29941 | RP11-155G14.6 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-625-5p | 13 | MDM4 | Sponge network | 2.483 | 0 | 0.345 | 0.00762 | 0.347 |
| 29942 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-27b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NEDD4 | Sponge network | 1.308 | 0 | 0.02 | 0.89834 | 0.347 |
| 29943 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-655-3p | 27 | DMD | Sponge network | 0.349 | 0.01695 | -1.506 | 0 | 0.347 |
| 29944 | H1FX-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 10 | RHOB | Sponge network | -0.206 | 0.12942 | -1.052 | 0 | 0.347 |
| 29945 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | RSPO2 | Sponge network | 0.572 | 0.01722 | -3.038 | 0 | 0.347 |
| 29946 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-940;hsa-miR-96-5p | 22 | CRTC3 | Sponge network | 0.246 | 0.59912 | 0.164 | 0.16786 | 0.347 |
| 29947 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 23 | PRDM2 | Sponge network | 0.532 | 0.00505 | -0.258 | 0.00721 | 0.347 |
| 29948 | CTA-445C9.14 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 13 | GPAM | Sponge network | 0.718 | 4.0E-5 | 0.142 | 0.29732 | 0.347 |
| 29949 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p | 57 | KIF1B | Sponge network | 0.494 | 0.00339 | -0.118 | 0.32258 | 0.347 |
| 29950 | RP11-401P9.4 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-511-5p;hsa-miR-590-5p | 12 | CELF4 | Sponge network | -0.384 | 0.40652 | -1.135 | 0.00344 | 0.347 |
| 29951 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p | 16 | DMD | Sponge network | -0.206 | 0.12942 | -1.506 | 0 | 0.347 |
| 29952 | RP11-1006G14.4 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | 0.559 | 0.00026 | -0.704 | 0.00106 | 0.347 |
| 29953 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | GALNT13 | Sponge network | -0.342 | 0.02288 | -0.857 | 0.07439 | 0.347 |
| 29954 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-940 | 24 | HLF | Sponge network | 0.174 | 0.30034 | -2.443 | 0 | 0.347 |
| 29955 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p | 12 | TMTC3 | Sponge network | 1.316 | 0 | 0.123 | 0.22189 | 0.347 |
| 29956 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-744-3p | 19 | PTPN14 | Sponge network | 1.302 | 7.0E-5 | 0.144 | 0.34595 | 0.347 |
| 29957 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-93-5p | 23 | LRIG1 | Sponge network | -0.384 | 0.40652 | -0.537 | 0.00588 | 0.347 |
| 29958 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | LIFR | Sponge network | -0.125 | 0.49414 | -1.794 | 0 | 0.347 |
| 29959 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p | 16 | FGFR1 | Sponge network | 1.302 | 7.0E-5 | -0.509 | 0.03957 | 0.347 |
| 29960 | RP11-996F15.2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-424-5p | 11 | ABL2 | Sponge network | 0.983 | 0.00048 | 0.82 | 0 | 0.347 |
| 29961 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | ST6GALNAC3 | Sponge network | 0.4 | 0.14611 | -0.987 | 0 | 0.347 |
| 29962 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-429 | 11 | RICTOR | Sponge network | 2.364 | 0.00134 | 0.388 | 0.00188 | 0.347 |
| 29963 | BOLA3-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p | 11 | ID4 | Sponge network | -0.246 | 0.35034 | -1.644 | 0 | 0.347 |
| 29964 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | HOOK3 | Sponge network | 0.61 | 0.01593 | 0.273 | 0.09957 | 0.347 |
| 29965 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -0.465 | 0.04703 | -0.354 | 0.14694 | 0.347 |
| 29966 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | DPYD | Sponge network | -0.376 | 0.00337 | -0.736 | 0.00076 | 0.347 |
| 29967 | LINC00476 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-877-5p | 16 | INPP4A | Sponge network | -0.615 | 0.00015 | 0.07 | 0.53563 | 0.347 |
| 29968 | RP11-53O19.3 |
hsa-miR-101-3p;hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | GSK3B | Sponge network | 1.205 | 0 | 0.266 | 0.00045 | 0.347 |
| 29969 | RP11-589P10.7 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-215-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-505-3p | 14 | DST | Sponge network | -2.044 | 0.00066 | -0.713 | 0.00096 | 0.347 |
| 29970 | RP5-1139B12.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-502-5p | 15 | IL17RD | Sponge network | 0.598 | 0.38481 | -0.019 | 0.94873 | 0.347 |
| 29971 | RP11-707P17.2 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-495-3p;hsa-miR-654-3p | 14 | PHF6 | Sponge network | 1.877 | 0 | 0.785 | 0 | 0.347 |
| 29972 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 26 | SEMA3C | Sponge network | 0.101 | 0.60919 | -0.272 | 0.31153 | 0.347 |
| 29973 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p | 11 | TSC22D1 | Sponge network | -0.615 | 0.00015 | -0.529 | 2.0E-5 | 0.347 |
| 29974 | MAGI2-AS3 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 55 | PPM1L | Sponge network | -0.129 | 0.57177 | -0.537 | 0.00616 | 0.347 |
| 29975 | AC006116.24 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-93-5p | 21 | PTPRD | Sponge network | -0.473 | 0.07602 | -1.005 | 0.00064 | 0.347 |
| 29976 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-424-5p | 19 | MYBL1 | Sponge network | 0.4 | 0.14611 | 0.494 | 0.02152 | 0.347 |
| 29977 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-96-5p | 22 | SOX5 | Sponge network | -0.326 | 0.14314 | -1.091 | 4.0E-5 | 0.347 |
| 29978 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-93-5p | 17 | TGFBR2 | Sponge network | 0.056 | 0.81195 | -0.243 | 0.11408 | 0.347 |
| 29979 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-421;hsa-miR-625-5p | 15 | RSPO2 | Sponge network | -4.477 | 0 | -3.038 | 0 | 0.347 |
| 29980 | RP11-434H6.7 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-132-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-409-3p | 13 | SMAD2 | Sponge network | 0.358 | 0.14302 | -0.128 | 0.12732 | 0.347 |
| 29981 | CTC-429P9.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-409-3p | 18 | BCL2 | Sponge network | 0.497 | 0.01451 | -0.899 | 0 | 0.347 |
| 29982 | FAM157C |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | PIK3C2A | Sponge network | 0.91 | 0 | 0.061 | 0.53776 | 0.347 |
| 29983 | AC003090.1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p | 19 | CTIF | Sponge network | -2.117 | 0.00161 | -0.389 | 0.00549 | 0.347 |
| 29984 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | ABCA10 | Sponge network | -0.739 | 0.00044 | -0.571 | 0.03674 | 0.347 |
| 29985 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 19 | PGR | Sponge network | -0.031 | 0.89063 | -1.704 | 0 | 0.347 |
| 29986 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-769-5p;hsa-miR-93-5p | 29 | HBP1 | Sponge network | -2.69 | 0.0001 | -0.454 | 0 | 0.347 |
| 29987 | RAMP2-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-629-5p | 13 | WNK2 | Sponge network | -0.513 | 0.04703 | -0.352 | 0.31066 | 0.347 |
| 29988 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -1.439 | 4.0E-5 | -0.966 | 0.00035 | 0.347 |
| 29989 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-654-3p | 20 | RICTOR | Sponge network | 1.344 | 0 | 0.388 | 0.00188 | 0.347 |
| 29990 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-874-3p | 21 | NR2C2 | Sponge network | 0.082 | 0.55654 | 0.21 | 0.03224 | 0.347 |
| 29991 | LINC00094 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p | 11 | LHFP | Sponge network | 0.253 | 0.09565 | -0.167 | 0.41371 | 0.347 |
| 29992 | HOTAIRM1 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | RIMS2 | Sponge network | -0.053 | 0.80685 | -1.365 | 0.00208 | 0.347 |
| 29993 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p | 10 | NCAM2 | Sponge network | -0.465 | 0.04703 | -0.482 | 0.22565 | 0.347 |
| 29994 | MIR497HG |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | EYA4 | Sponge network | -1.439 | 4.0E-5 | 0.3 | 0.36876 | 0.347 |
| 29995 | BCYRN1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 21 | ATM | Sponge network | 0.311 | 0.10344 | 0.178 | 0.21568 | 0.347 |
| 29996 | MIR22HG |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-3913-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 36 | AFF1 | Sponge network | -1.047 | 0 | -0.384 | 0.00053 | 0.347 |
| 29997 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | PNRC1 | Sponge network | -2.117 | 0.00161 | -0.646 | 0 | 0.347 |
| 29998 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | ITCH | Sponge network | 0.873 | 0 | 0.084 | 0.34058 | 0.347 |
| 29999 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | 0.572 | 0.01722 | -0.252 | 0.15511 | 0.347 |
| 30000 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | MLLT10 | Sponge network | -0.612 | 0.00094 | 0.105 | 0.33292 | 0.347 |
| 30001 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-589-3p | 12 | ESR1 | Sponge network | -4.463 | 0.00025 | -1.035 | 3.0E-5 | 0.347 |
| 30002 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-590-3p | 12 | AKAP11 | Sponge network | 0.8 | 0 | 0.196 | 0.09825 | 0.347 |
| 30003 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-362-3p;hsa-miR-93-5p | 15 | EPHA7 | Sponge network | -1.255 | 0.00981 | -2.197 | 3.0E-5 | 0.347 |
| 30004 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | CUL5 | Sponge network | 1.601 | 0 | 0.026 | 0.77301 | 0.347 |
| 30005 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | MED12L | Sponge network | -0.739 | 0.0574 | -0.194 | 0.55299 | 0.347 |
| 30006 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | RABEP1 | Sponge network | -0.129 | 0.57177 | -0.066 | 0.43142 | 0.347 |
| 30007 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-744-3p | 19 | HIP1 | Sponge network | 0.051 | 0.83388 | 0.774 | 0 | 0.347 |
| 30008 | U91328.19 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 47 | PPM1L | Sponge network | -0.303 | 0.21002 | -0.537 | 0.00616 | 0.347 |
| 30009 | SERTAD4-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-424-5p | 22 | NOTCH2 | Sponge network | -0.932 | 0.00329 | 0.138 | 0.35163 | 0.347 |
| 30010 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-495-3p | 10 | ATRX | Sponge network | 0.752 | 0 | 0.261 | 0.04408 | 0.347 |
| 30011 | AC004893.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p | 10 | CEP170 | Sponge network | 1.317 | 0 | 0.943 | 0 | 0.347 |
| 30012 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-493-5p | 12 | SYNE1 | Sponge network | -1.942 | 0 | -0.738 | 0.00316 | 0.347 |
| 30013 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 51 | DTNA | Sponge network | 0.083 | 0.66405 | -1.648 | 1.0E-5 | 0.347 |
| 30014 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | ZMYM2 | Sponge network | 0.79 | 0 | 0.279 | 0.01273 | 0.347 |
| 30015 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TBX18 | Sponge network | -0.487 | 0.02157 | 0.843 | 0.02945 | 0.347 |
| 30016 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-450b-5p | 11 | GRIA3 | Sponge network | -0.517 | 0.02935 | -1.956 | 0 | 0.347 |
| 30017 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 20 | CNTN1 | Sponge network | -0.227 | 0.31657 | -2.594 | 0 | 0.347 |
| 30018 | RP11-38L15.3 |
hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 17 | IL6ST | Sponge network | 0.344 | 0.3127 | -0.402 | 0.00676 | 0.347 |
| 30019 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 11 | FAM188A | Sponge network | -0.031 | 0.89063 | -0.44 | 0.00018 | 0.347 |
| 30020 | PART1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | RYR2 | Sponge network | -2.925 | 0 | -1.466 | 0.00031 | 0.347 |
| 30021 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | CXXC4 | Sponge network | -2.117 | 0.00161 | -0.433 | 0.21055 | 0.347 |
| 30022 | FAM13A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-766-3p;hsa-miR-93-5p | 14 | EZH1 | Sponge network | -0.83 | 0 | -0.564 | 1.0E-5 | 0.347 |
| 30023 | RP11-25K21.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p | 16 | BMPR2 | Sponge network | 0.489 | 0.25786 | 0.206 | 0.0635 | 0.347 |
| 30024 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 36 | SASH1 | Sponge network | -0.355 | 0.0077 | -0.502 | 0.00175 | 0.347 |
| 30025 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | CNTN1 | Sponge network | -0.663 | 0.10887 | -2.594 | 0 | 0.347 |
| 30026 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-625-5p | 14 | LHX6 | Sponge network | -0.323 | 0.01372 | -0.087 | 0.69193 | 0.347 |
| 30027 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p | 16 | HLF | Sponge network | 0.37 | 0.27614 | -2.443 | 0 | 0.347 |
| 30028 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CLIP1 | Sponge network | 0.421 | 0.00084 | -0.215 | 0.08684 | 0.347 |
| 30029 | RP11-597D13.9 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | ERG | Sponge network | 1.302 | 7.0E-5 | 0.002 | 0.99011 | 0.347 |
| 30030 | RP11-130L8.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p | 13 | CBFA2T3 | Sponge network | -0.663 | 0.10887 | -1.221 | 1.0E-5 | 0.347 |
| 30031 | RP1-122P22.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-532-5p;hsa-miR-629-3p | 20 | AR | Sponge network | 0.065 | 0.73468 | -1.554 | 0 | 0.347 |
| 30032 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | NALCN | Sponge network | -2.46 | 0 | 1.068 | 0.00237 | 0.347 |
| 30033 | RP4-714D9.5 |
hsa-miR-125b-2-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-143-5p;hsa-miR-23b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30c-2-3p;hsa-miR-30e-3p;hsa-miR-374b-5p | 10 | DIS3 | Sponge network | 1.015 | 0 | 0.39 | 8.0E-5 | 0.347 |
| 30034 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-500a-3p;hsa-miR-93-3p | 13 | ERC1 | Sponge network | 0.598 | 0.38481 | -0.108 | 0.38794 | 0.347 |
| 30035 | RP11-212P7.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 27 | SETBP1 | Sponge network | 0.219 | 0.1516 | -1.302 | 1.0E-5 | 0.347 |
| 30036 | ADAMTS9-AS2 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 22 | IRS1 | Sponge network | -2.452 | 0 | -0.026 | 0.88855 | 0.347 |
| 30037 | LINC00957 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | KLF10 | Sponge network | -0.421 | 0.0114 | -0.783 | 0 | 0.347 |
| 30038 | HAS2-AS1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-30b-3p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-942-5p | 14 | IGFBP5 | Sponge network | -0.011 | 0.967 | -0.806 | 0.00105 | 0.347 |
| 30039 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-29b-1-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | KAT6B | Sponge network | -0.021 | 0.93598 | -0.478 | 0.00018 | 0.347 |
| 30040 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 32 | ADARB1 | Sponge network | 0.056 | 0.81195 | -0.335 | 0.06631 | 0.347 |
| 30041 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | MYO9A | Sponge network | -0.739 | 0.00044 | -0.147 | 0.32373 | 0.347 |
| 30042 | C4B-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-486-5p;hsa-miR-96-5p | 15 | PRRX1 | Sponge network | 2.306 | 9.0E-5 | 1.316 | 0 | 0.347 |
| 30043 | RP11-2H8.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | KDSR | Sponge network | 1.089 | 0 | 0.239 | 0.02216 | 0.347 |
| 30044 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 18 | LRRC55 | Sponge network | 1.302 | 7.0E-5 | -0.026 | 0.93163 | 0.347 |
| 30045 | ADIRF-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | PLAG1 | Sponge network | -0.223 | 0.47816 | -0.315 | 0.26532 | 0.347 |
| 30046 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 30 | CBL | Sponge network | -0.323 | 0.01372 | 0.291 | 0.01267 | 0.347 |
| 30047 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-625-5p | 14 | LHX6 | Sponge network | 0.064 | 0.72934 | -0.087 | 0.69193 | 0.347 |
| 30048 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-335-3p | 13 | DLC1 | Sponge network | 0.497 | 0.01451 | -0.382 | 0.05038 | 0.347 |
| 30049 | FAM66C |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 40 | PIK3R1 | Sponge network | -0.901 | 0.00019 | -0.133 | 0.36854 | 0.347 |
| 30050 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 32 | ERC1 | Sponge network | 0.494 | 0.00339 | -0.108 | 0.38794 | 0.347 |
| 30051 | RP11-473I1.10 |
hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | KIF5B | Sponge network | 0.673 | 0 | 0.362 | 0.00066 | 0.347 |
| 30052 | LPP-AS2 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | PPM1L | Sponge network | -0.18 | 0.20343 | -0.537 | 0.00616 | 0.347 |
| 30053 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | TSHZ3 | Sponge network | 0.4 | 0.14611 | -0.556 | 0.01516 | 0.347 |
| 30054 | NNT-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p | 19 | TRIM33 | Sponge network | 0.799 | 1.0E-5 | 0.402 | 7.0E-5 | 0.347 |
| 30055 | AC005618.6 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ITGB1 | Sponge network | 0.862 | 0.06213 | 0.524 | 0.00017 | 0.347 |
| 30056 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 31 | TGFBR3 | Sponge network | 0.72 | 0.0001 | -1.173 | 1.0E-5 | 0.347 |
| 30057 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 19 | BNC2 | Sponge network | 0.37 | 0.27614 | -0.491 | 0.09988 | 0.347 |
| 30058 | FGF14-AS2 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-660-5p | 14 | LIMCH1 | Sponge network | -1.582 | 8.0E-5 | -0.915 | 1.0E-5 | 0.347 |
| 30059 | RP11-65J21.3 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-542-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 17 | DTNA | Sponge network | -0.557 | 0.11135 | -1.648 | 1.0E-5 | 0.347 |
| 30060 | LINC00654 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-3p | 15 | FMNL3 | Sponge network | 0.374 | 0.20054 | 0.555 | 4.0E-5 | 0.347 |
| 30061 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 15 | TBL1XR1 | Sponge network | 1.163 | 0.00018 | 0.578 | 0 | 0.347 |
| 30062 | DNAJC27-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-942-5p;hsa-miR-96-5p | 13 | ZBTB20 | Sponge network | -0.969 | 2.0E-5 | -0.122 | 0.57569 | 0.347 |
| 30063 | CTD-3138B18.6 |
hsa-miR-125a-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-590-3p | 10 | AKAP11 | Sponge network | 2.248 | 0 | 0.196 | 0.09825 | 0.347 |
| 30064 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TBX18 | Sponge network | 0.374 | 0.20054 | 0.843 | 0.02945 | 0.347 |
| 30065 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 32 | FBXO32 | Sponge network | 0.219 | 0.1516 | -0.495 | 0.07561 | 0.347 |
| 30066 | RP11-244H3.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZMYND11 | Sponge network | 0.659 | 3.0E-5 | -0.2 | 0.05449 | 0.347 |
| 30067 | LINC00865 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-935 | 32 | ABI2 | Sponge network | -1.249 | 7.0E-5 | 0.175 | 0.14787 | 0.347 |
| 30068 | LINC00092 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 13 | TET1 | Sponge network | -1.886 | 0 | -0.135 | 0.52138 | 0.347 |
| 30069 | FAM66C |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-5p | 22 | RNASEL | Sponge network | -0.901 | 0.00019 | -0.272 | 0.01796 | 0.347 |
| 30070 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | ST6GALNAC3 | Sponge network | -4.477 | 0 | -0.987 | 0 | 0.347 |
| 30071 | RP11-182J1.1 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | AKAP11 | Sponge network | -0.187 | 0.23787 | 0.196 | 0.09825 | 0.347 |
| 30072 | RP5-1139B12.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p | 13 | ITPKB | Sponge network | 0.598 | 0.38481 | -0.704 | 0.00106 | 0.347 |
| 30073 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-942-5p | 28 | HLF | Sponge network | 0.494 | 0.00339 | -2.443 | 0 | 0.347 |
| 30074 | LINC00473 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935 | 42 | PTPRD | Sponge network | -1.203 | 0.03881 | -1.005 | 0.00064 | 0.347 |
| 30075 | RP11-1094M14.11 |
hsa-miR-101-3p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 10 | PHF6 | Sponge network | 0.586 | 2.0E-5 | 0.785 | 0 | 0.347 |
| 30076 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-493-5p | 11 | NRXN3 | Sponge network | 1.46 | 1.0E-5 | -0.893 | 0.04186 | 0.347 |
| 30077 | CTC-487M23.5 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-92a-3p | 11 | ATM | Sponge network | 0.912 | 0.00014 | 0.178 | 0.21568 | 0.347 |
| 30078 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 29 | FAT3 | Sponge network | -0.206 | 0.12942 | -1.601 | 0.00051 | 0.347 |
| 30079 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-744-3p | 22 | PTPN14 | Sponge network | -0.896 | 0.00099 | 0.144 | 0.34595 | 0.347 |
| 30080 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | NCAM2 | Sponge network | -0.942 | 0 | -0.482 | 0.22565 | 0.347 |
| 30081 | RP11-428J1.5 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | RASAL2 | Sponge network | 0.538 | 0.00076 | 0.869 | 0 | 0.347 |
| 30082 | A2M-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-484 | 12 | ITGA5 | Sponge network | -0.08 | 0.90092 | 0.452 | 0.03524 | 0.347 |
| 30083 | FAM66C |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | -0.901 | 0.00019 | -0.001 | 0.99669 | 0.347 |
| 30084 | LINC00174 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 13 | PHC3 | Sponge network | 1.253 | 0 | 0.212 | 0.0499 | 0.347 |
| 30085 | RP11-554A11.4 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-505-5p | 10 | WNK2 | Sponge network | -0.583 | 0.14589 | -0.352 | 0.31066 | 0.347 |
| 30086 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p | 30 | PDGFRA | Sponge network | -4.546 | 0 | -0.372 | 0.0037 | 0.347 |
| 30087 | RP5-994D16.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | BRD3 | Sponge network | 0.252 | 0.08508 | 0.37 | 0.00013 | 0.347 |
| 30088 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 0.194 | 0.64278 | 0.114 | 0.30638 | 0.347 |
| 30089 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-93-3p | 25 | NCOA1 | Sponge network | -2.69 | 0.0001 | -0.432 | 0 | 0.347 |
| 30090 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 37 | CBL | Sponge network | -0.129 | 0.57177 | 0.291 | 0.01267 | 0.347 |
| 30091 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 41 | CBX5 | Sponge network | -0.405 | 0.22013 | 0.165 | 0.24446 | 0.347 |
| 30092 | AC006116.24 |
hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p | 10 | KCNJ5 | Sponge network | -0.473 | 0.07602 | -0.281 | 0.3339 | 0.347 |
| 30093 | RP4-758J18.10 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-450b-5p | 15 | IL17RD | Sponge network | -0.091 | 0.71468 | -0.019 | 0.94873 | 0.347 |
| 30094 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p | 13 | FOXO3 | Sponge network | 1.11 | 1.0E-5 | -0.04 | 0.72028 | 0.347 |
| 30095 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p | 14 | CHD1 | Sponge network | 0.75 | 0.00054 | 0.322 | 0.00215 | 0.347 |
| 30096 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | PHC3 | Sponge network | 1.317 | 0 | 0.212 | 0.0499 | 0.347 |
| 30097 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p | 15 | ARNT2 | Sponge network | 0.214 | 0.18246 | -0.332 | 0.25851 | 0.347 |
| 30098 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DACT1 | Sponge network | -2.117 | 0.00161 | 0.783 | 0.00072 | 0.347 |
| 30099 | AC005154.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 27 | ABI2 | Sponge network | 0.848 | 0 | 0.175 | 0.14787 | 0.347 |
| 30100 | LINC00680 |
hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-497-5p;hsa-miR-582-5p | 15 | BRD3 | Sponge network | 0.884 | 0 | 0.37 | 0.00013 | 0.347 |
| 30101 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-96-5p | 19 | SCN9A | Sponge network | 0.622 | 0.00015 | -0.525 | 0.10593 | 0.347 |
| 30102 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 41 | NOTCH2 | Sponge network | -1.575 | 6.0E-5 | 0.138 | 0.35163 | 0.347 |
| 30103 | LINC00578 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 23 | PTPRD | Sponge network | 0.811 | 0.03228 | -1.005 | 0.00064 | 0.347 |
| 30104 | RP5-855D21.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 12 | CREB1 | Sponge network | 1.728 | 0 | 0.203 | 0.00537 | 0.347 |
| 30105 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | HECA | Sponge network | 0.559 | 0.00026 | -0.217 | 0.03463 | 0.347 |
| 30106 | LINC00641 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-98-5p | 17 | ITGA5 | Sponge network | -0.222 | 0.32445 | 0.452 | 0.03524 | 0.347 |
| 30107 | TPTEP1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | LIMCH1 | Sponge network | -1.216 | 0 | -0.915 | 1.0E-5 | 0.347 |
| 30108 | THAP9-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p | 15 | CREB1 | Sponge network | 1.079 | 0 | 0.203 | 0.00537 | 0.347 |
| 30109 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 13 | GPAM | Sponge network | 0.663 | 0.00073 | 0.142 | 0.29732 | 0.347 |
| 30110 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-942-5p | 19 | NOTCH2 | Sponge network | -0.693 | 0.05629 | 0.138 | 0.35163 | 0.347 |
| 30111 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-590-3p | 10 | JAKMIP2 | Sponge network | 0.199 | 0.38015 | -1.388 | 0 | 0.346 |
| 30112 | AC006116.24 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-539-5p | 16 | BTG2 | Sponge network | -0.473 | 0.07602 | -1.763 | 0 | 0.346 |
| 30113 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SLC6A15 | Sponge network | -0.053 | 0.80685 | -2.373 | 2.0E-5 | 0.346 |
| 30114 | RP11-774O3.3 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 26 | LIMCH1 | Sponge network | -0.487 | 0.02157 | -0.915 | 1.0E-5 | 0.346 |
| 30115 | AC106786.1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-629-3p | 11 | ESR1 | Sponge network | 1.122 | 0.02989 | -1.035 | 3.0E-5 | 0.346 |
| 30116 | SNHG14 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-942-5p | 37 | OPCML | Sponge network | -1.111 | 0.0003 | -2.498 | 0 | 0.346 |
| 30117 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-877-5p | 25 | GRIN2A | Sponge network | -0.371 | 0.24604 | -2.161 | 0 | 0.346 |
| 30118 | RP11-175O19.4 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p | 21 | CREB1 | Sponge network | 0.917 | 0 | 0.203 | 0.00537 | 0.346 |
| 30119 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 18 | ZFP36L1 | Sponge network | 0.194 | 0.64278 | -0.505 | 8.0E-5 | 0.346 |
| 30120 | BOLA3-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | NEDD4 | Sponge network | -0.246 | 0.35034 | 0.02 | 0.89834 | 0.346 |
| 30121 | RP11-517B11.7 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-495-3p | 17 | ZFX | Sponge network | 0.706 | 0 | 0.09 | 0.42563 | 0.346 |
| 30122 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 45 | THRB | Sponge network | -0.611 | 0.09607 | -1.117 | 0.0004 | 0.346 |
| 30123 | RP11-305E6.4 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-766-3p | 10 | ATM | Sponge network | 1.317 | 0 | 0.178 | 0.21568 | 0.346 |
| 30124 | TP73-AS1 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-93-5p;hsa-miR-935 | 26 | ACSL6 | Sponge network | -0.563 | 0.02545 | -1.593 | 0 | 0.346 |
| 30125 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-629-3p | 14 | PSIP1 | Sponge network | 0.174 | 0.30034 | -0.005 | 0.96897 | 0.346 |
| 30126 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 2.094 | 0 | 0.259 | 0.00962 | 0.346 |
| 30127 | GNG12-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 20 | TET1 | Sponge network | -0.793 | 7.0E-5 | -0.135 | 0.52138 | 0.346 |
| 30128 | LENG8-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-539-5p | 12 | ABI2 | Sponge network | 1.471 | 0 | 0.175 | 0.14787 | 0.346 |
| 30129 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | CXXC4 | Sponge network | 0.411 | 0.1335 | -0.433 | 0.21055 | 0.346 |
| 30130 | LINC00909 |
hsa-miR-125a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-411-5p;hsa-miR-484;hsa-miR-493-5p | 10 | SMAD2 | Sponge network | 0.363 | 0.00601 | -0.128 | 0.12732 | 0.346 |
| 30131 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 45 | NTRK3 | Sponge network | -0.342 | 0.02288 | -1.77 | 3.0E-5 | 0.346 |
| 30132 | ARHGEF26-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-628-5p;hsa-miR-708-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 28 | TOM1L2 | Sponge network | -2.46 | 0 | -0.994 | 0 | 0.346 |
| 30133 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374b-5p | 15 | CDC73 | Sponge network | 1.266 | 0 | 0.094 | 0.20463 | 0.346 |
| 30134 | LINC00957 |
hsa-let-7d-5p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | THBS1 | Sponge network | -0.421 | 0.0114 | 0.741 | 0.00386 | 0.346 |
| 30135 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-7-1-3p | 14 | PLSCR4 | Sponge network | 0.389 | 0.04293 | -0.474 | 0.03833 | 0.346 |
| 30136 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 14 | SPTBN4 | Sponge network | -0.769 | 0.00035 | -0.786 | 0.01281 | 0.346 |
| 30137 | RP11-37B2.1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p | 10 | ATM | Sponge network | 1.03 | 0 | 0.178 | 0.21568 | 0.346 |
| 30138 | FGD5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PDE4DIP | Sponge network | 0.401 | 0.00066 | -0.282 | 0.0257 | 0.346 |
| 30139 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | TBX18 | Sponge network | 0.299 | 0.57722 | 0.843 | 0.02945 | 0.346 |
| 30140 | RP11-497H16.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | KDSR | Sponge network | 0.545 | 0.00064 | 0.239 | 0.02216 | 0.346 |
| 30141 | LINC00092 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-25-3p;hsa-miR-362-3p;hsa-miR-542-3p | 11 | ST6GAL2 | Sponge network | -1.886 | 0 | -1.201 | 0.00597 | 0.346 |
| 30142 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-493-5p | 21 | KIF1B | Sponge network | 1.46 | 1.0E-5 | -0.118 | 0.32258 | 0.346 |
| 30143 | RP11-701H24.3 |
hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 11 | RAP1A | Sponge network | -1.425 | 0.04204 | -0.929 | 0 | 0.346 |
| 30144 | RP11-35G9.3 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TLL1 | Sponge network | 0.657 | 4.0E-5 | -1.195 | 3.0E-5 | 0.346 |
| 30145 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ATRX | Sponge network | 0.986 | 0.01022 | 0.261 | 0.04408 | 0.346 |
| 30146 | RP11-35G9.3 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p | 36 | RUNX1T1 | Sponge network | 0.657 | 4.0E-5 | -0.344 | 0.2374 | 0.346 |
| 30147 | RP11-426C22.5 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | ABL2 | Sponge network | 0.749 | 0 | 0.82 | 0 | 0.346 |
| 30148 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 29 | USP12 | Sponge network | 0.249 | 0.35902 | -0.102 | 0.40768 | 0.346 |
| 30149 | RP11-216B9.6 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | ASTN1 | Sponge network | 0.174 | 0.30034 | -2.389 | 2.0E-5 | 0.346 |
| 30150 | LINC00641 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | NOTCH2NL | Sponge network | -0.222 | 0.32445 | 0.024 | 0.84307 | 0.346 |
| 30151 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-5p | 10 | DPYD | Sponge network | -1.047 | 0 | -0.736 | 0.00076 | 0.346 |
| 30152 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ARID5B | Sponge network | -0.031 | 0.89063 | 0.342 | 0.01676 | 0.346 |
| 30153 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 42 | PDGFRA | Sponge network | -0.323 | 0.01372 | -0.372 | 0.0037 | 0.346 |
| 30154 | AC016747.3 |
hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | GJA1 | Sponge network | 0.199 | 0.38015 | -0.485 | 0.02179 | 0.346 |
| 30155 | RP11-2H8.2 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 19 | FOXO3 | Sponge network | 1.089 | 0 | -0.04 | 0.72028 | 0.346 |
| 30156 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-654-3p | 20 | NR2C2 | Sponge network | 0.385 | 0.10067 | 0.21 | 0.03224 | 0.346 |
| 30157 | PART1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | FAT4 | Sponge network | -2.925 | 0 | -0.354 | 0.14694 | 0.346 |
| 30158 | RP1-122P22.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-629-3p | 11 | BHLHE41 | Sponge network | 0.065 | 0.73468 | 0.353 | 0.12753 | 0.346 |
| 30159 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-29a-5p | 10 | RBMS3 | Sponge network | 1.46 | 1.0E-5 | -0.519 | 0.03551 | 0.346 |
| 30160 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 31 | PRDM2 | Sponge network | -0.777 | 0.1134 | -0.258 | 0.00721 | 0.346 |
| 30161 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-375;hsa-miR-590-3p | 18 | BMPR2 | Sponge network | 0.545 | 0.05789 | 0.206 | 0.0635 | 0.346 |
| 30162 | RP11-158K1.3 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-450b-5p | 10 | MN1 | Sponge network | 0.349 | 0.01695 | -0.358 | 0.24172 | 0.346 |
| 30163 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 35 | CREB3L2 | Sponge network | -0.487 | 0.02157 | -0.525 | 2.0E-5 | 0.346 |
| 30164 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 24 | ARHGAP28 | Sponge network | 0.572 | 0.01722 | -0.911 | 0.00013 | 0.346 |
| 30165 | TP73-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429 | 14 | KLF10 | Sponge network | -0.563 | 0.02545 | -0.783 | 0 | 0.346 |
| 30166 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | ST6GAL2 | Sponge network | -1.249 | 7.0E-5 | -1.201 | 0.00597 | 0.346 |
| 30167 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | THBS1 | Sponge network | -2.117 | 0.00161 | 0.741 | 0.00386 | 0.346 |
| 30168 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p | 10 | PSIP1 | Sponge network | 0.594 | 0.00064 | -0.005 | 0.96897 | 0.346 |
| 30169 | BVES-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-576-5p | 19 | PDE4DIP | Sponge network | -2.62 | 0 | -0.282 | 0.0257 | 0.346 |
| 30170 | LINC00092 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-590-3p | 11 | MAP3K12 | Sponge network | -1.886 | 0 | -0.057 | 0.59369 | 0.346 |
| 30171 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 15 | GAS7 | Sponge network | -7.551 | 0 | -0.555 | 0.01602 | 0.346 |
| 30172 | RP4-613B23.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-589-5p;hsa-miR-877-5p | 12 | SUFU | Sponge network | -0.309 | 0.1145 | -0.04 | 0.65489 | 0.346 |
| 30173 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 63 | NTRK3 | Sponge network | 0.101 | 0.60919 | -1.77 | 3.0E-5 | 0.346 |
| 30174 | TRIM52-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-577;hsa-miR-7-1-3p | 27 | DTNA | Sponge network | -0.418 | 0.00068 | -1.648 | 1.0E-5 | 0.346 |
| 30175 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-3p | 29 | FOXO3 | Sponge network | 0.559 | 0.00026 | -0.04 | 0.72028 | 0.346 |
| 30176 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 16 | PTGFR | Sponge network | -0.342 | 0.02288 | -0.233 | 0.45421 | 0.346 |
| 30177 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ITGB3 | Sponge network | 0.225 | 0.30644 | -0.011 | 0.95751 | 0.346 |
| 30178 | PART1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 17 | KLF10 | Sponge network | -2.925 | 0 | -0.783 | 0 | 0.346 |
| 30179 | AP001055.6 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-455-3p | 13 | GRIK3 | Sponge network | 0.693 | 0.00076 | -3.208 | 0 | 0.346 |
| 30180 | CTC-429P9.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-940 | 20 | CREB1 | Sponge network | 0.082 | 0.55397 | 0.203 | 0.00537 | 0.346 |
| 30181 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | BMPR2 | Sponge network | 0.953 | 0 | 0.206 | 0.0635 | 0.346 |
| 30182 | WDR86-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-429 | 10 | MAFB | Sponge network | 0.348 | 0.32912 | -0.023 | 0.89865 | 0.346 |
| 30183 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-93-5p | 23 | SASH1 | Sponge network | -2.69 | 0.0001 | -0.502 | 0.00175 | 0.346 |
| 30184 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | ATRX | Sponge network | 0.219 | 0.1516 | 0.261 | 0.04408 | 0.346 |
| 30185 | RP11-390K5.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 11 | BRD3 | Sponge network | 1.148 | 2.0E-5 | 0.37 | 0.00013 | 0.346 |
| 30186 | ZNF582-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-93-5p | 21 | ACSL6 | Sponge network | -1.349 | 0.00017 | -1.593 | 0 | 0.346 |
| 30187 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 19 | RICTOR | Sponge network | 0.796 | 4.0E-5 | 0.388 | 0.00188 | 0.346 |
| 30188 | SNX29P2 |
hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | PCDH18 | Sponge network | 0.4 | 0.14611 | 0.216 | 0.24645 | 0.346 |
| 30189 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p | 18 | NOTCH2 | Sponge network | 0.898 | 1.0E-5 | 0.138 | 0.35163 | 0.346 |
| 30190 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 23 | KANK1 | Sponge network | -2.46 | 0 | -0.55 | 0.00134 | 0.346 |
| 30191 | AC108676.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | NF1 | Sponge network | 4.346 | 2.0E-5 | 0.51 | 0 | 0.346 |
| 30192 | RP11-344B5.2 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p | 10 | EPHA3 | Sponge network | -0.055 | 0.88482 | 0.185 | 0.53588 | 0.346 |
| 30193 | BDNF-AS |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 12 | RIMS1 | Sponge network | -0.668 | 3.0E-5 | -3.143 | 0 | 0.346 |
| 30194 | PWAR6 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 16 | SHPRH | Sponge network | -1.575 | 6.0E-5 | 0.098 | 0.40994 | 0.346 |
| 30195 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-421 | 18 | MITF | Sponge network | -1.645 | 0 | -0.769 | 0.00022 | 0.346 |
| 30196 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-5p;hsa-miR-708-3p | 24 | PRKAB2 | Sponge network | -0.384 | 0.40652 | -0.367 | 0.01371 | 0.346 |
| 30197 | GNG12-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | ERRFI1 | Sponge network | -0.793 | 7.0E-5 | -0.798 | 1.0E-5 | 0.346 |
| 30198 | RP11-798M19.6 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SH3GLB1 | Sponge network | -0.612 | 0.00094 | -0.115 | 0.14773 | 0.346 |
| 30199 | RP4-717I23.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 10 | PHF6 | Sponge network | 0.637 | 0.00069 | 0.785 | 0 | 0.346 |
| 30200 | HCG18 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 30 | ZFX | Sponge network | 1.063 | 0 | 0.09 | 0.42563 | 0.346 |
| 30201 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-590-3p | 39 | ADARB1 | Sponge network | -0.125 | 0.49414 | -0.335 | 0.06631 | 0.346 |
| 30202 | CTD-3064M3.3 |
hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-7-5p | 11 | IRS1 | Sponge network | -0.469 | 0.08721 | -0.026 | 0.88855 | 0.346 |
| 30203 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-502-5p | 13 | PTPN14 | Sponge network | 0.912 | 0.00014 | 0.144 | 0.34595 | 0.346 |
| 30204 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | KAT6B | Sponge network | 0.532 | 0.00505 | -0.478 | 0.00018 | 0.346 |
| 30205 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 14 | TXNIP | Sponge network | 0.4 | 0.14611 | -1.277 | 0 | 0.346 |
| 30206 | RP11-981G7.6 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-766-3p | 13 | ATM | Sponge network | 0.986 | 0.01022 | 0.178 | 0.21568 | 0.346 |
| 30207 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-5p | 16 | CLASP2 | Sponge network | 1.04 | 0.00023 | -0.038 | 0.71 | 0.346 |
| 30208 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-629-3p | 14 | USP12 | Sponge network | 0.637 | 0.00069 | -0.102 | 0.40768 | 0.346 |
| 30209 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 42 | ERBB4 | Sponge network | -0.899 | 6.0E-5 | -1.975 | 2.0E-5 | 0.346 |
| 30210 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-93-3p | 58 | IL6ST | Sponge network | 0.056 | 0.81195 | -0.402 | 0.00676 | 0.346 |
| 30211 | AC011526.1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | PCDH9 | Sponge network | 0.342 | 0.03129 | -1.823 | 4.0E-5 | 0.346 |
| 30212 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-590-5p | 14 | BTG2 | Sponge network | -0.72 | 0.24239 | -1.763 | 0 | 0.346 |
| 30213 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 28 | RNASEL | Sponge network | -1.575 | 6.0E-5 | -0.272 | 0.01796 | 0.346 |
| 30214 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-582-3p | 12 | FOXO3 | Sponge network | 1.361 | 0 | -0.04 | 0.72028 | 0.346 |
| 30215 | CTD-2554C21.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-96-5p | 13 | UNC5C | Sponge network | -1.654 | 0.00144 | -0.178 | 0.40214 | 0.346 |
| 30216 | MAGI2-AS3 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-942-5p | 34 | OPCML | Sponge network | -0.129 | 0.57177 | -2.498 | 0 | 0.346 |
| 30217 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-378a-5p | 15 | PDS5B | Sponge network | 0.792 | 0.0049 | 0.259 | 0.00962 | 0.346 |
| 30218 | MEG3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-451a;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 38 | BCL2 | Sponge network | 0.249 | 0.35902 | -0.899 | 0 | 0.346 |
| 30219 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 13 | LRRC55 | Sponge network | 1.153 | 0.00114 | -0.026 | 0.93163 | 0.346 |
| 30220 | FAM66C |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 37 | NOTCH2 | Sponge network | -0.901 | 0.00019 | 0.138 | 0.35163 | 0.346 |
| 30221 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | SON | Sponge network | 1.163 | 0.00018 | 0.168 | 0.04406 | 0.346 |
| 30222 | RASSF8-AS1 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-629-3p | 10 | SPTBN4 | Sponge network | 0.335 | 0.20596 | -0.786 | 0.01281 | 0.346 |
| 30223 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | RAP1A | Sponge network | -1.057 | 0.00029 | -0.929 | 0 | 0.346 |
| 30224 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 27 | FAT3 | Sponge network | 1.757 | 0 | -1.601 | 0.00051 | 0.346 |
| 30225 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 22 | CDHR3 | Sponge network | -1.697 | 0.00889 | -0.977 | 4.0E-5 | 0.346 |
| 30226 | OIP5-AS1 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | 0.421 | 0.00084 | -0.366 | 0.01036 | 0.346 |
| 30227 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-590-3p | 16 | DDX5 | Sponge network | 0.083 | 0.66405 | -0.099 | 0.17428 | 0.346 |
| 30228 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-655-3p | 18 | SON | Sponge network | 0.594 | 0.41522 | 0.168 | 0.04406 | 0.346 |
| 30229 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SLC6A15 | Sponge network | -0.513 | 0.04703 | -2.373 | 2.0E-5 | 0.346 |
| 30230 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-502-5p;hsa-miR-590-3p | 14 | FOXO1 | Sponge network | -0.096 | 0.67958 | -0.141 | 0.33874 | 0.346 |
| 30231 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | AHNAK | Sponge network | -0.899 | 6.0E-5 | -1.207 | 0 | 0.346 |
| 30232 | RP11-401P9.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-539-5p | 10 | TSC1 | Sponge network | 1.524 | 0.1098 | 0.103 | 0.26071 | 0.346 |
| 30233 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p | 10 | PTPRB | Sponge network | -0.517 | 0.02935 | 0.105 | 0.47122 | 0.346 |
| 30234 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-7-5p | 13 | ATRX | Sponge network | -1.331 | 3.0E-5 | 0.261 | 0.04408 | 0.346 |
| 30235 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-942-5p | 18 | PCDH17 | Sponge network | -0.777 | 0.16667 | 0.731 | 2.0E-5 | 0.346 |
| 30236 | FENDRR |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PTCH1 | Sponge network | -1.281 | 0.00012 | -0.1 | 0.6179 | 0.346 |
| 30237 | FAM215B |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 10 | CEP170 | Sponge network | 0.817 | 3.0E-5 | 0.943 | 0 | 0.346 |
| 30238 | RP11-686D22.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p | 14 | RBMS3 | Sponge network | 0.793 | 5.0E-5 | -0.519 | 0.03551 | 0.346 |
| 30239 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-425-5p | 26 | ERC1 | Sponge network | 0.225 | 0.30644 | -0.108 | 0.38794 | 0.346 |
| 30240 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-24-3p | 10 | ARID5B | Sponge network | 1.969 | 0.00316 | 0.342 | 0.01676 | 0.346 |
| 30241 | BZRAP1-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-671-5p;hsa-miR-7-5p | 11 | EHD3 | Sponge network | -0.465 | 0.04703 | -0.484 | 0.01747 | 0.346 |
| 30242 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-93-5p;hsa-miR-942-5p | 12 | FBXO31 | Sponge network | 0.246 | 0.59912 | -0.085 | 0.42389 | 0.346 |
| 30243 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | AHNAK | Sponge network | 0.249 | 0.35902 | -1.207 | 0 | 0.346 |
| 30244 | RP11-6O2.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p | 17 | ERCC4 | Sponge network | 0.331 | 0.57247 | 0.126 | 0.15266 | 0.346 |
| 30245 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p | 18 | KIT | Sponge network | 0.176 | 0.37402 | -2.184 | 0 | 0.346 |
| 30246 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-874-3p | 10 | ST5 | Sponge network | -1.645 | 0 | -0.78 | 0 | 0.346 |
| 30247 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 32 | SASH1 | Sponge network | -0.563 | 0.02545 | -0.502 | 0.00175 | 0.346 |
| 30248 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 32 | MXI1 | Sponge network | -0.942 | 0 | -0.987 | 0 | 0.346 |
| 30249 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 13 | PRKAB2 | Sponge network | 1.162 | 5.0E-5 | -0.367 | 0.01371 | 0.346 |
| 30250 | MIR22HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 16 | CDH23 | Sponge network | -1.047 | 0 | -0.974 | 0.00034 | 0.346 |
| 30251 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-940 | 15 | GALNT13 | Sponge network | -0.738 | 0.21233 | -0.857 | 0.07439 | 0.346 |
| 30252 | LINC00641 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 13 | HMCN1 | Sponge network | -0.222 | 0.32445 | 0.809 | 0.00544 | 0.346 |
| 30253 | LINC00641 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-93-3p;hsa-miR-940 | 29 | TOM1L2 | Sponge network | -0.222 | 0.32445 | -0.994 | 0 | 0.346 |
| 30254 | RP11-403I13.8 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | DTNA | Sponge network | 0.619 | 0.00157 | -1.648 | 1.0E-5 | 0.346 |
| 30255 | RP11-403I13.8 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | PIK3CA | Sponge network | 0.619 | 0.00157 | 0.195 | 0.06908 | 0.346 |
| 30256 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | HDAC9 | Sponge network | -0.901 | 0.00019 | -0.755 | 0.00495 | 0.346 |
| 30257 | RP11-400K9.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | TET1 | Sponge network | -0.611 | 0.09607 | -0.135 | 0.52138 | 0.346 |
| 30258 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | HECA | Sponge network | 1.153 | 0 | -0.217 | 0.03463 | 0.346 |
| 30259 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-577 | 12 | AKAP12 | Sponge network | -0.465 | 0.04703 | -0.932 | 0.0028 | 0.346 |
| 30260 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 30 | FOXO3 | Sponge network | 0.083 | 0.66405 | -0.04 | 0.72028 | 0.346 |
| 30261 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429 | 17 | EYA4 | Sponge network | -1.654 | 0.00144 | 0.3 | 0.36876 | 0.346 |
| 30262 | CTA-941F9.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-576-5p | 10 | TRPS1 | Sponge network | -0.344 | 0.49624 | -0.191 | 0.44109 | 0.346 |
| 30263 | TRIM52-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-7-1-3p | 20 | ANK2 | Sponge network | -0.418 | 0.00068 | -1.603 | 1.0E-5 | 0.346 |
| 30264 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-7-1-3p | 18 | RARB | Sponge network | 0.4 | 0.14611 | -0.825 | 2.0E-5 | 0.346 |
| 30265 | AGAP11 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-5p | 11 | RTN4 | Sponge network | -1.645 | 0 | -0.412 | 0.00014 | 0.346 |
| 30266 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 14 | PDZD2 | Sponge network | 0.064 | 0.72934 | -0.909 | 3.0E-5 | 0.346 |
| 30267 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-766-3p | 31 | IL6ST | Sponge network | -0.517 | 0.02935 | -0.402 | 0.00676 | 0.346 |
| 30268 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-5p | 10 | LRRK2 | Sponge network | 0.395 | 0.15451 | -1.136 | 1.0E-5 | 0.346 |
| 30269 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-502-5p | 20 | IL17RD | Sponge network | -0.469 | 0.08721 | -0.019 | 0.94873 | 0.346 |
| 30270 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 38 | ETV1 | Sponge network | -1.697 | 0.00889 | -0.096 | 0.67674 | 0.346 |
| 30271 | HNRNPU-AS1 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 10 | ERCC4 | Sponge network | 1.601 | 0 | 0.126 | 0.15266 | 0.346 |
| 30272 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | CNTN1 | Sponge network | 0.619 | 0.00157 | -2.594 | 0 | 0.346 |
| 30273 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-98-5p | 13 | LRIG1 | Sponge network | -0.295 | 0.02604 | -0.537 | 0.00588 | 0.346 |
| 30274 | RP11-35G9.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | SETBP1 | Sponge network | 0.657 | 4.0E-5 | -1.302 | 1.0E-5 | 0.346 |
| 30275 | CTC-457L16.2 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p | 11 | ABL2 | Sponge network | 1.153 | 0.00114 | 0.82 | 0 | 0.346 |
| 30276 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | ZNF292 | Sponge network | 0.174 | 0.30034 | 0.276 | 0.04148 | 0.346 |
| 30277 | RP11-144G6.12 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | USP9X | Sponge network | 0.826 | 1.0E-5 | 0.222 | 0.01689 | 0.346 |
| 30278 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-93-3p | 18 | RPS6KA2 | Sponge network | 0.997 | 0.00066 | -0.57 | 0.00013 | 0.346 |
| 30279 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p | 29 | PPP1R9A | Sponge network | -2.69 | 0.0001 | -0.903 | 0.04254 | 0.346 |
| 30280 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 25 | PHC3 | Sponge network | -0.611 | 0.09607 | 0.212 | 0.0499 | 0.346 |
| 30281 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | ZFHX4 | Sponge network | 0.853 | 0.15352 | 0.018 | 0.95574 | 0.346 |
| 30282 | RP11-967K21.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-92a-3p | 32 | AFF1 | Sponge network | 0.056 | 0.81195 | -0.384 | 0.00053 | 0.346 |
| 30283 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p | 13 | MYCBP2 | Sponge network | 0.532 | 0.00505 | -0.027 | 0.82016 | 0.346 |
| 30284 | RP11-216B9.6 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 16 | GLIPR1 | Sponge network | 0.174 | 0.30034 | 0.146 | 0.3276 | 0.346 |
| 30285 | BCYRN1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | ACVR2A | Sponge network | 0.311 | 0.10344 | -0.409 | 0.00039 | 0.346 |
| 30286 | AC090587.5 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-589-3p | 10 | AFF1 | Sponge network | -0.088 | 0.65011 | -0.384 | 0.00053 | 0.346 |
| 30287 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-511-5p;hsa-miR-582-5p | 13 | ZFHX3 | Sponge network | 1.093 | 0 | 0.389 | 0.01363 | 0.346 |
| 30288 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 34 | FOXP1 | Sponge network | -0.227 | 0.31657 | 0.042 | 0.74368 | 0.346 |
| 30289 | C4A-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-486-5p | 11 | NRK | Sponge network | 3.345 | 0 | 0.656 | 0.19217 | 0.346 |
| 30290 | A2M-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | GALNT13 | Sponge network | -0.08 | 0.90092 | -0.857 | 0.07439 | 0.346 |
| 30291 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 16 | PIK3C2A | Sponge network | 0.178 | 0.29106 | 0.061 | 0.53776 | 0.346 |
| 30292 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | CDH10 | Sponge network | -1.216 | 0 | -2.397 | 0 | 0.346 |
| 30293 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | NEDD4 | Sponge network | 0.311 | 0.10344 | 0.02 | 0.89834 | 0.346 |
| 30294 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | HECA | Sponge network | 0.225 | 0.30644 | -0.217 | 0.03463 | 0.346 |
| 30295 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-590-3p | 14 | SMARCA2 | Sponge network | -0.896 | 0.00099 | -0.529 | 0.00014 | 0.346 |
| 30296 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NR4A3 | Sponge network | -1.161 | 0.00591 | -1.385 | 0 | 0.346 |
| 30297 | BVES-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p;hsa-miR-493-5p | 24 | NCOA1 | Sponge network | -2.62 | 0 | -0.432 | 0 | 0.346 |
| 30298 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | LSAMP | Sponge network | 0.082 | 0.55654 | -0.066 | 0.81708 | 0.346 |
| 30299 | RP11-20I23.11 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | ZFHX3 | Sponge network | 0.875 | 0.00019 | 0.389 | 0.01363 | 0.346 |
| 30300 | AC108488.4 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p | 10 | PTGFR | Sponge network | 0.259 | 0.12542 | -0.233 | 0.45421 | 0.346 |
| 30301 | LINC00702 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 15 | ACTB | Sponge network | -0.737 | 0.06182 | -0.247 | 0.01736 | 0.346 |
| 30302 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p | 44 | FAT3 | Sponge network | -0.465 | 0.04703 | -1.601 | 0.00051 | 0.346 |
| 30303 | RP11-473I1.10 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p | 10 | ABL1 | Sponge network | 0.673 | 0 | -0.215 | 0.07804 | 0.346 |
| 30304 | RP11-13K12.1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-628-5p | 23 | RIMS2 | Sponge network | -0.154 | 0.78939 | -1.365 | 0.00208 | 0.346 |
| 30305 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-5p | 54 | TCF4 | Sponge network | 0.494 | 0.00339 | 0.016 | 0.92271 | 0.346 |
| 30306 | LINC00403 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-424-5p | 19 | AR | Sponge network | -3.586 | 0.00032 | -1.554 | 0 | 0.346 |
| 30307 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-625-5p | 14 | LHX6 | Sponge network | -0.487 | 0.02157 | -0.087 | 0.69193 | 0.346 |
| 30308 | RP11-597D13.8 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-26b-5p;hsa-miR-27a-3p | 11 | ZFHX4 | Sponge network | 0.936 | 0.03889 | 0.018 | 0.95574 | 0.346 |
| 30309 | LINC00472 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-335-3p | 11 | ELMO1 | Sponge network | -0.842 | 0.01361 | 0.04 | 0.83147 | 0.346 |
| 30310 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935 | 42 | PTPRD | Sponge network | -0.513 | 0.04703 | -1.005 | 0.00064 | 0.346 |
| 30311 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-582-5p | 13 | BMPR2 | Sponge network | 0.757 | 0 | 0.206 | 0.0635 | 0.346 |
| 30312 | FAM157B |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p | 15 | BRD3 | Sponge network | 0.92 | 0 | 0.37 | 0.00013 | 0.346 |
| 30313 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | AKAP11 | Sponge network | -0.176 | 0.59692 | 0.196 | 0.09825 | 0.346 |
| 30314 | RP11-254F7.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | LUZP2 | Sponge network | 0.176 | 0.37402 | -0.056 | 0.89416 | 0.346 |
| 30315 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p | 18 | CREBBP | Sponge network | -1.575 | 6.0E-5 | 0.126 | 0.15186 | 0.346 |
| 30316 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p | 13 | FGFR1 | Sponge network | 0.331 | 0.57247 | -0.509 | 0.03957 | 0.346 |
| 30317 | RP1-59D14.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-326 | 12 | USP6 | Sponge network | 1.162 | 5.0E-5 | 0.188 | 0.3871 | 0.346 |
| 30318 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-93-5p | 30 | ZBTB4 | Sponge network | -0.969 | 2.0E-5 | -0.739 | 0 | 0.346 |
| 30319 | RASSF8-AS1 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | CSDE1 | Sponge network | 0.335 | 0.20596 | -0.493 | 0 | 0.346 |
| 30320 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-589-3p | 12 | RELN | Sponge network | -0.318 | 0.65847 | -2.403 | 0 | 0.346 |
| 30321 | RP11-35G9.3 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 14 | PTGFR | Sponge network | 0.657 | 4.0E-5 | -0.233 | 0.45421 | 0.346 |
| 30322 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TBX18 | Sponge network | 0.619 | 0.00157 | 0.843 | 0.02945 | 0.346 |
| 30323 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 40 | AKAP11 | Sponge network | -0.129 | 0.57177 | 0.196 | 0.09825 | 0.346 |
| 30324 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-744-3p | 16 | KALRN | Sponge network | -0.793 | 7.0E-5 | -0.819 | 1.0E-5 | 0.346 |
| 30325 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ITGAV | Sponge network | 0.194 | 0.64278 | 0.337 | 0.01002 | 0.346 |
| 30326 | NR2F1-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 19 | CELF4 | Sponge network | -0.49 | 0.04946 | -1.135 | 0.00344 | 0.346 |
| 30327 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | NEDD4 | Sponge network | 0.603 | 0.00127 | 0.02 | 0.89834 | 0.346 |
| 30328 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-5p | 24 | MOB1B | Sponge network | 0.214 | 0.18246 | 0.197 | 0.15266 | 0.346 |
| 30329 | RP11-1246C19.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-625-5p | 14 | ABI2 | Sponge network | 1.352 | 0 | 0.175 | 0.14787 | 0.346 |
| 30330 | RP11-967K21.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-5p | 21 | RYR2 | Sponge network | 0.056 | 0.81195 | -1.466 | 0.00031 | 0.346 |
| 30331 | AC012360.6 |
hsa-miR-106a-5p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | GNAQ | Sponge network | 0.624 | 0.02176 | -0.367 | 0.00102 | 0.346 |
| 30332 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 48 | CUX1 | Sponge network | 0.494 | 0.00339 | -0.039 | 0.6705 | 0.346 |
| 30333 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-425-5p | 19 | CBL | Sponge network | 0.082 | 0.55397 | 0.291 | 0.01267 | 0.346 |
| 30334 | RP11-130L8.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p | 12 | SYNM | Sponge network | -0.663 | 0.10887 | -2.309 | 1.0E-5 | 0.346 |
| 30335 | RP11-226L15.5 |
hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | SHPRH | Sponge network | 0.8 | 0 | 0.098 | 0.40994 | 0.346 |
| 30336 | FENDRR |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 30 | CTIF | Sponge network | -1.281 | 0.00012 | -0.389 | 0.00549 | 0.345 |
| 30337 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | TCF4 | Sponge network | 0.959 | 0 | 0.016 | 0.92271 | 0.345 |
| 30338 | ERVH48-1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | ABI2 | Sponge network | 1.04 | 0.00023 | 0.175 | 0.14787 | 0.345 |
| 30339 | OSER1-AS1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | PLSCR4 | Sponge network | -0.13 | 0.48734 | -0.474 | 0.03833 | 0.345 |
| 30340 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | NRXN3 | Sponge network | 0.222 | 0.29133 | -0.893 | 0.04186 | 0.345 |
| 30341 | RP5-1074L1.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 20 | CCDC6 | Sponge network | 1.923 | 0 | 0.27 | 0.02555 | 0.345 |
| 30342 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | USP12 | Sponge network | 1.153 | 0 | -0.102 | 0.40768 | 0.345 |
| 30343 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-93-5p | 18 | LRRC55 | Sponge network | 0.199 | 0.38015 | -0.026 | 0.93163 | 0.345 |
| 30344 | LINC00667 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p | 32 | DMD | Sponge network | -0.235 | 0.15142 | -1.506 | 0 | 0.345 |
| 30345 | BDNF-AS |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 10 | MAPK10 | Sponge network | -0.668 | 3.0E-5 | -1.774 | 0 | 0.345 |
| 30346 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 30 | GPC6 | Sponge network | -0.942 | 0 | -0.054 | 0.81465 | 0.345 |
| 30347 | FAM66C |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | RTN4 | Sponge network | -0.901 | 0.00019 | -0.412 | 0.00014 | 0.345 |
| 30348 | RP5-994D16.9 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | WDFY3 | Sponge network | 0.252 | 0.08508 | -0.201 | 0.0967 | 0.345 |
| 30349 | LINC00672 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-503-5p | 16 | ABL1 | Sponge network | -0.227 | 0.31657 | -0.215 | 0.07804 | 0.345 |
| 30350 | TRAM2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p | 21 | ABI2 | Sponge network | 0.336 | 0.00739 | 0.175 | 0.14787 | 0.345 |
| 30351 | ZSCAN16-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | CADM2 | Sponge network | -0.342 | 0.02288 | -3.782 | 0 | 0.345 |
| 30352 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-940 | 33 | NTRK2 | Sponge network | 0.331 | 0.57247 | -0.736 | 0.06495 | 0.345 |
| 30353 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p | 51 | KIF1B | Sponge network | 0.401 | 0.00066 | -0.118 | 0.32258 | 0.345 |
| 30354 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 42 | UNC80 | Sponge network | -0.487 | 0.02157 | -1.934 | 0 | 0.345 |
| 30355 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-590-3p | 11 | RBM5 | Sponge network | 0.494 | 0.00339 | -0.205 | 0.0129 | 0.345 |
| 30356 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p | 23 | AKAP11 | Sponge network | 0.569 | 0.00067 | 0.196 | 0.09825 | 0.345 |
| 30357 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-590-3p | 11 | CHD1 | Sponge network | -0.031 | 0.89063 | 0.322 | 0.00215 | 0.345 |
| 30358 | CTC-490E21.10 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p | 11 | CCDC6 | Sponge network | 1.46 | 1.0E-5 | 0.27 | 0.02555 | 0.345 |
| 30359 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 35 | NOTCH2 | Sponge network | 0.101 | 0.60919 | 0.138 | 0.35163 | 0.345 |
| 30360 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | TCF12 | Sponge network | 0.683 | 1.0E-5 | 0.174 | 0.13271 | 0.345 |
| 30361 | KB-1572G7.2 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | ERCC6 | Sponge network | 0.924 | 0 | 0.23 | 0.015 | 0.345 |
| 30362 | RP11-395A13.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p | 17 | IL17RD | Sponge network | 0.057 | 0.61225 | -0.019 | 0.94873 | 0.345 |
| 30363 | LINC00648 |
hsa-miR-140-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 10 | BCL9 | Sponge network | 0.633 | 0.51678 | 0.321 | 0.00267 | 0.345 |
| 30364 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-942-5p | 38 | NOTCH2 | Sponge network | -1.439 | 4.0E-5 | 0.138 | 0.35163 | 0.345 |
| 30365 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-33a-3p | 10 | RARB | Sponge network | -0.693 | 0.05629 | -0.825 | 2.0E-5 | 0.345 |
| 30366 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 17 | CDC73 | Sponge network | 1.317 | 0 | 0.094 | 0.20463 | 0.345 |
| 30367 | RP11-384O8.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-92a-3p | 27 | IGFBP5 | Sponge network | -0.738 | 0.21233 | -0.806 | 0.00105 | 0.345 |
| 30368 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 20 | SCN9A | Sponge network | 0.657 | 4.0E-5 | -0.525 | 0.10593 | 0.345 |
| 30369 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 16 | PKNOX1 | Sponge network | 0.194 | 0.64278 | 0.085 | 0.28917 | 0.345 |
| 30370 | CTC-429P9.5 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 20 | AFF1 | Sponge network | 0.178 | 0.29106 | -0.384 | 0.00053 | 0.345 |
| 30371 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-654-3p | 12 | MYBL1 | Sponge network | 1.157 | 0.00455 | 0.494 | 0.02152 | 0.345 |
| 30372 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DACT1 | Sponge network | -0.08 | 0.90092 | 0.783 | 0.00072 | 0.345 |
| 30373 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-940 | 22 | MCC | Sponge network | 0.622 | 0.00015 | -1.005 | 0 | 0.345 |
| 30374 | CTC-490E21.10 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p | 13 | AFF1 | Sponge network | 1.46 | 1.0E-5 | -0.384 | 0.00053 | 0.345 |
| 30375 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-744-3p | 17 | HIP1 | Sponge network | 0.603 | 0.00127 | 0.774 | 0 | 0.345 |
| 30376 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-197-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-628-5p | 19 | TCF4 | Sponge network | -1.116 | 0.23686 | 0.016 | 0.92271 | 0.345 |
| 30377 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 15 | TP53INP1 | Sponge network | -1.057 | 0.00029 | -0.496 | 0.00064 | 0.345 |
| 30378 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-93-5p | 13 | TRIM3 | Sponge network | -1.697 | 0.00889 | -0.577 | 0 | 0.345 |
| 30379 | RP11-497H16.9 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-218-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-653-5p | 12 | UBR3 | Sponge network | 0.545 | 0.00064 | -0.021 | 0.82774 | 0.345 |
| 30380 | LINC00476 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-542-3p | 11 | PRKD1 | Sponge network | -0.615 | 0.00015 | -0.466 | 0.03583 | 0.345 |
| 30381 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 37 | GAS7 | Sponge network | -0.405 | 0.22013 | -0.555 | 0.01602 | 0.345 |
| 30382 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-628-5p | 13 | EPHA3 | Sponge network | 0.792 | 0.0049 | 0.185 | 0.53588 | 0.345 |
| 30383 | AC025171.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 0.998 | 0 | 0.51 | 0 | 0.345 |
| 30384 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | EPAS1 | Sponge network | -0.777 | 0.1134 | -0.184 | 0.14418 | 0.345 |
| 30385 | ACTA2-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 25 | CTIF | Sponge network | 0.194 | 0.64278 | -0.389 | 0.00549 | 0.345 |
| 30386 | RP11-282O18.3 |
hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p | 10 | DICER1 | Sponge network | 0.757 | 0 | 0.042 | 0.674 | 0.345 |
| 30387 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 19 | CSMD1 | Sponge network | -0.246 | 0.35034 | -0.63 | 0.18881 | 0.345 |
| 30388 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | TACC1 | Sponge network | 0.603 | 0.00127 | -0.362 | 0.05406 | 0.345 |
| 30389 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | NEDD4 | Sponge network | -0.303 | 0.21002 | 0.02 | 0.89834 | 0.345 |
| 30390 | AC099850.1 |
hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 11 | MLLT10 | Sponge network | 0.803 | 0 | 0.105 | 0.33292 | 0.345 |
| 30391 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-590-3p | 11 | JAKMIP2 | Sponge network | 0.225 | 0.30644 | -1.388 | 0 | 0.345 |
| 30392 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p | 14 | ATRX | Sponge network | 1.206 | 0 | 0.261 | 0.04408 | 0.345 |
| 30393 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-93-3p | 14 | SUFU | Sponge network | -0.154 | 0.53954 | -0.04 | 0.65489 | 0.345 |
| 30394 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | HECA | Sponge network | 0.997 | 0.00066 | -0.217 | 0.03463 | 0.345 |
| 30395 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p | 12 | DKK3 | Sponge network | 1.757 | 0 | -0.052 | 0.77783 | 0.345 |
| 30396 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-7-1-3p | 17 | GNAQ | Sponge network | -0.285 | 0.2172 | -0.367 | 0.00102 | 0.345 |
| 30397 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-550a-3p | 13 | ITGAV | Sponge network | 0.331 | 0.57247 | 0.337 | 0.01002 | 0.345 |
| 30398 | FAM66C |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 17 | ITGA5 | Sponge network | -0.901 | 0.00019 | 0.452 | 0.03524 | 0.345 |
| 30399 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 33 | FOXP1 | Sponge network | -0.031 | 0.89063 | 0.042 | 0.74368 | 0.345 |
| 30400 | LINC00865 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p | 12 | ABCA10 | Sponge network | -1.249 | 7.0E-5 | -0.571 | 0.03674 | 0.345 |
| 30401 | AC108488.4 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-577 | 13 | GREM1 | Sponge network | 0.259 | 0.12542 | 0.902 | 0.01691 | 0.345 |
| 30402 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 32 | TGFBR3 | Sponge network | 0.532 | 0.00505 | -1.173 | 1.0E-5 | 0.345 |
| 30403 | RP11-728F11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 27 | RUNX1T1 | Sponge network | 0.238 | 0.70544 | -0.344 | 0.2374 | 0.345 |
| 30404 | MIR22HG |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 12 | RHOB | Sponge network | -1.047 | 0 | -1.052 | 0 | 0.345 |
| 30405 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p | 15 | MOB1B | Sponge network | 0.93 | 0 | 0.197 | 0.15266 | 0.345 |
| 30406 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | NEDD4 | Sponge network | -0.031 | 0.89063 | 0.02 | 0.89834 | 0.345 |
| 30407 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 17 | AKAP12 | Sponge network | 1.302 | 7.0E-5 | -0.932 | 0.0028 | 0.345 |
| 30408 | GHRLOS |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-99b-3p | 14 | GRIK3 | Sponge network | -1.942 | 0 | -3.208 | 0 | 0.345 |
| 30409 | RP11-440L14.1 |
hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-150-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-625-5p | 15 | MDM4 | Sponge network | 0.599 | 0.0001 | 0.345 | 0.00762 | 0.345 |
| 30410 | RP11-157J24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-576-5p | 16 | PBX1 | Sponge network | -1.664 | 0.05165 | -1.206 | 0 | 0.345 |
| 30411 | LINC00173 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | TRPS1 | Sponge network | 0.056 | 0.8494 | -0.191 | 0.44109 | 0.345 |
| 30412 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-877-5p;hsa-miR-942-5p | 16 | GLIPR1 | Sponge network | -0.777 | 0.16667 | 0.146 | 0.3276 | 0.345 |
| 30413 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-96-5p | 12 | FNBP1 | Sponge network | 0.083 | 0.66405 | -0.969 | 4.0E-5 | 0.345 |
| 30414 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-450b-5p | 10 | NAV3 | Sponge network | 0.912 | 0.00014 | 0.212 | 0.47729 | 0.345 |
| 30415 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 17 | GPAM | Sponge network | 0.537 | 0.00139 | 0.142 | 0.29732 | 0.345 |
| 30416 | RP11-166D19.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | BTG2 | Sponge network | -0.576 | 0.10121 | -1.763 | 0 | 0.345 |
| 30417 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p | 14 | UBE4B | Sponge network | 0.082 | 0.55397 | -0.235 | 0.007 | 0.345 |
| 30418 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p | 26 | FBXO32 | Sponge network | 0.051 | 0.83388 | -0.495 | 0.07561 | 0.345 |
| 30419 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | PRKAA2 | Sponge network | 0.082 | 0.55654 | -2.462 | 0 | 0.345 |
| 30420 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 37 | MAF | Sponge network | 0.335 | 0.20596 | -1.257 | 0 | 0.345 |
| 30421 | RP11-35G9.3 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | TAL1 | Sponge network | 0.657 | 4.0E-5 | -0.462 | 0.00574 | 0.345 |
| 30422 | HOXB-AS2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p | 16 | CBL | Sponge network | 1.316 | 0.01106 | 0.291 | 0.01267 | 0.345 |
| 30423 | AC108488.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 12 | NCAM2 | Sponge network | 0.259 | 0.12542 | -0.482 | 0.22565 | 0.345 |
| 30424 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 56 | THRB | Sponge network | -0.487 | 0.02157 | -1.117 | 0.0004 | 0.345 |
| 30425 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 34 | HLF | Sponge network | 0.249 | 0.35902 | -2.443 | 0 | 0.345 |
| 30426 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-625-5p | 19 | YAP1 | Sponge network | 0.078 | 0.55803 | 0.22 | 0.11836 | 0.345 |
| 30427 | LINC00854 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 11 | NAV3 | Sponge network | 1.18 | 0 | 0.212 | 0.47729 | 0.345 |
| 30428 | CTC-297N7.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-590-5p | 12 | RSPO2 | Sponge network | -2.069 | 0 | -3.038 | 0 | 0.345 |
| 30429 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 11 | ZNF770 | Sponge network | 1.4 | 0 | 0.206 | 0.06751 | 0.345 |
| 30430 | CTC-429P9.5 |
hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | PIK3CA | Sponge network | 0.178 | 0.29106 | 0.195 | 0.06908 | 0.345 |
| 30431 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-93-3p | 11 | SSBP2 | Sponge network | -1.717 | 0.01522 | -1.58 | 0 | 0.345 |
| 30432 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-940 | 35 | NFIB | Sponge network | 0.082 | 0.55397 | 0.023 | 0.90795 | 0.345 |
| 30433 | FAM157B |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 10 | PHF6 | Sponge network | 0.92 | 0 | 0.785 | 0 | 0.345 |
| 30434 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PTGFR | Sponge network | 1.757 | 0 | -0.233 | 0.45421 | 0.345 |
| 30435 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-423-3p;hsa-miR-454-3p;hsa-miR-486-5p | 12 | ROBO1 | Sponge network | 0.853 | 0.15352 | -0.009 | 0.96154 | 0.345 |
| 30436 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 20 | UNC5C | Sponge network | -1.111 | 0.0003 | -0.178 | 0.40214 | 0.345 |
| 30437 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 28 | CXXC4 | Sponge network | -2.452 | 0 | -0.433 | 0.21055 | 0.345 |
| 30438 | CTC-487M23.5 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-450b-5p | 11 | ERCC4 | Sponge network | 0.912 | 0.00014 | 0.126 | 0.15266 | 0.345 |
| 30439 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-7-5p;hsa-miR-93-5p | 19 | ZBTB4 | Sponge network | 0.056 | 0.8494 | -0.739 | 0 | 0.345 |
| 30440 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | PHC3 | Sponge network | -1.697 | 0.00889 | 0.212 | 0.0499 | 0.345 |
| 30441 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-7-5p | 15 | CRTC3 | Sponge network | 0.051 | 0.83388 | 0.164 | 0.16786 | 0.345 |
| 30442 | AP001347.6 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p | 13 | ITGA5 | Sponge network | -1.161 | 0.00591 | 0.452 | 0.03524 | 0.345 |
| 30443 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | SSBP2 | Sponge network | -0.351 | 0.11358 | -1.58 | 0 | 0.345 |
| 30444 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | NEDD4 | Sponge network | -1.439 | 4.0E-5 | 0.02 | 0.89834 | 0.345 |
| 30445 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 28 | PBX1 | Sponge network | 0.222 | 0.29133 | -1.206 | 0 | 0.345 |
| 30446 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | FOXO3 | Sponge network | 0.813 | 1.0E-5 | -0.04 | 0.72028 | 0.345 |
| 30447 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p | 19 | APC | Sponge network | 0.331 | 0.57247 | -0.255 | 0.04051 | 0.345 |
| 30448 | RP11-283I3.6 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | 0.614 | 1.0E-5 | 0.524 | 0.00017 | 0.345 |
| 30449 | TRIM52-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421 | 10 | DYNC1I1 | Sponge network | -0.418 | 0.00068 | -1.12 | 0.00107 | 0.345 |
| 30450 | LINC00969 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 18 | NF1 | Sponge network | 0.683 | 1.0E-5 | 0.51 | 0 | 0.345 |
| 30451 | PART1 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | HOMER2 | Sponge network | -2.925 | 0 | -2.698 | 0 | 0.345 |
| 30452 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 12 | PLA2R1 | Sponge network | -1.697 | 0.00889 | -0.265 | 0.24676 | 0.345 |
| 30453 | LINC00654 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p | 16 | IGF2 | Sponge network | 0.374 | 0.20054 | 0.848 | 0.03842 | 0.345 |
| 30454 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 30 | ADARB1 | Sponge network | 0.659 | 3.0E-5 | -0.335 | 0.06631 | 0.345 |
| 30455 | LINC00621 |
hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | EYA4 | Sponge network | 0.131 | 0.8436 | 0.3 | 0.36876 | 0.345 |
| 30456 | RP11-384O8.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 24 | ASTN1 | Sponge network | -0.738 | 0.21233 | -2.389 | 2.0E-5 | 0.345 |
| 30457 | NR2F1-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | BTG2 | Sponge network | -0.49 | 0.04946 | -1.763 | 0 | 0.345 |
| 30458 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-93-5p | 29 | JAZF1 | Sponge network | 0.385 | 0.10067 | -0.377 | 0.02677 | 0.345 |
| 30459 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 16 | GPAM | Sponge network | -0.222 | 0.32445 | 0.142 | 0.29732 | 0.345 |
| 30460 | RP11-59H7.3 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-576-5p | 15 | UBR3 | Sponge network | 2.163 | 0 | -0.021 | 0.82774 | 0.345 |
| 30461 | LINC00863 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-342-3p;hsa-miR-423-5p | 16 | ABL1 | Sponge network | 0.189 | 0.13981 | -0.215 | 0.07804 | 0.345 |
| 30462 | FAM157B |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-3614-5p | 10 | PHC3 | Sponge network | 0.92 | 0 | 0.212 | 0.0499 | 0.345 |
| 30463 | LINC00094 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 34 | ERC1 | Sponge network | 0.253 | 0.09565 | -0.108 | 0.38794 | 0.345 |
| 30464 | H1FX-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PRKCB | Sponge network | -0.206 | 0.12942 | -1.313 | 2.0E-5 | 0.345 |
| 30465 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-5p | 16 | SPG20 | Sponge network | -0.384 | 0.40652 | -1.16 | 0 | 0.345 |
| 30466 | BVES-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-935 | 20 | ACSL6 | Sponge network | -2.62 | 0 | -1.593 | 0 | 0.345 |
| 30467 | FAM13A-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 16 | PRUNE2 | Sponge network | -0.83 | 0 | -1.733 | 0.00022 | 0.345 |
| 30468 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 11 | FAM172A | Sponge network | -0.668 | 3.0E-5 | -0.57 | 0 | 0.345 |
| 30469 | RP11-360F5.3 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 11 | SNX29 | Sponge network | 1.867 | 0 | -0.34 | 0.01371 | 0.345 |
| 30470 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-532-3p | 11 | CDH23 | Sponge network | 0.225 | 0.30644 | -0.974 | 0.00034 | 0.345 |
| 30471 | LINC00672 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-98-5p | 23 | KCNA6 | Sponge network | -0.227 | 0.31657 | -1.531 | 0.00013 | 0.345 |
| 30472 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MLLT10 | Sponge network | 0.91 | 0 | 0.105 | 0.33292 | 0.345 |
| 30473 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 18 | LIFR | Sponge network | -0.811 | 2.0E-5 | -1.794 | 0 | 0.345 |
| 30474 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-654-3p | 21 | NR2C2 | Sponge network | -0.709 | 0.18168 | 0.21 | 0.03224 | 0.345 |
| 30475 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p | 15 | AKAP13 | Sponge network | -0.18 | 0.20343 | 0.028 | 0.7729 | 0.345 |
| 30476 | JAZF1-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | PPM1L | Sponge network | -0.769 | 0.00035 | -0.537 | 0.00616 | 0.345 |
| 30477 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | PGR | Sponge network | 0.381 | 0.25967 | -1.704 | 0 | 0.345 |
| 30478 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PTPRB | Sponge network | 0.335 | 0.20596 | 0.105 | 0.47122 | 0.345 |
| 30479 | ZNF32-AS1 |
hsa-let-7g-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-576-5p | 10 | RICTOR | Sponge network | 2.413 | 1.0E-5 | 0.388 | 0.00188 | 0.345 |
| 30480 | RP11-672A2.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p | 13 | TACC1 | Sponge network | 1.344 | 0 | -0.362 | 0.05406 | 0.345 |
| 30481 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | TMEM132B | Sponge network | -1.886 | 0 | -0.83 | 0.01084 | 0.345 |
| 30482 | CTA-204B4.2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-590-3p | 12 | PIK3CA | Sponge network | 0.765 | 7.0E-5 | 0.195 | 0.06908 | 0.345 |
| 30483 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | RET | Sponge network | -0.811 | 2.0E-5 | -0.833 | 0.03517 | 0.345 |
| 30484 | ACTA2-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 23 | SIRT1 | Sponge network | 0.194 | 0.64278 | 0.109 | 0.2593 | 0.345 |
| 30485 | RP11-758N13.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p;hsa-miR-33a-5p;hsa-miR-424-5p | 14 | ABL2 | Sponge network | 2.939 | 3.0E-5 | 0.82 | 0 | 0.345 |
| 30486 | PCA3 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 13 | ID4 | Sponge network | -0.709 | 0.18168 | -1.644 | 0 | 0.345 |
| 30487 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CLIP1 | Sponge network | 0.225 | 0.30644 | -0.215 | 0.08684 | 0.345 |
| 30488 | RP11-326G21.1 |
hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | IGSF10 | Sponge network | -0.021 | 0.93598 | -1.751 | 1.0E-5 | 0.345 |
| 30489 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | NOTCH2NL | Sponge network | 0.705 | 0.00014 | 0.024 | 0.84307 | 0.345 |
| 30490 | ZNF582-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | PCDH18 | Sponge network | -1.349 | 0.00017 | 0.216 | 0.24645 | 0.345 |
| 30491 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | LRRC7 | Sponge network | -0.053 | 0.80685 | -1.341 | 0.01193 | 0.345 |
| 30492 | HAND2-AS1 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 14 | ACTB | Sponge network | -1.697 | 0.00889 | -0.247 | 0.01736 | 0.345 |
| 30493 | SERHL |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-421 | 12 | AKAP6 | Sponge network | -0.602 | 0.00331 | -1.586 | 3.0E-5 | 0.345 |
| 30494 | CTD-2314G24.2 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CTGF | Sponge network | 0.246 | 0.59912 | 0.321 | 0.11682 | 0.345 |
| 30495 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 10 | ZMYM2 | Sponge network | 2.368 | 2.0E-5 | 0.279 | 0.01273 | 0.345 |
| 30496 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 40 | UNC80 | Sponge network | -0.096 | 0.67958 | -1.934 | 0 | 0.345 |
| 30497 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 12 | TBL1XR1 | Sponge network | 0.545 | 0.00064 | 0.578 | 0 | 0.345 |
| 30498 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p | 15 | ABCB5 | Sponge network | -0.421 | 0.0114 | -0.956 | 0.04077 | 0.345 |
| 30499 | HAR1A |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-93-3p | 11 | EPHA3 | Sponge network | -0.672 | 0.19447 | 0.185 | 0.53588 | 0.345 |
| 30500 | ADAMTS9-AS2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 21 | CELF4 | Sponge network | -2.452 | 0 | -1.135 | 0.00344 | 0.345 |
| 30501 | TINCR |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-671-5p;hsa-miR-7-5p | 11 | EHD3 | Sponge network | -0.664 | 0.14543 | -0.484 | 0.01747 | 0.345 |
| 30502 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 15 | BHLHE41 | Sponge network | 0.998 | 0 | 0.353 | 0.12753 | 0.345 |
| 30503 | LINC00621 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 11 | MYO9A | Sponge network | 0.131 | 0.8436 | -0.147 | 0.32373 | 0.345 |
| 30504 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 24 | RSPO2 | Sponge network | 0.249 | 0.35902 | -3.038 | 0 | 0.345 |
| 30505 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-93-5p | 38 | PDGFRA | Sponge network | -0.154 | 0.78939 | -0.372 | 0.0037 | 0.345 |
| 30506 | RP4-669H2.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c | 16 | BRD3 | Sponge network | 1.711 | 0.00019 | 0.37 | 0.00013 | 0.345 |
| 30507 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p | 31 | HBP1 | Sponge network | -0.899 | 6.0E-5 | -0.454 | 0 | 0.345 |
| 30508 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 13 | EYA4 | Sponge network | 0.419 | 0.0319 | 0.3 | 0.36876 | 0.345 |
| 30509 | CTC-471J1.2 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-766-3p | 10 | ATM | Sponge network | 1.11 | 1.0E-5 | 0.178 | 0.21568 | 0.345 |
| 30510 | RP1-302G2.5 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-7-5p | 14 | TCF4 | Sponge network | -1.483 | 9.0E-5 | 0.016 | 0.92271 | 0.345 |
| 30511 | AC156455.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-502-5p | 17 | BNC2 | Sponge network | 1.154 | 0.00041 | -0.491 | 0.09988 | 0.345 |
| 30512 | AC108488.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-577 | 19 | SYNPO2 | Sponge network | 0.259 | 0.12542 | -2.342 | 0 | 0.345 |
| 30513 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-150-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 11 | FOXO3 | Sponge network | 1.597 | 0 | -0.04 | 0.72028 | 0.345 |
| 30514 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-942-5p | 19 | PCDH17 | Sponge network | 1.757 | 0 | 0.731 | 2.0E-5 | 0.345 |
| 30515 | C4A-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-486-5p;hsa-miR-96-5p | 14 | PRRX1 | Sponge network | 3.345 | 0 | 1.316 | 0 | 0.345 |
| 30516 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p | 19 | MYBL1 | Sponge network | 0.569 | 0.00067 | 0.494 | 0.02152 | 0.345 |
| 30517 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-539-5p;hsa-miR-582-3p | 15 | CLASP2 | Sponge network | 0.858 | 3.0E-5 | -0.038 | 0.71 | 0.345 |
| 30518 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 38 | FZD3 | Sponge network | 0.225 | 0.30644 | 0.104 | 0.60316 | 0.345 |
| 30519 | EMX2OS |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 21 | GLIPR1 | Sponge network | -0.777 | 0.1134 | 0.146 | 0.3276 | 0.345 |
| 30520 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | PEG3 | Sponge network | 0.222 | 0.29133 | -1.74 | 0 | 0.345 |
| 30521 | FAM13A-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 17 | ABCA10 | Sponge network | -0.83 | 0 | -0.571 | 0.03674 | 0.345 |
| 30522 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-590-3p | 23 | PRKCE | Sponge network | -0.421 | 0.0114 | -0.403 | 0.00056 | 0.345 |
| 30523 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-590-3p | 23 | HBP1 | Sponge network | -1.161 | 0.00591 | -0.454 | 0 | 0.345 |
| 30524 | RP11-458D21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 23 | CREB1 | Sponge network | 0.663 | 0.00073 | 0.203 | 0.00537 | 0.345 |
| 30525 | ERVH48-1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 11 | TSC1 | Sponge network | 1.04 | 0.00023 | 0.103 | 0.26071 | 0.345 |
| 30526 | AGAP11 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-455-5p | 12 | MACF1 | Sponge network | -1.645 | 0 | 0.019 | 0.90335 | 0.345 |
| 30527 | NR2F1-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 31 | OPCML | Sponge network | -0.49 | 0.04946 | -2.498 | 0 | 0.345 |
| 30528 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 22 | GRIN2A | Sponge network | 0.41 | 0.02214 | -2.161 | 0 | 0.345 |
| 30529 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p | 21 | PRDM2 | Sponge network | 0.335 | 0.20596 | -0.258 | 0.00721 | 0.345 |
| 30530 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | YAP1 | Sponge network | 0.194 | 0.64278 | 0.22 | 0.11836 | 0.345 |
| 30531 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | HDAC9 | Sponge network | -0.737 | 0.06182 | -0.755 | 0.00495 | 0.345 |
| 30532 | FENDRR |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | IL17RD | Sponge network | -1.281 | 0.00012 | -0.019 | 0.94873 | 0.345 |
| 30533 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p | 25 | ZFHX3 | Sponge network | 1.11 | 0 | 0.389 | 0.01363 | 0.345 |
| 30534 | RP11-53O19.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p | 13 | ROBO1 | Sponge network | -0.405 | 0.22013 | -0.009 | 0.96154 | 0.345 |
| 30535 | LIPE-AS1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-7-5p;hsa-miR-93-3p | 24 | ERC1 | Sponge network | 0.078 | 0.55803 | -0.108 | 0.38794 | 0.345 |
| 30536 | SNX29P2 |
hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 13 | GLI3 | Sponge network | 0.4 | 0.14611 | -0.615 | 0.01482 | 0.345 |
| 30537 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 38 | ADARB1 | Sponge network | 0.349 | 0.01695 | -0.335 | 0.06631 | 0.345 |
| 30538 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-500a-3p | 11 | TSHZ3 | Sponge network | -7.551 | 0 | -0.556 | 0.01516 | 0.345 |
| 30539 | LINC00621 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p | 20 | NOTCH2 | Sponge network | 0.131 | 0.8436 | 0.138 | 0.35163 | 0.345 |
| 30540 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | USP25 | Sponge network | -0.323 | 0.01372 | -0.242 | 0.01397 | 0.345 |
| 30541 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 52 | WDFY3 | Sponge network | -0.563 | 0.02545 | -0.201 | 0.0967 | 0.345 |
| 30542 | LINC00854 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-625-5p | 10 | ABI2 | Sponge network | 1.18 | 0 | 0.175 | 0.14787 | 0.345 |
| 30543 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | FREM2 | Sponge network | -0.901 | 0.00019 | -0.437 | 0.46353 | 0.345 |
| 30544 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p | 10 | PRDM5 | Sponge network | 0.16 | 0.50785 | -1.09 | 0.00014 | 0.345 |
| 30545 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | CRTC1 | Sponge network | -0.576 | 0.10121 | -0.116 | 0.28412 | 0.345 |
| 30546 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 19 | PRKAR1A | Sponge network | 0.082 | 0.55654 | -0.063 | 0.50314 | 0.345 |
| 30547 | RP11-347P5.1 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-940 | 12 | CREB1 | Sponge network | 1.364 | 0 | 0.203 | 0.00537 | 0.345 |
| 30548 | RP11-276H19.2 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-96-5p | 11 | CADM1 | Sponge network | -0.472 | 0.48851 | -0.9 | 0.00084 | 0.345 |
| 30549 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-542-3p | 18 | LRP1B | Sponge network | -0.517 | 0.02935 | -2.163 | 0 | 0.344 |
| 30550 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 30 | LRIG1 | Sponge network | -1.216 | 0 | -0.537 | 0.00588 | 0.344 |
| 30551 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-671-5p | 22 | KCNA6 | Sponge network | -0.738 | 0.21233 | -1.531 | 0.00013 | 0.344 |
| 30552 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-590-3p | 10 | PTPN11 | Sponge network | 0.539 | 0.03722 | 0.431 | 2.0E-5 | 0.344 |
| 30553 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | CBL | Sponge network | 0.222 | 0.29133 | 0.291 | 0.01267 | 0.344 |
| 30554 | DNM3OS |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 12 | ID4 | Sponge network | 0.572 | 0.01722 | -1.644 | 0 | 0.344 |
| 30555 | LINC00473 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | GALNT13 | Sponge network | -1.203 | 0.03881 | -0.857 | 0.07439 | 0.344 |
| 30556 | LINC00472 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 26 | EYA4 | Sponge network | -0.842 | 0.01361 | 0.3 | 0.36876 | 0.344 |
| 30557 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p | 13 | MAP3K12 | Sponge network | -0.901 | 0.00019 | -0.057 | 0.59369 | 0.344 |
| 30558 | RP11-254F7.2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-454-3p;hsa-miR-484 | 11 | IGF2 | Sponge network | 0.176 | 0.37402 | 0.848 | 0.03842 | 0.344 |
| 30559 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | NOTCH2NL | Sponge network | 0.61 | 0.01593 | 0.024 | 0.84307 | 0.344 |
| 30560 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | NEDD4 | Sponge network | 0.619 | 0.00157 | 0.02 | 0.89834 | 0.344 |
| 30561 | GAS6-AS2 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | CSDE1 | Sponge network | 0.16 | 0.50785 | -0.493 | 0 | 0.344 |
| 30562 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-942-5p | 36 | HLF | Sponge network | 0.335 | 0.20596 | -2.443 | 0 | 0.344 |
| 30563 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 27 | EPHA5 | Sponge network | -1.654 | 0.00144 | -0.957 | 0.01733 | 0.344 |
| 30564 | CTC-510F12.2 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-34a-5p | 12 | BCL2 | Sponge network | -0.691 | 0.00146 | -0.899 | 0 | 0.344 |
| 30565 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PHC3 | Sponge network | 0.91 | 0 | 0.212 | 0.0499 | 0.344 |
| 30566 | BDNF-AS |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | ID4 | Sponge network | -0.668 | 3.0E-5 | -1.644 | 0 | 0.344 |
| 30567 | OSER1-AS1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | MED12L | Sponge network | -0.13 | 0.48734 | -0.194 | 0.55299 | 0.344 |
| 30568 | RP11-434B12.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 27 | NTRK2 | Sponge network | -0.031 | 0.89063 | -0.736 | 0.06495 | 0.344 |
| 30569 | LINC00094 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | 0.253 | 0.09565 | -0.564 | 1.0E-5 | 0.344 |
| 30570 | ZNF582-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | TRPS1 | Sponge network | -1.349 | 0.00017 | -0.191 | 0.44109 | 0.344 |
| 30571 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | ATRX | Sponge network | -0.285 | 0.2172 | 0.261 | 0.04408 | 0.344 |
| 30572 | RP1-151F17.2 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | CSDE1 | Sponge network | 0.222 | 0.29133 | -0.493 | 0 | 0.344 |
| 30573 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 13 | ZNF292 | Sponge network | 0.683 | 1.0E-5 | 0.276 | 0.04148 | 0.344 |
| 30574 | RP3-402G11.26 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-500a-3p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-940 | 13 | CHD5 | Sponge network | -0.254 | 0.19303 | -0.922 | 0.01521 | 0.344 |
| 30575 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | PHC3 | Sponge network | 0.415 | 0.14657 | 0.212 | 0.0499 | 0.344 |
| 30576 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 28 | MITF | Sponge network | -0.563 | 0.02545 | -0.769 | 0.00022 | 0.344 |
| 30577 | RP11-53O19.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-628-5p | 14 | CDH23 | Sponge network | -0.405 | 0.22013 | -0.974 | 0.00034 | 0.344 |
| 30578 | TPTEP1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | SYT4 | Sponge network | -1.216 | 0 | -3.207 | 0 | 0.344 |
| 30579 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-590-3p | 14 | DDX5 | Sponge network | 0.101 | 0.60919 | -0.099 | 0.17428 | 0.344 |
| 30580 | MYLK-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-550a-5p | 10 | RIMBP2 | Sponge network | -1.331 | 3.0E-5 | -1.968 | 5.0E-5 | 0.344 |
| 30581 | SNX29P2 |
hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-3p | 13 | RELN | Sponge network | 0.4 | 0.14611 | -2.403 | 0 | 0.344 |
| 30582 | TTC28-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 21 | TBL1XR1 | Sponge network | 0.373 | 0.00068 | 0.578 | 0 | 0.344 |
| 30583 | PGM5-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-378c;hsa-miR-450b-5p | 17 | ERC1 | Sponge network | -4.546 | 0 | -0.108 | 0.38794 | 0.344 |
| 30584 | KCNJ2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 12 | KANK1 | Sponge network | -0.436 | 0.05325 | -0.55 | 0.00134 | 0.344 |
| 30585 | CTA-363E19.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-625-5p | 12 | ABI2 | Sponge network | 1.055 | 0 | 0.175 | 0.14787 | 0.344 |
| 30586 | LINC00641 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-625-5p | 10 | ARID2 | Sponge network | -0.222 | 0.32445 | 0.235 | 0.02697 | 0.344 |
| 30587 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | NCAM2 | Sponge network | 0.082 | 0.55654 | -0.482 | 0.22565 | 0.344 |
| 30588 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | HACE1 | Sponge network | 0.34 | 0.00273 | -0.072 | 0.55239 | 0.344 |
| 30589 | TINCR |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-940;hsa-miR-96-5p | 16 | MYO9A | Sponge network | -0.664 | 0.14543 | -0.147 | 0.32373 | 0.344 |
| 30590 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-98-5p | 30 | ZFHX3 | Sponge network | -0.18 | 0.20343 | 0.389 | 0.01363 | 0.344 |
| 30591 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p | 16 | SPG20 | Sponge network | 0.16 | 0.50785 | -1.16 | 0 | 0.344 |
| 30592 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 20 | FAT4 | Sponge network | 0.72 | 0.0001 | -0.354 | 0.14694 | 0.344 |
| 30593 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | RBL2 | Sponge network | 0.659 | 3.0E-5 | -0.282 | 0.00254 | 0.344 |
| 30594 | AC005562.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-3p | 33 | ABL2 | Sponge network | 0.488 | 0.00084 | 0.82 | 0 | 0.344 |
| 30595 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | CACNA2D1 | Sponge network | -0.235 | 0.15142 | -0.846 | 0.00675 | 0.344 |
| 30596 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p | 14 | TSHZ3 | Sponge network | -0.829 | 0.01343 | -0.556 | 0.01516 | 0.344 |
| 30597 | AC007879.7 |
hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-338-3p;hsa-miR-338-5p | 10 | CBL | Sponge network | 4.3 | 0 | 0.291 | 0.01267 | 0.344 |
| 30598 | U91328.19 |
hsa-miR-126-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-660-5p;hsa-miR-7-5p | 15 | IRS1 | Sponge network | -0.303 | 0.21002 | -0.026 | 0.88855 | 0.344 |
| 30599 | RP11-195F19.9 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | ETV1 | Sponge network | -0.726 | 0.00132 | -0.096 | 0.67674 | 0.344 |
| 30600 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | QKI | Sponge network | -0.053 | 0.80685 | -0.333 | 0.04111 | 0.344 |
| 30601 | RP11-1006G14.4 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 26 | ERC1 | Sponge network | 0.559 | 0.00026 | -0.108 | 0.38794 | 0.344 |
| 30602 | USP3-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p | 11 | USP25 | Sponge network | -0.162 | 0.47687 | -0.242 | 0.01397 | 0.344 |
| 30603 | RP11-13K12.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-629-3p | 22 | RYR2 | Sponge network | -0.154 | 0.78939 | -1.466 | 0.00031 | 0.344 |
| 30604 | HAND2-AS1 |
hsa-miR-10b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 10 | EXTL2 | Sponge network | -1.697 | 0.00889 | 0.109 | 0.37375 | 0.344 |
| 30605 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | AKAP11 | Sponge network | 1.308 | 0 | 0.196 | 0.09825 | 0.344 |
| 30606 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-423-5p;hsa-miR-455-5p | 11 | PNRC1 | Sponge network | 0.056 | 0.8494 | -0.646 | 0 | 0.344 |
| 30607 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p | 12 | RYR2 | Sponge network | 0.497 | 0.01451 | -1.466 | 0.00031 | 0.344 |
| 30608 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 25 | LIFR | Sponge network | -1.349 | 0.00017 | -1.794 | 0 | 0.344 |
| 30609 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-96-5p | 30 | JAZF1 | Sponge network | 0.331 | 0.57247 | -0.377 | 0.02677 | 0.344 |
| 30610 | CALML3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p | 20 | PTPN14 | Sponge network | 0.95 | 0.15943 | 0.144 | 0.34595 | 0.344 |
| 30611 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-27b-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-497-5p | 14 | SOX11 | Sponge network | 1.352 | 0 | 2.68 | 0 | 0.344 |
| 30612 | LINC00094 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p | 14 | FMNL3 | Sponge network | 0.253 | 0.09565 | 0.555 | 4.0E-5 | 0.344 |
| 30613 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-342-3p | 12 | TMTC3 | Sponge network | 1.344 | 0 | 0.123 | 0.22189 | 0.344 |
| 30614 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 26 | BMPR2 | Sponge network | 0.796 | 4.0E-5 | 0.206 | 0.0635 | 0.344 |
| 30615 | CALML3-AS1 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-7-5p | 11 | MLLT11 | Sponge network | 0.95 | 0.15943 | -0.661 | 0.01733 | 0.344 |
| 30616 | RP11-474O21.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378a-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p | 20 | NTRK3 | Sponge network | -0.023 | 0.95109 | -1.77 | 3.0E-5 | 0.344 |
| 30617 | AC005618.6 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | MACF1 | Sponge network | 0.862 | 0.06213 | 0.019 | 0.90335 | 0.344 |
| 30618 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TBX18 | Sponge network | 0.385 | 0.10067 | 0.843 | 0.02945 | 0.344 |
| 30619 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | -0.323 | 0.01372 | -3.143 | 0 | 0.344 |
| 30620 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 13 | CNTNAP3 | Sponge network | -1.057 | 0.00029 | -1.465 | 0.00033 | 0.344 |
| 30621 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-493-3p;hsa-miR-505-3p | 18 | JAKMIP2 | Sponge network | -0.303 | 0.21002 | -1.388 | 0 | 0.344 |
| 30622 | MIR22HG |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p | 24 | SETBP1 | Sponge network | -1.047 | 0 | -1.302 | 1.0E-5 | 0.344 |
| 30623 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-411-5p;hsa-miR-582-5p | 18 | USP6 | Sponge network | 0.792 | 0.0049 | 0.188 | 0.3871 | 0.344 |
| 30624 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-93-3p | 24 | FOXO3 | Sponge network | 0.75 | 0.00054 | -0.04 | 0.72028 | 0.344 |
| 30625 | FAM157B |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 12 | UBR3 | Sponge network | 0.92 | 0 | -0.021 | 0.82774 | 0.344 |
| 30626 | RP11-532F6.3 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p | 16 | DOCK4 | Sponge network | -0.896 | 0.00099 | 0.502 | 0.00108 | 0.344 |
| 30627 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | DKK3 | Sponge network | -0.513 | 0.04703 | -0.052 | 0.77783 | 0.344 |
| 30628 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 24 | SCN9A | Sponge network | -1.161 | 0.00591 | -0.525 | 0.10593 | 0.344 |
| 30629 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-345-5p;hsa-miR-450b-5p | 11 | CDHR3 | Sponge network | -0.583 | 0.14589 | -0.977 | 4.0E-5 | 0.344 |
| 30630 | RP1-102E24.8 |
hsa-miR-101-3p;hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p | 10 | GSK3B | Sponge network | 1.737 | 0 | 0.266 | 0.00045 | 0.344 |
| 30631 | CTC-429P9.3 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p | 10 | NGFR | Sponge network | 0.082 | 0.55397 | -1.271 | 0.01058 | 0.344 |
| 30632 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-92a-3p | 23 | ST6GAL2 | Sponge network | -0.738 | 0.21233 | -1.201 | 0.00597 | 0.344 |
| 30633 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-532-5p | 39 | BMPR2 | Sponge network | 0.082 | 0.55397 | 0.206 | 0.0635 | 0.344 |
| 30634 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-590-3p | 15 | STARD13 | Sponge network | -0.739 | 0.00044 | -0.187 | 0.27383 | 0.344 |
| 30635 | DNM3OS |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | TMEFF2 | Sponge network | 0.572 | 0.01722 | -2.426 | 0 | 0.344 |
| 30636 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 21 | GRIA3 | Sponge network | -1.057 | 0.00029 | -1.956 | 0 | 0.344 |
| 30637 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 48 | PPM1A | Sponge network | 0.064 | 0.72934 | -0.623 | 0 | 0.344 |
| 30638 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-324-3p | 17 | ZBTB4 | Sponge network | -4.477 | 0 | -0.739 | 0 | 0.344 |
| 30639 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-539-5p;hsa-miR-582-5p | 19 | MOB1B | Sponge network | 1.294 | 0 | 0.197 | 0.15266 | 0.344 |
| 30640 | RP11-274B21.10 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | ABL2 | Sponge network | 1.073 | 0.00216 | 0.82 | 0 | 0.344 |
| 30641 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-500a-5p | 47 | KIF1B | Sponge network | 0.082 | 0.55397 | -0.118 | 0.32258 | 0.344 |
| 30642 | CTC-457L16.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-425-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940 | 17 | PTPRT | Sponge network | 1.153 | 0.00114 | -0.907 | 0.03727 | 0.344 |
| 30643 | MEG3 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p;hsa-miR-96-5p | 11 | ZYX | Sponge network | 0.249 | 0.35902 | 0.019 | 0.89763 | 0.344 |
| 30644 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-625-5p | 24 | YAP1 | Sponge network | 0.95 | 0.15943 | 0.22 | 0.11836 | 0.344 |
| 30645 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 11 | RARB | Sponge network | -1.057 | 0.00029 | -0.825 | 2.0E-5 | 0.344 |
| 30646 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-654-3p | 16 | MYBL1 | Sponge network | 0.439 | 0.00313 | 0.494 | 0.02152 | 0.344 |
| 30647 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-769-5p | 11 | RCAN2 | Sponge network | -0.773 | 0.04073 | -0.33 | 0.15172 | 0.344 |
| 30648 | WDFY3-AS2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-331-5p;hsa-miR-429 | 11 | DUSP1 | Sponge network | -0.942 | 0 | -1.754 | 0 | 0.344 |
| 30649 | DNM1P35 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-625-5p | 21 | ABI2 | Sponge network | 0.051 | 0.83388 | 0.175 | 0.14787 | 0.344 |
| 30650 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p | 10 | PDZD2 | Sponge network | -0.246 | 0.35034 | -0.909 | 3.0E-5 | 0.344 |
| 30651 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-582-5p | 13 | KAT6B | Sponge network | 0.792 | 0.0049 | -0.478 | 0.00018 | 0.344 |
| 30652 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 32 | GAS7 | Sponge network | 0.225 | 0.30644 | -0.555 | 0.01602 | 0.344 |
| 30653 | PCA3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PDE4DIP | Sponge network | -0.709 | 0.18168 | -0.282 | 0.0257 | 0.344 |
| 30654 | LINC00957 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 11 | SEPT6 | Sponge network | -0.421 | 0.0114 | -0.598 | 0.00181 | 0.344 |
| 30655 | PROSER2-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p | 15 | ABL2 | Sponge network | 1.825 | 0 | 0.82 | 0 | 0.344 |
| 30656 | LINC00886 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-5p | 21 | AKAP6 | Sponge network | -0.371 | 0.24604 | -1.586 | 3.0E-5 | 0.344 |
| 30657 | LINC00578 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-500a-3p;hsa-miR-577 | 12 | AFF3 | Sponge network | 0.811 | 0.03228 | -2.373 | 0 | 0.344 |
| 30658 | CTC-457L16.2 |
hsa-miR-130b-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-940 | 10 | SORCS1 | Sponge network | 1.153 | 0.00114 | -3.492 | 0 | 0.344 |
| 30659 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 1.163 | 0.00018 | 0.322 | 0.00215 | 0.344 |
| 30660 | MIR600HG |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-625-3p | 23 | ABL2 | Sponge network | 0.594 | 0.41522 | 0.82 | 0 | 0.344 |
| 30661 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | CACNA2D1 | Sponge network | -0.83 | 0 | -0.846 | 0.00675 | 0.344 |
| 30662 | RP5-1139B12.3 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-500a-3p;hsa-miR-502-5p | 21 | DST | Sponge network | 0.598 | 0.38481 | -0.713 | 0.00096 | 0.344 |
| 30663 | CASC2 |
hsa-miR-10b-3p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p | 16 | CTDSPL | Sponge network | 0.385 | 0.01306 | 0.085 | 0.45121 | 0.344 |
| 30664 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-590-5p | 11 | ABCA10 | Sponge network | -1.349 | 0.00017 | -0.571 | 0.03674 | 0.344 |
| 30665 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | ZMYM2 | Sponge network | 0.083 | 0.66405 | 0.279 | 0.01273 | 0.344 |
| 30666 | LINC00854 |
hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p | 11 | TBL1XR1 | Sponge network | 1.18 | 0 | 0.578 | 0 | 0.344 |
| 30667 | RP11-504P24.8 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-497-5p | 10 | BRD3 | Sponge network | 1.178 | 0 | 0.37 | 0.00013 | 0.344 |
| 30668 | RP11-344B5.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-320b | 15 | RASSF2 | Sponge network | -0.055 | 0.88482 | -0.273 | 0.21961 | 0.344 |
| 30669 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 1.205 | 0 | 0.322 | 0.00215 | 0.344 |
| 30670 | AC009948.5 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-744-3p | 31 | IGFBP5 | Sponge network | 0.532 | 0.00505 | -0.806 | 0.00105 | 0.344 |
| 30671 | RP11-389C8.2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 31 | BCL2 | Sponge network | 0.727 | 3.0E-5 | -0.899 | 0 | 0.344 |
| 30672 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 34 | ERBB4 | Sponge network | -0.246 | 0.35034 | -1.975 | 2.0E-5 | 0.344 |
| 30673 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-629-3p;hsa-miR-940 | 18 | BHLHE41 | Sponge network | -0.154 | 0.53954 | 0.353 | 0.12753 | 0.344 |
| 30674 | RP11-526I2.5 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-495-3p | 13 | CCDC6 | Sponge network | 1.054 | 1.0E-5 | 0.27 | 0.02555 | 0.344 |
| 30675 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-590-3p | 18 | CNTN1 | Sponge network | 0.051 | 0.83388 | -2.594 | 0 | 0.344 |
| 30676 | NDUFA6-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 57 | DST | Sponge network | -0.355 | 0.0077 | -0.713 | 0.00096 | 0.344 |
| 30677 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p | 25 | MAF | Sponge network | -0.246 | 0.35034 | -1.257 | 0 | 0.344 |
| 30678 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | CDHR3 | Sponge network | -0.615 | 0.00015 | -0.977 | 4.0E-5 | 0.344 |
| 30679 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | FGF5 | Sponge network | 0.61 | 0.01593 | 0.549 | 0.28659 | 0.344 |
| 30680 | USP3-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-940 | 16 | CELF4 | Sponge network | -0.162 | 0.47687 | -1.135 | 0.00344 | 0.344 |
| 30681 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-671-5p | 11 | PIK3IP1 | Sponge network | 0.572 | 0.01722 | -0.187 | 0.19945 | 0.344 |
| 30682 | DIO3OS |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 11 | ALDH1A2 | Sponge network | -0.773 | 0.04073 | -2.411 | 0 | 0.344 |
| 30683 | DNM1P35 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 21 | RASSF8 | Sponge network | 0.051 | 0.83388 | -0.122 | 0.65591 | 0.344 |
| 30684 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 30 | ATRX | Sponge network | 0.082 | 0.55654 | 0.261 | 0.04408 | 0.344 |
| 30685 | RP11-155G14.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-378c | 18 | ABL2 | Sponge network | 2.483 | 0 | 0.82 | 0 | 0.344 |
| 30686 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p | 18 | AFF4 | Sponge network | 1.106 | 0 | -0.266 | 0.01565 | 0.344 |
| 30687 | CTD-2354A18.1 |
hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-664a-5p | 10 | CBFB | Sponge network | 4.745 | 0 | 1.061 | 0 | 0.344 |
| 30688 | SNX29P2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | ROBO1 | Sponge network | 0.4 | 0.14611 | -0.009 | 0.96154 | 0.344 |
| 30689 | FGD5-AS1 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p | 19 | DICER1 | Sponge network | 0.401 | 0.00066 | 0.042 | 0.674 | 0.344 |
| 30690 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | SON | Sponge network | 1.317 | 0 | 0.168 | 0.04406 | 0.344 |
| 30691 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | EYA4 | Sponge network | 0.559 | 0.00026 | 0.3 | 0.36876 | 0.344 |
| 30692 | KB-1572G7.2 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 13 | RASAL2 | Sponge network | 0.924 | 0 | 0.869 | 0 | 0.344 |
| 30693 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-590-5p | 14 | SPG20 | Sponge network | 0.395 | 0.15451 | -1.16 | 0 | 0.344 |
| 30694 | U91328.19 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 26 | RNF144A | Sponge network | -0.303 | 0.21002 | 0.594 | 0.0009 | 0.344 |
| 30695 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-940 | 31 | AKAP11 | Sponge network | -0.162 | 0.47687 | 0.196 | 0.09825 | 0.344 |
| 30696 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-940 | 19 | NTRK3 | Sponge network | -0.421 | 0.12556 | -1.77 | 3.0E-5 | 0.344 |
| 30697 | LINC00957 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | NR3C2 | Sponge network | -0.421 | 0.0114 | -1.56 | 0 | 0.344 |
| 30698 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 45 | CREB3L2 | Sponge network | 0.997 | 0.00066 | -0.525 | 2.0E-5 | 0.344 |
| 30699 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 42 | MEF2C | Sponge network | -0.899 | 6.0E-5 | -0.499 | 0.01448 | 0.344 |
| 30700 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 16 | ERCC4 | Sponge network | 0.225 | 0.30644 | 0.126 | 0.15266 | 0.344 |
| 30701 | MIR497HG |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-96-5p | 26 | CADM1 | Sponge network | -1.439 | 4.0E-5 | -0.9 | 0.00084 | 0.344 |
| 30702 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | ABCB5 | Sponge network | -0.513 | 0.04703 | -0.956 | 0.04077 | 0.344 |
| 30703 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-590-3p | 16 | RAP1A | Sponge network | -0.739 | 0.00044 | -0.929 | 0 | 0.344 |
| 30704 | AC092301.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-378a-5p | 11 | CREB1 | Sponge network | 4.497 | 0 | 0.203 | 0.00537 | 0.344 |
| 30705 | CTC-471J1.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-7-1-3p | 14 | BRD3 | Sponge network | 1.11 | 1.0E-5 | 0.37 | 0.00013 | 0.344 |
| 30706 | MAP3K14 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p | 22 | LPP | Sponge network | -0.295 | 0.02604 | -0.459 | 0.04475 | 0.344 |
| 30707 | GAS6-AS2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p | 17 | RYR2 | Sponge network | 0.16 | 0.50785 | -1.466 | 0.00031 | 0.344 |
| 30708 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p | 11 | TSLP | Sponge network | -1.697 | 0.00889 | -2.209 | 0 | 0.344 |
| 30709 | RP11-359E10.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | PRKCB | Sponge network | 0.299 | 0.57722 | -1.313 | 2.0E-5 | 0.344 |
| 30710 | HHIP-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | PREX2 | Sponge network | -0.737 | 0.10822 | -0.272 | 0.25382 | 0.344 |
| 30711 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 41 | MOB1B | Sponge network | -0.49 | 0.04946 | 0.197 | 0.15266 | 0.344 |
| 30712 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-454-3p | 11 | RARB | Sponge network | -0.663 | 0.10887 | -0.825 | 2.0E-5 | 0.344 |
| 30713 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CXXC4 | Sponge network | -0.053 | 0.80685 | -0.433 | 0.21055 | 0.344 |
| 30714 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-942-5p | 13 | RB1CC1 | Sponge network | 0.663 | 0.00073 | 0.175 | 0.12559 | 0.344 |
| 30715 | LINC00092 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-940 | 12 | CSRNP1 | Sponge network | -1.886 | 0 | -1.448 | 0 | 0.344 |
| 30716 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | EPAS1 | Sponge network | -1.349 | 0.00017 | -0.184 | 0.14418 | 0.344 |
| 30717 | RP4-669L17.10 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p | 20 | CREB1 | Sponge network | 1.093 | 0 | 0.203 | 0.00537 | 0.344 |
| 30718 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | PIK3C2A | Sponge network | 1.163 | 0.00018 | 0.061 | 0.53776 | 0.344 |
| 30719 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-214-3p;hsa-miR-214-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 46 | PPM1A | Sponge network | 0.421 | 0.00084 | -0.623 | 0 | 0.344 |
| 30720 | SNHG16 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-497-5p | 14 | PAFAH1B2 | Sponge network | 0.961 | 0 | 0.318 | 0.0001 | 0.344 |
| 30721 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | PHC3 | Sponge network | 1.122 | 0 | 0.212 | 0.0499 | 0.344 |
| 30722 | RASSF8-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PIK3CA | Sponge network | 0.335 | 0.20596 | 0.195 | 0.06908 | 0.344 |
| 30723 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-93-5p | 15 | FAT4 | Sponge network | -0.384 | 0.40652 | -0.354 | 0.14694 | 0.344 |
| 30724 | PSMG3-AS1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p | 10 | DNMT3A | Sponge network | -0.125 | 0.49414 | 0.302 | 0.0262 | 0.344 |
| 30725 | OIP5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | PRRX1 | Sponge network | 0.421 | 0.00084 | 1.316 | 0 | 0.344 |
| 30726 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-98-5p | 43 | CADM2 | Sponge network | -0.154 | 0.78939 | -3.782 | 0 | 0.344 |
| 30727 | RP11-554A11.4 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 13 | LIMCH1 | Sponge network | -0.583 | 0.14589 | -0.915 | 1.0E-5 | 0.344 |
| 30728 | HOXB-AS3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-550a-3p;hsa-miR-629-3p | 21 | EPHA7 | Sponge network | 0.343 | 0.28887 | -2.197 | 3.0E-5 | 0.344 |
| 30729 | RP5-1198O20.4 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p | 16 | PLXNC1 | Sponge network | 0.997 | 0.00066 | 0.569 | 0.00329 | 0.344 |
| 30730 | PGM5-AS1 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 18 | OPCML | Sponge network | -4.546 | 0 | -2.498 | 0 | 0.344 |
| 30731 | RP11-196G18.23 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZFHX3 | Sponge network | 0.724 | 0.00039 | 0.389 | 0.01363 | 0.344 |
| 30732 | FLNB-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | UBR3 | Sponge network | 1.163 | 0.00018 | -0.021 | 0.82774 | 0.344 |
| 30733 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-199b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-339-5p | 13 | TSC1 | Sponge network | 0.542 | 0.00054 | 0.103 | 0.26071 | 0.344 |
| 30734 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-5p | 24 | JAZF1 | Sponge network | -0.246 | 0.35034 | -0.377 | 0.02677 | 0.344 |
| 30735 | RP11-384K6.6 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p | 11 | UBE4B | Sponge network | 0.946 | 0 | -0.235 | 0.007 | 0.344 |
| 30736 | MEG3 |
hsa-miR-107;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 21 | ZFP36L1 | Sponge network | 0.249 | 0.35902 | -0.505 | 8.0E-5 | 0.344 |
| 30737 | U91328.19 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-940 | 39 | NTRK2 | Sponge network | -0.303 | 0.21002 | -0.736 | 0.06495 | 0.344 |
| 30738 | RP4-717I23.3 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-576-5p | 18 | TSC1 | Sponge network | 0.637 | 0.00069 | 0.103 | 0.26071 | 0.344 |
| 30739 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | ESR1 | Sponge network | 0.898 | 1.0E-5 | -1.035 | 3.0E-5 | 0.344 |
| 30740 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 11 | SEMA3E | Sponge network | -0.376 | 0.00337 | -1.717 | 0.00137 | 0.344 |
| 30741 | AC108488.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 12 | SETBP1 | Sponge network | 0.259 | 0.12542 | -1.302 | 1.0E-5 | 0.344 |
| 30742 | RP11-473I1.5 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374b-5p | 11 | EPHA3 | Sponge network | 0.542 | 0.00054 | 0.185 | 0.53588 | 0.344 |
| 30743 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-590-3p | 15 | GPAM | Sponge network | 0.919 | 0 | 0.142 | 0.29732 | 0.344 |
| 30744 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-374b-5p | 16 | NOTCH2 | Sponge network | 1.46 | 1.0E-5 | 0.138 | 0.35163 | 0.344 |
| 30745 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 33 | CNTN1 | Sponge network | -0.222 | 0.32445 | -2.594 | 0 | 0.344 |
| 30746 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p | 22 | ST6GALNAC3 | Sponge network | -0.693 | 0.05629 | -0.987 | 0 | 0.344 |
| 30747 | CTC-429P9.5 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p | 11 | PDE4DIP | Sponge network | 0.178 | 0.29106 | -0.282 | 0.0257 | 0.344 |
| 30748 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-484;hsa-miR-550a-5p | 15 | CHD5 | Sponge network | 1.316 | 0.01106 | -0.922 | 0.01521 | 0.344 |
| 30749 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 35 | ETV1 | Sponge network | -0.737 | 0.06182 | -0.096 | 0.67674 | 0.344 |
| 30750 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 17 | MOB1B | Sponge network | 0.336 | 0.00739 | 0.197 | 0.15266 | 0.344 |
| 30751 | AC018890.6 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | MDM4 | Sponge network | 1.61 | 0.00961 | 0.345 | 0.00762 | 0.344 |
| 30752 | RP11-67L2.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | ROBO1 | Sponge network | 0.72 | 0.0001 | -0.009 | 0.96154 | 0.343 |
| 30753 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p | 11 | PDGFC | Sponge network | 0.082 | 0.55654 | -0.33 | 0.06483 | 0.343 |
| 30754 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-940 | 40 | PTPRT | Sponge network | -0.576 | 0.10121 | -0.907 | 0.03727 | 0.343 |
| 30755 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | PLSCR4 | Sponge network | 0.4 | 0.14611 | -0.474 | 0.03833 | 0.343 |
| 30756 | FGD5-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | RPS6KA3 | Sponge network | 0.401 | 0.00066 | 0.256 | 0.02419 | 0.343 |
| 30757 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ADAMTSL3 | Sponge network | -0.668 | 3.0E-5 | -1.559 | 0 | 0.343 |
| 30758 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-340-5p | 15 | MXI1 | Sponge network | -0.693 | 0.05629 | -0.987 | 0 | 0.343 |
| 30759 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-708-5p;hsa-miR-935 | 18 | MTSS1 | Sponge network | -2.62 | 0 | -0.81 | 0.00035 | 0.343 |
| 30760 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | ABCA10 | Sponge network | 0.997 | 0.00066 | -0.571 | 0.03674 | 0.343 |
| 30761 | RP11-6O2.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-590-5p | 13 | ITGB3 | Sponge network | 0.331 | 0.57247 | -0.011 | 0.95751 | 0.343 |
| 30762 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 35 | ADARB1 | Sponge network | 0.603 | 0.00127 | -0.335 | 0.06631 | 0.343 |
| 30763 | HHIP-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | LIMCH1 | Sponge network | -0.737 | 0.10822 | -0.915 | 1.0E-5 | 0.343 |
| 30764 | LINC00094 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-93-3p;hsa-miR-93-5p | 12 | FYN | Sponge network | 0.253 | 0.09565 | -0.131 | 0.37051 | 0.343 |
| 30765 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ZNF536 | Sponge network | -0.487 | 0.02157 | -3.115 | 0 | 0.343 |
| 30766 | RP11-434B12.1 |
hsa-miR-10b-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-653-5p | 19 | UBR3 | Sponge network | -0.031 | 0.89063 | -0.021 | 0.82774 | 0.343 |
| 30767 | RP11-2H8.2 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | NF1 | Sponge network | 1.089 | 0 | 0.51 | 0 | 0.343 |
| 30768 | CTD-3099C6.9 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 14 | CREB1 | Sponge network | 1.361 | 0 | 0.203 | 0.00537 | 0.343 |
| 30769 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | TSHZ3 | Sponge network | -0.355 | 0.0077 | -0.556 | 0.01516 | 0.343 |
| 30770 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b | 10 | MIER3 | Sponge network | 0.545 | 0.00064 | -0.212 | 0.04957 | 0.343 |
| 30771 | FZD10-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p | 22 | WWTR1 | Sponge network | -0.727 | 0.04726 | -0.345 | 0.11997 | 0.343 |
| 30772 | H1FX-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 12 | SPG20 | Sponge network | -0.206 | 0.12942 | -1.16 | 0 | 0.343 |
| 30773 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-940 | 31 | NTRK2 | Sponge network | -0.465 | 0.04703 | -0.736 | 0.06495 | 0.343 |
| 30774 | FAM215B |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-7-1-3p | 17 | ZFX | Sponge network | 0.817 | 3.0E-5 | 0.09 | 0.42563 | 0.343 |
| 30775 | RP11-488C13.5 |
hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-150-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-497-5p | 11 | MDM4 | Sponge network | 0.753 | 0 | 0.345 | 0.00762 | 0.343 |
| 30776 | RP5-1139B12.3 |
hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-671-5p | 11 | KIT | Sponge network | 0.598 | 0.38481 | -2.184 | 0 | 0.343 |
| 30777 | RP13-1039J1.4 |
hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-589-3p | 10 | MED12L | Sponge network | 0.356 | 0.33467 | -0.194 | 0.55299 | 0.343 |
| 30778 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-455-3p | 15 | MAF | Sponge network | 0.693 | 0.00076 | -1.257 | 0 | 0.343 |
| 30779 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-590-3p | 26 | SOX5 | Sponge network | 0.051 | 0.83388 | -1.091 | 4.0E-5 | 0.343 |
| 30780 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 21 | PPP1R9A | Sponge network | -1.092 | 4.0E-5 | -0.903 | 0.04254 | 0.343 |
| 30781 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | CLASP2 | Sponge network | -0.176 | 0.59692 | -0.038 | 0.71 | 0.343 |
| 30782 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p | 10 | DMTF1 | Sponge network | 0.556 | 0.00298 | 0.248 | 0.02726 | 0.343 |
| 30783 | RP11-403I13.7 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-335-5p | 15 | PLXNC1 | Sponge network | 0.395 | 0.15451 | 0.569 | 0.00329 | 0.343 |
| 30784 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | PRKAB2 | Sponge network | -0.709 | 0.18168 | -0.367 | 0.01371 | 0.343 |
| 30785 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-590-3p | 10 | RBM5 | Sponge network | 0.657 | 4.0E-5 | -0.205 | 0.0129 | 0.343 |
| 30786 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | APC | Sponge network | -0.737 | 0.06182 | -0.255 | 0.04051 | 0.343 |
| 30787 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p | 31 | RICTOR | Sponge network | 0.385 | 0.10067 | 0.388 | 0.00188 | 0.343 |
| 30788 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRIM58 | Sponge network | -0.942 | 0 | -1.692 | 0 | 0.343 |
| 30789 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 36 | TGFBR3 | Sponge network | 0.082 | 0.55654 | -1.173 | 1.0E-5 | 0.343 |
| 30790 | FZD10-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | HOMER2 | Sponge network | -0.727 | 0.04726 | -2.698 | 0 | 0.343 |
| 30791 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-503-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 38 | RICTOR | Sponge network | 0.064 | 0.72934 | 0.388 | 0.00188 | 0.343 |
| 30792 | RP11-767N6.7 |
hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-214-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-590-5p | 11 | MLLT10 | Sponge network | 0.466 | 0.00155 | 0.105 | 0.33292 | 0.343 |
| 30793 | ACTA2-AS1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 28 | RIMS2 | Sponge network | 0.194 | 0.64278 | -1.365 | 0.00208 | 0.343 |
| 30794 | PWAR6 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 22 | PRKAR1A | Sponge network | -1.575 | 6.0E-5 | -0.063 | 0.50314 | 0.343 |
| 30795 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 28 | KIF1B | Sponge network | 0.972 | 0 | -0.118 | 0.32258 | 0.343 |
| 30796 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-874-3p | 16 | CRTC3 | Sponge network | 0.683 | 1.0E-5 | 0.164 | 0.16786 | 0.343 |
| 30797 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-5p | 10 | ZNF208 | Sponge network | 0.395 | 0.15451 | -0.4 | 0.22381 | 0.343 |
| 30798 | MRVI1-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-539-5p | 13 | ACSL6 | Sponge network | -1.092 | 4.0E-5 | -1.593 | 0 | 0.343 |
| 30799 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-625-3p | 27 | ZFHX3 | Sponge network | 0.637 | 0.00069 | 0.389 | 0.01363 | 0.343 |
| 30800 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | CCND2 | Sponge network | -2.117 | 0.00161 | -0.267 | 0.3112 | 0.343 |
| 30801 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-576-5p;hsa-miR-625-3p | 17 | CBX5 | Sponge network | 0.537 | 0.0006 | 0.165 | 0.24446 | 0.343 |
| 30802 | CTC-296K1.3 |
hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-582-5p | 14 | GJA1 | Sponge network | -2.095 | 0.00112 | -0.485 | 0.02179 | 0.343 |
| 30803 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 15 | FREM2 | Sponge network | -0.563 | 0.02545 | -0.437 | 0.46353 | 0.343 |
| 30804 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | ETV1 | Sponge network | 0.559 | 0.00026 | -0.096 | 0.67674 | 0.343 |
| 30805 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-629-5p | 36 | SPRY3 | Sponge network | 0.494 | 0.00339 | 0.016 | 0.91286 | 0.343 |
| 30806 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 19 | BMPR2 | Sponge network | 2.094 | 0 | 0.206 | 0.0635 | 0.343 |
| 30807 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-3613-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 51 | SMAD2 | Sponge network | -0.222 | 0.32445 | -0.128 | 0.12732 | 0.343 |
| 30808 | MIR143HG |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CTNNA2 | Sponge network | -0.739 | 0.0574 | -2.547 | 0 | 0.343 |
| 30809 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | CLASP2 | Sponge network | 0.349 | 0.01695 | -0.038 | 0.71 | 0.343 |
| 30810 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TRIP11 | Sponge network | 0.873 | 0 | -0.158 | 0.06384 | 0.343 |
| 30811 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 30 | CNTN1 | Sponge network | 1.302 | 7.0E-5 | -2.594 | 0 | 0.343 |
| 30812 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 15 | CHD5 | Sponge network | -1.255 | 0.00981 | -0.922 | 0.01521 | 0.343 |
| 30813 | CTC-429P9.1 |
hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p | 11 | NEDD4 | Sponge network | 0.497 | 0.01451 | 0.02 | 0.89834 | 0.343 |
| 30814 | RP11-33B1.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | UBR3 | Sponge network | 0.826 | 0 | -0.021 | 0.82774 | 0.343 |
| 30815 | RP11-13J8.1 |
hsa-let-7a-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 14 | RICTOR | Sponge network | 3.03 | 0 | 0.388 | 0.00188 | 0.343 |
| 30816 | RP11-403I13.7 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | PIK3CA | Sponge network | 0.395 | 0.15451 | 0.195 | 0.06908 | 0.343 |
| 30817 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-92a-3p | 14 | SPTBN4 | Sponge network | -0.323 | 0.01372 | -0.786 | 0.01281 | 0.343 |
| 30818 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-935 | 14 | RNF144A | Sponge network | -0.376 | 0.00337 | 0.594 | 0.0009 | 0.343 |
| 30819 | ACTA2-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | PPM1L | Sponge network | 0.194 | 0.64278 | -0.537 | 0.00616 | 0.343 |
| 30820 | RP11-686D22.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-590-3p | 11 | ZFHX4 | Sponge network | 0.793 | 5.0E-5 | 0.018 | 0.95574 | 0.343 |
| 30821 | AGAP11 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p | 25 | PCDH9 | Sponge network | -1.645 | 0 | -1.823 | 4.0E-5 | 0.343 |
| 30822 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p | 20 | SON | Sponge network | 0.401 | 0.00066 | 0.168 | 0.04406 | 0.343 |
| 30823 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 16 | CLIP1 | Sponge network | 0.75 | 0.00054 | -0.215 | 0.08684 | 0.343 |
| 30824 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 25 | ZFHX4 | Sponge network | 0.419 | 0.0319 | 0.018 | 0.95574 | 0.343 |
| 30825 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TRIP11 | Sponge network | 0.083 | 0.66405 | -0.158 | 0.06384 | 0.343 |
| 30826 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-501-3p | 22 | APC | Sponge network | -1.645 | 0 | -0.255 | 0.04051 | 0.343 |
| 30827 | RP11-356J5.12 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | KANK1 | Sponge network | -0.899 | 6.0E-5 | -0.55 | 0.00134 | 0.343 |
| 30828 | AC009948.5 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | KLF10 | Sponge network | 0.532 | 0.00505 | -0.783 | 0 | 0.343 |
| 30829 | FAM215B |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p | 12 | NF1 | Sponge network | 0.817 | 3.0E-5 | 0.51 | 0 | 0.343 |
| 30830 | LINC01018 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-93-5p | 54 | DST | Sponge network | -2.915 | 0.00015 | -0.713 | 0.00096 | 0.343 |
| 30831 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-744-3p;hsa-miR-96-5p | 13 | HIP1 | Sponge network | 0.614 | 1.0E-5 | 0.774 | 0 | 0.343 |
| 30832 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 41 | PHC3 | Sponge network | 0.311 | 0.10344 | 0.212 | 0.0499 | 0.343 |
| 30833 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | PDE4DIP | Sponge network | 0.395 | 0.15451 | -0.282 | 0.0257 | 0.343 |
| 30834 | LINC00702 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NRG3 | Sponge network | -0.737 | 0.06182 | -1.836 | 4.0E-5 | 0.343 |
| 30835 | RP11-890B15.3 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-7-1-3p | 13 | RASSF3 | Sponge network | 0.101 | 0.60919 | -0.226 | 0.11691 | 0.343 |
| 30836 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 15 | SPTBN4 | Sponge network | 0.246 | 0.59912 | -0.786 | 0.01281 | 0.343 |
| 30837 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p | 25 | CBL | Sponge network | 0.637 | 0.00069 | 0.291 | 0.01267 | 0.343 |
| 30838 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-424-5p | 19 | RASSF8 | Sponge network | -0.465 | 0.04703 | -0.122 | 0.65591 | 0.343 |
| 30839 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 11 | MED12L | Sponge network | 0.131 | 0.8436 | -0.194 | 0.55299 | 0.343 |
| 30840 | PWAR6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | CSMD1 | Sponge network | -1.575 | 6.0E-5 | -0.63 | 0.18881 | 0.343 |
| 30841 | FZD10-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | CDH10 | Sponge network | -0.727 | 0.04726 | -2.397 | 0 | 0.343 |
| 30842 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ABCC9 | Sponge network | 0.862 | 0.06213 | -0.466 | 0.14121 | 0.343 |
| 30843 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 0.832 | 0 | 0.51 | 0 | 0.343 |
| 30844 | RP11-35G9.5 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-96-5p | 16 | RASSF8 | Sponge network | 0.622 | 0.00015 | -0.122 | 0.65591 | 0.343 |
| 30845 | NR2F1-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | LIMCH1 | Sponge network | -0.49 | 0.04946 | -0.915 | 1.0E-5 | 0.343 |
| 30846 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 29 | TCF4 | Sponge network | -0.663 | 0.10887 | 0.016 | 0.92271 | 0.343 |
| 30847 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM13 | Sponge network | 0.225 | 0.30644 | 0.183 | 0.06968 | 0.343 |
| 30848 | LINC00957 |
hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-532-3p | 11 | PIM1 | Sponge network | -0.421 | 0.0114 | -1.114 | 0 | 0.343 |
| 30849 | AC006116.24 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-577 | 15 | CSMD1 | Sponge network | -0.473 | 0.07602 | -0.63 | 0.18881 | 0.343 |
| 30850 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MLF1 | Sponge network | -1.216 | 0 | -0.893 | 0.00727 | 0.343 |
| 30851 | RP11-439A17.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-5p | 10 | SOX5 | Sponge network | 0.051 | 0.95756 | -1.091 | 4.0E-5 | 0.343 |
| 30852 | FZD10-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TAL1 | Sponge network | -0.727 | 0.04726 | -0.462 | 0.00574 | 0.343 |
| 30853 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | CUL5 | Sponge network | 0.225 | 0.30644 | 0.026 | 0.77301 | 0.343 |
| 30854 | CTD-3025N20.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-590-3p;hsa-miR-654-3p | 11 | MYBL1 | Sponge network | 1.046 | 0 | 0.494 | 0.02152 | 0.343 |
| 30855 | U91328.19 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 19 | SETBP1 | Sponge network | -0.303 | 0.21002 | -1.302 | 1.0E-5 | 0.343 |
| 30856 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 21 | KAT6B | Sponge network | -0.405 | 0.22013 | -0.478 | 0.00018 | 0.343 |
| 30857 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-93-3p | 19 | BNC2 | Sponge network | 0.912 | 0.00014 | -0.491 | 0.09988 | 0.343 |
| 30858 | RP11-166D19.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 43 | AFF1 | Sponge network | -0.576 | 0.10121 | -0.384 | 0.00053 | 0.343 |
| 30859 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-582-3p | 10 | PRDM2 | Sponge network | 0.993 | 0 | -0.258 | 0.00721 | 0.343 |
| 30860 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-582-5p | 11 | CXXC4 | Sponge network | -0.469 | 0.08721 | -0.433 | 0.21055 | 0.343 |
| 30861 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-655-3p;hsa-miR-664a-5p | 10 | TET2 | Sponge network | 0.683 | 1.0E-5 | -0.222 | 0.04665 | 0.343 |
| 30862 | AC141928.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 15 | AKAP6 | Sponge network | 0.853 | 0.15352 | -1.586 | 3.0E-5 | 0.343 |
| 30863 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | DDX5 | Sponge network | 0.532 | 0.00505 | -0.099 | 0.17428 | 0.343 |
| 30864 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-5p | 47 | NFIB | Sponge network | -0.18 | 0.20343 | 0.023 | 0.90795 | 0.343 |
| 30865 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 32 | KCNA6 | Sponge network | -0.942 | 0 | -1.531 | 0.00013 | 0.343 |
| 30866 | BDNF-AS |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | DTNA | Sponge network | -0.668 | 3.0E-5 | -1.648 | 1.0E-5 | 0.343 |
| 30867 | FAM157B |
hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | PIK3C2A | Sponge network | 0.92 | 0 | 0.061 | 0.53776 | 0.343 |
| 30868 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | PPP1R9A | Sponge network | 0.41 | 0.02214 | -0.903 | 0.04254 | 0.343 |
| 30869 | RP11-158K1.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | AFF1 | Sponge network | 0.349 | 0.01695 | -0.384 | 0.00053 | 0.343 |
| 30870 | TPTEP1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 46 | NTRK2 | Sponge network | -1.216 | 0 | -0.736 | 0.06495 | 0.343 |
| 30871 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | TMEM132B | Sponge network | -0.739 | 0.00044 | -0.83 | 0.01084 | 0.343 |
| 30872 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 14 | KCNAB1 | Sponge network | 0.064 | 0.72934 | -0.755 | 2.0E-5 | 0.343 |
| 30873 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 15 | MCC | Sponge network | 0.131 | 0.8436 | -1.005 | 0 | 0.343 |
| 30874 | RP5-994D16.9 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZFHX3 | Sponge network | 0.252 | 0.08508 | 0.389 | 0.01363 | 0.343 |
| 30875 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 12 | GIGYF2 | Sponge network | 0.72 | 0.0001 | 0.136 | 0.04403 | 0.343 |
| 30876 | AC009120.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | CLASP2 | Sponge network | 0.919 | 0 | -0.038 | 0.71 | 0.343 |
| 30877 | LINC01018 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | RIMBP2 | Sponge network | -2.915 | 0.00015 | -1.968 | 5.0E-5 | 0.343 |
| 30878 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | PGR | Sponge network | 0.727 | 3.0E-5 | -1.704 | 0 | 0.343 |
| 30879 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p | 11 | ERCC4 | Sponge network | 1.46 | 1.0E-5 | 0.126 | 0.15266 | 0.343 |
| 30880 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-589-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | CXXC4 | Sponge network | -0.303 | 0.21002 | -0.433 | 0.21055 | 0.343 |
| 30881 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-590-3p | 12 | CNTNAP3 | Sponge network | 0.349 | 0.01695 | -1.465 | 0.00033 | 0.343 |
| 30882 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | FGF5 | Sponge network | -1.216 | 0 | 0.549 | 0.28659 | 0.343 |
| 30883 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p | 10 | CDH23 | Sponge network | 0.727 | 3.0E-5 | -0.974 | 0.00034 | 0.343 |
| 30884 | LINC00205 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-30a-5p;hsa-miR-628-5p | 10 | SLC5A3 | Sponge network | 1.271 | 0 | 0.493 | 5.0E-5 | 0.343 |
| 30885 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | NOTCH2NL | Sponge network | 0.389 | 0.04293 | 0.024 | 0.84307 | 0.343 |
| 30886 | LINC00472 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-877-5p | 11 | FLI1 | Sponge network | -0.842 | 0.01361 | -0.294 | 0.10905 | 0.343 |
| 30887 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-92a-3p | 12 | NRXN3 | Sponge network | 0.4 | 0.14611 | -0.893 | 0.04186 | 0.343 |
| 30888 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p | 10 | HIP1 | Sponge network | 1.153 | 0.00114 | 0.774 | 0 | 0.343 |
| 30889 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 25 | MOB1B | Sponge network | 0.569 | 0.00067 | 0.197 | 0.15266 | 0.343 |
| 30890 | HOXD-AS2 |
hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-338-3p | 11 | EYA4 | Sponge network | -0.208 | 0.65592 | 0.3 | 0.36876 | 0.343 |
| 30891 | THAP9-AS1 |
hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 15 | SLC5A3 | Sponge network | 1.079 | 0 | 0.493 | 5.0E-5 | 0.343 |
| 30892 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-421 | 14 | APC | Sponge network | -1.654 | 0.00144 | -0.255 | 0.04051 | 0.343 |
| 30893 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 19 | RCAN2 | Sponge network | -0.769 | 0.00035 | -0.33 | 0.15172 | 0.343 |
| 30894 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-940 | 22 | AFF4 | Sponge network | 0.993 | 0 | -0.266 | 0.01565 | 0.343 |
| 30895 | MALAT1 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-328-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-550a-5p;hsa-miR-628-5p | 20 | VPS53 | Sponge network | 0.782 | 0.00016 | 0.103 | 0.22667 | 0.343 |
| 30896 | LINC00094 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-92a-3p | 11 | CSDE1 | Sponge network | 0.253 | 0.09565 | -0.493 | 0 | 0.343 |
| 30897 | JAZF1-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | JDP2 | Sponge network | -0.769 | 0.00035 | -0.967 | 0 | 0.343 |
| 30898 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-582-5p | 13 | PIK3C2A | Sponge network | 1.093 | 0 | 0.061 | 0.53776 | 0.343 |
| 30899 | RP1-257A7.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-511-5p | 16 | LPP | Sponge network | 0.849 | 0.00178 | -0.459 | 0.04475 | 0.343 |
| 30900 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p | 10 | RBM5 | Sponge network | -0.567 | 0 | -0.205 | 0.0129 | 0.343 |
| 30901 | RP11-693J15.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-93-3p | 16 | UNC5C | Sponge network | -0.72 | 0.24239 | -0.178 | 0.40214 | 0.343 |
| 30902 | RP11-571M6.17 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-3p | 17 | FZD3 | Sponge network | 1.013 | 0 | 0.104 | 0.60316 | 0.343 |
| 30903 | RP1-151F17.2 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 15 | INPP4A | Sponge network | 0.222 | 0.29133 | 0.07 | 0.53563 | 0.343 |
| 30904 | GABPB1-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | CEP170 | Sponge network | 1.11 | 0 | 0.943 | 0 | 0.343 |
| 30905 | BZRAP1-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p | 11 | PTCH1 | Sponge network | -0.465 | 0.04703 | -0.1 | 0.6179 | 0.343 |
| 30906 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | FSTL5 | Sponge network | -0.355 | 0.0077 | -1.3 | 0.01619 | 0.343 |
| 30907 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 30 | CCND2 | Sponge network | -0.811 | 2.0E-5 | -0.267 | 0.3112 | 0.343 |
| 30908 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | EPAS1 | Sponge network | -0.563 | 0.02545 | -0.184 | 0.14418 | 0.343 |
| 30909 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 41 | PIK3R1 | Sponge network | -0.777 | 0.1134 | -0.133 | 0.36854 | 0.343 |
| 30910 | RP11-798G7.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-375 | 15 | NOTCH2 | Sponge network | 0.858 | 3.0E-5 | 0.138 | 0.35163 | 0.343 |
| 30911 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-7-1-3p | 12 | CLIP1 | Sponge network | 0.056 | 0.8494 | -0.215 | 0.08684 | 0.343 |
| 30912 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-214-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MXI1 | Sponge network | -0.031 | 0.89063 | -0.987 | 0 | 0.343 |
| 30913 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-590-3p | 14 | AKAP11 | Sponge network | 1.148 | 2.0E-5 | 0.196 | 0.09825 | 0.343 |
| 30914 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-590-3p | 20 | MYBL1 | Sponge network | 0.214 | 0.18246 | 0.494 | 0.02152 | 0.343 |
| 30915 | DNAJC27-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 39 | TGFBR3 | Sponge network | -0.969 | 2.0E-5 | -1.173 | 1.0E-5 | 0.343 |
| 30916 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 19 | AKAP6 | Sponge network | -0.125 | 0.49414 | -1.586 | 3.0E-5 | 0.343 |
| 30917 | AC005618.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-92a-3p | 41 | DST | Sponge network | 0.862 | 0.06213 | -0.713 | 0.00096 | 0.343 |
| 30918 | ACTA2-AS1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | SH3GLB1 | Sponge network | 0.194 | 0.64278 | -0.115 | 0.14773 | 0.343 |
| 30919 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 17 | TGFBR2 | Sponge network | -2.117 | 0.00161 | -0.243 | 0.11408 | 0.343 |
| 30920 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 33 | FOXO3 | Sponge network | 0.421 | 0.00084 | -0.04 | 0.72028 | 0.343 |
| 30921 | AC108488.4 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 34 | QKI | Sponge network | 0.259 | 0.12542 | -0.333 | 0.04111 | 0.343 |
| 30922 | DNAJC27-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 13 | DCN | Sponge network | -0.969 | 2.0E-5 | -1.288 | 0 | 0.343 |
| 30923 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 51 | WDFY3 | Sponge network | -0.727 | 0.04726 | -0.201 | 0.0967 | 0.343 |
| 30924 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -0.227 | 0.31657 | -0.243 | 0.11408 | 0.343 |
| 30925 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 15 | PHC3 | Sponge network | 2.368 | 2.0E-5 | 0.212 | 0.0499 | 0.343 |
| 30926 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p | 17 | RIMBP2 | Sponge network | -0.405 | 0.22013 | -1.968 | 5.0E-5 | 0.343 |
| 30927 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 38 | PDGFRA | Sponge network | -0.096 | 0.67958 | -0.372 | 0.0037 | 0.343 |
| 30928 | RP11-819C21.1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-379-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | SH3GLB1 | Sponge network | 0.537 | 0.00139 | -0.115 | 0.14773 | 0.343 |
| 30929 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p | 18 | TGFBR2 | Sponge network | 0.194 | 0.64278 | -0.243 | 0.11408 | 0.343 |
| 30930 | HCG11 |
hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | GLI3 | Sponge network | 0.385 | 0.10067 | -0.615 | 0.01482 | 0.343 |
| 30931 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-542-3p;hsa-miR-93-5p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | -1.281 | 0.00012 | -0.085 | 0.42389 | 0.343 |
| 30932 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-590-3p | 12 | RBM5 | Sponge network | 0.222 | 0.29133 | -0.205 | 0.0129 | 0.343 |
| 30933 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 36 | AKAP11 | Sponge network | 0.997 | 0.00066 | 0.196 | 0.09825 | 0.343 |
| 30934 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-744-5p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | 0.4 | 0.14611 | -0.509 | 0.03957 | 0.343 |
| 30935 | RP11-359E10.1 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-3p;hsa-miR-582-5p | 16 | AFF3 | Sponge network | 0.299 | 0.57722 | -2.373 | 0 | 0.343 |
| 30936 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 24 | ARNT2 | Sponge network | -0.576 | 0.10121 | -0.332 | 0.25851 | 0.343 |
| 30937 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-362-3p | 11 | DLC1 | Sponge network | -2.549 | 0.00528 | -0.382 | 0.05038 | 0.343 |
| 30938 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | AKAP6 | Sponge network | 0.349 | 0.01695 | -1.586 | 3.0E-5 | 0.343 |
| 30939 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-93-5p | 11 | TRIM3 | Sponge network | -0.18 | 0.20343 | -0.577 | 0 | 0.343 |
| 30940 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 25 | ST6GAL2 | Sponge network | 0.385 | 0.10067 | -1.201 | 0.00597 | 0.343 |
| 30941 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-654-3p | 22 | MYBL1 | Sponge network | 0.385 | 0.10067 | 0.494 | 0.02152 | 0.343 |
| 30942 | NR2F1-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p | 10 | PXDN | Sponge network | -0.49 | 0.04946 | 1.038 | 0 | 0.343 |
| 30943 | TINCR |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-454-3p | 10 | TP63 | Sponge network | -0.664 | 0.14543 | -0.363 | 0.38191 | 0.343 |
| 30944 | RP11-597D13.9 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-429;hsa-miR-455-5p | 16 | PLXNC1 | Sponge network | 1.302 | 7.0E-5 | 0.569 | 0.00329 | 0.343 |
| 30945 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p | 20 | QKI | Sponge network | 0.497 | 0.01451 | -0.333 | 0.04111 | 0.343 |
| 30946 | ADAMTS9-AS2 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-96-5p | 13 | PRKACA | Sponge network | -2.452 | 0 | -0.255 | 0.0012 | 0.343 |
| 30947 | CTC-429P9.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 39 | DST | Sponge network | 0.082 | 0.55397 | -0.713 | 0.00096 | 0.343 |
| 30948 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-629-3p | 23 | ZBTB4 | Sponge network | -0.141 | 0.26434 | -0.739 | 0 | 0.343 |
| 30949 | RP11-119F7.5 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | SFRP1 | Sponge network | 0.225 | 0.30644 | -3.566 | 0 | 0.343 |
| 30950 | RP11-418J17.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 14 | RICTOR | Sponge network | 0.998 | 0 | 0.388 | 0.00188 | 0.343 |
| 30951 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | TLL1 | Sponge network | -0.08 | 0.90092 | -1.195 | 3.0E-5 | 0.343 |
| 30952 | CTD-2260A17.1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-3607-3p;hsa-miR-940 | 10 | AFF4 | Sponge network | 0.518 | 0.01048 | -0.266 | 0.01565 | 0.343 |
| 30953 | CTC-296K1.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-493-5p | 16 | LRIG1 | Sponge network | -2.095 | 0.00112 | -0.537 | 0.00588 | 0.343 |
| 30954 | ARHGAP5-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-21-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 16 | AKAP6 | Sponge network | -0.644 | 0.00021 | -1.586 | 3.0E-5 | 0.343 |
| 30955 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | TRPS1 | Sponge network | 0.299 | 0.57722 | -0.191 | 0.44109 | 0.342 |
| 30956 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 26 | DIP2C | Sponge network | 0.603 | 0.00127 | -0.436 | 0.01713 | 0.342 |
| 30957 | RP11-822E23.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p | 18 | ARNT2 | Sponge network | -0.777 | 0.16667 | -0.332 | 0.25851 | 0.342 |
| 30958 | RP11-356J5.12 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | MED12L | Sponge network | -0.899 | 6.0E-5 | -0.194 | 0.55299 | 0.342 |
| 30959 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 22 | ABI2 | Sponge network | 1.089 | 0 | 0.175 | 0.14787 | 0.342 |
| 30960 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-501-5p;hsa-miR-589-3p | 17 | MAFB | Sponge network | -0.777 | 0.16667 | -0.023 | 0.89865 | 0.342 |
| 30961 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-93-5p | 23 | MAF | Sponge network | -0.318 | 0.65847 | -1.257 | 0 | 0.342 |
| 30962 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-192-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 12 | CHD2 | Sponge network | 0.174 | 0.30034 | 0.049 | 0.55371 | 0.342 |
| 30963 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 19 | THBS1 | Sponge network | 0.619 | 0.00157 | 0.741 | 0.00386 | 0.342 |
| 30964 | MALAT1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | ERCC4 | Sponge network | 0.782 | 0.00016 | 0.126 | 0.15266 | 0.342 |
| 30965 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p | 23 | CREB3L2 | Sponge network | -4.477 | 0 | -0.525 | 2.0E-5 | 0.342 |
| 30966 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 24 | FAT3 | Sponge network | 0.497 | 0.01451 | -1.601 | 0.00051 | 0.342 |
| 30967 | RP11-98I9.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | BRD3 | Sponge network | 0.67 | 0.00048 | 0.37 | 0.00013 | 0.342 |
| 30968 | CTD-3064M3.3 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-629-3p | 10 | ID4 | Sponge network | -0.469 | 0.08721 | -1.644 | 0 | 0.342 |
| 30969 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 16 | CXXC4 | Sponge network | -0.358 | 0.17255 | -0.433 | 0.21055 | 0.342 |
| 30970 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 10 | ADARB1 | Sponge network | 1.46 | 1.0E-5 | -0.335 | 0.06631 | 0.342 |
| 30971 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-655-3p | 12 | ARID2 | Sponge network | 1.601 | 0 | 0.235 | 0.02697 | 0.342 |
| 30972 | RP11-57H14.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-424-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | WWTR1 | Sponge network | 0.083 | 0.66405 | -0.345 | 0.11997 | 0.342 |
| 30973 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p | 25 | PRDM2 | Sponge network | -0.563 | 0.02545 | -0.258 | 0.00721 | 0.342 |
| 30974 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-424-3p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-942-5p | 28 | NRXN1 | Sponge network | -1.047 | 0 | -3.778 | 0 | 0.342 |
| 30975 | DNM1P35 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-628-5p | 34 | DTNA | Sponge network | 0.051 | 0.83388 | -1.648 | 1.0E-5 | 0.342 |
| 30976 | RP11-767N6.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-590-5p | 14 | ACVR2A | Sponge network | 0.466 | 0.00155 | -0.409 | 0.00039 | 0.342 |
| 30977 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | DMTF1 | Sponge network | -0.612 | 0.00094 | 0.248 | 0.02726 | 0.342 |
| 30978 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-589-3p | 13 | RELN | Sponge network | 0.056 | 0.81195 | -2.403 | 0 | 0.342 |
| 30979 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-7-1-3p | 16 | CLIP1 | Sponge network | -0.842 | 0.01361 | -0.215 | 0.08684 | 0.342 |
| 30980 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-671-5p;hsa-miR-96-5p | 39 | FOXP1 | Sponge network | 0.331 | 0.57247 | 0.042 | 0.74368 | 0.342 |
| 30981 | LINC00094 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-92a-3p | 11 | PREX2 | Sponge network | 0.253 | 0.09565 | -0.272 | 0.25382 | 0.342 |
| 30982 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p | 26 | ZFHX3 | Sponge network | -2.69 | 0.0001 | 0.389 | 0.01363 | 0.342 |
| 30983 | MRVI1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p | 16 | ERCC4 | Sponge network | -1.092 | 4.0E-5 | 0.126 | 0.15266 | 0.342 |
| 30984 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ABCC9 | Sponge network | 0.222 | 0.29133 | -0.466 | 0.14121 | 0.342 |
| 30985 | OIP5-AS1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 21 | RPS6KA3 | Sponge network | 0.421 | 0.00084 | 0.256 | 0.02419 | 0.342 |
| 30986 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-5p;hsa-miR-629-3p;hsa-miR-652-3p | 14 | ZBTB4 | Sponge network | -1.375 | 0.08642 | -0.739 | 0 | 0.342 |
| 30987 | PWAR6 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-629-3p | 15 | ITGA5 | Sponge network | -1.575 | 6.0E-5 | 0.452 | 0.03524 | 0.342 |
| 30988 | HOXD-AS2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-455-3p | 12 | PLSCR4 | Sponge network | -0.208 | 0.65592 | -0.474 | 0.03833 | 0.342 |
| 30989 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | 0.219 | 0.1516 | -1.751 | 1.0E-5 | 0.342 |
| 30990 | RP4-613B23.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-539-5p | 18 | BTG2 | Sponge network | -0.309 | 0.1145 | -1.763 | 0 | 0.342 |
| 30991 | AGAP11 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p | 10 | ALDH1A2 | Sponge network | -1.645 | 0 | -2.411 | 0 | 0.342 |
| 30992 | DLGAP1-AS2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p | 13 | RICTOR | Sponge network | 2.406 | 0 | 0.388 | 0.00188 | 0.342 |
| 30993 | GNG12-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 11 | NRG3 | Sponge network | -0.793 | 7.0E-5 | -1.836 | 4.0E-5 | 0.342 |
| 30994 | CTB-179K24.3 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 16 | TBL1XR1 | Sponge network | 0.841 | 0.04055 | 0.578 | 0 | 0.342 |
| 30995 | RP11-356J5.12 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | LIMCH1 | Sponge network | -0.899 | 6.0E-5 | -0.915 | 1.0E-5 | 0.342 |
| 30996 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | ACVR1 | Sponge network | -0.769 | 0.00035 | 0.279 | 0.00234 | 0.342 |
| 30997 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-96-5p | 32 | IL17RD | Sponge network | 0.311 | 0.10344 | -0.019 | 0.94873 | 0.342 |
| 30998 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-93-5p | 23 | AR | Sponge network | 0.497 | 0.01451 | -1.554 | 0 | 0.342 |
| 30999 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-93-5p | 26 | LRP1B | Sponge network | 0.056 | 0.81195 | -2.163 | 0 | 0.342 |
| 31000 | RP11-473I1.5 |
hsa-miR-125a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p | 15 | CCDC6 | Sponge network | 0.542 | 0.00054 | 0.27 | 0.02555 | 0.342 |
| 31001 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 19 | LRRC7 | Sponge network | -0.842 | 0.01361 | -1.341 | 0.01193 | 0.342 |
| 31002 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | MED12L | Sponge network | -0.222 | 0.32445 | -0.194 | 0.55299 | 0.342 |
| 31003 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-98-5p | 21 | USP12 | Sponge network | 0.082 | 0.55397 | -0.102 | 0.40768 | 0.342 |
| 31004 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p | 11 | GLI3 | Sponge network | -0.513 | 0.04703 | -0.615 | 0.01482 | 0.342 |
| 31005 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | LRRC55 | Sponge network | 0.246 | 0.59912 | -0.026 | 0.93163 | 0.342 |
| 31006 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | GALNT13 | Sponge network | -0.969 | 2.0E-5 | -0.857 | 0.07439 | 0.342 |
| 31007 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-424-5p;hsa-miR-7-1-3p | 13 | USP12 | Sponge network | -0.187 | 0.23787 | -0.102 | 0.40768 | 0.342 |
| 31008 | AC156455.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p | 25 | RUNX1T1 | Sponge network | 1.154 | 0.00041 | -0.344 | 0.2374 | 0.342 |
| 31009 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-935 | 23 | EBF1 | Sponge network | -0.738 | 0.21233 | -0.384 | 0.08856 | 0.342 |
| 31010 | TP73-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 16 | POU2F2 | Sponge network | -0.563 | 0.02545 | -0.233 | 0.32214 | 0.342 |
| 31011 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-93-5p | 19 | SLITRK3 | Sponge network | -0.303 | 0.21002 | -3.328 | 0 | 0.342 |
| 31012 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p | 10 | HERC2 | Sponge network | -0.162 | 0.47687 | 0.114 | 0.30638 | 0.342 |
| 31013 | LINC00476 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 11 | NRG3 | Sponge network | -0.615 | 0.00015 | -1.836 | 4.0E-5 | 0.342 |
| 31014 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 15 | FGF5 | Sponge network | -0.611 | 0.09607 | 0.549 | 0.28659 | 0.342 |
| 31015 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-671-5p | 20 | KIT | Sponge network | -0.465 | 0.04703 | -2.184 | 0 | 0.342 |
| 31016 | U91328.19 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p | 24 | DLC1 | Sponge network | -0.303 | 0.21002 | -0.382 | 0.05038 | 0.342 |
| 31017 | AC074289.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-940;hsa-miR-96-5p | 24 | SNX29 | Sponge network | -0.326 | 0.14314 | -0.34 | 0.01371 | 0.342 |
| 31018 | LINC00702 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | FAM117A | Sponge network | -0.737 | 0.06182 | -1.379 | 0 | 0.342 |
| 31019 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 22 | LIFR | Sponge network | 0.4 | 0.14611 | -1.794 | 0 | 0.342 |
| 31020 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-5p | 13 | FAM188A | Sponge network | -1.645 | 0 | -0.44 | 0.00018 | 0.342 |
| 31021 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 17 | SYNE1 | Sponge network | 0.614 | 1.0E-5 | -0.738 | 0.00316 | 0.342 |
| 31022 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 16 | BCL11A | Sponge network | -0.246 | 0.35034 | -0.932 | 0.0063 | 0.342 |
| 31023 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | PRRX1 | Sponge network | -0.421 | 0.0114 | 1.316 | 0 | 0.342 |
| 31024 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | 0.082 | 0.55654 | -1.751 | 1.0E-5 | 0.342 |
| 31025 | RP11-774O3.3 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-590-3p | 10 | RBM6 | Sponge network | -0.487 | 0.02157 | -0.137 | 0.15663 | 0.342 |
| 31026 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-96-5p | 25 | PRKCE | Sponge network | -1.439 | 4.0E-5 | -0.403 | 0.00056 | 0.342 |
| 31027 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-145-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 12 | MOB1B | Sponge network | 1.11 | 1.0E-5 | 0.197 | 0.15266 | 0.342 |
| 31028 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 12 | DKK3 | Sponge network | -0.141 | 0.26434 | -0.052 | 0.77783 | 0.342 |
| 31029 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p | 14 | TRIM33 | Sponge network | 0.643 | 0 | 0.402 | 7.0E-5 | 0.342 |
| 31030 | GNG12-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 12 | UVRAG | Sponge network | -0.793 | 7.0E-5 | -0.017 | 0.83865 | 0.342 |
| 31031 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 15 | ITGB3 | Sponge network | -0.899 | 6.0E-5 | -0.011 | 0.95751 | 0.342 |
| 31032 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 40 | ETV1 | Sponge network | -2.46 | 0 | -0.096 | 0.67674 | 0.342 |
| 31033 | OIP5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p | 14 | EZH1 | Sponge network | 0.421 | 0.00084 | -0.564 | 1.0E-5 | 0.342 |
| 31034 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | RBL2 | Sponge network | -0.421 | 0.0114 | -0.282 | 0.00254 | 0.342 |
| 31035 | AGAP11 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-30d-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-629-3p | 12 | RHOB | Sponge network | -1.645 | 0 | -1.052 | 0 | 0.342 |
| 31036 | LPP-AS2 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p | 21 | PCDH9 | Sponge network | -0.18 | 0.20343 | -1.823 | 4.0E-5 | 0.342 |
| 31037 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | APC | Sponge network | 0.545 | 0.00064 | -0.255 | 0.04051 | 0.342 |
| 31038 | AC009948.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | GALNT13 | Sponge network | 0.532 | 0.00505 | -0.857 | 0.07439 | 0.342 |
| 31039 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-539-5p | 19 | PBX1 | Sponge network | 0.422 | 0.27021 | -1.206 | 0 | 0.342 |
| 31040 | RAMP2-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | INPP4A | Sponge network | -0.513 | 0.04703 | 0.07 | 0.53563 | 0.342 |
| 31041 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | NEDD4 | Sponge network | -0.405 | 0.22013 | 0.02 | 0.89834 | 0.342 |
| 31042 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-5p | 17 | RNASEL | Sponge network | 1.302 | 7.0E-5 | -0.272 | 0.01796 | 0.342 |
| 31043 | AC159540.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-3607-3p | 11 | MOB1B | Sponge network | 1.634 | 0 | 0.197 | 0.15266 | 0.342 |
| 31044 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 41 | NTRK3 | Sponge network | 0.657 | 4.0E-5 | -1.77 | 3.0E-5 | 0.342 |
| 31045 | LINC00672 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | SIRT1 | Sponge network | -0.227 | 0.31657 | 0.109 | 0.2593 | 0.342 |
| 31046 | HCG11 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | NCAM2 | Sponge network | 0.385 | 0.10067 | -0.482 | 0.22565 | 0.342 |
| 31047 | RP3-402G11.26 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 15 | DTNA | Sponge network | -0.254 | 0.19303 | -1.648 | 1.0E-5 | 0.342 |
| 31048 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | FGFR1 | Sponge network | 0.727 | 3.0E-5 | -0.509 | 0.03957 | 0.342 |
| 31049 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-590-3p | 13 | JAZF1 | Sponge network | 1.757 | 0 | -0.377 | 0.02677 | 0.342 |
| 31050 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 15 | PRKAA1 | Sponge network | 0.959 | 0 | 0.317 | 0.0031 | 0.342 |
| 31051 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | ABI2 | Sponge network | -0.206 | 0.12942 | 0.175 | 0.14787 | 0.342 |
| 31052 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-96-5p | 16 | MITF | Sponge network | -1.654 | 0.00144 | -0.769 | 0.00022 | 0.342 |
| 31053 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p | 13 | PRDM5 | Sponge network | 0.225 | 0.30644 | -1.09 | 0.00014 | 0.342 |
| 31054 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 44 | ZFHX4 | Sponge network | -0.303 | 0.21002 | 0.018 | 0.95574 | 0.342 |
| 31055 | RAB30-AS1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-342-3p | 11 | BRD3 | Sponge network | 0.752 | 0 | 0.37 | 0.00013 | 0.342 |
| 31056 | A2M-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 11 | KLF10 | Sponge network | -0.08 | 0.90092 | -0.783 | 0 | 0.342 |
| 31057 | LIPE-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-625-5p | 16 | PTPN14 | Sponge network | 0.078 | 0.55803 | 0.144 | 0.34595 | 0.342 |
| 31058 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 38 | KIF1B | Sponge network | 0.889 | 9.0E-5 | -0.118 | 0.32258 | 0.342 |
| 31059 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 11 | ABCA10 | Sponge network | 0.494 | 0.00339 | -0.571 | 0.03674 | 0.342 |
| 31060 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 23 | CDHR3 | Sponge network | -0.49 | 0.04946 | -0.977 | 4.0E-5 | 0.342 |
| 31061 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 14 | EPAS1 | Sponge network | -0.487 | 0.02157 | -0.184 | 0.14418 | 0.342 |
| 31062 | GS1-124K5.11 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 10 | PREX2 | Sponge network | 0.494 | 0.00339 | -0.272 | 0.25382 | 0.342 |
| 31063 | WDFY3-AS2 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | JDP2 | Sponge network | -0.942 | 0 | -0.967 | 0 | 0.342 |
| 31064 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-940 | 13 | FAM172A | Sponge network | -0.563 | 0.02545 | -0.57 | 0 | 0.342 |
| 31065 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 50 | PAFAH1B1 | Sponge network | -1.697 | 0.00889 | -0.429 | 0 | 0.342 |
| 31066 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 43 | EPHA7 | Sponge network | 0.219 | 0.1516 | -2.197 | 3.0E-5 | 0.342 |
| 31067 | LINC00476 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | GLI3 | Sponge network | -0.615 | 0.00015 | -0.615 | 0.01482 | 0.342 |
| 31068 | CTD-2245F17.3 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-339-5p;hsa-miR-539-5p;hsa-miR-942-5p | 17 | THRB | Sponge network | -0.421 | 0.12556 | -1.117 | 0.0004 | 0.342 |
| 31069 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 16 | MYO9A | Sponge network | 0.476 | 0.19203 | -0.147 | 0.32373 | 0.342 |
| 31070 | RP5-998N21.10 |
hsa-let-7g-3p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-671-5p | 11 | GAS7 | Sponge network | -0.098 | 0.76298 | -0.555 | 0.01602 | 0.342 |
| 31071 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 36 | SASH1 | Sponge network | 0.064 | 0.72934 | -0.502 | 0.00175 | 0.342 |
| 31072 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | TP53INP1 | Sponge network | -0.487 | 0.02157 | -0.496 | 0.00064 | 0.342 |
| 31073 | RP11-535M15.1 |
hsa-miR-10a-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-362-3p | 11 | RASSF2 | Sponge network | -1.14 | 0.03513 | -0.273 | 0.21961 | 0.342 |
| 31074 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-454-3p;hsa-miR-590-3p | 10 | RB1CC1 | Sponge network | 0.336 | 0.00739 | 0.175 | 0.12559 | 0.342 |
| 31075 | RP5-994D16.9 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 11 | EPHA3 | Sponge network | 0.252 | 0.08508 | 0.185 | 0.53588 | 0.342 |
| 31076 | RP11-48B3.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-590-3p | 12 | ASTN1 | Sponge network | 1.757 | 0 | -2.389 | 2.0E-5 | 0.342 |
| 31077 | PART1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | RET | Sponge network | -2.925 | 0 | -0.833 | 0.03517 | 0.342 |
| 31078 | ADIRF-AS1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 42 | DTNA | Sponge network | -0.223 | 0.47816 | -1.648 | 1.0E-5 | 0.342 |
| 31079 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 39 | ZFHX3 | Sponge network | -0.355 | 0.0077 | 0.389 | 0.01363 | 0.342 |
| 31080 | RP5-1198O20.4 |
hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SH3GLB1 | Sponge network | 0.997 | 0.00066 | -0.115 | 0.14773 | 0.342 |
| 31081 | GABPB1-AS1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-363-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p | 14 | TRIM13 | Sponge network | 1.11 | 0 | 0.183 | 0.06968 | 0.342 |
| 31082 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 38 | PRKAB2 | Sponge network | -0.49 | 0.04946 | -0.367 | 0.01371 | 0.342 |
| 31083 | RP11-798G7.7 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-484;hsa-miR-625-5p | 24 | CBX5 | Sponge network | 0.858 | 3.0E-5 | 0.165 | 0.24446 | 0.342 |
| 31084 | RP11-473I1.5 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p | 19 | ZFHX3 | Sponge network | 0.542 | 0.00054 | 0.389 | 0.01363 | 0.342 |
| 31085 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 58 | MDM4 | Sponge network | 0.72 | 0.0001 | 0.345 | 0.00762 | 0.342 |
| 31086 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-7-1-3p | 21 | PRKAB2 | Sponge network | 0.389 | 0.04293 | -0.367 | 0.01371 | 0.342 |
| 31087 | AC008746.12 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p | 12 | RICTOR | Sponge network | 2.279 | 0 | 0.388 | 0.00188 | 0.342 |
| 31088 | RP11-506M13.3 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 11 | CDK6 | Sponge network | 0.769 | 0 | 0.991 | 0 | 0.342 |
| 31089 | BCYRN1 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-5p | 10 | MLLT11 | Sponge network | 0.311 | 0.10344 | -0.661 | 0.01733 | 0.342 |
| 31090 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | PAFAH1B1 | Sponge network | -0.726 | 0.00132 | -0.429 | 0 | 0.342 |
| 31091 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-940;hsa-miR-96-5p | 39 | QKI | Sponge network | 0.331 | 0.57247 | -0.333 | 0.04111 | 0.342 |
| 31092 | FAM157B |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-5p | 14 | WDFY3 | Sponge network | 0.92 | 0 | -0.201 | 0.0967 | 0.342 |
| 31093 | RP11-327P2.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-628-5p | 44 | DTNA | Sponge network | -0.147 | 0.40452 | -1.648 | 1.0E-5 | 0.342 |
| 31094 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-29b-1-5p;hsa-miR-331-5p | 11 | WDFY3 | Sponge network | -0.326 | 0.28809 | -0.201 | 0.0967 | 0.342 |
| 31095 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 22 | ZFHX3 | Sponge network | -4.546 | 0 | 0.389 | 0.01363 | 0.342 |
| 31096 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-576-5p | 21 | CACNA2D1 | Sponge network | 0.253 | 0.09565 | -0.846 | 0.00675 | 0.342 |
| 31097 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | CBX5 | Sponge network | 0.349 | 0.01695 | 0.165 | 0.24446 | 0.342 |
| 31098 | RP11-83A24.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 14 | ZMYND11 | Sponge network | 0.417 | 0.00445 | -0.2 | 0.05449 | 0.342 |
| 31099 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 31 | ADARB1 | Sponge network | -0.031 | 0.89063 | -0.335 | 0.06631 | 0.342 |
| 31100 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-92a-3p | 14 | ITGA5 | Sponge network | -0.323 | 0.01372 | 0.452 | 0.03524 | 0.342 |
| 31101 | LINC00641 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | -0.222 | 0.32445 | -1.628 | 0 | 0.342 |
| 31102 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CLIP1 | Sponge network | 1.153 | 0 | -0.215 | 0.08684 | 0.342 |
| 31103 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 15 | MAP3K12 | Sponge network | -0.769 | 0.00035 | -0.057 | 0.59369 | 0.342 |
| 31104 | PCA3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ZMYND11 | Sponge network | -0.709 | 0.18168 | -0.2 | 0.05449 | 0.342 |
| 31105 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 20 | MYO9A | Sponge network | -0.223 | 0.47816 | -0.147 | 0.32373 | 0.342 |
| 31106 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-629-3p | 26 | AR | Sponge network | 2.306 | 9.0E-5 | -1.554 | 0 | 0.342 |
| 31107 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-629-3p | 18 | AR | Sponge network | -1.054 | 0.0706 | -1.554 | 0 | 0.342 |
| 31108 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 22 | LRRC7 | Sponge network | -0.83 | 0 | -1.341 | 0.01193 | 0.342 |
| 31109 | MIR600HG |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-655-3p | 16 | UBE4B | Sponge network | 0.594 | 0.41522 | -0.235 | 0.007 | 0.342 |
| 31110 | RP11-244H3.1 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 11 | CNTNAP1 | Sponge network | 0.659 | 3.0E-5 | 0.358 | 0.13727 | 0.342 |
| 31111 | EMX2OS |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | PRKCB | Sponge network | -0.777 | 0.1134 | -1.313 | 2.0E-5 | 0.342 |
| 31112 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-96-5p | 21 | IL17RD | Sponge network | 0.619 | 0.00157 | -0.019 | 0.94873 | 0.342 |
| 31113 | MIR497HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-96-5p | 17 | RIMBP2 | Sponge network | -1.439 | 4.0E-5 | -1.968 | 5.0E-5 | 0.342 |
| 31114 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 12 | GIGYF2 | Sponge network | 0.349 | 0.01695 | 0.136 | 0.04403 | 0.342 |
| 31115 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | PRDM2 | Sponge network | -0.355 | 0.0077 | -0.258 | 0.00721 | 0.342 |
| 31116 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 17 | SOX11 | Sponge network | 1.757 | 0 | 2.68 | 0 | 0.342 |
| 31117 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-486-5p | 12 | DACT1 | Sponge network | -2.62 | 0 | 0.783 | 0.00072 | 0.342 |
| 31118 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p | 17 | CNTNAP3 | Sponge network | -0.323 | 0.01372 | -1.465 | 0.00033 | 0.342 |
| 31119 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-7-5p | 23 | ATRX | Sponge network | -0.384 | 0.40652 | 0.261 | 0.04408 | 0.342 |
| 31120 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DACT1 | Sponge network | 0.619 | 0.00157 | 0.783 | 0.00072 | 0.342 |
| 31121 | RP11-67L2.2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 27 | DLC1 | Sponge network | 0.72 | 0.0001 | -0.382 | 0.05038 | 0.342 |
| 31122 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | MLLT10 | Sponge network | 1.04 | 0.00023 | 0.105 | 0.33292 | 0.342 |
| 31123 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 47 | NOTCH2 | Sponge network | -1.697 | 0.00889 | 0.138 | 0.35163 | 0.342 |
| 31124 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-93-5p | 30 | TACC1 | Sponge network | -0.663 | 0.10887 | -0.362 | 0.05406 | 0.342 |
| 31125 | LINC00672 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | PDE4DIP | Sponge network | -0.227 | 0.31657 | -0.282 | 0.0257 | 0.342 |
| 31126 | LINC00094 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-93-5p | 44 | PPM1L | Sponge network | 0.253 | 0.09565 | -0.537 | 0.00616 | 0.342 |
| 31127 | CTA-363E19.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-495-3p;hsa-miR-550a-5p | 18 | CCDC6 | Sponge network | 1.055 | 0 | 0.27 | 0.02555 | 0.342 |
| 31128 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-942-5p | 31 | NOTCH2 | Sponge network | 0.082 | 0.55654 | 0.138 | 0.35163 | 0.342 |
| 31129 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-7-1-3p | 41 | DTNA | Sponge network | 0.853 | 0.15352 | -1.648 | 1.0E-5 | 0.342 |
| 31130 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | DAB2 | Sponge network | -0.901 | 0.00019 | 0.431 | 0.0113 | 0.342 |
| 31131 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-577 | 11 | SYNE1 | Sponge network | 1.294 | 0.0031 | -0.738 | 0.00316 | 0.342 |
| 31132 | TP73-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-92a-3p | 27 | BTG2 | Sponge network | -0.563 | 0.02545 | -1.763 | 0 | 0.342 |
| 31133 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 41 | CADM2 | Sponge network | 0.299 | 0.57722 | -3.782 | 0 | 0.342 |
| 31134 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-375;hsa-miR-424-5p | 17 | MYBL1 | Sponge network | 0.622 | 0.00015 | 0.494 | 0.02152 | 0.342 |
| 31135 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 12 | NALCN | Sponge network | 0.064 | 0.72934 | 1.068 | 0.00237 | 0.342 |
| 31136 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-96-5p | 27 | NRXN1 | Sponge network | -0.18 | 0.20343 | -3.778 | 0 | 0.342 |
| 31137 | RP11-384K6.6 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p;hsa-miR-654-3p | 11 | PHF6 | Sponge network | 0.946 | 0 | 0.785 | 0 | 0.342 |
| 31138 | TRAF3IP2-AS1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PHF3 | Sponge network | -0.323 | 0.01372 | 0.126 | 0.19652 | 0.342 |
| 31139 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p | 21 | HECA | Sponge network | 0.331 | 0.57247 | -0.217 | 0.03463 | 0.342 |
| 31140 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-450b-5p | 12 | JDP2 | Sponge network | -0.583 | 0.14589 | -0.967 | 0 | 0.342 |
| 31141 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | MXI1 | Sponge network | 0.101 | 0.60919 | -0.987 | 0 | 0.342 |
| 31142 | RP4-613B23.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p | 10 | HIP1 | Sponge network | -0.309 | 0.1145 | 0.774 | 0 | 0.342 |
| 31143 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p | 10 | SASH1 | Sponge network | 1.344 | 0 | -0.502 | 0.00175 | 0.342 |
| 31144 | RP11-403I13.8 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-96-5p | 32 | GAS7 | Sponge network | 0.619 | 0.00157 | -0.555 | 0.01602 | 0.342 |
| 31145 | RP5-1198O20.4 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 19 | IRS1 | Sponge network | 0.997 | 0.00066 | -0.026 | 0.88855 | 0.342 |
| 31146 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 15 | CRTC3 | Sponge network | 0.556 | 0.00298 | 0.164 | 0.16786 | 0.342 |
| 31147 | RP11-13K12.5 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-629-3p | 22 | PTPN14 | Sponge network | -0.318 | 0.65847 | 0.144 | 0.34595 | 0.342 |
| 31148 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-503-5p | 24 | RICTOR | Sponge network | 0.622 | 0.00015 | 0.388 | 0.00188 | 0.342 |
| 31149 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 40 | BMPR2 | Sponge network | 0.705 | 0.00014 | 0.206 | 0.0635 | 0.342 |
| 31150 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 47 | ZFHX4 | Sponge network | -0.222 | 0.32445 | 0.018 | 0.95574 | 0.342 |
| 31151 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | ERG | Sponge network | -2.46 | 0 | 0.002 | 0.99011 | 0.342 |
| 31152 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p | 23 | EPHA7 | Sponge network | 1.757 | 0 | -2.197 | 3.0E-5 | 0.342 |
| 31153 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p | 18 | MAFB | Sponge network | 0.194 | 0.64278 | -0.023 | 0.89865 | 0.342 |
| 31154 | AC005154.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 33 | ARHGEF12 | Sponge network | 0.848 | 0 | 0.09 | 0.35693 | 0.342 |
| 31155 | LINC01018 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PTGFR | Sponge network | -2.915 | 0.00015 | -0.233 | 0.45421 | 0.342 |
| 31156 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 21 | MDM4 | Sponge network | 1.46 | 1.0E-5 | 0.345 | 0.00762 | 0.342 |
| 31157 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 22 | ST6GAL2 | Sponge network | 0.862 | 0.06213 | -1.201 | 0.00597 | 0.342 |
| 31158 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-5p | 21 | ESR1 | Sponge network | -0.469 | 0.08721 | -1.035 | 3.0E-5 | 0.342 |
| 31159 | MRVI1-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 14 | PIK3CA | Sponge network | -1.092 | 4.0E-5 | 0.195 | 0.06908 | 0.341 |
| 31160 | TPTEP1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | CSDE1 | Sponge network | -1.216 | 0 | -0.493 | 0 | 0.341 |
| 31161 | AC034220.3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | ARHGEF12 | Sponge network | 0.309 | 0.07304 | 0.09 | 0.35693 | 0.341 |
| 31162 | RP11-678G14.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-7-1-3p | 17 | DIP2C | Sponge network | -4.477 | 0 | -0.436 | 0.01713 | 0.341 |
| 31163 | MAP3K14 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-7-5p | 13 | BTG2 | Sponge network | -0.295 | 0.02604 | -1.763 | 0 | 0.341 |
| 31164 | CTD-2245F17.3 |
hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-26b-3p;hsa-miR-539-5p;hsa-miR-629-3p | 10 | EPHA7 | Sponge network | -0.421 | 0.12556 | -2.197 | 3.0E-5 | 0.341 |
| 31165 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | PLA2R1 | Sponge network | -1.216 | 0 | -0.265 | 0.24676 | 0.341 |
| 31166 | LINC00473 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 22 | NAV3 | Sponge network | -1.203 | 0.03881 | 0.212 | 0.47729 | 0.341 |
| 31167 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-501-5p;hsa-miR-93-3p | 21 | EBF3 | Sponge network | -0.405 | 0.22013 | -0.058 | 0.81437 | 0.341 |
| 31168 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | JAZF1 | Sponge network | -0.969 | 2.0E-5 | -0.377 | 0.02677 | 0.341 |
| 31169 | ACTA2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | CREB1 | Sponge network | 0.194 | 0.64278 | 0.203 | 0.00537 | 0.341 |
| 31170 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-3p | 21 | RICTOR | Sponge network | 0.368 | 0.20093 | 0.388 | 0.00188 | 0.341 |
| 31171 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 27 | IGF1 | Sponge network | -0.738 | 0.21233 | -0.204 | 0.5898 | 0.341 |
| 31172 | MEG3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 14 | EHD3 | Sponge network | 0.249 | 0.35902 | -0.484 | 0.01747 | 0.341 |
| 31173 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CHD2 | Sponge network | 0.67 | 0.00048 | 0.049 | 0.55371 | 0.341 |
| 31174 | RP11-999E24.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-425-5p | 15 | LRIG1 | Sponge network | -0.693 | 0.05629 | -0.537 | 0.00588 | 0.341 |
| 31175 | RP11-156E6.1 |
hsa-miR-101-3p;hsa-miR-139-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | GSK3B | Sponge network | 1.266 | 0 | 0.266 | 0.00045 | 0.341 |
| 31176 | LINC00476 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | PDE4DIP | Sponge network | -0.615 | 0.00015 | -0.282 | 0.0257 | 0.341 |
| 31177 | FENDRR |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MLF1 | Sponge network | -1.281 | 0.00012 | -0.893 | 0.00727 | 0.341 |
| 31178 | RP11-53O19.3 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-9-5p | 12 | CDK6 | Sponge network | 1.205 | 0 | 0.991 | 0 | 0.341 |
| 31179 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-874-3p | 10 | BCLAF1 | Sponge network | 1.093 | 0 | 0.211 | 0.00684 | 0.341 |
| 31180 | LINC00654 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 19 | SFRP1 | Sponge network | 0.374 | 0.20054 | -3.566 | 0 | 0.341 |
| 31181 | RP11-6O2.3 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-96-5p | 10 | ZBTB20 | Sponge network | 0.331 | 0.57247 | -0.122 | 0.57569 | 0.341 |
| 31182 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p | 11 | CHD2 | Sponge network | 1.317 | 0 | 0.049 | 0.55371 | 0.341 |
| 31183 | HAND2-AS1 |
hsa-miR-148b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p | 10 | GNAI2 | Sponge network | -1.697 | 0.00889 | -0.063 | 0.54532 | 0.341 |
| 31184 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-9-5p | 17 | YAP1 | Sponge network | 0.873 | 0 | 0.22 | 0.11836 | 0.341 |
| 31185 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p | 24 | PTPN14 | Sponge network | -0.563 | 0.02545 | 0.144 | 0.34595 | 0.341 |
| 31186 | LINC00654 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-935 | 15 | RELN | Sponge network | 0.374 | 0.20054 | -2.403 | 0 | 0.341 |
| 31187 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | 0.782 | 0.00016 | 0.323 | 0.00792 | 0.341 |
| 31188 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 13 | SUFU | Sponge network | -0.187 | 0.23787 | -0.04 | 0.65489 | 0.341 |
| 31189 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 20 | NCOA1 | Sponge network | -0.358 | 0.17255 | -0.432 | 0 | 0.341 |
| 31190 | CTD-2303H24.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | CREB1 | Sponge network | 0.415 | 0.14657 | 0.203 | 0.00537 | 0.341 |
| 31191 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | LRRC4 | Sponge network | -2.117 | 0.00161 | -1.774 | 0 | 0.341 |
| 31192 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | CADM2 | Sponge network | -0.72 | 0.24239 | -3.782 | 0 | 0.341 |
| 31193 | SNX29P2 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 19 | PCDH17 | Sponge network | 0.4 | 0.14611 | 0.731 | 2.0E-5 | 0.341 |
| 31194 | LINC00472 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p | 13 | UNC5C | Sponge network | -0.842 | 0.01361 | -0.178 | 0.40214 | 0.341 |
| 31195 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 14 | SEPT6 | Sponge network | 0.101 | 0.60919 | -0.598 | 0.00181 | 0.341 |
| 31196 | RP11-254F7.2 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-590-5p | 10 | FMNL3 | Sponge network | 0.176 | 0.37402 | 0.555 | 4.0E-5 | 0.341 |
| 31197 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | FSTL5 | Sponge network | -0.942 | 0 | -1.3 | 0.01619 | 0.341 |
| 31198 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 18 | SLC5A3 | Sponge network | 1.601 | 0 | 0.493 | 5.0E-5 | 0.341 |
| 31199 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 17 | BRD3 | Sponge network | 0.529 | 0.00552 | 0.37 | 0.00013 | 0.341 |
| 31200 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-582-3p | 14 | RBMS3 | Sponge network | -0.31 | 0.33871 | -0.519 | 0.03551 | 0.341 |
| 31201 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-582-3p | 12 | CBL | Sponge network | 1.4 | 0 | 0.291 | 0.01267 | 0.341 |
| 31202 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-744-3p | 18 | HIP1 | Sponge network | -0.777 | 0.16667 | 0.774 | 0 | 0.341 |
| 31203 | FZD10-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | TPO | Sponge network | -0.727 | 0.04726 | -1.87 | 2.0E-5 | 0.341 |
| 31204 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SON | Sponge network | 0.659 | 3.0E-5 | 0.168 | 0.04406 | 0.341 |
| 31205 | RP11-6O2.3 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-590-5p | 30 | ARHGEF12 | Sponge network | 0.331 | 0.57247 | 0.09 | 0.35693 | 0.341 |
| 31206 | RP11-464F9.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-655-3p | 11 | ARID2 | Sponge network | 0.959 | 0 | 0.235 | 0.02697 | 0.341 |
| 31207 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-93-3p | 10 | EPHA3 | Sponge network | 1.46 | 1.0E-5 | 0.185 | 0.53588 | 0.341 |
| 31208 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-93-5p | 18 | PPM1A | Sponge network | 0.497 | 0.01451 | -0.623 | 0 | 0.341 |
| 31209 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 24 | CDHR3 | Sponge network | -1.111 | 0.0003 | -0.977 | 4.0E-5 | 0.341 |
| 31210 | PWAR6 |
hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 14 | FBXW7 | Sponge network | -1.575 | 6.0E-5 | -0.418 | 2.0E-5 | 0.341 |
| 31211 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-34a-5p | 19 | RNASEL | Sponge network | -1.331 | 3.0E-5 | -0.272 | 0.01796 | 0.341 |
| 31212 | TTC28-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-625-5p | 19 | ABI2 | Sponge network | 0.373 | 0.00068 | 0.175 | 0.14787 | 0.341 |
| 31213 | CTD-3138B18.6 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ABI2 | Sponge network | 2.248 | 0 | 0.175 | 0.14787 | 0.341 |
| 31214 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 22 | RASSF8 | Sponge network | -0.206 | 0.12942 | -0.122 | 0.65591 | 0.341 |
| 31215 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-664a-3p;hsa-miR-766-3p | 26 | AFF4 | Sponge network | 0.706 | 0 | -0.266 | 0.01565 | 0.341 |
| 31216 | AC108676.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-511-5p | 11 | ATRX | Sponge network | 4.346 | 2.0E-5 | 0.261 | 0.04408 | 0.341 |
| 31217 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PDS5B | Sponge network | 1.757 | 0 | 0.259 | 0.00962 | 0.341 |
| 31218 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 32 | MYO9A | Sponge network | -1.216 | 0 | -0.147 | 0.32373 | 0.341 |
| 31219 | CTC-429P9.5 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | RPS6KA6 | Sponge network | 0.178 | 0.29106 | -2.989 | 0 | 0.341 |
| 31220 | RP1-59D14.5 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 14 | FOXO3 | Sponge network | 1.162 | 5.0E-5 | -0.04 | 0.72028 | 0.341 |
| 31221 | HAR1A |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-940 | 24 | GAS7 | Sponge network | -0.672 | 0.19447 | -0.555 | 0.01602 | 0.341 |
| 31222 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PIK3CA | Sponge network | 0.61 | 0.01593 | 0.195 | 0.06908 | 0.341 |
| 31223 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | RET | Sponge network | -2.117 | 0.00161 | -0.833 | 0.03517 | 0.341 |
| 31224 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-455-5p;hsa-miR-590-3p | 22 | JAZF1 | Sponge network | -0.737 | 0.10822 | -0.377 | 0.02677 | 0.341 |
| 31225 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-940 | 24 | PBX1 | Sponge network | 0.392 | 0.0278 | -1.206 | 0 | 0.341 |
| 31226 | LINC00702 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 19 | ERRFI1 | Sponge network | -0.737 | 0.06182 | -0.798 | 1.0E-5 | 0.341 |
| 31227 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 30 | RBMS3 | Sponge network | -0.322 | 0.25945 | -0.519 | 0.03551 | 0.341 |
| 31228 | LINC00476 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | TLE4 | Sponge network | -0.615 | 0.00015 | -1.157 | 0 | 0.341 |
| 31229 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 10 | SEPT6 | Sponge network | 0.082 | 0.55654 | -0.598 | 0.00181 | 0.341 |
| 31230 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | TET1 | Sponge network | 0.238 | 0.70544 | -0.135 | 0.52138 | 0.341 |
| 31231 | CTC-296K1.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-708-5p | 13 | TOM1L2 | Sponge network | -2.076 | 0.00548 | -0.994 | 0 | 0.341 |
| 31232 | CTA-204B4.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 13 | HECA | Sponge network | 0.765 | 7.0E-5 | -0.217 | 0.03463 | 0.341 |
| 31233 | MIR22HG |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-708-3p | 11 | UNC5D | Sponge network | -1.047 | 0 | -3.646 | 0 | 0.341 |
| 31234 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-484 | 22 | CADM1 | Sponge network | -0.777 | 0.16667 | -0.9 | 0.00084 | 0.341 |
| 31235 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p | 20 | HIP1 | Sponge network | 0.421 | 0.00084 | 0.774 | 0 | 0.341 |
| 31236 | RP11-6N17.3 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-331-3p | 10 | ABI2 | Sponge network | 0.108 | 0.69556 | 0.175 | 0.14787 | 0.341 |
| 31237 | RP11-53O19.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 12 | INPP4A | Sponge network | -0.405 | 0.22013 | 0.07 | 0.53563 | 0.341 |
| 31238 | LINC00641 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 15 | SEPT6 | Sponge network | -0.222 | 0.32445 | -0.598 | 0.00181 | 0.341 |
| 31239 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-7-1-3p | 23 | RBMS3 | Sponge network | -4.477 | 0 | -0.519 | 0.03551 | 0.341 |
| 31240 | LINC01018 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CDH10 | Sponge network | -2.915 | 0.00015 | -2.397 | 0 | 0.341 |
| 31241 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 51 | WDFY3 | Sponge network | 0.385 | 0.10067 | -0.201 | 0.0967 | 0.341 |
| 31242 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-744-3p | 16 | IGF2 | Sponge network | -1.697 | 0.00889 | 0.848 | 0.03842 | 0.341 |
| 31243 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-625-5p | 15 | TMTC2 | Sponge network | -0.162 | 0.47687 | -0.215 | 0.18248 | 0.341 |
| 31244 | LINC00403 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-452-5p | 12 | CADM2 | Sponge network | -3.586 | 0.00032 | -3.782 | 0 | 0.341 |
| 31245 | RP11-35G9.3 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | PRKCB | Sponge network | 0.657 | 4.0E-5 | -1.313 | 2.0E-5 | 0.341 |
| 31246 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | CCND2 | Sponge network | 0.246 | 0.59912 | -0.267 | 0.3112 | 0.341 |
| 31247 | LINC00265 |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-539-5p | 15 | DDX6 | Sponge network | 1.07 | 0 | 0.091 | 0.36428 | 0.341 |
| 31248 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-93-5p | 20 | AFF4 | Sponge network | -2.039 | 0.03447 | -0.266 | 0.01565 | 0.341 |
| 31249 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-625-5p | 17 | CADM1 | Sponge network | -0.318 | 0.65847 | -0.9 | 0.00084 | 0.341 |
| 31250 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | PRKAA1 | Sponge network | 0.769 | 0 | 0.317 | 0.0031 | 0.341 |
| 31251 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3613-5p;hsa-miR-411-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 16 | SMAD2 | Sponge network | 0.673 | 0 | -0.128 | 0.12732 | 0.341 |
| 31252 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 26 | ZFP36L1 | Sponge network | -1.111 | 0.0003 | -0.505 | 8.0E-5 | 0.341 |
| 31253 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-935 | 17 | RARB | Sponge network | -0.513 | 0.04703 | -0.825 | 2.0E-5 | 0.341 |
| 31254 | RP11-571M6.17 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-5p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | CCDC6 | Sponge network | 1.013 | 0 | 0.27 | 0.02555 | 0.341 |
| 31255 | RAB30-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 0.752 | 0 | 0.578 | 0 | 0.341 |
| 31256 | CTD-2336O2.1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-532-3p | 11 | SYNM | Sponge network | -0.141 | 0.26434 | -2.309 | 1.0E-5 | 0.341 |
| 31257 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p | 31 | UNC80 | Sponge network | 0.532 | 0.00505 | -1.934 | 0 | 0.341 |
| 31258 | RP11-159D12.2 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-550a-5p;hsa-miR-628-5p | 26 | VPS53 | Sponge network | 1.254 | 0 | 0.103 | 0.22667 | 0.341 |
| 31259 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 34 | PRKAB2 | Sponge network | 0.249 | 0.35902 | -0.367 | 0.01371 | 0.341 |
| 31260 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-625-5p;hsa-miR-744-3p | 16 | PTPN14 | Sponge network | 0.614 | 1.0E-5 | 0.144 | 0.34595 | 0.341 |
| 31261 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-93-5p | 20 | IL17RD | Sponge network | -0.206 | 0.12942 | -0.019 | 0.94873 | 0.341 |
| 31262 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p | 23 | CBL | Sponge network | 1.757 | 0 | 0.291 | 0.01267 | 0.341 |
| 31263 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | RCAN2 | Sponge network | 0.953 | 0 | -0.33 | 0.15172 | 0.341 |
| 31264 | ARHGEF26-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | HOMER2 | Sponge network | -2.46 | 0 | -2.698 | 0 | 0.341 |
| 31265 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | ZMYM2 | Sponge network | 0.439 | 0.00313 | 0.279 | 0.01273 | 0.341 |
| 31266 | MRVI1-AS1 |
hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-5p | 10 | CRTC1 | Sponge network | -1.092 | 4.0E-5 | -0.116 | 0.28412 | 0.341 |
| 31267 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 27 | NCOA1 | Sponge network | -0.842 | 0.01361 | -0.432 | 0 | 0.341 |
| 31268 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 27 | PHC3 | Sponge network | -0.842 | 0.01361 | 0.212 | 0.0499 | 0.341 |
| 31269 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-940 | 58 | NFIB | Sponge network | 0.101 | 0.60919 | 0.023 | 0.90795 | 0.341 |
| 31270 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | APC | Sponge network | -1.697 | 0.00889 | -0.255 | 0.04051 | 0.341 |
| 31271 | AC006116.24 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-5p | 10 | IRS1 | Sponge network | -0.473 | 0.07602 | -0.026 | 0.88855 | 0.341 |
| 31272 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 37 | HLF | Sponge network | -0.777 | 0.16667 | -2.443 | 0 | 0.341 |
| 31273 | SH3BP5-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 26 | EPS15 | Sponge network | 0.267 | 0.2937 | -0.052 | 0.53998 | 0.341 |
| 31274 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 32 | SPRY3 | Sponge network | 0.537 | 0.00139 | 0.016 | 0.91286 | 0.341 |
| 31275 | GAS5 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 12 | TBL1XR1 | Sponge network | 0.728 | 0 | 0.578 | 0 | 0.341 |
| 31276 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-362-3p | 12 | MAF | Sponge network | -2.549 | 0.00528 | -1.257 | 0 | 0.341 |
| 31277 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 27 | QKI | Sponge network | -0.663 | 0.10887 | -0.333 | 0.04111 | 0.341 |
| 31278 | RAMP2-AS1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 16 | ATM | Sponge network | -0.513 | 0.04703 | 0.178 | 0.21568 | 0.341 |
| 31279 | CTD-3092A11.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | TCF12 | Sponge network | 0.873 | 0 | 0.174 | 0.13271 | 0.341 |
| 31280 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | BCLAF1 | Sponge network | 0.439 | 0.00313 | 0.211 | 0.00684 | 0.341 |
| 31281 | RP11-67L2.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 13 | ZBTB20 | Sponge network | 0.72 | 0.0001 | -0.122 | 0.57569 | 0.341 |
| 31282 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | HACE1 | Sponge network | 0.401 | 0.00066 | -0.072 | 0.55239 | 0.341 |
| 31283 | AC006116.24 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-550a-3p | 16 | PTPRT | Sponge network | -0.473 | 0.07602 | -0.907 | 0.03727 | 0.341 |
| 31284 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-485-3p;hsa-miR-495-3p | 13 | PRKAB2 | Sponge network | 1.055 | 0 | -0.367 | 0.01371 | 0.341 |
| 31285 | NR2F1-AS1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | GATA2 | Sponge network | -0.49 | 0.04946 | -0.654 | 0.0016 | 0.341 |
| 31286 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | FREM2 | Sponge network | 1.757 | 0 | -0.437 | 0.46353 | 0.341 |
| 31287 | CTD-2291D10.4 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-96-5p | 12 | NAV3 | Sponge network | 3.21 | 0 | 0.212 | 0.47729 | 0.341 |
| 31288 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-3p | 17 | PTPN11 | Sponge network | 0.194 | 0.64278 | 0.431 | 2.0E-5 | 0.341 |
| 31289 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-940 | 32 | NOTCH2 | Sponge network | -0.896 | 0.00099 | 0.138 | 0.35163 | 0.341 |
| 31290 | RP11-890B15.3 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | SH3GLB1 | Sponge network | 0.101 | 0.60919 | -0.115 | 0.14773 | 0.341 |
| 31291 | RP11-244H3.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ITGB1 | Sponge network | 0.659 | 3.0E-5 | 0.524 | 0.00017 | 0.341 |
| 31292 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-93-3p | 19 | PTPN11 | Sponge network | 0.997 | 0.00066 | 0.431 | 2.0E-5 | 0.341 |
| 31293 | RP11-305E6.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | ABI2 | Sponge network | 1.317 | 0 | 0.175 | 0.14787 | 0.341 |
| 31294 | RP11-401P9.4 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p | 16 | KLF10 | Sponge network | -0.384 | 0.40652 | -0.783 | 0 | 0.341 |
| 31295 | RP11-305E6.4 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p | 10 | PAFAH1B2 | Sponge network | 1.317 | 0 | 0.318 | 0.0001 | 0.341 |
| 31296 | LINC00654 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | PRUNE2 | Sponge network | 0.374 | 0.20054 | -1.733 | 0.00022 | 0.341 |
| 31297 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 27 | PRKAB2 | Sponge network | -0.147 | 0.40452 | -0.367 | 0.01371 | 0.341 |
| 31298 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 54 | AFF4 | Sponge network | 0.082 | 0.55654 | -0.266 | 0.01565 | 0.341 |
| 31299 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577 | 23 | IGF1 | Sponge network | -0.18 | 0.20343 | -0.204 | 0.5898 | 0.341 |
| 31300 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 13 | LRP1B | Sponge network | -2.039 | 0.03447 | -2.163 | 0 | 0.341 |
| 31301 | RP11-401P9.4 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-93-3p | 22 | CTIF | Sponge network | -0.384 | 0.40652 | -0.389 | 0.00549 | 0.341 |
| 31302 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -1.349 | 0.00017 | -0.243 | 0.11408 | 0.341 |
| 31303 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-93-3p | 48 | BMPR2 | Sponge network | -2.69 | 0.0001 | 0.206 | 0.0635 | 0.341 |
| 31304 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | AFF3 | Sponge network | -0.125 | 0.49414 | -2.373 | 0 | 0.341 |
| 31305 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 20 | ADAMTSL3 | Sponge network | 0.395 | 0.15451 | -1.559 | 0 | 0.341 |
| 31306 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 17 | GREM1 | Sponge network | 0.385 | 0.10067 | 0.902 | 0.01691 | 0.341 |
| 31307 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 15 | TBL1XR1 | Sponge network | 0.594 | 0.00064 | 0.578 | 0 | 0.341 |
| 31308 | RP11-161M6.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-96-5p | 15 | IL17RD | Sponge network | 0.61 | 0.06611 | -0.019 | 0.94873 | 0.341 |
| 31309 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-484 | 11 | GREM1 | Sponge network | -1.886 | 0 | 0.902 | 0.01691 | 0.341 |
| 31310 | LPP-AS2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-454-3p | 18 | SPG20 | Sponge network | -0.18 | 0.20343 | -1.16 | 0 | 0.341 |
| 31311 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-7-1-3p | 13 | ZMYND11 | Sponge network | 0.299 | 0.57722 | -0.2 | 0.05449 | 0.341 |
| 31312 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p | 11 | CBL | Sponge network | 0.79 | 0 | 0.291 | 0.01267 | 0.341 |
| 31313 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 22 | ZFHX4 | Sponge network | 1.153 | 0.00114 | 0.018 | 0.95574 | 0.341 |
| 31314 | SH3RF3-AS1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-486-5p | 11 | DNMT3A | Sponge network | 0.889 | 9.0E-5 | 0.302 | 0.0262 | 0.341 |
| 31315 | FAM13A-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 11 | PHOX2B | Sponge network | -0.83 | 0 | -2.68 | 0 | 0.341 |
| 31316 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-7-5p | 22 | ZBTB4 | Sponge network | -0.125 | 0.49414 | -0.739 | 0 | 0.341 |
| 31317 | RP11-13K12.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-502-5p | 15 | FMNL3 | Sponge network | -0.154 | 0.78939 | 0.555 | 4.0E-5 | 0.341 |
| 31318 | NEAT1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PHF3 | Sponge network | 0.705 | 0.00014 | 0.126 | 0.19652 | 0.341 |
| 31319 | RP11-283I3.6 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-96-5p | 57 | LPP | Sponge network | 0.614 | 1.0E-5 | -0.459 | 0.04475 | 0.341 |
| 31320 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-7-1-3p | 19 | MLLT10 | Sponge network | 0.817 | 3.0E-5 | 0.105 | 0.33292 | 0.341 |
| 31321 | LINC01018 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | HOMER2 | Sponge network | -2.915 | 0.00015 | -2.698 | 0 | 0.341 |
| 31322 | HAND2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-7-5p | 22 | KCNJ5 | Sponge network | -1.697 | 0.00889 | -0.281 | 0.3339 | 0.341 |
| 31323 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p | 36 | ESR1 | Sponge network | -0.615 | 0.00015 | -1.035 | 3.0E-5 | 0.341 |
| 31324 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 22 | MYBL1 | Sponge network | -0.096 | 0.67958 | 0.494 | 0.02152 | 0.341 |
| 31325 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 22 | PDGFRA | Sponge network | -0.351 | 0.11358 | -0.372 | 0.0037 | 0.341 |
| 31326 | TMEM9B-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p | 13 | ABL1 | Sponge network | 0.529 | 0.00552 | -0.215 | 0.07804 | 0.341 |
| 31327 | EMX2OS |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p | 12 | CITED2 | Sponge network | -0.777 | 0.1134 | -1.545 | 0 | 0.341 |
| 31328 | AC009120.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-3p;hsa-miR-590-3p | 15 | MYBL1 | Sponge network | 0.919 | 0 | 0.494 | 0.02152 | 0.341 |
| 31329 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374a-3p | 14 | TMTC3 | Sponge network | 0.706 | 0 | 0.123 | 0.22189 | 0.341 |
| 31330 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 28 | WDFY3 | Sponge network | -0.376 | 0.00337 | -0.201 | 0.0967 | 0.341 |
| 31331 | RP11-35G9.5 |
hsa-let-7d-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-191-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-96-5p | 11 | CBFA2T3 | Sponge network | 0.622 | 0.00015 | -1.221 | 1.0E-5 | 0.341 |
| 31332 | SNHG6 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-486-5p | 10 | CBFB | Sponge network | 0.668 | 0 | 1.061 | 0 | 0.341 |
| 31333 | SOX2-OT |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 26 | TET1 | Sponge network | -1.264 | 6.0E-5 | -0.135 | 0.52138 | 0.341 |
| 31334 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | EPAS1 | Sponge network | -0.421 | 0.0114 | -0.184 | 0.14418 | 0.341 |
| 31335 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-27a-5p;hsa-miR-3607-3p;hsa-miR-93-5p;hsa-miR-940 | 21 | HLF | Sponge network | 0.056 | 0.8494 | -2.443 | 0 | 0.341 |
| 31336 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | PAFAH1B1 | Sponge network | -1.654 | 0.00144 | -0.429 | 0 | 0.341 |
| 31337 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-590-3p | 22 | DMD | Sponge network | 0.199 | 0.38015 | -1.506 | 0 | 0.341 |
| 31338 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p | 11 | PDS5B | Sponge network | 0.917 | 0 | 0.259 | 0.00962 | 0.341 |
| 31339 | FGD5-AS1 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | ERCC4 | Sponge network | 0.401 | 0.00066 | 0.126 | 0.15266 | 0.341 |
| 31340 | RP11-166D19.1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 28 | EFNA5 | Sponge network | -0.576 | 0.10121 | -0.421 | 0.17868 | 0.341 |
| 31341 | RP11-680C21.1 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-940 | 11 | CELF4 | Sponge network | -1.424 | 0.04346 | -1.135 | 0.00344 | 0.341 |
| 31342 | RP11-47A8.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-744-5p | 10 | FGFR1 | Sponge network | 0.103 | 0.43169 | -0.509 | 0.03957 | 0.341 |
| 31343 | RP11-968O1.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-484 | 18 | SOX5 | Sponge network | -0.829 | 0.01343 | -1.091 | 4.0E-5 | 0.341 |
| 31344 | AC005154.6 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-363-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TRIM13 | Sponge network | 0.848 | 0 | 0.183 | 0.06968 | 0.341 |
| 31345 | RP11-890B15.3 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | CXCL12 | Sponge network | 0.101 | 0.60919 | -1.421 | 0 | 0.341 |
| 31346 | LENG8-AS1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-338-5p;hsa-miR-539-5p | 12 | TSC1 | Sponge network | 1.471 | 0 | 0.103 | 0.26071 | 0.341 |
| 31347 | RP11-384K6.6 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | USP9X | Sponge network | 0.946 | 0 | 0.222 | 0.01689 | 0.341 |
| 31348 | JAZF1-AS1 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 10 | EPB41L3 | Sponge network | -0.769 | 0.00035 | -1.193 | 0 | 0.341 |
| 31349 | PWAR6 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | PTCH1 | Sponge network | -1.575 | 6.0E-5 | -0.1 | 0.6179 | 0.341 |
| 31350 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 13 | UBE4B | Sponge network | -0.318 | 0.65847 | -0.235 | 0.007 | 0.341 |
| 31351 | RP11-983P16.4 |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-532-3p;hsa-miR-625-5p | 10 | CTNNA2 | Sponge network | 0.41 | 0.02214 | -2.547 | 0 | 0.341 |
| 31352 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-877-5p | 12 | CBFA2T3 | Sponge network | -0.309 | 0.1145 | -1.221 | 1.0E-5 | 0.341 |
| 31353 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p | 10 | GPAM | Sponge network | 1.134 | 0 | 0.142 | 0.29732 | 0.341 |
| 31354 | RP11-345P4.7 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-495-3p | 14 | ZFX | Sponge network | 1.597 | 0 | 0.09 | 0.42563 | 0.341 |
| 31355 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-181b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-7-1-3p | 12 | PTPRB | Sponge network | -4.546 | 0 | 0.105 | 0.47122 | 0.341 |
| 31356 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | SOX5 | Sponge network | -0.663 | 0.10887 | -1.091 | 4.0E-5 | 0.341 |
| 31357 | CTC-429P9.2 |
hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-539-5p | 12 | TMEM132B | Sponge network | 0.043 | 0.81628 | -0.83 | 0.01084 | 0.341 |
| 31358 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ZMYND11 | Sponge network | -0.355 | 0.0077 | -0.2 | 0.05449 | 0.341 |
| 31359 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-766-3p | 34 | MAPK1 | Sponge network | 1.254 | 0 | -0.017 | 0.81465 | 0.341 |
| 31360 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-33a-5p;hsa-miR-539-5p | 13 | PGR | Sponge network | -0.214 | 0.44248 | -1.704 | 0 | 0.341 |
| 31361 | HAND2-AS1 |
hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-590-3p | 11 | EGR1 | Sponge network | -1.697 | 0.00889 | -1.195 | 0 | 0.341 |
| 31362 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 39 | CBL | Sponge network | -1.111 | 0.0003 | 0.291 | 0.01267 | 0.341 |
| 31363 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 41 | BMPR2 | Sponge network | -0.18 | 0.20343 | 0.206 | 0.0635 | 0.341 |
| 31364 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p | 10 | PHC3 | Sponge network | 0.813 | 1.0E-5 | 0.212 | 0.0499 | 0.341 |
| 31365 | AC025171.1 |
hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 0.998 | 0 | 0.259 | 0.00962 | 0.341 |
| 31366 | LENG8-AS1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-374a-3p;hsa-miR-511-5p | 15 | CREB1 | Sponge network | 1.471 | 0 | 0.203 | 0.00537 | 0.341 |
| 31367 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | ATRX | Sponge network | 0.603 | 0.00127 | 0.261 | 0.04408 | 0.341 |
| 31368 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-590-5p | 10 | TGFBR2 | Sponge network | -0.733 | 0.19469 | -0.243 | 0.11408 | 0.341 |
| 31369 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | ST6GALNAC3 | Sponge network | 0.532 | 0.00505 | -0.987 | 0 | 0.341 |
| 31370 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 18 | KAT6B | Sponge network | -0.896 | 0.00099 | -0.478 | 0.00018 | 0.341 |
| 31371 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | NOTCH2 | Sponge network | -0.842 | 0.01361 | 0.138 | 0.35163 | 0.341 |
| 31372 | RP11-798M19.6 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 16 | INPP4A | Sponge network | -0.612 | 0.00094 | 0.07 | 0.53563 | 0.341 |
| 31373 | TP73-AS1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-96-5p | 11 | DNMT3A | Sponge network | -0.563 | 0.02545 | 0.302 | 0.0262 | 0.341 |
| 31374 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | TMTC3 | Sponge network | 1.063 | 0 | 0.123 | 0.22189 | 0.341 |
| 31375 | RP11-156E6.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-374b-5p | 10 | KIAA1024 | Sponge network | 1.266 | 0 | 1.083 | 0 | 0.341 |
| 31376 | RP11-53O19.1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-7-1-3p | 15 | SH3GLB1 | Sponge network | -0.405 | 0.22013 | -0.115 | 0.14773 | 0.341 |
| 31377 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 29 | NTRK2 | Sponge network | 0.559 | 0.00026 | -0.736 | 0.06495 | 0.341 |
| 31378 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CACNA2D1 | Sponge network | 0.349 | 0.01695 | -0.846 | 0.00675 | 0.341 |
| 31379 | CTC-559E9.6 |
hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-654-3p;hsa-miR-940 | 10 | BAZ2B | Sponge network | 0.601 | 1.0E-5 | -0.276 | 0.02833 | 0.341 |
| 31380 | LINC00957 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 12 | FYN | Sponge network | -0.421 | 0.0114 | -0.131 | 0.37051 | 0.341 |
| 31381 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-101-3p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 15 | TMTC3 | Sponge network | 1.055 | 0 | 0.123 | 0.22189 | 0.341 |
| 31382 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-7-1-3p | 14 | DACT1 | Sponge network | -0.405 | 0.22013 | 0.783 | 0.00072 | 0.34 |
| 31383 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 24 | RIMBP2 | Sponge network | -0.576 | 0.10121 | -1.968 | 5.0E-5 | 0.34 |
| 31384 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | LRP1B | Sponge network | 0.335 | 0.20596 | -2.163 | 0 | 0.34 |
| 31385 | TINCR |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-93-3p;hsa-miR-96-5p | 25 | FOXO3 | Sponge network | -0.664 | 0.14543 | -0.04 | 0.72028 | 0.34 |
| 31386 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-495-3p | 10 | CUL5 | Sponge network | 0.997 | 0 | 0.026 | 0.77301 | 0.34 |
| 31387 | RP11-344B5.2 |
hsa-let-7d-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-664a-3p | 10 | THBS1 | Sponge network | -0.055 | 0.88482 | 0.741 | 0.00386 | 0.34 |
| 31388 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-7-1-3p | 19 | PRKAB2 | Sponge network | 0.419 | 0.0319 | -0.367 | 0.01371 | 0.34 |
| 31389 | RP11-499P20.2 |
hsa-let-7d-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-493-5p | 12 | ACVR2A | Sponge network | 0.187 | 0.44121 | -0.409 | 0.00039 | 0.34 |
| 31390 | CTC-444N24.11 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-744-3p | 18 | PTPN14 | Sponge network | 1.153 | 0 | 0.144 | 0.34595 | 0.34 |
| 31391 | RP11-418J17.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 0.998 | 0 | 0.51 | 0 | 0.34 |
| 31392 | HOXA-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p | 10 | THSD7A | Sponge network | 0.61 | 0.01593 | 0.242 | 0.25154 | 0.34 |
| 31393 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p | 12 | NRK | Sponge network | 0.497 | 0.01451 | 0.656 | 0.19217 | 0.34 |
| 31394 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-935 | 12 | DAB2 | Sponge network | -1.249 | 7.0E-5 | 0.431 | 0.0113 | 0.34 |
| 31395 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-576-5p | 19 | CACNA2D1 | Sponge network | -1.645 | 0 | -0.846 | 0.00675 | 0.34 |
| 31396 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p | 12 | PIK3R1 | Sponge network | 1.294 | 0.0031 | -0.133 | 0.36854 | 0.34 |
| 31397 | RP11-403I13.8 |
hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | RIMS2 | Sponge network | 0.619 | 0.00157 | -1.365 | 0.00208 | 0.34 |
| 31398 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-93-5p | 37 | EPHA7 | Sponge network | -0.465 | 0.04703 | -2.197 | 3.0E-5 | 0.34 |
| 31399 | RP11-589P10.7 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-338-3p | 12 | TGFBR3 | Sponge network | -2.044 | 0.00066 | -1.173 | 1.0E-5 | 0.34 |
| 31400 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p | 13 | TMTC3 | Sponge network | 0.545 | 0.00064 | 0.123 | 0.22189 | 0.34 |
| 31401 | AC005618.6 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | BCL9 | Sponge network | 0.862 | 0.06213 | 0.321 | 0.00267 | 0.34 |
| 31402 | PTOV1-AS1 |
hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-664a-5p | 13 | SOX11 | Sponge network | 0.87 | 0 | 2.68 | 0 | 0.34 |
| 31403 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-582-5p | 12 | KAT6B | Sponge network | -0.469 | 0.08721 | -0.478 | 0.00018 | 0.34 |
| 31404 | MIR143HG |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 18 | POU2F2 | Sponge network | -0.739 | 0.0574 | -0.233 | 0.32214 | 0.34 |
| 31405 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p | 31 | CBX5 | Sponge network | 0.176 | 0.37402 | 0.165 | 0.24446 | 0.34 |
| 31406 | SNX29P2 |
hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | FLNA | Sponge network | 0.4 | 0.14611 | -0.771 | 0.01377 | 0.34 |
| 31407 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p | 10 | RB1CC1 | Sponge network | 0.614 | 1.0E-5 | 0.175 | 0.12559 | 0.34 |
| 31408 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-96-5p | 14 | ELMO1 | Sponge network | 0.997 | 0.00066 | 0.04 | 0.83147 | 0.34 |
| 31409 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 16 | WWTR1 | Sponge network | 0.559 | 0.00026 | -0.345 | 0.11997 | 0.34 |
| 31410 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-450b-5p;hsa-miR-576-5p | 14 | ST6GAL2 | Sponge network | 0.176 | 0.37402 | -1.201 | 0.00597 | 0.34 |
| 31411 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | GRIA3 | Sponge network | -0.83 | 0 | -1.956 | 0 | 0.34 |
| 31412 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | FAT4 | Sponge network | 1.153 | 0.00114 | -0.354 | 0.14694 | 0.34 |
| 31413 | RP11-263K19.4 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-429;hsa-miR-625-5p | 22 | ABI2 | Sponge network | 0.859 | 0.00331 | 0.175 | 0.14787 | 0.34 |
| 31414 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 28 | AR | Sponge network | -0.72 | 0.24239 | -1.554 | 0 | 0.34 |
| 31415 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-96-5p | 18 | ARHGAP29 | Sponge network | -0.563 | 0.02545 | 0.131 | 0.42953 | 0.34 |
| 31416 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-874-3p | 10 | ST5 | Sponge network | -0.563 | 0.02545 | -0.78 | 0 | 0.34 |
| 31417 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | TSHZ3 | Sponge network | -0.342 | 0.02288 | -0.556 | 0.01516 | 0.34 |
| 31418 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-93-5p | 21 | RNASEL | Sponge network | -0.421 | 0.0114 | -0.272 | 0.01796 | 0.34 |
| 31419 | FENDRR |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-940;hsa-miR-96-5p | 10 | FOXO4 | Sponge network | -1.281 | 0.00012 | -0.516 | 2.0E-5 | 0.34 |
| 31420 | CTD-2184D3.6 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-590-3p | 19 | DST | Sponge network | 0.272 | 0.44692 | -0.713 | 0.00096 | 0.34 |
| 31421 | RP11-403I13.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | SNX29 | Sponge network | 0.395 | 0.15451 | -0.34 | 0.01371 | 0.34 |
| 31422 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | BCL2 | Sponge network | -0.021 | 0.93598 | -0.899 | 0 | 0.34 |
| 31423 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | FOXO1 | Sponge network | 0.249 | 0.35902 | -0.141 | 0.33874 | 0.34 |
| 31424 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-93-5p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | -0.421 | 0.0114 | -0.085 | 0.42389 | 0.34 |
| 31425 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-93-5p | 10 | EDA2R | Sponge network | 0.253 | 0.09565 | -0.587 | 0.01598 | 0.34 |
| 31426 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 41 | THRB | Sponge network | -0.318 | 0.65847 | -1.117 | 0.0004 | 0.34 |
| 31427 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-3p;hsa-miR-501-5p;hsa-miR-7-1-3p | 17 | UBE4B | Sponge network | 0.194 | 0.64278 | -0.235 | 0.007 | 0.34 |
| 31428 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-590-3p | 10 | RIMS1 | Sponge network | -0.739 | 0.00044 | -3.143 | 0 | 0.34 |
| 31429 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | NEDD4 | Sponge network | 0.349 | 0.01695 | 0.02 | 0.89834 | 0.34 |
| 31430 | RP11-180O5.2 |
hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 12 | MDM4 | Sponge network | 3.607 | 0.0001 | 0.345 | 0.00762 | 0.34 |
| 31431 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | SOX5 | Sponge network | -0.342 | 0.02288 | -1.091 | 4.0E-5 | 0.34 |
| 31432 | RP11-517B11.7 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-9-5p | 11 | CDK6 | Sponge network | 0.706 | 0 | 0.991 | 0 | 0.34 |
| 31433 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-323a-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ATRX | Sponge network | 0.91 | 0 | 0.261 | 0.04408 | 0.34 |
| 31434 | AC010226.4 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | FMNL3 | Sponge network | -0.811 | 2.0E-5 | 0.555 | 4.0E-5 | 0.34 |
| 31435 | LINC00654 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | ERG | Sponge network | 0.374 | 0.20054 | 0.002 | 0.99011 | 0.34 |
| 31436 | CTA-228A9.3 |
hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-654-3p | 11 | PHF6 | Sponge network | 2.466 | 0 | 0.785 | 0 | 0.34 |
| 31437 | CKMT2-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p | 25 | BRD3 | Sponge network | 0.082 | 0.55654 | 0.37 | 0.00013 | 0.34 |
| 31438 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | JAZF1 | Sponge network | -0.351 | 0.11358 | -0.377 | 0.02677 | 0.34 |
| 31439 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 26 | NCOA1 | Sponge network | -0.769 | 0.00035 | -0.432 | 0 | 0.34 |
| 31440 | AC004893.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | ABI2 | Sponge network | 1.317 | 0 | 0.175 | 0.14787 | 0.34 |
| 31441 | CTD-2314G24.2 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 11 | NGFR | Sponge network | 0.246 | 0.59912 | -1.271 | 0.01058 | 0.34 |
| 31442 | RP11-181G12.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-331-3p | 14 | ABI2 | Sponge network | 0.556 | 0.00298 | 0.175 | 0.14787 | 0.34 |
| 31443 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-590-3p | 21 | BRD3 | Sponge network | 0.919 | 0 | 0.37 | 0.00013 | 0.34 |
| 31444 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-874-3p | 10 | NR2C2 | Sponge network | 1.056 | 0 | 0.21 | 0.03224 | 0.34 |
| 31445 | LINC00092 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | PRDM2 | Sponge network | -1.886 | 0 | -0.258 | 0.00721 | 0.34 |
| 31446 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 25 | USP12 | Sponge network | 0.374 | 0.20054 | -0.102 | 0.40768 | 0.34 |
| 31447 | AP001347.6 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-335-3p;hsa-miR-338-3p | 10 | ELMO1 | Sponge network | -1.161 | 0.00591 | 0.04 | 0.83147 | 0.34 |
| 31448 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 20 | RBL2 | Sponge network | -2.46 | 0 | -0.282 | 0.00254 | 0.34 |
| 31449 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p | 25 | EPHA5 | Sponge network | 0.174 | 0.30034 | -0.957 | 0.01733 | 0.34 |
| 31450 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 22 | ARHGAP29 | Sponge network | 0.72 | 0.0001 | 0.131 | 0.42953 | 0.34 |
| 31451 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-590-3p | 12 | ZFP36L1 | Sponge network | -0.739 | 0.00044 | -0.505 | 8.0E-5 | 0.34 |
| 31452 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | BNC2 | Sponge network | 0.657 | 4.0E-5 | -0.491 | 0.09988 | 0.34 |
| 31453 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-3p | 17 | RUNX1T1 | Sponge network | -0.295 | 0.02604 | -0.344 | 0.2374 | 0.34 |
| 31454 | RP11-686D22.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 13 | IGF1 | Sponge network | 0.793 | 5.0E-5 | -0.204 | 0.5898 | 0.34 |
| 31455 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-940 | 14 | CRTC3 | Sponge network | -1.886 | 0 | 0.164 | 0.16786 | 0.34 |
| 31456 | RP11-736K20.5 |
hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | OPCML | Sponge network | -0.358 | 0.17255 | -2.498 | 0 | 0.34 |
| 31457 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-625-5p;hsa-miR-629-3p | 13 | PTPN14 | Sponge network | 0.898 | 1.0E-5 | 0.144 | 0.34595 | 0.34 |
| 31458 | RP11-356J5.12 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | PDE4DIP | Sponge network | -0.899 | 6.0E-5 | -0.282 | 0.0257 | 0.34 |
| 31459 | PART1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | TAL1 | Sponge network | -2.925 | 0 | -0.462 | 0.00574 | 0.34 |
| 31460 | RP11-458F8.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p | 19 | CCDC6 | Sponge network | 1.518 | 0 | 0.27 | 0.02555 | 0.34 |
| 31461 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-450b-5p | 15 | NIN | Sponge network | -0.517 | 0.02935 | -0.253 | 0.13794 | 0.34 |
| 31462 | SNHG16 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-495-3p | 12 | DDX6 | Sponge network | 0.961 | 0 | 0.091 | 0.36428 | 0.34 |
| 31463 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | THRB | Sponge network | -0.031 | 0.89063 | -1.117 | 0.0004 | 0.34 |
| 31464 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-629-3p | 15 | DDX6 | Sponge network | 1.122 | 0 | 0.091 | 0.36428 | 0.34 |
| 31465 | RP11-53O19.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | ZFX | Sponge network | 1.205 | 0 | 0.09 | 0.42563 | 0.34 |
| 31466 | OIP5-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZNF844 | Sponge network | 0.421 | 0.00084 | -0.966 | 0.00035 | 0.34 |
| 31467 | SOX2-OT |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 39 | CHD5 | Sponge network | -1.264 | 6.0E-5 | -0.922 | 0.01521 | 0.34 |
| 31468 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-5p | 15 | EPHA5 | Sponge network | -1.255 | 0.00981 | -0.957 | 0.01733 | 0.34 |
| 31469 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | MYO9A | Sponge network | 1.317 | 0 | -0.147 | 0.32373 | 0.34 |
| 31470 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-493-5p | 11 | MLLT10 | Sponge network | 0.909 | 0 | 0.105 | 0.33292 | 0.34 |
| 31471 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p | 24 | ABL2 | Sponge network | 0.214 | 0.18246 | 0.82 | 0 | 0.34 |
| 31472 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | MTSS1 | Sponge network | -1.575 | 6.0E-5 | -0.81 | 0.00035 | 0.34 |
| 31473 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p | 29 | SPRY3 | Sponge network | -0.176 | 0.59692 | 0.016 | 0.91286 | 0.34 |
| 31474 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 17 | BMPR2 | Sponge network | 1.148 | 2.0E-5 | 0.206 | 0.0635 | 0.34 |
| 31475 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484 | 14 | RET | Sponge network | 0.299 | 0.57722 | -0.833 | 0.03517 | 0.34 |
| 31476 | AC009120.5 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 12 | NFIB | Sponge network | 1.383 | 0 | 0.023 | 0.90795 | 0.34 |
| 31477 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 25 | SUFU | Sponge network | -0.777 | 0.1134 | -0.04 | 0.65489 | 0.34 |
| 31478 | TRAF3IP2-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-744-3p;hsa-miR-744-5p | 16 | IGF2 | Sponge network | -0.323 | 0.01372 | 0.848 | 0.03842 | 0.34 |
| 31479 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p | 16 | DMXL1 | Sponge network | 0.594 | 0.00064 | -0.426 | 0.00056 | 0.34 |
| 31480 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-505-3p | 11 | SON | Sponge network | 0.453 | 0.01839 | 0.168 | 0.04406 | 0.34 |
| 31481 | SERHL |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-320b;hsa-miR-539-5p | 19 | DMD | Sponge network | -0.602 | 0.00331 | -1.506 | 0 | 0.34 |
| 31482 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 18 | HECA | Sponge network | -0.227 | 0.31657 | -0.217 | 0.03463 | 0.34 |
| 31483 | PWAR6 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | RTN4 | Sponge network | -1.575 | 6.0E-5 | -0.412 | 0.00014 | 0.34 |
| 31484 | LPP-AS2 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421 | 10 | RHOB | Sponge network | -0.18 | 0.20343 | -1.052 | 0 | 0.34 |
| 31485 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p | 16 | EBF3 | Sponge network | 0.657 | 4.0E-5 | -0.058 | 0.81437 | 0.34 |
| 31486 | EMX2OS |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 31 | TOM1L2 | Sponge network | -0.777 | 0.1134 | -0.994 | 0 | 0.34 |
| 31487 | LINC00641 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | -0.222 | 0.32445 | -0.001 | 0.99669 | 0.34 |
| 31488 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-93-5p | 47 | CADM2 | Sponge network | -0.384 | 0.40652 | -3.782 | 0 | 0.34 |
| 31489 | LINC00863 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 38 | BMPR2 | Sponge network | 0.189 | 0.13981 | 0.206 | 0.0635 | 0.34 |
| 31490 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | MAF | Sponge network | -1.216 | 0 | -1.257 | 0 | 0.34 |
| 31491 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | CLASP2 | Sponge network | -0.031 | 0.89063 | -0.038 | 0.71 | 0.34 |
| 31492 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 33 | CHD5 | Sponge network | -0.611 | 0.09607 | -0.922 | 0.01521 | 0.34 |
| 31493 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | TSC1 | Sponge network | 0.683 | 1.0E-5 | 0.103 | 0.26071 | 0.34 |
| 31494 | RP11-428J1.5 |
hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | PIK3CA | Sponge network | 0.538 | 0.00076 | 0.195 | 0.06908 | 0.34 |
| 31495 | RP11-983P16.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 15 | WWTR1 | Sponge network | 0.41 | 0.02214 | -0.345 | 0.11997 | 0.34 |
| 31496 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | PRKCE | Sponge network | -0.487 | 0.02157 | -0.403 | 0.00056 | 0.34 |
| 31497 | RP11-283I3.6 |
hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 14 | TMEM30A | Sponge network | 0.614 | 1.0E-5 | 0.096 | 0.31031 | 0.34 |
| 31498 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 12 | LHX6 | Sponge network | 1.153 | 0.00114 | -0.087 | 0.69193 | 0.34 |
| 31499 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 64 | WDFY3 | Sponge network | -2.46 | 0 | -0.201 | 0.0967 | 0.34 |
| 31500 | RP11-458D21.1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-766-3p | 22 | CHD5 | Sponge network | 0.663 | 0.00073 | -0.922 | 0.01521 | 0.34 |
| 31501 | RP11-48B3.4 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | PREX2 | Sponge network | 1.757 | 0 | -0.272 | 0.25382 | 0.34 |
| 31502 | RP11-686D22.8 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ATM | Sponge network | 0.898 | 1.0E-5 | 0.178 | 0.21568 | 0.34 |
| 31503 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-451a;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 37 | BCL2 | Sponge network | 0.299 | 0.57722 | -0.899 | 0 | 0.34 |
| 31504 | RP11-384K6.6 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p | 12 | HECA | Sponge network | 0.946 | 0 | -0.217 | 0.03463 | 0.34 |
| 31505 | RP11-67L2.2 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 17 | PRKAR1A | Sponge network | 0.72 | 0.0001 | -0.063 | 0.50314 | 0.34 |
| 31506 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-511-5p;hsa-miR-590-3p | 17 | BCL6 | Sponge network | -0.342 | 0.02288 | -0.211 | 0.19489 | 0.34 |
| 31507 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 46 | CBX5 | Sponge network | 0.532 | 0.00505 | 0.165 | 0.24446 | 0.34 |
| 31508 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | GRIN2A | Sponge network | -0.08 | 0.90092 | -2.161 | 0 | 0.34 |
| 31509 | AC025171.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 39 | MDM4 | Sponge network | 0.998 | 0 | 0.345 | 0.00762 | 0.34 |
| 31510 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 36 | PRKAA2 | Sponge network | 0.199 | 0.38015 | -2.462 | 0 | 0.34 |
| 31511 | LINC00621 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 18 | ASTN1 | Sponge network | 0.131 | 0.8436 | -2.389 | 2.0E-5 | 0.34 |
| 31512 | RP11-473I1.9 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | INPP4A | Sponge network | 0.683 | 1.0E-5 | 0.07 | 0.53563 | 0.34 |
| 31513 | MIR143HG |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | GATA2 | Sponge network | -0.739 | 0.0574 | -0.654 | 0.0016 | 0.34 |
| 31514 | H1FX-AS1 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 14 | PTGFR | Sponge network | -0.206 | 0.12942 | -0.233 | 0.45421 | 0.34 |
| 31515 | RP5-855D21.1 |
hsa-miR-10b-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-29a-5p | 10 | ARHGEF12 | Sponge network | 1.115 | 0.00188 | 0.09 | 0.35693 | 0.34 |
| 31516 | MRPL23-AS1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p | 13 | BRD3 | Sponge network | -0.278 | 0.76559 | 0.37 | 0.00013 | 0.34 |
| 31517 | RP11-1006G14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PRRX1 | Sponge network | 0.559 | 0.00026 | 1.316 | 0 | 0.34 |
| 31518 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 30 | BHLHE41 | Sponge network | -0.901 | 0.00019 | 0.353 | 0.12753 | 0.34 |
| 31519 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-590-3p | 12 | GPAM | Sponge network | 0.72 | 0.0001 | 0.142 | 0.29732 | 0.34 |
| 31520 | RP11-166D19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 25 | SUFU | Sponge network | -0.576 | 0.10121 | -0.04 | 0.65489 | 0.34 |
| 31521 | NUTM2A-AS1 |
hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | ARHGEF12 | Sponge network | 0.643 | 0 | 0.09 | 0.35693 | 0.34 |
| 31522 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | FYN | Sponge network | 0.657 | 4.0E-5 | -0.131 | 0.37051 | 0.34 |
| 31523 | RP11-102N12.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 15 | NRXN1 | Sponge network | -0.351 | 0.11358 | -3.778 | 0 | 0.34 |
| 31524 | CTC-297N7.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-590-5p | 13 | RECK | Sponge network | -2.069 | 0 | -1.043 | 0 | 0.34 |
| 31525 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 27 | ETV1 | Sponge network | -0.976 | 0.00025 | -0.096 | 0.67674 | 0.34 |
| 31526 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 12 | KIAA1024 | Sponge network | 0.683 | 1.0E-5 | 1.083 | 0 | 0.34 |
| 31527 | BCYRN1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | RNF180 | Sponge network | 0.311 | 0.10344 | -1.127 | 1.0E-5 | 0.34 |
| 31528 | LINC00641 |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CTNNA2 | Sponge network | -0.222 | 0.32445 | -2.547 | 0 | 0.34 |
| 31529 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-758-3p | 15 | CDC73 | Sponge network | 0.706 | 0 | 0.094 | 0.20463 | 0.34 |
| 31530 | LINC00963 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-96-5p | 33 | TACC1 | Sponge network | 0.523 | 9.0E-5 | -0.362 | 0.05406 | 0.34 |
| 31531 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p | 11 | FSTL5 | Sponge network | -0.18 | 0.20343 | -1.3 | 0.01619 | 0.34 |
| 31532 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-589-3p | 12 | MAFB | Sponge network | 0.622 | 0.00015 | -0.023 | 0.89865 | 0.34 |
| 31533 | AC025171.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | CBL | Sponge network | 0.998 | 0 | 0.291 | 0.01267 | 0.34 |
| 31534 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-590-3p | 28 | CREB1 | Sponge network | 1.089 | 0 | 0.203 | 0.00537 | 0.34 |
| 31535 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | FLI1 | Sponge network | 0.246 | 0.59912 | -0.294 | 0.10905 | 0.34 |
| 31536 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-582-5p | 16 | ZFHX3 | Sponge network | 1.294 | 0 | 0.389 | 0.01363 | 0.34 |
| 31537 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 18 | BCL11A | Sponge network | 0.225 | 0.30644 | -0.932 | 0.0063 | 0.34 |
| 31538 | C20orf166-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-769-5p;hsa-miR-93-5p | 45 | WDFY3 | Sponge network | -2.69 | 0.0001 | -0.201 | 0.0967 | 0.34 |
| 31539 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p | 11 | TMTC3 | Sponge network | 0.959 | 0 | 0.123 | 0.22189 | 0.34 |
| 31540 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-590-3p | 13 | FGF5 | Sponge network | 0.381 | 0.25967 | 0.549 | 0.28659 | 0.34 |
| 31541 | AGAP11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-5p | 14 | ZNF208 | Sponge network | -1.645 | 0 | -0.4 | 0.22381 | 0.34 |
| 31542 | RP11-66B24.2 |
hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p | 10 | EYA4 | Sponge network | 0.57 | 0.1866 | 0.3 | 0.36876 | 0.34 |
| 31543 | RASSF8-AS1 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | LIMCH1 | Sponge network | 0.335 | 0.20596 | -0.915 | 1.0E-5 | 0.34 |
| 31544 | RP1-122P22.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 10 | DCLK1 | Sponge network | 0.065 | 0.73468 | -1.324 | 0.00014 | 0.34 |
| 31545 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 0.539 | 0.03722 | 0.259 | 0.00962 | 0.34 |
| 31546 | BOLA3-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 44 | WDFY3 | Sponge network | -0.246 | 0.35034 | -0.201 | 0.0967 | 0.34 |
| 31547 | H19 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-664a-3p | 11 | GREM1 | Sponge network | 2.439 | 0 | 0.902 | 0.01691 | 0.34 |
| 31548 | TUG1 |
hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | DDX3X | Sponge network | 0.972 | 0 | -0.152 | 0.07686 | 0.34 |
| 31549 | CTD-2528L19.6 |
hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | PIK3R1 | Sponge network | 0.953 | 0 | -0.133 | 0.36854 | 0.34 |
| 31550 | MIR22HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-5p | 31 | DMD | Sponge network | -1.047 | 0 | -1.506 | 0 | 0.34 |
| 31551 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 36 | TMEM132B | Sponge network | -2.915 | 0.00015 | -0.83 | 0.01084 | 0.34 |
| 31552 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-5p | 19 | PPM1A | Sponge network | -4.463 | 0.00025 | -0.623 | 0 | 0.34 |
| 31553 | RP11-379F4.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | RPS6KA3 | Sponge network | 1.316 | 0 | 0.256 | 0.02419 | 0.34 |
| 31554 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-429 | 14 | IL6ST | Sponge network | 2.364 | 0.00134 | -0.402 | 0.00676 | 0.34 |
| 31555 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 19 | NUAK1 | Sponge network | 0.299 | 0.57722 | 0.904 | 0 | 0.34 |
| 31556 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 23 | TBL1XR1 | Sponge network | 0.856 | 0 | 0.578 | 0 | 0.34 |
| 31557 | RP11-403I13.8 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PCDH9 | Sponge network | 0.619 | 0.00157 | -1.823 | 4.0E-5 | 0.34 |
| 31558 | CD27-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 70 | LPP | Sponge network | 0.177 | 0.16681 | -0.459 | 0.04475 | 0.34 |
| 31559 | PCA3 |
hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 19 | EMP2 | Sponge network | -0.709 | 0.18168 | -0.423 | 0.00985 | 0.34 |
| 31560 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p | 27 | PTPN14 | Sponge network | -0.899 | 6.0E-5 | 0.144 | 0.34595 | 0.34 |
| 31561 | GHRLOS |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-940 | 16 | MEF2C | Sponge network | -1.942 | 0 | -0.499 | 0.01448 | 0.34 |
| 31562 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-503-5p;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | 0.219 | 0.1516 | -0.684 | 0.00159 | 0.34 |
| 31563 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-96-5p | 36 | FAT3 | Sponge network | 0.619 | 0.00157 | -1.601 | 0.00051 | 0.34 |
| 31564 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 14 | MLLT10 | Sponge network | 0.178 | 0.29106 | 0.105 | 0.33292 | 0.34 |
| 31565 | AC093627.8 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-222-5p | 13 | NOTCH2 | Sponge network | -0.787 | 0.3928 | 0.138 | 0.35163 | 0.34 |
| 31566 | LINC00621 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | ABI2 | Sponge network | 0.131 | 0.8436 | 0.175 | 0.14787 | 0.34 |
| 31567 | AC074289.1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-940 | 20 | MCC | Sponge network | -0.326 | 0.14314 | -1.005 | 0 | 0.34 |
| 31568 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 10 | RBL2 | Sponge network | 0.657 | 4.0E-5 | -0.282 | 0.00254 | 0.34 |
| 31569 | CALML3-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-5p | 30 | ERC1 | Sponge network | 0.95 | 0.15943 | -0.108 | 0.38794 | 0.34 |
| 31570 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-27a-5p | 11 | TSHZ3 | Sponge network | -0.214 | 0.44248 | -0.556 | 0.01516 | 0.34 |
| 31571 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | TMTC2 | Sponge network | 0.72 | 0.0001 | -0.215 | 0.18248 | 0.34 |
| 31572 | RP11-474O21.5 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-7-1-3p | 11 | TRPS1 | Sponge network | -0.023 | 0.95109 | -0.191 | 0.44109 | 0.34 |
| 31573 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 19 | HLF | Sponge network | 0.497 | 0.01451 | -2.443 | 0 | 0.34 |
| 31574 | LINC00702 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 31 | PDE4DIP | Sponge network | -0.737 | 0.06182 | -0.282 | 0.0257 | 0.34 |
| 31575 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-493-5p | 14 | CDC73 | Sponge network | 0.909 | 0 | 0.094 | 0.20463 | 0.34 |
| 31576 | CTD-2002H8.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NIN | Sponge network | 0.931 | 1.0E-5 | -0.253 | 0.13794 | 0.34 |
| 31577 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CACNA2D1 | Sponge network | -0.355 | 0.0077 | -0.846 | 0.00675 | 0.34 |
| 31578 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | 0.532 | 0.00505 | -1.3 | 0.01619 | 0.34 |
| 31579 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-940 | 33 | TOM1L2 | Sponge network | -0.576 | 0.10121 | -0.994 | 0 | 0.34 |
| 31580 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-874-3p | 10 | BCLAF1 | Sponge network | 0.817 | 3.0E-5 | 0.211 | 0.00684 | 0.34 |
| 31581 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | EPHA3 | Sponge network | 0.381 | 0.25967 | 0.185 | 0.53588 | 0.34 |
| 31582 | CTD-3157E16.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-484 | 11 | DST | Sponge network | -0.326 | 0.28809 | -0.713 | 0.00096 | 0.34 |
| 31583 | RP11-981G7.6 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 16 | KIF5B | Sponge network | 0.986 | 0.01022 | 0.362 | 0.00066 | 0.34 |
| 31584 | RP4-669P10.16 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-342-5p;hsa-miR-542-3p;hsa-miR-769-5p | 14 | SYNPO2 | Sponge network | -0.301 | 0.06597 | -2.342 | 0 | 0.34 |
| 31585 | RP3-402G11.26 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-590-3p | 10 | SOX5 | Sponge network | -0.254 | 0.19303 | -1.091 | 4.0E-5 | 0.34 |
| 31586 | MEG3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 20 | RPS6KA2 | Sponge network | 0.249 | 0.35902 | -0.57 | 0.00013 | 0.34 |
| 31587 | BOLA3-AS1 |
hsa-miR-126-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-660-5p | 13 | IRS1 | Sponge network | -0.246 | 0.35034 | -0.026 | 0.88855 | 0.34 |
| 31588 | C4A-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-486-5p;hsa-miR-590-5p | 10 | NRXN3 | Sponge network | 3.345 | 0 | -0.893 | 0.04186 | 0.34 |
| 31589 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-455-3p | 11 | CSRNP1 | Sponge network | -2.69 | 0.0001 | -1.448 | 0 | 0.34 |
| 31590 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 19 | LRRC7 | Sponge network | -0.08 | 0.90092 | -1.341 | 0.01193 | 0.339 |
| 31591 | KB-1460A1.5 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SEMA3C | Sponge network | 0.603 | 0.00127 | -0.272 | 0.31153 | 0.339 |
| 31592 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MED12L | Sponge network | -0.206 | 0.12942 | -0.194 | 0.55299 | 0.339 |
| 31593 | MEG3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-5p | 13 | FBXO11 | Sponge network | 0.249 | 0.35902 | 0.142 | 0.04938 | 0.339 |
| 31594 | TRIM52-AS1 |
hsa-miR-128-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-502-5p;hsa-miR-96-5p | 11 | PAIP2 | Sponge network | -0.418 | 0.00068 | -0.515 | 0 | 0.339 |
| 31595 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ITGAV | Sponge network | -0.222 | 0.32445 | 0.337 | 0.01002 | 0.339 |
| 31596 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | CBL | Sponge network | 0.889 | 9.0E-5 | 0.291 | 0.01267 | 0.339 |
| 31597 | AC004540.4 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110 | 10 | MRVI1 | Sponge network | -2.549 | 0.00528 | -1.051 | 0.00022 | 0.339 |
| 31598 | AC106786.1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-576-5p | 13 | QKI | Sponge network | 1.122 | 0.02989 | -0.333 | 0.04111 | 0.339 |
| 31599 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-5p | 22 | BTG2 | Sponge network | -1.349 | 0.00017 | -1.763 | 0 | 0.339 |
| 31600 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 18 | DCLK1 | Sponge network | -0.303 | 0.21002 | -1.324 | 0.00014 | 0.339 |
| 31601 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-942-5p | 13 | CREBBP | Sponge network | 0.594 | 0.41522 | 0.126 | 0.15186 | 0.339 |
| 31602 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-93-5p | 14 | LRRC55 | Sponge network | -0.465 | 0.04703 | -0.026 | 0.93163 | 0.339 |
| 31603 | RP11-48B3.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 19 | CADM1 | Sponge network | 1.757 | 0 | -0.9 | 0.00084 | 0.339 |
| 31604 | RP13-516M14.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | SETBP1 | Sponge network | 0.389 | 0.04293 | -1.302 | 1.0E-5 | 0.339 |
| 31605 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 10 | FLI1 | Sponge network | -2.117 | 0.00161 | -0.294 | 0.10905 | 0.339 |
| 31606 | LINC00094 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429 | 17 | ITPKB | Sponge network | 0.253 | 0.09565 | -0.704 | 0.00106 | 0.339 |
| 31607 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | AFF3 | Sponge network | -0.355 | 0.0077 | -2.373 | 0 | 0.339 |
| 31608 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | EPAS1 | Sponge network | 0.194 | 0.64278 | -0.184 | 0.14418 | 0.339 |
| 31609 | GNG12-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-486-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-942-5p | 20 | GLIPR1 | Sponge network | -0.793 | 7.0E-5 | 0.146 | 0.3276 | 0.339 |
| 31610 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 17 | DKK3 | Sponge network | 0.101 | 0.60919 | -0.052 | 0.77783 | 0.339 |
| 31611 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 18 | PIK3C2A | Sponge network | 0.765 | 7.0E-5 | 0.061 | 0.53776 | 0.339 |
| 31612 | PGM5-AS1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p | 12 | PAIP2 | Sponge network | -4.546 | 0 | -0.515 | 0 | 0.339 |
| 31613 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | ETV1 | Sponge network | -0.323 | 0.01372 | -0.096 | 0.67674 | 0.339 |
| 31614 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | EPHA5 | Sponge network | -1.349 | 0.00017 | -0.957 | 0.01733 | 0.339 |
| 31615 | LINC00472 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | DOCK4 | Sponge network | -0.842 | 0.01361 | 0.502 | 0.00108 | 0.339 |
| 31616 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 34 | RBMS3 | Sponge network | 0.41 | 0.02214 | -0.519 | 0.03551 | 0.339 |
| 31617 | PART1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 11 | ZNF382 | Sponge network | -2.925 | 0 | -0.494 | 0.03831 | 0.339 |
| 31618 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-495-3p | 12 | MYO9A | Sponge network | 1.093 | 0 | -0.147 | 0.32373 | 0.339 |
| 31619 | NDUFA6-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 16 | SORCS1 | Sponge network | -0.355 | 0.0077 | -3.492 | 0 | 0.339 |
| 31620 | RP11-504P24.8 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p | 11 | CBFB | Sponge network | 1.178 | 0 | 1.061 | 0 | 0.339 |
| 31621 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 23 | EBF3 | Sponge network | -2.915 | 0.00015 | -0.058 | 0.81437 | 0.339 |
| 31622 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | GIGYF2 | Sponge network | 0.222 | 0.29133 | 0.136 | 0.04403 | 0.339 |
| 31623 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p | 14 | AFF3 | Sponge network | 0.082 | 0.55397 | -2.373 | 0 | 0.339 |
| 31624 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p | 11 | ESR1 | Sponge network | 0.796 | 4.0E-5 | -1.035 | 3.0E-5 | 0.339 |
| 31625 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 32 | CBX5 | Sponge network | 1.153 | 0 | 0.165 | 0.24446 | 0.339 |
| 31626 | SNHG14 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RBM6 | Sponge network | -1.111 | 0.0003 | -0.137 | 0.15663 | 0.339 |
| 31627 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 40 | SSBP2 | Sponge network | -0.668 | 3.0E-5 | -1.58 | 0 | 0.339 |
| 31628 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 18 | TET1 | Sponge network | 0.921 | 0.00118 | -0.135 | 0.52138 | 0.339 |
| 31629 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p | 13 | PDZD2 | Sponge network | 0.72 | 0.0001 | -0.909 | 3.0E-5 | 0.339 |
| 31630 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | TLE4 | Sponge network | -0.942 | 0 | -1.157 | 0 | 0.339 |
| 31631 | RP11-130L8.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 18 | DIP2C | Sponge network | -0.663 | 0.10887 | -0.436 | 0.01713 | 0.339 |
| 31632 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p | 28 | ST6GAL2 | Sponge network | -0.793 | 7.0E-5 | -1.201 | 0.00597 | 0.339 |
| 31633 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-429 | 14 | ZFHX3 | Sponge network | 1.361 | 0 | 0.389 | 0.01363 | 0.339 |
| 31634 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p | 32 | CBL | Sponge network | 0.253 | 0.09565 | 0.291 | 0.01267 | 0.339 |
| 31635 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | DMTF1 | Sponge network | 1.205 | 0 | 0.248 | 0.02726 | 0.339 |
| 31636 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | ERCC4 | Sponge network | -0.323 | 0.01372 | 0.126 | 0.15266 | 0.339 |
| 31637 | DIO3OS |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-550a-3p | 13 | PAK3 | Sponge network | -0.773 | 0.04073 | -0.628 | 0.07253 | 0.339 |
| 31638 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-942-5p | 15 | FBXO31 | Sponge network | -0.49 | 0.04946 | -0.085 | 0.42389 | 0.339 |
| 31639 | RP11-403I13.7 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ITGB1 | Sponge network | 0.395 | 0.15451 | 0.524 | 0.00017 | 0.339 |
| 31640 | RP11-5C23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-452-5p;hsa-miR-590-3p | 22 | ADARB1 | Sponge network | 0.274 | 0.07681 | -0.335 | 0.06631 | 0.339 |
| 31641 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 30 | TACC1 | Sponge network | 0.392 | 0.0278 | -0.362 | 0.05406 | 0.339 |
| 31642 | CD27-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-671-5p | 39 | ADARB1 | Sponge network | 0.177 | 0.16681 | -0.335 | 0.06631 | 0.339 |
| 31643 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-582-5p | 17 | SOX5 | Sponge network | 0.792 | 0.0049 | -1.091 | 4.0E-5 | 0.339 |
| 31644 | TRIM52-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | RECK | Sponge network | -0.418 | 0.00068 | -1.043 | 0 | 0.339 |
| 31645 | FAM215B |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 11 | TRIM13 | Sponge network | 0.817 | 3.0E-5 | 0.183 | 0.06968 | 0.339 |
| 31646 | SERHL |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p | 28 | PRKAA2 | Sponge network | -0.602 | 0.00331 | -2.462 | 0 | 0.339 |
| 31647 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 13 | AHNAK | Sponge network | 0.056 | 0.81195 | -1.207 | 0 | 0.339 |
| 31648 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | EPHA3 | Sponge network | -0.351 | 0.11358 | 0.185 | 0.53588 | 0.339 |
| 31649 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-935 | 24 | FBXO32 | Sponge network | -0.141 | 0.26434 | -0.495 | 0.07561 | 0.339 |
| 31650 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-7-1-3p | 21 | TIMP3 | Sponge network | -0.405 | 0.22013 | -0.546 | 0.02307 | 0.339 |
| 31651 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-942-5p | 24 | GPC6 | Sponge network | -2.62 | 0 | -0.054 | 0.81465 | 0.339 |
| 31652 | AC083843.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p | 14 | SLC5A3 | Sponge network | 1.4 | 0 | 0.493 | 5.0E-5 | 0.339 |
| 31653 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p | 19 | DMXL1 | Sponge network | 0.817 | 3.0E-5 | -0.426 | 0.00056 | 0.339 |
| 31654 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | MGA | Sponge network | 1.03 | 0 | 0.323 | 0.00792 | 0.339 |
| 31655 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 37 | CLASP2 | Sponge network | -0.739 | 0.0574 | -0.038 | 0.71 | 0.339 |
| 31656 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 55 | THRB | Sponge network | -0.096 | 0.67958 | -1.117 | 0.0004 | 0.339 |
| 31657 | RP11-212P7.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p | 24 | ABL2 | Sponge network | 0.219 | 0.1516 | 0.82 | 0 | 0.339 |
| 31658 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 18 | USP12 | Sponge network | 0.05 | 0.95483 | -0.102 | 0.40768 | 0.339 |
| 31659 | AGAP11 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-93-3p | 25 | PCDH10 | Sponge network | -1.645 | 0 | -1.644 | 0.0078 | 0.339 |
| 31660 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-93-3p | 21 | PAFAH1B1 | Sponge network | -4.463 | 0.00025 | -0.429 | 0 | 0.339 |
| 31661 | RP13-516M14.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | WWTR1 | Sponge network | 0.389 | 0.04293 | -0.345 | 0.11997 | 0.339 |
| 31662 | RP4-791M13.3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-425-5p | 24 | ADARB1 | Sponge network | 0.419 | 0.0319 | -0.335 | 0.06631 | 0.339 |
| 31663 | LINC00957 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-590-3p | 13 | STARD13 | Sponge network | -0.421 | 0.0114 | -0.187 | 0.27383 | 0.339 |
| 31664 | RP11-968O1.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-505-5p | 40 | LPP | Sponge network | -0.829 | 0.01343 | -0.459 | 0.04475 | 0.339 |
| 31665 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429 | 16 | STARD13 | Sponge network | -1.654 | 0.00144 | -0.187 | 0.27383 | 0.339 |
| 31666 | RP11-359B12.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-942-5p;hsa-miR-96-5p | 12 | ZBTB20 | Sponge network | 0.064 | 0.72934 | -0.122 | 0.57569 | 0.339 |
| 31667 | RP11-758N13.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p | 12 | FAT4 | Sponge network | 2.939 | 3.0E-5 | -0.354 | 0.14694 | 0.339 |
| 31668 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 21 | SLITRK6 | Sponge network | -0.129 | 0.57177 | -2.121 | 0 | 0.339 |
| 31669 | AC009120.6 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 14 | RASAL2 | Sponge network | 0.919 | 0 | 0.869 | 0 | 0.339 |
| 31670 | RP4-714D9.5 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-497-5p;hsa-miR-9-5p | 15 | CCDC6 | Sponge network | 1.015 | 0 | 0.27 | 0.02555 | 0.339 |
| 31671 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 25 | APC | Sponge network | 0.083 | 0.66405 | -0.255 | 0.04051 | 0.339 |
| 31672 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 27 | ARHGAP28 | Sponge network | -0.576 | 0.10121 | -0.911 | 0.00013 | 0.339 |
| 31673 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | ARID5B | Sponge network | 0.614 | 1.0E-5 | 0.342 | 0.01676 | 0.339 |
| 31674 | RP11-894P9.1 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-550a-5p | 19 | CCDC6 | Sponge network | 0.993 | 0 | 0.27 | 0.02555 | 0.339 |
| 31675 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 32 | FAT3 | Sponge network | -0.031 | 0.89063 | -1.601 | 0.00051 | 0.339 |
| 31676 | RP11-38L15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-590-3p | 12 | PIK3R1 | Sponge network | 0.344 | 0.3127 | -0.133 | 0.36854 | 0.339 |
| 31677 | RP11-401P9.4 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-93-3p;hsa-miR-935 | 36 | GRIK3 | Sponge network | -0.384 | 0.40652 | -3.208 | 0 | 0.339 |
| 31678 | RP5-1085F17.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-940 | 19 | SIRT1 | Sponge network | 0.541 | 0.00125 | 0.109 | 0.2593 | 0.339 |
| 31679 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | ITGAV | Sponge network | 0.889 | 9.0E-5 | 0.337 | 0.01002 | 0.339 |
| 31680 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 13 | CNTNAP1 | Sponge network | -0.125 | 0.49414 | 0.358 | 0.13727 | 0.339 |
| 31681 | CTC-471J1.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 1.11 | 1.0E-5 | 1.083 | 0 | 0.339 |
| 31682 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3913-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 28 | ZBTB4 | Sponge network | -1.203 | 0.03881 | -0.739 | 0 | 0.339 |
| 31683 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | PDGFRA | Sponge network | -1.161 | 0.00591 | -0.372 | 0.0037 | 0.339 |
| 31684 | CTC-490E21.10 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-93-3p | 11 | AKAP11 | Sponge network | 1.46 | 1.0E-5 | 0.196 | 0.09825 | 0.339 |
| 31685 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p | 14 | ERG | Sponge network | -0.246 | 0.35034 | 0.002 | 0.99011 | 0.339 |
| 31686 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-590-3p | 17 | BCL6 | Sponge network | -0.709 | 0.18168 | -0.211 | 0.19489 | 0.339 |
| 31687 | LINC00641 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | LHFP | Sponge network | -0.222 | 0.32445 | -0.167 | 0.41371 | 0.339 |
| 31688 | CTD-3025N20.3 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-590-3p | 12 | CREB1 | Sponge network | 1.046 | 0 | 0.203 | 0.00537 | 0.339 |
| 31689 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-629-5p;hsa-miR-942-5p | 39 | UNC80 | Sponge network | -1.249 | 7.0E-5 | -1.934 | 0 | 0.339 |
| 31690 | RP11-384K6.6 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-629-3p;hsa-miR-654-3p | 17 | PIK3R1 | Sponge network | 0.946 | 0 | -0.133 | 0.36854 | 0.339 |
| 31691 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-550a-3p | 11 | PAK3 | Sponge network | -0.465 | 0.04703 | -0.628 | 0.07253 | 0.339 |
| 31692 | LINC00969 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 14 | KIF5B | Sponge network | 0.683 | 1.0E-5 | 0.362 | 0.00066 | 0.339 |
| 31693 | AC007566.10 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-497-5p | 13 | ARHGEF12 | Sponge network | 2.368 | 2.0E-5 | 0.09 | 0.35693 | 0.339 |
| 31694 | GNG12-AS1 |
hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940;hsa-miR-942-5p | 20 | EMP2 | Sponge network | -0.793 | 7.0E-5 | -0.423 | 0.00985 | 0.339 |
| 31695 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 12 | TBX18 | Sponge network | -1.349 | 0.00017 | 0.843 | 0.02945 | 0.339 |
| 31696 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | CRTC1 | Sponge network | -1.111 | 0.0003 | -0.116 | 0.28412 | 0.339 |
| 31697 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-940 | 26 | HLF | Sponge network | 0.259 | 0.12542 | -2.443 | 0 | 0.339 |
| 31698 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 50 | QKI | Sponge network | 0.72 | 0.0001 | -0.333 | 0.04111 | 0.339 |
| 31699 | RP11-327P2.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-542-3p;hsa-miR-590-5p | 14 | KANK1 | Sponge network | -0.147 | 0.40452 | -0.55 | 0.00134 | 0.339 |
| 31700 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-582-5p | 14 | PIK3C2A | Sponge network | 1.294 | 0 | 0.061 | 0.53776 | 0.339 |
| 31701 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 52 | CBX5 | Sponge network | -1.111 | 0.0003 | 0.165 | 0.24446 | 0.339 |
| 31702 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 32 | FOXO3 | Sponge network | 1.302 | 7.0E-5 | -0.04 | 0.72028 | 0.339 |
| 31703 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | LRRC55 | Sponge network | -0.351 | 0.11358 | -0.026 | 0.93163 | 0.339 |
| 31704 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MLLT10 | Sponge network | 0.705 | 0.00014 | 0.105 | 0.33292 | 0.339 |
| 31705 | RP11-244H3.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-940 | 17 | SIRT1 | Sponge network | 0.659 | 3.0E-5 | 0.109 | 0.2593 | 0.339 |
| 31706 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-93-5p | 22 | NIN | Sponge network | -0.384 | 0.40652 | -0.253 | 0.13794 | 0.339 |
| 31707 | C4B-AS1 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p | 11 | ZFHX3 | Sponge network | 2.306 | 9.0E-5 | 0.389 | 0.01363 | 0.339 |
| 31708 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-93-5p | 15 | SASH1 | Sponge network | 0.497 | 0.01451 | -0.502 | 0.00175 | 0.339 |
| 31709 | CD27-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p | 10 | FLNA | Sponge network | 0.177 | 0.16681 | -0.771 | 0.01377 | 0.339 |
| 31710 | CKMT2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | MSN | Sponge network | 0.082 | 0.55654 | -0.023 | 0.88422 | 0.339 |
| 31711 | PCA3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 43 | MDM4 | Sponge network | -0.709 | 0.18168 | 0.345 | 0.00762 | 0.339 |
| 31712 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | FLI1 | Sponge network | -1.216 | 0 | -0.294 | 0.10905 | 0.339 |
| 31713 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 31 | ESR1 | Sponge network | -0.773 | 0.04073 | -1.035 | 3.0E-5 | 0.339 |
| 31714 | RP11-686D22.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | MEF2C | Sponge network | 0.793 | 5.0E-5 | -0.499 | 0.01448 | 0.339 |
| 31715 | NNT-AS1 |
hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | TAF1 | Sponge network | 0.799 | 1.0E-5 | 0.39 | 3.0E-5 | 0.339 |
| 31716 | LINC00865 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-744-3p | 20 | HIP1 | Sponge network | -1.249 | 7.0E-5 | 0.774 | 0 | 0.339 |
| 31717 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 14 | STARD13 | Sponge network | -0.031 | 0.89063 | -0.187 | 0.27383 | 0.339 |
| 31718 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p | 17 | BCL6 | Sponge network | -1.264 | 6.0E-5 | -0.211 | 0.19489 | 0.339 |
| 31719 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-93-5p | 18 | PGR | Sponge network | 0.056 | 0.8494 | -1.704 | 0 | 0.339 |
| 31720 | RP11-620J15.3 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | RPS6KA6 | Sponge network | -0.739 | 0.00044 | -2.989 | 0 | 0.339 |
| 31721 | RP11-119F7.5 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | KLF10 | Sponge network | 0.225 | 0.30644 | -0.783 | 0 | 0.339 |
| 31722 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 43 | BNC2 | Sponge network | -0.303 | 0.21002 | -0.491 | 0.09988 | 0.339 |
| 31723 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | CRTC1 | Sponge network | 0.064 | 0.72934 | -0.116 | 0.28412 | 0.339 |
| 31724 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 10 | DPYD | Sponge network | -0.421 | 0.0114 | -0.736 | 0.00076 | 0.339 |
| 31725 | FAM215B |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-7-1-3p | 24 | ARHGEF12 | Sponge network | 0.817 | 3.0E-5 | 0.09 | 0.35693 | 0.339 |
| 31726 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 23 | EBF3 | Sponge network | 0.494 | 0.00339 | -0.058 | 0.81437 | 0.339 |
| 31727 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-93-5p | 20 | PRRX1 | Sponge network | -2.531 | 0 | 1.316 | 0 | 0.339 |
| 31728 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | FOXO3 | Sponge network | 0.225 | 0.30644 | -0.04 | 0.72028 | 0.339 |
| 31729 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 37 | ARID5B | Sponge network | -1.111 | 0.0003 | 0.342 | 0.01676 | 0.339 |
| 31730 | RP11-35G9.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 18 | RASSF8 | Sponge network | 0.657 | 4.0E-5 | -0.122 | 0.65591 | 0.339 |
| 31731 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-98-5p | 16 | ITGA5 | Sponge network | 0.101 | 0.60919 | 0.452 | 0.03524 | 0.339 |
| 31732 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p | 12 | RELN | Sponge network | -0.726 | 0.00132 | -2.403 | 0 | 0.339 |
| 31733 | USP3-AS1 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-589-5p | 12 | ARID1B | Sponge network | -0.162 | 0.47687 | 0.003 | 0.97503 | 0.339 |
| 31734 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | RBL2 | Sponge network | -0.83 | 0 | -0.282 | 0.00254 | 0.339 |
| 31735 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429 | 18 | MYO9A | Sponge network | 0.859 | 0.00331 | -0.147 | 0.32373 | 0.339 |
| 31736 | FGF14-AS2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p | 13 | CRTC1 | Sponge network | -1.582 | 8.0E-5 | -0.116 | 0.28412 | 0.339 |
| 31737 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 44 | THRB | Sponge network | -0.13 | 0.48734 | -1.117 | 0.0004 | 0.339 |
| 31738 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 13 | KCNAB1 | Sponge network | 0.72 | 0.0001 | -0.755 | 2.0E-5 | 0.339 |
| 31739 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | LATS2 | Sponge network | -0.08 | 0.90092 | 0.159 | 0.2304 | 0.339 |
| 31740 | OIP5-AS1 |
hsa-miR-125a-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-195-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-590-3p | 10 | SS18 | Sponge network | 0.421 | 0.00084 | 0.067 | 0.39667 | 0.339 |
| 31741 | RP11-25K19.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | ROBO1 | Sponge network | -1.057 | 0.00029 | -0.009 | 0.96154 | 0.339 |
| 31742 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | 1.254 | 0 | 0.323 | 0.00792 | 0.339 |
| 31743 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 20 | ZMYND11 | Sponge network | 0.532 | 0.00505 | -0.2 | 0.05449 | 0.339 |
| 31744 | RP11-894P9.1 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p | 16 | PIK3R1 | Sponge network | 0.993 | 0 | -0.133 | 0.36854 | 0.339 |
| 31745 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-33a-3p;hsa-miR-96-5p | 10 | LATS2 | Sponge network | -0.517 | 0.02935 | 0.159 | 0.2304 | 0.339 |
| 31746 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 21 | BMPR2 | Sponge network | 0.643 | 0 | 0.206 | 0.0635 | 0.339 |
| 31747 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p | 14 | RET | Sponge network | 0.811 | 0.03228 | -0.833 | 0.03517 | 0.339 |
| 31748 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 15 | RBL2 | Sponge network | -0.096 | 0.67958 | -0.282 | 0.00254 | 0.339 |
| 31749 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 34 | PBX1 | Sponge network | -0.371 | 0.24604 | -1.206 | 0 | 0.339 |
| 31750 | RP11-464F9.1 |
hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-369-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | RNF111 | Sponge network | 0.959 | 0 | 0.098 | 0.21093 | 0.339 |
| 31751 | RASSF8-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NRG3 | Sponge network | 0.335 | 0.20596 | -1.836 | 4.0E-5 | 0.339 |
| 31752 | NR2F1-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429 | 15 | PLXNC1 | Sponge network | -0.49 | 0.04946 | 0.569 | 0.00329 | 0.339 |
| 31753 | RP1-59D14.5 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 22 | CCDC6 | Sponge network | 1.162 | 5.0E-5 | 0.27 | 0.02555 | 0.339 |
| 31754 | HCG11 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 15 | SHPRH | Sponge network | 0.385 | 0.10067 | 0.098 | 0.40994 | 0.339 |
| 31755 | RP11-121C2.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 11 | TCF12 | Sponge network | 1.147 | 0 | 0.174 | 0.13271 | 0.339 |
| 31756 | SERHL |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-539-5p | 10 | PRUNE2 | Sponge network | -0.602 | 0.00331 | -1.733 | 0.00022 | 0.339 |
| 31757 | C20orf166-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-671-5p | 11 | EHD3 | Sponge network | -2.69 | 0.0001 | -0.484 | 0.01747 | 0.339 |
| 31758 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 16 | RARB | Sponge network | -0.72 | 0.24239 | -0.825 | 2.0E-5 | 0.339 |
| 31759 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-590-5p | 25 | CBL | Sponge network | 1.11 | 0 | 0.291 | 0.01267 | 0.339 |
| 31760 | RP11-6N17.3 |
hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-455-5p | 17 | IL6ST | Sponge network | 0.108 | 0.69556 | -0.402 | 0.00676 | 0.339 |
| 31761 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-7-1-3p | 19 | EBF1 | Sponge network | 0.389 | 0.04293 | -0.384 | 0.08856 | 0.339 |
| 31762 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-629-3p | 17 | GPC6 | Sponge network | -0.246 | 0.35034 | -0.054 | 0.81465 | 0.339 |
| 31763 | RP11-212P7.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | BRD3 | Sponge network | 0.219 | 0.1516 | 0.37 | 0.00013 | 0.339 |
| 31764 | PRKAG2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | THRB | Sponge network | -0.622 | 0.02807 | -1.117 | 0.0004 | 0.339 |
| 31765 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-935 | 17 | RAP1A | Sponge network | -0.376 | 0.00337 | -0.929 | 0 | 0.339 |
| 31766 | LINC00969 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p | 23 | ARHGEF12 | Sponge network | 0.683 | 1.0E-5 | 0.09 | 0.35693 | 0.339 |
| 31767 | RP11-344B5.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-505-3p | 17 | AR | Sponge network | -0.055 | 0.88482 | -1.554 | 0 | 0.339 |
| 31768 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | IGSF10 | Sponge network | -0.663 | 0.10887 | -1.751 | 1.0E-5 | 0.339 |
| 31769 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | 0.392 | 0.0278 | -1.548 | 0 | 0.339 |
| 31770 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-96-5p | 11 | JAZF1 | Sponge network | -0.214 | 0.44248 | -0.377 | 0.02677 | 0.339 |
| 31771 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 25 | PHC3 | Sponge network | -0.896 | 0.00099 | 0.212 | 0.0499 | 0.339 |
| 31772 | FZD10-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 18 | RELN | Sponge network | -0.727 | 0.04726 | -2.403 | 0 | 0.339 |
| 31773 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-940 | 37 | NOTCH2 | Sponge network | -1.349 | 0.00017 | 0.138 | 0.35163 | 0.339 |
| 31774 | RP11-701H24.3 |
hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-215-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | BCL2 | Sponge network | -1.425 | 0.04204 | -0.899 | 0 | 0.339 |
| 31775 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | SLITRK6 | Sponge network | -0.358 | 0.17255 | -2.121 | 0 | 0.339 |
| 31776 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | FNBP1 | Sponge network | 0.331 | 0.57247 | -0.969 | 4.0E-5 | 0.339 |
| 31777 | DNM1P35 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 27 | DMD | Sponge network | 0.051 | 0.83388 | -1.506 | 0 | 0.339 |
| 31778 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-590-5p | 12 | DMD | Sponge network | -0.72 | 0.24239 | -1.506 | 0 | 0.339 |
| 31779 | AC011526.1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-590-3p | 10 | DYNC1I1 | Sponge network | 0.342 | 0.03129 | -1.12 | 0.00107 | 0.339 |
| 31780 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-33a-5p;hsa-miR-539-5p | 16 | DST | Sponge network | -0.214 | 0.44248 | -0.713 | 0.00096 | 0.339 |
| 31781 | U91328.19 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | SFRP1 | Sponge network | -0.303 | 0.21002 | -3.566 | 0 | 0.339 |
| 31782 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 23 | PDGFRA | Sponge network | -0.811 | 2.0E-5 | -0.372 | 0.0037 | 0.339 |
| 31783 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-671-5p;hsa-miR-940 | 12 | CRTC3 | Sponge network | -2.095 | 0.00112 | 0.164 | 0.16786 | 0.339 |
| 31784 | AC093627.8 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p | 10 | PLSCR4 | Sponge network | -0.787 | 0.3928 | -0.474 | 0.03833 | 0.339 |
| 31785 | U91328.21 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-629-3p | 22 | DST | Sponge network | -0.735 | 0.15983 | -0.713 | 0.00096 | 0.339 |
| 31786 | RP11-2H8.2 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-590-3p | 23 | SPRY3 | Sponge network | 1.089 | 0 | 0.016 | 0.91286 | 0.339 |
| 31787 | RP13-516M14.1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | 0.389 | 0.04293 | -1.74 | 0 | 0.339 |
| 31788 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 56 | BMPR2 | Sponge network | -0.901 | 0.00019 | 0.206 | 0.0635 | 0.339 |
| 31789 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | MLLT10 | Sponge network | 0.72 | 0.0001 | 0.105 | 0.33292 | 0.339 |
| 31790 | RP11-166D19.1 |
hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | GUCY1A2 | Sponge network | -0.576 | 0.10121 | 1.213 | 0 | 0.339 |
| 31791 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-331-5p | 12 | STARD13 | Sponge network | -0.693 | 0.05629 | -0.187 | 0.27383 | 0.339 |
| 31792 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | CXXC4 | Sponge network | -0.976 | 0.00025 | -0.433 | 0.21055 | 0.339 |
| 31793 | BCYRN1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p | 37 | ABL2 | Sponge network | 0.311 | 0.10344 | 0.82 | 0 | 0.339 |
| 31794 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | LIMD1 | Sponge network | 0.972 | 0 | 0.103 | 0.28978 | 0.339 |
| 31795 | AGAP11 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p | 18 | PPP2R5C | Sponge network | -1.645 | 0 | -0.314 | 3.0E-5 | 0.339 |
| 31796 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | PPM1A | Sponge network | -0.187 | 0.23787 | -0.623 | 0 | 0.339 |
| 31797 | NUTM2A-AS1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 16 | SLC5A3 | Sponge network | 0.643 | 0 | 0.493 | 5.0E-5 | 0.339 |
| 31798 | HOTAIRM1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-590-3p | 10 | MAPK10 | Sponge network | -0.053 | 0.80685 | -1.774 | 0 | 0.339 |
| 31799 | AC004540.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-5p | 19 | NTRK2 | Sponge network | -2.549 | 0.00528 | -0.736 | 0.06495 | 0.339 |
| 31800 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | PLA2R1 | Sponge network | -0.358 | 0.17255 | -0.265 | 0.24676 | 0.339 |
| 31801 | PART1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | ZDBF2 | Sponge network | -2.925 | 0 | -0.998 | 0.00025 | 0.339 |
| 31802 | LINC00094 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-378a-5p | 11 | CACNA1E | Sponge network | 0.253 | 0.09565 | 1.752 | 0 | 0.339 |
| 31803 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940 | 13 | CRTC3 | Sponge network | 0.225 | 0.30644 | 0.164 | 0.16786 | 0.339 |
| 31804 | FGD5-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 16 | ZMYND11 | Sponge network | 0.401 | 0.00066 | -0.2 | 0.05449 | 0.339 |
| 31805 | JAZF1-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-421 | 12 | PLXNC1 | Sponge network | -0.769 | 0.00035 | 0.569 | 0.00329 | 0.339 |
| 31806 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-337-3p;hsa-miR-550a-5p;hsa-miR-940 | 11 | HOOK3 | Sponge network | 0.267 | 0.2937 | 0.273 | 0.09957 | 0.339 |
| 31807 | CTD-2314G24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 48 | PTPRD | Sponge network | 0.246 | 0.59912 | -1.005 | 0.00064 | 0.339 |
| 31808 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 30 | PRKAB2 | Sponge network | 0.214 | 0.18246 | -0.367 | 0.01371 | 0.339 |
| 31809 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-337-3p;hsa-miR-374b-5p | 10 | CBL | Sponge network | 1.46 | 1.0E-5 | 0.291 | 0.01267 | 0.339 |
| 31810 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-5p | 17 | USP6 | Sponge network | 0.817 | 3.0E-5 | 0.188 | 0.3871 | 0.339 |
| 31811 | CTC-487M23.5 |
hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-450b-5p | 12 | NEDD4 | Sponge network | 0.912 | 0.00014 | 0.02 | 0.89834 | 0.339 |
| 31812 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-590-3p | 11 | HERC2 | Sponge network | -0.355 | 0.0077 | 0.114 | 0.30638 | 0.339 |
| 31813 | RP11-384O8.1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | TPO | Sponge network | -0.738 | 0.21233 | -1.87 | 2.0E-5 | 0.339 |
| 31814 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | AR | Sponge network | 0.727 | 3.0E-5 | -1.554 | 0 | 0.339 |
| 31815 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p | 32 | ST6GALNAC3 | Sponge network | -0.162 | 0.47687 | -0.987 | 0 | 0.339 |
| 31816 | ZNF32-AS2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p | 13 | CREB1 | Sponge network | 1.552 | 7.0E-5 | 0.203 | 0.00537 | 0.339 |
| 31817 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-96-5p | 13 | BNC2 | Sponge network | -0.214 | 0.44248 | -0.491 | 0.09988 | 0.339 |
| 31818 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | HACE1 | Sponge network | 1.601 | 0 | -0.072 | 0.55239 | 0.339 |
| 31819 | CTD-3099C6.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-429;hsa-miR-576-5p | 12 | ABI2 | Sponge network | 1.361 | 0 | 0.175 | 0.14787 | 0.339 |
| 31820 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 27 | SON | Sponge network | 0.311 | 0.10344 | 0.168 | 0.04406 | 0.339 |
| 31821 | FGD5-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 15 | PICALM | Sponge network | 0.401 | 0.00066 | 0.086 | 0.23523 | 0.338 |
| 31822 | LINC00672 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 11 | GLI3 | Sponge network | -0.227 | 0.31657 | -0.615 | 0.01482 | 0.338 |
| 31823 | ATP1B3-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-19b-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 13 | EPS15 | Sponge network | 1.2 | 0.01813 | -0.052 | 0.53998 | 0.338 |
| 31824 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-25-3p | 11 | ST6GAL2 | Sponge network | -0.811 | 2.0E-5 | -1.201 | 0.00597 | 0.338 |
| 31825 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 23 | MOB1B | Sponge network | 0.381 | 0.25967 | 0.197 | 0.15266 | 0.338 |
| 31826 | RP11-181G12.2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 14 | IL17RD | Sponge network | 0.556 | 0.00298 | -0.019 | 0.94873 | 0.338 |
| 31827 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-590-5p | 20 | PIK3C2A | Sponge network | 0.267 | 0.2937 | 0.061 | 0.53776 | 0.338 |
| 31828 | ARHGEF26-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 43 | AFF1 | Sponge network | -2.46 | 0 | -0.384 | 0.00053 | 0.338 |
| 31829 | RP11-104L21.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p | 10 | TCF4 | Sponge network | 0.527 | 0.16506 | 0.016 | 0.92271 | 0.338 |
| 31830 | RP11-403I13.8 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | DCLK1 | Sponge network | 0.619 | 0.00157 | -1.324 | 0.00014 | 0.338 |
| 31831 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-411-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p | 31 | SMAD2 | Sponge network | 1.254 | 0 | -0.128 | 0.12732 | 0.338 |
| 31832 | TPTEP1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | LUZP2 | Sponge network | -1.216 | 0 | -0.056 | 0.89416 | 0.338 |
| 31833 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 13 | MOB1B | Sponge network | 1.46 | 1.0E-5 | 0.197 | 0.15266 | 0.338 |
| 31834 | MYLK-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-424-5p;hsa-miR-940 | 19 | AKAP11 | Sponge network | -1.331 | 3.0E-5 | 0.196 | 0.09825 | 0.338 |
| 31835 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 28 | ZFX | Sponge network | 0.225 | 0.30644 | 0.09 | 0.42563 | 0.338 |
| 31836 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p | 15 | TXNIP | Sponge network | -1.249 | 7.0E-5 | -1.277 | 0 | 0.338 |
| 31837 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-940 | 64 | NFIB | Sponge network | -1.697 | 0.00889 | 0.023 | 0.90795 | 0.338 |
| 31838 | CTC-457L16.2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3613-5p;hsa-miR-589-3p | 12 | KCNJ5 | Sponge network | 1.153 | 0.00114 | -0.281 | 0.3339 | 0.338 |
| 31839 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | PRKAB2 | Sponge network | 0.782 | 0.00016 | -0.367 | 0.01371 | 0.338 |
| 31840 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-671-5p | 12 | SEPT6 | Sponge network | -2.69 | 0.0001 | -0.598 | 0.00181 | 0.338 |
| 31841 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-5p;hsa-miR-423-5p | 12 | ERRFI1 | Sponge network | 0.299 | 0.57722 | -0.798 | 1.0E-5 | 0.338 |
| 31842 | TP73-AS1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-503-5p | 18 | ABL1 | Sponge network | -0.563 | 0.02545 | -0.215 | 0.07804 | 0.338 |
| 31843 | BZRAP1-AS1 |
hsa-miR-128-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-375 | 14 | PAIP2 | Sponge network | -0.465 | 0.04703 | -0.515 | 0 | 0.338 |
| 31844 | AC009948.5 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | GJA1 | Sponge network | 0.532 | 0.00505 | -0.485 | 0.02179 | 0.338 |
| 31845 | CTA-14H9.5 |
hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-508-3p;hsa-miR-582-5p | 11 | FBXO32 | Sponge network | 0.145 | 0.53343 | -0.495 | 0.07561 | 0.338 |
| 31846 | RP11-822E23.8 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-629-3p | 20 | RYR2 | Sponge network | -0.777 | 0.16667 | -1.466 | 0.00031 | 0.338 |
| 31847 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-93-5p | 18 | LRRC55 | Sponge network | -0.777 | 0.16667 | -0.026 | 0.93163 | 0.338 |
| 31848 | HOXB-AS3 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-330-5p | 10 | ABL1 | Sponge network | 0.343 | 0.28887 | -0.215 | 0.07804 | 0.338 |
| 31849 | RP11-678G14.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | PTPRT | Sponge network | -4.477 | 0 | -0.907 | 0.03727 | 0.338 |
| 31850 | RP11-473I1.5 |
hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p | 10 | CHD1 | Sponge network | 0.542 | 0.00054 | 0.322 | 0.00215 | 0.338 |
| 31851 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 20 | RBL2 | Sponge network | -1.697 | 0.00889 | -0.282 | 0.00254 | 0.338 |
| 31852 | AC106786.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-942-5p | 17 | NOTCH2 | Sponge network | 1.122 | 0.02989 | 0.138 | 0.35163 | 0.338 |
| 31853 | RP11-395G23.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 23 | DMD | Sponge network | 0.381 | 0.25967 | -1.506 | 0 | 0.338 |
| 31854 | LINC00969 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 29 | CCDC6 | Sponge network | 0.683 | 1.0E-5 | 0.27 | 0.02555 | 0.338 |
| 31855 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-484 | 17 | MED12L | Sponge network | -1.331 | 3.0E-5 | -0.194 | 0.55299 | 0.338 |
| 31856 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 57 | IL6ST | Sponge network | -2.915 | 0.00015 | -0.402 | 0.00676 | 0.338 |
| 31857 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | NTRK3 | Sponge network | -4.477 | 0 | -1.77 | 3.0E-5 | 0.338 |
| 31858 | RP1-151F17.2 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | UBR3 | Sponge network | 0.222 | 0.29133 | -0.021 | 0.82774 | 0.338 |
| 31859 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 18 | NOTCH2 | Sponge network | -1.582 | 8.0E-5 | 0.138 | 0.35163 | 0.338 |
| 31860 | RP11-736K20.5 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 17 | RIMS2 | Sponge network | -0.358 | 0.17255 | -1.365 | 0.00208 | 0.338 |
| 31861 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-629-3p | 17 | LIFR | Sponge network | -0.583 | 0.14589 | -1.794 | 0 | 0.338 |
| 31862 | HOTAIRM1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 12 | SPG20 | Sponge network | -0.053 | 0.80685 | -1.16 | 0 | 0.338 |
| 31863 | RP11-53O19.3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | ARHGEF12 | Sponge network | 1.205 | 0 | 0.09 | 0.35693 | 0.338 |
| 31864 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | EDA2R | Sponge network | 0.374 | 0.20054 | -0.587 | 0.01598 | 0.338 |
| 31865 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p | 22 | PRKCE | Sponge network | -0.162 | 0.47687 | -0.403 | 0.00056 | 0.338 |
| 31866 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 14 | SOX11 | Sponge network | 0.559 | 0.00026 | 2.68 | 0 | 0.338 |
| 31867 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 51 | PAFAH1B1 | Sponge network | -1.216 | 0 | -0.429 | 0 | 0.338 |
| 31868 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CXXC4 | Sponge network | 0.476 | 0.19203 | -0.433 | 0.21055 | 0.338 |
| 31869 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p | 14 | DACT1 | Sponge network | -0.793 | 7.0E-5 | 0.783 | 0.00072 | 0.338 |
| 31870 | RP1-122P22.2 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-625-5p | 12 | ANK2 | Sponge network | 0.065 | 0.73468 | -1.603 | 1.0E-5 | 0.338 |
| 31871 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 14 | GPAM | Sponge network | 0.799 | 1.0E-5 | 0.142 | 0.29732 | 0.338 |
| 31872 | PART1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p;hsa-miR-624-5p | 13 | SMARCA1 | Sponge network | -2.925 | 0 | -0.684 | 0.00159 | 0.338 |
| 31873 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | AKAP11 | Sponge network | 0.796 | 4.0E-5 | 0.196 | 0.09825 | 0.338 |
| 31874 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p | 11 | TRIM3 | Sponge network | -0.517 | 0.02935 | -0.577 | 0 | 0.338 |
| 31875 | AC007566.10 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-455-3p;hsa-miR-582-5p | 10 | CREB1 | Sponge network | 2.368 | 2.0E-5 | 0.203 | 0.00537 | 0.338 |
| 31876 | FZD10-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | CDH19 | Sponge network | -0.727 | 0.04726 | -3.434 | 0 | 0.338 |
| 31877 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p | 32 | FOXP1 | Sponge network | -0.896 | 0.00099 | 0.042 | 0.74368 | 0.338 |
| 31878 | GS1-124K5.11 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-361-3p;hsa-miR-625-5p | 10 | BCORL1 | Sponge network | 0.494 | 0.00339 | 0.354 | 0.01214 | 0.338 |
| 31879 | RP11-283I3.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NAV3 | Sponge network | 0.614 | 1.0E-5 | 0.212 | 0.47729 | 0.338 |
| 31880 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-3p | 24 | GRIA3 | Sponge network | 0.335 | 0.20596 | -1.956 | 0 | 0.338 |
| 31881 | AC108488.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p | 11 | ACVR1 | Sponge network | 0.259 | 0.12542 | 0.279 | 0.00234 | 0.338 |
| 31882 | RP11-307B6.3 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p | 18 | BTG2 | Sponge network | -4.463 | 0.00025 | -1.763 | 0 | 0.338 |
| 31883 | CTA-204B4.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-590-3p | 21 | NTRK2 | Sponge network | 0.765 | 7.0E-5 | -0.736 | 0.06495 | 0.338 |
| 31884 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-532-3p | 18 | PBX1 | Sponge network | 0.765 | 7.0E-5 | -1.206 | 0 | 0.338 |
| 31885 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | PLSCR4 | Sponge network | 1.153 | 0.00114 | -0.474 | 0.03833 | 0.338 |
| 31886 | RP11-395G23.3 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p | 10 | KCNAB1 | Sponge network | 0.381 | 0.25967 | -0.755 | 2.0E-5 | 0.338 |
| 31887 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-421 | 18 | RNASEL | Sponge network | -0.583 | 0.14589 | -0.272 | 0.01796 | 0.338 |
| 31888 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | LRRC7 | Sponge network | 0.559 | 0.00026 | -1.341 | 0.01193 | 0.338 |
| 31889 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 16 | EPHA3 | Sponge network | 0.765 | 7.0E-5 | 0.185 | 0.53588 | 0.338 |
| 31890 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p | 12 | SNX29 | Sponge network | 1.122 | 0.02989 | -0.34 | 0.01371 | 0.338 |
| 31891 | LINC01018 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-590-3p | 14 | KLF10 | Sponge network | -2.915 | 0.00015 | -0.783 | 0 | 0.338 |
| 31892 | RP11-98I9.4 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-199b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-495-3p | 10 | PHF6 | Sponge network | 0.67 | 0.00048 | 0.785 | 0 | 0.338 |
| 31893 | PCA3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 10 | PLA2R1 | Sponge network | -0.709 | 0.18168 | -0.265 | 0.24676 | 0.338 |
| 31894 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 13 | CHD2 | Sponge network | 0.657 | 4.0E-5 | 0.049 | 0.55371 | 0.338 |
| 31895 | LINC00654 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | NCAM2 | Sponge network | 0.374 | 0.20054 | -0.482 | 0.22565 | 0.338 |
| 31896 | RP11-798M19.6 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | ZDBF2 | Sponge network | -0.612 | 0.00094 | -0.998 | 0.00025 | 0.338 |
| 31897 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | CDYL | Sponge network | 0.72 | 0.0001 | 0.12 | 0.12425 | 0.338 |
| 31898 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-429 | 10 | FLI1 | Sponge network | -0.469 | 0.08721 | -0.294 | 0.10905 | 0.338 |
| 31899 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | CBFA2T3 | Sponge network | -0.125 | 0.49414 | -1.221 | 1.0E-5 | 0.338 |
| 31900 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-96-5p | 42 | QKI | Sponge network | -0.18 | 0.20343 | -0.333 | 0.04111 | 0.338 |
| 31901 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-7-1-3p | 13 | PDGFC | Sponge network | -0.405 | 0.22013 | -0.33 | 0.06483 | 0.338 |
| 31902 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 18 | DYNC1I1 | Sponge network | -0.125 | 0.49414 | -1.12 | 0.00107 | 0.338 |
| 31903 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | PDGFRA | Sponge network | -0.18 | 0.20343 | -0.372 | 0.0037 | 0.338 |
| 31904 | FGD5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | CEP170 | Sponge network | 0.401 | 0.00066 | 0.943 | 0 | 0.338 |
| 31905 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 15 | LHX6 | Sponge network | -0.421 | 0.0114 | -0.087 | 0.69193 | 0.338 |
| 31906 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 16 | DKK3 | Sponge network | 0.064 | 0.72934 | -0.052 | 0.77783 | 0.338 |
| 31907 | RP11-10N23.2 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-590-5p | 10 | UBR3 | Sponge network | 1.134 | 0 | -0.021 | 0.82774 | 0.338 |
| 31908 | RP11-13K12.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-629-5p | 25 | CRTC1 | Sponge network | -0.154 | 0.78939 | -0.116 | 0.28412 | 0.338 |
| 31909 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-942-5p | 11 | FBXO31 | Sponge network | -0.517 | 0.02935 | -0.085 | 0.42389 | 0.338 |
| 31910 | LINC00473 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | CSDE1 | Sponge network | -1.203 | 0.03881 | -0.493 | 0 | 0.338 |
| 31911 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 40 | CBX5 | Sponge network | 0.335 | 0.20596 | 0.165 | 0.24446 | 0.338 |
| 31912 | HHIP-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p | 29 | GAS7 | Sponge network | -0.737 | 0.10822 | -0.555 | 0.01602 | 0.338 |
| 31913 | RP11-157J24.2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-769-5p | 24 | NTRK3 | Sponge network | -1.664 | 0.05165 | -1.77 | 3.0E-5 | 0.338 |
| 31914 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | LRRC55 | Sponge network | 0.101 | 0.60919 | -0.026 | 0.93163 | 0.338 |
| 31915 | AC074289.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-96-5p | 18 | CYLD | Sponge network | -0.326 | 0.14314 | -0.367 | 0.00171 | 0.338 |
| 31916 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-93-5p | 16 | PLSCR4 | Sponge network | -0.785 | 0.00035 | -0.474 | 0.03833 | 0.338 |
| 31917 | TRIM52-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-7-1-3p | 27 | AR | Sponge network | -0.418 | 0.00068 | -1.554 | 0 | 0.338 |
| 31918 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | APC | Sponge network | -2.452 | 0 | -0.255 | 0.04051 | 0.338 |
| 31919 | RP11-20I23.13 |
hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-511-5p | 20 | ZFHX3 | Sponge network | 0.505 | 0.0014 | 0.389 | 0.01363 | 0.338 |
| 31920 | RP11-283I3.6 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 11 | KCNAB1 | Sponge network | 0.614 | 1.0E-5 | -0.755 | 2.0E-5 | 0.338 |
| 31921 | DNAJC27-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-96-5p | 16 | FNBP1 | Sponge network | -0.969 | 2.0E-5 | -0.969 | 4.0E-5 | 0.338 |
| 31922 | RP11-400K9.4 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 17 | RIMS2 | Sponge network | -0.611 | 0.09607 | -1.365 | 0.00208 | 0.338 |
| 31923 | BCYRN1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 50 | RUNX1T1 | Sponge network | 0.311 | 0.10344 | -0.344 | 0.2374 | 0.338 |
| 31924 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-7-5p | 34 | ESR1 | Sponge network | -0.162 | 0.47687 | -1.035 | 3.0E-5 | 0.338 |
| 31925 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | SASH1 | Sponge network | 0.16 | 0.50785 | -0.502 | 0.00175 | 0.338 |
| 31926 | DNAJC27-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-98-5p | 11 | SCN11A | Sponge network | -0.969 | 2.0E-5 | -1.15 | 0.00299 | 0.338 |
| 31927 | LINC00473 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | ROBO1 | Sponge network | -1.203 | 0.03881 | -0.009 | 0.96154 | 0.338 |
| 31928 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-3p | 24 | PRKAB2 | Sponge network | -0.235 | 0.15142 | -0.367 | 0.01371 | 0.338 |
| 31929 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-628-5p;hsa-miR-940 | 14 | TRIM33 | Sponge network | 0.792 | 0.0049 | 0.402 | 7.0E-5 | 0.338 |
| 31930 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 39 | ERBB4 | Sponge network | -0.976 | 0.00025 | -1.975 | 2.0E-5 | 0.338 |
| 31931 | RP5-1139B12.3 |
hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p | 10 | LIFR | Sponge network | 0.598 | 0.38481 | -1.794 | 0 | 0.338 |
| 31932 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-7-1-3p | 12 | CBL | Sponge network | -0.187 | 0.23787 | 0.291 | 0.01267 | 0.338 |
| 31933 | LINC00863 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-362-5p;hsa-miR-576-5p | 17 | ERC1 | Sponge network | 0.189 | 0.13981 | -0.108 | 0.38794 | 0.338 |
| 31934 | RP11-48B3.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 31 | TRPS1 | Sponge network | 1.757 | 0 | -0.191 | 0.44109 | 0.338 |
| 31935 | A2M-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 10 | PARK2 | Sponge network | -0.08 | 0.90092 | -1.892 | 0 | 0.338 |
| 31936 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-93-5p | 15 | CNTNAP3 | Sponge network | -1.349 | 0.00017 | -1.465 | 0.00033 | 0.338 |
| 31937 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 31 | KCNA6 | Sponge network | -0.769 | 0.00035 | -1.531 | 0.00013 | 0.338 |
| 31938 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-93-5p | 27 | PRKAA2 | Sponge network | 0.497 | 0.01451 | -2.462 | 0 | 0.338 |
| 31939 | RP13-225O21.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 12 | MDM4 | Sponge network | 1.279 | 8.0E-5 | 0.345 | 0.00762 | 0.338 |
| 31940 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 28 | IGF1 | Sponge network | -0.612 | 0.00094 | -0.204 | 0.5898 | 0.338 |
| 31941 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-935;hsa-miR-96-5p | 28 | FBXO32 | Sponge network | 0.537 | 0.00139 | -0.495 | 0.07561 | 0.338 |
| 31942 | ZNF667-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-590-5p | 13 | PTCH1 | Sponge network | -0.976 | 0.00025 | -0.1 | 0.6179 | 0.338 |
| 31943 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p | 24 | USP6 | Sponge network | 0.873 | 0 | 0.188 | 0.3871 | 0.338 |
| 31944 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-589-3p | 24 | RNASEL | Sponge network | 0.194 | 0.64278 | -0.272 | 0.01796 | 0.338 |
| 31945 | KCNJ2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 11 | LRIG1 | Sponge network | -0.436 | 0.05325 | -0.537 | 0.00588 | 0.338 |
| 31946 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | LRIG1 | Sponge network | 0.174 | 0.30034 | -0.537 | 0.00588 | 0.338 |
| 31947 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 12 | SPG20 | Sponge network | 0.727 | 3.0E-5 | -1.16 | 0 | 0.338 |
| 31948 | SOX2-OT |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 11 | RNF180 | Sponge network | -1.264 | 6.0E-5 | -1.127 | 1.0E-5 | 0.338 |
| 31949 | RP11-119F7.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | LATS2 | Sponge network | 0.225 | 0.30644 | 0.159 | 0.2304 | 0.338 |
| 31950 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 18 | NOTCH2NL | Sponge network | -0.096 | 0.67958 | 0.024 | 0.84307 | 0.338 |
| 31951 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | -0.668 | 3.0E-5 | -1.751 | 1.0E-5 | 0.338 |
| 31952 | AC156455.1 |
hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-942-5p | 11 | GPC6 | Sponge network | 1.154 | 0.00041 | -0.054 | 0.81465 | 0.338 |
| 31953 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 43 | CBX5 | Sponge network | 0.75 | 0.00054 | 0.165 | 0.24446 | 0.338 |
| 31954 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | TET1 | Sponge network | 0.299 | 0.57722 | -0.135 | 0.52138 | 0.338 |
| 31955 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-501-5p | 18 | CXXC4 | Sponge network | -0.777 | 0.16667 | -0.433 | 0.21055 | 0.338 |
| 31956 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p | 14 | CBL | Sponge network | 1.093 | 0 | 0.291 | 0.01267 | 0.338 |
| 31957 | RP11-119F7.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | ERG | Sponge network | 0.225 | 0.30644 | 0.002 | 0.99011 | 0.338 |
| 31958 | RP11-458F8.4 |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-539-5p | 13 | DDX6 | Sponge network | 1.518 | 0 | 0.091 | 0.36428 | 0.338 |
| 31959 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-589-3p | 32 | CCND2 | Sponge network | -0.154 | 0.53954 | -0.267 | 0.3112 | 0.338 |
| 31960 | CTD-2666L21.1 |
hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-629-3p | 19 | PIK3R1 | Sponge network | 0.368 | 0.20093 | -0.133 | 0.36854 | 0.338 |
| 31961 | RP11-67L2.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PRUNE2 | Sponge network | 0.72 | 0.0001 | -1.733 | 0.00022 | 0.338 |
| 31962 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p | 22 | APC | Sponge network | -0.096 | 0.67958 | -0.255 | 0.04051 | 0.338 |
| 31963 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | LATS1 | Sponge network | 1.089 | 0 | 0.109 | 0.34208 | 0.338 |
| 31964 | HOXB-AS3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p | 10 | SCN9A | Sponge network | 0.343 | 0.28887 | -0.525 | 0.10593 | 0.338 |
| 31965 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-942-5p | 12 | GLIPR1 | Sponge network | 0.082 | 0.55397 | 0.146 | 0.3276 | 0.338 |
| 31966 | RP11-35G9.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | MED12L | Sponge network | 0.657 | 4.0E-5 | -0.194 | 0.55299 | 0.338 |
| 31967 | SH3BP5-AS1 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 20 | DICER1 | Sponge network | 0.267 | 0.2937 | 0.042 | 0.674 | 0.338 |
| 31968 | RP11-38L15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-25-3p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | NOTCH2 | Sponge network | 0.344 | 0.3127 | 0.138 | 0.35163 | 0.338 |
| 31969 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | CBX5 | Sponge network | 0.538 | 0.00076 | 0.165 | 0.24446 | 0.338 |
| 31970 | RP5-1198O20.4 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-365a-3p;hsa-miR-378a-5p | 14 | CACNA1E | Sponge network | 0.997 | 0.00066 | 1.752 | 0 | 0.338 |
| 31971 | BDNF-AS |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | RBMS3 | Sponge network | -0.668 | 3.0E-5 | -0.519 | 0.03551 | 0.338 |
| 31972 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ABCC9 | Sponge network | 0.657 | 4.0E-5 | -0.466 | 0.14121 | 0.338 |
| 31973 | LINC00476 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 45 | NTRK2 | Sponge network | -0.615 | 0.00015 | -0.736 | 0.06495 | 0.338 |
| 31974 | DIO3OS |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-671-5p;hsa-miR-7-5p | 13 | EHD3 | Sponge network | -0.773 | 0.04073 | -0.484 | 0.01747 | 0.338 |
| 31975 | BDNF-AS |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | PRKAA2 | Sponge network | -0.668 | 3.0E-5 | -2.462 | 0 | 0.338 |
| 31976 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | RCAN2 | Sponge network | 0.782 | 0.00016 | -0.33 | 0.15172 | 0.338 |
| 31977 | RP11-13K12.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-500a-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-877-5p | 22 | CRTC1 | Sponge network | -0.318 | 0.65847 | -0.116 | 0.28412 | 0.338 |
| 31978 | RP11-345P4.7 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | USP9X | Sponge network | 1.597 | 0 | 0.222 | 0.01689 | 0.338 |
| 31979 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | KAT6B | Sponge network | -0.358 | 0.17255 | -0.478 | 0.00018 | 0.338 |
| 31980 | CTC-559E9.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-539-5p;hsa-miR-576-5p | 25 | ARHGEF12 | Sponge network | 0.594 | 0.00064 | 0.09 | 0.35693 | 0.338 |
| 31981 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-338-3p | 11 | ELMO1 | Sponge network | 0.214 | 0.18246 | 0.04 | 0.83147 | 0.338 |
| 31982 | RP11-6N17.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-331-3p | 11 | SNX29 | Sponge network | 0.108 | 0.69556 | -0.34 | 0.01371 | 0.338 |
| 31983 | LINC00957 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 24 | ACSL6 | Sponge network | -0.421 | 0.0114 | -1.593 | 0 | 0.338 |
| 31984 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p | 16 | MCC | Sponge network | -2.549 | 0.00528 | -1.005 | 0 | 0.338 |
| 31985 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p | 32 | CBX5 | Sponge network | 0.056 | 0.81195 | 0.165 | 0.24446 | 0.338 |
| 31986 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-92a-3p | 21 | GPC6 | Sponge network | 0.056 | 0.81195 | -0.054 | 0.81465 | 0.338 |
| 31987 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p | 30 | PBX1 | Sponge network | 0.253 | 0.09565 | -1.206 | 0 | 0.338 |
| 31988 | RP11-25K19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 33 | TGFBR3 | Sponge network | -1.057 | 0.00029 | -1.173 | 1.0E-5 | 0.338 |
| 31989 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SEMA3E | Sponge network | -1.057 | 0.00029 | -1.717 | 0.00137 | 0.338 |
| 31990 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 13 | PICALM | Sponge network | 1.089 | 0 | 0.086 | 0.23523 | 0.338 |
| 31991 | CTC-510F12.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-3614-5p | 16 | GAS7 | Sponge network | -0.691 | 0.00146 | -0.555 | 0.01602 | 0.338 |
| 31992 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-93-5p | 37 | GAS7 | Sponge network | -0.206 | 0.12942 | -0.555 | 0.01602 | 0.338 |
| 31993 | RP11-439A17.10 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | SSBP2 | Sponge network | 0.051 | 0.95756 | -1.58 | 0 | 0.338 |
| 31994 | RP11-819C21.1 |
hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p | 15 | CSDE1 | Sponge network | 0.537 | 0.00139 | -0.493 | 0 | 0.338 |
| 31995 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | RBL2 | Sponge network | -0.576 | 0.10121 | -0.282 | 0.00254 | 0.338 |
| 31996 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 31 | SOX5 | Sponge network | -0.322 | 0.25945 | -1.091 | 4.0E-5 | 0.338 |
| 31997 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TBX18 | Sponge network | -1.216 | 0 | 0.843 | 0.02945 | 0.338 |
| 31998 | ARHGEF26-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | CRTC1 | Sponge network | -2.46 | 0 | -0.116 | 0.28412 | 0.338 |
| 31999 | BVES-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p | 12 | LUZP2 | Sponge network | -2.62 | 0 | -0.056 | 0.89416 | 0.338 |
| 32000 | RP11-360F5.3 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-629-3p | 11 | RASSF2 | Sponge network | 1.867 | 0 | -0.273 | 0.21961 | 0.338 |
| 32001 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | AFF3 | Sponge network | 0.214 | 0.18246 | -2.373 | 0 | 0.338 |
| 32002 | RP11-401P9.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-589-5p | 13 | ROBO1 | Sponge network | -0.384 | 0.40652 | -0.009 | 0.96154 | 0.338 |
| 32003 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 35 | THRB | Sponge network | 0.419 | 0.0319 | -1.117 | 0.0004 | 0.338 |
| 32004 | LINC00173 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 14 | CDH19 | Sponge network | 0.056 | 0.8494 | -3.434 | 0 | 0.338 |
| 32005 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 12 | CLASP2 | Sponge network | 1.055 | 0 | -0.038 | 0.71 | 0.338 |
| 32006 | LINC01006 |
hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-576-5p | 28 | PBX1 | Sponge network | -0.702 | 0.04814 | -1.206 | 0 | 0.338 |
| 32007 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-93-3p | 10 | PCDH10 | Sponge network | 0.598 | 0.38481 | -1.644 | 0.0078 | 0.338 |
| 32008 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-339-5p;hsa-miR-33a-3p | 12 | SON | Sponge network | 0.792 | 0.0049 | 0.168 | 0.04406 | 0.338 |
| 32009 | LINC01018 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 13 | KCNAB1 | Sponge network | -2.915 | 0.00015 | -0.755 | 2.0E-5 | 0.338 |
| 32010 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-92a-3p | 20 | GRIN2A | Sponge network | 0.214 | 0.18246 | -2.161 | 0 | 0.338 |
| 32011 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 28 | PBX1 | Sponge network | 1.302 | 7.0E-5 | -1.206 | 0 | 0.338 |
| 32012 | AC010226.4 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 14 | OPCML | Sponge network | -0.811 | 2.0E-5 | -2.498 | 0 | 0.338 |
| 32013 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-532-5p | 17 | ZMYND11 | Sponge network | -0.384 | 0.40652 | -0.2 | 0.05449 | 0.338 |
| 32014 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-424-5p | 16 | UNC80 | Sponge network | -3.586 | 0.00032 | -1.934 | 0 | 0.338 |
| 32015 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-93-3p | 24 | SUFU | Sponge network | -0.154 | 0.78939 | -0.04 | 0.65489 | 0.338 |
| 32016 | DNAJC27-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484 | 14 | FLNA | Sponge network | -0.969 | 2.0E-5 | -0.771 | 0.01377 | 0.338 |
| 32017 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 24 | ZMYM2 | Sponge network | 0.532 | 0.00505 | 0.279 | 0.01273 | 0.338 |
| 32018 | LINC00403 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-299-5p | 11 | DIP2C | Sponge network | -3.586 | 0.00032 | -0.436 | 0.01713 | 0.338 |
| 32019 | RP11-401P9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-877-5p | 21 | NAV3 | Sponge network | -0.384 | 0.40652 | 0.212 | 0.47729 | 0.338 |
| 32020 | RP11-254F7.2 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-5p | 10 | TLL1 | Sponge network | 0.176 | 0.37402 | -1.195 | 3.0E-5 | 0.338 |
| 32021 | TMEM9B-AS1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | ERCC6 | Sponge network | 0.529 | 0.00552 | 0.23 | 0.015 | 0.338 |
| 32022 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-539-5p | 11 | NEDD4 | Sponge network | -0.309 | 0.1145 | 0.02 | 0.89834 | 0.338 |
| 32023 | RP11-373D23.3 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | UBE4B | Sponge network | -0.285 | 0.2172 | -0.235 | 0.007 | 0.338 |
| 32024 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-590-3p | 10 | ZNF536 | Sponge network | -0.899 | 6.0E-5 | -3.115 | 0 | 0.338 |
| 32025 | LINC00702 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | TMTC2 | Sponge network | -0.737 | 0.06182 | -0.215 | 0.18248 | 0.338 |
| 32026 | CTA-29F11.1 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p;hsa-miR-664a-5p | 11 | CBFB | Sponge network | 0.583 | 0.00027 | 1.061 | 0 | 0.338 |
| 32027 | CTC-444N24.11 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | DDX3X | Sponge network | 1.153 | 0 | -0.152 | 0.07686 | 0.338 |
| 32028 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484 | 14 | GREM1 | Sponge network | -0.08 | 0.90092 | 0.902 | 0.01691 | 0.338 |
| 32029 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-3p | 29 | FOXO3 | Sponge network | 0.494 | 0.00339 | -0.04 | 0.72028 | 0.338 |
| 32030 | RP1-151F17.2 |
hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | HERC1 | Sponge network | 0.222 | 0.29133 | -0.196 | 0.08544 | 0.338 |
| 32031 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | HBP1 | Sponge network | -2.117 | 0.00161 | -0.454 | 0 | 0.338 |
| 32032 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-345-5p | 12 | ARHGAP28 | Sponge network | -2.095 | 0.00112 | -0.911 | 0.00013 | 0.338 |
| 32033 | RP11-620J15.3 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-744-5p | 10 | PRKACA | Sponge network | -0.739 | 0.00044 | -0.255 | 0.0012 | 0.338 |
| 32034 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-940 | 26 | CRTC3 | Sponge network | 0.421 | 0.00084 | 0.164 | 0.16786 | 0.338 |
| 32035 | RP11-2H8.2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p | 12 | ITGB1 | Sponge network | 1.089 | 0 | 0.524 | 0.00017 | 0.337 |
| 32036 | MIR497HG |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 24 | TET1 | Sponge network | -1.439 | 4.0E-5 | -0.135 | 0.52138 | 0.337 |
| 32037 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 21 | GREM1 | Sponge network | -0.405 | 0.22013 | 0.902 | 0.01691 | 0.337 |
| 32038 | C4B-AS1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-629-5p;hsa-miR-96-5p | 10 | PCDH17 | Sponge network | 2.306 | 9.0E-5 | 0.731 | 2.0E-5 | 0.337 |
| 32039 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-7-1-3p | 13 | DNAJB4 | Sponge network | 0.16 | 0.50785 | -0.7 | 1.0E-5 | 0.337 |
| 32040 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | HDAC9 | Sponge network | -0.976 | 0.00025 | -0.755 | 0.00495 | 0.337 |
| 32041 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 25 | HBP1 | Sponge network | -0.487 | 0.02157 | -0.454 | 0 | 0.337 |
| 32042 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 55 | KIF1B | Sponge network | 0.603 | 0.00127 | -0.118 | 0.32258 | 0.337 |
| 32043 | AP001055.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-98-5p | 11 | DMD | Sponge network | 0.693 | 0.00076 | -1.506 | 0 | 0.337 |
| 32044 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 22 | FOXP1 | Sponge network | 1.153 | 0 | 0.042 | 0.74368 | 0.337 |
| 32045 | RP1-151F17.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p | 12 | DST | Sponge network | -0.321 | 0.22761 | -0.713 | 0.00096 | 0.337 |
| 32046 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 22 | PRKCE | Sponge network | -0.793 | 7.0E-5 | -0.403 | 0.00056 | 0.337 |
| 32047 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | QKI | Sponge network | 0.796 | 4.0E-5 | -0.333 | 0.04111 | 0.337 |
| 32048 | LINC00957 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | CSRNP1 | Sponge network | -0.421 | 0.0114 | -1.448 | 0 | 0.337 |
| 32049 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-576-5p | 15 | GREM1 | Sponge network | -0.053 | 0.80685 | 0.902 | 0.01691 | 0.337 |
| 32050 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | USP25 | Sponge network | 0.064 | 0.72934 | -0.242 | 0.01397 | 0.337 |
| 32051 | AC108488.4 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | RECK | Sponge network | 0.259 | 0.12542 | -1.043 | 0 | 0.337 |
| 32052 | RP11-390K5.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-484;hsa-miR-590-3p | 12 | ZFX | Sponge network | 1.148 | 2.0E-5 | 0.09 | 0.42563 | 0.337 |
| 32053 | RP11-264I13.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p | 11 | FOXP1 | Sponge network | 0.421 | 0.01652 | 0.042 | 0.74368 | 0.337 |
| 32054 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-93-5p | 25 | SYNE1 | Sponge network | -1.047 | 0 | -0.738 | 0.00316 | 0.337 |
| 32055 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 22 | ANK2 | Sponge network | 0.537 | 0.00139 | -1.603 | 1.0E-5 | 0.337 |
| 32056 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | EPAS1 | Sponge network | -1.216 | 0 | -0.184 | 0.14418 | 0.337 |
| 32057 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 36 | TMEM132B | Sponge network | -0.738 | 0.21233 | -0.83 | 0.01084 | 0.337 |
| 32058 | LIPE-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-942-5p | 27 | NOTCH2 | Sponge network | 0.078 | 0.55803 | 0.138 | 0.35163 | 0.337 |
| 32059 | RASSF8-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | HERC1 | Sponge network | 0.335 | 0.20596 | -0.196 | 0.08544 | 0.337 |
| 32060 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 10 | PDGFC | Sponge network | 0.61 | 0.01593 | -0.33 | 0.06483 | 0.337 |
| 32061 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 40 | TSHZ3 | Sponge network | 0.083 | 0.66405 | -0.556 | 0.01516 | 0.337 |
| 32062 | CTD-2184D3.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZFX | Sponge network | 0.272 | 0.44692 | 0.09 | 0.42563 | 0.337 |
| 32063 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 14 | ACTB | Sponge network | -0.769 | 0.00035 | -0.247 | 0.01736 | 0.337 |
| 32064 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 31 | USP12 | Sponge network | -1.697 | 0.00889 | -0.102 | 0.40768 | 0.337 |
| 32065 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | 0.199 | 0.38015 | -1.751 | 1.0E-5 | 0.337 |
| 32066 | MIR22HG |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 52 | CADM2 | Sponge network | -1.047 | 0 | -3.782 | 0 | 0.337 |
| 32067 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 18 | KANK1 | Sponge network | 0.214 | 0.18246 | -0.55 | 0.00134 | 0.337 |
| 32068 | RP11-360F5.3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-590-3p;hsa-miR-877-5p | 10 | ANK2 | Sponge network | 1.867 | 0 | -1.603 | 1.0E-5 | 0.337 |
| 32069 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 56 | WDFY3 | Sponge network | 1.302 | 7.0E-5 | -0.201 | 0.0967 | 0.337 |
| 32070 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | EBF1 | Sponge network | 0.889 | 9.0E-5 | -0.384 | 0.08856 | 0.337 |
| 32071 | ZSCAN16-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-942-5p | 31 | IGFBP5 | Sponge network | -0.342 | 0.02288 | -0.806 | 0.00105 | 0.337 |
| 32072 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 46 | WDFY3 | Sponge network | 0.349 | 0.01695 | -0.201 | 0.0967 | 0.337 |
| 32073 | RP11-434B12.1 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 19 | SYNPO2 | Sponge network | -0.031 | 0.89063 | -2.342 | 0 | 0.337 |
| 32074 | AC003090.1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-361-5p | 15 | ABL1 | Sponge network | -2.117 | 0.00161 | -0.215 | 0.07804 | 0.337 |
| 32075 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NCAM2 | Sponge network | -0.737 | 0.10822 | -0.482 | 0.22565 | 0.337 |
| 32076 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | ITGAV | Sponge network | 1.302 | 7.0E-5 | 0.337 | 0.01002 | 0.337 |
| 32077 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-93-5p | 15 | CNTNAP3 | Sponge network | -0.384 | 0.40652 | -1.465 | 0.00033 | 0.337 |
| 32078 | RP1-302G2.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-93-3p | 11 | BNC2 | Sponge network | -1.483 | 9.0E-5 | -0.491 | 0.09988 | 0.337 |
| 32079 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-590-3p | 14 | PTPN11 | Sponge network | 1.317 | 0 | 0.431 | 2.0E-5 | 0.337 |
| 32080 | DNAJC27-AS1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p | 22 | MRVI1 | Sponge network | -0.969 | 2.0E-5 | -1.051 | 0.00022 | 0.337 |
| 32081 | KB-1460A1.5 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 13 | UBE4B | Sponge network | 0.603 | 0.00127 | -0.235 | 0.007 | 0.337 |
| 32082 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 36 | MCC | Sponge network | 0.374 | 0.20054 | -1.005 | 0 | 0.337 |
| 32083 | C4B-AS1 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 11 | NAV3 | Sponge network | 2.306 | 9.0E-5 | 0.212 | 0.47729 | 0.337 |
| 32084 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 48 | EPHA5 | Sponge network | -2.925 | 0 | -0.957 | 0.01733 | 0.337 |
| 32085 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p | 10 | FBXO31 | Sponge network | -0.896 | 0.00099 | -0.085 | 0.42389 | 0.337 |
| 32086 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PLSCR4 | Sponge network | 0.998 | 0 | -0.474 | 0.03833 | 0.337 |
| 32087 | PSMD5-AS1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 16 | ATM | Sponge network | 0.569 | 0.00067 | 0.178 | 0.21568 | 0.337 |
| 32088 | RP11-458D21.1 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-766-3p | 13 | ATM | Sponge network | 0.663 | 0.00073 | 0.178 | 0.21568 | 0.337 |
| 32089 | PART1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p | 32 | CTIF | Sponge network | -2.925 | 0 | -0.389 | 0.00549 | 0.337 |
| 32090 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 25 | YAP1 | Sponge network | -0.222 | 0.32445 | 0.22 | 0.11836 | 0.337 |
| 32091 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | GRIN2A | Sponge network | 0.064 | 0.72934 | -2.161 | 0 | 0.337 |
| 32092 | HNRNPU-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-337-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | RPS6KA3 | Sponge network | 1.601 | 0 | 0.256 | 0.02419 | 0.337 |
| 32093 | RP11-290F24.6 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-337-3p | 16 | MDM4 | Sponge network | 2.052 | 0 | 0.345 | 0.00762 | 0.337 |
| 32094 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 15 | EBF3 | Sponge network | -0.206 | 0.12942 | -0.058 | 0.81437 | 0.337 |
| 32095 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p | 18 | PTPRT | Sponge network | -1.375 | 0.08642 | -0.907 | 0.03727 | 0.337 |
| 32096 | TPTEP1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | JDP2 | Sponge network | -1.216 | 0 | -0.967 | 0 | 0.337 |
| 32097 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-590-3p | 14 | SAMHD1 | Sponge network | -0.096 | 0.67958 | 0.072 | 0.62895 | 0.337 |
| 32098 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | TMTC2 | Sponge network | -0.487 | 0.02157 | -0.215 | 0.18248 | 0.337 |
| 32099 | HAND2-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 26 | JDP2 | Sponge network | -1.697 | 0.00889 | -0.967 | 0 | 0.337 |
| 32100 | RP11-822E23.8 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | EGR2 | Sponge network | -0.777 | 0.16667 | 0.309 | 0.17079 | 0.337 |
| 32101 | RP3-402G11.26 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-22-3p;hsa-miR-505-5p;hsa-miR-590-3p | 15 | AR | Sponge network | -0.254 | 0.19303 | -1.554 | 0 | 0.337 |
| 32102 | RP11-180O5.2 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-23a-5p;hsa-miR-495-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | DDX6 | Sponge network | 3.607 | 0.0001 | 0.091 | 0.36428 | 0.337 |
| 32103 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484 | 16 | CRTC3 | Sponge network | -0.227 | 0.31657 | 0.164 | 0.16786 | 0.337 |
| 32104 | RP11-359E10.1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 13 | PCDH17 | Sponge network | 0.299 | 0.57722 | 0.731 | 2.0E-5 | 0.337 |
| 32105 | AF131215.9 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p | 12 | ATM | Sponge network | -0.322 | 0.25945 | 0.178 | 0.21568 | 0.337 |
| 32106 | RP1-302G2.5 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-935 | 10 | TSHZ3 | Sponge network | -1.483 | 9.0E-5 | -0.556 | 0.01516 | 0.337 |
| 32107 | RP11-67L2.2 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | PPM1L | Sponge network | 0.72 | 0.0001 | -0.537 | 0.00616 | 0.337 |
| 32108 | RP11-400K9.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-940 | 18 | CADM2 | Sponge network | -0.733 | 0.19469 | -3.782 | 0 | 0.337 |
| 32109 | TRAF3IP2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 36 | AFF1 | Sponge network | -0.323 | 0.01372 | -0.384 | 0.00053 | 0.337 |
| 32110 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p | 12 | MYCBP2 | Sponge network | 0.539 | 0.03722 | -0.027 | 0.82016 | 0.337 |
| 32111 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-324-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-98-5p | 27 | USP12 | Sponge network | -0.227 | 0.31657 | -0.102 | 0.40768 | 0.337 |
| 32112 | RP11-296I10.3 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-7-5p | 17 | ERC1 | Sponge network | 0.592 | 0.00014 | -0.108 | 0.38794 | 0.337 |
| 32113 | MEG3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-7-1-3p | 24 | ZMYND11 | Sponge network | 0.249 | 0.35902 | -0.2 | 0.05449 | 0.337 |
| 32114 | CTD-2336O2.1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 15 | PCDH10 | Sponge network | -0.141 | 0.26434 | -1.644 | 0.0078 | 0.337 |
| 32115 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p | 37 | EPHA5 | Sponge network | -1.249 | 7.0E-5 | -0.957 | 0.01733 | 0.337 |
| 32116 | EMX2OS |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 18 | ERRFI1 | Sponge network | -0.777 | 0.1134 | -0.798 | 1.0E-5 | 0.337 |
| 32117 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-495-3p | 11 | MYO9A | Sponge network | 1.597 | 0 | -0.147 | 0.32373 | 0.337 |
| 32118 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | KAT6B | Sponge network | 0.311 | 0.10344 | -0.478 | 0.00018 | 0.337 |
| 32119 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-590-3p | 21 | AKAP11 | Sponge network | 1.04 | 0.00023 | 0.196 | 0.09825 | 0.337 |
| 32120 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-454-3p;hsa-miR-590-3p | 15 | CXXC4 | Sponge network | 0.765 | 7.0E-5 | -0.433 | 0.21055 | 0.337 |
| 32121 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p | 12 | ERC1 | Sponge network | 0.959 | 0 | -0.108 | 0.38794 | 0.337 |
| 32122 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 19 | ITGB3 | Sponge network | -2.46 | 0 | -0.011 | 0.95751 | 0.337 |
| 32123 | BOLA3-AS1 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-429 | 13 | AFF3 | Sponge network | -0.246 | 0.35034 | -2.373 | 0 | 0.337 |
| 32124 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-628-5p | 15 | TRIM33 | Sponge network | 1.073 | 0.00216 | 0.402 | 7.0E-5 | 0.337 |
| 32125 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | AHNAK | Sponge network | 0.572 | 0.01722 | -1.207 | 0 | 0.337 |
| 32126 | LINC00173 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-493-5p;hsa-miR-539-5p | 22 | DMD | Sponge network | 0.056 | 0.8494 | -1.506 | 0 | 0.337 |
| 32127 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | GALNT13 | Sponge network | -0.096 | 0.67958 | -0.857 | 0.07439 | 0.337 |
| 32128 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | MTSS1 | Sponge network | -0.323 | 0.01372 | -0.81 | 0.00035 | 0.337 |
| 32129 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p | 15 | DIP2C | Sponge network | 1.46 | 1.0E-5 | -0.436 | 0.01713 | 0.337 |
| 32130 | FAM13A-AS1 |
hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | BTG2 | Sponge network | -0.83 | 0 | -1.763 | 0 | 0.337 |
| 32131 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-92a-3p | 26 | GRIA3 | Sponge network | -0.563 | 0.02545 | -1.956 | 0 | 0.337 |
| 32132 | RP1-151F17.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-330-3p | 12 | QKI | Sponge network | -0.321 | 0.22761 | -0.333 | 0.04111 | 0.337 |
| 32133 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p | 12 | CREB1 | Sponge network | 0.37 | 0.27614 | 0.203 | 0.00537 | 0.337 |
| 32134 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-3p | 15 | UNC80 | Sponge network | -2.549 | 0.00528 | -1.934 | 0 | 0.337 |
| 32135 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 20 | STARD13 | Sponge network | -0.942 | 0 | -0.187 | 0.27383 | 0.337 |
| 32136 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 23 | MYO9A | Sponge network | -0.176 | 0.59692 | -0.147 | 0.32373 | 0.337 |
| 32137 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 12 | ABCA10 | Sponge network | -1.057 | 0.00029 | -0.571 | 0.03674 | 0.337 |
| 32138 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | PPM1L | Sponge network | -1.654 | 0.00144 | -0.537 | 0.00616 | 0.337 |
| 32139 | RP11-195F19.9 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-539-5p;hsa-miR-7-5p | 22 | BTG2 | Sponge network | -0.726 | 0.00132 | -1.763 | 0 | 0.337 |
| 32140 | RP11-274B21.10 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | ABI2 | Sponge network | 1.073 | 0.00216 | 0.175 | 0.14787 | 0.337 |
| 32141 | RP11-57H14.4 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | AFF1 | Sponge network | 0.083 | 0.66405 | -0.384 | 0.00053 | 0.337 |
| 32142 | RP11-532F6.3 |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PAIP2 | Sponge network | -0.896 | 0.00099 | -0.515 | 0 | 0.337 |
| 32143 | RP11-141C7.4 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 10 | PHF6 | Sponge network | 2.412 | 0 | 0.785 | 0 | 0.337 |
| 32144 | AC010226.4 |
hsa-miR-1266-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-7-5p | 11 | KCNJ5 | Sponge network | -0.811 | 2.0E-5 | -0.281 | 0.3339 | 0.337 |
| 32145 | RP11-196G18.23 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 19 | ABI2 | Sponge network | 0.724 | 0.00039 | 0.175 | 0.14787 | 0.337 |
| 32146 | CTD-2006H14.2 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | UBR3 | Sponge network | 1.378 | 0 | -0.021 | 0.82774 | 0.337 |
| 32147 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p | 13 | ANK2 | Sponge network | -0.214 | 0.44248 | -1.603 | 1.0E-5 | 0.337 |
| 32148 | MYLK-AS1 |
hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-340-5p | 10 | INPP4B | Sponge network | -1.331 | 3.0E-5 | -0.097 | 0.60503 | 0.337 |
| 32149 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-93-5p | 28 | LRP1B | Sponge network | -0.154 | 0.78939 | -2.163 | 0 | 0.337 |
| 32150 | ZNF667-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 24 | SUFU | Sponge network | -0.976 | 0.00025 | -0.04 | 0.65489 | 0.337 |
| 32151 | ZNF582-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | EGR2 | Sponge network | -1.349 | 0.00017 | 0.309 | 0.17079 | 0.337 |
| 32152 | RP11-263K19.4 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-429 | 10 | DYNC1I1 | Sponge network | 0.859 | 0.00331 | -1.12 | 0.00107 | 0.337 |
| 32153 | RP11-359B12.2 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | GJA1 | Sponge network | 0.064 | 0.72934 | -0.485 | 0.02179 | 0.337 |
| 32154 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | ZMYM2 | Sponge network | 0.559 | 0.00026 | 0.279 | 0.01273 | 0.337 |
| 32155 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 28 | MAF | Sponge network | 0.199 | 0.38015 | -1.257 | 0 | 0.337 |
| 32156 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 11 | GPAM | Sponge network | 0.917 | 0 | 0.142 | 0.29732 | 0.337 |
| 32157 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | ST6GAL2 | Sponge network | -0.727 | 0.04726 | -1.201 | 0.00597 | 0.337 |
| 32158 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-766-3p | 11 | EZH1 | Sponge network | 0.657 | 4.0E-5 | -0.564 | 1.0E-5 | 0.337 |
| 32159 | CD27-AS1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 18 | ITPKB | Sponge network | 0.177 | 0.16681 | -0.704 | 0.00106 | 0.337 |
| 32160 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-539-5p | 13 | DDX6 | Sponge network | 0.917 | 0 | 0.091 | 0.36428 | 0.337 |
| 32161 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-708-5p | 29 | MTSS1 | Sponge network | -1.645 | 0 | -0.81 | 0.00035 | 0.337 |
| 32162 | RP11-305E6.4 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | DICER1 | Sponge network | 1.317 | 0 | 0.042 | 0.674 | 0.337 |
| 32163 | CTC-457L16.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | RASSF8 | Sponge network | 1.153 | 0.00114 | -0.122 | 0.65591 | 0.337 |
| 32164 | FENDRR |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | ZYX | Sponge network | -1.281 | 0.00012 | 0.019 | 0.89763 | 0.337 |
| 32165 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-940 | 10 | FAM172A | Sponge network | -0.777 | 0.16667 | -0.57 | 0 | 0.337 |
| 32166 | RP11-411K7.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CLIP1 | Sponge network | 0.155 | 0.81273 | -0.215 | 0.08684 | 0.337 |
| 32167 | LINC01018 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 28 | KCNA6 | Sponge network | -2.915 | 0.00015 | -1.531 | 0.00013 | 0.337 |
| 32168 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-942-5p | 23 | GPC6 | Sponge network | 0.082 | 0.55654 | -0.054 | 0.81465 | 0.337 |
| 32169 | H19 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p | 16 | CHD5 | Sponge network | 2.439 | 0 | -0.922 | 0.01521 | 0.337 |
| 32170 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p | 13 | PNRC1 | Sponge network | 0.335 | 0.20596 | -0.646 | 0 | 0.337 |
| 32171 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-324-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 26 | EPHA7 | Sponge network | -4.477 | 0 | -2.197 | 3.0E-5 | 0.337 |
| 32172 | ACTA2-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZNF844 | Sponge network | 0.194 | 0.64278 | -0.966 | 0.00035 | 0.337 |
| 32173 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | PIK3C2A | Sponge network | 1.317 | 0 | 0.061 | 0.53776 | 0.337 |
| 32174 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-93-5p | 29 | TSHZ3 | Sponge network | 0.253 | 0.09565 | -0.556 | 0.01516 | 0.337 |
| 32175 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | PDGFRA | Sponge network | 0.532 | 0.00505 | -0.372 | 0.0037 | 0.337 |
| 32176 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-26b-5p | 10 | USP6 | Sponge network | 0.858 | 3.0E-5 | 0.188 | 0.3871 | 0.337 |
| 32177 | AC127904.2 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 29 | LPP | Sponge network | 1.308 | 0 | -0.459 | 0.04475 | 0.337 |
| 32178 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 12 | SLC6A15 | Sponge network | -0.246 | 0.35034 | -2.373 | 2.0E-5 | 0.337 |
| 32179 | RP11-434H6.7 |
hsa-miR-132-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p | 14 | UBR3 | Sponge network | 0.358 | 0.14302 | -0.021 | 0.82774 | 0.337 |
| 32180 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | BNC2 | Sponge network | 0.537 | 0.00139 | -0.491 | 0.09988 | 0.337 |
| 32181 | RP11-597D13.9 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p | 20 | SYNPO2 | Sponge network | 1.302 | 7.0E-5 | -2.342 | 0 | 0.337 |
| 32182 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | KAT6B | Sponge network | -0.737 | 0.06182 | -0.478 | 0.00018 | 0.337 |
| 32183 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 49 | PPM1A | Sponge network | -0.576 | 0.10121 | -0.623 | 0 | 0.337 |
| 32184 | CTD-2089N3.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-455-5p | 10 | AKAP6 | Sponge network | -0.314 | 0.62033 | -1.586 | 3.0E-5 | 0.337 |
| 32185 | AC007879.7 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | CREB1 | Sponge network | 4.3 | 0 | 0.203 | 0.00537 | 0.337 |
| 32186 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | YAP1 | Sponge network | 1.317 | 0 | 0.22 | 0.11836 | 0.337 |
| 32187 | RP11-166D19.1 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | EDNRB | Sponge network | -0.576 | 0.10121 | -0.682 | 0.00138 | 0.337 |
| 32188 | RP11-725P16.2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | BCL9 | Sponge network | 0.422 | 0.27021 | 0.321 | 0.00267 | 0.337 |
| 32189 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | EBF1 | Sponge network | -0.663 | 0.10887 | -0.384 | 0.08856 | 0.337 |
| 32190 | CTD-2089N3.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-149-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-455-5p | 13 | NFIB | Sponge network | -0.314 | 0.62033 | 0.023 | 0.90795 | 0.337 |
| 32191 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-582-5p | 11 | DDX6 | Sponge network | 2.368 | 2.0E-5 | 0.091 | 0.36428 | 0.337 |
| 32192 | A1BG-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-589-5p | 14 | SOX5 | Sponge network | -0.164 | 0.45106 | -1.091 | 4.0E-5 | 0.337 |
| 32193 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p | 13 | RNASEL | Sponge network | -1.054 | 0.0706 | -0.272 | 0.01796 | 0.337 |
| 32194 | RP11-195B17.1 |
hsa-miR-141-3p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p | 10 | PCDH9 | Sponge network | 0.37 | 0.27614 | -1.823 | 4.0E-5 | 0.337 |
| 32195 | RP11-894P9.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-532-3p | 14 | FOXP1 | Sponge network | 0.993 | 0 | 0.042 | 0.74368 | 0.337 |
| 32196 | RP11-888D10.4 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 14 | UBR3 | Sponge network | 0.702 | 7.0E-5 | -0.021 | 0.82774 | 0.337 |
| 32197 | RP11-404E16.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-532-5p | 10 | DST | Sponge network | 0.097 | 0.91311 | -0.713 | 0.00096 | 0.337 |
| 32198 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 28 | RNF144A | Sponge network | 0.619 | 0.00157 | 0.594 | 0.0009 | 0.337 |
| 32199 | RP1-102E24.8 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p | 13 | RICTOR | Sponge network | 1.737 | 0 | 0.388 | 0.00188 | 0.337 |
| 32200 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | SASH1 | Sponge network | 0.906 | 0.31715 | -0.502 | 0.00175 | 0.337 |
| 32201 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 35 | ZFHX4 | Sponge network | 0.559 | 0.00026 | 0.018 | 0.95574 | 0.337 |
| 32202 | RP11-815I9.4 |
hsa-let-7g-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30c-5p;hsa-miR-378a-5p;hsa-miR-576-5p | 12 | CREB1 | Sponge network | 0.537 | 0.0006 | 0.203 | 0.00537 | 0.337 |
| 32203 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 16 | SOX5 | Sponge network | 0.497 | 0.01451 | -1.091 | 4.0E-5 | 0.337 |
| 32204 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 12 | ZFHX3 | Sponge network | 1.2 | 0.01813 | 0.389 | 0.01363 | 0.337 |
| 32205 | C4A-AS1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p | 12 | ABL2 | Sponge network | 3.345 | 0 | 0.82 | 0 | 0.337 |
| 32206 | CTC-296K1.3 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p | 11 | ABL1 | Sponge network | -2.095 | 0.00112 | -0.215 | 0.07804 | 0.337 |
| 32207 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-625-3p;hsa-miR-625-5p | 24 | CBX5 | Sponge network | 1.352 | 0 | 0.165 | 0.24446 | 0.337 |
| 32208 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 37 | BNC2 | Sponge network | 0.222 | 0.29133 | -0.491 | 0.09988 | 0.337 |
| 32209 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-511-5p | 16 | BCL6 | Sponge network | 0.331 | 0.57247 | -0.211 | 0.19489 | 0.337 |
| 32210 | LINC00957 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3C | Sponge network | -0.421 | 0.0114 | -0.272 | 0.31153 | 0.337 |
| 32211 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 15 | CSRNP1 | Sponge network | -0.976 | 0.00025 | -1.448 | 0 | 0.337 |
| 32212 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-7-1-3p | 12 | TBX18 | Sponge network | -0.611 | 0.09607 | 0.843 | 0.02945 | 0.337 |
| 32213 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-450b-5p;hsa-miR-93-5p | 15 | CDHR3 | Sponge network | -0.18 | 0.20343 | -0.977 | 4.0E-5 | 0.337 |
| 32214 | DNAJC27-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 42 | GRIK3 | Sponge network | -0.969 | 2.0E-5 | -3.208 | 0 | 0.337 |
| 32215 | RP11-263K19.4 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-378c | 16 | BRD3 | Sponge network | 0.859 | 0.00331 | 0.37 | 0.00013 | 0.337 |
| 32216 | LINC00094 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p | 18 | ERCC4 | Sponge network | 0.253 | 0.09565 | 0.126 | 0.15266 | 0.337 |
| 32217 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-942-5p | 14 | RB1CC1 | Sponge network | 0.101 | 0.60919 | 0.175 | 0.12559 | 0.337 |
| 32218 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 13 | MCC | Sponge network | 0.497 | 0.01451 | -1.005 | 0 | 0.337 |
| 32219 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 22 | TACC1 | Sponge network | 0.419 | 0.0319 | -0.362 | 0.05406 | 0.337 |
| 32220 | MEG3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 28 | BRD3 | Sponge network | 0.249 | 0.35902 | 0.37 | 0.00013 | 0.337 |
| 32221 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p | 16 | PBX1 | Sponge network | 0.368 | 0.20093 | -1.206 | 0 | 0.337 |
| 32222 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 75 | AFF4 | Sponge network | -0.737 | 0.06182 | -0.266 | 0.01565 | 0.337 |
| 32223 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p | 32 | MAPK1 | Sponge network | 0.401 | 0.00066 | -0.017 | 0.81465 | 0.337 |
| 32224 | LINC00957 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 15 | CSDE1 | Sponge network | -0.421 | 0.0114 | -0.493 | 0 | 0.337 |
| 32225 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 23 | ZBTB4 | Sponge network | 0.253 | 0.09565 | -0.739 | 0 | 0.337 |
| 32226 | USP3-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p | 18 | PIK3CA | Sponge network | -0.162 | 0.47687 | 0.195 | 0.06908 | 0.337 |
| 32227 | U91328.19 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-500a-3p;hsa-miR-7-5p;hsa-miR-93-3p | 28 | ERC1 | Sponge network | -0.303 | 0.21002 | -0.108 | 0.38794 | 0.337 |
| 32228 | AP000487.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p | 15 | ABL2 | Sponge network | 1.987 | 0 | 0.82 | 0 | 0.337 |
| 32229 | LINC00641 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 22 | CDH23 | Sponge network | -0.222 | 0.32445 | -0.974 | 0.00034 | 0.337 |
| 32230 | RP11-244O19.1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | RIMS2 | Sponge network | -0.096 | 0.67958 | -1.365 | 0.00208 | 0.337 |
| 32231 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 21 | NIN | Sponge network | 0.75 | 0.00054 | -0.253 | 0.13794 | 0.337 |
| 32232 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-7-5p;hsa-miR-93-5p | 27 | ZBTB4 | Sponge network | -0.384 | 0.40652 | -0.739 | 0 | 0.337 |
| 32233 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 12 | PDS5B | Sponge network | 0.663 | 0.00073 | 0.259 | 0.00962 | 0.337 |
| 32234 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | PGR | Sponge network | 0.131 | 0.8436 | -1.704 | 0 | 0.337 |
| 32235 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-93-3p | 24 | FOXO3 | Sponge network | -0.154 | 0.78939 | -0.04 | 0.72028 | 0.337 |
| 32236 | RP4-669L17.10 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-625-5p | 16 | ABI2 | Sponge network | 1.093 | 0 | 0.175 | 0.14787 | 0.337 |
| 32237 | OIP5-AS1 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 15 | RAPGEF5 | Sponge network | 0.421 | 0.00084 | 0.536 | 4.0E-5 | 0.337 |
| 32238 | RP11-159D12.2 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-590-3p | 12 | ITGB1 | Sponge network | 1.254 | 0 | 0.524 | 0.00017 | 0.337 |
| 32239 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 25 | CUL5 | Sponge network | 0.848 | 0 | 0.026 | 0.77301 | 0.337 |
| 32240 | CALML3-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p | 16 | KLF10 | Sponge network | 0.95 | 0.15943 | -0.783 | 0 | 0.337 |
| 32241 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 21 | MOB1B | Sponge network | 0.411 | 0.1335 | 0.197 | 0.15266 | 0.337 |
| 32242 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 12 | GPAM | Sponge network | 0.826 | 1.0E-5 | 0.142 | 0.29732 | 0.337 |
| 32243 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 22 | NAV3 | Sponge network | -0.738 | 0.21233 | 0.212 | 0.47729 | 0.337 |
| 32244 | PART1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-935 | 15 | STAT5B | Sponge network | -2.925 | 0 | -0.182 | 0.1082 | 0.337 |
| 32245 | LINC00173 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-940 | 11 | PEG3 | Sponge network | 0.056 | 0.8494 | -1.74 | 0 | 0.337 |
| 32246 | LINC00863 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 26 | CBL | Sponge network | 0.189 | 0.13981 | 0.291 | 0.01267 | 0.337 |
| 32247 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | ANK2 | Sponge network | -0.023 | 0.95109 | -1.603 | 1.0E-5 | 0.337 |
| 32248 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-5p | 15 | PDGFRA | Sponge network | -2.076 | 0.00548 | -0.372 | 0.0037 | 0.336 |
| 32249 | LINC00654 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | CTNNA3 | Sponge network | 0.374 | 0.20054 | -2.046 | 0.00019 | 0.336 |
| 32250 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 12 | ADARB1 | Sponge network | -2.039 | 0.03447 | -0.335 | 0.06631 | 0.336 |
| 32251 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PRRX1 | Sponge network | 0.61 | 0.01593 | 1.316 | 0 | 0.336 |
| 32252 | FAM66C |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PTCH1 | Sponge network | -0.901 | 0.00019 | -0.1 | 0.6179 | 0.336 |
| 32253 | AC108488.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p | 17 | DLC1 | Sponge network | 0.259 | 0.12542 | -0.382 | 0.05038 | 0.336 |
| 32254 | PGM5-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 17 | EYA4 | Sponge network | -4.546 | 0 | 0.3 | 0.36876 | 0.336 |
| 32255 | TP73-AS1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-92a-3p | 18 | CSDE1 | Sponge network | -0.563 | 0.02545 | -0.493 | 0 | 0.336 |
| 32256 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-361-3p | 10 | NCOA1 | Sponge network | -7.551 | 0 | -0.432 | 0 | 0.336 |
| 32257 | HOXB-AS2 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-628-5p | 18 | ERC1 | Sponge network | 1.316 | 0.01106 | -0.108 | 0.38794 | 0.336 |
| 32258 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 21 | CDHR3 | Sponge network | 0.219 | 0.1516 | -0.977 | 4.0E-5 | 0.336 |
| 32259 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-590-3p | 13 | PNRC1 | Sponge network | -0.342 | 0.02288 | -0.646 | 0 | 0.336 |
| 32260 | RP11-254F7.2 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-590-5p | 13 | LIMCH1 | Sponge network | 0.176 | 0.37402 | -0.915 | 1.0E-5 | 0.336 |
| 32261 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PDS5B | Sponge network | 0.401 | 0.00066 | 0.259 | 0.00962 | 0.336 |
| 32262 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-338-5p | 13 | MYBL1 | Sponge network | 0.594 | 0.00064 | 0.494 | 0.02152 | 0.336 |
| 32263 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-625-5p;hsa-miR-96-5p | 19 | NUAK1 | Sponge network | 0.898 | 1.0E-5 | 0.904 | 0 | 0.336 |
| 32264 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | SLITRK3 | Sponge network | 0.101 | 0.60919 | -3.328 | 0 | 0.336 |
| 32265 | RP11-46C24.7 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-590-3p | 11 | DNMT3A | Sponge network | 0.392 | 0.0278 | 0.302 | 0.0262 | 0.336 |
| 32266 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-493-5p | 17 | CREBBP | Sponge network | 0.75 | 0.00054 | 0.126 | 0.15186 | 0.336 |
| 32267 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-590-3p | 11 | NEDD4 | Sponge network | 0.178 | 0.29106 | 0.02 | 0.89834 | 0.336 |
| 32268 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SLC6A15 | Sponge network | -0.709 | 0.18168 | -2.373 | 2.0E-5 | 0.336 |
| 32269 | RP11-678G14.3 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p | 10 | CDH23 | Sponge network | -4.477 | 0 | -0.974 | 0.00034 | 0.336 |
| 32270 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p | 11 | GREM1 | Sponge network | 0.497 | 0.01451 | 0.902 | 0.01691 | 0.336 |
| 32271 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 20 | CUL5 | Sponge network | 0.417 | 0.00445 | 0.026 | 0.77301 | 0.336 |
| 32272 | RP11-262H14.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | MOB1B | Sponge network | -0.121 | 0.53195 | 0.197 | 0.15266 | 0.336 |
| 32273 | PSMG3-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 13 | CXCL12 | Sponge network | -0.125 | 0.49414 | -1.421 | 0 | 0.336 |
| 32274 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p | 24 | ERC1 | Sponge network | -0.246 | 0.35034 | -0.108 | 0.38794 | 0.336 |
| 32275 | RP11-326G21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | NFIB | Sponge network | -0.021 | 0.93598 | 0.023 | 0.90795 | 0.336 |
| 32276 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 17 | SLC5A3 | Sponge network | 0.72 | 0.0001 | 0.493 | 5.0E-5 | 0.336 |
| 32277 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 28 | BHLHE41 | Sponge network | -0.793 | 7.0E-5 | 0.353 | 0.12753 | 0.336 |
| 32278 | RP4-669P10.16 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-424-5p;hsa-miR-542-3p | 10 | DYNC1I1 | Sponge network | -0.301 | 0.06597 | -1.12 | 0.00107 | 0.336 |
| 32279 | PWAR6 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | ITGB1 | Sponge network | -1.575 | 6.0E-5 | 0.524 | 0.00017 | 0.336 |
| 32280 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | CREM | Sponge network | -2.46 | 0 | -0.345 | 0.00258 | 0.336 |
| 32281 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 30 | ARID5B | Sponge network | 0.72 | 0.0001 | 0.342 | 0.01676 | 0.336 |
| 32282 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 29 | CTIF | Sponge network | -2.46 | 0 | -0.389 | 0.00549 | 0.336 |
| 32283 | RP11-822E23.8 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p | 11 | ITGB1 | Sponge network | -0.777 | 0.16667 | 0.524 | 0.00017 | 0.336 |
| 32284 | GS1-124K5.11 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-744-3p | 11 | KIF5B | Sponge network | 0.494 | 0.00339 | 0.362 | 0.00066 | 0.336 |
| 32285 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-590-3p | 14 | MCC | Sponge network | -1.375 | 0.08642 | -1.005 | 0 | 0.336 |
| 32286 | LINC01018 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | FYN | Sponge network | -2.915 | 0.00015 | -0.131 | 0.37051 | 0.336 |
| 32287 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | MAF | Sponge network | -1.654 | 0.00144 | -1.257 | 0 | 0.336 |
| 32288 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 14 | CHD2 | Sponge network | 1.317 | 0 | 0.049 | 0.55371 | 0.336 |
| 32289 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p | 21 | SLC6A15 | Sponge network | -0.162 | 0.47687 | -2.373 | 2.0E-5 | 0.336 |
| 32290 | RP11-67L2.2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ITPKB | Sponge network | 0.72 | 0.0001 | -0.704 | 0.00106 | 0.336 |
| 32291 | RP5-1198O20.4 |
hsa-miR-142-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 10 | TTL | Sponge network | 0.997 | 0.00066 | 0.494 | 1.0E-5 | 0.336 |
| 32292 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CXXC4 | Sponge network | 0.246 | 0.59912 | -0.433 | 0.21055 | 0.336 |
| 32293 | RP11-755F10.1 |
hsa-miR-103a-2-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-34a-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-940 | 12 | AFF4 | Sponge network | 0.952 | 0 | -0.266 | 0.01565 | 0.336 |
| 32294 | LINC00969 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-625-3p | 24 | ABL2 | Sponge network | 0.683 | 1.0E-5 | 0.82 | 0 | 0.336 |
| 32295 | MIR22HG |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-940 | 11 | NR3C2 | Sponge network | -1.047 | 0 | -1.56 | 0 | 0.336 |
| 32296 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 48 | IL6ST | Sponge network | 0.199 | 0.38015 | -0.402 | 0.00676 | 0.336 |
| 32297 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 46 | RUNX1T1 | Sponge network | -0.125 | 0.49414 | -0.344 | 0.2374 | 0.336 |
| 32298 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | RBMS3 | Sponge network | 0.898 | 1.0E-5 | -0.519 | 0.03551 | 0.336 |
| 32299 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | PRKCE | Sponge network | -0.355 | 0.0077 | -0.403 | 0.00056 | 0.336 |
| 32300 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 18 | KIT | Sponge network | 0.199 | 0.38015 | -2.184 | 0 | 0.336 |
| 32301 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 10 | UBE4B | Sponge network | 0.559 | 0.00026 | -0.235 | 0.007 | 0.336 |
| 32302 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429 | 12 | ABCB5 | Sponge network | -0.246 | 0.35034 | -0.956 | 0.04077 | 0.336 |
| 32303 | U91328.19 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | -0.303 | 0.21002 | 0.783 | 0.00072 | 0.336 |
| 32304 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-590-3p | 26 | SPRY3 | Sponge network | 0.765 | 7.0E-5 | 0.016 | 0.91286 | 0.336 |
| 32305 | A2M-AS1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p | 12 | CSDE1 | Sponge network | -0.08 | 0.90092 | -0.493 | 0 | 0.336 |
| 32306 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-493-5p | 23 | USP6 | Sponge network | 0.75 | 0.00054 | 0.188 | 0.3871 | 0.336 |
| 32307 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-5p | 24 | MOB1B | Sponge network | 0.176 | 0.37402 | 0.197 | 0.15266 | 0.336 |
| 32308 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 26 | FOXO3 | Sponge network | 0.782 | 0.00016 | -0.04 | 0.72028 | 0.336 |
| 32309 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-625-3p | 29 | TMTC3 | Sponge network | 0.537 | 0.00139 | 0.123 | 0.22189 | 0.336 |
| 32310 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p | 15 | DKK3 | Sponge network | -0.18 | 0.20343 | -0.052 | 0.77783 | 0.336 |
| 32311 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-19b-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 11 | EBF1 | Sponge network | 0.199 | 0.52763 | -0.384 | 0.08856 | 0.336 |
| 32312 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-877-5p | 23 | CRTC3 | Sponge network | -0.615 | 0.00015 | 0.164 | 0.16786 | 0.336 |
| 32313 | RP13-516M14.1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-582-5p | 12 | STARD13 | Sponge network | 0.389 | 0.04293 | -0.187 | 0.27383 | 0.336 |
| 32314 | RP11-1006G14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 45 | RUNX1T1 | Sponge network | 0.559 | 0.00026 | -0.344 | 0.2374 | 0.336 |
| 32315 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-877-5p | 21 | GRIN2A | Sponge network | 0.494 | 0.00339 | -2.161 | 0 | 0.336 |
| 32316 | KB-431C1.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p | 13 | SLC5A3 | Sponge network | 0.909 | 0 | 0.493 | 5.0E-5 | 0.336 |
| 32317 | HAR1A |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-362-3p;hsa-miR-877-5p | 16 | AR | Sponge network | -0.672 | 0.19447 | -1.554 | 0 | 0.336 |
| 32318 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CXXC4 | Sponge network | 0.422 | 0.27021 | -0.433 | 0.21055 | 0.336 |
| 32319 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 41 | ETV1 | Sponge network | -1.111 | 0.0003 | -0.096 | 0.67674 | 0.336 |
| 32320 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 42 | CUX1 | Sponge network | -0.612 | 0.00094 | -0.039 | 0.6705 | 0.336 |
| 32321 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | RUNX1T1 | Sponge network | 0.131 | 0.8436 | -0.344 | 0.2374 | 0.336 |
| 32322 | AC016747.3 |
hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | NRXN3 | Sponge network | 0.199 | 0.38015 | -0.893 | 0.04186 | 0.336 |
| 32323 | RP11-53O19.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-874-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 33 | GRIK3 | Sponge network | -0.405 | 0.22013 | -3.208 | 0 | 0.336 |
| 32324 | RP5-894A10.6 |
hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 11 | PHF6 | Sponge network | 0.697 | 9.0E-5 | 0.785 | 0 | 0.336 |
| 32325 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 47 | DTNA | Sponge network | 1.302 | 7.0E-5 | -1.648 | 1.0E-5 | 0.336 |
| 32326 | LPP-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-589-3p | 16 | TAL1 | Sponge network | -0.18 | 0.20343 | -0.462 | 0.00574 | 0.336 |
| 32327 | RP11-166D19.1 |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 20 | JUN | Sponge network | -0.576 | 0.10121 | -0.932 | 0 | 0.336 |
| 32328 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-942-5p | 23 | ARHGAP28 | Sponge network | -0.777 | 0.16667 | -0.911 | 0.00013 | 0.336 |
| 32329 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577 | 20 | GREM1 | Sponge network | -0.769 | 0.00035 | 0.902 | 0.01691 | 0.336 |
| 32330 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-497-5p | 18 | TBL1XR1 | Sponge network | 0.903 | 0 | 0.578 | 0 | 0.336 |
| 32331 | CTD-2002H8.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | NFIB | Sponge network | 0.931 | 1.0E-5 | 0.023 | 0.90795 | 0.336 |
| 32332 | LINC00476 |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-590-5p | 11 | PACRG | Sponge network | -0.615 | 0.00015 | -2.248 | 0 | 0.336 |
| 32333 | DIO3OS |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-5p | 11 | TLL1 | Sponge network | -0.773 | 0.04073 | -1.195 | 3.0E-5 | 0.336 |
| 32334 | RP5-1092A3.4 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-940 | 13 | CREB1 | Sponge network | 1.495 | 0 | 0.203 | 0.00537 | 0.336 |
| 32335 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | TMEM132B | Sponge network | -2.117 | 0.00161 | -0.83 | 0.01084 | 0.336 |
| 32336 | LINC00476 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 14 | THSD7A | Sponge network | -0.615 | 0.00015 | 0.242 | 0.25154 | 0.336 |
| 32337 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 23 | SETBP1 | Sponge network | 0.537 | 0.00139 | -1.302 | 1.0E-5 | 0.336 |
| 32338 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-93-5p | 13 | RBL2 | Sponge network | -2.69 | 0.0001 | -0.282 | 0.00254 | 0.336 |
| 32339 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-576-5p | 16 | PBX1 | Sponge network | 0.131 | 0.8436 | -1.206 | 0 | 0.336 |
| 32340 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 32 | SASH1 | Sponge network | -0.727 | 0.04726 | -0.502 | 0.00175 | 0.336 |
| 32341 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 15 | MAP3K12 | Sponge network | 0.246 | 0.59912 | -0.057 | 0.59369 | 0.336 |
| 32342 | FENDRR |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | DACT1 | Sponge network | -1.281 | 0.00012 | 0.783 | 0.00072 | 0.336 |
| 32343 | LINC00957 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 10 | KCNAB1 | Sponge network | -0.421 | 0.0114 | -0.755 | 2.0E-5 | 0.336 |
| 32344 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SYNE1 | Sponge network | 0.537 | 0.00139 | -0.738 | 0.00316 | 0.336 |
| 32345 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | EPAS1 | Sponge network | -0.18 | 0.20343 | -0.184 | 0.14418 | 0.336 |
| 32346 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 33 | USP12 | Sponge network | -0.576 | 0.10121 | -0.102 | 0.40768 | 0.336 |
| 32347 | CTC-297N7.9 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 10 | ST6GALNAC3 | Sponge network | -2.069 | 0 | -0.987 | 0 | 0.336 |
| 32348 | EMX2OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | -0.777 | 0.1134 | -0.001 | 0.99669 | 0.336 |
| 32349 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 15 | EPHA5 | Sponge network | -1.375 | 0.08642 | -0.957 | 0.01733 | 0.336 |
| 32350 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 39 | BCL2 | Sponge network | -0.899 | 6.0E-5 | -0.899 | 0 | 0.336 |
| 32351 | RP11-327P2.5 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92a-1-5p | 21 | AKAP6 | Sponge network | -0.147 | 0.40452 | -1.586 | 3.0E-5 | 0.336 |
| 32352 | RP11-359E10.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-671-5p | 12 | CDH23 | Sponge network | 0.299 | 0.57722 | -0.974 | 0.00034 | 0.336 |
| 32353 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 10 | SASH1 | Sponge network | 0.542 | 0.00054 | -0.502 | 0.00175 | 0.336 |
| 32354 | RP11-672A2.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-505-5p | 26 | LPP | Sponge network | 1.344 | 0 | -0.459 | 0.04475 | 0.336 |
| 32355 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-942-5p | 33 | DDX6 | Sponge network | -0.615 | 0.00015 | 0.091 | 0.36428 | 0.336 |
| 32356 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 10 | PLA2R1 | Sponge network | 0.72 | 0.0001 | -0.265 | 0.24676 | 0.336 |
| 32357 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | AFF3 | Sponge network | 0.225 | 0.30644 | -2.373 | 0 | 0.336 |
| 32358 | RP11-999E24.3 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-340-5p | 11 | CSDE1 | Sponge network | -0.693 | 0.05629 | -0.493 | 0 | 0.336 |
| 32359 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 11 | KIAA1024 | Sponge network | 1.254 | 0 | 1.083 | 0 | 0.336 |
| 32360 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-577 | 17 | SCN9A | Sponge network | -0.465 | 0.04703 | -0.525 | 0.10593 | 0.336 |
| 32361 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-7-1-3p | 10 | RCAN2 | Sponge network | 0.056 | 0.8494 | -0.33 | 0.15172 | 0.336 |
| 32362 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | PRKAB2 | Sponge network | 0.572 | 0.01722 | -0.367 | 0.01371 | 0.336 |
| 32363 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | RBL2 | Sponge network | -0.899 | 6.0E-5 | -0.282 | 0.00254 | 0.336 |
| 32364 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | PLSCR4 | Sponge network | -0.355 | 0.0077 | -0.474 | 0.03833 | 0.336 |
| 32365 | RP11-156P1.3 |
hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-940 | 13 | SFRP1 | Sponge network | 0.214 | 0.18246 | -3.566 | 0 | 0.336 |
| 32366 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-5p;hsa-miR-624-5p | 18 | LATS1 | Sponge network | 0.331 | 0.57247 | 0.109 | 0.34208 | 0.336 |
| 32367 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p | 10 | RBM5 | Sponge network | -1.645 | 0 | -0.205 | 0.0129 | 0.336 |
| 32368 | AC108488.4 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-1-3p | 18 | ADAMTSL3 | Sponge network | 0.259 | 0.12542 | -1.559 | 0 | 0.336 |
| 32369 | FAM13A-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p | 13 | CITED2 | Sponge network | -0.83 | 0 | -1.545 | 0 | 0.336 |
| 32370 | LINC00865 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 17 | SEMA3C | Sponge network | -1.249 | 7.0E-5 | -0.272 | 0.31153 | 0.336 |
| 32371 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 11 | SCN11A | Sponge network | 1.302 | 7.0E-5 | -1.15 | 0.00299 | 0.336 |
| 32372 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 21 | SSBP2 | Sponge network | 0.342 | 0.03129 | -1.58 | 0 | 0.336 |
| 32373 | A2M-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-96-5p | 13 | FNBP1 | Sponge network | -0.08 | 0.90092 | -0.969 | 4.0E-5 | 0.336 |
| 32374 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p | 12 | ZBTB4 | Sponge network | 1.344 | 0 | -0.739 | 0 | 0.336 |
| 32375 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 30 | NRXN1 | Sponge network | -0.668 | 3.0E-5 | -3.778 | 0 | 0.336 |
| 32376 | LINC00473 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | -1.203 | 0.03881 | -2.417 | 0 | 0.336 |
| 32377 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-3607-3p;hsa-miR-365a-3p;hsa-miR-576-5p | 15 | RICTOR | Sponge network | 0.537 | 0.0006 | 0.388 | 0.00188 | 0.336 |
| 32378 | AF131215.2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-671-5p | 13 | SORCS1 | Sponge network | -0.176 | 0.59692 | -3.492 | 0 | 0.336 |
| 32379 | FZD10-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -0.727 | 0.04726 | -0.252 | 0.15511 | 0.336 |
| 32380 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-7-1-3p | 10 | NEDD4 | Sponge network | -0.557 | 0.11135 | 0.02 | 0.89834 | 0.336 |
| 32381 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-495-3p | 12 | PRKAB2 | Sponge network | 0.993 | 0 | -0.367 | 0.01371 | 0.336 |
| 32382 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 18 | PRUNE2 | Sponge network | 1.302 | 7.0E-5 | -1.733 | 0.00022 | 0.336 |
| 32383 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | KANK1 | Sponge network | 0.225 | 0.30644 | -0.55 | 0.00134 | 0.336 |
| 32384 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | EPHA3 | Sponge network | -0.053 | 0.80685 | 0.185 | 0.53588 | 0.336 |
| 32385 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-629-3p | 25 | LIFR | Sponge network | -1.645 | 0 | -1.794 | 0 | 0.336 |
| 32386 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-629-3p;hsa-miR-7-5p | 28 | ZBTB4 | Sponge network | -0.773 | 0.04073 | -0.739 | 0 | 0.336 |
| 32387 | RP11-148O21.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-671-5p | 13 | GAS7 | Sponge network | 0.233 | 0.81029 | -0.555 | 0.01602 | 0.336 |
| 32388 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | KAT6B | Sponge network | 0.817 | 3.0E-5 | -0.478 | 0.00018 | 0.336 |
| 32389 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | SUFU | Sponge network | -0.358 | 0.17255 | -0.04 | 0.65489 | 0.336 |
| 32390 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-409-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 38 | TRPS1 | Sponge network | 0.559 | 0.00026 | -0.191 | 0.44109 | 0.336 |
| 32391 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PTPRB | Sponge network | 0.532 | 0.00505 | 0.105 | 0.47122 | 0.336 |
| 32392 | LINC00667 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | PTPRT | Sponge network | -0.235 | 0.15142 | -0.907 | 0.03727 | 0.336 |
| 32393 | AC159540.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 18 | TBL1XR1 | Sponge network | 1.122 | 0 | 0.578 | 0 | 0.336 |
| 32394 | MEG3 |
hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | PAK3 | Sponge network | 0.249 | 0.35902 | -0.628 | 0.07253 | 0.336 |
| 32395 | RP11-597D13.9 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | BCL9 | Sponge network | 1.302 | 7.0E-5 | 0.321 | 0.00267 | 0.336 |
| 32396 | DIO3OS |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-93-3p | 15 | PCDH10 | Sponge network | -0.773 | 0.04073 | -1.644 | 0.0078 | 0.336 |
| 32397 | AC109309.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 13 | CADM2 | Sponge network | 0.163 | 0.87219 | -3.782 | 0 | 0.336 |
| 32398 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PDE4DIP | Sponge network | 0.683 | 1.0E-5 | -0.282 | 0.0257 | 0.336 |
| 32399 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-942-5p | 10 | RB1CC1 | Sponge network | 0.921 | 0.00118 | 0.175 | 0.12559 | 0.336 |
| 32400 | AP001055.6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | NOTCH2 | Sponge network | 0.693 | 0.00076 | 0.138 | 0.35163 | 0.336 |
| 32401 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 11 | CDH10 | Sponge network | -0.405 | 0.22013 | -2.397 | 0 | 0.336 |
| 32402 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-590-5p | 11 | TMEFF2 | Sponge network | -0.384 | 0.40652 | -2.426 | 0 | 0.336 |
| 32403 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-590-3p | 22 | EYA4 | Sponge network | -0.896 | 0.00099 | 0.3 | 0.36876 | 0.336 |
| 32404 | RP4-605O3.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 11 | EBF1 | Sponge network | 0.262 | 0.0475 | -0.384 | 0.08856 | 0.336 |
| 32405 | RP11-2H8.2 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | UBR3 | Sponge network | 1.089 | 0 | -0.021 | 0.82774 | 0.336 |
| 32406 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-629-3p;hsa-miR-96-5p | 18 | EBF3 | Sponge network | 0.331 | 0.57247 | -0.058 | 0.81437 | 0.336 |
| 32407 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | ZFHX3 | Sponge network | -2.117 | 0.00161 | 0.389 | 0.01363 | 0.336 |
| 32408 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p | 10 | STARD13 | Sponge network | -0.465 | 0.04703 | -0.187 | 0.27383 | 0.336 |
| 32409 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b | 14 | ERCC4 | Sponge network | 0.37 | 0.27614 | 0.126 | 0.15266 | 0.336 |
| 32410 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 13 | RARB | Sponge network | 0.225 | 0.30644 | -0.825 | 2.0E-5 | 0.336 |
| 32411 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 21 | MCC | Sponge network | 1.153 | 0.00114 | -1.005 | 0 | 0.336 |
| 32412 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 13 | ST6GAL2 | Sponge network | -0.726 | 0.00132 | -1.201 | 0.00597 | 0.336 |
| 32413 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-328-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-590-3p | 16 | TMTC2 | Sponge network | 0.194 | 0.64278 | -0.215 | 0.18248 | 0.336 |
| 32414 | LINC00654 |
hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 11 | LZTS1 | Sponge network | 0.374 | 0.20054 | 1.664 | 0 | 0.336 |
| 32415 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TBX18 | Sponge network | -1.161 | 0.00591 | 0.843 | 0.02945 | 0.336 |
| 32416 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 31 | MCC | Sponge network | 0.395 | 0.15451 | -1.005 | 0 | 0.336 |
| 32417 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-429 | 25 | PRDM2 | Sponge network | -2.69 | 0.0001 | -0.258 | 0.00721 | 0.336 |
| 32418 | RP5-1139B12.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-502-5p | 25 | DTNA | Sponge network | 0.598 | 0.38481 | -1.648 | 1.0E-5 | 0.336 |
| 32419 | KCNJ2-AS1 |
hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-590-3p | 14 | DMD | Sponge network | -0.436 | 0.05325 | -1.506 | 0 | 0.336 |
| 32420 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-455-3p | 12 | TLE4 | Sponge network | -1.654 | 0.00144 | -1.157 | 0 | 0.336 |
| 32421 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | TP53INP1 | Sponge network | -2.117 | 0.00161 | -0.496 | 0.00064 | 0.336 |
| 32422 | AC141928.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 10 | CNTNAP1 | Sponge network | 0.853 | 0.15352 | 0.358 | 0.13727 | 0.336 |
| 32423 | EMX2OS |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | -0.777 | 0.1134 | 0.524 | 0.00017 | 0.336 |
| 32424 | RP4-717I23.3 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-485-3p;hsa-miR-550a-3p;hsa-miR-576-5p | 16 | HECA | Sponge network | 0.637 | 0.00069 | -0.217 | 0.03463 | 0.336 |
| 32425 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-590-5p | 22 | GRIN2A | Sponge network | 0.331 | 0.57247 | -2.161 | 0 | 0.336 |
| 32426 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | EDA2R | Sponge network | 0.246 | 0.59912 | -0.587 | 0.01598 | 0.336 |
| 32427 | RP11-384O8.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-484;hsa-miR-539-5p;hsa-miR-589-5p;hsa-miR-590-3p | 25 | PRKCB | Sponge network | -0.738 | 0.21233 | -1.313 | 2.0E-5 | 0.336 |
| 32428 | RP11-13K12.5 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-940 | 38 | WDFY3 | Sponge network | -0.318 | 0.65847 | -0.201 | 0.0967 | 0.336 |
| 32429 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | NEDD4 | Sponge network | 0.219 | 0.1516 | 0.02 | 0.89834 | 0.336 |
| 32430 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b | 13 | USP12 | Sponge network | 0.594 | 0.00064 | -0.102 | 0.40768 | 0.336 |
| 32431 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 18 | NOTCH2 | Sponge network | -0.726 | 0.00132 | 0.138 | 0.35163 | 0.336 |
| 32432 | PSMG3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-484 | 15 | EZH1 | Sponge network | -0.125 | 0.49414 | -0.564 | 1.0E-5 | 0.336 |
| 32433 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p | 14 | MOB1B | Sponge network | 0.752 | 0 | 0.197 | 0.15266 | 0.336 |
| 32434 | PART1 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -2.925 | 0 | -1.692 | 0 | 0.336 |
| 32435 | RAMP2-AS1 |
hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PIK3CA | Sponge network | -0.513 | 0.04703 | 0.195 | 0.06908 | 0.336 |
| 32436 | TP73-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-96-5p | 17 | UNC5C | Sponge network | -0.563 | 0.02545 | -0.178 | 0.40214 | 0.336 |
| 32437 | RP11-806O11.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | NFIB | Sponge network | 0.284 | 0.52463 | 0.023 | 0.90795 | 0.336 |
| 32438 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | FGF5 | Sponge network | -2.925 | 0 | 0.549 | 0.28659 | 0.336 |
| 32439 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 12 | NAV3 | Sponge network | -0.72 | 0.24239 | 0.212 | 0.47729 | 0.336 |
| 32440 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 18 | PHC3 | Sponge network | -1.092 | 4.0E-5 | 0.212 | 0.0499 | 0.336 |
| 32441 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ANK2 | Sponge network | 0.349 | 0.01695 | -1.603 | 1.0E-5 | 0.336 |
| 32442 | RP11-589P10.7 |
hsa-let-7a-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p | 16 | THRB | Sponge network | -2.044 | 0.00066 | -1.117 | 0.0004 | 0.336 |
| 32443 | GNG12-AS1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | PAIP2 | Sponge network | -0.793 | 7.0E-5 | -0.515 | 0 | 0.336 |
| 32444 | CKMT2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-96-5p | 25 | ASTN1 | Sponge network | 0.082 | 0.55654 | -2.389 | 2.0E-5 | 0.336 |
| 32445 | TPTEP1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 32 | ERC1 | Sponge network | -1.216 | 0 | -0.108 | 0.38794 | 0.336 |
| 32446 | LINC00578 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-501-5p | 24 | WDFY3 | Sponge network | 0.811 | 0.03228 | -0.201 | 0.0967 | 0.336 |
| 32447 | AC138035.2 |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p | 13 | DDX6 | Sponge network | 1.073 | 0 | 0.091 | 0.36428 | 0.336 |
| 32448 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-361-5p;hsa-miR-590-3p | 10 | AKAP12 | Sponge network | -0.668 | 3.0E-5 | -0.932 | 0.0028 | 0.336 |
| 32449 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TIMP3 | Sponge network | 0.61 | 0.01593 | -0.546 | 0.02307 | 0.336 |
| 32450 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-484;hsa-miR-590-3p | 11 | PKNOX1 | Sponge network | 0.214 | 0.18246 | 0.085 | 0.28917 | 0.336 |
| 32451 | PSMG3-AS1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-455-3p;hsa-miR-484 | 15 | ABL1 | Sponge network | -0.125 | 0.49414 | -0.215 | 0.07804 | 0.336 |
| 32452 | AC141928.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-362-3p | 20 | DMD | Sponge network | 0.853 | 0.15352 | -1.506 | 0 | 0.336 |
| 32453 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 21 | ZMYM2 | Sponge network | 0.799 | 1.0E-5 | 0.279 | 0.01273 | 0.336 |
| 32454 | NDUFA6-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | BCL9 | Sponge network | -0.355 | 0.0077 | 0.321 | 0.00267 | 0.336 |
| 32455 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SMARCA2 | Sponge network | 0.083 | 0.66405 | -0.529 | 0.00014 | 0.336 |
| 32456 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | TRPS1 | Sponge network | -0.246 | 0.35034 | -0.191 | 0.44109 | 0.336 |
| 32457 | RP11-736K20.5 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 26 | PPM1L | Sponge network | -0.358 | 0.17255 | -0.537 | 0.00616 | 0.336 |
| 32458 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-940 | 17 | NEDD4 | Sponge network | -0.777 | 0.16667 | 0.02 | 0.89834 | 0.336 |
| 32459 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-942-5p | 22 | ARHGAP28 | Sponge network | -0.842 | 0.01361 | -0.911 | 0.00013 | 0.336 |
| 32460 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-511-5p;hsa-miR-7-5p | 12 | RBMS3 | Sponge network | -1.054 | 0.0706 | -0.519 | 0.03551 | 0.336 |
| 32461 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 38 | QKI | Sponge network | -1.057 | 0.00029 | -0.333 | 0.04111 | 0.336 |
| 32462 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 42 | CUX1 | Sponge network | 0.083 | 0.66405 | -0.039 | 0.6705 | 0.336 |
| 32463 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-625-5p | 13 | LHX6 | Sponge network | 0.214 | 0.18246 | -0.087 | 0.69193 | 0.336 |
| 32464 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -0.769 | 0.00035 | 0.309 | 0.17079 | 0.336 |
| 32465 | USP3-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-942-5p | 22 | PCDH17 | Sponge network | -0.162 | 0.47687 | 0.731 | 2.0E-5 | 0.336 |
| 32466 | RP4-714D9.5 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-497-5p | 10 | PRKAA1 | Sponge network | 1.015 | 0 | 0.317 | 0.0031 | 0.335 |
| 32467 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p | 14 | JAKMIP2 | Sponge network | 0.056 | 0.81195 | -1.388 | 0 | 0.335 |
| 32468 | RP11-195F19.9 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-539-5p | 10 | GRIK3 | Sponge network | -0.726 | 0.00132 | -3.208 | 0 | 0.335 |
| 32469 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-93-5p | 15 | PGR | Sponge network | -0.023 | 0.95109 | -1.704 | 0 | 0.335 |
| 32470 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-98-5p | 19 | ARHGAP28 | Sponge network | -0.154 | 0.78939 | -0.911 | 0.00013 | 0.335 |
| 32471 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | PRRX1 | Sponge network | 0.727 | 3.0E-5 | 1.316 | 0 | 0.335 |
| 32472 | SNX29P2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-96-5p | 16 | UNC5C | Sponge network | 0.4 | 0.14611 | -0.178 | 0.40214 | 0.335 |
| 32473 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429 | 17 | EYA4 | Sponge network | -0.469 | 0.08721 | 0.3 | 0.36876 | 0.335 |
| 32474 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | CBL | Sponge network | 1.352 | 0 | 0.291 | 0.01267 | 0.335 |
| 32475 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-342-3p;hsa-miR-654-3p;hsa-miR-664a-3p | 12 | ITCH | Sponge network | 0.909 | 0 | 0.084 | 0.34058 | 0.335 |
| 32476 | RP11-705C15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | BCL2 | Sponge network | 0.796 | 4.0E-5 | -0.899 | 0 | 0.335 |
| 32477 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p | 12 | DACT1 | Sponge network | 0.056 | 0.81195 | 0.783 | 0.00072 | 0.335 |
| 32478 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | ERCC4 | Sponge network | 0.349 | 0.01695 | 0.126 | 0.15266 | 0.335 |
| 32479 | LINC00473 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p | 11 | ELMO1 | Sponge network | -1.203 | 0.03881 | 0.04 | 0.83147 | 0.335 |
| 32480 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | GRIN2A | Sponge network | -0.342 | 0.02288 | -2.161 | 0 | 0.335 |
| 32481 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 39 | AR | Sponge network | 0.619 | 0.00157 | -1.554 | 0 | 0.335 |
| 32482 | RP11-798G7.7 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p | 12 | DMXL1 | Sponge network | 0.858 | 3.0E-5 | -0.426 | 0.00056 | 0.335 |
| 32483 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PDGFC | Sponge network | -2.915 | 0.00015 | -0.33 | 0.06483 | 0.335 |
| 32484 | RP11-244O19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | ABCB5 | Sponge network | -0.096 | 0.67958 | -0.956 | 0.04077 | 0.335 |
| 32485 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | HDAC9 | Sponge network | -2.46 | 0 | -0.755 | 0.00495 | 0.335 |
| 32486 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 37 | NOTCH2 | Sponge network | 1.302 | 7.0E-5 | 0.138 | 0.35163 | 0.335 |
| 32487 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 15 | LHX6 | Sponge network | 0.253 | 0.09565 | -0.087 | 0.69193 | 0.335 |
| 32488 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | RELN | Sponge network | -0.342 | 0.02288 | -2.403 | 0 | 0.335 |
| 32489 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-628-5p | 16 | ACVR1 | Sponge network | -0.162 | 0.47687 | 0.279 | 0.00234 | 0.335 |
| 32490 | LINC00472 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 17 | NCAM2 | Sponge network | -0.842 | 0.01361 | -0.482 | 0.22565 | 0.335 |
| 32491 | AC108488.4 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-940 | 12 | MAPK10 | Sponge network | 0.259 | 0.12542 | -1.774 | 0 | 0.335 |
| 32492 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 16 | ERBB4 | Sponge network | -0.376 | 0.00337 | -1.975 | 2.0E-5 | 0.335 |
| 32493 | C4B-AS1 |
hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-744-3p | 13 | ADAMTSL3 | Sponge network | 2.306 | 9.0E-5 | -1.559 | 0 | 0.335 |
| 32494 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p | 12 | ZFHX3 | Sponge network | 1.157 | 0.00455 | 0.389 | 0.01363 | 0.335 |
| 32495 | TTC25 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-5p | 13 | TCF4 | Sponge network | -0.208 | 0.38261 | 0.016 | 0.92271 | 0.335 |
| 32496 | MIR497HG |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | MED12L | Sponge network | -1.439 | 4.0E-5 | -0.194 | 0.55299 | 0.335 |
| 32497 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 16 | CHD2 | Sponge network | 1.063 | 0 | 0.049 | 0.55371 | 0.335 |
| 32498 | MALAT1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | PICALM | Sponge network | 0.782 | 0.00016 | 0.086 | 0.23523 | 0.335 |
| 32499 | NDUFA6-AS1 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | CSDE1 | Sponge network | -0.355 | 0.0077 | -0.493 | 0 | 0.335 |
| 32500 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-625-5p;hsa-miR-98-5p | 21 | RSPO2 | Sponge network | -0.773 | 0.04073 | -3.038 | 0 | 0.335 |
| 32501 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 10 | YAP1 | Sponge network | -0.517 | 0.02935 | 0.22 | 0.11836 | 0.335 |
| 32502 | AP001347.6 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | GLIPR1 | Sponge network | -1.161 | 0.00591 | 0.146 | 0.3276 | 0.335 |
| 32503 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 24 | CDH19 | Sponge network | -0.355 | 0.0077 | -3.434 | 0 | 0.335 |
| 32504 | CTC-559E9.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-576-5p | 13 | TCF12 | Sponge network | 0.594 | 0.00064 | 0.174 | 0.13271 | 0.335 |
| 32505 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-7-1-3p | 16 | PLSCR4 | Sponge network | -0.72 | 0.24239 | -0.474 | 0.03833 | 0.335 |
| 32506 | EMX2OS |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 15 | EHD3 | Sponge network | -0.777 | 0.1134 | -0.484 | 0.01747 | 0.335 |
| 32507 | MIR22HG |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-590-5p;hsa-miR-708-5p | 18 | FAM135B | Sponge network | -1.047 | 0 | -3.978 | 0 | 0.335 |
| 32508 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-429 | 18 | CHD5 | Sponge network | -0.031 | 0.89063 | -0.922 | 0.01521 | 0.335 |
| 32509 | CTD-2089N3.1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-3065-3p | 21 | LPP | Sponge network | -0.314 | 0.62033 | -0.459 | 0.04475 | 0.335 |
| 32510 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | LRRC7 | Sponge network | -0.769 | 0.00035 | -1.341 | 0.01193 | 0.335 |
| 32511 | RP11-13K12.5 |
hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 10 | FGF5 | Sponge network | -0.318 | 0.65847 | 0.549 | 0.28659 | 0.335 |
| 32512 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 31 | DDX6 | Sponge network | -0.487 | 0.02157 | 0.091 | 0.36428 | 0.335 |
| 32513 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p | 12 | ERG | Sponge network | 0.082 | 0.55397 | 0.002 | 0.99011 | 0.335 |
| 32514 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 18 | SLC6A15 | Sponge network | 0.082 | 0.55654 | -2.373 | 2.0E-5 | 0.335 |
| 32515 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-484;hsa-miR-503-5p | 14 | ABL1 | Sponge network | -0.342 | 0.02288 | -0.215 | 0.07804 | 0.335 |
| 32516 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-582-5p | 13 | DDX6 | Sponge network | 0.757 | 0 | 0.091 | 0.36428 | 0.335 |
| 32517 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 13 | ABCA10 | Sponge network | 0.246 | 0.59912 | -0.571 | 0.03674 | 0.335 |
| 32518 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 11 | EYA4 | Sponge network | -0.187 | 0.23787 | 0.3 | 0.36876 | 0.335 |
| 32519 | LINC00476 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ARNT | Sponge network | -0.615 | 0.00015 | -0.08 | 0.26073 | 0.335 |
| 32520 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p | 28 | NTRK2 | Sponge network | -0.125 | 0.49414 | -0.736 | 0.06495 | 0.335 |
| 32521 | RP11-181G12.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-550a-5p;hsa-miR-744-3p | 26 | DST | Sponge network | 0.556 | 0.00298 | -0.713 | 0.00096 | 0.335 |
| 32522 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 26 | SUFU | Sponge network | -0.49 | 0.04946 | -0.04 | 0.65489 | 0.335 |
| 32523 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 26 | AFF4 | Sponge network | -0.376 | 0.00337 | -0.266 | 0.01565 | 0.335 |
| 32524 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 17 | TET1 | Sponge network | -0.322 | 0.25945 | -0.135 | 0.52138 | 0.335 |
| 32525 | AGAP11 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-93-3p | 26 | ERC1 | Sponge network | -1.645 | 0 | -0.108 | 0.38794 | 0.335 |
| 32526 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | PPP1R9A | Sponge network | -0.976 | 0.00025 | -0.903 | 0.04254 | 0.335 |
| 32527 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p | 20 | USP12 | Sponge network | 0.214 | 0.18246 | -0.102 | 0.40768 | 0.335 |
| 32528 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | TGFBR2 | Sponge network | 0.335 | 0.20596 | -0.243 | 0.11408 | 0.335 |
| 32529 | MALAT1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | BRD3 | Sponge network | 0.782 | 0.00016 | 0.37 | 0.00013 | 0.335 |
| 32530 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 16 | NEDD4 | Sponge network | 0.253 | 0.09565 | 0.02 | 0.89834 | 0.335 |
| 32531 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | PRKAB2 | Sponge network | 0.8 | 0 | -0.367 | 0.01371 | 0.335 |
| 32532 | MEG3 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 33 | CTIF | Sponge network | 0.249 | 0.35902 | -0.389 | 0.00549 | 0.335 |
| 32533 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-511-5p;hsa-miR-539-5p | 27 | NRXN1 | Sponge network | -0.384 | 0.40652 | -3.778 | 0 | 0.335 |
| 32534 | AC009948.5 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 0.532 | 0.00505 | 0.321 | 0.00267 | 0.335 |
| 32535 | AC003090.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | EYA4 | Sponge network | -2.117 | 0.00161 | 0.3 | 0.36876 | 0.335 |
| 32536 | RP11-384O8.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 35 | CCND2 | Sponge network | -0.738 | 0.21233 | -0.267 | 0.3112 | 0.335 |
| 32537 | RP11-155G14.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 11 | CBL | Sponge network | 2.483 | 0 | 0.291 | 0.01267 | 0.335 |
| 32538 | RP11-728F11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-582-3p | 14 | PBX1 | Sponge network | 0.238 | 0.70544 | -1.206 | 0 | 0.335 |
| 32539 | RP11-983P16.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 27 | TGFBR3 | Sponge network | 0.41 | 0.02214 | -1.173 | 1.0E-5 | 0.335 |
| 32540 | MAGI2-AS3 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | LIMCH1 | Sponge network | -0.129 | 0.57177 | -0.915 | 1.0E-5 | 0.335 |
| 32541 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p | 12 | GLI3 | Sponge network | -0.342 | 0.02288 | -0.615 | 0.01482 | 0.335 |
| 32542 | RP13-516M14.1 |
hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-532-3p | 11 | SYNM | Sponge network | 0.389 | 0.04293 | -2.309 | 1.0E-5 | 0.335 |
| 32543 | RP11-611E13.2 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p | 11 | PHF6 | Sponge network | 0.822 | 0 | 0.785 | 0 | 0.335 |
| 32544 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | CBX5 | Sponge network | 0.101 | 0.60919 | 0.165 | 0.24446 | 0.335 |
| 32545 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 26 | EBF1 | Sponge network | 0.537 | 0.00139 | -0.384 | 0.08856 | 0.335 |
| 32546 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-96-5p | 14 | CRTC3 | Sponge network | -1.582 | 8.0E-5 | 0.164 | 0.16786 | 0.335 |
| 32547 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SLC6A15 | Sponge network | 0.194 | 0.64278 | -2.373 | 2.0E-5 | 0.335 |
| 32548 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 28 | ADARB1 | Sponge network | 0.051 | 0.83388 | -0.335 | 0.06631 | 0.335 |
| 32549 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | TSC1 | Sponge network | 0.222 | 0.29133 | 0.103 | 0.26071 | 0.335 |
| 32550 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-590-3p | 12 | AKAP12 | Sponge network | 0.537 | 0.00139 | -0.932 | 0.0028 | 0.335 |
| 32551 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 25 | CADM1 | Sponge network | -0.154 | 0.78939 | -0.9 | 0.00084 | 0.335 |
| 32552 | RP11-798M19.6 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-942-5p | 26 | IGFBP5 | Sponge network | -0.612 | 0.00094 | -0.806 | 0.00105 | 0.335 |
| 32553 | RP11-46C24.7 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | DTNA | Sponge network | 0.392 | 0.0278 | -1.648 | 1.0E-5 | 0.335 |
| 32554 | LINC01018 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | LUZP2 | Sponge network | -2.915 | 0.00015 | -0.056 | 0.89416 | 0.335 |
| 32555 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-539-5p | 46 | KIF1B | Sponge network | -0.384 | 0.40652 | -0.118 | 0.32258 | 0.335 |
| 32556 | WDFY3-AS2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ITPKB | Sponge network | -0.942 | 0 | -0.704 | 0.00106 | 0.335 |
| 32557 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-590-3p;hsa-miR-655-3p | 21 | PIK3C2A | Sponge network | 0.083 | 0.66405 | 0.061 | 0.53776 | 0.335 |
| 32558 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | -0.227 | 0.31657 | -1.751 | 1.0E-5 | 0.335 |
| 32559 | LINC00654 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-5p | 11 | PDZD2 | Sponge network | 0.374 | 0.20054 | -0.909 | 3.0E-5 | 0.335 |
| 32560 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | CHD2 | Sponge network | 0.422 | 0.27021 | 0.049 | 0.55371 | 0.335 |
| 32561 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-577;hsa-miR-590-3p | 10 | NALCN | Sponge network | 0.392 | 0.0278 | 1.068 | 0.00237 | 0.335 |
| 32562 | SNHG14 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 26 | KCNJ5 | Sponge network | -1.111 | 0.0003 | -0.281 | 0.3339 | 0.335 |
| 32563 | RP11-395G23.3 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-93-5p | 10 | PRUNE2 | Sponge network | 0.381 | 0.25967 | -1.733 | 0.00022 | 0.335 |
| 32564 | RP11-968O1.5 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-493-5p | 14 | AFF3 | Sponge network | -0.829 | 0.01343 | -2.373 | 0 | 0.335 |
| 32565 | CTD-2245F17.3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-539-5p | 12 | SSBP2 | Sponge network | -0.421 | 0.12556 | -1.58 | 0 | 0.335 |
| 32566 | GS1-124K5.3 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | TBL1XR1 | Sponge network | 1.501 | 0 | 0.578 | 0 | 0.335 |
| 32567 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | SLC6A15 | Sponge network | -0.49 | 0.04946 | -2.373 | 2.0E-5 | 0.335 |
| 32568 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-3p | 14 | RET | Sponge network | -0.206 | 0.12942 | -0.833 | 0.03517 | 0.335 |
| 32569 | DNM3OS |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | LUZP2 | Sponge network | 0.572 | 0.01722 | -0.056 | 0.89416 | 0.335 |
| 32570 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-200a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-455-3p | 12 | MAF | Sponge network | -1.942 | 0 | -1.257 | 0 | 0.335 |
| 32571 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-93-5p | 26 | MAF | Sponge network | -0.842 | 0.01361 | -1.257 | 0 | 0.335 |
| 32572 | LINC00476 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 32 | SNX29 | Sponge network | -0.615 | 0.00015 | -0.34 | 0.01371 | 0.335 |
| 32573 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | CUL5 | Sponge network | 0.72 | 0.0001 | 0.026 | 0.77301 | 0.335 |
| 32574 | DNM1P35 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-93-3p | 15 | PCDH10 | Sponge network | 0.051 | 0.83388 | -1.644 | 0.0078 | 0.335 |
| 32575 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-96-5p | 12 | SETBP1 | Sponge network | 0.37 | 0.27614 | -1.302 | 1.0E-5 | 0.335 |
| 32576 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p | 19 | PHC3 | Sponge network | 1.03 | 0 | 0.212 | 0.0499 | 0.335 |
| 32577 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | PIK3C2A | Sponge network | 0.401 | 0.00066 | 0.061 | 0.53776 | 0.335 |
| 32578 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | PIK3R1 | Sponge network | 0.246 | 0.59912 | -0.133 | 0.36854 | 0.335 |
| 32579 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-5p | 17 | NEDD4 | Sponge network | 0.176 | 0.37402 | 0.02 | 0.89834 | 0.335 |
| 32580 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p | 21 | CBL | Sponge network | 0.622 | 0.00015 | 0.291 | 0.01267 | 0.335 |
| 32581 | RP11-389C8.2 |
hsa-miR-103a-2-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-532-3p | 11 | SYNM | Sponge network | 0.727 | 3.0E-5 | -2.309 | 1.0E-5 | 0.335 |
| 32582 | A2M-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | EYA4 | Sponge network | -0.08 | 0.90092 | 0.3 | 0.36876 | 0.335 |
| 32583 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | PRKAA2 | Sponge network | -0.811 | 2.0E-5 | -2.462 | 0 | 0.335 |
| 32584 | OIP5-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | NAV3 | Sponge network | 0.421 | 0.00084 | 0.212 | 0.47729 | 0.335 |
| 32585 | RP11-244H3.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ITPKB | Sponge network | 0.659 | 3.0E-5 | -0.704 | 0.00106 | 0.335 |
| 32586 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-500a-5p;hsa-miR-590-5p | 22 | SASH1 | Sponge network | 0.078 | 0.55803 | -0.502 | 0.00175 | 0.335 |
| 32587 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 34 | HLF | Sponge network | -1.264 | 6.0E-5 | -2.443 | 0 | 0.335 |
| 32588 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p | 16 | USP6 | Sponge network | 0.657 | 4.0E-5 | 0.188 | 0.3871 | 0.335 |
| 32589 | RP11-802E16.3 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-337-3p;hsa-miR-497-5p | 11 | SLC5A3 | Sponge network | 2.048 | 0 | 0.493 | 5.0E-5 | 0.335 |
| 32590 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | BTG2 | Sponge network | -0.096 | 0.67958 | -1.763 | 0 | 0.335 |
| 32591 | HCG11 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | RNF144A | Sponge network | 0.385 | 0.10067 | 0.594 | 0.0009 | 0.335 |
| 32592 | AC159540.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-424-5p | 13 | ABL2 | Sponge network | 1.122 | 0 | 0.82 | 0 | 0.335 |
| 32593 | NR2F1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 41 | AFF1 | Sponge network | -0.49 | 0.04946 | -0.384 | 0.00053 | 0.335 |
| 32594 | RP11-806L2.6 |
hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p | 10 | RICTOR | Sponge network | 2.103 | 0.0002 | 0.388 | 0.00188 | 0.335 |
| 32595 | CTA-445C9.14 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-582-3p | 11 | BRD3 | Sponge network | 0.718 | 4.0E-5 | 0.37 | 0.00013 | 0.335 |
| 32596 | LINC00472 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-374a-3p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-628-5p | 29 | ERC1 | Sponge network | -0.842 | 0.01361 | -0.108 | 0.38794 | 0.335 |
| 32597 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 53 | FOXP1 | Sponge network | 0.311 | 0.10344 | 0.042 | 0.74368 | 0.335 |
| 32598 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-7-1-3p | 11 | PEG3 | Sponge network | 0.395 | 0.15451 | -1.74 | 0 | 0.335 |
| 32599 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-3p | 22 | PCDH10 | Sponge network | 0.494 | 0.00339 | -1.644 | 0.0078 | 0.335 |
| 32600 | AC108488.4 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577 | 17 | HIP1 | Sponge network | 0.259 | 0.12542 | 0.774 | 0 | 0.335 |
| 32601 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | CADM2 | Sponge network | -0.726 | 0.00132 | -3.782 | 0 | 0.335 |
| 32602 | FAM13A-AS1 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-361-5p;hsa-miR-590-3p | 15 | FAM135B | Sponge network | -0.83 | 0 | -3.978 | 0 | 0.335 |
| 32603 | RP11-384O8.1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p | 15 | IRS1 | Sponge network | -0.738 | 0.21233 | -0.026 | 0.88855 | 0.335 |
| 32604 | NUTM2A-AS1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | GSK3B | Sponge network | 0.643 | 0 | 0.266 | 0.00045 | 0.335 |
| 32605 | RP11-574K11.29 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | BRD3 | Sponge network | 0.997 | 0 | 0.37 | 0.00013 | 0.335 |
| 32606 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-5p | 35 | PIK3R1 | Sponge network | -0.154 | 0.78939 | -0.133 | 0.36854 | 0.335 |
| 32607 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-7-1-3p | 22 | RARB | Sponge network | 0.997 | 0.00066 | -0.825 | 2.0E-5 | 0.335 |
| 32608 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-654-3p | 10 | USP25 | Sponge network | 0.083 | 0.66405 | -0.242 | 0.01397 | 0.335 |
| 32609 | RP11-13K12.5 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | MOB1B | Sponge network | -0.318 | 0.65847 | 0.197 | 0.15266 | 0.335 |
| 32610 | AC034220.3 |
hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 0.309 | 0.07304 | 0.322 | 0.00215 | 0.335 |
| 32611 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p | 13 | USP12 | Sponge network | 1.317 | 0 | -0.102 | 0.40768 | 0.335 |
| 32612 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-550a-3p;hsa-miR-628-5p;hsa-miR-940 | 32 | MEF2C | Sponge network | -0.773 | 0.04073 | -0.499 | 0.01448 | 0.335 |
| 32613 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | BCL11A | Sponge network | -0.513 | 0.04703 | -0.932 | 0.0063 | 0.335 |
| 32614 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 42 | SSBP2 | Sponge network | -0.227 | 0.31657 | -1.58 | 0 | 0.335 |
| 32615 | AGAP11 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-421 | 11 | ZNF844 | Sponge network | -1.645 | 0 | -0.966 | 0.00035 | 0.335 |
| 32616 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | CDHR3 | Sponge network | -0.737 | 0.10822 | -0.977 | 4.0E-5 | 0.335 |
| 32617 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | ATRX | Sponge network | 0.683 | 1.0E-5 | 0.261 | 0.04408 | 0.335 |
| 32618 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 18 | CNTNAP3 | Sponge network | 0.082 | 0.55654 | -1.465 | 0.00033 | 0.335 |
| 32619 | LINC01018 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 10 | SPOP | Sponge network | -2.915 | 0.00015 | -0.419 | 1.0E-5 | 0.335 |
| 32620 | RP11-403I13.8 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FMNL3 | Sponge network | 0.619 | 0.00157 | 0.555 | 4.0E-5 | 0.335 |
| 32621 | CTD-3099C6.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p | 12 | BRD3 | Sponge network | 1.361 | 0 | 0.37 | 0.00013 | 0.335 |
| 32622 | RP11-1006G14.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ARHGEF12 | Sponge network | 0.559 | 0.00026 | 0.09 | 0.35693 | 0.335 |
| 32623 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-451a;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 36 | BCL2 | Sponge network | -0.125 | 0.49414 | -0.899 | 0 | 0.335 |
| 32624 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 15 | CNTNAP3 | Sponge network | -0.727 | 0.04726 | -1.465 | 0.00033 | 0.335 |
| 32625 | SNX29P2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 10 | RABEP1 | Sponge network | 0.4 | 0.14611 | -0.066 | 0.43142 | 0.335 |
| 32626 | CTD-2314G24.2 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-589-5p | 10 | SORCS2 | Sponge network | 0.246 | 0.59912 | -0.223 | 0.4253 | 0.335 |
| 32627 | MAP3K14 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-503-5p;hsa-miR-93-3p | 11 | TOM1L2 | Sponge network | -0.295 | 0.02604 | -0.994 | 0 | 0.335 |
| 32628 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 12 | RBMS3 | Sponge network | -0.557 | 0.11135 | -0.519 | 0.03551 | 0.335 |
| 32629 | RP11-290F24.6 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 12 | PHF6 | Sponge network | 2.052 | 0 | 0.785 | 0 | 0.335 |
| 32630 | MYLK-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-7-5p | 15 | KCNJ5 | Sponge network | -1.331 | 3.0E-5 | -0.281 | 0.3339 | 0.335 |
| 32631 | PCA3 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | PAK3 | Sponge network | -0.709 | 0.18168 | -0.628 | 0.07253 | 0.335 |
| 32632 | MIR22HG |
hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-7-5p | 11 | PIM1 | Sponge network | -1.047 | 0 | -1.114 | 0 | 0.335 |
| 32633 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-7-1-3p | 10 | MLF1 | Sponge network | -0.405 | 0.22013 | -0.893 | 0.00727 | 0.335 |
| 32634 | RP11-248G5.8 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p | 22 | ARHGEF12 | Sponge network | 1.294 | 0 | 0.09 | 0.35693 | 0.335 |
| 32635 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | 0.335 | 0.20596 | -1.277 | 0 | 0.335 |
| 32636 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PBRM1 | Sponge network | 0.873 | 0 | -0.029 | 0.78601 | 0.335 |
| 32637 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 35 | PAFAH1B1 | Sponge network | -0.611 | 0.09607 | -0.429 | 0 | 0.335 |
| 32638 | SERTAD4-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-424-5p | 15 | CBFA2T3 | Sponge network | -0.932 | 0.00329 | -1.221 | 1.0E-5 | 0.335 |
| 32639 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-532-3p | 13 | RPS6KA2 | Sponge network | -1.057 | 0.00029 | -0.57 | 0.00013 | 0.335 |
| 32640 | TRAF3IP2-AS1 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | RTN4 | Sponge network | -0.323 | 0.01372 | -0.412 | 0.00014 | 0.335 |
| 32641 | TINCR |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-5p | 12 | KANK1 | Sponge network | -0.664 | 0.14543 | -0.55 | 0.00134 | 0.335 |
| 32642 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 46 | PRKAA2 | Sponge network | 0.083 | 0.66405 | -2.462 | 0 | 0.335 |
| 32643 | CTD-2002H8.2 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | IL6ST | Sponge network | 0.931 | 1.0E-5 | -0.402 | 0.00676 | 0.335 |
| 32644 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p | 10 | TBRG1 | Sponge network | 0.311 | 0.10344 | -0.071 | 0.37324 | 0.335 |
| 32645 | RP11-156P1.3 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-874-3p | 10 | ARID1B | Sponge network | 0.214 | 0.18246 | 0.003 | 0.97503 | 0.335 |
| 32646 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-5p | 21 | USP12 | Sponge network | -0.896 | 0.00099 | -0.102 | 0.40768 | 0.335 |
| 32647 | LINC00863 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ZFHX3 | Sponge network | 0.189 | 0.13981 | 0.389 | 0.01363 | 0.335 |
| 32648 | RP11-458F8.4 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-539-5p | 10 | MOB1B | Sponge network | 1.518 | 0 | 0.197 | 0.15266 | 0.335 |
| 32649 | TPT1-AS1 |
hsa-miR-129-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-664a-3p | 14 | RASAL2 | Sponge network | 0.903 | 0 | 0.869 | 0 | 0.335 |
| 32650 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | FOXO1 | Sponge network | -0.129 | 0.57177 | -0.141 | 0.33874 | 0.335 |
| 32651 | LINC00173 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 36 | CADM2 | Sponge network | 0.056 | 0.8494 | -3.782 | 0 | 0.335 |
| 32652 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | ARHGAP29 | Sponge network | 0.381 | 0.25967 | 0.131 | 0.42953 | 0.335 |
| 32653 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-93-5p | 22 | LRIG1 | Sponge network | -0.777 | 0.16667 | -0.537 | 0.00588 | 0.335 |
| 32654 | A2M-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 10 | MACF1 | Sponge network | -0.08 | 0.90092 | 0.019 | 0.90335 | 0.335 |
| 32655 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-942-5p;hsa-miR-98-5p | 24 | ARHGAP28 | Sponge network | -2.62 | 0 | -0.911 | 0.00013 | 0.335 |
| 32656 | ZNF667-AS1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-671-5p | 11 | GATA2 | Sponge network | -0.976 | 0.00025 | -0.654 | 0.0016 | 0.335 |
| 32657 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-654-3p | 24 | BMPR2 | Sponge network | 0.972 | 0 | 0.206 | 0.0635 | 0.335 |
| 32658 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 10 | DACH2 | Sponge network | -1.582 | 8.0E-5 | -1.635 | 0.00034 | 0.335 |
| 32659 | RP11-728F11.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p | 19 | ZFHX4 | Sponge network | 0.238 | 0.70544 | 0.018 | 0.95574 | 0.335 |
| 32660 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 43 | EPHA7 | Sponge network | -1.047 | 0 | -2.197 | 3.0E-5 | 0.335 |
| 32661 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-96-5p | 24 | CLASP2 | Sponge network | 0.331 | 0.57247 | -0.038 | 0.71 | 0.335 |
| 32662 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | SETBP1 | Sponge network | 0.238 | 0.70544 | -1.302 | 1.0E-5 | 0.335 |
| 32663 | RP11-2B6.2 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 19 | UBR3 | Sponge network | 0.539 | 0.03722 | -0.021 | 0.82774 | 0.335 |
| 32664 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-7-5p;hsa-miR-93-3p | 20 | ERC1 | Sponge network | -0.693 | 0.05629 | -0.108 | 0.38794 | 0.335 |
| 32665 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | SLITRK6 | Sponge network | -1.216 | 0 | -2.121 | 0 | 0.335 |
| 32666 | AC005562.1 |
hsa-miR-125a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | ERCC6 | Sponge network | 0.488 | 0.00084 | 0.23 | 0.015 | 0.335 |
| 32667 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-33a-5p | 12 | SOX11 | Sponge network | 0.331 | 0.57247 | 2.68 | 0 | 0.335 |
| 32668 | LINC01018 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-877-5p | 22 | CRTC1 | Sponge network | -2.915 | 0.00015 | -0.116 | 0.28412 | 0.335 |
| 32669 | FAM215B |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p | 19 | PIK3R1 | Sponge network | 0.817 | 3.0E-5 | -0.133 | 0.36854 | 0.335 |
| 32670 | AC141928.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p | 23 | ABL2 | Sponge network | 0.853 | 0.15352 | 0.82 | 0 | 0.335 |
| 32671 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-744-5p | 31 | NOTCH2 | Sponge network | -0.18 | 0.20343 | 0.138 | 0.35163 | 0.335 |
| 32672 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-92a-3p | 10 | PDE4DIP | Sponge network | 0.912 | 0.00014 | -0.282 | 0.0257 | 0.335 |
| 32673 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RCAN2 | Sponge network | 0.538 | 0.00076 | -0.33 | 0.15172 | 0.335 |
| 32674 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | PIK3R1 | Sponge network | -2.117 | 0.00161 | -0.133 | 0.36854 | 0.335 |
| 32675 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | ZNF521 | Sponge network | 0.056 | 0.81195 | -0.111 | 0.58068 | 0.335 |
| 32676 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 11 | CLIP1 | Sponge network | -0.318 | 0.65847 | -0.215 | 0.08684 | 0.335 |
| 32677 | RP11-274B21.10 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-654-3p | 10 | BAZ2B | Sponge network | 1.073 | 0.00216 | -0.276 | 0.02833 | 0.335 |
| 32678 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 34 | EBF1 | Sponge network | -1.249 | 7.0E-5 | -0.384 | 0.08856 | 0.335 |
| 32679 | FAM157B |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-338-3p | 12 | ABI2 | Sponge network | 0.92 | 0 | 0.175 | 0.14787 | 0.335 |
| 32680 | RP11-531A24.5 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-1-3p | 13 | ITPKB | Sponge network | 0.75 | 0.00054 | -0.704 | 0.00106 | 0.335 |
| 32681 | RP11-641D5.2 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-223-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-9-5p | 10 | NFIB | Sponge network | -1.403 | 0.02373 | 0.023 | 0.90795 | 0.335 |
| 32682 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 44 | MEF2C | Sponge network | 0.064 | 0.72934 | -0.499 | 0.01448 | 0.335 |
| 32683 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 30 | EBF1 | Sponge network | 0.083 | 0.66405 | -0.384 | 0.08856 | 0.335 |
| 32684 | AC009948.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p | 15 | STARD13 | Sponge network | 0.532 | 0.00505 | -0.187 | 0.27383 | 0.335 |
| 32685 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 34 | IGF1 | Sponge network | -2.925 | 0 | -0.204 | 0.5898 | 0.335 |
| 32686 | PSMG3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p | 16 | SYNM | Sponge network | -0.125 | 0.49414 | -2.309 | 1.0E-5 | 0.335 |
| 32687 | DNAJC27-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 14 | NGFR | Sponge network | -0.969 | 2.0E-5 | -1.271 | 0.01058 | 0.335 |
| 32688 | LINC01018 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 25 | OPCML | Sponge network | -2.915 | 0.00015 | -2.498 | 0 | 0.335 |
| 32689 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 31 | HLF | Sponge network | 0.61 | 0.01593 | -2.443 | 0 | 0.335 |
| 32690 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 30 | SCN9A | Sponge network | 1.302 | 7.0E-5 | -0.525 | 0.10593 | 0.335 |
| 32691 | LINC00092 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-590-3p | 11 | FMNL3 | Sponge network | -1.886 | 0 | 0.555 | 4.0E-5 | 0.335 |
| 32692 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p | 18 | NCOA1 | Sponge network | -0.517 | 0.02935 | -0.432 | 0 | 0.335 |
| 32693 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | RIMBP2 | Sponge network | -0.49 | 0.04946 | -1.968 | 5.0E-5 | 0.335 |
| 32694 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CLASP2 | Sponge network | 2.094 | 0 | -0.038 | 0.71 | 0.334 |
| 32695 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | CDH10 | Sponge network | -0.899 | 6.0E-5 | -2.397 | 0 | 0.334 |
| 32696 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 25 | EIF4A2 | Sponge network | -0.222 | 0.32445 | -0.383 | 0.00046 | 0.334 |
| 32697 | MIR497HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-7-5p;hsa-miR-93-3p | 27 | ERC1 | Sponge network | -1.439 | 4.0E-5 | -0.108 | 0.38794 | 0.334 |
| 32698 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | SOX5 | Sponge network | -1.264 | 6.0E-5 | -1.091 | 4.0E-5 | 0.334 |
| 32699 | AC009948.5 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 19 | TMEM30A | Sponge network | 0.532 | 0.00505 | 0.096 | 0.31031 | 0.334 |
| 32700 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | PRKAB2 | Sponge network | -1.161 | 0.00591 | -0.367 | 0.01371 | 0.334 |
| 32701 | RP11-428J1.5 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-429 | 11 | SYNM | Sponge network | 0.538 | 0.00076 | -2.309 | 1.0E-5 | 0.334 |
| 32702 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 25 | PRKAB2 | Sponge network | -1.645 | 0 | -0.367 | 0.01371 | 0.334 |
| 32703 | RP11-439L18.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-454-3p;hsa-miR-484 | 13 | KIF1B | Sponge network | 0.224 | 0.48719 | -0.118 | 0.32258 | 0.334 |
| 32704 | TP73-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429 | 14 | STARD13 | Sponge network | -0.563 | 0.02545 | -0.187 | 0.27383 | 0.334 |
| 32705 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 24 | CBX5 | Sponge network | 1.153 | 0.00114 | 0.165 | 0.24446 | 0.334 |
| 32706 | MIR22HG |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-5p | 11 | RTN4 | Sponge network | -1.047 | 0 | -0.412 | 0.00014 | 0.334 |
| 32707 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 22 | IL6ST | Sponge network | 1.153 | 0.00114 | -0.402 | 0.00676 | 0.334 |
| 32708 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-5p | 32 | MOB1B | Sponge network | 0.95 | 0.15943 | 0.197 | 0.15266 | 0.334 |
| 32709 | RP11-296I10.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-539-5p | 16 | ABI2 | Sponge network | 0.592 | 0.00014 | 0.175 | 0.14787 | 0.334 |
| 32710 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 40 | NTRK3 | Sponge network | 0.889 | 9.0E-5 | -1.77 | 3.0E-5 | 0.334 |
| 32711 | RP11-327P2.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p | 31 | CHD5 | Sponge network | -0.147 | 0.40452 | -0.922 | 0.01521 | 0.334 |
| 32712 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577 | 30 | NOTCH2 | Sponge network | -2.69 | 0.0001 | 0.138 | 0.35163 | 0.334 |
| 32713 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 18 | NCOA1 | Sponge network | 0.453 | 0.01839 | -0.432 | 0 | 0.334 |
| 32714 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | DAB2 | Sponge network | 0.4 | 0.14611 | 0.431 | 0.0113 | 0.334 |
| 32715 | EMX2OS |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p | 23 | HIP1 | Sponge network | -0.777 | 0.1134 | 0.774 | 0 | 0.334 |
| 32716 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TBX18 | Sponge network | -0.942 | 0 | 0.843 | 0.02945 | 0.334 |
| 32717 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-625-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 52 | BMPR2 | Sponge network | 0.253 | 0.09565 | 0.206 | 0.0635 | 0.334 |
| 32718 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 53 | CCND2 | Sponge network | -0.49 | 0.04946 | -0.267 | 0.3112 | 0.334 |
| 32719 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 11 | DSE | Sponge network | -0.942 | 0 | -0.231 | 0.05347 | 0.334 |
| 32720 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | USP12 | Sponge network | 0.765 | 7.0E-5 | -0.102 | 0.40768 | 0.334 |
| 32721 | RP11-400K9.4 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 19 | JDP2 | Sponge network | -0.611 | 0.09607 | -0.967 | 0 | 0.334 |
| 32722 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 14 | CDHR3 | Sponge network | 0.174 | 0.30034 | -0.977 | 4.0E-5 | 0.334 |
| 32723 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | AKAP12 | Sponge network | -0.969 | 2.0E-5 | -0.932 | 0.0028 | 0.334 |
| 32724 | LIPE-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-96-5p | 12 | LATS2 | Sponge network | 0.078 | 0.55803 | 0.159 | 0.2304 | 0.334 |
| 32725 | RP11-400K9.4 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 22 | SYT4 | Sponge network | -0.611 | 0.09607 | -3.207 | 0 | 0.334 |
| 32726 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | RBL2 | Sponge network | -0.358 | 0.17255 | -0.282 | 0.00254 | 0.334 |
| 32727 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 36 | FAT3 | Sponge network | 0.559 | 0.00026 | -1.601 | 0.00051 | 0.334 |
| 32728 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TET1 | Sponge network | 0.272 | 0.44692 | -0.135 | 0.52138 | 0.334 |
| 32729 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-590-3p | 22 | IGF1 | Sponge network | -0.663 | 0.10887 | -0.204 | 0.5898 | 0.334 |
| 32730 | AF131215.9 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-940 | 14 | SORCS1 | Sponge network | -0.322 | 0.25945 | -3.492 | 0 | 0.334 |
| 32731 | FLNB-AS1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p | 10 | RASAL2 | Sponge network | 1.163 | 0.00018 | 0.869 | 0 | 0.334 |
| 32732 | DNAJC27-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 15 | RHOB | Sponge network | -0.969 | 2.0E-5 | -1.052 | 0 | 0.334 |
| 32733 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-340-5p | 24 | GNAQ | Sponge network | -1.331 | 3.0E-5 | -0.367 | 0.00102 | 0.334 |
| 32734 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p | 24 | HLF | Sponge network | -0.663 | 0.10887 | -2.443 | 0 | 0.334 |
| 32735 | FZD10-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-589-5p | 16 | RPS6KA2 | Sponge network | -0.727 | 0.04726 | -0.57 | 0.00013 | 0.334 |
| 32736 | MIR497HG |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577 | 15 | CSDE1 | Sponge network | -1.439 | 4.0E-5 | -0.493 | 0 | 0.334 |
| 32737 | RP11-244H3.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | CREB1 | Sponge network | 0.659 | 3.0E-5 | 0.203 | 0.00537 | 0.334 |
| 32738 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-539-5p | 10 | MGA | Sponge network | -0.154 | 0.78939 | 0.323 | 0.00792 | 0.334 |
| 32739 | RP11-701H24.3 |
hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-590-3p | 11 | ADARB1 | Sponge network | -1.425 | 0.04204 | -0.335 | 0.06631 | 0.334 |
| 32740 | XXYLT1-AS2 |
hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-744-3p | 10 | MEF2C | Sponge network | -0.764 | 0.05257 | -0.499 | 0.01448 | 0.334 |
| 32741 | HAND2-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 19 | CELF4 | Sponge network | -1.697 | 0.00889 | -1.135 | 0.00344 | 0.334 |
| 32742 | RP11-57H14.4 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | RYR2 | Sponge network | 0.083 | 0.66405 | -1.466 | 0.00031 | 0.334 |
| 32743 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-7-1-3p | 18 | ZFHX4 | Sponge network | -0.557 | 0.11135 | 0.018 | 0.95574 | 0.334 |
| 32744 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-96-5p | 12 | ZFP36L1 | Sponge network | -0.517 | 0.02935 | -0.505 | 8.0E-5 | 0.334 |
| 32745 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p | 10 | PRKAA2 | Sponge network | -1.054 | 0.0706 | -2.462 | 0 | 0.334 |
| 32746 | RP11-983P16.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | PDE4DIP | Sponge network | 0.41 | 0.02214 | -0.282 | 0.0257 | 0.334 |
| 32747 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-486-5p | 13 | EPHA3 | Sponge network | 0.556 | 0.00298 | 0.185 | 0.53588 | 0.334 |
| 32748 | RP1-151F17.2 |
hsa-miR-10a-3p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-589-5p | 10 | MN1 | Sponge network | 0.222 | 0.29133 | -0.358 | 0.24172 | 0.334 |
| 32749 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-425-5p;hsa-miR-7-1-3p | 16 | PTPRT | Sponge network | -0.187 | 0.23787 | -0.907 | 0.03727 | 0.334 |
| 32750 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-942-5p | 13 | RB1CC1 | Sponge network | 0.494 | 0.00339 | 0.175 | 0.12559 | 0.334 |
| 32751 | FZD10-AS1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | GATA2 | Sponge network | -0.727 | 0.04726 | -0.654 | 0.0016 | 0.334 |
| 32752 | CTD-2528L19.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | ARHGEF12 | Sponge network | 0.953 | 0 | 0.09 | 0.35693 | 0.334 |
| 32753 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 24 | CNTN1 | Sponge network | 0.174 | 0.30034 | -2.594 | 0 | 0.334 |
| 32754 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p | 21 | CREB1 | Sponge network | 1.46 | 1.0E-5 | 0.203 | 0.00537 | 0.334 |
| 32755 | CKMT2-AS1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | PPM1L | Sponge network | 0.082 | 0.55654 | -0.537 | 0.00616 | 0.334 |
| 32756 | RP11-83A24.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-409-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 14 | BAZ2B | Sponge network | 0.417 | 0.00445 | -0.276 | 0.02833 | 0.334 |
| 32757 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-185-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p | 10 | PLXNA2 | Sponge network | -0.829 | 0.01343 | -0.395 | 0.00779 | 0.334 |
| 32758 | LINC00865 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-942-5p | 25 | OPCML | Sponge network | -1.249 | 7.0E-5 | -2.498 | 0 | 0.334 |
| 32759 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 26 | AR | Sponge network | 0.131 | 0.8436 | -1.554 | 0 | 0.334 |
| 32760 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | PHC3 | Sponge network | 0.385 | 0.10067 | 0.212 | 0.0499 | 0.334 |
| 32761 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429 | 19 | NCOA1 | Sponge network | -1.654 | 0.00144 | -0.432 | 0 | 0.334 |
| 32762 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-429 | 10 | NEDD4 | Sponge network | -1.654 | 0.00144 | 0.02 | 0.89834 | 0.334 |
| 32763 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p | 13 | CLASP2 | Sponge network | 0.768 | 7.0E-5 | -0.038 | 0.71 | 0.334 |
| 32764 | SOX2-OT |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p | 30 | PRDM2 | Sponge network | -1.264 | 6.0E-5 | -0.258 | 0.00721 | 0.334 |
| 32765 | RP11-182J1.1 |
hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | MED12L | Sponge network | -0.187 | 0.23787 | -0.194 | 0.55299 | 0.334 |
| 32766 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 33 | BNC2 | Sponge network | 0.862 | 0.06213 | -0.491 | 0.09988 | 0.334 |
| 32767 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 38 | HBP1 | Sponge network | -1.111 | 0.0003 | -0.454 | 0 | 0.334 |
| 32768 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 18 | ARHGAP28 | Sponge network | 0.225 | 0.30644 | -0.911 | 0.00013 | 0.334 |
| 32769 | RP11-458F8.4 |
hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p | 11 | BRD3 | Sponge network | 1.518 | 0 | 0.37 | 0.00013 | 0.334 |
| 32770 | RP4-669P10.16 |
hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-505-5p;hsa-miR-769-5p;hsa-miR-93-3p | 16 | GRIK3 | Sponge network | -0.301 | 0.06597 | -3.208 | 0 | 0.334 |
| 32771 | LINC00957 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | ZNF208 | Sponge network | -0.421 | 0.0114 | -0.4 | 0.22381 | 0.334 |
| 32772 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-708-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 31 | RAP1A | Sponge network | -0.096 | 0.67958 | -0.929 | 0 | 0.334 |
| 32773 | HAND2-AS1 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | DIRAS1 | Sponge network | -1.697 | 0.00889 | -2.518 | 0 | 0.334 |
| 32774 | CTC-296K1.4 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-629-5p | 11 | CTIF | Sponge network | -2.076 | 0.00548 | -0.389 | 0.00549 | 0.334 |
| 32775 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | CDH19 | Sponge network | 0.16 | 0.50785 | -3.434 | 0 | 0.334 |
| 32776 | RP11-535M15.1 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-219a-1-3p;hsa-miR-320b | 10 | JAZF1 | Sponge network | -1.14 | 0.03513 | -0.377 | 0.02677 | 0.334 |
| 32777 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p | 12 | SMARCA1 | Sponge network | 0.997 | 0.00066 | -0.684 | 0.00159 | 0.334 |
| 32778 | TP73-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | PCDH17 | Sponge network | -0.563 | 0.02545 | 0.731 | 2.0E-5 | 0.334 |
| 32779 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 29 | AR | Sponge network | 1.757 | 0 | -1.554 | 0 | 0.334 |
| 32780 | LINC00865 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-629-3p | 15 | CXCL12 | Sponge network | -1.249 | 7.0E-5 | -1.421 | 0 | 0.334 |
| 32781 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 25 | ZMYM2 | Sponge network | 0.541 | 0.00125 | 0.279 | 0.01273 | 0.334 |
| 32782 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 49 | AR | Sponge network | 0.083 | 0.66405 | -1.554 | 0 | 0.334 |
| 32783 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429 | 10 | TSC22D1 | Sponge network | -2.62 | 0 | -0.529 | 2.0E-5 | 0.334 |
| 32784 | RP11-181G12.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-378a-5p;hsa-miR-940 | 14 | CREB1 | Sponge network | 0.556 | 0.00298 | 0.203 | 0.00537 | 0.334 |
| 32785 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-877-5p | 27 | GRIN2A | Sponge network | -0.303 | 0.21002 | -2.161 | 0 | 0.334 |
| 32786 | PART1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-624-5p | 10 | SCN11A | Sponge network | -2.925 | 0 | -1.15 | 0.00299 | 0.334 |
| 32787 | AC003090.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 10 | UVRAG | Sponge network | -2.117 | 0.00161 | -0.017 | 0.83865 | 0.334 |
| 32788 | FAM157C |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p | 12 | UBR3 | Sponge network | 0.91 | 0 | -0.021 | 0.82774 | 0.334 |
| 32789 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 19 | ZFHX3 | Sponge network | 0.693 | 0.00076 | 0.389 | 0.01363 | 0.334 |
| 32790 | RP5-1139B12.3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-502-5p | 10 | PRKCB | Sponge network | 0.598 | 0.38481 | -1.313 | 2.0E-5 | 0.334 |
| 32791 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 24 | SUFU | Sponge network | -1.439 | 4.0E-5 | -0.04 | 0.65489 | 0.334 |
| 32792 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | JAZF1 | Sponge network | -2.915 | 0.00015 | -0.377 | 0.02677 | 0.334 |
| 32793 | GNG12-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-590-3p | 24 | NCOA1 | Sponge network | -0.793 | 7.0E-5 | -0.432 | 0 | 0.334 |
| 32794 | RP11-98I9.4 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZMYND11 | Sponge network | 0.67 | 0.00048 | -0.2 | 0.05449 | 0.334 |
| 32795 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-93-3p | 31 | BMPR2 | Sponge network | -0.469 | 0.08721 | 0.206 | 0.0635 | 0.334 |
| 32796 | LINC01006 |
hsa-miR-103a-3p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-429 | 13 | DYNC1I1 | Sponge network | -0.702 | 0.04814 | -1.12 | 0.00107 | 0.334 |
| 32797 | RP11-59H7.3 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-455-5p;hsa-miR-576-5p | 10 | ARHGEF12 | Sponge network | 2.163 | 0 | 0.09 | 0.35693 | 0.334 |
| 32798 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 11 | ZNF292 | Sponge network | 0.569 | 0.00067 | 0.276 | 0.04148 | 0.334 |
| 32799 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 26 | ARHGAP28 | Sponge network | -0.737 | 0.06182 | -0.911 | 0.00013 | 0.334 |
| 32800 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 21 | BCL6 | Sponge network | -0.793 | 7.0E-5 | -0.211 | 0.19489 | 0.334 |
| 32801 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 38 | FOXP1 | Sponge network | 0.335 | 0.20596 | 0.042 | 0.74368 | 0.334 |
| 32802 | AC005618.6 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PIK3CA | Sponge network | 0.862 | 0.06213 | 0.195 | 0.06908 | 0.334 |
| 32803 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | PEG3 | Sponge network | 0.219 | 0.1516 | -1.74 | 0 | 0.334 |
| 32804 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-365a-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 32 | RICTOR | Sponge network | 0.082 | 0.55654 | 0.388 | 0.00188 | 0.334 |
| 32805 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-7-1-3p | 12 | FAT4 | Sponge network | -0.405 | 0.22013 | -0.354 | 0.14694 | 0.334 |
| 32806 | LINC00702 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3613-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | GATA2 | Sponge network | -0.737 | 0.06182 | -0.654 | 0.0016 | 0.334 |
| 32807 | RP11-119F7.5 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | LUZP2 | Sponge network | 0.225 | 0.30644 | -0.056 | 0.89416 | 0.334 |
| 32808 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-577 | 17 | PGR | Sponge network | 0.389 | 0.04293 | -1.704 | 0 | 0.334 |
| 32809 | HOXB-AS2 |
hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-429 | 16 | AFF1 | Sponge network | 1.316 | 0.01106 | -0.384 | 0.00053 | 0.334 |
| 32810 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | ZFHX4 | Sponge network | 0.131 | 0.8436 | 0.018 | 0.95574 | 0.334 |
| 32811 | TPTEP1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | TAL1 | Sponge network | -1.216 | 0 | -0.462 | 0.00574 | 0.334 |
| 32812 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | AFF4 | Sponge network | -1.654 | 0.00144 | -0.266 | 0.01565 | 0.334 |
| 32813 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 22 | BMPR2 | Sponge network | 1.308 | 0 | 0.206 | 0.0635 | 0.334 |
| 32814 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-628-5p | 13 | CDC73 | Sponge network | 1.344 | 0 | 0.094 | 0.20463 | 0.334 |
| 32815 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | SCN9A | Sponge network | -0.668 | 3.0E-5 | -0.525 | 0.10593 | 0.334 |
| 32816 | RP11-38L15.3 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-590-3p | 10 | BNC2 | Sponge network | 0.344 | 0.3127 | -0.491 | 0.09988 | 0.334 |
| 32817 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 23 | TET1 | Sponge network | 0.395 | 0.15451 | -0.135 | 0.52138 | 0.334 |
| 32818 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-511-5p | 12 | BCL6 | Sponge network | -0.326 | 0.14314 | -0.211 | 0.19489 | 0.334 |
| 32819 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-33a-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 17 | SOX11 | Sponge network | 0.603 | 0.00127 | 2.68 | 0 | 0.334 |
| 32820 | RP4-639F20.1 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-7-5p | 13 | PRKCB | Sponge network | -0.517 | 0.02935 | -1.313 | 2.0E-5 | 0.334 |
| 32821 | RP11-672A2.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-505-5p;hsa-miR-671-5p | 10 | GRIK3 | Sponge network | 1.344 | 0 | -3.208 | 0 | 0.334 |
| 32822 | RP11-404E16.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p | 12 | ZFHX4 | Sponge network | 0.097 | 0.91311 | 0.018 | 0.95574 | 0.334 |
| 32823 | LINC00476 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | NR3C2 | Sponge network | -0.615 | 0.00015 | -1.56 | 0 | 0.334 |
| 32824 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 17 | TBX18 | Sponge network | -0.563 | 0.02545 | 0.843 | 0.02945 | 0.334 |
| 32825 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | ZFHX4 | Sponge network | 1.61 | 0.00961 | 0.018 | 0.95574 | 0.334 |
| 32826 | AF131215.9 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-450b-5p | 10 | ZBTB20 | Sponge network | -0.322 | 0.25945 | -0.122 | 0.57569 | 0.334 |
| 32827 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | MOB1B | Sponge network | 1.308 | 0 | 0.197 | 0.15266 | 0.334 |
| 32828 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 17 | PIK3C2A | Sponge network | 0.539 | 0.03722 | 0.061 | 0.53776 | 0.334 |
| 32829 | CTC-296K1.3 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-582-5p | 18 | BTG2 | Sponge network | -2.095 | 0.00112 | -1.763 | 0 | 0.334 |
| 32830 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-96-5p | 19 | CADM1 | Sponge network | -1.654 | 0.00144 | -0.9 | 0.00084 | 0.334 |
| 32831 | TINCR |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-874-3p | 13 | NR2C2 | Sponge network | -0.664 | 0.14543 | 0.21 | 0.03224 | 0.334 |
| 32832 | RP11-326C3.7 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-424-5p | 10 | ZFHX4 | Sponge network | 0.295 | 0.28897 | 0.018 | 0.95574 | 0.334 |
| 32833 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | PDGFRA | Sponge network | -0.733 | 0.19469 | -0.372 | 0.0037 | 0.334 |
| 32834 | A2M-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | SETBP1 | Sponge network | -0.08 | 0.90092 | -1.302 | 1.0E-5 | 0.334 |
| 32835 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | 0.246 | 0.59912 | -1.751 | 1.0E-5 | 0.334 |
| 32836 | HOTAIRM1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NCAM2 | Sponge network | -0.053 | 0.80685 | -0.482 | 0.22565 | 0.334 |
| 32837 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | IGSF10 | Sponge network | 0.131 | 0.8436 | -1.751 | 1.0E-5 | 0.334 |
| 32838 | LINC00403 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-219a-1-3p | 12 | EPHA5 | Sponge network | -3.586 | 0.00032 | -0.957 | 0.01733 | 0.334 |
| 32839 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-495-3p | 10 | TP53INP1 | Sponge network | 0.817 | 3.0E-5 | -0.496 | 0.00064 | 0.334 |
| 32840 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-3615;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | DMTF1 | Sponge network | 0.083 | 0.66405 | 0.248 | 0.02726 | 0.334 |
| 32841 | LINC00702 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 30 | CYLD | Sponge network | -0.737 | 0.06182 | -0.367 | 0.00171 | 0.334 |
| 32842 | NDUFA6-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | -0.355 | 0.0077 | -0.974 | 0.00034 | 0.334 |
| 32843 | LINC00854 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p;hsa-miR-629-3p | 11 | PTPN14 | Sponge network | 1.18 | 0 | 0.144 | 0.34595 | 0.334 |
| 32844 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 15 | UVRAG | Sponge network | 0.101 | 0.60919 | -0.017 | 0.83865 | 0.334 |
| 32845 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 19 | RET | Sponge network | -0.487 | 0.02157 | -0.833 | 0.03517 | 0.334 |
| 32846 | RP1-302G2.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p | 10 | PLSCR4 | Sponge network | -1.483 | 9.0E-5 | -0.474 | 0.03833 | 0.334 |
| 32847 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | PTPRB | Sponge network | -1.886 | 0 | 0.105 | 0.47122 | 0.334 |
| 32848 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 16 | PDS5B | Sponge network | 0.959 | 0 | 0.259 | 0.00962 | 0.334 |
| 32849 | RP11-526I2.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-539-5p | 11 | MOB1B | Sponge network | 1.054 | 1.0E-5 | 0.197 | 0.15266 | 0.334 |
| 32850 | CTC-301O7.4 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 10 | IL6ST | Sponge network | 0.292 | 0.43384 | -0.402 | 0.00676 | 0.334 |
| 32851 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 21 | MYBL1 | Sponge network | 0.603 | 0.00127 | 0.494 | 0.02152 | 0.334 |
| 32852 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 21 | SOX11 | Sponge network | 1.266 | 0 | 2.68 | 0 | 0.334 |
| 32853 | TINCR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-96-5p | 33 | PRKAA2 | Sponge network | -0.664 | 0.14543 | -2.462 | 0 | 0.334 |
| 32854 | RP13-516M14.1 |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-7-1-3p | 11 | ZMYND11 | Sponge network | 0.389 | 0.04293 | -0.2 | 0.05449 | 0.334 |
| 32855 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 27 | PCDH17 | Sponge network | 0.101 | 0.60919 | 0.731 | 2.0E-5 | 0.334 |
| 32856 | RP11-77H9.2 |
hsa-let-7a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-376c-3p | 10 | TRIM33 | Sponge network | 1.677 | 0 | 0.402 | 7.0E-5 | 0.334 |
| 32857 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | NRXN1 | Sponge network | -0.355 | 0.0077 | -3.778 | 0 | 0.334 |
| 32858 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-655-3p | 22 | LATS1 | Sponge network | 0.532 | 0.00505 | 0.109 | 0.34208 | 0.334 |
| 32859 | LINC00094 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-625-5p | 14 | CEP170 | Sponge network | 0.253 | 0.09565 | 0.943 | 0 | 0.334 |
| 32860 | AC009948.5 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p;hsa-miR-93-5p | 53 | DST | Sponge network | 0.532 | 0.00505 | -0.713 | 0.00096 | 0.334 |
| 32861 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p;hsa-miR-940 | 10 | FAM117A | Sponge network | 0.214 | 0.18246 | -1.379 | 0 | 0.334 |
| 32862 | HOTAIRM1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | -0.053 | 0.80685 | -0.704 | 0.00106 | 0.334 |
| 32863 | AC010226.4 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 18 | PTPRT | Sponge network | -0.811 | 2.0E-5 | -0.907 | 0.03727 | 0.334 |
| 32864 | LINC00622 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | RICTOR | Sponge network | 1.169 | 0.00028 | 0.388 | 0.00188 | 0.334 |
| 32865 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 27 | PDGFRA | Sponge network | -0.739 | 0.00044 | -0.372 | 0.0037 | 0.334 |
| 32866 | RP11-403I13.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 16 | AKAP6 | Sponge network | 0.395 | 0.15451 | -1.586 | 3.0E-5 | 0.334 |
| 32867 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-495-3p | 13 | RPS6KA3 | Sponge network | 0.614 | 1.0E-5 | 0.256 | 0.02419 | 0.334 |
| 32868 | RP11-426C22.5 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 10 | ZFHX4 | Sponge network | 0.749 | 0 | 0.018 | 0.95574 | 0.334 |
| 32869 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 10 | CDH10 | Sponge network | -0.513 | 0.04703 | -2.397 | 0 | 0.334 |
| 32870 | RP11-1006G14.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PRUNE2 | Sponge network | 0.559 | 0.00026 | -1.733 | 0.00022 | 0.334 |
| 32871 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-500a-5p | 11 | MAF | Sponge network | -0.011 | 0.967 | -1.257 | 0 | 0.334 |
| 32872 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b | 10 | PTPRB | Sponge network | 0.001 | 0.99753 | 0.105 | 0.47122 | 0.334 |
| 32873 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | KAT6B | Sponge network | 0.083 | 0.66405 | -0.478 | 0.00018 | 0.334 |
| 32874 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-96-5p | 40 | BCL2 | Sponge network | 0.082 | 0.55654 | -0.899 | 0 | 0.334 |
| 32875 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | HDAC9 | Sponge network | -1.249 | 7.0E-5 | -0.755 | 0.00495 | 0.334 |
| 32876 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p | 27 | PRDM2 | Sponge network | 0.082 | 0.55654 | -0.258 | 0.00721 | 0.334 |
| 32877 | RP1-302G2.5 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-940 | 13 | MEF2C | Sponge network | -1.483 | 9.0E-5 | -0.499 | 0.01448 | 0.334 |
| 32878 | LINC00476 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-935 | 10 | LRRN3 | Sponge network | -0.615 | 0.00015 | -0.393 | 0.16293 | 0.334 |
| 32879 | HAR1A |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-940 | 25 | NTRK3 | Sponge network | -0.672 | 0.19447 | -1.77 | 3.0E-5 | 0.334 |
| 32880 | RP11-305E6.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | ZFX | Sponge network | 1.317 | 0 | 0.09 | 0.42563 | 0.334 |
| 32881 | PCA3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | NIN | Sponge network | -0.709 | 0.18168 | -0.253 | 0.13794 | 0.334 |
| 32882 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | CCND2 | Sponge network | -0.976 | 0.00025 | -0.267 | 0.3112 | 0.334 |
| 32883 | LINC00854 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-654-3p;hsa-miR-940 | 11 | AKAP11 | Sponge network | 1.18 | 0 | 0.196 | 0.09825 | 0.334 |
| 32884 | RP11-53O19.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 28 | PDE4DIP | Sponge network | -0.405 | 0.22013 | -0.282 | 0.0257 | 0.334 |
| 32885 | RP11-774O3.3 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-590-3p | 14 | FMNL3 | Sponge network | -0.487 | 0.02157 | 0.555 | 4.0E-5 | 0.334 |
| 32886 | RP11-983P16.4 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-7-1-3p | 18 | ITPKB | Sponge network | 0.41 | 0.02214 | -0.704 | 0.00106 | 0.334 |
| 32887 | RP11-389C8.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-942-5p | 17 | IGFBP5 | Sponge network | 0.727 | 3.0E-5 | -0.806 | 0.00105 | 0.334 |
| 32888 | CTC-490E21.10 |
hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-493-5p | 10 | NEDD4 | Sponge network | 1.46 | 1.0E-5 | 0.02 | 0.89834 | 0.334 |
| 32889 | RP11-344B5.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p | 10 | EBF1 | Sponge network | -0.055 | 0.88482 | -0.384 | 0.08856 | 0.334 |
| 32890 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p | 30 | TACC1 | Sponge network | 0.177 | 0.16681 | -0.362 | 0.05406 | 0.334 |
| 32891 | LINC00641 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 36 | SNX29 | Sponge network | -0.222 | 0.32445 | -0.34 | 0.01371 | 0.334 |
| 32892 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | RBL2 | Sponge network | 0.349 | 0.01695 | -0.282 | 0.00254 | 0.334 |
| 32893 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | USP12 | Sponge network | -0.358 | 0.17255 | -0.102 | 0.40768 | 0.334 |
| 32894 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | EPAS1 | Sponge network | -0.727 | 0.04726 | -0.184 | 0.14418 | 0.334 |
| 32895 | RP11-182L21.6 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-625-5p | 14 | ABI2 | Sponge network | 0.353 | 0.00175 | 0.175 | 0.14787 | 0.334 |
| 32896 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p | 21 | JAZF1 | Sponge network | 0.659 | 3.0E-5 | -0.377 | 0.02677 | 0.334 |
| 32897 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 13 | HIP1 | Sponge network | 0.174 | 0.30034 | 0.774 | 0 | 0.334 |
| 32898 | KB-431C1.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-625-5p;hsa-miR-766-3p | 25 | MDM4 | Sponge network | 0.909 | 0 | 0.345 | 0.00762 | 0.334 |
| 32899 | LPP-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-96-5p | 15 | UNC5C | Sponge network | -0.18 | 0.20343 | -0.178 | 0.40214 | 0.334 |
| 32900 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 13 | GIGYF2 | Sponge network | 1.11 | 0 | 0.136 | 0.04403 | 0.334 |
| 32901 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-335-5p;hsa-miR-455-3p | 10 | TLE4 | Sponge network | -0.465 | 0.04703 | -1.157 | 0 | 0.334 |
| 32902 | RP11-196G18.24 |
hsa-miR-101-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | UBR3 | Sponge network | 1.262 | 0 | -0.021 | 0.82774 | 0.334 |
| 32903 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-375 | 15 | ERC1 | Sponge network | 0.858 | 3.0E-5 | -0.108 | 0.38794 | 0.334 |
| 32904 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 33 | PBX1 | Sponge network | -1.047 | 0 | -1.206 | 0 | 0.334 |
| 32905 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-577;hsa-miR-7-1-3p | 10 | ABCC9 | Sponge network | 0.419 | 0.0319 | -0.466 | 0.14121 | 0.334 |
| 32906 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | MED12L | Sponge network | -0.421 | 0.0114 | -0.194 | 0.55299 | 0.334 |
| 32907 | SNX29P2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-532-3p | 13 | SYNM | Sponge network | 0.4 | 0.14611 | -2.309 | 1.0E-5 | 0.334 |
| 32908 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 23 | MYO9A | Sponge network | 0.401 | 0.00066 | -0.147 | 0.32373 | 0.334 |
| 32909 | GS1-124K5.11 |
hsa-miR-126-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p | 13 | IRS1 | Sponge network | 0.494 | 0.00339 | -0.026 | 0.88855 | 0.334 |
| 32910 | ANKRD10-IT1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-7-1-3p | 10 | UBE2D1 | Sponge network | 1.739 | 0 | 0.468 | 0 | 0.334 |
| 32911 | RP11-473I1.5 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p | 11 | FOXO3 | Sponge network | 0.542 | 0.00054 | -0.04 | 0.72028 | 0.334 |
| 32912 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p | 10 | SMARCA2 | Sponge network | 0.178 | 0.29106 | -0.529 | 0.00014 | 0.334 |
| 32913 | MIR143HG |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 28 | EFNA5 | Sponge network | -0.739 | 0.0574 | -0.421 | 0.17868 | 0.334 |
| 32914 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.439 | 0.00313 | 0.323 | 0.00792 | 0.334 |
| 32915 | TMEM9B-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-324-5p | 18 | ZFX | Sponge network | 0.529 | 0.00552 | 0.09 | 0.42563 | 0.334 |
| 32916 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | PCDH9 | Sponge network | 0.381 | 0.25967 | -1.823 | 4.0E-5 | 0.334 |
| 32917 | RP11-517B11.7 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-766-3p | 10 | ATM | Sponge network | 0.706 | 0 | 0.178 | 0.21568 | 0.334 |
| 32918 | KB-1460A1.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p | 66 | LPP | Sponge network | 0.603 | 0.00127 | -0.459 | 0.04475 | 0.334 |
| 32919 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-500a-5p | 17 | PRKAB2 | Sponge network | -0.932 | 0.00329 | -0.367 | 0.01371 | 0.334 |
| 32920 | RP11-156P1.3 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p | 14 | IRS1 | Sponge network | 0.214 | 0.18246 | -0.026 | 0.88855 | 0.334 |
| 32921 | FENDRR |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 14 | SEPT6 | Sponge network | -1.281 | 0.00012 | -0.598 | 0.00181 | 0.334 |
| 32922 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | MCC | Sponge network | -0.023 | 0.95109 | -1.005 | 0 | 0.333 |
| 32923 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-590-3p | 10 | RBM5 | Sponge network | 0.415 | 0.14657 | -0.205 | 0.0129 | 0.333 |
| 32924 | AC116366.6 |
hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-299-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-577 | 13 | PHC3 | Sponge network | 0.329 | 0.11122 | 0.212 | 0.0499 | 0.333 |
| 32925 | TMEM9B-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-582-5p | 28 | BMPR2 | Sponge network | 0.529 | 0.00552 | 0.206 | 0.0635 | 0.333 |
| 32926 | IGF2-AS |
hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-500a-3p | 13 | ZFHX4 | Sponge network | 3.339 | 0.00237 | 0.018 | 0.95574 | 0.333 |
| 32927 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | EPHA3 | Sponge network | 0.853 | 0.15352 | 0.185 | 0.53588 | 0.333 |
| 32928 | RP11-434B12.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | BCL9 | Sponge network | -0.031 | 0.89063 | 0.321 | 0.00267 | 0.333 |
| 32929 | HAND2-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ZNF536 | Sponge network | -1.697 | 0.00889 | -3.115 | 0 | 0.333 |
| 32930 | RP4-669P10.16 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p | 12 | ZBTB4 | Sponge network | -0.301 | 0.06597 | -0.739 | 0 | 0.333 |
| 32931 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 37 | ERC1 | Sponge network | -2.46 | 0 | -0.108 | 0.38794 | 0.333 |
| 32932 | BCYRN1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 17 | SHPRH | Sponge network | 0.311 | 0.10344 | 0.098 | 0.40994 | 0.333 |
| 32933 | H19 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-589-3p | 12 | ZFHX4 | Sponge network | 2.439 | 0 | 0.018 | 0.95574 | 0.333 |
| 32934 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | MYO9A | Sponge network | 0.663 | 0.00073 | -0.147 | 0.32373 | 0.333 |
| 32935 | RP11-983P16.4 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p | 14 | MRVI1 | Sponge network | 0.41 | 0.02214 | -1.051 | 0.00022 | 0.333 |
| 32936 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 39 | TMEM132B | Sponge network | -0.227 | 0.31657 | -0.83 | 0.01084 | 0.333 |
| 32937 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-7-1-3p | 14 | LRIG1 | Sponge network | 0.889 | 9.0E-5 | -0.537 | 0.00588 | 0.333 |
| 32938 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-539-5p;hsa-miR-625-5p | 14 | RSPO2 | Sponge network | 0.299 | 0.57722 | -3.038 | 0 | 0.333 |
| 32939 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p | 18 | NEDD4 | Sponge network | -0.227 | 0.31657 | 0.02 | 0.89834 | 0.333 |
| 32940 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-590-3p | 10 | GPAM | Sponge network | -0.513 | 0.04703 | 0.142 | 0.29732 | 0.333 |
| 32941 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-421;hsa-miR-590-3p | 13 | MITF | Sponge network | 0.178 | 0.29106 | -0.769 | 0.00022 | 0.333 |
| 32942 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 21 | LRRC55 | Sponge network | -0.421 | 0.0114 | -0.026 | 0.93163 | 0.333 |
| 32943 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | BNC2 | Sponge network | 0.559 | 0.00026 | -0.491 | 0.09988 | 0.333 |
| 32944 | OIP5-AS1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 21 | KIF5B | Sponge network | 0.421 | 0.00084 | 0.362 | 0.00066 | 0.333 |
| 32945 | AC084219.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 1.296 | 0.0054 | 0.578 | 0 | 0.333 |
| 32946 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 54 | IL6ST | Sponge network | 0.494 | 0.00339 | -0.402 | 0.00676 | 0.333 |
| 32947 | AC090587.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-589-3p | 15 | ADARB1 | Sponge network | -0.088 | 0.65011 | -0.335 | 0.06631 | 0.333 |
| 32948 | HOTAIRM1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | NAV3 | Sponge network | -0.053 | 0.80685 | 0.212 | 0.47729 | 0.333 |
| 32949 | LINC00680 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-625-5p | 15 | ABI2 | Sponge network | 0.884 | 0 | 0.175 | 0.14787 | 0.333 |
| 32950 | RP11-130L8.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 30 | CADM2 | Sponge network | -0.663 | 0.10887 | -3.782 | 0 | 0.333 |
| 32951 | RP4-613B23.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-539-5p | 10 | TMEM132B | Sponge network | -0.309 | 0.1145 | -0.83 | 0.01084 | 0.333 |
| 32952 | CTD-3110H11.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 12 | CREB1 | Sponge network | 1.735 | 0 | 0.203 | 0.00537 | 0.333 |
| 32953 | RP1-257A7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-191-5p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-629-3p | 10 | DDX6 | Sponge network | 0.849 | 0.00178 | 0.091 | 0.36428 | 0.333 |
| 32954 | RP11-981G7.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-486-5p | 10 | PHF6 | Sponge network | 0.986 | 0.01022 | 0.785 | 0 | 0.333 |
| 32955 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 39 | ERBB4 | Sponge network | -1.249 | 7.0E-5 | -1.975 | 2.0E-5 | 0.333 |
| 32956 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-577 | 10 | AMPH | Sponge network | -0.899 | 6.0E-5 | -0.916 | 5.0E-5 | 0.333 |
| 32957 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-628-5p | 17 | CDC73 | Sponge network | 0.401 | 0.00066 | 0.094 | 0.20463 | 0.333 |
| 32958 | RP11-403I13.7 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 11 | ZBTB20 | Sponge network | 0.395 | 0.15451 | -0.122 | 0.57569 | 0.333 |
| 32959 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | BNC2 | Sponge network | 0.219 | 0.1516 | -0.491 | 0.09988 | 0.333 |
| 32960 | BDNF-AS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p | 30 | DMD | Sponge network | -0.668 | 3.0E-5 | -1.506 | 0 | 0.333 |
| 32961 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-625-3p | 25 | ZFHX3 | Sponge network | 0.594 | 0.41522 | 0.389 | 0.01363 | 0.333 |
| 32962 | RP11-439L18.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-454-3p | 14 | WDFY3 | Sponge network | 0.224 | 0.48719 | -0.201 | 0.0967 | 0.333 |
| 32963 | PAN3-AS1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-369-3p | 11 | ABI2 | Sponge network | 0.062 | 0.55274 | 0.175 | 0.14787 | 0.333 |
| 32964 | HNRNPU-AS1 |
hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DOCK4 | Sponge network | 1.601 | 0 | 0.502 | 0.00108 | 0.333 |
| 32965 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 10 | SETBP1 | Sponge network | -0.285 | 0.2172 | -1.302 | 1.0E-5 | 0.333 |
| 32966 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 34 | MOB1B | Sponge network | 1.302 | 7.0E-5 | 0.197 | 0.15266 | 0.333 |
| 32967 | WDFY3-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | HOMER2 | Sponge network | -0.942 | 0 | -2.698 | 0 | 0.333 |
| 32968 | AC010226.4 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-508-3p | 10 | RASSF3 | Sponge network | -0.811 | 2.0E-5 | -0.226 | 0.11691 | 0.333 |
| 32969 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | ZFHX4 | Sponge network | -0.08 | 0.90092 | 0.018 | 0.95574 | 0.333 |
| 32970 | RP11-693J15.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-502-5p;hsa-miR-7-1-3p | 28 | DTNA | Sponge network | -0.72 | 0.24239 | -1.648 | 1.0E-5 | 0.333 |
| 32971 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -0.615 | 0.00015 | -0.354 | 0.14694 | 0.333 |
| 32972 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 38 | WDFY3 | Sponge network | 0.683 | 1.0E-5 | -0.201 | 0.0967 | 0.333 |
| 32973 | FENDRR |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | JDP2 | Sponge network | -1.281 | 0.00012 | -0.967 | 0 | 0.333 |
| 32974 | AC074289.1 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p | 16 | ANK2 | Sponge network | -0.326 | 0.14314 | -1.603 | 1.0E-5 | 0.333 |
| 32975 | RP11-685N10.1 |
hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p | 11 | CREB1 | Sponge network | 2.743 | 0 | 0.203 | 0.00537 | 0.333 |
| 32976 | AC005618.6 |
hsa-let-7b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 40 | AR | Sponge network | 0.862 | 0.06213 | -1.554 | 0 | 0.333 |
| 32977 | RP5-998N21.10 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-502-5p;hsa-miR-505-5p | 10 | AR | Sponge network | -0.098 | 0.76298 | -1.554 | 0 | 0.333 |
| 32978 | LINC00957 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | PDGFRA | Sponge network | -0.421 | 0.0114 | -0.372 | 0.0037 | 0.333 |
| 32979 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | KAT6B | Sponge network | -2.117 | 0.00161 | -0.478 | 0.00018 | 0.333 |
| 32980 | CTD-2528L19.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ZFX | Sponge network | 0.953 | 0 | 0.09 | 0.42563 | 0.333 |
| 32981 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-590-3p | 18 | PPP2R5C | Sponge network | -0.342 | 0.02288 | -0.314 | 3.0E-5 | 0.333 |
| 32982 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-744-5p | 14 | IGF2 | Sponge network | -0.469 | 0.08721 | 0.848 | 0.03842 | 0.333 |
| 32983 | RP11-403I13.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p | 38 | RUNX1T1 | Sponge network | 0.619 | 0.00157 | -0.344 | 0.2374 | 0.333 |
| 32984 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-664a-3p | 20 | FZD3 | Sponge network | 1.294 | 0 | 0.104 | 0.60316 | 0.333 |
| 32985 | RP11-356J5.12 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 49 | PPM1L | Sponge network | -0.899 | 6.0E-5 | -0.537 | 0.00616 | 0.333 |
| 32986 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 19 | FAT4 | Sponge network | 0.083 | 0.66405 | -0.354 | 0.14694 | 0.333 |
| 32987 | TUG1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-654-3p | 18 | DDX6 | Sponge network | 0.972 | 0 | 0.091 | 0.36428 | 0.333 |
| 32988 | A2M-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | NIN | Sponge network | -0.08 | 0.90092 | -0.253 | 0.13794 | 0.333 |
| 32989 | RAD51-AS1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p | 10 | MAT2A | Sponge network | 0.768 | 7.0E-5 | -0.073 | 0.36195 | 0.333 |
| 32990 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b | 11 | SMARCA2 | Sponge network | 0.082 | 0.55397 | -0.529 | 0.00014 | 0.333 |
| 32991 | RP11-474O21.5 |
hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-7-1-3p | 13 | IL6ST | Sponge network | -0.023 | 0.95109 | -0.402 | 0.00676 | 0.333 |
| 32992 | RP5-1047A19.4 |
hsa-miR-148a-3p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p | 10 | KIF1B | Sponge network | 0.745 | 1.0E-5 | -0.118 | 0.32258 | 0.333 |
| 32993 | HCG11 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PHF3 | Sponge network | 0.385 | 0.10067 | 0.126 | 0.19652 | 0.333 |
| 32994 | LPP-AS2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-769-5p | 15 | KALRN | Sponge network | -0.18 | 0.20343 | -0.819 | 1.0E-5 | 0.333 |
| 32995 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-769-5p | 14 | RCAN2 | Sponge network | -1.645 | 0 | -0.33 | 0.15172 | 0.333 |
| 32996 | RP5-1139B12.3 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 17 | WDFY3 | Sponge network | 0.598 | 0.38481 | -0.201 | 0.0967 | 0.333 |
| 32997 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | EBF1 | Sponge network | -0.053 | 0.80685 | -0.384 | 0.08856 | 0.333 |
| 32998 | BOLA3-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-935 | 19 | GJA1 | Sponge network | -0.246 | 0.35034 | -0.485 | 0.02179 | 0.333 |
| 32999 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 20 | RUNX1T1 | Sponge network | 0.912 | 0.00014 | -0.344 | 0.2374 | 0.333 |
| 33000 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | LATS2 | Sponge network | -0.739 | 0.00044 | 0.159 | 0.2304 | 0.333 |
| 33001 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 13 | CDHR3 | Sponge network | 0.274 | 0.07681 | -0.977 | 4.0E-5 | 0.333 |
| 33002 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | NR4A3 | Sponge network | -0.737 | 0.06182 | -1.385 | 0 | 0.333 |
| 33003 | ERVK3-1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p | 15 | PHF6 | Sponge network | 0.58 | 0 | 0.785 | 0 | 0.333 |
| 33004 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-7-5p | 25 | BCL2 | Sponge network | -0.693 | 0.05629 | -0.899 | 0 | 0.333 |
| 33005 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | IGSF10 | Sponge network | 0.61 | 0.01593 | -1.751 | 1.0E-5 | 0.333 |
| 33006 | RP11-196G18.23 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-590-3p | 11 | MYBL1 | Sponge network | 0.724 | 0.00039 | 0.494 | 0.02152 | 0.333 |
| 33007 | LINC00094 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p | 16 | MRVI1 | Sponge network | 0.253 | 0.09565 | -1.051 | 0.00022 | 0.333 |
| 33008 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 11 | INPP4A | Sponge network | -0.318 | 0.65847 | 0.07 | 0.53563 | 0.333 |
| 33009 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-671-5p | 17 | KIT | Sponge network | 0.727 | 3.0E-5 | -2.184 | 0 | 0.333 |
| 33010 | CTD-2303H24.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | UVRAG | Sponge network | 0.415 | 0.14657 | -0.017 | 0.83865 | 0.333 |
| 33011 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 34 | GAS7 | Sponge network | -0.08 | 0.90092 | -0.555 | 0.01602 | 0.333 |
| 33012 | AC005562.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 14 | SLC5A3 | Sponge network | 0.488 | 0.00084 | 0.493 | 5.0E-5 | 0.333 |
| 33013 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-589-3p | 22 | ADARB1 | Sponge network | 0.622 | 0.00015 | -0.335 | 0.06631 | 0.333 |
| 33014 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p | 16 | ANK2 | Sponge network | 2.306 | 9.0E-5 | -1.603 | 1.0E-5 | 0.333 |
| 33015 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | ARHGAP29 | Sponge network | -1.582 | 8.0E-5 | 0.131 | 0.42953 | 0.333 |
| 33016 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-629-3p | 15 | PTPN14 | Sponge network | 0.453 | 0.01839 | 0.144 | 0.34595 | 0.333 |
| 33017 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 16 | CREBBP | Sponge network | 0.765 | 7.0E-5 | 0.126 | 0.15186 | 0.333 |
| 33018 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | TMEM132B | Sponge network | -4.546 | 0 | -0.83 | 0.01084 | 0.333 |
| 33019 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-539-5p | 10 | PRKAA1 | Sponge network | 0.594 | 0.00064 | 0.317 | 0.0031 | 0.333 |
| 33020 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | HDAC9 | Sponge network | -0.323 | 0.01372 | -0.755 | 0.00495 | 0.333 |
| 33021 | KB-1410C5.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | QKI | Sponge network | 0.959 | 0 | -0.333 | 0.04111 | 0.333 |
| 33022 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 32 | PCDH10 | Sponge network | -0.355 | 0.0077 | -1.644 | 0.0078 | 0.333 |
| 33023 | RP13-516M14.1 |
hsa-let-7b-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 36 | AR | Sponge network | 0.389 | 0.04293 | -1.554 | 0 | 0.333 |
| 33024 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 41 | SOX5 | Sponge network | -0.612 | 0.00094 | -1.091 | 4.0E-5 | 0.333 |
| 33025 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 13 | RPS6KA2 | Sponge network | 0.246 | 0.59912 | -0.57 | 0.00013 | 0.333 |
| 33026 | CKMT2-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | GJA1 | Sponge network | 0.082 | 0.55654 | -0.485 | 0.02179 | 0.333 |
| 33027 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-942-5p | 12 | CREBBP | Sponge network | 0.078 | 0.55803 | 0.126 | 0.15186 | 0.333 |
| 33028 | HCG11 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-942-5p | 12 | ZBTB20 | Sponge network | 0.385 | 0.10067 | -0.122 | 0.57569 | 0.333 |
| 33029 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 14 | TBX18 | Sponge network | -1.439 | 4.0E-5 | 0.843 | 0.02945 | 0.333 |
| 33030 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-429 | 17 | ZMYND11 | Sponge network | -0.154 | 0.53954 | -0.2 | 0.05449 | 0.333 |
| 33031 | CTD-2245F17.3 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-539-5p;hsa-miR-940 | 12 | MEF2C | Sponge network | -0.421 | 0.12556 | -0.499 | 0.01448 | 0.333 |
| 33032 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p | 11 | ZMYM2 | Sponge network | 0.542 | 0.00054 | 0.279 | 0.01273 | 0.333 |
| 33033 | ARHGEF26-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-5p | 13 | EHD3 | Sponge network | -2.46 | 0 | -0.484 | 0.01747 | 0.333 |
| 33034 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-326;hsa-miR-330-5p | 10 | TBX18 | Sponge network | 0.176 | 0.37402 | 0.843 | 0.02945 | 0.333 |
| 33035 | EIF3J-AS1 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | RCAN2 | Sponge network | 0.331 | 0.00509 | -0.33 | 0.15172 | 0.333 |
| 33036 | RP11-384O8.1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | HERC1 | Sponge network | -0.738 | 0.21233 | -0.196 | 0.08544 | 0.333 |
| 33037 | RP11-359E10.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 33 | CBX5 | Sponge network | 0.299 | 0.57722 | 0.165 | 0.24446 | 0.333 |
| 33038 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 12 | ALDH1A2 | Sponge network | 0.194 | 0.64278 | -2.411 | 0 | 0.333 |
| 33039 | AGAP11 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p | 19 | CSNK1A1 | Sponge network | -1.645 | 0 | -0.19 | 0.01687 | 0.333 |
| 33040 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-582-3p | 13 | JAZF1 | Sponge network | -2.076 | 0.00548 | -0.377 | 0.02677 | 0.333 |
| 33041 | RP11-395G23.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 15 | AKAP6 | Sponge network | 0.381 | 0.25967 | -1.586 | 3.0E-5 | 0.333 |
| 33042 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 14 | CNTNAP3 | Sponge network | 0.72 | 0.0001 | -1.465 | 0.00033 | 0.333 |
| 33043 | RP11-473I1.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p | 12 | SCN9A | Sponge network | 0.542 | 0.00054 | -0.525 | 0.10593 | 0.333 |
| 33044 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 19 | MTSS1 | Sponge network | -0.318 | 0.65847 | -0.81 | 0.00035 | 0.333 |
| 33045 | LINC00621 |
hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | MOB1B | Sponge network | 0.131 | 0.8436 | 0.197 | 0.15266 | 0.333 |
| 33046 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 15 | PICALM | Sponge network | 0.997 | 0.00066 | 0.086 | 0.23523 | 0.333 |
| 33047 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | ARHGEF12 | Sponge network | 1.316 | 0 | 0.09 | 0.35693 | 0.333 |
| 33048 | RP11-490M8.1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-539-5p;hsa-miR-96-5p | 16 | FAT3 | Sponge network | -0.214 | 0.44248 | -1.601 | 0.00051 | 0.333 |
| 33049 | TP73-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | LUZP2 | Sponge network | -0.563 | 0.02545 | -0.056 | 0.89416 | 0.333 |
| 33050 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | FGF5 | Sponge network | -1.575 | 6.0E-5 | 0.549 | 0.28659 | 0.333 |
| 33051 | LINC00672 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | LATS2 | Sponge network | -0.227 | 0.31657 | 0.159 | 0.2304 | 0.333 |
| 33052 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 33 | ATRX | Sponge network | 0.572 | 0.01722 | 0.261 | 0.04408 | 0.333 |
| 33053 | RP11-116O18.1 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ACSL6 | Sponge network | 0.05 | 0.95483 | -1.593 | 0 | 0.333 |
| 33054 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p | 23 | TIMP3 | Sponge network | 0.056 | 0.81195 | -0.546 | 0.02307 | 0.333 |
| 33055 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-155-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-26b-3p | 10 | RSPO2 | Sponge network | -0.326 | 0.28809 | -3.038 | 0 | 0.333 |
| 33056 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 17 | YAP1 | Sponge network | -0.611 | 0.09607 | 0.22 | 0.11836 | 0.333 |
| 33057 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | MED12L | Sponge network | 0.272 | 0.44692 | -0.194 | 0.55299 | 0.333 |
| 33058 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ZMYND11 | Sponge network | -2.117 | 0.00161 | -0.2 | 0.05449 | 0.333 |
| 33059 | DNM1P35 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 39 | RUNX1T1 | Sponge network | 0.051 | 0.83388 | -0.344 | 0.2374 | 0.333 |
| 33060 | NEAT1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | CREB1 | Sponge network | 0.705 | 0.00014 | 0.203 | 0.00537 | 0.333 |
| 33061 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-93-5p | 24 | GAS7 | Sponge network | -0.663 | 0.10887 | -0.555 | 0.01602 | 0.333 |
| 33062 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 38 | KIF1B | Sponge network | -0.318 | 0.65847 | -0.118 | 0.32258 | 0.333 |
| 33063 | AC141928.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-7-1-3p | 24 | PPP1R9A | Sponge network | 0.853 | 0.15352 | -0.903 | 0.04254 | 0.333 |
| 33064 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | AKAP11 | Sponge network | 1.122 | 0 | 0.196 | 0.09825 | 0.333 |
| 33065 | CTB-51J22.1 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-92a-3p;hsa-miR-940 | 13 | PTPRT | Sponge network | -0.803 | 0.23737 | -0.907 | 0.03727 | 0.333 |
| 33066 | MIR497HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-93-5p | 21 | ARNT2 | Sponge network | -1.439 | 4.0E-5 | -0.332 | 0.25851 | 0.333 |
| 33067 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p | 10 | ABCC9 | Sponge network | 0.368 | 0.20093 | -0.466 | 0.14121 | 0.333 |
| 33068 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-5p | 14 | SOX11 | Sponge network | 0.299 | 0.57722 | 2.68 | 0 | 0.333 |
| 33069 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | -2.915 | 0.00015 | -0.354 | 0.14694 | 0.333 |
| 33070 | LINC00657 |
hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | NUAK1 | Sponge network | 0.91 | 0 | 0.904 | 0 | 0.333 |
| 33071 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 22 | PCDH17 | Sponge network | -0.323 | 0.01372 | 0.731 | 2.0E-5 | 0.333 |
| 33072 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-30b-5p;hsa-miR-30c-5p | 13 | CLASP2 | Sponge network | 1.46 | 1.0E-5 | -0.038 | 0.71 | 0.333 |
| 33073 | LINC00957 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 14 | TLE4 | Sponge network | -0.421 | 0.0114 | -1.157 | 0 | 0.333 |
| 33074 | RP11-672A2.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 10 | ABI2 | Sponge network | 1.344 | 0 | 0.175 | 0.14787 | 0.333 |
| 33075 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-212-3p;hsa-miR-32-5p;hsa-miR-493-5p;hsa-miR-577 | 12 | MYCBP2 | Sponge network | 0.637 | 0.00069 | -0.027 | 0.82016 | 0.333 |
| 33076 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 29 | KCNA6 | Sponge network | -0.355 | 0.0077 | -1.531 | 0.00013 | 0.333 |
| 33077 | SOX2-OT |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 48 | DTNA | Sponge network | -1.264 | 6.0E-5 | -1.648 | 1.0E-5 | 0.333 |
| 33078 | RP11-46C24.7 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 21 | ABL2 | Sponge network | 0.392 | 0.0278 | 0.82 | 0 | 0.333 |
| 33079 | LINC00702 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 43 | PTPRT | Sponge network | -0.737 | 0.06182 | -0.907 | 0.03727 | 0.333 |
| 33080 | BCYRN1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | NRXN3 | Sponge network | 0.311 | 0.10344 | -0.893 | 0.04186 | 0.333 |
| 33081 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-744-3p | 17 | ADAMTSL3 | Sponge network | -0.785 | 0.00035 | -1.559 | 0 | 0.333 |
| 33082 | RP11-57H14.4 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-625-5p | 12 | NGFR | Sponge network | 0.083 | 0.66405 | -1.271 | 0.01058 | 0.333 |
| 33083 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-93-3p;hsa-miR-93-5p | 40 | PAFAH1B1 | Sponge network | -0.162 | 0.47687 | -0.429 | 0 | 0.333 |
| 33084 | AC005562.1 |
hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 18 | BCL9 | Sponge network | 0.488 | 0.00084 | 0.321 | 0.00267 | 0.333 |
| 33085 | MIR22HG |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-5p | 11 | KCTD8 | Sponge network | -1.047 | 0 | -3.359 | 0 | 0.333 |
| 33086 | RP11-403I13.7 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-532-3p | 14 | SYNM | Sponge network | 0.395 | 0.15451 | -2.309 | 1.0E-5 | 0.333 |
| 33087 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | PHC3 | Sponge network | 0.796 | 4.0E-5 | 0.212 | 0.0499 | 0.333 |
| 33088 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 22 | KIT | Sponge network | 0.101 | 0.60919 | -2.184 | 0 | 0.333 |
| 33089 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 16 | DOCK4 | Sponge network | 0.559 | 0.00026 | 0.502 | 0.00108 | 0.333 |
| 33090 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-96-5p | 10 | CAV1 | Sponge network | 0.331 | 0.57247 | -1.013 | 6.0E-5 | 0.333 |
| 33091 | TINCR |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 34 | TRPS1 | Sponge network | -0.664 | 0.14543 | -0.191 | 0.44109 | 0.333 |
| 33092 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-590-3p | 13 | HACE1 | Sponge network | -0.096 | 0.67958 | -0.072 | 0.55239 | 0.333 |
| 33093 | MIR143HG |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 46 | PPP1R9A | Sponge network | -0.739 | 0.0574 | -0.903 | 0.04254 | 0.333 |
| 33094 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | ADAMTSL3 | Sponge network | 1.153 | 0.00114 | -1.559 | 0 | 0.333 |
| 33095 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | PPP1R9A | Sponge network | -2.117 | 0.00161 | -0.903 | 0.04254 | 0.333 |
| 33096 | RP11-968O1.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p | 10 | CDH19 | Sponge network | -0.829 | 0.01343 | -3.434 | 0 | 0.333 |
| 33097 | RP11-532F6.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | RABEP1 | Sponge network | -0.896 | 0.00099 | -0.066 | 0.43142 | 0.333 |
| 33098 | CTA-363E19.2 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 10 | ERCC6 | Sponge network | 1.055 | 0 | 0.23 | 0.015 | 0.333 |
| 33099 | PCA3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | ACVR2A | Sponge network | -0.709 | 0.18168 | -0.409 | 0.00039 | 0.333 |
| 33100 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-455-3p;hsa-miR-96-5p | 14 | CRTC3 | Sponge network | 0.614 | 1.0E-5 | 0.164 | 0.16786 | 0.333 |
| 33101 | RP4-791M13.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-582-5p;hsa-miR-940 | 10 | BAZ2B | Sponge network | 0.419 | 0.0319 | -0.276 | 0.02833 | 0.333 |
| 33102 | CTD-2583A14.8 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 10 | CDK6 | Sponge network | 1.134 | 0 | 0.991 | 0 | 0.333 |
| 33103 | PART1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | ERG | Sponge network | -2.925 | 0 | 0.002 | 0.99011 | 0.333 |
| 33104 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 12 | HMCN1 | Sponge network | -0.355 | 0.0077 | 0.809 | 0.00544 | 0.333 |
| 33105 | KB-1460A1.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-766-3p | 27 | IL17RD | Sponge network | 0.603 | 0.00127 | -0.019 | 0.94873 | 0.333 |
| 33106 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-5p | 27 | GRIA3 | Sponge network | -0.969 | 2.0E-5 | -1.956 | 0 | 0.333 |
| 33107 | RP13-225O21.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 10 | CCDC6 | Sponge network | 1.279 | 8.0E-5 | 0.27 | 0.02555 | 0.333 |
| 33108 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-96-5p | 19 | TCF4 | Sponge network | 3.21 | 0 | 0.016 | 0.92271 | 0.333 |
| 33109 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 24 | SCN9A | Sponge network | 0.083 | 0.66405 | -0.525 | 0.10593 | 0.333 |
| 33110 | CTA-14H9.5 |
hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-7-1-3p | 11 | EYA4 | Sponge network | 0.145 | 0.53343 | 0.3 | 0.36876 | 0.333 |
| 33111 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 16 | ABL2 | Sponge network | 0.898 | 1.0E-5 | 0.82 | 0 | 0.333 |
| 33112 | LINC00094 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 26 | CADM1 | Sponge network | 0.253 | 0.09565 | -0.9 | 0.00084 | 0.333 |
| 33113 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | YAP1 | Sponge network | 0.75 | 0.00054 | 0.22 | 0.11836 | 0.333 |
| 33114 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-629-3p | 13 | PTPN14 | Sponge network | 1.162 | 5.0E-5 | 0.144 | 0.34595 | 0.333 |
| 33115 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 14 | TP53INP1 | Sponge network | -0.358 | 0.17255 | -0.496 | 0.00064 | 0.333 |
| 33116 | LIPE-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p | 12 | FLNA | Sponge network | 0.078 | 0.55803 | -0.771 | 0.01377 | 0.333 |
| 33117 | CTB-31O20.4 |
hsa-miR-10b-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-497-5p | 13 | ARHGEF12 | Sponge network | 1.619 | 0 | 0.09 | 0.35693 | 0.333 |
| 33118 | CTC-429P9.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 16 | MED12L | Sponge network | 0.178 | 0.29106 | -0.194 | 0.55299 | 0.333 |
| 33119 | RP11-540B6.6 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | DDX3X | Sponge network | 0.93 | 0 | -0.152 | 0.07686 | 0.333 |
| 33120 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 30 | PRKCE | Sponge network | -0.769 | 0.00035 | -0.403 | 0.00056 | 0.333 |
| 33121 | PART1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-889-3p;hsa-miR-93-5p;hsa-miR-942-5p | 53 | UNC80 | Sponge network | -2.925 | 0 | -1.934 | 0 | 0.333 |
| 33122 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p | 13 | DIP2C | Sponge network | -1.054 | 0.0706 | -0.436 | 0.01713 | 0.333 |
| 33123 | MALAT1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 14 | ABCA10 | Sponge network | 0.782 | 0.00016 | -0.571 | 0.03674 | 0.333 |
| 33124 | RP11-283I3.6 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-455-3p | 15 | ABL1 | Sponge network | 0.614 | 1.0E-5 | -0.215 | 0.07804 | 0.333 |
| 33125 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-942-5p | 15 | ERRFI1 | Sponge network | -0.769 | 0.00035 | -0.798 | 1.0E-5 | 0.333 |
| 33126 | CKMT2-AS1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 13 | TAF1 | Sponge network | 0.082 | 0.55654 | 0.39 | 3.0E-5 | 0.333 |
| 33127 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-942-5p | 52 | THRB | Sponge network | 0.494 | 0.00339 | -1.117 | 0.0004 | 0.333 |
| 33128 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 21 | CDC73 | Sponge network | 0.417 | 0.00445 | 0.094 | 0.20463 | 0.333 |
| 33129 | AC009948.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | ABCA10 | Sponge network | 0.532 | 0.00505 | -0.571 | 0.03674 | 0.333 |
| 33130 | AC004893.11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p | 13 | TRIM33 | Sponge network | 1.317 | 0 | 0.402 | 7.0E-5 | 0.333 |
| 33131 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-590-3p;hsa-miR-96-5p | 17 | ABI2 | Sponge network | -1.582 | 8.0E-5 | 0.175 | 0.14787 | 0.333 |
| 33132 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-493-5p;hsa-miR-654-3p;hsa-miR-874-3p | 20 | NR2C2 | Sponge network | -0.405 | 0.22013 | 0.21 | 0.03224 | 0.333 |
| 33133 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 23 | ANK2 | Sponge network | 0.381 | 0.25967 | -1.603 | 1.0E-5 | 0.333 |
| 33134 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p | 28 | CHD5 | Sponge network | 0.051 | 0.83388 | -0.922 | 0.01521 | 0.333 |
| 33135 | RP11-672A2.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p | 10 | RET | Sponge network | 1.344 | 0 | -0.833 | 0.03517 | 0.333 |
| 33136 | TP73-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-421;hsa-miR-429 | 14 | PLXNC1 | Sponge network | -0.563 | 0.02545 | 0.569 | 0.00329 | 0.333 |
| 33137 | RP11-400K9.3 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 14 | THRB | Sponge network | -0.733 | 0.19469 | -1.117 | 0.0004 | 0.333 |
| 33138 | CTA-14H9.5 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 20 | LPP | Sponge network | 0.145 | 0.53343 | -0.459 | 0.04475 | 0.333 |
| 33139 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 19 | CDC73 | Sponge network | 1.601 | 0 | 0.094 | 0.20463 | 0.333 |
| 33140 | RP11-403I13.7 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-532-5p;hsa-miR-582-3p | 16 | RYR2 | Sponge network | 0.395 | 0.15451 | -1.466 | 0.00031 | 0.333 |
| 33141 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PDE4DIP | Sponge network | 0.919 | 0 | -0.282 | 0.0257 | 0.333 |
| 33142 | RP11-38L15.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | IGF1 | Sponge network | 0.344 | 0.3127 | -0.204 | 0.5898 | 0.333 |
| 33143 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 22 | CRTC3 | Sponge network | -0.773 | 0.04073 | 0.164 | 0.16786 | 0.333 |
| 33144 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-7-1-3p | 11 | SMARCA2 | Sponge network | -0.611 | 0.09607 | -0.529 | 0.00014 | 0.333 |
| 33145 | RP11-822E23.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p | 13 | ZNF208 | Sponge network | -0.777 | 0.16667 | -0.4 | 0.22381 | 0.333 |
| 33146 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-338-3p | 14 | RBMS3 | Sponge network | 0.065 | 0.73468 | -0.519 | 0.03551 | 0.333 |
| 33147 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 16 | PDS5B | Sponge network | 1.04 | 0.00023 | 0.259 | 0.00962 | 0.333 |
| 33148 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 36 | NTRK2 | Sponge network | 0.72 | 0.0001 | -0.736 | 0.06495 | 0.333 |
| 33149 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-625-5p | 18 | RSPO2 | Sponge network | -0.465 | 0.04703 | -3.038 | 0 | 0.333 |
| 33150 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | DMXL1 | Sponge network | 0.683 | 1.0E-5 | -0.426 | 0.00056 | 0.332 |
| 33151 | RP11-1277A3.1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-505-3p;hsa-miR-629-3p;hsa-miR-93-3p | 29 | IL6ST | Sponge network | 0.453 | 0.01839 | -0.402 | 0.00676 | 0.332 |
| 33152 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 12 | TBX18 | Sponge network | 0.199 | 0.38015 | 0.843 | 0.02945 | 0.332 |
| 33153 | RP4-669P10.16 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-769-5p | 18 | WDFY3 | Sponge network | -0.301 | 0.06597 | -0.201 | 0.0967 | 0.332 |
| 33154 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-3p | 18 | EPHA3 | Sponge network | 0.274 | 0.07681 | 0.185 | 0.53588 | 0.332 |
| 33155 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-7-1-3p | 12 | DKK3 | Sponge network | 0.853 | 0.15352 | -0.052 | 0.77783 | 0.332 |
| 33156 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | NR4A3 | Sponge network | -0.376 | 0.00337 | -1.385 | 0 | 0.332 |
| 33157 | GHRLOS |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-589-5p | 14 | SOX5 | Sponge network | -1.942 | 0 | -1.091 | 4.0E-5 | 0.332 |
| 33158 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-7-1-3p | 20 | KCNA6 | Sponge network | -0.418 | 0.00068 | -1.531 | 0.00013 | 0.332 |
| 33159 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-424-5p | 12 | NR2C2 | Sponge network | 1.153 | 0.00114 | 0.21 | 0.03224 | 0.332 |
| 33160 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | TET1 | Sponge network | -0.942 | 0 | -0.135 | 0.52138 | 0.332 |
| 33161 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DACH2 | Sponge network | 0.246 | 0.59912 | -1.635 | 0.00034 | 0.332 |
| 33162 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-624-5p | 24 | FOXP1 | Sponge network | -0.358 | 0.17255 | 0.042 | 0.74368 | 0.332 |
| 33163 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 17 | FAT4 | Sponge network | 0.082 | 0.55654 | -0.354 | 0.14694 | 0.332 |
| 33164 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429 | 16 | USP6 | Sponge network | 1.106 | 0 | 0.188 | 0.3871 | 0.332 |
| 33165 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p | 20 | CLIP1 | Sponge network | 0.082 | 0.55654 | -0.215 | 0.08684 | 0.332 |
| 33166 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-671-5p;hsa-miR-98-5p | 14 | CREB3L2 | Sponge network | 1.344 | 0 | -0.525 | 2.0E-5 | 0.332 |
| 33167 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | THBS1 | Sponge network | -0.513 | 0.04703 | 0.741 | 0.00386 | 0.332 |
| 33168 | RP4-669P10.16 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-409-3p;hsa-miR-629-5p | 13 | DST | Sponge network | -0.301 | 0.06597 | -0.713 | 0.00096 | 0.332 |
| 33169 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 33 | TGFBR3 | Sponge network | -0.739 | 0.00044 | -1.173 | 1.0E-5 | 0.332 |
| 33170 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429 | 12 | MAP3K12 | Sponge network | -0.154 | 0.53954 | -0.057 | 0.59369 | 0.332 |
| 33171 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-590-3p | 21 | ZFHX3 | Sponge network | 1.089 | 0 | 0.389 | 0.01363 | 0.332 |
| 33172 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-1266-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-502-5p | 10 | ANK2 | Sponge network | -0.164 | 0.45106 | -1.603 | 1.0E-5 | 0.332 |
| 33173 | ZNF667-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | LIMCH1 | Sponge network | -0.976 | 0.00025 | -0.915 | 1.0E-5 | 0.332 |
| 33174 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | MCC | Sponge network | -4.477 | 0 | -1.005 | 0 | 0.332 |
| 33175 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-589-3p | 10 | RELN | Sponge network | 0.176 | 0.37402 | -2.403 | 0 | 0.332 |
| 33176 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-323a-3p | 14 | GNA13 | Sponge network | -0.465 | 0.04703 | 0.007 | 0.93884 | 0.332 |
| 33177 | RP11-967K21.1 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | RIMS2 | Sponge network | 0.056 | 0.81195 | -1.365 | 0.00208 | 0.332 |
| 33178 | AGAP11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p | 19 | KANK1 | Sponge network | -1.645 | 0 | -0.55 | 0.00134 | 0.332 |
| 33179 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-655-3p | 20 | LATS1 | Sponge network | 0.385 | 0.10067 | 0.109 | 0.34208 | 0.332 |
| 33180 | OSER1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-576-5p | 27 | NTRK2 | Sponge network | -0.13 | 0.48734 | -0.736 | 0.06495 | 0.332 |
| 33181 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | RNF144A | Sponge network | 1.308 | 0 | 0.594 | 0.0009 | 0.332 |
| 33182 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | DOCK4 | Sponge network | -0.323 | 0.01372 | 0.502 | 0.00108 | 0.332 |
| 33183 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 40 | NTRK2 | Sponge network | -0.899 | 6.0E-5 | -0.736 | 0.06495 | 0.332 |
| 33184 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-576-5p | 12 | TRIP11 | Sponge network | 0.637 | 0.00069 | -0.158 | 0.06384 | 0.332 |
| 33185 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-874-3p | 25 | CREB3L2 | Sponge network | -0.469 | 0.08721 | -0.525 | 2.0E-5 | 0.332 |
| 33186 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 21 | CDHR3 | Sponge network | -0.096 | 0.67958 | -0.977 | 4.0E-5 | 0.332 |
| 33187 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p | 10 | BCLAF1 | Sponge network | 1.262 | 0 | 0.211 | 0.00684 | 0.332 |
| 33188 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-93-5p | 12 | PGR | Sponge network | -0.351 | 0.11358 | -1.704 | 0 | 0.332 |
| 33189 | RP11-389C8.2 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 15 | WWTR1 | Sponge network | 0.727 | 3.0E-5 | -0.345 | 0.11997 | 0.332 |
| 33190 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 10 | TRIM3 | Sponge network | -2.095 | 0.00112 | -0.577 | 0 | 0.332 |
| 33191 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-96-5p | 12 | PIK3CA | Sponge network | 0.37 | 0.27614 | 0.195 | 0.06908 | 0.332 |
| 33192 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-582-5p | 14 | CLASP2 | Sponge network | 0.792 | 0.0049 | -0.038 | 0.71 | 0.332 |
| 33193 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-7-1-3p | 34 | ZFHX3 | Sponge network | -1.439 | 4.0E-5 | 0.389 | 0.01363 | 0.332 |
| 33194 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 51 | AR | Sponge network | 0.72 | 0.0001 | -1.554 | 0 | 0.332 |
| 33195 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 12 | ITCH | Sponge network | 0.539 | 0.03722 | 0.084 | 0.34058 | 0.332 |
| 33196 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-93-5p | 46 | PTEN | Sponge network | -1.645 | 0 | -0.187 | 0.07748 | 0.332 |
| 33197 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 39 | ERBB4 | Sponge network | -1.161 | 0.00591 | -1.975 | 2.0E-5 | 0.332 |
| 33198 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | PCDH8 | Sponge network | -2.117 | 0.00161 | -0.997 | 0.05366 | 0.332 |
| 33199 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-501-5p | 12 | TXNIP | Sponge network | -0.738 | 0.21233 | -1.277 | 0 | 0.332 |
| 33200 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 24 | EBF3 | Sponge network | 0.537 | 0.00139 | -0.058 | 0.81437 | 0.332 |
| 33201 | LINC00473 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | HERC1 | Sponge network | -1.203 | 0.03881 | -0.196 | 0.08544 | 0.332 |
| 33202 | HCG11 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | SPG20 | Sponge network | 0.385 | 0.10067 | -1.16 | 0 | 0.332 |
| 33203 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-93-5p | 12 | TRIM3 | Sponge network | -0.162 | 0.47687 | -0.577 | 0 | 0.332 |
| 33204 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | EYA4 | Sponge network | 1.308 | 0 | 0.3 | 0.36876 | 0.332 |
| 33205 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 35 | SPRY3 | Sponge network | 0.782 | 0.00016 | 0.016 | 0.91286 | 0.332 |
| 33206 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | PPP1R9A | Sponge network | -0.246 | 0.35034 | -0.903 | 0.04254 | 0.332 |
| 33207 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 27 | ADAMTSL3 | Sponge network | 0.219 | 0.1516 | -1.559 | 0 | 0.332 |
| 33208 | PSMG3-AS1 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p | 10 | FHL1 | Sponge network | -0.125 | 0.49414 | -2.687 | 0 | 0.332 |
| 33209 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421 | 12 | SMARCA2 | Sponge network | -2.62 | 0 | -0.529 | 0.00014 | 0.332 |
| 33210 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-942-5p | 21 | GPC6 | Sponge network | -0.08 | 0.90092 | -0.054 | 0.81465 | 0.332 |
| 33211 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-7-5p | 17 | ESR1 | Sponge network | 0.299 | 0.57722 | -1.035 | 3.0E-5 | 0.332 |
| 33212 | LINC00641 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | CDYL | Sponge network | -0.222 | 0.32445 | 0.12 | 0.12425 | 0.332 |
| 33213 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-25-3p | 10 | MYCBP2 | Sponge network | 0.622 | 0.00015 | -0.027 | 0.82016 | 0.332 |
| 33214 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-486-5p;hsa-miR-495-3p | 12 | MYO9A | Sponge network | 0.924 | 0 | -0.147 | 0.32373 | 0.332 |
| 33215 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | MYO9A | Sponge network | 0.853 | 0.15352 | -0.147 | 0.32373 | 0.332 |
| 33216 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | AKAP11 | Sponge network | 0.832 | 0 | 0.196 | 0.09825 | 0.332 |
| 33217 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-455-5p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | JAZF1 | Sponge network | 0.056 | 0.8494 | -0.377 | 0.02677 | 0.332 |
| 33218 | MALAT1 |
hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | HERC1 | Sponge network | 0.782 | 0.00016 | -0.196 | 0.08544 | 0.332 |
| 33219 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 45 | THRB | Sponge network | -0.053 | 0.80685 | -1.117 | 0.0004 | 0.332 |
| 33220 | USP3-AS1 |
hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-653-5p | 25 | UBR3 | Sponge network | -0.162 | 0.47687 | -0.021 | 0.82774 | 0.332 |
| 33221 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-3p | 14 | SOX11 | Sponge network | 2.163 | 0 | 2.68 | 0 | 0.332 |
| 33222 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-5p | 26 | RICTOR | Sponge network | 0.389 | 0.04293 | 0.388 | 0.00188 | 0.332 |
| 33223 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-421 | 14 | RET | Sponge network | -0.351 | 0.11358 | -0.833 | 0.03517 | 0.332 |
| 33224 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 19 | PKNOX1 | Sponge network | -0.739 | 0.0574 | 0.085 | 0.28917 | 0.332 |
| 33225 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 23 | NUAK1 | Sponge network | -0.611 | 0.09607 | 0.904 | 0 | 0.332 |
| 33226 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | RNF144A | Sponge network | 0.538 | 0.00076 | 0.594 | 0.0009 | 0.332 |
| 33227 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 27 | GRIN2A | Sponge network | 0.532 | 0.00505 | -2.161 | 0 | 0.332 |
| 33228 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 13 | ERCC4 | Sponge network | -0.465 | 0.04703 | 0.126 | 0.15266 | 0.332 |
| 33229 | DNM3OS |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SYT4 | Sponge network | 0.572 | 0.01722 | -3.207 | 0 | 0.332 |
| 33230 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-3615;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | DMTF1 | Sponge network | 0.603 | 0.00127 | 0.248 | 0.02726 | 0.332 |
| 33231 | GS1-124K5.11 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-874-3p;hsa-miR-98-5p | 10 | HSPB7 | Sponge network | 0.494 | 0.00339 | -2.268 | 1.0E-5 | 0.332 |
| 33232 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-532-5p;hsa-miR-935;hsa-miR-96-5p | 25 | FBXO32 | Sponge network | 0.395 | 0.15451 | -0.495 | 0.07561 | 0.332 |
| 33233 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p | 13 | CREB1 | Sponge network | 0.912 | 0.00014 | 0.203 | 0.00537 | 0.332 |
| 33234 | AF131215.2 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-7-1-3p | 16 | LIMCH1 | Sponge network | -0.176 | 0.59692 | -0.915 | 1.0E-5 | 0.332 |
| 33235 | LINC00654 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p | 12 | AMPH | Sponge network | 0.374 | 0.20054 | -0.916 | 5.0E-5 | 0.332 |
| 33236 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-505-5p;hsa-miR-590-3p | 12 | RBM5 | Sponge network | -0.096 | 0.67958 | -0.205 | 0.0129 | 0.332 |
| 33237 | BZRAP1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-671-5p | 13 | CDH23 | Sponge network | -0.465 | 0.04703 | -0.974 | 0.00034 | 0.332 |
| 33238 | AC159540.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | TBL1XR1 | Sponge network | 1.634 | 0 | 0.578 | 0 | 0.332 |
| 33239 | CTC-444N24.11 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZMYND11 | Sponge network | 1.153 | 0 | -0.2 | 0.05449 | 0.332 |
| 33240 | LINC00174 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-582-5p | 16 | DDX6 | Sponge network | 1.253 | 0 | 0.091 | 0.36428 | 0.332 |
| 33241 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p | 15 | LRRC4 | Sponge network | -0.615 | 0.00015 | -1.774 | 0 | 0.332 |
| 33242 | RP4-791M13.3 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 11 | PCDH9 | Sponge network | 0.419 | 0.0319 | -1.823 | 4.0E-5 | 0.332 |
| 33243 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p | 52 | BMPR2 | Sponge network | 0.61 | 0.01593 | 0.206 | 0.0635 | 0.332 |
| 33244 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-3913-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-5p | 32 | LPP | Sponge network | -1.054 | 0.0706 | -0.459 | 0.04475 | 0.332 |
| 33245 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 41 | TACC1 | Sponge network | 0.311 | 0.10344 | -0.362 | 0.05406 | 0.332 |
| 33246 | LINC00865 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p | 16 | ITGB3 | Sponge network | -1.249 | 7.0E-5 | -0.011 | 0.95751 | 0.332 |
| 33247 | PGM5-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-7-1-3p | 15 | TET1 | Sponge network | -4.546 | 0 | -0.135 | 0.52138 | 0.332 |
| 33248 | TPTEP1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | EYA4 | Sponge network | -1.216 | 0 | 0.3 | 0.36876 | 0.332 |
| 33249 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 49 | TMEM132B | Sponge network | -1.697 | 0.00889 | -0.83 | 0.01084 | 0.332 |
| 33250 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-485-3p | 18 | CACNA2D1 | Sponge network | -0.465 | 0.04703 | -0.846 | 0.00675 | 0.332 |
| 33251 | RP11-822E23.8 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-484 | 16 | ABL1 | Sponge network | -0.777 | 0.16667 | -0.215 | 0.07804 | 0.332 |
| 33252 | H1FX-AS1 |
hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-532-3p | 10 | SYNM | Sponge network | -0.206 | 0.12942 | -2.309 | 1.0E-5 | 0.332 |
| 33253 | CTC-297N7.9 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 14 | GRIK3 | Sponge network | -2.069 | 0 | -3.208 | 0 | 0.332 |
| 33254 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-335-5p;hsa-miR-7-5p | 11 | KCNA6 | Sponge network | -0.421 | 0.12556 | -1.531 | 0.00013 | 0.332 |
| 33255 | RP11-532F6.3 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | ZFP36L1 | Sponge network | -0.896 | 0.00099 | -0.505 | 8.0E-5 | 0.332 |
| 33256 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-93-5p | 13 | ACSL6 | Sponge network | -0.351 | 0.11358 | -1.593 | 0 | 0.332 |
| 33257 | LPP-AS2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-625-5p | 22 | PTPN14 | Sponge network | -0.18 | 0.20343 | 0.144 | 0.34595 | 0.332 |
| 33258 | FAM66C |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | EFNA5 | Sponge network | -0.901 | 0.00019 | -0.421 | 0.17868 | 0.332 |
| 33259 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 32 | DDX6 | Sponge network | 0.659 | 3.0E-5 | 0.091 | 0.36428 | 0.332 |
| 33260 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | FREM2 | Sponge network | -1.111 | 0.0003 | -0.437 | 0.46353 | 0.332 |
| 33261 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 22 | YAP1 | Sponge network | 0.083 | 0.66405 | 0.22 | 0.11836 | 0.332 |
| 33262 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 46 | SSBP2 | Sponge network | -0.83 | 0 | -1.58 | 0 | 0.332 |
| 33263 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-589-5p | 15 | RPS6KA2 | Sponge network | -0.738 | 0.21233 | -0.57 | 0.00013 | 0.332 |
| 33264 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | NAV3 | Sponge network | 0.998 | 0 | 0.212 | 0.47729 | 0.332 |
| 33265 | FAM66C |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CDH11 | Sponge network | -0.901 | 0.00019 | 1.427 | 0 | 0.332 |
| 33266 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 16 | PNRC1 | Sponge network | 0.194 | 0.64278 | -0.646 | 0 | 0.332 |
| 33267 | RP11-38L15.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 11 | ESR1 | Sponge network | 0.344 | 0.3127 | -1.035 | 3.0E-5 | 0.332 |
| 33268 | WDFY3-AS2 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 18 | IRS1 | Sponge network | -0.942 | 0 | -0.026 | 0.88855 | 0.332 |
| 33269 | AF131215.9 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-940 | 21 | PTPRT | Sponge network | -0.322 | 0.25945 | -0.907 | 0.03727 | 0.332 |
| 33270 | SNHG10 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 10 | CBFB | Sponge network | 0.931 | 0 | 1.061 | 0 | 0.332 |
| 33271 | HOXD-AS2 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-3607-3p | 11 | CDH19 | Sponge network | -0.208 | 0.65592 | -3.434 | 0 | 0.332 |
| 33272 | CTD-2002H8.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TCF4 | Sponge network | 0.931 | 1.0E-5 | 0.016 | 0.92271 | 0.332 |
| 33273 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | LATS1 | Sponge network | 0.569 | 0.00067 | 0.109 | 0.34208 | 0.332 |
| 33274 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-942-5p;hsa-miR-98-5p | 17 | ARHGAP28 | Sponge network | -0.773 | 0.04073 | -0.911 | 0.00013 | 0.332 |
| 33275 | DNM3OS |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-3p | 21 | CTIF | Sponge network | 0.572 | 0.01722 | -0.389 | 0.00549 | 0.332 |
| 33276 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 19 | RBL2 | Sponge network | -0.49 | 0.04946 | -0.282 | 0.00254 | 0.332 |
| 33277 | FZD10-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p | 27 | ASTN1 | Sponge network | -0.727 | 0.04726 | -2.389 | 2.0E-5 | 0.332 |
| 33278 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-93-3p | 10 | FYN | Sponge network | -0.693 | 0.05629 | -0.131 | 0.37051 | 0.332 |
| 33279 | RP11-25K19.1 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | ID4 | Sponge network | -1.057 | 0.00029 | -1.644 | 0 | 0.332 |
| 33280 | RP11-35G9.3 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-940 | 20 | SNX29 | Sponge network | 0.657 | 4.0E-5 | -0.34 | 0.01371 | 0.332 |
| 33281 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 45 | CCND2 | Sponge network | -0.612 | 0.00094 | -0.267 | 0.3112 | 0.332 |
| 33282 | RP11-35G9.3 |
hsa-let-7b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | INPP4A | Sponge network | 0.657 | 4.0E-5 | 0.07 | 0.53563 | 0.332 |
| 33283 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 30 | TGFBR3 | Sponge network | 0.61 | 0.01593 | -1.173 | 1.0E-5 | 0.332 |
| 33284 | RP11-834C11.4 |
hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-940;hsa-miR-96-5p | 11 | LIFR | Sponge network | -0.883 | 0.01839 | -1.794 | 0 | 0.332 |
| 33285 | RP11-2B6.2 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ARHGEF12 | Sponge network | 0.539 | 0.03722 | 0.09 | 0.35693 | 0.332 |
| 33286 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 28 | PRKAB2 | Sponge network | -0.842 | 0.01361 | -0.367 | 0.01371 | 0.332 |
| 33287 | NDUFA6-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | -0.355 | 0.0077 | -0.001 | 0.99669 | 0.332 |
| 33288 | DNAJC27-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-660-5p;hsa-miR-93-3p | 57 | RUNX1T1 | Sponge network | -0.969 | 2.0E-5 | -0.344 | 0.2374 | 0.332 |
| 33289 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | EPHA5 | Sponge network | 0.559 | 0.00026 | -0.957 | 0.01733 | 0.332 |
| 33290 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PTPN11 | Sponge network | 1.11 | 0 | 0.431 | 2.0E-5 | 0.332 |
| 33291 | JAZF1-AS1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | RPS6KA6 | Sponge network | -0.769 | 0.00035 | -2.989 | 0 | 0.332 |
| 33292 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-942-5p | 23 | GPC6 | Sponge network | -1.161 | 0.00591 | -0.054 | 0.81465 | 0.332 |
| 33293 | RP4-639F20.1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-7-5p | 13 | RNF144A | Sponge network | -0.517 | 0.02935 | 0.594 | 0.0009 | 0.332 |
| 33294 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-7-1-3p | 29 | PRKAA2 | Sponge network | -0.285 | 0.2172 | -2.462 | 0 | 0.332 |
| 33295 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-582-3p | 13 | CBL | Sponge network | 0.909 | 0 | 0.291 | 0.01267 | 0.332 |
| 33296 | ACTA2-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p | 17 | IGF2 | Sponge network | 0.194 | 0.64278 | 0.848 | 0.03842 | 0.332 |
| 33297 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 24 | IGF1 | Sponge network | 0.174 | 0.30034 | -0.204 | 0.5898 | 0.332 |
| 33298 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 16 | ACTB | Sponge network | -0.576 | 0.10121 | -0.247 | 0.01736 | 0.332 |
| 33299 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | AHNAK | Sponge network | -0.227 | 0.31657 | -1.207 | 0 | 0.332 |
| 33300 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-5p | 18 | RSPO2 | Sponge network | 0.176 | 0.37402 | -3.038 | 0 | 0.332 |
| 33301 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-577 | 12 | MYCBP2 | Sponge network | 0.267 | 0.2937 | -0.027 | 0.82016 | 0.332 |
| 33302 | LINC00672 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p | 13 | DNAJB4 | Sponge network | -0.227 | 0.31657 | -0.7 | 1.0E-5 | 0.332 |
| 33303 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-337-3p;hsa-miR-590-3p | 11 | PBRM1 | Sponge network | -0.096 | 0.67958 | -0.029 | 0.78601 | 0.332 |
| 33304 | LINC00641 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | TLL1 | Sponge network | -0.222 | 0.32445 | -1.195 | 3.0E-5 | 0.332 |
| 33305 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | ADAMTSL3 | Sponge network | 0.381 | 0.25967 | -1.559 | 0 | 0.332 |
| 33306 | TRAF3IP2-AS1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | RBBP6 | Sponge network | -0.323 | 0.01372 | 0.163 | 0.05101 | 0.332 |
| 33307 | LINC00476 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | ZDBF2 | Sponge network | -0.615 | 0.00015 | -0.998 | 0.00025 | 0.332 |
| 33308 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 51 | SMAD4 | Sponge network | -0.709 | 0.18168 | -0.313 | 0.00413 | 0.332 |
| 33309 | RP11-158K1.3 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | INPP4A | Sponge network | 0.349 | 0.01695 | 0.07 | 0.53563 | 0.332 |
| 33310 | MIR181A2HG |
hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 15 | RICTOR | Sponge network | 1.822 | 1.0E-5 | 0.388 | 0.00188 | 0.332 |
| 33311 | PART1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 37 | RASSF8 | Sponge network | -2.925 | 0 | -0.122 | 0.65591 | 0.332 |
| 33312 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-629-3p | 21 | GPC6 | Sponge network | -1.349 | 0.00017 | -0.054 | 0.81465 | 0.332 |
| 33313 | AC003090.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-576-5p | 11 | EHD3 | Sponge network | -2.117 | 0.00161 | -0.484 | 0.01747 | 0.332 |
| 33314 | LINC00957 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-362-5p | 16 | KCNJ5 | Sponge network | -0.421 | 0.0114 | -0.281 | 0.3339 | 0.332 |
| 33315 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 20 | USP12 | Sponge network | 0.619 | 0.00157 | -0.102 | 0.40768 | 0.332 |
| 33316 | AC009948.5 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p | 12 | ITGA5 | Sponge network | 0.532 | 0.00505 | 0.452 | 0.03524 | 0.332 |
| 33317 | DNAJC27-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | WWTR1 | Sponge network | -0.969 | 2.0E-5 | -0.345 | 0.11997 | 0.332 |
| 33318 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | MXI1 | Sponge network | -0.896 | 0.00099 | -0.987 | 0 | 0.332 |
| 33319 | GABPB1-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | UBR3 | Sponge network | 1.11 | 0 | -0.021 | 0.82774 | 0.332 |
| 33320 | SOX2-OT |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 17 | SORCS1 | Sponge network | -1.264 | 6.0E-5 | -3.492 | 0 | 0.332 |
| 33321 | RP11-758N13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p | 15 | EPHA5 | Sponge network | 2.939 | 3.0E-5 | -0.957 | 0.01733 | 0.332 |
| 33322 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 12 | THBS1 | Sponge network | -2.039 | 0.03447 | 0.741 | 0.00386 | 0.332 |
| 33323 | LINC00843 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-505-5p | 25 | LPP | Sponge network | 1.106 | 0 | -0.459 | 0.04475 | 0.332 |
| 33324 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-577 | 24 | TMEM132B | Sponge network | -0.473 | 0.07602 | -0.83 | 0.01084 | 0.332 |
| 33325 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | TCF12 | Sponge network | 0.101 | 0.60919 | 0.174 | 0.13271 | 0.332 |
| 33326 | RP11-344B5.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-664a-3p | 10 | AKAP6 | Sponge network | -0.055 | 0.88482 | -1.586 | 3.0E-5 | 0.332 |
| 33327 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-590-3p | 11 | MAF | Sponge network | -1.375 | 0.08642 | -1.257 | 0 | 0.332 |
| 33328 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 13 | MYO9A | Sponge network | -0.932 | 0.00329 | -0.147 | 0.32373 | 0.332 |
| 33329 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | DICER1 | Sponge network | -0.487 | 0.02157 | 0.042 | 0.674 | 0.332 |
| 33330 | RP11-37B2.1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-495-3p | 10 | STAG2 | Sponge network | 1.03 | 0 | 0.252 | 0.00438 | 0.332 |
| 33331 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p | 28 | TGFBR3 | Sponge network | -0.246 | 0.35034 | -1.173 | 1.0E-5 | 0.332 |
| 33332 | ZNF32-AS1 |
hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | MLLT10 | Sponge network | 2.413 | 1.0E-5 | 0.105 | 0.33292 | 0.332 |
| 33333 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | ETV1 | Sponge network | -0.942 | 0 | -0.096 | 0.67674 | 0.332 |
| 33334 | LINC01006 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-154-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-582-5p | 12 | GRIN2A | Sponge network | -0.702 | 0.04814 | -2.161 | 0 | 0.332 |
| 33335 | HOXB-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-625-5p | 14 | CRTC1 | Sponge network | -0.154 | 0.53954 | -0.116 | 0.28412 | 0.332 |
| 33336 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-378a-3p;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-769-5p;hsa-miR-93-5p | 41 | NTRK3 | Sponge network | 0.381 | 0.25967 | -1.77 | 3.0E-5 | 0.332 |
| 33337 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | 0.299 | 0.57722 | -1.751 | 1.0E-5 | 0.332 |
| 33338 | SNX29P2 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 38 | GRIK3 | Sponge network | 0.4 | 0.14611 | -3.208 | 0 | 0.332 |
| 33339 | LINC00926 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-940 | 11 | IGF1 | Sponge network | 0.756 | 0.00739 | -0.204 | 0.5898 | 0.332 |
| 33340 | MRVI1-AS1 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p | 11 | PTCH1 | Sponge network | -1.092 | 4.0E-5 | -0.1 | 0.6179 | 0.332 |
| 33341 | TP73-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-3p | 18 | MAFB | Sponge network | -0.563 | 0.02545 | -0.023 | 0.89865 | 0.332 |
| 33342 | RP11-282O18.3 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 12 | ZFX | Sponge network | 0.757 | 0 | 0.09 | 0.42563 | 0.332 |
| 33343 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-582-5p | 16 | ACVR1 | Sponge network | 0.889 | 9.0E-5 | 0.279 | 0.00234 | 0.332 |
| 33344 | HCG18 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | TET1 | Sponge network | 1.063 | 0 | -0.135 | 0.52138 | 0.332 |
| 33345 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429 | 31 | NOTCH2 | Sponge network | 0.95 | 0.15943 | 0.138 | 0.35163 | 0.332 |
| 33346 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | SASH1 | Sponge network | 0.381 | 0.25967 | -0.502 | 0.00175 | 0.332 |
| 33347 | RP11-967K21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-582-5p | 14 | BRD3 | Sponge network | 0.056 | 0.81195 | 0.37 | 0.00013 | 0.332 |
| 33348 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 57 | WDFY3 | Sponge network | -1.216 | 0 | -0.201 | 0.0967 | 0.332 |
| 33349 | TP73-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 13 | HOMER2 | Sponge network | -0.563 | 0.02545 | -2.698 | 0 | 0.332 |
| 33350 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-664a-3p | 22 | FZD3 | Sponge network | 1.147 | 0 | 0.104 | 0.60316 | 0.332 |
| 33351 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p | 26 | GRIA3 | Sponge network | -0.487 | 0.02157 | -1.956 | 0 | 0.332 |
| 33352 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 16 | LHX6 | Sponge network | -1.216 | 0 | -0.087 | 0.69193 | 0.332 |
| 33353 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-3613-5p | 11 | AFF3 | Sponge network | 1.153 | 0.00114 | -2.373 | 0 | 0.332 |
| 33354 | RP11-212P7.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | MED12L | Sponge network | 0.219 | 0.1516 | -0.194 | 0.55299 | 0.332 |
| 33355 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 34 | MAF | Sponge network | -0.615 | 0.00015 | -1.257 | 0 | 0.332 |
| 33356 | LINC00403 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-452-5p | 13 | RBMS3 | Sponge network | -3.586 | 0.00032 | -0.519 | 0.03551 | 0.332 |
| 33357 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 34 | DIP2C | Sponge network | 0.537 | 0.00139 | -0.436 | 0.01713 | 0.332 |
| 33358 | CTC-429P9.5 |
hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-29b-1-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-3p | 18 | CUX1 | Sponge network | 0.178 | 0.29106 | -0.039 | 0.6705 | 0.332 |
| 33359 | RP11-57H14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | NCOA1 | Sponge network | 0.083 | 0.66405 | -0.432 | 0 | 0.332 |
| 33360 | RP11-705C15.3 |
hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | SYNE1 | Sponge network | 0.796 | 4.0E-5 | -0.738 | 0.00316 | 0.332 |
| 33361 | LINC00621 |
hsa-miR-10a-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | CADM1 | Sponge network | 0.131 | 0.8436 | -0.9 | 0.00084 | 0.332 |
| 33362 | AGAP11 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-455-5p | 10 | PRDM5 | Sponge network | -1.645 | 0 | -1.09 | 0.00014 | 0.332 |
| 33363 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-98-5p | 16 | AKAP6 | Sponge network | -0.773 | 0.04073 | -1.586 | 3.0E-5 | 0.332 |
| 33364 | RP11-244H3.1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TSC1 | Sponge network | 0.659 | 3.0E-5 | 0.103 | 0.26071 | 0.332 |
| 33365 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p | 15 | NUAK1 | Sponge network | 0.056 | 0.81195 | 0.904 | 0 | 0.332 |
| 33366 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-629-3p | 11 | SPTBN4 | Sponge network | -0.739 | 0.00044 | -0.786 | 0.01281 | 0.332 |
| 33367 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-550a-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-940 | 32 | PTPRT | Sponge network | -0.793 | 7.0E-5 | -0.907 | 0.03727 | 0.332 |
| 33368 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-874-3p | 20 | NR2C2 | Sponge network | 0.249 | 0.35902 | 0.21 | 0.03224 | 0.332 |
| 33369 | RP11-736K20.5 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-625-3p | 21 | ABL2 | Sponge network | -0.358 | 0.17255 | 0.82 | 0 | 0.332 |
| 33370 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-93-5p | 16 | CHD5 | Sponge network | -0.726 | 0.00132 | -0.922 | 0.01521 | 0.332 |
| 33371 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-625-5p | 23 | ABI2 | Sponge network | -0.469 | 0.08721 | 0.175 | 0.14787 | 0.332 |
| 33372 | AC034220.3 |
hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | APC | Sponge network | 0.309 | 0.07304 | -0.255 | 0.04051 | 0.332 |
| 33373 | AGAP11 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-3p;hsa-miR-374b-5p | 10 | USP25 | Sponge network | -1.645 | 0 | -0.242 | 0.01397 | 0.332 |
| 33374 | RP11-554A11.4 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p | 12 | HIP1 | Sponge network | -0.583 | 0.14589 | 0.774 | 0 | 0.332 |
| 33375 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 35 | TMEM132B | Sponge network | -0.08 | 0.90092 | -0.83 | 0.01084 | 0.332 |
| 33376 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 51 | CBX5 | Sponge network | 0.997 | 0.00066 | 0.165 | 0.24446 | 0.332 |
| 33377 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 19 | MCC | Sponge network | 0.853 | 0.15352 | -1.005 | 0 | 0.332 |
| 33378 | RP11-760H22.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p | 11 | ZFX | Sponge network | 0.001 | 0.99753 | 0.09 | 0.42563 | 0.332 |
| 33379 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p | 22 | ARNT2 | Sponge network | -0.49 | 0.04946 | -0.332 | 0.25851 | 0.332 |
| 33380 | RP11-758N13.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p | 24 | DST | Sponge network | 2.939 | 3.0E-5 | -0.713 | 0.00096 | 0.331 |
| 33381 | RP11-166D19.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 19 | CELF4 | Sponge network | -0.576 | 0.10121 | -1.135 | 0.00344 | 0.331 |
| 33382 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 36 | CBL | Sponge network | 0.572 | 0.01722 | 0.291 | 0.01267 | 0.331 |
| 33383 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-378a-5p | 15 | NUAK1 | Sponge network | -0.777 | 0.16667 | 0.904 | 0 | 0.331 |
| 33384 | LL22NC03-N64E9.1 | hsa-miR-101-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-486-5p | 10 | CBFB | Sponge network | 7.139 | 0 | 1.061 | 0 | 0.331 |
| 33385 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 23 | CUL5 | Sponge network | 0.101 | 0.60919 | 0.026 | 0.77301 | 0.331 |
| 33386 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 28 | THBS1 | Sponge network | -0.901 | 0.00019 | 0.741 | 0.00386 | 0.331 |
| 33387 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 24 | CDHR3 | Sponge network | -2.925 | 0 | -0.977 | 4.0E-5 | 0.331 |
| 33388 | LINC00173 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-7-1-3p | 14 | PCDH9 | Sponge network | 0.056 | 0.8494 | -1.823 | 4.0E-5 | 0.331 |
| 33389 | LINC00094 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p;hsa-miR-92a-3p | 13 | SPTBN4 | Sponge network | 0.253 | 0.09565 | -0.786 | 0.01281 | 0.331 |
| 33390 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 21 | MYBL1 | Sponge network | -0.612 | 0.00094 | 0.494 | 0.02152 | 0.331 |
| 33391 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-497-5p | 18 | SOX11 | Sponge network | 0.946 | 0 | 2.68 | 0 | 0.331 |
| 33392 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-629-5p;hsa-miR-7-5p | 13 | CRTC1 | Sponge network | -0.517 | 0.02935 | -0.116 | 0.28412 | 0.331 |
| 33393 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 31 | SOX5 | Sponge network | -0.176 | 0.59692 | -1.091 | 4.0E-5 | 0.331 |
| 33394 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | PDGFRA | Sponge network | 0.225 | 0.30644 | -0.372 | 0.0037 | 0.331 |
| 33395 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-940 | 38 | CADM2 | Sponge network | 0.174 | 0.30034 | -3.782 | 0 | 0.331 |
| 33396 | MEG3 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p | 11 | ADH1B | Sponge network | 0.249 | 0.35902 | -3.716 | 0 | 0.331 |
| 33397 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 22 | USP6 | Sponge network | 0.532 | 0.00505 | 0.188 | 0.3871 | 0.331 |
| 33398 | RP11-20I23.11 |
hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | ABL2 | Sponge network | 0.875 | 0.00019 | 0.82 | 0 | 0.331 |
| 33399 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 10 | ABCB5 | Sponge network | -1.057 | 0.00029 | -0.956 | 0.04077 | 0.331 |
| 33400 | RP11-571M6.17 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | ARHGEF12 | Sponge network | 1.013 | 0 | 0.09 | 0.35693 | 0.331 |
| 33401 | AC016747.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 10 | CADM3 | Sponge network | 0.199 | 0.38015 | -3.663 | 0 | 0.331 |
| 33402 | RP11-66B24.2 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-375 | 12 | RASSF8 | Sponge network | 0.57 | 0.1866 | -0.122 | 0.65591 | 0.331 |
| 33403 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 65 | AFF4 | Sponge network | 0.997 | 0.00066 | -0.266 | 0.01565 | 0.331 |
| 33404 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-93-5p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | 0.082 | 0.55397 | -0.085 | 0.42389 | 0.331 |
| 33405 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-96-5p | 30 | FBXO32 | Sponge network | 1.302 | 7.0E-5 | -0.495 | 0.07561 | 0.331 |
| 33406 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-425-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | SOX5 | Sponge network | 0.41 | 0.02214 | -1.091 | 4.0E-5 | 0.331 |
| 33407 | LPP-AS2 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-96-5p | 15 | TLL1 | Sponge network | -0.18 | 0.20343 | -1.195 | 3.0E-5 | 0.331 |
| 33408 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-502-5p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-93-5p | 30 | ANK2 | Sponge network | 0.253 | 0.09565 | -1.603 | 1.0E-5 | 0.331 |
| 33409 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p | 17 | BHLHE41 | Sponge network | -1.092 | 4.0E-5 | 0.353 | 0.12753 | 0.331 |
| 33410 | RP11-359B12.2 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 37 | ARHGEF12 | Sponge network | 0.064 | 0.72934 | 0.09 | 0.35693 | 0.331 |
| 33411 | LINC00094 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 42 | TRPS1 | Sponge network | 0.253 | 0.09565 | -0.191 | 0.44109 | 0.331 |
| 33412 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 33 | ARID5B | Sponge network | 0.311 | 0.10344 | 0.342 | 0.01676 | 0.331 |
| 33413 | HAR1A |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-671-5p;hsa-miR-93-3p | 14 | BNC2 | Sponge network | -0.672 | 0.19447 | -0.491 | 0.09988 | 0.331 |
| 33414 | LIPE-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | NF1 | Sponge network | 0.078 | 0.55803 | 0.51 | 0 | 0.331 |
| 33415 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p | 11 | CRTC3 | Sponge network | 0.592 | 0.00014 | 0.164 | 0.16786 | 0.331 |
| 33416 | MIR7-3HG |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p | 11 | CADM2 | Sponge network | -2.688 | 0.00397 | -3.782 | 0 | 0.331 |
| 33417 | CALML3-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p | 11 | HMCN1 | Sponge network | 0.95 | 0.15943 | 0.809 | 0.00544 | 0.331 |
| 33418 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 14 | CREB3L2 | Sponge network | -0.295 | 0.02604 | -0.525 | 2.0E-5 | 0.331 |
| 33419 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 26 | HECA | Sponge network | -0.615 | 0.00015 | -0.217 | 0.03463 | 0.331 |
| 33420 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-935 | 19 | EBF1 | Sponge network | 0.637 | 0.00069 | -0.384 | 0.08856 | 0.331 |
| 33421 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 28 | PBX1 | Sponge network | 0.72 | 0.0001 | -1.206 | 0 | 0.331 |
| 33422 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PTPN13 | Sponge network | 0.082 | 0.55654 | -1.208 | 0 | 0.331 |
| 33423 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 53 | SMAD4 | Sponge network | -0.096 | 0.67958 | -0.313 | 0.00413 | 0.331 |
| 33424 | HNRNPU-AS1 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ITGB1 | Sponge network | 1.601 | 0 | 0.524 | 0.00017 | 0.331 |
| 33425 | FAM83H-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 10 | TBL1XR1 | Sponge network | 3.083 | 0 | 0.578 | 0 | 0.331 |
| 33426 | LINC01001 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-486-5p | 10 | MYO9A | Sponge network | 1.055 | 0 | -0.147 | 0.32373 | 0.331 |
| 33427 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | EBF1 | Sponge network | -2.915 | 0.00015 | -0.384 | 0.08856 | 0.331 |
| 33428 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-493-5p;hsa-miR-93-5p | 10 | ANK2 | Sponge network | -0.31 | 0.33871 | -1.603 | 1.0E-5 | 0.331 |
| 33429 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PRKAA2 | Sponge network | 0.272 | 0.44692 | -2.462 | 0 | 0.331 |
| 33430 | AC025171.1 |
hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | NUAK1 | Sponge network | 0.998 | 0 | 0.904 | 0 | 0.331 |
| 33431 | PGM5-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-484 | 10 | ITGA5 | Sponge network | -4.546 | 0 | 0.452 | 0.03524 | 0.331 |
| 33432 | RP11-356J5.12 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p | 17 | CDH23 | Sponge network | -0.899 | 6.0E-5 | -0.974 | 0.00034 | 0.331 |
| 33433 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-5p | 33 | MOB1B | Sponge network | 0.253 | 0.09565 | 0.197 | 0.15266 | 0.331 |
| 33434 | LINC00476 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p | 12 | UVRAG | Sponge network | -0.615 | 0.00015 | -0.017 | 0.83865 | 0.331 |
| 33435 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-450b-5p | 19 | KAT6B | Sponge network | 0.331 | 0.57247 | -0.478 | 0.00018 | 0.331 |
| 33436 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 30 | CBX5 | Sponge network | 1.04 | 0.00023 | 0.165 | 0.24446 | 0.331 |
| 33437 | CTD-2528L19.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 13 | ERCC4 | Sponge network | 0.953 | 0 | 0.126 | 0.15266 | 0.331 |
| 33438 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-429;hsa-miR-577;hsa-miR-93-5p | 21 | NIN | Sponge network | -2.69 | 0.0001 | -0.253 | 0.13794 | 0.331 |
| 33439 | AC006547.13 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-654-3p | 19 | RICTOR | Sponge network | 1.159 | 0 | 0.388 | 0.00188 | 0.331 |
| 33440 | CTC-429P9.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-362-5p;hsa-miR-877-5p;hsa-miR-940 | 11 | PTPRT | Sponge network | 0.043 | 0.81628 | -0.907 | 0.03727 | 0.331 |
| 33441 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 35 | ADARB1 | Sponge network | 0.392 | 0.0278 | -0.335 | 0.06631 | 0.331 |
| 33442 | GS1-124K5.11 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.494 | 0.00339 | -0.57 | 0 | 0.331 |
| 33443 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | RECK | Sponge network | -0.969 | 2.0E-5 | -1.043 | 0 | 0.331 |
| 33444 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 37 | TMEM132B | Sponge network | -0.611 | 0.09607 | -0.83 | 0.01084 | 0.331 |
| 33445 | PSMD5-AS1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-495-3p | 15 | STAG2 | Sponge network | 0.569 | 0.00067 | 0.252 | 0.00438 | 0.331 |
| 33446 | HCG11 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 16 | RARB | Sponge network | 0.385 | 0.10067 | -0.825 | 2.0E-5 | 0.331 |
| 33447 | LINC00403 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-30d-3p | 11 | NTRK3 | Sponge network | -3.586 | 0.00032 | -1.77 | 3.0E-5 | 0.331 |
| 33448 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-374a-3p | 17 | GREM1 | Sponge network | 0.331 | 0.57247 | 0.902 | 0.01691 | 0.331 |
| 33449 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 25 | TMTC3 | Sponge network | 0.72 | 0.0001 | 0.123 | 0.22189 | 0.331 |
| 33450 | SNHG1 |
hsa-miR-125a-5p;hsa-miR-150-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 10 | RAD23B | Sponge network | 1.265 | 0 | 0.108 | 0.17414 | 0.331 |
| 33451 | RP11-693J15.4 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p | 10 | EGR2 | Sponge network | -0.72 | 0.24239 | 0.309 | 0.17079 | 0.331 |
| 33452 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | FZD3 | Sponge network | 0.545 | 0.00064 | 0.104 | 0.60316 | 0.331 |
| 33453 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p | 11 | KIT | Sponge network | -1.255 | 0.00981 | -2.184 | 0 | 0.331 |
| 33454 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p | 42 | FOXP1 | Sponge network | -0.793 | 7.0E-5 | 0.042 | 0.74368 | 0.331 |
| 33455 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p | 33 | RASSF8 | Sponge network | -0.615 | 0.00015 | -0.122 | 0.65591 | 0.331 |
| 33456 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 18 | TMEFF2 | Sponge network | 0.532 | 0.00505 | -2.426 | 0 | 0.331 |
| 33457 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 50 | BMPR2 | Sponge network | 0.415 | 0.14657 | 0.206 | 0.0635 | 0.331 |
| 33458 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-629-3p | 17 | USP12 | Sponge network | 0.811 | 0.03228 | -0.102 | 0.40768 | 0.331 |
| 33459 | FZD10-AS1 |
hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -0.727 | 0.04726 | -1.692 | 0 | 0.331 |
| 33460 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | PAFAH1B1 | Sponge network | -2.46 | 0 | -0.429 | 0 | 0.331 |
| 33461 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-590-3p | 17 | NTRK2 | Sponge network | 0.381 | 0.25967 | -0.736 | 0.06495 | 0.331 |
| 33462 | RP11-400K9.4 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-374b-3p | 13 | RPS6KA6 | Sponge network | -0.611 | 0.09607 | -2.989 | 0 | 0.331 |
| 33463 | LINC00174 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p;hsa-miR-582-5p | 15 | BRD3 | Sponge network | 1.253 | 0 | 0.37 | 0.00013 | 0.331 |
| 33464 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p | 21 | AKAP11 | Sponge network | 1.044 | 2.0E-5 | 0.196 | 0.09825 | 0.331 |
| 33465 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p | 11 | SPG20 | Sponge network | 0.4 | 0.14611 | -1.16 | 0 | 0.331 |
| 33466 | GAS6-AS2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 27 | IGFBP5 | Sponge network | 0.16 | 0.50785 | -0.806 | 0.00105 | 0.331 |
| 33467 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | FZD3 | Sponge network | 1.205 | 0 | 0.104 | 0.60316 | 0.331 |
| 33468 | CTB-133G6.2 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p | 16 | LPP | Sponge network | 0.4 | 0.39299 | -0.459 | 0.04475 | 0.331 |
| 33469 | RP11-218M22.1 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 18 | CADM1 | Sponge network | 0.197 | 0.25618 | -0.9 | 0.00084 | 0.331 |
| 33470 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-212-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | FGF5 | Sponge network | 0.559 | 0.00026 | 0.549 | 0.28659 | 0.331 |
| 33471 | CTA-14H9.5 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | TCF4 | Sponge network | 0.145 | 0.53343 | 0.016 | 0.92271 | 0.331 |
| 33472 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-589-5p;hsa-miR-93-5p;hsa-miR-98-5p | 38 | PRKAA2 | Sponge network | 0.082 | 0.55397 | -2.462 | 0 | 0.331 |
| 33473 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ZFX | Sponge network | 0.194 | 0.64278 | 0.09 | 0.42563 | 0.331 |
| 33474 | RP11-35G9.3 |
hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 10 | RNF180 | Sponge network | 0.657 | 4.0E-5 | -1.127 | 1.0E-5 | 0.331 |
| 33475 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 18 | TSHZ3 | Sponge network | 1.757 | 0 | -0.556 | 0.01516 | 0.331 |
| 33476 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-589-3p | 29 | TACC1 | Sponge network | -0.773 | 0.04073 | -0.362 | 0.05406 | 0.331 |
| 33477 | MEG3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 33 | ETV1 | Sponge network | 0.249 | 0.35902 | -0.096 | 0.67674 | 0.331 |
| 33478 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p | 16 | FOXO3 | Sponge network | 0.178 | 0.29106 | -0.04 | 0.72028 | 0.331 |
| 33479 | RP11-728F11.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 14 | CNTN1 | Sponge network | 0.238 | 0.70544 | -2.594 | 0 | 0.331 |
| 33480 | MIR497HG |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p | 22 | RIMS2 | Sponge network | -1.439 | 4.0E-5 | -1.365 | 0.00208 | 0.331 |
| 33481 | RP11-196G18.23 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PRKAB2 | Sponge network | 0.724 | 0.00039 | -0.367 | 0.01371 | 0.331 |
| 33482 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 34 | LIFR | Sponge network | -0.222 | 0.32445 | -1.794 | 0 | 0.331 |
| 33483 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-590-3p | 14 | PKNOX1 | Sponge network | 0.683 | 1.0E-5 | 0.085 | 0.28917 | 0.331 |
| 33484 | GAS6-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-5p | 10 | ABCA10 | Sponge network | 0.16 | 0.50785 | -0.571 | 0.03674 | 0.331 |
| 33485 | CTC-487M23.5 |
hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-92a-3p | 12 | THBS1 | Sponge network | 0.912 | 0.00014 | 0.741 | 0.00386 | 0.331 |
| 33486 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p | 16 | LATS2 | Sponge network | -0.405 | 0.22013 | 0.159 | 0.2304 | 0.331 |
| 33487 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 37 | DMXL1 | Sponge network | -0.129 | 0.57177 | -0.426 | 0.00056 | 0.331 |
| 33488 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 39 | EPHA5 | Sponge network | 0.349 | 0.01695 | -0.957 | 0.01733 | 0.331 |
| 33489 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 37 | MOB1B | Sponge network | 0.219 | 0.1516 | 0.197 | 0.15266 | 0.331 |
| 33490 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p | 16 | ITGB3 | Sponge network | 0.199 | 0.38015 | -0.011 | 0.95751 | 0.331 |
| 33491 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 21 | APC | Sponge network | 0.683 | 1.0E-5 | -0.255 | 0.04051 | 0.331 |
| 33492 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 23 | SOX5 | Sponge network | -4.477 | 0 | -1.091 | 4.0E-5 | 0.331 |
| 33493 | RP5-1021I20.1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-532-3p | 15 | SYNM | Sponge network | -0.785 | 0.00035 | -2.309 | 1.0E-5 | 0.331 |
| 33494 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TBX18 | Sponge network | -0.901 | 0.00019 | 0.843 | 0.02945 | 0.331 |
| 33495 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-30a-3p;hsa-miR-582-5p | 13 | CLASP2 | Sponge network | 0.529 | 0.00552 | -0.038 | 0.71 | 0.331 |
| 33496 | RP11-983P16.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 15 | PRUNE2 | Sponge network | 0.41 | 0.02214 | -1.733 | 0.00022 | 0.331 |
| 33497 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-942-5p | 12 | RB1CC1 | Sponge network | -1.645 | 0 | 0.175 | 0.12559 | 0.331 |
| 33498 | CTD-3126B10.4 |
hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | ABL2 | Sponge network | 0.778 | 0.00101 | 0.82 | 0 | 0.331 |
| 33499 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | LATS2 | Sponge network | -1.216 | 0 | 0.159 | 0.2304 | 0.331 |
| 33500 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PTPRB | Sponge network | -0.323 | 0.01372 | 0.105 | 0.47122 | 0.331 |
| 33501 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 48 | CHD5 | Sponge network | 0.249 | 0.35902 | -0.922 | 0.01521 | 0.331 |
| 33502 | H1FX-AS1 |
hsa-let-7g-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p | 13 | NRK | Sponge network | -0.206 | 0.12942 | 0.656 | 0.19217 | 0.331 |
| 33503 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 29 | SSBP2 | Sponge network | -0.141 | 0.26434 | -1.58 | 0 | 0.331 |
| 33504 | LIPE-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-625-5p;hsa-miR-93-3p | 16 | SUFU | Sponge network | 0.078 | 0.55803 | -0.04 | 0.65489 | 0.331 |
| 33505 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | FOXO1 | Sponge network | -0.72 | 0.24239 | -0.141 | 0.33874 | 0.331 |
| 33506 | CTB-179K24.3 |
hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450a-5p;hsa-miR-497-5p | 10 | CHD2 | Sponge network | 0.841 | 0.04055 | 0.049 | 0.55371 | 0.331 |
| 33507 | RP11-356J5.12 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | NAV3 | Sponge network | -0.899 | 6.0E-5 | 0.212 | 0.47729 | 0.331 |
| 33508 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | PRKCE | Sponge network | -0.125 | 0.49414 | -0.403 | 0.00056 | 0.331 |
| 33509 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 18 | ARHGAP29 | Sponge network | 0.222 | 0.29133 | 0.131 | 0.42953 | 0.331 |
| 33510 | ARHGEF26-AS1 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | RIT1 | Sponge network | -2.46 | 0 | -0.296 | 0.00054 | 0.331 |
| 33511 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-339-5p | 11 | ZNF521 | Sponge network | -0.405 | 0.22013 | -0.111 | 0.58068 | 0.331 |
| 33512 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-664a-3p;hsa-miR-92a-3p | 16 | ST6GAL2 | Sponge network | 0.214 | 0.18246 | -1.201 | 0.00597 | 0.331 |
| 33513 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-199b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-539-5p;hsa-miR-542-3p | 23 | TSC1 | Sponge network | 0.078 | 0.55803 | 0.103 | 0.26071 | 0.331 |
| 33514 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-93-5p | 15 | LRIG1 | Sponge network | -2.531 | 0 | -0.537 | 0.00588 | 0.331 |
| 33515 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-877-5p;hsa-miR-93-3p | 21 | SUFU | Sponge network | -0.465 | 0.04703 | -0.04 | 0.65489 | 0.331 |
| 33516 | CTD-2666L21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-92a-1-5p | 12 | AKAP6 | Sponge network | 0.368 | 0.20093 | -1.586 | 3.0E-5 | 0.331 |
| 33517 | RP11-35G9.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | TET1 | Sponge network | 0.657 | 4.0E-5 | -0.135 | 0.52138 | 0.331 |
| 33518 | RP11-867G23.10 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 12 | SPTBN4 | Sponge network | -2.531 | 0 | -0.786 | 0.01281 | 0.331 |
| 33519 | AC084219.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 13 | GPAM | Sponge network | 1.296 | 0.0054 | 0.142 | 0.29732 | 0.331 |
| 33520 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-664a-3p | 19 | NUAK1 | Sponge network | 0.214 | 0.18246 | 0.904 | 0 | 0.331 |
| 33521 | RP11-822E23.8 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-93-5p;hsa-miR-940 | 10 | FAM117A | Sponge network | -0.777 | 0.16667 | -1.379 | 0 | 0.331 |
| 33522 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | WWTR1 | Sponge network | -0.737 | 0.10822 | -0.345 | 0.11997 | 0.331 |
| 33523 | RP11-815I9.4 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-576-5p | 10 | TSC1 | Sponge network | 0.537 | 0.0006 | 0.103 | 0.26071 | 0.331 |
| 33524 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 17 | NEDD4 | Sponge network | 0.614 | 1.0E-5 | 0.02 | 0.89834 | 0.331 |
| 33525 | RP11-686O6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-324-5p;hsa-miR-455-3p;hsa-miR-577 | 11 | PRKAB2 | Sponge network | 0.453 | 0.01971 | -0.367 | 0.01371 | 0.331 |
| 33526 | AC005562.1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | CREB1 | Sponge network | 0.488 | 0.00084 | 0.203 | 0.00537 | 0.331 |
| 33527 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-93-5p | 16 | FGFR1 | Sponge network | 0.61 | 0.01593 | -0.509 | 0.03957 | 0.331 |
| 33528 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-362-3p;hsa-miR-7-5p | 11 | CRTC3 | Sponge network | -0.693 | 0.05629 | 0.164 | 0.16786 | 0.331 |
| 33529 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 54 | WDFY3 | Sponge network | 0.572 | 0.01722 | -0.201 | 0.0967 | 0.331 |
| 33530 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | NUAK1 | Sponge network | 0.194 | 0.64278 | 0.904 | 0 | 0.331 |
| 33531 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 15 | ZFHX3 | Sponge network | -0.376 | 0.00337 | 0.389 | 0.01363 | 0.331 |
| 33532 | RP11-395G23.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 31 | RUNX1T1 | Sponge network | 0.381 | 0.25967 | -0.344 | 0.2374 | 0.331 |
| 33533 | AC127904.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | NFIB | Sponge network | 1.308 | 0 | 0.023 | 0.90795 | 0.331 |
| 33534 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429 | 11 | ABCB5 | Sponge network | -0.469 | 0.08721 | -0.956 | 0.04077 | 0.331 |
| 33535 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 30 | FOXO3 | Sponge network | 0.249 | 0.35902 | -0.04 | 0.72028 | 0.331 |
| 33536 | LINC00963 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | DNMT3A | Sponge network | 0.523 | 9.0E-5 | 0.302 | 0.0262 | 0.331 |
| 33537 | RP11-678G14.3 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-625-5p | 17 | CADM1 | Sponge network | -4.477 | 0 | -0.9 | 0.00084 | 0.331 |
| 33538 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 24 | PRKCE | Sponge network | 0.219 | 0.1516 | -0.403 | 0.00056 | 0.331 |
| 33539 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-629-5p | 15 | TCF4 | Sponge network | 1.122 | 0.02989 | 0.016 | 0.92271 | 0.331 |
| 33540 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 57 | PTPRD | Sponge network | 0.997 | 0.00066 | -1.005 | 0.00064 | 0.331 |
| 33541 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-93-5p | 18 | PLSCR4 | Sponge network | 0.253 | 0.09565 | -0.474 | 0.03833 | 0.331 |
| 33542 | RP11-116O18.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RET | Sponge network | 0.05 | 0.95483 | -0.833 | 0.03517 | 0.331 |
| 33543 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-582-5p | 28 | BTG2 | Sponge network | -2.69 | 0.0001 | -1.763 | 0 | 0.331 |
| 33544 | RP11-216B9.6 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p | 11 | KLF10 | Sponge network | 0.174 | 0.30034 | -0.783 | 0 | 0.331 |
| 33545 | RP4-613B23.1 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-877-5p | 12 | AFF1 | Sponge network | -0.309 | 0.1145 | -0.384 | 0.00053 | 0.331 |
| 33546 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | IGSF10 | Sponge network | 0.657 | 4.0E-5 | -1.751 | 1.0E-5 | 0.331 |
| 33547 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 11 | MYBL1 | Sponge network | 1.923 | 0 | 0.494 | 0.02152 | 0.331 |
| 33548 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-93-5p | 22 | SYNE1 | Sponge network | 0.253 | 0.09565 | -0.738 | 0.00316 | 0.331 |
| 33549 | DIO3OS |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-625-5p | 20 | CADM1 | Sponge network | -0.773 | 0.04073 | -0.9 | 0.00084 | 0.331 |
| 33550 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 57 | AFF4 | Sponge network | -0.83 | 0 | -0.266 | 0.01565 | 0.331 |
| 33551 | RP11-13K12.1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-629-3p | 11 | RHOB | Sponge network | -0.154 | 0.78939 | -1.052 | 0 | 0.331 |
| 33552 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 15 | IGSF10 | Sponge network | 0.395 | 0.15451 | -1.751 | 1.0E-5 | 0.331 |
| 33553 | RP11-798M19.6 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | SIRT1 | Sponge network | -0.612 | 0.00094 | 0.109 | 0.2593 | 0.331 |
| 33554 | RP11-983P16.4 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-766-3p | 15 | ATM | Sponge network | 0.41 | 0.02214 | 0.178 | 0.21568 | 0.331 |
| 33555 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-429;hsa-miR-484;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 33 | SOX5 | Sponge network | -0.125 | 0.49414 | -1.091 | 4.0E-5 | 0.331 |
| 33556 | FAM13A-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | RECK | Sponge network | -0.83 | 0 | -1.043 | 0 | 0.331 |
| 33557 | A2M-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 60 | LPP | Sponge network | -0.08 | 0.90092 | -0.459 | 0.04475 | 0.331 |
| 33558 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p | 12 | MYCBP2 | Sponge network | 0.657 | 4.0E-5 | -0.027 | 0.82016 | 0.331 |
| 33559 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-625-5p;hsa-miR-93-5p | 13 | LHX6 | Sponge network | -0.793 | 7.0E-5 | -0.087 | 0.69193 | 0.331 |
| 33560 | RP11-35G9.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | ASTN1 | Sponge network | 0.657 | 4.0E-5 | -2.389 | 2.0E-5 | 0.331 |
| 33561 | RP4-669P10.16 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-542-3p | 31 | LPP | Sponge network | -0.301 | 0.06597 | -0.459 | 0.04475 | 0.331 |
| 33562 | RP11-212P7.2 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-874-3p | 10 | HSPB7 | Sponge network | 0.219 | 0.1516 | -2.268 | 1.0E-5 | 0.331 |
| 33563 | HCG11 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 14 | CSDE1 | Sponge network | 0.385 | 0.10067 | -0.493 | 0 | 0.331 |
| 33564 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-625-5p | 14 | RSPO2 | Sponge network | -0.318 | 0.65847 | -3.038 | 0 | 0.331 |
| 33565 | LINC00094 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 29 | SNX29 | Sponge network | 0.253 | 0.09565 | -0.34 | 0.01371 | 0.331 |
| 33566 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | SOX5 | Sponge network | -0.969 | 2.0E-5 | -1.091 | 4.0E-5 | 0.331 |
| 33567 | RP11-356J5.12 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | HERC1 | Sponge network | -0.899 | 6.0E-5 | -0.196 | 0.08544 | 0.331 |
| 33568 | CTD-3092A11.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-7-1-3p | 12 | VEZT | Sponge network | 0.873 | 0 | 0.259 | 0.00296 | 0.331 |
| 33569 | RP11-359E10.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-7-5p | 12 | IRS1 | Sponge network | 0.299 | 0.57722 | -0.026 | 0.88855 | 0.331 |
| 33570 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | SYNPO2 | Sponge network | 0.219 | 0.1516 | -2.342 | 0 | 0.331 |
| 33571 | USP3-AS1 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-7-5p | 14 | IRS1 | Sponge network | -0.162 | 0.47687 | -0.026 | 0.88855 | 0.331 |
| 33572 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | KANK1 | Sponge network | 0.101 | 0.60919 | -0.55 | 0.00134 | 0.331 |
| 33573 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | MYBL1 | Sponge network | 1.294 | 0 | 0.494 | 0.02152 | 0.331 |
| 33574 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 16 | INPP4A | Sponge network | -0.355 | 0.0077 | 0.07 | 0.53563 | 0.331 |
| 33575 | ACTA2-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 13 | EHD3 | Sponge network | 0.194 | 0.64278 | -0.484 | 0.01747 | 0.331 |
| 33576 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | CCND2 | Sponge network | -0.376 | 0.00337 | -0.267 | 0.3112 | 0.331 |
| 33577 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-5p | 26 | BCL2 | Sponge network | -0.469 | 0.08721 | -0.899 | 0 | 0.33 |
| 33578 | C4A-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 14 | EBF3 | Sponge network | 3.345 | 0 | -0.058 | 0.81437 | 0.33 |
| 33579 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 30 | LIFR | Sponge network | 0.101 | 0.60919 | -1.794 | 0 | 0.33 |
| 33580 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | MYO9A | Sponge network | 0.619 | 0.00157 | -0.147 | 0.32373 | 0.33 |
| 33581 | KB-1572G7.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p | 19 | ARHGEF12 | Sponge network | 0.924 | 0 | 0.09 | 0.35693 | 0.33 |
| 33582 | MYLK-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-766-3p | 10 | ATM | Sponge network | -1.331 | 3.0E-5 | 0.178 | 0.21568 | 0.33 |
| 33583 | RAB30-AS1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-495-3p | 11 | STAG2 | Sponge network | 0.752 | 0 | 0.252 | 0.00438 | 0.33 |
| 33584 | CALML3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-542-3p | 16 | SYNM | Sponge network | 0.95 | 0.15943 | -2.309 | 1.0E-5 | 0.33 |
| 33585 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NUAK1 | Sponge network | 0.72 | 0.0001 | 0.904 | 0 | 0.33 |
| 33586 | RP11-25K21.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p | 10 | RASSF2 | Sponge network | 0.489 | 0.25786 | -0.273 | 0.21961 | 0.33 |
| 33587 | LINC00654 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-624-5p | 11 | SCN11A | Sponge network | 0.374 | 0.20054 | -1.15 | 0.00299 | 0.33 |
| 33588 | LINC00957 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | TAL1 | Sponge network | -0.421 | 0.0114 | -0.462 | 0.00574 | 0.33 |
| 33589 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | PRKAA2 | Sponge network | -0.726 | 0.00132 | -2.462 | 0 | 0.33 |
| 33590 | IQCH-AS1 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-9-5p | 10 | SPTBN1 | Sponge network | 0.929 | 0 | 0.561 | 1.0E-5 | 0.33 |
| 33591 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | DDX6 | Sponge network | 0.385 | 0.10067 | 0.091 | 0.36428 | 0.33 |
| 33592 | RP11-38L15.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p | 10 | KCNJ5 | Sponge network | 0.344 | 0.3127 | -0.281 | 0.3339 | 0.33 |
| 33593 | GAS6-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 69 | LPP | Sponge network | 0.16 | 0.50785 | -0.459 | 0.04475 | 0.33 |
| 33594 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 16 | RBL2 | Sponge network | -1.654 | 0.00144 | -0.282 | 0.00254 | 0.33 |
| 33595 | DNM3OS |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | RPS6KA6 | Sponge network | 0.572 | 0.01722 | -2.989 | 0 | 0.33 |
| 33596 | CTB-133G6.2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-940 | 10 | CADM2 | Sponge network | 0.4 | 0.39299 | -3.782 | 0 | 0.33 |
| 33597 | PART1 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | CSDE1 | Sponge network | -2.925 | 0 | -0.493 | 0 | 0.33 |
| 33598 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 56 | PTEN | Sponge network | -0.222 | 0.32445 | -0.187 | 0.07748 | 0.33 |
| 33599 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 27 | PTPRT | Sponge network | -0.739 | 0.00044 | -0.907 | 0.03727 | 0.33 |
| 33600 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | ITGAV | Sponge network | 0.72 | 0.0001 | 0.337 | 0.01002 | 0.33 |
| 33601 | RP11-356J5.12 |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | PACRG | Sponge network | -0.899 | 6.0E-5 | -2.248 | 0 | 0.33 |
| 33602 | RP11-305E6.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 16 | UBR3 | Sponge network | 1.317 | 0 | -0.021 | 0.82774 | 0.33 |
| 33603 | HHIP-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | CBFA2T3 | Sponge network | -0.737 | 0.10822 | -1.221 | 1.0E-5 | 0.33 |
| 33604 | PCA3 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-3p | 23 | CTIF | Sponge network | -0.709 | 0.18168 | -0.389 | 0.00549 | 0.33 |
| 33605 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-942-5p | 24 | COPS2 | Sponge network | -0.612 | 0.00094 | -0.178 | 0.04974 | 0.33 |
| 33606 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-96-5p | 25 | HBP1 | Sponge network | -1.654 | 0.00144 | -0.454 | 0 | 0.33 |
| 33607 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | HOOK3 | Sponge network | -0.899 | 6.0E-5 | 0.273 | 0.09957 | 0.33 |
| 33608 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p | 11 | EZH1 | Sponge network | -1.375 | 0.08642 | -0.564 | 1.0E-5 | 0.33 |
| 33609 | AC003090.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | GJA1 | Sponge network | -2.117 | 0.00161 | -0.485 | 0.02179 | 0.33 |
| 33610 | LPP-AS2 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p | 13 | PRDM5 | Sponge network | -0.18 | 0.20343 | -1.09 | 0.00014 | 0.33 |
| 33611 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p | 12 | ERCC4 | Sponge network | -0.583 | 0.14589 | 0.126 | 0.15266 | 0.33 |
| 33612 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 46 | FOXP1 | Sponge network | -0.83 | 0 | 0.042 | 0.74368 | 0.33 |
| 33613 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p | 15 | DDX5 | Sponge network | 0.75 | 0.00054 | -0.099 | 0.17428 | 0.33 |
| 33614 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p | 12 | PTPN11 | Sponge network | 0.594 | 0.00064 | 0.431 | 2.0E-5 | 0.33 |
| 33615 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 35 | TSHZ3 | Sponge network | -0.303 | 0.21002 | -0.556 | 0.01516 | 0.33 |
| 33616 | RP11-401P9.4 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-542-3p | 14 | SFRP1 | Sponge network | -0.384 | 0.40652 | -3.566 | 0 | 0.33 |
| 33617 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-582-3p | 13 | PRDM2 | Sponge network | 0.614 | 1.0E-5 | -0.258 | 0.00721 | 0.33 |
| 33618 | RP5-1198O20.4 |
hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | EMP1 | Sponge network | 0.997 | 0.00066 | -0.942 | 5.0E-5 | 0.33 |
| 33619 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92b-3p | 20 | AFF3 | Sponge network | -0.322 | 0.25945 | -2.373 | 0 | 0.33 |
| 33620 | FAM66C |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | FMNL3 | Sponge network | -0.901 | 0.00019 | 0.555 | 4.0E-5 | 0.33 |
| 33621 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421 | 12 | CNTNAP3 | Sponge network | -0.285 | 0.2172 | -1.465 | 0.00033 | 0.33 |
| 33622 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 28 | CSMD1 | Sponge network | -2.46 | 0 | -0.63 | 0.18881 | 0.33 |
| 33623 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | MGA | Sponge network | 1.302 | 7.0E-5 | 0.323 | 0.00792 | 0.33 |
| 33624 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-5p | 16 | CSMD1 | Sponge network | 0.176 | 0.37402 | -0.63 | 0.18881 | 0.33 |
| 33625 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-493-5p | 12 | KAT6B | Sponge network | 0.368 | 0.20093 | -0.478 | 0.00018 | 0.33 |
| 33626 | LA16c-321D4.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 17 | MDM4 | Sponge network | 2.459 | 5.0E-5 | 0.345 | 0.00762 | 0.33 |
| 33627 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-942-5p | 37 | UNC80 | Sponge network | 0.335 | 0.20596 | -1.934 | 0 | 0.33 |
| 33628 | DIO3OS |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-660-5p | 19 | EYA4 | Sponge network | -0.773 | 0.04073 | 0.3 | 0.36876 | 0.33 |
| 33629 | MIR22HG |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 29 | OPCML | Sponge network | -1.047 | 0 | -2.498 | 0 | 0.33 |
| 33630 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | EPHA5 | Sponge network | 0.219 | 0.1516 | -0.957 | 0.01733 | 0.33 |
| 33631 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-628-5p | 19 | FBXO32 | Sponge network | 0.859 | 0.00331 | -0.495 | 0.07561 | 0.33 |
| 33632 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-93-3p | 20 | FOXO3 | Sponge network | 0.056 | 0.81195 | -0.04 | 0.72028 | 0.33 |
| 33633 | SERTAD4-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-93-3p | 16 | SNX29 | Sponge network | -0.932 | 0.00329 | -0.34 | 0.01371 | 0.33 |
| 33634 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-935;hsa-miR-96-5p | 16 | PCDH10 | Sponge network | 0.37 | 0.27614 | -1.644 | 0.0078 | 0.33 |
| 33635 | OSER1-AS1 |
hsa-miR-106a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | IGSF10 | Sponge network | -0.13 | 0.48734 | -1.751 | 1.0E-5 | 0.33 |
| 33636 | AC141928.1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-27a-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-96-5p | 10 | WNK2 | Sponge network | 0.853 | 0.15352 | -0.352 | 0.31066 | 0.33 |
| 33637 | LINC00641 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 24 | GJA1 | Sponge network | -0.222 | 0.32445 | -0.485 | 0.02179 | 0.33 |
| 33638 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p | 20 | DLC1 | Sponge network | -0.206 | 0.12942 | -0.382 | 0.05038 | 0.33 |
| 33639 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-766-3p | 25 | MAPK1 | Sponge network | 0.972 | 0 | -0.017 | 0.81465 | 0.33 |
| 33640 | LINC00702 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | EDNRB | Sponge network | -0.737 | 0.06182 | -0.682 | 0.00138 | 0.33 |
| 33641 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-625-5p | 28 | PTPRT | Sponge network | -0.154 | 0.78939 | -0.907 | 0.03727 | 0.33 |
| 33642 | TTC28-AS1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-5p;hsa-miR-432-5p;hsa-miR-495-3p | 12 | STAG2 | Sponge network | 0.373 | 0.00068 | 0.252 | 0.00438 | 0.33 |
| 33643 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | 0.392 | 0.0278 | -0.085 | 0.42389 | 0.33 |
| 33644 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 36 | ATRX | Sponge network | -0.129 | 0.57177 | 0.261 | 0.04408 | 0.33 |
| 33645 | LINC00909 |
hsa-miR-125a-3p;hsa-miR-154-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-409-5p;hsa-miR-539-5p | 11 | LIMD1 | Sponge network | 0.363 | 0.00601 | 0.103 | 0.28978 | 0.33 |
| 33646 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | RBL2 | Sponge network | 0.532 | 0.00505 | -0.282 | 0.00254 | 0.33 |
| 33647 | RP11-403I13.7 |
hsa-miR-126-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | IRS1 | Sponge network | 0.395 | 0.15451 | -0.026 | 0.88855 | 0.33 |
| 33648 | LINC00173 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | PRUNE2 | Sponge network | 0.056 | 0.8494 | -1.733 | 0.00022 | 0.33 |
| 33649 | CTD-2002H8.2 |
hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-374b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | APC | Sponge network | 0.931 | 1.0E-5 | -0.255 | 0.04051 | 0.33 |
| 33650 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ABCC9 | Sponge network | 0.392 | 0.0278 | -0.466 | 0.14121 | 0.33 |
| 33651 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | AKAP11 | Sponge network | 0.419 | 0.0319 | 0.196 | 0.09825 | 0.33 |
| 33652 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p | 25 | AFF4 | Sponge network | 1.46 | 1.0E-5 | -0.266 | 0.01565 | 0.33 |
| 33653 | EMX2OS |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-590-3p | 17 | STARD13 | Sponge network | -0.777 | 0.1134 | -0.187 | 0.27383 | 0.33 |
| 33654 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-589-5p;hsa-miR-7-1-3p | 44 | PRKAA2 | Sponge network | 0.16 | 0.50785 | -2.462 | 0 | 0.33 |
| 33655 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ARID5B | Sponge network | 0.657 | 4.0E-5 | 0.342 | 0.01676 | 0.33 |
| 33656 | RP11-13K12.5 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | LUZP2 | Sponge network | -0.318 | 0.65847 | -0.056 | 0.89416 | 0.33 |
| 33657 | CTD-2554C21.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-5p | 25 | BTG2 | Sponge network | -1.654 | 0.00144 | -1.763 | 0 | 0.33 |
| 33658 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p | 21 | THBS1 | Sponge network | -0.842 | 0.01361 | 0.741 | 0.00386 | 0.33 |
| 33659 | RP11-57H14.4 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 46 | PPM1L | Sponge network | 0.083 | 0.66405 | -0.537 | 0.00616 | 0.33 |
| 33660 | RP11-479G22.8 |
hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 13 | UBE2D1 | Sponge network | 1.308 | 0 | 0.468 | 0 | 0.33 |
| 33661 | FENDRR |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | PCLO | Sponge network | -1.281 | 0.00012 | -1.843 | 0.00064 | 0.33 |
| 33662 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 23 | NOTCH2 | Sponge network | 0.727 | 3.0E-5 | 0.138 | 0.35163 | 0.33 |
| 33663 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-671-5p;hsa-miR-874-3p | 20 | ARID5B | Sponge network | -0.465 | 0.04703 | 0.342 | 0.01676 | 0.33 |
| 33664 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 14 | DDX5 | Sponge network | 0.225 | 0.30644 | -0.099 | 0.17428 | 0.33 |
| 33665 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | NIN | Sponge network | 0.727 | 3.0E-5 | -0.253 | 0.13794 | 0.33 |
| 33666 | RP11-473I1.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 18 | SNX29 | Sponge network | 0.683 | 1.0E-5 | -0.34 | 0.01371 | 0.33 |
| 33667 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-942-5p | 24 | NRXN1 | Sponge network | -0.773 | 0.04073 | -3.778 | 0 | 0.33 |
| 33668 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-96-5p | 12 | TP53INP1 | Sponge network | 0.622 | 0.00015 | -0.496 | 0.00064 | 0.33 |
| 33669 | MAGI2-AS3 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | SH3GLB1 | Sponge network | -0.129 | 0.57177 | -0.115 | 0.14773 | 0.33 |
| 33670 | RP11-13K12.5 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p | 11 | IGF2 | Sponge network | -0.318 | 0.65847 | 0.848 | 0.03842 | 0.33 |
| 33671 | DNM3OS |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 29 | ZFX | Sponge network | 0.572 | 0.01722 | 0.09 | 0.42563 | 0.33 |
| 33672 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-577 | 19 | BCL2 | Sponge network | 0.811 | 0.03228 | -0.899 | 0 | 0.33 |
| 33673 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 11 | NALCN | Sponge network | -0.793 | 7.0E-5 | 1.068 | 0.00237 | 0.33 |
| 33674 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | DAB2 | Sponge network | -1.575 | 6.0E-5 | 0.431 | 0.0113 | 0.33 |
| 33675 | LINC00963 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 25 | EPHA3 | Sponge network | 0.523 | 9.0E-5 | 0.185 | 0.53588 | 0.33 |
| 33676 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | SON | Sponge network | 1.03 | 0 | 0.168 | 0.04406 | 0.33 |
| 33677 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-589-3p | 12 | MCC | Sponge network | -0.164 | 0.45106 | -1.005 | 0 | 0.33 |
| 33678 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-374b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-577 | 10 | HACE1 | Sponge network | 0.921 | 0.00118 | -0.072 | 0.55239 | 0.33 |
| 33679 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 16 | DCLK1 | Sponge network | -1.047 | 0 | -1.324 | 0.00014 | 0.33 |
| 33680 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-539-5p | 20 | PDE4DIP | Sponge network | -0.154 | 0.78939 | -0.282 | 0.0257 | 0.33 |
| 33681 | RP11-999E24.3 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 13 | LIMCH1 | Sponge network | -0.693 | 0.05629 | -0.915 | 1.0E-5 | 0.33 |
| 33682 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-424-5p | 11 | BHLHE41 | Sponge network | -0.932 | 0.00329 | 0.353 | 0.12753 | 0.33 |
| 33683 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 17 | RBMS3 | Sponge network | 0.131 | 0.8436 | -0.519 | 0.03551 | 0.33 |
| 33684 | KB-1460A1.5 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p | 10 | ABL1 | Sponge network | 0.603 | 0.00127 | -0.215 | 0.07804 | 0.33 |
| 33685 | RP4-584D14.7 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-577 | 14 | BNC2 | Sponge network | 1.294 | 0.0031 | -0.491 | 0.09988 | 0.33 |
| 33686 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-625-5p | 10 | MCC | Sponge network | -1.054 | 0.0706 | -1.005 | 0 | 0.33 |
| 33687 | BCYRN1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | BRD3 | Sponge network | 0.311 | 0.10344 | 0.37 | 0.00013 | 0.33 |
| 33688 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 29 | ARID5B | Sponge network | 0.083 | 0.66405 | 0.342 | 0.01676 | 0.33 |
| 33689 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | ERBB4 | Sponge network | -0.323 | 0.01372 | -1.975 | 2.0E-5 | 0.33 |
| 33690 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 52 | PTPRD | Sponge network | -2.46 | 0 | -1.005 | 0.00064 | 0.33 |
| 33691 | U91328.19 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -0.303 | 0.21002 | -0.966 | 0.00035 | 0.33 |
| 33692 | RP11-458D21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-500b-5p;hsa-miR-624-5p | 11 | HMCN1 | Sponge network | 0.663 | 0.00073 | 0.809 | 0.00544 | 0.33 |
| 33693 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 14 | ZMYM2 | Sponge network | 1.266 | 0 | 0.279 | 0.01273 | 0.33 |
| 33694 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 14 | TBX18 | Sponge network | 0.16 | 0.50785 | 0.843 | 0.02945 | 0.33 |
| 33695 | KB-1460A1.5 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 22 | RASSF8 | Sponge network | 0.603 | 0.00127 | -0.122 | 0.65591 | 0.33 |
| 33696 | AC009120.6 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | UBR3 | Sponge network | 0.919 | 0 | -0.021 | 0.82774 | 0.33 |
| 33697 | CTD-2303H24.2 |
hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | RABEP1 | Sponge network | 0.415 | 0.14657 | -0.066 | 0.43142 | 0.33 |
| 33698 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-345-5p;hsa-miR-484;hsa-miR-539-5p | 28 | KIF1B | Sponge network | 0.858 | 3.0E-5 | -0.118 | 0.32258 | 0.33 |
| 33699 | BZRAP1-AS1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-942-5p | 11 | ZBTB20 | Sponge network | -0.465 | 0.04703 | -0.122 | 0.57569 | 0.33 |
| 33700 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-590-3p | 11 | PCDH17 | Sponge network | 0.178 | 0.29106 | 0.731 | 2.0E-5 | 0.33 |
| 33701 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p | 18 | CYLD | Sponge network | 0.727 | 3.0E-5 | -0.367 | 0.00171 | 0.33 |
| 33702 | RP11-56L13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-345-5p;hsa-miR-432-5p;hsa-miR-485-3p | 14 | IKZF2 | Sponge network | -0.992 | 0.18158 | -0.284 | 0.1942 | 0.33 |
| 33703 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SMARCA2 | Sponge network | -1.697 | 0.00889 | -0.529 | 0.00014 | 0.33 |
| 33704 | CTD-2002H8.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-2355-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | AFF4 | Sponge network | 0.931 | 1.0E-5 | -0.266 | 0.01565 | 0.33 |
| 33705 | RP11-6O2.3 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-624-5p | 14 | GJA1 | Sponge network | 0.331 | 0.57247 | -0.485 | 0.02179 | 0.33 |
| 33706 | RP11-21A7A.2 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-628-5p | 10 | RIMS2 | Sponge network | -1.116 | 0.23686 | -1.365 | 0.00208 | 0.33 |
| 33707 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | EPHA5 | Sponge network | 0.659 | 3.0E-5 | -0.957 | 0.01733 | 0.33 |
| 33708 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-940 | 26 | PBX1 | Sponge network | 0.75 | 0.00054 | -1.206 | 0 | 0.33 |
| 33709 | NDUFA6-AS1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-629-5p | 14 | WNK2 | Sponge network | -0.355 | 0.0077 | -0.352 | 0.31066 | 0.33 |
| 33710 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-590-3p | 13 | CDC73 | Sponge network | 0.67 | 0.00048 | 0.094 | 0.20463 | 0.33 |
| 33711 | RP11-403I13.7 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 22 | SETBP1 | Sponge network | 0.395 | 0.15451 | -1.302 | 1.0E-5 | 0.33 |
| 33712 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 12 | PARK2 | Sponge network | 0.374 | 0.20054 | -1.892 | 0 | 0.33 |
| 33713 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p | 11 | PDGFC | Sponge network | 0.056 | 0.81195 | -0.33 | 0.06483 | 0.33 |
| 33714 | RP4-791M13.3 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-582-5p | 27 | FAT3 | Sponge network | 0.419 | 0.0319 | -1.601 | 0.00051 | 0.33 |
| 33715 | FAM66C |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 30 | OPCML | Sponge network | -0.901 | 0.00019 | -2.498 | 0 | 0.33 |
| 33716 | EMX2OS |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | -0.777 | 0.1134 | -2.417 | 0 | 0.33 |
| 33717 | RP11-758N13.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-93-3p | 25 | NTRK3 | Sponge network | 2.939 | 3.0E-5 | -1.77 | 3.0E-5 | 0.33 |
| 33718 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | FGF5 | Sponge network | -0.323 | 0.01372 | 0.549 | 0.28659 | 0.33 |
| 33719 | HOTAIRM1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | EYA4 | Sponge network | -0.053 | 0.80685 | 0.3 | 0.36876 | 0.33 |
| 33720 | CTD-3092A11.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | BRD3 | Sponge network | 0.873 | 0 | 0.37 | 0.00013 | 0.33 |
| 33721 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-935;hsa-miR-96-5p | 29 | FBXO32 | Sponge network | -0.83 | 0 | -0.495 | 0.07561 | 0.33 |
| 33722 | CROCCP2 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 11 | CDK6 | Sponge network | 0.79 | 0 | 0.991 | 0 | 0.33 |
| 33723 | H1FX-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | EPHA7 | Sponge network | -0.206 | 0.12942 | -2.197 | 3.0E-5 | 0.33 |
| 33724 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 19 | MYBL1 | Sponge network | 1.153 | 0 | 0.494 | 0.02152 | 0.33 |
| 33725 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p | 11 | ZNF208 | Sponge network | -0.405 | 0.22013 | -0.4 | 0.22381 | 0.33 |
| 33726 | PWAR6 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FBXO11 | Sponge network | -1.575 | 6.0E-5 | 0.142 | 0.04938 | 0.33 |
| 33727 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 52 | SSBP2 | Sponge network | -0.969 | 2.0E-5 | -1.58 | 0 | 0.33 |
| 33728 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-93-3p;hsa-miR-93-5p | 40 | SSBP2 | Sponge network | -0.154 | 0.78939 | -1.58 | 0 | 0.33 |
| 33729 | OIP5-AS1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | USP9X | Sponge network | 0.421 | 0.00084 | 0.222 | 0.01689 | 0.33 |
| 33730 | RP11-999E24.3 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-374a-5p;hsa-miR-744-3p | 14 | PTPN14 | Sponge network | -0.693 | 0.05629 | 0.144 | 0.34595 | 0.33 |
| 33731 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 19 | NEDD4 | Sponge network | -0.896 | 0.00099 | 0.02 | 0.89834 | 0.33 |
| 33732 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-582-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 18 | GAS7 | Sponge network | -0.31 | 0.33871 | -0.555 | 0.01602 | 0.33 |
| 33733 | FGD5-AS1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-664a-3p | 16 | RASAL2 | Sponge network | 0.401 | 0.00066 | 0.869 | 0 | 0.33 |
| 33734 | LINC00473 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 52 | DST | Sponge network | -1.203 | 0.03881 | -0.713 | 0.00096 | 0.33 |
| 33735 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | 0.862 | 0.06213 | 0.783 | 0.00072 | 0.33 |
| 33736 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 37 | TMEM132B | Sponge network | -0.563 | 0.02545 | -0.83 | 0.01084 | 0.33 |
| 33737 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p | 21 | RASSF2 | Sponge network | 0.178 | 0.29106 | -0.273 | 0.21961 | 0.33 |
| 33738 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-93-5p | 32 | LRP1B | Sponge network | -0.18 | 0.20343 | -2.163 | 0 | 0.33 |
| 33739 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DNAJB4 | Sponge network | -0.513 | 0.04703 | -0.7 | 1.0E-5 | 0.33 |
| 33740 | CTC-559E9.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-576-5p | 12 | BRD3 | Sponge network | 0.594 | 0.00064 | 0.37 | 0.00013 | 0.33 |
| 33741 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-502-5p;hsa-miR-625-5p | 11 | ANK2 | Sponge network | 1.294 | 0.0031 | -1.603 | 1.0E-5 | 0.33 |
| 33742 | DIO3OS |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-98-5p | 14 | SEMA3C | Sponge network | -0.773 | 0.04073 | -0.272 | 0.31153 | 0.33 |
| 33743 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 22 | LATS1 | Sponge network | 0.488 | 0.00084 | 0.109 | 0.34208 | 0.33 |
| 33744 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-744-5p;hsa-miR-942-5p | 23 | NOTCH2 | Sponge network | -4.463 | 0.00025 | 0.138 | 0.35163 | 0.33 |
| 33745 | LINC00092 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | FBXO32 | Sponge network | -1.886 | 0 | -0.495 | 0.07561 | 0.33 |
| 33746 | RP4-717I23.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p | 25 | ZFX | Sponge network | 0.637 | 0.00069 | 0.09 | 0.42563 | 0.33 |
| 33747 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-671-5p | 12 | PIK3IP1 | Sponge network | 0.997 | 0.00066 | -0.187 | 0.19945 | 0.33 |
| 33748 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | FOXO3 | Sponge network | 0.272 | 0.44692 | -0.04 | 0.72028 | 0.33 |
| 33749 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-7-1-3p | 18 | ERBB4 | Sponge network | -4.477 | 0 | -1.975 | 2.0E-5 | 0.33 |
| 33750 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | ZNF292 | Sponge network | 0.349 | 0.01695 | 0.276 | 0.04148 | 0.33 |
| 33751 | CTD-2184D3.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | BRD3 | Sponge network | 0.272 | 0.44692 | 0.37 | 0.00013 | 0.33 |
| 33752 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-589-5p | 11 | GRIA3 | Sponge network | -0.309 | 0.1145 | -1.956 | 0 | 0.33 |
| 33753 | RP11-203M5.7 |
hsa-miR-101-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-590-5p | 12 | UBR3 | Sponge network | 1.684 | 1.0E-5 | -0.021 | 0.82774 | 0.33 |
| 33754 | RP11-517B11.7 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 14 | DICER1 | Sponge network | 0.706 | 0 | 0.042 | 0.674 | 0.33 |
| 33755 | U91328.19 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 30 | ETV1 | Sponge network | -0.303 | 0.21002 | -0.096 | 0.67674 | 0.33 |
| 33756 | RP11-130L8.1 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PRKCB | Sponge network | -0.663 | 0.10887 | -1.313 | 2.0E-5 | 0.33 |
| 33757 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-5p | 26 | BHLHE41 | Sponge network | 0.395 | 0.15451 | 0.353 | 0.12753 | 0.33 |
| 33758 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 27 | MED12L | Sponge network | 0.082 | 0.55654 | -0.194 | 0.55299 | 0.33 |
| 33759 | RP11-434H6.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-132-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p | 11 | FBXO32 | Sponge network | 0.358 | 0.14302 | -0.495 | 0.07561 | 0.33 |
| 33760 | RP11-359E10.1 |
hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | RNF144A | Sponge network | 0.299 | 0.57722 | 0.594 | 0.0009 | 0.33 |
| 33761 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-940 | 24 | MEF2C | Sponge network | -4.477 | 0 | -0.499 | 0.01448 | 0.33 |
| 33762 | LINC00654 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 21 | ARNT2 | Sponge network | 0.374 | 0.20054 | -0.332 | 0.25851 | 0.33 |
| 33763 | RP11-426C22.5 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | NAV3 | Sponge network | 0.749 | 0 | 0.212 | 0.47729 | 0.33 |
| 33764 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-671-5p | 15 | CADM3 | Sponge network | 0.532 | 0.00505 | -3.663 | 0 | 0.33 |
| 33765 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 12 | EPAS1 | Sponge network | -1.161 | 0.00591 | -0.184 | 0.14418 | 0.33 |
| 33766 | LINC00473 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 20 | TET1 | Sponge network | -1.203 | 0.03881 | -0.135 | 0.52138 | 0.33 |
| 33767 | LINC00472 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-7-1-3p | 16 | PIK3CA | Sponge network | -0.842 | 0.01361 | 0.195 | 0.06908 | 0.33 |
| 33768 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 33 | MOB1B | Sponge network | 0.177 | 0.16681 | 0.197 | 0.15266 | 0.33 |
| 33769 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | FSTL5 | Sponge network | 0.385 | 0.10067 | -1.3 | 0.01619 | 0.33 |
| 33770 | SNX29P2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 13 | EGR2 | Sponge network | 0.4 | 0.14611 | 0.309 | 0.17079 | 0.33 |
| 33771 | RP11-67L2.2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 17 | KLF10 | Sponge network | 0.72 | 0.0001 | -0.783 | 0 | 0.33 |
| 33772 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | MOB1B | Sponge network | 0.8 | 0 | 0.197 | 0.15266 | 0.33 |
| 33773 | PCA3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | BRD3 | Sponge network | -0.709 | 0.18168 | 0.37 | 0.00013 | 0.33 |
| 33774 | CTD-3157E16.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-21-5p | 11 | CADM2 | Sponge network | -0.326 | 0.28809 | -3.782 | 0 | 0.33 |
| 33775 | RP4-639F20.1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-629-5p;hsa-miR-96-5p | 13 | WNK2 | Sponge network | -0.517 | 0.02935 | -0.352 | 0.31066 | 0.33 |
| 33776 | RP11-359B12.2 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZNF844 | Sponge network | 0.064 | 0.72934 | -0.966 | 0.00035 | 0.33 |
| 33777 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | SLITRK3 | Sponge network | 0.051 | 0.83388 | -3.328 | 0 | 0.33 |
| 33778 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | SYNE1 | Sponge network | 0.619 | 0.00157 | -0.738 | 0.00316 | 0.33 |
| 33779 | RP11-410L14.2 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 11 | BRD3 | Sponge network | -0.052 | 0.76839 | 0.37 | 0.00013 | 0.33 |
| 33780 | AC116366.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-577 | 14 | BCL2 | Sponge network | 0.329 | 0.11122 | -0.899 | 0 | 0.33 |
| 33781 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p | 50 | NTRK3 | Sponge network | 0.385 | 0.10067 | -1.77 | 3.0E-5 | 0.33 |
| 33782 | RP11-686D22.8 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 24 | RUNX1T1 | Sponge network | 0.898 | 1.0E-5 | -0.344 | 0.2374 | 0.33 |
| 33783 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | NEDD4 | Sponge network | 0.083 | 0.66405 | 0.02 | 0.89834 | 0.33 |
| 33784 | NNT-AS1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 31 | CREB1 | Sponge network | 0.799 | 1.0E-5 | 0.203 | 0.00537 | 0.33 |
| 33785 | ANKRD10-IT1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-7-1-3p | 14 | ZFX | Sponge network | 1.739 | 0 | 0.09 | 0.42563 | 0.33 |
| 33786 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-92a-3p | 28 | CNTN1 | Sponge network | 0.253 | 0.09565 | -2.594 | 0 | 0.33 |
| 33787 | RP11-218M22.1 |
hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | MED12L | Sponge network | 0.197 | 0.25618 | -0.194 | 0.55299 | 0.33 |
| 33788 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | CAMTA1 | Sponge network | 0.541 | 0.00125 | -0.436 | 1.0E-5 | 0.33 |
| 33789 | HNRNPU-AS1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-9-5p | 15 | CDK6 | Sponge network | 1.601 | 0 | 0.991 | 0 | 0.33 |
| 33790 | AC003090.1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-590-3p | 17 | GLIPR1 | Sponge network | -2.117 | 0.00161 | 0.146 | 0.3276 | 0.33 |
| 33791 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-93-5p | 11 | DAB2 | Sponge network | -0.465 | 0.04703 | 0.431 | 0.0113 | 0.33 |
| 33792 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-7-1-3p | 27 | BNC2 | Sponge network | 0.389 | 0.04293 | -0.491 | 0.09988 | 0.33 |
| 33793 | RP5-1139B12.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p | 11 | SCN9A | Sponge network | 0.598 | 0.38481 | -0.525 | 0.10593 | 0.33 |
| 33794 | FAM66C |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | BTG2 | Sponge network | -0.901 | 0.00019 | -1.763 | 0 | 0.33 |
| 33795 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-3p | 29 | AKAP11 | Sponge network | 0.253 | 0.09565 | 0.196 | 0.09825 | 0.33 |
| 33796 | LINC00672 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-590-3p | 32 | NTRK2 | Sponge network | -0.227 | 0.31657 | -0.736 | 0.06495 | 0.33 |
| 33797 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p | 17 | MLLT10 | Sponge network | 0.331 | 0.57247 | 0.105 | 0.33292 | 0.33 |
| 33798 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 22 | LRIG1 | Sponge network | 0.222 | 0.29133 | -0.537 | 0.00588 | 0.33 |
| 33799 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | ABCB5 | Sponge network | -0.811 | 2.0E-5 | -0.956 | 0.04077 | 0.33 |
| 33800 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-625-5p;hsa-miR-93-5p | 28 | JAZF1 | Sponge network | 0.056 | 0.81195 | -0.377 | 0.02677 | 0.33 |
| 33801 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | EPHA3 | Sponge network | -0.663 | 0.10887 | 0.185 | 0.53588 | 0.33 |
| 33802 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-93-5p | 33 | NTRK3 | Sponge network | -0.663 | 0.10887 | -1.77 | 3.0E-5 | 0.33 |
| 33803 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | FAM188A | Sponge network | -2.46 | 0 | -0.44 | 0.00018 | 0.33 |
| 33804 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 17 | GPAM | Sponge network | 0.064 | 0.72934 | 0.142 | 0.29732 | 0.33 |
| 33805 | LPP-AS2 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-625-5p;hsa-miR-96-5p | 13 | SFRP1 | Sponge network | -0.18 | 0.20343 | -3.566 | 0 | 0.33 |
| 33806 | LINC00672 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | RADIL | Sponge network | -0.227 | 0.31657 | -1.628 | 0 | 0.33 |
| 33807 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-502-3p | 18 | ARID5B | Sponge network | 0.082 | 0.55397 | 0.342 | 0.01676 | 0.33 |
| 33808 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | PSIP1 | Sponge network | -1.575 | 6.0E-5 | -0.005 | 0.96897 | 0.33 |
| 33809 | HOXD-AS2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-3607-3p;hsa-miR-362-5p | 12 | IGF1 | Sponge network | -0.208 | 0.65592 | -0.204 | 0.5898 | 0.33 |
| 33810 | LINC00476 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-214-5p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-708-3p;hsa-miR-874-3p | 33 | MXI1 | Sponge network | -0.615 | 0.00015 | -0.987 | 0 | 0.33 |
| 33811 | RP11-967K21.1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | CSDE1 | Sponge network | 0.056 | 0.81195 | -0.493 | 0 | 0.33 |
| 33812 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p | 18 | SYNPO2 | Sponge network | -0.125 | 0.49414 | -2.342 | 0 | 0.33 |
| 33813 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-664a-3p | 19 | CNTNAP3 | Sponge network | 0.16 | 0.50785 | -1.465 | 0.00033 | 0.33 |
| 33814 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p | 11 | TGFBR2 | Sponge network | -1.582 | 8.0E-5 | -0.243 | 0.11408 | 0.33 |
| 33815 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -0.513 | 0.04703 | -0.243 | 0.11408 | 0.33 |
| 33816 | WDFY3-AS2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 26 | RYR2 | Sponge network | -0.942 | 0 | -1.466 | 0.00031 | 0.329 |
| 33817 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-93-5p | 22 | PLSCR4 | Sponge network | -1.645 | 0 | -0.474 | 0.03833 | 0.329 |
| 33818 | LINC00094 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-589-3p | 13 | PEG3 | Sponge network | 0.253 | 0.09565 | -1.74 | 0 | 0.329 |
| 33819 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p | 10 | FBXO31 | Sponge network | -0.222 | 0.32445 | -0.085 | 0.42389 | 0.329 |
| 33820 | RP11-967K21.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p | 17 | GJA1 | Sponge network | 0.056 | 0.81195 | -0.485 | 0.02179 | 0.329 |
| 33821 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-577 | 18 | PRKAB2 | Sponge network | 0.811 | 0.03228 | -0.367 | 0.01371 | 0.329 |
| 33822 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PTPN11 | Sponge network | 0.569 | 0.00067 | 0.431 | 2.0E-5 | 0.329 |
| 33823 | RP11-46C24.7 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MED12L | Sponge network | 0.392 | 0.0278 | -0.194 | 0.55299 | 0.329 |
| 33824 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p | 20 | BCL6 | Sponge network | 0.056 | 0.81195 | -0.211 | 0.19489 | 0.329 |
| 33825 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-98-5p | 27 | KCNA6 | Sponge network | -0.154 | 0.78939 | -1.531 | 0.00013 | 0.329 |
| 33826 | RP11-2H8.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-744-3p | 48 | LPP | Sponge network | 1.089 | 0 | -0.459 | 0.04475 | 0.329 |
| 33827 | NR2F1-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-92a-1-5p | 13 | PRKACA | Sponge network | -0.49 | 0.04946 | -0.255 | 0.0012 | 0.329 |
| 33828 | RP11-1006G14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | TGFBR3 | Sponge network | 0.559 | 0.00026 | -1.173 | 1.0E-5 | 0.329 |
| 33829 | RP11-403I13.8 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 24 | PCDH10 | Sponge network | 0.619 | 0.00157 | -1.644 | 0.0078 | 0.329 |
| 33830 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | RECK | Sponge network | 0.395 | 0.15451 | -1.043 | 0 | 0.329 |
| 33831 | H1FX-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 27 | BNC2 | Sponge network | -0.206 | 0.12942 | -0.491 | 0.09988 | 0.329 |
| 33832 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-93-5p | 14 | EPAS1 | Sponge network | -0.793 | 7.0E-5 | -0.184 | 0.14418 | 0.329 |
| 33833 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | GRIN2A | Sponge network | -0.668 | 3.0E-5 | -2.161 | 0 | 0.329 |
| 33834 | LINC00173 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-455-5p | 10 | ABCA10 | Sponge network | 0.056 | 0.8494 | -0.571 | 0.03674 | 0.329 |
| 33835 | RP11-479G22.8 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 13 | PRRX1 | Sponge network | 1.308 | 0 | 1.316 | 0 | 0.329 |
| 33836 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 41 | TRPS1 | Sponge network | -0.969 | 2.0E-5 | -0.191 | 0.44109 | 0.329 |
| 33837 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | PHC3 | Sponge network | -0.83 | 0 | 0.212 | 0.0499 | 0.329 |
| 33838 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | PDGFRA | Sponge network | -0.318 | 0.65847 | -0.372 | 0.0037 | 0.329 |
| 33839 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 14 | TRIM3 | Sponge network | -0.769 | 0.00035 | -0.577 | 0 | 0.329 |
| 33840 | OIP5-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 50 | PPM1L | Sponge network | 0.421 | 0.00084 | -0.537 | 0.00616 | 0.329 |
| 33841 | CALML3-AS1 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-625-5p | 11 | PPP2R2C | Sponge network | 0.95 | 0.15943 | -0.959 | 0.07428 | 0.329 |
| 33842 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | HOOK3 | Sponge network | -0.896 | 0.00099 | 0.273 | 0.09957 | 0.329 |
| 33843 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-590-3p | 20 | FOXO3 | Sponge network | 0.422 | 0.27021 | -0.04 | 0.72028 | 0.329 |
| 33844 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p | 47 | PAFAH1B1 | Sponge network | -0.615 | 0.00015 | -0.429 | 0 | 0.329 |
| 33845 | RP11-403I13.7 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p | 11 | MACF1 | Sponge network | 0.395 | 0.15451 | 0.019 | 0.90335 | 0.329 |
| 33846 | GAS6-AS2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-324-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-874-3p;hsa-miR-940 | 15 | SORCS1 | Sponge network | 0.16 | 0.50785 | -3.492 | 0 | 0.329 |
| 33847 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 29 | CADM1 | Sponge network | -0.942 | 0 | -0.9 | 0.00084 | 0.329 |
| 33848 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-582-5p | 15 | ZMYM2 | Sponge network | 0.389 | 0.04293 | 0.279 | 0.01273 | 0.329 |
| 33849 | RP5-994D16.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CBX5 | Sponge network | 0.252 | 0.08508 | 0.165 | 0.24446 | 0.329 |
| 33850 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-590-3p | 11 | DMD | Sponge network | 0.252 | 0.08508 | -1.506 | 0 | 0.329 |
| 33851 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-96-5p | 13 | CREB3L2 | Sponge network | -0.31 | 0.33871 | -0.525 | 2.0E-5 | 0.329 |
| 33852 | TINCR |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p | 15 | TMTC3 | Sponge network | -0.664 | 0.14543 | 0.123 | 0.22189 | 0.329 |
| 33853 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-935 | 29 | MTSS1 | Sponge network | -1.249 | 7.0E-5 | -0.81 | 0.00035 | 0.329 |
| 33854 | RP11-373D23.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-421;hsa-miR-7-1-3p | 10 | PEG3 | Sponge network | -0.285 | 0.2172 | -1.74 | 0 | 0.329 |
| 33855 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | FOXP1 | Sponge network | -0.976 | 0.00025 | 0.042 | 0.74368 | 0.329 |
| 33856 | RP11-53O19.1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-655-3p | 15 | RPS6KA6 | Sponge network | -0.405 | 0.22013 | -2.989 | 0 | 0.329 |
| 33857 | AC005618.6 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 25 | PCDH10 | Sponge network | 0.862 | 0.06213 | -1.644 | 0.0078 | 0.329 |
| 33858 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-942-5p | 34 | DDX6 | Sponge network | 0.401 | 0.00066 | 0.091 | 0.36428 | 0.329 |
| 33859 | LINC00641 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | NF1 | Sponge network | -0.222 | 0.32445 | 0.51 | 0 | 0.329 |
| 33860 | RP11-535M15.1 |
hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 12 | ESR1 | Sponge network | -1.14 | 0.03513 | -1.035 | 3.0E-5 | 0.329 |
| 33861 | RP11-46C24.7 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | INPP4A | Sponge network | 0.392 | 0.0278 | 0.07 | 0.53563 | 0.329 |
| 33862 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 15 | CHD1 | Sponge network | 1.063 | 0 | 0.322 | 0.00215 | 0.329 |
| 33863 | RP11-701H24.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 14 | SNX29 | Sponge network | -1.425 | 0.04204 | -0.34 | 0.01371 | 0.329 |
| 33864 | A1BG-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p | 10 | EBF1 | Sponge network | -0.164 | 0.45106 | -0.384 | 0.08856 | 0.329 |
| 33865 | AC074289.1 |
hsa-let-7d-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-942-5p | 11 | GLIPR1 | Sponge network | -0.326 | 0.14314 | 0.146 | 0.3276 | 0.329 |
| 33866 | RP11-35G9.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-940 | 18 | CREB1 | Sponge network | 0.622 | 0.00015 | 0.203 | 0.00537 | 0.329 |
| 33867 | MEG3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 27 | LRIG1 | Sponge network | 0.249 | 0.35902 | -0.537 | 0.00588 | 0.329 |
| 33868 | MIR143HG |
hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-7-5p | 10 | CMKLR1 | Sponge network | -0.739 | 0.0574 | 0.288 | 0.17621 | 0.329 |
| 33869 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p | 15 | AHNAK | Sponge network | -0.738 | 0.21233 | -1.207 | 0 | 0.329 |
| 33870 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-577 | 17 | GREM1 | Sponge network | -1.161 | 0.00591 | 0.902 | 0.01691 | 0.329 |
| 33871 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577 | 20 | GREM1 | Sponge network | -0.323 | 0.01372 | 0.902 | 0.01691 | 0.329 |
| 33872 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | ANK2 | Sponge network | 0.41 | 0.02214 | -1.603 | 1.0E-5 | 0.329 |
| 33873 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p | 13 | EBF1 | Sponge network | -1.116 | 0.23686 | -0.384 | 0.08856 | 0.329 |
| 33874 | RP11-693J15.4 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-942-5p | 10 | GLIPR1 | Sponge network | -0.72 | 0.24239 | 0.146 | 0.3276 | 0.329 |
| 33875 | MIR143HG |
hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-590-3p | 11 | EGR1 | Sponge network | -0.739 | 0.0574 | -1.195 | 0 | 0.329 |
| 33876 | AC010226.4 |
hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | CSDE1 | Sponge network | -0.811 | 2.0E-5 | -0.493 | 0 | 0.329 |
| 33877 | DNM3OS |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 31 | RIMS2 | Sponge network | 0.572 | 0.01722 | -1.365 | 0.00208 | 0.329 |
| 33878 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p | 20 | MYBL1 | Sponge network | 1.11 | 0 | 0.494 | 0.02152 | 0.329 |
| 33879 | CTC-444N24.11 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-379-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SH3GLB1 | Sponge network | 1.153 | 0 | -0.115 | 0.14773 | 0.329 |
| 33880 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | GALNT13 | Sponge network | -0.318 | 0.65847 | -0.857 | 0.07439 | 0.329 |
| 33881 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-500a-5p;hsa-miR-935 | 15 | DMD | Sponge network | 1.153 | 0.00114 | -1.506 | 0 | 0.329 |
| 33882 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-28-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-766-3p | 10 | PHACTR4 | Sponge network | 0.946 | 0 | 0.069 | 0.45203 | 0.329 |
| 33883 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | LATS1 | Sponge network | 0.643 | 0 | 0.109 | 0.34208 | 0.329 |
| 33884 | LINC00476 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 12 | ZNF844 | Sponge network | -0.615 | 0.00015 | -0.966 | 0.00035 | 0.329 |
| 33885 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | NIN | Sponge network | 0.222 | 0.29133 | -0.253 | 0.13794 | 0.329 |
| 33886 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | NR4A3 | Sponge network | -0.793 | 7.0E-5 | -1.385 | 0 | 0.329 |
| 33887 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 19 | CSMD1 | Sponge network | -1.349 | 0.00017 | -0.63 | 0.18881 | 0.329 |
| 33888 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b | 10 | MYCBP2 | Sponge network | 0.594 | 0.00064 | -0.027 | 0.82016 | 0.329 |
| 33889 | CTD-2336O2.1 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | SFRP1 | Sponge network | -0.141 | 0.26434 | -3.566 | 0 | 0.329 |
| 33890 | A2M-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TET1 | Sponge network | -0.08 | 0.90092 | -0.135 | 0.52138 | 0.329 |
| 33891 | AC025335.1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 16 | TBL1XR1 | Sponge network | 0.783 | 0 | 0.578 | 0 | 0.329 |
| 33892 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-33a-3p | 10 | SCN9A | Sponge network | -1.255 | 0.00981 | -0.525 | 0.10593 | 0.329 |
| 33893 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 15 | CRTC3 | Sponge network | 0.453 | 0.01839 | 0.164 | 0.16786 | 0.329 |
| 33894 | RP11-195F19.9 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-7-5p | 17 | IGFBP5 | Sponge network | -0.726 | 0.00132 | -0.806 | 0.00105 | 0.329 |
| 33895 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 50 | BMPR2 | Sponge network | 0.392 | 0.0278 | 0.206 | 0.0635 | 0.329 |
| 33896 | RP4-717I23.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-577 | 15 | NIN | Sponge network | 0.637 | 0.00069 | -0.253 | 0.13794 | 0.329 |
| 33897 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | PRDM5 | Sponge network | -0.612 | 0.00094 | -1.09 | 0.00014 | 0.329 |
| 33898 | LPP-AS2 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 10 | TPO | Sponge network | -0.18 | 0.20343 | -1.87 | 2.0E-5 | 0.329 |
| 33899 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ZFHX3 | Sponge network | -0.053 | 0.80685 | 0.389 | 0.01363 | 0.329 |
| 33900 | RP11-263K19.4 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-629-3p | 12 | RASSF8 | Sponge network | 0.859 | 0.00331 | -0.122 | 0.65591 | 0.329 |
| 33901 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-30b-3p | 13 | GPC6 | Sponge network | -0.726 | 0.00132 | -0.054 | 0.81465 | 0.329 |
| 33902 | RP5-1198O20.4 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 23 | ATM | Sponge network | 0.997 | 0.00066 | 0.178 | 0.21568 | 0.329 |
| 33903 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 16 | TCF4 | Sponge network | 1.969 | 0.00316 | 0.016 | 0.92271 | 0.329 |
| 33904 | RP11-2H8.2 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p | 14 | RASAL2 | Sponge network | 1.089 | 0 | 0.869 | 0 | 0.329 |
| 33905 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 32 | SASH1 | Sponge network | -0.793 | 7.0E-5 | -0.502 | 0.00175 | 0.329 |
| 33906 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 19 | IGF2 | Sponge network | 0.997 | 0.00066 | 0.848 | 0.03842 | 0.329 |
| 33907 | CTD-2583A14.8 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 11 | PHF6 | Sponge network | 1.134 | 0 | 0.785 | 0 | 0.329 |
| 33908 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | USP12 | Sponge network | 0.91 | 0 | -0.102 | 0.40768 | 0.329 |
| 33909 | RP11-400K9.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-940 | 16 | AFF4 | Sponge network | -0.733 | 0.19469 | -0.266 | 0.01565 | 0.329 |
| 33910 | FZD10-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 26 | TET1 | Sponge network | -0.727 | 0.04726 | -0.135 | 0.52138 | 0.329 |
| 33911 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 25 | CYLD | Sponge network | -0.576 | 0.10121 | -0.367 | 0.00171 | 0.329 |
| 33912 | CTA-941F9.9 |
hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p | 10 | ZFHX3 | Sponge network | -0.344 | 0.49624 | 0.389 | 0.01363 | 0.329 |
| 33913 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | SETBP1 | Sponge network | 0.72 | 0.0001 | -1.302 | 1.0E-5 | 0.329 |
| 33914 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-7-1-3p | 14 | DACH2 | Sponge network | -0.563 | 0.02545 | -1.635 | 0.00034 | 0.329 |
| 33915 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 40 | SSBP2 | Sponge network | 0.4 | 0.14611 | -1.58 | 0 | 0.329 |
| 33916 | RP11-927P21.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-629-3p | 15 | PTPN14 | Sponge network | 0.921 | 0.00118 | 0.144 | 0.34595 | 0.329 |
| 33917 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | EPHA5 | Sponge network | -0.342 | 0.02288 | -0.957 | 0.01733 | 0.329 |
| 33918 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | ST5 | Sponge network | -0.222 | 0.32445 | -0.78 | 0 | 0.329 |
| 33919 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | AKAP12 | Sponge network | 0.657 | 4.0E-5 | -0.932 | 0.0028 | 0.329 |
| 33920 | LINC00092 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-21-5p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | NIN | Sponge network | -1.886 | 0 | -0.253 | 0.13794 | 0.329 |
| 33921 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-424-5p;hsa-miR-96-5p | 10 | LATS2 | Sponge network | -0.187 | 0.23787 | 0.159 | 0.2304 | 0.329 |
| 33922 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 11 | CAV1 | Sponge network | -0.342 | 0.02288 | -1.013 | 6.0E-5 | 0.329 |
| 33923 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 13 | FREM2 | Sponge network | 0.853 | 0.15352 | -0.437 | 0.46353 | 0.329 |
| 33924 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-5p;hsa-miR-877-5p;hsa-miR-940 | 21 | CRTC3 | Sponge network | -0.612 | 0.00094 | 0.164 | 0.16786 | 0.329 |
| 33925 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | MCC | Sponge network | 0.083 | 0.66405 | -1.005 | 0 | 0.329 |
| 33926 | RP4-639F20.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p | 14 | BRD3 | Sponge network | -0.517 | 0.02935 | 0.37 | 0.00013 | 0.329 |
| 33927 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 26 | CBL | Sponge network | 0.657 | 4.0E-5 | 0.291 | 0.01267 | 0.329 |
| 33928 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | SLITRK3 | Sponge network | 0.246 | 0.59912 | -3.328 | 0 | 0.329 |
| 33929 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 34 | TMEM132B | Sponge network | -1.057 | 0.00029 | -0.83 | 0.01084 | 0.329 |
| 33930 | RP11-5C23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-429;hsa-miR-590-3p | 12 | RBL2 | Sponge network | 0.274 | 0.07681 | -0.282 | 0.00254 | 0.329 |
| 33931 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-30e-5p;hsa-miR-532-3p;hsa-miR-93-3p | 13 | RPS6KA2 | Sponge network | -0.513 | 0.04703 | -0.57 | 0.00013 | 0.329 |
| 33932 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-339-5p | 12 | MLLT10 | Sponge network | 0.373 | 0.00068 | 0.105 | 0.33292 | 0.329 |
| 33933 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p | 31 | FAT3 | Sponge network | 0.389 | 0.04293 | -1.601 | 0.00051 | 0.329 |
| 33934 | AC009948.5 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 17 | IRS1 | Sponge network | 0.532 | 0.00505 | -0.026 | 0.88855 | 0.329 |
| 33935 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | NEDD4 | Sponge network | -0.487 | 0.02157 | 0.02 | 0.89834 | 0.329 |
| 33936 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 24 | CNTN1 | Sponge network | 0.4 | 0.14611 | -2.594 | 0 | 0.329 |
| 33937 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 41 | AR | Sponge network | -0.125 | 0.49414 | -1.554 | 0 | 0.329 |
| 33938 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | SLC6A15 | Sponge network | -0.129 | 0.57177 | -2.373 | 2.0E-5 | 0.329 |
| 33939 | RP11-967K21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-940 | 24 | CREB1 | Sponge network | 0.056 | 0.81195 | 0.203 | 0.00537 | 0.329 |
| 33940 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | CREB3L2 | Sponge network | 0.572 | 0.01722 | -0.525 | 2.0E-5 | 0.329 |
| 33941 | SH3RF3-AS1 |
hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | PIK3CA | Sponge network | 0.889 | 9.0E-5 | 0.195 | 0.06908 | 0.329 |
| 33942 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 30 | BHLHE41 | Sponge network | -2.452 | 0 | 0.353 | 0.12753 | 0.329 |
| 33943 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | ST6GAL2 | Sponge network | -0.096 | 0.67958 | -1.201 | 0.00597 | 0.329 |
| 33944 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-181b-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-484;hsa-miR-590-3p | 11 | PKNOX1 | Sponge network | -0.342 | 0.02288 | 0.085 | 0.28917 | 0.329 |
| 33945 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577 | 22 | DMXL1 | Sponge network | 0.267 | 0.2937 | -0.426 | 0.00056 | 0.329 |
| 33946 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | LRRC55 | Sponge network | -2.915 | 0.00015 | -0.026 | 0.93163 | 0.329 |
| 33947 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-511-5p | 16 | EYA4 | Sponge network | 0.622 | 0.00015 | 0.3 | 0.36876 | 0.329 |
| 33948 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-576-5p | 26 | EPHA5 | Sponge network | 0.176 | 0.37402 | -0.957 | 0.01733 | 0.329 |
| 33949 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | FYN | Sponge network | -0.08 | 0.90092 | -0.131 | 0.37051 | 0.329 |
| 33950 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-342-5p;hsa-miR-374a-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 17 | BMPR2 | Sponge network | -0.557 | 0.11135 | 0.206 | 0.0635 | 0.329 |
| 33951 | LINC00094 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429 | 20 | SOX11 | Sponge network | 0.253 | 0.09565 | 2.68 | 0 | 0.329 |
| 33952 | CTC-510F12.2 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-484 | 10 | CTIF | Sponge network | -0.691 | 0.00146 | -0.389 | 0.00549 | 0.329 |
| 33953 | BDNF-AS |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | SLITRK3 | Sponge network | -0.668 | 3.0E-5 | -3.328 | 0 | 0.329 |
| 33954 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 10 | ZNF208 | Sponge network | -0.08 | 0.90092 | -0.4 | 0.22381 | 0.329 |
| 33955 | RP11-867G23.10 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p | 15 | PTPN14 | Sponge network | -2.531 | 0 | 0.144 | 0.34595 | 0.329 |
| 33956 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p | 24 | FOXO3 | Sponge network | 0.862 | 0.06213 | -0.04 | 0.72028 | 0.329 |
| 33957 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 13 | GLI3 | Sponge network | -1.057 | 0.00029 | -0.615 | 0.01482 | 0.329 |
| 33958 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 34 | NOTCH2 | Sponge network | 0.349 | 0.01695 | 0.138 | 0.35163 | 0.329 |
| 33959 | RP11-506M13.3 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 0.769 | 0 | 0.51 | 0 | 0.329 |
| 33960 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | DMXL1 | Sponge network | 0.873 | 0 | -0.426 | 0.00056 | 0.329 |
| 33961 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p | 11 | HECA | Sponge network | 0.453 | 0.01839 | -0.217 | 0.03463 | 0.329 |
| 33962 | BDNF-AS |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 52 | CADM2 | Sponge network | -0.668 | 3.0E-5 | -3.782 | 0 | 0.329 |
| 33963 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p | 39 | TMEM132B | Sponge network | -0.303 | 0.21002 | -0.83 | 0.01084 | 0.329 |
| 33964 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 24 | NAV3 | Sponge network | -0.355 | 0.0077 | 0.212 | 0.47729 | 0.329 |
| 33965 | RP1-102E24.8 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-664a-5p | 16 | SOX11 | Sponge network | 1.737 | 0 | 2.68 | 0 | 0.329 |
| 33966 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PBRM1 | Sponge network | 0.539 | 0.03722 | -0.029 | 0.78601 | 0.329 |
| 33967 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | NCOA1 | Sponge network | 0.349 | 0.01695 | -0.432 | 0 | 0.329 |
| 33968 | LINC00957 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | PPM1A | Sponge network | -0.421 | 0.0114 | -0.623 | 0 | 0.329 |
| 33969 | RP11-48B3.4 |
hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 11 | GLI3 | Sponge network | 1.757 | 0 | -0.615 | 0.01482 | 0.329 |
| 33970 | SNHG14 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-708-5p | 21 | FAM135B | Sponge network | -1.111 | 0.0003 | -3.978 | 0 | 0.329 |
| 33971 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 21 | PRKAB2 | Sponge network | 0.663 | 0.00073 | -0.367 | 0.01371 | 0.329 |
| 33972 | EIF3J-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ZFHX3 | Sponge network | 0.331 | 0.00509 | 0.389 | 0.01363 | 0.329 |
| 33973 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 33 | RBMS3 | Sponge network | -0.176 | 0.59692 | -0.519 | 0.03551 | 0.329 |
| 33974 | RP11-403I13.7 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-532-3p | 16 | MRVI1 | Sponge network | 0.395 | 0.15451 | -1.051 | 0.00022 | 0.329 |
| 33975 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 56 | NFIB | Sponge network | 0.311 | 0.10344 | 0.023 | 0.90795 | 0.329 |
| 33976 | RP11-686D22.4 |
hsa-miR-106a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-582-3p;hsa-miR-590-3p | 11 | JAZF1 | Sponge network | 0.793 | 5.0E-5 | -0.377 | 0.02677 | 0.329 |
| 33977 | MEG3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 57 | NFIB | Sponge network | 0.249 | 0.35902 | 0.023 | 0.90795 | 0.329 |
| 33978 | RP11-356J5.12 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | PRKCB | Sponge network | -0.899 | 6.0E-5 | -1.313 | 2.0E-5 | 0.329 |
| 33979 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 18 | PRKAR1A | Sponge network | -0.323 | 0.01372 | -0.063 | 0.50314 | 0.329 |
| 33980 | RP11-401P9.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-539-5p;hsa-miR-625-5p | 12 | ABI2 | Sponge network | 1.524 | 0.1098 | 0.175 | 0.14787 | 0.329 |
| 33981 | HOXD-AS2 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-338-3p | 14 | ZFHX4 | Sponge network | -0.208 | 0.65592 | 0.018 | 0.95574 | 0.329 |
| 33982 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 11 | KALRN | Sponge network | -1.092 | 4.0E-5 | -0.819 | 1.0E-5 | 0.329 |
| 33983 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-590-3p | 22 | ABL2 | Sponge network | -0.323 | 0.01372 | 0.82 | 0 | 0.329 |
| 33984 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-370-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | JAZF1 | Sponge network | -0.663 | 0.10887 | -0.377 | 0.02677 | 0.329 |
| 33985 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | GNA13 | Sponge network | 0.873 | 0 | 0.007 | 0.93884 | 0.329 |
| 33986 | A2M-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | PAK3 | Sponge network | -0.08 | 0.90092 | -0.628 | 0.07253 | 0.329 |
| 33987 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | SASH1 | Sponge network | -2.117 | 0.00161 | -0.502 | 0.00175 | 0.329 |
| 33988 | EMX2OS |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | SYT4 | Sponge network | -0.777 | 0.1134 | -3.207 | 0 | 0.329 |
| 33989 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-96-5p;hsa-miR-98-5p | 41 | CREB3L2 | Sponge network | 0.101 | 0.60919 | -0.525 | 2.0E-5 | 0.329 |
| 33990 | RP11-401P9.4 |
hsa-let-7f-1-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-590-5p;hsa-miR-93-3p | 11 | EMP2 | Sponge network | -0.384 | 0.40652 | -0.423 | 0.00985 | 0.329 |
| 33991 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-93-5p;hsa-miR-98-5p | 23 | PRRX1 | Sponge network | -0.154 | 0.78939 | 1.316 | 0 | 0.329 |
| 33992 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 23 | NF1 | Sponge network | 1.11 | 0 | 0.51 | 0 | 0.329 |
| 33993 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | USP12 | Sponge network | 1.254 | 0 | -0.102 | 0.40768 | 0.329 |
| 33994 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | MLLT10 | Sponge network | 0.919 | 0 | 0.105 | 0.33292 | 0.329 |
| 33995 | GNG12-AS1 |
hsa-miR-127-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 14 | JUN | Sponge network | -0.793 | 7.0E-5 | -0.932 | 0 | 0.329 |
| 33996 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-452-5p | 18 | IL6ST | Sponge network | -3.586 | 0.00032 | -0.402 | 0.00676 | 0.329 |
| 33997 | LINC00702 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | HECA | Sponge network | -0.737 | 0.06182 | -0.217 | 0.03463 | 0.329 |
| 33998 | CTD-2336O2.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | FAM172A | Sponge network | -0.141 | 0.26434 | -0.57 | 0 | 0.329 |
| 33999 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 38 | AKAP11 | Sponge network | -0.739 | 0.0574 | 0.196 | 0.09825 | 0.329 |
| 34000 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p | 16 | NTRK2 | Sponge network | 0.37 | 0.27614 | -0.736 | 0.06495 | 0.329 |
| 34001 | ZNF582-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-7-1-3p | 15 | JDP2 | Sponge network | -1.349 | 0.00017 | -0.967 | 0 | 0.329 |
| 34002 | AC016747.3 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p | 19 | DOCK4 | Sponge network | 0.199 | 0.38015 | 0.502 | 0.00108 | 0.329 |
| 34003 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-493-5p | 17 | PHC3 | Sponge network | 0.368 | 0.20093 | 0.212 | 0.0499 | 0.329 |
| 34004 | LINC01001 |
hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a | 11 | FOXO3 | Sponge network | 1.055 | 0 | -0.04 | 0.72028 | 0.329 |
| 34005 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-629-3p | 16 | NTRK3 | Sponge network | 1.969 | 0.00316 | -1.77 | 3.0E-5 | 0.329 |
| 34006 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | TCF12 | Sponge network | 0.559 | 0.00026 | 0.174 | 0.13271 | 0.329 |
| 34007 | PWAR6 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 12 | FAM133A | Sponge network | -1.575 | 6.0E-5 | 0.549 | 0.29009 | 0.329 |
| 34008 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | LRIG1 | Sponge network | 0.532 | 0.00505 | -0.537 | 0.00588 | 0.329 |
| 34009 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | IGSF10 | Sponge network | -0.141 | 0.26434 | -1.751 | 1.0E-5 | 0.329 |
| 34010 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-370-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-93-5p | 35 | JAZF1 | Sponge network | -1.047 | 0 | -0.377 | 0.02677 | 0.329 |
| 34011 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-671-5p | 11 | PIK3IP1 | Sponge network | -2.69 | 0.0001 | -0.187 | 0.19945 | 0.329 |
| 34012 | THAP9-AS1 |
hsa-miR-100-5p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p | 12 | RB1 | Sponge network | 1.079 | 0 | 0.382 | 0.0017 | 0.329 |
| 34013 | CTD-3157E16.1 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-29b-1-5p | 10 | LPP | Sponge network | -0.326 | 0.28809 | -0.459 | 0.04475 | 0.329 |
| 34014 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 14 | DKK3 | Sponge network | 0.385 | 0.10067 | -0.052 | 0.77783 | 0.329 |
| 34015 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-5p | 20 | NIN | Sponge network | 0.331 | 0.57247 | -0.253 | 0.13794 | 0.329 |
| 34016 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 28 | SSBP2 | Sponge network | -0.206 | 0.12942 | -1.58 | 0 | 0.329 |
| 34017 | AC093627.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26b-5p | 10 | CACNA2D1 | Sponge network | -0.787 | 0.3928 | -0.846 | 0.00675 | 0.329 |
| 34018 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-629-3p | 15 | CHD2 | Sponge network | 1.03 | 0 | 0.049 | 0.55371 | 0.329 |
| 34019 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 19 | RPS6KA2 | Sponge network | -2.46 | 0 | -0.57 | 0.00013 | 0.329 |
| 34020 | HOXA-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | 0.61 | 0.01593 | -0.564 | 1.0E-5 | 0.329 |
| 34021 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p | 11 | PBRM1 | Sponge network | 0.799 | 1.0E-5 | -0.029 | 0.78601 | 0.329 |
| 34022 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-7-5p | 17 | SNX29 | Sponge network | -0.726 | 0.00132 | -0.34 | 0.01371 | 0.329 |
| 34023 | RP11-410L14.2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-497-5p | 10 | PRKAB2 | Sponge network | -0.052 | 0.76839 | -0.367 | 0.01371 | 0.329 |
| 34024 | MIR22HG |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-3p;hsa-miR-532-3p;hsa-miR-542-3p | 18 | SYNM | Sponge network | -1.047 | 0 | -2.309 | 1.0E-5 | 0.329 |
| 34025 | LINC00654 |
hsa-miR-126-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | CXCL12 | Sponge network | 0.374 | 0.20054 | -1.421 | 0 | 0.329 |
| 34026 | AC034220.3 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 11 | RAPGEF5 | Sponge network | 0.309 | 0.07304 | 0.536 | 4.0E-5 | 0.329 |
| 34027 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p | 21 | MAF | Sponge network | -0.693 | 0.05629 | -1.257 | 0 | 0.329 |
| 34028 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TSC1 | Sponge network | 0.225 | 0.30644 | 0.103 | 0.26071 | 0.329 |
| 34029 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | MAF | Sponge network | -2.117 | 0.00161 | -1.257 | 0 | 0.329 |
| 34030 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-96-5p | 23 | CADM1 | Sponge network | 0.41 | 0.02214 | -0.9 | 0.00084 | 0.329 |
| 34031 | RP11-855A2.3 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 22 | LPP | Sponge network | 2.094 | 0 | -0.459 | 0.04475 | 0.329 |
| 34032 | RP11-150O12.1 |
hsa-let-7g-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p | 11 | BRD3 | Sponge network | 2.605 | 2.0E-5 | 0.37 | 0.00013 | 0.329 |
| 34033 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p | 29 | NTRK2 | Sponge network | -1.654 | 0.00144 | -0.736 | 0.06495 | 0.329 |
| 34034 | LINC00578 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p | 10 | ZMYND11 | Sponge network | 0.811 | 0.03228 | -0.2 | 0.05449 | 0.329 |
| 34035 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 12 | HECA | Sponge network | -1.331 | 3.0E-5 | -0.217 | 0.03463 | 0.329 |
| 34036 | AC109309.4 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-3p | 17 | DST | Sponge network | 0.163 | 0.87219 | -0.713 | 0.00096 | 0.329 |
| 34037 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | CHD2 | Sponge network | -0.227 | 0.31657 | 0.049 | 0.55371 | 0.329 |
| 34038 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 23 | ACVR1 | Sponge network | -1.697 | 0.00889 | 0.279 | 0.00234 | 0.329 |
| 34039 | RP11-356J5.12 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | RASSF3 | Sponge network | -0.899 | 6.0E-5 | -0.226 | 0.11691 | 0.329 |
| 34040 | MIR143HG |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-493-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-874-3p | 17 | ARID1B | Sponge network | -0.739 | 0.0574 | 0.003 | 0.97503 | 0.329 |
| 34041 | RP1-302G2.5 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-224-5p;hsa-miR-26b-3p | 12 | ZFHX4 | Sponge network | -1.483 | 9.0E-5 | 0.018 | 0.95574 | 0.329 |
| 34042 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p | 10 | SASH1 | Sponge network | 1.316 | 0.01106 | -0.502 | 0.00175 | 0.329 |
| 34043 | C4A-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 20 | BNC2 | Sponge network | 3.345 | 0 | -0.491 | 0.09988 | 0.329 |
| 34044 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 46 | SASH1 | Sponge network | -1.697 | 0.00889 | -0.502 | 0.00175 | 0.329 |
| 34045 | AC074289.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p | 13 | FMNL3 | Sponge network | -0.326 | 0.14314 | 0.555 | 4.0E-5 | 0.329 |
| 34046 | TTC28-AS1 |
hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-655-3p;hsa-miR-664a-5p | 10 | SHPRH | Sponge network | 0.373 | 0.00068 | 0.098 | 0.40994 | 0.329 |
| 34047 | CTB-133G6.2 |
hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p | 11 | ZFHX3 | Sponge network | 0.4 | 0.39299 | 0.389 | 0.01363 | 0.329 |
| 34048 | RP11-21A7A.2 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-625-5p | 11 | MCC | Sponge network | -1.116 | 0.23686 | -1.005 | 0 | 0.328 |
| 34049 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | EPAS1 | Sponge network | 0.335 | 0.20596 | -0.184 | 0.14418 | 0.328 |
| 34050 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-98-5p | 13 | ITGA5 | Sponge network | 0.246 | 0.59912 | 0.452 | 0.03524 | 0.328 |
| 34051 | RP11-434B12.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DNAJB4 | Sponge network | -0.031 | 0.89063 | -0.7 | 1.0E-5 | 0.328 |
| 34052 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | AKAP6 | Sponge network | -0.206 | 0.12942 | -1.586 | 3.0E-5 | 0.328 |
| 34053 | RP11-693J15.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-7-1-3p | 16 | SYT4 | Sponge network | -0.72 | 0.24239 | -3.207 | 0 | 0.328 |
| 34054 | RP11-46C24.7 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PIK3CA | Sponge network | 0.392 | 0.0278 | 0.195 | 0.06908 | 0.328 |
| 34055 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | PREX2 | Sponge network | -0.355 | 0.0077 | -0.272 | 0.25382 | 0.328 |
| 34056 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 24 | SCN9A | Sponge network | 0.494 | 0.00339 | -0.525 | 0.10593 | 0.328 |
| 34057 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p | 11 | CHD1 | Sponge network | 0.817 | 3.0E-5 | 0.322 | 0.00215 | 0.328 |
| 34058 | GNG12-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | ITGB1 | Sponge network | -0.793 | 7.0E-5 | 0.524 | 0.00017 | 0.328 |
| 34059 | LINC00641 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | EPG5 | Sponge network | -0.222 | 0.32445 | 0.101 | 0.3563 | 0.328 |
| 34060 | RP11-305E6.4 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 24 | CCDC6 | Sponge network | 1.317 | 0 | 0.27 | 0.02555 | 0.328 |
| 34061 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 14 | NF1 | Sponge network | 0.702 | 7.0E-5 | 0.51 | 0 | 0.328 |
| 34062 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p | 15 | AFF3 | Sponge network | 0.622 | 0.00015 | -2.373 | 0 | 0.328 |
| 34063 | LINC00173 |
hsa-miR-126-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-625-5p;hsa-miR-96-5p | 12 | CXCL12 | Sponge network | 0.056 | 0.8494 | -1.421 | 0 | 0.328 |
| 34064 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-877-5p | 18 | PGR | Sponge network | -0.125 | 0.49414 | -1.704 | 0 | 0.328 |
| 34065 | HNRNPU-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-655-3p;hsa-miR-7-1-3p | 12 | UBE4B | Sponge network | 1.601 | 0 | -0.235 | 0.007 | 0.328 |
| 34066 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | HACE1 | Sponge network | -0.612 | 0.00094 | -0.072 | 0.55239 | 0.328 |
| 34067 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | MITF | Sponge network | 1.253 | 0 | -0.769 | 0.00022 | 0.328 |
| 34068 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | 0.349 | 0.01695 | -0.466 | 0.14121 | 0.328 |
| 34069 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 30 | KIF1B | Sponge network | 0.826 | 1.0E-5 | -0.118 | 0.32258 | 0.328 |
| 34070 | RP11-166D19.1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 53 | PPM1L | Sponge network | -0.576 | 0.10121 | -0.537 | 0.00616 | 0.328 |
| 34071 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-5p | 19 | EPHA3 | Sponge network | 0.921 | 0.00118 | 0.185 | 0.53588 | 0.328 |
| 34072 | EMX2OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 12 | PLA2R1 | Sponge network | -0.777 | 0.1134 | -0.265 | 0.24676 | 0.328 |
| 34073 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 15 | USP6 | Sponge network | 0.174 | 0.30034 | 0.188 | 0.3871 | 0.328 |
| 34074 | FGD5-AS1 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | TMEM30A | Sponge network | 0.401 | 0.00066 | 0.096 | 0.31031 | 0.328 |
| 34075 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p | 26 | NCOA1 | Sponge network | 0.494 | 0.00339 | -0.432 | 0 | 0.328 |
| 34076 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | MTSS1 | Sponge network | 0.225 | 0.30644 | -0.81 | 0.00035 | 0.328 |
| 34077 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-92b-3p | 16 | AKAP12 | Sponge network | -0.355 | 0.0077 | -0.932 | 0.0028 | 0.328 |
| 34078 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p | 17 | KIT | Sponge network | -0.322 | 0.25945 | -2.184 | 0 | 0.328 |
| 34079 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-584-5p | 23 | ST6GAL2 | Sponge network | 0.331 | 0.57247 | -1.201 | 0.00597 | 0.328 |
| 34080 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-5p | 18 | ZBTB4 | Sponge network | -0.663 | 0.10887 | -0.739 | 0 | 0.328 |
| 34081 | DNAJC3-AS1 |
hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p | 15 | TBL1XR1 | Sponge network | 0.753 | 0.00014 | 0.578 | 0 | 0.328 |
| 34082 | CTD-2303H24.2 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | TSC1 | Sponge network | 0.415 | 0.14657 | 0.103 | 0.26071 | 0.328 |
| 34083 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-429 | 12 | LHX6 | Sponge network | 1.302 | 7.0E-5 | -0.087 | 0.69193 | 0.328 |
| 34084 | OIP5-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 31 | RASSF8 | Sponge network | 0.421 | 0.00084 | -0.122 | 0.65591 | 0.328 |
| 34085 | LINC00092 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-940 | 25 | TRPS1 | Sponge network | -1.886 | 0 | -0.191 | 0.44109 | 0.328 |
| 34086 | RP4-717I23.3 |
hsa-miR-10b-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-485-3p;hsa-miR-576-5p | 22 | UBR3 | Sponge network | 0.637 | 0.00069 | -0.021 | 0.82774 | 0.328 |
| 34087 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-940 | 25 | PBX1 | Sponge network | 0.537 | 0.00139 | -1.206 | 0 | 0.328 |
| 34088 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-590-3p | 14 | RBM5 | Sponge network | -0.615 | 0.00015 | -0.205 | 0.0129 | 0.328 |
| 34089 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-335-5p;hsa-miR-339-5p | 17 | RUNX1T1 | Sponge network | -0.421 | 0.12556 | -0.344 | 0.2374 | 0.328 |
| 34090 | LINC00654 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 14 | ITGA5 | Sponge network | 0.374 | 0.20054 | 0.452 | 0.03524 | 0.328 |
| 34091 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 34 | ESR1 | Sponge network | -0.709 | 0.18168 | -1.035 | 3.0E-5 | 0.328 |
| 34092 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SEMA3C | Sponge network | -0.487 | 0.02157 | -0.272 | 0.31153 | 0.328 |
| 34093 | RP11-359B12.2 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | PCM1 | Sponge network | 0.064 | 0.72934 | 0.164 | 0.20307 | 0.328 |
| 34094 | RP11-967K21.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-940 | 11 | SORCS1 | Sponge network | 0.056 | 0.81195 | -3.492 | 0 | 0.328 |
| 34095 | RP11-760H22.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-7-5p | 34 | LPP | Sponge network | 0.001 | 0.99753 | -0.459 | 0.04475 | 0.328 |
| 34096 | RP11-678G14.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p | 11 | RUNX1T1 | Sponge network | -1.765 | 0.0778 | -0.344 | 0.2374 | 0.328 |
| 34097 | CTC-457L16.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-671-5p | 14 | CDH23 | Sponge network | 1.153 | 0.00114 | -0.974 | 0.00034 | 0.328 |
| 34098 | RP11-13K12.5 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-629-3p | 11 | SPTBN4 | Sponge network | -0.318 | 0.65847 | -0.786 | 0.01281 | 0.328 |
| 34099 | RP11-384K6.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-744-3p | 14 | PTPN14 | Sponge network | 0.946 | 0 | 0.144 | 0.34595 | 0.328 |
| 34100 | RP11-890B15.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 34 | IGFBP5 | Sponge network | 0.101 | 0.60919 | -0.806 | 0.00105 | 0.328 |
| 34101 | RP11-999E24.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p | 12 | LATS2 | Sponge network | -0.693 | 0.05629 | 0.159 | 0.2304 | 0.328 |
| 34102 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | NEDD4 | Sponge network | 0.75 | 0.00054 | 0.02 | 0.89834 | 0.328 |
| 34103 | CTC-429P9.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p | 11 | ARID5B | Sponge network | 0.497 | 0.01451 | 0.342 | 0.01676 | 0.328 |
| 34104 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 18 | NEDD4 | Sponge network | 0.082 | 0.55654 | 0.02 | 0.89834 | 0.328 |
| 34105 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 40 | QKI | Sponge network | 0.659 | 3.0E-5 | -0.333 | 0.04111 | 0.328 |
| 34106 | HOXB-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-429;hsa-miR-93-3p | 14 | DOCK4 | Sponge network | -0.154 | 0.53954 | 0.502 | 0.00108 | 0.328 |
| 34107 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | ABI2 | Sponge network | 0.998 | 0 | 0.175 | 0.14787 | 0.328 |
| 34108 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-362-5p;hsa-miR-590-3p | 10 | TLE4 | Sponge network | -0.663 | 0.10887 | -1.157 | 0 | 0.328 |
| 34109 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-629-5p;hsa-miR-93-5p | 31 | MAF | Sponge network | -1.047 | 0 | -1.257 | 0 | 0.328 |
| 34110 | RP11-890B15.3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | BCL9 | Sponge network | 0.101 | 0.60919 | 0.321 | 0.00267 | 0.328 |
| 34111 | RP11-571M6.17 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ZFX | Sponge network | 1.013 | 0 | 0.09 | 0.42563 | 0.328 |
| 34112 | AC141928.1 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-7-1-3p | 18 | PCDH9 | Sponge network | 0.853 | 0.15352 | -1.823 | 4.0E-5 | 0.328 |
| 34113 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | CXXC4 | Sponge network | 0.311 | 0.10344 | -0.433 | 0.21055 | 0.328 |
| 34114 | RP5-994D16.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SMAD4 | Sponge network | 0.252 | 0.08508 | -0.313 | 0.00413 | 0.328 |
| 34115 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-539-5p | 11 | MGA | Sponge network | 0.41 | 0.02214 | 0.323 | 0.00792 | 0.328 |
| 34116 | BVES-AS1 |
hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-330-5p;hsa-miR-877-5p | 13 | CRTC1 | Sponge network | -2.62 | 0 | -0.116 | 0.28412 | 0.328 |
| 34117 | U91328.19 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 23 | PDE4DIP | Sponge network | -0.303 | 0.21002 | -0.282 | 0.0257 | 0.328 |
| 34118 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 22 | ACVR1 | Sponge network | -0.737 | 0.06182 | 0.279 | 0.00234 | 0.328 |
| 34119 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 15 | PAFAH1B1 | Sponge network | -0.295 | 0.02604 | -0.429 | 0 | 0.328 |
| 34120 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-628-5p | 26 | BMPR2 | Sponge network | 0.792 | 0.0049 | 0.206 | 0.0635 | 0.328 |
| 34121 | RP11-705C15.3 |
hsa-let-7g-3p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p | 14 | ABL2 | Sponge network | 0.796 | 4.0E-5 | 0.82 | 0 | 0.328 |
| 34122 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | YAP1 | Sponge network | 0.673 | 0 | 0.22 | 0.11836 | 0.328 |
| 34123 | RP11-152F13.8 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p | 13 | KIF1B | Sponge network | 0.843 | 0.01775 | -0.118 | 0.32258 | 0.328 |
| 34124 | AP001347.6 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PCDH18 | Sponge network | -1.161 | 0.00591 | 0.216 | 0.24645 | 0.328 |
| 34125 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | MAF | Sponge network | -2.915 | 0.00015 | -1.257 | 0 | 0.328 |
| 34126 | RP11-38L15.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 12 | NTRK2 | Sponge network | 0.344 | 0.3127 | -0.736 | 0.06495 | 0.328 |
| 34127 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | AR | Sponge network | -0.351 | 0.11358 | -1.554 | 0 | 0.328 |
| 34128 | CTB-179K24.3 |
hsa-miR-125b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | ZMYM2 | Sponge network | 0.841 | 0.04055 | 0.279 | 0.01273 | 0.328 |
| 34129 | DNM3OS |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | BRD3 | Sponge network | 0.572 | 0.01722 | 0.37 | 0.00013 | 0.328 |
| 34130 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 56 | THRB | Sponge network | 0.194 | 0.64278 | -1.117 | 0.0004 | 0.328 |
| 34131 | RP11-326G21.1 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DIP2C | Sponge network | -0.021 | 0.93598 | -0.436 | 0.01713 | 0.328 |
| 34132 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 13 | FAM188A | Sponge network | -0.323 | 0.01372 | -0.44 | 0.00018 | 0.328 |
| 34133 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-454-3p | 20 | HECA | Sponge network | -2.69 | 0.0001 | -0.217 | 0.03463 | 0.328 |
| 34134 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-98-5p | 12 | ARHGAP28 | Sponge network | 0.342 | 0.03129 | -0.911 | 0.00013 | 0.328 |
| 34135 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | PRRX1 | Sponge network | -0.738 | 0.21233 | 1.316 | 0 | 0.328 |
| 34136 | RP11-798M19.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-375;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 45 | GAS7 | Sponge network | -0.612 | 0.00094 | -0.555 | 0.01602 | 0.328 |
| 34137 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 15 | GPC6 | Sponge network | 0.727 | 3.0E-5 | -0.054 | 0.81465 | 0.328 |
| 34138 | HAR1A |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 14 | ESR1 | Sponge network | -0.672 | 0.19447 | -1.035 | 3.0E-5 | 0.328 |
| 34139 | AC003090.1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | IRS1 | Sponge network | -2.117 | 0.00161 | -0.026 | 0.88855 | 0.328 |
| 34140 | CTD-2314G24.2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | 0.246 | 0.59912 | 0.524 | 0.00017 | 0.328 |
| 34141 | OIP5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p | 29 | PRDM2 | Sponge network | 0.421 | 0.00084 | -0.258 | 0.00721 | 0.328 |
| 34142 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p | 10 | SPRY3 | Sponge network | 1.134 | 0 | 0.016 | 0.91286 | 0.328 |
| 34143 | TINCR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p | 11 | CNTNAP3 | Sponge network | -0.664 | 0.14543 | -1.465 | 0.00033 | 0.328 |
| 34144 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-629-3p;hsa-miR-940 | 16 | GAS7 | Sponge network | -0.421 | 0.12556 | -0.555 | 0.01602 | 0.328 |
| 34145 | LINC00667 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-3p | 29 | GRIK3 | Sponge network | -0.235 | 0.15142 | -3.208 | 0 | 0.328 |
| 34146 | AC108488.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577 | 18 | SCN9A | Sponge network | 0.259 | 0.12542 | -0.525 | 0.10593 | 0.328 |
| 34147 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 21 | LIFR | Sponge network | -0.322 | 0.25945 | -1.794 | 0 | 0.328 |
| 34148 | RP11-326G21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TRPS1 | Sponge network | -0.021 | 0.93598 | -0.191 | 0.44109 | 0.328 |
| 34149 | LINC01082 |
hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-29a-5p | 11 | SSBP2 | Sponge network | -3.322 | 8.0E-5 | -1.58 | 0 | 0.328 |
| 34150 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-625-5p | 13 | JAZF1 | Sponge network | -1.054 | 0.0706 | -0.377 | 0.02677 | 0.328 |
| 34151 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-7-1-3p | 29 | DDX6 | Sponge network | 0.572 | 0.01722 | 0.091 | 0.36428 | 0.328 |
| 34152 | RP11-867G23.10 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-942-5p | 18 | TMEM132B | Sponge network | -2.531 | 0 | -0.83 | 0.01084 | 0.328 |
| 34153 | RP11-723D22.3 |
hsa-miR-10a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-500a-5p | 13 | LPP | Sponge network | 0.562 | 0.20117 | -0.459 | 0.04475 | 0.328 |
| 34154 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 48 | TCF4 | Sponge network | 0.16 | 0.50785 | 0.016 | 0.92271 | 0.328 |
| 34155 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-93-5p | 14 | AHNAK | Sponge network | -0.777 | 0.16667 | -1.207 | 0 | 0.328 |
| 34156 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429 | 15 | SPRY3 | Sponge network | 1.106 | 0 | 0.016 | 0.91286 | 0.328 |
| 34157 | LINC00621 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-19b-1-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | FREM2 | Sponge network | 0.131 | 0.8436 | -0.437 | 0.46353 | 0.328 |
| 34158 | BDNF-AS |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | SYNPO2 | Sponge network | -0.668 | 3.0E-5 | -2.342 | 0 | 0.328 |
| 34159 | RP11-326G21.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 14 | GJA1 | Sponge network | -0.021 | 0.93598 | -0.485 | 0.02179 | 0.328 |
| 34160 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-590-3p | 15 | TES | Sponge network | 0.194 | 0.64278 | -0.348 | 0.00639 | 0.328 |
| 34161 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 10 | CREB1 | Sponge network | -0.187 | 0.23787 | 0.203 | 0.00537 | 0.328 |
| 34162 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 41 | RBMS3 | Sponge network | 0.395 | 0.15451 | -0.519 | 0.03551 | 0.328 |
| 34163 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | LATS2 | Sponge network | 0.101 | 0.60919 | 0.159 | 0.2304 | 0.328 |
| 34164 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p | 14 | BCL11A | Sponge network | 0.389 | 0.04293 | -0.932 | 0.0063 | 0.328 |
| 34165 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378c | 19 | LPP | Sponge network | -1.14 | 0.03513 | -0.459 | 0.04475 | 0.328 |
| 34166 | LINC00173 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-532-3p | 13 | MRVI1 | Sponge network | 0.056 | 0.8494 | -1.051 | 0.00022 | 0.328 |
| 34167 | AC005154.6 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TSC1 | Sponge network | 0.848 | 0 | 0.103 | 0.26071 | 0.328 |
| 34168 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | TACC1 | Sponge network | -0.125 | 0.49414 | -0.362 | 0.05406 | 0.328 |
| 34169 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SMARCA2 | Sponge network | -0.49 | 0.04946 | -0.529 | 0.00014 | 0.328 |
| 34170 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-7-5p | 13 | EPHA3 | Sponge network | 0.592 | 0.00014 | 0.185 | 0.53588 | 0.328 |
| 34171 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-93-5p | 45 | FAT3 | Sponge network | -1.645 | 0 | -1.601 | 0.00051 | 0.328 |
| 34172 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-425-5p;hsa-miR-511-5p | 18 | ATRX | Sponge network | 0.622 | 0.00015 | 0.261 | 0.04408 | 0.328 |
| 34173 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 32 | CLASP2 | Sponge network | -0.405 | 0.22013 | -0.038 | 0.71 | 0.328 |
| 34174 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-590-3p | 12 | USP6 | Sponge network | 0.178 | 0.29106 | 0.188 | 0.3871 | 0.328 |
| 34175 | CTC-296K1.4 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-5p | 15 | NTRK2 | Sponge network | -2.076 | 0.00548 | -0.736 | 0.06495 | 0.328 |
| 34176 | RP11-35G9.3 |
hsa-let-7d-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-191-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-92a-3p | 12 | CBFA2T3 | Sponge network | 0.657 | 4.0E-5 | -1.221 | 1.0E-5 | 0.328 |
| 34177 | DNM3OS |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CDH10 | Sponge network | 0.572 | 0.01722 | -2.397 | 0 | 0.328 |
| 34178 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p | 11 | RB1CC1 | Sponge network | -0.303 | 0.21002 | 0.175 | 0.12559 | 0.328 |
| 34179 | BDNF-AS |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | RPS6KA6 | Sponge network | -0.668 | 3.0E-5 | -2.989 | 0 | 0.328 |
| 34180 | RP11-403I13.7 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-5p | 16 | FMNL3 | Sponge network | 0.395 | 0.15451 | 0.555 | 4.0E-5 | 0.328 |
| 34181 | MYLK-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-7-5p | 20 | MOB1B | Sponge network | -1.331 | 3.0E-5 | 0.197 | 0.15266 | 0.328 |
| 34182 | AC004540.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-629-3p | 11 | ASTN1 | Sponge network | -2.549 | 0.00528 | -2.389 | 2.0E-5 | 0.328 |
| 34183 | CTD-2666L21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-493-5p | 10 | NRXN3 | Sponge network | 0.368 | 0.20093 | -0.893 | 0.04186 | 0.328 |
| 34184 | RP11-434B12.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ITGB1 | Sponge network | -0.031 | 0.89063 | 0.524 | 0.00017 | 0.328 |
| 34185 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 30 | RBMS3 | Sponge network | 0.75 | 0.00054 | -0.519 | 0.03551 | 0.328 |
| 34186 | TRIM52-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-196a-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-577 | 10 | NRK | Sponge network | -0.418 | 0.00068 | 0.656 | 0.19217 | 0.328 |
| 34187 | SOX2-OT |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 26 | ANK2 | Sponge network | -1.264 | 6.0E-5 | -1.603 | 1.0E-5 | 0.328 |
| 34188 | GUSBP11 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-495-3p | 19 | CREB1 | Sponge network | 0.856 | 0 | 0.203 | 0.00537 | 0.328 |
| 34189 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | TP53INP1 | Sponge network | -0.615 | 0.00015 | -0.496 | 0.00064 | 0.328 |
| 34190 | RP11-13K12.5 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | GJA1 | Sponge network | -0.318 | 0.65847 | -0.485 | 0.02179 | 0.328 |
| 34191 | RP11-195B17.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p | 11 | MRVI1 | Sponge network | 0.37 | 0.27614 | -1.051 | 0.00022 | 0.328 |
| 34192 | THAP9-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-374b-5p;hsa-miR-664a-3p | 12 | UBE2D1 | Sponge network | 1.079 | 0 | 0.468 | 0 | 0.328 |
| 34193 | RP1-59D14.5 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-424-5p | 10 | UBE4B | Sponge network | 1.162 | 5.0E-5 | -0.235 | 0.007 | 0.328 |
| 34194 | RP11-611E13.2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326 | 13 | CBFB | Sponge network | 0.822 | 0 | 1.061 | 0 | 0.328 |
| 34195 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-3p | 16 | NCAM2 | Sponge network | 0.174 | 0.30034 | -0.482 | 0.22565 | 0.328 |
| 34196 | RP5-1021I20.1 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-532-3p | 13 | MRVI1 | Sponge network | -0.785 | 0.00035 | -1.051 | 0.00022 | 0.328 |
| 34197 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | FGF5 | Sponge network | -0.487 | 0.02157 | 0.549 | 0.28659 | 0.328 |
| 34198 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 19 | YAP1 | Sponge network | 0.559 | 0.00026 | 0.22 | 0.11836 | 0.328 |
| 34199 | LINC01018 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | PRKCB | Sponge network | -2.915 | 0.00015 | -1.313 | 2.0E-5 | 0.328 |
| 34200 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 0.539 | 0.03722 | 0.322 | 0.00215 | 0.328 |
| 34201 | EIF3J-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | ZMYM2 | Sponge network | 0.331 | 0.00509 | 0.279 | 0.01273 | 0.328 |
| 34202 | RP11-589P10.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-93-3p | 12 | NCOA1 | Sponge network | -2.044 | 0.00066 | -0.432 | 0 | 0.328 |
| 34203 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-214-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-93-5p | 41 | PPM1A | Sponge network | -1.047 | 0 | -0.623 | 0 | 0.328 |
| 34204 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p | 17 | KAT6B | Sponge network | 0.637 | 0.00069 | -0.478 | 0.00018 | 0.328 |
| 34205 | RP11-360F5.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | AKAP11 | Sponge network | 1.867 | 0 | 0.196 | 0.09825 | 0.328 |
| 34206 | KB-1410C5.5 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 11 | ARHGEF12 | Sponge network | 0.959 | 0 | 0.09 | 0.35693 | 0.328 |
| 34207 | CTC-429P9.5 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p | 18 | ASTN1 | Sponge network | 0.178 | 0.29106 | -2.389 | 2.0E-5 | 0.328 |
| 34208 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-495-3p | 12 | SLC5A3 | Sponge network | 0.614 | 1.0E-5 | 0.493 | 5.0E-5 | 0.328 |
| 34209 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-493-5p;hsa-miR-664a-3p | 29 | KIF1B | Sponge network | 0.706 | 0 | -0.118 | 0.32258 | 0.328 |
| 34210 | TUG1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | RPS6KA3 | Sponge network | 0.972 | 0 | 0.256 | 0.02419 | 0.328 |
| 34211 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 32 | EBF1 | Sponge network | -0.899 | 6.0E-5 | -0.384 | 0.08856 | 0.328 |
| 34212 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | FREM2 | Sponge network | -1.281 | 0.00012 | -0.437 | 0.46353 | 0.328 |
| 34213 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 15 | GALNT13 | Sponge network | 0.214 | 0.18246 | -0.857 | 0.07439 | 0.328 |
| 34214 | NEAT1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | DDX6 | Sponge network | 0.705 | 0.00014 | 0.091 | 0.36428 | 0.328 |
| 34215 | RP1-228H13.5 |
hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p | 11 | CDK6 | Sponge network | 1.397 | 0 | 0.991 | 0 | 0.328 |
| 34216 | AC016747.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-877-5p | 15 | TET1 | Sponge network | 0.199 | 0.38015 | -0.135 | 0.52138 | 0.328 |
| 34217 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-708-5p | 25 | SPRY3 | Sponge network | 0.705 | 0.00014 | 0.016 | 0.91286 | 0.328 |
| 34218 | LINC00094 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 26 | NAV3 | Sponge network | 0.253 | 0.09565 | 0.212 | 0.47729 | 0.328 |
| 34219 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577 | 19 | GREM1 | Sponge network | -0.342 | 0.02288 | 0.902 | 0.01691 | 0.328 |
| 34220 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 31 | MEF2C | Sponge network | 0.659 | 3.0E-5 | -0.499 | 0.01448 | 0.328 |
| 34221 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-590-5p | 14 | RECK | Sponge network | -0.384 | 0.40652 | -1.043 | 0 | 0.328 |
| 34222 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-96-5p | 22 | CRTC3 | Sponge network | 0.078 | 0.55803 | 0.164 | 0.16786 | 0.328 |
| 34223 | AC141928.1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-577 | 10 | MACF1 | Sponge network | 0.853 | 0.15352 | 0.019 | 0.90335 | 0.328 |
| 34224 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | TMTC2 | Sponge network | -1.697 | 0.00889 | -0.215 | 0.18248 | 0.328 |
| 34225 | RP11-597D13.9 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | PCDH9 | Sponge network | 1.302 | 7.0E-5 | -1.823 | 4.0E-5 | 0.328 |
| 34226 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | GRIN2A | Sponge network | 0.16 | 0.50785 | -2.161 | 0 | 0.328 |
| 34227 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p | 17 | UNC80 | Sponge network | -2.076 | 0.00548 | -1.934 | 0 | 0.328 |
| 34228 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-942-5p | 31 | NOTCH2 | Sponge network | -0.318 | 0.65847 | 0.138 | 0.35163 | 0.328 |
| 34229 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 39 | DDX6 | Sponge network | -1.111 | 0.0003 | 0.091 | 0.36428 | 0.328 |
| 34230 | LINC01018 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p | 11 | AMPH | Sponge network | -2.915 | 0.00015 | -0.916 | 5.0E-5 | 0.328 |
| 34231 | RP11-464F9.1 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374b-5p;hsa-miR-376c-3p | 12 | PAWR | Sponge network | 0.959 | 0 | 0.636 | 0 | 0.328 |
| 34232 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-744-3p | 19 | CUL5 | Sponge network | 0.614 | 1.0E-5 | 0.026 | 0.77301 | 0.328 |
| 34233 | MEG3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-96-5p | 14 | ELMO1 | Sponge network | 0.249 | 0.35902 | 0.04 | 0.83147 | 0.328 |
| 34234 | KB-1460A1.5 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | 0.603 | 0.00127 | -0.226 | 0.11691 | 0.328 |
| 34235 | SH3BP5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | ABI2 | Sponge network | 0.267 | 0.2937 | 0.175 | 0.14787 | 0.328 |
| 34236 | U91328.19 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-362-3p;hsa-miR-484 | 12 | IGF2 | Sponge network | -0.303 | 0.21002 | 0.848 | 0.03842 | 0.328 |
| 34237 | CTC-429P9.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-744-3p;hsa-miR-93-3p | 19 | TOM1L2 | Sponge network | 0.178 | 0.29106 | -0.994 | 0 | 0.328 |
| 34238 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-942-5p | 25 | HLF | Sponge network | 0.41 | 0.02214 | -2.443 | 0 | 0.328 |
| 34239 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-625-5p | 20 | ZMYM2 | Sponge network | 1.317 | 0 | 0.279 | 0.01273 | 0.328 |
| 34240 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | ESR1 | Sponge network | -0.021 | 0.93598 | -1.035 | 3.0E-5 | 0.328 |
| 34241 | ERVH48-1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 10 | BCL9 | Sponge network | 1.04 | 0.00023 | 0.321 | 0.00267 | 0.328 |
| 34242 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | HBP1 | Sponge network | -1.582 | 8.0E-5 | -0.454 | 0 | 0.328 |
| 34243 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-628-5p | 30 | CUX1 | Sponge network | -0.583 | 0.14589 | -0.039 | 0.6705 | 0.328 |
| 34244 | RP11-728F11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p | 15 | DMD | Sponge network | 0.238 | 0.70544 | -1.506 | 0 | 0.328 |
| 34245 | RP11-428J1.5 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429 | 16 | PTPN14 | Sponge network | 0.538 | 0.00076 | 0.144 | 0.34595 | 0.328 |
| 34246 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p | 15 | PTGFR | Sponge network | 0.537 | 0.00139 | -0.233 | 0.45421 | 0.328 |
| 34247 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-96-5p | 18 | IL17RD | Sponge network | -0.555 | 0.06156 | -0.019 | 0.94873 | 0.328 |
| 34248 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 16 | LHX6 | Sponge network | -0.465 | 0.04703 | -0.087 | 0.69193 | 0.328 |
| 34249 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | PHC3 | Sponge network | 0.556 | 0.00298 | 0.212 | 0.0499 | 0.328 |
| 34250 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-532-3p | 12 | CBL | Sponge network | 1.055 | 0 | 0.291 | 0.01267 | 0.328 |
| 34251 | PSMD5-AS1 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | MAP3K4 | Sponge network | 0.569 | 0.00067 | 0.058 | 0.54229 | 0.328 |
| 34252 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -1.654 | 0.00144 | -0.243 | 0.11408 | 0.328 |
| 34253 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-93-3p;hsa-miR-96-5p | 12 | EBF3 | Sponge network | -0.31 | 0.33871 | -0.058 | 0.81437 | 0.328 |
| 34254 | LINC00476 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-508-3p | 13 | RASSF3 | Sponge network | -0.615 | 0.00015 | -0.226 | 0.11691 | 0.328 |
| 34255 | KB-1410C5.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 11 | FOXP1 | Sponge network | 0.959 | 0 | 0.042 | 0.74368 | 0.328 |
| 34256 | RP11-121C2.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p | 16 | ABI2 | Sponge network | 1.147 | 0 | 0.175 | 0.14787 | 0.328 |
| 34257 | HNRNPU-AS1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 17 | BAZ2B | Sponge network | 1.601 | 0 | -0.276 | 0.02833 | 0.328 |
| 34258 | RP11-822E23.8 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -0.777 | 0.16667 | -0.564 | 1.0E-5 | 0.328 |
| 34259 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p | 12 | SASH1 | Sponge network | -2.076 | 0.00548 | -0.502 | 0.00175 | 0.328 |
| 34260 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-655-3p;hsa-miR-93-5p | 46 | PRKAA2 | Sponge network | 0.253 | 0.09565 | -2.462 | 0 | 0.328 |
| 34261 | PCA3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 33 | ETV1 | Sponge network | -0.709 | 0.18168 | -0.096 | 0.67674 | 0.328 |
| 34262 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 28 | YAP1 | Sponge network | -0.739 | 0.0574 | 0.22 | 0.11836 | 0.328 |
| 34263 | RP11-439L18.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 14 | GAS7 | Sponge network | 0.224 | 0.48719 | -0.555 | 0.01602 | 0.328 |
| 34264 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 17 | MED12L | Sponge network | 0.921 | 0.00118 | -0.194 | 0.55299 | 0.328 |
| 34265 | TMEM51-AS1 |
hsa-let-7i-5p;hsa-miR-127-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-432-5p | 13 | PLXNA2 | Sponge network | 0.23 | 0.38667 | -0.395 | 0.00779 | 0.328 |
| 34266 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | ST6GALNAC3 | Sponge network | 0.889 | 9.0E-5 | -0.987 | 0 | 0.328 |
| 34267 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-455-5p | 10 | TSC22D1 | Sponge network | -0.162 | 0.47687 | -0.529 | 2.0E-5 | 0.328 |
| 34268 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-190a-5p;hsa-miR-223-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | CXXC4 | Sponge network | 1.61 | 0.00961 | -0.433 | 0.21055 | 0.327 |
| 34269 | RP11-46C24.7 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 16 | CSDE1 | Sponge network | 0.392 | 0.0278 | -0.493 | 0 | 0.327 |
| 34270 | CTD-2528L19.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-429;hsa-miR-7-1-3p | 10 | VEZT | Sponge network | 0.953 | 0 | 0.259 | 0.00296 | 0.327 |
| 34271 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | CADM2 | Sponge network | -0.351 | 0.11358 | -3.782 | 0 | 0.327 |
| 34272 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-935;hsa-miR-96-5p | 14 | EBF1 | Sponge network | 0.37 | 0.27614 | -0.384 | 0.08856 | 0.327 |
| 34273 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429 | 12 | SLC6A15 | Sponge network | -1.654 | 0.00144 | -2.373 | 2.0E-5 | 0.327 |
| 34274 | MRPL23-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-3p;hsa-miR-625-5p | 10 | MCC | Sponge network | -0.278 | 0.76559 | -1.005 | 0 | 0.327 |
| 34275 | RP11-706C16.7 |
hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577 | 11 | ZFHX3 | Sponge network | 0.357 | 0.72407 | 0.389 | 0.01363 | 0.327 |
| 34276 | CD27-AS1 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-7-1-3p | 14 | RASSF3 | Sponge network | 0.177 | 0.16681 | -0.226 | 0.11691 | 0.327 |
| 34277 | MIR143HG |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | CREM | Sponge network | -0.739 | 0.0574 | -0.345 | 0.00258 | 0.327 |
| 34278 | LINC00265 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 11 | PIK3C2A | Sponge network | 1.07 | 0 | 0.061 | 0.53776 | 0.327 |
| 34279 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | PPM1A | Sponge network | -0.342 | 0.02288 | -0.623 | 0 | 0.327 |
| 34280 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-7-1-3p | 14 | SYNE1 | Sponge network | 0.419 | 0.0319 | -0.738 | 0.00316 | 0.327 |
| 34281 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | MED12L | Sponge network | 0.101 | 0.60919 | -0.194 | 0.55299 | 0.327 |
| 34282 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-935 | 41 | FAT3 | Sponge network | 0.392 | 0.0278 | -1.601 | 0.00051 | 0.327 |
| 34283 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-92a-3p | 10 | EGR2 | Sponge network | 0.214 | 0.18246 | 0.309 | 0.17079 | 0.327 |
| 34284 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-5p | 15 | TET1 | Sponge network | 0.331 | 0.57247 | -0.135 | 0.52138 | 0.327 |
| 34285 | CTC-296K1.4 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-590-5p;hsa-miR-629-5p | 15 | BCL2 | Sponge network | -2.076 | 0.00548 | -0.899 | 0 | 0.327 |
| 34286 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-532-3p;hsa-miR-940 | 25 | PBX1 | Sponge network | 0.259 | 0.12542 | -1.206 | 0 | 0.327 |
| 34287 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 28 | EPHA7 | Sponge network | 0.853 | 0.15352 | -2.197 | 3.0E-5 | 0.327 |
| 34288 | RP11-798M19.6 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 46 | PPM1L | Sponge network | -0.612 | 0.00094 | -0.537 | 0.00616 | 0.327 |
| 34289 | RP11-356J5.12 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 14 | CXCL12 | Sponge network | -0.899 | 6.0E-5 | -1.421 | 0 | 0.327 |
| 34290 | AC159540.2 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-214-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-328-3p;hsa-miR-628-5p | 13 | VPS53 | Sponge network | 1.634 | 0 | 0.103 | 0.22667 | 0.327 |
| 34291 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 29 | CLASP2 | Sponge network | 0.494 | 0.00339 | -0.038 | 0.71 | 0.327 |
| 34292 | RP11-360F5.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 16 | TRPS1 | Sponge network | 1.867 | 0 | -0.191 | 0.44109 | 0.327 |
| 34293 | FENDRR |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-3127-5p;hsa-miR-421;hsa-miR-629-3p;hsa-miR-708-5p;hsa-miR-744-3p | 10 | RHOBTB2 | Sponge network | -1.281 | 0.00012 | -0.167 | 0.19499 | 0.327 |
| 34294 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | MED12L | Sponge network | 0.853 | 0.15352 | -0.194 | 0.55299 | 0.327 |
| 34295 | RP11-517B11.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p | 10 | KDSR | Sponge network | 0.706 | 0 | 0.239 | 0.02216 | 0.327 |
| 34296 | MIR143HG |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | NR3C2 | Sponge network | -0.739 | 0.0574 | -1.56 | 0 | 0.327 |
| 34297 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-625-5p | 18 | ZMYM2 | Sponge network | 1.117 | 0 | 0.279 | 0.01273 | 0.327 |
| 34298 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p | 11 | PARK2 | Sponge network | 0.331 | 0.57247 | -1.892 | 0 | 0.327 |
| 34299 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | 0.537 | 0.00139 | -1.74 | 0 | 0.327 |
| 34300 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-500a-5p | 17 | ZFHX3 | Sponge network | -0.932 | 0.00329 | 0.389 | 0.01363 | 0.327 |
| 34301 | RP11-5C23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 14 | SYNE1 | Sponge network | 0.274 | 0.07681 | -0.738 | 0.00316 | 0.327 |
| 34302 | RP11-540B6.6 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | USP9X | Sponge network | 0.93 | 0 | 0.222 | 0.01689 | 0.327 |
| 34303 | RP11-999E24.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-362-3p | 14 | CADM1 | Sponge network | -0.693 | 0.05629 | -0.9 | 0.00084 | 0.327 |
| 34304 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | NEDD4 | Sponge network | 0.389 | 0.04293 | 0.02 | 0.89834 | 0.327 |
| 34305 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-942-5p | 15 | GPC6 | Sponge network | -4.546 | 0 | -0.054 | 0.81465 | 0.327 |
| 34306 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SLC6A15 | Sponge network | -0.899 | 6.0E-5 | -2.373 | 2.0E-5 | 0.327 |
| 34307 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | CLASP2 | Sponge network | 0.415 | 0.14657 | -0.038 | 0.71 | 0.327 |
| 34308 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | LATS2 | Sponge network | -0.738 | 0.21233 | 0.159 | 0.2304 | 0.327 |
| 34309 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-629-3p | 27 | GPC6 | Sponge network | -0.154 | 0.78939 | -0.054 | 0.81465 | 0.327 |
| 34310 | CTD-3099C6.9 |
hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-766-3p | 27 | MDM4 | Sponge network | 1.361 | 0 | 0.345 | 0.00762 | 0.327 |
| 34311 | RP11-254F7.2 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 21 | PRKCB | Sponge network | 0.176 | 0.37402 | -1.313 | 2.0E-5 | 0.327 |
| 34312 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | AFF3 | Sponge network | 0.342 | 0.03129 | -2.373 | 0 | 0.327 |
| 34313 | RP11-6O2.3 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-590-5p;hsa-miR-766-3p | 14 | ATM | Sponge network | 0.331 | 0.57247 | 0.178 | 0.21568 | 0.327 |
| 34314 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | SLITRK3 | Sponge network | -0.053 | 0.80685 | -3.328 | 0 | 0.327 |
| 34315 | RP11-815I9.4 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30c-5p;hsa-miR-34a-5p;hsa-miR-590-5p | 11 | NOTCH2 | Sponge network | 0.537 | 0.0006 | 0.138 | 0.35163 | 0.327 |
| 34316 | TP73-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p | 12 | PTCH1 | Sponge network | -0.563 | 0.02545 | -0.1 | 0.6179 | 0.327 |
| 34317 | RP11-347P5.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-26b-5p;hsa-miR-374a-3p | 10 | IL6ST | Sponge network | 1.364 | 0 | -0.402 | 0.00676 | 0.327 |
| 34318 | DNM3OS |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p | 18 | CSDE1 | Sponge network | 0.572 | 0.01722 | -0.493 | 0 | 0.327 |
| 34319 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | TAL1 | Sponge network | -0.942 | 0 | -0.462 | 0.00574 | 0.327 |
| 34320 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p;hsa-miR-505-3p | 14 | JAKMIP2 | Sponge network | -0.969 | 2.0E-5 | -1.388 | 0 | 0.327 |
| 34321 | CTD-3064M3.3 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-93-3p | 15 | DOCK4 | Sponge network | -0.469 | 0.08721 | 0.502 | 0.00108 | 0.327 |
| 34322 | MRPL23-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-629-3p | 26 | AR | Sponge network | -0.278 | 0.76559 | -1.554 | 0 | 0.327 |
| 34323 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | DYNC1I1 | Sponge network | 0.083 | 0.66405 | -1.12 | 0.00107 | 0.327 |
| 34324 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | SASH1 | Sponge network | 0.537 | 0.00139 | -0.502 | 0.00175 | 0.327 |
| 34325 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p | 15 | CXXC4 | Sponge network | -0.693 | 0.05629 | -0.433 | 0.21055 | 0.327 |
| 34326 | GUSBP11 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p | 12 | PIK3C2A | Sponge network | 0.856 | 0 | 0.061 | 0.53776 | 0.327 |
| 34327 | LINC01018 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | MXI1 | Sponge network | -2.915 | 0.00015 | -0.987 | 0 | 0.327 |
| 34328 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | PIK3C2A | Sponge network | 0.415 | 0.14657 | 0.061 | 0.53776 | 0.327 |
| 34329 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 41 | AR | Sponge network | 0.4 | 0.14611 | -1.554 | 0 | 0.327 |
| 34330 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-3607-3p | 23 | AR | Sponge network | 0.37 | 0.27614 | -1.554 | 0 | 0.327 |
| 34331 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | GALNT13 | Sponge network | 0.194 | 0.64278 | -0.857 | 0.07439 | 0.327 |
| 34332 | RP11-21A7A.2 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-628-5p | 16 | MEF2C | Sponge network | -1.116 | 0.23686 | -0.499 | 0.01448 | 0.327 |
| 34333 | RP11-244O19.1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-502-5p;hsa-miR-590-3p | 19 | PAIP2 | Sponge network | -0.096 | 0.67958 | -0.515 | 0 | 0.327 |
| 34334 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-532-3p | 18 | USP12 | Sponge network | -2.69 | 0.0001 | -0.102 | 0.40768 | 0.327 |
| 34335 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-10a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p | 19 | RASSF2 | Sponge network | -0.932 | 0.00329 | -0.273 | 0.21961 | 0.327 |
| 34336 | LINC00702 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | ERCC4 | Sponge network | -0.737 | 0.06182 | 0.126 | 0.15266 | 0.327 |
| 34337 | RP11-181G12.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p | 11 | TSC1 | Sponge network | 0.556 | 0.00298 | 0.103 | 0.26071 | 0.327 |
| 34338 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | TBX18 | Sponge network | 0.389 | 0.04293 | 0.843 | 0.02945 | 0.327 |
| 34339 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | SASH1 | Sponge network | 0.727 | 3.0E-5 | -0.502 | 0.00175 | 0.327 |
| 34340 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-96-5p | 20 | SCN9A | Sponge network | 0.614 | 1.0E-5 | -0.525 | 0.10593 | 0.327 |
| 34341 | PSMD5-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | BCL9 | Sponge network | 0.569 | 0.00067 | 0.321 | 0.00267 | 0.327 |
| 34342 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-7-1-3p | 10 | ARID5B | Sponge network | 0.693 | 0.00076 | 0.342 | 0.01676 | 0.327 |
| 34343 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-629-3p | 29 | PIK3R1 | Sponge network | -2.62 | 0 | -0.133 | 0.36854 | 0.327 |
| 34344 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-582-3p | 20 | ABL2 | Sponge network | -0.318 | 0.65847 | 0.82 | 0 | 0.327 |
| 34345 | RP11-13K12.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 14 | NCAM2 | Sponge network | -0.154 | 0.78939 | -0.482 | 0.22565 | 0.327 |
| 34346 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 14 | GPAM | Sponge network | 0.253 | 0.09565 | 0.142 | 0.29732 | 0.327 |
| 34347 | TUG1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | TSC1 | Sponge network | 0.972 | 0 | 0.103 | 0.26071 | 0.327 |
| 34348 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | ERCC4 | Sponge network | 1.089 | 0 | 0.126 | 0.15266 | 0.327 |
| 34349 | RP11-48B3.4 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | INPP4A | Sponge network | 1.757 | 0 | 0.07 | 0.53563 | 0.327 |
| 34350 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 15 | CLIP1 | Sponge network | -0.227 | 0.31657 | -0.215 | 0.08684 | 0.327 |
| 34351 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 28 | ARID5B | Sponge network | -0.323 | 0.01372 | 0.342 | 0.01676 | 0.327 |
| 34352 | RP4-791M13.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-577 | 10 | NRK | Sponge network | 0.419 | 0.0319 | 0.656 | 0.19217 | 0.327 |
| 34353 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-96-5p | 10 | ZHX2 | Sponge network | -0.942 | 0 | -0.192 | 0.18459 | 0.327 |
| 34354 | CTC-444N24.11 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | BAZ2B | Sponge network | 1.153 | 0 | -0.276 | 0.02833 | 0.327 |
| 34355 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLASP2 | Sponge network | 0.539 | 0.03722 | -0.038 | 0.71 | 0.327 |
| 34356 | RP11-497H16.9 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 10 | PHF6 | Sponge network | 0.545 | 0.00064 | 0.785 | 0 | 0.327 |
| 34357 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935 | 28 | FBXO32 | Sponge network | 0.225 | 0.30644 | -0.495 | 0.07561 | 0.327 |
| 34358 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-589-3p | 28 | MOB1B | Sponge network | -0.154 | 0.78939 | 0.197 | 0.15266 | 0.327 |
| 34359 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 16 | ARID5B | Sponge network | 0.594 | 0.00064 | 0.342 | 0.01676 | 0.327 |
| 34360 | LINC00854 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-664a-5p | 14 | SOX11 | Sponge network | 1.18 | 0 | 2.68 | 0 | 0.327 |
| 34361 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | NEDD4 | Sponge network | 0.16 | 0.50785 | 0.02 | 0.89834 | 0.327 |
| 34362 | RP11-119F7.5 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | TPO | Sponge network | 0.225 | 0.30644 | -1.87 | 2.0E-5 | 0.327 |
| 34363 | ZSCAN16-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | NAV3 | Sponge network | -0.342 | 0.02288 | 0.212 | 0.47729 | 0.327 |
| 34364 | PGM5-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-660-5p | 12 | RIMBP2 | Sponge network | -4.546 | 0 | -1.968 | 5.0E-5 | 0.327 |
| 34365 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CLIP1 | Sponge network | 0.873 | 0 | -0.215 | 0.08684 | 0.327 |
| 34366 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 36 | EPHA7 | Sponge network | -0.125 | 0.49414 | -2.197 | 3.0E-5 | 0.327 |
| 34367 | CTC-559E9.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-338-5p | 13 | PRDM2 | Sponge network | 0.594 | 0.00064 | -0.258 | 0.00721 | 0.327 |
| 34368 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 14 | ITGB3 | Sponge network | 0.72 | 0.0001 | -0.011 | 0.95751 | 0.327 |
| 34369 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-93-3p;hsa-miR-940 | 24 | TOM1L2 | Sponge network | -0.769 | 0.00035 | -0.994 | 0 | 0.327 |
| 34370 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 25 | BCL11A | Sponge network | -2.925 | 0 | -0.932 | 0.0063 | 0.327 |
| 34371 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p | 30 | MAF | Sponge network | -0.727 | 0.04726 | -1.257 | 0 | 0.327 |
| 34372 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | ZFHX3 | Sponge network | 1.063 | 0 | 0.389 | 0.01363 | 0.327 |
| 34373 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | HECA | Sponge network | 0.862 | 0.06213 | -0.217 | 0.03463 | 0.327 |
| 34374 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p | 10 | ZYX | Sponge network | -0.777 | 0.16667 | 0.019 | 0.89763 | 0.327 |
| 34375 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 25 | PRKAB2 | Sponge network | -0.421 | 0.0114 | -0.367 | 0.01371 | 0.327 |
| 34376 | LINC00654 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | PCDH17 | Sponge network | 0.374 | 0.20054 | 0.731 | 2.0E-5 | 0.327 |
| 34377 | CTC-429P9.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-320b | 10 | WWTR1 | Sponge network | 0.082 | 0.55397 | -0.345 | 0.11997 | 0.327 |
| 34378 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p;hsa-miR-98-5p | 30 | MITF | Sponge network | -0.942 | 0 | -0.769 | 0.00022 | 0.327 |
| 34379 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-655-3p | 12 | NF1 | Sponge network | 0.214 | 0.18246 | 0.51 | 0 | 0.327 |
| 34380 | RP11-226L15.5 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 10 | MED12L | Sponge network | 0.8 | 0 | -0.194 | 0.55299 | 0.327 |
| 34381 | CTD-2647L4.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-590-5p | 15 | SMAD4 | Sponge network | 0.555 | 0.00197 | -0.313 | 0.00413 | 0.327 |
| 34382 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | DAB2 | Sponge network | -1.654 | 0.00144 | 0.431 | 0.0113 | 0.327 |
| 34383 | ARHGEF26-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-940 | 24 | BAZ2B | Sponge network | -2.46 | 0 | -0.276 | 0.02833 | 0.327 |
| 34384 | RP11-6N17.3 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p | 12 | ZBTB4 | Sponge network | 0.108 | 0.69556 | -0.739 | 0 | 0.327 |
| 34385 | C20orf166-AS1 |
hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-874-3p | 10 | AJAP1 | Sponge network | -2.69 | 0.0001 | -0.561 | 0.04832 | 0.327 |
| 34386 | PART1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | GATA2 | Sponge network | -2.925 | 0 | -0.654 | 0.0016 | 0.327 |
| 34387 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-511-5p | 12 | ZFHX3 | Sponge network | 0.997 | 0 | 0.389 | 0.01363 | 0.327 |
| 34388 | AF131215.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 11 | SPG20 | Sponge network | -0.322 | 0.25945 | -1.16 | 0 | 0.327 |
| 34389 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421 | 15 | CITED2 | Sponge network | -0.162 | 0.47687 | -1.545 | 0 | 0.327 |
| 34390 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | TSHZ3 | Sponge network | -0.72 | 0.24239 | -0.556 | 0.01516 | 0.327 |
| 34391 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-484 | 11 | TRIM3 | Sponge network | -4.546 | 0 | -0.577 | 0 | 0.327 |
| 34392 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | MYO9A | Sponge network | -0.811 | 2.0E-5 | -0.147 | 0.32373 | 0.327 |
| 34393 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-590-3p | 12 | PKNOX1 | Sponge network | 0.225 | 0.30644 | 0.085 | 0.28917 | 0.327 |
| 34394 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | RNF144A | Sponge network | 0.727 | 3.0E-5 | 0.594 | 0.0009 | 0.327 |
| 34395 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 27 | TBL1XR1 | Sponge network | 0.569 | 0.00067 | 0.578 | 0 | 0.327 |
| 34396 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | FAT3 | Sponge network | 0.4 | 0.14611 | -1.601 | 0.00051 | 0.327 |
| 34397 | FGF14-AS2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-874-3p | 13 | MXI1 | Sponge network | -1.582 | 8.0E-5 | -0.987 | 0 | 0.327 |
| 34398 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 21 | EBF1 | Sponge network | 0.659 | 3.0E-5 | -0.384 | 0.08856 | 0.327 |
| 34399 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 16 | RARB | Sponge network | -0.405 | 0.22013 | -0.825 | 2.0E-5 | 0.327 |
| 34400 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 29 | FOXO3 | Sponge network | -1.111 | 0.0003 | -0.04 | 0.72028 | 0.327 |
| 34401 | TRIM52-AS1 |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-92a-1-5p | 18 | GRIK3 | Sponge network | -0.418 | 0.00068 | -3.208 | 0 | 0.327 |
| 34402 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-877-5p | 23 | CRTC1 | Sponge network | -0.976 | 0.00025 | -0.116 | 0.28412 | 0.327 |
| 34403 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 20 | BMPR2 | Sponge network | 0.476 | 0.19203 | 0.206 | 0.0635 | 0.327 |
| 34404 | RP11-216B9.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | MED12L | Sponge network | 0.174 | 0.30034 | -0.194 | 0.55299 | 0.327 |
| 34405 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DACH2 | Sponge network | -0.737 | 0.10822 | -1.635 | 0.00034 | 0.327 |
| 34406 | CTC-429P9.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p | 15 | CYLD | Sponge network | 0.178 | 0.29106 | -0.367 | 0.00171 | 0.327 |
| 34407 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-93-5p | 28 | RNASEL | Sponge network | -0.129 | 0.57177 | -0.272 | 0.01796 | 0.327 |
| 34408 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-590-3p | 33 | PRKAA2 | Sponge network | 0.174 | 0.30034 | -2.462 | 0 | 0.327 |
| 34409 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SAMHD1 | Sponge network | 0.997 | 0.00066 | 0.072 | 0.62895 | 0.327 |
| 34410 | RP5-1021I20.1 |
hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429 | 13 | DYNC1I1 | Sponge network | -0.785 | 0.00035 | -1.12 | 0.00107 | 0.327 |
| 34411 | RP11-464F9.1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-320a;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-655-3p | 11 | HECA | Sponge network | 0.959 | 0 | -0.217 | 0.03463 | 0.327 |
| 34412 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 20 | AKAP11 | Sponge network | 0.411 | 0.1335 | 0.196 | 0.09825 | 0.327 |
| 34413 | MIR143HG |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 52 | GNAQ | Sponge network | -0.739 | 0.0574 | -0.367 | 0.00102 | 0.327 |
| 34414 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | SMARCA2 | Sponge network | -2.117 | 0.00161 | -0.529 | 0.00014 | 0.327 |
| 34415 | RP11-35G9.3 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | MACF1 | Sponge network | 0.657 | 4.0E-5 | 0.019 | 0.90335 | 0.327 |
| 34416 | PART1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-874-3p | 28 | MXI1 | Sponge network | -2.925 | 0 | -0.987 | 0 | 0.327 |
| 34417 | CKMT2-AS1 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-421;hsa-miR-423-5p;hsa-miR-940 | 12 | SPOP | Sponge network | 0.082 | 0.55654 | -0.419 | 1.0E-5 | 0.327 |
| 34418 | AC007277.3 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-342-3p;hsa-miR-576-5p | 10 | NTRK2 | Sponge network | 2.377 | 0.00108 | -0.736 | 0.06495 | 0.327 |
| 34419 | ZNF32-AS2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p | 12 | UBR3 | Sponge network | 1.552 | 7.0E-5 | -0.021 | 0.82774 | 0.327 |
| 34420 | DNAJC27-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-532-5p | 10 | THSD7B | Sponge network | -0.969 | 2.0E-5 | 0.154 | 0.74723 | 0.327 |
| 34421 | LINC00963 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RCAN2 | Sponge network | 0.523 | 9.0E-5 | -0.33 | 0.15172 | 0.327 |
| 34422 | RP11-479G22.8 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PIK3CA | Sponge network | 1.308 | 0 | 0.195 | 0.06908 | 0.327 |
| 34423 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p | 35 | ESR1 | Sponge network | 0.101 | 0.60919 | -1.035 | 3.0E-5 | 0.327 |
| 34424 | RP11-428J1.5 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | PPM1L | Sponge network | 0.538 | 0.00076 | -0.537 | 0.00616 | 0.327 |
| 34425 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 21 | DMXL1 | Sponge network | -0.896 | 0.00099 | -0.426 | 0.00056 | 0.327 |
| 34426 | RP11-401P9.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-590-5p | 13 | LUZP2 | Sponge network | -0.384 | 0.40652 | -0.056 | 0.89416 | 0.327 |
| 34427 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-382-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | HDAC9 | Sponge network | -1.264 | 6.0E-5 | -0.755 | 0.00495 | 0.327 |
| 34428 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-493-5p;hsa-miR-505-3p | 22 | USP6 | Sponge network | 0.614 | 1.0E-5 | 0.188 | 0.3871 | 0.327 |
| 34429 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 24 | LPP | Sponge network | 0.673 | 0 | -0.459 | 0.04475 | 0.327 |
| 34430 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TBX18 | Sponge network | 1.308 | 0 | 0.843 | 0.02945 | 0.327 |
| 34431 | H1FX-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-942-5p | 24 | IGFBP5 | Sponge network | -0.206 | 0.12942 | -0.806 | 0.00105 | 0.327 |
| 34432 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-7-1-3p;hsa-miR-744-5p | 13 | NOTCH2 | Sponge network | -0.023 | 0.95109 | 0.138 | 0.35163 | 0.327 |
| 34433 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | PRKAA2 | Sponge network | 0.395 | 0.15451 | -2.462 | 0 | 0.327 |
| 34434 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 16 | ETV1 | Sponge network | -0.517 | 0.02935 | -0.096 | 0.67674 | 0.327 |
| 34435 | GABPB1-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 18 | RASAL2 | Sponge network | 1.11 | 0 | 0.869 | 0 | 0.327 |
| 34436 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p | 10 | GPAM | Sponge network | 1.106 | 0 | 0.142 | 0.29732 | 0.327 |
| 34437 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PTPRB | Sponge network | -0.83 | 0 | 0.105 | 0.47122 | 0.327 |
| 34438 | AC016747.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-486-5p;hsa-miR-532-3p | 16 | SYNM | Sponge network | 0.199 | 0.38015 | -2.309 | 1.0E-5 | 0.327 |
| 34439 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-382-5p;hsa-miR-589-3p;hsa-miR-590-5p | 11 | ABCA10 | Sponge network | 0.331 | 0.57247 | -0.571 | 0.03674 | 0.327 |
| 34440 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 13 | DNAJB4 | Sponge network | 0.214 | 0.18246 | -0.7 | 1.0E-5 | 0.327 |
| 34441 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | AKAP11 | Sponge network | 0.924 | 0 | 0.196 | 0.09825 | 0.327 |
| 34442 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 29 | BHLHE41 | Sponge network | -0.976 | 0.00025 | 0.353 | 0.12753 | 0.327 |
| 34443 | CKMT2-AS1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p | 28 | RIMS2 | Sponge network | 0.082 | 0.55654 | -1.365 | 0.00208 | 0.327 |
| 34444 | ZNF667-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 30 | TOM1L2 | Sponge network | -0.976 | 0.00025 | -0.994 | 0 | 0.327 |
| 34445 | RP11-498P14.3 | hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 11 | CBFB | Sponge network | 0.864 | 0 | 1.061 | 0 | 0.327 |
| 34446 | TRAF3IP2-AS1 |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-7-1-3p | 11 | PACRG | Sponge network | -0.323 | 0.01372 | -2.248 | 0 | 0.327 |
| 34447 | RP11-728F11.4 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 18 | PTPRT | Sponge network | 0.238 | 0.70544 | -0.907 | 0.03727 | 0.327 |
| 34448 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-652-3p;hsa-miR-7-1-3p | 40 | SMAD2 | Sponge network | 0.72 | 0.0001 | -0.128 | 0.12732 | 0.327 |
| 34449 | LINC00957 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SLC6A15 | Sponge network | -0.421 | 0.0114 | -2.373 | 2.0E-5 | 0.327 |
| 34450 | LINC00476 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | FAM117A | Sponge network | -0.615 | 0.00015 | -1.379 | 0 | 0.327 |
| 34451 | RP11-195F19.9 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-425-5p;hsa-miR-539-5p | 12 | RYR2 | Sponge network | -0.726 | 0.00132 | -1.466 | 0.00031 | 0.327 |
| 34452 | RP11-894P9.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-582-5p | 16 | SPRY3 | Sponge network | 0.993 | 0 | 0.016 | 0.91286 | 0.327 |
| 34453 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 35 | CREB3L2 | Sponge network | 0.335 | 0.20596 | -0.525 | 2.0E-5 | 0.327 |
| 34454 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p | 28 | JAZF1 | Sponge network | 0.494 | 0.00339 | -0.377 | 0.02677 | 0.327 |
| 34455 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | CDHR3 | Sponge network | -0.811 | 2.0E-5 | -0.977 | 4.0E-5 | 0.327 |
| 34456 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-629-5p | 52 | KIF1B | Sponge network | -0.154 | 0.78939 | -0.118 | 0.32258 | 0.327 |
| 34457 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | CSMD1 | Sponge network | -2.452 | 0 | -0.63 | 0.18881 | 0.327 |
| 34458 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-589-3p | 21 | PRDM2 | Sponge network | -0.154 | 0.53954 | -0.258 | 0.00721 | 0.327 |
| 34459 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | AHNAK | Sponge network | -0.563 | 0.02545 | -1.207 | 0 | 0.327 |
| 34460 | RP11-473I1.9 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | SIRT1 | Sponge network | 0.683 | 1.0E-5 | 0.109 | 0.2593 | 0.327 |
| 34461 | RP11-819C21.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | AFF1 | Sponge network | 0.537 | 0.00139 | -0.384 | 0.00053 | 0.327 |
| 34462 | LINC00473 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-708-5p | 16 | FAM135B | Sponge network | -1.203 | 0.03881 | -3.978 | 0 | 0.327 |
| 34463 | AC025171.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | AKAP11 | Sponge network | 0.998 | 0 | 0.196 | 0.09825 | 0.327 |
| 34464 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 49 | PTPRD | Sponge network | -0.323 | 0.01372 | -1.005 | 0.00064 | 0.327 |
| 34465 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-455-5p | 12 | PRDM5 | Sponge network | -0.969 | 2.0E-5 | -1.09 | 0.00014 | 0.327 |
| 34466 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | USP12 | Sponge network | 0.556 | 0.00298 | -0.102 | 0.40768 | 0.327 |
| 34467 | CTD-2002H8.2 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ARID5B | Sponge network | 0.931 | 1.0E-5 | 0.342 | 0.01676 | 0.327 |
| 34468 | RP11-57H14.4 |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | CTNNA2 | Sponge network | 0.083 | 0.66405 | -2.547 | 0 | 0.327 |
| 34469 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-191-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-495-3p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 24 | DDX6 | Sponge network | -0.031 | 0.89063 | 0.091 | 0.36428 | 0.327 |
| 34470 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 10 | CNTNAP3 | Sponge network | 0.559 | 0.00026 | -1.465 | 0.00033 | 0.327 |
| 34471 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-96-5p | 15 | PCDH8 | Sponge network | -2.452 | 0 | -0.997 | 0.05366 | 0.327 |
| 34472 | RP11-535M15.1 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-362-3p | 13 | IL6ST | Sponge network | -1.14 | 0.03513 | -0.402 | 0.00676 | 0.327 |
| 34473 | AC083843.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-582-3p | 12 | BRD3 | Sponge network | 1.4 | 0 | 0.37 | 0.00013 | 0.327 |
| 34474 | RP11-150O12.1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-30e-5p;hsa-miR-326 | 10 | SOX11 | Sponge network | 2.605 | 2.0E-5 | 2.68 | 0 | 0.327 |
| 34475 | RP11-403I13.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 0.395 | 0.15451 | 0.943 | 0 | 0.327 |
| 34476 | HAR1A |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-362-3p | 10 | GREM1 | Sponge network | -0.672 | 0.19447 | 0.902 | 0.01691 | 0.327 |
| 34477 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 18 | JAZF1 | Sponge network | 0.381 | 0.25967 | -0.377 | 0.02677 | 0.327 |
| 34478 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 18 | AR | Sponge network | -0.295 | 0.02604 | -1.554 | 0 | 0.327 |
| 34479 | RP11-77H9.2 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | NF1 | Sponge network | 1.677 | 0 | 0.51 | 0 | 0.327 |
| 34480 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-940 | 50 | NTRK3 | Sponge network | 0.214 | 0.18246 | -1.77 | 3.0E-5 | 0.327 |
| 34481 | FAM13A-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DNAJB4 | Sponge network | -0.83 | 0 | -0.7 | 1.0E-5 | 0.327 |
| 34482 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b | 14 | DMD | Sponge network | -0.351 | 0.11358 | -1.506 | 0 | 0.327 |
| 34483 | GABPB1-AS1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | EPG5 | Sponge network | 1.11 | 0 | 0.101 | 0.3563 | 0.327 |
| 34484 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-629-3p | 32 | ESR1 | Sponge network | -0.154 | 0.78939 | -1.035 | 3.0E-5 | 0.327 |
| 34485 | CALML3-AS1 |
hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 10 | LZTS1 | Sponge network | 0.95 | 0.15943 | 1.664 | 0 | 0.327 |
| 34486 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | USP6 | Sponge network | -0.222 | 0.32445 | 0.188 | 0.3871 | 0.327 |
| 34487 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-582-5p | 22 | EPHA5 | Sponge network | -2.095 | 0.00112 | -0.957 | 0.01733 | 0.327 |
| 34488 | LINC01018 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-5p | 28 | BTG2 | Sponge network | -2.915 | 0.00015 | -1.763 | 0 | 0.327 |
| 34489 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | RET | Sponge network | -2.46 | 0 | -0.833 | 0.03517 | 0.327 |
| 34490 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-590-3p | 31 | PPM1A | Sponge network | -0.738 | 0.21233 | -0.623 | 0 | 0.327 |
| 34491 | AC108488.4 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | BCL9 | Sponge network | 0.259 | 0.12542 | 0.321 | 0.00267 | 0.327 |
| 34492 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-93-5p | 18 | EPHA5 | Sponge network | -2.039 | 0.03447 | -0.957 | 0.01733 | 0.327 |
| 34493 | RP11-360F5.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-877-5p | 13 | FAT3 | Sponge network | 1.867 | 0 | -1.601 | 0.00051 | 0.327 |
| 34494 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 28 | PCDH9 | Sponge network | 0.219 | 0.1516 | -1.823 | 4.0E-5 | 0.327 |
| 34495 | RP11-384K6.6 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 11 | CDK6 | Sponge network | 0.946 | 0 | 0.991 | 0 | 0.327 |
| 34496 | PWAR6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 12 | PLA2R1 | Sponge network | -1.575 | 6.0E-5 | -0.265 | 0.24676 | 0.327 |
| 34497 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-550a-5p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | 0.41 | 0.02214 | 0.273 | 0.09957 | 0.326 |
| 34498 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-877-5p | 28 | TET1 | Sponge network | -0.615 | 0.00015 | -0.135 | 0.52138 | 0.326 |
| 34499 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CLIP1 | Sponge network | -0.053 | 0.80685 | -0.215 | 0.08684 | 0.326 |
| 34500 | LINC00672 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 27 | PPP1R9A | Sponge network | -0.227 | 0.31657 | -0.903 | 0.04254 | 0.326 |
| 34501 | WDFY3-AS2 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 14 | FOXO4 | Sponge network | -0.942 | 0 | -0.516 | 2.0E-5 | 0.326 |
| 34502 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | CHD2 | Sponge network | -1.575 | 6.0E-5 | 0.049 | 0.55371 | 0.326 |
| 34503 | RP4-668J24.2 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-24-2-5p;hsa-miR-424-5p | 10 | PCDH9 | Sponge network | -1.054 | 0.0706 | -1.823 | 4.0E-5 | 0.326 |
| 34504 | HOTAIRM1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PCDH10 | Sponge network | -0.053 | 0.80685 | -1.644 | 0.0078 | 0.326 |
| 34505 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | APC | Sponge network | 0.997 | 0.00066 | -0.255 | 0.04051 | 0.326 |
| 34506 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 15 | UVRAG | Sponge network | -0.942 | 0 | -0.017 | 0.83865 | 0.326 |
| 34507 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 24 | ZFP36L1 | Sponge network | -0.942 | 0 | -0.505 | 8.0E-5 | 0.326 |
| 34508 | KB-431C1.4 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p | 10 | KIF5B | Sponge network | 0.909 | 0 | 0.362 | 0.00066 | 0.326 |
| 34509 | DNAJC27-AS1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 22 | ITPKB | Sponge network | -0.969 | 2.0E-5 | -0.704 | 0.00106 | 0.326 |
| 34510 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | CLASP2 | Sponge network | -0.187 | 0.23787 | -0.038 | 0.71 | 0.326 |
| 34511 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 29 | PGR | Sponge network | 0.537 | 0.00139 | -1.704 | 0 | 0.326 |
| 34512 | MALAT1 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-590-3p | 12 | MACF1 | Sponge network | 0.782 | 0.00016 | 0.019 | 0.90335 | 0.326 |
| 34513 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 34 | IL6ST | Sponge network | -0.726 | 0.00132 | -0.402 | 0.00676 | 0.326 |
| 34514 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | KAT6B | Sponge network | 1.04 | 0.00023 | -0.478 | 0.00018 | 0.326 |
| 34515 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | CADM2 | Sponge network | -0.206 | 0.12942 | -3.782 | 0 | 0.326 |
| 34516 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p | 18 | ERC1 | Sponge network | -0.739 | 0.00044 | -0.108 | 0.38794 | 0.326 |
| 34517 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 33 | EBF1 | Sponge network | 1.302 | 7.0E-5 | -0.384 | 0.08856 | 0.326 |
| 34518 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-5p | 21 | SOX11 | Sponge network | 0.873 | 0 | 2.68 | 0 | 0.326 |
| 34519 | FENDRR |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-96-5p | 27 | CYLD | Sponge network | -1.281 | 0.00012 | -0.367 | 0.00171 | 0.326 |
| 34520 | RP11-428J1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | FGFR1 | Sponge network | 0.538 | 0.00076 | -0.509 | 0.03957 | 0.326 |
| 34521 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-942-5p | 15 | GPC6 | Sponge network | 0.238 | 0.70544 | -0.054 | 0.81465 | 0.326 |
| 34522 | RP11-158K1.3 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | DOCK4 | Sponge network | 0.349 | 0.01695 | 0.502 | 0.00108 | 0.326 |
| 34523 | RP4-717I23.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 10 | ABCA10 | Sponge network | 0.637 | 0.00069 | -0.571 | 0.03674 | 0.326 |
| 34524 | CTC-296K1.4 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-424-5p | 11 | LRIG1 | Sponge network | -2.076 | 0.00548 | -0.537 | 0.00588 | 0.326 |
| 34525 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-940 | 30 | TRPS1 | Sponge network | 0.16 | 0.50785 | -0.191 | 0.44109 | 0.326 |
| 34526 | HHIP-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-502-5p;hsa-miR-92a-3p | 22 | IL17RD | Sponge network | -0.737 | 0.10822 | -0.019 | 0.94873 | 0.326 |
| 34527 | RP11-819C21.1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p | 15 | RASAL2 | Sponge network | 0.537 | 0.00139 | 0.869 | 0 | 0.326 |
| 34528 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 21 | LRRC7 | Sponge network | 0.395 | 0.15451 | -1.341 | 0.01193 | 0.326 |
| 34529 | AC009120.6 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-590-3p | 11 | ZMYND11 | Sponge network | 0.919 | 0 | -0.2 | 0.05449 | 0.326 |
| 34530 | CTB-31O20.4 |
hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p | 11 | BRD3 | Sponge network | 1.619 | 0 | 0.37 | 0.00013 | 0.326 |
| 34531 | RP3-402G11.26 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-500a-3p;hsa-miR-590-3p | 14 | ZFHX4 | Sponge network | -0.254 | 0.19303 | 0.018 | 0.95574 | 0.326 |
| 34532 | GNG12-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p | 16 | ROBO1 | Sponge network | -0.793 | 7.0E-5 | -0.009 | 0.96154 | 0.326 |
| 34533 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 57 | TCF4 | Sponge network | -2.915 | 0.00015 | 0.016 | 0.92271 | 0.326 |
| 34534 | NEAT1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 17 | KIF5B | Sponge network | 0.705 | 0.00014 | 0.362 | 0.00066 | 0.326 |
| 34535 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PLSCR4 | Sponge network | 0.272 | 0.44692 | -0.474 | 0.03833 | 0.326 |
| 34536 | RP11-21A7A.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-576-5p;hsa-miR-93-3p | 16 | SSBP2 | Sponge network | -1.116 | 0.23686 | -1.58 | 0 | 0.326 |
| 34537 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429 | 15 | LHX6 | Sponge network | -1.249 | 7.0E-5 | -0.087 | 0.69193 | 0.326 |
| 34538 | CTC-490E21.10 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-493-5p | 19 | FAT3 | Sponge network | 1.46 | 1.0E-5 | -1.601 | 0.00051 | 0.326 |
| 34539 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 45 | TRPS1 | Sponge network | 0.083 | 0.66405 | -0.191 | 0.44109 | 0.326 |
| 34540 | RP11-53O19.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 21 | GJA1 | Sponge network | -0.405 | 0.22013 | -0.485 | 0.02179 | 0.326 |
| 34541 | AC003090.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | NCOA1 | Sponge network | -2.117 | 0.00161 | -0.432 | 0 | 0.326 |
| 34542 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | LRRC4 | Sponge network | -2.915 | 0.00015 | -1.774 | 0 | 0.326 |
| 34543 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-766-3p | 20 | TBL1XR1 | Sponge network | 0.75 | 0.00054 | 0.578 | 0 | 0.326 |
| 34544 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | FOXP1 | Sponge network | 0.532 | 0.00505 | 0.042 | 0.74368 | 0.326 |
| 34545 | LINC00473 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 16 | GJA1 | Sponge network | -1.203 | 0.03881 | -0.485 | 0.02179 | 0.326 |
| 34546 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | PREX2 | Sponge network | 0.219 | 0.1516 | -0.272 | 0.25382 | 0.326 |
| 34547 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | SON | Sponge network | 0.494 | 0.00339 | 0.168 | 0.04406 | 0.326 |
| 34548 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-654-3p | 16 | DDX6 | Sponge network | 1.923 | 0 | 0.091 | 0.36428 | 0.326 |
| 34549 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-5p | 12 | SMARCA2 | Sponge network | -1.645 | 0 | -0.529 | 0.00014 | 0.326 |
| 34550 | AC016747.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 17 | CDH23 | Sponge network | 0.199 | 0.38015 | -0.974 | 0.00034 | 0.326 |
| 34551 | HCG11 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DNAJB4 | Sponge network | 0.385 | 0.10067 | -0.7 | 1.0E-5 | 0.326 |
| 34552 | AC005618.6 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | 0.862 | 0.06213 | -0.998 | 0.00025 | 0.326 |
| 34553 | ZNF667-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-942-5p | 23 | OPCML | Sponge network | -0.976 | 0.00025 | -2.498 | 0 | 0.326 |
| 34554 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-362-3p | 12 | IGF2 | Sponge network | -0.154 | 0.53954 | 0.848 | 0.03842 | 0.326 |
| 34555 | FZD10-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TLL1 | Sponge network | -0.727 | 0.04726 | -1.195 | 3.0E-5 | 0.326 |
| 34556 | CTD-3092A11.2 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | KIF5B | Sponge network | 0.873 | 0 | 0.362 | 0.00066 | 0.326 |
| 34557 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 16 | TET1 | Sponge network | 0.811 | 0.03228 | -0.135 | 0.52138 | 0.326 |
| 34558 | RP11-359E10.1 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-326;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p | 12 | CSDE1 | Sponge network | 0.299 | 0.57722 | -0.493 | 0 | 0.326 |
| 34559 | AC003090.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-940 | 22 | TOM1L2 | Sponge network | -2.117 | 0.00161 | -0.994 | 0 | 0.326 |
| 34560 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p;hsa-miR-655-3p | 14 | ARID2 | Sponge network | 0.614 | 1.0E-5 | 0.235 | 0.02697 | 0.326 |
| 34561 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-345-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-664a-5p | 35 | LPP | Sponge network | 0.858 | 3.0E-5 | -0.459 | 0.04475 | 0.326 |
| 34562 | TRAM2-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | PIK3CA | Sponge network | 0.336 | 0.00739 | 0.195 | 0.06908 | 0.326 |
| 34563 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | ITCH | Sponge network | 1.317 | 0 | 0.084 | 0.34058 | 0.326 |
| 34564 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 22 | GPC6 | Sponge network | -2.915 | 0.00015 | -0.054 | 0.81465 | 0.326 |
| 34565 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p | 17 | CRTC3 | Sponge network | 0.603 | 0.00127 | 0.164 | 0.16786 | 0.326 |
| 34566 | RP11-597D13.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 1.302 | 7.0E-5 | 0.943 | 0 | 0.326 |
| 34567 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-628-5p | 11 | BCL6 | Sponge network | -0.376 | 0.00337 | -0.211 | 0.19489 | 0.326 |
| 34568 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | SLITRK3 | Sponge network | 0.532 | 0.00505 | -3.328 | 0 | 0.326 |
| 34569 | DOCK9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p | 13 | HBP1 | Sponge network | -0.231 | 0.28214 | -0.454 | 0 | 0.326 |
| 34570 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PBRM1 | Sponge network | 0.101 | 0.60919 | -0.029 | 0.78601 | 0.326 |
| 34571 | SH3RF3-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 18 | LUZP2 | Sponge network | 0.889 | 9.0E-5 | -0.056 | 0.89416 | 0.326 |
| 34572 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-625-5p | 21 | ABI2 | Sponge network | -2.531 | 0 | 0.175 | 0.14787 | 0.326 |
| 34573 | GABPB1-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | TCF12 | Sponge network | 1.11 | 0 | 0.174 | 0.13271 | 0.326 |
| 34574 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-5p | 11 | SMARCA2 | Sponge network | -1.047 | 0 | -0.529 | 0.00014 | 0.326 |
| 34575 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-5p | 20 | LRP1B | Sponge network | 0.176 | 0.37402 | -2.163 | 0 | 0.326 |
| 34576 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 32 | SOX5 | Sponge network | 0.75 | 0.00054 | -1.091 | 4.0E-5 | 0.326 |
| 34577 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 38 | TCF4 | Sponge network | 1.61 | 0.00961 | 0.016 | 0.92271 | 0.326 |
| 34578 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | PRKCE | Sponge network | -0.323 | 0.01372 | -0.403 | 0.00056 | 0.326 |
| 34579 | RP11-181G12.2 |
hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-744-3p | 30 | LPP | Sponge network | 0.556 | 0.00298 | -0.459 | 0.04475 | 0.326 |
| 34580 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 58 | NTRK3 | Sponge network | -1.047 | 0 | -1.77 | 3.0E-5 | 0.326 |
| 34581 | HOXB-AS1 |
hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-589-5p | 11 | ERRFI1 | Sponge network | -0.154 | 0.53954 | -0.798 | 1.0E-5 | 0.326 |
| 34582 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | IL6ST | Sponge network | -0.737 | 0.10822 | -0.402 | 0.00676 | 0.326 |
| 34583 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 28 | SNX29 | Sponge network | 0.72 | 0.0001 | -0.34 | 0.01371 | 0.326 |
| 34584 | LINC01006 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-539-5p;hsa-miR-624-5p | 14 | BCL11A | Sponge network | -0.702 | 0.04814 | -0.932 | 0.0063 | 0.326 |
| 34585 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 27 | DMXL1 | Sponge network | 0.782 | 0.00016 | -0.426 | 0.00056 | 0.326 |
| 34586 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 26 | UNC80 | Sponge network | 0.051 | 0.83388 | -1.934 | 0 | 0.326 |
| 34587 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 11 | GPAM | Sponge network | 1.316 | 0 | 0.142 | 0.29732 | 0.326 |
| 34588 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-93-5p | 14 | TXNIP | Sponge network | 0.532 | 0.00505 | -1.277 | 0 | 0.326 |
| 34589 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p | 12 | ABCB5 | Sponge network | 0.176 | 0.37402 | -0.956 | 0.04077 | 0.326 |
| 34590 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PLSCR4 | Sponge network | 0.559 | 0.00026 | -0.474 | 0.03833 | 0.326 |
| 34591 | GHRLOS |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-98-5p | 25 | NTRK3 | Sponge network | -1.942 | 0 | -1.77 | 3.0E-5 | 0.326 |
| 34592 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 24 | MCC | Sponge network | -0.322 | 0.25945 | -1.005 | 0 | 0.326 |
| 34593 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-577 | 14 | NOTCH2NL | Sponge network | -0.18 | 0.20343 | 0.024 | 0.84307 | 0.326 |
| 34594 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 30 | RNF144A | Sponge network | -0.942 | 0 | 0.594 | 0.0009 | 0.326 |
| 34595 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | CLIP1 | Sponge network | -2.095 | 0.00112 | -0.215 | 0.08684 | 0.326 |
| 34596 | CTB-31O20.2 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-299-5p | 10 | MAT2A | Sponge network | 0.608 | 3.0E-5 | -0.073 | 0.36195 | 0.326 |
| 34597 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TMEFF2 | Sponge network | -0.668 | 3.0E-5 | -2.426 | 0 | 0.326 |
| 34598 | CTD-3064M3.3 |
hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429 | 15 | OPCML | Sponge network | -0.469 | 0.08721 | -2.498 | 0 | 0.326 |
| 34599 | DNM1P35 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | PEG3 | Sponge network | 0.051 | 0.83388 | -1.74 | 0 | 0.326 |
| 34600 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 13 | RAPGEF5 | Sponge network | 0.683 | 1.0E-5 | 0.536 | 4.0E-5 | 0.326 |
| 34601 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | LRP1B | Sponge network | -1.349 | 0.00017 | -2.163 | 0 | 0.326 |
| 34602 | RP1-39G22.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p | 27 | MDM4 | Sponge network | 0.439 | 0.00313 | 0.345 | 0.00762 | 0.326 |
| 34603 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 23 | CREB3L2 | Sponge network | -0.726 | 0.00132 | -0.525 | 2.0E-5 | 0.326 |
| 34604 | C4B-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-330-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 13 | EBF1 | Sponge network | 2.306 | 9.0E-5 | -0.384 | 0.08856 | 0.326 |
| 34605 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-942-5p | 48 | THRB | Sponge network | -0.147 | 0.40452 | -1.117 | 0.0004 | 0.326 |
| 34606 | RP11-395G23.3 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 33 | DST | Sponge network | 0.381 | 0.25967 | -0.713 | 0.00096 | 0.326 |
| 34607 | RP11-400K9.3 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-576-5p | 11 | TMEM132B | Sponge network | -0.733 | 0.19469 | -0.83 | 0.01084 | 0.326 |
| 34608 | RP11-725P16.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | BRD3 | Sponge network | 0.422 | 0.27021 | 0.37 | 0.00013 | 0.326 |
| 34609 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429 | 12 | CLIP1 | Sponge network | -1.654 | 0.00144 | -0.215 | 0.08684 | 0.326 |
| 34610 | RP11-356J5.12 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p | 19 | ABL1 | Sponge network | -0.899 | 6.0E-5 | -0.215 | 0.07804 | 0.326 |
| 34611 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-5p | 17 | RET | Sponge network | -1.349 | 0.00017 | -0.833 | 0.03517 | 0.326 |
| 34612 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | FREM2 | Sponge network | -1.264 | 6.0E-5 | -0.437 | 0.46353 | 0.326 |
| 34613 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-501-5p;hsa-miR-590-3p | 15 | EBF3 | Sponge network | -0.663 | 0.10887 | -0.058 | 0.81437 | 0.326 |
| 34614 | RP11-1006G14.4 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | BAZ2B | Sponge network | 0.559 | 0.00026 | -0.276 | 0.02833 | 0.326 |
| 34615 | ZNF667-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 19 | UNC5C | Sponge network | -0.976 | 0.00025 | -0.178 | 0.40214 | 0.326 |
| 34616 | RP11-57H14.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-708-5p;hsa-miR-93-3p;hsa-miR-940 | 31 | TOM1L2 | Sponge network | 0.083 | 0.66405 | -0.994 | 0 | 0.326 |
| 34617 | U91328.19 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-660-5p;hsa-miR-93-3p | 56 | RUNX1T1 | Sponge network | -0.303 | 0.21002 | -0.344 | 0.2374 | 0.326 |
| 34618 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-628-5p | 22 | GRIA3 | Sponge network | -0.896 | 0.00099 | -1.956 | 0 | 0.326 |
| 34619 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 30 | ST6GAL2 | Sponge network | 1.302 | 7.0E-5 | -1.201 | 0.00597 | 0.326 |
| 34620 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-628-5p;hsa-miR-7-5p | 20 | TCF4 | Sponge network | -1.054 | 0.0706 | 0.016 | 0.92271 | 0.326 |
| 34621 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 31 | PPM1A | Sponge network | -0.031 | 0.89063 | -0.623 | 0 | 0.326 |
| 34622 | KB-431C1.4 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-664a-3p | 16 | RASAL2 | Sponge network | 0.909 | 0 | 0.869 | 0 | 0.326 |
| 34623 | RP11-360F5.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p | 15 | AR | Sponge network | 1.867 | 0 | -1.554 | 0 | 0.326 |
| 34624 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-93-5p | 12 | JAK1 | Sponge network | -0.129 | 0.57177 | 0.001 | 0.98982 | 0.326 |
| 34625 | RP1-302G2.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-766-3p | 12 | IL17RD | Sponge network | -1.483 | 9.0E-5 | -0.019 | 0.94873 | 0.326 |
| 34626 | SOX2-OT |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | SPOP | Sponge network | -1.264 | 6.0E-5 | -0.419 | 1.0E-5 | 0.326 |
| 34627 | RP11-693J15.4 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-7-1-3p;hsa-miR-93-3p | 14 | DOCK4 | Sponge network | -0.72 | 0.24239 | 0.502 | 0.00108 | 0.326 |
| 34628 | H19 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-664a-3p | 12 | RASAL2 | Sponge network | 2.439 | 0 | 0.869 | 0 | 0.326 |
| 34629 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 51 | TCF4 | Sponge network | -0.405 | 0.22013 | 0.016 | 0.92271 | 0.326 |
| 34630 | KB-1460A1.5 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PDE4DIP | Sponge network | 0.603 | 0.00127 | -0.282 | 0.0257 | 0.326 |
| 34631 | RP11-251G23.5 |
hsa-miR-10b-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p;hsa-miR-539-5p | 14 | ARHGEF12 | Sponge network | 1.344 | 0 | 0.09 | 0.35693 | 0.326 |
| 34632 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 22 | EGFR | Sponge network | -1.092 | 4.0E-5 | -0.175 | 0.48451 | 0.326 |
| 34633 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | CLASP2 | Sponge network | 1.317 | 0 | -0.038 | 0.71 | 0.326 |
| 34634 | AGAP11 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-455-5p | 10 | AMPH | Sponge network | -1.645 | 0 | -0.916 | 5.0E-5 | 0.326 |
| 34635 | LINC00672 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 11 | PRDM5 | Sponge network | -0.227 | 0.31657 | -1.09 | 0.00014 | 0.326 |
| 34636 | RP11-927P21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-590-5p | 16 | AKAP6 | Sponge network | 0.921 | 0.00118 | -1.586 | 3.0E-5 | 0.326 |
| 34637 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | CREBBP | Sponge network | -0.777 | 0.1134 | 0.126 | 0.15186 | 0.326 |
| 34638 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | GRIN2A | Sponge network | -0.663 | 0.10887 | -2.161 | 0 | 0.326 |
| 34639 | RP4-584D14.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-577 | 22 | RUNX1T1 | Sponge network | 1.294 | 0.0031 | -0.344 | 0.2374 | 0.326 |
| 34640 | RP11-723D22.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-500a-5p | 10 | RICTOR | Sponge network | 0.562 | 0.20117 | 0.388 | 0.00188 | 0.326 |
| 34641 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-493-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 24 | ZFHX3 | Sponge network | 0.056 | 0.8494 | 0.389 | 0.01363 | 0.326 |
| 34642 | A1BG-AS1 |
hsa-miR-10a-3p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-532-5p | 15 | IGFBP5 | Sponge network | -0.164 | 0.45106 | -0.806 | 0.00105 | 0.326 |
| 34643 | WDFY3-AS2 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p | 22 | CSDE1 | Sponge network | -0.942 | 0 | -0.493 | 0 | 0.326 |
| 34644 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 28 | GRIN2A | Sponge network | -1.047 | 0 | -2.161 | 0 | 0.326 |
| 34645 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-424-5p | 14 | DYNC1I1 | Sponge network | -0.465 | 0.04703 | -1.12 | 0.00107 | 0.326 |
| 34646 | U91328.19 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 22 | CDH19 | Sponge network | -0.303 | 0.21002 | -3.434 | 0 | 0.326 |
| 34647 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 11 | ZMYM2 | Sponge network | 0.832 | 0 | 0.279 | 0.01273 | 0.326 |
| 34648 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-942-5p | 13 | RB1CC1 | Sponge network | -0.223 | 0.47816 | 0.175 | 0.12559 | 0.326 |
| 34649 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-96-5p | 23 | MYO9A | Sponge network | 0.078 | 0.55803 | -0.147 | 0.32373 | 0.326 |
| 34650 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | RBMS3 | Sponge network | 0.349 | 0.01695 | -0.519 | 0.03551 | 0.326 |
| 34651 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p | 19 | IGSF10 | Sponge network | -0.738 | 0.21233 | -1.751 | 1.0E-5 | 0.326 |
| 34652 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-628-5p;hsa-miR-940 | 24 | MEF2C | Sponge network | 0.637 | 0.00069 | -0.499 | 0.01448 | 0.326 |
| 34653 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p | 15 | PTPRB | Sponge network | 0.056 | 0.81195 | 0.105 | 0.47122 | 0.326 |
| 34654 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DACT1 | Sponge network | -0.323 | 0.01372 | 0.783 | 0.00072 | 0.326 |
| 34655 | RP11-867G23.10 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-501-5p;hsa-miR-589-3p | 11 | RNF144A | Sponge network | -2.531 | 0 | 0.594 | 0.0009 | 0.326 |
| 34656 | BZRAP1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-550a-3p;hsa-miR-584-5p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 28 | PTPRT | Sponge network | -0.465 | 0.04703 | -0.907 | 0.03727 | 0.326 |
| 34657 | AC009948.5 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 17 | SHPRH | Sponge network | 0.532 | 0.00505 | 0.098 | 0.40994 | 0.326 |
| 34658 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 13 | GPAM | Sponge network | 1.352 | 0 | 0.142 | 0.29732 | 0.326 |
| 34659 | RP11-53O19.1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 23 | LIMCH1 | Sponge network | -0.405 | 0.22013 | -0.915 | 1.0E-5 | 0.326 |
| 34660 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p | 13 | PSIP1 | Sponge network | 0.389 | 0.04293 | -0.005 | 0.96897 | 0.326 |
| 34661 | GABPB1-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PTPN12 | Sponge network | 1.11 | 0 | 0.768 | 0 | 0.326 |
| 34662 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-93-5p | 49 | AR | Sponge network | 0.253 | 0.09565 | -1.554 | 0 | 0.326 |
| 34663 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-361-3p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-708-5p;hsa-miR-935 | 26 | MTSS1 | Sponge network | -0.384 | 0.40652 | -0.81 | 0.00035 | 0.326 |
| 34664 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 54 | TCF4 | Sponge network | 0.083 | 0.66405 | 0.016 | 0.92271 | 0.326 |
| 34665 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-940;hsa-miR-96-5p | 18 | CRTC3 | Sponge network | -0.08 | 0.90092 | 0.164 | 0.16786 | 0.326 |
| 34666 | RP5-1085F17.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 18 | TSC1 | Sponge network | 0.541 | 0.00125 | 0.103 | 0.26071 | 0.326 |
| 34667 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-500b-5p | 15 | SLITRK6 | Sponge network | -1.645 | 0 | -2.121 | 0 | 0.326 |
| 34668 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p | 28 | PRDM2 | Sponge network | -1.216 | 0 | -0.258 | 0.00721 | 0.326 |
| 34669 | CTD-2314G24.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 31 | ETV1 | Sponge network | 0.246 | 0.59912 | -0.096 | 0.67674 | 0.326 |
| 34670 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | PBRM1 | Sponge network | 0.953 | 0 | -0.029 | 0.78601 | 0.326 |
| 34671 | RP11-47A8.5 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-92a-1-5p | 18 | CUX1 | Sponge network | 0.103 | 0.43169 | -0.039 | 0.6705 | 0.326 |
| 34672 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-486-5p | 14 | NR2C2 | Sponge network | 1.61 | 0.00961 | 0.21 | 0.03224 | 0.326 |
| 34673 | RP11-25K19.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 13 | UNC5D | Sponge network | -1.057 | 0.00029 | -3.646 | 0 | 0.326 |
| 34674 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 17 | FGF5 | Sponge network | -0.405 | 0.22013 | 0.549 | 0.28659 | 0.326 |
| 34675 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-3p | 22 | PRDM2 | Sponge network | -0.246 | 0.35034 | -0.258 | 0.00721 | 0.326 |
| 34676 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-7-1-3p | 10 | DACT1 | Sponge network | -4.546 | 0 | 0.783 | 0.00072 | 0.326 |
| 34677 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 19 | MYBL1 | Sponge network | 0.862 | 0.06213 | 0.494 | 0.02152 | 0.326 |
| 34678 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p | 10 | ABCB5 | Sponge network | 0.238 | 0.70544 | -0.956 | 0.04077 | 0.326 |
| 34679 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-96-5p | 37 | FAT3 | Sponge network | 0.41 | 0.02214 | -1.601 | 0.00051 | 0.326 |
| 34680 | MIR600HG |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-505-5p | 14 | DDX3X | Sponge network | 0.594 | 0.41522 | -0.152 | 0.07686 | 0.326 |
| 34681 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 22 | LRRC55 | Sponge network | -1.281 | 0.00012 | -0.026 | 0.93163 | 0.326 |
| 34682 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-7-1-3p | 13 | GRIN2A | Sponge network | -0.351 | 0.11358 | -2.161 | 0 | 0.326 |
| 34683 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-423-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 15 | SUFU | Sponge network | 0.299 | 0.57722 | -0.04 | 0.65489 | 0.326 |
| 34684 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-3607-3p | 10 | AFAP1L2 | Sponge network | -0.611 | 0.09607 | -0.284 | 0.15434 | 0.326 |
| 34685 | MIR497HG |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 12 | FOXO4 | Sponge network | -1.439 | 4.0E-5 | -0.516 | 2.0E-5 | 0.326 |
| 34686 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 28 | DOCK4 | Sponge network | -1.697 | 0.00889 | 0.502 | 0.00108 | 0.326 |
| 34687 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 17 | LRRC7 | Sponge network | 0.214 | 0.18246 | -1.341 | 0.01193 | 0.326 |
| 34688 | CTD-2336O2.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-940 | 26 | NTRK2 | Sponge network | -0.141 | 0.26434 | -0.736 | 0.06495 | 0.326 |
| 34689 | MIR22HG |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 16 | CXCL12 | Sponge network | -1.047 | 0 | -1.421 | 0 | 0.326 |
| 34690 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-654-3p | 11 | PSIP1 | Sponge network | 0.657 | 4.0E-5 | -0.005 | 0.96897 | 0.326 |
| 34691 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-874-3p;hsa-miR-940 | 17 | CRTC3 | Sponge network | 0.392 | 0.0278 | 0.164 | 0.16786 | 0.326 |
| 34692 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ZFHX3 | Sponge network | 0.998 | 0 | 0.389 | 0.01363 | 0.326 |
| 34693 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-93-3p | 18 | EBF3 | Sponge network | 0.559 | 0.00026 | -0.058 | 0.81437 | 0.326 |
| 34694 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-7-1-3p | 14 | DKK3 | Sponge network | 0.41 | 0.02214 | -0.052 | 0.77783 | 0.326 |
| 34695 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | AR | Sponge network | -0.023 | 0.95109 | -1.554 | 0 | 0.326 |
| 34696 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | PDE4DIP | Sponge network | 0.219 | 0.1516 | -0.282 | 0.0257 | 0.326 |
| 34697 | RP11-327P2.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-532-5p | 26 | IL17RD | Sponge network | -0.147 | 0.40452 | -0.019 | 0.94873 | 0.326 |
| 34698 | CTD-2303H24.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p | 11 | FBXO11 | Sponge network | 0.415 | 0.14657 | 0.142 | 0.04938 | 0.326 |
| 34699 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-3p | 33 | KIF1B | Sponge network | 0.765 | 7.0E-5 | -0.118 | 0.32258 | 0.326 |
| 34700 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 12 | DDX6 | Sponge network | 0.272 | 0.44692 | 0.091 | 0.36428 | 0.326 |
| 34701 | RP11-65J21.3 |
hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 29 | LPP | Sponge network | -0.557 | 0.11135 | -0.459 | 0.04475 | 0.326 |
| 34702 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | THBS1 | Sponge network | -0.487 | 0.02157 | 0.741 | 0.00386 | 0.326 |
| 34703 | RP11-244H3.1 |
hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-505-3p;hsa-miR-7-5p | 12 | FNBP1 | Sponge network | 0.659 | 3.0E-5 | -0.969 | 4.0E-5 | 0.326 |
| 34704 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | LATS2 | Sponge network | -0.793 | 7.0E-5 | 0.159 | 0.2304 | 0.326 |
| 34705 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 28 | SASH1 | Sponge network | 0.214 | 0.18246 | -0.502 | 0.00175 | 0.326 |
| 34706 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-423-5p | 10 | CCND2 | Sponge network | -1.717 | 0.01522 | -0.267 | 0.3112 | 0.326 |
| 34707 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 35 | MOB1B | Sponge network | -0.901 | 0.00019 | 0.197 | 0.15266 | 0.326 |
| 34708 | LINC00861 |
hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-582-3p;hsa-miR-877-5p;hsa-miR-940;hsa-miR-96-5p | 13 | GAS7 | Sponge network | 0.911 | 0.00854 | -0.555 | 0.01602 | 0.326 |
| 34709 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | BCL2 | Sponge network | 0.225 | 0.30644 | -0.899 | 0 | 0.326 |
| 34710 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 33 | AKAP11 | Sponge network | 0.415 | 0.14657 | 0.196 | 0.09825 | 0.326 |
| 34711 | RP11-474O21.5 |
hsa-miR-130b-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-424-3p;hsa-miR-7-1-3p | 12 | WWTR1 | Sponge network | -0.023 | 0.95109 | -0.345 | 0.11997 | 0.326 |
| 34712 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | BHLHE41 | Sponge network | -0.737 | 0.10822 | 0.353 | 0.12753 | 0.326 |
| 34713 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 14 | LATS1 | Sponge network | 1.317 | 0 | 0.109 | 0.34208 | 0.326 |
| 34714 | THAP9-AS1 |
hsa-miR-125a-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | ERCC6 | Sponge network | 1.079 | 0 | 0.23 | 0.015 | 0.326 |
| 34715 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-93-5p | 36 | RASSF2 | Sponge network | -0.615 | 0.00015 | -0.273 | 0.21961 | 0.326 |
| 34716 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 25 | GPC6 | Sponge network | -0.793 | 7.0E-5 | -0.054 | 0.81465 | 0.326 |
| 34717 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 15 | AKAP11 | Sponge network | 2.094 | 0 | 0.196 | 0.09825 | 0.326 |
| 34718 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 51 | THRB | Sponge network | -0.235 | 0.15142 | -1.117 | 0.0004 | 0.326 |
| 34719 | CD27-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 15 | NRXN3 | Sponge network | 0.177 | 0.16681 | -0.893 | 0.04186 | 0.326 |
| 34720 | RP11-157J24.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-576-5p | 16 | AR | Sponge network | -1.664 | 0.05165 | -1.554 | 0 | 0.326 |
| 34721 | CTA-14H9.5 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 10 | ABI2 | Sponge network | 0.145 | 0.53343 | 0.175 | 0.14787 | 0.326 |
| 34722 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | TIMP3 | Sponge network | 0.064 | 0.72934 | -0.546 | 0.02307 | 0.326 |
| 34723 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | MCC | Sponge network | -0.206 | 0.12942 | -1.005 | 0 | 0.326 |
| 34724 | HAND2-AS1 |
hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-660-5p | 10 | DNER | Sponge network | -1.697 | 0.00889 | -3.787 | 0 | 0.326 |
| 34725 | MIR143HG |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-940 | 24 | BAZ2B | Sponge network | -0.739 | 0.0574 | -0.276 | 0.02833 | 0.326 |
| 34726 | RP11-800A3.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-330-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | RASSF8 | Sponge network | 0.18 | 0.7147 | -0.122 | 0.65591 | 0.326 |
| 34727 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 21 | PIK3R1 | Sponge network | 0.921 | 0.00118 | -0.133 | 0.36854 | 0.326 |
| 34728 | CTB-179K24.3 |
hsa-let-7d-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | RICTOR | Sponge network | 0.841 | 0.04055 | 0.388 | 0.00188 | 0.326 |
| 34729 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -2.69 | 0.0001 | -0.243 | 0.11408 | 0.326 |
| 34730 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p | 16 | CAV1 | Sponge network | -0.615 | 0.00015 | -1.013 | 6.0E-5 | 0.326 |
| 34731 | RP1-122P22.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-629-3p;hsa-miR-942-5p | 10 | GPC6 | Sponge network | 0.065 | 0.73468 | -0.054 | 0.81465 | 0.326 |
| 34732 | GNG12-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p | 23 | CRTC1 | Sponge network | -0.793 | 7.0E-5 | -0.116 | 0.28412 | 0.326 |
| 34733 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | JAZF1 | Sponge network | -0.08 | 0.90092 | -0.377 | 0.02677 | 0.326 |
| 34734 | MIR497HG |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | RNF144A | Sponge network | -1.439 | 4.0E-5 | 0.594 | 0.0009 | 0.326 |
| 34735 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 21 | EPHA3 | Sponge network | 0.637 | 0.00069 | 0.185 | 0.53588 | 0.325 |
| 34736 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MYO9A | Sponge network | -0.342 | 0.02288 | -0.147 | 0.32373 | 0.325 |
| 34737 | RP11-253E3.3 |
hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-497-5p | 10 | TBL1XR1 | Sponge network | 1.764 | 0 | 0.578 | 0 | 0.325 |
| 34738 | CTC-510F12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-361-3p;hsa-miR-376c-3p | 11 | NCOA1 | Sponge network | -0.691 | 0.00146 | -0.432 | 0 | 0.325 |
| 34739 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 17 | HIP1 | Sponge network | 0.619 | 0.00157 | 0.774 | 0 | 0.325 |
| 34740 | MRPL23-AS1 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-625-5p | 19 | ANK2 | Sponge network | -0.278 | 0.76559 | -1.603 | 1.0E-5 | 0.325 |
| 34741 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 18 | ARHGAP29 | Sponge network | 0.537 | 0.00139 | 0.131 | 0.42953 | 0.325 |
| 34742 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-590-3p | 11 | MAP3K12 | Sponge network | -0.737 | 0.10822 | -0.057 | 0.59369 | 0.325 |
| 34743 | RP11-158K1.3 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p | 17 | ABL1 | Sponge network | 0.349 | 0.01695 | -0.215 | 0.07804 | 0.325 |
| 34744 | RP11-705C15.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 13 | ABI2 | Sponge network | 0.796 | 4.0E-5 | 0.175 | 0.14787 | 0.325 |
| 34745 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 55 | WDFY3 | Sponge network | 0.421 | 0.00084 | -0.201 | 0.0967 | 0.325 |
| 34746 | TRAF3IP2-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 17 | FMNL3 | Sponge network | -0.323 | 0.01372 | 0.555 | 4.0E-5 | 0.325 |
| 34747 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 29 | RBMS3 | Sponge network | 0.614 | 1.0E-5 | -0.519 | 0.03551 | 0.325 |
| 34748 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-7-5p | 14 | CRTC3 | Sponge network | 0.663 | 0.00073 | 0.164 | 0.16786 | 0.325 |
| 34749 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 25 | SCN9A | Sponge network | 0.219 | 0.1516 | -0.525 | 0.10593 | 0.325 |
| 34750 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p | 17 | FREM2 | Sponge network | -0.162 | 0.47687 | -0.437 | 0.46353 | 0.325 |
| 34751 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-484 | 11 | PKNOX1 | Sponge network | -1.331 | 3.0E-5 | 0.085 | 0.28917 | 0.325 |
| 34752 | MEG3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | MED12L | Sponge network | 0.249 | 0.35902 | -0.194 | 0.55299 | 0.325 |
| 34753 | HOXB-AS3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-339-5p | 10 | SOX11 | Sponge network | 0.343 | 0.28887 | 2.68 | 0 | 0.325 |
| 34754 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 19 | TACC1 | Sponge network | -0.285 | 0.2172 | -0.362 | 0.05406 | 0.325 |
| 34755 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 13 | ABCB5 | Sponge network | -2.915 | 0.00015 | -0.956 | 0.04077 | 0.325 |
| 34756 | ADAMTS9-AS2 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 20 | ATM | Sponge network | -2.452 | 0 | 0.178 | 0.21568 | 0.325 |
| 34757 | LINC00662 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-223-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | MTSS1 | Sponge network | 0.213 | 0.32297 | -0.81 | 0.00035 | 0.325 |
| 34758 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-495-3p | 25 | RICTOR | Sponge network | 0.331 | 0.57247 | 0.388 | 0.00188 | 0.325 |
| 34759 | PART1 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-708-5p | 20 | FAM135B | Sponge network | -2.925 | 0 | -3.978 | 0 | 0.325 |
| 34760 | RP11-867G23.10 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-450b-5p;hsa-miR-93-5p | 12 | CDHR3 | Sponge network | -2.531 | 0 | -0.977 | 4.0E-5 | 0.325 |
| 34761 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-505-5p | 13 | KCNA6 | Sponge network | -0.829 | 0.01343 | -1.531 | 0.00013 | 0.325 |
| 34762 | CTD-2089N3.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p | 11 | PIK3R1 | Sponge network | -0.314 | 0.62033 | -0.133 | 0.36854 | 0.325 |
| 34763 | AC016747.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p | 11 | RELN | Sponge network | 0.199 | 0.38015 | -2.403 | 0 | 0.325 |
| 34764 | RP11-819C21.1 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 15 | GLIPR1 | Sponge network | 0.537 | 0.00139 | 0.146 | 0.3276 | 0.325 |
| 34765 | HAND2-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-429 | 11 | DUSP1 | Sponge network | -1.697 | 0.00889 | -1.754 | 0 | 0.325 |
| 34766 | DNM1P35 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | ARNT2 | Sponge network | 0.051 | 0.83388 | -0.332 | 0.25851 | 0.325 |
| 34767 | MIR143HG |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-92a-1-5p | 13 | PRKACA | Sponge network | -0.739 | 0.0574 | -0.255 | 0.0012 | 0.325 |
| 34768 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429 | 13 | CNTN1 | Sponge network | -0.785 | 0.00035 | -2.594 | 0 | 0.325 |
| 34769 | RP1-151F17.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PHF3 | Sponge network | 0.222 | 0.29133 | 0.126 | 0.19652 | 0.325 |
| 34770 | GNG12-AS1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | SH3GLB1 | Sponge network | -0.793 | 7.0E-5 | -0.115 | 0.14773 | 0.325 |
| 34771 | AC010226.4 |
hsa-let-7a-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-330-3p;hsa-miR-33a-3p | 10 | MAFB | Sponge network | -0.811 | 2.0E-5 | -0.023 | 0.89865 | 0.325 |
| 34772 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-93-5p | 27 | LRP1B | Sponge network | 0.082 | 0.55654 | -2.163 | 0 | 0.325 |
| 34773 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 20 | ARHGAP28 | Sponge network | -1.057 | 0.00029 | -0.911 | 0.00013 | 0.325 |
| 34774 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 38 | EPHA7 | Sponge network | 0.199 | 0.38015 | -2.197 | 3.0E-5 | 0.325 |
| 34775 | RP13-516M14.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-582-5p | 39 | RUNX1T1 | Sponge network | 0.389 | 0.04293 | -0.344 | 0.2374 | 0.325 |
| 34776 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | SYNE1 | Sponge network | -0.83 | 0 | -0.738 | 0.00316 | 0.325 |
| 34777 | LINC01018 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 33 | PTPRT | Sponge network | -2.915 | 0.00015 | -0.907 | 0.03727 | 0.325 |
| 34778 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | RBMS3 | Sponge network | 0.392 | 0.0278 | -0.519 | 0.03551 | 0.325 |
| 34779 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | CUL5 | Sponge network | 0.539 | 0.03722 | 0.026 | 0.77301 | 0.325 |
| 34780 | RP11-59H7.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-744-3p | 27 | LPP | Sponge network | 2.163 | 0 | -0.459 | 0.04475 | 0.325 |
| 34781 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-744-3p | 21 | PTPN14 | Sponge network | -1.654 | 0.00144 | 0.144 | 0.34595 | 0.325 |
| 34782 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 19 | LRRC7 | Sponge network | 0.537 | 0.00139 | -1.341 | 0.01193 | 0.325 |
| 34783 | CTC-296K1.4 |
hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-576-5p | 18 | TMEM132B | Sponge network | -2.076 | 0.00548 | -0.83 | 0.01084 | 0.325 |
| 34784 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p | 25 | PRDM2 | Sponge network | -0.405 | 0.22013 | -0.258 | 0.00721 | 0.325 |
| 34785 | RP11-473I1.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p | 11 | BRD3 | Sponge network | 0.542 | 0.00054 | 0.37 | 0.00013 | 0.325 |
| 34786 | RP4-758J18.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-330-5p;hsa-miR-450b-5p | 14 | DMD | Sponge network | -0.091 | 0.71468 | -1.506 | 0 | 0.325 |
| 34787 | GAS6-AS2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p | 22 | ERC1 | Sponge network | 0.16 | 0.50785 | -0.108 | 0.38794 | 0.325 |
| 34788 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-7-1-3p | 16 | FGF5 | Sponge network | 0.395 | 0.15451 | 0.549 | 0.28659 | 0.325 |
| 34789 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 29 | THBS1 | Sponge network | 0.101 | 0.60919 | 0.741 | 0.00386 | 0.325 |
| 34790 | RP11-464F9.1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-664a-5p | 15 | CBFB | Sponge network | 0.959 | 0 | 1.061 | 0 | 0.325 |
| 34791 | LINC01018 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-942-5p;hsa-miR-96-5p | 14 | ZBTB20 | Sponge network | -2.915 | 0.00015 | -0.122 | 0.57569 | 0.325 |
| 34792 | RP11-13K12.1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-766-3p | 13 | ATM | Sponge network | -0.154 | 0.78939 | 0.178 | 0.21568 | 0.325 |
| 34793 | KCNJ2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3913-5p;hsa-miR-590-3p | 19 | HLF | Sponge network | -0.436 | 0.05325 | -2.443 | 0 | 0.325 |
| 34794 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-942-5p | 17 | CREBBP | Sponge network | 0.249 | 0.35902 | 0.126 | 0.15186 | 0.325 |
| 34795 | RP11-774O3.3 |
hsa-miR-141-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | RANBP9 | Sponge network | -0.487 | 0.02157 | -0.191 | 0.04628 | 0.325 |
| 34796 | CTC-297N7.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-671-5p | 10 | FAT4 | Sponge network | -2.069 | 0 | -0.354 | 0.14694 | 0.325 |
| 34797 | LINC00854 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-5p | 11 | ZFHX4 | Sponge network | 1.18 | 0 | 0.018 | 0.95574 | 0.325 |
| 34798 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-455-5p | 15 | HDAC9 | Sponge network | -0.465 | 0.04703 | -0.755 | 0.00495 | 0.325 |
| 34799 | DIO3OS |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-671-5p | 25 | DLC1 | Sponge network | -0.773 | 0.04073 | -0.382 | 0.05038 | 0.325 |
| 34800 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-671-5p | 36 | FOXP1 | Sponge network | -2.69 | 0.0001 | 0.042 | 0.74368 | 0.325 |
| 34801 | AC108488.4 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | DCLK1 | Sponge network | 0.259 | 0.12542 | -1.324 | 0.00014 | 0.325 |
| 34802 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-93-3p | 19 | BMPR2 | Sponge network | 2.163 | 0 | 0.206 | 0.0635 | 0.325 |
| 34803 | LINC00886 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-532-5p | 12 | PARK2 | Sponge network | -0.371 | 0.24604 | -1.892 | 0 | 0.325 |
| 34804 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 37 | FZD3 | Sponge network | -0.323 | 0.01372 | 0.104 | 0.60316 | 0.325 |
| 34805 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 19 | TSC1 | Sponge network | 0.389 | 0.04293 | 0.103 | 0.26071 | 0.325 |
| 34806 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | YAP1 | Sponge network | 1.153 | 0 | 0.22 | 0.11836 | 0.325 |
| 34807 | MIR22HG |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p | 16 | RPS6KA6 | Sponge network | -1.047 | 0 | -2.989 | 0 | 0.325 |
| 34808 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PDS5B | Sponge network | 0.101 | 0.60919 | 0.259 | 0.00962 | 0.325 |
| 34809 | LINC00863 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 43 | WDFY3 | Sponge network | 0.189 | 0.13981 | -0.201 | 0.0967 | 0.325 |
| 34810 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-423-5p | 13 | SUFU | Sponge network | -0.583 | 0.14589 | -0.04 | 0.65489 | 0.325 |
| 34811 | GHRLOS |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-98-5p | 10 | RSPO2 | Sponge network | -1.942 | 0 | -3.038 | 0 | 0.325 |
| 34812 | LINC00957 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | KAT6B | Sponge network | -0.421 | 0.0114 | -0.478 | 0.00018 | 0.325 |
| 34813 | RP1-151F17.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | WWTR1 | Sponge network | 0.222 | 0.29133 | -0.345 | 0.11997 | 0.325 |
| 34814 | LPP-AS2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p | 14 | RELN | Sponge network | -0.18 | 0.20343 | -2.403 | 0 | 0.325 |
| 34815 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 44 | ZFHX4 | Sponge network | -0.355 | 0.0077 | 0.018 | 0.95574 | 0.325 |
| 34816 | SNHG14 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | USP25 | Sponge network | -1.111 | 0.0003 | -0.242 | 0.01397 | 0.325 |
| 34817 | RP11-13K12.5 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p | 12 | FMNL3 | Sponge network | -0.318 | 0.65847 | 0.555 | 4.0E-5 | 0.325 |
| 34818 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 29 | MITF | Sponge network | -0.615 | 0.00015 | -0.769 | 0.00022 | 0.325 |
| 34819 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-539-5p | 11 | PDE4DIP | Sponge network | -0.465 | 0.04703 | -0.282 | 0.0257 | 0.325 |
| 34820 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p | 14 | CITED2 | Sponge network | -0.323 | 0.01372 | -1.545 | 0 | 0.325 |
| 34821 | LINC00174 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-486-5p | 10 | MYO9A | Sponge network | 1.253 | 0 | -0.147 | 0.32373 | 0.325 |
| 34822 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p | 15 | MYO9A | Sponge network | 0.93 | 0 | -0.147 | 0.32373 | 0.325 |
| 34823 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | RABEP1 | Sponge network | -0.031 | 0.89063 | -0.066 | 0.43142 | 0.325 |
| 34824 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p | 27 | RBMS3 | Sponge network | 0.921 | 0.00118 | -0.519 | 0.03551 | 0.325 |
| 34825 | RP4-798A10.7 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p | 16 | MDM4 | Sponge network | 1.757 | 0 | 0.345 | 0.00762 | 0.325 |
| 34826 | SNHG16 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-495-3p | 18 | MAPK1 | Sponge network | 0.961 | 0 | -0.017 | 0.81465 | 0.325 |
| 34827 | AC093627.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110 | 17 | AR | Sponge network | -0.787 | 0.3928 | -1.554 | 0 | 0.325 |
| 34828 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-508-3p | 23 | MAF | Sponge network | -0.154 | 0.53954 | -1.257 | 0 | 0.325 |
| 34829 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-93-5p | 23 | ZBTB4 | Sponge network | -0.08 | 0.90092 | -0.739 | 0 | 0.325 |
| 34830 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | TMTC3 | Sponge network | 0.401 | 0.00066 | 0.123 | 0.22189 | 0.325 |
| 34831 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 23 | LRRC7 | Sponge network | -0.969 | 2.0E-5 | -1.341 | 0.01193 | 0.325 |
| 34832 | H1FX-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | PRKACA | Sponge network | -0.206 | 0.12942 | -0.255 | 0.0012 | 0.325 |
| 34833 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p | 15 | UNC5C | Sponge network | 0.335 | 0.20596 | -0.178 | 0.40214 | 0.325 |
| 34834 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500b-5p | 15 | GRIA3 | Sponge network | -0.246 | 0.35034 | -1.956 | 0 | 0.325 |
| 34835 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429 | 14 | DMXL1 | Sponge network | 1.106 | 0 | -0.426 | 0.00056 | 0.325 |
| 34836 | RP11-46C24.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | WWTR1 | Sponge network | 0.392 | 0.0278 | -0.345 | 0.11997 | 0.325 |
| 34837 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 40 | PTEN | Sponge network | -0.83 | 0 | -0.187 | 0.07748 | 0.325 |
| 34838 | MIR22HG |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-502-5p;hsa-miR-940 | 10 | FAM172A | Sponge network | -1.047 | 0 | -0.57 | 0 | 0.325 |
| 34839 | GNG12-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-3913-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p | 38 | AFF1 | Sponge network | -0.793 | 7.0E-5 | -0.384 | 0.00053 | 0.325 |
| 34840 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429 | 15 | ABCB5 | Sponge network | -1.249 | 7.0E-5 | -0.956 | 0.04077 | 0.325 |
| 34841 | CD27-AS1 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | 0.177 | 0.16681 | 0.358 | 0.13727 | 0.325 |
| 34842 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p | 20 | RARB | Sponge network | -1.047 | 0 | -0.825 | 2.0E-5 | 0.325 |
| 34843 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-93-5p | 25 | LRIG1 | Sponge network | -2.69 | 0.0001 | -0.537 | 0.00588 | 0.325 |
| 34844 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 26 | SCN9A | Sponge network | 0.72 | 0.0001 | -0.525 | 0.10593 | 0.325 |
| 34845 | RP11-359E10.1 |
hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-500a-3p;hsa-miR-582-5p | 12 | ARHGAP29 | Sponge network | 0.299 | 0.57722 | 0.131 | 0.42953 | 0.325 |
| 34846 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ABCC9 | Sponge network | -0.031 | 0.89063 | -0.466 | 0.14121 | 0.325 |
| 34847 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | EDA2R | Sponge network | 0.082 | 0.55654 | -0.587 | 0.01598 | 0.325 |
| 34848 | RP11-262H14.4 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | PIK3CA | Sponge network | -0.121 | 0.53195 | 0.195 | 0.06908 | 0.325 |
| 34849 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 33 | EPHA3 | Sponge network | -0.969 | 2.0E-5 | 0.185 | 0.53588 | 0.325 |
| 34850 | RP11-678G14.3 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-484;hsa-miR-7-1-3p | 12 | MED12L | Sponge network | -4.477 | 0 | -0.194 | 0.55299 | 0.325 |
| 34851 | RP11-774O3.3 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITGB1 | Sponge network | -0.487 | 0.02157 | 0.524 | 0.00017 | 0.325 |
| 34852 | RP11-983P16.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-576-5p | 14 | ERCC4 | Sponge network | 0.41 | 0.02214 | 0.126 | 0.15266 | 0.325 |
| 34853 | RP11-999E24.3 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p | 17 | RIMS2 | Sponge network | -0.693 | 0.05629 | -1.365 | 0.00208 | 0.325 |
| 34854 | RP1-257A7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-511-5p | 10 | RBMS3 | Sponge network | 0.849 | 0.00178 | -0.519 | 0.03551 | 0.325 |
| 34855 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 27 | YAP1 | Sponge network | -0.576 | 0.10121 | 0.22 | 0.11836 | 0.325 |
| 34856 | MIR143HG |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | MLF1 | Sponge network | -0.739 | 0.0574 | -0.893 | 0.00727 | 0.325 |
| 34857 | AC108488.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | PRUNE2 | Sponge network | 0.259 | 0.12542 | -1.733 | 0.00022 | 0.325 |
| 34858 | CTA-14H9.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-33a-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | CADM2 | Sponge network | 0.145 | 0.53343 | -3.782 | 0 | 0.325 |
| 34859 | RP11-53O19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 34 | CREB1 | Sponge network | -0.405 | 0.22013 | 0.203 | 0.00537 | 0.325 |
| 34860 | AC090587.5 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-501-5p | 10 | WDFY3 | Sponge network | -0.088 | 0.65011 | -0.201 | 0.0967 | 0.325 |
| 34861 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-223-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-708-5p | 16 | MTSS1 | Sponge network | -1.886 | 0 | -0.81 | 0.00035 | 0.325 |
| 34862 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-7-1-3p | 27 | ZFHX3 | Sponge network | -0.611 | 0.09607 | 0.389 | 0.01363 | 0.325 |
| 34863 | LINC00654 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | BRD3 | Sponge network | 0.374 | 0.20054 | 0.37 | 0.00013 | 0.325 |
| 34864 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-3p;hsa-miR-93-5p | 12 | MAP3K12 | Sponge network | -0.206 | 0.12942 | -0.057 | 0.59369 | 0.325 |
| 34865 | RP11-532F6.3 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p | 10 | RTN4 | Sponge network | -0.896 | 0.00099 | -0.412 | 0.00014 | 0.325 |
| 34866 | LINC00173 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-222-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | RECK | Sponge network | 0.056 | 0.8494 | -1.043 | 0 | 0.325 |
| 34867 | CTC-559E9.5 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-337-3p;hsa-miR-33a-3p | 11 | RPS6KA3 | Sponge network | 0.594 | 0.00064 | 0.256 | 0.02419 | 0.325 |
| 34868 | CTD-2184D3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | PRDM2 | Sponge network | 0.272 | 0.44692 | -0.258 | 0.00721 | 0.325 |
| 34869 | RP5-998N21.10 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p | 10 | NOTCH2 | Sponge network | -0.098 | 0.76298 | 0.138 | 0.35163 | 0.325 |
| 34870 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | FGFR1 | Sponge network | -0.351 | 0.11358 | -0.509 | 0.03957 | 0.325 |
| 34871 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 21 | EPHA3 | Sponge network | 0.056 | 0.8494 | 0.185 | 0.53588 | 0.325 |
| 34872 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | DDX5 | Sponge network | -0.615 | 0.00015 | -0.099 | 0.17428 | 0.325 |
| 34873 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | MLF1 | Sponge network | -2.117 | 0.00161 | -0.893 | 0.00727 | 0.325 |
| 34874 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 30 | SNX29 | Sponge network | 0.494 | 0.00339 | -0.34 | 0.01371 | 0.325 |
| 34875 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-98-5p | 19 | SEMA3C | Sponge network | 0.494 | 0.00339 | -0.272 | 0.31153 | 0.325 |
| 34876 | HAND2-AS1 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-625-5p;hsa-miR-7-5p | 10 | LZTS1 | Sponge network | -1.697 | 0.00889 | 1.664 | 0 | 0.325 |
| 34877 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 27 | SOX11 | Sponge network | 1.302 | 7.0E-5 | 2.68 | 0 | 0.325 |
| 34878 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 42 | CCND2 | Sponge network | -0.227 | 0.31657 | -0.267 | 0.3112 | 0.325 |
| 34879 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MLLT10 | Sponge network | 0.194 | 0.64278 | 0.105 | 0.33292 | 0.325 |
| 34880 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-425-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p | 27 | SOX5 | Sponge network | 0.174 | 0.30034 | -1.091 | 4.0E-5 | 0.325 |
| 34881 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | YAP1 | Sponge network | 0.953 | 0 | 0.22 | 0.11836 | 0.325 |
| 34882 | RP11-98I9.4 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-32-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | KIF5B | Sponge network | 0.67 | 0.00048 | 0.362 | 0.00066 | 0.325 |
| 34883 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429 | 24 | IL17RD | Sponge network | -0.031 | 0.89063 | -0.019 | 0.94873 | 0.325 |
| 34884 | RP11-21A7A.2 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-98-5p | 11 | ITGA5 | Sponge network | -1.116 | 0.23686 | 0.452 | 0.03524 | 0.325 |
| 34885 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-29b-3p | 13 | ESR1 | Sponge network | -4.477 | 0 | -1.035 | 3.0E-5 | 0.325 |
| 34886 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940 | 23 | HLF | Sponge network | -0.141 | 0.26434 | -2.443 | 0 | 0.325 |
| 34887 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-411-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | SMAD2 | Sponge network | 0.683 | 1.0E-5 | -0.128 | 0.12732 | 0.325 |
| 34888 | BOLA3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-660-5p;hsa-miR-877-5p | 19 | CRTC1 | Sponge network | -0.246 | 0.35034 | -0.116 | 0.28412 | 0.325 |
| 34889 | AP001055.6 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 15 | LRIG1 | Sponge network | 0.693 | 0.00076 | -0.537 | 0.00588 | 0.325 |
| 34890 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-589-3p;hsa-miR-96-5p | 26 | MOB1B | Sponge network | 0.594 | 0.41522 | 0.197 | 0.15266 | 0.325 |
| 34891 | HAND2-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p | 20 | PLXNC1 | Sponge network | -1.697 | 0.00889 | 0.569 | 0.00329 | 0.325 |
| 34892 | TRIM52-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-421;hsa-miR-577 | 12 | PGR | Sponge network | -0.418 | 0.00068 | -1.704 | 0 | 0.325 |
| 34893 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p | 11 | WWTR1 | Sponge network | 0.37 | 0.27614 | -0.345 | 0.11997 | 0.325 |
| 34894 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 36 | FZD3 | Sponge network | 0.541 | 0.00125 | 0.104 | 0.60316 | 0.325 |
| 34895 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-424-5p | 33 | ZFHX4 | Sponge network | -0.465 | 0.04703 | 0.018 | 0.95574 | 0.325 |
| 34896 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 24 | PTPN14 | Sponge network | -0.793 | 7.0E-5 | 0.144 | 0.34595 | 0.325 |
| 34897 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-708-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | RAP1A | Sponge network | -1.047 | 0 | -0.929 | 0 | 0.325 |
| 34898 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 26 | AFF3 | Sponge network | 0.064 | 0.72934 | -2.373 | 0 | 0.325 |
| 34899 | H19 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-589-3p | 20 | ADARB1 | Sponge network | 2.439 | 0 | -0.335 | 0.06631 | 0.325 |
| 34900 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 11 | DNAJB4 | Sponge network | 0.395 | 0.15451 | -0.7 | 1.0E-5 | 0.325 |
| 34901 | DNM1P35 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | PPP1R9A | Sponge network | 0.051 | 0.83388 | -0.903 | 0.04254 | 0.325 |
| 34902 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | RABEP1 | Sponge network | -1.697 | 0.00889 | -0.066 | 0.43142 | 0.325 |
| 34903 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 16 | PCDH17 | Sponge network | -0.896 | 0.00099 | 0.731 | 2.0E-5 | 0.325 |
| 34904 | RP11-38L15.3 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-590-3p | 10 | SNX29 | Sponge network | 0.344 | 0.3127 | -0.34 | 0.01371 | 0.325 |
| 34905 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | HECA | Sponge network | -2.117 | 0.00161 | -0.217 | 0.03463 | 0.325 |
| 34906 | LINC00476 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 27 | CTIF | Sponge network | -0.615 | 0.00015 | -0.389 | 0.00549 | 0.325 |
| 34907 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | KAT6B | Sponge network | -1.697 | 0.00889 | -0.478 | 0.00018 | 0.325 |
| 34908 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-7-1-3p | 14 | LRRC55 | Sponge network | -0.125 | 0.49414 | -0.026 | 0.93163 | 0.325 |
| 34909 | RP11-20I23.11 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | NOTCH2 | Sponge network | 0.875 | 0.00019 | 0.138 | 0.35163 | 0.325 |
| 34910 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p | 12 | KAT6B | Sponge network | 0.37 | 0.27614 | -0.478 | 0.00018 | 0.325 |
| 34911 | TMEM9B-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-362-5p | 12 | NIN | Sponge network | 0.529 | 0.00552 | -0.253 | 0.13794 | 0.325 |
| 34912 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -0.737 | 0.06182 | 0.309 | 0.17079 | 0.325 |
| 34913 | RP11-148O21.2 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-589-3p;hsa-miR-671-5p | 11 | KCNA6 | Sponge network | 0.233 | 0.81029 | -1.531 | 0.00013 | 0.325 |
| 34914 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 22 | TMEM132B | Sponge network | -0.811 | 2.0E-5 | -0.83 | 0.01084 | 0.325 |
| 34915 | LINC00265 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 18 | CCDC6 | Sponge network | 1.07 | 0 | 0.27 | 0.02555 | 0.325 |
| 34916 | ACTA2-AS1 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-874-3p | 11 | ARID1B | Sponge network | 0.194 | 0.64278 | 0.003 | 0.97503 | 0.325 |
| 34917 | RP11-983P16.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p | 20 | DLC1 | Sponge network | 0.41 | 0.02214 | -0.382 | 0.05038 | 0.325 |
| 34918 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 14 | MEF2C | Sponge network | 0.199 | 0.52763 | -0.499 | 0.01448 | 0.325 |
| 34919 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | ABI2 | Sponge network | -0.769 | 0.00035 | 0.175 | 0.14787 | 0.325 |
| 34920 | BOLA3-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -0.246 | 0.35034 | -0.966 | 0.00035 | 0.325 |
| 34921 | HCG11 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | AHNAK | Sponge network | 0.385 | 0.10067 | -1.207 | 0 | 0.325 |
| 34922 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PRKAB2 | Sponge network | 0.67 | 0.00048 | -0.367 | 0.01371 | 0.325 |
| 34923 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 47 | CBX5 | Sponge network | -1.575 | 6.0E-5 | 0.165 | 0.24446 | 0.325 |
| 34924 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-5p | 11 | RNF144A | Sponge network | -1.582 | 8.0E-5 | 0.594 | 0.0009 | 0.325 |
| 34925 | DIO3OS |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 13 | FLNA | Sponge network | -0.773 | 0.04073 | -0.771 | 0.01377 | 0.325 |
| 34926 | EMX2OS |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 13 | FYN | Sponge network | -0.777 | 0.1134 | -0.131 | 0.37051 | 0.325 |
| 34927 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRIP11 | Sponge network | 0.539 | 0.03722 | -0.158 | 0.06384 | 0.325 |
| 34928 | FAM157A |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p | 15 | AFF4 | Sponge network | 0.813 | 1.0E-5 | -0.266 | 0.01565 | 0.325 |
| 34929 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-629-3p;hsa-miR-942-5p | 18 | DDX6 | Sponge network | 1.056 | 0 | 0.091 | 0.36428 | 0.325 |
| 34930 | RP11-21A7A.2 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-500a-5p | 10 | PRKAB2 | Sponge network | -1.116 | 0.23686 | -0.367 | 0.01371 | 0.325 |
| 34931 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-98-5p | 17 | ITGA5 | Sponge network | -0.154 | 0.78939 | 0.452 | 0.03524 | 0.325 |
| 34932 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-576-5p | 22 | PGR | Sponge network | 0.41 | 0.02214 | -1.704 | 0 | 0.325 |
| 34933 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 37 | CHD5 | Sponge network | 0.219 | 0.1516 | -0.922 | 0.01521 | 0.325 |
| 34934 | CTD-2666L21.1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p | 17 | CHD5 | Sponge network | 0.368 | 0.20093 | -0.922 | 0.01521 | 0.325 |
| 34935 | RP11-53O19.1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-501-5p | 12 | TXNIP | Sponge network | -0.405 | 0.22013 | -1.277 | 0 | 0.325 |
| 34936 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH11 | Sponge network | -0.769 | 0.00035 | 1.427 | 0 | 0.325 |
| 34937 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-589-3p | 33 | PDGFRA | Sponge network | -0.154 | 0.53954 | -0.372 | 0.0037 | 0.325 |
| 34938 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | EPHA5 | Sponge network | -1.886 | 0 | -0.957 | 0.01733 | 0.325 |
| 34939 | RP11-13K12.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p | 18 | GJA1 | Sponge network | -0.154 | 0.78939 | -0.485 | 0.02179 | 0.325 |
| 34940 | TMEM9B-AS1 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-497-5p | 23 | ARHGEF12 | Sponge network | 0.529 | 0.00552 | 0.09 | 0.35693 | 0.325 |
| 34941 | RP11-384O8.1 |
hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 20 | KIT | Sponge network | -0.738 | 0.21233 | -2.184 | 0 | 0.325 |
| 34942 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 18 | BCL11A | Sponge network | -0.727 | 0.04726 | -0.932 | 0.0063 | 0.325 |
| 34943 | AF131215.9 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-590-3p | 17 | SETBP1 | Sponge network | -0.322 | 0.25945 | -1.302 | 1.0E-5 | 0.325 |
| 34944 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-629-5p | 21 | IGF1 | Sponge network | -0.125 | 0.49414 | -0.204 | 0.5898 | 0.325 |
| 34945 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 15 | NUAK1 | Sponge network | 0.16 | 0.50785 | 0.904 | 0 | 0.325 |
| 34946 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p | 25 | PBX1 | Sponge network | -0.223 | 0.47816 | -1.206 | 0 | 0.325 |
| 34947 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | HIP1 | Sponge network | 0.082 | 0.55654 | 0.774 | 0 | 0.325 |
| 34948 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-455-3p;hsa-miR-7-1-3p | 14 | NRXN1 | Sponge network | 0.693 | 0.00076 | -3.778 | 0 | 0.325 |
| 34949 | TP73-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 36 | PPP1R9A | Sponge network | -0.563 | 0.02545 | -0.903 | 0.04254 | 0.325 |
| 34950 | RP1-59D14.5 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | KIF5B | Sponge network | 1.162 | 5.0E-5 | 0.362 | 0.00066 | 0.325 |
| 34951 | CALML3-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 19 | ABL1 | Sponge network | 0.95 | 0.15943 | -0.215 | 0.07804 | 0.325 |
| 34952 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-96-5p | 11 | HOOK3 | Sponge network | 0.853 | 0.15352 | 0.273 | 0.09957 | 0.325 |
| 34953 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | MITF | Sponge network | -1.161 | 0.00591 | -0.769 | 0.00022 | 0.325 |
| 34954 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 33 | MEF2C | Sponge network | -0.125 | 0.49414 | -0.499 | 0.01448 | 0.325 |
| 34955 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 36 | PAFAH1B1 | Sponge network | -0.612 | 0.00094 | -0.429 | 0 | 0.325 |
| 34956 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ZNF208 | Sponge network | 0.619 | 0.00157 | -0.4 | 0.22381 | 0.325 |
| 34957 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p | 15 | MYCBP2 | Sponge network | -0.323 | 0.01372 | -0.027 | 0.82016 | 0.325 |
| 34958 | HOTAIRM1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | GJA1 | Sponge network | -0.053 | 0.80685 | -0.485 | 0.02179 | 0.325 |
| 34959 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 16 | PSIP1 | Sponge network | -0.709 | 0.18168 | -0.005 | 0.96897 | 0.325 |
| 34960 | CTD-2089N3.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p | 11 | PTPRD | Sponge network | -0.314 | 0.62033 | -1.005 | 0.00064 | 0.325 |
| 34961 | PCA3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 17 | UBE4B | Sponge network | -0.709 | 0.18168 | -0.235 | 0.007 | 0.325 |
| 34962 | LINC00094 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 24 | CTIF | Sponge network | 0.253 | 0.09565 | -0.389 | 0.00549 | 0.325 |
| 34963 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | PRKAA1 | Sponge network | 0.997 | 0 | 0.317 | 0.0031 | 0.325 |
| 34964 | TINCR |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-495-3p | 17 | DICER1 | Sponge network | -0.664 | 0.14543 | 0.042 | 0.674 | 0.325 |
| 34965 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 13 | SPTBN4 | Sponge network | -1.057 | 0.00029 | -0.786 | 0.01281 | 0.325 |
| 34966 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | BCL2 | Sponge network | -1.161 | 0.00591 | -0.899 | 0 | 0.325 |
| 34967 | CALML3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-92a-1-5p | 27 | SNX29 | Sponge network | 0.95 | 0.15943 | -0.34 | 0.01371 | 0.325 |
| 34968 | AC004540.4 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-324-5p | 14 | QKI | Sponge network | -2.549 | 0.00528 | -0.333 | 0.04111 | 0.325 |
| 34969 | ADIRF-AS1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | MED12L | Sponge network | -0.223 | 0.47816 | -0.194 | 0.55299 | 0.325 |
| 34970 | AC127904.2 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 14 | DICER1 | Sponge network | 1.308 | 0 | 0.042 | 0.674 | 0.325 |
| 34971 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429 | 10 | CSMD1 | Sponge network | -1.654 | 0.00144 | -0.63 | 0.18881 | 0.325 |
| 34972 | PSMG3-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | LRIG1 | Sponge network | -0.125 | 0.49414 | -0.537 | 0.00588 | 0.325 |
| 34973 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | MAF | Sponge network | 0.374 | 0.20054 | -1.257 | 0 | 0.325 |
| 34974 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-654-3p | 16 | ITCH | Sponge network | 0.417 | 0.00445 | 0.084 | 0.34058 | 0.325 |
| 34975 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 24 | PTGFR | Sponge network | -0.942 | 0 | -0.233 | 0.45421 | 0.325 |
| 34976 | RP5-1021I20.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429 | 12 | AKAP6 | Sponge network | -0.785 | 0.00035 | -1.586 | 3.0E-5 | 0.325 |
| 34977 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-7-1-3p | 18 | TBX18 | Sponge network | -0.405 | 0.22013 | 0.843 | 0.02945 | 0.325 |
| 34978 | RP3-402G11.26 |
hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-590-3p | 10 | MED12L | Sponge network | -0.254 | 0.19303 | -0.194 | 0.55299 | 0.325 |
| 34979 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-93-5p | 11 | LATS2 | Sponge network | -2.531 | 0 | 0.159 | 0.2304 | 0.325 |
| 34980 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | LRRK2 | Sponge network | -0.899 | 6.0E-5 | -1.136 | 1.0E-5 | 0.324 |
| 34981 | RP11-536O18.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p | 11 | ESR1 | Sponge network | -0.074 | 0.90758 | -1.035 | 3.0E-5 | 0.324 |
| 34982 | FGD5-AS1 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | MAP3K4 | Sponge network | 0.401 | 0.00066 | 0.058 | 0.54229 | 0.324 |
| 34983 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | CDHR3 | Sponge network | -0.709 | 0.18168 | -0.977 | 4.0E-5 | 0.324 |
| 34984 | MYLK-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-424-5p | 10 | KALRN | Sponge network | -1.331 | 3.0E-5 | -0.819 | 1.0E-5 | 0.324 |
| 34985 | CTA-363E19.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p | 22 | AFF4 | Sponge network | 1.055 | 0 | -0.266 | 0.01565 | 0.324 |
| 34986 | SNHG14 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 13 | LZTS1 | Sponge network | -1.111 | 0.0003 | 1.664 | 0 | 0.324 |
| 34987 | LINC00865 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 38 | CREB3L2 | Sponge network | -1.249 | 7.0E-5 | -0.525 | 2.0E-5 | 0.324 |
| 34988 | RP11-686D22.8 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 13 | IGF1 | Sponge network | 0.898 | 1.0E-5 | -0.204 | 0.5898 | 0.324 |
| 34989 | RP11-728F11.4 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p | 11 | IGF1 | Sponge network | 0.238 | 0.70544 | -0.204 | 0.5898 | 0.324 |
| 34990 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-96-5p | 14 | RIMBP2 | Sponge network | -0.555 | 0.06156 | -1.968 | 5.0E-5 | 0.324 |
| 34991 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-93-5p | 15 | CNTNAP3 | Sponge network | -0.777 | 0.16667 | -1.465 | 0.00033 | 0.324 |
| 34992 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | LRP1B | Sponge network | 0.532 | 0.00505 | -2.163 | 0 | 0.324 |
| 34993 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 28 | LRRC7 | Sponge network | -2.925 | 0 | -1.341 | 0.01193 | 0.324 |
| 34994 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 12 | BCLAF1 | Sponge network | 0.705 | 0.00014 | 0.211 | 0.00684 | 0.324 |
| 34995 | AF131215.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 11 | FAM172A | Sponge network | -0.322 | 0.25945 | -0.57 | 0 | 0.324 |
| 34996 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 33 | EPHA5 | Sponge network | -0.18 | 0.20343 | -0.957 | 0.01733 | 0.324 |
| 34997 | RP11-102N12.3 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-505-5p | 12 | MRVI1 | Sponge network | -0.351 | 0.11358 | -1.051 | 0.00022 | 0.324 |
| 34998 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | PRKAA2 | Sponge network | -0.141 | 0.26434 | -2.462 | 0 | 0.324 |
| 34999 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 11 | FSTL5 | Sponge network | 0.174 | 0.30034 | -1.3 | 0.01619 | 0.324 |
| 35000 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 32 | CADM1 | Sponge network | -0.323 | 0.01372 | -0.9 | 0.00084 | 0.324 |
| 35001 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 31 | NCOA1 | Sponge network | 0.997 | 0.00066 | -0.432 | 0 | 0.324 |
| 35002 | H1FX-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-590-3p | 17 | HIP1 | Sponge network | -0.206 | 0.12942 | 0.774 | 0 | 0.324 |
| 35003 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-590-3p | 12 | RBM5 | Sponge network | -0.355 | 0.0077 | -0.205 | 0.0129 | 0.324 |
| 35004 | AC010226.4 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-96-5p | 12 | CSRNP1 | Sponge network | -0.811 | 2.0E-5 | -1.448 | 0 | 0.324 |
| 35005 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-577 | 10 | ZNF844 | Sponge network | -2.69 | 0.0001 | -0.966 | 0.00035 | 0.324 |
| 35006 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-27b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-664a-5p | 12 | SOX11 | Sponge network | 0.757 | 0 | 2.68 | 0 | 0.324 |
| 35007 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p | 22 | SCN9A | Sponge network | -0.031 | 0.89063 | -0.525 | 0.10593 | 0.324 |
| 35008 | LINC00654 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 40 | GRIK3 | Sponge network | 0.374 | 0.20054 | -3.208 | 0 | 0.324 |
| 35009 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-942-5p | 12 | HLF | Sponge network | -0.326 | 0.28809 | -2.443 | 0 | 0.324 |
| 35010 | LINC00472 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 21 | PCDH17 | Sponge network | -0.842 | 0.01361 | 0.731 | 2.0E-5 | 0.324 |
| 35011 | USP3-AS1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-940 | 16 | NR3C2 | Sponge network | -0.162 | 0.47687 | -1.56 | 0 | 0.324 |
| 35012 | TPTEP1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 33 | ETV1 | Sponge network | -1.216 | 0 | -0.096 | 0.67674 | 0.324 |
| 35013 | LINC00957 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484 | 13 | ITGA5 | Sponge network | -0.421 | 0.0114 | 0.452 | 0.03524 | 0.324 |
| 35014 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SON | Sponge network | 0.222 | 0.29133 | 0.168 | 0.04406 | 0.324 |
| 35015 | HOXA-AS2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-590-3p | 19 | RYR2 | Sponge network | 0.61 | 0.01593 | -1.466 | 0.00031 | 0.324 |
| 35016 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | PRKAA2 | Sponge network | -0.08 | 0.90092 | -2.462 | 0 | 0.324 |
| 35017 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-940 | 35 | QKI | Sponge network | 0.082 | 0.55397 | -0.333 | 0.04111 | 0.324 |
| 35018 | GHRLOS |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-93-3p;hsa-miR-940 | 10 | POU2F2 | Sponge network | -1.942 | 0 | -0.233 | 0.32214 | 0.324 |
| 35019 | MYLK-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-935 | 17 | ACSL6 | Sponge network | -1.331 | 3.0E-5 | -1.593 | 0 | 0.324 |
| 35020 | JAZF1-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | RIMS2 | Sponge network | -0.769 | 0.00035 | -1.365 | 0.00208 | 0.324 |
| 35021 | RP11-890B15.3 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-7-5p | 10 | LZTS1 | Sponge network | 0.101 | 0.60919 | 1.664 | 0 | 0.324 |
| 35022 | RP11-35G9.3 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 15 | NRK | Sponge network | 0.657 | 4.0E-5 | 0.656 | 0.19217 | 0.324 |
| 35023 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | MED12L | Sponge network | -0.322 | 0.25945 | -0.194 | 0.55299 | 0.324 |
| 35024 | RP11-21A7A.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 19 | QKI | Sponge network | -1.116 | 0.23686 | -0.333 | 0.04111 | 0.324 |
| 35025 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-625-5p | 13 | CBX5 | Sponge network | 1.134 | 0 | 0.165 | 0.24446 | 0.324 |
| 35026 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-654-3p | 12 | DDX6 | Sponge network | 1.344 | 0 | 0.091 | 0.36428 | 0.324 |
| 35027 | A2M-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 16 | NRK | Sponge network | -0.08 | 0.90092 | 0.656 | 0.19217 | 0.324 |
| 35028 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | APC | Sponge network | 0.782 | 0.00016 | -0.255 | 0.04051 | 0.324 |
| 35029 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 19 | WWTR1 | Sponge network | 0.619 | 0.00157 | -0.345 | 0.11997 | 0.324 |
| 35030 | AF131215.9 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-940 | 38 | MDM4 | Sponge network | -0.322 | 0.25945 | 0.345 | 0.00762 | 0.324 |
| 35031 | LINC00473 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 25 | SNX29 | Sponge network | -1.203 | 0.03881 | -0.34 | 0.01371 | 0.324 |
| 35032 | FAM215B |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 43 | LPP | Sponge network | 0.817 | 3.0E-5 | -0.459 | 0.04475 | 0.324 |
| 35033 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ZMYND11 | Sponge network | 0.335 | 0.20596 | -0.2 | 0.05449 | 0.324 |
| 35034 | RP11-175O19.4 |
hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-664a-3p | 10 | PICALM | Sponge network | 0.917 | 0 | 0.086 | 0.23523 | 0.324 |
| 35035 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-654-3p | 18 | PSIP1 | Sponge network | 0.101 | 0.60919 | -0.005 | 0.96897 | 0.324 |
| 35036 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | FOXO1 | Sponge network | 0.614 | 1.0E-5 | -0.141 | 0.33874 | 0.324 |
| 35037 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-590-3p | 10 | USP6 | Sponge network | 1.148 | 2.0E-5 | 0.188 | 0.3871 | 0.324 |
| 35038 | RP11-967K21.1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p | 11 | FNBP1 | Sponge network | 0.056 | 0.81195 | -0.969 | 4.0E-5 | 0.324 |
| 35039 | U91328.19 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p | 16 | SEMA3C | Sponge network | -0.303 | 0.21002 | -0.272 | 0.31153 | 0.324 |
| 35040 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 11 | NALCN | Sponge network | -0.612 | 0.00094 | 1.068 | 0.00237 | 0.324 |
| 35041 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 43 | TMEM132B | Sponge network | -0.323 | 0.01372 | -0.83 | 0.01084 | 0.324 |
| 35042 | PGM5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p | 21 | NCOA1 | Sponge network | -4.546 | 0 | -0.432 | 0 | 0.324 |
| 35043 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-450b-5p | 16 | APC | Sponge network | -0.583 | 0.14589 | -0.255 | 0.04051 | 0.324 |
| 35044 | CD27-AS1 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-542-3p | 18 | SYNM | Sponge network | 0.177 | 0.16681 | -2.309 | 1.0E-5 | 0.324 |
| 35045 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 20 | CREB3L2 | Sponge network | -1.375 | 0.08642 | -0.525 | 2.0E-5 | 0.324 |
| 35046 | RP11-203M5.7 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-590-5p | 12 | ARHGEF12 | Sponge network | 1.684 | 1.0E-5 | 0.09 | 0.35693 | 0.324 |
| 35047 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 12 | FAM133A | Sponge network | -2.452 | 0 | 0.549 | 0.29009 | 0.324 |
| 35048 | RP11-35G9.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-96-5p | 16 | MED12L | Sponge network | 0.622 | 0.00015 | -0.194 | 0.55299 | 0.324 |
| 35049 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | SSBP2 | Sponge network | 0.532 | 0.00505 | -1.58 | 0 | 0.324 |
| 35050 | A2M-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-942-5p | 29 | IGFBP5 | Sponge network | -0.08 | 0.90092 | -0.806 | 0.00105 | 0.324 |
| 35051 | RP11-53O19.1 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 22 | NUAK1 | Sponge network | -0.405 | 0.22013 | 0.904 | 0 | 0.324 |
| 35052 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | CBL | Sponge network | 0.603 | 0.00127 | 0.291 | 0.01267 | 0.324 |
| 35053 | LINC00667 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | ADCY1 | Sponge network | -0.235 | 0.15142 | 0.362 | 0.16982 | 0.324 |
| 35054 | RP11-307B6.3 |
hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p | 19 | PPM1L | Sponge network | -4.463 | 0.00025 | -0.537 | 0.00616 | 0.324 |
| 35055 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | PTPRB | Sponge network | -0.318 | 0.65847 | 0.105 | 0.47122 | 0.324 |
| 35056 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 40 | SSBP2 | Sponge network | -0.513 | 0.04703 | -1.58 | 0 | 0.324 |
| 35057 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | MOB1B | Sponge network | 1.073 | 0.00216 | 0.197 | 0.15266 | 0.324 |
| 35058 | LINC01004 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-582-3p | 12 | ABL2 | Sponge network | 1.115 | 0 | 0.82 | 0 | 0.324 |
| 35059 | LINC00702 |
hsa-miR-10b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 10 | EXTL2 | Sponge network | -0.737 | 0.06182 | 0.109 | 0.37375 | 0.324 |
| 35060 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 39 | JAZF1 | Sponge network | -2.925 | 0 | -0.377 | 0.02677 | 0.324 |
| 35061 | ADIRF-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p | 17 | DYNC1I1 | Sponge network | -0.223 | 0.47816 | -1.12 | 0.00107 | 0.324 |
| 35062 | RP11-379F4.7 |
hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | KIF5B | Sponge network | 1.316 | 0 | 0.362 | 0.00066 | 0.324 |
| 35063 | CTD-2314G24.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.246 | 0.59912 | -0.57 | 0 | 0.324 |
| 35064 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-374b-3p;hsa-miR-539-5p | 11 | TP53INP1 | Sponge network | -1.092 | 4.0E-5 | -0.496 | 0.00064 | 0.324 |
| 35065 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-629-3p | 17 | EBF3 | Sponge network | 0.368 | 0.20093 | -0.058 | 0.81437 | 0.324 |
| 35066 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 65 | TCF4 | Sponge network | -0.942 | 0 | 0.016 | 0.92271 | 0.324 |
| 35067 | AC016747.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 39 | FAT3 | Sponge network | 0.199 | 0.38015 | -1.601 | 0.00051 | 0.324 |
| 35068 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-7-5p | 38 | BMPR2 | Sponge network | 0.176 | 0.37402 | 0.206 | 0.0635 | 0.324 |
| 35069 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-590-3p | 15 | EBF3 | Sponge network | -0.737 | 0.10822 | -0.058 | 0.81437 | 0.324 |
| 35070 | RP11-395G23.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 49 | LPP | Sponge network | 0.381 | 0.25967 | -0.459 | 0.04475 | 0.324 |
| 35071 | MRVI1-AS1 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-708-5p | 11 | FAM135B | Sponge network | -1.092 | 4.0E-5 | -3.978 | 0 | 0.324 |
| 35072 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 62 | NTRK3 | Sponge network | -0.355 | 0.0077 | -1.77 | 3.0E-5 | 0.324 |
| 35073 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-508-3p;hsa-miR-590-3p | 17 | MYO9A | Sponge network | 0.051 | 0.83388 | -0.147 | 0.32373 | 0.324 |
| 35074 | LINC00578 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-576-5p;hsa-miR-577 | 12 | LUZP2 | Sponge network | 0.811 | 0.03228 | -0.056 | 0.89416 | 0.324 |
| 35075 | LINC00174 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 10 | CDK6 | Sponge network | 1.253 | 0 | 0.991 | 0 | 0.324 |
| 35076 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 11 | ZHX2 | Sponge network | 0.997 | 0.00066 | -0.192 | 0.18459 | 0.324 |
| 35077 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p | 10 | MNT | Sponge network | -1.645 | 0 | -0.157 | 0.06598 | 0.324 |
| 35078 | RP4-584D14.7 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-362-3p | 13 | ZFHX4 | Sponge network | 1.294 | 0.0031 | 0.018 | 0.95574 | 0.324 |
| 35079 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-484;hsa-miR-590-3p | 13 | MYCBP2 | Sponge network | 0.194 | 0.64278 | -0.027 | 0.82016 | 0.324 |
| 35080 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-576-5p | 40 | AR | Sponge network | 0.95 | 0.15943 | -1.554 | 0 | 0.324 |
| 35081 | RP11-5C23.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 18 | LRIG1 | Sponge network | 0.274 | 0.07681 | -0.537 | 0.00588 | 0.324 |
| 35082 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-429;hsa-miR-590-3p | 19 | USP6 | Sponge network | 0.953 | 0 | 0.188 | 0.3871 | 0.324 |
| 35083 | RP11-597D13.9 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 35 | DIP2C | Sponge network | 1.302 | 7.0E-5 | -0.436 | 0.01713 | 0.324 |
| 35084 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | HDAC9 | Sponge network | -0.811 | 2.0E-5 | -0.755 | 0.00495 | 0.324 |
| 35085 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p | 14 | ADAMTSL3 | Sponge network | 1.757 | 0 | -1.559 | 0 | 0.324 |
| 35086 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p | 10 | PDS5B | Sponge network | 1.344 | 0 | 0.259 | 0.00962 | 0.324 |
| 35087 | RP11-532F6.3 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | ATM | Sponge network | -0.896 | 0.00099 | 0.178 | 0.21568 | 0.324 |
| 35088 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-629-3p | 22 | ZBTB4 | Sponge network | -0.246 | 0.35034 | -0.739 | 0 | 0.324 |
| 35089 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p | 17 | ABCB5 | Sponge network | -0.969 | 2.0E-5 | -0.956 | 0.04077 | 0.324 |
| 35090 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 27 | MXI1 | Sponge network | 0.194 | 0.64278 | -0.987 | 0 | 0.324 |
| 35091 | AC034220.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PRKAB2 | Sponge network | 0.309 | 0.07304 | -0.367 | 0.01371 | 0.324 |
| 35092 | LINC00865 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p | 18 | TLE4 | Sponge network | -1.249 | 7.0E-5 | -1.157 | 0 | 0.324 |
| 35093 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-5p;hsa-miR-96-5p | 12 | PCDH8 | Sponge network | -1.439 | 4.0E-5 | -0.997 | 0.05366 | 0.324 |
| 35094 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | FGF5 | Sponge network | 0.214 | 0.18246 | 0.549 | 0.28659 | 0.324 |
| 35095 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 13 | SPG20 | Sponge network | -0.08 | 0.90092 | -1.16 | 0 | 0.324 |
| 35096 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 49 | FAT3 | Sponge network | -0.355 | 0.0077 | -1.601 | 0.00051 | 0.324 |
| 35097 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-577;hsa-miR-96-5p | 21 | CNTN1 | Sponge network | -0.18 | 0.20343 | -2.594 | 0 | 0.324 |
| 35098 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 18 | SNX29 | Sponge network | 0.453 | 0.01839 | -0.34 | 0.01371 | 0.324 |
| 35099 | RP11-53O19.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 21 | PIK3CA | Sponge network | -0.405 | 0.22013 | 0.195 | 0.06908 | 0.324 |
| 35100 | CTD-2666L21.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-708-5p | 10 | EZH1 | Sponge network | 0.368 | 0.20093 | -0.564 | 1.0E-5 | 0.324 |
| 35101 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | PRKAB2 | Sponge network | -0.738 | 0.21233 | -0.367 | 0.01371 | 0.324 |
| 35102 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 49 | NTRK3 | Sponge network | 0.225 | 0.30644 | -1.77 | 3.0E-5 | 0.324 |
| 35103 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-342-5p;hsa-miR-495-3p;hsa-miR-511-5p | 14 | ATRX | Sponge network | 1.117 | 0 | 0.261 | 0.04408 | 0.324 |
| 35104 | MIR143HG |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 22 | NOTCH2NL | Sponge network | -0.739 | 0.0574 | 0.024 | 0.84307 | 0.324 |
| 35105 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 13 | PAK3 | Sponge network | -0.738 | 0.21233 | -0.628 | 0.07253 | 0.324 |
| 35106 | THAP9-AS1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 20 | MDM4 | Sponge network | 1.079 | 0 | 0.345 | 0.00762 | 0.324 |
| 35107 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p | 15 | HIP1 | Sponge network | -1.092 | 4.0E-5 | 0.774 | 0 | 0.324 |
| 35108 | RP5-1198O20.4 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | RASSF3 | Sponge network | 0.997 | 0.00066 | -0.226 | 0.11691 | 0.324 |
| 35109 | HOXA-AS2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | BRD3 | Sponge network | 0.61 | 0.01593 | 0.37 | 0.00013 | 0.324 |
| 35110 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p | 10 | PCDH8 | Sponge network | -0.162 | 0.47687 | -0.997 | 0.05366 | 0.324 |
| 35111 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-342-3p | 16 | TMTC3 | Sponge network | 0.972 | 0 | 0.123 | 0.22189 | 0.324 |
| 35112 | RP11-195B17.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-5p | 20 | CBX5 | Sponge network | 0.37 | 0.27614 | 0.165 | 0.24446 | 0.324 |
| 35113 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 19 | EBF3 | Sponge network | 0.174 | 0.30034 | -0.058 | 0.81437 | 0.324 |
| 35114 | RP1-122P22.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-942-5p | 21 | IGFBP5 | Sponge network | 0.065 | 0.73468 | -0.806 | 0.00105 | 0.324 |
| 35115 | FGD5-AS1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-708-5p | 32 | CCDC6 | Sponge network | 0.401 | 0.00066 | 0.27 | 0.02555 | 0.324 |
| 35116 | FAM13A-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 19 | EPS15 | Sponge network | -0.83 | 0 | -0.052 | 0.53998 | 0.324 |
| 35117 | RP1-59D14.5 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | ERCC6 | Sponge network | 1.162 | 5.0E-5 | 0.23 | 0.015 | 0.324 |
| 35118 | CALML3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p | 20 | PRDM2 | Sponge network | 0.95 | 0.15943 | -0.258 | 0.00721 | 0.324 |
| 35119 | FGD5-AS1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p | 10 | EPG5 | Sponge network | 0.401 | 0.00066 | 0.101 | 0.3563 | 0.324 |
| 35120 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 41 | CADM2 | Sponge network | -0.318 | 0.65847 | -3.782 | 0 | 0.324 |
| 35121 | LINC00854 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-5p | 25 | LPP | Sponge network | 1.18 | 0 | -0.459 | 0.04475 | 0.324 |
| 35122 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | GNA13 | Sponge network | 0.101 | 0.60919 | 0.007 | 0.93884 | 0.324 |
| 35123 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-940 | 13 | GAS7 | Sponge network | 0.959 | 0 | -0.555 | 0.01602 | 0.324 |
| 35124 | PART1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ZNF208 | Sponge network | -2.925 | 0 | -0.4 | 0.22381 | 0.324 |
| 35125 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 14 | ARID5B | Sponge network | 1.601 | 0 | 0.342 | 0.01676 | 0.324 |
| 35126 | RP11-403I13.8 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 0.619 | 0.00157 | 0.321 | 0.00267 | 0.324 |
| 35127 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-940 | 51 | WDFY3 | Sponge network | 0.249 | 0.35902 | -0.201 | 0.0967 | 0.324 |
| 35128 | LINC00926 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-877-5p | 12 | RBMS3 | Sponge network | 0.756 | 0.00739 | -0.519 | 0.03551 | 0.324 |
| 35129 | FAM157A |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326 | 12 | BRD3 | Sponge network | 0.813 | 1.0E-5 | 0.37 | 0.00013 | 0.324 |
| 35130 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 18 | THRB | Sponge network | -2.039 | 0.03447 | -1.117 | 0.0004 | 0.324 |
| 35131 | C4B-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-486-5p;hsa-miR-744-3p;hsa-miR-96-5p | 20 | FAT3 | Sponge network | 2.306 | 9.0E-5 | -1.601 | 0.00051 | 0.324 |
| 35132 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 17 | LHX6 | Sponge network | -0.777 | 0.1134 | -0.087 | 0.69193 | 0.324 |
| 35133 | RP11-216B9.6 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-940 | 11 | SIRT1 | Sponge network | 0.174 | 0.30034 | 0.109 | 0.2593 | 0.324 |
| 35134 | TINCR |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-539-5p | 19 | RIMS2 | Sponge network | -0.664 | 0.14543 | -1.365 | 0.00208 | 0.324 |
| 35135 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-98-5p | 16 | MITF | Sponge network | 0.082 | 0.55397 | -0.769 | 0.00022 | 0.324 |
| 35136 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | LRRK2 | Sponge network | 0.082 | 0.55654 | -1.136 | 1.0E-5 | 0.324 |
| 35137 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-486-5p | 15 | PRRX1 | Sponge network | -0.309 | 0.1145 | 1.316 | 0 | 0.324 |
| 35138 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-424-3p;hsa-miR-7-1-3p | 16 | BMPR2 | Sponge network | -0.023 | 0.95109 | 0.206 | 0.0635 | 0.324 |
| 35139 | RP4-717I23.3 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-532-3p | 12 | TRIM13 | Sponge network | 0.637 | 0.00069 | 0.183 | 0.06968 | 0.324 |
| 35140 | TP73-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 22 | RIMBP2 | Sponge network | -0.563 | 0.02545 | -1.968 | 5.0E-5 | 0.324 |
| 35141 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 18 | GPAM | Sponge network | 0.421 | 0.00084 | 0.142 | 0.29732 | 0.324 |
| 35142 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-455-3p | 15 | PRKAB2 | Sponge network | 0.792 | 0.0049 | -0.367 | 0.01371 | 0.324 |
| 35143 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CACNA2D1 | Sponge network | -0.727 | 0.04726 | -0.846 | 0.00675 | 0.324 |
| 35144 | DNAJC27-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-5p | 11 | KCTD8 | Sponge network | -0.969 | 2.0E-5 | -3.359 | 0 | 0.324 |
| 35145 | LINC00092 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p | 14 | MED12L | Sponge network | -1.886 | 0 | -0.194 | 0.55299 | 0.324 |
| 35146 | ADIRF-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-92a-3p | 47 | DST | Sponge network | -0.223 | 0.47816 | -0.713 | 0.00096 | 0.324 |
| 35147 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-625-5p | 32 | ABI2 | Sponge network | -2.69 | 0.0001 | 0.175 | 0.14787 | 0.324 |
| 35148 | HHIP-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ERG | Sponge network | -0.737 | 0.10822 | 0.002 | 0.99011 | 0.324 |
| 35149 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 18 | ITGB3 | Sponge network | 1.302 | 7.0E-5 | -0.011 | 0.95751 | 0.324 |
| 35150 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-7-5p | 23 | CRTC1 | Sponge network | -0.487 | 0.02157 | -0.116 | 0.28412 | 0.324 |
| 35151 | RP11-48B3.4 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 12 | IGSF10 | Sponge network | 1.757 | 0 | -1.751 | 1.0E-5 | 0.324 |
| 35152 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 26 | CBL | Sponge network | 0.4 | 0.14611 | 0.291 | 0.01267 | 0.324 |
| 35153 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429 | 16 | LHX6 | Sponge network | -0.942 | 0 | -0.087 | 0.69193 | 0.324 |
| 35154 | CTC-429P9.5 |
hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-590-3p | 10 | DICER1 | Sponge network | 0.178 | 0.29106 | 0.042 | 0.674 | 0.324 |
| 35155 | RASSF8-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-92a-1-5p | 22 | CTIF | Sponge network | 0.335 | 0.20596 | -0.389 | 0.00549 | 0.324 |
| 35156 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 17 | POU2F2 | Sponge network | -0.576 | 0.10121 | -0.233 | 0.32214 | 0.324 |
| 35157 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p | 21 | FGF5 | Sponge network | -0.303 | 0.21002 | 0.549 | 0.28659 | 0.324 |
| 35158 | RP11-283I3.6 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-664a-3p | 18 | RASAL2 | Sponge network | 0.614 | 1.0E-5 | 0.869 | 0 | 0.324 |
| 35159 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | ABCB5 | Sponge network | -1.886 | 0 | -0.956 | 0.04077 | 0.324 |
| 35160 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | DDX6 | Sponge network | 0.267 | 0.2937 | 0.091 | 0.36428 | 0.324 |
| 35161 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p | 10 | PTPN11 | Sponge network | 1.11 | 1.0E-5 | 0.431 | 2.0E-5 | 0.324 |
| 35162 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | ERBB4 | Sponge network | -0.563 | 0.02545 | -1.975 | 2.0E-5 | 0.324 |
| 35163 | AC018890.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-96-5p | 12 | IL17RD | Sponge network | 1.61 | 0.00961 | -0.019 | 0.94873 | 0.324 |
| 35164 | RP11-195B17.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-96-5p | 19 | FOXP1 | Sponge network | 0.37 | 0.27614 | 0.042 | 0.74368 | 0.324 |
| 35165 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-590-3p | 17 | CDC73 | Sponge network | 0.959 | 0 | 0.094 | 0.20463 | 0.324 |
| 35166 | LINC00092 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | PDE4DIP | Sponge network | -1.886 | 0 | -0.282 | 0.0257 | 0.324 |
| 35167 | RP11-536O18.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-502-5p | 14 | IL17RD | Sponge network | -0.074 | 0.90758 | -0.019 | 0.94873 | 0.324 |
| 35168 | TRAM2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-655-3p | 10 | ZMYND11 | Sponge network | 0.336 | 0.00739 | -0.2 | 0.05449 | 0.324 |
| 35169 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 20 | PDS5B | Sponge network | 0.569 | 0.00067 | 0.259 | 0.00962 | 0.324 |
| 35170 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 15 | MED12L | Sponge network | 0.497 | 0.01451 | -0.194 | 0.55299 | 0.324 |
| 35171 | RP11-401P9.4 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-369-3p;hsa-miR-542-3p;hsa-miR-590-5p | 11 | ITGB1 | Sponge network | -0.384 | 0.40652 | 0.524 | 0.00017 | 0.324 |
| 35172 | RP11-389C8.2 |
hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PIK3CA | Sponge network | 0.727 | 3.0E-5 | 0.195 | 0.06908 | 0.324 |
| 35173 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-5p | 16 | PTGFR | Sponge network | 0.494 | 0.00339 | -0.233 | 0.45421 | 0.324 |
| 35174 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | -0.727 | 0.04726 | 0.024 | 0.89889 | 0.324 |
| 35175 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-940 | 37 | QKI | Sponge network | 0.174 | 0.30034 | -0.333 | 0.04111 | 0.324 |
| 35176 | RP11-57H14.4 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 15 | MAPK10 | Sponge network | 0.083 | 0.66405 | -1.774 | 0 | 0.324 |
| 35177 | RP11-254F7.2 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p | 14 | GJA1 | Sponge network | 0.176 | 0.37402 | -0.485 | 0.02179 | 0.324 |
| 35178 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 14 | GPAM | Sponge network | 0.706 | 0 | 0.142 | 0.29732 | 0.324 |
| 35179 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p | 15 | PIK3C2A | Sponge network | 0.93 | 0 | 0.061 | 0.53776 | 0.324 |
| 35180 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-577 | 18 | PRKAB2 | Sponge network | 0.637 | 0.00069 | -0.367 | 0.01371 | 0.324 |
| 35181 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 39 | SMAD2 | Sponge network | 0.75 | 0.00054 | -0.128 | 0.12732 | 0.324 |
| 35182 | RP4-614O4.11 |
hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-424-5p | 10 | ABL2 | Sponge network | 1.042 | 0.00177 | 0.82 | 0 | 0.324 |
| 35183 | RP11-506M13.3 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 17 | CCDC6 | Sponge network | 0.769 | 0 | 0.27 | 0.02555 | 0.324 |
| 35184 | RP11-473I1.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 19 | SMAD2 | Sponge network | 0.542 | 0.00054 | -0.128 | 0.12732 | 0.324 |
| 35185 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-183-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-454-3p | 14 | ABI2 | Sponge network | 0.598 | 0.38481 | 0.175 | 0.14787 | 0.324 |
| 35186 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p | 10 | PRRX1 | Sponge network | 0.043 | 0.81628 | 1.316 | 0 | 0.324 |
| 35187 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-550a-5p;hsa-miR-766-3p;hsa-miR-93-5p | 14 | CHD5 | Sponge network | 0.131 | 0.8436 | -0.922 | 0.01521 | 0.324 |
| 35188 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 24 | LRRC55 | Sponge network | 0.374 | 0.20054 | -0.026 | 0.93163 | 0.324 |
| 35189 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | DPYD | Sponge network | 0.572 | 0.01722 | -0.736 | 0.00076 | 0.324 |
| 35190 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | LRIG1 | Sponge network | -0.739 | 0.00044 | -0.537 | 0.00588 | 0.324 |
| 35191 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-744-5p;hsa-miR-93-5p | 14 | CHD5 | Sponge network | -0.023 | 0.95109 | -0.922 | 0.01521 | 0.324 |
| 35192 | LINC00476 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-877-5p | 24 | NRK | Sponge network | -0.615 | 0.00015 | 0.656 | 0.19217 | 0.324 |
| 35193 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 24 | ZFHX3 | Sponge network | -0.13 | 0.48734 | 0.389 | 0.01363 | 0.324 |
| 35194 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p | 22 | TSC1 | Sponge network | 0.253 | 0.09565 | 0.103 | 0.26071 | 0.324 |
| 35195 | FAM66C |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | HOMER2 | Sponge network | -0.901 | 0.00019 | -2.698 | 0 | 0.324 |
| 35196 | FGF14-AS2 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-7-5p | 16 | ERC1 | Sponge network | -1.582 | 8.0E-5 | -0.108 | 0.38794 | 0.324 |
| 35197 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-629-3p | 27 | CBX5 | Sponge network | 0.946 | 0 | 0.165 | 0.24446 | 0.324 |
| 35198 | TRAF3IP2-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ZNF844 | Sponge network | -0.323 | 0.01372 | -0.966 | 0.00035 | 0.324 |
| 35199 | FAM13A-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | PRKCB | Sponge network | -0.83 | 0 | -1.313 | 2.0E-5 | 0.324 |
| 35200 | RP11-819C21.1 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PCDH9 | Sponge network | 0.537 | 0.00139 | -1.823 | 4.0E-5 | 0.324 |
| 35201 | RP11-244O19.1 |
hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-629-5p | 25 | UBR3 | Sponge network | -0.096 | 0.67958 | -0.021 | 0.82774 | 0.324 |
| 35202 | PRKAG2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ST6GALNAC3 | Sponge network | -0.622 | 0.02807 | -0.987 | 0 | 0.324 |
| 35203 | HOXB-AS1 |
hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-508-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-93-3p | 30 | CUX1 | Sponge network | -0.154 | 0.53954 | -0.039 | 0.6705 | 0.324 |
| 35204 | DIO3OS |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-98-5p | 20 | LRIG1 | Sponge network | -0.773 | 0.04073 | -0.537 | 0.00588 | 0.324 |
| 35205 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 18 | RCAN2 | Sponge network | 0.421 | 0.00084 | -0.33 | 0.15172 | 0.324 |
| 35206 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-96-5p | 22 | PDGFRA | Sponge network | -0.517 | 0.02935 | -0.372 | 0.0037 | 0.324 |
| 35207 | FAM13A-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-769-5p | 23 | SYNPO2 | Sponge network | -0.83 | 0 | -2.342 | 0 | 0.324 |
| 35208 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | PAK3 | Sponge network | 0.194 | 0.64278 | -0.628 | 0.07253 | 0.324 |
| 35209 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 16 | CXXC4 | Sponge network | -0.18 | 0.20343 | -0.433 | 0.21055 | 0.324 |
| 35210 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 46 | AR | Sponge network | -0.668 | 3.0E-5 | -1.554 | 0 | 0.324 |
| 35211 | RP11-400K9.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-576-5p | 11 | ABI2 | Sponge network | -0.733 | 0.19469 | 0.175 | 0.14787 | 0.324 |
| 35212 | RP11-65J21.3 |
hsa-let-7d-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-628-5p | 12 | RUNX1T1 | Sponge network | -0.557 | 0.11135 | -0.344 | 0.2374 | 0.324 |
| 35213 | LINC00657 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 26 | SOX11 | Sponge network | 0.91 | 0 | 2.68 | 0 | 0.324 |
| 35214 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-7-5p | 12 | FNBP1 | Sponge network | 0.494 | 0.00339 | -0.969 | 4.0E-5 | 0.324 |
| 35215 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-7-1-3p | 20 | RNF144A | Sponge network | 0.853 | 0.15352 | 0.594 | 0.0009 | 0.324 |
| 35216 | SNHG5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-485-3p;hsa-miR-495-3p | 13 | HACE1 | Sponge network | 0.13 | 0.47174 | -0.072 | 0.55239 | 0.324 |
| 35217 | HNRNPU-AS1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 19 | DDX3X | Sponge network | 1.601 | 0 | -0.152 | 0.07686 | 0.324 |
| 35218 | BMS1P20 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p | 17 | DDX3X | Sponge network | 0.34 | 0.00273 | -0.152 | 0.07686 | 0.324 |
| 35219 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | CACNA2D1 | Sponge network | 0.395 | 0.15451 | -0.846 | 0.00675 | 0.324 |
| 35220 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | RAP1A | Sponge network | -0.901 | 0.00019 | -0.929 | 0 | 0.324 |
| 35221 | RP11-77H9.2 |
hsa-miR-129-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-376c-3p | 10 | RASAL2 | Sponge network | 1.677 | 0 | 0.869 | 0 | 0.323 |
| 35222 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 18 | SCN9A | Sponge network | -0.737 | 0.10822 | -0.525 | 0.10593 | 0.323 |
| 35223 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-940 | 50 | WDFY3 | Sponge network | -0.777 | 0.16667 | -0.201 | 0.0967 | 0.323 |
| 35224 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | USP12 | Sponge network | 0.174 | 0.30034 | -0.102 | 0.40768 | 0.323 |
| 35225 | CTD-2336O2.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | SPG20 | Sponge network | -0.141 | 0.26434 | -1.16 | 0 | 0.323 |
| 35226 | RP11-2B6.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 20 | ZFX | Sponge network | 0.539 | 0.03722 | 0.09 | 0.42563 | 0.323 |
| 35227 | CROCCP2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 14 | ABL2 | Sponge network | 0.79 | 0 | 0.82 | 0 | 0.323 |
| 35228 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429 | 10 | SCN5A | Sponge network | -2.46 | 0 | -0.259 | 0.48224 | 0.323 |
| 35229 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 33 | ETV1 | Sponge network | 0.101 | 0.60919 | -0.096 | 0.67674 | 0.323 |
| 35230 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 20 | SCN9A | Sponge network | 0.392 | 0.0278 | -0.525 | 0.10593 | 0.323 |
| 35231 | RP11-130L8.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | CDH19 | Sponge network | -0.663 | 0.10887 | -3.434 | 0 | 0.323 |
| 35232 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PCDH17 | Sponge network | 0.657 | 4.0E-5 | 0.731 | 2.0E-5 | 0.323 |
| 35233 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p | 17 | ACVR1 | Sponge network | -0.611 | 0.09607 | 0.279 | 0.00234 | 0.323 |
| 35234 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-409-3p | 16 | AFF3 | Sponge network | -0.326 | 0.14314 | -2.373 | 0 | 0.323 |
| 35235 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-590-3p | 15 | RIMBP2 | Sponge network | -1.057 | 0.00029 | -1.968 | 5.0E-5 | 0.323 |
| 35236 | PSMG3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SAMHD1 | Sponge network | -0.125 | 0.49414 | 0.072 | 0.62895 | 0.323 |
| 35237 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-450b-5p | 11 | SPTBN4 | Sponge network | -0.517 | 0.02935 | -0.786 | 0.01281 | 0.323 |
| 35238 | LINC00621 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 23 | EPHA7 | Sponge network | 0.131 | 0.8436 | -2.197 | 3.0E-5 | 0.323 |
| 35239 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 10 | CSRNP1 | Sponge network | -0.726 | 0.00132 | -1.448 | 0 | 0.323 |
| 35240 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | PDS5B | Sponge network | 0.826 | 1.0E-5 | 0.259 | 0.00962 | 0.323 |
| 35241 | TPTEP1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | PCDH18 | Sponge network | -1.216 | 0 | 0.216 | 0.24645 | 0.323 |
| 35242 | HOXA-AS2 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | RADIL | Sponge network | 0.61 | 0.01593 | -1.628 | 0 | 0.323 |
| 35243 | CTD-3092A11.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | KDSR | Sponge network | 0.873 | 0 | 0.239 | 0.02216 | 0.323 |
| 35244 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-493-5p;hsa-miR-590-3p | 12 | CDC73 | Sponge network | 0.826 | 1.0E-5 | 0.094 | 0.20463 | 0.323 |
| 35245 | LINC00863 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-93-5p | 28 | CHD5 | Sponge network | 0.189 | 0.13981 | -0.922 | 0.01521 | 0.323 |
| 35246 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | CNTN1 | Sponge network | -0.72 | 0.24239 | -2.594 | 0 | 0.323 |
| 35247 | CTC-487M23.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-92a-3p | 20 | FAT3 | Sponge network | 0.912 | 0.00014 | -1.601 | 0.00051 | 0.323 |
| 35248 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | SON | Sponge network | 0.385 | 0.10067 | 0.168 | 0.04406 | 0.323 |
| 35249 | AC005618.6 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 20 | NRK | Sponge network | 0.862 | 0.06213 | 0.656 | 0.19217 | 0.323 |
| 35250 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p | 13 | HECA | Sponge network | 0.37 | 0.27614 | -0.217 | 0.03463 | 0.323 |
| 35251 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | CLASP2 | Sponge network | 1.205 | 0 | -0.038 | 0.71 | 0.323 |
| 35252 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p | 22 | CBL | Sponge network | 0.921 | 0.00118 | 0.291 | 0.01267 | 0.323 |
| 35253 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 12 | GPAM | Sponge network | 1.4 | 0 | 0.142 | 0.29732 | 0.323 |
| 35254 | C4A-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-744-3p;hsa-miR-96-5p | 14 | HIP1 | Sponge network | 3.345 | 0 | 0.774 | 0 | 0.323 |
| 35255 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 56 | PRKAA2 | Sponge network | 0.246 | 0.59912 | -2.462 | 0 | 0.323 |
| 35256 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3614-5p | 12 | EPHA3 | Sponge network | 0.529 | 0.00552 | 0.185 | 0.53588 | 0.323 |
| 35257 | RP11-119F7.5 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-935;hsa-miR-940 | 33 | GRIK3 | Sponge network | 0.225 | 0.30644 | -3.208 | 0 | 0.323 |
| 35258 | CTD-2314G24.2 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-96-5p | 18 | IRS1 | Sponge network | 0.246 | 0.59912 | -0.026 | 0.88855 | 0.323 |
| 35259 | SERHL |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-539-5p | 17 | DIP2C | Sponge network | -0.602 | 0.00331 | -0.436 | 0.01713 | 0.323 |
| 35260 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ZMYM2 | Sponge network | -0.031 | 0.89063 | 0.279 | 0.01273 | 0.323 |
| 35261 | RP11-2H8.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 17 | RASSF8 | Sponge network | 1.089 | 0 | -0.122 | 0.65591 | 0.323 |
| 35262 | AC159540.1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p | 23 | CREB1 | Sponge network | 1.122 | 0 | 0.203 | 0.00537 | 0.323 |
| 35263 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 33 | BMPR2 | Sponge network | 0.663 | 0.00073 | 0.206 | 0.0635 | 0.323 |
| 35264 | RP11-597D13.9 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 12 | KIF5B | Sponge network | 1.302 | 7.0E-5 | 0.362 | 0.00066 | 0.323 |
| 35265 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p | 12 | LHX6 | Sponge network | -1.057 | 0.00029 | -0.087 | 0.69193 | 0.323 |
| 35266 | PSMG3-AS1 |
hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | RNF180 | Sponge network | -0.125 | 0.49414 | -1.127 | 1.0E-5 | 0.323 |
| 35267 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 29 | ZBTB4 | Sponge network | 0.246 | 0.59912 | -0.739 | 0 | 0.323 |
| 35268 | RP11-392P7.6 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 10 | CBFB | Sponge network | 0.853 | 0 | 1.061 | 0 | 0.323 |
| 35269 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 14 | TXNIP | Sponge network | 0.997 | 0.00066 | -1.277 | 0 | 0.323 |
| 35270 | FAM157C |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | FOXO3 | Sponge network | 0.91 | 0 | -0.04 | 0.72028 | 0.323 |
| 35271 | AC103563.8 |
hsa-miR-10a-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p | 11 | IGFBP5 | Sponge network | -7.551 | 0 | -0.806 | 0.00105 | 0.323 |
| 35272 | RP11-25K19.1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 25 | RIMS2 | Sponge network | -1.057 | 0.00029 | -1.365 | 0.00208 | 0.323 |
| 35273 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | EPAS1 | Sponge network | -0.942 | 0 | -0.184 | 0.14418 | 0.323 |
| 35274 | RP11-6O2.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-2355-5p;hsa-miR-326;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-624-5p;hsa-miR-940 | 11 | SIRT1 | Sponge network | 0.331 | 0.57247 | 0.109 | 0.2593 | 0.323 |
| 35275 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 43 | GAS7 | Sponge network | 0.61 | 0.01593 | -0.555 | 0.01602 | 0.323 |
| 35276 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | IGF1 | Sponge network | 0.385 | 0.10067 | -0.204 | 0.5898 | 0.323 |
| 35277 | RP11-61L19.3 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 10 | PAFAH1B2 | Sponge network | 1.411 | 0 | 0.318 | 0.0001 | 0.323 |
| 35278 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p | 10 | PCDH8 | Sponge network | 0.051 | 0.83388 | -0.997 | 0.05366 | 0.323 |
| 35279 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | SASH1 | Sponge network | 0.299 | 0.57722 | -0.502 | 0.00175 | 0.323 |
| 35280 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | ZNF521 | Sponge network | -2.925 | 0 | -0.111 | 0.58068 | 0.323 |
| 35281 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | PDGFC | Sponge network | -1.203 | 0.03881 | -0.33 | 0.06483 | 0.323 |
| 35282 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-942-5p | 14 | RB1CC1 | Sponge network | 0.385 | 0.10067 | 0.175 | 0.12559 | 0.323 |
| 35283 | RP11-686D22.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-3p | 15 | TCF4 | Sponge network | 0.793 | 5.0E-5 | 0.016 | 0.92271 | 0.323 |
| 35284 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-7-1-3p | 14 | IGSF10 | Sponge network | -0.72 | 0.24239 | -1.751 | 1.0E-5 | 0.323 |
| 35285 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | RASSF8 | Sponge network | -0.285 | 0.2172 | -0.122 | 0.65591 | 0.323 |
| 35286 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-500a-3p;hsa-miR-7-1-3p | 14 | ADAMTSL3 | Sponge network | 0.238 | 0.70544 | -1.559 | 0 | 0.323 |
| 35287 | CTA-363E19.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-374b-5p;hsa-miR-539-5p | 20 | KIF1B | Sponge network | 1.055 | 0 | -0.118 | 0.32258 | 0.323 |
| 35288 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 29 | MTSS1 | Sponge network | -0.976 | 0.00025 | -0.81 | 0.00035 | 0.323 |
| 35289 | RP11-359B12.2 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TPO | Sponge network | 0.064 | 0.72934 | -1.87 | 2.0E-5 | 0.323 |
| 35290 | RP11-359B12.2 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 22 | CTIF | Sponge network | 0.064 | 0.72934 | -0.389 | 0.00549 | 0.323 |
| 35291 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p | 12 | BCL6 | Sponge network | -0.246 | 0.35034 | -0.211 | 0.19489 | 0.323 |
| 35292 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-450b-5p | 20 | CADM1 | Sponge network | -0.405 | 0.22013 | -0.9 | 0.00084 | 0.323 |
| 35293 | RP11-473I1.9 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 12 | ROBO1 | Sponge network | 0.683 | 1.0E-5 | -0.009 | 0.96154 | 0.323 |
| 35294 | CTD-2314G24.2 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | NR3C2 | Sponge network | 0.246 | 0.59912 | -1.56 | 0 | 0.323 |
| 35295 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-93-5p | 13 | FGFR1 | Sponge network | 1.153 | 0.00114 | -0.509 | 0.03957 | 0.323 |
| 35296 | SH3BP5-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 19 | HECA | Sponge network | 0.267 | 0.2937 | -0.217 | 0.03463 | 0.323 |
| 35297 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 42 | KIF1B | Sponge network | -0.611 | 0.09607 | -0.118 | 0.32258 | 0.323 |
| 35298 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-33a-5p | 19 | RUNX1T1 | Sponge network | -2.549 | 0.00528 | -0.344 | 0.2374 | 0.323 |
| 35299 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 13 | PRKAB2 | Sponge network | 1.11 | 1.0E-5 | -0.367 | 0.01371 | 0.323 |
| 35300 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-625-3p | 21 | TMTC3 | Sponge network | 1.11 | 1.0E-5 | 0.123 | 0.22189 | 0.323 |
| 35301 | RP11-46C24.7 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-5p | 48 | DST | Sponge network | 0.392 | 0.0278 | -0.713 | 0.00096 | 0.323 |
| 35302 | AC005562.1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 14 | RASAL2 | Sponge network | 0.488 | 0.00084 | 0.869 | 0 | 0.323 |
| 35303 | RP11-890B15.3 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | RPS6KA6 | Sponge network | 0.101 | 0.60919 | -2.989 | 0 | 0.323 |
| 35304 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 23 | KIT | Sponge network | -0.615 | 0.00015 | -2.184 | 0 | 0.323 |
| 35305 | EMX2OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 10 | FAM133A | Sponge network | -0.777 | 0.1134 | 0.549 | 0.29009 | 0.323 |
| 35306 | ERVH48-1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | DMD | Sponge network | 1.04 | 0.00023 | -1.506 | 0 | 0.323 |
| 35307 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 29 | SOX11 | Sponge network | -0.129 | 0.57177 | 2.68 | 0 | 0.323 |
| 35308 | RP11-597D13.8 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p | 10 | MEF2C | Sponge network | 0.936 | 0.03889 | -0.499 | 0.01448 | 0.323 |
| 35309 | RP11-401P9.4 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-877-5p | 13 | INPP4A | Sponge network | -0.384 | 0.40652 | 0.07 | 0.53563 | 0.323 |
| 35310 | BZRAP1-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 21 | EPS15 | Sponge network | -0.465 | 0.04703 | -0.052 | 0.53998 | 0.323 |
| 35311 | RP11-315O6.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-940 | 10 | MDM4 | Sponge network | 0.552 | 0.07637 | 0.345 | 0.00762 | 0.323 |
| 35312 | RP11-290F24.6 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p | 18 | CCDC6 | Sponge network | 2.052 | 0 | 0.27 | 0.02555 | 0.323 |
| 35313 | MIR497HG |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-96-5p | 20 | HIP1 | Sponge network | -1.439 | 4.0E-5 | 0.774 | 0 | 0.323 |
| 35314 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-484;hsa-miR-576-5p | 19 | GREM1 | Sponge network | 0.082 | 0.55654 | 0.902 | 0.01691 | 0.323 |
| 35315 | RP11-98I9.4 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 20 | ABL2 | Sponge network | 0.67 | 0.00048 | 0.82 | 0 | 0.323 |
| 35316 | LINC00265 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-3607-3p | 11 | FOXO3 | Sponge network | 1.07 | 0 | -0.04 | 0.72028 | 0.323 |
| 35317 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 11 | FYN | Sponge network | -0.154 | 0.78939 | -0.131 | 0.37051 | 0.323 |
| 35318 | BVES-AS1 |
hsa-let-7b-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-98-5p | 13 | TES | Sponge network | -2.62 | 0 | -0.348 | 0.00639 | 0.323 |
| 35319 | SNHG14 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-940 | 24 | BAZ2B | Sponge network | -1.111 | 0.0003 | -0.276 | 0.02833 | 0.323 |
| 35320 | RP11-359E10.1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-7-1-3p | 19 | RIMS2 | Sponge network | 0.299 | 0.57722 | -1.365 | 0.00208 | 0.323 |
| 35321 | RP11-597D13.9 |
hsa-miR-142-3p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-577 | 11 | AMPH | Sponge network | 1.302 | 7.0E-5 | -0.916 | 5.0E-5 | 0.323 |
| 35322 | RP11-66B24.2 |
hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-378c | 14 | TACC1 | Sponge network | 0.57 | 0.1866 | -0.362 | 0.05406 | 0.323 |
| 35323 | LINC00648 |
hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | PLAG1 | Sponge network | 0.633 | 0.51678 | -0.315 | 0.26532 | 0.323 |
| 35324 | RP11-890B15.3 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 36 | ARHGEF12 | Sponge network | 0.101 | 0.60919 | 0.09 | 0.35693 | 0.323 |
| 35325 | RP11-277P12.20 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 16 | NAV3 | Sponge network | 0.687 | 0.02753 | 0.212 | 0.47729 | 0.323 |
| 35326 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-940 | 11 | CRTC3 | Sponge network | -0.141 | 0.26434 | 0.164 | 0.16786 | 0.323 |
| 35327 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-7-1-3p | 11 | LRRC55 | Sponge network | -1.057 | 0.00029 | -0.026 | 0.93163 | 0.323 |
| 35328 | PART1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p | 35 | CADM1 | Sponge network | -2.925 | 0 | -0.9 | 0.00084 | 0.323 |
| 35329 | SH3RF3-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-874-3p;hsa-miR-877-5p | 17 | ASTN1 | Sponge network | 0.889 | 9.0E-5 | -2.389 | 2.0E-5 | 0.323 |
| 35330 | RP11-707P17.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | CCDC6 | Sponge network | 1.877 | 0 | 0.27 | 0.02555 | 0.323 |
| 35331 | MALAT1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | NOTCH2NL | Sponge network | 0.782 | 0.00016 | 0.024 | 0.84307 | 0.323 |
| 35332 | RP11-46C24.7 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PRUNE2 | Sponge network | 0.392 | 0.0278 | -1.733 | 0.00022 | 0.323 |
| 35333 | RP11-968O1.5 |
hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-493-5p | 13 | GRIA3 | Sponge network | -0.829 | 0.01343 | -1.956 | 0 | 0.323 |
| 35334 | RP11-152F13.8 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | UBR3 | Sponge network | 0.843 | 0.01775 | -0.021 | 0.82774 | 0.323 |
| 35335 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | LRRC7 | Sponge network | 0.61 | 0.01593 | -1.341 | 0.01193 | 0.323 |
| 35336 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | SMAD2 | Sponge network | 0.545 | 0.00064 | -0.128 | 0.12732 | 0.323 |
| 35337 | RP11-403I13.8 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 21 | ASTN1 | Sponge network | 0.619 | 0.00157 | -2.389 | 2.0E-5 | 0.323 |
| 35338 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 13 | LRRC7 | Sponge network | -0.318 | 0.65847 | -1.341 | 0.01193 | 0.323 |
| 35339 | GNG12-AS1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-940 | 12 | CSRNP1 | Sponge network | -0.793 | 7.0E-5 | -1.448 | 0 | 0.323 |
| 35340 | FENDRR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | -1.281 | 0.00012 | 0.126 | 0.15266 | 0.323 |
| 35341 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 17 | FOXO3 | Sponge network | 0.197 | 0.25618 | -0.04 | 0.72028 | 0.323 |
| 35342 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 47 | SSBP2 | Sponge network | 0.246 | 0.59912 | -1.58 | 0 | 0.323 |
| 35343 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PDS5B | Sponge network | 0.349 | 0.01695 | 0.259 | 0.00962 | 0.323 |
| 35344 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 31 | ETV1 | Sponge network | -0.487 | 0.02157 | -0.096 | 0.67674 | 0.323 |
| 35345 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-93-3p | 29 | AKAP11 | Sponge network | -0.227 | 0.31657 | 0.196 | 0.09825 | 0.323 |
| 35346 | NEAT1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-708-5p | 39 | CCDC6 | Sponge network | 0.705 | 0.00014 | 0.27 | 0.02555 | 0.323 |
| 35347 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 31 | DMXL1 | Sponge network | 0.72 | 0.0001 | -0.426 | 0.00056 | 0.323 |
| 35348 | PCA3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 24 | ACSL6 | Sponge network | -0.709 | 0.18168 | -1.593 | 0 | 0.323 |
| 35349 | CTC-429P9.3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-589-5p | 16 | PRKCB | Sponge network | 0.082 | 0.55397 | -1.313 | 2.0E-5 | 0.323 |
| 35350 | LINC00865 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 14 | GABRB2 | Sponge network | -1.249 | 7.0E-5 | -1.841 | 0.00029 | 0.323 |
| 35351 | FZD10-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p | 13 | ELMO1 | Sponge network | -0.727 | 0.04726 | 0.04 | 0.83147 | 0.323 |
| 35352 | RP11-156P1.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p | 18 | CDH23 | Sponge network | 0.214 | 0.18246 | -0.974 | 0.00034 | 0.323 |
| 35353 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 23 | THBS1 | Sponge network | 0.385 | 0.10067 | 0.741 | 0.00386 | 0.323 |
| 35354 | TINCR |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-199b-5p;hsa-miR-200b-3p;hsa-miR-214-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-874-3p | 19 | MXI1 | Sponge network | -0.664 | 0.14543 | -0.987 | 0 | 0.323 |
| 35355 | ZNF582-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -1.349 | 0.00017 | -0.252 | 0.15511 | 0.323 |
| 35356 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p | 10 | TBRG1 | Sponge network | 0.101 | 0.60919 | -0.071 | 0.37324 | 0.323 |
| 35357 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 29 | SOX5 | Sponge network | 0.419 | 0.0319 | -1.091 | 4.0E-5 | 0.323 |
| 35358 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | DACH2 | Sponge network | -1.575 | 6.0E-5 | -1.635 | 0.00034 | 0.323 |
| 35359 | LINC00654 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 37 | CREB3L2 | Sponge network | 0.374 | 0.20054 | -0.525 | 2.0E-5 | 0.323 |
| 35360 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-590-3p | 12 | HOOK3 | Sponge network | -0.342 | 0.02288 | 0.273 | 0.09957 | 0.323 |
| 35361 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p | 14 | TBX18 | Sponge network | 0.214 | 0.18246 | 0.843 | 0.02945 | 0.323 |
| 35362 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-98-5p | 17 | KCNA6 | Sponge network | -1.116 | 0.23686 | -1.531 | 0.00013 | 0.323 |
| 35363 | CTB-31O20.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 20 | CCDC6 | Sponge network | 1.619 | 0 | 0.27 | 0.02555 | 0.323 |
| 35364 | LINC00476 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 25 | CYLD | Sponge network | -0.615 | 0.00015 | -0.367 | 0.00171 | 0.323 |
| 35365 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-484 | 15 | GRIA3 | Sponge network | -4.477 | 0 | -1.956 | 0 | 0.323 |
| 35366 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-96-5p;hsa-miR-98-5p | 39 | CREB3L2 | Sponge network | 0.249 | 0.35902 | -0.525 | 2.0E-5 | 0.323 |
| 35367 | BMS1P20 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | KDSR | Sponge network | 0.34 | 0.00273 | 0.239 | 0.02216 | 0.323 |
| 35368 | LINC00854 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26b-3p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-940 | 13 | EPS15 | Sponge network | 1.18 | 0 | -0.052 | 0.53998 | 0.323 |
| 35369 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | SLITRK6 | Sponge network | 0.225 | 0.30644 | -2.121 | 0 | 0.323 |
| 35370 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940;hsa-miR-96-5p | 38 | GAS7 | Sponge network | 0.72 | 0.0001 | -0.555 | 0.01602 | 0.323 |
| 35371 | RP11-57H14.4 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | KLF10 | Sponge network | 0.083 | 0.66405 | -0.783 | 0 | 0.323 |
| 35372 | LINC00854 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-5p;hsa-miR-940 | 21 | NFIB | Sponge network | 1.18 | 0 | 0.023 | 0.90795 | 0.323 |
| 35373 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 28 | BHLHE41 | Sponge network | 0.61 | 0.01593 | 0.353 | 0.12753 | 0.323 |
| 35374 | RP11-774O3.3 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 14 | NR3C2 | Sponge network | -0.487 | 0.02157 | -1.56 | 0 | 0.323 |
| 35375 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-495-3p | 10 | TBL1XR1 | Sponge network | 0.336 | 0.00739 | 0.578 | 0 | 0.323 |
| 35376 | LINC01006 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p | 24 | ST6GALNAC3 | Sponge network | -0.702 | 0.04814 | -0.987 | 0 | 0.323 |
| 35377 | AGAP11 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-590-5p | 11 | FBXO11 | Sponge network | -1.645 | 0 | 0.142 | 0.04938 | 0.323 |
| 35378 | RP11-53O19.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | ABI2 | Sponge network | 1.205 | 0 | 0.175 | 0.14787 | 0.323 |
| 35379 | RP4-639F20.1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 27 | CREB3L2 | Sponge network | -0.517 | 0.02935 | -0.525 | 2.0E-5 | 0.323 |
| 35380 | LINC00702 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-3913-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 40 | EGFR | Sponge network | -0.737 | 0.06182 | -0.175 | 0.48451 | 0.323 |
| 35381 | CTD-2666L21.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-744-3p | 16 | DIP2C | Sponge network | 0.368 | 0.20093 | -0.436 | 0.01713 | 0.323 |
| 35382 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | FGF5 | Sponge network | 0.619 | 0.00157 | 0.549 | 0.28659 | 0.323 |
| 35383 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p | 17 | PPP1R9A | Sponge network | -2.531 | 0 | -0.903 | 0.04254 | 0.323 |
| 35384 | AC005618.6 |
hsa-miR-132-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 20 | PCDH9 | Sponge network | 0.862 | 0.06213 | -1.823 | 4.0E-5 | 0.323 |
| 35385 | RP11-819C21.1 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p | 12 | SYNM | Sponge network | 0.537 | 0.00139 | -2.309 | 1.0E-5 | 0.323 |
| 35386 | RP5-994D16.9 |
hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-22-3p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PPM1L | Sponge network | 0.252 | 0.08508 | -0.537 | 0.00616 | 0.323 |
| 35387 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 17 | ESR1 | Sponge network | 0.178 | 0.29106 | -1.035 | 3.0E-5 | 0.323 |
| 35388 | RP11-798G7.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-29b-3p | 15 | EPS15 | Sponge network | 0.858 | 3.0E-5 | -0.052 | 0.53998 | 0.323 |
| 35389 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | CRTC1 | Sponge network | -1.697 | 0.00889 | -0.116 | 0.28412 | 0.323 |
| 35390 | BDNF-AS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 18 | AKAP6 | Sponge network | -0.668 | 3.0E-5 | -1.586 | 3.0E-5 | 0.323 |
| 35391 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-940;hsa-miR-96-5p | 16 | MYO9A | Sponge network | -0.326 | 0.14314 | -0.147 | 0.32373 | 0.323 |
| 35392 | AGAP11 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-942-5p | 12 | ZBTB20 | Sponge network | -1.645 | 0 | -0.122 | 0.57569 | 0.323 |
| 35393 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 23 | PRKAB2 | Sponge network | -0.358 | 0.17255 | -0.367 | 0.01371 | 0.323 |
| 35394 | HAND2-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 54 | PPM1L | Sponge network | -1.697 | 0.00889 | -0.537 | 0.00616 | 0.323 |
| 35395 | RP11-983P16.4 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-484;hsa-miR-7-5p | 10 | FLNA | Sponge network | 0.41 | 0.02214 | -0.771 | 0.01377 | 0.323 |
| 35396 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-495-3p | 20 | MYO9A | Sponge network | 1.03 | 0 | -0.147 | 0.32373 | 0.323 |
| 35397 | RP11-180O5.2 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-495-3p | 13 | RICTOR | Sponge network | 3.607 | 0.0001 | 0.388 | 0.00188 | 0.323 |
| 35398 | RP11-21A7A.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-5p | 20 | DST | Sponge network | -1.116 | 0.23686 | -0.713 | 0.00096 | 0.323 |
| 35399 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 10 | PRKAB2 | Sponge network | 1.2 | 0.01813 | -0.367 | 0.01371 | 0.323 |
| 35400 | RP11-421L21.3 |
hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-497-5p;hsa-miR-766-3p | 11 | TBL1XR1 | Sponge network | 0.962 | 0 | 0.578 | 0 | 0.323 |
| 35401 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-576-5p | 19 | ARHGAP28 | Sponge network | -1.349 | 0.00017 | -0.911 | 0.00013 | 0.323 |
| 35402 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 30 | CBL | Sponge network | -0.487 | 0.02157 | 0.291 | 0.01267 | 0.323 |
| 35403 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-577 | 16 | MYBL1 | Sponge network | 0.921 | 0.00118 | 0.494 | 0.02152 | 0.323 |
| 35404 | AC074289.1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-5p | 10 | PTGFR | Sponge network | -0.326 | 0.14314 | -0.233 | 0.45421 | 0.323 |
| 35405 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-629-5p | 22 | CRTC1 | Sponge network | -2.69 | 0.0001 | -0.116 | 0.28412 | 0.323 |
| 35406 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 10 | FAM117A | Sponge network | -0.421 | 0.0114 | -1.379 | 0 | 0.323 |
| 35407 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | IGSF10 | Sponge network | -0.342 | 0.02288 | -1.751 | 1.0E-5 | 0.323 |
| 35408 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-26a-5p;hsa-miR-26b-3p | 12 | CBX5 | Sponge network | 0.912 | 0.00014 | 0.165 | 0.24446 | 0.323 |
| 35409 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | TSHZ3 | Sponge network | 0.72 | 0.0001 | -0.556 | 0.01516 | 0.323 |
| 35410 | FAM215B |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-495-3p | 13 | VPS53 | Sponge network | 0.817 | 3.0E-5 | 0.103 | 0.22667 | 0.323 |
| 35411 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | LRP1B | Sponge network | -1.216 | 0 | -2.163 | 0 | 0.323 |
| 35412 | RP11-973H7.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-33a-3p | 16 | MDM4 | Sponge network | 0.901 | 0 | 0.345 | 0.00762 | 0.323 |
| 35413 | RP11-620J15.3 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | HOMER2 | Sponge network | -0.739 | 0.00044 | -2.698 | 0 | 0.323 |
| 35414 | PART1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 19 | CELF4 | Sponge network | -2.925 | 0 | -1.135 | 0.00344 | 0.323 |
| 35415 | RP11-890B15.3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-98-5p | 18 | KLF10 | Sponge network | 0.101 | 0.60919 | -0.783 | 0 | 0.323 |
| 35416 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-93-5p | 10 | TCF4 | Sponge network | -0.295 | 0.02604 | 0.016 | 0.92271 | 0.323 |
| 35417 | RP11-277P12.20 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-877-5p | 25 | GAS7 | Sponge network | 0.687 | 0.02753 | -0.555 | 0.01602 | 0.323 |
| 35418 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | NIN | Sponge network | -0.726 | 0.00132 | -0.253 | 0.13794 | 0.323 |
| 35419 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | HDAC9 | Sponge network | -2.117 | 0.00161 | -0.755 | 0.00495 | 0.323 |
| 35420 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-942-5p | 20 | ARHGAP28 | Sponge network | -4.546 | 0 | -0.911 | 0.00013 | 0.323 |
| 35421 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-493-5p | 17 | AFF3 | Sponge network | 0.056 | 0.8494 | -2.373 | 0 | 0.323 |
| 35422 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -0.709 | 0.18168 | -0.087 | 0.69193 | 0.323 |
| 35423 | OIP5-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 17 | ROBO1 | Sponge network | 0.421 | 0.00084 | -0.009 | 0.96154 | 0.323 |
| 35424 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PTPRB | Sponge network | -0.487 | 0.02157 | 0.105 | 0.47122 | 0.323 |
| 35425 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 18 | AFF3 | Sponge network | -0.08 | 0.90092 | -2.373 | 0 | 0.323 |
| 35426 | LINC00963 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 25 | PTPN14 | Sponge network | 0.523 | 9.0E-5 | 0.144 | 0.34595 | 0.323 |
| 35427 | RP5-1139B12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p | 10 | RET | Sponge network | 0.598 | 0.38481 | -0.833 | 0.03517 | 0.323 |
| 35428 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-654-3p;hsa-miR-942-5p | 30 | DDX6 | Sponge network | 0.078 | 0.55803 | 0.091 | 0.36428 | 0.323 |
| 35429 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SEMA3C | Sponge network | 0.862 | 0.06213 | -0.272 | 0.31153 | 0.323 |
| 35430 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | USP12 | Sponge network | 0.541 | 0.00125 | -0.102 | 0.40768 | 0.323 |
| 35431 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p | 19 | PRKAB2 | Sponge network | -0.469 | 0.08721 | -0.367 | 0.01371 | 0.323 |
| 35432 | AC156455.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-330-3p | 14 | ZFHX4 | Sponge network | 1.154 | 0.00041 | 0.018 | 0.95574 | 0.323 |
| 35433 | SNHG14 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p | 19 | PLXNC1 | Sponge network | -1.111 | 0.0003 | 0.569 | 0.00329 | 0.323 |
| 35434 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-25-3p | 10 | ST6GAL2 | Sponge network | -2.095 | 0.00112 | -1.201 | 0.00597 | 0.323 |
| 35435 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 24 | KCNA6 | Sponge network | 0.101 | 0.60919 | -1.531 | 0.00013 | 0.322 |
| 35436 | LINC00672 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | RPS6KA6 | Sponge network | -0.227 | 0.31657 | -2.989 | 0 | 0.322 |
| 35437 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p | 11 | RCAN2 | Sponge network | -0.465 | 0.04703 | -0.33 | 0.15172 | 0.322 |
| 35438 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 24 | ACVR2A | Sponge network | 0.082 | 0.55397 | -0.409 | 0.00039 | 0.322 |
| 35439 | RP11-968O1.5 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-532-5p | 21 | IGFBP5 | Sponge network | -0.829 | 0.01343 | -0.806 | 0.00105 | 0.322 |
| 35440 | GHRLOS |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-455-3p | 14 | ACSL6 | Sponge network | -1.942 | 0 | -1.593 | 0 | 0.322 |
| 35441 | CTB-51J22.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-92a-3p;hsa-miR-940 | 13 | NFIB | Sponge network | -0.803 | 0.23737 | 0.023 | 0.90795 | 0.322 |
| 35442 | RP11-37B2.1 |
hsa-miR-129-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-664a-3p | 17 | RASAL2 | Sponge network | 1.03 | 0 | 0.869 | 0 | 0.322 |
| 35443 | AC005062.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | EIF4A2 | Sponge network | -0.019 | 0.81504 | -0.383 | 0.00046 | 0.322 |
| 35444 | SNHG16 |
hsa-let-7a-3p;hsa-miR-154-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374a-3p | 14 | TMTC3 | Sponge network | 0.961 | 0 | 0.123 | 0.22189 | 0.322 |
| 35445 | DNM3OS |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3E | Sponge network | 0.572 | 0.01722 | -1.717 | 0.00137 | 0.322 |
| 35446 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 27 | JAZF1 | Sponge network | -0.322 | 0.25945 | -0.377 | 0.02677 | 0.322 |
| 35447 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 28 | LATS1 | Sponge network | 0.782 | 0.00016 | 0.109 | 0.34208 | 0.322 |
| 35448 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-590-3p | 20 | GRIA3 | Sponge network | -0.342 | 0.02288 | -1.956 | 0 | 0.322 |
| 35449 | LINC00403 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-452-5p | 25 | LPP | Sponge network | -3.586 | 0.00032 | -0.459 | 0.04475 | 0.322 |
| 35450 | EMX2OS |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 13 | PAK3 | Sponge network | -0.777 | 0.1134 | -0.628 | 0.07253 | 0.322 |
| 35451 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 42 | BNC2 | Sponge network | 0.72 | 0.0001 | -0.491 | 0.09988 | 0.322 |
| 35452 | OIP5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | NUAK1 | Sponge network | 0.421 | 0.00084 | 0.904 | 0 | 0.322 |
| 35453 | LINC00641 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | NOTCH2 | Sponge network | -0.222 | 0.32445 | 0.138 | 0.35163 | 0.322 |
| 35454 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p | 14 | RBL2 | Sponge network | 0.214 | 0.18246 | -0.282 | 0.00254 | 0.322 |
| 35455 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SUFU | Sponge network | -0.942 | 0 | -0.04 | 0.65489 | 0.322 |
| 35456 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | FAT4 | Sponge network | 0.349 | 0.01695 | -0.354 | 0.14694 | 0.322 |
| 35457 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-664a-5p;hsa-miR-92a-3p | 20 | DST | Sponge network | 0.912 | 0.00014 | -0.713 | 0.00096 | 0.322 |
| 35458 | RP11-531A24.5 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | INPP4A | Sponge network | 0.75 | 0.00054 | 0.07 | 0.53563 | 0.322 |
| 35459 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 51 | TCF4 | Sponge network | 0.056 | 0.81195 | 0.016 | 0.92271 | 0.322 |
| 35460 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-7-1-3p | 24 | ADAMTSL3 | Sponge network | -0.303 | 0.21002 | -1.559 | 0 | 0.322 |
| 35461 | RP11-758N13.1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-532-3p | 10 | SYNM | Sponge network | 2.939 | 3.0E-5 | -2.309 | 1.0E-5 | 0.322 |
| 35462 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p | 14 | PTGFR | Sponge network | 0.75 | 0.00054 | -0.233 | 0.45421 | 0.322 |
| 35463 | MEG3 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 12 | PEA15 | Sponge network | 0.249 | 0.35902 | 0.009 | 0.93318 | 0.322 |
| 35464 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | PRRX1 | Sponge network | -1.582 | 8.0E-5 | 1.316 | 0 | 0.322 |
| 35465 | MIR143HG |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | USP25 | Sponge network | -0.739 | 0.0574 | -0.242 | 0.01397 | 0.322 |
| 35466 | AC074289.1 |
hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-34a-5p;hsa-miR-375 | 10 | ITPKB | Sponge network | -0.326 | 0.14314 | -0.704 | 0.00106 | 0.322 |
| 35467 | PCA3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | ABI2 | Sponge network | -0.709 | 0.18168 | 0.175 | 0.14787 | 0.322 |
| 35468 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-503-5p | 27 | RICTOR | Sponge network | 0.082 | 0.55397 | 0.388 | 0.00188 | 0.322 |
| 35469 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p | 12 | CXXC4 | Sponge network | -1.331 | 3.0E-5 | -0.433 | 0.21055 | 0.322 |
| 35470 | LINC00473 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 19 | RIMBP2 | Sponge network | -1.203 | 0.03881 | -1.968 | 5.0E-5 | 0.322 |
| 35471 | AC093627.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p | 10 | RECK | Sponge network | -0.787 | 0.3928 | -1.043 | 0 | 0.322 |
| 35472 | BCYRN1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.311 | 0.10344 | -0.57 | 0 | 0.322 |
| 35473 | SNHG16 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p | 10 | CHD1 | Sponge network | 0.961 | 0 | 0.322 | 0.00215 | 0.322 |
| 35474 | RP11-571M6.17 |
hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-590-3p;hsa-miR-625-3p | 11 | TMTC3 | Sponge network | 1.013 | 0 | 0.123 | 0.22189 | 0.322 |
| 35475 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 0.101 | 0.60919 | 0.943 | 0 | 0.322 |
| 35476 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 18 | SSBP2 | Sponge network | 0.693 | 0.00076 | -1.58 | 0 | 0.322 |
| 35477 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | TET1 | Sponge network | -0.031 | 0.89063 | -0.135 | 0.52138 | 0.322 |
| 35478 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-590-3p | 14 | FGF5 | Sponge network | 0.082 | 0.55654 | 0.549 | 0.28659 | 0.322 |
| 35479 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-93-3p | 22 | SSBP2 | Sponge network | -1.942 | 0 | -1.58 | 0 | 0.322 |
| 35480 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-495-3p | 10 | CUL5 | Sponge network | 0.909 | 0 | 0.026 | 0.77301 | 0.322 |
| 35481 | C20orf166-AS1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-486-5p | 11 | RASSF3 | Sponge network | -2.69 | 0.0001 | -0.226 | 0.11691 | 0.322 |
| 35482 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-331-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p | 14 | SLITRK6 | Sponge network | 0.056 | 0.81195 | -2.121 | 0 | 0.322 |
| 35483 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 14 | PRKAR1A | Sponge network | -0.739 | 0.00044 | -0.063 | 0.50314 | 0.322 |
| 35484 | GHRLOS |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-493-5p;hsa-miR-98-5p | 12 | DMD | Sponge network | -1.942 | 0 | -1.506 | 0 | 0.322 |
| 35485 | BOLA3-AS1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | MEF2C | Sponge network | -0.246 | 0.35034 | -0.499 | 0.01448 | 0.322 |
| 35486 | RP11-401P9.4 |
hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p | 16 | CSDE1 | Sponge network | -0.384 | 0.40652 | -0.493 | 0 | 0.322 |
| 35487 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 27 | FOXO3 | Sponge network | 0.101 | 0.60919 | -0.04 | 0.72028 | 0.322 |
| 35488 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | PNRC1 | Sponge network | -1.057 | 0.00029 | -0.646 | 0 | 0.322 |
| 35489 | AC006116.24 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-33a-5p | 12 | KANK1 | Sponge network | -0.473 | 0.07602 | -0.55 | 0.00134 | 0.322 |
| 35490 | ZNF582-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | GJA1 | Sponge network | -1.349 | 0.00017 | -0.485 | 0.02179 | 0.322 |
| 35491 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-590-3p | 12 | HERC2 | Sponge network | -0.323 | 0.01372 | 0.114 | 0.30638 | 0.322 |
| 35492 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-590-3p | 20 | USP6 | Sponge network | 0.559 | 0.00026 | 0.188 | 0.3871 | 0.322 |
| 35493 | RP11-67L2.2 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p | 19 | DDX3X | Sponge network | 0.72 | 0.0001 | -0.152 | 0.07686 | 0.322 |
| 35494 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 33 | AKAP11 | Sponge network | -0.487 | 0.02157 | 0.196 | 0.09825 | 0.322 |
| 35495 | RP11-835E18.5 |
hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PTPRT | Sponge network | -0.462 | 0.01226 | -0.907 | 0.03727 | 0.322 |
| 35496 | RP11-25K19.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 16 | CYLD | Sponge network | -1.057 | 0.00029 | -0.367 | 0.00171 | 0.322 |
| 35497 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 12 | IGSF10 | Sponge network | 0.622 | 0.00015 | -1.751 | 1.0E-5 | 0.322 |
| 35498 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | ZFHX4 | Sponge network | 0.064 | 0.72934 | 0.018 | 0.95574 | 0.322 |
| 35499 | LPP-AS2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p | 11 | FAM172A | Sponge network | -0.18 | 0.20343 | -0.57 | 0 | 0.322 |
| 35500 | WDFY3-AS2 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | FAM117A | Sponge network | -0.942 | 0 | -1.379 | 0 | 0.322 |
| 35501 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SLC6A15 | Sponge network | -2.915 | 0.00015 | -2.373 | 2.0E-5 | 0.322 |
| 35502 | RP4-758J18.10 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-370-3p | 20 | CHD5 | Sponge network | -0.091 | 0.71468 | -0.922 | 0.01521 | 0.322 |
| 35503 | RP11-384O8.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-501-3p | 13 | ZBTB20 | Sponge network | -0.738 | 0.21233 | -0.122 | 0.57569 | 0.322 |
| 35504 | WDR86-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-96-5p | 13 | HIP1 | Sponge network | 0.348 | 0.32912 | 0.774 | 0 | 0.322 |
| 35505 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p | 20 | CXXC4 | Sponge network | -0.147 | 0.40452 | -0.433 | 0.21055 | 0.322 |
| 35506 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SOX5 | Sponge network | 0.272 | 0.44692 | -1.091 | 4.0E-5 | 0.322 |
| 35507 | RP11-156E6.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-495-3p | 17 | CREB1 | Sponge network | 1.266 | 0 | 0.203 | 0.00537 | 0.322 |
| 35508 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TSHZ3 | Sponge network | 1.253 | 0 | -0.556 | 0.01516 | 0.322 |
| 35509 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 13 | DAB2 | Sponge network | -0.421 | 0.0114 | 0.431 | 0.0113 | 0.322 |
| 35510 | LINC00843 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | BRD3 | Sponge network | 1.106 | 0 | 0.37 | 0.00013 | 0.322 |
| 35511 | AP001258.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-486-5p | 13 | CBFB | Sponge network | 0.931 | 0 | 1.061 | 0 | 0.322 |
| 35512 | CTC-487M23.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-92a-3p | 15 | ZFHX4 | Sponge network | 0.912 | 0.00014 | 0.018 | 0.95574 | 0.322 |
| 35513 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p | 11 | DNAJB4 | Sponge network | 0.176 | 0.37402 | -0.7 | 1.0E-5 | 0.322 |
| 35514 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 36 | PRKAB2 | Sponge network | 0.997 | 0.00066 | -0.367 | 0.01371 | 0.322 |
| 35515 | SH3RF3-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-877-5p | 15 | GLIPR1 | Sponge network | 0.889 | 9.0E-5 | 0.146 | 0.3276 | 0.322 |
| 35516 | TAPT1-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 11 | PRKAA1 | Sponge network | 0.403 | 2.0E-5 | 0.317 | 0.0031 | 0.322 |
| 35517 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429 | 20 | AFF3 | Sponge network | -0.969 | 2.0E-5 | -2.373 | 0 | 0.322 |
| 35518 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | CREBBP | Sponge network | -1.697 | 0.00889 | 0.126 | 0.15186 | 0.322 |
| 35519 | RP11-182J1.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 16 | TMEM132B | Sponge network | -0.187 | 0.23787 | -0.83 | 0.01084 | 0.322 |
| 35520 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-455-5p | 13 | FGF5 | Sponge network | -0.154 | 0.78939 | 0.549 | 0.28659 | 0.322 |
| 35521 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 12 | GPAM | Sponge network | 0.392 | 0.0278 | 0.142 | 0.29732 | 0.322 |
| 35522 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 41 | BMPR2 | Sponge network | 0.657 | 4.0E-5 | 0.206 | 0.0635 | 0.322 |
| 35523 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-93-5p | 16 | RBL2 | Sponge network | 0.056 | 0.81195 | -0.282 | 0.00254 | 0.322 |
| 35524 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PRKAB2 | Sponge network | -0.053 | 0.80685 | -0.367 | 0.01371 | 0.322 |
| 35525 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 14 | FOXP1 | Sponge network | 1.308 | 0 | 0.042 | 0.74368 | 0.322 |
| 35526 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-493-5p;hsa-miR-532-5p | 15 | FBXO32 | Sponge network | -0.829 | 0.01343 | -0.495 | 0.07561 | 0.322 |
| 35527 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p | 17 | CDHR3 | Sponge network | -1.249 | 7.0E-5 | -0.977 | 4.0E-5 | 0.322 |
| 35528 | AC074289.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-940;hsa-miR-96-5p | 27 | GAS7 | Sponge network | -0.326 | 0.14314 | -0.555 | 0.01602 | 0.322 |
| 35529 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 38 | GAS7 | Sponge network | -1.645 | 0 | -0.555 | 0.01602 | 0.322 |
| 35530 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p | 26 | CRTC1 | Sponge network | -0.942 | 0 | -0.116 | 0.28412 | 0.322 |
| 35531 | RP11-46C24.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 27 | NTRK2 | Sponge network | 0.392 | 0.0278 | -0.736 | 0.06495 | 0.322 |
| 35532 | RP11-102N12.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | TET1 | Sponge network | -0.351 | 0.11358 | -0.135 | 0.52138 | 0.322 |
| 35533 | HCG11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | ZFX | Sponge network | 0.385 | 0.10067 | 0.09 | 0.42563 | 0.322 |
| 35534 | CROCCP2 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-339-5p | 11 | RASAL2 | Sponge network | 0.79 | 0 | 0.869 | 0 | 0.322 |
| 35535 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p | 15 | FOXO3 | Sponge network | 1.093 | 0 | -0.04 | 0.72028 | 0.322 |
| 35536 | RP11-65J21.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-374a-3p;hsa-miR-7-1-3p | 10 | MCC | Sponge network | -0.557 | 0.11135 | -1.005 | 0 | 0.322 |
| 35537 | LINC00957 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 40 | WDFY3 | Sponge network | -0.421 | 0.0114 | -0.201 | 0.0967 | 0.322 |
| 35538 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-511-5p | 21 | ADAMTSL3 | Sponge network | 0.331 | 0.57247 | -1.559 | 0 | 0.322 |
| 35539 | CTD-3138B18.6 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | CCDC6 | Sponge network | 2.248 | 0 | 0.27 | 0.02555 | 0.322 |
| 35540 | SNHG16 |
hsa-let-7a-3p;hsa-miR-154-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 10 | GPAM | Sponge network | 0.961 | 0 | 0.142 | 0.29732 | 0.322 |
| 35541 | RP11-725P16.2 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | KIF5B | Sponge network | 0.422 | 0.27021 | 0.362 | 0.00066 | 0.322 |
| 35542 | CTC-490E21.10 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-493-5p | 11 | SYNM | Sponge network | 1.46 | 1.0E-5 | -2.309 | 1.0E-5 | 0.322 |
| 35543 | RP11-65J21.3 |
hsa-let-7d-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-374a-3p;hsa-miR-532-3p | 15 | GAS7 | Sponge network | -0.557 | 0.11135 | -0.555 | 0.01602 | 0.322 |
| 35544 | RP11-13K12.5 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 34 | CBX5 | Sponge network | -0.318 | 0.65847 | 0.165 | 0.24446 | 0.322 |
| 35545 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 22 | PHC3 | Sponge network | -0.176 | 0.59692 | 0.212 | 0.0499 | 0.322 |
| 35546 | CTD-2336O2.1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-590-3p | 10 | MRVI1 | Sponge network | -0.141 | 0.26434 | -1.051 | 0.00022 | 0.322 |
| 35547 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 54 | IL6ST | Sponge network | 0.385 | 0.10067 | -0.402 | 0.00676 | 0.322 |
| 35548 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-671-5p | 45 | ADARB1 | Sponge network | -1.249 | 7.0E-5 | -0.335 | 0.06631 | 0.322 |
| 35549 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-582-5p | 17 | MOB1B | Sponge network | 0.792 | 0.0049 | 0.197 | 0.15266 | 0.322 |
| 35550 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-7-1-3p | 28 | ATRX | Sponge network | 0.177 | 0.16681 | 0.261 | 0.04408 | 0.322 |
| 35551 | CTC-296K1.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-625-5p | 13 | CSMD1 | Sponge network | -2.095 | 0.00112 | -0.63 | 0.18881 | 0.322 |
| 35552 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-532-5p;hsa-miR-576-5p | 17 | NUAK1 | Sponge network | -0.739 | 0.00044 | 0.904 | 0 | 0.322 |
| 35553 | RP11-166D19.1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | LIMCH1 | Sponge network | -0.576 | 0.10121 | -0.915 | 1.0E-5 | 0.322 |
| 35554 | LINC00963 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-96-5p | 25 | LRIG1 | Sponge network | 0.523 | 9.0E-5 | -0.537 | 0.00588 | 0.322 |
| 35555 | LINC00173 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p | 12 | CADM3 | Sponge network | 0.056 | 0.8494 | -3.663 | 0 | 0.322 |
| 35556 | AF131215.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 22 | SNX29 | Sponge network | -0.176 | 0.59692 | -0.34 | 0.01371 | 0.322 |
| 35557 | RP11-798G7.7 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p | 10 | RASAL2 | Sponge network | 0.858 | 3.0E-5 | 0.869 | 0 | 0.322 |
| 35558 | RP11-216B9.6 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-590-3p;hsa-miR-93-3p | 14 | DOCK4 | Sponge network | 0.174 | 0.30034 | 0.502 | 0.00108 | 0.322 |
| 35559 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p | 11 | CHD2 | Sponge network | 1.147 | 0 | 0.049 | 0.55371 | 0.322 |
| 35560 | LIPE-AS1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p | 11 | RASAL2 | Sponge network | 0.078 | 0.55803 | 0.869 | 0 | 0.322 |
| 35561 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 14 | FOXP1 | Sponge network | 1.2 | 0.01813 | 0.042 | 0.74368 | 0.322 |
| 35562 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-96-5p | 11 | SETBP1 | Sponge network | 0.453 | 0.01839 | -1.302 | 1.0E-5 | 0.322 |
| 35563 | KB-1410C5.5 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 16 | IL6ST | Sponge network | 0.959 | 0 | -0.402 | 0.00676 | 0.322 |
| 35564 | RP5-1061H20.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 11 | CBFB | Sponge network | 1.921 | 0 | 1.061 | 0 | 0.322 |
| 35565 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | PREX2 | Sponge network | -0.899 | 6.0E-5 | -0.272 | 0.25382 | 0.322 |
| 35566 | RP4-669P10.16 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1271-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-93-3p | 10 | EPHA3 | Sponge network | -0.301 | 0.06597 | 0.185 | 0.53588 | 0.322 |
| 35567 | CTD-2291D10.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-96-5p | 10 | PRRX1 | Sponge network | 3.21 | 0 | 1.316 | 0 | 0.322 |
| 35568 | LINC00863 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | PPM1L | Sponge network | 0.189 | 0.13981 | -0.537 | 0.00616 | 0.322 |
| 35569 | CTD-2528L19.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | DLG1 | Sponge network | 0.953 | 0 | -0.014 | 0.8926 | 0.322 |
| 35570 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | DMTF1 | Sponge network | -1.575 | 6.0E-5 | 0.248 | 0.02726 | 0.322 |
| 35571 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | KDSR | Sponge network | 0.603 | 0.00127 | 0.239 | 0.02216 | 0.322 |
| 35572 | RP11-158K1.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PRUNE2 | Sponge network | 0.349 | 0.01695 | -1.733 | 0.00022 | 0.322 |
| 35573 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 36 | AKAP11 | Sponge network | -0.709 | 0.18168 | 0.196 | 0.09825 | 0.322 |
| 35574 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 17 | ZFHX3 | Sponge network | 1.153 | 0.00114 | 0.389 | 0.01363 | 0.322 |
| 35575 | RP11-33B1.4 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p | 10 | CBX5 | Sponge network | 0.826 | 0 | 0.165 | 0.24446 | 0.322 |
| 35576 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 27 | USP12 | Sponge network | -0.487 | 0.02157 | -0.102 | 0.40768 | 0.322 |
| 35577 | C4A-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-744-3p;hsa-miR-744-5p | 10 | IGF2 | Sponge network | 3.345 | 0 | 0.848 | 0.03842 | 0.322 |
| 35578 | RP1-302G2.5 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-29b-1-5p;hsa-miR-935 | 10 | OPCML | Sponge network | -1.483 | 9.0E-5 | -2.498 | 0 | 0.322 |
| 35579 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 46 | ZFHX4 | Sponge network | -0.615 | 0.00015 | 0.018 | 0.95574 | 0.322 |
| 35580 | RP11-35G9.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PIK3CA | Sponge network | 0.657 | 4.0E-5 | 0.195 | 0.06908 | 0.322 |
| 35581 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-590-3p | 12 | EDA2R | Sponge network | -1.057 | 0.00029 | -0.587 | 0.01598 | 0.322 |
| 35582 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ADAMTSL3 | Sponge network | -0.235 | 0.15142 | -1.559 | 0 | 0.322 |
| 35583 | LINC00957 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 14 | RIMS1 | Sponge network | -0.421 | 0.0114 | -3.143 | 0 | 0.322 |
| 35584 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | LRP1B | Sponge network | 0.572 | 0.01722 | -2.163 | 0 | 0.322 |
| 35585 | HAND2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 40 | PTPRT | Sponge network | -1.697 | 0.00889 | -0.907 | 0.03727 | 0.322 |
| 35586 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | CBL | Sponge network | 0.856 | 0 | 0.291 | 0.01267 | 0.322 |
| 35587 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-493-5p;hsa-miR-576-5p | 16 | FREM2 | Sponge network | -2.62 | 0 | -0.437 | 0.46353 | 0.322 |
| 35588 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-33a-3p | 12 | SYNE1 | Sponge network | 0.542 | 0.00054 | -0.738 | 0.00316 | 0.322 |
| 35589 | CTD-2666L21.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p | 13 | PTGFR | Sponge network | 0.368 | 0.20093 | -0.233 | 0.45421 | 0.322 |
| 35590 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940 | 31 | HLF | Sponge network | 0.219 | 0.1516 | -2.443 | 0 | 0.322 |
| 35591 | RP11-305E6.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | TCF12 | Sponge network | 1.317 | 0 | 0.174 | 0.13271 | 0.322 |
| 35592 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p | 10 | PIK3C2A | Sponge network | 0.453 | 0.01839 | 0.061 | 0.53776 | 0.322 |
| 35593 | AC025171.1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | CHD2 | Sponge network | 0.998 | 0 | 0.049 | 0.55371 | 0.322 |
| 35594 | DNM1P35 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | PRUNE2 | Sponge network | 0.051 | 0.83388 | -1.733 | 0.00022 | 0.322 |
| 35595 | RP4-669L17.10 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 13 | UBR3 | Sponge network | 1.093 | 0 | -0.021 | 0.82774 | 0.322 |
| 35596 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-532-3p | 14 | RPS6KA2 | Sponge network | -0.899 | 6.0E-5 | -0.57 | 0.00013 | 0.322 |
| 35597 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | FOXO1 | Sponge network | -0.222 | 0.32445 | -0.141 | 0.33874 | 0.322 |
| 35598 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-590-3p | 22 | RBMS3 | Sponge network | 0.274 | 0.07681 | -0.519 | 0.03551 | 0.322 |
| 35599 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | SON | Sponge network | 0.79 | 0 | 0.168 | 0.04406 | 0.322 |
| 35600 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | FREM2 | Sponge network | -0.739 | 0.00044 | -0.437 | 0.46353 | 0.322 |
| 35601 | TRAF3IP2-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-550a-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PCDH18 | Sponge network | -0.323 | 0.01372 | 0.216 | 0.24645 | 0.322 |
| 35602 | CALML3-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-625-5p | 10 | NGFR | Sponge network | 0.95 | 0.15943 | -1.271 | 0.01058 | 0.322 |
| 35603 | RP11-110I1.11 |
hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p | 17 | LPP | Sponge network | 1.034 | 0 | -0.459 | 0.04475 | 0.322 |
| 35604 | LINC00963 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 19 | NEDD4 | Sponge network | 0.523 | 9.0E-5 | 0.02 | 0.89834 | 0.322 |
| 35605 | RP11-728F11.4 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p | 15 | HIP1 | Sponge network | 0.238 | 0.70544 | 0.774 | 0 | 0.322 |
| 35606 | RP11-750H9.5 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-224-5p;hsa-miR-3065-3p;hsa-miR-877-5p | 11 | AFF1 | Sponge network | 0.349 | 0.13152 | -0.384 | 0.00053 | 0.322 |
| 35607 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 15 | FREM2 | Sponge network | 0.051 | 0.83388 | -0.437 | 0.46353 | 0.322 |
| 35608 | RP11-968O1.5 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p | 11 | RASSF8 | Sponge network | -0.829 | 0.01343 | -0.122 | 0.65591 | 0.322 |
| 35609 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p | 12 | TSHZ3 | Sponge network | 0.912 | 0.00014 | -0.556 | 0.01516 | 0.322 |
| 35610 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-93-5p | 13 | MCC | Sponge network | 0.348 | 0.32912 | -1.005 | 0 | 0.322 |
| 35611 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-214-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 28 | LRP1B | Sponge network | -1.047 | 0 | -2.163 | 0 | 0.322 |
| 35612 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940 | 44 | MEF2C | Sponge network | -0.405 | 0.22013 | -0.499 | 0.01448 | 0.322 |
| 35613 | HAR1A |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-550a-5p | 12 | TSHZ3 | Sponge network | -0.672 | 0.19447 | -0.556 | 0.01516 | 0.322 |
| 35614 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-429 | 16 | LRIG1 | Sponge network | -0.154 | 0.53954 | -0.537 | 0.00588 | 0.322 |
| 35615 | RP11-244H3.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | 0.659 | 3.0E-5 | -0.001 | 0.99669 | 0.322 |
| 35616 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | SSBP2 | Sponge network | -0.053 | 0.80685 | -1.58 | 0 | 0.322 |
| 35617 | PWAR6 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-429 | 10 | DUSP1 | Sponge network | -1.575 | 6.0E-5 | -1.754 | 0 | 0.322 |
| 35618 | RP11-707P17.2 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | ARHGEF12 | Sponge network | 1.877 | 0 | 0.09 | 0.35693 | 0.322 |
| 35619 | LIPE-AS1 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p | 15 | ERCC4 | Sponge network | 0.078 | 0.55803 | 0.126 | 0.15266 | 0.322 |
| 35620 | LINC00092 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | HOMER2 | Sponge network | -1.886 | 0 | -2.698 | 0 | 0.322 |
| 35621 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 29 | MEF2C | Sponge network | -0.322 | 0.25945 | -0.499 | 0.01448 | 0.322 |
| 35622 | CTD-2002H8.2 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | RBMS3 | Sponge network | 0.931 | 1.0E-5 | -0.519 | 0.03551 | 0.322 |
| 35623 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 27 | BHLHE41 | Sponge network | -0.777 | 0.16667 | 0.353 | 0.12753 | 0.322 |
| 35624 | TPTEP1 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 11 | DSE | Sponge network | -1.216 | 0 | -0.231 | 0.05347 | 0.322 |
| 35625 | CTD-2666L21.1 |
hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p | 16 | MED12L | Sponge network | 0.368 | 0.20093 | -0.194 | 0.55299 | 0.322 |
| 35626 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 13 | RNF144A | Sponge network | -0.141 | 0.26434 | 0.594 | 0.0009 | 0.322 |
| 35627 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-451a;hsa-miR-7-5p | 33 | BCL2 | Sponge network | -0.777 | 0.16667 | -0.899 | 0 | 0.322 |
| 35628 | LINC00886 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-5p | 12 | RIT1 | Sponge network | -0.371 | 0.24604 | -0.296 | 0.00054 | 0.322 |
| 35629 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | NEDD4 | Sponge network | 0.177 | 0.16681 | 0.02 | 0.89834 | 0.322 |
| 35630 | RP11-218M22.1 |
hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-96-5p | 18 | IL17RD | Sponge network | 0.197 | 0.25618 | -0.019 | 0.94873 | 0.322 |
| 35631 | RP11-226L15.5 |
hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-374b-5p | 10 | CDK6 | Sponge network | 0.8 | 0 | 0.991 | 0 | 0.322 |
| 35632 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | GAS7 | Sponge network | 0.131 | 0.8436 | -0.555 | 0.01602 | 0.322 |
| 35633 | RP11-532F6.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-505-3p | 16 | KCNJ5 | Sponge network | -0.896 | 0.00099 | -0.281 | 0.3339 | 0.322 |
| 35634 | PWAR6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 48 | GNAQ | Sponge network | -1.575 | 6.0E-5 | -0.367 | 0.00102 | 0.322 |
| 35635 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-93-3p | 13 | WWTR1 | Sponge network | 1.46 | 1.0E-5 | -0.345 | 0.11997 | 0.322 |
| 35636 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 13 | TRIM3 | Sponge network | -0.355 | 0.0077 | -0.577 | 0 | 0.322 |
| 35637 | LINC00476 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935 | 29 | ACSL6 | Sponge network | -0.615 | 0.00015 | -1.593 | 0 | 0.322 |
| 35638 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 21 | PRKCE | Sponge network | 0.4 | 0.14611 | -0.403 | 0.00056 | 0.322 |
| 35639 | MALAT1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 22 | RPS6KA3 | Sponge network | 0.782 | 0.00016 | 0.256 | 0.02419 | 0.322 |
| 35640 | SH3BP5-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | PIK3CA | Sponge network | 0.267 | 0.2937 | 0.195 | 0.06908 | 0.322 |
| 35641 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 35 | THRB | Sponge network | -4.477 | 0 | -1.117 | 0.0004 | 0.322 |
| 35642 | RP11-439A17.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-20a-3p;hsa-miR-323a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-98-5p | 12 | CADM2 | Sponge network | 0.051 | 0.95756 | -3.782 | 0 | 0.322 |
| 35643 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 24 | SNX29 | Sponge network | 0.176 | 0.37402 | -0.34 | 0.01371 | 0.322 |
| 35644 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-539-5p | 15 | MOB1B | Sponge network | 0.924 | 0 | 0.197 | 0.15266 | 0.322 |
| 35645 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 21 | CHD2 | Sponge network | 0.421 | 0.00084 | 0.049 | 0.55371 | 0.322 |
| 35646 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 10 | DACT1 | Sponge network | 0.174 | 0.30034 | 0.783 | 0.00072 | 0.322 |
| 35647 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 22 | TP53INP1 | Sponge network | -2.452 | 0 | -0.496 | 0.00064 | 0.322 |
| 35648 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | FSTL5 | Sponge network | 0.219 | 0.1516 | -1.3 | 0.01619 | 0.322 |
| 35649 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 35 | QKI | Sponge network | -0.125 | 0.49414 | -0.333 | 0.04111 | 0.322 |
| 35650 | RP11-536O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p | 14 | ZBTB4 | Sponge network | -0.074 | 0.90758 | -0.739 | 0 | 0.322 |
| 35651 | LINC00863 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-200b-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | MYO9A | Sponge network | 0.189 | 0.13981 | -0.147 | 0.32373 | 0.322 |
| 35652 | LINC00865 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-582-5p | 17 | STARD13 | Sponge network | -1.249 | 7.0E-5 | -0.187 | 0.27383 | 0.322 |
| 35653 | CTC-429P9.1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-29a-5p | 14 | SPRY3 | Sponge network | 0.497 | 0.01451 | 0.016 | 0.91286 | 0.322 |
| 35654 | RP5-994D16.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | LPP | Sponge network | 0.252 | 0.08508 | -0.459 | 0.04475 | 0.322 |
| 35655 | RP11-439A17.10 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-5p | 16 | IL6ST | Sponge network | 0.051 | 0.95756 | -0.402 | 0.00676 | 0.322 |
| 35656 | AP001055.6 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-7-1-3p | 10 | ABI2 | Sponge network | 0.693 | 0.00076 | 0.175 | 0.14787 | 0.322 |
| 35657 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p | 14 | IL17RD | Sponge network | 0.919 | 0 | -0.019 | 0.94873 | 0.322 |
| 35658 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p | 13 | PTPN11 | Sponge network | 1.044 | 2.0E-5 | 0.431 | 2.0E-5 | 0.322 |
| 35659 | RP11-152F13.8 |
hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | CREB1 | Sponge network | 0.843 | 0.01775 | 0.203 | 0.00537 | 0.322 |
| 35660 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 20 | MYBL1 | Sponge network | 0.222 | 0.29133 | 0.494 | 0.02152 | 0.322 |
| 35661 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-429 | 24 | SOX11 | Sponge network | 0.75 | 0.00054 | 2.68 | 0 | 0.322 |
| 35662 | CTC-429P9.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-33a-5p | 10 | CNTN1 | Sponge network | 0.497 | 0.01451 | -2.594 | 0 | 0.322 |
| 35663 | OIP5-AS1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 14 | GSK3B | Sponge network | 0.421 | 0.00084 | 0.266 | 0.00045 | 0.322 |
| 35664 | RP11-166D19.1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 10 | PKD1L2 | Sponge network | -0.576 | 0.10121 | -0.971 | 0.00438 | 0.322 |
| 35665 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 38 | TMEM132B | Sponge network | -1.161 | 0.00591 | -0.83 | 0.01084 | 0.322 |
| 35666 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p | 11 | TSC22D1 | Sponge network | -0.576 | 0.10121 | -0.529 | 2.0E-5 | 0.322 |
| 35667 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 16 | ZMYND11 | Sponge network | 0.41 | 0.02214 | -0.2 | 0.05449 | 0.322 |
| 35668 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | FREM2 | Sponge network | -2.915 | 0.00015 | -0.437 | 0.46353 | 0.322 |
| 35669 | RP11-440L14.4 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-30e-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | NFIB | Sponge network | 0.545 | 0.05789 | 0.023 | 0.90795 | 0.322 |
| 35670 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p | 36 | ADARB1 | Sponge network | 0.523 | 9.0E-5 | -0.335 | 0.06631 | 0.322 |
| 35671 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PBRM1 | Sponge network | 0.349 | 0.01695 | -0.029 | 0.78601 | 0.322 |
| 35672 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p | 11 | ZFHX3 | Sponge network | 0.062 | 0.55274 | 0.389 | 0.01363 | 0.322 |
| 35673 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | CACNA2D1 | Sponge network | 0.199 | 0.38015 | -0.846 | 0.00675 | 0.322 |
| 35674 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-338-3p | 12 | ELMO1 | Sponge network | -0.154 | 0.78939 | 0.04 | 0.83147 | 0.322 |
| 35675 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-34a-5p | 10 | TXNIP | Sponge network | -0.342 | 0.02288 | -1.277 | 0 | 0.322 |
| 35676 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429 | 10 | SCN5A | Sponge network | -0.49 | 0.04946 | -0.259 | 0.48224 | 0.322 |
| 35677 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-374b-5p | 11 | SOX11 | Sponge network | 1.46 | 1.0E-5 | 2.68 | 0 | 0.322 |
| 35678 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 14 | TXNIP | Sponge network | 0.249 | 0.35902 | -1.277 | 0 | 0.322 |
| 35679 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | DACH2 | Sponge network | -2.46 | 0 | -1.635 | 0.00034 | 0.322 |
| 35680 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 39 | PPM1A | Sponge network | -0.405 | 0.22013 | -0.623 | 0 | 0.322 |
| 35681 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 21 | PDE4DIP | Sponge network | 0.614 | 1.0E-5 | -0.282 | 0.0257 | 0.322 |
| 35682 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | MYO9A | Sponge network | 0.953 | 0 | -0.147 | 0.32373 | 0.322 |
| 35683 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-301a-3p | 10 | PGR | Sponge network | 0.238 | 0.70544 | -1.704 | 0 | 0.322 |
| 35684 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 29 | DDX6 | Sponge network | -0.176 | 0.59692 | 0.091 | 0.36428 | 0.322 |
| 35685 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-7-1-3p | 11 | MITF | Sponge network | -0.72 | 0.24239 | -0.769 | 0.00022 | 0.322 |
| 35686 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-5p | 23 | ESR1 | Sponge network | 0.176 | 0.37402 | -1.035 | 3.0E-5 | 0.322 |
| 35687 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | PCDH8 | Sponge network | -0.709 | 0.18168 | -0.997 | 0.05366 | 0.322 |
| 35688 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577 | 17 | SLC6A15 | Sponge network | 0.056 | 0.81195 | -2.373 | 2.0E-5 | 0.322 |
| 35689 | RP11-999E24.3 |
hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-7-5p | 14 | CRTC1 | Sponge network | -0.693 | 0.05629 | -0.116 | 0.28412 | 0.322 |
| 35690 | RP11-47A8.5 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-15b-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-423-5p;hsa-miR-501-5p;hsa-miR-625-5p;hsa-miR-92a-1-5p | 10 | CTIF | Sponge network | 0.103 | 0.43169 | -0.389 | 0.00549 | 0.322 |
| 35691 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | TSC1 | Sponge network | 0.101 | 0.60919 | 0.103 | 0.26071 | 0.322 |
| 35692 | RP1-302G2.5 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-935 | 16 | FAT3 | Sponge network | -1.483 | 9.0E-5 | -1.601 | 0.00051 | 0.322 |
| 35693 | LINC00473 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p | 15 | IRS1 | Sponge network | -1.203 | 0.03881 | -0.026 | 0.88855 | 0.322 |
| 35694 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p | 18 | FGFR1 | Sponge network | 0.392 | 0.0278 | -0.509 | 0.03957 | 0.322 |
| 35695 | KB-1460A1.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PRUNE2 | Sponge network | 0.603 | 0.00127 | -1.733 | 0.00022 | 0.322 |
| 35696 | RP11-1006G14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-582-3p;hsa-miR-590-3p | 16 | PRDM2 | Sponge network | 0.559 | 0.00026 | -0.258 | 0.00721 | 0.322 |
| 35697 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | MYO9A | Sponge network | 1.308 | 0 | -0.147 | 0.32373 | 0.322 |
| 35698 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | ITCH | Sponge network | 1.205 | 0 | 0.084 | 0.34058 | 0.322 |
| 35699 | RP11-244O19.1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 10 | SH3GLB1 | Sponge network | -0.096 | 0.67958 | -0.115 | 0.14773 | 0.321 |
| 35700 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | CDHR3 | Sponge network | 0.051 | 0.83388 | -0.977 | 4.0E-5 | 0.321 |
| 35701 | FZD10-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | FLNA | Sponge network | -0.727 | 0.04726 | -0.771 | 0.01377 | 0.321 |
| 35702 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | NR4A3 | Sponge network | -0.323 | 0.01372 | -1.385 | 0 | 0.321 |
| 35703 | KB-1460A1.5 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RBBP6 | Sponge network | 0.603 | 0.00127 | 0.163 | 0.05101 | 0.321 |
| 35704 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-30d-5p;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 29 | ERBB4 | Sponge network | -0.739 | 0.00044 | -1.975 | 2.0E-5 | 0.321 |
| 35705 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | BMPR2 | Sponge network | 0.67 | 0.00048 | 0.206 | 0.0635 | 0.321 |
| 35706 | RP11-356J5.12 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 24 | NRK | Sponge network | -0.899 | 6.0E-5 | 0.656 | 0.19217 | 0.321 |
| 35707 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-508-3p;hsa-miR-589-3p | 26 | SCN9A | Sponge network | 0.253 | 0.09565 | -0.525 | 0.10593 | 0.321 |
| 35708 | RP11-6O2.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-505-5p;hsa-miR-940 | 20 | TOM1L2 | Sponge network | 0.331 | 0.57247 | -0.994 | 0 | 0.321 |
| 35709 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 26 | PDGFRA | Sponge network | 0.176 | 0.37402 | -0.372 | 0.0037 | 0.321 |
| 35710 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 21 | PHC3 | Sponge network | 0.419 | 0.0319 | 0.212 | 0.0499 | 0.321 |
| 35711 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 25 | MYBL1 | Sponge network | 0.064 | 0.72934 | 0.494 | 0.02152 | 0.321 |
| 35712 | LINC00654 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p | 10 | UVRAG | Sponge network | 0.374 | 0.20054 | -0.017 | 0.83865 | 0.321 |
| 35713 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | MAF | Sponge network | -0.709 | 0.18168 | -1.257 | 0 | 0.321 |
| 35714 | CTA-14H9.5 |
hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-7-1-3p | 10 | SYNE1 | Sponge network | 0.145 | 0.53343 | -0.738 | 0.00316 | 0.321 |
| 35715 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | PBRM1 | Sponge network | 0.222 | 0.29133 | -0.029 | 0.78601 | 0.321 |
| 35716 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-338-3p | 11 | HDAC9 | Sponge network | -0.777 | 0.16667 | -0.755 | 0.00495 | 0.321 |
| 35717 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 17 | TP53INP1 | Sponge network | -0.322 | 0.25945 | -0.496 | 0.00064 | 0.321 |
| 35718 | RP11-53O19.1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-576-5p | 18 | ATM | Sponge network | -0.405 | 0.22013 | 0.178 | 0.21568 | 0.321 |
| 35719 | FZD10-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-1-5p | 29 | CTIF | Sponge network | -0.727 | 0.04726 | -0.389 | 0.00549 | 0.321 |
| 35720 | AC005618.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 20 | SOX11 | Sponge network | 0.862 | 0.06213 | 2.68 | 0 | 0.321 |
| 35721 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p | 16 | PRDM2 | Sponge network | -2.531 | 0 | -0.258 | 0.00721 | 0.321 |
| 35722 | RP4-669P10.16 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-93-3p | 11 | SUFU | Sponge network | -0.301 | 0.06597 | -0.04 | 0.65489 | 0.321 |
| 35723 | GS1-124K5.11 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 25 | DLC1 | Sponge network | 0.494 | 0.00339 | -0.382 | 0.05038 | 0.321 |
| 35724 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577 | 12 | AMPH | Sponge network | 0.101 | 0.60919 | -0.916 | 5.0E-5 | 0.321 |
| 35725 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 19 | ANK2 | Sponge network | -0.141 | 0.26434 | -1.603 | 1.0E-5 | 0.321 |
| 35726 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 20 | EBF1 | Sponge network | -0.322 | 0.25945 | -0.384 | 0.08856 | 0.321 |
| 35727 | CTD-2089N3.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-590-3p | 12 | EYA4 | Sponge network | -1.375 | 0.08642 | 0.3 | 0.36876 | 0.321 |
| 35728 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 19 | RICTOR | Sponge network | 1.352 | 0 | 0.388 | 0.00188 | 0.321 |
| 35729 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 21 | MYBL1 | Sponge network | 0.488 | 0.00084 | 0.494 | 0.02152 | 0.321 |
| 35730 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-93-5p | 26 | GAS7 | Sponge network | -0.726 | 0.00132 | -0.555 | 0.01602 | 0.321 |
| 35731 | AP001347.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 15 | KANK1 | Sponge network | -1.161 | 0.00591 | -0.55 | 0.00134 | 0.321 |
| 35732 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-424-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | NRXN1 | Sponge network | 0.225 | 0.30644 | -3.778 | 0 | 0.321 |
| 35733 | RP11-196G18.22 |
hsa-miR-101-3p;hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 12 | UBR3 | Sponge network | 1.206 | 0 | -0.021 | 0.82774 | 0.321 |
| 35734 | MIR143HG |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 23 | CELF4 | Sponge network | -0.739 | 0.0574 | -1.135 | 0.00344 | 0.321 |
| 35735 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p | 12 | FOXO3 | Sponge network | 0.917 | 0 | -0.04 | 0.72028 | 0.321 |
| 35736 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-539-5p | 20 | EPHA7 | Sponge network | 0.37 | 0.27614 | -2.197 | 3.0E-5 | 0.321 |
| 35737 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-93-5p;hsa-miR-96-5p | 10 | DAB2 | Sponge network | -0.351 | 0.11358 | 0.431 | 0.0113 | 0.321 |
| 35738 | RP11-144G6.12 |
hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-590-3p | 10 | TMEM30A | Sponge network | 0.826 | 1.0E-5 | 0.096 | 0.31031 | 0.321 |
| 35739 | PCA3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 15 | PKNOX1 | Sponge network | -0.709 | 0.18168 | 0.085 | 0.28917 | 0.321 |
| 35740 | RP1-151F17.2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | KLF10 | Sponge network | 0.222 | 0.29133 | -0.783 | 0 | 0.321 |
| 35741 | RP11-244O19.1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-629-5p | 15 | WNK2 | Sponge network | -0.096 | 0.67958 | -0.352 | 0.31066 | 0.321 |
| 35742 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-660-5p | 16 | CXXC4 | Sponge network | 0.197 | 0.25618 | -0.433 | 0.21055 | 0.321 |
| 35743 | LPP-AS2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-577 | 11 | AKAP12 | Sponge network | -0.18 | 0.20343 | -0.932 | 0.0028 | 0.321 |
| 35744 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-429 | 13 | ARNT2 | Sponge network | -0.154 | 0.53954 | -0.332 | 0.25851 | 0.321 |
| 35745 | AC006547.13 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-324-3p;hsa-miR-340-3p;hsa-miR-455-3p;hsa-miR-654-3p;hsa-miR-940 | 12 | NFIB | Sponge network | 1.159 | 0 | 0.023 | 0.90795 | 0.321 |
| 35746 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-214-5p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-93-5p;hsa-miR-935 | 60 | SMAD4 | Sponge network | -0.615 | 0.00015 | -0.313 | 0.00413 | 0.321 |
| 35747 | RP11-13K12.1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p | 14 | ID4 | Sponge network | -0.154 | 0.78939 | -1.644 | 0 | 0.321 |
| 35748 | CALML3-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-375;hsa-miR-429;hsa-miR-542-3p | 12 | ITGB1 | Sponge network | 0.95 | 0.15943 | 0.524 | 0.00017 | 0.321 |
| 35749 | RP11-48B3.4 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 10 | FMNL3 | Sponge network | 1.757 | 0 | 0.555 | 4.0E-5 | 0.321 |
| 35750 | PSMD5-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-485-3p;hsa-miR-590-3p | 18 | SIRT1 | Sponge network | 0.569 | 0.00067 | 0.109 | 0.2593 | 0.321 |
| 35751 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 30 | NCOA1 | Sponge network | 0.72 | 0.0001 | -0.432 | 0 | 0.321 |
| 35752 | RP11-212P7.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p | 14 | KANK1 | Sponge network | 0.219 | 0.1516 | -0.55 | 0.00134 | 0.321 |
| 35753 | RP11-158K1.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 20 | BAZ2B | Sponge network | 0.349 | 0.01695 | -0.276 | 0.02833 | 0.321 |
| 35754 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | ARHGAP29 | Sponge network | -0.793 | 7.0E-5 | 0.131 | 0.42953 | 0.321 |
| 35755 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p | 38 | RUNX1T1 | Sponge network | -0.031 | 0.89063 | -0.344 | 0.2374 | 0.321 |
| 35756 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-940 | 26 | HLF | Sponge network | 0.75 | 0.00054 | -2.443 | 0 | 0.321 |
| 35757 | OIP5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p | 86 | LPP | Sponge network | 0.421 | 0.00084 | -0.459 | 0.04475 | 0.321 |
| 35758 | HOXA-AS2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | LRIG1 | Sponge network | 0.61 | 0.01593 | -0.537 | 0.00588 | 0.321 |
| 35759 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | ANK3 | Sponge network | 0.222 | 0.29133 | -0.55 | 0.00086 | 0.321 |
| 35760 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | TSC1 | Sponge network | 0.082 | 0.55654 | 0.103 | 0.26071 | 0.321 |
| 35761 | TUG1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 18 | ZFX | Sponge network | 0.972 | 0 | 0.09 | 0.42563 | 0.321 |
| 35762 | LINC00622 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-200b-5p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MOB1B | Sponge network | 1.169 | 0.00028 | 0.197 | 0.15266 | 0.321 |
| 35763 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | SPG20 | Sponge network | 0.174 | 0.30034 | -1.16 | 0 | 0.321 |
| 35764 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | 0.619 | 0.00157 | -1.047 | 0.00364 | 0.321 |
| 35765 | LINC01018 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 10 | PREX2 | Sponge network | -2.915 | 0.00015 | -0.272 | 0.25382 | 0.321 |
| 35766 | C4B-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-744-3p | 13 | DIP2C | Sponge network | 2.306 | 9.0E-5 | -0.436 | 0.01713 | 0.321 |
| 35767 | RP11-434B12.1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 14 | DYNC1I1 | Sponge network | -0.031 | 0.89063 | -1.12 | 0.00107 | 0.321 |
| 35768 | CTC-457L16.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-93-5p | 18 | IL17RD | Sponge network | 1.153 | 0.00114 | -0.019 | 0.94873 | 0.321 |
| 35769 | CTD-3025N20.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-654-3p | 11 | AKAP11 | Sponge network | 1.046 | 0 | 0.196 | 0.09825 | 0.321 |
| 35770 | RP11-244H3.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 22 | RASSF8 | Sponge network | 0.659 | 3.0E-5 | -0.122 | 0.65591 | 0.321 |
| 35771 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | GPC6 | Sponge network | -0.738 | 0.21233 | -0.054 | 0.81465 | 0.321 |
| 35772 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-577;hsa-miR-7-1-3p | 16 | FGF5 | Sponge network | -1.349 | 0.00017 | 0.549 | 0.28659 | 0.321 |
| 35773 | LINC00173 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-454-3p | 14 | MAFB | Sponge network | 0.056 | 0.8494 | -0.023 | 0.89865 | 0.321 |
| 35774 | NDUFA6-AS1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 13 | PHOX2B | Sponge network | -0.355 | 0.0077 | -2.68 | 0 | 0.321 |
| 35775 | RP11-894P9.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-532-3p;hsa-miR-550a-5p | 17 | DST | Sponge network | 0.993 | 0 | -0.713 | 0.00096 | 0.321 |
| 35776 | MALAT1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-379-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SH3GLB1 | Sponge network | 0.782 | 0.00016 | -0.115 | 0.14773 | 0.321 |
| 35777 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.559 | 0.00026 | 0.323 | 0.00792 | 0.321 |
| 35778 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p | 14 | LRRC4 | Sponge network | -0.612 | 0.00094 | -1.774 | 0 | 0.321 |
| 35779 | HAND2-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 23 | ERRFI1 | Sponge network | -1.697 | 0.00889 | -0.798 | 1.0E-5 | 0.321 |
| 35780 | RP11-597D13.9 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RET | Sponge network | 1.302 | 7.0E-5 | -0.833 | 0.03517 | 0.321 |
| 35781 | RP11-155G14.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-326 | 11 | RICTOR | Sponge network | 2.483 | 0 | 0.388 | 0.00188 | 0.321 |
| 35782 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 30 | ST6GALNAC3 | Sponge network | 0.572 | 0.01722 | -0.987 | 0 | 0.321 |
| 35783 | RP1-151F17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p | 13 | GAS7 | Sponge network | -0.321 | 0.22761 | -0.555 | 0.01602 | 0.321 |
| 35784 | LINC00654 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-7-5p | 10 | COL1A2 | Sponge network | 0.374 | 0.20054 | 1.991 | 0 | 0.321 |
| 35785 | RP11-116O18.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | MED12L | Sponge network | 0.05 | 0.95483 | -0.194 | 0.55299 | 0.321 |
| 35786 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-5p | 19 | ESR1 | Sponge network | -0.693 | 0.05629 | -1.035 | 3.0E-5 | 0.321 |
| 35787 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | SON | Sponge network | 1.262 | 0 | 0.168 | 0.04406 | 0.321 |
| 35788 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-590-3p | 25 | ZFX | Sponge network | 1.089 | 0 | 0.09 | 0.42563 | 0.321 |
| 35789 | ZSCAN16-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p | 18 | CBFA2T3 | Sponge network | -0.342 | 0.02288 | -1.221 | 1.0E-5 | 0.321 |
| 35790 | RP11-503E24.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | PRKAA1 | Sponge network | 2.218 | 0 | 0.317 | 0.0031 | 0.321 |
| 35791 | PART1 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | RIMS2 | Sponge network | -2.925 | 0 | -1.365 | 0.00208 | 0.321 |
| 35792 | LINC00473 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | GABRB2 | Sponge network | -1.203 | 0.03881 | -1.841 | 0.00029 | 0.321 |
| 35793 | FAM157A |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 12 | DDX6 | Sponge network | 0.813 | 1.0E-5 | 0.091 | 0.36428 | 0.321 |
| 35794 | RP11-473I1.9 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-484 | 16 | ABL1 | Sponge network | 0.683 | 1.0E-5 | -0.215 | 0.07804 | 0.321 |
| 35795 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p | 15 | IL17RD | Sponge network | 1.04 | 0.00023 | -0.019 | 0.94873 | 0.321 |
| 35796 | BVES-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-429 | 15 | KANK1 | Sponge network | -2.62 | 0 | -0.55 | 0.00134 | 0.321 |
| 35797 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-532-5p | 10 | LRRC4 | Sponge network | -0.154 | 0.78939 | -1.774 | 0 | 0.321 |
| 35798 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p | 13 | PDS5B | Sponge network | 1.044 | 2.0E-5 | 0.259 | 0.00962 | 0.321 |
| 35799 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | TRIM3 | Sponge network | -1.349 | 0.00017 | -0.577 | 0 | 0.321 |
| 35800 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | NEDD4 | Sponge network | 0.419 | 0.0319 | 0.02 | 0.89834 | 0.321 |
| 35801 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 55 | WDFY3 | Sponge network | -0.355 | 0.0077 | -0.201 | 0.0967 | 0.321 |
| 35802 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | HDAC9 | Sponge network | -0.563 | 0.02545 | -0.755 | 0.00495 | 0.321 |
| 35803 | LINC00173 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | MED12L | Sponge network | 0.056 | 0.8494 | -0.194 | 0.55299 | 0.321 |
| 35804 | LINC00641 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 14 | SPTBN4 | Sponge network | -0.222 | 0.32445 | -0.786 | 0.01281 | 0.321 |
| 35805 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-935 | 17 | RARB | Sponge network | -0.246 | 0.35034 | -0.825 | 2.0E-5 | 0.321 |
| 35806 | RP11-411K7.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 52 | DST | Sponge network | 0.155 | 0.81273 | -0.713 | 0.00096 | 0.321 |
| 35807 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | ABI2 | Sponge network | 0.246 | 0.59912 | 0.175 | 0.14787 | 0.321 |
| 35808 | RP11-401P9.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-629-3p | 23 | KIF1B | Sponge network | 1.524 | 0.1098 | -0.118 | 0.32258 | 0.321 |
| 35809 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-577 | 13 | PGR | Sponge network | 0.419 | 0.0319 | -1.704 | 0 | 0.321 |
| 35810 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 30 | USP12 | Sponge network | -0.709 | 0.18168 | -0.102 | 0.40768 | 0.321 |
| 35811 | OIP5-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-940 | 16 | DLG1 | Sponge network | 0.421 | 0.00084 | -0.014 | 0.8926 | 0.321 |
| 35812 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | UNC80 | Sponge network | 0.225 | 0.30644 | -1.934 | 0 | 0.321 |
| 35813 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | MYBL1 | Sponge network | 1.254 | 0 | 0.494 | 0.02152 | 0.321 |
| 35814 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450a-5p;hsa-miR-532-3p;hsa-miR-576-5p | 19 | PBX1 | Sponge network | 1.757 | 0 | -1.206 | 0 | 0.321 |
| 35815 | HAR1A |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 11 | RASSF8 | Sponge network | -0.672 | 0.19447 | -0.122 | 0.65591 | 0.321 |
| 35816 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 23 | PGR | Sponge network | 0.392 | 0.0278 | -1.704 | 0 | 0.321 |
| 35817 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-92a-3p | 28 | CRTC1 | Sponge network | -0.563 | 0.02545 | -0.116 | 0.28412 | 0.321 |
| 35818 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 20 | YAP1 | Sponge network | -0.096 | 0.67958 | 0.22 | 0.11836 | 0.321 |
| 35819 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p | 10 | MNT | Sponge network | -0.421 | 0.0114 | -0.157 | 0.06598 | 0.321 |
| 35820 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-7-1-3p | 24 | ZMYND11 | Sponge network | -0.969 | 2.0E-5 | -0.2 | 0.05449 | 0.321 |
| 35821 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | NEDD4 | Sponge network | -0.777 | 0.1134 | 0.02 | 0.89834 | 0.321 |
| 35822 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TMEFF2 | Sponge network | -0.342 | 0.02288 | -2.426 | 0 | 0.321 |
| 35823 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-550a-5p;hsa-miR-766-3p;hsa-miR-940 | 28 | CHD5 | Sponge network | 0.349 | 0.01695 | -0.922 | 0.01521 | 0.321 |
| 35824 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-5p | 13 | LRRC55 | Sponge network | 2.306 | 9.0E-5 | -0.026 | 0.93163 | 0.321 |
| 35825 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-582-5p | 27 | FBXO32 | Sponge network | -1.047 | 0 | -0.495 | 0.07561 | 0.321 |
| 35826 | HAND2-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | AFAP1L2 | Sponge network | -1.697 | 0.00889 | -0.284 | 0.15434 | 0.321 |
| 35827 | RP11-400K9.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-576-5p | 11 | INPP4B | Sponge network | -0.611 | 0.09607 | -0.097 | 0.60503 | 0.321 |
| 35828 | BCYRN1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | RASSF8 | Sponge network | 0.311 | 0.10344 | -0.122 | 0.65591 | 0.321 |
| 35829 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 24 | BMPR2 | Sponge network | 0.342 | 0.03129 | 0.206 | 0.0635 | 0.321 |
| 35830 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-429 | 20 | MYBL1 | Sponge network | 0.663 | 0.00073 | 0.494 | 0.02152 | 0.321 |
| 35831 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 38 | ERBB4 | Sponge network | -0.513 | 0.04703 | -1.975 | 2.0E-5 | 0.321 |
| 35832 | HCG18 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 34 | CCDC6 | Sponge network | 1.063 | 0 | 0.27 | 0.02555 | 0.321 |
| 35833 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | PBRM1 | Sponge network | 0.34 | 0.00273 | -0.029 | 0.78601 | 0.321 |
| 35834 | LINC00863 |
hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PIK3CA | Sponge network | 0.189 | 0.13981 | 0.195 | 0.06908 | 0.321 |
| 35835 | SOX2-OT |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | PACRG | Sponge network | -1.264 | 6.0E-5 | -2.248 | 0 | 0.321 |
| 35836 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | IGSF10 | Sponge network | 1.302 | 7.0E-5 | -1.751 | 1.0E-5 | 0.321 |
| 35837 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | MED12L | Sponge network | -0.235 | 0.15142 | -0.194 | 0.55299 | 0.321 |
| 35838 | RP11-21A7A.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 10 | TACC1 | Sponge network | -1.116 | 0.23686 | -0.362 | 0.05406 | 0.321 |
| 35839 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-877-5p | 22 | GAS7 | Sponge network | -1.116 | 0.23686 | -0.555 | 0.01602 | 0.321 |
| 35840 | GS1-124K5.11 |
hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 12 | LZTS1 | Sponge network | 0.494 | 0.00339 | 1.664 | 0 | 0.321 |
| 35841 | CTC-429P9.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-3p | 24 | CADM2 | Sponge network | 0.178 | 0.29106 | -3.782 | 0 | 0.321 |
| 35842 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 30 | CXXC4 | Sponge network | -0.129 | 0.57177 | -0.433 | 0.21055 | 0.321 |
| 35843 | MIR143HG |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RBM6 | Sponge network | -0.739 | 0.0574 | -0.137 | 0.15663 | 0.321 |
| 35844 | H1FX-AS1 |
hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | LIFR | Sponge network | -0.206 | 0.12942 | -1.794 | 0 | 0.321 |
| 35845 | KTN1-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 12 | TBL1XR1 | Sponge network | 0.927 | 0 | 0.578 | 0 | 0.321 |
| 35846 | LINC00476 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p | 33 | RIMS2 | Sponge network | -0.615 | 0.00015 | -1.365 | 0.00208 | 0.321 |
| 35847 | GAS6-AS2 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-5p | 12 | FNBP1 | Sponge network | 0.16 | 0.50785 | -0.969 | 4.0E-5 | 0.321 |
| 35848 | RP3-402G11.26 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-671-5p | 17 | ADARB1 | Sponge network | -0.254 | 0.19303 | -0.335 | 0.06631 | 0.321 |
| 35849 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-7-5p | 23 | ERBB4 | Sponge network | -1.331 | 3.0E-5 | -1.975 | 2.0E-5 | 0.321 |
| 35850 | RP11-344B5.2 |
hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-30b-3p;hsa-miR-505-3p | 11 | SOX5 | Sponge network | -0.055 | 0.88482 | -1.091 | 4.0E-5 | 0.321 |
| 35851 | RP11-305E6.4 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p | 10 | CDK6 | Sponge network | 1.317 | 0 | 0.991 | 0 | 0.321 |
| 35852 | HNRNPU-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-429;hsa-miR-590-3p | 13 | PICALM | Sponge network | 1.601 | 0 | 0.086 | 0.23523 | 0.321 |
| 35853 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | RICTOR | Sponge network | 0.476 | 0.19203 | 0.388 | 0.00188 | 0.321 |
| 35854 | MIR497HG |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-532-5p;hsa-miR-7-1-3p | 21 | ZMYND11 | Sponge network | -1.439 | 4.0E-5 | -0.2 | 0.05449 | 0.321 |
| 35855 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-455-3p | 12 | DKK3 | Sponge network | 0.051 | 0.83388 | -0.052 | 0.77783 | 0.321 |
| 35856 | PSMD5-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | UBR3 | Sponge network | 0.569 | 0.00067 | -0.021 | 0.82774 | 0.321 |
| 35857 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 20 | ZFHX3 | Sponge network | -0.206 | 0.12942 | 0.389 | 0.01363 | 0.321 |
| 35858 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-92a-3p | 22 | ADAMTSL3 | Sponge network | 0.253 | 0.09565 | -1.559 | 0 | 0.321 |
| 35859 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 15 | NTRK2 | Sponge network | -0.187 | 0.23787 | -0.736 | 0.06495 | 0.321 |
| 35860 | MYLK-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-664a-5p;hsa-miR-935 | 33 | SMAD4 | Sponge network | -1.331 | 3.0E-5 | -0.313 | 0.00413 | 0.321 |
| 35861 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | LATS1 | Sponge network | 0.34 | 0.00273 | 0.109 | 0.34208 | 0.321 |
| 35862 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 15 | LATS1 | Sponge network | 0.594 | 0.00064 | 0.109 | 0.34208 | 0.321 |
| 35863 | AC016747.3 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 14 | GLIPR1 | Sponge network | 0.199 | 0.38015 | 0.146 | 0.3276 | 0.321 |
| 35864 | SERTAD4-AS1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 11 | CSRNP1 | Sponge network | -0.932 | 0.00329 | -1.448 | 0 | 0.321 |
| 35865 | RP11-760H22.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-502-3p;hsa-miR-655-3p | 11 | SHPRH | Sponge network | 0.001 | 0.99753 | 0.098 | 0.40994 | 0.321 |
| 35866 | RP11-116O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | HECA | Sponge network | 0.05 | 0.95483 | -0.217 | 0.03463 | 0.321 |
| 35867 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | LRIG1 | Sponge network | 0.299 | 0.57722 | -0.537 | 0.00588 | 0.321 |
| 35868 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-576-5p | 22 | ITGAV | Sponge network | -0.162 | 0.47687 | 0.337 | 0.01002 | 0.321 |
| 35869 | CALML3-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 23 | NAV3 | Sponge network | 0.95 | 0.15943 | 0.212 | 0.47729 | 0.321 |
| 35870 | LINC00641 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | RPS6KA6 | Sponge network | -0.222 | 0.32445 | -2.989 | 0 | 0.321 |
| 35871 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DACT1 | Sponge network | 0.101 | 0.60919 | 0.783 | 0.00072 | 0.321 |
| 35872 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 20 | EPHA3 | Sponge network | 0.051 | 0.83388 | 0.185 | 0.53588 | 0.321 |
| 35873 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 11 | PDGFC | Sponge network | 0.222 | 0.29133 | -0.33 | 0.06483 | 0.321 |
| 35874 | FZD10-AS1 |
hsa-miR-126-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 14 | CXCL12 | Sponge network | -0.727 | 0.04726 | -1.421 | 0 | 0.321 |
| 35875 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | RAP1A | Sponge network | -1.349 | 0.00017 | -0.929 | 0 | 0.321 |
| 35876 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | CDHR3 | Sponge network | 0.194 | 0.64278 | -0.977 | 4.0E-5 | 0.321 |
| 35877 | ZNF667-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 15 | POU2F2 | Sponge network | -0.976 | 0.00025 | -0.233 | 0.32214 | 0.321 |
| 35878 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940 | 34 | NOTCH2 | Sponge network | -0.738 | 0.21233 | 0.138 | 0.35163 | 0.321 |
| 35879 | AC141928.1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-486-5p;hsa-miR-96-5p | 10 | DNMT3A | Sponge network | 0.853 | 0.15352 | 0.302 | 0.0262 | 0.321 |
| 35880 | RP11-46C24.7 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 23 | PCDH10 | Sponge network | 0.392 | 0.0278 | -1.644 | 0.0078 | 0.321 |
| 35881 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | AHNAK | Sponge network | 0.064 | 0.72934 | -1.207 | 0 | 0.321 |
| 35882 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 22 | EYA4 | Sponge network | 0.299 | 0.57722 | 0.3 | 0.36876 | 0.321 |
| 35883 | SNX29P2 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-424-5p | 12 | DYNC1I1 | Sponge network | 0.4 | 0.14611 | -1.12 | 0.00107 | 0.321 |
| 35884 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p | 12 | FGFR1 | Sponge network | -0.72 | 0.24239 | -0.509 | 0.03957 | 0.321 |
| 35885 | RP11-73M18.10 |
hsa-miR-129-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-342-3p;hsa-miR-497-5p | 12 | CCDC6 | Sponge network | 0.85 | 0 | 0.27 | 0.02555 | 0.321 |
| 35886 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 21 | ACVR1 | Sponge network | 0.335 | 0.20596 | 0.279 | 0.00234 | 0.321 |
| 35887 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | AFAP1L2 | Sponge network | 0.194 | 0.64278 | -0.284 | 0.15434 | 0.321 |
| 35888 | AC009948.5 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 14 | RET | Sponge network | 0.532 | 0.00505 | -0.833 | 0.03517 | 0.321 |
| 35889 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | LRRC7 | Sponge network | 0.174 | 0.30034 | -1.341 | 0.01193 | 0.321 |
| 35890 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 13 | GIGYF2 | Sponge network | 0.311 | 0.10344 | 0.136 | 0.04403 | 0.321 |
| 35891 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-374a-3p | 15 | TMTC3 | Sponge network | 0.331 | 0.57247 | 0.123 | 0.22189 | 0.321 |
| 35892 | LINC00476 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | TLL1 | Sponge network | -0.615 | 0.00015 | -1.195 | 3.0E-5 | 0.321 |
| 35893 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | DMTF1 | Sponge network | -0.227 | 0.31657 | 0.248 | 0.02726 | 0.321 |
| 35894 | CTC-429P9.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-502-5p;hsa-miR-532-5p | 13 | IGFBP5 | Sponge network | 0.043 | 0.81628 | -0.806 | 0.00105 | 0.321 |
| 35895 | RP11-890B15.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-450b-5p | 10 | SPTBN4 | Sponge network | 0.101 | 0.60919 | -0.786 | 0.01281 | 0.321 |
| 35896 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SON | Sponge network | 0.415 | 0.14657 | 0.168 | 0.04406 | 0.321 |
| 35897 | RP11-379F4.7 |
hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-654-3p | 11 | PIK3R1 | Sponge network | 1.316 | 0 | -0.133 | 0.36854 | 0.321 |
| 35898 | RP11-434B12.1 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | RYR2 | Sponge network | -0.031 | 0.89063 | -1.466 | 0.00031 | 0.321 |
| 35899 | LINC00657 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TET1 | Sponge network | 0.91 | 0 | -0.135 | 0.52138 | 0.321 |
| 35900 | RP1-228H13.5 |
hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p | 12 | SLC5A3 | Sponge network | 1.397 | 0 | 0.493 | 5.0E-5 | 0.321 |
| 35901 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -0.08 | 0.90092 | -3.328 | 0 | 0.321 |
| 35902 | RP1-151F17.2 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | INPP4B | Sponge network | 0.222 | 0.29133 | -0.097 | 0.60503 | 0.321 |
| 35903 | AC156455.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-378a-5p | 13 | NUAK1 | Sponge network | 1.154 | 0.00041 | 0.904 | 0 | 0.321 |
| 35904 | PGM5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p | 13 | PTPN14 | Sponge network | -4.546 | 0 | 0.144 | 0.34595 | 0.321 |
| 35905 | LINC00667 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 15 | CDH23 | Sponge network | -0.235 | 0.15142 | -0.974 | 0.00034 | 0.321 |
| 35906 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 47 | SSBP2 | Sponge network | -0.727 | 0.04726 | -1.58 | 0 | 0.321 |
| 35907 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-940 | 27 | CRTC3 | Sponge network | -0.899 | 6.0E-5 | 0.164 | 0.16786 | 0.321 |
| 35908 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-93-5p | 10 | FBXO31 | Sponge network | 0.572 | 0.01722 | -0.085 | 0.42389 | 0.321 |
| 35909 | RP11-701H24.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 11 | LRIG1 | Sponge network | -1.425 | 0.04204 | -0.537 | 0.00588 | 0.321 |
| 35910 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-376c-3p;hsa-miR-411-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p | 35 | SMAD2 | Sponge network | 0.401 | 0.00066 | -0.128 | 0.12732 | 0.321 |
| 35911 | RP4-669P10.16 |
hsa-miR-103a-2-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-629-5p;hsa-miR-93-3p | 21 | BMPR2 | Sponge network | -0.301 | 0.06597 | 0.206 | 0.0635 | 0.321 |
| 35912 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-93-5p;hsa-miR-942-5p | 14 | FBXO31 | Sponge network | -0.976 | 0.00025 | -0.085 | 0.42389 | 0.321 |
| 35913 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 15 | SUFU | Sponge network | -0.726 | 0.00132 | -0.04 | 0.65489 | 0.321 |
| 35914 | CTB-133G6.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 12 | DST | Sponge network | 0.4 | 0.39299 | -0.713 | 0.00096 | 0.321 |
| 35915 | TMEM9B-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-582-5p | 17 | ABI2 | Sponge network | 0.529 | 0.00552 | 0.175 | 0.14787 | 0.321 |
| 35916 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 24 | SCN9A | Sponge network | 0.862 | 0.06213 | -0.525 | 0.10593 | 0.321 |
| 35917 | AP001055.6 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 17 | DTNA | Sponge network | 0.693 | 0.00076 | -1.648 | 1.0E-5 | 0.321 |
| 35918 | RP11-38L15.3 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | RBMS3 | Sponge network | 0.344 | 0.3127 | -0.519 | 0.03551 | 0.321 |
| 35919 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 25 | LRP1B | Sponge network | -0.246 | 0.35034 | -2.163 | 0 | 0.321 |
| 35920 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 19 | PRKAB2 | Sponge network | 0.299 | 0.57722 | -0.367 | 0.01371 | 0.321 |
| 35921 | U91328.19 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-3607-3p | 10 | AFAP1L2 | Sponge network | -0.303 | 0.21002 | -0.284 | 0.15434 | 0.321 |
| 35922 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429 | 14 | CBFA2T3 | Sponge network | -0.206 | 0.12942 | -1.221 | 1.0E-5 | 0.321 |
| 35923 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | WWTR1 | Sponge network | 0.349 | 0.01695 | -0.345 | 0.11997 | 0.321 |
| 35924 | CTC-297N7.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-34a-5p;hsa-miR-877-5p | 15 | CBFA2T3 | Sponge network | -2.069 | 0 | -1.221 | 1.0E-5 | 0.321 |
| 35925 | LINC00969 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 14 | ZNF770 | Sponge network | 0.683 | 1.0E-5 | 0.206 | 0.06751 | 0.321 |
| 35926 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-671-5p | 20 | KIT | Sponge network | -0.154 | 0.53954 | -2.184 | 0 | 0.321 |
| 35927 | KTN1-AS1 |
hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-664a-5p | 11 | CBFB | Sponge network | 0.927 | 0 | 1.061 | 0 | 0.321 |
| 35928 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 27 | ARHGAP28 | Sponge network | -0.942 | 0 | -0.911 | 0.00013 | 0.321 |
| 35929 | RP11-121C2.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-664a-3p | 30 | CCDC6 | Sponge network | 1.147 | 0 | 0.27 | 0.02555 | 0.321 |
| 35930 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-424-5p | 13 | MYBL1 | Sponge network | 1.153 | 0.00114 | 0.494 | 0.02152 | 0.321 |
| 35931 | RP11-473I1.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 13 | HMCN1 | Sponge network | 0.683 | 1.0E-5 | 0.809 | 0.00544 | 0.321 |
| 35932 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 23 | CDC73 | Sponge network | 0.683 | 1.0E-5 | 0.094 | 0.20463 | 0.321 |
| 35933 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-590-3p | 19 | USP6 | Sponge network | 1.254 | 0 | 0.188 | 0.3871 | 0.321 |
| 35934 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 19 | LRRC7 | Sponge network | -0.513 | 0.04703 | -1.341 | 0.01193 | 0.321 |
| 35935 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-98-5p | 24 | LRIG1 | Sponge network | 0.494 | 0.00339 | -0.537 | 0.00588 | 0.321 |
| 35936 | HOXB-AS2 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p | 15 | EGFR | Sponge network | 1.316 | 0.01106 | -0.175 | 0.48451 | 0.321 |
| 35937 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-671-5p | 14 | FAT4 | Sponge network | 0.331 | 0.57247 | -0.354 | 0.14694 | 0.321 |
| 35938 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 20 | DOCK4 | Sponge network | -0.769 | 0.00035 | 0.502 | 0.00108 | 0.321 |
| 35939 | LINC00473 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-423-3p;hsa-miR-511-5p;hsa-miR-590-3p | 12 | CELF4 | Sponge network | -1.203 | 0.03881 | -1.135 | 0.00344 | 0.321 |
| 35940 | RP11-5C23.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-429;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | ABI2 | Sponge network | 0.274 | 0.07681 | 0.175 | 0.14787 | 0.321 |
| 35941 | CTC-559E9.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-1271-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-628-5p;hsa-miR-7-5p | 10 | EPHA3 | Sponge network | 0.601 | 1.0E-5 | 0.185 | 0.53588 | 0.321 |
| 35942 | LINC00173 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | RASSF8 | Sponge network | 0.056 | 0.8494 | -0.122 | 0.65591 | 0.321 |
| 35943 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | MITF | Sponge network | 0.214 | 0.18246 | -0.769 | 0.00022 | 0.321 |
| 35944 | RP11-531A24.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p | 11 | UVRAG | Sponge network | 0.75 | 0.00054 | -0.017 | 0.83865 | 0.321 |
| 35945 | PSMG3-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | EGR2 | Sponge network | -0.125 | 0.49414 | 0.309 | 0.17079 | 0.321 |
| 35946 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLI1 | Sponge network | -0.323 | 0.01372 | -0.294 | 0.10905 | 0.321 |
| 35947 | RP11-130L8.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-590-3p | 19 | TRPS1 | Sponge network | -0.663 | 0.10887 | -0.191 | 0.44109 | 0.321 |
| 35948 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 10 | TMTC2 | Sponge network | 0.542 | 0.00054 | -0.215 | 0.18248 | 0.321 |
| 35949 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | PCDH10 | Sponge network | 0.559 | 0.00026 | -1.644 | 0.0078 | 0.321 |
| 35950 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 23 | HBP1 | Sponge network | -0.668 | 3.0E-5 | -0.454 | 0 | 0.321 |
| 35951 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-493-5p | 23 | AFF3 | Sponge network | -0.405 | 0.22013 | -2.373 | 0 | 0.321 |
| 35952 | AC005562.1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p | 14 | STAG2 | Sponge network | 0.488 | 0.00084 | 0.252 | 0.00438 | 0.321 |
| 35953 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-625-5p | 24 | JAZF1 | Sponge network | -0.773 | 0.04073 | -0.377 | 0.02677 | 0.321 |
| 35954 | FAM13A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 19 | RYR2 | Sponge network | -0.83 | 0 | -1.466 | 0.00031 | 0.321 |
| 35955 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.083 | 0.66405 | 1.083 | 0 | 0.32 |
| 35956 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | PEG3 | Sponge network | 0.72 | 0.0001 | -1.74 | 0 | 0.32 |
| 35957 | LINC00173 |
hsa-let-7d-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-96-5p | 15 | CBFA2T3 | Sponge network | 0.056 | 0.8494 | -1.221 | 1.0E-5 | 0.32 |
| 35958 | TINCR |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-495-3p | 11 | CLIP1 | Sponge network | -0.664 | 0.14543 | -0.215 | 0.08684 | 0.32 |
| 35959 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p | 10 | GIGYF2 | Sponge network | 0.101 | 0.60919 | 0.136 | 0.04403 | 0.32 |
| 35960 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p | 12 | APC | Sponge network | -0.376 | 0.00337 | -0.255 | 0.04051 | 0.32 |
| 35961 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | CCND2 | Sponge network | -1.439 | 4.0E-5 | -0.267 | 0.3112 | 0.32 |
| 35962 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-744-3p | 16 | HIP1 | Sponge network | -0.693 | 0.05629 | 0.774 | 0 | 0.32 |
| 35963 | RP11-13K12.5 |
hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-576-5p | 11 | PTCH1 | Sponge network | -0.318 | 0.65847 | -0.1 | 0.6179 | 0.32 |
| 35964 | AC005562.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 13 | IRS1 | Sponge network | 0.488 | 0.00084 | -0.026 | 0.88855 | 0.32 |
| 35965 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | ABCA10 | Sponge network | 0.174 | 0.30034 | -0.571 | 0.03674 | 0.32 |
| 35966 | FAM66C |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | FLI1 | Sponge network | -0.901 | 0.00019 | -0.294 | 0.10905 | 0.32 |
| 35967 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p | 10 | GPAM | Sponge network | 0.793 | 0 | 0.142 | 0.29732 | 0.32 |
| 35968 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p | 11 | MGA | Sponge network | 0.594 | 0.41522 | 0.323 | 0.00792 | 0.32 |
| 35969 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | CACNA2D1 | Sponge network | 0.381 | 0.25967 | -0.846 | 0.00675 | 0.32 |
| 35970 | KB-1460A1.5 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 16 | ATM | Sponge network | 0.603 | 0.00127 | 0.178 | 0.21568 | 0.32 |
| 35971 | RP11-277P12.20 |
hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 11 | NRXN3 | Sponge network | 0.687 | 0.02753 | -0.893 | 0.04186 | 0.32 |
| 35972 | RP11-571M6.17 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ERCC6 | Sponge network | 1.013 | 0 | 0.23 | 0.015 | 0.32 |
| 35973 | SOX2-OT |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-10a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-940 | 11 | ABCC5 | Sponge network | -1.264 | 6.0E-5 | -0.698 | 0 | 0.32 |
| 35974 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p | 17 | RIMBP2 | Sponge network | -0.154 | 0.78939 | -1.968 | 5.0E-5 | 0.32 |
| 35975 | CTC-444N24.11 |
hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-628-5p | 21 | VPS53 | Sponge network | 1.153 | 0 | 0.103 | 0.22667 | 0.32 |
| 35976 | CTA-204B4.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-629-3p;hsa-miR-744-3p | 13 | PTPN14 | Sponge network | 0.765 | 7.0E-5 | 0.144 | 0.34595 | 0.32 |
| 35977 | RP11-458D21.1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | 0.663 | 0.00073 | 0.098 | 0.40994 | 0.32 |
| 35978 | RP11-130L8.1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-454-3p;hsa-miR-590-3p | 12 | ITPKB | Sponge network | -0.663 | 0.10887 | -0.704 | 0.00106 | 0.32 |
| 35979 | RAD51-AS1 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | DICER1 | Sponge network | 0.768 | 7.0E-5 | 0.042 | 0.674 | 0.32 |
| 35980 | AC138035.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-193b-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-505-5p;hsa-miR-539-5p | 15 | ARHGEF12 | Sponge network | 1.073 | 0 | 0.09 | 0.35693 | 0.32 |
| 35981 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | PICALM | Sponge network | 0.559 | 0.00026 | 0.086 | 0.23523 | 0.32 |
| 35982 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-98-5p | 13 | ITGB3 | Sponge network | 0.082 | 0.55397 | -0.011 | 0.95751 | 0.32 |
| 35983 | TPTEP1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ABCA10 | Sponge network | -1.216 | 0 | -0.571 | 0.03674 | 0.32 |
| 35984 | TINCR |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-940;hsa-miR-96-5p | 11 | HOOK3 | Sponge network | -0.664 | 0.14543 | 0.273 | 0.09957 | 0.32 |
| 35985 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-7-1-3p | 21 | CACNA2D1 | Sponge network | 0.16 | 0.50785 | -0.846 | 0.00675 | 0.32 |
| 35986 | LINC00926 |
hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 19 | MDM4 | Sponge network | 0.756 | 0.00739 | 0.345 | 0.00762 | 0.32 |
| 35987 | FZD10-AS1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p | 14 | ZNF208 | Sponge network | -0.727 | 0.04726 | -0.4 | 0.22381 | 0.32 |
| 35988 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NR4A3 | Sponge network | 0.335 | 0.20596 | -1.385 | 0 | 0.32 |
| 35989 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-590-3p | 11 | DACT1 | Sponge network | 0.051 | 0.83388 | 0.783 | 0.00072 | 0.32 |
| 35990 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 17 | SASH1 | Sponge network | -1.886 | 0 | -0.502 | 0.00175 | 0.32 |
| 35991 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 30 | ZFHX3 | Sponge network | 0.683 | 1.0E-5 | 0.389 | 0.01363 | 0.32 |
| 35992 | TPTEP1 |
hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-590-3p | 10 | EGR1 | Sponge network | -1.216 | 0 | -1.195 | 0 | 0.32 |
| 35993 | AF131215.2 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | INPP4A | Sponge network | -0.176 | 0.59692 | 0.07 | 0.53563 | 0.32 |
| 35994 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 23 | EPHA3 | Sponge network | 0.619 | 0.00157 | 0.185 | 0.53588 | 0.32 |
| 35995 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 22 | HECA | Sponge network | 0.75 | 0.00054 | -0.217 | 0.03463 | 0.32 |
| 35996 | RP11-48B3.4 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 13 | MRVI1 | Sponge network | 1.757 | 0 | -1.051 | 0.00022 | 0.32 |
| 35997 | RP11-798G7.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-539-5p | 11 | NIN | Sponge network | 0.858 | 3.0E-5 | -0.253 | 0.13794 | 0.32 |
| 35998 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | RNF144A | Sponge network | -0.726 | 0.00132 | 0.594 | 0.0009 | 0.32 |
| 35999 | LINC00865 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-429 | 12 | DUSP1 | Sponge network | -1.249 | 7.0E-5 | -1.754 | 0 | 0.32 |
| 36000 | KTN1-AS1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-495-3p | 10 | PHF6 | Sponge network | 0.927 | 0 | 0.785 | 0 | 0.32 |
| 36001 | MIR22HG |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-582-5p | 48 | DTNA | Sponge network | -1.047 | 0 | -1.648 | 1.0E-5 | 0.32 |
| 36002 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-429 | 10 | ZNF521 | Sponge network | 0.064 | 0.72934 | -0.111 | 0.58068 | 0.32 |
| 36003 | ARHGAP5-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 22 | PBX1 | Sponge network | -0.644 | 0.00021 | -1.206 | 0 | 0.32 |
| 36004 | LINC00863 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p | 24 | ABL2 | Sponge network | 0.189 | 0.13981 | 0.82 | 0 | 0.32 |
| 36005 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | DACH2 | Sponge network | -1.439 | 4.0E-5 | -1.635 | 0.00034 | 0.32 |
| 36006 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 48 | ZFHX3 | Sponge network | 0.421 | 0.00084 | 0.389 | 0.01363 | 0.32 |
| 36007 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-454-3p | 17 | PDGFRA | Sponge network | 0.598 | 0.38481 | -0.372 | 0.0037 | 0.32 |
| 36008 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 47 | TMEM132B | Sponge network | -0.942 | 0 | -0.83 | 0.01084 | 0.32 |
| 36009 | RP11-686D22.4 |
hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-577;hsa-miR-590-3p | 10 | NRK | Sponge network | 0.793 | 5.0E-5 | 0.656 | 0.19217 | 0.32 |
| 36010 | RP11-25K19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | ARNT2 | Sponge network | -1.057 | 0.00029 | -0.332 | 0.25851 | 0.32 |
| 36011 | HAND2-AS1 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-7-5p | 11 | COL1A2 | Sponge network | -1.697 | 0.00889 | 1.991 | 0 | 0.32 |
| 36012 | AC103563.9 |
hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577 | 14 | DTNA | Sponge network | -6.057 | 0 | -1.648 | 1.0E-5 | 0.32 |
| 36013 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 18 | JAZF1 | Sponge network | 0.898 | 1.0E-5 | -0.377 | 0.02677 | 0.32 |
| 36014 | ACTA2-AS1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 18 | BAZ2B | Sponge network | 0.194 | 0.64278 | -0.276 | 0.02833 | 0.32 |
| 36015 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-590-3p | 13 | PHC3 | Sponge network | -0.376 | 0.00337 | 0.212 | 0.0499 | 0.32 |
| 36016 | PSMG3-AS1 |
hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-93-3p | 10 | PEA15 | Sponge network | -0.125 | 0.49414 | 0.009 | 0.93318 | 0.32 |
| 36017 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 20 | CHD2 | Sponge network | -0.487 | 0.02157 | 0.049 | 0.55371 | 0.32 |
| 36018 | LINC00662 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 0.213 | 0.32297 | 0.321 | 0.00267 | 0.32 |
| 36019 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-501-5p | 10 | UBE2QL1 | Sponge network | 0.395 | 0.15451 | -2.417 | 0 | 0.32 |
| 36020 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p | 17 | ESR1 | Sponge network | -0.663 | 0.10887 | -1.035 | 3.0E-5 | 0.32 |
| 36021 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-7-1-3p | 16 | RARB | Sponge network | -0.206 | 0.12942 | -0.825 | 2.0E-5 | 0.32 |
| 36022 | ZNF582-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-450b-5p | 22 | PRDM2 | Sponge network | -1.349 | 0.00017 | -0.258 | 0.00721 | 0.32 |
| 36023 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-940 | 27 | GAS7 | Sponge network | -4.477 | 0 | -0.555 | 0.01602 | 0.32 |
| 36024 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 27 | ADARB1 | Sponge network | 0.381 | 0.25967 | -0.335 | 0.06631 | 0.32 |
| 36025 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CSMD1 | Sponge network | 0.335 | 0.20596 | -0.63 | 0.18881 | 0.32 |
| 36026 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-424-5p | 10 | PLSCR4 | Sponge network | 0.622 | 0.00015 | -0.474 | 0.03833 | 0.32 |
| 36027 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-942-5p | 19 | ARHGAP28 | Sponge network | -0.693 | 0.05629 | -0.911 | 0.00013 | 0.32 |
| 36028 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 18 | IGF2 | Sponge network | 1.302 | 7.0E-5 | 0.848 | 0.03842 | 0.32 |
| 36029 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-424-5p | 18 | PHC3 | Sponge network | 0.622 | 0.00015 | 0.212 | 0.0499 | 0.32 |
| 36030 | CTD-2666L21.1 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-493-5p | 15 | ANK2 | Sponge network | 0.368 | 0.20093 | -1.603 | 1.0E-5 | 0.32 |
| 36031 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-629-5p | 35 | SOX5 | Sponge network | -0.773 | 0.04073 | -1.091 | 4.0E-5 | 0.32 |
| 36032 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | PHC3 | Sponge network | -0.612 | 0.00094 | 0.212 | 0.0499 | 0.32 |
| 36033 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-93-5p | 29 | JAZF1 | Sponge network | 0.253 | 0.09565 | -0.377 | 0.02677 | 0.32 |
| 36034 | RAD51-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | BRD3 | Sponge network | 0.768 | 7.0E-5 | 0.37 | 0.00013 | 0.32 |
| 36035 | LINC00473 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NCAM2 | Sponge network | -1.203 | 0.03881 | -0.482 | 0.22565 | 0.32 |
| 36036 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 16 | MAF | Sponge network | -2.095 | 0.00112 | -1.257 | 0 | 0.32 |
| 36037 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 29 | ST6GAL2 | Sponge network | -0.969 | 2.0E-5 | -1.201 | 0.00597 | 0.32 |
| 36038 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p;hsa-miR-98-5p | 21 | PICALM | Sponge network | -0.129 | 0.57177 | 0.086 | 0.23523 | 0.32 |
| 36039 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-744-5p | 20 | EIF4A2 | Sponge network | -0.323 | 0.01372 | -0.383 | 0.00046 | 0.32 |
| 36040 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-190a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-429;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-582-5p | 19 | TCF4 | Sponge network | 1.157 | 0.00455 | 0.016 | 0.92271 | 0.32 |
| 36041 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-493-5p | 19 | AKAP11 | Sponge network | 0.368 | 0.20093 | 0.196 | 0.09825 | 0.32 |
| 36042 | RP11-386G11.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-365a-3p | 12 | RICTOR | Sponge network | 2.031 | 0.00154 | 0.388 | 0.00188 | 0.32 |
| 36043 | LINC00472 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p | 26 | BTG2 | Sponge network | -0.842 | 0.01361 | -1.763 | 0 | 0.32 |
| 36044 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-98-5p | 28 | THBS1 | Sponge network | 0.056 | 0.81195 | 0.741 | 0.00386 | 0.32 |
| 36045 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p | 44 | KIF1B | Sponge network | 1.063 | 0 | -0.118 | 0.32258 | 0.32 |
| 36046 | LPP-AS2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-493-5p | 13 | CREBBP | Sponge network | -0.18 | 0.20343 | 0.126 | 0.15186 | 0.32 |
| 36047 | HOXD-AS2 |
hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p | 11 | EBF1 | Sponge network | -0.208 | 0.65592 | -0.384 | 0.08856 | 0.32 |
| 36048 | C4B-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 20 | QKI | Sponge network | 2.306 | 9.0E-5 | -0.333 | 0.04111 | 0.32 |
| 36049 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 38 | RBMS3 | Sponge network | -0.303 | 0.21002 | -0.519 | 0.03551 | 0.32 |
| 36050 | LINC00865 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | KCNAB1 | Sponge network | -1.249 | 7.0E-5 | -0.755 | 2.0E-5 | 0.32 |
| 36051 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SEMA3E | Sponge network | -0.668 | 3.0E-5 | -1.717 | 0.00137 | 0.32 |
| 36052 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | ERG | Sponge network | 0.064 | 0.72934 | 0.002 | 0.99011 | 0.32 |
| 36053 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-96-5p | 20 | CLASP2 | Sponge network | 0.594 | 0.41522 | -0.038 | 0.71 | 0.32 |
| 36054 | RP11-927P21.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-940 | 11 | SIRT1 | Sponge network | 0.921 | 0.00118 | 0.109 | 0.2593 | 0.32 |
| 36055 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 47 | PTPRD | Sponge network | -0.487 | 0.02157 | -1.005 | 0.00064 | 0.32 |
| 36056 | PCA3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 17 | CELF4 | Sponge network | -0.709 | 0.18168 | -1.135 | 0.00344 | 0.32 |
| 36057 | RP11-539I5.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | HECA | Sponge network | -0.468 | 0.32587 | -0.217 | 0.03463 | 0.32 |
| 36058 | AC103563.8 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p | 24 | LPP | Sponge network | -7.551 | 0 | -0.459 | 0.04475 | 0.32 |
| 36059 | SERTAD4-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-576-5p | 17 | PTPRT | Sponge network | -0.932 | 0.00329 | -0.907 | 0.03727 | 0.32 |
| 36060 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-590-3p;hsa-miR-654-3p | 28 | RICTOR | Sponge network | -0.227 | 0.31657 | 0.388 | 0.00188 | 0.32 |
| 36061 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 24 | EBF3 | Sponge network | 0.659 | 3.0E-5 | -0.058 | 0.81437 | 0.32 |
| 36062 | AC003090.1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | MXI1 | Sponge network | -2.117 | 0.00161 | -0.987 | 0 | 0.32 |
| 36063 | AC010226.4 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-485-3p;hsa-miR-590-3p | 20 | AFF1 | Sponge network | -0.811 | 2.0E-5 | -0.384 | 0.00053 | 0.32 |
| 36064 | LINC00886 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p | 19 | SYNM | Sponge network | -0.371 | 0.24604 | -2.309 | 1.0E-5 | 0.32 |
| 36065 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 18 | PIK3C2A | Sponge network | 0.494 | 0.00339 | 0.061 | 0.53776 | 0.32 |
| 36066 | LINC00265 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 10 | CDK6 | Sponge network | 1.07 | 0 | 0.991 | 0 | 0.32 |
| 36067 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | TET1 | Sponge network | -0.235 | 0.15142 | -0.135 | 0.52138 | 0.32 |
| 36068 | RP11-373D23.3 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p | 11 | PTPN14 | Sponge network | -0.285 | 0.2172 | 0.144 | 0.34595 | 0.32 |
| 36069 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 46 | EPHA5 | Sponge network | 0.421 | 0.00084 | -0.957 | 0.01733 | 0.32 |
| 36070 | CTB-133G6.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p | 16 | RUNX1T1 | Sponge network | 0.4 | 0.39299 | -0.344 | 0.2374 | 0.32 |
| 36071 | RP11-226L15.5 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | ATM | Sponge network | 0.8 | 0 | 0.178 | 0.21568 | 0.32 |
| 36072 | LINC00403 |
hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-324-3p | 12 | PBX1 | Sponge network | -3.586 | 0.00032 | -1.206 | 0 | 0.32 |
| 36073 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p | 11 | WDFY3 | Sponge network | 0.062 | 0.55274 | -0.201 | 0.0967 | 0.32 |
| 36074 | AC009120.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ARHGEF12 | Sponge network | 0.919 | 0 | 0.09 | 0.35693 | 0.32 |
| 36075 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 23 | ACVR1 | Sponge network | 0.532 | 0.00505 | 0.279 | 0.00234 | 0.32 |
| 36076 | GS1-124K5.11 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 14 | SHPRH | Sponge network | 0.494 | 0.00339 | 0.098 | 0.40994 | 0.32 |
| 36077 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 15 | MAP3K12 | Sponge network | -0.709 | 0.18168 | -0.057 | 0.59369 | 0.32 |
| 36078 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | FAM172A | Sponge network | -0.08 | 0.90092 | -0.57 | 0 | 0.32 |
| 36079 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 15 | GLI3 | Sponge network | 0.083 | 0.66405 | -0.615 | 0.01482 | 0.32 |
| 36080 | HOXD-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-3607-3p;hsa-miR-455-3p | 13 | KCNA6 | Sponge network | -0.208 | 0.65592 | -1.531 | 0.00013 | 0.32 |
| 36081 | ZSCAN16-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TPO | Sponge network | -0.342 | 0.02288 | -1.87 | 2.0E-5 | 0.32 |
| 36082 | LINC00476 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-874-3p | 15 | ARID1B | Sponge network | -0.615 | 0.00015 | 0.003 | 0.97503 | 0.32 |
| 36083 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-505-5p | 11 | HBP1 | Sponge network | -0.932 | 0.00329 | -0.454 | 0 | 0.32 |
| 36084 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p | 11 | DNAJB4 | Sponge network | 0.082 | 0.55397 | -0.7 | 1.0E-5 | 0.32 |
| 36085 | LINC00969 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | UBR3 | Sponge network | 0.683 | 1.0E-5 | -0.021 | 0.82774 | 0.32 |
| 36086 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | TSHZ3 | Sponge network | 0.219 | 0.1516 | -0.556 | 0.01516 | 0.32 |
| 36087 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-940 | 33 | MEF2C | Sponge network | 0.214 | 0.18246 | -0.499 | 0.01448 | 0.32 |
| 36088 | CTB-31O20.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | UBR3 | Sponge network | 1.619 | 0 | -0.021 | 0.82774 | 0.32 |
| 36089 | RP11-158K1.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-450b-5p | 10 | ZBTB20 | Sponge network | 0.349 | 0.01695 | -0.122 | 0.57569 | 0.32 |
| 36090 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 15 | FREM2 | Sponge network | -0.358 | 0.17255 | -0.437 | 0.46353 | 0.32 |
| 36091 | TPTEP1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | SPOP | Sponge network | -1.216 | 0 | -0.419 | 1.0E-5 | 0.32 |
| 36092 | RP1-151F17.2 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p | 17 | SYNM | Sponge network | 0.222 | 0.29133 | -2.309 | 1.0E-5 | 0.32 |
| 36093 | RP11-38L15.3 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-590-3p;hsa-miR-935 | 12 | TSHZ3 | Sponge network | 0.344 | 0.3127 | -0.556 | 0.01516 | 0.32 |
| 36094 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | FAT4 | Sponge network | -0.125 | 0.49414 | -0.354 | 0.14694 | 0.32 |
| 36095 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-96-5p | 28 | FOXO3 | Sponge network | 0.614 | 1.0E-5 | -0.04 | 0.72028 | 0.32 |
| 36096 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 27 | RNASEL | Sponge network | -0.737 | 0.06182 | -0.272 | 0.01796 | 0.32 |
| 36097 | SOX2-OT |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | IGSF10 | Sponge network | -1.264 | 6.0E-5 | -1.751 | 1.0E-5 | 0.32 |
| 36098 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 19 | GLIPR1 | Sponge network | 0.082 | 0.55654 | 0.146 | 0.3276 | 0.32 |
| 36099 | HOXB-AS3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-877-5p | 18 | CCND2 | Sponge network | 0.343 | 0.28887 | -0.267 | 0.3112 | 0.32 |
| 36100 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | NUAK1 | Sponge network | 0.559 | 0.00026 | 0.904 | 0 | 0.32 |
| 36101 | CTD-2528L19.6 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-664a-5p | 11 | CBFB | Sponge network | 0.953 | 0 | 1.061 | 0 | 0.32 |
| 36102 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | FAT4 | Sponge network | -4.477 | 0 | -0.354 | 0.14694 | 0.32 |
| 36103 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-485-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SAMHD1 | Sponge network | -0.739 | 0.0574 | 0.072 | 0.62895 | 0.32 |
| 36104 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SYNE1 | Sponge network | 0.349 | 0.01695 | -0.738 | 0.00316 | 0.32 |
| 36105 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | HDAC9 | Sponge network | -1.349 | 0.00017 | -0.755 | 0.00495 | 0.32 |
| 36106 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | PHC3 | Sponge network | 1.923 | 0 | 0.212 | 0.0499 | 0.32 |
| 36107 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PDGFC | Sponge network | -0.323 | 0.01372 | -0.33 | 0.06483 | 0.32 |
| 36108 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 21 | CDHR3 | Sponge network | -0.737 | 0.06182 | -0.977 | 4.0E-5 | 0.32 |
| 36109 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 27 | KIF1B | Sponge network | 1.317 | 0 | -0.118 | 0.32258 | 0.32 |
| 36110 | RP11-122K13.12 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p | 10 | KIF1B | Sponge network | 1.219 | 0 | -0.118 | 0.32258 | 0.32 |
| 36111 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 53 | FAT3 | Sponge network | 0.219 | 0.1516 | -1.601 | 0.00051 | 0.32 |
| 36112 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p | 16 | MYBL1 | Sponge network | 0.799 | 1.0E-5 | 0.494 | 0.02152 | 0.32 |
| 36113 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 51 | SSBP2 | Sponge network | -0.355 | 0.0077 | -1.58 | 0 | 0.32 |
| 36114 | LINC00094 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p | 13 | ITGB3 | Sponge network | 0.253 | 0.09565 | -0.011 | 0.95751 | 0.32 |
| 36115 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-93-5p | 14 | LRRC55 | Sponge network | -0.18 | 0.20343 | -0.026 | 0.93163 | 0.32 |
| 36116 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-625-5p | 24 | CHD5 | Sponge network | -0.342 | 0.02288 | -0.922 | 0.01521 | 0.32 |
| 36117 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-93-5p | 17 | CDHR3 | Sponge network | -0.969 | 2.0E-5 | -0.977 | 4.0E-5 | 0.32 |
| 36118 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429 | 12 | BCL6 | Sponge network | -1.654 | 0.00144 | -0.211 | 0.19489 | 0.32 |
| 36119 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | DACH2 | Sponge network | -0.942 | 0 | -1.635 | 0.00034 | 0.32 |
| 36120 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | PHC3 | Sponge network | 0.392 | 0.0278 | 0.212 | 0.0499 | 0.32 |
| 36121 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DNAJB4 | Sponge network | 0.559 | 0.00026 | -0.7 | 1.0E-5 | 0.32 |
| 36122 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | -0.465 | 0.04703 | -1.751 | 1.0E-5 | 0.32 |
| 36123 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 39 | EBF1 | Sponge network | -2.925 | 0 | -0.384 | 0.08856 | 0.32 |
| 36124 | GHRLOS |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-421;hsa-miR-942-5p | 25 | UNC80 | Sponge network | -1.942 | 0 | -1.934 | 0 | 0.32 |
| 36125 | RP11-403I13.8 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p | 44 | DST | Sponge network | 0.619 | 0.00157 | -0.713 | 0.00096 | 0.32 |
| 36126 | RP11-225B17.2 |
hsa-miR-129-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-590-3p | 14 | CCDC6 | Sponge network | 1.055 | 0 | 0.27 | 0.02555 | 0.32 |
| 36127 | RP3-402G11.26 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29b-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | HLF | Sponge network | -0.254 | 0.19303 | -2.443 | 0 | 0.32 |
| 36128 | RP11-305E6.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | PIK3CA | Sponge network | 1.317 | 0 | 0.195 | 0.06908 | 0.32 |
| 36129 | LINC00702 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 26 | RIMBP2 | Sponge network | -0.737 | 0.06182 | -1.968 | 5.0E-5 | 0.32 |
| 36130 | PART1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-629-5p | 14 | WNK2 | Sponge network | -2.925 | 0 | -0.352 | 0.31066 | 0.32 |
| 36131 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 22 | ZFHX4 | Sponge network | -0.72 | 0.24239 | 0.018 | 0.95574 | 0.32 |
| 36132 | XXyac-YX65C7_A.2 |
hsa-miR-1266-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p | 12 | ABL2 | Sponge network | 0.885 | 1.0E-5 | 0.82 | 0 | 0.32 |
| 36133 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-590-3p | 15 | AKAP6 | Sponge network | 0.199 | 0.38015 | -1.586 | 3.0E-5 | 0.32 |
| 36134 | RP11-727A23.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 1.117 | 0 | 1.083 | 0 | 0.32 |
| 36135 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | TGFBR3 | Sponge network | 0.889 | 9.0E-5 | -1.173 | 1.0E-5 | 0.32 |
| 36136 | HOXA-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CLIP1 | Sponge network | 0.61 | 0.01593 | -0.215 | 0.08684 | 0.32 |
| 36137 | HCG11 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | RET | Sponge network | 0.385 | 0.10067 | -0.833 | 0.03517 | 0.32 |
| 36138 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SEMA3E | Sponge network | 0.61 | 0.01593 | -1.717 | 0.00137 | 0.32 |
| 36139 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | 0.199 | 0.38015 | -1.379 | 0 | 0.32 |
| 36140 | ANKRD10-IT1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 15 | CCDC6 | Sponge network | 1.739 | 0 | 0.27 | 0.02555 | 0.32 |
| 36141 | RP11-527J8.1 |
hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-664a-5p | 11 | SOX11 | Sponge network | 1.324 | 0 | 2.68 | 0 | 0.32 |
| 36142 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | ARID5B | Sponge network | 1.253 | 0 | 0.342 | 0.01676 | 0.32 |
| 36143 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 18 | MLLT10 | Sponge network | -0.227 | 0.31657 | 0.105 | 0.33292 | 0.32 |
| 36144 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PLSCR4 | Sponge network | 0.222 | 0.29133 | -0.474 | 0.03833 | 0.32 |
| 36145 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-5p | 18 | PTPRB | Sponge network | -0.371 | 0.24604 | 0.105 | 0.47122 | 0.32 |
| 36146 | RP11-728F11.4 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-7-1-3p | 12 | ZFHX3 | Sponge network | 0.238 | 0.70544 | 0.389 | 0.01363 | 0.32 |
| 36147 | LIPE-AS1 |
hsa-miR-125a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | ERCC6 | Sponge network | 0.078 | 0.55803 | 0.23 | 0.015 | 0.32 |
| 36148 | LINC00174 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-582-5p | 11 | PIK3C2A | Sponge network | 1.253 | 0 | 0.061 | 0.53776 | 0.32 |
| 36149 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p | 27 | RASSF8 | Sponge network | -1.645 | 0 | -0.122 | 0.65591 | 0.32 |
| 36150 | MIR143HG |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | PAIP2 | Sponge network | -0.739 | 0.0574 | -0.515 | 0 | 0.32 |
| 36151 | RP4-668J24.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-532-3p;hsa-miR-625-5p | 17 | PTPRT | Sponge network | -1.054 | 0.0706 | -0.907 | 0.03727 | 0.32 |
| 36152 | LINC00174 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-582-3p;hsa-miR-582-5p | 18 | BMPR2 | Sponge network | 1.253 | 0 | 0.206 | 0.0635 | 0.32 |
| 36153 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-582-5p;hsa-miR-590-3p | 27 | SPRY3 | Sponge network | 0.225 | 0.30644 | 0.016 | 0.91286 | 0.32 |
| 36154 | PWAR6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 33 | ACSL6 | Sponge network | -1.575 | 6.0E-5 | -1.593 | 0 | 0.32 |
| 36155 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-874-3p | 10 | ST5 | Sponge network | 0.331 | 0.57247 | -0.78 | 0 | 0.32 |
| 36156 | HOTAIRM1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SEMA3C | Sponge network | -0.053 | 0.80685 | -0.272 | 0.31153 | 0.32 |
| 36157 | RP4-758J18.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-330-5p | 11 | AKAP6 | Sponge network | -0.091 | 0.71468 | -1.586 | 3.0E-5 | 0.32 |
| 36158 | EMX2OS |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p | 10 | DUSP1 | Sponge network | -0.777 | 0.1134 | -1.754 | 0 | 0.32 |
| 36159 | PSMG3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-3p | 14 | UNC5C | Sponge network | -0.125 | 0.49414 | -0.178 | 0.40214 | 0.32 |
| 36160 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 30 | CNTN1 | Sponge network | 0.219 | 0.1516 | -2.594 | 0 | 0.32 |
| 36161 | LINC00472 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | -0.842 | 0.01361 | -0.366 | 0.01036 | 0.32 |
| 36162 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | PHC3 | Sponge network | 1.153 | 0.00114 | 0.212 | 0.0499 | 0.32 |
| 36163 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-935 | 14 | PTPRD | Sponge network | -1.483 | 9.0E-5 | -1.005 | 0.00064 | 0.32 |
| 36164 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 23 | IL17RD | Sponge network | -0.223 | 0.47816 | -0.019 | 0.94873 | 0.32 |
| 36165 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-429 | 14 | MYBL1 | Sponge network | -0.246 | 0.35034 | 0.494 | 0.02152 | 0.32 |
| 36166 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-660-5p | 17 | ZFP36L1 | Sponge network | -0.487 | 0.02157 | -0.505 | 8.0E-5 | 0.32 |
| 36167 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 29 | SCN9A | Sponge network | 0.222 | 0.29133 | -0.525 | 0.10593 | 0.32 |
| 36168 | LINC00473 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-505-5p;hsa-miR-629-5p | 12 | WNK2 | Sponge network | -1.203 | 0.03881 | -0.352 | 0.31066 | 0.32 |
| 36169 | RP11-410L14.2 |
hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p | 22 | LPP | Sponge network | -0.052 | 0.76839 | -0.459 | 0.04475 | 0.32 |
| 36170 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 43 | EPHA5 | Sponge network | 0.72 | 0.0001 | -0.957 | 0.01733 | 0.32 |
| 36171 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 12 | ST5 | Sponge network | -1.216 | 0 | -0.78 | 0 | 0.32 |
| 36172 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 16 | SOX11 | Sponge network | 0.619 | 0.00157 | 2.68 | 0 | 0.32 |
| 36173 | LINC00621 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-590-3p | 12 | MRVI1 | Sponge network | 0.131 | 0.8436 | -1.051 | 0.00022 | 0.32 |
| 36174 | CTD-3092A11.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 12 | RPS6KA3 | Sponge network | 0.873 | 0 | 0.256 | 0.02419 | 0.32 |
| 36175 | HAR1A |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b | 11 | MRVI1 | Sponge network | -0.672 | 0.19447 | -1.051 | 0.00022 | 0.32 |
| 36176 | A2M-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | ERG | Sponge network | -0.08 | 0.90092 | 0.002 | 0.99011 | 0.32 |
| 36177 | MIR497HG |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-7-5p | 10 | LZTS1 | Sponge network | -1.439 | 4.0E-5 | 1.664 | 0 | 0.32 |
| 36178 | RP11-867G23.10 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-93-3p;hsa-miR-93-5p | 24 | PAFAH1B1 | Sponge network | -2.531 | 0 | -0.429 | 0 | 0.32 |
| 36179 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-326 | 11 | NF1 | Sponge network | 0.592 | 0.00014 | 0.51 | 0 | 0.32 |
| 36180 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-550a-5p;hsa-miR-96-5p | 11 | HOOK3 | Sponge network | 0.395 | 0.15451 | 0.273 | 0.09957 | 0.32 |
| 36181 | RP11-53O19.1 |
hsa-miR-141-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-940 | 13 | RANBP9 | Sponge network | -0.405 | 0.22013 | -0.191 | 0.04628 | 0.32 |
| 36182 | KB-431C1.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p | 16 | UBR3 | Sponge network | 0.909 | 0 | -0.021 | 0.82774 | 0.32 |
| 36183 | RP11-158K1.3 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | CNTNAP1 | Sponge network | 0.349 | 0.01695 | 0.358 | 0.13727 | 0.32 |
| 36184 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p | 14 | TBX18 | Sponge network | -0.096 | 0.67958 | 0.843 | 0.02945 | 0.32 |
| 36185 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 23 | ZFHX4 | Sponge network | -0.141 | 0.26434 | 0.018 | 0.95574 | 0.32 |
| 36186 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-501-5p | 15 | KIT | Sponge network | -0.405 | 0.22013 | -2.184 | 0 | 0.32 |
| 36187 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p | 12 | MAFB | Sponge network | -2.69 | 0.0001 | -0.023 | 0.89865 | 0.32 |
| 36188 | RP4-669P10.16 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-34a-5p;hsa-miR-424-5p | 12 | ANK2 | Sponge network | -0.301 | 0.06597 | -1.603 | 1.0E-5 | 0.32 |
| 36189 | LINC00865 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429 | 17 | FAM135B | Sponge network | -1.249 | 7.0E-5 | -3.978 | 0 | 0.32 |
| 36190 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-664a-3p | 19 | ITCH | Sponge network | 0.569 | 0.00067 | 0.084 | 0.34058 | 0.32 |
| 36191 | RP1-122P22.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-500a-3p | 10 | ADAMTSL3 | Sponge network | 0.065 | 0.73468 | -1.559 | 0 | 0.32 |
| 36192 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SEMA3E | Sponge network | 0.194 | 0.64278 | -1.717 | 0.00137 | 0.32 |
| 36193 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-455-3p | 19 | RICTOR | Sponge network | 0.453 | 0.01839 | 0.388 | 0.00188 | 0.32 |
| 36194 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-33a-3p | 11 | RSPO2 | Sponge network | -2.039 | 0.03447 | -3.038 | 0 | 0.319 |
| 36195 | LINC00173 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | TAL1 | Sponge network | 0.056 | 0.8494 | -0.462 | 0.00574 | 0.319 |
| 36196 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | EBF3 | Sponge network | 0.72 | 0.0001 | -0.058 | 0.81437 | 0.319 |
| 36197 | RP11-1006G14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-590-3p | 24 | DMD | Sponge network | 0.559 | 0.00026 | -1.506 | 0 | 0.319 |
| 36198 | MAGI2-AS3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 26 | SUFU | Sponge network | -0.129 | 0.57177 | -0.04 | 0.65489 | 0.319 |
| 36199 | RP11-83A24.2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | UBR3 | Sponge network | 0.417 | 0.00445 | -0.021 | 0.82774 | 0.319 |
| 36200 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3E | Sponge network | 0.997 | 0.00066 | -1.717 | 0.00137 | 0.319 |
| 36201 | TMEM9B-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-940 | 12 | SIRT1 | Sponge network | 0.529 | 0.00552 | 0.109 | 0.2593 | 0.319 |
| 36202 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | USP12 | Sponge network | -0.021 | 0.93598 | -0.102 | 0.40768 | 0.319 |
| 36203 | FAM157A |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-326 | 10 | SLC5A3 | Sponge network | 0.813 | 1.0E-5 | 0.493 | 5.0E-5 | 0.319 |
| 36204 | BOLA3-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-7-1-3p | 10 | PCDH18 | Sponge network | -0.246 | 0.35034 | 0.216 | 0.24645 | 0.319 |
| 36205 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-589-5p | 22 | CXXC4 | Sponge network | -0.154 | 0.78939 | -0.433 | 0.21055 | 0.319 |
| 36206 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-651-5p | 29 | PRDM2 | Sponge network | 0.249 | 0.35902 | -0.258 | 0.00721 | 0.319 |
| 36207 | RP11-218M22.1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-96-5p | 11 | WNK2 | Sponge network | 0.197 | 0.25618 | -0.352 | 0.31066 | 0.319 |
| 36208 | CKMT2-AS1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | PAIP2 | Sponge network | 0.082 | 0.55654 | -0.515 | 0 | 0.319 |
| 36209 | WAC-AS1 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-497-5p | 11 | CDK6 | Sponge network | 0.821 | 0 | 0.991 | 0 | 0.319 |
| 36210 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | PRKAA2 | Sponge network | 0.559 | 0.00026 | -2.462 | 0 | 0.319 |
| 36211 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 51 | CUX1 | Sponge network | -1.697 | 0.00889 | -0.039 | 0.6705 | 0.319 |
| 36212 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | DCLK1 | Sponge network | 0.381 | 0.25967 | -1.324 | 0.00014 | 0.319 |
| 36213 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | MYO9A | Sponge network | -2.915 | 0.00015 | -0.147 | 0.32373 | 0.319 |
| 36214 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 11 | PIK3C2A | Sponge network | 1.597 | 0 | 0.061 | 0.53776 | 0.319 |
| 36215 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p | 10 | RARB | Sponge network | -2.076 | 0.00548 | -0.825 | 2.0E-5 | 0.319 |
| 36216 | PART1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NRG3 | Sponge network | -2.925 | 0 | -1.836 | 4.0E-5 | 0.319 |
| 36217 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 17 | NR4A3 | Sponge network | -0.709 | 0.18168 | -1.385 | 0 | 0.319 |
| 36218 | RP11-254F7.2 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-501-5p | 17 | CTIF | Sponge network | 0.176 | 0.37402 | -0.389 | 0.00549 | 0.319 |
| 36219 | LPP-AS2 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-3p | 11 | STAT5B | Sponge network | -0.18 | 0.20343 | -0.182 | 0.1082 | 0.319 |
| 36220 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-539-5p | 24 | KIF1B | Sponge network | -0.473 | 0.07602 | -0.118 | 0.32258 | 0.319 |
| 36221 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 12 | DCN | Sponge network | -0.405 | 0.22013 | -1.288 | 0 | 0.319 |
| 36222 | RP11-384O8.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 20 | GJA1 | Sponge network | -0.738 | 0.21233 | -0.485 | 0.02179 | 0.319 |
| 36223 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-223-3p;hsa-miR-323a-3p;hsa-miR-582-5p | 11 | MTSS1 | Sponge network | 0.792 | 0.0049 | -0.81 | 0.00035 | 0.319 |
| 36224 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-93-5p;hsa-miR-940 | 40 | CADM2 | Sponge network | -0.465 | 0.04703 | -3.782 | 0 | 0.319 |
| 36225 | EMX2OS |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p | 12 | PDZD2 | Sponge network | -0.777 | 0.1134 | -0.909 | 3.0E-5 | 0.319 |
| 36226 | LINC00963 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 55 | BMPR2 | Sponge network | 0.523 | 9.0E-5 | 0.206 | 0.0635 | 0.319 |
| 36227 | GHRLOS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-493-5p | 11 | CITED2 | Sponge network | -1.942 | 0 | -1.545 | 0 | 0.319 |
| 36228 | RP11-337C18.8 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 11 | ABL2 | Sponge network | 0.496 | 0.0011 | 0.82 | 0 | 0.319 |
| 36229 | CTD-3092A11.2 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DOCK4 | Sponge network | 0.873 | 0 | 0.502 | 0.00108 | 0.319 |
| 36230 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 18 | LRRC7 | Sponge network | -0.738 | 0.21233 | -1.341 | 0.01193 | 0.319 |
| 36231 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 32 | BHLHE41 | Sponge network | -0.709 | 0.18168 | 0.353 | 0.12753 | 0.319 |
| 36232 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-454-3p | 17 | CXXC4 | Sponge network | -0.473 | 0.07602 | -0.433 | 0.21055 | 0.319 |
| 36233 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PLSCR4 | Sponge network | -0.031 | 0.89063 | -0.474 | 0.03833 | 0.319 |
| 36234 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p | 17 | ZFHX3 | Sponge network | -2.531 | 0 | 0.389 | 0.01363 | 0.319 |
| 36235 | AC108488.4 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p | 12 | KLF10 | Sponge network | 0.259 | 0.12542 | -0.783 | 0 | 0.319 |
| 36236 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TSC1 | Sponge network | -0.487 | 0.02157 | 0.103 | 0.26071 | 0.319 |
| 36237 | RP11-758N13.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-455-3p | 24 | TCF4 | Sponge network | 2.939 | 3.0E-5 | 0.016 | 0.92271 | 0.319 |
| 36238 | PART1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | GJA1 | Sponge network | -2.925 | 0 | -0.485 | 0.02179 | 0.319 |
| 36239 | SERTAD4-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-505-5p | 13 | ETV1 | Sponge network | -0.932 | 0.00329 | -0.096 | 0.67674 | 0.319 |
| 36240 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p | 10 | PHACTR4 | Sponge network | 0.826 | 1.0E-5 | 0.069 | 0.45203 | 0.319 |
| 36241 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-590-3p | 13 | STARD13 | Sponge network | -1.161 | 0.00591 | -0.187 | 0.27383 | 0.319 |
| 36242 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 24 | PRRX1 | Sponge network | 0.214 | 0.18246 | 1.316 | 0 | 0.319 |
| 36243 | PSMG3-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | PRKCB | Sponge network | -0.125 | 0.49414 | -1.313 | 2.0E-5 | 0.319 |
| 36244 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 0.559 | 0.00026 | 0.51 | 0 | 0.319 |
| 36245 | AC103563.8 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-323a-3p | 10 | MXI1 | Sponge network | -7.551 | 0 | -0.987 | 0 | 0.319 |
| 36246 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-655-3p | 17 | KANK1 | Sponge network | 0.75 | 0.00054 | -0.55 | 0.00134 | 0.319 |
| 36247 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | RCAN2 | Sponge network | 0.619 | 0.00157 | -0.33 | 0.15172 | 0.319 |
| 36248 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | BCL11A | Sponge network | -0.72 | 0.24239 | -0.932 | 0.0063 | 0.319 |
| 36249 | CTA-204B4.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | RASSF8 | Sponge network | 0.765 | 7.0E-5 | -0.122 | 0.65591 | 0.319 |
| 36250 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ZFHX3 | Sponge network | 1.308 | 0 | 0.389 | 0.01363 | 0.319 |
| 36251 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 20 | TIMP3 | Sponge network | -0.141 | 0.26434 | -0.546 | 0.02307 | 0.319 |
| 36252 | HOTAIRM1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 39 | RUNX1T1 | Sponge network | -0.053 | 0.80685 | -0.344 | 0.2374 | 0.319 |
| 36253 | LINC01018 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | ERG | Sponge network | -2.915 | 0.00015 | 0.002 | 0.99011 | 0.319 |
| 36254 | RP11-999E24.3 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p | 12 | SYT4 | Sponge network | -0.693 | 0.05629 | -3.207 | 0 | 0.319 |
| 36255 | LINC01001 |
hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p | 10 | AKAP11 | Sponge network | 1.055 | 0 | 0.196 | 0.09825 | 0.319 |
| 36256 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | NALCN | Sponge network | 0.101 | 0.60919 | 1.068 | 0.00237 | 0.319 |
| 36257 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-96-5p | 18 | NTRK3 | Sponge network | -0.214 | 0.44248 | -1.77 | 3.0E-5 | 0.319 |
| 36258 | RP11-158K1.3 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-940 | 41 | PPARA | Sponge network | 0.349 | 0.01695 | -0.379 | 0.00548 | 0.319 |
| 36259 | SH3RF3-AS1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 17 | BRD3 | Sponge network | 0.889 | 9.0E-5 | 0.37 | 0.00013 | 0.319 |
| 36260 | RP11-158K1.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | 0.349 | 0.01695 | -0.704 | 0.00106 | 0.319 |
| 36261 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-769-5p | 16 | SYNPO2 | Sponge network | 0.381 | 0.25967 | -2.342 | 0 | 0.319 |
| 36262 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TBX18 | Sponge network | 0.72 | 0.0001 | 0.843 | 0.02945 | 0.319 |
| 36263 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | DAB2 | Sponge network | -1.582 | 8.0E-5 | 0.431 | 0.0113 | 0.319 |
| 36264 | RP11-6O2.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-940 | 25 | CREB1 | Sponge network | 0.331 | 0.57247 | 0.203 | 0.00537 | 0.319 |
| 36265 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-93-5p | 16 | NIN | Sponge network | -1.654 | 0.00144 | -0.253 | 0.13794 | 0.319 |
| 36266 | FAM13A-AS1 |
hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 11 | AHCYL1 | Sponge network | -0.83 | 0 | -0.479 | 0 | 0.319 |
| 36267 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | NEDD4 | Sponge network | 0.272 | 0.44692 | 0.02 | 0.89834 | 0.319 |
| 36268 | CTD-2666L21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-486-5p | 11 | NRK | Sponge network | 0.368 | 0.20093 | 0.656 | 0.19217 | 0.319 |
| 36269 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 15 | IGSF10 | Sponge network | 0.174 | 0.30034 | -1.751 | 1.0E-5 | 0.319 |
| 36270 | WDFY3-AS2 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | PTCH1 | Sponge network | -0.942 | 0 | -0.1 | 0.6179 | 0.319 |
| 36271 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PBRM1 | Sponge network | 0.532 | 0.00505 | -0.029 | 0.78601 | 0.319 |
| 36272 | CTD-2314G24.2 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | RPS6KA6 | Sponge network | 0.246 | 0.59912 | -2.989 | 0 | 0.319 |
| 36273 | CROCCP2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p | 18 | ARHGEF12 | Sponge network | 0.79 | 0 | 0.09 | 0.35693 | 0.319 |
| 36274 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 14 | RCAN2 | Sponge network | 0.614 | 1.0E-5 | -0.33 | 0.15172 | 0.319 |
| 36275 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 28 | CYLD | Sponge network | -1.697 | 0.00889 | -0.367 | 0.00171 | 0.319 |
| 36276 | RP11-597D13.9 |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-5p | 11 | MLLT11 | Sponge network | 1.302 | 7.0E-5 | -0.661 | 0.01733 | 0.319 |
| 36277 | AC005562.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | NOTCH2 | Sponge network | 0.488 | 0.00084 | 0.138 | 0.35163 | 0.319 |
| 36278 | LINC00173 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-7-1-3p | 10 | INPP4A | Sponge network | 0.056 | 0.8494 | 0.07 | 0.53563 | 0.319 |
| 36279 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-7-1-3p;hsa-miR-940 | 36 | WDFY3 | Sponge network | -4.546 | 0 | -0.201 | 0.0967 | 0.319 |
| 36280 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-331-3p;hsa-miR-375 | 14 | QKI | Sponge network | 1.969 | 0.00316 | -0.333 | 0.04111 | 0.319 |
| 36281 | RP1-122P22.2 |
hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-532-5p | 11 | FBXO32 | Sponge network | 0.065 | 0.73468 | -0.495 | 0.07561 | 0.319 |
| 36282 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-671-5p | 15 | RBMS3 | Sponge network | 0.453 | 0.01839 | -0.519 | 0.03551 | 0.319 |
| 36283 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-5p | 21 | TMTC3 | Sponge network | 1.147 | 0 | 0.123 | 0.22189 | 0.319 |
| 36284 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-29a-5p | 10 | SYNE1 | Sponge network | 0.453 | 0.01839 | -0.738 | 0.00316 | 0.319 |
| 36285 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 14 | NALCN | Sponge network | 0.72 | 0.0001 | 1.068 | 0.00237 | 0.319 |
| 36286 | AC034220.3 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-655-3p | 20 | LATS1 | Sponge network | 0.309 | 0.07304 | 0.109 | 0.34208 | 0.319 |
| 36287 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 26 | PDGFRA | Sponge network | -0.246 | 0.35034 | -0.372 | 0.0037 | 0.319 |
| 36288 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-429;hsa-miR-550a-3p | 10 | AFF4 | Sponge network | 2.364 | 0.00134 | -0.266 | 0.01565 | 0.319 |
| 36289 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-664a-3p | 13 | SOX11 | Sponge network | 1.316 | 0 | 2.68 | 0 | 0.319 |
| 36290 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-96-5p | 13 | FBXO32 | Sponge network | -0.418 | 0.00068 | -0.495 | 0.07561 | 0.319 |
| 36291 | HOXA-AS2 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | RIT1 | Sponge network | 0.61 | 0.01593 | -0.296 | 0.00054 | 0.319 |
| 36292 | FZD10-AS1 |
hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SEMA3E | Sponge network | -0.727 | 0.04726 | -1.717 | 0.00137 | 0.319 |
| 36293 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 28 | TACC1 | Sponge network | 0.862 | 0.06213 | -0.362 | 0.05406 | 0.319 |
| 36294 | PART1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | CBFA2T3 | Sponge network | -2.925 | 0 | -1.221 | 1.0E-5 | 0.319 |
| 36295 | RP4-605O3.4 |
hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-590-3p | 10 | ZMYND11 | Sponge network | 0.262 | 0.0475 | -0.2 | 0.05449 | 0.319 |
| 36296 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p | 19 | JAZF1 | Sponge network | 0.389 | 0.04293 | -0.377 | 0.02677 | 0.319 |
| 36297 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | FOXO1 | Sponge network | -1.203 | 0.03881 | -0.141 | 0.33874 | 0.319 |
| 36298 | U91328.19 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | GALNT13 | Sponge network | -0.303 | 0.21002 | -0.857 | 0.07439 | 0.319 |
| 36299 | MIR181A2HG |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-582-5p | 11 | CREB1 | Sponge network | 1.822 | 1.0E-5 | 0.203 | 0.00537 | 0.319 |
| 36300 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-589-5p | 13 | MN1 | Sponge network | -0.969 | 2.0E-5 | -0.358 | 0.24172 | 0.319 |
| 36301 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 44 | ERBB4 | Sponge network | -0.49 | 0.04946 | -1.975 | 2.0E-5 | 0.319 |
| 36302 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 53 | WDFY3 | Sponge network | 0.335 | 0.20596 | -0.201 | 0.0967 | 0.319 |
| 36303 | FAM13A-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-96-5p | 22 | CYLD | Sponge network | -0.83 | 0 | -0.367 | 0.00171 | 0.319 |
| 36304 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 18 | LATS2 | Sponge network | -1.203 | 0.03881 | 0.159 | 0.2304 | 0.319 |
| 36305 | MEG3 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 11 | UBE2QL1 | Sponge network | 0.249 | 0.35902 | -2.417 | 0 | 0.319 |
| 36306 | LIPE-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-96-5p | 27 | ABI2 | Sponge network | 0.078 | 0.55803 | 0.175 | 0.14787 | 0.319 |
| 36307 | RP11-326G21.1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p | 11 | CHD5 | Sponge network | -0.021 | 0.93598 | -0.922 | 0.01521 | 0.319 |
| 36308 | RP11-77H9.2 |
hsa-let-7a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | CHD2 | Sponge network | 1.677 | 0 | 0.049 | 0.55371 | 0.319 |
| 36309 | C20orf166-AS1 |
hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-5p | 14 | ARHGAP29 | Sponge network | -2.69 | 0.0001 | 0.131 | 0.42953 | 0.319 |
| 36310 | LINC00926 |
hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-96-5p | 10 | MED12L | Sponge network | 0.756 | 0.00739 | -0.194 | 0.55299 | 0.319 |
| 36311 | U91328.19 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p | 16 | SPG20 | Sponge network | -0.303 | 0.21002 | -1.16 | 0 | 0.319 |
| 36312 | RP11-497H16.9 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PIK3CA | Sponge network | 0.545 | 0.00064 | 0.195 | 0.06908 | 0.319 |
| 36313 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PRKAB2 | Sponge network | 0.415 | 0.14657 | -0.367 | 0.01371 | 0.319 |
| 36314 | LINC00957 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-3p | 22 | GRIA3 | Sponge network | -0.421 | 0.0114 | -1.956 | 0 | 0.319 |
| 36315 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 15 | ZFHX3 | Sponge network | -0.557 | 0.11135 | 0.389 | 0.01363 | 0.319 |
| 36316 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-576-5p | 18 | NCAM2 | Sponge network | 0.056 | 0.81195 | -0.482 | 0.22565 | 0.319 |
| 36317 | RP11-156P1.3 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | TPO | Sponge network | 0.214 | 0.18246 | -1.87 | 2.0E-5 | 0.319 |
| 36318 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TBX18 | Sponge network | -0.727 | 0.04726 | 0.843 | 0.02945 | 0.319 |
| 36319 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-942-5p;hsa-miR-96-5p | 14 | PCDH17 | Sponge network | 0.622 | 0.00015 | 0.731 | 2.0E-5 | 0.319 |
| 36320 | MAGI2-AS3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | CREB1 | Sponge network | -0.129 | 0.57177 | 0.203 | 0.00537 | 0.319 |
| 36321 | AC009948.5 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 15 | CELF4 | Sponge network | 0.532 | 0.00505 | -1.135 | 0.00344 | 0.319 |
| 36322 | RP11-59H7.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p | 10 | BAZ2B | Sponge network | 2.163 | 0 | -0.276 | 0.02833 | 0.319 |
| 36323 | LINC01006 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-5p | 26 | ERBB4 | Sponge network | -0.702 | 0.04814 | -1.975 | 2.0E-5 | 0.319 |
| 36324 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 33 | EPHA5 | Sponge network | 0.395 | 0.15451 | -0.957 | 0.01733 | 0.319 |
| 36325 | AC108488.4 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-484 | 19 | ABL1 | Sponge network | 0.259 | 0.12542 | -0.215 | 0.07804 | 0.319 |
| 36326 | RP11-389C8.2 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-671-5p | 23 | GRIK3 | Sponge network | 0.727 | 3.0E-5 | -3.208 | 0 | 0.319 |
| 36327 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 15 | GALNT13 | Sponge network | 0.082 | 0.55654 | -0.857 | 0.07439 | 0.319 |
| 36328 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p | 10 | CSRNP1 | Sponge network | -0.583 | 0.14589 | -1.448 | 0 | 0.319 |
| 36329 | RP11-6N17.3 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-5p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-455-5p | 19 | KIF1B | Sponge network | 0.108 | 0.69556 | -0.118 | 0.32258 | 0.319 |
| 36330 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-590-3p | 11 | DAB2 | Sponge network | -0.323 | 0.01372 | 0.431 | 0.0113 | 0.319 |
| 36331 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p | 13 | ABCA10 | Sponge network | -0.611 | 0.09607 | -0.571 | 0.03674 | 0.319 |
| 36332 | AC009120.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-625-5p;hsa-miR-629-3p | 18 | PTPN14 | Sponge network | 0.919 | 0 | 0.144 | 0.34595 | 0.319 |
| 36333 | RP11-166D19.1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ZNF844 | Sponge network | -0.576 | 0.10121 | -0.966 | 0.00035 | 0.319 |
| 36334 | HOTAIRM1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 20 | CADM1 | Sponge network | -0.053 | 0.80685 | -0.9 | 0.00084 | 0.319 |
| 36335 | RP11-434H6.7 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26b-5p | 10 | CTDSPL | Sponge network | 0.358 | 0.14302 | 0.085 | 0.45121 | 0.319 |
| 36336 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 17 | GRIN2A | Sponge network | -0.13 | 0.48734 | -2.161 | 0 | 0.319 |
| 36337 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | MLLT10 | Sponge network | 0.643 | 0 | 0.105 | 0.33292 | 0.319 |
| 36338 | RP11-244H3.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | LATS2 | Sponge network | 0.659 | 3.0E-5 | 0.159 | 0.2304 | 0.319 |
| 36339 | MIR600HG |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-3p | 13 | BCL9 | Sponge network | 0.594 | 0.41522 | 0.321 | 0.00267 | 0.319 |
| 36340 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | ITCH | Sponge network | 0.67 | 0.00048 | 0.084 | 0.34058 | 0.319 |
| 36341 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | NUAK1 | Sponge network | -0.563 | 0.02545 | 0.904 | 0 | 0.319 |
| 36342 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | RBL2 | Sponge network | 0.082 | 0.55654 | -0.282 | 0.00254 | 0.319 |
| 36343 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 23 | FOXP1 | Sponge network | 0.411 | 0.1335 | 0.042 | 0.74368 | 0.319 |
| 36344 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-590-3p | 11 | LRIG1 | Sponge network | -1.375 | 0.08642 | -0.537 | 0.00588 | 0.319 |
| 36345 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 38 | CHD5 | Sponge network | 0.532 | 0.00505 | -0.922 | 0.01521 | 0.319 |
| 36346 | RAMP2-AS1 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | GABRB2 | Sponge network | -0.513 | 0.04703 | -1.841 | 0.00029 | 0.319 |
| 36347 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | AFAP1L2 | Sponge network | -0.487 | 0.02157 | -0.284 | 0.15434 | 0.319 |
| 36348 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PCDH17 | Sponge network | 0.225 | 0.30644 | 0.731 | 2.0E-5 | 0.319 |
| 36349 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 14 | CNTNAP1 | Sponge network | -0.72 | 0.24239 | 0.358 | 0.13727 | 0.319 |
| 36350 | AGAP11 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-590-5p | 15 | CSMD1 | Sponge network | -1.645 | 0 | -0.63 | 0.18881 | 0.319 |
| 36351 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 20 | TMTC3 | Sponge network | 0.953 | 0 | 0.123 | 0.22189 | 0.319 |
| 36352 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 11 | GPAM | Sponge network | 0.79 | 0 | 0.142 | 0.29732 | 0.319 |
| 36353 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-93-3p | 25 | NCOA1 | Sponge network | -0.896 | 0.00099 | -0.432 | 0 | 0.319 |
| 36354 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-624-5p | 12 | PCDH8 | Sponge network | 0.253 | 0.09565 | -0.997 | 0.05366 | 0.319 |
| 36355 | BCYRN1 |
hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | ZNF382 | Sponge network | 0.311 | 0.10344 | -0.494 | 0.03831 | 0.319 |
| 36356 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | PRKAR1A | Sponge network | -0.487 | 0.02157 | -0.063 | 0.50314 | 0.319 |
| 36357 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 18 | RARB | Sponge network | -0.125 | 0.49414 | -0.825 | 2.0E-5 | 0.319 |
| 36358 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-93-5p | 13 | TRIM3 | Sponge network | -0.096 | 0.67958 | -0.577 | 0 | 0.319 |
| 36359 | RP5-1085F17.3 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 13 | ATM | Sponge network | 0.541 | 0.00125 | 0.178 | 0.21568 | 0.319 |
| 36360 | FAM66C |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 30 | ETV1 | Sponge network | -0.901 | 0.00019 | -0.096 | 0.67674 | 0.319 |
| 36361 | RP4-791M13.3 |
hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 10 | LRRC55 | Sponge network | 0.419 | 0.0319 | -0.026 | 0.93163 | 0.319 |
| 36362 | RP11-531A24.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p | 11 | THSD7A | Sponge network | 0.75 | 0.00054 | 0.242 | 0.25154 | 0.319 |
| 36363 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 31 | PRKAA2 | Sponge network | 0.197 | 0.25618 | -2.462 | 0 | 0.319 |
| 36364 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-455-5p;hsa-miR-93-3p | 18 | DOCK4 | Sponge network | -0.162 | 0.47687 | 0.502 | 0.00108 | 0.319 |
| 36365 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 40 | CBX5 | Sponge network | -0.513 | 0.04703 | 0.165 | 0.24446 | 0.319 |
| 36366 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-590-3p | 15 | DICER1 | Sponge network | 1.089 | 0 | 0.042 | 0.674 | 0.319 |
| 36367 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-629-3p | 26 | EBF1 | Sponge network | -1.645 | 0 | -0.384 | 0.08856 | 0.319 |
| 36368 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | RABEP1 | Sponge network | -2.452 | 0 | -0.066 | 0.43142 | 0.319 |
| 36369 | RP4-791M13.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 36 | DTNA | Sponge network | 0.419 | 0.0319 | -1.648 | 1.0E-5 | 0.319 |
| 36370 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-3127-5p;hsa-miR-532-3p;hsa-miR-589-5p | 15 | RPS6KA2 | Sponge network | 0.101 | 0.60919 | -0.57 | 0.00013 | 0.319 |
| 36371 | MIR143HG |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | INPP4B | Sponge network | -0.739 | 0.0574 | -0.097 | 0.60503 | 0.319 |
| 36372 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 41 | CHD5 | Sponge network | 0.72 | 0.0001 | -0.922 | 0.01521 | 0.319 |
| 36373 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 17 | MOB1B | Sponge network | 1.317 | 0 | 0.197 | 0.15266 | 0.319 |
| 36374 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DACT1 | Sponge network | 0.225 | 0.30644 | 0.783 | 0.00072 | 0.319 |
| 36375 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-942-5p | 36 | UNC80 | Sponge network | -0.777 | 0.16667 | -1.934 | 0 | 0.319 |
| 36376 | MIR143HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ARNT | Sponge network | -0.739 | 0.0574 | -0.08 | 0.26073 | 0.319 |
| 36377 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 19 | NRXN1 | Sponge network | 0.174 | 0.30034 | -3.778 | 0 | 0.319 |
| 36378 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-337-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-628-5p | 16 | BMPR2 | Sponge network | 2.368 | 2.0E-5 | 0.206 | 0.0635 | 0.319 |
| 36379 | CD27-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-624-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | WWTR1 | Sponge network | 0.177 | 0.16681 | -0.345 | 0.11997 | 0.319 |
| 36380 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | EPHA5 | Sponge network | -2.915 | 0.00015 | -0.957 | 0.01733 | 0.319 |
| 36381 | AC034220.3 |
hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 12 | ITCH | Sponge network | 0.309 | 0.07304 | 0.084 | 0.34058 | 0.319 |
| 36382 | BOLA3-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-429 | 12 | STARD13 | Sponge network | -0.246 | 0.35034 | -0.187 | 0.27383 | 0.319 |
| 36383 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | PEG3 | Sponge network | 0.75 | 0.00054 | -1.74 | 0 | 0.319 |
| 36384 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 38 | SSBP2 | Sponge network | -0.246 | 0.35034 | -1.58 | 0 | 0.319 |
| 36385 | MIR22HG |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 16 | FLNA | Sponge network | -1.047 | 0 | -0.771 | 0.01377 | 0.319 |
| 36386 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-27a-3p | 11 | EBF3 | Sponge network | -0.726 | 0.00132 | -0.058 | 0.81437 | 0.319 |
| 36387 | FAM13A-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | TPO | Sponge network | -0.83 | 0 | -1.87 | 2.0E-5 | 0.319 |
| 36388 | CTD-2245F17.3 |
hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-502-5p | 12 | BNC2 | Sponge network | -0.421 | 0.12556 | -0.491 | 0.09988 | 0.319 |
| 36389 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-484 | 11 | ITGA5 | Sponge network | -0.384 | 0.40652 | 0.452 | 0.03524 | 0.319 |
| 36390 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | ARNT2 | Sponge network | 0.174 | 0.30034 | -0.332 | 0.25851 | 0.319 |
| 36391 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 10 | SPTBN4 | Sponge network | 0.331 | 0.57247 | -0.786 | 0.01281 | 0.319 |
| 36392 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p | 23 | HECA | Sponge network | 0.95 | 0.15943 | -0.217 | 0.03463 | 0.319 |
| 36393 | LINC00648 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | PRKAA2 | Sponge network | 0.633 | 0.51678 | -2.462 | 0 | 0.319 |
| 36394 | PGM5-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 11 | NIN | Sponge network | -4.546 | 0 | -0.253 | 0.13794 | 0.319 |
| 36395 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 51 | FOXP1 | Sponge network | -0.49 | 0.04946 | 0.042 | 0.74368 | 0.319 |
| 36396 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | FBXO32 | Sponge network | -1.425 | 0.04204 | -0.495 | 0.07561 | 0.319 |
| 36397 | MIR22HG |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p | 11 | PRDM5 | Sponge network | -1.047 | 0 | -1.09 | 0.00014 | 0.319 |
| 36398 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-629-3p | 24 | EBF1 | Sponge network | -0.773 | 0.04073 | -0.384 | 0.08856 | 0.319 |
| 36399 | PCA3 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -0.709 | 0.18168 | -2.417 | 0 | 0.319 |
| 36400 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-214-5p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-935 | 51 | SMAD4 | Sponge network | 0.083 | 0.66405 | -0.313 | 0.00413 | 0.319 |
| 36401 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p | 11 | DCLK1 | Sponge network | 0.331 | 0.57247 | -1.324 | 0.00014 | 0.319 |
| 36402 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 32 | NOTCH2 | Sponge network | 0.494 | 0.00339 | 0.138 | 0.35163 | 0.319 |
| 36403 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | SLC6A15 | Sponge network | -1.697 | 0.00889 | -2.373 | 2.0E-5 | 0.319 |
| 36404 | PSMG3-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 12 | SORCS1 | Sponge network | -0.125 | 0.49414 | -3.492 | 0 | 0.319 |
| 36405 | RP11-497H16.9 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | PIK3R1 | Sponge network | 0.545 | 0.00064 | -0.133 | 0.36854 | 0.319 |
| 36406 | HCG11 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p | 11 | FNBP1 | Sponge network | 0.385 | 0.10067 | -0.969 | 4.0E-5 | 0.319 |
| 36407 | PCA3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DACT1 | Sponge network | -0.709 | 0.18168 | 0.783 | 0.00072 | 0.319 |
| 36408 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | AHNAK | Sponge network | -1.161 | 0.00591 | -1.207 | 0 | 0.319 |
| 36409 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-664a-3p | 12 | ITCH | Sponge network | 0.706 | 0 | 0.084 | 0.34058 | 0.319 |
| 36410 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p | 17 | TIMP3 | Sponge network | 0.082 | 0.55397 | -0.546 | 0.02307 | 0.319 |
| 36411 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-484 | 12 | GREM1 | Sponge network | 0.687 | 0.02753 | 0.902 | 0.01691 | 0.319 |
| 36412 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-455-3p;hsa-miR-484 | 18 | ZFX | Sponge network | 0.858 | 3.0E-5 | 0.09 | 0.42563 | 0.319 |
| 36413 | RP11-758N13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-424-5p | 15 | ANK2 | Sponge network | 2.939 | 3.0E-5 | -1.603 | 1.0E-5 | 0.319 |
| 36414 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-93-5p | 30 | TACC1 | Sponge network | 0.381 | 0.25967 | -0.362 | 0.05406 | 0.319 |
| 36415 | MALAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p | 17 | PRKAA1 | Sponge network | 0.782 | 0.00016 | 0.317 | 0.0031 | 0.319 |
| 36416 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-590-3p | 19 | MLLT10 | Sponge network | 0.174 | 0.30034 | 0.105 | 0.33292 | 0.319 |
| 36417 | NNT-AS1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-409-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 13 | BAZ2B | Sponge network | 0.799 | 1.0E-5 | -0.276 | 0.02833 | 0.319 |
| 36418 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 20 | SETBP1 | Sponge network | 0.222 | 0.29133 | -1.302 | 1.0E-5 | 0.319 |
| 36419 | CTD-2554C21.2 |
hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-96-5p | 10 | ARHGAP29 | Sponge network | -1.654 | 0.00144 | 0.131 | 0.42953 | 0.319 |
| 36420 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 21 | DOCK4 | Sponge network | 0.395 | 0.15451 | 0.502 | 0.00108 | 0.319 |
| 36421 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | SEMA3E | Sponge network | 1.757 | 0 | -1.717 | 0.00137 | 0.319 |
| 36422 | ACTA2-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-484;hsa-miR-590-3p | 11 | FBXO11 | Sponge network | 0.194 | 0.64278 | 0.142 | 0.04938 | 0.319 |
| 36423 | CTB-113P19.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-629-3p | 11 | IKZF2 | Sponge network | 0.906 | 0.31715 | -0.284 | 0.1942 | 0.318 |
| 36424 | DNM1P35 |
hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-532-3p | 11 | SYNM | Sponge network | 0.051 | 0.83388 | -2.309 | 1.0E-5 | 0.318 |
| 36425 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-7-1-3p | 30 | CREB3L2 | Sponge network | -0.246 | 0.35034 | -0.525 | 2.0E-5 | 0.318 |
| 36426 | ERVH48-1 |
hsa-let-7d-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p | 10 | PRDM2 | Sponge network | 1.04 | 0.00023 | -0.258 | 0.00721 | 0.318 |
| 36427 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-577 | 14 | HIP1 | Sponge network | 0.389 | 0.04293 | 0.774 | 0 | 0.318 |
| 36428 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 41 | NTRK2 | Sponge network | 0.082 | 0.55654 | -0.736 | 0.06495 | 0.318 |
| 36429 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 49 | IL6ST | Sponge network | -0.031 | 0.89063 | -0.402 | 0.00676 | 0.318 |
| 36430 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | ATRX | Sponge network | 1.317 | 0 | 0.261 | 0.04408 | 0.318 |
| 36431 | RP4-613B23.1 |
hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-27a-3p;hsa-miR-33a-3p | 10 | MED12L | Sponge network | -0.309 | 0.1145 | -0.194 | 0.55299 | 0.318 |
| 36432 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 56 | NTRK3 | Sponge network | 0.61 | 0.01593 | -1.77 | 3.0E-5 | 0.318 |
| 36433 | SH3BP5-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-589-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.267 | 0.2937 | -0.57 | 0 | 0.318 |
| 36434 | RP11-473I1.9 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p | 10 | MAT2A | Sponge network | 0.683 | 1.0E-5 | -0.073 | 0.36195 | 0.318 |
| 36435 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | EPHA5 | Sponge network | -1.203 | 0.03881 | -0.957 | 0.01733 | 0.318 |
| 36436 | RP11-434B12.1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 14 | KLF10 | Sponge network | -0.031 | 0.89063 | -0.783 | 0 | 0.318 |
| 36437 | CTC-296K1.4 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-5p | 15 | PAFAH1B1 | Sponge network | -2.076 | 0.00548 | -0.429 | 0 | 0.318 |
| 36438 | MYLK-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-301a-3p | 13 | SUFU | Sponge network | -1.331 | 3.0E-5 | -0.04 | 0.65489 | 0.318 |
| 36439 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p | 39 | BMPR2 | Sponge network | -0.318 | 0.65847 | 0.206 | 0.0635 | 0.318 |
| 36440 | RP11-244H3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 13 | HMCN1 | Sponge network | 0.659 | 3.0E-5 | 0.809 | 0.00544 | 0.318 |
| 36441 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 60 | NFIB | Sponge network | 0.997 | 0.00066 | 0.023 | 0.90795 | 0.318 |
| 36442 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 26 | TCF4 | Sponge network | 0.796 | 4.0E-5 | 0.016 | 0.92271 | 0.318 |
| 36443 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577 | 18 | GREM1 | Sponge network | 1.757 | 0 | 0.902 | 0.01691 | 0.318 |
| 36444 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p | 26 | PBX1 | Sponge network | -0.176 | 0.59692 | -1.206 | 0 | 0.318 |
| 36445 | RP11-104L21.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p | 12 | RUNX1T1 | Sponge network | 0.527 | 0.16506 | -0.344 | 0.2374 | 0.318 |
| 36446 | AC006116.24 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-577 | 18 | RIMS2 | Sponge network | -0.473 | 0.07602 | -1.365 | 0.00208 | 0.318 |
| 36447 | LINC00092 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-503-5p | 11 | ABL1 | Sponge network | -1.886 | 0 | -0.215 | 0.07804 | 0.318 |
| 36448 | RP11-53O19.3 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-497-5p | 11 | PAFAH1B2 | Sponge network | 1.205 | 0 | 0.318 | 0.0001 | 0.318 |
| 36449 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 37 | ST6GAL2 | Sponge network | -2.925 | 0 | -1.201 | 0.00597 | 0.318 |
| 36450 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 25 | ESR1 | Sponge network | 0.657 | 4.0E-5 | -1.035 | 3.0E-5 | 0.318 |
| 36451 | LINC00963 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 21 | YAP1 | Sponge network | 0.523 | 9.0E-5 | 0.22 | 0.11836 | 0.318 |
| 36452 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p | 25 | CADM1 | Sponge network | -0.342 | 0.02288 | -0.9 | 0.00084 | 0.318 |
| 36453 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-671-5p | 21 | PPP2R5C | Sponge network | 0.101 | 0.60919 | -0.314 | 3.0E-5 | 0.318 |
| 36454 | RP11-1006G14.4 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | DICER1 | Sponge network | 0.559 | 0.00026 | 0.042 | 0.674 | 0.318 |
| 36455 | RP4-669H2.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p | 14 | RICTOR | Sponge network | 1.711 | 0.00019 | 0.388 | 0.00188 | 0.318 |
| 36456 | RP5-1021I20.1 |
hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-539-5p | 12 | SYNPO2 | Sponge network | -0.785 | 0.00035 | -2.342 | 0 | 0.318 |
| 36457 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 10 | FAM133A | Sponge network | -0.323 | 0.01372 | 0.549 | 0.29009 | 0.318 |
| 36458 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | SPG20 | Sponge network | -0.355 | 0.0077 | -1.16 | 0 | 0.318 |
| 36459 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | EDA2R | Sponge network | -0.727 | 0.04726 | -0.587 | 0.01598 | 0.318 |
| 36460 | RP11-706C16.7 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577 | 19 | DTNA | Sponge network | 0.357 | 0.72407 | -1.648 | 1.0E-5 | 0.318 |
| 36461 | RP1-39G22.7 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | BCL9 | Sponge network | 0.439 | 0.00313 | 0.321 | 0.00267 | 0.318 |
| 36462 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-590-3p | 11 | ABCA10 | Sponge network | 0.083 | 0.66405 | -0.571 | 0.03674 | 0.318 |
| 36463 | PSMD5-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | PHF3 | Sponge network | 0.569 | 0.00067 | 0.126 | 0.19652 | 0.318 |
| 36464 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | ACVR2A | Sponge network | -1.575 | 6.0E-5 | -0.409 | 0.00039 | 0.318 |
| 36465 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-93-5p | 29 | PDGFRA | Sponge network | 0.056 | 0.81195 | -0.372 | 0.0037 | 0.318 |
| 36466 | RP11-182J1.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-424-5p;hsa-miR-7-5p | 15 | BTG2 | Sponge network | -0.187 | 0.23787 | -1.763 | 0 | 0.318 |
| 36467 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | HIP1 | Sponge network | -1.582 | 8.0E-5 | 0.774 | 0 | 0.318 |
| 36468 | AC108488.4 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-93-5p | 13 | CDHR3 | Sponge network | 0.259 | 0.12542 | -0.977 | 4.0E-5 | 0.318 |
| 36469 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-3614-5p;hsa-miR-629-3p | 19 | GAS7 | Sponge network | -2.549 | 0.00528 | -0.555 | 0.01602 | 0.318 |
| 36470 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | FGF5 | Sponge network | 0.862 | 0.06213 | 0.549 | 0.28659 | 0.318 |
| 36471 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-576-5p | 10 | EPHA5 | Sponge network | -2.076 | 0.00548 | -0.957 | 0.01733 | 0.318 |
| 36472 | RP11-517B11.7 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-339-5p;hsa-miR-495-3p | 12 | TSC1 | Sponge network | 0.706 | 0 | 0.103 | 0.26071 | 0.318 |
| 36473 | SH3BP5-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | DLG1 | Sponge network | 0.267 | 0.2937 | -0.014 | 0.8926 | 0.318 |
| 36474 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p | 18 | ABCC9 | Sponge network | 0.177 | 0.16681 | -0.466 | 0.14121 | 0.318 |
| 36475 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 22 | THBS1 | Sponge network | 0.331 | 0.57247 | 0.741 | 0.00386 | 0.318 |
| 36476 | RP11-672A2.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-339-5p | 12 | NTRK2 | Sponge network | 1.344 | 0 | -0.736 | 0.06495 | 0.318 |
| 36477 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p | 14 | SLC6A15 | Sponge network | -0.154 | 0.78939 | -2.373 | 2.0E-5 | 0.318 |
| 36478 | CTC-490E21.10 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-93-3p | 22 | IL6ST | Sponge network | 1.46 | 1.0E-5 | -0.402 | 0.00676 | 0.318 |
| 36479 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-624-5p | 11 | HMCN1 | Sponge network | -0.899 | 6.0E-5 | 0.809 | 0.00544 | 0.318 |
| 36480 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | CDH10 | Sponge network | -0.738 | 0.21233 | -2.397 | 0 | 0.318 |
| 36481 | LINC00886 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-500a-5p;hsa-miR-590-5p | 13 | ABCA10 | Sponge network | -0.371 | 0.24604 | -0.571 | 0.03674 | 0.318 |
| 36482 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p | 14 | SLITRK3 | Sponge network | 0.331 | 0.57247 | -3.328 | 0 | 0.318 |
| 36483 | GHRLOS |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-493-5p | 15 | ASTN1 | Sponge network | -1.942 | 0 | -2.389 | 2.0E-5 | 0.318 |
| 36484 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-5p | 12 | LRRK2 | Sponge network | 0.331 | 0.57247 | -1.136 | 1.0E-5 | 0.318 |
| 36485 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 21 | KAT6B | Sponge network | -0.842 | 0.01361 | -0.478 | 0.00018 | 0.318 |
| 36486 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | ERBB4 | Sponge network | -0.576 | 0.10121 | -1.975 | 2.0E-5 | 0.318 |
| 36487 | ADAMTS9-AS2 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-940 | 21 | BAZ2B | Sponge network | -2.452 | 0 | -0.276 | 0.02833 | 0.318 |
| 36488 | FAM83H-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-497-5p | 10 | CCDC6 | Sponge network | 3.083 | 0 | 0.27 | 0.02555 | 0.318 |
| 36489 | AC009948.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p | 18 | NF1 | Sponge network | 0.532 | 0.00505 | 0.51 | 0 | 0.318 |
| 36490 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 44 | CBX5 | Sponge network | -0.727 | 0.04726 | 0.165 | 0.24446 | 0.318 |
| 36491 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-454-3p | 25 | RASSF2 | Sponge network | 0.176 | 0.37402 | -0.273 | 0.21961 | 0.318 |
| 36492 | AC108488.4 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 42 | DST | Sponge network | 0.259 | 0.12542 | -0.713 | 0.00096 | 0.318 |
| 36493 | PCBP1-AS1 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-654-3p;hsa-miR-92b-3p;hsa-miR-940 | 11 | BAZ2B | Sponge network | -0.567 | 0 | -0.276 | 0.02833 | 0.318 |
| 36494 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-624-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 61 | NTRK3 | Sponge network | -0.303 | 0.21002 | -1.77 | 3.0E-5 | 0.318 |
| 36495 | RP11-531A24.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p | 19 | NRK | Sponge network | 0.75 | 0.00054 | 0.656 | 0.19217 | 0.318 |
| 36496 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 28 | FOXO3 | Sponge network | -1.575 | 6.0E-5 | -0.04 | 0.72028 | 0.318 |
| 36497 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 19 | RET | Sponge network | 0.214 | 0.18246 | -0.833 | 0.03517 | 0.318 |
| 36498 | RP11-822E23.8 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-877-5p | 19 | TET1 | Sponge network | -0.777 | 0.16667 | -0.135 | 0.52138 | 0.318 |
| 36499 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SEMA3C | Sponge network | -0.737 | 0.10822 | -0.272 | 0.31153 | 0.318 |
| 36500 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 17 | TES | Sponge network | -2.46 | 0 | -0.348 | 0.00639 | 0.318 |
| 36501 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-497-5p;hsa-miR-664a-3p;hsa-miR-664a-5p | 24 | SOX11 | Sponge network | 1.294 | 0 | 2.68 | 0 | 0.318 |
| 36502 | HOXB-AS3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-550a-3p;hsa-miR-744-3p | 10 | DIP2C | Sponge network | 0.343 | 0.28887 | -0.436 | 0.01713 | 0.318 |
| 36503 | SNHG14 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 29 | RIMBP2 | Sponge network | -1.111 | 0.0003 | -1.968 | 5.0E-5 | 0.318 |
| 36504 | HAND2-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PIK3CA | Sponge network | -1.697 | 0.00889 | 0.195 | 0.06908 | 0.318 |
| 36505 | RP11-327P2.5 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-744-5p | 15 | FGFR1 | Sponge network | -0.147 | 0.40452 | -0.509 | 0.03957 | 0.318 |
| 36506 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 10 | FREM2 | Sponge network | -0.326 | 0.28809 | -0.437 | 0.46353 | 0.318 |
| 36507 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p | 13 | MYCBP2 | Sponge network | 0.494 | 0.00339 | -0.027 | 0.82016 | 0.318 |
| 36508 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 14 | CAV1 | Sponge network | 0.082 | 0.55654 | -1.013 | 6.0E-5 | 0.318 |
| 36509 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-590-3p | 14 | LRRK2 | Sponge network | 0.72 | 0.0001 | -1.136 | 1.0E-5 | 0.318 |
| 36510 | RP11-539I5.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p | 30 | PPARA | Sponge network | -0.468 | 0.32587 | -0.379 | 0.00548 | 0.318 |
| 36511 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | TCF12 | Sponge network | 1.254 | 0 | 0.174 | 0.13271 | 0.318 |
| 36512 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 44 | SMAD4 | Sponge network | 0.683 | 1.0E-5 | -0.313 | 0.00413 | 0.318 |
| 36513 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p | 10 | MGA | Sponge network | 0.95 | 0.15943 | 0.323 | 0.00792 | 0.318 |
| 36514 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | KAT6B | Sponge network | -1.057 | 0.00029 | -0.478 | 0.00018 | 0.318 |
| 36515 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | SCN9A | Sponge network | 0.61 | 0.01593 | -0.525 | 0.10593 | 0.318 |
| 36516 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 12 | BCLAF1 | Sponge network | 0.373 | 0.00068 | 0.211 | 0.00684 | 0.318 |
| 36517 | RP11-46C24.7 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 14 | SHPRH | Sponge network | 0.392 | 0.0278 | 0.098 | 0.40994 | 0.318 |
| 36518 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | THBS1 | Sponge network | -1.161 | 0.00591 | 0.741 | 0.00386 | 0.318 |
| 36519 | CTB-113P19.5 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 10 | PRKCB | Sponge network | 0.199 | 0.52763 | -1.313 | 2.0E-5 | 0.318 |
| 36520 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | SUFU | Sponge network | -0.563 | 0.02545 | -0.04 | 0.65489 | 0.318 |
| 36521 | RP11-13K12.1 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p | 10 | PHOX2B | Sponge network | -0.154 | 0.78939 | -2.68 | 0 | 0.318 |
| 36522 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p | 27 | PDGFRA | Sponge network | -2.095 | 0.00112 | -0.372 | 0.0037 | 0.318 |
| 36523 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 34 | NCOA1 | Sponge network | -0.576 | 0.10121 | -0.432 | 0 | 0.318 |
| 36524 | LINC00094 |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p | 11 | CADM4 | Sponge network | 0.253 | 0.09565 | -0.635 | 0.00305 | 0.318 |
| 36525 | MIR22HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 37 | CREB3L2 | Sponge network | -1.047 | 0 | -0.525 | 2.0E-5 | 0.318 |
| 36526 | RP11-73M18.8 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | ARHGEF12 | Sponge network | 0.832 | 0 | 0.09 | 0.35693 | 0.318 |
| 36527 | SH3RF3-AS1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p | 22 | GRIK3 | Sponge network | 0.889 | 9.0E-5 | -3.208 | 0 | 0.318 |
| 36528 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 23 | DMXL1 | Sponge network | 0.349 | 0.01695 | -0.426 | 0.00056 | 0.318 |
| 36529 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 51 | TCF4 | Sponge network | -0.612 | 0.00094 | 0.016 | 0.92271 | 0.318 |
| 36530 | SH3RF3-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | TMEFF2 | Sponge network | 0.889 | 9.0E-5 | -2.426 | 0 | 0.318 |
| 36531 | DOCK9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375 | 14 | AKAP6 | Sponge network | -0.231 | 0.28214 | -1.586 | 3.0E-5 | 0.318 |
| 36532 | BZRAP1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p | 65 | LPP | Sponge network | -0.465 | 0.04703 | -0.459 | 0.04475 | 0.318 |
| 36533 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | AFF3 | Sponge network | 0.657 | 4.0E-5 | -2.373 | 0 | 0.318 |
| 36534 | RP11-798M19.6 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PDE4DIP | Sponge network | -0.612 | 0.00094 | -0.282 | 0.0257 | 0.318 |
| 36535 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | LATS2 | Sponge network | -0.323 | 0.01372 | 0.159 | 0.2304 | 0.318 |
| 36536 | PWAR6 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-940 | 64 | NFIB | Sponge network | -1.575 | 6.0E-5 | 0.023 | 0.90795 | 0.318 |
| 36537 | LINC00578 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p | 13 | ERCC4 | Sponge network | 0.811 | 0.03228 | 0.126 | 0.15266 | 0.318 |
| 36538 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-877-5p;hsa-miR-93-5p | 14 | PGR | Sponge network | -0.785 | 0.00035 | -1.704 | 0 | 0.318 |
| 36539 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | MITF | Sponge network | 0.064 | 0.72934 | -0.769 | 0.00022 | 0.318 |
| 36540 | C4A-AS1 |
hsa-let-7d-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 12 | THBS1 | Sponge network | 3.345 | 0 | 0.741 | 0.00386 | 0.318 |
| 36541 | RP11-428J1.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 12 | LATS2 | Sponge network | 0.538 | 0.00076 | 0.159 | 0.2304 | 0.318 |
| 36542 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-940 | 27 | PBX1 | Sponge network | -0.08 | 0.90092 | -1.206 | 0 | 0.318 |
| 36543 | FENDRR |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p | 27 | CRTC1 | Sponge network | -1.281 | 0.00012 | -0.116 | 0.28412 | 0.318 |
| 36544 | RP11-158K1.3 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | ERCC6 | Sponge network | 0.349 | 0.01695 | 0.23 | 0.015 | 0.318 |
| 36545 | MEG3 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p | 15 | PLXNC1 | Sponge network | 0.249 | 0.35902 | 0.569 | 0.00329 | 0.318 |
| 36546 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 16 | NR2C2 | Sponge network | -0.154 | 0.78939 | 0.21 | 0.03224 | 0.318 |
| 36547 | RP11-119F7.5 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | CXCL12 | Sponge network | 0.225 | 0.30644 | -1.421 | 0 | 0.318 |
| 36548 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-590-3p | 10 | TBX18 | Sponge network | 0.906 | 0.31715 | 0.843 | 0.02945 | 0.318 |
| 36549 | LINC00473 |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 11 | FLI1 | Sponge network | -1.203 | 0.03881 | -0.294 | 0.10905 | 0.318 |
| 36550 | HCG11 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | ZMYND11 | Sponge network | 0.385 | 0.10067 | -0.2 | 0.05449 | 0.318 |
| 36551 | RP11-6O2.3 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-5p | 10 | RTN4 | Sponge network | 0.331 | 0.57247 | -0.412 | 0.00014 | 0.318 |
| 36552 | AC013271.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-19b-1-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-582-5p;hsa-miR-589-3p | 10 | THRB | Sponge network | 0.297 | 0.35401 | -1.117 | 0.0004 | 0.318 |
| 36553 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-93-3p | 21 | FOXO3 | Sponge network | -0.223 | 0.47816 | -0.04 | 0.72028 | 0.318 |
| 36554 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | UBE2QL1 | Sponge network | -2.117 | 0.00161 | -2.417 | 0 | 0.318 |
| 36555 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-96-5p | 17 | CBFA2T3 | Sponge network | 0.532 | 0.00505 | -1.221 | 1.0E-5 | 0.318 |
| 36556 | LINC00476 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-942-5p | 28 | OPCML | Sponge network | -0.615 | 0.00015 | -2.498 | 0 | 0.318 |
| 36557 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p | 10 | YAP1 | Sponge network | 0.946 | 0 | 0.22 | 0.11836 | 0.318 |
| 36558 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CBX5 | Sponge network | 0.272 | 0.44692 | 0.165 | 0.24446 | 0.318 |
| 36559 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-629-5p;hsa-miR-93-5p | 30 | MAF | Sponge network | -0.154 | 0.78939 | -1.257 | 0 | 0.318 |
| 36560 | PART1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-624-5p | 12 | HMCN1 | Sponge network | -2.925 | 0 | 0.809 | 0.00544 | 0.318 |
| 36561 | RP11-37B2.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-375 | 12 | TET1 | Sponge network | 1.03 | 0 | -0.135 | 0.52138 | 0.318 |
| 36562 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 15 | LHX6 | Sponge network | 0.4 | 0.14611 | -0.087 | 0.69193 | 0.318 |
| 36563 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-5p | 12 | PTGFR | Sponge network | -0.176 | 0.59692 | -0.233 | 0.45421 | 0.318 |
| 36564 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p | 10 | MAP3K12 | Sponge network | -0.693 | 0.05629 | -0.057 | 0.59369 | 0.318 |
| 36565 | LPP-AS2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-421 | 12 | STARD13 | Sponge network | -0.18 | 0.20343 | -0.187 | 0.27383 | 0.318 |
| 36566 | EMX2OS |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | ABCA10 | Sponge network | -0.777 | 0.1134 | -0.571 | 0.03674 | 0.318 |
| 36567 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | TSC1 | Sponge network | 0.194 | 0.64278 | 0.103 | 0.26071 | 0.318 |
| 36568 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | PRKAB2 | Sponge network | -0.376 | 0.00337 | -0.367 | 0.01371 | 0.318 |
| 36569 | RP11-540B6.6 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-484 | 17 | ZFX | Sponge network | 0.93 | 0 | 0.09 | 0.42563 | 0.318 |
| 36570 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-93-5p | 17 | JAZF1 | Sponge network | 0.497 | 0.01451 | -0.377 | 0.02677 | 0.318 |
| 36571 | U62631.5 |
hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-577 | 13 | IL6ST | Sponge network | 3.115 | 0.00463 | -0.402 | 0.00676 | 0.318 |
| 36572 | RP11-13K12.1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 30 | CTIF | Sponge network | -0.154 | 0.78939 | -0.389 | 0.00549 | 0.318 |
| 36573 | EIF3J-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-766-3p | 15 | ATM | Sponge network | 0.331 | 0.00509 | 0.178 | 0.21568 | 0.318 |
| 36574 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 15 | CREBBP | Sponge network | -1.281 | 0.00012 | 0.126 | 0.15186 | 0.318 |
| 36575 | RP11-725P16.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TET1 | Sponge network | 0.422 | 0.27021 | -0.135 | 0.52138 | 0.318 |
| 36576 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | HBP1 | Sponge network | -0.737 | 0.06182 | -0.454 | 0 | 0.318 |
| 36577 | DOCK9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p | 15 | IL17RD | Sponge network | -0.231 | 0.28214 | -0.019 | 0.94873 | 0.318 |
| 36578 | ARHGEF26-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 30 | PDE4DIP | Sponge network | -2.46 | 0 | -0.282 | 0.0257 | 0.318 |
| 36579 | RP11-359E10.1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-484 | 11 | IGF2 | Sponge network | 0.299 | 0.57722 | 0.848 | 0.03842 | 0.318 |
| 36580 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-378a-5p | 12 | PDS5B | Sponge network | 1.117 | 0 | 0.259 | 0.00962 | 0.318 |
| 36581 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-96-5p | 19 | ARID5B | Sponge network | 0.622 | 0.00015 | 0.342 | 0.01676 | 0.318 |
| 36582 | CTD-2089N3.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 24 | DST | Sponge network | -1.375 | 0.08642 | -0.713 | 0.00096 | 0.318 |
| 36583 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | SSBP2 | Sponge network | -0.72 | 0.24239 | -1.58 | 0 | 0.318 |
| 36584 | HOTAIRM1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | LATS2 | Sponge network | -0.053 | 0.80685 | 0.159 | 0.2304 | 0.318 |
| 36585 | AC141928.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 14 | FAT4 | Sponge network | 0.853 | 0.15352 | -0.354 | 0.14694 | 0.318 |
| 36586 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-590-3p | 16 | FREM2 | Sponge network | -1.161 | 0.00591 | -0.437 | 0.46353 | 0.318 |
| 36587 | CALML3-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p | 15 | NRK | Sponge network | 0.95 | 0.15943 | 0.656 | 0.19217 | 0.318 |
| 36588 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-429 | 14 | ZMYND11 | Sponge network | -0.469 | 0.08721 | -0.2 | 0.05449 | 0.318 |
| 36589 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | CUL5 | Sponge network | 0.541 | 0.00125 | 0.026 | 0.77301 | 0.318 |
| 36590 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | WDFY3 | Sponge network | 0.91 | 0 | -0.201 | 0.0967 | 0.318 |
| 36591 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-93-5p | 35 | SSBP2 | Sponge network | 0.082 | 0.55397 | -1.58 | 0 | 0.318 |
| 36592 | RP11-798G7.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-28-3p;hsa-miR-30e-3p;hsa-miR-766-3p | 10 | TBL1XR1 | Sponge network | 0.858 | 3.0E-5 | 0.578 | 0 | 0.318 |
| 36593 | LINC00863 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | LATS1 | Sponge network | 0.189 | 0.13981 | 0.109 | 0.34208 | 0.318 |
| 36594 | LINC00173 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 29 | GAS7 | Sponge network | 0.056 | 0.8494 | -0.555 | 0.01602 | 0.318 |
| 36595 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CDH10 | Sponge network | 0.335 | 0.20596 | -2.397 | 0 | 0.318 |
| 36596 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | MCC | Sponge network | 0.657 | 4.0E-5 | -1.005 | 0 | 0.318 |
| 36597 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-93-5p | 12 | EDA2R | Sponge network | 0.056 | 0.81195 | -0.587 | 0.01598 | 0.318 |
| 36598 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 18 | MAP3K12 | Sponge network | -1.111 | 0.0003 | -0.057 | 0.59369 | 0.318 |
| 36599 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 17 | RBL2 | Sponge network | -0.793 | 7.0E-5 | -0.282 | 0.00254 | 0.318 |
| 36600 | LINC00854 |
hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-940 | 11 | SIRT1 | Sponge network | 1.18 | 0 | 0.109 | 0.2593 | 0.318 |
| 36601 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p | 16 | ARNT2 | Sponge network | -0.125 | 0.49414 | -0.332 | 0.25851 | 0.318 |
| 36602 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 33 | CBL | Sponge network | 0.395 | 0.15451 | 0.291 | 0.01267 | 0.318 |
| 36603 | SERTAD4-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p | 13 | CADM1 | Sponge network | -0.932 | 0.00329 | -0.9 | 0.00084 | 0.318 |
| 36604 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-7-5p;hsa-miR-93-3p | 26 | ERC1 | Sponge network | -0.465 | 0.04703 | -0.108 | 0.38794 | 0.318 |
| 36605 | AC141928.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | EYA4 | Sponge network | 0.853 | 0.15352 | 0.3 | 0.36876 | 0.318 |
| 36606 | MIR143HG |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | UBE4B | Sponge network | -0.739 | 0.0574 | -0.235 | 0.007 | 0.318 |
| 36607 | TRIM52-AS1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-7-1-3p | 10 | MXI1 | Sponge network | -0.418 | 0.00068 | -0.987 | 0 | 0.318 |
| 36608 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-424-5p | 13 | ABCB5 | Sponge network | 0.299 | 0.57722 | -0.956 | 0.04077 | 0.318 |
| 36609 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3614-5p | 21 | TCF4 | Sponge network | -2.549 | 0.00528 | 0.016 | 0.92271 | 0.318 |
| 36610 | RP11-21A7A.2 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-484;hsa-miR-505-5p | 10 | WNK2 | Sponge network | -1.116 | 0.23686 | -0.352 | 0.31066 | 0.318 |
| 36611 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | EYA4 | Sponge network | 0.657 | 4.0E-5 | 0.3 | 0.36876 | 0.318 |
| 36612 | RP4-791M13.3 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-495-3p;hsa-miR-532-3p | 12 | SYNM | Sponge network | 0.419 | 0.0319 | -2.309 | 1.0E-5 | 0.318 |
| 36613 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | EPAS1 | Sponge network | -1.111 | 0.0003 | -0.184 | 0.14418 | 0.318 |
| 36614 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | NUAK1 | Sponge network | -0.08 | 0.90092 | 0.904 | 0 | 0.318 |
| 36615 | CTD-3126B10.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 12 | MYO9A | Sponge network | 0.778 | 0.00101 | -0.147 | 0.32373 | 0.318 |
| 36616 | RP11-359B12.2 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 40 | GRIK3 | Sponge network | 0.064 | 0.72934 | -3.208 | 0 | 0.318 |
| 36617 | BOLA3-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 37 | CBX5 | Sponge network | -0.246 | 0.35034 | 0.165 | 0.24446 | 0.318 |
| 36618 | RP11-244H3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | NIN | Sponge network | 0.659 | 3.0E-5 | -0.253 | 0.13794 | 0.318 |
| 36619 | AC093627.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-625-5p | 10 | RSPO2 | Sponge network | -0.787 | 0.3928 | -3.038 | 0 | 0.318 |
| 36620 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-942-5p | 10 | GLIPR1 | Sponge network | -0.517 | 0.02935 | 0.146 | 0.3276 | 0.318 |
| 36621 | NDUFA6-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 31 | SNX29 | Sponge network | -0.355 | 0.0077 | -0.34 | 0.01371 | 0.318 |
| 36622 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-628-5p | 15 | ANK3 | Sponge network | -0.83 | 0 | -0.55 | 0.00086 | 0.318 |
| 36623 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p | 36 | BMPR2 | Sponge network | 0.259 | 0.12542 | 0.206 | 0.0635 | 0.318 |
| 36624 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429 | 15 | CREBBP | Sponge network | -0.563 | 0.02545 | 0.126 | 0.15186 | 0.318 |
| 36625 | FAM13A-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 20 | SEMA3C | Sponge network | -0.83 | 0 | -0.272 | 0.31153 | 0.318 |
| 36626 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 13 | TMEFF2 | Sponge network | -0.738 | 0.21233 | -2.426 | 0 | 0.318 |
| 36627 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-493-5p | 12 | LRIG1 | Sponge network | -0.829 | 0.01343 | -0.537 | 0.00588 | 0.318 |
| 36628 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-96-5p | 22 | MED12L | Sponge network | -0.18 | 0.20343 | -0.194 | 0.55299 | 0.318 |
| 36629 | CTC-429P9.3 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-532-5p | 12 | RPS6KA6 | Sponge network | 0.082 | 0.55397 | -2.989 | 0 | 0.318 |
| 36630 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-324-5p | 12 | TSHZ3 | Sponge network | -2.549 | 0.00528 | -0.556 | 0.01516 | 0.318 |
| 36631 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374a-3p;hsa-miR-374b-5p | 14 | TMTC3 | Sponge network | 0.542 | 0.00054 | 0.123 | 0.22189 | 0.318 |
| 36632 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 33 | CBL | Sponge network | 0.415 | 0.14657 | 0.291 | 0.01267 | 0.318 |
| 36633 | RP11-156P1.3 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 19 | PRKCB | Sponge network | 0.214 | 0.18246 | -1.313 | 2.0E-5 | 0.318 |
| 36634 | CTD-2647L4.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-495-3p | 10 | FZD3 | Sponge network | 0.555 | 0.00197 | 0.104 | 0.60316 | 0.318 |
| 36635 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | BCL2 | Sponge network | -2.915 | 0.00015 | -0.899 | 0 | 0.318 |
| 36636 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | CBX5 | Sponge network | -0.125 | 0.49414 | 0.165 | 0.24446 | 0.318 |
| 36637 | RP11-6O2.3 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 15 | SORCS1 | Sponge network | 0.331 | 0.57247 | -3.492 | 0 | 0.318 |
| 36638 | RP11-728F11.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-7-1-3p | 10 | NRXN3 | Sponge network | 0.238 | 0.70544 | -0.893 | 0.04186 | 0.318 |
| 36639 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 35 | CBL | Sponge network | -1.575 | 6.0E-5 | 0.291 | 0.01267 | 0.318 |
| 36640 | LINC00926 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p | 10 | CDH23 | Sponge network | 0.756 | 0.00739 | -0.974 | 0.00034 | 0.318 |
| 36641 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | NEDD4 | Sponge network | 0.91 | 0 | 0.02 | 0.89834 | 0.318 |
| 36642 | OIP5-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | AFF1 | Sponge network | 0.421 | 0.00084 | -0.384 | 0.00053 | 0.318 |
| 36643 | HOXA-AS2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PDE4DIP | Sponge network | 0.61 | 0.01593 | -0.282 | 0.0257 | 0.318 |
| 36644 | RP11-218M22.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 32 | DST | Sponge network | 0.197 | 0.25618 | -0.713 | 0.00096 | 0.318 |
| 36645 | RP11-395G23.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-671-5p | 19 | DLC1 | Sponge network | 0.381 | 0.25967 | -0.382 | 0.05038 | 0.318 |
| 36646 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 17 | NUAK1 | Sponge network | 0.657 | 4.0E-5 | 0.904 | 0 | 0.318 |
| 36647 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | MYO9A | Sponge network | -1.582 | 8.0E-5 | -0.147 | 0.32373 | 0.318 |
| 36648 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 37 | RASSF2 | Sponge network | 0.374 | 0.20054 | -0.273 | 0.21961 | 0.318 |
| 36649 | HOXA-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | MED12L | Sponge network | 0.61 | 0.01593 | -0.194 | 0.55299 | 0.318 |
| 36650 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-93-5p;hsa-miR-942-5p | 12 | FBXO31 | Sponge network | -0.465 | 0.04703 | -0.085 | 0.42389 | 0.318 |
| 36651 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p | 43 | BCL2 | Sponge network | -0.793 | 7.0E-5 | -0.899 | 0 | 0.317 |
| 36652 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 33 | NCOA1 | Sponge network | -1.697 | 0.00889 | -0.432 | 0 | 0.317 |
| 36653 | CD27-AS1 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-7-1-3p | 11 | CAV1 | Sponge network | 0.177 | 0.16681 | -1.013 | 6.0E-5 | 0.317 |
| 36654 | LINC00621 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | NAV3 | Sponge network | 0.131 | 0.8436 | 0.212 | 0.47729 | 0.317 |
| 36655 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | CSMD1 | Sponge network | -0.323 | 0.01372 | -0.63 | 0.18881 | 0.317 |
| 36656 | PRKAG2-AS1 |
hsa-let-7b-3p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 10 | PBX1 | Sponge network | -0.622 | 0.02807 | -1.206 | 0 | 0.317 |
| 36657 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 46 | PRKAA2 | Sponge network | -0.18 | 0.20343 | -2.462 | 0 | 0.317 |
| 36658 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | USP12 | Sponge network | 1.055 | 0 | -0.102 | 0.40768 | 0.317 |
| 36659 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 33 | BCL2 | Sponge network | 0.537 | 0.00139 | -0.899 | 0 | 0.317 |
| 36660 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429 | 18 | ZFHX3 | Sponge network | -1.654 | 0.00144 | 0.389 | 0.01363 | 0.317 |
| 36661 | SNHG14 |
hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-7-5p | 10 | CMKLR1 | Sponge network | -1.111 | 0.0003 | 0.288 | 0.17621 | 0.317 |
| 36662 | HAND2-AS1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-940 | 11 | FOXO4 | Sponge network | -1.697 | 0.00889 | -0.516 | 2.0E-5 | 0.317 |
| 36663 | RP11-517B11.7 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p | 12 | MAT2A | Sponge network | 0.706 | 0 | -0.073 | 0.36195 | 0.317 |
| 36664 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-331-5p | 13 | MEF2C | Sponge network | -0.326 | 0.28809 | -0.499 | 0.01448 | 0.317 |
| 36665 | LINC00654 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | INPP4A | Sponge network | 0.374 | 0.20054 | 0.07 | 0.53563 | 0.317 |
| 36666 | RP11-130L8.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PTGFR | Sponge network | -0.663 | 0.10887 | -0.233 | 0.45421 | 0.317 |
| 36667 | AC005562.1 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 23 | DOCK4 | Sponge network | 0.488 | 0.00084 | 0.502 | 0.00108 | 0.317 |
| 36668 | HOTAIRM1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-590-3p | 10 | NRK | Sponge network | -0.053 | 0.80685 | 0.656 | 0.19217 | 0.317 |
| 36669 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TIMP3 | Sponge network | 0.385 | 0.10067 | -0.546 | 0.02307 | 0.317 |
| 36670 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 23 | LRRC55 | Sponge network | -2.46 | 0 | -0.026 | 0.93163 | 0.317 |
| 36671 | RP4-669P10.16 |
hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-196b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-34a-5p | 10 | ZMYND11 | Sponge network | -0.301 | 0.06597 | -0.2 | 0.05449 | 0.317 |
| 36672 | RP11-48B3.4 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 26 | PPP1R9A | Sponge network | 1.757 | 0 | -0.903 | 0.04254 | 0.317 |
| 36673 | USP46-AS1 |
hsa-miR-106a-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p | 16 | PPM1L | Sponge network | 0.102 | 0.51464 | -0.537 | 0.00616 | 0.317 |
| 36674 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p | 23 | MAF | Sponge network | -0.342 | 0.02288 | -1.257 | 0 | 0.317 |
| 36675 | RP11-403I13.7 |
hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-7-1-3p | 20 | RIMS2 | Sponge network | 0.395 | 0.15451 | -1.365 | 0.00208 | 0.317 |
| 36676 | RP13-516M14.1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577 | 15 | NRK | Sponge network | 0.389 | 0.04293 | 0.656 | 0.19217 | 0.317 |
| 36677 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-577;hsa-miR-582-5p | 16 | KAT6B | Sponge network | -2.69 | 0.0001 | -0.478 | 0.00018 | 0.317 |
| 36678 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-624-5p | 10 | HMCN1 | Sponge network | 0.41 | 0.02214 | 0.809 | 0.00544 | 0.317 |
| 36679 | NR2F1-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | PTCH1 | Sponge network | -0.49 | 0.04946 | -0.1 | 0.6179 | 0.317 |
| 36680 | RP1-122P22.2 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p | 12 | EGFR | Sponge network | 0.065 | 0.73468 | -0.175 | 0.48451 | 0.317 |
| 36681 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-577 | 13 | ZBTB4 | Sponge network | 1.294 | 0.0031 | -0.739 | 0 | 0.317 |
| 36682 | EMX2OS |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | -0.777 | 0.1134 | -1.379 | 0 | 0.317 |
| 36683 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 28 | TACC1 | Sponge network | -1.057 | 0.00029 | -0.362 | 0.05406 | 0.317 |
| 36684 | CTD-2528L19.6 |
hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | UBE2D1 | Sponge network | 0.953 | 0 | 0.468 | 0 | 0.317 |
| 36685 | NDUFA6-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-5p;hsa-miR-7-5p | 15 | FNBP1 | Sponge network | -0.355 | 0.0077 | -0.969 | 4.0E-5 | 0.317 |
| 36686 | AC012360.6 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 10 | BAZ2B | Sponge network | 0.624 | 0.02176 | -0.276 | 0.02833 | 0.317 |
| 36687 | LINC00648 |
hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-96-5p | 11 | IL17RD | Sponge network | 0.633 | 0.51678 | -0.019 | 0.94873 | 0.317 |
| 36688 | SOX2-OT |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | ZNF536 | Sponge network | -1.264 | 6.0E-5 | -3.115 | 0 | 0.317 |
| 36689 | RP1-151F17.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | BRD3 | Sponge network | 0.222 | 0.29133 | 0.37 | 0.00013 | 0.317 |
| 36690 | BCYRN1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 37 | NTRK2 | Sponge network | 0.311 | 0.10344 | -0.736 | 0.06495 | 0.317 |
| 36691 | RP11-6N17.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p | 11 | EPHA7 | Sponge network | 0.108 | 0.69556 | -2.197 | 3.0E-5 | 0.317 |
| 36692 | BCYRN1 |
hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p | 29 | UBR3 | Sponge network | 0.311 | 0.10344 | -0.021 | 0.82774 | 0.317 |
| 36693 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-935 | 27 | MTSS1 | Sponge network | -0.147 | 0.40452 | -0.81 | 0.00035 | 0.317 |
| 36694 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-582-5p | 10 | ZMYM2 | Sponge network | 1.157 | 0.00455 | 0.279 | 0.01273 | 0.317 |
| 36695 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940;hsa-miR-96-5p | 43 | GAS7 | Sponge network | 0.083 | 0.66405 | -0.555 | 0.01602 | 0.317 |
| 36696 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-590-3p | 13 | SEMA3C | Sponge network | 0.174 | 0.30034 | -0.272 | 0.31153 | 0.317 |
| 36697 | DNM1P35 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-590-3p | 13 | RIMBP2 | Sponge network | 0.051 | 0.83388 | -1.968 | 5.0E-5 | 0.317 |
| 36698 | OIP5-AS1 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-942-5p | 38 | NOTCH2 | Sponge network | 0.421 | 0.00084 | 0.138 | 0.35163 | 0.317 |
| 36699 | RP11-195B17.1 |
hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b | 10 | MLLT10 | Sponge network | 0.37 | 0.27614 | 0.105 | 0.33292 | 0.317 |
| 36700 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-590-3p | 16 | CREBBP | Sponge network | 0.572 | 0.01722 | 0.126 | 0.15186 | 0.317 |
| 36701 | DLEU2 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-497-5p | 12 | PAFAH1B2 | Sponge network | 1.831 | 0 | 0.318 | 0.0001 | 0.317 |
| 36702 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 13 | DIP2C | Sponge network | 0.542 | 0.00054 | -0.436 | 0.01713 | 0.317 |
| 36703 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | RPS6KA3 | Sponge network | 1.153 | 0 | 0.256 | 0.02419 | 0.317 |
| 36704 | RP11-517B11.7 |
hsa-miR-100-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 12 | RB1 | Sponge network | 0.706 | 0 | 0.382 | 0.0017 | 0.317 |
| 36705 | RP11-403I13.8 |
hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | GLIPR1 | Sponge network | 0.619 | 0.00157 | 0.146 | 0.3276 | 0.317 |
| 36706 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-93-5p | 21 | SYNE1 | Sponge network | -0.384 | 0.40652 | -0.738 | 0.00316 | 0.317 |
| 36707 | DNM1P35 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-590-3p | 19 | BRD3 | Sponge network | 0.051 | 0.83388 | 0.37 | 0.00013 | 0.317 |
| 36708 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-365a-3p;hsa-miR-455-3p;hsa-miR-582-5p | 27 | RICTOR | Sponge network | 0.792 | 0.0049 | 0.388 | 0.00188 | 0.317 |
| 36709 | CTD-2017D11.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-582-5p;hsa-miR-940 | 20 | PPP1R9A | Sponge network | 0.792 | 0.0049 | -0.903 | 0.04254 | 0.317 |
| 36710 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | GRIN2A | Sponge network | -0.72 | 0.24239 | -2.161 | 0 | 0.317 |
| 36711 | LINC00173 |
hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-532-3p | 12 | SYNM | Sponge network | 0.056 | 0.8494 | -2.309 | 1.0E-5 | 0.317 |
| 36712 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-96-5p | 27 | IL17RD | Sponge network | 0.523 | 9.0E-5 | -0.019 | 0.94873 | 0.317 |
| 36713 | LINC00403 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-181b-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-33a-5p | 10 | FOXP1 | Sponge network | -3.586 | 0.00032 | 0.042 | 0.74368 | 0.317 |
| 36714 | RP11-158K1.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | LATS2 | Sponge network | 0.349 | 0.01695 | 0.159 | 0.2304 | 0.317 |
| 36715 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | RBMS3 | Sponge network | 0.238 | 0.70544 | -0.519 | 0.03551 | 0.317 |
| 36716 | PRKAG2-AS1 |
hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | MED12L | Sponge network | -0.622 | 0.02807 | -0.194 | 0.55299 | 0.317 |
| 36717 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 19 | SYNE1 | Sponge network | -0.176 | 0.59692 | -0.738 | 0.00316 | 0.317 |
| 36718 | TUG1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-582-5p | 13 | PIK3C2A | Sponge network | 0.972 | 0 | 0.061 | 0.53776 | 0.317 |
| 36719 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | ARHGAP29 | Sponge network | -0.513 | 0.04703 | 0.131 | 0.42953 | 0.317 |
| 36720 | THAP9-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-664a-3p | 15 | SOX11 | Sponge network | 1.079 | 0 | 2.68 | 0 | 0.317 |
| 36721 | LINC00672 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-550a-5p;hsa-miR-584-5p;hsa-miR-590-3p | 11 | PCDH18 | Sponge network | -0.227 | 0.31657 | 0.216 | 0.24645 | 0.317 |
| 36722 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 42 | PPP1R9A | Sponge network | -0.49 | 0.04946 | -0.903 | 0.04254 | 0.317 |
| 36723 | CTC-429P9.1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-511-5p | 10 | PCDH10 | Sponge network | 0.497 | 0.01451 | -1.644 | 0.0078 | 0.317 |
| 36724 | GS1-124K5.11 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 10 | LHFP | Sponge network | 0.494 | 0.00339 | -0.167 | 0.41371 | 0.317 |
| 36725 | RP1-122P22.2 |
hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 11 | TSHZ3 | Sponge network | 0.065 | 0.73468 | -0.556 | 0.01516 | 0.317 |
| 36726 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 24 | DMXL1 | Sponge network | -0.709 | 0.18168 | -0.426 | 0.00056 | 0.317 |
| 36727 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-7-1-3p | 14 | CLIP1 | Sponge network | 0.16 | 0.50785 | -0.215 | 0.08684 | 0.317 |
| 36728 | RP5-1198O20.4 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | RPS6KA6 | Sponge network | 0.997 | 0.00066 | -2.989 | 0 | 0.317 |
| 36729 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940 | 13 | SNX29 | Sponge network | -1.483 | 9.0E-5 | -0.34 | 0.01371 | 0.317 |
| 36730 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-33a-5p;hsa-miR-495-3p;hsa-miR-628-5p | 14 | BMPR2 | Sponge network | 1.597 | 0 | 0.206 | 0.0635 | 0.317 |
| 36731 | GNG12-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | GALNT13 | Sponge network | -0.793 | 7.0E-5 | -0.857 | 0.07439 | 0.317 |
| 36732 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-3p | 23 | ABL2 | Sponge network | -0.246 | 0.35034 | 0.82 | 0 | 0.317 |
| 36733 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SYNE1 | Sponge network | 0.272 | 0.44692 | -0.738 | 0.00316 | 0.317 |
| 36734 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 15 | NEDD4 | Sponge network | 0.381 | 0.25967 | 0.02 | 0.89834 | 0.317 |
| 36735 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p | 25 | FOXO3 | Sponge network | -0.323 | 0.01372 | -0.04 | 0.72028 | 0.317 |
| 36736 | H1FX-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-3p | 11 | POU2F2 | Sponge network | -0.206 | 0.12942 | -0.233 | 0.32214 | 0.317 |
| 36737 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | THBS1 | Sponge network | -0.08 | 0.90092 | 0.741 | 0.00386 | 0.317 |
| 36738 | RP11-48B3.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | SIRT1 | Sponge network | 1.757 | 0 | 0.109 | 0.2593 | 0.317 |
| 36739 | LINC00863 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TET1 | Sponge network | 0.189 | 0.13981 | -0.135 | 0.52138 | 0.317 |
| 36740 | PART1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 23 | PTGFR | Sponge network | -2.925 | 0 | -0.233 | 0.45421 | 0.317 |
| 36741 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | CREBBP | Sponge network | -0.576 | 0.10121 | 0.126 | 0.15186 | 0.317 |
| 36742 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p | 19 | FOXO3 | Sponge network | 0.946 | 0 | -0.04 | 0.72028 | 0.317 |
| 36743 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-96-5p | 14 | PCDH8 | Sponge network | -1.281 | 0.00012 | -0.997 | 0.05366 | 0.317 |
| 36744 | ADIRF-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | UBE4B | Sponge network | -0.223 | 0.47816 | -0.235 | 0.007 | 0.317 |
| 36745 | HAR1A |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-671-5p;hsa-miR-877-5p | 15 | RBMS3 | Sponge network | -0.672 | 0.19447 | -0.519 | 0.03551 | 0.317 |
| 36746 | RP11-819C21.1 |
hsa-miR-135b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | 0.537 | 0.00139 | 0.358 | 0.13727 | 0.317 |
| 36747 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | LRIG1 | Sponge network | 0.335 | 0.20596 | -0.537 | 0.00588 | 0.317 |
| 36748 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | GPC6 | Sponge network | 0.381 | 0.25967 | -0.054 | 0.81465 | 0.317 |
| 36749 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | NOTCH2NL | Sponge network | -0.323 | 0.01372 | 0.024 | 0.84307 | 0.317 |
| 36750 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 22 | CLASP2 | Sponge network | 1.11 | 0 | -0.038 | 0.71 | 0.317 |
| 36751 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p | 15 | PRKAB2 | Sponge network | 0.594 | 0.00064 | -0.367 | 0.01371 | 0.317 |
| 36752 | KB-1460A1.5 |
hsa-miR-101-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-30e-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 12 | PKNOX1 | Sponge network | 0.603 | 0.00127 | 0.085 | 0.28917 | 0.317 |
| 36753 | LINC00865 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 36 | TGFBR3 | Sponge network | -1.249 | 7.0E-5 | -1.173 | 1.0E-5 | 0.317 |
| 36754 | RP11-770J1.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-577 | 11 | DTNA | Sponge network | -2.011 | 0.0005 | -1.648 | 1.0E-5 | 0.317 |
| 36755 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 33 | SASH1 | Sponge network | -1.047 | 0 | -0.502 | 0.00175 | 0.317 |
| 36756 | AC007566.10 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 2.368 | 2.0E-5 | 0.51 | 0 | 0.317 |
| 36757 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940 | 30 | HLF | Sponge network | 0.537 | 0.00139 | -2.443 | 0 | 0.317 |
| 36758 | BVES-AS1 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-576-5p | 10 | PTCH1 | Sponge network | -2.62 | 0 | -0.1 | 0.6179 | 0.317 |
| 36759 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 27 | THBS1 | Sponge network | 0.72 | 0.0001 | 0.741 | 0.00386 | 0.317 |
| 36760 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 24 | GRIN2A | Sponge network | -0.727 | 0.04726 | -2.161 | 0 | 0.317 |
| 36761 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 17 | CNTNAP3 | Sponge network | 0.61 | 0.01593 | -1.465 | 0.00033 | 0.317 |
| 36762 | LINC00672 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-577;hsa-miR-590-3p | 12 | TMEFF2 | Sponge network | -0.227 | 0.31657 | -2.426 | 0 | 0.317 |
| 36763 | NEAT1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p | 12 | STAG2 | Sponge network | 0.705 | 0.00014 | 0.252 | 0.00438 | 0.317 |
| 36764 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PDS5B | Sponge network | 0.532 | 0.00505 | 0.259 | 0.00962 | 0.317 |
| 36765 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 25 | CYLD | Sponge network | 0.572 | 0.01722 | -0.367 | 0.00171 | 0.317 |
| 36766 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 41 | EPHA5 | Sponge network | 0.083 | 0.66405 | -0.957 | 0.01733 | 0.317 |
| 36767 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 55 | WDFY3 | Sponge network | -0.777 | 0.1134 | -0.201 | 0.0967 | 0.317 |
| 36768 | HOXD-AS2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 10 | ST6GALNAC3 | Sponge network | -0.208 | 0.65592 | -0.987 | 0 | 0.317 |
| 36769 | TRIM52-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 19 | PLSCR4 | Sponge network | -0.418 | 0.00068 | -0.474 | 0.03833 | 0.317 |
| 36770 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 30 | CLASP2 | Sponge network | -0.709 | 0.18168 | -0.038 | 0.71 | 0.317 |
| 36771 | RP11-400K9.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-940 | 11 | NTRK2 | Sponge network | -0.733 | 0.19469 | -0.736 | 0.06495 | 0.317 |
| 36772 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 31 | PIK3C2A | Sponge network | 0.421 | 0.00084 | 0.061 | 0.53776 | 0.317 |
| 36773 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | GALNT13 | Sponge network | -0.612 | 0.00094 | -0.857 | 0.07439 | 0.317 |
| 36774 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-335-5p | 11 | KCNA6 | Sponge network | 0.043 | 0.81628 | -1.531 | 0.00013 | 0.317 |
| 36775 | FENDRR |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -1.281 | 0.00012 | -2.417 | 0 | 0.317 |
| 36776 | LINC00578 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-664a-3p | 34 | KIF1B | Sponge network | 0.811 | 0.03228 | -0.118 | 0.32258 | 0.317 |
| 36777 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 42 | PRKAA2 | Sponge network | 0.392 | 0.0278 | -2.462 | 0 | 0.317 |
| 36778 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 13 | CHD1 | Sponge network | 0.541 | 0.00125 | 0.322 | 0.00215 | 0.317 |
| 36779 | RP11-464F9.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | ABI2 | Sponge network | 0.959 | 0 | 0.175 | 0.14787 | 0.317 |
| 36780 | RP11-13K12.5 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | NCAM2 | Sponge network | -0.318 | 0.65847 | -0.482 | 0.22565 | 0.317 |
| 36781 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | ANK2 | Sponge network | -0.011 | 0.967 | -1.603 | 1.0E-5 | 0.317 |
| 36782 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 18 | COPS2 | Sponge network | 0.559 | 0.00026 | -0.178 | 0.04974 | 0.317 |
| 36783 | RP11-890B15.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 20 | BAZ2B | Sponge network | 0.101 | 0.60919 | -0.276 | 0.02833 | 0.317 |
| 36784 | RP11-983P16.4 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | RASSF8 | Sponge network | 0.41 | 0.02214 | -0.122 | 0.65591 | 0.317 |
| 36785 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 16 | MYO9A | Sponge network | 0.381 | 0.25967 | -0.147 | 0.32373 | 0.317 |
| 36786 | AC074289.1 |
hsa-let-7a-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-96-5p | 13 | HDAC9 | Sponge network | -0.326 | 0.14314 | -0.755 | 0.00495 | 0.317 |
| 36787 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | PPP1R9A | Sponge network | 0.101 | 0.60919 | -0.903 | 0.04254 | 0.317 |
| 36788 | MIR143HG |
hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | GUCY1A2 | Sponge network | -0.739 | 0.0574 | 1.213 | 0 | 0.317 |
| 36789 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374a-5p | 14 | PRKAB2 | Sponge network | -0.693 | 0.05629 | -0.367 | 0.01371 | 0.317 |
| 36790 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484 | 18 | ARNT2 | Sponge network | -0.773 | 0.04073 | -0.332 | 0.25851 | 0.317 |
| 36791 | CTD-2666L21.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-629-3p | 13 | RASSF8 | Sponge network | 0.368 | 0.20093 | -0.122 | 0.65591 | 0.317 |
| 36792 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-501-3p | 14 | TGFBR3 | Sponge network | 0.37 | 0.27614 | -1.173 | 1.0E-5 | 0.317 |
| 36793 | TP73-AS1 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-429 | 15 | FAM135B | Sponge network | -0.563 | 0.02545 | -3.978 | 0 | 0.317 |
| 36794 | RP11-890B15.3 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-3127-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ADH1B | Sponge network | 0.101 | 0.60919 | -3.716 | 0 | 0.317 |
| 36795 | RP11-6O2.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 39 | MDM4 | Sponge network | 0.331 | 0.57247 | 0.345 | 0.00762 | 0.317 |
| 36796 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-532-5p | 12 | FGFR1 | Sponge network | 0.395 | 0.15451 | -0.509 | 0.03957 | 0.317 |
| 36797 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 19 | ARHGAP29 | Sponge network | -0.942 | 0 | 0.131 | 0.42953 | 0.317 |
| 36798 | LINC00641 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 13 | DLG1 | Sponge network | -0.222 | 0.32445 | -0.014 | 0.8926 | 0.317 |
| 36799 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TET1 | Sponge network | -0.021 | 0.93598 | -0.135 | 0.52138 | 0.317 |
| 36800 | LINC00963 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | AKAP6 | Sponge network | 0.523 | 9.0E-5 | -1.586 | 3.0E-5 | 0.317 |
| 36801 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | DMTF1 | Sponge network | 0.392 | 0.0278 | 0.248 | 0.02726 | 0.317 |
| 36802 | AC005618.6 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-590-3p | 12 | DNMT3A | Sponge network | 0.862 | 0.06213 | 0.302 | 0.0262 | 0.317 |
| 36803 | PGM5-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-511-5p;hsa-miR-7-1-3p | 16 | MXI1 | Sponge network | -4.546 | 0 | -0.987 | 0 | 0.317 |
| 36804 | WDR86-AS1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-429 | 10 | DLC1 | Sponge network | 0.348 | 0.32912 | -0.382 | 0.05038 | 0.317 |
| 36805 | RP11-48B3.4 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p | 14 | PCDH10 | Sponge network | 1.757 | 0 | -1.644 | 0.0078 | 0.317 |
| 36806 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p | 20 | LIFR | Sponge network | -0.176 | 0.59692 | -1.794 | 0 | 0.317 |
| 36807 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 38 | AR | Sponge network | -0.031 | 0.89063 | -1.554 | 0 | 0.317 |
| 36808 | PPP1R26-AS1 |
hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-378c | 12 | ABL2 | Sponge network | 1.454 | 0 | 0.82 | 0 | 0.317 |
| 36809 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | SASH1 | Sponge network | 0.385 | 0.10067 | -0.502 | 0.00175 | 0.317 |
| 36810 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | CAV1 | Sponge network | 0.532 | 0.00505 | -1.013 | 6.0E-5 | 0.317 |
| 36811 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | FOXO1 | Sponge network | -1.111 | 0.0003 | -0.141 | 0.33874 | 0.317 |
| 36812 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 24 | CUL5 | Sponge network | 0.311 | 0.10344 | 0.026 | 0.77301 | 0.317 |
| 36813 | C4A-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-629-3p | 10 | GPC6 | Sponge network | 3.345 | 0 | -0.054 | 0.81465 | 0.317 |
| 36814 | RP13-516M14.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-3p | 16 | MRVI1 | Sponge network | 0.389 | 0.04293 | -1.051 | 0.00022 | 0.317 |
| 36815 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | PTPRB | Sponge network | 0.176 | 0.37402 | 0.105 | 0.47122 | 0.317 |
| 36816 | RP11-181G12.2 |
hsa-miR-127-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-584-5p | 16 | PIK3R1 | Sponge network | 0.556 | 0.00298 | -0.133 | 0.36854 | 0.317 |
| 36817 | HCG18 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p;hsa-miR-664a-5p | 16 | CBFB | Sponge network | 1.063 | 0 | 1.061 | 0 | 0.317 |
| 36818 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | LRRC55 | Sponge network | -0.709 | 0.18168 | -0.026 | 0.93163 | 0.317 |
| 36819 | RP11-968O1.5 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-338-3p;hsa-miR-493-5p | 12 | LIMCH1 | Sponge network | -0.829 | 0.01343 | -0.915 | 1.0E-5 | 0.317 |
| 36820 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 12 | EBF3 | Sponge network | 0.898 | 1.0E-5 | -0.058 | 0.81437 | 0.317 |
| 36821 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 12 | TES | Sponge network | -0.896 | 0.00099 | -0.348 | 0.00639 | 0.317 |
| 36822 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | ITGAV | Sponge network | -0.358 | 0.17255 | 0.337 | 0.01002 | 0.317 |
| 36823 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 23 | CDHR3 | Sponge network | -0.576 | 0.10121 | -0.977 | 4.0E-5 | 0.317 |
| 36824 | LINC00702 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MLF1 | Sponge network | -0.737 | 0.06182 | -0.893 | 0.00727 | 0.317 |
| 36825 | HHIP-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | CRTC1 | Sponge network | -0.737 | 0.10822 | -0.116 | 0.28412 | 0.317 |
| 36826 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-577 | 18 | GREM1 | Sponge network | -0.303 | 0.21002 | 0.902 | 0.01691 | 0.317 |
| 36827 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p | 11 | SAMHD1 | Sponge network | 0.214 | 0.18246 | 0.072 | 0.62895 | 0.317 |
| 36828 | RP11-119F7.5 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FMNL3 | Sponge network | 0.225 | 0.30644 | 0.555 | 4.0E-5 | 0.317 |
| 36829 | JAZF1-AS1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | GATA2 | Sponge network | -0.769 | 0.00035 | -0.654 | 0.0016 | 0.317 |
| 36830 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | CEP170 | Sponge network | 0.214 | 0.18246 | 0.943 | 0 | 0.317 |
| 36831 | RP11-216B9.6 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | RYR2 | Sponge network | 0.174 | 0.30034 | -1.466 | 0.00031 | 0.317 |
| 36832 | RP11-506M13.3 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-326 | 11 | CBFB | Sponge network | 0.769 | 0 | 1.061 | 0 | 0.317 |
| 36833 | RP11-251G23.5 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p | 11 | CDK6 | Sponge network | 1.344 | 0 | 0.991 | 0 | 0.317 |
| 36834 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p | 11 | BHLHE41 | Sponge network | -0.309 | 0.1145 | 0.353 | 0.12753 | 0.317 |
| 36835 | CTD-2089N3.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 17 | CCND2 | Sponge network | -1.375 | 0.08642 | -0.267 | 0.3112 | 0.317 |
| 36836 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ZMYND11 | Sponge network | 0.246 | 0.59912 | -0.2 | 0.05449 | 0.317 |
| 36837 | KB-1460A1.5 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 10 | CNTNAP1 | Sponge network | 0.603 | 0.00127 | 0.358 | 0.13727 | 0.317 |
| 36838 | RBPMS-AS1 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | PRKAA2 | Sponge network | 0.144 | 0.64989 | -2.462 | 0 | 0.317 |
| 36839 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | MLLT10 | Sponge network | 0.415 | 0.14657 | 0.105 | 0.33292 | 0.317 |
| 36840 | SOX2-OT |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 27 | AKAP6 | Sponge network | -1.264 | 6.0E-5 | -1.586 | 3.0E-5 | 0.317 |
| 36841 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | HIP1 | Sponge network | 0.349 | 0.01695 | 0.774 | 0 | 0.317 |
| 36842 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 27 | RSPO2 | Sponge network | -0.096 | 0.67958 | -3.038 | 0 | 0.317 |
| 36843 | RP11-21A7A.2 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-576-5p | 10 | MED12L | Sponge network | -1.116 | 0.23686 | -0.194 | 0.55299 | 0.317 |
| 36844 | RP11-571M6.17 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-495-3p | 12 | TBL1XR1 | Sponge network | 1.013 | 0 | 0.578 | 0 | 0.317 |
| 36845 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-3p | 14 | DKK3 | Sponge network | 0.082 | 0.55654 | -0.052 | 0.77783 | 0.317 |
| 36846 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p | 29 | CADM1 | Sponge network | -0.899 | 6.0E-5 | -0.9 | 0.00084 | 0.317 |
| 36847 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-511-5p | 15 | ZFHX3 | Sponge network | 0.453 | 0.01839 | 0.389 | 0.01363 | 0.317 |
| 36848 | LINC00672 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 41 | TRPS1 | Sponge network | -0.227 | 0.31657 | -0.191 | 0.44109 | 0.317 |
| 36849 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-3607-3p;hsa-miR-425-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 17 | SOX5 | Sponge network | 0.342 | 0.03129 | -1.091 | 4.0E-5 | 0.317 |
| 36850 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | RABEP1 | Sponge network | -0.227 | 0.31657 | -0.066 | 0.43142 | 0.317 |
| 36851 | JAZF1-AS1 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RTN4 | Sponge network | -0.769 | 0.00035 | -0.412 | 0.00014 | 0.317 |
| 36852 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-505-5p;hsa-miR-769-5p;hsa-miR-93-5p | 15 | HBP1 | Sponge network | -2.531 | 0 | -0.454 | 0 | 0.317 |
| 36853 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 42 | SSBP2 | Sponge network | 0.056 | 0.81195 | -1.58 | 0 | 0.317 |
| 36854 | DIO3OS |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-93-3p | 10 | FYN | Sponge network | -0.773 | 0.04073 | -0.131 | 0.37051 | 0.317 |
| 36855 | RP11-890B15.3 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940 | 12 | RHOB | Sponge network | 0.101 | 0.60919 | -1.052 | 0 | 0.317 |
| 36856 | RP11-130L8.1 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-671-5p | 17 | KCNA6 | Sponge network | -0.663 | 0.10887 | -1.531 | 0.00013 | 0.317 |
| 36857 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 15 | PRKAA1 | Sponge network | 1.262 | 0 | 0.317 | 0.0031 | 0.317 |
| 36858 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p | 22 | ZFX | Sponge network | 0.214 | 0.18246 | 0.09 | 0.42563 | 0.317 |
| 36859 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-93-5p | 20 | HBP1 | Sponge network | -0.473 | 0.07602 | -0.454 | 0 | 0.317 |
| 36860 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | PAFAH1B1 | Sponge network | 0.064 | 0.72934 | -0.429 | 0 | 0.317 |
| 36861 | CTA-941F9.9 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-223-3p | 11 | TGFBR3 | Sponge network | -0.344 | 0.49624 | -1.173 | 1.0E-5 | 0.317 |
| 36862 | RAMP2-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-590-3p | 13 | STARD13 | Sponge network | -0.513 | 0.04703 | -0.187 | 0.27383 | 0.317 |
| 36863 | RP11-439L18.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-484;hsa-miR-532-5p | 17 | DST | Sponge network | 0.224 | 0.48719 | -0.713 | 0.00096 | 0.317 |
| 36864 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-3p | 17 | EBF3 | Sponge network | -0.125 | 0.49414 | -0.058 | 0.81437 | 0.317 |
| 36865 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-589-5p | 21 | RARB | Sponge network | -0.154 | 0.78939 | -0.825 | 2.0E-5 | 0.317 |
| 36866 | CTB-31O20.4 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 11 | CDK6 | Sponge network | 1.619 | 0 | 0.991 | 0 | 0.317 |
| 36867 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p | 23 | MEF2C | Sponge network | 1.757 | 0 | -0.499 | 0.01448 | 0.317 |
| 36868 | CTC-490E21.10 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-193a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p | 11 | NF1 | Sponge network | 1.46 | 1.0E-5 | 0.51 | 0 | 0.317 |
| 36869 | RP11-736K20.5 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-375 | 12 | CACNA1E | Sponge network | -0.358 | 0.17255 | 1.752 | 0 | 0.317 |
| 36870 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577 | 25 | NRXN1 | Sponge network | 0.056 | 0.81195 | -3.778 | 0 | 0.317 |
| 36871 | C20orf166-AS1 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-582-5p | 12 | HDAC9 | Sponge network | -2.69 | 0.0001 | -0.755 | 0.00495 | 0.317 |
| 36872 | MEG3 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-935 | 15 | SEPT6 | Sponge network | 0.249 | 0.35902 | -0.598 | 0.00181 | 0.317 |
| 36873 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | BCL11A | Sponge network | 0.541 | 0.00125 | -0.932 | 0.0063 | 0.317 |
| 36874 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LRRK2 | Sponge network | 1.302 | 7.0E-5 | -1.136 | 1.0E-5 | 0.317 |
| 36875 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | RCAN2 | Sponge network | 0.374 | 0.20054 | -0.33 | 0.15172 | 0.317 |
| 36876 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-940 | 26 | THBS1 | Sponge network | 0.082 | 0.55654 | 0.741 | 0.00386 | 0.317 |
| 36877 | BZRAP1-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-577;hsa-miR-7-5p;hsa-miR-877-5p | 21 | TET1 | Sponge network | -0.465 | 0.04703 | -0.135 | 0.52138 | 0.317 |
| 36878 | TPT1-AS1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 10 | STAG2 | Sponge network | 0.903 | 0 | 0.252 | 0.00438 | 0.317 |
| 36879 | CTD-2089N3.1 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29a-5p | 19 | THRB | Sponge network | -0.314 | 0.62033 | -1.117 | 0.0004 | 0.317 |
| 36880 | PGM5-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p | 13 | CYLD | Sponge network | -4.546 | 0 | -0.367 | 0.00171 | 0.317 |
| 36881 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-223-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-450b-5p | 13 | DMD | Sponge network | 0.912 | 0.00014 | -1.506 | 0 | 0.317 |
| 36882 | LINC00672 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | TLE4 | Sponge network | -0.227 | 0.31657 | -1.157 | 0 | 0.317 |
| 36883 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p | 13 | MLLT10 | Sponge network | 2.163 | 0 | 0.105 | 0.33292 | 0.317 |
| 36884 | RP11-158K1.3 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | DCLK1 | Sponge network | 0.349 | 0.01695 | -1.324 | 0.00014 | 0.317 |
| 36885 | NDUFA6-AS1 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | PTCH1 | Sponge network | -0.355 | 0.0077 | -0.1 | 0.6179 | 0.317 |
| 36886 | LINC00621 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 12 | PIK3CA | Sponge network | 0.131 | 0.8436 | 0.195 | 0.06908 | 0.317 |
| 36887 | RP11-359B12.2 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 19 | FMNL3 | Sponge network | 0.064 | 0.72934 | 0.555 | 4.0E-5 | 0.317 |
| 36888 | GHRLOS |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-93-3p | 11 | UNC5C | Sponge network | -1.942 | 0 | -0.178 | 0.40214 | 0.317 |
| 36889 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 32 | AKAP11 | Sponge network | 0.659 | 3.0E-5 | 0.196 | 0.09825 | 0.317 |
| 36890 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-590-3p | 23 | MYBL1 | Sponge network | 0.494 | 0.00339 | 0.494 | 0.02152 | 0.316 |
| 36891 | RP11-395A13.2 |
hsa-miR-106a-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-500a-5p | 12 | TACC1 | Sponge network | 0.057 | 0.61225 | -0.362 | 0.05406 | 0.316 |
| 36892 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-423-5p;hsa-miR-7-5p;hsa-miR-96-5p | 10 | ZYX | Sponge network | -1.349 | 0.00017 | 0.019 | 0.89763 | 0.316 |
| 36893 | LINC01006 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p | 16 | CXXC4 | Sponge network | -0.702 | 0.04814 | -0.433 | 0.21055 | 0.316 |
| 36894 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-7-5p | 15 | KCNA6 | Sponge network | -1.483 | 9.0E-5 | -1.531 | 0.00013 | 0.316 |
| 36895 | MALAT1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | KCNAB1 | Sponge network | 0.782 | 0.00016 | -0.755 | 2.0E-5 | 0.316 |
| 36896 | HOXB-AS1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-3p | 15 | BRD3 | Sponge network | -0.154 | 0.53954 | 0.37 | 0.00013 | 0.316 |
| 36897 | RP11-701H24.7 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-532-5p;hsa-miR-576-5p | 10 | NTRK2 | Sponge network | -2.039 | 0.03447 | -0.736 | 0.06495 | 0.316 |
| 36898 | RP4-717I23.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-5p | 15 | TET1 | Sponge network | 0.637 | 0.00069 | -0.135 | 0.52138 | 0.316 |
| 36899 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 29 | HBP1 | Sponge network | -0.49 | 0.04946 | -0.454 | 0 | 0.316 |
| 36900 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b | 14 | CDH19 | Sponge network | 0.082 | 0.55397 | -3.434 | 0 | 0.316 |
| 36901 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-5p | 14 | PDZD2 | Sponge network | -0.355 | 0.0077 | -0.909 | 3.0E-5 | 0.316 |
| 36902 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 17 | ACVR1 | Sponge network | 0.194 | 0.64278 | 0.279 | 0.00234 | 0.316 |
| 36903 | AC016747.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 17 | RPS6KA2 | Sponge network | 0.199 | 0.38015 | -0.57 | 0.00013 | 0.316 |
| 36904 | HOXD-AS2 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-502-5p;hsa-miR-625-5p | 15 | ANK2 | Sponge network | -0.208 | 0.65592 | -1.603 | 1.0E-5 | 0.316 |
| 36905 | H19 |
hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-576-5p | 19 | RUNX1T1 | Sponge network | 2.439 | 0 | -0.344 | 0.2374 | 0.316 |
| 36906 | PSMD5-AS1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 33 | CREB1 | Sponge network | 0.569 | 0.00067 | 0.203 | 0.00537 | 0.316 |
| 36907 | RP11-401P9.4 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-93-3p | 16 | DOCK4 | Sponge network | -0.384 | 0.40652 | 0.502 | 0.00108 | 0.316 |
| 36908 | AC108676.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | ZMYM2 | Sponge network | 4.346 | 2.0E-5 | 0.279 | 0.01273 | 0.316 |
| 36909 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-940 | 23 | LIFR | Sponge network | -0.465 | 0.04703 | -1.794 | 0 | 0.316 |
| 36910 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | TP53INP1 | Sponge network | 0.225 | 0.30644 | -0.496 | 0.00064 | 0.316 |
| 36911 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 18 | PRKAA1 | Sponge network | 1.254 | 0 | 0.317 | 0.0031 | 0.316 |
| 36912 | RP4-791M13.3 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-382-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 44 | LPP | Sponge network | 0.419 | 0.0319 | -0.459 | 0.04475 | 0.316 |
| 36913 | PCA3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | TET1 | Sponge network | -0.709 | 0.18168 | -0.135 | 0.52138 | 0.316 |
| 36914 | CKMT2-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | RHOB | Sponge network | 0.082 | 0.55654 | -1.052 | 0 | 0.316 |
| 36915 | LINC00641 |
hsa-miR-130b-3p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RASA1 | Sponge network | -0.222 | 0.32445 | -0.002 | 0.98204 | 0.316 |
| 36916 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 11 | FGF5 | Sponge network | -4.546 | 0 | 0.549 | 0.28659 | 0.316 |
| 36917 | CKMT2-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p | 14 | ROBO1 | Sponge network | 0.082 | 0.55654 | -0.009 | 0.96154 | 0.316 |
| 36918 | LINC00476 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 50 | TRPS1 | Sponge network | -0.615 | 0.00015 | -0.191 | 0.44109 | 0.316 |
| 36919 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 38 | ERBB4 | Sponge network | -1.203 | 0.03881 | -1.975 | 2.0E-5 | 0.316 |
| 36920 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | CXXC4 | Sponge network | -0.709 | 0.18168 | -0.433 | 0.21055 | 0.316 |
| 36921 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p | 24 | CNTN1 | Sponge network | 0.494 | 0.00339 | -2.594 | 0 | 0.316 |
| 36922 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429 | 10 | TSC22D1 | Sponge network | -0.49 | 0.04946 | -0.529 | 2.0E-5 | 0.316 |
| 36923 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 16 | MOB1B | Sponge network | 0.796 | 4.0E-5 | 0.197 | 0.15266 | 0.316 |
| 36924 | PSMG3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-93-3p | 17 | RPS6KA2 | Sponge network | -0.125 | 0.49414 | -0.57 | 0.00013 | 0.316 |
| 36925 | RP11-395G23.3 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-625-5p | 10 | NGFR | Sponge network | 0.381 | 0.25967 | -1.271 | 0.01058 | 0.316 |
| 36926 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-26b-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | ZMYM2 | Sponge network | 0.545 | 0.00064 | 0.279 | 0.01273 | 0.316 |
| 36927 | RP11-158K1.3 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TSC1 | Sponge network | 0.349 | 0.01695 | 0.103 | 0.26071 | 0.316 |
| 36928 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-590-3p | 10 | FGFR1 | Sponge network | 0.559 | 0.00026 | -0.509 | 0.03957 | 0.316 |
| 36929 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-454-3p | 15 | CLIP1 | Sponge network | -0.154 | 0.78939 | -0.215 | 0.08684 | 0.316 |
| 36930 | RP11-758N13.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p | 10 | IGF1 | Sponge network | 2.939 | 3.0E-5 | -0.204 | 0.5898 | 0.316 |
| 36931 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | ARHGAP28 | Sponge network | -0.739 | 0.00044 | -0.911 | 0.00013 | 0.316 |
| 36932 | RP11-728F11.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 13 | NAV3 | Sponge network | 0.238 | 0.70544 | 0.212 | 0.47729 | 0.316 |
| 36933 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 61 | IL6ST | Sponge network | 1.302 | 7.0E-5 | -0.402 | 0.00676 | 0.316 |
| 36934 | CTA-204B4.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | 0.765 | 7.0E-5 | 0.809 | 0.00544 | 0.316 |
| 36935 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-628-5p;hsa-miR-629-3p | 15 | CHD2 | Sponge network | 0.594 | 0.41522 | 0.049 | 0.55371 | 0.316 |
| 36936 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | KALRN | Sponge network | -2.452 | 0 | -0.819 | 1.0E-5 | 0.316 |
| 36937 | RP11-968O1.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p | 16 | EGFR | Sponge network | -0.829 | 0.01343 | -0.175 | 0.48451 | 0.316 |
| 36938 | GAS6-AS2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | BRD3 | Sponge network | 0.16 | 0.50785 | 0.37 | 0.00013 | 0.316 |
| 36939 | KB-1410C5.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 12 | NOTCH2 | Sponge network | 0.959 | 0 | 0.138 | 0.35163 | 0.316 |
| 36940 | RP11-35G9.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.657 | 4.0E-5 | 1.083 | 0 | 0.316 |
| 36941 | AGAP11 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 39 | GRIK3 | Sponge network | -1.645 | 0 | -3.208 | 0 | 0.316 |
| 36942 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-93-5p | 13 | TP53INP1 | Sponge network | -0.473 | 0.07602 | -0.496 | 0.00064 | 0.316 |
| 36943 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-629-3p | 10 | EPHA7 | Sponge network | -1.717 | 0.01522 | -2.197 | 3.0E-5 | 0.316 |
| 36944 | AF131215.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 11 | PHF3 | Sponge network | -0.176 | 0.59692 | 0.126 | 0.19652 | 0.316 |
| 36945 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | USP12 | Sponge network | 1.205 | 0 | -0.102 | 0.40768 | 0.316 |
| 36946 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p | 10 | MAFB | Sponge network | 0.906 | 0.31715 | -0.023 | 0.89865 | 0.316 |
| 36947 | MIR497HG |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-7-1-3p | 12 | SAMHD1 | Sponge network | -1.439 | 4.0E-5 | 0.072 | 0.62895 | 0.316 |
| 36948 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-3p;hsa-miR-92a-3p | 44 | ZFHX4 | Sponge network | 0.253 | 0.09565 | 0.018 | 0.95574 | 0.316 |
| 36949 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | LRIG1 | Sponge network | 0.225 | 0.30644 | -0.537 | 0.00588 | 0.316 |
| 36950 | LINC00969 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 18 | PIK3C2A | Sponge network | 0.683 | 1.0E-5 | 0.061 | 0.53776 | 0.316 |
| 36951 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3E | Sponge network | -0.323 | 0.01372 | -1.717 | 0.00137 | 0.316 |
| 36952 | TPT1-AS1 |
hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | SLC5A3 | Sponge network | 0.903 | 0 | 0.493 | 5.0E-5 | 0.316 |
| 36953 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p | 23 | FOXP1 | Sponge network | -0.693 | 0.05629 | 0.042 | 0.74368 | 0.316 |
| 36954 | RP11-983P16.4 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-361-3p;hsa-miR-625-5p | 10 | BCORL1 | Sponge network | 0.41 | 0.02214 | 0.354 | 0.01214 | 0.316 |
| 36955 | RP11-116O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 19 | MAF | Sponge network | 0.05 | 0.95483 | -1.257 | 0 | 0.316 |
| 36956 | RP11-758N13.1 |
hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-424-5p | 11 | EYA4 | Sponge network | 2.939 | 3.0E-5 | 0.3 | 0.36876 | 0.316 |
| 36957 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-874-3p | 16 | NR2C2 | Sponge network | 0.422 | 0.27021 | 0.21 | 0.03224 | 0.316 |
| 36958 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | ZFHX4 | Sponge network | 0.537 | 0.00139 | 0.018 | 0.95574 | 0.316 |
| 36959 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 34 | USP12 | Sponge network | -0.49 | 0.04946 | -0.102 | 0.40768 | 0.316 |
| 36960 | RP11-13K12.5 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-500a-5p;hsa-miR-582-3p;hsa-miR-629-3p | 16 | RYR2 | Sponge network | -0.318 | 0.65847 | -1.466 | 0.00031 | 0.316 |
| 36961 | CTC-490E21.10 |
hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p | 11 | GJA1 | Sponge network | 1.46 | 1.0E-5 | -0.485 | 0.02179 | 0.316 |
| 36962 | RP11-345P4.7 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 10 | GSK3B | Sponge network | 1.597 | 0 | 0.266 | 0.00045 | 0.316 |
| 36963 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 35 | PRKCE | Sponge network | -1.111 | 0.0003 | -0.403 | 0.00056 | 0.316 |
| 36964 | ZRANB2-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-93-5p | 18 | PPM1L | Sponge network | -0.376 | 0.00337 | -0.537 | 0.00616 | 0.316 |
| 36965 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 15 | KAT6B | Sponge network | -4.546 | 0 | -0.478 | 0.00018 | 0.316 |
| 36966 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 20 | RNF144A | Sponge network | 0.056 | 0.81195 | 0.594 | 0.0009 | 0.316 |
| 36967 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | FGF5 | Sponge network | 0.246 | 0.59912 | 0.549 | 0.28659 | 0.316 |
| 36968 | AF131215.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 24 | MED12L | Sponge network | -0.176 | 0.59692 | -0.194 | 0.55299 | 0.316 |
| 36969 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-93-3p | 22 | NCOA1 | Sponge network | -0.469 | 0.08721 | -0.432 | 0 | 0.316 |
| 36970 | C4A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-629-5p | 16 | IGF1 | Sponge network | 3.345 | 0 | -0.204 | 0.5898 | 0.316 |
| 36971 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p | 14 | SMARCA2 | Sponge network | -0.162 | 0.47687 | -0.529 | 0.00014 | 0.316 |
| 36972 | RP11-999E24.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-7-5p;hsa-miR-93-3p | 19 | SNX29 | Sponge network | -0.693 | 0.05629 | -0.34 | 0.01371 | 0.316 |
| 36973 | LINC00472 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-942-5p | 12 | ERRFI1 | Sponge network | -0.842 | 0.01361 | -0.798 | 1.0E-5 | 0.316 |
| 36974 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 21 | EPS15 | Sponge network | 1.153 | 0 | -0.052 | 0.53998 | 0.316 |
| 36975 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | MOB1B | Sponge network | 0.538 | 0.00076 | 0.197 | 0.15266 | 0.316 |
| 36976 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-874-3p | 10 | BCLAF1 | Sponge network | 0.72 | 0.0001 | 0.211 | 0.00684 | 0.316 |
| 36977 | ZNF833P |
hsa-let-7a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-766-3p | 12 | TBL1XR1 | Sponge network | 0.688 | 1.0E-5 | 0.578 | 0 | 0.316 |
| 36978 | RP3-402G11.26 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | CADM1 | Sponge network | -0.254 | 0.19303 | -0.9 | 0.00084 | 0.316 |
| 36979 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-652-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 50 | QKI | Sponge network | 0.374 | 0.20054 | -0.333 | 0.04111 | 0.316 |
| 36980 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PLSCR4 | Sponge network | 0.657 | 4.0E-5 | -0.474 | 0.03833 | 0.316 |
| 36981 | U91328.19 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 47 | CADM2 | Sponge network | -0.303 | 0.21002 | -3.782 | 0 | 0.316 |
| 36982 | LINC00667 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TAL1 | Sponge network | -0.235 | 0.15142 | -0.462 | 0.00574 | 0.316 |
| 36983 | SNHG1 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-654-3p | 11 | RICTOR | Sponge network | 1.265 | 0 | 0.388 | 0.00188 | 0.316 |
| 36984 | MIR22HG |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 15 | GLI3 | Sponge network | -1.047 | 0 | -0.615 | 0.01482 | 0.316 |
| 36985 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | NF1 | Sponge network | 1.178 | 0 | 0.51 | 0 | 0.316 |
| 36986 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-766-3p;hsa-miR-940 | 17 | CADM2 | Sponge network | 0.043 | 0.81628 | -3.782 | 0 | 0.316 |
| 36987 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 16 | TMEFF2 | Sponge network | -0.355 | 0.0077 | -2.426 | 0 | 0.316 |
| 36988 | RP11-725P16.2 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PHF3 | Sponge network | 0.422 | 0.27021 | 0.126 | 0.19652 | 0.316 |
| 36989 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | ZMYM2 | Sponge network | 1.089 | 0 | 0.279 | 0.01273 | 0.316 |
| 36990 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-576-5p | 18 | CDHR3 | Sponge network | -2.62 | 0 | -0.977 | 4.0E-5 | 0.316 |
| 36991 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-628-5p;hsa-miR-93-3p | 18 | EPHA3 | Sponge network | 0.859 | 0.00331 | 0.185 | 0.53588 | 0.316 |
| 36992 | BDNF-AS |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p | 11 | CDH23 | Sponge network | -0.668 | 3.0E-5 | -0.974 | 0.00034 | 0.316 |
| 36993 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 31 | FOXP1 | Sponge network | 0.659 | 3.0E-5 | 0.042 | 0.74368 | 0.316 |
| 36994 | CTC-444N24.11 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-628-5p | 11 | MAT2A | Sponge network | 1.153 | 0 | -0.073 | 0.36195 | 0.316 |
| 36995 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-590-3p | 27 | JAZF1 | Sponge network | -0.125 | 0.49414 | -0.377 | 0.02677 | 0.316 |
| 36996 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-93-5p | 15 | RBL2 | Sponge network | -0.842 | 0.01361 | -0.282 | 0.00254 | 0.316 |
| 36997 | RP11-244O19.1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 19 | LIMCH1 | Sponge network | -0.096 | 0.67958 | -0.915 | 1.0E-5 | 0.316 |
| 36998 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 36 | SSBP2 | Sponge network | -0.08 | 0.90092 | -1.58 | 0 | 0.316 |
| 36999 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p | 12 | NCOA1 | Sponge network | 0.858 | 3.0E-5 | -0.432 | 0 | 0.316 |
| 37000 | IQCH-AS1 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-9-5p | 14 | PAWR | Sponge network | 0.929 | 0 | 0.636 | 0 | 0.316 |
| 37001 | ZRANB2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | ETV1 | Sponge network | -0.376 | 0.00337 | -0.096 | 0.67674 | 0.316 |
| 37002 | CTC-457L16.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200c-3p;hsa-miR-671-5p | 12 | DLC1 | Sponge network | 1.153 | 0.00114 | -0.382 | 0.05038 | 0.316 |
| 37003 | MIR600HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p | 17 | AKAP6 | Sponge network | 0.594 | 0.41522 | -1.586 | 3.0E-5 | 0.316 |
| 37004 | RP11-359B12.2 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | 0.064 | 0.72934 | 0.154 | 0.74723 | 0.316 |
| 37005 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-342-3p | 11 | ZNRF3 | Sponge network | 1.344 | 0 | 0.637 | 0.00172 | 0.316 |
| 37006 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-590-3p | 11 | HOOK3 | Sponge network | -0.663 | 0.10887 | 0.273 | 0.09957 | 0.316 |
| 37007 | RP1-59D14.5 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-629-3p | 15 | PIK3R1 | Sponge network | 1.162 | 5.0E-5 | -0.133 | 0.36854 | 0.316 |
| 37008 | PART1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-708-5p;hsa-miR-744-3p;hsa-miR-940 | 36 | TOM1L2 | Sponge network | -2.925 | 0 | -0.994 | 0 | 0.316 |
| 37009 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p | 18 | LRP1B | Sponge network | 0.178 | 0.29106 | -2.163 | 0 | 0.316 |
| 37010 | LINC01001 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-539-5p | 10 | ABI2 | Sponge network | 1.055 | 0 | 0.175 | 0.14787 | 0.316 |
| 37011 | AC108488.4 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-940 | 12 | GLI3 | Sponge network | 0.259 | 0.12542 | -0.615 | 0.01482 | 0.316 |
| 37012 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | WDFY3 | Sponge network | -1.161 | 0.00591 | -0.201 | 0.0967 | 0.316 |
| 37013 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-93-5p | 53 | LPP | Sponge network | 0.051 | 0.83388 | -0.459 | 0.04475 | 0.316 |
| 37014 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | RABEP1 | Sponge network | 0.72 | 0.0001 | -0.066 | 0.43142 | 0.316 |
| 37015 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 19 | CRTC3 | Sponge network | 0.222 | 0.29133 | 0.164 | 0.16786 | 0.316 |
| 37016 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 20 | CDH19 | Sponge network | 0.395 | 0.15451 | -3.434 | 0 | 0.316 |
| 37017 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-654-3p;hsa-miR-664a-3p | 10 | ITCH | Sponge network | 1.344 | 0 | 0.084 | 0.34058 | 0.316 |
| 37018 | SH3BP5-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 13 | PHF3 | Sponge network | 0.267 | 0.2937 | 0.126 | 0.19652 | 0.316 |
| 37019 | PCA3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | INPP4A | Sponge network | -0.709 | 0.18168 | 0.07 | 0.53563 | 0.316 |
| 37020 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-335-5p | 13 | FOXO3 | Sponge network | 0.997 | 0 | -0.04 | 0.72028 | 0.316 |
| 37021 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-590-3p | 11 | TP53INP1 | Sponge network | 0.178 | 0.29106 | -0.496 | 0.00064 | 0.316 |
| 37022 | RP11-33B1.4 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 17 | KIF1B | Sponge network | 0.826 | 0 | -0.118 | 0.32258 | 0.316 |
| 37023 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-671-5p | 20 | RBMS3 | Sponge network | 0.765 | 7.0E-5 | -0.519 | 0.03551 | 0.316 |
| 37024 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 25 | TSHZ3 | Sponge network | 0.051 | 0.83388 | -0.556 | 0.01516 | 0.316 |
| 37025 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-940 | 23 | PBX1 | Sponge network | 0.4 | 0.14611 | -1.206 | 0 | 0.316 |
| 37026 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p | 10 | GPAM | Sponge network | 1.344 | 0 | 0.142 | 0.29732 | 0.316 |
| 37027 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p | 14 | MYO9A | Sponge network | 0.768 | 7.0E-5 | -0.147 | 0.32373 | 0.316 |
| 37028 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 13 | TRIM33 | Sponge network | 1.757 | 0 | 0.402 | 7.0E-5 | 0.316 |
| 37029 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-7-1-3p | 18 | LRP1B | Sponge network | -0.72 | 0.24239 | -2.163 | 0 | 0.316 |
| 37030 | RP11-244H3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-940 | 27 | NTRK2 | Sponge network | 0.659 | 3.0E-5 | -0.736 | 0.06495 | 0.316 |
| 37031 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 38 | PAFAH1B1 | Sponge network | -0.421 | 0.0114 | -0.429 | 0 | 0.316 |
| 37032 | AC034220.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 47 | MDM4 | Sponge network | 0.309 | 0.07304 | 0.345 | 0.00762 | 0.316 |
| 37033 | RP11-195F19.9 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | PPM1L | Sponge network | -0.726 | 0.00132 | -0.537 | 0.00616 | 0.316 |
| 37034 | RP5-1085F17.3 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-664a-5p | 50 | LPP | Sponge network | 0.541 | 0.00125 | -0.459 | 0.04475 | 0.316 |
| 37035 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 32 | AKAP11 | Sponge network | 0.572 | 0.01722 | 0.196 | 0.09825 | 0.316 |
| 37036 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-501-5p;hsa-miR-942-5p | 13 | ERRFI1 | Sponge network | -0.777 | 0.16667 | -0.798 | 1.0E-5 | 0.316 |
| 37037 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | KAT6B | Sponge network | -0.769 | 0.00035 | -0.478 | 0.00018 | 0.316 |
| 37038 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-485-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 30 | PPP1R9A | Sponge network | -0.896 | 0.00099 | -0.903 | 0.04254 | 0.316 |
| 37039 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | BCL6 | Sponge network | 0.101 | 0.60919 | -0.211 | 0.19489 | 0.316 |
| 37040 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | 0.083 | 0.66405 | -1.3 | 0.01619 | 0.316 |
| 37041 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p | 12 | CSRNP1 | Sponge network | -0.469 | 0.08721 | -1.448 | 0 | 0.316 |
| 37042 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-425-5p;hsa-miR-590-5p | 14 | CBL | Sponge network | 0.706 | 0 | 0.291 | 0.01267 | 0.316 |
| 37043 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-942-5p | 25 | NRXN1 | Sponge network | 0.214 | 0.18246 | -3.778 | 0 | 0.316 |
| 37044 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p | 35 | CREB3L2 | Sponge network | -1.057 | 0.00029 | -0.525 | 2.0E-5 | 0.316 |
| 37045 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 13 | FOXP1 | Sponge network | 1.969 | 0.00316 | 0.042 | 0.74368 | 0.316 |
| 37046 | RP11-158K1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | NAV3 | Sponge network | 0.349 | 0.01695 | 0.212 | 0.47729 | 0.316 |
| 37047 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-671-5p | 26 | CREB3L2 | Sponge network | -0.154 | 0.53954 | -0.525 | 2.0E-5 | 0.316 |
| 37048 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ARID5B | Sponge network | 1.308 | 0 | 0.342 | 0.01676 | 0.316 |
| 37049 | RP11-67L2.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 22 | NRK | Sponge network | 0.72 | 0.0001 | 0.656 | 0.19217 | 0.316 |
| 37050 | LINC01006 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p | 34 | PRKAA2 | Sponge network | -0.702 | 0.04814 | -2.462 | 0 | 0.316 |
| 37051 | GNG12-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 43 | PIK3R1 | Sponge network | -0.793 | 7.0E-5 | -0.133 | 0.36854 | 0.316 |
| 37052 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-7-1-3p | 18 | ANK2 | Sponge network | 0.389 | 0.04293 | -1.603 | 1.0E-5 | 0.316 |
| 37053 | CTC-296K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 12 | TGFBR2 | Sponge network | -2.095 | 0.00112 | -0.243 | 0.11408 | 0.316 |
| 37054 | RP11-531A24.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p | 19 | PRDM2 | Sponge network | 0.75 | 0.00054 | -0.258 | 0.00721 | 0.316 |
| 37055 | FAM66C |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZNF844 | Sponge network | -0.901 | 0.00019 | -0.966 | 0.00035 | 0.316 |
| 37056 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p | 14 | KIT | Sponge network | 0.178 | 0.29106 | -2.184 | 0 | 0.316 |
| 37057 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | ZMYM2 | Sponge network | 0.919 | 0 | 0.279 | 0.01273 | 0.316 |
| 37058 | LINC00473 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 11 | EGR2 | Sponge network | -1.203 | 0.03881 | 0.309 | 0.17079 | 0.316 |
| 37059 | FGD5-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 12 | BCL9 | Sponge network | 0.401 | 0.00066 | 0.321 | 0.00267 | 0.316 |
| 37060 | RP5-894A10.6 |
hsa-let-7a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p | 15 | KIF1B | Sponge network | 0.697 | 9.0E-5 | -0.118 | 0.32258 | 0.316 |
| 37061 | PRKAG2-AS1 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PTPRT | Sponge network | -0.622 | 0.02807 | -0.907 | 0.03727 | 0.316 |
| 37062 | CTD-3025N20.3 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 11 | ABL2 | Sponge network | 1.046 | 0 | 0.82 | 0 | 0.316 |
| 37063 | LIPE-AS1 |
hsa-miR-125a-5p;hsa-miR-150-5p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-34c-5p;hsa-miR-497-5p | 10 | RAD23B | Sponge network | 0.078 | 0.55803 | 0.108 | 0.17414 | 0.316 |
| 37064 | RP11-890B15.3 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 21 | GLIPR1 | Sponge network | 0.101 | 0.60919 | 0.146 | 0.3276 | 0.316 |
| 37065 | RP11-395G23.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | NCAM2 | Sponge network | 0.381 | 0.25967 | -0.482 | 0.22565 | 0.316 |
| 37066 | NR2F1-AS1 |
hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-7-1-3p | 10 | PACRG | Sponge network | -0.49 | 0.04946 | -2.248 | 0 | 0.316 |
| 37067 | AC016747.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 29 | NTRK2 | Sponge network | 0.199 | 0.38015 | -0.736 | 0.06495 | 0.316 |
| 37068 | LINC00843 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429 | 11 | PTPN14 | Sponge network | 1.106 | 0 | 0.144 | 0.34595 | 0.316 |
| 37069 | MRPL23-AS1 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p | 13 | PCDH9 | Sponge network | -0.278 | 0.76559 | -1.823 | 4.0E-5 | 0.316 |
| 37070 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-660-5p | 20 | TIMP3 | Sponge network | 0.253 | 0.09565 | -0.546 | 0.02307 | 0.316 |
| 37071 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 38 | CBX5 | Sponge network | 0.619 | 0.00157 | 0.165 | 0.24446 | 0.316 |
| 37072 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 18 | MYBL1 | Sponge network | 0.946 | 0 | 0.494 | 0.02152 | 0.316 |
| 37073 | RP11-77H9.2 |
hsa-miR-125a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 20 | MDM4 | Sponge network | 1.677 | 0 | 0.345 | 0.00762 | 0.316 |
| 37074 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-455-3p | 11 | TLE4 | Sponge network | 0.082 | 0.55397 | -1.157 | 0 | 0.316 |
| 37075 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 21 | RSPO2 | Sponge network | -0.342 | 0.02288 | -3.038 | 0 | 0.316 |
| 37076 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-942-5p | 24 | NOTCH2 | Sponge network | 0.998 | 0 | 0.138 | 0.35163 | 0.316 |
| 37077 | RP11-404E16.1 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-532-5p | 12 | IL6ST | Sponge network | 0.097 | 0.91311 | -0.402 | 0.00676 | 0.316 |
| 37078 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-493-5p | 23 | NFIB | Sponge network | 1.46 | 1.0E-5 | 0.023 | 0.90795 | 0.316 |
| 37079 | RP11-119F7.5 |
hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RET | Sponge network | 0.225 | 0.30644 | -0.833 | 0.03517 | 0.316 |
| 37080 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | CLASP2 | Sponge network | -0.376 | 0.00337 | -0.038 | 0.71 | 0.316 |
| 37081 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-590-5p | 10 | NOTCH2NL | Sponge network | 0.331 | 0.57247 | 0.024 | 0.84307 | 0.316 |
| 37082 | RP11-359B12.2 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 13 | DNMT3A | Sponge network | 0.064 | 0.72934 | 0.302 | 0.0262 | 0.316 |
| 37083 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-629-3p | 17 | IL6ST | Sponge network | 1.969 | 0.00316 | -0.402 | 0.00676 | 0.316 |
| 37084 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 12 | ARHGAP29 | Sponge network | -1.161 | 0.00591 | 0.131 | 0.42953 | 0.316 |
| 37085 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 22 | NAV3 | Sponge network | 0.419 | 0.0319 | 0.212 | 0.47729 | 0.316 |
| 37086 | MRPL23-AS1 |
hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-484 | 10 | IGF2 | Sponge network | -0.278 | 0.76559 | 0.848 | 0.03842 | 0.316 |
| 37087 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | NEDD4 | Sponge network | -0.376 | 0.00337 | 0.02 | 0.89834 | 0.316 |
| 37088 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 21 | CUL5 | Sponge network | 0.415 | 0.14657 | 0.026 | 0.77301 | 0.316 |
| 37089 | RP11-130L8.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | NAV3 | Sponge network | -0.663 | 0.10887 | 0.212 | 0.47729 | 0.316 |
| 37090 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 23 | BMPR2 | Sponge network | 0.131 | 0.8436 | 0.206 | 0.0635 | 0.316 |
| 37091 | RP11-597D13.9 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | 1.302 | 7.0E-5 | -0.704 | 0.00106 | 0.316 |
| 37092 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p | 23 | PIK3C2A | Sponge network | 0.532 | 0.00505 | 0.061 | 0.53776 | 0.316 |
| 37093 | RP11-401P9.4 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-550a-5p | 10 | PCDH18 | Sponge network | -0.384 | 0.40652 | 0.216 | 0.24645 | 0.316 |
| 37094 | AC009948.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-671-5p | 17 | CDH23 | Sponge network | 0.532 | 0.00505 | -0.974 | 0.00034 | 0.316 |
| 37095 | ADAMTS9-AS2 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | GABRB2 | Sponge network | -2.452 | 0 | -1.841 | 0.00029 | 0.316 |
| 37096 | H19 |
hsa-miR-146a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-625-5p | 10 | ADCY1 | Sponge network | 2.439 | 0 | 0.362 | 0.16982 | 0.316 |
| 37097 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | MTSS1 | Sponge network | -0.351 | 0.11358 | -0.81 | 0.00035 | 0.316 |
| 37098 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 52 | IL6ST | Sponge network | 0.222 | 0.29133 | -0.402 | 0.00676 | 0.316 |
| 37099 | CTB-174D11.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p | 11 | ERBB4 | Sponge network | -2.1 | 0.04665 | -1.975 | 2.0E-5 | 0.316 |
| 37100 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-92b-3p | 18 | DMXL1 | Sponge network | -0.567 | 0 | -0.426 | 0.00056 | 0.316 |
| 37101 | RP11-755F10.1 |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | WDFY3 | Sponge network | 0.952 | 0 | -0.201 | 0.0967 | 0.316 |
| 37102 | AC074289.1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-629-3p;hsa-miR-96-5p | 12 | CXCL12 | Sponge network | -0.326 | 0.14314 | -1.421 | 0 | 0.316 |
| 37103 | RP11-127B20.2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-655-3p | 16 | DMD | Sponge network | 0.411 | 0.1335 | -1.506 | 0 | 0.316 |
| 37104 | AC004893.11 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 11 | BCL9 | Sponge network | 1.317 | 0 | 0.321 | 0.00267 | 0.316 |
| 37105 | MIR143HG |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | PRKAR1A | Sponge network | -0.739 | 0.0574 | -0.063 | 0.50314 | 0.316 |
| 37106 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PRKAB2 | Sponge network | 1.378 | 0 | -0.367 | 0.01371 | 0.316 |
| 37107 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 25 | PRRX1 | Sponge network | -0.487 | 0.02157 | 1.316 | 0 | 0.316 |
| 37108 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | AFF3 | Sponge network | 0.199 | 0.38015 | -2.373 | 0 | 0.316 |
| 37109 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-34a-5p;hsa-miR-484 | 11 | IGF2 | Sponge network | -1.331 | 3.0E-5 | 0.848 | 0.03842 | 0.316 |
| 37110 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 19 | ERRFI1 | Sponge network | -2.46 | 0 | -0.798 | 1.0E-5 | 0.316 |
| 37111 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | PDGFRA | Sponge network | -0.187 | 0.23787 | -0.372 | 0.0037 | 0.316 |
| 37112 | HOXA-AS2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | KLF10 | Sponge network | 0.61 | 0.01593 | -0.783 | 0 | 0.315 |
| 37113 | RP11-53O19.1 |
hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 40 | PPM1L | Sponge network | -0.405 | 0.22013 | -0.537 | 0.00616 | 0.315 |
| 37114 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-454-3p;hsa-miR-493-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 29 | DDX6 | Sponge network | 0.603 | 0.00127 | 0.091 | 0.36428 | 0.315 |
| 37115 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 13 | DKK3 | Sponge network | -0.421 | 0.0114 | -0.052 | 0.77783 | 0.315 |
| 37116 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-484 | 10 | TRIM3 | Sponge network | -0.829 | 0.01343 | -0.577 | 0 | 0.315 |
| 37117 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZNF844 | Sponge network | -0.49 | 0.04946 | -0.966 | 0.00035 | 0.315 |
| 37118 | LINC00472 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-874-3p | 21 | CTIF | Sponge network | -0.842 | 0.01361 | -0.389 | 0.00549 | 0.315 |
| 37119 | RP11-65J21.3 |
hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-7-1-3p | 12 | SSBP2 | Sponge network | -0.557 | 0.11135 | -1.58 | 0 | 0.315 |
| 37120 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | PCDH9 | Sponge network | -0.125 | 0.49414 | -1.823 | 4.0E-5 | 0.315 |
| 37121 | AC010226.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | ZMYND11 | Sponge network | -0.811 | 2.0E-5 | -0.2 | 0.05449 | 0.315 |
| 37122 | RP11-597D13.9 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | IRS1 | Sponge network | 1.302 | 7.0E-5 | -0.026 | 0.88855 | 0.315 |
| 37123 | RP11-686D22.8 |
hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 13 | THBS1 | Sponge network | 0.898 | 1.0E-5 | 0.741 | 0.00386 | 0.315 |
| 37124 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 36 | BHLHE41 | Sponge network | -1.111 | 0.0003 | 0.353 | 0.12753 | 0.315 |
| 37125 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-654-3p | 31 | RICTOR | Sponge network | -0.176 | 0.59692 | 0.388 | 0.00188 | 0.315 |
| 37126 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-93-5p | 14 | LRRC55 | Sponge network | 0.082 | 0.55397 | -0.026 | 0.93163 | 0.315 |
| 37127 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 25 | ETV1 | Sponge network | -0.738 | 0.21233 | -0.096 | 0.67674 | 0.315 |
| 37128 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p | 17 | DYNC1I1 | Sponge network | 0.395 | 0.15451 | -1.12 | 0.00107 | 0.315 |
| 37129 | RP11-531A24.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 44 | RUNX1T1 | Sponge network | 0.75 | 0.00054 | -0.344 | 0.2374 | 0.315 |
| 37130 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-590-3p | 16 | GPAM | Sponge network | 1.153 | 0 | 0.142 | 0.29732 | 0.315 |
| 37131 | RP11-38L15.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 14 | QKI | Sponge network | 0.344 | 0.3127 | -0.333 | 0.04111 | 0.315 |
| 37132 | AC074289.1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | SETBP1 | Sponge network | -0.326 | 0.14314 | -1.302 | 1.0E-5 | 0.315 |
| 37133 | RP11-130L8.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-874-3p | 16 | ASTN1 | Sponge network | -0.663 | 0.10887 | -2.389 | 2.0E-5 | 0.315 |
| 37134 | RP11-890B15.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-708-5p;hsa-miR-744-3p;hsa-miR-940 | 24 | TOM1L2 | Sponge network | 0.101 | 0.60919 | -0.994 | 0 | 0.315 |
| 37135 | RP11-867G23.10 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-93-5p | 18 | ACSL6 | Sponge network | -2.531 | 0 | -1.593 | 0 | 0.315 |
| 37136 | H1FX-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ERG | Sponge network | -0.206 | 0.12942 | 0.002 | 0.99011 | 0.315 |
| 37137 | DNM1P35 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-5p | 14 | PTGFR | Sponge network | 0.051 | 0.83388 | -0.233 | 0.45421 | 0.315 |
| 37138 | DNAJC27-AS1 |
hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-96-5p | 22 | PAIP2 | Sponge network | -0.969 | 2.0E-5 | -0.515 | 0 | 0.315 |
| 37139 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | FGF5 | Sponge network | -0.053 | 0.80685 | 0.549 | 0.28659 | 0.315 |
| 37140 | FENDRR |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 12 | UVRAG | Sponge network | -1.281 | 0.00012 | -0.017 | 0.83865 | 0.315 |
| 37141 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | RSPO2 | Sponge network | -0.615 | 0.00015 | -3.038 | 0 | 0.315 |
| 37142 | SNHG14 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | DDX5 | Sponge network | -1.111 | 0.0003 | -0.099 | 0.17428 | 0.315 |
| 37143 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 42 | CUX1 | Sponge network | -1.092 | 4.0E-5 | -0.039 | 0.6705 | 0.315 |
| 37144 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 10 | FNBP1 | Sponge network | 0.131 | 0.8436 | -0.969 | 4.0E-5 | 0.315 |
| 37145 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | MLF1 | Sponge network | -0.899 | 6.0E-5 | -0.893 | 0.00727 | 0.315 |
| 37146 | LPP-AS2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-577 | 11 | INPP4A | Sponge network | -0.18 | 0.20343 | 0.07 | 0.53563 | 0.315 |
| 37147 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 29 | SSBP2 | Sponge network | -0.285 | 0.2172 | -1.58 | 0 | 0.315 |
| 37148 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | CREB1 | Sponge network | 0.419 | 0.0319 | 0.203 | 0.00537 | 0.315 |
| 37149 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 17 | SCN9A | Sponge network | 0.272 | 0.44692 | -0.525 | 0.10593 | 0.315 |
| 37150 | PART1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 12 | ALDH1A2 | Sponge network | -2.925 | 0 | -2.411 | 0 | 0.315 |
| 37151 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 21 | MOB1B | Sponge network | 0.368 | 0.20093 | 0.197 | 0.15266 | 0.315 |
| 37152 | RP11-758N13.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-505-5p | 33 | LPP | Sponge network | 2.939 | 3.0E-5 | -0.459 | 0.04475 | 0.315 |
| 37153 | RP11-686D22.8 |
hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-590-3p | 11 | NRK | Sponge network | 0.898 | 1.0E-5 | 0.656 | 0.19217 | 0.315 |
| 37154 | HAR1A |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 10 | SETBP1 | Sponge network | -0.672 | 0.19447 | -1.302 | 1.0E-5 | 0.315 |
| 37155 | CTB-113P19.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | LPP | Sponge network | 0.199 | 0.52763 | -0.459 | 0.04475 | 0.315 |
| 37156 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 18 | BCL11A | Sponge network | -0.355 | 0.0077 | -0.932 | 0.0063 | 0.315 |
| 37157 | LPP-AS2 |
hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-98-5p | 12 | KLF10 | Sponge network | -0.18 | 0.20343 | -0.783 | 0 | 0.315 |
| 37158 | RP11-532F6.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 16 | PIK3CA | Sponge network | -0.896 | 0.00099 | 0.195 | 0.06908 | 0.315 |
| 37159 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-98-5p | 11 | ITGA5 | Sponge network | -0.405 | 0.22013 | 0.452 | 0.03524 | 0.315 |
| 37160 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p | 45 | BMPR2 | Sponge network | -0.777 | 0.16667 | 0.206 | 0.0635 | 0.315 |
| 37161 | RP11-83A24.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 14 | SHPRH | Sponge network | 0.417 | 0.00445 | 0.098 | 0.40994 | 0.315 |
| 37162 | CTC-510F12.2 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-2355-3p;hsa-miR-3614-5p | 10 | TSC1 | Sponge network | -0.691 | 0.00146 | 0.103 | 0.26071 | 0.315 |
| 37163 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | HECA | Sponge network | -0.901 | 0.00019 | -0.217 | 0.03463 | 0.315 |
| 37164 | DNM1P35 |
hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-590-3p | 10 | MAPK10 | Sponge network | 0.051 | 0.83388 | -1.774 | 0 | 0.315 |
| 37165 | AC003090.1 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | NR3C2 | Sponge network | -2.117 | 0.00161 | -1.56 | 0 | 0.315 |
| 37166 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 13 | NR4A3 | Sponge network | -1.349 | 0.00017 | -1.385 | 0 | 0.315 |
| 37167 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 25 | EBF1 | Sponge network | -0.246 | 0.35034 | -0.384 | 0.08856 | 0.315 |
| 37168 | RP11-983P16.4 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | GALNT13 | Sponge network | 0.41 | 0.02214 | -0.857 | 0.07439 | 0.315 |
| 37169 | LINC00863 |
hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ERCC6 | Sponge network | 0.189 | 0.13981 | 0.23 | 0.015 | 0.315 |
| 37170 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | CLIP1 | Sponge network | -0.896 | 0.00099 | -0.215 | 0.08684 | 0.315 |
| 37171 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-935 | 34 | HBP1 | Sponge network | -2.925 | 0 | -0.454 | 0 | 0.315 |
| 37172 | RP11-333A23.4 |
hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SLC6A15 | Sponge network | -1.791 | 0.04269 | -2.373 | 2.0E-5 | 0.315 |
| 37173 | LINC00472 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-3p | 17 | TLE4 | Sponge network | -0.842 | 0.01361 | -1.157 | 0 | 0.315 |
| 37174 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 25 | LATS1 | Sponge network | 0.064 | 0.72934 | 0.109 | 0.34208 | 0.315 |
| 37175 | RP1-59D14.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p;hsa-miR-940 | 24 | AFF4 | Sponge network | 1.162 | 5.0E-5 | -0.266 | 0.01565 | 0.315 |
| 37176 | RP11-67L2.2 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 24 | SYNPO2 | Sponge network | 0.72 | 0.0001 | -2.342 | 0 | 0.315 |
| 37177 | HOXB-AS3 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p | 13 | CHD5 | Sponge network | 0.343 | 0.28887 | -0.922 | 0.01521 | 0.315 |
| 37178 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | NIN | Sponge network | 0.395 | 0.15451 | -0.253 | 0.13794 | 0.315 |
| 37179 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-576-5p | 24 | IGF1 | Sponge network | 0.395 | 0.15451 | -0.204 | 0.5898 | 0.315 |
| 37180 | GAS6-AS2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-369-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | PREX2 | Sponge network | 0.16 | 0.50785 | -0.272 | 0.25382 | 0.315 |
| 37181 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-495-3p | 18 | CUL5 | Sponge network | 0.373 | 0.00068 | 0.026 | 0.77301 | 0.315 |
| 37182 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-589-3p | 19 | PDGFRA | Sponge network | -4.463 | 0.00025 | -0.372 | 0.0037 | 0.315 |
| 37183 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p | 48 | NTRK3 | Sponge network | -0.384 | 0.40652 | -1.77 | 3.0E-5 | 0.315 |
| 37184 | LINC01018 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-340-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-590-3p | 16 | CSDE1 | Sponge network | -2.915 | 0.00015 | -0.493 | 0 | 0.315 |
| 37185 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 18 | ARID5B | Sponge network | 1.153 | 0.00114 | 0.342 | 0.01676 | 0.315 |
| 37186 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 36 | ABI2 | Sponge network | -0.899 | 6.0E-5 | 0.175 | 0.14787 | 0.315 |
| 37187 | RP11-244H3.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | BRD3 | Sponge network | 0.659 | 3.0E-5 | 0.37 | 0.00013 | 0.315 |
| 37188 | GS1-124K5.11 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-590-3p | 14 | FMNL3 | Sponge network | 0.494 | 0.00339 | 0.555 | 4.0E-5 | 0.315 |
| 37189 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 19 | PIK3R1 | Sponge network | 0.898 | 1.0E-5 | -0.133 | 0.36854 | 0.315 |
| 37190 | RP11-102N12.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-96-5p | 11 | CRTC3 | Sponge network | -0.351 | 0.11358 | 0.164 | 0.16786 | 0.315 |
| 37191 | AC006547.13 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-424-5p | 11 | PHC3 | Sponge network | 1.159 | 0 | 0.212 | 0.0499 | 0.315 |
| 37192 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 12 | NAV3 | Sponge network | 0.37 | 0.27614 | 0.212 | 0.47729 | 0.315 |
| 37193 | RP11-498C9.15 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p | 10 | PAFAH1B2 | Sponge network | 1.087 | 0 | 0.318 | 0.0001 | 0.315 |
| 37194 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 54 | PAFAH1B1 | Sponge network | -0.49 | 0.04946 | -0.429 | 0 | 0.315 |
| 37195 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-744-5p | 11 | USP12 | Sponge network | -0.309 | 0.1145 | -0.102 | 0.40768 | 0.315 |
| 37196 | RP11-296I10.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-34a-5p | 10 | ZMYND11 | Sponge network | 0.592 | 0.00014 | -0.2 | 0.05449 | 0.315 |
| 37197 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429 | 17 | LATS2 | Sponge network | -1.249 | 7.0E-5 | 0.159 | 0.2304 | 0.315 |
| 37198 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 14 | IGF1 | Sponge network | 0.497 | 0.01451 | -0.204 | 0.5898 | 0.315 |
| 37199 | LINC01001 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | UBR3 | Sponge network | 1.055 | 0 | -0.021 | 0.82774 | 0.315 |
| 37200 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 21 | TRPS1 | Sponge network | -0.278 | 0.76559 | -0.191 | 0.44109 | 0.315 |
| 37201 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-628-5p | 11 | FOXP1 | Sponge network | 1.122 | 0 | 0.042 | 0.74368 | 0.315 |
| 37202 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SAMHD1 | Sponge network | -0.576 | 0.10121 | 0.072 | 0.62895 | 0.315 |
| 37203 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 13 | IGSF10 | Sponge network | 0.389 | 0.04293 | -1.751 | 1.0E-5 | 0.315 |
| 37204 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | MED12L | Sponge network | -0.737 | 0.06182 | -0.194 | 0.55299 | 0.315 |
| 37205 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TBX18 | Sponge network | 0.219 | 0.1516 | 0.843 | 0.02945 | 0.315 |
| 37206 | CTD-2314G24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 26 | PRRX1 | Sponge network | 0.246 | 0.59912 | 1.316 | 0 | 0.315 |
| 37207 | RP11-758N13.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 10 | RBMS3 | Sponge network | 2.939 | 3.0E-5 | -0.519 | 0.03551 | 0.315 |
| 37208 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-96-5p | 18 | CLASP2 | Sponge network | 0.37 | 0.27614 | -0.038 | 0.71 | 0.315 |
| 37209 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | TGFBR2 | Sponge network | 0.898 | 1.0E-5 | -0.243 | 0.11408 | 0.315 |
| 37210 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 15 | FREM2 | Sponge network | -0.737 | 0.10822 | -0.437 | 0.46353 | 0.315 |
| 37211 | LIPE-AS1 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-708-5p | 20 | CCDC6 | Sponge network | 0.078 | 0.55803 | 0.27 | 0.02555 | 0.315 |
| 37212 | CTA-445C9.14 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-376c-3p | 10 | TRIM33 | Sponge network | 0.718 | 4.0E-5 | 0.402 | 7.0E-5 | 0.315 |
| 37213 | RP11-35G9.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-589-3p | 14 | TET1 | Sponge network | 0.622 | 0.00015 | -0.135 | 0.52138 | 0.315 |
| 37214 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p | 13 | CXXC4 | Sponge network | 0.368 | 0.20093 | -0.433 | 0.21055 | 0.315 |
| 37215 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | ACVR2A | Sponge network | -2.452 | 0 | -0.409 | 0.00039 | 0.315 |
| 37216 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | CAV1 | Sponge network | 0.101 | 0.60919 | -1.013 | 6.0E-5 | 0.315 |
| 37217 | CTC-444N24.11 |
hsa-let-7e-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 17 | COPS2 | Sponge network | 1.153 | 0 | -0.178 | 0.04974 | 0.315 |
| 37218 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-96-5p | 12 | NUAK1 | Sponge network | 0.622 | 0.00015 | 0.904 | 0 | 0.315 |
| 37219 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | MYO9A | Sponge network | -2.925 | 0 | -0.147 | 0.32373 | 0.315 |
| 37220 | RP11-384O8.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-940 | 45 | WDFY3 | Sponge network | -0.738 | 0.21233 | -0.201 | 0.0967 | 0.315 |
| 37221 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | LATS2 | Sponge network | -2.46 | 0 | 0.159 | 0.2304 | 0.315 |
| 37222 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p | 12 | ABCB5 | Sponge network | -0.405 | 0.22013 | -0.956 | 0.04077 | 0.315 |
| 37223 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | ZNF292 | Sponge network | 0.101 | 0.60919 | 0.276 | 0.04148 | 0.315 |
| 37224 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p;hsa-miR-590-3p | 18 | BRD3 | Sponge network | 1.254 | 0 | 0.37 | 0.00013 | 0.315 |
| 37225 | LINC00476 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-92a-1-5p | 12 | PRKACA | Sponge network | -0.615 | 0.00015 | -0.255 | 0.0012 | 0.315 |
| 37226 | OSER1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 35 | DTNA | Sponge network | -0.13 | 0.48734 | -1.648 | 1.0E-5 | 0.315 |
| 37227 | RASSF8-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 20 | LUZP2 | Sponge network | 0.335 | 0.20596 | -0.056 | 0.89416 | 0.315 |
| 37228 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p | 11 | GAS1 | Sponge network | -1.281 | 0.00012 | -1.047 | 0.00364 | 0.315 |
| 37229 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-493-3p;hsa-miR-590-3p | 14 | JAKMIP2 | Sponge network | -0.355 | 0.0077 | -1.388 | 0 | 0.315 |
| 37230 | NR2F1-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PIK3CA | Sponge network | -0.49 | 0.04946 | 0.195 | 0.06908 | 0.315 |
| 37231 | CTD-2666L21.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p | 11 | ZMYND11 | Sponge network | 0.368 | 0.20093 | -0.2 | 0.05449 | 0.315 |
| 37232 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-7-1-3p | 13 | DKK3 | Sponge network | -0.13 | 0.48734 | -0.052 | 0.77783 | 0.315 |
| 37233 | RAB30-AS1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 18 | CREB1 | Sponge network | 0.752 | 0 | 0.203 | 0.00537 | 0.315 |
| 37234 | SNHG16 |
hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p | 14 | ABL2 | Sponge network | 0.961 | 0 | 0.82 | 0 | 0.315 |
| 37235 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-93-5p | 17 | NIN | Sponge network | -2.531 | 0 | -0.253 | 0.13794 | 0.315 |
| 37236 | RP11-2B6.2 |
hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | PIK3R1 | Sponge network | 0.539 | 0.03722 | -0.133 | 0.36854 | 0.315 |
| 37237 | TAPT1-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-628-5p | 13 | TRIM33 | Sponge network | 0.403 | 2.0E-5 | 0.402 | 7.0E-5 | 0.315 |
| 37238 | RP11-356J5.12 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 36 | IGFBP5 | Sponge network | -0.899 | 6.0E-5 | -0.806 | 0.00105 | 0.315 |
| 37239 | RP11-1277A3.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | BMPR2 | Sponge network | 0.453 | 0.01839 | 0.206 | 0.0635 | 0.315 |
| 37240 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-655-3p | 20 | ZMYND11 | Sponge network | 0.214 | 0.18246 | -0.2 | 0.05449 | 0.315 |
| 37241 | IQCH-AS1 |
hsa-miR-125a-3p;hsa-miR-143-3p;hsa-miR-154-5p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | LIMD1 | Sponge network | 0.929 | 0 | 0.103 | 0.28978 | 0.315 |
| 37242 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | RNF144A | Sponge network | 0.421 | 0.00084 | 0.594 | 0.0009 | 0.315 |
| 37243 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-455-3p;hsa-miR-590-3p | 13 | RELN | Sponge network | 0.101 | 0.60919 | -2.403 | 0 | 0.315 |
| 37244 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-96-5p | 32 | SPRY3 | Sponge network | 0.614 | 1.0E-5 | 0.016 | 0.91286 | 0.315 |
| 37245 | CTD-2002H8.2 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | BRD3 | Sponge network | 0.931 | 1.0E-5 | 0.37 | 0.00013 | 0.315 |
| 37246 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 42 | NTRK3 | Sponge network | 0.174 | 0.30034 | -1.77 | 3.0E-5 | 0.315 |
| 37247 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 26 | ACSL6 | Sponge network | -0.769 | 0.00035 | -1.593 | 0 | 0.315 |
| 37248 | RP11-819C21.1 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-550a-5p;hsa-miR-590-3p | 35 | CCDC6 | Sponge network | 0.537 | 0.00139 | 0.27 | 0.02555 | 0.315 |
| 37249 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-935 | 20 | RARB | Sponge network | -0.096 | 0.67958 | -0.825 | 2.0E-5 | 0.315 |
| 37250 | CALML3-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-542-3p | 14 | TP63 | Sponge network | 0.95 | 0.15943 | -0.363 | 0.38191 | 0.315 |
| 37251 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 33 | AFF4 | Sponge network | 0.453 | 0.01839 | -0.266 | 0.01565 | 0.315 |
| 37252 | MALAT1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ZNF208 | Sponge network | 0.782 | 0.00016 | -0.4 | 0.22381 | 0.315 |
| 37253 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429 | 21 | MOB1B | Sponge network | 0.859 | 0.00331 | 0.197 | 0.15266 | 0.315 |
| 37254 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p | 29 | ADARB1 | Sponge network | 0.657 | 4.0E-5 | -0.335 | 0.06631 | 0.315 |
| 37255 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-5p | 41 | CBX5 | Sponge network | -0.384 | 0.40652 | 0.165 | 0.24446 | 0.315 |
| 37256 | FAM66C |
hsa-miR-107;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | SEPT6 | Sponge network | -0.901 | 0.00019 | -0.598 | 0.00181 | 0.315 |
| 37257 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 16 | PNRC1 | Sponge network | 0.572 | 0.01722 | -0.646 | 0 | 0.315 |
| 37258 | HOXD-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-500a-5p | 19 | CADM2 | Sponge network | -0.208 | 0.65592 | -3.782 | 0 | 0.315 |
| 37259 | LINC00173 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | NIN | Sponge network | 0.056 | 0.8494 | -0.253 | 0.13794 | 0.315 |
| 37260 | RP11-251G23.5 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p | 14 | PAFAH1B2 | Sponge network | 1.344 | 0 | 0.318 | 0.0001 | 0.315 |
| 37261 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | EZH1 | Sponge network | -0.206 | 0.12942 | -0.564 | 1.0E-5 | 0.315 |
| 37262 | A2M-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-935;hsa-miR-940 | 32 | GRIK3 | Sponge network | -0.08 | 0.90092 | -3.208 | 0 | 0.315 |
| 37263 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 14 | DKK3 | Sponge network | 0.392 | 0.0278 | -0.052 | 0.77783 | 0.315 |
| 37264 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 28 | ERBB4 | Sponge network | -0.737 | 0.10822 | -1.975 | 2.0E-5 | 0.315 |
| 37265 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | CADM2 | Sponge network | 0.385 | 0.10067 | -3.782 | 0 | 0.315 |
| 37266 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-671-5p | 25 | KIT | Sponge network | 0.249 | 0.35902 | -2.184 | 0 | 0.315 |
| 37267 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 54 | NTRK3 | Sponge network | -0.125 | 0.49414 | -1.77 | 3.0E-5 | 0.315 |
| 37268 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p | 11 | ABCB5 | Sponge network | -4.477 | 0 | -0.956 | 0.04077 | 0.315 |
| 37269 | RP11-59H7.3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-576-5p | 13 | ABI2 | Sponge network | 2.163 | 0 | 0.175 | 0.14787 | 0.315 |
| 37270 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 16 | RBL2 | Sponge network | 0.572 | 0.01722 | -0.282 | 0.00254 | 0.315 |
| 37271 | AF131215.2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 12 | RABEP1 | Sponge network | -0.176 | 0.59692 | -0.066 | 0.43142 | 0.315 |
| 37272 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p | 15 | ARNT2 | Sponge network | -0.465 | 0.04703 | -0.332 | 0.25851 | 0.315 |
| 37273 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p | 13 | PTGFR | Sponge network | 0.056 | 0.81195 | -0.233 | 0.45421 | 0.315 |
| 37274 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-93-5p | 35 | RASSF2 | Sponge network | -0.154 | 0.78939 | -0.273 | 0.21961 | 0.315 |
| 37275 | AC074289.1 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p | 11 | FGFR1 | Sponge network | -0.326 | 0.14314 | -0.509 | 0.03957 | 0.315 |
| 37276 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-590-3p | 11 | PARK2 | Sponge network | 0.532 | 0.00505 | -1.892 | 0 | 0.315 |
| 37277 | RP11-296I10.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-539-5p | 10 | PDE4DIP | Sponge network | 0.592 | 0.00014 | -0.282 | 0.0257 | 0.315 |
| 37278 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-576-5p | 10 | DACH2 | Sponge network | -2.62 | 0 | -1.635 | 0.00034 | 0.315 |
| 37279 | BMS1P20 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | RICTOR | Sponge network | 0.34 | 0.00273 | 0.388 | 0.00188 | 0.315 |
| 37280 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577 | 11 | CAV1 | Sponge network | 0.056 | 0.81195 | -1.013 | 6.0E-5 | 0.315 |
| 37281 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 13 | FYN | Sponge network | -0.611 | 0.09607 | -0.131 | 0.37051 | 0.315 |
| 37282 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 23 | DMD | Sponge network | -0.125 | 0.49414 | -1.506 | 0 | 0.315 |
| 37283 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-452-5p | 16 | BCL2 | Sponge network | -3.586 | 0.00032 | -0.899 | 0 | 0.315 |
| 37284 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | GRIA3 | Sponge network | -0.709 | 0.18168 | -1.956 | 0 | 0.315 |
| 37285 | GUSBP11 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p | 14 | RASAL2 | Sponge network | 0.856 | 0 | 0.869 | 0 | 0.315 |
| 37286 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-424-5p | 15 | EPHA7 | Sponge network | -0.301 | 0.06597 | -2.197 | 3.0E-5 | 0.315 |
| 37287 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-942-5p | 17 | GPC6 | Sponge network | -0.405 | 0.22013 | -0.054 | 0.81465 | 0.315 |
| 37288 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-654-3p;hsa-miR-96-5p | 25 | FBXO32 | Sponge network | 0.078 | 0.55803 | -0.495 | 0.07561 | 0.315 |
| 37289 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | EPAS1 | Sponge network | -2.117 | 0.00161 | -0.184 | 0.14418 | 0.315 |
| 37290 | TPTEP1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | HERC1 | Sponge network | -1.216 | 0 | -0.196 | 0.08544 | 0.315 |
| 37291 | RP11-1246C19.1 |
hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | ERCC6 | Sponge network | 1.352 | 0 | 0.23 | 0.015 | 0.315 |
| 37292 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-542-3p | 10 | KANK1 | Sponge network | -1.092 | 4.0E-5 | -0.55 | 0.00134 | 0.315 |
| 37293 | LINC00662 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DIP2C | Sponge network | 0.213 | 0.32297 | -0.436 | 0.01713 | 0.315 |
| 37294 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | TACC1 | Sponge network | 0.659 | 3.0E-5 | -0.362 | 0.05406 | 0.315 |
| 37295 | LINC00963 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 31 | NTRK2 | Sponge network | 0.523 | 9.0E-5 | -0.736 | 0.06495 | 0.315 |
| 37296 | TINCR |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p | 18 | UBE4B | Sponge network | -0.664 | 0.14543 | -0.235 | 0.007 | 0.315 |
| 37297 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 12 | GNA13 | Sponge network | 0.594 | 0.00064 | 0.007 | 0.93884 | 0.315 |
| 37298 | RP11-156E6.1 |
hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-497-5p | 10 | RAD23B | Sponge network | 1.266 | 0 | 0.108 | 0.17414 | 0.315 |
| 37299 | EIF3J-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-500b-5p;hsa-miR-590-3p | 19 | SCN9A | Sponge network | 0.331 | 0.00509 | -0.525 | 0.10593 | 0.315 |
| 37300 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-5p | 12 | ITGB3 | Sponge network | 0.16 | 0.50785 | -0.011 | 0.95751 | 0.315 |
| 37301 | THAP9-AS1 |
hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-5p | 13 | TMTC3 | Sponge network | 1.079 | 0 | 0.123 | 0.22189 | 0.315 |
| 37302 | RP11-531A24.5 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-671-5p | 22 | DLC1 | Sponge network | 0.75 | 0.00054 | -0.382 | 0.05038 | 0.315 |
| 37303 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-671-5p;hsa-miR-940 | 15 | THBS1 | Sponge network | -2.095 | 0.00112 | 0.741 | 0.00386 | 0.315 |
| 37304 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 15 | GPAM | Sponge network | 0.946 | 0 | 0.142 | 0.29732 | 0.315 |
| 37305 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-942-5p | 37 | UNC80 | Sponge network | -0.405 | 0.22013 | -1.934 | 0 | 0.315 |
| 37306 | LINC00173 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | PCDH10 | Sponge network | 0.056 | 0.8494 | -1.644 | 0.0078 | 0.315 |
| 37307 | RP11-428J1.5 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-505-5p;hsa-miR-96-5p | 12 | WNK2 | Sponge network | 0.538 | 0.00076 | -0.352 | 0.31066 | 0.315 |
| 37308 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-93-5p | 33 | JAZF1 | Sponge network | -1.645 | 0 | -0.377 | 0.02677 | 0.315 |
| 37309 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | FZD3 | Sponge network | 0.252 | 0.08508 | 0.104 | 0.60316 | 0.315 |
| 37310 | WDFY3-AS2 |
hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | RIT1 | Sponge network | -0.942 | 0 | -0.296 | 0.00054 | 0.315 |
| 37311 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | MOB1B | Sponge network | -0.517 | 0.02935 | 0.197 | 0.15266 | 0.315 |
| 37312 | HOXB-AS2 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-484 | 10 | ABL1 | Sponge network | 1.316 | 0.01106 | -0.215 | 0.07804 | 0.315 |
| 37313 | RP11-403I13.7 |
hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-590-5p;hsa-miR-935 | 12 | MAPK10 | Sponge network | 0.395 | 0.15451 | -1.774 | 0 | 0.315 |
| 37314 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-582-5p | 10 | RELN | Sponge network | 0.889 | 9.0E-5 | -2.403 | 0 | 0.315 |
| 37315 | RP5-1021I20.1 |
hsa-miR-1266-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p | 13 | KCNJ5 | Sponge network | -0.785 | 0.00035 | -0.281 | 0.3339 | 0.315 |
| 37316 | LINC00672 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p | 14 | NOTCH2NL | Sponge network | -0.227 | 0.31657 | 0.024 | 0.84307 | 0.315 |
| 37317 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-96-5p | 11 | MYO9A | Sponge network | 3.21 | 0 | -0.147 | 0.32373 | 0.315 |
| 37318 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | RAP1A | Sponge network | -2.117 | 0.00161 | -0.929 | 0 | 0.315 |
| 37319 | AP001347.6 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p | 16 | ABL1 | Sponge network | -1.161 | 0.00591 | -0.215 | 0.07804 | 0.315 |
| 37320 | CTD-2303H24.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 14 | SHPRH | Sponge network | 0.415 | 0.14657 | 0.098 | 0.40994 | 0.315 |
| 37321 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-7-1-3p | 16 | DKK3 | Sponge network | -0.303 | 0.21002 | -0.052 | 0.77783 | 0.315 |
| 37322 | LINC00472 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | GALNT13 | Sponge network | -0.842 | 0.01361 | -0.857 | 0.07439 | 0.315 |
| 37323 | SH3RF3-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | PTPRT | Sponge network | 0.889 | 9.0E-5 | -0.907 | 0.03727 | 0.315 |
| 37324 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TSC1 | Sponge network | 0.603 | 0.00127 | 0.103 | 0.26071 | 0.315 |
| 37325 | DNAJC27-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 26 | TET1 | Sponge network | -0.969 | 2.0E-5 | -0.135 | 0.52138 | 0.315 |
| 37326 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 21 | ETV1 | Sponge network | 0.299 | 0.57722 | -0.096 | 0.67674 | 0.315 |
| 37327 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-940 | 23 | WDFY3 | Sponge network | 0.529 | 0.00552 | -0.201 | 0.0967 | 0.315 |
| 37328 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | KDSR | Sponge network | 0.782 | 0.00016 | 0.239 | 0.02216 | 0.315 |
| 37329 | GAS6-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-590-5p | 14 | KCNJ5 | Sponge network | 0.16 | 0.50785 | -0.281 | 0.3339 | 0.315 |
| 37330 | LA16c-321D4.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | BRD3 | Sponge network | 2.459 | 5.0E-5 | 0.37 | 0.00013 | 0.315 |
| 37331 | H1FX-AS1 |
hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DACT1 | Sponge network | -0.206 | 0.12942 | 0.783 | 0.00072 | 0.315 |
| 37332 | SERHL |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484 | 11 | NRXN3 | Sponge network | -0.602 | 0.00331 | -0.893 | 0.04186 | 0.315 |
| 37333 | OIP5-AS1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-485-3p;hsa-miR-655-3p;hsa-miR-664a-5p | 13 | TET2 | Sponge network | 0.421 | 0.00084 | -0.222 | 0.04665 | 0.315 |
| 37334 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | WDFY3 | Sponge network | -4.477 | 0 | -0.201 | 0.0967 | 0.315 |
| 37335 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-652-3p;hsa-miR-93-3p | 49 | QKI | Sponge network | 0.253 | 0.09565 | -0.333 | 0.04111 | 0.315 |
| 37336 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 43 | BMPR2 | Sponge network | -1.161 | 0.00591 | 0.206 | 0.0635 | 0.315 |
| 37337 | TAPT1-AS1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p;hsa-miR-664a-5p | 14 | CBFB | Sponge network | 0.403 | 2.0E-5 | 1.061 | 0 | 0.315 |
| 37338 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | SLITRK6 | Sponge network | -1.697 | 0.00889 | -2.121 | 0 | 0.315 |
| 37339 | LINC00865 |
hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-429 | 10 | PPP2R2C | Sponge network | -1.249 | 7.0E-5 | -0.959 | 0.07428 | 0.315 |
| 37340 | DIO3OS |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-940 | 15 | TAL1 | Sponge network | -0.773 | 0.04073 | -0.462 | 0.00574 | 0.315 |
| 37341 | TRIM52-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-3p | 13 | RELN | Sponge network | -0.418 | 0.00068 | -2.403 | 0 | 0.315 |
| 37342 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 19 | ST6GAL2 | Sponge network | 0.889 | 9.0E-5 | -1.201 | 0.00597 | 0.315 |
| 37343 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p | 10 | ELMO1 | Sponge network | 0.299 | 0.57722 | 0.04 | 0.83147 | 0.315 |
| 37344 | MIR600HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-451a;hsa-miR-96-5p | 23 | BCL2 | Sponge network | 0.594 | 0.41522 | -0.899 | 0 | 0.315 |
| 37345 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-5p | 24 | ESR1 | Sponge network | -0.318 | 0.65847 | -1.035 | 3.0E-5 | 0.315 |
| 37346 | LINC00641 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | COPS2 | Sponge network | -0.222 | 0.32445 | -0.178 | 0.04974 | 0.315 |
| 37347 | RP11-1006G14.4 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | ERCC6 | Sponge network | 0.559 | 0.00026 | 0.23 | 0.015 | 0.315 |
| 37348 | GHRLOS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-421 | 12 | RECK | Sponge network | -1.942 | 0 | -1.043 | 0 | 0.315 |
| 37349 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 60 | TRPS1 | Sponge network | 0.421 | 0.00084 | -0.191 | 0.44109 | 0.315 |
| 37350 | FENDRR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 10 | CHD3 | Sponge network | -1.281 | 0.00012 | -0.001 | 0.99669 | 0.315 |
| 37351 | PGM5-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-484 | 15 | CADM1 | Sponge network | -4.546 | 0 | -0.9 | 0.00084 | 0.315 |
| 37352 | DNM3OS |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-375;hsa-miR-378a-5p | 11 | CACNA1E | Sponge network | 0.572 | 0.01722 | 1.752 | 0 | 0.315 |
| 37353 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 33 | EPHA5 | Sponge network | 0.614 | 1.0E-5 | -0.957 | 0.01733 | 0.315 |
| 37354 | LINC00342 |
hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 10 | RICTOR | Sponge network | 0.297 | 0.19933 | 0.388 | 0.00188 | 0.315 |
| 37355 | RP11-6O2.3 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-382-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-590-5p | 18 | INPP4B | Sponge network | 0.331 | 0.57247 | -0.097 | 0.60503 | 0.315 |
| 37356 | CTC-457L16.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p | 11 | EZH1 | Sponge network | 1.153 | 0.00114 | -0.564 | 1.0E-5 | 0.315 |
| 37357 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-324-3p | 12 | ZBTB4 | Sponge network | 0.37 | 0.27614 | -0.739 | 0 | 0.315 |
| 37358 | CTA-363E19.2 |
hsa-miR-142-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 10 | CDC73 | Sponge network | 1.055 | 0 | 0.094 | 0.20463 | 0.315 |
| 37359 | LINC00622 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p | 11 | MYBL1 | Sponge network | 1.169 | 0.00028 | 0.494 | 0.02152 | 0.315 |
| 37360 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577 | 21 | SCN9A | Sponge network | 0.389 | 0.04293 | -0.525 | 0.10593 | 0.315 |
| 37361 | DIO3OS |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p | 16 | RPS6KA6 | Sponge network | -0.773 | 0.04073 | -2.989 | 0 | 0.315 |
| 37362 | RP11-244O19.1 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-625-5p | 10 | LZTS1 | Sponge network | -0.096 | 0.67958 | 1.664 | 0 | 0.315 |
| 37363 | RP11-345P4.7 |
hsa-miR-10b-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 16 | ARHGEF12 | Sponge network | 1.597 | 0 | 0.09 | 0.35693 | 0.315 |
| 37364 | LINC00702 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 11 | FOXO4 | Sponge network | -0.737 | 0.06182 | -0.516 | 2.0E-5 | 0.315 |
| 37365 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SUFU | Sponge network | 0.727 | 3.0E-5 | -0.04 | 0.65489 | 0.315 |
| 37366 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ABCC9 | Sponge network | -0.141 | 0.26434 | -0.466 | 0.14121 | 0.315 |
| 37367 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p | 10 | RBMS3 | Sponge network | 2.364 | 0.00134 | -0.519 | 0.03551 | 0.315 |
| 37368 | RP11-20I23.13 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-92a-3p | 16 | LRIG1 | Sponge network | 0.505 | 0.0014 | -0.537 | 0.00588 | 0.315 |
| 37369 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | GALNT13 | Sponge network | 0.395 | 0.15451 | -0.857 | 0.07439 | 0.315 |
| 37370 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-766-3p | 26 | CHD5 | Sponge network | -0.602 | 0.00331 | -0.922 | 0.01521 | 0.315 |
| 37371 | RP11-554A11.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-421;hsa-miR-450b-5p | 13 | PRKCE | Sponge network | -0.583 | 0.14589 | -0.403 | 0.00056 | 0.315 |
| 37372 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 21 | ADAMTSL3 | Sponge network | 0.41 | 0.02214 | -1.559 | 0 | 0.315 |
| 37373 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 16 | SUFU | Sponge network | -0.206 | 0.12942 | -0.04 | 0.65489 | 0.314 |
| 37374 | BCYRN1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p | 60 | DST | Sponge network | 0.311 | 0.10344 | -0.713 | 0.00096 | 0.314 |
| 37375 | RP11-307B6.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-532-5p | 13 | MAF | Sponge network | -4.463 | 0.00025 | -1.257 | 0 | 0.314 |
| 37376 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-629-5p | 35 | BCL2 | Sponge network | -0.154 | 0.78939 | -0.899 | 0 | 0.314 |
| 37377 | LINC00865 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-7-1-3p | 10 | NRG3 | Sponge network | -1.249 | 7.0E-5 | -1.836 | 4.0E-5 | 0.314 |
| 37378 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 30 | ETV1 | Sponge network | 0.532 | 0.00505 | -0.096 | 0.67674 | 0.314 |
| 37379 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-96-5p | 16 | BMPR2 | Sponge network | 3.21 | 0 | 0.206 | 0.0635 | 0.314 |
| 37380 | LINC00963 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | NF1 | Sponge network | 0.523 | 9.0E-5 | 0.51 | 0 | 0.314 |
| 37381 | RP1-302G2.5 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-29b-1-5p;hsa-miR-766-3p;hsa-miR-93-3p | 14 | IL6ST | Sponge network | -1.483 | 9.0E-5 | -0.402 | 0.00676 | 0.314 |
| 37382 | THAP9-AS1 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 12 | NF1 | Sponge network | 1.079 | 0 | 0.51 | 0 | 0.314 |
| 37383 | BVES-AS1 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-877-5p | 27 | AFF1 | Sponge network | -2.62 | 0 | -0.384 | 0.00053 | 0.314 |
| 37384 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 25 | MCC | Sponge network | -0.227 | 0.31657 | -1.005 | 0 | 0.314 |
| 37385 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-96-5p | 14 | ELMO1 | Sponge network | 0.194 | 0.64278 | 0.04 | 0.83147 | 0.314 |
| 37386 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | PRRX1 | Sponge network | 0.898 | 1.0E-5 | 1.316 | 0 | 0.314 |
| 37387 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | CLASP2 | Sponge network | 0.659 | 3.0E-5 | -0.038 | 0.71 | 0.314 |
| 37388 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p | 12 | TSHZ3 | Sponge network | 1.46 | 1.0E-5 | -0.556 | 0.01516 | 0.314 |
| 37389 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 18 | RBL2 | Sponge network | -1.281 | 0.00012 | -0.282 | 0.00254 | 0.314 |
| 37390 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-940 | 35 | MEF2C | Sponge network | 0.494 | 0.00339 | -0.499 | 0.01448 | 0.314 |
| 37391 | LINC00672 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 17 | ERCC4 | Sponge network | -0.227 | 0.31657 | 0.126 | 0.15266 | 0.314 |
| 37392 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-29a-5p | 16 | JAZF1 | Sponge network | 0.419 | 0.0319 | -0.377 | 0.02677 | 0.314 |
| 37393 | LINC00094 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p | 38 | NTRK2 | Sponge network | 0.253 | 0.09565 | -0.736 | 0.06495 | 0.314 |
| 37394 | RP5-994D16.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PPM1A | Sponge network | 0.252 | 0.08508 | -0.623 | 0 | 0.314 |
| 37395 | RP11-360F5.1 |
hsa-miR-10a-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429 | 16 | LPP | Sponge network | 2.364 | 0.00134 | -0.459 | 0.04475 | 0.314 |
| 37396 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-877-5p | 43 | BMPR2 | Sponge network | -2.62 | 0 | 0.206 | 0.0635 | 0.314 |
| 37397 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-3p | 10 | ATRX | Sponge network | 1.4 | 0 | 0.261 | 0.04408 | 0.314 |
| 37398 | RP11-597D13.9 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 10 | PXDN | Sponge network | 1.302 | 7.0E-5 | 1.038 | 0 | 0.314 |
| 37399 | AC083843.1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p | 15 | CBFB | Sponge network | 1.4 | 0 | 1.061 | 0 | 0.314 |
| 37400 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p | 16 | EYA4 | Sponge network | -0.517 | 0.02935 | 0.3 | 0.36876 | 0.314 |
| 37401 | EMX2OS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p | 13 | ELMO1 | Sponge network | -0.777 | 0.1134 | 0.04 | 0.83147 | 0.314 |
| 37402 | DNAJC27-AS1 |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | PACRG | Sponge network | -0.969 | 2.0E-5 | -2.248 | 0 | 0.314 |
| 37403 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 43 | TCF4 | Sponge network | -0.737 | 0.10822 | 0.016 | 0.92271 | 0.314 |
| 37404 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 37 | ZFHX3 | Sponge network | -0.303 | 0.21002 | 0.389 | 0.01363 | 0.314 |
| 37405 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p | 10 | FOXO3 | Sponge network | 0.793 | 0 | -0.04 | 0.72028 | 0.314 |
| 37406 | RP11-526I2.5 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | CLASP2 | Sponge network | 1.054 | 1.0E-5 | -0.038 | 0.71 | 0.314 |
| 37407 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SOX5 | Sponge network | -0.021 | 0.93598 | -1.091 | 4.0E-5 | 0.314 |
| 37408 | RP11-967K21.1 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-3p | 20 | LIMCH1 | Sponge network | 0.056 | 0.81195 | -0.915 | 1.0E-5 | 0.314 |
| 37409 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 17 | MYBL1 | Sponge network | 1.757 | 0 | 0.494 | 0.02152 | 0.314 |
| 37410 | LINC00863 |
hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ABI2 | Sponge network | 0.189 | 0.13981 | 0.175 | 0.14787 | 0.314 |
| 37411 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | ZNF521 | Sponge network | -1.047 | 0 | -0.111 | 0.58068 | 0.314 |
| 37412 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-744-3p | 20 | ADAMTSL3 | Sponge network | 0.853 | 0.15352 | -1.559 | 0 | 0.314 |
| 37413 | RP11-983P16.4 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-576-5p | 10 | HERC1 | Sponge network | 0.41 | 0.02214 | -0.196 | 0.08544 | 0.314 |
| 37414 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 70 | KIF1B | Sponge network | -0.129 | 0.57177 | -0.118 | 0.32258 | 0.314 |
| 37415 | FAM13A-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 14 | ZBTB20 | Sponge network | -0.83 | 0 | -0.122 | 0.57569 | 0.314 |
| 37416 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p | 21 | NRK | Sponge network | 0.083 | 0.66405 | 0.656 | 0.19217 | 0.314 |
| 37417 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-93-5p | 12 | TRIM3 | Sponge network | -0.83 | 0 | -0.577 | 0 | 0.314 |
| 37418 | AC138035.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-539-5p | 19 | KIF1B | Sponge network | 1.073 | 0 | -0.118 | 0.32258 | 0.314 |
| 37419 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-96-5p | 51 | FAT3 | Sponge network | 1.302 | 7.0E-5 | -1.601 | 0.00051 | 0.314 |
| 37420 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-196a-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500b-5p | 15 | RSPO2 | Sponge network | -0.418 | 0.00068 | -3.038 | 0 | 0.314 |
| 37421 | RP11-1006G14.4 |
hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PCDH9 | Sponge network | 0.559 | 0.00026 | -1.823 | 4.0E-5 | 0.314 |
| 37422 | RP3-475N16.1 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-3614-5p;hsa-miR-495-3p | 12 | TSC1 | Sponge network | 1.056 | 0 | 0.103 | 0.26071 | 0.314 |
| 37423 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p | 26 | ARID5B | Sponge network | 0.494 | 0.00339 | 0.342 | 0.01676 | 0.314 |
| 37424 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-582-5p | 18 | RBMS3 | Sponge network | 0.792 | 0.0049 | -0.519 | 0.03551 | 0.314 |
| 37425 | MEG3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | CREB1 | Sponge network | 0.249 | 0.35902 | 0.203 | 0.00537 | 0.314 |
| 37426 | LINC00094 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 14 | DACT1 | Sponge network | 0.253 | 0.09565 | 0.783 | 0.00072 | 0.314 |
| 37427 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 15 | GPAM | Sponge network | 0.924 | 0 | 0.142 | 0.29732 | 0.314 |
| 37428 | RP11-855A2.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ZFX | Sponge network | 2.094 | 0 | 0.09 | 0.42563 | 0.314 |
| 37429 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3613-5p | 16 | GPAM | Sponge network | 0.614 | 1.0E-5 | 0.142 | 0.29732 | 0.314 |
| 37430 | RP11-728F11.4 |
hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 11 | EPHA3 | Sponge network | 0.238 | 0.70544 | 0.185 | 0.53588 | 0.314 |
| 37431 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 25 | NRXN1 | Sponge network | 0.395 | 0.15451 | -3.778 | 0 | 0.314 |
| 37432 | CD27-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-625-5p | 27 | JAZF1 | Sponge network | 0.177 | 0.16681 | -0.377 | 0.02677 | 0.314 |
| 37433 | AC106786.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p | 13 | FAT4 | Sponge network | 1.122 | 0.02989 | -0.354 | 0.14694 | 0.314 |
| 37434 | RP11-13K12.5 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p | 12 | CSDE1 | Sponge network | -0.318 | 0.65847 | -0.493 | 0 | 0.314 |
| 37435 | GS1-124K5.11 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-942-5p | 29 | ARHGEF12 | Sponge network | 0.494 | 0.00339 | 0.09 | 0.35693 | 0.314 |
| 37436 | TPTEP1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 19 | GLIPR1 | Sponge network | -1.216 | 0 | 0.146 | 0.3276 | 0.314 |
| 37437 | LINC00865 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-582-5p | 14 | PTCH1 | Sponge network | -1.249 | 7.0E-5 | -0.1 | 0.6179 | 0.314 |
| 37438 | AC108488.4 |
hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-7-1-3p | 10 | FGF5 | Sponge network | 0.259 | 0.12542 | 0.549 | 0.28659 | 0.314 |
| 37439 | SH3RF3-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | AFF1 | Sponge network | 0.889 | 9.0E-5 | -0.384 | 0.00053 | 0.314 |
| 37440 | LINC00094 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p | 49 | DTNA | Sponge network | 0.253 | 0.09565 | -1.648 | 1.0E-5 | 0.314 |
| 37441 | EIF3J-AS1 |
hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 0.331 | 0.00509 | 0.321 | 0.00267 | 0.314 |
| 37442 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-93-3p | 16 | RPS6KA2 | Sponge network | 0.083 | 0.66405 | -0.57 | 0.00013 | 0.314 |
| 37443 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | PHC3 | Sponge network | 0.856 | 0 | 0.212 | 0.0499 | 0.314 |
| 37444 | RP5-994D16.9 |
hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 16 | IL6ST | Sponge network | 0.252 | 0.08508 | -0.402 | 0.00676 | 0.314 |
| 37445 | TRIM52-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-362-5p;hsa-miR-532-3p | 13 | MRVI1 | Sponge network | -0.418 | 0.00068 | -1.051 | 0.00022 | 0.314 |
| 37446 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 12 | CHD1 | Sponge network | 0.72 | 0.0001 | 0.322 | 0.00215 | 0.314 |
| 37447 | RP11-3P17.4 |
hsa-miR-125a-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-34c-5p;hsa-miR-497-5p | 13 | RAD23B | Sponge network | 0.144 | 0.07137 | 0.108 | 0.17414 | 0.314 |
| 37448 | RP11-395G23.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 23 | TGFBR3 | Sponge network | 0.381 | 0.25967 | -1.173 | 1.0E-5 | 0.314 |
| 37449 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p | 31 | RICTOR | Sponge network | 0.194 | 0.64278 | 0.388 | 0.00188 | 0.314 |
| 37450 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p | 20 | NTRK2 | Sponge network | -0.726 | 0.00132 | -0.736 | 0.06495 | 0.314 |
| 37451 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | PSIP1 | Sponge network | -0.096 | 0.67958 | -0.005 | 0.96897 | 0.314 |
| 37452 | HOXB-AS2 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-429 | 10 | AKAP11 | Sponge network | 1.316 | 0.01106 | 0.196 | 0.09825 | 0.314 |
| 37453 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 24 | CYLD | Sponge network | -1.439 | 4.0E-5 | -0.367 | 0.00171 | 0.314 |
| 37454 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-874-3p | 26 | CREB3L2 | Sponge network | -0.465 | 0.04703 | -0.525 | 2.0E-5 | 0.314 |
| 37455 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 33 | UNC80 | Sponge network | 0.176 | 0.37402 | -1.934 | 0 | 0.314 |
| 37456 | AGAP11 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p | 17 | EFNA5 | Sponge network | -1.645 | 0 | -0.421 | 0.17868 | 0.314 |
| 37457 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 38 | EBF1 | Sponge network | -0.942 | 0 | -0.384 | 0.08856 | 0.314 |
| 37458 | LINC00863 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | BRD3 | Sponge network | 0.189 | 0.13981 | 0.37 | 0.00013 | 0.314 |
| 37459 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | SON | Sponge network | 0.983 | 0.00048 | 0.168 | 0.04406 | 0.314 |
| 37460 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | 0.494 | 0.00339 | -0.001 | 0.99669 | 0.314 |
| 37461 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 31 | PRKAB2 | Sponge network | -0.83 | 0 | -0.367 | 0.01371 | 0.314 |
| 37462 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 15 | BCL6 | Sponge network | 1.757 | 0 | -0.211 | 0.19489 | 0.314 |
| 37463 | GAS6-AS2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | ROBO1 | Sponge network | 0.16 | 0.50785 | -0.009 | 0.96154 | 0.314 |
| 37464 | CD27-AS1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-655-3p;hsa-miR-940 | 11 | TAF1 | Sponge network | 0.177 | 0.16681 | 0.39 | 3.0E-5 | 0.314 |
| 37465 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | NFIB | Sponge network | -0.285 | 0.2172 | 0.023 | 0.90795 | 0.314 |
| 37466 | HHIP-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | DIP2C | Sponge network | -0.737 | 0.10822 | -0.436 | 0.01713 | 0.314 |
| 37467 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 15 | IGSF10 | Sponge network | 0.494 | 0.00339 | -1.751 | 1.0E-5 | 0.314 |
| 37468 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30e-5p;hsa-miR-532-3p;hsa-miR-93-3p | 11 | RPS6KA2 | Sponge network | -0.405 | 0.22013 | -0.57 | 0.00013 | 0.314 |
| 37469 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p | 26 | TMEM132B | Sponge network | 0.178 | 0.29106 | -0.83 | 0.01084 | 0.314 |
| 37470 | RP11-403I13.7 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-484 | 13 | EZH1 | Sponge network | 0.395 | 0.15451 | -0.564 | 1.0E-5 | 0.314 |
| 37471 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 53 | TCF4 | Sponge network | 0.537 | 0.00139 | 0.016 | 0.92271 | 0.314 |
| 37472 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 14 | DACT1 | Sponge network | 1.757 | 0 | 0.783 | 0.00072 | 0.314 |
| 37473 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 25 | ARHGAP28 | Sponge network | -0.769 | 0.00035 | -0.911 | 0.00013 | 0.314 |
| 37474 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-7-1-3p | 14 | SLC6A15 | Sponge network | 0.056 | 0.8494 | -2.373 | 2.0E-5 | 0.314 |
| 37475 | RP1-59D14.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-337-3p | 15 | ZFX | Sponge network | 1.162 | 5.0E-5 | 0.09 | 0.42563 | 0.314 |
| 37476 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-629-5p | 33 | KIF1B | Sponge network | 0.001 | 0.99753 | -0.118 | 0.32258 | 0.314 |
| 37477 | FAM13A-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 16 | SORCS1 | Sponge network | -0.83 | 0 | -3.492 | 0 | 0.314 |
| 37478 | CTD-3092A11.2 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-664a-5p | 10 | CBFB | Sponge network | 0.873 | 0 | 1.061 | 0 | 0.314 |
| 37479 | PCA3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 12 | PIK3IP1 | Sponge network | -0.709 | 0.18168 | -0.187 | 0.19945 | 0.314 |
| 37480 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 13 | NUAK1 | Sponge network | -0.737 | 0.10822 | 0.904 | 0 | 0.314 |
| 37481 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CAV1 | Sponge network | 0.199 | 0.38015 | -1.013 | 6.0E-5 | 0.314 |
| 37482 | GAS6-AS2 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-940 | 10 | RHOB | Sponge network | 0.16 | 0.50785 | -1.052 | 0 | 0.314 |
| 37483 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-940 | 47 | NTRK3 | Sponge network | -0.738 | 0.21233 | -1.77 | 3.0E-5 | 0.314 |
| 37484 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 37 | RASSF2 | Sponge network | 1.302 | 7.0E-5 | -0.273 | 0.21961 | 0.314 |
| 37485 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIP11 | Sponge network | 1.601 | 0 | -0.158 | 0.06384 | 0.314 |
| 37486 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-589-5p | 16 | ARHGAP29 | Sponge network | -0.154 | 0.78939 | 0.131 | 0.42953 | 0.314 |
| 37487 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-223-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p | 10 | PDS5B | Sponge network | 1.262 | 0 | 0.259 | 0.00962 | 0.314 |
| 37488 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-590-3p | 11 | AKAP11 | Sponge network | 0.702 | 7.0E-5 | 0.196 | 0.09825 | 0.314 |
| 37489 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 11 | ACTB | Sponge network | -1.092 | 4.0E-5 | -0.247 | 0.01736 | 0.314 |
| 37490 | LINC00449 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p | 11 | GSK3B | Sponge network | 1.525 | 0 | 0.266 | 0.00045 | 0.314 |
| 37491 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | THBS1 | Sponge network | -0.021 | 0.93598 | 0.741 | 0.00386 | 0.314 |
| 37492 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-532-5p | 13 | MAF | Sponge network | -0.164 | 0.45106 | -1.257 | 0 | 0.314 |
| 37493 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | PTPN11 | Sponge network | 0.909 | 0 | 0.431 | 2.0E-5 | 0.314 |
| 37494 | RP11-707P17.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | PRKAA1 | Sponge network | 1.877 | 0 | 0.317 | 0.0031 | 0.314 |
| 37495 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b | 10 | RBM5 | Sponge network | 0.614 | 1.0E-5 | -0.205 | 0.0129 | 0.314 |
| 37496 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | PDS5B | Sponge network | 0.873 | 0 | 0.259 | 0.00962 | 0.314 |
| 37497 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-3p | 12 | CREBBP | Sponge network | 0.331 | 0.57247 | 0.126 | 0.15186 | 0.314 |
| 37498 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-339-5p | 10 | ZNF521 | Sponge network | 0.395 | 0.15451 | -0.111 | 0.58068 | 0.314 |
| 37499 | RP11-356J5.12 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | LUZP2 | Sponge network | -0.899 | 6.0E-5 | -0.056 | 0.89416 | 0.314 |
| 37500 | U91328.19 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 42 | PPARA | Sponge network | -0.303 | 0.21002 | -0.379 | 0.00548 | 0.314 |
| 37501 | SOX2-OT |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 37 | NTRK2 | Sponge network | -1.264 | 6.0E-5 | -0.736 | 0.06495 | 0.314 |
| 37502 | KB-1572G7.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p | 17 | ZFX | Sponge network | 0.924 | 0 | 0.09 | 0.42563 | 0.314 |
| 37503 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p | 42 | PRKAA2 | Sponge network | 0.95 | 0.15943 | -2.462 | 0 | 0.314 |
| 37504 | AC109309.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-193b-5p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b | 10 | DMD | Sponge network | 0.163 | 0.87219 | -1.506 | 0 | 0.314 |
| 37505 | RP11-395G23.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-769-5p | 15 | SETBP1 | Sponge network | 0.381 | 0.25967 | -1.302 | 1.0E-5 | 0.314 |
| 37506 | CTC-444N24.11 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-299-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-369-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-590-3p | 11 | MACF1 | Sponge network | 1.153 | 0 | 0.019 | 0.90335 | 0.314 |
| 37507 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-7-1-3p | 12 | ZMYND11 | Sponge network | -0.285 | 0.2172 | -0.2 | 0.05449 | 0.314 |
| 37508 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 18 | TSHZ3 | Sponge network | 1.153 | 0.00114 | -0.556 | 0.01516 | 0.314 |
| 37509 | RP11-693J15.4 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-590-5p | 10 | CITED2 | Sponge network | -0.72 | 0.24239 | -1.545 | 0 | 0.314 |
| 37510 | DNM1P35 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 11 | BCL9 | Sponge network | 0.051 | 0.83388 | 0.321 | 0.00267 | 0.314 |
| 37511 | RP11-620J15.3 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | OPCML | Sponge network | -0.739 | 0.00044 | -2.498 | 0 | 0.314 |
| 37512 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p | 16 | HECA | Sponge network | 0.921 | 0.00118 | -0.217 | 0.03463 | 0.314 |
| 37513 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-664a-3p;hsa-miR-93-5p | 22 | PRRX1 | Sponge network | -2.69 | 0.0001 | 1.316 | 0 | 0.314 |
| 37514 | LINC00969 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 13 | KIAA1024 | Sponge network | 0.683 | 1.0E-5 | 1.083 | 0 | 0.314 |
| 37515 | RP11-254F7.2 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-539-5p | 18 | RIMS2 | Sponge network | 0.176 | 0.37402 | -1.365 | 0.00208 | 0.314 |
| 37516 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | RNF144A | Sponge network | 0.61 | 0.01593 | 0.594 | 0.0009 | 0.314 |
| 37517 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-93-5p | 18 | CDHR3 | Sponge network | -0.842 | 0.01361 | -0.977 | 4.0E-5 | 0.314 |
| 37518 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZMYND11 | Sponge network | -1.161 | 0.00591 | -0.2 | 0.05449 | 0.314 |
| 37519 | RP11-13K12.1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p | 16 | CSDE1 | Sponge network | -0.154 | 0.78939 | -0.493 | 0 | 0.314 |
| 37520 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | MAF | Sponge network | -0.811 | 2.0E-5 | -1.257 | 0 | 0.314 |
| 37521 | RP11-434B12.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PEG3 | Sponge network | -0.031 | 0.89063 | -1.74 | 0 | 0.314 |
| 37522 | LINC00641 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | THSD7B | Sponge network | -0.222 | 0.32445 | 0.154 | 0.74723 | 0.314 |
| 37523 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-484 | 13 | GREM1 | Sponge network | 0.4 | 0.14611 | 0.902 | 0.01691 | 0.314 |
| 37524 | RP11-401P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326 | 11 | AKAP6 | Sponge network | 1.524 | 0.1098 | -1.586 | 3.0E-5 | 0.314 |
| 37525 | FAM13A-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 35 | BCL2 | Sponge network | -0.83 | 0 | -0.899 | 0 | 0.314 |
| 37526 | PART1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 34 | KCNA6 | Sponge network | -2.925 | 0 | -1.531 | 0.00013 | 0.314 |
| 37527 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 16 | CREBBP | Sponge network | -2.452 | 0 | 0.126 | 0.15186 | 0.314 |
| 37528 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p | 16 | USP12 | Sponge network | 0.594 | 0.41522 | -0.102 | 0.40768 | 0.314 |
| 37529 | LINC00654 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 15 | CELF4 | Sponge network | 0.374 | 0.20054 | -1.135 | 0.00344 | 0.314 |
| 37530 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-589-3p | 12 | ABCA10 | Sponge network | 0.056 | 0.81195 | -0.571 | 0.03674 | 0.314 |
| 37531 | RP11-1277A3.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-629-3p;hsa-miR-942-5p | 20 | DDX6 | Sponge network | 0.453 | 0.01839 | 0.091 | 0.36428 | 0.314 |
| 37532 | RP11-479G22.8 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 13 | RASAL2 | Sponge network | 1.308 | 0 | 0.869 | 0 | 0.314 |
| 37533 | RP11-819C21.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p | 23 | DLC1 | Sponge network | 0.537 | 0.00139 | -0.382 | 0.05038 | 0.314 |
| 37534 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-660-5p;hsa-miR-96-5p | 15 | LRRC4 | Sponge network | 0.249 | 0.35902 | -1.774 | 0 | 0.314 |
| 37535 | LINC00578 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p | 14 | ETV1 | Sponge network | 0.811 | 0.03228 | -0.096 | 0.67674 | 0.314 |
| 37536 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-5p | 11 | LRRC55 | Sponge network | 0.131 | 0.8436 | -0.026 | 0.93163 | 0.314 |
| 37537 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 50 | CUX1 | Sponge network | 0.194 | 0.64278 | -0.039 | 0.6705 | 0.314 |
| 37538 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | HDAC9 | Sponge network | -1.281 | 0.00012 | -0.755 | 0.00495 | 0.314 |
| 37539 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-590-3p | 17 | ACVR1 | Sponge network | 0.199 | 0.38015 | 0.279 | 0.00234 | 0.314 |
| 37540 | RP11-758N13.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p | 13 | HIP1 | Sponge network | 2.939 | 3.0E-5 | 0.774 | 0 | 0.314 |
| 37541 | RP11-536O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p | 13 | SSBP2 | Sponge network | -0.074 | 0.90758 | -1.58 | 0 | 0.314 |
| 37542 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 18 | MAPK10 | Sponge network | 1.302 | 7.0E-5 | -1.774 | 0 | 0.314 |
| 37543 | MIR22HG |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-93-5p | 15 | PRUNE2 | Sponge network | -1.047 | 0 | -1.733 | 0.00022 | 0.314 |
| 37544 | AP001347.6 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | LIMCH1 | Sponge network | -1.161 | 0.00591 | -0.915 | 1.0E-5 | 0.314 |
| 37545 | FAM66C |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-744-5p | 15 | IGF2 | Sponge network | -0.901 | 0.00019 | 0.848 | 0.03842 | 0.314 |
| 37546 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TIMP3 | Sponge network | -1.057 | 0.00029 | -0.546 | 0.02307 | 0.314 |
| 37547 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p | 16 | RET | Sponge network | -0.773 | 0.04073 | -0.833 | 0.03517 | 0.314 |
| 37548 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p | 10 | GAS1 | Sponge network | -0.405 | 0.22013 | -1.047 | 0.00364 | 0.314 |
| 37549 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-96-5p | 22 | CBFA2T3 | Sponge network | 0.194 | 0.64278 | -1.221 | 1.0E-5 | 0.314 |
| 37550 | RP5-1074L1.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p | 14 | ABI2 | Sponge network | 1.923 | 0 | 0.175 | 0.14787 | 0.314 |
| 37551 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-590-3p | 11 | ANK3 | Sponge network | 0.959 | 0 | -0.55 | 0.00086 | 0.314 |
| 37552 | ADAMTS9-AS2 |
hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 13 | PIM1 | Sponge network | -2.452 | 0 | -1.114 | 0 | 0.314 |
| 37553 | RP5-1139B12.3 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-942-5p | 18 | IGFBP5 | Sponge network | 0.598 | 0.38481 | -0.806 | 0.00105 | 0.314 |
| 37554 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-7-1-3p | 23 | KAT6B | Sponge network | 0.249 | 0.35902 | -0.478 | 0.00018 | 0.314 |
| 37555 | LINC00092 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-362-3p;hsa-miR-590-3p | 10 | BCL6 | Sponge network | -1.886 | 0 | -0.211 | 0.19489 | 0.314 |
| 37556 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 18 | SLC5A3 | Sponge network | 0.537 | 0.00139 | 0.493 | 5.0E-5 | 0.314 |
| 37557 | FAM66C |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | PIK3CA | Sponge network | -0.901 | 0.00019 | 0.195 | 0.06908 | 0.314 |
| 37558 | RP11-35G9.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-375;hsa-miR-940;hsa-miR-96-5p | 19 | GAS7 | Sponge network | 0.622 | 0.00015 | -0.555 | 0.01602 | 0.314 |
| 37559 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-5p | 10 | RSPO2 | Sponge network | -0.72 | 0.24239 | -3.038 | 0 | 0.314 |
| 37560 | AC005618.6 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | DIP2C | Sponge network | 0.862 | 0.06213 | -0.436 | 0.01713 | 0.314 |
| 37561 | AC011526.1 |
hsa-let-7b-3p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | ARID5B | Sponge network | 0.342 | 0.03129 | 0.342 | 0.01676 | 0.314 |
| 37562 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-550a-3p;hsa-miR-577 | 14 | ITGAV | Sponge network | -0.473 | 0.07602 | 0.337 | 0.01002 | 0.314 |
| 37563 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p | 12 | JAZF1 | Sponge network | 1.46 | 1.0E-5 | -0.377 | 0.02677 | 0.314 |
| 37564 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-361-5p;hsa-miR-429 | 10 | EPAS1 | Sponge network | -2.62 | 0 | -0.184 | 0.14418 | 0.314 |
| 37565 | PWAR6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 21 | ZFP36L1 | Sponge network | -1.575 | 6.0E-5 | -0.505 | 8.0E-5 | 0.314 |
| 37566 | IQCH-AS1 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 12 | PRKAA1 | Sponge network | 0.929 | 0 | 0.317 | 0.0031 | 0.314 |
| 37567 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 24 | EBF1 | Sponge network | 0.385 | 0.10067 | -0.384 | 0.08856 | 0.314 |
| 37568 | RP11-195B17.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-96-5p | 14 | SCN9A | Sponge network | 0.37 | 0.27614 | -0.525 | 0.10593 | 0.314 |
| 37569 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-935 | 23 | ACSL6 | Sponge network | -0.896 | 0.00099 | -1.593 | 0 | 0.314 |
| 37570 | RP11-356J5.12 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-940 | 28 | TOM1L2 | Sponge network | -0.899 | 6.0E-5 | -0.994 | 0 | 0.314 |
| 37571 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 11 | NALCN | Sponge network | 0.657 | 4.0E-5 | 1.068 | 0.00237 | 0.314 |
| 37572 | RP11-121C2.2 |
hsa-miR-100-5p;hsa-miR-101-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 10 | ATP2A2 | Sponge network | 1.147 | 0 | 0.604 | 0 | 0.314 |
| 37573 | NEAT1 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 21 | DICER1 | Sponge network | 0.705 | 0.00014 | 0.042 | 0.674 | 0.314 |
| 37574 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 24 | ATRX | Sponge network | -0.405 | 0.22013 | 0.261 | 0.04408 | 0.314 |
| 37575 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 10 | THBS1 | Sponge network | -0.557 | 0.11135 | 0.741 | 0.00386 | 0.314 |
| 37576 | RP11-389C8.2 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-375;hsa-miR-550a-3p | 10 | CACNA1E | Sponge network | 0.727 | 3.0E-5 | 1.752 | 0 | 0.314 |
| 37577 | AC011526.1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 23 | FAT3 | Sponge network | 0.342 | 0.03129 | -1.601 | 0.00051 | 0.314 |
| 37578 | RP11-428J1.5 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 31 | DST | Sponge network | 0.538 | 0.00076 | -0.713 | 0.00096 | 0.314 |
| 37579 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | LPP | Sponge network | 0.272 | 0.44692 | -0.459 | 0.04475 | 0.314 |
| 37580 | MIR143HG |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | JDP2 | Sponge network | -0.739 | 0.0574 | -0.967 | 0 | 0.314 |
| 37581 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-429 | 10 | CREBBP | Sponge network | -0.154 | 0.53954 | 0.126 | 0.15186 | 0.314 |
| 37582 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-93-3p | 18 | BNC2 | Sponge network | 1.46 | 1.0E-5 | -0.491 | 0.09988 | 0.314 |
| 37583 | RP11-225B17.2 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 13 | ZFX | Sponge network | 1.055 | 0 | 0.09 | 0.42563 | 0.314 |
| 37584 | AC005062.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-940 | 25 | MDM4 | Sponge network | -0.019 | 0.81504 | 0.345 | 0.00762 | 0.314 |
| 37585 | THAP9-AS1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 14 | GSK3B | Sponge network | 1.079 | 0 | 0.266 | 0.00045 | 0.314 |
| 37586 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-331-5p;hsa-miR-539-5p | 12 | BCL11A | Sponge network | -0.473 | 0.07602 | -0.932 | 0.0063 | 0.314 |
| 37587 | CALML3-AS1 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-542-3p | 11 | CSDE1 | Sponge network | 0.95 | 0.15943 | -0.493 | 0 | 0.314 |
| 37588 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PDS5B | Sponge network | 0.385 | 0.10067 | 0.259 | 0.00962 | 0.314 |
| 37589 | LINC00265 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-484 | 16 | CBX5 | Sponge network | 1.07 | 0 | 0.165 | 0.24446 | 0.314 |
| 37590 | LPP-AS2 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-577;hsa-miR-96-5p | 13 | PRKAR1A | Sponge network | -0.18 | 0.20343 | -0.063 | 0.50314 | 0.314 |
| 37591 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-93-3p | 34 | PAFAH1B1 | Sponge network | -0.896 | 0.00099 | -0.429 | 0 | 0.314 |
| 37592 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 44 | CADM2 | Sponge network | 0.61 | 0.01593 | -3.782 | 0 | 0.314 |
| 37593 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-25-3p | 10 | DMXL1 | Sponge network | 0.693 | 0.00076 | -0.426 | 0.00056 | 0.314 |
| 37594 | RP11-968O1.5 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-532-5p | 14 | RYR2 | Sponge network | -0.829 | 0.01343 | -1.466 | 0.00031 | 0.314 |
| 37595 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 54 | PAFAH1B1 | Sponge network | -0.576 | 0.10121 | -0.429 | 0 | 0.314 |
| 37596 | RP11-473I1.9 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | KIF5B | Sponge network | 0.683 | 1.0E-5 | 0.362 | 0.00066 | 0.314 |
| 37597 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p | 10 | SPTBN4 | Sponge network | 0.082 | 0.55654 | -0.786 | 0.01281 | 0.313 |
| 37598 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p | 12 | IGF1 | Sponge network | 0.348 | 0.32912 | -0.204 | 0.5898 | 0.313 |
| 37599 | MAGI2-AS3 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | GATA2 | Sponge network | -0.129 | 0.57177 | -0.654 | 0.0016 | 0.313 |
| 37600 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-98-5p | 18 | CLASP2 | Sponge network | -0.473 | 0.07602 | -0.038 | 0.71 | 0.313 |
| 37601 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 18 | PTGFR | Sponge network | -0.899 | 6.0E-5 | -0.233 | 0.45421 | 0.313 |
| 37602 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 0.953 | 0 | 0.259 | 0.00962 | 0.313 |
| 37603 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-582-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | JAZF1 | Sponge network | -0.31 | 0.33871 | -0.377 | 0.02677 | 0.313 |
| 37604 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 18 | LRRC7 | Sponge network | 0.16 | 0.50785 | -1.341 | 0.01193 | 0.313 |
| 37605 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | BCL11A | Sponge network | -2.117 | 0.00161 | -0.932 | 0.0063 | 0.313 |
| 37606 | SERTAD4-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-93-3p | 13 | TOM1L2 | Sponge network | -0.932 | 0.00329 | -0.994 | 0 | 0.313 |
| 37607 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 25 | RSPO2 | Sponge network | -0.899 | 6.0E-5 | -3.038 | 0 | 0.313 |
| 37608 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-429 | 13 | LATS2 | Sponge network | -0.246 | 0.35034 | 0.159 | 0.2304 | 0.313 |
| 37609 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | LRRC7 | Sponge network | -0.031 | 0.89063 | -1.341 | 0.01193 | 0.313 |
| 37610 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | NTRK2 | Sponge network | 1.04 | 0.00023 | -0.736 | 0.06495 | 0.313 |
| 37611 | LINC00657 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ERCC4 | Sponge network | 0.91 | 0 | 0.126 | 0.15266 | 0.313 |
| 37612 | ARHGEF26-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 30 | EFNA5 | Sponge network | -2.46 | 0 | -0.421 | 0.17868 | 0.313 |
| 37613 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | AFF3 | Sponge network | 0.056 | 0.81195 | -2.373 | 0 | 0.313 |
| 37614 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-339-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | USP25 | Sponge network | -0.612 | 0.00094 | -0.242 | 0.01397 | 0.313 |
| 37615 | MIR22HG |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | CSDE1 | Sponge network | -1.047 | 0 | -0.493 | 0 | 0.313 |
| 37616 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | MXI1 | Sponge network | -0.141 | 0.26434 | -0.987 | 0 | 0.313 |
| 37617 | C4B-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-629-3p | 18 | ESR1 | Sponge network | 2.306 | 9.0E-5 | -1.035 | 3.0E-5 | 0.313 |
| 37618 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-708-3p | 32 | GRIA3 | Sponge network | -0.615 | 0.00015 | -1.956 | 0 | 0.313 |
| 37619 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-3p | 17 | SEMA3C | Sponge network | 0.214 | 0.18246 | -0.272 | 0.31153 | 0.313 |
| 37620 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | SLITRK6 | Sponge network | -0.739 | 0.00044 | -2.121 | 0 | 0.313 |
| 37621 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-218-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | PTPN11 | Sponge network | 0.702 | 7.0E-5 | 0.431 | 2.0E-5 | 0.313 |
| 37622 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-940 | 13 | NR4A3 | Sponge network | -0.162 | 0.47687 | -1.385 | 0 | 0.313 |
| 37623 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-940 | 27 | PBX1 | Sponge network | -0.141 | 0.26434 | -1.206 | 0 | 0.313 |
| 37624 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-7-5p;hsa-miR-93-3p | 24 | ERC1 | Sponge network | -0.513 | 0.04703 | -0.108 | 0.38794 | 0.313 |
| 37625 | RP11-888D10.4 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | EPS15 | Sponge network | 0.702 | 7.0E-5 | -0.052 | 0.53998 | 0.313 |
| 37626 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 16 | BNC2 | Sponge network | 0.348 | 0.32912 | -0.491 | 0.09988 | 0.313 |
| 37627 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | KAT6B | Sponge network | -1.161 | 0.00591 | -0.478 | 0.00018 | 0.313 |
| 37628 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | MITF | Sponge network | -0.738 | 0.21233 | -0.769 | 0.00022 | 0.313 |
| 37629 | RP11-927P21.1 |
hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-5p | 18 | UBR3 | Sponge network | 0.921 | 0.00118 | -0.021 | 0.82774 | 0.313 |
| 37630 | MAP3K14 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-3127-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | SUFU | Sponge network | -0.295 | 0.02604 | -0.04 | 0.65489 | 0.313 |
| 37631 | RP11-834C11.4 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-96-5p | 10 | SOX5 | Sponge network | -0.883 | 0.01839 | -1.091 | 4.0E-5 | 0.313 |
| 37632 | AGAP11 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-93-3p | 10 | ZNF382 | Sponge network | -1.645 | 0 | -0.494 | 0.03831 | 0.313 |
| 37633 | RP11-458D21.1 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-365a-3p;hsa-miR-495-3p | 11 | ERCC6 | Sponge network | 0.663 | 0.00073 | 0.23 | 0.015 | 0.313 |
| 37634 | DOCK9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 20 | PBX1 | Sponge network | -0.231 | 0.28214 | -1.206 | 0 | 0.313 |
| 37635 | LINC00667 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 24 | SETBP1 | Sponge network | -0.235 | 0.15142 | -1.302 | 1.0E-5 | 0.313 |
| 37636 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 24 | ARHGAP28 | Sponge network | -0.323 | 0.01372 | -0.911 | 0.00013 | 0.313 |
| 37637 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p | 14 | MYCBP2 | Sponge network | 0.064 | 0.72934 | -0.027 | 0.82016 | 0.313 |
| 37638 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 14 | TRIM3 | Sponge network | -0.421 | 0.0114 | -0.577 | 0 | 0.313 |
| 37639 | LINC00173 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-940 | 11 | BAZ2B | Sponge network | 0.056 | 0.8494 | -0.276 | 0.02833 | 0.313 |
| 37640 | LPP-AS2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-769-5p;hsa-miR-93-5p | 13 | PRUNE2 | Sponge network | -0.18 | 0.20343 | -1.733 | 0.00022 | 0.313 |
| 37641 | LPP-AS2 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-96-5p | 12 | CSRNP1 | Sponge network | -0.18 | 0.20343 | -1.448 | 0 | 0.313 |
| 37642 | RP11-890B15.3 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-92a-1-5p | 24 | CTIF | Sponge network | 0.101 | 0.60919 | -0.389 | 0.00549 | 0.313 |
| 37643 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-374b-3p | 20 | SPRY3 | Sponge network | 0.946 | 0 | 0.016 | 0.91286 | 0.313 |
| 37644 | RP11-620J15.3 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 18 | LIMCH1 | Sponge network | -0.739 | 0.00044 | -0.915 | 1.0E-5 | 0.313 |
| 37645 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PLSCR4 | Sponge network | 0.311 | 0.10344 | -0.474 | 0.03833 | 0.313 |
| 37646 | RP11-983P16.4 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-5p | 13 | RET | Sponge network | 0.41 | 0.02214 | -0.833 | 0.03517 | 0.313 |
| 37647 | LINC00578 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-576-5p | 15 | PDE4DIP | Sponge network | 0.811 | 0.03228 | -0.282 | 0.0257 | 0.313 |
| 37648 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | NIN | Sponge network | 0.862 | 0.06213 | -0.253 | 0.13794 | 0.313 |
| 37649 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p | 24 | SCN9A | Sponge network | 0.75 | 0.00054 | -0.525 | 0.10593 | 0.313 |
| 37650 | RP11-305E6.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p | 19 | ARHGEF12 | Sponge network | 1.317 | 0 | 0.09 | 0.35693 | 0.313 |
| 37651 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | HDAC9 | Sponge network | 0.532 | 0.00505 | -0.755 | 0.00495 | 0.313 |
| 37652 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 19 | FAT4 | Sponge network | 0.16 | 0.50785 | -0.354 | 0.14694 | 0.313 |
| 37653 | AC141928.1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-486-5p | 16 | DYNC1I1 | Sponge network | 0.853 | 0.15352 | -1.12 | 0.00107 | 0.313 |
| 37654 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-96-5p | 13 | DOCK4 | Sponge network | -0.517 | 0.02935 | 0.502 | 0.00108 | 0.313 |
| 37655 | RP11-284N8.3 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-224-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 11 | SNX29 | Sponge network | -0.31 | 0.33871 | -0.34 | 0.01371 | 0.313 |
| 37656 | LINC00476 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-379-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 16 | SH3GLB1 | Sponge network | -0.615 | 0.00015 | -0.115 | 0.14773 | 0.313 |
| 37657 | RP11-175O19.4 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-497-5p;hsa-miR-505-5p | 19 | DDX3X | Sponge network | 0.917 | 0 | -0.152 | 0.07686 | 0.313 |
| 37658 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p | 11 | PDZD2 | Sponge network | 0.083 | 0.66405 | -0.909 | 3.0E-5 | 0.313 |
| 37659 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 30 | AR | Sponge network | 0.419 | 0.0319 | -1.554 | 0 | 0.313 |
| 37660 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | TACC1 | Sponge network | -0.303 | 0.21002 | -0.362 | 0.05406 | 0.313 |
| 37661 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | HACE1 | Sponge network | 0.91 | 0 | -0.072 | 0.55239 | 0.313 |
| 37662 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p | 14 | LRRC4 | Sponge network | 0.335 | 0.20596 | -1.774 | 0 | 0.313 |
| 37663 | RP5-1085F17.3 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 15 | ZMYND11 | Sponge network | 0.541 | 0.00125 | -0.2 | 0.05449 | 0.313 |
| 37664 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-590-5p | 19 | BHLHE41 | Sponge network | 0.889 | 9.0E-5 | 0.353 | 0.12753 | 0.313 |
| 37665 | LINC00205 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 10 | PHF6 | Sponge network | 1.271 | 0 | 0.785 | 0 | 0.313 |
| 37666 | RP11-760H22.2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-629-5p | 11 | UBR3 | Sponge network | 0.001 | 0.99753 | -0.021 | 0.82774 | 0.313 |
| 37667 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-940 | 20 | TOM1L2 | Sponge network | -0.154 | 0.53954 | -0.994 | 0 | 0.313 |
| 37668 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-940 | 11 | HOOK3 | Sponge network | -1.249 | 7.0E-5 | 0.273 | 0.09957 | 0.313 |
| 37669 | LINC00854 |
hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-625-5p | 11 | ZMYM2 | Sponge network | 1.18 | 0 | 0.279 | 0.01273 | 0.313 |
| 37670 | FAM66C |
hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-625-5p | 13 | PPP2R2C | Sponge network | -0.901 | 0.00019 | -0.959 | 0.07428 | 0.313 |
| 37671 | RP11-212P7.2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 29 | DLC1 | Sponge network | 0.219 | 0.1516 | -0.382 | 0.05038 | 0.313 |
| 37672 | TRAF3IP2-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PIK3CA | Sponge network | -0.323 | 0.01372 | 0.195 | 0.06908 | 0.313 |
| 37673 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-582-5p | 14 | PRKCE | Sponge network | -0.469 | 0.08721 | -0.403 | 0.00056 | 0.313 |
| 37674 | KB-1572G7.2 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 0.924 | 0 | 0.362 | 0.00066 | 0.313 |
| 37675 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374b-5p | 14 | PPM1A | Sponge network | 0.542 | 0.00054 | -0.623 | 0 | 0.313 |
| 37676 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -0.739 | 0.0574 | 0.309 | 0.17079 | 0.313 |
| 37677 | RP11-13K12.1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-93-3p | 21 | DOCK4 | Sponge network | -0.154 | 0.78939 | 0.502 | 0.00108 | 0.313 |
| 37678 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-96-5p | 30 | FBXO32 | Sponge network | 0.374 | 0.20054 | -0.495 | 0.07561 | 0.313 |
| 37679 | LINC00265 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-486-5p | 11 | CBFB | Sponge network | 1.07 | 0 | 1.061 | 0 | 0.313 |
| 37680 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | EPHA3 | Sponge network | -0.326 | 0.14314 | 0.185 | 0.53588 | 0.313 |
| 37681 | HOTAIRM1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-424-5p | 18 | PTPN14 | Sponge network | -0.053 | 0.80685 | 0.144 | 0.34595 | 0.313 |
| 37682 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | PCDH8 | Sponge network | -0.08 | 0.90092 | -0.997 | 0.05366 | 0.313 |
| 37683 | RP4-668J24.2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-3913-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-942-5p | 18 | IGFBP5 | Sponge network | -1.054 | 0.0706 | -0.806 | 0.00105 | 0.313 |
| 37684 | LPP-AS2 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 12 | TLE4 | Sponge network | -0.18 | 0.20343 | -1.157 | 0 | 0.313 |
| 37685 | BMS1P20 |
hsa-let-7g-3p;hsa-miR-10a-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CREB1 | Sponge network | 0.34 | 0.00273 | 0.203 | 0.00537 | 0.313 |
| 37686 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 56 | NTRK3 | Sponge network | -0.83 | 0 | -1.77 | 3.0E-5 | 0.313 |
| 37687 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 30 | ABL2 | Sponge network | 0.064 | 0.72934 | 0.82 | 0 | 0.313 |
| 37688 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-3p | 17 | ADAMTSL3 | Sponge network | 0.051 | 0.83388 | -1.559 | 0 | 0.313 |
| 37689 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | LSAMP | Sponge network | -0.942 | 0 | -0.066 | 0.81708 | 0.313 |
| 37690 | DNAJC3-AS1 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 13 | ABL2 | Sponge network | 0.753 | 0.00014 | 0.82 | 0 | 0.313 |
| 37691 | LINC00641 |
hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 13 | FBXW7 | Sponge network | -0.222 | 0.32445 | -0.418 | 2.0E-5 | 0.313 |
| 37692 | TP73-AS1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-660-5p;hsa-miR-96-5p | 15 | IRS1 | Sponge network | -0.563 | 0.02545 | -0.026 | 0.88855 | 0.313 |
| 37693 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-500a-5p | 11 | PKNOX1 | Sponge network | 0.614 | 1.0E-5 | 0.085 | 0.28917 | 0.313 |
| 37694 | RP11-307B6.3 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-424-5p | 13 | CADM1 | Sponge network | -4.463 | 0.00025 | -0.9 | 0.00084 | 0.313 |
| 37695 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-589-3p;hsa-miR-93-5p | 18 | RNASEL | Sponge network | 0.4 | 0.14611 | -0.272 | 0.01796 | 0.313 |
| 37696 | LINC00641 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | TCF12 | Sponge network | -0.222 | 0.32445 | 0.174 | 0.13271 | 0.313 |
| 37697 | PAN3-AS1 |
hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p | 10 | ABL2 | Sponge network | 0.062 | 0.55274 | 0.82 | 0 | 0.313 |
| 37698 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 43 | PTPRD | Sponge network | 0.194 | 0.64278 | -1.005 | 0.00064 | 0.313 |
| 37699 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p | 12 | FSTL5 | Sponge network | -0.727 | 0.04726 | -1.3 | 0.01619 | 0.313 |
| 37700 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p | 14 | FAT4 | Sponge network | 0.494 | 0.00339 | -0.354 | 0.14694 | 0.313 |
| 37701 | RP11-46C24.7 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p | 15 | MRVI1 | Sponge network | 0.392 | 0.0278 | -1.051 | 0.00022 | 0.313 |
| 37702 | LINC00654 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | TLL1 | Sponge network | 0.374 | 0.20054 | -1.195 | 3.0E-5 | 0.313 |
| 37703 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-362-3p | 11 | AHNAK | Sponge network | 0.078 | 0.55803 | -1.207 | 0 | 0.313 |
| 37704 | RP11-359E10.1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-629-5p | 18 | CTIF | Sponge network | 0.299 | 0.57722 | -0.389 | 0.00549 | 0.313 |
| 37705 | AC093627.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-20a-5p | 12 | TSHZ3 | Sponge network | -0.787 | 0.3928 | -0.556 | 0.01516 | 0.313 |
| 37706 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-664a-5p | 28 | PPM1L | Sponge network | -0.583 | 0.14589 | -0.537 | 0.00616 | 0.313 |
| 37707 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | TRIM33 | Sponge network | 0.415 | 0.14657 | 0.402 | 7.0E-5 | 0.313 |
| 37708 | GS1-124K5.11 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-877-5p | 30 | AFF1 | Sponge network | 0.494 | 0.00339 | -0.384 | 0.00053 | 0.313 |
| 37709 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 40 | ADARB1 | Sponge network | 0.374 | 0.20054 | -0.335 | 0.06631 | 0.313 |
| 37710 | OIP5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p | 38 | IL17RD | Sponge network | 0.421 | 0.00084 | -0.019 | 0.94873 | 0.313 |
| 37711 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 34 | MXI1 | Sponge network | -0.129 | 0.57177 | -0.987 | 0 | 0.313 |
| 37712 | DTX2P1-UPK3BP1-PMS2P11 | hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-497-5p | 10 | CCDC6 | Sponge network | 0.828 | 0 | 0.27 | 0.02555 | 0.313 |
| 37713 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | ATRX | Sponge network | 0.335 | 0.20596 | 0.261 | 0.04408 | 0.313 |
| 37714 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 55 | NTRK3 | Sponge network | 0.199 | 0.38015 | -1.77 | 3.0E-5 | 0.313 |
| 37715 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | JDP2 | Sponge network | -0.421 | 0.0114 | -0.967 | 0 | 0.313 |
| 37716 | RP11-158K1.3 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 39 | PPM1L | Sponge network | 0.349 | 0.01695 | -0.537 | 0.00616 | 0.313 |
| 37717 | MYLK-AS1 |
hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-484 | 12 | YAP1 | Sponge network | -1.331 | 3.0E-5 | 0.22 | 0.11836 | 0.313 |
| 37718 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-505-5p | 18 | HBP1 | Sponge network | -4.546 | 0 | -0.454 | 0 | 0.313 |
| 37719 | RP11-21A7A.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-576-5p | 16 | NTRK2 | Sponge network | -1.116 | 0.23686 | -0.736 | 0.06495 | 0.313 |
| 37720 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-576-5p | 10 | GREM1 | Sponge network | -2.039 | 0.03447 | 0.902 | 0.01691 | 0.313 |
| 37721 | BMS1P20 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | AKAP11 | Sponge network | 0.34 | 0.00273 | 0.196 | 0.09825 | 0.313 |
| 37722 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p | 16 | LIMD1 | Sponge network | 0.569 | 0.00067 | 0.103 | 0.28978 | 0.313 |
| 37723 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-590-3p | 14 | RBM5 | Sponge network | -1.111 | 0.0003 | -0.205 | 0.0129 | 0.313 |
| 37724 | GS1-124K5.11 |
hsa-miR-146b-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-574-3p;hsa-miR-590-3p | 10 | FHL1 | Sponge network | 0.494 | 0.00339 | -2.687 | 0 | 0.313 |
| 37725 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 33 | PAFAH1B1 | Sponge network | -0.227 | 0.31657 | -0.429 | 0 | 0.313 |
| 37726 | LINC00672 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p | 15 | PIK3CA | Sponge network | -0.227 | 0.31657 | 0.195 | 0.06908 | 0.313 |
| 37727 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-877-5p | 10 | FLI1 | Sponge network | -1.654 | 0.00144 | -0.294 | 0.10905 | 0.313 |
| 37728 | RP3-475N16.1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-625-5p | 21 | MDM4 | Sponge network | 1.056 | 0 | 0.345 | 0.00762 | 0.313 |
| 37729 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | ARID5B | Sponge network | 0.222 | 0.29133 | 0.342 | 0.01676 | 0.313 |
| 37730 | RP11-536O18.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-5p | 15 | TRPS1 | Sponge network | -0.074 | 0.90758 | -0.191 | 0.44109 | 0.313 |
| 37731 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-940 | 56 | WDFY3 | Sponge network | -0.303 | 0.21002 | -0.201 | 0.0967 | 0.313 |
| 37732 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 31 | LIFR | Sponge network | -1.216 | 0 | -1.794 | 0 | 0.313 |
| 37733 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p | 25 | PRKAB2 | Sponge network | -0.896 | 0.00099 | -0.367 | 0.01371 | 0.313 |
| 37734 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | PPP1R9A | Sponge network | -0.358 | 0.17255 | -0.903 | 0.04254 | 0.313 |
| 37735 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | RBL2 | Sponge network | -0.355 | 0.0077 | -0.282 | 0.00254 | 0.313 |
| 37736 | HCG22 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 10 | GJA1 | Sponge network | 0.816 | 0.32368 | -0.485 | 0.02179 | 0.313 |
| 37737 | RP11-203M5.7 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-495-3p | 11 | CCDC6 | Sponge network | 1.684 | 1.0E-5 | 0.27 | 0.02555 | 0.313 |
| 37738 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-539-5p | 13 | ITCH | Sponge network | 0.75 | 0.00054 | 0.084 | 0.34058 | 0.313 |
| 37739 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-96-5p | 27 | CADM1 | Sponge network | -0.969 | 2.0E-5 | -0.9 | 0.00084 | 0.313 |
| 37740 | RP11-774O3.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | CREB1 | Sponge network | -0.487 | 0.02157 | 0.203 | 0.00537 | 0.313 |
| 37741 | AGAP11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-590-5p | 17 | NRXN3 | Sponge network | -1.645 | 0 | -0.893 | 0.04186 | 0.313 |
| 37742 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-589-5p;hsa-miR-590-3p | 18 | GRIA3 | Sponge network | -0.811 | 2.0E-5 | -1.956 | 0 | 0.313 |
| 37743 | GAS6-AS2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | ITGB1 | Sponge network | 0.16 | 0.50785 | 0.524 | 0.00017 | 0.313 |
| 37744 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 31 | TGFBR3 | Sponge network | -0.318 | 0.65847 | -1.173 | 1.0E-5 | 0.313 |
| 37745 | CTD-2336O2.1 |
hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 11 | PCDH9 | Sponge network | -0.141 | 0.26434 | -1.823 | 4.0E-5 | 0.313 |
| 37746 | HCG22 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-501-5p;hsa-miR-511-5p | 10 | BCL6 | Sponge network | 0.816 | 0.32368 | -0.211 | 0.19489 | 0.313 |
| 37747 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-30d-3p | 12 | IL17RD | Sponge network | -2.076 | 0.00548 | -0.019 | 0.94873 | 0.313 |
| 37748 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-340-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | EDA2R | Sponge network | 0.051 | 0.83388 | -0.587 | 0.01598 | 0.313 |
| 37749 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | MED12L | Sponge network | 0.311 | 0.10344 | -0.194 | 0.55299 | 0.313 |
| 37750 | RP11-141C7.4 |
hsa-let-7a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 10 | RICTOR | Sponge network | 2.412 | 0 | 0.388 | 0.00188 | 0.313 |
| 37751 | RP11-403I13.8 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CREB1 | Sponge network | 0.619 | 0.00157 | 0.203 | 0.00537 | 0.313 |
| 37752 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-511-5p | 24 | ADAMTSL3 | Sponge network | -1.645 | 0 | -1.559 | 0 | 0.313 |
| 37753 | LINC00094 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-582-5p | 17 | STARD13 | Sponge network | 0.253 | 0.09565 | -0.187 | 0.27383 | 0.313 |
| 37754 | LIPE-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 14 | TBL1XR1 | Sponge network | 0.078 | 0.55803 | 0.578 | 0 | 0.313 |
| 37755 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 17 | CADM3 | Sponge network | 0.101 | 0.60919 | -3.663 | 0 | 0.313 |
| 37756 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-154-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-98-5p | 15 | PRKAA2 | Sponge network | -0.301 | 0.06597 | -2.462 | 0 | 0.313 |
| 37757 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 53 | MDM4 | Sponge network | 0.064 | 0.72934 | 0.345 | 0.00762 | 0.313 |
| 37758 | DNM3OS |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | SPOP | Sponge network | 0.572 | 0.01722 | -0.419 | 1.0E-5 | 0.313 |
| 37759 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | RNF144A | Sponge network | 0.349 | 0.01695 | 0.594 | 0.0009 | 0.313 |
| 37760 | AF131215.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-589-3p | 10 | FAM172A | Sponge network | -0.176 | 0.59692 | -0.57 | 0 | 0.313 |
| 37761 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | ANK2 | Sponge network | -0.83 | 0 | -1.603 | 1.0E-5 | 0.313 |
| 37762 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-576-5p | 21 | HDAC9 | Sponge network | -0.162 | 0.47687 | -0.755 | 0.00495 | 0.313 |
| 37763 | LINC00702 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 14 | UVRAG | Sponge network | -0.737 | 0.06182 | -0.017 | 0.83865 | 0.313 |
| 37764 | CTD-2314G24.2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 13 | CELF4 | Sponge network | 0.246 | 0.59912 | -1.135 | 0.00344 | 0.313 |
| 37765 | CTD-2314G24.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 41 | TGFBR3 | Sponge network | 0.246 | 0.59912 | -1.173 | 1.0E-5 | 0.313 |
| 37766 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-582-3p | 13 | CBL | Sponge network | 0.903 | 0 | 0.291 | 0.01267 | 0.313 |
| 37767 | MYLK-AS1 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-340-5p | 12 | FAM135B | Sponge network | -1.331 | 3.0E-5 | -3.978 | 0 | 0.313 |
| 37768 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 26 | AFF4 | Sponge network | -0.285 | 0.2172 | -0.266 | 0.01565 | 0.313 |
| 37769 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 19 | FOXO3 | Sponge network | 0.539 | 0.03722 | -0.04 | 0.72028 | 0.313 |
| 37770 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | LATS1 | Sponge network | 1.03 | 0 | 0.109 | 0.34208 | 0.313 |
| 37771 | RP11-195B17.1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p | 23 | IL6ST | Sponge network | 0.37 | 0.27614 | -0.402 | 0.00676 | 0.313 |
| 37772 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-942-5p | 20 | ARHGAP28 | Sponge network | -0.611 | 0.09607 | -0.911 | 0.00013 | 0.313 |
| 37773 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-93-3p | 21 | PTPN11 | Sponge network | -0.162 | 0.47687 | 0.431 | 2.0E-5 | 0.313 |
| 37774 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-455-5p | 18 | GRIA3 | Sponge network | -0.473 | 0.07602 | -1.956 | 0 | 0.313 |
| 37775 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | SETBP1 | Sponge network | -0.031 | 0.89063 | -1.302 | 1.0E-5 | 0.313 |
| 37776 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p | 20 | CBFA2T3 | Sponge network | -0.318 | 0.65847 | -1.221 | 1.0E-5 | 0.313 |
| 37777 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-935 | 11 | ARNT2 | Sponge network | 0.225 | 0.30644 | -0.332 | 0.25851 | 0.313 |
| 37778 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p | 19 | RNF144A | Sponge network | 0.199 | 0.38015 | 0.594 | 0.0009 | 0.313 |
| 37779 | C4A-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-628-5p;hsa-miR-96-5p | 17 | FBXO32 | Sponge network | 3.345 | 0 | -0.495 | 0.07561 | 0.313 |
| 37780 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | GALNT13 | Sponge network | 0.299 | 0.57722 | -0.857 | 0.07439 | 0.313 |
| 37781 | AC016747.3 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 14 | ITGA5 | Sponge network | 0.199 | 0.38015 | 0.452 | 0.03524 | 0.313 |
| 37782 | LINC00957 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 27 | TOM1L2 | Sponge network | -0.421 | 0.0114 | -0.994 | 0 | 0.313 |
| 37783 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ATRX | Sponge network | 0.67 | 0.00048 | 0.261 | 0.04408 | 0.313 |
| 37784 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p | 14 | CLASP2 | Sponge network | 1.294 | 0 | -0.038 | 0.71 | 0.313 |
| 37785 | RP4-669P10.16 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-542-3p | 13 | ADAMTSL3 | Sponge network | -0.301 | 0.06597 | -1.559 | 0 | 0.313 |
| 37786 | RP11-379F4.7 |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-654-3p | 10 | DICER1 | Sponge network | 1.316 | 0 | 0.042 | 0.674 | 0.313 |
| 37787 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 12 | LIFR | Sponge network | -0.726 | 0.00132 | -1.794 | 0 | 0.313 |
| 37788 | CTB-113P19.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-93-3p | 13 | RUNX1T1 | Sponge network | 0.199 | 0.52763 | -0.344 | 0.2374 | 0.313 |
| 37789 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-369-3p | 15 | CDC73 | Sponge network | 1.262 | 0 | 0.094 | 0.20463 | 0.313 |
| 37790 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-628-5p | 12 | SLC5A3 | Sponge network | 0.559 | 0.00026 | 0.493 | 5.0E-5 | 0.313 |
| 37791 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | CXXC4 | Sponge network | -1.439 | 4.0E-5 | -0.433 | 0.21055 | 0.313 |
| 37792 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-629-3p | 16 | BHLHE41 | Sponge network | 0.811 | 0.03228 | 0.353 | 0.12753 | 0.313 |
| 37793 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p | 16 | ITGA5 | Sponge network | 0.064 | 0.72934 | 0.452 | 0.03524 | 0.313 |
| 37794 | BVES-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-212-3p | 11 | JDP2 | Sponge network | -2.62 | 0 | -0.967 | 0 | 0.313 |
| 37795 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-629-3p | 29 | ESR1 | Sponge network | -1.249 | 7.0E-5 | -1.035 | 3.0E-5 | 0.313 |
| 37796 | AC009948.5 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | KCTD8 | Sponge network | 0.532 | 0.00505 | -3.359 | 0 | 0.313 |
| 37797 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p | 13 | EDA2R | Sponge network | -0.125 | 0.49414 | -0.587 | 0.01598 | 0.313 |
| 37798 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | CDHR3 | Sponge network | -0.901 | 0.00019 | -0.977 | 4.0E-5 | 0.313 |
| 37799 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | SYNE1 | Sponge network | 0.998 | 0 | -0.738 | 0.00316 | 0.313 |
| 37800 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-5p | 11 | USP6 | Sponge network | 2.368 | 2.0E-5 | 0.188 | 0.3871 | 0.313 |
| 37801 | A2M-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | DOCK4 | Sponge network | -0.08 | 0.90092 | 0.502 | 0.00108 | 0.313 |
| 37802 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p | 12 | CHD1 | Sponge network | 0.267 | 0.2937 | 0.322 | 0.00215 | 0.313 |
| 37803 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-362-5p;hsa-miR-500b-5p | 11 | SLITRK6 | Sponge network | 0.176 | 0.37402 | -2.121 | 0 | 0.313 |
| 37804 | LINC00672 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374a-5p;hsa-miR-584-5p;hsa-miR-590-3p | 16 | GLIPR1 | Sponge network | -0.227 | 0.31657 | 0.146 | 0.3276 | 0.313 |
| 37805 | RP11-226L15.5 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 12 | RASAL2 | Sponge network | 0.8 | 0 | 0.869 | 0 | 0.313 |
| 37806 | RP11-927P21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-582-3p;hsa-miR-589-3p | 13 | PRDM2 | Sponge network | 0.921 | 0.00118 | -0.258 | 0.00721 | 0.313 |
| 37807 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 33 | NCOA1 | Sponge network | -2.46 | 0 | -0.432 | 0 | 0.313 |
| 37808 | LINC00863 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | 0.189 | 0.13981 | 0.809 | 0.00544 | 0.313 |
| 37809 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 10 | CNTNAP1 | Sponge network | -0.351 | 0.11358 | 0.358 | 0.13727 | 0.313 |
| 37810 | RP11-356J5.12 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-539-5p | 10 | PIK3IP1 | Sponge network | -0.899 | 6.0E-5 | -0.187 | 0.19945 | 0.313 |
| 37811 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-484;hsa-miR-539-5p | 15 | DDX6 | Sponge network | 0.79 | 0 | 0.091 | 0.36428 | 0.313 |
| 37812 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TIMP3 | Sponge network | -0.206 | 0.12942 | -0.546 | 0.02307 | 0.313 |
| 37813 | RP11-822E23.8 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-671-5p;hsa-miR-7-5p | 14 | EHD3 | Sponge network | -0.777 | 0.16667 | -0.484 | 0.01747 | 0.313 |
| 37814 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-331-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-3p | 10 | POU2F2 | Sponge network | -2.531 | 0 | -0.233 | 0.32214 | 0.313 |
| 37815 | CTC-297N7.9 |
hsa-let-7d-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-590-5p | 13 | ACSL6 | Sponge network | -2.069 | 0 | -1.593 | 0 | 0.313 |
| 37816 | HOXB-AS2 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-429 | 13 | DIP2C | Sponge network | 1.316 | 0.01106 | -0.436 | 0.01713 | 0.313 |
| 37817 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-628-5p | 19 | CDC73 | Sponge network | 0.373 | 0.00068 | 0.094 | 0.20463 | 0.313 |
| 37818 | LINC00473 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-629-3p;hsa-miR-92a-3p | 14 | SPTBN4 | Sponge network | -1.203 | 0.03881 | -0.786 | 0.01281 | 0.313 |
| 37819 | EMX2OS |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 30 | MXI1 | Sponge network | -0.777 | 0.1134 | -0.987 | 0 | 0.313 |
| 37820 | RP11-473I1.5 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p | 10 | RASSF8 | Sponge network | 0.542 | 0.00054 | -0.122 | 0.65591 | 0.313 |
| 37821 | AF131215.9 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-424-5p | 13 | PLAG1 | Sponge network | -0.322 | 0.25945 | -0.315 | 0.26532 | 0.313 |
| 37822 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-3p | 19 | PDE4DIP | Sponge network | 0.494 | 0.00339 | -0.282 | 0.0257 | 0.313 |
| 37823 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 40 | PTPRT | Sponge network | -0.49 | 0.04946 | -0.907 | 0.03727 | 0.313 |
| 37824 | LINC00865 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-429 | 12 | ELMO1 | Sponge network | -1.249 | 7.0E-5 | 0.04 | 0.83147 | 0.313 |
| 37825 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-628-5p;hsa-miR-935;hsa-miR-96-5p | 29 | FBXO32 | Sponge network | 0.41 | 0.02214 | -0.495 | 0.07561 | 0.313 |
| 37826 | CTD-3138B18.5 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 21 | FOXP1 | Sponge network | 0.379 | 0.03091 | 0.042 | 0.74368 | 0.313 |
| 37827 | RP11-517B11.7 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-5p | 15 | PIK3R1 | Sponge network | 0.706 | 0 | -0.133 | 0.36854 | 0.313 |
| 37828 | USP3-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-502-5p | 13 | FMNL3 | Sponge network | -0.162 | 0.47687 | 0.555 | 4.0E-5 | 0.313 |
| 37829 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 34 | IL6ST | Sponge network | -0.663 | 0.10887 | -0.402 | 0.00676 | 0.313 |
| 37830 | RP11-359E10.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p | 12 | TES | Sponge network | 0.299 | 0.57722 | -0.348 | 0.00639 | 0.313 |
| 37831 | RP11-418J17.1 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p | 11 | RASAL2 | Sponge network | 0.998 | 0 | 0.869 | 0 | 0.313 |
| 37832 | TP73-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | CSMD1 | Sponge network | -0.563 | 0.02545 | -0.63 | 0.18881 | 0.313 |
| 37833 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-7-1-3p | 14 | CBL | Sponge network | 2.094 | 0 | 0.291 | 0.01267 | 0.313 |
| 37834 | RP11-686O6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-27a-3p | 12 | MOB1B | Sponge network | 0.453 | 0.01971 | 0.197 | 0.15266 | 0.313 |
| 37835 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-93-5p | 15 | ARNT2 | Sponge network | 0.299 | 0.57722 | -0.332 | 0.25851 | 0.313 |
| 37836 | RP11-67L2.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-96-5p | 23 | CYLD | Sponge network | 0.72 | 0.0001 | -0.367 | 0.00171 | 0.313 |
| 37837 | RP11-434B12.1 |
hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-505-5p;hsa-miR-590-3p | 16 | MRVI1 | Sponge network | -0.031 | 0.89063 | -1.051 | 0.00022 | 0.313 |
| 37838 | LINC00957 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p | 14 | IRS1 | Sponge network | -0.421 | 0.0114 | -0.026 | 0.88855 | 0.313 |
| 37839 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-942-5p | 26 | COPS2 | Sponge network | 0.064 | 0.72934 | -0.178 | 0.04974 | 0.313 |
| 37840 | FZD10-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 18 | IGF2 | Sponge network | -0.727 | 0.04726 | 0.848 | 0.03842 | 0.313 |
| 37841 | LINC00863 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 44 | KIF1B | Sponge network | 0.189 | 0.13981 | -0.118 | 0.32258 | 0.313 |
| 37842 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | RPS6KA3 | Sponge network | 0.349 | 0.01695 | 0.256 | 0.02419 | 0.313 |
| 37843 | HOXB-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-589-3p | 24 | AFF1 | Sponge network | -0.154 | 0.53954 | -0.384 | 0.00053 | 0.313 |
| 37844 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-664a-3p | 18 | CNTNAP3 | Sponge network | 0.214 | 0.18246 | -1.465 | 0.00033 | 0.313 |
| 37845 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-5p | 12 | TIMP3 | Sponge network | 2.306 | 9.0E-5 | -0.546 | 0.02307 | 0.313 |
| 37846 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-495-3p | 12 | TXNIP | Sponge network | 0.064 | 0.72934 | -1.277 | 0 | 0.313 |
| 37847 | FAM215B |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-7-1-3p | 13 | NIN | Sponge network | 0.817 | 3.0E-5 | -0.253 | 0.13794 | 0.312 |
| 37848 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p | 15 | ABCB5 | Sponge network | -0.18 | 0.20343 | -0.956 | 0.04077 | 0.312 |
| 37849 | RP11-571M6.17 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | RPS6KA3 | Sponge network | 1.013 | 0 | 0.256 | 0.02419 | 0.312 |
| 37850 | MIR497HG |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-577 | 10 | PLA2R1 | Sponge network | -1.439 | 4.0E-5 | -0.265 | 0.24676 | 0.312 |
| 37851 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 39 | AFF4 | Sponge network | 0.91 | 0 | -0.266 | 0.01565 | 0.312 |
| 37852 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 24 | THBS1 | Sponge network | 0.16 | 0.50785 | 0.741 | 0.00386 | 0.312 |
| 37853 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 52 | AFF4 | Sponge network | 0.222 | 0.29133 | -0.266 | 0.01565 | 0.312 |
| 37854 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | TCF12 | Sponge network | 1.153 | 0 | 0.174 | 0.13271 | 0.312 |
| 37855 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | EYA4 | Sponge network | 0.05 | 0.95483 | 0.3 | 0.36876 | 0.312 |
| 37856 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 37 | ZFHX3 | Sponge network | -0.563 | 0.02545 | 0.389 | 0.01363 | 0.312 |
| 37857 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-335-3p | 11 | ELMO1 | Sponge network | -0.487 | 0.02157 | 0.04 | 0.83147 | 0.312 |
| 37858 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 23 | ZBTB4 | Sponge network | 0.051 | 0.83388 | -0.739 | 0 | 0.312 |
| 37859 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-577;hsa-miR-7-1-3p | 15 | SLC6A15 | Sponge network | -1.439 | 4.0E-5 | -2.373 | 2.0E-5 | 0.312 |
| 37860 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 33 | USP12 | Sponge network | -2.46 | 0 | -0.102 | 0.40768 | 0.312 |
| 37861 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p | 34 | NTRK3 | Sponge network | -0.785 | 0.00035 | -1.77 | 3.0E-5 | 0.312 |
| 37862 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 27 | RASSF2 | Sponge network | 0.659 | 3.0E-5 | -0.273 | 0.21961 | 0.312 |
| 37863 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ABCC9 | Sponge network | 0.619 | 0.00157 | -0.466 | 0.14121 | 0.312 |
| 37864 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-576-5p | 20 | TMTC3 | Sponge network | -1.645 | 0 | 0.123 | 0.22189 | 0.312 |
| 37865 | RP11-597D13.9 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CTGF | Sponge network | 1.302 | 7.0E-5 | 0.321 | 0.11682 | 0.312 |
| 37866 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p | 13 | CRTC3 | Sponge network | -0.785 | 0.00035 | 0.164 | 0.16786 | 0.312 |
| 37867 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 46 | TMEM132B | Sponge network | -0.612 | 0.00094 | -0.83 | 0.01084 | 0.312 |
| 37868 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-942-5p | 13 | CREBBP | Sponge network | 0.4 | 0.14611 | 0.126 | 0.15186 | 0.312 |
| 37869 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SEMA3C | Sponge network | 0.72 | 0.0001 | -0.272 | 0.31153 | 0.312 |
| 37870 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | ARHGAP29 | Sponge network | -1.203 | 0.03881 | 0.131 | 0.42953 | 0.312 |
| 37871 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-7-1-3p | 29 | IL6ST | Sponge network | -4.477 | 0 | -0.402 | 0.00676 | 0.312 |
| 37872 | RP4-613B23.1 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-455-3p | 13 | PAFAH1B1 | Sponge network | -0.309 | 0.1145 | -0.429 | 0 | 0.312 |
| 37873 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 47 | BCL2 | Sponge network | -0.942 | 0 | -0.899 | 0 | 0.312 |
| 37874 | MRPL23-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-30b-3p;hsa-miR-484 | 14 | SOX5 | Sponge network | -0.278 | 0.76559 | -1.091 | 4.0E-5 | 0.312 |
| 37875 | RP11-356J5.12 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 28 | OPCML | Sponge network | -0.899 | 6.0E-5 | -2.498 | 0 | 0.312 |
| 37876 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-940;hsa-miR-96-5p | 27 | CRTC3 | Sponge network | -0.969 | 2.0E-5 | 0.164 | 0.16786 | 0.312 |
| 37877 | RP11-2B6.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CEP170 | Sponge network | 0.539 | 0.03722 | 0.943 | 0 | 0.312 |
| 37878 | MIR497HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | NIN | Sponge network | -1.439 | 4.0E-5 | -0.253 | 0.13794 | 0.312 |
| 37879 | PART1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 24 | ABL1 | Sponge network | -2.925 | 0 | -0.215 | 0.07804 | 0.312 |
| 37880 | RP11-307B6.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-378c;hsa-miR-93-3p | 13 | ERC1 | Sponge network | -4.463 | 0.00025 | -0.108 | 0.38794 | 0.312 |
| 37881 | LINC00173 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-940;hsa-miR-96-5p | 22 | SNX29 | Sponge network | 0.056 | 0.8494 | -0.34 | 0.01371 | 0.312 |
| 37882 | LINC01006 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p | 14 | FREM2 | Sponge network | -0.702 | 0.04814 | -0.437 | 0.46353 | 0.312 |
| 37883 | RP11-967K21.1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | TPO | Sponge network | 0.056 | 0.81195 | -1.87 | 2.0E-5 | 0.312 |
| 37884 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-671-5p;hsa-miR-7-1-3p | 20 | BNC2 | Sponge network | -0.285 | 0.2172 | -0.491 | 0.09988 | 0.312 |
| 37885 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p | 11 | ST5 | Sponge network | -0.615 | 0.00015 | -0.78 | 0 | 0.312 |
| 37886 | RP11-156E6.1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 22 | CCDC6 | Sponge network | 1.266 | 0 | 0.27 | 0.02555 | 0.312 |
| 37887 | NDUFA6-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 25 | DLC1 | Sponge network | -0.355 | 0.0077 | -0.382 | 0.05038 | 0.312 |
| 37888 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | SEMA3E | Sponge network | -0.738 | 0.21233 | -1.717 | 0.00137 | 0.312 |
| 37889 | RP11-411K7.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-550a-5p | 10 | FAT2 | Sponge network | 0.155 | 0.81273 | -0.278 | 0.44877 | 0.312 |
| 37890 | RP11-359B12.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 26 | ASTN1 | Sponge network | 0.064 | 0.72934 | -2.389 | 2.0E-5 | 0.312 |
| 37891 | H1FX-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 14 | RELN | Sponge network | -0.206 | 0.12942 | -2.403 | 0 | 0.312 |
| 37892 | RP11-1277A3.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-940 | 12 | SIRT1 | Sponge network | 0.453 | 0.01839 | 0.109 | 0.2593 | 0.312 |
| 37893 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 26 | CBL | Sponge network | 0.727 | 3.0E-5 | 0.291 | 0.01267 | 0.312 |
| 37894 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 34 | DDX6 | Sponge network | 0.997 | 0.00066 | 0.091 | 0.36428 | 0.312 |
| 37895 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 25 | TP53INP1 | Sponge network | -0.222 | 0.32445 | -0.496 | 0.00064 | 0.312 |
| 37896 | FENDRR |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 28 | ACSL6 | Sponge network | -1.281 | 0.00012 | -1.593 | 0 | 0.312 |
| 37897 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | EDA2R | Sponge network | 0.101 | 0.60919 | -0.587 | 0.01598 | 0.312 |
| 37898 | LINC01018 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-590-3p | 11 | MACF1 | Sponge network | -2.915 | 0.00015 | 0.019 | 0.90335 | 0.312 |
| 37899 | RP11-244O19.1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-1-5p | 12 | PRKACA | Sponge network | -0.096 | 0.67958 | -0.255 | 0.0012 | 0.312 |
| 37900 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p | 12 | TSC22D1 | Sponge network | -0.942 | 0 | -0.529 | 2.0E-5 | 0.312 |
| 37901 | RP5-1085F17.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p | 11 | IGF2 | Sponge network | 0.541 | 0.00125 | 0.848 | 0.03842 | 0.312 |
| 37902 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484 | 15 | TRIM3 | Sponge network | -0.576 | 0.10121 | -0.577 | 0 | 0.312 |
| 37903 | RP11-384K6.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 13 | RPS6KA3 | Sponge network | 0.946 | 0 | 0.256 | 0.02419 | 0.312 |
| 37904 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | PRDM2 | Sponge network | -0.739 | 0.00044 | -0.258 | 0.00721 | 0.312 |
| 37905 | RP11-148O21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-942-5p | 11 | UNC80 | Sponge network | 0.233 | 0.81029 | -1.934 | 0 | 0.312 |
| 37906 | AGAP11 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-653-5p;hsa-miR-93-5p | 41 | PPM1L | Sponge network | -1.645 | 0 | -0.537 | 0.00616 | 0.312 |
| 37907 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-378a-3p | 10 | MLF1 | Sponge network | -0.162 | 0.47687 | -0.893 | 0.00727 | 0.312 |
| 37908 | USP3-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-766-3p | 17 | ATM | Sponge network | -0.162 | 0.47687 | 0.178 | 0.21568 | 0.312 |
| 37909 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 31 | ERC1 | Sponge network | -0.405 | 0.22013 | -0.108 | 0.38794 | 0.312 |
| 37910 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 11 | CREBBP | Sponge network | 1.317 | 0 | 0.126 | 0.15186 | 0.312 |
| 37911 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-582-3p | 10 | CLIP1 | Sponge network | 0.921 | 0.00118 | -0.215 | 0.08684 | 0.312 |
| 37912 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 34 | MEF2C | Sponge network | 0.16 | 0.50785 | -0.499 | 0.01448 | 0.312 |
| 37913 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 11 | USP6 | Sponge network | 1.055 | 0 | 0.188 | 0.3871 | 0.312 |
| 37914 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | SMARCA2 | Sponge network | 0.572 | 0.01722 | -0.529 | 0.00014 | 0.312 |
| 37915 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 23 | GAS7 | Sponge network | 0.898 | 1.0E-5 | -0.555 | 0.01602 | 0.312 |
| 37916 | CTC-429P9.5 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-744-3p | 16 | EPS15 | Sponge network | 0.178 | 0.29106 | -0.052 | 0.53998 | 0.312 |
| 37917 | PART1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZNF844 | Sponge network | -2.925 | 0 | -0.966 | 0.00035 | 0.312 |
| 37918 | CALML3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p | 15 | EZH1 | Sponge network | 0.95 | 0.15943 | -0.564 | 1.0E-5 | 0.312 |
| 37919 | MALAT1 |
hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | UBE2D1 | Sponge network | 0.782 | 0.00016 | 0.468 | 0 | 0.312 |
| 37920 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-143-3p;hsa-miR-154-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | LIMD1 | Sponge network | 1.4 | 0 | 0.103 | 0.28978 | 0.312 |
| 37921 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-532-5p | 12 | LRRC4 | Sponge network | -0.384 | 0.40652 | -1.774 | 0 | 0.312 |
| 37922 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p | 14 | TRIP11 | Sponge network | 0.594 | 0.00064 | -0.158 | 0.06384 | 0.312 |
| 37923 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 33 | WDFY3 | Sponge network | 0.401 | 0.00066 | -0.201 | 0.0967 | 0.312 |
| 37924 | MIR497HG |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-590-5p;hsa-miR-708-5p | 17 | FAM135B | Sponge network | -1.439 | 4.0E-5 | -3.978 | 0 | 0.312 |
| 37925 | RP11-597D13.9 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | MED12L | Sponge network | 1.302 | 7.0E-5 | -0.194 | 0.55299 | 0.312 |
| 37926 | RP11-575A19.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-629-3p | 11 | AR | Sponge network | 1.969 | 0.00316 | -1.554 | 0 | 0.312 |
| 37927 | CALML3-AS1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-7-5p | 10 | SH3GLB1 | Sponge network | 0.95 | 0.15943 | -0.115 | 0.14773 | 0.312 |
| 37928 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p | 18 | BCL11A | Sponge network | -0.154 | 0.78939 | -0.932 | 0.0063 | 0.312 |
| 37929 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -0.227 | 0.31657 | -3.328 | 0 | 0.312 |
| 37930 | RP11-121C2.2 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-497-5p | 15 | PAFAH1B2 | Sponge network | 1.147 | 0 | 0.318 | 0.0001 | 0.312 |
| 37931 | TRAF3IP2-AS1 |
hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 25 | UBR3 | Sponge network | -0.323 | 0.01372 | -0.021 | 0.82774 | 0.312 |
| 37932 | RP11-395G23.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 32 | GAS7 | Sponge network | 0.381 | 0.25967 | -0.555 | 0.01602 | 0.312 |
| 37933 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-500a-5p;hsa-miR-93-5p | 21 | SASH1 | Sponge network | 0.082 | 0.55397 | -0.502 | 0.00175 | 0.312 |
| 37934 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p | 11 | LHX6 | Sponge network | -0.737 | 0.10822 | -0.087 | 0.69193 | 0.312 |
| 37935 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 36 | EPHA7 | Sponge network | 0.16 | 0.50785 | -2.197 | 3.0E-5 | 0.312 |
| 37936 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-590-3p;hsa-miR-628-5p | 33 | BMPR2 | Sponge network | 1.089 | 0 | 0.206 | 0.0635 | 0.312 |
| 37937 | GABPB1-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 25 | ABI2 | Sponge network | 1.11 | 0 | 0.175 | 0.14787 | 0.312 |
| 37938 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-378c;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p | 27 | MED12L | Sponge network | -0.096 | 0.67958 | -0.194 | 0.55299 | 0.312 |
| 37939 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 23 | BHLHE41 | Sponge network | 0.657 | 4.0E-5 | 0.353 | 0.12753 | 0.312 |
| 37940 | RP1-122P22.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p | 11 | CDH23 | Sponge network | 0.065 | 0.73468 | -0.974 | 0.00034 | 0.312 |
| 37941 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ARNT | Sponge network | 0.421 | 0.00084 | -0.08 | 0.26073 | 0.312 |
| 37942 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-493-5p | 20 | MYBL1 | Sponge network | 0.614 | 1.0E-5 | 0.494 | 0.02152 | 0.312 |
| 37943 | DNAJC27-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 13 | NRG3 | Sponge network | -0.969 | 2.0E-5 | -1.836 | 4.0E-5 | 0.312 |
| 37944 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-96-5p | 25 | CYLD | Sponge network | 0.997 | 0.00066 | -0.367 | 0.00171 | 0.312 |
| 37945 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 25 | GRIA3 | Sponge network | -0.738 | 0.21233 | -1.956 | 0 | 0.312 |
| 37946 | LINC00403 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-30d-3p | 10 | AFF1 | Sponge network | -3.586 | 0.00032 | -0.384 | 0.00053 | 0.312 |
| 37947 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 28 | MEF2C | Sponge network | -0.176 | 0.59692 | -0.499 | 0.01448 | 0.312 |
| 37948 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-154-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 31 | PBX1 | Sponge network | -1.264 | 6.0E-5 | -1.206 | 0 | 0.312 |
| 37949 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 20 | KIF1B | Sponge network | 1.352 | 0 | -0.118 | 0.32258 | 0.312 |
| 37950 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | UVRAG | Sponge network | -0.487 | 0.02157 | -0.017 | 0.83865 | 0.312 |
| 37951 | HCG18 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | TCF12 | Sponge network | 1.063 | 0 | 0.174 | 0.13271 | 0.312 |
| 37952 | LINC00403 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p | 10 | MCC | Sponge network | -3.586 | 0.00032 | -1.005 | 0 | 0.312 |
| 37953 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | CHD2 | Sponge network | -0.612 | 0.00094 | 0.049 | 0.55371 | 0.312 |
| 37954 | RP4-669P10.16 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-766-3p | 14 | IL17RD | Sponge network | -0.301 | 0.06597 | -0.019 | 0.94873 | 0.312 |
| 37955 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | PHC3 | Sponge network | 1.163 | 0.00018 | 0.212 | 0.0499 | 0.312 |
| 37956 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 17 | NF1 | Sponge network | 0.349 | 0.01695 | 0.51 | 0 | 0.312 |
| 37957 | AC034220.3 |
hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 13 | CHD2 | Sponge network | 0.309 | 0.07304 | 0.049 | 0.55371 | 0.312 |
| 37958 | LINC00865 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-935 | 19 | GJA1 | Sponge network | -1.249 | 7.0E-5 | -0.485 | 0.02179 | 0.312 |
| 37959 | EIF3J-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PRKAB2 | Sponge network | 0.331 | 0.00509 | -0.367 | 0.01371 | 0.312 |
| 37960 | RP11-403I13.8 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | BRD3 | Sponge network | 0.619 | 0.00157 | 0.37 | 0.00013 | 0.312 |
| 37961 | RP11-53O19.1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -0.405 | 0.22013 | -0.966 | 0.00035 | 0.312 |
| 37962 | RP11-927P21.1 |
hsa-let-7d-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-454-3p | 10 | DICER1 | Sponge network | 0.921 | 0.00118 | 0.042 | 0.674 | 0.312 |
| 37963 | RP11-274B21.10 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-590-3p | 11 | SIRT1 | Sponge network | 1.073 | 0.00216 | 0.109 | 0.2593 | 0.312 |
| 37964 | CTC-487M23.5 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-34a-5p | 11 | ITPKB | Sponge network | 0.912 | 0.00014 | -0.704 | 0.00106 | 0.312 |
| 37965 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 26 | SOX11 | Sponge network | 0.537 | 0.00139 | 2.68 | 0 | 0.312 |
| 37966 | RP11-282O18.3 |
hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-505-5p | 14 | DDX3X | Sponge network | 0.757 | 0 | -0.152 | 0.07686 | 0.312 |
| 37967 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 18 | ARID5B | Sponge network | -0.227 | 0.31657 | 0.342 | 0.01676 | 0.312 |
| 37968 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | AR | Sponge network | 0.056 | 0.8494 | -1.554 | 0 | 0.312 |
| 37969 | AC106786.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-877-5p | 16 | ASTN1 | Sponge network | 1.122 | 0.02989 | -2.389 | 2.0E-5 | 0.312 |
| 37970 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 33 | CLASP2 | Sponge network | 0.222 | 0.29133 | -0.038 | 0.71 | 0.312 |
| 37971 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-942-5p | 32 | HLF | Sponge network | 0.311 | 0.10344 | -2.443 | 0 | 0.312 |
| 37972 | LINC01004 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 1.115 | 0 | 0.51 | 0 | 0.312 |
| 37973 | AC034220.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ABI2 | Sponge network | 0.309 | 0.07304 | 0.175 | 0.14787 | 0.312 |
| 37974 | RP11-13K12.1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p | 16 | RPS6KA6 | Sponge network | -0.154 | 0.78939 | -2.989 | 0 | 0.312 |
| 37975 | CTD-2528L19.6 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | ITGB1 | Sponge network | 0.953 | 0 | 0.524 | 0.00017 | 0.312 |
| 37976 | AC108488.4 |
hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | MYO9A | Sponge network | 0.259 | 0.12542 | -0.147 | 0.32373 | 0.312 |
| 37977 | RP11-155G14.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-625-5p | 11 | ZMYM2 | Sponge network | 2.483 | 0 | 0.279 | 0.01273 | 0.312 |
| 37978 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | KAT6B | Sponge network | -0.576 | 0.10121 | -0.478 | 0.00018 | 0.312 |
| 37979 | DIO3OS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-455-5p | 10 | THSD7A | Sponge network | -0.773 | 0.04073 | 0.242 | 0.25154 | 0.312 |
| 37980 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-23a-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p | 11 | CNTN1 | Sponge network | -0.278 | 0.76559 | -2.594 | 0 | 0.312 |
| 37981 | RP11-802E16.3 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-497-5p | 13 | SOX11 | Sponge network | 2.048 | 0 | 2.68 | 0 | 0.312 |
| 37982 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p | 40 | ERBB4 | Sponge network | -0.793 | 7.0E-5 | -1.975 | 2.0E-5 | 0.312 |
| 37983 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 14 | SLC6A15 | Sponge network | -0.563 | 0.02545 | -2.373 | 2.0E-5 | 0.312 |
| 37984 | SH3BP5-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 63 | LPP | Sponge network | 0.267 | 0.2937 | -0.459 | 0.04475 | 0.312 |
| 37985 | HCG22 |
hsa-let-7a-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p | 15 | ST6GAL2 | Sponge network | 0.816 | 0.32368 | -1.201 | 0.00597 | 0.312 |
| 37986 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-93-5p | 12 | TRIM3 | Sponge network | -2.117 | 0.00161 | -0.577 | 0 | 0.312 |
| 37987 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-744-5p | 11 | FGFR1 | Sponge network | -4.477 | 0 | -0.509 | 0.03957 | 0.312 |
| 37988 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-708-3p | 22 | GRIA3 | Sponge network | -0.384 | 0.40652 | -1.956 | 0 | 0.312 |
| 37989 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p | 13 | JAZF1 | Sponge network | -2.549 | 0.00528 | -0.377 | 0.02677 | 0.312 |
| 37990 | RP4-605O3.4 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | PHC3 | Sponge network | 0.262 | 0.0475 | 0.212 | 0.0499 | 0.312 |
| 37991 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 12 | SYT4 | Sponge network | -0.187 | 0.23787 | -3.207 | 0 | 0.312 |
| 37992 | RP11-403I13.8 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 11 | MACF1 | Sponge network | 0.619 | 0.00157 | 0.019 | 0.90335 | 0.312 |
| 37993 | RP4-584D14.7 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-362-3p | 11 | EPHA7 | Sponge network | 1.294 | 0.0031 | -2.197 | 3.0E-5 | 0.312 |
| 37994 | HOXB-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429 | 16 | MXI1 | Sponge network | -0.154 | 0.53954 | -0.987 | 0 | 0.312 |
| 37995 | RP3-402G11.26 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | ABI2 | Sponge network | -0.254 | 0.19303 | 0.175 | 0.14787 | 0.312 |
| 37996 | MRVI1-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-452-5p | 10 | INPP4B | Sponge network | -1.092 | 4.0E-5 | -0.097 | 0.60503 | 0.312 |
| 37997 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | BHLHE41 | Sponge network | -1.375 | 0.08642 | 0.353 | 0.12753 | 0.312 |
| 37998 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p | 12 | DDX6 | Sponge network | 0.537 | 0.0006 | 0.091 | 0.36428 | 0.312 |
| 37999 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 22 | EPHA3 | Sponge network | -0.031 | 0.89063 | 0.185 | 0.53588 | 0.312 |
| 38000 | RP11-77H9.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | TBL1XR1 | Sponge network | 1.677 | 0 | 0.578 | 0 | 0.312 |
| 38001 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-940 | 16 | BHLHE41 | Sponge network | -0.829 | 0.01343 | 0.353 | 0.12753 | 0.312 |
| 38002 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-7-1-3p | 10 | EGR2 | Sponge network | 0.395 | 0.15451 | 0.309 | 0.17079 | 0.312 |
| 38003 | MYLK-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 10 | UNC5C | Sponge network | -1.331 | 3.0E-5 | -0.178 | 0.40214 | 0.312 |
| 38004 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-590-3p | 10 | NALCN | Sponge network | -0.08 | 0.90092 | 1.068 | 0.00237 | 0.312 |
| 38005 | KB-1572G7.2 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | UBR3 | Sponge network | 0.924 | 0 | -0.021 | 0.82774 | 0.312 |
| 38006 | RP11-1006G14.4 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-454-3p;hsa-miR-744-3p | 11 | IGF2 | Sponge network | 0.559 | 0.00026 | 0.848 | 0.03842 | 0.312 |
| 38007 | RP1-257A7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-484 | 10 | CRTC3 | Sponge network | 0.849 | 0.00178 | 0.164 | 0.16786 | 0.312 |
| 38008 | CTA-204B4.2 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-590-3p | 11 | DOCK4 | Sponge network | 0.765 | 7.0E-5 | 0.502 | 0.00108 | 0.312 |
| 38009 | ERVK3-1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-376c-3p;hsa-miR-495-3p | 13 | TBL1XR1 | Sponge network | 0.58 | 0 | 0.578 | 0 | 0.312 |
| 38010 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-877-5p | 25 | GRIN2A | Sponge network | 0.082 | 0.55654 | -2.161 | 0 | 0.312 |
| 38011 | RP11-226L15.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | PIK3CA | Sponge network | 0.8 | 0 | 0.195 | 0.06908 | 0.312 |
| 38012 | RP11-434B12.1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 11 | SH3GLB1 | Sponge network | -0.031 | 0.89063 | -0.115 | 0.14773 | 0.312 |
| 38013 | RP11-747H7.3 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-497-5p;hsa-miR-9-5p | 13 | CCDC6 | Sponge network | 1.379 | 0 | 0.27 | 0.02555 | 0.312 |
| 38014 | HAND2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | CREM | Sponge network | -1.697 | 0.00889 | -0.345 | 0.00258 | 0.312 |
| 38015 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | ZMYM2 | Sponge network | 1.316 | 0 | 0.279 | 0.01273 | 0.312 |
| 38016 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | PRKAB2 | Sponge network | 0.768 | 7.0E-5 | -0.367 | 0.01371 | 0.312 |
| 38017 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-93-5p | 34 | TSHZ3 | Sponge network | -1.047 | 0 | -0.556 | 0.01516 | 0.312 |
| 38018 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-342-3p | 12 | SLC6A15 | Sponge network | -2.095 | 0.00112 | -2.373 | 2.0E-5 | 0.312 |
| 38019 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-455-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | NRXN1 | Sponge network | -0.418 | 0.00068 | -3.778 | 0 | 0.312 |
| 38020 | OIP5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940 | 35 | SNX29 | Sponge network | 0.421 | 0.00084 | -0.34 | 0.01371 | 0.312 |
| 38021 | RP11-927P21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p | 21 | DMD | Sponge network | 0.921 | 0.00118 | -1.506 | 0 | 0.312 |
| 38022 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | SMARCA2 | Sponge network | -0.576 | 0.10121 | -0.529 | 0.00014 | 0.312 |
| 38023 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ABCC9 | Sponge network | 0.659 | 3.0E-5 | -0.466 | 0.14121 | 0.312 |
| 38024 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 22 | PHC3 | Sponge network | 0.389 | 0.04293 | 0.212 | 0.0499 | 0.312 |
| 38025 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | -0.206 | 0.12942 | 0.309 | 0.17079 | 0.312 |
| 38026 | AF131215.9 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-940 | 15 | BAZ2B | Sponge network | -0.322 | 0.25945 | -0.276 | 0.02833 | 0.312 |
| 38027 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-629-3p | 33 | EPHA7 | Sponge network | -0.773 | 0.04073 | -2.197 | 3.0E-5 | 0.312 |
| 38028 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | CBX5 | Sponge network | 0.392 | 0.0278 | 0.165 | 0.24446 | 0.312 |
| 38029 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-589-5p | 11 | TCF4 | Sponge network | -7.551 | 0 | 0.016 | 0.92271 | 0.312 |
| 38030 | NDUFA6-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-590-3p | 13 | CXCL12 | Sponge network | -0.355 | 0.0077 | -1.421 | 0 | 0.312 |
| 38031 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 10 | PAK3 | Sponge network | -0.342 | 0.02288 | -0.628 | 0.07253 | 0.312 |
| 38032 | ZSCAN16-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-942-5p | 10 | ZBTB20 | Sponge network | -0.342 | 0.02288 | -0.122 | 0.57569 | 0.312 |
| 38033 | LINC01018 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 16 | INPP4A | Sponge network | -2.915 | 0.00015 | 0.07 | 0.53563 | 0.312 |
| 38034 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-590-5p | 22 | SCN9A | Sponge network | -0.384 | 0.40652 | -0.525 | 0.10593 | 0.312 |
| 38035 | GHRLOS |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-493-5p | 14 | LRP1B | Sponge network | -1.942 | 0 | -2.163 | 0 | 0.312 |
| 38036 | CTB-113P19.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | MCC | Sponge network | 0.906 | 0.31715 | -1.005 | 0 | 0.312 |
| 38037 | H1FX-AS1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | ZNF208 | Sponge network | -0.206 | 0.12942 | -0.4 | 0.22381 | 0.312 |
| 38038 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p | 15 | HIP1 | Sponge network | -0.405 | 0.22013 | 0.774 | 0 | 0.312 |
| 38039 | CKMT2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | ARNT2 | Sponge network | 0.082 | 0.55654 | -0.332 | 0.25851 | 0.312 |
| 38040 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SETBP1 | Sponge network | 0.349 | 0.01695 | -1.302 | 1.0E-5 | 0.312 |
| 38041 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | PRDM2 | Sponge network | -0.358 | 0.17255 | -0.258 | 0.00721 | 0.312 |
| 38042 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-590-5p | 22 | CBL | Sponge network | 0.331 | 0.57247 | 0.291 | 0.01267 | 0.312 |
| 38043 | SH3BP5-AS1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-654-3p | 32 | PIK3R1 | Sponge network | 0.267 | 0.2937 | -0.133 | 0.36854 | 0.312 |
| 38044 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 22 | ARID5B | Sponge network | 0.889 | 9.0E-5 | 0.342 | 0.01676 | 0.312 |
| 38045 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-877-5p | 16 | NOTCH2NL | Sponge network | 0.494 | 0.00339 | 0.024 | 0.84307 | 0.312 |
| 38046 | RP5-1198O20.4 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | BCL9 | Sponge network | 0.997 | 0.00066 | 0.321 | 0.00267 | 0.312 |
| 38047 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | TAL1 | Sponge network | 0.214 | 0.18246 | -0.462 | 0.00574 | 0.312 |
| 38048 | RP11-359E10.1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 11 | RASSF3 | Sponge network | 0.299 | 0.57722 | -0.226 | 0.11691 | 0.312 |
| 38049 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | NRXN3 | Sponge network | 0.349 | 0.01695 | -0.893 | 0.04186 | 0.312 |
| 38050 | RP11-473I1.10 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | SCN9A | Sponge network | 0.673 | 0 | -0.525 | 0.10593 | 0.312 |
| 38051 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-744-3p | 21 | PTPN14 | Sponge network | -0.223 | 0.47816 | 0.144 | 0.34595 | 0.312 |
| 38052 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | TACC1 | Sponge network | -2.915 | 0.00015 | -0.362 | 0.05406 | 0.312 |
| 38053 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-7-1-3p | 23 | EPHA7 | Sponge network | -0.72 | 0.24239 | -2.197 | 3.0E-5 | 0.312 |
| 38054 | GAS6-AS2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 20 | MXI1 | Sponge network | 0.16 | 0.50785 | -0.987 | 0 | 0.312 |
| 38055 | CTD-2314G24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | ARNT2 | Sponge network | 0.246 | 0.59912 | -0.332 | 0.25851 | 0.312 |
| 38056 | RP5-994D16.9 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PAFAH1B1 | Sponge network | 0.252 | 0.08508 | -0.429 | 0 | 0.312 |
| 38057 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-98-5p | 30 | PBX1 | Sponge network | -0.83 | 0 | -1.206 | 0 | 0.312 |
| 38058 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p | 18 | ZBTB4 | Sponge network | 0.225 | 0.30644 | -0.739 | 0 | 0.312 |
| 38059 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 33 | TRPS1 | Sponge network | -1.057 | 0.00029 | -0.191 | 0.44109 | 0.311 |
| 38060 | LINC00654 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3C | Sponge network | 0.374 | 0.20054 | -0.272 | 0.31153 | 0.311 |
| 38061 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | AKAP13 | Sponge network | 0.727 | 3.0E-5 | 0.028 | 0.7729 | 0.311 |
| 38062 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-940 | 12 | TRIM33 | Sponge network | 0.082 | 0.55397 | 0.402 | 7.0E-5 | 0.311 |
| 38063 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 26 | DMXL1 | Sponge network | 0.082 | 0.55654 | -0.426 | 0.00056 | 0.311 |
| 38064 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 19 | HECA | Sponge network | 0.4 | 0.14611 | -0.217 | 0.03463 | 0.311 |
| 38065 | CTC-490E21.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p | 12 | PRKAB2 | Sponge network | 1.46 | 1.0E-5 | -0.367 | 0.01371 | 0.311 |
| 38066 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-93-5p | 33 | RASSF2 | Sponge network | -0.162 | 0.47687 | -0.273 | 0.21961 | 0.311 |
| 38067 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 10 | LRRK2 | Sponge network | 0.385 | 0.10067 | -1.136 | 1.0E-5 | 0.311 |
| 38068 | BCYRN1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | UBE2D1 | Sponge network | 0.311 | 0.10344 | 0.468 | 0 | 0.311 |
| 38069 | RP11-212P7.2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ITPKB | Sponge network | 0.219 | 0.1516 | -0.704 | 0.00106 | 0.311 |
| 38070 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-590-3p | 17 | CLASP2 | Sponge network | 1.089 | 0 | -0.038 | 0.71 | 0.311 |
| 38071 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-628-5p | 18 | CDC73 | Sponge network | 0.93 | 0 | 0.094 | 0.20463 | 0.311 |
| 38072 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 15 | GPAM | Sponge network | 0.765 | 7.0E-5 | 0.142 | 0.29732 | 0.311 |
| 38073 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | CXXC4 | Sponge network | 0.389 | 0.04293 | -0.433 | 0.21055 | 0.311 |
| 38074 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | EPS15 | Sponge network | 0.683 | 1.0E-5 | -0.052 | 0.53998 | 0.311 |
| 38075 | RP11-166D19.1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | GATA2 | Sponge network | -0.576 | 0.10121 | -0.654 | 0.0016 | 0.311 |
| 38076 | LPP-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-96-5p | 15 | ERG | Sponge network | -0.18 | 0.20343 | 0.002 | 0.99011 | 0.311 |
| 38077 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 24 | YAP1 | Sponge network | -0.737 | 0.06182 | 0.22 | 0.11836 | 0.311 |
| 38078 | C4A-AS1 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-511-5p | 13 | ZFHX3 | Sponge network | 3.345 | 0 | 0.389 | 0.01363 | 0.311 |
| 38079 | AC004893.11 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | 1.317 | 0 | 0.142 | 0.04938 | 0.311 |
| 38080 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-3p | 29 | SMAD4 | Sponge network | 0.178 | 0.29106 | -0.313 | 0.00413 | 0.311 |
| 38081 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | DCLK1 | Sponge network | -4.477 | 0 | -1.324 | 0.00014 | 0.311 |
| 38082 | MEG3 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PRDM11 | Sponge network | 0.249 | 0.35902 | -0.252 | 0.15511 | 0.311 |
| 38083 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 23 | ACSL6 | Sponge network | -2.69 | 0.0001 | -1.593 | 0 | 0.311 |
| 38084 | XXYLT1-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p | 11 | RASSF2 | Sponge network | -0.764 | 0.05257 | -0.273 | 0.21961 | 0.311 |
| 38085 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-7-1-3p | 13 | GRIN2A | Sponge network | -0.285 | 0.2172 | -2.161 | 0 | 0.311 |
| 38086 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p | 20 | CBFA2T3 | Sponge network | -0.773 | 0.04073 | -1.221 | 1.0E-5 | 0.311 |
| 38087 | CTD-2089N3.2 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 13 | OPCML | Sponge network | -1.375 | 0.08642 | -2.498 | 0 | 0.311 |
| 38088 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p | 11 | LIMD1 | Sponge network | 1.294 | 0 | 0.103 | 0.28978 | 0.311 |
| 38089 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 15 | HERC2 | Sponge network | -0.739 | 0.0574 | 0.114 | 0.30638 | 0.311 |
| 38090 | BZRAP1-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-375;hsa-miR-409-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-577;hsa-miR-93-3p;hsa-miR-940 | 35 | TRPS1 | Sponge network | -0.465 | 0.04703 | -0.191 | 0.44109 | 0.311 |
| 38091 | FAM13A-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-671-5p | 14 | CADM3 | Sponge network | -0.83 | 0 | -3.663 | 0 | 0.311 |
| 38092 | DIO3OS |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-660-5p | 22 | JDP2 | Sponge network | -0.773 | 0.04073 | -0.967 | 0 | 0.311 |
| 38093 | FAM66C |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p | 17 | MAFB | Sponge network | -0.901 | 0.00019 | -0.023 | 0.89865 | 0.311 |
| 38094 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-425-5p | 15 | ERC1 | Sponge network | 0.37 | 0.27614 | -0.108 | 0.38794 | 0.311 |
| 38095 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 43 | EPHA7 | Sponge network | -1.264 | 6.0E-5 | -2.197 | 3.0E-5 | 0.311 |
| 38096 | H1FX-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 18 | EPHA3 | Sponge network | -0.206 | 0.12942 | 0.185 | 0.53588 | 0.311 |
| 38097 | U91328.21 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-629-3p | 21 | AR | Sponge network | -0.735 | 0.15983 | -1.554 | 0 | 0.311 |
| 38098 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-940 | 36 | PTPRT | Sponge network | -0.096 | 0.67958 | -0.907 | 0.03727 | 0.311 |
| 38099 | HCG11 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p | 19 | ASTN1 | Sponge network | 0.385 | 0.10067 | -2.389 | 2.0E-5 | 0.311 |
| 38100 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577 | 21 | KIT | Sponge network | 0.056 | 0.81195 | -2.184 | 0 | 0.311 |
| 38101 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 23 | HECA | Sponge network | -0.303 | 0.21002 | -0.217 | 0.03463 | 0.311 |
| 38102 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | RBMS3 | Sponge network | 0.272 | 0.44692 | -0.519 | 0.03551 | 0.311 |
| 38103 | RP11-540B6.6 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-3p;hsa-miR-424-5p | 10 | UBE4B | Sponge network | 0.93 | 0 | -0.235 | 0.007 | 0.311 |
| 38104 | CTC-338M12.4 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-582-3p | 17 | ABL2 | Sponge network | 0.638 | 0 | 0.82 | 0 | 0.311 |
| 38105 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 11 | MYO9A | Sponge network | 1.11 | 1.0E-5 | -0.147 | 0.32373 | 0.311 |
| 38106 | ARHGEF26-AS1 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | NR3C2 | Sponge network | -2.46 | 0 | -1.56 | 0 | 0.311 |
| 38107 | MCM3AP-AS1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 10 | CBFB | Sponge network | 0.331 | 1.0E-5 | 1.061 | 0 | 0.311 |
| 38108 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 19 | PGR | Sponge network | 1.757 | 0 | -1.704 | 0 | 0.311 |
| 38109 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | 0.374 | 0.20054 | 0.309 | 0.17079 | 0.311 |
| 38110 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-361-3p | 12 | TGFBR3 | Sponge network | 0.598 | 0.38481 | -1.173 | 1.0E-5 | 0.311 |
| 38111 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30b-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-5p | 12 | CXXC4 | Sponge network | -1.116 | 0.23686 | -0.433 | 0.21055 | 0.311 |
| 38112 | TRAF3IP2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p | 22 | CYLD | Sponge network | -0.323 | 0.01372 | -0.367 | 0.00171 | 0.311 |
| 38113 | RP1-122P22.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-744-5p;hsa-miR-942-5p | 16 | NOTCH2 | Sponge network | 0.065 | 0.73468 | 0.138 | 0.35163 | 0.311 |
| 38114 | RP11-701H24.3 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 10 | MDM4 | Sponge network | -1.425 | 0.04204 | 0.345 | 0.00762 | 0.311 |
| 38115 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | USP12 | Sponge network | -0.793 | 7.0E-5 | -0.102 | 0.40768 | 0.311 |
| 38116 | RP5-1074L1.4 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | BRD3 | Sponge network | 1.923 | 0 | 0.37 | 0.00013 | 0.311 |
| 38117 | PWAR6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | BRD3 | Sponge network | -1.575 | 6.0E-5 | 0.37 | 0.00013 | 0.311 |
| 38118 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 20 | NRXN1 | Sponge network | -0.053 | 0.80685 | -3.778 | 0 | 0.311 |
| 38119 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-942-5p | 25 | GPC6 | Sponge network | 0.101 | 0.60919 | -0.054 | 0.81465 | 0.311 |
| 38120 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FAM188A | Sponge network | -0.615 | 0.00015 | -0.44 | 0.00018 | 0.311 |
| 38121 | LINC00667 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-590-3p | 11 | HERC1 | Sponge network | -0.235 | 0.15142 | -0.196 | 0.08544 | 0.311 |
| 38122 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p | 16 | PBX1 | Sponge network | 0.001 | 0.99753 | -1.206 | 0 | 0.311 |
| 38123 | MALAT1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ITGB1 | Sponge network | 0.782 | 0.00016 | 0.524 | 0.00017 | 0.311 |
| 38124 | PART1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-940 | 45 | PTPRT | Sponge network | -2.925 | 0 | -0.907 | 0.03727 | 0.311 |
| 38125 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-576-5p | 15 | PTGFR | Sponge network | -0.405 | 0.22013 | -0.233 | 0.45421 | 0.311 |
| 38126 | AC018890.6 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-96-5p | 10 | DNMT3A | Sponge network | 1.61 | 0.00961 | 0.302 | 0.0262 | 0.311 |
| 38127 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-671-3p;hsa-miR-93-5p | 13 | MAP3K12 | Sponge network | -0.842 | 0.01361 | -0.057 | 0.59369 | 0.311 |
| 38128 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | HIP1 | Sponge network | -0.053 | 0.80685 | 0.774 | 0 | 0.311 |
| 38129 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p | 12 | GREM1 | Sponge network | -0.726 | 0.00132 | 0.902 | 0.01691 | 0.311 |
| 38130 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 30 | ZFHX3 | Sponge network | 0.4 | 0.14611 | 0.389 | 0.01363 | 0.311 |
| 38131 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NOTCH2NL | Sponge network | 0.559 | 0.00026 | 0.024 | 0.84307 | 0.311 |
| 38132 | LINC00621 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 22 | QKI | Sponge network | 0.131 | 0.8436 | -0.333 | 0.04111 | 0.311 |
| 38133 | TRIM52-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-24-2-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-7-1-3p | 12 | PCDH9 | Sponge network | -0.418 | 0.00068 | -1.823 | 4.0E-5 | 0.311 |
| 38134 | RP1-102E24.8 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | FOXO3 | Sponge network | 1.737 | 0 | -0.04 | 0.72028 | 0.311 |
| 38135 | RP4-545C24.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-424-5p | 10 | ABL2 | Sponge network | 0.363 | 0.02231 | 0.82 | 0 | 0.311 |
| 38136 | HAS2-AS1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-625-5p | 11 | CADM1 | Sponge network | -0.011 | 0.967 | -0.9 | 0.00084 | 0.311 |
| 38137 | AC016747.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 38 | DTNA | Sponge network | 0.199 | 0.38015 | -1.648 | 1.0E-5 | 0.311 |
| 38138 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | PRKCE | Sponge network | -0.739 | 0.0574 | -0.403 | 0.00056 | 0.311 |
| 38139 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 37 | PHC3 | Sponge network | -2.452 | 0 | 0.212 | 0.0499 | 0.311 |
| 38140 | ERVK3-1 |
hsa-let-7c-5p;hsa-miR-100-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-363-3p | 11 | ATP2A2 | Sponge network | 0.58 | 0 | 0.604 | 0 | 0.311 |
| 38141 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-671-5p | 24 | BMPR2 | Sponge network | 0.556 | 0.00298 | 0.206 | 0.0635 | 0.311 |
| 38142 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 36 | EPHA5 | Sponge network | 0.392 | 0.0278 | -0.957 | 0.01733 | 0.311 |
| 38143 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | PHC3 | Sponge network | -0.737 | 0.06182 | 0.212 | 0.0499 | 0.311 |
| 38144 | RP11-798G7.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-582-3p | 16 | FOXO3 | Sponge network | 0.858 | 3.0E-5 | -0.04 | 0.72028 | 0.311 |
| 38145 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ZNF208 | Sponge network | 0.082 | 0.55654 | -0.4 | 0.22381 | 0.311 |
| 38146 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-93-5p | 12 | MAP3K12 | Sponge network | -0.777 | 0.16667 | -0.057 | 0.59369 | 0.311 |
| 38147 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-654-3p | 11 | TP53INP1 | Sponge network | 1.157 | 0.00455 | -0.496 | 0.00064 | 0.311 |
| 38148 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 31 | NTRK2 | Sponge network | 0.537 | 0.00139 | -0.736 | 0.06495 | 0.311 |
| 38149 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p | 25 | ADARB1 | Sponge network | -0.602 | 0.00331 | -0.335 | 0.06631 | 0.311 |
| 38150 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 41 | TMEM132B | Sponge network | 0.572 | 0.01722 | -0.83 | 0.01084 | 0.311 |
| 38151 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 0.768 | 7.0E-5 | 0.114 | 0.30638 | 0.311 |
| 38152 | MIR143HG |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-7-5p | 11 | COL1A2 | Sponge network | -0.739 | 0.0574 | 1.991 | 0 | 0.311 |
| 38153 | AC005618.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | EYA4 | Sponge network | 0.862 | 0.06213 | 0.3 | 0.36876 | 0.311 |
| 38154 | OIP5-AS1 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-5p | 12 | RANBP6 | Sponge network | 0.421 | 0.00084 | 0.175 | 0.19756 | 0.311 |
| 38155 | MIR143HG |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | SH3GLB1 | Sponge network | -0.739 | 0.0574 | -0.115 | 0.14773 | 0.311 |
| 38156 | LINC00476 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | ABCB5 | Sponge network | -0.615 | 0.00015 | -0.956 | 0.04077 | 0.311 |
| 38157 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-3613-5p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | TACC1 | Sponge network | 0.056 | 0.8494 | -0.362 | 0.05406 | 0.311 |
| 38158 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-1-3p | 27 | TGFBR3 | Sponge network | -4.477 | 0 | -1.173 | 1.0E-5 | 0.311 |
| 38159 | SERTAD4-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-576-5p | 15 | ABI2 | Sponge network | -0.932 | 0.00329 | 0.175 | 0.14787 | 0.311 |
| 38160 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-582-5p | 17 | PIK3C2A | Sponge network | 0.792 | 0.0049 | 0.061 | 0.53776 | 0.311 |
| 38161 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 25 | TCF4 | Sponge network | 0.131 | 0.8436 | 0.016 | 0.92271 | 0.311 |
| 38162 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | TP53INP1 | Sponge network | 0.101 | 0.60919 | -0.496 | 0.00064 | 0.311 |
| 38163 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 31 | LIFR | Sponge network | 0.246 | 0.59912 | -1.794 | 0 | 0.311 |
| 38164 | WDFY3-AS2 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-423-5p | 19 | ABL1 | Sponge network | -0.942 | 0 | -0.215 | 0.07804 | 0.311 |
| 38165 | AC084219.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p | 11 | CDC73 | Sponge network | 1.296 | 0.0054 | 0.094 | 0.20463 | 0.311 |
| 38166 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p | 12 | THBS1 | Sponge network | 0.348 | 0.32912 | 0.741 | 0.00386 | 0.311 |
| 38167 | RP11-175O19.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p;hsa-miR-664a-5p | 17 | CBFB | Sponge network | 0.917 | 0 | 1.061 | 0 | 0.311 |
| 38168 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | TACC1 | Sponge network | -0.206 | 0.12942 | -0.362 | 0.05406 | 0.311 |
| 38169 | RP11-379F4.7 |
hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | DDX3X | Sponge network | 1.316 | 0 | -0.152 | 0.07686 | 0.311 |
| 38170 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | RBMS3 | Sponge network | 0.411 | 0.1335 | -0.519 | 0.03551 | 0.311 |
| 38171 | RP11-473I1.10 |
hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-590-3p;hsa-miR-628-5p | 10 | CDC73 | Sponge network | 0.673 | 0 | 0.094 | 0.20463 | 0.311 |
| 38172 | CTC-297N7.9 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-590-5p;hsa-miR-744-3p | 15 | MEF2C | Sponge network | -2.069 | 0 | -0.499 | 0.01448 | 0.311 |
| 38173 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 12 | PTPRB | Sponge network | -0.693 | 0.05629 | 0.105 | 0.47122 | 0.311 |
| 38174 | KTN1-AS1 |
hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-664a-3p;hsa-miR-664a-5p | 12 | SOX11 | Sponge network | 0.927 | 0 | 2.68 | 0 | 0.311 |
| 38175 | LINC00672 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-98-5p | 14 | SEMA3C | Sponge network | -0.227 | 0.31657 | -0.272 | 0.31153 | 0.311 |
| 38176 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 44 | PRKAA2 | Sponge network | 0.222 | 0.29133 | -2.462 | 0 | 0.311 |
| 38177 | RP11-403I13.7 |
hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-7-1-3p | 14 | ITPKB | Sponge network | 0.395 | 0.15451 | -0.704 | 0.00106 | 0.311 |
| 38178 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3C | Sponge network | 0.537 | 0.00139 | -0.272 | 0.31153 | 0.311 |
| 38179 | DNAJC27-AS1 |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-542-3p | 11 | CADM4 | Sponge network | -0.969 | 2.0E-5 | -0.635 | 0.00305 | 0.311 |
| 38180 | OSER1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | NRXN3 | Sponge network | -0.13 | 0.48734 | -0.893 | 0.04186 | 0.311 |
| 38181 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | PRKAR1A | Sponge network | 0.997 | 0.00066 | -0.063 | 0.50314 | 0.311 |
| 38182 | RP11-760H22.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p | 10 | NR3C2 | Sponge network | 0.001 | 0.99753 | -1.56 | 0 | 0.311 |
| 38183 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p | 36 | RBMS3 | Sponge network | -1.047 | 0 | -0.519 | 0.03551 | 0.311 |
| 38184 | WDFY3-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | MED12L | Sponge network | -0.942 | 0 | -0.194 | 0.55299 | 0.311 |
| 38185 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | CHD2 | Sponge network | 0.603 | 0.00127 | 0.049 | 0.55371 | 0.311 |
| 38186 | RP11-212P7.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | STARD13 | Sponge network | 0.219 | 0.1516 | -0.187 | 0.27383 | 0.311 |
| 38187 | LINC00865 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p | 19 | ERG | Sponge network | -1.249 | 7.0E-5 | 0.002 | 0.99011 | 0.311 |
| 38188 | RAMP2-AS1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | BRD3 | Sponge network | -0.513 | 0.04703 | 0.37 | 0.00013 | 0.311 |
| 38189 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-96-5p | 17 | RIMBP2 | Sponge network | -0.668 | 3.0E-5 | -1.968 | 5.0E-5 | 0.311 |
| 38190 | RP11-473I1.9 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-5p | 10 | RANBP6 | Sponge network | 0.683 | 1.0E-5 | 0.175 | 0.19756 | 0.311 |
| 38191 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | NAV3 | Sponge network | -1.057 | 0.00029 | 0.212 | 0.47729 | 0.311 |
| 38192 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | EPHA7 | Sponge network | 0.392 | 0.0278 | -2.197 | 3.0E-5 | 0.311 |
| 38193 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-7-1-3p | 15 | AHNAK | Sponge network | -1.249 | 7.0E-5 | -1.207 | 0 | 0.311 |
| 38194 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-532-5p | 14 | PTPRB | Sponge network | 0.082 | 0.55397 | 0.105 | 0.47122 | 0.311 |
| 38195 | CTC-429P9.3 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p | 10 | PTCH1 | Sponge network | 0.082 | 0.55397 | -0.1 | 0.6179 | 0.311 |
| 38196 | FAM157B |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-378a-5p | 14 | BMPR2 | Sponge network | 0.92 | 0 | 0.206 | 0.0635 | 0.311 |
| 38197 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | PDE4DIP | Sponge network | 0.299 | 0.57722 | -0.282 | 0.0257 | 0.311 |
| 38198 | LINC00526 |
hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-590-5p | 16 | PTPRT | Sponge network | -0.377 | 0.0384 | -0.907 | 0.03727 | 0.311 |
| 38199 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 19 | SOX11 | Sponge network | 0.222 | 0.29133 | 2.68 | 0 | 0.311 |
| 38200 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 32 | HLF | Sponge network | -1.057 | 0.00029 | -2.443 | 0 | 0.311 |
| 38201 | EMX2OS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | CSMD1 | Sponge network | -0.777 | 0.1134 | -0.63 | 0.18881 | 0.311 |
| 38202 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | NF1 | Sponge network | 0.8 | 0 | 0.51 | 0 | 0.311 |
| 38203 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p | 28 | FOXP1 | Sponge network | 0.921 | 0.00118 | 0.042 | 0.74368 | 0.311 |
| 38204 | AC009948.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 23 | SOX11 | Sponge network | 0.532 | 0.00505 | 2.68 | 0 | 0.311 |
| 38205 | FAM157B |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 11 | DDX6 | Sponge network | 0.92 | 0 | 0.091 | 0.36428 | 0.311 |
| 38206 | RP11-83A24.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 12 | DLG1 | Sponge network | 0.417 | 0.00445 | -0.014 | 0.8926 | 0.311 |
| 38207 | RP11-67L2.2 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | INPP4B | Sponge network | 0.72 | 0.0001 | -0.097 | 0.60503 | 0.311 |
| 38208 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ARNT | Sponge network | -0.576 | 0.10121 | -0.08 | 0.26073 | 0.311 |
| 38209 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-654-3p | 17 | ITCH | Sponge network | 0.311 | 0.10344 | 0.084 | 0.34058 | 0.311 |
| 38210 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 22 | SEMA3C | Sponge network | -0.405 | 0.22013 | -0.272 | 0.31153 | 0.311 |
| 38211 | PSMD5-AS1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p | 18 | KIF5B | Sponge network | 0.569 | 0.00067 | 0.362 | 0.00066 | 0.311 |
| 38212 | RP11-536O18.1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-330-3p | 11 | QKI | Sponge network | -0.074 | 0.90758 | -0.333 | 0.04111 | 0.311 |
| 38213 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p | 23 | CREB3L2 | Sponge network | -0.829 | 0.01343 | -0.525 | 2.0E-5 | 0.311 |
| 38214 | CTC-510F12.2 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-3913-5p | 17 | IGFBP5 | Sponge network | -0.691 | 0.00146 | -0.806 | 0.00105 | 0.311 |
| 38215 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-590-5p | 13 | KCNJ5 | Sponge network | 0.898 | 1.0E-5 | -0.281 | 0.3339 | 0.311 |
| 38216 | USP3-AS1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-7-5p | 12 | SH3GLB1 | Sponge network | -0.162 | 0.47687 | -0.115 | 0.14773 | 0.311 |
| 38217 | HOXB-AS3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-339-5p | 12 | PDE4DIP | Sponge network | 0.343 | 0.28887 | -0.282 | 0.0257 | 0.311 |
| 38218 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RCAN2 | Sponge network | -0.141 | 0.26434 | -0.33 | 0.15172 | 0.311 |
| 38219 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-7-1-3p | 12 | TLL1 | Sponge network | -0.246 | 0.35034 | -1.195 | 3.0E-5 | 0.311 |
| 38220 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p | 45 | CBX5 | Sponge network | 0.95 | 0.15943 | 0.165 | 0.24446 | 0.311 |
| 38221 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | FGF5 | Sponge network | 1.302 | 7.0E-5 | 0.549 | 0.28659 | 0.311 |
| 38222 | BVES-AS1 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-628-5p | 14 | IRS1 | Sponge network | -2.62 | 0 | -0.026 | 0.88855 | 0.311 |
| 38223 | HOTAIRM1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | CDH19 | Sponge network | -0.053 | 0.80685 | -3.434 | 0 | 0.311 |
| 38224 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-576-5p | 12 | PCDH17 | Sponge network | 0.176 | 0.37402 | 0.731 | 2.0E-5 | 0.311 |
| 38225 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 11 | CNTNAP3 | Sponge network | 0.199 | 0.38015 | -1.465 | 0.00033 | 0.311 |
| 38226 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-374a-3p | 14 | GREM1 | Sponge network | 0.174 | 0.30034 | 0.902 | 0.01691 | 0.311 |
| 38227 | RP13-516M14.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-940 | 13 | SIRT1 | Sponge network | 0.389 | 0.04293 | 0.109 | 0.2593 | 0.311 |
| 38228 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-miR-128-3p;hsa-miR-151a-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-98-5p | 11 | MPL | Sponge network | -1.216 | 0 | -0.624 | 0.00133 | 0.311 |
| 38229 | HOXB-AS2 |
hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p | 10 | WNK2 | Sponge network | 1.316 | 0.01106 | -0.352 | 0.31066 | 0.311 |
| 38230 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | TMTC3 | Sponge network | 0.702 | 7.0E-5 | 0.123 | 0.22189 | 0.311 |
| 38231 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 15 | ADAMTSL3 | Sponge network | 0.056 | 0.8494 | -1.559 | 0 | 0.311 |
| 38232 | LINC00702 |
hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | SH3GLB1 | Sponge network | -0.737 | 0.06182 | -0.115 | 0.14773 | 0.311 |
| 38233 | CKMT2-AS1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-550a-3p | 12 | RASSF3 | Sponge network | 0.082 | 0.55654 | -0.226 | 0.11691 | 0.311 |
| 38234 | HOTAIRM1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-671-5p | 23 | DLC1 | Sponge network | -0.053 | 0.80685 | -0.382 | 0.05038 | 0.311 |
| 38235 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-577;hsa-miR-625-5p | 22 | CADM1 | Sponge network | -2.69 | 0.0001 | -0.9 | 0.00084 | 0.311 |
| 38236 | RP11-356J5.12 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | TET1 | Sponge network | -0.899 | 6.0E-5 | -0.135 | 0.52138 | 0.311 |
| 38237 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-411-5p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | USP6 | Sponge network | 1.317 | 0 | 0.188 | 0.3871 | 0.311 |
| 38238 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 14 | FOXP1 | Sponge network | 2.094 | 0 | 0.042 | 0.74368 | 0.311 |
| 38239 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-411-5p;hsa-miR-505-3p | 15 | USP6 | Sponge network | 0.453 | 0.01839 | 0.188 | 0.3871 | 0.311 |
| 38240 | RP11-473I1.9 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SH3GLB1 | Sponge network | 0.683 | 1.0E-5 | -0.115 | 0.14773 | 0.311 |
| 38241 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | FREM2 | Sponge network | -0.323 | 0.01372 | -0.437 | 0.46353 | 0.311 |
| 38242 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p | 33 | ST6GALNAC3 | Sponge network | 0.056 | 0.81195 | -0.987 | 0 | 0.311 |
| 38243 | U91328.19 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p | 18 | UBR3 | Sponge network | -0.303 | 0.21002 | -0.021 | 0.82774 | 0.311 |
| 38244 | LPP-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-532-3p | 14 | CDH23 | Sponge network | -0.18 | 0.20343 | -0.974 | 0.00034 | 0.311 |
| 38245 | RP13-516M14.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-7-1-3p | 12 | ITGB1 | Sponge network | 0.389 | 0.04293 | 0.524 | 0.00017 | 0.311 |
| 38246 | RASSF8-AS1 |
hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-590-3p | 10 | DNMT3A | Sponge network | 0.335 | 0.20596 | 0.302 | 0.0262 | 0.311 |
| 38247 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 10 | BNIP3L | Sponge network | -0.612 | 0.00094 | -0.167 | 0.14549 | 0.311 |
| 38248 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 14 | PDZD2 | Sponge network | -0.612 | 0.00094 | -0.909 | 3.0E-5 | 0.311 |
| 38249 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-766-3p | 15 | AFF4 | Sponge network | 1.163 | 0.00018 | -0.266 | 0.01565 | 0.311 |
| 38250 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | SCN9A | Sponge network | 0.683 | 1.0E-5 | -0.525 | 0.10593 | 0.311 |
| 38251 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -1.264 | 6.0E-5 | -3.328 | 0 | 0.311 |
| 38252 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-455-3p;hsa-miR-590-3p | 18 | NRXN1 | Sponge network | 0.051 | 0.83388 | -3.778 | 0 | 0.311 |
| 38253 | RP11-127B20.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 11 | RASSF8 | Sponge network | 0.411 | 0.1335 | -0.122 | 0.65591 | 0.311 |
| 38254 | RP11-6O2.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-539-5p | 16 | PDE4DIP | Sponge network | 0.331 | 0.57247 | -0.282 | 0.0257 | 0.311 |
| 38255 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | MYO9A | Sponge network | -1.886 | 0 | -0.147 | 0.32373 | 0.311 |
| 38256 | AC005562.1 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | INPP4A | Sponge network | 0.488 | 0.00084 | 0.07 | 0.53563 | 0.311 |
| 38257 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-576-5p | 15 | CBL | Sponge network | 2.163 | 0 | 0.291 | 0.01267 | 0.311 |
| 38258 | RP11-254F7.2 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-484 | 11 | WNK2 | Sponge network | 0.176 | 0.37402 | -0.352 | 0.31066 | 0.311 |
| 38259 | RP11-46C24.7 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | UBR3 | Sponge network | 0.392 | 0.0278 | -0.021 | 0.82774 | 0.311 |
| 38260 | FENDRR |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | EYA4 | Sponge network | -1.281 | 0.00012 | 0.3 | 0.36876 | 0.311 |
| 38261 | FAM13A-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 16 | MRVI1 | Sponge network | -0.83 | 0 | -1.051 | 0.00022 | 0.311 |
| 38262 | FAM215B |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-330-5p;hsa-miR-7-1-3p | 14 | NCOA1 | Sponge network | 0.817 | 3.0E-5 | -0.432 | 0 | 0.311 |
| 38263 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p | 12 | PDS5B | Sponge network | 1.206 | 0 | 0.259 | 0.00962 | 0.311 |
| 38264 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-369-3p | 20 | PRDM2 | Sponge network | -0.611 | 0.09607 | -0.258 | 0.00721 | 0.311 |
| 38265 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 37 | PTPRT | Sponge network | -0.769 | 0.00035 | -0.907 | 0.03727 | 0.311 |
| 38266 | SNHG1 |
hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | MDM4 | Sponge network | 1.265 | 0 | 0.345 | 0.00762 | 0.311 |
| 38267 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 34 | ABI2 | Sponge network | -0.793 | 7.0E-5 | 0.175 | 0.14787 | 0.311 |
| 38268 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | TSC1 | Sponge network | -1.575 | 6.0E-5 | 0.103 | 0.26071 | 0.311 |
| 38269 | FAM13A-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -0.83 | 0 | -0.354 | 0.14694 | 0.311 |
| 38270 | RP11-798G7.7 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p | 11 | CYLD | Sponge network | 0.858 | 3.0E-5 | -0.367 | 0.00171 | 0.311 |
| 38271 | RP11-798M19.6 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 10 | PRKACA | Sponge network | -0.612 | 0.00094 | -0.255 | 0.0012 | 0.311 |
| 38272 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | ARID5B | Sponge network | -0.376 | 0.00337 | 0.342 | 0.01676 | 0.311 |
| 38273 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | FAM188A | Sponge network | -2.452 | 0 | -0.44 | 0.00018 | 0.311 |
| 38274 | LA16c-321D4.2 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 13 | UBR3 | Sponge network | 2.459 | 5.0E-5 | -0.021 | 0.82774 | 0.311 |
| 38275 | AC016747.3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p | 18 | EYA4 | Sponge network | 0.199 | 0.38015 | 0.3 | 0.36876 | 0.311 |
| 38276 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | FLI1 | Sponge network | 0.997 | 0.00066 | -0.294 | 0.10905 | 0.311 |
| 38277 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-539-5p;hsa-miR-576-5p | 15 | CDHR3 | Sponge network | 0.41 | 0.02214 | -0.977 | 4.0E-5 | 0.311 |
| 38278 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 47 | FOXP1 | Sponge network | -0.901 | 0.00019 | 0.042 | 0.74368 | 0.311 |
| 38279 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | IGF1 | Sponge network | 0.131 | 0.8436 | -0.204 | 0.5898 | 0.311 |
| 38280 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-98-5p | 17 | ARHGAP28 | Sponge network | -0.18 | 0.20343 | -0.911 | 0.00013 | 0.311 |
| 38281 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-362-3p;hsa-miR-429;hsa-miR-940 | 12 | CRTC3 | Sponge network | 0.389 | 0.04293 | 0.164 | 0.16786 | 0.311 |
| 38282 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 33 | CBL | Sponge network | 0.782 | 0.00016 | 0.291 | 0.01267 | 0.311 |
| 38283 | RP4-669P10.16 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-424-5p | 13 | TACC1 | Sponge network | -0.301 | 0.06597 | -0.362 | 0.05406 | 0.311 |
| 38284 | SNHG14 |
hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-590-3p | 11 | EGR1 | Sponge network | -1.111 | 0.0003 | -1.195 | 0 | 0.311 |
| 38285 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 60 | TCF4 | Sponge network | 0.219 | 0.1516 | 0.016 | 0.92271 | 0.311 |
| 38286 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-502-5p;hsa-miR-590-3p | 11 | TGFBR2 | Sponge network | -0.739 | 0.00044 | -0.243 | 0.11408 | 0.311 |
| 38287 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | TSHZ3 | Sponge network | 0.056 | 0.8494 | -0.556 | 0.01516 | 0.311 |
| 38288 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | LRP1B | Sponge network | 0.101 | 0.60919 | -2.163 | 0 | 0.311 |
| 38289 | AC005618.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PRRX1 | Sponge network | 0.862 | 0.06213 | 1.316 | 0 | 0.311 |
| 38290 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | MAF | Sponge network | -0.351 | 0.11358 | -1.257 | 0 | 0.311 |
| 38291 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-493-5p | 14 | ANK2 | Sponge network | -1.942 | 0 | -1.603 | 1.0E-5 | 0.311 |
| 38292 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-145-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 11 | MOB1B | Sponge network | 1.157 | 0.00455 | 0.197 | 0.15266 | 0.311 |
| 38293 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | HACE1 | Sponge network | -0.405 | 0.22013 | -0.072 | 0.55239 | 0.311 |
| 38294 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 27 | USP12 | Sponge network | -0.901 | 0.00019 | -0.102 | 0.40768 | 0.311 |
| 38295 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-93-5p | 11 | LHX6 | Sponge network | 0.199 | 0.38015 | -0.087 | 0.69193 | 0.311 |
| 38296 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 19 | ACVR1 | Sponge network | 0.246 | 0.59912 | 0.279 | 0.00234 | 0.311 |
| 38297 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 19 | USP12 | Sponge network | -0.421 | 0.0114 | -0.102 | 0.40768 | 0.311 |
| 38298 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p | 17 | CDC73 | Sponge network | 1.294 | 0 | 0.094 | 0.20463 | 0.311 |
| 38299 | DNAJC27-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-629-3p | 23 | RYR2 | Sponge network | -0.969 | 2.0E-5 | -1.466 | 0.00031 | 0.311 |
| 38300 | GNG12-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-628-5p | 29 | ERC1 | Sponge network | -0.793 | 7.0E-5 | -0.108 | 0.38794 | 0.311 |
| 38301 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | EXOC8 | Sponge network | 1.11 | 0 | 0.216 | 0.00598 | 0.311 |
| 38302 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SAMHD1 | Sponge network | 0.572 | 0.01722 | 0.072 | 0.62895 | 0.311 |
| 38303 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 35 | CLASP2 | Sponge network | -0.129 | 0.57177 | -0.038 | 0.71 | 0.311 |
| 38304 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-3p | 19 | FOXO3 | Sponge network | -0.513 | 0.04703 | -0.04 | 0.72028 | 0.311 |
| 38305 | AC159540.1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-326 | 10 | EPS15 | Sponge network | 1.122 | 0 | -0.052 | 0.53998 | 0.311 |
| 38306 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-330-5p;hsa-miR-34a-5p | 10 | ZMYND11 | Sponge network | -2.531 | 0 | -0.2 | 0.05449 | 0.311 |
| 38307 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 16 | NR2C2 | Sponge network | 0.214 | 0.18246 | 0.21 | 0.03224 | 0.311 |
| 38308 | RP11-216B9.6 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p | 17 | PDE4DIP | Sponge network | 0.174 | 0.30034 | -0.282 | 0.0257 | 0.311 |
| 38309 | FGD5-AS1 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-432-5p;hsa-miR-495-3p | 10 | STAG2 | Sponge network | 0.401 | 0.00066 | 0.252 | 0.00438 | 0.311 |
| 38310 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-96-5p | 18 | ZFHX4 | Sponge network | 0.37 | 0.27614 | 0.018 | 0.95574 | 0.311 |
| 38311 | LINC01018 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | FAM133A | Sponge network | -2.915 | 0.00015 | 0.549 | 0.29009 | 0.311 |
| 38312 | AC006129.2 |
hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-940 | 11 | GAS7 | Sponge network | 0.44 | 0.15392 | -0.555 | 0.01602 | 0.311 |
| 38313 | LINC00476 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-340-5p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-935 | 57 | NFIB | Sponge network | -0.615 | 0.00015 | 0.023 | 0.90795 | 0.311 |
| 38314 | FAM157B |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-766-3p | 16 | AFF4 | Sponge network | 0.92 | 0 | -0.266 | 0.01565 | 0.311 |
| 38315 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | ZNF208 | Sponge network | 0.101 | 0.60919 | -0.4 | 0.22381 | 0.311 |
| 38316 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | HECA | Sponge network | -1.161 | 0.00591 | -0.217 | 0.03463 | 0.31 |
| 38317 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484 | 14 | ZFP36L1 | Sponge network | -2.69 | 0.0001 | -0.505 | 8.0E-5 | 0.31 |
| 38318 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | EPAS1 | Sponge network | 0.572 | 0.01722 | -0.184 | 0.14418 | 0.31 |
| 38319 | CTD-2666L21.1 |
hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-423-5p;hsa-miR-455-3p | 12 | ABL1 | Sponge network | 0.368 | 0.20093 | -0.215 | 0.07804 | 0.31 |
| 38320 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 46 | ACVR2A | Sponge network | -2.46 | 0 | -0.409 | 0.00039 | 0.31 |
| 38321 | LINC00657 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 14 | HECA | Sponge network | 0.91 | 0 | -0.217 | 0.03463 | 0.31 |
| 38322 | RP11-890B15.3 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | NR3C2 | Sponge network | 0.101 | 0.60919 | -1.56 | 0 | 0.31 |
| 38323 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | EPHA5 | Sponge network | 0.385 | 0.10067 | -0.957 | 0.01733 | 0.31 |
| 38324 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | KAT6B | Sponge network | 0.349 | 0.01695 | -0.478 | 0.00018 | 0.31 |
| 38325 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | TACC1 | Sponge network | 0.131 | 0.8436 | -0.362 | 0.05406 | 0.31 |
| 38326 | IGF2-AS |
hsa-let-7g-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-3p;hsa-miR-361-3p | 12 | BRD3 | Sponge network | 3.339 | 0.00237 | 0.37 | 0.00013 | 0.31 |
| 38327 | RP11-35G9.5 |
hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-940 | 10 | GLI3 | Sponge network | 0.622 | 0.00015 | -0.615 | 0.01482 | 0.31 |
| 38328 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 16 | ABCB5 | Sponge network | -0.769 | 0.00035 | -0.956 | 0.04077 | 0.31 |
| 38329 | RP11-2B6.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TET1 | Sponge network | 0.539 | 0.03722 | -0.135 | 0.52138 | 0.31 |
| 38330 | RP1-122P22.2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p | 11 | PDGFRA | Sponge network | 0.065 | 0.73468 | -0.372 | 0.0037 | 0.31 |
| 38331 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | CBL | Sponge network | 0.272 | 0.44692 | 0.291 | 0.01267 | 0.31 |
| 38332 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 25 | PCDH10 | Sponge network | 0.083 | 0.66405 | -1.644 | 0.0078 | 0.31 |
| 38333 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | NR2C2 | Sponge network | 0.643 | 0 | 0.21 | 0.03224 | 0.31 |
| 38334 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-7-5p | 11 | PTGFR | Sponge network | 0.419 | 0.0319 | -0.233 | 0.45421 | 0.31 |
| 38335 | PWAR6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | ZFX | Sponge network | -1.575 | 6.0E-5 | 0.09 | 0.42563 | 0.31 |
| 38336 | LINC00648 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 12 | GPAM | Sponge network | 0.633 | 0.51678 | 0.142 | 0.29732 | 0.31 |
| 38337 | RP11-359B12.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p | 62 | DST | Sponge network | 0.064 | 0.72934 | -0.713 | 0.00096 | 0.31 |
| 38338 | RP11-35G9.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | WWTR1 | Sponge network | 0.657 | 4.0E-5 | -0.345 | 0.11997 | 0.31 |
| 38339 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 24 | YAP1 | Sponge network | 0.249 | 0.35902 | 0.22 | 0.11836 | 0.31 |
| 38340 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 30 | USP12 | Sponge network | 0.219 | 0.1516 | -0.102 | 0.40768 | 0.31 |
| 38341 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-505-5p | 10 | RBM5 | Sponge network | 0.056 | 0.8494 | -0.205 | 0.0129 | 0.31 |
| 38342 | LINC01006 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p | 35 | THRB | Sponge network | -0.702 | 0.04814 | -1.117 | 0.0004 | 0.31 |
| 38343 | RP11-6N17.3 |
hsa-miR-103a-2-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-455-5p | 14 | ARHGEF12 | Sponge network | 0.108 | 0.69556 | 0.09 | 0.35693 | 0.31 |
| 38344 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-576-5p | 20 | PRRX1 | Sponge network | 0.176 | 0.37402 | 1.316 | 0 | 0.31 |
| 38345 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-629-3p | 24 | GPC6 | Sponge network | -0.487 | 0.02157 | -0.054 | 0.81465 | 0.31 |
| 38346 | RP11-356J5.12 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 16 | CELF4 | Sponge network | -0.899 | 6.0E-5 | -1.135 | 0.00344 | 0.31 |
| 38347 | NEAT1 |
hsa-let-7a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 0.705 | 0.00014 | 0.114 | 0.30638 | 0.31 |
| 38348 | RP1-102E24.8 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-5p | 12 | PHF6 | Sponge network | 1.737 | 0 | 0.785 | 0 | 0.31 |
| 38349 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-7-1-3p | 14 | TBX18 | Sponge network | 0.614 | 1.0E-5 | 0.843 | 0.02945 | 0.31 |
| 38350 | MRPL23-AS1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-629-3p;hsa-miR-664a-5p | 31 | DST | Sponge network | -0.278 | 0.76559 | -0.713 | 0.00096 | 0.31 |
| 38351 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | ADAMTSL3 | Sponge network | 0.72 | 0.0001 | -1.559 | 0 | 0.31 |
| 38352 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | ARID5B | Sponge network | 1.2 | 0.01813 | 0.342 | 0.01676 | 0.31 |
| 38353 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-330-5p;hsa-miR-424-5p | 11 | ABCB5 | Sponge network | -0.777 | 0.16667 | -0.956 | 0.04077 | 0.31 |
| 38354 | AC141928.1 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | DOCK4 | Sponge network | 0.853 | 0.15352 | 0.502 | 0.00108 | 0.31 |
| 38355 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-654-3p | 15 | AKAP11 | Sponge network | 0.439 | 0.00313 | 0.196 | 0.09825 | 0.31 |
| 38356 | USP3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-93-5p | 41 | GNAQ | Sponge network | -0.162 | 0.47687 | -0.367 | 0.00102 | 0.31 |
| 38357 | LINC00957 |
hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | SLITRK6 | Sponge network | -0.421 | 0.0114 | -2.121 | 0 | 0.31 |
| 38358 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-629-3p;hsa-miR-7-5p | 23 | ZBTB4 | Sponge network | 0.659 | 3.0E-5 | -0.739 | 0 | 0.31 |
| 38359 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 21 | ZMYM2 | Sponge network | 0.415 | 0.14657 | 0.279 | 0.01273 | 0.31 |
| 38360 | RP11-264B14.1 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | MLLT10 | Sponge network | 0.557 | 0.00564 | 0.105 | 0.33292 | 0.31 |
| 38361 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p | 12 | LRRK2 | Sponge network | 0.056 | 0.81195 | -1.136 | 1.0E-5 | 0.31 |
| 38362 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p | 28 | MCC | Sponge network | -0.18 | 0.20343 | -1.005 | 0 | 0.31 |
| 38363 | C4B-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-486-5p | 19 | DTNA | Sponge network | 2.306 | 9.0E-5 | -1.648 | 1.0E-5 | 0.31 |
| 38364 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NEDD4 | Sponge network | -0.053 | 0.80685 | 0.02 | 0.89834 | 0.31 |
| 38365 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-629-5p;hsa-miR-940 | 15 | IGF1 | Sponge network | -4.477 | 0 | -0.204 | 0.5898 | 0.31 |
| 38366 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p | 10 | USP6 | Sponge network | 1.117 | 0 | 0.188 | 0.3871 | 0.31 |
| 38367 | RP11-767N6.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-3607-3p | 10 | USP6 | Sponge network | 0.466 | 0.00155 | 0.188 | 0.3871 | 0.31 |
| 38368 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 27 | NCOA1 | Sponge network | 0.064 | 0.72934 | -0.432 | 0 | 0.31 |
| 38369 | PART1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 19 | ABCB5 | Sponge network | -2.925 | 0 | -0.956 | 0.04077 | 0.31 |
| 38370 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-7-1-3p | 10 | EBF1 | Sponge network | 0.411 | 0.1335 | -0.384 | 0.08856 | 0.31 |
| 38371 | CTD-2528L19.6 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 39 | LPP | Sponge network | 0.953 | 0 | -0.459 | 0.04475 | 0.31 |
| 38372 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p | 25 | PGR | Sponge network | -0.176 | 0.59692 | -1.704 | 0 | 0.31 |
| 38373 | RP11-867G23.10 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-93-5p | 23 | PPM1A | Sponge network | -2.531 | 0 | -0.623 | 0 | 0.31 |
| 38374 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-409-3p | 18 | SCN9A | Sponge network | 0.594 | 0.00064 | -0.525 | 0.10593 | 0.31 |
| 38375 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-671-5p | 24 | DLC1 | Sponge network | 0.61 | 0.01593 | -0.382 | 0.05038 | 0.31 |
| 38376 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 32 | TACC1 | Sponge network | -0.342 | 0.02288 | -0.362 | 0.05406 | 0.31 |
| 38377 | LINC00969 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 16 | RPS6KA3 | Sponge network | 0.683 | 1.0E-5 | 0.256 | 0.02419 | 0.31 |
| 38378 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p | 20 | CADM1 | Sponge network | -0.896 | 0.00099 | -0.9 | 0.00084 | 0.31 |
| 38379 | RP11-554A11.4 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-3p | 17 | PPP1R9A | Sponge network | -0.583 | 0.14589 | -0.903 | 0.04254 | 0.31 |
| 38380 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | MYO9A | Sponge network | 1.61 | 0.00961 | -0.147 | 0.32373 | 0.31 |
| 38381 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 45 | KIF1B | Sponge network | 0.299 | 0.57722 | -0.118 | 0.32258 | 0.31 |
| 38382 | PART1 |
hsa-miR-107;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 11 | RNF103 | Sponge network | -2.925 | 0 | -0.341 | 1.0E-5 | 0.31 |
| 38383 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-7-1-3p | 28 | RBMS3 | Sponge network | 0.853 | 0.15352 | -0.519 | 0.03551 | 0.31 |
| 38384 | RP11-774O3.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-5p | 31 | BTG2 | Sponge network | -0.487 | 0.02157 | -1.763 | 0 | 0.31 |
| 38385 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-374b-3p | 19 | FAT3 | Sponge network | 0.065 | 0.73468 | -1.601 | 0.00051 | 0.31 |
| 38386 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-744-3p | 12 | HIP1 | Sponge network | 0.368 | 0.20093 | 0.774 | 0 | 0.31 |
| 38387 | AGAP11 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-590-5p | 10 | RBM6 | Sponge network | -1.645 | 0 | -0.137 | 0.15663 | 0.31 |
| 38388 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 28 | PBX1 | Sponge network | 0.311 | 0.10344 | -1.206 | 0 | 0.31 |
| 38389 | RP11-25K19.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 16 | STARD13 | Sponge network | -1.057 | 0.00029 | -0.187 | 0.27383 | 0.31 |
| 38390 | AC074289.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p | 14 | ST6GAL2 | Sponge network | -0.326 | 0.14314 | -1.201 | 0.00597 | 0.31 |
| 38391 | RP11-464F9.1 |
hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-655-3p | 14 | TMEM30A | Sponge network | 0.959 | 0 | 0.096 | 0.31031 | 0.31 |
| 38392 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-93-5p;hsa-miR-935 | 27 | HBP1 | Sponge network | -0.162 | 0.47687 | -0.454 | 0 | 0.31 |
| 38393 | PART1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940 | 12 | FOXO4 | Sponge network | -2.925 | 0 | -0.516 | 2.0E-5 | 0.31 |
| 38394 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | NUAK1 | Sponge network | -1.111 | 0.0003 | 0.904 | 0 | 0.31 |
| 38395 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | APC | Sponge network | -1.281 | 0.00012 | -0.255 | 0.04051 | 0.31 |
| 38396 | LINC00667 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-96-5p | 11 | WNK2 | Sponge network | -0.235 | 0.15142 | -0.352 | 0.31066 | 0.31 |
| 38397 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 27 | EPHA5 | Sponge network | 0.419 | 0.0319 | -0.957 | 0.01733 | 0.31 |
| 38398 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | SASH1 | Sponge network | 0.225 | 0.30644 | -0.502 | 0.00175 | 0.31 |
| 38399 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-93-5p | 32 | AR | Sponge network | -0.785 | 0.00035 | -1.554 | 0 | 0.31 |
| 38400 | LINC00957 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-455-5p | 13 | PLXNC1 | Sponge network | -0.421 | 0.0114 | 0.569 | 0.00329 | 0.31 |
| 38401 | RP5-1085F17.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 19 | MED12L | Sponge network | 0.541 | 0.00125 | -0.194 | 0.55299 | 0.31 |
| 38402 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TBX18 | Sponge network | 0.559 | 0.00026 | 0.843 | 0.02945 | 0.31 |
| 38403 | RP11-490M8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-96-5p | 14 | QKI | Sponge network | -0.214 | 0.44248 | -0.333 | 0.04111 | 0.31 |
| 38404 | AGAP11 |
hsa-miR-142-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-629-3p | 12 | ZDBF2 | Sponge network | -1.645 | 0 | -0.998 | 0.00025 | 0.31 |
| 38405 | RP4-639F20.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-502-5p | 11 | FMNL3 | Sponge network | -0.517 | 0.02935 | 0.555 | 4.0E-5 | 0.31 |
| 38406 | MIR600HG |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-369-3p | 13 | NIN | Sponge network | 0.594 | 0.41522 | -0.253 | 0.13794 | 0.31 |
| 38407 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-589-5p;hsa-miR-93-3p | 14 | RPS6KA2 | Sponge network | -0.318 | 0.65847 | -0.57 | 0.00013 | 0.31 |
| 38408 | OSER1-AS1 |
hsa-miR-106a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-93-5p | 13 | FGFR1 | Sponge network | -0.13 | 0.48734 | -0.509 | 0.03957 | 0.31 |
| 38409 | RP11-1277A3.1 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-34a-5p | 17 | ANK2 | Sponge network | 0.453 | 0.01839 | -1.603 | 1.0E-5 | 0.31 |
| 38410 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 24 | LATS1 | Sponge network | 0.848 | 0 | 0.109 | 0.34208 | 0.31 |
| 38411 | SNHG12 |
hsa-miR-125b-2-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-9-5p | 11 | PAWR | Sponge network | 1.103 | 0 | 0.636 | 0 | 0.31 |
| 38412 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 21 | PTPN11 | Sponge network | 0.572 | 0.01722 | 0.431 | 2.0E-5 | 0.31 |
| 38413 | RP11-999E24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-362-3p | 17 | PRDM2 | Sponge network | -0.693 | 0.05629 | -0.258 | 0.00721 | 0.31 |
| 38414 | LINC00654 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 14 | SORCS1 | Sponge network | 0.374 | 0.20054 | -3.492 | 0 | 0.31 |
| 38415 | TMEM51-AS1 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 23 | KIF1B | Sponge network | 0.23 | 0.38667 | -0.118 | 0.32258 | 0.31 |
| 38416 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | AKAP11 | Sponge network | 0.705 | 0.00014 | 0.196 | 0.09825 | 0.31 |
| 38417 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | AFF4 | Sponge network | 1.601 | 0 | -0.266 | 0.01565 | 0.31 |
| 38418 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-93-3p | 27 | SNX29 | Sponge network | -0.384 | 0.40652 | -0.34 | 0.01371 | 0.31 |
| 38419 | RP4-584D14.7 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-577 | 11 | SCN9A | Sponge network | 1.294 | 0.0031 | -0.525 | 0.10593 | 0.31 |
| 38420 | CKMT2-AS1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 11 | SHPRH | Sponge network | 0.082 | 0.55654 | 0.098 | 0.40994 | 0.31 |
| 38421 | A2M-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p | 13 | CADM3 | Sponge network | -0.08 | 0.90092 | -3.663 | 0 | 0.31 |
| 38422 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 16 | ARNT2 | Sponge network | -0.896 | 0.00099 | -0.332 | 0.25851 | 0.31 |
| 38423 | FENDRR |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 20 | UBE4B | Sponge network | -1.281 | 0.00012 | -0.235 | 0.007 | 0.31 |
| 38424 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-539-5p | 14 | NEDD4 | Sponge network | 0.594 | 0.00064 | 0.02 | 0.89834 | 0.31 |
| 38425 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p | 14 | SMARCA2 | Sponge network | -0.227 | 0.31657 | -0.529 | 0.00014 | 0.31 |
| 38426 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p | 15 | CUL5 | Sponge network | 0.637 | 0.00069 | 0.026 | 0.77301 | 0.31 |
| 38427 | RP5-1139B12.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-671-5p | 22 | ADARB1 | Sponge network | 0.598 | 0.38481 | -0.335 | 0.06631 | 0.31 |
| 38428 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3614-5p;hsa-miR-424-5p | 18 | ABL2 | Sponge network | 0.529 | 0.00552 | 0.82 | 0 | 0.31 |
| 38429 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 36 | PPM1A | Sponge network | 0.537 | 0.00139 | -0.623 | 0 | 0.31 |
| 38430 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | ITCH | Sponge network | 0.953 | 0 | 0.084 | 0.34058 | 0.31 |
| 38431 | GABPB1-AS1 |
hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | DOCK4 | Sponge network | 1.11 | 0 | 0.502 | 0.00108 | 0.31 |
| 38432 | RP11-25K19.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 14 | LUZP2 | Sponge network | -1.057 | 0.00029 | -0.056 | 0.89416 | 0.31 |
| 38433 | LINC01018 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | ABI2 | Sponge network | -2.915 | 0.00015 | 0.175 | 0.14787 | 0.31 |
| 38434 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | LATS1 | Sponge network | -0.323 | 0.01372 | 0.109 | 0.34208 | 0.31 |
| 38435 | C4B-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 10 | UNC5C | Sponge network | 2.306 | 9.0E-5 | -0.178 | 0.40214 | 0.31 |
| 38436 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-576-5p | 16 | DMXL1 | Sponge network | -0.176 | 0.59692 | -0.426 | 0.00056 | 0.31 |
| 38437 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | TACC1 | Sponge network | -0.08 | 0.90092 | -0.362 | 0.05406 | 0.31 |
| 38438 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | LRP1B | Sponge network | -0.318 | 0.65847 | -2.163 | 0 | 0.31 |
| 38439 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-671-5p | 29 | FOXP1 | Sponge network | -0.154 | 0.53954 | 0.042 | 0.74368 | 0.31 |
| 38440 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | SYNE1 | Sponge network | -0.303 | 0.21002 | -0.738 | 0.00316 | 0.31 |
| 38441 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | SASH1 | Sponge network | 0.572 | 0.01722 | -0.502 | 0.00175 | 0.31 |
| 38442 | A2M-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 32 | ABI2 | Sponge network | -0.08 | 0.90092 | 0.175 | 0.14787 | 0.31 |
| 38443 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-577 | 11 | AMPH | Sponge network | 0.064 | 0.72934 | -0.916 | 5.0E-5 | 0.31 |
| 38444 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 14 | GNA13 | Sponge network | 0.817 | 3.0E-5 | 0.007 | 0.93884 | 0.31 |
| 38445 | LINC00476 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p | 20 | ROBO1 | Sponge network | -0.615 | 0.00015 | -0.009 | 0.96154 | 0.31 |
| 38446 | H19 |
hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-625-5p | 12 | YAP1 | Sponge network | 2.439 | 0 | 0.22 | 0.11836 | 0.31 |
| 38447 | PART1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 17 | ITGA5 | Sponge network | -2.925 | 0 | 0.452 | 0.03524 | 0.31 |
| 38448 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-3p;hsa-miR-590-3p | 23 | ABL2 | Sponge network | 0.082 | 0.55654 | 0.82 | 0 | 0.31 |
| 38449 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | HECA | Sponge network | -0.223 | 0.47816 | -0.217 | 0.03463 | 0.31 |
| 38450 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 32 | SNX29 | Sponge network | -0.18 | 0.20343 | -0.34 | 0.01371 | 0.31 |
| 38451 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | GALNT13 | Sponge network | 0.219 | 0.1516 | -0.857 | 0.07439 | 0.31 |
| 38452 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 40 | MOB1B | Sponge network | -2.46 | 0 | 0.197 | 0.15266 | 0.31 |
| 38453 | TPTEP1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-671-5p | 11 | PIK3IP1 | Sponge network | -1.216 | 0 | -0.187 | 0.19945 | 0.31 |
| 38454 | LPP-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-5p | 14 | CDH19 | Sponge network | -0.18 | 0.20343 | -3.434 | 0 | 0.31 |
| 38455 | RP11-283I3.6 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-7-1-3p | 26 | AFF1 | Sponge network | 0.614 | 1.0E-5 | -0.384 | 0.00053 | 0.31 |
| 38456 | AC141928.1 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | NUAK1 | Sponge network | 0.853 | 0.15352 | 0.904 | 0 | 0.31 |
| 38457 | LINC00476 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-942-5p | 35 | IGFBP5 | Sponge network | -0.615 | 0.00015 | -0.806 | 0.00105 | 0.31 |
| 38458 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-361-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PAK3 | Sponge network | -0.612 | 0.00094 | -0.628 | 0.07253 | 0.31 |
| 38459 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-590-3p | 34 | EPHA5 | Sponge network | 0.494 | 0.00339 | -0.957 | 0.01733 | 0.31 |
| 38460 | RP11-182J1.1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-24-3p;hsa-miR-484;hsa-miR-7-1-3p | 10 | RIMS2 | Sponge network | -0.187 | 0.23787 | -1.365 | 0.00208 | 0.31 |
| 38461 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-493-5p;hsa-miR-582-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | SRGAP3 | Sponge network | -0.31 | 0.33871 | -0.202 | 0.29297 | 0.31 |
| 38462 | AF131215.9 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | ABCA10 | Sponge network | -0.322 | 0.25945 | -0.571 | 0.03674 | 0.31 |
| 38463 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-511-5p | 12 | EYA4 | Sponge network | 0.497 | 0.01451 | 0.3 | 0.36876 | 0.31 |
| 38464 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p | 14 | SMARCA2 | Sponge network | -0.793 | 7.0E-5 | -0.529 | 0.00014 | 0.31 |
| 38465 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | EPHA5 | Sponge network | -0.563 | 0.02545 | -0.957 | 0.01733 | 0.31 |
| 38466 | RP11-298J20.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-671-5p | 13 | ADARB1 | Sponge network | 0.002 | 0.99057 | -0.335 | 0.06631 | 0.31 |
| 38467 | FZD10-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 25 | DLC1 | Sponge network | -0.727 | 0.04726 | -0.382 | 0.05038 | 0.31 |
| 38468 | RP11-59H7.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-576-5p | 11 | TSC1 | Sponge network | 2.163 | 0 | 0.103 | 0.26071 | 0.31 |
| 38469 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | CBFA2T3 | Sponge network | -0.096 | 0.67958 | -1.221 | 1.0E-5 | 0.31 |
| 38470 | AC009120.6 |
hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | PIK3R1 | Sponge network | 0.919 | 0 | -0.133 | 0.36854 | 0.31 |
| 38471 | LINC00702 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 24 | SUFU | Sponge network | -0.737 | 0.06182 | -0.04 | 0.65489 | 0.31 |
| 38472 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-874-3p | 17 | CRTC3 | Sponge network | 0.401 | 0.00066 | 0.164 | 0.16786 | 0.31 |
| 38473 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | YAP1 | Sponge network | 0.683 | 1.0E-5 | 0.22 | 0.11836 | 0.31 |
| 38474 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | EBF1 | Sponge network | 0.214 | 0.18246 | -0.384 | 0.08856 | 0.31 |
| 38475 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 17 | MOB1B | Sponge network | 0.373 | 0.00068 | 0.197 | 0.15266 | 0.31 |
| 38476 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PRKAB2 | Sponge network | 0.673 | 0 | -0.367 | 0.01371 | 0.31 |
| 38477 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MYO9A | Sponge network | 0.67 | 0.00048 | -0.147 | 0.32373 | 0.31 |
| 38478 | SERTAD4-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 23 | PTPRD | Sponge network | -0.932 | 0.00329 | -1.005 | 0.00064 | 0.31 |
| 38479 | CTA-363E19.2 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-382-5p;hsa-miR-485-3p | 13 | DDX3X | Sponge network | 1.055 | 0 | -0.152 | 0.07686 | 0.31 |
| 38480 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-7-1-3p | 21 | MED12L | Sponge network | 0.299 | 0.57722 | -0.194 | 0.55299 | 0.31 |
| 38481 | WDFY3-AS2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 14 | PIK3IP1 | Sponge network | -0.942 | 0 | -0.187 | 0.19945 | 0.31 |
| 38482 | RP11-473I1.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 12 | LATS2 | Sponge network | 0.542 | 0.00054 | 0.159 | 0.2304 | 0.31 |
| 38483 | BZRAP1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-877-5p | 28 | AFF1 | Sponge network | -0.465 | 0.04703 | -0.384 | 0.00053 | 0.31 |
| 38484 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | NIN | Sponge network | -0.769 | 0.00035 | -0.253 | 0.13794 | 0.31 |
| 38485 | AC141928.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 35 | TRPS1 | Sponge network | 0.853 | 0.15352 | -0.191 | 0.44109 | 0.31 |
| 38486 | AC002117.1 |
hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | LPP | Sponge network | 0.476 | 0.19203 | -0.459 | 0.04475 | 0.31 |
| 38487 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-589-3p | 34 | PDGFRA | Sponge network | -0.773 | 0.04073 | -0.372 | 0.0037 | 0.31 |
| 38488 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-223-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | CXXC4 | Sponge network | 0.799 | 1.0E-5 | -0.433 | 0.21055 | 0.31 |
| 38489 | TTC25 |
hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-5p | 10 | BNC2 | Sponge network | -0.208 | 0.38261 | -0.491 | 0.09988 | 0.31 |
| 38490 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | IGSF10 | Sponge network | 0.349 | 0.01695 | -1.751 | 1.0E-5 | 0.31 |
| 38491 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-576-5p | 19 | CDH19 | Sponge network | 0.056 | 0.81195 | -3.434 | 0 | 0.31 |
| 38492 | A2M-AS1 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | IRS1 | Sponge network | -0.08 | 0.90092 | -0.026 | 0.88855 | 0.31 |
| 38493 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 43 | NTRK3 | Sponge network | 0.056 | 0.8494 | -1.77 | 3.0E-5 | 0.31 |
| 38494 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 17 | HIP1 | Sponge network | 0.537 | 0.00139 | 0.774 | 0 | 0.31 |
| 38495 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 25 | PRDM2 | Sponge network | 0.064 | 0.72934 | -0.258 | 0.00721 | 0.31 |
| 38496 | RP11-531A24.5 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | WWTR1 | Sponge network | 0.75 | 0.00054 | -0.345 | 0.11997 | 0.31 |
| 38497 | NUTM2A-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 11 | KIAA1024 | Sponge network | 0.643 | 0 | 1.083 | 0 | 0.31 |
| 38498 | RP11-33B1.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-342-3p | 10 | ZFX | Sponge network | 0.826 | 0 | 0.09 | 0.42563 | 0.31 |
| 38499 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 21 | ATRX | Sponge network | -0.322 | 0.25945 | 0.261 | 0.04408 | 0.31 |
| 38500 | RP11-98I9.4 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TSC1 | Sponge network | 0.67 | 0.00048 | 0.103 | 0.26071 | 0.31 |
| 38501 | LINC00657 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p | 12 | PAWR | Sponge network | 0.91 | 0 | 0.636 | 0 | 0.31 |
| 38502 | AC108488.4 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-664a-5p | 14 | SOX11 | Sponge network | 0.259 | 0.12542 | 2.68 | 0 | 0.31 |
| 38503 | RP11-403I13.8 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | ANK2 | Sponge network | 0.619 | 0.00157 | -1.603 | 1.0E-5 | 0.31 |
| 38504 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 40 | CBL | Sponge network | -0.739 | 0.0574 | 0.291 | 0.01267 | 0.31 |
| 38505 | RP4-613B23.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-539-5p | 13 | ETV1 | Sponge network | -0.309 | 0.1145 | -0.096 | 0.67674 | 0.31 |
| 38506 | MIR22HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-590-5p | 13 | ABCA10 | Sponge network | -1.047 | 0 | -0.571 | 0.03674 | 0.31 |
| 38507 | RP11-195F19.9 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 10 | CDH19 | Sponge network | -0.726 | 0.00132 | -3.434 | 0 | 0.31 |
| 38508 | FAM13A-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ZMYND11 | Sponge network | -0.83 | 0 | -0.2 | 0.05449 | 0.31 |
| 38509 | RP11-999E24.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p | 13 | ARNT2 | Sponge network | -0.693 | 0.05629 | -0.332 | 0.25851 | 0.31 |
| 38510 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p | 25 | PTPRT | Sponge network | 0.176 | 0.37402 | -0.907 | 0.03727 | 0.31 |
| 38511 | DOCK9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-375 | 16 | TACC1 | Sponge network | -0.231 | 0.28214 | -0.362 | 0.05406 | 0.31 |
| 38512 | SERHL |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-214-5p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421 | 14 | MXI1 | Sponge network | -0.602 | 0.00331 | -0.987 | 0 | 0.31 |
| 38513 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 29 | ERBB4 | Sponge network | -1.092 | 4.0E-5 | -1.975 | 2.0E-5 | 0.31 |
| 38514 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-3p;hsa-miR-339-5p | 15 | MYBL1 | Sponge network | 0.817 | 3.0E-5 | 0.494 | 0.02152 | 0.31 |
| 38515 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-93-5p;hsa-miR-96-5p | 14 | DPYD | Sponge network | -0.976 | 0.00025 | -0.736 | 0.00076 | 0.31 |
| 38516 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | MED12L | Sponge network | 0.395 | 0.15451 | -0.194 | 0.55299 | 0.31 |
| 38517 | SNHG14 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 40 | ZFX | Sponge network | -1.111 | 0.0003 | 0.09 | 0.42563 | 0.31 |
| 38518 | CTA-29F11.1 |
hsa-let-7a-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-628-5p | 10 | CDC73 | Sponge network | 0.583 | 0.00027 | 0.094 | 0.20463 | 0.31 |
| 38519 | LINC00476 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | ERG | Sponge network | -0.615 | 0.00015 | 0.002 | 0.99011 | 0.31 |
| 38520 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 0.603 | 0.00127 | 0.259 | 0.00962 | 0.31 |
| 38521 | CTB-179K24.3 |
hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 11 | ABL2 | Sponge network | 0.841 | 0.04055 | 0.82 | 0 | 0.31 |
| 38522 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 43 | CUX1 | Sponge network | -0.323 | 0.01372 | -0.039 | 0.6705 | 0.31 |
| 38523 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-628-5p | 31 | TMEM132B | Sponge network | -0.154 | 0.53954 | -0.83 | 0.01084 | 0.31 |
| 38524 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-7-5p | 13 | FNBP1 | Sponge network | 0.253 | 0.09565 | -0.969 | 4.0E-5 | 0.31 |
| 38525 | FGD5-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-500a-5p | 14 | UBE4B | Sponge network | 0.401 | 0.00066 | -0.235 | 0.007 | 0.31 |
| 38526 | LINC00854 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-654-3p | 10 | DICER1 | Sponge network | 1.18 | 0 | 0.042 | 0.674 | 0.31 |
| 38527 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-590-3p | 10 | RBM5 | Sponge network | -0.612 | 0.00094 | -0.205 | 0.0129 | 0.31 |
| 38528 | LINC01004 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p | 11 | PIK3C2A | Sponge network | 1.115 | 0 | 0.061 | 0.53776 | 0.31 |
| 38529 | PWAR6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-940 | 26 | TOM1L2 | Sponge network | -1.575 | 6.0E-5 | -0.994 | 0 | 0.31 |
| 38530 | THAP9-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-664a-3p;hsa-miR-9-5p | 23 | CCDC6 | Sponge network | 1.079 | 0 | 0.27 | 0.02555 | 0.31 |
| 38531 | BCYRN1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-940 | 14 | TAF1 | Sponge network | 0.311 | 0.10344 | 0.39 | 3.0E-5 | 0.31 |
| 38532 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-3065-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-7-1-3p | 23 | CBL | Sponge network | 0.299 | 0.57722 | 0.291 | 0.01267 | 0.31 |
| 38533 | RP11-244O19.1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 11 | PIK3IP1 | Sponge network | -0.096 | 0.67958 | -0.187 | 0.19945 | 0.31 |
| 38534 | LINC00672 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-590-3p | 30 | AFF1 | Sponge network | -0.227 | 0.31657 | -0.384 | 0.00053 | 0.31 |
| 38535 | LINC00865 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p | 20 | CSMD1 | Sponge network | -1.249 | 7.0E-5 | -0.63 | 0.18881 | 0.31 |
| 38536 | RP11-517B11.7 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-495-3p | 22 | CREB1 | Sponge network | 0.706 | 0 | 0.203 | 0.00537 | 0.31 |
| 38537 | LINC00957 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-423-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 13 | CELF4 | Sponge network | -0.421 | 0.0114 | -1.135 | 0.00344 | 0.31 |
| 38538 | PWAR6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 22 | HIP1 | Sponge network | -1.575 | 6.0E-5 | 0.774 | 0 | 0.31 |
| 38539 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | ARNT2 | Sponge network | -0.323 | 0.01372 | -0.332 | 0.25851 | 0.31 |
| 38540 | RP11-67L2.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 17 | SUFU | Sponge network | 0.72 | 0.0001 | -0.04 | 0.65489 | 0.31 |
| 38541 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-539-5p | 43 | FAT3 | Sponge network | 0.177 | 0.16681 | -1.601 | 0.00051 | 0.31 |
| 38542 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p | 17 | CNTNAP3 | Sponge network | 0.064 | 0.72934 | -1.465 | 0.00033 | 0.31 |
| 38543 | AC010226.4 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 14 | CTIF | Sponge network | -0.811 | 2.0E-5 | -0.389 | 0.00549 | 0.31 |
| 38544 | DNM1P35 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p | 14 | RECK | Sponge network | 0.051 | 0.83388 | -1.043 | 0 | 0.31 |
| 38545 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 22 | TSC1 | Sponge network | 0.41 | 0.02214 | 0.103 | 0.26071 | 0.31 |
| 38546 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-628-5p | 18 | BCL6 | Sponge network | 0.4 | 0.14611 | -0.211 | 0.19489 | 0.31 |
| 38547 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-331-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 18 | ABI2 | Sponge network | 0.898 | 1.0E-5 | 0.175 | 0.14787 | 0.31 |
| 38548 | BAIAP2-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c | 13 | ABL2 | Sponge network | 0.702 | 6.0E-5 | 0.82 | 0 | 0.31 |
| 38549 | RP11-102N12.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-7-1-3p | 14 | SYT4 | Sponge network | -0.351 | 0.11358 | -3.207 | 0 | 0.31 |
| 38550 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p | 10 | DAB2 | Sponge network | 0.657 | 4.0E-5 | 0.431 | 0.0113 | 0.31 |
| 38551 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | RB1CC1 | Sponge network | 0.538 | 0.00076 | 0.175 | 0.12559 | 0.31 |
| 38552 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 36 | EPHA5 | Sponge network | -0.031 | 0.89063 | -0.957 | 0.01733 | 0.31 |
| 38553 | RP11-196G11.4 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 10 | UBR3 | Sponge network | 0.875 | 0 | -0.021 | 0.82774 | 0.31 |
| 38554 | C4B-AS1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-629-3p | 21 | EPHA7 | Sponge network | 2.306 | 9.0E-5 | -2.197 | 3.0E-5 | 0.31 |
| 38555 | HOTAIRM1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | BRD3 | Sponge network | -0.053 | 0.80685 | 0.37 | 0.00013 | 0.31 |
| 38556 | RP11-61L19.3 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-497-5p | 12 | TBL1XR1 | Sponge network | 1.411 | 0 | 0.578 | 0 | 0.31 |
| 38557 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 21 | LRRC7 | Sponge network | 0.494 | 0.00339 | -1.341 | 0.01193 | 0.31 |
| 38558 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-744-3p | 11 | HIP1 | Sponge network | 0.792 | 0.0049 | 0.774 | 0 | 0.31 |
| 38559 | LINC00886 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-877-5p;hsa-miR-93-5p | 27 | PGR | Sponge network | -0.371 | 0.24604 | -1.704 | 0 | 0.31 |
| 38560 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 46 | IL6ST | Sponge network | -0.318 | 0.65847 | -0.402 | 0.00676 | 0.31 |
| 38561 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | AKAP11 | Sponge network | 1.153 | 0.00114 | 0.196 | 0.09825 | 0.31 |
| 38562 | RP11-13K12.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484 | 18 | RET | Sponge network | -0.154 | 0.78939 | -0.833 | 0.03517 | 0.31 |
| 38563 | LINC00476 |
hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-653-5p | 28 | UBR3 | Sponge network | -0.615 | 0.00015 | -0.021 | 0.82774 | 0.31 |
| 38564 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SLC6A15 | Sponge network | 0.385 | 0.10067 | -2.373 | 2.0E-5 | 0.31 |
| 38565 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429 | 11 | TSC22D1 | Sponge network | -0.976 | 0.00025 | -0.529 | 2.0E-5 | 0.31 |
| 38566 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-577 | 10 | NALCN | Sponge network | 0.853 | 0.15352 | 1.068 | 0.00237 | 0.31 |
| 38567 | U91328.19 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | SH3GLB1 | Sponge network | -0.303 | 0.21002 | -0.115 | 0.14773 | 0.309 |
| 38568 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-590-5p | 15 | MLLT10 | Sponge network | -0.567 | 0 | 0.105 | 0.33292 | 0.309 |
| 38569 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | SNX29 | Sponge network | 0.673 | 0 | -0.34 | 0.01371 | 0.309 |
| 38570 | RP11-855A2.5 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-93-3p | 10 | TRPS1 | Sponge network | -1.717 | 0.01522 | -0.191 | 0.44109 | 0.309 |
| 38571 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3E | Sponge network | -0.355 | 0.0077 | -1.717 | 0.00137 | 0.309 |
| 38572 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 16 | CSRNP1 | Sponge network | -0.576 | 0.10121 | -1.448 | 0 | 0.309 |
| 38573 | RP11-760H22.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-92a-1-5p | 12 | AKAP6 | Sponge network | 0.001 | 0.99753 | -1.586 | 3.0E-5 | 0.309 |
| 38574 | GABPB1-AS1 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | MAP3K4 | Sponge network | 1.11 | 0 | 0.058 | 0.54229 | 0.309 |
| 38575 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-590-3p | 16 | TGFBR2 | Sponge network | -0.487 | 0.02157 | -0.243 | 0.11408 | 0.309 |
| 38576 | RP4-791M13.3 |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-7-1-3p | 10 | MLLT10 | Sponge network | 0.419 | 0.0319 | 0.105 | 0.33292 | 0.309 |
| 38577 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-5p | 24 | CBL | Sponge network | 0.569 | 0.00067 | 0.291 | 0.01267 | 0.309 |
| 38578 | LINC00654 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 17 | EZH1 | Sponge network | 0.374 | 0.20054 | -0.564 | 1.0E-5 | 0.309 |
| 38579 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 27 | CBL | Sponge network | -0.176 | 0.59692 | 0.291 | 0.01267 | 0.309 |
| 38580 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-411-5p;hsa-miR-493-5p;hsa-miR-590-3p | 22 | USP6 | Sponge network | -0.615 | 0.00015 | 0.188 | 0.3871 | 0.309 |
| 38581 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | EPHA5 | Sponge network | 0.259 | 0.12542 | -0.957 | 0.01733 | 0.309 |
| 38582 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-96-5p | 12 | CSRNP1 | Sponge network | -1.654 | 0.00144 | -1.448 | 0 | 0.309 |
| 38583 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CHD2 | Sponge network | 0.72 | 0.0001 | 0.049 | 0.55371 | 0.309 |
| 38584 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | USP6 | Sponge network | -0.342 | 0.02288 | 0.188 | 0.3871 | 0.309 |
| 38585 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | MCC | Sponge network | -0.141 | 0.26434 | -1.005 | 0 | 0.309 |
| 38586 | AP001347.6 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-590-3p | 11 | PAK3 | Sponge network | -1.161 | 0.00591 | -0.628 | 0.07253 | 0.309 |
| 38587 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | BCL2 | Sponge network | 0.614 | 1.0E-5 | -0.899 | 0 | 0.309 |
| 38588 | FENDRR |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-98-5p | 14 | TES | Sponge network | -1.281 | 0.00012 | -0.348 | 0.00639 | 0.309 |
| 38589 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-5p | 10 | FAM133A | Sponge network | -0.969 | 2.0E-5 | 0.549 | 0.29009 | 0.309 |
| 38590 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DACH2 | Sponge network | -0.727 | 0.04726 | -1.635 | 0.00034 | 0.309 |
| 38591 | CKMT2-AS1 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 15 | IRS1 | Sponge network | 0.082 | 0.55654 | -0.026 | 0.88855 | 0.309 |
| 38592 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 17 | CHD1 | Sponge network | 1.11 | 0 | 0.322 | 0.00215 | 0.309 |
| 38593 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-589-3p | 23 | PRDM2 | Sponge network | 0.331 | 0.57247 | -0.258 | 0.00721 | 0.309 |
| 38594 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-940 | 30 | MCC | Sponge network | 0.331 | 0.57247 | -1.005 | 0 | 0.309 |
| 38595 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940 | 36 | HLF | Sponge network | 0.374 | 0.20054 | -2.443 | 0 | 0.309 |
| 38596 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-655-3p | 27 | LRP1B | Sponge network | 0.214 | 0.18246 | -2.163 | 0 | 0.309 |
| 38597 | RP11-452L6.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-484 | 15 | CBX5 | Sponge network | 0.665 | 0.00061 | 0.165 | 0.24446 | 0.309 |
| 38598 | CTD-2184D3.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SETBP1 | Sponge network | 0.272 | 0.44692 | -1.302 | 1.0E-5 | 0.309 |
| 38599 | RP11-48B3.4 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 14 | CSMD1 | Sponge network | 1.757 | 0 | -0.63 | 0.18881 | 0.309 |
| 38600 | FZD10-AS1 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | RIMS2 | Sponge network | -0.727 | 0.04726 | -1.365 | 0.00208 | 0.309 |
| 38601 | RP11-212P7.2 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 18 | ATM | Sponge network | 0.219 | 0.1516 | 0.178 | 0.21568 | 0.309 |
| 38602 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | ZNF292 | Sponge network | 0.392 | 0.0278 | 0.276 | 0.04148 | 0.309 |
| 38603 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 19 | SOX5 | Sponge network | 0.368 | 0.20093 | -1.091 | 4.0E-5 | 0.309 |
| 38604 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-582-3p | 12 | NAV3 | Sponge network | 0.858 | 3.0E-5 | 0.212 | 0.47729 | 0.309 |
| 38605 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-93-5p | 33 | TCF4 | Sponge network | 0.381 | 0.25967 | 0.016 | 0.92271 | 0.309 |
| 38606 | RP11-373D23.3 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p | 12 | RPS6KA6 | Sponge network | -0.285 | 0.2172 | -2.989 | 0 | 0.309 |
| 38607 | CTD-2245F17.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-940 | 14 | TRPS1 | Sponge network | -0.421 | 0.12556 | -0.191 | 0.44109 | 0.309 |
| 38608 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-484 | 10 | ARNT2 | Sponge network | -0.187 | 0.23787 | -0.332 | 0.25851 | 0.309 |
| 38609 | BZRAP1-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-940 | 10 | FAM172A | Sponge network | -0.465 | 0.04703 | -0.57 | 0 | 0.309 |
| 38610 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-495-3p | 10 | SLC5A3 | Sponge network | 0.993 | 0 | 0.493 | 5.0E-5 | 0.309 |
| 38611 | TP73-AS1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 16 | CSRNP1 | Sponge network | -0.563 | 0.02545 | -1.448 | 0 | 0.309 |
| 38612 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p | 11 | PCDH8 | Sponge network | -0.096 | 0.67958 | -0.997 | 0.05366 | 0.309 |
| 38613 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-93-5p | 13 | MAP3K12 | Sponge network | -1.349 | 0.00017 | -0.057 | 0.59369 | 0.309 |
| 38614 | AC005562.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p | 18 | TBL1XR1 | Sponge network | 0.488 | 0.00084 | 0.578 | 0 | 0.309 |
| 38615 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | -0.08 | 0.90092 | -0.7 | 1.0E-5 | 0.309 |
| 38616 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | EPHA3 | Sponge network | 0.177 | 0.16681 | 0.185 | 0.53588 | 0.309 |
| 38617 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p | 26 | SSBP2 | Sponge network | 0.178 | 0.29106 | -1.58 | 0 | 0.309 |
| 38618 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 37 | MEF2C | Sponge network | 1.302 | 7.0E-5 | -0.499 | 0.01448 | 0.309 |
| 38619 | RP11-597D13.9 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 50 | TRPS1 | Sponge network | 1.302 | 7.0E-5 | -0.191 | 0.44109 | 0.309 |
| 38620 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p | 17 | LRRC4 | Sponge network | -0.222 | 0.32445 | -1.774 | 0 | 0.309 |
| 38621 | RP11-21A7A.2 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-942-5p | 19 | IGFBP5 | Sponge network | -1.116 | 0.23686 | -0.806 | 0.00105 | 0.309 |
| 38622 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 18 | LRRC7 | Sponge network | -0.246 | 0.35034 | -1.341 | 0.01193 | 0.309 |
| 38623 | RP11-473I1.9 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RBBP6 | Sponge network | 0.683 | 1.0E-5 | 0.163 | 0.05101 | 0.309 |
| 38624 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | EBF1 | Sponge network | 0.064 | 0.72934 | -0.384 | 0.08856 | 0.309 |
| 38625 | RP11-35G9.5 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-589-3p | 16 | KCNA6 | Sponge network | 0.622 | 0.00015 | -1.531 | 0.00013 | 0.309 |
| 38626 | CROCCP2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-378a-5p | 19 | CREB1 | Sponge network | 0.79 | 0 | 0.203 | 0.00537 | 0.309 |
| 38627 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | CACNA2D1 | Sponge network | 0.614 | 1.0E-5 | -0.846 | 0.00675 | 0.309 |
| 38628 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 57 | AFF4 | Sponge network | 0.782 | 0.00016 | -0.266 | 0.01565 | 0.309 |
| 38629 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-935 | 46 | SSBP2 | Sponge network | -0.303 | 0.21002 | -1.58 | 0 | 0.309 |
| 38630 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-629-3p | 13 | RBL2 | Sponge network | 0.331 | 0.57247 | -0.282 | 0.00254 | 0.309 |
| 38631 | SNHG14 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 53 | GNAQ | Sponge network | -1.111 | 0.0003 | -0.367 | 0.00102 | 0.309 |
| 38632 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 31 | IGF1 | Sponge network | 0.494 | 0.00339 | -0.204 | 0.5898 | 0.309 |
| 38633 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-455-5p | 11 | THSD7A | Sponge network | -0.465 | 0.04703 | 0.242 | 0.25154 | 0.309 |
| 38634 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-671-5p;hsa-miR-93-5p | 47 | ADARB1 | Sponge network | -0.969 | 2.0E-5 | -0.335 | 0.06631 | 0.309 |
| 38635 | LIPE-AS1 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-654-3p | 17 | DICER1 | Sponge network | 0.078 | 0.55803 | 0.042 | 0.674 | 0.309 |
| 38636 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | ACVR2A | Sponge network | -0.376 | 0.00337 | -0.409 | 0.00039 | 0.309 |
| 38637 | HAND2-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZNF844 | Sponge network | -1.697 | 0.00889 | -0.966 | 0.00035 | 0.309 |
| 38638 | RP11-53O19.1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-629-5p | 14 | CRTC1 | Sponge network | -0.405 | 0.22013 | -0.116 | 0.28412 | 0.309 |
| 38639 | FGF14-AS2 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-590-3p | 14 | FAM135B | Sponge network | -1.582 | 8.0E-5 | -3.978 | 0 | 0.309 |
| 38640 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p | 12 | ABCB5 | Sponge network | -0.738 | 0.21233 | -0.956 | 0.04077 | 0.309 |
| 38641 | RP11-155G14.6 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 11 | UBR3 | Sponge network | 2.483 | 0 | -0.021 | 0.82774 | 0.309 |
| 38642 | RP11-203M5.7 |
hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-429 | 13 | ABL2 | Sponge network | 1.684 | 1.0E-5 | 0.82 | 0 | 0.309 |
| 38643 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-744-3p | 39 | FAT3 | Sponge network | 0.603 | 0.00127 | -1.601 | 0.00051 | 0.309 |
| 38644 | RP5-994D16.9 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-590-3p | 13 | SPRY3 | Sponge network | 0.252 | 0.08508 | 0.016 | 0.91286 | 0.309 |
| 38645 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PBRM1 | Sponge network | 0.385 | 0.10067 | -0.029 | 0.78601 | 0.309 |
| 38646 | BMS1P20 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-148a-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | UBE2D1 | Sponge network | 0.34 | 0.00273 | 0.468 | 0 | 0.309 |
| 38647 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 50 | CUX1 | Sponge network | 0.997 | 0.00066 | -0.039 | 0.6705 | 0.309 |
| 38648 | RP11-46C24.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SETBP1 | Sponge network | 0.392 | 0.0278 | -1.302 | 1.0E-5 | 0.309 |
| 38649 | CTD-2245F17.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-539-5p;hsa-miR-942-5p | 12 | NRXN1 | Sponge network | -0.421 | 0.12556 | -3.778 | 0 | 0.309 |
| 38650 | RP5-1198O20.4 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | PTCH1 | Sponge network | 0.997 | 0.00066 | -0.1 | 0.6179 | 0.309 |
| 38651 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | ARHGAP29 | Sponge network | 0.082 | 0.55654 | 0.131 | 0.42953 | 0.309 |
| 38652 | LINC00342 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 12 | PRKAA1 | Sponge network | 0.297 | 0.19933 | 0.317 | 0.0031 | 0.309 |
| 38653 | FLNB-AS1 |
hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-99b-5p | 13 | CTDSPL | Sponge network | 1.163 | 0.00018 | 0.085 | 0.45121 | 0.309 |
| 38654 | RP3-402G11.26 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | ZFHX3 | Sponge network | -0.254 | 0.19303 | 0.389 | 0.01363 | 0.309 |
| 38655 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | MLLT10 | Sponge network | 0.799 | 1.0E-5 | 0.105 | 0.33292 | 0.309 |
| 38656 | LINC00174 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | AKAP11 | Sponge network | 1.253 | 0 | 0.196 | 0.09825 | 0.309 |
| 38657 | RP11-736K20.5 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-511-5p;hsa-miR-590-3p | 11 | CELF4 | Sponge network | -0.358 | 0.17255 | -1.135 | 0.00344 | 0.309 |
| 38658 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | UBE4B | Sponge network | -0.187 | 0.23787 | -0.235 | 0.007 | 0.309 |
| 38659 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 2.094 | 0 | 0.51 | 0 | 0.309 |
| 38660 | RP11-403I13.7 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 11 | NGFR | Sponge network | 0.395 | 0.15451 | -1.271 | 0.01058 | 0.309 |
| 38661 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-5p | 18 | TET1 | Sponge network | -0.18 | 0.20343 | -0.135 | 0.52138 | 0.309 |
| 38662 | AC034220.3 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 16 | SHPRH | Sponge network | 0.309 | 0.07304 | 0.098 | 0.40994 | 0.309 |
| 38663 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 16 | ZMYND11 | Sponge network | -0.738 | 0.21233 | -0.2 | 0.05449 | 0.309 |
| 38664 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-582-5p | 14 | PIK3C2A | Sponge network | 1.262 | 0 | 0.061 | 0.53776 | 0.309 |
| 38665 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | GNA13 | Sponge network | 0.782 | 0.00016 | 0.007 | 0.93884 | 0.309 |
| 38666 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-362-3p | 11 | GRIA3 | Sponge network | -1.255 | 0.00981 | -1.956 | 0 | 0.309 |
| 38667 | RP11-13K12.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-502-5p;hsa-miR-625-5p | 13 | CELF4 | Sponge network | -0.154 | 0.78939 | -1.135 | 0.00344 | 0.309 |
| 38668 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-93-5p | 25 | ETV1 | Sponge network | -2.69 | 0.0001 | -0.096 | 0.67674 | 0.309 |
| 38669 | RP11-686D22.8 |
hsa-miR-101-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 10 | NF1 | Sponge network | 0.898 | 1.0E-5 | 0.51 | 0 | 0.309 |
| 38670 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 37 | EPHA5 | Sponge network | -0.738 | 0.21233 | -0.957 | 0.01733 | 0.309 |
| 38671 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 63 | AFF4 | Sponge network | -0.323 | 0.01372 | -0.266 | 0.01565 | 0.309 |
| 38672 | FAM157A |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 12 | WDFY3 | Sponge network | 0.813 | 1.0E-5 | -0.201 | 0.0967 | 0.309 |
| 38673 | RP11-981G7.6 |
hsa-let-7g-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-766-3p | 23 | PPARA | Sponge network | 0.986 | 0.01022 | -0.379 | 0.00548 | 0.309 |
| 38674 | RP11-1100L3.8 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-625-5p | 13 | MDM4 | Sponge network | 1.072 | 0.11718 | 0.345 | 0.00762 | 0.309 |
| 38675 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p | 17 | MYO9A | Sponge network | -0.602 | 0.00331 | -0.147 | 0.32373 | 0.309 |
| 38676 | RP11-504P24.4 |
hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | PHC3 | Sponge network | 0.624 | 0.0004 | 0.212 | 0.0499 | 0.309 |
| 38677 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-452-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | INPP4B | Sponge network | -0.129 | 0.57177 | -0.097 | 0.60503 | 0.309 |
| 38678 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | CUL5 | Sponge network | 0.401 | 0.00066 | 0.026 | 0.77301 | 0.309 |
| 38679 | RP11-428J1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-766-3p | 25 | CHD5 | Sponge network | 0.538 | 0.00076 | -0.922 | 0.01521 | 0.309 |
| 38680 | RP11-6O2.3 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-5p | 11 | UBE4B | Sponge network | 0.331 | 0.57247 | -0.235 | 0.007 | 0.309 |
| 38681 | RP4-584D14.7 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-577;hsa-miR-940 | 15 | NOTCH2 | Sponge network | 1.294 | 0.0031 | 0.138 | 0.35163 | 0.309 |
| 38682 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DACT1 | Sponge network | 0.657 | 4.0E-5 | 0.783 | 0.00072 | 0.309 |
| 38683 | RP11-473I1.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p | 21 | TRPS1 | Sponge network | 0.542 | 0.00054 | -0.191 | 0.44109 | 0.309 |
| 38684 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-96-5p | 20 | PRKAR1A | Sponge network | -0.976 | 0.00025 | -0.063 | 0.50314 | 0.309 |
| 38685 | RP11-98I9.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | TCF12 | Sponge network | 0.67 | 0.00048 | 0.174 | 0.13271 | 0.309 |
| 38686 | RP11-835E18.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | ADAMTSL3 | Sponge network | -0.462 | 0.01226 | -1.559 | 0 | 0.309 |
| 38687 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | GPC6 | Sponge network | -0.709 | 0.18168 | -0.054 | 0.81465 | 0.309 |
| 38688 | MIR600HG |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-940 | 21 | SIRT1 | Sponge network | 0.594 | 0.41522 | 0.109 | 0.2593 | 0.309 |
| 38689 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | 0.051 | 0.83388 | -0.354 | 0.14694 | 0.309 |
| 38690 | RP11-819C21.1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | DDX3X | Sponge network | 0.537 | 0.00139 | -0.152 | 0.07686 | 0.309 |
| 38691 | RP11-403I13.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | AKAP6 | Sponge network | 0.619 | 0.00157 | -1.586 | 3.0E-5 | 0.309 |
| 38692 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-671-5p | 10 | SEPT6 | Sponge network | -0.469 | 0.08721 | -0.598 | 0.00181 | 0.309 |
| 38693 | LINC01004 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-495-3p | 14 | CREB1 | Sponge network | 1.115 | 0 | 0.203 | 0.00537 | 0.309 |
| 38694 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320b | 13 | DYNC1I1 | Sponge network | -0.72 | 0.24239 | -1.12 | 0.00107 | 0.309 |
| 38695 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-589-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 29 | TCF4 | Sponge network | -0.726 | 0.00132 | 0.016 | 0.92271 | 0.309 |
| 38696 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-877-5p | 40 | EGFR | Sponge network | -0.129 | 0.57177 | -0.175 | 0.48451 | 0.309 |
| 38697 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-664a-3p | 12 | PRKAA1 | Sponge network | 0.706 | 0 | 0.317 | 0.0031 | 0.309 |
| 38698 | CTD-3064M3.3 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-423-5p;hsa-miR-424-5p | 11 | CSDE1 | Sponge network | -0.469 | 0.08721 | -0.493 | 0 | 0.309 |
| 38699 | HOTAIRM1 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | RIT1 | Sponge network | -0.053 | 0.80685 | -0.296 | 0.00054 | 0.309 |
| 38700 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | TET1 | Sponge network | -0.769 | 0.00035 | -0.135 | 0.52138 | 0.309 |
| 38701 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-877-5p;hsa-miR-93-5p | 39 | CCND2 | Sponge network | -0.465 | 0.04703 | -0.267 | 0.3112 | 0.309 |
| 38702 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 45 | KIF1B | Sponge network | 1.254 | 0 | -0.118 | 0.32258 | 0.309 |
| 38703 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-577 | 10 | DNAJB4 | Sponge network | -0.465 | 0.04703 | -0.7 | 1.0E-5 | 0.309 |
| 38704 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 31 | ETV1 | Sponge network | -0.777 | 0.1134 | -0.096 | 0.67674 | 0.309 |
| 38705 | BDNF-AS |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 13 | ZBTB20 | Sponge network | -0.668 | 3.0E-5 | -0.122 | 0.57569 | 0.309 |
| 38706 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p | 14 | PBX1 | Sponge network | -0.351 | 0.11358 | -1.206 | 0 | 0.309 |
| 38707 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-874-3p | 22 | MXI1 | Sponge network | 0.331 | 0.57247 | -0.987 | 0 | 0.309 |
| 38708 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-625-5p;hsa-miR-940 | 16 | SNX29 | Sponge network | -4.477 | 0 | -0.34 | 0.01371 | 0.309 |
| 38709 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-98-5p | 17 | PNRC1 | Sponge network | -1.216 | 0 | -0.646 | 0 | 0.309 |
| 38710 | LINC00886 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-940 | 44 | PPARA | Sponge network | -0.371 | 0.24604 | -0.379 | 0.00548 | 0.309 |
| 38711 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SAMHD1 | Sponge network | -0.737 | 0.06182 | 0.072 | 0.62895 | 0.309 |
| 38712 | PART1 |
hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | PPP2R2C | Sponge network | -2.925 | 0 | -0.959 | 0.07428 | 0.309 |
| 38713 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-744-5p | 17 | USP12 | Sponge network | -0.469 | 0.08721 | -0.102 | 0.40768 | 0.309 |
| 38714 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-33a-3p | 10 | RIMBP2 | Sponge network | -0.583 | 0.14589 | -1.968 | 5.0E-5 | 0.309 |
| 38715 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | EPHA3 | Sponge network | 0.476 | 0.19203 | 0.185 | 0.53588 | 0.309 |
| 38716 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | ZFX | Sponge network | -0.129 | 0.57177 | 0.09 | 0.42563 | 0.309 |
| 38717 | RP11-35G9.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 19 | DMD | Sponge network | 0.657 | 4.0E-5 | -1.506 | 0 | 0.309 |
| 38718 | FENDRR |
hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-590-3p | 10 | EGR1 | Sponge network | -1.281 | 0.00012 | -1.195 | 0 | 0.309 |
| 38719 | AC156455.1 |
hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484 | 13 | ABL1 | Sponge network | 1.154 | 0.00041 | -0.215 | 0.07804 | 0.309 |
| 38720 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-96-5p | 12 | HDAC9 | Sponge network | -1.582 | 8.0E-5 | -0.755 | 0.00495 | 0.309 |
| 38721 | RP11-48B3.4 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-429;hsa-miR-590-3p | 11 | ITPKB | Sponge network | 1.757 | 0 | -0.704 | 0.00106 | 0.309 |
| 38722 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-744-3p | 23 | CUL5 | Sponge network | 0.267 | 0.2937 | 0.026 | 0.77301 | 0.309 |
| 38723 | ADARB2-AS1 |
hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p | 11 | HIP1 | Sponge network | 0.876 | 0.37332 | 0.774 | 0 | 0.309 |
| 38724 | LINC00476 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-671-5p | 26 | KCNA6 | Sponge network | -0.615 | 0.00015 | -1.531 | 0.00013 | 0.309 |
| 38725 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-493-5p;hsa-miR-940 | 10 | HOOK3 | Sponge network | -2.095 | 0.00112 | 0.273 | 0.09957 | 0.309 |
| 38726 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 20 | GNA13 | Sponge network | 0.614 | 1.0E-5 | 0.007 | 0.93884 | 0.309 |
| 38727 | RP11-2B6.2 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-590-3p | 12 | DDX3X | Sponge network | 0.539 | 0.03722 | -0.152 | 0.07686 | 0.309 |
| 38728 | A2M-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | SORCS1 | Sponge network | -0.08 | 0.90092 | -3.492 | 0 | 0.309 |
| 38729 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577 | 40 | TMEM132B | Sponge network | 0.056 | 0.81195 | -0.83 | 0.01084 | 0.309 |
| 38730 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577 | 10 | HACE1 | Sponge network | 0.637 | 0.00069 | -0.072 | 0.55239 | 0.309 |
| 38731 | CTD-2303H24.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 13 | DDX5 | Sponge network | 0.415 | 0.14657 | -0.099 | 0.17428 | 0.309 |
| 38732 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-429 | 22 | PTPN14 | Sponge network | -0.738 | 0.21233 | 0.144 | 0.34595 | 0.309 |
| 38733 | ZRANB2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-3p | 19 | NTRK2 | Sponge network | -0.376 | 0.00337 | -0.736 | 0.06495 | 0.309 |
| 38734 | CALML3-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-429 | 11 | DNAJB4 | Sponge network | 0.95 | 0.15943 | -0.7 | 1.0E-5 | 0.309 |
| 38735 | LINC00865 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | ETV1 | Sponge network | -1.249 | 7.0E-5 | -0.096 | 0.67674 | 0.309 |
| 38736 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | MED12L | Sponge network | 0.72 | 0.0001 | -0.194 | 0.55299 | 0.309 |
| 38737 | LINC00963 |
hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 13 | TAF1 | Sponge network | 0.523 | 9.0E-5 | 0.39 | 3.0E-5 | 0.309 |
| 38738 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-500a-3p | 11 | TSHZ3 | Sponge network | -0.164 | 0.45106 | -0.556 | 0.01516 | 0.309 |
| 38739 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | CBL | Sponge network | 0.67 | 0.00048 | 0.291 | 0.01267 | 0.309 |
| 38740 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421 | 10 | DPYD | Sponge network | -2.62 | 0 | -0.736 | 0.00076 | 0.309 |
| 38741 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 15 | TACC1 | Sponge network | 0.238 | 0.70544 | -0.362 | 0.05406 | 0.309 |
| 38742 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-454-3p;hsa-miR-590-3p | 21 | PIK3C2A | Sponge network | 0.559 | 0.00026 | 0.061 | 0.53776 | 0.309 |
| 38743 | LINC00473 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 25 | ACSL6 | Sponge network | -1.203 | 0.03881 | -1.593 | 0 | 0.309 |
| 38744 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-582-3p | 12 | CLIP1 | Sponge network | -0.473 | 0.07602 | -0.215 | 0.08684 | 0.309 |
| 38745 | AC034220.3 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | MOB1B | Sponge network | 0.309 | 0.07304 | 0.197 | 0.15266 | 0.309 |
| 38746 | HCG11 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p | 28 | NTRK2 | Sponge network | 0.385 | 0.10067 | -0.736 | 0.06495 | 0.309 |
| 38747 | HHIP-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 17 | TET1 | Sponge network | -0.737 | 0.10822 | -0.135 | 0.52138 | 0.309 |
| 38748 | RP11-706C16.7 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-625-5p;hsa-miR-940 | 10 | CHD5 | Sponge network | 0.357 | 0.72407 | -0.922 | 0.01521 | 0.309 |
| 38749 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ZFHX4 | Sponge network | 0.392 | 0.0278 | 0.018 | 0.95574 | 0.309 |
| 38750 | RP11-359B12.2 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | PRKCB | Sponge network | 0.064 | 0.72934 | -1.313 | 2.0E-5 | 0.309 |
| 38751 | RP11-758N13.1 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484 | 12 | CRTC3 | Sponge network | 2.939 | 3.0E-5 | 0.164 | 0.16786 | 0.309 |
| 38752 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 29 | DDX6 | Sponge network | 0.488 | 0.00084 | 0.091 | 0.36428 | 0.309 |
| 38753 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 26 | ATRX | Sponge network | -0.842 | 0.01361 | 0.261 | 0.04408 | 0.309 |
| 38754 | CTD-2619J13.9 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-628-5p | 10 | TCF4 | Sponge network | 0.576 | 0.0022 | 0.016 | 0.92271 | 0.309 |
| 38755 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p | 10 | CUL5 | Sponge network | 1.117 | 0 | 0.026 | 0.77301 | 0.309 |
| 38756 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ARID5B | Sponge network | 0.873 | 0 | 0.342 | 0.01676 | 0.309 |
| 38757 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 27 | EPHA7 | Sponge network | 0.342 | 0.03129 | -2.197 | 3.0E-5 | 0.309 |
| 38758 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | NEDD4 | Sponge network | 1.134 | 0 | 0.02 | 0.89834 | 0.309 |
| 38759 | MIR22HG |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-582-5p | 19 | GJA1 | Sponge network | -1.047 | 0 | -0.485 | 0.02179 | 0.309 |
| 38760 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-5p | 25 | ADARB1 | Sponge network | 1.153 | 0.00114 | -0.335 | 0.06631 | 0.309 |
| 38761 | RP4-717I23.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 18 | MED12L | Sponge network | 0.637 | 0.00069 | -0.194 | 0.55299 | 0.309 |
| 38762 | TINCR |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-942-5p | 12 | RND3 | Sponge network | -0.664 | 0.14543 | -0.254 | 0.11478 | 0.309 |
| 38763 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | FAT3 | Sponge network | 0.131 | 0.8436 | -1.601 | 0.00051 | 0.309 |
| 38764 | RP11-890B15.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-96-5p | 24 | ASTN1 | Sponge network | 0.101 | 0.60919 | -2.389 | 2.0E-5 | 0.309 |
| 38765 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 41 | PPM1L | Sponge network | -2.69 | 0.0001 | -0.537 | 0.00616 | 0.309 |
| 38766 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-590-3p | 14 | CREBBP | Sponge network | 0.659 | 3.0E-5 | 0.126 | 0.15186 | 0.309 |
| 38767 | LINC00865 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-7-1-3p | 13 | INPP4A | Sponge network | -1.249 | 7.0E-5 | 0.07 | 0.53563 | 0.309 |
| 38768 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | WDFY3 | Sponge network | 0.538 | 0.00076 | -0.201 | 0.0967 | 0.309 |
| 38769 | LINC01018 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 40 | TGFBR3 | Sponge network | -2.915 | 0.00015 | -1.173 | 1.0E-5 | 0.309 |
| 38770 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 10 | CHD3 | Sponge network | -2.46 | 0 | -0.001 | 0.99669 | 0.309 |
| 38771 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ITGAV | Sponge network | 0.335 | 0.20596 | 0.337 | 0.01002 | 0.309 |
| 38772 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p | 13 | CSMD1 | Sponge network | -0.351 | 0.11358 | -0.63 | 0.18881 | 0.309 |
| 38773 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-96-5p | 43 | FAT3 | Sponge network | 0.222 | 0.29133 | -1.601 | 0.00051 | 0.309 |
| 38774 | RP11-13K12.5 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-940 | 12 | CELF4 | Sponge network | -0.318 | 0.65847 | -1.135 | 0.00344 | 0.309 |
| 38775 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 23 | NRK | Sponge network | 0.199 | 0.38015 | 0.656 | 0.19217 | 0.309 |
| 38776 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-532-5p | 10 | LRRC55 | Sponge network | 0.174 | 0.30034 | -0.026 | 0.93163 | 0.309 |
| 38777 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429 | 18 | HECA | Sponge network | -1.654 | 0.00144 | -0.217 | 0.03463 | 0.309 |
| 38778 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | CLASP2 | Sponge network | 1.308 | 0 | -0.038 | 0.71 | 0.309 |
| 38779 | NDUFA6-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -0.355 | 0.0077 | -0.966 | 0.00035 | 0.309 |
| 38780 | RP11-434B12.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PIK3CA | Sponge network | -0.031 | 0.89063 | 0.195 | 0.06908 | 0.309 |
| 38781 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 51 | QKI | Sponge network | 0.083 | 0.66405 | -0.333 | 0.04111 | 0.309 |
| 38782 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-940 | 20 | BHLHE41 | Sponge network | -1.331 | 3.0E-5 | 0.353 | 0.12753 | 0.309 |
| 38783 | RP11-356J5.12 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | ROBO1 | Sponge network | -0.899 | 6.0E-5 | -0.009 | 0.96154 | 0.309 |
| 38784 | RP11-59H7.3 |
hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-502-3p;hsa-miR-576-5p | 10 | NUAK1 | Sponge network | 2.163 | 0 | 0.904 | 0 | 0.309 |
| 38785 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p | 12 | RBM5 | Sponge network | 0.331 | 0.57247 | -0.205 | 0.0129 | 0.309 |
| 38786 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 20 | PCDH17 | Sponge network | -0.611 | 0.09607 | 0.731 | 2.0E-5 | 0.309 |
| 38787 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-9-5p | 11 | YAP1 | Sponge network | 0.706 | 0 | 0.22 | 0.11836 | 0.309 |
| 38788 | TINCR |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-629-3p;hsa-miR-664a-5p | 48 | DST | Sponge network | -0.664 | 0.14543 | -0.713 | 0.00096 | 0.309 |
| 38789 | USP3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-935 | 19 | ARNT2 | Sponge network | -0.162 | 0.47687 | -0.332 | 0.25851 | 0.309 |
| 38790 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | PLSCR4 | Sponge network | 0.056 | 0.8494 | -0.474 | 0.03833 | 0.309 |
| 38791 | GS1-124K5.11 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | TAL1 | Sponge network | 0.494 | 0.00339 | -0.462 | 0.00574 | 0.309 |
| 38792 | RP11-5C23.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p | 32 | RUNX1T1 | Sponge network | 0.274 | 0.07681 | -0.344 | 0.2374 | 0.309 |
| 38793 | CTC-301O7.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-539-5p | 11 | DIP2C | Sponge network | 0.292 | 0.43384 | -0.436 | 0.01713 | 0.309 |
| 38794 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 21 | PCDH10 | Sponge network | 0.41 | 0.02214 | -1.644 | 0.0078 | 0.309 |
| 38795 | PART1 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | GABRB2 | Sponge network | -2.925 | 0 | -1.841 | 0.00029 | 0.309 |
| 38796 | CTC-297N7.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p | 11 | LRP1B | Sponge network | -2.069 | 0 | -2.163 | 0 | 0.309 |
| 38797 | RP11-83A24.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 19 | SIRT1 | Sponge network | 0.417 | 0.00445 | 0.109 | 0.2593 | 0.309 |
| 38798 | SNX29P2 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 17 | SFRP1 | Sponge network | 0.4 | 0.14611 | -3.566 | 0 | 0.309 |
| 38799 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 33 | PTPRD | Sponge network | -0.896 | 0.00099 | -1.005 | 0.00064 | 0.309 |
| 38800 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-5p | 20 | SPG20 | Sponge network | -0.969 | 2.0E-5 | -1.16 | 0 | 0.309 |
| 38801 | RP11-46C24.7 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 13 | CELF4 | Sponge network | 0.392 | 0.0278 | -1.135 | 0.00344 | 0.309 |
| 38802 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-942-5p;hsa-miR-96-5p | 19 | NRXN1 | Sponge network | 0.622 | 0.00015 | -3.778 | 0 | 0.309 |
| 38803 | RP5-1074L1.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 18 | ARHGEF12 | Sponge network | 1.923 | 0 | 0.09 | 0.35693 | 0.309 |
| 38804 | RP11-996F15.2 |
hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 0.983 | 0.00048 | 0.362 | 0.00066 | 0.309 |
| 38805 | CTD-2314G24.2 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NRG3 | Sponge network | 0.246 | 0.59912 | -1.836 | 4.0E-5 | 0.309 |
| 38806 | MIR22HG |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | TPO | Sponge network | -1.047 | 0 | -1.87 | 2.0E-5 | 0.309 |
| 38807 | ACTA2-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 28 | EPS15 | Sponge network | 0.194 | 0.64278 | -0.052 | 0.53998 | 0.309 |
| 38808 | RP11-254F7.2 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29b-2-5p;hsa-miR-424-5p | 10 | DOCK4 | Sponge network | 0.176 | 0.37402 | 0.502 | 0.00108 | 0.309 |
| 38809 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p | 11 | PHC3 | Sponge network | 0.912 | 0.00014 | 0.212 | 0.0499 | 0.308 |
| 38810 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ABCA10 | Sponge network | 1.04 | 0.00023 | -0.571 | 0.03674 | 0.308 |
| 38811 | RAMP2-AS1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p | 10 | GATA2 | Sponge network | -0.513 | 0.04703 | -0.654 | 0.0016 | 0.308 |
| 38812 | CTD-2314G24.2 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-374b-3p;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | 0.246 | 0.59912 | -0.366 | 0.01036 | 0.308 |
| 38813 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-590-3p | 10 | RBM5 | Sponge network | 0.848 | 0 | -0.205 | 0.0129 | 0.308 |
| 38814 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p | 53 | IL6ST | Sponge network | 0.349 | 0.01695 | -0.402 | 0.00676 | 0.308 |
| 38815 | AC034220.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 29 | RICTOR | Sponge network | 0.309 | 0.07304 | 0.388 | 0.00188 | 0.308 |
| 38816 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ST6GALNAC3 | Sponge network | 0.225 | 0.30644 | -0.987 | 0 | 0.308 |
| 38817 | MRPL23-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p | 10 | RIMBP2 | Sponge network | -0.278 | 0.76559 | -1.968 | 5.0E-5 | 0.308 |
| 38818 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-5p | 25 | RSPO2 | Sponge network | -0.777 | 0.1134 | -3.038 | 0 | 0.308 |
| 38819 | LINC00654 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | KCNA6 | Sponge network | 0.374 | 0.20054 | -1.531 | 0.00013 | 0.308 |
| 38820 | DNM3OS |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 14 | POU2F2 | Sponge network | 0.572 | 0.01722 | -0.233 | 0.32214 | 0.308 |
| 38821 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | GRIN2A | Sponge network | -0.053 | 0.80685 | -2.161 | 0 | 0.308 |
| 38822 | CTC-301O7.4 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-511-5p;hsa-miR-539-5p | 16 | LPP | Sponge network | 0.292 | 0.43384 | -0.459 | 0.04475 | 0.308 |
| 38823 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 17 | ARNT2 | Sponge network | -0.08 | 0.90092 | -0.332 | 0.25851 | 0.308 |
| 38824 | RP11-770J1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-142-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-532-3p | 12 | PBX1 | Sponge network | -2.011 | 0.0005 | -1.206 | 0 | 0.308 |
| 38825 | RP11-479G22.8 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-497-5p | 11 | SLC5A3 | Sponge network | 1.308 | 0 | 0.493 | 5.0E-5 | 0.308 |
| 38826 | RP11-967K21.1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | IGFBP5 | Sponge network | 0.056 | 0.81195 | -0.806 | 0.00105 | 0.308 |
| 38827 | AP001055.6 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-7-1-3p | 18 | AFF1 | Sponge network | 0.693 | 0.00076 | -0.384 | 0.00053 | 0.308 |
| 38828 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-150-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-532-3p | 13 | PBX1 | Sponge network | 0.252 | 0.08508 | -1.206 | 0 | 0.308 |
| 38829 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429 | 21 | PPP1R9A | Sponge network | -1.654 | 0.00144 | -0.903 | 0.04254 | 0.308 |
| 38830 | RP11-307B6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-942-5p | 13 | ARHGAP28 | Sponge network | -4.463 | 0.00025 | -0.911 | 0.00013 | 0.308 |
| 38831 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 15 | MYCBP2 | Sponge network | -1.575 | 6.0E-5 | -0.027 | 0.82016 | 0.308 |
| 38832 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | USP12 | Sponge network | 1.601 | 0 | -0.102 | 0.40768 | 0.308 |
| 38833 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 34 | MEF2C | Sponge network | -0.08 | 0.90092 | -0.499 | 0.01448 | 0.308 |
| 38834 | RP11-21A7A.2 |
hsa-miR-103a-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p | 12 | BCL2 | Sponge network | -1.116 | 0.23686 | -0.899 | 0 | 0.308 |
| 38835 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | NR4A3 | Sponge network | -0.901 | 0.00019 | -1.385 | 0 | 0.308 |
| 38836 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 53 | ADARB1 | Sponge network | -2.925 | 0 | -0.335 | 0.06631 | 0.308 |
| 38837 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-301a-3p | 15 | ZBTB4 | Sponge network | -0.72 | 0.24239 | -0.739 | 0 | 0.308 |
| 38838 | RP11-67L2.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-3p | 15 | STARD13 | Sponge network | 0.72 | 0.0001 | -0.187 | 0.27383 | 0.308 |
| 38839 | RP11-815I9.4 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-576-5p;hsa-miR-590-5p | 10 | UBR3 | Sponge network | 0.537 | 0.0006 | -0.021 | 0.82774 | 0.308 |
| 38840 | RP11-212P7.2 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-505-5p;hsa-miR-629-5p;hsa-miR-96-5p | 15 | WNK2 | Sponge network | 0.219 | 0.1516 | -0.352 | 0.31066 | 0.308 |
| 38841 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | MEF2C | Sponge network | 0.796 | 4.0E-5 | -0.499 | 0.01448 | 0.308 |
| 38842 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p | 10 | ZYX | Sponge network | 0.299 | 0.57722 | 0.019 | 0.89763 | 0.308 |
| 38843 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 31 | ABI2 | Sponge network | -0.738 | 0.21233 | 0.175 | 0.14787 | 0.308 |
| 38844 | CTD-3064M3.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-429 | 10 | DUSP1 | Sponge network | -0.469 | 0.08721 | -1.754 | 0 | 0.308 |
| 38845 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-940 | 15 | HLF | Sponge network | -1.483 | 9.0E-5 | -2.443 | 0 | 0.308 |
| 38846 | CTC-487M23.5 |
hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-93-3p | 27 | IL6ST | Sponge network | 0.912 | 0.00014 | -0.402 | 0.00676 | 0.308 |
| 38847 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 16 | KANK1 | Sponge network | -0.513 | 0.04703 | -0.55 | 0.00134 | 0.308 |
| 38848 | AC009948.5 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PHF3 | Sponge network | 0.532 | 0.00505 | 0.126 | 0.19652 | 0.308 |
| 38849 | RP11-440L14.4 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 13 | TSC1 | Sponge network | 0.545 | 0.05789 | 0.103 | 0.26071 | 0.308 |
| 38850 | RP11-384K6.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-744-3p;hsa-miR-940 | 19 | EPS15 | Sponge network | 0.946 | 0 | -0.052 | 0.53998 | 0.308 |
| 38851 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-323a-3p;hsa-miR-455-5p;hsa-miR-93-5p | 13 | SLITRK3 | Sponge network | -0.18 | 0.20343 | -3.328 | 0 | 0.308 |
| 38852 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 39 | CCND2 | Sponge network | -1.203 | 0.03881 | -0.267 | 0.3112 | 0.308 |
| 38853 | PSMG3-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 22 | TET1 | Sponge network | -0.125 | 0.49414 | -0.135 | 0.52138 | 0.308 |
| 38854 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p | 11 | PRKAR1A | Sponge network | -1.092 | 4.0E-5 | -0.063 | 0.50314 | 0.308 |
| 38855 | AC093110.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 14 | PRKAA1 | Sponge network | 1.044 | 2.0E-5 | 0.317 | 0.0031 | 0.308 |
| 38856 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 26 | CBX5 | Sponge network | 0.817 | 3.0E-5 | 0.165 | 0.24446 | 0.308 |
| 38857 | ARHGEF26-AS1 |
hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 12 | PPP2R2C | Sponge network | -2.46 | 0 | -0.959 | 0.07428 | 0.308 |
| 38858 | RP11-158K1.3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 20 | PTPN14 | Sponge network | 0.349 | 0.01695 | 0.144 | 0.34595 | 0.308 |
| 38859 | RP11-148O21.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-500a-5p;hsa-miR-589-3p | 13 | CADM2 | Sponge network | 0.233 | 0.81029 | -3.782 | 0 | 0.308 |
| 38860 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | LRP1B | Sponge network | 0.056 | 0.8494 | -2.163 | 0 | 0.308 |
| 38861 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | NUAK1 | Sponge network | 0.101 | 0.60919 | 0.904 | 0 | 0.308 |
| 38862 | RP11-98I9.4 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-590-3p | 11 | BAZ2B | Sponge network | 0.67 | 0.00048 | -0.276 | 0.02833 | 0.308 |
| 38863 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-421;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 12 | CAV1 | Sponge network | -1.057 | 0.00029 | -1.013 | 6.0E-5 | 0.308 |
| 38864 | RP11-720L2.4 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p | 11 | DIP2C | Sponge network | -1.255 | 0.00981 | -0.436 | 0.01713 | 0.308 |
| 38865 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-96-5p | 30 | PIK3R1 | Sponge network | 0.078 | 0.55803 | -0.133 | 0.36854 | 0.308 |
| 38866 | SNX29P2 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 34 | IGFBP5 | Sponge network | 0.4 | 0.14611 | -0.806 | 0.00105 | 0.308 |
| 38867 | GHRLOS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-7-5p | 18 | ERBB4 | Sponge network | -1.942 | 0 | -1.975 | 2.0E-5 | 0.308 |
| 38868 | FZD10-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | -0.727 | 0.04726 | -3.143 | 0 | 0.308 |
| 38869 | AC025335.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-495-3p | 10 | SLC5A3 | Sponge network | 0.783 | 0 | 0.493 | 5.0E-5 | 0.308 |
| 38870 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 52 | TCF4 | Sponge network | 0.659 | 3.0E-5 | 0.016 | 0.92271 | 0.308 |
| 38871 | RP11-356J5.12 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | RABEP1 | Sponge network | -0.899 | 6.0E-5 | -0.066 | 0.43142 | 0.308 |
| 38872 | LINC00242 |
hsa-miR-182-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-590-3p | 10 | CRTC1 | Sponge network | 0.745 | 0.00347 | -0.116 | 0.28412 | 0.308 |
| 38873 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-660-5p | 16 | ETV1 | Sponge network | -0.555 | 0.06156 | -0.096 | 0.67674 | 0.308 |
| 38874 | RP11-686D22.8 |
hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-96-5p | 18 | MEF2C | Sponge network | 0.898 | 1.0E-5 | -0.499 | 0.01448 | 0.308 |
| 38875 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 29 | PRKAB2 | Sponge network | 1.302 | 7.0E-5 | -0.367 | 0.01371 | 0.308 |
| 38876 | PART1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 20 | CITED2 | Sponge network | -2.925 | 0 | -1.545 | 0 | 0.308 |
| 38877 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 19 | MDM4 | Sponge network | 0.769 | 0 | 0.345 | 0.00762 | 0.308 |
| 38878 | DOCK9-AS2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-629-5p | 31 | DST | Sponge network | -0.231 | 0.28214 | -0.713 | 0.00096 | 0.308 |
| 38879 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | PDGFRA | Sponge network | 0.385 | 0.10067 | -0.372 | 0.0037 | 0.308 |
| 38880 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | AR | Sponge network | 0.559 | 0.00026 | -1.554 | 0 | 0.308 |
| 38881 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-940 | 23 | SNX29 | Sponge network | 0.082 | 0.55397 | -0.34 | 0.01371 | 0.308 |
| 38882 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 19 | KCNA6 | Sponge network | -0.405 | 0.22013 | -1.531 | 0.00013 | 0.308 |
| 38883 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | SMARCA2 | Sponge network | -0.899 | 6.0E-5 | -0.529 | 0.00014 | 0.308 |
| 38884 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 30 | MAF | Sponge network | -0.513 | 0.04703 | -1.257 | 0 | 0.308 |
| 38885 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p | 21 | DDX6 | Sponge network | 1.03 | 0 | 0.091 | 0.36428 | 0.308 |
| 38886 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | PCDH9 | Sponge network | -0.355 | 0.0077 | -1.823 | 4.0E-5 | 0.308 |
| 38887 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | PRRX1 | Sponge network | 0.348 | 0.32912 | 1.316 | 0 | 0.308 |
| 38888 | AC141928.1 |
hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-486-5p;hsa-miR-532-3p | 11 | SYNM | Sponge network | 0.853 | 0.15352 | -2.309 | 1.0E-5 | 0.308 |
| 38889 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 27 | HECA | Sponge network | 0.249 | 0.35902 | -0.217 | 0.03463 | 0.308 |
| 38890 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 16 | CHD2 | Sponge network | -0.405 | 0.22013 | 0.049 | 0.55371 | 0.308 |
| 38891 | WAC-AS1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | SOX11 | Sponge network | 0.821 | 0 | 2.68 | 0 | 0.308 |
| 38892 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-874-3p | 11 | DCUN1D3 | Sponge network | 0.683 | 1.0E-5 | 0.152 | 0.1769 | 0.308 |
| 38893 | RP11-274B21.10 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 21 | CREB1 | Sponge network | 1.073 | 0.00216 | 0.203 | 0.00537 | 0.308 |
| 38894 | RP11-527J8.1 |
hsa-let-7g-3p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p | 11 | ZFX | Sponge network | 1.324 | 0 | 0.09 | 0.42563 | 0.308 |
| 38895 | AGAP11 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-369-3p | 14 | DICER1 | Sponge network | -1.645 | 0 | 0.042 | 0.674 | 0.308 |
| 38896 | FZD10-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | DOCK4 | Sponge network | -0.727 | 0.04726 | 0.502 | 0.00108 | 0.308 |
| 38897 | RP4-669P10.16 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-151a-5p;hsa-miR-185-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p | 12 | NTRK2 | Sponge network | -0.301 | 0.06597 | -0.736 | 0.06495 | 0.308 |
| 38898 | FENDRR |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-98-5p | 20 | PNRC1 | Sponge network | -1.281 | 0.00012 | -0.646 | 0 | 0.308 |
| 38899 | RP11-6O2.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-33a-5p;hsa-miR-3614-5p | 17 | ABL2 | Sponge network | 0.331 | 0.57247 | 0.82 | 0 | 0.308 |
| 38900 | BDNF-AS |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | SORCS1 | Sponge network | -0.668 | 3.0E-5 | -3.492 | 0 | 0.308 |
| 38901 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-550a-3p | 10 | PCDH17 | Sponge network | -2.095 | 0.00112 | 0.731 | 2.0E-5 | 0.308 |
| 38902 | TPT1-AS1 |
hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | ERCC6 | Sponge network | 0.903 | 0 | 0.23 | 0.015 | 0.308 |
| 38903 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-628-5p | 39 | FZD3 | Sponge network | -0.096 | 0.67958 | 0.104 | 0.60316 | 0.308 |
| 38904 | AC005154.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | ZFX | Sponge network | 0.848 | 0 | 0.09 | 0.42563 | 0.308 |
| 38905 | RP11-175O19.4 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-497-5p | 16 | PAFAH1B2 | Sponge network | 0.917 | 0 | 0.318 | 0.0001 | 0.308 |
| 38906 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | PTPRB | Sponge network | -0.222 | 0.32445 | 0.105 | 0.47122 | 0.308 |
| 38907 | TPTEP1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | NR3C2 | Sponge network | -1.216 | 0 | -1.56 | 0 | 0.308 |
| 38908 | AC034220.3 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p | 15 | ANK3 | Sponge network | 0.309 | 0.07304 | -0.55 | 0.00086 | 0.308 |
| 38909 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 26 | NR2C2 | Sponge network | -0.739 | 0.0574 | 0.21 | 0.03224 | 0.308 |
| 38910 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p | 17 | PRKAA1 | Sponge network | 1.153 | 0 | 0.317 | 0.0031 | 0.308 |
| 38911 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b | 16 | APC | Sponge network | 0.594 | 0.00064 | -0.255 | 0.04051 | 0.308 |
| 38912 | RP11-403I13.8 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p | 12 | DYNC1I1 | Sponge network | 0.619 | 0.00157 | -1.12 | 0.00107 | 0.308 |
| 38913 | RP11-34P13.14 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | UBR3 | Sponge network | 1.888 | 0 | -0.021 | 0.82774 | 0.308 |
| 38914 | RP11-263K19.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429 | 18 | DMD | Sponge network | 0.859 | 0.00331 | -1.506 | 0 | 0.308 |
| 38915 | RP11-152F13.8 |
hsa-miR-145-3p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-328-3p;hsa-miR-411-5p | 10 | VPS53 | Sponge network | 0.843 | 0.01775 | 0.103 | 0.22667 | 0.308 |
| 38916 | RP11-473I1.10 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | WDFY3 | Sponge network | 0.673 | 0 | -0.201 | 0.0967 | 0.308 |
| 38917 | RP11-890B15.3 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p | 16 | FMNL3 | Sponge network | 0.101 | 0.60919 | 0.555 | 4.0E-5 | 0.308 |
| 38918 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-503-5p;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | 0.494 | 0.00339 | -0.684 | 0.00159 | 0.308 |
| 38919 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-590-3p | 13 | FAM135B | Sponge network | -0.513 | 0.04703 | -3.978 | 0 | 0.308 |
| 38920 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | MYO9A | Sponge network | -0.663 | 0.10887 | -0.147 | 0.32373 | 0.308 |
| 38921 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MLLT10 | Sponge network | 0.532 | 0.00505 | 0.105 | 0.33292 | 0.308 |
| 38922 | RP11-725P16.2 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-429 | 19 | IL17RD | Sponge network | 0.422 | 0.27021 | -0.019 | 0.94873 | 0.308 |
| 38923 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-582-5p | 14 | PHC3 | Sponge network | 0.792 | 0.0049 | 0.212 | 0.0499 | 0.308 |
| 38924 | TMEM161B-AS1 |
hsa-let-7e-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | COPS2 | Sponge network | -0.04 | 0.7606 | -0.178 | 0.04974 | 0.308 |
| 38925 | PTOV1-AS1 |
hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p | 11 | TMTC3 | Sponge network | 0.87 | 0 | 0.123 | 0.22189 | 0.308 |
| 38926 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 17 | ZFP36L1 | Sponge network | 0.572 | 0.01722 | -0.505 | 8.0E-5 | 0.308 |
| 38927 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-940 | 17 | AKAP11 | Sponge network | 0.792 | 0.0049 | 0.196 | 0.09825 | 0.308 |
| 38928 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-940 | 19 | BHLHE41 | Sponge network | 0.622 | 0.00015 | 0.353 | 0.12753 | 0.308 |
| 38929 | CTD-2303H24.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | ABI2 | Sponge network | 0.415 | 0.14657 | 0.175 | 0.14787 | 0.308 |
| 38930 | AC159540.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p | 10 | BRD3 | Sponge network | 1.122 | 0 | 0.37 | 0.00013 | 0.308 |
| 38931 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-93-5p | 30 | JAZF1 | Sponge network | -0.154 | 0.78939 | -0.377 | 0.02677 | 0.308 |
| 38932 | FGD5-AS1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 29 | PIK3R1 | Sponge network | 0.401 | 0.00066 | -0.133 | 0.36854 | 0.308 |
| 38933 | RP11-156P1.3 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | CSDE1 | Sponge network | 0.214 | 0.18246 | -0.493 | 0 | 0.308 |
| 38934 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-196a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | BCL11A | Sponge network | -0.663 | 0.10887 | -0.932 | 0.0063 | 0.308 |
| 38935 | CTD-3092A11.2 |
hsa-let-7e-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | COPS2 | Sponge network | 0.873 | 0 | -0.178 | 0.04974 | 0.308 |
| 38936 | CTD-2314G24.2 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 25 | OPCML | Sponge network | 0.246 | 0.59912 | -2.498 | 0 | 0.308 |
| 38937 | CALML3-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-5p | 25 | TET1 | Sponge network | 0.95 | 0.15943 | -0.135 | 0.52138 | 0.308 |
| 38938 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | ZYX | Sponge network | -1.111 | 0.0003 | 0.019 | 0.89763 | 0.308 |
| 38939 | RP11-701H24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-33a-3p | 14 | CREB3L2 | Sponge network | -2.039 | 0.03447 | -0.525 | 2.0E-5 | 0.308 |
| 38940 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | PDGFRA | Sponge network | -0.727 | 0.04726 | -0.372 | 0.0037 | 0.308 |
| 38941 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 16 | SASH1 | Sponge network | 0.342 | 0.03129 | -0.502 | 0.00175 | 0.308 |
| 38942 | CKMT2-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-361-3p | 10 | NGFR | Sponge network | 0.082 | 0.55654 | -1.271 | 0.01058 | 0.308 |
| 38943 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-338-3p;hsa-miR-429 | 10 | ELMO1 | Sponge network | -0.246 | 0.35034 | 0.04 | 0.83147 | 0.308 |
| 38944 | MIR7-3HG |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-324-3p | 10 | EPHA7 | Sponge network | -2.688 | 0.00397 | -2.197 | 3.0E-5 | 0.308 |
| 38945 | AC093627.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-625-5p | 16 | ANK2 | Sponge network | -0.787 | 0.3928 | -1.603 | 1.0E-5 | 0.308 |
| 38946 | RP11-428J1.5 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | 0.538 | 0.00076 | 0.524 | 0.00017 | 0.308 |
| 38947 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-424-5p | 24 | FAT3 | Sponge network | -4.477 | 0 | -1.601 | 0.00051 | 0.308 |
| 38948 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-590-3p | 20 | AFF3 | Sponge network | -0.668 | 3.0E-5 | -2.373 | 0 | 0.308 |
| 38949 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 36 | PIK3R1 | Sponge network | 0.782 | 0.00016 | -0.133 | 0.36854 | 0.308 |
| 38950 | FZD10-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | EYA4 | Sponge network | -0.727 | 0.04726 | 0.3 | 0.36876 | 0.308 |
| 38951 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-542-3p | 13 | ABCB5 | Sponge network | -0.611 | 0.09607 | -0.956 | 0.04077 | 0.308 |
| 38952 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 12 | FAT4 | Sponge network | 0.898 | 1.0E-5 | -0.354 | 0.14694 | 0.308 |
| 38953 | LINC01018 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-93-3p | 27 | CTIF | Sponge network | -2.915 | 0.00015 | -0.389 | 0.00549 | 0.308 |
| 38954 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | PTPRT | Sponge network | -0.206 | 0.12942 | -0.907 | 0.03727 | 0.308 |
| 38955 | AC016747.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-3p | 13 | UNC5C | Sponge network | 0.199 | 0.38015 | -0.178 | 0.40214 | 0.308 |
| 38956 | CD27-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | 0.177 | 0.16681 | 0.524 | 0.00017 | 0.308 |
| 38957 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | EGLN1 | Sponge network | 0.101 | 0.60919 | -0.127 | 0.1536 | 0.308 |
| 38958 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | EYA4 | Sponge network | 0.082 | 0.55654 | 0.3 | 0.36876 | 0.308 |
| 38959 | RP11-158K1.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | ROBO1 | Sponge network | 0.349 | 0.01695 | -0.009 | 0.96154 | 0.308 |
| 38960 | HCG11 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-93-5p | 11 | LHX6 | Sponge network | 0.385 | 0.10067 | -0.087 | 0.69193 | 0.308 |
| 38961 | RP11-755F10.1 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-29b-1-5p;hsa-miR-589-3p | 12 | LPP | Sponge network | 0.952 | 0 | -0.459 | 0.04475 | 0.308 |
| 38962 | HAR1A |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-942-5p | 11 | GPC6 | Sponge network | -0.672 | 0.19447 | -0.054 | 0.81465 | 0.308 |
| 38963 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-34a-5p;hsa-miR-502-5p | 12 | ANK2 | Sponge network | 0.912 | 0.00014 | -1.603 | 1.0E-5 | 0.308 |
| 38964 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | LRRC7 | Sponge network | 0.693 | 0.00076 | -1.341 | 0.01193 | 0.308 |
| 38965 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-214-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-708-3p | 16 | MXI1 | Sponge network | -0.285 | 0.2172 | -0.987 | 0 | 0.308 |
| 38966 | CTD-2303H24.2 |
hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | HERC1 | Sponge network | 0.415 | 0.14657 | -0.196 | 0.08544 | 0.308 |
| 38967 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-455-5p | 12 | ZNF521 | Sponge network | -0.615 | 0.00015 | -0.111 | 0.58068 | 0.308 |
| 38968 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 30 | ANK2 | Sponge network | 0.72 | 0.0001 | -1.603 | 1.0E-5 | 0.308 |
| 38969 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | AHNAK | Sponge network | -0.355 | 0.0077 | -1.207 | 0 | 0.308 |
| 38970 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p | 11 | ZMYM2 | Sponge network | 1.073 | 0.00216 | 0.279 | 0.01273 | 0.308 |
| 38971 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-192-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | FZD3 | Sponge network | 1.378 | 0 | 0.104 | 0.60316 | 0.308 |
| 38972 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-940 | 19 | PBX1 | Sponge network | 0.556 | 0.00298 | -1.206 | 0 | 0.308 |
| 38973 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-539-5p | 15 | ARHGEF12 | Sponge network | 0.37 | 0.27614 | 0.09 | 0.35693 | 0.308 |
| 38974 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -1.161 | 0.00591 | -2.417 | 0 | 0.308 |
| 38975 | RP11-46C24.7 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PCDH9 | Sponge network | 0.392 | 0.0278 | -1.823 | 4.0E-5 | 0.308 |
| 38976 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-3p | 22 | FOXO3 | Sponge network | -1.645 | 0 | -0.04 | 0.72028 | 0.308 |
| 38977 | TINCR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-940 | 24 | MCC | Sponge network | -0.664 | 0.14543 | -1.005 | 0 | 0.308 |
| 38978 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-940 | 36 | PPP1R9A | Sponge network | -0.096 | 0.67958 | -0.903 | 0.04254 | 0.308 |
| 38979 | MIR22HG |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-5p | 23 | PRKCB | Sponge network | -1.047 | 0 | -1.313 | 2.0E-5 | 0.308 |
| 38980 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-374a-3p;hsa-miR-505-5p;hsa-miR-766-3p | 11 | TBL1XR1 | Sponge network | 0.765 | 7.0E-5 | 0.578 | 0 | 0.308 |
| 38981 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | SSBP2 | Sponge network | 0.889 | 9.0E-5 | -1.58 | 0 | 0.308 |
| 38982 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-744-3p | 16 | CUL5 | Sponge network | 1.089 | 0 | 0.026 | 0.77301 | 0.308 |
| 38983 | PART1 |
hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-942-5p | 25 | EMP2 | Sponge network | -2.925 | 0 | -0.423 | 0.00985 | 0.308 |
| 38984 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | PHC3 | Sponge network | 1.11 | 0 | 0.212 | 0.0499 | 0.308 |
| 38985 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 20 | DOCK4 | Sponge network | 0.222 | 0.29133 | 0.502 | 0.00108 | 0.308 |
| 38986 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 41 | CCND2 | Sponge network | -0.793 | 7.0E-5 | -0.267 | 0.3112 | 0.308 |
| 38987 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-5p | 27 | ESR1 | Sponge network | -0.18 | 0.20343 | -1.035 | 3.0E-5 | 0.308 |
| 38988 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-576-5p | 12 | FOXO3 | Sponge network | 1.044 | 2.0E-5 | -0.04 | 0.72028 | 0.308 |
| 38989 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p | 13 | CLIP1 | Sponge network | 0.906 | 0.31715 | -0.215 | 0.08684 | 0.308 |
| 38990 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | PRKAB2 | Sponge network | 1.063 | 0 | -0.367 | 0.01371 | 0.308 |
| 38991 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 18 | MYBL1 | Sponge network | 0.953 | 0 | 0.494 | 0.02152 | 0.308 |
| 38992 | H1FX-AS1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 11 | FREM2 | Sponge network | -0.206 | 0.12942 | -0.437 | 0.46353 | 0.308 |
| 38993 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-5p | 12 | RB1CC1 | Sponge network | -0.384 | 0.40652 | 0.175 | 0.12559 | 0.308 |
| 38994 | RP11-196G11.4 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | CREB1 | Sponge network | 0.875 | 0 | 0.203 | 0.00537 | 0.308 |
| 38995 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 49 | PPM1A | Sponge network | -0.49 | 0.04946 | -0.623 | 0 | 0.308 |
| 38996 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 11 | FLI1 | Sponge network | -0.513 | 0.04703 | -0.294 | 0.10905 | 0.308 |
| 38997 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 33 | AR | Sponge network | 0.174 | 0.30034 | -1.554 | 0 | 0.308 |
| 38998 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-93-5p | 27 | SMAD4 | Sponge network | 0.497 | 0.01451 | -0.313 | 0.00413 | 0.308 |
| 38999 | RP11-121C2.2 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-655-3p | 10 | UBE4B | Sponge network | 1.147 | 0 | -0.235 | 0.007 | 0.308 |
| 39000 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-542-3p | 12 | SLITRK6 | Sponge network | -0.611 | 0.09607 | -2.121 | 0 | 0.308 |
| 39001 | AC109309.4 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-320a;hsa-miR-320b | 10 | NTRK2 | Sponge network | 0.163 | 0.87219 | -0.736 | 0.06495 | 0.308 |
| 39002 | AC034220.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 38 | BMPR2 | Sponge network | 0.309 | 0.07304 | 0.206 | 0.0635 | 0.308 |
| 39003 | MIR181A2HG |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p | 11 | ABL2 | Sponge network | 1.822 | 1.0E-5 | 0.82 | 0 | 0.308 |
| 39004 | HOXD-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-500a-5p | 13 | DMD | Sponge network | -0.208 | 0.65592 | -1.506 | 0 | 0.308 |
| 39005 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 47 | BMPR2 | Sponge network | 0.078 | 0.55803 | 0.206 | 0.0635 | 0.308 |
| 39006 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-935 | 23 | RNF144A | Sponge network | -0.162 | 0.47687 | 0.594 | 0.0009 | 0.308 |
| 39007 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p | 17 | PTPRB | Sponge network | -2.62 | 0 | 0.105 | 0.47122 | 0.308 |
| 39008 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | EPHA3 | Sponge network | 0.663 | 0.00073 | 0.185 | 0.53588 | 0.308 |
| 39009 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p | 23 | BCL6 | Sponge network | -0.942 | 0 | -0.211 | 0.19489 | 0.308 |
| 39010 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 13 | CREBBP | Sponge network | 1.04 | 0.00023 | 0.126 | 0.15186 | 0.308 |
| 39011 | RP11-48B3.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | CREB1 | Sponge network | 1.757 | 0 | 0.203 | 0.00537 | 0.308 |
| 39012 | MIR4458HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-223-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p | 10 | FAM188A | Sponge network | -0.427 | 0.0623 | -0.44 | 0.00018 | 0.308 |
| 39013 | CASC2 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | KIF5B | Sponge network | 0.385 | 0.01306 | 0.362 | 0.00066 | 0.308 |
| 39014 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | TMEM132B | Sponge network | 0.225 | 0.30644 | -0.83 | 0.01084 | 0.308 |
| 39015 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-505-5p;hsa-miR-589-5p | 10 | RBM5 | Sponge network | -0.465 | 0.04703 | -0.205 | 0.0129 | 0.308 |
| 39016 | RP11-359E10.1 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-301a-3p;hsa-miR-338-5p | 10 | MAFB | Sponge network | 0.299 | 0.57722 | -0.023 | 0.89865 | 0.308 |
| 39017 | RP11-597D13.9 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | CREB1 | Sponge network | 1.302 | 7.0E-5 | 0.203 | 0.00537 | 0.308 |
| 39018 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 11 | FSTL5 | Sponge network | 0.494 | 0.00339 | -1.3 | 0.01619 | 0.308 |
| 39019 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | CADM2 | Sponge network | 0.131 | 0.8436 | -3.782 | 0 | 0.308 |
| 39020 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-92a-3p | 16 | LATS2 | Sponge network | 0.16 | 0.50785 | 0.159 | 0.2304 | 0.308 |
| 39021 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 19 | MYO9A | Sponge network | 0.056 | 0.8494 | -0.147 | 0.32373 | 0.308 |
| 39022 | RP11-53O19.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | TCF12 | Sponge network | 1.205 | 0 | 0.174 | 0.13271 | 0.308 |
| 39023 | RP11-33B1.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | CCDC6 | Sponge network | 0.826 | 0 | 0.27 | 0.02555 | 0.308 |
| 39024 | CTC-378H22.2 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-652-3p | 12 | QKI | Sponge network | 0.001 | 0.99663 | -0.333 | 0.04111 | 0.308 |
| 39025 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 51 | CUX1 | Sponge network | -0.576 | 0.10121 | -0.039 | 0.6705 | 0.308 |
| 39026 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 31 | CREB3L2 | Sponge network | -0.513 | 0.04703 | -0.525 | 2.0E-5 | 0.308 |
| 39027 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p | 24 | PRKAA2 | Sponge network | -0.829 | 0.01343 | -2.462 | 0 | 0.308 |
| 39028 | RP4-613B23.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p | 13 | WDFY3 | Sponge network | -0.309 | 0.1145 | -0.201 | 0.0967 | 0.308 |
| 39029 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p | 10 | GPAM | Sponge network | 0.997 | 0 | 0.142 | 0.29732 | 0.308 |
| 39030 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-7-1-3p | 14 | ABCC9 | Sponge network | 0.614 | 1.0E-5 | -0.466 | 0.14121 | 0.308 |
| 39031 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-93-5p | 12 | FBXO31 | Sponge network | -0.303 | 0.21002 | -0.085 | 0.42389 | 0.308 |
| 39032 | CTB-113P19.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-338-5p;hsa-miR-455-5p | 14 | JAZF1 | Sponge network | 0.199 | 0.52763 | -0.377 | 0.02677 | 0.308 |
| 39033 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-539-5p | 21 | TMEM132B | Sponge network | -0.469 | 0.08721 | -0.83 | 0.01084 | 0.308 |
| 39034 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 32 | ERBB4 | Sponge network | 0.225 | 0.30644 | -1.975 | 2.0E-5 | 0.308 |
| 39035 | RP11-21A7A.2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 12 | JAZF1 | Sponge network | -1.116 | 0.23686 | -0.377 | 0.02677 | 0.308 |
| 39036 | RP11-473I1.9 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-629-3p | 15 | PTPN14 | Sponge network | 0.683 | 1.0E-5 | 0.144 | 0.34595 | 0.308 |
| 39037 | RP11-65J21.3 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-501-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 11 | RIMS2 | Sponge network | -0.557 | 0.11135 | -1.365 | 0.00208 | 0.308 |
| 39038 | OIP5-AS1 |
hsa-miR-125a-5p;hsa-miR-141-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | RANBP9 | Sponge network | 0.421 | 0.00084 | -0.191 | 0.04628 | 0.308 |
| 39039 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-505-5p | 22 | AR | Sponge network | -0.301 | 0.06597 | -1.554 | 0 | 0.308 |
| 39040 | CTA-204B4.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-590-3p | 11 | KANK1 | Sponge network | 0.765 | 7.0E-5 | -0.55 | 0.00134 | 0.308 |
| 39041 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-3p | 23 | FOXO3 | Sponge network | -0.031 | 0.89063 | -0.04 | 0.72028 | 0.308 |
| 39042 | RP11-262H14.3 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p | 21 | TBL1XR1 | Sponge network | -0.106 | 0.63074 | 0.578 | 0 | 0.308 |
| 39043 | LINC00621 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | NTRK2 | Sponge network | 0.131 | 0.8436 | -0.736 | 0.06495 | 0.308 |
| 39044 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-590-5p | 19 | PIK3C2A | Sponge network | 0.331 | 0.57247 | 0.061 | 0.53776 | 0.308 |
| 39045 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 45 | CBX5 | Sponge network | 0.064 | 0.72934 | 0.165 | 0.24446 | 0.308 |
| 39046 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p | 14 | ESR1 | Sponge network | 0.687 | 0.02753 | -1.035 | 3.0E-5 | 0.308 |
| 39047 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-629-3p | 13 | AR | Sponge network | -1.717 | 0.01522 | -1.554 | 0 | 0.308 |
| 39048 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-590-3p | 14 | BCL11A | Sponge network | -0.739 | 0.00044 | -0.932 | 0.0063 | 0.308 |
| 39049 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-1-3p | 27 | RASSF2 | Sponge network | -0.176 | 0.59692 | -0.273 | 0.21961 | 0.308 |
| 39050 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-450b-5p | 16 | PRDM2 | Sponge network | -0.517 | 0.02935 | -0.258 | 0.00721 | 0.308 |
| 39051 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-5p | 13 | ITGB3 | Sponge network | -0.384 | 0.40652 | -0.011 | 0.95751 | 0.308 |
| 39052 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | PIK3C2A | Sponge network | -0.323 | 0.01372 | 0.061 | 0.53776 | 0.308 |
| 39053 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-539-5p;hsa-miR-93-5p | 15 | CDHR3 | Sponge network | -0.726 | 0.00132 | -0.977 | 4.0E-5 | 0.308 |
| 39054 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-323a-3p | 10 | LATS1 | Sponge network | -0.473 | 0.07602 | 0.109 | 0.34208 | 0.308 |
| 39055 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 42 | CBX5 | Sponge network | -0.563 | 0.02545 | 0.165 | 0.24446 | 0.308 |
| 39056 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-93-3p | 10 | WWTR1 | Sponge network | 0.912 | 0.00014 | -0.345 | 0.11997 | 0.308 |
| 39057 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-629-5p | 12 | MAF | Sponge network | -4.477 | 0 | -1.257 | 0 | 0.308 |
| 39058 | PART1 |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p | 12 | CADM4 | Sponge network | -2.925 | 0 | -0.635 | 0.00305 | 0.308 |
| 39059 | LINC00926 |
hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-629-3p;hsa-miR-940 | 11 | BHLHE41 | Sponge network | 0.756 | 0.00739 | 0.353 | 0.12753 | 0.308 |
| 39060 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p | 13 | TIMP3 | Sponge network | -0.785 | 0.00035 | -0.546 | 0.02307 | 0.308 |
| 39061 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-493-5p | 12 | MYCBP2 | Sponge network | 0.056 | 0.8494 | -0.027 | 0.82016 | 0.308 |
| 39062 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 12 | KCNAB1 | Sponge network | -0.769 | 0.00035 | -0.755 | 2.0E-5 | 0.308 |
| 39063 | RP11-283I3.6 |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 10 | DCLK1 | Sponge network | 0.614 | 1.0E-5 | -1.324 | 0.00014 | 0.308 |
| 39064 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 37 | SASH1 | Sponge network | 0.056 | 0.81195 | -0.502 | 0.00175 | 0.308 |
| 39065 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 36 | SASH1 | Sponge network | -0.769 | 0.00035 | -0.502 | 0.00175 | 0.308 |
| 39066 | PCA3 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 18 | ATM | Sponge network | -0.709 | 0.18168 | 0.178 | 0.21568 | 0.308 |
| 39067 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-369-3p | 10 | FAT4 | Sponge network | 0.542 | 0.00054 | -0.354 | 0.14694 | 0.308 |
| 39068 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 31 | SOX5 | Sponge network | -0.13 | 0.48734 | -1.091 | 4.0E-5 | 0.308 |
| 39069 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p | 20 | CREB3L2 | Sponge network | -0.309 | 0.1145 | -0.525 | 2.0E-5 | 0.308 |
| 39070 | RP11-736K20.5 |
hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | RABEP1 | Sponge network | -0.358 | 0.17255 | -0.066 | 0.43142 | 0.308 |
| 39071 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 39 | PPP1R9A | Sponge network | -0.323 | 0.01372 | -0.903 | 0.04254 | 0.308 |
| 39072 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | EBF1 | Sponge network | -0.727 | 0.04726 | -0.384 | 0.08856 | 0.308 |
| 39073 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 28 | PGR | Sponge network | 0.72 | 0.0001 | -1.704 | 0 | 0.308 |
| 39074 | MIR600HG |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-484 | 18 | ABL1 | Sponge network | 0.594 | 0.41522 | -0.215 | 0.07804 | 0.308 |
| 39075 | LINC00094 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p | 11 | GALNT13 | Sponge network | 0.253 | 0.09565 | -0.857 | 0.07439 | 0.308 |
| 39076 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-5p | 14 | ATRX | Sponge network | 0.983 | 0.00048 | 0.261 | 0.04408 | 0.308 |
| 39077 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | BCL6 | Sponge network | 0.381 | 0.25967 | -0.211 | 0.19489 | 0.308 |
| 39078 | AC006116.24 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-577 | 11 | LUZP2 | Sponge network | -0.473 | 0.07602 | -0.056 | 0.89416 | 0.308 |
| 39079 | RP4-584D14.7 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-577;hsa-miR-940 | 15 | TRPS1 | Sponge network | 1.294 | 0.0031 | -0.191 | 0.44109 | 0.308 |
| 39080 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 35 | CUX1 | Sponge network | 0.299 | 0.57722 | -0.039 | 0.6705 | 0.308 |
| 39081 | CTD-2336O2.1 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | SYNPO2 | Sponge network | -0.141 | 0.26434 | -2.342 | 0 | 0.308 |
| 39082 | MALAT1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 18 | RASAL2 | Sponge network | 0.782 | 0.00016 | 0.869 | 0 | 0.308 |
| 39083 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 11 | FAT4 | Sponge network | -0.285 | 0.2172 | -0.354 | 0.14694 | 0.308 |
| 39084 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-429 | 12 | DDX6 | Sponge network | 2.364 | 0.00134 | 0.091 | 0.36428 | 0.308 |
| 39085 | CTC-297N7.9 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-590-5p | 24 | THRB | Sponge network | -2.069 | 0 | -1.117 | 0.0004 | 0.308 |
| 39086 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 15 | RARB | Sponge network | -4.477 | 0 | -0.825 | 2.0E-5 | 0.308 |
| 39087 | MIR143HG |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | BRD3 | Sponge network | -0.739 | 0.0574 | 0.37 | 0.00013 | 0.308 |
| 39088 | LINC00174 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 19 | CCDC6 | Sponge network | 1.253 | 0 | 0.27 | 0.02555 | 0.308 |
| 39089 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 24 | FOXP1 | Sponge network | 0.873 | 0 | 0.042 | 0.74368 | 0.308 |
| 39090 | RASSF8-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | ID4 | Sponge network | 0.335 | 0.20596 | -1.644 | 0 | 0.308 |
| 39091 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PBRM1 | Sponge network | 0.67 | 0.00048 | -0.029 | 0.78601 | 0.307 |
| 39092 | RP11-159D12.2 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | ERCC4 | Sponge network | 1.254 | 0 | 0.126 | 0.15266 | 0.307 |
| 39093 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 39 | WDFY3 | Sponge network | -0.896 | 0.00099 | -0.201 | 0.0967 | 0.307 |
| 39094 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-582-5p | 11 | RB1CC1 | Sponge network | 0.056 | 0.81195 | 0.175 | 0.12559 | 0.307 |
| 39095 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 44 | NTRK3 | Sponge network | -0.053 | 0.80685 | -1.77 | 3.0E-5 | 0.307 |
| 39096 | HCG11 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 41 | TRPS1 | Sponge network | 0.385 | 0.10067 | -0.191 | 0.44109 | 0.307 |
| 39097 | USP3-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-744-3p | 14 | IGF2 | Sponge network | -0.162 | 0.47687 | 0.848 | 0.03842 | 0.307 |
| 39098 | LINC00657 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-766-3p | 15 | IL17RD | Sponge network | 0.91 | 0 | -0.019 | 0.94873 | 0.307 |
| 39099 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 25 | PRRX1 | Sponge network | -0.08 | 0.90092 | 1.316 | 0 | 0.307 |
| 39100 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p | 12 | LHX6 | Sponge network | -1.161 | 0.00591 | -0.087 | 0.69193 | 0.307 |
| 39101 | FAM157C |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-539-5p | 12 | ARHGEF12 | Sponge network | 0.91 | 0 | 0.09 | 0.35693 | 0.307 |
| 39102 | EIF3J-AS1 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-671-5p | 11 | CRTC3 | Sponge network | 0.331 | 0.00509 | 0.164 | 0.16786 | 0.307 |
| 39103 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PRKAB2 | Sponge network | 0.986 | 0.01022 | -0.367 | 0.01371 | 0.307 |
| 39104 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | TP53INP1 | Sponge network | -0.663 | 0.10887 | -0.496 | 0.00064 | 0.307 |
| 39105 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | INPP4B | Sponge network | -0.942 | 0 | -0.097 | 0.60503 | 0.307 |
| 39106 | EMX2OS |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | DIRAS1 | Sponge network | -0.777 | 0.1134 | -2.518 | 0 | 0.307 |
| 39107 | HOXB-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-582-3p | 18 | ABL2 | Sponge network | -0.154 | 0.53954 | 0.82 | 0 | 0.307 |
| 39108 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | MYO9A | Sponge network | 0.796 | 4.0E-5 | -0.147 | 0.32373 | 0.307 |
| 39109 | CTC-296K1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p | 13 | RNASEL | Sponge network | -2.095 | 0.00112 | -0.272 | 0.01796 | 0.307 |
| 39110 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p | 49 | BMPR2 | Sponge network | -0.384 | 0.40652 | 0.206 | 0.0635 | 0.307 |
| 39111 | RP11-59H7.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p | 13 | TCF12 | Sponge network | 2.163 | 0 | 0.174 | 0.13271 | 0.307 |
| 39112 | LINC00174 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 14 | ARHGEF12 | Sponge network | 1.253 | 0 | 0.09 | 0.35693 | 0.307 |
| 39113 | RP11-212P7.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 46 | TRPS1 | Sponge network | 0.219 | 0.1516 | -0.191 | 0.44109 | 0.307 |
| 39114 | RP11-212P7.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | LRIG1 | Sponge network | 0.219 | 0.1516 | -0.537 | 0.00588 | 0.307 |
| 39115 | RP11-263K19.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-744-3p | 17 | DIP2C | Sponge network | 0.859 | 0.00331 | -0.436 | 0.01713 | 0.307 |
| 39116 | RP11-83A24.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | CREB1 | Sponge network | 0.417 | 0.00445 | 0.203 | 0.00537 | 0.307 |
| 39117 | PCA3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | RNF144A | Sponge network | -0.709 | 0.18168 | 0.594 | 0.0009 | 0.307 |
| 39118 | RP11-490M8.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-539-5p | 13 | TMEM132B | Sponge network | -0.214 | 0.44248 | -0.83 | 0.01084 | 0.307 |
| 39119 | RP11-473I1.9 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PREX2 | Sponge network | 0.683 | 1.0E-5 | -0.272 | 0.25382 | 0.307 |
| 39120 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 24 | DMXL1 | Sponge network | 0.532 | 0.00505 | -0.426 | 0.00056 | 0.307 |
| 39121 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-877-5p | 18 | CRTC1 | Sponge network | -0.612 | 0.00094 | -0.116 | 0.28412 | 0.307 |
| 39122 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-940 | 30 | LIFR | Sponge network | -0.793 | 7.0E-5 | -1.794 | 0 | 0.307 |
| 39123 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-744-5p | 15 | USP12 | Sponge network | -0.285 | 0.2172 | -0.102 | 0.40768 | 0.307 |
| 39124 | TRIM52-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-342-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-577 | 10 | SYNPO2 | Sponge network | -0.418 | 0.00068 | -2.342 | 0 | 0.307 |
| 39125 | RBPMS-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | AR | Sponge network | 0.144 | 0.64989 | -1.554 | 0 | 0.307 |
| 39126 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 14 | PRKAB2 | Sponge network | -0.726 | 0.00132 | -0.367 | 0.01371 | 0.307 |
| 39127 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | KANK1 | Sponge network | 0.082 | 0.55654 | -0.55 | 0.00134 | 0.307 |
| 39128 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | NIN | Sponge network | 0.4 | 0.14611 | -0.253 | 0.13794 | 0.307 |
| 39129 | RP11-890B15.3 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | WAC | Sponge network | 0.101 | 0.60919 | -0.054 | 0.43688 | 0.307 |
| 39130 | RP11-182J1.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-96-5p | 10 | PRKCE | Sponge network | -0.187 | 0.23787 | -0.403 | 0.00056 | 0.307 |
| 39131 | SOX2-OT |
hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-96-5p | 31 | CADM1 | Sponge network | -1.264 | 6.0E-5 | -0.9 | 0.00084 | 0.307 |
| 39132 | LINC00865 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 11 | SPTBN4 | Sponge network | -1.249 | 7.0E-5 | -0.786 | 0.01281 | 0.307 |
| 39133 | AC074117.10 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | CBFB | Sponge network | 1.481 | 0 | 1.061 | 0 | 0.307 |
| 39134 | RP11-244H3.1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | RBBP6 | Sponge network | 0.659 | 3.0E-5 | 0.163 | 0.05101 | 0.307 |
| 39135 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | FOXO3 | Sponge network | 0.959 | 0 | -0.04 | 0.72028 | 0.307 |
| 39136 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | RAP1A | Sponge network | 0.064 | 0.72934 | -0.929 | 0 | 0.307 |
| 39137 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | RNF144A | Sponge network | -0.737 | 0.10822 | 0.594 | 0.0009 | 0.307 |
| 39138 | WDR86-AS1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-7-5p | 13 | IGFBP5 | Sponge network | 0.348 | 0.32912 | -0.806 | 0.00105 | 0.307 |
| 39139 | CTC-429P9.1 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 21 | AFF1 | Sponge network | 0.497 | 0.01451 | -0.384 | 0.00053 | 0.307 |
| 39140 | AC025171.1 |
hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | LRRC55 | Sponge network | 0.998 | 0 | -0.026 | 0.93163 | 0.307 |
| 39141 | RP1-39G22.7 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | KIF5B | Sponge network | 0.439 | 0.00313 | 0.362 | 0.00066 | 0.307 |
| 39142 | RP11-458D21.1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 18 | MED12L | Sponge network | 0.663 | 0.00073 | -0.194 | 0.55299 | 0.307 |
| 39143 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 17 | GPAM | Sponge network | 0.8 | 0 | 0.142 | 0.29732 | 0.307 |
| 39144 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | USP12 | Sponge network | 0.539 | 0.03722 | -0.102 | 0.40768 | 0.307 |
| 39145 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 26 | TSHZ3 | Sponge network | 0.619 | 0.00157 | -0.556 | 0.01516 | 0.307 |
| 39146 | RP11-166D19.1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 18 | ATM | Sponge network | -0.576 | 0.10121 | 0.178 | 0.21568 | 0.307 |
| 39147 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 28 | LRP1B | Sponge network | 0.083 | 0.66405 | -2.163 | 0 | 0.307 |
| 39148 | RP11-403I13.8 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-589-5p;hsa-miR-590-3p | 11 | ROBO1 | Sponge network | 0.619 | 0.00157 | -0.009 | 0.96154 | 0.307 |
| 39149 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 50 | PAFAH1B1 | Sponge network | 0.72 | 0.0001 | -0.429 | 0 | 0.307 |
| 39150 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 12 | PPP1R9A | Sponge network | -1.116 | 0.23686 | -0.903 | 0.04254 | 0.307 |
| 39151 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 21 | PBX1 | Sponge network | 0.559 | 0.00026 | -1.206 | 0 | 0.307 |
| 39152 | RP11-305E6.4 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | GSK3B | Sponge network | 1.317 | 0 | 0.266 | 0.00045 | 0.307 |
| 39153 | RP11-597D13.9 |
hsa-miR-140-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | NOV | Sponge network | 1.302 | 7.0E-5 | -0.58 | 0.04676 | 0.307 |
| 39154 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-935 | 30 | LIFR | Sponge network | -0.615 | 0.00015 | -1.794 | 0 | 0.307 |
| 39155 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | HACE1 | Sponge network | -0.709 | 0.18168 | -0.072 | 0.55239 | 0.307 |
| 39156 | LINC00672 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-532-3p;hsa-miR-93-3p | 14 | RPS6KA2 | Sponge network | -0.227 | 0.31657 | -0.57 | 0.00013 | 0.307 |
| 39157 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30b-3p;hsa-miR-370-3p;hsa-miR-589-5p | 11 | RPS6KA2 | Sponge network | -0.187 | 0.23787 | -0.57 | 0.00013 | 0.307 |
| 39158 | LINC00667 |
hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | PAK3 | Sponge network | -0.235 | 0.15142 | -0.628 | 0.07253 | 0.307 |
| 39159 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-590-3p | 10 | RB1CC1 | Sponge network | 0.174 | 0.30034 | 0.175 | 0.12559 | 0.307 |
| 39160 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 24 | MYO9A | Sponge network | -0.08 | 0.90092 | -0.147 | 0.32373 | 0.307 |
| 39161 | RP11-1277A3.1 |
hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-92a-3p | 13 | ADAMTSL3 | Sponge network | 0.453 | 0.01839 | -1.559 | 0 | 0.307 |
| 39162 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | FAM172A | Sponge network | 0.61 | 0.01593 | -0.57 | 0 | 0.307 |
| 39163 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | MYO9A | Sponge network | -1.057 | 0.00029 | -0.147 | 0.32373 | 0.307 |
| 39164 | BOLA3-AS1 |
hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-429 | 13 | ARHGAP29 | Sponge network | -0.246 | 0.35034 | 0.131 | 0.42953 | 0.307 |
| 39165 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CACNA2D1 | Sponge network | -0.125 | 0.49414 | -0.846 | 0.00675 | 0.307 |
| 39166 | RP11-760H22.2 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-454-3p | 22 | DIP2C | Sponge network | 0.001 | 0.99753 | -0.436 | 0.01713 | 0.307 |
| 39167 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | NF1 | Sponge network | 0.603 | 0.00127 | 0.51 | 0 | 0.307 |
| 39168 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 15 | AKAP6 | Sponge network | -0.418 | 0.00068 | -1.586 | 3.0E-5 | 0.307 |
| 39169 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-590-3p;hsa-miR-654-3p | 10 | MYBL1 | Sponge network | 2.094 | 0 | 0.494 | 0.02152 | 0.307 |
| 39170 | RP11-490M8.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-539-5p;hsa-miR-96-5p | 21 | LPP | Sponge network | -0.214 | 0.44248 | -0.459 | 0.04475 | 0.307 |
| 39171 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-455-3p | 19 | PRKAB2 | Sponge network | 0.594 | 0.41522 | -0.367 | 0.01371 | 0.307 |
| 39172 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-429 | 12 | PIK3C2A | Sponge network | 1.361 | 0 | 0.061 | 0.53776 | 0.307 |
| 39173 | RP11-13K12.5 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p | 10 | STARD13 | Sponge network | -0.318 | 0.65847 | -0.187 | 0.27383 | 0.307 |
| 39174 | CTC-471J1.2 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p | 11 | SIRT1 | Sponge network | 1.11 | 1.0E-5 | 0.109 | 0.2593 | 0.307 |
| 39175 | LIPE-AS1 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-942-5p | 37 | ARHGEF12 | Sponge network | 0.078 | 0.55803 | 0.09 | 0.35693 | 0.307 |
| 39176 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | EYA4 | Sponge network | 0.349 | 0.01695 | 0.3 | 0.36876 | 0.307 |
| 39177 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TRIP11 | Sponge network | 1.308 | 0 | -0.158 | 0.06384 | 0.307 |
| 39178 | CTD-2554C21.2 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-502-5p | 10 | FMNL3 | Sponge network | -1.654 | 0.00144 | 0.555 | 4.0E-5 | 0.307 |
| 39179 | RP11-656D10.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-7-1-3p | 16 | TACC1 | Sponge network | 0.406 | 0.26236 | -0.362 | 0.05406 | 0.307 |
| 39180 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | ABCC9 | Sponge network | 0.199 | 0.52763 | -0.466 | 0.14121 | 0.307 |
| 39181 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-374b-3p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 18 | TP53INP1 | Sponge network | -0.896 | 0.00099 | -0.496 | 0.00064 | 0.307 |
| 39182 | RP11-968O1.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-484 | 17 | SPRY3 | Sponge network | -0.829 | 0.01343 | 0.016 | 0.91286 | 0.307 |
| 39183 | AC034220.3 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 12 | TAF1 | Sponge network | 0.309 | 0.07304 | 0.39 | 3.0E-5 | 0.307 |
| 39184 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 22 | PRKAB2 | Sponge network | -0.13 | 0.48734 | -0.367 | 0.01371 | 0.307 |
| 39185 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | SOX5 | Sponge network | -0.235 | 0.15142 | -1.091 | 4.0E-5 | 0.307 |
| 39186 | RASSF8-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | FAM117A | Sponge network | 0.335 | 0.20596 | -1.379 | 0 | 0.307 |
| 39187 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 10 | ALDH1A2 | Sponge network | -0.969 | 2.0E-5 | -2.411 | 0 | 0.307 |
| 39188 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-654-3p;hsa-miR-940;hsa-miR-942-5p | 30 | HLF | Sponge network | 0.177 | 0.16681 | -2.443 | 0 | 0.307 |
| 39189 | LINC01018 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-423-5p;hsa-miR-455-3p | 15 | ABL1 | Sponge network | -2.915 | 0.00015 | -0.215 | 0.07804 | 0.307 |
| 39190 | AC004540.4 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-362-3p | 15 | ZFHX4 | Sponge network | -2.549 | 0.00528 | 0.018 | 0.95574 | 0.307 |
| 39191 | TRAF3IP2-AS1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | SIAH1 | Sponge network | -0.323 | 0.01372 | -0.057 | 0.41415 | 0.307 |
| 39192 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p | 41 | IL6ST | Sponge network | 0.921 | 0.00118 | -0.402 | 0.00676 | 0.307 |
| 39193 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | ZFP36L1 | Sponge network | -0.323 | 0.01372 | -0.505 | 8.0E-5 | 0.307 |
| 39194 | AC005618.6 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-5p | 12 | FGFR1 | Sponge network | 0.862 | 0.06213 | -0.509 | 0.03957 | 0.307 |
| 39195 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p | 18 | ETV1 | Sponge network | -0.583 | 0.14589 | -0.096 | 0.67674 | 0.307 |
| 39196 | IQCH-AS1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-9-5p | 16 | CDK6 | Sponge network | 0.929 | 0 | 0.991 | 0 | 0.307 |
| 39197 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 15 | YAP1 | Sponge network | 0.214 | 0.18246 | 0.22 | 0.11836 | 0.307 |
| 39198 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 13 | LRRC7 | Sponge network | 0.225 | 0.30644 | -1.341 | 0.01193 | 0.307 |
| 39199 | RP11-212P7.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-744-3p;hsa-miR-744-5p | 18 | IGF2 | Sponge network | 0.219 | 0.1516 | 0.848 | 0.03842 | 0.307 |
| 39200 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 17 | TGFBR2 | Sponge network | 1.302 | 7.0E-5 | -0.243 | 0.11408 | 0.307 |
| 39201 | RP1-59D14.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-345-5p | 15 | ADARB1 | Sponge network | 1.162 | 5.0E-5 | -0.335 | 0.06631 | 0.307 |
| 39202 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | NCAM2 | Sponge network | -0.899 | 6.0E-5 | -0.482 | 0.22565 | 0.307 |
| 39203 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p | 16 | GPC6 | Sponge network | 0.299 | 0.57722 | -0.054 | 0.81465 | 0.307 |
| 39204 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | EGFR | Sponge network | 0.72 | 0.0001 | -0.175 | 0.48451 | 0.307 |
| 39205 | KB-1460A1.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-362-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | NRXN3 | Sponge network | 0.603 | 0.00127 | -0.893 | 0.04186 | 0.307 |
| 39206 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 44 | AFF4 | Sponge network | -0.176 | 0.59692 | -0.266 | 0.01565 | 0.307 |
| 39207 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | RELN | Sponge network | -0.899 | 6.0E-5 | -2.403 | 0 | 0.307 |
| 39208 | RP4-605O3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | RICTOR | Sponge network | 0.262 | 0.0475 | 0.388 | 0.00188 | 0.307 |
| 39209 | RP11-620J15.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 29 | CBX5 | Sponge network | -0.739 | 0.00044 | 0.165 | 0.24446 | 0.307 |
| 39210 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-378a-5p | 10 | PDS5B | Sponge network | 0.79 | 0 | 0.259 | 0.00962 | 0.307 |
| 39211 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 30 | AKAP11 | Sponge network | -0.303 | 0.21002 | 0.196 | 0.09825 | 0.307 |
| 39212 | RP11-686D22.8 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | PHC3 | Sponge network | 0.898 | 1.0E-5 | 0.212 | 0.0499 | 0.307 |
| 39213 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-577 | 18 | KIT | Sponge network | -0.18 | 0.20343 | -2.184 | 0 | 0.307 |
| 39214 | CALML3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 19 | WWTR1 | Sponge network | 0.95 | 0.15943 | -0.345 | 0.11997 | 0.307 |
| 39215 | RP11-758N13.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-342-5p | 17 | DTNA | Sponge network | 2.939 | 3.0E-5 | -1.648 | 1.0E-5 | 0.307 |
| 39216 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-93-3p;hsa-miR-96-5p | 18 | EBF3 | Sponge network | -0.18 | 0.20343 | -0.058 | 0.81437 | 0.307 |
| 39217 | TPTEP1 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | EDNRB | Sponge network | -1.216 | 0 | -0.682 | 0.00138 | 0.307 |
| 39218 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-455-3p | 11 | TRIM3 | Sponge network | -0.896 | 0.00099 | -0.577 | 0 | 0.307 |
| 39219 | RP11-25K19.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-3913-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | AFF1 | Sponge network | -1.057 | 0.00029 | -0.384 | 0.00053 | 0.307 |
| 39220 | LINC00865 |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-940 | 13 | JUN | Sponge network | -1.249 | 7.0E-5 | -0.932 | 0 | 0.307 |
| 39221 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-5p | 14 | CAV1 | Sponge network | 0.214 | 0.18246 | -1.013 | 6.0E-5 | 0.307 |
| 39222 | RP11-473I1.5 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p | 26 | IL6ST | Sponge network | 0.542 | 0.00054 | -0.402 | 0.00676 | 0.307 |
| 39223 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | CACNA2D1 | Sponge network | -1.349 | 0.00017 | -0.846 | 0.00675 | 0.307 |
| 39224 | RP11-46C24.7 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-942-5p | 11 | ZBTB20 | Sponge network | 0.392 | 0.0278 | -0.122 | 0.57569 | 0.307 |
| 39225 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p | 20 | RICTOR | Sponge network | 0.903 | 0 | 0.388 | 0.00188 | 0.307 |
| 39226 | ZNF667-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | PPM1L | Sponge network | -0.976 | 0.00025 | -0.537 | 0.00616 | 0.307 |
| 39227 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p | 10 | ZYX | Sponge network | 0.214 | 0.18246 | 0.019 | 0.89763 | 0.307 |
| 39228 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 18 | GRIN2A | Sponge network | 0.199 | 0.38015 | -2.161 | 0 | 0.307 |
| 39229 | ARHGAP5-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-542-3p;hsa-miR-942-5p | 25 | HLF | Sponge network | -0.644 | 0.00021 | -2.443 | 0 | 0.307 |
| 39230 | RP11-620J15.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-361-5p;hsa-miR-590-3p | 30 | CREB3L2 | Sponge network | -0.739 | 0.00044 | -0.525 | 2.0E-5 | 0.307 |
| 39231 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 14 | FREM2 | Sponge network | 0.765 | 7.0E-5 | -0.437 | 0.46353 | 0.307 |
| 39232 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-96-5p | 23 | BNC2 | Sponge network | -0.326 | 0.14314 | -0.491 | 0.09988 | 0.307 |
| 39233 | AF131215.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 22 | ZFX | Sponge network | -0.176 | 0.59692 | 0.09 | 0.42563 | 0.307 |
| 39234 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-7-1-3p | 10 | CNTN5 | Sponge network | -1.264 | 6.0E-5 | -0.219 | 0.61957 | 0.307 |
| 39235 | MIR22HG |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-590-5p | 14 | TMEFF2 | Sponge network | -1.047 | 0 | -2.426 | 0 | 0.307 |
| 39236 | PART1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | PRKCB | Sponge network | -2.925 | 0 | -1.313 | 2.0E-5 | 0.307 |
| 39237 | PART1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 13 | THSD7A | Sponge network | -2.925 | 0 | 0.242 | 0.25154 | 0.307 |
| 39238 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-874-3p | 18 | ARID5B | Sponge network | 0.946 | 0 | 0.342 | 0.01676 | 0.307 |
| 39239 | BMS1P20 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | UBR3 | Sponge network | 0.34 | 0.00273 | -0.021 | 0.82774 | 0.307 |
| 39240 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p | 27 | SNX29 | Sponge network | -0.513 | 0.04703 | -0.34 | 0.01371 | 0.307 |
| 39241 | CTC-296K1.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-345-5p | 18 | PRDM2 | Sponge network | -2.095 | 0.00112 | -0.258 | 0.00721 | 0.307 |
| 39242 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PTPRB | Sponge network | 0.657 | 4.0E-5 | 0.105 | 0.47122 | 0.307 |
| 39243 | TRAM2-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 17 | TCF12 | Sponge network | 0.336 | 0.00739 | 0.174 | 0.13271 | 0.307 |
| 39244 | ZNF667-AS1 |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429 | 10 | CADM4 | Sponge network | -0.976 | 0.00025 | -0.635 | 0.00305 | 0.307 |
| 39245 | LINC00969 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | AKAP11 | Sponge network | 0.683 | 1.0E-5 | 0.196 | 0.09825 | 0.307 |
| 39246 | C20orf166-AS1 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429 | 16 | FAM135B | Sponge network | -2.69 | 0.0001 | -3.978 | 0 | 0.307 |
| 39247 | LINC00476 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | LIMCH1 | Sponge network | -0.615 | 0.00015 | -0.915 | 1.0E-5 | 0.307 |
| 39248 | RP4-717I23.3 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-654-3p | 17 | DICER1 | Sponge network | 0.637 | 0.00069 | 0.042 | 0.674 | 0.307 |
| 39249 | RP11-983P16.4 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | PEG3 | Sponge network | 0.41 | 0.02214 | -1.74 | 0 | 0.307 |
| 39250 | TRIM52-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-629-5p | 12 | CRTC1 | Sponge network | -0.418 | 0.00068 | -0.116 | 0.28412 | 0.307 |
| 39251 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | SCN9A | Sponge network | -1.886 | 0 | -0.525 | 0.10593 | 0.307 |
| 39252 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p | 12 | ARID2 | Sponge network | 1.254 | 0 | 0.235 | 0.02697 | 0.307 |
| 39253 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 29 | THBS1 | Sponge network | -0.323 | 0.01372 | 0.741 | 0.00386 | 0.307 |
| 39254 | ARHGEF26-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | JDP2 | Sponge network | -2.46 | 0 | -0.967 | 0 | 0.307 |
| 39255 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 25 | HECA | Sponge network | 0.532 | 0.00505 | -0.217 | 0.03463 | 0.307 |
| 39256 | RP11-195F19.9 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p | 11 | PTPN14 | Sponge network | -0.726 | 0.00132 | 0.144 | 0.34595 | 0.307 |
| 39257 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | ZFHX4 | Sponge network | 0.998 | 0 | 0.018 | 0.95574 | 0.307 |
| 39258 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 11 | LHFP | Sponge network | 0.083 | 0.66405 | -0.167 | 0.41371 | 0.307 |
| 39259 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | BMPR2 | Sponge network | 0.272 | 0.44692 | 0.206 | 0.0635 | 0.307 |
| 39260 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | USP12 | Sponge network | 0.545 | 0.00064 | -0.102 | 0.40768 | 0.307 |
| 39261 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ADAMTSL3 | Sponge network | -0.053 | 0.80685 | -1.559 | 0 | 0.307 |
| 39262 | SNHG5 |
hsa-let-7e-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-452-5p | 16 | COPS2 | Sponge network | 0.13 | 0.47174 | -0.178 | 0.04974 | 0.307 |
| 39263 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-935 | 45 | PTPRD | Sponge network | -0.738 | 0.21233 | -1.005 | 0.00064 | 0.307 |
| 39264 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | EBF3 | Sponge network | -0.727 | 0.04726 | -0.058 | 0.81437 | 0.307 |
| 39265 | FENDRR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 52 | PTPRD | Sponge network | -1.281 | 0.00012 | -1.005 | 0.00064 | 0.307 |
| 39266 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | ARID5B | Sponge network | 0.064 | 0.72934 | 0.342 | 0.01676 | 0.307 |
| 39267 | C4B-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-96-5p | 13 | ABI2 | Sponge network | 2.306 | 9.0E-5 | 0.175 | 0.14787 | 0.307 |
| 39268 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | LRRC55 | Sponge network | -0.187 | 0.23787 | -0.026 | 0.93163 | 0.307 |
| 39269 | RP11-298J20.4 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p | 11 | IL17RD | Sponge network | 0.002 | 0.99057 | -0.019 | 0.94873 | 0.307 |
| 39270 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ZFHX4 | Sponge network | 0.727 | 3.0E-5 | 0.018 | 0.95574 | 0.307 |
| 39271 | RP11-212P7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 28 | NCOA1 | Sponge network | 0.219 | 0.1516 | -0.432 | 0 | 0.307 |
| 39272 | RP11-35G9.3 |
hsa-let-7d-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | PRDM2 | Sponge network | 0.657 | 4.0E-5 | -0.258 | 0.00721 | 0.307 |
| 39273 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-744-3p | 15 | CUL5 | Sponge network | 0.765 | 7.0E-5 | 0.026 | 0.77301 | 0.307 |
| 39274 | LINC00476 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 12 | FYN | Sponge network | -0.615 | 0.00015 | -0.131 | 0.37051 | 0.307 |
| 39275 | AC103563.8 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-500a-3p | 18 | DST | Sponge network | -7.551 | 0 | -0.713 | 0.00096 | 0.307 |
| 39276 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-3607-3p | 13 | USP6 | Sponge network | 0.411 | 0.1335 | 0.188 | 0.3871 | 0.307 |
| 39277 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 24 | PRKAB2 | Sponge network | 0.41 | 0.02214 | -0.367 | 0.01371 | 0.307 |
| 39278 | RP11-597D13.8 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p | 11 | LPP | Sponge network | 0.936 | 0.03889 | -0.459 | 0.04475 | 0.307 |
| 39279 | RP5-1198O20.4 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | ID4 | Sponge network | 0.997 | 0.00066 | -1.644 | 0 | 0.307 |
| 39280 | SNHG1 |
hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-497-5p | 10 | CDK6 | Sponge network | 1.265 | 0 | 0.991 | 0 | 0.307 |
| 39281 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-671-5p | 39 | ADARB1 | Sponge network | 0.395 | 0.15451 | -0.335 | 0.06631 | 0.307 |
| 39282 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p | 50 | AFF4 | Sponge network | -0.842 | 0.01361 | -0.266 | 0.01565 | 0.307 |
| 39283 | FAM66C |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 37 | AFF1 | Sponge network | -0.901 | 0.00019 | -0.384 | 0.00053 | 0.307 |
| 39284 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NOTCH2NL | Sponge network | -1.161 | 0.00591 | 0.024 | 0.84307 | 0.307 |
| 39285 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | IL6ST | Sponge network | 0.545 | 0.00064 | -0.402 | 0.00676 | 0.307 |
| 39286 | RP11-767N6.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-576-5p | 15 | RICTOR | Sponge network | 0.466 | 0.00155 | 0.388 | 0.00188 | 0.307 |
| 39287 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-576-5p | 17 | PBX1 | Sponge network | 0.663 | 0.00073 | -1.206 | 0 | 0.307 |
| 39288 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-326;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TBX18 | Sponge network | -0.053 | 0.80685 | 0.843 | 0.02945 | 0.307 |
| 39289 | CTD-3020H12.4 |
hsa-miR-10a-3p;hsa-miR-1266-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-628-5p | 10 | LPP | Sponge network | 1.874 | 0.00389 | -0.459 | 0.04475 | 0.307 |
| 39290 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | NIN | Sponge network | 0.919 | 0 | -0.253 | 0.13794 | 0.307 |
| 39291 | LINC00886 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | -0.371 | 0.24604 | -1.751 | 1.0E-5 | 0.307 |
| 39292 | RP11-195F19.9 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-539-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | PRKCB | Sponge network | -0.726 | 0.00132 | -1.313 | 2.0E-5 | 0.307 |
| 39293 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p | 17 | LRRC7 | Sponge network | 0.082 | 0.55397 | -1.341 | 0.01193 | 0.307 |
| 39294 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 32 | KIF1B | Sponge network | 0.959 | 0 | -0.118 | 0.32258 | 0.307 |
| 39295 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 58 | KIF1B | Sponge network | -0.303 | 0.21002 | -0.118 | 0.32258 | 0.307 |
| 39296 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | PTGFR | Sponge network | 0.862 | 0.06213 | -0.233 | 0.45421 | 0.307 |
| 39297 | SOX2-OT |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 14 | SPG20 | Sponge network | -1.264 | 6.0E-5 | -1.16 | 0 | 0.307 |
| 39298 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p | 27 | MCC | Sponge network | 0.95 | 0.15943 | -1.005 | 0 | 0.307 |
| 39299 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 11 | DAB2 | Sponge network | -0.513 | 0.04703 | 0.431 | 0.0113 | 0.307 |
| 39300 | CTD-2017D11.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-628-5p | 27 | DTNA | Sponge network | 0.792 | 0.0049 | -1.648 | 1.0E-5 | 0.307 |
| 39301 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 18 | GPC6 | Sponge network | -1.886 | 0 | -0.054 | 0.81465 | 0.307 |
| 39302 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-664a-5p;hsa-miR-940 | 28 | TRPS1 | Sponge network | 0.082 | 0.55397 | -0.191 | 0.44109 | 0.307 |
| 39303 | AGAP11 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-5p | 23 | CYLD | Sponge network | -1.645 | 0 | -0.367 | 0.00171 | 0.307 |
| 39304 | RP11-701H24.3 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 13 | THRB | Sponge network | -1.425 | 0.04204 | -1.117 | 0.0004 | 0.307 |
| 39305 | LINC00092 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-323a-3p;hsa-miR-345-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 32 | DST | Sponge network | -1.886 | 0 | -0.713 | 0.00096 | 0.307 |
| 39306 | KB-1410C5.5 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 13 | DTNA | Sponge network | 0.959 | 0 | -1.648 | 1.0E-5 | 0.307 |
| 39307 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | TCF4 | Sponge network | 0.614 | 1.0E-5 | 0.016 | 0.92271 | 0.307 |
| 39308 | FAM157C |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p | 10 | GSK3B | Sponge network | 0.91 | 0 | 0.266 | 0.00045 | 0.307 |
| 39309 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | YAP1 | Sponge network | 0.538 | 0.00076 | 0.22 | 0.11836 | 0.307 |
| 39310 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-429 | 14 | PRKAR1A | Sponge network | -0.246 | 0.35034 | -0.063 | 0.50314 | 0.307 |
| 39311 | RP11-680C21.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 32 | PPP1R9A | Sponge network | -1.424 | 0.04346 | -0.903 | 0.04254 | 0.307 |
| 39312 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 22 | NUAK1 | Sponge network | -0.727 | 0.04726 | 0.904 | 0 | 0.307 |
| 39313 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 20 | EPHA3 | Sponge network | 0.401 | 0.00066 | 0.185 | 0.53588 | 0.307 |
| 39314 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 28 | TSC1 | Sponge network | -0.615 | 0.00015 | 0.103 | 0.26071 | 0.307 |
| 39315 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p | 13 | ZNF536 | Sponge network | -0.969 | 2.0E-5 | -3.115 | 0 | 0.307 |
| 39316 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p | 10 | MAFB | Sponge network | 0.796 | 4.0E-5 | -0.023 | 0.89865 | 0.307 |
| 39317 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | -0.942 | 0 | 0.783 | 0.00072 | 0.307 |
| 39318 | RP11-158K1.3 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 23 | DLC1 | Sponge network | 0.349 | 0.01695 | -0.382 | 0.05038 | 0.306 |
| 39319 | LINC01018 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | ABCA10 | Sponge network | -2.915 | 0.00015 | -0.571 | 0.03674 | 0.306 |
| 39320 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p | 25 | DTNA | Sponge network | -0.285 | 0.2172 | -1.648 | 1.0E-5 | 0.306 |
| 39321 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | HECA | Sponge network | 0.657 | 4.0E-5 | -0.217 | 0.03463 | 0.306 |
| 39322 | RP11-389C8.2 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-590-3p | 11 | PTCH1 | Sponge network | 0.727 | 3.0E-5 | -0.1 | 0.6179 | 0.306 |
| 39323 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | NR4A3 | Sponge network | -2.46 | 0 | -1.385 | 0 | 0.306 |
| 39324 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p | 36 | RASSF2 | Sponge network | -0.222 | 0.32445 | -0.273 | 0.21961 | 0.306 |
| 39325 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | EPHA3 | Sponge network | 0.336 | 0.00739 | 0.185 | 0.53588 | 0.306 |
| 39326 | MRVI1-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p | 19 | CADM1 | Sponge network | -1.092 | 4.0E-5 | -0.9 | 0.00084 | 0.306 |
| 39327 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 26 | LRRC7 | Sponge network | 0.083 | 0.66405 | -1.341 | 0.01193 | 0.306 |
| 39328 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-877-5p;hsa-miR-96-5p | 20 | CBFA2T3 | Sponge network | 0.101 | 0.60919 | -1.221 | 1.0E-5 | 0.306 |
| 39329 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p | 13 | TMTC3 | Sponge network | 1.923 | 0 | 0.123 | 0.22189 | 0.306 |
| 39330 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p | 13 | PPP2R5C | Sponge network | -0.726 | 0.00132 | -0.314 | 3.0E-5 | 0.306 |
| 39331 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | PRKAA1 | Sponge network | 0.768 | 7.0E-5 | 0.317 | 0.0031 | 0.306 |
| 39332 | RP11-46C24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p | 43 | RUNX1T1 | Sponge network | 0.392 | 0.0278 | -0.344 | 0.2374 | 0.306 |
| 39333 | MIR600HG |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-96-5p | 13 | DOCK4 | Sponge network | 0.594 | 0.41522 | 0.502 | 0.00108 | 0.306 |
| 39334 | AC127904.2 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 16 | UBR3 | Sponge network | 1.308 | 0 | -0.021 | 0.82774 | 0.306 |
| 39335 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | TRPS1 | Sponge network | -0.899 | 6.0E-5 | -0.191 | 0.44109 | 0.306 |
| 39336 | LINC00094 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 53 | RUNX1T1 | Sponge network | 0.253 | 0.09565 | -0.344 | 0.2374 | 0.306 |
| 39337 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-708-3p | 16 | PRKAB2 | Sponge network | -0.285 | 0.2172 | -0.367 | 0.01371 | 0.306 |
| 39338 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-935 | 11 | ACSL6 | Sponge network | -1.483 | 9.0E-5 | -1.593 | 0 | 0.306 |
| 39339 | RP11-65J21.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-628-5p | 13 | ERC1 | Sponge network | -0.557 | 0.11135 | -0.108 | 0.38794 | 0.306 |
| 39340 | LINC00476 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-766-3p | 48 | MDM4 | Sponge network | -0.615 | 0.00015 | 0.345 | 0.00762 | 0.306 |
| 39341 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-590-3p | 15 | ARHGAP29 | Sponge network | 0.559 | 0.00026 | 0.131 | 0.42953 | 0.306 |
| 39342 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | THRB | Sponge network | -1.057 | 0.00029 | -1.117 | 0.0004 | 0.306 |
| 39343 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | MOB1B | Sponge network | -0.342 | 0.02288 | 0.197 | 0.15266 | 0.306 |
| 39344 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-361-3p;hsa-miR-590-3p | 13 | BCL6 | Sponge network | -0.053 | 0.80685 | -0.211 | 0.19489 | 0.306 |
| 39345 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-500a-5p | 12 | MAF | Sponge network | 0.811 | 0.03228 | -1.257 | 0 | 0.306 |
| 39346 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 51 | RUNX1T1 | Sponge network | 0.72 | 0.0001 | -0.344 | 0.2374 | 0.306 |
| 39347 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | 0.374 | 0.20054 | -1.047 | 0.00364 | 0.306 |
| 39348 | RP11-867G23.10 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-502-5p;hsa-miR-589-3p | 11 | TET1 | Sponge network | -2.531 | 0 | -0.135 | 0.52138 | 0.306 |
| 39349 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-942-5p | 20 | GPC6 | Sponge network | -0.842 | 0.01361 | -0.054 | 0.81465 | 0.306 |
| 39350 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-874-3p;hsa-miR-93-3p | 23 | CTIF | Sponge network | -0.469 | 0.08721 | -0.389 | 0.00549 | 0.306 |
| 39351 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | MCC | Sponge network | 1.757 | 0 | -1.005 | 0 | 0.306 |
| 39352 | ARHGAP5-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-369-3p;hsa-miR-532-5p | 27 | DST | Sponge network | -0.644 | 0.00021 | -0.713 | 0.00096 | 0.306 |
| 39353 | MAGI2-AS3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-940 | 31 | EPS15 | Sponge network | -0.129 | 0.57177 | -0.052 | 0.53998 | 0.306 |
| 39354 | CTD-2260A17.1 |
hsa-miR-125a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-550a-5p | 10 | CCDC6 | Sponge network | 0.518 | 0.01048 | 0.27 | 0.02555 | 0.306 |
| 39355 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 53 | KIF1B | Sponge network | 0.61 | 0.01593 | -0.118 | 0.32258 | 0.306 |
| 39356 | RP11-389C8.2 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PCDH9 | Sponge network | 0.727 | 3.0E-5 | -1.823 | 4.0E-5 | 0.306 |
| 39357 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | DIP2C | Sponge network | -0.206 | 0.12942 | -0.436 | 0.01713 | 0.306 |
| 39358 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 25 | AKAP11 | Sponge network | 0.619 | 0.00157 | 0.196 | 0.09825 | 0.306 |
| 39359 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 15 | MITF | Sponge network | 0.497 | 0.01451 | -0.769 | 0.00022 | 0.306 |
| 39360 | RP11-574K11.29 |
hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-654-3p | 10 | BAZ2B | Sponge network | 0.997 | 0 | -0.276 | 0.02833 | 0.306 |
| 39361 | C20orf166-AS1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-577 | 16 | RNF144A | Sponge network | -2.69 | 0.0001 | 0.594 | 0.0009 | 0.306 |
| 39362 | OSER1-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-582-5p | 10 | CELF4 | Sponge network | -0.13 | 0.48734 | -1.135 | 0.00344 | 0.306 |
| 39363 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ZFHX3 | Sponge network | 0.67 | 0.00048 | 0.389 | 0.01363 | 0.306 |
| 39364 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-940 | 31 | MCC | Sponge network | 0.082 | 0.55654 | -1.005 | 0 | 0.306 |
| 39365 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | CACNA2D1 | Sponge network | 0.174 | 0.30034 | -0.846 | 0.00675 | 0.306 |
| 39366 | AC005154.6 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 19 | DICER1 | Sponge network | 0.848 | 0 | 0.042 | 0.674 | 0.306 |
| 39367 | RP11-428J1.5 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | ABI2 | Sponge network | 0.538 | 0.00076 | 0.175 | 0.14787 | 0.306 |
| 39368 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p | 23 | HECA | Sponge network | -1.047 | 0 | -0.217 | 0.03463 | 0.306 |
| 39369 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-93-5p | 13 | LHX6 | Sponge network | 0.101 | 0.60919 | -0.087 | 0.69193 | 0.306 |
| 39370 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-330-3p;hsa-miR-33a-3p | 13 | CLASP2 | Sponge network | 2.163 | 0 | -0.038 | 0.71 | 0.306 |
| 39371 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | EBF3 | Sponge network | -0.899 | 6.0E-5 | -0.058 | 0.81437 | 0.306 |
| 39372 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 23 | ADAMTSL3 | Sponge network | 0.61 | 0.01593 | -1.559 | 0 | 0.306 |
| 39373 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | DCLK1 | Sponge network | 0.219 | 0.1516 | -1.324 | 0.00014 | 0.306 |
| 39374 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-25-3p | 10 | MYCBP2 | Sponge network | -0.611 | 0.09607 | -0.027 | 0.82016 | 0.306 |
| 39375 | PART1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 24 | KANK1 | Sponge network | -2.925 | 0 | -0.55 | 0.00134 | 0.306 |
| 39376 | AC018890.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 1.61 | 0.00961 | 0.943 | 0 | 0.306 |
| 39377 | DNM3OS |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 12 | EHD3 | Sponge network | 0.572 | 0.01722 | -0.484 | 0.01747 | 0.306 |
| 39378 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 22 | DOCK4 | Sponge network | 0.082 | 0.55654 | 0.502 | 0.00108 | 0.306 |
| 39379 | RP11-497H16.9 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | CCDC6 | Sponge network | 0.545 | 0.00064 | 0.27 | 0.02555 | 0.306 |
| 39380 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-429 | 12 | KIF1B | Sponge network | 2.364 | 0.00134 | -0.118 | 0.32258 | 0.306 |
| 39381 | RP11-464F9.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | BRD3 | Sponge network | 0.959 | 0 | 0.37 | 0.00013 | 0.306 |
| 39382 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-96-5p | 20 | HIP1 | Sponge network | -1.349 | 0.00017 | 0.774 | 0 | 0.306 |
| 39383 | LINC01006 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-624-5p | 11 | PCDH8 | Sponge network | -0.702 | 0.04814 | -0.997 | 0.05366 | 0.306 |
| 39384 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 27 | ST6GAL2 | Sponge network | 0.056 | 0.81195 | -1.201 | 0.00597 | 0.306 |
| 39385 | EMX2OS |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-7-5p | 10 | COL1A2 | Sponge network | -0.777 | 0.1134 | 1.991 | 0 | 0.306 |
| 39386 | CTC-487M23.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-92a-3p | 18 | IL17RD | Sponge network | 0.912 | 0.00014 | -0.019 | 0.94873 | 0.306 |
| 39387 | RP11-57H14.4 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 17 | SORCS1 | Sponge network | 0.083 | 0.66405 | -3.492 | 0 | 0.306 |
| 39388 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 18 | ARID5B | Sponge network | 1.04 | 0.00023 | 0.342 | 0.01676 | 0.306 |
| 39389 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429 | 15 | RNASEL | Sponge network | -1.057 | 0.00029 | -0.272 | 0.01796 | 0.306 |
| 39390 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | WDFY3 | Sponge network | -0.726 | 0.00132 | -0.201 | 0.0967 | 0.306 |
| 39391 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 21 | ST6GALNAC3 | Sponge network | 0.199 | 0.38015 | -0.987 | 0 | 0.306 |
| 39392 | AC016747.3 |
hsa-miR-103a-2-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | EFNA5 | Sponge network | 0.199 | 0.38015 | -0.421 | 0.17868 | 0.306 |
| 39393 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 18 | CXXC4 | Sponge network | -0.227 | 0.31657 | -0.433 | 0.21055 | 0.306 |
| 39394 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | TACC1 | Sponge network | -0.13 | 0.48734 | -0.362 | 0.05406 | 0.306 |
| 39395 | RP5-1085F17.3 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 23 | CHD5 | Sponge network | 0.541 | 0.00125 | -0.922 | 0.01521 | 0.306 |
| 39396 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 30 | MYO9A | Sponge network | -0.769 | 0.00035 | -0.147 | 0.32373 | 0.306 |
| 39397 | AGAP11 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p | 13 | BAZ2B | Sponge network | -1.645 | 0 | -0.276 | 0.02833 | 0.306 |
| 39398 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-340-5p | 14 | SLC6A15 | Sponge network | 0.331 | 0.57247 | -2.373 | 2.0E-5 | 0.306 |
| 39399 | CTD-3138B18.6 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | ARHGEF12 | Sponge network | 2.248 | 0 | 0.09 | 0.35693 | 0.306 |
| 39400 | RP11-216B9.6 |
hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 15 | GLI3 | Sponge network | 0.174 | 0.30034 | -0.615 | 0.01482 | 0.306 |
| 39401 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | FGF5 | Sponge network | -0.222 | 0.32445 | 0.549 | 0.28659 | 0.306 |
| 39402 | RP11-597D13.9 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZMYND11 | Sponge network | 1.302 | 7.0E-5 | -0.2 | 0.05449 | 0.306 |
| 39403 | TINCR |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-942-5p | 10 | RB1CC1 | Sponge network | -0.664 | 0.14543 | 0.175 | 0.12559 | 0.306 |
| 39404 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 15 | PKNOX1 | Sponge network | 0.082 | 0.55654 | 0.085 | 0.28917 | 0.306 |
| 39405 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p | 13 | PREX2 | Sponge network | -0.405 | 0.22013 | -0.272 | 0.25382 | 0.306 |
| 39406 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-93-5p | 27 | BNC2 | Sponge network | -0.663 | 0.10887 | -0.491 | 0.09988 | 0.306 |
| 39407 | CTD-2006H14.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | KDSR | Sponge network | 1.378 | 0 | 0.239 | 0.02216 | 0.306 |
| 39408 | PSMG3-AS1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | NRXN3 | Sponge network | -0.125 | 0.49414 | -0.893 | 0.04186 | 0.306 |
| 39409 | FGD5-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p | 23 | AFF1 | Sponge network | 0.401 | 0.00066 | -0.384 | 0.00053 | 0.306 |
| 39410 | ARHGEF26-AS1 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 10 | EPB41L3 | Sponge network | -2.46 | 0 | -1.193 | 0 | 0.306 |
| 39411 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-214-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 14 | TBL1XR1 | Sponge network | 0.222 | 0.29133 | 0.578 | 0 | 0.306 |
| 39412 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-539-5p | 18 | MITF | Sponge network | -0.465 | 0.04703 | -0.769 | 0.00022 | 0.306 |
| 39413 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LRRK2 | Sponge network | 0.064 | 0.72934 | -1.136 | 1.0E-5 | 0.306 |
| 39414 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | SCN9A | Sponge network | 0.603 | 0.00127 | -0.525 | 0.10593 | 0.306 |
| 39415 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-375;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 29 | THBS1 | Sponge network | 0.246 | 0.59912 | 0.741 | 0.00386 | 0.306 |
| 39416 | FENDRR |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | MED12L | Sponge network | -1.281 | 0.00012 | -0.194 | 0.55299 | 0.306 |
| 39417 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 14 | TP53INP1 | Sponge network | 0.727 | 3.0E-5 | -0.496 | 0.00064 | 0.306 |
| 39418 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-93-5p | 10 | PGR | Sponge network | 1.153 | 0.00114 | -1.704 | 0 | 0.306 |
| 39419 | RP11-104L21.2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-539-5p | 13 | LPP | Sponge network | 0.527 | 0.16506 | -0.459 | 0.04475 | 0.306 |
| 39420 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p | 19 | CADM1 | Sponge network | -1.057 | 0.00029 | -0.9 | 0.00084 | 0.306 |
| 39421 | LINC00702 |
hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30c-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH13 | Sponge network | -0.737 | 0.06182 | 0.634 | 0.00094 | 0.306 |
| 39422 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-582-5p | 19 | ZFHX4 | Sponge network | 0.792 | 0.0049 | 0.018 | 0.95574 | 0.306 |
| 39423 | RP11-727A23.5 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-766-3p | 11 | ATM | Sponge network | 1.117 | 0 | 0.178 | 0.21568 | 0.306 |
| 39424 | CTC-559E9.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p | 13 | NIN | Sponge network | 0.594 | 0.00064 | -0.253 | 0.13794 | 0.306 |
| 39425 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 29 | PRKAB2 | Sponge network | 0.335 | 0.20596 | -0.367 | 0.01371 | 0.306 |
| 39426 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 14 | ABCB5 | Sponge network | 0.082 | 0.55654 | -0.956 | 0.04077 | 0.306 |
| 39427 | LINC00476 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 17 | PDZD2 | Sponge network | -0.615 | 0.00015 | -0.909 | 3.0E-5 | 0.306 |
| 39428 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-3p | 20 | DOCK4 | Sponge network | 0.494 | 0.00339 | 0.502 | 0.00108 | 0.306 |
| 39429 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 32 | MCC | Sponge network | 0.61 | 0.01593 | -1.005 | 0 | 0.306 |
| 39430 | LLNLF-65H9.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-454-3p | 12 | PRKAA2 | Sponge network | -0.033 | 0.89251 | -2.462 | 0 | 0.306 |
| 39431 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p | 11 | ZNF208 | Sponge network | 0.056 | 0.81195 | -0.4 | 0.22381 | 0.306 |
| 39432 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p | 52 | IL6ST | Sponge network | -0.738 | 0.21233 | -0.402 | 0.00676 | 0.306 |
| 39433 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | LRRC7 | Sponge network | 0.064 | 0.72934 | -1.341 | 0.01193 | 0.306 |
| 39434 | CTA-363E19.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-374b-5p | 10 | EPS15 | Sponge network | 1.055 | 0 | -0.052 | 0.53998 | 0.306 |
| 39435 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 50 | THRB | Sponge network | -0.405 | 0.22013 | -1.117 | 0.0004 | 0.306 |
| 39436 | SOX2-OT |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-935 | 38 | DMD | Sponge network | -1.264 | 6.0E-5 | -1.506 | 0 | 0.306 |
| 39437 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 19 | RIMBP2 | Sponge network | -0.738 | 0.21233 | -1.968 | 5.0E-5 | 0.306 |
| 39438 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-940 | 22 | NTRK2 | Sponge network | 0.419 | 0.0319 | -0.736 | 0.06495 | 0.306 |
| 39439 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-590-3p | 16 | DOCK4 | Sponge network | -0.738 | 0.21233 | 0.502 | 0.00108 | 0.306 |
| 39440 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-590-5p;hsa-miR-935 | 23 | EBF1 | Sponge network | -0.384 | 0.40652 | -0.384 | 0.08856 | 0.306 |
| 39441 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-629-5p | 14 | MYO9A | Sponge network | 0.001 | 0.99753 | -0.147 | 0.32373 | 0.306 |
| 39442 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p | 10 | ARID2 | Sponge network | 0.919 | 0 | 0.235 | 0.02697 | 0.306 |
| 39443 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-940 | 17 | THBS1 | Sponge network | -4.546 | 0 | 0.741 | 0.00386 | 0.306 |
| 39444 | RP4-758J18.10 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-378a-5p;hsa-miR-450b-5p | 22 | ADARB1 | Sponge network | -0.091 | 0.71468 | -0.335 | 0.06631 | 0.306 |
| 39445 | HCG11 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | TCF12 | Sponge network | 0.385 | 0.10067 | 0.174 | 0.13271 | 0.306 |
| 39446 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p | 37 | THRB | Sponge network | 0.259 | 0.12542 | -1.117 | 0.0004 | 0.306 |
| 39447 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-501-5p;hsa-miR-93-5p | 12 | TXNIP | Sponge network | -0.668 | 3.0E-5 | -1.277 | 0 | 0.306 |
| 39448 | MAGI2-AS3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | JDP2 | Sponge network | -0.129 | 0.57177 | -0.967 | 0 | 0.306 |
| 39449 | RP11-56L13.1 |
hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-664a-3p | 10 | RASAL2 | Sponge network | -0.992 | 0.18158 | 0.869 | 0 | 0.306 |
| 39450 | HHIP-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 38 | DST | Sponge network | -0.737 | 0.10822 | -0.713 | 0.00096 | 0.306 |
| 39451 | RP11-175O19.4 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p | 12 | MAT2A | Sponge network | 0.917 | 0 | -0.073 | 0.36195 | 0.306 |
| 39452 | RP4-605O3.4 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | EPHA3 | Sponge network | 0.262 | 0.0475 | 0.185 | 0.53588 | 0.306 |
| 39453 | TRAF3IP2-AS1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | UBE2QL1 | Sponge network | -0.323 | 0.01372 | -2.417 | 0 | 0.306 |
| 39454 | LINC00578 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577 | 24 | CREB3L2 | Sponge network | 0.811 | 0.03228 | -0.525 | 2.0E-5 | 0.306 |
| 39455 | RP11-597D13.9 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 21 | ATM | Sponge network | 1.302 | 7.0E-5 | 0.178 | 0.21568 | 0.306 |
| 39456 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-324-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | HACE1 | Sponge network | 0.082 | 0.55654 | -0.072 | 0.55239 | 0.306 |
| 39457 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-590-5p | 11 | PBRM1 | Sponge network | 0.331 | 0.57247 | -0.029 | 0.78601 | 0.306 |
| 39458 | AC141928.1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-484 | 10 | FLNA | Sponge network | 0.853 | 0.15352 | -0.771 | 0.01377 | 0.306 |
| 39459 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | CACNA2D1 | Sponge network | -0.176 | 0.59692 | -0.846 | 0.00675 | 0.306 |
| 39460 | DIO3OS |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-7-5p | 21 | PRKCB | Sponge network | -0.773 | 0.04073 | -1.313 | 2.0E-5 | 0.306 |
| 39461 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 20 | NOTCH2NL | Sponge network | 0.311 | 0.10344 | 0.024 | 0.84307 | 0.306 |
| 39462 | MAP3K14 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 11 | PRKACA | Sponge network | -0.295 | 0.02604 | -0.255 | 0.0012 | 0.306 |
| 39463 | CTD-3138B18.5 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p | 10 | KAT6B | Sponge network | 0.379 | 0.03091 | -0.478 | 0.00018 | 0.306 |
| 39464 | LINC00173 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | TLL1 | Sponge network | 0.056 | 0.8494 | -1.195 | 3.0E-5 | 0.306 |
| 39465 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 39 | TSHZ3 | Sponge network | -0.969 | 2.0E-5 | -0.556 | 0.01516 | 0.306 |
| 39466 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | SOX5 | Sponge network | -0.053 | 0.80685 | -1.091 | 4.0E-5 | 0.306 |
| 39467 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | GRIN2A | Sponge network | 0.61 | 0.01593 | -2.161 | 0 | 0.306 |
| 39468 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-577 | 23 | PHC3 | Sponge network | -0.465 | 0.04703 | 0.212 | 0.0499 | 0.306 |
| 39469 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CXXC4 | Sponge network | 0.392 | 0.0278 | -0.433 | 0.21055 | 0.306 |
| 39470 | AC016747.3 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 18 | PRKAR1A | Sponge network | 0.199 | 0.38015 | -0.063 | 0.50314 | 0.306 |
| 39471 | RP11-373D23.3 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 10 | SH3GLB1 | Sponge network | -0.285 | 0.2172 | -0.115 | 0.14773 | 0.306 |
| 39472 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p | 16 | CDHR3 | Sponge network | 0.176 | 0.37402 | -0.977 | 4.0E-5 | 0.306 |
| 39473 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-96-5p | 25 | ABI2 | Sponge network | -1.654 | 0.00144 | 0.175 | 0.14787 | 0.306 |
| 39474 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-532-5p | 12 | PTPRB | Sponge network | -0.465 | 0.04703 | 0.105 | 0.47122 | 0.306 |
| 39475 | HOTAIRM1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PCDH9 | Sponge network | -0.053 | 0.80685 | -1.823 | 4.0E-5 | 0.306 |
| 39476 | LINC00963 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-940;hsa-miR-96-5p | 23 | MYO9A | Sponge network | 0.523 | 9.0E-5 | -0.147 | 0.32373 | 0.306 |
| 39477 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p | 14 | MYCBP2 | Sponge network | -0.709 | 0.18168 | -0.027 | 0.82016 | 0.306 |
| 39478 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | -0.487 | 0.02157 | 0.783 | 0.00072 | 0.306 |
| 39479 | RP13-516M14.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 21 | NAV3 | Sponge network | 0.389 | 0.04293 | 0.212 | 0.47729 | 0.306 |
| 39480 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 12 | PHC3 | Sponge network | 1.11 | 1.0E-5 | 0.212 | 0.0499 | 0.306 |
| 39481 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p | 25 | ZBTB4 | Sponge network | 0.494 | 0.00339 | -0.739 | 0 | 0.306 |
| 39482 | RP13-516M14.1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 20 | PCDH10 | Sponge network | 0.389 | 0.04293 | -1.644 | 0.0078 | 0.306 |
| 39483 | RP11-65J21.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-532-3p;hsa-miR-629-5p | 20 | DST | Sponge network | -0.557 | 0.11135 | -0.713 | 0.00096 | 0.306 |
| 39484 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-5p | 10 | AKAP12 | Sponge network | 0.41 | 0.02214 | -0.932 | 0.0028 | 0.306 |
| 39485 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | RNF144A | Sponge network | 0.72 | 0.0001 | 0.594 | 0.0009 | 0.306 |
| 39486 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 27 | ZFHX3 | Sponge network | 0.422 | 0.27021 | 0.389 | 0.01363 | 0.306 |
| 39487 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p | 16 | TACC1 | Sponge network | -1.054 | 0.0706 | -0.362 | 0.05406 | 0.306 |
| 39488 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 16 | CREBBP | Sponge network | 0.335 | 0.20596 | 0.126 | 0.15186 | 0.306 |
| 39489 | LINC00861 |
hsa-miR-130b-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-940 | 11 | MDM4 | Sponge network | 0.911 | 0.00854 | 0.345 | 0.00762 | 0.306 |
| 39490 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | TAL1 | Sponge network | -0.612 | 0.00094 | -0.462 | 0.00574 | 0.306 |
| 39491 | KB-1410C5.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 10 | EPS15 | Sponge network | 0.959 | 0 | -0.052 | 0.53998 | 0.306 |
| 39492 | CTD-2314G24.2 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 17 | TES | Sponge network | 0.246 | 0.59912 | -0.348 | 0.00639 | 0.306 |
| 39493 | RP11-35G9.5 |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-96-5p | 10 | RECK | Sponge network | 0.622 | 0.00015 | -1.043 | 0 | 0.306 |
| 39494 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 45 | ERBB4 | Sponge network | -1.439 | 4.0E-5 | -1.975 | 2.0E-5 | 0.306 |
| 39495 | CTB-51J22.1 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-628-5p;hsa-miR-92a-3p | 24 | LPP | Sponge network | -0.803 | 0.23737 | -0.459 | 0.04475 | 0.306 |
| 39496 | HCG11 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | GALNT13 | Sponge network | 0.385 | 0.10067 | -0.857 | 0.07439 | 0.306 |
| 39497 | U91328.19 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 23 | EYA4 | Sponge network | -0.303 | 0.21002 | 0.3 | 0.36876 | 0.306 |
| 39498 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p | 20 | ZFHX3 | Sponge network | -0.663 | 0.10887 | 0.389 | 0.01363 | 0.306 |
| 39499 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | LRRC7 | Sponge network | -4.477 | 0 | -1.341 | 0.01193 | 0.306 |
| 39500 | LINC00672 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p | 17 | NRK | Sponge network | -0.227 | 0.31657 | 0.656 | 0.19217 | 0.306 |
| 39501 | RP11-344B5.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-505-3p;hsa-miR-664a-3p | 13 | ASTN1 | Sponge network | -0.055 | 0.88482 | -2.389 | 2.0E-5 | 0.306 |
| 39502 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 53 | FAT3 | Sponge network | 0.72 | 0.0001 | -1.601 | 0.00051 | 0.306 |
| 39503 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-940 | 15 | HOOK3 | Sponge network | 0.177 | 0.16681 | 0.273 | 0.09957 | 0.306 |
| 39504 | RP11-725P16.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p | 12 | KANK1 | Sponge network | 0.422 | 0.27021 | -0.55 | 0.00134 | 0.306 |
| 39505 | GNG12-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p | 36 | CUX1 | Sponge network | -0.793 | 7.0E-5 | -0.039 | 0.6705 | 0.306 |
| 39506 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 38 | RASSF2 | Sponge network | -0.727 | 0.04726 | -0.273 | 0.21961 | 0.306 |
| 39507 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-708-3p | 17 | MXI1 | Sponge network | 0.001 | 0.99753 | -0.987 | 0 | 0.306 |
| 39508 | HAR1A |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-744-3p;hsa-miR-877-5p | 23 | FAT3 | Sponge network | -0.672 | 0.19447 | -1.601 | 0.00051 | 0.306 |
| 39509 | HCG11 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PTGFR | Sponge network | 0.385 | 0.10067 | -0.233 | 0.45421 | 0.306 |
| 39510 | AF131215.2 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 13 | TAL1 | Sponge network | -0.176 | 0.59692 | -0.462 | 0.00574 | 0.306 |
| 39511 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-940 | 13 | NOTCH2 | Sponge network | -1.255 | 0.00981 | 0.138 | 0.35163 | 0.306 |
| 39512 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3613-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 28 | NTRK3 | Sponge network | 1.153 | 0.00114 | -1.77 | 3.0E-5 | 0.306 |
| 39513 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | FOXO1 | Sponge network | 0.572 | 0.01722 | -0.141 | 0.33874 | 0.306 |
| 39514 | CTC-297N7.9 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p | 10 | MRVI1 | Sponge network | -2.069 | 0 | -1.051 | 0.00022 | 0.306 |
| 39515 | RP11-798M19.6 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | NR3C2 | Sponge network | -0.612 | 0.00094 | -1.56 | 0 | 0.306 |
| 39516 | CALML3-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-664a-3p | 15 | PRUNE2 | Sponge network | 0.95 | 0.15943 | -1.733 | 0.00022 | 0.306 |
| 39517 | DIO3OS |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p | 12 | ERG | Sponge network | -0.773 | 0.04073 | 0.002 | 0.99011 | 0.306 |
| 39518 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | RIMBP2 | Sponge network | -0.096 | 0.67958 | -1.968 | 5.0E-5 | 0.306 |
| 39519 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | MXI1 | Sponge network | -0.738 | 0.21233 | -0.987 | 0 | 0.306 |
| 39520 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 43 | DTNA | Sponge network | -0.125 | 0.49414 | -1.648 | 1.0E-5 | 0.306 |
| 39521 | RP11-35G9.5 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 11 | PIK3CA | Sponge network | 0.622 | 0.00015 | 0.195 | 0.06908 | 0.306 |
| 39522 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | ANK2 | Sponge network | 0.222 | 0.29133 | -1.603 | 1.0E-5 | 0.306 |
| 39523 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-493-5p | 20 | CACNA2D1 | Sponge network | -0.371 | 0.24604 | -0.846 | 0.00675 | 0.306 |
| 39524 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p | 11 | TGFBR2 | Sponge network | -0.176 | 0.59692 | -0.243 | 0.11408 | 0.306 |
| 39525 | RP11-37B2.1 |
hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | ERCC6 | Sponge network | 1.03 | 0 | 0.23 | 0.015 | 0.306 |
| 39526 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-940 | 18 | MCC | Sponge network | 2.306 | 9.0E-5 | -1.005 | 0 | 0.306 |
| 39527 | LINC00621 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | WWTR1 | Sponge network | 0.131 | 0.8436 | -0.345 | 0.11997 | 0.306 |
| 39528 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-93-3p | 20 | SUFU | Sponge network | -2.69 | 0.0001 | -0.04 | 0.65489 | 0.306 |
| 39529 | HOXA-AS2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | SLITRK3 | Sponge network | 0.61 | 0.01593 | -3.328 | 0 | 0.306 |
| 39530 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | PPM1A | Sponge network | 0.559 | 0.00026 | -0.623 | 0 | 0.306 |
| 39531 | AC025171.1 |
hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ATRX | Sponge network | 0.998 | 0 | 0.261 | 0.04408 | 0.306 |
| 39532 | BCYRN1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | BCL9 | Sponge network | 0.311 | 0.10344 | 0.321 | 0.00267 | 0.306 |
| 39533 | FAM13A-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-942-5p | 41 | UNC80 | Sponge network | -0.83 | 0 | -1.934 | 0 | 0.306 |
| 39534 | NOP14-AS1 |
hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-7-5p | 17 | EGFR | Sponge network | 0.077 | 0.52917 | -0.175 | 0.48451 | 0.306 |
| 39535 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p | 11 | FNBP1 | Sponge network | -0.322 | 0.25945 | -0.969 | 4.0E-5 | 0.306 |
| 39536 | RP11-283I3.6 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-766-3p;hsa-miR-96-5p | 22 | IL17RD | Sponge network | 0.614 | 1.0E-5 | -0.019 | 0.94873 | 0.306 |
| 39537 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-429;hsa-miR-629-3p | 18 | PTPN14 | Sponge network | 0.727 | 3.0E-5 | 0.144 | 0.34595 | 0.306 |
| 39538 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p | 11 | PCDH8 | Sponge network | -0.773 | 0.04073 | -0.997 | 0.05366 | 0.306 |
| 39539 | CALML3-AS1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p | 44 | DTNA | Sponge network | 0.95 | 0.15943 | -1.648 | 1.0E-5 | 0.306 |
| 39540 | LIPE-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-500a-5p | 26 | AFF1 | Sponge network | 0.078 | 0.55803 | -0.384 | 0.00053 | 0.306 |
| 39541 | CTD-2666L21.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p | 12 | ERC1 | Sponge network | 0.368 | 0.20093 | -0.108 | 0.38794 | 0.306 |
| 39542 | ERVH48-1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-624-5p | 17 | SIRT1 | Sponge network | 1.04 | 0.00023 | 0.109 | 0.2593 | 0.306 |
| 39543 | CTC-301O7.4 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p | 10 | PIK3R1 | Sponge network | 0.292 | 0.43384 | -0.133 | 0.36854 | 0.306 |
| 39544 | FAM66C |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | JDP2 | Sponge network | -0.901 | 0.00019 | -0.967 | 0 | 0.306 |
| 39545 | RP5-1021I20.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-539-5p | 19 | GRIK3 | Sponge network | -0.785 | 0.00035 | -3.208 | 0 | 0.306 |
| 39546 | PART1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 52 | TRPS1 | Sponge network | -2.925 | 0 | -0.191 | 0.44109 | 0.306 |
| 39547 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | CNTN1 | Sponge network | -0.125 | 0.49414 | -2.594 | 0 | 0.306 |
| 39548 | RP11-536O18.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-330-3p | 21 | LPP | Sponge network | -0.074 | 0.90758 | -0.459 | 0.04475 | 0.306 |
| 39549 | AP001055.6 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | MDM4 | Sponge network | 0.693 | 0.00076 | 0.345 | 0.00762 | 0.306 |
| 39550 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-576-5p | 10 | RBL2 | Sponge network | -0.405 | 0.22013 | -0.282 | 0.00254 | 0.306 |
| 39551 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p | 16 | PHC3 | Sponge network | 1.147 | 0 | 0.212 | 0.0499 | 0.306 |
| 39552 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-505-5p;hsa-miR-590-3p | 12 | RBM5 | Sponge network | 0.064 | 0.72934 | -0.205 | 0.0129 | 0.306 |
| 39553 | MIR22HG |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3913-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 39 | IGFBP5 | Sponge network | -1.047 | 0 | -0.806 | 0.00105 | 0.306 |
| 39554 | AC004893.11 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | UBR3 | Sponge network | 1.317 | 0 | -0.021 | 0.82774 | 0.306 |
| 39555 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-654-3p | 12 | BMPR2 | Sponge network | 1.157 | 0.00455 | 0.206 | 0.0635 | 0.306 |
| 39556 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | SLC6A15 | Sponge network | 0.532 | 0.00505 | -2.373 | 2.0E-5 | 0.306 |
| 39557 | LINC00621 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-323a-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p | 29 | DST | Sponge network | 0.131 | 0.8436 | -0.713 | 0.00096 | 0.306 |
| 39558 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 17 | ABCB5 | Sponge network | 0.214 | 0.18246 | -0.956 | 0.04077 | 0.306 |
| 39559 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 12 | TP53INP1 | Sponge network | -1.654 | 0.00144 | -0.496 | 0.00064 | 0.305 |
| 39560 | LINC00094 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-5p | 13 | RET | Sponge network | 0.253 | 0.09565 | -0.833 | 0.03517 | 0.305 |
| 39561 | AC007566.10 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-455-3p | 12 | ZFX | Sponge network | 2.368 | 2.0E-5 | 0.09 | 0.42563 | 0.305 |
| 39562 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | IGSF10 | Sponge network | 0.083 | 0.66405 | -1.751 | 1.0E-5 | 0.305 |
| 39563 | OIP5-AS1 |
hsa-miR-106b-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | RABEP1 | Sponge network | 0.421 | 0.00084 | -0.066 | 0.43142 | 0.305 |
| 39564 | RP5-994D16.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | EPS15 | Sponge network | 0.252 | 0.08508 | -0.052 | 0.53998 | 0.305 |
| 39565 | RP11-707P17.2 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | UBR3 | Sponge network | 1.877 | 0 | -0.021 | 0.82774 | 0.305 |
| 39566 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p | 11 | ABCC9 | Sponge network | 0.274 | 0.07681 | -0.466 | 0.14121 | 0.305 |
| 39567 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p | 15 | MOB1B | Sponge network | 0.706 | 0 | 0.197 | 0.15266 | 0.305 |
| 39568 | PWAR6 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-589-3p | 21 | MAFB | Sponge network | -1.575 | 6.0E-5 | -0.023 | 0.89865 | 0.305 |
| 39569 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p | 11 | MAP3K12 | Sponge network | 0.176 | 0.37402 | -0.057 | 0.59369 | 0.305 |
| 39570 | RP11-819C21.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 14 | ROBO1 | Sponge network | 0.537 | 0.00139 | -0.009 | 0.96154 | 0.305 |
| 39571 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ITGAV | Sponge network | 0.401 | 0.00066 | 0.337 | 0.01002 | 0.305 |
| 39572 | RP5-1074L1.4 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | USP9X | Sponge network | 1.923 | 0 | 0.222 | 0.01689 | 0.305 |
| 39573 | SNHG14 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 18 | NR3C2 | Sponge network | -1.111 | 0.0003 | -1.56 | 0 | 0.305 |
| 39574 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 24 | EBF1 | Sponge network | 0.782 | 0.00016 | -0.384 | 0.08856 | 0.305 |
| 39575 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-96-5p | 27 | TCF4 | Sponge network | 3.345 | 0 | 0.016 | 0.92271 | 0.305 |
| 39576 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 61 | TCF4 | Sponge network | 0.395 | 0.15451 | 0.016 | 0.92271 | 0.305 |
| 39577 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-589-3p | 22 | SCN9A | Sponge network | -0.773 | 0.04073 | -0.525 | 0.10593 | 0.305 |
| 39578 | RP11-888D10.4 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374b-3p;hsa-miR-590-3p | 19 | CCDC6 | Sponge network | 0.702 | 7.0E-5 | 0.27 | 0.02555 | 0.305 |
| 39579 | OSER1-AS1 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 36 | AR | Sponge network | -0.13 | 0.48734 | -1.554 | 0 | 0.305 |
| 39580 | SH3BP5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | NIN | Sponge network | 0.267 | 0.2937 | -0.253 | 0.13794 | 0.305 |
| 39581 | USP46-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-429 | 18 | WDFY3 | Sponge network | 0.102 | 0.51464 | -0.201 | 0.0967 | 0.305 |
| 39582 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 12 | FYN | Sponge network | 0.395 | 0.15451 | -0.131 | 0.37051 | 0.305 |
| 39583 | KB-1460A1.5 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p | 20 | SYNM | Sponge network | 0.603 | 0.00127 | -2.309 | 1.0E-5 | 0.305 |
| 39584 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | PRRX1 | Sponge network | -0.896 | 0.00099 | 1.316 | 0 | 0.305 |
| 39585 | RP11-57H14.4 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | 0.083 | 0.66405 | -0.966 | 0.00035 | 0.305 |
| 39586 | GNG12-AS1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-629-5p | 11 | WNK2 | Sponge network | -0.793 | 7.0E-5 | -0.352 | 0.31066 | 0.305 |
| 39587 | RP11-701H24.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 17 | SOX5 | Sponge network | -1.425 | 0.04204 | -1.091 | 4.0E-5 | 0.305 |
| 39588 | USP3-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-942-5p | 14 | ERRFI1 | Sponge network | -0.162 | 0.47687 | -0.798 | 1.0E-5 | 0.305 |
| 39589 | LINC00969 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p | 17 | PRKAB2 | Sponge network | 0.683 | 1.0E-5 | -0.367 | 0.01371 | 0.305 |
| 39590 | RBPMS-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-629-3p;hsa-miR-93-5p | 17 | DST | Sponge network | 0.144 | 0.64989 | -0.713 | 0.00096 | 0.305 |
| 39591 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | PTGFR | Sponge network | 0.083 | 0.66405 | -0.233 | 0.45421 | 0.305 |
| 39592 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-629-5p | 39 | SOX5 | Sponge network | -1.047 | 0 | -1.091 | 4.0E-5 | 0.305 |
| 39593 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-374b-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p | 15 | TP53INP1 | Sponge network | -2.62 | 0 | -0.496 | 0.00064 | 0.305 |
| 39594 | AC156455.1 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p | 13 | PIK3R1 | Sponge network | 1.154 | 0.00041 | -0.133 | 0.36854 | 0.305 |
| 39595 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-654-3p | 12 | USP25 | Sponge network | 0.101 | 0.60919 | -0.242 | 0.01397 | 0.305 |
| 39596 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | -0.235 | 0.15142 | -1.751 | 1.0E-5 | 0.305 |
| 39597 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | MYO9A | Sponge network | 0.177 | 0.16681 | -0.147 | 0.32373 | 0.305 |
| 39598 | LINC00969 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p | 11 | BAZ2B | Sponge network | 0.683 | 1.0E-5 | -0.276 | 0.02833 | 0.305 |
| 39599 | CD27-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-455-5p | 10 | THSD7A | Sponge network | 0.177 | 0.16681 | 0.242 | 0.25154 | 0.305 |
| 39600 | HAND2-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 24 | ATM | Sponge network | -1.697 | 0.00889 | 0.178 | 0.21568 | 0.305 |
| 39601 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-766-3p | 11 | PHACTR4 | Sponge network | 0.848 | 0 | 0.069 | 0.45203 | 0.305 |
| 39602 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | EPAS1 | Sponge network | 1.302 | 7.0E-5 | -0.184 | 0.14418 | 0.305 |
| 39603 | RP11-395G23.3 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | CSDE1 | Sponge network | 0.381 | 0.25967 | -0.493 | 0 | 0.305 |
| 39604 | GABPB1-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 16 | SLC5A3 | Sponge network | 1.11 | 0 | 0.493 | 5.0E-5 | 0.305 |
| 39605 | FAM66C |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | EGR2 | Sponge network | -0.901 | 0.00019 | 0.309 | 0.17079 | 0.305 |
| 39606 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-7-1-3p | 19 | APC | Sponge network | 0.817 | 3.0E-5 | -0.255 | 0.04051 | 0.305 |
| 39607 | RP11-927P21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p | 15 | BRD3 | Sponge network | 0.921 | 0.00118 | 0.37 | 0.00013 | 0.305 |
| 39608 | RP11-686D22.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p | 11 | EBF1 | Sponge network | 0.793 | 5.0E-5 | -0.384 | 0.08856 | 0.305 |
| 39609 | AGAP11 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p | 21 | NAV3 | Sponge network | -1.645 | 0 | 0.212 | 0.47729 | 0.305 |
| 39610 | RP11-798M19.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | PRRX1 | Sponge network | -0.612 | 0.00094 | 1.316 | 0 | 0.305 |
| 39611 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | TCF12 | Sponge network | 0.919 | 0 | 0.174 | 0.13271 | 0.305 |
| 39612 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-93-3p | 12 | ERC1 | Sponge network | 1.46 | 1.0E-5 | -0.108 | 0.38794 | 0.305 |
| 39613 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | CBL | Sponge network | 0.556 | 0.00298 | 0.291 | 0.01267 | 0.305 |
| 39614 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-629-3p | 20 | DST | Sponge network | -1.054 | 0.0706 | -0.713 | 0.00096 | 0.305 |
| 39615 | SNX29P2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 23 | PCDH10 | Sponge network | 0.4 | 0.14611 | -1.644 | 0.0078 | 0.305 |
| 39616 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-628-5p | 20 | CDC73 | Sponge network | 0.75 | 0.00054 | 0.094 | 0.20463 | 0.305 |
| 39617 | RP11-545G3.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p | 11 | RICTOR | Sponge network | 2.622 | 0.00025 | 0.388 | 0.00188 | 0.305 |
| 39618 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | PIK3R1 | Sponge network | -0.769 | 0.00035 | -0.133 | 0.36854 | 0.305 |
| 39619 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-3614-5p | 11 | MOB1B | Sponge network | 0.909 | 0 | 0.197 | 0.15266 | 0.305 |
| 39620 | RP11-156P1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-942-5p | 37 | UNC80 | Sponge network | 0.214 | 0.18246 | -1.934 | 0 | 0.305 |
| 39621 | HOXA-AS2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | GALNT13 | Sponge network | 0.61 | 0.01593 | -0.857 | 0.07439 | 0.305 |
| 39622 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | CLASP2 | Sponge network | -0.896 | 0.00099 | -0.038 | 0.71 | 0.305 |
| 39623 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 13 | EBF1 | Sponge network | 0.817 | 3.0E-5 | -0.384 | 0.08856 | 0.305 |
| 39624 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p | 10 | CNTNAP3 | Sponge network | 0.178 | 0.29106 | -1.465 | 0.00033 | 0.305 |
| 39625 | RP11-67L2.2 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | WAC | Sponge network | 0.72 | 0.0001 | -0.054 | 0.43688 | 0.305 |
| 39626 | AC006116.24 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 14 | ERCC4 | Sponge network | -0.473 | 0.07602 | 0.126 | 0.15266 | 0.305 |
| 39627 | CTD-3025N20.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-323a-3p;hsa-miR-590-3p | 11 | ABI2 | Sponge network | 1.046 | 0 | 0.175 | 0.14787 | 0.305 |
| 39628 | LINC00957 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PTCH1 | Sponge network | -0.421 | 0.0114 | -0.1 | 0.6179 | 0.305 |
| 39629 | LINC00863 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | FGFR1 | Sponge network | 0.189 | 0.13981 | -0.509 | 0.03957 | 0.305 |
| 39630 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 37 | BCL2 | Sponge network | -0.842 | 0.01361 | -0.899 | 0 | 0.305 |
| 39631 | ARHGEF26-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-584-5p;hsa-miR-590-3p | 18 | GLIPR1 | Sponge network | -2.46 | 0 | 0.146 | 0.3276 | 0.305 |
| 39632 | CTC-457L16.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-940 | 16 | NTRK2 | Sponge network | 1.153 | 0.00114 | -0.736 | 0.06495 | 0.305 |
| 39633 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 11 | LRRC4 | Sponge network | -1.057 | 0.00029 | -1.774 | 0 | 0.305 |
| 39634 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 19 | ERCC4 | Sponge network | -0.842 | 0.01361 | 0.126 | 0.15266 | 0.305 |
| 39635 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | ABI2 | Sponge network | -0.355 | 0.0077 | 0.175 | 0.14787 | 0.305 |
| 39636 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | MED12L | Sponge network | -0.487 | 0.02157 | -0.194 | 0.55299 | 0.305 |
| 39637 | LINC00957 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-671-5p | 18 | CYLD | Sponge network | -0.421 | 0.0114 | -0.367 | 0.00171 | 0.305 |
| 39638 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-501-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 20 | UBE4B | Sponge network | -0.405 | 0.22013 | -0.235 | 0.007 | 0.305 |
| 39639 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p | 46 | KIF1B | Sponge network | 0.056 | 0.81195 | -0.118 | 0.32258 | 0.305 |
| 39640 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-96-5p | 19 | DMXL1 | Sponge network | 0.594 | 0.41522 | -0.426 | 0.00056 | 0.305 |
| 39641 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-582-5p | 14 | NEDD4 | Sponge network | 1.03 | 0 | 0.02 | 0.89834 | 0.305 |
| 39642 | HOTAIRM1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-766-3p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -0.053 | 0.80685 | -0.564 | 1.0E-5 | 0.305 |
| 39643 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | CUL5 | Sponge network | 0.569 | 0.00067 | 0.026 | 0.77301 | 0.305 |
| 39644 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-654-3p | 13 | ITCH | Sponge network | 0.439 | 0.00313 | 0.084 | 0.34058 | 0.305 |
| 39645 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 30 | PDGFRA | Sponge network | -0.176 | 0.59692 | -0.372 | 0.0037 | 0.305 |
| 39646 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | RNF144A | Sponge network | -2.452 | 0 | 0.594 | 0.0009 | 0.305 |
| 39647 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 13 | USP12 | Sponge network | 0.817 | 3.0E-5 | -0.102 | 0.40768 | 0.305 |
| 39648 | MEG3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577 | 18 | CSDE1 | Sponge network | 0.249 | 0.35902 | -0.493 | 0 | 0.305 |
| 39649 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p | 13 | DCLK1 | Sponge network | -0.384 | 0.40652 | -1.324 | 0.00014 | 0.305 |
| 39650 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-338-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | SOX11 | Sponge network | 1.378 | 0 | 2.68 | 0 | 0.305 |
| 39651 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-7-1-3p | 19 | TBX18 | Sponge network | -0.842 | 0.01361 | 0.843 | 0.02945 | 0.305 |
| 39652 | RP11-401P9.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-539-5p;hsa-miR-542-3p | 20 | TSC1 | Sponge network | -0.384 | 0.40652 | 0.103 | 0.26071 | 0.305 |
| 39653 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TSHZ3 | Sponge network | 0.862 | 0.06213 | -0.556 | 0.01516 | 0.305 |
| 39654 | RP11-158K1.3 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-532-3p | 17 | SYNM | Sponge network | 0.349 | 0.01695 | -2.309 | 1.0E-5 | 0.305 |
| 39655 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 32 | RICTOR | Sponge network | 0.488 | 0.00084 | 0.388 | 0.00188 | 0.305 |
| 39656 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | CACNA2D1 | Sponge network | 0.219 | 0.1516 | -0.846 | 0.00675 | 0.305 |
| 39657 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-874-3p | 15 | CRTC3 | Sponge network | 0.381 | 0.25967 | 0.164 | 0.16786 | 0.305 |
| 39658 | RP11-390K5.6 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-365a-3p;hsa-miR-590-3p | 10 | ERCC6 | Sponge network | 1.148 | 2.0E-5 | 0.23 | 0.015 | 0.305 |
| 39659 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 47 | PAFAH1B1 | Sponge network | -0.901 | 0.00019 | -0.429 | 0 | 0.305 |
| 39660 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 44 | KIF1B | Sponge network | 0.225 | 0.30644 | -0.118 | 0.32258 | 0.305 |
| 39661 | RP11-401P9.4 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-5p | 19 | LIMCH1 | Sponge network | -0.384 | 0.40652 | -0.915 | 1.0E-5 | 0.305 |
| 39662 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-940 | 25 | MYO9A | Sponge network | -0.777 | 0.16667 | -0.147 | 0.32373 | 0.305 |
| 39663 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-96-5p | 39 | KIF1B | Sponge network | -1.654 | 0.00144 | -0.118 | 0.32258 | 0.305 |
| 39664 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p | 12 | ELMO1 | Sponge network | -0.318 | 0.65847 | 0.04 | 0.83147 | 0.305 |
| 39665 | AC003090.1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | EFNA5 | Sponge network | -2.117 | 0.00161 | -0.421 | 0.17868 | 0.305 |
| 39666 | RP11-46C24.7 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-484 | 11 | WNK2 | Sponge network | 0.392 | 0.0278 | -0.352 | 0.31066 | 0.305 |
| 39667 | BMS1P20 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-194-3p;hsa-miR-214-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | MLLT10 | Sponge network | 0.34 | 0.00273 | 0.105 | 0.33292 | 0.305 |
| 39668 | RP5-1139B12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-29a-3p;hsa-miR-29b-3p | 11 | PIK3R1 | Sponge network | 0.598 | 0.38481 | -0.133 | 0.36854 | 0.305 |
| 39669 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 26 | EPHA7 | Sponge network | 0.389 | 0.04293 | -2.197 | 3.0E-5 | 0.305 |
| 39670 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 32 | AFF4 | Sponge network | 0.873 | 0 | -0.266 | 0.01565 | 0.305 |
| 39671 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | KIF1B | Sponge network | 0.873 | 0 | -0.118 | 0.32258 | 0.305 |
| 39672 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-577;hsa-miR-940 | 13 | ETV1 | Sponge network | 1.294 | 0.0031 | -0.096 | 0.67674 | 0.305 |
| 39673 | SNX29P2 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | 0.4 | 0.14611 | -0.966 | 0.00035 | 0.305 |
| 39674 | RP11-1277A3.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p | 24 | ADARB1 | Sponge network | 0.453 | 0.01839 | -0.335 | 0.06631 | 0.305 |
| 39675 | RP11-474O21.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | PRKAA2 | Sponge network | -0.023 | 0.95109 | -2.462 | 0 | 0.305 |
| 39676 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-93-3p | 23 | NTRK3 | Sponge network | -0.164 | 0.45106 | -1.77 | 3.0E-5 | 0.305 |
| 39677 | U91328.19 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-769-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940 | 34 | GRIK3 | Sponge network | -0.303 | 0.21002 | -3.208 | 0 | 0.305 |
| 39678 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-940 | 39 | TRPS1 | Sponge network | 0.349 | 0.01695 | -0.191 | 0.44109 | 0.305 |
| 39679 | A2M-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | ASTN1 | Sponge network | -0.08 | 0.90092 | -2.389 | 2.0E-5 | 0.305 |
| 39680 | TP73-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 21 | SEMA3C | Sponge network | -0.563 | 0.02545 | -0.272 | 0.31153 | 0.305 |
| 39681 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | THBS1 | Sponge network | 0.61 | 0.01593 | 0.741 | 0.00386 | 0.305 |
| 39682 | AC009948.5 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 15 | CSDE1 | Sponge network | 0.532 | 0.00505 | -0.493 | 0 | 0.305 |
| 39683 | MEG3 |
hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-429 | 10 | DUSP1 | Sponge network | 0.249 | 0.35902 | -1.754 | 0 | 0.305 |
| 39684 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-98-5p | 31 | TGFBR3 | Sponge network | 0.056 | 0.81195 | -1.173 | 1.0E-5 | 0.305 |
| 39685 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 16 | TXNIP | Sponge network | -0.969 | 2.0E-5 | -1.277 | 0 | 0.305 |
| 39686 | RP11-119F7.5 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SYT4 | Sponge network | 0.225 | 0.30644 | -3.207 | 0 | 0.305 |
| 39687 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 38 | MOB1B | Sponge network | -0.709 | 0.18168 | 0.197 | 0.15266 | 0.305 |
| 39688 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-497-5p | 15 | ZMYM2 | Sponge network | 0.917 | 0 | 0.279 | 0.01273 | 0.305 |
| 39689 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 17 | TET1 | Sponge network | 0.051 | 0.83388 | -0.135 | 0.52138 | 0.305 |
| 39690 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-7-1-3p | 15 | AKAP11 | Sponge network | 1.11 | 1.0E-5 | 0.196 | 0.09825 | 0.305 |
| 39691 | RP11-428J1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | AR | Sponge network | 0.538 | 0.00076 | -1.554 | 0 | 0.305 |
| 39692 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-501-3p | 12 | KAT6B | Sponge network | -0.154 | 0.53954 | -0.478 | 0.00018 | 0.305 |
| 39693 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 10 | NUAK1 | Sponge network | -1.255 | 0.00981 | 0.904 | 0 | 0.305 |
| 39694 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-365a-3p;hsa-miR-375;hsa-miR-550a-3p | 13 | CACNA1E | Sponge network | 0.082 | 0.55654 | 1.752 | 0 | 0.305 |
| 39695 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-93-5p | 38 | PIK3R1 | Sponge network | -2.69 | 0.0001 | -0.133 | 0.36854 | 0.305 |
| 39696 | RP11-195B17.1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-425-5p | 19 | DTNA | Sponge network | 0.37 | 0.27614 | -1.648 | 1.0E-5 | 0.305 |
| 39697 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 21 | EPHA3 | Sponge network | 1.61 | 0.00961 | 0.185 | 0.53588 | 0.305 |
| 39698 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | LRP1B | Sponge network | 0.4 | 0.14611 | -2.163 | 0 | 0.305 |
| 39699 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-671-5p | 35 | ADARB1 | Sponge network | 0.95 | 0.15943 | -0.335 | 0.06631 | 0.305 |
| 39700 | LA16c-321D4.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-590-5p;hsa-miR-654-3p | 13 | BMPR2 | Sponge network | 2.459 | 5.0E-5 | 0.206 | 0.0635 | 0.305 |
| 39701 | RP11-13K12.1 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-625-5p | 11 | ADCY1 | Sponge network | -0.154 | 0.78939 | 0.362 | 0.16982 | 0.305 |
| 39702 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 32 | ST6GALNAC3 | Sponge network | -0.342 | 0.02288 | -0.987 | 0 | 0.305 |
| 39703 | RP11-411K7.1 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 16 | RND3 | Sponge network | 0.155 | 0.81273 | -0.254 | 0.11478 | 0.305 |
| 39704 | RP11-736K20.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-589-3p | 20 | RNASEL | Sponge network | -0.358 | 0.17255 | -0.272 | 0.01796 | 0.305 |
| 39705 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p | 12 | FGFR1 | Sponge network | 0.95 | 0.15943 | -0.509 | 0.03957 | 0.305 |
| 39706 | ADIRF-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-590-3p | 22 | ABL2 | Sponge network | -0.223 | 0.47816 | 0.82 | 0 | 0.305 |
| 39707 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | SOX5 | Sponge network | 0.537 | 0.00139 | -1.091 | 4.0E-5 | 0.305 |
| 39708 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-500a-5p | 22 | PPM1A | Sponge network | -0.932 | 0.00329 | -0.623 | 0 | 0.305 |
| 39709 | RP11-244H3.1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PHF3 | Sponge network | 0.659 | 3.0E-5 | 0.126 | 0.19652 | 0.305 |
| 39710 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p | 15 | HACE1 | Sponge network | -0.567 | 0 | -0.072 | 0.55239 | 0.305 |
| 39711 | LA16c-321D4.2 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-511-5p | 10 | ZFHX3 | Sponge network | 2.459 | 5.0E-5 | 0.389 | 0.01363 | 0.305 |
| 39712 | RP5-855D21.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-30a-5p;hsa-miR-495-3p | 11 | SLC5A3 | Sponge network | 1.728 | 0 | 0.493 | 5.0E-5 | 0.305 |
| 39713 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p | 11 | RET | Sponge network | -0.726 | 0.00132 | -0.833 | 0.03517 | 0.305 |
| 39714 | FZD10-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | HERC1 | Sponge network | -0.727 | 0.04726 | -0.196 | 0.08544 | 0.305 |
| 39715 | AC005618.6 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-484;hsa-miR-505-5p | 10 | WNK2 | Sponge network | 0.862 | 0.06213 | -0.352 | 0.31066 | 0.305 |
| 39716 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 20 | KCNA6 | Sponge network | -0.246 | 0.35034 | -1.531 | 0.00013 | 0.305 |
| 39717 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-7-1-3p | 18 | USP12 | Sponge network | 0.389 | 0.04293 | -0.102 | 0.40768 | 0.305 |
| 39718 | H19 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p | 16 | BNC2 | Sponge network | 2.439 | 0 | -0.491 | 0.09988 | 0.305 |
| 39719 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 42 | TMEM132B | Sponge network | 0.249 | 0.35902 | -0.83 | 0.01084 | 0.305 |
| 39720 | LINC00641 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-577 | 13 | AMPH | Sponge network | -0.222 | 0.32445 | -0.916 | 5.0E-5 | 0.305 |
| 39721 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 35 | BNC2 | Sponge network | 0.75 | 0.00054 | -0.491 | 0.09988 | 0.305 |
| 39722 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | NFIB | Sponge network | -1.375 | 0.08642 | 0.023 | 0.90795 | 0.305 |
| 39723 | RP11-144G6.12 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | BAZ2B | Sponge network | 0.826 | 1.0E-5 | -0.276 | 0.02833 | 0.305 |
| 39724 | RP11-181G12.2 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-503-5p | 12 | ABL1 | Sponge network | 0.556 | 0.00298 | -0.215 | 0.07804 | 0.305 |
| 39725 | HCG18 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | USP9X | Sponge network | 1.063 | 0 | 0.222 | 0.01689 | 0.305 |
| 39726 | FENDRR |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | UNC5D | Sponge network | -1.281 | 0.00012 | -3.646 | 0 | 0.305 |
| 39727 | AC108488.4 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 11 | FREM2 | Sponge network | 0.259 | 0.12542 | -0.437 | 0.46353 | 0.305 |
| 39728 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-550a-5p;hsa-miR-590-5p | 10 | TSHZ3 | Sponge network | 3.345 | 0 | -0.556 | 0.01516 | 0.305 |
| 39729 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | CUL5 | Sponge network | 1.308 | 0 | 0.026 | 0.77301 | 0.305 |
| 39730 | BOLA3-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-589-3p | 10 | RPS6KA6 | Sponge network | -0.246 | 0.35034 | -2.989 | 0 | 0.305 |
| 39731 | MIR600HG |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-625-5p;hsa-miR-96-5p | 24 | ABI2 | Sponge network | 0.594 | 0.41522 | 0.175 | 0.14787 | 0.305 |
| 39732 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-96-5p | 17 | CBFA2T3 | Sponge network | 0.572 | 0.01722 | -1.221 | 1.0E-5 | 0.305 |
| 39733 | AC005562.1 |
hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | PPM1L | Sponge network | 0.488 | 0.00084 | -0.537 | 0.00616 | 0.305 |
| 39734 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p | 25 | CRTC1 | Sponge network | 0.194 | 0.64278 | -0.116 | 0.28412 | 0.305 |
| 39735 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-501-3p;hsa-miR-93-5p | 12 | SLITRK3 | Sponge network | -0.351 | 0.11358 | -3.328 | 0 | 0.305 |
| 39736 | LINC00963 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 22 | SOX11 | Sponge network | 0.523 | 9.0E-5 | 2.68 | 0 | 0.305 |
| 39737 | AC009948.5 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 17 | CXCL12 | Sponge network | 0.532 | 0.00505 | -1.421 | 0 | 0.305 |
| 39738 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | ST6GAL2 | Sponge network | 0.225 | 0.30644 | -1.201 | 0.00597 | 0.305 |
| 39739 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DACT1 | Sponge network | -0.021 | 0.93598 | 0.783 | 0.00072 | 0.305 |
| 39740 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-429 | 11 | AKAP11 | Sponge network | 2.364 | 0.00134 | 0.196 | 0.09825 | 0.305 |
| 39741 | ZNF667-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-92a-1-5p;hsa-miR-96-5p | 11 | PRKACA | Sponge network | -0.976 | 0.00025 | -0.255 | 0.0012 | 0.305 |
| 39742 | RP11-705C15.3 |
hsa-miR-130b-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 10 | IKZF2 | Sponge network | 0.796 | 4.0E-5 | -0.284 | 0.1942 | 0.305 |
| 39743 | DIO3OS |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-629-3p;hsa-miR-629-5p | 49 | DST | Sponge network | -0.773 | 0.04073 | -0.713 | 0.00096 | 0.305 |
| 39744 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | RIMBP2 | Sponge network | 0.335 | 0.20596 | -1.968 | 5.0E-5 | 0.305 |
| 39745 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-93-5p | 27 | ZBTB4 | Sponge network | 0.259 | 0.12542 | -0.739 | 0 | 0.305 |
| 39746 | PDXDC2P |
hsa-miR-139-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-497-5p | 10 | SUZ12 | Sponge network | 0.756 | 0 | 0.619 | 0 | 0.305 |
| 39747 | OIP5-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 24 | NF1 | Sponge network | 0.421 | 0.00084 | 0.51 | 0 | 0.305 |
| 39748 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-93-3p | 18 | ST6GAL2 | Sponge network | -0.465 | 0.04703 | -1.201 | 0.00597 | 0.305 |
| 39749 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 20 | BCL6 | Sponge network | -0.355 | 0.0077 | -0.211 | 0.19489 | 0.305 |
| 39750 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | FAT4 | Sponge network | -0.206 | 0.12942 | -0.354 | 0.14694 | 0.305 |
| 39751 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-96-5p | 26 | ZFHX4 | Sponge network | 0.622 | 0.00015 | 0.018 | 0.95574 | 0.305 |
| 39752 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | SLC6A15 | Sponge network | -0.739 | 0.00044 | -2.373 | 2.0E-5 | 0.305 |
| 39753 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p | 20 | MLLT10 | Sponge network | 0.95 | 0.15943 | 0.105 | 0.33292 | 0.305 |
| 39754 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 27 | ETV1 | Sponge network | 0.082 | 0.55654 | -0.096 | 0.67674 | 0.305 |
| 39755 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NCAM2 | Sponge network | 0.222 | 0.29133 | -0.482 | 0.22565 | 0.305 |
| 39756 | RP11-867G23.10 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-502-5p | 10 | FMNL3 | Sponge network | -2.531 | 0 | 0.555 | 4.0E-5 | 0.305 |
| 39757 | PART1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | PREX2 | Sponge network | -2.925 | 0 | -0.272 | 0.25382 | 0.305 |
| 39758 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-5p | 17 | PDZD2 | Sponge network | -0.969 | 2.0E-5 | -0.909 | 3.0E-5 | 0.305 |
| 39759 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-7-1-3p | 15 | GRIN2A | Sponge network | 0.853 | 0.15352 | -2.161 | 0 | 0.305 |
| 39760 | AC025171.1 |
hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | MED12L | Sponge network | 0.998 | 0 | -0.194 | 0.55299 | 0.305 |
| 39761 | RP11-13K12.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-93-3p | 18 | FOXO3 | Sponge network | -0.318 | 0.65847 | -0.04 | 0.72028 | 0.305 |
| 39762 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-96-5p | 24 | CBFA2T3 | Sponge network | -0.942 | 0 | -1.221 | 1.0E-5 | 0.305 |
| 39763 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-423-5p;hsa-miR-429 | 10 | PNRC1 | Sponge network | -0.154 | 0.53954 | -0.646 | 0 | 0.305 |
| 39764 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-128-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-942-5p | 11 | ARHGAP28 | Sponge network | -0.351 | 0.11358 | -0.911 | 0.00013 | 0.305 |
| 39765 | CTD-2517M22.14 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-376c-3p;hsa-miR-497-5p | 11 | TBL1XR1 | Sponge network | 3.997 | 0 | 0.578 | 0 | 0.305 |
| 39766 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | CEP170 | Sponge network | 0.174 | 0.30034 | 0.943 | 0 | 0.305 |
| 39767 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | LRP1B | Sponge network | 0.299 | 0.57722 | -2.163 | 0 | 0.305 |
| 39768 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577;hsa-miR-940 | 20 | NTRK2 | Sponge network | 0.357 | 0.72407 | -0.736 | 0.06495 | 0.305 |
| 39769 | AC012360.6 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-550a-5p;hsa-miR-590-3p | 14 | CCDC6 | Sponge network | 0.624 | 0.02176 | 0.27 | 0.02555 | 0.305 |
| 39770 | RP11-359B12.2 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 19 | SFRP1 | Sponge network | 0.064 | 0.72934 | -3.566 | 0 | 0.305 |
| 39771 | LINC00854 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p | 11 | ARID5B | Sponge network | 1.18 | 0 | 0.342 | 0.01676 | 0.305 |
| 39772 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-409-3p;hsa-miR-539-5p | 16 | MITF | Sponge network | 0.176 | 0.37402 | -0.769 | 0.00022 | 0.305 |
| 39773 | AC006262.5 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c | 16 | ABL2 | Sponge network | 5.342 | 0 | 0.82 | 0 | 0.305 |
| 39774 | MIR22HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-590-5p | 19 | AKAP6 | Sponge network | -1.047 | 0 | -1.586 | 3.0E-5 | 0.305 |
| 39775 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p | 20 | PPP2R5C | Sponge network | -0.323 | 0.01372 | -0.314 | 3.0E-5 | 0.305 |
| 39776 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | ARNT2 | Sponge network | 0.335 | 0.20596 | -0.332 | 0.25851 | 0.305 |
| 39777 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-942-5p | 27 | DDX6 | Sponge network | 0.214 | 0.18246 | 0.091 | 0.36428 | 0.305 |
| 39778 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 16 | ITGA5 | Sponge network | -2.46 | 0 | 0.452 | 0.03524 | 0.305 |
| 39779 | CTC-559E9.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-654-3p | 20 | RICTOR | Sponge network | 0.601 | 1.0E-5 | 0.388 | 0.00188 | 0.305 |
| 39780 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-590-3p | 14 | CDC73 | Sponge network | -0.222 | 0.32445 | 0.094 | 0.20463 | 0.305 |
| 39781 | FAM66C |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | CREM | Sponge network | -0.901 | 0.00019 | -0.345 | 0.00258 | 0.305 |
| 39782 | RP11-98I9.4 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 0.67 | 0.00048 | 0.51 | 0 | 0.305 |
| 39783 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-590-3p | 30 | TSHZ3 | Sponge network | 0.494 | 0.00339 | -0.556 | 0.01516 | 0.305 |
| 39784 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-582-5p | 17 | DLC1 | Sponge network | 0.419 | 0.0319 | -0.382 | 0.05038 | 0.305 |
| 39785 | RP11-53O19.3 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | DDX3X | Sponge network | 1.205 | 0 | -0.152 | 0.07686 | 0.305 |
| 39786 | LINC00476 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p | 16 | TRIM13 | Sponge network | -0.615 | 0.00015 | 0.183 | 0.06968 | 0.305 |
| 39787 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TRIP11 | Sponge network | -0.487 | 0.02157 | -0.158 | 0.06384 | 0.304 |
| 39788 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 11 | KIAA1024 | Sponge network | 0.349 | 0.01695 | 1.083 | 0 | 0.304 |
| 39789 | RP11-373D23.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-421 | 10 | SPG20 | Sponge network | -0.285 | 0.2172 | -1.16 | 0 | 0.304 |
| 39790 | CTB-113P19.4 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | GJA1 | Sponge network | 0.906 | 0.31715 | -0.485 | 0.02179 | 0.304 |
| 39791 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 66 | NTRK3 | Sponge network | -0.969 | 2.0E-5 | -1.77 | 3.0E-5 | 0.304 |
| 39792 | RP11-401P9.4 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-539-5p | 22 | RIMS2 | Sponge network | -0.384 | 0.40652 | -1.365 | 0.00208 | 0.304 |
| 39793 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | DPYD | Sponge network | 0.997 | 0.00066 | -0.736 | 0.00076 | 0.304 |
| 39794 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | RBL2 | Sponge network | 0.494 | 0.00339 | -0.282 | 0.00254 | 0.304 |
| 39795 | NDUFA6-AS1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | NR3C2 | Sponge network | -0.355 | 0.0077 | -1.56 | 0 | 0.304 |
| 39796 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ERG | Sponge network | -0.125 | 0.49414 | 0.002 | 0.99011 | 0.304 |
| 39797 | LINC00473 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 34 | GNAQ | Sponge network | -1.203 | 0.03881 | -0.367 | 0.00102 | 0.304 |
| 39798 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | KDSR | Sponge network | 0.75 | 0.00054 | 0.239 | 0.02216 | 0.304 |
| 39799 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 18 | MED12L | Sponge network | -0.583 | 0.14589 | -0.194 | 0.55299 | 0.304 |
| 39800 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-93-5p | 14 | PRRX1 | Sponge network | 0.497 | 0.01451 | 1.316 | 0 | 0.304 |
| 39801 | RP5-1021I20.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-320a;hsa-miR-320b | 10 | WWTR1 | Sponge network | -0.785 | 0.00035 | -0.345 | 0.11997 | 0.304 |
| 39802 | A2M-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-96-5p | 11 | ELMO1 | Sponge network | -0.08 | 0.90092 | 0.04 | 0.83147 | 0.304 |
| 39803 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | SMARCA2 | Sponge network | -0.358 | 0.17255 | -0.529 | 0.00014 | 0.304 |
| 39804 | CTD-2291D10.4 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-96-5p | 10 | IL17RD | Sponge network | 3.21 | 0 | -0.019 | 0.94873 | 0.304 |
| 39805 | OIP5-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | TET1 | Sponge network | 0.421 | 0.00084 | -0.135 | 0.52138 | 0.304 |
| 39806 | HOTAIRM1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | SFRP1 | Sponge network | -0.053 | 0.80685 | -3.566 | 0 | 0.304 |
| 39807 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 14 | CEP170 | Sponge network | -0.576 | 0.10121 | 0.943 | 0 | 0.304 |
| 39808 | RP11-798M19.6 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 25 | OPCML | Sponge network | -0.612 | 0.00094 | -2.498 | 0 | 0.304 |
| 39809 | RP11-490M8.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-223-5p;hsa-miR-24-3p | 16 | DTNA | Sponge network | -0.214 | 0.44248 | -1.648 | 1.0E-5 | 0.304 |
| 39810 | RP11-6N17.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-301a-3p;hsa-miR-455-5p | 12 | RUNX1T1 | Sponge network | 0.108 | 0.69556 | -0.344 | 0.2374 | 0.304 |
| 39811 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 13 | ABCA10 | Sponge network | -0.668 | 3.0E-5 | -0.571 | 0.03674 | 0.304 |
| 39812 | H19 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-629-3p | 17 | AR | Sponge network | 2.439 | 0 | -1.554 | 0 | 0.304 |
| 39813 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | MOB1B | Sponge network | -0.576 | 0.10121 | 0.197 | 0.15266 | 0.304 |
| 39814 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 10 | PRKAB2 | Sponge network | 0.545 | 0.05789 | -0.367 | 0.01371 | 0.304 |
| 39815 | IQCH-AS1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-9-5p | 23 | CCDC6 | Sponge network | 0.929 | 0 | 0.27 | 0.02555 | 0.304 |
| 39816 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-455-3p | 11 | RELN | Sponge network | 0.082 | 0.55397 | -2.403 | 0 | 0.304 |
| 39817 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-378a-5p | 12 | ARHGAP29 | Sponge network | -0.611 | 0.09607 | 0.131 | 0.42953 | 0.304 |
| 39818 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-7-1-3p | 21 | BNC2 | Sponge network | -4.477 | 0 | -0.491 | 0.09988 | 0.304 |
| 39819 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 0.559 | 0.00026 | 0.322 | 0.00215 | 0.304 |
| 39820 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-7-5p | 25 | RBMS3 | Sponge network | -0.326 | 0.14314 | -0.519 | 0.03551 | 0.304 |
| 39821 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p | 19 | PRDM2 | Sponge network | 0.299 | 0.57722 | -0.258 | 0.00721 | 0.304 |
| 39822 | RP11-404E16.1 |
hsa-miR-1271-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-199b-3p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-532-5p | 11 | PPM1L | Sponge network | 0.097 | 0.91311 | -0.537 | 0.00616 | 0.304 |
| 39823 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 29 | TACC1 | Sponge network | -0.176 | 0.59692 | -0.362 | 0.05406 | 0.304 |
| 39824 | RP11-373D23.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p | 14 | BAZ2B | Sponge network | -0.285 | 0.2172 | -0.276 | 0.02833 | 0.304 |
| 39825 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 11 | FREM2 | Sponge network | -1.582 | 8.0E-5 | -0.437 | 0.46353 | 0.304 |
| 39826 | RP11-296I10.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-539-5p | 13 | NIN | Sponge network | 0.592 | 0.00014 | -0.253 | 0.13794 | 0.304 |
| 39827 | CTD-3138B18.6 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 12 | UBR3 | Sponge network | 2.248 | 0 | -0.021 | 0.82774 | 0.304 |
| 39828 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 26 | CBX5 | Sponge network | 0.539 | 0.03722 | 0.165 | 0.24446 | 0.304 |
| 39829 | CALML3-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p | 13 | WNK2 | Sponge network | 0.95 | 0.15943 | -0.352 | 0.31066 | 0.304 |
| 39830 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | CBL | Sponge network | 0.848 | 0 | 0.291 | 0.01267 | 0.304 |
| 39831 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p | 10 | PCDH8 | Sponge network | -0.777 | 0.16667 | -0.997 | 0.05366 | 0.304 |
| 39832 | AC025171.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 16 | SCN9A | Sponge network | 0.998 | 0 | -0.525 | 0.10593 | 0.304 |
| 39833 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-532-3p;hsa-miR-935 | 11 | DAB2 | Sponge network | -0.246 | 0.35034 | 0.431 | 0.0113 | 0.304 |
| 39834 | RP11-473I1.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p | 35 | LPP | Sponge network | 0.542 | 0.00054 | -0.459 | 0.04475 | 0.304 |
| 39835 | RP11-156P1.3 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-92a-3p | 12 | RASSF3 | Sponge network | 0.214 | 0.18246 | -0.226 | 0.11691 | 0.304 |
| 39836 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p | 15 | FBXO32 | Sponge network | 0.556 | 0.00298 | -0.495 | 0.07561 | 0.304 |
| 39837 | CTB-51J22.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-424-5p | 10 | TGFBR3 | Sponge network | -0.803 | 0.23737 | -1.173 | 1.0E-5 | 0.304 |
| 39838 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-577;hsa-miR-7-1-3p | 14 | SLC6A15 | Sponge network | 0.389 | 0.04293 | -2.373 | 2.0E-5 | 0.304 |
| 39839 | EIF3J-AS1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 16 | HIP1 | Sponge network | 0.331 | 0.00509 | 0.774 | 0 | 0.304 |
| 39840 | AC084219.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-374b-5p | 15 | TMTC3 | Sponge network | 1.296 | 0.0054 | 0.123 | 0.22189 | 0.304 |
| 39841 | RP11-655M14.13 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-454-3p;hsa-miR-590-3p | 11 | PIK3C2A | Sponge network | 1.473 | 0.0057 | 0.061 | 0.53776 | 0.304 |
| 39842 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 36 | TMEM132B | Sponge network | -0.513 | 0.04703 | -0.83 | 0.01084 | 0.304 |
| 39843 | RP11-216B9.6 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | CXCL12 | Sponge network | 0.174 | 0.30034 | -1.421 | 0 | 0.304 |
| 39844 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | HECA | Sponge network | -0.899 | 6.0E-5 | -0.217 | 0.03463 | 0.304 |
| 39845 | CKMT2-AS1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 43 | MDM4 | Sponge network | 0.082 | 0.55654 | 0.345 | 0.00762 | 0.304 |
| 39846 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | FAM172A | Sponge network | 0.219 | 0.1516 | -0.57 | 0 | 0.304 |
| 39847 | RP1-122P22.2 |
hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 12 | NUAK1 | Sponge network | 0.065 | 0.73468 | 0.904 | 0 | 0.304 |
| 39848 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 23 | BCL11A | Sponge network | -1.264 | 6.0E-5 | -0.932 | 0.0063 | 0.304 |
| 39849 | AC011526.1 |
hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | NCAM2 | Sponge network | 0.342 | 0.03129 | -0.482 | 0.22565 | 0.304 |
| 39850 | AGAP11 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p | 10 | PLA2R1 | Sponge network | -1.645 | 0 | -0.265 | 0.24676 | 0.304 |
| 39851 | U91328.19 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-940 | 14 | RHOB | Sponge network | -0.303 | 0.21002 | -1.052 | 0 | 0.304 |
| 39852 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-940 | 19 | HLF | Sponge network | -0.829 | 0.01343 | -2.443 | 0 | 0.304 |
| 39853 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-369-3p | 10 | PPM1L | Sponge network | 1.46 | 1.0E-5 | -0.537 | 0.00616 | 0.304 |
| 39854 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CLASP2 | Sponge network | 0.91 | 0 | -0.038 | 0.71 | 0.304 |
| 39855 | RP11-384K6.6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-629-3p | 12 | TCF12 | Sponge network | 0.946 | 0 | 0.174 | 0.13271 | 0.304 |
| 39856 | KB-431C1.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-495-3p;hsa-miR-664a-3p | 24 | CCDC6 | Sponge network | 0.909 | 0 | 0.27 | 0.02555 | 0.304 |
| 39857 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | PHC3 | Sponge network | 0.603 | 0.00127 | 0.212 | 0.0499 | 0.304 |
| 39858 | RP11-384O8.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 10 | ITGB1 | Sponge network | -0.738 | 0.21233 | 0.524 | 0.00017 | 0.304 |
| 39859 | RP11-159D12.2 |
hsa-miR-130b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-590-3p | 11 | RASA1 | Sponge network | 1.254 | 0 | -0.002 | 0.98204 | 0.304 |
| 39860 | RP4-669H2.1 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-628-5p;hsa-miR-629-3p | 10 | CHD2 | Sponge network | 1.711 | 0.00019 | 0.049 | 0.55371 | 0.304 |
| 39861 | AC034220.3 |
hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | RNF111 | Sponge network | 0.309 | 0.07304 | 0.098 | 0.21093 | 0.304 |
| 39862 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | FAM188A | Sponge network | -0.709 | 0.18168 | -0.44 | 0.00018 | 0.304 |
| 39863 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 53 | NTRK3 | Sponge network | -0.18 | 0.20343 | -1.77 | 3.0E-5 | 0.304 |
| 39864 | CTC-490E21.10 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p | 13 | ARID5B | Sponge network | 1.46 | 1.0E-5 | 0.342 | 0.01676 | 0.304 |
| 39865 | RP4-605O3.4 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 12 | MLLT10 | Sponge network | 0.262 | 0.0475 | 0.105 | 0.33292 | 0.304 |
| 39866 | RP11-545G3.1 |
hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p | 11 | BRD3 | Sponge network | 2.622 | 0.00025 | 0.37 | 0.00013 | 0.304 |
| 39867 | PWAR6 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | WAC | Sponge network | -1.575 | 6.0E-5 | -0.054 | 0.43688 | 0.304 |
| 39868 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 10 | SKIL | Sponge network | 0.705 | 0.00014 | 0.559 | 8.0E-5 | 0.304 |
| 39869 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-214-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 29 | MXI1 | Sponge network | -0.355 | 0.0077 | -0.987 | 0 | 0.304 |
| 39870 | LINC00094 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-582-5p | 10 | THSD7B | Sponge network | 0.253 | 0.09565 | 0.154 | 0.74723 | 0.304 |
| 39871 | RP11-822E23.8 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p | 13 | GJA1 | Sponge network | -0.777 | 0.16667 | -0.485 | 0.02179 | 0.304 |
| 39872 | RP11-53O19.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | TSC1 | Sponge network | 1.205 | 0 | 0.103 | 0.26071 | 0.304 |
| 39873 | RP11-166D19.1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-96-5p | 13 | PRKACA | Sponge network | -0.576 | 0.10121 | -0.255 | 0.0012 | 0.304 |
| 39874 | HHIP-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | TPO | Sponge network | -0.737 | 0.10822 | -1.87 | 2.0E-5 | 0.304 |
| 39875 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-369-3p;hsa-miR-500a-5p | 23 | WDFY3 | Sponge network | -0.932 | 0.00329 | -0.201 | 0.0967 | 0.304 |
| 39876 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-942-5p | 14 | RB1CC1 | Sponge network | 0.401 | 0.00066 | 0.175 | 0.12559 | 0.304 |
| 39877 | CTD-2666L21.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-744-3p | 17 | PTPN14 | Sponge network | 0.368 | 0.20093 | 0.144 | 0.34595 | 0.304 |
| 39878 | RP4-717I23.3 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577 | 10 | INPP4A | Sponge network | 0.637 | 0.00069 | 0.07 | 0.53563 | 0.304 |
| 39879 | RP11-359E10.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-338-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 10 | BCL9 | Sponge network | 0.299 | 0.57722 | 0.321 | 0.00267 | 0.304 |
| 39880 | SNHG12 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 12 | RICTOR | Sponge network | 1.103 | 0 | 0.388 | 0.00188 | 0.304 |
| 39881 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | 0.083 | 0.66405 | 0.206 | 0.06751 | 0.304 |
| 39882 | PWAR6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | PRRX1 | Sponge network | -1.575 | 6.0E-5 | 1.316 | 0 | 0.304 |
| 39883 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-590-3p | 14 | TGFBR2 | Sponge network | 0.494 | 0.00339 | -0.243 | 0.11408 | 0.304 |
| 39884 | RP11-25K19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | PTPRD | Sponge network | -1.057 | 0.00029 | -1.005 | 0.00064 | 0.304 |
| 39885 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DNAJB4 | Sponge network | -0.727 | 0.04726 | -0.7 | 1.0E-5 | 0.304 |
| 39886 | HOXB-AS2 |
hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-505-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-942-5p | 14 | ARHGEF12 | Sponge network | 1.316 | 0.01106 | 0.09 | 0.35693 | 0.304 |
| 39887 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p | 10 | TRIM33 | Sponge network | 0.542 | 0.00054 | 0.402 | 7.0E-5 | 0.304 |
| 39888 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | SMAD2 | Sponge network | 1.205 | 0 | -0.128 | 0.12732 | 0.304 |
| 39889 | RP11-156P1.3 |
hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 12 | RHOB | Sponge network | 0.214 | 0.18246 | -1.052 | 0 | 0.304 |
| 39890 | SOX2-OT |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 20 | CDH19 | Sponge network | -1.264 | 6.0E-5 | -3.434 | 0 | 0.304 |
| 39891 | RP11-276H19.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-660-5p;hsa-miR-96-5p | 15 | ERBB4 | Sponge network | -0.472 | 0.48851 | -1.975 | 2.0E-5 | 0.304 |
| 39892 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-582-5p | 18 | PRKCE | Sponge network | -2.69 | 0.0001 | -0.403 | 0.00056 | 0.304 |
| 39893 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-664a-3p | 19 | SOX11 | Sponge network | 1.344 | 0 | 2.68 | 0 | 0.304 |
| 39894 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 40 | CBX5 | Sponge network | 0.862 | 0.06213 | 0.165 | 0.24446 | 0.304 |
| 39895 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SLC6A15 | Sponge network | 0.997 | 0.00066 | -2.373 | 2.0E-5 | 0.304 |
| 39896 | CTBP1-AS2 |
hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 11 | ABL2 | Sponge network | 0.623 | 0 | 0.82 | 0 | 0.304 |
| 39897 | LPP-AS2 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-625-5p;hsa-miR-96-5p | 10 | CXCL12 | Sponge network | -0.18 | 0.20343 | -1.421 | 0 | 0.304 |
| 39898 | RP5-855D21.3 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-495-3p | 11 | ARHGEF12 | Sponge network | 1.728 | 0 | 0.09 | 0.35693 | 0.304 |
| 39899 | LINC00886 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-628-5p | 48 | DTNA | Sponge network | -0.371 | 0.24604 | -1.648 | 1.0E-5 | 0.304 |
| 39900 | RP11-400K9.4 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 18 | OPCML | Sponge network | -0.611 | 0.09607 | -2.498 | 0 | 0.304 |
| 39901 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-942-5p | 37 | UNC80 | Sponge network | -1.161 | 0.00591 | -1.934 | 0 | 0.304 |
| 39902 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 15 | USP12 | Sponge network | 0.419 | 0.0319 | -0.102 | 0.40768 | 0.304 |
| 39903 | AC002117.1 |
hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | BAZ2B | Sponge network | 0.476 | 0.19203 | -0.276 | 0.02833 | 0.304 |
| 39904 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | AFF4 | Sponge network | 0.545 | 0.00064 | -0.266 | 0.01565 | 0.304 |
| 39905 | RP5-1074L1.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 17 | ZFX | Sponge network | 1.923 | 0 | 0.09 | 0.42563 | 0.304 |
| 39906 | LINC00472 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p | 13 | FMNL3 | Sponge network | -0.842 | 0.01361 | 0.555 | 4.0E-5 | 0.304 |
| 39907 | AC005618.6 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | ATM | Sponge network | 0.862 | 0.06213 | 0.178 | 0.21568 | 0.304 |
| 39908 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p | 27 | DTNA | Sponge network | 0.497 | 0.01451 | -1.648 | 1.0E-5 | 0.304 |
| 39909 | RP11-5C23.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | BCL2 | Sponge network | 0.274 | 0.07681 | -0.899 | 0 | 0.304 |
| 39910 | AC002117.1 |
hsa-let-7d-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935 | 14 | DMD | Sponge network | 0.476 | 0.19203 | -1.506 | 0 | 0.304 |
| 39911 | LIPE-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 22 | BRD3 | Sponge network | 0.078 | 0.55803 | 0.37 | 0.00013 | 0.304 |
| 39912 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | RIMBP2 | Sponge network | -0.899 | 6.0E-5 | -1.968 | 5.0E-5 | 0.304 |
| 39913 | CTC-457L16.2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 12 | TET1 | Sponge network | 1.153 | 0.00114 | -0.135 | 0.52138 | 0.304 |
| 39914 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 26 | ANK2 | Sponge network | 0.659 | 3.0E-5 | -1.603 | 1.0E-5 | 0.304 |
| 39915 | AC108488.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 19 | EYA4 | Sponge network | 0.259 | 0.12542 | 0.3 | 0.36876 | 0.304 |
| 39916 | RP11-98I9.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ZFX | Sponge network | 0.67 | 0.00048 | 0.09 | 0.42563 | 0.304 |
| 39917 | RP1-122P22.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-629-3p;hsa-miR-629-5p | 10 | RASSF8 | Sponge network | 0.065 | 0.73468 | -0.122 | 0.65591 | 0.304 |
| 39918 | PTOV1-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-664a-5p | 15 | CBFB | Sponge network | 0.87 | 0 | 1.061 | 0 | 0.304 |
| 39919 | RAD51-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 20 | ARHGEF12 | Sponge network | 0.768 | 7.0E-5 | 0.09 | 0.35693 | 0.304 |
| 39920 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-450b-5p | 24 | PIK3R1 | Sponge network | -4.546 | 0 | -0.133 | 0.36854 | 0.304 |
| 39921 | LINC00957 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PIK3CA | Sponge network | -0.421 | 0.0114 | 0.195 | 0.06908 | 0.304 |
| 39922 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-942-5p | 17 | FBXO31 | Sponge network | -0.777 | 0.1134 | -0.085 | 0.42389 | 0.304 |
| 39923 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 14 | LRRC7 | Sponge network | -0.726 | 0.00132 | -1.341 | 0.01193 | 0.304 |
| 39924 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-590-5p | 10 | THSD7A | Sponge network | 0.395 | 0.15451 | 0.242 | 0.25154 | 0.304 |
| 39925 | RAD51-AS1 |
hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | ZFX | Sponge network | 0.768 | 7.0E-5 | 0.09 | 0.42563 | 0.304 |
| 39926 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | CXXC4 | Sponge network | -0.576 | 0.10121 | -0.433 | 0.21055 | 0.304 |
| 39927 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-590-3p | 11 | HERC2 | Sponge network | -0.709 | 0.18168 | 0.114 | 0.30638 | 0.304 |
| 39928 | CTB-113P19.4 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-744-3p | 20 | PTPN14 | Sponge network | 0.906 | 0.31715 | 0.144 | 0.34595 | 0.304 |
| 39929 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | PTPN11 | Sponge network | 0.799 | 1.0E-5 | 0.431 | 2.0E-5 | 0.304 |
| 39930 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 45 | KIF1B | Sponge network | -0.246 | 0.35034 | -0.118 | 0.32258 | 0.304 |
| 39931 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 28 | ESR1 | Sponge network | -0.513 | 0.04703 | -1.035 | 3.0E-5 | 0.304 |
| 39932 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | 0.214 | 0.18246 | 0.323 | 0.00792 | 0.304 |
| 39933 | LINC00641 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 18 | ABCB5 | Sponge network | -0.222 | 0.32445 | -0.956 | 0.04077 | 0.304 |
| 39934 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p | 20 | SOX11 | Sponge network | 0.335 | 0.20596 | 2.68 | 0 | 0.304 |
| 39935 | RP11-767N6.7 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-3607-3p;hsa-miR-539-5p | 10 | DDX6 | Sponge network | 0.466 | 0.00155 | 0.091 | 0.36428 | 0.304 |
| 39936 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 17 | GRIN2A | Sponge network | 0.889 | 9.0E-5 | -2.161 | 0 | 0.304 |
| 39937 | LINC00854 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-26a-2-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-628-5p;hsa-miR-629-3p | 18 | IL6ST | Sponge network | 1.18 | 0 | -0.402 | 0.00676 | 0.304 |
| 39938 | MIR143HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 26 | RIMBP2 | Sponge network | -0.739 | 0.0574 | -1.968 | 5.0E-5 | 0.304 |
| 39939 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | MYBL1 | Sponge network | 1.063 | 0 | 0.494 | 0.02152 | 0.304 |
| 39940 | KB-1460A1.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 37 | CUX1 | Sponge network | 0.603 | 0.00127 | -0.039 | 0.6705 | 0.304 |
| 39941 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 16 | SYNPO2 | Sponge network | 0.395 | 0.15451 | -2.342 | 0 | 0.304 |
| 39942 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-590-3p | 10 | SEPT6 | Sponge network | 0.064 | 0.72934 | -0.598 | 0.00181 | 0.304 |
| 39943 | TUG1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-628-5p | 16 | CDC73 | Sponge network | 0.972 | 0 | 0.094 | 0.20463 | 0.304 |
| 39944 | CTD-2303H24.2 |
hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-96-5p | 66 | LPP | Sponge network | 0.415 | 0.14657 | -0.459 | 0.04475 | 0.304 |
| 39945 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p | 28 | GRIA3 | Sponge network | 0.572 | 0.01722 | -1.956 | 0 | 0.304 |
| 39946 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-5p | 12 | PTPN13 | Sponge network | -0.969 | 2.0E-5 | -1.208 | 0 | 0.304 |
| 39947 | OSER1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | SETBP1 | Sponge network | -0.13 | 0.48734 | -1.302 | 1.0E-5 | 0.304 |
| 39948 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | NEDD4 | Sponge network | -0.615 | 0.00015 | 0.02 | 0.89834 | 0.304 |
| 39949 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FAM133A | Sponge network | -0.942 | 0 | 0.549 | 0.29009 | 0.304 |
| 39950 | AC090587.5 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p | 11 | RASSF2 | Sponge network | -0.088 | 0.65011 | -0.273 | 0.21961 | 0.304 |
| 39951 | RP11-403I13.5 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-766-3p | 12 | IL17RD | Sponge network | 1.062 | 0.00512 | -0.019 | 0.94873 | 0.304 |
| 39952 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 35 | DMXL1 | Sponge network | -0.737 | 0.06182 | -0.426 | 0.00056 | 0.304 |
| 39953 | RP11-428J1.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p | 12 | ROBO1 | Sponge network | 0.538 | 0.00076 | -0.009 | 0.96154 | 0.304 |
| 39954 | RP11-21A7A.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-500a-5p | 10 | WDFY3 | Sponge network | -1.116 | 0.23686 | -0.201 | 0.0967 | 0.304 |
| 39955 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 51 | WDFY3 | Sponge network | 0.219 | 0.1516 | -0.201 | 0.0967 | 0.304 |
| 39956 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p | 11 | LRRC7 | Sponge network | -0.469 | 0.08721 | -1.341 | 0.01193 | 0.304 |
| 39957 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-628-5p | 31 | TCF4 | Sponge network | 0.792 | 0.0049 | 0.016 | 0.92271 | 0.304 |
| 39958 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-590-3p | 21 | MAF | Sponge network | -0.737 | 0.10822 | -1.257 | 0 | 0.304 |
| 39959 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p | 18 | KAT6B | Sponge network | 0.056 | 0.81195 | -0.478 | 0.00018 | 0.304 |
| 39960 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | CREBBP | Sponge network | -0.355 | 0.0077 | 0.126 | 0.15186 | 0.304 |
| 39961 | RP11-611E13.2 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-497-5p | 18 | MDM4 | Sponge network | 0.822 | 0 | 0.345 | 0.00762 | 0.304 |
| 39962 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 11 | TCF12 | Sponge network | 1.055 | 0 | 0.174 | 0.13271 | 0.304 |
| 39963 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-93-5p | 10 | IGSF10 | Sponge network | 0.082 | 0.55397 | -1.751 | 1.0E-5 | 0.304 |
| 39964 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-362-3p;hsa-miR-7-1-3p | 13 | DKK3 | Sponge network | -0.342 | 0.02288 | -0.052 | 0.77783 | 0.304 |
| 39965 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | PPP1R9A | Sponge network | -1.057 | 0.00029 | -0.903 | 0.04254 | 0.304 |
| 39966 | LINC00957 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | GLIPR1 | Sponge network | -0.421 | 0.0114 | 0.146 | 0.3276 | 0.304 |
| 39967 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | EPHA5 | Sponge network | -0.726 | 0.00132 | -0.957 | 0.01733 | 0.304 |
| 39968 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429 | 11 | LATS2 | Sponge network | -2.62 | 0 | 0.159 | 0.2304 | 0.304 |
| 39969 | LINC00641 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 20 | CELF4 | Sponge network | -0.222 | 0.32445 | -1.135 | 0.00344 | 0.304 |
| 39970 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | PRKAA2 | Sponge network | -0.235 | 0.15142 | -2.462 | 0 | 0.304 |
| 39971 | RP5-1021I20.1 |
hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429 | 10 | CITED2 | Sponge network | -0.785 | 0.00035 | -1.545 | 0 | 0.304 |
| 39972 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-940 | 16 | PBX1 | Sponge network | 0.792 | 0.0049 | -1.206 | 0 | 0.304 |
| 39973 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 15 | SLITRK6 | Sponge network | -0.513 | 0.04703 | -2.121 | 0 | 0.304 |
| 39974 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | FGF5 | Sponge network | -1.281 | 0.00012 | 0.549 | 0.28659 | 0.304 |
| 39975 | RP11-25K19.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | RYR2 | Sponge network | -1.057 | 0.00029 | -1.466 | 0.00031 | 0.304 |
| 39976 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 21 | SEMA3C | Sponge network | -0.899 | 6.0E-5 | -0.272 | 0.31153 | 0.304 |
| 39977 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-374a-3p | 14 | GAS7 | Sponge network | 0.542 | 0.00054 | -0.555 | 0.01602 | 0.304 |
| 39978 | CTD-3064M3.3 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-708-5p | 13 | FAM135B | Sponge network | -0.469 | 0.08721 | -3.978 | 0 | 0.304 |
| 39979 | FAM215B |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-505-5p | 19 | TBL1XR1 | Sponge network | 0.817 | 3.0E-5 | 0.578 | 0 | 0.304 |
| 39980 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SYNE1 | Sponge network | 0.862 | 0.06213 | -0.738 | 0.00316 | 0.304 |
| 39981 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | SMARCA2 | Sponge network | -1.439 | 4.0E-5 | -0.529 | 0.00014 | 0.304 |
| 39982 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-96-5p | 10 | ELMO1 | Sponge network | 0.395 | 0.15451 | 0.04 | 0.83147 | 0.304 |
| 39983 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p | 23 | MOB1B | Sponge network | -0.154 | 0.53954 | 0.197 | 0.15266 | 0.304 |
| 39984 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-502-5p;hsa-miR-539-5p | 16 | BCL11A | Sponge network | -0.162 | 0.47687 | -0.932 | 0.0063 | 0.304 |
| 39985 | RP11-1006G14.4 |
hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 12 | GLI3 | Sponge network | 0.559 | 0.00026 | -0.615 | 0.01482 | 0.304 |
| 39986 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 17 | CDHR3 | Sponge network | 0.392 | 0.0278 | -0.977 | 4.0E-5 | 0.304 |
| 39987 | LINC00173 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 12 | SFRP1 | Sponge network | 0.056 | 0.8494 | -3.566 | 0 | 0.304 |
| 39988 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | TCF4 | Sponge network | 0.998 | 0 | 0.016 | 0.92271 | 0.304 |
| 39989 | HCG22 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 15 | MCC | Sponge network | 0.816 | 0.32368 | -1.005 | 0 | 0.304 |
| 39990 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-576-5p | 17 | SSBP2 | Sponge network | 1.122 | 0.02989 | -1.58 | 0 | 0.304 |
| 39991 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-940 | 11 | FAM172A | Sponge network | 0.4 | 0.14611 | -0.57 | 0 | 0.304 |
| 39992 | RP11-48B3.4 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | GALNT13 | Sponge network | 1.757 | 0 | -0.857 | 0.07439 | 0.304 |
| 39993 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-664a-3p | 17 | FZD3 | Sponge network | 0.706 | 0 | 0.104 | 0.60316 | 0.304 |
| 39994 | RP5-1085F17.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-940 | 28 | CREB1 | Sponge network | 0.541 | 0.00125 | 0.203 | 0.00537 | 0.304 |
| 39995 | AC090587.5 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-501-5p;hsa-miR-93-3p | 10 | GRIK3 | Sponge network | -0.088 | 0.65011 | -3.208 | 0 | 0.304 |
| 39996 | AC093110.3 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p | 10 | PIK3R1 | Sponge network | 1.044 | 2.0E-5 | -0.133 | 0.36854 | 0.304 |
| 39997 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 22 | USP12 | Sponge network | 0.415 | 0.14657 | -0.102 | 0.40768 | 0.304 |
| 39998 | CTC-429P9.3 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-424-5p | 12 | DOCK4 | Sponge network | 0.082 | 0.55397 | 0.502 | 0.00108 | 0.304 |
| 39999 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-455-3p;hsa-miR-7-1-3p | 10 | DKK3 | Sponge network | 0.693 | 0.00076 | -0.052 | 0.77783 | 0.304 |
| 40000 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-98-5p | 10 | PBX1 | Sponge network | 1.344 | 0 | -1.206 | 0 | 0.304 |
| 40001 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 11 | FREM2 | Sponge network | -1.886 | 0 | -0.437 | 0.46353 | 0.304 |
| 40002 | FGD5-AS1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | CSDE1 | Sponge network | 0.401 | 0.00066 | -0.493 | 0 | 0.304 |
| 40003 | RP5-894A10.6 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-495-3p | 12 | BMPR2 | Sponge network | 0.697 | 9.0E-5 | 0.206 | 0.0635 | 0.304 |
| 40004 | RP11-181G12.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-671-5p | 21 | ADARB1 | Sponge network | 0.556 | 0.00298 | -0.335 | 0.06631 | 0.304 |
| 40005 | AC141928.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | SETBP1 | Sponge network | 0.853 | 0.15352 | -1.302 | 1.0E-5 | 0.304 |
| 40006 | NEAT1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 25 | ZFHX3 | Sponge network | 0.705 | 0.00014 | 0.389 | 0.01363 | 0.304 |
| 40007 | RP11-182J1.1 |
hsa-let-7g-5p;hsa-miR-146a-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-96-5p | 10 | HIP1 | Sponge network | -0.187 | 0.23787 | 0.774 | 0 | 0.304 |
| 40008 | CTD-3110H11.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-3607-3p;hsa-miR-629-3p | 11 | DDX6 | Sponge network | 1.735 | 0 | 0.091 | 0.36428 | 0.304 |
| 40009 | AC106786.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-877-5p | 10 | SUFU | Sponge network | 1.122 | 0.02989 | -0.04 | 0.65489 | 0.304 |
| 40010 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | PRKAA2 | Sponge network | 1.302 | 7.0E-5 | -2.462 | 0 | 0.304 |
| 40011 | CTD-2303H24.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 29 | ABL2 | Sponge network | 0.415 | 0.14657 | 0.82 | 0 | 0.304 |
| 40012 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | SYNE1 | Sponge network | 0.219 | 0.1516 | -0.738 | 0.00316 | 0.304 |
| 40013 | HAR1A |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-93-3p;hsa-miR-940 | 20 | QKI | Sponge network | -0.672 | 0.19447 | -0.333 | 0.04111 | 0.304 |
| 40014 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 36 | RAP1A | Sponge network | -1.216 | 0 | -0.929 | 0 | 0.304 |
| 40015 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 12 | SPG20 | Sponge network | 1.757 | 0 | -1.16 | 0 | 0.304 |
| 40016 | NDUFA6-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-98-5p | 14 | SCN11A | Sponge network | -0.355 | 0.0077 | -1.15 | 0.00299 | 0.304 |
| 40017 | EMX2OS |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 16 | JUN | Sponge network | -0.777 | 0.1134 | -0.932 | 0 | 0.304 |
| 40018 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SEMA3C | Sponge network | -1.161 | 0.00591 | -0.272 | 0.31153 | 0.304 |
| 40019 | SNX29P2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 16 | DOCK4 | Sponge network | 0.4 | 0.14611 | 0.502 | 0.00108 | 0.304 |
| 40020 | RP4-668J24.2 |
hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CADM1 | Sponge network | -1.054 | 0.0706 | -0.9 | 0.00084 | 0.304 |
| 40021 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p;hsa-miR-93-3p | 27 | AKAP11 | Sponge network | 0.078 | 0.55803 | 0.196 | 0.09825 | 0.304 |
| 40022 | RP11-283I3.6 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-500a-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 13 | UBE4B | Sponge network | 0.614 | 1.0E-5 | -0.235 | 0.007 | 0.304 |
| 40023 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-539-5p;hsa-miR-96-5p | 13 | TP53INP1 | Sponge network | -0.326 | 0.14314 | -0.496 | 0.00064 | 0.304 |
| 40024 | CTD-2554C21.2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-744-3p;hsa-miR-96-5p | 19 | HIP1 | Sponge network | -1.654 | 0.00144 | 0.774 | 0 | 0.304 |
| 40025 | LINC00173 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | CNTNAP1 | Sponge network | 0.056 | 0.8494 | 0.358 | 0.13727 | 0.304 |
| 40026 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-500a-5p;hsa-miR-629-5p | 10 | MAF | Sponge network | -2.076 | 0.00548 | -1.257 | 0 | 0.304 |
| 40027 | CTD-2139B15.2 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p | 10 | RASAL2 | Sponge network | 0.286 | 0.00612 | 0.869 | 0 | 0.304 |
| 40028 | RP11-479G22.8 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-2355-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ETV1 | Sponge network | 1.308 | 0 | -0.096 | 0.67674 | 0.304 |
| 40029 | PART1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | LUZP2 | Sponge network | -2.925 | 0 | -0.056 | 0.89416 | 0.304 |
| 40030 | CTC-559E9.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-877-5p | 14 | NAV3 | Sponge network | 0.594 | 0.00064 | 0.212 | 0.47729 | 0.304 |
| 40031 | RP11-102N12.3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-96-5p | 12 | HIP1 | Sponge network | -0.351 | 0.11358 | 0.774 | 0 | 0.304 |
| 40032 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 28 | CXXC4 | Sponge network | -0.222 | 0.32445 | -0.433 | 0.21055 | 0.304 |
| 40033 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 14 | PSIP1 | Sponge network | 0.659 | 3.0E-5 | -0.005 | 0.96897 | 0.304 |
| 40034 | LINC00865 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p | 14 | CSDE1 | Sponge network | -1.249 | 7.0E-5 | -0.493 | 0 | 0.304 |
| 40035 | RP1-151F17.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.222 | 0.29133 | -0.57 | 0 | 0.304 |
| 40036 | RP11-38L15.3 |
hsa-miR-128-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-935 | 10 | RNF144A | Sponge network | 0.344 | 0.3127 | 0.594 | 0.0009 | 0.304 |
| 40037 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | LATS1 | Sponge network | 0.556 | 0.00298 | 0.109 | 0.34208 | 0.304 |
| 40038 | AC006116.24 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-550a-3p | 11 | RASSF3 | Sponge network | -0.473 | 0.07602 | -0.226 | 0.11691 | 0.304 |
| 40039 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-5p | 36 | ERBB4 | Sponge network | 0.176 | 0.37402 | -1.975 | 2.0E-5 | 0.304 |
| 40040 | HOXB-AS3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-92a-1-5p | 14 | PRKAB2 | Sponge network | 0.343 | 0.28887 | -0.367 | 0.01371 | 0.304 |
| 40041 | SERTAD4-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-93-3p | 14 | ERC1 | Sponge network | -0.932 | 0.00329 | -0.108 | 0.38794 | 0.304 |
| 40042 | RP11-248G5.8 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 14 | RPS6KA3 | Sponge network | 1.294 | 0 | 0.256 | 0.02419 | 0.304 |
| 40043 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-7-1-3p | 17 | CXXC4 | Sponge network | -1.092 | 4.0E-5 | -0.433 | 0.21055 | 0.304 |
| 40044 | MIR143HG |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | PTCH1 | Sponge network | -0.739 | 0.0574 | -0.1 | 0.6179 | 0.304 |
| 40045 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-93-5p | 18 | PLSCR4 | Sponge network | -0.465 | 0.04703 | -0.474 | 0.03833 | 0.304 |
| 40046 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-590-3p | 10 | FAM188A | Sponge network | -0.342 | 0.02288 | -0.44 | 0.00018 | 0.304 |
| 40047 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | SLITRK6 | Sponge network | -0.322 | 0.25945 | -2.121 | 0 | 0.304 |
| 40048 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 10 | ARHGAP28 | Sponge network | -0.829 | 0.01343 | -0.911 | 0.00013 | 0.303 |
| 40049 | LINC01018 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZNF844 | Sponge network | -2.915 | 0.00015 | -0.966 | 0.00035 | 0.303 |
| 40050 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | KCNAB1 | Sponge network | 0.421 | 0.00084 | -0.755 | 2.0E-5 | 0.303 |
| 40051 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | SON | Sponge network | 1.294 | 0 | 0.168 | 0.04406 | 0.303 |
| 40052 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | -0.738 | 0.21233 | -3.143 | 0 | 0.303 |
| 40053 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | RECK | Sponge network | -0.355 | 0.0077 | -1.043 | 0 | 0.303 |
| 40054 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 23 | ADARB1 | Sponge network | -0.663 | 0.10887 | -0.335 | 0.06631 | 0.303 |
| 40055 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | CLASP2 | Sponge network | 1.923 | 0 | -0.038 | 0.71 | 0.303 |
| 40056 | CTC-296K1.4 |
hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-5p | 11 | LIFR | Sponge network | -2.076 | 0.00548 | -1.794 | 0 | 0.303 |
| 40057 | PSMD5-AS1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | EPG5 | Sponge network | 0.569 | 0.00067 | 0.101 | 0.3563 | 0.303 |
| 40058 | LINC00957 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-942-5p | 21 | OPCML | Sponge network | -0.421 | 0.0114 | -2.498 | 0 | 0.303 |
| 40059 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 23 | SOX5 | Sponge network | 0.381 | 0.25967 | -1.091 | 4.0E-5 | 0.303 |
| 40060 | LINC00667 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | PRUNE2 | Sponge network | -0.235 | 0.15142 | -1.733 | 0.00022 | 0.303 |
| 40061 | PART1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | JDP2 | Sponge network | -2.925 | 0 | -0.967 | 0 | 0.303 |
| 40062 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SEMA3E | Sponge network | -0.612 | 0.00094 | -1.717 | 0.00137 | 0.303 |
| 40063 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-5p | 17 | CRTC1 | Sponge network | -0.342 | 0.02288 | -0.116 | 0.28412 | 0.303 |
| 40064 | AC024560.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-331-3p;hsa-miR-342-5p | 12 | ATRX | Sponge network | 2.166 | 0.00228 | 0.261 | 0.04408 | 0.303 |
| 40065 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-625-5p | 20 | ZMYM2 | Sponge network | 0.72 | 0.0001 | 0.279 | 0.01273 | 0.303 |
| 40066 | RP11-983P16.4 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ITGB1 | Sponge network | 0.41 | 0.02214 | 0.524 | 0.00017 | 0.303 |
| 40067 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | ERRFI1 | Sponge network | 0.381 | 0.25967 | -0.798 | 1.0E-5 | 0.303 |
| 40068 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | ARNT2 | Sponge network | -0.487 | 0.02157 | -0.332 | 0.25851 | 0.303 |
| 40069 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 10 | EDA2R | Sponge network | -0.342 | 0.02288 | -0.587 | 0.01598 | 0.303 |
| 40070 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | FOXO1 | Sponge network | 0.101 | 0.60919 | -0.141 | 0.33874 | 0.303 |
| 40071 | AGAP11 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-590-5p | 23 | LIMCH1 | Sponge network | -1.645 | 0 | -0.915 | 1.0E-5 | 0.303 |
| 40072 | RP11-13K12.5 |
hsa-miR-183-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-7-1-3p | 10 | ITGB1 | Sponge network | -0.318 | 0.65847 | 0.524 | 0.00017 | 0.303 |
| 40073 | ACTA2-AS1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 14 | SHPRH | Sponge network | 0.194 | 0.64278 | 0.098 | 0.40994 | 0.303 |
| 40074 | LINC00957 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935 | 31 | ABI2 | Sponge network | -0.421 | 0.0114 | 0.175 | 0.14787 | 0.303 |
| 40075 | CKMT2-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 45 | GRIK3 | Sponge network | 0.082 | 0.55654 | -3.208 | 0 | 0.303 |
| 40076 | BOLA3-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p | 18 | CTIF | Sponge network | -0.246 | 0.35034 | -0.389 | 0.00549 | 0.303 |
| 40077 | RP11-473I1.10 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 16 | FOXP1 | Sponge network | 0.673 | 0 | 0.042 | 0.74368 | 0.303 |
| 40078 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 68 | KIF1B | Sponge network | -1.575 | 6.0E-5 | -0.118 | 0.32258 | 0.303 |
| 40079 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-5p | 35 | WDFY3 | Sponge network | 0.176 | 0.37402 | -0.201 | 0.0967 | 0.303 |
| 40080 | AC156455.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326 | 10 | ABI2 | Sponge network | 1.154 | 0.00041 | 0.175 | 0.14787 | 0.303 |
| 40081 | GNG12-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 14 | SPTBN4 | Sponge network | -0.793 | 7.0E-5 | -0.786 | 0.01281 | 0.303 |
| 40082 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | SOX5 | Sponge network | -0.668 | 3.0E-5 | -1.091 | 4.0E-5 | 0.303 |
| 40083 | GNG12-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p | 36 | CREB3L2 | Sponge network | -0.793 | 7.0E-5 | -0.525 | 2.0E-5 | 0.303 |
| 40084 | RP5-1085F17.3 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-34a-5p | 11 | STAG2 | Sponge network | 0.541 | 0.00125 | 0.252 | 0.00438 | 0.303 |
| 40085 | HOXD-AS2 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-455-3p;hsa-miR-502-5p | 22 | DST | Sponge network | -0.208 | 0.65592 | -0.713 | 0.00096 | 0.303 |
| 40086 | AGAP11 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-629-3p | 15 | CXCL12 | Sponge network | -1.645 | 0 | -1.421 | 0 | 0.303 |
| 40087 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 16 | DKK3 | Sponge network | -2.117 | 0.00161 | -0.052 | 0.77783 | 0.303 |
| 40088 | PWAR6 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PHF3 | Sponge network | -1.575 | 6.0E-5 | 0.126 | 0.19652 | 0.303 |
| 40089 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PCDH17 | Sponge network | -0.487 | 0.02157 | 0.731 | 2.0E-5 | 0.303 |
| 40090 | RP5-855D21.1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-29a-5p | 11 | KIF1B | Sponge network | 1.115 | 0.00188 | -0.118 | 0.32258 | 0.303 |
| 40091 | RP11-384K6.6 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 10 | PICALM | Sponge network | 0.946 | 0 | 0.086 | 0.23523 | 0.303 |
| 40092 | RP11-927P21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p | 11 | ZNF208 | Sponge network | 0.921 | 0.00118 | -0.4 | 0.22381 | 0.303 |
| 40093 | RP11-212P7.2 |
hsa-miR-126-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | IRS1 | Sponge network | 0.219 | 0.1516 | -0.026 | 0.88855 | 0.303 |
| 40094 | AF131215.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-429 | 16 | KANK1 | Sponge network | -0.176 | 0.59692 | -0.55 | 0.00134 | 0.303 |
| 40095 | RP11-1006G14.4 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 12 | CELF4 | Sponge network | 0.559 | 0.00026 | -1.135 | 0.00344 | 0.303 |
| 40096 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-629-3p | 33 | KIF1B | Sponge network | -0.583 | 0.14589 | -0.118 | 0.32258 | 0.303 |
| 40097 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p | 11 | DAB2 | Sponge network | -0.125 | 0.49414 | 0.431 | 0.0113 | 0.303 |
| 40098 | AP001469.9 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 11 | TBL1XR1 | Sponge network | 1.096 | 0 | 0.578 | 0 | 0.303 |
| 40099 | PWAR6 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | PCDH17 | Sponge network | -1.575 | 6.0E-5 | 0.731 | 2.0E-5 | 0.303 |
| 40100 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-625-3p | 18 | FZD3 | Sponge network | 0.752 | 0 | 0.104 | 0.60316 | 0.303 |
| 40101 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-493-5p | 19 | MYBL1 | Sponge network | 0.368 | 0.20093 | 0.494 | 0.02152 | 0.303 |
| 40102 | MEG3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-629-5p | 13 | UVRAG | Sponge network | 0.249 | 0.35902 | -0.017 | 0.83865 | 0.303 |
| 40103 | RP11-890B15.3 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | LIMCH1 | Sponge network | 0.101 | 0.60919 | -0.915 | 1.0E-5 | 0.303 |
| 40104 | H19 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-484 | 13 | ABL1 | Sponge network | 2.439 | 0 | -0.215 | 0.07804 | 0.303 |
| 40105 | RP4-605O3.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-493-3p;hsa-miR-590-3p | 10 | PRUNE2 | Sponge network | 0.262 | 0.0475 | -1.733 | 0.00022 | 0.303 |
| 40106 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p | 22 | CRTC1 | Sponge network | 0.083 | 0.66405 | -0.116 | 0.28412 | 0.303 |
| 40107 | ZNF667-AS1 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-93-5p;hsa-miR-940 | 13 | JUN | Sponge network | -0.976 | 0.00025 | -0.932 | 0 | 0.303 |
| 40108 | CTD-2089N3.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p | 14 | SSBP2 | Sponge network | -0.314 | 0.62033 | -1.58 | 0 | 0.303 |
| 40109 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-935 | 32 | RAP1A | Sponge network | -0.969 | 2.0E-5 | -0.929 | 0 | 0.303 |
| 40110 | LPP-AS2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-769-5p | 12 | RCAN2 | Sponge network | -0.18 | 0.20343 | -0.33 | 0.15172 | 0.303 |
| 40111 | FZD10-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 31 | DIP2C | Sponge network | -0.727 | 0.04726 | -0.436 | 0.01713 | 0.303 |
| 40112 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 31 | LIFR | Sponge network | -0.899 | 6.0E-5 | -1.794 | 0 | 0.303 |
| 40113 | CTC-471J1.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 11 | ABI2 | Sponge network | 1.11 | 1.0E-5 | 0.175 | 0.14787 | 0.303 |
| 40114 | RP11-13K12.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-589-3p | 28 | AFF1 | Sponge network | -0.154 | 0.78939 | -0.384 | 0.00053 | 0.303 |
| 40115 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-331-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-93-5p | 33 | WDFY3 | Sponge network | 0.381 | 0.25967 | -0.201 | 0.0967 | 0.303 |
| 40116 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-24-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-671-5p;hsa-miR-744-5p | 12 | CHD5 | Sponge network | -0.285 | 0.2172 | -0.922 | 0.01521 | 0.303 |
| 40117 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-376c-3p;hsa-miR-411-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | SMAD2 | Sponge network | 0.559 | 0.00026 | -0.128 | 0.12732 | 0.303 |
| 40118 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 22 | TSHZ3 | Sponge network | -0.141 | 0.26434 | -0.556 | 0.01516 | 0.303 |
| 40119 | HHIP-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 20 | LRIG1 | Sponge network | -0.737 | 0.10822 | -0.537 | 0.00588 | 0.303 |
| 40120 | KB-1410C5.5 |
hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | UBR3 | Sponge network | 0.959 | 0 | -0.021 | 0.82774 | 0.303 |
| 40121 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-409-3p;hsa-miR-935 | 24 | NFIB | Sponge network | 0.37 | 0.27614 | 0.023 | 0.90795 | 0.303 |
| 40122 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-5p | 32 | PIK3R1 | Sponge network | 0.259 | 0.12542 | -0.133 | 0.36854 | 0.303 |
| 40123 | LINC00578 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577 | 13 | SEMA3E | Sponge network | 0.811 | 0.03228 | -1.717 | 0.00137 | 0.303 |
| 40124 | AC034220.3 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | AKAP11 | Sponge network | 0.309 | 0.07304 | 0.196 | 0.09825 | 0.303 |
| 40125 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-940 | 14 | AKAP11 | Sponge network | 0.983 | 0.00048 | 0.196 | 0.09825 | 0.303 |
| 40126 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | USP6 | Sponge network | 0.8 | 0 | 0.188 | 0.3871 | 0.303 |
| 40127 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-493-5p;hsa-miR-654-3p | 21 | MYBL1 | Sponge network | -0.405 | 0.22013 | 0.494 | 0.02152 | 0.303 |
| 40128 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | BCL2 | Sponge network | 0.16 | 0.50785 | -0.899 | 0 | 0.303 |
| 40129 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 17 | SLITRK6 | Sponge network | -2.915 | 0.00015 | -2.121 | 0 | 0.303 |
| 40130 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-301a-3p | 12 | PBX1 | Sponge network | 0.598 | 0.38481 | -1.206 | 0 | 0.303 |
| 40131 | RP11-686D22.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 14 | BNC2 | Sponge network | 0.793 | 5.0E-5 | -0.491 | 0.09988 | 0.303 |
| 40132 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p | 10 | ITCH | Sponge network | 1.044 | 2.0E-5 | 0.084 | 0.34058 | 0.303 |
| 40133 | DNM1P35 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p | 19 | CADM1 | Sponge network | 0.051 | 0.83388 | -0.9 | 0.00084 | 0.303 |
| 40134 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 14 | ERRFI1 | Sponge network | -0.739 | 0.00044 | -0.798 | 1.0E-5 | 0.303 |
| 40135 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-625-5p | 17 | ZMYM2 | Sponge network | 0.909 | 0 | 0.279 | 0.01273 | 0.303 |
| 40136 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p | 26 | TMTC3 | Sponge network | -0.222 | 0.32445 | 0.123 | 0.22189 | 0.303 |
| 40137 | RP11-212P7.2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | KLF10 | Sponge network | 0.219 | 0.1516 | -0.783 | 0 | 0.303 |
| 40138 | CTC-559E9.6 |
hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-7-5p | 13 | MOB1B | Sponge network | 0.601 | 1.0E-5 | 0.197 | 0.15266 | 0.303 |
| 40139 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 36 | BCL2 | Sponge network | 0.421 | 0.00084 | -0.899 | 0 | 0.303 |
| 40140 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-92a-3p | 24 | CRTC1 | Sponge network | 0.253 | 0.09565 | -0.116 | 0.28412 | 0.303 |
| 40141 | HOXB-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-664a-5p | 36 | LPP | Sponge network | 1.316 | 0.01106 | -0.459 | 0.04475 | 0.303 |
| 40142 | RP11-244H3.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | TGFBR3 | Sponge network | 0.659 | 3.0E-5 | -1.173 | 1.0E-5 | 0.303 |
| 40143 | RP11-434B12.1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | ZDBF2 | Sponge network | -0.031 | 0.89063 | -0.998 | 0.00025 | 0.303 |
| 40144 | RP11-535M15.1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-942-5p | 11 | UNC80 | Sponge network | -1.14 | 0.03513 | -1.934 | 0 | 0.303 |
| 40145 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30b-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p | 15 | CXXC4 | Sponge network | -0.555 | 0.06156 | -0.433 | 0.21055 | 0.303 |
| 40146 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 20 | NTRK3 | Sponge network | -0.31 | 0.33871 | -1.77 | 3.0E-5 | 0.303 |
| 40147 | HOXA-AS2 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | GJA1 | Sponge network | 0.61 | 0.01593 | -0.485 | 0.02179 | 0.303 |
| 40148 | RP11-156P1.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-942-5p | 31 | IGFBP5 | Sponge network | 0.214 | 0.18246 | -0.806 | 0.00105 | 0.303 |
| 40149 | RP11-421L21.3 |
hsa-miR-101-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 11 | CBFB | Sponge network | 0.962 | 0 | 1.061 | 0 | 0.303 |
| 40150 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 12 | FAT4 | Sponge network | 0.238 | 0.70544 | -0.354 | 0.14694 | 0.303 |
| 40151 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p | 32 | RASSF2 | Sponge network | -0.773 | 0.04073 | -0.273 | 0.21961 | 0.303 |
| 40152 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p | 15 | ATRX | Sponge network | 1.03 | 0 | 0.261 | 0.04408 | 0.303 |
| 40153 | SERHL |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p | 11 | WNK2 | Sponge network | -0.602 | 0.00331 | -0.352 | 0.31066 | 0.303 |
| 40154 | LINC00886 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-532-5p | 16 | ZMYND11 | Sponge network | -0.371 | 0.24604 | -0.2 | 0.05449 | 0.303 |
| 40155 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-451a;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | BCL2 | Sponge network | -0.318 | 0.65847 | -0.899 | 0 | 0.303 |
| 40156 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | NF1 | Sponge network | 1.308 | 0 | 0.51 | 0 | 0.303 |
| 40157 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | SLITRK6 | Sponge network | -0.896 | 0.00099 | -2.121 | 0 | 0.303 |
| 40158 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 22 | GPC6 | Sponge network | -0.612 | 0.00094 | -0.054 | 0.81465 | 0.303 |
| 40159 | RP11-806O11.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-455-3p;hsa-miR-590-3p | 14 | RICTOR | Sponge network | 0.284 | 0.52463 | 0.388 | 0.00188 | 0.303 |
| 40160 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | UBE4B | Sponge network | -0.899 | 6.0E-5 | -0.235 | 0.007 | 0.303 |
| 40161 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-708-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 37 | TOM1L2 | Sponge network | -1.111 | 0.0003 | -0.994 | 0 | 0.303 |
| 40162 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PRKAB2 | Sponge network | 0.539 | 0.03722 | -0.367 | 0.01371 | 0.303 |
| 40163 | CTD-3064M3.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-625-5p | 10 | CELF4 | Sponge network | -0.469 | 0.08721 | -1.135 | 0.00344 | 0.303 |
| 40164 | EIF3J-AS1 |
hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | CHD2 | Sponge network | 0.331 | 0.00509 | 0.049 | 0.55371 | 0.303 |
| 40165 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | 0.083 | 0.66405 | -0.7 | 1.0E-5 | 0.303 |
| 40166 | DNM3OS |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 16 | ERRFI1 | Sponge network | 0.572 | 0.01722 | -0.798 | 1.0E-5 | 0.303 |
| 40167 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-589-3p | 20 | ERBB4 | Sponge network | -4.463 | 0.00025 | -1.975 | 2.0E-5 | 0.303 |
| 40168 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | PEG3 | Sponge network | 0.349 | 0.01695 | -1.74 | 0 | 0.303 |
| 40169 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-628-5p | 28 | GRIA3 | Sponge network | 0.249 | 0.35902 | -1.956 | 0 | 0.303 |
| 40170 | RP11-439L18.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p | 11 | TSHZ3 | Sponge network | 0.224 | 0.48719 | -0.556 | 0.01516 | 0.303 |
| 40171 | DNM1P35 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 31 | TRPS1 | Sponge network | 0.051 | 0.83388 | -0.191 | 0.44109 | 0.303 |
| 40172 | LINC00621 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | GRIN2A | Sponge network | 0.131 | 0.8436 | -2.161 | 0 | 0.303 |
| 40173 | MAGI2-AS3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 20 | CELF4 | Sponge network | -0.129 | 0.57177 | -1.135 | 0.00344 | 0.303 |
| 40174 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 21 | PRKAB2 | Sponge network | 0.05 | 0.95483 | -0.367 | 0.01371 | 0.303 |
| 40175 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-940 | 24 | QKI | Sponge network | -4.477 | 0 | -0.333 | 0.04111 | 0.303 |
| 40176 | AC108488.4 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 19 | BRD3 | Sponge network | 0.259 | 0.12542 | 0.37 | 0.00013 | 0.303 |
| 40177 | RP11-535M15.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p | 13 | FBXO32 | Sponge network | -1.14 | 0.03513 | -0.495 | 0.07561 | 0.303 |
| 40178 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-940 | 18 | AKAP11 | Sponge network | 0.556 | 0.00298 | 0.196 | 0.09825 | 0.303 |
| 40179 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 24 | LRRC55 | Sponge network | -1.216 | 0 | -0.026 | 0.93163 | 0.303 |
| 40180 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | EDA2R | Sponge network | -0.709 | 0.18168 | -0.587 | 0.01598 | 0.303 |
| 40181 | NEAT1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.705 | 0.00014 | 0.943 | 0 | 0.303 |
| 40182 | CTD-3064M3.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 11 | FREM2 | Sponge network | -0.469 | 0.08721 | -0.437 | 0.46353 | 0.303 |
| 40183 | RP5-1074L1.4 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-374b-5p | 13 | UBE2D1 | Sponge network | 1.923 | 0 | 0.468 | 0 | 0.303 |
| 40184 | FAM157C |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c | 14 | BRD3 | Sponge network | 0.91 | 0 | 0.37 | 0.00013 | 0.303 |
| 40185 | RP11-248G5.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 20 | ZFX | Sponge network | 1.294 | 0 | 0.09 | 0.42563 | 0.303 |
| 40186 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | GLIPR1 | Sponge network | 0.064 | 0.72934 | 0.146 | 0.3276 | 0.303 |
| 40187 | RP5-1139B12.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-93-3p | 10 | UNC5C | Sponge network | 0.598 | 0.38481 | -0.178 | 0.40214 | 0.303 |
| 40188 | CTD-2017D11.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-582-5p | 12 | NRXN3 | Sponge network | 0.792 | 0.0049 | -0.893 | 0.04186 | 0.303 |
| 40189 | C1RL-AS1 |
hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-376c-3p | 10 | TBL1XR1 | Sponge network | 1.26 | 0 | 0.578 | 0 | 0.303 |
| 40190 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 39 | TMEM132B | Sponge network | -0.235 | 0.15142 | -0.83 | 0.01084 | 0.303 |
| 40191 | WDFY3-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-708-5p;hsa-miR-744-3p;hsa-miR-940 | 29 | TOM1L2 | Sponge network | -0.942 | 0 | -0.994 | 0 | 0.303 |
| 40192 | RP11-686D22.8 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-96-5p | 16 | ZFHX4 | Sponge network | 0.898 | 1.0E-5 | 0.018 | 0.95574 | 0.303 |
| 40193 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-629-3p | 55 | KIF1B | Sponge network | 0.253 | 0.09565 | -0.118 | 0.32258 | 0.303 |
| 40194 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-93-5p | 13 | TRIM3 | Sponge network | -0.129 | 0.57177 | -0.577 | 0 | 0.303 |
| 40195 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 39 | CBX5 | Sponge network | 0.082 | 0.55654 | 0.165 | 0.24446 | 0.303 |
| 40196 | RP5-1074L1.4 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-214-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-628-5p | 11 | MAT2A | Sponge network | 1.923 | 0 | -0.073 | 0.36195 | 0.303 |
| 40197 | C4B-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-486-5p;hsa-miR-590-5p | 11 | AKAP6 | Sponge network | 2.306 | 9.0E-5 | -1.586 | 3.0E-5 | 0.303 |
| 40198 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 13 | BCLAF1 | Sponge network | 0.569 | 0.00067 | 0.211 | 0.00684 | 0.303 |
| 40199 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-374b-5p | 10 | ABCC9 | Sponge network | 0.001 | 0.99753 | -0.466 | 0.14121 | 0.303 |
| 40200 | RP11-999E24.3 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 16 | MED12L | Sponge network | -0.693 | 0.05629 | -0.194 | 0.55299 | 0.303 |
| 40201 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p | 11 | ELMO1 | Sponge network | -0.096 | 0.67958 | 0.04 | 0.83147 | 0.303 |
| 40202 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | NR4A3 | Sponge network | -1.249 | 7.0E-5 | -1.385 | 0 | 0.303 |
| 40203 | RP11-148O21.2 |
hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-361-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p | 10 | QKI | Sponge network | 0.233 | 0.81029 | -0.333 | 0.04111 | 0.303 |
| 40204 | RP11-175O19.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-497-5p | 18 | UBR3 | Sponge network | 0.917 | 0 | -0.021 | 0.82774 | 0.303 |
| 40205 | RP11-155G14.6 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-326 | 16 | CCDC6 | Sponge network | 2.483 | 0 | 0.27 | 0.02555 | 0.303 |
| 40206 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | GRIN2A | Sponge network | 0.385 | 0.10067 | -2.161 | 0 | 0.303 |
| 40207 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 18 | MAP3K12 | Sponge network | -1.575 | 6.0E-5 | -0.057 | 0.59369 | 0.303 |
| 40208 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | MED12L | Sponge network | 0.41 | 0.02214 | -0.194 | 0.55299 | 0.303 |
| 40209 | RP11-1006G14.4 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-590-3p | 14 | GLIPR1 | Sponge network | 0.559 | 0.00026 | 0.146 | 0.3276 | 0.303 |
| 40210 | RP11-798G7.7 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-766-3p | 22 | IL6ST | Sponge network | 0.858 | 3.0E-5 | -0.402 | 0.00676 | 0.303 |
| 40211 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 31 | ZFHX4 | Sponge network | 0.657 | 4.0E-5 | 0.018 | 0.95574 | 0.303 |
| 40212 | LINC00173 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | ERG | Sponge network | 0.056 | 0.8494 | 0.002 | 0.99011 | 0.303 |
| 40213 | CTC-297N7.9 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-26b-3p | 14 | EPHA7 | Sponge network | -2.069 | 0 | -2.197 | 3.0E-5 | 0.303 |
| 40214 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-744-5p;hsa-miR-942-5p | 25 | NOTCH2 | Sponge network | -2.531 | 0 | 0.138 | 0.35163 | 0.303 |
| 40215 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | KANK1 | Sponge network | -2.452 | 0 | -0.55 | 0.00134 | 0.303 |
| 40216 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 51 | SMAD4 | Sponge network | -0.899 | 6.0E-5 | -0.313 | 0.00413 | 0.303 |
| 40217 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-5p | 15 | PTGFR | Sponge network | 0.395 | 0.15451 | -0.233 | 0.45421 | 0.303 |
| 40218 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 13 | TMTC3 | Sponge network | 0.993 | 0 | 0.123 | 0.22189 | 0.303 |
| 40219 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 29 | ZMYM2 | Sponge network | 0.569 | 0.00067 | 0.279 | 0.01273 | 0.303 |
| 40220 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-28-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 12 | HACE1 | Sponge network | 0.541 | 0.00125 | -0.072 | 0.55239 | 0.303 |
| 40221 | RP11-263K19.4 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 16 | PTPN14 | Sponge network | 0.859 | 0.00331 | 0.144 | 0.34595 | 0.303 |
| 40222 | GNG12-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-671-5p | 11 | PIK3IP1 | Sponge network | -0.793 | 7.0E-5 | -0.187 | 0.19945 | 0.303 |
| 40223 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-671-5p | 25 | KCNA6 | Sponge network | 0.331 | 0.57247 | -1.531 | 0.00013 | 0.303 |
| 40224 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-935;hsa-miR-96-5p | 32 | FBXO32 | Sponge network | -0.969 | 2.0E-5 | -0.495 | 0.07561 | 0.303 |
| 40225 | FENDRR |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 28 | PRDM2 | Sponge network | -1.281 | 0.00012 | -0.258 | 0.00721 | 0.303 |
| 40226 | LINC00963 |
hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 11 | LZTS1 | Sponge network | 0.523 | 9.0E-5 | 1.664 | 0 | 0.303 |
| 40227 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-590-3p | 17 | NIN | Sponge network | 1.254 | 0 | -0.253 | 0.13794 | 0.303 |
| 40228 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-96-5p | 45 | FOXP1 | Sponge network | 0.082 | 0.55654 | 0.042 | 0.74368 | 0.303 |
| 40229 | AC004893.11 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 13 | PTPN14 | Sponge network | 1.317 | 0 | 0.144 | 0.34595 | 0.303 |
| 40230 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-93-5p | 16 | CDHR3 | Sponge network | -0.303 | 0.21002 | -0.977 | 4.0E-5 | 0.303 |
| 40231 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-7-5p | 17 | ESR1 | Sponge network | 0.348 | 0.32912 | -1.035 | 3.0E-5 | 0.303 |
| 40232 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-3913-5p;hsa-miR-452-5p;hsa-miR-550a-3p | 18 | RAP1A | Sponge network | -2.095 | 0.00112 | -0.929 | 0 | 0.303 |
| 40233 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-93-3p | 19 | SNX29 | Sponge network | -0.206 | 0.12942 | -0.34 | 0.01371 | 0.303 |
| 40234 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | RARB | Sponge network | -0.612 | 0.00094 | -0.825 | 2.0E-5 | 0.303 |
| 40235 | RP11-182J1.1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-484;hsa-miR-589-5p | 16 | CTIF | Sponge network | -0.187 | 0.23787 | -0.389 | 0.00549 | 0.303 |
| 40236 | LPP-AS2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-454-3p | 10 | PIK3IP1 | Sponge network | -0.18 | 0.20343 | -0.187 | 0.19945 | 0.303 |
| 40237 | AP001347.6 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-425-5p;hsa-miR-590-3p | 11 | RIT1 | Sponge network | -1.161 | 0.00591 | -0.296 | 0.00054 | 0.303 |
| 40238 | HOXB-AS2 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-576-5p | 13 | ZFHX4 | Sponge network | 1.316 | 0.01106 | 0.018 | 0.95574 | 0.303 |
| 40239 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-628-5p | 14 | SLC5A3 | Sponge network | 0.75 | 0.00054 | 0.493 | 5.0E-5 | 0.303 |
| 40240 | GUSBP11 |
hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 11 | KIF5B | Sponge network | 0.856 | 0 | 0.362 | 0.00066 | 0.303 |
| 40241 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 11 | EPAS1 | Sponge network | -4.546 | 0 | -0.184 | 0.14418 | 0.303 |
| 40242 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 17 | PDS5B | Sponge network | 0.064 | 0.72934 | 0.259 | 0.00962 | 0.303 |
| 40243 | GABPB1-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-940 | 19 | SIRT1 | Sponge network | 1.11 | 0 | 0.109 | 0.2593 | 0.303 |
| 40244 | RP11-490M8.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-5p | 15 | RUNX1T1 | Sponge network | -0.214 | 0.44248 | -0.344 | 0.2374 | 0.303 |
| 40245 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 21 | JAZF1 | Sponge network | 0.537 | 0.00139 | -0.377 | 0.02677 | 0.303 |
| 40246 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | PLSCR4 | Sponge network | 0.898 | 1.0E-5 | -0.474 | 0.03833 | 0.303 |
| 40247 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 32 | NTRK2 | Sponge network | -0.342 | 0.02288 | -0.736 | 0.06495 | 0.303 |
| 40248 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p | 27 | PRDM2 | Sponge network | -0.769 | 0.00035 | -0.258 | 0.00721 | 0.303 |
| 40249 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 50 | IL6ST | Sponge network | 0.16 | 0.50785 | -0.402 | 0.00676 | 0.303 |
| 40250 | CTD-2184D3.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NIN | Sponge network | 0.272 | 0.44692 | -0.253 | 0.13794 | 0.303 |
| 40251 | RP11-21A7A.2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-625-5p | 10 | ST6GALNAC3 | Sponge network | -1.116 | 0.23686 | -0.987 | 0 | 0.303 |
| 40252 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CLASP2 | Sponge network | -0.612 | 0.00094 | -0.038 | 0.71 | 0.303 |
| 40253 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | ZFHX3 | Sponge network | 1.205 | 0 | 0.389 | 0.01363 | 0.303 |
| 40254 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-590-3p | 13 | ARHGAP29 | Sponge network | 0.174 | 0.30034 | 0.131 | 0.42953 | 0.303 |
| 40255 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-3p | 31 | FZD3 | Sponge network | 1.063 | 0 | 0.104 | 0.60316 | 0.303 |
| 40256 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-629-3p;hsa-miR-7-5p | 18 | ZBTB4 | Sponge network | 0.174 | 0.30034 | -0.739 | 0 | 0.303 |
| 40257 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | ABCB5 | Sponge network | 0.101 | 0.60919 | -0.956 | 0.04077 | 0.303 |
| 40258 | NR2F1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-5p | 23 | KCNJ5 | Sponge network | -0.49 | 0.04946 | -0.281 | 0.3339 | 0.303 |
| 40259 | AC003090.1 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 15 | CELF4 | Sponge network | -2.117 | 0.00161 | -1.135 | 0.00344 | 0.303 |
| 40260 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-92a-1-5p | 27 | PRKAB2 | Sponge network | 0.95 | 0.15943 | -0.367 | 0.01371 | 0.303 |
| 40261 | LINC00621 |
hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-590-3p | 10 | ADAMTSL3 | Sponge network | 0.131 | 0.8436 | -1.559 | 0 | 0.303 |
| 40262 | HHIP-AS1 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 10 | KCNAB1 | Sponge network | -0.737 | 0.10822 | -0.755 | 2.0E-5 | 0.303 |
| 40263 | RP11-296I10.3 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-450b-5p | 15 | IL17RD | Sponge network | 0.592 | 0.00014 | -0.019 | 0.94873 | 0.303 |
| 40264 | TUG1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 15 | MYBL1 | Sponge network | 0.972 | 0 | 0.494 | 0.02152 | 0.303 |
| 40265 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-485-3p;hsa-miR-532-5p | 13 | PPM1L | Sponge network | 0.043 | 0.81628 | -0.537 | 0.00616 | 0.303 |
| 40266 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-96-5p | 25 | SOX5 | Sponge network | 0.622 | 0.00015 | -1.091 | 4.0E-5 | 0.303 |
| 40267 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-877-5p | 14 | GRIN2A | Sponge network | -0.785 | 0.00035 | -2.161 | 0 | 0.303 |
| 40268 | RP11-428J1.5 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 14 | ZMYND11 | Sponge network | 0.538 | 0.00076 | -0.2 | 0.05449 | 0.303 |
| 40269 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-542-3p | 18 | DYNC1I1 | Sponge network | -0.773 | 0.04073 | -1.12 | 0.00107 | 0.303 |
| 40270 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | EYA4 | Sponge network | -1.057 | 0.00029 | 0.3 | 0.36876 | 0.303 |
| 40271 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | DAB2 | Sponge network | 0.61 | 0.01593 | 0.431 | 0.0113 | 0.303 |
| 40272 | RP5-894A10.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-485-3p;hsa-miR-495-3p | 10 | MYO9A | Sponge network | 0.697 | 9.0E-5 | -0.147 | 0.32373 | 0.303 |
| 40273 | PART1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 29 | NAV3 | Sponge network | -2.925 | 0 | 0.212 | 0.47729 | 0.303 |
| 40274 | MIR22HG |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-3913-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 76 | LPP | Sponge network | -1.047 | 0 | -0.459 | 0.04475 | 0.303 |
| 40275 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-455-5p | 12 | MAPK10 | Sponge network | 0.853 | 0.15352 | -1.774 | 0 | 0.303 |
| 40276 | C4B-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-589-3p;hsa-miR-590-5p | 11 | TET1 | Sponge network | 2.306 | 9.0E-5 | -0.135 | 0.52138 | 0.303 |
| 40277 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | BCL2 | Sponge network | -0.726 | 0.00132 | -0.899 | 0 | 0.303 |
| 40278 | AC093627.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-625-5p | 12 | SUFU | Sponge network | -0.787 | 0.3928 | -0.04 | 0.65489 | 0.303 |
| 40279 | RP11-672A2.4 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p | 14 | AFF1 | Sponge network | 1.344 | 0 | -0.384 | 0.00053 | 0.303 |
| 40280 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 39 | PPP1R9A | Sponge network | 0.532 | 0.00505 | -0.903 | 0.04254 | 0.303 |
| 40281 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 32 | PRKAB2 | Sponge network | 0.253 | 0.09565 | -0.367 | 0.01371 | 0.303 |
| 40282 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | -1.216 | 0 | 0.273 | 0.09957 | 0.303 |
| 40283 | MRPL23-AS1 |
hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-93-3p | 11 | FOXO3 | Sponge network | -0.278 | 0.76559 | -0.04 | 0.72028 | 0.303 |
| 40284 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-335-5p | 13 | FOXO1 | Sponge network | -0.465 | 0.04703 | -0.141 | 0.33874 | 0.303 |
| 40285 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | ACVR1 | Sponge network | 0.222 | 0.29133 | 0.279 | 0.00234 | 0.303 |
| 40286 | RP11-102N12.3 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | ERBB4 | Sponge network | -0.351 | 0.11358 | -1.975 | 2.0E-5 | 0.303 |
| 40287 | FZD10-AS1 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | FAM172A | Sponge network | -0.727 | 0.04726 | -0.57 | 0 | 0.303 |
| 40288 | RP11-620J15.3 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-590-3p | 12 | CSDE1 | Sponge network | -0.739 | 0.00044 | -0.493 | 0 | 0.303 |
| 40289 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 15 | IGF1 | Sponge network | -0.726 | 0.00132 | -0.204 | 0.5898 | 0.303 |
| 40290 | RP11-35G9.5 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-93-3p;hsa-miR-96-5p | 14 | PCDH10 | Sponge network | 0.622 | 0.00015 | -1.644 | 0.0078 | 0.303 |
| 40291 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p | 35 | FZD3 | Sponge network | 1.757 | 0 | 0.104 | 0.60316 | 0.303 |
| 40292 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-655-3p | 22 | EPHA5 | Sponge network | 0.542 | 0.00054 | -0.957 | 0.01733 | 0.303 |
| 40293 | AC006116.24 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-454-3p | 17 | NCOA1 | Sponge network | -0.473 | 0.07602 | -0.432 | 0 | 0.303 |
| 40294 | AC016747.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-744-3p | 17 | PTPN14 | Sponge network | 0.199 | 0.38015 | 0.144 | 0.34595 | 0.303 |
| 40295 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 17 | TES | Sponge network | -0.129 | 0.57177 | -0.348 | 0.00639 | 0.303 |
| 40296 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 12 | AFAP1L2 | Sponge network | 0.335 | 0.20596 | -0.284 | 0.15434 | 0.303 |
| 40297 | RP11-686D22.4 |
hsa-let-7a-3p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-577;hsa-miR-590-3p | 10 | HIP1 | Sponge network | 0.793 | 5.0E-5 | 0.774 | 0 | 0.303 |
| 40298 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-96-5p | 16 | EBF3 | Sponge network | 0.614 | 1.0E-5 | -0.058 | 0.81437 | 0.303 |
| 40299 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-501-3p | 14 | CLASP2 | Sponge network | -2.039 | 0.03447 | -0.038 | 0.71 | 0.303 |
| 40300 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-93-5p | 39 | PDGFRA | Sponge network | -0.162 | 0.47687 | -0.372 | 0.0037 | 0.303 |
| 40301 | CTD-2017D11.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-582-5p | 18 | ABI2 | Sponge network | 0.792 | 0.0049 | 0.175 | 0.14787 | 0.303 |
| 40302 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | LATS2 | Sponge network | -0.942 | 0 | 0.159 | 0.2304 | 0.303 |
| 40303 | EIF3J-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | AKAP6 | Sponge network | 0.331 | 0.00509 | -1.586 | 3.0E-5 | 0.303 |
| 40304 | CTD-3064M3.3 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-3614-5p;hsa-miR-378a-5p | 10 | CACNA1E | Sponge network | -0.469 | 0.08721 | 1.752 | 0 | 0.303 |
| 40305 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-942-5p | 43 | UNC80 | Sponge network | 0.194 | 0.64278 | -1.934 | 0 | 0.302 |
| 40306 | RP3-402G11.26 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | MEF2C | Sponge network | -0.254 | 0.19303 | -0.499 | 0.01448 | 0.302 |
| 40307 | HCG11 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TAL1 | Sponge network | 0.385 | 0.10067 | -0.462 | 0.00574 | 0.302 |
| 40308 | DNAJC27-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-542-3p | 12 | PRKD1 | Sponge network | -0.969 | 2.0E-5 | -0.466 | 0.03583 | 0.302 |
| 40309 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 26 | USP12 | Sponge network | 0.862 | 0.06213 | -0.102 | 0.40768 | 0.302 |
| 40310 | PCA3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-96-5p | 11 | ELMO1 | Sponge network | -0.709 | 0.18168 | 0.04 | 0.83147 | 0.302 |
| 40311 | RP11-21A7A.2 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-98-5p | 10 | LIFR | Sponge network | -1.116 | 0.23686 | -1.794 | 0 | 0.302 |
| 40312 | CTD-3092A11.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-625-5p | 13 | PTPN14 | Sponge network | 0.873 | 0 | 0.144 | 0.34595 | 0.302 |
| 40313 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | HACE1 | Sponge network | 0.349 | 0.01695 | -0.072 | 0.55239 | 0.302 |
| 40314 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-625-5p | 28 | PTPRT | Sponge network | -0.18 | 0.20343 | -0.907 | 0.03727 | 0.302 |
| 40315 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-664a-3p | 13 | ITCH | Sponge network | 0.95 | 0.15943 | 0.084 | 0.34058 | 0.302 |
| 40316 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p | 13 | FBXO32 | Sponge network | 2.163 | 0 | -0.495 | 0.07561 | 0.302 |
| 40317 | TMEM161B-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-214-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p | 25 | PPM1A | Sponge network | -0.04 | 0.7606 | -0.623 | 0 | 0.302 |
| 40318 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | ETV1 | Sponge network | 0.064 | 0.72934 | -0.096 | 0.67674 | 0.302 |
| 40319 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-550a-3p;hsa-miR-577 | 40 | DTNA | Sponge network | -0.465 | 0.04703 | -1.648 | 1.0E-5 | 0.302 |
| 40320 | MAGI2-AS3 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | PTCH1 | Sponge network | -0.129 | 0.57177 | -0.1 | 0.6179 | 0.302 |
| 40321 | RP11-802E16.3 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-497-5p | 14 | MDM4 | Sponge network | 2.048 | 0 | 0.345 | 0.00762 | 0.302 |
| 40322 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 35 | EPHA7 | Sponge network | 0.253 | 0.09565 | -2.197 | 3.0E-5 | 0.302 |
| 40323 | CTD-2245F17.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-629-3p | 20 | DST | Sponge network | -0.421 | 0.12556 | -0.713 | 0.00096 | 0.302 |
| 40324 | CTB-31O20.4 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-628-5p | 11 | BMPR2 | Sponge network | 1.619 | 0 | 0.206 | 0.0635 | 0.302 |
| 40325 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 29 | BNC2 | Sponge network | -0.141 | 0.26434 | -0.491 | 0.09988 | 0.302 |
| 40326 | MIR22HG |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p | 19 | KLF6 | Sponge network | -1.047 | 0 | -0.479 | 0.00015 | 0.302 |
| 40327 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p | 12 | EDA2R | Sponge network | 0.064 | 0.72934 | -0.587 | 0.01598 | 0.302 |
| 40328 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 50 | BMPR2 | Sponge network | -0.176 | 0.59692 | 0.206 | 0.0635 | 0.302 |
| 40329 | AC109309.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p | 16 | PBX1 | Sponge network | 0.163 | 0.87219 | -1.206 | 0 | 0.302 |
| 40330 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-505-5p;hsa-miR-93-5p | 24 | HBP1 | Sponge network | -0.842 | 0.01361 | -0.454 | 0 | 0.302 |
| 40331 | AC005562.1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p | 14 | PHF6 | Sponge network | 0.488 | 0.00084 | 0.785 | 0 | 0.302 |
| 40332 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 15 | GPAM | Sponge network | 0.488 | 0.00084 | 0.142 | 0.29732 | 0.302 |
| 40333 | AC093110.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p | 10 | BAZ2B | Sponge network | 1.044 | 2.0E-5 | -0.276 | 0.02833 | 0.302 |
| 40334 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-590-5p | 19 | KANK1 | Sponge network | 0.331 | 0.57247 | -0.55 | 0.00134 | 0.302 |
| 40335 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-7-1-3p | 15 | PDGFRA | Sponge network | 0.238 | 0.70544 | -0.372 | 0.0037 | 0.302 |
| 40336 | AC108488.4 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | MEF2C | Sponge network | 0.259 | 0.12542 | -0.499 | 0.01448 | 0.302 |
| 40337 | RP11-359E10.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | CCND2 | Sponge network | 0.299 | 0.57722 | -0.267 | 0.3112 | 0.302 |
| 40338 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | SASH1 | Sponge network | -0.612 | 0.00094 | -0.502 | 0.00175 | 0.302 |
| 40339 | IQCH-AS1 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | RICTOR | Sponge network | 0.929 | 0 | 0.388 | 0.00188 | 0.302 |
| 40340 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | GRIA3 | Sponge network | -0.727 | 0.04726 | -1.956 | 0 | 0.302 |
| 40341 | RP11-384O8.1 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 11 | PPP2R2C | Sponge network | -0.738 | 0.21233 | -0.959 | 0.07428 | 0.302 |
| 40342 | RP11-67L2.2 |
hsa-miR-126-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 14 | IRS1 | Sponge network | 0.72 | 0.0001 | -0.026 | 0.88855 | 0.302 |
| 40343 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | MTSS1 | Sponge network | -2.915 | 0.00015 | -0.81 | 0.00035 | 0.302 |
| 40344 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-16-5p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-654-3p | 15 | DDX6 | Sponge network | 0.909 | 0 | 0.091 | 0.36428 | 0.302 |
| 40345 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-454-3p | 18 | PRKAA2 | Sponge network | 0.598 | 0.38481 | -2.462 | 0 | 0.302 |
| 40346 | EMX2OS |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 28 | LRIG1 | Sponge network | -0.777 | 0.1134 | -0.537 | 0.00588 | 0.302 |
| 40347 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 17 | GPAM | Sponge network | 1.11 | 0 | 0.142 | 0.29732 | 0.302 |
| 40348 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p | 13 | CBX5 | Sponge network | 0.793 | 0 | 0.165 | 0.24446 | 0.302 |
| 40349 | HOXB-AS2 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-576-5p | 15 | PHC3 | Sponge network | 1.316 | 0.01106 | 0.212 | 0.0499 | 0.302 |
| 40350 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-577 | 20 | SCN9A | Sponge network | 0.637 | 0.00069 | -0.525 | 0.10593 | 0.302 |
| 40351 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 11 | ZNF770 | Sponge network | 1.178 | 0 | 0.206 | 0.06751 | 0.302 |
| 40352 | RP11-725P16.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZMYND11 | Sponge network | 0.422 | 0.27021 | -0.2 | 0.05449 | 0.302 |
| 40353 | LINC00969 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-629-3p | 21 | DDX6 | Sponge network | 0.683 | 1.0E-5 | 0.091 | 0.36428 | 0.302 |
| 40354 | OIP5-AS1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | RNF180 | Sponge network | 0.421 | 0.00084 | -1.127 | 1.0E-5 | 0.302 |
| 40355 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-421;hsa-miR-93-5p | 10 | JAK1 | Sponge network | 0.532 | 0.00505 | 0.001 | 0.98982 | 0.302 |
| 40356 | ACTA2-AS1 |
hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 22 | UBR3 | Sponge network | 0.194 | 0.64278 | -0.021 | 0.82774 | 0.302 |
| 40357 | NEAT1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-942-5p | 15 | PICALM | Sponge network | 0.705 | 0.00014 | 0.086 | 0.23523 | 0.302 |
| 40358 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-940;hsa-miR-942-5p | 31 | HLF | Sponge network | -0.773 | 0.04073 | -2.443 | 0 | 0.302 |
| 40359 | NR2F1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-93-5p | 28 | RNASEL | Sponge network | -0.49 | 0.04946 | -0.272 | 0.01796 | 0.302 |
| 40360 | ZNF667-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 17 | IGF2 | Sponge network | -0.976 | 0.00025 | 0.848 | 0.03842 | 0.302 |
| 40361 | CTD-2017D11.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-339-5p | 15 | IL17RD | Sponge network | 0.792 | 0.0049 | -0.019 | 0.94873 | 0.302 |
| 40362 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-127-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | PER2 | Sponge network | -0.222 | 0.32445 | -0.396 | 0.00269 | 0.302 |
| 40363 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 14 | CRTC3 | Sponge network | 0.488 | 0.00084 | 0.164 | 0.16786 | 0.302 |
| 40364 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-576-5p | 23 | TMTC3 | Sponge network | 0.95 | 0.15943 | 0.123 | 0.22189 | 0.302 |
| 40365 | OIP5-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 28 | CYLD | Sponge network | 0.421 | 0.00084 | -0.367 | 0.00171 | 0.302 |
| 40366 | LINC00672 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | ASTN1 | Sponge network | -0.227 | 0.31657 | -2.389 | 2.0E-5 | 0.302 |
| 40367 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 38 | NTRK2 | Sponge network | -0.738 | 0.21233 | -0.736 | 0.06495 | 0.302 |
| 40368 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | PHC3 | Sponge network | -0.901 | 0.00019 | 0.212 | 0.0499 | 0.302 |
| 40369 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | AHNAK | Sponge network | -2.925 | 0 | -1.207 | 0 | 0.302 |
| 40370 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-96-5p | 15 | JAZF1 | Sponge network | 0.37 | 0.27614 | -0.377 | 0.02677 | 0.302 |
| 40371 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-455-3p | 12 | FBXO32 | Sponge network | 0.693 | 0.00076 | -0.495 | 0.07561 | 0.302 |
| 40372 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-940 | 10 | NR4A3 | Sponge network | -0.096 | 0.67958 | -1.385 | 0 | 0.302 |
| 40373 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-539-5p | 26 | PBX1 | Sponge network | -0.125 | 0.49414 | -1.206 | 0 | 0.302 |
| 40374 | GNG12-AS1 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-590-3p | 15 | FAM135B | Sponge network | -0.793 | 7.0E-5 | -3.978 | 0 | 0.302 |
| 40375 | U91328.19 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 44 | TRPS1 | Sponge network | -0.303 | 0.21002 | -0.191 | 0.44109 | 0.302 |
| 40376 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 13 | TIMP3 | Sponge network | 0.174 | 0.30034 | -0.546 | 0.02307 | 0.302 |
| 40377 | AC007566.10 |
hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | CCDC6 | Sponge network | 2.368 | 2.0E-5 | 0.27 | 0.02555 | 0.302 |
| 40378 | TPTEP1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p | 21 | STARD13 | Sponge network | -1.216 | 0 | -0.187 | 0.27383 | 0.302 |
| 40379 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-576-5p | 11 | PTGFR | Sponge network | 0.614 | 1.0E-5 | -0.233 | 0.45421 | 0.302 |
| 40380 | RP11-48B3.4 |
hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | PCDH9 | Sponge network | 1.757 | 0 | -1.823 | 4.0E-5 | 0.302 |
| 40381 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-874-3p | 11 | DCUN1D3 | Sponge network | 0.523 | 9.0E-5 | 0.152 | 0.1769 | 0.302 |
| 40382 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p | 26 | BMPR2 | Sponge network | 0.336 | 0.00739 | 0.206 | 0.0635 | 0.302 |
| 40383 | HCG11 |
hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-590-3p | 11 | PAK3 | Sponge network | 0.385 | 0.10067 | -0.628 | 0.07253 | 0.302 |
| 40384 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-532-3p;hsa-miR-539-5p | 30 | PBX1 | Sponge network | -0.235 | 0.15142 | -1.206 | 0 | 0.302 |
| 40385 | LIPE-AS1 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-542-3p;hsa-miR-758-3p | 14 | CSDE1 | Sponge network | 0.078 | 0.55803 | -0.493 | 0 | 0.302 |
| 40386 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p | 15 | MOB1B | Sponge network | 0.856 | 0 | 0.197 | 0.15266 | 0.302 |
| 40387 | RP11-166D19.1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | PTCH1 | Sponge network | -0.576 | 0.10121 | -0.1 | 0.6179 | 0.302 |
| 40388 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | MCC | Sponge network | 0.959 | 0 | -1.005 | 0 | 0.302 |
| 40389 | RP11-373D23.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-7-1-3p | 29 | IL6ST | Sponge network | -0.285 | 0.2172 | -0.402 | 0.00676 | 0.302 |
| 40390 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-93-5p | 10 | JAK1 | Sponge network | -0.611 | 0.09607 | 0.001 | 0.98982 | 0.302 |
| 40391 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 29 | EPHA7 | Sponge network | 0.693 | 0.00076 | -2.197 | 3.0E-5 | 0.302 |
| 40392 | CALML3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p | 29 | TGFBR3 | Sponge network | 0.95 | 0.15943 | -1.173 | 1.0E-5 | 0.302 |
| 40393 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-942-5p | 22 | TMEM132B | Sponge network | -0.376 | 0.00337 | -0.83 | 0.01084 | 0.302 |
| 40394 | RP11-35G9.3 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRIM13 | Sponge network | 0.657 | 4.0E-5 | 0.183 | 0.06968 | 0.302 |
| 40395 | AGAP11 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-328-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p | 14 | TMTC2 | Sponge network | -1.645 | 0 | -0.215 | 0.18248 | 0.302 |
| 40396 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | SON | Sponge network | 1.757 | 0 | 0.168 | 0.04406 | 0.302 |
| 40397 | AC005154.6 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 15 | ZMYND11 | Sponge network | 0.848 | 0 | -0.2 | 0.05449 | 0.302 |
| 40398 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | LRRC55 | Sponge network | -0.726 | 0.00132 | -0.026 | 0.93163 | 0.302 |
| 40399 | RP11-121C2.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | RPS6KA3 | Sponge network | 1.147 | 0 | 0.256 | 0.02419 | 0.302 |
| 40400 | RASSF8-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3E | Sponge network | 0.335 | 0.20596 | -1.717 | 0.00137 | 0.302 |
| 40401 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-590-3p | 16 | ARHGAP29 | Sponge network | -0.612 | 0.00094 | 0.131 | 0.42953 | 0.302 |
| 40402 | ZNF667-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-421;hsa-miR-429 | 17 | PLXNC1 | Sponge network | -0.976 | 0.00025 | 0.569 | 0.00329 | 0.302 |
| 40403 | RP11-983P16.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-590-5p | 12 | ITGB3 | Sponge network | 0.41 | 0.02214 | -0.011 | 0.95751 | 0.302 |
| 40404 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | HACE1 | Sponge network | 0.439 | 0.00313 | -0.072 | 0.55239 | 0.302 |
| 40405 | CTD-3099C6.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-429 | 11 | NOTCH2 | Sponge network | 1.361 | 0 | 0.138 | 0.35163 | 0.302 |
| 40406 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-628-5p;hsa-miR-7-5p | 26 | ERC1 | Sponge network | 0.537 | 0.00139 | -0.108 | 0.38794 | 0.302 |
| 40407 | RP11-327P2.5 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-542-3p | 17 | DYNC1I1 | Sponge network | -0.147 | 0.40452 | -1.12 | 0.00107 | 0.302 |
| 40408 | AP001347.6 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | INPP4A | Sponge network | -1.161 | 0.00591 | 0.07 | 0.53563 | 0.302 |
| 40409 | RP11-155G14.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-378a-5p | 13 | CREB1 | Sponge network | 2.483 | 0 | 0.203 | 0.00537 | 0.302 |
| 40410 | ERVH48-1 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | ERCC4 | Sponge network | 1.04 | 0.00023 | 0.126 | 0.15266 | 0.302 |
| 40411 | RP11-2H8.2 |
hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-493-5p;hsa-miR-590-3p | 16 | MLLT10 | Sponge network | 1.089 | 0 | 0.105 | 0.33292 | 0.302 |
| 40412 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | TET1 | Sponge network | -0.223 | 0.47816 | -0.135 | 0.52138 | 0.302 |
| 40413 | CTC-429P9.5 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-744-3p | 16 | IGFBP5 | Sponge network | 0.178 | 0.29106 | -0.806 | 0.00105 | 0.302 |
| 40414 | TAPT1-AS1 |
hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p;hsa-miR-495-3p | 11 | PHF6 | Sponge network | 0.403 | 2.0E-5 | 0.785 | 0 | 0.302 |
| 40415 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p | 10 | EPHA3 | Sponge network | 0.858 | 3.0E-5 | 0.185 | 0.53588 | 0.302 |
| 40416 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-5p | 32 | ESR1 | Sponge network | -0.727 | 0.04726 | -1.035 | 3.0E-5 | 0.302 |
| 40417 | FAM13A-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | LRP1B | Sponge network | -0.83 | 0 | -2.163 | 0 | 0.302 |
| 40418 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 33 | ETV1 | Sponge network | 0.997 | 0.00066 | -0.096 | 0.67674 | 0.302 |
| 40419 | RP11-798M19.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 26 | ERC1 | Sponge network | -0.612 | 0.00094 | -0.108 | 0.38794 | 0.302 |
| 40420 | H1FX-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 22 | NTRK2 | Sponge network | -0.206 | 0.12942 | -0.736 | 0.06495 | 0.302 |
| 40421 | RP11-403I13.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-7-1-3p | 13 | MOB1B | Sponge network | 1.062 | 0.00512 | 0.197 | 0.15266 | 0.302 |
| 40422 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-424-5p | 11 | ZFHX3 | Sponge network | 0.924 | 0 | 0.389 | 0.01363 | 0.302 |
| 40423 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p | 13 | PGR | Sponge network | 0.912 | 0.00014 | -1.704 | 0 | 0.302 |
| 40424 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p | 16 | MOB1B | Sponge network | 0.556 | 0.00298 | 0.197 | 0.15266 | 0.302 |
| 40425 | RP11-855A2.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | EPS15 | Sponge network | 2.094 | 0 | -0.052 | 0.53998 | 0.302 |
| 40426 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-485-3p;hsa-miR-542-3p | 10 | UNC80 | Sponge network | 0.043 | 0.81628 | -1.934 | 0 | 0.302 |
| 40427 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 46 | CUX1 | Sponge network | -0.899 | 6.0E-5 | -0.039 | 0.6705 | 0.302 |
| 40428 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | EPHA5 | Sponge network | -0.83 | 0 | -0.957 | 0.01733 | 0.302 |
| 40429 | LINC00854 |
hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-628-5p;hsa-miR-940 | 11 | MEF2C | Sponge network | 1.18 | 0 | -0.499 | 0.01448 | 0.302 |
| 40430 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 20 | FOXO3 | Sponge network | 1.153 | 0 | -0.04 | 0.72028 | 0.302 |
| 40431 | RP11-21A7A.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 15 | ERC1 | Sponge network | -1.116 | 0.23686 | -0.108 | 0.38794 | 0.302 |
| 40432 | AC016747.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 27 | IL17RD | Sponge network | 0.199 | 0.38015 | -0.019 | 0.94873 | 0.302 |
| 40433 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-93-5p | 18 | TGFBR2 | Sponge network | -0.162 | 0.47687 | -0.243 | 0.11408 | 0.302 |
| 40434 | RP11-1277A3.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-339-5p | 11 | TSC1 | Sponge network | 0.453 | 0.01839 | 0.103 | 0.26071 | 0.302 |
| 40435 | CTC-559E9.6 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p | 16 | BRD3 | Sponge network | 0.601 | 1.0E-5 | 0.37 | 0.00013 | 0.302 |
| 40436 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-590-3p | 12 | RBM5 | Sponge network | 0.659 | 3.0E-5 | -0.205 | 0.0129 | 0.302 |
| 40437 | RP11-1006G14.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 22 | DLC1 | Sponge network | 0.559 | 0.00026 | -0.382 | 0.05038 | 0.302 |
| 40438 | BDNF-AS |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | RECK | Sponge network | -0.668 | 3.0E-5 | -1.043 | 0 | 0.302 |
| 40439 | HCG18 |
hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p | 16 | NUAK1 | Sponge network | 1.063 | 0 | 0.904 | 0 | 0.302 |
| 40440 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p | 16 | TGFBR2 | Sponge network | -1.249 | 7.0E-5 | -0.243 | 0.11408 | 0.302 |
| 40441 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | TACC1 | Sponge network | -0.969 | 2.0E-5 | -0.362 | 0.05406 | 0.302 |
| 40442 | GAS6-AS2 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | SPOP | Sponge network | 0.16 | 0.50785 | -0.419 | 1.0E-5 | 0.302 |
| 40443 | RP11-440L14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-590-3p | 15 | DST | Sponge network | 0.545 | 0.05789 | -0.713 | 0.00096 | 0.302 |
| 40444 | LINC00954 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-429 | 10 | EYA4 | Sponge network | 1.157 | 0.00455 | 0.3 | 0.36876 | 0.302 |
| 40445 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p | 38 | RASSF2 | Sponge network | -1.249 | 7.0E-5 | -0.273 | 0.21961 | 0.302 |
| 40446 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b | 13 | NR2C2 | Sponge network | 0.001 | 0.99753 | 0.21 | 0.03224 | 0.302 |
| 40447 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p | 20 | RASSF2 | Sponge network | -2.076 | 0.00548 | -0.273 | 0.21961 | 0.302 |
| 40448 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-93-5p | 24 | RASSF2 | Sponge network | -0.663 | 0.10887 | -0.273 | 0.21961 | 0.302 |
| 40449 | AC010226.4 |
hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-590-3p;hsa-miR-7-5p | 14 | CRTC1 | Sponge network | -0.811 | 2.0E-5 | -0.116 | 0.28412 | 0.302 |
| 40450 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-93-5p | 36 | EPHA5 | Sponge network | -0.384 | 0.40652 | -0.957 | 0.01733 | 0.302 |
| 40451 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 13 | FYN | Sponge network | 0.194 | 0.64278 | -0.131 | 0.37051 | 0.302 |
| 40452 | LINC00473 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DOCK4 | Sponge network | -1.203 | 0.03881 | 0.502 | 0.00108 | 0.302 |
| 40453 | TRIM52-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-7-1-3p | 13 | ITPKB | Sponge network | -0.418 | 0.00068 | -0.704 | 0.00106 | 0.302 |
| 40454 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-93-5p | 26 | SYNE1 | Sponge network | -1.645 | 0 | -0.738 | 0.00316 | 0.302 |
| 40455 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SLC6A15 | Sponge network | -0.487 | 0.02157 | -2.373 | 2.0E-5 | 0.302 |
| 40456 | HCG11 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | LUZP2 | Sponge network | 0.385 | 0.10067 | -0.056 | 0.89416 | 0.302 |
| 40457 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 11 | PTPN13 | Sponge network | -0.899 | 6.0E-5 | -1.208 | 0 | 0.302 |
| 40458 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SASH1 | Sponge network | 0.538 | 0.00076 | -0.502 | 0.00175 | 0.302 |
| 40459 | RP11-203M5.7 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-5p | 12 | BMPR2 | Sponge network | 1.684 | 1.0E-5 | 0.206 | 0.0635 | 0.302 |
| 40460 | RP11-283I3.6 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 13 | PICALM | Sponge network | 0.614 | 1.0E-5 | 0.086 | 0.23523 | 0.302 |
| 40461 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-96-5p | 21 | SCN9A | Sponge network | 0.4 | 0.14611 | -0.525 | 0.10593 | 0.302 |
| 40462 | AC141928.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-628-5p | 42 | RUNX1T1 | Sponge network | 0.853 | 0.15352 | -0.344 | 0.2374 | 0.302 |
| 40463 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 41 | ZFHX3 | Sponge network | -0.942 | 0 | 0.389 | 0.01363 | 0.302 |
| 40464 | RP11-235E17.6 | hsa-miR-129-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326 | 11 | CCDC6 | Sponge network | 1.596 | 0 | 0.27 | 0.02555 | 0.302 |
| 40465 | CTA-363E19.2 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-495-3p;hsa-miR-539-5p | 12 | TSC1 | Sponge network | 1.055 | 0 | 0.103 | 0.26071 | 0.302 |
| 40466 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 23 | CREB1 | Sponge network | 0.214 | 0.18246 | 0.203 | 0.00537 | 0.302 |
| 40467 | RP11-736K20.5 |
hsa-let-7d-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-590-3p | 15 | ACSL6 | Sponge network | -0.358 | 0.17255 | -1.593 | 0 | 0.302 |
| 40468 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p | 25 | FBXO32 | Sponge network | -0.223 | 0.47816 | -0.495 | 0.07561 | 0.302 |
| 40469 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-655-3p | 15 | KANK1 | Sponge network | 0.41 | 0.02214 | -0.55 | 0.00134 | 0.302 |
| 40470 | RP1-151F17.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-590-3p | 16 | MRVI1 | Sponge network | 0.222 | 0.29133 | -1.051 | 0.00022 | 0.302 |
| 40471 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | ARID5B | Sponge network | 0.101 | 0.60919 | 0.342 | 0.01676 | 0.302 |
| 40472 | LPP-AS2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-589-3p | 14 | SORCS1 | Sponge network | -0.18 | 0.20343 | -3.492 | 0 | 0.302 |
| 40473 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-577;hsa-miR-590-3p | 19 | GRIN2A | Sponge network | -0.322 | 0.25945 | -2.161 | 0 | 0.302 |
| 40474 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TMEFF2 | Sponge network | -0.08 | 0.90092 | -2.426 | 0 | 0.302 |
| 40475 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 10 | DDX5 | Sponge network | -0.612 | 0.00094 | -0.099 | 0.17428 | 0.302 |
| 40476 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p;hsa-miR-655-3p | 14 | ARID2 | Sponge network | 0.421 | 0.00084 | 0.235 | 0.02697 | 0.302 |
| 40477 | LENG8-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-374a-3p | 11 | CBL | Sponge network | 1.471 | 0 | 0.291 | 0.01267 | 0.302 |
| 40478 | RP11-973H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p | 10 | TMTC3 | Sponge network | 0.901 | 0 | 0.123 | 0.22189 | 0.302 |
| 40479 | RP11-327P2.5 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-486-5p;hsa-miR-542-3p | 13 | SYNM | Sponge network | -0.147 | 0.40452 | -2.309 | 1.0E-5 | 0.302 |
| 40480 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p | 10 | USP25 | Sponge network | -0.355 | 0.0077 | -0.242 | 0.01397 | 0.302 |
| 40481 | RP5-994D16.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NOTCH2 | Sponge network | 0.252 | 0.08508 | 0.138 | 0.35163 | 0.302 |
| 40482 | LINC00094 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-93-5p | 13 | PRUNE2 | Sponge network | 0.253 | 0.09565 | -1.733 | 0.00022 | 0.302 |
| 40483 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ANK2 | Sponge network | 0.392 | 0.0278 | -1.603 | 1.0E-5 | 0.302 |
| 40484 | OSER1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 12 | WWTR1 | Sponge network | -0.13 | 0.48734 | -0.345 | 0.11997 | 0.302 |
| 40485 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-654-3p | 12 | NR2C2 | Sponge network | 0.411 | 0.1335 | 0.21 | 0.03224 | 0.302 |
| 40486 | RP11-6O2.3 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-495-3p | 11 | TMEM30A | Sponge network | 0.331 | 0.57247 | 0.096 | 0.31031 | 0.302 |
| 40487 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | IGSF10 | Sponge network | 0.051 | 0.83388 | -1.751 | 1.0E-5 | 0.302 |
| 40488 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | ST6GALNAC3 | Sponge network | -0.668 | 3.0E-5 | -0.987 | 0 | 0.302 |
| 40489 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | ABCB5 | Sponge network | -0.487 | 0.02157 | -0.956 | 0.04077 | 0.302 |
| 40490 | RP11-983P16.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | ZNF208 | Sponge network | 0.41 | 0.02214 | -0.4 | 0.22381 | 0.302 |
| 40491 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p | 17 | TBX18 | Sponge network | -0.162 | 0.47687 | 0.843 | 0.02945 | 0.302 |
| 40492 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 16 | FOXP1 | Sponge network | -0.932 | 0.00329 | 0.042 | 0.74368 | 0.302 |
| 40493 | GS1-124K5.11 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | MAPK10 | Sponge network | 0.494 | 0.00339 | -1.774 | 0 | 0.302 |
| 40494 | TINCR |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-454-3p | 36 | DTNA | Sponge network | -0.664 | 0.14543 | -1.648 | 1.0E-5 | 0.302 |
| 40495 | RP11-13J8.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-495-3p | 10 | PHC3 | Sponge network | 3.03 | 0 | 0.212 | 0.0499 | 0.302 |
| 40496 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 25 | MED12L | Sponge network | 0.214 | 0.18246 | -0.194 | 0.55299 | 0.302 |
| 40497 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p | 10 | SEPT6 | Sponge network | -0.125 | 0.49414 | -0.598 | 0.00181 | 0.302 |
| 40498 | RP1-68D18.4 |
hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-455-3p | 12 | CREB1 | Sponge network | 2.811 | 0 | 0.203 | 0.00537 | 0.302 |
| 40499 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-935;hsa-miR-96-5p | 33 | FBXO32 | Sponge network | 0.222 | 0.29133 | -0.495 | 0.07561 | 0.302 |
| 40500 | LINC00672 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | ABCB5 | Sponge network | -0.227 | 0.31657 | -0.956 | 0.04077 | 0.302 |
| 40501 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-589-3p | 13 | SUFU | Sponge network | 0.811 | 0.03228 | -0.04 | 0.65489 | 0.302 |
| 40502 | RP11-38L15.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-345-5p;hsa-miR-590-3p | 14 | TRPS1 | Sponge network | 0.344 | 0.3127 | -0.191 | 0.44109 | 0.302 |
| 40503 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | SOX5 | Sponge network | 0.657 | 4.0E-5 | -1.091 | 4.0E-5 | 0.302 |
| 40504 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 28 | AKAP11 | Sponge network | 1.302 | 7.0E-5 | 0.196 | 0.09825 | 0.302 |
| 40505 | C4B-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 26 | NTRK3 | Sponge network | 2.306 | 9.0E-5 | -1.77 | 3.0E-5 | 0.302 |
| 40506 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-577;hsa-miR-582-5p | 13 | EBF3 | Sponge network | 0.419 | 0.0319 | -0.058 | 0.81437 | 0.302 |
| 40507 | RP11-66B24.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-502-5p | 18 | AR | Sponge network | 0.57 | 0.1866 | -1.554 | 0 | 0.302 |
| 40508 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-5p | 25 | ZBTB4 | Sponge network | -0.176 | 0.59692 | -0.739 | 0 | 0.302 |
| 40509 | RP5-855D21.3 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 14 | CCDC6 | Sponge network | 1.728 | 0 | 0.27 | 0.02555 | 0.302 |
| 40510 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | EPHA7 | Sponge network | 0.537 | 0.00139 | -2.197 | 3.0E-5 | 0.302 |
| 40511 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-324-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | HACE1 | Sponge network | -0.487 | 0.02157 | -0.072 | 0.55239 | 0.302 |
| 40512 | LIPE-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-542-3p | 18 | PDE4DIP | Sponge network | 0.078 | 0.55803 | -0.282 | 0.0257 | 0.302 |
| 40513 | RP11-531A24.5 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-493-5p | 17 | DDX3X | Sponge network | 0.75 | 0.00054 | -0.152 | 0.07686 | 0.302 |
| 40514 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | GRIN2A | Sponge network | 0.392 | 0.0278 | -2.161 | 0 | 0.302 |
| 40515 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 10 | ZNF770 | Sponge network | -0.567 | 0 | 0.206 | 0.06751 | 0.302 |
| 40516 | PWAR6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | RNF144A | Sponge network | -1.575 | 6.0E-5 | 0.594 | 0.0009 | 0.302 |
| 40517 | UBA6-AS1 |
hsa-miR-125a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | ERCC6 | Sponge network | 0.312 | 0.00725 | 0.23 | 0.015 | 0.302 |
| 40518 | OSER1-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-744-3p;hsa-miR-93-5p | 37 | DST | Sponge network | -0.13 | 0.48734 | -0.713 | 0.00096 | 0.302 |
| 40519 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | DMTF1 | Sponge network | 0.421 | 0.00084 | 0.248 | 0.02726 | 0.302 |
| 40520 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 15 | SYNE1 | Sponge network | 0.131 | 0.8436 | -0.738 | 0.00316 | 0.302 |
| 40521 | RP4-758J18.10 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-450b-5p | 14 | PRKAB2 | Sponge network | -0.091 | 0.71468 | -0.367 | 0.01371 | 0.302 |
| 40522 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 15 | BCL11A | Sponge network | -1.092 | 4.0E-5 | -0.932 | 0.0063 | 0.302 |
| 40523 | HOXB-AS2 |
hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p | 15 | RICTOR | Sponge network | 1.316 | 0.01106 | 0.388 | 0.00188 | 0.302 |
| 40524 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 31 | EBF1 | Sponge network | 0.374 | 0.20054 | -0.384 | 0.08856 | 0.302 |
| 40525 | RP11-159D12.2 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 16 | UBE2D1 | Sponge network | 1.254 | 0 | 0.468 | 0 | 0.302 |
| 40526 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 33 | PDGFRA | Sponge network | 0.214 | 0.18246 | -0.372 | 0.0037 | 0.302 |
| 40527 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-501-3p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | 0.299 | 0.57722 | -0.243 | 0.11408 | 0.302 |
| 40528 | RASSF8-AS1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 14 | RPS6KA6 | Sponge network | 0.335 | 0.20596 | -2.989 | 0 | 0.302 |
| 40529 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 25 | SASH1 | Sponge network | 0.614 | 1.0E-5 | -0.502 | 0.00175 | 0.302 |
| 40530 | RP13-516M14.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 23 | TGFBR3 | Sponge network | 0.389 | 0.04293 | -1.173 | 1.0E-5 | 0.302 |
| 40531 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 27 | CTDSPL | Sponge network | 0.72 | 0.0001 | 0.085 | 0.45121 | 0.302 |
| 40532 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | PRKAB2 | Sponge network | 0.862 | 0.06213 | -0.367 | 0.01371 | 0.302 |
| 40533 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-33a-3p | 12 | PCDH17 | Sponge network | 0.368 | 0.20093 | 0.731 | 2.0E-5 | 0.302 |
| 40534 | NDUFA6-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 30 | WWTR1 | Sponge network | -0.355 | 0.0077 | -0.345 | 0.11997 | 0.302 |
| 40535 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | LRP1B | Sponge network | -0.663 | 0.10887 | -2.163 | 0 | 0.302 |
| 40536 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-942-5p | 24 | DDX6 | Sponge network | 1.317 | 0 | 0.091 | 0.36428 | 0.302 |
| 40537 | RP4-669P10.16 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p | 17 | ADARB1 | Sponge network | -0.301 | 0.06597 | -0.335 | 0.06631 | 0.302 |
| 40538 | RP11-473I1.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p | 13 | PRKAB2 | Sponge network | 0.542 | 0.00054 | -0.367 | 0.01371 | 0.302 |
| 40539 | AC010226.4 |
hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | UNC5C | Sponge network | -0.811 | 2.0E-5 | -0.178 | 0.40214 | 0.302 |
| 40540 | RP11-356J5.12 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 19 | GJA1 | Sponge network | -0.899 | 6.0E-5 | -0.485 | 0.02179 | 0.302 |
| 40541 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 39 | TMEM132B | Sponge network | 0.349 | 0.01695 | -0.83 | 0.01084 | 0.302 |
| 40542 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429 | 14 | CREBBP | Sponge network | 0.95 | 0.15943 | 0.126 | 0.15186 | 0.302 |
| 40543 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 17 | SCN9A | Sponge network | 0.67 | 0.00048 | -0.525 | 0.10593 | 0.302 |
| 40544 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 56 | FOXP1 | Sponge network | -2.46 | 0 | 0.042 | 0.74368 | 0.302 |
| 40545 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p | 31 | FOXP1 | Sponge network | 0.637 | 0.00069 | 0.042 | 0.74368 | 0.302 |
| 40546 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-3p | 14 | AKAP11 | Sponge network | -0.376 | 0.00337 | 0.196 | 0.09825 | 0.302 |
| 40547 | AF131215.2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 14 | PIK3CA | Sponge network | -0.176 | 0.59692 | 0.195 | 0.06908 | 0.302 |
| 40548 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-5p | 19 | GRIN2A | Sponge network | 0.174 | 0.30034 | -2.161 | 0 | 0.302 |
| 40549 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 26 | MOB1B | Sponge network | -1.092 | 4.0E-5 | 0.197 | 0.15266 | 0.302 |
| 40550 | LIPE-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-5p | 29 | EGFR | Sponge network | 0.078 | 0.55803 | -0.175 | 0.48451 | 0.302 |
| 40551 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 19 | MDM4 | Sponge network | 0.252 | 0.08508 | 0.345 | 0.00762 | 0.302 |
| 40552 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 19 | SUFU | Sponge network | 0.064 | 0.72934 | -0.04 | 0.65489 | 0.302 |
| 40553 | WDFY3-AS2 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | PPP2R2C | Sponge network | -0.942 | 0 | -0.959 | 0.07428 | 0.302 |
| 40554 | U91328.21 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-589-5p | 14 | SOX5 | Sponge network | -0.735 | 0.15983 | -1.091 | 4.0E-5 | 0.302 |
| 40555 | NDUFA6-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | TPO | Sponge network | -0.355 | 0.0077 | -1.87 | 2.0E-5 | 0.302 |
| 40556 | AC006547.13 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-375 | 10 | MLLT10 | Sponge network | 1.159 | 0 | 0.105 | 0.33292 | 0.302 |
| 40557 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-550a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | CCND2 | Sponge network | -0.611 | 0.09607 | -0.267 | 0.3112 | 0.301 |
| 40558 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | LRP1B | Sponge network | 0.246 | 0.59912 | -2.163 | 0 | 0.301 |
| 40559 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | EBF1 | Sponge network | 0.559 | 0.00026 | -0.384 | 0.08856 | 0.301 |
| 40560 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 20 | AKAP11 | Sponge network | 1.03 | 0 | 0.196 | 0.09825 | 0.301 |
| 40561 | RP11-503E24.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-374b-5p | 10 | SOX11 | Sponge network | 2.218 | 0 | 2.68 | 0 | 0.301 |
| 40562 | CTD-2017D11.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-582-5p | 12 | SETBP1 | Sponge network | 0.792 | 0.0049 | -1.302 | 1.0E-5 | 0.301 |
| 40563 | LINC00621 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-511-5p;hsa-miR-590-3p | 19 | ZFHX3 | Sponge network | 0.131 | 0.8436 | 0.389 | 0.01363 | 0.301 |
| 40564 | RP4-613B23.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p | 20 | CUX1 | Sponge network | -0.309 | 0.1145 | -0.039 | 0.6705 | 0.301 |
| 40565 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577 | 12 | KIT | Sponge network | 0.811 | 0.03228 | -2.184 | 0 | 0.301 |
| 40566 | RP4-794H19.1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-345-5p;hsa-miR-539-5p;hsa-miR-92a-3p | 11 | CDHR3 | Sponge network | -0.825 | 1.0E-5 | -0.977 | 4.0E-5 | 0.301 |
| 40567 | LINC00648 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 14 | NRK | Sponge network | 0.633 | 0.51678 | 0.656 | 0.19217 | 0.301 |
| 40568 | RP11-326G21.1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | MED12L | Sponge network | -0.021 | 0.93598 | -0.194 | 0.55299 | 0.301 |
| 40569 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | LATS1 | Sponge network | 0.222 | 0.29133 | 0.109 | 0.34208 | 0.301 |
| 40570 | RP11-927P21.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-940 | 18 | EPS15 | Sponge network | 0.921 | 0.00118 | -0.052 | 0.53998 | 0.301 |
| 40571 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | AHNAK | Sponge network | -0.318 | 0.65847 | -1.207 | 0 | 0.301 |
| 40572 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-34c-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p | 17 | ANK3 | Sponge network | -0.567 | 0 | -0.55 | 0.00086 | 0.301 |
| 40573 | AC005618.6 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p | 11 | DYNC1I1 | Sponge network | 0.862 | 0.06213 | -1.12 | 0.00107 | 0.301 |
| 40574 | AP001347.6 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-486-5p;hsa-miR-590-3p | 10 | DNMT3A | Sponge network | -1.161 | 0.00591 | 0.302 | 0.0262 | 0.301 |
| 40575 | AP001347.6 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PIK3CA | Sponge network | -1.161 | 0.00591 | 0.195 | 0.06908 | 0.301 |
| 40576 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-92b-3p | 12 | AKAP12 | Sponge network | -0.125 | 0.49414 | -0.932 | 0.0028 | 0.301 |
| 40577 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -0.811 | 2.0E-5 | -0.243 | 0.11408 | 0.301 |
| 40578 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 12 | PDE4DIP | Sponge network | 0.4 | 0.14611 | -0.282 | 0.0257 | 0.301 |
| 40579 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-5p | 26 | NOTCH2 | Sponge network | -0.739 | 0.00044 | 0.138 | 0.35163 | 0.301 |
| 40580 | CTC-429P9.3 |
hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-653-5p | 19 | UBR3 | Sponge network | 0.082 | 0.55397 | -0.021 | 0.82774 | 0.301 |
| 40581 | RP11-46C24.7 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 46 | MDM4 | Sponge network | 0.392 | 0.0278 | 0.345 | 0.00762 | 0.301 |
| 40582 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-582-5p | 14 | FBXO32 | Sponge network | 0.419 | 0.0319 | -0.495 | 0.07561 | 0.301 |
| 40583 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | CAV1 | Sponge network | -2.915 | 0.00015 | -1.013 | 6.0E-5 | 0.301 |
| 40584 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-590-3p | 14 | MLLT10 | Sponge network | 1.317 | 0 | 0.105 | 0.33292 | 0.301 |
| 40585 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p | 10 | PDS5B | Sponge network | 0.389 | 0.04293 | 0.259 | 0.00962 | 0.301 |
| 40586 | RP11-855A2.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-29b-1-5p;hsa-miR-450b-5p;hsa-miR-505-5p | 18 | LPP | Sponge network | -1.717 | 0.01522 | -0.459 | 0.04475 | 0.301 |
| 40587 | CTA-204B4.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-942-5p | 21 | ARHGEF12 | Sponge network | 0.765 | 7.0E-5 | 0.09 | 0.35693 | 0.301 |
| 40588 | RP11-244O19.1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 12 | SHPRH | Sponge network | -0.096 | 0.67958 | 0.098 | 0.40994 | 0.301 |
| 40589 | LINC00963 |
hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-375;hsa-miR-590-3p | 12 | THRA | Sponge network | 0.523 | 9.0E-5 | -0.307 | 0.05099 | 0.301 |
| 40590 | ACTA2-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | BCL9 | Sponge network | 0.194 | 0.64278 | 0.321 | 0.00267 | 0.301 |
| 40591 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | PTPRB | Sponge network | -0.899 | 6.0E-5 | 0.105 | 0.47122 | 0.301 |
| 40592 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-942-5p | 32 | UNC80 | Sponge network | -0.465 | 0.04703 | -1.934 | 0 | 0.301 |
| 40593 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-18a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | ARHGAP28 | Sponge network | 0.131 | 0.8436 | -0.911 | 0.00013 | 0.301 |
| 40594 | RP11-597D13.9 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | RADIL | Sponge network | 1.302 | 7.0E-5 | -1.628 | 0 | 0.301 |
| 40595 | MIR22HG |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-940 | 12 | TAL1 | Sponge network | -1.047 | 0 | -0.462 | 0.00574 | 0.301 |
| 40596 | MRPL23-AS1 |
hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-629-3p | 22 | CBX5 | Sponge network | -0.278 | 0.76559 | 0.165 | 0.24446 | 0.301 |
| 40597 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-629-3p | 23 | AR | Sponge network | 3.345 | 0 | -1.554 | 0 | 0.301 |
| 40598 | RP11-373D23.3 |
hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-671-5p | 24 | PPARA | Sponge network | -0.285 | 0.2172 | -0.379 | 0.00548 | 0.301 |
| 40599 | RP11-686D22.4 |
hsa-let-7a-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-323a-3p;hsa-miR-577;hsa-miR-590-3p | 11 | IL6ST | Sponge network | 0.793 | 5.0E-5 | -0.402 | 0.00676 | 0.301 |
| 40600 | RP11-750H9.5 |
hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-24-2-5p;hsa-miR-877-5p | 10 | NTRK3 | Sponge network | 0.349 | 0.13152 | -1.77 | 3.0E-5 | 0.301 |
| 40601 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PRKAB2 | Sponge network | 1.153 | 0 | -0.367 | 0.01371 | 0.301 |
| 40602 | AF131215.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 25 | DMD | Sponge network | -0.176 | 0.59692 | -1.506 | 0 | 0.301 |
| 40603 | FAM13A-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | TLL1 | Sponge network | -0.83 | 0 | -1.195 | 3.0E-5 | 0.301 |
| 40604 | RP11-181G12.2 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 12 | DICER1 | Sponge network | 0.556 | 0.00298 | 0.042 | 0.674 | 0.301 |
| 40605 | RP11-67L2.2 |
hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-940;hsa-miR-96-5p | 10 | FOXO4 | Sponge network | 0.72 | 0.0001 | -0.516 | 2.0E-5 | 0.301 |
| 40606 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p | 23 | PBX1 | Sponge network | 0.197 | 0.25618 | -1.206 | 0 | 0.301 |
| 40607 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-940 | 28 | WDFY3 | Sponge network | -2.095 | 0.00112 | -0.201 | 0.0967 | 0.301 |
| 40608 | LINC00863 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | EPHA3 | Sponge network | 0.189 | 0.13981 | 0.185 | 0.53588 | 0.301 |
| 40609 | CTD-2336O2.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-935 | 22 | DMD | Sponge network | -0.141 | 0.26434 | -1.506 | 0 | 0.301 |
| 40610 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 42 | RASSF2 | Sponge network | -0.777 | 0.1134 | -0.273 | 0.21961 | 0.301 |
| 40611 | MALAT1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | FAM172A | Sponge network | 0.782 | 0.00016 | -0.57 | 0 | 0.301 |
| 40612 | CTC-471J1.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 10 | PLAG1 | Sponge network | 1.11 | 1.0E-5 | -0.315 | 0.26532 | 0.301 |
| 40613 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 58 | SMAD4 | Sponge network | 0.064 | 0.72934 | -0.313 | 0.00413 | 0.301 |
| 40614 | ZNF32-AS1 |
hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-33a-5p | 10 | AKAP11 | Sponge network | 2.413 | 1.0E-5 | 0.196 | 0.09825 | 0.301 |
| 40615 | PPP1R26-AS1 |
hsa-miR-129-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326 | 10 | CCDC6 | Sponge network | 1.454 | 0 | 0.27 | 0.02555 | 0.301 |
| 40616 | MIR22HG |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | BTG2 | Sponge network | -1.047 | 0 | -1.763 | 0 | 0.301 |
| 40617 | HOXB-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-3p | 15 | TET1 | Sponge network | -0.154 | 0.53954 | -0.135 | 0.52138 | 0.301 |
| 40618 | LINC00094 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-660-5p | 24 | EYA4 | Sponge network | 0.253 | 0.09565 | 0.3 | 0.36876 | 0.301 |
| 40619 | MIR497HG |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-339-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 15 | LUZP2 | Sponge network | -1.439 | 4.0E-5 | -0.056 | 0.89416 | 0.301 |
| 40620 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-27a-3p | 16 | CBX5 | Sponge network | 3.21 | 0 | 0.165 | 0.24446 | 0.301 |
| 40621 | RP11-999E24.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-425-5p | 10 | ITGB3 | Sponge network | -0.693 | 0.05629 | -0.011 | 0.95751 | 0.301 |
| 40622 | RP4-584D14.7 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-500a-5p | 13 | RYR2 | Sponge network | 1.294 | 0.0031 | -1.466 | 0.00031 | 0.301 |
| 40623 | PART1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 31 | ZMYND11 | Sponge network | -2.925 | 0 | -0.2 | 0.05449 | 0.301 |
| 40624 | AC074289.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-96-5p | 12 | RECK | Sponge network | -0.326 | 0.14314 | -1.043 | 0 | 0.301 |
| 40625 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TBX18 | Sponge network | -0.021 | 0.93598 | 0.843 | 0.02945 | 0.301 |
| 40626 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 50 | ERBB4 | Sponge network | -1.697 | 0.00889 | -1.975 | 2.0E-5 | 0.301 |
| 40627 | CTD-2314G24.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | IL17RD | Sponge network | 0.246 | 0.59912 | -0.019 | 0.94873 | 0.301 |
| 40628 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | EPHA3 | Sponge network | 0.862 | 0.06213 | 0.185 | 0.53588 | 0.301 |
| 40629 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-935 | 36 | PTPRD | Sponge network | -0.246 | 0.35034 | -1.005 | 0.00064 | 0.301 |
| 40630 | MAGI2-AS3 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-625-5p | 19 | TMTC2 | Sponge network | -0.129 | 0.57177 | -0.215 | 0.18248 | 0.301 |
| 40631 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | JAKMIP2 | Sponge network | 0.083 | 0.66405 | -1.388 | 0 | 0.301 |
| 40632 | RP11-390K5.6 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-590-3p | 14 | CCDC6 | Sponge network | 1.148 | 2.0E-5 | 0.27 | 0.02555 | 0.301 |
| 40633 | AF131215.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-93-3p | 12 | UNC5C | Sponge network | -0.176 | 0.59692 | -0.178 | 0.40214 | 0.301 |
| 40634 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p | 40 | WDFY3 | Sponge network | -2.62 | 0 | -0.201 | 0.0967 | 0.301 |
| 40635 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 26 | CADM1 | Sponge network | -0.096 | 0.67958 | -0.9 | 0.00084 | 0.301 |
| 40636 | CTD-2017D11.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-484 | 11 | CADM1 | Sponge network | 0.792 | 0.0049 | -0.9 | 0.00084 | 0.301 |
| 40637 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 27 | HBP1 | Sponge network | -0.83 | 0 | -0.454 | 0 | 0.301 |
| 40638 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-429 | 10 | SCN5A | Sponge network | -0.942 | 0 | -0.259 | 0.48224 | 0.301 |
| 40639 | RAMP2-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-942-5p | 10 | ZBTB20 | Sponge network | -0.513 | 0.04703 | -0.122 | 0.57569 | 0.301 |
| 40640 | AC090587.4 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-424-5p;hsa-miR-484 | 13 | KIF1B | Sponge network | -0.125 | 0.60166 | -0.118 | 0.32258 | 0.301 |
| 40641 | CTD-2260A17.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-940 | 14 | MDM4 | Sponge network | 0.518 | 0.01048 | 0.345 | 0.00762 | 0.301 |
| 40642 | RP11-158K1.3 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-342-5p | 10 | WNK2 | Sponge network | 0.349 | 0.01695 | -0.352 | 0.31066 | 0.301 |
| 40643 | AC108488.4 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-629-3p | 13 | ITGA5 | Sponge network | 0.259 | 0.12542 | 0.452 | 0.03524 | 0.301 |
| 40644 | MIR143HG |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 24 | KCNJ5 | Sponge network | -0.739 | 0.0574 | -0.281 | 0.3339 | 0.301 |
| 40645 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-628-5p | 23 | RUNX1T1 | Sponge network | -1.054 | 0.0706 | -0.344 | 0.2374 | 0.301 |
| 40646 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | FGF5 | Sponge network | 0.603 | 0.00127 | 0.549 | 0.28659 | 0.301 |
| 40647 | RP11-428J1.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SUFU | Sponge network | 0.538 | 0.00076 | -0.04 | 0.65489 | 0.301 |
| 40648 | RP11-867G23.10 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 31 | CUX1 | Sponge network | -2.531 | 0 | -0.039 | 0.6705 | 0.301 |
| 40649 | KB-431C1.4 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-582-3p;hsa-miR-625-3p | 20 | ABL2 | Sponge network | 0.909 | 0 | 0.82 | 0 | 0.301 |
| 40650 | RP11-6N17.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-18a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-455-5p | 10 | TRPS1 | Sponge network | 0.108 | 0.69556 | -0.191 | 0.44109 | 0.301 |
| 40651 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | MOB1B | Sponge network | -1.161 | 0.00591 | 0.197 | 0.15266 | 0.301 |
| 40652 | RP11-158K1.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-628-5p | 14 | CDH23 | Sponge network | 0.349 | 0.01695 | -0.974 | 0.00034 | 0.301 |
| 40653 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-455-5p | 11 | TBX18 | Sponge network | 2.163 | 0 | 0.843 | 0.02945 | 0.301 |
| 40654 | TMEM161B-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 11 | CHD1 | Sponge network | -0.04 | 0.7606 | 0.322 | 0.00215 | 0.301 |
| 40655 | AF131215.2 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30e-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 20 | RASSF8 | Sponge network | -0.176 | 0.59692 | -0.122 | 0.65591 | 0.301 |
| 40656 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p | 20 | ZFHX3 | Sponge network | 1.254 | 0 | 0.389 | 0.01363 | 0.301 |
| 40657 | AC010226.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | LRP1B | Sponge network | -0.811 | 2.0E-5 | -2.163 | 0 | 0.301 |
| 40658 | RP11-65J21.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-501-5p | 15 | NTRK2 | Sponge network | -0.557 | 0.11135 | -0.736 | 0.06495 | 0.301 |
| 40659 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | HECA | Sponge network | -0.358 | 0.17255 | -0.217 | 0.03463 | 0.301 |
| 40660 | TPTEP1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-96-5p | 14 | ELMO1 | Sponge network | -1.216 | 0 | 0.04 | 0.83147 | 0.301 |
| 40661 | RP11-967K21.1 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-582-5p | 10 | PTCH1 | Sponge network | 0.056 | 0.81195 | -0.1 | 0.6179 | 0.301 |
| 40662 | RP11-981G7.6 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-590-3p | 18 | UBR3 | Sponge network | 0.986 | 0.01022 | -0.021 | 0.82774 | 0.301 |
| 40663 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-940 | 10 | MYO9A | Sponge network | -1.255 | 0.00981 | -0.147 | 0.32373 | 0.301 |
| 40664 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 24 | KCNA6 | Sponge network | 0.16 | 0.50785 | -1.531 | 0.00013 | 0.301 |
| 40665 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | TGFBR2 | Sponge network | -0.737 | 0.10822 | -0.243 | 0.11408 | 0.301 |
| 40666 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-577;hsa-miR-7-1-3p | 17 | SYT4 | Sponge network | -0.418 | 0.00068 | -3.207 | 0 | 0.301 |
| 40667 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 38 | ETV1 | Sponge network | -1.575 | 6.0E-5 | -0.096 | 0.67674 | 0.301 |
| 40668 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 37 | DDX6 | Sponge network | -0.129 | 0.57177 | 0.091 | 0.36428 | 0.301 |
| 40669 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-940 | 18 | CRTC3 | Sponge network | 0.657 | 4.0E-5 | 0.164 | 0.16786 | 0.301 |
| 40670 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 35 | RICTOR | Sponge network | 1.302 | 7.0E-5 | 0.388 | 0.00188 | 0.301 |
| 40671 | ZNF582-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 13 | SPTBN4 | Sponge network | -1.349 | 0.00017 | -0.786 | 0.01281 | 0.301 |
| 40672 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-671-3p;hsa-miR-93-5p | 15 | MAP3K12 | Sponge network | -0.563 | 0.02545 | -0.057 | 0.59369 | 0.301 |
| 40673 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-542-3p | 10 | FBXO31 | Sponge network | -0.487 | 0.02157 | -0.085 | 0.42389 | 0.301 |
| 40674 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | PDS5B | Sponge network | 0.311 | 0.10344 | 0.259 | 0.00962 | 0.301 |
| 40675 | RAMP2-AS1 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 20 | LIMCH1 | Sponge network | -0.513 | 0.04703 | -0.915 | 1.0E-5 | 0.301 |
| 40676 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p | 23 | TSC1 | Sponge network | -0.162 | 0.47687 | 0.103 | 0.26071 | 0.301 |
| 40677 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-511-5p | 11 | CADM2 | Sponge network | -1.054 | 0.0706 | -3.782 | 0 | 0.301 |
| 40678 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | PCDH8 | Sponge network | 0.101 | 0.60919 | -0.997 | 0.05366 | 0.301 |
| 40679 | CROCCP2 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p | 12 | GSK3B | Sponge network | 0.79 | 0 | 0.266 | 0.00045 | 0.301 |
| 40680 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | CHD2 | Sponge network | 0.417 | 0.00445 | 0.049 | 0.55371 | 0.301 |
| 40681 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-93-5p;hsa-miR-942-5p | 37 | UNC80 | Sponge network | -0.842 | 0.01361 | -1.934 | 0 | 0.301 |
| 40682 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | ARHGAP28 | Sponge network | -0.896 | 0.00099 | -0.911 | 0.00013 | 0.301 |
| 40683 | FAM66C |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | NIN | Sponge network | -0.901 | 0.00019 | -0.253 | 0.13794 | 0.301 |
| 40684 | MIR143HG |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-3913-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-877-5p | 43 | EGFR | Sponge network | -0.739 | 0.0574 | -0.175 | 0.48451 | 0.301 |
| 40685 | PWAR6 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 21 | UBE4B | Sponge network | -1.575 | 6.0E-5 | -0.235 | 0.007 | 0.301 |
| 40686 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-744-3p | 10 | HIP1 | Sponge network | 1.153 | 0 | 0.774 | 0 | 0.301 |
| 40687 | DNM3OS |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | AFF1 | Sponge network | 0.572 | 0.01722 | -0.384 | 0.00053 | 0.301 |
| 40688 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | PRKAA2 | Sponge network | 0.862 | 0.06213 | -2.462 | 0 | 0.301 |
| 40689 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-93-5p | 29 | LRP1B | Sponge network | -0.777 | 0.16667 | -2.163 | 0 | 0.301 |
| 40690 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-493-5p;hsa-miR-654-3p | 10 | MYBL1 | Sponge network | 0.909 | 0 | 0.494 | 0.02152 | 0.301 |
| 40691 | HCG11 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | RYR2 | Sponge network | 0.385 | 0.10067 | -1.466 | 0.00031 | 0.301 |
| 40692 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-455-3p | 14 | DKK3 | Sponge network | 0.082 | 0.55397 | -0.052 | 0.77783 | 0.301 |
| 40693 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CACNA2D1 | Sponge network | 0.657 | 4.0E-5 | -0.846 | 0.00675 | 0.301 |
| 40694 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-93-5p | 12 | PGR | Sponge network | -0.31 | 0.33871 | -1.704 | 0 | 0.301 |
| 40695 | AC025171.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 26 | RUNX1T1 | Sponge network | 0.998 | 0 | -0.344 | 0.2374 | 0.301 |
| 40696 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p | 24 | SPRY3 | Sponge network | -0.227 | 0.31657 | 0.016 | 0.91286 | 0.301 |
| 40697 | RP11-967K21.1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-940 | 11 | RHOB | Sponge network | 0.056 | 0.81195 | -1.052 | 0 | 0.301 |
| 40698 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | KANK1 | Sponge network | 0.194 | 0.64278 | -0.55 | 0.00134 | 0.301 |
| 40699 | LINC00863 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-590-3p | 11 | FMNL3 | Sponge network | 0.189 | 0.13981 | 0.555 | 4.0E-5 | 0.301 |
| 40700 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-590-3p | 13 | MYCBP2 | Sponge network | 0.349 | 0.01695 | -0.027 | 0.82016 | 0.301 |
| 40701 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | SYNE1 | Sponge network | 0.051 | 0.83388 | -0.738 | 0.00316 | 0.301 |
| 40702 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 19 | RNF144A | Sponge network | 0.225 | 0.30644 | 0.594 | 0.0009 | 0.301 |
| 40703 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 11 | TMEFF2 | Sponge network | 0.214 | 0.18246 | -2.426 | 0 | 0.301 |
| 40704 | HAS2-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-30b-3p;hsa-miR-7-1-3p | 12 | SOX5 | Sponge network | -0.011 | 0.967 | -1.091 | 4.0E-5 | 0.301 |
| 40705 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | MLLT10 | Sponge network | 0.417 | 0.00445 | 0.105 | 0.33292 | 0.301 |
| 40706 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-654-3p | 12 | ITCH | Sponge network | -0.567 | 0 | 0.084 | 0.34058 | 0.301 |
| 40707 | AC108488.4 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 10 | FYN | Sponge network | 0.259 | 0.12542 | -0.131 | 0.37051 | 0.301 |
| 40708 | RP11-13K12.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | NIN | Sponge network | -0.318 | 0.65847 | -0.253 | 0.13794 | 0.301 |
| 40709 | RP11-35G9.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-940 | 23 | TRPS1 | Sponge network | 0.622 | 0.00015 | -0.191 | 0.44109 | 0.301 |
| 40710 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 13 | MLLT10 | Sponge network | 0.556 | 0.00298 | 0.105 | 0.33292 | 0.301 |
| 40711 | GAS6-AS2 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 19 | GJA1 | Sponge network | 0.16 | 0.50785 | -0.485 | 0.02179 | 0.301 |
| 40712 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 34 | CREB3L2 | Sponge network | -0.154 | 0.78939 | -0.525 | 2.0E-5 | 0.301 |
| 40713 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p | 13 | ANK3 | Sponge network | 0.705 | 0.00014 | -0.55 | 0.00086 | 0.301 |
| 40714 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935 | 33 | PTPRD | Sponge network | 0.225 | 0.30644 | -1.005 | 0.00064 | 0.301 |
| 40715 | CALML3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-582-5p | 29 | DMD | Sponge network | 0.95 | 0.15943 | -1.506 | 0 | 0.301 |
| 40716 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | LATS2 | Sponge network | 0.082 | 0.55654 | 0.159 | 0.2304 | 0.301 |
| 40717 | MIR22HG |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-625-5p | 12 | TMTC2 | Sponge network | -1.047 | 0 | -0.215 | 0.18248 | 0.301 |
| 40718 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-539-5p | 10 | DDX5 | Sponge network | 0.078 | 0.55803 | -0.099 | 0.17428 | 0.301 |
| 40719 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | CHD2 | Sponge network | 1.294 | 0 | 0.049 | 0.55371 | 0.301 |
| 40720 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TBX18 | Sponge network | 0.101 | 0.60919 | 0.843 | 0.02945 | 0.301 |
| 40721 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-5p;hsa-miR-942-5p | 28 | NOTCH2 | Sponge network | 0.381 | 0.25967 | 0.138 | 0.35163 | 0.301 |
| 40722 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 30 | AKAP11 | Sponge network | 0.177 | 0.16681 | 0.196 | 0.09825 | 0.301 |
| 40723 | GAS6-AS2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-942-5p | 11 | ZBTB20 | Sponge network | 0.16 | 0.50785 | -0.122 | 0.57569 | 0.301 |
| 40724 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 16 | RARB | Sponge network | -0.318 | 0.65847 | -0.825 | 2.0E-5 | 0.301 |
| 40725 | RP11-6O2.3 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-660-5p;hsa-miR-96-5p | 13 | IRS1 | Sponge network | 0.331 | 0.57247 | -0.026 | 0.88855 | 0.301 |
| 40726 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | LATS2 | Sponge network | -1.349 | 0.00017 | 0.159 | 0.2304 | 0.301 |
| 40727 | WDFY3-AS2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | ROBO1 | Sponge network | -0.942 | 0 | -0.009 | 0.96154 | 0.301 |
| 40728 | RP1-59D14.5 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-874-3p | 11 | ARID5B | Sponge network | 1.162 | 5.0E-5 | 0.342 | 0.01676 | 0.301 |
| 40729 | HAS2-AS1 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 11 | CNTN1 | Sponge network | -0.011 | 0.967 | -2.594 | 0 | 0.301 |
| 40730 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-708-5p | 20 | MTSS1 | Sponge network | -1.092 | 4.0E-5 | -0.81 | 0.00035 | 0.301 |
| 40731 | RP11-305E6.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | RPS6KA3 | Sponge network | 1.317 | 0 | 0.256 | 0.02419 | 0.301 |
| 40732 | MIR600HG |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-625-5p;hsa-miR-655-3p | 10 | ARID2 | Sponge network | 0.594 | 0.41522 | 0.235 | 0.02697 | 0.301 |
| 40733 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p | 31 | CBX5 | Sponge network | 1.11 | 0 | 0.165 | 0.24446 | 0.301 |
| 40734 | RP11-473I1.10 |
hsa-miR-125a-5p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-628-5p | 11 | VPS53 | Sponge network | 0.673 | 0 | 0.103 | 0.22667 | 0.301 |
| 40735 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | ATRX | Sponge network | -0.176 | 0.59692 | 0.261 | 0.04408 | 0.301 |
| 40736 | RP11-403I13.8 |
hsa-miR-15b-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-335-5p | 10 | PLXNC1 | Sponge network | 0.619 | 0.00157 | 0.569 | 0.00329 | 0.301 |
| 40737 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-7-1-3p | 17 | DDX6 | Sponge network | 0.693 | 0.00076 | 0.091 | 0.36428 | 0.301 |
| 40738 | RP11-473I1.10 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 12 | EPHA3 | Sponge network | 0.673 | 0 | 0.185 | 0.53588 | 0.301 |
| 40739 | RP11-497H16.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 30 | LPP | Sponge network | 0.545 | 0.00064 | -0.459 | 0.04475 | 0.301 |
| 40740 | RP11-59H7.3 |
hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-33a-3p | 11 | RASAL2 | Sponge network | 2.163 | 0 | 0.869 | 0 | 0.301 |
| 40741 | WDR86-AS1 |
hsa-let-7f-1-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-96-5p | 10 | NAV3 | Sponge network | 0.348 | 0.32912 | 0.212 | 0.47729 | 0.301 |
| 40742 | RP11-156P1.3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 10 | ZNF844 | Sponge network | 0.214 | 0.18246 | -0.966 | 0.00035 | 0.301 |
| 40743 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RCAN2 | Sponge network | 0.873 | 0 | -0.33 | 0.15172 | 0.301 |
| 40744 | CTB-31O20.2 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-376c-3p;hsa-miR-497-5p | 14 | TBL1XR1 | Sponge network | 0.608 | 3.0E-5 | 0.578 | 0 | 0.301 |
| 40745 | U91328.19 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 25 | TET1 | Sponge network | -0.303 | 0.21002 | -0.135 | 0.52138 | 0.301 |
| 40746 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | NEDD4 | Sponge network | 0.765 | 7.0E-5 | 0.02 | 0.89834 | 0.301 |
| 40747 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 46 | AR | Sponge network | 0.349 | 0.01695 | -1.554 | 0 | 0.301 |
| 40748 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 0.986 | 0.01022 | 0.259 | 0.00962 | 0.301 |
| 40749 | RP11-166D19.1 |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-542-3p | 13 | CADM4 | Sponge network | -0.576 | 0.10121 | -0.635 | 0.00305 | 0.301 |
| 40750 | ARHGEF26-AS1 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | RASSF3 | Sponge network | -2.46 | 0 | -0.226 | 0.11691 | 0.301 |
| 40751 | NPPA-AS1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p;hsa-miR-940 | 15 | MDM4 | Sponge network | 2.169 | 0.00056 | 0.345 | 0.00762 | 0.301 |
| 40752 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-942-5p | 31 | TMEM132B | Sponge network | 0.082 | 0.55397 | -0.83 | 0.01084 | 0.301 |
| 40753 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | TCF4 | Sponge network | 0.259 | 0.12542 | 0.016 | 0.92271 | 0.301 |
| 40754 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | 1.308 | 0 | 0.323 | 0.00792 | 0.301 |
| 40755 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 20 | ARNT2 | Sponge network | -2.452 | 0 | -0.332 | 0.25851 | 0.301 |
| 40756 | LINC00472 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-503-5p | 20 | ABL1 | Sponge network | -0.842 | 0.01361 | -0.215 | 0.07804 | 0.301 |
| 40757 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PDE4DIP | Sponge network | -0.08 | 0.90092 | -0.282 | 0.0257 | 0.301 |
| 40758 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | PDGFRA | Sponge network | -0.72 | 0.24239 | -0.372 | 0.0037 | 0.301 |
| 40759 | GS1-124K5.3 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 12 | CCDC6 | Sponge network | 1.501 | 0 | 0.27 | 0.02555 | 0.301 |
| 40760 | CTC-296K1.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-671-5p | 25 | CREB3L2 | Sponge network | -2.095 | 0.00112 | -0.525 | 2.0E-5 | 0.301 |
| 40761 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 48 | ERBB4 | Sponge network | -0.737 | 0.06182 | -1.975 | 2.0E-5 | 0.301 |
| 40762 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-577 | 14 | GREM1 | Sponge network | -1.057 | 0.00029 | 0.902 | 0.01691 | 0.301 |
| 40763 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 11 | CEP170 | Sponge network | -0.323 | 0.01372 | 0.943 | 0 | 0.301 |
| 40764 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ZNF770 | Sponge network | 0.72 | 0.0001 | 0.206 | 0.06751 | 0.301 |
| 40765 | TRIM52-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | CAV1 | Sponge network | -0.418 | 0.00068 | -1.013 | 6.0E-5 | 0.301 |
| 40766 | MIR22HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-671-5p | 26 | CYLD | Sponge network | -1.047 | 0 | -0.367 | 0.00171 | 0.301 |
| 40767 | ZNF32-AS2 |
hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-654-3p | 13 | AKAP11 | Sponge network | 1.552 | 7.0E-5 | 0.196 | 0.09825 | 0.301 |
| 40768 | RP11-155G14.6 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-326 | 10 | CBFB | Sponge network | 2.483 | 0 | 1.061 | 0 | 0.301 |
| 40769 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-7-1-3p | 29 | DTNA | Sponge network | 0.238 | 0.70544 | -1.648 | 1.0E-5 | 0.301 |
| 40770 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-96-5p | 36 | MTSS1 | Sponge network | -2.452 | 0 | -0.81 | 0.00035 | 0.301 |
| 40771 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | ERBB4 | Sponge network | -0.668 | 3.0E-5 | -1.975 | 2.0E-5 | 0.301 |
| 40772 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 30 | CHD5 | Sponge network | -0.08 | 0.90092 | -0.922 | 0.01521 | 0.301 |
| 40773 | CTD-2336O2.1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | RASSF8 | Sponge network | -0.141 | 0.26434 | -0.122 | 0.65591 | 0.301 |
| 40774 | RP11-774O3.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 27 | EPS15 | Sponge network | -0.487 | 0.02157 | -0.052 | 0.53998 | 0.301 |
| 40775 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ABCA10 | Sponge network | 0.222 | 0.29133 | -0.571 | 0.03674 | 0.301 |
| 40776 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 15 | CSRNP1 | Sponge network | -2.46 | 0 | -1.448 | 0 | 0.301 |
| 40777 | RP11-967K21.1 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-3p | 18 | DOCK4 | Sponge network | 0.056 | 0.81195 | 0.502 | 0.00108 | 0.301 |
| 40778 | RP11-400K9.3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p | 11 | CREB3L2 | Sponge network | -0.733 | 0.19469 | -0.525 | 2.0E-5 | 0.301 |
| 40779 | AC127904.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ABI2 | Sponge network | 1.308 | 0 | 0.175 | 0.14787 | 0.301 |
| 40780 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-5p | 10 | MCC | Sponge network | -0.214 | 0.44248 | -1.005 | 0 | 0.301 |
| 40781 | RP11-373D23.3 |
hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p | 15 | RAP1A | Sponge network | -0.285 | 0.2172 | -0.929 | 0 | 0.301 |
| 40782 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 21 | CLASP2 | Sponge network | 1.757 | 0 | -0.038 | 0.71 | 0.301 |
| 40783 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p | 10 | LATS2 | Sponge network | -4.546 | 0 | 0.159 | 0.2304 | 0.301 |
| 40784 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-942-5p | 26 | HLF | Sponge network | -0.053 | 0.80685 | -2.443 | 0 | 0.301 |
| 40785 | AC003090.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | INPP4A | Sponge network | -2.117 | 0.00161 | 0.07 | 0.53563 | 0.301 |
| 40786 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 38 | FAT3 | Sponge network | 0.862 | 0.06213 | -1.601 | 0.00051 | 0.301 |
| 40787 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | SLC6A15 | Sponge network | -0.222 | 0.32445 | -2.373 | 2.0E-5 | 0.301 |
| 40788 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DNAJB4 | Sponge network | -0.737 | 0.10822 | -0.7 | 1.0E-5 | 0.301 |
| 40789 | THAP9-AS1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-628-5p | 12 | MAT2A | Sponge network | 1.079 | 0 | -0.073 | 0.36195 | 0.301 |
| 40790 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-98-5p | 13 | TGFBR3 | Sponge network | -0.295 | 0.02604 | -1.173 | 1.0E-5 | 0.301 |
| 40791 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 26 | IL6ST | Sponge network | 0.594 | 0.00064 | -0.402 | 0.00676 | 0.301 |
| 40792 | AC004540.4 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-362-3p | 16 | MEF2C | Sponge network | -2.549 | 0.00528 | -0.499 | 0.01448 | 0.301 |
| 40793 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | DACH2 | Sponge network | -2.925 | 0 | -1.635 | 0.00034 | 0.301 |
| 40794 | SH3BP5-AS1 |
hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-5p | 23 | UBR3 | Sponge network | 0.267 | 0.2937 | -0.021 | 0.82774 | 0.301 |
| 40795 | BVES-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-429 | 20 | EFNA5 | Sponge network | -2.62 | 0 | -0.421 | 0.17868 | 0.301 |
| 40796 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 57 | KIF1B | Sponge network | -0.323 | 0.01372 | -0.118 | 0.32258 | 0.301 |
| 40797 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-7-1-3p | 15 | MCC | Sponge network | -0.285 | 0.2172 | -1.005 | 0 | 0.301 |
| 40798 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-409-3p | 10 | SASH1 | Sponge network | -0.309 | 0.1145 | -0.502 | 0.00175 | 0.301 |
| 40799 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p | 17 | RARB | Sponge network | 0.082 | 0.55654 | -0.825 | 2.0E-5 | 0.3 |
| 40800 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-3p | 14 | PTPRB | Sponge network | 0.174 | 0.30034 | 0.105 | 0.47122 | 0.3 |
| 40801 | HAR1A |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-671-5p | 12 | CADM3 | Sponge network | -0.672 | 0.19447 | -3.663 | 0 | 0.3 |
| 40802 | LINC00472 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-493-3p | 18 | CSDE1 | Sponge network | -0.842 | 0.01361 | -0.493 | 0 | 0.3 |
| 40803 | LINC00667 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | RIMBP2 | Sponge network | -0.235 | 0.15142 | -1.968 | 5.0E-5 | 0.3 |
| 40804 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 23 | CRTC3 | Sponge network | 0.4 | 0.14611 | 0.164 | 0.16786 | 0.3 |
| 40805 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-590-3p | 29 | RICTOR | Sponge network | -0.096 | 0.67958 | 0.388 | 0.00188 | 0.3 |
| 40806 | RP11-401P9.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-590-5p | 25 | MED12L | Sponge network | -0.384 | 0.40652 | -0.194 | 0.55299 | 0.3 |
| 40807 | RP11-156P1.3 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-92a-3p | 20 | RIMS2 | Sponge network | 0.214 | 0.18246 | -1.365 | 0.00208 | 0.3 |
| 40808 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-539-5p;hsa-miR-766-3p | 11 | AFF4 | Sponge network | 0.793 | 0 | -0.266 | 0.01565 | 0.3 |
| 40809 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-501-3p | 22 | CLASP2 | Sponge network | -1.645 | 0 | -0.038 | 0.71 | 0.3 |
| 40810 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-210-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p | 12 | APC | Sponge network | -0.285 | 0.2172 | -0.255 | 0.04051 | 0.3 |
| 40811 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p | 19 | CACNA2D1 | Sponge network | -0.384 | 0.40652 | -0.846 | 0.00675 | 0.3 |
| 40812 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p | 19 | FBXO32 | Sponge network | 0.001 | 0.99753 | -0.495 | 0.07561 | 0.3 |
| 40813 | RP13-516M14.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-940 | 10 | CELF4 | Sponge network | 0.389 | 0.04293 | -1.135 | 0.00344 | 0.3 |
| 40814 | HAR1A |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-744-3p;hsa-miR-940 | 18 | MEF2C | Sponge network | -0.672 | 0.19447 | -0.499 | 0.01448 | 0.3 |
| 40815 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 18 | EIF4A2 | Sponge network | -0.612 | 0.00094 | -0.383 | 0.00046 | 0.3 |
| 40816 | RP11-119F7.5 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | RYR2 | Sponge network | 0.225 | 0.30644 | -1.466 | 0.00031 | 0.3 |
| 40817 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p | 17 | NR2C2 | Sponge network | -0.303 | 0.21002 | 0.21 | 0.03224 | 0.3 |
| 40818 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p | 10 | PDS5B | Sponge network | 1.093 | 0 | 0.259 | 0.00962 | 0.3 |
| 40819 | AC006116.24 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-582-3p | 18 | PPP1R9A | Sponge network | -0.473 | 0.07602 | -0.903 | 0.04254 | 0.3 |
| 40820 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-539-5p | 15 | PBX1 | Sponge network | 0.37 | 0.27614 | -1.206 | 0 | 0.3 |
| 40821 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | MOB1B | Sponge network | 0.997 | 0.00066 | 0.197 | 0.15266 | 0.3 |
| 40822 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 14 | RNF144A | Sponge network | 0.197 | 0.25618 | 0.594 | 0.0009 | 0.3 |
| 40823 | GUSBP11 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | ERCC6 | Sponge network | 0.856 | 0 | 0.23 | 0.015 | 0.3 |
| 40824 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p;hsa-miR-655-3p | 13 | ARID2 | Sponge network | 0.75 | 0.00054 | 0.235 | 0.02697 | 0.3 |
| 40825 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-5p | 12 | TBX18 | Sponge network | 0.622 | 0.00015 | 0.843 | 0.02945 | 0.3 |
| 40826 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 31 | CCND2 | Sponge network | -4.546 | 0 | -0.267 | 0.3112 | 0.3 |
| 40827 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 21 | CDC73 | Sponge network | 0.91 | 0 | 0.094 | 0.20463 | 0.3 |
| 40828 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-589-3p | 11 | SCN9A | Sponge network | 1.153 | 0.00114 | -0.525 | 0.10593 | 0.3 |
| 40829 | MIR497HG |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | EFNA5 | Sponge network | -1.439 | 4.0E-5 | -0.421 | 0.17868 | 0.3 |
| 40830 | CTD-3064M3.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-429 | 10 | BCL6 | Sponge network | -0.469 | 0.08721 | -0.211 | 0.19489 | 0.3 |
| 40831 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | HACE1 | Sponge network | 0.417 | 0.00445 | -0.072 | 0.55239 | 0.3 |
| 40832 | AF131215.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10a-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 28 | CREB1 | Sponge network | -0.176 | 0.59692 | 0.203 | 0.00537 | 0.3 |
| 40833 | HOTAIRM1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | LIMCH1 | Sponge network | -0.053 | 0.80685 | -0.915 | 1.0E-5 | 0.3 |
| 40834 | RP11-774O3.3 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | PTCH1 | Sponge network | -0.487 | 0.02157 | -0.1 | 0.6179 | 0.3 |
| 40835 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | DMXL1 | Sponge network | -0.899 | 6.0E-5 | -0.426 | 0.00056 | 0.3 |
| 40836 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | ST6GAL2 | Sponge network | -0.737 | 0.10822 | -1.201 | 0.00597 | 0.3 |
| 40837 | LINC00173 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-625-5p | 18 | PTPN14 | Sponge network | 0.056 | 0.8494 | 0.144 | 0.34595 | 0.3 |
| 40838 | AC141928.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 13 | LUZP2 | Sponge network | 0.853 | 0.15352 | -0.056 | 0.89416 | 0.3 |
| 40839 | DIO3OS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p | 62 | LPP | Sponge network | -0.773 | 0.04073 | -0.459 | 0.04475 | 0.3 |
| 40840 | SNHG12 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 13 | PAFAH1B2 | Sponge network | 1.103 | 0 | 0.318 | 0.0001 | 0.3 |
| 40841 | SH3RF3-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 12 | SEMA3C | Sponge network | 0.889 | 9.0E-5 | -0.272 | 0.31153 | 0.3 |
| 40842 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-7-1-3p | 12 | DACT1 | Sponge network | 0.853 | 0.15352 | 0.783 | 0.00072 | 0.3 |
| 40843 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577;hsa-miR-582-5p | 15 | KAT6B | Sponge network | 0.921 | 0.00118 | -0.478 | 0.00018 | 0.3 |
| 40844 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-590-3p | 11 | JAKMIP2 | Sponge network | -0.727 | 0.04726 | -1.388 | 0 | 0.3 |
| 40845 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 16 | MLLT10 | Sponge network | 0.594 | 0.41522 | 0.105 | 0.33292 | 0.3 |
| 40846 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | TRIM33 | Sponge network | 0.998 | 0 | 0.402 | 7.0E-5 | 0.3 |
| 40847 | HOXB-AS2 |
hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p | 12 | RNF144A | Sponge network | 1.316 | 0.01106 | 0.594 | 0.0009 | 0.3 |
| 40848 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-374b-5p | 11 | CBL | Sponge network | 0.912 | 0.00014 | 0.291 | 0.01267 | 0.3 |
| 40849 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p | 15 | HECA | Sponge network | -0.583 | 0.14589 | -0.217 | 0.03463 | 0.3 |
| 40850 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p | 12 | HOOK3 | Sponge network | -0.223 | 0.47816 | 0.273 | 0.09957 | 0.3 |
| 40851 | RP11-161M6.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-589-3p | 13 | PPP1R9A | Sponge network | 0.61 | 0.06611 | -0.903 | 0.04254 | 0.3 |
| 40852 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | TP53INP1 | Sponge network | -0.376 | 0.00337 | -0.496 | 0.00064 | 0.3 |
| 40853 | DNM1P35 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-93-3p | 16 | WWTR1 | Sponge network | 0.051 | 0.83388 | -0.345 | 0.11997 | 0.3 |
| 40854 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | LIFR | Sponge network | 0.727 | 3.0E-5 | -1.794 | 0 | 0.3 |
| 40855 | SOX2-OT |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 24 | SETBP1 | Sponge network | -1.264 | 6.0E-5 | -1.302 | 1.0E-5 | 0.3 |
| 40856 | CTD-2666L21.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-629-3p | 11 | CDH23 | Sponge network | 0.368 | 0.20093 | -0.974 | 0.00034 | 0.3 |
| 40857 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 12 | EBF3 | Sponge network | 0.381 | 0.25967 | -0.058 | 0.81437 | 0.3 |
| 40858 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-93-5p | 28 | CHD5 | Sponge network | -0.206 | 0.12942 | -0.922 | 0.01521 | 0.3 |
| 40859 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-93-5p;hsa-miR-96-5p | 12 | ST6GALNAC3 | Sponge network | -0.31 | 0.33871 | -0.987 | 0 | 0.3 |
| 40860 | RP5-855D21.1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p | 10 | CCDC6 | Sponge network | 1.115 | 0.00188 | 0.27 | 0.02555 | 0.3 |
| 40861 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | ZNF770 | Sponge network | 0.222 | 0.29133 | 0.206 | 0.06751 | 0.3 |
| 40862 | LINC00969 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-495-3p | 23 | ZFX | Sponge network | 0.683 | 1.0E-5 | 0.09 | 0.42563 | 0.3 |
| 40863 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 28 | TIMP3 | Sponge network | -0.303 | 0.21002 | -0.546 | 0.02307 | 0.3 |
| 40864 | LINC00886 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-5p | 17 | LIMCH1 | Sponge network | -0.371 | 0.24604 | -0.915 | 1.0E-5 | 0.3 |
| 40865 | CTD-2336O2.1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | -0.141 | 0.26434 | -1.09 | 0.00014 | 0.3 |
| 40866 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-495-3p | 11 | MYO9A | Sponge network | 0.856 | 0 | -0.147 | 0.32373 | 0.3 |
| 40867 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p | 20 | MYBL1 | Sponge network | 0.335 | 0.20596 | 0.494 | 0.02152 | 0.3 |
| 40868 | AC090587.5 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-93-3p | 14 | QKI | Sponge network | -0.088 | 0.65011 | -0.333 | 0.04111 | 0.3 |
| 40869 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-454-3p;hsa-miR-93-5p | 19 | PAFAH1B1 | Sponge network | -0.473 | 0.07602 | -0.429 | 0 | 0.3 |
| 40870 | LINC00472 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 25 | RIMS2 | Sponge network | -0.842 | 0.01361 | -1.365 | 0.00208 | 0.3 |
| 40871 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 11 | ITCH | Sponge network | 0.765 | 7.0E-5 | 0.084 | 0.34058 | 0.3 |
| 40872 | AC004540.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-484 | 14 | GRIA3 | Sponge network | -2.549 | 0.00528 | -1.956 | 0 | 0.3 |
| 40873 | CTD-2528L19.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | EPS15 | Sponge network | 0.953 | 0 | -0.052 | 0.53998 | 0.3 |
| 40874 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-582-3p | 11 | CLASP2 | Sponge network | 0.909 | 0 | -0.038 | 0.71 | 0.3 |
| 40875 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-940 | 29 | CHD5 | Sponge network | 0.523 | 9.0E-5 | -0.922 | 0.01521 | 0.3 |
| 40876 | CD27-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-93-3p | 50 | RUNX1T1 | Sponge network | 0.177 | 0.16681 | -0.344 | 0.2374 | 0.3 |
| 40877 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | TP53INP1 | Sponge network | -0.513 | 0.04703 | -0.496 | 0.00064 | 0.3 |
| 40878 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | IGSF10 | Sponge network | -0.031 | 0.89063 | -1.751 | 1.0E-5 | 0.3 |
| 40879 | RP11-264I13.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-30a-3p;hsa-miR-576-5p | 11 | CREB1 | Sponge network | 0.421 | 0.01652 | 0.203 | 0.00537 | 0.3 |
| 40880 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-590-3p | 15 | SPG20 | Sponge network | 0.083 | 0.66405 | -1.16 | 0 | 0.3 |
| 40881 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | PTPRB | Sponge network | -0.811 | 2.0E-5 | 0.105 | 0.47122 | 0.3 |
| 40882 | LENG8-AS1 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-338-5p;hsa-miR-374a-3p;hsa-miR-625-3p;hsa-miR-628-5p | 16 | BMPR2 | Sponge network | 1.471 | 0 | 0.206 | 0.0635 | 0.3 |
| 40883 | RP11-38L15.3 |
hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 11 | CNTN1 | Sponge network | 0.344 | 0.3127 | -2.594 | 0 | 0.3 |
| 40884 | BDNF-AS |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | -0.668 | 3.0E-5 | -1.12 | 0.00107 | 0.3 |
| 40885 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-590-3p | 15 | CXXC4 | Sponge network | 0.051 | 0.83388 | -0.433 | 0.21055 | 0.3 |
| 40886 | RP11-890B15.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 48 | PPARA | Sponge network | 0.101 | 0.60919 | -0.379 | 0.00548 | 0.3 |
| 40887 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | CNTNAP3 | Sponge network | 0.219 | 0.1516 | -1.465 | 0.00033 | 0.3 |
| 40888 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 18 | SIK1 | Sponge network | -2.452 | 0 | -0.98 | 0 | 0.3 |
| 40889 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p | 14 | MOB1B | Sponge network | 1.266 | 0 | 0.197 | 0.15266 | 0.3 |
| 40890 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p | 13 | MYBL1 | Sponge network | 1.106 | 0 | 0.494 | 0.02152 | 0.3 |
| 40891 | AC156455.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p | 12 | GAS7 | Sponge network | 1.154 | 0.00041 | -0.555 | 0.01602 | 0.3 |
| 40892 | ZNF32-AS1 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-576-5p | 10 | ZFX | Sponge network | 2.413 | 1.0E-5 | 0.09 | 0.42563 | 0.3 |
| 40893 | LINC00861 |
hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 15 | NTRK3 | Sponge network | 0.911 | 0.00854 | -1.77 | 3.0E-5 | 0.3 |
| 40894 | LINC00094 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-508-3p | 17 | RECK | Sponge network | 0.253 | 0.09565 | -1.043 | 0 | 0.3 |
| 40895 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 14 | FGF5 | Sponge network | -0.777 | 0.16667 | 0.549 | 0.28659 | 0.3 |
| 40896 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 15 | ACVR1 | Sponge network | -0.737 | 0.10822 | 0.279 | 0.00234 | 0.3 |
| 40897 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 29 | NOTCH2 | Sponge network | -0.125 | 0.49414 | 0.138 | 0.35163 | 0.3 |
| 40898 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | MITF | Sponge network | 0.225 | 0.30644 | -0.769 | 0.00022 | 0.3 |
| 40899 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p | 31 | PIK3R1 | Sponge network | -1.161 | 0.00591 | -0.133 | 0.36854 | 0.3 |
| 40900 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 39 | FZD3 | Sponge network | 0.614 | 1.0E-5 | 0.104 | 0.60316 | 0.3 |
| 40901 | PCA3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | LATS2 | Sponge network | -0.709 | 0.18168 | 0.159 | 0.2304 | 0.3 |
| 40902 | CTB-31O20.4 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | PIK3C2A | Sponge network | 1.619 | 0 | 0.061 | 0.53776 | 0.3 |
| 40903 | RP11-102N12.3 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-7-5p;hsa-miR-942-5p | 21 | IGFBP5 | Sponge network | -0.351 | 0.11358 | -0.806 | 0.00105 | 0.3 |
| 40904 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 23 | SOX11 | Sponge network | 0.72 | 0.0001 | 2.68 | 0 | 0.3 |
| 40905 | RP11-822E23.8 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-362-5p | 15 | TLE4 | Sponge network | -0.777 | 0.16667 | -1.157 | 0 | 0.3 |
| 40906 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 25 | CDHR3 | Sponge network | -0.129 | 0.57177 | -0.977 | 4.0E-5 | 0.3 |
| 40907 | CTC-296K1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-744-5p;hsa-miR-940 | 16 | NOTCH2 | Sponge network | -2.095 | 0.00112 | 0.138 | 0.35163 | 0.3 |
| 40908 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 53 | PTEN | Sponge network | -0.487 | 0.02157 | -0.187 | 0.07748 | 0.3 |
| 40909 | RP11-6O2.3 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-495-3p | 13 | DICER1 | Sponge network | 0.331 | 0.57247 | 0.042 | 0.674 | 0.3 |
| 40910 | LINC00886 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-940 | 49 | CADM2 | Sponge network | -0.371 | 0.24604 | -3.782 | 0 | 0.3 |
| 40911 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-877-5p | 21 | RBMS3 | Sponge network | 0.594 | 0.00064 | -0.519 | 0.03551 | 0.3 |
| 40912 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 31 | FZD3 | Sponge network | 1.153 | 0 | 0.104 | 0.60316 | 0.3 |
| 40913 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 45 | CCND2 | Sponge network | 0.335 | 0.20596 | -0.267 | 0.3112 | 0.3 |
| 40914 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 52 | SMAD4 | Sponge network | -0.83 | 0 | -0.313 | 0.00413 | 0.3 |
| 40915 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NOTCH2NL | Sponge network | 0.537 | 0.00139 | 0.024 | 0.84307 | 0.3 |
| 40916 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-664a-3p | 29 | KIF1B | Sponge network | 0.917 | 0 | -0.118 | 0.32258 | 0.3 |
| 40917 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | CXXC4 | Sponge network | 1.302 | 7.0E-5 | -0.433 | 0.21055 | 0.3 |
| 40918 | RAB30-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p | 12 | ABI2 | Sponge network | 0.752 | 0 | 0.175 | 0.14787 | 0.3 |
| 40919 | LINC00476 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 48 | CUX1 | Sponge network | -0.615 | 0.00015 | -0.039 | 0.6705 | 0.3 |
| 40920 | LINC00865 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-935 | 24 | RNF144A | Sponge network | -1.249 | 7.0E-5 | 0.594 | 0.0009 | 0.3 |
| 40921 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-324-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-589-3p | 22 | PPM1L | Sponge network | 0.529 | 0.00552 | -0.537 | 0.00616 | 0.3 |
| 40922 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 14 | UBE4B | Sponge network | -0.612 | 0.00094 | -0.235 | 0.007 | 0.3 |
| 40923 | LINC00662 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PLSCR4 | Sponge network | 0.213 | 0.32297 | -0.474 | 0.03833 | 0.3 |
| 40924 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-340-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-940 | 54 | NFIB | Sponge network | -0.096 | 0.67958 | 0.023 | 0.90795 | 0.3 |
| 40925 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-629-3p | 32 | KIF1B | Sponge network | 1.04 | 0.00023 | -0.118 | 0.32258 | 0.3 |
| 40926 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 0.919 | 0 | 0.322 | 0.00215 | 0.3 |
| 40927 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-501-5p | 13 | MAFB | Sponge network | 0.619 | 0.00157 | -0.023 | 0.89865 | 0.3 |
| 40928 | RP11-275I14.4 | hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-9-5p | 12 | CCDC6 | Sponge network | 2.165 | 4.0E-5 | 0.27 | 0.02555 | 0.3 |
| 40929 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | TMTC3 | Sponge network | 1.205 | 0 | 0.123 | 0.22189 | 0.3 |
| 40930 | GNG12-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 13 | UNC5C | Sponge network | -0.793 | 7.0E-5 | -0.178 | 0.40214 | 0.3 |
| 40931 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-590-5p | 12 | ZNF208 | Sponge network | -0.384 | 0.40652 | -0.4 | 0.22381 | 0.3 |
| 40932 | LINC00094 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 12 | PPP2R2C | Sponge network | 0.253 | 0.09565 | -0.959 | 0.07428 | 0.3 |
| 40933 | AP001347.6 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | SYT4 | Sponge network | -1.161 | 0.00591 | -3.207 | 0 | 0.3 |
| 40934 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p | 23 | SASH1 | Sponge network | -0.693 | 0.05629 | -0.502 | 0.00175 | 0.3 |
| 40935 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | PAFAH1B1 | Sponge network | 0.537 | 0.00139 | -0.429 | 0 | 0.3 |
| 40936 | RASSF8-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 18 | PRKAR1A | Sponge network | 0.335 | 0.20596 | -0.063 | 0.50314 | 0.3 |
| 40937 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | NOTCH2NL | Sponge network | 0.997 | 0.00066 | 0.024 | 0.84307 | 0.3 |
| 40938 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-940;hsa-miR-96-5p | 13 | CRTC3 | Sponge network | 0.622 | 0.00015 | 0.164 | 0.16786 | 0.3 |
| 40939 | RP1-39G22.7 |
hsa-miR-101-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 13 | UBR3 | Sponge network | 0.439 | 0.00313 | -0.021 | 0.82774 | 0.3 |
| 40940 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p | 11 | RCAN2 | Sponge network | 0.274 | 0.07681 | -0.33 | 0.15172 | 0.3 |
| 40941 | DIO3OS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p | 16 | RELN | Sponge network | -0.773 | 0.04073 | -2.403 | 0 | 0.3 |
| 40942 | H1FX-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TLL1 | Sponge network | -0.206 | 0.12942 | -1.195 | 3.0E-5 | 0.3 |
| 40943 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-550a-5p | 11 | HOOK3 | Sponge network | -0.611 | 0.09607 | 0.273 | 0.09957 | 0.3 |
| 40944 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-577 | 24 | IGF1 | Sponge network | 0.853 | 0.15352 | -0.204 | 0.5898 | 0.3 |
| 40945 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p | 20 | CDH19 | Sponge network | 0.082 | 0.55654 | -3.434 | 0 | 0.3 |
| 40946 | RP11-20I23.13 |
hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-486-5p | 12 | MYO9A | Sponge network | 0.505 | 0.0014 | -0.147 | 0.32373 | 0.3 |
| 40947 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p | 25 | JAZF1 | Sponge network | -0.031 | 0.89063 | -0.377 | 0.02677 | 0.3 |
| 40948 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | ANK2 | Sponge network | -0.176 | 0.59692 | -1.603 | 1.0E-5 | 0.3 |
| 40949 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | USP6 | Sponge network | 1.063 | 0 | 0.188 | 0.3871 | 0.3 |
| 40950 | CTD-3157E16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-942-5p | 10 | TMEM132B | Sponge network | -0.326 | 0.28809 | -0.83 | 0.01084 | 0.3 |
| 40951 | CTD-2303H24.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ZMYND11 | Sponge network | 0.415 | 0.14657 | -0.2 | 0.05449 | 0.3 |
| 40952 | AGAP11 |
hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 15 | HERC1 | Sponge network | -1.645 | 0 | -0.196 | 0.08544 | 0.3 |
| 40953 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | ZNF770 | Sponge network | 0.848 | 0 | 0.206 | 0.06751 | 0.3 |
| 40954 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-495-3p | 12 | ATRX | Sponge network | 1.056 | 0 | 0.261 | 0.04408 | 0.3 |
| 40955 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-744-3p | 37 | LPP | Sponge network | 1.04 | 0.00023 | -0.459 | 0.04475 | 0.3 |
| 40956 | CTD-3025N20.3 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-30a-3p | 12 | TBL1XR1 | Sponge network | 1.046 | 0 | 0.578 | 0 | 0.3 |
| 40957 | RP11-34P13.14 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p | 12 | CCDC6 | Sponge network | 1.888 | 0 | 0.27 | 0.02555 | 0.3 |
| 40958 | TRIM52-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-577 | 14 | ZBTB4 | Sponge network | -0.418 | 0.00068 | -0.739 | 0 | 0.3 |
| 40959 | FAM66C |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | CSRNP1 | Sponge network | -0.901 | 0.00019 | -1.448 | 0 | 0.3 |
| 40960 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | CREBBP | Sponge network | 0.603 | 0.00127 | 0.126 | 0.15186 | 0.3 |
| 40961 | CTD-2336O2.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PRRX1 | Sponge network | -0.141 | 0.26434 | 1.316 | 0 | 0.3 |
| 40962 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-744-3p;hsa-miR-93-5p | 22 | HLF | Sponge network | -0.785 | 0.00035 | -2.443 | 0 | 0.3 |
| 40963 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p;hsa-miR-942-5p | 18 | GPC6 | Sponge network | -0.318 | 0.65847 | -0.054 | 0.81465 | 0.3 |
| 40964 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 22 | TP53INP1 | Sponge network | -1.281 | 0.00012 | -0.496 | 0.00064 | 0.3 |
| 40965 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p | 29 | AKAP11 | Sponge network | 0.95 | 0.15943 | 0.196 | 0.09825 | 0.3 |
| 40966 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p | 13 | MYCBP2 | Sponge network | 0.214 | 0.18246 | -0.027 | 0.82016 | 0.3 |
| 40967 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30c-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 19 | AFF4 | Sponge network | 0.537 | 0.0006 | -0.266 | 0.01565 | 0.3 |
| 40968 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | LIFR | Sponge network | -0.737 | 0.10822 | -1.794 | 0 | 0.3 |
| 40969 | HOXA-AS3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 36 | DST | Sponge network | -0.555 | 0.06156 | -0.713 | 0.00096 | 0.3 |
| 40970 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 32 | HECA | Sponge network | -2.46 | 0 | -0.217 | 0.03463 | 0.3 |
| 40971 | PART1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 21 | CDH23 | Sponge network | -2.925 | 0 | -0.974 | 0.00034 | 0.3 |
| 40972 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p | 36 | SMAD2 | Sponge network | -1.645 | 0 | -0.128 | 0.12732 | 0.3 |
| 40973 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | BCL11A | Sponge network | -1.439 | 4.0E-5 | -0.932 | 0.0063 | 0.3 |
| 40974 | RP11-439A17.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-576-5p | 12 | TRPS1 | Sponge network | 0.051 | 0.95756 | -0.191 | 0.44109 | 0.3 |
| 40975 | FAM13A-AS1 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | PCDH9 | Sponge network | -0.83 | 0 | -1.823 | 4.0E-5 | 0.3 |
| 40976 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-493-5p;hsa-miR-93-3p | 17 | RUNX1T1 | Sponge network | 1.46 | 1.0E-5 | -0.344 | 0.2374 | 0.3 |
| 40977 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-93-5p | 10 | FBXO31 | Sponge network | -1.349 | 0.00017 | -0.085 | 0.42389 | 0.3 |
| 40978 | RP11-327P2.5 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-361-3p;hsa-miR-625-5p | 10 | BCORL1 | Sponge network | -0.147 | 0.40452 | 0.354 | 0.01214 | 0.3 |
| 40979 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | WWTR1 | Sponge network | -1.057 | 0.00029 | -0.345 | 0.11997 | 0.3 |
| 40980 | AC005154.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p;hsa-miR-655-3p | 14 | ARID2 | Sponge network | 0.848 | 0 | 0.235 | 0.02697 | 0.3 |
| 40981 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 28 | EPHA5 | Sponge network | -0.465 | 0.04703 | -0.957 | 0.01733 | 0.3 |
| 40982 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 54 | AFF4 | Sponge network | 0.385 | 0.10067 | -0.266 | 0.01565 | 0.3 |
| 40983 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 63 | THRB | Sponge network | -1.216 | 0 | -1.117 | 0.0004 | 0.3 |
| 40984 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p | 25 | CBL | Sponge network | 0.056 | 0.81195 | 0.291 | 0.01267 | 0.3 |
| 40985 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-214-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-589-5p | 34 | SMAD4 | Sponge network | 0.001 | 0.99753 | -0.313 | 0.00413 | 0.3 |
| 40986 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-93-5p | 15 | MAP3K12 | Sponge network | 0.249 | 0.35902 | -0.057 | 0.59369 | 0.3 |
| 40987 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-500a-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CSMD1 | Sponge network | 0.238 | 0.70544 | -0.63 | 0.18881 | 0.3 |
| 40988 | RP11-48B3.4 |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 10 | ZMYND11 | Sponge network | 1.757 | 0 | -0.2 | 0.05449 | 0.3 |
| 40989 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 24 | KIT | Sponge network | -0.222 | 0.32445 | -2.184 | 0 | 0.3 |
| 40990 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | NIN | Sponge network | -1.349 | 0.00017 | -0.253 | 0.13794 | 0.3 |
| 40991 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-340-5p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-940 | 26 | NOTCH2 | Sponge network | 0.331 | 0.57247 | 0.138 | 0.35163 | 0.3 |
| 40992 | FZD10-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-590-3p | 13 | PAK3 | Sponge network | -0.727 | 0.04726 | -0.628 | 0.07253 | 0.3 |
| 40993 | RP11-6N17.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p | 10 | IL17RD | Sponge network | 0.108 | 0.69556 | -0.019 | 0.94873 | 0.3 |
| 40994 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-454-3p | 13 | SPG20 | Sponge network | 0.853 | 0.15352 | -1.16 | 0 | 0.3 |
| 40995 | RP11-166D19.1 |
hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | PACRG | Sponge network | -0.576 | 0.10121 | -2.248 | 0 | 0.3 |
| 40996 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 19 | USP6 | Sponge network | 1.757 | 0 | 0.188 | 0.3871 | 0.3 |
| 40997 | LINC00654 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | HERC1 | Sponge network | 0.374 | 0.20054 | -0.196 | 0.08544 | 0.3 |
| 40998 | AC009948.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | CSMD1 | Sponge network | 0.532 | 0.00505 | -0.63 | 0.18881 | 0.3 |
| 40999 | RP11-981G7.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-378c;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | BRD3 | Sponge network | 0.986 | 0.01022 | 0.37 | 0.00013 | 0.3 |
| 41000 | RP11-701H24.3 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | NTRK2 | Sponge network | -1.425 | 0.04204 | -0.736 | 0.06495 | 0.3 |
| 41001 | RP11-156E6.1 |
hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p | 11 | PAWR | Sponge network | 1.266 | 0 | 0.636 | 0 | 0.3 |
| 41002 | MIR600HG |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p | 10 | DDX5 | Sponge network | 0.594 | 0.41522 | -0.099 | 0.17428 | 0.3 |
| 41003 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p | 23 | NTRK2 | Sponge network | -0.469 | 0.08721 | -0.736 | 0.06495 | 0.3 |
| 41004 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | 0.253 | 0.09565 | -0.354 | 0.14694 | 0.3 |
| 41005 | RP11-967K21.1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-92a-3p | 17 | ATM | Sponge network | 0.056 | 0.81195 | 0.178 | 0.21568 | 0.3 |
| 41006 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-374b-3p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-942-5p | 33 | UNC80 | Sponge network | -0.611 | 0.09607 | -1.934 | 0 | 0.3 |
| 41007 | PCA3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 44 | PTPRD | Sponge network | -0.709 | 0.18168 | -1.005 | 0.00064 | 0.3 |
| 41008 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 20 | PRKAA1 | Sponge network | 1.147 | 0 | 0.317 | 0.0031 | 0.3 |
| 41009 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | EGR2 | Sponge network | -2.452 | 0 | 0.309 | 0.17079 | 0.3 |
| 41010 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-93-3p;hsa-miR-940 | 44 | QKI | Sponge network | -1.047 | 0 | -0.333 | 0.04111 | 0.3 |
| 41011 | AC127904.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | BRD3 | Sponge network | 1.308 | 0 | 0.37 | 0.00013 | 0.3 |
| 41012 | RP11-490M8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-33a-5p;hsa-miR-539-5p;hsa-miR-96-5p | 11 | SETBP1 | Sponge network | -0.214 | 0.44248 | -1.302 | 1.0E-5 | 0.3 |
| 41013 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-590-3p | 10 | SLITRK3 | Sponge network | 0.178 | 0.29106 | -3.328 | 0 | 0.3 |
| 41014 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-589-5p | 16 | ZFHX3 | Sponge network | 0.001 | 0.99753 | 0.389 | 0.01363 | 0.3 |
| 41015 | RP11-999E24.3 |
hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-942-5p | 12 | ERRFI1 | Sponge network | -0.693 | 0.05629 | -0.798 | 1.0E-5 | 0.3 |
| 41016 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | SUFU | Sponge network | 0.083 | 0.66405 | -0.04 | 0.65489 | 0.3 |
| 41017 | RP11-1348G14.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-495-3p;hsa-miR-7-1-3p | 13 | PHC3 | Sponge network | 0.597 | 0.00037 | 0.212 | 0.0499 | 0.3 |
| 41018 | RP11-440L14.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-375;hsa-miR-590-3p | 11 | ABI2 | Sponge network | 0.545 | 0.05789 | 0.175 | 0.14787 | 0.3 |
| 41019 | CTC-378H22.2 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p | 10 | BNC2 | Sponge network | 0.001 | 0.99663 | -0.491 | 0.09988 | 0.3 |
| 41020 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | EYA4 | Sponge network | 0.272 | 0.44692 | 0.3 | 0.36876 | 0.3 |
| 41021 | RP11-85G18.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29b-2-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-5p | 10 | BRD3 | Sponge network | 3.073 | 0 | 0.37 | 0.00013 | 0.3 |
| 41022 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NEDD4 | Sponge network | 1.378 | 0 | 0.02 | 0.89834 | 0.3 |
| 41023 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 12 | PTPN11 | Sponge network | 1.308 | 0 | 0.431 | 2.0E-5 | 0.3 |
| 41024 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 48 | WDFY3 | Sponge network | -0.769 | 0.00035 | -0.201 | 0.0967 | 0.3 |
| 41025 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-505-3p | 15 | USP6 | Sponge network | 0.622 | 0.00015 | 0.188 | 0.3871 | 0.3 |
| 41026 | HCG11 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | KLF10 | Sponge network | 0.385 | 0.10067 | -0.783 | 0 | 0.3 |
| 41027 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | SOX5 | Sponge network | 0.395 | 0.15451 | -1.091 | 4.0E-5 | 0.3 |
| 41028 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | PDGFRA | Sponge network | -0.08 | 0.90092 | -0.372 | 0.0037 | 0.3 |
| 41029 | MIR600HG |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-589-3p;hsa-miR-877-5p | 20 | AFF1 | Sponge network | 0.594 | 0.41522 | -0.384 | 0.00053 | 0.3 |
| 41030 | RP11-251G23.5 |
hsa-miR-125a-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-411-5p | 10 | RAD23B | Sponge network | 1.344 | 0 | 0.108 | 0.17414 | 0.3 |
| 41031 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | PCDH8 | Sponge network | -0.235 | 0.15142 | -0.997 | 0.05366 | 0.3 |
| 41032 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | ABCA10 | Sponge network | 0.349 | 0.01695 | -0.571 | 0.03674 | 0.3 |
| 41033 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 38 | ERBB4 | Sponge network | -0.405 | 0.22013 | -1.975 | 2.0E-5 | 0.3 |
| 41034 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | AKAP11 | Sponge network | 0.986 | 0.01022 | 0.196 | 0.09825 | 0.3 |
| 41035 | C4B-AS1 |
hsa-miR-107;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-629-3p | 11 | GPC6 | Sponge network | 2.306 | 9.0E-5 | -0.054 | 0.81465 | 0.3 |
| 41036 | CTA-14H9.5 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | QKI | Sponge network | 0.145 | 0.53343 | -0.333 | 0.04111 | 0.3 |
| 41037 | RP3-467N11.1 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-497-5p | 10 | RICTOR | Sponge network | 0.953 | 0 | 0.388 | 0.00188 | 0.3 |
| 41038 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 29 | LRIG1 | Sponge network | -0.969 | 2.0E-5 | -0.537 | 0.00588 | 0.3 |
| 41039 | RP11-526I2.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 13 | TBL1XR1 | Sponge network | 1.054 | 1.0E-5 | 0.578 | 0 | 0.3 |
| 41040 | PWAR6 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | OPCML | Sponge network | -1.575 | 6.0E-5 | -2.498 | 0 | 0.3 |
| 41041 | RP11-597D13.9 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | KLF10 | Sponge network | 1.302 | 7.0E-5 | -0.783 | 0 | 0.3 |
| 41042 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | FAT4 | Sponge network | 0.174 | 0.30034 | -0.354 | 0.14694 | 0.3 |
| 41043 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p | 13 | PCDH8 | Sponge network | 0.225 | 0.30644 | -0.997 | 0.05366 | 0.3 |
| 41044 | TRIM52-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-501-5p | 10 | TXNIP | Sponge network | -0.418 | 0.00068 | -1.277 | 0 | 0.3 |
| 41045 | PSMG3-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 16 | SPG20 | Sponge network | -0.125 | 0.49414 | -1.16 | 0 | 0.3 |
| 41046 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-222-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | NTRK3 | Sponge network | -0.011 | 0.967 | -1.77 | 3.0E-5 | 0.3 |
| 41047 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p | 14 | MYCBP2 | Sponge network | 0.082 | 0.55654 | -0.027 | 0.82016 | 0.3 |
| 41048 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429 | 21 | UNC80 | Sponge network | -0.469 | 0.08721 | -1.934 | 0 | 0.3 |
| 41049 | HOXA-AS2 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH11 | Sponge network | 0.61 | 0.01593 | 1.427 | 0 | 0.3 |
| 41050 | GAS6-AS2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | TAL1 | Sponge network | 0.16 | 0.50785 | -0.462 | 0.00574 | 0.3 |
| 41051 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 24 | MAF | Sponge network | -1.057 | 0.00029 | -1.257 | 0 | 0.3 |
| 41052 | IGF2-AS |
hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-500a-3p;hsa-miR-660-5p | 17 | RUNX1T1 | Sponge network | 3.339 | 0.00237 | -0.344 | 0.2374 | 0.3 |
| 41053 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-582-5p | 13 | PRKAA1 | Sponge network | 0.8 | 0 | 0.317 | 0.0031 | 0.3 |
| 41054 | DNM1P35 |
hsa-let-7a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-766-3p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | 0.051 | 0.83388 | -0.564 | 1.0E-5 | 0.3 |
| 41055 | TRIM52-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | RASSF8 | Sponge network | -0.418 | 0.00068 | -0.122 | 0.65591 | 0.3 |
| 41056 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p | 13 | JAKMIP2 | Sponge network | 0.374 | 0.20054 | -1.388 | 0 | 0.3 |
| 41057 | RP11-968O1.5 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p | 15 | NRXN1 | Sponge network | -0.829 | 0.01343 | -3.778 | 0 | 0.3 |
| 41058 | LINC00472 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p | 16 | CITED2 | Sponge network | -0.842 | 0.01361 | -1.545 | 0 | 0.3 |
| 41059 | IQCH-AS1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-495-3p | 11 | ZNF770 | Sponge network | 0.929 | 0 | 0.206 | 0.06751 | 0.3 |
| 41060 | SH3BP5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 29 | ZFX | Sponge network | 0.267 | 0.2937 | 0.09 | 0.42563 | 0.3 |
| 41061 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p | 18 | KANK1 | Sponge network | 0.537 | 0.00139 | -0.55 | 0.00134 | 0.3 |
| 41062 | MRPL23-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-503-5p | 18 | RICTOR | Sponge network | -0.278 | 0.76559 | 0.388 | 0.00188 | 0.3 |
| 41063 | RP11-389C8.2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TET1 | Sponge network | 0.727 | 3.0E-5 | -0.135 | 0.52138 | 0.3 |
| 41064 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-539-5p | 12 | MOB1B | Sponge network | 1.597 | 0 | 0.197 | 0.15266 | 0.3 |
| 41065 | LINC00865 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | SPOP | Sponge network | -1.249 | 7.0E-5 | -0.419 | 1.0E-5 | 0.3 |
| 41066 | RP11-672A2.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-766-3p | 13 | EZH1 | Sponge network | 1.344 | 0 | -0.564 | 1.0E-5 | 0.3 |
| 41067 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-5p;hsa-miR-511-5p;hsa-miR-576-5p | 13 | ATRX | Sponge network | 0.48 | 0.05578 | 0.261 | 0.04408 | 0.3 |
| 41068 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 10 | BCLAF1 | Sponge network | 0.986 | 0.01022 | 0.211 | 0.00684 | 0.3 |
| 41069 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-589-5p;hsa-miR-98-5p | 10 | ST5 | Sponge network | -0.154 | 0.78939 | -0.78 | 0 | 0.3 |
| 41070 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-93-5p | 27 | PGR | Sponge network | 0.253 | 0.09565 | -1.704 | 0 | 0.3 |
| 41071 | U91328.21 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-320b | 15 | FAT3 | Sponge network | -0.735 | 0.15983 | -1.601 | 0.00051 | 0.3 |
| 41072 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p | 11 | PDS5B | Sponge network | 0.411 | 0.1335 | 0.259 | 0.00962 | 0.3 |
| 41073 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 48 | TCF4 | Sponge network | 0.559 | 0.00026 | 0.016 | 0.92271 | 0.3 |
| 41074 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p | 12 | FLI1 | Sponge network | -0.487 | 0.02157 | -0.294 | 0.10905 | 0.3 |
| 41075 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-503-5p;hsa-miR-576-5p;hsa-miR-582-5p | 28 | RICTOR | Sponge network | 0.95 | 0.15943 | 0.388 | 0.00188 | 0.3 |
| 41076 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | MLLT10 | Sponge network | 0.796 | 4.0E-5 | 0.105 | 0.33292 | 0.3 |
| 41077 | KB-1460A1.5 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p | 46 | MDM4 | Sponge network | 0.603 | 0.00127 | 0.345 | 0.00762 | 0.299 |
| 41078 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-625-3p | 23 | CBX5 | Sponge network | 1.317 | 0 | 0.165 | 0.24446 | 0.299 |
| 41079 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-362-3p | 21 | BMPR2 | Sponge network | -1.255 | 0.00981 | 0.206 | 0.0635 | 0.299 |
| 41080 | RP11-25K21.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-374a-3p;hsa-miR-589-3p | 15 | MCC | Sponge network | 0.489 | 0.25786 | -1.005 | 0 | 0.299 |
| 41081 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | TIMP3 | Sponge network | -0.969 | 2.0E-5 | -0.546 | 0.02307 | 0.299 |
| 41082 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-505-5p;hsa-miR-935 | 21 | HBP1 | Sponge network | -2.62 | 0 | -0.454 | 0 | 0.299 |
| 41083 | OIP5-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-5p | 13 | THSD7A | Sponge network | 0.421 | 0.00084 | 0.242 | 0.25154 | 0.299 |
| 41084 | RP11-244H3.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-624-5p | 11 | SCN11A | Sponge network | 0.659 | 3.0E-5 | -1.15 | 0.00299 | 0.299 |
| 41085 | AC018890.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | NAV3 | Sponge network | 1.61 | 0.00961 | 0.212 | 0.47729 | 0.299 |
| 41086 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-455-3p;hsa-miR-96-5p | 12 | CRTC3 | Sponge network | -0.418 | 0.00068 | 0.164 | 0.16786 | 0.299 |
| 41087 | AC006547.13 |
hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p | 10 | BRD3 | Sponge network | 1.159 | 0 | 0.37 | 0.00013 | 0.299 |
| 41088 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-577 | 19 | EFNA5 | Sponge network | -2.69 | 0.0001 | -0.421 | 0.17868 | 0.299 |
| 41089 | HCG11 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 13 | PKNOX1 | Sponge network | 0.385 | 0.10067 | 0.085 | 0.28917 | 0.299 |
| 41090 | ZNF582-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-577 | 13 | CSDE1 | Sponge network | -1.349 | 0.00017 | -0.493 | 0 | 0.299 |
| 41091 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-7-1-3p | 18 | KAT6B | Sponge network | 0.267 | 0.2937 | -0.478 | 0.00018 | 0.299 |
| 41092 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-505-5p;hsa-miR-590-3p | 11 | RBM5 | Sponge network | -0.83 | 0 | -0.205 | 0.0129 | 0.299 |
| 41093 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 22 | EPHA3 | Sponge network | -0.176 | 0.59692 | 0.185 | 0.53588 | 0.299 |
| 41094 | RP11-248G5.8 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | UBR3 | Sponge network | 1.294 | 0 | -0.021 | 0.82774 | 0.299 |
| 41095 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | MTSS1 | Sponge network | -0.663 | 0.10887 | -0.81 | 0.00035 | 0.299 |
| 41096 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 15 | NR4A3 | Sponge network | 0.194 | 0.64278 | -1.385 | 0 | 0.299 |
| 41097 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | ABCC9 | Sponge network | -0.969 | 2.0E-5 | -0.466 | 0.14121 | 0.299 |
| 41098 | RP11-67L2.2 |
hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 17 | GLI3 | Sponge network | 0.72 | 0.0001 | -0.615 | 0.01482 | 0.299 |
| 41099 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ITGAV | Sponge network | -0.612 | 0.00094 | 0.337 | 0.01002 | 0.299 |
| 41100 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 21 | PAFAH1B1 | Sponge network | 0.529 | 0.00552 | -0.429 | 0 | 0.299 |
| 41101 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | HACE1 | Sponge network | 0.064 | 0.72934 | -0.072 | 0.55239 | 0.299 |
| 41102 | RP1-151F17.2 |
hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | AFAP1L2 | Sponge network | 0.222 | 0.29133 | -0.284 | 0.15434 | 0.299 |
| 41103 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-7-5p | 23 | MOB1B | Sponge network | -0.693 | 0.05629 | 0.197 | 0.15266 | 0.299 |
| 41104 | RP11-597D13.9 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | TAL1 | Sponge network | 1.302 | 7.0E-5 | -0.462 | 0.00574 | 0.299 |
| 41105 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-93-5p | 19 | TP53INP1 | Sponge network | -0.611 | 0.09607 | -0.496 | 0.00064 | 0.299 |
| 41106 | CTC-297N7.9 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-34a-5p;hsa-miR-877-5p | 10 | ANK2 | Sponge network | -2.069 | 0 | -1.603 | 1.0E-5 | 0.299 |
| 41107 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-370-3p;hsa-miR-625-5p | 19 | JAZF1 | Sponge network | -4.477 | 0 | -0.377 | 0.02677 | 0.299 |
| 41108 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-98-5p | 14 | ITGB3 | Sponge network | -0.727 | 0.04726 | -0.011 | 0.95751 | 0.299 |
| 41109 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | 1.302 | 7.0E-5 | -0.974 | 0.00034 | 0.299 |
| 41110 | CTD-2002H8.2 |
hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TACC1 | Sponge network | 0.931 | 1.0E-5 | -0.362 | 0.05406 | 0.299 |
| 41111 | BOLA3-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429 | 12 | FMNL3 | Sponge network | -0.246 | 0.35034 | 0.555 | 4.0E-5 | 0.299 |
| 41112 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SON | Sponge network | -0.323 | 0.01372 | 0.168 | 0.04406 | 0.299 |
| 41113 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PICALM | Sponge network | 0.349 | 0.01695 | 0.086 | 0.23523 | 0.299 |
| 41114 | RP11-360F5.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | DIP2C | Sponge network | 1.867 | 0 | -0.436 | 0.01713 | 0.299 |
| 41115 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-500a-5p | 13 | RUNX1T1 | Sponge network | -0.011 | 0.967 | -0.344 | 0.2374 | 0.299 |
| 41116 | AP001347.6 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | -1.161 | 0.00591 | 0.321 | 0.00267 | 0.299 |
| 41117 | MEG3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-5p | 10 | ARNT | Sponge network | 0.249 | 0.35902 | -0.08 | 0.26073 | 0.299 |
| 41118 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b | 13 | CLIP1 | Sponge network | 0.082 | 0.55397 | -0.215 | 0.08684 | 0.299 |
| 41119 | RP5-1198O20.4 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-3p | 15 | TMTC2 | Sponge network | 0.997 | 0.00066 | -0.215 | 0.18248 | 0.299 |
| 41120 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-744-5p | 22 | USP12 | Sponge network | -0.147 | 0.40452 | -0.102 | 0.40768 | 0.299 |
| 41121 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-374a-5p | 10 | PNRC1 | Sponge network | -0.693 | 0.05629 | -0.646 | 0 | 0.299 |
| 41122 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-421 | 10 | SLC6A15 | Sponge network | -0.583 | 0.14589 | -2.373 | 2.0E-5 | 0.299 |
| 41123 | LINC00886 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-93-5p | 16 | PRUNE2 | Sponge network | -0.371 | 0.24604 | -1.733 | 0.00022 | 0.299 |
| 41124 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | RB1CC1 | Sponge network | 0.862 | 0.06213 | 0.175 | 0.12559 | 0.299 |
| 41125 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | RBMS3 | Sponge network | 0.199 | 0.52763 | -0.519 | 0.03551 | 0.299 |
| 41126 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 40 | CBX5 | Sponge network | 0.853 | 0.15352 | 0.165 | 0.24446 | 0.299 |
| 41127 | CTC-297N7.9 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p | 20 | RBMS3 | Sponge network | -2.069 | 0 | -0.519 | 0.03551 | 0.299 |
| 41128 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 45 | THRB | Sponge network | -0.737 | 0.10822 | -1.117 | 0.0004 | 0.299 |
| 41129 | HOTAIRM1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | ETV1 | Sponge network | -0.053 | 0.80685 | -0.096 | 0.67674 | 0.299 |
| 41130 | RP11-439L18.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-454-3p | 11 | SASH1 | Sponge network | 0.224 | 0.48719 | -0.502 | 0.00175 | 0.299 |
| 41131 | TP73-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935 | 26 | OPCML | Sponge network | -0.563 | 0.02545 | -2.498 | 0 | 0.299 |
| 41132 | RP11-344B5.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-423-5p | 11 | CBFA2T3 | Sponge network | -0.055 | 0.88482 | -1.221 | 1.0E-5 | 0.299 |
| 41133 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-940 | 36 | CHD5 | Sponge network | 1.302 | 7.0E-5 | -0.922 | 0.01521 | 0.299 |
| 41134 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 19 | RBL2 | Sponge network | 0.997 | 0.00066 | -0.282 | 0.00254 | 0.299 |
| 41135 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 24 | TACC1 | Sponge network | 0.921 | 0.00118 | -0.362 | 0.05406 | 0.299 |
| 41136 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-629-3p | 18 | USP12 | Sponge network | 0.368 | 0.20093 | -0.102 | 0.40768 | 0.299 |
| 41137 | RP11-116O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | IGSF10 | Sponge network | 0.05 | 0.95483 | -1.751 | 1.0E-5 | 0.299 |
| 41138 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-874-3p;hsa-miR-96-5p | 23 | TACC1 | Sponge network | -0.326 | 0.14314 | -0.362 | 0.05406 | 0.299 |
| 41139 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | IGSF10 | Sponge network | -0.726 | 0.00132 | -1.751 | 1.0E-5 | 0.299 |
| 41140 | CTD-2017D11.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484 | 14 | ZFX | Sponge network | 0.792 | 0.0049 | 0.09 | 0.42563 | 0.299 |
| 41141 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-425-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 39 | ADARB1 | Sponge network | 1.302 | 7.0E-5 | -0.335 | 0.06631 | 0.299 |
| 41142 | JAZF1-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | LIMCH1 | Sponge network | -0.769 | 0.00035 | -0.915 | 1.0E-5 | 0.299 |
| 41143 | OSER1-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 13 | PLAG1 | Sponge network | -0.13 | 0.48734 | -0.315 | 0.26532 | 0.299 |
| 41144 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | RET | Sponge network | -0.355 | 0.0077 | -0.833 | 0.03517 | 0.299 |
| 41145 | RP11-968O1.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-532-5p | 25 | BMPR2 | Sponge network | -0.829 | 0.01343 | 0.206 | 0.0635 | 0.299 |
| 41146 | RP11-21A7A.2 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-5p | 12 | PRKCB | Sponge network | -1.116 | 0.23686 | -1.313 | 2.0E-5 | 0.299 |
| 41147 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 41 | TMEM132B | Sponge network | 0.72 | 0.0001 | -0.83 | 0.01084 | 0.299 |
| 41148 | LINC00094 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 25 | PCDH9 | Sponge network | 0.253 | 0.09565 | -1.823 | 4.0E-5 | 0.299 |
| 41149 | GAS6-AS2 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-942-5p | 13 | GLIPR1 | Sponge network | 0.16 | 0.50785 | 0.146 | 0.3276 | 0.299 |
| 41150 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 22 | YAP1 | Sponge network | 0.488 | 0.00084 | 0.22 | 0.11836 | 0.299 |
| 41151 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | CDH10 | Sponge network | -0.096 | 0.67958 | -2.397 | 0 | 0.299 |
| 41152 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 28 | PRKCE | Sponge network | -0.942 | 0 | -0.403 | 0.00056 | 0.299 |
| 41153 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-455-5p | 11 | GALNT13 | Sponge network | -2.69 | 0.0001 | -0.857 | 0.07439 | 0.299 |
| 41154 | RP11-276H19.2 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-96-5p | 14 | SOX5 | Sponge network | -0.472 | 0.48851 | -1.091 | 4.0E-5 | 0.299 |
| 41155 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429 | 15 | LATS1 | Sponge network | 0.056 | 0.81195 | 0.109 | 0.34208 | 0.299 |
| 41156 | AC141928.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NAV3 | Sponge network | 0.853 | 0.15352 | 0.212 | 0.47729 | 0.299 |
| 41157 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p | 22 | RASSF2 | Sponge network | -0.693 | 0.05629 | -0.273 | 0.21961 | 0.299 |
| 41158 | DNM1P35 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 13 | SORCS1 | Sponge network | 0.051 | 0.83388 | -3.492 | 0 | 0.299 |
| 41159 | RP11-479G22.8 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p | 13 | PTPN14 | Sponge network | 1.308 | 0 | 0.144 | 0.34595 | 0.299 |
| 41160 | RP11-574K11.29 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-495-3p | 13 | ZFX | Sponge network | 0.997 | 0 | 0.09 | 0.42563 | 0.299 |
| 41161 | NEAT1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-940 | 42 | AFF4 | Sponge network | 0.705 | 0.00014 | -0.266 | 0.01565 | 0.299 |
| 41162 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p | 11 | KCNAB1 | Sponge network | 1.089 | 0 | -0.755 | 2.0E-5 | 0.299 |
| 41163 | CTC-378H22.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-331-3p | 15 | EPHA7 | Sponge network | 0.001 | 0.99663 | -2.197 | 3.0E-5 | 0.299 |
| 41164 | RP11-2H8.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p | 27 | ARHGEF12 | Sponge network | 1.089 | 0 | 0.09 | 0.35693 | 0.299 |
| 41165 | RP11-693J15.4 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b | 12 | SYNM | Sponge network | -0.72 | 0.24239 | -2.309 | 1.0E-5 | 0.299 |
| 41166 | RP5-1139B12.3 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 10 | GJA1 | Sponge network | 0.598 | 0.38481 | -0.485 | 0.02179 | 0.299 |
| 41167 | CTD-3064M3.3 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-539-5p | 11 | RIMS2 | Sponge network | -0.469 | 0.08721 | -1.365 | 0.00208 | 0.299 |
| 41168 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 34 | HBP1 | Sponge network | -0.969 | 2.0E-5 | -0.454 | 0 | 0.299 |
| 41169 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | 0.541 | 0.00125 | 0.206 | 0.06751 | 0.299 |
| 41170 | RP1-151F17.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | ERCC4 | Sponge network | 0.222 | 0.29133 | 0.126 | 0.15266 | 0.299 |
| 41171 | SH3BP5-AS1 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 10 | MAP3K4 | Sponge network | 0.267 | 0.2937 | 0.058 | 0.54229 | 0.299 |
| 41172 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-625-5p | 20 | YAP1 | Sponge network | 0.056 | 0.81195 | 0.22 | 0.11836 | 0.299 |
| 41173 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | GRIN2A | Sponge network | -0.223 | 0.47816 | -2.161 | 0 | 0.299 |
| 41174 | RP11-535M15.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-330-3p | 11 | CREB3L2 | Sponge network | -1.14 | 0.03513 | -0.525 | 2.0E-5 | 0.299 |
| 41175 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p | 14 | ADARB1 | Sponge network | 0.542 | 0.00054 | -0.335 | 0.06631 | 0.299 |
| 41176 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | TRIM33 | Sponge network | 0.659 | 3.0E-5 | 0.402 | 7.0E-5 | 0.299 |
| 41177 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 25 | YAP1 | Sponge network | -0.777 | 0.1134 | 0.22 | 0.11836 | 0.299 |
| 41178 | MIR7-3HG |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 11 | ASTN1 | Sponge network | -2.688 | 0.00397 | -2.389 | 2.0E-5 | 0.299 |
| 41179 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-590-3p | 17 | FREM2 | Sponge network | -0.49 | 0.04946 | -0.437 | 0.46353 | 0.299 |
| 41180 | RP11-968O1.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-362-3p | 12 | GPC6 | Sponge network | -0.829 | 0.01343 | -0.054 | 0.81465 | 0.299 |
| 41181 | C4B-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 18 | TRPS1 | Sponge network | 2.306 | 9.0E-5 | -0.191 | 0.44109 | 0.299 |
| 41182 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-429 | 10 | MAP3K12 | Sponge network | -2.62 | 0 | -0.057 | 0.59369 | 0.299 |
| 41183 | CTB-174D11.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-3p | 11 | IL17RD | Sponge network | -2.1 | 0.04665 | -0.019 | 0.94873 | 0.299 |
| 41184 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p | 12 | DDX6 | Sponge network | 0.702 | 7.0E-5 | 0.091 | 0.36428 | 0.299 |
| 41185 | RP11-867G23.10 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-93-5p | 26 | CCND2 | Sponge network | -2.531 | 0 | -0.267 | 0.3112 | 0.299 |
| 41186 | CKMT2-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-3p | 13 | PHF3 | Sponge network | 0.082 | 0.55654 | 0.126 | 0.19652 | 0.299 |
| 41187 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 27 | BHLHE41 | Sponge network | -1.439 | 4.0E-5 | 0.353 | 0.12753 | 0.299 |
| 41188 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-511-5p | 16 | BCL6 | Sponge network | -0.384 | 0.40652 | -0.211 | 0.19489 | 0.299 |
| 41189 | OSER1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-486-5p;hsa-miR-532-3p | 13 | SYNM | Sponge network | -0.13 | 0.48734 | -2.309 | 1.0E-5 | 0.299 |
| 41190 | NDUFA6-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 58 | CADM2 | Sponge network | -0.355 | 0.0077 | -3.782 | 0 | 0.299 |
| 41191 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 10 | NF1 | Sponge network | 1.757 | 0 | 0.51 | 0 | 0.299 |
| 41192 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-624-5p;hsa-miR-655-3p | 19 | LATS1 | Sponge network | 0.253 | 0.09565 | 0.109 | 0.34208 | 0.299 |
| 41193 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 0.986 | 0.01022 | 0.51 | 0 | 0.299 |
| 41194 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-2355-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-654-3p | 27 | DDX6 | Sponge network | -0.567 | 0 | 0.091 | 0.36428 | 0.299 |
| 41195 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-331-3p;hsa-miR-7-1-3p | 10 | ATRX | Sponge network | 0.693 | 0.00076 | 0.261 | 0.04408 | 0.299 |
| 41196 | GS1-124K5.11 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | TPO | Sponge network | 0.494 | 0.00339 | -1.87 | 2.0E-5 | 0.299 |
| 41197 | RP11-473I1.10 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | RASSF8 | Sponge network | 0.673 | 0 | -0.122 | 0.65591 | 0.299 |
| 41198 | LINC00473 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | JDP2 | Sponge network | -1.203 | 0.03881 | -0.967 | 0 | 0.299 |
| 41199 | BDNF-AS |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p | 14 | SYNM | Sponge network | -0.668 | 3.0E-5 | -2.309 | 1.0E-5 | 0.299 |
| 41200 | AC004893.11 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 13 | TET1 | Sponge network | 1.317 | 0 | -0.135 | 0.52138 | 0.299 |
| 41201 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 17 | RBL2 | Sponge network | -2.117 | 0.00161 | -0.282 | 0.00254 | 0.299 |
| 41202 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-653-5p;hsa-miR-96-5p | 24 | DMXL1 | Sponge network | 0.331 | 0.57247 | -0.426 | 0.00056 | 0.299 |
| 41203 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | USP6 | Sponge network | 0.222 | 0.29133 | 0.188 | 0.3871 | 0.299 |
| 41204 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 20 | MXI1 | Sponge network | -0.342 | 0.02288 | -0.987 | 0 | 0.299 |
| 41205 | AP000487.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-376c-3p;hsa-miR-505-5p;hsa-miR-766-3p | 18 | TBL1XR1 | Sponge network | 1.987 | 0 | 0.578 | 0 | 0.299 |
| 41206 | FAM157A |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326 | 13 | ZFX | Sponge network | 0.813 | 1.0E-5 | 0.09 | 0.42563 | 0.299 |
| 41207 | RP11-686D22.8 |
hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-375;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | MITF | Sponge network | 0.898 | 1.0E-5 | -0.769 | 0.00022 | 0.299 |
| 41208 | LINC00578 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-577 | 11 | PRKAR1A | Sponge network | 0.811 | 0.03228 | -0.063 | 0.50314 | 0.299 |
| 41209 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | KDSR | Sponge network | 0.673 | 0 | 0.239 | 0.02216 | 0.299 |
| 41210 | RP11-244H3.1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-501-5p | 11 | MN1 | Sponge network | 0.659 | 3.0E-5 | -0.358 | 0.24172 | 0.299 |
| 41211 | LINC00957 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-744-3p | 24 | PTPN14 | Sponge network | -0.421 | 0.0114 | 0.144 | 0.34595 | 0.299 |
| 41212 | RAD51-AS1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | ERCC6 | Sponge network | 0.768 | 7.0E-5 | 0.23 | 0.015 | 0.299 |
| 41213 | RP11-66B24.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3607-3p | 16 | PPP1R9A | Sponge network | 0.57 | 0.1866 | -0.903 | 0.04254 | 0.299 |
| 41214 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p | 16 | FOXO3 | Sponge network | 0.792 | 0.0049 | -0.04 | 0.72028 | 0.299 |
| 41215 | MIR497HG |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-940 | 52 | NFIB | Sponge network | -1.439 | 4.0E-5 | 0.023 | 0.90795 | 0.299 |
| 41216 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | RNF144A | Sponge network | 0.246 | 0.59912 | 0.594 | 0.0009 | 0.299 |
| 41217 | LINC00578 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-628-5p | 27 | TMEM132B | Sponge network | 0.811 | 0.03228 | -0.83 | 0.01084 | 0.299 |
| 41218 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p | 15 | GREM1 | Sponge network | 0.082 | 0.55397 | 0.902 | 0.01691 | 0.299 |
| 41219 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 12 | ITGB3 | Sponge network | -0.125 | 0.49414 | -0.011 | 0.95751 | 0.299 |
| 41220 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | ZFHX3 | Sponge network | 0.657 | 4.0E-5 | 0.389 | 0.01363 | 0.299 |
| 41221 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 11 | LRRK2 | Sponge network | -0.125 | 0.49414 | -1.136 | 1.0E-5 | 0.299 |
| 41222 | RP11-888D10.4 |
hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | TMEM30A | Sponge network | 0.702 | 7.0E-5 | 0.096 | 0.31031 | 0.299 |
| 41223 | LINC00648 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 18 | CADM1 | Sponge network | 0.633 | 0.51678 | -0.9 | 0.00084 | 0.299 |
| 41224 | HOXA-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | ABCA10 | Sponge network | 0.61 | 0.01593 | -0.571 | 0.03674 | 0.299 |
| 41225 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p | 11 | MLLT10 | Sponge network | 0.752 | 0 | 0.105 | 0.33292 | 0.299 |
| 41226 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SLC6A15 | Sponge network | -0.612 | 0.00094 | -2.373 | 2.0E-5 | 0.299 |
| 41227 | PART1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 35 | BTG2 | Sponge network | -2.925 | 0 | -1.763 | 0 | 0.299 |
| 41228 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-93-5p | 21 | NIN | Sponge network | -0.154 | 0.78939 | -0.253 | 0.13794 | 0.299 |
| 41229 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | MAP3K12 | Sponge network | -0.615 | 0.00015 | -0.057 | 0.59369 | 0.299 |
| 41230 | RP11-356J5.12 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | LSAMP | Sponge network | -0.899 | 6.0E-5 | -0.066 | 0.81708 | 0.299 |
| 41231 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-326 | 14 | FAT3 | Sponge network | -0.011 | 0.967 | -1.601 | 0.00051 | 0.299 |
| 41232 | XXYLT1-AS2 |
hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-93-3p | 14 | QKI | Sponge network | -0.764 | 0.05257 | -0.333 | 0.04111 | 0.299 |
| 41233 | LINC00839 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p | 10 | TCF4 | Sponge network | 0.522 | 0.08221 | 0.016 | 0.92271 | 0.299 |
| 41234 | RP1-302G2.5 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-5p | 14 | BCL2 | Sponge network | -1.483 | 9.0E-5 | -0.899 | 0 | 0.299 |
| 41235 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 72 | TCF4 | Sponge network | -2.925 | 0 | 0.016 | 0.92271 | 0.299 |
| 41236 | AP001469.9 |
hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 11 | MDM4 | Sponge network | 1.096 | 0 | 0.345 | 0.00762 | 0.299 |
| 41237 | RP11-535M15.1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p | 11 | NTRK2 | Sponge network | -1.14 | 0.03513 | -0.736 | 0.06495 | 0.299 |
| 41238 | DNAJC27-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 82 | LPP | Sponge network | -0.969 | 2.0E-5 | -0.459 | 0.04475 | 0.299 |
| 41239 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ZFHX3 | Sponge network | 0.873 | 0 | 0.389 | 0.01363 | 0.299 |
| 41240 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | PAFAH1B1 | Sponge network | -1.439 | 4.0E-5 | -0.429 | 0 | 0.299 |
| 41241 | CTB-133G6.2 |
hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-424-5p | 10 | UNC80 | Sponge network | 0.4 | 0.39299 | -1.934 | 0 | 0.299 |
| 41242 | RP11-244H3.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | LRIG1 | Sponge network | 0.659 | 3.0E-5 | -0.537 | 0.00588 | 0.299 |
| 41243 | RP11-967K21.1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p | 11 | ZBTB20 | Sponge network | 0.056 | 0.81195 | -0.122 | 0.57569 | 0.299 |
| 41244 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | HACE1 | Sponge network | -1.575 | 6.0E-5 | -0.072 | 0.55239 | 0.299 |
| 41245 | RP11-539I5.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p | 11 | CDH23 | Sponge network | -0.468 | 0.32587 | -0.974 | 0.00034 | 0.299 |
| 41246 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | RELN | Sponge network | -0.355 | 0.0077 | -2.403 | 0 | 0.299 |
| 41247 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 27 | HLF | Sponge network | -0.737 | 0.10822 | -2.443 | 0 | 0.299 |
| 41248 | RP11-890B15.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-7-5p | 27 | BTG2 | Sponge network | 0.101 | 0.60919 | -1.763 | 0 | 0.299 |
| 41249 | RP11-276H19.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-550a-3p | 21 | DTNA | Sponge network | -0.472 | 0.48851 | -1.648 | 1.0E-5 | 0.299 |
| 41250 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | PDS5B | Sponge network | 0.222 | 0.29133 | 0.259 | 0.00962 | 0.299 |
| 41251 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p | 20 | RYR2 | Sponge network | -0.738 | 0.21233 | -1.466 | 0.00031 | 0.299 |
| 41252 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-96-5p;hsa-miR-98-5p | 10 | PLAGL1 | Sponge network | -0.969 | 2.0E-5 | -0.588 | 0.00912 | 0.299 |
| 41253 | RP11-597D13.9 |
hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 11 | PAK3 | Sponge network | 1.302 | 7.0E-5 | -0.628 | 0.07253 | 0.299 |
| 41254 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-935;hsa-miR-96-5p | 32 | MTSS1 | Sponge network | 0.083 | 0.66405 | -0.81 | 0.00035 | 0.299 |
| 41255 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-96-5p | 17 | NAV3 | Sponge network | 0.453 | 0.01839 | 0.212 | 0.47729 | 0.299 |
| 41256 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | HACE1 | Sponge network | 0.311 | 0.10344 | -0.072 | 0.55239 | 0.299 |
| 41257 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p | 13 | TCF12 | Sponge network | 0.082 | 0.55397 | 0.174 | 0.13271 | 0.299 |
| 41258 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | LRRC55 | Sponge network | 0.064 | 0.72934 | -0.026 | 0.93163 | 0.299 |
| 41259 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-93-5p | 18 | RNASEL | Sponge network | -0.668 | 3.0E-5 | -0.272 | 0.01796 | 0.299 |
| 41260 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | PHC3 | Sponge network | -1.281 | 0.00012 | 0.212 | 0.0499 | 0.299 |
| 41261 | CTB-51J22.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-940 | 11 | MEF2C | Sponge network | -0.803 | 0.23737 | -0.499 | 0.01448 | 0.299 |
| 41262 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429 | 12 | QKI | Sponge network | 2.364 | 0.00134 | -0.333 | 0.04111 | 0.299 |
| 41263 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 23 | RSPO2 | Sponge network | -0.83 | 0 | -3.038 | 0 | 0.299 |
| 41264 | RP11-38L15.3 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 18 | RUNX1T1 | Sponge network | 0.344 | 0.3127 | -0.344 | 0.2374 | 0.299 |
| 41265 | U91328.21 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b | 13 | HBP1 | Sponge network | -0.735 | 0.15983 | -0.454 | 0 | 0.299 |
| 41266 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TACC1 | Sponge network | -0.141 | 0.26434 | -0.362 | 0.05406 | 0.299 |
| 41267 | AC005618.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | NAV3 | Sponge network | 0.862 | 0.06213 | 0.212 | 0.47729 | 0.299 |
| 41268 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-625-3p;hsa-miR-664a-3p | 27 | FZD3 | Sponge network | 1.03 | 0 | 0.104 | 0.60316 | 0.299 |
| 41269 | RP11-20I23.13 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-486-5p | 14 | EZH1 | Sponge network | 0.505 | 0.0014 | -0.564 | 1.0E-5 | 0.299 |
| 41270 | RP11-473I1.9 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 14 | PRKAR1A | Sponge network | 0.683 | 1.0E-5 | -0.063 | 0.50314 | 0.299 |
| 41271 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 53 | SMAD4 | Sponge network | 0.194 | 0.64278 | -0.313 | 0.00413 | 0.299 |
| 41272 | RP11-888D10.4 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p | 10 | CBFB | Sponge network | 0.702 | 7.0E-5 | 1.061 | 0 | 0.299 |
| 41273 | RP11-46C24.7 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | CREB1 | Sponge network | 0.392 | 0.0278 | 0.203 | 0.00537 | 0.299 |
| 41274 | BDNF-AS |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | SETBP1 | Sponge network | -0.668 | 3.0E-5 | -1.302 | 1.0E-5 | 0.299 |
| 41275 | RP11-532F6.3 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p | 18 | LIMCH1 | Sponge network | -0.896 | 0.00099 | -0.915 | 1.0E-5 | 0.299 |
| 41276 | AC034220.3 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p | 18 | NF1 | Sponge network | 0.309 | 0.07304 | 0.51 | 0 | 0.299 |
| 41277 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 29 | IGF1 | Sponge network | 0.083 | 0.66405 | -0.204 | 0.5898 | 0.299 |
| 41278 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CLIP1 | Sponge network | 0.953 | 0 | -0.215 | 0.08684 | 0.299 |
| 41279 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-429;hsa-miR-93-5p | 10 | CNTNAP3 | Sponge network | -0.785 | 0.00035 | -1.465 | 0.00033 | 0.299 |
| 41280 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-429 | 19 | ZFHX3 | Sponge network | 0.859 | 0.00331 | 0.389 | 0.01363 | 0.299 |
| 41281 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 42 | AR | Sponge network | 0.41 | 0.02214 | -1.554 | 0 | 0.299 |
| 41282 | SH3RF3-AS1 |
hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-5p | 13 | CADM1 | Sponge network | 0.889 | 9.0E-5 | -0.9 | 0.00084 | 0.299 |
| 41283 | HOXB-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 11 | POU2F2 | Sponge network | -0.154 | 0.53954 | -0.233 | 0.32214 | 0.299 |
| 41284 | OSER1-AS1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-877-5p | 13 | CRTC1 | Sponge network | -0.13 | 0.48734 | -0.116 | 0.28412 | 0.299 |
| 41285 | CTC-297N7.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-590-5p | 15 | UNC80 | Sponge network | -2.069 | 0 | -1.934 | 0 | 0.299 |
| 41286 | DNM3OS |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 19 | PRKAR1A | Sponge network | 0.572 | 0.01722 | -0.063 | 0.50314 | 0.299 |
| 41287 | TPTEP1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | PAIP2 | Sponge network | -1.216 | 0 | -0.515 | 0 | 0.299 |
| 41288 | RP11-344B5.2 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b | 18 | FAT3 | Sponge network | -0.055 | 0.88482 | -1.601 | 0.00051 | 0.299 |
| 41289 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p | 18 | MXI1 | Sponge network | -1.654 | 0.00144 | -0.987 | 0 | 0.299 |
| 41290 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-378a-5p | 11 | NUAK1 | Sponge network | -1.375 | 0.08642 | 0.904 | 0 | 0.299 |
| 41291 | RP11-67L2.2 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 29 | PCDH9 | Sponge network | 0.72 | 0.0001 | -1.823 | 4.0E-5 | 0.299 |
| 41292 | AC108488.4 |
hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-577 | 17 | MYBL1 | Sponge network | 0.259 | 0.12542 | 0.494 | 0.02152 | 0.299 |
| 41293 | RP11-35G9.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-193b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 10 | SORCS1 | Sponge network | 0.657 | 4.0E-5 | -3.492 | 0 | 0.299 |
| 41294 | RP11-283I3.6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 14 | ZMYND11 | Sponge network | 0.614 | 1.0E-5 | -0.2 | 0.05449 | 0.299 |
| 41295 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 28 | KCNA6 | Sponge network | -0.899 | 6.0E-5 | -1.531 | 0.00013 | 0.299 |
| 41296 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-590-3p | 10 | DLC1 | Sponge network | -0.295 | 0.02604 | -0.382 | 0.05038 | 0.299 |
| 41297 | AC003090.1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -2.117 | 0.00161 | -0.966 | 0.00035 | 0.299 |
| 41298 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-576-5p | 13 | CXXC4 | Sponge network | -0.932 | 0.00329 | -0.433 | 0.21055 | 0.299 |
| 41299 | PSMG3-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | DIP2C | Sponge network | -0.125 | 0.49414 | -0.436 | 0.01713 | 0.299 |
| 41300 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-505-3p;hsa-miR-590-3p | 12 | JAKMIP2 | Sponge network | 0.619 | 0.00157 | -1.388 | 0 | 0.299 |
| 41301 | LINC00578 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-577 | 14 | CSDE1 | Sponge network | 0.811 | 0.03228 | -0.493 | 0 | 0.299 |
| 41302 | H1FX-AS1 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TAL1 | Sponge network | -0.206 | 0.12942 | -0.462 | 0.00574 | 0.299 |
| 41303 | USP3-AS1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-576-5p | 21 | PAIP2 | Sponge network | -0.162 | 0.47687 | -0.515 | 0 | 0.299 |
| 41304 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p | 18 | RICTOR | Sponge network | 1.361 | 0 | 0.388 | 0.00188 | 0.299 |
| 41305 | LIPE-AS1 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-590-5p | 15 | INPP4B | Sponge network | 0.078 | 0.55803 | -0.097 | 0.60503 | 0.299 |
| 41306 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 30 | EPHA7 | Sponge network | 0.75 | 0.00054 | -2.197 | 3.0E-5 | 0.299 |
| 41307 | RP11-774O3.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 26 | JDP2 | Sponge network | -0.487 | 0.02157 | -0.967 | 0 | 0.299 |
| 41308 | RP13-516M14.1 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 11 | ABL1 | Sponge network | 0.389 | 0.04293 | -0.215 | 0.07804 | 0.299 |
| 41309 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-429 | 14 | RNASEL | Sponge network | -0.469 | 0.08721 | -0.272 | 0.01796 | 0.299 |
| 41310 | CTD-2089N3.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p | 11 | IL17RD | Sponge network | -1.375 | 0.08642 | -0.019 | 0.94873 | 0.299 |
| 41311 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-5p | 21 | JAZF1 | Sponge network | -0.141 | 0.26434 | -0.377 | 0.02677 | 0.299 |
| 41312 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | NIN | Sponge network | 0.889 | 9.0E-5 | -0.253 | 0.13794 | 0.299 |
| 41313 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | CREBBP | Sponge network | -0.896 | 0.00099 | 0.126 | 0.15186 | 0.299 |
| 41314 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-877-5p | 12 | CRTC1 | Sponge network | 0.889 | 9.0E-5 | -0.116 | 0.28412 | 0.299 |
| 41315 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PTPRB | Sponge network | 0.61 | 0.01593 | 0.105 | 0.47122 | 0.299 |
| 41316 | FZD10-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 18 | LUZP2 | Sponge network | -0.727 | 0.04726 | -0.056 | 0.89416 | 0.299 |
| 41317 | LINC00667 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | ZDBF2 | Sponge network | -0.235 | 0.15142 | -0.998 | 0.00025 | 0.299 |
| 41318 | FAM66C |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-590-5p;hsa-miR-7-5p | 22 | KCNJ5 | Sponge network | -0.901 | 0.00019 | -0.281 | 0.3339 | 0.298 |
| 41319 | RP11-458D21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-744-3p | 30 | DST | Sponge network | 0.663 | 0.00073 | -0.713 | 0.00096 | 0.298 |
| 41320 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 40 | AFF4 | Sponge network | -0.358 | 0.17255 | -0.266 | 0.01565 | 0.298 |
| 41321 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 20 | HECA | Sponge network | -0.405 | 0.22013 | -0.217 | 0.03463 | 0.298 |
| 41322 | LINC00969 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p | 17 | PDS5B | Sponge network | 0.683 | 1.0E-5 | 0.259 | 0.00962 | 0.298 |
| 41323 | RP11-473I1.9 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-328-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-628-5p | 23 | VPS53 | Sponge network | 0.683 | 1.0E-5 | 0.103 | 0.22667 | 0.298 |
| 41324 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 20 | SEMA3C | Sponge network | -0.769 | 0.00035 | -0.272 | 0.31153 | 0.298 |
| 41325 | RP11-159D12.2 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 26 | PIK3R1 | Sponge network | 1.254 | 0 | -0.133 | 0.36854 | 0.298 |
| 41326 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | LATS2 | Sponge network | -2.117 | 0.00161 | 0.159 | 0.2304 | 0.298 |
| 41327 | HNRNPU-AS1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p | 10 | MAT2A | Sponge network | 1.601 | 0 | -0.073 | 0.36195 | 0.298 |
| 41328 | AC016747.3 |
hsa-let-7a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 21 | ST6GAL2 | Sponge network | 0.199 | 0.38015 | -1.201 | 0.00597 | 0.298 |
| 41329 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-590-3p | 15 | GPAM | Sponge network | 0.559 | 0.00026 | 0.142 | 0.29732 | 0.298 |
| 41330 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 10 | TSLP | Sponge network | -0.615 | 0.00015 | -2.209 | 0 | 0.298 |
| 41331 | RP11-359B12.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FBXO11 | Sponge network | 0.064 | 0.72934 | 0.142 | 0.04938 | 0.298 |
| 41332 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-590-3p;hsa-miR-590-5p | 34 | WDFY3 | Sponge network | 0.906 | 0.31715 | -0.201 | 0.0967 | 0.298 |
| 41333 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p | 12 | ALDH1A2 | Sponge network | -0.154 | 0.78939 | -2.411 | 0 | 0.298 |
| 41334 | AC090587.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p | 14 | KIF1B | Sponge network | -0.088 | 0.65011 | -0.118 | 0.32258 | 0.298 |
| 41335 | DNAJC27-AS1 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 19 | CXCL12 | Sponge network | -0.969 | 2.0E-5 | -1.421 | 0 | 0.298 |
| 41336 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 14 | DYNC1I1 | Sponge network | -0.351 | 0.11358 | -1.12 | 0.00107 | 0.298 |
| 41337 | CTA-228A9.3 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-342-3p | 13 | CCDC6 | Sponge network | 2.466 | 0 | 0.27 | 0.02555 | 0.298 |
| 41338 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | RASSF2 | Sponge network | 0.056 | 0.8494 | -0.273 | 0.21961 | 0.298 |
| 41339 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-493-5p | 13 | LATS1 | Sponge network | 0.368 | 0.20093 | 0.109 | 0.34208 | 0.298 |
| 41340 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-942-5p | 12 | CREBBP | Sponge network | 0.082 | 0.55397 | 0.126 | 0.15186 | 0.298 |
| 41341 | RP11-119F7.5 |
hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-324-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | PRKAR1A | Sponge network | 0.225 | 0.30644 | -0.063 | 0.50314 | 0.298 |
| 41342 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-502-5p | 12 | TGFBR2 | Sponge network | -0.517 | 0.02935 | -0.243 | 0.11408 | 0.298 |
| 41343 | RP11-25K19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-628-5p | 21 | ERC1 | Sponge network | -1.057 | 0.00029 | -0.108 | 0.38794 | 0.298 |
| 41344 | LINC00702 |
hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-874-3p;hsa-miR-96-5p | 10 | AJAP1 | Sponge network | -0.737 | 0.06182 | -0.561 | 0.04832 | 0.298 |
| 41345 | RP11-403I13.7 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-877-5p;hsa-miR-96-5p | 33 | IL17RD | Sponge network | 0.395 | 0.15451 | -0.019 | 0.94873 | 0.298 |
| 41346 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-629-3p | 10 | POU2F2 | Sponge network | 0.727 | 3.0E-5 | -0.233 | 0.32214 | 0.298 |
| 41347 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-576-5p | 18 | FREM2 | Sponge network | -0.405 | 0.22013 | -0.437 | 0.46353 | 0.298 |
| 41348 | LINC00680 |
hsa-let-7a-3p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-664a-3p | 16 | SOX11 | Sponge network | 0.884 | 0 | 2.68 | 0 | 0.298 |
| 41349 | SNHG14 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-550a-3p | 15 | CACNA1E | Sponge network | -1.111 | 0.0003 | 1.752 | 0 | 0.298 |
| 41350 | MIR497HG |
hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-874-3p;hsa-miR-96-5p | 10 | AJAP1 | Sponge network | -1.439 | 4.0E-5 | -0.561 | 0.04832 | 0.298 |
| 41351 | RP11-359E10.1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-671-5p | 13 | KIT | Sponge network | 0.299 | 0.57722 | -2.184 | 0 | 0.298 |
| 41352 | HCG11 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-629-3p | 10 | ITGA5 | Sponge network | 0.385 | 0.10067 | 0.452 | 0.03524 | 0.298 |
| 41353 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 27 | RIMBP2 | Sponge network | -0.129 | 0.57177 | -1.968 | 5.0E-5 | 0.298 |
| 41354 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | PRKAA2 | Sponge network | 0.259 | 0.12542 | -2.462 | 0 | 0.298 |
| 41355 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-550a-3p | 22 | RAP1A | Sponge network | 0.082 | 0.55397 | -0.929 | 0 | 0.298 |
| 41356 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 30 | MXI1 | Sponge network | -0.323 | 0.01372 | -0.987 | 0 | 0.298 |
| 41357 | SERHL |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-342-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p | 11 | SYNPO2 | Sponge network | -0.602 | 0.00331 | -2.342 | 0 | 0.298 |
| 41358 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-96-5p | 36 | MEF2C | Sponge network | 0.374 | 0.20054 | -0.499 | 0.01448 | 0.298 |
| 41359 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 34 | ERBB4 | Sponge network | -0.842 | 0.01361 | -1.975 | 2.0E-5 | 0.298 |
| 41360 | KB-1410C5.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | CUX1 | Sponge network | 0.959 | 0 | -0.039 | 0.6705 | 0.298 |
| 41361 | HCG22 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-501-5p | 10 | MAFB | Sponge network | 0.816 | 0.32368 | -0.023 | 0.89865 | 0.298 |
| 41362 | HAND2-AS1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | PAIP2 | Sponge network | -1.697 | 0.00889 | -0.515 | 0 | 0.298 |
| 41363 | AC009120.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 19 | PIK3C2A | Sponge network | 0.919 | 0 | 0.061 | 0.53776 | 0.298 |
| 41364 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 12 | ZNF208 | Sponge network | -1.057 | 0.00029 | -0.4 | 0.22381 | 0.298 |
| 41365 | LINC00476 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-93-3p | 28 | NCOA1 | Sponge network | -0.615 | 0.00015 | -0.432 | 0 | 0.298 |
| 41366 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-589-5p | 17 | ARHGAP29 | Sponge network | -2.62 | 0 | 0.131 | 0.42953 | 0.298 |
| 41367 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MED12L | Sponge network | 0.559 | 0.00026 | -0.194 | 0.55299 | 0.298 |
| 41368 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 51 | THRB | Sponge network | -0.342 | 0.02288 | -1.117 | 0.0004 | 0.298 |
| 41369 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-582-5p | 24 | SPRY3 | Sponge network | 0.817 | 3.0E-5 | 0.016 | 0.91286 | 0.298 |
| 41370 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 19 | PDE4DIP | Sponge network | -1.349 | 0.00017 | -0.282 | 0.0257 | 0.298 |
| 41371 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-493-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 37 | DDX6 | Sponge network | -0.739 | 0.0574 | 0.091 | 0.36428 | 0.298 |
| 41372 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | PRKAB2 | Sponge network | 0.848 | 0 | -0.367 | 0.01371 | 0.298 |
| 41373 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-942-5p | 22 | UNC80 | Sponge network | -4.463 | 0.00025 | -1.934 | 0 | 0.298 |
| 41374 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-877-5p | 21 | CRTC1 | Sponge network | 0.082 | 0.55654 | -0.116 | 0.28412 | 0.298 |
| 41375 | RP5-1198O20.4 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | PPM1L | Sponge network | 0.997 | 0.00066 | -0.537 | 0.00616 | 0.298 |
| 41376 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-582-5p | 11 | RB1CC1 | Sponge network | 0.817 | 3.0E-5 | 0.175 | 0.12559 | 0.298 |
| 41377 | SERHL |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-628-5p | 29 | DTNA | Sponge network | -0.602 | 0.00331 | -1.648 | 1.0E-5 | 0.298 |
| 41378 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | FYN | Sponge network | -0.342 | 0.02288 | -0.131 | 0.37051 | 0.298 |
| 41379 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 23 | ST6GAL2 | Sponge network | 0.95 | 0.15943 | -1.201 | 0.00597 | 0.298 |
| 41380 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-93-5p | 18 | TP53INP1 | Sponge network | -0.842 | 0.01361 | -0.496 | 0.00064 | 0.298 |
| 41381 | PDXDC2P |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 14 | ARHGEF12 | Sponge network | 0.756 | 0 | 0.09 | 0.35693 | 0.298 |
| 41382 | RP11-384O8.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | HIP1 | Sponge network | -0.738 | 0.21233 | 0.774 | 0 | 0.298 |
| 41383 | TPT1-AS1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p | 14 | PHF6 | Sponge network | 0.903 | 0 | 0.785 | 0 | 0.298 |
| 41384 | LINC00963 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 27 | DMD | Sponge network | 0.523 | 9.0E-5 | -1.506 | 0 | 0.298 |
| 41385 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 17 | LRRC55 | Sponge network | 0.16 | 0.50785 | -0.026 | 0.93163 | 0.298 |
| 41386 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 26 | EYA4 | Sponge network | -0.612 | 0.00094 | 0.3 | 0.36876 | 0.298 |
| 41387 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429 | 13 | TBX18 | Sponge network | -1.654 | 0.00144 | 0.843 | 0.02945 | 0.298 |
| 41388 | HCG11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | 0.385 | 0.10067 | -0.243 | 0.11408 | 0.298 |
| 41389 | RP11-38L15.3 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 15 | WDFY3 | Sponge network | 0.344 | 0.3127 | -0.201 | 0.0967 | 0.298 |
| 41390 | MIR143HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 26 | SUFU | Sponge network | -0.739 | 0.0574 | -0.04 | 0.65489 | 0.298 |
| 41391 | LENG8-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-5p | 10 | MYBL1 | Sponge network | 1.471 | 0 | 0.494 | 0.02152 | 0.298 |
| 41392 | FAM13A-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 23 | CDH19 | Sponge network | -0.83 | 0 | -3.434 | 0 | 0.298 |
| 41393 | RP11-531A24.5 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-539-5p | 16 | RYR2 | Sponge network | 0.75 | 0.00054 | -1.466 | 0.00031 | 0.298 |
| 41394 | RP11-434B12.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-629-3p | 19 | PTPN14 | Sponge network | -0.031 | 0.89063 | 0.144 | 0.34595 | 0.298 |
| 41395 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | SLC6A15 | Sponge network | -2.925 | 0 | -2.373 | 2.0E-5 | 0.298 |
| 41396 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | ANK2 | Sponge network | 0.75 | 0.00054 | -1.603 | 1.0E-5 | 0.298 |
| 41397 | CTC-296K1.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p | 10 | NIN | Sponge network | -2.095 | 0.00112 | -0.253 | 0.13794 | 0.298 |
| 41398 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p | 20 | TBX18 | Sponge network | -1.249 | 7.0E-5 | 0.843 | 0.02945 | 0.298 |
| 41399 | LINC00672 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | -0.227 | 0.31657 | 0.142 | 0.04938 | 0.298 |
| 41400 | DNM3OS |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-7-5p | 22 | KCNJ5 | Sponge network | 0.572 | 0.01722 | -0.281 | 0.3339 | 0.298 |
| 41401 | FAM66C |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | GATA2 | Sponge network | -0.901 | 0.00019 | -0.654 | 0.0016 | 0.298 |
| 41402 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-577 | 21 | MYBL1 | Sponge network | -0.465 | 0.04703 | 0.494 | 0.02152 | 0.298 |
| 41403 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-93-5p | 12 | DPYD | Sponge network | -2.69 | 0.0001 | -0.736 | 0.00076 | 0.298 |
| 41404 | RP11-181G12.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-5p;hsa-miR-424-5p | 12 | RASSF8 | Sponge network | 0.556 | 0.00298 | -0.122 | 0.65591 | 0.298 |
| 41405 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | USP12 | Sponge network | 1.044 | 2.0E-5 | -0.102 | 0.40768 | 0.298 |
| 41406 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 41 | ERBB4 | Sponge network | -0.096 | 0.67958 | -1.975 | 2.0E-5 | 0.298 |
| 41407 | WAC-AS1 |
hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | RICTOR | Sponge network | 0.821 | 0 | 0.388 | 0.00188 | 0.298 |
| 41408 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-590-5p | 14 | TGFBR2 | Sponge network | 0.176 | 0.37402 | -0.243 | 0.11408 | 0.298 |
| 41409 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-590-3p | 10 | CACNA2D1 | Sponge network | -0.663 | 0.10887 | -0.846 | 0.00675 | 0.298 |
| 41410 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-501-3p;hsa-miR-590-3p | 19 | KAT6B | Sponge network | 0.082 | 0.55654 | -0.478 | 0.00018 | 0.298 |
| 41411 | RP11-73M18.10 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-497-5p | 12 | UBR3 | Sponge network | 0.85 | 0 | -0.021 | 0.82774 | 0.298 |
| 41412 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 18 | GPAM | Sponge network | 1.254 | 0 | 0.142 | 0.29732 | 0.298 |
| 41413 | KB-1460A1.5 |
hsa-miR-10b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | CSDE1 | Sponge network | 0.603 | 0.00127 | -0.493 | 0 | 0.298 |
| 41414 | CTA-29F11.1 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-30a-5p;hsa-miR-664a-5p | 12 | SOX11 | Sponge network | 0.583 | 0.00027 | 2.68 | 0 | 0.298 |
| 41415 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p | 19 | AFF3 | Sponge network | -0.176 | 0.59692 | -2.373 | 0 | 0.298 |
| 41416 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-5p | 33 | ERBB4 | Sponge network | -2.69 | 0.0001 | -1.975 | 2.0E-5 | 0.298 |
| 41417 | AC009948.5 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | SYT4 | Sponge network | 0.532 | 0.00505 | -3.207 | 0 | 0.298 |
| 41418 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 49 | SSBP2 | Sponge network | 0.064 | 0.72934 | -1.58 | 0 | 0.298 |
| 41419 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-942-5p | 26 | TMEM132B | Sponge network | 0.381 | 0.25967 | -0.83 | 0.01084 | 0.298 |
| 41420 | RP11-439A17.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-98-5p | 10 | PBX1 | Sponge network | 0.051 | 0.95756 | -1.206 | 0 | 0.298 |
| 41421 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | 0.082 | 0.55654 | 0.024 | 0.89889 | 0.298 |
| 41422 | AC108488.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-940 | 28 | NTRK2 | Sponge network | 0.259 | 0.12542 | -0.736 | 0.06495 | 0.298 |
| 41423 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-628-5p | 21 | BCL6 | Sponge network | -0.154 | 0.78939 | -0.211 | 0.19489 | 0.298 |
| 41424 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-324-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 22 | USP12 | Sponge network | -0.176 | 0.59692 | -0.102 | 0.40768 | 0.298 |
| 41425 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-511-5p | 11 | LRRC4 | Sponge network | -0.777 | 0.16667 | -1.774 | 0 | 0.298 |
| 41426 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-940 | 39 | PTPRT | Sponge network | -0.942 | 0 | -0.907 | 0.03727 | 0.298 |
| 41427 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 22 | PRKAR1A | Sponge network | -2.452 | 0 | -0.063 | 0.50314 | 0.298 |
| 41428 | WDFY3-AS2 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -0.942 | 0 | -0.966 | 0.00035 | 0.298 |
| 41429 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 48 | PRKAA2 | Sponge network | 0.72 | 0.0001 | -2.462 | 0 | 0.298 |
| 41430 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-940;hsa-miR-96-5p | 41 | GAS7 | Sponge network | 0.331 | 0.57247 | -0.555 | 0.01602 | 0.298 |
| 41431 | EMX2OS |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 20 | FMNL3 | Sponge network | -0.777 | 0.1134 | 0.555 | 4.0E-5 | 0.298 |
| 41432 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 25 | ZBTB4 | Sponge network | 0.4 | 0.14611 | -0.739 | 0 | 0.298 |
| 41433 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NOTCH2NL | Sponge network | 0.873 | 0 | 0.024 | 0.84307 | 0.298 |
| 41434 | HAR1A |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-744-3p;hsa-miR-942-5p | 19 | IGFBP5 | Sponge network | -0.672 | 0.19447 | -0.806 | 0.00105 | 0.298 |
| 41435 | RP11-678G14.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-339-5p | 12 | RYR2 | Sponge network | -4.477 | 0 | -1.466 | 0.00031 | 0.298 |
| 41436 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 23 | CRTC1 | Sponge network | -0.513 | 0.04703 | -0.116 | 0.28412 | 0.298 |
| 41437 | EMX2OS |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | AFAP1L2 | Sponge network | -0.777 | 0.1134 | -0.284 | 0.15434 | 0.298 |
| 41438 | WDFY3-AS2 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | EDNRB | Sponge network | -0.942 | 0 | -0.682 | 0.00138 | 0.298 |
| 41439 | RP11-404E16.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 16 | KIF1B | Sponge network | 0.097 | 0.91311 | -0.118 | 0.32258 | 0.298 |
| 41440 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | FAT3 | Sponge network | 0.056 | 0.8494 | -1.601 | 0.00051 | 0.298 |
| 41441 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | RNF144A | Sponge network | -0.611 | 0.09607 | 0.594 | 0.0009 | 0.298 |
| 41442 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-96-5p | 22 | KIF1B | Sponge network | 1.2 | 0.01813 | -0.118 | 0.32258 | 0.298 |
| 41443 | RAMP2-AS1 |
hsa-miR-126-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p | 15 | IRS1 | Sponge network | -0.513 | 0.04703 | -0.026 | 0.88855 | 0.298 |
| 41444 | ATP1B3-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p | 11 | NFIB | Sponge network | 1.2 | 0.01813 | 0.023 | 0.90795 | 0.298 |
| 41445 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-511-5p | 13 | ADAMTSL3 | Sponge network | -0.326 | 0.14314 | -1.559 | 0 | 0.298 |
| 41446 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-539-5p | 14 | MOB1B | Sponge network | 0.983 | 0.00048 | 0.197 | 0.15266 | 0.298 |
| 41447 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-93-5p | 31 | ST6GALNAC3 | Sponge network | -1.047 | 0 | -0.987 | 0 | 0.298 |
| 41448 | ZNF582-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-877-5p | 20 | CRTC1 | Sponge network | -1.349 | 0.00017 | -0.116 | 0.28412 | 0.298 |
| 41449 | USP3-AS1 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b | 14 | PRKAR1A | Sponge network | -0.162 | 0.47687 | -0.063 | 0.50314 | 0.298 |
| 41450 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-374b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-629-3p | 23 | KIF1B | Sponge network | 1.148 | 2.0E-5 | -0.118 | 0.32258 | 0.298 |
| 41451 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 11 | KCNAB1 | Sponge network | 0.16 | 0.50785 | -0.755 | 2.0E-5 | 0.298 |
| 41452 | ERVH48-1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 17 | MED12L | Sponge network | 1.04 | 0.00023 | -0.194 | 0.55299 | 0.298 |
| 41453 | RP11-526I2.5 |
hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-365a-3p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-628-5p | 15 | VPS53 | Sponge network | 1.054 | 1.0E-5 | 0.103 | 0.22667 | 0.298 |
| 41454 | NEAT1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p;hsa-miR-708-5p | 12 | MAT2A | Sponge network | 0.705 | 0.00014 | -0.073 | 0.36195 | 0.298 |
| 41455 | OIP5-AS1 |
hsa-miR-141-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | STAT5B | Sponge network | 0.421 | 0.00084 | -0.182 | 0.1082 | 0.298 |
| 41456 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-628-5p | 22 | CDC73 | Sponge network | 0.782 | 0.00016 | 0.094 | 0.20463 | 0.298 |
| 41457 | RP11-244H3.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p | 41 | RUNX1T1 | Sponge network | 0.659 | 3.0E-5 | -0.344 | 0.2374 | 0.298 |
| 41458 | TRAF3IP2-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 31 | OPCML | Sponge network | -0.323 | 0.01372 | -2.498 | 0 | 0.298 |
| 41459 | SOX2-OT |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-940 | 16 | MAPK10 | Sponge network | -1.264 | 6.0E-5 | -1.774 | 0 | 0.298 |
| 41460 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 11 | LRRK2 | Sponge network | -0.08 | 0.90092 | -1.136 | 1.0E-5 | 0.298 |
| 41461 | RP11-981G7.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ZFX | Sponge network | 0.986 | 0.01022 | 0.09 | 0.42563 | 0.298 |
| 41462 | AC108676.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p | 11 | SON | Sponge network | 4.346 | 2.0E-5 | 0.168 | 0.04406 | 0.298 |
| 41463 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 43 | KIF1B | Sponge network | -0.031 | 0.89063 | -0.118 | 0.32258 | 0.298 |
| 41464 | CTD-2336O2.1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ITPKB | Sponge network | -0.141 | 0.26434 | -0.704 | 0.00106 | 0.298 |
| 41465 | LINC00094 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-625-5p | 14 | CELF4 | Sponge network | 0.253 | 0.09565 | -1.135 | 0.00344 | 0.298 |
| 41466 | U91328.19 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 14 | ITGB1 | Sponge network | -0.303 | 0.21002 | 0.524 | 0.00017 | 0.298 |
| 41467 | AC009948.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 18 | TRIM33 | Sponge network | 0.532 | 0.00505 | 0.402 | 7.0E-5 | 0.298 |
| 41468 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | FREM2 | Sponge network | -1.575 | 6.0E-5 | -0.437 | 0.46353 | 0.298 |
| 41469 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | IGSF10 | Sponge network | 1.153 | 0.00114 | -1.751 | 1.0E-5 | 0.298 |
| 41470 | RP11-2H8.2 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-590-3p | 16 | TSC1 | Sponge network | 1.089 | 0 | 0.103 | 0.26071 | 0.298 |
| 41471 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p | 24 | RNASEL | Sponge network | -0.222 | 0.32445 | -0.272 | 0.01796 | 0.298 |
| 41472 | LIPE-AS1 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-362-3p | 12 | TMEM30A | Sponge network | 0.078 | 0.55803 | 0.096 | 0.31031 | 0.298 |
| 41473 | MIR600HG |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-940 | 24 | CREB1 | Sponge network | 0.594 | 0.41522 | 0.203 | 0.00537 | 0.298 |
| 41474 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429 | 11 | ABCA10 | Sponge network | 0.75 | 0.00054 | -0.571 | 0.03674 | 0.298 |
| 41475 | CTD-2002H8.2 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TMEM30A | Sponge network | 0.931 | 1.0E-5 | 0.096 | 0.31031 | 0.298 |
| 41476 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-654-3p;hsa-miR-664a-3p | 10 | ITCH | Sponge network | 1.03 | 0 | 0.084 | 0.34058 | 0.298 |
| 41477 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 10 | RIMS1 | Sponge network | -0.384 | 0.40652 | -3.143 | 0 | 0.298 |
| 41478 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-5p | 38 | SOX5 | Sponge network | 0.494 | 0.00339 | -1.091 | 4.0E-5 | 0.298 |
| 41479 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429 | 10 | TRIM3 | Sponge network | -0.154 | 0.53954 | -0.577 | 0 | 0.298 |
| 41480 | RP5-1085F17.3 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-664a-5p | 11 | SHPRH | Sponge network | 0.541 | 0.00125 | 0.098 | 0.40994 | 0.298 |
| 41481 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 17 | CNTNAP3 | Sponge network | -0.668 | 3.0E-5 | -1.465 | 0.00033 | 0.298 |
| 41482 | DIO3OS |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-3p | 10 | STAT5B | Sponge network | -0.773 | 0.04073 | -0.182 | 0.1082 | 0.298 |
| 41483 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-766-3p | 18 | TBL1XR1 | Sponge network | 0.986 | 0.01022 | 0.578 | 0 | 0.298 |
| 41484 | HOXB-AS3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-92a-1-5p | 17 | AKAP6 | Sponge network | 0.343 | 0.28887 | -1.586 | 3.0E-5 | 0.298 |
| 41485 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p | 17 | EYA4 | Sponge network | -0.555 | 0.06156 | 0.3 | 0.36876 | 0.298 |
| 41486 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | PPM1A | Sponge network | -0.811 | 2.0E-5 | -0.623 | 0 | 0.298 |
| 41487 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-93-5p;hsa-miR-942-5p | 14 | FBXO31 | Sponge network | -0.615 | 0.00015 | -0.085 | 0.42389 | 0.298 |
| 41488 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ARID5B | Sponge network | 0.953 | 0 | 0.342 | 0.01676 | 0.298 |
| 41489 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 41 | PDGFRA | Sponge network | -0.222 | 0.32445 | -0.372 | 0.0037 | 0.298 |
| 41490 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 33 | CNTN1 | Sponge network | 0.101 | 0.60919 | -2.594 | 0 | 0.298 |
| 41491 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | PDGFRA | Sponge network | -0.726 | 0.00132 | -0.372 | 0.0037 | 0.298 |
| 41492 | TPTEP1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NRG3 | Sponge network | -1.216 | 0 | -1.836 | 4.0E-5 | 0.298 |
| 41493 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 17 | ZFHX3 | Sponge network | 1.317 | 0 | 0.389 | 0.01363 | 0.298 |
| 41494 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | CBL | Sponge network | 0.953 | 0 | 0.291 | 0.01267 | 0.298 |
| 41495 | PCA3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | LUZP2 | Sponge network | -0.709 | 0.18168 | -0.056 | 0.89416 | 0.298 |
| 41496 | RP11-403I13.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | KCNA6 | Sponge network | 0.395 | 0.15451 | -1.531 | 0.00013 | 0.298 |
| 41497 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-589-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 41 | SMAD4 | Sponge network | 0.082 | 0.55397 | -0.313 | 0.00413 | 0.298 |
| 41498 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FSTL5 | Sponge network | -1.264 | 6.0E-5 | -1.3 | 0.01619 | 0.298 |
| 41499 | LINC00473 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SEMA3C | Sponge network | -1.203 | 0.03881 | -0.272 | 0.31153 | 0.298 |
| 41500 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-93-3p | 37 | QKI | Sponge network | -0.384 | 0.40652 | -0.333 | 0.04111 | 0.298 |
| 41501 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-98-5p | 27 | LIFR | Sponge network | -0.405 | 0.22013 | -1.794 | 0 | 0.298 |
| 41502 | MIR497HG |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-942-5p | 16 | ERRFI1 | Sponge network | -1.439 | 4.0E-5 | -0.798 | 1.0E-5 | 0.298 |
| 41503 | CTD-2583A14.8 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 11 | CBFB | Sponge network | 1.134 | 0 | 1.061 | 0 | 0.298 |
| 41504 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | MOB1B | Sponge network | 0.657 | 4.0E-5 | 0.197 | 0.15266 | 0.298 |
| 41505 | RP11-57H14.4 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p | 14 | FMNL3 | Sponge network | 0.083 | 0.66405 | 0.555 | 4.0E-5 | 0.298 |
| 41506 | DNM1P35 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-629-3p | 11 | SPTBN4 | Sponge network | 0.051 | 0.83388 | -0.786 | 0.01281 | 0.298 |
| 41507 | RP5-1085F17.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-766-3p | 21 | IL17RD | Sponge network | 0.541 | 0.00125 | -0.019 | 0.94873 | 0.298 |
| 41508 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p | 12 | SASH1 | Sponge network | 0.946 | 0 | -0.502 | 0.00175 | 0.298 |
| 41509 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | USP25 | Sponge network | -0.576 | 0.10121 | -0.242 | 0.01397 | 0.298 |
| 41510 | RP11-686D22.8 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | PCDH17 | Sponge network | 0.898 | 1.0E-5 | 0.731 | 2.0E-5 | 0.298 |
| 41511 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | TMEFF2 | Sponge network | 0.082 | 0.55654 | -2.426 | 0 | 0.298 |
| 41512 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p | 10 | CREBBP | Sponge network | 2.163 | 0 | 0.126 | 0.15186 | 0.298 |
| 41513 | RP1-302G2.5 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p | 14 | DST | Sponge network | -1.483 | 9.0E-5 | -0.713 | 0.00096 | 0.298 |
| 41514 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 55 | QKI | Sponge network | 0.246 | 0.59912 | -0.333 | 0.04111 | 0.298 |
| 41515 | LINC00173 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 29 | DTNA | Sponge network | 0.056 | 0.8494 | -1.648 | 1.0E-5 | 0.298 |
| 41516 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | BNC2 | Sponge network | 0.898 | 1.0E-5 | -0.491 | 0.09988 | 0.298 |
| 41517 | RP11-344B5.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p | 14 | KCNA6 | Sponge network | -0.055 | 0.88482 | -1.531 | 0.00013 | 0.298 |
| 41518 | RP4-669H2.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-625-5p;hsa-miR-629-3p | 17 | CBX5 | Sponge network | 1.711 | 0.00019 | 0.165 | 0.24446 | 0.298 |
| 41519 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 14 | CLASP2 | Sponge network | 0.368 | 0.20093 | -0.038 | 0.71 | 0.298 |
| 41520 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-942-5p | 22 | HLF | Sponge network | 0.381 | 0.25967 | -2.443 | 0 | 0.298 |
| 41521 | RP11-57H14.4 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | 0.083 | 0.66405 | -2.633 | 0 | 0.298 |
| 41522 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-425-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p | 39 | SOX5 | Sponge network | -1.645 | 0 | -1.091 | 4.0E-5 | 0.298 |
| 41523 | DLGAP1-AS2 |
hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p | 15 | MDM4 | Sponge network | 2.406 | 0 | 0.345 | 0.00762 | 0.298 |
| 41524 | RP11-927P21.1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p | 20 | IL17RD | Sponge network | 0.921 | 0.00118 | -0.019 | 0.94873 | 0.298 |
| 41525 | RP11-127B20.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 22 | NFIB | Sponge network | 0.411 | 0.1335 | 0.023 | 0.90795 | 0.298 |
| 41526 | RP11-53O19.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | RPS6KA3 | Sponge network | 1.205 | 0 | 0.256 | 0.02419 | 0.298 |
| 41527 | GAS6-AS2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-421 | 11 | CDH23 | Sponge network | 0.16 | 0.50785 | -0.974 | 0.00034 | 0.298 |
| 41528 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FOXO3 | Sponge network | 1.205 | 0 | -0.04 | 0.72028 | 0.298 |
| 41529 | U91328.19 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-577 | 18 | HIP1 | Sponge network | -0.303 | 0.21002 | 0.774 | 0 | 0.298 |
| 41530 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 20 | SUFU | Sponge network | -0.318 | 0.65847 | -0.04 | 0.65489 | 0.298 |
| 41531 | RP11-283I3.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-550a-3p | 13 | NRK | Sponge network | 0.614 | 1.0E-5 | 0.656 | 0.19217 | 0.298 |
| 41532 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-326 | 15 | RICTOR | Sponge network | 3.21 | 0 | 0.388 | 0.00188 | 0.298 |
| 41533 | RP1-39G22.7 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | PIK3CA | Sponge network | 0.439 | 0.00313 | 0.195 | 0.06908 | 0.298 |
| 41534 | ARHGAP5-AS1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 10 | MXI1 | Sponge network | -0.644 | 0.00021 | -0.987 | 0 | 0.298 |
| 41535 | KCNJ2-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-330-5p | 11 | ABL1 | Sponge network | -0.436 | 0.05325 | -0.215 | 0.07804 | 0.298 |
| 41536 | AC011526.1 |
hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p | 10 | CTIF | Sponge network | 0.342 | 0.03129 | -0.389 | 0.00549 | 0.298 |
| 41537 | RP11-532F6.3 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 24 | RIMS2 | Sponge network | -0.896 | 0.00099 | -1.365 | 0.00208 | 0.298 |
| 41538 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-93-5p | 11 | DAB2 | Sponge network | 0.259 | 0.12542 | 0.431 | 0.0113 | 0.298 |
| 41539 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-5p | 10 | THSD7A | Sponge network | -0.384 | 0.40652 | 0.242 | 0.25154 | 0.298 |
| 41540 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-96-5p | 11 | ELMO1 | Sponge network | -1.654 | 0.00144 | 0.04 | 0.83147 | 0.298 |
| 41541 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p | 15 | CLIP1 | Sponge network | 0.078 | 0.55803 | -0.215 | 0.08684 | 0.298 |
| 41542 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-5p | 21 | ESR1 | Sponge network | -0.737 | 0.10822 | -1.035 | 3.0E-5 | 0.298 |
| 41543 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 51 | TMEM132B | Sponge network | 0.064 | 0.72934 | -0.83 | 0.01084 | 0.298 |
| 41544 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 15 | SEMA3C | Sponge network | -1.582 | 8.0E-5 | -0.272 | 0.31153 | 0.298 |
| 41545 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | SASH1 | Sponge network | 0.335 | 0.20596 | -0.502 | 0.00175 | 0.298 |
| 41546 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-154-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-539-5p | 11 | LIMD1 | Sponge network | 0.917 | 0 | 0.103 | 0.28978 | 0.298 |
| 41547 | MRVI1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-450b-5p | 10 | ARNT2 | Sponge network | -1.092 | 4.0E-5 | -0.332 | 0.25851 | 0.298 |
| 41548 | KB-1460A1.5 |
hsa-miR-101-3p;hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-125a-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p | 28 | UBR3 | Sponge network | 0.603 | 0.00127 | -0.021 | 0.82774 | 0.298 |
| 41549 | SNX29P2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-940 | 44 | NFIB | Sponge network | 0.4 | 0.14611 | 0.023 | 0.90795 | 0.298 |
| 41550 | AGAP11 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p | 12 | CSRNP1 | Sponge network | -1.645 | 0 | -1.448 | 0 | 0.298 |
| 41551 | RP11-474O21.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | CADM2 | Sponge network | -0.023 | 0.95109 | -3.782 | 0 | 0.298 |
| 41552 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | NOTCH2NL | Sponge network | -2.46 | 0 | 0.024 | 0.84307 | 0.298 |
| 41553 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | EPHA5 | Sponge network | -0.125 | 0.49414 | -0.957 | 0.01733 | 0.298 |
| 41554 | MIR7-3HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p | 14 | PRKAA2 | Sponge network | -2.688 | 0.00397 | -2.462 | 0 | 0.298 |
| 41555 | LINC00641 |
hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ADH1B | Sponge network | -0.222 | 0.32445 | -3.716 | 0 | 0.298 |
| 41556 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p | 10 | CUL5 | Sponge network | 1.122 | 0 | 0.026 | 0.77301 | 0.298 |
| 41557 | C20orf166-AS1 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p | 13 | TBX18 | Sponge network | -2.69 | 0.0001 | 0.843 | 0.02945 | 0.298 |
| 41558 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-744-5p | 15 | FGFR1 | Sponge network | 0.494 | 0.00339 | -0.509 | 0.03957 | 0.298 |
| 41559 | LIPE-AS1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | USP9X | Sponge network | 0.078 | 0.55803 | 0.222 | 0.01689 | 0.298 |
| 41560 | MIR600HG |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-877-5p | 16 | TET1 | Sponge network | 0.594 | 0.41522 | -0.135 | 0.52138 | 0.298 |
| 41561 | RP11-218M22.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p | 14 | PLSCR4 | Sponge network | 0.197 | 0.25618 | -0.474 | 0.03833 | 0.298 |
| 41562 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | BNC2 | Sponge network | 0.349 | 0.01695 | -0.491 | 0.09988 | 0.298 |
| 41563 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-93-5p | 14 | EPAS1 | Sponge network | -0.154 | 0.78939 | -0.184 | 0.14418 | 0.298 |
| 41564 | AC109309.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-182-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b | 10 | FBXO32 | Sponge network | 0.163 | 0.87219 | -0.495 | 0.07561 | 0.298 |
| 41565 | MRPL23-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-378a-5p;hsa-miR-532-3p | 14 | PBX1 | Sponge network | -0.278 | 0.76559 | -1.206 | 0 | 0.298 |
| 41566 | ZSCAN16-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 11 | ROBO1 | Sponge network | -0.342 | 0.02288 | -0.009 | 0.96154 | 0.298 |
| 41567 | SNX29P2 |
hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p | 15 | RET | Sponge network | 0.4 | 0.14611 | -0.833 | 0.03517 | 0.298 |
| 41568 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 23 | PIK3R1 | Sponge network | 0.174 | 0.30034 | -0.133 | 0.36854 | 0.298 |
| 41569 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-654-3p | 24 | MYBL1 | Sponge network | 0.267 | 0.2937 | 0.494 | 0.02152 | 0.298 |
| 41570 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-339-5p | 12 | MYBL1 | Sponge network | -0.517 | 0.02935 | 0.494 | 0.02152 | 0.298 |
| 41571 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | YAP1 | Sponge network | 0.299 | 0.57722 | 0.22 | 0.11836 | 0.298 |
| 41572 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-96-5p | 11 | LRRC7 | Sponge network | 0.622 | 0.00015 | -1.341 | 0.01193 | 0.298 |
| 41573 | RP11-725P16.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | LUZP2 | Sponge network | 0.422 | 0.27021 | -0.056 | 0.89416 | 0.298 |
| 41574 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-421;hsa-miR-629-3p | 14 | RBL2 | Sponge network | -0.583 | 0.14589 | -0.282 | 0.00254 | 0.298 |
| 41575 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-940 | 16 | NEDD4 | Sponge network | 0.082 | 0.55397 | 0.02 | 0.89834 | 0.298 |
| 41576 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429 | 11 | PTPRB | Sponge network | -0.154 | 0.53954 | 0.105 | 0.47122 | 0.298 |
| 41577 | AC074289.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-935 | 10 | ARNT2 | Sponge network | -0.326 | 0.14314 | -0.332 | 0.25851 | 0.298 |
| 41578 | FAM157C |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-539-5p | 17 | KIF1B | Sponge network | 0.91 | 0 | -0.118 | 0.32258 | 0.298 |
| 41579 | RP11-25K19.1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 14 | CSDE1 | Sponge network | -1.057 | 0.00029 | -0.493 | 0 | 0.298 |
| 41580 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-30b-3p;hsa-miR-582-5p | 13 | CXXC4 | Sponge network | 0.792 | 0.0049 | -0.433 | 0.21055 | 0.298 |
| 41581 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-429 | 12 | KANK1 | Sponge network | 0.389 | 0.04293 | -0.55 | 0.00134 | 0.298 |
| 41582 | FAM66C |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 18 | ATM | Sponge network | -0.901 | 0.00019 | 0.178 | 0.21568 | 0.298 |
| 41583 | HNRNPU-AS1 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 20 | PIK3R1 | Sponge network | 1.601 | 0 | -0.133 | 0.36854 | 0.298 |
| 41584 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-424-5p | 20 | CBL | Sponge network | 0.368 | 0.20093 | 0.291 | 0.01267 | 0.297 |
| 41585 | RP11-264B14.1 |
hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-181a-2-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | PIK3C2A | Sponge network | 0.557 | 0.00564 | 0.061 | 0.53776 | 0.297 |
| 41586 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p | 31 | PTPRT | Sponge network | 0.335 | 0.20596 | -0.907 | 0.03727 | 0.297 |
| 41587 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-424-5p | 17 | FAT3 | Sponge network | -1.054 | 0.0706 | -1.601 | 0.00051 | 0.297 |
| 41588 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-590-3p | 11 | LRRK2 | Sponge network | -0.83 | 0 | -1.136 | 1.0E-5 | 0.297 |
| 41589 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | FAM188A | Sponge network | -1.575 | 6.0E-5 | -0.44 | 0.00018 | 0.297 |
| 41590 | CTD-3064M3.3 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-582-5p | 12 | PTCH1 | Sponge network | -0.469 | 0.08721 | -0.1 | 0.6179 | 0.297 |
| 41591 | RP11-216B9.6 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 17 | TSC1 | Sponge network | 0.174 | 0.30034 | 0.103 | 0.26071 | 0.297 |
| 41592 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-500a-3p;hsa-miR-7-1-3p | 19 | TMEM132B | Sponge network | -4.477 | 0 | -0.83 | 0.01084 | 0.297 |
| 41593 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-93-5p | 25 | JAZF1 | Sponge network | -0.318 | 0.65847 | -0.377 | 0.02677 | 0.297 |
| 41594 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | SRGAP3 | Sponge network | -0.487 | 0.02157 | -0.202 | 0.29297 | 0.297 |
| 41595 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 27 | SOX5 | Sponge network | 0.727 | 3.0E-5 | -1.091 | 4.0E-5 | 0.297 |
| 41596 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-5p | 27 | NOTCH2 | Sponge network | 0.176 | 0.37402 | 0.138 | 0.35163 | 0.297 |
| 41597 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-497-5p | 13 | SOX11 | Sponge network | 1.162 | 5.0E-5 | 2.68 | 0 | 0.297 |
| 41598 | HCG18 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p | 15 | HECA | Sponge network | 1.063 | 0 | -0.217 | 0.03463 | 0.297 |
| 41599 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 18 | PKNOX1 | Sponge network | -0.129 | 0.57177 | 0.085 | 0.28917 | 0.297 |
| 41600 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 21 | TSHZ3 | Sponge network | -0.785 | 0.00035 | -0.556 | 0.01516 | 0.297 |
| 41601 | ADAMTS9-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-7-5p | 23 | KCNJ5 | Sponge network | -2.452 | 0 | -0.281 | 0.3339 | 0.297 |
| 41602 | RP11-554A11.4 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 10 | CYLD | Sponge network | -0.583 | 0.14589 | -0.367 | 0.00171 | 0.297 |
| 41603 | LINC00963 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p | 16 | DYNC1I1 | Sponge network | 0.523 | 9.0E-5 | -1.12 | 0.00107 | 0.297 |
| 41604 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 18 | GLI3 | Sponge network | 0.219 | 0.1516 | -0.615 | 0.01482 | 0.297 |
| 41605 | RP11-25K21.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p | 14 | TCF4 | Sponge network | 0.489 | 0.25786 | 0.016 | 0.92271 | 0.297 |
| 41606 | RP11-701H24.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 13 | PTPRD | Sponge network | -1.425 | 0.04204 | -1.005 | 0.00064 | 0.297 |
| 41607 | RP11-798M19.6 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 45 | MDM4 | Sponge network | -0.612 | 0.00094 | 0.345 | 0.00762 | 0.297 |
| 41608 | AP001347.6 |
hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p | 15 | CSDE1 | Sponge network | -1.161 | 0.00591 | -0.493 | 0 | 0.297 |
| 41609 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | BNC2 | Sponge network | -0.969 | 2.0E-5 | -0.491 | 0.09988 | 0.297 |
| 41610 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-7-1-3p | 28 | CBL | Sponge network | 0.862 | 0.06213 | 0.291 | 0.01267 | 0.297 |
| 41611 | PAXIP1-AS1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p | 11 | PHF6 | Sponge network | 0.66 | 1.0E-5 | 0.785 | 0 | 0.297 |
| 41612 | RP11-119F7.5 |
hsa-miR-146a-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | ADCY1 | Sponge network | 0.225 | 0.30644 | 0.362 | 0.16982 | 0.297 |
| 41613 | ADIRF-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | -0.223 | 0.47816 | 0.321 | 0.00267 | 0.297 |
| 41614 | AGAP11 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-501-5p | 21 | RIMS2 | Sponge network | -1.645 | 0 | -1.365 | 0.00208 | 0.297 |
| 41615 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | TLL1 | Sponge network | 0.225 | 0.30644 | -1.195 | 3.0E-5 | 0.297 |
| 41616 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-590-3p | 19 | ZMYM2 | Sponge network | 0.986 | 0.01022 | 0.279 | 0.01273 | 0.297 |
| 41617 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 17 | SOX11 | Sponge network | 0.61 | 0.01593 | 2.68 | 0 | 0.297 |
| 41618 | LINC00702 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | JDP2 | Sponge network | -0.737 | 0.06182 | -0.967 | 0 | 0.297 |
| 41619 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p | 27 | APC | Sponge network | -0.162 | 0.47687 | -0.255 | 0.04051 | 0.297 |
| 41620 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-7-5p | 49 | BMPR2 | Sponge network | 0.95 | 0.15943 | 0.206 | 0.0635 | 0.297 |
| 41621 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-654-3p | 32 | FBXO32 | Sponge network | -0.371 | 0.24604 | -0.495 | 0.07561 | 0.297 |
| 41622 | A2M-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | NAV3 | Sponge network | -0.08 | 0.90092 | 0.212 | 0.47729 | 0.297 |
| 41623 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p | 13 | HIP1 | Sponge network | 1.122 | 0.02989 | 0.774 | 0 | 0.297 |
| 41624 | RP11-802E16.3 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 11 | RICTOR | Sponge network | 2.048 | 0 | 0.388 | 0.00188 | 0.297 |
| 41625 | AGAP11 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-769-5p | 31 | EGFR | Sponge network | -1.645 | 0 | -0.175 | 0.48451 | 0.297 |
| 41626 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | PRKAA2 | Sponge network | 0.619 | 0.00157 | -2.462 | 0 | 0.297 |
| 41627 | HOXB-AS3 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-452-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-625-5p;hsa-miR-744-3p | 36 | LPP | Sponge network | 0.343 | 0.28887 | -0.459 | 0.04475 | 0.297 |
| 41628 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | RASSF2 | Sponge network | 0.299 | 0.57722 | -0.273 | 0.21961 | 0.297 |
| 41629 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p | 14 | KANK1 | Sponge network | 0.494 | 0.00339 | -0.55 | 0.00134 | 0.297 |
| 41630 | RP11-283I3.6 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 12 | PRUNE2 | Sponge network | 0.614 | 1.0E-5 | -1.733 | 0.00022 | 0.297 |
| 41631 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p | 25 | KIF1B | Sponge network | 0.997 | 0 | -0.118 | 0.32258 | 0.297 |
| 41632 | ACTA2-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-590-3p | 10 | PXDN | Sponge network | 0.194 | 0.64278 | 1.038 | 0 | 0.297 |
| 41633 | CD27-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-655-3p | 28 | DMD | Sponge network | 0.177 | 0.16681 | -1.506 | 0 | 0.297 |
| 41634 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 27 | FBXO32 | Sponge network | 0.862 | 0.06213 | -0.495 | 0.07561 | 0.297 |
| 41635 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-96-5p | 12 | NUAK1 | Sponge network | -1.582 | 8.0E-5 | 0.904 | 0 | 0.297 |
| 41636 | FENDRR |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | PLA2R1 | Sponge network | -1.281 | 0.00012 | -0.265 | 0.24676 | 0.297 |
| 41637 | LINC00654 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 18 | ATM | Sponge network | 0.374 | 0.20054 | 0.178 | 0.21568 | 0.297 |
| 41638 | GHRLOS |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-361-5p | 11 | GRIN2A | Sponge network | -1.942 | 0 | -2.161 | 0 | 0.297 |
| 41639 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-369-3p | 10 | CUL5 | Sponge network | 1.262 | 0 | 0.026 | 0.77301 | 0.297 |
| 41640 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p | 11 | CLIP1 | Sponge network | 0.594 | 0.00064 | -0.215 | 0.08684 | 0.297 |
| 41641 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p | 21 | ANK2 | Sponge network | -0.322 | 0.25945 | -1.603 | 1.0E-5 | 0.297 |
| 41642 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-409-3p;hsa-miR-500a-3p;hsa-miR-96-5p | 15 | MOB1B | Sponge network | 0.453 | 0.01839 | 0.197 | 0.15266 | 0.297 |
| 41643 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p | 37 | CREB3L2 | Sponge network | -0.421 | 0.0114 | -0.525 | 2.0E-5 | 0.297 |
| 41644 | RP11-894P9.1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-505-5p | 12 | DDX3X | Sponge network | 0.993 | 0 | -0.152 | 0.07686 | 0.297 |
| 41645 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 36 | MOB1B | Sponge network | -0.615 | 0.00015 | 0.197 | 0.15266 | 0.297 |
| 41646 | RP11-144G6.12 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-582-5p;hsa-miR-590-3p | 14 | ABI2 | Sponge network | 0.826 | 1.0E-5 | 0.175 | 0.14787 | 0.297 |
| 41647 | TRIM52-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-361-5p;hsa-miR-96-5p | 10 | FNBP1 | Sponge network | -0.418 | 0.00068 | -0.969 | 4.0E-5 | 0.297 |
| 41648 | AC016747.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-744-3p | 24 | DIP2C | Sponge network | 0.199 | 0.38015 | -0.436 | 0.01713 | 0.297 |
| 41649 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 46 | ZFHX4 | Sponge network | 0.72 | 0.0001 | 0.018 | 0.95574 | 0.297 |
| 41650 | CTD-2245F17.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-339-5p | 10 | PDGFRA | Sponge network | -0.421 | 0.12556 | -0.372 | 0.0037 | 0.297 |
| 41651 | FAM66C |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | PAIP2 | Sponge network | -0.901 | 0.00019 | -0.515 | 0 | 0.297 |
| 41652 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | CDHR3 | Sponge network | 0.222 | 0.29133 | -0.977 | 4.0E-5 | 0.297 |
| 41653 | MEG3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | NOTCH2NL | Sponge network | 0.249 | 0.35902 | 0.024 | 0.84307 | 0.297 |
| 41654 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 27 | RNASEL | Sponge network | -2.46 | 0 | -0.272 | 0.01796 | 0.297 |
| 41655 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 36 | DMXL1 | Sponge network | -1.697 | 0.00889 | -0.426 | 0.00056 | 0.297 |
| 41656 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | ARNT2 | Sponge network | 0.101 | 0.60919 | -0.332 | 0.25851 | 0.297 |
| 41657 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-590-5p;hsa-miR-96-5p | 20 | SCN9A | Sponge network | 0.41 | 0.02214 | -0.525 | 0.10593 | 0.297 |
| 41658 | CTD-3092A11.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PRRX1 | Sponge network | 0.873 | 0 | 1.316 | 0 | 0.297 |
| 41659 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 17 | NIN | Sponge network | 0.174 | 0.30034 | -0.253 | 0.13794 | 0.297 |
| 41660 | CALML3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-589-3p | 28 | DIP2C | Sponge network | 0.95 | 0.15943 | -0.436 | 0.01713 | 0.297 |
| 41661 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 18 | LATS1 | Sponge network | 0.559 | 0.00026 | 0.109 | 0.34208 | 0.297 |
| 41662 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-455-5p | 10 | ABCC9 | Sponge network | 0.859 | 0.00331 | -0.466 | 0.14121 | 0.297 |
| 41663 | RP11-20I23.13 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-744-5p;hsa-miR-92a-3p | 23 | NOTCH2 | Sponge network | 0.505 | 0.0014 | 0.138 | 0.35163 | 0.297 |
| 41664 | CALML3-AS1 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-429 | 13 | RND3 | Sponge network | 0.95 | 0.15943 | -0.254 | 0.11478 | 0.297 |
| 41665 | AC016747.3 |
hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 12 | ITPKB | Sponge network | 0.199 | 0.38015 | -0.704 | 0.00106 | 0.297 |
| 41666 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 26 | IL17RD | Sponge network | 0.657 | 4.0E-5 | -0.019 | 0.94873 | 0.297 |
| 41667 | RP5-994D16.9 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DIP2C | Sponge network | 0.252 | 0.08508 | -0.436 | 0.01713 | 0.297 |
| 41668 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-374b-5p | 19 | EPHA5 | Sponge network | 0.912 | 0.00014 | -0.957 | 0.01733 | 0.297 |
| 41669 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-501-5p | 14 | UBE4B | Sponge network | 0.082 | 0.55654 | -0.235 | 0.007 | 0.297 |
| 41670 | U91328.19 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 12 | PPP2R2C | Sponge network | -0.303 | 0.21002 | -0.959 | 0.07428 | 0.297 |
| 41671 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p | 13 | BCL11A | Sponge network | 0.082 | 0.55397 | -0.932 | 0.0063 | 0.297 |
| 41672 | RP11-983P16.4 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-935;hsa-miR-940 | 32 | GRIK3 | Sponge network | 0.41 | 0.02214 | -3.208 | 0 | 0.297 |
| 41673 | CTC-429P9.3 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-505-5p | 11 | WNK2 | Sponge network | 0.082 | 0.55397 | -0.352 | 0.31066 | 0.297 |
| 41674 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | SASH1 | Sponge network | -0.141 | 0.26434 | -0.502 | 0.00175 | 0.297 |
| 41675 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-484;hsa-miR-590-3p | 13 | PKNOX1 | Sponge network | 0.72 | 0.0001 | 0.085 | 0.28917 | 0.297 |
| 41676 | RP11-6O2.3 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-3614-5p | 11 | NGFR | Sponge network | 0.331 | 0.57247 | -1.271 | 0.01058 | 0.297 |
| 41677 | RP11-13K12.1 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-375 | 10 | CTGF | Sponge network | -0.154 | 0.78939 | 0.321 | 0.11682 | 0.297 |
| 41678 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TBX18 | Sponge network | -0.323 | 0.01372 | 0.843 | 0.02945 | 0.297 |
| 41679 | RP11-1277A3.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-96-5p | 18 | SOX5 | Sponge network | 0.453 | 0.01839 | -1.091 | 4.0E-5 | 0.297 |
| 41680 | CD27-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-542-3p | 16 | MRVI1 | Sponge network | 0.177 | 0.16681 | -1.051 | 0.00022 | 0.297 |
| 41681 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | PIK3R1 | Sponge network | 0.056 | 0.8494 | -0.133 | 0.36854 | 0.297 |
| 41682 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p | 11 | CITED2 | Sponge network | -0.777 | 0.16667 | -1.545 | 0 | 0.297 |
| 41683 | RP11-218M22.1 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 11 | BCL9 | Sponge network | 0.197 | 0.25618 | 0.321 | 0.00267 | 0.297 |
| 41684 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | AKAP11 | Sponge network | 0.389 | 0.04293 | 0.196 | 0.09825 | 0.297 |
| 41685 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | LATS2 | Sponge network | -0.18 | 0.20343 | 0.159 | 0.2304 | 0.297 |
| 41686 | RP1-59D14.5 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p | 14 | AFF1 | Sponge network | 1.162 | 5.0E-5 | -0.384 | 0.00053 | 0.297 |
| 41687 | LINC01018 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | -2.915 | 0.00015 | 0.309 | 0.17079 | 0.297 |
| 41688 | RP11-195F19.9 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-628-5p | 10 | TOM1L2 | Sponge network | -0.726 | 0.00132 | -0.994 | 0 | 0.297 |
| 41689 | CTC-301O7.4 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p | 15 | GAS7 | Sponge network | 0.292 | 0.43384 | -0.555 | 0.01602 | 0.297 |
| 41690 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | PRRX1 | Sponge network | -0.563 | 0.02545 | 1.316 | 0 | 0.297 |
| 41691 | RP11-158K1.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | MED12L | Sponge network | 0.349 | 0.01695 | -0.194 | 0.55299 | 0.297 |
| 41692 | CTC-429P9.5 |
hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-93-3p | 14 | CTIF | Sponge network | 0.178 | 0.29106 | -0.389 | 0.00549 | 0.297 |
| 41693 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SEMA3E | Sponge network | 0.219 | 0.1516 | -1.717 | 0.00137 | 0.297 |
| 41694 | RP11-276H19.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-501-5p | 12 | NTRK2 | Sponge network | -0.472 | 0.48851 | -0.736 | 0.06495 | 0.297 |
| 41695 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-940 | 27 | BHLHE41 | Sponge network | 0.494 | 0.00339 | 0.353 | 0.12753 | 0.297 |
| 41696 | LINC00886 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-590-5p | 10 | PCLO | Sponge network | -0.371 | 0.24604 | -1.843 | 0.00064 | 0.297 |
| 41697 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-629-3p | 11 | ZBTB4 | Sponge network | -1.717 | 0.01522 | -0.739 | 0 | 0.297 |
| 41698 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-484;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | PPM1A | Sponge network | 0.082 | 0.55654 | -0.623 | 0 | 0.297 |
| 41699 | RP11-110I1.11 |
hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 12 | BMPR2 | Sponge network | 1.034 | 0 | 0.206 | 0.0635 | 0.297 |
| 41700 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 23 | LRIG1 | Sponge network | -0.405 | 0.22013 | -0.537 | 0.00588 | 0.297 |
| 41701 | DLG5-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p | 11 | DST | Sponge network | 0.556 | 0.07377 | -0.713 | 0.00096 | 0.297 |
| 41702 | CALML3-AS1 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p | 11 | CNTNAP1 | Sponge network | 0.95 | 0.15943 | 0.358 | 0.13727 | 0.297 |
| 41703 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | SLITRK3 | Sponge network | 0.219 | 0.1516 | -3.328 | 0 | 0.297 |
| 41704 | LINC00578 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-628-5p | 20 | FOXP1 | Sponge network | 0.811 | 0.03228 | 0.042 | 0.74368 | 0.297 |
| 41705 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p | 24 | USP6 | Sponge network | -1.575 | 6.0E-5 | 0.188 | 0.3871 | 0.297 |
| 41706 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-942-5p | 29 | NOTCH2 | Sponge network | -4.546 | 0 | 0.138 | 0.35163 | 0.297 |
| 41707 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | PHC3 | Sponge network | 0.93 | 0 | 0.212 | 0.0499 | 0.297 |
| 41708 | RP11-21A7A.2 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 24 | IL6ST | Sponge network | -1.116 | 0.23686 | -0.402 | 0.00676 | 0.297 |
| 41709 | RP11-532F6.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | MED12L | Sponge network | -0.896 | 0.00099 | -0.194 | 0.55299 | 0.297 |
| 41710 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | TMEFF2 | Sponge network | 0.064 | 0.72934 | -2.426 | 0 | 0.297 |
| 41711 | SNHG14 |
hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-874-3p;hsa-miR-96-5p | 11 | AJAP1 | Sponge network | -1.111 | 0.0003 | -0.561 | 0.04832 | 0.297 |
| 41712 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-5p | 16 | SYNE1 | Sponge network | 0.921 | 0.00118 | -0.738 | 0.00316 | 0.297 |
| 41713 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-450b-5p | 24 | GNAQ | Sponge network | -0.583 | 0.14589 | -0.367 | 0.00102 | 0.297 |
| 41714 | H19 |
hsa-miR-101-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-576-5p | 16 | FOXO3 | Sponge network | 2.439 | 0 | -0.04 | 0.72028 | 0.297 |
| 41715 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-671-5p;hsa-miR-874-3p | 18 | CREB3L2 | Sponge network | 0.598 | 0.38481 | -0.525 | 2.0E-5 | 0.297 |
| 41716 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-590-3p | 13 | CHD1 | Sponge network | 0.222 | 0.29133 | 0.322 | 0.00215 | 0.297 |
| 41717 | PSMG3-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PRUNE2 | Sponge network | -0.125 | 0.49414 | -1.733 | 0.00022 | 0.297 |
| 41718 | RP11-65J21.3 |
hsa-let-7d-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-500a-5p;hsa-miR-542-3p | 14 | DMD | Sponge network | -0.557 | 0.11135 | -1.506 | 0 | 0.297 |
| 41719 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | NTRK3 | Sponge network | -0.726 | 0.00132 | -1.77 | 3.0E-5 | 0.297 |
| 41720 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-93-3p | 23 | BMPR2 | Sponge network | -0.932 | 0.00329 | 0.206 | 0.0635 | 0.297 |
| 41721 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p | 14 | CXXC4 | Sponge network | 0.178 | 0.29106 | -0.433 | 0.21055 | 0.297 |
| 41722 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-455-5p;hsa-miR-96-5p | 17 | JAZF1 | Sponge network | 0.622 | 0.00015 | -0.377 | 0.02677 | 0.297 |
| 41723 | CALML3-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 18 | GJA1 | Sponge network | 0.95 | 0.15943 | -0.485 | 0.02179 | 0.297 |
| 41724 | RASSF8-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-7-5p | 20 | KCNJ5 | Sponge network | 0.335 | 0.20596 | -0.281 | 0.3339 | 0.297 |
| 41725 | RP3-402G11.26 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-590-3p | 10 | FAT3 | Sponge network | -0.254 | 0.19303 | -1.601 | 0.00051 | 0.297 |
| 41726 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 22 | GRIA3 | Sponge network | -0.513 | 0.04703 | -1.956 | 0 | 0.297 |
| 41727 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-497-5p | 11 | PRKAB2 | Sponge network | 2.368 | 2.0E-5 | -0.367 | 0.01371 | 0.297 |
| 41728 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 24 | CADM1 | Sponge network | 0.532 | 0.00505 | -0.9 | 0.00084 | 0.297 |
| 41729 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421 | 12 | PTPRB | Sponge network | -0.18 | 0.20343 | 0.105 | 0.47122 | 0.297 |
| 41730 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-532-3p | 10 | DAB2 | Sponge network | -0.773 | 0.04073 | 0.431 | 0.0113 | 0.297 |
| 41731 | DOCK9-AS2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-484;hsa-miR-589-5p;hsa-miR-629-3p | 16 | CBX5 | Sponge network | -0.231 | 0.28214 | 0.165 | 0.24446 | 0.297 |
| 41732 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-98-5p | 24 | KCNA6 | Sponge network | 0.082 | 0.55397 | -1.531 | 0.00013 | 0.297 |
| 41733 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-340-5p | 12 | DNAJB4 | Sponge network | -0.384 | 0.40652 | -0.7 | 1.0E-5 | 0.297 |
| 41734 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-5p;hsa-miR-625-5p | 19 | ZMYM2 | Sponge network | 0.267 | 0.2937 | 0.279 | 0.01273 | 0.297 |
| 41735 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-5p;hsa-miR-93-5p | 39 | PRKAA2 | Sponge network | -0.465 | 0.04703 | -2.462 | 0 | 0.297 |
| 41736 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-539-5p | 14 | MOB1B | Sponge network | 1.344 | 0 | 0.197 | 0.15266 | 0.297 |
| 41737 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p | 18 | DMXL1 | Sponge network | -0.358 | 0.17255 | -0.426 | 0.00056 | 0.297 |
| 41738 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p | 10 | HIP1 | Sponge network | 0.529 | 0.00552 | 0.774 | 0 | 0.297 |
| 41739 | RP11-159D12.2 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-9-5p | 15 | CDK6 | Sponge network | 1.254 | 0 | 0.991 | 0 | 0.297 |
| 41740 | RP11-182J1.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-7-5p | 18 | IGFBP5 | Sponge network | -0.187 | 0.23787 | -0.806 | 0.00105 | 0.297 |
| 41741 | SOX2-OT |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | -1.264 | 6.0E-5 | -3.143 | 0 | 0.297 |
| 41742 | LINC00578 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-653-5p;hsa-miR-664a-3p | 24 | PPM1L | Sponge network | 0.811 | 0.03228 | -0.537 | 0.00616 | 0.297 |
| 41743 | RP5-994D16.9 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | EPHA5 | Sponge network | 0.252 | 0.08508 | -0.957 | 0.01733 | 0.297 |
| 41744 | RP11-66B24.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-652-3p;hsa-miR-93-3p | 20 | QKI | Sponge network | 0.57 | 0.1866 | -0.333 | 0.04111 | 0.297 |
| 41745 | RP11-390K5.6 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 19 | LPP | Sponge network | 1.148 | 2.0E-5 | -0.459 | 0.04475 | 0.297 |
| 41746 | LINC00969 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-495-3p | 15 | MYO9A | Sponge network | 0.683 | 1.0E-5 | -0.147 | 0.32373 | 0.297 |
| 41747 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 50 | WDFY3 | Sponge network | -0.793 | 7.0E-5 | -0.201 | 0.0967 | 0.297 |
| 41748 | RP11-503E24.2 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-505-5p | 11 | ARHGEF12 | Sponge network | 2.218 | 0 | 0.09 | 0.35693 | 0.297 |
| 41749 | HCG11 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PCDH18 | Sponge network | 0.385 | 0.10067 | 0.216 | 0.24645 | 0.297 |
| 41750 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-940;hsa-miR-96-5p | 24 | SNX29 | Sponge network | -0.08 | 0.90092 | -0.34 | 0.01371 | 0.297 |
| 41751 | KB-1460A1.5 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | PCDH10 | Sponge network | 0.603 | 0.00127 | -1.644 | 0.0078 | 0.297 |
| 41752 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 17 | LRRC4 | Sponge network | 0.997 | 0.00066 | -1.774 | 0 | 0.297 |
| 41753 | CTD-2002H8.2 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | CCDC6 | Sponge network | 0.931 | 1.0E-5 | 0.27 | 0.02555 | 0.297 |
| 41754 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-25-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-942-5p | 18 | NOTCH2 | Sponge network | -0.351 | 0.11358 | 0.138 | 0.35163 | 0.297 |
| 41755 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 21 | SYT4 | Sponge network | -0.738 | 0.21233 | -3.207 | 0 | 0.297 |
| 41756 | HOXA-AS3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-96-5p | 22 | FBXO32 | Sponge network | -0.555 | 0.06156 | -0.495 | 0.07561 | 0.297 |
| 41757 | HCG11 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p | 14 | NF1 | Sponge network | 0.385 | 0.10067 | 0.51 | 0 | 0.297 |
| 41758 | RP1-302G2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-93-3p;hsa-miR-935 | 17 | SSBP2 | Sponge network | -1.483 | 9.0E-5 | -1.58 | 0 | 0.297 |
| 41759 | RP11-5C23.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 12 | MRVI1 | Sponge network | 0.274 | 0.07681 | -1.051 | 0.00022 | 0.297 |
| 41760 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-766-3p;hsa-miR-93-5p | 33 | CHD5 | Sponge network | -0.053 | 0.80685 | -0.922 | 0.01521 | 0.297 |
| 41761 | RP11-2H8.2 |
hsa-let-7g-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-766-3p | 25 | PPARA | Sponge network | 1.089 | 0 | -0.379 | 0.00548 | 0.297 |
| 41762 | CTC-297N7.9 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-301a-3p | 10 | MAF | Sponge network | -2.069 | 0 | -1.257 | 0 | 0.297 |
| 41763 | RP11-344B5.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p | 13 | ADARB1 | Sponge network | -0.055 | 0.88482 | -0.335 | 0.06631 | 0.297 |
| 41764 | CTD-3092A11.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 14 | BAZ2B | Sponge network | 0.873 | 0 | -0.276 | 0.02833 | 0.297 |
| 41765 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | FAT4 | Sponge network | -0.355 | 0.0077 | -0.354 | 0.14694 | 0.297 |
| 41766 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | SSBP2 | Sponge network | 0.056 | 0.8494 | -1.58 | 0 | 0.297 |
| 41767 | RP1-151F17.2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | BCL9 | Sponge network | 0.222 | 0.29133 | 0.321 | 0.00267 | 0.297 |
| 41768 | RP11-21A7A.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-942-5p | 12 | GPC6 | Sponge network | -1.116 | 0.23686 | -0.054 | 0.81465 | 0.297 |
| 41769 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 39 | BNC2 | Sponge network | 0.311 | 0.10344 | -0.491 | 0.09988 | 0.297 |
| 41770 | RP11-66B24.2 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-378c;hsa-miR-502-5p | 22 | DST | Sponge network | 0.57 | 0.1866 | -0.713 | 0.00096 | 0.297 |
| 41771 | RP11-344B5.2 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b | 13 | DTNA | Sponge network | -0.055 | 0.88482 | -1.648 | 1.0E-5 | 0.297 |
| 41772 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 25 | MCC | Sponge network | -0.342 | 0.02288 | -1.005 | 0 | 0.297 |
| 41773 | AF131215.9 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | PRDM5 | Sponge network | -0.322 | 0.25945 | -1.09 | 0.00014 | 0.297 |
| 41774 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | ROBO1 | Sponge network | 1.302 | 7.0E-5 | -0.009 | 0.96154 | 0.297 |
| 41775 | GS1-124K5.11 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 23 | RYR2 | Sponge network | 0.494 | 0.00339 | -1.466 | 0.00031 | 0.297 |
| 41776 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 37 | RBMS3 | Sponge network | -0.83 | 0 | -0.519 | 0.03551 | 0.297 |
| 41777 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 32 | KIF1B | Sponge network | 1.294 | 0 | -0.118 | 0.32258 | 0.297 |
| 41778 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p | 11 | TSC22D1 | Sponge network | -0.739 | 0.0574 | -0.529 | 2.0E-5 | 0.297 |
| 41779 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p | 20 | MXI1 | Sponge network | 0.056 | 0.81195 | -0.987 | 0 | 0.297 |
| 41780 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 51 | THRB | Sponge network | 1.302 | 7.0E-5 | -1.117 | 0.0004 | 0.297 |
| 41781 | RP11-254F7.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p | 10 | HOMER2 | Sponge network | 0.176 | 0.37402 | -2.698 | 0 | 0.297 |
| 41782 | TRAF3IP2-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-942-5p | 17 | ERRFI1 | Sponge network | -0.323 | 0.01372 | -0.798 | 1.0E-5 | 0.297 |
| 41783 | KB-1572G7.2 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-497-5p | 11 | CDK6 | Sponge network | 0.924 | 0 | 0.991 | 0 | 0.297 |
| 41784 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-877-5p | 23 | PGR | Sponge network | 0.659 | 3.0E-5 | -1.704 | 0 | 0.297 |
| 41785 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-942-5p | 10 | CREBBP | Sponge network | 1.106 | 0 | 0.126 | 0.15186 | 0.297 |
| 41786 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p | 10 | AMPH | Sponge network | 0.331 | 0.57247 | -0.916 | 5.0E-5 | 0.297 |
| 41787 | RP11-244O19.1 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | EPB41L3 | Sponge network | -0.096 | 0.67958 | -1.193 | 0 | 0.297 |
| 41788 | RP11-37B2.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p | 11 | ARID2 | Sponge network | 1.03 | 0 | 0.235 | 0.02697 | 0.297 |
| 41789 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 58 | NTRK3 | Sponge network | 0.082 | 0.55654 | -1.77 | 3.0E-5 | 0.297 |
| 41790 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p | 18 | FOXO3 | Sponge network | 0.368 | 0.20093 | -0.04 | 0.72028 | 0.297 |
| 41791 | C4B-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-590-5p | 12 | EYA4 | Sponge network | 2.306 | 9.0E-5 | 0.3 | 0.36876 | 0.297 |
| 41792 | TRAF3IP2-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 27 | CTIF | Sponge network | -0.323 | 0.01372 | -0.389 | 0.00549 | 0.297 |
| 41793 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-629-3p | 19 | PIK3R1 | Sponge network | -0.154 | 0.53954 | -0.133 | 0.36854 | 0.297 |
| 41794 | SOX2-OT |
hsa-miR-130b-5p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 12 | TPO | Sponge network | -1.264 | 6.0E-5 | -1.87 | 2.0E-5 | 0.297 |
| 41795 | RP11-999E24.3 |
hsa-let-7g-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 10 | ZMYND11 | Sponge network | -0.693 | 0.05629 | -0.2 | 0.05449 | 0.297 |
| 41796 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 20 | PIK3C2A | Sponge network | 1.205 | 0 | 0.061 | 0.53776 | 0.297 |
| 41797 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 20 | MYBL1 | Sponge network | 0.848 | 0 | 0.494 | 0.02152 | 0.297 |
| 41798 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | SMAD2 | Sponge network | 0.537 | 0.00139 | -0.128 | 0.12732 | 0.297 |
| 41799 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 26 | MYBL1 | Sponge network | 0.415 | 0.14657 | 0.494 | 0.02152 | 0.297 |
| 41800 | LINC00473 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-629-5p | 10 | UVRAG | Sponge network | -1.203 | 0.03881 | -0.017 | 0.83865 | 0.297 |
| 41801 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DACT1 | Sponge network | -1.161 | 0.00591 | 0.783 | 0.00072 | 0.297 |
| 41802 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-589-3p | 17 | ABCC9 | Sponge network | 0.95 | 0.15943 | -0.466 | 0.14121 | 0.297 |
| 41803 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p | 33 | ADARB1 | Sponge network | 0.222 | 0.29133 | -0.335 | 0.06631 | 0.297 |
| 41804 | SH3BP5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | ZNF208 | Sponge network | 0.267 | 0.2937 | -0.4 | 0.22381 | 0.297 |
| 41805 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429 | 19 | ERC1 | Sponge network | 0.727 | 3.0E-5 | -0.108 | 0.38794 | 0.297 |
| 41806 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-655-3p | 15 | LATS1 | Sponge network | -0.567 | 0 | 0.109 | 0.34208 | 0.297 |
| 41807 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 49 | PTPRD | Sponge network | 0.532 | 0.00505 | -1.005 | 0.00064 | 0.297 |
| 41808 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 14 | IGSF10 | Sponge network | -0.176 | 0.59692 | -1.751 | 1.0E-5 | 0.297 |
| 41809 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-376c-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 23 | IL6ST | Sponge network | 0.796 | 4.0E-5 | -0.402 | 0.00676 | 0.297 |
| 41810 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-96-5p | 16 | PAFAH1B1 | Sponge network | 0.37 | 0.27614 | -0.429 | 0 | 0.297 |
| 41811 | RP11-723D22.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-500a-5p | 10 | RUNX1T1 | Sponge network | 0.562 | 0.20117 | -0.344 | 0.2374 | 0.297 |
| 41812 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 45 | KIF1B | Sponge network | 0.16 | 0.50785 | -0.118 | 0.32258 | 0.297 |
| 41813 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p | 11 | ABCB5 | Sponge network | 0.056 | 0.81195 | -0.956 | 0.04077 | 0.297 |
| 41814 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | ABCC9 | Sponge network | 0.421 | 0.00084 | -0.466 | 0.14121 | 0.297 |
| 41815 | POLR2J4 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | HBP1 | Sponge network | -0.041 | 0.69032 | -0.454 | 0 | 0.297 |
| 41816 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 22 | ADAMTSL3 | Sponge network | -1.264 | 6.0E-5 | -1.559 | 0 | 0.297 |
| 41817 | RP11-326C3.7 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-324-3p;hsa-miR-424-5p | 10 | PTPN14 | Sponge network | 0.295 | 0.28897 | 0.144 | 0.34595 | 0.297 |
| 41818 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 25 | KIT | Sponge network | -0.777 | 0.1134 | -2.184 | 0 | 0.297 |
| 41819 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-590-3p | 14 | PNRC1 | Sponge network | -0.096 | 0.67958 | -0.646 | 0 | 0.297 |
| 41820 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 36 | ERBB4 | Sponge network | -1.349 | 0.00017 | -1.975 | 2.0E-5 | 0.297 |
| 41821 | AC025171.1 |
hsa-miR-135b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 10 | CNTNAP1 | Sponge network | 0.998 | 0 | 0.358 | 0.13727 | 0.297 |
| 41822 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LRRK2 | Sponge network | -0.355 | 0.0077 | -1.136 | 1.0E-5 | 0.297 |
| 41823 | LINC00702 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-421;hsa-miR-455-5p | 18 | PLXNC1 | Sponge network | -0.737 | 0.06182 | 0.569 | 0.00329 | 0.297 |
| 41824 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 15 | FYN | Sponge network | 0.997 | 0.00066 | -0.131 | 0.37051 | 0.297 |
| 41825 | RP11-531A24.5 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-486-5p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | 0.75 | 0.00054 | -0.226 | 0.11691 | 0.297 |
| 41826 | RP11-767N6.7 |
hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | UBR3 | Sponge network | 0.466 | 0.00155 | -0.021 | 0.82774 | 0.297 |
| 41827 | MAGI2-AS3 |
hsa-miR-140-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p | 10 | CHD6 | Sponge network | -0.129 | 0.57177 | 0.32 | 0.01326 | 0.297 |
| 41828 | PCA3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p | 12 | MNT | Sponge network | -0.709 | 0.18168 | -0.157 | 0.06598 | 0.297 |
| 41829 | AGAP11 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p | 13 | TAL1 | Sponge network | -1.645 | 0 | -0.462 | 0.00574 | 0.297 |
| 41830 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 43 | EPHA5 | Sponge network | -0.969 | 2.0E-5 | -0.957 | 0.01733 | 0.297 |
| 41831 | GAS6-AS2 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-5p | 15 | FMNL3 | Sponge network | 0.16 | 0.50785 | 0.555 | 4.0E-5 | 0.297 |
| 41832 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-222-3p;hsa-miR-339-5p | 12 | MYBL1 | Sponge network | 2.163 | 0 | 0.494 | 0.02152 | 0.297 |
| 41833 | HAR1A |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-940 | 16 | CADM2 | Sponge network | -0.672 | 0.19447 | -3.782 | 0 | 0.297 |
| 41834 | RP4-668J24.2 |
hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3913-5p;hsa-miR-511-5p | 11 | WWTR1 | Sponge network | -1.054 | 0.0706 | -0.345 | 0.11997 | 0.297 |
| 41835 | RP11-48B3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | PRRX1 | Sponge network | 1.757 | 0 | 1.316 | 0 | 0.297 |
| 41836 | HCG11 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-942-5p | 30 | IGFBP5 | Sponge network | 0.385 | 0.10067 | -0.806 | 0.00105 | 0.297 |
| 41837 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326 | 11 | HIP1 | Sponge network | 0.912 | 0.00014 | 0.774 | 0 | 0.297 |
| 41838 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-5p | 13 | SPRY3 | Sponge network | 0.997 | 0 | 0.016 | 0.91286 | 0.297 |
| 41839 | RP11-6O2.3 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-940 | 11 | MAPK10 | Sponge network | 0.331 | 0.57247 | -1.774 | 0 | 0.297 |
| 41840 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 20 | CADM1 | Sponge network | 0.299 | 0.57722 | -0.9 | 0.00084 | 0.297 |
| 41841 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | EPHA7 | Sponge network | 0.056 | 0.8494 | -2.197 | 3.0E-5 | 0.297 |
| 41842 | RP11-360F5.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-3607-3p;hsa-miR-877-5p | 13 | IL17RD | Sponge network | 1.867 | 0 | -0.019 | 0.94873 | 0.296 |
| 41843 | RP11-206L10.11 |
hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-664a-5p | 11 | LPP | Sponge network | 0.315 | 0.01647 | -0.459 | 0.04475 | 0.296 |
| 41844 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429 | 20 | ESR1 | Sponge network | -0.785 | 0.00035 | -1.035 | 3.0E-5 | 0.296 |
| 41845 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | -0.222 | 0.32445 | 0.206 | 0.06751 | 0.296 |
| 41846 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 24 | LRRC7 | Sponge network | -0.563 | 0.02545 | -1.341 | 0.01193 | 0.296 |
| 41847 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | CNTNAP3 | Sponge network | -0.227 | 0.31657 | -1.465 | 0.00033 | 0.296 |
| 41848 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | EPHA3 | Sponge network | 1.2 | 0.01813 | 0.185 | 0.53588 | 0.296 |
| 41849 | GAS6-AS2 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-590-5p;hsa-miR-874-3p;hsa-miR-935;hsa-miR-940 | 30 | GRIK3 | Sponge network | 0.16 | 0.50785 | -3.208 | 0 | 0.296 |
| 41850 | FAM215B |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | DOCK4 | Sponge network | 0.817 | 3.0E-5 | 0.502 | 0.00108 | 0.296 |
| 41851 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-330-3p;hsa-miR-508-3p;hsa-miR-590-3p | 13 | CACNA2D1 | Sponge network | 0.051 | 0.83388 | -0.846 | 0.00675 | 0.296 |
| 41852 | NNT-AS1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-766-3p | 33 | MDM4 | Sponge network | 0.799 | 1.0E-5 | 0.345 | 0.00762 | 0.296 |
| 41853 | RP11-545G3.1 |
hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-511-5p | 10 | CREB1 | Sponge network | 2.622 | 0.00025 | 0.203 | 0.00537 | 0.296 |
| 41854 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p | 12 | PTPN11 | Sponge network | 0.594 | 0.41522 | 0.431 | 2.0E-5 | 0.296 |
| 41855 | FENDRR |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | -1.281 | 0.00012 | 0.524 | 0.00017 | 0.296 |
| 41856 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 60 | TCF4 | Sponge network | 0.72 | 0.0001 | 0.016 | 0.92271 | 0.296 |
| 41857 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-935 | 21 | RARB | Sponge network | 0.572 | 0.01722 | -0.825 | 2.0E-5 | 0.296 |
| 41858 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 28 | NRXN1 | Sponge network | 0.16 | 0.50785 | -3.778 | 0 | 0.296 |
| 41859 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-93-3p;hsa-miR-96-5p | 34 | BMPR2 | Sponge network | 0.622 | 0.00015 | 0.206 | 0.0635 | 0.296 |
| 41860 | RP11-195B17.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-96-5p | 10 | HIP1 | Sponge network | 0.37 | 0.27614 | 0.774 | 0 | 0.296 |
| 41861 | HOXA-AS2 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-484 | 13 | ITGA5 | Sponge network | 0.61 | 0.01593 | 0.452 | 0.03524 | 0.296 |
| 41862 | CTD-2666L21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-3p | 15 | NAV3 | Sponge network | 0.368 | 0.20093 | 0.212 | 0.47729 | 0.296 |
| 41863 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 16 | CDH19 | Sponge network | 0.214 | 0.18246 | -3.434 | 0 | 0.296 |
| 41864 | ZNF582-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | AHNAK | Sponge network | -1.349 | 0.00017 | -1.207 | 0 | 0.296 |
| 41865 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | ARHGAP28 | Sponge network | -0.663 | 0.10887 | -0.911 | 0.00013 | 0.296 |
| 41866 | BCYRN1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ABCA10 | Sponge network | 0.311 | 0.10344 | -0.571 | 0.03674 | 0.296 |
| 41867 | AC005154.6 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | HECA | Sponge network | 0.848 | 0 | -0.217 | 0.03463 | 0.296 |
| 41868 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 25 | SOX11 | Sponge network | 0.997 | 0.00066 | 2.68 | 0 | 0.296 |
| 41869 | MEG3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 22 | PDE4DIP | Sponge network | 0.249 | 0.35902 | -0.282 | 0.0257 | 0.296 |
| 41870 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-495-3p | 16 | EGLN1 | Sponge network | 1.055 | 0 | -0.127 | 0.1536 | 0.296 |
| 41871 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p | 10 | PER2 | Sponge network | -0.285 | 0.2172 | -0.396 | 0.00269 | 0.296 |
| 41872 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 12 | PSIP1 | Sponge network | 1.11 | 0 | -0.005 | 0.96897 | 0.296 |
| 41873 | TPTEP1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | CSMD1 | Sponge network | -1.216 | 0 | -0.63 | 0.18881 | 0.296 |
| 41874 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 22 | PRKCE | Sponge network | 0.214 | 0.18246 | -0.403 | 0.00056 | 0.296 |
| 41875 | HOXA-AS2 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p | 10 | FHL1 | Sponge network | 0.61 | 0.01593 | -2.687 | 0 | 0.296 |
| 41876 | ACTA2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-375 | 12 | CACNA1E | Sponge network | 0.194 | 0.64278 | 1.752 | 0 | 0.296 |
| 41877 | MIR143HG |
hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30c-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH13 | Sponge network | -0.739 | 0.0574 | 0.634 | 0.00094 | 0.296 |
| 41878 | MAGI2-AS3 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-365a-3p;hsa-miR-375;hsa-miR-378a-5p | 13 | CACNA1E | Sponge network | -0.129 | 0.57177 | 1.752 | 0 | 0.296 |
| 41879 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-214-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 16 | PPM1A | Sponge network | 1.2 | 0.01813 | -0.623 | 0 | 0.296 |
| 41880 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 23 | LRIG1 | Sponge network | -1.654 | 0.00144 | -0.537 | 0.00588 | 0.296 |
| 41881 | LINC00094 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p | 14 | PTCH1 | Sponge network | 0.253 | 0.09565 | -0.1 | 0.6179 | 0.296 |
| 41882 | AC016747.3 |
hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | MACF1 | Sponge network | 0.199 | 0.38015 | 0.019 | 0.90335 | 0.296 |
| 41883 | RP1-102E24.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p | 14 | KIF1B | Sponge network | 1.737 | 0 | -0.118 | 0.32258 | 0.296 |
| 41884 | RP11-774O3.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | GNAQ | Sponge network | -0.487 | 0.02157 | -0.367 | 0.00102 | 0.296 |
| 41885 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | PRKAB2 | Sponge network | 0.799 | 1.0E-5 | -0.367 | 0.01371 | 0.296 |
| 41886 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 23 | HIP1 | Sponge network | -0.487 | 0.02157 | 0.774 | 0 | 0.296 |
| 41887 | SH3BP5-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-589-3p;hsa-miR-590-5p | 12 | ABCA10 | Sponge network | 0.267 | 0.2937 | -0.571 | 0.03674 | 0.296 |
| 41888 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 35 | LIFR | Sponge network | -1.697 | 0.00889 | -1.794 | 0 | 0.296 |
| 41889 | CTC-429P9.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b | 13 | KANK1 | Sponge network | 0.082 | 0.55397 | -0.55 | 0.00134 | 0.296 |
| 41890 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p | 36 | FBXO32 | Sponge network | 0.421 | 0.00084 | -0.495 | 0.07561 | 0.296 |
| 41891 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-590-3p | 13 | CNTNAP3 | Sponge network | 0.392 | 0.0278 | -1.465 | 0.00033 | 0.296 |
| 41892 | CTD-2303H24.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 13 | ZBTB20 | Sponge network | 0.415 | 0.14657 | -0.122 | 0.57569 | 0.296 |
| 41893 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-542-3p | 11 | TXNIP | Sponge network | 0.214 | 0.18246 | -1.277 | 0 | 0.296 |
| 41894 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p | 12 | CITED2 | Sponge network | -1.161 | 0.00591 | -1.545 | 0 | 0.296 |
| 41895 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 14 | SPG20 | Sponge network | 0.494 | 0.00339 | -1.16 | 0 | 0.296 |
| 41896 | AC074289.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-539-5p | 15 | PGR | Sponge network | -0.326 | 0.14314 | -1.704 | 0 | 0.296 |
| 41897 | RP11-110I1.12 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a | 11 | DDX6 | Sponge network | 0.639 | 0.00028 | 0.091 | 0.36428 | 0.296 |
| 41898 | MEG3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 19 | POU2F2 | Sponge network | 0.249 | 0.35902 | -0.233 | 0.32214 | 0.296 |
| 41899 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PBRM1 | Sponge network | 1.11 | 0 | -0.029 | 0.78601 | 0.296 |
| 41900 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-660-5p | 21 | TIMP3 | Sponge network | 0.331 | 0.57247 | -0.546 | 0.02307 | 0.296 |
| 41901 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-93-5p | 14 | CDHR3 | Sponge network | -2.69 | 0.0001 | -0.977 | 4.0E-5 | 0.296 |
| 41902 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 23 | SOX11 | Sponge network | 1.601 | 0 | 2.68 | 0 | 0.296 |
| 41903 | RP11-403I13.5 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-7-1-3p | 11 | KIF5B | Sponge network | 1.062 | 0.00512 | 0.362 | 0.00066 | 0.296 |
| 41904 | RP11-13K12.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-375 | 11 | ITGB1 | Sponge network | -0.154 | 0.78939 | 0.524 | 0.00017 | 0.296 |
| 41905 | MYLK-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 13 | FREM2 | Sponge network | -1.331 | 3.0E-5 | -0.437 | 0.46353 | 0.296 |
| 41906 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 40 | CBX5 | Sponge network | 0.41 | 0.02214 | 0.165 | 0.24446 | 0.296 |
| 41907 | HHIP-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MED12L | Sponge network | -0.737 | 0.10822 | -0.194 | 0.55299 | 0.296 |
| 41908 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 19 | PLSCR4 | Sponge network | 0.41 | 0.02214 | -0.474 | 0.03833 | 0.296 |
| 41909 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-664a-5p | 16 | SOX11 | Sponge network | 0.41 | 0.02214 | 2.68 | 0 | 0.296 |
| 41910 | RP11-158K1.3 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | USP9X | Sponge network | 0.349 | 0.01695 | 0.222 | 0.01689 | 0.296 |
| 41911 | AC009948.5 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CTGF | Sponge network | 0.532 | 0.00505 | 0.321 | 0.11682 | 0.296 |
| 41912 | RP11-263K19.4 |
hsa-let-7d-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-766-3p | 29 | MDM4 | Sponge network | 0.859 | 0.00331 | 0.345 | 0.00762 | 0.296 |
| 41913 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | ZNF292 | Sponge network | 0.657 | 4.0E-5 | 0.276 | 0.04148 | 0.296 |
| 41914 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-940 | 34 | PPP1R9A | Sponge network | -0.969 | 2.0E-5 | -0.903 | 0.04254 | 0.296 |
| 41915 | RAMP2-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | RIMS2 | Sponge network | -0.513 | 0.04703 | -1.365 | 0.00208 | 0.296 |
| 41916 | AC127904.2 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ARHGEF12 | Sponge network | 1.308 | 0 | 0.09 | 0.35693 | 0.296 |
| 41917 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 20 | ERRFI1 | Sponge network | -0.942 | 0 | -0.798 | 1.0E-5 | 0.296 |
| 41918 | CTD-2666L21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-493-5p | 13 | PDE4DIP | Sponge network | 0.368 | 0.20093 | -0.282 | 0.0257 | 0.296 |
| 41919 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-629-5p;hsa-miR-664a-3p | 47 | KIF1B | Sponge network | -2.69 | 0.0001 | -0.118 | 0.32258 | 0.296 |
| 41920 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-590-5p | 15 | ABCC9 | Sponge network | -1.047 | 0 | -0.466 | 0.14121 | 0.296 |
| 41921 | CALML3-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429 | 14 | ZMYND11 | Sponge network | 0.95 | 0.15943 | -0.2 | 0.05449 | 0.296 |
| 41922 | RP11-890B15.3 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-940;hsa-miR-96-5p | 10 | FOXO4 | Sponge network | 0.101 | 0.60919 | -0.516 | 2.0E-5 | 0.296 |
| 41923 | THAP9-AS1 |
hsa-miR-126-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-337-3p;hsa-miR-374b-5p | 11 | MOB1B | Sponge network | 1.079 | 0 | 0.197 | 0.15266 | 0.296 |
| 41924 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | CBL | Sponge network | 1.923 | 0 | 0.291 | 0.01267 | 0.296 |
| 41925 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-29a-5p | 11 | ITGAV | Sponge network | 0.078 | 0.55803 | 0.337 | 0.01002 | 0.296 |
| 41926 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | LATS1 | Sponge network | 0.702 | 7.0E-5 | 0.109 | 0.34208 | 0.296 |
| 41927 | OIP5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | AKAP6 | Sponge network | 0.421 | 0.00084 | -1.586 | 3.0E-5 | 0.296 |
| 41928 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | EDA2R | Sponge network | -2.925 | 0 | -0.587 | 0.01598 | 0.296 |
| 41929 | DNM1P35 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | ASTN1 | Sponge network | 0.051 | 0.83388 | -2.389 | 2.0E-5 | 0.296 |
| 41930 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 17 | THBS1 | Sponge network | 0.683 | 1.0E-5 | 0.741 | 0.00386 | 0.296 |
| 41931 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-576-5p | 10 | SLC6A15 | Sponge network | -0.932 | 0.00329 | -2.373 | 2.0E-5 | 0.296 |
| 41932 | ARHGEF26-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | HERC1 | Sponge network | -2.46 | 0 | -0.196 | 0.08544 | 0.296 |
| 41933 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | MCC | Sponge network | 0.72 | 0.0001 | -1.005 | 0 | 0.296 |
| 41934 | NEAT1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | ERCC6 | Sponge network | 0.705 | 0.00014 | 0.23 | 0.015 | 0.296 |
| 41935 | RP11-277P12.20 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | SYNE1 | Sponge network | 0.687 | 0.02753 | -0.738 | 0.00316 | 0.296 |
| 41936 | RP11-410L14.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-3p | 11 | AKAP6 | Sponge network | -0.052 | 0.76839 | -1.586 | 3.0E-5 | 0.296 |
| 41937 | RP11-158K1.3 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-590-3p | 16 | MRVI1 | Sponge network | 0.349 | 0.01695 | -1.051 | 0.00022 | 0.296 |
| 41938 | RP11-181G12.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-940 | 11 | SIRT1 | Sponge network | 0.556 | 0.00298 | 0.109 | 0.2593 | 0.296 |
| 41939 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-629-5p;hsa-miR-93-5p | 23 | MAF | Sponge network | 0.299 | 0.57722 | -1.257 | 0 | 0.296 |
| 41940 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ITGAV | Sponge network | 0.421 | 0.00084 | 0.337 | 0.01002 | 0.296 |
| 41941 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-92a-3p | 13 | SPTBN4 | Sponge network | -0.355 | 0.0077 | -0.786 | 0.01281 | 0.296 |
| 41942 | EMX2OS |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | CSDE1 | Sponge network | -0.777 | 0.1134 | -0.493 | 0 | 0.296 |
| 41943 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p | 13 | ATRX | Sponge network | 1.294 | 0 | 0.261 | 0.04408 | 0.296 |
| 41944 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PLSCR4 | Sponge network | 0.537 | 0.00139 | -0.474 | 0.03833 | 0.296 |
| 41945 | RP11-2B6.2 |
hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DOCK4 | Sponge network | 0.539 | 0.03722 | 0.502 | 0.00108 | 0.296 |
| 41946 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FSTL5 | Sponge network | 1.302 | 7.0E-5 | -1.3 | 0.01619 | 0.296 |
| 41947 | EIF3J-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ETV1 | Sponge network | 0.331 | 0.00509 | -0.096 | 0.67674 | 0.296 |
| 41948 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 17 | GLI3 | Sponge network | -0.355 | 0.0077 | -0.615 | 0.01482 | 0.296 |
| 41949 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-624-5p | 22 | LATS1 | Sponge network | -0.487 | 0.02157 | 0.109 | 0.34208 | 0.296 |
| 41950 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-628-5p | 15 | CDC73 | Sponge network | 0.559 | 0.00026 | 0.094 | 0.20463 | 0.296 |
| 41951 | AC025171.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | BCL9 | Sponge network | 0.998 | 0 | 0.321 | 0.00267 | 0.296 |
| 41952 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | EPHA7 | Sponge network | 0.61 | 0.01593 | -2.197 | 3.0E-5 | 0.296 |
| 41953 | CTD-2089N3.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-185-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p | 10 | PBX1 | Sponge network | -0.314 | 0.62033 | -1.206 | 0 | 0.296 |
| 41954 | AC005154.6 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ITGB1 | Sponge network | 0.848 | 0 | 0.524 | 0.00017 | 0.296 |
| 41955 | CTA-445C9.14 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-3607-3p | 10 | CUL5 | Sponge network | 0.718 | 4.0E-5 | 0.026 | 0.77301 | 0.296 |
| 41956 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 36 | PTPRD | Sponge network | 0.199 | 0.38015 | -1.005 | 0.00064 | 0.296 |
| 41957 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | DDX5 | Sponge network | 0.782 | 0.00016 | -0.099 | 0.17428 | 0.296 |
| 41958 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-877-5p | 24 | CRTC1 | Sponge network | 0.494 | 0.00339 | -0.116 | 0.28412 | 0.296 |
| 41959 | CTC-296K1.3 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-330-5p;hsa-miR-452-5p;hsa-miR-653-5p | 20 | PPM1L | Sponge network | -2.095 | 0.00112 | -0.537 | 0.00616 | 0.296 |
| 41960 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p | 12 | AKAP11 | Sponge network | 1.093 | 0 | 0.196 | 0.09825 | 0.296 |
| 41961 | TMEM161B-AS1 |
hsa-let-7e-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-3p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-590-3p;hsa-miR-628-5p | 13 | MAPK9 | Sponge network | -0.04 | 0.7606 | -0.054 | 0.54209 | 0.296 |
| 41962 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | CXXC4 | Sponge network | -2.925 | 0 | -0.433 | 0.21055 | 0.296 |
| 41963 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p | 15 | CCND2 | Sponge network | 1.969 | 0.00316 | -0.267 | 0.3112 | 0.296 |
| 41964 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-5p;hsa-miR-708-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | GRIA3 | Sponge network | -1.047 | 0 | -1.956 | 0 | 0.296 |
| 41965 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 13 | KAT6B | Sponge network | 1.757 | 0 | -0.478 | 0.00018 | 0.296 |
| 41966 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | LATS1 | Sponge network | -0.376 | 0.00337 | 0.109 | 0.34208 | 0.296 |
| 41967 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-539-5p | 14 | DDX6 | Sponge network | 1.4 | 0 | 0.091 | 0.36428 | 0.296 |
| 41968 | RP11-620J15.3 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 12 | IRS1 | Sponge network | -0.739 | 0.00044 | -0.026 | 0.88855 | 0.296 |
| 41969 | RP11-157J24.2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-576-5p | 16 | RBMS3 | Sponge network | -1.664 | 0.05165 | -0.519 | 0.03551 | 0.296 |
| 41970 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | MTSS1 | Sponge network | 0.197 | 0.25618 | -0.81 | 0.00035 | 0.296 |
| 41971 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | SLC6A15 | Sponge network | -1.281 | 0.00012 | -2.373 | 2.0E-5 | 0.296 |
| 41972 | MIR22HG |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 39 | GAS7 | Sponge network | -1.047 | 0 | -0.555 | 0.01602 | 0.296 |
| 41973 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-629-3p | 22 | EPHA7 | Sponge network | 0.368 | 0.20093 | -2.197 | 3.0E-5 | 0.296 |
| 41974 | RP3-467N11.1 |
hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-497-5p | 10 | PRKAA1 | Sponge network | 0.953 | 0 | 0.317 | 0.0031 | 0.296 |
| 41975 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 37 | ATRX | Sponge network | -2.46 | 0 | 0.261 | 0.04408 | 0.296 |
| 41976 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-493-5p;hsa-miR-495-3p | 12 | TRIM33 | Sponge network | 0.909 | 0 | 0.402 | 7.0E-5 | 0.296 |
| 41977 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 17 | CDH23 | Sponge network | 0.853 | 0.15352 | -0.974 | 0.00034 | 0.296 |
| 41978 | AC006116.24 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 20 | UNC80 | Sponge network | -0.473 | 0.07602 | -1.934 | 0 | 0.296 |
| 41979 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | LRRC4 | Sponge network | -0.738 | 0.21233 | -1.774 | 0 | 0.296 |
| 41980 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 39 | PPM1L | Sponge network | -0.421 | 0.0114 | -0.537 | 0.00616 | 0.296 |
| 41981 | RP11-226L15.5 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 24 | CREB1 | Sponge network | 0.8 | 0 | 0.203 | 0.00537 | 0.296 |
| 41982 | HOXB-AS1 |
hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p | 20 | EGFR | Sponge network | -0.154 | 0.53954 | -0.175 | 0.48451 | 0.296 |
| 41983 | RP11-35G9.3 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | CXCL12 | Sponge network | 0.657 | 4.0E-5 | -1.421 | 0 | 0.296 |
| 41984 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ZFHX3 | Sponge network | 0.619 | 0.00157 | 0.389 | 0.01363 | 0.296 |
| 41985 | RP11-48B3.4 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 16 | RIMS2 | Sponge network | 1.757 | 0 | -1.365 | 0.00208 | 0.296 |
| 41986 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SLC6A15 | Sponge network | -0.769 | 0.00035 | -2.373 | 2.0E-5 | 0.296 |
| 41987 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p | 18 | MOB1B | Sponge network | 1.117 | 0 | 0.197 | 0.15266 | 0.296 |
| 41988 | RP11-536O18.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3065-3p | 12 | CCND2 | Sponge network | -0.074 | 0.90758 | -0.267 | 0.3112 | 0.296 |
| 41989 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 11 | PDGFRA | Sponge network | 0.043 | 0.81628 | -0.372 | 0.0037 | 0.296 |
| 41990 | RP11-6O2.3 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-495-3p | 20 | CCDC6 | Sponge network | 0.331 | 0.57247 | 0.27 | 0.02555 | 0.296 |
| 41991 | CD27-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 17 | PRUNE2 | Sponge network | 0.177 | 0.16681 | -1.733 | 0.00022 | 0.296 |
| 41992 | AC034220.3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | NEDD4 | Sponge network | 0.309 | 0.07304 | 0.02 | 0.89834 | 0.296 |
| 41993 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 20 | SETBP1 | Sponge network | 0.41 | 0.02214 | -1.302 | 1.0E-5 | 0.296 |
| 41994 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 49 | BMPR2 | Sponge network | 0.4 | 0.14611 | 0.206 | 0.0635 | 0.296 |
| 41995 | LINC00863 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | ERCC4 | Sponge network | 0.189 | 0.13981 | 0.126 | 0.15266 | 0.296 |
| 41996 | CTD-2002H8.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p | 13 | FOXO3 | Sponge network | 0.931 | 1.0E-5 | -0.04 | 0.72028 | 0.296 |
| 41997 | RP11-968O1.5 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-362-3p | 27 | DTNA | Sponge network | -0.829 | 0.01343 | -1.648 | 1.0E-5 | 0.296 |
| 41998 | RP11-218M22.1 |
hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | ROBO2 | Sponge network | 0.197 | 0.25618 | 0.092 | 0.78768 | 0.296 |
| 41999 | RP11-473I1.9 |
hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ITGB3 | Sponge network | 0.683 | 1.0E-5 | -0.011 | 0.95751 | 0.296 |
| 42000 | LOXL1-AS1 |
hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | DIP2C | Sponge network | 0.487 | 0.04053 | -0.436 | 0.01713 | 0.296 |
| 42001 | PGM5-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 16 | JDP2 | Sponge network | -4.546 | 0 | -0.967 | 0 | 0.296 |
| 42002 | RP11-2B6.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-629-3p | 11 | PTPN14 | Sponge network | 0.539 | 0.03722 | 0.144 | 0.34595 | 0.296 |
| 42003 | RP11-25K19.1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 13 | KLF10 | Sponge network | -1.057 | 0.00029 | -0.783 | 0 | 0.296 |
| 42004 | LINC00473 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -1.203 | 0.03881 | -0.966 | 0.00035 | 0.296 |
| 42005 | BCYRN1 |
hsa-miR-10b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 10 | EXTL2 | Sponge network | 0.311 | 0.10344 | 0.109 | 0.37375 | 0.296 |
| 42006 | LINC00173 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p | 10 | SPG20 | Sponge network | 0.056 | 0.8494 | -1.16 | 0 | 0.296 |
| 42007 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 26 | ZFX | Sponge network | 0.082 | 0.55654 | 0.09 | 0.42563 | 0.296 |
| 42008 | HOXD-AS2 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-502-5p | 20 | TCF4 | Sponge network | -0.208 | 0.65592 | 0.016 | 0.92271 | 0.296 |
| 42009 | CTC-471J1.2 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p | 13 | UBR3 | Sponge network | 1.11 | 1.0E-5 | -0.021 | 0.82774 | 0.296 |
| 42010 | RP4-758J18.10 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p | 12 | BRD3 | Sponge network | -0.091 | 0.71468 | 0.37 | 0.00013 | 0.296 |
| 42011 | RP13-516M14.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p | 15 | PRDM2 | Sponge network | 0.389 | 0.04293 | -0.258 | 0.00721 | 0.296 |
| 42012 | PCA3 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -0.709 | 0.18168 | 0.024 | 0.89889 | 0.296 |
| 42013 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-769-5p | 23 | EGFR | Sponge network | -0.583 | 0.14589 | -0.175 | 0.48451 | 0.296 |
| 42014 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | DDX5 | Sponge network | 0.559 | 0.00026 | -0.099 | 0.17428 | 0.296 |
| 42015 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 34 | CBX5 | Sponge network | 0.889 | 9.0E-5 | 0.165 | 0.24446 | 0.296 |
| 42016 | SNHG12 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p | 10 | UBE2D1 | Sponge network | 1.103 | 0 | 0.468 | 0 | 0.296 |
| 42017 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | MITF | Sponge network | -1.349 | 0.00017 | -0.769 | 0.00022 | 0.296 |
| 42018 | AC141928.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-532-3p | 13 | MRVI1 | Sponge network | 0.853 | 0.15352 | -1.051 | 0.00022 | 0.296 |
| 42019 | LINC00173 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 15 | RIMS2 | Sponge network | 0.056 | 0.8494 | -1.365 | 0.00208 | 0.296 |
| 42020 | FAM13A-AS1 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-328-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | TMTC2 | Sponge network | -0.83 | 0 | -0.215 | 0.18248 | 0.296 |
| 42021 | RP4-717I23.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-671-5p | 15 | CYLD | Sponge network | 0.637 | 0.00069 | -0.367 | 0.00171 | 0.296 |
| 42022 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-590-3p | 13 | MAP3K12 | Sponge network | -1.161 | 0.00591 | -0.057 | 0.59369 | 0.296 |
| 42023 | AC005154.6 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ERCC4 | Sponge network | 0.848 | 0 | 0.126 | 0.15266 | 0.296 |
| 42024 | PART1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | ITGB3 | Sponge network | -2.925 | 0 | -0.011 | 0.95751 | 0.296 |
| 42025 | HCG11 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 12 | PLAG1 | Sponge network | 0.385 | 0.10067 | -0.315 | 0.26532 | 0.296 |
| 42026 | DNM3OS |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | MED12L | Sponge network | 0.572 | 0.01722 | -0.194 | 0.55299 | 0.296 |
| 42027 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-5p | 13 | CRTC3 | Sponge network | 1.04 | 0.00023 | 0.164 | 0.16786 | 0.296 |
| 42028 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-550a-3p | 16 | CBX5 | Sponge network | 1.157 | 0.00455 | 0.165 | 0.24446 | 0.296 |
| 42029 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 48 | NTRK3 | Sponge network | -0.08 | 0.90092 | -1.77 | 3.0E-5 | 0.296 |
| 42030 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | SEMA3E | Sponge network | -0.969 | 2.0E-5 | -1.717 | 0.00137 | 0.296 |
| 42031 | RP4-584D14.7 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-577 | 15 | IL17RD | Sponge network | 1.294 | 0.0031 | -0.019 | 0.94873 | 0.296 |
| 42032 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | MOB1B | Sponge network | 0.34 | 0.00273 | 0.197 | 0.15266 | 0.296 |
| 42033 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | AHNAK | Sponge network | 0.349 | 0.01695 | -1.207 | 0 | 0.296 |
| 42034 | AC005618.6 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 44 | DTNA | Sponge network | 0.862 | 0.06213 | -1.648 | 1.0E-5 | 0.296 |
| 42035 | AC006262.5 |
hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-497-5p | 10 | BRD3 | Sponge network | 5.342 | 0 | 0.37 | 0.00013 | 0.296 |
| 42036 | RP11-212P7.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | PARK2 | Sponge network | 0.219 | 0.1516 | -1.892 | 0 | 0.296 |
| 42037 | RP11-395A13.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-29a-5p;hsa-miR-301a-3p | 10 | ABI2 | Sponge network | 0.057 | 0.61225 | 0.175 | 0.14787 | 0.296 |
| 42038 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-3615;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-5p | 11 | DMTF1 | Sponge network | 0.249 | 0.35902 | 0.248 | 0.02726 | 0.296 |
| 42039 | RP11-983P16.4 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | TAL1 | Sponge network | 0.41 | 0.02214 | -0.462 | 0.00574 | 0.296 |
| 42040 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-660-5p;hsa-miR-940;hsa-miR-98-5p | 18 | ZFP36L1 | Sponge network | -0.773 | 0.04073 | -0.505 | 8.0E-5 | 0.296 |
| 42041 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-940 | 12 | CHD5 | Sponge network | 0.272 | 0.44692 | -0.922 | 0.01521 | 0.296 |
| 42042 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 14 | TMEFF2 | Sponge network | 0.16 | 0.50785 | -2.426 | 0 | 0.296 |
| 42043 | AC002117.1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CREB1 | Sponge network | 0.476 | 0.19203 | 0.203 | 0.00537 | 0.296 |
| 42044 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-590-3p | 12 | PKNOX1 | Sponge network | -0.487 | 0.02157 | 0.085 | 0.28917 | 0.296 |
| 42045 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p | 14 | SPG20 | Sponge network | 0.61 | 0.01593 | -1.16 | 0 | 0.296 |
| 42046 | LINC00403 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-424-5p | 12 | KCNA6 | Sponge network | -3.586 | 0.00032 | -1.531 | 0.00013 | 0.296 |
| 42047 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 24 | RAP1A | Sponge network | -1.161 | 0.00591 | -0.929 | 0 | 0.296 |
| 42048 | RP11-428J1.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NRXN3 | Sponge network | 0.538 | 0.00076 | -0.893 | 0.04186 | 0.296 |
| 42049 | LINC00472 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -0.842 | 0.01361 | -0.966 | 0.00035 | 0.296 |
| 42050 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-7-5p;hsa-miR-98-5p | 13 | KCNA6 | Sponge network | -0.295 | 0.02604 | -1.531 | 0.00013 | 0.296 |
| 42051 | RP11-693J15.4 |
hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | NRXN3 | Sponge network | -0.72 | 0.24239 | -0.893 | 0.04186 | 0.296 |
| 42052 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-590-5p | 17 | PRKAB2 | Sponge network | 0.921 | 0.00118 | -0.367 | 0.01371 | 0.296 |
| 42053 | LINC00654 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 23 | CDH19 | Sponge network | 0.374 | 0.20054 | -3.434 | 0 | 0.296 |
| 42054 | CTD-3064M3.3 |
hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-708-5p | 18 | RAP1A | Sponge network | -0.469 | 0.08721 | -0.929 | 0 | 0.296 |
| 42055 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | DAB2 | Sponge network | 0.214 | 0.18246 | 0.431 | 0.0113 | 0.296 |
| 42056 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-96-5p;hsa-miR-98-5p | 33 | CREB3L2 | Sponge network | -0.18 | 0.20343 | -0.525 | 2.0E-5 | 0.296 |
| 42057 | LINC00957 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | CCND2 | Sponge network | -0.421 | 0.0114 | -0.267 | 0.3112 | 0.296 |
| 42058 | NR2F1-AS1 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 19 | JUN | Sponge network | -0.49 | 0.04946 | -0.932 | 0 | 0.296 |
| 42059 | KB-1410C5.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p | 10 | SPRY3 | Sponge network | 0.959 | 0 | 0.016 | 0.91286 | 0.296 |
| 42060 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 22 | ARNT2 | Sponge network | 0.572 | 0.01722 | -0.332 | 0.25851 | 0.296 |
| 42061 | RP11-206L10.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 13 | CCDC6 | Sponge network | 0.793 | 0 | 0.27 | 0.02555 | 0.296 |
| 42062 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 18 | MAP3K12 | Sponge network | -2.925 | 0 | -0.057 | 0.59369 | 0.296 |
| 42063 | RP11-434B12.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 37 | CADM2 | Sponge network | -0.031 | 0.89063 | -3.782 | 0 | 0.296 |
| 42064 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-7-5p | 39 | RBMS3 | Sponge network | 0.253 | 0.09565 | -0.519 | 0.03551 | 0.296 |
| 42065 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-5p | 29 | USP12 | Sponge network | -0.615 | 0.00015 | -0.102 | 0.40768 | 0.296 |
| 42066 | HHIP-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 34 | CREB3L2 | Sponge network | -0.737 | 0.10822 | -0.525 | 2.0E-5 | 0.296 |
| 42067 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-628-5p;hsa-miR-758-3p | 14 | CDC73 | Sponge network | 1.122 | 0 | 0.094 | 0.20463 | 0.296 |
| 42068 | RP11-395G23.3 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | CADM1 | Sponge network | 0.381 | 0.25967 | -0.9 | 0.00084 | 0.296 |
| 42069 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 14 | FREM2 | Sponge network | -1.203 | 0.03881 | -0.437 | 0.46353 | 0.296 |
| 42070 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p | 10 | NIN | Sponge network | 0.912 | 0.00014 | -0.253 | 0.13794 | 0.296 |
| 42071 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | CDC73 | Sponge network | 0.34 | 0.00273 | 0.094 | 0.20463 | 0.296 |
| 42072 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CLIP1 | Sponge network | 0.603 | 0.00127 | -0.215 | 0.08684 | 0.296 |
| 42073 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 30 | NOTCH2 | Sponge network | 0.199 | 0.38015 | 0.138 | 0.35163 | 0.296 |
| 42074 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PRKAB2 | Sponge network | 0.272 | 0.44692 | -0.367 | 0.01371 | 0.296 |
| 42075 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | CDHR3 | Sponge network | -0.942 | 0 | -0.977 | 4.0E-5 | 0.296 |
| 42076 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-134-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 11 | PIK3C2A | Sponge network | 0.545 | 0.05789 | 0.061 | 0.53776 | 0.296 |
| 42077 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-338-5p;hsa-miR-590-3p | 10 | SPRY3 | Sponge network | 1.378 | 0 | 0.016 | 0.91286 | 0.296 |
| 42078 | CTC-559E9.5 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p | 11 | DOCK4 | Sponge network | 0.594 | 0.00064 | 0.502 | 0.00108 | 0.296 |
| 42079 | AP001347.6 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-501-5p;hsa-miR-590-3p | 18 | CTIF | Sponge network | -1.161 | 0.00591 | -0.389 | 0.00549 | 0.296 |
| 42080 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p | 10 | PDS5B | Sponge network | 0.556 | 0.00298 | 0.259 | 0.00962 | 0.296 |
| 42081 | AC005562.1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | USP9X | Sponge network | 0.488 | 0.00084 | 0.222 | 0.01689 | 0.296 |
| 42082 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | BCLAF1 | Sponge network | 1.03 | 0 | 0.211 | 0.00684 | 0.295 |
| 42083 | EMX2OS |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | PAIP2 | Sponge network | -0.777 | 0.1134 | -0.515 | 0 | 0.295 |
| 42084 | KB-1572G7.2 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p | 11 | DICER1 | Sponge network | 0.924 | 0 | 0.042 | 0.674 | 0.295 |
| 42085 | RP11-158K1.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | STARD13 | Sponge network | 0.349 | 0.01695 | -0.187 | 0.27383 | 0.295 |
| 42086 | LINC00865 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 27 | PRKCB | Sponge network | -1.249 | 7.0E-5 | -1.313 | 2.0E-5 | 0.295 |
| 42087 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CLIP1 | Sponge network | 0.545 | 0.00064 | -0.215 | 0.08684 | 0.295 |
| 42088 | MEG3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 31 | AFF1 | Sponge network | 0.249 | 0.35902 | -0.384 | 0.00053 | 0.295 |
| 42089 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p | 15 | SON | Sponge network | 0.082 | 0.55397 | 0.168 | 0.04406 | 0.295 |
| 42090 | FENDRR |
hsa-miR-136-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374a-5p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-7-5p | 10 | MLLT11 | Sponge network | -1.281 | 0.00012 | -0.661 | 0.01733 | 0.295 |
| 42091 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | RCAN2 | Sponge network | 0.395 | 0.15451 | -0.33 | 0.15172 | 0.295 |
| 42092 | A1BG-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-3p;hsa-miR-34a-5p | 14 | NOTCH2 | Sponge network | -0.164 | 0.45106 | 0.138 | 0.35163 | 0.295 |
| 42093 | CTD-2554C21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-93-5p | 11 | FBXO31 | Sponge network | -1.654 | 0.00144 | -0.085 | 0.42389 | 0.295 |
| 42094 | RP11-464F9.1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | USP9X | Sponge network | 0.959 | 0 | 0.222 | 0.01689 | 0.295 |
| 42095 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 30 | CXXC4 | Sponge network | -0.739 | 0.0574 | -0.433 | 0.21055 | 0.295 |
| 42096 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 33 | TMEM132B | Sponge network | -0.727 | 0.04726 | -0.83 | 0.01084 | 0.295 |
| 42097 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p | 26 | FOXO3 | Sponge network | 0.61 | 0.01593 | -0.04 | 0.72028 | 0.295 |
| 42098 | U91328.19 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p;hsa-miR-935 | 18 | ARNT2 | Sponge network | -0.303 | 0.21002 | -0.332 | 0.25851 | 0.295 |
| 42099 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 24 | MOB1B | Sponge network | 0.197 | 0.25618 | 0.197 | 0.15266 | 0.295 |
| 42100 | A2M-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | PCDH18 | Sponge network | -0.08 | 0.90092 | 0.216 | 0.24645 | 0.295 |
| 42101 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | SON | Sponge network | 0.8 | 0 | 0.168 | 0.04406 | 0.295 |
| 42102 | RP11-499P20.2 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-337-3p;hsa-miR-409-3p;hsa-miR-539-5p | 11 | MOB1B | Sponge network | 0.187 | 0.44121 | 0.197 | 0.15266 | 0.295 |
| 42103 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-93-3p;hsa-miR-96-5p | 18 | PCDH10 | Sponge network | -0.18 | 0.20343 | -1.644 | 0.0078 | 0.295 |
| 42104 | AC003090.1 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RTN4 | Sponge network | -2.117 | 0.00161 | -0.412 | 0.00014 | 0.295 |
| 42105 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p | 15 | CREB3L2 | Sponge network | -1.483 | 9.0E-5 | -0.525 | 2.0E-5 | 0.295 |
| 42106 | RP11-83A24.2 |
hsa-let-7g-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-409-3p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-940 | 29 | PPARA | Sponge network | 0.417 | 0.00445 | -0.379 | 0.00548 | 0.295 |
| 42107 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 25 | MXI1 | Sponge network | -0.969 | 2.0E-5 | -0.987 | 0 | 0.295 |
| 42108 | RP11-359B12.2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-429;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 15 | TES | Sponge network | 0.064 | 0.72934 | -0.348 | 0.00639 | 0.295 |
| 42109 | RP11-927P21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p | 16 | SETBP1 | Sponge network | 0.921 | 0.00118 | -1.302 | 1.0E-5 | 0.295 |
| 42110 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PBRM1 | Sponge network | 0.569 | 0.00067 | -0.029 | 0.78601 | 0.295 |
| 42111 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p | 38 | THRB | Sponge network | 0.051 | 0.83388 | -1.117 | 0.0004 | 0.295 |
| 42112 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | YAP1 | Sponge network | -0.031 | 0.89063 | 0.22 | 0.11836 | 0.295 |
| 42113 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-653-5p | 30 | DMXL1 | Sponge network | -0.615 | 0.00015 | -0.426 | 0.00056 | 0.295 |
| 42114 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-590-3p | 15 | ANK3 | Sponge network | 0.72 | 0.0001 | -0.55 | 0.00086 | 0.295 |
| 42115 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | MITF | Sponge network | 0.395 | 0.15451 | -0.769 | 0.00022 | 0.295 |
| 42116 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 28 | CADM1 | Sponge network | -0.769 | 0.00035 | -0.9 | 0.00084 | 0.295 |
| 42117 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 15 | LRRC55 | Sponge network | -0.405 | 0.22013 | -0.026 | 0.93163 | 0.295 |
| 42118 | RP11-428J1.5 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 14 | DYNC1I1 | Sponge network | 0.538 | 0.00076 | -1.12 | 0.00107 | 0.295 |
| 42119 | AC156455.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-500b-5p | 14 | SCN9A | Sponge network | 1.154 | 0.00041 | -0.525 | 0.10593 | 0.295 |
| 42120 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 22 | CBX5 | Sponge network | 0.411 | 0.1335 | 0.165 | 0.24446 | 0.295 |
| 42121 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-96-5p | 14 | DOCK4 | Sponge network | 0.331 | 0.57247 | 0.502 | 0.00108 | 0.295 |
| 42122 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-582-3p;hsa-miR-590-3p | 22 | PRDM2 | Sponge network | 0.214 | 0.18246 | -0.258 | 0.00721 | 0.295 |
| 42123 | CKMT2-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 14 | ZBTB20 | Sponge network | 0.082 | 0.55654 | -0.122 | 0.57569 | 0.295 |
| 42124 | LINC00641 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p | 34 | BTG2 | Sponge network | -0.222 | 0.32445 | -1.763 | 0 | 0.295 |
| 42125 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | MLLT10 | Sponge network | 0.657 | 4.0E-5 | 0.105 | 0.33292 | 0.295 |
| 42126 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 10 | EBF1 | Sponge network | 0.545 | 0.05789 | -0.384 | 0.08856 | 0.295 |
| 42127 | U91328.19 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-940 | 13 | SORCS1 | Sponge network | -0.303 | 0.21002 | -3.492 | 0 | 0.295 |
| 42128 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 10 | TSLP | Sponge network | -0.777 | 0.1134 | -2.209 | 0 | 0.295 |
| 42129 | MALAT1 |
hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-214-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-652-3p;hsa-miR-940 | 11 | SLC12A6 | Sponge network | 0.782 | 0.00016 | -0.343 | 0.00043 | 0.295 |
| 42130 | CTD-2666L21.1 |
hsa-miR-142-3p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-455-3p | 10 | SIRT1 | Sponge network | 0.368 | 0.20093 | 0.109 | 0.2593 | 0.295 |
| 42131 | FAM66C |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 27 | CTIF | Sponge network | -0.901 | 0.00019 | -0.389 | 0.00549 | 0.295 |
| 42132 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-5p | 29 | ESR1 | Sponge network | 0.395 | 0.15451 | -1.035 | 3.0E-5 | 0.295 |
| 42133 | RP11-597D13.9 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-584-5p;hsa-miR-942-5p | 10 | BMPR1A | Sponge network | 1.302 | 7.0E-5 | -0.366 | 0.01036 | 0.295 |
| 42134 | HHIP-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 51 | LPP | Sponge network | -0.737 | 0.10822 | -0.459 | 0.04475 | 0.295 |
| 42135 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-576-5p | 27 | ZFX | Sponge network | -0.162 | 0.47687 | 0.09 | 0.42563 | 0.295 |
| 42136 | KB-1572G7.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-539-5p | 12 | ABI2 | Sponge network | 0.924 | 0 | 0.175 | 0.14787 | 0.295 |
| 42137 | A2M-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | INPP4A | Sponge network | -0.08 | 0.90092 | 0.07 | 0.53563 | 0.295 |
| 42138 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-7-1-3p | 14 | NCOA1 | Sponge network | -4.477 | 0 | -0.432 | 0 | 0.295 |
| 42139 | RP11-395G23.3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-625-5p | 10 | SFRP1 | Sponge network | 0.381 | 0.25967 | -3.566 | 0 | 0.295 |
| 42140 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | KANK1 | Sponge network | -0.739 | 0.0574 | -0.55 | 0.00134 | 0.295 |
| 42141 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421 | 10 | CITED2 | Sponge network | -0.351 | 0.11358 | -1.545 | 0 | 0.295 |
| 42142 | ERVK3-1 |
hsa-miR-101-5p;hsa-miR-139-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p;hsa-miR-655-3p | 10 | NF1 | Sponge network | 0.58 | 0 | 0.51 | 0 | 0.295 |
| 42143 | RP4-668J24.2 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-942-5p | 11 | OPCML | Sponge network | -1.054 | 0.0706 | -2.498 | 0 | 0.295 |
| 42144 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-590-3p | 10 | FGF5 | Sponge network | 0.342 | 0.03129 | 0.549 | 0.28659 | 0.295 |
| 42145 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-98-5p | 19 | KCNA6 | Sponge network | 1.344 | 0 | -1.531 | 0.00013 | 0.295 |
| 42146 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 12 | BHLHE41 | Sponge network | -0.021 | 0.93598 | 0.353 | 0.12753 | 0.295 |
| 42147 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 55 | THRB | Sponge network | -1.264 | 6.0E-5 | -1.117 | 0.0004 | 0.295 |
| 42148 | RP4-639F20.1 |
hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-500a-3p;hsa-miR-96-5p | 12 | ARHGAP29 | Sponge network | -0.517 | 0.02935 | 0.131 | 0.42953 | 0.295 |
| 42149 | LENG8-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p | 10 | PIK3C2A | Sponge network | 1.471 | 0 | 0.061 | 0.53776 | 0.295 |
| 42150 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | KIF1B | Sponge network | -0.187 | 0.23787 | -0.118 | 0.32258 | 0.295 |
| 42151 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-582-3p;hsa-miR-582-5p | 24 | TCF4 | Sponge network | 0.993 | 0 | 0.016 | 0.92271 | 0.295 |
| 42152 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 36 | PTPRT | Sponge network | 0.997 | 0.00066 | -0.907 | 0.03727 | 0.295 |
| 42153 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 55 | THRB | Sponge network | -0.355 | 0.0077 | -1.117 | 0.0004 | 0.295 |
| 42154 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-629-3p | 38 | KIF1B | Sponge network | -0.154 | 0.53954 | -0.118 | 0.32258 | 0.295 |
| 42155 | RAMP2-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-629-3p;hsa-miR-92a-3p | 10 | SPTBN4 | Sponge network | -0.513 | 0.04703 | -0.786 | 0.01281 | 0.295 |
| 42156 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 33 | SPRY3 | Sponge network | 0.349 | 0.01695 | 0.016 | 0.91286 | 0.295 |
| 42157 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 16 | CREBBP | Sponge network | -0.235 | 0.15142 | 0.126 | 0.15186 | 0.295 |
| 42158 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p | 11 | ZYX | Sponge network | -0.469 | 0.08721 | 0.019 | 0.89763 | 0.295 |
| 42159 | RP11-359E10.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 37 | WDFY3 | Sponge network | 0.299 | 0.57722 | -0.201 | 0.0967 | 0.295 |
| 42160 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | APC | Sponge network | 0.572 | 0.01722 | -0.255 | 0.04051 | 0.295 |
| 42161 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-590-3p | 17 | KIT | Sponge network | -0.227 | 0.31657 | -2.184 | 0 | 0.295 |
| 42162 | RP11-428J1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p | 15 | ERCC4 | Sponge network | 0.538 | 0.00076 | 0.126 | 0.15266 | 0.295 |
| 42163 | LINC00657 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p | 13 | PAFAH1B2 | Sponge network | 0.91 | 0 | 0.318 | 0.0001 | 0.295 |
| 42164 | AC108488.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p | 11 | INPP4B | Sponge network | 0.259 | 0.12542 | -0.097 | 0.60503 | 0.295 |
| 42165 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p | 12 | PBRM1 | Sponge network | 0.494 | 0.00339 | -0.029 | 0.78601 | 0.295 |
| 42166 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p | 10 | USP6 | Sponge network | 0.983 | 0.00048 | 0.188 | 0.3871 | 0.295 |
| 42167 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p | 29 | PTPRD | Sponge network | 0.176 | 0.37402 | -1.005 | 0.00064 | 0.295 |
| 42168 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 21 | ST6GAL2 | Sponge network | -0.342 | 0.02288 | -1.201 | 0.00597 | 0.295 |
| 42169 | CTD-2554C21.2 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-629-5p;hsa-miR-96-5p | 17 | PCDH17 | Sponge network | -1.654 | 0.00144 | 0.731 | 2.0E-5 | 0.295 |
| 42170 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-96-5p | 27 | MTSS1 | Sponge network | -0.235 | 0.15142 | -0.81 | 0.00035 | 0.295 |
| 42171 | RP11-490M8.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-539-5p | 18 | THRB | Sponge network | -0.214 | 0.44248 | -1.117 | 0.0004 | 0.295 |
| 42172 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-342-3p | 10 | ZNRF3 | Sponge network | 0.569 | 0.00067 | 0.637 | 0.00172 | 0.295 |
| 42173 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-425-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ATRX | Sponge network | 0.862 | 0.06213 | 0.261 | 0.04408 | 0.295 |
| 42174 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 13 | PIK3CA | Sponge network | 0.299 | 0.57722 | 0.195 | 0.06908 | 0.295 |
| 42175 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-493-5p | 10 | LATS1 | Sponge network | 1.46 | 1.0E-5 | 0.109 | 0.34208 | 0.295 |
| 42176 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 25 | MED12L | Sponge network | 0.75 | 0.00054 | -0.194 | 0.55299 | 0.295 |
| 42177 | BCYRN1 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PHF3 | Sponge network | 0.311 | 0.10344 | 0.126 | 0.19652 | 0.295 |
| 42178 | KB-1460A1.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | FAM172A | Sponge network | 0.603 | 0.00127 | -0.57 | 0 | 0.295 |
| 42179 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 15 | PTPN11 | Sponge network | 0.494 | 0.00339 | 0.431 | 2.0E-5 | 0.295 |
| 42180 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | NEDD4 | Sponge network | -0.563 | 0.02545 | 0.02 | 0.89834 | 0.295 |
| 42181 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 30 | LIFR | Sponge network | -0.83 | 0 | -1.794 | 0 | 0.295 |
| 42182 | RP11-67L2.2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-590-3p | 23 | RYR2 | Sponge network | 0.72 | 0.0001 | -1.466 | 0.00031 | 0.295 |
| 42183 | RP11-73M18.8 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p | 10 | UBE2D1 | Sponge network | 0.832 | 0 | 0.468 | 0 | 0.295 |
| 42184 | NR2F1-AS1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 12 | DNMT3A | Sponge network | -0.49 | 0.04946 | 0.302 | 0.0262 | 0.295 |
| 42185 | TRIM52-AS1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-96-5p | 24 | GAS7 | Sponge network | -0.418 | 0.00068 | -0.555 | 0.01602 | 0.295 |
| 42186 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 29 | USP12 | Sponge network | -0.976 | 0.00025 | -0.102 | 0.40768 | 0.295 |
| 42187 | H1FX-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | RCAN2 | Sponge network | -0.206 | 0.12942 | -0.33 | 0.15172 | 0.295 |
| 42188 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZNF844 | Sponge network | 0.997 | 0.00066 | -0.966 | 0.00035 | 0.295 |
| 42189 | AGAP11 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-5p | 16 | CITED2 | Sponge network | -1.645 | 0 | -1.545 | 0 | 0.295 |
| 42190 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-664a-3p | 17 | KIF1B | Sponge network | 1.316 | 0 | -0.118 | 0.32258 | 0.295 |
| 42191 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-590-5p | 12 | ACSL6 | Sponge network | -2.076 | 0.00548 | -1.593 | 0 | 0.295 |
| 42192 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 63 | THRB | Sponge network | -0.777 | 0.1134 | -1.117 | 0.0004 | 0.295 |
| 42193 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | 0.421 | 0.00084 | -0.7 | 1.0E-5 | 0.295 |
| 42194 | FAM215B |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-7-1-3p | 16 | MED12L | Sponge network | 0.817 | 3.0E-5 | -0.194 | 0.55299 | 0.295 |
| 42195 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 22 | LRRC7 | Sponge network | 0.95 | 0.15943 | -1.341 | 0.01193 | 0.295 |
| 42196 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 12 | GPAM | Sponge network | 0.813 | 1.0E-5 | 0.142 | 0.29732 | 0.295 |
| 42197 | CTC-559E9.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p | 14 | AKAP6 | Sponge network | 0.594 | 0.00064 | -1.586 | 3.0E-5 | 0.295 |
| 42198 | AGAP11 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p | 18 | PIK3CA | Sponge network | -1.645 | 0 | 0.195 | 0.06908 | 0.295 |
| 42199 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-214-5p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-5p | 48 | SMAD4 | Sponge network | -1.645 | 0 | -0.313 | 0.00413 | 0.295 |
| 42200 | RP11-196G11.4 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-365a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | ARHGEF12 | Sponge network | 0.875 | 0 | 0.09 | 0.35693 | 0.295 |
| 42201 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ATRX | Sponge network | 0.997 | 0.00066 | 0.261 | 0.04408 | 0.295 |
| 42202 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577 | 24 | ERBB4 | Sponge network | -0.473 | 0.07602 | -1.975 | 2.0E-5 | 0.295 |
| 42203 | AP000487.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-664a-5p | 22 | SOX11 | Sponge network | 1.987 | 0 | 2.68 | 0 | 0.295 |
| 42204 | HCG11 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p | 10 | AMPH | Sponge network | 0.385 | 0.10067 | -0.916 | 5.0E-5 | 0.295 |
| 42205 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-877-5p;hsa-miR-93-5p | 34 | GAS7 | Sponge network | -0.384 | 0.40652 | -0.555 | 0.01602 | 0.295 |
| 42206 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 12 | GAS1 | Sponge network | -2.925 | 0 | -1.047 | 0.00364 | 0.295 |
| 42207 | RP11-13K12.1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | EGR2 | Sponge network | -0.154 | 0.78939 | 0.309 | 0.17079 | 0.295 |
| 42208 | KB-1460A1.5 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-744-3p | 20 | PTPN14 | Sponge network | 0.603 | 0.00127 | 0.144 | 0.34595 | 0.295 |
| 42209 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-577 | 12 | AFF3 | Sponge network | -0.418 | 0.00068 | -2.373 | 0 | 0.295 |
| 42210 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.083 | 0.66405 | 0.943 | 0 | 0.295 |
| 42211 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | KAT6B | Sponge network | -1.216 | 0 | -0.478 | 0.00018 | 0.295 |
| 42212 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p | 18 | TCF12 | Sponge network | 0.75 | 0.00054 | 0.174 | 0.13271 | 0.295 |
| 42213 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-589-5p | 15 | ARHGAP29 | Sponge network | -0.162 | 0.47687 | 0.131 | 0.42953 | 0.295 |
| 42214 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 17 | ARHGAP28 | Sponge network | 0.727 | 3.0E-5 | -0.911 | 0.00013 | 0.295 |
| 42215 | GS1-124K5.11 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-590-3p | 14 | ROBO1 | Sponge network | 0.494 | 0.00339 | -0.009 | 0.96154 | 0.295 |
| 42216 | MIR600HG |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-511-5p;hsa-miR-628-5p;hsa-miR-940 | 16 | TRIM33 | Sponge network | 0.594 | 0.41522 | 0.402 | 7.0E-5 | 0.295 |
| 42217 | RP3-475N16.1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-3607-3p | 10 | NAV3 | Sponge network | 1.056 | 0 | 0.212 | 0.47729 | 0.295 |
| 42218 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CACNA2D1 | Sponge network | 0.05 | 0.95483 | -0.846 | 0.00675 | 0.295 |
| 42219 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p | 29 | WDFY3 | Sponge network | 1.757 | 0 | -0.201 | 0.0967 | 0.295 |
| 42220 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-590-3p | 10 | HOOK3 | Sponge network | -0.513 | 0.04703 | 0.273 | 0.09957 | 0.295 |
| 42221 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-532-3p | 21 | TRPS1 | Sponge network | 0.497 | 0.01451 | -0.191 | 0.44109 | 0.295 |
| 42222 | CTC-471J1.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 18 | ZFX | Sponge network | 1.11 | 1.0E-5 | 0.09 | 0.42563 | 0.295 |
| 42223 | SOX2-OT |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-889-3p;hsa-miR-96-5p | 17 | RPS6KA6 | Sponge network | -1.264 | 6.0E-5 | -2.989 | 0 | 0.295 |
| 42224 | AP001055.6 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 17 | EYA4 | Sponge network | 0.693 | 0.00076 | 0.3 | 0.36876 | 0.295 |
| 42225 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-577;hsa-miR-590-3p | 19 | APC | Sponge network | -0.896 | 0.00099 | -0.255 | 0.04051 | 0.295 |
| 42226 | RP4-669P10.16 |
hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-98-5p | 10 | ZFHX3 | Sponge network | -0.301 | 0.06597 | 0.389 | 0.01363 | 0.295 |
| 42227 | PCA3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | PRRX1 | Sponge network | -0.709 | 0.18168 | 1.316 | 0 | 0.295 |
| 42228 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 38 | MCC | Sponge network | -0.969 | 2.0E-5 | -1.005 | 0 | 0.295 |
| 42229 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-98-5p | 15 | ITGA5 | Sponge network | 0.056 | 0.81195 | 0.452 | 0.03524 | 0.295 |
| 42230 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-590-5p | 20 | CCND2 | Sponge network | -2.076 | 0.00548 | -0.267 | 0.3112 | 0.295 |
| 42231 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 22 | CADM3 | Sponge network | -0.969 | 2.0E-5 | -3.663 | 0 | 0.295 |
| 42232 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 19 | LATS2 | Sponge network | 0.064 | 0.72934 | 0.159 | 0.2304 | 0.295 |
| 42233 | FGD5-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | SLC2A13 | Sponge network | 0.401 | 0.00066 | -0.331 | 0.03658 | 0.295 |
| 42234 | RP11-890B15.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 30 | EPS15 | Sponge network | 0.101 | 0.60919 | -0.052 | 0.53998 | 0.295 |
| 42235 | LINC00654 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | ADCY1 | Sponge network | 0.374 | 0.20054 | 0.362 | 0.16982 | 0.295 |
| 42236 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-500a-5p | 17 | ZFX | Sponge network | 0.082 | 0.55397 | 0.09 | 0.42563 | 0.295 |
| 42237 | LINC00667 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | AKAP6 | Sponge network | -0.235 | 0.15142 | -1.586 | 3.0E-5 | 0.295 |
| 42238 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-93-5p | 21 | RNASEL | Sponge network | 0.101 | 0.60919 | -0.272 | 0.01796 | 0.295 |
| 42239 | LINC00854 |
hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-654-3p | 14 | PIK3R1 | Sponge network | 1.18 | 0 | -0.133 | 0.36854 | 0.295 |
| 42240 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-550a-3p | 10 | PCDH8 | Sponge network | -2.62 | 0 | -0.997 | 0.05366 | 0.295 |
| 42241 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | USP12 | Sponge network | 0.222 | 0.29133 | -0.102 | 0.40768 | 0.295 |
| 42242 | AC010226.4 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-590-3p | 16 | RIMS2 | Sponge network | -0.811 | 2.0E-5 | -1.365 | 0.00208 | 0.295 |
| 42243 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-664a-3p | 14 | SOX11 | Sponge network | 0.909 | 0 | 2.68 | 0 | 0.295 |
| 42244 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | PPM1A | Sponge network | -0.355 | 0.0077 | -0.623 | 0 | 0.295 |
| 42245 | RP11-53O19.1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-576-5p | 10 | HERC1 | Sponge network | -0.405 | 0.22013 | -0.196 | 0.08544 | 0.295 |
| 42246 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 25 | CBL | Sponge network | -0.513 | 0.04703 | 0.291 | 0.01267 | 0.295 |
| 42247 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 46 | FOXP1 | Sponge network | 0.72 | 0.0001 | 0.042 | 0.74368 | 0.295 |
| 42248 | RP11-102N12.3 |
hsa-miR-10a-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-96-5p | 17 | CADM1 | Sponge network | -0.351 | 0.11358 | -0.9 | 0.00084 | 0.295 |
| 42249 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-5p | 37 | ERBB4 | Sponge network | -0.384 | 0.40652 | -1.975 | 2.0E-5 | 0.295 |
| 42250 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-935 | 35 | HBP1 | Sponge network | -0.615 | 0.00015 | -0.454 | 0 | 0.295 |
| 42251 | RP5-1085F17.3 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | PHF3 | Sponge network | 0.541 | 0.00125 | 0.126 | 0.19652 | 0.295 |
| 42252 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 16 | WWTR1 | Sponge network | -0.223 | 0.47816 | -0.345 | 0.11997 | 0.295 |
| 42253 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-577;hsa-miR-7-1-3p | 28 | SOX5 | Sponge network | 0.259 | 0.12542 | -1.091 | 4.0E-5 | 0.295 |
| 42254 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 32 | PAFAH1B1 | Sponge network | -0.738 | 0.21233 | -0.429 | 0 | 0.295 |
| 42255 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | ZMYM2 | Sponge network | 0.385 | 0.10067 | 0.279 | 0.01273 | 0.295 |
| 42256 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-874-3p | 11 | CRTC3 | Sponge network | -1.057 | 0.00029 | 0.164 | 0.16786 | 0.295 |
| 42257 | RP11-474O21.5 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | BNC2 | Sponge network | -0.023 | 0.95109 | -0.491 | 0.09988 | 0.295 |
| 42258 | RP11-474O21.5 |
hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 11 | QKI | Sponge network | -0.023 | 0.95109 | -0.333 | 0.04111 | 0.295 |
| 42259 | CTD-2017D11.1 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p | 12 | ANK2 | Sponge network | 0.792 | 0.0049 | -1.603 | 1.0E-5 | 0.295 |
| 42260 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-331-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | CDHR3 | Sponge network | 0.603 | 0.00127 | -0.977 | 4.0E-5 | 0.295 |
| 42261 | SNHG14 |
hsa-miR-148b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30d-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | GNAI2 | Sponge network | -1.111 | 0.0003 | -0.063 | 0.54532 | 0.295 |
| 42262 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | PGR | Sponge network | 0.559 | 0.00026 | -1.704 | 0 | 0.295 |
| 42263 | SOX2-OT |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 12 | CDH10 | Sponge network | -1.264 | 6.0E-5 | -2.397 | 0 | 0.295 |
| 42264 | CTD-2245F17.3 |
hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-219a-1-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 11 | JAZF1 | Sponge network | -0.421 | 0.12556 | -0.377 | 0.02677 | 0.295 |
| 42265 | RP11-359B12.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 23 | CYLD | Sponge network | 0.064 | 0.72934 | -0.367 | 0.00171 | 0.295 |
| 42266 | EMX2OS |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 23 | SOX11 | Sponge network | -0.777 | 0.1134 | 2.68 | 0 | 0.295 |
| 42267 | BDNF-AS |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-758-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 34 | HLF | Sponge network | -0.668 | 3.0E-5 | -2.443 | 0 | 0.295 |
| 42268 | RASSF8-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | AHNAK | Sponge network | 0.335 | 0.20596 | -1.207 | 0 | 0.295 |
| 42269 | FAM66C |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MXI1 | Sponge network | -0.901 | 0.00019 | -0.987 | 0 | 0.295 |
| 42270 | CTD-2184D3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-30b-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | DMD | Sponge network | 0.272 | 0.44692 | -1.506 | 0 | 0.295 |
| 42271 | RP11-531A24.5 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 10 | RNF180 | Sponge network | 0.75 | 0.00054 | -1.127 | 1.0E-5 | 0.295 |
| 42272 | TPTEP1 |
hsa-miR-107;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | CDKN1C | Sponge network | -1.216 | 0 | -0.929 | 0.00033 | 0.295 |
| 42273 | TTC25 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-629-5p | 10 | UNC80 | Sponge network | -0.208 | 0.38261 | -1.934 | 0 | 0.295 |
| 42274 | LINC00621 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | MTSS1 | Sponge network | 0.131 | 0.8436 | -0.81 | 0.00035 | 0.295 |
| 42275 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p | 20 | PIK3R1 | Sponge network | -0.693 | 0.05629 | -0.133 | 0.36854 | 0.295 |
| 42276 | AC159540.1 |
hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-214-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-328-3p;hsa-miR-365a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-628-5p;hsa-miR-758-3p | 19 | VPS53 | Sponge network | 1.122 | 0 | 0.103 | 0.22667 | 0.295 |
| 42277 | RP11-116O18.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-92a-1-5p | 33 | PPARA | Sponge network | 0.05 | 0.95483 | -0.379 | 0.00548 | 0.295 |
| 42278 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | FREM2 | Sponge network | -2.452 | 0 | -0.437 | 0.46353 | 0.295 |
| 42279 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-590-3p | 14 | LRP1B | Sponge network | -1.375 | 0.08642 | -2.163 | 0 | 0.295 |
| 42280 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 22 | TP53INP1 | Sponge network | -0.323 | 0.01372 | -0.496 | 0.00064 | 0.295 |
| 42281 | RP13-1039J1.4 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-34a-5p;hsa-miR-582-5p;hsa-miR-589-3p | 10 | ZFHX4 | Sponge network | 0.356 | 0.33467 | 0.018 | 0.95574 | 0.295 |
| 42282 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -0.384 | 0.40652 | -0.087 | 0.69193 | 0.295 |
| 42283 | AC009948.5 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-940 | 32 | GRIK3 | Sponge network | 0.532 | 0.00505 | -3.208 | 0 | 0.295 |
| 42284 | LINC00621 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 16 | ABL2 | Sponge network | 0.131 | 0.8436 | 0.82 | 0 | 0.295 |
| 42285 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | SMARCA2 | Sponge network | 0.249 | 0.35902 | -0.529 | 0.00014 | 0.295 |
| 42286 | RP4-614O4.11 |
hsa-miR-103a-2-5p;hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-628-5p | 11 | BMPR2 | Sponge network | 1.042 | 0.00177 | 0.206 | 0.0635 | 0.295 |
| 42287 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 64 | KIF1B | Sponge network | -0.576 | 0.10121 | -0.118 | 0.32258 | 0.295 |
| 42288 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-378a-5p | 18 | NUAK1 | Sponge network | 0.542 | 0.00054 | 0.904 | 0 | 0.295 |
| 42289 | SOX2-OT |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 20 | RELN | Sponge network | -1.264 | 6.0E-5 | -2.403 | 0 | 0.295 |
| 42290 | LINC00957 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CXXC4 | Sponge network | -0.421 | 0.0114 | -0.433 | 0.21055 | 0.295 |
| 42291 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-577;hsa-miR-590-3p | 21 | HIP1 | Sponge network | -1.161 | 0.00591 | 0.774 | 0 | 0.295 |
| 42292 | RP4-605O3.4 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-590-3p | 13 | RBMS3 | Sponge network | 0.262 | 0.0475 | -0.519 | 0.03551 | 0.295 |
| 42293 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 15 | CREBBP | Sponge network | -0.901 | 0.00019 | 0.126 | 0.15186 | 0.295 |
| 42294 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-497-5p | 14 | SOX11 | Sponge network | 1.262 | 0 | 2.68 | 0 | 0.295 |
| 42295 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-5p | 20 | KANK1 | Sponge network | -0.976 | 0.00025 | -0.55 | 0.00134 | 0.295 |
| 42296 | RP11-434B12.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | EPS15 | Sponge network | -0.031 | 0.89063 | -0.052 | 0.53998 | 0.295 |
| 42297 | C4A-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-24-3p | 12 | DLC1 | Sponge network | 3.345 | 0 | -0.382 | 0.05038 | 0.295 |
| 42298 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | APC | Sponge network | -0.358 | 0.17255 | -0.255 | 0.04051 | 0.295 |
| 42299 | MALAT1 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 12 | RAPGEF5 | Sponge network | 0.782 | 0.00016 | 0.536 | 4.0E-5 | 0.295 |
| 42300 | CTB-133G6.2 |
hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p | 10 | TACC1 | Sponge network | 0.4 | 0.39299 | -0.362 | 0.05406 | 0.295 |
| 42301 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 34 | ZFX | Sponge network | -0.487 | 0.02157 | 0.09 | 0.42563 | 0.295 |
| 42302 | LINC00657 |
hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-363-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 10 | RAPGEF5 | Sponge network | 0.91 | 0 | 0.536 | 4.0E-5 | 0.295 |
| 42303 | AC103563.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p | 15 | CREB3L2 | Sponge network | -7.551 | 0 | -0.525 | 2.0E-5 | 0.295 |
| 42304 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | 0.064 | 0.72934 | -1.047 | 0.00364 | 0.295 |
| 42305 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 20 | USP12 | Sponge network | 0.395 | 0.15451 | -0.102 | 0.40768 | 0.295 |
| 42306 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-5p;hsa-miR-362-3p | 15 | PTPRD | Sponge network | -2.549 | 0.00528 | -1.005 | 0.00064 | 0.295 |
| 42307 | RP11-37B2.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-629-3p | 13 | PTPN14 | Sponge network | 1.03 | 0 | 0.144 | 0.34595 | 0.295 |
| 42308 | RP11-216B9.6 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | ERG | Sponge network | 0.174 | 0.30034 | 0.002 | 0.99011 | 0.295 |
| 42309 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-3p;hsa-miR-590-3p | 10 | CREBBP | Sponge network | 0.539 | 0.03722 | 0.126 | 0.15186 | 0.295 |
| 42310 | MAGI2-AS3 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 23 | PAIP2 | Sponge network | -0.129 | 0.57177 | -0.515 | 0 | 0.295 |
| 42311 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 16 | RB1CC1 | Sponge network | 0.064 | 0.72934 | 0.175 | 0.12559 | 0.295 |
| 42312 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | ARHGEF12 | Sponge network | 0.769 | 0 | 0.09 | 0.35693 | 0.295 |
| 42313 | AC018890.6 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | RASSF8 | Sponge network | 1.61 | 0.00961 | -0.122 | 0.65591 | 0.295 |
| 42314 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p | 11 | RARB | Sponge network | 1.344 | 0 | -0.825 | 2.0E-5 | 0.295 |
| 42315 | MIR7-3HG |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-30b-3p | 10 | SOX5 | Sponge network | -2.688 | 0.00397 | -1.091 | 4.0E-5 | 0.295 |
| 42316 | CD27-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p | 11 | TES | Sponge network | 0.177 | 0.16681 | -0.348 | 0.00639 | 0.295 |
| 42317 | RP11-356J5.12 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 16 | TES | Sponge network | -0.899 | 6.0E-5 | -0.348 | 0.00639 | 0.295 |
| 42318 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 25 | NCOA1 | Sponge network | 0.537 | 0.00139 | -0.432 | 0 | 0.295 |
| 42319 | SNHG14 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 62 | PPM1L | Sponge network | -1.111 | 0.0003 | -0.537 | 0.00616 | 0.295 |
| 42320 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-93-3p | 18 | PTPN11 | Sponge network | -0.405 | 0.22013 | 0.431 | 2.0E-5 | 0.295 |
| 42321 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 12 | CRTC3 | Sponge network | 0.342 | 0.03129 | 0.164 | 0.16786 | 0.295 |
| 42322 | BZRAP1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p | 22 | RYR2 | Sponge network | -0.465 | 0.04703 | -1.466 | 0.00031 | 0.295 |
| 42323 | H1FX-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p | 12 | KIT | Sponge network | -0.206 | 0.12942 | -2.184 | 0 | 0.295 |
| 42324 | ADIRF-AS1 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-625-5p | 10 | NGFR | Sponge network | -0.223 | 0.47816 | -1.271 | 0.01058 | 0.295 |
| 42325 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p | 13 | PCDH8 | Sponge network | -1.697 | 0.00889 | -0.997 | 0.05366 | 0.295 |
| 42326 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -0.612 | 0.00094 | -0.252 | 0.15511 | 0.295 |
| 42327 | RP11-384K6.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p | 15 | SLC5A3 | Sponge network | 0.946 | 0 | 0.493 | 5.0E-5 | 0.295 |
| 42328 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p | 21 | PIK3C2A | Sponge network | -0.176 | 0.59692 | 0.061 | 0.53776 | 0.295 |
| 42329 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-766-3p | 45 | MDM4 | Sponge network | -0.487 | 0.02157 | 0.345 | 0.00762 | 0.295 |
| 42330 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | HECA | Sponge network | -0.355 | 0.0077 | -0.217 | 0.03463 | 0.295 |
| 42331 | BDNF-AS |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 39 | TRPS1 | Sponge network | -0.668 | 3.0E-5 | -0.191 | 0.44109 | 0.295 |
| 42332 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | FOXP1 | Sponge network | -2.117 | 0.00161 | 0.042 | 0.74368 | 0.295 |
| 42333 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-93-3p | 52 | QKI | Sponge network | -0.615 | 0.00015 | -0.333 | 0.04111 | 0.295 |
| 42334 | OIP5-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 26 | PTPN14 | Sponge network | 0.421 | 0.00084 | 0.144 | 0.34595 | 0.295 |
| 42335 | CTD-2089N3.1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-455-5p | 14 | TGFBR3 | Sponge network | -0.314 | 0.62033 | -1.173 | 1.0E-5 | 0.295 |
| 42336 | DNM1P35 |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | ZMYND11 | Sponge network | 0.051 | 0.83388 | -0.2 | 0.05449 | 0.295 |
| 42337 | RP11-927P21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-582-5p | 16 | NAV3 | Sponge network | 0.921 | 0.00118 | 0.212 | 0.47729 | 0.295 |
| 42338 | RP4-639F20.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p | 18 | PTPRT | Sponge network | -0.517 | 0.02935 | -0.907 | 0.03727 | 0.295 |
| 42339 | MIR143HG |
hsa-miR-132-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | PACRG | Sponge network | -0.739 | 0.0574 | -2.248 | 0 | 0.295 |
| 42340 | RP11-359B12.2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-539-5p | 11 | PIK3IP1 | Sponge network | 0.064 | 0.72934 | -0.187 | 0.19945 | 0.295 |
| 42341 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 14 | ST6GALNAC3 | Sponge network | -1.942 | 0 | -0.987 | 0 | 0.295 |
| 42342 | LINC00865 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-942-5p | 15 | ERRFI1 | Sponge network | -1.249 | 7.0E-5 | -0.798 | 1.0E-5 | 0.295 |
| 42343 | AC093627.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-625-5p | 21 | NTRK3 | Sponge network | -0.787 | 0.3928 | -1.77 | 3.0E-5 | 0.295 |
| 42344 | LINC00476 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p | 14 | WNK2 | Sponge network | -0.615 | 0.00015 | -0.352 | 0.31066 | 0.295 |
| 42345 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | TGFBR3 | Sponge network | -0.08 | 0.90092 | -1.173 | 1.0E-5 | 0.295 |
| 42346 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-96-5p | 53 | BMPR2 | Sponge network | -1.439 | 4.0E-5 | 0.206 | 0.0635 | 0.295 |
| 42347 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p | 10 | KCNAB1 | Sponge network | -0.777 | 0.16667 | -0.755 | 2.0E-5 | 0.295 |
| 42348 | RP11-244H3.1 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | PRKCB | Sponge network | 0.659 | 3.0E-5 | -1.313 | 2.0E-5 | 0.295 |
| 42349 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TRIP11 | Sponge network | 0.101 | 0.60919 | -0.158 | 0.06384 | 0.295 |
| 42350 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-7-1-3p | 12 | EPHA3 | Sponge network | 0.817 | 3.0E-5 | 0.185 | 0.53588 | 0.295 |
| 42351 | RP11-620J15.3 |
hsa-miR-1266-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-589-3p | 13 | KCNJ5 | Sponge network | -0.739 | 0.00044 | -0.281 | 0.3339 | 0.295 |
| 42352 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | MLLT10 | Sponge network | -1.575 | 6.0E-5 | 0.105 | 0.33292 | 0.295 |
| 42353 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-7-1-3p | 29 | PRKAB2 | Sponge network | 0.177 | 0.16681 | -0.367 | 0.01371 | 0.295 |
| 42354 | RP11-968O1.5 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-505-5p;hsa-miR-940 | 27 | PPARA | Sponge network | -0.829 | 0.01343 | -0.379 | 0.00548 | 0.295 |
| 42355 | AC093627.8 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p | 10 | FAT4 | Sponge network | -0.787 | 0.3928 | -0.354 | 0.14694 | 0.295 |
| 42356 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 43 | WDFY3 | Sponge network | 0.16 | 0.50785 | -0.201 | 0.0967 | 0.295 |
| 42357 | LINC00854 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-654-3p | 13 | FOXP1 | Sponge network | 1.18 | 0 | 0.042 | 0.74368 | 0.295 |
| 42358 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-877-5p | 13 | GLIPR1 | Sponge network | -1.654 | 0.00144 | 0.146 | 0.3276 | 0.295 |
| 42359 | CTA-204B4.2 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-454-3p;hsa-miR-590-3p | 10 | ITPKB | Sponge network | 0.765 | 7.0E-5 | -0.704 | 0.00106 | 0.294 |
| 42360 | RP11-53O19.1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-450b-5p | 14 | CXCL12 | Sponge network | -0.405 | 0.22013 | -1.421 | 0 | 0.294 |
| 42361 | AP001258.4 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 13 | CREB1 | Sponge network | 0.931 | 0 | 0.203 | 0.00537 | 0.294 |
| 42362 | LINC00641 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PCDH17 | Sponge network | -0.222 | 0.32445 | 0.731 | 2.0E-5 | 0.294 |
| 42363 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | AKAP11 | Sponge network | -0.513 | 0.04703 | 0.196 | 0.09825 | 0.294 |
| 42364 | RP11-981G7.6 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 10 | PLAG1 | Sponge network | 0.986 | 0.01022 | -0.315 | 0.26532 | 0.294 |
| 42365 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p | 13 | KDSR | Sponge network | 0.331 | 0.57247 | 0.239 | 0.02216 | 0.294 |
| 42366 | RP11-307B6.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-589-3p | 17 | PRDM2 | Sponge network | -4.463 | 0.00025 | -0.258 | 0.00721 | 0.294 |
| 42367 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 33 | EBF1 | Sponge network | 0.311 | 0.10344 | -0.384 | 0.08856 | 0.294 |
| 42368 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-495-3p | 10 | GNA13 | Sponge network | 1.106 | 0 | 0.007 | 0.93884 | 0.294 |
| 42369 | RP11-116O18.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 22 | KCNA6 | Sponge network | 0.05 | 0.95483 | -1.531 | 0.00013 | 0.294 |
| 42370 | RP11-263K19.4 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-378a-5p | 30 | ADARB1 | Sponge network | 0.859 | 0.00331 | -0.335 | 0.06631 | 0.294 |
| 42371 | LINC01023 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-93-3p | 16 | NTRK3 | Sponge network | -0.45 | 0.00486 | -1.77 | 3.0E-5 | 0.294 |
| 42372 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-500b-5p | 22 | GRIA3 | Sponge network | -0.777 | 0.16667 | -1.956 | 0 | 0.294 |
| 42373 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-502-5p | 15 | CHD5 | Sponge network | 0.912 | 0.00014 | -0.922 | 0.01521 | 0.294 |
| 42374 | RP11-48B3.4 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | GAS7 | Sponge network | 1.757 | 0 | -0.555 | 0.01602 | 0.294 |
| 42375 | RP11-53O19.3 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ATM | Sponge network | 1.205 | 0 | 0.178 | 0.21568 | 0.294 |
| 42376 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-224-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 11 | AKAP13 | Sponge network | -0.176 | 0.59692 | 0.028 | 0.7729 | 0.294 |
| 42377 | LINC00174 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | UBR3 | Sponge network | 1.253 | 0 | -0.021 | 0.82774 | 0.294 |
| 42378 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | SASH1 | Sponge network | 0.349 | 0.01695 | -0.502 | 0.00175 | 0.294 |
| 42379 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-5p | 22 | CXXC4 | Sponge network | -0.371 | 0.24604 | -0.433 | 0.21055 | 0.294 |
| 42380 | MEG3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | FLI1 | Sponge network | 0.249 | 0.35902 | -0.294 | 0.10905 | 0.294 |
| 42381 | RP13-516M14.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p | 13 | PRUNE2 | Sponge network | 0.389 | 0.04293 | -1.733 | 0.00022 | 0.294 |
| 42382 | RP11-13K12.5 |
hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 12 | PIK3CA | Sponge network | -0.318 | 0.65847 | 0.195 | 0.06908 | 0.294 |
| 42383 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-409-3p | 21 | SMAD2 | Sponge network | 0.497 | 0.01451 | -0.128 | 0.12732 | 0.294 |
| 42384 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 19 | JUN | Sponge network | -0.129 | 0.57177 | -0.932 | 0 | 0.294 |
| 42385 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 17 | SOX11 | Sponge network | 0.702 | 7.0E-5 | 2.68 | 0 | 0.294 |
| 42386 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-5p;hsa-miR-940;hsa-miR-942-5p | 12 | HLF | Sponge network | -0.421 | 0.12556 | -2.443 | 0 | 0.294 |
| 42387 | RP11-38L15.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 12 | ZFHX3 | Sponge network | 0.344 | 0.3127 | 0.389 | 0.01363 | 0.294 |
| 42388 | RP11-728F11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 11 | AKAP6 | Sponge network | 0.238 | 0.70544 | -1.586 | 3.0E-5 | 0.294 |
| 42389 | BZRAP1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-93-5p | 14 | AHNAK | Sponge network | -0.465 | 0.04703 | -1.207 | 0 | 0.294 |
| 42390 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 31 | MCC | Sponge network | 0.349 | 0.01695 | -1.005 | 0 | 0.294 |
| 42391 | RP11-46C24.7 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | KIF5B | Sponge network | 0.392 | 0.0278 | 0.362 | 0.00066 | 0.294 |
| 42392 | RP11-819C21.1 |
hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | GSK3B | Sponge network | 0.537 | 0.00139 | 0.266 | 0.00045 | 0.294 |
| 42393 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 26 | CRTC1 | Sponge network | 0.374 | 0.20054 | -0.116 | 0.28412 | 0.294 |
| 42394 | RP1-102E24.8 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-330-5p | 14 | CCDC6 | Sponge network | 1.737 | 0 | 0.27 | 0.02555 | 0.294 |
| 42395 | HOXA-AS3 |
hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | EMP2 | Sponge network | -0.555 | 0.06156 | -0.423 | 0.00985 | 0.294 |
| 42396 | MIR497HG |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 21 | DOCK4 | Sponge network | -1.439 | 4.0E-5 | 0.502 | 0.00108 | 0.294 |
| 42397 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-625-5p | 17 | TMTC2 | Sponge network | -2.452 | 0 | -0.215 | 0.18248 | 0.294 |
| 42398 | LINC00094 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-7-5p | 26 | SETBP1 | Sponge network | 0.253 | 0.09565 | -1.302 | 1.0E-5 | 0.294 |
| 42399 | TMEM9B-AS1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 29 | KIF1B | Sponge network | 0.529 | 0.00552 | -0.118 | 0.32258 | 0.294 |
| 42400 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940 | 23 | HLF | Sponge network | 0.559 | 0.00026 | -2.443 | 0 | 0.294 |
| 42401 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | GRIN2A | Sponge network | -0.176 | 0.59692 | -2.161 | 0 | 0.294 |
| 42402 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-940 | 34 | NOTCH2 | Sponge network | -0.246 | 0.35034 | 0.138 | 0.35163 | 0.294 |
| 42403 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 34 | TMEM132B | Sponge network | 0.41 | 0.02214 | -0.83 | 0.01084 | 0.294 |
| 42404 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DACT1 | Sponge network | 0.392 | 0.0278 | 0.783 | 0.00072 | 0.294 |
| 42405 | CTD-2184D3.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ABI2 | Sponge network | 0.272 | 0.44692 | 0.175 | 0.14787 | 0.294 |
| 42406 | RP11-728F11.4 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-7-5p | 10 | CRTC3 | Sponge network | 0.238 | 0.70544 | 0.164 | 0.16786 | 0.294 |
| 42407 | RP1-302G2.5 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-7-5p | 13 | BTG2 | Sponge network | -1.483 | 9.0E-5 | -1.763 | 0 | 0.294 |
| 42408 | RP11-212P7.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 26 | PTPN14 | Sponge network | 0.219 | 0.1516 | 0.144 | 0.34595 | 0.294 |
| 42409 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | HIVEP1 | Sponge network | 0.659 | 3.0E-5 | 0.368 | 0.00064 | 0.294 |
| 42410 | PCA3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 16 | RPS6KA2 | Sponge network | -0.709 | 0.18168 | -0.57 | 0.00013 | 0.294 |
| 42411 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p | 14 | CADM3 | Sponge network | -0.18 | 0.20343 | -3.663 | 0 | 0.294 |
| 42412 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-486-5p;hsa-miR-584-5p;hsa-miR-671-5p | 17 | RBMS3 | Sponge network | 0.556 | 0.00298 | -0.519 | 0.03551 | 0.294 |
| 42413 | LINC00094 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-655-3p;hsa-miR-92a-3p | 19 | UBE4B | Sponge network | 0.253 | 0.09565 | -0.235 | 0.007 | 0.294 |
| 42414 | RP11-464F9.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-320a;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940 | 19 | EPS15 | Sponge network | 0.959 | 0 | -0.052 | 0.53998 | 0.294 |
| 42415 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TBX18 | Sponge network | -2.915 | 0.00015 | 0.843 | 0.02945 | 0.294 |
| 42416 | LINC00578 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-33a-3p;hsa-miR-3614-5p | 17 | ABL2 | Sponge network | 0.811 | 0.03228 | 0.82 | 0 | 0.294 |
| 42417 | RP11-47A8.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-362-5p;hsa-miR-500a-5p | 15 | PPM1L | Sponge network | 0.103 | 0.43169 | -0.537 | 0.00616 | 0.294 |
| 42418 | AC016747.3 |
hsa-miR-107;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 11 | TAL1 | Sponge network | 0.199 | 0.38015 | -0.462 | 0.00574 | 0.294 |
| 42419 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | SMARCA2 | Sponge network | -1.216 | 0 | -0.529 | 0.00014 | 0.294 |
| 42420 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-92a-3p | 19 | CBFA2T3 | Sponge network | -0.738 | 0.21233 | -1.221 | 1.0E-5 | 0.294 |
| 42421 | RP11-867G23.10 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-92a-1-5p | 11 | PRKACA | Sponge network | -2.531 | 0 | -0.255 | 0.0012 | 0.294 |
| 42422 | RP11-571M6.17 |
hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-30a-5p;hsa-miR-590-3p | 13 | SOX11 | Sponge network | 1.013 | 0 | 2.68 | 0 | 0.294 |
| 42423 | WDFY3-AS2 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 15 | SPTBN4 | Sponge network | -0.942 | 0 | -0.786 | 0.01281 | 0.294 |
| 42424 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 13 | RBL2 | Sponge network | 0.385 | 0.10067 | -0.282 | 0.00254 | 0.294 |
| 42425 | AC016747.3 |
hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p | 16 | IRS1 | Sponge network | 0.199 | 0.38015 | -0.026 | 0.88855 | 0.294 |
| 42426 | SERHL |
hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-532-3p | 10 | SYNM | Sponge network | -0.602 | 0.00331 | -2.309 | 1.0E-5 | 0.294 |
| 42427 | RP11-848P1.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | LPP | Sponge network | 1.253 | 0 | -0.459 | 0.04475 | 0.294 |
| 42428 | LINC00957 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 38 | CBX5 | Sponge network | -0.421 | 0.0114 | 0.165 | 0.24446 | 0.294 |
| 42429 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CXXC4 | Sponge network | 0.101 | 0.60919 | -0.433 | 0.21055 | 0.294 |
| 42430 | BDNF-AS |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 14 | SFRP1 | Sponge network | -0.668 | 3.0E-5 | -3.566 | 0 | 0.294 |
| 42431 | GS1-124K5.11 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | 0.494 | 0.00339 | -2.633 | 0 | 0.294 |
| 42432 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 38 | PHC3 | Sponge network | -0.576 | 0.10121 | 0.212 | 0.0499 | 0.294 |
| 42433 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 19 | JAZF1 | Sponge network | 0.619 | 0.00157 | -0.377 | 0.02677 | 0.294 |
| 42434 | ADIRF-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 18 | AKAP6 | Sponge network | -0.223 | 0.47816 | -1.586 | 3.0E-5 | 0.294 |
| 42435 | LINC00476 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | CELF4 | Sponge network | -0.615 | 0.00015 | -1.135 | 0.00344 | 0.294 |
| 42436 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-93-5p | 10 | EDA2R | Sponge network | 0.259 | 0.12542 | -0.587 | 0.01598 | 0.294 |
| 42437 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 24 | KCNA6 | Sponge network | 0.532 | 0.00505 | -1.531 | 0.00013 | 0.294 |
| 42438 | CTD-2336O2.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | ETV1 | Sponge network | -0.141 | 0.26434 | -0.096 | 0.67674 | 0.294 |
| 42439 | RP11-283I3.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-96-5p | 17 | CYLD | Sponge network | 0.614 | 1.0E-5 | -0.367 | 0.00171 | 0.294 |
| 42440 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | ERBB4 | Sponge network | -1.057 | 0.00029 | -1.975 | 2.0E-5 | 0.294 |
| 42441 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-450a-5p;hsa-miR-590-3p;hsa-miR-628-5p | 13 | CHD2 | Sponge network | 1.757 | 0 | 0.049 | 0.55371 | 0.294 |
| 42442 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-576-5p | 10 | AKAP12 | Sponge network | 0.75 | 0.00054 | -0.932 | 0.0028 | 0.294 |
| 42443 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | TMEM132B | Sponge network | 0.538 | 0.00076 | -0.83 | 0.01084 | 0.294 |
| 42444 | RP11-130L8.1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p | 21 | GRIK3 | Sponge network | -0.663 | 0.10887 | -3.208 | 0 | 0.294 |
| 42445 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PLSCR4 | Sponge network | -0.125 | 0.49414 | -0.474 | 0.03833 | 0.294 |
| 42446 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | PRKAA2 | Sponge network | -0.223 | 0.47816 | -2.462 | 0 | 0.294 |
| 42447 | EIF3J-AS1 |
hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | RB1CC1 | Sponge network | 0.331 | 0.00509 | 0.175 | 0.12559 | 0.294 |
| 42448 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 21 | LATS1 | Sponge network | 1.11 | 0 | 0.109 | 0.34208 | 0.294 |
| 42449 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-539-5p | 16 | DDX6 | Sponge network | 0.924 | 0 | 0.091 | 0.36428 | 0.294 |
| 42450 | SNHG14 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 15 | FOXO4 | Sponge network | -1.111 | 0.0003 | -0.516 | 2.0E-5 | 0.294 |
| 42451 | LINC00662 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | DTNA | Sponge network | 0.213 | 0.32297 | -1.648 | 1.0E-5 | 0.294 |
| 42452 | SH3BP5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-582-3p;hsa-miR-589-3p | 21 | PRDM2 | Sponge network | 0.267 | 0.2937 | -0.258 | 0.00721 | 0.294 |
| 42453 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 27 | PRKAB2 | Sponge network | -0.223 | 0.47816 | -0.367 | 0.01371 | 0.294 |
| 42454 | FZD10-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | FAM117A | Sponge network | -0.727 | 0.04726 | -1.379 | 0 | 0.294 |
| 42455 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-590-5p | 11 | LHFP | Sponge network | -0.969 | 2.0E-5 | -0.167 | 0.41371 | 0.294 |
| 42456 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p | 10 | CLIP1 | Sponge network | 0.541 | 0.00125 | -0.215 | 0.08684 | 0.294 |
| 42457 | RP11-400K9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 15 | PRRX1 | Sponge network | -0.733 | 0.19469 | 1.316 | 0 | 0.294 |
| 42458 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-450a-5p;hsa-miR-505-3p | 11 | CHD2 | Sponge network | 0.622 | 0.00015 | 0.049 | 0.55371 | 0.294 |
| 42459 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PLSCR4 | Sponge network | 0.659 | 3.0E-5 | -0.474 | 0.03833 | 0.294 |
| 42460 | RP1-151F17.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 14 | DLG1 | Sponge network | 0.222 | 0.29133 | -0.014 | 0.8926 | 0.294 |
| 42461 | RP11-254F7.2 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-589-3p | 17 | SYT4 | Sponge network | 0.176 | 0.37402 | -3.207 | 0 | 0.294 |
| 42462 | LINC00963 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-744-3p | 15 | IGF2 | Sponge network | 0.523 | 9.0E-5 | 0.848 | 0.03842 | 0.294 |
| 42463 | CTD-3126B10.4 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | NOTCH2 | Sponge network | 0.778 | 0.00101 | 0.138 | 0.35163 | 0.294 |
| 42464 | GHRLOS |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-421 | 10 | CDH23 | Sponge network | -1.942 | 0 | -0.974 | 0.00034 | 0.294 |
| 42465 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 12 | EBF3 | Sponge network | 0.998 | 0 | -0.058 | 0.81437 | 0.294 |
| 42466 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-502-3p | 11 | ARID5B | Sponge network | -1.255 | 0.00981 | 0.342 | 0.01676 | 0.294 |
| 42467 | RP11-968O1.5 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-493-5p;hsa-miR-532-5p | 21 | TMEM132B | Sponge network | -0.829 | 0.01343 | -0.83 | 0.01084 | 0.294 |
| 42468 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-550a-5p | 14 | FAT2 | Sponge network | -0.154 | 0.78939 | -0.278 | 0.44877 | 0.294 |
| 42469 | FENDRR |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | NRG3 | Sponge network | -1.281 | 0.00012 | -1.836 | 4.0E-5 | 0.294 |
| 42470 | TMEM161B-AS1 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | PKNOX1 | Sponge network | -0.04 | 0.7606 | 0.085 | 0.28917 | 0.294 |
| 42471 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-589-5p | 13 | ERBB4 | Sponge network | -1.14 | 0.03513 | -1.975 | 2.0E-5 | 0.294 |
| 42472 | RP11-395G23.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 25 | PTPRD | Sponge network | 0.381 | 0.25967 | -1.005 | 0.00064 | 0.294 |
| 42473 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | KIF1B | Sponge network | 0.931 | 1.0E-5 | -0.118 | 0.32258 | 0.294 |
| 42474 | HCG11 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | PPP1R9A | Sponge network | 0.385 | 0.10067 | -0.903 | 0.04254 | 0.294 |
| 42475 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-30d-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | JDP2 | Sponge network | -0.739 | 0.00044 | -0.967 | 0 | 0.294 |
| 42476 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 19 | PAFAH1B1 | Sponge network | -0.285 | 0.2172 | -0.429 | 0 | 0.294 |
| 42477 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-942-5p | 20 | DDX6 | Sponge network | 1.757 | 0 | 0.091 | 0.36428 | 0.294 |
| 42478 | RP11-276H19.2 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-96-5p | 14 | ZFHX4 | Sponge network | -0.472 | 0.48851 | 0.018 | 0.95574 | 0.294 |
| 42479 | RP4-669P10.16 |
hsa-miR-103a-2-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-214-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-505-5p | 21 | ARHGEF12 | Sponge network | -0.301 | 0.06597 | 0.09 | 0.35693 | 0.294 |
| 42480 | MIR4458HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 28 | CAMTA1 | Sponge network | -0.427 | 0.0623 | -0.436 | 1.0E-5 | 0.294 |
| 42481 | EMX2OS |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-378a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | MLF1 | Sponge network | -0.777 | 0.1134 | -0.893 | 0.00727 | 0.294 |
| 42482 | NDUFA6-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 13 | DCN | Sponge network | -0.355 | 0.0077 | -1.288 | 0 | 0.294 |
| 42483 | CTD-2002H8.2 |
hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p | 19 | DST | Sponge network | 0.931 | 1.0E-5 | -0.713 | 0.00096 | 0.294 |
| 42484 | RP11-359B12.2 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-429;hsa-miR-625-5p | 11 | NGFR | Sponge network | 0.064 | 0.72934 | -1.271 | 0.01058 | 0.294 |
| 42485 | MRPL23-AS1 |
hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26a-2-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-34a-5p | 19 | FAT3 | Sponge network | -0.278 | 0.76559 | -1.601 | 0.00051 | 0.294 |
| 42486 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 23 | EBF1 | Sponge network | -0.031 | 0.89063 | -0.384 | 0.08856 | 0.294 |
| 42487 | HAND2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 29 | ACSL6 | Sponge network | -1.697 | 0.00889 | -1.593 | 0 | 0.294 |
| 42488 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | IL6ST | Sponge network | 0.199 | 0.52763 | -0.402 | 0.00676 | 0.294 |
| 42489 | RP11-517B11.7 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 10 | AFF1 | Sponge network | 0.706 | 0 | -0.384 | 0.00053 | 0.294 |
| 42490 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-93-5p | 25 | LRP1B | Sponge network | -0.384 | 0.40652 | -2.163 | 0 | 0.294 |
| 42491 | PCA3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | MED12L | Sponge network | -0.709 | 0.18168 | -0.194 | 0.55299 | 0.294 |
| 42492 | AC034220.3 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-766-3p | 18 | ATM | Sponge network | 0.309 | 0.07304 | 0.178 | 0.21568 | 0.294 |
| 42493 | PCA3 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | SIAH1 | Sponge network | -0.709 | 0.18168 | -0.057 | 0.41415 | 0.294 |
| 42494 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-582-5p | 14 | TBX18 | Sponge network | 0.253 | 0.09565 | 0.843 | 0.02945 | 0.294 |
| 42495 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 15 | LRRC55 | Sponge network | 0.41 | 0.02214 | -0.026 | 0.93163 | 0.294 |
| 42496 | RP11-326G21.1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | PIK3R1 | Sponge network | -0.021 | 0.93598 | -0.133 | 0.36854 | 0.294 |
| 42497 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | FREM2 | Sponge network | -2.117 | 0.00161 | -0.437 | 0.46353 | 0.294 |
| 42498 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-629-3p | 16 | EBF1 | Sponge network | 0.368 | 0.20093 | -0.384 | 0.08856 | 0.294 |
| 42499 | MIR4435-1HG |
hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 10 | PAFAH1B2 | Sponge network | 2.275 | 0 | 0.318 | 0.0001 | 0.294 |
| 42500 | CTD-2336O2.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DNAJB4 | Sponge network | -0.141 | 0.26434 | -0.7 | 1.0E-5 | 0.294 |
| 42501 | LINC00886 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p | 17 | SETBP1 | Sponge network | -0.371 | 0.24604 | -1.302 | 1.0E-5 | 0.294 |
| 42502 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-375 | 11 | MYBL1 | Sponge network | 0.858 | 3.0E-5 | 0.494 | 0.02152 | 0.294 |
| 42503 | RP11-283I3.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | SETBP1 | Sponge network | 0.614 | 1.0E-5 | -1.302 | 1.0E-5 | 0.294 |
| 42504 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 55 | QKI | Sponge network | 0.421 | 0.00084 | -0.333 | 0.04111 | 0.294 |
| 42505 | RP11-244O19.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-362-5p;hsa-miR-589-3p | 17 | KCNJ5 | Sponge network | -0.096 | 0.67958 | -0.281 | 0.3339 | 0.294 |
| 42506 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-452-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | INPP4B | Sponge network | -0.576 | 0.10121 | -0.097 | 0.60503 | 0.294 |
| 42507 | FAM215B |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 15 | UBR3 | Sponge network | 0.817 | 3.0E-5 | -0.021 | 0.82774 | 0.294 |
| 42508 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | ZNF208 | Sponge network | 0.222 | 0.29133 | -0.4 | 0.22381 | 0.294 |
| 42509 | RP11-798M19.6 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 14 | CELF4 | Sponge network | -0.612 | 0.00094 | -1.135 | 0.00344 | 0.294 |
| 42510 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-424-5p | 14 | RET | Sponge network | 0.082 | 0.55397 | -0.833 | 0.03517 | 0.294 |
| 42511 | RP11-21A7A.2 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-942-5p | 23 | THRB | Sponge network | -1.116 | 0.23686 | -1.117 | 0.0004 | 0.294 |
| 42512 | RP11-244O19.1 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | ZNF536 | Sponge network | -0.096 | 0.67958 | -3.115 | 0 | 0.294 |
| 42513 | RP11-400K9.3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1301-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p | 10 | KCNA6 | Sponge network | -0.733 | 0.19469 | -1.531 | 0.00013 | 0.294 |
| 42514 | RP11-439L18.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-3065-3p;hsa-miR-454-3p;hsa-miR-484 | 12 | CHD5 | Sponge network | 0.224 | 0.48719 | -0.922 | 0.01521 | 0.294 |
| 42515 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CLIP1 | Sponge network | 0.683 | 1.0E-5 | -0.215 | 0.08684 | 0.294 |
| 42516 | CTD-2554C21.2 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429 | 14 | OPCML | Sponge network | -1.654 | 0.00144 | -2.498 | 0 | 0.294 |
| 42517 | HHIP-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | JDP2 | Sponge network | -0.737 | 0.10822 | -0.967 | 0 | 0.294 |
| 42518 | LINC00621 |
hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 14 | LIFR | Sponge network | 0.131 | 0.8436 | -1.794 | 0 | 0.294 |
| 42519 | RASSF8-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | EFNA5 | Sponge network | 0.335 | 0.20596 | -0.421 | 0.17868 | 0.294 |
| 42520 | KB-1572G7.3 |
hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 10 | RICTOR | Sponge network | 0.786 | 1.0E-5 | 0.388 | 0.00188 | 0.294 |
| 42521 | RP11-434B12.1 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | INPP4A | Sponge network | -0.031 | 0.89063 | 0.07 | 0.53563 | 0.294 |
| 42522 | PDXDC2P |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-497-5p | 12 | UBR3 | Sponge network | 0.756 | 0 | -0.021 | 0.82774 | 0.294 |
| 42523 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | TSHZ3 | Sponge network | -0.125 | 0.49414 | -0.556 | 0.01516 | 0.294 |
| 42524 | RP11-798M19.6 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 38 | AFF1 | Sponge network | -0.612 | 0.00094 | -0.384 | 0.00053 | 0.294 |
| 42525 | LINC01018 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | PNRC1 | Sponge network | -2.915 | 0.00015 | -0.646 | 0 | 0.294 |
| 42526 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-590-5p | 10 | PIK3R1 | Sponge network | -0.733 | 0.19469 | -0.133 | 0.36854 | 0.294 |
| 42527 | AC093627.8 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-7-5p | 11 | RBMS3 | Sponge network | -0.787 | 0.3928 | -0.519 | 0.03551 | 0.294 |
| 42528 | RP11-531A24.5 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-486-5p | 15 | DYNC1I1 | Sponge network | 0.75 | 0.00054 | -1.12 | 0.00107 | 0.294 |
| 42529 | RP11-216B9.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | DDX5 | Sponge network | 0.174 | 0.30034 | -0.099 | 0.17428 | 0.294 |
| 42530 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p | 11 | CHD1 | Sponge network | 1.344 | 0 | 0.322 | 0.00215 | 0.294 |
| 42531 | CTD-2528L19.6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | TCF12 | Sponge network | 0.953 | 0 | 0.174 | 0.13271 | 0.294 |
| 42532 | CROCCP2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-484 | 10 | CUL5 | Sponge network | 0.79 | 0 | 0.026 | 0.77301 | 0.294 |
| 42533 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 28 | PIK3C2A | Sponge network | 0.064 | 0.72934 | 0.061 | 0.53776 | 0.294 |
| 42534 | CKMT2-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-671-5p | 10 | PIK3IP1 | Sponge network | 0.082 | 0.55654 | -0.187 | 0.19945 | 0.294 |
| 42535 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-671-5p | 16 | DLC1 | Sponge network | -0.285 | 0.2172 | -0.382 | 0.05038 | 0.294 |
| 42536 | SERHL |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-339-5p | 10 | FNBP1 | Sponge network | -0.602 | 0.00331 | -0.969 | 4.0E-5 | 0.294 |
| 42537 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 18 | SOX11 | Sponge network | 0.919 | 0 | 2.68 | 0 | 0.294 |
| 42538 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | RARB | Sponge network | 0.299 | 0.57722 | -0.825 | 2.0E-5 | 0.294 |
| 42539 | LINC00473 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | MED12L | Sponge network | -1.203 | 0.03881 | -0.194 | 0.55299 | 0.294 |
| 42540 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-5p | 24 | PRKAB2 | Sponge network | 0.176 | 0.37402 | -0.367 | 0.01371 | 0.294 |
| 42541 | LINC00662 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p | 18 | FOXO3 | Sponge network | 0.213 | 0.32297 | -0.04 | 0.72028 | 0.294 |
| 42542 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 30 | AKAP11 | Sponge network | 0.374 | 0.20054 | 0.196 | 0.09825 | 0.294 |
| 42543 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-93-3p | 51 | RUNX1T1 | Sponge network | -1.645 | 0 | -0.344 | 0.2374 | 0.294 |
| 42544 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-500a-3p;hsa-miR-590-3p | 15 | ARHGAP29 | Sponge network | 0.61 | 0.01593 | 0.131 | 0.42953 | 0.294 |
| 42545 | RP11-283I3.6 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p | 16 | PCDH9 | Sponge network | 0.614 | 1.0E-5 | -1.823 | 4.0E-5 | 0.294 |
| 42546 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | ETV1 | Sponge network | 0.225 | 0.30644 | -0.096 | 0.67674 | 0.294 |
| 42547 | RP11-35G9.5 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-500a-5p | 18 | DMD | Sponge network | 0.622 | 0.00015 | -1.506 | 0 | 0.294 |
| 42548 | GNG12-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | KANK1 | Sponge network | -0.793 | 7.0E-5 | -0.55 | 0.00134 | 0.294 |
| 42549 | LINC01006 |
hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-429 | 10 | IGSF10 | Sponge network | -0.702 | 0.04814 | -1.751 | 1.0E-5 | 0.294 |
| 42550 | MIR497HG |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 33 | AFF1 | Sponge network | -1.439 | 4.0E-5 | -0.384 | 0.00053 | 0.294 |
| 42551 | LINC00926 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-629-3p | 12 | ESR1 | Sponge network | 0.756 | 0.00739 | -1.035 | 3.0E-5 | 0.294 |
| 42552 | CTB-133G6.2 |
hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-654-3p | 10 | NR2C2 | Sponge network | 0.4 | 0.39299 | 0.21 | 0.03224 | 0.294 |
| 42553 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | CACNA2D1 | Sponge network | 0.401 | 0.00066 | -0.846 | 0.00675 | 0.294 |
| 42554 | RP11-2H8.2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | NEDD4 | Sponge network | 1.089 | 0 | 0.02 | 0.89834 | 0.294 |
| 42555 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | CXXC4 | Sponge network | -0.615 | 0.00015 | -0.433 | 0.21055 | 0.294 |
| 42556 | AC159540.1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p;hsa-miR-664a-5p | 16 | CBFB | Sponge network | 1.122 | 0 | 1.061 | 0 | 0.294 |
| 42557 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p | 18 | FOXO3 | Sponge network | -0.246 | 0.35034 | -0.04 | 0.72028 | 0.294 |
| 42558 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CLIP1 | Sponge network | 0.705 | 0.00014 | -0.215 | 0.08684 | 0.294 |
| 42559 | HOXD-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p | 11 | DLC1 | Sponge network | -0.208 | 0.65592 | -0.382 | 0.05038 | 0.294 |
| 42560 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | EPHA5 | Sponge network | -0.513 | 0.04703 | -0.957 | 0.01733 | 0.294 |
| 42561 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-539-5p | 18 | MOB1B | Sponge network | -0.473 | 0.07602 | 0.197 | 0.15266 | 0.294 |
| 42562 | RP11-13K12.5 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 10 | NR2C2 | Sponge network | -0.318 | 0.65847 | 0.21 | 0.03224 | 0.294 |
| 42563 | RP11-254F7.2 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | PTCH1 | Sponge network | 0.176 | 0.37402 | -0.1 | 0.6179 | 0.294 |
| 42564 | LINC00641 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 21 | CADM3 | Sponge network | -0.222 | 0.32445 | -3.663 | 0 | 0.294 |
| 42565 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-3127-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p | 12 | CRTC1 | Sponge network | -0.295 | 0.02604 | -0.116 | 0.28412 | 0.294 |
| 42566 | CTC-429P9.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-532-5p | 14 | CDH23 | Sponge network | 0.082 | 0.55397 | -0.974 | 0.00034 | 0.294 |
| 42567 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p | 19 | PPP2R5C | Sponge network | -0.096 | 0.67958 | -0.314 | 3.0E-5 | 0.294 |
| 42568 | RP11-66B24.2 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-3607-3p | 10 | CNTN1 | Sponge network | 0.57 | 0.1866 | -2.594 | 0 | 0.294 |
| 42569 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-590-3p | 13 | RBM5 | Sponge network | 0.532 | 0.00505 | -0.205 | 0.0129 | 0.294 |
| 42570 | AC116366.6 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 28 | LPP | Sponge network | 0.329 | 0.11122 | -0.459 | 0.04475 | 0.294 |
| 42571 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-493-5p;hsa-miR-539-5p | 13 | TAL1 | Sponge network | -0.384 | 0.40652 | -0.462 | 0.00574 | 0.294 |
| 42572 | RP11-536O18.1 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p | 11 | PDGFRA | Sponge network | -0.074 | 0.90758 | -0.372 | 0.0037 | 0.294 |
| 42573 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 38 | PIK3R1 | Sponge network | 0.61 | 0.01593 | -0.133 | 0.36854 | 0.294 |
| 42574 | PCA3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p | 11 | DUSP1 | Sponge network | -0.709 | 0.18168 | -1.754 | 0 | 0.294 |
| 42575 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-3p;hsa-miR-576-5p | 37 | RUNX1T1 | Sponge network | 0.614 | 1.0E-5 | -0.344 | 0.2374 | 0.294 |
| 42576 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-550a-5p;hsa-miR-7-1-3p | 19 | PDGFRA | Sponge network | 0.693 | 0.00076 | -0.372 | 0.0037 | 0.294 |
| 42577 | BOLA3-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 12 | HOMER2 | Sponge network | -0.246 | 0.35034 | -2.698 | 0 | 0.294 |
| 42578 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 44 | WDFY3 | Sponge network | -1.047 | 0 | -0.201 | 0.0967 | 0.294 |
| 42579 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 24 | BCL6 | Sponge network | -0.615 | 0.00015 | -0.211 | 0.19489 | 0.294 |
| 42580 | RP11-344B5.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-320a;hsa-miR-320b | 15 | TACC1 | Sponge network | -0.055 | 0.88482 | -0.362 | 0.05406 | 0.294 |
| 42581 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ERG | Sponge network | -0.342 | 0.02288 | 0.002 | 0.99011 | 0.294 |
| 42582 | MIR600HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-655-3p | 12 | KANK1 | Sponge network | 0.594 | 0.41522 | -0.55 | 0.00134 | 0.294 |
| 42583 | RP11-678G14.2 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30d-3p | 11 | RASSF2 | Sponge network | -1.765 | 0.0778 | -0.273 | 0.21961 | 0.294 |
| 42584 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-96-5p | 32 | PPM1A | Sponge network | 0.331 | 0.57247 | -0.623 | 0 | 0.294 |
| 42585 | AC127904.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TET1 | Sponge network | 1.308 | 0 | -0.135 | 0.52138 | 0.294 |
| 42586 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-664a-3p | 12 | GREM1 | Sponge network | 1.308 | 0 | 0.902 | 0.01691 | 0.294 |
| 42587 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-378a-5p | 11 | PDS5B | Sponge network | 1.352 | 0 | 0.259 | 0.00962 | 0.294 |
| 42588 | RP11-575A19.2 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p | 11 | MDM4 | Sponge network | 1.969 | 0.00316 | 0.345 | 0.00762 | 0.294 |
| 42589 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-654-3p | 16 | PSIP1 | Sponge network | 0.194 | 0.64278 | -0.005 | 0.96897 | 0.294 |
| 42590 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 10 | RB1CC1 | Sponge network | 1.308 | 0 | 0.175 | 0.12559 | 0.294 |
| 42591 | CTC-429P9.5 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500b-5p;hsa-miR-590-3p | 17 | ACVR2A | Sponge network | 0.178 | 0.29106 | -0.409 | 0.00039 | 0.294 |
| 42592 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-485-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SAMHD1 | Sponge network | -1.697 | 0.00889 | 0.072 | 0.62895 | 0.294 |
| 42593 | RP11-360F5.1 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429 | 13 | MDM4 | Sponge network | 2.364 | 0.00134 | 0.345 | 0.00762 | 0.294 |
| 42594 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-502-5p;hsa-miR-92a-3p | 27 | TCF4 | Sponge network | 0.912 | 0.00014 | 0.016 | 0.92271 | 0.294 |
| 42595 | RP11-67L2.2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 28 | PCDH10 | Sponge network | 0.72 | 0.0001 | -1.644 | 0.0078 | 0.294 |
| 42596 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | KIT | Sponge network | -0.612 | 0.00094 | -2.184 | 0 | 0.294 |
| 42597 | MEG3 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-7-1-3p | 27 | RIMS2 | Sponge network | 0.249 | 0.35902 | -1.365 | 0.00208 | 0.294 |
| 42598 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-98-5p | 51 | NTRK3 | Sponge network | -0.227 | 0.31657 | -1.77 | 3.0E-5 | 0.294 |
| 42599 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 22 | CDH19 | Sponge network | 0.61 | 0.01593 | -3.434 | 0 | 0.294 |
| 42600 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 17 | MYCBP2 | Sponge network | -0.739 | 0.0574 | -0.027 | 0.82016 | 0.294 |
| 42601 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-664a-3p | 12 | CNTNAP3 | Sponge network | 0.95 | 0.15943 | -1.465 | 0.00033 | 0.294 |
| 42602 | RP11-67L2.2 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 14 | FMNL3 | Sponge network | 0.72 | 0.0001 | 0.555 | 4.0E-5 | 0.294 |
| 42603 | RP11-2H8.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 15 | APC | Sponge network | 1.089 | 0 | -0.255 | 0.04051 | 0.294 |
| 42604 | USP3-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p | 37 | CREB3L2 | Sponge network | -0.162 | 0.47687 | -0.525 | 2.0E-5 | 0.294 |
| 42605 | RP11-157J24.2 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-769-5p | 10 | SYNPO2 | Sponge network | -1.664 | 0.05165 | -2.342 | 0 | 0.294 |
| 42606 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-625-5p | 19 | ANK2 | Sponge network | -0.602 | 0.00331 | -1.603 | 1.0E-5 | 0.294 |
| 42607 | LINC00863 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | PTCH1 | Sponge network | 0.189 | 0.13981 | -0.1 | 0.6179 | 0.294 |
| 42608 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p | 13 | HLF | Sponge network | -2.549 | 0.00528 | -2.443 | 0 | 0.294 |
| 42609 | RBPMS-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | CADM2 | Sponge network | 0.144 | 0.64989 | -3.782 | 0 | 0.294 |
| 42610 | NDUFA6-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | BTG2 | Sponge network | -0.355 | 0.0077 | -1.763 | 0 | 0.294 |
| 42611 | CTD-2245F17.3 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-629-3p | 10 | EBF1 | Sponge network | -0.421 | 0.12556 | -0.384 | 0.08856 | 0.294 |
| 42612 | ARHGEF26-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935 | 34 | OPCML | Sponge network | -2.46 | 0 | -2.498 | 0 | 0.294 |
| 42613 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p | 16 | ARHGAP28 | Sponge network | 0.889 | 9.0E-5 | -0.911 | 0.00013 | 0.294 |
| 42614 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | -0.668 | 3.0E-5 | -0.509 | 0.03957 | 0.294 |
| 42615 | SERHL |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p | 12 | DYNC1I1 | Sponge network | -0.602 | 0.00331 | -1.12 | 0.00107 | 0.294 |
| 42616 | RP5-994D16.9 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-590-3p | 13 | ADARB1 | Sponge network | 0.252 | 0.08508 | -0.335 | 0.06631 | 0.294 |
| 42617 | CTC-429P9.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-940 | 20 | TOM1L2 | Sponge network | 0.082 | 0.55397 | -0.994 | 0 | 0.294 |
| 42618 | AC109309.4 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b | 14 | RUNX1T1 | Sponge network | 0.163 | 0.87219 | -0.344 | 0.2374 | 0.294 |
| 42619 | RP4-605O3.4 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 11 | MOB1B | Sponge network | 0.262 | 0.0475 | 0.197 | 0.15266 | 0.294 |
| 42620 | RP11-244H3.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-339-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | STARD13 | Sponge network | 0.659 | 3.0E-5 | -0.187 | 0.27383 | 0.294 |
| 42621 | RP5-1021I20.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-93-5p | 27 | CADM2 | Sponge network | -0.785 | 0.00035 | -3.782 | 0 | 0.294 |
| 42622 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 40 | CBX5 | Sponge network | 0.219 | 0.1516 | 0.165 | 0.24446 | 0.294 |
| 42623 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-655-3p;hsa-miR-92a-3p | 14 | UBE4B | Sponge network | 0.214 | 0.18246 | -0.235 | 0.007 | 0.294 |
| 42624 | RP1-151F17.2 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p | 11 | DYNC1I1 | Sponge network | 0.222 | 0.29133 | -1.12 | 0.00107 | 0.294 |
| 42625 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 12 | ITGB3 | Sponge network | 0.349 | 0.01695 | -0.011 | 0.95751 | 0.294 |
| 42626 | RP11-531A24.5 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429 | 13 | MRVI1 | Sponge network | 0.75 | 0.00054 | -1.051 | 0.00022 | 0.294 |
| 42627 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-590-5p | 17 | YAP1 | Sponge network | -1.645 | 0 | 0.22 | 0.11836 | 0.293 |
| 42628 | U91328.19 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-629-3p;hsa-miR-877-5p | 22 | ASTN1 | Sponge network | -0.303 | 0.21002 | -2.389 | 2.0E-5 | 0.293 |
| 42629 | RP11-678G14.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-339-5p;hsa-miR-7-1-3p | 10 | LUZP2 | Sponge network | -4.477 | 0 | -0.056 | 0.89416 | 0.293 |
| 42630 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p | 16 | BMPR2 | Sponge network | 0.924 | 0 | 0.206 | 0.0635 | 0.293 |
| 42631 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | MLLT10 | Sponge network | 0.603 | 0.00127 | 0.105 | 0.33292 | 0.293 |
| 42632 | RP11-35G9.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 16 | BAZ2B | Sponge network | 0.657 | 4.0E-5 | -0.276 | 0.02833 | 0.293 |
| 42633 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 48 | WDFY3 | Sponge network | -1.439 | 4.0E-5 | -0.201 | 0.0967 | 0.293 |
| 42634 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-582-5p | 13 | RELN | Sponge network | 0.299 | 0.57722 | -2.403 | 0 | 0.293 |
| 42635 | LINC00667 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 45 | DTNA | Sponge network | -0.235 | 0.15142 | -1.648 | 1.0E-5 | 0.293 |
| 42636 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DACH2 | Sponge network | 0.374 | 0.20054 | -1.635 | 0.00034 | 0.293 |
| 42637 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | RASSF2 | Sponge network | -0.206 | 0.12942 | -0.273 | 0.21961 | 0.293 |
| 42638 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-96-5p | 21 | ARID5B | Sponge network | 0.331 | 0.57247 | 0.342 | 0.01676 | 0.293 |
| 42639 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | -0.513 | 0.04703 | -1.047 | 0.00364 | 0.293 |
| 42640 | KB-431C1.4 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-3614-5p;hsa-miR-495-3p | 13 | TSC1 | Sponge network | 0.909 | 0 | 0.103 | 0.26071 | 0.293 |
| 42641 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-577 | 14 | MYCBP2 | Sponge network | 0.921 | 0.00118 | -0.027 | 0.82016 | 0.293 |
| 42642 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-766-3p | 30 | MAPK1 | Sponge network | 0.559 | 0.00026 | -0.017 | 0.81465 | 0.293 |
| 42643 | LINC00476 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 18 | ID4 | Sponge network | -0.615 | 0.00015 | -1.644 | 0 | 0.293 |
| 42644 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-589-5p | 15 | ADARB1 | Sponge network | -7.551 | 0 | -0.335 | 0.06631 | 0.293 |
| 42645 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 19 | ARHGAP29 | Sponge network | -0.777 | 0.1134 | 0.131 | 0.42953 | 0.293 |
| 42646 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-501-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 24 | TGFBR3 | Sponge network | 0.614 | 1.0E-5 | -1.173 | 1.0E-5 | 0.293 |
| 42647 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-375;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-5p | 11 | PDGFC | Sponge network | 0.214 | 0.18246 | -0.33 | 0.06483 | 0.293 |
| 42648 | LINC00702 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 18 | PICALM | Sponge network | -0.737 | 0.06182 | 0.086 | 0.23523 | 0.293 |
| 42649 | LINC00957 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 15 | UBE4B | Sponge network | -0.421 | 0.0114 | -0.235 | 0.007 | 0.293 |
| 42650 | RP4-613B23.1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-409-3p | 10 | OPCML | Sponge network | -0.309 | 0.1145 | -2.498 | 0 | 0.293 |
| 42651 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 31 | THRB | Sponge network | -0.285 | 0.2172 | -1.117 | 0.0004 | 0.293 |
| 42652 | BCYRN1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p;hsa-miR-940 | 13 | DLG1 | Sponge network | 0.311 | 0.10344 | -0.014 | 0.8926 | 0.293 |
| 42653 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | AKAP6 | Sponge network | 0.727 | 3.0E-5 | -1.586 | 3.0E-5 | 0.293 |
| 42654 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | PIK3C2A | Sponge network | 1.04 | 0.00023 | 0.061 | 0.53776 | 0.293 |
| 42655 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-590-5p;hsa-miR-942-5p | 12 | RB1CC1 | Sponge network | 0.267 | 0.2937 | 0.175 | 0.12559 | 0.293 |
| 42656 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-3614-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 10 | NUAK1 | Sponge network | -0.011 | 0.967 | 0.904 | 0 | 0.293 |
| 42657 | DNM1P35 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | SYNPO2 | Sponge network | 0.051 | 0.83388 | -2.342 | 0 | 0.293 |
| 42658 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | PCDH8 | Sponge network | -0.576 | 0.10121 | -0.997 | 0.05366 | 0.293 |
| 42659 | RP11-819C21.1 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | USP9X | Sponge network | 0.537 | 0.00139 | 0.222 | 0.01689 | 0.293 |
| 42660 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-628-5p | 10 | TRIM33 | Sponge network | 1.361 | 0 | 0.402 | 7.0E-5 | 0.293 |
| 42661 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-874-3p | 23 | NR2C2 | Sponge network | -0.615 | 0.00015 | 0.21 | 0.03224 | 0.293 |
| 42662 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-874-3p | 12 | NR2C2 | Sponge network | 0.859 | 0.00331 | 0.21 | 0.03224 | 0.293 |
| 42663 | RP11-344B5.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-24-3p | 13 | PTPRD | Sponge network | -0.055 | 0.88482 | -1.005 | 0.00064 | 0.293 |
| 42664 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 12 | SLITRK6 | Sponge network | -0.612 | 0.00094 | -2.121 | 0 | 0.293 |
| 42665 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 19 | CCND2 | Sponge network | 0.497 | 0.01451 | -0.267 | 0.3112 | 0.293 |
| 42666 | AC109309.4 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p | 12 | THRB | Sponge network | 0.163 | 0.87219 | -1.117 | 0.0004 | 0.293 |
| 42667 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | -0.096 | 0.67958 | 0.323 | 0.00792 | 0.293 |
| 42668 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-628-5p | 21 | CDC73 | Sponge network | 1.11 | 0 | 0.094 | 0.20463 | 0.293 |
| 42669 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p | 15 | PRKAA1 | Sponge network | 1.206 | 0 | 0.317 | 0.0031 | 0.293 |
| 42670 | LINC00672 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-1-5p | 14 | LUZP2 | Sponge network | -0.227 | 0.31657 | -0.056 | 0.89416 | 0.293 |
| 42671 | ADAMTS9-AS2 |
hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CDH11 | Sponge network | -2.452 | 0 | 1.427 | 0 | 0.293 |
| 42672 | LINC00265 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-539-5p | 23 | KIF1B | Sponge network | 1.07 | 0 | -0.118 | 0.32258 | 0.293 |
| 42673 | RP11-473I1.9 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 13 | LATS2 | Sponge network | 0.683 | 1.0E-5 | 0.159 | 0.2304 | 0.293 |
| 42674 | RP11-531A24.5 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p | 14 | PREX2 | Sponge network | 0.75 | 0.00054 | -0.272 | 0.25382 | 0.293 |
| 42675 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p | 14 | RET | Sponge network | -0.611 | 0.09607 | -0.833 | 0.03517 | 0.293 |
| 42676 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 24 | ST6GAL2 | Sponge network | 0.222 | 0.29133 | -1.201 | 0.00597 | 0.293 |
| 42677 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-93-5p | 19 | AHNAK | Sponge network | -0.154 | 0.78939 | -1.207 | 0 | 0.293 |
| 42678 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-7-5p | 26 | CLASP2 | Sponge network | -0.384 | 0.40652 | -0.038 | 0.71 | 0.293 |
| 42679 | CTD-2017D11.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p | 11 | PRDM2 | Sponge network | 0.792 | 0.0049 | -0.258 | 0.00721 | 0.293 |
| 42680 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 10 | PDZD2 | Sponge network | 0.349 | 0.01695 | -0.909 | 3.0E-5 | 0.293 |
| 42681 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | RSPO2 | Sponge network | 0.199 | 0.38015 | -3.038 | 0 | 0.293 |
| 42682 | FAM215B |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-320a;hsa-miR-320b | 13 | CYLD | Sponge network | 0.817 | 3.0E-5 | -0.367 | 0.00171 | 0.293 |
| 42683 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-877-5p | 14 | NOTCH2NL | Sponge network | -0.162 | 0.47687 | 0.024 | 0.84307 | 0.293 |
| 42684 | CTD-2303H24.2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-940 | 20 | SIRT1 | Sponge network | 0.415 | 0.14657 | 0.109 | 0.2593 | 0.293 |
| 42685 | RP11-46C24.7 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 21 | SYNPO2 | Sponge network | 0.392 | 0.0278 | -2.342 | 0 | 0.293 |
| 42686 | WDFY3-AS2 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | GABRB2 | Sponge network | -0.942 | 0 | -1.841 | 0.00029 | 0.293 |
| 42687 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-550a-5p | 11 | FAT2 | Sponge network | 0.72 | 0.0001 | -0.278 | 0.44877 | 0.293 |
| 42688 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 23 | PGR | Sponge network | 0.349 | 0.01695 | -1.704 | 0 | 0.293 |
| 42689 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | ERG | Sponge network | 0.214 | 0.18246 | 0.002 | 0.99011 | 0.293 |
| 42690 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 74 | KIF1B | Sponge network | -1.111 | 0.0003 | -0.118 | 0.32258 | 0.293 |
| 42691 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TIMP3 | Sponge network | -0.053 | 0.80685 | -0.546 | 0.02307 | 0.293 |
| 42692 | RP11-678G14.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-532-3p | 11 | GAS7 | Sponge network | -1.765 | 0.0778 | -0.555 | 0.01602 | 0.293 |
| 42693 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | TACC1 | Sponge network | 0.411 | 0.1335 | -0.362 | 0.05406 | 0.293 |
| 42694 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ZFHX3 | Sponge network | 0.727 | 3.0E-5 | 0.389 | 0.01363 | 0.293 |
| 42695 | PART1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 26 | ARNT2 | Sponge network | -2.925 | 0 | -0.332 | 0.25851 | 0.293 |
| 42696 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-342-3p | 11 | UBE4B | Sponge network | -0.793 | 7.0E-5 | -0.235 | 0.007 | 0.293 |
| 42697 | RP11-113K21.4 |
hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p | 10 | CREB1 | Sponge network | 0.676 | 0 | 0.203 | 0.00537 | 0.293 |
| 42698 | SH3BP5-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-671-5p | 17 | CYLD | Sponge network | 0.267 | 0.2937 | -0.367 | 0.00171 | 0.293 |
| 42699 | RP11-212P7.2 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 14 | KIF5B | Sponge network | 0.219 | 0.1516 | 0.362 | 0.00066 | 0.293 |
| 42700 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | PRKAB2 | Sponge network | 0.683 | 1.0E-5 | -0.367 | 0.01371 | 0.293 |
| 42701 | LINC00667 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 18 | DYNC1I1 | Sponge network | -0.235 | 0.15142 | -1.12 | 0.00107 | 0.293 |
| 42702 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | FZD3 | Sponge network | 0.222 | 0.29133 | 0.104 | 0.60316 | 0.293 |
| 42703 | RP11-66B24.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p | 10 | CXXC4 | Sponge network | 0.57 | 0.1866 | -0.433 | 0.21055 | 0.293 |
| 42704 | RP11-574K11.29 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | SLC5A3 | Sponge network | 0.997 | 0 | 0.493 | 5.0E-5 | 0.293 |
| 42705 | AC009948.5 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-532-5p;hsa-miR-940;hsa-miR-96-5p | 11 | POU2F2 | Sponge network | 0.532 | 0.00505 | -0.233 | 0.32214 | 0.293 |
| 42706 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 25 | CREB3L2 | Sponge network | -0.72 | 0.24239 | -0.525 | 2.0E-5 | 0.293 |
| 42707 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p | 13 | FREM2 | Sponge network | -0.13 | 0.48734 | -0.437 | 0.46353 | 0.293 |
| 42708 | AC009120.6 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | KIF5B | Sponge network | 0.919 | 0 | 0.362 | 0.00066 | 0.293 |
| 42709 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-143-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p | 17 | CLASP2 | Sponge network | 1.044 | 2.0E-5 | -0.038 | 0.71 | 0.293 |
| 42710 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | ABCC9 | Sponge network | 0.311 | 0.10344 | -0.466 | 0.14121 | 0.293 |
| 42711 | LIPE-AS1 |
hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-328-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-625-5p | 13 | TMTC2 | Sponge network | 0.078 | 0.55803 | -0.215 | 0.18248 | 0.293 |
| 42712 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 21 | TSHZ3 | Sponge network | 0.389 | 0.04293 | -0.556 | 0.01516 | 0.293 |
| 42713 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | TIMP3 | Sponge network | -2.925 | 0 | -0.546 | 0.02307 | 0.293 |
| 42714 | FENDRR |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 23 | DOCK4 | Sponge network | -1.281 | 0.00012 | 0.502 | 0.00108 | 0.293 |
| 42715 | ADIRF-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | -0.223 | 0.47816 | 0.154 | 0.74723 | 0.293 |
| 42716 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 30 | PRKCE | Sponge network | -2.46 | 0 | -0.403 | 0.00056 | 0.293 |
| 42717 | GS1-124K5.11 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-450b-5p | 10 | PRKD1 | Sponge network | 0.494 | 0.00339 | -0.466 | 0.03583 | 0.293 |
| 42718 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p | 27 | PHC3 | Sponge network | 0.056 | 0.81195 | 0.212 | 0.0499 | 0.293 |
| 42719 | CTD-2017D11.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-628-5p | 11 | RASSF8 | Sponge network | 0.792 | 0.0049 | -0.122 | 0.65591 | 0.293 |
| 42720 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-5p | 19 | EGLN1 | Sponge network | 0.706 | 0 | -0.127 | 0.1536 | 0.293 |
| 42721 | RP1-122P22.2 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p | 10 | KCNA6 | Sponge network | 0.065 | 0.73468 | -1.531 | 0.00013 | 0.293 |
| 42722 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | NOTCH2NL | Sponge network | 0.532 | 0.00505 | 0.024 | 0.84307 | 0.293 |
| 42723 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-3p | 15 | PTPN11 | Sponge network | -0.031 | 0.89063 | 0.431 | 2.0E-5 | 0.293 |
| 42724 | LINC00094 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-92a-3p | 10 | PTPN13 | Sponge network | 0.253 | 0.09565 | -1.208 | 0 | 0.293 |
| 42725 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | HECA | Sponge network | 0.603 | 0.00127 | -0.217 | 0.03463 | 0.293 |
| 42726 | RP11-755F10.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-223-3p;hsa-miR-940 | 10 | MEF2C | Sponge network | 0.952 | 0 | -0.499 | 0.01448 | 0.293 |
| 42727 | EMX2OS |
hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-5p | 14 | PIM1 | Sponge network | -0.777 | 0.1134 | -1.114 | 0 | 0.293 |
| 42728 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 12 | KCNAB1 | Sponge network | 0.75 | 0.00054 | -0.755 | 2.0E-5 | 0.293 |
| 42729 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | CEP170 | Sponge network | 0.494 | 0.00339 | 0.943 | 0 | 0.293 |
| 42730 | LINC01006 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p | 10 | CDHR3 | Sponge network | -0.702 | 0.04814 | -0.977 | 4.0E-5 | 0.293 |
| 42731 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ZFHX4 | Sponge network | 0.272 | 0.44692 | 0.018 | 0.95574 | 0.293 |
| 42732 | LINC00863 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 14 | SNX29 | Sponge network | 0.189 | 0.13981 | -0.34 | 0.01371 | 0.293 |
| 42733 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 15 | EYA4 | Sponge network | 0.238 | 0.70544 | 0.3 | 0.36876 | 0.293 |
| 42734 | HAND2-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 14 | USP25 | Sponge network | -1.697 | 0.00889 | -0.242 | 0.01397 | 0.293 |
| 42735 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-424-5p | 21 | PHC3 | Sponge network | 0.594 | 0.41522 | 0.212 | 0.0499 | 0.293 |
| 42736 | CTD-2017D11.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-628-5p | 23 | RUNX1T1 | Sponge network | 0.792 | 0.0049 | -0.344 | 0.2374 | 0.293 |
| 42737 | RP11-474O21.5 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-744-3p;hsa-miR-93-5p | 16 | DST | Sponge network | -0.023 | 0.95109 | -0.713 | 0.00096 | 0.293 |
| 42738 | RP11-384K6.6 |
hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-411-5p | 10 | TMEM30A | Sponge network | 0.946 | 0 | 0.096 | 0.31031 | 0.293 |
| 42739 | ARHGEF26-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | BCL9 | Sponge network | -2.46 | 0 | 0.321 | 0.00267 | 0.293 |
| 42740 | AC034220.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | BAZ2B | Sponge network | 0.309 | 0.07304 | -0.276 | 0.02833 | 0.293 |
| 42741 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-590-5p | 19 | RNASEL | Sponge network | 0.331 | 0.57247 | -0.272 | 0.01796 | 0.293 |
| 42742 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 40 | TMEM132B | Sponge network | -0.896 | 0.00099 | -0.83 | 0.01084 | 0.293 |
| 42743 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 15 | PIK3C2A | Sponge network | 1.089 | 0 | 0.061 | 0.53776 | 0.293 |
| 42744 | AC016747.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p | 10 | KCTD8 | Sponge network | 0.199 | 0.38015 | -3.359 | 0 | 0.293 |
| 42745 | RP11-38L15.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 11 | ARID5B | Sponge network | 0.344 | 0.3127 | 0.342 | 0.01676 | 0.293 |
| 42746 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 29 | IGF1 | Sponge network | 0.064 | 0.72934 | -0.204 | 0.5898 | 0.293 |
| 42747 | RP11-307B6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-93-3p | 17 | NCOA1 | Sponge network | -4.463 | 0.00025 | -0.432 | 0 | 0.293 |
| 42748 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 16 | PGR | Sponge network | -0.141 | 0.26434 | -1.704 | 0 | 0.293 |
| 42749 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | APC | Sponge network | 0.873 | 0 | -0.255 | 0.04051 | 0.293 |
| 42750 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-582-5p | 16 | PIK3C2A | Sponge network | 1.03 | 0 | 0.061 | 0.53776 | 0.293 |
| 42751 | WDFY3-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 37 | AFF1 | Sponge network | -0.942 | 0 | -0.384 | 0.00053 | 0.293 |
| 42752 | DNM1P35 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 13 | SPG20 | Sponge network | 0.051 | 0.83388 | -1.16 | 0 | 0.293 |
| 42753 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | HECA | Sponge network | 0.998 | 0 | -0.217 | 0.03463 | 0.293 |
| 42754 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-625-5p | 18 | ZMYM2 | Sponge network | 0.594 | 0.41522 | 0.279 | 0.01273 | 0.293 |
| 42755 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p | 17 | DYNC1I1 | Sponge network | -1.047 | 0 | -1.12 | 0.00107 | 0.293 |
| 42756 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-766-3p | 15 | IL17RD | Sponge network | 0.238 | 0.70544 | -0.019 | 0.94873 | 0.293 |
| 42757 | DNAJC27-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 28 | NAV3 | Sponge network | -0.969 | 2.0E-5 | 0.212 | 0.47729 | 0.293 |
| 42758 | AF131215.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-671-5p | 12 | CDH23 | Sponge network | -0.176 | 0.59692 | -0.974 | 0.00034 | 0.293 |
| 42759 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-96-5p | 17 | RIMBP2 | Sponge network | -0.976 | 0.00025 | -1.968 | 5.0E-5 | 0.293 |
| 42760 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p | 11 | FGFR1 | Sponge network | 0.238 | 0.70544 | -0.509 | 0.03957 | 0.293 |
| 42761 | LINC00473 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p | 10 | PRDM11 | Sponge network | -1.203 | 0.03881 | -0.252 | 0.15511 | 0.293 |
| 42762 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-362-3p | 12 | GRIN2A | Sponge network | -1.255 | 0.00981 | -2.161 | 0 | 0.293 |
| 42763 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | AHNAK | Sponge network | -1.203 | 0.03881 | -1.207 | 0 | 0.293 |
| 42764 | RP11-35G9.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 24 | NTRK2 | Sponge network | 0.657 | 4.0E-5 | -0.736 | 0.06495 | 0.293 |
| 42765 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-93-5p | 22 | ETV1 | Sponge network | -1.654 | 0.00144 | -0.096 | 0.67674 | 0.293 |
| 42766 | SNHG14 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | UBE4B | Sponge network | -1.111 | 0.0003 | -0.235 | 0.007 | 0.293 |
| 42767 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p | 22 | SCN9A | Sponge network | 0.267 | 0.2937 | -0.525 | 0.10593 | 0.293 |
| 42768 | AC108488.4 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p | 10 | GLIPR1 | Sponge network | 0.259 | 0.12542 | 0.146 | 0.3276 | 0.293 |
| 42769 | RP3-337H4.8 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-9-5p | 13 | CCDC6 | Sponge network | 1.527 | 0 | 0.27 | 0.02555 | 0.293 |
| 42770 | AC018890.6 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | PIK3CA | Sponge network | 1.61 | 0.00961 | 0.195 | 0.06908 | 0.293 |
| 42771 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-877-5p | 22 | GRIN2A | Sponge network | -0.465 | 0.04703 | -2.161 | 0 | 0.293 |
| 42772 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p | 17 | RUNX1T1 | Sponge network | 0.348 | 0.32912 | -0.344 | 0.2374 | 0.293 |
| 42773 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-93-5p | 14 | IGSF10 | Sponge network | 0.497 | 0.01451 | -1.751 | 1.0E-5 | 0.293 |
| 42774 | RP11-21A7A.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-589-5p | 11 | EYA4 | Sponge network | -1.116 | 0.23686 | 0.3 | 0.36876 | 0.293 |
| 42775 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 12 | SOX11 | Sponge network | 0.673 | 0 | 2.68 | 0 | 0.293 |
| 42776 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TBX18 | Sponge network | 0.421 | 0.00084 | 0.843 | 0.02945 | 0.293 |
| 42777 | HOXB-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-429 | 17 | IL17RD | Sponge network | 1.316 | 0.01106 | -0.019 | 0.94873 | 0.293 |
| 42778 | A2M-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-935 | 12 | RELN | Sponge network | -0.08 | 0.90092 | -2.403 | 0 | 0.293 |
| 42779 | AC018890.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 11 | PLAG1 | Sponge network | 1.61 | 0.00961 | -0.315 | 0.26532 | 0.293 |
| 42780 | MRVI1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 14 | EFNA5 | Sponge network | -1.092 | 4.0E-5 | -0.421 | 0.17868 | 0.293 |
| 42781 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 51 | TRPS1 | Sponge network | 0.311 | 0.10344 | -0.191 | 0.44109 | 0.293 |
| 42782 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 25 | CYLD | Sponge network | -0.49 | 0.04946 | -0.367 | 0.00171 | 0.293 |
| 42783 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-98-5p | 35 | CADM2 | Sponge network | -0.773 | 0.04073 | -3.782 | 0 | 0.293 |
| 42784 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b | 12 | MYCBP2 | Sponge network | 0.331 | 0.57247 | -0.027 | 0.82016 | 0.293 |
| 42785 | RP11-244H3.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-940 | 44 | NFIB | Sponge network | 0.659 | 3.0E-5 | 0.023 | 0.90795 | 0.293 |
| 42786 | RAMP2-AS1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | RPS6KA6 | Sponge network | -0.513 | 0.04703 | -2.989 | 0 | 0.293 |
| 42787 | LINC00702 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 17 | POU2F2 | Sponge network | -0.737 | 0.06182 | -0.233 | 0.32214 | 0.293 |
| 42788 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 12 | GPAM | Sponge network | 1.597 | 0 | 0.142 | 0.29732 | 0.293 |
| 42789 | AGAP11 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 11 | PHOX2B | Sponge network | -1.645 | 0 | -2.68 | 0 | 0.293 |
| 42790 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-93-5p | 14 | CNTNAP3 | Sponge network | 0.253 | 0.09565 | -1.465 | 0.00033 | 0.293 |
| 42791 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-374a-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 30 | USP12 | Sponge network | -2.452 | 0 | -0.102 | 0.40768 | 0.293 |
| 42792 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 29 | QKI | Sponge network | -0.141 | 0.26434 | -0.333 | 0.04111 | 0.293 |
| 42793 | TPTEP1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-942-5p | 33 | OPCML | Sponge network | -1.216 | 0 | -2.498 | 0 | 0.293 |
| 42794 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-376c-3p;hsa-miR-590-3p | 15 | CREBBP | Sponge network | 0.532 | 0.00505 | 0.126 | 0.15186 | 0.293 |
| 42795 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 34 | ARID5B | Sponge network | -0.737 | 0.06182 | 0.342 | 0.01676 | 0.293 |
| 42796 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-505-3p;hsa-miR-590-3p | 12 | ZNF292 | Sponge network | 0.415 | 0.14657 | 0.276 | 0.04148 | 0.293 |
| 42797 | RP11-77H9.2 |
hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | BRD3 | Sponge network | 1.677 | 0 | 0.37 | 0.00013 | 0.293 |
| 42798 | RP11-890B15.3 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | PPM1L | Sponge network | 0.101 | 0.60919 | -0.537 | 0.00616 | 0.293 |
| 42799 | RP11-195B17.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b | 10 | NRK | Sponge network | 0.37 | 0.27614 | 0.656 | 0.19217 | 0.293 |
| 42800 | RP11-277P12.20 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | SETBP1 | Sponge network | 0.687 | 0.02753 | -1.302 | 1.0E-5 | 0.293 |
| 42801 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-378a-5p | 10 | CACNA1E | Sponge network | -1.161 | 0.00591 | 1.752 | 0 | 0.293 |
| 42802 | PCA3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | GNAQ | Sponge network | -0.709 | 0.18168 | -0.367 | 0.00102 | 0.293 |
| 42803 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-940 | 12 | HOOK3 | Sponge network | -0.303 | 0.21002 | 0.273 | 0.09957 | 0.293 |
| 42804 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p | 17 | EBF3 | Sponge network | 0.389 | 0.04293 | -0.058 | 0.81437 | 0.293 |
| 42805 | SNHG5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-23b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | BCLAF1 | Sponge network | 0.13 | 0.47174 | 0.211 | 0.00684 | 0.293 |
| 42806 | CTC-490E21.10 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p | 21 | DST | Sponge network | 1.46 | 1.0E-5 | -0.713 | 0.00096 | 0.293 |
| 42807 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-93-5p | 14 | LRRC55 | Sponge network | -0.793 | 7.0E-5 | -0.026 | 0.93163 | 0.293 |
| 42808 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | ITGAV | Sponge network | -1.697 | 0.00889 | 0.337 | 0.01002 | 0.293 |
| 42809 | PGM5-AS1 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-23a-3p | 10 | FAM135B | Sponge network | -4.546 | 0 | -3.978 | 0 | 0.293 |
| 42810 | RP11-6N17.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-331-3p | 11 | DDX6 | Sponge network | 0.108 | 0.69556 | 0.091 | 0.36428 | 0.293 |
| 42811 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-93-5p | 10 | EDA2R | Sponge network | -0.351 | 0.11358 | -0.587 | 0.01598 | 0.293 |
| 42812 | RP11-21A7A.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-455-3p | 12 | PIK3R1 | Sponge network | -1.116 | 0.23686 | -0.133 | 0.36854 | 0.293 |
| 42813 | RP11-166D19.1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SH3GLB1 | Sponge network | -0.576 | 0.10121 | -0.115 | 0.14773 | 0.293 |
| 42814 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 15 | CDC73 | Sponge network | 0.439 | 0.00313 | 0.094 | 0.20463 | 0.293 |
| 42815 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-3p | 17 | EPHA5 | Sponge network | 0.37 | 0.27614 | -0.957 | 0.01733 | 0.293 |
| 42816 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-455-5p | 10 | THSD7A | Sponge network | 0.219 | 0.1516 | 0.242 | 0.25154 | 0.293 |
| 42817 | AC005154.6 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-654-3p | 27 | PIK3R1 | Sponge network | 0.848 | 0 | -0.133 | 0.36854 | 0.293 |
| 42818 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 18 | CNTN1 | Sponge network | 0.622 | 0.00015 | -2.594 | 0 | 0.293 |
| 42819 | ZNF667-AS1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-96-5p | 12 | DNMT3A | Sponge network | -0.976 | 0.00025 | 0.302 | 0.0262 | 0.293 |
| 42820 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-450b-5p;hsa-miR-501-5p | 12 | BCL6 | Sponge network | 0.176 | 0.37402 | -0.211 | 0.19489 | 0.293 |
| 42821 | RP11-589P10.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-671-5p | 10 | PTPRD | Sponge network | -2.044 | 0.00066 | -1.005 | 0.00064 | 0.293 |
| 42822 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | RCAN2 | Sponge network | 0.267 | 0.2937 | -0.33 | 0.15172 | 0.293 |
| 42823 | RP11-983P16.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 21 | PTPN14 | Sponge network | 0.41 | 0.02214 | 0.144 | 0.34595 | 0.293 |
| 42824 | CTC-559E9.6 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-654-3p | 16 | DDX6 | Sponge network | 0.601 | 1.0E-5 | 0.091 | 0.36428 | 0.293 |
| 42825 | CTA-204B4.2 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-374a-3p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | TRIM13 | Sponge network | 0.765 | 7.0E-5 | 0.183 | 0.06968 | 0.293 |
| 42826 | AP001347.6 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CTGF | Sponge network | -1.161 | 0.00591 | 0.321 | 0.11682 | 0.293 |
| 42827 | RP11-344B5.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-664a-3p | 10 | CDH19 | Sponge network | -0.055 | 0.88482 | -3.434 | 0 | 0.293 |
| 42828 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | NEDD4 | Sponge network | 0.826 | 1.0E-5 | 0.02 | 0.89834 | 0.293 |
| 42829 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-625-5p;hsa-miR-655-3p | 11 | ARID2 | Sponge network | 0.083 | 0.66405 | 0.235 | 0.02697 | 0.293 |
| 42830 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | SASH1 | Sponge network | -0.053 | 0.80685 | -0.502 | 0.00175 | 0.293 |
| 42831 | AC018890.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | EYA4 | Sponge network | 1.61 | 0.00961 | 0.3 | 0.36876 | 0.293 |
| 42832 | ADIRF-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | BRD3 | Sponge network | -0.223 | 0.47816 | 0.37 | 0.00013 | 0.293 |
| 42833 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 28 | CBX5 | Sponge network | 0.91 | 0 | 0.165 | 0.24446 | 0.293 |
| 42834 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | 0.853 | 0.15352 | -0.466 | 0.14121 | 0.293 |
| 42835 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | USP12 | Sponge network | 1.063 | 0 | -0.102 | 0.40768 | 0.293 |
| 42836 | A2M-AS1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-93-5p | 45 | DST | Sponge network | -0.08 | 0.90092 | -0.713 | 0.00096 | 0.293 |
| 42837 | RP4-791M13.3 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p | 21 | PIK3R1 | Sponge network | 0.419 | 0.0319 | -0.133 | 0.36854 | 0.293 |
| 42838 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p | 15 | TACC1 | Sponge network | 1.361 | 0 | -0.362 | 0.05406 | 0.293 |
| 42839 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 37 | TRPS1 | Sponge network | -0.513 | 0.04703 | -0.191 | 0.44109 | 0.293 |
| 42840 | MIR22HG |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-590-5p;hsa-miR-7-5p | 13 | FNBP1 | Sponge network | -1.047 | 0 | -0.969 | 4.0E-5 | 0.293 |
| 42841 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 22 | RICTOR | Sponge network | 0.529 | 0.00552 | 0.388 | 0.00188 | 0.293 |
| 42842 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | PNRC1 | Sponge network | -0.358 | 0.17255 | -0.646 | 0 | 0.293 |
| 42843 | CTA-204B4.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-3p | 10 | PLAG1 | Sponge network | 0.765 | 7.0E-5 | -0.315 | 0.26532 | 0.293 |
| 42844 | TRAF3IP2-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | LUZP2 | Sponge network | -0.323 | 0.01372 | -0.056 | 0.89416 | 0.293 |
| 42845 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-365a-3p;hsa-miR-378a-5p | 12 | CACNA1E | Sponge network | 0.083 | 0.66405 | 1.752 | 0 | 0.293 |
| 42846 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PBRM1 | Sponge network | 0.214 | 0.18246 | -0.029 | 0.78601 | 0.293 |
| 42847 | LINC00622 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 12 | ABI2 | Sponge network | 1.169 | 0.00028 | 0.175 | 0.14787 | 0.293 |
| 42848 | RP11-181G12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p | 13 | DMD | Sponge network | 0.556 | 0.00298 | -1.506 | 0 | 0.293 |
| 42849 | RP11-253E3.3 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-497-5p;hsa-miR-9-5p | 13 | CDK6 | Sponge network | 1.764 | 0 | 0.991 | 0 | 0.293 |
| 42850 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 18 | KCNA6 | Sponge network | -4.477 | 0 | -1.531 | 0.00013 | 0.293 |
| 42851 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 18 | SOX11 | Sponge network | 0.082 | 0.55654 | 2.68 | 0 | 0.293 |
| 42852 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-93-5p | 45 | ADARB1 | Sponge network | 0.219 | 0.1516 | -0.335 | 0.06631 | 0.293 |
| 42853 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-7-1-3p | 20 | MLLT10 | Sponge network | 0.177 | 0.16681 | 0.105 | 0.33292 | 0.293 |
| 42854 | CTC-429P9.5 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p | 14 | KCNA6 | Sponge network | 0.178 | 0.29106 | -1.531 | 0.00013 | 0.293 |
| 42855 | FAM66C |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | AFAP1L2 | Sponge network | -0.901 | 0.00019 | -0.284 | 0.15434 | 0.293 |
| 42856 | GNG12-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-590-3p | 25 | EYA4 | Sponge network | -0.793 | 7.0E-5 | 0.3 | 0.36876 | 0.293 |
| 42857 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 41 | SMAD4 | Sponge network | 0.614 | 1.0E-5 | -0.313 | 0.00413 | 0.293 |
| 42858 | RP4-669P10.16 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-629-5p | 19 | BCL2 | Sponge network | -0.301 | 0.06597 | -0.899 | 0 | 0.293 |
| 42859 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 19 | RASSF2 | Sponge network | 0.497 | 0.01451 | -0.273 | 0.21961 | 0.293 |
| 42860 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-625-5p;hsa-miR-98-5p | 24 | RSPO2 | Sponge network | -0.18 | 0.20343 | -3.038 | 0 | 0.293 |
| 42861 | CTD-2528L19.6 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | BCL9 | Sponge network | 0.953 | 0 | 0.321 | 0.00267 | 0.293 |
| 42862 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-3614-5p;hsa-miR-590-5p | 10 | EPHA3 | Sponge network | 1.134 | 0 | 0.185 | 0.53588 | 0.293 |
| 42863 | FAM215B |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p | 11 | RASAL2 | Sponge network | 0.817 | 3.0E-5 | 0.869 | 0 | 0.293 |
| 42864 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SEMA3E | Sponge network | 1.302 | 7.0E-5 | -1.717 | 0.00137 | 0.293 |
| 42865 | RP11-848P1.3 |
hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | MDM4 | Sponge network | 1.253 | 0 | 0.345 | 0.00762 | 0.293 |
| 42866 | BVES-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-935 | 14 | ARNT2 | Sponge network | -2.62 | 0 | -0.332 | 0.25851 | 0.293 |
| 42867 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p | 20 | ERBB4 | Sponge network | -2.531 | 0 | -1.975 | 2.0E-5 | 0.293 |
| 42868 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | CSMD1 | Sponge network | -0.358 | 0.17255 | -0.63 | 0.18881 | 0.293 |
| 42869 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | TACC1 | Sponge network | 0.853 | 0.15352 | -0.362 | 0.05406 | 0.293 |
| 42870 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | CUL5 | Sponge network | 0.799 | 1.0E-5 | 0.026 | 0.77301 | 0.293 |
| 42871 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 28 | MOB1B | Sponge network | 0.41 | 0.02214 | 0.197 | 0.15266 | 0.293 |
| 42872 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | PIK3C2A | Sponge network | 0.659 | 3.0E-5 | 0.061 | 0.53776 | 0.293 |
| 42873 | CALML3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-625-5p | 16 | SUFU | Sponge network | 0.95 | 0.15943 | -0.04 | 0.65489 | 0.293 |
| 42874 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | EPHA3 | Sponge network | 0.538 | 0.00076 | 0.185 | 0.53588 | 0.293 |
| 42875 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-539-5p | 12 | MGA | Sponge network | 0.389 | 0.04293 | 0.323 | 0.00792 | 0.293 |
| 42876 | MIR143HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 25 | ARNT2 | Sponge network | -0.739 | 0.0574 | -0.332 | 0.25851 | 0.293 |
| 42877 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-3p | 31 | ERC1 | Sponge network | -0.355 | 0.0077 | -0.108 | 0.38794 | 0.293 |
| 42878 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-214-3p;hsa-miR-301a-3p;hsa-miR-30a-3p | 12 | MLLT10 | Sponge network | 0.529 | 0.00552 | 0.105 | 0.33292 | 0.293 |
| 42879 | RP11-327P2.5 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-450b-5p | 10 | IGSF10 | Sponge network | -0.147 | 0.40452 | -1.751 | 1.0E-5 | 0.293 |
| 42880 | JAZF1-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 59 | DST | Sponge network | -0.769 | 0.00035 | -0.713 | 0.00096 | 0.293 |
| 42881 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p | 13 | EPHA3 | Sponge network | 0.993 | 0 | 0.185 | 0.53588 | 0.293 |
| 42882 | AC141928.1 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 12 | IGSF10 | Sponge network | 0.853 | 0.15352 | -1.751 | 1.0E-5 | 0.293 |
| 42883 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 31 | MOB1B | Sponge network | -0.842 | 0.01361 | 0.197 | 0.15266 | 0.293 |
| 42884 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 11 | CHD2 | Sponge network | 0.799 | 1.0E-5 | 0.049 | 0.55371 | 0.293 |
| 42885 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | HACE1 | Sponge network | 0.848 | 0 | -0.072 | 0.55239 | 0.293 |
| 42886 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | BCL2 | Sponge network | 0.889 | 9.0E-5 | -0.899 | 0 | 0.293 |
| 42887 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-484 | 13 | ITGA5 | Sponge network | 0.082 | 0.55654 | 0.452 | 0.03524 | 0.293 |
| 42888 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PCDH8 | Sponge network | -0.739 | 0.0574 | -0.997 | 0.05366 | 0.293 |
| 42889 | RP11-166D19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | ERCC4 | Sponge network | -0.576 | 0.10121 | 0.126 | 0.15266 | 0.293 |
| 42890 | ERVH48-1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | UBR3 | Sponge network | 1.04 | 0.00023 | -0.021 | 0.82774 | 0.293 |
| 42891 | RP11-212P7.2 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-503-5p | 16 | ABL1 | Sponge network | 0.219 | 0.1516 | -0.215 | 0.07804 | 0.293 |
| 42892 | HNRNPU-AS1 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-328-3p;hsa-miR-335-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-495-3p | 21 | VPS53 | Sponge network | 1.601 | 0 | 0.103 | 0.22667 | 0.293 |
| 42893 | RP11-798G7.7 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-30d-3p | 13 | AFF1 | Sponge network | 0.858 | 3.0E-5 | -0.384 | 0.00053 | 0.293 |
| 42894 | PSMD5-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 13 | KIAA1024 | Sponge network | 0.569 | 0.00067 | 1.083 | 0 | 0.293 |
| 42895 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-326;hsa-miR-338-5p | 12 | SOX11 | Sponge network | 1.093 | 0 | 2.68 | 0 | 0.293 |
| 42896 | RP11-119F7.5 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | PRKCB | Sponge network | 0.225 | 0.30644 | -1.313 | 2.0E-5 | 0.293 |
| 42897 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-500a-3p;hsa-miR-628-5p;hsa-miR-7-5p | 28 | ERC1 | Sponge network | -0.899 | 6.0E-5 | -0.108 | 0.38794 | 0.293 |
| 42898 | LINC00621 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p | 16 | RIMS2 | Sponge network | 0.131 | 0.8436 | -1.365 | 0.00208 | 0.293 |
| 42899 | RP11-822E23.8 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-362-5p | 26 | ABI2 | Sponge network | -0.777 | 0.16667 | 0.175 | 0.14787 | 0.293 |
| 42900 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-590-3p | 19 | SMAD2 | Sponge network | 1.2 | 0.01813 | -0.128 | 0.12732 | 0.293 |
| 42901 | LINC00476 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 18 | RPS6KA2 | Sponge network | -0.615 | 0.00015 | -0.57 | 0.00013 | 0.293 |
| 42902 | RP11-395G23.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-766-3p;hsa-miR-93-5p | 14 | EZH1 | Sponge network | 0.381 | 0.25967 | -0.564 | 1.0E-5 | 0.293 |
| 42903 | RP5-1198O20.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 30 | CREB1 | Sponge network | 0.997 | 0.00066 | 0.203 | 0.00537 | 0.293 |
| 42904 | RP11-212P7.2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | PIK3CA | Sponge network | 0.219 | 0.1516 | 0.195 | 0.06908 | 0.293 |
| 42905 | CTD-3099C6.11 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-766-3p | 10 | MDM4 | Sponge network | 0.926 | 0.00148 | 0.345 | 0.00762 | 0.293 |
| 42906 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PHC3 | Sponge network | -0.021 | 0.93598 | 0.212 | 0.0499 | 0.293 |
| 42907 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-3613-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 43 | SMAD2 | Sponge network | -0.899 | 6.0E-5 | -0.128 | 0.12732 | 0.293 |
| 42908 | MAGI2-AS3 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 30 | EFNA5 | Sponge network | -0.129 | 0.57177 | -0.421 | 0.17868 | 0.293 |
| 42909 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-450b-5p | 12 | ADARB1 | Sponge network | -1.717 | 0.01522 | -0.335 | 0.06631 | 0.293 |
| 42910 | RP11-175O19.4 |
hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-497-5p | 11 | BRD3 | Sponge network | 0.917 | 0 | 0.37 | 0.00013 | 0.293 |
| 42911 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429 | 13 | ERCC4 | Sponge network | -1.654 | 0.00144 | 0.126 | 0.15266 | 0.292 |
| 42912 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | BMPR2 | Sponge network | 0.538 | 0.00076 | 0.206 | 0.0635 | 0.292 |
| 42913 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | CRTC1 | Sponge network | -0.777 | 0.1134 | -0.116 | 0.28412 | 0.292 |
| 42914 | BOLA3-AS1 |
hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p | 11 | PTCH1 | Sponge network | -0.246 | 0.35034 | -0.1 | 0.6179 | 0.292 |
| 42915 | RP11-867G23.10 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p | 11 | PDE4DIP | Sponge network | -2.531 | 0 | -0.282 | 0.0257 | 0.292 |
| 42916 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-29b-1-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-708-3p | 14 | PLXNA2 | Sponge network | -0.285 | 0.2172 | -0.395 | 0.00779 | 0.292 |
| 42917 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-877-5p | 16 | SUFU | Sponge network | -1.654 | 0.00144 | -0.04 | 0.65489 | 0.292 |
| 42918 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 18 | ZMYM2 | Sponge network | 0.34 | 0.00273 | 0.279 | 0.01273 | 0.292 |
| 42919 | RP11-46C24.7 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p | 13 | PKNOX1 | Sponge network | 0.392 | 0.0278 | 0.085 | 0.28917 | 0.292 |
| 42920 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | CUX1 | Sponge network | -1.161 | 0.00591 | -0.039 | 0.6705 | 0.292 |
| 42921 | RP11-46C24.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | FAM172A | Sponge network | 0.392 | 0.0278 | -0.57 | 0 | 0.292 |
| 42922 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 10 | ANK2 | Sponge network | 0.348 | 0.32912 | -1.603 | 1.0E-5 | 0.292 |
| 42923 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | NUAK1 | Sponge network | -0.246 | 0.35034 | 0.904 | 0 | 0.292 |
| 42924 | CTD-3092A11.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | HECA | Sponge network | 0.873 | 0 | -0.217 | 0.03463 | 0.292 |
| 42925 | RP4-584D14.7 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-577 | 17 | BMPR2 | Sponge network | 1.294 | 0.0031 | 0.206 | 0.0635 | 0.292 |
| 42926 | PCA3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 17 | POU2F2 | Sponge network | -0.709 | 0.18168 | -0.233 | 0.32214 | 0.292 |
| 42927 | RP11-57H14.4 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-590-3p | 14 | DDX3X | Sponge network | 0.083 | 0.66405 | -0.152 | 0.07686 | 0.292 |
| 42928 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 20 | NR2C2 | Sponge network | 0.862 | 0.06213 | 0.21 | 0.03224 | 0.292 |
| 42929 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | TBL1XR1 | Sponge network | 1.352 | 0 | 0.578 | 0 | 0.292 |
| 42930 | RP3-402G11.26 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-590-3p | 18 | TCF4 | Sponge network | -0.254 | 0.19303 | 0.016 | 0.92271 | 0.292 |
| 42931 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-7-1-3p | 24 | PDGFRA | Sponge network | -4.477 | 0 | -0.372 | 0.0037 | 0.292 |
| 42932 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-96-5p | 12 | ARHGAP29 | Sponge network | 0.331 | 0.57247 | 0.131 | 0.42953 | 0.292 |
| 42933 | H1FX-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-3p | 16 | FMNL3 | Sponge network | -0.206 | 0.12942 | 0.555 | 4.0E-5 | 0.292 |
| 42934 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 12 | BCL11A | Sponge network | 0.4 | 0.14611 | -0.932 | 0.0063 | 0.292 |
| 42935 | PART1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 46 | PPP1R9A | Sponge network | -2.925 | 0 | -0.903 | 0.04254 | 0.292 |
| 42936 | RP11-262H14.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 18 | SCN9A | Sponge network | -0.121 | 0.53195 | -0.525 | 0.10593 | 0.292 |
| 42937 | MIR600HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-655-3p | 21 | NCOA1 | Sponge network | 0.594 | 0.41522 | -0.432 | 0 | 0.292 |
| 42938 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | RASSF8 | Sponge network | -0.969 | 2.0E-5 | -0.122 | 0.65591 | 0.292 |
| 42939 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-98-5p | 20 | RSPO2 | Sponge network | -0.227 | 0.31657 | -3.038 | 0 | 0.292 |
| 42940 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 13 | PRKAA2 | Sponge network | 1.157 | 0.00455 | -2.462 | 0 | 0.292 |
| 42941 | RP11-434B12.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 13 | SPG20 | Sponge network | -0.031 | 0.89063 | -1.16 | 0 | 0.292 |
| 42942 | RP11-999E24.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-744-3p | 25 | PTPRT | Sponge network | -0.693 | 0.05629 | -0.907 | 0.03727 | 0.292 |
| 42943 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PHC3 | Sponge network | 0.67 | 0.00048 | 0.212 | 0.0499 | 0.292 |
| 42944 | CTA-363E19.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p | 12 | FOXP1 | Sponge network | 1.055 | 0 | 0.042 | 0.74368 | 0.292 |
| 42945 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-539-5p | 16 | MOB1B | Sponge network | 1.044 | 2.0E-5 | 0.197 | 0.15266 | 0.292 |
| 42946 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 38 | MCC | Sponge network | -0.355 | 0.0077 | -1.005 | 0 | 0.292 |
| 42947 | RP11-83A24.2 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p | 13 | MAT2A | Sponge network | 0.417 | 0.00445 | -0.073 | 0.36195 | 0.292 |
| 42948 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p | 40 | BMPR2 | Sponge network | 1.254 | 0 | 0.206 | 0.0635 | 0.292 |
| 42949 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-625-5p | 24 | LPP | Sponge network | 1.055 | 0 | -0.459 | 0.04475 | 0.292 |
| 42950 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | HECA | Sponge network | -0.053 | 0.80685 | -0.217 | 0.03463 | 0.292 |
| 42951 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-629-3p | 16 | RYR2 | Sponge network | 0.259 | 0.12542 | -1.466 | 0.00031 | 0.292 |
| 42952 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 20 | PGR | Sponge network | -0.322 | 0.25945 | -1.704 | 0 | 0.292 |
| 42953 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | LRRC55 | Sponge network | 0.259 | 0.12542 | -0.026 | 0.93163 | 0.292 |
| 42954 | AP001056.1 |
hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-429;hsa-miR-96-5p | 11 | JAZF1 | Sponge network | 1.062 | 0.00912 | -0.377 | 0.02677 | 0.292 |
| 42955 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p | 11 | PBRM1 | Sponge network | 0.95 | 0.15943 | -0.029 | 0.78601 | 0.292 |
| 42956 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-96-5p | 31 | NRXN1 | Sponge network | 0.082 | 0.55654 | -3.778 | 0 | 0.292 |
| 42957 | LINC00473 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-590-3p | 10 | UNC5D | Sponge network | -1.203 | 0.03881 | -3.646 | 0 | 0.292 |
| 42958 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p | 12 | TSC22D1 | Sponge network | -1.264 | 6.0E-5 | -0.529 | 2.0E-5 | 0.292 |
| 42959 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-93-5p | 18 | SLITRK3 | Sponge network | -0.465 | 0.04703 | -3.328 | 0 | 0.292 |
| 42960 | RP4-717I23.3 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p | 11 | KIF5B | Sponge network | 0.637 | 0.00069 | 0.362 | 0.00066 | 0.292 |
| 42961 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | SASH1 | Sponge network | 0.683 | 1.0E-5 | -0.502 | 0.00175 | 0.292 |
| 42962 | ZNF582-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | ABI2 | Sponge network | -1.349 | 0.00017 | 0.175 | 0.14787 | 0.292 |
| 42963 | NNT-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | TCF12 | Sponge network | 0.799 | 1.0E-5 | 0.174 | 0.13271 | 0.292 |
| 42964 | RP11-37B2.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p | 10 | ZMYND11 | Sponge network | 1.03 | 0 | -0.2 | 0.05449 | 0.292 |
| 42965 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 10 | GPAM | Sponge network | 1.055 | 0 | 0.142 | 0.29732 | 0.292 |
| 42966 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-421 | 11 | CADM3 | Sponge network | 0.16 | 0.50785 | -3.663 | 0 | 0.292 |
| 42967 | CTC-559E9.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p | 15 | ACVR2A | Sponge network | 0.594 | 0.00064 | -0.409 | 0.00039 | 0.292 |
| 42968 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-5p | 34 | ERC1 | Sponge network | -0.942 | 0 | -0.108 | 0.38794 | 0.292 |
| 42969 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 33 | TMEM132B | Sponge network | -0.668 | 3.0E-5 | -0.83 | 0.01084 | 0.292 |
| 42970 | GS1-124K5.11 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p | 10 | WAC | Sponge network | 0.494 | 0.00339 | -0.054 | 0.43688 | 0.292 |
| 42971 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 11 | CREBBP | Sponge network | 0.727 | 3.0E-5 | 0.126 | 0.15186 | 0.292 |
| 42972 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | MOB1B | Sponge network | -0.176 | 0.59692 | 0.197 | 0.15266 | 0.292 |
| 42973 | RP11-158K1.3 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 12 | RAPGEF5 | Sponge network | 0.349 | 0.01695 | 0.536 | 4.0E-5 | 0.292 |
| 42974 | RP11-981G7.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CREB1 | Sponge network | 0.986 | 0.01022 | 0.203 | 0.00537 | 0.292 |
| 42975 | RP11-822E23.8 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-501-5p | 20 | RIMS2 | Sponge network | -0.777 | 0.16667 | -1.365 | 0.00208 | 0.292 |
| 42976 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 13 | HECA | Sponge network | 1.153 | 0.00114 | -0.217 | 0.03463 | 0.292 |
| 42977 | ARHGEF26-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 52 | PIK3R1 | Sponge network | -2.46 | 0 | -0.133 | 0.36854 | 0.292 |
| 42978 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-497-5p;hsa-miR-766-3p | 11 | TBL1XR1 | Sponge network | 1.308 | 0 | 0.578 | 0 | 0.292 |
| 42979 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p | 10 | THBS1 | Sponge network | -0.187 | 0.23787 | 0.741 | 0.00386 | 0.292 |
| 42980 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-628-5p | 24 | FOXP1 | Sponge network | 1.254 | 0 | 0.042 | 0.74368 | 0.292 |
| 42981 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-3607-3p | 10 | NEDD4 | Sponge network | 1.056 | 0 | 0.02 | 0.89834 | 0.292 |
| 42982 | MIR22HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-7-5p | 22 | KCNA6 | Sponge network | -1.047 | 0 | -1.531 | 0.00013 | 0.292 |
| 42983 | LINC01023 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-93-3p | 17 | SSBP2 | Sponge network | -0.45 | 0.00486 | -1.58 | 0 | 0.292 |
| 42984 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 20 | NR2C2 | Sponge network | -0.355 | 0.0077 | 0.21 | 0.03224 | 0.292 |
| 42985 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | PAFAH1B1 | Sponge network | 0.082 | 0.55654 | -0.429 | 0 | 0.292 |
| 42986 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 43 | AR | Sponge network | -0.83 | 0 | -1.554 | 0 | 0.292 |
| 42987 | AC093110.3 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p | 30 | LPP | Sponge network | 1.044 | 2.0E-5 | -0.459 | 0.04475 | 0.292 |
| 42988 | HOTAIRM1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | RPS6KA6 | Sponge network | -0.053 | 0.80685 | -2.989 | 0 | 0.292 |
| 42989 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MYO9A | Sponge network | -0.727 | 0.04726 | -0.147 | 0.32373 | 0.292 |
| 42990 | CTA-941F9.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-502-5p | 10 | IL17RD | Sponge network | -0.344 | 0.49624 | -0.019 | 0.94873 | 0.292 |
| 42991 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 25 | EPHA5 | Sponge network | 0.411 | 0.1335 | -0.957 | 0.01733 | 0.292 |
| 42992 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-93-5p | 15 | SLITRK3 | Sponge network | 0.082 | 0.55397 | -3.328 | 0 | 0.292 |
| 42993 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-505-3p | 10 | PTPN11 | Sponge network | 0.373 | 0.00068 | 0.431 | 2.0E-5 | 0.292 |
| 42994 | RP11-5C23.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p | 20 | AFF1 | Sponge network | 0.274 | 0.07681 | -0.384 | 0.00053 | 0.292 |
| 42995 | MIR497HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p | 28 | PRDM2 | Sponge network | -1.439 | 4.0E-5 | -0.258 | 0.00721 | 0.292 |
| 42996 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-3p;hsa-miR-590-3p | 12 | MYBL1 | Sponge network | 0.8 | 0 | 0.494 | 0.02152 | 0.292 |
| 42997 | RP11-819C21.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | WWTR1 | Sponge network | 0.537 | 0.00139 | -0.345 | 0.11997 | 0.292 |
| 42998 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-654-3p | 14 | ITCH | Sponge network | 0.078 | 0.55803 | 0.084 | 0.34058 | 0.292 |
| 42999 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | RASSF2 | Sponge network | -0.318 | 0.65847 | -0.273 | 0.21961 | 0.292 |
| 43000 | CTD-3092A11.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 42 | LPP | Sponge network | 0.873 | 0 | -0.459 | 0.04475 | 0.292 |
| 43001 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-33a-3p | 15 | BNC2 | Sponge network | 0.368 | 0.20093 | -0.491 | 0.09988 | 0.292 |
| 43002 | MIR497HG |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p | 14 | IGF2 | Sponge network | -1.439 | 4.0E-5 | 0.848 | 0.03842 | 0.292 |
| 43003 | RP11-384K6.6 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-744-3p | 11 | KIF5B | Sponge network | 0.946 | 0 | 0.362 | 0.00066 | 0.292 |
| 43004 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | RET | Sponge network | 0.219 | 0.1516 | -0.833 | 0.03517 | 0.292 |
| 43005 | RP11-384K6.6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p | 10 | TET1 | Sponge network | 0.946 | 0 | -0.135 | 0.52138 | 0.292 |
| 43006 | RP11-434B12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | NCOA1 | Sponge network | -0.031 | 0.89063 | -0.432 | 0 | 0.292 |
| 43007 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-539-5p | 14 | DDX6 | Sponge network | 1.178 | 0 | 0.091 | 0.36428 | 0.292 |
| 43008 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ABCA10 | Sponge network | -0.355 | 0.0077 | -0.571 | 0.03674 | 0.292 |
| 43009 | PART1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 25 | NCAM2 | Sponge network | -2.925 | 0 | -0.482 | 0.22565 | 0.292 |
| 43010 | RP11-401P9.4 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-369-3p;hsa-miR-7-5p | 10 | CAV1 | Sponge network | -0.384 | 0.40652 | -1.013 | 6.0E-5 | 0.292 |
| 43011 | LINC00641 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 19 | RPS6KA2 | Sponge network | -0.222 | 0.32445 | -0.57 | 0.00013 | 0.292 |
| 43012 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 39 | SMAD2 | Sponge network | -0.303 | 0.21002 | -0.128 | 0.12732 | 0.292 |
| 43013 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p | 12 | ALDH1A2 | Sponge network | 0.997 | 0.00066 | -2.411 | 0 | 0.292 |
| 43014 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | NR4A3 | Sponge network | -0.777 | 0.1134 | -1.385 | 0 | 0.292 |
| 43015 | MIR600HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 29 | ADARB1 | Sponge network | 0.594 | 0.41522 | -0.335 | 0.06631 | 0.292 |
| 43016 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 12 | GPAM | Sponge network | 0.538 | 0.00076 | 0.142 | 0.29732 | 0.292 |
| 43017 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-93-5p | 17 | CNTNAP3 | Sponge network | -0.355 | 0.0077 | -1.465 | 0.00033 | 0.292 |
| 43018 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 14 | YAP1 | Sponge network | 0.705 | 0.00014 | 0.22 | 0.11836 | 0.292 |
| 43019 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p | 20 | TIMP3 | Sponge network | -0.18 | 0.20343 | -0.546 | 0.02307 | 0.292 |
| 43020 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-375 | 10 | MLLT10 | Sponge network | 0.903 | 0 | 0.105 | 0.33292 | 0.292 |
| 43021 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-877-5p;hsa-miR-935 | 30 | PTPRT | Sponge network | -0.384 | 0.40652 | -0.907 | 0.03727 | 0.292 |
| 43022 | KB-1460A1.5 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PHF3 | Sponge network | 0.603 | 0.00127 | 0.126 | 0.19652 | 0.292 |
| 43023 | CTC-487M23.5 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-92a-3p | 10 | ITGA5 | Sponge network | 0.912 | 0.00014 | 0.452 | 0.03524 | 0.292 |
| 43024 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p | 36 | CBX5 | Sponge network | -0.358 | 0.17255 | 0.165 | 0.24446 | 0.292 |
| 43025 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 25 | EPHA5 | Sponge network | 0.381 | 0.25967 | -0.957 | 0.01733 | 0.292 |
| 43026 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ZNF770 | Sponge network | 0.67 | 0.00048 | 0.206 | 0.06751 | 0.292 |
| 43027 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 21 | BCL11A | Sponge network | -0.899 | 6.0E-5 | -0.932 | 0.0063 | 0.292 |
| 43028 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | FAM188A | Sponge network | -1.111 | 0.0003 | -0.44 | 0.00018 | 0.292 |
| 43029 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-671-5p | 24 | KCNA6 | Sponge network | 0.082 | 0.55654 | -1.531 | 0.00013 | 0.292 |
| 43030 | SNHG12 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-497-5p;hsa-miR-9-5p | 13 | CDK6 | Sponge network | 1.103 | 0 | 0.991 | 0 | 0.292 |
| 43031 | NEAT1 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p | 10 | RAPGEF5 | Sponge network | 0.705 | 0.00014 | 0.536 | 4.0E-5 | 0.292 |
| 43032 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p | 11 | RSPO2 | Sponge network | -0.829 | 0.01343 | -3.038 | 0 | 0.292 |
| 43033 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-500a-3p | 13 | CSMD1 | Sponge network | -0.517 | 0.02935 | -0.63 | 0.18881 | 0.292 |
| 43034 | TMEM9B-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-589-3p | 10 | YAP1 | Sponge network | 0.529 | 0.00552 | 0.22 | 0.11836 | 0.292 |
| 43035 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 16 | ZFX | Sponge network | 0.174 | 0.30034 | 0.09 | 0.42563 | 0.292 |
| 43036 | RP11-13K12.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-539-5p | 18 | TSC1 | Sponge network | -0.154 | 0.78939 | 0.103 | 0.26071 | 0.292 |
| 43037 | RP11-575A19.2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-629-3p | 16 | DST | Sponge network | 1.969 | 0.00316 | -0.713 | 0.00096 | 0.292 |
| 43038 | AC004540.4 |
hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-484 | 12 | RIMS2 | Sponge network | -2.549 | 0.00528 | -1.365 | 0.00208 | 0.292 |
| 43039 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | FGF5 | Sponge network | 0.385 | 0.10067 | 0.549 | 0.28659 | 0.292 |
| 43040 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-532-5p | 11 | LRRC4 | Sponge network | -0.465 | 0.04703 | -1.774 | 0 | 0.292 |
| 43041 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 17 | ADAMTSL3 | Sponge network | -0.13 | 0.48734 | -1.559 | 0 | 0.292 |
| 43042 | RP11-620J15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | SUFU | Sponge network | -0.739 | 0.00044 | -0.04 | 0.65489 | 0.292 |
| 43043 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 15 | EPHA5 | Sponge network | -0.557 | 0.11135 | -0.957 | 0.01733 | 0.292 |
| 43044 | CTC-429P9.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-590-3p | 22 | PPARA | Sponge network | 0.178 | 0.29106 | -0.379 | 0.00548 | 0.292 |
| 43045 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-495-3p | 13 | TXNIP | Sponge network | -0.125 | 0.49414 | -1.277 | 0 | 0.292 |
| 43046 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-7-5p | 42 | TCF4 | Sponge network | 0.637 | 0.00069 | 0.016 | 0.92271 | 0.292 |
| 43047 | RP11-345P4.7 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 21 | CCDC6 | Sponge network | 1.597 | 0 | 0.27 | 0.02555 | 0.292 |
| 43048 | LINC00886 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-93-5p | 17 | CNTNAP3 | Sponge network | -0.371 | 0.24604 | -1.465 | 0.00033 | 0.292 |
| 43049 | AC003090.1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 14 | CSRNP1 | Sponge network | -2.117 | 0.00161 | -1.448 | 0 | 0.292 |
| 43050 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-96-5p | 53 | FAT3 | Sponge network | 0.311 | 0.10344 | -1.601 | 0.00051 | 0.292 |
| 43051 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-653-5p | 26 | DMXL1 | Sponge network | -0.162 | 0.47687 | -0.426 | 0.00056 | 0.292 |
| 43052 | U91328.19 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-500a-5p | 11 | ZNF208 | Sponge network | -0.303 | 0.21002 | -0.4 | 0.22381 | 0.292 |
| 43053 | BVES-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429 | 13 | BAZ2B | Sponge network | -2.62 | 0 | -0.276 | 0.02833 | 0.292 |
| 43054 | AC156455.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-345-5p | 23 | FAT3 | Sponge network | 1.154 | 0.00041 | -1.601 | 0.00051 | 0.292 |
| 43055 | HOXD-AS2 |
hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p | 17 | RBMS3 | Sponge network | -0.208 | 0.65592 | -0.519 | 0.03551 | 0.292 |
| 43056 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | FGF5 | Sponge network | -0.141 | 0.26434 | 0.549 | 0.28659 | 0.292 |
| 43057 | FAM157A |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326 | 16 | CCDC6 | Sponge network | 0.813 | 1.0E-5 | 0.27 | 0.02555 | 0.292 |
| 43058 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429 | 14 | CNTNAP3 | Sponge network | -0.176 | 0.59692 | -1.465 | 0.00033 | 0.292 |
| 43059 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-940 | 51 | WDFY3 | Sponge network | -0.323 | 0.01372 | -0.201 | 0.0967 | 0.292 |
| 43060 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p | 10 | RNF144A | Sponge network | 0.622 | 0.00015 | 0.594 | 0.0009 | 0.292 |
| 43061 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | BNC2 | Sponge network | 0.199 | 0.52763 | -0.491 | 0.09988 | 0.292 |
| 43062 | MEG3 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-96-5p | 18 | RPS6KA6 | Sponge network | 0.249 | 0.35902 | -2.989 | 0 | 0.292 |
| 43063 | WDFY3-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-590-5p;hsa-miR-7-5p | 23 | KCNJ5 | Sponge network | -0.942 | 0 | -0.281 | 0.3339 | 0.292 |
| 43064 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-23a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | BCL11A | Sponge network | -0.031 | 0.89063 | -0.932 | 0.0063 | 0.292 |
| 43065 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-625-3p;hsa-miR-93-5p | 26 | NTRK3 | Sponge network | 0.497 | 0.01451 | -1.77 | 3.0E-5 | 0.292 |
| 43066 | RP4-791M13.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-940 | 11 | CELF4 | Sponge network | 0.419 | 0.0319 | -1.135 | 0.00344 | 0.292 |
| 43067 | RP5-1198O20.4 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-874-3p | 12 | ARID1B | Sponge network | 0.997 | 0.00066 | 0.003 | 0.97503 | 0.292 |
| 43068 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 43 | EPHA5 | Sponge network | -0.355 | 0.0077 | -0.957 | 0.01733 | 0.292 |
| 43069 | AC116366.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-577;hsa-miR-877-5p | 10 | RBMS3 | Sponge network | 0.329 | 0.11122 | -0.519 | 0.03551 | 0.292 |
| 43070 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | CNTN1 | Sponge network | -1.047 | 0 | -2.594 | 0 | 0.292 |
| 43071 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-589-3p | 12 | MAFB | Sponge network | 1.153 | 0.00114 | -0.023 | 0.89865 | 0.292 |
| 43072 | FGF14-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-660-5p | 20 | ETV1 | Sponge network | -1.582 | 8.0E-5 | -0.096 | 0.67674 | 0.292 |
| 43073 | LINC00702 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | INPP4B | Sponge network | -0.737 | 0.06182 | -0.097 | 0.60503 | 0.292 |
| 43074 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 24 | CREB3L2 | Sponge network | 0.727 | 3.0E-5 | -0.525 | 2.0E-5 | 0.292 |
| 43075 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-98-5p | 50 | NTRK3 | Sponge network | 0.494 | 0.00339 | -1.77 | 3.0E-5 | 0.292 |
| 43076 | RP11-822E23.8 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378c;hsa-miR-484;hsa-miR-589-3p | 26 | MED12L | Sponge network | -0.777 | 0.16667 | -0.194 | 0.55299 | 0.292 |
| 43077 | LINC00702 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 29 | EFNA5 | Sponge network | -0.737 | 0.06182 | -0.421 | 0.17868 | 0.292 |
| 43078 | BVES-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-493-5p | 25 | ACVR2A | Sponge network | -2.62 | 0 | -0.409 | 0.00039 | 0.292 |
| 43079 | LINC00854 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484 | 14 | ZFX | Sponge network | 1.18 | 0 | 0.09 | 0.42563 | 0.292 |
| 43080 | RP11-159D12.2 |
hsa-let-7g-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | PPP2R5C | Sponge network | 1.254 | 0 | -0.314 | 3.0E-5 | 0.292 |
| 43081 | HCG11 |
hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | DAB2 | Sponge network | 0.385 | 0.10067 | 0.431 | 0.0113 | 0.292 |
| 43082 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TBX18 | Sponge network | 0.349 | 0.01695 | 0.843 | 0.02945 | 0.292 |
| 43083 | SNX29P2 |
hsa-let-7d-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-3p | 10 | TLE4 | Sponge network | 0.4 | 0.14611 | -1.157 | 0 | 0.292 |
| 43084 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 20 | CTDSPL | Sponge network | 0.222 | 0.29133 | 0.085 | 0.45121 | 0.292 |
| 43085 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | EYA4 | Sponge network | 0.219 | 0.1516 | 0.3 | 0.36876 | 0.292 |
| 43086 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-589-5p | 18 | CXXC4 | Sponge network | -2.69 | 0.0001 | -0.433 | 0.21055 | 0.292 |
| 43087 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 46 | IL6ST | Sponge network | -0.08 | 0.90092 | -0.402 | 0.00676 | 0.292 |
| 43088 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | ZFHX4 | Sponge network | -0.969 | 2.0E-5 | 0.018 | 0.95574 | 0.292 |
| 43089 | RP11-403I13.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 23 | SETBP1 | Sponge network | 0.619 | 0.00157 | -1.302 | 1.0E-5 | 0.292 |
| 43090 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | DACH2 | Sponge network | -1.264 | 6.0E-5 | -1.635 | 0.00034 | 0.292 |
| 43091 | RP11-6O2.3 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p | 23 | RIMS2 | Sponge network | 0.331 | 0.57247 | -1.365 | 0.00208 | 0.292 |
| 43092 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-493-5p;hsa-miR-590-3p | 20 | USP6 | Sponge network | 1.089 | 0 | 0.188 | 0.3871 | 0.292 |
| 43093 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | CNTNAP3 | Sponge network | -0.811 | 2.0E-5 | -1.465 | 0.00033 | 0.292 |
| 43094 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p | 19 | UNC80 | Sponge network | 0.178 | 0.29106 | -1.934 | 0 | 0.292 |
| 43095 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p | 15 | TRPS1 | Sponge network | 0.065 | 0.73468 | -0.191 | 0.44109 | 0.292 |
| 43096 | CTD-2184D3.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | CREB1 | Sponge network | 0.272 | 0.44692 | 0.203 | 0.00537 | 0.292 |
| 43097 | RP11-326G21.1 |
hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | OPCML | Sponge network | -0.021 | 0.93598 | -2.498 | 0 | 0.292 |
| 43098 | LINC00094 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p | 11 | GLI3 | Sponge network | 0.253 | 0.09565 | -0.615 | 0.01482 | 0.292 |
| 43099 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | HECA | Sponge network | 0.659 | 3.0E-5 | -0.217 | 0.03463 | 0.292 |
| 43100 | NEAT1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 54 | LPP | Sponge network | 0.705 | 0.00014 | -0.459 | 0.04475 | 0.292 |
| 43101 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-3614-5p;hsa-miR-424-5p | 17 | ABL2 | Sponge network | 0.082 | 0.55397 | 0.82 | 0 | 0.292 |
| 43102 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | THBS1 | Sponge network | 0.673 | 0 | 0.741 | 0.00386 | 0.292 |
| 43103 | SOX2-OT |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | SYT4 | Sponge network | -1.264 | 6.0E-5 | -3.207 | 0 | 0.292 |
| 43104 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | GNA13 | Sponge network | 0.545 | 0.00064 | 0.007 | 0.93884 | 0.292 |
| 43105 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | LRP1B | Sponge network | -0.295 | 0.02604 | -2.163 | 0 | 0.292 |
| 43106 | DIO3OS |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p | 14 | IRS1 | Sponge network | -0.773 | 0.04073 | -0.026 | 0.88855 | 0.292 |
| 43107 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-96-5p | 16 | TP53INP1 | Sponge network | 0.614 | 1.0E-5 | -0.496 | 0.00064 | 0.292 |
| 43108 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | PRDM5 | Sponge network | 0.219 | 0.1516 | -1.09 | 0.00014 | 0.292 |
| 43109 | RP11-158K1.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | LUZP2 | Sponge network | 0.349 | 0.01695 | -0.056 | 0.89416 | 0.292 |
| 43110 | MIR22HG |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-5p | 19 | LIMCH1 | Sponge network | -1.047 | 0 | -0.915 | 1.0E-5 | 0.292 |
| 43111 | PSMD5-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 18 | SLC5A3 | Sponge network | 0.569 | 0.00067 | 0.493 | 5.0E-5 | 0.292 |
| 43112 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ZNF208 | Sponge network | 0.998 | 0 | -0.4 | 0.22381 | 0.292 |
| 43113 | CTC-510F12.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-376c-3p | 15 | WDFY3 | Sponge network | -0.691 | 0.00146 | -0.201 | 0.0967 | 0.292 |
| 43114 | RP11-166D19.1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 15 | FOXO4 | Sponge network | -0.576 | 0.10121 | -0.516 | 2.0E-5 | 0.292 |
| 43115 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ZMYND11 | Sponge network | -0.342 | 0.02288 | -0.2 | 0.05449 | 0.292 |
| 43116 | LINC00476 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | -0.615 | 0.00015 | 0.809 | 0.00544 | 0.292 |
| 43117 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-92a-3p | 33 | RASSF2 | Sponge network | -0.738 | 0.21233 | -0.273 | 0.21961 | 0.292 |
| 43118 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SYNE1 | Sponge network | 0.392 | 0.0278 | -0.738 | 0.00316 | 0.292 |
| 43119 | NDUFA6-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-877-5p | 27 | ASTN1 | Sponge network | -0.355 | 0.0077 | -2.389 | 2.0E-5 | 0.292 |
| 43120 | RP11-774O3.3 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-362-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | ARHGEF12 | Sponge network | -0.487 | 0.02157 | 0.09 | 0.35693 | 0.292 |
| 43121 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | NTRK3 | Sponge network | 0.727 | 3.0E-5 | -1.77 | 3.0E-5 | 0.292 |
| 43122 | RP11-384O8.1 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | CDH11 | Sponge network | -0.738 | 0.21233 | 1.427 | 0 | 0.292 |
| 43123 | CTD-2002H8.2 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PDE4DIP | Sponge network | 0.931 | 1.0E-5 | -0.282 | 0.0257 | 0.292 |
| 43124 | HAR1A |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-3p | 18 | SSBP2 | Sponge network | -0.672 | 0.19447 | -1.58 | 0 | 0.292 |
| 43125 | AP001347.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-628-5p | 18 | TOM1L2 | Sponge network | -1.161 | 0.00591 | -0.994 | 0 | 0.292 |
| 43126 | OSER1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | TET1 | Sponge network | -0.13 | 0.48734 | -0.135 | 0.52138 | 0.292 |
| 43127 | RP11-38L15.3 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 12 | HIP1 | Sponge network | 0.344 | 0.3127 | 0.774 | 0 | 0.292 |
| 43128 | SOX2-OT |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 39 | TRPS1 | Sponge network | -1.264 | 6.0E-5 | -0.191 | 0.44109 | 0.292 |
| 43129 | RP11-144G6.12 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-590-3p | 14 | EPS15 | Sponge network | 0.826 | 1.0E-5 | -0.052 | 0.53998 | 0.292 |
| 43130 | ANKRD10-IT1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-7-1-3p | 10 | EPS15 | Sponge network | 1.739 | 0 | -0.052 | 0.53998 | 0.292 |
| 43131 | RP1-151F17.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 44 | RUNX1T1 | Sponge network | 0.222 | 0.29133 | -0.344 | 0.2374 | 0.292 |
| 43132 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p | 11 | GPAM | Sponge network | 0.594 | 0.00064 | 0.142 | 0.29732 | 0.292 |
| 43133 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | CBL | Sponge network | 0.267 | 0.2937 | 0.291 | 0.01267 | 0.292 |
| 43134 | RP11-38L15.3 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 16 | ZFHX4 | Sponge network | 0.344 | 0.3127 | 0.018 | 0.95574 | 0.292 |
| 43135 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | BCL2 | Sponge network | -0.663 | 0.10887 | -0.899 | 0 | 0.292 |
| 43136 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ARID5B | Sponge network | 0.476 | 0.19203 | 0.342 | 0.01676 | 0.292 |
| 43137 | RP11-401P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-539-5p | 13 | DMD | Sponge network | 1.524 | 0.1098 | -1.506 | 0 | 0.292 |
| 43138 | H1FX-AS1 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-421 | 16 | BTG2 | Sponge network | -0.206 | 0.12942 | -1.763 | 0 | 0.292 |
| 43139 | LINC00578 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 14 | ARID5B | Sponge network | 0.811 | 0.03228 | 0.342 | 0.01676 | 0.292 |
| 43140 | TINCR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-503-5p | 24 | RICTOR | Sponge network | -0.664 | 0.14543 | 0.388 | 0.00188 | 0.292 |
| 43141 | LINC00654 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | CDH10 | Sponge network | 0.374 | 0.20054 | -2.397 | 0 | 0.292 |
| 43142 | FZD10-AS1 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 19 | ABL1 | Sponge network | -0.727 | 0.04726 | -0.215 | 0.07804 | 0.292 |
| 43143 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | TP53INP1 | Sponge network | -0.83 | 0 | -0.496 | 0.00064 | 0.292 |
| 43144 | RP11-373D23.3 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-421;hsa-miR-7-1-3p | 13 | LIMCH1 | Sponge network | -0.285 | 0.2172 | -0.915 | 1.0E-5 | 0.292 |
| 43145 | LINC00472 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | -0.842 | 0.01361 | -0.226 | 0.11691 | 0.292 |
| 43146 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 44 | WDFY3 | Sponge network | 0.537 | 0.00139 | -0.201 | 0.0967 | 0.292 |
| 43147 | RP11-21A7A.2 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p | 13 | PTPN14 | Sponge network | -1.116 | 0.23686 | 0.144 | 0.34595 | 0.292 |
| 43148 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 13 | GPAM | Sponge network | 1.923 | 0 | 0.142 | 0.29732 | 0.292 |
| 43149 | HOXA-AS3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | SCN9A | Sponge network | -0.555 | 0.06156 | -0.525 | 0.10593 | 0.292 |
| 43150 | AC138035.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 11 | GPAM | Sponge network | 1.073 | 0 | 0.142 | 0.29732 | 0.292 |
| 43151 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | PTPRB | Sponge network | -0.942 | 0 | 0.105 | 0.47122 | 0.292 |
| 43152 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 23 | MYBL1 | Sponge network | 0.572 | 0.01722 | 0.494 | 0.02152 | 0.292 |
| 43153 | CD27-AS1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | PCDH10 | Sponge network | 0.177 | 0.16681 | -1.644 | 0.0078 | 0.292 |
| 43154 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-7-5p | 15 | ERC1 | Sponge network | -0.285 | 0.2172 | -0.108 | 0.38794 | 0.292 |
| 43155 | FAM13A-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | -0.83 | 0 | -3.143 | 0 | 0.292 |
| 43156 | CTD-3138B18.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-3607-3p | 11 | SIRT1 | Sponge network | 0.379 | 0.03091 | 0.109 | 0.2593 | 0.292 |
| 43157 | LINC00863 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CACNA2D1 | Sponge network | 0.189 | 0.13981 | -0.846 | 0.00675 | 0.292 |
| 43158 | LINC00648 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | SOX5 | Sponge network | 0.633 | 0.51678 | -1.091 | 4.0E-5 | 0.292 |
| 43159 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | MED12L | Sponge network | 0.889 | 9.0E-5 | -0.194 | 0.55299 | 0.292 |
| 43160 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 34 | ZFHX4 | Sponge network | 0.222 | 0.29133 | 0.018 | 0.95574 | 0.292 |
| 43161 | RP11-798M19.6 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ATM | Sponge network | -0.612 | 0.00094 | 0.178 | 0.21568 | 0.292 |
| 43162 | RP11-307C18.1 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-629-5p;hsa-miR-7-5p | 11 | BMPR2 | Sponge network | 1.082 | 0.00023 | 0.206 | 0.0635 | 0.292 |
| 43163 | RP11-13K12.5 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-93-3p | 20 | CTIF | Sponge network | -0.318 | 0.65847 | -0.389 | 0.00549 | 0.292 |
| 43164 | LINC00641 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 41 | GNAQ | Sponge network | -0.222 | 0.32445 | -0.367 | 0.00102 | 0.292 |
| 43165 | PART1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-942-5p | 35 | OPCML | Sponge network | -2.925 | 0 | -2.498 | 0 | 0.292 |
| 43166 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-940 | 48 | NFIB | Sponge network | -2.117 | 0.00161 | 0.023 | 0.90795 | 0.292 |
| 43167 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-582-5p | 14 | PHC3 | Sponge network | 1.093 | 0 | 0.212 | 0.0499 | 0.292 |
| 43168 | ZNF888 |
hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-149-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-337-3p | 10 | RB1 | Sponge network | 1.328 | 0 | 0.382 | 0.0017 | 0.292 |
| 43169 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b | 11 | FAM188A | Sponge network | -0.405 | 0.22013 | -0.44 | 0.00018 | 0.292 |
| 43170 | LINC00702 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 24 | ATM | Sponge network | -0.737 | 0.06182 | 0.178 | 0.21568 | 0.292 |
| 43171 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-3607-3p | 13 | IL17RD | Sponge network | 0.993 | 0 | -0.019 | 0.94873 | 0.292 |
| 43172 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-590-3p | 30 | PRKAA2 | Sponge network | 0.765 | 7.0E-5 | -2.462 | 0 | 0.292 |
| 43173 | SOX2-OT |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 22 | RARB | Sponge network | -1.264 | 6.0E-5 | -0.825 | 2.0E-5 | 0.292 |
| 43174 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 23 | ARHGAP28 | Sponge network | 0.194 | 0.64278 | -0.911 | 0.00013 | 0.292 |
| 43175 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p;hsa-miR-98-5p | 14 | MITF | Sponge network | 0.342 | 0.03129 | -0.769 | 0.00022 | 0.292 |
| 43176 | PWAR6 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 18 | NR3C2 | Sponge network | -1.575 | 6.0E-5 | -1.56 | 0 | 0.292 |
| 43177 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | DACH2 | Sponge network | -0.976 | 0.00025 | -1.635 | 0.00034 | 0.292 |
| 43178 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 48 | THRB | Sponge network | 0.572 | 0.01722 | -1.117 | 0.0004 | 0.292 |
| 43179 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 14 | THBS1 | Sponge network | -0.726 | 0.00132 | 0.741 | 0.00386 | 0.292 |
| 43180 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p | 15 | DLC1 | Sponge network | 0.37 | 0.27614 | -0.382 | 0.05038 | 0.292 |
| 43181 | SERHL |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-532-3p;hsa-miR-539-5p | 26 | PBX1 | Sponge network | -0.602 | 0.00331 | -1.206 | 0 | 0.292 |
| 43182 | RP4-584D14.7 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 10 | ASTN1 | Sponge network | 1.294 | 0.0031 | -2.389 | 2.0E-5 | 0.292 |
| 43183 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | RABEP1 | Sponge network | 0.494 | 0.00339 | -0.066 | 0.43142 | 0.292 |
| 43184 | LINC00622 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | AKAP11 | Sponge network | 1.169 | 0.00028 | 0.196 | 0.09825 | 0.292 |
| 43185 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 30 | BHLHE41 | Sponge network | 0.246 | 0.59912 | 0.353 | 0.12753 | 0.292 |
| 43186 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | USP12 | Sponge network | 0.826 | 1.0E-5 | -0.102 | 0.40768 | 0.292 |
| 43187 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | HECA | Sponge network | -0.83 | 0 | -0.217 | 0.03463 | 0.292 |
| 43188 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-93-5p | 11 | DAB2 | Sponge network | -2.69 | 0.0001 | 0.431 | 0.0113 | 0.292 |
| 43189 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | LSAMP | Sponge network | 0.064 | 0.72934 | -0.066 | 0.81708 | 0.292 |
| 43190 | RP11-531A24.5 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-940 | 18 | BAZ2B | Sponge network | 0.75 | 0.00054 | -0.276 | 0.02833 | 0.292 |
| 43191 | RP4-605O3.4 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | MYO9A | Sponge network | 0.262 | 0.0475 | -0.147 | 0.32373 | 0.292 |
| 43192 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 35 | SOX5 | Sponge network | 0.331 | 0.57247 | -1.091 | 4.0E-5 | 0.292 |
| 43193 | HHIP-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | CADM1 | Sponge network | -0.737 | 0.10822 | -0.9 | 0.00084 | 0.292 |
| 43194 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-542-3p | 18 | JDP2 | Sponge network | -2.69 | 0.0001 | -0.967 | 0 | 0.291 |
| 43195 | PCA3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | ERCC4 | Sponge network | -0.709 | 0.18168 | 0.126 | 0.15266 | 0.291 |
| 43196 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p | 32 | SSBP2 | Sponge network | 0.051 | 0.83388 | -1.58 | 0 | 0.291 |
| 43197 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p | 10 | ARHGAP28 | Sponge network | -1.255 | 0.00981 | -0.911 | 0.00013 | 0.291 |
| 43198 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 12 | EDA2R | Sponge network | -1.047 | 0 | -0.587 | 0.01598 | 0.291 |
| 43199 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-92a-3p;hsa-miR-940 | 50 | AFF4 | Sponge network | 0.214 | 0.18246 | -0.266 | 0.01565 | 0.291 |
| 43200 | RP11-404E16.1 |
hsa-miR-103a-2-5p;hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193b-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-5p;hsa-miR-532-5p | 11 | BMPR2 | Sponge network | 0.097 | 0.91311 | 0.206 | 0.0635 | 0.291 |
| 43201 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-450b-5p | 26 | ERC1 | Sponge network | 0.331 | 0.57247 | -0.108 | 0.38794 | 0.291 |
| 43202 | RP11-400K9.4 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-576-5p | 12 | ATM | Sponge network | -0.611 | 0.09607 | 0.178 | 0.21568 | 0.291 |
| 43203 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 18 | PTPRB | Sponge network | -1.249 | 7.0E-5 | 0.105 | 0.47122 | 0.291 |
| 43204 | RP11-276H19.2 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-550a-3p | 10 | PGR | Sponge network | -0.472 | 0.48851 | -1.704 | 0 | 0.291 |
| 43205 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-3p | 28 | PIK3R1 | Sponge network | 0.559 | 0.00026 | -0.133 | 0.36854 | 0.291 |
| 43206 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 19 | LATS2 | Sponge network | -1.047 | 0 | 0.159 | 0.2304 | 0.291 |
| 43207 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-708-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 36 | HLF | Sponge network | -1.047 | 0 | -2.443 | 0 | 0.291 |
| 43208 | HOTAIRM1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | TGFBR3 | Sponge network | -0.053 | 0.80685 | -1.173 | 1.0E-5 | 0.291 |
| 43209 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-93-5p | 17 | ARNT2 | Sponge network | -0.611 | 0.09607 | -0.332 | 0.25851 | 0.291 |
| 43210 | IQCH-AS1 |
hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | BCLAF1 | Sponge network | 0.929 | 0 | 0.211 | 0.00684 | 0.291 |
| 43211 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-424-5p;hsa-miR-455-3p | 10 | CRTC3 | Sponge network | 0.368 | 0.20093 | 0.164 | 0.16786 | 0.291 |
| 43212 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-589-3p | 12 | PGR | Sponge network | 2.306 | 9.0E-5 | -1.704 | 0 | 0.291 |
| 43213 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 32 | TMEM132B | Sponge network | -0.246 | 0.35034 | -0.83 | 0.01084 | 0.291 |
| 43214 | CKMT2-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-590-5p | 25 | BTG2 | Sponge network | 0.082 | 0.55654 | -1.763 | 0 | 0.291 |
| 43215 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-429 | 13 | PRKAB2 | Sponge network | -0.154 | 0.53954 | -0.367 | 0.01371 | 0.291 |
| 43216 | RP11-770J1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-625-3p | 23 | NTRK3 | Sponge network | -2.011 | 0.0005 | -1.77 | 3.0E-5 | 0.291 |
| 43217 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 50 | WDFY3 | Sponge network | -0.405 | 0.22013 | -0.201 | 0.0967 | 0.291 |
| 43218 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | AKAP11 | Sponge network | 1.352 | 0 | 0.196 | 0.09825 | 0.291 |
| 43219 | RP11-404E16.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-342-5p;hsa-miR-424-5p;hsa-miR-940 | 11 | TOM1L2 | Sponge network | 0.097 | 0.91311 | -0.994 | 0 | 0.291 |
| 43220 | RP11-641D5.2 |
hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-223-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | ACVR2A | Sponge network | -1.403 | 0.02373 | -0.409 | 0.00039 | 0.291 |
| 43221 | FAM66C |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 19 | JUN | Sponge network | -0.901 | 0.00019 | -0.932 | 0 | 0.291 |
| 43222 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | ZNF521 | Sponge network | -0.899 | 6.0E-5 | -0.111 | 0.58068 | 0.291 |
| 43223 | RP11-6N17.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-455-5p | 12 | TGFBR3 | Sponge network | 0.108 | 0.69556 | -1.173 | 1.0E-5 | 0.291 |
| 43224 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-629-3p | 36 | PIK3R1 | Sponge network | -0.773 | 0.04073 | -0.133 | 0.36854 | 0.291 |
| 43225 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-342-3p | 13 | CBX5 | Sponge network | 0.917 | 0 | 0.165 | 0.24446 | 0.291 |
| 43226 | AC116366.6 |
hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-577;hsa-miR-940 | 10 | IGF1 | Sponge network | 0.329 | 0.11122 | -0.204 | 0.5898 | 0.291 |
| 43227 | RP5-1021I20.1 |
hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429 | 13 | PCDH10 | Sponge network | -0.785 | 0.00035 | -1.644 | 0.0078 | 0.291 |
| 43228 | RP11-46C24.7 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 17 | NRK | Sponge network | 0.392 | 0.0278 | 0.656 | 0.19217 | 0.291 |
| 43229 | CKMT2-AS1 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-940;hsa-miR-96-5p | 18 | SFRP1 | Sponge network | 0.082 | 0.55654 | -3.566 | 0 | 0.291 |
| 43230 | RBPMS-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p | 11 | HIP1 | Sponge network | 0.144 | 0.64989 | 0.774 | 0 | 0.291 |
| 43231 | RP11-701H24.3 |
hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 11 | FOXP1 | Sponge network | -1.425 | 0.04204 | 0.042 | 0.74368 | 0.291 |
| 43232 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-500a-3p;hsa-miR-7-1-3p | 18 | ADAMTSL3 | Sponge network | -0.176 | 0.59692 | -1.559 | 0 | 0.291 |
| 43233 | KB-1572G7.2 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p | 14 | BRD3 | Sponge network | 0.924 | 0 | 0.37 | 0.00013 | 0.291 |
| 43234 | RP11-6O2.3 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-455-3p | 12 | KCNAB1 | Sponge network | 0.331 | 0.57247 | -0.755 | 2.0E-5 | 0.291 |
| 43235 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-940 | 24 | IGF1 | Sponge network | -0.08 | 0.90092 | -0.204 | 0.5898 | 0.291 |
| 43236 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 17 | NR4A3 | Sponge network | -0.942 | 0 | -1.385 | 0 | 0.291 |
| 43237 | AC009014.3 |
hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p | 10 | PBX1 | Sponge network | -0.917 | 0.24351 | -1.206 | 0 | 0.291 |
| 43238 | LINC00863 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | DOCK4 | Sponge network | 0.189 | 0.13981 | 0.502 | 0.00108 | 0.291 |
| 43239 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 34 | EPHA5 | Sponge network | 0.41 | 0.02214 | -0.957 | 0.01733 | 0.291 |
| 43240 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-577 | 18 | PPM1L | Sponge network | 1.294 | 0.0031 | -0.537 | 0.00616 | 0.291 |
| 43241 | LINC00886 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-708-5p;hsa-miR-93-5p | 18 | EZH1 | Sponge network | -0.371 | 0.24604 | -0.564 | 1.0E-5 | 0.291 |
| 43242 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-3913-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p | 26 | ESR1 | Sponge network | -0.342 | 0.02288 | -1.035 | 3.0E-5 | 0.291 |
| 43243 | RP11-251G23.5 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-495-3p | 17 | CREB1 | Sponge network | 1.344 | 0 | 0.203 | 0.00537 | 0.291 |
| 43244 | OIP5-AS1 |
hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-214-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 13 | SLC12A6 | Sponge network | 0.421 | 0.00084 | -0.343 | 0.00043 | 0.291 |
| 43245 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-93-3p | 16 | PTPN11 | Sponge network | 0.056 | 0.81195 | 0.431 | 2.0E-5 | 0.291 |
| 43246 | RP11-725P16.2 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 12 | UBE4B | Sponge network | 0.422 | 0.27021 | -0.235 | 0.007 | 0.291 |
| 43247 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429 | 21 | RNASEL | Sponge network | -1.249 | 7.0E-5 | -0.272 | 0.01796 | 0.291 |
| 43248 | RP11-57H14.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 21 | CDH23 | Sponge network | 0.083 | 0.66405 | -0.974 | 0.00034 | 0.291 |
| 43249 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -0.663 | 0.10887 | -0.354 | 0.14694 | 0.291 |
| 43250 | MALAT1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | ERCC6 | Sponge network | 0.782 | 0.00016 | 0.23 | 0.015 | 0.291 |
| 43251 | PCA3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | NCOA1 | Sponge network | -0.709 | 0.18168 | -0.432 | 0 | 0.291 |
| 43252 | ADIRF-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 24 | ABI2 | Sponge network | -0.223 | 0.47816 | 0.175 | 0.14787 | 0.291 |
| 43253 | RP11-571M6.17 |
hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-625-3p | 16 | CBX5 | Sponge network | 1.013 | 0 | 0.165 | 0.24446 | 0.291 |
| 43254 | RP11-283I3.6 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-493-5p | 16 | DDX3X | Sponge network | 0.614 | 1.0E-5 | -0.152 | 0.07686 | 0.291 |
| 43255 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-7-1-3p | 11 | FOXO1 | Sponge network | -1.092 | 4.0E-5 | -0.141 | 0.33874 | 0.291 |
| 43256 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-5p | 18 | PCDH17 | Sponge network | 0.174 | 0.30034 | 0.731 | 2.0E-5 | 0.291 |
| 43257 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-30a-3p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 13 | CBL | Sponge network | 1.205 | 0 | 0.291 | 0.01267 | 0.291 |
| 43258 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | TET1 | Sponge network | 0.998 | 0 | -0.135 | 0.52138 | 0.291 |
| 43259 | FZD10-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | MACF1 | Sponge network | -0.727 | 0.04726 | 0.019 | 0.90335 | 0.291 |
| 43260 | FGD5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p | 17 | NUAK1 | Sponge network | 0.401 | 0.00066 | 0.904 | 0 | 0.291 |
| 43261 | USP3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-93-5p | 23 | RNASEL | Sponge network | -0.162 | 0.47687 | -0.272 | 0.01796 | 0.291 |
| 43262 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-493-5p | 10 | PGR | Sponge network | -1.942 | 0 | -1.704 | 0 | 0.291 |
| 43263 | AC108488.4 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-7-1-3p | 15 | CNTN1 | Sponge network | 0.259 | 0.12542 | -2.594 | 0 | 0.291 |
| 43264 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 29 | JAZF1 | Sponge network | 0.72 | 0.0001 | -0.377 | 0.02677 | 0.291 |
| 43265 | TP73-AS1 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 13 | GABRB2 | Sponge network | -0.563 | 0.02545 | -1.841 | 0.00029 | 0.291 |
| 43266 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-340-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | FGF5 | Sponge network | -2.915 | 0.00015 | 0.549 | 0.28659 | 0.291 |
| 43267 | PVT1 |
hsa-miR-129-5p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-497-5p;hsa-miR-9-5p | 13 | CCDC6 | Sponge network | 1.985 | 0 | 0.27 | 0.02555 | 0.291 |
| 43268 | RP5-894A10.6 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-485-3p | 11 | DDX3X | Sponge network | 0.697 | 9.0E-5 | -0.152 | 0.07686 | 0.291 |
| 43269 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-590-3p | 14 | DDX5 | Sponge network | 0.194 | 0.64278 | -0.099 | 0.17428 | 0.291 |
| 43270 | CTD-2336O2.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | NRK | Sponge network | -0.141 | 0.26434 | 0.656 | 0.19217 | 0.291 |
| 43271 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 16 | PRKAA1 | Sponge network | 0.401 | 0.00066 | 0.317 | 0.0031 | 0.291 |
| 43272 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-940 | 31 | NTRK2 | Sponge network | -0.668 | 3.0E-5 | -0.736 | 0.06495 | 0.291 |
| 43273 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-628-5p | 14 | CDC73 | Sponge network | 1.206 | 0 | 0.094 | 0.20463 | 0.291 |
| 43274 | PCA3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 34 | NOTCH2 | Sponge network | -0.709 | 0.18168 | 0.138 | 0.35163 | 0.291 |
| 43275 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-744-3p | 12 | HIP1 | Sponge network | 1.04 | 0.00023 | 0.774 | 0 | 0.291 |
| 43276 | RP11-983P16.4 |
hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | PAK3 | Sponge network | 0.41 | 0.02214 | -0.628 | 0.07253 | 0.291 |
| 43277 | RP11-389C8.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PTPRT | Sponge network | 0.727 | 3.0E-5 | -0.907 | 0.03727 | 0.291 |
| 43278 | RP11-158K1.3 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p | 10 | PARK2 | Sponge network | 0.349 | 0.01695 | -1.892 | 0 | 0.291 |
| 43279 | RP13-516M14.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-940 | 34 | TRPS1 | Sponge network | 0.389 | 0.04293 | -0.191 | 0.44109 | 0.291 |
| 43280 | AC141928.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-425-5p;hsa-miR-500a-3p;hsa-miR-628-5p | 23 | ERC1 | Sponge network | 0.853 | 0.15352 | -0.108 | 0.38794 | 0.291 |
| 43281 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-484 | 13 | GREM1 | Sponge network | 0.494 | 0.00339 | 0.902 | 0.01691 | 0.291 |
| 43282 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-96-5p | 13 | FAM188A | Sponge network | 0.101 | 0.60919 | -0.44 | 0.00018 | 0.291 |
| 43283 | RP11-403I13.7 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-365a-3p;hsa-miR-378a-5p | 12 | CACNA1E | Sponge network | 0.395 | 0.15451 | 1.752 | 0 | 0.291 |
| 43284 | LINC00863 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 0.189 | 0.13981 | 0.51 | 0 | 0.291 |
| 43285 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-3p | 14 | BNC2 | Sponge network | -0.164 | 0.45106 | -0.491 | 0.09988 | 0.291 |
| 43286 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 14 | DACT1 | Sponge network | 0.494 | 0.00339 | 0.783 | 0.00072 | 0.291 |
| 43287 | RP11-701H24.7 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-576-5p | 10 | PPP1R9A | Sponge network | -2.039 | 0.03447 | -0.903 | 0.04254 | 0.291 |
| 43288 | ZSCAN16-AS1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 18 | RPS6KA6 | Sponge network | -0.342 | 0.02288 | -2.989 | 0 | 0.291 |
| 43289 | CTC-550B14.6 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | RICTOR | Sponge network | 3.885 | 0 | 0.388 | 0.00188 | 0.291 |
| 43290 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-5p | 13 | SCN9A | Sponge network | 0.796 | 4.0E-5 | -0.525 | 0.10593 | 0.291 |
| 43291 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-942-5p | 33 | NOTCH2 | Sponge network | -1.645 | 0 | 0.138 | 0.35163 | 0.291 |
| 43292 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH10 | Sponge network | 0.194 | 0.64278 | -2.397 | 0 | 0.291 |
| 43293 | KB-1460A1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 20 | ERCC4 | Sponge network | 0.603 | 0.00127 | 0.126 | 0.15266 | 0.291 |
| 43294 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 22 | SPRY3 | Sponge network | 0.539 | 0.03722 | 0.016 | 0.91286 | 0.291 |
| 43295 | RP13-1039J1.4 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27b-5p;hsa-miR-34a-5p;hsa-miR-582-5p | 12 | PRKAA2 | Sponge network | 0.356 | 0.33467 | -2.462 | 0 | 0.291 |
| 43296 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-624-5p | 10 | PCDH8 | Sponge network | -0.303 | 0.21002 | -0.997 | 0.05366 | 0.291 |
| 43297 | AC156455.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p | 14 | ZFHX3 | Sponge network | 1.154 | 0.00041 | 0.389 | 0.01363 | 0.291 |
| 43298 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | TGFBR2 | Sponge network | 0.064 | 0.72934 | -0.243 | 0.11408 | 0.291 |
| 43299 | RP11-798G7.7 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-582-3p | 18 | BRD3 | Sponge network | 0.858 | 3.0E-5 | 0.37 | 0.00013 | 0.291 |
| 43300 | TMEM161B-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-411-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-889-3p | 32 | SMAD2 | Sponge network | -0.04 | 0.7606 | -0.128 | 0.12732 | 0.291 |
| 43301 | EMX2OS |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p | 19 | IRS1 | Sponge network | -0.777 | 0.1134 | -0.026 | 0.88855 | 0.291 |
| 43302 | SNX29P2 |
hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | LUZP2 | Sponge network | 0.4 | 0.14611 | -0.056 | 0.89416 | 0.291 |
| 43303 | CTD-2260A17.1 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-550a-5p | 10 | ARHGEF12 | Sponge network | 0.518 | 0.01048 | 0.09 | 0.35693 | 0.291 |
| 43304 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 16 | CLIP1 | Sponge network | 0.614 | 1.0E-5 | -0.215 | 0.08684 | 0.291 |
| 43305 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | CCND2 | Sponge network | -0.726 | 0.00132 | -0.267 | 0.3112 | 0.291 |
| 43306 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 51 | WDFY3 | Sponge network | -2.117 | 0.00161 | -0.201 | 0.0967 | 0.291 |
| 43307 | NNT-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-3p | 18 | ETV1 | Sponge network | 0.799 | 1.0E-5 | -0.096 | 0.67674 | 0.291 |
| 43308 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p | 11 | LATS1 | Sponge network | -0.611 | 0.09607 | 0.109 | 0.34208 | 0.291 |
| 43309 | AP001055.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 12 | RARB | Sponge network | 0.693 | 0.00076 | -0.825 | 2.0E-5 | 0.291 |
| 43310 | LINC00094 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p | 13 | ADCY1 | Sponge network | 0.253 | 0.09565 | 0.362 | 0.16982 | 0.291 |
| 43311 | TP73-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-335-3p | 15 | GLIPR1 | Sponge network | -0.563 | 0.02545 | 0.146 | 0.3276 | 0.291 |
| 43312 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p | 17 | MYBL1 | Sponge network | 0.176 | 0.37402 | 0.494 | 0.02152 | 0.291 |
| 43313 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | TRIP11 | Sponge network | 0.921 | 0.00118 | -0.158 | 0.06384 | 0.291 |
| 43314 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 16 | PRKAA1 | Sponge network | 1.317 | 0 | 0.317 | 0.0031 | 0.291 |
| 43315 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | PIK3R1 | Sponge network | -1.886 | 0 | -0.133 | 0.36854 | 0.291 |
| 43316 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p | 20 | DDX6 | Sponge network | 1.044 | 2.0E-5 | 0.091 | 0.36428 | 0.291 |
| 43317 | RP11-473I1.9 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 12 | PAWR | Sponge network | 0.683 | 1.0E-5 | 0.636 | 0 | 0.291 |
| 43318 | RP5-855D21.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p | 12 | SOX11 | Sponge network | 1.728 | 0 | 2.68 | 0 | 0.291 |
| 43319 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181a-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MYO9A | Sponge network | 0.545 | 0.00064 | -0.147 | 0.32373 | 0.291 |
| 43320 | RP11-527J8.1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p | 10 | KIF1B | Sponge network | 1.324 | 0 | -0.118 | 0.32258 | 0.291 |
| 43321 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-935 | 30 | HBP1 | Sponge network | -0.303 | 0.21002 | -0.454 | 0 | 0.291 |
| 43322 | RP11-819C21.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PDE4DIP | Sponge network | 0.537 | 0.00139 | -0.282 | 0.0257 | 0.291 |
| 43323 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-942-5p | 15 | RB1CC1 | Sponge network | -0.405 | 0.22013 | 0.175 | 0.12559 | 0.291 |
| 43324 | AC074289.1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-96-5p | 19 | CADM1 | Sponge network | -0.326 | 0.14314 | -0.9 | 0.00084 | 0.291 |
| 43325 | RP11-774O3.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-589-3p;hsa-miR-7-5p | 19 | KCNJ5 | Sponge network | -0.487 | 0.02157 | -0.281 | 0.3339 | 0.291 |
| 43326 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p | 13 | HECA | Sponge network | -0.829 | 0.01343 | -0.217 | 0.03463 | 0.291 |
| 43327 | RP11-620J15.3 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p | 12 | CSRNP1 | Sponge network | -0.739 | 0.00044 | -1.448 | 0 | 0.291 |
| 43328 | SNX29P2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-877-5p | 19 | NRK | Sponge network | 0.4 | 0.14611 | 0.656 | 0.19217 | 0.291 |
| 43329 | RP11-244O19.1 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | PHF3 | Sponge network | -0.096 | 0.67958 | 0.126 | 0.19652 | 0.291 |
| 43330 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-93-5p | 14 | LATS2 | Sponge network | -0.384 | 0.40652 | 0.159 | 0.2304 | 0.291 |
| 43331 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | ZFHX3 | Sponge network | 0.643 | 0 | 0.389 | 0.01363 | 0.291 |
| 43332 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | GRIN2A | Sponge network | 0.083 | 0.66405 | -2.161 | 0 | 0.291 |
| 43333 | CTD-2528L19.6 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-409-5p;hsa-miR-495-3p;hsa-miR-590-3p | 23 | CCDC6 | Sponge network | 0.953 | 0 | 0.27 | 0.02555 | 0.291 |
| 43334 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-671-5p | 40 | ADARB1 | Sponge network | -0.773 | 0.04073 | -0.335 | 0.06631 | 0.291 |
| 43335 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 12 | RCAN2 | Sponge network | -0.513 | 0.04703 | -0.33 | 0.15172 | 0.291 |
| 43336 | LINC01006 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-935 | 21 | DMD | Sponge network | -0.702 | 0.04814 | -1.506 | 0 | 0.291 |
| 43337 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | NIN | Sponge network | -2.46 | 0 | -0.253 | 0.13794 | 0.291 |
| 43338 | AC093627.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p | 18 | PTPRD | Sponge network | -0.787 | 0.3928 | -1.005 | 0.00064 | 0.291 |
| 43339 | LINC00863 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | AKAP6 | Sponge network | 0.189 | 0.13981 | -1.586 | 3.0E-5 | 0.291 |
| 43340 | RP1-302G2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p | 13 | RASSF2 | Sponge network | -1.483 | 9.0E-5 | -0.273 | 0.21961 | 0.291 |
| 43341 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p | 15 | PRKAA1 | Sponge network | 1.4 | 0 | 0.317 | 0.0031 | 0.291 |
| 43342 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 50 | TCF4 | Sponge network | 0.862 | 0.06213 | 0.016 | 0.92271 | 0.291 |
| 43343 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | NEDD4 | Sponge network | 0.663 | 0.00073 | 0.02 | 0.89834 | 0.291 |
| 43344 | CTD-2314G24.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 17 | IGF2 | Sponge network | 0.246 | 0.59912 | 0.848 | 0.03842 | 0.291 |
| 43345 | RP11-37B2.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | SLC5A3 | Sponge network | 1.03 | 0 | 0.493 | 5.0E-5 | 0.291 |
| 43346 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 21 | CLASP2 | Sponge network | -1.092 | 4.0E-5 | -0.038 | 0.71 | 0.291 |
| 43347 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p | 35 | TMEM132B | Sponge network | -0.384 | 0.40652 | -0.83 | 0.01084 | 0.291 |
| 43348 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | EPHA5 | Sponge network | -0.08 | 0.90092 | -0.957 | 0.01733 | 0.291 |
| 43349 | CTD-3099C6.9 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-766-3p | 10 | ATM | Sponge network | 1.361 | 0 | 0.178 | 0.21568 | 0.291 |
| 43350 | ERVK3-1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-486-5p | 14 | CBFB | Sponge network | 0.58 | 0 | 1.061 | 0 | 0.291 |
| 43351 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 68 | TCF4 | Sponge network | 0.421 | 0.00084 | 0.016 | 0.92271 | 0.291 |
| 43352 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | RASSF2 | Sponge network | -0.709 | 0.18168 | -0.273 | 0.21961 | 0.291 |
| 43353 | RP11-166D19.1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PCLO | Sponge network | -0.576 | 0.10121 | -1.843 | 0.00064 | 0.291 |
| 43354 | RP11-344B5.2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p | 10 | HIP1 | Sponge network | -0.055 | 0.88482 | 0.774 | 0 | 0.291 |
| 43355 | PCA3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3E | Sponge network | -0.709 | 0.18168 | -1.717 | 0.00137 | 0.291 |
| 43356 | CTC-429P9.3 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p | 11 | PHF3 | Sponge network | 0.082 | 0.55397 | 0.126 | 0.19652 | 0.291 |
| 43357 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-589-3p | 13 | ABCA10 | Sponge network | -0.154 | 0.78939 | -0.571 | 0.03674 | 0.291 |
| 43358 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | BCLAF1 | Sponge network | 0.903 | 0 | 0.211 | 0.00684 | 0.291 |
| 43359 | CROCCP2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | UBR3 | Sponge network | 0.79 | 0 | -0.021 | 0.82774 | 0.291 |
| 43360 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577 | 24 | GREM1 | Sponge network | 0.101 | 0.60919 | 0.902 | 0.01691 | 0.291 |
| 43361 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 31 | BMPR2 | Sponge network | -0.663 | 0.10887 | 0.206 | 0.0635 | 0.291 |
| 43362 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 10 | PTPN13 | Sponge network | 0.064 | 0.72934 | -1.208 | 0 | 0.291 |
| 43363 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 21 | USP6 | Sponge network | -1.264 | 6.0E-5 | 0.188 | 0.3871 | 0.291 |
| 43364 | RP4-791M13.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 13 | ITPKB | Sponge network | 0.419 | 0.0319 | -0.704 | 0.00106 | 0.291 |
| 43365 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 10 | PBRM1 | Sponge network | 1.317 | 0 | -0.029 | 0.78601 | 0.291 |
| 43366 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | FNBP1 | Sponge network | 0.395 | 0.15451 | -0.969 | 4.0E-5 | 0.291 |
| 43367 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | NCOA1 | Sponge network | -0.318 | 0.65847 | -0.432 | 0 | 0.291 |
| 43368 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-766-3p | 27 | MAPK1 | Sponge network | 1.147 | 0 | -0.017 | 0.81465 | 0.291 |
| 43369 | MIR22HG |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p | 12 | KCNAB1 | Sponge network | -1.047 | 0 | -0.755 | 2.0E-5 | 0.291 |
| 43370 | U91328.19 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | NIN | Sponge network | -0.303 | 0.21002 | -0.253 | 0.13794 | 0.291 |
| 43371 | FGD5-AS1 |
hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | HERC1 | Sponge network | 0.401 | 0.00066 | -0.196 | 0.08544 | 0.291 |
| 43372 | BCYRN1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | PIK3CA | Sponge network | 0.311 | 0.10344 | 0.195 | 0.06908 | 0.291 |
| 43373 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-5p | 16 | ZFP36L1 | Sponge network | -1.645 | 0 | -0.505 | 8.0E-5 | 0.291 |
| 43374 | GAS5-AS1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 11 | TBL1XR1 | Sponge network | 0.598 | 0 | 0.578 | 0 | 0.291 |
| 43375 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | DPYD | Sponge network | 0.532 | 0.00505 | -0.736 | 0.00076 | 0.291 |
| 43376 | RP11-720L2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-93-5p | 12 | TGFBR2 | Sponge network | -1.255 | 0.00981 | -0.243 | 0.11408 | 0.291 |
| 43377 | CTC-296K1.4 |
hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-708-5p | 14 | RAP1A | Sponge network | -2.076 | 0.00548 | -0.929 | 0 | 0.291 |
| 43378 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-98-5p | 21 | CLASP2 | Sponge network | -2.62 | 0 | -0.038 | 0.71 | 0.291 |
| 43379 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-935;hsa-miR-96-5p | 38 | MTSS1 | Sponge network | -0.942 | 0 | -0.81 | 0.00035 | 0.291 |
| 43380 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-7-1-3p | 21 | CXXC4 | Sponge network | 0.853 | 0.15352 | -0.433 | 0.21055 | 0.291 |
| 43381 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 22 | PRKAB2 | Sponge network | 0.541 | 0.00125 | -0.367 | 0.01371 | 0.291 |
| 43382 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-590-3p | 18 | EBF3 | Sponge network | 0.349 | 0.01695 | -0.058 | 0.81437 | 0.291 |
| 43383 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 53 | IL6ST | Sponge network | -0.342 | 0.02288 | -0.402 | 0.00676 | 0.291 |
| 43384 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-484 | 17 | CUL5 | Sponge network | 0.594 | 0.41522 | 0.026 | 0.77301 | 0.291 |
| 43385 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | MYO9A | Sponge network | 0.537 | 0.0006 | -0.147 | 0.32373 | 0.291 |
| 43386 | RP11-159D12.2 |
hsa-miR-100-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-1301-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p | 14 | RB1 | Sponge network | 1.254 | 0 | 0.382 | 0.0017 | 0.291 |
| 43387 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p | 40 | UNC80 | Sponge network | 0.572 | 0.01722 | -1.934 | 0 | 0.291 |
| 43388 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-93-5p | 19 | LRRC55 | Sponge network | 0.056 | 0.81195 | -0.026 | 0.93163 | 0.291 |
| 43389 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PTPRB | Sponge network | 0.349 | 0.01695 | 0.105 | 0.47122 | 0.291 |
| 43390 | DNM1P35 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-374a-5p;hsa-miR-590-3p | 12 | SEMA3C | Sponge network | 0.051 | 0.83388 | -0.272 | 0.31153 | 0.291 |
| 43391 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-493-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 14 | NRXN3 | Sponge network | 0.614 | 1.0E-5 | -0.893 | 0.04186 | 0.291 |
| 43392 | RP11-597D13.9 |
hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-574-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | FHL1 | Sponge network | 1.302 | 7.0E-5 | -2.687 | 0 | 0.291 |
| 43393 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-590-3p | 12 | CITED2 | Sponge network | -0.096 | 0.67958 | -1.545 | 0 | 0.291 |
| 43394 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | HACE1 | Sponge network | 0.705 | 0.00014 | -0.072 | 0.55239 | 0.291 |
| 43395 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-5p;hsa-miR-940 | 33 | CHD5 | Sponge network | -0.769 | 0.00035 | -0.922 | 0.01521 | 0.291 |
| 43396 | RP11-326C3.7 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 14 | LPP | Sponge network | 0.295 | 0.28897 | -0.459 | 0.04475 | 0.291 |
| 43397 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-539-5p | 14 | GRIN2A | Sponge network | -0.602 | 0.00331 | -2.161 | 0 | 0.291 |
| 43398 | AC016747.3 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-942-5p | 16 | PCDH17 | Sponge network | 0.199 | 0.38015 | 0.731 | 2.0E-5 | 0.291 |
| 43399 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CLIP1 | Sponge network | -1.216 | 0 | -0.215 | 0.08684 | 0.291 |
| 43400 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | IGSF10 | Sponge network | -0.322 | 0.25945 | -1.751 | 1.0E-5 | 0.291 |
| 43401 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p | 40 | FOXP1 | Sponge network | 0.494 | 0.00339 | 0.042 | 0.74368 | 0.291 |
| 43402 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-708-3p | 19 | HLF | Sponge network | 0.001 | 0.99753 | -2.443 | 0 | 0.291 |
| 43403 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30b-3p | 11 | CXXC4 | Sponge network | -0.583 | 0.14589 | -0.433 | 0.21055 | 0.291 |
| 43404 | RP11-53O19.1 |
hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-452-5p;hsa-miR-7-1-3p | 10 | RABEP1 | Sponge network | -0.405 | 0.22013 | -0.066 | 0.43142 | 0.291 |
| 43405 | MIR181A2HG |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 11 | NR2C2 | Sponge network | 1.822 | 1.0E-5 | 0.21 | 0.03224 | 0.291 |
| 43406 | BDNF-AS |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 16 | MRVI1 | Sponge network | -0.668 | 3.0E-5 | -1.051 | 0.00022 | 0.291 |
| 43407 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-582-5p | 24 | SPRY3 | Sponge network | 0.792 | 0.0049 | 0.016 | 0.91286 | 0.291 |
| 43408 | RP11-678G14.2 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-29b-1-5p | 11 | IL6ST | Sponge network | -1.765 | 0.0778 | -0.402 | 0.00676 | 0.291 |
| 43409 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326 | 10 | ABCB5 | Sponge network | 0.331 | 0.57247 | -0.956 | 0.04077 | 0.291 |
| 43410 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | -1.575 | 6.0E-5 | 0.309 | 0.17079 | 0.291 |
| 43411 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 21 | ARID5B | Sponge network | -1.092 | 4.0E-5 | 0.342 | 0.01676 | 0.291 |
| 43412 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | ST6GAL2 | Sponge network | -0.303 | 0.21002 | -1.201 | 0.00597 | 0.291 |
| 43413 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-708-5p | 23 | MTSS1 | Sponge network | 0.056 | 0.81195 | -0.81 | 0.00035 | 0.291 |
| 43414 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-495-3p | 10 | ZNF770 | Sponge network | 1.597 | 0 | 0.206 | 0.06751 | 0.291 |
| 43415 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-577 | 32 | TMEM132B | Sponge network | -0.18 | 0.20343 | -0.83 | 0.01084 | 0.291 |
| 43416 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-3p;hsa-miR-590-3p | 11 | CLIP1 | Sponge network | -1.582 | 8.0E-5 | -0.215 | 0.08684 | 0.291 |
| 43417 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | ERRFI1 | Sponge network | -0.896 | 0.00099 | -0.798 | 1.0E-5 | 0.291 |
| 43418 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | RBMS3 | Sponge network | 0.998 | 0 | -0.519 | 0.03551 | 0.291 |
| 43419 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | CDHR3 | Sponge network | 0.101 | 0.60919 | -0.977 | 4.0E-5 | 0.291 |
| 43420 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | KAT6B | Sponge network | 0.765 | 7.0E-5 | -0.478 | 0.00018 | 0.291 |
| 43421 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-374b-3p;hsa-miR-629-5p | 13 | IGF1 | Sponge network | 0.065 | 0.73468 | -0.204 | 0.5898 | 0.291 |
| 43422 | AF131215.9 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 16 | RASSF8 | Sponge network | -0.322 | 0.25945 | -0.122 | 0.65591 | 0.291 |
| 43423 | HHIP-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 16 | RIMBP2 | Sponge network | -0.737 | 0.10822 | -1.968 | 5.0E-5 | 0.291 |
| 43424 | MEG3 |
hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-7-5p | 10 | CMKLR1 | Sponge network | 0.249 | 0.35902 | 0.288 | 0.17621 | 0.291 |
| 43425 | CALML3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-576-5p | 23 | TSC1 | Sponge network | 0.95 | 0.15943 | 0.103 | 0.26071 | 0.291 |
| 43426 | RP11-384K6.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-539-5p | 16 | NIN | Sponge network | 0.946 | 0 | -0.253 | 0.13794 | 0.291 |
| 43427 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | AR | Sponge network | 0.392 | 0.0278 | -1.554 | 0 | 0.291 |
| 43428 | RP11-473I1.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 11 | EPS15 | Sponge network | 0.542 | 0.00054 | -0.052 | 0.53998 | 0.291 |
| 43429 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 32 | PGR | Sponge network | 0.311 | 0.10344 | -1.704 | 0 | 0.291 |
| 43430 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 37 | BCL2 | Sponge network | -0.405 | 0.22013 | -0.899 | 0 | 0.291 |
| 43431 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-98-5p | 25 | LIFR | Sponge network | -0.227 | 0.31657 | -1.794 | 0 | 0.291 |
| 43432 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p | 13 | ACVR1 | Sponge network | -0.469 | 0.08721 | 0.279 | 0.00234 | 0.291 |
| 43433 | RP11-800A3.7 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | QKI | Sponge network | 0.18 | 0.7147 | -0.333 | 0.04111 | 0.291 |
| 43434 | NEAT1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PBRM1 | Sponge network | 0.705 | 0.00014 | -0.029 | 0.78601 | 0.291 |
| 43435 | SNHG14 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 18 | ADCY1 | Sponge network | -1.111 | 0.0003 | 0.362 | 0.16982 | 0.291 |
| 43436 | RP11-344B5.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-320a;hsa-miR-320b | 12 | BMPR2 | Sponge network | -0.055 | 0.88482 | 0.206 | 0.0635 | 0.291 |
| 43437 | RP11-98I9.4 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRIM13 | Sponge network | 0.67 | 0.00048 | 0.183 | 0.06968 | 0.291 |
| 43438 | LINC00886 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-93-5p | 50 | AR | Sponge network | -0.371 | 0.24604 | -1.554 | 0 | 0.291 |
| 43439 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 15 | SLITRK6 | Sponge network | 0.194 | 0.64278 | -2.121 | 0 | 0.291 |
| 43440 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 17 | WWTR1 | Sponge network | 0.4 | 0.14611 | -0.345 | 0.11997 | 0.291 |
| 43441 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 39 | PRKAA2 | Sponge network | -0.147 | 0.40452 | -2.462 | 0 | 0.291 |
| 43442 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | FZD3 | Sponge network | 0.67 | 0.00048 | 0.104 | 0.60316 | 0.291 |
| 43443 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-940 | 10 | ZFP36L1 | Sponge network | 0.299 | 0.57722 | -0.505 | 8.0E-5 | 0.291 |
| 43444 | AC108488.4 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-93-5p | 14 | LATS2 | Sponge network | 0.259 | 0.12542 | 0.159 | 0.2304 | 0.291 |
| 43445 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | FOXO1 | Sponge network | 0.335 | 0.20596 | -0.141 | 0.33874 | 0.291 |
| 43446 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 36 | RICTOR | Sponge network | -1.575 | 6.0E-5 | 0.388 | 0.00188 | 0.291 |
| 43447 | MIR600HG |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-484 | 10 | FBXO11 | Sponge network | 0.594 | 0.41522 | 0.142 | 0.04938 | 0.291 |
| 43448 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PLSCR4 | Sponge network | 0.349 | 0.01695 | -0.474 | 0.03833 | 0.291 |
| 43449 | CTD-2666L21.1 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-493-5p | 11 | SYNM | Sponge network | 0.368 | 0.20093 | -2.309 | 1.0E-5 | 0.291 |
| 43450 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 46 | AFF4 | Sponge network | 0.559 | 0.00026 | -0.266 | 0.01565 | 0.29 |
| 43451 | CTA-445C9.14 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-664a-3p | 14 | RASAL2 | Sponge network | 0.718 | 4.0E-5 | 0.869 | 0 | 0.29 |
| 43452 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 33 | PCDH10 | Sponge network | 0.222 | 0.29133 | -1.644 | 0.0078 | 0.29 |
| 43453 | MAP3K14 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 18 | EPHA7 | Sponge network | -0.295 | 0.02604 | -2.197 | 3.0E-5 | 0.29 |
| 43454 | AC007193.6 | hsa-let-7g-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p | 10 | BRD3 | Sponge network | 3.047 | 0.00303 | 0.37 | 0.00013 | 0.29 |
| 43455 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | PAK3 | Sponge network | -0.899 | 6.0E-5 | -0.628 | 0.07253 | 0.29 |
| 43456 | GAS6-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-664a-3p;hsa-miR-874-3p | 24 | ASTN1 | Sponge network | 0.16 | 0.50785 | -2.389 | 2.0E-5 | 0.29 |
| 43457 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | CTDSPL | Sponge network | -0.612 | 0.00094 | 0.085 | 0.45121 | 0.29 |
| 43458 | PCBP1-AS1 |
hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 10 | HERC1 | Sponge network | -0.567 | 0 | -0.196 | 0.08544 | 0.29 |
| 43459 | HCG11 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | SIRT1 | Sponge network | 0.385 | 0.10067 | 0.109 | 0.2593 | 0.29 |
| 43460 | CTD-3110H11.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | CBX5 | Sponge network | 1.735 | 0 | 0.165 | 0.24446 | 0.29 |
| 43461 | RP11-927P21.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-590-5p | 10 | ITGB1 | Sponge network | 0.921 | 0.00118 | 0.524 | 0.00017 | 0.29 |
| 43462 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-505-5p;hsa-miR-940 | 17 | SNX29 | Sponge network | -0.829 | 0.01343 | -0.34 | 0.01371 | 0.29 |
| 43463 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p | 25 | GRIA3 | Sponge network | -2.915 | 0.00015 | -1.956 | 0 | 0.29 |
| 43464 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-98-5p | 18 | NTRK3 | Sponge network | -0.295 | 0.02604 | -1.77 | 3.0E-5 | 0.29 |
| 43465 | RP11-554A11.4 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-874-3p | 15 | ARID5B | Sponge network | -0.583 | 0.14589 | 0.342 | 0.01676 | 0.29 |
| 43466 | CTD-2666L21.1 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-29a-5p | 10 | TSC1 | Sponge network | 0.368 | 0.20093 | 0.103 | 0.26071 | 0.29 |
| 43467 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-877-5p | 28 | EGFR | Sponge network | -1.654 | 0.00144 | -0.175 | 0.48451 | 0.29 |
| 43468 | RP11-283I3.6 |
hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p | 10 | CDK6 | Sponge network | 0.614 | 1.0E-5 | 0.991 | 0 | 0.29 |
| 43469 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | AR | Sponge network | 0.272 | 0.44692 | -1.554 | 0 | 0.29 |
| 43470 | CTC-297N7.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-590-5p | 11 | DMD | Sponge network | -2.069 | 0 | -1.506 | 0 | 0.29 |
| 43471 | U91328.19 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-940 | 16 | GLI3 | Sponge network | -0.303 | 0.21002 | -0.615 | 0.01482 | 0.29 |
| 43472 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | AKAP11 | Sponge network | 1.262 | 0 | 0.196 | 0.09825 | 0.29 |
| 43473 | AC005618.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-501-5p;hsa-miR-590-3p | 14 | PRDM2 | Sponge network | 0.862 | 0.06213 | -0.258 | 0.00721 | 0.29 |
| 43474 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | FOXP1 | Sponge network | -0.342 | 0.02288 | 0.042 | 0.74368 | 0.29 |
| 43475 | RP11-212P7.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | ROBO1 | Sponge network | 0.219 | 0.1516 | -0.009 | 0.96154 | 0.29 |
| 43476 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429 | 20 | WDFY3 | Sponge network | 1.316 | 0.01106 | -0.201 | 0.0967 | 0.29 |
| 43477 | ATP1B3-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 14 | CCDC6 | Sponge network | 1.2 | 0.01813 | 0.27 | 0.02555 | 0.29 |
| 43478 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 32 | ST6GALNAC3 | Sponge network | -0.487 | 0.02157 | -0.987 | 0 | 0.29 |
| 43479 | DNAJC27-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-877-5p | 23 | NRK | Sponge network | -0.969 | 2.0E-5 | 0.656 | 0.19217 | 0.29 |
| 43480 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-940 | 27 | BHLHE41 | Sponge network | -0.738 | 0.21233 | 0.353 | 0.12753 | 0.29 |
| 43481 | TINCR |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p | 10 | CSDE1 | Sponge network | -0.664 | 0.14543 | -0.493 | 0 | 0.29 |
| 43482 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-576-5p | 15 | GREM1 | Sponge network | 0.559 | 0.00026 | 0.902 | 0.01691 | 0.29 |
| 43483 | AC108488.4 |
hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 13 | MLLT10 | Sponge network | 0.259 | 0.12542 | 0.105 | 0.33292 | 0.29 |
| 43484 | RP11-384K6.6 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-335-5p | 10 | RB1 | Sponge network | 0.946 | 0 | 0.382 | 0.0017 | 0.29 |
| 43485 | LINC00094 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-625-5p | 11 | NGFR | Sponge network | 0.253 | 0.09565 | -1.271 | 0.01058 | 0.29 |
| 43486 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 61 | TCF4 | Sponge network | -0.899 | 6.0E-5 | 0.016 | 0.92271 | 0.29 |
| 43487 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-98-5p | 18 | ZFP36L1 | Sponge network | -0.899 | 6.0E-5 | -0.505 | 8.0E-5 | 0.29 |
| 43488 | AC156455.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p | 10 | EPHA5 | Sponge network | 1.154 | 0.00041 | -0.957 | 0.01733 | 0.29 |
| 43489 | AC141928.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-7-1-3p | 35 | PTPRD | Sponge network | 0.853 | 0.15352 | -1.005 | 0.00064 | 0.29 |
| 43490 | BCYRN1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-96-5p | 32 | NAV3 | Sponge network | 0.311 | 0.10344 | 0.212 | 0.47729 | 0.29 |
| 43491 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | EPHA3 | Sponge network | 0.272 | 0.44692 | 0.185 | 0.53588 | 0.29 |
| 43492 | CD27-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.177 | 0.16681 | -0.57 | 0 | 0.29 |
| 43493 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-376c-3p;hsa-miR-590-3p | 13 | CREBBP | Sponge network | 0.559 | 0.00026 | 0.126 | 0.15186 | 0.29 |
| 43494 | C4A-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-629-3p | 16 | EPHA7 | Sponge network | 3.345 | 0 | -2.197 | 3.0E-5 | 0.29 |
| 43495 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-942-5p | 21 | GPC6 | Sponge network | 0.61 | 0.01593 | -0.054 | 0.81465 | 0.29 |
| 43496 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | HECA | Sponge network | -1.281 | 0.00012 | -0.217 | 0.03463 | 0.29 |
| 43497 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p | 11 | LRRK2 | Sponge network | 0.659 | 3.0E-5 | -1.136 | 1.0E-5 | 0.29 |
| 43498 | CKMT2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 24 | CYLD | Sponge network | 0.082 | 0.55654 | -0.367 | 0.00171 | 0.29 |
| 43499 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 32 | SOX5 | Sponge network | -0.223 | 0.47816 | -1.091 | 4.0E-5 | 0.29 |
| 43500 | LINC00641 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | SLC2A13 | Sponge network | -0.222 | 0.32445 | -0.331 | 0.03658 | 0.29 |
| 43501 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-93-5p | 27 | HBP1 | Sponge network | -0.227 | 0.31657 | -0.454 | 0 | 0.29 |
| 43502 | RP11-479G22.8 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-101-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | CDYL | Sponge network | 1.308 | 0 | 0.12 | 0.12425 | 0.29 |
| 43503 | AC074289.1 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-96-5p | 19 | NAV3 | Sponge network | -0.326 | 0.14314 | 0.212 | 0.47729 | 0.29 |
| 43504 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-337-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-5p | 17 | CUL5 | Sponge network | 0.331 | 0.57247 | 0.026 | 0.77301 | 0.29 |
| 43505 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | -0.487 | 0.02157 | -2.417 | 0 | 0.29 |
| 43506 | AF131215.2 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-582-5p | 10 | PTCH1 | Sponge network | -0.176 | 0.59692 | -0.1 | 0.6179 | 0.29 |
| 43507 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 37 | ZFHX4 | Sponge network | -0.125 | 0.49414 | 0.018 | 0.95574 | 0.29 |
| 43508 | HOXB-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-628-5p | 16 | RIMS2 | Sponge network | -0.154 | 0.53954 | -1.365 | 0.00208 | 0.29 |
| 43509 | DNM1P35 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-5p | 14 | SETBP1 | Sponge network | 0.051 | 0.83388 | -1.302 | 1.0E-5 | 0.29 |
| 43510 | AC005618.6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ZMYND11 | Sponge network | 0.862 | 0.06213 | -0.2 | 0.05449 | 0.29 |
| 43511 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | THBS1 | Sponge network | 0.559 | 0.00026 | 0.741 | 0.00386 | 0.29 |
| 43512 | KB-1460A1.5 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 11 | STARD13 | Sponge network | 0.603 | 0.00127 | -0.187 | 0.27383 | 0.29 |
| 43513 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-590-3p | 14 | LATS1 | Sponge network | 1.317 | 0 | 0.109 | 0.34208 | 0.29 |
| 43514 | ANKRD10-IT1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 12 | TMTC3 | Sponge network | 1.739 | 0 | 0.123 | 0.22189 | 0.29 |
| 43515 | LINC00667 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | NRXN3 | Sponge network | -0.235 | 0.15142 | -0.893 | 0.04186 | 0.29 |
| 43516 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-590-3p | 22 | AFF3 | Sponge network | 0.082 | 0.55654 | -2.373 | 0 | 0.29 |
| 43517 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-93-5p;hsa-miR-942-5p | 14 | FBXO31 | Sponge network | -0.08 | 0.90092 | -0.085 | 0.42389 | 0.29 |
| 43518 | RP11-967K21.1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-539-5p;hsa-miR-582-5p | 10 | PHF3 | Sponge network | 0.056 | 0.81195 | 0.126 | 0.19652 | 0.29 |
| 43519 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-484 | 10 | EBF3 | Sponge network | -4.477 | 0 | -0.058 | 0.81437 | 0.29 |
| 43520 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p | 17 | NTRK2 | Sponge network | 0.497 | 0.01451 | -0.736 | 0.06495 | 0.29 |
| 43521 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-450b-5p;hsa-miR-455-3p | 13 | DKK3 | Sponge network | 0.331 | 0.57247 | -0.052 | 0.77783 | 0.29 |
| 43522 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-93-5p | 33 | RASSF2 | Sponge network | 0.082 | 0.55654 | -0.273 | 0.21961 | 0.29 |
| 43523 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | TCF4 | Sponge network | 0.272 | 0.44692 | 0.016 | 0.92271 | 0.29 |
| 43524 | FAM13A-AS1 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 15 | SFRP1 | Sponge network | -0.83 | 0 | -3.566 | 0 | 0.29 |
| 43525 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 24 | ARHGAP28 | Sponge network | -0.777 | 0.1134 | -0.911 | 0.00013 | 0.29 |
| 43526 | RP11-216B9.6 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-940 | 23 | GRIK3 | Sponge network | 0.174 | 0.30034 | -3.208 | 0 | 0.29 |
| 43527 | RP11-890B15.3 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NRG3 | Sponge network | 0.101 | 0.60919 | -1.836 | 4.0E-5 | 0.29 |
| 43528 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | PAFAH1B1 | Sponge network | -0.976 | 0.00025 | -0.429 | 0 | 0.29 |
| 43529 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-935 | 19 | RARB | Sponge network | -1.203 | 0.03881 | -0.825 | 2.0E-5 | 0.29 |
| 43530 | RP11-403I13.8 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 35 | TRPS1 | Sponge network | 0.619 | 0.00157 | -0.191 | 0.44109 | 0.29 |
| 43531 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | ATRX | Sponge network | 0.385 | 0.10067 | 0.261 | 0.04408 | 0.29 |
| 43532 | RP11-401P9.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-532-5p;hsa-miR-7-5p | 24 | IGFBP5 | Sponge network | -0.384 | 0.40652 | -0.806 | 0.00105 | 0.29 |
| 43533 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 0.34 | 0.00273 | 0.51 | 0 | 0.29 |
| 43534 | CTB-31O20.4 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-629-3p | 18 | KIF1B | Sponge network | 1.619 | 0 | -0.118 | 0.32258 | 0.29 |
| 43535 | FGD5-AS1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-628-5p;hsa-miR-708-5p | 12 | MAT2A | Sponge network | 0.401 | 0.00066 | -0.073 | 0.36195 | 0.29 |
| 43536 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-576-5p | 19 | CACNA2D1 | Sponge network | -0.147 | 0.40452 | -0.846 | 0.00675 | 0.29 |
| 43537 | LINC00886 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p | 17 | RARB | Sponge network | -0.371 | 0.24604 | -0.825 | 2.0E-5 | 0.29 |
| 43538 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 20 | TSC1 | Sponge network | -0.227 | 0.31657 | 0.103 | 0.26071 | 0.29 |
| 43539 | RP13-1039J1.4 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-589-3p;hsa-miR-625-5p | 12 | NTRK3 | Sponge network | 0.356 | 0.33467 | -1.77 | 3.0E-5 | 0.29 |
| 43540 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CACNA2D1 | Sponge network | 0.61 | 0.01593 | -0.846 | 0.00675 | 0.29 |
| 43541 | GABPB1-AS1 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p | 17 | DICER1 | Sponge network | 1.11 | 0 | 0.042 | 0.674 | 0.29 |
| 43542 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-935 | 33 | HBP1 | Sponge network | -0.129 | 0.57177 | -0.454 | 0 | 0.29 |
| 43543 | RP11-263K19.4 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-93-3p | 12 | WWTR1 | Sponge network | 0.859 | 0.00331 | -0.345 | 0.11997 | 0.29 |
| 43544 | RP11-5C23.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-2355-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | SIRT1 | Sponge network | 0.274 | 0.07681 | 0.109 | 0.2593 | 0.29 |
| 43545 | LINC00957 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZNF844 | Sponge network | -0.421 | 0.0114 | -0.966 | 0.00035 | 0.29 |
| 43546 | RP5-998N21.10 |
hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-3613-5p;hsa-miR-502-5p | 10 | NTRK3 | Sponge network | -0.098 | 0.76298 | -1.77 | 3.0E-5 | 0.29 |
| 43547 | RP4-613B23.1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p | 11 | MOB1B | Sponge network | -0.309 | 0.1145 | 0.197 | 0.15266 | 0.29 |
| 43548 | CKMT2-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-940 | 23 | EPS15 | Sponge network | 0.082 | 0.55654 | -0.052 | 0.53998 | 0.29 |
| 43549 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-539-5p | 14 | CDHR3 | Sponge network | -0.602 | 0.00331 | -0.977 | 4.0E-5 | 0.29 |
| 43550 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577 | 21 | DST | Sponge network | 0.357 | 0.72407 | -0.713 | 0.00096 | 0.29 |
| 43551 | RP11-855A2.3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-590-3p | 16 | UBR3 | Sponge network | 2.094 | 0 | -0.021 | 0.82774 | 0.29 |
| 43552 | RBPMS-AS1 |
hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-7-1-3p | 10 | EFNA5 | Sponge network | 0.144 | 0.64989 | -0.421 | 0.17868 | 0.29 |
| 43553 | AC005618.6 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | STARD13 | Sponge network | 0.862 | 0.06213 | -0.187 | 0.27383 | 0.29 |
| 43554 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 18 | ZMYND11 | Sponge network | 0.16 | 0.50785 | -0.2 | 0.05449 | 0.29 |
| 43555 | RP11-401P9.4 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-7-5p | 11 | IRS1 | Sponge network | -0.384 | 0.40652 | -0.026 | 0.88855 | 0.29 |
| 43556 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 36 | NTRK2 | Sponge network | -0.896 | 0.00099 | -0.736 | 0.06495 | 0.29 |
| 43557 | MRVI1-AS1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-338-5p;hsa-miR-542-3p | 15 | PAIP2 | Sponge network | -1.092 | 4.0E-5 | -0.515 | 0 | 0.29 |
| 43558 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-421 | 12 | PHC3 | Sponge network | -0.583 | 0.14589 | 0.212 | 0.0499 | 0.29 |
| 43559 | RP11-6O2.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-590-5p | 10 | PTGFR | Sponge network | 0.331 | 0.57247 | -0.233 | 0.45421 | 0.29 |
| 43560 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 32 | IL6ST | Sponge network | 0.687 | 0.02753 | -0.402 | 0.00676 | 0.29 |
| 43561 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | EPHA7 | Sponge network | 0.349 | 0.01695 | -2.197 | 3.0E-5 | 0.29 |
| 43562 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZNF844 | Sponge network | 0.572 | 0.01722 | -0.966 | 0.00035 | 0.29 |
| 43563 | ADAMTS9-AS2 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 18 | TES | Sponge network | -2.452 | 0 | -0.348 | 0.00639 | 0.29 |
| 43564 | RP11-327P2.5 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p | 16 | UBE4B | Sponge network | -0.147 | 0.40452 | -0.235 | 0.007 | 0.29 |
| 43565 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-590-3p;hsa-miR-98-5p | 15 | ITGB3 | Sponge network | 0.494 | 0.00339 | -0.011 | 0.95751 | 0.29 |
| 43566 | HOXB-AS3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-671-5p | 12 | DLC1 | Sponge network | 0.343 | 0.28887 | -0.382 | 0.05038 | 0.29 |
| 43567 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-664a-5p | 12 | SOX11 | Sponge network | 0.813 | 1.0E-5 | 2.68 | 0 | 0.29 |
| 43568 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-590-5p | 11 | THSD7A | Sponge network | -0.899 | 6.0E-5 | 0.242 | 0.25154 | 0.29 |
| 43569 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-501-5p | 12 | FGF5 | Sponge network | 0.176 | 0.37402 | 0.549 | 0.28659 | 0.29 |
| 43570 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-576-5p;hsa-miR-590-3p | 13 | HIP1 | Sponge network | 0.401 | 0.00066 | 0.774 | 0 | 0.29 |
| 43571 | MRVI1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-505-3p | 17 | KCNJ5 | Sponge network | -1.092 | 4.0E-5 | -0.281 | 0.3339 | 0.29 |
| 43572 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 66 | IL6ST | Sponge network | 0.421 | 0.00084 | -0.402 | 0.00676 | 0.29 |
| 43573 | RP11-798M19.6 |
hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-653-5p | 26 | UBR3 | Sponge network | -0.612 | 0.00094 | -0.021 | 0.82774 | 0.29 |
| 43574 | SOX2-OT |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PCLO | Sponge network | -1.264 | 6.0E-5 | -1.843 | 0.00064 | 0.29 |
| 43575 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | PTPN11 | Sponge network | 0.8 | 0 | 0.431 | 2.0E-5 | 0.29 |
| 43576 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p | 12 | TSC22D1 | Sponge network | -2.46 | 0 | -0.529 | 2.0E-5 | 0.29 |
| 43577 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 16 | SCN9A | Sponge network | 1.308 | 0 | -0.525 | 0.10593 | 0.29 |
| 43578 | RP11-1006G14.4 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | GALNT13 | Sponge network | 0.559 | 0.00026 | -0.857 | 0.07439 | 0.29 |
| 43579 | HOTAIRM1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | KLF10 | Sponge network | -0.053 | 0.80685 | -0.783 | 0 | 0.29 |
| 43580 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 38 | SSBP2 | Sponge network | 0.214 | 0.18246 | -1.58 | 0 | 0.29 |
| 43581 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PHC3 | Sponge network | 0.986 | 0.01022 | 0.212 | 0.0499 | 0.29 |
| 43582 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | TCF12 | Sponge network | 0.214 | 0.18246 | 0.174 | 0.13271 | 0.29 |
| 43583 | SNHG14 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 16 | SHPRH | Sponge network | -1.111 | 0.0003 | 0.098 | 0.40994 | 0.29 |
| 43584 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 28 | PPM1A | Sponge network | -1.654 | 0.00144 | -0.623 | 0 | 0.29 |
| 43585 | RP11-3P17.4 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p | 12 | RB1 | Sponge network | 0.144 | 0.07137 | 0.382 | 0.0017 | 0.29 |
| 43586 | LINC00654 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-7-1-3p | 11 | CNTN5 | Sponge network | 0.374 | 0.20054 | -0.219 | 0.61957 | 0.29 |
| 43587 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 23 | HIP1 | Sponge network | 0.246 | 0.59912 | 0.774 | 0 | 0.29 |
| 43588 | AC156455.1 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 11 | ERCC4 | Sponge network | 1.154 | 0.00041 | 0.126 | 0.15266 | 0.29 |
| 43589 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 0.349 | 0.01695 | 0.578 | 0 | 0.29 |
| 43590 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 18 | POU2F2 | Sponge network | -0.49 | 0.04946 | -0.233 | 0.32214 | 0.29 |
| 43591 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92b-3p | 39 | TCF4 | Sponge network | -0.322 | 0.25945 | 0.016 | 0.92271 | 0.29 |
| 43592 | RP11-458D21.1 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p | 12 | PTCH1 | Sponge network | 0.663 | 0.00073 | -0.1 | 0.6179 | 0.29 |
| 43593 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 18 | SASH1 | Sponge network | -0.517 | 0.02935 | -0.502 | 0.00175 | 0.29 |
| 43594 | MYLK-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-671-5p | 15 | ARID5B | Sponge network | -1.331 | 3.0E-5 | 0.342 | 0.01676 | 0.29 |
| 43595 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-877-5p | 21 | NRK | Sponge network | -0.465 | 0.04703 | 0.656 | 0.19217 | 0.29 |
| 43596 | BVES-AS1 |
hsa-miR-132-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p | 13 | RIT1 | Sponge network | -2.62 | 0 | -0.296 | 0.00054 | 0.29 |
| 43597 | HOXA-AS3 |
hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-5p | 11 | RNF144A | Sponge network | -0.555 | 0.06156 | 0.594 | 0.0009 | 0.29 |
| 43598 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | ACVR2A | Sponge network | 0.537 | 0.00139 | -0.409 | 0.00039 | 0.29 |
| 43599 | NEAT1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TRIP11 | Sponge network | 0.705 | 0.00014 | -0.158 | 0.06384 | 0.29 |
| 43600 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-98-5p | 15 | ITGB3 | Sponge network | -0.405 | 0.22013 | -0.011 | 0.95751 | 0.29 |
| 43601 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-505-5p | 18 | HBP1 | Sponge network | -0.602 | 0.00331 | -0.454 | 0 | 0.29 |
| 43602 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 39 | RASSF2 | Sponge network | -1.047 | 0 | -0.273 | 0.21961 | 0.29 |
| 43603 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p | 19 | DMD | Sponge network | 1.46 | 1.0E-5 | -1.506 | 0 | 0.29 |
| 43604 | LINC01018 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | CREB3L2 | Sponge network | -2.915 | 0.00015 | -0.525 | 2.0E-5 | 0.29 |
| 43605 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 35 | TMEM132B | Sponge network | 0.559 | 0.00026 | -0.83 | 0.01084 | 0.29 |
| 43606 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CDH19 | Sponge network | -0.668 | 3.0E-5 | -3.434 | 0 | 0.29 |
| 43607 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-532-3p;hsa-miR-532-5p | 11 | LRRC55 | Sponge network | -0.829 | 0.01343 | -0.026 | 0.93163 | 0.29 |
| 43608 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p | 13 | HIVEP1 | Sponge network | -0.129 | 0.57177 | 0.368 | 0.00064 | 0.29 |
| 43609 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p | 20 | NR2C2 | Sponge network | -0.162 | 0.47687 | 0.21 | 0.03224 | 0.29 |
| 43610 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 11 | CSRNP1 | Sponge network | -0.611 | 0.09607 | -1.448 | 0 | 0.29 |
| 43611 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-486-5p | 12 | GLIPR1 | Sponge network | 0.176 | 0.37402 | 0.146 | 0.3276 | 0.29 |
| 43612 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ZNF208 | Sponge network | 0.064 | 0.72934 | -0.4 | 0.22381 | 0.29 |
| 43613 | LPP-AS2 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-96-5p | 11 | PRKACA | Sponge network | -0.18 | 0.20343 | -0.255 | 0.0012 | 0.29 |
| 43614 | AGAP11 |
hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 11 | PPP2R2C | Sponge network | -1.645 | 0 | -0.959 | 0.07428 | 0.29 |
| 43615 | RASSF8-AS1 |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | PAIP2 | Sponge network | 0.335 | 0.20596 | -0.515 | 0 | 0.29 |
| 43616 | AC010226.4 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | LIMCH1 | Sponge network | -0.811 | 2.0E-5 | -0.915 | 1.0E-5 | 0.29 |
| 43617 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p | 33 | GAS7 | Sponge network | -0.342 | 0.02288 | -0.555 | 0.01602 | 0.29 |
| 43618 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 11 | EZH1 | Sponge network | 0.131 | 0.8436 | -0.564 | 1.0E-5 | 0.29 |
| 43619 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PLSCR4 | Sponge network | 0.603 | 0.00127 | -0.474 | 0.03833 | 0.29 |
| 43620 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p | 18 | MOB1B | Sponge network | -0.583 | 0.14589 | 0.197 | 0.15266 | 0.29 |
| 43621 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-590-3p | 10 | ANK3 | Sponge network | -0.031 | 0.89063 | -0.55 | 0.00086 | 0.29 |
| 43622 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-93-5p | 46 | PRKAA2 | Sponge network | -0.371 | 0.24604 | -2.462 | 0 | 0.29 |
| 43623 | AC108488.4 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-511-5p | 12 | BCL6 | Sponge network | 0.259 | 0.12542 | -0.211 | 0.19489 | 0.29 |
| 43624 | RP11-983P16.4 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 45 | DTNA | Sponge network | 0.41 | 0.02214 | -1.648 | 1.0E-5 | 0.29 |
| 43625 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p | 18 | APC | Sponge network | 0.082 | 0.55397 | -0.255 | 0.04051 | 0.29 |
| 43626 | RP11-203M5.7 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-495-3p | 14 | AFF4 | Sponge network | 1.684 | 1.0E-5 | -0.266 | 0.01565 | 0.29 |
| 43627 | LIPE-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-34c-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | SLC5A3 | Sponge network | 0.078 | 0.55803 | 0.493 | 5.0E-5 | 0.29 |
| 43628 | RP11-403I13.8 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p | 34 | ADARB1 | Sponge network | 0.619 | 0.00157 | -0.335 | 0.06631 | 0.29 |
| 43629 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | 1.302 | 7.0E-5 | -0.684 | 0.00159 | 0.29 |
| 43630 | AC018890.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | ABI2 | Sponge network | 1.61 | 0.00961 | 0.175 | 0.14787 | 0.29 |
| 43631 | RP11-760H22.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-454-3p | 23 | MDM4 | Sponge network | 0.001 | 0.99753 | 0.345 | 0.00762 | 0.29 |
| 43632 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | DPYD | Sponge network | -1.111 | 0.0003 | -0.736 | 0.00076 | 0.29 |
| 43633 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TBX18 | Sponge network | -0.612 | 0.00094 | 0.843 | 0.02945 | 0.29 |
| 43634 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | SYNE1 | Sponge network | 0.381 | 0.25967 | -0.738 | 0.00316 | 0.29 |
| 43635 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-93-5p | 16 | CNTNAP3 | Sponge network | -0.18 | 0.20343 | -1.465 | 0.00033 | 0.29 |
| 43636 | AC005618.6 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | 0.862 | 0.06213 | 0.142 | 0.04938 | 0.29 |
| 43637 | LINC00578 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 34 | THRB | Sponge network | 0.811 | 0.03228 | -1.117 | 0.0004 | 0.29 |
| 43638 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-654-3p | 14 | ITCH | Sponge network | 0.083 | 0.66405 | 0.084 | 0.34058 | 0.29 |
| 43639 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-628-5p | 23 | FBXO32 | Sponge network | 1.089 | 0 | -0.495 | 0.07561 | 0.29 |
| 43640 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | ARHGAP29 | Sponge network | 0.311 | 0.10344 | 0.131 | 0.42953 | 0.29 |
| 43641 | RP11-798G7.7 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p | 18 | WDFY3 | Sponge network | 0.858 | 3.0E-5 | -0.201 | 0.0967 | 0.29 |
| 43642 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 43 | PPM1A | Sponge network | -0.901 | 0.00019 | -0.623 | 0 | 0.29 |
| 43643 | AC011526.1 |
hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-92a-3p | 13 | ST6GAL2 | Sponge network | 0.342 | 0.03129 | -1.201 | 0.00597 | 0.29 |
| 43644 | RP11-212P7.2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 17 | CELF4 | Sponge network | 0.219 | 0.1516 | -1.135 | 0.00344 | 0.29 |
| 43645 | A2M-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | TAL1 | Sponge network | -0.08 | 0.90092 | -0.462 | 0.00574 | 0.29 |
| 43646 | RP1-228H13.5 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-654-3p | 11 | RICTOR | Sponge network | 1.397 | 0 | 0.388 | 0.00188 | 0.29 |
| 43647 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p | 26 | SPRY3 | Sponge network | 1.04 | 0.00023 | 0.016 | 0.91286 | 0.29 |
| 43648 | AC012360.6 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | PHC3 | Sponge network | 0.624 | 0.02176 | 0.212 | 0.0499 | 0.29 |
| 43649 | LINC00672 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-629-3p | 13 | SPTBN4 | Sponge network | -0.227 | 0.31657 | -0.786 | 0.01281 | 0.29 |
| 43650 | EMX2OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | CBFA2T3 | Sponge network | -0.777 | 0.1134 | -1.221 | 1.0E-5 | 0.29 |
| 43651 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p | 13 | EPHA5 | Sponge network | -2.549 | 0.00528 | -0.957 | 0.01733 | 0.29 |
| 43652 | AC108676.1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 11 | PHF6 | Sponge network | 4.346 | 2.0E-5 | 0.785 | 0 | 0.29 |
| 43653 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 53 | CCND2 | Sponge network | -0.777 | 0.1134 | -0.267 | 0.3112 | 0.29 |
| 43654 | RP11-1006G14.4 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 12 | DYNC1I1 | Sponge network | 0.559 | 0.00026 | -1.12 | 0.00107 | 0.29 |
| 43655 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 23 | PRKCE | Sponge network | 0.083 | 0.66405 | -0.403 | 0.00056 | 0.29 |
| 43656 | LINC00094 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-374a-5p;hsa-miR-429 | 15 | SPG20 | Sponge network | 0.253 | 0.09565 | -1.16 | 0 | 0.29 |
| 43657 | LINC00641 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | PRDM5 | Sponge network | -0.222 | 0.32445 | -1.09 | 0.00014 | 0.29 |
| 43658 | LINC00621 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 22 | WDFY3 | Sponge network | 0.131 | 0.8436 | -0.201 | 0.0967 | 0.29 |
| 43659 | RP13-1039J1.4 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p | 13 | DST | Sponge network | 0.356 | 0.33467 | -0.713 | 0.00096 | 0.29 |
| 43660 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | FGF5 | Sponge network | -0.899 | 6.0E-5 | 0.549 | 0.28659 | 0.29 |
| 43661 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 62 | BMPR2 | Sponge network | -0.612 | 0.00094 | 0.206 | 0.0635 | 0.29 |
| 43662 | RP11-686D22.8 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | ERG | Sponge network | 0.898 | 1.0E-5 | 0.002 | 0.99011 | 0.29 |
| 43663 | RP11-574K11.29 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-495-3p | 10 | ABI2 | Sponge network | 0.997 | 0 | 0.175 | 0.14787 | 0.29 |
| 43664 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 25 | YAP1 | Sponge network | -0.129 | 0.57177 | 0.22 | 0.11836 | 0.29 |
| 43665 | SOX2-OT |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 11 | PARK2 | Sponge network | -1.264 | 6.0E-5 | -1.892 | 0 | 0.29 |
| 43666 | LINC01018 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | EYA4 | Sponge network | -2.915 | 0.00015 | 0.3 | 0.36876 | 0.29 |
| 43667 | RP3-402G11.26 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-500a-3p;hsa-miR-590-3p | 11 | TET1 | Sponge network | -0.254 | 0.19303 | -0.135 | 0.52138 | 0.29 |
| 43668 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-501-3p | 11 | TET1 | Sponge network | 0.37 | 0.27614 | -0.135 | 0.52138 | 0.29 |
| 43669 | ACTA2-AS1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-940 | 10 | TAF1 | Sponge network | 0.194 | 0.64278 | 0.39 | 3.0E-5 | 0.29 |
| 43670 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-5p | 14 | HDAC9 | Sponge network | -1.047 | 0 | -0.755 | 0.00495 | 0.29 |
| 43671 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-411-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 38 | SMAD2 | Sponge network | 0.614 | 1.0E-5 | -0.128 | 0.12732 | 0.29 |
| 43672 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-577 | 19 | GREM1 | Sponge network | -0.227 | 0.31657 | 0.902 | 0.01691 | 0.29 |
| 43673 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 18 | ITGAV | Sponge network | -0.096 | 0.67958 | 0.337 | 0.01002 | 0.29 |
| 43674 | AC074289.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-511-5p | 13 | WWTR1 | Sponge network | -0.326 | 0.14314 | -0.345 | 0.11997 | 0.29 |
| 43675 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-339-5p | 18 | DMXL1 | Sponge network | 0.946 | 0 | -0.426 | 0.00056 | 0.29 |
| 43676 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-500a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 31 | MOB1B | Sponge network | 0.395 | 0.15451 | 0.197 | 0.15266 | 0.29 |
| 43677 | SNHG14 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 16 | CSRNP1 | Sponge network | -1.111 | 0.0003 | -1.448 | 0 | 0.29 |
| 43678 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-92a-1-5p | 17 | PRKAB2 | Sponge network | -2.531 | 0 | -0.367 | 0.01371 | 0.29 |
| 43679 | AC141928.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484 | 15 | ARNT2 | Sponge network | 0.853 | 0.15352 | -0.332 | 0.25851 | 0.29 |
| 43680 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 35 | TCF4 | Sponge network | 0.419 | 0.0319 | 0.016 | 0.92271 | 0.29 |
| 43681 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | DMXL1 | Sponge network | -0.811 | 2.0E-5 | -0.426 | 0.00056 | 0.29 |
| 43682 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 41 | EPHA5 | Sponge network | 0.311 | 0.10344 | -0.957 | 0.01733 | 0.29 |
| 43683 | RP11-428J1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PLSCR4 | Sponge network | 0.538 | 0.00076 | -0.474 | 0.03833 | 0.29 |
| 43684 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-629-3p | 20 | DDX6 | Sponge network | 0.174 | 0.30034 | 0.091 | 0.36428 | 0.29 |
| 43685 | RP4-717I23.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-3607-3p;hsa-miR-582-3p | 11 | PLAG1 | Sponge network | 0.637 | 0.00069 | -0.315 | 0.26532 | 0.29 |
| 43686 | RP1-43E13.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-21-5p;hsa-miR-590-5p | 10 | CXXC4 | Sponge network | -0.034 | 0.83228 | -0.433 | 0.21055 | 0.29 |
| 43687 | PCA3 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | PCDH17 | Sponge network | -0.709 | 0.18168 | 0.731 | 2.0E-5 | 0.29 |
| 43688 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p | 14 | CLIP1 | Sponge network | 0.214 | 0.18246 | -0.215 | 0.08684 | 0.29 |
| 43689 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | -1.111 | 0.0003 | 0.323 | 0.00792 | 0.29 |
| 43690 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 11 | MITF | Sponge network | 0.199 | 0.52763 | -0.769 | 0.00022 | 0.29 |
| 43691 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | BNC2 | Sponge network | -0.418 | 0.00068 | -0.491 | 0.09988 | 0.29 |
| 43692 | RP11-400K9.4 |
hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-339-5p;hsa-miR-542-3p | 11 | TP63 | Sponge network | -0.611 | 0.09607 | -0.363 | 0.38191 | 0.29 |
| 43693 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 17 | KIT | Sponge network | 0.214 | 0.18246 | -2.184 | 0 | 0.29 |
| 43694 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 25 | ACSL6 | Sponge network | -0.323 | 0.01372 | -1.593 | 0 | 0.29 |
| 43695 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p | 10 | ZFHX3 | Sponge network | 1.969 | 0.00316 | 0.389 | 0.01363 | 0.29 |
| 43696 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-766-3p | 11 | PHACTR4 | Sponge network | 1.254 | 0 | 0.069 | 0.45203 | 0.29 |
| 43697 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 13 | PIK3C2A | Sponge network | 1.178 | 0 | 0.061 | 0.53776 | 0.29 |
| 43698 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | USP12 | Sponge network | 0.299 | 0.57722 | -0.102 | 0.40768 | 0.29 |
| 43699 | RP4-758J18.10 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-450b-5p | 30 | LPP | Sponge network | -0.091 | 0.71468 | -0.459 | 0.04475 | 0.29 |
| 43700 | RP11-283I3.6 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-2-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-664a-5p | 14 | RAPGEF5 | Sponge network | 0.614 | 1.0E-5 | 0.536 | 4.0E-5 | 0.29 |
| 43701 | RP11-344B5.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-320b | 10 | JAZF1 | Sponge network | -0.055 | 0.88482 | -0.377 | 0.02677 | 0.29 |
| 43702 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | CSRNP1 | Sponge network | -0.129 | 0.57177 | -1.448 | 0 | 0.29 |
| 43703 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-98-5p | 10 | PICALM | Sponge network | 0.056 | 0.81195 | 0.086 | 0.23523 | 0.29 |
| 43704 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-331-5p;hsa-miR-532-3p | 12 | USP12 | Sponge network | 0.792 | 0.0049 | -0.102 | 0.40768 | 0.29 |
| 43705 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-96-5p | 29 | HBP1 | Sponge network | 0.194 | 0.64278 | -0.454 | 0 | 0.29 |
| 43706 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 35 | NOTCH2 | Sponge network | 0.61 | 0.01593 | 0.138 | 0.35163 | 0.29 |
| 43707 | RP11-359B12.2 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-96-5p | 14 | WNK2 | Sponge network | 0.064 | 0.72934 | -0.352 | 0.31066 | 0.29 |
| 43708 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 37 | KIF1B | Sponge network | 0.539 | 0.03722 | -0.118 | 0.32258 | 0.29 |
| 43709 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | CDYL | Sponge network | 0.225 | 0.30644 | 0.12 | 0.12425 | 0.29 |
| 43710 | TTC25 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | MEF2C | Sponge network | -0.208 | 0.38261 | -0.499 | 0.01448 | 0.29 |
| 43711 | RP11-384O8.1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-940 | 10 | HOOK3 | Sponge network | -0.738 | 0.21233 | 0.273 | 0.09957 | 0.29 |
| 43712 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | NOTCH2 | Sponge network | 0.395 | 0.15451 | 0.138 | 0.35163 | 0.29 |
| 43713 | LINC00476 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p | 11 | SCN11A | Sponge network | -0.615 | 0.00015 | -1.15 | 0.00299 | 0.29 |
| 43714 | RP4-639F20.1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-33a-3p | 13 | ABL2 | Sponge network | -0.517 | 0.02935 | 0.82 | 0 | 0.29 |
| 43715 | CTD-2336O2.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 12 | MAPK10 | Sponge network | -0.141 | 0.26434 | -1.774 | 0 | 0.29 |
| 43716 | RP1-257A7.5 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p | 10 | NAV3 | Sponge network | 0.849 | 0.00178 | 0.212 | 0.47729 | 0.29 |
| 43717 | AF131215.2 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-654-3p | 12 | BAZ2B | Sponge network | -0.176 | 0.59692 | -0.276 | 0.02833 | 0.29 |
| 43718 | LINC00886 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-493-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-542-3p | 16 | MRVI1 | Sponge network | -0.371 | 0.24604 | -1.051 | 0.00022 | 0.29 |
| 43719 | RP11-2H8.2 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-5p;hsa-miR-590-3p | 30 | CCDC6 | Sponge network | 1.089 | 0 | 0.27 | 0.02555 | 0.29 |
| 43720 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 62 | BMPR2 | Sponge network | -0.777 | 0.1134 | 0.206 | 0.0635 | 0.29 |
| 43721 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-27a-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 10 | TP53INP1 | Sponge network | -0.811 | 2.0E-5 | -0.496 | 0.00064 | 0.29 |
| 43722 | CTD-2303H24.2 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | ZDBF2 | Sponge network | 0.415 | 0.14657 | -0.998 | 0.00025 | 0.29 |
| 43723 | SERHL |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p | 16 | LRIG1 | Sponge network | -0.602 | 0.00331 | -0.537 | 0.00588 | 0.29 |
| 43724 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p | 12 | MAPK1 | Sponge network | 1.316 | 0 | -0.017 | 0.81465 | 0.29 |
| 43725 | ZNF582-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 17 | PTPN14 | Sponge network | -1.349 | 0.00017 | 0.144 | 0.34595 | 0.29 |
| 43726 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-3p;hsa-miR-874-3p | 20 | ARID5B | Sponge network | -0.384 | 0.40652 | 0.342 | 0.01676 | 0.29 |
| 43727 | HAR1A |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-93-3p | 24 | RUNX1T1 | Sponge network | -0.672 | 0.19447 | -0.344 | 0.2374 | 0.29 |
| 43728 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TAL1 | Sponge network | -0.342 | 0.02288 | -0.462 | 0.00574 | 0.29 |
| 43729 | C20orf166-AS1 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-582-5p | 13 | PTCH1 | Sponge network | -2.69 | 0.0001 | -0.1 | 0.6179 | 0.29 |
| 43730 | RP11-116O18.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ZNF208 | Sponge network | 0.05 | 0.95483 | -0.4 | 0.22381 | 0.289 |
| 43731 | ACTA2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-96-5p | 41 | CREB3L2 | Sponge network | 0.194 | 0.64278 | -0.525 | 2.0E-5 | 0.289 |
| 43732 | TRAM2-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-590-3p | 18 | ABL2 | Sponge network | 0.336 | 0.00739 | 0.82 | 0 | 0.289 |
| 43733 | ERVH48-1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-550a-5p;hsa-miR-625-5p | 19 | CHD5 | Sponge network | 1.04 | 0.00023 | -0.922 | 0.01521 | 0.289 |
| 43734 | TRIM52-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | SETBP1 | Sponge network | -0.418 | 0.00068 | -1.302 | 1.0E-5 | 0.289 |
| 43735 | TINCR |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-940 | 36 | WDFY3 | Sponge network | -0.664 | 0.14543 | -0.201 | 0.0967 | 0.289 |
| 43736 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 17 | APC | Sponge network | -0.176 | 0.59692 | -0.255 | 0.04051 | 0.289 |
| 43737 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | LIFR | Sponge network | -0.342 | 0.02288 | -1.794 | 0 | 0.289 |
| 43738 | BDNF-AS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DCN | Sponge network | -0.668 | 3.0E-5 | -1.288 | 0 | 0.289 |
| 43739 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-93-3p | 21 | NCOA1 | Sponge network | -0.154 | 0.53954 | -0.432 | 0 | 0.289 |
| 43740 | LINC00963 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-590-3p | 10 | PXDN | Sponge network | 0.523 | 9.0E-5 | 1.038 | 0 | 0.289 |
| 43741 | MIR143HG |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p | 34 | ABL2 | Sponge network | -0.739 | 0.0574 | 0.82 | 0 | 0.289 |
| 43742 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-532-5p;hsa-miR-940 | 10 | BHLHE41 | Sponge network | 0.043 | 0.81628 | 0.353 | 0.12753 | 0.289 |
| 43743 | RP11-760H22.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p | 24 | ADARB1 | Sponge network | 0.001 | 0.99753 | -0.335 | 0.06631 | 0.289 |
| 43744 | ZNF32-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-378a-5p;hsa-miR-576-5p | 11 | CREB1 | Sponge network | 2.413 | 1.0E-5 | 0.203 | 0.00537 | 0.289 |
| 43745 | RP11-67L2.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-5p | 12 | UVRAG | Sponge network | 0.72 | 0.0001 | -0.017 | 0.83865 | 0.289 |
| 43746 | LINC00473 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | CITED2 | Sponge network | -1.203 | 0.03881 | -1.545 | 0 | 0.289 |
| 43747 | AF131215.2 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 12 | GABRB2 | Sponge network | -0.176 | 0.59692 | -1.841 | 0.00029 | 0.289 |
| 43748 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | ZNF521 | Sponge network | -0.125 | 0.49414 | -0.111 | 0.58068 | 0.289 |
| 43749 | RP11-166D19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | HECA | Sponge network | -0.576 | 0.10121 | -0.217 | 0.03463 | 0.289 |
| 43750 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 25 | PRKAA2 | Sponge network | -0.72 | 0.24239 | -2.462 | 0 | 0.289 |
| 43751 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-539-5p | 12 | PDZD2 | Sponge network | 0.219 | 0.1516 | -0.909 | 3.0E-5 | 0.289 |
| 43752 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-577 | 18 | PGR | Sponge network | 0.853 | 0.15352 | -1.704 | 0 | 0.289 |
| 43753 | MALAT1 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | PRDM2 | Sponge network | 0.782 | 0.00016 | -0.258 | 0.00721 | 0.289 |
| 43754 | RP11-981G7.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-484;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-744-3p | 27 | DST | Sponge network | 0.986 | 0.01022 | -0.713 | 0.00096 | 0.289 |
| 43755 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 50 | ERBB4 | Sponge network | -0.129 | 0.57177 | -1.975 | 2.0E-5 | 0.289 |
| 43756 | SNX29P2 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-335-5p | 11 | PLXNC1 | Sponge network | 0.4 | 0.14611 | 0.569 | 0.00329 | 0.289 |
| 43757 | RP11-403I13.8 |
hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | PGR | Sponge network | 0.619 | 0.00157 | -1.704 | 0 | 0.289 |
| 43758 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-93-5p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | 0.082 | 0.55654 | -0.085 | 0.42389 | 0.289 |
| 43759 | RP11-474O21.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-30b-5p;hsa-miR-30d-5p | 10 | DMD | Sponge network | -0.023 | 0.95109 | -1.506 | 0 | 0.289 |
| 43760 | GAS6-AS2 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | EYA4 | Sponge network | 0.16 | 0.50785 | 0.3 | 0.36876 | 0.289 |
| 43761 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | NEDD4 | Sponge network | 0.421 | 0.00084 | 0.02 | 0.89834 | 0.289 |
| 43762 | LINC00680 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-625-5p | 16 | ZMYM2 | Sponge network | 0.884 | 0 | 0.279 | 0.01273 | 0.289 |
| 43763 | LINC00667 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 10 | PCLO | Sponge network | -0.235 | 0.15142 | -1.843 | 0.00064 | 0.289 |
| 43764 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | MYO9A | Sponge network | 0.488 | 0.00084 | -0.147 | 0.32373 | 0.289 |
| 43765 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | TET1 | Sponge network | 0.683 | 1.0E-5 | -0.135 | 0.52138 | 0.289 |
| 43766 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 34 | RBMS3 | Sponge network | 0.619 | 0.00157 | -0.519 | 0.03551 | 0.289 |
| 43767 | RP11-156E6.1 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 13 | CDK6 | Sponge network | 1.266 | 0 | 0.991 | 0 | 0.289 |
| 43768 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | FOXO3 | Sponge network | 0.194 | 0.64278 | -0.04 | 0.72028 | 0.289 |
| 43769 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 10 | NALCN | Sponge network | -0.709 | 0.18168 | 1.068 | 0.00237 | 0.289 |
| 43770 | OIP5-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p | 27 | NRK | Sponge network | 0.421 | 0.00084 | 0.656 | 0.19217 | 0.289 |
| 43771 | RP11-77H9.2 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p | 11 | DDX6 | Sponge network | 1.677 | 0 | 0.091 | 0.36428 | 0.289 |
| 43772 | AC004893.11 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | DICER1 | Sponge network | 1.317 | 0 | 0.042 | 0.674 | 0.289 |
| 43773 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-582-5p | 15 | PIK3C2A | Sponge network | 1.206 | 0 | 0.061 | 0.53776 | 0.289 |
| 43774 | RP11-890B15.3 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ID4 | Sponge network | 0.101 | 0.60919 | -1.644 | 0 | 0.289 |
| 43775 | WDFY3-AS2 |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-940 | 16 | JUN | Sponge network | -0.942 | 0 | -0.932 | 0 | 0.289 |
| 43776 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-493-5p | 10 | SYNE1 | Sponge network | 1.46 | 1.0E-5 | -0.738 | 0.00316 | 0.289 |
| 43777 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-34a-5p;hsa-miR-34c-5p;hsa-miR-424-5p;hsa-miR-766-3p | 28 | MAPK1 | Sponge network | 0.078 | 0.55803 | -0.017 | 0.81465 | 0.289 |
| 43778 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | TACC1 | Sponge network | 1.153 | 0.00114 | -0.362 | 0.05406 | 0.289 |
| 43779 | RP11-686D22.8 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | ARID5B | Sponge network | 0.898 | 1.0E-5 | 0.342 | 0.01676 | 0.289 |
| 43780 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | GRIN2A | Sponge network | -0.235 | 0.15142 | -2.161 | 0 | 0.289 |
| 43781 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | 0.335 | 0.20596 | -0.966 | 0.00035 | 0.289 |
| 43782 | RP11-53O19.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-450b-5p | 10 | SPTBN4 | Sponge network | -0.405 | 0.22013 | -0.786 | 0.01281 | 0.289 |
| 43783 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 15 | CEP170 | Sponge network | 0.064 | 0.72934 | 0.943 | 0 | 0.289 |
| 43784 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 11 | DMXL1 | Sponge network | 1.2 | 0.01813 | -0.426 | 0.00056 | 0.289 |
| 43785 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 12 | EBF1 | Sponge network | 0.476 | 0.19203 | -0.384 | 0.08856 | 0.289 |
| 43786 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-199b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 18 | SMAD4 | Sponge network | 0.673 | 0 | -0.313 | 0.00413 | 0.289 |
| 43787 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | EPHA5 | Sponge network | 0.61 | 0.01593 | -0.957 | 0.01733 | 0.289 |
| 43788 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-511-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ADAMTSL3 | Sponge network | 0.392 | 0.0278 | -1.559 | 0 | 0.289 |
| 43789 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-429 | 11 | FOXP1 | Sponge network | 1.106 | 0 | 0.042 | 0.74368 | 0.289 |
| 43790 | RP3-402G11.26 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-590-3p | 19 | THRB | Sponge network | -0.254 | 0.19303 | -1.117 | 0.0004 | 0.289 |
| 43791 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-93-5p | 26 | DST | Sponge network | -0.663 | 0.10887 | -0.713 | 0.00096 | 0.289 |
| 43792 | RP11-244H3.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-5p | 13 | PTGFR | Sponge network | 0.659 | 3.0E-5 | -0.233 | 0.45421 | 0.289 |
| 43793 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484 | 16 | RET | Sponge network | -4.477 | 0 | -0.833 | 0.03517 | 0.289 |
| 43794 | RP11-760H22.2 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-454-3p | 10 | ZNF844 | Sponge network | 0.001 | 0.99753 | -0.966 | 0.00035 | 0.289 |
| 43795 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 51 | AFF4 | Sponge network | 0.75 | 0.00054 | -0.266 | 0.01565 | 0.289 |
| 43796 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 52 | BMPR2 | Sponge network | -2.117 | 0.00161 | 0.206 | 0.0635 | 0.289 |
| 43797 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p | 13 | UBE4B | Sponge network | -0.384 | 0.40652 | -0.235 | 0.007 | 0.289 |
| 43798 | LINC00476 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | THSD7B | Sponge network | -0.615 | 0.00015 | 0.154 | 0.74723 | 0.289 |
| 43799 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 11 | FYN | Sponge network | -0.384 | 0.40652 | -0.131 | 0.37051 | 0.289 |
| 43800 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | MED12L | Sponge network | 0.997 | 0.00066 | -0.194 | 0.55299 | 0.289 |
| 43801 | RP11-195F19.9 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 12 | SYT4 | Sponge network | -0.726 | 0.00132 | -3.207 | 0 | 0.289 |
| 43802 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 0.659 | 3.0E-5 | 0.114 | 0.30638 | 0.289 |
| 43803 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-5p | 37 | ERBB4 | Sponge network | -0.773 | 0.04073 | -1.975 | 2.0E-5 | 0.289 |
| 43804 | SOX2-OT |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NRG3 | Sponge network | -1.264 | 6.0E-5 | -1.836 | 4.0E-5 | 0.289 |
| 43805 | RP11-251G23.5 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-664a-3p | 24 | CCDC6 | Sponge network | 1.344 | 0 | 0.27 | 0.02555 | 0.289 |
| 43806 | RP11-428J1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-429 | 10 | LHX6 | Sponge network | 0.538 | 0.00076 | -0.087 | 0.69193 | 0.289 |
| 43807 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 14 | GPAM | Sponge network | 0.078 | 0.55803 | 0.142 | 0.29732 | 0.289 |
| 43808 | DNM3OS |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 15 | PICALM | Sponge network | 0.572 | 0.01722 | 0.086 | 0.23523 | 0.289 |
| 43809 | PART1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | DACT1 | Sponge network | -2.925 | 0 | 0.783 | 0.00072 | 0.289 |
| 43810 | RP11-535M15.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-942-5p | 12 | TMEM132B | Sponge network | -1.14 | 0.03513 | -0.83 | 0.01084 | 0.289 |
| 43811 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-625-5p | 20 | CSMD1 | Sponge network | -2.69 | 0.0001 | -0.63 | 0.18881 | 0.289 |
| 43812 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-7-5p | 25 | BTG2 | Sponge network | 0.246 | 0.59912 | -1.763 | 0 | 0.289 |
| 43813 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p | 12 | PCDH8 | Sponge network | 0.335 | 0.20596 | -0.997 | 0.05366 | 0.289 |
| 43814 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 22 | HBP1 | Sponge network | -0.053 | 0.80685 | -0.454 | 0 | 0.289 |
| 43815 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 11 | CREM | Sponge network | -0.899 | 6.0E-5 | -0.345 | 0.00258 | 0.289 |
| 43816 | HAND2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 27 | RIMBP2 | Sponge network | -1.697 | 0.00889 | -1.968 | 5.0E-5 | 0.289 |
| 43817 | LINC00662 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | BRD3 | Sponge network | 0.213 | 0.32297 | 0.37 | 0.00013 | 0.289 |
| 43818 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TGFBR3 | Sponge network | 0.252 | 0.08508 | -1.173 | 1.0E-5 | 0.289 |
| 43819 | AF131215.9 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | RPS6KA6 | Sponge network | -0.322 | 0.25945 | -2.989 | 0 | 0.289 |
| 43820 | XXbac-B444P24.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | FAT4 | Sponge network | 2.264 | 0.0012 | -0.354 | 0.14694 | 0.289 |
| 43821 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-532-3p | 14 | PBX1 | Sponge network | -0.557 | 0.11135 | -1.206 | 0 | 0.289 |
| 43822 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SEMA3E | Sponge network | 0.222 | 0.29133 | -1.717 | 0.00137 | 0.289 |
| 43823 | GS1-124K5.11 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p | 29 | RIMS2 | Sponge network | 0.494 | 0.00339 | -1.365 | 0.00208 | 0.289 |
| 43824 | FENDRR |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | EFNA5 | Sponge network | -1.281 | 0.00012 | -0.421 | 0.17868 | 0.289 |
| 43825 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-362-3p | 10 | BCL6 | Sponge network | -1.255 | 0.00981 | -0.211 | 0.19489 | 0.289 |
| 43826 | AC025171.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-590-3p | 12 | NRK | Sponge network | 0.998 | 0 | 0.656 | 0.19217 | 0.289 |
| 43827 | DIO3OS |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-532-5p;hsa-miR-671-5p | 10 | PIK3IP1 | Sponge network | -0.773 | 0.04073 | -0.187 | 0.19945 | 0.289 |
| 43828 | BDNF-AS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-96-5p | 30 | IL17RD | Sponge network | -0.668 | 3.0E-5 | -0.019 | 0.94873 | 0.289 |
| 43829 | RP5-1198O20.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | PDE4DIP | Sponge network | 0.997 | 0.00066 | -0.282 | 0.0257 | 0.289 |
| 43830 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-29a-5p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-577 | 14 | SLC6A15 | Sponge network | -0.18 | 0.20343 | -2.373 | 2.0E-5 | 0.289 |
| 43831 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-744-3p | 19 | HIP1 | Sponge network | -0.223 | 0.47816 | 0.774 | 0 | 0.289 |
| 43832 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-93-5p | 17 | IGSF10 | Sponge network | -1.047 | 0 | -1.751 | 1.0E-5 | 0.289 |
| 43833 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 39 | EPHA5 | Sponge network | 0.75 | 0.00054 | -0.957 | 0.01733 | 0.289 |
| 43834 | CTC-296K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-493-5p | 23 | NCOA1 | Sponge network | -2.095 | 0.00112 | -0.432 | 0 | 0.289 |
| 43835 | AC141928.1 |
hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-625-5p | 12 | LHX6 | Sponge network | 0.853 | 0.15352 | -0.087 | 0.69193 | 0.289 |
| 43836 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 34 | BNC2 | Sponge network | 0.659 | 3.0E-5 | -0.491 | 0.09988 | 0.289 |
| 43837 | AC093627.8 |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-766-3p | 15 | IL6ST | Sponge network | -0.787 | 0.3928 | -0.402 | 0.00676 | 0.289 |
| 43838 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | LRP1B | Sponge network | -0.351 | 0.11358 | -2.163 | 0 | 0.289 |
| 43839 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 29 | ARHGAP28 | Sponge network | -1.697 | 0.00889 | -0.911 | 0.00013 | 0.289 |
| 43840 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 20 | PRKAR1A | Sponge network | 0.064 | 0.72934 | -0.063 | 0.50314 | 0.289 |
| 43841 | U91328.19 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-424-5p | 17 | PPP2R5C | Sponge network | -0.303 | 0.21002 | -0.314 | 3.0E-5 | 0.289 |
| 43842 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 25 | TACC1 | Sponge network | 0.197 | 0.25618 | -0.362 | 0.05406 | 0.289 |
| 43843 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-505-3p | 21 | USP6 | Sponge network | 0.41 | 0.02214 | 0.188 | 0.3871 | 0.289 |
| 43844 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-450b-5p;hsa-miR-539-5p | 16 | CDHR3 | Sponge network | 0.331 | 0.57247 | -0.977 | 4.0E-5 | 0.289 |
| 43845 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p | 12 | CHD1 | Sponge network | 1.03 | 0 | 0.322 | 0.00215 | 0.289 |
| 43846 | ZNF582-AS1 |
hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-423-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 11 | CELF4 | Sponge network | -1.349 | 0.00017 | -1.135 | 0.00344 | 0.289 |
| 43847 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-590-3p;hsa-miR-744-3p | 10 | CUL5 | Sponge network | 0.178 | 0.29106 | 0.026 | 0.77301 | 0.289 |
| 43848 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 0.349 | 0.01695 | 0.322 | 0.00215 | 0.289 |
| 43849 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 12 | NEDD4 | Sponge network | -0.726 | 0.00132 | 0.02 | 0.89834 | 0.289 |
| 43850 | SERHL |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-501-5p | 25 | NTRK2 | Sponge network | -0.602 | 0.00331 | -0.736 | 0.06495 | 0.289 |
| 43851 | EMX2OS |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 37 | AFF1 | Sponge network | -0.777 | 0.1134 | -0.384 | 0.00053 | 0.289 |
| 43852 | RP11-25K21.6 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p | 11 | WDFY3 | Sponge network | 0.489 | 0.25786 | -0.201 | 0.0967 | 0.289 |
| 43853 | RP1-151F17.2 |
hsa-miR-10a-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 16 | GLI3 | Sponge network | 0.222 | 0.29133 | -0.615 | 0.01482 | 0.289 |
| 43854 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 17 | PICALM | Sponge network | 0.101 | 0.60919 | 0.086 | 0.23523 | 0.289 |
| 43855 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-93-3p | 13 | CUX1 | Sponge network | 1.46 | 1.0E-5 | -0.039 | 0.6705 | 0.289 |
| 43856 | LINC00473 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 35 | TRPS1 | Sponge network | -1.203 | 0.03881 | -0.191 | 0.44109 | 0.289 |
| 43857 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p | 14 | FAT4 | Sponge network | -0.176 | 0.59692 | -0.354 | 0.14694 | 0.289 |
| 43858 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | PRUNE2 | Sponge network | 0.727 | 3.0E-5 | -1.733 | 0.00022 | 0.289 |
| 43859 | RP11-822E23.8 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-7-5p;hsa-miR-93-3p | 30 | ERC1 | Sponge network | -0.777 | 0.16667 | -0.108 | 0.38794 | 0.289 |
| 43860 | MALAT1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | NIN | Sponge network | 0.782 | 0.00016 | -0.253 | 0.13794 | 0.289 |
| 43861 | H1FX-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p | 11 | IGF2 | Sponge network | -0.206 | 0.12942 | 0.848 | 0.03842 | 0.289 |
| 43862 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | HBP1 | Sponge network | -0.709 | 0.18168 | -0.454 | 0 | 0.289 |
| 43863 | LINC00654 |
hsa-let-7b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484 | 22 | ABL1 | Sponge network | 0.374 | 0.20054 | -0.215 | 0.07804 | 0.289 |
| 43864 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | CXXC4 | Sponge network | 0.349 | 0.01695 | -0.433 | 0.21055 | 0.289 |
| 43865 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | EPHA5 | Sponge network | -0.053 | 0.80685 | -0.957 | 0.01733 | 0.289 |
| 43866 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-589-5p | 20 | CXXC4 | Sponge network | -0.162 | 0.47687 | -0.433 | 0.21055 | 0.289 |
| 43867 | RP11-157J24.2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-589-5p | 19 | PRKAA2 | Sponge network | -1.664 | 0.05165 | -2.462 | 0 | 0.289 |
| 43868 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 15 | LRRC55 | Sponge network | 0.659 | 3.0E-5 | -0.026 | 0.93163 | 0.289 |
| 43869 | HOXA-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-877-5p | 26 | ASTN1 | Sponge network | 0.61 | 0.01593 | -2.389 | 2.0E-5 | 0.289 |
| 43870 | RP11-474O21.5 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | WDFY3 | Sponge network | -0.023 | 0.95109 | -0.201 | 0.0967 | 0.289 |
| 43871 | FAM13A-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-942-5p | 15 | ERRFI1 | Sponge network | -0.83 | 0 | -0.798 | 1.0E-5 | 0.289 |
| 43872 | ARHGEF26-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p | 12 | PIK3IP1 | Sponge network | -2.46 | 0 | -0.187 | 0.19945 | 0.289 |
| 43873 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-362-3p | 17 | HBP1 | Sponge network | -0.693 | 0.05629 | -0.454 | 0 | 0.289 |
| 43874 | RP1-151F17.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 23 | NRK | Sponge network | 0.222 | 0.29133 | 0.656 | 0.19217 | 0.289 |
| 43875 | RP11-13K12.5 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 13 | DOCK4 | Sponge network | -0.318 | 0.65847 | 0.502 | 0.00108 | 0.289 |
| 43876 | ADIRF-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-766-3p | 13 | EZH1 | Sponge network | -0.223 | 0.47816 | -0.564 | 1.0E-5 | 0.289 |
| 43877 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 30 | WDFY3 | Sponge network | 0.663 | 0.00073 | -0.201 | 0.0967 | 0.289 |
| 43878 | RP11-867G23.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-93-3p | 16 | NCOA1 | Sponge network | -2.531 | 0 | -0.432 | 0 | 0.289 |
| 43879 | PWAR6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CEP170 | Sponge network | -1.575 | 6.0E-5 | 0.943 | 0 | 0.289 |
| 43880 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 38 | ZFHX3 | Sponge network | -0.727 | 0.04726 | 0.389 | 0.01363 | 0.289 |
| 43881 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-33a-5p | 21 | ABL2 | Sponge network | -0.384 | 0.40652 | 0.82 | 0 | 0.289 |
| 43882 | HOXD-AS2 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-500a-5p | 19 | GRIK3 | Sponge network | -0.208 | 0.65592 | -3.208 | 0 | 0.289 |
| 43883 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | BRD3 | Sponge network | 1.316 | 0 | 0.37 | 0.00013 | 0.289 |
| 43884 | CTA-445C9.14 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-342-3p | 10 | ABI2 | Sponge network | 0.718 | 4.0E-5 | 0.175 | 0.14787 | 0.289 |
| 43885 | CTC-559E9.5 |
hsa-let-7d-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p | 11 | DICER1 | Sponge network | 0.594 | 0.00064 | 0.042 | 0.674 | 0.289 |
| 43886 | RP4-798A10.7 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p | 11 | PRKAA1 | Sponge network | 1.757 | 0 | 0.317 | 0.0031 | 0.289 |
| 43887 | TPT1-AS1 |
hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-664a-3p | 10 | NUAK1 | Sponge network | 0.903 | 0 | 0.904 | 0 | 0.289 |
| 43888 | AC103563.9 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-455-5p;hsa-miR-577 | 15 | NTRK3 | Sponge network | -6.057 | 0 | -1.77 | 3.0E-5 | 0.289 |
| 43889 | CROCCP2 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p | 10 | DICER1 | Sponge network | 0.79 | 0 | 0.042 | 0.674 | 0.289 |
| 43890 | RP11-404E16.1 |
hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-370-3p;hsa-miR-424-5p | 15 | SSBP2 | Sponge network | 0.097 | 0.91311 | -1.58 | 0 | 0.289 |
| 43891 | SNX29P2 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 11 | CNN1 | Sponge network | 0.4 | 0.14611 | -2.633 | 0 | 0.289 |
| 43892 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-582-3p | 14 | FOXO3 | Sponge network | 1.4 | 0 | -0.04 | 0.72028 | 0.289 |
| 43893 | RP11-48B3.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 13 | PRUNE2 | Sponge network | 1.757 | 0 | -1.733 | 0.00022 | 0.289 |
| 43894 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | FGFR1 | Sponge network | -0.295 | 0.02604 | -0.509 | 0.03957 | 0.289 |
| 43895 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | SOX5 | Sponge network | 0.862 | 0.06213 | -1.091 | 4.0E-5 | 0.289 |
| 43896 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | ZFHX4 | Sponge network | 0.687 | 0.02753 | 0.018 | 0.95574 | 0.289 |
| 43897 | LINC00472 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 35 | AFF1 | Sponge network | -0.842 | 0.01361 | -0.384 | 0.00053 | 0.289 |
| 43898 | SNHG14 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-940 | 71 | NFIB | Sponge network | -1.111 | 0.0003 | 0.023 | 0.90795 | 0.289 |
| 43899 | RP11-59H7.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p | 19 | NFIB | Sponge network | 2.163 | 0 | 0.023 | 0.90795 | 0.289 |
| 43900 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 10 | AKAP12 | Sponge network | 0.95 | 0.15943 | -0.932 | 0.0028 | 0.289 |
| 43901 | CTA-204B4.2 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 15 | DICER1 | Sponge network | 0.765 | 7.0E-5 | 0.042 | 0.674 | 0.289 |
| 43902 | RP11-48B3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ARNT2 | Sponge network | 1.757 | 0 | -0.332 | 0.25851 | 0.289 |
| 43903 | HCG11 |
hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | LRRC55 | Sponge network | 0.385 | 0.10067 | -0.026 | 0.93163 | 0.289 |
| 43904 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-511-5p;hsa-miR-940 | 11 | TRIM33 | Sponge network | 0.453 | 0.01839 | 0.402 | 7.0E-5 | 0.289 |
| 43905 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | PHC3 | Sponge network | 0.41 | 0.02214 | 0.212 | 0.0499 | 0.289 |
| 43906 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | FLI1 | Sponge network | 0.194 | 0.64278 | -0.294 | 0.10905 | 0.289 |
| 43907 | HOXB-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p | 23 | AFF4 | Sponge network | 1.316 | 0.01106 | -0.266 | 0.01565 | 0.289 |
| 43908 | RP11-25K21.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p | 12 | RUNX1T1 | Sponge network | 0.489 | 0.25786 | -0.344 | 0.2374 | 0.289 |
| 43909 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p | 12 | RBM5 | Sponge network | -2.62 | 0 | -0.205 | 0.0129 | 0.289 |
| 43910 | HHIP-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-455-3p;hsa-miR-590-3p | 10 | TLE4 | Sponge network | -0.737 | 0.10822 | -1.157 | 0 | 0.289 |
| 43911 | MIR600HG |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p | 15 | UBR3 | Sponge network | 0.594 | 0.41522 | -0.021 | 0.82774 | 0.289 |
| 43912 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | TRIP11 | Sponge network | 0.75 | 0.00054 | -0.158 | 0.06384 | 0.289 |
| 43913 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-7-1-3p | 10 | AHNAK | Sponge network | -4.477 | 0 | -1.207 | 0 | 0.289 |
| 43914 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p | 10 | CUL5 | Sponge network | 0.643 | 0 | 0.026 | 0.77301 | 0.289 |
| 43915 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-532-5p | 17 | NUAK1 | Sponge network | 0.178 | 0.29106 | 0.904 | 0 | 0.289 |
| 43916 | LINC00963 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 12 | INPP4A | Sponge network | 0.523 | 9.0E-5 | 0.07 | 0.53563 | 0.289 |
| 43917 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ABCA10 | Sponge network | -0.769 | 0.00035 | -0.571 | 0.03674 | 0.289 |
| 43918 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-590-3p | 21 | FBXO32 | Sponge network | -0.737 | 0.10822 | -0.495 | 0.07561 | 0.289 |
| 43919 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-7-1-3p | 24 | KCNA6 | Sponge network | -0.842 | 0.01361 | -1.531 | 0.00013 | 0.289 |
| 43920 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | TCF4 | Sponge network | -0.053 | 0.80685 | 0.016 | 0.92271 | 0.289 |
| 43921 | RP11-356J5.12 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 18 | ATM | Sponge network | -0.899 | 6.0E-5 | 0.178 | 0.21568 | 0.289 |
| 43922 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p | 57 | BMPR2 | Sponge network | -0.793 | 7.0E-5 | 0.206 | 0.0635 | 0.289 |
| 43923 | MIR22HG |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-539-5p | 11 | DDX5 | Sponge network | -1.047 | 0 | -0.099 | 0.17428 | 0.289 |
| 43924 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | PRKAB2 | Sponge network | 1.317 | 0 | -0.367 | 0.01371 | 0.289 |
| 43925 | H1FX-AS1 |
hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | RIMS2 | Sponge network | -0.206 | 0.12942 | -1.365 | 0.00208 | 0.289 |
| 43926 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-93-5p | 11 | DAB2 | Sponge network | -0.842 | 0.01361 | 0.431 | 0.0113 | 0.289 |
| 43927 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | LATS1 | Sponge network | 0.194 | 0.64278 | 0.109 | 0.34208 | 0.289 |
| 43928 | RP11-999E24.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3614-5p | 25 | CREB3L2 | Sponge network | -0.693 | 0.05629 | -0.525 | 2.0E-5 | 0.289 |
| 43929 | RP11-686D22.8 |
hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 10 | DACT1 | Sponge network | 0.898 | 1.0E-5 | 0.783 | 0.00072 | 0.289 |
| 43930 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | TRIP11 | Sponge network | 0.267 | 0.2937 | -0.158 | 0.06384 | 0.289 |
| 43931 | DIO3OS |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-93-3p;hsa-miR-940 | 38 | GRIK3 | Sponge network | -0.773 | 0.04073 | -3.208 | 0 | 0.289 |
| 43932 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | EPHA5 | Sponge network | 1.253 | 0 | -0.957 | 0.01733 | 0.289 |
| 43933 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-942-5p | 33 | TMEM132B | Sponge network | -0.777 | 0.16667 | -0.83 | 0.01084 | 0.289 |
| 43934 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-500a-3p;hsa-miR-590-3p | 17 | ARHGAP29 | Sponge network | 0.494 | 0.00339 | 0.131 | 0.42953 | 0.289 |
| 43935 | RP11-554A11.4 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p | 12 | MNT | Sponge network | -0.583 | 0.14589 | -0.157 | 0.06598 | 0.289 |
| 43936 | LINC00403 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-424-5p | 12 | CBFA2T3 | Sponge network | -3.586 | 0.00032 | -1.221 | 1.0E-5 | 0.289 |
| 43937 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 14 | DMXL1 | Sponge network | 0.453 | 0.01839 | -0.426 | 0.00056 | 0.289 |
| 43938 | U91328.19 |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 14 | PHOX2B | Sponge network | -0.303 | 0.21002 | -2.68 | 0 | 0.289 |
| 43939 | HAS2-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p | 12 | ASTN1 | Sponge network | -0.011 | 0.967 | -2.389 | 2.0E-5 | 0.289 |
| 43940 | RP11-426C22.5 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | TCF4 | Sponge network | 0.749 | 0 | 0.016 | 0.92271 | 0.289 |
| 43941 | CTC-550B14.7 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-365a-3p;hsa-miR-374a-3p | 10 | ARHGEF12 | Sponge network | 1.161 | 0 | 0.09 | 0.35693 | 0.289 |
| 43942 | RP5-1085F17.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 18 | RASSF8 | Sponge network | 0.541 | 0.00125 | -0.122 | 0.65591 | 0.289 |
| 43943 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p | 17 | ABL2 | Sponge network | 0.176 | 0.37402 | 0.82 | 0 | 0.289 |
| 43944 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 28 | TMEM132B | Sponge network | 0.299 | 0.57722 | -0.83 | 0.01084 | 0.289 |
| 43945 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p | 10 | DACH2 | Sponge network | -1.092 | 4.0E-5 | -1.635 | 0.00034 | 0.289 |
| 43946 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-93-5p | 24 | BHLHE41 | Sponge network | -2.69 | 0.0001 | 0.353 | 0.12753 | 0.289 |
| 43947 | RP4-714D9.5 |
hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-497-5p | 10 | UBR3 | Sponge network | 1.015 | 0 | -0.021 | 0.82774 | 0.289 |
| 43948 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | PDGFRA | Sponge network | -2.46 | 0 | -0.372 | 0.0037 | 0.289 |
| 43949 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | ATRX | Sponge network | -0.342 | 0.02288 | 0.261 | 0.04408 | 0.289 |
| 43950 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 46 | DST | Sponge network | 0.199 | 0.38015 | -0.713 | 0.00096 | 0.289 |
| 43951 | RP11-890B15.3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 13 | EHD3 | Sponge network | 0.101 | 0.60919 | -0.484 | 0.01747 | 0.289 |
| 43952 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 33 | KIF1B | Sponge network | 0.919 | 0 | -0.118 | 0.32258 | 0.289 |
| 43953 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p | 19 | NTRK2 | Sponge network | 0.238 | 0.70544 | -0.736 | 0.06495 | 0.289 |
| 43954 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 25 | EPHA3 | Sponge network | 0.683 | 1.0E-5 | 0.185 | 0.53588 | 0.289 |
| 43955 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p | 11 | FREM2 | Sponge network | -0.932 | 0.00329 | -0.437 | 0.46353 | 0.289 |
| 43956 | RP11-981G7.6 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 27 | ARHGEF12 | Sponge network | 0.986 | 0.01022 | 0.09 | 0.35693 | 0.289 |
| 43957 | CTC-429P9.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-940 | 11 | WDFY3 | Sponge network | 0.043 | 0.81628 | -0.201 | 0.0967 | 0.289 |
| 43958 | RP13-516M14.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | CSMD1 | Sponge network | 0.389 | 0.04293 | -0.63 | 0.18881 | 0.289 |
| 43959 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | FOXO1 | Sponge network | -0.358 | 0.17255 | -0.141 | 0.33874 | 0.289 |
| 43960 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-653-5p | 18 | DMXL1 | Sponge network | 1.254 | 0 | -0.426 | 0.00056 | 0.289 |
| 43961 | LINC00578 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-589-3p | 19 | AFF1 | Sponge network | 0.811 | 0.03228 | -0.384 | 0.00053 | 0.289 |
| 43962 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-30b-3p;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | AR | Sponge network | -0.011 | 0.967 | -1.554 | 0 | 0.289 |
| 43963 | PSMG3-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | ROBO1 | Sponge network | -0.125 | 0.49414 | -0.009 | 0.96154 | 0.289 |
| 43964 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | ARHGAP29 | Sponge network | -0.896 | 0.00099 | 0.131 | 0.42953 | 0.289 |
| 43965 | RP11-35G9.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 26 | TRPS1 | Sponge network | 0.657 | 4.0E-5 | -0.191 | 0.44109 | 0.289 |
| 43966 | LINC01018 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 24 | PTPN14 | Sponge network | -2.915 | 0.00015 | 0.144 | 0.34595 | 0.289 |
| 43967 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 30 | MAF | Sponge network | -0.793 | 7.0E-5 | -1.257 | 0 | 0.289 |
| 43968 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-532-5p;hsa-miR-629-5p | 17 | BMPR2 | Sponge network | 0.065 | 0.73468 | 0.206 | 0.0635 | 0.289 |
| 43969 | FZD10-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 24 | JDP2 | Sponge network | -0.727 | 0.04726 | -0.967 | 0 | 0.289 |
| 43970 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935 | 32 | MTSS1 | Sponge network | -0.615 | 0.00015 | -0.81 | 0.00035 | 0.289 |
| 43971 | CTC-296K1.4 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-590-5p | 18 | PPM1A | Sponge network | -2.076 | 0.00548 | -0.623 | 0 | 0.289 |
| 43972 | DNM1P35 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-93-5p | 44 | DST | Sponge network | 0.051 | 0.83388 | -0.713 | 0.00096 | 0.289 |
| 43973 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p | 12 | MYCBP2 | Sponge network | 0.659 | 3.0E-5 | -0.027 | 0.82016 | 0.289 |
| 43974 | RP5-894A10.6 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-485-3p | 13 | PIK3C2A | Sponge network | 0.697 | 9.0E-5 | 0.061 | 0.53776 | 0.289 |
| 43975 | BZRAP1-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 27 | IGFBP5 | Sponge network | -0.465 | 0.04703 | -0.806 | 0.00105 | 0.289 |
| 43976 | RP11-277P12.20 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 14 | IKZF2 | Sponge network | 0.687 | 0.02753 | -0.284 | 0.1942 | 0.289 |
| 43977 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3614-5p;hsa-miR-484 | 14 | GREM1 | Sponge network | 0.862 | 0.06213 | 0.902 | 0.01691 | 0.289 |
| 43978 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p | 11 | PCDH8 | Sponge network | -0.727 | 0.04726 | -0.997 | 0.05366 | 0.289 |
| 43979 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | FGF5 | Sponge network | -1.161 | 0.00591 | 0.549 | 0.28659 | 0.289 |
| 43980 | RP11-327P2.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-935 | 28 | DIP2C | Sponge network | -0.147 | 0.40452 | -0.436 | 0.01713 | 0.289 |
| 43981 | ARHGEF26-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p;hsa-miR-98-5p | 49 | CREB3L2 | Sponge network | -2.46 | 0 | -0.525 | 2.0E-5 | 0.289 |
| 43982 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 49 | PPM1A | Sponge network | -1.216 | 0 | -0.623 | 0 | 0.289 |
| 43983 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-550a-3p | 11 | MEF2C | Sponge network | 2.364 | 0.00134 | -0.499 | 0.01448 | 0.289 |
| 43984 | CTC-457L16.2 |
hsa-miR-130b-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-7-1-3p | 10 | AKAP6 | Sponge network | 1.153 | 0.00114 | -1.586 | 3.0E-5 | 0.289 |
| 43985 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | PTPRB | Sponge network | -0.769 | 0.00035 | 0.105 | 0.47122 | 0.289 |
| 43986 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-590-5p | 15 | ATRX | Sponge network | 0.706 | 0 | 0.261 | 0.04408 | 0.289 |
| 43987 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-766-3p | 17 | TBL1XR1 | Sponge network | 0.559 | 0.00026 | 0.578 | 0 | 0.289 |
| 43988 | TPTEP1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PCLO | Sponge network | -1.216 | 0 | -1.843 | 0.00064 | 0.289 |
| 43989 | CTA-204B4.2 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | PIK3R1 | Sponge network | 0.765 | 7.0E-5 | -0.133 | 0.36854 | 0.289 |
| 43990 | SOX2-OT |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-625-5p | 13 | NGFR | Sponge network | -1.264 | 6.0E-5 | -1.271 | 0.01058 | 0.289 |
| 43991 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 14 | FREM2 | Sponge network | 0.219 | 0.1516 | -0.437 | 0.46353 | 0.289 |
| 43992 | AC116366.6 |
hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-877-5p | 16 | CCND2 | Sponge network | 0.329 | 0.11122 | -0.267 | 0.3112 | 0.289 |
| 43993 | NDUFA6-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 20 | ITGB3 | Sponge network | -0.355 | 0.0077 | -0.011 | 0.95751 | 0.289 |
| 43994 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-590-5p | 10 | FSTL5 | Sponge network | 0.395 | 0.15451 | -1.3 | 0.01619 | 0.289 |
| 43995 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 37 | ZFHX4 | Sponge network | 0.311 | 0.10344 | 0.018 | 0.95574 | 0.289 |
| 43996 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-7-1-3p | 11 | RCAN2 | Sponge network | -0.13 | 0.48734 | -0.33 | 0.15172 | 0.289 |
| 43997 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | KCNA6 | Sponge network | 1.302 | 7.0E-5 | -1.531 | 0.00013 | 0.289 |
| 43998 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-502-5p | 13 | SETBP1 | Sponge network | -0.278 | 0.76559 | -1.302 | 1.0E-5 | 0.289 |
| 43999 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 11 | NIN | Sponge network | -1.582 | 8.0E-5 | -0.253 | 0.13794 | 0.289 |
| 44000 | WDFY3-AS2 |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-542-3p | 12 | CADM4 | Sponge network | -0.942 | 0 | -0.635 | 0.00305 | 0.289 |
| 44001 | ERVK3-1 |
hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374b-5p;hsa-miR-376c-3p | 10 | PAWR | Sponge network | 0.58 | 0 | 0.636 | 0 | 0.289 |
| 44002 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 35 | BNC2 | Sponge network | 0.392 | 0.0278 | -0.491 | 0.09988 | 0.288 |
| 44003 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PRKAB2 | Sponge network | 1.205 | 0 | -0.367 | 0.01371 | 0.288 |
| 44004 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | CDH19 | Sponge network | 1.302 | 7.0E-5 | -3.434 | 0 | 0.288 |
| 44005 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-7-1-3p | 27 | ZFHX3 | Sponge network | -1.349 | 0.00017 | 0.389 | 0.01363 | 0.288 |
| 44006 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-96-5p | 14 | LRIG1 | Sponge network | 0.37 | 0.27614 | -0.537 | 0.00588 | 0.288 |
| 44007 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-454-3p | 22 | AKAP11 | Sponge network | 0.001 | 0.99753 | 0.196 | 0.09825 | 0.288 |
| 44008 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 40 | BMPR2 | Sponge network | -0.421 | 0.0114 | 0.206 | 0.0635 | 0.288 |
| 44009 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | NRXN1 | Sponge network | 0.657 | 4.0E-5 | -3.778 | 0 | 0.288 |
| 44010 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-590-5p | 11 | AKAP12 | Sponge network | -0.371 | 0.24604 | -0.932 | 0.0028 | 0.288 |
| 44011 | RP11-798M19.6 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | LIMCH1 | Sponge network | -0.612 | 0.00094 | -0.915 | 1.0E-5 | 0.288 |
| 44012 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 37 | WDFY3 | Sponge network | 0.614 | 1.0E-5 | -0.201 | 0.0967 | 0.288 |
| 44013 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 19 | ARID5B | Sponge network | 1.153 | 0 | 0.342 | 0.01676 | 0.288 |
| 44014 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p | 28 | KIT | Sponge network | 0.997 | 0.00066 | -2.184 | 0 | 0.288 |
| 44015 | CTC-429P9.3 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-942-5p | 26 | ARHGEF12 | Sponge network | 0.082 | 0.55397 | 0.09 | 0.35693 | 0.288 |
| 44016 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | ZNF770 | Sponge network | 1.308 | 0 | 0.206 | 0.06751 | 0.288 |
| 44017 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-652-3p;hsa-miR-7-1-3p | 41 | QKI | Sponge network | -0.727 | 0.04726 | -0.333 | 0.04111 | 0.288 |
| 44018 | CTD-3092A11.2 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | DDX3X | Sponge network | 0.873 | 0 | -0.152 | 0.07686 | 0.288 |
| 44019 | AC011526.1 |
hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 17 | BCL2 | Sponge network | 0.342 | 0.03129 | -0.899 | 0 | 0.288 |
| 44020 | AC108488.4 |
hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-330-5p | 11 | THRA | Sponge network | 0.259 | 0.12542 | -0.307 | 0.05099 | 0.288 |
| 44021 | RP11-20I23.13 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-93-3p | 14 | EPHA3 | Sponge network | 0.505 | 0.0014 | 0.185 | 0.53588 | 0.288 |
| 44022 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | AFF4 | Sponge network | 1.2 | 0.01813 | -0.266 | 0.01565 | 0.288 |
| 44023 | CTD-2666L21.1 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p | 17 | LRIG1 | Sponge network | 0.368 | 0.20093 | -0.537 | 0.00588 | 0.288 |
| 44024 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-653-5p;hsa-miR-7-1-3p | 31 | EGLN1 | Sponge network | -0.222 | 0.32445 | -0.127 | 0.1536 | 0.288 |
| 44025 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-942-5p | 31 | UNC80 | Sponge network | 0.41 | 0.02214 | -1.934 | 0 | 0.288 |
| 44026 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p | 15 | ABCA10 | Sponge network | -0.738 | 0.21233 | -0.571 | 0.03674 | 0.288 |
| 44027 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | RB1CC1 | Sponge network | -0.053 | 0.80685 | 0.175 | 0.12559 | 0.288 |
| 44028 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-766-3p | 10 | TBL1XR1 | Sponge network | 1.361 | 0 | 0.578 | 0 | 0.288 |
| 44029 | RP11-244O19.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 15 | CELF4 | Sponge network | -0.096 | 0.67958 | -1.135 | 0.00344 | 0.288 |
| 44030 | CTB-113P19.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 10 | CNTNAP3 | Sponge network | 0.906 | 0.31715 | -1.465 | 0.00033 | 0.288 |
| 44031 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 46 | BNC2 | Sponge network | 0.421 | 0.00084 | -0.491 | 0.09988 | 0.288 |
| 44032 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | TACC1 | Sponge network | 1.253 | 0 | -0.362 | 0.05406 | 0.288 |
| 44033 | SOX2-OT |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 22 | DYNC1I1 | Sponge network | -1.264 | 6.0E-5 | -1.12 | 0.00107 | 0.288 |
| 44034 | CALML3-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p | 12 | ROBO1 | Sponge network | 0.95 | 0.15943 | -0.009 | 0.96154 | 0.288 |
| 44035 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 16 | PIK3C2A | Sponge network | 1.073 | 0.00216 | 0.061 | 0.53776 | 0.288 |
| 44036 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p | 19 | CBFA2T3 | Sponge network | -0.154 | 0.78939 | -1.221 | 1.0E-5 | 0.288 |
| 44037 | RP4-613B23.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-877-5p | 14 | PTPRT | Sponge network | -0.309 | 0.1145 | -0.907 | 0.03727 | 0.288 |
| 44038 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | CUL5 | Sponge network | 0.439 | 0.00313 | 0.026 | 0.77301 | 0.288 |
| 44039 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p | 32 | PBX1 | Sponge network | -0.342 | 0.02288 | -1.206 | 0 | 0.288 |
| 44040 | RP4-791M13.3 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-425-5p;hsa-miR-7-1-3p | 17 | LRIG1 | Sponge network | 0.419 | 0.0319 | -0.537 | 0.00588 | 0.288 |
| 44041 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | FOXO3 | Sponge network | 0.997 | 0.00066 | -0.04 | 0.72028 | 0.288 |
| 44042 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-532-3p | 13 | LATS1 | Sponge network | 0.792 | 0.0049 | 0.109 | 0.34208 | 0.288 |
| 44043 | RP11-819C21.1 |
hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 12 | CDK6 | Sponge network | 0.537 | 0.00139 | 0.991 | 0 | 0.288 |
| 44044 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | CNTN1 | Sponge network | 0.853 | 0.15352 | -2.594 | 0 | 0.288 |
| 44045 | KTN1-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 12 | RASAL2 | Sponge network | 0.927 | 0 | 0.869 | 0 | 0.288 |
| 44046 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-96-5p | 15 | NUAK1 | Sponge network | 0.131 | 0.8436 | 0.904 | 0 | 0.288 |
| 44047 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | PAFAH1B1 | Sponge network | 0.683 | 1.0E-5 | -0.429 | 0 | 0.288 |
| 44048 | MYLK-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-671-5p | 14 | PPP2R5C | Sponge network | -1.331 | 3.0E-5 | -0.314 | 3.0E-5 | 0.288 |
| 44049 | RP11-6O2.3 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-539-5p | 13 | PHF3 | Sponge network | 0.331 | 0.57247 | 0.126 | 0.19652 | 0.288 |
| 44050 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 27 | PIK3C2A | Sponge network | -1.575 | 6.0E-5 | 0.061 | 0.53776 | 0.288 |
| 44051 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 36 | ZFHX4 | Sponge network | 0.603 | 0.00127 | 0.018 | 0.95574 | 0.288 |
| 44052 | RP11-244H3.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 35 | TRPS1 | Sponge network | 0.659 | 3.0E-5 | -0.191 | 0.44109 | 0.288 |
| 44053 | H1FX-AS1 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CELF4 | Sponge network | -0.206 | 0.12942 | -1.135 | 0.00344 | 0.288 |
| 44054 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-940 | 25 | PBX1 | Sponge network | 0.594 | 0.41522 | -1.206 | 0 | 0.288 |
| 44055 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p | 11 | ZYX | Sponge network | -0.777 | 0.1134 | 0.019 | 0.89763 | 0.288 |
| 44056 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-590-3p | 12 | ZFHX3 | Sponge network | 0.768 | 7.0E-5 | 0.389 | 0.01363 | 0.288 |
| 44057 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 37 | ERBB4 | Sponge network | 0.056 | 0.81195 | -1.975 | 2.0E-5 | 0.288 |
| 44058 | RP11-327P2.5 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-935 | 31 | DMD | Sponge network | -0.147 | 0.40452 | -1.506 | 0 | 0.288 |
| 44059 | HOTAIRM1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 36 | GAS7 | Sponge network | -0.053 | 0.80685 | -0.555 | 0.01602 | 0.288 |
| 44060 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | LRP1B | Sponge network | -0.021 | 0.93598 | -2.163 | 0 | 0.288 |
| 44061 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 13 | GALNT13 | Sponge network | -0.611 | 0.09607 | -0.857 | 0.07439 | 0.288 |
| 44062 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | RASSF2 | Sponge network | 0.225 | 0.30644 | -0.273 | 0.21961 | 0.288 |
| 44063 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 53 | TCF4 | Sponge network | -0.125 | 0.49414 | 0.016 | 0.92271 | 0.288 |
| 44064 | A1BG-AS1 |
hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-219a-1-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-93-3p | 25 | IL6ST | Sponge network | -0.164 | 0.45106 | -0.402 | 0.00676 | 0.288 |
| 44065 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-223-3p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | CXXC4 | Sponge network | 0.8 | 0 | -0.433 | 0.21055 | 0.288 |
| 44066 | LOXL1-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FREM2 | Sponge network | 0.487 | 0.04053 | -0.437 | 0.46353 | 0.288 |
| 44067 | RP13-516M14.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-7-1-3p | 15 | PIK3CA | Sponge network | 0.389 | 0.04293 | 0.195 | 0.06908 | 0.288 |
| 44068 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-375 | 12 | TACC1 | Sponge network | 0.858 | 3.0E-5 | -0.362 | 0.05406 | 0.288 |
| 44069 | FGD5-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p | 11 | FBXO11 | Sponge network | 0.401 | 0.00066 | 0.142 | 0.04938 | 0.288 |
| 44070 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | CNTNAP1 | Sponge network | -4.477 | 0 | 0.358 | 0.13727 | 0.288 |
| 44071 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-3p | 63 | BMPR2 | Sponge network | -0.615 | 0.00015 | 0.206 | 0.0635 | 0.288 |
| 44072 | AC116366.6 |
hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-629-3p | 10 | ESR1 | Sponge network | 0.329 | 0.11122 | -1.035 | 3.0E-5 | 0.288 |
| 44073 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | AKAP11 | Sponge network | 0.657 | 4.0E-5 | 0.196 | 0.09825 | 0.288 |
| 44074 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | RNF144A | Sponge network | 0.657 | 4.0E-5 | 0.594 | 0.0009 | 0.288 |
| 44075 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | PRKAA2 | Sponge network | 1.61 | 0.00961 | -2.462 | 0 | 0.288 |
| 44076 | RP11-439L18.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-93-3p | 11 | TRPS1 | Sponge network | 0.224 | 0.48719 | -0.191 | 0.44109 | 0.288 |
| 44077 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | NEDD4 | Sponge network | -1.349 | 0.00017 | 0.02 | 0.89834 | 0.288 |
| 44078 | AC005154.6 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 38 | CCDC6 | Sponge network | 0.848 | 0 | 0.27 | 0.02555 | 0.288 |
| 44079 | HOXB-AS3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-452-5p;hsa-miR-671-5p | 20 | ADARB1 | Sponge network | 0.343 | 0.28887 | -0.335 | 0.06631 | 0.288 |
| 44080 | ZNF667-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 24 | CYLD | Sponge network | -0.976 | 0.00025 | -0.367 | 0.00171 | 0.288 |
| 44081 | PRKAG2-AS1 |
hsa-miR-106a-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PTPRD | Sponge network | -0.622 | 0.02807 | -1.005 | 0.00064 | 0.288 |
| 44082 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 14 | GPAM | Sponge network | 0.214 | 0.18246 | 0.142 | 0.29732 | 0.288 |
| 44083 | LINC00641 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TRIM13 | Sponge network | -0.222 | 0.32445 | 0.183 | 0.06968 | 0.288 |
| 44084 | CTC-429P9.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-18a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-485-3p;hsa-miR-542-3p | 10 | ERBB4 | Sponge network | 0.043 | 0.81628 | -1.975 | 2.0E-5 | 0.288 |
| 44085 | LINC00702 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | ARNT2 | Sponge network | -0.737 | 0.06182 | -0.332 | 0.25851 | 0.288 |
| 44086 | ZRANB2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 12 | CREB1 | Sponge network | -0.376 | 0.00337 | 0.203 | 0.00537 | 0.288 |
| 44087 | LINC00865 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-629-3p | 14 | ITGA5 | Sponge network | -1.249 | 7.0E-5 | 0.452 | 0.03524 | 0.288 |
| 44088 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940 | 63 | WDFY3 | Sponge network | -1.697 | 0.00889 | -0.201 | 0.0967 | 0.288 |
| 44089 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-5p | 32 | PIK3R1 | Sponge network | 0.16 | 0.50785 | -0.133 | 0.36854 | 0.288 |
| 44090 | CALML3-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429 | 13 | LATS2 | Sponge network | 0.95 | 0.15943 | 0.159 | 0.2304 | 0.288 |
| 44091 | RP11-57H14.4 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 28 | CTIF | Sponge network | 0.083 | 0.66405 | -0.389 | 0.00549 | 0.288 |
| 44092 | LINC01018 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-96-5p | 12 | ELMO1 | Sponge network | -2.915 | 0.00015 | 0.04 | 0.83147 | 0.288 |
| 44093 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | NEDD4 | Sponge network | -0.355 | 0.0077 | 0.02 | 0.89834 | 0.288 |
| 44094 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-708-5p;hsa-miR-96-5p | 37 | SPRY3 | Sponge network | 0.101 | 0.60919 | 0.016 | 0.91286 | 0.288 |
| 44095 | AC093627.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p | 11 | SYNE1 | Sponge network | -0.787 | 0.3928 | -0.738 | 0.00316 | 0.288 |
| 44096 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SEMA3C | Sponge network | -0.08 | 0.90092 | -0.272 | 0.31153 | 0.288 |
| 44097 | WAC-AS1 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FOXO3 | Sponge network | 0.821 | 0 | -0.04 | 0.72028 | 0.288 |
| 44098 | RAMP2-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | LUZP2 | Sponge network | -0.513 | 0.04703 | -0.056 | 0.89416 | 0.288 |
| 44099 | LINC00672 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 12 | TLL1 | Sponge network | -0.227 | 0.31657 | -1.195 | 3.0E-5 | 0.288 |
| 44100 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 53 | PTEN | Sponge network | -0.323 | 0.01372 | -0.187 | 0.07748 | 0.288 |
| 44101 | HOXA-AS2 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 12 | NGFR | Sponge network | 0.61 | 0.01593 | -1.271 | 0.01058 | 0.288 |
| 44102 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-625-5p;hsa-miR-7-5p | 15 | CRTC1 | Sponge network | 0.238 | 0.70544 | -0.116 | 0.28412 | 0.288 |
| 44103 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | MAP3K12 | Sponge network | -2.117 | 0.00161 | -0.057 | 0.59369 | 0.288 |
| 44104 | RP11-2H8.2 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | USP9X | Sponge network | 1.089 | 0 | 0.222 | 0.01689 | 0.288 |
| 44105 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-584-5p;hsa-miR-877-5p;hsa-miR-942-5p | 16 | GLIPR1 | Sponge network | 0.395 | 0.15451 | 0.146 | 0.3276 | 0.288 |
| 44106 | LINC00672 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 32 | BCL2 | Sponge network | -0.227 | 0.31657 | -0.899 | 0 | 0.288 |
| 44107 | RP1-228H13.5 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-374b-5p | 12 | CREB1 | Sponge network | 1.397 | 0 | 0.203 | 0.00537 | 0.288 |
| 44108 | RP11-284N8.3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-93-3p | 20 | RUNX1T1 | Sponge network | -0.31 | 0.33871 | -0.344 | 0.2374 | 0.288 |
| 44109 | RP11-531A24.5 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-199b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 35 | PPM1L | Sponge network | 0.75 | 0.00054 | -0.537 | 0.00616 | 0.288 |
| 44110 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-576-5p | 13 | ST6GAL2 | Sponge network | 0.906 | 0.31715 | -1.201 | 0.00597 | 0.288 |
| 44111 | NR2F1-AS1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92a-3p | 16 | ATM | Sponge network | -0.49 | 0.04946 | 0.178 | 0.21568 | 0.288 |
| 44112 | CTC-429P9.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p | 17 | TMEM132B | Sponge network | 0.497 | 0.01451 | -0.83 | 0.01084 | 0.288 |
| 44113 | RP11-3P17.4 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p | 14 | CBFB | Sponge network | 0.144 | 0.07137 | 1.061 | 0 | 0.288 |
| 44114 | RP11-25K19.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-3p | 30 | PIK3R1 | Sponge network | -1.057 | 0.00029 | -0.133 | 0.36854 | 0.288 |
| 44115 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 12 | PDGFC | Sponge network | 0.101 | 0.60919 | -0.33 | 0.06483 | 0.288 |
| 44116 | CTC-297N7.9 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-590-5p | 12 | MCC | Sponge network | -2.069 | 0 | -1.005 | 0 | 0.288 |
| 44117 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-5p | 16 | MOB1B | Sponge network | 0.592 | 0.00014 | 0.197 | 0.15266 | 0.288 |
| 44118 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-455-5p | 12 | AMPH | Sponge network | -0.969 | 2.0E-5 | -0.916 | 5.0E-5 | 0.288 |
| 44119 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-935 | 18 | ARNT2 | Sponge network | 0.16 | 0.50785 | -0.332 | 0.25851 | 0.288 |
| 44120 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | ST6GALNAC3 | Sponge network | -0.08 | 0.90092 | -0.987 | 0 | 0.288 |
| 44121 | MCM3AP-AS1 |
hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-2-3p | 12 | VPS53 | Sponge network | 0.331 | 1.0E-5 | 0.103 | 0.22667 | 0.288 |
| 44122 | SOX2-OT |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | TLE4 | Sponge network | -1.264 | 6.0E-5 | -1.157 | 0 | 0.288 |
| 44123 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | LRP1B | Sponge network | -0.726 | 0.00132 | -2.163 | 0 | 0.288 |
| 44124 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-33a-3p | 10 | CLASP2 | Sponge network | 1.134 | 0 | -0.038 | 0.71 | 0.288 |
| 44125 | DNM1P35 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-590-3p | 16 | ITPKB | Sponge network | 0.051 | 0.83388 | -0.704 | 0.00106 | 0.288 |
| 44126 | RP11-672A2.4 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p | 11 | PTPRT | Sponge network | 1.344 | 0 | -0.907 | 0.03727 | 0.288 |
| 44127 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 51 | FAT3 | Sponge network | -0.83 | 0 | -1.601 | 0.00051 | 0.288 |
| 44128 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-493-5p | 14 | PGR | Sponge network | 1.46 | 1.0E-5 | -1.704 | 0 | 0.288 |
| 44129 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | CCND2 | Sponge network | -0.08 | 0.90092 | -0.267 | 0.3112 | 0.288 |
| 44130 | RP11-359E10.1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-7-1-3p | 10 | PCDH18 | Sponge network | 0.299 | 0.57722 | 0.216 | 0.24645 | 0.288 |
| 44131 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-628-5p | 27 | GRIA3 | Sponge network | 0.194 | 0.64278 | -1.956 | 0 | 0.288 |
| 44132 | RP11-701H24.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 11 | NFIB | Sponge network | -2.039 | 0.03447 | 0.023 | 0.90795 | 0.288 |
| 44133 | CTD-2314G24.2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 16 | POU2F2 | Sponge network | 0.246 | 0.59912 | -0.233 | 0.32214 | 0.288 |
| 44134 | LINC00672 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | BCL9 | Sponge network | -0.227 | 0.31657 | 0.321 | 0.00267 | 0.288 |
| 44135 | LINC00094 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 14 | DCLK1 | Sponge network | 0.253 | 0.09565 | -1.324 | 0.00014 | 0.288 |
| 44136 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-653-5p | 23 | DMXL1 | Sponge network | -0.612 | 0.00094 | -0.426 | 0.00056 | 0.288 |
| 44137 | RP11-66B24.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-425-5p;hsa-miR-550a-3p | 11 | DIP2C | Sponge network | 0.57 | 0.1866 | -0.436 | 0.01713 | 0.288 |
| 44138 | RP11-244H3.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | NRXN3 | Sponge network | 0.659 | 3.0E-5 | -0.893 | 0.04186 | 0.288 |
| 44139 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-98-5p | 15 | SEMA3C | Sponge network | 0.056 | 0.81195 | -0.272 | 0.31153 | 0.288 |
| 44140 | PCBP1-AS1 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-654-3p | 16 | DICER1 | Sponge network | -0.567 | 0 | 0.042 | 0.674 | 0.288 |
| 44141 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | ERCC4 | Sponge network | 0.898 | 1.0E-5 | 0.126 | 0.15266 | 0.288 |
| 44142 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 19 | ERG | Sponge network | -0.405 | 0.22013 | 0.002 | 0.99011 | 0.288 |
| 44143 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-96-5p | 12 | CACNA2D1 | Sponge network | 0.622 | 0.00015 | -0.846 | 0.00675 | 0.288 |
| 44144 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-744-3p | 15 | ADAMTSL3 | Sponge network | 0.622 | 0.00015 | -1.559 | 0 | 0.288 |
| 44145 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 38 | FOXP1 | Sponge network | 0.4 | 0.14611 | 0.042 | 0.74368 | 0.288 |
| 44146 | RP11-158K1.3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | KLF10 | Sponge network | 0.349 | 0.01695 | -0.783 | 0 | 0.288 |
| 44147 | FAM66C |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PRRX1 | Sponge network | -0.901 | 0.00019 | 1.316 | 0 | 0.288 |
| 44148 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p | 11 | FBXO32 | Sponge network | -0.301 | 0.06597 | -0.495 | 0.07561 | 0.288 |
| 44149 | AC010226.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | NIN | Sponge network | -0.811 | 2.0E-5 | -0.253 | 0.13794 | 0.288 |
| 44150 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 23 | RASSF2 | Sponge network | -4.477 | 0 | -0.273 | 0.21961 | 0.288 |
| 44151 | CTA-445C9.14 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p | 10 | NF1 | Sponge network | 0.718 | 4.0E-5 | 0.51 | 0 | 0.288 |
| 44152 | SNHG14 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-874-3p | 17 | ARID1B | Sponge network | -1.111 | 0.0003 | 0.003 | 0.97503 | 0.288 |
| 44153 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | LRRK2 | Sponge network | -2.925 | 0 | -1.136 | 1.0E-5 | 0.288 |
| 44154 | HOXD-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-500a-5p | 25 | THRB | Sponge network | -0.208 | 0.65592 | -1.117 | 0.0004 | 0.288 |
| 44155 | LINC01018 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p | 10 | DUSP1 | Sponge network | -2.915 | 0.00015 | -1.754 | 0 | 0.288 |
| 44156 | RAB30-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.752 | 0 | 1.083 | 0 | 0.288 |
| 44157 | SNHG14 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PHF3 | Sponge network | -1.111 | 0.0003 | 0.126 | 0.19652 | 0.288 |
| 44158 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-374b-5p | 11 | SASH1 | Sponge network | 1.46 | 1.0E-5 | -0.502 | 0.00175 | 0.288 |
| 44159 | NUTM2A-AS1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | ABI2 | Sponge network | 0.643 | 0 | 0.175 | 0.14787 | 0.288 |
| 44160 | RP11-458D21.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 12 | PIK3CA | Sponge network | 0.663 | 0.00073 | 0.195 | 0.06908 | 0.288 |
| 44161 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | ST6GALNAC3 | Sponge network | -0.418 | 0.00068 | -0.987 | 0 | 0.288 |
| 44162 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-590-5p | 17 | FREM2 | Sponge network | -0.976 | 0.00025 | -0.437 | 0.46353 | 0.288 |
| 44163 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-374a-3p | 12 | SLC5A3 | Sponge network | 0.919 | 0 | 0.493 | 5.0E-5 | 0.288 |
| 44164 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-654-3p | 22 | RICTOR | Sponge network | 0.411 | 0.1335 | 0.388 | 0.00188 | 0.288 |
| 44165 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-655-3p | 12 | NCOA1 | Sponge network | 0.542 | 0.00054 | -0.432 | 0 | 0.288 |
| 44166 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-423-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | SMARCA2 | Sponge network | -0.976 | 0.00025 | -0.529 | 0.00014 | 0.288 |
| 44167 | LINC00886 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-940 | 16 | NR3C2 | Sponge network | -0.371 | 0.24604 | -1.56 | 0 | 0.288 |
| 44168 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | PSIP1 | Sponge network | 0.572 | 0.01722 | -0.005 | 0.96897 | 0.288 |
| 44169 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | TSHZ3 | Sponge network | 0.41 | 0.02214 | -0.556 | 0.01516 | 0.288 |
| 44170 | PDXDC2P |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-9-5p | 19 | CCDC6 | Sponge network | 0.756 | 0 | 0.27 | 0.02555 | 0.288 |
| 44171 | CTD-2002H8.2 |
hsa-let-7g-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | MEF2C | Sponge network | 0.931 | 1.0E-5 | -0.499 | 0.01448 | 0.288 |
| 44172 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SLC6A15 | Sponge network | 0.219 | 0.1516 | -2.373 | 2.0E-5 | 0.288 |
| 44173 | RP11-254F7.2 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-590-5p | 11 | PRKAR1A | Sponge network | 0.176 | 0.37402 | -0.063 | 0.50314 | 0.288 |
| 44174 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-935 | 16 | RELN | Sponge network | -0.969 | 2.0E-5 | -2.403 | 0 | 0.288 |
| 44175 | RP11-254F7.2 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p | 14 | CSDE1 | Sponge network | 0.176 | 0.37402 | -0.493 | 0 | 0.288 |
| 44176 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-337-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | PICALM | Sponge network | 0.194 | 0.64278 | 0.086 | 0.23523 | 0.288 |
| 44177 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-93-5p | 47 | WDFY3 | Sponge network | -1.645 | 0 | -0.201 | 0.0967 | 0.288 |
| 44178 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-590-3p | 16 | SLITRK3 | Sponge network | 0.064 | 0.72934 | -3.328 | 0 | 0.288 |
| 44179 | RP11-426C22.5 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | QKI | Sponge network | 0.749 | 0 | -0.333 | 0.04111 | 0.288 |
| 44180 | RP11-98I9.4 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | HECA | Sponge network | 0.67 | 0.00048 | -0.217 | 0.03463 | 0.288 |
| 44181 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-511-5p;hsa-miR-942-5p | 10 | NRXN1 | Sponge network | -1.054 | 0.0706 | -3.778 | 0 | 0.288 |
| 44182 | RP11-25K19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SIK1 | Sponge network | -1.057 | 0.00029 | -0.98 | 0 | 0.288 |
| 44183 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-93-3p | 14 | AKAP11 | Sponge network | 2.163 | 0 | 0.196 | 0.09825 | 0.288 |
| 44184 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-93-5p | 19 | ARNT2 | Sponge network | -0.18 | 0.20343 | -0.332 | 0.25851 | 0.288 |
| 44185 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-577 | 16 | GREM1 | Sponge network | -0.465 | 0.04703 | 0.902 | 0.01691 | 0.288 |
| 44186 | RP11-212P7.2 |
hsa-miR-146b-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | FHL1 | Sponge network | 0.219 | 0.1516 | -2.687 | 0 | 0.288 |
| 44187 | RP11-855A2.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 19 | NFIB | Sponge network | 2.094 | 0 | 0.023 | 0.90795 | 0.288 |
| 44188 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-539-5p | 13 | DMD | Sponge network | -0.214 | 0.44248 | -1.506 | 0 | 0.288 |
| 44189 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PDE4DIP | Sponge network | 0.673 | 0 | -0.282 | 0.0257 | 0.288 |
| 44190 | RP11-35G9.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-940 | 16 | NTRK2 | Sponge network | 0.622 | 0.00015 | -0.736 | 0.06495 | 0.288 |
| 44191 | LPP-AS2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-335-3p;hsa-miR-96-5p | 11 | ELMO1 | Sponge network | -0.18 | 0.20343 | 0.04 | 0.83147 | 0.288 |
| 44192 | RP11-403I13.7 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 32 | IGFBP5 | Sponge network | 0.395 | 0.15451 | -0.806 | 0.00105 | 0.288 |
| 44193 | AC083843.1 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 10 | PAWR | Sponge network | 1.4 | 0 | 0.636 | 0 | 0.288 |
| 44194 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-5p | 26 | CLASP2 | Sponge network | 0.174 | 0.30034 | -0.038 | 0.71 | 0.288 |
| 44195 | RP11-244H3.1 |
hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-877-5p | 13 | GLIPR1 | Sponge network | 0.659 | 3.0E-5 | 0.146 | 0.3276 | 0.288 |
| 44196 | PCA3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 26 | CYLD | Sponge network | -0.709 | 0.18168 | -0.367 | 0.00171 | 0.288 |
| 44197 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-93-5p | 12 | DPYD | Sponge network | -1.645 | 0 | -0.736 | 0.00076 | 0.288 |
| 44198 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-590-3p | 12 | TIMP3 | Sponge network | 0.178 | 0.29106 | -0.546 | 0.02307 | 0.288 |
| 44199 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-502-5p;hsa-miR-590-5p | 15 | TGFBR2 | Sponge network | -0.72 | 0.24239 | -0.243 | 0.11408 | 0.288 |
| 44200 | RP11-686D22.8 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 11 | PRKAR1A | Sponge network | 0.898 | 1.0E-5 | -0.063 | 0.50314 | 0.288 |
| 44201 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | TMTC3 | Sponge network | 0.311 | 0.10344 | 0.123 | 0.22189 | 0.288 |
| 44202 | RP11-428J1.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p | 12 | KANK1 | Sponge network | 0.538 | 0.00076 | -0.55 | 0.00134 | 0.288 |
| 44203 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 17 | MYCBP2 | Sponge network | -1.111 | 0.0003 | -0.027 | 0.82016 | 0.288 |
| 44204 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | DOCK4 | Sponge network | 0.41 | 0.02214 | 0.502 | 0.00108 | 0.288 |
| 44205 | RP11-490M8.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-33a-5p;hsa-miR-96-5p | 13 | ZFHX4 | Sponge network | -0.214 | 0.44248 | 0.018 | 0.95574 | 0.288 |
| 44206 | RP11-48B3.4 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 11 | RECK | Sponge network | 1.757 | 0 | -1.043 | 0 | 0.288 |
| 44207 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 49 | MEF2C | Sponge network | -0.942 | 0 | -0.499 | 0.01448 | 0.288 |
| 44208 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-590-3p | 14 | MITF | Sponge network | 0.381 | 0.25967 | -0.769 | 0.00022 | 0.288 |
| 44209 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 26 | NCOA1 | Sponge network | -0.513 | 0.04703 | -0.432 | 0 | 0.288 |
| 44210 | PWAR6 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 18 | FAM135B | Sponge network | -1.575 | 6.0E-5 | -3.978 | 0 | 0.288 |
| 44211 | BMS1P20 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | RB1 | Sponge network | 0.34 | 0.00273 | 0.382 | 0.0017 | 0.288 |
| 44212 | AC010226.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-7-5p | 10 | ERC1 | Sponge network | -0.811 | 2.0E-5 | -0.108 | 0.38794 | 0.288 |
| 44213 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | FOXO1 | Sponge network | -0.563 | 0.02545 | -0.141 | 0.33874 | 0.288 |
| 44214 | CTD-2017D11.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-376c-3p | 10 | HECA | Sponge network | 0.792 | 0.0049 | -0.217 | 0.03463 | 0.288 |
| 44215 | BZRAP1-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-539-5p | 12 | DDX5 | Sponge network | -0.465 | 0.04703 | -0.099 | 0.17428 | 0.288 |
| 44216 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 39 | AKAP11 | Sponge network | 0.311 | 0.10344 | 0.196 | 0.09825 | 0.288 |
| 44217 | RP4-639F20.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p | 14 | ACSL6 | Sponge network | -0.517 | 0.02935 | -1.593 | 0 | 0.288 |
| 44218 | ADAMTS9-AS2 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 51 | PPM1L | Sponge network | -2.452 | 0 | -0.537 | 0.00616 | 0.288 |
| 44219 | HOXB-AS1 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-501-3p;hsa-miR-589-3p | 11 | JDP2 | Sponge network | -0.154 | 0.53954 | -0.967 | 0 | 0.288 |
| 44220 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-590-3p | 12 | RBMS3 | Sponge network | 0.545 | 0.05789 | -0.519 | 0.03551 | 0.288 |
| 44221 | CTA-445C9.14 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-338-3p;hsa-miR-664a-3p | 16 | SOX11 | Sponge network | 0.718 | 4.0E-5 | 2.68 | 0 | 0.288 |
| 44222 | TMEM161B-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-199a-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-27b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-628-5p | 17 | MIER3 | Sponge network | -0.04 | 0.7606 | -0.212 | 0.04957 | 0.288 |
| 44223 | PDXDC2P |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-495-3p | 12 | GSK3B | Sponge network | 0.756 | 0 | 0.266 | 0.00045 | 0.288 |
| 44224 | LINC00854 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-628-5p;hsa-miR-629-3p | 10 | RASSF8 | Sponge network | 1.18 | 0 | -0.122 | 0.65591 | 0.288 |
| 44225 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-590-5p;hsa-miR-93-5p | 14 | RNASEL | Sponge network | -0.663 | 0.10887 | -0.272 | 0.01796 | 0.288 |
| 44226 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-3p;hsa-miR-455-3p | 10 | DKK3 | Sponge network | 0.197 | 0.25618 | -0.052 | 0.77783 | 0.288 |
| 44227 | NDUFA6-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 27 | CTIF | Sponge network | -0.355 | 0.0077 | -0.389 | 0.00549 | 0.288 |
| 44228 | LINC00472 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-877-5p;hsa-miR-942-5p | 17 | GLIPR1 | Sponge network | -0.842 | 0.01361 | 0.146 | 0.3276 | 0.288 |
| 44229 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 26 | EBF1 | Sponge network | 0.16 | 0.50785 | -0.384 | 0.08856 | 0.288 |
| 44230 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-940 | 38 | WDFY3 | Sponge network | -0.154 | 0.53954 | -0.201 | 0.0967 | 0.288 |
| 44231 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-942-5p | 15 | RB1CC1 | Sponge network | -0.323 | 0.01372 | 0.175 | 0.12559 | 0.288 |
| 44232 | AC106786.1 |
hsa-miR-107;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-576-5p | 16 | DTNA | Sponge network | 1.122 | 0.02989 | -1.648 | 1.0E-5 | 0.288 |
| 44233 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-590-5p | 20 | GRIA3 | Sponge network | -1.645 | 0 | -1.956 | 0 | 0.288 |
| 44234 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193b-3p;hsa-miR-590-3p;hsa-miR-654-3p | 13 | MYBL1 | Sponge network | 1.073 | 0.00216 | 0.494 | 0.02152 | 0.288 |
| 44235 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-5p | 15 | ABCA10 | Sponge network | -0.969 | 2.0E-5 | -0.571 | 0.03674 | 0.288 |
| 44236 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577;hsa-miR-7-1-3p | 17 | SYNE1 | Sponge network | 0.389 | 0.04293 | -0.738 | 0.00316 | 0.288 |
| 44237 | FAM13A-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-532-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | PIK3R1 | Sponge network | -0.83 | 0 | -0.133 | 0.36854 | 0.288 |
| 44238 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-590-3p | 14 | TGFBR2 | Sponge network | 0.178 | 0.29106 | -0.243 | 0.11408 | 0.288 |
| 44239 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | ABCC9 | Sponge network | 0.603 | 0.00127 | -0.466 | 0.14121 | 0.288 |
| 44240 | LINC00476 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 13 | PIK3IP1 | Sponge network | -0.615 | 0.00015 | -0.187 | 0.19945 | 0.288 |
| 44241 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 11 | AKAP11 | Sponge network | 0.856 | 0 | 0.196 | 0.09825 | 0.288 |
| 44242 | LINC00957 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | FAM133A | Sponge network | -0.421 | 0.0114 | 0.549 | 0.29009 | 0.288 |
| 44243 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | SLITRK3 | Sponge network | 0.374 | 0.20054 | -3.328 | 0 | 0.288 |
| 44244 | RAMP2-AS1 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | CSDE1 | Sponge network | -0.513 | 0.04703 | -0.493 | 0 | 0.288 |
| 44245 | PCA3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | EYA4 | Sponge network | -0.709 | 0.18168 | 0.3 | 0.36876 | 0.288 |
| 44246 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p | 24 | BCL2 | Sponge network | 0.174 | 0.30034 | -0.899 | 0 | 0.288 |
| 44247 | BCYRN1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 25 | PTPN14 | Sponge network | 0.311 | 0.10344 | 0.144 | 0.34595 | 0.288 |
| 44248 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p;hsa-miR-935 | 37 | PTPRD | Sponge network | -0.384 | 0.40652 | -1.005 | 0.00064 | 0.288 |
| 44249 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | RASSF8 | Sponge network | -0.72 | 0.24239 | -0.122 | 0.65591 | 0.287 |
| 44250 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 14 | AFF4 | Sponge network | 0.959 | 0 | -0.266 | 0.01565 | 0.287 |
| 44251 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | ANK2 | Sponge network | 0.559 | 0.00026 | -1.603 | 1.0E-5 | 0.287 |
| 44252 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p | 17 | CLASP2 | Sponge network | 1.317 | 0 | -0.038 | 0.71 | 0.287 |
| 44253 | PART1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | PAK3 | Sponge network | -2.925 | 0 | -0.628 | 0.07253 | 0.287 |
| 44254 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p | 18 | CADM3 | Sponge network | 0.214 | 0.18246 | -3.663 | 0 | 0.287 |
| 44255 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | SMARCA2 | Sponge network | -0.901 | 0.00019 | -0.529 | 0.00014 | 0.287 |
| 44256 | RP11-102N12.3 |
hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 11 | ITPKB | Sponge network | -0.351 | 0.11358 | -0.704 | 0.00106 | 0.287 |
| 44257 | RP11-539I5.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PPP1R9A | Sponge network | -0.468 | 0.32587 | -0.903 | 0.04254 | 0.287 |
| 44258 | RP11-798G7.7 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-455-3p;hsa-miR-484 | 16 | SPRY3 | Sponge network | 0.858 | 3.0E-5 | 0.016 | 0.91286 | 0.287 |
| 44259 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 29 | THBS1 | Sponge network | -0.222 | 0.32445 | 0.741 | 0.00386 | 0.287 |
| 44260 | RP11-474O21.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-744-3p;hsa-miR-93-5p | 15 | HLF | Sponge network | -0.023 | 0.95109 | -2.443 | 0 | 0.287 |
| 44261 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-93-3p | 31 | AKAP11 | Sponge network | -0.154 | 0.78939 | 0.196 | 0.09825 | 0.287 |
| 44262 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-5p | 22 | GRIA3 | Sponge network | 0.176 | 0.37402 | -1.956 | 0 | 0.287 |
| 44263 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 25 | USP12 | Sponge network | 1.302 | 7.0E-5 | -0.102 | 0.40768 | 0.287 |
| 44264 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p | 13 | TSC22D1 | Sponge network | -1.111 | 0.0003 | -0.529 | 2.0E-5 | 0.287 |
| 44265 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TBX18 | Sponge network | -0.709 | 0.18168 | 0.843 | 0.02945 | 0.287 |
| 44266 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 23 | NEDD4 | Sponge network | 0.862 | 0.06213 | 0.02 | 0.89834 | 0.287 |
| 44267 | SNHG14 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | CEP170 | Sponge network | -1.111 | 0.0003 | 0.943 | 0 | 0.287 |
| 44268 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 15 | EDA2R | Sponge network | -0.222 | 0.32445 | -0.587 | 0.01598 | 0.287 |
| 44269 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p | 12 | NALCN | Sponge network | -0.899 | 6.0E-5 | 1.068 | 0.00237 | 0.287 |
| 44270 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-93-5p | 38 | ADARB1 | Sponge network | -0.18 | 0.20343 | -0.335 | 0.06631 | 0.287 |
| 44271 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 25 | LRRC7 | Sponge network | -0.355 | 0.0077 | -1.341 | 0.01193 | 0.287 |
| 44272 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | PRDM5 | Sponge network | 0.064 | 0.72934 | -1.09 | 0.00014 | 0.287 |
| 44273 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 28 | PTPRT | Sponge network | -0.611 | 0.09607 | -0.907 | 0.03727 | 0.287 |
| 44274 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b | 10 | KAT6B | Sponge network | -1.654 | 0.00144 | -0.478 | 0.00018 | 0.287 |
| 44275 | RP11-276H19.2 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-5p | 12 | IGF1 | Sponge network | -0.472 | 0.48851 | -0.204 | 0.5898 | 0.287 |
| 44276 | RP11-440L14.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-511-5p | 11 | CREB1 | Sponge network | 0.599 | 0.0001 | 0.203 | 0.00537 | 0.287 |
| 44277 | FAM83H-AS1 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-497-5p | 11 | PAFAH1B2 | Sponge network | 3.083 | 0 | 0.318 | 0.0001 | 0.287 |
| 44278 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-511-5p | 11 | BCL6 | Sponge network | 0.622 | 0.00015 | -0.211 | 0.19489 | 0.287 |
| 44279 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-629-3p;hsa-miR-93-5p | 15 | RBL2 | Sponge network | -0.303 | 0.21002 | -0.282 | 0.00254 | 0.287 |
| 44280 | PWAR6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p | 24 | CRTC1 | Sponge network | -1.575 | 6.0E-5 | -0.116 | 0.28412 | 0.287 |
| 44281 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 29 | MAF | Sponge network | -0.738 | 0.21233 | -1.257 | 0 | 0.287 |
| 44282 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | TIMP3 | Sponge network | -0.355 | 0.0077 | -0.546 | 0.02307 | 0.287 |
| 44283 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ZNF770 | Sponge network | 0.401 | 0.00066 | 0.206 | 0.06751 | 0.287 |
| 44284 | RP11-181G12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-486-5p | 14 | AKAP6 | Sponge network | 0.556 | 0.00298 | -1.586 | 3.0E-5 | 0.287 |
| 44285 | RP11-359B12.2 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 17 | IRS1 | Sponge network | 0.064 | 0.72934 | -0.026 | 0.88855 | 0.287 |
| 44286 | RP11-531A24.5 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p | 19 | SYNPO2 | Sponge network | 0.75 | 0.00054 | -2.342 | 0 | 0.287 |
| 44287 | RP11-758N13.1 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-93-3p | 11 | DOCK4 | Sponge network | 2.939 | 3.0E-5 | 0.502 | 0.00108 | 0.287 |
| 44288 | HCG18 |
hsa-miR-142-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p | 15 | TMEM30A | Sponge network | 1.063 | 0 | 0.096 | 0.31031 | 0.287 |
| 44289 | RP11-37B2.1 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 13 | PHF6 | Sponge network | 1.03 | 0 | 0.785 | 0 | 0.287 |
| 44290 | RP11-410L14.2 |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-497-5p | 15 | AKAP11 | Sponge network | -0.052 | 0.76839 | 0.196 | 0.09825 | 0.287 |
| 44291 | RP4-613B23.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-33a-3p | 10 | AKAP11 | Sponge network | -0.309 | 0.1145 | 0.196 | 0.09825 | 0.287 |
| 44292 | AC005154.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | RPS6KA3 | Sponge network | 0.848 | 0 | 0.256 | 0.02419 | 0.287 |
| 44293 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-874-3p;hsa-miR-93-5p | 39 | TACC1 | Sponge network | -1.645 | 0 | -0.362 | 0.05406 | 0.287 |
| 44294 | AC009120.6 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | SIRT1 | Sponge network | 0.919 | 0 | 0.109 | 0.2593 | 0.287 |
| 44295 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 26 | JAZF1 | Sponge network | 0.61 | 0.01593 | -0.377 | 0.02677 | 0.287 |
| 44296 | RP11-212P7.2 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 12 | NGFR | Sponge network | 0.219 | 0.1516 | -1.271 | 0.01058 | 0.287 |
| 44297 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 33 | CUX1 | Sponge network | 0.559 | 0.00026 | -0.039 | 0.6705 | 0.287 |
| 44298 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-542-3p | 10 | USP25 | Sponge network | -1.092 | 4.0E-5 | -0.242 | 0.01397 | 0.287 |
| 44299 | RP11-38L15.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 11 | RASSF8 | Sponge network | 0.344 | 0.3127 | -0.122 | 0.65591 | 0.287 |
| 44300 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-450b-5p | 23 | ESR1 | Sponge network | 0.16 | 0.50785 | -1.035 | 3.0E-5 | 0.287 |
| 44301 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p | 23 | JAZF1 | Sponge network | 0.657 | 4.0E-5 | -0.377 | 0.02677 | 0.287 |
| 44302 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-493-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-942-5p | 30 | DDX6 | Sponge network | -0.162 | 0.47687 | 0.091 | 0.36428 | 0.287 |
| 44303 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-942-5p | 30 | HLF | Sponge network | 0.385 | 0.10067 | -2.443 | 0 | 0.287 |
| 44304 | BVES-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-653-5p | 31 | PPM1L | Sponge network | -2.62 | 0 | -0.537 | 0.00616 | 0.287 |
| 44305 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-629-5p | 13 | SOX5 | Sponge network | 0.065 | 0.73468 | -1.091 | 4.0E-5 | 0.287 |
| 44306 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-93-5p | 15 | TXNIP | Sponge network | -0.08 | 0.90092 | -1.277 | 0 | 0.287 |
| 44307 | RP11-983P16.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-628-5p | 42 | RUNX1T1 | Sponge network | 0.41 | 0.02214 | -0.344 | 0.2374 | 0.287 |
| 44308 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SEMA3E | Sponge network | 0.225 | 0.30644 | -1.717 | 0.00137 | 0.287 |
| 44309 | RP13-516M14.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | LUZP2 | Sponge network | 0.389 | 0.04293 | -0.056 | 0.89416 | 0.287 |
| 44310 | HOXB-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-429 | 16 | CBFA2T3 | Sponge network | -0.154 | 0.53954 | -1.221 | 1.0E-5 | 0.287 |
| 44311 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-3913-5p;hsa-miR-577;hsa-miR-7-5p | 24 | ZBTB4 | Sponge network | -0.342 | 0.02288 | -0.739 | 0 | 0.287 |
| 44312 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-429;hsa-miR-590-3p | 17 | DMXL1 | Sponge network | 0.274 | 0.07681 | -0.426 | 0.00056 | 0.287 |
| 44313 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | ZNF208 | Sponge network | 0.72 | 0.0001 | -0.4 | 0.22381 | 0.287 |
| 44314 | CTA-29F11.1 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-143-3p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-486-5p | 14 | PHF6 | Sponge network | 0.583 | 0.00027 | 0.785 | 0 | 0.287 |
| 44315 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 13 | BCL11A | Sponge network | -0.021 | 0.93598 | -0.932 | 0.0063 | 0.287 |
| 44316 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-200c-3p;hsa-miR-21-5p | 10 | FREM2 | Sponge network | -0.473 | 0.07602 | -0.437 | 0.46353 | 0.287 |
| 44317 | ARHGEF26-AS1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | INPP4A | Sponge network | -2.46 | 0 | 0.07 | 0.53563 | 0.287 |
| 44318 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 19 | PTPN14 | Sponge network | 0.619 | 0.00157 | 0.144 | 0.34595 | 0.287 |
| 44319 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-629-3p | 17 | RASSF2 | Sponge network | 2.306 | 9.0E-5 | -0.273 | 0.21961 | 0.287 |
| 44320 | LINC00648 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 14 | UNC80 | Sponge network | 0.633 | 0.51678 | -1.934 | 0 | 0.287 |
| 44321 | TPTEP1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 25 | KCNJ5 | Sponge network | -1.216 | 0 | -0.281 | 0.3339 | 0.287 |
| 44322 | RP11-395A13.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-500a-5p | 16 | PRKAA2 | Sponge network | 0.057 | 0.61225 | -2.462 | 0 | 0.287 |
| 44323 | U91328.21 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-629-3p | 17 | EPHA7 | Sponge network | -0.735 | 0.15983 | -2.197 | 3.0E-5 | 0.287 |
| 44324 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | MTSS1 | Sponge network | -2.117 | 0.00161 | -0.81 | 0.00035 | 0.287 |
| 44325 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 23 | MOB1B | Sponge network | 0.051 | 0.83388 | 0.197 | 0.15266 | 0.287 |
| 44326 | CTC-510F12.2 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p | 12 | PAFAH1B1 | Sponge network | -0.691 | 0.00146 | -0.429 | 0 | 0.287 |
| 44327 | SNHG16 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-497-5p | 10 | SON | Sponge network | 0.961 | 0 | 0.168 | 0.04406 | 0.287 |
| 44328 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | LRP1B | Sponge network | -0.342 | 0.02288 | -2.163 | 0 | 0.287 |
| 44329 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | AHNAK | Sponge network | 0.532 | 0.00505 | -1.207 | 0 | 0.287 |
| 44330 | LINC00680 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-664a-3p | 15 | FZD3 | Sponge network | 0.884 | 0 | 0.104 | 0.60316 | 0.287 |
| 44331 | RP11-968O1.5 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-493-5p | 12 | SYT4 | Sponge network | -0.829 | 0.01343 | -3.207 | 0 | 0.287 |
| 44332 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429 | 16 | SCN9A | Sponge network | 0.859 | 0.00331 | -0.525 | 0.10593 | 0.287 |
| 44333 | PWAR6 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-3p | 28 | CTIF | Sponge network | -1.575 | 6.0E-5 | -0.389 | 0.00549 | 0.287 |
| 44334 | LINC01018 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SEMA3C | Sponge network | -2.915 | 0.00015 | -0.272 | 0.31153 | 0.287 |
| 44335 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 12 | EPHA3 | Sponge network | 0.796 | 4.0E-5 | 0.185 | 0.53588 | 0.287 |
| 44336 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 18 | PRKAB2 | Sponge network | 0.919 | 0 | -0.367 | 0.01371 | 0.287 |
| 44337 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 18 | CNTN1 | Sponge network | 1.757 | 0 | -2.594 | 0 | 0.287 |
| 44338 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-590-3p | 10 | NALCN | Sponge network | 0.659 | 3.0E-5 | 1.068 | 0.00237 | 0.287 |
| 44339 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 38 | PPP1R9A | Sponge network | 0.311 | 0.10344 | -0.903 | 0.04254 | 0.287 |
| 44340 | CTC-378H22.2 |
hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-338-3p | 10 | ZFHX4 | Sponge network | 0.001 | 0.99663 | 0.018 | 0.95574 | 0.287 |
| 44341 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30e-5p | 16 | SOX11 | Sponge network | 1.206 | 0 | 2.68 | 0 | 0.287 |
| 44342 | RP5-1085F17.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 11 | DDX5 | Sponge network | 0.541 | 0.00125 | -0.099 | 0.17428 | 0.287 |
| 44343 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 30 | BNC2 | Sponge network | -0.031 | 0.89063 | -0.491 | 0.09988 | 0.287 |
| 44344 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-214-5p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935 | 50 | SMAD4 | Sponge network | 0.537 | 0.00139 | -0.313 | 0.00413 | 0.287 |
| 44345 | PCA3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ZNF844 | Sponge network | -0.709 | 0.18168 | -0.966 | 0.00035 | 0.287 |
| 44346 | RP11-535M15.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p | 18 | WDFY3 | Sponge network | -1.14 | 0.03513 | -0.201 | 0.0967 | 0.287 |
| 44347 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 16 | TGFBR2 | Sponge network | -1.057 | 0.00029 | -0.243 | 0.11408 | 0.287 |
| 44348 | CTA-941F9.9 |
hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-26b-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-935 | 11 | GRIK3 | Sponge network | -0.344 | 0.49624 | -3.208 | 0 | 0.287 |
| 44349 | RP11-1006G14.4 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 38 | DTNA | Sponge network | 0.559 | 0.00026 | -1.648 | 1.0E-5 | 0.287 |
| 44350 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-5p | 17 | CUL5 | Sponge network | 0.921 | 0.00118 | 0.026 | 0.77301 | 0.287 |
| 44351 | RP11-2H8.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-744-3p | 16 | PTPN14 | Sponge network | 1.089 | 0 | 0.144 | 0.34595 | 0.287 |
| 44352 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 30 | SCN9A | Sponge network | 0.311 | 0.10344 | -0.525 | 0.10593 | 0.287 |
| 44353 | ERVH48-1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | AKAP6 | Sponge network | 1.04 | 0.00023 | -1.586 | 3.0E-5 | 0.287 |
| 44354 | NUTM2A-AS1 |
hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | CCDC6 | Sponge network | 0.643 | 0 | 0.27 | 0.02555 | 0.287 |
| 44355 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-7-5p | 30 | RBMS3 | Sponge network | 0.637 | 0.00069 | -0.519 | 0.03551 | 0.287 |
| 44356 | RP11-705C15.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 14 | RASSF8 | Sponge network | 0.796 | 4.0E-5 | -0.122 | 0.65591 | 0.287 |
| 44357 | RP11-333A23.4 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CNTN1 | Sponge network | -1.791 | 0.04269 | -2.594 | 0 | 0.287 |
| 44358 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-5p | 16 | LATS1 | Sponge network | 0.889 | 9.0E-5 | 0.109 | 0.34208 | 0.287 |
| 44359 | RP11-307B6.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 15 | PTPRT | Sponge network | -4.463 | 0.00025 | -0.907 | 0.03727 | 0.287 |
| 44360 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p | 15 | GPAM | Sponge network | 1.117 | 0 | 0.142 | 0.29732 | 0.287 |
| 44361 | RP11-325K4.2 |
hsa-let-7a-3p;hsa-miR-143-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p | 10 | RICTOR | Sponge network | 0.198 | 0.27624 | 0.388 | 0.00188 | 0.287 |
| 44362 | RP11-56L13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-485-3p;hsa-miR-590-5p | 11 | SASH1 | Sponge network | -0.992 | 0.18158 | -0.502 | 0.00175 | 0.287 |
| 44363 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 38 | ZFHX3 | Sponge network | -1.249 | 7.0E-5 | 0.389 | 0.01363 | 0.287 |
| 44364 | MYLK-AS1 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-484 | 16 | ZFX | Sponge network | -1.331 | 3.0E-5 | 0.09 | 0.42563 | 0.287 |
| 44365 | NDUFA6-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 19 | ATM | Sponge network | -0.355 | 0.0077 | 0.178 | 0.21568 | 0.287 |
| 44366 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-96-5p | 12 | PIK3CA | Sponge network | -0.517 | 0.02935 | 0.195 | 0.06908 | 0.287 |
| 44367 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ABCC9 | Sponge network | 0.272 | 0.44692 | -0.466 | 0.14121 | 0.287 |
| 44368 | AC024560.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-940 | 30 | MDM4 | Sponge network | 2.166 | 0.00228 | 0.345 | 0.00762 | 0.287 |
| 44369 | RP11-244H3.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PDE4DIP | Sponge network | 0.659 | 3.0E-5 | -0.282 | 0.0257 | 0.287 |
| 44370 | AC127904.2 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-664a-5p | 11 | CBFB | Sponge network | 1.308 | 0 | 1.061 | 0 | 0.287 |
| 44371 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 35 | ST6GALNAC3 | Sponge network | -0.709 | 0.18168 | -0.987 | 0 | 0.287 |
| 44372 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | SOX5 | Sponge network | 0.559 | 0.00026 | -1.091 | 4.0E-5 | 0.287 |
| 44373 | LINC00648 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | GALNT13 | Sponge network | 0.633 | 0.51678 | -0.857 | 0.07439 | 0.287 |
| 44374 | MIR600HG |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-532-3p | 11 | TRIM13 | Sponge network | 0.594 | 0.41522 | 0.183 | 0.06968 | 0.287 |
| 44375 | OIP5-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | PRUNE2 | Sponge network | 0.421 | 0.00084 | -1.733 | 0.00022 | 0.287 |
| 44376 | LINC00094 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 13 | MAPK10 | Sponge network | 0.253 | 0.09565 | -1.774 | 0 | 0.287 |
| 44377 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 10 | NF1 | Sponge network | 0.796 | 4.0E-5 | 0.51 | 0 | 0.287 |
| 44378 | RP11-819C21.1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-550a-3p | 12 | CACNA1E | Sponge network | 0.537 | 0.00139 | 1.752 | 0 | 0.287 |
| 44379 | GNG12-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p | 23 | EFNA5 | Sponge network | -0.793 | 7.0E-5 | -0.421 | 0.17868 | 0.287 |
| 44380 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 32 | LIFR | Sponge network | -0.969 | 2.0E-5 | -1.794 | 0 | 0.287 |
| 44381 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | LRRK2 | Sponge network | 0.421 | 0.00084 | -1.136 | 1.0E-5 | 0.287 |
| 44382 | LINC00865 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-7-1-3p | 25 | LRIG1 | Sponge network | -1.249 | 7.0E-5 | -0.537 | 0.00588 | 0.287 |
| 44383 | AC006547.13 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-324-3p;hsa-miR-424-5p | 11 | ZFHX3 | Sponge network | 1.159 | 0 | 0.389 | 0.01363 | 0.287 |
| 44384 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-7-1-3p | 14 | BCL11A | Sponge network | -0.13 | 0.48734 | -0.932 | 0.0063 | 0.287 |
| 44385 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EPAS1 | Sponge network | 0.299 | 0.57722 | -0.184 | 0.14418 | 0.287 |
| 44386 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-452-5p;hsa-miR-590-3p | 29 | PAFAH1B1 | Sponge network | -0.358 | 0.17255 | -0.429 | 0 | 0.287 |
| 44387 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 31 | KIF1B | Sponge network | 1.147 | 0 | -0.118 | 0.32258 | 0.287 |
| 44388 | LINC00654 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 19 | LUZP2 | Sponge network | 0.374 | 0.20054 | -0.056 | 0.89416 | 0.287 |
| 44389 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 23 | GRIN2A | Sponge network | -0.125 | 0.49414 | -2.161 | 0 | 0.287 |
| 44390 | RP11-6O2.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 30 | GRIK3 | Sponge network | 0.331 | 0.57247 | -3.208 | 0 | 0.287 |
| 44391 | RP11-122K13.12 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-15b-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | AFF4 | Sponge network | 1.219 | 0 | -0.266 | 0.01565 | 0.287 |
| 44392 | RP11-440L14.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CBX5 | Sponge network | 0.599 | 0.0001 | 0.165 | 0.24446 | 0.287 |
| 44393 | CTD-2139B15.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-766-3p | 11 | TBL1XR1 | Sponge network | 0.286 | 0.00612 | 0.578 | 0 | 0.287 |
| 44394 | LINC00654 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 35 | TGFBR3 | Sponge network | 0.374 | 0.20054 | -1.173 | 1.0E-5 | 0.287 |
| 44395 | MRPL23-AS1 |
hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p | 13 | MTSS1 | Sponge network | -0.278 | 0.76559 | -0.81 | 0.00035 | 0.287 |
| 44396 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-7-1-3p | 15 | PDE4DIP | Sponge network | 0.853 | 0.15352 | -0.282 | 0.0257 | 0.287 |
| 44397 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-421 | 10 | JAK1 | Sponge network | -0.487 | 0.02157 | 0.001 | 0.98982 | 0.287 |
| 44398 | RP11-157J24.2 |
hsa-miR-130b-3p;hsa-miR-181a-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p | 12 | ZBTB4 | Sponge network | -1.664 | 0.05165 | -0.739 | 0 | 0.287 |
| 44399 | RP11-532F6.3 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-590-3p | 14 | FAM135B | Sponge network | -0.896 | 0.00099 | -3.978 | 0 | 0.287 |
| 44400 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 18 | PTPRB | Sponge network | -0.563 | 0.02545 | 0.105 | 0.47122 | 0.287 |
| 44401 | RP11-411K7.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 35 | SPRY3 | Sponge network | 0.155 | 0.81273 | 0.016 | 0.91286 | 0.287 |
| 44402 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | LRRC7 | Sponge network | -1.249 | 7.0E-5 | -1.341 | 0.01193 | 0.287 |
| 44403 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577 | 22 | EBF1 | Sponge network | 0.056 | 0.81195 | -0.384 | 0.08856 | 0.287 |
| 44404 | LINC00957 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-935 | 21 | GJA1 | Sponge network | -0.421 | 0.0114 | -0.485 | 0.02179 | 0.287 |
| 44405 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DIP2C | Sponge network | 0.272 | 0.44692 | -0.436 | 0.01713 | 0.287 |
| 44406 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-93-5p | 14 | BHLHE41 | Sponge network | -2.039 | 0.03447 | 0.353 | 0.12753 | 0.287 |
| 44407 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | PPP1R9A | Sponge network | 0.064 | 0.72934 | -0.903 | 0.04254 | 0.287 |
| 44408 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-369-3p | 10 | MOB1B | Sponge network | 0.062 | 0.55274 | 0.197 | 0.15266 | 0.287 |
| 44409 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-654-3p | 25 | MYBL1 | Sponge network | 0.177 | 0.16681 | 0.494 | 0.02152 | 0.287 |
| 44410 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | SOX11 | Sponge network | 1.205 | 0 | 2.68 | 0 | 0.287 |
| 44411 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-374b-3p;hsa-miR-421 | 13 | TP53INP1 | Sponge network | -4.546 | 0 | -0.496 | 0.00064 | 0.287 |
| 44412 | USP3-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-335-3p;hsa-miR-338-3p | 12 | ELMO1 | Sponge network | -0.162 | 0.47687 | 0.04 | 0.83147 | 0.287 |
| 44413 | SNHG16 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-497-5p | 13 | CDK6 | Sponge network | 0.961 | 0 | 0.991 | 0 | 0.287 |
| 44414 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-5p | 17 | AR | Sponge network | -0.164 | 0.45106 | -1.554 | 0 | 0.287 |
| 44415 | RP11-130L8.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ERG | Sponge network | -0.663 | 0.10887 | 0.002 | 0.99011 | 0.287 |
| 44416 | HCG11 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-590-3p | 15 | PRKAR1A | Sponge network | 0.385 | 0.10067 | -0.063 | 0.50314 | 0.287 |
| 44417 | AC003090.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | CREB3L2 | Sponge network | -2.117 | 0.00161 | -0.525 | 2.0E-5 | 0.287 |
| 44418 | LINC00476 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-424-5p | 18 | UBE4B | Sponge network | -0.615 | 0.00015 | -0.235 | 0.007 | 0.287 |
| 44419 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-655-3p | 24 | PIK3C2A | Sponge network | 0.349 | 0.01695 | 0.061 | 0.53776 | 0.287 |
| 44420 | RP11-774O3.3 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 10 | PRKACA | Sponge network | -0.487 | 0.02157 | -0.255 | 0.0012 | 0.287 |
| 44421 | WDFY3-AS2 |
hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | PCDH18 | Sponge network | -0.942 | 0 | 0.216 | 0.24645 | 0.287 |
| 44422 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-532-3p | 17 | WDFY3 | Sponge network | 1.361 | 0 | -0.201 | 0.0967 | 0.287 |
| 44423 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 41 | KIF1B | Sponge network | 1.089 | 0 | -0.118 | 0.32258 | 0.287 |
| 44424 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 32 | NOTCH2 | Sponge network | -2.117 | 0.00161 | 0.138 | 0.35163 | 0.287 |
| 44425 | RP11-403I13.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | 0.619 | 0.00157 | -0.001 | 0.99669 | 0.287 |
| 44426 | AC009014.3 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p | 10 | BHLHE41 | Sponge network | -0.917 | 0.24351 | 0.353 | 0.12753 | 0.287 |
| 44427 | GAS6-AS2 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | INPP4A | Sponge network | 0.16 | 0.50785 | 0.07 | 0.53563 | 0.287 |
| 44428 | RP11-473I1.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-339-5p | 15 | IL17RD | Sponge network | 0.542 | 0.00054 | -0.019 | 0.94873 | 0.287 |
| 44429 | RP11-67L2.2 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 12 | RAPGEF5 | Sponge network | 0.72 | 0.0001 | 0.536 | 4.0E-5 | 0.287 |
| 44430 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-382-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 28 | ACSL6 | Sponge network | -2.117 | 0.00161 | -1.593 | 0 | 0.287 |
| 44431 | DNM3OS |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-331-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CREM | Sponge network | 0.572 | 0.01722 | -0.345 | 0.00258 | 0.287 |
| 44432 | RP11-680C21.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | FREM2 | Sponge network | -1.424 | 0.04346 | -0.437 | 0.46353 | 0.287 |
| 44433 | CTD-2314G24.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p | 18 | ROBO1 | Sponge network | 0.246 | 0.59912 | -0.009 | 0.96154 | 0.287 |
| 44434 | AC103563.9 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p | 11 | AR | Sponge network | -6.057 | 0 | -1.554 | 0 | 0.287 |
| 44435 | RP11-46C24.7 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | PEG3 | Sponge network | 0.392 | 0.0278 | -1.74 | 0 | 0.287 |
| 44436 | PWAR6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 23 | RIMBP2 | Sponge network | -1.575 | 6.0E-5 | -1.968 | 5.0E-5 | 0.287 |
| 44437 | DNM3OS |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 10 | PTPN12 | Sponge network | 0.572 | 0.01722 | 0.768 | 0 | 0.287 |
| 44438 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-532-3p;hsa-miR-93-3p | 15 | RPS6KA2 | Sponge network | -0.18 | 0.20343 | -0.57 | 0.00013 | 0.287 |
| 44439 | LINC00641 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 52 | PPARA | Sponge network | -0.222 | 0.32445 | -0.379 | 0.00548 | 0.287 |
| 44440 | RP11-244H3.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p;hsa-miR-940 | 29 | GAS7 | Sponge network | 0.659 | 3.0E-5 | -0.555 | 0.01602 | 0.287 |
| 44441 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | MEF2C | Sponge network | -0.737 | 0.10822 | -0.499 | 0.01448 | 0.287 |
| 44442 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-5p | 12 | ZNF208 | Sponge network | 0.16 | 0.50785 | -0.4 | 0.22381 | 0.287 |
| 44443 | RP11-46C24.7 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | NAV3 | Sponge network | 0.392 | 0.0278 | 0.212 | 0.47729 | 0.287 |
| 44444 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-33a-3p;hsa-miR-424-5p | 11 | SON | Sponge network | 0.368 | 0.20093 | 0.168 | 0.04406 | 0.287 |
| 44445 | CD27-AS1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-493-3p | 11 | JAKMIP2 | Sponge network | 0.177 | 0.16681 | -1.388 | 0 | 0.287 |
| 44446 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 24 | MAF | Sponge network | 0.225 | 0.30644 | -1.257 | 0 | 0.287 |
| 44447 | DNAJC27-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 27 | SYT4 | Sponge network | -0.969 | 2.0E-5 | -3.207 | 0 | 0.287 |
| 44448 | ERVH48-1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-744-3p | 10 | KIF5B | Sponge network | 1.04 | 0.00023 | 0.362 | 0.00066 | 0.287 |
| 44449 | SNX29P2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-339-5p | 10 | STARD13 | Sponge network | 0.4 | 0.14611 | -0.187 | 0.27383 | 0.287 |
| 44450 | LINC00662 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PRKAB2 | Sponge network | 0.213 | 0.32297 | -0.367 | 0.01371 | 0.287 |
| 44451 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 30 | EPHA5 | Sponge network | 1.757 | 0 | -0.957 | 0.01733 | 0.287 |
| 44452 | U91328.21 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p | 13 | ANK2 | Sponge network | -0.735 | 0.15983 | -1.603 | 1.0E-5 | 0.287 |
| 44453 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 43 | ZFHX4 | Sponge network | 0.083 | 0.66405 | 0.018 | 0.95574 | 0.287 |
| 44454 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 13 | FZD3 | Sponge network | 1.266 | 0 | 0.104 | 0.60316 | 0.287 |
| 44455 | RP11-216B9.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-940 | 25 | CREB1 | Sponge network | 0.174 | 0.30034 | 0.203 | 0.00537 | 0.287 |
| 44456 | RP11-575A19.2 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | 1.969 | 0.00316 | -0.133 | 0.36854 | 0.287 |
| 44457 | FENDRR |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-744-3p | 28 | PTPN14 | Sponge network | -1.281 | 0.00012 | 0.144 | 0.34595 | 0.287 |
| 44458 | HCG11 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SUFU | Sponge network | 0.385 | 0.10067 | -0.04 | 0.65489 | 0.287 |
| 44459 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p | 25 | RUNX1T1 | Sponge network | -0.285 | 0.2172 | -0.344 | 0.2374 | 0.287 |
| 44460 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-671-5p | 10 | HIC1 | Sponge network | 0.101 | 0.60919 | 0.024 | 0.89889 | 0.287 |
| 44461 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 28 | TACC1 | Sponge network | -0.223 | 0.47816 | -0.362 | 0.05406 | 0.287 |
| 44462 | LINC00861 |
hsa-miR-130b-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-93-3p | 13 | RUNX1T1 | Sponge network | 0.911 | 0.00854 | -0.344 | 0.2374 | 0.287 |
| 44463 | RP11-116O18.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 23 | PTPRT | Sponge network | 0.05 | 0.95483 | -0.907 | 0.03727 | 0.287 |
| 44464 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | YAP1 | Sponge network | 0.225 | 0.30644 | 0.22 | 0.11836 | 0.287 |
| 44465 | RP11-244H3.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | AKAP6 | Sponge network | 0.659 | 3.0E-5 | -1.586 | 3.0E-5 | 0.287 |
| 44466 | RP11-798G7.7 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30e-3p | 14 | CCDC6 | Sponge network | 0.858 | 3.0E-5 | 0.27 | 0.02555 | 0.287 |
| 44467 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | 0.199 | 0.38015 | -3.328 | 0 | 0.287 |
| 44468 | RP11-401P9.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-874-3p;hsa-miR-877-5p | 22 | ASTN1 | Sponge network | -0.384 | 0.40652 | -2.389 | 2.0E-5 | 0.287 |
| 44469 | CALML3-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p | 25 | CREB1 | Sponge network | 0.95 | 0.15943 | 0.203 | 0.00537 | 0.287 |
| 44470 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 31 | CREB3L2 | Sponge network | 0.214 | 0.18246 | -0.525 | 2.0E-5 | 0.287 |
| 44471 | LINC00621 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | SYNPO2 | Sponge network | 0.131 | 0.8436 | -2.342 | 0 | 0.287 |
| 44472 | RP11-384O8.1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p | 11 | GATA2 | Sponge network | -0.738 | 0.21233 | -0.654 | 0.0016 | 0.287 |
| 44473 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | HECA | Sponge network | 1.254 | 0 | -0.217 | 0.03463 | 0.287 |
| 44474 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 13 | PTPN14 | Sponge network | 1.04 | 0.00023 | 0.144 | 0.34595 | 0.287 |
| 44475 | RP11-359B12.2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 15 | SORCS1 | Sponge network | 0.064 | 0.72934 | -3.492 | 0 | 0.287 |
| 44476 | TTC25 |
hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-590-5p;hsa-miR-629-5p | 12 | DST | Sponge network | -0.208 | 0.38261 | -0.713 | 0.00096 | 0.287 |
| 44477 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p | 11 | ITCH | Sponge network | 1.11 | 1.0E-5 | 0.084 | 0.34058 | 0.287 |
| 44478 | RP11-5C23.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 17 | PRKCB | Sponge network | 0.274 | 0.07681 | -1.313 | 2.0E-5 | 0.287 |
| 44479 | RP11-464F9.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | TCF12 | Sponge network | 0.959 | 0 | 0.174 | 0.13271 | 0.287 |
| 44480 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-508-3p;hsa-miR-96-5p | 28 | FBXO32 | Sponge network | -0.18 | 0.20343 | -0.495 | 0.07561 | 0.287 |
| 44481 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-744-3p | 17 | HIP1 | Sponge network | 0.859 | 0.00331 | 0.774 | 0 | 0.287 |
| 44482 | RP11-356J5.12 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-940 | 22 | BAZ2B | Sponge network | -0.899 | 6.0E-5 | -0.276 | 0.02833 | 0.287 |
| 44483 | HOTAIRM1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ITGB1 | Sponge network | -0.053 | 0.80685 | 0.524 | 0.00017 | 0.287 |
| 44484 | HOXB-AS3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-550a-3p | 13 | BRD3 | Sponge network | 0.343 | 0.28887 | 0.37 | 0.00013 | 0.287 |
| 44485 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | AFF4 | Sponge network | 0.817 | 3.0E-5 | -0.266 | 0.01565 | 0.287 |
| 44486 | ACTA2-AS1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-96-5p | 18 | IRS1 | Sponge network | 0.194 | 0.64278 | -0.026 | 0.88855 | 0.287 |
| 44487 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 15 | PRKAA2 | Sponge network | -0.557 | 0.11135 | -2.462 | 0 | 0.287 |
| 44488 | C4A-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 11 | NAV3 | Sponge network | 3.345 | 0 | 0.212 | 0.47729 | 0.287 |
| 44489 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-493-5p | 17 | ZFX | Sponge network | 1.46 | 1.0E-5 | 0.09 | 0.42563 | 0.287 |
| 44490 | TUG1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-664a-3p | 24 | CCDC6 | Sponge network | 0.972 | 0 | 0.27 | 0.02555 | 0.287 |
| 44491 | SNHG16 |
hsa-miR-100-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-337-3p | 14 | RB1 | Sponge network | 0.961 | 0 | 0.382 | 0.0017 | 0.287 |
| 44492 | RP11-693J15.4 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-484;hsa-miR-590-5p | 14 | CADM1 | Sponge network | -0.72 | 0.24239 | -0.9 | 0.00084 | 0.287 |
| 44493 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-362-3p | 13 | DKK3 | Sponge network | -0.227 | 0.31657 | -0.052 | 0.77783 | 0.287 |
| 44494 | SNHG14 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 24 | ERRFI1 | Sponge network | -1.111 | 0.0003 | -0.798 | 1.0E-5 | 0.287 |
| 44495 | CTD-2184D3.6 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PIK3CA | Sponge network | 0.272 | 0.44692 | 0.195 | 0.06908 | 0.287 |
| 44496 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 20 | AKAP6 | Sponge network | 0.614 | 1.0E-5 | -1.586 | 3.0E-5 | 0.287 |
| 44497 | CTC-471J1.2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 10 | PIK3CA | Sponge network | 1.11 | 1.0E-5 | 0.195 | 0.06908 | 0.287 |
| 44498 | RP11-540B6.6 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-331-3p | 12 | ABI2 | Sponge network | 0.93 | 0 | 0.175 | 0.14787 | 0.287 |
| 44499 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3913-5p;hsa-miR-7-5p;hsa-miR-93-5p | 26 | ZBTB4 | Sponge network | -0.83 | 0 | -0.739 | 0 | 0.287 |
| 44500 | FAM157C |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-539-5p | 12 | PRKAA1 | Sponge network | 0.91 | 0 | 0.317 | 0.0031 | 0.287 |
| 44501 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-30b-5p;hsa-miR-409-5p;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | LIMD1 | Sponge network | 0.064 | 0.72934 | 0.103 | 0.28978 | 0.287 |
| 44502 | MIR600HG |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-96-5p | 12 | WNK2 | Sponge network | 0.594 | 0.41522 | -0.352 | 0.31066 | 0.287 |
| 44503 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-7-5p | 29 | MOB1B | Sponge network | -0.384 | 0.40652 | 0.197 | 0.15266 | 0.287 |
| 44504 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | YAP1 | Sponge network | -0.487 | 0.02157 | 0.22 | 0.11836 | 0.287 |
| 44505 | RP11-1277A3.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p | 14 | CDH23 | Sponge network | 0.453 | 0.01839 | -0.974 | 0.00034 | 0.287 |
| 44506 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-93-3p | 13 | RPS6KA2 | Sponge network | 0.056 | 0.81195 | -0.57 | 0.00013 | 0.287 |
| 44507 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | TRIP11 | Sponge network | 0.919 | 0 | -0.158 | 0.06384 | 0.287 |
| 44508 | RP11-35G9.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PRUNE2 | Sponge network | 0.657 | 4.0E-5 | -1.733 | 0.00022 | 0.287 |
| 44509 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-577;hsa-miR-7-1-3p | 22 | EPHA3 | Sponge network | 0.389 | 0.04293 | 0.185 | 0.53588 | 0.287 |
| 44510 | RP4-791M13.3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | PCDH10 | Sponge network | 0.419 | 0.0319 | -1.644 | 0.0078 | 0.287 |
| 44511 | CTC-297N7.9 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-34a-5p | 10 | SYT4 | Sponge network | -2.069 | 0 | -3.207 | 0 | 0.287 |
| 44512 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | PDGFC | Sponge network | -2.925 | 0 | -0.33 | 0.06483 | 0.287 |
| 44513 | CTD-2314G24.2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | EGR2 | Sponge network | 0.246 | 0.59912 | 0.309 | 0.17079 | 0.287 |
| 44514 | RP11-400K9.4 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-542-3p;hsa-miR-7-1-3p | 15 | EFNA5 | Sponge network | -0.611 | 0.09607 | -0.421 | 0.17868 | 0.287 |
| 44515 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | SLITRK3 | Sponge network | -0.513 | 0.04703 | -3.328 | 0 | 0.287 |
| 44516 | LINC01018 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-671-5p | 12 | EHD3 | Sponge network | -2.915 | 0.00015 | -0.484 | 0.01747 | 0.287 |
| 44517 | LINC01082 |
hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p | 11 | THRB | Sponge network | -3.322 | 8.0E-5 | -1.117 | 0.0004 | 0.287 |
| 44518 | AC003090.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 24 | CYLD | Sponge network | -2.117 | 0.00161 | -0.367 | 0.00171 | 0.287 |
| 44519 | A2M-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | UNC5C | Sponge network | -0.08 | 0.90092 | -0.178 | 0.40214 | 0.287 |
| 44520 | RP11-218M22.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | BRD3 | Sponge network | 0.197 | 0.25618 | 0.37 | 0.00013 | 0.287 |
| 44521 | EMX2OS |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 13 | RIMS1 | Sponge network | -0.777 | 0.1134 | -3.143 | 0 | 0.287 |
| 44522 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p;hsa-miR-98-5p | 29 | LRIG1 | Sponge network | -0.942 | 0 | -0.537 | 0.00588 | 0.286 |
| 44523 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | APC | Sponge network | 0.826 | 1.0E-5 | -0.255 | 0.04051 | 0.286 |
| 44524 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | SLITRK3 | Sponge network | -0.663 | 0.10887 | -3.328 | 0 | 0.286 |
| 44525 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p | 12 | JAZF1 | Sponge network | -0.164 | 0.45106 | -0.377 | 0.02677 | 0.286 |
| 44526 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p | 18 | ERCC4 | Sponge network | 0.889 | 9.0E-5 | 0.126 | 0.15266 | 0.286 |
| 44527 | ADIRF-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | CSMD1 | Sponge network | -0.223 | 0.47816 | -0.63 | 0.18881 | 0.286 |
| 44528 | SOX2-OT |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SEMA3E | Sponge network | -1.264 | 6.0E-5 | -1.717 | 0.00137 | 0.286 |
| 44529 | RP11-890B15.3 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | SPOP | Sponge network | 0.101 | 0.60919 | -0.419 | 1.0E-5 | 0.286 |
| 44530 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | ANK2 | Sponge network | -0.235 | 0.15142 | -1.603 | 1.0E-5 | 0.286 |
| 44531 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-942-5p | 30 | HLF | Sponge network | -0.727 | 0.04726 | -2.443 | 0 | 0.286 |
| 44532 | FZD10-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-378c;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-5p | 30 | ERC1 | Sponge network | -0.727 | 0.04726 | -0.108 | 0.38794 | 0.286 |
| 44533 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | ERG | Sponge network | -0.612 | 0.00094 | 0.002 | 0.99011 | 0.286 |
| 44534 | GABPB1-AS1 |
hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-369-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p | 10 | RNF111 | Sponge network | 1.11 | 0 | 0.098 | 0.21093 | 0.286 |
| 44535 | GUSBP11 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p | 17 | ARHGEF12 | Sponge network | 0.856 | 0 | 0.09 | 0.35693 | 0.286 |
| 44536 | LINC00578 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-500a-3p;hsa-miR-589-3p | 19 | MOB1B | Sponge network | 0.811 | 0.03228 | 0.197 | 0.15266 | 0.286 |
| 44537 | AC005562.1 |
hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 16 | PRKAR1A | Sponge network | 0.488 | 0.00084 | -0.063 | 0.50314 | 0.286 |
| 44538 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | ST6GALNAC3 | Sponge network | 0.194 | 0.64278 | -0.987 | 0 | 0.286 |
| 44539 | AC127904.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-590-3p | 22 | CCDC6 | Sponge network | 1.308 | 0 | 0.27 | 0.02555 | 0.286 |
| 44540 | RP11-410L14.2 |
hsa-let-7g-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-497-5p | 13 | PPM1A | Sponge network | -0.052 | 0.76839 | -0.623 | 0 | 0.286 |
| 44541 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-93-5p | 14 | TRIM3 | Sponge network | -1.203 | 0.03881 | -0.577 | 0 | 0.286 |
| 44542 | LINC00641 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940 | 14 | FOXO4 | Sponge network | -0.222 | 0.32445 | -0.516 | 2.0E-5 | 0.286 |
| 44543 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-331-5p | 17 | BHLHE41 | Sponge network | -0.693 | 0.05629 | 0.353 | 0.12753 | 0.286 |
| 44544 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 31 | PAFAH1B1 | Sponge network | -0.176 | 0.59692 | -0.429 | 0 | 0.286 |
| 44545 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 28 | SPRY3 | Sponge network | 0.848 | 0 | 0.016 | 0.91286 | 0.286 |
| 44546 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429 | 21 | RNASEL | Sponge network | -0.125 | 0.49414 | -0.272 | 0.01796 | 0.286 |
| 44547 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-744-3p;hsa-miR-96-5p | 38 | FAT3 | Sponge network | 0.853 | 0.15352 | -1.601 | 0.00051 | 0.286 |
| 44548 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p | 24 | MED12L | Sponge network | 0.253 | 0.09565 | -0.194 | 0.55299 | 0.286 |
| 44549 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 16 | TXNIP | Sponge network | 0.082 | 0.55654 | -1.277 | 0 | 0.286 |
| 44550 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SYNE1 | Sponge network | -0.031 | 0.89063 | -0.738 | 0.00316 | 0.286 |
| 44551 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | EBF1 | Sponge network | 0.765 | 7.0E-5 | -0.384 | 0.08856 | 0.286 |
| 44552 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-199a-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-34c-5p;hsa-miR-493-5p | 11 | MIER3 | Sponge network | -0.567 | 0 | -0.212 | 0.04957 | 0.286 |
| 44553 | RP11-326G21.1 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | LIMCH1 | Sponge network | -0.021 | 0.93598 | -0.915 | 1.0E-5 | 0.286 |
| 44554 | A2M-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZMYND11 | Sponge network | -0.08 | 0.90092 | -0.2 | 0.05449 | 0.286 |
| 44555 | RP11-244H3.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | DMD | Sponge network | 0.659 | 3.0E-5 | -1.506 | 0 | 0.286 |
| 44556 | RP11-1277A3.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-409-3p;hsa-miR-505-3p;hsa-miR-92a-3p | 20 | AFF1 | Sponge network | 0.453 | 0.01839 | -0.384 | 0.00053 | 0.286 |
| 44557 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 38 | TCF4 | Sponge network | 0.389 | 0.04293 | 0.016 | 0.92271 | 0.286 |
| 44558 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 15 | ERCC4 | Sponge network | 0.727 | 3.0E-5 | 0.126 | 0.15266 | 0.286 |
| 44559 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p | 18 | CUL5 | Sponge network | 1.044 | 2.0E-5 | 0.026 | 0.77301 | 0.286 |
| 44560 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p | 35 | BNC2 | Sponge network | 0.051 | 0.83388 | -0.491 | 0.09988 | 0.286 |
| 44561 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-93-5p | 27 | LRP1B | Sponge network | -0.465 | 0.04703 | -2.163 | 0 | 0.286 |
| 44562 | RP11-1277A3.1 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-96-5p | 13 | HIP1 | Sponge network | 0.453 | 0.01839 | 0.774 | 0 | 0.286 |
| 44563 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 36 | MCC | Sponge network | -0.303 | 0.21002 | -1.005 | 0 | 0.286 |
| 44564 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-5p | 12 | PDZD2 | Sponge network | 0.082 | 0.55654 | -0.909 | 3.0E-5 | 0.286 |
| 44565 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | CCND2 | Sponge network | -0.737 | 0.10822 | -0.267 | 0.3112 | 0.286 |
| 44566 | RP11-384K6.6 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-654-3p;hsa-miR-940 | 14 | BAZ2B | Sponge network | 0.946 | 0 | -0.276 | 0.02833 | 0.286 |
| 44567 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TBX18 | Sponge network | 0.537 | 0.00139 | 0.843 | 0.02945 | 0.286 |
| 44568 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-7-1-3p | 15 | RAP1A | Sponge network | 0.199 | 0.52763 | -0.929 | 0 | 0.286 |
| 44569 | RP1-257A7.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-5p | 13 | RUNX1T1 | Sponge network | 0.849 | 0.00178 | -0.344 | 0.2374 | 0.286 |
| 44570 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | FZD3 | Sponge network | 0.101 | 0.60919 | 0.104 | 0.60316 | 0.286 |
| 44571 | GNG12-AS1 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | GABRB2 | Sponge network | -0.793 | 7.0E-5 | -1.841 | 0.00029 | 0.286 |
| 44572 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-502-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 24 | NTRK3 | Sponge network | 0.912 | 0.00014 | -1.77 | 3.0E-5 | 0.286 |
| 44573 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-940;hsa-miR-96-5p | 14 | LIFR | Sponge network | 0.622 | 0.00015 | -1.794 | 0 | 0.286 |
| 44574 | CTC-429P9.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-532-5p;hsa-miR-942-5p | 28 | IGFBP5 | Sponge network | 0.082 | 0.55397 | -0.806 | 0.00105 | 0.286 |
| 44575 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p | 19 | LRRC55 | Sponge network | -0.773 | 0.04073 | -0.026 | 0.93163 | 0.286 |
| 44576 | PWAR6 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CDH11 | Sponge network | -1.575 | 6.0E-5 | 1.427 | 0 | 0.286 |
| 44577 | RP11-5C23.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 11 | STARD13 | Sponge network | 0.274 | 0.07681 | -0.187 | 0.27383 | 0.286 |
| 44578 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 13 | ARHGAP29 | Sponge network | -0.227 | 0.31657 | 0.131 | 0.42953 | 0.286 |
| 44579 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-500a-3p | 30 | RUNX1T1 | Sponge network | -4.477 | 0 | -0.344 | 0.2374 | 0.286 |
| 44580 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CACNA2D1 | Sponge network | -0.021 | 0.93598 | -0.846 | 0.00675 | 0.286 |
| 44581 | RP11-244H3.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | DDX5 | Sponge network | 0.659 | 3.0E-5 | -0.099 | 0.17428 | 0.286 |
| 44582 | RASSF8-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p | 16 | ZFP36L1 | Sponge network | 0.335 | 0.20596 | -0.505 | 8.0E-5 | 0.286 |
| 44583 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | ERCC4 | Sponge network | -0.738 | 0.21233 | 0.126 | 0.15266 | 0.286 |
| 44584 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 15 | CRTC1 | Sponge network | -0.358 | 0.17255 | -0.116 | 0.28412 | 0.286 |
| 44585 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-454-3p | 13 | CLIP1 | Sponge network | -0.384 | 0.40652 | -0.215 | 0.08684 | 0.286 |
| 44586 | IQCH-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-486-5p;hsa-miR-664a-5p | 17 | CBFB | Sponge network | 0.929 | 0 | 1.061 | 0 | 0.286 |
| 44587 | THAP9-AS1 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-654-3p | 10 | DICER1 | Sponge network | 1.079 | 0 | 0.042 | 0.674 | 0.286 |
| 44588 | AC074289.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-375;hsa-miR-409-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 31 | TRPS1 | Sponge network | -0.326 | 0.14314 | -0.191 | 0.44109 | 0.286 |
| 44589 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-629-5p;hsa-miR-92a-1-5p | 19 | CTIF | Sponge network | -0.418 | 0.00068 | -0.389 | 0.00549 | 0.286 |
| 44590 | DNAJC27-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 23 | CDH23 | Sponge network | -0.969 | 2.0E-5 | -0.974 | 0.00034 | 0.286 |
| 44591 | TTC28-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-628-5p | 15 | TRIM33 | Sponge network | 0.373 | 0.00068 | 0.402 | 7.0E-5 | 0.286 |
| 44592 | MIR143HG |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | CSRNP1 | Sponge network | -0.739 | 0.0574 | -1.448 | 0 | 0.286 |
| 44593 | CTA-14H9.5 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 11 | PPP1R9A | Sponge network | 0.145 | 0.53343 | -0.903 | 0.04254 | 0.286 |
| 44594 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-362-3p | 25 | SSBP2 | Sponge network | -0.829 | 0.01343 | -1.58 | 0 | 0.286 |
| 44595 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | USP6 | Sponge network | 0.476 | 0.19203 | 0.188 | 0.3871 | 0.286 |
| 44596 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | SLITRK6 | Sponge network | -0.769 | 0.00035 | -2.121 | 0 | 0.286 |
| 44597 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | NEDD4 | Sponge network | -4.546 | 0 | 0.02 | 0.89834 | 0.286 |
| 44598 | CTD-2303H24.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | HECA | Sponge network | 0.415 | 0.14657 | -0.217 | 0.03463 | 0.286 |
| 44599 | DIO3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-671-5p;hsa-miR-98-5p | 31 | CREB3L2 | Sponge network | -0.773 | 0.04073 | -0.525 | 2.0E-5 | 0.286 |
| 44600 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-92a-3p | 14 | DMXL1 | Sponge network | 0.912 | 0.00014 | -0.426 | 0.00056 | 0.286 |
| 44601 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-744-3p | 20 | PTPN14 | Sponge network | 0.395 | 0.15451 | 0.144 | 0.34595 | 0.286 |
| 44602 | RP11-2B6.2 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 39 | LPP | Sponge network | 0.539 | 0.03722 | -0.459 | 0.04475 | 0.286 |
| 44603 | DIO3OS |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p | 10 | KCNAB1 | Sponge network | -0.773 | 0.04073 | -0.755 | 2.0E-5 | 0.286 |
| 44604 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-589-3p;hsa-miR-590-3p | 31 | CCND2 | Sponge network | -0.358 | 0.17255 | -0.267 | 0.3112 | 0.286 |
| 44605 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 16 | PIK3C2A | Sponge network | 0.67 | 0.00048 | 0.061 | 0.53776 | 0.286 |
| 44606 | RP11-225B17.2 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p | 17 | LPP | Sponge network | 1.055 | 0 | -0.459 | 0.04475 | 0.286 |
| 44607 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-335-5p | 13 | IL17RD | Sponge network | -0.285 | 0.2172 | -0.019 | 0.94873 | 0.286 |
| 44608 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 17 | KIF1B | Sponge network | 0.832 | 0 | -0.118 | 0.32258 | 0.286 |
| 44609 | LINC00094 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-92a-3p | 12 | RASSF3 | Sponge network | 0.253 | 0.09565 | -0.226 | 0.11691 | 0.286 |
| 44610 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421 | 19 | RASSF2 | Sponge network | -1.942 | 0 | -0.273 | 0.21961 | 0.286 |
| 44611 | PART1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p | 10 | PEA15 | Sponge network | -2.925 | 0 | 0.009 | 0.93318 | 0.286 |
| 44612 | CTC-297N7.9 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-590-5p;hsa-miR-7-5p | 12 | BTG2 | Sponge network | -2.069 | 0 | -1.763 | 0 | 0.286 |
| 44613 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p;hsa-miR-98-5p | 28 | PRRX1 | Sponge network | 0.101 | 0.60919 | 1.316 | 0 | 0.286 |
| 44614 | RP11-774O3.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SEMA3E | Sponge network | -0.487 | 0.02157 | -1.717 | 0.00137 | 0.286 |
| 44615 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | -0.405 | 0.22013 | 0.943 | 0 | 0.286 |
| 44616 | AC007277.3 |
hsa-let-7a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-625-3p | 10 | ZFHX3 | Sponge network | 2.377 | 0.00108 | 0.389 | 0.01363 | 0.286 |
| 44617 | RP11-356J5.12 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-423-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 44 | GNAQ | Sponge network | -0.899 | 6.0E-5 | -0.367 | 0.00102 | 0.286 |
| 44618 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 20 | BCL11A | Sponge network | 0.374 | 0.20054 | -0.932 | 0.0063 | 0.286 |
| 44619 | RP3-430N8.11 |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-539-5p | 11 | KIF1B | Sponge network | 0.973 | 0 | -0.118 | 0.32258 | 0.286 |
| 44620 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p | 20 | ERBB4 | Sponge network | 0.178 | 0.29106 | -1.975 | 2.0E-5 | 0.286 |
| 44621 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-935 | 25 | FBXO32 | Sponge network | -0.513 | 0.04703 | -0.495 | 0.07561 | 0.286 |
| 44622 | RP11-360F5.3 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-877-5p | 11 | AFF1 | Sponge network | 1.867 | 0 | -0.384 | 0.00053 | 0.286 |
| 44623 | RP11-262H14.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | NIN | Sponge network | -0.121 | 0.53195 | -0.253 | 0.13794 | 0.286 |
| 44624 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-590-3p | 18 | ACSL6 | Sponge network | -1.582 | 8.0E-5 | -1.593 | 0 | 0.286 |
| 44625 | KB-1572G7.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | RPS6KA3 | Sponge network | 0.924 | 0 | 0.256 | 0.02419 | 0.286 |
| 44626 | FAM66C |
hsa-miR-130b-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p | 11 | ROBO2 | Sponge network | -0.901 | 0.00019 | 0.092 | 0.78768 | 0.286 |
| 44627 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-7-1-3p | 16 | EBF1 | Sponge network | 0.419 | 0.0319 | -0.384 | 0.08856 | 0.286 |
| 44628 | RP11-307B6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p | 20 | PTPRD | Sponge network | -4.463 | 0.00025 | -1.005 | 0.00064 | 0.286 |
| 44629 | RP11-116O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PLSCR4 | Sponge network | 0.05 | 0.95483 | -0.474 | 0.03833 | 0.286 |
| 44630 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-629-3p | 20 | RASSF2 | Sponge network | -1.054 | 0.0706 | -0.273 | 0.21961 | 0.286 |
| 44631 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p | 10 | CAV1 | Sponge network | 0.051 | 0.83388 | -1.013 | 6.0E-5 | 0.286 |
| 44632 | RP11-999E24.3 |
hsa-let-7d-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-33a-3p | 10 | TLE4 | Sponge network | -0.693 | 0.05629 | -1.157 | 0 | 0.286 |
| 44633 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p | 14 | NEDD4 | Sponge network | 0.368 | 0.20093 | 0.02 | 0.89834 | 0.286 |
| 44634 | JAZF1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 25 | RNASEL | Sponge network | -0.769 | 0.00035 | -0.272 | 0.01796 | 0.286 |
| 44635 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-935 | 19 | LRRC55 | Sponge network | -1.249 | 7.0E-5 | -0.026 | 0.93163 | 0.286 |
| 44636 | AC009948.5 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 19 | BAZ2B | Sponge network | 0.532 | 0.00505 | -0.276 | 0.02833 | 0.286 |
| 44637 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p | 17 | SCN9A | Sponge network | 0.921 | 0.00118 | -0.525 | 0.10593 | 0.286 |
| 44638 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 21 | CUL5 | Sponge network | 0.064 | 0.72934 | 0.026 | 0.77301 | 0.286 |
| 44639 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-424-5p | 11 | UBE4B | Sponge network | 1.089 | 0 | -0.235 | 0.007 | 0.286 |
| 44640 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 15 | ACTB | Sponge network | -0.129 | 0.57177 | -0.247 | 0.01736 | 0.286 |
| 44641 | OIP5-AS1 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | INPP4A | Sponge network | 0.421 | 0.00084 | 0.07 | 0.53563 | 0.286 |
| 44642 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-589-5p;hsa-miR-590-3p | 31 | ZFHX3 | Sponge network | -0.738 | 0.21233 | 0.389 | 0.01363 | 0.286 |
| 44643 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p | 10 | ROBO2 | Sponge network | 0.889 | 9.0E-5 | 0.092 | 0.78768 | 0.286 |
| 44644 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-424-5p | 10 | USP12 | Sponge network | -2.076 | 0.00548 | -0.102 | 0.40768 | 0.286 |
| 44645 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-335-3p;hsa-miR-96-5p | 12 | ELMO1 | Sponge network | 0.101 | 0.60919 | 0.04 | 0.83147 | 0.286 |
| 44646 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | NUAK1 | Sponge network | 0.61 | 0.01593 | 0.904 | 0 | 0.286 |
| 44647 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-5p | 19 | PIK3R1 | Sponge network | 1.153 | 0.00114 | -0.133 | 0.36854 | 0.286 |
| 44648 | PSMD5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 25 | ABI2 | Sponge network | 0.569 | 0.00067 | 0.175 | 0.14787 | 0.286 |
| 44649 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 22 | BMPR2 | Sponge network | 1.153 | 0.00114 | 0.206 | 0.0635 | 0.286 |
| 44650 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 18 | PRKAA1 | Sponge network | 1.601 | 0 | 0.317 | 0.0031 | 0.286 |
| 44651 | A2M-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-425-5p | 21 | ERC1 | Sponge network | -0.08 | 0.90092 | -0.108 | 0.38794 | 0.286 |
| 44652 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | RPS6KA3 | Sponge network | 1.089 | 0 | 0.256 | 0.02419 | 0.286 |
| 44653 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 29 | AKAP11 | Sponge network | 0.4 | 0.14611 | 0.196 | 0.09825 | 0.286 |
| 44654 | RP11-458D21.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 19 | DIP2C | Sponge network | 0.663 | 0.00073 | -0.436 | 0.01713 | 0.286 |
| 44655 | BOLA3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-629-3p | 18 | RYR2 | Sponge network | -0.246 | 0.35034 | -1.466 | 0.00031 | 0.286 |
| 44656 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-93-5p | 30 | PTPRD | Sponge network | -1.654 | 0.00144 | -1.005 | 0.00064 | 0.286 |
| 44657 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | HDAC9 | Sponge network | -0.021 | 0.93598 | -0.755 | 0.00495 | 0.286 |
| 44658 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-486-5p | 14 | IGF1 | Sponge network | 1.61 | 0.00961 | -0.204 | 0.5898 | 0.286 |
| 44659 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-369-3p | 17 | SMAD4 | Sponge network | 0.062 | 0.55274 | -0.313 | 0.00413 | 0.286 |
| 44660 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 11 | PTPN11 | Sponge network | 0.538 | 0.00076 | 0.431 | 2.0E-5 | 0.286 |
| 44661 | RP11-3P17.4 |
hsa-let-7a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p | 11 | ZNF770 | Sponge network | 0.144 | 0.07137 | 0.206 | 0.06751 | 0.286 |
| 44662 | FENDRR |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 11 | PIK3IP1 | Sponge network | -1.281 | 0.00012 | -0.187 | 0.19945 | 0.286 |
| 44663 | LINC00680 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 0.884 | 0 | 0.362 | 0.00066 | 0.286 |
| 44664 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 37 | PTPRT | Sponge network | 0.194 | 0.64278 | -0.907 | 0.03727 | 0.286 |
| 44665 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-338-5p | 10 | MYBL1 | Sponge network | 0.752 | 0 | 0.494 | 0.02152 | 0.286 |
| 44666 | U91328.19 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-532-3p | 17 | CADM3 | Sponge network | -0.303 | 0.21002 | -3.663 | 0 | 0.286 |
| 44667 | RP11-981G7.6 |
hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-590-3p | 10 | BAZ2B | Sponge network | 0.986 | 0.01022 | -0.276 | 0.02833 | 0.286 |
| 44668 | KB-1460A1.5 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | 0.603 | 0.00127 | 0.098 | 0.40994 | 0.286 |
| 44669 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TRIM13 | Sponge network | 0.101 | 0.60919 | 0.183 | 0.06968 | 0.286 |
| 44670 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-493-5p | 18 | NEDD4 | Sponge network | -2.62 | 0 | 0.02 | 0.89834 | 0.286 |
| 44671 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 10 | HERC2 | Sponge network | 1.757 | 0 | 0.114 | 0.30638 | 0.286 |
| 44672 | A2M-AS1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-484 | 15 | ABL1 | Sponge network | -0.08 | 0.90092 | -0.215 | 0.07804 | 0.286 |
| 44673 | A1BG-AS1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-671-5p | 13 | RBMS3 | Sponge network | -0.164 | 0.45106 | -0.519 | 0.03551 | 0.286 |
| 44674 | MIR4458HG |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-19a-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 10 | CELF4 | Sponge network | -0.427 | 0.0623 | -1.135 | 0.00344 | 0.286 |
| 44675 | AC127904.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-539-5p | 11 | PRKAA1 | Sponge network | 1.308 | 0 | 0.317 | 0.0031 | 0.286 |
| 44676 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | RNF144A | Sponge network | -1.349 | 0.00017 | 0.594 | 0.0009 | 0.286 |
| 44677 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-590-3p | 10 | RB1CC1 | Sponge network | 0.272 | 0.44692 | 0.175 | 0.12559 | 0.286 |
| 44678 | CTC-296K1.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-493-5p;hsa-miR-582-5p | 16 | PIK3R1 | Sponge network | -2.095 | 0.00112 | -0.133 | 0.36854 | 0.286 |
| 44679 | RP11-359B12.2 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SH3GLB1 | Sponge network | 0.064 | 0.72934 | -0.115 | 0.14773 | 0.286 |
| 44680 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NOTCH2NL | Sponge network | -0.031 | 0.89063 | 0.024 | 0.84307 | 0.286 |
| 44681 | RP11-536O18.1 |
hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-362-5p | 17 | IL6ST | Sponge network | -0.074 | 0.90758 | -0.402 | 0.00676 | 0.286 |
| 44682 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 11 | BCLAF1 | Sponge network | 0.799 | 1.0E-5 | 0.211 | 0.00684 | 0.286 |
| 44683 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 21 | PRKAR1A | Sponge network | -2.46 | 0 | -0.063 | 0.50314 | 0.286 |
| 44684 | RP11-798M19.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 29 | SNX29 | Sponge network | -0.612 | 0.00094 | -0.34 | 0.01371 | 0.286 |
| 44685 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 14 | SON | Sponge network | -0.031 | 0.89063 | 0.168 | 0.04406 | 0.286 |
| 44686 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-708-5p;hsa-miR-96-5p | 36 | SPRY3 | Sponge network | 0.194 | 0.64278 | 0.016 | 0.91286 | 0.286 |
| 44687 | RP11-264B17.2 |
hsa-miR-129-5p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 10 | CCDC6 | Sponge network | 1.82 | 0 | 0.27 | 0.02555 | 0.286 |
| 44688 | LINC00092 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-425-5p | 13 | ERC1 | Sponge network | -1.886 | 0 | -0.108 | 0.38794 | 0.286 |
| 44689 | RP11-404E16.1 |
hsa-miR-125a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-940 | 14 | MDM4 | Sponge network | 0.097 | 0.91311 | 0.345 | 0.00762 | 0.286 |
| 44690 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-671-5p | 13 | CRTC3 | Sponge network | -0.278 | 0.76559 | 0.164 | 0.16786 | 0.286 |
| 44691 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-942-5p | 38 | TMEM132B | Sponge network | -1.047 | 0 | -0.83 | 0.01084 | 0.286 |
| 44692 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-590-5p;hsa-miR-93-5p | 17 | TGFBR2 | Sponge network | -0.384 | 0.40652 | -0.243 | 0.11408 | 0.286 |
| 44693 | CTD-2336O2.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NAV3 | Sponge network | -0.141 | 0.26434 | 0.212 | 0.47729 | 0.286 |
| 44694 | CTC-297N7.9 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-590-5p | 14 | PDGFRA | Sponge network | -2.069 | 0 | -0.372 | 0.0037 | 0.286 |
| 44695 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 17 | MITF | Sponge network | 0.687 | 0.02753 | -0.769 | 0.00022 | 0.286 |
| 44696 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p | 19 | CLASP2 | Sponge network | 1.03 | 0 | -0.038 | 0.71 | 0.286 |
| 44697 | LINC01018 |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | PAIP2 | Sponge network | -2.915 | 0.00015 | -0.515 | 0 | 0.286 |
| 44698 | RP11-66B24.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b | 12 | ZFHX3 | Sponge network | 0.57 | 0.1866 | 0.389 | 0.01363 | 0.286 |
| 44699 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 27 | PRKCE | Sponge network | -2.117 | 0.00161 | -0.403 | 0.00056 | 0.286 |
| 44700 | RP11-395G23.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-361-3p;hsa-miR-590-3p | 15 | ITPKB | Sponge network | 0.381 | 0.25967 | -0.704 | 0.00106 | 0.286 |
| 44701 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PRKAB2 | Sponge network | 0.538 | 0.00076 | -0.367 | 0.01371 | 0.286 |
| 44702 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-3p | 15 | PTPN11 | Sponge network | -0.323 | 0.01372 | 0.431 | 2.0E-5 | 0.286 |
| 44703 | C4B-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-744-3p;hsa-miR-744-5p | 11 | IGF2 | Sponge network | 2.306 | 9.0E-5 | 0.848 | 0.03842 | 0.286 |
| 44704 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-655-3p | 16 | KANK1 | Sponge network | 0.253 | 0.09565 | -0.55 | 0.00134 | 0.286 |
| 44705 | HOTAIRM1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PIK3CA | Sponge network | -0.053 | 0.80685 | 0.195 | 0.06908 | 0.286 |
| 44706 | AC074289.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-375 | 25 | ADARB1 | Sponge network | -0.326 | 0.14314 | -0.335 | 0.06631 | 0.286 |
| 44707 | RP11-800A3.7 |
hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | AR | Sponge network | 0.18 | 0.7147 | -1.554 | 0 | 0.286 |
| 44708 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-340-5p;hsa-miR-495-3p | 12 | NF1 | Sponge network | 0.331 | 0.57247 | 0.51 | 0 | 0.286 |
| 44709 | HCG22 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-501-5p | 15 | PRDM2 | Sponge network | 0.816 | 0.32368 | -0.258 | 0.00721 | 0.286 |
| 44710 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p | 16 | SLC6A15 | Sponge network | -0.384 | 0.40652 | -2.373 | 2.0E-5 | 0.286 |
| 44711 | POLR2J4 |
hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92b-3p | 11 | TP63 | Sponge network | -0.041 | 0.69032 | -0.363 | 0.38191 | 0.286 |
| 44712 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | NUAK1 | Sponge network | 0.349 | 0.01695 | 0.904 | 0 | 0.286 |
| 44713 | AC016747.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ABI2 | Sponge network | 0.199 | 0.38015 | 0.175 | 0.14787 | 0.286 |
| 44714 | HCG11 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-93-5p | 36 | PIK3R1 | Sponge network | 0.385 | 0.10067 | -0.133 | 0.36854 | 0.286 |
| 44715 | PSMG3-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-877-5p | 22 | ASTN1 | Sponge network | -0.125 | 0.49414 | -2.389 | 2.0E-5 | 0.286 |
| 44716 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-369-3p | 10 | JAKMIP2 | Sponge network | -0.384 | 0.40652 | -1.388 | 0 | 0.286 |
| 44717 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-501-3p;hsa-miR-589-5p | 16 | SLITRK3 | Sponge network | 0.395 | 0.15451 | -3.328 | 0 | 0.286 |
| 44718 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-93-5p | 11 | LHX6 | Sponge network | -0.235 | 0.15142 | -0.087 | 0.69193 | 0.286 |
| 44719 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p | 16 | RELN | Sponge network | -0.125 | 0.49414 | -2.403 | 0 | 0.286 |
| 44720 | RP4-669P10.16 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 12 | ABI2 | Sponge network | -0.301 | 0.06597 | 0.175 | 0.14787 | 0.286 |
| 44721 | DNAJC27-AS1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-93-3p | 31 | CTIF | Sponge network | -0.969 | 2.0E-5 | -0.389 | 0.00549 | 0.286 |
| 44722 | LINC00957 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 26 | AKAP11 | Sponge network | -0.421 | 0.0114 | 0.196 | 0.09825 | 0.286 |
| 44723 | RP11-434B12.1 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PCDH9 | Sponge network | -0.031 | 0.89063 | -1.823 | 4.0E-5 | 0.286 |
| 44724 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | PDGFRA | Sponge network | -0.777 | 0.1134 | -0.372 | 0.0037 | 0.286 |
| 44725 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-590-5p | 13 | SYNE1 | Sponge network | 3.345 | 0 | -0.738 | 0.00316 | 0.286 |
| 44726 | CKMT2-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 55 | DST | Sponge network | 0.082 | 0.55654 | -0.713 | 0.00096 | 0.286 |
| 44727 | CTC-457L16.2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p;hsa-miR-935 | 12 | ARNT2 | Sponge network | 1.153 | 0.00114 | -0.332 | 0.25851 | 0.286 |
| 44728 | CTA-14H9.5 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 10 | NEDD4 | Sponge network | 0.145 | 0.53343 | 0.02 | 0.89834 | 0.286 |
| 44729 | AC006116.24 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-93-5p | 17 | PPM1L | Sponge network | -0.473 | 0.07602 | -0.537 | 0.00616 | 0.286 |
| 44730 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | MAP3K12 | Sponge network | -0.811 | 2.0E-5 | -0.057 | 0.59369 | 0.286 |
| 44731 | CTC-429P9.2 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-24-3p;hsa-miR-532-5p;hsa-miR-539-5p | 10 | RIMS2 | Sponge network | 0.043 | 0.81628 | -1.365 | 0.00208 | 0.286 |
| 44732 | RP11-218M22.1 |
hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 16 | TET1 | Sponge network | 0.197 | 0.25618 | -0.135 | 0.52138 | 0.286 |
| 44733 | U91328.19 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p | 12 | ID4 | Sponge network | -0.303 | 0.21002 | -1.644 | 0 | 0.286 |
| 44734 | BMS1P20 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | CBL | Sponge network | 0.34 | 0.00273 | 0.291 | 0.01267 | 0.286 |
| 44735 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 58 | KIF1B | Sponge network | -0.777 | 0.1134 | -0.118 | 0.32258 | 0.286 |
| 44736 | RP4-639F20.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p | 23 | CBX5 | Sponge network | -0.517 | 0.02935 | 0.165 | 0.24446 | 0.286 |
| 44737 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 25 | CBL | Sponge network | -0.611 | 0.09607 | 0.291 | 0.01267 | 0.286 |
| 44738 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-5p | 18 | ARHGAP29 | Sponge network | -1.249 | 7.0E-5 | 0.131 | 0.42953 | 0.286 |
| 44739 | RP11-680C21.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | LUZP2 | Sponge network | -1.424 | 0.04346 | -0.056 | 0.89416 | 0.286 |
| 44740 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | DDX5 | Sponge network | -0.096 | 0.67958 | -0.099 | 0.17428 | 0.286 |
| 44741 | LINC00863 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 35 | ADARB1 | Sponge network | 0.189 | 0.13981 | -0.335 | 0.06631 | 0.286 |
| 44742 | CTC-457L16.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-505-5p | 10 | MRVI1 | Sponge network | 1.153 | 0.00114 | -1.051 | 0.00022 | 0.286 |
| 44743 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-654-3p | 13 | AKAP11 | Sponge network | 1.157 | 0.00455 | 0.196 | 0.09825 | 0.286 |
| 44744 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-5p | 36 | PIK3R1 | Sponge network | -0.777 | 0.16667 | -0.133 | 0.36854 | 0.286 |
| 44745 | RP11-248G5.8 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p | 11 | PAWR | Sponge network | 1.294 | 0 | 0.636 | 0 | 0.286 |
| 44746 | RP11-620J15.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-361-5p;hsa-miR-590-3p | 19 | ACSL6 | Sponge network | -0.739 | 0.00044 | -1.593 | 0 | 0.286 |
| 44747 | NDUFA6-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | ROBO1 | Sponge network | -0.355 | 0.0077 | -0.009 | 0.96154 | 0.286 |
| 44748 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | LRRC4 | Sponge network | 0.101 | 0.60919 | -1.774 | 0 | 0.286 |
| 44749 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 21 | MYO9A | Sponge network | 0.274 | 0.07681 | -0.147 | 0.32373 | 0.286 |
| 44750 | LINC00648 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-190a-5p;hsa-miR-223-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CXXC4 | Sponge network | 0.633 | 0.51678 | -0.433 | 0.21055 | 0.286 |
| 44751 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 24 | NUAK1 | Sponge network | -0.421 | 0.0114 | 0.904 | 0 | 0.286 |
| 44752 | RP11-770J1.3 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-505-5p | 12 | ASTN1 | Sponge network | -2.011 | 0.0005 | -2.389 | 2.0E-5 | 0.286 |
| 44753 | AC083843.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p | 11 | RPS6KA3 | Sponge network | 1.4 | 0 | 0.256 | 0.02419 | 0.286 |
| 44754 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-93-3p | 16 | ST6GAL2 | Sponge network | 0.622 | 0.00015 | -1.201 | 0.00597 | 0.286 |
| 44755 | GS1-124K5.11 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 14 | NGFR | Sponge network | 0.494 | 0.00339 | -1.271 | 0.01058 | 0.286 |
| 44756 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-589-3p | 11 | JDP2 | Sponge network | -4.463 | 0.00025 | -0.967 | 0 | 0.286 |
| 44757 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-576-5p | 18 | GREM1 | Sponge network | -0.727 | 0.04726 | 0.902 | 0.01691 | 0.286 |
| 44758 | CTD-3064M3.3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-5p | 12 | EHD3 | Sponge network | -0.469 | 0.08721 | -0.484 | 0.01747 | 0.286 |
| 44759 | FZD10-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 16 | ITGA5 | Sponge network | -0.727 | 0.04726 | 0.452 | 0.03524 | 0.286 |
| 44760 | ZNF582-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-7-1-3p | 10 | ZNF844 | Sponge network | -1.349 | 0.00017 | -0.966 | 0.00035 | 0.286 |
| 44761 | HOXB-AS3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p | 11 | DYNC1I1 | Sponge network | 0.343 | 0.28887 | -1.12 | 0.00107 | 0.286 |
| 44762 | CTB-133G6.2 |
hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-424-5p | 11 | RASSF2 | Sponge network | 0.4 | 0.39299 | -0.273 | 0.21961 | 0.286 |
| 44763 | RP11-678G14.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | TRPS1 | Sponge network | -4.477 | 0 | -0.191 | 0.44109 | 0.286 |
| 44764 | MYLK-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p | 15 | EFNA5 | Sponge network | -1.331 | 3.0E-5 | -0.421 | 0.17868 | 0.286 |
| 44765 | U91328.19 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-7-1-3p | 16 | TMEFF2 | Sponge network | -0.303 | 0.21002 | -2.426 | 0 | 0.286 |
| 44766 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 30 | CBX5 | Sponge network | 0.197 | 0.25618 | 0.165 | 0.24446 | 0.286 |
| 44767 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | PDS5B | Sponge network | 0.417 | 0.00445 | 0.259 | 0.00962 | 0.286 |
| 44768 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-485-3p;hsa-miR-7-1-3p | 15 | PHC3 | Sponge network | 0.199 | 0.52763 | 0.212 | 0.0499 | 0.286 |
| 44769 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 19 | BHLHE41 | Sponge network | -0.811 | 2.0E-5 | 0.353 | 0.12753 | 0.286 |
| 44770 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p | 12 | FREM2 | Sponge network | 0.419 | 0.0319 | -0.437 | 0.46353 | 0.286 |
| 44771 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-96-5p | 16 | RASSF8 | Sponge network | 0.898 | 1.0E-5 | -0.122 | 0.65591 | 0.286 |
| 44772 | TINCR |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-96-5p | 50 | KIF1B | Sponge network | -0.664 | 0.14543 | -0.118 | 0.32258 | 0.286 |
| 44773 | RP11-296I10.3 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-502-3p | 14 | NUAK1 | Sponge network | 0.592 | 0.00014 | 0.904 | 0 | 0.286 |
| 44774 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 14 | MYBL1 | Sponge network | 0.373 | 0.00068 | 0.494 | 0.02152 | 0.286 |
| 44775 | FZD10-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -0.727 | 0.04726 | -0.704 | 0.00106 | 0.286 |
| 44776 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-455-3p | 11 | DKK3 | Sponge network | 0.368 | 0.20093 | -0.052 | 0.77783 | 0.286 |
| 44777 | TP73-AS1 |
hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-429 | 10 | EGR1 | Sponge network | -0.563 | 0.02545 | -1.195 | 0 | 0.286 |
| 44778 | LINC00621 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 20 | BNC2 | Sponge network | 0.131 | 0.8436 | -0.491 | 0.09988 | 0.285 |
| 44779 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p | 15 | BCL11A | Sponge network | -2.69 | 0.0001 | -0.932 | 0.0063 | 0.285 |
| 44780 | RP11-967K21.1 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577 | 10 | CDH11 | Sponge network | 0.056 | 0.81195 | 1.427 | 0 | 0.285 |
| 44781 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-744-3p | 14 | HIP1 | Sponge network | 0.693 | 0.00076 | 0.774 | 0 | 0.285 |
| 44782 | FAM66C |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PRKAR1A | Sponge network | -0.901 | 0.00019 | -0.063 | 0.50314 | 0.285 |
| 44783 | GAS6-AS2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-3p | 16 | UBE4B | Sponge network | 0.16 | 0.50785 | -0.235 | 0.007 | 0.285 |
| 44784 | OR2A1-AS1 |
hsa-let-7a-3p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-511-5p;hsa-miR-577 | 12 | ATRX | Sponge network | 0.542 | 0.00241 | 0.261 | 0.04408 | 0.285 |
| 44785 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 47 | CBX5 | Sponge network | 0.374 | 0.20054 | 0.165 | 0.24446 | 0.285 |
| 44786 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-940 | 42 | CADM2 | Sponge network | 0.214 | 0.18246 | -3.782 | 0 | 0.285 |
| 44787 | FGD5-AS1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 12 | SH3GLB1 | Sponge network | 0.401 | 0.00066 | -0.115 | 0.14773 | 0.285 |
| 44788 | PCBP1-AS1 |
hsa-miR-16-2-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-5p | 10 | RIT1 | Sponge network | -0.567 | 0 | -0.296 | 0.00054 | 0.285 |
| 44789 | RP11-758N13.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-375;hsa-miR-93-3p | 18 | QKI | Sponge network | 2.939 | 3.0E-5 | -0.333 | 0.04111 | 0.285 |
| 44790 | RP11-46C24.7 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-484 | 13 | IGF2 | Sponge network | 0.392 | 0.0278 | 0.848 | 0.03842 | 0.285 |
| 44791 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 18 | CADM3 | Sponge network | -0.355 | 0.0077 | -3.663 | 0 | 0.285 |
| 44792 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-940 | 19 | CRTC3 | Sponge network | 0.75 | 0.00054 | 0.164 | 0.16786 | 0.285 |
| 44793 | H19 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 11 | WWTR1 | Sponge network | 2.439 | 0 | -0.345 | 0.11997 | 0.285 |
| 44794 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-495-3p | 11 | ZNF292 | Sponge network | 0.331 | 0.57247 | 0.276 | 0.04148 | 0.285 |
| 44795 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-7-1-3p | 21 | EBF1 | Sponge network | -0.176 | 0.59692 | -0.384 | 0.08856 | 0.285 |
| 44796 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-199a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-766-3p | 31 | MAPK1 | Sponge network | -0.222 | 0.32445 | -0.017 | 0.81465 | 0.285 |
| 44797 | RP11-597D13.9 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | GALNT13 | Sponge network | 1.302 | 7.0E-5 | -0.857 | 0.07439 | 0.285 |
| 44798 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 11 | PDGFC | Sponge network | 0.374 | 0.20054 | -0.33 | 0.06483 | 0.285 |
| 44799 | OIP5-AS1 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | PREX2 | Sponge network | 0.421 | 0.00084 | -0.272 | 0.25382 | 0.285 |
| 44800 | MYLK-AS1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-484 | 11 | WNK2 | Sponge network | -1.331 | 3.0E-5 | -0.352 | 0.31066 | 0.285 |
| 44801 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-590-3p | 13 | RBM5 | Sponge network | -0.739 | 0.0574 | -0.205 | 0.0129 | 0.285 |
| 44802 | RP11-345P4.7 |
hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | DDX3X | Sponge network | 1.597 | 0 | -0.152 | 0.07686 | 0.285 |
| 44803 | DNM1P35 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-590-3p | 13 | KLF10 | Sponge network | 0.051 | 0.83388 | -0.783 | 0 | 0.285 |
| 44804 | NNT-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CEP170 | Sponge network | 0.799 | 1.0E-5 | 0.943 | 0 | 0.285 |
| 44805 | HOXB-AS2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-429 | 10 | PRKAB2 | Sponge network | 1.316 | 0.01106 | -0.367 | 0.01371 | 0.285 |
| 44806 | DNAJC3-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 11 | GPAM | Sponge network | 0.753 | 0.00014 | 0.142 | 0.29732 | 0.285 |
| 44807 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p | 11 | CREB3L2 | Sponge network | -0.164 | 0.45106 | -0.525 | 2.0E-5 | 0.285 |
| 44808 | RP11-327P2.5 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-93-3p;hsa-miR-935 | 10 | STAT5B | Sponge network | -0.147 | 0.40452 | -0.182 | 0.1082 | 0.285 |
| 44809 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-874-3p | 18 | NR2C2 | Sponge network | -1.645 | 0 | 0.21 | 0.03224 | 0.285 |
| 44810 | RP11-536O18.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p | 15 | CADM2 | Sponge network | -0.074 | 0.90758 | -3.782 | 0 | 0.285 |
| 44811 | AP000487.5 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 11 | PHF6 | Sponge network | 1.987 | 0 | 0.785 | 0 | 0.285 |
| 44812 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | FOXO3 | Sponge network | 1.04 | 0.00023 | -0.04 | 0.72028 | 0.285 |
| 44813 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-96-5p | 13 | MTSS1 | Sponge network | -1.582 | 8.0E-5 | -0.81 | 0.00035 | 0.285 |
| 44814 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-539-5p | 12 | PDZD2 | Sponge network | 0.61 | 0.01593 | -0.909 | 3.0E-5 | 0.285 |
| 44815 | RP11-13K12.5 |
hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 17 | LIMCH1 | Sponge network | -0.318 | 0.65847 | -0.915 | 1.0E-5 | 0.285 |
| 44816 | RP4-717I23.3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-942-5p | 25 | ARHGEF12 | Sponge network | 0.637 | 0.00069 | 0.09 | 0.35693 | 0.285 |
| 44817 | NDUFA6-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | CBFA2T3 | Sponge network | -0.355 | 0.0077 | -1.221 | 1.0E-5 | 0.285 |
| 44818 | AC005562.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | ERCC4 | Sponge network | 0.488 | 0.00084 | 0.126 | 0.15266 | 0.285 |
| 44819 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-331-5p;hsa-miR-935 | 11 | RELN | Sponge network | -0.384 | 0.40652 | -2.403 | 0 | 0.285 |
| 44820 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 10 | FREM2 | Sponge network | 0.8 | 0 | -0.437 | 0.46353 | 0.285 |
| 44821 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | RASSF2 | Sponge network | -0.08 | 0.90092 | -0.273 | 0.21961 | 0.285 |
| 44822 | MIR600HG |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-484 | 25 | ZFX | Sponge network | 0.594 | 0.41522 | 0.09 | 0.42563 | 0.285 |
| 44823 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-539-5p | 30 | KIF1B | Sponge network | 0.592 | 0.00014 | -0.118 | 0.32258 | 0.285 |
| 44824 | A1BG-AS1 |
hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-589-3p | 11 | TET1 | Sponge network | -0.164 | 0.45106 | -0.135 | 0.52138 | 0.285 |
| 44825 | LINC00472 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-7-1-3p | 12 | ITGB1 | Sponge network | -0.842 | 0.01361 | 0.524 | 0.00017 | 0.285 |
| 44826 | RP4-614O4.11 |
hsa-miR-103a-2-5p;hsa-miR-1271-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-424-5p | 11 | ARHGEF12 | Sponge network | 1.042 | 0.00177 | 0.09 | 0.35693 | 0.285 |
| 44827 | LINC00476 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 39 | ACVR2A | Sponge network | -0.615 | 0.00015 | -0.409 | 0.00039 | 0.285 |
| 44828 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p | 30 | ZFHX3 | Sponge network | -0.777 | 0.16667 | 0.389 | 0.01363 | 0.285 |
| 44829 | LINC00648 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | DTNA | Sponge network | 0.633 | 0.51678 | -1.648 | 1.0E-5 | 0.285 |
| 44830 | RP11-195F19.9 |
hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | CUX1 | Sponge network | -0.726 | 0.00132 | -0.039 | 0.6705 | 0.285 |
| 44831 | AGAP11 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-629-3p;hsa-miR-93-3p | 17 | IKZF2 | Sponge network | -1.645 | 0 | -0.284 | 0.1942 | 0.285 |
| 44832 | SOX2-OT |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 32 | PCDH9 | Sponge network | -1.264 | 6.0E-5 | -1.823 | 4.0E-5 | 0.285 |
| 44833 | HAND2-AS1 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 18 | JUN | Sponge network | -1.697 | 0.00889 | -0.932 | 0 | 0.285 |
| 44834 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 34 | PTPRT | Sponge network | -0.487 | 0.02157 | -0.907 | 0.03727 | 0.285 |
| 44835 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-940;hsa-miR-98-5p | 27 | LIFR | Sponge network | 0.056 | 0.81195 | -1.794 | 0 | 0.285 |
| 44836 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 45 | PTPRD | Sponge network | 0.101 | 0.60919 | -1.005 | 0.00064 | 0.285 |
| 44837 | LINC00957 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | MTSS1 | Sponge network | -0.421 | 0.0114 | -0.81 | 0.00035 | 0.285 |
| 44838 | RP13-225O21.2 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 11 | CREB1 | Sponge network | 1.279 | 8.0E-5 | 0.203 | 0.00537 | 0.285 |
| 44839 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 17 | HECA | Sponge network | 0.395 | 0.15451 | -0.217 | 0.03463 | 0.285 |
| 44840 | LINC01004 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 13 | ARHGEF12 | Sponge network | 1.115 | 0 | 0.09 | 0.35693 | 0.285 |
| 44841 | RP11-506M13.3 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-582-3p | 12 | FOXO3 | Sponge network | 0.769 | 0 | -0.04 | 0.72028 | 0.285 |
| 44842 | RP5-1085F17.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 17 | TET1 | Sponge network | 0.541 | 0.00125 | -0.135 | 0.52138 | 0.285 |
| 44843 | CTD-2528L19.6 |
hsa-let-7d-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DICER1 | Sponge network | 0.953 | 0 | 0.042 | 0.674 | 0.285 |
| 44844 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p | 23 | PRDM2 | Sponge network | -0.513 | 0.04703 | -0.258 | 0.00721 | 0.285 |
| 44845 | USP3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-576-5p | 15 | RIMBP2 | Sponge network | -0.162 | 0.47687 | -1.968 | 5.0E-5 | 0.285 |
| 44846 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429 | 13 | ITGAV | Sponge network | -0.154 | 0.53954 | 0.337 | 0.01002 | 0.285 |
| 44847 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-493-5p | 13 | MLLT10 | Sponge network | 0.368 | 0.20093 | 0.105 | 0.33292 | 0.285 |
| 44848 | LINC00654 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 27 | SNX29 | Sponge network | 0.374 | 0.20054 | -0.34 | 0.01371 | 0.285 |
| 44849 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 10 | CHD2 | Sponge network | 0.796 | 4.0E-5 | 0.049 | 0.55371 | 0.285 |
| 44850 | RP11-458F8.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-539-5p | 14 | ARHGEF12 | Sponge network | 1.518 | 0 | 0.09 | 0.35693 | 0.285 |
| 44851 | ZRANB2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | EFNA5 | Sponge network | -0.376 | 0.00337 | -0.421 | 0.17868 | 0.285 |
| 44852 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-7-1-3p | 11 | AHNAK | Sponge network | -0.405 | 0.22013 | -1.207 | 0 | 0.285 |
| 44853 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-5p | 30 | ZBTB4 | Sponge network | -0.303 | 0.21002 | -0.739 | 0 | 0.285 |
| 44854 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 10 | ZNF770 | Sponge network | 0.594 | 0.41522 | 0.206 | 0.06751 | 0.285 |
| 44855 | RP11-655M14.13 |
hsa-let-7d-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-940 | 19 | MDM4 | Sponge network | 1.473 | 0.0057 | 0.345 | 0.00762 | 0.285 |
| 44856 | AC009948.5 |
hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940 | 11 | RHOB | Sponge network | 0.532 | 0.00505 | -1.052 | 0 | 0.285 |
| 44857 | HAND2-AS1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MLF1 | Sponge network | -1.697 | 0.00889 | -0.893 | 0.00727 | 0.285 |
| 44858 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 18 | ITGB3 | Sponge network | -0.942 | 0 | -0.011 | 0.95751 | 0.285 |
| 44859 | RP11-1348G14.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-940 | 12 | NTRK2 | Sponge network | 0.597 | 0.00037 | -0.736 | 0.06495 | 0.285 |
| 44860 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | LRRC4 | Sponge network | -0.355 | 0.0077 | -1.774 | 0 | 0.285 |
| 44861 | RP11-597D13.9 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | INPP4B | Sponge network | 1.302 | 7.0E-5 | -0.097 | 0.60503 | 0.285 |
| 44862 | RP11-967K21.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-452-5p;hsa-miR-576-5p | 18 | INPP4B | Sponge network | 0.056 | 0.81195 | -0.097 | 0.60503 | 0.285 |
| 44863 | RP11-20I23.13 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-92a-3p | 24 | ZFHX4 | Sponge network | 0.505 | 0.0014 | 0.018 | 0.95574 | 0.285 |
| 44864 | HCG11 |
hsa-miR-106b-5p;hsa-miR-1468-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | EPAS1 | Sponge network | 0.385 | 0.10067 | -0.184 | 0.14418 | 0.285 |
| 44865 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-9-5p | 12 | YAP1 | Sponge network | 1.601 | 0 | 0.22 | 0.11836 | 0.285 |
| 44866 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 11 | FYN | Sponge network | -0.318 | 0.65847 | -0.131 | 0.37051 | 0.285 |
| 44867 | AF131215.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 22 | DMD | Sponge network | -0.322 | 0.25945 | -1.506 | 0 | 0.285 |
| 44868 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | HACE1 | Sponge network | 0.532 | 0.00505 | -0.072 | 0.55239 | 0.285 |
| 44869 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 29 | PTPRT | Sponge network | -0.246 | 0.35034 | -0.907 | 0.03727 | 0.285 |
| 44870 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 55 | SMAD4 | Sponge network | -0.487 | 0.02157 | -0.313 | 0.00413 | 0.285 |
| 44871 | AC005562.1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 12 | MACF1 | Sponge network | 0.488 | 0.00084 | 0.019 | 0.90335 | 0.285 |
| 44872 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | SOX5 | Sponge network | 0.614 | 1.0E-5 | -1.091 | 4.0E-5 | 0.285 |
| 44873 | LINC00473 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 37 | BCL2 | Sponge network | -1.203 | 0.03881 | -0.899 | 0 | 0.285 |
| 44874 | LIPE-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-5p | 20 | NIN | Sponge network | 0.078 | 0.55803 | -0.253 | 0.13794 | 0.285 |
| 44875 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | ZFHX4 | Sponge network | 0.349 | 0.01695 | 0.018 | 0.95574 | 0.285 |
| 44876 | LINC00654 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-671-5p | 19 | CADM3 | Sponge network | 0.374 | 0.20054 | -3.663 | 0 | 0.285 |
| 44877 | RP11-403I13.7 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-671-5p | 21 | DLC1 | Sponge network | 0.395 | 0.15451 | -0.382 | 0.05038 | 0.285 |
| 44878 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 13 | FBXO32 | Sponge network | -0.206 | 0.12942 | -0.495 | 0.07561 | 0.285 |
| 44879 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-935;hsa-miR-940 | 27 | LIFR | Sponge network | -0.162 | 0.47687 | -1.794 | 0 | 0.285 |
| 44880 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-654-3p;hsa-miR-942-5p | 20 | DDX6 | Sponge network | 0.799 | 1.0E-5 | 0.091 | 0.36428 | 0.285 |
| 44881 | LINC00672 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | KIF5B | Sponge network | -0.227 | 0.31657 | 0.362 | 0.00066 | 0.285 |
| 44882 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-93-5p | 13 | EPAS1 | Sponge network | 0.056 | 0.81195 | -0.184 | 0.14418 | 0.285 |
| 44883 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 42 | IL6ST | Sponge network | -0.125 | 0.49414 | -0.402 | 0.00676 | 0.285 |
| 44884 | AP001347.6 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-7-1-3p | 12 | CSRNP1 | Sponge network | -1.161 | 0.00591 | -1.448 | 0 | 0.285 |
| 44885 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | HACE1 | Sponge network | 0.222 | 0.29133 | -0.072 | 0.55239 | 0.285 |
| 44886 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-192-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ARID5B | Sponge network | 1.378 | 0 | 0.342 | 0.01676 | 0.285 |
| 44887 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-455-3p | 10 | KCNAB1 | Sponge network | 0.082 | 0.55654 | -0.755 | 2.0E-5 | 0.285 |
| 44888 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 46 | FZD3 | Sponge network | 0.064 | 0.72934 | 0.104 | 0.60316 | 0.285 |
| 44889 | CTC-510F12.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-484 | 15 | CUX1 | Sponge network | -0.691 | 0.00146 | -0.039 | 0.6705 | 0.285 |
| 44890 | TPTEP1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 13 | ZBTB20 | Sponge network | -1.216 | 0 | -0.122 | 0.57569 | 0.285 |
| 44891 | OIP5-AS1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 43 | DIP2C | Sponge network | 0.421 | 0.00084 | -0.436 | 0.01713 | 0.285 |
| 44892 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 15 | GPAM | Sponge network | 0.349 | 0.01695 | 0.142 | 0.29732 | 0.285 |
| 44893 | WDR86-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 10 | FYN | Sponge network | 0.348 | 0.32912 | -0.131 | 0.37051 | 0.285 |
| 44894 | CTD-2303H24.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 20 | BAZ2B | Sponge network | 0.415 | 0.14657 | -0.276 | 0.02833 | 0.285 |
| 44895 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 13 | PPP1R9A | Sponge network | -0.187 | 0.23787 | -0.903 | 0.04254 | 0.285 |
| 44896 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-495-3p | 10 | SLC5A3 | Sponge network | 1.055 | 0 | 0.493 | 5.0E-5 | 0.285 |
| 44897 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 19 | ARID5B | Sponge network | -0.358 | 0.17255 | 0.342 | 0.01676 | 0.285 |
| 44898 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-624-5p;hsa-miR-942-5p | 23 | NOTCH2 | Sponge network | -0.517 | 0.02935 | 0.138 | 0.35163 | 0.285 |
| 44899 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SON | Sponge network | 0.272 | 0.44692 | 0.168 | 0.04406 | 0.285 |
| 44900 | RP11-401P9.4 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-5p | 12 | GABRB2 | Sponge network | -0.384 | 0.40652 | -1.841 | 0.00029 | 0.285 |
| 44901 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | PCDH8 | Sponge network | -0.737 | 0.06182 | -0.997 | 0.05366 | 0.285 |
| 44902 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-7-1-3p | 11 | BCL11A | Sponge network | 0.419 | 0.0319 | -0.932 | 0.0063 | 0.285 |
| 44903 | RP4-668J24.2 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-628-5p;hsa-miR-629-3p | 24 | IL6ST | Sponge network | -1.054 | 0.0706 | -0.402 | 0.00676 | 0.285 |
| 44904 | LINC00843 |
hsa-miR-127-5p;hsa-miR-143-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p | 10 | DDX3X | Sponge network | 1.106 | 0 | -0.152 | 0.07686 | 0.285 |
| 44905 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p | 12 | JAKMIP2 | Sponge network | 0.16 | 0.50785 | -1.388 | 0 | 0.285 |
| 44906 | CTD-2089N3.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3065-3p | 13 | ADARB1 | Sponge network | -0.314 | 0.62033 | -0.335 | 0.06631 | 0.285 |
| 44907 | C4B-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-425-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-96-5p | 17 | SOX5 | Sponge network | 2.306 | 9.0E-5 | -1.091 | 4.0E-5 | 0.285 |
| 44908 | LINC00472 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-877-5p | 21 | NRK | Sponge network | -0.842 | 0.01361 | 0.656 | 0.19217 | 0.285 |
| 44909 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-766-3p | 15 | TBL1XR1 | Sponge network | 0.663 | 0.00073 | 0.578 | 0 | 0.285 |
| 44910 | TUG1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-664a-5p | 15 | CBFB | Sponge network | 0.972 | 0 | 1.061 | 0 | 0.285 |
| 44911 | RP11-5C23.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-1-5p | 17 | AKAP6 | Sponge network | 0.274 | 0.07681 | -1.586 | 3.0E-5 | 0.285 |
| 44912 | AF131215.9 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | TAL1 | Sponge network | -0.322 | 0.25945 | -0.462 | 0.00574 | 0.285 |
| 44913 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 16 | SUFU | Sponge network | -0.246 | 0.35034 | -0.04 | 0.65489 | 0.285 |
| 44914 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | PRKAB2 | Sponge network | 1.044 | 2.0E-5 | -0.367 | 0.01371 | 0.285 |
| 44915 | AC005618.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 30 | CREB1 | Sponge network | 0.862 | 0.06213 | 0.203 | 0.00537 | 0.285 |
| 44916 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-940 | 30 | ETV1 | Sponge network | 0.194 | 0.64278 | -0.096 | 0.67674 | 0.285 |
| 44917 | RP11-822E23.8 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 25 | CTIF | Sponge network | -0.777 | 0.16667 | -0.389 | 0.00549 | 0.285 |
| 44918 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-654-3p | 20 | MYBL1 | Sponge network | -0.031 | 0.89063 | 0.494 | 0.02152 | 0.285 |
| 44919 | LINC00473 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 23 | RNF144A | Sponge network | -1.203 | 0.03881 | 0.594 | 0.0009 | 0.285 |
| 44920 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | ZYX | Sponge network | -1.216 | 0 | 0.019 | 0.89763 | 0.285 |
| 44921 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 15 | ZFHX3 | Sponge network | 1.11 | 1.0E-5 | 0.389 | 0.01363 | 0.285 |
| 44922 | RP11-855A2.5 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-93-3p | 12 | GRIK3 | Sponge network | -1.717 | 0.01522 | -3.208 | 0 | 0.285 |
| 44923 | RP11-464F9.20 |
hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-625-5p | 10 | MDM4 | Sponge network | 0.643 | 0.00136 | 0.345 | 0.00762 | 0.285 |
| 44924 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | BCL11A | Sponge network | 0.083 | 0.66405 | -0.932 | 0.0063 | 0.285 |
| 44925 | CTB-113P19.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 12 | PRRX1 | Sponge network | 0.199 | 0.52763 | 1.316 | 0 | 0.285 |
| 44926 | JAZF1-AS1 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 16 | IRS1 | Sponge network | -0.769 | 0.00035 | -0.026 | 0.88855 | 0.285 |
| 44927 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 15 | LHX6 | Sponge network | 0.056 | 0.81195 | -0.087 | 0.69193 | 0.285 |
| 44928 | BDNF-AS |
hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | PAK3 | Sponge network | -0.668 | 3.0E-5 | -0.628 | 0.07253 | 0.285 |
| 44929 | CTC-297N7.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-671-5p;hsa-miR-7-5p | 14 | KCNA6 | Sponge network | -2.069 | 0 | -1.531 | 0.00013 | 0.285 |
| 44930 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 0.214 | 0.18246 | 0.259 | 0.00962 | 0.285 |
| 44931 | RP11-157J24.2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 12 | MCC | Sponge network | -1.664 | 0.05165 | -1.005 | 0 | 0.285 |
| 44932 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-539-5p | 13 | NIN | Sponge network | 0.37 | 0.27614 | -0.253 | 0.13794 | 0.285 |
| 44933 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-671-5p;hsa-miR-7-1-3p | 19 | FOXP1 | Sponge network | -0.285 | 0.2172 | 0.042 | 0.74368 | 0.285 |
| 44934 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 17 | DAB2 | Sponge network | -1.216 | 0 | 0.431 | 0.0113 | 0.285 |
| 44935 | C20orf166-AS1 |
hsa-let-7b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-5p | 13 | NEDD4 | Sponge network | -2.69 | 0.0001 | 0.02 | 0.89834 | 0.285 |
| 44936 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p | 19 | NUAK1 | Sponge network | -0.738 | 0.21233 | 0.904 | 0 | 0.285 |
| 44937 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 34 | AKAP11 | Sponge network | 0.267 | 0.2937 | 0.196 | 0.09825 | 0.285 |
| 44938 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p | 17 | LRRC7 | Sponge network | 0.056 | 0.81195 | -1.341 | 0.01193 | 0.285 |
| 44939 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-590-3p | 16 | ZFHX3 | Sponge network | -1.582 | 8.0E-5 | 0.389 | 0.01363 | 0.285 |
| 44940 | RP4-639F20.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-542-3p | 11 | RAP1A | Sponge network | -0.517 | 0.02935 | -0.929 | 0 | 0.285 |
| 44941 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 50 | AFF4 | Sponge network | 0.056 | 0.81195 | -0.266 | 0.01565 | 0.285 |
| 44942 | AC009120.6 |
hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-625-5p | 13 | NUAK1 | Sponge network | 0.919 | 0 | 0.904 | 0 | 0.285 |
| 44943 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-7-1-3p | 15 | KIF1B | Sponge network | 0.959 | 0 | -0.118 | 0.32258 | 0.285 |
| 44944 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-3p | 17 | CBX5 | Sponge network | 1.205 | 0 | 0.165 | 0.24446 | 0.285 |
| 44945 | LINC00094 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-93-5p | 26 | NIN | Sponge network | 0.253 | 0.09565 | -0.253 | 0.13794 | 0.285 |
| 44946 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | TIMP3 | Sponge network | 0.051 | 0.83388 | -0.546 | 0.02307 | 0.285 |
| 44947 | AC005154.6 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | MAP3K4 | Sponge network | 0.848 | 0 | 0.058 | 0.54229 | 0.285 |
| 44948 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 27 | USP12 | Sponge network | -0.563 | 0.02545 | -0.102 | 0.40768 | 0.285 |
| 44949 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 10 | RB1CC1 | Sponge network | 1.317 | 0 | 0.175 | 0.12559 | 0.285 |
| 44950 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-708-3p | 18 | ANK3 | Sponge network | -0.899 | 6.0E-5 | -0.55 | 0.00086 | 0.285 |
| 44951 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-502-3p | 28 | RUNX1T1 | Sponge network | -0.351 | 0.11358 | -0.344 | 0.2374 | 0.285 |
| 44952 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 31 | ARID5B | Sponge network | 0.335 | 0.20596 | 0.342 | 0.01676 | 0.285 |
| 44953 | RP11-5C23.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 10 | PRUNE2 | Sponge network | 0.274 | 0.07681 | -1.733 | 0.00022 | 0.285 |
| 44954 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-214-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | MLLT10 | Sponge network | -1.264 | 6.0E-5 | 0.105 | 0.33292 | 0.285 |
| 44955 | FLNB-AS1 |
hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-766-3p | 13 | PPARA | Sponge network | 1.163 | 0.00018 | -0.379 | 0.00548 | 0.285 |
| 44956 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-5p | 17 | ZMYM2 | Sponge network | 0.706 | 0 | 0.279 | 0.01273 | 0.285 |
| 44957 | LINC00865 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-629-5p | 18 | CRTC1 | Sponge network | -1.249 | 7.0E-5 | -0.116 | 0.28412 | 0.285 |
| 44958 | RP4-669H2.1 |
hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p | 15 | MDM4 | Sponge network | 1.711 | 0.00019 | 0.345 | 0.00762 | 0.285 |
| 44959 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PBRM1 | Sponge network | 1.308 | 0 | -0.029 | 0.78601 | 0.285 |
| 44960 | LINC00476 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | ERCC4 | Sponge network | -0.615 | 0.00015 | 0.126 | 0.15266 | 0.285 |
| 44961 | FGD5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-590-3p | 14 | PKNOX1 | Sponge network | 0.401 | 0.00066 | 0.085 | 0.28917 | 0.285 |
| 44962 | RP11-535M15.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-532-3p | 11 | TRPS1 | Sponge network | -1.14 | 0.03513 | -0.191 | 0.44109 | 0.285 |
| 44963 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-654-3p | 23 | RICTOR | Sponge network | 0.373 | 0.00068 | 0.388 | 0.00188 | 0.285 |
| 44964 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 36 | THRB | Sponge network | 0.619 | 0.00157 | -1.117 | 0.0004 | 0.285 |
| 44965 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-935 | 27 | FAT3 | Sponge network | -0.141 | 0.26434 | -1.601 | 0.00051 | 0.285 |
| 44966 | RP11-999E24.3 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-7-5p | 17 | PRKCB | Sponge network | -0.693 | 0.05629 | -1.313 | 2.0E-5 | 0.285 |
| 44967 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p | 17 | PTPRB | Sponge network | -0.842 | 0.01361 | 0.105 | 0.47122 | 0.285 |
| 44968 | LINC00476 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p | 52 | GAS7 | Sponge network | -0.615 | 0.00015 | -0.555 | 0.01602 | 0.285 |
| 44969 | AC108488.4 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-874-3p;hsa-miR-93-3p | 22 | CTIF | Sponge network | 0.259 | 0.12542 | -0.389 | 0.00549 | 0.285 |
| 44970 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-590-3p | 15 | DMXL1 | Sponge network | 0.959 | 0 | -0.426 | 0.00056 | 0.285 |
| 44971 | RP11-532F6.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | JDP2 | Sponge network | -0.896 | 0.00099 | -0.967 | 0 | 0.285 |
| 44972 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | TSHZ3 | Sponge network | 0.559 | 0.00026 | -0.556 | 0.01516 | 0.285 |
| 44973 | CTD-2291D10.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-532-3p | 12 | TRPS1 | Sponge network | 3.21 | 0 | -0.191 | 0.44109 | 0.285 |
| 44974 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | NEDD4 | Sponge network | -1.203 | 0.03881 | 0.02 | 0.89834 | 0.285 |
| 44975 | RP11-1006G14.4 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-376c-3p | 14 | RASAL2 | Sponge network | 0.559 | 0.00026 | 0.869 | 0 | 0.285 |
| 44976 | RP11-327P2.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-671-5p | 14 | CDH23 | Sponge network | -0.147 | 0.40452 | -0.974 | 0.00034 | 0.285 |
| 44977 | WDFY3-AS2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 19 | CELF4 | Sponge network | -0.942 | 0 | -1.135 | 0.00344 | 0.285 |
| 44978 | AC004540.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-5p | 11 | PPP1R9A | Sponge network | -2.549 | 0.00528 | -0.903 | 0.04254 | 0.285 |
| 44979 | RP11-403I13.8 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | GALNT13 | Sponge network | 0.619 | 0.00157 | -0.857 | 0.07439 | 0.285 |
| 44980 | RP11-274B21.10 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | PHF3 | Sponge network | 1.073 | 0.00216 | 0.126 | 0.19652 | 0.285 |
| 44981 | MRVI1-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-744-5p;hsa-miR-92a-1-5p | 10 | PRKACA | Sponge network | -1.092 | 4.0E-5 | -0.255 | 0.0012 | 0.285 |
| 44982 | NUTM2A-AS1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | TSC1 | Sponge network | 0.643 | 0 | 0.103 | 0.26071 | 0.285 |
| 44983 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 25 | BCL11A | Sponge network | -0.739 | 0.0574 | -0.932 | 0.0063 | 0.285 |
| 44984 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 29 | BMPR2 | Sponge network | 1.04 | 0.00023 | 0.206 | 0.0635 | 0.285 |
| 44985 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p | 22 | SMAD2 | Sponge network | 0.178 | 0.29106 | -0.128 | 0.12732 | 0.285 |
| 44986 | RAMP2-AS1 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | JUN | Sponge network | -0.513 | 0.04703 | -0.932 | 0 | 0.285 |
| 44987 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 24 | PCDH10 | Sponge network | 0.537 | 0.00139 | -1.644 | 0.0078 | 0.285 |
| 44988 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 21 | ERBB4 | Sponge network | -0.72 | 0.24239 | -1.975 | 2.0E-5 | 0.285 |
| 44989 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | ST6GALNAC3 | Sponge network | -0.206 | 0.12942 | -0.987 | 0 | 0.285 |
| 44990 | RP4-584D14.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-500a-5p | 14 | PRRX1 | Sponge network | 1.294 | 0.0031 | 1.316 | 0 | 0.285 |
| 44991 | RP11-356J5.12 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 44 | UNC80 | Sponge network | -0.899 | 6.0E-5 | -1.934 | 0 | 0.285 |
| 44992 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 28 | USP12 | Sponge network | -0.612 | 0.00094 | -0.102 | 0.40768 | 0.285 |
| 44993 | RP11-46C24.7 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 36 | TRPS1 | Sponge network | 0.392 | 0.0278 | -0.191 | 0.44109 | 0.285 |
| 44994 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-342-3p | 20 | ZBTB4 | Sponge network | 0.349 | 0.01695 | -0.739 | 0 | 0.285 |
| 44995 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 14 | ADAMTSL3 | Sponge network | 0.274 | 0.07681 | -1.559 | 0 | 0.285 |
| 44996 | AC074289.1 |
hsa-miR-10a-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-7-5p;hsa-miR-940 | 10 | GLI3 | Sponge network | -0.326 | 0.14314 | -0.615 | 0.01482 | 0.285 |
| 44997 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-655-3p | 14 | KANK1 | Sponge network | 0.385 | 0.10067 | -0.55 | 0.00134 | 0.285 |
| 44998 | RP11-325K4.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | CREB1 | Sponge network | 0.198 | 0.27624 | 0.203 | 0.00537 | 0.285 |
| 44999 | RP11-678G14.3 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 11 | ERG | Sponge network | -4.477 | 0 | 0.002 | 0.99011 | 0.285 |
| 45000 | LINC00473 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 13 | INPP4A | Sponge network | -1.203 | 0.03881 | 0.07 | 0.53563 | 0.285 |
| 45001 | RP11-196G18.23 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-590-3p | 14 | ABL2 | Sponge network | 0.724 | 0.00039 | 0.82 | 0 | 0.285 |
| 45002 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | RBMS3 | Sponge network | 0.476 | 0.19203 | -0.519 | 0.03551 | 0.285 |
| 45003 | HOTAIRM1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 27 | GRIK3 | Sponge network | -0.053 | 0.80685 | -3.208 | 0 | 0.285 |
| 45004 | RP11-540B6.6 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-664a-5p | 17 | CBFB | Sponge network | 0.93 | 0 | 1.061 | 0 | 0.285 |
| 45005 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p | 11 | ZFHX3 | Sponge network | 3.21 | 0 | 0.389 | 0.01363 | 0.285 |
| 45006 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | RBL2 | Sponge network | -0.08 | 0.90092 | -0.282 | 0.00254 | 0.285 |
| 45007 | LINC01018 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 17 | TLE4 | Sponge network | -2.915 | 0.00015 | -1.157 | 0 | 0.285 |
| 45008 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | KDSR | Sponge network | 0.101 | 0.60919 | 0.239 | 0.02216 | 0.285 |
| 45009 | AGAP11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p | 10 | DCN | Sponge network | -1.645 | 0 | -1.288 | 0 | 0.285 |
| 45010 | RP11-728F11.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-7-1-3p | 11 | PDE4DIP | Sponge network | 0.238 | 0.70544 | -0.282 | 0.0257 | 0.285 |
| 45011 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PTPRD | Sponge network | -0.021 | 0.93598 | -1.005 | 0.00064 | 0.285 |
| 45012 | AC156455.1 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484 | 10 | ITGA5 | Sponge network | 1.154 | 0.00041 | 0.452 | 0.03524 | 0.285 |
| 45013 | FZD10-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-455-3p | 10 | ALDH1A2 | Sponge network | -0.727 | 0.04726 | -2.411 | 0 | 0.285 |
| 45014 | CTD-2006H14.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | LPP | Sponge network | 1.378 | 0 | -0.459 | 0.04475 | 0.285 |
| 45015 | RP11-263K19.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-429 | 13 | ERCC4 | Sponge network | 0.859 | 0.00331 | 0.126 | 0.15266 | 0.285 |
| 45016 | LINC00173 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-874-3p;hsa-miR-940 | 24 | GRIK3 | Sponge network | 0.056 | 0.8494 | -3.208 | 0 | 0.285 |
| 45017 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-664a-3p | 23 | FZD3 | Sponge network | 1.317 | 0 | 0.104 | 0.60316 | 0.285 |
| 45018 | RP5-994D16.9 |
hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | ATRX | Sponge network | 0.252 | 0.08508 | 0.261 | 0.04408 | 0.285 |
| 45019 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TLL1 | Sponge network | -0.342 | 0.02288 | -1.195 | 3.0E-5 | 0.285 |
| 45020 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-500b-5p | 20 | GRIA3 | Sponge network | -0.611 | 0.09607 | -1.956 | 0 | 0.285 |
| 45021 | LINC00702 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 46 | GNAQ | Sponge network | -0.737 | 0.06182 | -0.367 | 0.00102 | 0.285 |
| 45022 | LINC00865 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 23 | EFNA5 | Sponge network | -1.249 | 7.0E-5 | -0.421 | 0.17868 | 0.285 |
| 45023 | RP11-35G9.3 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | KCNA6 | Sponge network | 0.657 | 4.0E-5 | -1.531 | 0.00013 | 0.285 |
| 45024 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | KALRN | Sponge network | -1.047 | 0 | -0.819 | 1.0E-5 | 0.285 |
| 45025 | AC005618.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 41 | RUNX1T1 | Sponge network | 0.862 | 0.06213 | -0.344 | 0.2374 | 0.285 |
| 45026 | RP13-1039J1.4 |
hsa-miR-1266-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p | 10 | ASTN1 | Sponge network | 0.356 | 0.33467 | -2.389 | 2.0E-5 | 0.285 |
| 45027 | CTD-3064M3.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-429 | 27 | WDFY3 | Sponge network | -0.469 | 0.08721 | -0.201 | 0.0967 | 0.285 |
| 45028 | AC138035.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326 | 12 | ZFX | Sponge network | 1.073 | 0 | 0.09 | 0.42563 | 0.285 |
| 45029 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-92a-3p | 18 | ARHGAP29 | Sponge network | 0.056 | 0.81195 | 0.131 | 0.42953 | 0.285 |
| 45030 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 0.101 | 0.60919 | 0.322 | 0.00215 | 0.285 |
| 45031 | CTA-941F9.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-576-5p | 13 | THRB | Sponge network | -0.344 | 0.49624 | -1.117 | 0.0004 | 0.285 |
| 45032 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | ZNF208 | Sponge network | -0.969 | 2.0E-5 | -0.4 | 0.22381 | 0.285 |
| 45033 | CTC-490E21.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-323a-3p | 13 | ZFHX4 | Sponge network | 1.46 | 1.0E-5 | 0.018 | 0.95574 | 0.285 |
| 45034 | RP11-506M13.3 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-374b-5p | 13 | UBE2D1 | Sponge network | 0.769 | 0 | 0.468 | 0 | 0.285 |
| 45035 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577 | 10 | TSLP | Sponge network | 0.056 | 0.81195 | -2.209 | 0 | 0.285 |
| 45036 | AC004893.11 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | TSC1 | Sponge network | 1.317 | 0 | 0.103 | 0.26071 | 0.285 |
| 45037 | RP5-1198O20.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | BRD3 | Sponge network | 0.997 | 0.00066 | 0.37 | 0.00013 | 0.284 |
| 45038 | TP73-AS1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-766-3p;hsa-miR-92a-3p | 13 | ATM | Sponge network | -0.563 | 0.02545 | 0.178 | 0.21568 | 0.284 |
| 45039 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | KDSR | Sponge network | 0.064 | 0.72934 | 0.239 | 0.02216 | 0.284 |
| 45040 | TPTEP1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 25 | RIMBP2 | Sponge network | -1.216 | 0 | -1.968 | 5.0E-5 | 0.284 |
| 45041 | MRVI1-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p | 10 | BAZ2B | Sponge network | -1.092 | 4.0E-5 | -0.276 | 0.02833 | 0.284 |
| 45042 | GAS6-AS2 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-7-1-3p | 10 | ZNF844 | Sponge network | 0.16 | 0.50785 | -0.966 | 0.00035 | 0.284 |
| 45043 | AC016747.3 |
hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 14 | POU2F2 | Sponge network | 0.199 | 0.38015 | -0.233 | 0.32214 | 0.284 |
| 45044 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | EGFR | Sponge network | -0.726 | 0.00132 | -0.175 | 0.48451 | 0.284 |
| 45045 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-98-5p | 61 | NTRK3 | Sponge network | -0.222 | 0.32445 | -1.77 | 3.0E-5 | 0.284 |
| 45046 | RP11-48B3.4 |
hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-455-3p | 10 | ABL1 | Sponge network | 1.757 | 0 | -0.215 | 0.07804 | 0.284 |
| 45047 | RP11-389C8.2 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-942-5p | 10 | ZBTB20 | Sponge network | 0.727 | 3.0E-5 | -0.122 | 0.57569 | 0.284 |
| 45048 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-93-5p | 14 | CDHR3 | Sponge network | 0.082 | 0.55397 | -0.977 | 4.0E-5 | 0.284 |
| 45049 | RP11-53O19.1 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | HOMER2 | Sponge network | -0.405 | 0.22013 | -2.698 | 0 | 0.284 |
| 45050 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CCND2 | Sponge network | -0.021 | 0.93598 | -0.267 | 0.3112 | 0.284 |
| 45051 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 31 | BMPR2 | Sponge network | -4.546 | 0 | 0.206 | 0.0635 | 0.284 |
| 45052 | LINC00654 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | LRIG1 | Sponge network | 0.374 | 0.20054 | -0.537 | 0.00588 | 0.284 |
| 45053 | RP11-48B3.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | LUZP2 | Sponge network | 1.757 | 0 | -0.056 | 0.89416 | 0.284 |
| 45054 | LINC00641 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | ERG | Sponge network | -0.222 | 0.32445 | 0.002 | 0.99011 | 0.284 |
| 45055 | RP1-43E13.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 15 | PRKAB2 | Sponge network | -0.034 | 0.83228 | -0.367 | 0.01371 | 0.284 |
| 45056 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-199a-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-493-5p | 12 | MIER3 | Sponge network | 0.706 | 0 | -0.212 | 0.04957 | 0.284 |
| 45057 | RP11-401P9.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-590-5p;hsa-miR-7-5p | 15 | PTGFR | Sponge network | -0.384 | 0.40652 | -0.233 | 0.45421 | 0.284 |
| 45058 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | RIMBP2 | Sponge network | -0.342 | 0.02288 | -1.968 | 5.0E-5 | 0.284 |
| 45059 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-744-3p | 22 | MEF2C | Sponge network | 0.368 | 0.20093 | -0.499 | 0.01448 | 0.284 |
| 45060 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-30b-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-629-3p | 10 | GAS7 | Sponge network | -0.011 | 0.967 | -0.555 | 0.01602 | 0.284 |
| 45061 | RP11-760H22.2 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-432-5p | 23 | PPM1L | Sponge network | 0.001 | 0.99753 | -0.537 | 0.00616 | 0.284 |
| 45062 | RP1-122P22.2 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-1976;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p | 12 | SNX29 | Sponge network | 0.065 | 0.73468 | -0.34 | 0.01371 | 0.284 |
| 45063 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-664a-5p | 13 | SOX11 | Sponge network | 0.912 | 0.00014 | 2.68 | 0 | 0.284 |
| 45064 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ZFX | Sponge network | 0.222 | 0.29133 | 0.09 | 0.42563 | 0.284 |
| 45065 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-628-5p;hsa-miR-940 | 17 | TRIM33 | Sponge network | 0.983 | 0.00048 | 0.402 | 7.0E-5 | 0.284 |
| 45066 | HOTAIRM1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-590-3p | 13 | STARD13 | Sponge network | -0.053 | 0.80685 | -0.187 | 0.27383 | 0.284 |
| 45067 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | ATRX | Sponge network | -0.899 | 6.0E-5 | 0.261 | 0.04408 | 0.284 |
| 45068 | RP11-156P1.3 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-340-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 18 | RNF144A | Sponge network | 0.214 | 0.18246 | 0.594 | 0.0009 | 0.284 |
| 45069 | HOXB-AS2 |
hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-629-3p | 24 | CBX5 | Sponge network | 1.316 | 0.01106 | 0.165 | 0.24446 | 0.284 |
| 45070 | C4B-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p | 10 | ARNT2 | Sponge network | 2.306 | 9.0E-5 | -0.332 | 0.25851 | 0.284 |
| 45071 | HOXA-AS2 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | 0.61 | 0.01593 | -0.704 | 0.00106 | 0.284 |
| 45072 | RP4-669P10.16 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-342-5p;hsa-miR-542-3p | 22 | DTNA | Sponge network | -0.301 | 0.06597 | -1.648 | 1.0E-5 | 0.284 |
| 45073 | AC108676.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-511-5p | 17 | CREB1 | Sponge network | 4.346 | 2.0E-5 | 0.203 | 0.00537 | 0.284 |
| 45074 | RP11-434B12.1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-505-5p;hsa-miR-629-5p | 12 | WNK2 | Sponge network | -0.031 | 0.89063 | -0.352 | 0.31066 | 0.284 |
| 45075 | RP11-290F24.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326 | 16 | TBL1XR1 | Sponge network | 2.052 | 0 | 0.578 | 0 | 0.284 |
| 45076 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | BCL6 | Sponge network | 0.225 | 0.30644 | -0.211 | 0.19489 | 0.284 |
| 45077 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 22 | JAZF1 | Sponge network | -0.829 | 0.01343 | -0.377 | 0.02677 | 0.284 |
| 45078 | FAM66C |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-942-5p | 15 | ERRFI1 | Sponge network | -0.901 | 0.00019 | -0.798 | 1.0E-5 | 0.284 |
| 45079 | TTC28-AS1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 13 | KIF5B | Sponge network | 0.373 | 0.00068 | 0.362 | 0.00066 | 0.284 |
| 45080 | RP5-1074L1.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | RPS6KA3 | Sponge network | 1.923 | 0 | 0.256 | 0.02419 | 0.284 |
| 45081 | SOX2-OT |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 17 | FNBP1 | Sponge network | -1.264 | 6.0E-5 | -0.969 | 4.0E-5 | 0.284 |
| 45082 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | GNA13 | Sponge network | 0.532 | 0.00505 | 0.007 | 0.93884 | 0.284 |
| 45083 | MIR22HG |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-424-3p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-651-5p;hsa-miR-93-3p | 22 | WWTR1 | Sponge network | -1.047 | 0 | -0.345 | 0.11997 | 0.284 |
| 45084 | RP11-403I13.8 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | INPP4A | Sponge network | 0.619 | 0.00157 | 0.07 | 0.53563 | 0.284 |
| 45085 | RP11-1006G14.4 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | INPP4A | Sponge network | 0.559 | 0.00026 | 0.07 | 0.53563 | 0.284 |
| 45086 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-590-3p | 30 | ZFX | Sponge network | 0.494 | 0.00339 | 0.09 | 0.42563 | 0.284 |
| 45087 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-766-3p;hsa-miR-93-5p | 36 | CHD5 | Sponge network | -1.645 | 0 | -0.922 | 0.01521 | 0.284 |
| 45088 | RP11-345P4.7 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-497-5p | 12 | CDK6 | Sponge network | 1.597 | 0 | 0.991 | 0 | 0.284 |
| 45089 | RP11-53O19.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-362-3p | 12 | IGF2 | Sponge network | -0.405 | 0.22013 | 0.848 | 0.03842 | 0.284 |
| 45090 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p | 10 | CREBBP | Sponge network | 0.299 | 0.57722 | 0.126 | 0.15186 | 0.284 |
| 45091 | AC005618.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 29 | ADARB1 | Sponge network | 0.862 | 0.06213 | -0.335 | 0.06631 | 0.284 |
| 45092 | AC003090.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 29 | ETV1 | Sponge network | -2.117 | 0.00161 | -0.096 | 0.67674 | 0.284 |
| 45093 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CXXC4 | Sponge network | -0.206 | 0.12942 | -0.433 | 0.21055 | 0.284 |
| 45094 | RP11-148O21.2 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-671-5p | 12 | BNC2 | Sponge network | 0.233 | 0.81029 | -0.491 | 0.09988 | 0.284 |
| 45095 | AC138035.2 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-378a-5p | 13 | BMPR2 | Sponge network | 1.073 | 0 | 0.206 | 0.0635 | 0.284 |
| 45096 | U91328.21 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-378c | 22 | WDFY3 | Sponge network | -0.735 | 0.15983 | -0.201 | 0.0967 | 0.284 |
| 45097 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p | 15 | NUAK1 | Sponge network | -1.331 | 3.0E-5 | 0.904 | 0 | 0.284 |
| 45098 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-769-5p;hsa-miR-940;hsa-miR-96-5p | 48 | NTRK3 | Sponge network | 0.331 | 0.57247 | -1.77 | 3.0E-5 | 0.284 |
| 45099 | RP11-13K12.5 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | BCL9 | Sponge network | -0.318 | 0.65847 | 0.321 | 0.00267 | 0.284 |
| 45100 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-7-1-3p | 15 | FGF5 | Sponge network | -0.842 | 0.01361 | 0.549 | 0.28659 | 0.284 |
| 45101 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-5p | 16 | KANK1 | Sponge network | 0.078 | 0.55803 | -0.55 | 0.00134 | 0.284 |
| 45102 | TRAF3IP2-AS1 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | PCLO | Sponge network | -0.323 | 0.01372 | -1.843 | 0.00064 | 0.284 |
| 45103 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-651-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 62 | AFF4 | Sponge network | 0.249 | 0.35902 | -0.266 | 0.01565 | 0.284 |
| 45104 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-345-5p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-664a-5p | 18 | TRPS1 | Sponge network | 0.858 | 3.0E-5 | -0.191 | 0.44109 | 0.284 |
| 45105 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-93-5p | 16 | CDHR3 | Sponge network | -0.976 | 0.00025 | -0.977 | 4.0E-5 | 0.284 |
| 45106 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-744-3p | 14 | HIP1 | Sponge network | 0.659 | 3.0E-5 | 0.774 | 0 | 0.284 |
| 45107 | RP11-439A17.10 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-5p | 13 | MEF2C | Sponge network | 0.051 | 0.95756 | -0.499 | 0.01448 | 0.284 |
| 45108 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p | 18 | HIP1 | Sponge network | 0.082 | 0.55397 | 0.774 | 0 | 0.284 |
| 45109 | HOXA-AS3 |
hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 16 | DIP2C | Sponge network | -0.555 | 0.06156 | -0.436 | 0.01713 | 0.284 |
| 45110 | AC141928.1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-628-5p;hsa-miR-96-5p | 11 | IRS1 | Sponge network | 0.853 | 0.15352 | -0.026 | 0.88855 | 0.284 |
| 45111 | LINC00662 |
hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DACT1 | Sponge network | 0.213 | 0.32297 | 0.783 | 0.00072 | 0.284 |
| 45112 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-362-3p | 17 | CACNA2D1 | Sponge network | -0.773 | 0.04073 | -0.846 | 0.00675 | 0.284 |
| 45113 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-375;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | QKI | Sponge network | 0.687 | 0.02753 | -0.333 | 0.04111 | 0.284 |
| 45114 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | HIP1 | Sponge network | -0.08 | 0.90092 | 0.774 | 0 | 0.284 |
| 45115 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 33 | SASH1 | Sponge network | 0.082 | 0.55654 | -0.502 | 0.00175 | 0.284 |
| 45116 | RP11-82L18.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-30a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-497-5p | 13 | SOX11 | Sponge network | 1.082 | 0.01453 | 2.68 | 0 | 0.284 |
| 45117 | RP11-760H22.2 |
hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-376c-3p | 13 | SYNM | Sponge network | 0.001 | 0.99753 | -2.309 | 1.0E-5 | 0.284 |
| 45118 | RP11-890B15.3 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | PTCH1 | Sponge network | 0.101 | 0.60919 | -0.1 | 0.6179 | 0.284 |
| 45119 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 11 | ITCH | Sponge network | 0.8 | 0 | 0.084 | 0.34058 | 0.284 |
| 45120 | RP13-516M14.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-429 | 10 | LATS2 | Sponge network | 0.389 | 0.04293 | 0.159 | 0.2304 | 0.284 |
| 45121 | RP11-680C21.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 19 | GRIN2A | Sponge network | -1.424 | 0.04346 | -2.161 | 0 | 0.284 |
| 45122 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-942-5p | 24 | HLF | Sponge network | -0.206 | 0.12942 | -2.443 | 0 | 0.284 |
| 45123 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 61 | KIF1B | Sponge network | -0.942 | 0 | -0.118 | 0.32258 | 0.284 |
| 45124 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | PAFAH1B1 | Sponge network | 0.614 | 1.0E-5 | -0.429 | 0 | 0.284 |
| 45125 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-654-3p | 27 | DDX6 | Sponge network | 0.569 | 0.00067 | 0.091 | 0.36428 | 0.284 |
| 45126 | RP11-736K20.5 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-362-3p | 10 | IGF2 | Sponge network | -0.358 | 0.17255 | 0.848 | 0.03842 | 0.284 |
| 45127 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 25 | MEF2C | Sponge network | 0.274 | 0.07681 | -0.499 | 0.01448 | 0.284 |
| 45128 | RP11-102N12.3 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | ZFHX4 | Sponge network | -0.351 | 0.11358 | 0.018 | 0.95574 | 0.284 |
| 45129 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 12 | UVRAG | Sponge network | 0.219 | 0.1516 | -0.017 | 0.83865 | 0.284 |
| 45130 | RP11-6N17.3 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p | 10 | UBR3 | Sponge network | 0.108 | 0.69556 | -0.021 | 0.82774 | 0.284 |
| 45131 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429 | 14 | GRIA3 | Sponge network | -0.469 | 0.08721 | -1.956 | 0 | 0.284 |
| 45132 | LINC00094 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-92a-3p | 16 | ROBO1 | Sponge network | 0.253 | 0.09565 | -0.009 | 0.96154 | 0.284 |
| 45133 | RP11-384O8.1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-590-3p | 15 | GLIPR1 | Sponge network | -0.738 | 0.21233 | 0.146 | 0.3276 | 0.284 |
| 45134 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | CACNA2D1 | Sponge network | 0.853 | 0.15352 | -0.846 | 0.00675 | 0.284 |
| 45135 | CTC-510F12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-34a-5p | 12 | SNX29 | Sponge network | -0.691 | 0.00146 | -0.34 | 0.01371 | 0.284 |
| 45136 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-590-3p | 18 | SLC6A15 | Sponge network | -0.096 | 0.67958 | -2.373 | 2.0E-5 | 0.284 |
| 45137 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500b-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 27 | RSPO2 | Sponge network | -0.969 | 2.0E-5 | -3.038 | 0 | 0.284 |
| 45138 | BCYRN1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p | 16 | IGF2 | Sponge network | 0.311 | 0.10344 | 0.848 | 0.03842 | 0.284 |
| 45139 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-96-5p | 13 | NRXN1 | Sponge network | -0.31 | 0.33871 | -3.778 | 0 | 0.284 |
| 45140 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-582-5p | 11 | HDAC9 | Sponge network | -2.095 | 0.00112 | -0.755 | 0.00495 | 0.284 |
| 45141 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | SPG20 | Sponge network | 0.222 | 0.29133 | -1.16 | 0 | 0.284 |
| 45142 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-590-3p | 23 | KAT6B | Sponge network | 0.494 | 0.00339 | -0.478 | 0.00018 | 0.284 |
| 45143 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 66 | SMAD4 | Sponge network | -0.739 | 0.0574 | -0.313 | 0.00413 | 0.284 |
| 45144 | GABPB1-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | ARHGEF12 | Sponge network | 1.11 | 0 | 0.09 | 0.35693 | 0.284 |
| 45145 | CASC2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 12 | YAP1 | Sponge network | 0.385 | 0.01306 | 0.22 | 0.11836 | 0.284 |
| 45146 | LINC00969 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 13 | CDK6 | Sponge network | 0.683 | 1.0E-5 | 0.991 | 0 | 0.284 |
| 45147 | AC003090.1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 10 | RASSF3 | Sponge network | -2.117 | 0.00161 | -0.226 | 0.11691 | 0.284 |
| 45148 | RP11-157J24.2 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-576-5p | 13 | IGF1 | Sponge network | -1.664 | 0.05165 | -0.204 | 0.5898 | 0.284 |
| 45149 | RP11-983P16.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 13 | LUZP2 | Sponge network | 0.41 | 0.02214 | -0.056 | 0.89416 | 0.284 |
| 45150 | HCG18 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p | 15 | TRIM13 | Sponge network | 1.063 | 0 | 0.183 | 0.06968 | 0.284 |
| 45151 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-96-5p | 12 | DOCK4 | Sponge network | 0.131 | 0.8436 | 0.502 | 0.00108 | 0.284 |
| 45152 | HOXA-AS3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-590-3p | 34 | DTNA | Sponge network | -0.555 | 0.06156 | -1.648 | 1.0E-5 | 0.284 |
| 45153 | ARHGEF26-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | UBE4B | Sponge network | -2.46 | 0 | -0.235 | 0.007 | 0.284 |
| 45154 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3913-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-769-5p | 28 | EGFR | Sponge network | -2.69 | 0.0001 | -0.175 | 0.48451 | 0.284 |
| 45155 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | BCL11A | Sponge network | 0.194 | 0.64278 | -0.932 | 0.0063 | 0.284 |
| 45156 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p | 29 | ST6GALNAC3 | Sponge network | -0.154 | 0.78939 | -0.987 | 0 | 0.284 |
| 45157 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-505-3p;hsa-miR-590-3p | 16 | KIT | Sponge network | 0.225 | 0.30644 | -2.184 | 0 | 0.284 |
| 45158 | CTD-2017D11.1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-744-3p | 37 | LPP | Sponge network | 0.792 | 0.0049 | -0.459 | 0.04475 | 0.284 |
| 45159 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-424-5p | 11 | RICTOR | Sponge network | -0.187 | 0.23787 | 0.388 | 0.00188 | 0.284 |
| 45160 | SNX29P2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | CREB1 | Sponge network | 0.4 | 0.14611 | 0.203 | 0.00537 | 0.284 |
| 45161 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | ARID5B | Sponge network | -0.576 | 0.10121 | 0.342 | 0.01676 | 0.284 |
| 45162 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484 | 13 | TRIM3 | Sponge network | -0.901 | 0.00019 | -0.577 | 0 | 0.284 |
| 45163 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-550a-3p | 35 | CUX1 | Sponge network | 0.082 | 0.55397 | -0.039 | 0.6705 | 0.284 |
| 45164 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PIK3C2A | Sponge network | 0.898 | 1.0E-5 | 0.061 | 0.53776 | 0.284 |
| 45165 | CTD-2314G24.2 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-671-5p | 12 | PIK3IP1 | Sponge network | 0.246 | 0.59912 | -0.187 | 0.19945 | 0.284 |
| 45166 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p | 19 | TACC1 | Sponge network | 0.368 | 0.20093 | -0.362 | 0.05406 | 0.284 |
| 45167 | HOXA-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DNAJB4 | Sponge network | 0.61 | 0.01593 | -0.7 | 1.0E-5 | 0.284 |
| 45168 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-503-5p;hsa-miR-590-3p | 30 | RICTOR | Sponge network | -0.223 | 0.47816 | 0.388 | 0.00188 | 0.284 |
| 45169 | OIP5-AS1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RBBP6 | Sponge network | 0.421 | 0.00084 | 0.163 | 0.05101 | 0.284 |
| 45170 | LINC00657 |
hsa-miR-142-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 16 | TMEM30A | Sponge network | 0.91 | 0 | 0.096 | 0.31031 | 0.284 |
| 45171 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | GALNT13 | Sponge network | -0.896 | 0.00099 | -0.857 | 0.07439 | 0.284 |
| 45172 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-629-3p | 12 | SPTBN4 | Sponge network | -1.654 | 0.00144 | -0.786 | 0.01281 | 0.284 |
| 45173 | RP11-2H8.2 |
hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-744-3p | 31 | DST | Sponge network | 1.089 | 0 | -0.713 | 0.00096 | 0.284 |
| 45174 | SERHL |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b | 11 | KANK1 | Sponge network | -0.602 | 0.00331 | -0.55 | 0.00134 | 0.284 |
| 45175 | AC156455.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p | 13 | NAV3 | Sponge network | 1.154 | 0.00041 | 0.212 | 0.47729 | 0.284 |
| 45176 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 19 | RNF144A | Sponge network | 0.614 | 1.0E-5 | 0.594 | 0.0009 | 0.284 |
| 45177 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | INPP4B | Sponge network | -0.612 | 0.00094 | -0.097 | 0.60503 | 0.284 |
| 45178 | RP11-66B24.2 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-330-5p;hsa-miR-378c;hsa-miR-550a-3p | 12 | BRD3 | Sponge network | 0.57 | 0.1866 | 0.37 | 0.00013 | 0.284 |
| 45179 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 11 | SASH1 | Sponge network | -0.557 | 0.11135 | -0.502 | 0.00175 | 0.284 |
| 45180 | RP11-439A17.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-590-5p | 10 | BMPR2 | Sponge network | 0.051 | 0.95756 | 0.206 | 0.0635 | 0.284 |
| 45181 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-532-3p;hsa-miR-629-3p | 18 | GAS7 | Sponge network | -1.054 | 0.0706 | -0.555 | 0.01602 | 0.284 |
| 45182 | RP11-693J15.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | MED12L | Sponge network | -0.72 | 0.24239 | -0.194 | 0.55299 | 0.284 |
| 45183 | ZNF32-AS2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-539-5p | 11 | ARHGEF12 | Sponge network | 1.552 | 7.0E-5 | 0.09 | 0.35693 | 0.284 |
| 45184 | RP11-384K6.6 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-328-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-550a-5p;hsa-miR-628-5p | 17 | VPS53 | Sponge network | 0.946 | 0 | 0.103 | 0.22667 | 0.284 |
| 45185 | RP11-736K20.5 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 19 | EFNA5 | Sponge network | -0.358 | 0.17255 | -0.421 | 0.17868 | 0.284 |
| 45186 | RP11-822E23.8 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-940 | 26 | ETV1 | Sponge network | -0.777 | 0.16667 | -0.096 | 0.67674 | 0.284 |
| 45187 | AC034220.3 |
hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | RB1CC1 | Sponge network | 0.309 | 0.07304 | 0.175 | 0.12559 | 0.284 |
| 45188 | LINC00969 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-628-5p | 16 | CDC73 | Sponge network | 0.683 | 1.0E-5 | 0.094 | 0.20463 | 0.284 |
| 45189 | KB-431C1.4 |
hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-582-3p | 12 | BRD3 | Sponge network | 0.909 | 0 | 0.37 | 0.00013 | 0.284 |
| 45190 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-654-3p | 13 | PSIP1 | Sponge network | 0.222 | 0.29133 | -0.005 | 0.96897 | 0.284 |
| 45191 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p | 11 | CACNA2D1 | Sponge network | 0.792 | 0.0049 | -0.846 | 0.00675 | 0.284 |
| 45192 | RP4-669H2.1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-628-5p | 12 | BMPR2 | Sponge network | 1.711 | 0.00019 | 0.206 | 0.0635 | 0.284 |
| 45193 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-744-5p | 25 | NOTCH2 | Sponge network | -0.469 | 0.08721 | 0.138 | 0.35163 | 0.284 |
| 45194 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p | 28 | ESR1 | Sponge network | -0.612 | 0.00094 | -1.035 | 3.0E-5 | 0.284 |
| 45195 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PTPRB | Sponge network | 0.72 | 0.0001 | 0.105 | 0.47122 | 0.284 |
| 45196 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-7-1-3p | 18 | ITGAV | Sponge network | -0.976 | 0.00025 | 0.337 | 0.01002 | 0.284 |
| 45197 | JAZF1-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | LUZP2 | Sponge network | -0.769 | 0.00035 | -0.056 | 0.89416 | 0.284 |
| 45198 | RP11-262H14.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | HACE1 | Sponge network | -0.106 | 0.63074 | -0.072 | 0.55239 | 0.284 |
| 45199 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374a-3p | 17 | GREM1 | Sponge network | 0.219 | 0.1516 | 0.902 | 0.01691 | 0.284 |
| 45200 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-7-1-3p | 21 | KAT6B | Sponge network | -0.969 | 2.0E-5 | -0.478 | 0.00018 | 0.284 |
| 45201 | LINC00173 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-940 | 37 | NFIB | Sponge network | 0.056 | 0.8494 | 0.023 | 0.90795 | 0.284 |
| 45202 | AF131215.2 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-3p | 12 | RELN | Sponge network | -0.176 | 0.59692 | -2.403 | 0 | 0.284 |
| 45203 | RP11-503E24.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-495-3p | 11 | ZFX | Sponge network | 2.218 | 0 | 0.09 | 0.42563 | 0.284 |
| 45204 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-93-5p | 20 | LRRC55 | Sponge network | 0.253 | 0.09565 | -0.026 | 0.93163 | 0.284 |
| 45205 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p | 17 | GNA13 | Sponge network | 0.082 | 0.55654 | 0.007 | 0.93884 | 0.284 |
| 45206 | CTD-2554C21.2 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-7-5p;hsa-miR-96-5p | 27 | ERBB4 | Sponge network | -1.654 | 0.00144 | -1.975 | 2.0E-5 | 0.284 |
| 45207 | AC074289.1 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-532-3p | 11 | CADM3 | Sponge network | -0.326 | 0.14314 | -3.663 | 0 | 0.284 |
| 45208 | CTD-2554C21.2 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-5p;hsa-miR-96-5p | 14 | IRS1 | Sponge network | -1.654 | 0.00144 | -0.026 | 0.88855 | 0.284 |
| 45209 | PPP1R26-AS1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-195-3p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-664a-5p | 10 | SOX11 | Sponge network | 1.454 | 0 | 2.68 | 0 | 0.284 |
| 45210 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p | 15 | ANK3 | Sponge network | 0.401 | 0.00066 | -0.55 | 0.00086 | 0.284 |
| 45211 | RP11-327P2.5 |
hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-625-5p | 11 | BCL9 | Sponge network | -0.147 | 0.40452 | 0.321 | 0.00267 | 0.284 |
| 45212 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | CSRNP1 | Sponge network | -0.49 | 0.04946 | -1.448 | 0 | 0.284 |
| 45213 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-429 | 12 | SLC6A15 | Sponge network | -0.154 | 0.53954 | -2.373 | 2.0E-5 | 0.284 |
| 45214 | RP11-411K7.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 34 | NTRK2 | Sponge network | 0.155 | 0.81273 | -0.736 | 0.06495 | 0.284 |
| 45215 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-484 | 14 | MED12L | Sponge network | 0.858 | 3.0E-5 | -0.194 | 0.55299 | 0.284 |
| 45216 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429 | 20 | RICTOR | Sponge network | 0.859 | 0.00331 | 0.388 | 0.00188 | 0.284 |
| 45217 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-34a-5p | 16 | ACSL6 | Sponge network | -4.477 | 0 | -1.593 | 0 | 0.284 |
| 45218 | FGF14-AS2 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | EFNA5 | Sponge network | -1.582 | 8.0E-5 | -0.421 | 0.17868 | 0.284 |
| 45219 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-3614-5p | 17 | MOB1B | Sponge network | 1.056 | 0 | 0.197 | 0.15266 | 0.284 |
| 45220 | RP11-770J1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-34a-5p | 11 | CBFA2T3 | Sponge network | -2.011 | 0.0005 | -1.221 | 1.0E-5 | 0.284 |
| 45221 | RP11-59H7.3 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 13 | PRRX1 | Sponge network | 2.163 | 0 | 1.316 | 0 | 0.284 |
| 45222 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | DACH2 | Sponge network | -0.576 | 0.10121 | -1.635 | 0.00034 | 0.284 |
| 45223 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p | 17 | EIF4A2 | Sponge network | -0.83 | 0 | -0.383 | 0.00046 | 0.284 |
| 45224 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PLSCR4 | Sponge network | 0.083 | 0.66405 | -0.474 | 0.03833 | 0.284 |
| 45225 | GS1-124K5.11 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-5p | 19 | UBR3 | Sponge network | 0.494 | 0.00339 | -0.021 | 0.82774 | 0.284 |
| 45226 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | ARHGAP29 | Sponge network | 0.199 | 0.38015 | 0.131 | 0.42953 | 0.284 |
| 45227 | AC005154.6 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p;hsa-miR-664a-5p | 13 | CBFB | Sponge network | 0.848 | 0 | 1.061 | 0 | 0.284 |
| 45228 | LINC00476 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | CITED2 | Sponge network | -0.615 | 0.00015 | -1.545 | 0 | 0.284 |
| 45229 | CTC-457L16.2 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-935;hsa-miR-940 | 21 | GRIK3 | Sponge network | 1.153 | 0.00114 | -3.208 | 0 | 0.284 |
| 45230 | LINC00641 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 19 | RPS6KA3 | Sponge network | -0.222 | 0.32445 | 0.256 | 0.02419 | 0.284 |
| 45231 | GNG12-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p | 25 | PDE4DIP | Sponge network | -0.793 | 7.0E-5 | -0.282 | 0.0257 | 0.284 |
| 45232 | AC138035.2 |
hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-33a-5p | 12 | SOX11 | Sponge network | 1.073 | 0 | 2.68 | 0 | 0.284 |
| 45233 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TRIP11 | Sponge network | -0.83 | 0 | -0.158 | 0.06384 | 0.284 |
| 45234 | RP11-35G9.3 |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | RECK | Sponge network | 0.657 | 4.0E-5 | -1.043 | 0 | 0.284 |
| 45235 | AC004893.11 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 21 | ARHGEF12 | Sponge network | 1.317 | 0 | 0.09 | 0.35693 | 0.284 |
| 45236 | AC156455.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-664a-5p | 22 | ADARB1 | Sponge network | 1.154 | 0.00041 | -0.335 | 0.06631 | 0.284 |
| 45237 | PSMG3-AS1 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PCDH18 | Sponge network | -0.125 | 0.49414 | 0.216 | 0.24645 | 0.284 |
| 45238 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | RB1CC1 | Sponge network | 0.532 | 0.00505 | 0.175 | 0.12559 | 0.284 |
| 45239 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p | 20 | PTPRD | Sponge network | -0.829 | 0.01343 | -1.005 | 0.00064 | 0.284 |
| 45240 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-590-3p | 14 | DACH2 | Sponge network | -0.793 | 7.0E-5 | -1.635 | 0.00034 | 0.284 |
| 45241 | RP11-815I9.4 |
hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30c-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 11 | NUAK1 | Sponge network | 0.537 | 0.0006 | 0.904 | 0 | 0.284 |
| 45242 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p | 13 | CUL5 | Sponge network | 0.082 | 0.55397 | 0.026 | 0.77301 | 0.284 |
| 45243 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ST6GALNAC3 | Sponge network | -0.125 | 0.49414 | -0.987 | 0 | 0.284 |
| 45244 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | CTDSPL | Sponge network | 0.782 | 0.00016 | 0.085 | 0.45121 | 0.284 |
| 45245 | TINCR |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-940 | 29 | NTRK2 | Sponge network | -0.664 | 0.14543 | -0.736 | 0.06495 | 0.284 |
| 45246 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-539-5p | 18 | PGR | Sponge network | -0.602 | 0.00331 | -1.704 | 0 | 0.284 |
| 45247 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | TACC1 | Sponge network | -0.726 | 0.00132 | -0.362 | 0.05406 | 0.284 |
| 45248 | AC025171.1 |
hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SHPRH | Sponge network | 0.998 | 0 | 0.098 | 0.40994 | 0.284 |
| 45249 | RP11-95D17.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-497-5p | 12 | BRD3 | Sponge network | 0.75 | 0 | 0.37 | 0.00013 | 0.284 |
| 45250 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p | 16 | SPG20 | Sponge network | -0.668 | 3.0E-5 | -1.16 | 0 | 0.284 |
| 45251 | LINC00702 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | NR3C2 | Sponge network | -0.737 | 0.06182 | -1.56 | 0 | 0.284 |
| 45252 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-935 | 36 | MTSS1 | Sponge network | -2.925 | 0 | -0.81 | 0.00035 | 0.284 |
| 45253 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | -0.421 | 0.0114 | 0.309 | 0.17079 | 0.284 |
| 45254 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 12 | UBE4B | Sponge network | 0.225 | 0.30644 | -0.235 | 0.007 | 0.284 |
| 45255 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b | 12 | MYCBP2 | Sponge network | -1.645 | 0 | -0.027 | 0.82016 | 0.284 |
| 45256 | AC003090.1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-501-5p | 17 | MAFB | Sponge network | -2.117 | 0.00161 | -0.023 | 0.89865 | 0.284 |
| 45257 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | GALNT13 | Sponge network | 0.101 | 0.60919 | -0.857 | 0.07439 | 0.284 |
| 45258 | RP11-46C24.7 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 10 | DCLK1 | Sponge network | 0.392 | 0.0278 | -1.324 | 0.00014 | 0.284 |
| 45259 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | CACNA2D1 | Sponge network | 0.538 | 0.00076 | -0.846 | 0.00675 | 0.284 |
| 45260 | KB-1460A1.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CEP170 | Sponge network | 0.603 | 0.00127 | 0.943 | 0 | 0.284 |
| 45261 | RP11-359B12.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | KCTD8 | Sponge network | 0.064 | 0.72934 | -3.359 | 0 | 0.284 |
| 45262 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p | 13 | GPAM | Sponge network | -0.384 | 0.40652 | 0.142 | 0.29732 | 0.284 |
| 45263 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 54 | TCF4 | Sponge network | 0.253 | 0.09565 | 0.016 | 0.92271 | 0.284 |
| 45264 | RAMP2-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-671-5p;hsa-miR-7-5p | 10 | EHD3 | Sponge network | -0.513 | 0.04703 | -0.484 | 0.01747 | 0.284 |
| 45265 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3C | Sponge network | 0.532 | 0.00505 | -0.272 | 0.31153 | 0.284 |
| 45266 | RP11-244H3.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 17 | TET1 | Sponge network | 0.659 | 3.0E-5 | -0.135 | 0.52138 | 0.284 |
| 45267 | RP11-276H19.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-550a-3p | 14 | PTPRT | Sponge network | -0.472 | 0.48851 | -0.907 | 0.03727 | 0.284 |
| 45268 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-424-5p | 16 | CBFA2T3 | Sponge network | 0.176 | 0.37402 | -1.221 | 1.0E-5 | 0.284 |
| 45269 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 35 | EGFR | Sponge network | -0.576 | 0.10121 | -0.175 | 0.48451 | 0.284 |
| 45270 | MIR143HG |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 56 | MDM4 | Sponge network | -0.739 | 0.0574 | 0.345 | 0.00762 | 0.284 |
| 45271 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | ST6GALNAC3 | Sponge network | 0.374 | 0.20054 | -0.987 | 0 | 0.284 |
| 45272 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | CREBBP | Sponge network | 2.094 | 0 | 0.126 | 0.15186 | 0.284 |
| 45273 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | DACH2 | Sponge network | -1.216 | 0 | -1.635 | 0.00034 | 0.284 |
| 45274 | MRVI1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-7-1-3p | 26 | GNAQ | Sponge network | -1.092 | 4.0E-5 | -0.367 | 0.00102 | 0.284 |
| 45275 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | BCLAF1 | Sponge network | 0.222 | 0.29133 | 0.211 | 0.00684 | 0.284 |
| 45276 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 27 | MTSS1 | Sponge network | -0.322 | 0.25945 | -0.81 | 0.00035 | 0.283 |
| 45277 | RP11-67L2.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | DCLK1 | Sponge network | 0.72 | 0.0001 | -1.324 | 0.00014 | 0.283 |
| 45278 | PWAR6 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-589-5p;hsa-miR-590-3p | 12 | ARID1B | Sponge network | -1.575 | 6.0E-5 | 0.003 | 0.97503 | 0.283 |
| 45279 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-33a-3p | 11 | NAV3 | Sponge network | 0.542 | 0.00054 | 0.212 | 0.47729 | 0.283 |
| 45280 | LINC00403 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-424-5p | 10 | ACVR2A | Sponge network | -3.586 | 0.00032 | -0.409 | 0.00039 | 0.283 |
| 45281 | CTA-204B4.2 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-590-3p | 11 | DYNC1I1 | Sponge network | 0.765 | 7.0E-5 | -1.12 | 0.00107 | 0.283 |
| 45282 | RP11-242D8.1 |
hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 10 | PHF6 | Sponge network | 1.404 | 0 | 0.785 | 0 | 0.283 |
| 45283 | LINC00472 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-7-1-3p | 19 | RNF144A | Sponge network | -0.842 | 0.01361 | 0.594 | 0.0009 | 0.283 |
| 45284 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-654-3p | 12 | USP25 | Sponge network | -0.405 | 0.22013 | -0.242 | 0.01397 | 0.283 |
| 45285 | OSER1-AS1 |
hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 10 | PEG3 | Sponge network | -0.13 | 0.48734 | -1.74 | 0 | 0.283 |
| 45286 | AC034220.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | ZMYM2 | Sponge network | 0.309 | 0.07304 | 0.279 | 0.01273 | 0.283 |
| 45287 | RP11-46C24.7 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 25 | DLC1 | Sponge network | 0.392 | 0.0278 | -0.382 | 0.05038 | 0.283 |
| 45288 | KB-1460A1.5 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | 0.603 | 0.00127 | -0.704 | 0.00106 | 0.283 |
| 45289 | OSER1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 15 | EZH1 | Sponge network | -0.13 | 0.48734 | -0.564 | 1.0E-5 | 0.283 |
| 45290 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p | 10 | HECA | Sponge network | 0.912 | 0.00014 | -0.217 | 0.03463 | 0.283 |
| 45291 | BDNF-AS |
hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 11 | PPP2R2C | Sponge network | -0.668 | 3.0E-5 | -0.959 | 0.07428 | 0.283 |
| 45292 | RP1-257A7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p | 11 | ARID5B | Sponge network | 0.849 | 0.00178 | 0.342 | 0.01676 | 0.283 |
| 45293 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 30 | ARID5B | Sponge network | 0.75 | 0.00054 | 0.342 | 0.01676 | 0.283 |
| 45294 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p | 15 | GPAM | Sponge network | -0.303 | 0.21002 | 0.142 | 0.29732 | 0.283 |
| 45295 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 33 | NCOA1 | Sponge network | -0.49 | 0.04946 | -0.432 | 0 | 0.283 |
| 45296 | AC025171.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SON | Sponge network | 0.998 | 0 | 0.168 | 0.04406 | 0.283 |
| 45297 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 58 | KIF1B | Sponge network | 0.488 | 0.00084 | -0.118 | 0.32258 | 0.283 |
| 45298 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 28 | PRDM2 | Sponge network | 0.72 | 0.0001 | -0.258 | 0.00721 | 0.283 |
| 45299 | RP4-613B23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-378a-5p | 10 | NUAK1 | Sponge network | -0.309 | 0.1145 | 0.904 | 0 | 0.283 |
| 45300 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | RAP1A | Sponge network | 0.274 | 0.07681 | -0.929 | 0 | 0.283 |
| 45301 | RP11-244O19.1 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-590-3p | 30 | ARHGEF12 | Sponge network | -0.096 | 0.67958 | 0.09 | 0.35693 | 0.283 |
| 45302 | RP11-25K19.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | ETV1 | Sponge network | -1.057 | 0.00029 | -0.096 | 0.67674 | 0.283 |
| 45303 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | SLITRK6 | Sponge network | -0.663 | 0.10887 | -2.121 | 0 | 0.283 |
| 45304 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PAFAH1B1 | Sponge network | 0.673 | 0 | -0.429 | 0 | 0.283 |
| 45305 | HOXB-AS3 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p | 10 | MRVI1 | Sponge network | 0.343 | 0.28887 | -1.051 | 0.00022 | 0.283 |
| 45306 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | PGR | Sponge network | -0.668 | 3.0E-5 | -1.704 | 0 | 0.283 |
| 45307 | A2M-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-590-3p | 10 | PTPN13 | Sponge network | -0.08 | 0.90092 | -1.208 | 0 | 0.283 |
| 45308 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-576-5p | 16 | ERCC4 | Sponge network | 0.299 | 0.57722 | 0.126 | 0.15266 | 0.283 |
| 45309 | BVES-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-429 | 11 | PRKAR1A | Sponge network | -2.62 | 0 | -0.063 | 0.50314 | 0.283 |
| 45310 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-98-5p | 17 | SEMA3C | Sponge network | 0.082 | 0.55397 | -0.272 | 0.31153 | 0.283 |
| 45311 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | TCF12 | Sponge network | 0.083 | 0.66405 | 0.174 | 0.13271 | 0.283 |
| 45312 | TP73-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-671-5p | 12 | PIK3IP1 | Sponge network | -0.563 | 0.02545 | -0.187 | 0.19945 | 0.283 |
| 45313 | RP11-244H3.1 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 28 | DIP2C | Sponge network | 0.659 | 3.0E-5 | -0.436 | 0.01713 | 0.283 |
| 45314 | DNM1P35 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-93-5p | 11 | GLI3 | Sponge network | 0.051 | 0.83388 | -0.615 | 0.01482 | 0.283 |
| 45315 | TINCR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-589-3p | 19 | PRDM2 | Sponge network | -0.664 | 0.14543 | -0.258 | 0.00721 | 0.283 |
| 45316 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-628-5p | 18 | BCL6 | Sponge network | -2.915 | 0.00015 | -0.211 | 0.19489 | 0.283 |
| 45317 | RP11-195F19.9 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | LRIG1 | Sponge network | -0.726 | 0.00132 | -0.537 | 0.00588 | 0.283 |
| 45318 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-671-5p | 20 | KIT | Sponge network | 0.4 | 0.14611 | -2.184 | 0 | 0.283 |
| 45319 | RP11-428J1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 19 | HECA | Sponge network | 0.538 | 0.00076 | -0.217 | 0.03463 | 0.283 |
| 45320 | AC005618.6 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | KIF5B | Sponge network | 0.862 | 0.06213 | 0.362 | 0.00066 | 0.283 |
| 45321 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | TET1 | Sponge network | 0.782 | 0.00016 | -0.135 | 0.52138 | 0.283 |
| 45322 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CTDSPL | Sponge network | 0.537 | 0.00139 | 0.085 | 0.45121 | 0.283 |
| 45323 | AF131215.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 30 | PPP1R9A | Sponge network | -0.176 | 0.59692 | -0.903 | 0.04254 | 0.283 |
| 45324 | DIO3OS |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-940 | 13 | CELF4 | Sponge network | -0.773 | 0.04073 | -1.135 | 0.00344 | 0.283 |
| 45325 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | AKAP13 | Sponge network | -0.842 | 0.01361 | 0.028 | 0.7729 | 0.283 |
| 45326 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 40 | CBX5 | Sponge network | -0.223 | 0.47816 | 0.165 | 0.24446 | 0.283 |
| 45327 | CTD-2303H24.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | ARHGEF12 | Sponge network | 0.415 | 0.14657 | 0.09 | 0.35693 | 0.283 |
| 45328 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 21 | SOX11 | Sponge network | 0.194 | 0.64278 | 2.68 | 0 | 0.283 |
| 45329 | AC009120.6 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-378c;hsa-miR-576-5p | 18 | ERC1 | Sponge network | 0.919 | 0 | -0.108 | 0.38794 | 0.283 |
| 45330 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 17 | NTRK2 | Sponge network | -0.021 | 0.93598 | -0.736 | 0.06495 | 0.283 |
| 45331 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | GPAM | Sponge network | 0.556 | 0.00298 | 0.142 | 0.29732 | 0.283 |
| 45332 | NNT-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | SIRT1 | Sponge network | 0.799 | 1.0E-5 | 0.109 | 0.2593 | 0.283 |
| 45333 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-93-3p | 23 | ST6GAL2 | Sponge network | 0.082 | 0.55654 | -1.201 | 0.00597 | 0.283 |
| 45334 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-629-3p | 32 | AR | Sponge network | -0.602 | 0.00331 | -1.554 | 0 | 0.283 |
| 45335 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p | 11 | ITCH | Sponge network | 0.752 | 0 | 0.084 | 0.34058 | 0.283 |
| 45336 | RP11-216B9.6 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | SORCS1 | Sponge network | 0.174 | 0.30034 | -3.492 | 0 | 0.283 |
| 45337 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 15 | SOX11 | Sponge network | 0.439 | 0.00313 | 2.68 | 0 | 0.283 |
| 45338 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-744-3p | 24 | HIP1 | Sponge network | -1.697 | 0.00889 | 0.774 | 0 | 0.283 |
| 45339 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-935 | 29 | FBXO32 | Sponge network | -0.727 | 0.04726 | -0.495 | 0.07561 | 0.283 |
| 45340 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-940 | 47 | WDFY3 | Sponge network | 0.494 | 0.00339 | -0.201 | 0.0967 | 0.283 |
| 45341 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | PHC3 | Sponge network | 0.219 | 0.1516 | 0.212 | 0.0499 | 0.283 |
| 45342 | RAMP2-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p | 10 | RHOB | Sponge network | -0.513 | 0.04703 | -1.052 | 0 | 0.283 |
| 45343 | GS1-124K5.11 |
hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 14 | GLI3 | Sponge network | 0.494 | 0.00339 | -0.615 | 0.01482 | 0.283 |
| 45344 | PDXDC2P |
hsa-miR-125b-2-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-376c-3p;hsa-miR-9-5p | 11 | PAWR | Sponge network | 0.756 | 0 | 0.636 | 0 | 0.283 |
| 45345 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 11 | NF1 | Sponge network | 1.302 | 7.0E-5 | 0.51 | 0 | 0.283 |
| 45346 | RP5-1021I20.1 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-744-3p | 22 | DIP2C | Sponge network | -0.785 | 0.00035 | -0.436 | 0.01713 | 0.283 |
| 45347 | FGD5-AS1 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | ATM | Sponge network | 0.401 | 0.00066 | 0.178 | 0.21568 | 0.283 |
| 45348 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 46 | CUX1 | Sponge network | 0.082 | 0.55654 | -0.039 | 0.6705 | 0.283 |
| 45349 | BDNF-AS |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | EFNA5 | Sponge network | -0.668 | 3.0E-5 | -0.421 | 0.17868 | 0.283 |
| 45350 | LINC00173 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p | 10 | FMNL3 | Sponge network | 0.056 | 0.8494 | 0.555 | 4.0E-5 | 0.283 |
| 45351 | HOXB-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-664a-5p | 23 | PPARA | Sponge network | 1.316 | 0.01106 | -0.379 | 0.00548 | 0.283 |
| 45352 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 26 | NUAK1 | Sponge network | -0.487 | 0.02157 | 0.904 | 0 | 0.283 |
| 45353 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-877-5p | 38 | RBMS3 | Sponge network | -0.371 | 0.24604 | -0.519 | 0.03551 | 0.283 |
| 45354 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 23 | TP53INP1 | Sponge network | -0.129 | 0.57177 | -0.496 | 0.00064 | 0.283 |
| 45355 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | 0.246 | 0.59912 | -0.7 | 1.0E-5 | 0.283 |
| 45356 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p | 24 | EYA4 | Sponge network | 0.214 | 0.18246 | 0.3 | 0.36876 | 0.283 |
| 45357 | BVES-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-5p | 11 | UNC5D | Sponge network | -2.62 | 0 | -3.646 | 0 | 0.283 |
| 45358 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-424-5p | 17 | LRRC7 | Sponge network | -0.777 | 0.16667 | -1.341 | 0.01193 | 0.283 |
| 45359 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-942-5p | 46 | UNC80 | Sponge network | 0.101 | 0.60919 | -1.934 | 0 | 0.283 |
| 45360 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | KDSR | Sponge network | 0.311 | 0.10344 | 0.239 | 0.02216 | 0.283 |
| 45361 | RP11-401P9.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-539-5p | 15 | PRKAA2 | Sponge network | 1.524 | 0.1098 | -2.462 | 0 | 0.283 |
| 45362 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 25 | CXXC4 | Sponge network | -0.727 | 0.04726 | -0.433 | 0.21055 | 0.283 |
| 45363 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p | 17 | FGFR1 | Sponge network | 0.72 | 0.0001 | -0.509 | 0.03957 | 0.283 |
| 45364 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-3065-3p;hsa-miR-339-5p;hsa-miR-542-3p;hsa-miR-942-5p | 11 | FBXO31 | Sponge network | 0.078 | 0.55803 | -0.085 | 0.42389 | 0.283 |
| 45365 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-590-3p | 13 | CLASP2 | Sponge network | 0.702 | 7.0E-5 | -0.038 | 0.71 | 0.283 |
| 45366 | LINC00472 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 19 | SEMA3C | Sponge network | -0.842 | 0.01361 | -0.272 | 0.31153 | 0.283 |
| 45367 | ZNF582-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p | 14 | TLE4 | Sponge network | -1.349 | 0.00017 | -1.157 | 0 | 0.283 |
| 45368 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 64 | KIF1B | Sponge network | -1.281 | 0.00012 | -0.118 | 0.32258 | 0.283 |
| 45369 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-582-5p | 15 | NAV3 | Sponge network | 0.993 | 0 | 0.212 | 0.47729 | 0.283 |
| 45370 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 18 | CCND2 | Sponge network | -2.039 | 0.03447 | -0.267 | 0.3112 | 0.283 |
| 45371 | RP11-428J1.5 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | UBR3 | Sponge network | 0.538 | 0.00076 | -0.021 | 0.82774 | 0.283 |
| 45372 | RP11-67L2.2 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | PTPN13 | Sponge network | 0.72 | 0.0001 | -1.208 | 0 | 0.283 |
| 45373 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-590-3p | 14 | DLC1 | Sponge network | 0.898 | 1.0E-5 | -0.382 | 0.05038 | 0.283 |
| 45374 | U91328.21 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 10 | PGR | Sponge network | -0.735 | 0.15983 | -1.704 | 0 | 0.283 |
| 45375 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | EPHA3 | Sponge network | 0.539 | 0.03722 | 0.185 | 0.53588 | 0.283 |
| 45376 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-139-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p | 15 | NF1 | Sponge network | 0.768 | 7.0E-5 | 0.51 | 0 | 0.283 |
| 45377 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-361-5p | 15 | DLC1 | Sponge network | -0.418 | 0.00068 | -0.382 | 0.05038 | 0.283 |
| 45378 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 35 | ADARB1 | Sponge network | 0.199 | 0.38015 | -0.335 | 0.06631 | 0.283 |
| 45379 | LINC00657 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | TCF12 | Sponge network | 0.91 | 0 | 0.174 | 0.13271 | 0.283 |
| 45380 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | SPG20 | Sponge network | 0.619 | 0.00157 | -1.16 | 0 | 0.283 |
| 45381 | RP11-968O1.5 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p | 11 | ERG | Sponge network | -0.829 | 0.01343 | 0.002 | 0.99011 | 0.283 |
| 45382 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | RABEP1 | Sponge network | -0.737 | 0.06182 | -0.066 | 0.43142 | 0.283 |
| 45383 | KB-1460A1.5 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | 0.603 | 0.00127 | -1.12 | 0.00107 | 0.283 |
| 45384 | BMS1P20 |
hsa-miR-136-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | USP9X | Sponge network | 0.34 | 0.00273 | 0.222 | 0.01689 | 0.283 |
| 45385 | RP11-635N19.1 |
hsa-miR-106a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-378a-3p;hsa-miR-455-3p | 10 | NTRK3 | Sponge network | -0.947 | 0.00455 | -1.77 | 3.0E-5 | 0.283 |
| 45386 | RP11-119F7.5 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | GABRB2 | Sponge network | 0.225 | 0.30644 | -1.841 | 0.00029 | 0.283 |
| 45387 | RP11-110I1.11 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p | 11 | UBR3 | Sponge network | 1.034 | 0 | -0.021 | 0.82774 | 0.283 |
| 45388 | RP11-130L8.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 25 | MDM4 | Sponge network | -0.663 | 0.10887 | 0.345 | 0.00762 | 0.283 |
| 45389 | RP11-13K12.5 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-505-5p | 11 | WNK2 | Sponge network | -0.318 | 0.65847 | -0.352 | 0.31066 | 0.283 |
| 45390 | RP11-983P16.4 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | CSMD1 | Sponge network | 0.41 | 0.02214 | -0.63 | 0.18881 | 0.283 |
| 45391 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p | 17 | KANK1 | Sponge network | -0.031 | 0.89063 | -0.55 | 0.00134 | 0.283 |
| 45392 | RP11-157J24.2 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-576-5p | 11 | NRXN1 | Sponge network | -1.664 | 0.05165 | -3.778 | 0 | 0.283 |
| 45393 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | NOTCH2NL | Sponge network | -0.612 | 0.00094 | 0.024 | 0.84307 | 0.283 |
| 45394 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-93-5p;hsa-miR-940 | 24 | BHLHE41 | Sponge network | 0.056 | 0.81195 | 0.353 | 0.12753 | 0.283 |
| 45395 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 46 | BMPR2 | Sponge network | -0.08 | 0.90092 | 0.206 | 0.0635 | 0.283 |
| 45396 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 19 | PHC3 | Sponge network | 0.001 | 0.99753 | 0.212 | 0.0499 | 0.283 |
| 45397 | RP11-6N17.3 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-199b-5p;hsa-miR-19b-1-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p | 10 | SMAD4 | Sponge network | 0.108 | 0.69556 | -0.313 | 0.00413 | 0.283 |
| 45398 | DNM3OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-651-5p | 20 | PRDM2 | Sponge network | 0.572 | 0.01722 | -0.258 | 0.00721 | 0.283 |
| 45399 | CTC-296K1.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-3913-5p;hsa-miR-454-3p | 20 | AFF1 | Sponge network | -2.095 | 0.00112 | -0.384 | 0.00053 | 0.283 |
| 45400 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 26 | LIFR | Sponge network | 0.194 | 0.64278 | -1.794 | 0 | 0.283 |
| 45401 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935;hsa-miR-940 | 48 | NFIB | Sponge network | 0.537 | 0.00139 | 0.023 | 0.90795 | 0.283 |
| 45402 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p | 14 | CREB3L2 | Sponge network | 0.043 | 0.81628 | -0.525 | 2.0E-5 | 0.283 |
| 45403 | RP11-386G11.5 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-511-5p | 15 | CREB1 | Sponge network | 2.031 | 0.00154 | 0.203 | 0.00537 | 0.283 |
| 45404 | RP11-531A24.5 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 44 | DTNA | Sponge network | 0.75 | 0.00054 | -1.648 | 1.0E-5 | 0.283 |
| 45405 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 13 | TMTC3 | Sponge network | 0.813 | 1.0E-5 | 0.123 | 0.22189 | 0.283 |
| 45406 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p | 13 | PDZD2 | Sponge network | -0.738 | 0.21233 | -0.909 | 3.0E-5 | 0.283 |
| 45407 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | PIK3R1 | Sponge network | -0.18 | 0.20343 | -0.133 | 0.36854 | 0.283 |
| 45408 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 30 | IGF1 | Sponge network | -0.899 | 6.0E-5 | -0.204 | 0.5898 | 0.283 |
| 45409 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 10 | ARNT | Sponge network | 0.997 | 0.00066 | -0.08 | 0.26073 | 0.283 |
| 45410 | EIF3J-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PIK3CA | Sponge network | 0.331 | 0.00509 | 0.195 | 0.06908 | 0.283 |
| 45411 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TRIP11 | Sponge network | 0.349 | 0.01695 | -0.158 | 0.06384 | 0.283 |
| 45412 | AC005618.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SETBP1 | Sponge network | 0.862 | 0.06213 | -1.302 | 1.0E-5 | 0.283 |
| 45413 | RP11-400K9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 43 | PTPRD | Sponge network | -0.611 | 0.09607 | -1.005 | 0.00064 | 0.283 |
| 45414 | RP11-37B2.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p;hsa-miR-582-5p | 23 | BRD3 | Sponge network | 1.03 | 0 | 0.37 | 0.00013 | 0.283 |
| 45415 | LINC00092 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 17 | PPP1R9A | Sponge network | -1.886 | 0 | -0.903 | 0.04254 | 0.283 |
| 45416 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SON | Sponge network | 0.34 | 0.00273 | 0.168 | 0.04406 | 0.283 |
| 45417 | RP11-196G18.22 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | TSC1 | Sponge network | 1.206 | 0 | 0.103 | 0.26071 | 0.283 |
| 45418 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 11 | HDAC9 | Sponge network | -4.546 | 0 | -0.755 | 0.00495 | 0.283 |
| 45419 | RP11-798M19.6 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-93-3p | 13 | RPS6KA2 | Sponge network | -0.612 | 0.00094 | -0.57 | 0.00013 | 0.283 |
| 45420 | RP11-384O8.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | NEDD4 | Sponge network | -0.738 | 0.21233 | 0.02 | 0.89834 | 0.283 |
| 45421 | HOXB-AS2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-484 | 10 | FBXO11 | Sponge network | 1.316 | 0.01106 | 0.142 | 0.04938 | 0.283 |
| 45422 | RP3-402G11.26 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | NTRK3 | Sponge network | -0.254 | 0.19303 | -1.77 | 3.0E-5 | 0.283 |
| 45423 | LINC00648 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p | 23 | ABL2 | Sponge network | 0.633 | 0.51678 | 0.82 | 0 | 0.283 |
| 45424 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 44 | CBX5 | Sponge network | 0.194 | 0.64278 | 0.165 | 0.24446 | 0.283 |
| 45425 | RP11-927P21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p | 29 | RUNX1T1 | Sponge network | 0.921 | 0.00118 | -0.344 | 0.2374 | 0.283 |
| 45426 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 12 | PARK2 | Sponge network | 0.72 | 0.0001 | -1.892 | 0 | 0.283 |
| 45427 | FZD10-AS1 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | ADCY1 | Sponge network | -0.727 | 0.04726 | 0.362 | 0.16982 | 0.283 |
| 45428 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p | 33 | SPRY3 | Sponge network | -0.899 | 6.0E-5 | 0.016 | 0.91286 | 0.283 |
| 45429 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p | 10 | KANK1 | Sponge network | 0.853 | 0.15352 | -0.55 | 0.00134 | 0.283 |
| 45430 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CEP170 | Sponge network | -0.096 | 0.67958 | 0.943 | 0 | 0.283 |
| 45431 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | TLL1 | Sponge network | 0.16 | 0.50785 | -1.195 | 3.0E-5 | 0.283 |
| 45432 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 19 | PCDH10 | Sponge network | -0.031 | 0.89063 | -1.644 | 0.0078 | 0.283 |
| 45433 | USP3-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-3065-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-664a-5p | 19 | SOX11 | Sponge network | -0.162 | 0.47687 | 2.68 | 0 | 0.283 |
| 45434 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 22 | BCL6 | Sponge network | -1.216 | 0 | -0.211 | 0.19489 | 0.283 |
| 45435 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 23 | EPHA3 | Sponge network | 0.848 | 0 | 0.185 | 0.53588 | 0.283 |
| 45436 | RP11-53O19.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p | 13 | PKNOX1 | Sponge network | -0.405 | 0.22013 | 0.085 | 0.28917 | 0.283 |
| 45437 | LINC00265 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-378a-5p | 14 | BMPR2 | Sponge network | 1.07 | 0 | 0.206 | 0.0635 | 0.283 |
| 45438 | BDNF-AS |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-484;hsa-miR-766-3p;hsa-miR-93-5p | 14 | EZH1 | Sponge network | -0.668 | 3.0E-5 | -0.564 | 1.0E-5 | 0.283 |
| 45439 | GS1-124K5.11 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 21 | CDH23 | Sponge network | 0.494 | 0.00339 | -0.974 | 0.00034 | 0.283 |
| 45440 | ARHGAP5-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 12 | GJA1 | Sponge network | -0.644 | 0.00021 | -0.485 | 0.02179 | 0.283 |
| 45441 | LINC00672 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-625-3p | 15 | NCAM2 | Sponge network | -0.227 | 0.31657 | -0.482 | 0.22565 | 0.283 |
| 45442 | NDUFA6-AS1 |
hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-877-5p | 13 | THRA | Sponge network | -0.355 | 0.0077 | -0.307 | 0.05099 | 0.283 |
| 45443 | DOCK9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-455-3p | 15 | FBXO32 | Sponge network | -0.231 | 0.28214 | -0.495 | 0.07561 | 0.283 |
| 45444 | AC074289.1 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-942-5p;hsa-miR-96-5p | 10 | ZBTB20 | Sponge network | -0.326 | 0.14314 | -0.122 | 0.57569 | 0.283 |
| 45445 | HOXB-AS2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-576-5p | 10 | BRD3 | Sponge network | 1.316 | 0.01106 | 0.37 | 0.00013 | 0.283 |
| 45446 | AC005154.6 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | GSK3B | Sponge network | 0.848 | 0 | 0.266 | 0.00045 | 0.283 |
| 45447 | RP5-994D16.9 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-2355-3p;hsa-miR-379-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 10 | SH3GLB1 | Sponge network | 0.252 | 0.08508 | -0.115 | 0.14773 | 0.283 |
| 45448 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-940 | 35 | NFIB | Sponge network | -0.154 | 0.53954 | 0.023 | 0.90795 | 0.283 |
| 45449 | HOXA-AS2 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | RECK | Sponge network | 0.61 | 0.01593 | -1.043 | 0 | 0.283 |
| 45450 | RP11-981G7.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PIK3CA | Sponge network | 0.986 | 0.01022 | 0.195 | 0.06908 | 0.283 |
| 45451 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p | 20 | CBX5 | Sponge network | 0.643 | 0 | 0.165 | 0.24446 | 0.283 |
| 45452 | TMEM51-AS1 |
hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-582-3p;hsa-miR-590-3p | 14 | FOXO3 | Sponge network | 0.23 | 0.38667 | -0.04 | 0.72028 | 0.283 |
| 45453 | AC004893.11 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | ERCC6 | Sponge network | 1.317 | 0 | 0.23 | 0.015 | 0.283 |
| 45454 | CTD-3025N20.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-590-3p | 11 | PIK3C2A | Sponge network | 1.046 | 0 | 0.061 | 0.53776 | 0.283 |
| 45455 | LINC00969 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-628-5p | 11 | ANK3 | Sponge network | 0.683 | 1.0E-5 | -0.55 | 0.00086 | 0.283 |
| 45456 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 21 | ACVR1 | Sponge network | -1.281 | 0.00012 | 0.279 | 0.00234 | 0.283 |
| 45457 | LINC00473 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-500a-3p;hsa-miR-628-5p;hsa-miR-7-5p | 24 | ERC1 | Sponge network | -1.203 | 0.03881 | -0.108 | 0.38794 | 0.283 |
| 45458 | RP11-327P2.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-324-5p;hsa-miR-502-5p;hsa-miR-576-5p | 15 | RIMBP2 | Sponge network | -0.147 | 0.40452 | -1.968 | 5.0E-5 | 0.283 |
| 45459 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 17 | PDGFRA | Sponge network | 2.306 | 9.0E-5 | -0.372 | 0.0037 | 0.283 |
| 45460 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-96-5p | 14 | PRKAR1A | Sponge network | -1.654 | 0.00144 | -0.063 | 0.50314 | 0.283 |
| 45461 | RP11-359B12.2 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p | 10 | LZTS1 | Sponge network | 0.064 | 0.72934 | 1.664 | 0 | 0.283 |
| 45462 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TBX18 | Sponge network | 0.657 | 4.0E-5 | 0.843 | 0.02945 | 0.283 |
| 45463 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | EPS15 | Sponge network | 0.225 | 0.30644 | -0.052 | 0.53998 | 0.283 |
| 45464 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 36 | CLASP2 | Sponge network | -1.697 | 0.00889 | -0.038 | 0.71 | 0.283 |
| 45465 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-146b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 13 | EIF4A2 | Sponge network | -0.031 | 0.89063 | -0.383 | 0.00046 | 0.283 |
| 45466 | RP1-39G22.7 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-3p | 22 | CREB1 | Sponge network | 0.439 | 0.00313 | 0.203 | 0.00537 | 0.283 |
| 45467 | HOTAIRM1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-590-3p | 18 | ASTN1 | Sponge network | -0.053 | 0.80685 | -2.389 | 2.0E-5 | 0.283 |
| 45468 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | KCNA6 | Sponge network | -0.342 | 0.02288 | -1.531 | 0.00013 | 0.283 |
| 45469 | LINC00641 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | NR3C2 | Sponge network | -0.222 | 0.32445 | -1.56 | 0 | 0.283 |
| 45470 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-628-5p | 11 | ANK3 | Sponge network | 0.537 | 0.00139 | -0.55 | 0.00086 | 0.283 |
| 45471 | RP11-458D21.1 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p | 11 | ERCC4 | Sponge network | 0.663 | 0.00073 | 0.126 | 0.15266 | 0.283 |
| 45472 | TINCR |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-376c-3p | 12 | RASAL2 | Sponge network | -0.664 | 0.14543 | 0.869 | 0 | 0.283 |
| 45473 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p | 11 | PCDH8 | Sponge network | -0.842 | 0.01361 | -0.997 | 0.05366 | 0.283 |
| 45474 | CTD-2554C21.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 11 | FYN | Sponge network | -1.654 | 0.00144 | -0.131 | 0.37051 | 0.283 |
| 45475 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | SYNE1 | Sponge network | 0.222 | 0.29133 | -0.738 | 0.00316 | 0.283 |
| 45476 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-590-3p | 13 | MYBL1 | Sponge network | 0.67 | 0.00048 | 0.494 | 0.02152 | 0.283 |
| 45477 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CXXC4 | Sponge network | 0.61 | 0.01593 | -0.433 | 0.21055 | 0.283 |
| 45478 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-7-1-3p | 12 | LRRC7 | Sponge network | -0.72 | 0.24239 | -1.341 | 0.01193 | 0.283 |
| 45479 | AP001347.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-502-5p;hsa-miR-590-3p | 16 | TGFBR2 | Sponge network | -1.161 | 0.00591 | -0.243 | 0.11408 | 0.283 |
| 45480 | NR2F1-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 54 | PPM1L | Sponge network | -0.49 | 0.04946 | -0.537 | 0.00616 | 0.283 |
| 45481 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p | 18 | CITED2 | Sponge network | -0.969 | 2.0E-5 | -1.545 | 0 | 0.283 |
| 45482 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 38 | AFF4 | Sponge network | 1.153 | 0 | -0.266 | 0.01565 | 0.283 |
| 45483 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 36 | MEF2C | Sponge network | -0.727 | 0.04726 | -0.499 | 0.01448 | 0.283 |
| 45484 | BMS1P20 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-134-5p;hsa-miR-16-2-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | PIK3C2A | Sponge network | 0.34 | 0.00273 | 0.061 | 0.53776 | 0.283 |
| 45485 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-93-5p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | -1.203 | 0.03881 | -0.085 | 0.42389 | 0.283 |
| 45486 | RP11-819C21.1 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 13 | PREX2 | Sponge network | 0.537 | 0.00139 | -0.272 | 0.25382 | 0.283 |
| 45487 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-625-5p;hsa-miR-96-5p | 16 | SNX29 | Sponge network | 0.614 | 1.0E-5 | -0.34 | 0.01371 | 0.283 |
| 45488 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-940 | 18 | CHD5 | Sponge network | 1.294 | 0.0031 | -0.922 | 0.01521 | 0.283 |
| 45489 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-497-5p | 12 | ZMYM2 | Sponge network | 0.48 | 0.05578 | 0.279 | 0.01273 | 0.283 |
| 45490 | XXYLT1-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p | 14 | JAZF1 | Sponge network | -0.764 | 0.05257 | -0.377 | 0.02677 | 0.283 |
| 45491 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p | 13 | TXNIP | Sponge network | -0.517 | 0.02935 | -1.277 | 0 | 0.283 |
| 45492 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-421 | 10 | CADM3 | Sponge network | 0.056 | 0.81195 | -3.663 | 0 | 0.283 |
| 45493 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | YAP1 | Sponge network | 0.572 | 0.01722 | 0.22 | 0.11836 | 0.283 |
| 45494 | AC005618.6 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PRUNE2 | Sponge network | 0.862 | 0.06213 | -1.733 | 0.00022 | 0.283 |
| 45495 | GABPB1-AS1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 17 | BAZ2B | Sponge network | 1.11 | 0 | -0.276 | 0.02833 | 0.283 |
| 45496 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-625-3p | 13 | CBX5 | Sponge network | 0.903 | 0 | 0.165 | 0.24446 | 0.283 |
| 45497 | HOXB-AS3 |
hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-452-5p;hsa-miR-532-3p | 10 | SYNM | Sponge network | 0.343 | 0.28887 | -2.309 | 1.0E-5 | 0.283 |
| 45498 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-940 | 19 | LIFR | Sponge network | 0.199 | 0.38015 | -1.794 | 0 | 0.283 |
| 45499 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-671-5p | 11 | HIC1 | Sponge network | -0.793 | 7.0E-5 | 0.024 | 0.89889 | 0.283 |
| 45500 | RP11-244O19.1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | TES | Sponge network | -0.096 | 0.67958 | -0.348 | 0.00639 | 0.283 |
| 45501 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-455-5p | 11 | ZNF521 | Sponge network | 0.659 | 3.0E-5 | -0.111 | 0.58068 | 0.283 |
| 45502 | RP3-475N16.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-495-3p | 14 | ZFX | Sponge network | 1.056 | 0 | 0.09 | 0.42563 | 0.283 |
| 45503 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-590-3p | 21 | USP6 | Sponge network | 0.083 | 0.66405 | 0.188 | 0.3871 | 0.283 |
| 45504 | RP11-400K9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-340-5p;hsa-miR-576-5p | 12 | EPHA5 | Sponge network | -0.733 | 0.19469 | -0.957 | 0.01733 | 0.283 |
| 45505 | MIR143HG |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-671-5p | 26 | PPP2R5C | Sponge network | -0.739 | 0.0574 | -0.314 | 3.0E-5 | 0.283 |
| 45506 | RP11-20I23.13 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-744-3p;hsa-miR-92a-3p | 47 | LPP | Sponge network | 0.505 | 0.0014 | -0.459 | 0.04475 | 0.283 |
| 45507 | FGF14-AS2 |
hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-660-5p | 10 | JDP2 | Sponge network | -1.582 | 8.0E-5 | -0.967 | 0 | 0.283 |
| 45508 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | CLIP1 | Sponge network | -2.117 | 0.00161 | -0.215 | 0.08684 | 0.283 |
| 45509 | RP11-403I13.7 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-671-5p | 14 | CADM3 | Sponge network | 0.395 | 0.15451 | -3.663 | 0 | 0.283 |
| 45510 | RP11-206L10.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-27b-3p;hsa-miR-338-3p;hsa-miR-339-5p | 13 | SOX11 | Sponge network | 0.793 | 0 | 2.68 | 0 | 0.283 |
| 45511 | HOXA-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p | 11 | PARK2 | Sponge network | 0.61 | 0.01593 | -1.892 | 0 | 0.283 |
| 45512 | LINC00672 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 11 | SHPRH | Sponge network | -0.227 | 0.31657 | 0.098 | 0.40994 | 0.283 |
| 45513 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | PDGFRA | Sponge network | 0.4 | 0.14611 | -0.372 | 0.0037 | 0.283 |
| 45514 | ZSCAN16-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | RYR2 | Sponge network | -0.342 | 0.02288 | -1.466 | 0.00031 | 0.283 |
| 45515 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p | 11 | FSTL5 | Sponge network | 0.75 | 0.00054 | -1.3 | 0.01619 | 0.283 |
| 45516 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | CLASP2 | Sponge network | -0.342 | 0.02288 | -0.038 | 0.71 | 0.283 |
| 45517 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | -0.563 | 0.02545 | 0.273 | 0.09957 | 0.283 |
| 45518 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-429;hsa-miR-629-3p | 17 | PIK3R1 | Sponge network | 1.316 | 0.01106 | -0.133 | 0.36854 | 0.283 |
| 45519 | BDNF-AS |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 14 | RPS6KA2 | Sponge network | -0.668 | 3.0E-5 | -0.57 | 0.00013 | 0.283 |
| 45520 | LINC00969 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | USP9X | Sponge network | 0.683 | 1.0E-5 | 0.222 | 0.01689 | 0.283 |
| 45521 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DNAJB4 | Sponge network | -0.668 | 3.0E-5 | -0.7 | 1.0E-5 | 0.283 |
| 45522 | RP11-400K9.4 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-7-5p | 25 | BTG2 | Sponge network | -0.611 | 0.09607 | -1.763 | 0 | 0.283 |
| 45523 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | PRRX1 | Sponge network | -0.323 | 0.01372 | 1.316 | 0 | 0.283 |
| 45524 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3913-5p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-590-3p | 15 | SLITRK6 | Sponge network | -0.342 | 0.02288 | -2.121 | 0 | 0.283 |
| 45525 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p | 19 | ABL2 | Sponge network | -0.031 | 0.89063 | 0.82 | 0 | 0.283 |
| 45526 | RP11-386G11.5 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p | 14 | TBL1XR1 | Sponge network | 2.031 | 0.00154 | 0.578 | 0 | 0.283 |
| 45527 | TP73-AS1 |
hsa-miR-146b-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | SPOP | Sponge network | -0.563 | 0.02545 | -0.419 | 1.0E-5 | 0.283 |
| 45528 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | -0.125 | 0.49414 | -1.3 | 0.01619 | 0.283 |
| 45529 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 16 | PICALM | Sponge network | -0.487 | 0.02157 | 0.086 | 0.23523 | 0.283 |
| 45530 | HAR1A |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-671-5p | 13 | DLC1 | Sponge network | -0.672 | 0.19447 | -0.382 | 0.05038 | 0.283 |
| 45531 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | MYBL1 | Sponge network | 0.924 | 0 | 0.494 | 0.02152 | 0.283 |
| 45532 | AF131215.2 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-539-5p | 17 | ACSL6 | Sponge network | -0.176 | 0.59692 | -1.593 | 0 | 0.283 |
| 45533 | RP11-1006G14.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-590-3p | 12 | STARD13 | Sponge network | 0.559 | 0.00026 | -0.187 | 0.27383 | 0.283 |
| 45534 | CTD-2089N3.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-378a-3p;hsa-miR-590-3p | 17 | TACC1 | Sponge network | -1.375 | 0.08642 | -0.362 | 0.05406 | 0.283 |
| 45535 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-940;hsa-miR-96-5p | 13 | HOOK3 | Sponge network | -1.439 | 4.0E-5 | 0.273 | 0.09957 | 0.283 |
| 45536 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-671-5p | 30 | PTPRD | Sponge network | -0.469 | 0.08721 | -1.005 | 0.00064 | 0.283 |
| 45537 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 49 | BMPR2 | Sponge network | 0.41 | 0.02214 | 0.206 | 0.0635 | 0.283 |
| 45538 | RP11-304M2.3 | hsa-let-7a-3p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p | 10 | KIF1B | Sponge network | 1.53 | 0 | -0.118 | 0.32258 | 0.283 |
| 45539 | RP11-597D13.9 |
hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30c-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 10 | CDH13 | Sponge network | 1.302 | 7.0E-5 | 0.634 | 0.00094 | 0.283 |
| 45540 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-429 | 12 | SOX11 | Sponge network | 0.389 | 0.04293 | 2.68 | 0 | 0.283 |
| 45541 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 29 | JAZF1 | Sponge network | -0.384 | 0.40652 | -0.377 | 0.02677 | 0.283 |
| 45542 | RP4-613B23.1 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-455-3p | 10 | CSRNP1 | Sponge network | -0.309 | 0.1145 | -1.448 | 0 | 0.283 |
| 45543 | RP5-1198O20.4 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | CSDE1 | Sponge network | 0.997 | 0.00066 | -0.493 | 0 | 0.283 |
| 45544 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | MOB1B | Sponge network | 1.61 | 0.00961 | 0.197 | 0.15266 | 0.283 |
| 45545 | RP11-680C21.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | MED12L | Sponge network | -1.424 | 0.04346 | -0.194 | 0.55299 | 0.283 |
| 45546 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 36 | MDM4 | Sponge network | 0.335 | 0.20596 | 0.345 | 0.00762 | 0.283 |
| 45547 | AC074138.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-200c-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577 | 10 | EPHA5 | Sponge network | -0.127 | 0.52761 | -0.957 | 0.01733 | 0.283 |
| 45548 | LINC00667 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | CSDE1 | Sponge network | -0.235 | 0.15142 | -0.493 | 0 | 0.283 |
| 45549 | RP11-410L14.2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-450b-5p | 13 | DTNA | Sponge network | -0.052 | 0.76839 | -1.648 | 1.0E-5 | 0.283 |
| 45550 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-942-5p | 16 | ERRFI1 | Sponge network | 0.194 | 0.64278 | -0.798 | 1.0E-5 | 0.283 |
| 45551 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | EPHA5 | Sponge network | -0.663 | 0.10887 | -0.957 | 0.01733 | 0.283 |
| 45552 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | MTSS1 | Sponge network | -0.739 | 0.00044 | -0.81 | 0.00035 | 0.283 |
| 45553 | RP11-157J24.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-301a-3p | 10 | IL17RD | Sponge network | -1.664 | 0.05165 | -0.019 | 0.94873 | 0.283 |
| 45554 | RP11-597D13.8 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p | 10 | NOTCH2 | Sponge network | 0.936 | 0.03889 | 0.138 | 0.35163 | 0.283 |
| 45555 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | TIMP3 | Sponge network | -0.222 | 0.32445 | -0.546 | 0.02307 | 0.283 |
| 45556 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940 | 59 | QKI | Sponge network | -2.925 | 0 | -0.333 | 0.04111 | 0.283 |
| 45557 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-590-3p | 14 | USP6 | Sponge network | 1.601 | 0 | 0.188 | 0.3871 | 0.283 |
| 45558 | PSMG3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ABI2 | Sponge network | -0.125 | 0.49414 | 0.175 | 0.14787 | 0.283 |
| 45559 | RP1-228H13.5 |
hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 11 | PAWR | Sponge network | 1.397 | 0 | 0.636 | 0 | 0.283 |
| 45560 | RP11-66B24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p | 16 | PBX1 | Sponge network | 0.57 | 0.1866 | -1.206 | 0 | 0.283 |
| 45561 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-493-5p | 12 | CACNA2D1 | Sponge network | 0.368 | 0.20093 | -0.846 | 0.00675 | 0.283 |
| 45562 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-576-5p | 13 | HIP1 | Sponge network | -0.932 | 0.00329 | 0.774 | 0 | 0.283 |
| 45563 | AC084219.4 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-539-5p | 11 | ITCH | Sponge network | 1.296 | 0.0054 | 0.084 | 0.34058 | 0.283 |
| 45564 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-577;hsa-miR-590-3p | 11 | TSLP | Sponge network | 0.997 | 0.00066 | -2.209 | 0 | 0.283 |
| 45565 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 26 | TSHZ3 | Sponge network | 0.75 | 0.00054 | -0.556 | 0.01516 | 0.283 |
| 45566 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | PTPN11 | Sponge network | 0.848 | 0 | 0.431 | 2.0E-5 | 0.283 |
| 45567 | RP11-25K21.6 |
hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p | 10 | RBMS3 | Sponge network | 0.489 | 0.25786 | -0.519 | 0.03551 | 0.283 |
| 45568 | RP11-5C23.1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 14 | SYNPO2 | Sponge network | 0.274 | 0.07681 | -2.342 | 0 | 0.283 |
| 45569 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-3p | 14 | SLITRK6 | Sponge network | -0.162 | 0.47687 | -2.121 | 0 | 0.283 |
| 45570 | HNRNPU-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p | 16 | PTPN14 | Sponge network | 1.601 | 0 | 0.144 | 0.34595 | 0.283 |
| 45571 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p | 13 | RIMBP2 | Sponge network | 0.419 | 0.0319 | -1.968 | 5.0E-5 | 0.283 |
| 45572 | TP73-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-629-3p | 21 | RYR2 | Sponge network | -0.563 | 0.02545 | -1.466 | 0.00031 | 0.283 |
| 45573 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-501-5p;hsa-miR-590-3p | 18 | KIT | Sponge network | -0.08 | 0.90092 | -2.184 | 0 | 0.283 |
| 45574 | RP11-15H20.7 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-34a-5p | 11 | PRKAA2 | Sponge network | 0.261 | 0.23673 | -2.462 | 0 | 0.283 |
| 45575 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484 | 24 | MED12L | Sponge network | -2.69 | 0.0001 | -0.194 | 0.55299 | 0.283 |
| 45576 | OIP5-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 41 | GAS7 | Sponge network | 0.421 | 0.00084 | -0.555 | 0.01602 | 0.283 |
| 45577 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | QKI | Sponge network | 0.614 | 1.0E-5 | -0.333 | 0.04111 | 0.283 |
| 45578 | DLGAP1-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-5p;hsa-miR-33a-3p | 10 | ABL2 | Sponge network | 0.755 | 3.0E-5 | 0.82 | 0 | 0.283 |
| 45579 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p | 15 | PCDH8 | Sponge network | -2.925 | 0 | -0.997 | 0.05366 | 0.283 |
| 45580 | AF131215.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ARNT2 | Sponge network | -0.322 | 0.25945 | -0.332 | 0.25851 | 0.283 |
| 45581 | LINC00662 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NRXN3 | Sponge network | 0.213 | 0.32297 | -0.893 | 0.04186 | 0.283 |
| 45582 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-455-3p | 13 | EZH1 | Sponge network | -0.418 | 0.00068 | -0.564 | 1.0E-5 | 0.283 |
| 45583 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-378a-3p;hsa-miR-378c | 19 | RBMS3 | Sponge network | 0.859 | 0.00331 | -0.519 | 0.03551 | 0.283 |
| 45584 | RP11-156P1.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-940 | 26 | ETV1 | Sponge network | 0.214 | 0.18246 | -0.096 | 0.67674 | 0.283 |
| 45585 | RP11-6N17.3 |
hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 10 | TMEM132B | Sponge network | 0.108 | 0.69556 | -0.83 | 0.01084 | 0.282 |
| 45586 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p | 24 | AKAP11 | Sponge network | -0.358 | 0.17255 | 0.196 | 0.09825 | 0.282 |
| 45587 | CTD-2336O2.1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-3p | 18 | CTIF | Sponge network | -0.141 | 0.26434 | -0.389 | 0.00549 | 0.282 |
| 45588 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-7-5p | 14 | ESR1 | Sponge network | 0.199 | 0.52763 | -1.035 | 3.0E-5 | 0.282 |
| 45589 | RP11-474O21.5 |
hsa-miR-106b-5p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-30b-3p;hsa-miR-744-3p;hsa-miR-93-5p | 12 | FAT3 | Sponge network | -0.023 | 0.95109 | -1.601 | 0.00051 | 0.282 |
| 45590 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 20 | TIMP3 | Sponge network | -0.513 | 0.04703 | -0.546 | 0.02307 | 0.282 |
| 45591 | RP11-403I13.8 |
hsa-miR-142-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-942-5p;hsa-miR-96-5p | 10 | ZBTB20 | Sponge network | 0.619 | 0.00157 | -0.122 | 0.57569 | 0.282 |
| 45592 | AC108488.4 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p | 11 | IRS1 | Sponge network | 0.259 | 0.12542 | -0.026 | 0.88855 | 0.282 |
| 45593 | CTC-429P9.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-505-5p | 18 | ASTN1 | Sponge network | 0.082 | 0.55397 | -2.389 | 2.0E-5 | 0.282 |
| 45594 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-421;hsa-miR-429;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92b-3p;hsa-miR-96-5p | 18 | LRRC4 | Sponge network | -1.264 | 6.0E-5 | -1.774 | 0 | 0.282 |
| 45595 | MIR22HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-493-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 28 | NCOA1 | Sponge network | -1.047 | 0 | -0.432 | 0 | 0.282 |
| 45596 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-590-3p | 17 | ACVR1 | Sponge network | -0.358 | 0.17255 | 0.279 | 0.00234 | 0.282 |
| 45597 | RP11-867G23.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-93-5p | 16 | ETV1 | Sponge network | -2.531 | 0 | -0.096 | 0.67674 | 0.282 |
| 45598 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-495-3p;hsa-miR-582-3p | 11 | CLIP1 | Sponge network | 0.637 | 0.00069 | -0.215 | 0.08684 | 0.282 |
| 45599 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ITGAV | Sponge network | 1.308 | 0 | 0.337 | 0.01002 | 0.282 |
| 45600 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | MED12L | Sponge network | -0.769 | 0.00035 | -0.194 | 0.55299 | 0.282 |
| 45601 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p | 20 | ACVR1 | Sponge network | -0.793 | 7.0E-5 | 0.279 | 0.00234 | 0.282 |
| 45602 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 25 | MLLT10 | Sponge network | -0.615 | 0.00015 | 0.105 | 0.33292 | 0.282 |
| 45603 | DNAJC27-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 14 | KCNAB1 | Sponge network | -0.969 | 2.0E-5 | -0.755 | 2.0E-5 | 0.282 |
| 45604 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-629-3p | 17 | EBF1 | Sponge network | 0.921 | 0.00118 | -0.384 | 0.08856 | 0.282 |
| 45605 | FZD10-AS1 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | KCNAB1 | Sponge network | -0.727 | 0.04726 | -0.755 | 2.0E-5 | 0.282 |
| 45606 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-93-5p | 42 | SMAD4 | Sponge network | -2.69 | 0.0001 | -0.313 | 0.00413 | 0.282 |
| 45607 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-940 | 29 | ETV1 | Sponge network | 0.331 | 0.57247 | -0.096 | 0.67674 | 0.282 |
| 45608 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-671-5p | 40 | ADARB1 | Sponge network | 0.078 | 0.55803 | -0.335 | 0.06631 | 0.282 |
| 45609 | RP11-403I13.8 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-582-3p;hsa-miR-590-3p | 28 | ABL2 | Sponge network | 0.619 | 0.00157 | 0.82 | 0 | 0.282 |
| 45610 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-532-5p;hsa-miR-766-3p | 18 | IL17RD | Sponge network | 0.065 | 0.73468 | -0.019 | 0.94873 | 0.282 |
| 45611 | RP11-473I1.9 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 21 | RASSF8 | Sponge network | 0.683 | 1.0E-5 | -0.122 | 0.65591 | 0.282 |
| 45612 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-577;hsa-miR-590-3p | 18 | PRKAB2 | Sponge network | 1.757 | 0 | -0.367 | 0.01371 | 0.282 |
| 45613 | CTD-2554C21.2 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-429 | 14 | FAM135B | Sponge network | -1.654 | 0.00144 | -3.978 | 0 | 0.282 |
| 45614 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 13 | NRXN3 | Sponge network | 1.04 | 0.00023 | -0.893 | 0.04186 | 0.282 |
| 45615 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | HDAC9 | Sponge network | -0.125 | 0.49414 | -0.755 | 0.00495 | 0.282 |
| 45616 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b | 11 | PTPN11 | Sponge network | 0.817 | 3.0E-5 | 0.431 | 2.0E-5 | 0.282 |
| 45617 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-766-3p | 12 | IL17RD | Sponge network | 1.162 | 5.0E-5 | -0.019 | 0.94873 | 0.282 |
| 45618 | RP11-283G6.3 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-3127-5p;hsa-miR-500a-3p;hsa-miR-502-5p | 12 | DST | Sponge network | 1.003 | 0.00053 | -0.713 | 0.00096 | 0.282 |
| 45619 | H19 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-576-5p | 18 | NTRK2 | Sponge network | 2.439 | 0 | -0.736 | 0.06495 | 0.282 |
| 45620 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | SLC2A13 | Sponge network | 0.683 | 1.0E-5 | -0.331 | 0.03658 | 0.282 |
| 45621 | NEAT1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-744-3p | 21 | CUL5 | Sponge network | 0.705 | 0.00014 | 0.026 | 0.77301 | 0.282 |
| 45622 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p | 12 | RB1CC1 | Sponge network | -0.154 | 0.78939 | 0.175 | 0.12559 | 0.282 |
| 45623 | SNHG12 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-9-5p;hsa-miR-99a-5p | 19 | CCDC6 | Sponge network | 1.103 | 0 | 0.27 | 0.02555 | 0.282 |
| 45624 | RP11-244H3.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-582-5p;hsa-miR-590-3p | 22 | DLC1 | Sponge network | 0.659 | 3.0E-5 | -0.382 | 0.05038 | 0.282 |
| 45625 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 17 | MYO9A | Sponge network | 0.727 | 3.0E-5 | -0.147 | 0.32373 | 0.282 |
| 45626 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p | 13 | SCN9A | Sponge network | 0.817 | 3.0E-5 | -0.525 | 0.10593 | 0.282 |
| 45627 | KCNJ2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-330-3p | 11 | IL17RD | Sponge network | -0.436 | 0.05325 | -0.019 | 0.94873 | 0.282 |
| 45628 | USP3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-877-5p | 19 | CBFA2T3 | Sponge network | -0.162 | 0.47687 | -1.221 | 1.0E-5 | 0.282 |
| 45629 | RP4-668J24.2 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-629-3p | 10 | CDH23 | Sponge network | -1.054 | 0.0706 | -0.974 | 0.00034 | 0.282 |
| 45630 | RP11-356J5.12 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-590-3p | 13 | PKNOX1 | Sponge network | -0.899 | 6.0E-5 | 0.085 | 0.28917 | 0.282 |
| 45631 | RP11-439A17.10 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-590-5p | 13 | WDFY3 | Sponge network | 0.051 | 0.95756 | -0.201 | 0.0967 | 0.282 |
| 45632 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MLLT10 | Sponge network | 0.101 | 0.60919 | 0.105 | 0.33292 | 0.282 |
| 45633 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-93-5p | 18 | PRRX1 | Sponge network | -0.465 | 0.04703 | 1.316 | 0 | 0.282 |
| 45634 | RP11-890B15.3 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 14 | KIF5B | Sponge network | 0.101 | 0.60919 | 0.362 | 0.00066 | 0.282 |
| 45635 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | EGLN1 | Sponge network | 1.601 | 0 | -0.127 | 0.1536 | 0.282 |
| 45636 | AC003090.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | PDE4DIP | Sponge network | -2.117 | 0.00161 | -0.282 | 0.0257 | 0.282 |
| 45637 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-629-3p | 13 | USP12 | Sponge network | 0.946 | 0 | -0.102 | 0.40768 | 0.282 |
| 45638 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 29 | CADM1 | Sponge network | 0.101 | 0.60919 | -0.9 | 0.00084 | 0.282 |
| 45639 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-766-3p | 39 | AFF4 | Sponge network | 1.089 | 0 | -0.266 | 0.01565 | 0.282 |
| 45640 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | KDSR | Sponge network | 0.299 | 0.57722 | 0.239 | 0.02216 | 0.282 |
| 45641 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | FOXP1 | Sponge network | 0.953 | 0 | 0.042 | 0.74368 | 0.282 |
| 45642 | HOXA-AS2 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 10 | STAT5B | Sponge network | 0.61 | 0.01593 | -0.182 | 0.1082 | 0.282 |
| 45643 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-362-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | PAK3 | Sponge network | 0.064 | 0.72934 | -0.628 | 0.07253 | 0.282 |
| 45644 | TUG1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p | 13 | CLASP2 | Sponge network | 0.972 | 0 | -0.038 | 0.71 | 0.282 |
| 45645 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p | 32 | RICTOR | Sponge network | 0.41 | 0.02214 | 0.388 | 0.00188 | 0.282 |
| 45646 | LPP-AS2 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-96-5p | 19 | RPS6KA6 | Sponge network | -0.18 | 0.20343 | -2.989 | 0 | 0.282 |
| 45647 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-93-5p | 47 | WDFY3 | Sponge network | -0.384 | 0.40652 | -0.201 | 0.0967 | 0.282 |
| 45648 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 14 | SNX29 | Sponge network | 0.898 | 1.0E-5 | -0.34 | 0.01371 | 0.282 |
| 45649 | RP11-999E24.3 |
hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-361-3p | 12 | JDP2 | Sponge network | -0.693 | 0.05629 | -0.967 | 0 | 0.282 |
| 45650 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 45 | FAT3 | Sponge network | 0.349 | 0.01695 | -1.601 | 0.00051 | 0.282 |
| 45651 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | ETV1 | Sponge network | 0.385 | 0.10067 | -0.096 | 0.67674 | 0.282 |
| 45652 | AC005154.6 |
hsa-miR-16-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-369-5p;hsa-miR-409-5p;hsa-miR-577;hsa-miR-590-3p | 10 | MACF1 | Sponge network | 0.848 | 0 | 0.019 | 0.90335 | 0.282 |
| 45653 | MIR22HG |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-5p | 17 | ERG | Sponge network | -1.047 | 0 | 0.002 | 0.99011 | 0.282 |
| 45654 | C4A-AS1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-629-5p;hsa-miR-96-5p | 10 | PCDH17 | Sponge network | 3.345 | 0 | 0.731 | 2.0E-5 | 0.282 |
| 45655 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TET1 | Sponge network | -0.421 | 0.0114 | -0.135 | 0.52138 | 0.282 |
| 45656 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | FOXP1 | Sponge network | 1.148 | 2.0E-5 | 0.042 | 0.74368 | 0.282 |
| 45657 | RP11-67L2.2 |
hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-369-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p | 10 | RNF111 | Sponge network | 0.72 | 0.0001 | 0.098 | 0.21093 | 0.282 |
| 45658 | EMX2OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | ARNT2 | Sponge network | -0.777 | 0.1134 | -0.332 | 0.25851 | 0.282 |
| 45659 | CD27-AS1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | 0.177 | 0.16681 | -2.633 | 0 | 0.282 |
| 45660 | RP11-156P1.3 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 18 | GJA1 | Sponge network | 0.214 | 0.18246 | -0.485 | 0.02179 | 0.282 |
| 45661 | RP11-532F6.3 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | PTCH1 | Sponge network | -0.896 | 0.00099 | -0.1 | 0.6179 | 0.282 |
| 45662 | HCG18 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-590-3p | 20 | DDX3X | Sponge network | 1.063 | 0 | -0.152 | 0.07686 | 0.282 |
| 45663 | SNHG14 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ZNF638 | Sponge network | -1.111 | 0.0003 | 0.22 | 0.01214 | 0.282 |
| 45664 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | PHC3 | Sponge network | 0.335 | 0.20596 | 0.212 | 0.0499 | 0.282 |
| 45665 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | LATS2 | Sponge network | -0.421 | 0.0114 | 0.159 | 0.2304 | 0.282 |
| 45666 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | ZNF208 | Sponge network | 0.349 | 0.01695 | -0.4 | 0.22381 | 0.282 |
| 45667 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374a-3p | 18 | CBL | Sponge network | 0.174 | 0.30034 | 0.291 | 0.01267 | 0.282 |
| 45668 | RP11-10N23.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p | 10 | ZFX | Sponge network | 1.134 | 0 | 0.09 | 0.42563 | 0.282 |
| 45669 | RP11-203M5.7 |
hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-5p | 10 | PIK3R1 | Sponge network | 1.684 | 1.0E-5 | -0.133 | 0.36854 | 0.282 |
| 45670 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | ST6GAL2 | Sponge network | 0.559 | 0.00026 | -1.201 | 0.00597 | 0.282 |
| 45671 | RASSF8-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-877-5p | 22 | CRTC1 | Sponge network | 0.335 | 0.20596 | -0.116 | 0.28412 | 0.282 |
| 45672 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 16 | PRKAB2 | Sponge network | -1.582 | 8.0E-5 | -0.367 | 0.01371 | 0.282 |
| 45673 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | PCDH8 | Sponge network | 0.246 | 0.59912 | -0.997 | 0.05366 | 0.282 |
| 45674 | MIR7-3HG |
hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 14 | DTNA | Sponge network | -2.688 | 0.00397 | -1.648 | 1.0E-5 | 0.282 |
| 45675 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | LRRC7 | Sponge network | 0.657 | 4.0E-5 | -1.341 | 0.01193 | 0.282 |
| 45676 | LINC00476 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-93-3p | 14 | POU2F2 | Sponge network | -0.615 | 0.00015 | -0.233 | 0.32214 | 0.282 |
| 45677 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-7-5p | 21 | CLASP2 | Sponge network | -0.465 | 0.04703 | -0.038 | 0.71 | 0.282 |
| 45678 | AC093627.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-5p | 15 | SSBP2 | Sponge network | -0.787 | 0.3928 | -1.58 | 0 | 0.282 |
| 45679 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-98-5p | 24 | RSPO2 | Sponge network | -0.154 | 0.78939 | -3.038 | 0 | 0.282 |
| 45680 | LINC00648 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | LRP1B | Sponge network | 0.633 | 0.51678 | -2.163 | 0 | 0.282 |
| 45681 | HOXB-AS3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-27a-5p;hsa-miR-324-5p | 10 | TSHZ3 | Sponge network | 0.343 | 0.28887 | -0.556 | 0.01516 | 0.282 |
| 45682 | CTC-296K1.4 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-331-3p;hsa-miR-590-5p;hsa-miR-708-5p | 10 | FAM135B | Sponge network | -2.076 | 0.00548 | -3.978 | 0 | 0.282 |
| 45683 | CTD-2554C21.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-503-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | CCND2 | Sponge network | -1.654 | 0.00144 | -0.267 | 0.3112 | 0.282 |
| 45684 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | LPP | Sponge network | 0.342 | 0.03129 | -0.459 | 0.04475 | 0.282 |
| 45685 | RP11-37B2.1 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | KIF5B | Sponge network | 1.03 | 0 | 0.362 | 0.00066 | 0.282 |
| 45686 | AC103563.9 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 11 | GAS7 | Sponge network | -6.057 | 0 | -0.555 | 0.01602 | 0.282 |
| 45687 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-7-1-3p | 15 | CLIP1 | Sponge network | 0.177 | 0.16681 | -0.215 | 0.08684 | 0.282 |
| 45688 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-409-3p;hsa-miR-7-1-3p | 11 | IGSF10 | Sponge network | 0.41 | 0.02214 | -1.751 | 1.0E-5 | 0.282 |
| 45689 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p | 11 | NALCN | Sponge network | 0.75 | 0.00054 | 1.068 | 0.00237 | 0.282 |
| 45690 | CKMT2-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FBXO11 | Sponge network | 0.082 | 0.55654 | 0.142 | 0.04938 | 0.282 |
| 45691 | TTC25 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-940 | 12 | GRIK3 | Sponge network | -0.208 | 0.38261 | -3.208 | 0 | 0.282 |
| 45692 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b | 14 | SON | Sponge network | 0.331 | 0.57247 | 0.168 | 0.04406 | 0.282 |
| 45693 | ACTA2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | SUFU | Sponge network | 0.194 | 0.64278 | -0.04 | 0.65489 | 0.282 |
| 45694 | AC016747.3 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 10 | TPO | Sponge network | 0.199 | 0.38015 | -1.87 | 2.0E-5 | 0.282 |
| 45695 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 15 | EBF1 | Sponge network | 0.594 | 0.00064 | -0.384 | 0.08856 | 0.282 |
| 45696 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 20 | NR2C2 | Sponge network | 0.572 | 0.01722 | 0.21 | 0.03224 | 0.282 |
| 45697 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | CITED2 | Sponge network | -0.355 | 0.0077 | -1.545 | 0 | 0.282 |
| 45698 | RP11-597D13.9 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 34 | IGFBP5 | Sponge network | 1.302 | 7.0E-5 | -0.806 | 0.00105 | 0.282 |
| 45699 | CTC-429P9.2 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-29b-1-5p;hsa-miR-502-5p;hsa-miR-532-5p | 15 | CUX1 | Sponge network | 0.043 | 0.81628 | -0.039 | 0.6705 | 0.282 |
| 45700 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p | 26 | FAT3 | Sponge network | -0.285 | 0.2172 | -1.601 | 0.00051 | 0.282 |
| 45701 | HOXB-AS2 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-429 | 13 | NCOA1 | Sponge network | 1.316 | 0.01106 | -0.432 | 0 | 0.282 |
| 45702 | RP5-887A10.1 |
hsa-let-7d-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 10 | NTRK3 | Sponge network | 0.546 | 0.4892 | -1.77 | 3.0E-5 | 0.282 |
| 45703 | AC108488.4 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p | 13 | TES | Sponge network | 0.259 | 0.12542 | -0.348 | 0.00639 | 0.282 |
| 45704 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ITGAV | Sponge network | 0.873 | 0 | 0.337 | 0.01002 | 0.282 |
| 45705 | CTC-487M23.5 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-301a-3p | 12 | IGF1 | Sponge network | 0.912 | 0.00014 | -0.204 | 0.5898 | 0.282 |
| 45706 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 15 | SRGAP3 | Sponge network | -1.057 | 0.00029 | -0.202 | 0.29297 | 0.282 |
| 45707 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-625-5p | 22 | YAP1 | Sponge network | 0.253 | 0.09565 | 0.22 | 0.11836 | 0.282 |
| 45708 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | GNA13 | Sponge network | -1.111 | 0.0003 | 0.007 | 0.93884 | 0.282 |
| 45709 | LINC00886 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-542-3p | 19 | SYNPO2 | Sponge network | -0.371 | 0.24604 | -2.342 | 0 | 0.282 |
| 45710 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | ETV1 | Sponge network | -0.206 | 0.12942 | -0.096 | 0.67674 | 0.282 |
| 45711 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p | 28 | CBX5 | Sponge network | 0.569 | 0.00067 | 0.165 | 0.24446 | 0.282 |
| 45712 | MIR143HG |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-629-5p | 15 | WNK2 | Sponge network | -0.739 | 0.0574 | -0.352 | 0.31066 | 0.282 |
| 45713 | SERHL |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-421 | 16 | ZMYND11 | Sponge network | -0.602 | 0.00331 | -0.2 | 0.05449 | 0.282 |
| 45714 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-5p | 10 | HIC1 | Sponge network | -2.46 | 0 | 0.024 | 0.89889 | 0.282 |
| 45715 | LINC00662 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | MYO9A | Sponge network | 0.213 | 0.32297 | -0.147 | 0.32373 | 0.282 |
| 45716 | CTA-445C9.14 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p | 13 | ZMYM2 | Sponge network | 0.718 | 4.0E-5 | 0.279 | 0.01273 | 0.282 |
| 45717 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p | 41 | CAMTA1 | Sponge network | -2.46 | 0 | -0.436 | 1.0E-5 | 0.282 |
| 45718 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-651-5p | 31 | PRDM2 | Sponge network | -0.942 | 0 | -0.258 | 0.00721 | 0.282 |
| 45719 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | NEDD4 | Sponge network | 0.659 | 3.0E-5 | 0.02 | 0.89834 | 0.282 |
| 45720 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-22-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a | 15 | AR | Sponge network | 0.357 | 0.72407 | -1.554 | 0 | 0.282 |
| 45721 | RP11-327P2.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-625-5p | 21 | CADM1 | Sponge network | -0.147 | 0.40452 | -0.9 | 0.00084 | 0.282 |
| 45722 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 25 | ZFHX4 | Sponge network | -0.663 | 0.10887 | 0.018 | 0.95574 | 0.282 |
| 45723 | LINC00473 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 18 | GLIPR1 | Sponge network | -1.203 | 0.03881 | 0.146 | 0.3276 | 0.282 |
| 45724 | RP11-98I9.4 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DICER1 | Sponge network | 0.67 | 0.00048 | 0.042 | 0.674 | 0.282 |
| 45725 | RP11-307B6.3 |
hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-532-5p;hsa-miR-93-3p | 21 | CUX1 | Sponge network | -4.463 | 0.00025 | -0.039 | 0.6705 | 0.282 |
| 45726 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-5p | 14 | FREM2 | Sponge network | 1.61 | 0.00961 | -0.437 | 0.46353 | 0.282 |
| 45727 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-493-5p | 18 | AFF3 | Sponge network | -0.384 | 0.40652 | -2.373 | 0 | 0.282 |
| 45728 | CTD-2336O2.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | HOMER2 | Sponge network | -0.141 | 0.26434 | -2.698 | 0 | 0.282 |
| 45729 | RP11-53O19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 14 | PLAG1 | Sponge network | -0.405 | 0.22013 | -0.315 | 0.26532 | 0.282 |
| 45730 | CTC-490E21.10 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-323a-3p | 13 | IL17RD | Sponge network | 1.46 | 1.0E-5 | -0.019 | 0.94873 | 0.282 |
| 45731 | RP11-6O2.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-550a-3p;hsa-miR-96-5p | 21 | PRRX1 | Sponge network | 0.331 | 0.57247 | 1.316 | 0 | 0.282 |
| 45732 | AC034220.3 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | UBR3 | Sponge network | 0.309 | 0.07304 | -0.021 | 0.82774 | 0.282 |
| 45733 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | PPP1R9A | Sponge network | -0.405 | 0.22013 | -0.903 | 0.04254 | 0.282 |
| 45734 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 22 | LRRC7 | Sponge network | 0.311 | 0.10344 | -1.341 | 0.01193 | 0.282 |
| 45735 | CTD-2139B15.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p | 11 | PHF6 | Sponge network | 0.286 | 0.00612 | 0.785 | 0 | 0.282 |
| 45736 | HOXA-AS2 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 12 | RASAL2 | Sponge network | 0.61 | 0.01593 | 0.869 | 0 | 0.282 |
| 45737 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p | 27 | NTRK2 | Sponge network | -0.223 | 0.47816 | -0.736 | 0.06495 | 0.282 |
| 45738 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p | 13 | PTCH1 | Sponge network | 0.419 | 0.0319 | -0.1 | 0.6179 | 0.282 |
| 45739 | LINC00854 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-940 | 11 | IGF1 | Sponge network | 1.18 | 0 | -0.204 | 0.5898 | 0.282 |
| 45740 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-3p | 12 | ACTB | Sponge network | -2.69 | 0.0001 | -0.247 | 0.01736 | 0.282 |
| 45741 | RP4-668J24.2 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-628-5p | 15 | QKI | Sponge network | -1.054 | 0.0706 | -0.333 | 0.04111 | 0.282 |
| 45742 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 35 | EPHA5 | Sponge network | 0.16 | 0.50785 | -0.957 | 0.01733 | 0.282 |
| 45743 | RP11-2B6.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | NIN | Sponge network | 0.539 | 0.03722 | -0.253 | 0.13794 | 0.282 |
| 45744 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-766-3p | 10 | PHACTR4 | Sponge network | 0.705 | 0.00014 | 0.069 | 0.45203 | 0.282 |
| 45745 | DNM3OS |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | USP25 | Sponge network | 0.572 | 0.01722 | -0.242 | 0.01397 | 0.282 |
| 45746 | RP11-46C24.7 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | 0.392 | 0.0278 | -0.704 | 0.00106 | 0.282 |
| 45747 | RP11-175O19.4 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-2-3p;hsa-miR-30e-3p;hsa-miR-328-3p;hsa-miR-342-3p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-411-5p | 25 | VPS53 | Sponge network | 0.917 | 0 | 0.103 | 0.22667 | 0.282 |
| 45748 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 16 | TRIM3 | Sponge network | -1.111 | 0.0003 | -0.577 | 0 | 0.282 |
| 45749 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p | 11 | NOTCH2 | Sponge network | 1.316 | 0 | 0.138 | 0.35163 | 0.282 |
| 45750 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 15 | CREBBP | Sponge network | 1.302 | 7.0E-5 | 0.126 | 0.15186 | 0.282 |
| 45751 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 55 | WDFY3 | Sponge network | 0.064 | 0.72934 | -0.201 | 0.0967 | 0.282 |
| 45752 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 16 | TRIM13 | Sponge network | 0.75 | 0.00054 | 0.183 | 0.06968 | 0.282 |
| 45753 | RP11-325K4.3 |
hsa-let-7a-3p;hsa-miR-1296-5p;hsa-miR-135b-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | MEF2C | Sponge network | 0.178 | 0.31659 | -0.499 | 0.01448 | 0.282 |
| 45754 | CROCCP2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-324-5p | 13 | PIK3C2A | Sponge network | 0.79 | 0 | 0.061 | 0.53776 | 0.282 |
| 45755 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-651-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 63 | AFF4 | Sponge network | -0.162 | 0.47687 | -0.266 | 0.01565 | 0.282 |
| 45756 | LINC00672 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 37 | GAS7 | Sponge network | -0.227 | 0.31657 | -0.555 | 0.01602 | 0.282 |
| 45757 | RP1-122P22.2 |
hsa-miR-1301-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-625-5p | 10 | SFRP1 | Sponge network | 0.065 | 0.73468 | -3.566 | 0 | 0.282 |
| 45758 | CTD-2017D11.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p | 10 | UBR3 | Sponge network | 0.792 | 0.0049 | -0.021 | 0.82774 | 0.282 |
| 45759 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | CDHR3 | Sponge network | 0.61 | 0.01593 | -0.977 | 4.0E-5 | 0.282 |
| 45760 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-582-3p | 12 | RBMS3 | Sponge network | 0.858 | 3.0E-5 | -0.519 | 0.03551 | 0.282 |
| 45761 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-664a-3p;hsa-miR-766-3p;hsa-miR-93-5p | 47 | AFF4 | Sponge network | -2.69 | 0.0001 | -0.266 | 0.01565 | 0.282 |
| 45762 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-629-3p | 18 | USP12 | Sponge network | 0.921 | 0.00118 | -0.102 | 0.40768 | 0.282 |
| 45763 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-502-3p;hsa-miR-576-5p | 17 | ZFHX4 | Sponge network | 2.163 | 0 | 0.018 | 0.95574 | 0.282 |
| 45764 | DNM3OS |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | TES | Sponge network | 0.572 | 0.01722 | -0.348 | 0.00639 | 0.282 |
| 45765 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p | 17 | BCL6 | Sponge network | 0.659 | 3.0E-5 | -0.211 | 0.19489 | 0.282 |
| 45766 | ASH1L-AS1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 11 | PRKAA1 | Sponge network | 0.948 | 0 | 0.317 | 0.0031 | 0.282 |
| 45767 | DNM1P35 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-590-3p | 10 | SEMA3E | Sponge network | 0.051 | 0.83388 | -1.717 | 0.00137 | 0.282 |
| 45768 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | PRKCE | Sponge network | -2.925 | 0 | -0.403 | 0.00056 | 0.282 |
| 45769 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | CACNA2D1 | Sponge network | -0.668 | 3.0E-5 | -0.846 | 0.00675 | 0.282 |
| 45770 | RP11-1277A3.1 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-500a-3p | 10 | FMNL3 | Sponge network | 0.453 | 0.01839 | 0.555 | 4.0E-5 | 0.282 |
| 45771 | RP11-395G23.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-874-3p | 17 | ASTN1 | Sponge network | 0.381 | 0.25967 | -2.389 | 2.0E-5 | 0.282 |
| 45772 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-421 | 11 | RNASEL | Sponge network | -0.285 | 0.2172 | -0.272 | 0.01796 | 0.282 |
| 45773 | CTD-3099C6.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-664a-5p | 23 | DST | Sponge network | 1.361 | 0 | -0.713 | 0.00096 | 0.282 |
| 45774 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-93-3p | 19 | QKI | Sponge network | 0.912 | 0.00014 | -0.333 | 0.04111 | 0.282 |
| 45775 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-96-5p | 17 | SNX29 | Sponge network | 0.131 | 0.8436 | -0.34 | 0.01371 | 0.282 |
| 45776 | RP11-37B2.1 |
hsa-miR-101-5p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p;hsa-miR-664a-5p | 11 | CBFB | Sponge network | 1.03 | 0 | 1.061 | 0 | 0.282 |
| 45777 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-7-1-3p | 17 | EYA4 | Sponge network | 0.389 | 0.04293 | 0.3 | 0.36876 | 0.282 |
| 45778 | CTA-204B4.2 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p | 10 | RASAL2 | Sponge network | 0.765 | 7.0E-5 | 0.869 | 0 | 0.282 |
| 45779 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p | 10 | ERG | Sponge network | 0.497 | 0.01451 | 0.002 | 0.99011 | 0.282 |
| 45780 | TMEM9B-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p;hsa-miR-940 | 31 | MDM4 | Sponge network | 0.529 | 0.00552 | 0.345 | 0.00762 | 0.282 |
| 45781 | RP11-479G22.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 14 | WWTR1 | Sponge network | 1.308 | 0 | -0.345 | 0.11997 | 0.282 |
| 45782 | RP11-46C24.7 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 29 | PPP1R9A | Sponge network | 0.392 | 0.0278 | -0.903 | 0.04254 | 0.282 |
| 45783 | HOXB-AS1 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p | 11 | BCL9 | Sponge network | -0.154 | 0.53954 | 0.321 | 0.00267 | 0.282 |
| 45784 | EIF3J-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | AKAP11 | Sponge network | 0.331 | 0.00509 | 0.196 | 0.09825 | 0.282 |
| 45785 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | -0.222 | 0.32445 | -1.047 | 0.00364 | 0.282 |
| 45786 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 12 | GPAM | Sponge network | 0.856 | 0 | 0.142 | 0.29732 | 0.282 |
| 45787 | RP11-326G21.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ACVR2A | Sponge network | -0.021 | 0.93598 | -0.409 | 0.00039 | 0.282 |
| 45788 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | INPP4A | Sponge network | -0.738 | 0.21233 | 0.07 | 0.53563 | 0.282 |
| 45789 | AC141928.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-96-5p | 28 | GAS7 | Sponge network | 0.853 | 0.15352 | -0.555 | 0.01602 | 0.282 |
| 45790 | AC090587.5 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-93-3p | 10 | EPHA3 | Sponge network | -0.088 | 0.65011 | 0.185 | 0.53588 | 0.282 |
| 45791 | RP11-473I1.9 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-328-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-590-3p | 14 | TMTC2 | Sponge network | 0.683 | 1.0E-5 | -0.215 | 0.18248 | 0.282 |
| 45792 | RP4-717I23.3 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-654-3p | 26 | PIK3R1 | Sponge network | 0.637 | 0.00069 | -0.133 | 0.36854 | 0.282 |
| 45793 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-628-5p | 15 | ACVR1 | Sponge network | -0.842 | 0.01361 | 0.279 | 0.00234 | 0.282 |
| 45794 | RP11-401P9.4 |
hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-532-5p | 14 | RPS6KA6 | Sponge network | -0.384 | 0.40652 | -2.989 | 0 | 0.282 |
| 45795 | RP11-37B2.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 10 | KIAA1024 | Sponge network | 1.03 | 0 | 1.083 | 0 | 0.282 |
| 45796 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-484;hsa-miR-550a-5p | 12 | FAT2 | Sponge network | -0.727 | 0.04726 | -0.278 | 0.44877 | 0.282 |
| 45797 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 20 | FOXO3 | Sponge network | -0.096 | 0.67958 | -0.04 | 0.72028 | 0.282 |
| 45798 | RP11-434B12.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 18 | WWTR1 | Sponge network | -0.031 | 0.89063 | -0.345 | 0.11997 | 0.282 |
| 45799 | RP11-439A17.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p | 17 | THRB | Sponge network | 0.051 | 0.95756 | -1.117 | 0.0004 | 0.282 |
| 45800 | DOCK9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-940 | 16 | CHD5 | Sponge network | -0.231 | 0.28214 | -0.922 | 0.01521 | 0.282 |
| 45801 | CTA-363E19.2 |
hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 10 | EPHA3 | Sponge network | 1.055 | 0 | 0.185 | 0.53588 | 0.282 |
| 45802 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 23 | BCL6 | Sponge network | -1.281 | 0.00012 | -0.211 | 0.19489 | 0.282 |
| 45803 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 20 | PRKAR1A | Sponge network | -0.576 | 0.10121 | -0.063 | 0.50314 | 0.282 |
| 45804 | RP11-728F11.4 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 26 | THRB | Sponge network | 0.238 | 0.70544 | -1.117 | 0.0004 | 0.282 |
| 45805 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-590-3p | 21 | HBP1 | Sponge network | -0.031 | 0.89063 | -0.454 | 0 | 0.282 |
| 45806 | RP4-758J18.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-331-3p | 12 | PRKAA2 | Sponge network | -0.091 | 0.71468 | -2.462 | 0 | 0.282 |
| 45807 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p | 24 | PBX1 | Sponge network | 0.395 | 0.15451 | -1.206 | 0 | 0.282 |
| 45808 | AGAP11 |
hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-653-5p | 22 | UBR3 | Sponge network | -1.645 | 0 | -0.021 | 0.82774 | 0.282 |
| 45809 | RP11-774O3.3 |
hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-653-5p | 23 | UBR3 | Sponge network | -0.487 | 0.02157 | -0.021 | 0.82774 | 0.282 |
| 45810 | EIF3J-AS1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-654-3p | 13 | NR2C2 | Sponge network | 0.331 | 0.00509 | 0.21 | 0.03224 | 0.282 |
| 45811 | RP11-263K19.4 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p | 43 | LPP | Sponge network | 0.859 | 0.00331 | -0.459 | 0.04475 | 0.282 |
| 45812 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-429;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-653-5p | 23 | DMXL1 | Sponge network | -2.62 | 0 | -0.426 | 0.00056 | 0.282 |
| 45813 | RP11-728F11.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p | 12 | NRK | Sponge network | 0.238 | 0.70544 | 0.656 | 0.19217 | 0.282 |
| 45814 | RP11-473I1.9 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 60 | LPP | Sponge network | 0.683 | 1.0E-5 | -0.459 | 0.04475 | 0.282 |
| 45815 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-629-3p | 10 | PSIP1 | Sponge network | -0.326 | 0.14314 | -0.005 | 0.96897 | 0.282 |
| 45816 | RP11-439A17.10 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-335-5p | 10 | ZBTB4 | Sponge network | 0.051 | 0.95756 | -0.739 | 0 | 0.282 |
| 45817 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 60 | BMPR2 | Sponge network | -0.355 | 0.0077 | 0.206 | 0.0635 | 0.282 |
| 45818 | SRP14-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-93-5p | 10 | CDHR3 | Sponge network | -0.061 | 0.70068 | -0.977 | 4.0E-5 | 0.281 |
| 45819 | RP11-403I13.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-935 | 43 | PTPRD | Sponge network | 0.395 | 0.15451 | -1.005 | 0.00064 | 0.281 |
| 45820 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | CNTN1 | Sponge network | 0.41 | 0.02214 | -2.594 | 0 | 0.281 |
| 45821 | BMS1P20 |
hsa-miR-1271-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-22-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 19 | PPM1L | Sponge network | 0.34 | 0.00273 | -0.537 | 0.00616 | 0.281 |
| 45822 | AC074289.1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-455-3p | 14 | PCDH9 | Sponge network | -0.326 | 0.14314 | -1.823 | 4.0E-5 | 0.281 |
| 45823 | LINC00578 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-589-3p | 17 | AKAP11 | Sponge network | 0.811 | 0.03228 | 0.196 | 0.09825 | 0.281 |
| 45824 | RP11-401P9.4 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-5p | 12 | ATM | Sponge network | -0.384 | 0.40652 | 0.178 | 0.21568 | 0.281 |
| 45825 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PCDH17 | Sponge network | 0.919 | 0 | 0.731 | 2.0E-5 | 0.281 |
| 45826 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 17 | CDHR3 | Sponge network | -0.355 | 0.0077 | -0.977 | 4.0E-5 | 0.281 |
| 45827 | CTA-29F11.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p | 11 | TBL1XR1 | Sponge network | 0.583 | 0.00027 | 0.578 | 0 | 0.281 |
| 45828 | TMEM161B-AS1 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p | 23 | ACVR2A | Sponge network | -0.04 | 0.7606 | -0.409 | 0.00039 | 0.281 |
| 45829 | SOX2-OT |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-484;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | NRXN3 | Sponge network | -1.264 | 6.0E-5 | -0.893 | 0.04186 | 0.281 |
| 45830 | RP11-77H9.2 |
hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | ERCC6 | Sponge network | 1.677 | 0 | 0.23 | 0.015 | 0.281 |
| 45831 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-98-5p | 23 | CLASP2 | Sponge network | 0.056 | 0.81195 | -0.038 | 0.71 | 0.281 |
| 45832 | MRPL23-AS1 |
hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-484 | 11 | MED12L | Sponge network | -0.278 | 0.76559 | -0.194 | 0.55299 | 0.281 |
| 45833 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p | 17 | BCL11A | Sponge network | -0.563 | 0.02545 | -0.932 | 0.0063 | 0.281 |
| 45834 | AC074289.1 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-539-5p;hsa-miR-940 | 12 | TAL1 | Sponge network | -0.326 | 0.14314 | -0.462 | 0.00574 | 0.281 |
| 45835 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-589-3p | 26 | SCN9A | Sponge network | 0.177 | 0.16681 | -0.525 | 0.10593 | 0.281 |
| 45836 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | PCDH8 | Sponge network | 0.219 | 0.1516 | -0.997 | 0.05366 | 0.281 |
| 45837 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-942-5p | 36 | THRB | Sponge network | 1.757 | 0 | -1.117 | 0.0004 | 0.281 |
| 45838 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p | 13 | SON | Sponge network | 1.073 | 0.00216 | 0.168 | 0.04406 | 0.281 |
| 45839 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-96-5p | 20 | ACVR1 | Sponge network | 0.249 | 0.35902 | 0.279 | 0.00234 | 0.281 |
| 45840 | RP11-389C8.2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | EGR2 | Sponge network | 0.727 | 3.0E-5 | 0.309 | 0.17079 | 0.281 |
| 45841 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 26 | RAP1A | Sponge network | 0.177 | 0.16681 | -0.929 | 0 | 0.281 |
| 45842 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | LATS1 | Sponge network | 2.368 | 2.0E-5 | 0.109 | 0.34208 | 0.281 |
| 45843 | RP11-798M19.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 14 | ITGB3 | Sponge network | -0.612 | 0.00094 | -0.011 | 0.95751 | 0.281 |
| 45844 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-577 | 20 | HIP1 | Sponge network | 0.177 | 0.16681 | 0.774 | 0 | 0.281 |
| 45845 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-424-5p;hsa-miR-590-5p | 15 | PIK3R1 | Sponge network | -2.076 | 0.00548 | -0.133 | 0.36854 | 0.281 |
| 45846 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 18 | SOX5 | Sponge network | 0.693 | 0.00076 | -1.091 | 4.0E-5 | 0.281 |
| 45847 | RP11-819C21.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | LRIG1 | Sponge network | 0.537 | 0.00139 | -0.537 | 0.00588 | 0.281 |
| 45848 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b | 11 | MOB1B | Sponge network | 0.357 | 0.72407 | 0.197 | 0.15266 | 0.281 |
| 45849 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 22 | ST6GAL2 | Sponge network | -0.842 | 0.01361 | -1.201 | 0.00597 | 0.281 |
| 45850 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-5p;hsa-miR-93-5p | 29 | HBP1 | Sponge network | -0.371 | 0.24604 | -0.454 | 0 | 0.281 |
| 45851 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 13 | CLIP1 | Sponge network | -0.738 | 0.21233 | -0.215 | 0.08684 | 0.281 |
| 45852 | TINCR |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-96-5p | 41 | AFF4 | Sponge network | -0.664 | 0.14543 | -0.266 | 0.01565 | 0.281 |
| 45853 | U91328.19 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p | 15 | PREX2 | Sponge network | -0.303 | 0.21002 | -0.272 | 0.25382 | 0.281 |
| 45854 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-3p | 22 | RICTOR | Sponge network | 1.61 | 0.00961 | 0.388 | 0.00188 | 0.281 |
| 45855 | LINC00265 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | UBR3 | Sponge network | 1.07 | 0 | -0.021 | 0.82774 | 0.281 |
| 45856 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-766-3p | 22 | CHD5 | Sponge network | 0.614 | 1.0E-5 | -0.922 | 0.01521 | 0.281 |
| 45857 | RP11-611E13.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-497-5p | 14 | TBL1XR1 | Sponge network | 0.822 | 0 | 0.578 | 0 | 0.281 |
| 45858 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p | 14 | CITED2 | Sponge network | -0.738 | 0.21233 | -1.545 | 0 | 0.281 |
| 45859 | TP73-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p | 15 | ABCB5 | Sponge network | -0.563 | 0.02545 | -0.956 | 0.04077 | 0.281 |
| 45860 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-3p | 15 | BCL11A | Sponge network | 0.494 | 0.00339 | -0.932 | 0.0063 | 0.281 |
| 45861 | EIF3J-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TET1 | Sponge network | 0.331 | 0.00509 | -0.135 | 0.52138 | 0.281 |
| 45862 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 45 | UNC80 | Sponge network | -0.222 | 0.32445 | -1.934 | 0 | 0.281 |
| 45863 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | EPHA5 | Sponge network | -1.264 | 6.0E-5 | -0.957 | 0.01733 | 0.281 |
| 45864 | AC109309.4 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-3p;hsa-miR-142-3p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-3p | 11 | WDFY3 | Sponge network | 0.163 | 0.87219 | -0.201 | 0.0967 | 0.281 |
| 45865 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 22 | NEDD4 | Sponge network | -0.709 | 0.18168 | 0.02 | 0.89834 | 0.281 |
| 45866 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-539-5p | 12 | EPHA7 | Sponge network | -0.214 | 0.44248 | -2.197 | 3.0E-5 | 0.281 |
| 45867 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-411-5p;hsa-miR-590-3p | 12 | USP6 | Sponge network | 0.272 | 0.44692 | 0.188 | 0.3871 | 0.281 |
| 45868 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-7-1-3p | 13 | DACH2 | Sponge network | -0.842 | 0.01361 | -1.635 | 0.00034 | 0.281 |
| 45869 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PTPRB | Sponge network | 0.537 | 0.00139 | 0.105 | 0.47122 | 0.281 |
| 45870 | KB-1460A1.5 |
hsa-miR-101-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | INPP4B | Sponge network | 0.603 | 0.00127 | -0.097 | 0.60503 | 0.281 |
| 45871 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | TMTC3 | Sponge network | 0.67 | 0.00048 | 0.123 | 0.22189 | 0.281 |
| 45872 | SERTAD4-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-93-3p | 20 | CUX1 | Sponge network | -0.932 | 0.00329 | -0.039 | 0.6705 | 0.281 |
| 45873 | U91328.19 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-629-3p | 16 | TCF12 | Sponge network | -0.303 | 0.21002 | 0.174 | 0.13271 | 0.281 |
| 45874 | CTB-113P19.4 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 27 | NOTCH2 | Sponge network | 0.906 | 0.31715 | 0.138 | 0.35163 | 0.281 |
| 45875 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 12 | DAB2 | Sponge network | -1.203 | 0.03881 | 0.431 | 0.0113 | 0.281 |
| 45876 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 22 | CLASP2 | Sponge network | 0.541 | 0.00125 | -0.038 | 0.71 | 0.281 |
| 45877 | RP11-1006G14.4 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TMEM30A | Sponge network | 0.559 | 0.00026 | 0.096 | 0.31031 | 0.281 |
| 45878 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 19 | PKNOX1 | Sponge network | -0.737 | 0.06182 | 0.085 | 0.28917 | 0.281 |
| 45879 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-429 | 12 | RNASEL | Sponge network | 1.316 | 0.01106 | -0.272 | 0.01796 | 0.281 |
| 45880 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SLC6A15 | Sponge network | 0.311 | 0.10344 | -2.373 | 2.0E-5 | 0.281 |
| 45881 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 15 | CDH19 | Sponge network | 0.4 | 0.14611 | -3.434 | 0 | 0.281 |
| 45882 | H19 |
hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-589-3p | 11 | MOB1B | Sponge network | 2.439 | 0 | 0.197 | 0.15266 | 0.281 |
| 45883 | RAD51-AS1 |
hsa-miR-10b-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-655-3p | 10 | ZMYND11 | Sponge network | 0.768 | 7.0E-5 | -0.2 | 0.05449 | 0.281 |
| 45884 | BZRAP1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-3p;hsa-miR-93-3p | 15 | WWTR1 | Sponge network | -0.465 | 0.04703 | -0.345 | 0.11997 | 0.281 |
| 45885 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | CACNA2D1 | Sponge network | 0.537 | 0.00139 | -0.846 | 0.00675 | 0.281 |
| 45886 | RP5-1198O20.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | ZFX | Sponge network | 0.997 | 0.00066 | 0.09 | 0.42563 | 0.281 |
| 45887 | AC009948.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3E | Sponge network | 0.532 | 0.00505 | -1.717 | 0.00137 | 0.281 |
| 45888 | RP11-21A7A.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-93-3p | 16 | SNX29 | Sponge network | -1.116 | 0.23686 | -0.34 | 0.01371 | 0.281 |
| 45889 | RP11-248G5.8 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p | 10 | DICER1 | Sponge network | 1.294 | 0 | 0.042 | 0.674 | 0.281 |
| 45890 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | CHD2 | Sponge network | -0.709 | 0.18168 | 0.049 | 0.55371 | 0.281 |
| 45891 | LINC00472 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-7-1-3p | 12 | DACT1 | Sponge network | -0.842 | 0.01361 | 0.783 | 0.00072 | 0.281 |
| 45892 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p | 20 | KIT | Sponge network | 0.16 | 0.50785 | -2.184 | 0 | 0.281 |
| 45893 | RP4-791M13.3 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | DOCK4 | Sponge network | 0.419 | 0.0319 | 0.502 | 0.00108 | 0.281 |
| 45894 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-93-5p | 11 | DAB2 | Sponge network | -0.154 | 0.78939 | 0.431 | 0.0113 | 0.281 |
| 45895 | FAM157B |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 16 | CBX5 | Sponge network | 0.92 | 0 | 0.165 | 0.24446 | 0.281 |
| 45896 | HOXD-AS2 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-455-3p | 14 | EPHA7 | Sponge network | -0.208 | 0.65592 | -2.197 | 3.0E-5 | 0.281 |
| 45897 | TINCR |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-96-5p | 26 | CADM1 | Sponge network | -0.664 | 0.14543 | -0.9 | 0.00084 | 0.281 |
| 45898 | RP4-714D9.5 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-497-5p | 12 | ARHGEF12 | Sponge network | 1.015 | 0 | 0.09 | 0.35693 | 0.281 |
| 45899 | RP11-890B15.3 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | RIMS2 | Sponge network | 0.101 | 0.60919 | -1.365 | 0.00208 | 0.281 |
| 45900 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 20 | KIF1B | Sponge network | 2.368 | 2.0E-5 | -0.118 | 0.32258 | 0.281 |
| 45901 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 23 | MYBL1 | Sponge network | 0.782 | 0.00016 | 0.494 | 0.02152 | 0.281 |
| 45902 | FAM66C |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | DOCK4 | Sponge network | -0.901 | 0.00019 | 0.502 | 0.00108 | 0.281 |
| 45903 | AC034220.3 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | PHC3 | Sponge network | 0.309 | 0.07304 | 0.212 | 0.0499 | 0.281 |
| 45904 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 46 | BMPR2 | Sponge network | 0.267 | 0.2937 | 0.206 | 0.0635 | 0.281 |
| 45905 | CTD-2245F17.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-335-5p;hsa-miR-539-5p | 13 | FAT3 | Sponge network | -0.421 | 0.12556 | -1.601 | 0.00051 | 0.281 |
| 45906 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 18 | TMTC3 | Sponge network | 1.308 | 0 | 0.123 | 0.22189 | 0.281 |
| 45907 | RP11-182J1.1 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | PAFAH1B1 | Sponge network | -0.187 | 0.23787 | -0.429 | 0 | 0.281 |
| 45908 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-625-5p | 24 | CADM1 | Sponge network | -0.162 | 0.47687 | -0.9 | 0.00084 | 0.281 |
| 45909 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940;hsa-miR-942-5p | 33 | HLF | Sponge network | 0.392 | 0.0278 | -2.443 | 0 | 0.281 |
| 45910 | AC006116.24 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p | 11 | LIMCH1 | Sponge network | -0.473 | 0.07602 | -0.915 | 1.0E-5 | 0.281 |
| 45911 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 14 | SMARCA2 | Sponge network | 0.214 | 0.18246 | -0.529 | 0.00014 | 0.281 |
| 45912 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 25 | ADAMTSL3 | Sponge network | 0.659 | 3.0E-5 | -1.559 | 0 | 0.281 |
| 45913 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | FOXO1 | Sponge network | -1.057 | 0.00029 | -0.141 | 0.33874 | 0.281 |
| 45914 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-93-5p;hsa-miR-96-5p | 14 | DPYD | Sponge network | 0.249 | 0.35902 | -0.736 | 0.00076 | 0.281 |
| 45915 | RP11-296I10.3 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-374b-3p | 11 | UBE4B | Sponge network | 0.592 | 0.00014 | -0.235 | 0.007 | 0.281 |
| 45916 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-582-5p | 26 | SPRY3 | Sponge network | 0.637 | 0.00069 | 0.016 | 0.91286 | 0.281 |
| 45917 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NIN | Sponge network | -0.021 | 0.93598 | -0.253 | 0.13794 | 0.281 |
| 45918 | RP11-48B3.4 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p | 11 | TAL1 | Sponge network | 1.757 | 0 | -0.462 | 0.00574 | 0.281 |
| 45919 | GNG12-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-421;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | HERC1 | Sponge network | -0.793 | 7.0E-5 | -0.196 | 0.08544 | 0.281 |
| 45920 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 17 | TGFBR2 | Sponge network | 0.72 | 0.0001 | -0.243 | 0.11408 | 0.281 |
| 45921 | HCG11 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | UBR3 | Sponge network | 0.385 | 0.10067 | -0.021 | 0.82774 | 0.281 |
| 45922 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 45 | CCND2 | Sponge network | -0.901 | 0.00019 | -0.267 | 0.3112 | 0.281 |
| 45923 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 11 | EPAS1 | Sponge network | -0.737 | 0.10822 | -0.184 | 0.14418 | 0.281 |
| 45924 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 36 | ATRX | Sponge network | -2.452 | 0 | 0.261 | 0.04408 | 0.281 |
| 45925 | RP11-35G9.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 12 | ABCB5 | Sponge network | 0.657 | 4.0E-5 | -0.956 | 0.04077 | 0.281 |
| 45926 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p | 11 | FGFR1 | Sponge network | 0.75 | 0.00054 | -0.509 | 0.03957 | 0.281 |
| 45927 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-505-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | KCNA6 | Sponge network | -0.031 | 0.89063 | -1.531 | 0.00013 | 0.281 |
| 45928 | LINC00472 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b | 17 | KCNJ5 | Sponge network | -0.842 | 0.01361 | -0.281 | 0.3339 | 0.281 |
| 45929 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 48 | IL6ST | Sponge network | 0.267 | 0.2937 | -0.402 | 0.00676 | 0.281 |
| 45930 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p | 13 | ABCC9 | Sponge network | 0.921 | 0.00118 | -0.466 | 0.14121 | 0.281 |
| 45931 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 24 | CRTC3 | Sponge network | -0.727 | 0.04726 | 0.164 | 0.16786 | 0.281 |
| 45932 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | APC | Sponge network | 0.858 | 3.0E-5 | -0.255 | 0.04051 | 0.281 |
| 45933 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-93-5p | 17 | JAZF1 | Sponge network | 1.153 | 0.00114 | -0.377 | 0.02677 | 0.281 |
| 45934 | LINC00648 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 19 | SCN9A | Sponge network | 0.633 | 0.51678 | -0.525 | 0.10593 | 0.281 |
| 45935 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 44 | QKI | Sponge network | 0.61 | 0.01593 | -0.333 | 0.04111 | 0.281 |
| 45936 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | NEDD4 | Sponge network | -0.612 | 0.00094 | 0.02 | 0.89834 | 0.281 |
| 45937 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | 0.539 | 0.03722 | 0.206 | 0.06751 | 0.281 |
| 45938 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-625-5p | 11 | JAZF1 | Sponge network | 1.294 | 0.0031 | -0.377 | 0.02677 | 0.281 |
| 45939 | SNHG14 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 32 | EFNA5 | Sponge network | -1.111 | 0.0003 | -0.421 | 0.17868 | 0.281 |
| 45940 | AC005618.6 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-940 | 29 | CHD5 | Sponge network | 0.862 | 0.06213 | -0.922 | 0.01521 | 0.281 |
| 45941 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-539-5p | 17 | RAP1A | Sponge network | -0.154 | 0.53954 | -0.929 | 0 | 0.281 |
| 45942 | RP11-157J24.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-576-5p | 16 | GAS7 | Sponge network | -1.664 | 0.05165 | -0.555 | 0.01602 | 0.281 |
| 45943 | RP11-1277A3.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-511-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 40 | LPP | Sponge network | 0.453 | 0.01839 | -0.459 | 0.04475 | 0.281 |
| 45944 | RP11-389C8.2 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CTGF | Sponge network | 0.727 | 3.0E-5 | 0.321 | 0.11682 | 0.281 |
| 45945 | RP11-798G7.7 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-375 | 11 | RASSF8 | Sponge network | 0.858 | 3.0E-5 | -0.122 | 0.65591 | 0.281 |
| 45946 | LINC00403 |
hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-452-5p | 10 | PAFAH1B1 | Sponge network | -3.586 | 0.00032 | -0.429 | 0 | 0.281 |
| 45947 | BCYRN1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 18 | CELF4 | Sponge network | 0.311 | 0.10344 | -1.135 | 0.00344 | 0.281 |
| 45948 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | STARD13 | Sponge network | -0.342 | 0.02288 | -0.187 | 0.27383 | 0.281 |
| 45949 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 17 | EYA4 | Sponge network | 0.614 | 1.0E-5 | 0.3 | 0.36876 | 0.281 |
| 45950 | RASSF8-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | BRD3 | Sponge network | 0.335 | 0.20596 | 0.37 | 0.00013 | 0.281 |
| 45951 | FENDRR |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-3p | 15 | FMNL3 | Sponge network | -1.281 | 0.00012 | 0.555 | 4.0E-5 | 0.281 |
| 45952 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CTDSPL | Sponge network | 0.559 | 0.00026 | 0.085 | 0.45121 | 0.281 |
| 45953 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 26 | EBF1 | Sponge network | -0.08 | 0.90092 | -0.384 | 0.08856 | 0.281 |
| 45954 | HAR1A |
hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-93-3p | 12 | ST6GAL2 | Sponge network | -0.672 | 0.19447 | -1.201 | 0.00597 | 0.281 |
| 45955 | CTD-2528L19.6 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | TET1 | Sponge network | 0.953 | 0 | -0.135 | 0.52138 | 0.281 |
| 45956 | MYLK-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p | 11 | MNT | Sponge network | -1.331 | 3.0E-5 | -0.157 | 0.06598 | 0.281 |
| 45957 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-654-3p | 33 | BMPR2 | Sponge network | 0.799 | 1.0E-5 | 0.206 | 0.0635 | 0.281 |
| 45958 | RP13-1039J1.4 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-582-5p | 12 | DTNA | Sponge network | 0.356 | 0.33467 | -1.648 | 1.0E-5 | 0.281 |
| 45959 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 38 | BMPR2 | Sponge network | 0.389 | 0.04293 | 0.206 | 0.0635 | 0.281 |
| 45960 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | PRKAR1A | Sponge network | 0.194 | 0.64278 | -0.063 | 0.50314 | 0.281 |
| 45961 | RP5-1139B12.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 17 | NTRK2 | Sponge network | 0.598 | 0.38481 | -0.736 | 0.06495 | 0.281 |
| 45962 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 21 | RSPO2 | Sponge network | 0.532 | 0.00505 | -3.038 | 0 | 0.281 |
| 45963 | HOTAIRM1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-590-3p | 14 | PRDM2 | Sponge network | -0.053 | 0.80685 | -0.258 | 0.00721 | 0.281 |
| 45964 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-590-3p | 17 | CDC73 | Sponge network | 0.222 | 0.29133 | 0.094 | 0.20463 | 0.281 |
| 45965 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 48 | IL6ST | Sponge network | 0.174 | 0.30034 | -0.402 | 0.00676 | 0.281 |
| 45966 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 17 | FREM2 | Sponge network | -0.793 | 7.0E-5 | -0.437 | 0.46353 | 0.281 |
| 45967 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p | 26 | RBMS3 | Sponge network | 0.906 | 0.31715 | -0.519 | 0.03551 | 0.281 |
| 45968 | LINC00476 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p | 31 | TOM1L2 | Sponge network | -0.615 | 0.00015 | -0.994 | 0 | 0.281 |
| 45969 | RP11-6O2.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-624-5p | 21 | ZFX | Sponge network | 0.331 | 0.57247 | 0.09 | 0.42563 | 0.281 |
| 45970 | CTC-471J1.2 |
hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 12 | RASAL2 | Sponge network | 1.11 | 1.0E-5 | 0.869 | 0 | 0.281 |
| 45971 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | NEDD4 | Sponge network | 0.392 | 0.0278 | 0.02 | 0.89834 | 0.281 |
| 45972 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 29 | USP12 | Sponge network | -0.899 | 6.0E-5 | -0.102 | 0.40768 | 0.281 |
| 45973 | RP11-13J8.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-766-3p | 12 | TBL1XR1 | Sponge network | 3.03 | 0 | 0.578 | 0 | 0.281 |
| 45974 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | MITF | Sponge network | -0.777 | 0.1134 | -0.769 | 0.00022 | 0.281 |
| 45975 | A2M-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-502-5p;hsa-miR-590-3p | 13 | FMNL3 | Sponge network | -0.08 | 0.90092 | 0.555 | 4.0E-5 | 0.281 |
| 45976 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-708-5p | 22 | SPRY3 | Sponge network | 0.594 | 0.00064 | 0.016 | 0.91286 | 0.281 |
| 45977 | AC013275.2 |
hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-370-3p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | JAZF1 | Sponge network | 0.488 | 0.5917 | -0.377 | 0.02677 | 0.281 |
| 45978 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | PDS5B | Sponge network | 0.415 | 0.14657 | 0.259 | 0.00962 | 0.281 |
| 45979 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-542-3p | 14 | ABCB5 | Sponge network | -0.773 | 0.04073 | -0.956 | 0.04077 | 0.281 |
| 45980 | RP11-401P9.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-550a-3p | 10 | BRD3 | Sponge network | 1.524 | 0.1098 | 0.37 | 0.00013 | 0.281 |
| 45981 | RP11-401P9.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-877-5p | 15 | NRK | Sponge network | -0.384 | 0.40652 | 0.656 | 0.19217 | 0.281 |
| 45982 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 34 | TACC1 | Sponge network | 0.349 | 0.01695 | -0.362 | 0.05406 | 0.281 |
| 45983 | LINC00265 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 11 | PRKAA1 | Sponge network | 1.07 | 0 | 0.317 | 0.0031 | 0.281 |
| 45984 | RP11-13K12.5 |
hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-335-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-589-3p | 13 | RPS6KA6 | Sponge network | -0.318 | 0.65847 | -2.989 | 0 | 0.281 |
| 45985 | RP11-693J15.4 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 14 | PCDH17 | Sponge network | -0.72 | 0.24239 | 0.731 | 2.0E-5 | 0.281 |
| 45986 | RP11-13K12.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 21 | BHLHE41 | Sponge network | -0.318 | 0.65847 | 0.353 | 0.12753 | 0.281 |
| 45987 | LINC00702 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-1-5p;hsa-miR-96-5p | 13 | PRKACA | Sponge network | -0.737 | 0.06182 | -0.255 | 0.0012 | 0.281 |
| 45988 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-96-5p | 14 | FOXO1 | Sponge network | 0.622 | 0.00015 | -0.141 | 0.33874 | 0.281 |
| 45989 | RP11-359B12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p | 22 | ARNT2 | Sponge network | 0.064 | 0.72934 | -0.332 | 0.25851 | 0.281 |
| 45990 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1976;hsa-miR-25-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-7-1-3p | 10 | DKK3 | Sponge network | -0.418 | 0.00068 | -0.052 | 0.77783 | 0.281 |
| 45991 | CTD-2139B15.2 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-539-5p | 13 | MOB1B | Sponge network | 0.286 | 0.00612 | 0.197 | 0.15266 | 0.281 |
| 45992 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-93-5p | 13 | EZH1 | Sponge network | -0.235 | 0.15142 | -0.564 | 1.0E-5 | 0.281 |
| 45993 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | NIN | Sponge network | -0.513 | 0.04703 | -0.253 | 0.13794 | 0.281 |
| 45994 | RP11-116O18.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-590-3p | 10 | ABCB5 | Sponge network | 0.05 | 0.95483 | -0.956 | 0.04077 | 0.281 |
| 45995 | RP11-116O18.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p | 10 | UNC5D | Sponge network | 0.05 | 0.95483 | -3.646 | 0 | 0.281 |
| 45996 | PCBP1-AS1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-193a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-339-5p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-505-5p | 12 | DDX3X | Sponge network | -0.567 | 0 | -0.152 | 0.07686 | 0.281 |
| 45997 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-28-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 19 | PTPN11 | Sponge network | 0.174 | 0.30034 | 0.431 | 2.0E-5 | 0.281 |
| 45998 | RP11-25K21.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-374a-3p;hsa-miR-484 | 11 | CBL | Sponge network | 0.489 | 0.25786 | 0.291 | 0.01267 | 0.281 |
| 45999 | RP11-196G18.23 |
hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | MOB1B | Sponge network | 0.724 | 0.00039 | 0.197 | 0.15266 | 0.281 |
| 46000 | RP11-531A24.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p | 26 | NAV3 | Sponge network | 0.75 | 0.00054 | 0.212 | 0.47729 | 0.281 |
| 46001 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 40 | TMEM132B | Sponge network | -0.777 | 0.1134 | -0.83 | 0.01084 | 0.281 |
| 46002 | RP11-497H16.9 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | ABL2 | Sponge network | 0.545 | 0.00064 | 0.82 | 0 | 0.281 |
| 46003 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | AFF3 | Sponge network | 0.385 | 0.10067 | -2.373 | 0 | 0.281 |
| 46004 | RP11-20I23.13 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-486-5p | 19 | PIK3R1 | Sponge network | 0.505 | 0.0014 | -0.133 | 0.36854 | 0.281 |
| 46005 | LOXL1-AS1 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | BCL9 | Sponge network | 0.487 | 0.04053 | 0.321 | 0.00267 | 0.281 |
| 46006 | RP1-151F17.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 19 | PRDM2 | Sponge network | 0.222 | 0.29133 | -0.258 | 0.00721 | 0.281 |
| 46007 | DLEU2 |
hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p | 10 | CDC73 | Sponge network | 1.831 | 0 | 0.094 | 0.20463 | 0.281 |
| 46008 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 39 | PRKAA2 | Sponge network | 0.75 | 0.00054 | -2.462 | 0 | 0.281 |
| 46009 | ACTA2-AS1 |
hsa-miR-142-3p;hsa-miR-183-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | CTNNA2 | Sponge network | 0.194 | 0.64278 | -2.547 | 0 | 0.281 |
| 46010 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 16 | TCF4 | Sponge network | -0.557 | 0.11135 | 0.016 | 0.92271 | 0.281 |
| 46011 | RP11-13J8.1 |
hsa-miR-10b-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-495-3p | 18 | CCDC6 | Sponge network | 3.03 | 0 | 0.27 | 0.02555 | 0.281 |
| 46012 | LINC00672 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | ROBO1 | Sponge network | -0.227 | 0.31657 | -0.009 | 0.96154 | 0.281 |
| 46013 | CTC-297N7.9 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-5p;hsa-miR-7-5p | 10 | PRKCB | Sponge network | -2.069 | 0 | -1.313 | 2.0E-5 | 0.281 |
| 46014 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 38 | NRXN1 | Sponge network | 0.064 | 0.72934 | -3.778 | 0 | 0.281 |
| 46015 | RP11-46C24.7 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PHF3 | Sponge network | 0.392 | 0.0278 | 0.126 | 0.19652 | 0.281 |
| 46016 | CTD-2554C21.2 |
hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429 | 10 | MAFB | Sponge network | -1.654 | 0.00144 | -0.023 | 0.89865 | 0.281 |
| 46017 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p | 18 | BCL6 | Sponge network | 0.385 | 0.10067 | -0.211 | 0.19489 | 0.281 |
| 46018 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 13 | EPHA3 | Sponge network | 0.453 | 0.01839 | 0.185 | 0.53588 | 0.281 |
| 46019 | RP11-620J15.3 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-369-3p;hsa-miR-590-3p | 13 | TLE4 | Sponge network | -0.739 | 0.00044 | -1.157 | 0 | 0.281 |
| 46020 | LINC00173 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CSMD1 | Sponge network | 0.056 | 0.8494 | -0.63 | 0.18881 | 0.281 |
| 46021 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p | 11 | PDS5B | Sponge network | 0.921 | 0.00118 | 0.259 | 0.00962 | 0.281 |
| 46022 | WAC-AS1 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | NF1 | Sponge network | 0.821 | 0 | 0.51 | 0 | 0.281 |
| 46023 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-7-1-3p | 19 | AKAP11 | Sponge network | 1.61 | 0.00961 | 0.196 | 0.09825 | 0.281 |
| 46024 | RP11-13K12.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484 | 18 | IGF2 | Sponge network | -0.154 | 0.78939 | 0.848 | 0.03842 | 0.281 |
| 46025 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | SON | Sponge network | 0.541 | 0.00125 | 0.168 | 0.04406 | 0.281 |
| 46026 | RP11-57H14.4 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p | 11 | RASAL2 | Sponge network | 0.083 | 0.66405 | 0.869 | 0 | 0.281 |
| 46027 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | KALRN | Sponge network | -0.769 | 0.00035 | -0.819 | 1.0E-5 | 0.281 |
| 46028 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITGAV | Sponge network | 1.153 | 0 | 0.337 | 0.01002 | 0.281 |
| 46029 | CTBP1-AS2 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-497-5p | 12 | TBL1XR1 | Sponge network | 0.623 | 0 | 0.578 | 0 | 0.281 |
| 46030 | LINC00622 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | MDM4 | Sponge network | 1.169 | 0.00028 | 0.345 | 0.00762 | 0.281 |
| 46031 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-576-5p | 12 | ARNT2 | Sponge network | 0.419 | 0.0319 | -0.332 | 0.25851 | 0.281 |
| 46032 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-5p;hsa-miR-96-5p | 17 | PAK3 | Sponge network | -0.969 | 2.0E-5 | -0.628 | 0.07253 | 0.281 |
| 46033 | SNHG16 |
hsa-let-7i-3p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-495-3p | 10 | STAG2 | Sponge network | 0.961 | 0 | 0.252 | 0.00438 | 0.281 |
| 46034 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429 | 15 | RBL2 | Sponge network | 0.16 | 0.50785 | -0.282 | 0.00254 | 0.281 |
| 46035 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p | 12 | THBS1 | Sponge network | 0.497 | 0.01451 | 0.741 | 0.00386 | 0.281 |
| 46036 | LINC01018 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | DNMT3A | Sponge network | -2.915 | 0.00015 | 0.302 | 0.0262 | 0.281 |
| 46037 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 25 | PCDH17 | Sponge network | 0.72 | 0.0001 | 0.731 | 2.0E-5 | 0.281 |
| 46038 | RP11-894P9.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p | 20 | CBX5 | Sponge network | 0.993 | 0 | 0.165 | 0.24446 | 0.281 |
| 46039 | AC025171.1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | RASSF8 | Sponge network | 0.998 | 0 | -0.122 | 0.65591 | 0.281 |
| 46040 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 26 | BNC2 | Sponge network | 0.419 | 0.0319 | -0.491 | 0.09988 | 0.281 |
| 46041 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DNAJB4 | Sponge network | 1.302 | 7.0E-5 | -0.7 | 1.0E-5 | 0.281 |
| 46042 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p | 19 | HIP1 | Sponge network | -0.147 | 0.40452 | 0.774 | 0 | 0.281 |
| 46043 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92b-3p | 18 | ADAMTSL3 | Sponge network | -0.322 | 0.25945 | -1.559 | 0 | 0.281 |
| 46044 | RP1-39G22.7 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p | 14 | TSC1 | Sponge network | 0.439 | 0.00313 | 0.103 | 0.26071 | 0.281 |
| 46045 | LINC00654 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | RIMS2 | Sponge network | 0.374 | 0.20054 | -1.365 | 0.00208 | 0.281 |
| 46046 | HCG11 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 17 | PICALM | Sponge network | 0.385 | 0.10067 | 0.086 | 0.23523 | 0.281 |
| 46047 | CTC-487M23.5 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-450b-5p;hsa-miR-502-5p | 26 | DTNA | Sponge network | 0.912 | 0.00014 | -1.648 | 1.0E-5 | 0.281 |
| 46048 | RP11-47A8.5 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-378a-5p | 14 | BMPR2 | Sponge network | 0.103 | 0.43169 | 0.206 | 0.0635 | 0.281 |
| 46049 | AC011526.1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-590-3p | 10 | EYA4 | Sponge network | 0.342 | 0.03129 | 0.3 | 0.36876 | 0.281 |
| 46050 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-628-5p | 14 | BMPR2 | Sponge network | 1.163 | 0.00018 | 0.206 | 0.0635 | 0.281 |
| 46051 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-539-5p | 16 | CDHR3 | Sponge network | 0.177 | 0.16681 | -0.977 | 4.0E-5 | 0.281 |
| 46052 | AC018890.6 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484 | 10 | CHD5 | Sponge network | 1.61 | 0.00961 | -0.922 | 0.01521 | 0.281 |
| 46053 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-378a-5p;hsa-miR-589-3p | 22 | ADARB1 | Sponge network | 0.529 | 0.00552 | -0.335 | 0.06631 | 0.281 |
| 46054 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 10 | USP12 | Sponge network | 1.147 | 0 | -0.102 | 0.40768 | 0.281 |
| 46055 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 30 | DDX6 | Sponge network | 0.392 | 0.0278 | 0.091 | 0.36428 | 0.281 |
| 46056 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-628-5p | 19 | BCL6 | Sponge network | -0.899 | 6.0E-5 | -0.211 | 0.19489 | 0.281 |
| 46057 | RP11-35G9.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-628-5p | 12 | CDH23 | Sponge network | 0.657 | 4.0E-5 | -0.974 | 0.00034 | 0.281 |
| 46058 | LINC00672 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p | 13 | CDH23 | Sponge network | -0.227 | 0.31657 | -0.974 | 0.00034 | 0.281 |
| 46059 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-7-1-3p | 12 | CLIP1 | Sponge network | -1.349 | 0.00017 | -0.215 | 0.08684 | 0.281 |
| 46060 | AC012360.6 |
hsa-miR-106b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-654-3p | 10 | PER2 | Sponge network | 0.624 | 0.02176 | -0.396 | 0.00269 | 0.281 |
| 46061 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 33 | EPHA7 | Sponge network | -0.83 | 0 | -2.197 | 3.0E-5 | 0.281 |
| 46062 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-7-1-3p | 13 | PRUNE2 | Sponge network | 0.853 | 0.15352 | -1.733 | 0.00022 | 0.281 |
| 46063 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 16 | MYCBP2 | Sponge network | -0.129 | 0.57177 | -0.027 | 0.82016 | 0.281 |
| 46064 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-744-3p | 16 | CUL5 | Sponge network | 0.663 | 0.00073 | 0.026 | 0.77301 | 0.281 |
| 46065 | USP46-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-429 | 19 | BMPR2 | Sponge network | 0.102 | 0.51464 | 0.206 | 0.0635 | 0.281 |
| 46066 | RP11-326G21.1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 14 | GRIK3 | Sponge network | -0.021 | 0.93598 | -3.208 | 0 | 0.281 |
| 46067 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 24 | TMTC3 | Sponge network | 1.254 | 0 | 0.123 | 0.22189 | 0.281 |
| 46068 | SNHG14 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | AFAP1L2 | Sponge network | -1.111 | 0.0003 | -0.284 | 0.15434 | 0.281 |
| 46069 | RP11-307B6.3 |
hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-589-3p | 11 | MOB1B | Sponge network | -4.463 | 0.00025 | 0.197 | 0.15266 | 0.281 |
| 46070 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p | 20 | ERCC4 | Sponge network | 0.249 | 0.35902 | 0.126 | 0.15266 | 0.281 |
| 46071 | CTC-297N7.9 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-7-5p | 12 | ESR1 | Sponge network | -2.069 | 0 | -1.035 | 3.0E-5 | 0.281 |
| 46072 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-3615;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p | 10 | DMTF1 | Sponge network | 0.373 | 0.00068 | 0.248 | 0.02726 | 0.281 |
| 46073 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-98-5p | 22 | KCNA6 | Sponge network | 0.056 | 0.81195 | -1.531 | 0.00013 | 0.281 |
| 46074 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p | 17 | SPRY3 | Sponge network | 0.476 | 0.19203 | 0.016 | 0.91286 | 0.281 |
| 46075 | RP11-672A2.4 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-30d-3p | 14 | BCL2 | Sponge network | 1.344 | 0 | -0.899 | 0 | 0.281 |
| 46076 | AC004893.11 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | SIRT1 | Sponge network | 1.317 | 0 | 0.109 | 0.2593 | 0.281 |
| 46077 | DNAJC27-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | -0.969 | 2.0E-5 | -0.382 | 0.05038 | 0.281 |
| 46078 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 26 | CXXC4 | Sponge network | 0.249 | 0.35902 | -0.433 | 0.21055 | 0.281 |
| 46079 | LINC00173 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | TET1 | Sponge network | 0.056 | 0.8494 | -0.135 | 0.52138 | 0.281 |
| 46080 | GABPB1-AS1 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 12 | CDK6 | Sponge network | 1.11 | 0 | 0.991 | 0 | 0.281 |
| 46081 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p | 21 | CBL | Sponge network | -0.358 | 0.17255 | 0.291 | 0.01267 | 0.281 |
| 46082 | RP11-1348G14.4 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 22 | MDM4 | Sponge network | 0.597 | 0.00037 | 0.345 | 0.00762 | 0.281 |
| 46083 | RBPMS-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-7-1-3p | 14 | MED12L | Sponge network | 0.144 | 0.64989 | -0.194 | 0.55299 | 0.281 |
| 46084 | TP73-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | PIK3CA | Sponge network | -0.563 | 0.02545 | 0.195 | 0.06908 | 0.281 |
| 46085 | LINC00476 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-93-3p | 36 | ERC1 | Sponge network | -0.615 | 0.00015 | -0.108 | 0.38794 | 0.281 |
| 46086 | U91328.19 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-589-3p | 14 | ABCA10 | Sponge network | -0.303 | 0.21002 | -0.571 | 0.03674 | 0.281 |
| 46087 | RP4-791M13.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 26 | ZFX | Sponge network | 0.419 | 0.0319 | 0.09 | 0.42563 | 0.28 |
| 46088 | HAND2-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | PTCH1 | Sponge network | -1.697 | 0.00889 | -0.1 | 0.6179 | 0.28 |
| 46089 | OIP5-AS1 |
hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | ZNF382 | Sponge network | 0.421 | 0.00084 | -0.494 | 0.03831 | 0.28 |
| 46090 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p | 12 | SLC5A3 | Sponge network | 0.862 | 0.06213 | 0.493 | 5.0E-5 | 0.28 |
| 46091 | PSMG3-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 31 | TRPS1 | Sponge network | -0.125 | 0.49414 | -0.191 | 0.44109 | 0.28 |
| 46092 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 31 | AKAP11 | Sponge network | 0.392 | 0.0278 | 0.196 | 0.09825 | 0.28 |
| 46093 | RP11-35G9.3 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 18 | PCDH10 | Sponge network | 0.657 | 4.0E-5 | -1.644 | 0.0078 | 0.28 |
| 46094 | CTC-523E23.11 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-429 | 14 | MEF2C | Sponge network | -0.235 | 0.35308 | -0.499 | 0.01448 | 0.28 |
| 46095 | PWAR6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 20 | UNC5C | Sponge network | -1.575 | 6.0E-5 | -0.178 | 0.40214 | 0.28 |
| 46096 | SNHG14 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | BRD3 | Sponge network | -1.111 | 0.0003 | 0.37 | 0.00013 | 0.28 |
| 46097 | AC003090.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 16 | ZFP36L1 | Sponge network | -2.117 | 0.00161 | -0.505 | 8.0E-5 | 0.28 |
| 46098 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-766-3p | 16 | CHD5 | Sponge network | 1.344 | 0 | -0.922 | 0.01521 | 0.28 |
| 46099 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p | 37 | CREB3L2 | Sponge network | -0.738 | 0.21233 | -0.525 | 2.0E-5 | 0.28 |
| 46100 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-500a-5p | 12 | MAF | Sponge network | -0.932 | 0.00329 | -1.257 | 0 | 0.28 |
| 46101 | AF131215.9 |
hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | RELN | Sponge network | -0.322 | 0.25945 | -2.403 | 0 | 0.28 |
| 46102 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-582-5p;hsa-miR-7-5p | 24 | CLASP2 | Sponge network | 0.253 | 0.09565 | -0.038 | 0.71 | 0.28 |
| 46103 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92b-3p | 16 | AKAP12 | Sponge network | -0.83 | 0 | -0.932 | 0.0028 | 0.28 |
| 46104 | ZRANB2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | CNTNAP3 | Sponge network | -0.376 | 0.00337 | -1.465 | 0.00033 | 0.28 |
| 46105 | RP11-678G14.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30d-3p | 11 | THRB | Sponge network | -1.765 | 0.0778 | -1.117 | 0.0004 | 0.28 |
| 46106 | CALML3-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p | 24 | NIN | Sponge network | 0.95 | 0.15943 | -0.253 | 0.13794 | 0.28 |
| 46107 | U91328.19 |
hsa-miR-126-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-7-1-3p | 13 | SEMA3E | Sponge network | -0.303 | 0.21002 | -1.717 | 0.00137 | 0.28 |
| 46108 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 12 | DNAJB4 | Sponge network | 0.199 | 0.38015 | -0.7 | 1.0E-5 | 0.28 |
| 46109 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | TOPORS | Sponge network | -0.615 | 0.00015 | -0.226 | 0.00524 | 0.28 |
| 46110 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p | 18 | FOXP1 | Sponge network | 1.04 | 0.00023 | 0.042 | 0.74368 | 0.28 |
| 46111 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p | 12 | KANK1 | Sponge network | 0.497 | 0.01451 | -0.55 | 0.00134 | 0.28 |
| 46112 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p | 13 | ARHGAP29 | Sponge network | 0.176 | 0.37402 | 0.131 | 0.42953 | 0.28 |
| 46113 | RP11-774O3.3 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | -0.487 | 0.02157 | -0.226 | 0.11691 | 0.28 |
| 46114 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-484 | 15 | GRIA3 | Sponge network | 0.299 | 0.57722 | -1.956 | 0 | 0.28 |
| 46115 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-940 | 10 | TAL1 | Sponge network | 0.056 | 0.81195 | -0.462 | 0.00574 | 0.28 |
| 46116 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 43 | CBX5 | Sponge network | -0.901 | 0.00019 | 0.165 | 0.24446 | 0.28 |
| 46117 | MIR497HG |
hsa-let-7g-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 11 | ACTB | Sponge network | -1.439 | 4.0E-5 | -0.247 | 0.01736 | 0.28 |
| 46118 | RP11-5C23.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 13 | NRK | Sponge network | 0.274 | 0.07681 | 0.656 | 0.19217 | 0.28 |
| 46119 | RP11-983P16.4 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p | 11 | HSPB7 | Sponge network | 0.41 | 0.02214 | -2.268 | 1.0E-5 | 0.28 |
| 46120 | RP11-53O19.1 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-93-3p | 21 | CTIF | Sponge network | -0.405 | 0.22013 | -0.389 | 0.00549 | 0.28 |
| 46121 | RP4-717I23.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-577 | 14 | SYNE1 | Sponge network | 0.637 | 0.00069 | -0.738 | 0.00316 | 0.28 |
| 46122 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 10 | PRRX1 | Sponge network | 1.46 | 1.0E-5 | 1.316 | 0 | 0.28 |
| 46123 | RP11-102N12.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-671-5p;hsa-miR-96-5p | 14 | CYLD | Sponge network | -0.351 | 0.11358 | -0.367 | 0.00171 | 0.28 |
| 46124 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-7-5p | 22 | ZBTB4 | Sponge network | -0.737 | 0.10822 | -0.739 | 0 | 0.28 |
| 46125 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-3p | 16 | MAFB | Sponge network | 0.657 | 4.0E-5 | -0.023 | 0.89865 | 0.28 |
| 46126 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577 | 17 | FGF5 | Sponge network | 0.056 | 0.81195 | 0.549 | 0.28659 | 0.28 |
| 46127 | RP4-717I23.3 |
hsa-let-7d-5p;hsa-miR-106b-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-582-3p | 14 | PRDM2 | Sponge network | 0.637 | 0.00069 | -0.258 | 0.00721 | 0.28 |
| 46128 | WAC-AS1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | MOB1B | Sponge network | 0.821 | 0 | 0.197 | 0.15266 | 0.28 |
| 46129 | DIO3OS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-877-5p | 19 | TET1 | Sponge network | -0.773 | 0.04073 | -0.135 | 0.52138 | 0.28 |
| 46130 | RP11-981G7.6 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 15 | RASAL2 | Sponge network | 0.986 | 0.01022 | 0.869 | 0 | 0.28 |
| 46131 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-425-5p;hsa-miR-532-3p | 12 | CBL | Sponge network | 3.21 | 0 | 0.291 | 0.01267 | 0.28 |
| 46132 | DNM1P35 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 29 | TGFBR3 | Sponge network | 0.051 | 0.83388 | -1.173 | 1.0E-5 | 0.28 |
| 46133 | MIR143HG |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 20 | BCL9 | Sponge network | -0.739 | 0.0574 | 0.321 | 0.00267 | 0.28 |
| 46134 | MRPL23-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-664a-5p;hsa-miR-671-5p | 20 | ADARB1 | Sponge network | -0.278 | 0.76559 | -0.335 | 0.06631 | 0.28 |
| 46135 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p | 27 | CBX5 | Sponge network | 0.174 | 0.30034 | 0.165 | 0.24446 | 0.28 |
| 46136 | RP11-127B20.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 20 | ABI2 | Sponge network | 0.411 | 0.1335 | 0.175 | 0.14787 | 0.28 |
| 46137 | HOXB-AS1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429 | 12 | SOX11 | Sponge network | -0.154 | 0.53954 | 2.68 | 0 | 0.28 |
| 46138 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 18 | AFF4 | Sponge network | 1.148 | 2.0E-5 | -0.266 | 0.01565 | 0.28 |
| 46139 | LINC00863 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PRKAB2 | Sponge network | 0.189 | 0.13981 | -0.367 | 0.01371 | 0.28 |
| 46140 | AC005618.6 |
hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-940 | 10 | GLI3 | Sponge network | 0.862 | 0.06213 | -0.615 | 0.01482 | 0.28 |
| 46141 | RP11-216B9.6 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 16 | PIK3CA | Sponge network | 0.174 | 0.30034 | 0.195 | 0.06908 | 0.28 |
| 46142 | LINC00667 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | PTCH1 | Sponge network | -0.235 | 0.15142 | -0.1 | 0.6179 | 0.28 |
| 46143 | GS1-124K5.11 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | RADIL | Sponge network | 0.494 | 0.00339 | -1.628 | 0 | 0.28 |
| 46144 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-628-5p | 12 | MEF2C | Sponge network | -1.054 | 0.0706 | -0.499 | 0.01448 | 0.28 |
| 46145 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | CSMD1 | Sponge network | -1.057 | 0.00029 | -0.63 | 0.18881 | 0.28 |
| 46146 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-339-5p;hsa-miR-425-5p;hsa-miR-590-5p | 13 | PAFAH1B1 | Sponge network | 0.706 | 0 | -0.429 | 0 | 0.28 |
| 46147 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 22 | ITGAV | Sponge network | 0.222 | 0.29133 | 0.337 | 0.01002 | 0.28 |
| 46148 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 24 | GPC6 | Sponge network | 0.253 | 0.09565 | -0.054 | 0.81465 | 0.28 |
| 46149 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 23 | ZMYM2 | Sponge network | 0.349 | 0.01695 | 0.279 | 0.01273 | 0.28 |
| 46150 | CTB-133G6.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p | 10 | ZFHX4 | Sponge network | 0.4 | 0.39299 | 0.018 | 0.95574 | 0.28 |
| 46151 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 18 | CBL | Sponge network | 1.308 | 0 | 0.291 | 0.01267 | 0.28 |
| 46152 | RP11-656D10.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-5p | 17 | SPRY3 | Sponge network | 0.406 | 0.26236 | 0.016 | 0.91286 | 0.28 |
| 46153 | RP11-392P7.6 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-766-3p | 11 | TBL1XR1 | Sponge network | 0.853 | 0 | 0.578 | 0 | 0.28 |
| 46154 | HCG18 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 17 | PICALM | Sponge network | 1.063 | 0 | 0.086 | 0.23523 | 0.28 |
| 46155 | MAP3K14 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | FAT3 | Sponge network | -0.295 | 0.02604 | -1.601 | 0.00051 | 0.28 |
| 46156 | LINC00648 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CDH19 | Sponge network | 0.633 | 0.51678 | -3.434 | 0 | 0.28 |
| 46157 | ERVH48-1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-624-5p | 26 | ZFX | Sponge network | 1.04 | 0.00023 | 0.09 | 0.42563 | 0.28 |
| 46158 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-708-5p | 30 | AFF4 | Sponge network | 0.594 | 0.00064 | -0.266 | 0.01565 | 0.28 |
| 46159 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-455-3p | 25 | SPRY3 | Sponge network | 0.082 | 0.55397 | 0.016 | 0.91286 | 0.28 |
| 46160 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-93-3p | 28 | BNC2 | Sponge network | 0.274 | 0.07681 | -0.491 | 0.09988 | 0.28 |
| 46161 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 23 | RARB | Sponge network | 0.249 | 0.35902 | -0.825 | 2.0E-5 | 0.28 |
| 46162 | XXYLT1-AS2 |
hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-93-3p | 14 | IL6ST | Sponge network | -0.764 | 0.05257 | -0.402 | 0.00676 | 0.28 |
| 46163 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-653-5p | 24 | DMXL1 | Sponge network | -0.031 | 0.89063 | -0.426 | 0.00056 | 0.28 |
| 46164 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-940 | 44 | PTPRT | Sponge network | -1.216 | 0 | -0.907 | 0.03727 | 0.28 |
| 46165 | CTD-2666L21.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p | 17 | IL17RD | Sponge network | 0.368 | 0.20093 | -0.019 | 0.94873 | 0.28 |
| 46166 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-33a-5p | 10 | SLITRK3 | Sponge network | -0.214 | 0.44248 | -3.328 | 0 | 0.28 |
| 46167 | RP11-158K1.3 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | DDX3X | Sponge network | 0.349 | 0.01695 | -0.152 | 0.07686 | 0.28 |
| 46168 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | EPHA5 | Sponge network | 0.619 | 0.00157 | -0.957 | 0.01733 | 0.28 |
| 46169 | RP11-98I9.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p | 11 | ERCC4 | Sponge network | 0.67 | 0.00048 | 0.126 | 0.15266 | 0.28 |
| 46170 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 15 | PRKAA1 | Sponge network | 0.537 | 0.00139 | 0.317 | 0.0031 | 0.28 |
| 46171 | CTC-429P9.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-532-3p | 21 | CUX1 | Sponge network | 0.497 | 0.01451 | -0.039 | 0.6705 | 0.28 |
| 46172 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | PIK3C2A | Sponge network | 0.643 | 0 | 0.061 | 0.53776 | 0.28 |
| 46173 | AC108488.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | PRRX1 | Sponge network | 0.259 | 0.12542 | 1.316 | 0 | 0.28 |
| 46174 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 21 | EPHA3 | Sponge network | 0.603 | 0.00127 | 0.185 | 0.53588 | 0.28 |
| 46175 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 31 | PPM1A | Sponge network | 0.75 | 0.00054 | -0.623 | 0 | 0.28 |
| 46176 | SERTAD4-AS1 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-576-5p | 13 | NUAK1 | Sponge network | -0.932 | 0.00329 | 0.904 | 0 | 0.28 |
| 46177 | RP11-218M22.1 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 14 | RASSF8 | Sponge network | 0.197 | 0.25618 | -0.122 | 0.65591 | 0.28 |
| 46178 | RP11-67L2.2 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 18 | IL13RA1 | Sponge network | 0.72 | 0.0001 | 0.019 | 0.86949 | 0.28 |
| 46179 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 51 | BMPR2 | Sponge network | 0.603 | 0.00127 | 0.206 | 0.0635 | 0.28 |
| 46180 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-421;hsa-miR-429;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | INPP4A | Sponge network | -1.057 | 0.00029 | 0.07 | 0.53563 | 0.28 |
| 46181 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MTSS1 | Sponge network | -0.223 | 0.47816 | -0.81 | 0.00035 | 0.28 |
| 46182 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | ITGAV | Sponge network | 0.082 | 0.55654 | 0.337 | 0.01002 | 0.28 |
| 46183 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | TAL1 | Sponge network | -0.405 | 0.22013 | -0.462 | 0.00574 | 0.28 |
| 46184 | AC108488.4 |
hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-1-3p | 15 | YAP1 | Sponge network | 0.259 | 0.12542 | 0.22 | 0.11836 | 0.28 |
| 46185 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 60 | THRB | Sponge network | -0.969 | 2.0E-5 | -1.117 | 0.0004 | 0.28 |
| 46186 | WDR86-AS1 |
hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-429 | 14 | DTNA | Sponge network | 0.348 | 0.32912 | -1.648 | 1.0E-5 | 0.28 |
| 46187 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | FGF5 | Sponge network | -2.117 | 0.00161 | 0.549 | 0.28659 | 0.28 |
| 46188 | CTD-2245F17.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-502-5p;hsa-miR-766-3p | 12 | IL17RD | Sponge network | -0.421 | 0.12556 | -0.019 | 0.94873 | 0.28 |
| 46189 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-940 | 19 | TRIM33 | Sponge network | 0.101 | 0.60919 | 0.402 | 7.0E-5 | 0.28 |
| 46190 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | FGF5 | Sponge network | 0.101 | 0.60919 | 0.549 | 0.28659 | 0.28 |
| 46191 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-766-3p;hsa-miR-93-3p | 50 | IL6ST | Sponge network | -0.322 | 0.25945 | -0.402 | 0.00676 | 0.28 |
| 46192 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-940 | 24 | BHLHE41 | Sponge network | 0.349 | 0.01695 | 0.353 | 0.12753 | 0.28 |
| 46193 | TUG1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-582-5p | 13 | ZFHX3 | Sponge network | 0.972 | 0 | 0.389 | 0.01363 | 0.28 |
| 46194 | DNM3OS |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-326;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 11 | ACTB | Sponge network | 0.572 | 0.01722 | -0.247 | 0.01736 | 0.28 |
| 46195 | RP4-668J24.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-629-3p | 15 | ASTN1 | Sponge network | -1.054 | 0.0706 | -2.389 | 2.0E-5 | 0.28 |
| 46196 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | AKAP11 | Sponge network | -0.899 | 6.0E-5 | 0.196 | 0.09825 | 0.28 |
| 46197 | OSER1-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p | 11 | WNK2 | Sponge network | -0.13 | 0.48734 | -0.352 | 0.31066 | 0.28 |
| 46198 | AC127904.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ZFX | Sponge network | 1.308 | 0 | 0.09 | 0.42563 | 0.28 |
| 46199 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | EYA4 | Sponge network | 0.392 | 0.0278 | 0.3 | 0.36876 | 0.28 |
| 46200 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | CNTN1 | Sponge network | 0.083 | 0.66405 | -2.594 | 0 | 0.28 |
| 46201 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | TGFBR2 | Sponge network | 0.174 | 0.30034 | -0.243 | 0.11408 | 0.28 |
| 46202 | RP11-218M22.1 |
hsa-miR-126-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 11 | IRS1 | Sponge network | 0.197 | 0.25618 | -0.026 | 0.88855 | 0.28 |
| 46203 | OIP5-AS1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 46 | PPARA | Sponge network | 0.421 | 0.00084 | -0.379 | 0.00548 | 0.28 |
| 46204 | LINC00886 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-940 | 13 | MAPK10 | Sponge network | -0.371 | 0.24604 | -1.774 | 0 | 0.28 |
| 46205 | U91328.19 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-625-5p | 10 | NGFR | Sponge network | -0.303 | 0.21002 | -1.271 | 0.01058 | 0.28 |
| 46206 | ZNF582-AS1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | IRS1 | Sponge network | -1.349 | 0.00017 | -0.026 | 0.88855 | 0.28 |
| 46207 | RP11-474O21.5 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-7-1-3p | 10 | PCDH9 | Sponge network | -0.023 | 0.95109 | -1.823 | 4.0E-5 | 0.28 |
| 46208 | RP11-3K16.2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-181a-2-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-664a-5p | 11 | LPP | Sponge network | 0.749 | 0.17788 | -0.459 | 0.04475 | 0.28 |
| 46209 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | YAP1 | Sponge network | 0.381 | 0.25967 | 0.22 | 0.11836 | 0.28 |
| 46210 | SERHL |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-532-3p | 13 | MRVI1 | Sponge network | -0.602 | 0.00331 | -1.051 | 0.00022 | 0.28 |
| 46211 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | HIVEP1 | Sponge network | 0.848 | 0 | 0.368 | 0.00064 | 0.28 |
| 46212 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p | 10 | FYN | Sponge network | -0.738 | 0.21233 | -0.131 | 0.37051 | 0.28 |
| 46213 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-188-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p | 43 | CAMTA1 | Sponge network | -0.615 | 0.00015 | -0.436 | 1.0E-5 | 0.28 |
| 46214 | LINC00472 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-93-5p | 38 | PIK3R1 | Sponge network | -0.842 | 0.01361 | -0.133 | 0.36854 | 0.28 |
| 46215 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p | 18 | CDH19 | Sponge network | 0.174 | 0.30034 | -3.434 | 0 | 0.28 |
| 46216 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p | 13 | ZMYM2 | Sponge network | 1.11 | 1.0E-5 | 0.279 | 0.01273 | 0.28 |
| 46217 | FGF14-AS2 |
hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 13 | ZMYND11 | Sponge network | -1.582 | 8.0E-5 | -0.2 | 0.05449 | 0.28 |
| 46218 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 16 | NOTCH2NL | Sponge network | -0.793 | 7.0E-5 | 0.024 | 0.84307 | 0.28 |
| 46219 | MIR497HG |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p | 11 | PLXNC1 | Sponge network | -1.439 | 4.0E-5 | 0.569 | 0.00329 | 0.28 |
| 46220 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 14 | FBXO31 | Sponge network | -0.899 | 6.0E-5 | -0.085 | 0.42389 | 0.28 |
| 46221 | RP11-307B6.3 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-424-5p | 24 | KIF1B | Sponge network | -4.463 | 0.00025 | -0.118 | 0.32258 | 0.28 |
| 46222 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-374a-5p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-96-5p | 27 | ABI2 | Sponge network | -0.18 | 0.20343 | 0.175 | 0.14787 | 0.28 |
| 46223 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 30 | MCC | Sponge network | 0.222 | 0.29133 | -1.005 | 0 | 0.28 |
| 46224 | RP11-770J1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-505-5p;hsa-miR-671-5p | 11 | KCNA6 | Sponge network | -2.011 | 0.0005 | -1.531 | 0.00013 | 0.28 |
| 46225 | RP11-158K1.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 20 | SIRT1 | Sponge network | 0.349 | 0.01695 | 0.109 | 0.2593 | 0.28 |
| 46226 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-625-3p | 37 | ERBB4 | Sponge network | -0.371 | 0.24604 | -1.975 | 2.0E-5 | 0.28 |
| 46227 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-194-5p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p | 15 | EPHA3 | Sponge network | 0.199 | 0.52763 | 0.185 | 0.53588 | 0.28 |
| 46228 | RP1-39G22.7 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-199b-5p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-590-3p | 11 | SIRT1 | Sponge network | 0.439 | 0.00313 | 0.109 | 0.2593 | 0.28 |
| 46229 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p | 18 | KANK1 | Sponge network | -0.777 | 0.1134 | -0.55 | 0.00134 | 0.28 |
| 46230 | SOX2-OT |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 15 | CELF4 | Sponge network | -1.264 | 6.0E-5 | -1.135 | 0.00344 | 0.28 |
| 46231 | RP11-1277A3.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-92a-3p | 12 | PCDH9 | Sponge network | 0.453 | 0.01839 | -1.823 | 4.0E-5 | 0.28 |
| 46232 | OIP5-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-940 | 34 | ETV1 | Sponge network | 0.421 | 0.00084 | -0.096 | 0.67674 | 0.28 |
| 46233 | LINC00667 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-3p | 17 | ITGB3 | Sponge network | -0.235 | 0.15142 | -0.011 | 0.95751 | 0.28 |
| 46234 | KB-1460A1.5 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-331-3p;hsa-miR-590-3p | 10 | SIRT1 | Sponge network | 0.603 | 0.00127 | 0.109 | 0.2593 | 0.28 |
| 46235 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | GRIN2A | Sponge network | -1.264 | 6.0E-5 | -2.161 | 0 | 0.28 |
| 46236 | LINC00662 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | EYA4 | Sponge network | 0.213 | 0.32297 | 0.3 | 0.36876 | 0.28 |
| 46237 | WDFY3-AS2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | LUZP2 | Sponge network | -0.942 | 0 | -0.056 | 0.89416 | 0.28 |
| 46238 | AC093627.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p | 13 | PDGFRA | Sponge network | -0.787 | 0.3928 | -0.372 | 0.0037 | 0.28 |
| 46239 | CTC-559E9.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-539-5p | 19 | DMD | Sponge network | 0.594 | 0.00064 | -1.506 | 0 | 0.28 |
| 46240 | AC016747.3 |
hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | LUZP2 | Sponge network | 0.199 | 0.38015 | -0.056 | 0.89416 | 0.28 |
| 46241 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 33 | QKI | Sponge network | 0.906 | 0.31715 | -0.333 | 0.04111 | 0.28 |
| 46242 | LINC00205 |
hsa-miR-101-5p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p | 11 | RICTOR | Sponge network | 1.271 | 0 | 0.388 | 0.00188 | 0.28 |
| 46243 | HCG18 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-655-3p | 10 | UBE4B | Sponge network | 1.063 | 0 | -0.235 | 0.007 | 0.28 |
| 46244 | LINC00886 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-5p | 14 | PDZD2 | Sponge network | -0.371 | 0.24604 | -0.909 | 3.0E-5 | 0.28 |
| 46245 | RP11-2H8.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | LATS2 | Sponge network | 1.089 | 0 | 0.159 | 0.2304 | 0.28 |
| 46246 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-590-3p | 13 | ABCB5 | Sponge network | -0.08 | 0.90092 | -0.956 | 0.04077 | 0.28 |
| 46247 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p | 10 | MNT | Sponge network | 0.101 | 0.60919 | -0.157 | 0.06598 | 0.28 |
| 46248 | AC010226.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | ABI2 | Sponge network | -0.811 | 2.0E-5 | 0.175 | 0.14787 | 0.28 |
| 46249 | ARHGAP5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-362-3p | 14 | HBP1 | Sponge network | -0.644 | 0.00021 | -0.454 | 0 | 0.28 |
| 46250 | RP11-774O3.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p | 21 | SIRT1 | Sponge network | -0.487 | 0.02157 | 0.109 | 0.2593 | 0.28 |
| 46251 | RP11-819C21.1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | 0.537 | 0.00139 | -1.12 | 0.00107 | 0.28 |
| 46252 | RP11-35G9.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-502-5p | 26 | DTNA | Sponge network | 0.622 | 0.00015 | -1.648 | 1.0E-5 | 0.28 |
| 46253 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-7-1-3p | 20 | EYA4 | Sponge network | -0.611 | 0.09607 | 0.3 | 0.36876 | 0.28 |
| 46254 | PSMG3-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 19 | DOCK4 | Sponge network | -0.125 | 0.49414 | 0.502 | 0.00108 | 0.28 |
| 46255 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | 0.199 | 0.38015 | -1.3 | 0.01619 | 0.28 |
| 46256 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | HECA | Sponge network | 1.04 | 0.00023 | -0.217 | 0.03463 | 0.28 |
| 46257 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-93-5p | 19 | RNASEL | Sponge network | 0.056 | 0.81195 | -0.272 | 0.01796 | 0.28 |
| 46258 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 22 | PIK3C2A | Sponge network | -0.227 | 0.31657 | 0.061 | 0.53776 | 0.28 |
| 46259 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-362-3p | 20 | TMTC3 | Sponge network | 0.078 | 0.55803 | 0.123 | 0.22189 | 0.28 |
| 46260 | RP1-151F17.2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 21 | DLC1 | Sponge network | 0.222 | 0.29133 | -0.382 | 0.05038 | 0.28 |
| 46261 | RP11-834C11.4 |
hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-940;hsa-miR-96-5p | 12 | QKI | Sponge network | -0.883 | 0.01839 | -0.333 | 0.04111 | 0.28 |
| 46262 | RP11-38L15.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 13 | CYLD | Sponge network | 0.344 | 0.3127 | -0.367 | 0.00171 | 0.28 |
| 46263 | KB-1460A1.5 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-590-3p | 15 | NRK | Sponge network | 0.603 | 0.00127 | 0.656 | 0.19217 | 0.28 |
| 46264 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 22 | EBF3 | Sponge network | 0.219 | 0.1516 | -0.058 | 0.81437 | 0.28 |
| 46265 | LINC00909 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-497-5p | 14 | TBL1XR1 | Sponge network | 0.363 | 0.00601 | 0.578 | 0 | 0.28 |
| 46266 | RP11-758N13.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-505-5p | 17 | KCNA6 | Sponge network | 2.939 | 3.0E-5 | -1.531 | 0.00013 | 0.28 |
| 46267 | CTC-429P9.3 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p | 11 | FMNL3 | Sponge network | 0.082 | 0.55397 | 0.555 | 4.0E-5 | 0.28 |
| 46268 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | PRKAA1 | Sponge network | 0.93 | 0 | 0.317 | 0.0031 | 0.28 |
| 46269 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 14 | ZNF770 | Sponge network | 0.75 | 0.00054 | 0.206 | 0.06751 | 0.28 |
| 46270 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 29 | DDX6 | Sponge network | -0.612 | 0.00094 | 0.091 | 0.36428 | 0.28 |
| 46271 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | CNTN1 | Sponge network | -0.555 | 0.06156 | -2.594 | 0 | 0.28 |
| 46272 | LINC00863 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-93-5p | 10 | FBXO31 | Sponge network | 0.189 | 0.13981 | -0.085 | 0.42389 | 0.28 |
| 46273 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p | 12 | PCDH8 | Sponge network | -0.355 | 0.0077 | -0.997 | 0.05366 | 0.28 |
| 46274 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 11 | PIK3C2A | Sponge network | 1.344 | 0 | 0.061 | 0.53776 | 0.28 |
| 46275 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-940 | 14 | IGF1 | Sponge network | -0.326 | 0.14314 | -0.204 | 0.5898 | 0.28 |
| 46276 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | MLLT10 | Sponge network | 1.344 | 0 | 0.105 | 0.33292 | 0.28 |
| 46277 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-143-3p;hsa-miR-154-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | LIMD1 | Sponge network | 1.344 | 0 | 0.103 | 0.28978 | 0.28 |
| 46278 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-450b-5p | 10 | UNC80 | Sponge network | -1.717 | 0.01522 | -1.934 | 0 | 0.28 |
| 46279 | CTD-2314G24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | TET1 | Sponge network | 0.246 | 0.59912 | -0.135 | 0.52138 | 0.28 |
| 46280 | RP5-855D21.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p | 11 | MOB1B | Sponge network | 1.728 | 0 | 0.197 | 0.15266 | 0.28 |
| 46281 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 50 | THRB | Sponge network | 0.532 | 0.00505 | -1.117 | 0.0004 | 0.28 |
| 46282 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p | 13 | PIK3C2A | Sponge network | 0.917 | 0 | 0.061 | 0.53776 | 0.28 |
| 46283 | WDFY3-AS2 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 31 | PRKCB | Sponge network | -0.942 | 0 | -1.313 | 2.0E-5 | 0.28 |
| 46284 | RP11-21A7A.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-877-5p | 10 | CBFA2T3 | Sponge network | -1.116 | 0.23686 | -1.221 | 1.0E-5 | 0.28 |
| 46285 | RP11-155G14.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 12 | TMTC3 | Sponge network | 2.483 | 0 | 0.123 | 0.22189 | 0.28 |
| 46286 | RP5-1198O20.4 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 35 | ARHGEF12 | Sponge network | 0.997 | 0.00066 | 0.09 | 0.35693 | 0.28 |
| 46287 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-212-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | KCNA6 | Sponge network | 0.419 | 0.0319 | -1.531 | 0.00013 | 0.28 |
| 46288 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | RBMS3 | Sponge network | 0.782 | 0.00016 | -0.519 | 0.03551 | 0.28 |
| 46289 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-940;hsa-miR-96-5p | 34 | AFF4 | Sponge network | 0.594 | 0.41522 | -0.266 | 0.01565 | 0.28 |
| 46290 | TP53TG1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-330-3p | 10 | RSPO2 | Sponge network | -0.574 | 0.00291 | -3.038 | 0 | 0.28 |
| 46291 | AC159540.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-378a-5p | 13 | CREB1 | Sponge network | 1.634 | 0 | 0.203 | 0.00537 | 0.28 |
| 46292 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-766-3p | 16 | IL17RD | Sponge network | 0.858 | 3.0E-5 | -0.019 | 0.94873 | 0.28 |
| 46293 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-628-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.494 | 0.00339 | 1.083 | 0 | 0.28 |
| 46294 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-589-3p | 38 | THRB | Sponge network | -0.154 | 0.53954 | -1.117 | 0.0004 | 0.28 |
| 46295 | RP11-196G11.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-590-3p | 11 | CCDC6 | Sponge network | 0.875 | 0 | 0.27 | 0.02555 | 0.28 |
| 46296 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PTPRB | Sponge network | -0.08 | 0.90092 | 0.105 | 0.47122 | 0.28 |
| 46297 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-331-5p;hsa-miR-590-3p | 13 | CUL5 | Sponge network | 1.073 | 0.00216 | 0.026 | 0.77301 | 0.28 |
| 46298 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-629-5p;hsa-miR-7-1-3p | 13 | GRIN2A | Sponge network | -0.418 | 0.00068 | -2.161 | 0 | 0.28 |
| 46299 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-3614-5p | 15 | CREB3L2 | Sponge network | -0.421 | 0.12556 | -0.525 | 2.0E-5 | 0.28 |
| 46300 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-7-5p | 14 | RBMS3 | Sponge network | 0.348 | 0.32912 | -0.519 | 0.03551 | 0.28 |
| 46301 | RP11-774O3.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-766-3p | 41 | PPARA | Sponge network | -0.487 | 0.02157 | -0.379 | 0.00548 | 0.28 |
| 46302 | RP11-389C8.2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-29b-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ITGB1 | Sponge network | 0.727 | 3.0E-5 | 0.524 | 0.00017 | 0.28 |
| 46303 | SNHG5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-5p | 17 | CUL5 | Sponge network | 0.13 | 0.47174 | 0.026 | 0.77301 | 0.28 |
| 46304 | RP11-212P7.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 26 | CADM1 | Sponge network | 0.219 | 0.1516 | -0.9 | 0.00084 | 0.28 |
| 46305 | AC108488.4 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-7-1-3p | 11 | RASSF3 | Sponge network | 0.259 | 0.12542 | -0.226 | 0.11691 | 0.28 |
| 46306 | RP11-927P21.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-5p | 13 | PTGFR | Sponge network | 0.921 | 0.00118 | -0.233 | 0.45421 | 0.28 |
| 46307 | TINCR |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-7-5p;hsa-miR-96-5p | 20 | CLASP2 | Sponge network | -0.664 | 0.14543 | -0.038 | 0.71 | 0.28 |
| 46308 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 22 | SOX11 | Sponge network | 0.541 | 0.00125 | 2.68 | 0 | 0.28 |
| 46309 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | MYO9A | Sponge network | 0.422 | 0.27021 | -0.147 | 0.32373 | 0.28 |
| 46310 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 21 | ADAMTSL3 | Sponge network | 0.222 | 0.29133 | -1.559 | 0 | 0.28 |
| 46311 | LINC01018 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-361-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | FOXO4 | Sponge network | -2.915 | 0.00015 | -0.516 | 2.0E-5 | 0.28 |
| 46312 | RP11-389C8.2 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TPO | Sponge network | 0.727 | 3.0E-5 | -1.87 | 2.0E-5 | 0.28 |
| 46313 | NEAT1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 22 | USP6 | Sponge network | 0.705 | 0.00014 | 0.188 | 0.3871 | 0.28 |
| 46314 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 14 | FOXP1 | Sponge network | 0.693 | 0.00076 | 0.042 | 0.74368 | 0.28 |
| 46315 | TIPARP-AS1 |
hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-154-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-199b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-214-3p;hsa-miR-2355-5p;hsa-miR-421 | 14 | BTG2 | Sponge network | -0.887 | 0 | -1.763 | 0 | 0.28 |
| 46316 | AC009948.5 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | PTCH1 | Sponge network | 0.532 | 0.00505 | -0.1 | 0.6179 | 0.28 |
| 46317 | RP11-680C21.1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | GRIA3 | Sponge network | -1.424 | 0.04346 | -1.956 | 0 | 0.28 |
| 46318 | CTC-297N7.9 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-590-5p | 16 | SOX5 | Sponge network | -2.069 | 0 | -1.091 | 4.0E-5 | 0.28 |
| 46319 | TUG1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-190a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c | 16 | WDFY3 | Sponge network | 0.972 | 0 | -0.201 | 0.0967 | 0.28 |
| 46320 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | DACH2 | Sponge network | -2.915 | 0.00015 | -1.635 | 0.00034 | 0.28 |
| 46321 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | 0.61 | 0.01593 | -1.3 | 0.01619 | 0.28 |
| 46322 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-5p | 27 | BCL2 | Sponge network | 0.176 | 0.37402 | -0.899 | 0 | 0.28 |
| 46323 | RP11-539I5.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | SMAD2 | Sponge network | -0.468 | 0.32587 | -0.128 | 0.12732 | 0.28 |
| 46324 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-625-5p | 14 | CADM1 | Sponge network | -0.278 | 0.76559 | -0.9 | 0.00084 | 0.28 |
| 46325 | TMEM51-AS1 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | PRKAB2 | Sponge network | 0.23 | 0.38667 | -0.367 | 0.01371 | 0.28 |
| 46326 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 41 | EPHA5 | Sponge network | -0.303 | 0.21002 | -0.957 | 0.01733 | 0.28 |
| 46327 | PART1 |
hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 31 | NRK | Sponge network | -2.925 | 0 | 0.656 | 0.19217 | 0.28 |
| 46328 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577 | 20 | DMXL1 | Sponge network | -2.69 | 0.0001 | -0.426 | 0.00056 | 0.28 |
| 46329 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | IGSF10 | Sponge network | 0.311 | 0.10344 | -1.751 | 1.0E-5 | 0.28 |
| 46330 | RP5-994D16.9 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-484;hsa-miR-766-3p | 12 | CHD5 | Sponge network | 0.252 | 0.08508 | -0.922 | 0.01521 | 0.28 |
| 46331 | AC005062.2 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | RB1 | Sponge network | -0.019 | 0.81504 | 0.382 | 0.0017 | 0.28 |
| 46332 | RP11-767N6.7 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-5p | 14 | FOXP1 | Sponge network | 0.466 | 0.00155 | 0.042 | 0.74368 | 0.28 |
| 46333 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-484;hsa-miR-582-5p;hsa-miR-93-3p | 20 | EBF3 | Sponge network | -0.176 | 0.59692 | -0.058 | 0.81437 | 0.28 |
| 46334 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-484 | 13 | BCL11A | Sponge network | 0.594 | 0.41522 | -0.932 | 0.0063 | 0.28 |
| 46335 | BOLA3-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | AKAP11 | Sponge network | -0.246 | 0.35034 | 0.196 | 0.09825 | 0.28 |
| 46336 | LINC00702 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | UBE4B | Sponge network | -0.737 | 0.06182 | -0.235 | 0.007 | 0.28 |
| 46337 | CTB-51J22.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-940 | 14 | CADM2 | Sponge network | -0.803 | 0.23737 | -3.782 | 0 | 0.28 |
| 46338 | RP11-981G7.6 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-590-3p | 16 | DDX3X | Sponge network | 0.986 | 0.01022 | -0.152 | 0.07686 | 0.28 |
| 46339 | EMX2OS |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 18 | INPP4A | Sponge network | -0.777 | 0.1134 | 0.07 | 0.53563 | 0.28 |
| 46340 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p | 24 | YAP1 | Sponge network | 0.494 | 0.00339 | 0.22 | 0.11836 | 0.28 |
| 46341 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-590-3p | 14 | CHD1 | Sponge network | 0.659 | 3.0E-5 | 0.322 | 0.00215 | 0.28 |
| 46342 | PART1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-708-3p | 15 | UNC5D | Sponge network | -2.925 | 0 | -3.646 | 0 | 0.28 |
| 46343 | LINC00476 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p | 10 | WAC | Sponge network | -0.615 | 0.00015 | -0.054 | 0.43688 | 0.28 |
| 46344 | SNX29P2 |
hsa-miR-10b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-92b-3p;hsa-miR-940 | 17 | BAZ2B | Sponge network | 0.4 | 0.14611 | -0.276 | 0.02833 | 0.28 |
| 46345 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-96-5p | 24 | SCN9A | Sponge network | -0.18 | 0.20343 | -0.525 | 0.10593 | 0.28 |
| 46346 | GABPB1-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | NIN | Sponge network | 1.11 | 0 | -0.253 | 0.13794 | 0.28 |
| 46347 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-625-5p | 13 | DCLK1 | Sponge network | -0.18 | 0.20343 | -1.324 | 0.00014 | 0.28 |
| 46348 | RP11-359E10.1 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | HDAC9 | Sponge network | 0.299 | 0.57722 | -0.755 | 0.00495 | 0.279 |
| 46349 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-653-5p | 11 | PDS5B | Sponge network | 0.545 | 0.00064 | 0.259 | 0.00962 | 0.279 |
| 46350 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-425-5p;hsa-miR-542-3p;hsa-miR-590-3p | 13 | SLITRK6 | Sponge network | -0.227 | 0.31657 | -2.121 | 0 | 0.279 |
| 46351 | RP11-305E6.4 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 11 | PICALM | Sponge network | 1.317 | 0 | 0.086 | 0.23523 | 0.279 |
| 46352 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | DPYD | Sponge network | -1.575 | 6.0E-5 | -0.736 | 0.00076 | 0.279 |
| 46353 | RP4-717I23.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-375;hsa-miR-486-5p;hsa-miR-495-3p | 13 | AKAP6 | Sponge network | 0.637 | 0.00069 | -1.586 | 3.0E-5 | 0.279 |
| 46354 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | NF1 | Sponge network | 0.659 | 3.0E-5 | 0.51 | 0 | 0.279 |
| 46355 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 38 | PTPRT | Sponge network | -0.355 | 0.0077 | -0.907 | 0.03727 | 0.279 |
| 46356 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | DAB2 | Sponge network | -2.117 | 0.00161 | 0.431 | 0.0113 | 0.279 |
| 46357 | CTB-133G6.2 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-424-5p | 13 | FAT3 | Sponge network | 0.4 | 0.39299 | -1.601 | 0.00051 | 0.279 |
| 46358 | CTD-2006H14.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZFX | Sponge network | 1.378 | 0 | 0.09 | 0.42563 | 0.279 |
| 46359 | RP11-57H14.4 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p | 11 | LZTS1 | Sponge network | 0.083 | 0.66405 | 1.664 | 0 | 0.279 |
| 46360 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PPM1A | Sponge network | 0.673 | 0 | -0.623 | 0 | 0.279 |
| 46361 | AC010226.4 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-224-5p;hsa-miR-331-3p;hsa-miR-590-3p | 10 | FAM135B | Sponge network | -0.811 | 2.0E-5 | -3.978 | 0 | 0.279 |
| 46362 | RP11-536O18.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-942-5p | 15 | HLF | Sponge network | -0.074 | 0.90758 | -2.443 | 0 | 0.279 |
| 46363 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | SYNE1 | Sponge network | 0.41 | 0.02214 | -0.738 | 0.00316 | 0.279 |
| 46364 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | USP12 | Sponge network | 0.873 | 0 | -0.102 | 0.40768 | 0.279 |
| 46365 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-628-5p | 26 | TCF4 | Sponge network | 1.361 | 0 | 0.016 | 0.92271 | 0.279 |
| 46366 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-378a-5p;hsa-miR-7-1-3p | 10 | NUAK1 | Sponge network | 0.673 | 0 | 0.904 | 0 | 0.279 |
| 46367 | RP11-473I1.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PRRX1 | Sponge network | 0.673 | 0 | 1.316 | 0 | 0.279 |
| 46368 | GNG12-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 41 | PTPRD | Sponge network | -0.793 | 7.0E-5 | -1.005 | 0.00064 | 0.279 |
| 46369 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SEMA3C | Sponge network | -2.117 | 0.00161 | -0.272 | 0.31153 | 0.279 |
| 46370 | RP4-758J18.10 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-5p | 11 | TSHZ3 | Sponge network | -0.091 | 0.71468 | -0.556 | 0.01516 | 0.279 |
| 46371 | BDNF-AS |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | CNTN1 | Sponge network | -0.668 | 3.0E-5 | -2.594 | 0 | 0.279 |
| 46372 | RP11-384O8.1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p | 22 | LIMCH1 | Sponge network | -0.738 | 0.21233 | -0.915 | 1.0E-5 | 0.279 |
| 46373 | LINC00954 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-375;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 10 | SETBP1 | Sponge network | 1.157 | 0.00455 | -1.302 | 1.0E-5 | 0.279 |
| 46374 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-671-5p | 20 | ARID5B | Sponge network | 0.765 | 7.0E-5 | 0.342 | 0.01676 | 0.279 |
| 46375 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | SASH1 | Sponge network | -0.726 | 0.00132 | -0.502 | 0.00175 | 0.279 |
| 46376 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p | 11 | BHLHE41 | Sponge network | -0.214 | 0.44248 | 0.353 | 0.12753 | 0.279 |
| 46377 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DNAJB4 | Sponge network | 0.222 | 0.29133 | -0.7 | 1.0E-5 | 0.279 |
| 46378 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 55 | PTEN | Sponge network | 0.194 | 0.64278 | -0.187 | 0.07748 | 0.279 |
| 46379 | PGM5-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 22 | AFF1 | Sponge network | -4.546 | 0 | -0.384 | 0.00053 | 0.279 |
| 46380 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | APC | Sponge network | -0.323 | 0.01372 | -0.255 | 0.04051 | 0.279 |
| 46381 | AC009948.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | CREB3L2 | Sponge network | 0.532 | 0.00505 | -0.525 | 2.0E-5 | 0.279 |
| 46382 | RP11-758N13.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-375 | 13 | RYR2 | Sponge network | 2.939 | 3.0E-5 | -1.466 | 0.00031 | 0.279 |
| 46383 | RP3-402G11.26 |
hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-505-5p;hsa-miR-590-3p | 10 | MRVI1 | Sponge network | -0.254 | 0.19303 | -1.051 | 0.00022 | 0.279 |
| 46384 | CTD-3138B18.6 |
hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p | 15 | LPP | Sponge network | 2.248 | 0 | -0.459 | 0.04475 | 0.279 |
| 46385 | RP11-35G9.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 26 | PTPRT | Sponge network | 0.657 | 4.0E-5 | -0.907 | 0.03727 | 0.279 |
| 46386 | LINC00702 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 19 | CELF4 | Sponge network | -0.737 | 0.06182 | -1.135 | 0.00344 | 0.279 |
| 46387 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-500a-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | ARHGAP29 | Sponge network | 0.101 | 0.60919 | 0.131 | 0.42953 | 0.279 |
| 46388 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | FSTL5 | Sponge network | -0.141 | 0.26434 | -1.3 | 0.01619 | 0.279 |
| 46389 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p | 20 | ERBB4 | Sponge network | -2.095 | 0.00112 | -1.975 | 2.0E-5 | 0.279 |
| 46390 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-877-5p | 40 | CCND2 | Sponge network | -0.773 | 0.04073 | -0.267 | 0.3112 | 0.279 |
| 46391 | RP11-706C16.7 |
hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-940 | 12 | PBX1 | Sponge network | 0.357 | 0.72407 | -1.206 | 0 | 0.279 |
| 46392 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 21 | ADAMTSL3 | Sponge network | 0.657 | 4.0E-5 | -1.559 | 0 | 0.279 |
| 46393 | CTD-2647L4.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-199a-3p;hsa-miR-199b-5p;hsa-miR-29a-5p;hsa-miR-495-3p | 11 | MAPK1 | Sponge network | 0.555 | 0.00197 | -0.017 | 0.81465 | 0.279 |
| 46394 | HOXB-AS3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-324-5p;hsa-miR-330-5p | 13 | CNTN1 | Sponge network | 0.343 | 0.28887 | -2.594 | 0 | 0.279 |
| 46395 | RP11-401P9.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-542-3p | 12 | ABCB5 | Sponge network | -0.384 | 0.40652 | -0.956 | 0.04077 | 0.279 |
| 46396 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-940 | 13 | CRTC3 | Sponge network | -4.477 | 0 | 0.164 | 0.16786 | 0.279 |
| 46397 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p | 45 | RUNX1T1 | Sponge network | 0.349 | 0.01695 | -0.344 | 0.2374 | 0.279 |
| 46398 | MEG3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 19 | UNC5C | Sponge network | 0.249 | 0.35902 | -0.178 | 0.40214 | 0.279 |
| 46399 | RP1-43E13.2 |
hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-378a-5p | 10 | DTNA | Sponge network | -0.034 | 0.83228 | -1.648 | 1.0E-5 | 0.279 |
| 46400 | DOCK9-AS2 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-629-3p | 13 | PTPN14 | Sponge network | -0.231 | 0.28214 | 0.144 | 0.34595 | 0.279 |
| 46401 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | MEF2C | Sponge network | 0.419 | 0.0319 | -0.499 | 0.01448 | 0.279 |
| 46402 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | KIF1B | Sponge network | 0.272 | 0.44692 | -0.118 | 0.32258 | 0.279 |
| 46403 | LINC00963 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-96-5p | 48 | KIF1B | Sponge network | 0.523 | 9.0E-5 | -0.118 | 0.32258 | 0.279 |
| 46404 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-942-5p | 16 | ARHGAP28 | Sponge network | -0.318 | 0.65847 | -0.911 | 0.00013 | 0.279 |
| 46405 | RP11-244H3.1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-3p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | WWTR1 | Sponge network | 0.659 | 3.0E-5 | -0.345 | 0.11997 | 0.279 |
| 46406 | AC156455.1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p | 11 | DOCK4 | Sponge network | 1.154 | 0.00041 | 0.502 | 0.00108 | 0.279 |
| 46407 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | TACC1 | Sponge network | -0.727 | 0.04726 | -0.362 | 0.05406 | 0.279 |
| 46408 | RP11-822E23.8 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-92a-1-5p | 12 | LUZP2 | Sponge network | -0.777 | 0.16667 | -0.056 | 0.89416 | 0.279 |
| 46409 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-532-3p | 18 | PBX1 | Sponge network | -0.418 | 0.00068 | -1.206 | 0 | 0.279 |
| 46410 | RP5-1198O20.4 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | DIRAS1 | Sponge network | 0.997 | 0.00066 | -2.518 | 0 | 0.279 |
| 46411 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-590-3p | 10 | PCDH8 | Sponge network | -0.053 | 0.80685 | -0.997 | 0.05366 | 0.279 |
| 46412 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 32 | FOXP1 | Sponge network | -0.611 | 0.09607 | 0.042 | 0.74368 | 0.279 |
| 46413 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-212-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-7-1-3p | 13 | FGF5 | Sponge network | 0.389 | 0.04293 | 0.549 | 0.28659 | 0.279 |
| 46414 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 15 | FYN | Sponge network | 1.302 | 7.0E-5 | -0.131 | 0.37051 | 0.279 |
| 46415 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-181a-2-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 22 | USP12 | Sponge network | -1.057 | 0.00029 | -0.102 | 0.40768 | 0.279 |
| 46416 | USP3-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p | 12 | MNT | Sponge network | -0.162 | 0.47687 | -0.157 | 0.06598 | 0.279 |
| 46417 | NUTM2A-AS1 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | DICER1 | Sponge network | 0.643 | 0 | 0.042 | 0.674 | 0.279 |
| 46418 | LINC00403 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-30d-3p;hsa-miR-33a-5p | 10 | TRPS1 | Sponge network | -3.586 | 0.00032 | -0.191 | 0.44109 | 0.279 |
| 46419 | NEAT1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-590-3p | 20 | FOXO3 | Sponge network | 0.705 | 0.00014 | -0.04 | 0.72028 | 0.279 |
| 46420 | PSMG3-AS1 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-484;hsa-miR-7-1-3p | 13 | SFRP1 | Sponge network | -0.125 | 0.49414 | -3.566 | 0 | 0.279 |
| 46421 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 26 | FOXO3 | Sponge network | 0.603 | 0.00127 | -0.04 | 0.72028 | 0.279 |
| 46422 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-96-5p | 18 | MTSS1 | Sponge network | -1.654 | 0.00144 | -0.81 | 0.00035 | 0.279 |
| 46423 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 22 | KAT6B | Sponge network | -1.249 | 7.0E-5 | -0.478 | 0.00018 | 0.279 |
| 46424 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 26 | HECA | Sponge network | 0.311 | 0.10344 | -0.217 | 0.03463 | 0.279 |
| 46425 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 22 | ZFP36L1 | Sponge network | 0.997 | 0.00066 | -0.505 | 8.0E-5 | 0.279 |
| 46426 | AC141928.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-96-5p | 14 | LATS2 | Sponge network | 0.853 | 0.15352 | 0.159 | 0.2304 | 0.279 |
| 46427 | RP11-539I5.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 28 | TMEM132B | Sponge network | -0.468 | 0.32587 | -0.83 | 0.01084 | 0.279 |
| 46428 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | NIN | Sponge network | -1.057 | 0.00029 | -0.253 | 0.13794 | 0.279 |
| 46429 | RP5-1085F17.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p | 15 | CEP170 | Sponge network | 0.541 | 0.00125 | 0.943 | 0 | 0.279 |
| 46430 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 17 | RB1CC1 | Sponge network | 0.082 | 0.55654 | 0.175 | 0.12559 | 0.279 |
| 46431 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 15 | GPC6 | Sponge network | 1.757 | 0 | -0.054 | 0.81465 | 0.279 |
| 46432 | MIR22HG |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 10 | PHOX2B | Sponge network | -1.047 | 0 | -2.68 | 0 | 0.279 |
| 46433 | RP11-401P9.4 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-7-5p;hsa-miR-93-3p | 23 | ERC1 | Sponge network | -0.384 | 0.40652 | -0.108 | 0.38794 | 0.279 |
| 46434 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p | 19 | PLSCR4 | Sponge network | 0.95 | 0.15943 | -0.474 | 0.03833 | 0.279 |
| 46435 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-93-5p | 30 | PIK3R1 | Sponge network | 0.056 | 0.81195 | -0.133 | 0.36854 | 0.279 |
| 46436 | MYLK-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-424-5p;hsa-miR-484 | 21 | CBL | Sponge network | -1.331 | 3.0E-5 | 0.291 | 0.01267 | 0.279 |
| 46437 | DHRS4-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 12 | TBL1XR1 | Sponge network | 0.198 | 0.20379 | 0.578 | 0 | 0.279 |
| 46438 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 19 | THBS1 | Sponge network | 0.862 | 0.06213 | 0.741 | 0.00386 | 0.279 |
| 46439 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p | 24 | FBXO32 | Sponge network | 0.659 | 3.0E-5 | -0.495 | 0.07561 | 0.279 |
| 46440 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p | 14 | TRPS1 | Sponge network | -1.054 | 0.0706 | -0.191 | 0.44109 | 0.279 |
| 46441 | LINC00863 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | DIP2C | Sponge network | 0.189 | 0.13981 | -0.436 | 0.01713 | 0.279 |
| 46442 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p | 16 | RET | Sponge network | 0.083 | 0.66405 | -0.833 | 0.03517 | 0.279 |
| 46443 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-96-5p | 24 | HIP1 | Sponge network | -0.942 | 0 | 0.774 | 0 | 0.279 |
| 46444 | AC003090.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p | 20 | CRTC1 | Sponge network | -2.117 | 0.00161 | -0.116 | 0.28412 | 0.279 |
| 46445 | BDNF-AS |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 20 | WWTR1 | Sponge network | -0.668 | 3.0E-5 | -0.345 | 0.11997 | 0.279 |
| 46446 | U91328.19 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p | 26 | RIMS2 | Sponge network | -0.303 | 0.21002 | -1.365 | 0.00208 | 0.279 |
| 46447 | FAM66C |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | ACSL6 | Sponge network | -0.901 | 0.00019 | -1.593 | 0 | 0.279 |
| 46448 | LINC01018 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 12 | UVRAG | Sponge network | -2.915 | 0.00015 | -0.017 | 0.83865 | 0.279 |
| 46449 | CTD-2336O2.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CDH19 | Sponge network | -0.141 | 0.26434 | -3.434 | 0 | 0.279 |
| 46450 | RP11-53O19.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | LUZP2 | Sponge network | -0.405 | 0.22013 | -0.056 | 0.89416 | 0.279 |
| 46451 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | MYBL1 | Sponge network | 1.089 | 0 | 0.494 | 0.02152 | 0.279 |
| 46452 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-188-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | CDHR3 | Sponge network | 0.311 | 0.10344 | -0.977 | 4.0E-5 | 0.279 |
| 46453 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-93-3p | 21 | QKI | Sponge network | 1.46 | 1.0E-5 | -0.333 | 0.04111 | 0.279 |
| 46454 | LINC00865 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-942-5p | 32 | NOTCH2 | Sponge network | -1.249 | 7.0E-5 | 0.138 | 0.35163 | 0.279 |
| 46455 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-577 | 34 | RUNX1T1 | Sponge network | -0.418 | 0.00068 | -0.344 | 0.2374 | 0.279 |
| 46456 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-3p | 15 | KAT6B | Sponge network | -0.693 | 0.05629 | -0.478 | 0.00018 | 0.279 |
| 46457 | RP11-359E10.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 19 | BRD3 | Sponge network | 0.299 | 0.57722 | 0.37 | 0.00013 | 0.279 |
| 46458 | RP11-25K21.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-589-3p | 11 | TACC1 | Sponge network | 0.489 | 0.25786 | -0.362 | 0.05406 | 0.279 |
| 46459 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 21 | JAZF1 | Sponge network | 0.051 | 0.83388 | -0.377 | 0.02677 | 0.279 |
| 46460 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-484;hsa-miR-576-5p | 11 | EDA2R | Sponge network | 0.395 | 0.15451 | -0.587 | 0.01598 | 0.279 |
| 46461 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 29 | MYBL1 | Sponge network | 0.421 | 0.00084 | 0.494 | 0.02152 | 0.279 |
| 46462 | RP11-359B12.2 |
hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-582-5p | 10 | PRICKLE1 | Sponge network | 0.064 | 0.72934 | -0.112 | 0.67088 | 0.279 |
| 46463 | RP11-597D13.9 |
hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | FNBP1 | Sponge network | 1.302 | 7.0E-5 | -0.969 | 4.0E-5 | 0.279 |
| 46464 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p | 10 | PBX1 | Sponge network | 0.062 | 0.55274 | -1.206 | 0 | 0.279 |
| 46465 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 12 | HOOK3 | Sponge network | -0.08 | 0.90092 | 0.273 | 0.09957 | 0.279 |
| 46466 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 13 | DDX6 | Sponge network | 1.266 | 0 | 0.091 | 0.36428 | 0.279 |
| 46467 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 13 | LHX6 | Sponge network | -0.206 | 0.12942 | -0.087 | 0.69193 | 0.279 |
| 46468 | A2M-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PRKCB | Sponge network | -0.08 | 0.90092 | -1.313 | 2.0E-5 | 0.279 |
| 46469 | AC016747.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-744-3p | 11 | IGF2 | Sponge network | 0.199 | 0.38015 | 0.848 | 0.03842 | 0.279 |
| 46470 | AC012309.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-455-3p | 13 | RUNX1T1 | Sponge network | 0.183 | 0.66792 | -0.344 | 0.2374 | 0.279 |
| 46471 | TTC28-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 16 | SLC5A3 | Sponge network | 0.373 | 0.00068 | 0.493 | 5.0E-5 | 0.279 |
| 46472 | GUSBP11 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-424-5p | 11 | ZFHX3 | Sponge network | 0.856 | 0 | 0.389 | 0.01363 | 0.279 |
| 46473 | RP4-613B23.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-539-5p | 22 | AFF4 | Sponge network | -0.309 | 0.1145 | -0.266 | 0.01565 | 0.279 |
| 46474 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p | 19 | DDX6 | Sponge network | 1.093 | 0 | 0.091 | 0.36428 | 0.279 |
| 46475 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-5p | 15 | PHC3 | Sponge network | 0.983 | 0.00048 | 0.212 | 0.0499 | 0.279 |
| 46476 | HOXA-AS2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-590-3p | 10 | PTPN13 | Sponge network | 0.61 | 0.01593 | -1.208 | 0 | 0.279 |
| 46477 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 13 | CNTNAP3 | Sponge network | 0.083 | 0.66405 | -1.465 | 0.00033 | 0.279 |
| 46478 | RP11-819C21.1 |
hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | UBE2D1 | Sponge network | 0.537 | 0.00139 | 0.468 | 0 | 0.279 |
| 46479 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-582-5p | 24 | DDX6 | Sponge network | 0.056 | 0.81195 | 0.091 | 0.36428 | 0.279 |
| 46480 | RP11-517B11.7 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-432-5p;hsa-miR-495-3p;hsa-miR-758-3p | 22 | VPS53 | Sponge network | 0.706 | 0 | 0.103 | 0.22667 | 0.279 |
| 46481 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 15 | WDFY3 | Sponge network | -2.039 | 0.03447 | -0.201 | 0.0967 | 0.279 |
| 46482 | DNM1P35 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-455-3p | 15 | ABL1 | Sponge network | 0.051 | 0.83388 | -0.215 | 0.07804 | 0.279 |
| 46483 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-744-3p | 45 | DST | Sponge network | 0.782 | 0.00016 | -0.713 | 0.00096 | 0.279 |
| 46484 | RP11-597D13.9 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | TPO | Sponge network | 1.302 | 7.0E-5 | -1.87 | 2.0E-5 | 0.279 |
| 46485 | AC034220.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-590-3p | 11 | FBXO11 | Sponge network | 0.309 | 0.07304 | 0.142 | 0.04938 | 0.279 |
| 46486 | RP11-156P1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-5p | 18 | LRRC55 | Sponge network | 0.214 | 0.18246 | -0.026 | 0.93163 | 0.279 |
| 46487 | MEG3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p | 12 | ZNF844 | Sponge network | 0.249 | 0.35902 | -0.966 | 0.00035 | 0.279 |
| 46488 | HOXB-AS3 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-629-3p | 10 | RYR2 | Sponge network | 0.343 | 0.28887 | -1.466 | 0.00031 | 0.279 |
| 46489 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SON | Sponge network | 0.67 | 0.00048 | 0.168 | 0.04406 | 0.279 |
| 46490 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 25 | MYBL1 | Sponge network | -2.46 | 0 | 0.494 | 0.02152 | 0.279 |
| 46491 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378c;hsa-miR-940 | 21 | WDFY3 | Sponge network | 0.946 | 0 | -0.201 | 0.0967 | 0.279 |
| 46492 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-500a-3p;hsa-miR-942-5p | 14 | HLF | Sponge network | 0.598 | 0.38481 | -2.443 | 0 | 0.279 |
| 46493 | PCA3 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | OPCML | Sponge network | -0.709 | 0.18168 | -2.498 | 0 | 0.279 |
| 46494 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-501-5p;hsa-miR-654-3p | 15 | PSIP1 | Sponge network | -0.405 | 0.22013 | -0.005 | 0.96897 | 0.279 |
| 46495 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378c;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p | 38 | LPP | Sponge network | 0.919 | 0 | -0.459 | 0.04475 | 0.279 |
| 46496 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-192-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | SASH1 | Sponge network | 0.34 | 0.00273 | -0.502 | 0.00175 | 0.279 |
| 46497 | GABPB1-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p;hsa-miR-664a-5p | 15 | CBFB | Sponge network | 1.11 | 0 | 1.061 | 0 | 0.279 |
| 46498 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-590-3p | 11 | RCAN2 | Sponge network | 1.254 | 0 | -0.33 | 0.15172 | 0.279 |
| 46499 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-93-5p | 19 | LRP1B | Sponge network | 0.082 | 0.55397 | -2.163 | 0 | 0.279 |
| 46500 | RP4-798A10.7 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p | 11 | TMTC3 | Sponge network | 1.757 | 0 | 0.123 | 0.22189 | 0.279 |
| 46501 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p | 15 | SPG20 | Sponge network | -0.176 | 0.59692 | -1.16 | 0 | 0.279 |
| 46502 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | HECA | Sponge network | 0.919 | 0 | -0.217 | 0.03463 | 0.279 |
| 46503 | U91328.21 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-629-3p | 10 | RASSF8 | Sponge network | -0.735 | 0.15983 | -0.122 | 0.65591 | 0.279 |
| 46504 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | NEDD4 | Sponge network | -0.513 | 0.04703 | 0.02 | 0.89834 | 0.279 |
| 46505 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-874-3p | 18 | NR2C2 | Sponge network | 0.4 | 0.14611 | 0.21 | 0.03224 | 0.279 |
| 46506 | RP11-166D19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 27 | CSMD1 | Sponge network | -0.576 | 0.10121 | -0.63 | 0.18881 | 0.279 |
| 46507 | RP11-822E23.8 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-484;hsa-miR-744-3p | 15 | IGF2 | Sponge network | -0.777 | 0.16667 | 0.848 | 0.03842 | 0.279 |
| 46508 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | PTPRB | Sponge network | -0.355 | 0.0077 | 0.105 | 0.47122 | 0.279 |
| 46509 | RP11-686D22.8 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-590-5p | 13 | CBL | Sponge network | 0.898 | 1.0E-5 | 0.291 | 0.01267 | 0.279 |
| 46510 | RP11-890B15.3 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | GJA1 | Sponge network | 0.101 | 0.60919 | -0.485 | 0.02179 | 0.279 |
| 46511 | CD27-AS1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577 | 13 | MACF1 | Sponge network | 0.177 | 0.16681 | 0.019 | 0.90335 | 0.279 |
| 46512 | CTD-3092A11.2 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-9-5p | 10 | CDK6 | Sponge network | 0.873 | 0 | 0.991 | 0 | 0.279 |
| 46513 | LINC00969 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | LATS1 | Sponge network | 0.683 | 1.0E-5 | 0.109 | 0.34208 | 0.279 |
| 46514 | LINC00621 |
hsa-let-7g-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | EPHA3 | Sponge network | 0.131 | 0.8436 | 0.185 | 0.53588 | 0.279 |
| 46515 | H1FX-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p | 21 | PTPN14 | Sponge network | -0.206 | 0.12942 | 0.144 | 0.34595 | 0.279 |
| 46516 | RP11-244O19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 31 | CREB1 | Sponge network | -0.096 | 0.67958 | 0.203 | 0.00537 | 0.279 |
| 46517 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p | 17 | FREM2 | Sponge network | 0.253 | 0.09565 | -0.437 | 0.46353 | 0.279 |
| 46518 | RP11-890B15.3 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CDH11 | Sponge network | 0.101 | 0.60919 | 1.427 | 0 | 0.279 |
| 46519 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 54 | KIF1B | Sponge network | 0.082 | 0.55654 | -0.118 | 0.32258 | 0.279 |
| 46520 | DNM1P35 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | LRIG1 | Sponge network | 0.051 | 0.83388 | -0.537 | 0.00588 | 0.279 |
| 46521 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-671-5p | 20 | PPP2R5C | Sponge network | -0.162 | 0.47687 | -0.314 | 3.0E-5 | 0.279 |
| 46522 | RP11-798M19.6 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-337-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | KDSR | Sponge network | -0.612 | 0.00094 | 0.239 | 0.02216 | 0.279 |
| 46523 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p | 18 | TBX18 | Sponge network | 0.494 | 0.00339 | 0.843 | 0.02945 | 0.279 |
| 46524 | BCYRN1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 24 | NRK | Sponge network | 0.311 | 0.10344 | 0.656 | 0.19217 | 0.279 |
| 46525 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-32-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PRKAB2 | Sponge network | 1.601 | 0 | -0.367 | 0.01371 | 0.279 |
| 46526 | TP73-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | SIK1 | Sponge network | -0.563 | 0.02545 | -0.98 | 0 | 0.279 |
| 46527 | RP11-806O11.1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | PHC3 | Sponge network | 0.284 | 0.52463 | 0.212 | 0.0499 | 0.279 |
| 46528 | AC016747.3 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-877-5p | 14 | CDH19 | Sponge network | 0.199 | 0.38015 | -3.434 | 0 | 0.279 |
| 46529 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-93-5p | 27 | RNASEL | Sponge network | 0.997 | 0.00066 | -0.272 | 0.01796 | 0.279 |
| 46530 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 13 | PRKAA1 | Sponge network | 1.178 | 0 | 0.317 | 0.0031 | 0.279 |
| 46531 | LENG8-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-539-5p;hsa-miR-629-3p | 11 | DDX6 | Sponge network | 1.471 | 0 | 0.091 | 0.36428 | 0.279 |
| 46532 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | GRIA3 | Sponge network | -0.021 | 0.93598 | -1.956 | 0 | 0.279 |
| 46533 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29b-3p | 11 | HLF | Sponge network | -0.214 | 0.44248 | -2.443 | 0 | 0.279 |
| 46534 | RP11-13K12.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 23 | CBL | Sponge network | -0.318 | 0.65847 | 0.291 | 0.01267 | 0.279 |
| 46535 | LINC00622 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 12 | GPAM | Sponge network | 1.169 | 0.00028 | 0.142 | 0.29732 | 0.279 |
| 46536 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 21 | APC | Sponge network | -0.031 | 0.89063 | -0.255 | 0.04051 | 0.279 |
| 46537 | RP11-428J1.5 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p | 10 | ABL1 | Sponge network | 0.538 | 0.00076 | -0.215 | 0.07804 | 0.279 |
| 46538 | RP11-728F11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-582-3p | 21 | GAS7 | Sponge network | 0.238 | 0.70544 | -0.555 | 0.01602 | 0.279 |
| 46539 | CTC-301O7.4 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-30b-3p | 10 | AR | Sponge network | 0.292 | 0.43384 | -1.554 | 0 | 0.279 |
| 46540 | LINC00662 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PPP1R9A | Sponge network | 0.213 | 0.32297 | -0.903 | 0.04254 | 0.279 |
| 46541 | AC005618.6 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b | 10 | SYNM | Sponge network | 0.862 | 0.06213 | -2.309 | 1.0E-5 | 0.279 |
| 46542 | RP13-516M14.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-577 | 14 | SYNPO2 | Sponge network | 0.389 | 0.04293 | -2.342 | 0 | 0.279 |
| 46543 | PSMD5-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-766-3p | 43 | MDM4 | Sponge network | 0.569 | 0.00067 | 0.345 | 0.00762 | 0.279 |
| 46544 | CTD-2017D11.1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-374a-3p;hsa-miR-532-3p | 10 | TRIM13 | Sponge network | 0.792 | 0.0049 | 0.183 | 0.06968 | 0.279 |
| 46545 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-96-5p | 26 | FBXO32 | Sponge network | 0.619 | 0.00157 | -0.495 | 0.07561 | 0.279 |
| 46546 | SNX29P2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-7-1-3p | 21 | EYA4 | Sponge network | 0.4 | 0.14611 | 0.3 | 0.36876 | 0.279 |
| 46547 | AC084219.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p | 16 | BRD3 | Sponge network | 1.296 | 0.0054 | 0.37 | 0.00013 | 0.279 |
| 46548 | FGD5-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | SIRT1 | Sponge network | 0.401 | 0.00066 | 0.109 | 0.2593 | 0.279 |
| 46549 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-210-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-590-3p | 13 | APC | Sponge network | 1.2 | 0.01813 | -0.255 | 0.04051 | 0.279 |
| 46550 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 17 | EPHA3 | Sponge network | 1.04 | 0.00023 | 0.185 | 0.53588 | 0.279 |
| 46551 | SNHG14 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | GABRB2 | Sponge network | -1.111 | 0.0003 | -1.841 | 0.00029 | 0.279 |
| 46552 | CTD-2314G24.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p;hsa-miR-93-5p | 56 | DST | Sponge network | 0.246 | 0.59912 | -0.713 | 0.00096 | 0.279 |
| 46553 | RP11-439L18.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p | 12 | AFF4 | Sponge network | 0.224 | 0.48719 | -0.266 | 0.01565 | 0.279 |
| 46554 | ERVK3-1 |
hsa-miR-139-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | GSK3B | Sponge network | 0.58 | 0 | 0.266 | 0.00045 | 0.279 |
| 46555 | CTD-2139B15.2 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-1301-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-337-3p | 13 | RB1 | Sponge network | 0.286 | 0.00612 | 0.382 | 0.0017 | 0.279 |
| 46556 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | SOX5 | Sponge network | 1.302 | 7.0E-5 | -1.091 | 4.0E-5 | 0.279 |
| 46557 | RP4-758J18.10 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-330-5p | 10 | CTIF | Sponge network | -0.091 | 0.71468 | -0.389 | 0.00549 | 0.279 |
| 46558 | SNX29P2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | LRIG1 | Sponge network | 0.4 | 0.14611 | -0.537 | 0.00588 | 0.279 |
| 46559 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-577 | 21 | DMXL1 | Sponge network | -0.465 | 0.04703 | -0.426 | 0.00056 | 0.279 |
| 46560 | HOXA-AS2 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | SFRP1 | Sponge network | 0.61 | 0.01593 | -3.566 | 0 | 0.279 |
| 46561 | RP11-927P21.1 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-629-3p | 16 | RASSF8 | Sponge network | 0.921 | 0.00118 | -0.122 | 0.65591 | 0.279 |
| 46562 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 24 | RARB | Sponge network | -0.969 | 2.0E-5 | -0.825 | 2.0E-5 | 0.279 |
| 46563 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 15 | CREBBP | Sponge network | 1.757 | 0 | 0.126 | 0.15186 | 0.279 |
| 46564 | OIP5-AS1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RBM6 | Sponge network | 0.421 | 0.00084 | -0.137 | 0.15663 | 0.279 |
| 46565 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-625-5p;hsa-miR-93-5p | 12 | LHX6 | Sponge network | 0.381 | 0.25967 | -0.087 | 0.69193 | 0.279 |
| 46566 | GUSBP11 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 12 | MYBL1 | Sponge network | 0.856 | 0 | 0.494 | 0.02152 | 0.279 |
| 46567 | EMX2OS |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 33 | CADM1 | Sponge network | -0.777 | 0.1134 | -0.9 | 0.00084 | 0.279 |
| 46568 | RP11-166D19.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | BRD3 | Sponge network | -0.576 | 0.10121 | 0.37 | 0.00013 | 0.279 |
| 46569 | AC034220.3 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MLLT10 | Sponge network | 0.309 | 0.07304 | 0.105 | 0.33292 | 0.279 |
| 46570 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 32 | CLASP2 | Sponge network | 0.572 | 0.01722 | -0.038 | 0.71 | 0.279 |
| 46571 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 26 | NCOA1 | Sponge network | 0.056 | 0.81195 | -0.432 | 0 | 0.279 |
| 46572 | OR2A1-AS1 |
hsa-let-7g-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-625-3p | 13 | ABL2 | Sponge network | 0.542 | 0.00241 | 0.82 | 0 | 0.279 |
| 46573 | RP1-122P22.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-629-5p | 15 | TCF4 | Sponge network | 0.065 | 0.73468 | 0.016 | 0.92271 | 0.279 |
| 46574 | AC090587.4 |
hsa-miR-103a-2-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-2355-5p;hsa-miR-424-5p;hsa-miR-532-3p | 10 | AFF4 | Sponge network | -0.125 | 0.60166 | -0.266 | 0.01565 | 0.279 |
| 46575 | RP11-536O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p | 10 | JAZF1 | Sponge network | -0.074 | 0.90758 | -0.377 | 0.02677 | 0.279 |
| 46576 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 32 | FOXP1 | Sponge network | -0.176 | 0.59692 | 0.042 | 0.74368 | 0.279 |
| 46577 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 21 | RECK | Sponge network | -0.303 | 0.21002 | -1.043 | 0 | 0.279 |
| 46578 | AF131215.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-454-3p | 11 | ROBO1 | Sponge network | -0.176 | 0.59692 | -0.009 | 0.96154 | 0.279 |
| 46579 | LPP-AS2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-361-3p;hsa-miR-450b-5p | 11 | SPTBN4 | Sponge network | -0.18 | 0.20343 | -0.786 | 0.01281 | 0.279 |
| 46580 | RP11-264I13.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-576-5p | 11 | RICTOR | Sponge network | 0.421 | 0.01652 | 0.388 | 0.00188 | 0.279 |
| 46581 | AGAP11 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-3p | 11 | ANK3 | Sponge network | -1.645 | 0 | -0.55 | 0.00086 | 0.279 |
| 46582 | OIP5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p | 13 | CNTNAP3 | Sponge network | 0.421 | 0.00084 | -1.465 | 0.00033 | 0.279 |
| 46583 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p | 44 | CUX1 | Sponge network | 0.101 | 0.60919 | -0.039 | 0.6705 | 0.279 |
| 46584 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SMARCA2 | Sponge network | 0.997 | 0.00066 | -0.529 | 0.00014 | 0.279 |
| 46585 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | BMPR2 | Sponge network | -0.021 | 0.93598 | 0.206 | 0.0635 | 0.279 |
| 46586 | RP11-290F24.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-326 | 10 | NF1 | Sponge network | 2.052 | 0 | 0.51 | 0 | 0.279 |
| 46587 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | ZNF770 | Sponge network | 0.8 | 0 | 0.206 | 0.06751 | 0.279 |
| 46588 | TRAF3IP2-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-92a-1-5p | 12 | PRKACA | Sponge network | -0.323 | 0.01372 | -0.255 | 0.0012 | 0.279 |
| 46589 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-942-5p | 30 | NOTCH2 | Sponge network | -0.08 | 0.90092 | 0.138 | 0.35163 | 0.279 |
| 46590 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 38 | BNC2 | Sponge network | 0.177 | 0.16681 | -0.491 | 0.09988 | 0.279 |
| 46591 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-23a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p | 10 | GPC6 | Sponge network | -0.557 | 0.11135 | -0.054 | 0.81465 | 0.279 |
| 46592 | RP11-5C23.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 36 | NFIB | Sponge network | 0.274 | 0.07681 | 0.023 | 0.90795 | 0.279 |
| 46593 | RP5-1085F17.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | NRXN3 | Sponge network | 0.541 | 0.00125 | -0.893 | 0.04186 | 0.279 |
| 46594 | RP1-151F17.2 |
hsa-miR-1301-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-589-5p;hsa-miR-590-3p | 13 | ERRFI1 | Sponge network | 0.222 | 0.29133 | -0.798 | 1.0E-5 | 0.279 |
| 46595 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | PBRM1 | Sponge network | 0.267 | 0.2937 | -0.029 | 0.78601 | 0.279 |
| 46596 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-96-5p | 13 | ELMO1 | Sponge network | -2.46 | 0 | 0.04 | 0.83147 | 0.279 |
| 46597 | CTA-204B4.2 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-455-3p | 11 | ABL1 | Sponge network | 0.765 | 7.0E-5 | -0.215 | 0.07804 | 0.279 |
| 46598 | HOTAIRM1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | RECK | Sponge network | -0.053 | 0.80685 | -1.043 | 0 | 0.279 |
| 46599 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 11 | PRKAA1 | Sponge network | 0.986 | 0.01022 | 0.317 | 0.0031 | 0.279 |
| 46600 | RP11-121C2.2 |
hsa-miR-142-5p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-655-3p | 15 | TMEM30A | Sponge network | 1.147 | 0 | 0.096 | 0.31031 | 0.279 |
| 46601 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 44 | AFF4 | Sponge network | -0.896 | 0.00099 | -0.266 | 0.01565 | 0.279 |
| 46602 | HOXA-AS2 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | PCLO | Sponge network | 0.61 | 0.01593 | -1.843 | 0.00064 | 0.279 |
| 46603 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | SLITRK6 | Sponge network | 0.246 | 0.59912 | -2.121 | 0 | 0.279 |
| 46604 | MRVI1-AS1 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p | 14 | NUAK1 | Sponge network | -1.092 | 4.0E-5 | 0.904 | 0 | 0.279 |
| 46605 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 21 | SETBP1 | Sponge network | 0.75 | 0.00054 | -1.302 | 1.0E-5 | 0.279 |
| 46606 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | TACC1 | Sponge network | 0.614 | 1.0E-5 | -0.362 | 0.05406 | 0.279 |
| 46607 | MIR600HG |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 10 | PIK3CA | Sponge network | 0.594 | 0.41522 | 0.195 | 0.06908 | 0.279 |
| 46608 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ITGAV | Sponge network | -0.487 | 0.02157 | 0.337 | 0.01002 | 0.279 |
| 46609 | OSER1-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | BRD3 | Sponge network | -0.13 | 0.48734 | 0.37 | 0.00013 | 0.279 |
| 46610 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-7-1-3p | 26 | PPM1A | Sponge network | -4.546 | 0 | -0.623 | 0 | 0.279 |
| 46611 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | LATS1 | Sponge network | 0.998 | 0 | 0.109 | 0.34208 | 0.279 |
| 46612 | RP11-48B3.4 |
hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 14 | PRKAR1A | Sponge network | 1.757 | 0 | -0.063 | 0.50314 | 0.279 |
| 46613 | LINC00092 |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-589-3p;hsa-miR-590-3p | 28 | CCND2 | Sponge network | -1.886 | 0 | -0.267 | 0.3112 | 0.279 |
| 46614 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | KAT6B | Sponge network | 0.174 | 0.30034 | -0.478 | 0.00018 | 0.279 |
| 46615 | RP11-434B12.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CDH19 | Sponge network | -0.031 | 0.89063 | -3.434 | 0 | 0.279 |
| 46616 | SOX2-OT |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-505-3p;hsa-miR-590-3p | 16 | JAKMIP2 | Sponge network | -1.264 | 6.0E-5 | -1.388 | 0 | 0.279 |
| 46617 | TUG1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 24 | MDM4 | Sponge network | 0.972 | 0 | 0.345 | 0.00762 | 0.279 |
| 46618 | RP11-53O19.1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-330-5p | 11 | DNMT3A | Sponge network | -0.405 | 0.22013 | 0.302 | 0.0262 | 0.279 |
| 46619 | LINC00473 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-744-3p | 21 | HIP1 | Sponge network | -1.203 | 0.03881 | 0.774 | 0 | 0.279 |
| 46620 | PWAR6 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | AFAP1L2 | Sponge network | -1.575 | 6.0E-5 | -0.284 | 0.15434 | 0.279 |
| 46621 | RP11-53O19.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-940 | 20 | SIRT1 | Sponge network | -0.405 | 0.22013 | 0.109 | 0.2593 | 0.279 |
| 46622 | RP11-968O1.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-362-3p | 18 | ZFHX4 | Sponge network | -0.829 | 0.01343 | 0.018 | 0.95574 | 0.279 |
| 46623 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-484;hsa-miR-93-5p | 11 | EDA2R | Sponge network | -0.384 | 0.40652 | -0.587 | 0.01598 | 0.279 |
| 46624 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | NTRK3 | Sponge network | 0.131 | 0.8436 | -1.77 | 3.0E-5 | 0.279 |
| 46625 | USP46-AS1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-199b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-409-5p;hsa-miR-429 | 20 | KIF1B | Sponge network | 0.102 | 0.51464 | -0.118 | 0.32258 | 0.279 |
| 46626 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p | 10 | AFAP1L2 | Sponge network | 0.395 | 0.15451 | -0.284 | 0.15434 | 0.279 |
| 46627 | HOTAIRM1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | LUZP2 | Sponge network | -0.053 | 0.80685 | -0.056 | 0.89416 | 0.279 |
| 46628 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | GRIN2A | Sponge network | 0.389 | 0.04293 | -2.161 | 0 | 0.279 |
| 46629 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | WDFY3 | Sponge network | -0.351 | 0.11358 | -0.201 | 0.0967 | 0.279 |
| 46630 | BMS1P20 |
hsa-miR-16-2-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | ITGB1 | Sponge network | 0.34 | 0.00273 | 0.524 | 0.00017 | 0.279 |
| 46631 | MAGI2-AS3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-503-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-940 | 30 | TOM1L2 | Sponge network | -0.129 | 0.57177 | -0.994 | 0 | 0.279 |
| 46632 | RP11-725P16.2 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | MED12L | Sponge network | 0.422 | 0.27021 | -0.194 | 0.55299 | 0.279 |
| 46633 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 16 | HBP1 | Sponge network | -0.726 | 0.00132 | -0.454 | 0 | 0.279 |
| 46634 | TTC28-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | UBR3 | Sponge network | 0.373 | 0.00068 | -0.021 | 0.82774 | 0.279 |
| 46635 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 14 | SPRY3 | Sponge network | 0.545 | 0.05789 | 0.016 | 0.91286 | 0.279 |
| 46636 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | AFAP1L2 | Sponge network | 0.997 | 0.00066 | -0.284 | 0.15434 | 0.279 |
| 46637 | LINC00476 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 13 | PRDM5 | Sponge network | -0.615 | 0.00015 | -1.09 | 0.00014 | 0.279 |
| 46638 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935 | 23 | ACSL6 | Sponge network | -0.976 | 0.00025 | -1.593 | 0 | 0.278 |
| 46639 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-625-5p | 15 | TGFBR3 | Sponge network | -0.421 | 0.12556 | -1.173 | 1.0E-5 | 0.278 |
| 46640 | AC005562.1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-96-5p | 24 | IL17RD | Sponge network | 0.488 | 0.00084 | -0.019 | 0.94873 | 0.278 |
| 46641 | GS1-124K5.11 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-940 | 17 | SORCS1 | Sponge network | 0.494 | 0.00339 | -3.492 | 0 | 0.278 |
| 46642 | RP11-428J1.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PRUNE2 | Sponge network | 0.538 | 0.00076 | -1.733 | 0.00022 | 0.278 |
| 46643 | FAM66C |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | -0.901 | 0.00019 | 0.126 | 0.15266 | 0.278 |
| 46644 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | PDZRN3 | Sponge network | 0.222 | 0.29133 | -1.548 | 0 | 0.278 |
| 46645 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 12 | AHNAK | Sponge network | 0.082 | 0.55654 | -1.207 | 0 | 0.278 |
| 46646 | JAZF1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-423-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | GNAQ | Sponge network | -0.769 | 0.00035 | -0.367 | 0.00102 | 0.278 |
| 46647 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-577;hsa-miR-93-5p | 22 | CCND2 | Sponge network | -0.473 | 0.07602 | -0.267 | 0.3112 | 0.278 |
| 46648 | FAM13A-AS1 |
hsa-miR-130b-3p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-590-3p | 10 | RASA1 | Sponge network | -0.83 | 0 | -0.002 | 0.98204 | 0.278 |
| 46649 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p | 19 | MAPK1 | Sponge network | 1.205 | 0 | -0.017 | 0.81465 | 0.278 |
| 46650 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484 | 14 | BCL11A | Sponge network | 0.95 | 0.15943 | -0.932 | 0.0063 | 0.278 |
| 46651 | LINC00662 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | MED12L | Sponge network | 0.213 | 0.32297 | -0.194 | 0.55299 | 0.278 |
| 46652 | RP11-890B15.3 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-3614-5p | 10 | CACNA1E | Sponge network | 0.101 | 0.60919 | 1.752 | 0 | 0.278 |
| 46653 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | NR2C2 | Sponge network | -0.513 | 0.04703 | 0.21 | 0.03224 | 0.278 |
| 46654 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-32-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-582-5p | 12 | ARHGAP29 | Sponge network | 0.637 | 0.00069 | 0.131 | 0.42953 | 0.278 |
| 46655 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | GRIN2A | Sponge network | -0.021 | 0.93598 | -2.161 | 0 | 0.278 |
| 46656 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | PRKCE | Sponge network | -0.727 | 0.04726 | -0.403 | 0.00056 | 0.278 |
| 46657 | FZD10-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ZNF844 | Sponge network | -0.727 | 0.04726 | -0.966 | 0.00035 | 0.278 |
| 46658 | CTD-2554C21.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-744-3p | 22 | TOM1L2 | Sponge network | -1.654 | 0.00144 | -0.994 | 0 | 0.278 |
| 46659 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-98-5p | 23 | RSPO2 | Sponge network | -0.405 | 0.22013 | -3.038 | 0 | 0.278 |
| 46660 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p | 10 | LHX6 | Sponge network | 0.178 | 0.29106 | -0.087 | 0.69193 | 0.278 |
| 46661 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-455-3p;hsa-miR-671-5p | 10 | CRTC3 | Sponge network | 0.765 | 7.0E-5 | 0.164 | 0.16786 | 0.278 |
| 46662 | RP11-390K5.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-5p;hsa-miR-374b-5p;hsa-miR-629-3p | 11 | PTPN14 | Sponge network | 1.148 | 2.0E-5 | 0.144 | 0.34595 | 0.278 |
| 46663 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 40 | PPP1R9A | Sponge network | -0.576 | 0.10121 | -0.903 | 0.04254 | 0.278 |
| 46664 | RP11-774O3.3 |
hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | PPP2R2C | Sponge network | -0.487 | 0.02157 | -0.959 | 0.07428 | 0.278 |
| 46665 | BCYRN1 |
hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p | 15 | SYNM | Sponge network | 0.311 | 0.10344 | -2.309 | 1.0E-5 | 0.278 |
| 46666 | AF131215.9 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | PTGFR | Sponge network | -0.322 | 0.25945 | -0.233 | 0.45421 | 0.278 |
| 46667 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 32 | TCF4 | Sponge network | 0.817 | 3.0E-5 | 0.016 | 0.92271 | 0.278 |
| 46668 | LINC00672 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-577;hsa-miR-590-3p | 11 | SEMA3E | Sponge network | -0.227 | 0.31657 | -1.717 | 0.00137 | 0.278 |
| 46669 | RP11-181G12.2 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 10 | CSDE1 | Sponge network | 0.556 | 0.00298 | -0.493 | 0 | 0.278 |
| 46670 | DNAJC27-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-877-5p | 24 | CRTC1 | Sponge network | -0.969 | 2.0E-5 | -0.116 | 0.28412 | 0.278 |
| 46671 | RP11-968O1.5 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-940 | 17 | GRIK3 | Sponge network | -0.829 | 0.01343 | -3.208 | 0 | 0.278 |
| 46672 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-590-5p | 16 | SCN9A | Sponge network | 0.687 | 0.02753 | -0.525 | 0.10593 | 0.278 |
| 46673 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 40 | QKI | Sponge network | -0.176 | 0.59692 | -0.333 | 0.04111 | 0.278 |
| 46674 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | 0.419 | 0.0319 | -1.74 | 0 | 0.278 |
| 46675 | RP11-996F15.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | BRD3 | Sponge network | 0.983 | 0.00048 | 0.37 | 0.00013 | 0.278 |
| 46676 | PRKAG2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-27b-5p;hsa-miR-590-3p | 11 | PPARA | Sponge network | -0.622 | 0.02807 | -0.379 | 0.00548 | 0.278 |
| 46677 | LINC00662 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | HIP1 | Sponge network | 0.213 | 0.32297 | 0.774 | 0 | 0.278 |
| 46678 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-188-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-33a-3p | 10 | LATS1 | Sponge network | 2.163 | 0 | 0.109 | 0.34208 | 0.278 |
| 46679 | PCBP1-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-940 | 19 | EPS15 | Sponge network | -0.567 | 0 | -0.052 | 0.53998 | 0.278 |
| 46680 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | TRIP11 | Sponge network | 0.953 | 0 | -0.158 | 0.06384 | 0.278 |
| 46681 | LINC00173 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-5p | 11 | ABCB5 | Sponge network | 0.056 | 0.8494 | -0.956 | 0.04077 | 0.278 |
| 46682 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-940 | 20 | GAS7 | Sponge network | -1.483 | 9.0E-5 | -0.555 | 0.01602 | 0.278 |
| 46683 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | TBX18 | Sponge network | 0.174 | 0.30034 | 0.843 | 0.02945 | 0.278 |
| 46684 | AC005062.2 |
hsa-let-7e-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | COPS2 | Sponge network | -0.019 | 0.81504 | -0.178 | 0.04974 | 0.278 |
| 46685 | GUSBP11 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p | 14 | BRD3 | Sponge network | 0.856 | 0 | 0.37 | 0.00013 | 0.278 |
| 46686 | RAMP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 21 | RYR2 | Sponge network | -0.513 | 0.04703 | -1.466 | 0.00031 | 0.278 |
| 46687 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 47 | BMPR2 | Sponge network | 0.862 | 0.06213 | 0.206 | 0.0635 | 0.278 |
| 46688 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-326 | 11 | SOX11 | Sponge network | 0.419 | 0.0319 | 2.68 | 0 | 0.278 |
| 46689 | ZNF582-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | OPCML | Sponge network | -1.349 | 0.00017 | -2.498 | 0 | 0.278 |
| 46690 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-93-3p;hsa-miR-96-5p | 22 | DOCK4 | Sponge network | 0.078 | 0.55803 | 0.502 | 0.00108 | 0.278 |
| 46691 | RP11-216B9.6 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | PKNOX1 | Sponge network | 0.174 | 0.30034 | 0.085 | 0.28917 | 0.278 |
| 46692 | RP11-390K5.6 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | EPS15 | Sponge network | 1.148 | 2.0E-5 | -0.052 | 0.53998 | 0.278 |
| 46693 | RP1-122P22.2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p | 23 | DTNA | Sponge network | 0.065 | 0.73468 | -1.648 | 1.0E-5 | 0.278 |
| 46694 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-214-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 41 | PPM1A | Sponge network | 0.349 | 0.01695 | -0.623 | 0 | 0.278 |
| 46695 | DIO3OS |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-629-3p;hsa-miR-940 | 11 | RHOB | Sponge network | -0.773 | 0.04073 | -1.052 | 0 | 0.278 |
| 46696 | C4A-AS1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-629-3p;hsa-miR-744-3p | 15 | IGFBP5 | Sponge network | 3.345 | 0 | -0.806 | 0.00105 | 0.278 |
| 46697 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-98-5p | 30 | CLASP2 | Sponge network | -0.899 | 6.0E-5 | -0.038 | 0.71 | 0.278 |
| 46698 | ZSCAN16-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-5p | 22 | BTG2 | Sponge network | -0.342 | 0.02288 | -1.763 | 0 | 0.278 |
| 46699 | HOXA-AS3 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-96-5p | 11 | WNK2 | Sponge network | -0.555 | 0.06156 | -0.352 | 0.31066 | 0.278 |
| 46700 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p | 48 | PTEN | Sponge network | -0.612 | 0.00094 | -0.187 | 0.07748 | 0.278 |
| 46701 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-532-3p | 10 | SYNM | Sponge network | 0.065 | 0.73468 | -2.309 | 1.0E-5 | 0.278 |
| 46702 | RP11-389C8.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | MED12L | Sponge network | 0.727 | 3.0E-5 | -0.194 | 0.55299 | 0.278 |
| 46703 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | PPP1R9A | Sponge network | 0.246 | 0.59912 | -0.903 | 0.04254 | 0.278 |
| 46704 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | NUAK1 | Sponge network | 0.683 | 1.0E-5 | 0.904 | 0 | 0.278 |
| 46705 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-7-1-3p | 35 | RBMS3 | Sponge network | 0.177 | 0.16681 | -0.519 | 0.03551 | 0.278 |
| 46706 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | MLLT10 | Sponge network | 0.422 | 0.27021 | 0.105 | 0.33292 | 0.278 |
| 46707 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 16 | GNA13 | Sponge network | 0.637 | 0.00069 | 0.007 | 0.93884 | 0.278 |
| 46708 | FAM66C |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | CSMD1 | Sponge network | -0.901 | 0.00019 | -0.63 | 0.18881 | 0.278 |
| 46709 | NUTM2A-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p | 12 | CBFB | Sponge network | 0.643 | 0 | 1.061 | 0 | 0.278 |
| 46710 | HCG18 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p | 10 | MAT2A | Sponge network | 1.063 | 0 | -0.073 | 0.36195 | 0.278 |
| 46711 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | LRRC4 | Sponge network | -0.125 | 0.49414 | -1.774 | 0 | 0.278 |
| 46712 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-582-3p;hsa-miR-589-3p | 27 | ADARB1 | Sponge network | 0.921 | 0.00118 | -0.335 | 0.06631 | 0.278 |
| 46713 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | HOOK3 | Sponge network | -0.322 | 0.25945 | 0.273 | 0.09957 | 0.278 |
| 46714 | RP11-403I13.7 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | CADM2 | Sponge network | 0.395 | 0.15451 | -3.782 | 0 | 0.278 |
| 46715 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 15 | CUL5 | Sponge network | 1.093 | 0 | 0.026 | 0.77301 | 0.278 |
| 46716 | RP11-686D22.8 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 17 | WWTR1 | Sponge network | 0.898 | 1.0E-5 | -0.345 | 0.11997 | 0.278 |
| 46717 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 28 | ST6GALNAC3 | Sponge network | -0.405 | 0.22013 | -0.987 | 0 | 0.278 |
| 46718 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-5p | 11 | LRRC55 | Sponge network | 3.345 | 0 | -0.026 | 0.93163 | 0.278 |
| 46719 | RP11-389C8.2 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | RABEP1 | Sponge network | 0.727 | 3.0E-5 | -0.066 | 0.43142 | 0.278 |
| 46720 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | RASSF8 | Sponge network | -0.351 | 0.11358 | -0.122 | 0.65591 | 0.278 |
| 46721 | CTD-2314G24.2 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 28 | RIMS2 | Sponge network | 0.246 | 0.59912 | -1.365 | 0.00208 | 0.278 |
| 46722 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 21 | SOX11 | Sponge network | 0.401 | 0.00066 | 2.68 | 0 | 0.278 |
| 46723 | AC005618.6 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-5p | 13 | IGF2 | Sponge network | 0.862 | 0.06213 | 0.848 | 0.03842 | 0.278 |
| 46724 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-3614-5p | 19 | RICTOR | Sponge network | 0.592 | 0.00014 | 0.388 | 0.00188 | 0.278 |
| 46725 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | NRXN1 | Sponge network | 0.083 | 0.66405 | -3.778 | 0 | 0.278 |
| 46726 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 14 | ACVR1 | Sponge network | 0.274 | 0.07681 | 0.279 | 0.00234 | 0.278 |
| 46727 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-33a-3p | 11 | AKAP11 | Sponge network | -2.039 | 0.03447 | 0.196 | 0.09825 | 0.278 |
| 46728 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 31 | CBX5 | Sponge network | 1.063 | 0 | 0.165 | 0.24446 | 0.278 |
| 46729 | RP11-254F7.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p | 23 | PPP1R9A | Sponge network | 0.176 | 0.37402 | -0.903 | 0.04254 | 0.278 |
| 46730 | RP11-403I13.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | KCNA6 | Sponge network | 0.619 | 0.00157 | -1.531 | 0.00013 | 0.278 |
| 46731 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-93-5p | 22 | PLSCR4 | Sponge network | -1.047 | 0 | -0.474 | 0.03833 | 0.278 |
| 46732 | SH3RF3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p | 17 | MAF | Sponge network | 0.889 | 9.0E-5 | -1.257 | 0 | 0.278 |
| 46733 | RP11-774O3.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-590-3p | 11 | FBXO11 | Sponge network | -0.487 | 0.02157 | 0.142 | 0.04938 | 0.278 |
| 46734 | AP001055.6 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-7-1-3p | 20 | NFIB | Sponge network | 0.693 | 0.00076 | 0.023 | 0.90795 | 0.278 |
| 46735 | RP11-597D13.9 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 13 | PTPN13 | Sponge network | 1.302 | 7.0E-5 | -1.208 | 0 | 0.278 |
| 46736 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | ADARB1 | Sponge network | 0.272 | 0.44692 | -0.335 | 0.06631 | 0.278 |
| 46737 | LINC00702 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 12 | STC1 | Sponge network | -0.737 | 0.06182 | 1.017 | 0 | 0.278 |
| 46738 | FGD5-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | TET1 | Sponge network | 0.401 | 0.00066 | -0.135 | 0.52138 | 0.278 |
| 46739 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 11 | RNF144A | Sponge network | -1.255 | 0.00981 | 0.594 | 0.0009 | 0.278 |
| 46740 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-935 | 18 | LRRC55 | Sponge network | -0.738 | 0.21233 | -0.026 | 0.93163 | 0.278 |
| 46741 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 12 | FBXO32 | Sponge network | -4.477 | 0 | -0.495 | 0.07561 | 0.278 |
| 46742 | LOXL1-AS1 |
hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 11 | RET | Sponge network | 0.487 | 0.04053 | -0.833 | 0.03517 | 0.278 |
| 46743 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p | 15 | NF1 | Sponge network | 0.953 | 0 | 0.51 | 0 | 0.278 |
| 46744 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p | 11 | RCAN2 | Sponge network | 0.663 | 0.00073 | -0.33 | 0.15172 | 0.278 |
| 46745 | RP11-296I10.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-502-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 22 | ADARB1 | Sponge network | 0.592 | 0.00014 | -0.335 | 0.06631 | 0.278 |
| 46746 | RP11-400K9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-93-3p | 11 | NCOA1 | Sponge network | -0.733 | 0.19469 | -0.432 | 0 | 0.278 |
| 46747 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 30 | LRP1B | Sponge network | 0.064 | 0.72934 | -2.163 | 0 | 0.278 |
| 46748 | MIR143HG |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-542-3p | 12 | CADM4 | Sponge network | -0.739 | 0.0574 | -0.635 | 0.00305 | 0.278 |
| 46749 | RP11-20I23.13 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-92a-3p | 18 | AFF1 | Sponge network | 0.505 | 0.0014 | -0.384 | 0.00053 | 0.278 |
| 46750 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-590-3p | 18 | USP6 | Sponge network | -0.227 | 0.31657 | 0.188 | 0.3871 | 0.278 |
| 46751 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 21 | PRKAA2 | Sponge network | -1.255 | 0.00981 | -2.462 | 0 | 0.278 |
| 46752 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-409-3p;hsa-miR-511-5p | 16 | ATRX | Sponge network | 0.594 | 0.41522 | 0.261 | 0.04408 | 0.278 |
| 46753 | CTC-296K1.4 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | MED12L | Sponge network | -2.076 | 0.00548 | -0.194 | 0.55299 | 0.278 |
| 46754 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p | 15 | CBX5 | Sponge network | 0.813 | 1.0E-5 | 0.165 | 0.24446 | 0.278 |
| 46755 | RP11-46C24.7 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TPO | Sponge network | 0.392 | 0.0278 | -1.87 | 2.0E-5 | 0.278 |
| 46756 | RP11-160O5.1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-3613-5p;hsa-miR-539-5p | 13 | DMD | Sponge network | -0.327 | 0.07353 | -1.506 | 0 | 0.278 |
| 46757 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-93-5p | 21 | TGFBR2 | Sponge network | 0.249 | 0.35902 | -0.243 | 0.11408 | 0.278 |
| 46758 | CTD-3099C6.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-550a-3p;hsa-miR-576-5p | 12 | PRRX1 | Sponge network | 1.361 | 0 | 1.316 | 0 | 0.278 |
| 46759 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-654-3p | 24 | MYBL1 | Sponge network | 0.194 | 0.64278 | 0.494 | 0.02152 | 0.278 |
| 46760 | PCA3 |
hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | SH3GLB1 | Sponge network | -0.709 | 0.18168 | -0.115 | 0.14773 | 0.278 |
| 46761 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 32 | RUNX1T1 | Sponge network | 0.453 | 0.01839 | -0.344 | 0.2374 | 0.278 |
| 46762 | CTD-2303H24.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 26 | EPS15 | Sponge network | 0.415 | 0.14657 | -0.052 | 0.53998 | 0.278 |
| 46763 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | CACNA2D1 | Sponge network | 0.494 | 0.00339 | -0.846 | 0.00675 | 0.278 |
| 46764 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 47 | MDM4 | Sponge network | -0.096 | 0.67958 | 0.345 | 0.00762 | 0.278 |
| 46765 | RP11-277P12.20 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 27 | RUNX1T1 | Sponge network | 0.687 | 0.02753 | -0.344 | 0.2374 | 0.278 |
| 46766 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p | 14 | NIN | Sponge network | -0.176 | 0.59692 | -0.253 | 0.13794 | 0.278 |
| 46767 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 44 | CCND2 | Sponge network | -0.355 | 0.0077 | -0.267 | 0.3112 | 0.278 |
| 46768 | RP11-973H7.3 |
hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-33a-3p | 13 | CREB1 | Sponge network | 0.901 | 0 | 0.203 | 0.00537 | 0.278 |
| 46769 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 35 | MEF2C | Sponge network | 0.385 | 0.10067 | -0.499 | 0.01448 | 0.278 |
| 46770 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p | 22 | MED12L | Sponge network | -0.227 | 0.31657 | -0.194 | 0.55299 | 0.278 |
| 46771 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-942-5p | 21 | DDX6 | Sponge network | 1.04 | 0.00023 | 0.091 | 0.36428 | 0.278 |
| 46772 | RP11-434B12.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SEMA3C | Sponge network | -0.031 | 0.89063 | -0.272 | 0.31153 | 0.278 |
| 46773 | LINC00648 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | EYA4 | Sponge network | 0.633 | 0.51678 | 0.3 | 0.36876 | 0.278 |
| 46774 | RP11-770J1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-584-5p | 16 | SSBP2 | Sponge network | -2.011 | 0.0005 | -1.58 | 0 | 0.278 |
| 46775 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | FAT4 | Sponge network | 0.259 | 0.12542 | -0.354 | 0.14694 | 0.278 |
| 46776 | LINC00473 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | LUZP2 | Sponge network | -1.203 | 0.03881 | -0.056 | 0.89416 | 0.278 |
| 46777 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-532-5p | 28 | SMAD2 | Sponge network | 0.082 | 0.55397 | -0.128 | 0.12732 | 0.278 |
| 46778 | RP11-401P9.1 |
hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-484 | 10 | PPM1L | Sponge network | 1.524 | 0.1098 | -0.537 | 0.00616 | 0.278 |
| 46779 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-744-3p;hsa-miR-96-5p | 18 | HIP1 | Sponge network | 0.4 | 0.14611 | 0.774 | 0 | 0.278 |
| 46780 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | -1.575 | 6.0E-5 | 0.323 | 0.00792 | 0.278 |
| 46781 | RP11-6O2.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-940 | 14 | BAZ2B | Sponge network | 0.331 | 0.57247 | -0.276 | 0.02833 | 0.278 |
| 46782 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-339-3p;hsa-miR-421 | 19 | PRKCE | Sponge network | -2.62 | 0 | -0.403 | 0.00056 | 0.278 |
| 46783 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 53 | KIF1B | Sponge network | 0.782 | 0.00016 | -0.118 | 0.32258 | 0.278 |
| 46784 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 15 | GPAM | Sponge network | 1.093 | 0 | 0.142 | 0.29732 | 0.278 |
| 46785 | ADIRF-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-874-3p | 23 | ASTN1 | Sponge network | -0.223 | 0.47816 | -2.389 | 2.0E-5 | 0.278 |
| 46786 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-454-3p | 12 | MLLT10 | Sponge network | 0.622 | 0.00015 | 0.105 | 0.33292 | 0.278 |
| 46787 | RP11-390K5.6 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | PIK3R1 | Sponge network | 1.148 | 2.0E-5 | -0.133 | 0.36854 | 0.278 |
| 46788 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-589-3p | 11 | ABCA10 | Sponge network | -0.176 | 0.59692 | -0.571 | 0.03674 | 0.278 |
| 46789 | GHRLOS |
hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-200a-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-93-3p | 24 | IL6ST | Sponge network | -1.942 | 0 | -0.402 | 0.00676 | 0.278 |
| 46790 | AC012360.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-331-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | BHLHE41 | Sponge network | 0.624 | 0.02176 | 0.353 | 0.12753 | 0.278 |
| 46791 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-625-3p | 13 | RIMBP2 | Sponge network | 0.174 | 0.30034 | -1.968 | 5.0E-5 | 0.278 |
| 46792 | MALAT1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 20 | PTPN14 | Sponge network | 0.782 | 0.00016 | 0.144 | 0.34595 | 0.278 |
| 46793 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | PICALM | Sponge network | 0.311 | 0.10344 | 0.086 | 0.23523 | 0.278 |
| 46794 | AP001347.6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-3p | 17 | RIMBP2 | Sponge network | -1.161 | 0.00591 | -1.968 | 5.0E-5 | 0.278 |
| 46795 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-224-5p;hsa-miR-335-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 11 | AKAP13 | Sponge network | 0.659 | 3.0E-5 | 0.028 | 0.7729 | 0.278 |
| 46796 | H1FX-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p | 14 | SCN9A | Sponge network | -0.206 | 0.12942 | -0.525 | 0.10593 | 0.278 |
| 46797 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429 | 20 | RNASEL | Sponge network | -2.62 | 0 | -0.272 | 0.01796 | 0.278 |
| 46798 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-93-5p | 12 | TRIM3 | Sponge network | -0.842 | 0.01361 | -0.577 | 0 | 0.278 |
| 46799 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 22 | MTSS1 | Sponge network | -0.176 | 0.59692 | -0.81 | 0.00035 | 0.278 |
| 46800 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 17 | RET | Sponge network | 0.064 | 0.72934 | -0.833 | 0.03517 | 0.278 |
| 46801 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-370-3p;hsa-miR-629-3p | 14 | CHD2 | Sponge network | 0.331 | 0.57247 | 0.049 | 0.55371 | 0.278 |
| 46802 | KB-1460A1.5 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | EPG5 | Sponge network | 0.603 | 0.00127 | 0.101 | 0.3563 | 0.278 |
| 46803 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-590-5p | 15 | TRIP11 | Sponge network | 0.331 | 0.57247 | -0.158 | 0.06384 | 0.278 |
| 46804 | CTD-2245F17.3 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-942-5p | 17 | IGFBP5 | Sponge network | -0.421 | 0.12556 | -0.806 | 0.00105 | 0.278 |
| 46805 | RP11-67L2.2 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-5p | 10 | RANBP6 | Sponge network | 0.72 | 0.0001 | 0.175 | 0.19756 | 0.278 |
| 46806 | U91328.19 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 20 | NCAM2 | Sponge network | -0.303 | 0.21002 | -0.482 | 0.22565 | 0.278 |
| 46807 | ARHGAP5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-625-5p | 18 | CHD5 | Sponge network | -0.644 | 0.00021 | -0.922 | 0.01521 | 0.278 |
| 46808 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 31 | WWTR1 | Sponge network | 0.421 | 0.00084 | -0.345 | 0.11997 | 0.278 |
| 46809 | LINC00654 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 49 | CADM2 | Sponge network | 0.374 | 0.20054 | -3.782 | 0 | 0.278 |
| 46810 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | CLASP2 | Sponge network | -0.303 | 0.21002 | -0.038 | 0.71 | 0.278 |
| 46811 | RP11-6N17.3 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-18a-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-744-3p | 13 | MEF2C | Sponge network | 0.108 | 0.69556 | -0.499 | 0.01448 | 0.278 |
| 46812 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-421 | 12 | AFF3 | Sponge network | -0.602 | 0.00331 | -2.373 | 0 | 0.278 |
| 46813 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-590-3p | 11 | GRIN2A | Sponge network | 0.381 | 0.25967 | -2.161 | 0 | 0.278 |
| 46814 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-655-3p;hsa-miR-7-1-3p | 17 | UBE4B | Sponge network | 0.41 | 0.02214 | -0.235 | 0.007 | 0.278 |
| 46815 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-744-3p | 24 | ADAMTSL3 | Sponge network | -0.371 | 0.24604 | -1.559 | 0 | 0.278 |
| 46816 | RP4-758J18.10 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p | 11 | HIP1 | Sponge network | -0.091 | 0.71468 | 0.774 | 0 | 0.278 |
| 46817 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | CREB3L2 | Sponge network | -0.351 | 0.11358 | -0.525 | 2.0E-5 | 0.278 |
| 46818 | RP11-13K12.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-93-5p | 21 | ARNT2 | Sponge network | -0.154 | 0.78939 | -0.332 | 0.25851 | 0.278 |
| 46819 | THAP9-AS1 |
hsa-let-7c-5p;hsa-miR-100-5p;hsa-miR-195-5p;hsa-miR-23b-3p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 11 | ATP2A2 | Sponge network | 1.079 | 0 | 0.604 | 0 | 0.278 |
| 46820 | NEAT1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-487b-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | KAT6B | Sponge network | 0.705 | 0.00014 | -0.478 | 0.00018 | 0.278 |
| 46821 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p | 12 | TGFBR3 | Sponge network | 1.46 | 1.0E-5 | -1.173 | 1.0E-5 | 0.278 |
| 46822 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-374b-5p | 10 | APC | Sponge network | 1.46 | 1.0E-5 | -0.255 | 0.04051 | 0.278 |
| 46823 | HOXB-AS2 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-629-3p | 12 | PTPN14 | Sponge network | 1.316 | 0.01106 | 0.144 | 0.34595 | 0.278 |
| 46824 | RP11-48B3.4 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-628-5p | 18 | ERC1 | Sponge network | 1.757 | 0 | -0.108 | 0.38794 | 0.278 |
| 46825 | RP11-728F11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-505-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 24 | CADM2 | Sponge network | 0.238 | 0.70544 | -3.782 | 0 | 0.278 |
| 46826 | LPP-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-96-5p | 15 | PIK3CA | Sponge network | -0.18 | 0.20343 | 0.195 | 0.06908 | 0.278 |
| 46827 | LINC00094 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 42 | GRIK3 | Sponge network | 0.253 | 0.09565 | -3.208 | 0 | 0.278 |
| 46828 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | EBF1 | Sponge network | 0.796 | 4.0E-5 | -0.384 | 0.08856 | 0.278 |
| 46829 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 17 | NEDD4 | Sponge network | 0.95 | 0.15943 | 0.02 | 0.89834 | 0.278 |
| 46830 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-501-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 21 | IL6ST | Sponge network | -0.557 | 0.11135 | -0.402 | 0.00676 | 0.278 |
| 46831 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | ARNT2 | Sponge network | -0.295 | 0.02604 | -0.332 | 0.25851 | 0.278 |
| 46832 | RP11-73M18.8 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | UBR3 | Sponge network | 0.832 | 0 | -0.021 | 0.82774 | 0.278 |
| 46833 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | CXXC4 | Sponge network | 0.05 | 0.95483 | -0.433 | 0.21055 | 0.278 |
| 46834 | USP46-AS1 |
hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p | 10 | HIP1 | Sponge network | 0.102 | 0.51464 | 0.774 | 0 | 0.278 |
| 46835 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | PTPRB | Sponge network | 0.05 | 0.95483 | 0.105 | 0.47122 | 0.278 |
| 46836 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-455-3p | 22 | BTG2 | Sponge network | -0.418 | 0.00068 | -1.763 | 0 | 0.278 |
| 46837 | HOTAIRM1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SEMA3E | Sponge network | -0.053 | 0.80685 | -1.717 | 0.00137 | 0.278 |
| 46838 | RP11-452L6.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-18a-5p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-409-3p | 10 | CLASP2 | Sponge network | 0.665 | 0.00061 | -0.038 | 0.71 | 0.278 |
| 46839 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 28 | SASH1 | Sponge network | -0.227 | 0.31657 | -0.502 | 0.00175 | 0.278 |
| 46840 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | MGA | Sponge network | 0.532 | 0.00505 | 0.323 | 0.00792 | 0.278 |
| 46841 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | GALNT13 | Sponge network | -0.899 | 6.0E-5 | -0.857 | 0.07439 | 0.278 |
| 46842 | HAR1A |
hsa-miR-103a-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-93-3p | 20 | IL6ST | Sponge network | -0.672 | 0.19447 | -0.402 | 0.00676 | 0.278 |
| 46843 | RP13-516M14.1 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-7-1-3p | 11 | INPP4A | Sponge network | 0.389 | 0.04293 | 0.07 | 0.53563 | 0.278 |
| 46844 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 12 | KIAA1024 | Sponge network | 0.75 | 0.00054 | 1.083 | 0 | 0.278 |
| 46845 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 14 | FGFR1 | Sponge network | 0.523 | 9.0E-5 | -0.509 | 0.03957 | 0.278 |
| 46846 | USP3-AS1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-421;hsa-miR-940 | 18 | BAZ2B | Sponge network | -0.162 | 0.47687 | -0.276 | 0.02833 | 0.278 |
| 46847 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p | 23 | CLIP1 | Sponge network | -0.615 | 0.00015 | -0.215 | 0.08684 | 0.278 |
| 46848 | RP11-35G9.3 |
hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 16 | GLI3 | Sponge network | 0.657 | 4.0E-5 | -0.615 | 0.01482 | 0.278 |
| 46849 | RP11-894P9.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-582-3p | 16 | ADARB1 | Sponge network | 0.993 | 0 | -0.335 | 0.06631 | 0.278 |
| 46850 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | HACE1 | Sponge network | 0.986 | 0.01022 | -0.072 | 0.55239 | 0.278 |
| 46851 | RP11-758N13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-375 | 21 | ADARB1 | Sponge network | 2.939 | 3.0E-5 | -0.335 | 0.06631 | 0.278 |
| 46852 | RP11-5C23.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | NRXN3 | Sponge network | 0.274 | 0.07681 | -0.893 | 0.04186 | 0.278 |
| 46853 | AC141928.1 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-342-3p;hsa-miR-423-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-625-5p | 11 | CELF4 | Sponge network | 0.853 | 0.15352 | -1.135 | 0.00344 | 0.278 |
| 46854 | RP11-5C23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-590-3p | 20 | JAZF1 | Sponge network | 0.274 | 0.07681 | -0.377 | 0.02677 | 0.278 |
| 46855 | AF131215.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-429 | 17 | CBFA2T3 | Sponge network | -0.176 | 0.59692 | -1.221 | 1.0E-5 | 0.278 |
| 46856 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | GNA13 | Sponge network | -0.323 | 0.01372 | 0.007 | 0.93884 | 0.278 |
| 46857 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-576-5p | 11 | TRPS1 | Sponge network | -2.076 | 0.00548 | -0.191 | 0.44109 | 0.278 |
| 46858 | THAP9-AS1 |
hsa-miR-125a-5p;hsa-miR-150-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-497-5p | 12 | RAD23B | Sponge network | 1.079 | 0 | 0.108 | 0.17414 | 0.278 |
| 46859 | RP11-531A24.5 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 24 | PDE4DIP | Sponge network | 0.75 | 0.00054 | -0.282 | 0.0257 | 0.278 |
| 46860 | CKMT2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 25 | PRRX1 | Sponge network | 0.082 | 0.55654 | 1.316 | 0 | 0.278 |
| 46861 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 31 | RICTOR | Sponge network | 0.862 | 0.06213 | 0.388 | 0.00188 | 0.278 |
| 46862 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-590-3p | 13 | TMTC3 | Sponge network | 0.826 | 1.0E-5 | 0.123 | 0.22189 | 0.278 |
| 46863 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p | 18 | EYA4 | Sponge network | 1.089 | 0 | 0.3 | 0.36876 | 0.278 |
| 46864 | RP11-25K19.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 10 | FMNL3 | Sponge network | -1.057 | 0.00029 | 0.555 | 4.0E-5 | 0.278 |
| 46865 | LINC01082 |
hsa-miR-103a-2-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-29b-1-5p | 14 | LPP | Sponge network | -3.322 | 8.0E-5 | -0.459 | 0.04475 | 0.278 |
| 46866 | AC124789.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 11 | NOTCH2 | Sponge network | 0.073 | 0.79848 | 0.138 | 0.35163 | 0.278 |
| 46867 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | MLLT10 | Sponge network | 0.392 | 0.0278 | 0.105 | 0.33292 | 0.278 |
| 46868 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | THBS1 | Sponge network | 0.421 | 0.00084 | 0.741 | 0.00386 | 0.278 |
| 46869 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | RICTOR | Sponge network | 0.336 | 0.00739 | 0.388 | 0.00188 | 0.278 |
| 46870 | LINC00957 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 16 | DOCK4 | Sponge network | -0.421 | 0.0114 | 0.502 | 0.00108 | 0.278 |
| 46871 | LINC00957 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | ERCC4 | Sponge network | -0.421 | 0.0114 | 0.126 | 0.15266 | 0.278 |
| 46872 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | PRRX1 | Sponge network | 0.385 | 0.10067 | 1.316 | 0 | 0.278 |
| 46873 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | SYNE1 | Sponge network | -0.023 | 0.95109 | -0.738 | 0.00316 | 0.278 |
| 46874 | WDR86-AS1 |
hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-7-5p | 10 | KCNJ5 | Sponge network | 0.348 | 0.32912 | -0.281 | 0.3339 | 0.278 |
| 46875 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577 | 23 | DMXL1 | Sponge network | 0.637 | 0.00069 | -0.426 | 0.00056 | 0.278 |
| 46876 | RP11-531A24.5 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-576-5p | 11 | PTCH1 | Sponge network | 0.75 | 0.00054 | -0.1 | 0.6179 | 0.278 |
| 46877 | TINCR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-92a-1-5p | 16 | AKAP6 | Sponge network | -0.664 | 0.14543 | -1.586 | 3.0E-5 | 0.278 |
| 46878 | RP11-158K1.3 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | KIF5B | Sponge network | 0.349 | 0.01695 | 0.362 | 0.00066 | 0.278 |
| 46879 | LINC00476 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | FAM135B | Sponge network | -0.615 | 0.00015 | -3.978 | 0 | 0.278 |
| 46880 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TBX18 | Sponge network | 0.538 | 0.00076 | 0.843 | 0.02945 | 0.278 |
| 46881 | RP11-130L8.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | INPP4A | Sponge network | -0.663 | 0.10887 | 0.07 | 0.53563 | 0.278 |
| 46882 | RP11-327P2.5 |
hsa-let-7b-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p | 39 | AR | Sponge network | -0.147 | 0.40452 | -1.554 | 0 | 0.278 |
| 46883 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-96-5p | 48 | TCF4 | Sponge network | 0.331 | 0.57247 | 0.016 | 0.92271 | 0.278 |
| 46884 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | RB1CC1 | Sponge network | 0.219 | 0.1516 | 0.175 | 0.12559 | 0.278 |
| 46885 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | KANK1 | Sponge network | 1.04 | 0.00023 | -0.55 | 0.00134 | 0.278 |
| 46886 | ACTA2-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | SYT4 | Sponge network | 0.194 | 0.64278 | -3.207 | 0 | 0.278 |
| 46887 | SNHG1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-664a-3p | 17 | CCDC6 | Sponge network | 1.265 | 0 | 0.27 | 0.02555 | 0.278 |
| 46888 | ACTA2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 32 | EGFR | Sponge network | 0.194 | 0.64278 | -0.175 | 0.48451 | 0.278 |
| 46889 | BZRAP1-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-550a-3p;hsa-miR-577 | 22 | SYT4 | Sponge network | -0.465 | 0.04703 | -3.207 | 0 | 0.278 |
| 46890 | RP11-57H14.4 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-96-5p | 19 | CXCL12 | Sponge network | 0.083 | 0.66405 | -1.421 | 0 | 0.278 |
| 46891 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 10 | HOOK3 | Sponge network | 0.374 | 0.20054 | 0.273 | 0.09957 | 0.278 |
| 46892 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b | 14 | ERCC4 | Sponge network | 0.082 | 0.55397 | 0.126 | 0.15266 | 0.278 |
| 46893 | AGAP11 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 11 | HOMER2 | Sponge network | -1.645 | 0 | -2.698 | 0 | 0.278 |
| 46894 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | 0.335 | 0.20596 | 0.126 | 0.15266 | 0.278 |
| 46895 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | NF1 | Sponge network | 0.415 | 0.14657 | 0.51 | 0 | 0.278 |
| 46896 | HHIP-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | PRKCB | Sponge network | -0.737 | 0.10822 | -1.313 | 2.0E-5 | 0.278 |
| 46897 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-92a-3p | 18 | RIMBP2 | Sponge network | 0.225 | 0.30644 | -1.968 | 5.0E-5 | 0.278 |
| 46898 | TUG1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p | 13 | ATRX | Sponge network | 0.972 | 0 | 0.261 | 0.04408 | 0.278 |
| 46899 | RP11-38L15.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 16 | BMPR2 | Sponge network | 0.344 | 0.3127 | 0.206 | 0.0635 | 0.278 |
| 46900 | RP11-38L15.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-345-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-935 | 18 | GRIK3 | Sponge network | 0.344 | 0.3127 | -3.208 | 0 | 0.278 |
| 46901 | RP11-428J1.5 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | DOCK4 | Sponge network | 0.538 | 0.00076 | 0.502 | 0.00108 | 0.278 |
| 46902 | RP11-5C23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-590-3p | 20 | PGR | Sponge network | 0.274 | 0.07681 | -1.704 | 0 | 0.278 |
| 46903 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | RBL2 | Sponge network | -0.031 | 0.89063 | -0.282 | 0.00254 | 0.278 |
| 46904 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | SPG20 | Sponge network | 1.302 | 7.0E-5 | -1.16 | 0 | 0.278 |
| 46905 | JAZF1-AS1 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | FAM135B | Sponge network | -0.769 | 0.00035 | -3.978 | 0 | 0.278 |
| 46906 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p | 16 | DDX5 | Sponge network | 0.72 | 0.0001 | -0.099 | 0.17428 | 0.278 |
| 46907 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-590-3p | 12 | PCDH8 | Sponge network | -0.129 | 0.57177 | -0.997 | 0.05366 | 0.278 |
| 46908 | RP11-822E23.8 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-589-3p;hsa-miR-7-5p | 21 | KCNJ5 | Sponge network | -0.777 | 0.16667 | -0.281 | 0.3339 | 0.278 |
| 46909 | RP11-678G14.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-484 | 12 | EZH1 | Sponge network | -4.477 | 0 | -0.564 | 1.0E-5 | 0.278 |
| 46910 | LINC00886 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p | 13 | KCNAB1 | Sponge network | -0.371 | 0.24604 | -0.755 | 2.0E-5 | 0.278 |
| 46911 | RP11-701H24.3 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 10 | PTPRT | Sponge network | -1.425 | 0.04204 | -0.907 | 0.03727 | 0.278 |
| 46912 | FAM215B |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 17 | AFF1 | Sponge network | 0.817 | 3.0E-5 | -0.384 | 0.00053 | 0.278 |
| 46913 | CTA-204B4.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-671-5p | 22 | ADARB1 | Sponge network | 0.765 | 7.0E-5 | -0.335 | 0.06631 | 0.278 |
| 46914 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 23 | TIMP3 | Sponge network | -0.727 | 0.04726 | -0.546 | 0.02307 | 0.278 |
| 46915 | RP4-669H2.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-629-3p | 16 | DST | Sponge network | 1.711 | 0.00019 | -0.713 | 0.00096 | 0.278 |
| 46916 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 30 | FOXP1 | Sponge network | 0.541 | 0.00125 | 0.042 | 0.74368 | 0.278 |
| 46917 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 11 | TRIM3 | Sponge network | -0.611 | 0.09607 | -0.577 | 0 | 0.278 |
| 46918 | RP11-894P9.1 |
hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p | 10 | TACC1 | Sponge network | 0.993 | 0 | -0.362 | 0.05406 | 0.278 |
| 46919 | RP11-38L15.3 |
hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-345-5p;hsa-miR-590-3p;hsa-miR-935 | 11 | PCDH10 | Sponge network | 0.344 | 0.3127 | -1.644 | 0.0078 | 0.278 |
| 46920 | RP11-212P7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 32 | TGFBR3 | Sponge network | 0.219 | 0.1516 | -1.173 | 1.0E-5 | 0.278 |
| 46921 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 66 | THRB | Sponge network | 0.997 | 0.00066 | -1.117 | 0.0004 | 0.278 |
| 46922 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-625-5p | 18 | RSPO2 | Sponge network | 0.4 | 0.14611 | -3.038 | 0 | 0.278 |
| 46923 | LINC00662 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p | 12 | RET | Sponge network | 0.213 | 0.32297 | -0.833 | 0.03517 | 0.278 |
| 46924 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-582-5p | 13 | NR2C2 | Sponge network | 0.056 | 0.81195 | 0.21 | 0.03224 | 0.278 |
| 46925 | RP11-46C24.7 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | STARD13 | Sponge network | 0.392 | 0.0278 | -0.187 | 0.27383 | 0.278 |
| 46926 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-940 | 58 | AFF4 | Sponge network | -0.405 | 0.22013 | -0.266 | 0.01565 | 0.278 |
| 46927 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 21 | CLASP2 | Sponge network | 0.569 | 0.00067 | -0.038 | 0.71 | 0.278 |
| 46928 | CROCCP2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-484 | 17 | ZFX | Sponge network | 0.79 | 0 | 0.09 | 0.42563 | 0.278 |
| 46929 | CTD-3064M3.3 |
hsa-let-7a-5p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-671-5p | 14 | THRA | Sponge network | -0.469 | 0.08721 | -0.307 | 0.05099 | 0.278 |
| 46930 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-654-3p | 27 | DDX6 | Sponge network | -0.227 | 0.31657 | 0.091 | 0.36428 | 0.278 |
| 46931 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TBX18 | Sponge network | 0.488 | 0.00084 | 0.843 | 0.02945 | 0.278 |
| 46932 | CROCCP2 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | SLC5A3 | Sponge network | 0.79 | 0 | 0.493 | 5.0E-5 | 0.278 |
| 46933 | RP11-973H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p | 10 | PTPN11 | Sponge network | 0.901 | 0 | 0.431 | 2.0E-5 | 0.278 |
| 46934 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p | 14 | PBX1 | Sponge network | 1.04 | 0.00023 | -1.206 | 0 | 0.278 |
| 46935 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 14 | MYO9A | Sponge network | 0.702 | 7.0E-5 | -0.147 | 0.32373 | 0.278 |
| 46936 | LINC00094 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 24 | PCDH10 | Sponge network | 0.253 | 0.09565 | -1.644 | 0.0078 | 0.278 |
| 46937 | MIR143HG |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 21 | JUN | Sponge network | -0.739 | 0.0574 | -0.932 | 0 | 0.277 |
| 46938 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-940 | 13 | EPS15 | Sponge network | 0.453 | 0.01839 | -0.052 | 0.53998 | 0.277 |
| 46939 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-421 | 16 | PRDM2 | Sponge network | -4.477 | 0 | -0.258 | 0.00721 | 0.277 |
| 46940 | RP11-701H24.7 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-93-5p | 11 | ACSL6 | Sponge network | -2.039 | 0.03447 | -1.593 | 0 | 0.277 |
| 46941 | USP3-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421 | 21 | LIMCH1 | Sponge network | -0.162 | 0.47687 | -0.915 | 1.0E-5 | 0.277 |
| 46942 | CTC-378H22.2 |
hsa-miR-107;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-224-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-502-5p | 13 | DTNA | Sponge network | 0.001 | 0.99663 | -1.648 | 1.0E-5 | 0.277 |
| 46943 | RP11-479G22.8 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | DST | Sponge network | 1.308 | 0 | -0.713 | 0.00096 | 0.277 |
| 46944 | RP11-888D10.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-30c-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | ABI2 | Sponge network | 0.702 | 7.0E-5 | 0.175 | 0.14787 | 0.277 |
| 46945 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 15 | ST6GAL2 | Sponge network | 0.056 | 0.8494 | -1.201 | 0.00597 | 0.277 |
| 46946 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | PTPN11 | Sponge network | 0.439 | 0.00313 | 0.431 | 2.0E-5 | 0.277 |
| 46947 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p | 14 | SUFU | Sponge network | -0.517 | 0.02935 | -0.04 | 0.65489 | 0.277 |
| 46948 | SOX2-OT |
hsa-miR-146b-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 10 | FHL1 | Sponge network | -1.264 | 6.0E-5 | -2.687 | 0 | 0.277 |
| 46949 | GNG12-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 27 | MED12L | Sponge network | -0.793 | 7.0E-5 | -0.194 | 0.55299 | 0.277 |
| 46950 | RP11-326G21.1 |
hsa-let-7b-5p;hsa-miR-106b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | PRDM2 | Sponge network | -0.021 | 0.93598 | -0.258 | 0.00721 | 0.277 |
| 46951 | RP11-434H6.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-146a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-29a-5p;hsa-miR-30a-3p | 16 | FZD3 | Sponge network | 0.358 | 0.14302 | 0.104 | 0.60316 | 0.277 |
| 46952 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p | 10 | USP6 | Sponge network | 0.997 | 0 | 0.188 | 0.3871 | 0.277 |
| 46953 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 14 | PRRX1 | Sponge network | -1.375 | 0.08642 | 1.316 | 0 | 0.277 |
| 46954 | SH3RF3-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-7-1-3p | 10 | SEMA3E | Sponge network | 0.889 | 9.0E-5 | -1.717 | 0.00137 | 0.277 |
| 46955 | AC025171.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | ITGB1 | Sponge network | 0.998 | 0 | 0.524 | 0.00017 | 0.277 |
| 46956 | RP11-428J1.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PDE4DIP | Sponge network | 0.538 | 0.00076 | -0.282 | 0.0257 | 0.277 |
| 46957 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p | 17 | LHX6 | Sponge network | -2.925 | 0 | -0.087 | 0.69193 | 0.277 |
| 46958 | MIR143HG |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 19 | UNC5C | Sponge network | -0.739 | 0.0574 | -0.178 | 0.40214 | 0.277 |
| 46959 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 72 | AFF4 | Sponge network | -1.281 | 0.00012 | -0.266 | 0.01565 | 0.277 |
| 46960 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-425-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p | 18 | SLITRK6 | Sponge network | -1.047 | 0 | -2.121 | 0 | 0.277 |
| 46961 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-362-3p | 18 | RAP1A | Sponge network | -0.693 | 0.05629 | -0.929 | 0 | 0.277 |
| 46962 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 22 | PHC3 | Sponge network | 1.757 | 0 | 0.212 | 0.0499 | 0.277 |
| 46963 | MIR143HG |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p | 33 | CADM1 | Sponge network | -0.739 | 0.0574 | -0.9 | 0.00084 | 0.277 |
| 46964 | DNM3OS |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 48 | MDM4 | Sponge network | 0.572 | 0.01722 | 0.345 | 0.00762 | 0.277 |
| 46965 | RP11-389C8.2 |
hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-7-1-3p | 12 | SFRP1 | Sponge network | 0.727 | 3.0E-5 | -3.566 | 0 | 0.277 |
| 46966 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 19 | UBE4B | Sponge network | -2.452 | 0 | -0.235 | 0.007 | 0.277 |
| 46967 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p | 22 | LATS1 | Sponge network | 0.997 | 0.00066 | 0.109 | 0.34208 | 0.277 |
| 46968 | FGF14-AS2 |
hsa-miR-1266-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-5p | 11 | KCNJ5 | Sponge network | -1.582 | 8.0E-5 | -0.281 | 0.3339 | 0.277 |
| 46969 | AC010761.8 |
hsa-miR-10b-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-582-5p | 10 | CREB1 | Sponge network | 1.638 | 0 | 0.203 | 0.00537 | 0.277 |
| 46970 | RP11-345P4.7 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-628-5p | 13 | VPS53 | Sponge network | 1.597 | 0 | 0.103 | 0.22667 | 0.277 |
| 46971 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 12 | ADAMTSL3 | Sponge network | -0.141 | 0.26434 | -1.559 | 0 | 0.277 |
| 46972 | GAS6-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-450b-5p | 12 | PKNOX1 | Sponge network | 0.16 | 0.50785 | 0.085 | 0.28917 | 0.277 |
| 46973 | AC084219.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 20 | RICTOR | Sponge network | 1.296 | 0.0054 | 0.388 | 0.00188 | 0.277 |
| 46974 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | LRRC7 | Sponge network | 0.051 | 0.83388 | -1.341 | 0.01193 | 0.277 |
| 46975 | RP11-426C22.5 |
hsa-miR-148a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-582-5p | 10 | RUNX1T1 | Sponge network | 0.749 | 0 | -0.344 | 0.2374 | 0.277 |
| 46976 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 43 | RASSF2 | Sponge network | -2.46 | 0 | -0.273 | 0.21961 | 0.277 |
| 46977 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | EPHA5 | Sponge network | 0.673 | 0 | -0.957 | 0.01733 | 0.277 |
| 46978 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-335-5p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 19 | RPS6KA2 | Sponge network | -0.154 | 0.78939 | -0.57 | 0.00013 | 0.277 |
| 46979 | RP11-760H22.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p | 21 | CHD5 | Sponge network | 0.001 | 0.99753 | -0.922 | 0.01521 | 0.277 |
| 46980 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p | 16 | ERCC4 | Sponge network | -0.18 | 0.20343 | 0.126 | 0.15266 | 0.277 |
| 46981 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | PICALM | Sponge network | 0.222 | 0.29133 | 0.086 | 0.23523 | 0.277 |
| 46982 | USP3-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-940 | 20 | SIRT1 | Sponge network | -0.162 | 0.47687 | 0.109 | 0.2593 | 0.277 |
| 46983 | CALML3-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p | 22 | DOCK4 | Sponge network | 0.95 | 0.15943 | 0.502 | 0.00108 | 0.277 |
| 46984 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | CXXC4 | Sponge network | 0.572 | 0.01722 | -0.433 | 0.21055 | 0.277 |
| 46985 | DNM1P35 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p | 25 | NTRK2 | Sponge network | 0.051 | 0.83388 | -0.736 | 0.06495 | 0.277 |
| 46986 | RP11-67L2.2 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | RBM6 | Sponge network | 0.72 | 0.0001 | -0.137 | 0.15663 | 0.277 |
| 46987 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 23 | LRRC7 | Sponge network | 0.082 | 0.55654 | -1.341 | 0.01193 | 0.277 |
| 46988 | HOXB-AS2 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p | 10 | HIP1 | Sponge network | 1.316 | 0.01106 | 0.774 | 0 | 0.277 |
| 46989 | RP5-994D16.9 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-532-3p | 10 | SYNM | Sponge network | 0.252 | 0.08508 | -2.309 | 1.0E-5 | 0.277 |
| 46990 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-942-5p | 19 | ARHGAP28 | Sponge network | 0.4 | 0.14611 | -0.911 | 0.00013 | 0.277 |
| 46991 | RP11-296I10.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-539-5p | 13 | TCF12 | Sponge network | 0.592 | 0.00014 | 0.174 | 0.13271 | 0.277 |
| 46992 | C20orf166-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-671-5p | 21 | CYLD | Sponge network | -2.69 | 0.0001 | -0.367 | 0.00171 | 0.277 |
| 46993 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 24 | PTPRD | Sponge network | -0.72 | 0.24239 | -1.005 | 0.00064 | 0.277 |
| 46994 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 26 | ARID5B | Sponge network | -0.611 | 0.09607 | 0.342 | 0.01676 | 0.277 |
| 46995 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-214-3p;hsa-miR-223-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-432-5p;hsa-miR-539-5p | 14 | SRGAP3 | Sponge network | -0.602 | 0.00331 | -0.202 | 0.29297 | 0.277 |
| 46996 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TMEFF2 | Sponge network | 0.101 | 0.60919 | -2.426 | 0 | 0.277 |
| 46997 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | RNF144A | Sponge network | 0.559 | 0.00026 | 0.594 | 0.0009 | 0.277 |
| 46998 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | EPHA7 | Sponge network | 0.727 | 3.0E-5 | -2.197 | 3.0E-5 | 0.277 |
| 46999 | ZNF833P |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-582-3p | 10 | ABL2 | Sponge network | 0.688 | 1.0E-5 | 0.82 | 0 | 0.277 |
| 47000 | RP11-373D23.3 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-335-5p;hsa-miR-421 | 11 | CTIF | Sponge network | -0.285 | 0.2172 | -0.389 | 0.00549 | 0.277 |
| 47001 | PCA3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p | 21 | PPP2R5C | Sponge network | -0.709 | 0.18168 | -0.314 | 3.0E-5 | 0.277 |
| 47002 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-340-3p;hsa-miR-369-3p;hsa-miR-532-3p;hsa-miR-940 | 23 | NFIB | Sponge network | 0.993 | 0 | 0.023 | 0.90795 | 0.277 |
| 47003 | RP11-326G21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CREB1 | Sponge network | -0.021 | 0.93598 | 0.203 | 0.00537 | 0.277 |
| 47004 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-342-3p | 21 | NCOA1 | Sponge network | 0.082 | 0.55397 | -0.432 | 0 | 0.277 |
| 47005 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-501-3p;hsa-miR-942-5p | 18 | UNC80 | Sponge network | 0.238 | 0.70544 | -1.934 | 0 | 0.277 |
| 47006 | RP11-65J21.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-505-5p | 12 | ASTN1 | Sponge network | -0.557 | 0.11135 | -2.389 | 2.0E-5 | 0.277 |
| 47007 | RP11-46C24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | TGFBR3 | Sponge network | 0.392 | 0.0278 | -1.173 | 1.0E-5 | 0.277 |
| 47008 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-550a-5p | 14 | FAT2 | Sponge network | 0.997 | 0.00066 | -0.278 | 0.44877 | 0.277 |
| 47009 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p | 27 | EPHA5 | Sponge network | 0.921 | 0.00118 | -0.957 | 0.01733 | 0.277 |
| 47010 | LINC00702 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 22 | PRKAR1A | Sponge network | -0.737 | 0.06182 | -0.063 | 0.50314 | 0.277 |
| 47011 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-589-3p | 12 | HECA | Sponge network | -4.463 | 0.00025 | -0.217 | 0.03463 | 0.277 |
| 47012 | RP11-158K1.3 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | PTCH1 | Sponge network | 0.349 | 0.01695 | -0.1 | 0.6179 | 0.277 |
| 47013 | HHIP-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-590-3p | 27 | NTRK2 | Sponge network | -0.737 | 0.10822 | -0.736 | 0.06495 | 0.277 |
| 47014 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | SOX11 | Sponge network | 0.659 | 3.0E-5 | 2.68 | 0 | 0.277 |
| 47015 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-7-5p | 26 | ERBB4 | Sponge network | -0.693 | 0.05629 | -1.975 | 2.0E-5 | 0.277 |
| 47016 | RP11-244H3.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-877-5p | 19 | NRK | Sponge network | 0.659 | 3.0E-5 | 0.656 | 0.19217 | 0.277 |
| 47017 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | PAFAH1B1 | Sponge network | -1.161 | 0.00591 | -0.429 | 0 | 0.277 |
| 47018 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 18 | SCN9A | Sponge network | 0.873 | 0 | -0.525 | 0.10593 | 0.277 |
| 47019 | A2M-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-96-5p | 20 | CADM1 | Sponge network | -0.08 | 0.90092 | -0.9 | 0.00084 | 0.277 |
| 47020 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378c;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | MED12L | Sponge network | 0.083 | 0.66405 | -0.194 | 0.55299 | 0.277 |
| 47021 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-671-5p | 14 | DLC1 | Sponge network | -0.351 | 0.11358 | -0.382 | 0.05038 | 0.277 |
| 47022 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PRDM11 | Sponge network | -0.235 | 0.15142 | -0.252 | 0.15511 | 0.277 |
| 47023 | ERVH48-1 |
hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 14 | DIP2C | Sponge network | 1.04 | 0.00023 | -0.436 | 0.01713 | 0.277 |
| 47024 | RP11-6N17.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 11 | TCF4 | Sponge network | 0.108 | 0.69556 | 0.016 | 0.92271 | 0.277 |
| 47025 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 36 | ZFHX3 | Sponge network | -0.969 | 2.0E-5 | 0.389 | 0.01363 | 0.277 |
| 47026 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 24 | ERBB4 | Sponge network | -0.811 | 2.0E-5 | -1.975 | 2.0E-5 | 0.277 |
| 47027 | BCYRN1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 50 | DTNA | Sponge network | 0.311 | 0.10344 | -1.648 | 1.0E-5 | 0.277 |
| 47028 | CTD-2089N3.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-5p | 13 | CHD5 | Sponge network | -1.375 | 0.08642 | -0.922 | 0.01521 | 0.277 |
| 47029 | RP11-819C21.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-96-5p | 20 | CYLD | Sponge network | 0.537 | 0.00139 | -0.367 | 0.00171 | 0.277 |
| 47030 | RP11-967K21.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-3p | 13 | SUFU | Sponge network | 0.056 | 0.81195 | -0.04 | 0.65489 | 0.277 |
| 47031 | AC007277.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-576-5p | 12 | TSC1 | Sponge network | 2.377 | 0.00108 | 0.103 | 0.26071 | 0.277 |
| 47032 | RP11-760H22.2 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-335-5p | 13 | ANK2 | Sponge network | 0.001 | 0.99753 | -1.603 | 1.0E-5 | 0.277 |
| 47033 | RP11-774O3.3 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | RIT1 | Sponge network | -0.487 | 0.02157 | -0.296 | 0.00054 | 0.277 |
| 47034 | FENDRR |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 39 | ABI2 | Sponge network | -1.281 | 0.00012 | 0.175 | 0.14787 | 0.277 |
| 47035 | RP11-571M6.17 |
hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-2355-3p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-3p | 15 | BMPR2 | Sponge network | 1.013 | 0 | 0.206 | 0.0635 | 0.277 |
| 47036 | RP11-345P4.7 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 16 | SLC5A3 | Sponge network | 1.597 | 0 | 0.493 | 5.0E-5 | 0.277 |
| 47037 | LINC00173 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-455-5p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | LATS2 | Sponge network | 0.056 | 0.8494 | 0.159 | 0.2304 | 0.277 |
| 47038 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | ITGAV | Sponge network | 0.101 | 0.60919 | 0.337 | 0.01002 | 0.277 |
| 47039 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-629-3p | 15 | ZBTB4 | Sponge network | -2.549 | 0.00528 | -0.739 | 0 | 0.277 |
| 47040 | ANKRD10-IT1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 10 | NEDD4 | Sponge network | 1.739 | 0 | 0.02 | 0.89834 | 0.277 |
| 47041 | RP4-717I23.3 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-375;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-3p | 14 | DOCK4 | Sponge network | 0.637 | 0.00069 | 0.502 | 0.00108 | 0.277 |
| 47042 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 19 | ARHGAP29 | Sponge network | 0.374 | 0.20054 | 0.131 | 0.42953 | 0.277 |
| 47043 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 23 | ACVR2A | Sponge network | -0.285 | 0.2172 | -0.409 | 0.00039 | 0.277 |
| 47044 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | PTPRB | Sponge network | -0.615 | 0.00015 | 0.105 | 0.47122 | 0.277 |
| 47045 | NR2F1-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 20 | PRKAR1A | Sponge network | -0.49 | 0.04946 | -0.063 | 0.50314 | 0.277 |
| 47046 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-653-5p | 13 | PDS5B | Sponge network | -0.031 | 0.89063 | 0.259 | 0.00962 | 0.277 |
| 47047 | LINC00662 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CXXC4 | Sponge network | 0.213 | 0.32297 | -0.433 | 0.21055 | 0.277 |
| 47048 | CTC-429P9.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-532-3p | 29 | NFIB | Sponge network | 0.497 | 0.01451 | 0.023 | 0.90795 | 0.277 |
| 47049 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 15 | GPAM | Sponge network | 0.373 | 0.00068 | 0.142 | 0.29732 | 0.277 |
| 47050 | PART1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940 | 53 | GAS7 | Sponge network | -2.925 | 0 | -0.555 | 0.01602 | 0.277 |
| 47051 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p | 11 | GNA13 | Sponge network | 0.826 | 1.0E-5 | 0.007 | 0.93884 | 0.277 |
| 47052 | RP11-1006G14.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-590-3p | 12 | ROBO1 | Sponge network | 0.559 | 0.00026 | -0.009 | 0.96154 | 0.277 |
| 47053 | EIF3J-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | DIP2C | Sponge network | 0.331 | 0.00509 | -0.436 | 0.01713 | 0.277 |
| 47054 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-7-1-3p | 25 | PHC3 | Sponge network | 0.056 | 0.8494 | 0.212 | 0.0499 | 0.277 |
| 47055 | ARHGEF26-AS1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-96-5p | 20 | IRS1 | Sponge network | -2.46 | 0 | -0.026 | 0.88855 | 0.277 |
| 47056 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 14 | PKNOX1 | Sponge network | 0.253 | 0.09565 | 0.085 | 0.28917 | 0.277 |
| 47057 | RP11-158K1.3 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TPO | Sponge network | 0.349 | 0.01695 | -1.87 | 2.0E-5 | 0.277 |
| 47058 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-942-5p | 35 | NOTCH2 | Sponge network | -0.727 | 0.04726 | 0.138 | 0.35163 | 0.277 |
| 47059 | FZD10-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | NIN | Sponge network | -0.727 | 0.04726 | -0.253 | 0.13794 | 0.277 |
| 47060 | RP11-276H19.2 |
hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-502-5p | 12 | ANK2 | Sponge network | -0.472 | 0.48851 | -1.603 | 1.0E-5 | 0.277 |
| 47061 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 18 | NEDD4 | Sponge network | -0.693 | 0.05629 | 0.02 | 0.89834 | 0.277 |
| 47062 | RP11-497H16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 11 | ERCC4 | Sponge network | 0.545 | 0.00064 | 0.126 | 0.15266 | 0.277 |
| 47063 | CTC-471J1.2 |
hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 10 | DICER1 | Sponge network | 1.11 | 1.0E-5 | 0.042 | 0.674 | 0.277 |
| 47064 | RP11-798G7.7 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-539-5p | 11 | TCF12 | Sponge network | 0.858 | 3.0E-5 | 0.174 | 0.13271 | 0.277 |
| 47065 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p | 12 | PAX6 | Sponge network | -0.342 | 0.02288 | -0.444 | 0.04689 | 0.277 |
| 47066 | ARHGEF26-AS1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | SH3GLB1 | Sponge network | -2.46 | 0 | -0.115 | 0.14773 | 0.277 |
| 47067 | RP11-655M14.13 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p | 10 | CUL5 | Sponge network | 1.473 | 0.0057 | 0.026 | 0.77301 | 0.277 |
| 47068 | ADIRF-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | SETBP1 | Sponge network | -0.223 | 0.47816 | -1.302 | 1.0E-5 | 0.277 |
| 47069 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | LRRC55 | Sponge network | -0.355 | 0.0077 | -0.026 | 0.93163 | 0.277 |
| 47070 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | PAFAH1B1 | Sponge network | 0.349 | 0.01695 | -0.429 | 0 | 0.277 |
| 47071 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-486-5p;hsa-miR-940 | 15 | IGF1 | Sponge network | 0.792 | 0.0049 | -0.204 | 0.5898 | 0.277 |
| 47072 | H19 |
hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p | 20 | CBL | Sponge network | 2.439 | 0 | 0.291 | 0.01267 | 0.277 |
| 47073 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | PIK3R1 | Sponge network | -1.349 | 0.00017 | -0.133 | 0.36854 | 0.277 |
| 47074 | RP11-2B6.2 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | KIF5B | Sponge network | 0.539 | 0.03722 | 0.362 | 0.00066 | 0.277 |
| 47075 | BDNF-AS |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 12 | DCLK1 | Sponge network | -0.668 | 3.0E-5 | -1.324 | 0.00014 | 0.277 |
| 47076 | RP4-717I23.3 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-375;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-935 | 19 | DMD | Sponge network | 0.637 | 0.00069 | -1.506 | 0 | 0.277 |
| 47077 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p | 16 | KANK1 | Sponge network | 0.72 | 0.0001 | -0.55 | 0.00134 | 0.277 |
| 47078 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-493-3p;hsa-miR-671-5p;hsa-miR-93-5p | 25 | CHD5 | Sponge network | -0.663 | 0.10887 | -0.922 | 0.01521 | 0.277 |
| 47079 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p | 15 | LRP1B | Sponge network | 0.174 | 0.30034 | -2.163 | 0 | 0.277 |
| 47080 | SERHL |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-628-5p | 17 | FBXO32 | Sponge network | -0.602 | 0.00331 | -0.495 | 0.07561 | 0.277 |
| 47081 | CTA-363E19.2 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-485-3p;hsa-miR-495-3p | 18 | SMAD2 | Sponge network | 1.055 | 0 | -0.128 | 0.12732 | 0.277 |
| 47082 | RP11-216B9.6 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | BAZ2B | Sponge network | 0.174 | 0.30034 | -0.276 | 0.02833 | 0.277 |
| 47083 | KCNJ2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | PRKAA2 | Sponge network | -0.436 | 0.05325 | -2.462 | 0 | 0.277 |
| 47084 | FAM66C |
hsa-miR-140-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | CHD6 | Sponge network | -0.901 | 0.00019 | 0.32 | 0.01326 | 0.277 |
| 47085 | RP11-389C8.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | ASTN1 | Sponge network | 0.727 | 3.0E-5 | -2.389 | 2.0E-5 | 0.277 |
| 47086 | HCG11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 23 | CADM1 | Sponge network | 0.385 | 0.10067 | -0.9 | 0.00084 | 0.277 |
| 47087 | MIR600HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-628-5p | 21 | ERC1 | Sponge network | 0.594 | 0.41522 | -0.108 | 0.38794 | 0.277 |
| 47088 | RP4-639F20.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-96-5p | 14 | MITF | Sponge network | -0.517 | 0.02935 | -0.769 | 0.00022 | 0.277 |
| 47089 | RP11-411K7.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-93-3p;hsa-miR-940 | 23 | TOM1L2 | Sponge network | 0.155 | 0.81273 | -0.994 | 0 | 0.277 |
| 47090 | AC106786.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-92a-1-5p | 11 | TRPS1 | Sponge network | 1.122 | 0.02989 | -0.191 | 0.44109 | 0.277 |
| 47091 | PWAR6 |
hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | RIT1 | Sponge network | -1.575 | 6.0E-5 | -0.296 | 0.00054 | 0.277 |
| 47092 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-625-5p | 10 | LHX6 | Sponge network | -4.477 | 0 | -0.087 | 0.69193 | 0.277 |
| 47093 | LINC00173 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-874-3p;hsa-miR-96-5p | 20 | ASTN1 | Sponge network | 0.056 | 0.8494 | -2.389 | 2.0E-5 | 0.277 |
| 47094 | RP11-359E10.1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 10 | EHD3 | Sponge network | 0.299 | 0.57722 | -0.484 | 0.01747 | 0.277 |
| 47095 | AC108488.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-625-5p | 16 | CRTC1 | Sponge network | 0.259 | 0.12542 | -0.116 | 0.28412 | 0.277 |
| 47096 | LINC00672 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-93-3p | 27 | NCOA1 | Sponge network | -0.227 | 0.31657 | -0.432 | 0 | 0.277 |
| 47097 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DACT1 | Sponge network | 0.05 | 0.95483 | 0.783 | 0.00072 | 0.277 |
| 47098 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | ZMYM2 | Sponge network | 1.122 | 0 | 0.279 | 0.01273 | 0.277 |
| 47099 | LA16c-321D4.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 10 | ZMYM2 | Sponge network | 2.459 | 5.0E-5 | 0.279 | 0.01273 | 0.277 |
| 47100 | GS1-124K5.11 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-877-5p | 28 | ASTN1 | Sponge network | 0.494 | 0.00339 | -2.389 | 2.0E-5 | 0.277 |
| 47101 | DNM3OS |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 26 | EPS15 | Sponge network | 0.572 | 0.01722 | -0.052 | 0.53998 | 0.277 |
| 47102 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | PRKAA1 | Sponge network | 1.163 | 0.00018 | 0.317 | 0.0031 | 0.277 |
| 47103 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-590-3p;hsa-miR-629-3p | 10 | RBL2 | Sponge network | 0.603 | 0.00127 | -0.282 | 0.00254 | 0.277 |
| 47104 | RP11-686D22.8 |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | RECK | Sponge network | 0.898 | 1.0E-5 | -1.043 | 0 | 0.277 |
| 47105 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-589-3p | 18 | USP12 | Sponge network | 0.176 | 0.37402 | -0.102 | 0.40768 | 0.277 |
| 47106 | RP11-6N17.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-455-5p;hsa-miR-744-3p | 15 | DST | Sponge network | 0.108 | 0.69556 | -0.713 | 0.00096 | 0.277 |
| 47107 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | FAT4 | Sponge network | 0.219 | 0.1516 | -0.354 | 0.14694 | 0.277 |
| 47108 | PCBP1-AS1 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 27 | ARHGEF12 | Sponge network | -0.567 | 0 | 0.09 | 0.35693 | 0.277 |
| 47109 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-93-5p | 18 | FGFR1 | Sponge network | -1.645 | 0 | -0.509 | 0.03957 | 0.277 |
| 47110 | LINC00654 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | PIK3CA | Sponge network | 0.374 | 0.20054 | 0.195 | 0.06908 | 0.277 |
| 47111 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | ZFHX3 | Sponge network | -0.125 | 0.49414 | 0.389 | 0.01363 | 0.277 |
| 47112 | JAZF1-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-942-5p | 30 | OPCML | Sponge network | -0.769 | 0.00035 | -2.498 | 0 | 0.277 |
| 47113 | RP11-403I13.8 |
hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | MED12L | Sponge network | 0.619 | 0.00157 | -0.194 | 0.55299 | 0.277 |
| 47114 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-486-5p;hsa-miR-495-3p | 10 | PHF6 | Sponge network | 0.75 | 0.00054 | 0.785 | 0 | 0.277 |
| 47115 | RP11-181G12.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p | 16 | ZFX | Sponge network | 0.556 | 0.00298 | 0.09 | 0.42563 | 0.277 |
| 47116 | LINC00094 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-629-3p;hsa-miR-874-3p | 24 | ASTN1 | Sponge network | 0.253 | 0.09565 | -2.389 | 2.0E-5 | 0.277 |
| 47117 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-654-3p | 27 | PIK3R1 | Sponge network | -0.176 | 0.59692 | -0.133 | 0.36854 | 0.277 |
| 47118 | LINC00969 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p | 13 | SIRT1 | Sponge network | 0.683 | 1.0E-5 | 0.109 | 0.2593 | 0.277 |
| 47119 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-877-5p | 17 | NRK | Sponge network | -0.125 | 0.49414 | 0.656 | 0.19217 | 0.277 |
| 47120 | HOXB-AS2 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-576-5p | 11 | WWTR1 | Sponge network | 1.316 | 0.01106 | -0.345 | 0.11997 | 0.277 |
| 47121 | RP11-57H14.4 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-629-3p | 12 | SPTBN4 | Sponge network | 0.083 | 0.66405 | -0.786 | 0.01281 | 0.277 |
| 47122 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 26 | PRKCE | Sponge network | 0.101 | 0.60919 | -0.403 | 0.00056 | 0.277 |
| 47123 | RP11-156P1.3 |
hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-7-5p | 10 | EHD3 | Sponge network | 0.214 | 0.18246 | -0.484 | 0.01747 | 0.277 |
| 47124 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p | 24 | RASSF8 | Sponge network | -1.047 | 0 | -0.122 | 0.65591 | 0.277 |
| 47125 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | GPC6 | Sponge network | 0.051 | 0.83388 | -0.054 | 0.81465 | 0.277 |
| 47126 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | 0.862 | 0.06213 | -1.74 | 0 | 0.277 |
| 47127 | TINCR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-940 | 24 | PBX1 | Sponge network | -0.664 | 0.14543 | -1.206 | 0 | 0.277 |
| 47128 | TP73-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-542-3p | 14 | ERRFI1 | Sponge network | -0.563 | 0.02545 | -0.798 | 1.0E-5 | 0.277 |
| 47129 | AF131215.2 |
hsa-miR-132-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p | 17 | PCDH9 | Sponge network | -0.176 | 0.59692 | -1.823 | 4.0E-5 | 0.277 |
| 47130 | RP11-119F7.5 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 10 | IRS1 | Sponge network | 0.225 | 0.30644 | -0.026 | 0.88855 | 0.277 |
| 47131 | DNAJC27-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-3p | 15 | ID4 | Sponge network | -0.969 | 2.0E-5 | -1.644 | 0 | 0.277 |
| 47132 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 50 | BMPR2 | Sponge network | -1.249 | 7.0E-5 | 0.206 | 0.0635 | 0.277 |
| 47133 | RP11-15H20.7 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-34a-5p | 13 | MDM4 | Sponge network | 0.261 | 0.23673 | 0.345 | 0.00762 | 0.277 |
| 47134 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 16 | ACTB | Sponge network | -0.49 | 0.04946 | -0.247 | 0.01736 | 0.277 |
| 47135 | RP11-967K21.1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 31 | GRIK3 | Sponge network | 0.056 | 0.81195 | -3.208 | 0 | 0.277 |
| 47136 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | SOX5 | Sponge network | 0.349 | 0.01695 | -1.091 | 4.0E-5 | 0.277 |
| 47137 | AC074289.1 |
hsa-let-7d-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-96-5p | 13 | CBFA2T3 | Sponge network | -0.326 | 0.14314 | -1.221 | 1.0E-5 | 0.277 |
| 47138 | PCA3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 10 | RABEP1 | Sponge network | -0.709 | 0.18168 | -0.066 | 0.43142 | 0.277 |
| 47139 | RP11-35G9.5 |
hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-455-5p | 12 | ITPKB | Sponge network | 0.622 | 0.00015 | -0.704 | 0.00106 | 0.277 |
| 47140 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | JAZF1 | Sponge network | 0.222 | 0.29133 | -0.377 | 0.02677 | 0.277 |
| 47141 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-93-5p | 13 | FBXO31 | Sponge network | -0.384 | 0.40652 | -0.085 | 0.42389 | 0.277 |
| 47142 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | NR4A3 | Sponge network | -0.222 | 0.32445 | -1.385 | 0 | 0.277 |
| 47143 | FAM66C |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | NUAK1 | Sponge network | -0.901 | 0.00019 | 0.904 | 0 | 0.277 |
| 47144 | SH3RF3-AS1 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | PRKCB | Sponge network | 0.889 | 9.0E-5 | -1.313 | 2.0E-5 | 0.277 |
| 47145 | LPP-AS2 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577 | 13 | FMNL3 | Sponge network | -0.18 | 0.20343 | 0.555 | 4.0E-5 | 0.277 |
| 47146 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-493-5p | 13 | MIER3 | Sponge network | 0.826 | 1.0E-5 | -0.212 | 0.04957 | 0.277 |
| 47147 | RP11-458D21.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-744-3p | 12 | IGF2 | Sponge network | 0.663 | 0.00073 | 0.848 | 0.03842 | 0.277 |
| 47148 | RP11-760H22.2 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b | 12 | MRVI1 | Sponge network | 0.001 | 0.99753 | -1.051 | 0.00022 | 0.277 |
| 47149 | A2M-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-590-3p;hsa-miR-942-5p | 14 | GLIPR1 | Sponge network | -0.08 | 0.90092 | 0.146 | 0.3276 | 0.277 |
| 47150 | AC007879.7 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 11 | CDK6 | Sponge network | 4.3 | 0 | 0.991 | 0 | 0.277 |
| 47151 | TMEM51-AS1 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-664a-3p | 10 | PRKAA1 | Sponge network | 0.23 | 0.38667 | 0.317 | 0.0031 | 0.277 |
| 47152 | CALML3-AS1 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p | 10 | PEA15 | Sponge network | 0.95 | 0.15943 | 0.009 | 0.93318 | 0.277 |
| 47153 | IQCH-AS1 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-655-3p | 14 | NF1 | Sponge network | 0.929 | 0 | 0.51 | 0 | 0.277 |
| 47154 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 32 | RICTOR | Sponge network | -0.615 | 0.00015 | 0.388 | 0.00188 | 0.277 |
| 47155 | CTC-487M23.5 |
hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-374b-5p | 11 | SYNPO2 | Sponge network | 0.912 | 0.00014 | -2.342 | 0 | 0.277 |
| 47156 | CTD-2303H24.2 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 11 | MACF1 | Sponge network | 0.415 | 0.14657 | 0.019 | 0.90335 | 0.277 |
| 47157 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-493-5p;hsa-miR-590-3p | 12 | DMXL1 | Sponge network | 0.826 | 1.0E-5 | -0.426 | 0.00056 | 0.277 |
| 47158 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-3127-5p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-532-3p | 14 | CADM3 | Sponge network | -0.384 | 0.40652 | -3.663 | 0 | 0.277 |
| 47159 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 17 | PAFAH1B1 | Sponge network | -2.039 | 0.03447 | -0.429 | 0 | 0.277 |
| 47160 | LINC00963 |
hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-7-5p | 11 | FLNA | Sponge network | 0.523 | 9.0E-5 | -0.771 | 0.01377 | 0.277 |
| 47161 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | EPHA5 | Sponge network | 0.131 | 0.8436 | -0.957 | 0.01733 | 0.277 |
| 47162 | HCG11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 17 | SOX11 | Sponge network | 0.385 | 0.10067 | 2.68 | 0 | 0.277 |
| 47163 | RP11-216B9.6 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-629-5p | 10 | WNK2 | Sponge network | 0.174 | 0.30034 | -0.352 | 0.31066 | 0.277 |
| 47164 | HAR1A |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-877-5p | 14 | GRIN2A | Sponge network | -0.672 | 0.19447 | -2.161 | 0 | 0.277 |
| 47165 | LINC00641 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 30 | CCDC6 | Sponge network | -0.222 | 0.32445 | 0.27 | 0.02555 | 0.277 |
| 47166 | OSER1-AS1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 14 | PCDH10 | Sponge network | -0.13 | 0.48734 | -1.644 | 0.0078 | 0.277 |
| 47167 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-93-3p | 36 | BMPR2 | Sponge network | 0.274 | 0.07681 | 0.206 | 0.0635 | 0.277 |
| 47168 | RP11-416N2.4 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-590-3p | 11 | CBX5 | Sponge network | 0.08 | 0.83013 | 0.165 | 0.24446 | 0.277 |
| 47169 | FAM13A-AS1 |
hsa-let-7d-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 11 | DCN | Sponge network | -0.83 | 0 | -1.288 | 0 | 0.277 |
| 47170 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | PRKAA2 | Sponge network | 0.538 | 0.00076 | -2.462 | 0 | 0.277 |
| 47171 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 10 | RCAN2 | Sponge network | 0.4 | 0.14611 | -0.33 | 0.15172 | 0.277 |
| 47172 | RP11-360F5.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-425-5p;hsa-miR-429 | 10 | ERC1 | Sponge network | 2.364 | 0.00134 | -0.108 | 0.38794 | 0.277 |
| 47173 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-539-5p | 12 | DMD | Sponge network | 0.858 | 3.0E-5 | -1.506 | 0 | 0.277 |
| 47174 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-484;hsa-miR-590-3p | 12 | PKNOX1 | Sponge network | -0.227 | 0.31657 | 0.085 | 0.28917 | 0.277 |
| 47175 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 26 | DMXL1 | Sponge network | -0.769 | 0.00035 | -0.426 | 0.00056 | 0.277 |
| 47176 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | PSIP1 | Sponge network | 0.765 | 7.0E-5 | -0.005 | 0.96897 | 0.277 |
| 47177 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 26 | ST6GALNAC3 | Sponge network | -0.227 | 0.31657 | -0.987 | 0 | 0.277 |
| 47178 | AC011526.1 |
hsa-let-7g-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-3613-5p;hsa-miR-532-3p;hsa-miR-590-3p | 12 | TRPS1 | Sponge network | 0.342 | 0.03129 | -0.191 | 0.44109 | 0.277 |
| 47179 | LINC00702 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 33 | BTG2 | Sponge network | -0.737 | 0.06182 | -1.763 | 0 | 0.277 |
| 47180 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-628-5p | 21 | QKI | Sponge network | 1.361 | 0 | -0.333 | 0.04111 | 0.277 |
| 47181 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-93-5p | 12 | TXNIP | Sponge network | 0.199 | 0.38015 | -1.277 | 0 | 0.276 |
| 47182 | MIR497HG |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | ABI2 | Sponge network | -1.439 | 4.0E-5 | 0.175 | 0.14787 | 0.276 |
| 47183 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-455-5p | 16 | TBX18 | Sponge network | -1.645 | 0 | 0.843 | 0.02945 | 0.276 |
| 47184 | CTB-113P19.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 34 | AR | Sponge network | 0.906 | 0.31715 | -1.554 | 0 | 0.276 |
| 47185 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 36 | ESR1 | Sponge network | -0.793 | 7.0E-5 | -1.035 | 3.0E-5 | 0.276 |
| 47186 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 16 | KIT | Sponge network | -0.663 | 0.10887 | -2.184 | 0 | 0.276 |
| 47187 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-7-1-3p | 19 | KAT6B | Sponge network | -1.439 | 4.0E-5 | -0.478 | 0.00018 | 0.276 |
| 47188 | RP11-244H3.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PRRX1 | Sponge network | 0.659 | 3.0E-5 | 1.316 | 0 | 0.276 |
| 47189 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 19 | LATS2 | Sponge network | -0.612 | 0.00094 | 0.159 | 0.2304 | 0.276 |
| 47190 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-769-5p;hsa-miR-935 | 29 | HBP1 | Sponge network | -1.249 | 7.0E-5 | -0.454 | 0 | 0.276 |
| 47191 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 13 | NALCN | Sponge network | 0.421 | 0.00084 | 1.068 | 0.00237 | 0.276 |
| 47192 | AC003090.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-576-5p | 28 | ERC1 | Sponge network | -2.117 | 0.00161 | -0.108 | 0.38794 | 0.276 |
| 47193 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-664a-5p | 14 | SOX11 | Sponge network | 0.174 | 0.30034 | 2.68 | 0 | 0.276 |
| 47194 | RP11-244O19.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-671-5p | 12 | EHD3 | Sponge network | -0.096 | 0.67958 | -0.484 | 0.01747 | 0.276 |
| 47195 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | PDGFRA | Sponge network | 0.131 | 0.8436 | -0.372 | 0.0037 | 0.276 |
| 47196 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p | 11 | EGR2 | Sponge network | 0.194 | 0.64278 | 0.309 | 0.17079 | 0.276 |
| 47197 | ARHGEF26-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | UNC5D | Sponge network | -2.46 | 0 | -3.646 | 0 | 0.276 |
| 47198 | ZRANB2-AS1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p | 13 | PPP1R9A | Sponge network | -0.376 | 0.00337 | -0.903 | 0.04254 | 0.276 |
| 47199 | KB-431C1.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-337-3p | 10 | KDSR | Sponge network | 0.909 | 0 | 0.239 | 0.02216 | 0.276 |
| 47200 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | -0.355 | 0.0077 | 0.783 | 0.00072 | 0.276 |
| 47201 | RP11-774O3.3 |
hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 19 | EMP2 | Sponge network | -0.487 | 0.02157 | -0.423 | 0.00985 | 0.276 |
| 47202 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-93-5p;hsa-miR-98-5p | 25 | LRIG1 | Sponge network | -0.154 | 0.78939 | -0.537 | 0.00588 | 0.276 |
| 47203 | CALML3-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-664a-3p | 11 | PEG3 | Sponge network | 0.95 | 0.15943 | -1.74 | 0 | 0.276 |
| 47204 | SOX2-OT |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 10 | FAM117A | Sponge network | -1.264 | 6.0E-5 | -1.379 | 0 | 0.276 |
| 47205 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p | 11 | NALCN | Sponge network | -0.227 | 0.31657 | 1.068 | 0.00237 | 0.276 |
| 47206 | RP1-151F17.2 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | RASSF3 | Sponge network | 0.222 | 0.29133 | -0.226 | 0.11691 | 0.276 |
| 47207 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 28 | FZD3 | Sponge network | 0.765 | 7.0E-5 | 0.104 | 0.60316 | 0.276 |
| 47208 | MIR22HG |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-214-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-708-3p | 17 | PLXNA2 | Sponge network | -1.047 | 0 | -0.395 | 0.00779 | 0.276 |
| 47209 | AC007879.7 |
hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-5p | 11 | BMPR2 | Sponge network | 4.3 | 0 | 0.206 | 0.0635 | 0.276 |
| 47210 | RP11-819C21.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 12 | ABCB5 | Sponge network | 0.537 | 0.00139 | -0.956 | 0.04077 | 0.276 |
| 47211 | ZNF667-AS1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-766-3p | 13 | ATM | Sponge network | -0.976 | 0.00025 | 0.178 | 0.21568 | 0.276 |
| 47212 | RP11-193H5.1 |
hsa-miR-10a-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-374b-3p | 17 | VPS53 | Sponge network | 1.279 | 2.0E-5 | 0.103 | 0.22667 | 0.276 |
| 47213 | EMX2OS |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | BCL9 | Sponge network | -0.777 | 0.1134 | 0.321 | 0.00267 | 0.276 |
| 47214 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 64 | THRB | Sponge network | -0.222 | 0.32445 | -1.117 | 0.0004 | 0.276 |
| 47215 | RP11-121C2.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | KDSR | Sponge network | 1.147 | 0 | 0.239 | 0.02216 | 0.276 |
| 47216 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | ZNF208 | Sponge network | -0.342 | 0.02288 | -0.4 | 0.22381 | 0.276 |
| 47217 | HHIP-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 20 | OPCML | Sponge network | -0.737 | 0.10822 | -2.498 | 0 | 0.276 |
| 47218 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | ERBB4 | Sponge network | -0.125 | 0.49414 | -1.975 | 2.0E-5 | 0.276 |
| 47219 | RP11-504P24.4 |
hsa-miR-10b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | ARHGEF12 | Sponge network | 0.624 | 0.0004 | 0.09 | 0.35693 | 0.276 |
| 47220 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-93-5p | 14 | SASH1 | Sponge network | -2.531 | 0 | -0.502 | 0.00175 | 0.276 |
| 47221 | RP4-614O4.11 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 12 | DST | Sponge network | 1.042 | 0.00177 | -0.713 | 0.00096 | 0.276 |
| 47222 | CTC-559E9.6 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p | 14 | ABL2 | Sponge network | 0.601 | 1.0E-5 | 0.82 | 0 | 0.276 |
| 47223 | AC005154.6 |
hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DOCK4 | Sponge network | 0.848 | 0 | 0.502 | 0.00108 | 0.276 |
| 47224 | LINC01006 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-935 | 24 | GRIK3 | Sponge network | -0.702 | 0.04814 | -3.208 | 0 | 0.276 |
| 47225 | RP4-798A10.7 |
hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p | 13 | RICTOR | Sponge network | 1.757 | 0 | 0.388 | 0.00188 | 0.276 |
| 47226 | CTC-296K1.3 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-30b-3p | 11 | EFNA5 | Sponge network | -2.095 | 0.00112 | -0.421 | 0.17868 | 0.276 |
| 47227 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FSTL5 | Sponge network | 0.603 | 0.00127 | -1.3 | 0.01619 | 0.276 |
| 47228 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-331-5p;hsa-miR-374a-5p | 14 | USP12 | Sponge network | -0.693 | 0.05629 | -0.102 | 0.40768 | 0.276 |
| 47229 | RP11-981G7.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-766-3p | 15 | IL17RD | Sponge network | 0.986 | 0.01022 | -0.019 | 0.94873 | 0.276 |
| 47230 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-590-3p | 12 | KAT6B | Sponge network | -1.582 | 8.0E-5 | -0.478 | 0.00018 | 0.276 |
| 47231 | LINC00648 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | CADM2 | Sponge network | 0.633 | 0.51678 | -3.782 | 0 | 0.276 |
| 47232 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | TRIP11 | Sponge network | 0.848 | 0 | -0.158 | 0.06384 | 0.276 |
| 47233 | RP11-244H3.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | ROBO1 | Sponge network | 0.659 | 3.0E-5 | -0.009 | 0.96154 | 0.276 |
| 47234 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p | 15 | ZFX | Sponge network | 0.37 | 0.27614 | 0.09 | 0.42563 | 0.276 |
| 47235 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | PDGFRA | Sponge network | -0.31 | 0.33871 | -0.372 | 0.0037 | 0.276 |
| 47236 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-874-3p | 16 | NR2C2 | Sponge network | -0.223 | 0.47816 | 0.21 | 0.03224 | 0.276 |
| 47237 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | SRGAP3 | Sponge network | 0.72 | 0.0001 | -0.202 | 0.29297 | 0.276 |
| 47238 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-96-5p | 16 | ACVR1 | Sponge network | 0.331 | 0.57247 | 0.279 | 0.00234 | 0.276 |
| 47239 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-590-3p | 19 | PRKCE | Sponge network | 0.494 | 0.00339 | -0.403 | 0.00056 | 0.276 |
| 47240 | FGF14-AS2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-582-3p;hsa-miR-590-3p | 16 | PRDM2 | Sponge network | -1.582 | 8.0E-5 | -0.258 | 0.00721 | 0.276 |
| 47241 | PART1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | LSAMP | Sponge network | -2.925 | 0 | -0.066 | 0.81708 | 0.276 |
| 47242 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | PTPRB | Sponge network | 0.41 | 0.02214 | 0.105 | 0.47122 | 0.276 |
| 47243 | LINC00621 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | BRD3 | Sponge network | 0.131 | 0.8436 | 0.37 | 0.00013 | 0.276 |
| 47244 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-96-5p | 22 | FBXO32 | Sponge network | 0.853 | 0.15352 | -0.495 | 0.07561 | 0.276 |
| 47245 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-625-5p | 21 | NUAK1 | Sponge network | -0.096 | 0.67958 | 0.904 | 0 | 0.276 |
| 47246 | RP11-10N23.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-590-5p;hsa-miR-744-3p | 16 | DST | Sponge network | 1.134 | 0 | -0.713 | 0.00096 | 0.276 |
| 47247 | CTA-14H9.5 |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-33a-5p;hsa-miR-7-1-3p | 11 | AKAP11 | Sponge network | 0.145 | 0.53343 | 0.196 | 0.09825 | 0.276 |
| 47248 | RP11-384O8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p | 34 | UNC80 | Sponge network | -0.738 | 0.21233 | -1.934 | 0 | 0.276 |
| 47249 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 14 | YAP1 | Sponge network | 0.401 | 0.00066 | 0.22 | 0.11836 | 0.276 |
| 47250 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-942-5p | 43 | TMEM132B | Sponge network | 0.082 | 0.55654 | -0.83 | 0.01084 | 0.276 |
| 47251 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CLIP1 | Sponge network | -1.281 | 0.00012 | -0.215 | 0.08684 | 0.276 |
| 47252 | CTC-297N7.9 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-877-5p | 18 | AR | Sponge network | -2.069 | 0 | -1.554 | 0 | 0.276 |
| 47253 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p | 20 | FOXO3 | Sponge network | 0.921 | 0.00118 | -0.04 | 0.72028 | 0.276 |
| 47254 | LINC00702 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-874-3p | 13 | ARID1B | Sponge network | -0.737 | 0.06182 | 0.003 | 0.97503 | 0.276 |
| 47255 | CTC-487M23.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-450b-5p | 14 | SOX5 | Sponge network | 0.912 | 0.00014 | -1.091 | 4.0E-5 | 0.276 |
| 47256 | RP11-473I1.9 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-550a-3p | 10 | CACNA1E | Sponge network | 0.683 | 1.0E-5 | 1.752 | 0 | 0.276 |
| 47257 | RP11-395G23.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | ARNT2 | Sponge network | 0.381 | 0.25967 | -0.332 | 0.25851 | 0.276 |
| 47258 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p | 10 | PRKAR1A | Sponge network | -0.583 | 0.14589 | -0.063 | 0.50314 | 0.276 |
| 47259 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 53 | KIF1B | Sponge network | 0.41 | 0.02214 | -0.118 | 0.32258 | 0.276 |
| 47260 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-455-3p;hsa-miR-93-5p | 13 | TSHZ3 | Sponge network | 0.348 | 0.32912 | -0.556 | 0.01516 | 0.276 |
| 47261 | AC074289.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p | 11 | SPG20 | Sponge network | -0.326 | 0.14314 | -1.16 | 0 | 0.276 |
| 47262 | RP11-2B6.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | AKAP6 | Sponge network | 0.539 | 0.03722 | -1.586 | 3.0E-5 | 0.276 |
| 47263 | AC006116.24 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-582-3p | 11 | ABL2 | Sponge network | -0.473 | 0.07602 | 0.82 | 0 | 0.276 |
| 47264 | GUSBP11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 16 | ZFX | Sponge network | 0.856 | 0 | 0.09 | 0.42563 | 0.276 |
| 47265 | MIR143HG |
hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p | 11 | EMP1 | Sponge network | -0.739 | 0.0574 | -0.942 | 5.0E-5 | 0.276 |
| 47266 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-421;hsa-miR-96-5p | 12 | PRKCE | Sponge network | -0.351 | 0.11358 | -0.403 | 0.00056 | 0.276 |
| 47267 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 11 | AHNAK | Sponge network | 0.16 | 0.50785 | -1.207 | 0 | 0.276 |
| 47268 | RP11-720L2.4 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-501-3p;hsa-miR-625-5p | 12 | ABI2 | Sponge network | -1.255 | 0.00981 | 0.175 | 0.14787 | 0.276 |
| 47269 | AC106786.1 |
hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-629-5p | 10 | SOX5 | Sponge network | 1.122 | 0.02989 | -1.091 | 4.0E-5 | 0.276 |
| 47270 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | LRRC7 | Sponge network | 0.385 | 0.10067 | -1.341 | 0.01193 | 0.276 |
| 47271 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 23 | PRKAB2 | Sponge network | 0.614 | 1.0E-5 | -0.367 | 0.01371 | 0.276 |
| 47272 | RP11-276H19.2 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-484;hsa-miR-96-5p | 15 | MED12L | Sponge network | -0.472 | 0.48851 | -0.194 | 0.55299 | 0.276 |
| 47273 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-92a-3p | 19 | ARHGAP29 | Sponge network | 0.253 | 0.09565 | 0.131 | 0.42953 | 0.276 |
| 47274 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | LIFR | Sponge network | -0.223 | 0.47816 | -1.794 | 0 | 0.276 |
| 47275 | RP11-119F7.5 |
hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 12 | RPS6KA6 | Sponge network | 0.225 | 0.30644 | -2.989 | 0 | 0.276 |
| 47276 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p | 24 | ESR1 | Sponge network | 0.082 | 0.55397 | -1.035 | 3.0E-5 | 0.276 |
| 47277 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-222-5p | 10 | CBL | Sponge network | -0.309 | 0.1145 | 0.291 | 0.01267 | 0.276 |
| 47278 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 15 | SASH1 | Sponge network | -1.255 | 0.00981 | -0.502 | 0.00175 | 0.276 |
| 47279 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3613-5p | 11 | GPAM | Sponge network | 1.055 | 0 | 0.142 | 0.29732 | 0.276 |
| 47280 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 50 | CUX1 | Sponge network | -2.452 | 0 | -0.039 | 0.6705 | 0.276 |
| 47281 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p | 23 | MYBL1 | Sponge network | 0.541 | 0.00125 | 0.494 | 0.02152 | 0.276 |
| 47282 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 28 | JAZF1 | Sponge network | -0.513 | 0.04703 | -0.377 | 0.02677 | 0.276 |
| 47283 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-93-5p | 14 | LRRC55 | Sponge network | 0.051 | 0.83388 | -0.026 | 0.93163 | 0.276 |
| 47284 | HOXA-AS2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | MAPK10 | Sponge network | 0.61 | 0.01593 | -1.774 | 0 | 0.276 |
| 47285 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-96-5p | 25 | JAZF1 | Sponge network | 0.395 | 0.15451 | -0.377 | 0.02677 | 0.276 |
| 47286 | RP1-68D18.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-375;hsa-miR-429;hsa-miR-628-5p | 10 | BMPR2 | Sponge network | 1.987 | 0 | 0.206 | 0.0635 | 0.276 |
| 47287 | MAP3K14 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | ASTN1 | Sponge network | -0.295 | 0.02604 | -2.389 | 2.0E-5 | 0.276 |
| 47288 | RP11-458D21.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 10 | PLAG1 | Sponge network | 0.663 | 0.00073 | -0.315 | 0.26532 | 0.276 |
| 47289 | FAM157C |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p | 12 | TMTC3 | Sponge network | 0.91 | 0 | 0.123 | 0.22189 | 0.276 |
| 47290 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-199a-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-374b-5p | 12 | MIER3 | Sponge network | 0.917 | 0 | -0.212 | 0.04957 | 0.276 |
| 47291 | GS1-124K5.11 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-671-5p | 12 | PIK3IP1 | Sponge network | 0.494 | 0.00339 | -0.187 | 0.19945 | 0.276 |
| 47292 | USP3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-935;hsa-miR-940 | 31 | PTPRT | Sponge network | -0.162 | 0.47687 | -0.907 | 0.03727 | 0.276 |
| 47293 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-342-3p | 18 | MXI1 | Sponge network | 0.082 | 0.55397 | -0.987 | 0 | 0.276 |
| 47294 | LINC00621 |
hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | CBX5 | Sponge network | 0.131 | 0.8436 | 0.165 | 0.24446 | 0.276 |
| 47295 | RP11-888D10.4 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | UBE2D1 | Sponge network | 0.702 | 7.0E-5 | 0.468 | 0 | 0.276 |
| 47296 | BCYRN1 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 22 | RYR2 | Sponge network | 0.311 | 0.10344 | -1.466 | 0.00031 | 0.276 |
| 47297 | AC005618.6 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p | 10 | COL1A2 | Sponge network | 0.862 | 0.06213 | 1.991 | 0 | 0.276 |
| 47298 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PTPRD | Sponge network | -0.351 | 0.11358 | -1.005 | 0.00064 | 0.276 |
| 47299 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 61 | NTRK3 | Sponge network | 0.064 | 0.72934 | -1.77 | 3.0E-5 | 0.276 |
| 47300 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 46 | QKI | Sponge network | 0.537 | 0.00139 | -0.333 | 0.04111 | 0.276 |
| 47301 | DNM1P35 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 25 | PTPN14 | Sponge network | 0.051 | 0.83388 | 0.144 | 0.34595 | 0.276 |
| 47302 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | PDGFRA | Sponge network | -2.915 | 0.00015 | -0.372 | 0.0037 | 0.276 |
| 47303 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-877-5p | 16 | PTPRT | Sponge network | -1.116 | 0.23686 | -0.907 | 0.03727 | 0.276 |
| 47304 | TINCR |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-942-5p | 31 | TMEM132B | Sponge network | -0.664 | 0.14543 | -0.83 | 0.01084 | 0.276 |
| 47305 | RP11-996F15.2 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-940 | 21 | CREB1 | Sponge network | 0.983 | 0.00048 | 0.203 | 0.00537 | 0.276 |
| 47306 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | PRDM5 | Sponge network | -0.223 | 0.47816 | -1.09 | 0.00014 | 0.276 |
| 47307 | RP3-402G11.26 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-590-3p;hsa-miR-671-5p | 14 | DLC1 | Sponge network | -0.254 | 0.19303 | -0.382 | 0.05038 | 0.276 |
| 47308 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | USP12 | Sponge network | 1.378 | 0 | -0.102 | 0.40768 | 0.276 |
| 47309 | RP11-890B15.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-877-5p | 25 | CRTC1 | Sponge network | 0.101 | 0.60919 | -0.116 | 0.28412 | 0.276 |
| 47310 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | HECA | Sponge network | 0.222 | 0.29133 | -0.217 | 0.03463 | 0.276 |
| 47311 | CTD-2260A17.1 |
hsa-let-7b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-30a-3p;hsa-miR-3607-3p | 10 | RICTOR | Sponge network | 0.518 | 0.01048 | 0.388 | 0.00188 | 0.276 |
| 47312 | AC116366.6 |
hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-577;hsa-miR-628-5p;hsa-miR-629-3p | 17 | IL6ST | Sponge network | 0.329 | 0.11122 | -0.402 | 0.00676 | 0.276 |
| 47313 | CTD-2666L21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-493-5p | 19 | NCOA1 | Sponge network | 0.368 | 0.20093 | -0.432 | 0 | 0.276 |
| 47314 | MALAT1 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 14 | GLIPR1 | Sponge network | 0.782 | 0.00016 | 0.146 | 0.3276 | 0.276 |
| 47315 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | MCC | Sponge network | -0.125 | 0.49414 | -1.005 | 0 | 0.276 |
| 47316 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-493-5p | 10 | DMXL1 | Sponge network | 0.706 | 0 | -0.426 | 0.00056 | 0.276 |
| 47317 | ACTA2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 35 | ARHGEF12 | Sponge network | 0.194 | 0.64278 | 0.09 | 0.35693 | 0.276 |
| 47318 | PCA3 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | MSN | Sponge network | -0.709 | 0.18168 | -0.023 | 0.88422 | 0.276 |
| 47319 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 16 | ITGA5 | Sponge network | -0.899 | 6.0E-5 | 0.452 | 0.03524 | 0.276 |
| 47320 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p | 11 | TSC22D1 | Sponge network | 0.083 | 0.66405 | -0.529 | 2.0E-5 | 0.276 |
| 47321 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | TSHZ3 | Sponge network | 0.349 | 0.01695 | -0.556 | 0.01516 | 0.276 |
| 47322 | RP11-5C23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-429 | 13 | ZBTB4 | Sponge network | 0.274 | 0.07681 | -0.739 | 0 | 0.276 |
| 47323 | RP11-705C15.3 |
hsa-let-7g-3p;hsa-miR-151a-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-590-5p;hsa-miR-940 | 14 | GAS7 | Sponge network | 0.796 | 4.0E-5 | -0.555 | 0.01602 | 0.276 |
| 47324 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-143-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 10 | CLASP2 | Sponge network | 0.912 | 0.00014 | -0.038 | 0.71 | 0.276 |
| 47325 | RP11-488L18.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 10 | TMTC3 | Sponge network | 1.392 | 0 | 0.123 | 0.22189 | 0.276 |
| 47326 | RP11-67L2.2 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 19 | DYNC1I1 | Sponge network | 0.72 | 0.0001 | -1.12 | 0.00107 | 0.276 |
| 47327 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | ZFHX3 | Sponge network | -2.915 | 0.00015 | 0.389 | 0.01363 | 0.276 |
| 47328 | RP11-760H22.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-501-3p | 23 | ABI2 | Sponge network | 0.001 | 0.99753 | 0.175 | 0.14787 | 0.276 |
| 47329 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p | 35 | SPRY3 | Sponge network | -0.323 | 0.01372 | 0.016 | 0.91286 | 0.276 |
| 47330 | LINC01018 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 16 | IRS1 | Sponge network | -2.915 | 0.00015 | -0.026 | 0.88855 | 0.276 |
| 47331 | TINCR |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-93-3p;hsa-miR-940 | 29 | TOM1L2 | Sponge network | -0.664 | 0.14543 | -0.994 | 0 | 0.276 |
| 47332 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p | 11 | DIP2C | Sponge network | 1.294 | 0.0031 | -0.436 | 0.01713 | 0.276 |
| 47333 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 27 | CBFA2T3 | Sponge network | 0.997 | 0.00066 | -1.221 | 1.0E-5 | 0.276 |
| 47334 | HOXD-AS2 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-502-5p | 12 | SYT4 | Sponge network | -0.208 | 0.65592 | -3.207 | 0 | 0.276 |
| 47335 | CTA-204B4.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-590-3p | 23 | ZFX | Sponge network | 0.765 | 7.0E-5 | 0.09 | 0.42563 | 0.276 |
| 47336 | RP11-531A24.5 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-486-5p | 13 | DNMT3A | Sponge network | 0.75 | 0.00054 | 0.302 | 0.0262 | 0.276 |
| 47337 | RP11-102N12.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-7-1-3p | 16 | EYA4 | Sponge network | -0.351 | 0.11358 | 0.3 | 0.36876 | 0.276 |
| 47338 | CTC-429P9.3 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-589-5p | 20 | CTIF | Sponge network | 0.082 | 0.55397 | -0.389 | 0.00549 | 0.276 |
| 47339 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-539-5p | 13 | CLASP2 | Sponge network | 1.122 | 0 | -0.038 | 0.71 | 0.276 |
| 47340 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-505-3p;hsa-miR-7-1-3p | 12 | SON | Sponge network | 0.389 | 0.04293 | 0.168 | 0.04406 | 0.276 |
| 47341 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-342-3p | 11 | AKAP11 | Sponge network | 0.752 | 0 | 0.196 | 0.09825 | 0.276 |
| 47342 | RP5-994D16.9 |
hsa-miR-103a-2-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SMAD2 | Sponge network | 0.252 | 0.08508 | -0.128 | 0.12732 | 0.276 |
| 47343 | CTC-490E21.10 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-337-3p;hsa-miR-374b-5p | 12 | FOXP1 | Sponge network | 1.46 | 1.0E-5 | 0.042 | 0.74368 | 0.276 |
| 47344 | RP11-20I23.13 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-424-5p;hsa-miR-582-3p | 21 | ABL2 | Sponge network | 0.505 | 0.0014 | 0.82 | 0 | 0.276 |
| 47345 | FAM13A-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 26 | TOM1L2 | Sponge network | -0.83 | 0 | -0.994 | 0 | 0.276 |
| 47346 | RP11-384K6.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-940 | 14 | NOTCH2 | Sponge network | 0.946 | 0 | 0.138 | 0.35163 | 0.276 |
| 47347 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-582-5p | 15 | SPRY3 | Sponge network | 2.368 | 2.0E-5 | 0.016 | 0.91286 | 0.276 |
| 47348 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MTSS1 | Sponge network | -1.057 | 0.00029 | -0.81 | 0.00035 | 0.276 |
| 47349 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p | 20 | BCL6 | Sponge network | 0.72 | 0.0001 | -0.211 | 0.19489 | 0.276 |
| 47350 | BVES-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-493-5p | 15 | PPP2R5C | Sponge network | -2.62 | 0 | -0.314 | 3.0E-5 | 0.276 |
| 47351 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-424-5p | 11 | EYA4 | Sponge network | -4.463 | 0.00025 | 0.3 | 0.36876 | 0.276 |
| 47352 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 26 | MTSS1 | Sponge network | 0.374 | 0.20054 | -0.81 | 0.00035 | 0.276 |
| 47353 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 55 | TCF4 | Sponge network | 0.75 | 0.00054 | 0.016 | 0.92271 | 0.276 |
| 47354 | RP11-48B3.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-628-5p | 10 | CDH23 | Sponge network | 1.757 | 0 | -0.974 | 0.00034 | 0.276 |
| 47355 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 44 | BMPR2 | Sponge network | -0.125 | 0.49414 | 0.206 | 0.0635 | 0.276 |
| 47356 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 19 | DKK3 | Sponge network | -0.222 | 0.32445 | -0.052 | 0.77783 | 0.276 |
| 47357 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | SIK1 | Sponge network | -0.769 | 0.00035 | -0.98 | 0 | 0.276 |
| 47358 | RP11-77H9.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p | 10 | MYO9A | Sponge network | 1.677 | 0 | -0.147 | 0.32373 | 0.276 |
| 47359 | RP11-150O12.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-5p | 10 | ZFHX3 | Sponge network | 2.605 | 2.0E-5 | 0.389 | 0.01363 | 0.276 |
| 47360 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429 | 12 | UBE4B | Sponge network | -0.154 | 0.53954 | -0.235 | 0.007 | 0.276 |
| 47361 | U91328.19 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 10 | CDH10 | Sponge network | -0.303 | 0.21002 | -2.397 | 0 | 0.276 |
| 47362 | RP11-119F7.5 |
hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-502-5p;hsa-miR-92a-3p | 26 | IGFBP5 | Sponge network | 0.225 | 0.30644 | -0.806 | 0.00105 | 0.276 |
| 47363 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-542-3p | 13 | RARB | Sponge network | -0.517 | 0.02935 | -0.825 | 2.0E-5 | 0.276 |
| 47364 | RP11-53O19.1 |
hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-942-5p | 22 | OPCML | Sponge network | -0.405 | 0.22013 | -2.498 | 0 | 0.276 |
| 47365 | LINC00641 |
hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-874-3p | 10 | AJAP1 | Sponge network | -0.222 | 0.32445 | -0.561 | 0.04832 | 0.276 |
| 47366 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-27a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | DMXL1 | Sponge network | 1.148 | 2.0E-5 | -0.426 | 0.00056 | 0.276 |
| 47367 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-625-5p | 14 | CBX5 | Sponge network | 0.983 | 0.00048 | 0.165 | 0.24446 | 0.276 |
| 47368 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-874-3p | 10 | ARID5B | Sponge network | 2.364 | 0.00134 | 0.342 | 0.01676 | 0.276 |
| 47369 | RP4-613B23.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-409-3p | 11 | PHC3 | Sponge network | -0.309 | 0.1145 | 0.212 | 0.0499 | 0.276 |
| 47370 | LINC00654 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 70 | LPP | Sponge network | 0.374 | 0.20054 | -0.459 | 0.04475 | 0.276 |
| 47371 | LINC00174 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-625-5p | 11 | ABI2 | Sponge network | 1.253 | 0 | 0.175 | 0.14787 | 0.276 |
| 47372 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | RNF144A | Sponge network | -0.053 | 0.80685 | 0.594 | 0.0009 | 0.276 |
| 47373 | DNAJC27-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-374a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 12 | HOMER2 | Sponge network | -0.969 | 2.0E-5 | -2.698 | 0 | 0.276 |
| 47374 | RP11-401P9.1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-511-5p | 11 | ATRX | Sponge network | 1.524 | 0.1098 | 0.261 | 0.04408 | 0.276 |
| 47375 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 39 | SMAD2 | Sponge network | 0.349 | 0.01695 | -0.128 | 0.12732 | 0.276 |
| 47376 | AC003090.1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-629-3p | 11 | ITGA5 | Sponge network | -2.117 | 0.00161 | 0.452 | 0.03524 | 0.276 |
| 47377 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-375;hsa-miR-484;hsa-miR-629-3p;hsa-miR-96-5p | 20 | EBF3 | Sponge network | 0.41 | 0.02214 | -0.058 | 0.81437 | 0.276 |
| 47378 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-7-1-3p | 16 | NUAK1 | Sponge network | -1.249 | 7.0E-5 | 0.904 | 0 | 0.276 |
| 47379 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-98-5p | 33 | TGFBR3 | Sponge network | -0.18 | 0.20343 | -1.173 | 1.0E-5 | 0.276 |
| 47380 | RP11-723D22.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-342-3p | 10 | DTNA | Sponge network | 0.562 | 0.20117 | -1.648 | 1.0E-5 | 0.276 |
| 47381 | LINC00667 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484 | 18 | ABL1 | Sponge network | -0.235 | 0.15142 | -0.215 | 0.07804 | 0.276 |
| 47382 | CTD-3064M3.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-429 | 21 | AFF1 | Sponge network | -0.469 | 0.08721 | -0.384 | 0.00053 | 0.276 |
| 47383 | RP1-39G22.7 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-495-3p | 11 | SLC5A3 | Sponge network | 0.439 | 0.00313 | 0.493 | 5.0E-5 | 0.276 |
| 47384 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-93-5p | 32 | FAT3 | Sponge network | -0.663 | 0.10887 | -1.601 | 0.00051 | 0.276 |
| 47385 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375 | 20 | MLLT10 | Sponge network | -0.162 | 0.47687 | 0.105 | 0.33292 | 0.276 |
| 47386 | FAM13A-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 19 | PEG3 | Sponge network | -0.83 | 0 | -1.74 | 0 | 0.276 |
| 47387 | CTC-297N7.9 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-7-5p;hsa-miR-744-3p | 16 | IGFBP5 | Sponge network | -2.069 | 0 | -0.806 | 0.00105 | 0.276 |
| 47388 | RP11-53O19.1 |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-576-5p | 17 | PAIP2 | Sponge network | -0.405 | 0.22013 | -0.515 | 0 | 0.276 |
| 47389 | FAM13A-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 12 | TES | Sponge network | -0.83 | 0 | -0.348 | 0.00639 | 0.276 |
| 47390 | FAM13A-AS1 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-382-5p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 24 | ACSL6 | Sponge network | -0.83 | 0 | -1.593 | 0 | 0.276 |
| 47391 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | TRPS1 | Sponge network | -0.342 | 0.02288 | -0.191 | 0.44109 | 0.276 |
| 47392 | ZNF582-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 20 | SUFU | Sponge network | -1.349 | 0.00017 | -0.04 | 0.65489 | 0.276 |
| 47393 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 10 | MLLT10 | Sponge network | 1.11 | 1.0E-5 | 0.105 | 0.33292 | 0.276 |
| 47394 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | TSHZ3 | Sponge network | -0.023 | 0.95109 | -0.556 | 0.01516 | 0.276 |
| 47395 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ABCC9 | Sponge network | -0.125 | 0.49414 | -0.466 | 0.14121 | 0.276 |
| 47396 | MIR497HG |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 21 | MXI1 | Sponge network | -1.439 | 4.0E-5 | -0.987 | 0 | 0.276 |
| 47397 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-432-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 36 | UNC80 | Sponge network | -0.227 | 0.31657 | -1.934 | 0 | 0.276 |
| 47398 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 30 | MYBL1 | Sponge network | 0.72 | 0.0001 | 0.494 | 0.02152 | 0.276 |
| 47399 | LINC00473 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-942-5p | 13 | ERRFI1 | Sponge network | -1.203 | 0.03881 | -0.798 | 1.0E-5 | 0.276 |
| 47400 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 34 | CLASP2 | Sponge network | 0.311 | 0.10344 | -0.038 | 0.71 | 0.276 |
| 47401 | MEG3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-96-5p | 29 | CADM1 | Sponge network | 0.249 | 0.35902 | -0.9 | 0.00084 | 0.276 |
| 47402 | PCA3 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 12 | PCDH18 | Sponge network | -0.709 | 0.18168 | 0.216 | 0.24645 | 0.276 |
| 47403 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CACNA2D1 | Sponge network | 0.272 | 0.44692 | -0.846 | 0.00675 | 0.276 |
| 47404 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | KDSR | Sponge network | 0.222 | 0.29133 | 0.239 | 0.02216 | 0.276 |
| 47405 | RP11-967K21.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-582-5p | 16 | STARD13 | Sponge network | 0.056 | 0.81195 | -0.187 | 0.27383 | 0.276 |
| 47406 | LINC00865 |
hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429 | 12 | RIT1 | Sponge network | -1.249 | 7.0E-5 | -0.296 | 0.00054 | 0.276 |
| 47407 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 18 | SOX11 | Sponge network | 0.392 | 0.0278 | 2.68 | 0 | 0.276 |
| 47408 | EIF3J-AS1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 14 | SHPRH | Sponge network | 0.331 | 0.00509 | 0.098 | 0.40994 | 0.276 |
| 47409 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-338-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-625-5p | 25 | JAZF1 | Sponge network | -0.176 | 0.59692 | -0.377 | 0.02677 | 0.276 |
| 47410 | FAM13A-AS1 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-769-5p;hsa-miR-874-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-940;hsa-miR-99b-3p | 41 | GRIK3 | Sponge network | -0.83 | 0 | -3.208 | 0 | 0.276 |
| 47411 | CD27-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-629-3p | 20 | RYR2 | Sponge network | 0.177 | 0.16681 | -1.466 | 0.00031 | 0.276 |
| 47412 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | FREM2 | Sponge network | -0.899 | 6.0E-5 | -0.437 | 0.46353 | 0.276 |
| 47413 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 13 | FGFR1 | Sponge network | -0.602 | 0.00331 | -0.509 | 0.03957 | 0.276 |
| 47414 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-429 | 13 | SOX11 | Sponge network | 0.859 | 0.00331 | 2.68 | 0 | 0.276 |
| 47415 | RP11-428J1.5 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | RASSF8 | Sponge network | 0.538 | 0.00076 | -0.122 | 0.65591 | 0.276 |
| 47416 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | TIMP3 | Sponge network | -0.615 | 0.00015 | -0.546 | 0.02307 | 0.276 |
| 47417 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-3p | 16 | CLASP2 | Sponge network | 1.361 | 0 | -0.038 | 0.71 | 0.276 |
| 47418 | RP3-402G11.26 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | NTRK2 | Sponge network | -0.254 | 0.19303 | -0.736 | 0.06495 | 0.276 |
| 47419 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-7-1-3p | 10 | PTPRB | Sponge network | 0.817 | 3.0E-5 | 0.105 | 0.47122 | 0.276 |
| 47420 | AC025171.1 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 29 | IL6ST | Sponge network | 0.998 | 0 | -0.402 | 0.00676 | 0.276 |
| 47421 | RP3-402G11.26 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-590-3p | 11 | EPHA7 | Sponge network | -0.254 | 0.19303 | -2.197 | 3.0E-5 | 0.276 |
| 47422 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-214-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 30 | PPM1A | Sponge network | 0.401 | 0.00066 | -0.623 | 0 | 0.276 |
| 47423 | CTC-487M23.5 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-92a-3p | 16 | FOXP1 | Sponge network | 0.912 | 0.00014 | 0.042 | 0.74368 | 0.276 |
| 47424 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SEMA3C | Sponge network | 0.998 | 0 | -0.272 | 0.31153 | 0.276 |
| 47425 | CD27-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-493-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 38 | CHD5 | Sponge network | 0.177 | 0.16681 | -0.922 | 0.01521 | 0.276 |
| 47426 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 14 | BCL6 | Sponge network | -0.141 | 0.26434 | -0.211 | 0.19489 | 0.276 |
| 47427 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-671-5p | 23 | ADARB1 | Sponge network | -0.164 | 0.45106 | -0.335 | 0.06631 | 0.276 |
| 47428 | RP11-983P16.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | NAV3 | Sponge network | 0.41 | 0.02214 | 0.212 | 0.47729 | 0.276 |
| 47429 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 41 | SOX5 | Sponge network | -0.355 | 0.0077 | -1.091 | 4.0E-5 | 0.276 |
| 47430 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-93-5p | 13 | MAP3K12 | Sponge network | 0.4 | 0.14611 | -0.057 | 0.59369 | 0.276 |
| 47431 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | RIMBP2 | Sponge network | -0.355 | 0.0077 | -1.968 | 5.0E-5 | 0.276 |
| 47432 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 14 | SEPT6 | Sponge network | 0.997 | 0.00066 | -0.598 | 0.00181 | 0.276 |
| 47433 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-501-5p | 11 | ERRFI1 | Sponge network | -0.246 | 0.35034 | -0.798 | 1.0E-5 | 0.276 |
| 47434 | PWAR6 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | GABRB2 | Sponge network | -1.575 | 6.0E-5 | -1.841 | 0.00029 | 0.276 |
| 47435 | USP3-AS1 |
hsa-let-7a-5p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-421;hsa-miR-671-5p;hsa-miR-877-5p | 16 | THRA | Sponge network | -0.162 | 0.47687 | -0.307 | 0.05099 | 0.276 |
| 47436 | AC004540.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-3614-5p;hsa-miR-629-3p | 17 | IGFBP5 | Sponge network | -2.549 | 0.00528 | -0.806 | 0.00105 | 0.276 |
| 47437 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 12 | PDGFRA | Sponge network | 1.122 | 0.02989 | -0.372 | 0.0037 | 0.276 |
| 47438 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 10 | FYN | Sponge network | 0.898 | 1.0E-5 | -0.131 | 0.37051 | 0.276 |
| 47439 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 27 | MTSS1 | Sponge network | -0.842 | 0.01361 | -0.81 | 0.00035 | 0.276 |
| 47440 | AC156455.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30e-3p;hsa-miR-335-5p | 21 | CCND2 | Sponge network | 1.154 | 0.00041 | -0.267 | 0.3112 | 0.276 |
| 47441 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 47 | THRB | Sponge network | -0.227 | 0.31657 | -1.117 | 0.0004 | 0.276 |
| 47442 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | ACVR2A | Sponge network | -0.487 | 0.02157 | -0.409 | 0.00039 | 0.276 |
| 47443 | LINC00865 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | EPAS1 | Sponge network | -1.249 | 7.0E-5 | -0.184 | 0.14418 | 0.276 |
| 47444 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CACNA2D1 | Sponge network | -0.053 | 0.80685 | -0.846 | 0.00675 | 0.276 |
| 47445 | ADAMTS9-AS2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | ITGB1 | Sponge network | -2.452 | 0 | 0.524 | 0.00017 | 0.276 |
| 47446 | CTC-559E9.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 13 | SYNE1 | Sponge network | 0.594 | 0.00064 | -0.738 | 0.00316 | 0.276 |
| 47447 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-655-3p | 30 | LATS1 | Sponge network | -1.111 | 0.0003 | 0.109 | 0.34208 | 0.276 |
| 47448 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-590-3p | 11 | CDHR3 | Sponge network | 0.178 | 0.29106 | -0.977 | 4.0E-5 | 0.276 |
| 47449 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-576-5p | 10 | PHC3 | Sponge network | 1.361 | 0 | 0.212 | 0.0499 | 0.276 |
| 47450 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 14 | MAP3K12 | Sponge network | -0.612 | 0.00094 | -0.057 | 0.59369 | 0.276 |
| 47451 | RP5-1021I20.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-429 | 15 | RECK | Sponge network | -0.785 | 0.00035 | -1.043 | 0 | 0.275 |
| 47452 | RP11-400K9.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | MED12L | Sponge network | -0.733 | 0.19469 | -0.194 | 0.55299 | 0.275 |
| 47453 | DIO3OS |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p | 19 | HIP1 | Sponge network | -0.773 | 0.04073 | 0.774 | 0 | 0.275 |
| 47454 | ACTA2-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-7-5p | 26 | BTG2 | Sponge network | 0.194 | 0.64278 | -1.763 | 0 | 0.275 |
| 47455 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 17 | MOB1B | Sponge network | 0.917 | 0 | 0.197 | 0.15266 | 0.275 |
| 47456 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | SON | Sponge network | -0.487 | 0.02157 | 0.168 | 0.04406 | 0.275 |
| 47457 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p | 26 | EBF3 | Sponge network | -2.925 | 0 | -0.058 | 0.81437 | 0.275 |
| 47458 | U91328.19 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-589-5p | 11 | ROBO1 | Sponge network | -0.303 | 0.21002 | -0.009 | 0.96154 | 0.275 |
| 47459 | RP11-263K19.4 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-429 | 10 | MRVI1 | Sponge network | 0.859 | 0.00331 | -1.051 | 0.00022 | 0.275 |
| 47460 | RP11-57H14.4 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 11 | KCTD8 | Sponge network | 0.083 | 0.66405 | -3.359 | 0 | 0.275 |
| 47461 | RP11-439L18.2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p | 10 | HIP1 | Sponge network | 0.224 | 0.48719 | 0.774 | 0 | 0.275 |
| 47462 | RP11-34P13.14 |
hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p | 12 | KIF1B | Sponge network | 1.888 | 0 | -0.118 | 0.32258 | 0.275 |
| 47463 | AC108676.1 |
hsa-miR-126-5p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-766-3p | 15 | TBL1XR1 | Sponge network | 4.346 | 2.0E-5 | 0.578 | 0 | 0.275 |
| 47464 | CTD-3064M3.3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p | 12 | BCL9 | Sponge network | -0.469 | 0.08721 | 0.321 | 0.00267 | 0.275 |
| 47465 | RP11-25K19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | -1.057 | 0.00029 | -0.684 | 0.00159 | 0.275 |
| 47466 | RP11-473I1.9 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | CSDE1 | Sponge network | 0.683 | 1.0E-5 | -0.493 | 0 | 0.275 |
| 47467 | RP11-968O1.5 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p | 15 | CTIF | Sponge network | -0.829 | 0.01343 | -0.389 | 0.00549 | 0.275 |
| 47468 | RP11-20I23.13 |
hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-92a-3p | 10 | FLNA | Sponge network | 0.505 | 0.0014 | -0.771 | 0.01377 | 0.275 |
| 47469 | RP11-554A11.9 |
hsa-let-7d-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-331-3p | 10 | CBFA2T3 | Sponge network | -2.279 | 0.00011 | -1.221 | 1.0E-5 | 0.275 |
| 47470 | FAM157B |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326 | 15 | ZFX | Sponge network | 0.92 | 0 | 0.09 | 0.42563 | 0.275 |
| 47471 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-664a-5p | 21 | ADARB1 | Sponge network | 1.316 | 0.01106 | -0.335 | 0.06631 | 0.275 |
| 47472 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-424-5p | 10 | RARB | Sponge network | -0.322 | 0.25945 | -0.825 | 2.0E-5 | 0.275 |
| 47473 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p | 20 | SMAD2 | Sponge network | 2.163 | 0 | -0.128 | 0.12732 | 0.275 |
| 47474 | BCYRN1 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | DNMT3A | Sponge network | 0.311 | 0.10344 | 0.302 | 0.0262 | 0.275 |
| 47475 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 27 | USP12 | Sponge network | 0.177 | 0.16681 | -0.102 | 0.40768 | 0.275 |
| 47476 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 40 | AR | Sponge network | 0.222 | 0.29133 | -1.554 | 0 | 0.275 |
| 47477 | RP5-994D16.9 |
hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ANK2 | Sponge network | 0.252 | 0.08508 | -1.603 | 1.0E-5 | 0.275 |
| 47478 | TINCR |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-5p;hsa-miR-96-5p | 24 | MOB1B | Sponge network | -0.664 | 0.14543 | 0.197 | 0.15266 | 0.275 |
| 47479 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | RIMBP2 | Sponge network | 0.083 | 0.66405 | -1.968 | 5.0E-5 | 0.275 |
| 47480 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-3p | 22 | DOCK4 | Sponge network | -0.615 | 0.00015 | 0.502 | 0.00108 | 0.275 |
| 47481 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 29 | DIP2C | Sponge network | 0.523 | 9.0E-5 | -0.436 | 0.01713 | 0.275 |
| 47482 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | RABEP1 | Sponge network | 0.782 | 0.00016 | -0.066 | 0.43142 | 0.275 |
| 47483 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -0.793 | 7.0E-5 | -0.243 | 0.11408 | 0.275 |
| 47484 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-744-3p | 10 | CUL5 | Sponge network | 2.163 | 0 | 0.026 | 0.77301 | 0.275 |
| 47485 | LINC00265 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-484 | 14 | ZFX | Sponge network | 1.07 | 0 | 0.09 | 0.42563 | 0.275 |
| 47486 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 22 | MXI1 | Sponge network | -0.611 | 0.09607 | -0.987 | 0 | 0.275 |
| 47487 | RP11-327P2.5 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-5p | 11 | RET | Sponge network | -0.147 | 0.40452 | -0.833 | 0.03517 | 0.275 |
| 47488 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 32 | PHC3 | Sponge network | -0.405 | 0.22013 | 0.212 | 0.0499 | 0.275 |
| 47489 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-625-5p | 17 | DCLK1 | Sponge network | -0.969 | 2.0E-5 | -1.324 | 0.00014 | 0.275 |
| 47490 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-96-5p | 19 | NUAK1 | Sponge network | 0.082 | 0.55654 | 0.904 | 0 | 0.275 |
| 47491 | RP11-344B5.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-361-3p;hsa-miR-423-5p | 11 | SUFU | Sponge network | -0.055 | 0.88482 | -0.04 | 0.65489 | 0.275 |
| 47492 | RP11-400K9.4 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-502-5p;hsa-miR-576-5p | 12 | PTCH1 | Sponge network | -0.611 | 0.09607 | -0.1 | 0.6179 | 0.275 |
| 47493 | RP11-359B12.2 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | ERCC6 | Sponge network | 0.064 | 0.72934 | 0.23 | 0.015 | 0.275 |
| 47494 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 26 | TMTC3 | Sponge network | 0.997 | 0.00066 | 0.123 | 0.22189 | 0.275 |
| 47495 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | PDGFRA | Sponge network | 0.082 | 0.55654 | -0.372 | 0.0037 | 0.275 |
| 47496 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 22 | TACC1 | Sponge network | -4.477 | 0 | -0.362 | 0.05406 | 0.275 |
| 47497 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p | 20 | KCNA6 | Sponge network | -0.206 | 0.12942 | -1.531 | 0.00013 | 0.275 |
| 47498 | CTD-3126B10.4 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | ZFHX4 | Sponge network | 0.778 | 0.00101 | 0.018 | 0.95574 | 0.275 |
| 47499 | LPP-AS2 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-493-5p | 19 | LIMCH1 | Sponge network | -0.18 | 0.20343 | -0.915 | 1.0E-5 | 0.275 |
| 47500 | AC025171.1 |
hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 10 | ZNF292 | Sponge network | 0.998 | 0 | 0.276 | 0.04148 | 0.275 |
| 47501 | TPTEP1 |
hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-7-5p | 13 | PIM1 | Sponge network | -1.216 | 0 | -1.114 | 0 | 0.275 |
| 47502 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p | 18 | BCL6 | Sponge network | 0.222 | 0.29133 | -0.211 | 0.19489 | 0.275 |
| 47503 | RP11-890B15.3 |
hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | PCDH18 | Sponge network | 0.101 | 0.60919 | 0.216 | 0.24645 | 0.275 |
| 47504 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | EGLN1 | Sponge network | 1.2 | 0.01813 | -0.127 | 0.1536 | 0.275 |
| 47505 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.541 | 0.00125 | 0.323 | 0.00792 | 0.275 |
| 47506 | AC005562.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | ZFX | Sponge network | 0.488 | 0.00084 | 0.09 | 0.42563 | 0.275 |
| 47507 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | PRKAB2 | Sponge network | 0.569 | 0.00067 | -0.367 | 0.01371 | 0.275 |
| 47508 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 12 | AKAP13 | Sponge network | -0.351 | 0.11358 | 0.028 | 0.7729 | 0.275 |
| 47509 | RP11-48B3.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 16 | SYNPO2 | Sponge network | 1.757 | 0 | -2.342 | 0 | 0.275 |
| 47510 | RP1-151F17.2 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RIT1 | Sponge network | 0.222 | 0.29133 | -0.296 | 0.00054 | 0.275 |
| 47511 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-92a-3p | 12 | ITGA5 | Sponge network | -0.513 | 0.04703 | 0.452 | 0.03524 | 0.275 |
| 47512 | RP5-1085F17.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-766-3p | 11 | KIAA1024 | Sponge network | 0.541 | 0.00125 | 1.083 | 0 | 0.275 |
| 47513 | RP11-686D22.8 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | LATS2 | Sponge network | 0.898 | 1.0E-5 | 0.159 | 0.2304 | 0.275 |
| 47514 | HOXB-AS3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-511-5p | 12 | EYA4 | Sponge network | 0.343 | 0.28887 | 0.3 | 0.36876 | 0.275 |
| 47515 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITGAV | Sponge network | 0.91 | 0 | 0.337 | 0.01002 | 0.275 |
| 47516 | RP11-212P7.2 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 34 | GRIK3 | Sponge network | 0.219 | 0.1516 | -3.208 | 0 | 0.275 |
| 47517 | AC034220.3 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ITGB1 | Sponge network | 0.309 | 0.07304 | 0.524 | 0.00017 | 0.275 |
| 47518 | C4B-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-191-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-96-5p | 11 | CBFA2T3 | Sponge network | 2.306 | 9.0E-5 | -1.221 | 1.0E-5 | 0.275 |
| 47519 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-320b | 10 | APC | Sponge network | 1.044 | 2.0E-5 | -0.255 | 0.04051 | 0.275 |
| 47520 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-629-5p | 27 | UNC80 | Sponge network | -4.477 | 0 | -1.934 | 0 | 0.275 |
| 47521 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-493-5p | 17 | AR | Sponge network | 1.46 | 1.0E-5 | -1.554 | 0 | 0.275 |
| 47522 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 20 | JDP2 | Sponge network | -0.668 | 3.0E-5 | -0.967 | 0 | 0.275 |
| 47523 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 10 | GPAM | Sponge network | 0.993 | 0 | 0.142 | 0.29732 | 0.275 |
| 47524 | RP11-1006G14.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | SETBP1 | Sponge network | 0.559 | 0.00026 | -1.302 | 1.0E-5 | 0.275 |
| 47525 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-660-5p;hsa-miR-96-5p | 20 | RIMBP2 | Sponge network | -0.969 | 2.0E-5 | -1.968 | 5.0E-5 | 0.275 |
| 47526 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | SOX11 | Sponge network | 0.381 | 0.25967 | 2.68 | 0 | 0.275 |
| 47527 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 19 | SOX11 | Sponge network | 0.8 | 0 | 2.68 | 0 | 0.275 |
| 47528 | RBPMS-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 19 | ZFHX4 | Sponge network | 0.144 | 0.64989 | 0.018 | 0.95574 | 0.275 |
| 47529 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p | 11 | PRKAA1 | Sponge network | 0.702 | 7.0E-5 | 0.317 | 0.0031 | 0.275 |
| 47530 | A1BG-AS1 |
hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-589-3p;hsa-miR-671-5p | 12 | THBS1 | Sponge network | -0.164 | 0.45106 | 0.741 | 0.00386 | 0.275 |
| 47531 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | TLL1 | Sponge network | 0.214 | 0.18246 | -1.195 | 3.0E-5 | 0.275 |
| 47532 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 33 | PHC3 | Sponge network | -0.899 | 6.0E-5 | 0.212 | 0.0499 | 0.275 |
| 47533 | RP11-156P1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-5p | 19 | KCNA6 | Sponge network | 0.214 | 0.18246 | -1.531 | 0.00013 | 0.275 |
| 47534 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TBX18 | Sponge network | 0.222 | 0.29133 | 0.843 | 0.02945 | 0.275 |
| 47535 | HCG11 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-664a-3p | 16 | RASAL2 | Sponge network | 0.385 | 0.10067 | 0.869 | 0 | 0.275 |
| 47536 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577 | 20 | GREM1 | Sponge network | 0.72 | 0.0001 | 0.902 | 0.01691 | 0.275 |
| 47537 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-7-1-3p | 13 | CBL | Sponge network | 1.378 | 0 | 0.291 | 0.01267 | 0.275 |
| 47538 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p | 10 | CLIP1 | Sponge network | 1.254 | 0 | -0.215 | 0.08684 | 0.275 |
| 47539 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | KIF1B | Sponge network | 0.538 | 0.00076 | -0.118 | 0.32258 | 0.275 |
| 47540 | CTD-2017D11.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-339-5p | 13 | TSC1 | Sponge network | 0.792 | 0.0049 | 0.103 | 0.26071 | 0.275 |
| 47541 | GS1-124K5.11 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 12 | LUZP2 | Sponge network | 0.494 | 0.00339 | -0.056 | 0.89416 | 0.275 |
| 47542 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 22 | DMXL1 | Sponge network | 0.657 | 4.0E-5 | -0.426 | 0.00056 | 0.275 |
| 47543 | LINC00578 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577 | 13 | PHC3 | Sponge network | 0.811 | 0.03228 | 0.212 | 0.0499 | 0.275 |
| 47544 | RP1-102E24.8 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326 | 10 | SLC5A3 | Sponge network | 1.737 | 0 | 0.493 | 5.0E-5 | 0.275 |
| 47545 | AC116366.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | RUNX1T1 | Sponge network | 0.329 | 0.11122 | -0.344 | 0.2374 | 0.275 |
| 47546 | RP11-47A8.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p | 10 | FAT4 | Sponge network | 0.103 | 0.43169 | -0.354 | 0.14694 | 0.275 |
| 47547 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p | 12 | LRRK2 | Sponge network | 0.083 | 0.66405 | -1.136 | 1.0E-5 | 0.275 |
| 47548 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 30 | ERC1 | Sponge network | 1.302 | 7.0E-5 | -0.108 | 0.38794 | 0.275 |
| 47549 | ARHGEF26-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 48 | CUX1 | Sponge network | -2.46 | 0 | -0.039 | 0.6705 | 0.275 |
| 47550 | RP11-2B6.2 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-550a-5p;hsa-miR-590-3p | 25 | CCDC6 | Sponge network | 0.539 | 0.03722 | 0.27 | 0.02555 | 0.275 |
| 47551 | RP11-327P2.5 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-486-5p | 15 | EZH1 | Sponge network | -0.147 | 0.40452 | -0.564 | 1.0E-5 | 0.275 |
| 47552 | RP11-21A7A.2 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-26a-2-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-877-5p;hsa-miR-93-3p | 13 | BMPR2 | Sponge network | -1.116 | 0.23686 | 0.206 | 0.0635 | 0.275 |
| 47553 | U91328.19 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p | 20 | PIK3CA | Sponge network | -0.303 | 0.21002 | 0.195 | 0.06908 | 0.275 |
| 47554 | TRAF3IP2-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 20 | UBE4B | Sponge network | -0.323 | 0.01372 | -0.235 | 0.007 | 0.275 |
| 47555 | CTC-378H22.2 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-324-3p;hsa-miR-378a-5p | 12 | ADARB1 | Sponge network | 0.001 | 0.99663 | -0.335 | 0.06631 | 0.275 |
| 47556 | RP11-47A8.5 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 14 | GAS7 | Sponge network | 0.103 | 0.43169 | -0.555 | 0.01602 | 0.275 |
| 47557 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-484;hsa-miR-493-5p;hsa-miR-590-3p | 15 | MYCBP2 | Sponge network | -0.615 | 0.00015 | -0.027 | 0.82016 | 0.275 |
| 47558 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-497-5p | 20 | AFF4 | Sponge network | 0.997 | 0 | -0.266 | 0.01565 | 0.275 |
| 47559 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-191-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-375;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TET1 | Sponge network | 0.673 | 0 | -0.135 | 0.52138 | 0.275 |
| 47560 | GUSBP11 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | UBR3 | Sponge network | 0.856 | 0 | -0.021 | 0.82774 | 0.275 |
| 47561 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-744-3p;hsa-miR-96-5p | 12 | HIP1 | Sponge network | 0.622 | 0.00015 | 0.774 | 0 | 0.275 |
| 47562 | RP11-411K7.1 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | KIF5B | Sponge network | 0.155 | 0.81273 | 0.362 | 0.00066 | 0.275 |
| 47563 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940;hsa-miR-98-5p | 23 | THBS1 | Sponge network | -0.773 | 0.04073 | 0.741 | 0.00386 | 0.275 |
| 47564 | MIR22HG |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | PCDH9 | Sponge network | -1.047 | 0 | -1.823 | 4.0E-5 | 0.275 |
| 47565 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-340-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-935 | 19 | RNF144A | Sponge network | -0.096 | 0.67958 | 0.594 | 0.0009 | 0.275 |
| 47566 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-3p | 13 | SUFU | Sponge network | -2.531 | 0 | -0.04 | 0.65489 | 0.275 |
| 47567 | CTD-3126B10.4 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | KIF1B | Sponge network | 0.778 | 0.00101 | -0.118 | 0.32258 | 0.275 |
| 47568 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | TGFBR2 | Sponge network | 0.523 | 9.0E-5 | -0.243 | 0.11408 | 0.275 |
| 47569 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 13 | HECA | Sponge network | -0.663 | 0.10887 | -0.217 | 0.03463 | 0.275 |
| 47570 | RP11-156E6.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p | 16 | ZFX | Sponge network | 1.266 | 0 | 0.09 | 0.42563 | 0.275 |
| 47571 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-582-5p | 14 | EBF3 | Sponge network | 0.792 | 0.0049 | -0.058 | 0.81437 | 0.275 |
| 47572 | HCG11 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | SEMA3E | Sponge network | 0.385 | 0.10067 | -1.717 | 0.00137 | 0.275 |
| 47573 | RP11-774O3.3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | BCL9 | Sponge network | -0.487 | 0.02157 | 0.321 | 0.00267 | 0.275 |
| 47574 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | EGLN1 | Sponge network | 0.349 | 0.01695 | -0.127 | 0.1536 | 0.275 |
| 47575 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | CREBBP | Sponge network | -0.513 | 0.04703 | 0.126 | 0.15186 | 0.275 |
| 47576 | ATP1B3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-577;hsa-miR-96-5p | 14 | IL17RD | Sponge network | 1.2 | 0.01813 | -0.019 | 0.94873 | 0.275 |
| 47577 | RP11-720L2.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-33a-3p | 10 | MED12L | Sponge network | -1.255 | 0.00981 | -0.194 | 0.55299 | 0.275 |
| 47578 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p | 11 | GPAM | Sponge network | 1.206 | 0 | 0.142 | 0.29732 | 0.275 |
| 47579 | LINC00621 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | TMEM132B | Sponge network | 0.131 | 0.8436 | -0.83 | 0.01084 | 0.275 |
| 47580 | RP11-927P21.1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-375;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-629-3p | 15 | RYR2 | Sponge network | 0.921 | 0.00118 | -1.466 | 0.00031 | 0.275 |
| 47581 | PDXDC2P |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-299-5p | 10 | MAT2A | Sponge network | 0.756 | 0 | -0.073 | 0.36195 | 0.275 |
| 47582 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 45 | NTRK3 | Sponge network | -0.223 | 0.47816 | -1.77 | 3.0E-5 | 0.275 |
| 47583 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p | 12 | PHC3 | Sponge network | 0.972 | 0 | 0.212 | 0.0499 | 0.275 |
| 47584 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | MCC | Sponge network | -0.223 | 0.47816 | -1.005 | 0 | 0.275 |
| 47585 | RBPMS-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | FAT4 | Sponge network | 0.144 | 0.64989 | -0.354 | 0.14694 | 0.275 |
| 47586 | AC006547.13 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-3607-3p | 14 | MOB1B | Sponge network | 1.159 | 0 | 0.197 | 0.15266 | 0.275 |
| 47587 | LPP-AS2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-7-5p;hsa-miR-93-3p | 23 | ERC1 | Sponge network | -0.18 | 0.20343 | -0.108 | 0.38794 | 0.275 |
| 47588 | RP11-362F19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-7-1-3p | 16 | RASSF8 | Sponge network | -0.012 | 0.98664 | -0.122 | 0.65591 | 0.275 |
| 47589 | RP11-479G22.8 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-766-3p | 16 | IL17RD | Sponge network | 1.308 | 0 | -0.019 | 0.94873 | 0.275 |
| 47590 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-96-5p | 18 | ST6GALNAC3 | Sponge network | -0.517 | 0.02935 | -0.987 | 0 | 0.275 |
| 47591 | LPP-AS2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p | 13 | ROBO1 | Sponge network | -0.18 | 0.20343 | -0.009 | 0.96154 | 0.275 |
| 47592 | RP11-20I23.13 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-486-5p | 11 | AKAP6 | Sponge network | 0.505 | 0.0014 | -1.586 | 3.0E-5 | 0.275 |
| 47593 | AP001055.6 |
hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-455-3p;hsa-miR-589-5p | 10 | CTIF | Sponge network | 0.693 | 0.00076 | -0.389 | 0.00549 | 0.275 |
| 47594 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 36 | CBL | Sponge network | -0.737 | 0.06182 | 0.291 | 0.01267 | 0.275 |
| 47595 | LINC00094 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-624-5p | 18 | GJA1 | Sponge network | 0.253 | 0.09565 | -0.485 | 0.02179 | 0.275 |
| 47596 | RP11-212P7.2 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | MAPK10 | Sponge network | 0.219 | 0.1516 | -1.774 | 0 | 0.275 |
| 47597 | LINC01001 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-539-5p | 15 | ARHGEF12 | Sponge network | 1.055 | 0 | 0.09 | 0.35693 | 0.275 |
| 47598 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | ARHGAP29 | Sponge network | 0.064 | 0.72934 | 0.131 | 0.42953 | 0.275 |
| 47599 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-7-1-3p | 23 | NTRK3 | Sponge network | -0.557 | 0.11135 | -1.77 | 3.0E-5 | 0.275 |
| 47600 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-590-3p | 16 | FOXO3 | Sponge network | -0.358 | 0.17255 | -0.04 | 0.72028 | 0.275 |
| 47601 | RP11-464F9.1 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | RB1 | Sponge network | 0.959 | 0 | 0.382 | 0.0017 | 0.275 |
| 47602 | RP11-439A17.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-194-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-576-5p | 10 | PTPRD | Sponge network | 0.051 | 0.95756 | -1.005 | 0.00064 | 0.275 |
| 47603 | CTC-487M23.5 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-326 | 15 | SPRY3 | Sponge network | 0.912 | 0.00014 | 0.016 | 0.91286 | 0.275 |
| 47604 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-93-5p | 21 | ANK2 | Sponge network | -0.785 | 0.00035 | -1.603 | 1.0E-5 | 0.275 |
| 47605 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-93-3p | 17 | EBF3 | Sponge network | 0.75 | 0.00054 | -0.058 | 0.81437 | 0.275 |
| 47606 | PSMD5-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p | 12 | UBE4B | Sponge network | 0.569 | 0.00067 | -0.235 | 0.007 | 0.275 |
| 47607 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CLASP2 | Sponge network | 0.986 | 0.01022 | -0.038 | 0.71 | 0.275 |
| 47608 | HCG22 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | QKI | Sponge network | 0.816 | 0.32368 | -0.333 | 0.04111 | 0.275 |
| 47609 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 23 | ARHGAP28 | Sponge network | 0.246 | 0.59912 | -0.911 | 0.00013 | 0.275 |
| 47610 | FAM13A-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p | 16 | SPG20 | Sponge network | -0.83 | 0 | -1.16 | 0 | 0.275 |
| 47611 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 49 | BMPR2 | Sponge network | 0.16 | 0.50785 | 0.206 | 0.0635 | 0.275 |
| 47612 | LINC00667 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | TGFBR3 | Sponge network | -0.235 | 0.15142 | -1.173 | 1.0E-5 | 0.275 |
| 47613 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p | 10 | NALCN | Sponge network | 0.537 | 0.00139 | 1.068 | 0.00237 | 0.275 |
| 47614 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 47 | WDFY3 | Sponge network | 0.225 | 0.30644 | -0.201 | 0.0967 | 0.275 |
| 47615 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 20 | TGFBR3 | Sponge network | -1.375 | 0.08642 | -1.173 | 1.0E-5 | 0.275 |
| 47616 | CTC-559E9.5 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-708-5p;hsa-miR-9-5p | 24 | CCDC6 | Sponge network | 0.594 | 0.00064 | 0.27 | 0.02555 | 0.275 |
| 47617 | USP3-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-935;hsa-miR-942-5p | 22 | OPCML | Sponge network | -0.162 | 0.47687 | -2.498 | 0 | 0.275 |
| 47618 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-96-5p | 23 | PRKAA2 | Sponge network | 0.37 | 0.27614 | -2.462 | 0 | 0.275 |
| 47619 | HCG11 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-378a-3p;hsa-miR-532-3p | 13 | RPS6KA2 | Sponge network | 0.385 | 0.10067 | -0.57 | 0.00013 | 0.275 |
| 47620 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | CLASP2 | Sponge network | -1.161 | 0.00591 | -0.038 | 0.71 | 0.275 |
| 47621 | CALML3-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-664a-3p | 39 | PPM1L | Sponge network | 0.95 | 0.15943 | -0.537 | 0.00616 | 0.275 |
| 47622 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p | 11 | GPC6 | Sponge network | 0.497 | 0.01451 | -0.054 | 0.81465 | 0.275 |
| 47623 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | CHD2 | Sponge network | 0.249 | 0.35902 | 0.049 | 0.55371 | 0.275 |
| 47624 | HHIP-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | LRP1B | Sponge network | -0.737 | 0.10822 | -2.163 | 0 | 0.275 |
| 47625 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PRRX1 | Sponge network | 0.225 | 0.30644 | 1.316 | 0 | 0.275 |
| 47626 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | MED12L | Sponge network | -0.031 | 0.89063 | -0.194 | 0.55299 | 0.275 |
| 47627 | CTD-2291D10.4 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-96-5p | 10 | ABI2 | Sponge network | 3.21 | 0 | 0.175 | 0.14787 | 0.275 |
| 47628 | RP11-403I13.7 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | TLL1 | Sponge network | 0.395 | 0.15451 | -1.195 | 3.0E-5 | 0.275 |
| 47629 | CTB-113P19.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-589-3p | 12 | PGR | Sponge network | 0.199 | 0.52763 | -1.704 | 0 | 0.275 |
| 47630 | RP11-403I13.8 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | LATS2 | Sponge network | 0.619 | 0.00157 | 0.159 | 0.2304 | 0.275 |
| 47631 | RP11-53O19.1 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-628-5p | 15 | IRS1 | Sponge network | -0.405 | 0.22013 | -0.026 | 0.88855 | 0.275 |
| 47632 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-511-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 15 | LRRC4 | Sponge network | -0.513 | 0.04703 | -1.774 | 0 | 0.275 |
| 47633 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-378a-5p | 14 | CACNA1E | Sponge network | -0.49 | 0.04946 | 1.752 | 0 | 0.275 |
| 47634 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-27b-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-7-1-3p | 18 | CNTN1 | Sponge network | -0.13 | 0.48734 | -2.594 | 0 | 0.275 |
| 47635 | AC005618.6 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | IGSF10 | Sponge network | 0.862 | 0.06213 | -1.751 | 1.0E-5 | 0.275 |
| 47636 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-940 | 13 | CSRNP1 | Sponge network | -0.896 | 0.00099 | -1.448 | 0 | 0.275 |
| 47637 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-151a-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 17 | GAS7 | Sponge network | -0.301 | 0.06597 | -0.555 | 0.01602 | 0.275 |
| 47638 | RP11-248G5.8 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-342-3p | 10 | TET1 | Sponge network | 1.294 | 0 | -0.135 | 0.52138 | 0.275 |
| 47639 | C20orf166-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577 | 16 | FMNL3 | Sponge network | -2.69 | 0.0001 | 0.555 | 4.0E-5 | 0.275 |
| 47640 | DNAJC3-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p | 10 | CBX5 | Sponge network | 0.753 | 0.00014 | 0.165 | 0.24446 | 0.275 |
| 47641 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | CDHR3 | Sponge network | 0.494 | 0.00339 | -0.977 | 4.0E-5 | 0.275 |
| 47642 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-5p | 15 | RET | Sponge network | 0.16 | 0.50785 | -0.833 | 0.03517 | 0.275 |
| 47643 | RP11-894P9.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-3614-5p;hsa-miR-495-3p | 12 | RPS6KA3 | Sponge network | 0.993 | 0 | 0.256 | 0.02419 | 0.275 |
| 47644 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | PRKAA2 | Sponge network | 0.056 | 0.8494 | -2.462 | 0 | 0.275 |
| 47645 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3614-5p;hsa-miR-370-3p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-96-5p | 23 | JAZF1 | Sponge network | 0.41 | 0.02214 | -0.377 | 0.02677 | 0.275 |
| 47646 | RAMP2-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 25 | PRKCB | Sponge network | -0.513 | 0.04703 | -1.313 | 2.0E-5 | 0.275 |
| 47647 | MAP3K14 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | ZFHX4 | Sponge network | -0.295 | 0.02604 | 0.018 | 0.95574 | 0.275 |
| 47648 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 14 | GPAM | Sponge network | 0.225 | 0.30644 | 0.142 | 0.29732 | 0.275 |
| 47649 | AC141928.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-671-5p | 24 | DLC1 | Sponge network | 0.853 | 0.15352 | -0.382 | 0.05038 | 0.275 |
| 47650 | ASH1L-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 12 | CBFB | Sponge network | 0.948 | 0 | 1.061 | 0 | 0.275 |
| 47651 | TRIM52-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-7-1-3p | 12 | IGSF10 | Sponge network | -0.418 | 0.00068 | -1.751 | 1.0E-5 | 0.275 |
| 47652 | RP13-516M14.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-429 | 11 | SPG20 | Sponge network | 0.389 | 0.04293 | -1.16 | 0 | 0.275 |
| 47653 | RP11-13K12.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 21 | AKAP11 | Sponge network | -0.318 | 0.65847 | 0.196 | 0.09825 | 0.275 |
| 47654 | RP11-973H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-33a-3p | 10 | MLLT10 | Sponge network | 0.901 | 0 | 0.105 | 0.33292 | 0.275 |
| 47655 | RP5-855D21.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-495-3p | 10 | PRKAA1 | Sponge network | 1.728 | 0 | 0.317 | 0.0031 | 0.275 |
| 47656 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-5p;hsa-miR-96-5p | 23 | CLASP2 | Sponge network | 0.078 | 0.55803 | -0.038 | 0.71 | 0.275 |
| 47657 | RP1-257A7.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p | 15 | NTRK2 | Sponge network | 0.849 | 0.00178 | -0.736 | 0.06495 | 0.275 |
| 47658 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-589-3p | 16 | HECA | Sponge network | 0.622 | 0.00015 | -0.217 | 0.03463 | 0.275 |
| 47659 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-590-5p | 13 | PBRM1 | Sponge network | 0.078 | 0.55803 | -0.029 | 0.78601 | 0.275 |
| 47660 | RP1-151F17.2 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TMEM30A | Sponge network | 0.222 | 0.29133 | 0.096 | 0.31031 | 0.275 |
| 47661 | AC006116.24 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 12 | CYLD | Sponge network | -0.473 | 0.07602 | -0.367 | 0.00171 | 0.275 |
| 47662 | AC124789.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p | 17 | DST | Sponge network | 0.073 | 0.79848 | -0.713 | 0.00096 | 0.275 |
| 47663 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-92a-3p | 26 | THBS1 | Sponge network | 0.253 | 0.09565 | 0.741 | 0.00386 | 0.275 |
| 47664 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-324-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | HACE1 | Sponge network | 0.194 | 0.64278 | -0.072 | 0.55239 | 0.275 |
| 47665 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 10 | JAKMIP2 | Sponge network | 0.862 | 0.06213 | -1.388 | 0 | 0.275 |
| 47666 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-590-3p | 12 | PRKAR1A | Sponge network | 0.381 | 0.25967 | -0.063 | 0.50314 | 0.275 |
| 47667 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 27 | ERBB4 | Sponge network | 0.051 | 0.83388 | -1.975 | 2.0E-5 | 0.275 |
| 47668 | LINC00672 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-590-3p | 16 | TMTC2 | Sponge network | -0.227 | 0.31657 | -0.215 | 0.18248 | 0.275 |
| 47669 | SH3RF3-AS1 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | UBE4B | Sponge network | 0.889 | 9.0E-5 | -0.235 | 0.007 | 0.275 |
| 47670 | RP4-668J24.2 |
hsa-miR-107;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-629-3p;hsa-miR-942-5p | 11 | GPC6 | Sponge network | -1.054 | 0.0706 | -0.054 | 0.81465 | 0.275 |
| 47671 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p | 32 | CBL | Sponge network | 0.95 | 0.15943 | 0.291 | 0.01267 | 0.275 |
| 47672 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 16 | PKNOX1 | Sponge network | -0.738 | 0.21233 | 0.085 | 0.28917 | 0.275 |
| 47673 | RP4-717I23.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-495-3p | 13 | RPS6KA3 | Sponge network | 0.637 | 0.00069 | 0.256 | 0.02419 | 0.275 |
| 47674 | RP11-35G9.5 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-96-5p | 10 | NCAM2 | Sponge network | 0.622 | 0.00015 | -0.482 | 0.22565 | 0.275 |
| 47675 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | PTPRB | Sponge network | 0.214 | 0.18246 | 0.105 | 0.47122 | 0.275 |
| 47676 | RP11-181G12.2 |
hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p | 12 | MED12L | Sponge network | 0.556 | 0.00298 | -0.194 | 0.55299 | 0.275 |
| 47677 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-942-5p | 15 | GPC6 | Sponge network | -0.72 | 0.24239 | -0.054 | 0.81465 | 0.275 |
| 47678 | RP11-359E10.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 11 | SEMA3E | Sponge network | 0.299 | 0.57722 | -1.717 | 0.00137 | 0.275 |
| 47679 | AC084219.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p | 14 | CREB1 | Sponge network | 1.296 | 0.0054 | 0.203 | 0.00537 | 0.275 |
| 47680 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 38 | THRB | Sponge network | 0.389 | 0.04293 | -1.117 | 0.0004 | 0.275 |
| 47681 | BOLA3-AS1 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-660-5p;hsa-miR-7-1-3p | 19 | LIMCH1 | Sponge network | -0.246 | 0.35034 | -0.915 | 1.0E-5 | 0.275 |
| 47682 | RP11-770J1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-34a-5p | 11 | ANK2 | Sponge network | -2.011 | 0.0005 | -1.603 | 1.0E-5 | 0.275 |
| 47683 | BCYRN1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-379-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | SH3GLB1 | Sponge network | 0.311 | 0.10344 | -0.115 | 0.14773 | 0.275 |
| 47684 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p | 20 | PPM1A | Sponge network | 0.001 | 0.99753 | -0.623 | 0 | 0.275 |
| 47685 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p | 12 | TXNIP | Sponge network | 0.659 | 3.0E-5 | -1.277 | 0 | 0.275 |
| 47686 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-671-5p | 10 | CRTC3 | Sponge network | -0.206 | 0.12942 | 0.164 | 0.16786 | 0.275 |
| 47687 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-425-5p;hsa-miR-7-1-3p | 14 | ATRX | Sponge network | 1.153 | 0.00114 | 0.261 | 0.04408 | 0.275 |
| 47688 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-942-5p | 12 | CREBBP | Sponge network | -0.465 | 0.04703 | 0.126 | 0.15186 | 0.275 |
| 47689 | PGM5-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p | 10 | KCNJ5 | Sponge network | -4.546 | 0 | -0.281 | 0.3339 | 0.275 |
| 47690 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 14 | FREM2 | Sponge network | 0.389 | 0.04293 | -0.437 | 0.46353 | 0.275 |
| 47691 | RP11-53O19.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-654-3p | 10 | ZNF638 | Sponge network | -0.405 | 0.22013 | 0.22 | 0.01214 | 0.275 |
| 47692 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 22 | MYO9A | Sponge network | 0.541 | 0.00125 | -0.147 | 0.32373 | 0.275 |
| 47693 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | EDA2R | Sponge network | 0.61 | 0.01593 | -0.587 | 0.01598 | 0.275 |
| 47694 | PGM5-AS1 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 22 | ABI2 | Sponge network | -4.546 | 0 | 0.175 | 0.14787 | 0.275 |
| 47695 | RP11-473I1.9 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-337-3p | 12 | RB1 | Sponge network | 0.683 | 1.0E-5 | 0.382 | 0.0017 | 0.275 |
| 47696 | RP11-6N17.3 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-455-5p | 10 | WDFY3 | Sponge network | 0.108 | 0.69556 | -0.201 | 0.0967 | 0.275 |
| 47697 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | CRTC1 | Sponge network | -0.737 | 0.06182 | -0.116 | 0.28412 | 0.275 |
| 47698 | HAR1A |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-940 | 15 | MCC | Sponge network | -0.672 | 0.19447 | -1.005 | 0 | 0.275 |
| 47699 | LINC00265 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | PHC3 | Sponge network | 1.07 | 0 | 0.212 | 0.0499 | 0.275 |
| 47700 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | TRIP11 | Sponge network | 1.063 | 0 | -0.158 | 0.06384 | 0.275 |
| 47701 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 26 | CBL | Sponge network | 0.663 | 0.00073 | 0.291 | 0.01267 | 0.275 |
| 47702 | AC004540.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-30b-3p | 13 | KCNA6 | Sponge network | -2.549 | 0.00528 | -1.531 | 0.00013 | 0.275 |
| 47703 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | PDGFRA | Sponge network | 0.101 | 0.60919 | -0.372 | 0.0037 | 0.275 |
| 47704 | FAM13A-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-874-3p;hsa-miR-96-5p | 27 | ASTN1 | Sponge network | -0.83 | 0 | -2.389 | 2.0E-5 | 0.275 |
| 47705 | SNHG12 |
hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 11 | CDC73 | Sponge network | 1.103 | 0 | 0.094 | 0.20463 | 0.275 |
| 47706 | DLGAP1-AS2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p | 11 | CREB1 | Sponge network | 2.406 | 0 | 0.203 | 0.00537 | 0.275 |
| 47707 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940 | 20 | LIFR | Sponge network | -0.246 | 0.35034 | -1.794 | 0 | 0.275 |
| 47708 | LINC00174 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p | 14 | PRKAA1 | Sponge network | 1.253 | 0 | 0.317 | 0.0031 | 0.275 |
| 47709 | AC004540.4 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-362-3p | 23 | THRB | Sponge network | -2.549 | 0.00528 | -1.117 | 0.0004 | 0.275 |
| 47710 | LINC00957 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p | 10 | LZTS1 | Sponge network | -0.421 | 0.0114 | 1.664 | 0 | 0.275 |
| 47711 | AC004893.11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | BRD3 | Sponge network | 1.317 | 0 | 0.37 | 0.00013 | 0.275 |
| 47712 | RP5-1061H20.4 |
hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 11 | CDC73 | Sponge network | 1.921 | 0 | 0.094 | 0.20463 | 0.275 |
| 47713 | OIP5-AS1 |
hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-7-5p | 10 | FLNA | Sponge network | 0.421 | 0.00084 | -0.771 | 0.01377 | 0.275 |
| 47714 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | GALNT13 | Sponge network | -0.487 | 0.02157 | -0.857 | 0.07439 | 0.275 |
| 47715 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 18 | IGF1 | Sponge network | 0.051 | 0.83388 | -0.204 | 0.5898 | 0.275 |
| 47716 | BCYRN1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-7-1-3p | 10 | VEZT | Sponge network | 0.311 | 0.10344 | 0.259 | 0.00296 | 0.275 |
| 47717 | RP11-57H14.4 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | 0.083 | 0.66405 | -0.226 | 0.11691 | 0.275 |
| 47718 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 15 | KIT | Sponge network | -0.384 | 0.40652 | -2.184 | 0 | 0.275 |
| 47719 | RP4-605O3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | AKAP6 | Sponge network | 0.262 | 0.0475 | -1.586 | 3.0E-5 | 0.275 |
| 47720 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | SEMA3C | Sponge network | -0.246 | 0.35034 | -0.272 | 0.31153 | 0.275 |
| 47721 | RAMP2-AS1 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 20 | ACSL6 | Sponge network | -0.513 | 0.04703 | -1.593 | 0 | 0.275 |
| 47722 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-484 | 11 | GREM1 | Sponge network | -0.72 | 0.24239 | 0.902 | 0.01691 | 0.275 |
| 47723 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p | 15 | PIK3C2A | Sponge network | 0.752 | 0 | 0.061 | 0.53776 | 0.274 |
| 47724 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-429 | 13 | LHX6 | Sponge network | 0.16 | 0.50785 | -0.087 | 0.69193 | 0.274 |
| 47725 | RP11-410L14.2 |
hsa-miR-101-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | UBR3 | Sponge network | -0.052 | 0.76839 | -0.021 | 0.82774 | 0.274 |
| 47726 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p | 13 | KANK1 | Sponge network | -0.285 | 0.2172 | -0.55 | 0.00134 | 0.274 |
| 47727 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 38 | CBX5 | Sponge network | 0.4 | 0.14611 | 0.165 | 0.24446 | 0.274 |
| 47728 | RP11-983P16.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p | 16 | SYNPO2 | Sponge network | 0.41 | 0.02214 | -2.342 | 0 | 0.274 |
| 47729 | H19 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p | 16 | FAT3 | Sponge network | 2.439 | 0 | -1.601 | 0.00051 | 0.274 |
| 47730 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 58 | AFF4 | Sponge network | -0.901 | 0.00019 | -0.266 | 0.01565 | 0.274 |
| 47731 | ZFAS1 |
hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 10 | PAFAH1B2 | Sponge network | 0.831 | 0 | 0.318 | 0.0001 | 0.274 |
| 47732 | LINC00094 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-671-5p;hsa-miR-93-5p | 42 | GAS7 | Sponge network | 0.253 | 0.09565 | -0.555 | 0.01602 | 0.274 |
| 47733 | RP11-758N13.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-93-3p | 12 | ERC1 | Sponge network | 2.939 | 3.0E-5 | -0.108 | 0.38794 | 0.274 |
| 47734 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-577 | 13 | AKAP12 | Sponge network | 0.177 | 0.16681 | -0.932 | 0.0028 | 0.274 |
| 47735 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | LRIG1 | Sponge network | -0.031 | 0.89063 | -0.537 | 0.00588 | 0.274 |
| 47736 | LINC01001 |
hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-539-5p | 13 | DDX6 | Sponge network | 1.055 | 0 | 0.091 | 0.36428 | 0.274 |
| 47737 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 45 | CBX5 | Sponge network | -0.709 | 0.18168 | 0.165 | 0.24446 | 0.274 |
| 47738 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-98-5p | 39 | CADM2 | Sponge network | 0.082 | 0.55397 | -3.782 | 0 | 0.274 |
| 47739 | RP11-360F5.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | DST | Sponge network | 1.867 | 0 | -0.713 | 0.00096 | 0.274 |
| 47740 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 21 | RBMS3 | Sponge network | 1.04 | 0.00023 | -0.519 | 0.03551 | 0.274 |
| 47741 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-671-5p;hsa-miR-877-5p | 18 | CRTC3 | Sponge network | -0.13 | 0.48734 | 0.164 | 0.16786 | 0.274 |
| 47742 | CTD-2245F17.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-7-5p | 12 | ERBB4 | Sponge network | -0.421 | 0.12556 | -1.975 | 2.0E-5 | 0.274 |
| 47743 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-505-3p;hsa-miR-590-3p | 11 | JAKMIP2 | Sponge network | -0.125 | 0.49414 | -1.388 | 0 | 0.274 |
| 47744 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p | 14 | CUL5 | Sponge network | 1.03 | 0 | 0.026 | 0.77301 | 0.274 |
| 47745 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 17 | ADAMTSL3 | Sponge network | 0.537 | 0.00139 | -1.559 | 0 | 0.274 |
| 47746 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-29a-5p | 10 | PBX1 | Sponge network | 1.46 | 1.0E-5 | -1.206 | 0 | 0.274 |
| 47747 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-493-5p;hsa-miR-590-3p | 14 | MYCBP2 | Sponge network | -0.355 | 0.0077 | -0.027 | 0.82016 | 0.274 |
| 47748 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 22 | MYO9A | Sponge network | -1.203 | 0.03881 | -0.147 | 0.32373 | 0.274 |
| 47749 | AC006116.24 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-331-5p;hsa-miR-455-5p | 10 | ERRFI1 | Sponge network | -0.473 | 0.07602 | -0.798 | 1.0E-5 | 0.274 |
| 47750 | RP11-760H22.2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-454-3p | 14 | DLC1 | Sponge network | 0.001 | 0.99753 | -0.382 | 0.05038 | 0.274 |
| 47751 | CD27-AS1 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p | 11 | DCLK1 | Sponge network | 0.177 | 0.16681 | -1.324 | 0.00014 | 0.274 |
| 47752 | H1FX-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 35 | THRB | Sponge network | -0.206 | 0.12942 | -1.117 | 0.0004 | 0.274 |
| 47753 | RP11-434B12.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 12 | PTGFR | Sponge network | -0.031 | 0.89063 | -0.233 | 0.45421 | 0.274 |
| 47754 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-935 | 18 | TSHZ3 | Sponge network | 0.37 | 0.27614 | -0.556 | 0.01516 | 0.274 |
| 47755 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-576-5p | 13 | TMEM132B | Sponge network | -2.039 | 0.03447 | -0.83 | 0.01084 | 0.274 |
| 47756 | MALAT1 |
hsa-miR-150-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | GSK3B | Sponge network | 0.782 | 0.00016 | 0.266 | 0.00045 | 0.274 |
| 47757 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-625-3p | 29 | TMTC3 | Sponge network | 0.75 | 0.00054 | 0.123 | 0.22189 | 0.274 |
| 47758 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-3913-5p;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-7-5p | 29 | ESR1 | Sponge network | -1.047 | 0 | -1.035 | 3.0E-5 | 0.274 |
| 47759 | LINC01004 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | UBR3 | Sponge network | 1.115 | 0 | -0.021 | 0.82774 | 0.274 |
| 47760 | CTC-429P9.3 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-550a-3p | 11 | RASSF3 | Sponge network | 0.082 | 0.55397 | -0.226 | 0.11691 | 0.274 |
| 47761 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p | 13 | TSC22D1 | Sponge network | -2.925 | 0 | -0.529 | 2.0E-5 | 0.274 |
| 47762 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | BNC2 | Sponge network | 0.853 | 0.15352 | -0.491 | 0.09988 | 0.274 |
| 47763 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 46 | FZD3 | Sponge network | 0.311 | 0.10344 | 0.104 | 0.60316 | 0.274 |
| 47764 | CKMT2-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | TES | Sponge network | 0.082 | 0.55654 | -0.348 | 0.00639 | 0.274 |
| 47765 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 12 | MYBL1 | Sponge network | 0.997 | 0 | 0.494 | 0.02152 | 0.274 |
| 47766 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-590-3p | 10 | AKAP11 | Sponge network | 0.545 | 0.05789 | 0.196 | 0.09825 | 0.274 |
| 47767 | RP11-116O18.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PDE4DIP | Sponge network | 0.05 | 0.95483 | -0.282 | 0.0257 | 0.274 |
| 47768 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 17 | GPAM | Sponge network | 0.439 | 0.00313 | 0.142 | 0.29732 | 0.274 |
| 47769 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-30b-5p | 11 | NOTCH2 | Sponge network | 0.37 | 0.27614 | 0.138 | 0.35163 | 0.274 |
| 47770 | RP1-257A7.5 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-629-3p | 10 | RASSF2 | Sponge network | 0.849 | 0.00178 | -0.273 | 0.21961 | 0.274 |
| 47771 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p | 17 | PKNOX1 | Sponge network | -0.576 | 0.10121 | 0.085 | 0.28917 | 0.274 |
| 47772 | CTD-2647L4.4 |
hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-495-3p | 10 | ABI2 | Sponge network | 0.555 | 0.00197 | 0.175 | 0.14787 | 0.274 |
| 47773 | CTC-429P9.3 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p | 11 | TLL1 | Sponge network | 0.082 | 0.55397 | -1.195 | 3.0E-5 | 0.274 |
| 47774 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | TMTC3 | Sponge network | 1.106 | 0 | 0.123 | 0.22189 | 0.274 |
| 47775 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p | 12 | MAP3K12 | Sponge network | -0.773 | 0.04073 | -0.057 | 0.59369 | 0.274 |
| 47776 | RP11-389C8.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 30 | TRPS1 | Sponge network | 0.727 | 3.0E-5 | -0.191 | 0.44109 | 0.274 |
| 47777 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-7-1-3p | 10 | CNTN5 | Sponge network | 0.41 | 0.02214 | -0.219 | 0.61957 | 0.274 |
| 47778 | HOXA-AS2 |
hsa-miR-107;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-940 | 13 | ZFP36L1 | Sponge network | 0.61 | 0.01593 | -0.505 | 8.0E-5 | 0.274 |
| 47779 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 12 | FBXO32 | Sponge network | 0.252 | 0.08508 | -0.495 | 0.07561 | 0.274 |
| 47780 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-628-5p | 11 | EPHA3 | Sponge network | 1.361 | 0 | 0.185 | 0.53588 | 0.274 |
| 47781 | AC009948.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | BRD3 | Sponge network | 0.532 | 0.00505 | 0.37 | 0.00013 | 0.274 |
| 47782 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | FGFR1 | Sponge network | -0.663 | 0.10887 | -0.509 | 0.03957 | 0.274 |
| 47783 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 25 | CBX5 | Sponge network | 1.601 | 0 | 0.165 | 0.24446 | 0.274 |
| 47784 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 14 | FOXO3 | Sponge network | 1.147 | 0 | -0.04 | 0.72028 | 0.274 |
| 47785 | AC003090.1 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | FAM135B | Sponge network | -2.117 | 0.00161 | -3.978 | 0 | 0.274 |
| 47786 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-7-5p;hsa-miR-93-5p | 16 | ZBTB4 | Sponge network | -0.351 | 0.11358 | -0.739 | 0 | 0.274 |
| 47787 | RP11-359E10.1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-500a-5p;hsa-miR-576-5p | 12 | ATM | Sponge network | 0.299 | 0.57722 | 0.178 | 0.21568 | 0.274 |
| 47788 | LPP-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | IL17RD | Sponge network | -0.18 | 0.20343 | -0.019 | 0.94873 | 0.274 |
| 47789 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CLASP2 | Sponge network | 0.848 | 0 | -0.038 | 0.71 | 0.274 |
| 47790 | LINC00657 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-337-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | KDSR | Sponge network | 0.91 | 0 | 0.239 | 0.02216 | 0.274 |
| 47791 | AC034220.3 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 23 | CCDC6 | Sponge network | 0.309 | 0.07304 | 0.27 | 0.02555 | 0.274 |
| 47792 | RP11-148O21.2 |
hsa-let-7d-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-500a-5p | 11 | PRKAA2 | Sponge network | 0.233 | 0.81029 | -2.462 | 0 | 0.274 |
| 47793 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | CCND2 | Sponge network | 0.614 | 1.0E-5 | -0.267 | 0.3112 | 0.274 |
| 47794 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-93-5p | 10 | EGR2 | Sponge network | -2.69 | 0.0001 | 0.309 | 0.17079 | 0.274 |
| 47795 | RP11-298J20.4 |
hsa-let-7b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p | 11 | PBX1 | Sponge network | 0.002 | 0.99057 | -1.206 | 0 | 0.274 |
| 47796 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | TAL1 | Sponge network | 0.082 | 0.55654 | -0.462 | 0.00574 | 0.274 |
| 47797 | RP11-597D13.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | BRD3 | Sponge network | 1.302 | 7.0E-5 | 0.37 | 0.00013 | 0.274 |
| 47798 | LIPE-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 22 | ABL2 | Sponge network | 0.078 | 0.55803 | 0.82 | 0 | 0.274 |
| 47799 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-653-5p | 20 | PDS5B | Sponge network | -1.575 | 6.0E-5 | 0.259 | 0.00962 | 0.274 |
| 47800 | SNHG14 |
hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 13 | FBXW7 | Sponge network | -1.111 | 0.0003 | -0.418 | 2.0E-5 | 0.274 |
| 47801 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | HDAC9 | Sponge network | 0.796 | 4.0E-5 | -0.755 | 0.00495 | 0.274 |
| 47802 | RP11-434B12.1 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-708-5p | 19 | CCDC6 | Sponge network | -0.031 | 0.89063 | 0.27 | 0.02555 | 0.274 |
| 47803 | HCG11 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | RASSF2 | Sponge network | 0.385 | 0.10067 | -0.273 | 0.21961 | 0.274 |
| 47804 | DNAJC27-AS1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 15 | FOXO4 | Sponge network | -0.969 | 2.0E-5 | -0.516 | 2.0E-5 | 0.274 |
| 47805 | ARHGEF26-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935 | 33 | ACSL6 | Sponge network | -2.46 | 0 | -1.593 | 0 | 0.274 |
| 47806 | LINC00672 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-577;hsa-miR-590-3p | 12 | ZNF844 | Sponge network | -0.227 | 0.31657 | -0.966 | 0.00035 | 0.274 |
| 47807 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-590-3p | 13 | RBM5 | Sponge network | 0.101 | 0.60919 | -0.205 | 0.0129 | 0.274 |
| 47808 | RP11-597D13.9 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | RTN4 | Sponge network | 1.302 | 7.0E-5 | -0.412 | 0.00014 | 0.274 |
| 47809 | PCA3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TSC1 | Sponge network | -0.709 | 0.18168 | 0.103 | 0.26071 | 0.274 |
| 47810 | RP11-815I9.4 |
hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-3607-3p;hsa-miR-664a-3p | 15 | CCDC6 | Sponge network | 0.537 | 0.0006 | 0.27 | 0.02555 | 0.274 |
| 47811 | RP11-720L2.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-33a-3p;hsa-miR-625-5p;hsa-miR-940 | 13 | PTPRT | Sponge network | -1.255 | 0.00981 | -0.907 | 0.03727 | 0.274 |
| 47812 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | RNF144A | Sponge network | 0.862 | 0.06213 | 0.594 | 0.0009 | 0.274 |
| 47813 | RP11-35G9.5 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-744-3p;hsa-miR-940 | 18 | PTPRT | Sponge network | 0.622 | 0.00015 | -0.907 | 0.03727 | 0.274 |
| 47814 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 11 | CNTNAP3 | Sponge network | 0.374 | 0.20054 | -1.465 | 0.00033 | 0.274 |
| 47815 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 25 | ARHGAP28 | Sponge network | 0.997 | 0.00066 | -0.911 | 0.00013 | 0.274 |
| 47816 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | GALNT13 | Sponge network | 0.889 | 9.0E-5 | -0.857 | 0.07439 | 0.274 |
| 47817 | AC116366.6 |
hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-375;hsa-miR-421;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | MITF | Sponge network | 0.329 | 0.11122 | -0.769 | 0.00022 | 0.274 |
| 47818 | HOXB-AS2 |
hsa-miR-101-5p;hsa-miR-125a-3p;hsa-miR-18a-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-576-5p | 10 | ATM | Sponge network | 1.316 | 0.01106 | 0.178 | 0.21568 | 0.274 |
| 47819 | AF131215.9 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-92b-3p | 18 | PCDH9 | Sponge network | -0.322 | 0.25945 | -1.823 | 4.0E-5 | 0.274 |
| 47820 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-34a-5p | 10 | CHD1 | Sponge network | 0.921 | 0.00118 | 0.322 | 0.00215 | 0.274 |
| 47821 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | TLL1 | Sponge network | 0.082 | 0.55654 | -1.195 | 3.0E-5 | 0.274 |
| 47822 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 30 | ST6GAL2 | Sponge network | 0.101 | 0.60919 | -1.201 | 0.00597 | 0.274 |
| 47823 | CTC-523E23.11 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-429 | 13 | GAS7 | Sponge network | -0.235 | 0.35308 | -0.555 | 0.01602 | 0.274 |
| 47824 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-484;hsa-miR-590-3p | 10 | MYCBP2 | Sponge network | 1.04 | 0.00023 | -0.027 | 0.82016 | 0.274 |
| 47825 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-96-5p | 13 | DOCK4 | Sponge network | 0.37 | 0.27614 | 0.502 | 0.00108 | 0.274 |
| 47826 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | TGFBR3 | Sponge network | 0.349 | 0.01695 | -1.173 | 1.0E-5 | 0.274 |
| 47827 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 18 | HIP1 | Sponge network | 0.222 | 0.29133 | 0.774 | 0 | 0.274 |
| 47828 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 14 | FAM188A | Sponge network | -2.925 | 0 | -0.44 | 0.00018 | 0.274 |
| 47829 | RP11-130L8.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p | 14 | DLC1 | Sponge network | -0.663 | 0.10887 | -0.382 | 0.05038 | 0.274 |
| 47830 | RP11-389C8.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 42 | LPP | Sponge network | 0.727 | 3.0E-5 | -0.459 | 0.04475 | 0.274 |
| 47831 | RP11-254F7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-5p | 23 | BHLHE41 | Sponge network | 0.176 | 0.37402 | 0.353 | 0.12753 | 0.274 |
| 47832 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 28 | AFF4 | Sponge network | 0.539 | 0.03722 | -0.266 | 0.01565 | 0.274 |
| 47833 | C4A-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-590-5p | 10 | ZNF208 | Sponge network | 3.345 | 0 | -0.4 | 0.22381 | 0.274 |
| 47834 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940;hsa-miR-96-5p | 71 | AFF4 | Sponge network | -0.576 | 0.10121 | -0.266 | 0.01565 | 0.274 |
| 47835 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-16-1-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-369-3p;hsa-miR-455-5p | 10 | PNRC1 | Sponge network | 0.199 | 0.52763 | -0.646 | 0 | 0.274 |
| 47836 | RP1-151F17.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | COPS2 | Sponge network | 0.222 | 0.29133 | -0.178 | 0.04974 | 0.274 |
| 47837 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 20 | PRKAB2 | Sponge network | 0.853 | 0.15352 | -0.367 | 0.01371 | 0.274 |
| 47838 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p | 10 | MGA | Sponge network | 0.249 | 0.35902 | 0.323 | 0.00792 | 0.274 |
| 47839 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | CDH10 | Sponge network | 0.174 | 0.30034 | -2.397 | 0 | 0.274 |
| 47840 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-374b-5p | 10 | TMTC3 | Sponge network | 1.46 | 1.0E-5 | 0.123 | 0.22189 | 0.274 |
| 47841 | RP11-968O1.5 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-940 | 30 | WDFY3 | Sponge network | -0.829 | 0.01343 | -0.201 | 0.0967 | 0.274 |
| 47842 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p | 10 | FBXO31 | Sponge network | -0.246 | 0.35034 | -0.085 | 0.42389 | 0.274 |
| 47843 | RP11-760H22.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-655-3p | 12 | KANK1 | Sponge network | 0.001 | 0.99753 | -0.55 | 0.00134 | 0.274 |
| 47844 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-98-5p | 50 | CADM2 | Sponge network | -0.727 | 0.04726 | -3.782 | 0 | 0.274 |
| 47845 | RP11-758N13.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-424-5p | 11 | ZFHX3 | Sponge network | 2.939 | 3.0E-5 | 0.389 | 0.01363 | 0.274 |
| 47846 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 29 | SMAD4 | Sponge network | -1.092 | 4.0E-5 | -0.313 | 0.00413 | 0.274 |
| 47847 | RP11-678G14.3 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-625-5p | 19 | IGFBP5 | Sponge network | -4.477 | 0 | -0.806 | 0.00105 | 0.274 |
| 47848 | RP11-597D13.9 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-942-5p | 19 | GLIPR1 | Sponge network | 1.302 | 7.0E-5 | 0.146 | 0.3276 | 0.274 |
| 47849 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 37 | MAF | Sponge network | -2.925 | 0 | -1.257 | 0 | 0.274 |
| 47850 | LINC01018 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | GJA1 | Sponge network | -2.915 | 0.00015 | -0.485 | 0.02179 | 0.274 |
| 47851 | TINCR |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-3p;hsa-miR-424-5p | 14 | UBR3 | Sponge network | -0.664 | 0.14543 | -0.021 | 0.82774 | 0.274 |
| 47852 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p | 12 | NIN | Sponge network | 0.898 | 1.0E-5 | -0.253 | 0.13794 | 0.274 |
| 47853 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 16 | HBP1 | Sponge network | -2.095 | 0.00112 | -0.454 | 0 | 0.274 |
| 47854 | RP11-774O3.3 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 22 | IKZF2 | Sponge network | -0.487 | 0.02157 | -0.284 | 0.1942 | 0.274 |
| 47855 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-624-5p | 13 | PCDH8 | Sponge network | -0.777 | 0.1134 | -0.997 | 0.05366 | 0.274 |
| 47856 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-7-1-3p | 20 | SYNE1 | Sponge network | 0.75 | 0.00054 | -0.738 | 0.00316 | 0.274 |
| 47857 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 36 | ATRX | Sponge network | -1.697 | 0.00889 | 0.261 | 0.04408 | 0.274 |
| 47858 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | PHC3 | Sponge network | 1.178 | 0 | 0.212 | 0.0499 | 0.274 |
| 47859 | HOTAIRM1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-335-3p | 10 | FNBP1 | Sponge network | -0.053 | 0.80685 | -0.969 | 4.0E-5 | 0.274 |
| 47860 | ZNF582-AS1 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-5p | 14 | PTCH1 | Sponge network | -1.349 | 0.00017 | -0.1 | 0.6179 | 0.274 |
| 47861 | RP4-758J18.10 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-503-5p | 11 | ABL1 | Sponge network | -0.091 | 0.71468 | -0.215 | 0.07804 | 0.274 |
| 47862 | U91328.19 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | PPP1R9A | Sponge network | -0.303 | 0.21002 | -0.903 | 0.04254 | 0.274 |
| 47863 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-154-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-940 | 27 | PBX1 | Sponge network | -0.465 | 0.04703 | -1.206 | 0 | 0.274 |
| 47864 | RP11-504P24.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-338-5p;hsa-miR-497-5p | 11 | MYBL1 | Sponge network | 1.178 | 0 | 0.494 | 0.02152 | 0.274 |
| 47865 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-590-3p | 13 | SPG20 | Sponge network | 0.219 | 0.1516 | -1.16 | 0 | 0.274 |
| 47866 | RP11-774O3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-361-3p | 11 | MNT | Sponge network | -0.487 | 0.02157 | -0.157 | 0.06598 | 0.274 |
| 47867 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 10 | PLA2R1 | Sponge network | 0.421 | 0.00084 | -0.265 | 0.24676 | 0.274 |
| 47868 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-493-5p | 20 | MYBL1 | Sponge network | -0.384 | 0.40652 | 0.494 | 0.02152 | 0.274 |
| 47869 | CTD-2303H24.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | ZNF208 | Sponge network | 0.415 | 0.14657 | -0.4 | 0.22381 | 0.274 |
| 47870 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 25 | SOX11 | Sponge network | 1.254 | 0 | 2.68 | 0 | 0.274 |
| 47871 | GNG12-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-34a-5p | 18 | CBFA2T3 | Sponge network | -0.793 | 7.0E-5 | -1.221 | 1.0E-5 | 0.274 |
| 47872 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 14 | KAT6B | Sponge network | 0.594 | 0.00064 | -0.478 | 0.00018 | 0.274 |
| 47873 | U91328.19 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-625-5p | 25 | CADM1 | Sponge network | -0.303 | 0.21002 | -0.9 | 0.00084 | 0.274 |
| 47874 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-5p | 19 | ERC1 | Sponge network | 0.176 | 0.37402 | -0.108 | 0.38794 | 0.274 |
| 47875 | MIR497HG |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-375;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | ITGB1 | Sponge network | -1.439 | 4.0E-5 | 0.524 | 0.00017 | 0.274 |
| 47876 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p | 12 | RBM5 | Sponge network | 0.249 | 0.35902 | -0.205 | 0.0129 | 0.274 |
| 47877 | AC024560.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3614-5p | 21 | ABL2 | Sponge network | 2.166 | 0.00228 | 0.82 | 0 | 0.274 |
| 47878 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 31 | DIP2C | Sponge network | -0.83 | 0 | -0.436 | 0.01713 | 0.274 |
| 47879 | PVT1 |
hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 11 | PAFAH1B2 | Sponge network | 1.985 | 0 | 0.318 | 0.0001 | 0.274 |
| 47880 | RP11-13K12.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-5p | 29 | PIK3R1 | Sponge network | -0.318 | 0.65847 | -0.133 | 0.36854 | 0.274 |
| 47881 | NDUFA6-AS1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-30c-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-940 | 12 | RHOB | Sponge network | -0.355 | 0.0077 | -1.052 | 0 | 0.274 |
| 47882 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 20 | CDHR3 | Sponge network | 0.064 | 0.72934 | -0.977 | 4.0E-5 | 0.274 |
| 47883 | RP11-517B11.7 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-664a-3p | 16 | RASAL2 | Sponge network | 0.706 | 0 | 0.869 | 0 | 0.274 |
| 47884 | CTA-445C9.14 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-337-3p | 12 | SLC5A3 | Sponge network | 0.718 | 4.0E-5 | 0.493 | 5.0E-5 | 0.274 |
| 47885 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-500a-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 14 | ARHGAP29 | Sponge network | 0.4 | 0.14611 | 0.131 | 0.42953 | 0.274 |
| 47886 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | FAM172A | Sponge network | -0.223 | 0.47816 | -0.57 | 0 | 0.274 |
| 47887 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-582-5p | 16 | DDX6 | Sponge network | 0.643 | 0 | 0.091 | 0.36428 | 0.274 |
| 47888 | RP11-428J1.5 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PHF3 | Sponge network | 0.538 | 0.00076 | 0.126 | 0.19652 | 0.274 |
| 47889 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p | 10 | FLI1 | Sponge network | -0.206 | 0.12942 | -0.294 | 0.10905 | 0.274 |
| 47890 | TPTEP1 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 12 | LZTS1 | Sponge network | -1.216 | 0 | 1.664 | 0 | 0.274 |
| 47891 | LINC00654 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | SYT4 | Sponge network | 0.374 | 0.20054 | -3.207 | 0 | 0.274 |
| 47892 | CTD-2554C21.2 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-508-3p | 10 | RASSF3 | Sponge network | -1.654 | 0.00144 | -0.226 | 0.11691 | 0.274 |
| 47893 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p | 20 | TCF4 | Sponge network | 1.056 | 0 | 0.016 | 0.92271 | 0.274 |
| 47894 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | NEDD4 | Sponge network | 0.41 | 0.02214 | 0.02 | 0.89834 | 0.274 |
| 47895 | TMEM51-AS1 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-708-5p | 19 | CCDC6 | Sponge network | 0.23 | 0.38667 | 0.27 | 0.02555 | 0.274 |
| 47896 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 27 | IL6ST | Sponge network | 0.817 | 3.0E-5 | -0.402 | 0.00676 | 0.274 |
| 47897 | RP5-1085F17.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-940 | 10 | FAM172A | Sponge network | 0.541 | 0.00125 | -0.57 | 0 | 0.274 |
| 47898 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-874-3p | 12 | BCLAF1 | Sponge network | 0.311 | 0.10344 | 0.211 | 0.00684 | 0.274 |
| 47899 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p | 10 | MGA | Sponge network | 0.373 | 0.00068 | 0.323 | 0.00792 | 0.274 |
| 47900 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 41 | BMPR2 | Sponge network | 0.619 | 0.00157 | 0.206 | 0.0635 | 0.274 |
| 47901 | RP11-248G5.8 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | USP9X | Sponge network | 1.294 | 0 | 0.222 | 0.01689 | 0.274 |
| 47902 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-942-5p | 29 | THRB | Sponge network | 0.859 | 0.00331 | -1.117 | 0.0004 | 0.274 |
| 47903 | MRPL23-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-223-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p | 13 | DMD | Sponge network | -0.278 | 0.76559 | -1.506 | 0 | 0.274 |
| 47904 | RP4-584D14.7 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-577 | 12 | HIP1 | Sponge network | 1.294 | 0.0031 | 0.774 | 0 | 0.274 |
| 47905 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | DMTF1 | Sponge network | 0.422 | 0.27021 | 0.248 | 0.02726 | 0.274 |
| 47906 | LINC00680 |
hsa-let-7a-3p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p | 14 | MOB1B | Sponge network | 0.884 | 0 | 0.197 | 0.15266 | 0.274 |
| 47907 | MALAT1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | USP9X | Sponge network | 0.782 | 0.00016 | 0.222 | 0.01689 | 0.274 |
| 47908 | RP11-127B20.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p | 15 | ABL2 | Sponge network | 0.411 | 0.1335 | 0.82 | 0 | 0.274 |
| 47909 | CROCCP2 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p | 12 | MAT2A | Sponge network | 0.79 | 0 | -0.073 | 0.36195 | 0.274 |
| 47910 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-93-3p | 14 | PTPN11 | Sponge network | 0.637 | 0.00069 | 0.431 | 2.0E-5 | 0.274 |
| 47911 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | EGR2 | Sponge network | 0.299 | 0.57722 | 0.309 | 0.17079 | 0.274 |
| 47912 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | RIMBP2 | Sponge network | 0.101 | 0.60919 | -1.968 | 5.0E-5 | 0.274 |
| 47913 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p | 18 | UNC80 | Sponge network | 0.174 | 0.30034 | -1.934 | 0 | 0.274 |
| 47914 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 28 | SSBP2 | Sponge network | -0.663 | 0.10887 | -1.58 | 0 | 0.274 |
| 47915 | LINC00476 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p | 27 | EPS15 | Sponge network | -0.615 | 0.00015 | -0.052 | 0.53998 | 0.274 |
| 47916 | RP11-35G9.5 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-455-5p | 10 | AKAP6 | Sponge network | 0.622 | 0.00015 | -1.586 | 3.0E-5 | 0.274 |
| 47917 | RP4-639F20.1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-629-5p;hsa-miR-942-5p;hsa-miR-96-5p | 16 | PCDH17 | Sponge network | -0.517 | 0.02935 | 0.731 | 2.0E-5 | 0.274 |
| 47918 | THAP9-AS1 |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-654-3p | 12 | DDX6 | Sponge network | 1.079 | 0 | 0.091 | 0.36428 | 0.274 |
| 47919 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-93-5p | 32 | RASSF2 | Sponge network | -0.227 | 0.31657 | -0.273 | 0.21961 | 0.274 |
| 47920 | RP11-48B3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p | 10 | PRDM2 | Sponge network | 1.757 | 0 | -0.258 | 0.00721 | 0.274 |
| 47921 | RP11-597D13.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p | 13 | NTRK3 | Sponge network | 0.936 | 0.03889 | -1.77 | 3.0E-5 | 0.274 |
| 47922 | RP11-400K9.3 |
hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-940 | 12 | PTPRT | Sponge network | -0.733 | 0.19469 | -0.907 | 0.03727 | 0.274 |
| 47923 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | RECK | Sponge network | 0.219 | 0.1516 | -1.043 | 0 | 0.274 |
| 47924 | RP11-822E23.8 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-942-5p | 21 | OPCML | Sponge network | -0.777 | 0.16667 | -2.498 | 0 | 0.274 |
| 47925 | HOTAIRM1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | GALNT13 | Sponge network | -0.053 | 0.80685 | -0.857 | 0.07439 | 0.274 |
| 47926 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-582-3p | 27 | PBX1 | Sponge network | 0.603 | 0.00127 | -1.206 | 0 | 0.274 |
| 47927 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-493-5p;hsa-miR-539-5p | 11 | DDX5 | Sponge network | 0.614 | 1.0E-5 | -0.099 | 0.17428 | 0.274 |
| 47928 | EMX2OS |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | BRD3 | Sponge network | -0.777 | 0.1134 | 0.37 | 0.00013 | 0.274 |
| 47929 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | SASH1 | Sponge network | 0.559 | 0.00026 | -0.502 | 0.00175 | 0.274 |
| 47930 | AC005618.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 0.862 | 0.06213 | 0.943 | 0 | 0.274 |
| 47931 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p | 12 | PHC3 | Sponge network | 1.4 | 0 | 0.212 | 0.0499 | 0.274 |
| 47932 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 67 | KIF1B | Sponge network | -1.697 | 0.00889 | -0.118 | 0.32258 | 0.274 |
| 47933 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-5p;hsa-miR-96-5p | 23 | FOXO3 | Sponge network | 0.331 | 0.57247 | -0.04 | 0.72028 | 0.274 |
| 47934 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-590-3p | 11 | CUL5 | Sponge network | 2.094 | 0 | 0.026 | 0.77301 | 0.274 |
| 47935 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 18 | UNC80 | Sponge network | 0.131 | 0.8436 | -1.934 | 0 | 0.274 |
| 47936 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | TCF12 | Sponge network | 0.72 | 0.0001 | 0.174 | 0.13271 | 0.274 |
| 47937 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-874-3p | 15 | CRTC3 | Sponge network | -0.663 | 0.10887 | 0.164 | 0.16786 | 0.274 |
| 47938 | ADAMTS9-AS2 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p | 16 | ACTB | Sponge network | -2.452 | 0 | -0.247 | 0.01736 | 0.274 |
| 47939 | LINC00662 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-532-3p;hsa-miR-576-5p | 19 | PBX1 | Sponge network | 0.213 | 0.32297 | -1.206 | 0 | 0.274 |
| 47940 | RP11-13K12.5 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 18 | BRD3 | Sponge network | -0.318 | 0.65847 | 0.37 | 0.00013 | 0.274 |
| 47941 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-7-1-3p | 11 | HACE1 | Sponge network | -0.303 | 0.21002 | -0.072 | 0.55239 | 0.274 |
| 47942 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-374b-5p | 17 | DMXL1 | Sponge network | 0.001 | 0.99753 | -0.426 | 0.00056 | 0.274 |
| 47943 | MIR143HG |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-335-5p;hsa-miR-429;hsa-miR-455-5p | 19 | PLXNC1 | Sponge network | -0.739 | 0.0574 | 0.569 | 0.00329 | 0.274 |
| 47944 | JAZF1-AS1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | NR3C2 | Sponge network | -0.769 | 0.00035 | -1.56 | 0 | 0.274 |
| 47945 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TIMP3 | Sponge network | 0.862 | 0.06213 | -0.546 | 0.02307 | 0.274 |
| 47946 | MAP3K14 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-3127-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p | 17 | THRB | Sponge network | -0.295 | 0.02604 | -1.117 | 0.0004 | 0.274 |
| 47947 | XXYLT1-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-432-5p;hsa-miR-500a-5p;hsa-miR-93-3p | 11 | IKZF2 | Sponge network | -0.764 | 0.05257 | -0.284 | 0.1942 | 0.274 |
| 47948 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-766-3p | 11 | EZH1 | Sponge network | 0.311 | 0.10344 | -0.564 | 1.0E-5 | 0.274 |
| 47949 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 21 | MYO9A | Sponge network | -0.125 | 0.49414 | -0.147 | 0.32373 | 0.274 |
| 47950 | LINC00672 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | ERG | Sponge network | -0.227 | 0.31657 | 0.002 | 0.99011 | 0.274 |
| 47951 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-7-1-3p | 26 | EPHA7 | Sponge network | -0.418 | 0.00068 | -2.197 | 3.0E-5 | 0.274 |
| 47952 | RP5-1085F17.3 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ITGB1 | Sponge network | 0.541 | 0.00125 | 0.524 | 0.00017 | 0.274 |
| 47953 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | SEMA3C | Sponge network | 0.082 | 0.55654 | -0.272 | 0.31153 | 0.274 |
| 47954 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 59 | THRB | Sponge network | 0.101 | 0.60919 | -1.117 | 0.0004 | 0.274 |
| 47955 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-532-3p | 19 | LATS1 | Sponge network | 0.637 | 0.00069 | 0.109 | 0.34208 | 0.274 |
| 47956 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 10 | JAZF1 | Sponge network | -0.011 | 0.967 | -0.377 | 0.02677 | 0.274 |
| 47957 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p | 11 | SPRY3 | Sponge network | 1.969 | 0.00316 | 0.016 | 0.91286 | 0.274 |
| 47958 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | HECA | Sponge network | 0.374 | 0.20054 | -0.217 | 0.03463 | 0.274 |
| 47959 | RP11-927P21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-590-5p | 11 | NRXN3 | Sponge network | 0.921 | 0.00118 | -0.893 | 0.04186 | 0.274 |
| 47960 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-590-3p | 10 | CHD2 | Sponge network | 0.274 | 0.07681 | 0.049 | 0.55371 | 0.274 |
| 47961 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 30 | PDGFRA | Sponge network | -0.322 | 0.25945 | -0.372 | 0.0037 | 0.274 |
| 47962 | RP11-20I23.13 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-671-5p;hsa-miR-93-3p | 18 | BNC2 | Sponge network | 0.505 | 0.0014 | -0.491 | 0.09988 | 0.274 |
| 47963 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | AFF3 | Sponge network | 0.16 | 0.50785 | -2.373 | 0 | 0.274 |
| 47964 | MIR143HG |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-361-3p | 13 | MNT | Sponge network | -0.739 | 0.0574 | -0.157 | 0.06598 | 0.274 |
| 47965 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 16 | FAT4 | Sponge network | 0.381 | 0.25967 | -0.354 | 0.14694 | 0.274 |
| 47966 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-577;hsa-miR-590-3p | 14 | MYBL1 | Sponge network | 0.539 | 0.03722 | 0.494 | 0.02152 | 0.274 |
| 47967 | CTC-429P9.2 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-24-3p;hsa-miR-335-5p;hsa-miR-766-3p;hsa-miR-940 | 16 | PPARA | Sponge network | 0.043 | 0.81628 | -0.379 | 0.00548 | 0.274 |
| 47968 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | PPM1A | Sponge network | 0.572 | 0.01722 | -0.623 | 0 | 0.274 |
| 47969 | LINC00657 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | TSC1 | Sponge network | 0.91 | 0 | 0.103 | 0.26071 | 0.274 |
| 47970 | HOXA-AS2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-423-3p;hsa-miR-590-3p | 14 | ROBO1 | Sponge network | 0.61 | 0.01593 | -0.009 | 0.96154 | 0.274 |
| 47971 | MAGI2-AS3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 29 | ACSL6 | Sponge network | -0.129 | 0.57177 | -1.593 | 0 | 0.274 |
| 47972 | LINC00654 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p | 15 | ABCA10 | Sponge network | 0.374 | 0.20054 | -0.571 | 0.03674 | 0.274 |
| 47973 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 38 | FOXP1 | Sponge network | 0.267 | 0.2937 | 0.042 | 0.74368 | 0.274 |
| 47974 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-1-3p | 14 | CLIP1 | Sponge network | -0.246 | 0.35034 | -0.215 | 0.08684 | 0.274 |
| 47975 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 14 | ARNT2 | Sponge network | -0.726 | 0.00132 | -0.332 | 0.25851 | 0.274 |
| 47976 | HOXD-AS2 |
hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-378c | 13 | MED12L | Sponge network | -0.208 | 0.65592 | -0.194 | 0.55299 | 0.274 |
| 47977 | CTD-2184D3.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PDE4DIP | Sponge network | 0.272 | 0.44692 | -0.282 | 0.0257 | 0.274 |
| 47978 | CTC-301O7.4 |
hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-484 | 11 | CBX5 | Sponge network | 0.292 | 0.43384 | 0.165 | 0.24446 | 0.274 |
| 47979 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-5p | 23 | HBP1 | Sponge network | 0.95 | 0.15943 | -0.454 | 0 | 0.274 |
| 47980 | LINC00843 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | SLC5A3 | Sponge network | 1.106 | 0 | 0.493 | 5.0E-5 | 0.274 |
| 47981 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p | 38 | FZD3 | Sponge network | 0.082 | 0.55654 | 0.104 | 0.60316 | 0.274 |
| 47982 | TAPT1-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-33a-3p | 12 | MLLT10 | Sponge network | 0.403 | 2.0E-5 | 0.105 | 0.33292 | 0.274 |
| 47983 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | PHC3 | Sponge network | 0.79 | 0 | 0.212 | 0.0499 | 0.274 |
| 47984 | LINC01018 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ITGB3 | Sponge network | -2.915 | 0.00015 | -0.011 | 0.95751 | 0.274 |
| 47985 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p | 10 | KAT6B | Sponge network | 2.163 | 0 | -0.478 | 0.00018 | 0.274 |
| 47986 | MIR600HG |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p | 14 | PDE4DIP | Sponge network | 0.594 | 0.41522 | -0.282 | 0.0257 | 0.274 |
| 47987 | CTB-113P19.5 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 11 | NAV3 | Sponge network | 0.199 | 0.52763 | 0.212 | 0.47729 | 0.274 |
| 47988 | RP11-473I1.5 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p | 10 | NEDD4 | Sponge network | 0.542 | 0.00054 | 0.02 | 0.89834 | 0.274 |
| 47989 | FAM215B |
hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | TAL1 | Sponge network | 0.817 | 3.0E-5 | -0.462 | 0.00574 | 0.274 |
| 47990 | RP11-401P9.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-7-5p;hsa-miR-877-5p | 21 | CRTC1 | Sponge network | -0.384 | 0.40652 | -0.116 | 0.28412 | 0.274 |
| 47991 | C20orf166-AS1 |
hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-625-5p | 11 | TMTC2 | Sponge network | -2.69 | 0.0001 | -0.215 | 0.18248 | 0.274 |
| 47992 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-550a-3p | 22 | RAP1A | Sponge network | -0.18 | 0.20343 | -0.929 | 0 | 0.274 |
| 47993 | LINC00403 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-452-5p | 12 | JAZF1 | Sponge network | -3.586 | 0.00032 | -0.377 | 0.02677 | 0.274 |
| 47994 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-362-3p | 16 | TACC1 | Sponge network | -0.829 | 0.01343 | -0.362 | 0.05406 | 0.274 |
| 47995 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 25 | EGLN1 | Sponge network | 0.078 | 0.55803 | -0.127 | 0.1536 | 0.274 |
| 47996 | TP73-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-542-3p | 19 | TLE4 | Sponge network | -0.563 | 0.02545 | -1.157 | 0 | 0.274 |
| 47997 | RP11-395A13.2 |
hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-744-5p | 12 | CHD5 | Sponge network | 0.057 | 0.61225 | -0.922 | 0.01521 | 0.274 |
| 47998 | AC003090.1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | JDP2 | Sponge network | -2.117 | 0.00161 | -0.967 | 0 | 0.274 |
| 47999 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | PPP1R9A | Sponge network | -1.161 | 0.00591 | -0.903 | 0.04254 | 0.274 |
| 48000 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3913-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 27 | ESR1 | Sponge network | -0.738 | 0.21233 | -1.035 | 3.0E-5 | 0.274 |
| 48001 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | WDFY3 | Sponge network | -0.141 | 0.26434 | -0.201 | 0.0967 | 0.273 |
| 48002 | AC016747.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-877-5p | 15 | ASTN1 | Sponge network | 0.199 | 0.38015 | -2.389 | 2.0E-5 | 0.273 |
| 48003 | KB-1460A1.5 |
hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-486-5p;hsa-miR-495-3p | 10 | PHF6 | Sponge network | 0.603 | 0.00127 | 0.785 | 0 | 0.273 |
| 48004 | BCYRN1 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RTN4 | Sponge network | 0.311 | 0.10344 | -0.412 | 0.00014 | 0.273 |
| 48005 | CTC-510F12.2 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p | 12 | GNAQ | Sponge network | -0.691 | 0.00146 | -0.367 | 0.00102 | 0.273 |
| 48006 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-3p | 20 | DOCK4 | Sponge network | -2.69 | 0.0001 | 0.502 | 0.00108 | 0.273 |
| 48007 | CTC-296K1.3 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-940 | 10 | ZFP36L1 | Sponge network | -2.095 | 0.00112 | -0.505 | 8.0E-5 | 0.273 |
| 48008 | RP13-1039J1.4 |
hsa-miR-106a-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-34a-5p | 12 | AR | Sponge network | 0.356 | 0.33467 | -1.554 | 0 | 0.273 |
| 48009 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-93-3p | 19 | EBF3 | Sponge network | -0.384 | 0.40652 | -0.058 | 0.81437 | 0.273 |
| 48010 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-5p | 13 | MLLT10 | Sponge network | 0.983 | 0.00048 | 0.105 | 0.33292 | 0.273 |
| 48011 | U91328.19 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p | 23 | SYT4 | Sponge network | -0.303 | 0.21002 | -3.207 | 0 | 0.273 |
| 48012 | ZRANB2-AS1 |
hsa-miR-106b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-590-3p;hsa-miR-629-5p | 12 | CRTC1 | Sponge network | -0.376 | 0.00337 | -0.116 | 0.28412 | 0.273 |
| 48013 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | SYNE1 | Sponge network | -0.053 | 0.80685 | -0.738 | 0.00316 | 0.273 |
| 48014 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-361-3p | 15 | NCOA1 | Sponge network | -0.309 | 0.1145 | -0.432 | 0 | 0.273 |
| 48015 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-5p | 14 | FREM2 | Sponge network | 0.176 | 0.37402 | -0.437 | 0.46353 | 0.273 |
| 48016 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DNAJB4 | Sponge network | -0.355 | 0.0077 | -0.7 | 1.0E-5 | 0.273 |
| 48017 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 27 | ST6GAL2 | Sponge network | 0.219 | 0.1516 | -1.201 | 0.00597 | 0.273 |
| 48018 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-940 | 30 | NFIB | Sponge network | -0.829 | 0.01343 | 0.023 | 0.90795 | 0.273 |
| 48019 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-503-5p | 29 | RICTOR | Sponge network | -0.303 | 0.21002 | 0.388 | 0.00188 | 0.273 |
| 48020 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-96-5p | 13 | ELMO1 | Sponge network | 0.064 | 0.72934 | 0.04 | 0.83147 | 0.273 |
| 48021 | LOXL1-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | BRD3 | Sponge network | 0.487 | 0.04053 | 0.37 | 0.00013 | 0.273 |
| 48022 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p | 22 | PGR | Sponge network | 0.862 | 0.06213 | -1.704 | 0 | 0.273 |
| 48023 | EIF3J-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-500b-5p;hsa-miR-590-3p | 11 | HMCN1 | Sponge network | 0.331 | 0.00509 | 0.809 | 0.00544 | 0.273 |
| 48024 | RP11-403I13.7 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-671-5p | 19 | CDH23 | Sponge network | 0.395 | 0.15451 | -0.974 | 0.00034 | 0.273 |
| 48025 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p | 10 | CDC73 | Sponge network | 0.757 | 0 | 0.094 | 0.20463 | 0.273 |
| 48026 | AC007566.10 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | RPS6KA3 | Sponge network | 2.368 | 2.0E-5 | 0.256 | 0.02419 | 0.273 |
| 48027 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 60 | KIF1B | Sponge network | 0.219 | 0.1516 | -0.118 | 0.32258 | 0.273 |
| 48028 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p;hsa-miR-98-5p | 29 | PRRX1 | Sponge network | -2.452 | 0 | 1.316 | 0 | 0.273 |
| 48029 | RP11-404E16.1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-199b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-942-5p | 11 | UNC80 | Sponge network | 0.097 | 0.91311 | -1.934 | 0 | 0.273 |
| 48030 | RP11-13K12.1 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-93-5p | 11 | JUN | Sponge network | -0.154 | 0.78939 | -0.932 | 0 | 0.273 |
| 48031 | CTD-2666L21.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-629-3p;hsa-miR-744-3p | 24 | DST | Sponge network | 0.368 | 0.20093 | -0.713 | 0.00096 | 0.273 |
| 48032 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NOTCH2NL | Sponge network | -0.223 | 0.47816 | 0.024 | 0.84307 | 0.273 |
| 48033 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 13 | HERC2 | Sponge network | -1.575 | 6.0E-5 | 0.114 | 0.30638 | 0.273 |
| 48034 | CTD-2666L21.1 |
hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-629-3p | 15 | ZBTB4 | Sponge network | 0.368 | 0.20093 | -0.739 | 0 | 0.273 |
| 48035 | FAM215B |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-7-1-3p | 11 | UBE4B | Sponge network | 0.817 | 3.0E-5 | -0.235 | 0.007 | 0.273 |
| 48036 | RP11-251G23.5 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-495-3p | 19 | MDM4 | Sponge network | 1.344 | 0 | 0.345 | 0.00762 | 0.273 |
| 48037 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | MLLT10 | Sponge network | 0.569 | 0.00067 | 0.105 | 0.33292 | 0.273 |
| 48038 | RP11-48B3.4 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-942-5p | 15 | GLIPR1 | Sponge network | 1.757 | 0 | 0.146 | 0.3276 | 0.273 |
| 48039 | FAM13A-AS1 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 11 | RTN4 | Sponge network | -0.83 | 0 | -0.412 | 0.00014 | 0.273 |
| 48040 | RP11-389C8.2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PCDH10 | Sponge network | 0.727 | 3.0E-5 | -1.644 | 0.0078 | 0.273 |
| 48041 | AC006129.2 |
hsa-let-7b-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-577;hsa-miR-940 | 11 | NTRK3 | Sponge network | 0.44 | 0.15392 | -1.77 | 3.0E-5 | 0.273 |
| 48042 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-96-5p | 36 | CCND2 | Sponge network | 0.331 | 0.57247 | -0.267 | 0.3112 | 0.273 |
| 48043 | AC141928.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577 | 14 | NRK | Sponge network | 0.853 | 0.15352 | 0.656 | 0.19217 | 0.273 |
| 48044 | RP11-35G9.3 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-429;hsa-miR-455-5p | 11 | PLXNC1 | Sponge network | 0.657 | 4.0E-5 | 0.569 | 0.00329 | 0.273 |
| 48045 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | FAT4 | Sponge network | 0.61 | 0.01593 | -0.354 | 0.14694 | 0.273 |
| 48046 | RP11-195B17.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-502-3p;hsa-miR-96-5p | 15 | ARID5B | Sponge network | 0.37 | 0.27614 | 0.342 | 0.01676 | 0.273 |
| 48047 | BMS1P20 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10a-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 23 | AFF4 | Sponge network | 0.34 | 0.00273 | -0.266 | 0.01565 | 0.273 |
| 48048 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | PRKAB2 | Sponge network | 0.532 | 0.00505 | -0.367 | 0.01371 | 0.273 |
| 48049 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 26 | BMPR2 | Sponge network | 1.61 | 0.00961 | 0.206 | 0.0635 | 0.273 |
| 48050 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-7-1-3p | 11 | CNTN5 | Sponge network | -0.942 | 0 | -0.219 | 0.61957 | 0.273 |
| 48051 | RP11-822E23.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-93-5p | 13 | TRIM3 | Sponge network | -0.777 | 0.16667 | -0.577 | 0 | 0.273 |
| 48052 | HAND2-AS1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | GATA2 | Sponge network | -1.697 | 0.00889 | -0.654 | 0.0016 | 0.273 |
| 48053 | AC034220.3 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | ERCC6 | Sponge network | 0.309 | 0.07304 | 0.23 | 0.015 | 0.273 |
| 48054 | RP11-6O2.3 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-584-5p | 15 | GLIPR1 | Sponge network | 0.331 | 0.57247 | 0.146 | 0.3276 | 0.273 |
| 48055 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-590-3p | 15 | CREBBP | Sponge network | 0.219 | 0.1516 | 0.126 | 0.15186 | 0.273 |
| 48056 | HAS2-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | PCDH17 | Sponge network | -0.011 | 0.967 | 0.731 | 2.0E-5 | 0.273 |
| 48057 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | FGF5 | Sponge network | 0.311 | 0.10344 | 0.549 | 0.28659 | 0.273 |
| 48058 | AC003090.1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | CSDE1 | Sponge network | -2.117 | 0.00161 | -0.493 | 0 | 0.273 |
| 48059 | HCG15 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 11 | ZFHX3 | Sponge network | 1.09 | 0.01914 | 0.389 | 0.01363 | 0.273 |
| 48060 | BCYRN1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 34 | TGFBR3 | Sponge network | 0.311 | 0.10344 | -1.173 | 1.0E-5 | 0.273 |
| 48061 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-29b-1-5p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | PHC3 | Sponge network | -0.295 | 0.02604 | 0.212 | 0.0499 | 0.273 |
| 48062 | CTD-2002H8.2 |
hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | ADARB1 | Sponge network | 0.931 | 1.0E-5 | -0.335 | 0.06631 | 0.273 |
| 48063 | AC007879.7 |
hsa-let-7g-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p | 12 | BRD3 | Sponge network | 4.3 | 0 | 0.37 | 0.00013 | 0.273 |
| 48064 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | LATS2 | Sponge network | 0.374 | 0.20054 | 0.159 | 0.2304 | 0.273 |
| 48065 | RP5-1074L1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-374b-5p;hsa-miR-497-5p | 14 | SOX11 | Sponge network | 1.923 | 0 | 2.68 | 0 | 0.273 |
| 48066 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 33 | MAF | Sponge network | 0.194 | 0.64278 | -1.257 | 0 | 0.273 |
| 48067 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p | 17 | ADARB1 | Sponge network | 1.122 | 0.02989 | -0.335 | 0.06631 | 0.273 |
| 48068 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-424-5p | 11 | RASSF8 | Sponge network | -3.586 | 0.00032 | -0.122 | 0.65591 | 0.273 |
| 48069 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 14 | CREBBP | Sponge network | -0.769 | 0.00035 | 0.126 | 0.15186 | 0.273 |
| 48070 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429 | 25 | MOB1B | Sponge network | -2.62 | 0 | 0.197 | 0.15266 | 0.273 |
| 48071 | RP11-439L18.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-454-3p;hsa-miR-484 | 11 | SOX5 | Sponge network | 0.224 | 0.48719 | -1.091 | 4.0E-5 | 0.273 |
| 48072 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-193b-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-5p | 11 | PTPN11 | Sponge network | 0.921 | 0.00118 | 0.431 | 2.0E-5 | 0.273 |
| 48073 | RP11-307B6.3 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-589-3p | 10 | ETV1 | Sponge network | -4.463 | 0.00025 | -0.096 | 0.67674 | 0.273 |
| 48074 | TPTEP1 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-96-5p | 18 | IRS1 | Sponge network | -1.216 | 0 | -0.026 | 0.88855 | 0.273 |
| 48075 | C4B-AS1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 11 | PCDH10 | Sponge network | 2.306 | 9.0E-5 | -1.644 | 0.0078 | 0.273 |
| 48076 | RP11-181G12.2 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-378a-5p | 12 | NUAK1 | Sponge network | 0.556 | 0.00298 | 0.904 | 0 | 0.273 |
| 48077 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 21 | ARNT2 | Sponge network | -0.727 | 0.04726 | -0.332 | 0.25851 | 0.273 |
| 48078 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 19 | SUFU | Sponge network | 0.374 | 0.20054 | -0.04 | 0.65489 | 0.273 |
| 48079 | RP11-680H20.2 | hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-664a-5p | 10 | CBFB | Sponge network | 6.264 | 0 | 1.061 | 0 | 0.273 |
| 48080 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 40 | TMEM132B | Sponge network | -0.355 | 0.0077 | -0.83 | 0.01084 | 0.273 |
| 48081 | ZNF582-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | LUZP2 | Sponge network | -1.349 | 0.00017 | -0.056 | 0.89416 | 0.273 |
| 48082 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | NEDD4 | Sponge network | 0.197 | 0.25618 | 0.02 | 0.89834 | 0.273 |
| 48083 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | FOXO1 | Sponge network | -1.216 | 0 | -0.141 | 0.33874 | 0.273 |
| 48084 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | SYNE1 | Sponge network | -0.285 | 0.2172 | -0.738 | 0.00316 | 0.273 |
| 48085 | RP11-822E23.8 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-424-5p | 13 | CSDE1 | Sponge network | -0.777 | 0.16667 | -0.493 | 0 | 0.273 |
| 48086 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-376c-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 61 | IL6ST | Sponge network | -0.355 | 0.0077 | -0.402 | 0.00676 | 0.273 |
| 48087 | RP11-181G12.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-940 | 22 | PPARA | Sponge network | 0.556 | 0.00298 | -0.379 | 0.00548 | 0.273 |
| 48088 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-7-1-3p;hsa-miR-940 | 46 | WDFY3 | Sponge network | 0.75 | 0.00054 | -0.201 | 0.0967 | 0.273 |
| 48089 | RP11-411K7.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 27 | HECA | Sponge network | 0.155 | 0.81273 | -0.217 | 0.03463 | 0.273 |
| 48090 | CTD-2554C21.2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-96-5p | 15 | DOCK4 | Sponge network | -1.654 | 0.00144 | 0.502 | 0.00108 | 0.273 |
| 48091 | CTD-2666L21.1 |
hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p | 10 | MAPK10 | Sponge network | 0.368 | 0.20093 | -1.774 | 0 | 0.273 |
| 48092 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | KDSR | Sponge network | 0.614 | 1.0E-5 | 0.239 | 0.02216 | 0.273 |
| 48093 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p | 11 | LRRK2 | Sponge network | 0.16 | 0.50785 | -1.136 | 1.0E-5 | 0.273 |
| 48094 | LINC00886 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 22 | CDH23 | Sponge network | -0.371 | 0.24604 | -0.974 | 0.00034 | 0.273 |
| 48095 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-429;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-93-3p | 22 | EBF3 | Sponge network | 0.253 | 0.09565 | -0.058 | 0.81437 | 0.273 |
| 48096 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 15 | SLITRK6 | Sponge network | -0.096 | 0.67958 | -2.121 | 0 | 0.273 |
| 48097 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | NALCN | Sponge network | 0.349 | 0.01695 | 1.068 | 0.00237 | 0.273 |
| 48098 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-590-3p | 13 | MYCBP2 | Sponge network | 0.559 | 0.00026 | -0.027 | 0.82016 | 0.273 |
| 48099 | RP11-706C16.7 |
hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b | 12 | MED12L | Sponge network | 0.357 | 0.72407 | -0.194 | 0.55299 | 0.273 |
| 48100 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 17 | PDS5B | Sponge network | 0.862 | 0.06213 | 0.259 | 0.00962 | 0.273 |
| 48101 | RP11-539I5.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | MED12L | Sponge network | -0.468 | 0.32587 | -0.194 | 0.55299 | 0.273 |
| 48102 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-93-5p | 12 | FBXO31 | Sponge network | -0.154 | 0.78939 | -0.085 | 0.42389 | 0.273 |
| 48103 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | LIMD1 | Sponge network | 0.267 | 0.2937 | 0.103 | 0.28978 | 0.273 |
| 48104 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p | 12 | TGFBR2 | Sponge network | 0.659 | 3.0E-5 | -0.243 | 0.11408 | 0.273 |
| 48105 | RP11-822E23.8 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-93-3p | 16 | DOCK4 | Sponge network | -0.777 | 0.16667 | 0.502 | 0.00108 | 0.273 |
| 48106 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-590-3p | 12 | RBM5 | Sponge network | -2.452 | 0 | -0.205 | 0.0129 | 0.273 |
| 48107 | RP11-298J20.4 |
hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-5p | 11 | CBX5 | Sponge network | 0.002 | 0.99057 | 0.165 | 0.24446 | 0.273 |
| 48108 | LINC00472 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 14 | CSRNP1 | Sponge network | -0.842 | 0.01361 | -1.448 | 0 | 0.273 |
| 48109 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | NCOA1 | Sponge network | 0.931 | 1.0E-5 | -0.432 | 0 | 0.273 |
| 48110 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p | 11 | CUL5 | Sponge network | 0.336 | 0.00739 | 0.026 | 0.77301 | 0.273 |
| 48111 | LINC00963 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-96-5p | 16 | ACVR1 | Sponge network | 0.523 | 9.0E-5 | 0.279 | 0.00234 | 0.273 |
| 48112 | HCG11 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-342-5p | 10 | WNK2 | Sponge network | 0.385 | 0.10067 | -0.352 | 0.31066 | 0.273 |
| 48113 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-576-5p | 13 | PCDH17 | Sponge network | -0.932 | 0.00329 | 0.731 | 2.0E-5 | 0.273 |
| 48114 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 16 | CHD2 | Sponge network | 0.194 | 0.64278 | 0.049 | 0.55371 | 0.273 |
| 48115 | RP11-46C24.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | ZFX | Sponge network | 0.392 | 0.0278 | 0.09 | 0.42563 | 0.273 |
| 48116 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 20 | PRRX1 | Sponge network | 0.174 | 0.30034 | 1.316 | 0 | 0.273 |
| 48117 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-629-3p | 16 | EPHA7 | Sponge network | 1.122 | 0.02989 | -2.197 | 3.0E-5 | 0.273 |
| 48118 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 49 | PRKAA2 | Sponge network | 0.311 | 0.10344 | -2.462 | 0 | 0.273 |
| 48119 | NDUFA6-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 27 | TET1 | Sponge network | -0.355 | 0.0077 | -0.135 | 0.52138 | 0.273 |
| 48120 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-495-3p | 12 | NF1 | Sponge network | 0.909 | 0 | 0.51 | 0 | 0.273 |
| 48121 | RP4-669P10.16 |
hsa-miR-148a-3p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-511-5p;hsa-miR-93-3p | 10 | PCDH10 | Sponge network | -0.301 | 0.06597 | -1.644 | 0.0078 | 0.273 |
| 48122 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | PTPRD | Sponge network | -0.726 | 0.00132 | -1.005 | 0.00064 | 0.273 |
| 48123 | PRKAG2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PRKAA2 | Sponge network | -0.622 | 0.02807 | -2.462 | 0 | 0.273 |
| 48124 | RP11-758N13.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-744-5p;hsa-miR-942-5p | 21 | NOTCH2 | Sponge network | 2.939 | 3.0E-5 | 0.138 | 0.35163 | 0.273 |
| 48125 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-7-5p;hsa-miR-96-5p | 10 | FNBP1 | Sponge network | 0.374 | 0.20054 | -0.969 | 4.0E-5 | 0.273 |
| 48126 | RP11-21A7A.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-18a-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-93-3p | 10 | TRPS1 | Sponge network | -1.116 | 0.23686 | -0.191 | 0.44109 | 0.273 |
| 48127 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 29 | ARID5B | Sponge network | -0.777 | 0.1134 | 0.342 | 0.01676 | 0.273 |
| 48128 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 53 | TCF4 | Sponge network | 0.349 | 0.01695 | 0.016 | 0.92271 | 0.273 |
| 48129 | TAPT1-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-766-3p | 19 | TBL1XR1 | Sponge network | 0.403 | 2.0E-5 | 0.578 | 0 | 0.273 |
| 48130 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | FOXO1 | Sponge network | -0.576 | 0.10121 | -0.141 | 0.33874 | 0.273 |
| 48131 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p | 34 | FAT3 | Sponge network | -0.13 | 0.48734 | -1.601 | 0.00051 | 0.273 |
| 48132 | MIR22HG |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 41 | GNAQ | Sponge network | -1.047 | 0 | -0.367 | 0.00102 | 0.273 |
| 48133 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 0.401 | 0.00066 | 0.322 | 0.00215 | 0.273 |
| 48134 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-299-5p;hsa-miR-493-5p | 14 | USP6 | Sponge network | 0.368 | 0.20093 | 0.188 | 0.3871 | 0.273 |
| 48135 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 27 | USP12 | Sponge network | 0.311 | 0.10344 | -0.102 | 0.40768 | 0.273 |
| 48136 | CTC-296K1.4 |
hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-590-5p | 11 | CRTC3 | Sponge network | -2.076 | 0.00548 | 0.164 | 0.16786 | 0.273 |
| 48137 | HOXB-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-628-5p | 22 | BMPR2 | Sponge network | 1.316 | 0.01106 | 0.206 | 0.0635 | 0.273 |
| 48138 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | ERCC4 | Sponge network | -1.697 | 0.00889 | 0.126 | 0.15266 | 0.273 |
| 48139 | FAM157C |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p | 17 | CCDC6 | Sponge network | 0.91 | 0 | 0.27 | 0.02555 | 0.273 |
| 48140 | LINC01018 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PDE4DIP | Sponge network | -2.915 | 0.00015 | -0.282 | 0.0257 | 0.273 |
| 48141 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-589-5p;hsa-miR-590-5p | 21 | CXXC4 | Sponge network | -0.384 | 0.40652 | -0.433 | 0.21055 | 0.273 |
| 48142 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p | 13 | MYBL1 | Sponge network | -0.021 | 0.93598 | 0.494 | 0.02152 | 0.273 |
| 48143 | CTD-2303H24.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 12 | ABCA10 | Sponge network | 0.415 | 0.14657 | -0.571 | 0.03674 | 0.273 |
| 48144 | AGAP11 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-629-5p;hsa-miR-93-5p | 30 | MAF | Sponge network | -1.645 | 0 | -1.257 | 0 | 0.273 |
| 48145 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | ITCH | Sponge network | 0.422 | 0.27021 | 0.084 | 0.34058 | 0.273 |
| 48146 | KB-1460A1.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FBXO11 | Sponge network | 0.603 | 0.00127 | 0.142 | 0.04938 | 0.273 |
| 48147 | RP11-574K11.29 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p | 13 | DDX3X | Sponge network | 0.997 | 0 | -0.152 | 0.07686 | 0.273 |
| 48148 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 11 | ST6GAL2 | Sponge network | 0.199 | 0.52763 | -1.201 | 0.00597 | 0.273 |
| 48149 | RP11-262H14.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | PIK3CA | Sponge network | -0.106 | 0.63074 | 0.195 | 0.06908 | 0.273 |
| 48150 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-625-5p | 12 | NUAK1 | Sponge network | 1.294 | 0.0031 | 0.904 | 0 | 0.273 |
| 48151 | H19 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-486-5p;hsa-miR-664a-3p | 11 | AKAP6 | Sponge network | 2.439 | 0 | -1.586 | 3.0E-5 | 0.273 |
| 48152 | NUTM2A-AS1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | KIF5B | Sponge network | 0.643 | 0 | 0.362 | 0.00066 | 0.273 |
| 48153 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429 | 11 | ERG | Sponge network | 0.056 | 0.81195 | 0.002 | 0.99011 | 0.273 |
| 48154 | AC108488.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 13 | CDH19 | Sponge network | 0.259 | 0.12542 | -3.434 | 0 | 0.273 |
| 48155 | H19 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-486-5p | 14 | CNTN1 | Sponge network | 2.439 | 0 | -2.594 | 0 | 0.273 |
| 48156 | BCYRN1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | PEG3 | Sponge network | 0.311 | 0.10344 | -1.74 | 0 | 0.273 |
| 48157 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | DMD | Sponge network | 0.931 | 1.0E-5 | -1.506 | 0 | 0.273 |
| 48158 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-342-3p | 10 | ZNRF3 | Sponge network | 1.11 | 0 | 0.637 | 0.00172 | 0.273 |
| 48159 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-335-5p;hsa-miR-421 | 11 | PRDM2 | Sponge network | -0.285 | 0.2172 | -0.258 | 0.00721 | 0.273 |
| 48160 | LINC00680 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 13 | RASAL2 | Sponge network | 0.884 | 0 | 0.869 | 0 | 0.273 |
| 48161 | PWAR6 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-653-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 53 | PPM1L | Sponge network | -1.575 | 6.0E-5 | -0.537 | 0.00616 | 0.273 |
| 48162 | MYLK-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-424-5p;hsa-miR-935 | 16 | MTSS1 | Sponge network | -1.331 | 3.0E-5 | -0.81 | 0.00035 | 0.273 |
| 48163 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | BNC2 | Sponge network | -0.31 | 0.33871 | -0.491 | 0.09988 | 0.273 |
| 48164 | AC016747.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 17 | NIN | Sponge network | 0.199 | 0.38015 | -0.253 | 0.13794 | 0.273 |
| 48165 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-671-5p | 36 | BNC2 | Sponge network | 0.95 | 0.15943 | -0.491 | 0.09988 | 0.273 |
| 48166 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-628-5p;hsa-miR-96-5p | 21 | EPHA3 | Sponge network | 0.594 | 0.41522 | 0.185 | 0.53588 | 0.273 |
| 48167 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | MLLT10 | Sponge network | 0.986 | 0.01022 | 0.105 | 0.33292 | 0.273 |
| 48168 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p | 21 | PDGFRA | Sponge network | 0.178 | 0.29106 | -0.372 | 0.0037 | 0.273 |
| 48169 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-744-3p | 11 | CUL5 | Sponge network | 2.368 | 2.0E-5 | 0.026 | 0.77301 | 0.273 |
| 48170 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | DOCK4 | Sponge network | -2.452 | 0 | 0.502 | 0.00108 | 0.273 |
| 48171 | MYLK-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-340-5p;hsa-miR-7-5p | 19 | EGFR | Sponge network | -1.331 | 3.0E-5 | -0.175 | 0.48451 | 0.273 |
| 48172 | MAGI2-AS3 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | NR3C2 | Sponge network | -0.129 | 0.57177 | -1.56 | 0 | 0.273 |
| 48173 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-940 | 30 | PTPRT | Sponge network | 0.225 | 0.30644 | -0.907 | 0.03727 | 0.273 |
| 48174 | NDUFA6-AS1 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ID4 | Sponge network | -0.355 | 0.0077 | -1.644 | 0 | 0.273 |
| 48175 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-21-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PBRM1 | Sponge network | -0.513 | 0.04703 | -0.029 | 0.78601 | 0.273 |
| 48176 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | ST6GALNAC3 | Sponge network | 0.131 | 0.8436 | -0.987 | 0 | 0.273 |
| 48177 | RP11-25K19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 25 | NCOA1 | Sponge network | -1.057 | 0.00029 | -0.432 | 0 | 0.273 |
| 48178 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 17 | AHNAK | Sponge network | -0.615 | 0.00015 | -1.207 | 0 | 0.273 |
| 48179 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 29 | TCF4 | Sponge network | 0.37 | 0.27614 | 0.016 | 0.92271 | 0.273 |
| 48180 | U91328.21 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p | 13 | CHD5 | Sponge network | -0.735 | 0.15983 | -0.922 | 0.01521 | 0.273 |
| 48181 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p | 47 | PAFAH1B1 | Sponge network | -0.777 | 0.1134 | -0.429 | 0 | 0.273 |
| 48182 | LINC00861 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-940 | 10 | MCC | Sponge network | 0.911 | 0.00854 | -1.005 | 0 | 0.273 |
| 48183 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-450b-5p | 11 | EBF1 | Sponge network | 0.912 | 0.00014 | -0.384 | 0.08856 | 0.273 |
| 48184 | ACTA2-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | INPP4B | Sponge network | 0.194 | 0.64278 | -0.097 | 0.60503 | 0.273 |
| 48185 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p | 11 | ABL1 | Sponge network | -0.031 | 0.89063 | -0.215 | 0.07804 | 0.273 |
| 48186 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | NEDD4 | Sponge network | 0.336 | 0.00739 | 0.02 | 0.89834 | 0.273 |
| 48187 | AP001347.6 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-590-3p | 11 | ZFP36L1 | Sponge network | -1.161 | 0.00591 | -0.505 | 8.0E-5 | 0.273 |
| 48188 | RP11-373D23.3 |
hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | CUX1 | Sponge network | -0.285 | 0.2172 | -0.039 | 0.6705 | 0.273 |
| 48189 | HOXB-AS3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-432-5p;hsa-miR-629-5p;hsa-miR-942-5p | 16 | UNC80 | Sponge network | 0.343 | 0.28887 | -1.934 | 0 | 0.273 |
| 48190 | NDUFA6-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | KCTD8 | Sponge network | -0.355 | 0.0077 | -3.359 | 0 | 0.273 |
| 48191 | AC009120.6 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-30e-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | RASSF8 | Sponge network | 0.919 | 0 | -0.122 | 0.65591 | 0.273 |
| 48192 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 19 | TCF4 | Sponge network | 0.476 | 0.19203 | 0.016 | 0.92271 | 0.273 |
| 48193 | RP11-428J1.5 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DNAJB4 | Sponge network | 0.538 | 0.00076 | -0.7 | 1.0E-5 | 0.273 |
| 48194 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-409-3p | 10 | AKAP6 | Sponge network | 0.453 | 0.01839 | -1.586 | 3.0E-5 | 0.273 |
| 48195 | RP11-822E23.8 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-93-5p | 23 | ACSL6 | Sponge network | -0.777 | 0.16667 | -1.593 | 0 | 0.273 |
| 48196 | RP11-798M19.6 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-485-3p;hsa-miR-590-3p | 16 | TMTC2 | Sponge network | -0.612 | 0.00094 | -0.215 | 0.18248 | 0.273 |
| 48197 | RP4-758J18.10 |
hsa-miR-10b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-450b-5p | 10 | TMEM132B | Sponge network | -0.091 | 0.71468 | -0.83 | 0.01084 | 0.273 |
| 48198 | MEG3 |
hsa-miR-140-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 18 | CELF4 | Sponge network | 0.249 | 0.35902 | -1.135 | 0.00344 | 0.273 |
| 48199 | RP11-894P9.1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-582-5p | 22 | IL6ST | Sponge network | 0.993 | 0 | -0.402 | 0.00676 | 0.273 |
| 48200 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 21 | CCND2 | Sponge network | -1.255 | 0.00981 | -0.267 | 0.3112 | 0.273 |
| 48201 | RP11-156P1.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 17 | LUZP2 | Sponge network | 0.214 | 0.18246 | -0.056 | 0.89416 | 0.273 |
| 48202 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | ADAMTSL3 | Sponge network | -0.83 | 0 | -1.559 | 0 | 0.273 |
| 48203 | RP11-403I13.8 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ITPKB | Sponge network | 0.619 | 0.00157 | -0.704 | 0.00106 | 0.273 |
| 48204 | LINC00173 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | KCNA6 | Sponge network | 0.056 | 0.8494 | -1.531 | 0.00013 | 0.273 |
| 48205 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | FOXO1 | Sponge network | -0.976 | 0.00025 | -0.141 | 0.33874 | 0.273 |
| 48206 | TMEM9B-AS1 |
hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 0.529 | 0.00552 | 0.51 | 0 | 0.273 |
| 48207 | HCG18 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | MED12L | Sponge network | 1.063 | 0 | -0.194 | 0.55299 | 0.273 |
| 48208 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | LRIG1 | Sponge network | -0.342 | 0.02288 | -0.537 | 0.00588 | 0.273 |
| 48209 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p | 10 | PRRX1 | Sponge network | -1.717 | 0.01522 | 1.316 | 0 | 0.273 |
| 48210 | TMEM9B-AS1 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-582-5p | 15 | ZFHX3 | Sponge network | 0.529 | 0.00552 | 0.389 | 0.01363 | 0.273 |
| 48211 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 26 | LIFR | Sponge network | -0.668 | 3.0E-5 | -1.794 | 0 | 0.273 |
| 48212 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | NIN | Sponge network | 1.089 | 0 | -0.253 | 0.13794 | 0.273 |
| 48213 | RP11-517B11.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-664a-5p | 11 | CSNK1A1 | Sponge network | 0.706 | 0 | -0.19 | 0.01687 | 0.273 |
| 48214 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 11 | CREBBP | Sponge network | 0.91 | 0 | 0.126 | 0.15186 | 0.273 |
| 48215 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-542-3p | 18 | SLITRK6 | Sponge network | -0.969 | 2.0E-5 | -2.121 | 0 | 0.273 |
| 48216 | CTD-3064M3.3 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-429 | 10 | DNMT3A | Sponge network | -0.469 | 0.08721 | 0.302 | 0.0262 | 0.273 |
| 48217 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | NALCN | Sponge network | -0.727 | 0.04726 | 1.068 | 0.00237 | 0.273 |
| 48218 | LLNLF-65H9.1 |
hsa-miR-103a-2-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-502-5p | 10 | IL17RD | Sponge network | -0.033 | 0.89251 | -0.019 | 0.94873 | 0.273 |
| 48219 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-589-3p | 14 | PGR | Sponge network | 0.622 | 0.00015 | -1.704 | 0 | 0.273 |
| 48220 | AP001347.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PDE4DIP | Sponge network | -1.161 | 0.00591 | -0.282 | 0.0257 | 0.273 |
| 48221 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | IGSF10 | Sponge network | -0.125 | 0.49414 | -1.751 | 1.0E-5 | 0.273 |
| 48222 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-708-5p;hsa-miR-935 | 27 | MTSS1 | Sponge network | -0.096 | 0.67958 | -0.81 | 0.00035 | 0.273 |
| 48223 | AC074289.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-96-5p | 13 | PRRX1 | Sponge network | -0.326 | 0.14314 | 1.316 | 0 | 0.273 |
| 48224 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 17 | TIMP3 | Sponge network | 0.4 | 0.14611 | -0.546 | 0.02307 | 0.273 |
| 48225 | AF131215.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-589-3p | 19 | PRDM2 | Sponge network | -0.176 | 0.59692 | -0.258 | 0.00721 | 0.273 |
| 48226 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | SETBP1 | Sponge network | 0.998 | 0 | -1.302 | 1.0E-5 | 0.273 |
| 48227 | AC011526.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 16 | TGFBR3 | Sponge network | 0.342 | 0.03129 | -1.173 | 1.0E-5 | 0.273 |
| 48228 | CKMT2-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 17 | LUZP2 | Sponge network | 0.082 | 0.55654 | -0.056 | 0.89416 | 0.273 |
| 48229 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 28 | SOX11 | Sponge network | 0.683 | 1.0E-5 | 2.68 | 0 | 0.273 |
| 48230 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-3p | 13 | MXI1 | Sponge network | -0.376 | 0.00337 | -0.987 | 0 | 0.273 |
| 48231 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | MYO9A | Sponge network | 0.336 | 0.00739 | -0.147 | 0.32373 | 0.273 |
| 48232 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | RARB | Sponge network | 0.259 | 0.12542 | -0.825 | 2.0E-5 | 0.273 |
| 48233 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-455-3p;hsa-miR-582-3p | 26 | TCF4 | Sponge network | 0.858 | 3.0E-5 | 0.016 | 0.92271 | 0.273 |
| 48234 | LINC00963 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | ERCC4 | Sponge network | 0.523 | 9.0E-5 | 0.126 | 0.15266 | 0.273 |
| 48235 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-93-5p | 10 | ARNT2 | Sponge network | -0.351 | 0.11358 | -0.332 | 0.25851 | 0.273 |
| 48236 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 22 | PTPN11 | Sponge network | 0.311 | 0.10344 | 0.431 | 2.0E-5 | 0.273 |
| 48237 | RP11-254F7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p | 21 | NCOA1 | Sponge network | 0.176 | 0.37402 | -0.432 | 0 | 0.273 |
| 48238 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-93-5p | 23 | RNASEL | Sponge network | -1.645 | 0 | -0.272 | 0.01796 | 0.273 |
| 48239 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-877-5p | 12 | CBFA2T3 | Sponge network | 0.889 | 9.0E-5 | -1.221 | 1.0E-5 | 0.273 |
| 48240 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-542-3p | 16 | RARB | Sponge network | -0.227 | 0.31657 | -0.825 | 2.0E-5 | 0.273 |
| 48241 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-330-5p;hsa-miR-590-3p | 14 | USP6 | Sponge network | 0.051 | 0.83388 | 0.188 | 0.3871 | 0.273 |
| 48242 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-130b-3p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-96-5p | 11 | FOXO3 | Sponge network | 3.21 | 0 | -0.04 | 0.72028 | 0.273 |
| 48243 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-5p | 20 | PRKCE | Sponge network | -0.147 | 0.40452 | -0.403 | 0.00056 | 0.273 |
| 48244 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-590-3p | 12 | GPAM | Sponge network | 0.34 | 0.00273 | 0.142 | 0.29732 | 0.273 |
| 48245 | FAM13A-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 45 | DTNA | Sponge network | -0.83 | 0 | -1.648 | 1.0E-5 | 0.273 |
| 48246 | ADIRF-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | HOMER2 | Sponge network | -0.223 | 0.47816 | -2.698 | 0 | 0.273 |
| 48247 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 48 | CBX5 | Sponge network | -0.777 | 0.1134 | 0.165 | 0.24446 | 0.273 |
| 48248 | BDNF-AS |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 25 | OPCML | Sponge network | -0.668 | 3.0E-5 | -2.498 | 0 | 0.273 |
| 48249 | HOXB-AS3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-629-3p;hsa-miR-629-5p | 12 | RASSF8 | Sponge network | 0.343 | 0.28887 | -0.122 | 0.65591 | 0.273 |
| 48250 | RP11-410L14.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-33a-5p | 10 | USP6 | Sponge network | -0.052 | 0.76839 | 0.188 | 0.3871 | 0.273 |
| 48251 | RP11-35G9.3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 35 | DTNA | Sponge network | 0.657 | 4.0E-5 | -1.648 | 1.0E-5 | 0.273 |
| 48252 | LINC01018 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | RNF144A | Sponge network | -2.915 | 0.00015 | 0.594 | 0.0009 | 0.273 |
| 48253 | LINC00654 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 39 | NOTCH2 | Sponge network | 0.374 | 0.20054 | 0.138 | 0.35163 | 0.273 |
| 48254 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-652-3p;hsa-miR-93-3p;hsa-miR-940 | 44 | QKI | Sponge network | 0.494 | 0.00339 | -0.333 | 0.04111 | 0.273 |
| 48255 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-502-5p | 10 | TGFBR2 | Sponge network | -0.164 | 0.45106 | -0.243 | 0.11408 | 0.273 |
| 48256 | RP11-428J1.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 23 | NTRK2 | Sponge network | 0.538 | 0.00076 | -0.736 | 0.06495 | 0.273 |
| 48257 | BDNF-AS |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-590-3p | 12 | RIT1 | Sponge network | -0.668 | 3.0E-5 | -0.296 | 0.00054 | 0.273 |
| 48258 | RP1-151F17.2 |
hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 19 | SYNPO2 | Sponge network | 0.222 | 0.29133 | -2.342 | 0 | 0.273 |
| 48259 | CTD-2619J13.14 |
hsa-miR-101-3p;hsa-miR-139-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 10 | GSK3B | Sponge network | 0.679 | 0 | 0.266 | 0.00045 | 0.273 |
| 48260 | RP11-327P2.5 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-664a-3p | 36 | PPM1L | Sponge network | -0.147 | 0.40452 | -0.537 | 0.00616 | 0.273 |
| 48261 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-582-5p | 25 | KIF1B | Sponge network | 1.093 | 0 | -0.118 | 0.32258 | 0.273 |
| 48262 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | IGSF10 | Sponge network | 0.381 | 0.25967 | -1.751 | 1.0E-5 | 0.273 |
| 48263 | AC005154.6 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | BCL9 | Sponge network | 0.848 | 0 | 0.321 | 0.00267 | 0.273 |
| 48264 | RP11-504P24.8 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-497-5p | 18 | CCDC6 | Sponge network | 1.178 | 0 | 0.27 | 0.02555 | 0.273 |
| 48265 | FAM66C |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940 | 12 | FOXO4 | Sponge network | -0.901 | 0.00019 | -0.516 | 2.0E-5 | 0.273 |
| 48266 | LINC00963 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 30 | PHC3 | Sponge network | 0.523 | 9.0E-5 | 0.212 | 0.0499 | 0.273 |
| 48267 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-93-3p | 16 | FOXO3 | Sponge network | -0.693 | 0.05629 | -0.04 | 0.72028 | 0.273 |
| 48268 | OIP5-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 24 | ABL1 | Sponge network | 0.421 | 0.00084 | -0.215 | 0.07804 | 0.272 |
| 48269 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p | 32 | PIK3R1 | Sponge network | -0.125 | 0.49414 | -0.133 | 0.36854 | 0.272 |
| 48270 | AF131215.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p | 22 | PRDM2 | Sponge network | -0.322 | 0.25945 | -0.258 | 0.00721 | 0.272 |
| 48271 | DNAJC3-AS1 |
hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-582-5p | 10 | PRKAA1 | Sponge network | 0.753 | 0.00014 | 0.317 | 0.0031 | 0.272 |
| 48272 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | ZNF292 | Sponge network | -0.615 | 0.00015 | 0.276 | 0.04148 | 0.272 |
| 48273 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-424-5p;hsa-miR-589-3p | 14 | TACC1 | Sponge network | 0.529 | 0.00552 | -0.362 | 0.05406 | 0.272 |
| 48274 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | NRXN1 | Sponge network | -0.141 | 0.26434 | -3.778 | 0 | 0.272 |
| 48275 | U91328.21 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 14 | TMEM132B | Sponge network | -0.735 | 0.15983 | -0.83 | 0.01084 | 0.272 |
| 48276 | LIPE-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-34a-5p;hsa-miR-625-5p | 11 | CEP170 | Sponge network | 0.078 | 0.55803 | 0.943 | 0 | 0.272 |
| 48277 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-942-5p | 12 | CREBBP | Sponge network | 0.538 | 0.00076 | 0.126 | 0.15186 | 0.272 |
| 48278 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-664a-5p | 26 | SMAD4 | Sponge network | -0.583 | 0.14589 | -0.313 | 0.00413 | 0.272 |
| 48279 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 31 | ST6GALNAC3 | Sponge network | -0.615 | 0.00015 | -0.987 | 0 | 0.272 |
| 48280 | FAM13A-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | CAV1 | Sponge network | -0.83 | 0 | -1.013 | 6.0E-5 | 0.272 |
| 48281 | AC093110.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 14 | RPS6KA3 | Sponge network | 1.044 | 2.0E-5 | 0.256 | 0.02419 | 0.272 |
| 48282 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | KAT6B | Sponge network | 0.385 | 0.10067 | -0.478 | 0.00018 | 0.272 |
| 48283 | AC034220.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | TCF12 | Sponge network | 0.309 | 0.07304 | 0.174 | 0.13271 | 0.272 |
| 48284 | RP5-998N21.10 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p | 10 | RASSF2 | Sponge network | -0.098 | 0.76298 | -0.273 | 0.21961 | 0.272 |
| 48285 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | ARNT2 | Sponge network | 0.385 | 0.10067 | -0.332 | 0.25851 | 0.272 |
| 48286 | TINCR |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-940 | 14 | TRIM33 | Sponge network | -0.664 | 0.14543 | 0.402 | 7.0E-5 | 0.272 |
| 48287 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | ST6GAL2 | Sponge network | -0.899 | 6.0E-5 | -1.201 | 0.00597 | 0.272 |
| 48288 | LINC00672 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 15 | CYLD | Sponge network | -0.227 | 0.31657 | -0.367 | 0.00171 | 0.272 |
| 48289 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 23 | PIK3C2A | Sponge network | 0.541 | 0.00125 | 0.061 | 0.53776 | 0.272 |
| 48290 | DOCK9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-539-5p;hsa-miR-589-5p | 19 | PRKAA2 | Sponge network | -0.231 | 0.28214 | -2.462 | 0 | 0.272 |
| 48291 | LPP-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-500a-5p | 11 | ZNF208 | Sponge network | -0.18 | 0.20343 | -0.4 | 0.22381 | 0.272 |
| 48292 | FAM13A-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-96-5p | 27 | SETBP1 | Sponge network | -0.83 | 0 | -1.302 | 1.0E-5 | 0.272 |
| 48293 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-629-3p | 13 | BHLHE41 | Sponge network | 1.122 | 0.02989 | 0.353 | 0.12753 | 0.272 |
| 48294 | FGD5-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 38 | CBX5 | Sponge network | 0.401 | 0.00066 | 0.165 | 0.24446 | 0.272 |
| 48295 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 16 | ADAMTSL3 | Sponge network | 0.619 | 0.00157 | -1.559 | 0 | 0.272 |
| 48296 | RP11-464F9.1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-342-3p;hsa-miR-590-3p | 12 | PICALM | Sponge network | 0.959 | 0 | 0.086 | 0.23523 | 0.272 |
| 48297 | PWAR6 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-93-3p | 10 | PEA15 | Sponge network | -1.575 | 6.0E-5 | 0.009 | 0.93318 | 0.272 |
| 48298 | LINC00672 |
hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 16 | LIMCH1 | Sponge network | -0.227 | 0.31657 | -0.915 | 1.0E-5 | 0.272 |
| 48299 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-542-3p | 10 | TSC22D1 | Sponge network | -1.439 | 4.0E-5 | -0.529 | 2.0E-5 | 0.272 |
| 48300 | RP5-1021I20.1 |
hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p | 10 | SEMA3C | Sponge network | -0.785 | 0.00035 | -0.272 | 0.31153 | 0.272 |
| 48301 | CTB-31O20.4 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | RPS6KA3 | Sponge network | 1.619 | 0 | 0.256 | 0.02419 | 0.272 |
| 48302 | H1FX-AS1 |
hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-93-3p | 12 | ST6GAL2 | Sponge network | -0.206 | 0.12942 | -1.201 | 0.00597 | 0.272 |
| 48303 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-26b-5p;hsa-miR-409-3p;hsa-miR-501-3p;hsa-miR-96-5p | 10 | CLASP2 | Sponge network | 0.453 | 0.01839 | -0.038 | 0.71 | 0.272 |
| 48304 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-7-1-3p | 16 | EYA4 | Sponge network | 0.663 | 0.00073 | 0.3 | 0.36876 | 0.272 |
| 48305 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 19 | USP12 | Sponge network | 0.267 | 0.2937 | -0.102 | 0.40768 | 0.272 |
| 48306 | NNT-AS1 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p | 13 | ATM | Sponge network | 0.799 | 1.0E-5 | 0.178 | 0.21568 | 0.272 |
| 48307 | NDUFA6-AS1 |
hsa-miR-106b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-377-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | PER2 | Sponge network | -0.355 | 0.0077 | -0.396 | 0.00269 | 0.272 |
| 48308 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-532-3p;hsa-miR-93-5p | 21 | PPM1L | Sponge network | 0.497 | 0.01451 | -0.537 | 0.00616 | 0.272 |
| 48309 | AGAP11 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-942-5p | 11 | RND3 | Sponge network | -1.645 | 0 | -0.254 | 0.11478 | 0.272 |
| 48310 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | PIK3C2A | Sponge network | 0.8 | 0 | 0.061 | 0.53776 | 0.272 |
| 48311 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 21 | RASSF8 | Sponge network | -0.125 | 0.49414 | -0.122 | 0.65591 | 0.272 |
| 48312 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 46 | PAFAH1B1 | Sponge network | 0.421 | 0.00084 | -0.429 | 0 | 0.272 |
| 48313 | ANKRD10-IT1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-505-3p;hsa-miR-7-1-3p | 13 | FZD3 | Sponge network | 1.739 | 0 | 0.104 | 0.60316 | 0.272 |
| 48314 | RP4-584D14.7 |
hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-577 | 13 | RBMS3 | Sponge network | 1.294 | 0.0031 | -0.519 | 0.03551 | 0.272 |
| 48315 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-501-3p;hsa-miR-590-3p | 10 | SMARCA1 | Sponge network | 0.559 | 0.00026 | -0.684 | 0.00159 | 0.272 |
| 48316 | AC005618.6 |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 18 | DOCK4 | Sponge network | 0.862 | 0.06213 | 0.502 | 0.00108 | 0.272 |
| 48317 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-708-5p;hsa-miR-96-5p | 34 | SPRY3 | Sponge network | -0.235 | 0.15142 | 0.016 | 0.91286 | 0.272 |
| 48318 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-345-5p;hsa-miR-625-5p | 12 | SNX29 | Sponge network | 0.858 | 3.0E-5 | -0.34 | 0.01371 | 0.272 |
| 48319 | LINC00472 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | SIK1 | Sponge network | -0.842 | 0.01361 | -0.98 | 0 | 0.272 |
| 48320 | RP11-13J8.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p | 10 | MOB1B | Sponge network | 3.03 | 0 | 0.197 | 0.15266 | 0.272 |
| 48321 | AC006116.24 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-550a-3p;hsa-miR-582-3p | 11 | BRD3 | Sponge network | -0.473 | 0.07602 | 0.37 | 0.00013 | 0.272 |
| 48322 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-429;hsa-miR-96-5p | 11 | CREB3L2 | Sponge network | 0.348 | 0.32912 | -0.525 | 2.0E-5 | 0.272 |
| 48323 | ADAMTS9-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | ETV1 | Sponge network | -2.452 | 0 | -0.096 | 0.67674 | 0.272 |
| 48324 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-664a-3p | 16 | AFF4 | Sponge network | 1.316 | 0 | -0.266 | 0.01565 | 0.272 |
| 48325 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-429 | 17 | ARHGAP28 | Sponge network | -0.154 | 0.53954 | -0.911 | 0.00013 | 0.272 |
| 48326 | MEG3 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-942-5p;hsa-miR-98-5p | 15 | PICALM | Sponge network | 0.249 | 0.35902 | 0.086 | 0.23523 | 0.272 |
| 48327 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 50 | FAT3 | Sponge network | -0.727 | 0.04726 | -1.601 | 0.00051 | 0.272 |
| 48328 | CALML3-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p | 18 | PIK3CA | Sponge network | 0.95 | 0.15943 | 0.195 | 0.06908 | 0.272 |
| 48329 | AC004540.4 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p | 13 | BNC2 | Sponge network | -2.549 | 0.00528 | -0.491 | 0.09988 | 0.272 |
| 48330 | HOXB-AS1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-362-3p | 11 | ATM | Sponge network | -0.154 | 0.53954 | 0.178 | 0.21568 | 0.272 |
| 48331 | RP11-434H6.7 |
hsa-let-7g-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-32-5p | 10 | BRD3 | Sponge network | 0.358 | 0.14302 | 0.37 | 0.00013 | 0.272 |
| 48332 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-940 | 24 | BHLHE41 | Sponge network | -0.896 | 0.00099 | 0.353 | 0.12753 | 0.272 |
| 48333 | CTC-487M23.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p | 14 | ARHGEF12 | Sponge network | 0.912 | 0.00014 | 0.09 | 0.35693 | 0.272 |
| 48334 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-340-5p;hsa-miR-374a-5p | 23 | PPM1A | Sponge network | -0.693 | 0.05629 | -0.623 | 0 | 0.272 |
| 48335 | KB-1572G7.2 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p | 11 | MAT2A | Sponge network | 0.924 | 0 | -0.073 | 0.36195 | 0.272 |
| 48336 | RP4-758J18.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p | 12 | HLF | Sponge network | -0.091 | 0.71468 | -2.443 | 0 | 0.272 |
| 48337 | CTC-296K1.3 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-671-5p | 14 | CYLD | Sponge network | -2.095 | 0.00112 | -0.367 | 0.00171 | 0.272 |
| 48338 | HCG11 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | CELF4 | Sponge network | 0.385 | 0.10067 | -1.135 | 0.00344 | 0.272 |
| 48339 | RP11-401P9.4 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-935 | 18 | GJA1 | Sponge network | -0.384 | 0.40652 | -0.485 | 0.02179 | 0.272 |
| 48340 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 30 | ZFHX3 | Sponge network | 0.488 | 0.00084 | 0.389 | 0.01363 | 0.272 |
| 48341 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | EYA4 | Sponge network | 0.998 | 0 | 0.3 | 0.36876 | 0.272 |
| 48342 | CTD-2184D3.6 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | DYNC1I1 | Sponge network | 0.272 | 0.44692 | -1.12 | 0.00107 | 0.272 |
| 48343 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p | 29 | MDM4 | Sponge network | 0.542 | 0.00054 | 0.345 | 0.00762 | 0.272 |
| 48344 | RP11-66B24.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-375 | 12 | ABI2 | Sponge network | 0.57 | 0.1866 | 0.175 | 0.14787 | 0.272 |
| 48345 | RP13-1039J1.4 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-589-3p | 15 | PTPRD | Sponge network | 0.356 | 0.33467 | -1.005 | 0.00064 | 0.272 |
| 48346 | LINC00954 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-375;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 10 | ABI2 | Sponge network | 1.157 | 0.00455 | 0.175 | 0.14787 | 0.272 |
| 48347 | RP11-758N13.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-375;hsa-miR-93-3p | 25 | BMPR2 | Sponge network | 2.939 | 3.0E-5 | 0.206 | 0.0635 | 0.272 |
| 48348 | RP11-452L6.1 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-664a-5p | 15 | LPP | Sponge network | 0.665 | 0.00061 | -0.459 | 0.04475 | 0.272 |
| 48349 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SEMA3C | Sponge network | 0.559 | 0.00026 | -0.272 | 0.31153 | 0.272 |
| 48350 | EIF3J-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 16 | ERCC4 | Sponge network | 0.331 | 0.00509 | 0.126 | 0.15266 | 0.272 |
| 48351 | HAND2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | MED12L | Sponge network | -1.697 | 0.00889 | -0.194 | 0.55299 | 0.272 |
| 48352 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-660-5p | 11 | EYA4 | Sponge network | 0.197 | 0.25618 | 0.3 | 0.36876 | 0.272 |
| 48353 | RP4-605O3.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 12 | SCN9A | Sponge network | 0.262 | 0.0475 | -0.525 | 0.10593 | 0.272 |
| 48354 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | NOTCH2NL | Sponge network | 0.225 | 0.30644 | 0.024 | 0.84307 | 0.272 |
| 48355 | LINC00672 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-629-3p | 13 | ID4 | Sponge network | -0.227 | 0.31657 | -1.644 | 0 | 0.272 |
| 48356 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-493-5p | 12 | CDC73 | Sponge network | 1.044 | 2.0E-5 | 0.094 | 0.20463 | 0.272 |
| 48357 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | LATS1 | Sponge network | 0.986 | 0.01022 | 0.109 | 0.34208 | 0.272 |
| 48358 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | TSHZ3 | Sponge network | 0.906 | 0.31715 | -0.556 | 0.01516 | 0.272 |
| 48359 | TP73-AS1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p | 11 | CACNA1E | Sponge network | -0.563 | 0.02545 | 1.752 | 0 | 0.272 |
| 48360 | DIO3OS |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-940 | 28 | PPP1R9A | Sponge network | -0.773 | 0.04073 | -0.903 | 0.04254 | 0.272 |
| 48361 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 22 | RARB | Sponge network | 0.101 | 0.60919 | -0.825 | 2.0E-5 | 0.272 |
| 48362 | ZNF667-AS1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 12 | FOXO4 | Sponge network | -0.976 | 0.00025 | -0.516 | 2.0E-5 | 0.272 |
| 48363 | CYP4F35P |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-17-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-331-3p;hsa-miR-96-5p;hsa-miR-98-5p | 10 | PRKAA2 | Sponge network | -0.798 | 0.18964 | -2.462 | 0 | 0.272 |
| 48364 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-125a-3p;hsa-miR-200c-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b | 10 | HIVEP1 | Sponge network | 0.75 | 0.00054 | 0.368 | 0.00064 | 0.272 |
| 48365 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-96-5p | 13 | FOXO3 | Sponge network | -1.654 | 0.00144 | -0.04 | 0.72028 | 0.272 |
| 48366 | LINC00854 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-455-3p;hsa-miR-628-5p | 15 | RUNX1T1 | Sponge network | 1.18 | 0 | -0.344 | 0.2374 | 0.272 |
| 48367 | LINC00926 |
hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-940 | 14 | GRIK3 | Sponge network | 0.756 | 0.00739 | -3.208 | 0 | 0.272 |
| 48368 | CTD-2528L19.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429 | 13 | PTPN14 | Sponge network | 0.953 | 0 | 0.144 | 0.34595 | 0.272 |
| 48369 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-5p | 17 | RBMS3 | Sponge network | 0.001 | 0.99753 | -0.519 | 0.03551 | 0.272 |
| 48370 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | SCN9A | Sponge network | -0.727 | 0.04726 | -0.525 | 0.10593 | 0.272 |
| 48371 | LINC00963 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-940 | 25 | PBX1 | Sponge network | 0.523 | 9.0E-5 | -1.206 | 0 | 0.272 |
| 48372 | OSER1-AS1 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 14 | LIMCH1 | Sponge network | -0.13 | 0.48734 | -0.915 | 1.0E-5 | 0.272 |
| 48373 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-589-3p | 21 | RNASEL | Sponge network | 0.72 | 0.0001 | -0.272 | 0.01796 | 0.272 |
| 48374 | BDNF-AS |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 42 | WDFY3 | Sponge network | -0.668 | 3.0E-5 | -0.201 | 0.0967 | 0.272 |
| 48375 | CTD-2336O2.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | ASTN1 | Sponge network | -0.141 | 0.26434 | -2.389 | 2.0E-5 | 0.272 |
| 48376 | RP11-686O6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-455-3p | 11 | RICTOR | Sponge network | 0.453 | 0.01971 | 0.388 | 0.00188 | 0.272 |
| 48377 | AF131215.9 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | HOMER2 | Sponge network | -0.322 | 0.25945 | -2.698 | 0 | 0.272 |
| 48378 | RP11-244H3.1 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | 0.659 | 3.0E-5 | 0.098 | 0.40994 | 0.272 |
| 48379 | CASC2 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-758-3p | 18 | CDC73 | Sponge network | 0.385 | 0.01306 | 0.094 | 0.20463 | 0.272 |
| 48380 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | BCL11A | Sponge network | -1.057 | 0.00029 | -0.932 | 0.0063 | 0.272 |
| 48381 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-93-5p;hsa-miR-940 | 25 | BHLHE41 | Sponge network | -0.162 | 0.47687 | 0.353 | 0.12753 | 0.272 |
| 48382 | TINCR |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-96-5p | 17 | PIK3CA | Sponge network | -0.664 | 0.14543 | 0.195 | 0.06908 | 0.272 |
| 48383 | RP11-2H8.2 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 16 | MED12L | Sponge network | 1.089 | 0 | -0.194 | 0.55299 | 0.272 |
| 48384 | PSMG3-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-532-3p | 17 | CADM3 | Sponge network | -0.125 | 0.49414 | -3.663 | 0 | 0.272 |
| 48385 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-590-3p | 20 | SON | Sponge network | 0.569 | 0.00067 | 0.168 | 0.04406 | 0.272 |
| 48386 | RP11-680C21.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 21 | TET1 | Sponge network | -1.424 | 0.04346 | -0.135 | 0.52138 | 0.272 |
| 48387 | RP11-967K21.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-3607-3p | 11 | PLAG1 | Sponge network | 0.056 | 0.81195 | -0.315 | 0.26532 | 0.272 |
| 48388 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429 | 12 | ABCB5 | Sponge network | 0.16 | 0.50785 | -0.956 | 0.04077 | 0.272 |
| 48389 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | LRRC7 | Sponge network | 0.72 | 0.0001 | -1.341 | 0.01193 | 0.272 |
| 48390 | RP11-158K1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | FGFR1 | Sponge network | 0.349 | 0.01695 | -0.509 | 0.03957 | 0.272 |
| 48391 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-484 | 11 | MYCBP2 | Sponge network | -0.176 | 0.59692 | -0.027 | 0.82016 | 0.272 |
| 48392 | TPTEP1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 14 | EGR2 | Sponge network | -1.216 | 0 | 0.309 | 0.17079 | 0.272 |
| 48393 | RP11-327P2.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-5p | 21 | MED12L | Sponge network | -0.147 | 0.40452 | -0.194 | 0.55299 | 0.272 |
| 48394 | CTD-3092A11.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 13 | CYLD | Sponge network | 0.873 | 0 | -0.367 | 0.00171 | 0.272 |
| 48395 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SYNE1 | Sponge network | 0.559 | 0.00026 | -0.738 | 0.00316 | 0.272 |
| 48396 | RP11-983P16.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-5p | 15 | PTGFR | Sponge network | 0.41 | 0.02214 | -0.233 | 0.45421 | 0.272 |
| 48397 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-500a-5p | 23 | CCND2 | Sponge network | -0.932 | 0.00329 | -0.267 | 0.3112 | 0.272 |
| 48398 | HOTAIRM1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 11 | ROBO1 | Sponge network | -0.053 | 0.80685 | -0.009 | 0.96154 | 0.272 |
| 48399 | CTC-296K1.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | JDP2 | Sponge network | -2.095 | 0.00112 | -0.967 | 0 | 0.272 |
| 48400 | GHRLOS |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-421;hsa-miR-589-5p;hsa-miR-98-5p | 21 | PRKAA2 | Sponge network | -1.942 | 0 | -2.462 | 0 | 0.272 |
| 48401 | RP4-791M13.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-382-5p;hsa-miR-576-5p;hsa-miR-577 | 13 | SYNPO2 | Sponge network | 0.419 | 0.0319 | -2.342 | 0 | 0.272 |
| 48402 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 37 | NTRK2 | Sponge network | 1.302 | 7.0E-5 | -0.736 | 0.06495 | 0.272 |
| 48403 | NNT-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 27 | ZFX | Sponge network | 0.799 | 1.0E-5 | 0.09 | 0.42563 | 0.272 |
| 48404 | CTD-2303H24.2 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 24 | JAZF1 | Sponge network | 0.415 | 0.14657 | -0.377 | 0.02677 | 0.272 |
| 48405 | RP11-464F9.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-655-3p | 12 | ZMYND11 | Sponge network | 0.959 | 0 | -0.2 | 0.05449 | 0.272 |
| 48406 | RP4-669H2.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-190a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c | 13 | WDFY3 | Sponge network | 1.711 | 0.00019 | -0.201 | 0.0967 | 0.272 |
| 48407 | CTA-941F9.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p | 11 | PIK3R1 | Sponge network | -0.344 | 0.49624 | -0.133 | 0.36854 | 0.272 |
| 48408 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | PRKAA1 | Sponge network | 1.117 | 0 | 0.317 | 0.0031 | 0.272 |
| 48409 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 14 | GPAM | Sponge network | 0.41 | 0.02214 | 0.142 | 0.29732 | 0.272 |
| 48410 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-628-5p;hsa-miR-7-5p | 20 | BMPR2 | Sponge network | -1.054 | 0.0706 | 0.206 | 0.0635 | 0.272 |
| 48411 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 16 | ARID5B | Sponge network | 2.094 | 0 | 0.342 | 0.01676 | 0.272 |
| 48412 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p | 12 | TBX18 | Sponge network | 0.082 | 0.55397 | 0.843 | 0.02945 | 0.272 |
| 48413 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-758-3p | 11 | CDC73 | Sponge network | -0.031 | 0.89063 | 0.094 | 0.20463 | 0.272 |
| 48414 | EIF3J-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ITGB1 | Sponge network | 0.331 | 0.00509 | 0.524 | 0.00017 | 0.272 |
| 48415 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | NOTCH2NL | Sponge network | 1.04 | 0.00023 | 0.024 | 0.84307 | 0.272 |
| 48416 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-532-3p;hsa-miR-93-5p | 22 | WDFY3 | Sponge network | 0.497 | 0.01451 | -0.201 | 0.0967 | 0.272 |
| 48417 | LINC00863 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NRXN3 | Sponge network | 0.189 | 0.13981 | -0.893 | 0.04186 | 0.272 |
| 48418 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | ZFHX4 | Sponge network | -0.031 | 0.89063 | 0.018 | 0.95574 | 0.272 |
| 48419 | CTB-31O20.4 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p | 12 | ZFX | Sponge network | 1.619 | 0 | 0.09 | 0.42563 | 0.272 |
| 48420 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-874-3p | 24 | ARID5B | Sponge network | 0.95 | 0.15943 | 0.342 | 0.01676 | 0.272 |
| 48421 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | LRRC4 | Sponge network | 0.572 | 0.01722 | -1.774 | 0 | 0.272 |
| 48422 | CTC-429P9.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-532-5p | 18 | PDE4DIP | Sponge network | 0.082 | 0.55397 | -0.282 | 0.0257 | 0.272 |
| 48423 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-576-5p | 19 | NUAK1 | Sponge network | 0.199 | 0.38015 | 0.904 | 0 | 0.272 |
| 48424 | RP11-244H3.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PRUNE2 | Sponge network | 0.659 | 3.0E-5 | -1.733 | 0.00022 | 0.272 |
| 48425 | BZRAP1-AS1 |
hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-874-3p;hsa-miR-93-3p | 22 | CTIF | Sponge network | -0.465 | 0.04703 | -0.389 | 0.00549 | 0.272 |
| 48426 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-96-5p | 23 | IL17RD | Sponge network | 0.622 | 0.00015 | -0.019 | 0.94873 | 0.272 |
| 48427 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 12 | GPAM | Sponge network | 0.67 | 0.00048 | 0.142 | 0.29732 | 0.272 |
| 48428 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 25 | YAP1 | Sponge network | 0.421 | 0.00084 | 0.22 | 0.11836 | 0.272 |
| 48429 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 30 | IGF1 | Sponge network | 0.082 | 0.55654 | -0.204 | 0.5898 | 0.272 |
| 48430 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-590-3p | 17 | SLITRK6 | Sponge network | 0.335 | 0.20596 | -2.121 | 0 | 0.272 |
| 48431 | OIP5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 18 | DDX5 | Sponge network | 0.421 | 0.00084 | -0.099 | 0.17428 | 0.272 |
| 48432 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-92a-3p | 11 | NRXN3 | Sponge network | 0.453 | 0.01839 | -0.893 | 0.04186 | 0.272 |
| 48433 | LINC00957 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-92a-1-5p | 13 | PRKACA | Sponge network | -0.421 | 0.0114 | -0.255 | 0.0012 | 0.272 |
| 48434 | TINCR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-96-5p | 19 | FBXO32 | Sponge network | -0.664 | 0.14543 | -0.495 | 0.07561 | 0.272 |
| 48435 | LINC00472 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | PTPRD | Sponge network | -0.842 | 0.01361 | -1.005 | 0.00064 | 0.272 |
| 48436 | RP11-2H8.2 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | ERCC6 | Sponge network | 1.089 | 0 | 0.23 | 0.015 | 0.272 |
| 48437 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p | 24 | TCF4 | Sponge network | 0.542 | 0.00054 | 0.016 | 0.92271 | 0.272 |
| 48438 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 37 | FOXP1 | Sponge network | 0.683 | 1.0E-5 | 0.042 | 0.74368 | 0.272 |
| 48439 | PCA3 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 24 | CTDSPL | Sponge network | -0.709 | 0.18168 | 0.085 | 0.45121 | 0.272 |
| 48440 | ADAMTS9-AS2 |
hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-7-5p | 10 | CMKLR1 | Sponge network | -2.452 | 0 | 0.288 | 0.17621 | 0.272 |
| 48441 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 26 | HECA | Sponge network | -0.793 | 7.0E-5 | -0.217 | 0.03463 | 0.272 |
| 48442 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-339-5p;hsa-miR-942-5p | 21 | THRB | Sponge network | 0.065 | 0.73468 | -1.117 | 0.0004 | 0.272 |
| 48443 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | SMAD4 | Sponge network | 0.545 | 0.00064 | -0.313 | 0.00413 | 0.272 |
| 48444 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | NCOA1 | Sponge network | 0.335 | 0.20596 | -0.432 | 0 | 0.272 |
| 48445 | DNM3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | HECA | Sponge network | 0.572 | 0.01722 | -0.217 | 0.03463 | 0.272 |
| 48446 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TRIP11 | Sponge network | 0.91 | 0 | -0.158 | 0.06384 | 0.272 |
| 48447 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 15 | USP6 | Sponge network | 0.986 | 0.01022 | 0.188 | 0.3871 | 0.272 |
| 48448 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 19 | ACVR1 | Sponge network | -0.942 | 0 | 0.279 | 0.00234 | 0.272 |
| 48449 | RP11-403I13.8 |
hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-532-3p | 10 | SYNM | Sponge network | 0.619 | 0.00157 | -2.309 | 1.0E-5 | 0.272 |
| 48450 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-940 | 25 | NTRK2 | Sponge network | 0.4 | 0.14611 | -0.736 | 0.06495 | 0.272 |
| 48451 | RP11-166D19.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-940 | 64 | NFIB | Sponge network | -0.576 | 0.10121 | 0.023 | 0.90795 | 0.272 |
| 48452 | LINC00963 |
hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ITGB3 | Sponge network | 0.523 | 9.0E-5 | -0.011 | 0.95751 | 0.272 |
| 48453 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 15 | PDS5B | Sponge network | -0.513 | 0.04703 | 0.259 | 0.00962 | 0.272 |
| 48454 | MAPKAPK5-AS1 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-30c-5p | 12 | PAFAH1B2 | Sponge network | 0.765 | 0 | 0.318 | 0.0001 | 0.272 |
| 48455 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-940 | 56 | WDFY3 | Sponge network | -0.709 | 0.18168 | -0.201 | 0.0967 | 0.272 |
| 48456 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-550a-3p | 14 | HECA | Sponge network | -0.473 | 0.07602 | -0.217 | 0.03463 | 0.272 |
| 48457 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | ZFHX4 | Sponge network | 0.659 | 3.0E-5 | 0.018 | 0.95574 | 0.272 |
| 48458 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 23 | PRRX1 | Sponge network | -0.246 | 0.35034 | 1.316 | 0 | 0.272 |
| 48459 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-625-3p | 25 | TMTC3 | Sponge network | 0.614 | 1.0E-5 | 0.123 | 0.22189 | 0.272 |
| 48460 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-660-5p | 25 | CXXC4 | Sponge network | 0.253 | 0.09565 | -0.433 | 0.21055 | 0.272 |
| 48461 | CTC-429P9.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-339-5p | 11 | LUZP2 | Sponge network | 0.082 | 0.55397 | -0.056 | 0.89416 | 0.272 |
| 48462 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 17 | EPHA3 | Sponge network | -0.141 | 0.26434 | 0.185 | 0.53588 | 0.272 |
| 48463 | RP11-798G7.7 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p | 14 | FOXP1 | Sponge network | 0.858 | 3.0E-5 | 0.042 | 0.74368 | 0.272 |
| 48464 | RP4-613B23.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-20a-5p | 11 | PRDM2 | Sponge network | -0.309 | 0.1145 | -0.258 | 0.00721 | 0.272 |
| 48465 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | SMARCA2 | Sponge network | -0.355 | 0.0077 | -0.529 | 0.00014 | 0.272 |
| 48466 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-539-5p | 18 | HECA | Sponge network | 0.078 | 0.55803 | -0.217 | 0.03463 | 0.272 |
| 48467 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | EPHA7 | Sponge network | -0.13 | 0.48734 | -2.197 | 3.0E-5 | 0.272 |
| 48468 | LINC00641 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | ZNF536 | Sponge network | -0.222 | 0.32445 | -3.115 | 0 | 0.272 |
| 48469 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ITGAV | Sponge network | -1.281 | 0.00012 | 0.337 | 0.01002 | 0.272 |
| 48470 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p | 11 | TACC1 | Sponge network | 0.912 | 0.00014 | -0.362 | 0.05406 | 0.272 |
| 48471 | LINC01001 |
hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-326 | 10 | SOX11 | Sponge network | 1.055 | 0 | 2.68 | 0 | 0.272 |
| 48472 | PAN3-AS1 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-655-3p | 13 | NCOA1 | Sponge network | 0.062 | 0.55274 | -0.432 | 0 | 0.272 |
| 48473 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 40 | WDFY3 | Sponge network | 0.559 | 0.00026 | -0.201 | 0.0967 | 0.272 |
| 48474 | RP11-254F7.2 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-539-5p | 15 | RYR2 | Sponge network | 0.176 | 0.37402 | -1.466 | 0.00031 | 0.272 |
| 48475 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-7-1-3p | 15 | DKK3 | Sponge network | -0.709 | 0.18168 | -0.052 | 0.77783 | 0.272 |
| 48476 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p | 14 | DMXL1 | Sponge network | 0.539 | 0.03722 | -0.426 | 0.00056 | 0.272 |
| 48477 | TPTEP1 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RTN4 | Sponge network | -1.216 | 0 | -0.412 | 0.00014 | 0.272 |
| 48478 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-5p | 25 | PRKCE | Sponge network | -0.154 | 0.78939 | -0.403 | 0.00056 | 0.272 |
| 48479 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-485-3p;hsa-miR-590-3p | 16 | TMTC2 | Sponge network | 0.101 | 0.60919 | -0.215 | 0.18248 | 0.272 |
| 48480 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | PRKAB2 | Sponge network | -2.117 | 0.00161 | -0.367 | 0.01371 | 0.272 |
| 48481 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | MAF | Sponge network | -0.663 | 0.10887 | -1.257 | 0 | 0.272 |
| 48482 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-34a-5p | 16 | AFF4 | Sponge network | 0.598 | 0.38481 | -0.266 | 0.01565 | 0.272 |
| 48483 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-30e-5p;hsa-miR-590-3p | 11 | ARID5B | Sponge network | 0.545 | 0.05789 | 0.342 | 0.01676 | 0.272 |
| 48484 | SERTAD4-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-362-5p | 15 | EGFR | Sponge network | -0.932 | 0.00329 | -0.175 | 0.48451 | 0.272 |
| 48485 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | EDA2R | Sponge network | -0.612 | 0.00094 | -0.587 | 0.01598 | 0.272 |
| 48486 | RP5-1085F17.3 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | UBR3 | Sponge network | 0.541 | 0.00125 | -0.021 | 0.82774 | 0.272 |
| 48487 | RP11-57H14.4 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | SFRP1 | Sponge network | 0.083 | 0.66405 | -3.566 | 0 | 0.272 |
| 48488 | AP001347.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p | 17 | ARNT2 | Sponge network | -1.161 | 0.00591 | -0.332 | 0.25851 | 0.272 |
| 48489 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-7-5p | 15 | ESR1 | Sponge network | -0.726 | 0.00132 | -1.035 | 3.0E-5 | 0.272 |
| 48490 | RP11-819C21.1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p | 11 | MAT2A | Sponge network | 0.537 | 0.00139 | -0.073 | 0.36195 | 0.272 |
| 48491 | MEG3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | RABEP1 | Sponge network | 0.249 | 0.35902 | -0.066 | 0.43142 | 0.272 |
| 48492 | AC083843.1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-99a-5p | 14 | DDX3X | Sponge network | 1.4 | 0 | -0.152 | 0.07686 | 0.272 |
| 48493 | CTD-2666L21.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-409-3p | 19 | BCL2 | Sponge network | 0.368 | 0.20093 | -0.899 | 0 | 0.272 |
| 48494 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | EGLN1 | Sponge network | 0.559 | 0.00026 | -0.127 | 0.1536 | 0.272 |
| 48495 | RP11-693J15.4 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-589-5p;hsa-miR-93-3p | 14 | RPS6KA2 | Sponge network | -0.72 | 0.24239 | -0.57 | 0.00013 | 0.272 |
| 48496 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 11 | PBRM1 | Sponge network | 1.757 | 0 | -0.029 | 0.78601 | 0.272 |
| 48497 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 19 | EBF1 | Sponge network | 0.541 | 0.00125 | -0.384 | 0.08856 | 0.272 |
| 48498 | RP11-686O6.2 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-577 | 11 | MDM4 | Sponge network | 0.453 | 0.01971 | 0.345 | 0.00762 | 0.272 |
| 48499 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421 | 22 | RBMS3 | Sponge network | -0.602 | 0.00331 | -0.519 | 0.03551 | 0.272 |
| 48500 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-940 | 33 | NTRK2 | Sponge network | -0.777 | 0.16667 | -0.736 | 0.06495 | 0.272 |
| 48501 | RP11-47A8.5 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-502-5p | 18 | DST | Sponge network | 0.103 | 0.43169 | -0.713 | 0.00096 | 0.272 |
| 48502 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-590-3p | 14 | SLITRK6 | Sponge network | -0.899 | 6.0E-5 | -2.121 | 0 | 0.272 |
| 48503 | RP11-439A17.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-335-5p;hsa-miR-98-5p | 10 | KCNA6 | Sponge network | 0.051 | 0.95756 | -1.531 | 0.00013 | 0.272 |
| 48504 | CKMT2-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | PRKCB | Sponge network | 0.082 | 0.55654 | -1.313 | 2.0E-5 | 0.272 |
| 48505 | A2M-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p | 12 | RPS6KA2 | Sponge network | -0.08 | 0.90092 | -0.57 | 0.00013 | 0.272 |
| 48506 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-93-5p | 19 | SLITRK3 | Sponge network | -0.371 | 0.24604 | -3.328 | 0 | 0.272 |
| 48507 | CTC-444N24.11 |
hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-369-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p | 11 | RNF111 | Sponge network | 1.153 | 0 | 0.098 | 0.21093 | 0.272 |
| 48508 | RP11-458D21.1 |
hsa-let-7g-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-7-1-3p | 12 | ZMYND11 | Sponge network | 0.663 | 0.00073 | -0.2 | 0.05449 | 0.272 |
| 48509 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-423-3p;hsa-miR-429 | 16 | UBE4B | Sponge network | -2.69 | 0.0001 | -0.235 | 0.007 | 0.272 |
| 48510 | MAPKAPK5-AS1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p | 11 | CDK6 | Sponge network | 0.765 | 0 | 0.991 | 0 | 0.272 |
| 48511 | RP11-116O18.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | SETBP1 | Sponge network | 0.05 | 0.95483 | -1.302 | 1.0E-5 | 0.272 |
| 48512 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-193a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | AHNAK | Sponge network | 0.683 | 1.0E-5 | -1.207 | 0 | 0.272 |
| 48513 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 23 | KIT | Sponge network | -0.83 | 0 | -2.184 | 0 | 0.272 |
| 48514 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 34 | EPHA5 | Sponge network | 0.199 | 0.38015 | -0.957 | 0.01733 | 0.272 |
| 48515 | HAND2-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | PRKACA | Sponge network | -1.697 | 0.00889 | -0.255 | 0.0012 | 0.272 |
| 48516 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | MYO9A | Sponge network | -0.235 | 0.15142 | -0.147 | 0.32373 | 0.272 |
| 48517 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TBX18 | Sponge network | 0.225 | 0.30644 | 0.843 | 0.02945 | 0.272 |
| 48518 | TMEM191A | hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 10 | PAFAH1B2 | Sponge network | 1.342 | 0 | 0.318 | 0.0001 | 0.272 |
| 48519 | RP11-277P12.20 |
hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-30b-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 11 | EFNA5 | Sponge network | 0.687 | 0.02753 | -0.421 | 0.17868 | 0.272 |
| 48520 | RP11-798G7.7 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-664a-5p | 28 | DST | Sponge network | 0.858 | 3.0E-5 | -0.713 | 0.00096 | 0.272 |
| 48521 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-744-5p;hsa-miR-93-5p;hsa-miR-940 | 44 | CHD5 | Sponge network | -1.216 | 0 | -0.922 | 0.01521 | 0.272 |
| 48522 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-576-5p | 16 | ARHGAP28 | Sponge network | 0.176 | 0.37402 | -0.911 | 0.00013 | 0.272 |
| 48523 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 14 | SLC6A15 | Sponge network | 1.757 | 0 | -2.373 | 2.0E-5 | 0.272 |
| 48524 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 10 | SYNE1 | Sponge network | 0.238 | 0.70544 | -0.738 | 0.00316 | 0.272 |
| 48525 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 30 | MYO9A | Sponge network | -0.83 | 0 | -0.147 | 0.32373 | 0.272 |
| 48526 | PGM5-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p | 14 | PDE4DIP | Sponge network | -4.546 | 0 | -0.282 | 0.0257 | 0.272 |
| 48527 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | TCF12 | Sponge network | 1.601 | 0 | 0.174 | 0.13271 | 0.272 |
| 48528 | GS1-124K5.11 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | RECK | Sponge network | 0.494 | 0.00339 | -1.043 | 0 | 0.272 |
| 48529 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p | 41 | BNC2 | Sponge network | 0.253 | 0.09565 | -0.491 | 0.09988 | 0.272 |
| 48530 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p | 13 | PLSCR4 | Sponge network | 0.921 | 0.00118 | -0.474 | 0.03833 | 0.272 |
| 48531 | RP11-620J15.3 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-590-3p | 13 | EFNA5 | Sponge network | -0.739 | 0.00044 | -0.421 | 0.17868 | 0.272 |
| 48532 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | MOB1B | Sponge network | -0.376 | 0.00337 | 0.197 | 0.15266 | 0.272 |
| 48533 | CASC2 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-212-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | UBR3 | Sponge network | 0.385 | 0.01306 | -0.021 | 0.82774 | 0.272 |
| 48534 | RP11-532F6.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 18 | TOM1L2 | Sponge network | -0.896 | 0.00099 | -0.994 | 0 | 0.272 |
| 48535 | LINC01018 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 20 | ZFP36L1 | Sponge network | -2.915 | 0.00015 | -0.505 | 8.0E-5 | 0.272 |
| 48536 | RP11-110I1.12 |
hsa-let-7a-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-500a-5p | 10 | RICTOR | Sponge network | 0.639 | 0.00028 | 0.388 | 0.00188 | 0.272 |
| 48537 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 22 | CBX5 | Sponge network | 0.8 | 0 | 0.165 | 0.24446 | 0.272 |
| 48538 | XXbac-B444P24.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | BHLHE41 | Sponge network | 2.264 | 0.0012 | 0.353 | 0.12753 | 0.272 |
| 48539 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 26 | FOXP1 | Sponge network | -4.546 | 0 | 0.042 | 0.74368 | 0.272 |
| 48540 | RP11-102N12.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 15 | TRPS1 | Sponge network | -0.351 | 0.11358 | -0.191 | 0.44109 | 0.272 |
| 48541 | MIR22HG |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-877-5p | 23 | ASTN1 | Sponge network | -1.047 | 0 | -2.389 | 2.0E-5 | 0.272 |
| 48542 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-493-5p | 19 | FBXO32 | Sponge network | 0.368 | 0.20093 | -0.495 | 0.07561 | 0.272 |
| 48543 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-5p | 18 | USP12 | Sponge network | 0.197 | 0.25618 | -0.102 | 0.40768 | 0.272 |
| 48544 | GHRLOS |
hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-493-5p | 22 | EPHA7 | Sponge network | -1.942 | 0 | -2.197 | 3.0E-5 | 0.272 |
| 48545 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | CXXC4 | Sponge network | 0.385 | 0.10067 | -0.433 | 0.21055 | 0.272 |
| 48546 | FAM66C |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | NOTCH2NL | Sponge network | -0.901 | 0.00019 | 0.024 | 0.84307 | 0.272 |
| 48547 | LINC00173 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-940 | 20 | NTRK2 | Sponge network | 0.056 | 0.8494 | -0.736 | 0.06495 | 0.272 |
| 48548 | FAM66C |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | BCL9 | Sponge network | -0.901 | 0.00019 | 0.321 | 0.00267 | 0.272 |
| 48549 | RP1-151F17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-29a-5p | 12 | PBX1 | Sponge network | -0.321 | 0.22761 | -1.206 | 0 | 0.272 |
| 48550 | FAM13A-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-940 | 13 | MAPK10 | Sponge network | -0.83 | 0 | -1.774 | 0 | 0.272 |
| 48551 | AC009120.5 |
hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-5p;hsa-miR-93-3p | 10 | BNC2 | Sponge network | 1.383 | 0 | -0.491 | 0.09988 | 0.272 |
| 48552 | NNT-AS1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-3607-3p;hsa-miR-766-3p | 11 | IL17RD | Sponge network | 0.799 | 1.0E-5 | -0.019 | 0.94873 | 0.272 |
| 48553 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | PIK3CA | Sponge network | -0.738 | 0.21233 | 0.195 | 0.06908 | 0.272 |
| 48554 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-539-5p | 25 | KIF1B | Sponge network | 0.79 | 0 | -0.118 | 0.32258 | 0.272 |
| 48555 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | CEP170 | Sponge network | 0.311 | 0.10344 | 0.943 | 0 | 0.272 |
| 48556 | BDNF-AS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-361-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | TGFBR3 | Sponge network | -0.668 | 3.0E-5 | -1.173 | 1.0E-5 | 0.271 |
| 48557 | FZD10-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-505-3p;hsa-miR-7-5p | 15 | FNBP1 | Sponge network | -0.727 | 0.04726 | -0.969 | 4.0E-5 | 0.271 |
| 48558 | RP11-155G14.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-539-5p | 14 | KIF1B | Sponge network | 2.483 | 0 | -0.118 | 0.32258 | 0.271 |
| 48559 | LINC00969 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | RB1 | Sponge network | 0.683 | 1.0E-5 | 0.382 | 0.0017 | 0.271 |
| 48560 | SH3RF3-AS1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-505-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 17 | ETV1 | Sponge network | 0.889 | 9.0E-5 | -0.096 | 0.67674 | 0.271 |
| 48561 | LINC00621 |
hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-30e-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | CXXC4 | Sponge network | 0.131 | 0.8436 | -0.433 | 0.21055 | 0.271 |
| 48562 | AF131215.9 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 12 | TMEFF2 | Sponge network | -0.322 | 0.25945 | -2.426 | 0 | 0.271 |
| 48563 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-671-5p | 17 | CRTC3 | Sponge network | -0.147 | 0.40452 | 0.164 | 0.16786 | 0.271 |
| 48564 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 34 | FOXP1 | Sponge network | 0.657 | 4.0E-5 | 0.042 | 0.74368 | 0.271 |
| 48565 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p | 23 | RIMBP2 | Sponge network | -0.487 | 0.02157 | -1.968 | 5.0E-5 | 0.271 |
| 48566 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-744-5p;hsa-miR-93-5p | 15 | FGFR1 | Sponge network | -0.785 | 0.00035 | -0.509 | 0.03957 | 0.271 |
| 48567 | CTD-2554C21.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-629-3p | 18 | USP12 | Sponge network | -1.654 | 0.00144 | -0.102 | 0.40768 | 0.271 |
| 48568 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DNAJB4 | Sponge network | 0.72 | 0.0001 | -0.7 | 1.0E-5 | 0.271 |
| 48569 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 37 | SOX5 | Sponge network | 0.16 | 0.50785 | -1.091 | 4.0E-5 | 0.271 |
| 48570 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 22 | BNC2 | Sponge network | -0.351 | 0.11358 | -0.491 | 0.09988 | 0.271 |
| 48571 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 12 | SOX11 | Sponge network | 2.094 | 0 | 2.68 | 0 | 0.271 |
| 48572 | WAC-AS1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RBBP6 | Sponge network | 0.821 | 0 | 0.163 | 0.05101 | 0.271 |
| 48573 | CTC-457L16.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-7-1-3p | 16 | MED12L | Sponge network | 1.153 | 0.00114 | -0.194 | 0.55299 | 0.271 |
| 48574 | RP11-182J1.1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 19 | ERBB4 | Sponge network | -0.187 | 0.23787 | -1.975 | 2.0E-5 | 0.271 |
| 48575 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 61 | SMAD4 | Sponge network | -1.575 | 6.0E-5 | -0.313 | 0.00413 | 0.271 |
| 48576 | CTC-559E9.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-576-5p | 28 | RUNX1T1 | Sponge network | 0.594 | 0.00064 | -0.344 | 0.2374 | 0.271 |
| 48577 | RP11-927P21.1 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-942-5p | 25 | ARHGEF12 | Sponge network | 0.921 | 0.00118 | 0.09 | 0.35693 | 0.271 |
| 48578 | PCBP1-AS1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-708-5p | 11 | MAT2A | Sponge network | -0.567 | 0 | -0.073 | 0.36195 | 0.271 |
| 48579 | PSMD5-AS1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | RBBP6 | Sponge network | 0.569 | 0.00067 | 0.163 | 0.05101 | 0.271 |
| 48580 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 41 | ERBB4 | Sponge network | 0.374 | 0.20054 | -1.975 | 2.0E-5 | 0.271 |
| 48581 | CTD-2314G24.2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-98-5p | 16 | KLF10 | Sponge network | 0.246 | 0.59912 | -0.783 | 0 | 0.271 |
| 48582 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-501-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | -1.047 | 0 | -3.328 | 0 | 0.271 |
| 48583 | AC005154.6 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 13 | CDK6 | Sponge network | 0.848 | 0 | 0.991 | 0 | 0.271 |
| 48584 | RP11-119F7.5 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | UBR3 | Sponge network | 0.225 | 0.30644 | -0.021 | 0.82774 | 0.271 |
| 48585 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 16 | NR2C2 | Sponge network | 0.538 | 0.00076 | 0.21 | 0.03224 | 0.271 |
| 48586 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-375;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-98-5p | 19 | THBS1 | Sponge network | 0.687 | 0.02753 | 0.741 | 0.00386 | 0.271 |
| 48587 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p | 12 | TACC1 | Sponge network | -2.549 | 0.00528 | -0.362 | 0.05406 | 0.271 |
| 48588 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-539-5p | 10 | PDE4DIP | Sponge network | 1.055 | 0 | -0.282 | 0.0257 | 0.271 |
| 48589 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 12 | HIP1 | Sponge network | 0.657 | 4.0E-5 | 0.774 | 0 | 0.271 |
| 48590 | RP11-360F5.3 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | KIF1B | Sponge network | 1.867 | 0 | -0.118 | 0.32258 | 0.271 |
| 48591 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | EGLN1 | Sponge network | 0.72 | 0.0001 | -0.127 | 0.1536 | 0.271 |
| 48592 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p | 12 | CBL | Sponge network | 0.768 | 7.0E-5 | 0.291 | 0.01267 | 0.271 |
| 48593 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p | 11 | DACH2 | Sponge network | -0.154 | 0.78939 | -1.635 | 0.00034 | 0.271 |
| 48594 | KB-1460A1.5 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 18 | WWTR1 | Sponge network | 0.603 | 0.00127 | -0.345 | 0.11997 | 0.271 |
| 48595 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-96-5p | 13 | EBF3 | Sponge network | 0.453 | 0.01839 | -0.058 | 0.81437 | 0.271 |
| 48596 | AF131215.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-582-3p | 17 | ABL2 | Sponge network | -0.176 | 0.59692 | 0.82 | 0 | 0.271 |
| 48597 | RP11-411K7.1 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-2-3p;hsa-miR-27b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 15 | RAPGEF5 | Sponge network | 0.155 | 0.81273 | 0.536 | 4.0E-5 | 0.271 |
| 48598 | RP11-144G6.12 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 13 | SLC5A3 | Sponge network | 0.826 | 1.0E-5 | 0.493 | 5.0E-5 | 0.271 |
| 48599 | C4B-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-3p | 22 | IL6ST | Sponge network | 2.306 | 9.0E-5 | -0.402 | 0.00676 | 0.271 |
| 48600 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-664a-3p | 21 | GREM1 | Sponge network | -0.942 | 0 | 0.902 | 0.01691 | 0.271 |
| 48601 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-874-3p | 23 | NR2C2 | Sponge network | 0.219 | 0.1516 | 0.21 | 0.03224 | 0.271 |
| 48602 | RP11-159D12.2 |
hsa-miR-10b-3p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | CTDSPL | Sponge network | 1.254 | 0 | 0.085 | 0.45121 | 0.271 |
| 48603 | RP4-791M13.3 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 10 | SHPRH | Sponge network | 0.419 | 0.0319 | 0.098 | 0.40994 | 0.271 |
| 48604 | TRIM52-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-577 | 18 | IGFBP5 | Sponge network | -0.418 | 0.00068 | -0.806 | 0.00105 | 0.271 |
| 48605 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-550a-5p | 11 | FAT2 | Sponge network | 0.194 | 0.64278 | -0.278 | 0.44877 | 0.271 |
| 48606 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 30 | BHLHE41 | Sponge network | -0.769 | 0.00035 | 0.353 | 0.12753 | 0.271 |
| 48607 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 32 | EPHA7 | Sponge network | 0.222 | 0.29133 | -2.197 | 3.0E-5 | 0.271 |
| 48608 | GHRLOS |
hsa-let-7d-5p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-942-5p;hsa-miR-98-5p | 12 | ARHGAP28 | Sponge network | -1.942 | 0 | -0.911 | 0.00013 | 0.271 |
| 48609 | RP11-25K21.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-589-3p | 12 | TRPS1 | Sponge network | 0.489 | 0.25786 | -0.191 | 0.44109 | 0.271 |
| 48610 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-484 | 11 | ARNT2 | Sponge network | -0.72 | 0.24239 | -0.332 | 0.25851 | 0.271 |
| 48611 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 36 | MEF2C | Sponge network | 0.083 | 0.66405 | -0.499 | 0.01448 | 0.271 |
| 48612 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-93-3p | 21 | FOXO3 | Sponge network | -1.249 | 7.0E-5 | -0.04 | 0.72028 | 0.271 |
| 48613 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 36 | CBL | Sponge network | 1.302 | 7.0E-5 | 0.291 | 0.01267 | 0.271 |
| 48614 | IQCH-AS1 |
hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p | 14 | ABL2 | Sponge network | 0.929 | 0 | 0.82 | 0 | 0.271 |
| 48615 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-424-5p;hsa-miR-7-1-3p | 19 | AKAP11 | Sponge network | 0.889 | 9.0E-5 | 0.196 | 0.09825 | 0.271 |
| 48616 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | NRXN3 | Sponge network | 0.998 | 0 | -0.893 | 0.04186 | 0.271 |
| 48617 | AC083843.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p | 12 | CLASP2 | Sponge network | 1.4 | 0 | -0.038 | 0.71 | 0.271 |
| 48618 | RP11-473I1.9 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-766-3p | 33 | PPARA | Sponge network | 0.683 | 1.0E-5 | -0.379 | 0.00548 | 0.271 |
| 48619 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-511-5p;hsa-miR-671-5p | 12 | RBMS3 | Sponge network | -0.278 | 0.76559 | -0.519 | 0.03551 | 0.271 |
| 48620 | AF131215.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 19 | EPS15 | Sponge network | -0.176 | 0.59692 | -0.052 | 0.53998 | 0.271 |
| 48621 | PSMG3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 13 | EPAS1 | Sponge network | -0.125 | 0.49414 | -0.184 | 0.14418 | 0.271 |
| 48622 | RP11-48B3.4 |
hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 13 | ERG | Sponge network | 1.757 | 0 | 0.002 | 0.99011 | 0.271 |
| 48623 | HAND2-AS1 |
hsa-miR-1976;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-7-1-3p | 10 | PACRG | Sponge network | -1.697 | 0.00889 | -2.248 | 0 | 0.271 |
| 48624 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | CXXC4 | Sponge network | -2.915 | 0.00015 | -0.433 | 0.21055 | 0.271 |
| 48625 | RP11-806O11.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-766-3p | 24 | MDM4 | Sponge network | 0.284 | 0.52463 | 0.345 | 0.00762 | 0.271 |
| 48626 | AGAP11 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-942-5p | 22 | OPCML | Sponge network | -1.645 | 0 | -2.498 | 0 | 0.271 |
| 48627 | SH3BP5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p | 15 | ERCC4 | Sponge network | 0.267 | 0.2937 | 0.126 | 0.15266 | 0.271 |
| 48628 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-493-5p | 24 | WDFY3 | Sponge network | 0.368 | 0.20093 | -0.201 | 0.0967 | 0.271 |
| 48629 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 38 | SSBP2 | Sponge network | 0.395 | 0.15451 | -1.58 | 0 | 0.271 |
| 48630 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-501-5p | 11 | PRKCE | Sponge network | -2.531 | 0 | -0.403 | 0.00056 | 0.271 |
| 48631 | PCA3 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TRIM13 | Sponge network | -0.709 | 0.18168 | 0.183 | 0.06968 | 0.271 |
| 48632 | RP11-822E23.8 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-940 | 13 | CSRNP1 | Sponge network | -0.777 | 0.16667 | -1.448 | 0 | 0.271 |
| 48633 | AC005562.1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-484 | 17 | ABL1 | Sponge network | 0.488 | 0.00084 | -0.215 | 0.07804 | 0.271 |
| 48634 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p | 14 | MYCBP2 | Sponge network | 0.997 | 0.00066 | -0.027 | 0.82016 | 0.271 |
| 48635 | LINC00654 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | PRRX1 | Sponge network | 0.374 | 0.20054 | 1.316 | 0 | 0.271 |
| 48636 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-7-1-3p | 18 | EPHA3 | Sponge network | -4.477 | 0 | 0.185 | 0.53588 | 0.271 |
| 48637 | HOXB-AS1 |
hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-628-5p | 10 | IRS1 | Sponge network | -0.154 | 0.53954 | -0.026 | 0.88855 | 0.271 |
| 48638 | RP11-6O2.3 |
hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-940;hsa-miR-96-5p | 14 | SFRP1 | Sponge network | 0.331 | 0.57247 | -3.566 | 0 | 0.271 |
| 48639 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-96-5p | 14 | RIMBP2 | Sponge network | 0.853 | 0.15352 | -1.968 | 5.0E-5 | 0.271 |
| 48640 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | MYO9A | Sponge network | 0.569 | 0.00067 | -0.147 | 0.32373 | 0.271 |
| 48641 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | SYNE1 | Sponge network | 0.61 | 0.01593 | -0.738 | 0.00316 | 0.271 |
| 48642 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | EDA2R | Sponge network | -0.663 | 0.10887 | -0.587 | 0.01598 | 0.271 |
| 48643 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DACH2 | Sponge network | -1.203 | 0.03881 | -1.635 | 0.00034 | 0.271 |
| 48644 | CALML3-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-374b-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-664a-3p | 23 | SYNPO2 | Sponge network | 0.95 | 0.15943 | -2.342 | 0 | 0.271 |
| 48645 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p | 10 | SPG20 | Sponge network | 0.37 | 0.27614 | -1.16 | 0 | 0.271 |
| 48646 | CTD-2291D10.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p | 16 | RUNX1T1 | Sponge network | 3.21 | 0 | -0.344 | 0.2374 | 0.271 |
| 48647 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-654-3p;hsa-miR-874-3p | 15 | NR2C2 | Sponge network | 0.373 | 0.00068 | 0.21 | 0.03224 | 0.271 |
| 48648 | HHIP-AS1 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-505-3p;hsa-miR-7-5p | 10 | FNBP1 | Sponge network | -0.737 | 0.10822 | -0.969 | 4.0E-5 | 0.271 |
| 48649 | ARHGEF26-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 24 | ARNT2 | Sponge network | -2.46 | 0 | -0.332 | 0.25851 | 0.271 |
| 48650 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-7-1-3p | 13 | TBX18 | Sponge network | -4.546 | 0 | 0.843 | 0.02945 | 0.271 |
| 48651 | RP11-218M22.1 |
hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-5p | 13 | IGF2 | Sponge network | 0.197 | 0.25618 | 0.848 | 0.03842 | 0.271 |
| 48652 | LINC00654 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 16 | KLF10 | Sponge network | 0.374 | 0.20054 | -0.783 | 0 | 0.271 |
| 48653 | LINC00476 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | SIRT1 | Sponge network | -0.615 | 0.00015 | 0.109 | 0.2593 | 0.271 |
| 48654 | CTD-2666L21.1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p | 10 | CRTC1 | Sponge network | 0.368 | 0.20093 | -0.116 | 0.28412 | 0.271 |
| 48655 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p | 21 | PRKCE | Sponge network | -0.842 | 0.01361 | -0.403 | 0.00056 | 0.271 |
| 48656 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | FOXP1 | Sponge network | 0.67 | 0.00048 | 0.042 | 0.74368 | 0.271 |
| 48657 | RP11-77H9.2 |
hsa-miR-10b-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | ARHGEF12 | Sponge network | 1.677 | 0 | 0.09 | 0.35693 | 0.271 |
| 48658 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-5p | 11 | PCDH8 | Sponge network | -0.384 | 0.40652 | -0.997 | 0.05366 | 0.271 |
| 48659 | LINC00662 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-424-5p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-590-3p | 11 | CSDE1 | Sponge network | 0.213 | 0.32297 | -0.493 | 0 | 0.271 |
| 48660 | RP11-20I23.13 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-671-5p | 20 | RBMS3 | Sponge network | 0.505 | 0.0014 | -0.519 | 0.03551 | 0.271 |
| 48661 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-7-1-3p | 30 | BCL2 | Sponge network | -0.246 | 0.35034 | -0.899 | 0 | 0.271 |
| 48662 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-590-3p | 16 | USP6 | Sponge network | -0.322 | 0.25945 | 0.188 | 0.3871 | 0.271 |
| 48663 | CTC-429P9.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-500a-5p | 16 | CYLD | Sponge network | 0.082 | 0.55397 | -0.367 | 0.00171 | 0.271 |
| 48664 | AF131215.9 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-3p | 16 | UNC5C | Sponge network | -0.322 | 0.25945 | -0.178 | 0.40214 | 0.271 |
| 48665 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 14 | LRRC7 | Sponge network | 0.419 | 0.0319 | -1.341 | 0.01193 | 0.271 |
| 48666 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935 | 48 | SMAD4 | Sponge network | 0.75 | 0.00054 | -0.313 | 0.00413 | 0.271 |
| 48667 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | NIN | Sponge network | -0.355 | 0.0077 | -0.253 | 0.13794 | 0.271 |
| 48668 | AC016747.3 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-5p | 31 | BCL2 | Sponge network | 0.199 | 0.38015 | -0.899 | 0 | 0.271 |
| 48669 | RP11-307B6.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-93-3p | 12 | SUFU | Sponge network | -4.463 | 0.00025 | -0.04 | 0.65489 | 0.271 |
| 48670 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p | 12 | MYO9A | Sponge network | 0.909 | 0 | -0.147 | 0.32373 | 0.271 |
| 48671 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-7-1-3p | 12 | LRRC55 | Sponge network | -4.546 | 0 | -0.026 | 0.93163 | 0.271 |
| 48672 | BMS1P20 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CLASP2 | Sponge network | 0.34 | 0.00273 | -0.038 | 0.71 | 0.271 |
| 48673 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-942-5p | 10 | CREBBP | Sponge network | -0.517 | 0.02935 | 0.126 | 0.15186 | 0.271 |
| 48674 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 23 | CBL | Sponge network | 0.538 | 0.00076 | 0.291 | 0.01267 | 0.271 |
| 48675 | RP11-395A13.2 |
hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-150-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p | 11 | PBX1 | Sponge network | 0.057 | 0.61225 | -1.206 | 0 | 0.271 |
| 48676 | RP11-497H16.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NIN | Sponge network | 0.545 | 0.00064 | -0.253 | 0.13794 | 0.271 |
| 48677 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-502-5p | 22 | AR | Sponge network | 1.294 | 0.0031 | -1.554 | 0 | 0.271 |
| 48678 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | BMPR2 | Sponge network | 0.545 | 0.00064 | 0.206 | 0.0635 | 0.271 |
| 48679 | RBPMS-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p | 13 | PBX1 | Sponge network | 0.144 | 0.64989 | -1.206 | 0 | 0.271 |
| 48680 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-654-3p;hsa-miR-7-1-3p | 29 | PPM1A | Sponge network | -0.176 | 0.59692 | -0.623 | 0 | 0.271 |
| 48681 | RP11-473I1.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 12 | SOX5 | Sponge network | 0.542 | 0.00054 | -1.091 | 4.0E-5 | 0.271 |
| 48682 | C4A-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-590-5p | 11 | DMD | Sponge network | 3.345 | 0 | -1.506 | 0 | 0.271 |
| 48683 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | EYA4 | Sponge network | 0.796 | 4.0E-5 | 0.3 | 0.36876 | 0.271 |
| 48684 | CTBP1-AS2 |
hsa-miR-100-5p;hsa-miR-101-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 10 | ATP2A2 | Sponge network | 0.623 | 0 | 0.604 | 0 | 0.271 |
| 48685 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | YAP1 | Sponge network | 0.16 | 0.50785 | 0.22 | 0.11836 | 0.271 |
| 48686 | FZD10-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-532-5p;hsa-miR-590-3p | 11 | PARK2 | Sponge network | -0.727 | 0.04726 | -1.892 | 0 | 0.271 |
| 48687 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-493-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | ANK2 | Sponge network | 0.614 | 1.0E-5 | -1.603 | 1.0E-5 | 0.271 |
| 48688 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p | 19 | CUL5 | Sponge network | 0.392 | 0.0278 | 0.026 | 0.77301 | 0.271 |
| 48689 | RP11-596D21.1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p | 14 | CCDC6 | Sponge network | 3.076 | 3.0E-5 | 0.27 | 0.02555 | 0.271 |
| 48690 | LIPE-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 23 | SNX29 | Sponge network | 0.078 | 0.55803 | -0.34 | 0.01371 | 0.271 |
| 48691 | SOX2-OT |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | -1.264 | 6.0E-5 | -0.57 | 0 | 0.271 |
| 48692 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 33 | DIP2C | Sponge network | 0.222 | 0.29133 | -0.436 | 0.01713 | 0.271 |
| 48693 | BDNF-AS |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-96-5p | 10 | LRRC4 | Sponge network | -0.668 | 3.0E-5 | -1.774 | 0 | 0.271 |
| 48694 | RP11-6O2.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-409-3p | 11 | SEMA3E | Sponge network | 0.331 | 0.57247 | -1.717 | 0.00137 | 0.271 |
| 48695 | RP11-110I1.12 |
hsa-let-7a-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p | 10 | NR2C2 | Sponge network | 0.639 | 0.00028 | 0.21 | 0.03224 | 0.271 |
| 48696 | RP11-473I1.9 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | FMNL3 | Sponge network | 0.683 | 1.0E-5 | 0.555 | 4.0E-5 | 0.271 |
| 48697 | TAPT1-AS1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-495-3p | 11 | ZMYM2 | Sponge network | 0.403 | 2.0E-5 | 0.279 | 0.01273 | 0.271 |
| 48698 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 32 | RASSF2 | Sponge network | -0.246 | 0.35034 | -0.273 | 0.21961 | 0.271 |
| 48699 | AC016747.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p | 14 | SAMHD1 | Sponge network | 0.199 | 0.38015 | 0.072 | 0.62895 | 0.271 |
| 48700 | AC005562.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | ARHGEF12 | Sponge network | 0.488 | 0.00084 | 0.09 | 0.35693 | 0.271 |
| 48701 | C4B-AS1 |
hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | SCN9A | Sponge network | 2.306 | 9.0E-5 | -0.525 | 0.10593 | 0.271 |
| 48702 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-497-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | PPM1A | Sponge network | 0.683 | 1.0E-5 | -0.623 | 0 | 0.271 |
| 48703 | ZSCAN16-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | GJA1 | Sponge network | -0.342 | 0.02288 | -0.485 | 0.02179 | 0.271 |
| 48704 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TIMP3 | Sponge network | 0.727 | 3.0E-5 | -0.546 | 0.02307 | 0.271 |
| 48705 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 16 | ARNT2 | Sponge network | 0.727 | 3.0E-5 | -0.332 | 0.25851 | 0.271 |
| 48706 | CTC-296K1.3 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 11 | OPCML | Sponge network | -2.095 | 0.00112 | -2.498 | 0 | 0.271 |
| 48707 | A2M-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 17 | RYR2 | Sponge network | -0.08 | 0.90092 | -1.466 | 0.00031 | 0.271 |
| 48708 | LINC00662 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 25 | TGFBR3 | Sponge network | 0.213 | 0.32297 | -1.173 | 1.0E-5 | 0.271 |
| 48709 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 28 | SPRY3 | Sponge network | 0.559 | 0.00026 | 0.016 | 0.91286 | 0.271 |
| 48710 | AGAP11 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p | 12 | PREX2 | Sponge network | -1.645 | 0 | -0.272 | 0.25382 | 0.271 |
| 48711 | HCG11 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 13 | TMEFF2 | Sponge network | 0.385 | 0.10067 | -2.426 | 0 | 0.271 |
| 48712 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-629-3p | 33 | KIF1B | Sponge network | 0.368 | 0.20093 | -0.118 | 0.32258 | 0.271 |
| 48713 | RP4-584D14.7 |
hsa-let-7d-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-940 | 12 | THBS1 | Sponge network | 1.294 | 0.0031 | 0.741 | 0.00386 | 0.271 |
| 48714 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-455-3p;hsa-miR-629-3p;hsa-miR-654-3p | 14 | PSIP1 | Sponge network | 0.177 | 0.16681 | -0.005 | 0.96897 | 0.271 |
| 48715 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | MAF | Sponge network | 0.997 | 0.00066 | -1.257 | 0 | 0.271 |
| 48716 | RP11-57H14.4 |
hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 10 | RADIL | Sponge network | 0.083 | 0.66405 | -1.628 | 0 | 0.271 |
| 48717 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-429;hsa-miR-576-5p | 11 | RBL2 | Sponge network | -0.176 | 0.59692 | -0.282 | 0.00254 | 0.271 |
| 48718 | LINC00654 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TMEFF2 | Sponge network | 0.374 | 0.20054 | -2.426 | 0 | 0.271 |
| 48719 | RP11-25K19.1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p | 44 | DST | Sponge network | -1.057 | 0.00029 | -0.713 | 0.00096 | 0.271 |
| 48720 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-501-5p;hsa-miR-590-3p | 27 | EGFR | Sponge network | -0.738 | 0.21233 | -0.175 | 0.48451 | 0.271 |
| 48721 | TRIM52-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-577;hsa-miR-7-1-3p | 13 | SYNE1 | Sponge network | -0.418 | 0.00068 | -0.738 | 0.00316 | 0.271 |
| 48722 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | USP12 | Sponge network | 0.986 | 0.01022 | -0.102 | 0.40768 | 0.271 |
| 48723 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-577 | 10 | NALCN | Sponge network | -0.303 | 0.21002 | 1.068 | 0.00237 | 0.271 |
| 48724 | RP11-404E16.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-942-5p | 10 | DDX6 | Sponge network | 0.097 | 0.91311 | 0.091 | 0.36428 | 0.271 |
| 48725 | RP3-402G11.26 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | CNTN1 | Sponge network | -0.254 | 0.19303 | -2.594 | 0 | 0.271 |
| 48726 | ADAMTS9-AS2 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | RBM6 | Sponge network | -2.452 | 0 | -0.137 | 0.15663 | 0.271 |
| 48727 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-539-5p | 22 | PBX1 | Sponge network | -0.785 | 0.00035 | -1.206 | 0 | 0.271 |
| 48728 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | HERC2 | Sponge network | 0.848 | 0 | 0.114 | 0.30638 | 0.271 |
| 48729 | AC009120.6 |
hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-590-3p | 18 | DIP2C | Sponge network | 0.919 | 0 | -0.436 | 0.01713 | 0.271 |
| 48730 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-539-5p;hsa-miR-590-3p | 11 | ITCH | Sponge network | 0.919 | 0 | 0.084 | 0.34058 | 0.271 |
| 48731 | RP11-130L8.1 |
hsa-miR-1271-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | RPS6KA6 | Sponge network | -0.663 | 0.10887 | -2.989 | 0 | 0.271 |
| 48732 | LINC00473 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 11 | RASSF3 | Sponge network | -1.203 | 0.03881 | -0.226 | 0.11691 | 0.271 |
| 48733 | RP11-156E6.1 |
hsa-miR-101-3p;hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-497-5p | 14 | UBR3 | Sponge network | 1.266 | 0 | -0.021 | 0.82774 | 0.271 |
| 48734 | H1FX-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30d-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | GJA1 | Sponge network | -0.206 | 0.12942 | -0.485 | 0.02179 | 0.271 |
| 48735 | RP11-327P2.5 |
hsa-let-7b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p | 18 | ABL1 | Sponge network | -0.147 | 0.40452 | -0.215 | 0.07804 | 0.271 |
| 48736 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | MCC | Sponge network | 1.302 | 7.0E-5 | -1.005 | 0 | 0.271 |
| 48737 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | RBMS3 | Sponge network | -0.141 | 0.26434 | -0.519 | 0.03551 | 0.271 |
| 48738 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-590-5p | 10 | RB1CC1 | Sponge network | 0.331 | 0.57247 | 0.175 | 0.12559 | 0.271 |
| 48739 | AC003090.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | AFF1 | Sponge network | -2.117 | 0.00161 | -0.384 | 0.00053 | 0.271 |
| 48740 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 60 | KIF1B | Sponge network | 0.249 | 0.35902 | -0.118 | 0.32258 | 0.271 |
| 48741 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 23 | ARHGAP29 | Sponge network | -2.46 | 0 | 0.131 | 0.42953 | 0.271 |
| 48742 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-484 | 12 | GREM1 | Sponge network | -0.141 | 0.26434 | 0.902 | 0.01691 | 0.271 |
| 48743 | DIO3OS |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-501-5p;hsa-miR-589-3p | 16 | MAFB | Sponge network | -0.773 | 0.04073 | -0.023 | 0.89865 | 0.271 |
| 48744 | RP11-490M8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-3p | 11 | EGFR | Sponge network | -0.214 | 0.44248 | -0.175 | 0.48451 | 0.271 |
| 48745 | MIR600HG |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-877-5p;hsa-miR-940 | 22 | EPS15 | Sponge network | 0.594 | 0.41522 | -0.052 | 0.53998 | 0.271 |
| 48746 | RP4-758J18.7 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p | 10 | DST | Sponge network | -1.32 | 0.02051 | -0.713 | 0.00096 | 0.271 |
| 48747 | CTC-487M23.5 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-450b-5p | 10 | RYR2 | Sponge network | 0.912 | 0.00014 | -1.466 | 0.00031 | 0.271 |
| 48748 | SH3BP5-AS1 |
hsa-miR-1296-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-577 | 16 | CSDE1 | Sponge network | 0.267 | 0.2937 | -0.493 | 0 | 0.271 |
| 48749 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 47 | CCND2 | Sponge network | -0.323 | 0.01372 | -0.267 | 0.3112 | 0.271 |
| 48750 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | MAF | Sponge network | 0.348 | 0.32912 | -1.257 | 0 | 0.271 |
| 48751 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p | 20 | CCND2 | Sponge network | 0.001 | 0.99753 | -0.267 | 0.3112 | 0.271 |
| 48752 | RP4-758J18.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-450b-5p | 10 | DLC1 | Sponge network | -0.091 | 0.71468 | -0.382 | 0.05038 | 0.271 |
| 48753 | BZRAP1-AS1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-577;hsa-miR-625-5p | 23 | CADM1 | Sponge network | -0.465 | 0.04703 | -0.9 | 0.00084 | 0.271 |
| 48754 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-628-5p | 12 | CHD2 | Sponge network | 0.792 | 0.0049 | 0.049 | 0.55371 | 0.271 |
| 48755 | LINC00865 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 21 | ZMYND11 | Sponge network | -1.249 | 7.0E-5 | -0.2 | 0.05449 | 0.271 |
| 48756 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-3p | 25 | FOXO3 | Sponge network | -0.487 | 0.02157 | -0.04 | 0.72028 | 0.271 |
| 48757 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 34 | MOB1B | Sponge network | -0.355 | 0.0077 | 0.197 | 0.15266 | 0.271 |
| 48758 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-96-5p | 16 | MEF2C | Sponge network | 0.348 | 0.32912 | -0.499 | 0.01448 | 0.271 |
| 48759 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | SASH1 | Sponge network | 0.61 | 0.01593 | -0.502 | 0.00175 | 0.271 |
| 48760 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 30 | BCL2 | Sponge network | 0.395 | 0.15451 | -0.899 | 0 | 0.271 |
| 48761 | RP11-758N13.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-424-5p;hsa-miR-455-3p | 18 | EPHA7 | Sponge network | 2.939 | 3.0E-5 | -2.197 | 3.0E-5 | 0.271 |
| 48762 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PTPRB | Sponge network | 0.083 | 0.66405 | 0.105 | 0.47122 | 0.271 |
| 48763 | TMEM51-AS1 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | CTDSPL | Sponge network | 0.23 | 0.38667 | 0.085 | 0.45121 | 0.271 |
| 48764 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | THBS1 | Sponge network | -0.612 | 0.00094 | 0.741 | 0.00386 | 0.271 |
| 48765 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 58 | BMPR2 | Sponge network | -0.899 | 6.0E-5 | 0.206 | 0.0635 | 0.271 |
| 48766 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 17 | ERBB4 | Sponge network | 0.131 | 0.8436 | -1.975 | 2.0E-5 | 0.271 |
| 48767 | LINC00403 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-452-5p | 13 | MEF2C | Sponge network | -3.586 | 0.00032 | -0.499 | 0.01448 | 0.271 |
| 48768 | HOXB-AS3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p | 16 | EPHA5 | Sponge network | 0.343 | 0.28887 | -0.957 | 0.01733 | 0.271 |
| 48769 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | TAL1 | Sponge network | -0.355 | 0.0077 | -0.462 | 0.00574 | 0.271 |
| 48770 | SNHG14 |
hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | PPP2R2C | Sponge network | -1.111 | 0.0003 | -0.959 | 0.07428 | 0.271 |
| 48771 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 23 | BCL11A | Sponge network | -0.777 | 0.1134 | -0.932 | 0.0063 | 0.271 |
| 48772 | RP11-2H8.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 18 | SCN9A | Sponge network | 1.089 | 0 | -0.525 | 0.10593 | 0.271 |
| 48773 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-744-5p;hsa-miR-93-5p | 17 | FGFR1 | Sponge network | -1.047 | 0 | -0.509 | 0.03957 | 0.271 |
| 48774 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-5p;hsa-miR-625-5p | 15 | ZMYM2 | Sponge network | 0.983 | 0.00048 | 0.279 | 0.01273 | 0.271 |
| 48775 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 28 | HLF | Sponge network | -0.08 | 0.90092 | -2.443 | 0 | 0.271 |
| 48776 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-375;hsa-miR-539-5p | 11 | DIP2C | Sponge network | 0.858 | 3.0E-5 | -0.436 | 0.01713 | 0.271 |
| 48777 | IQCH-AS1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 14 | GSK3B | Sponge network | 0.929 | 0 | 0.266 | 0.00045 | 0.271 |
| 48778 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 13 | SLITRK6 | Sponge network | 0.178 | 0.29106 | -2.121 | 0 | 0.271 |
| 48779 | RP11-855A2.3 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NAV3 | Sponge network | 2.094 | 0 | 0.212 | 0.47729 | 0.271 |
| 48780 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | DAB2 | Sponge network | 0.494 | 0.00339 | 0.431 | 0.0113 | 0.271 |
| 48781 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p | 18 | FOXO3 | Sponge network | 0.594 | 0.00064 | -0.04 | 0.72028 | 0.271 |
| 48782 | AF131215.9 |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | RECK | Sponge network | -0.322 | 0.25945 | -1.043 | 0 | 0.271 |
| 48783 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-34a-5p | 10 | RARB | Sponge network | 0.131 | 0.8436 | -0.825 | 2.0E-5 | 0.271 |
| 48784 | HHIP-AS1 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p | 18 | ACSL6 | Sponge network | -0.737 | 0.10822 | -1.593 | 0 | 0.271 |
| 48785 | SNHG14 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 15 | GATA2 | Sponge network | -1.111 | 0.0003 | -0.654 | 0.0016 | 0.271 |
| 48786 | FAM66C |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | GABRB2 | Sponge network | -0.901 | 0.00019 | -1.841 | 0.00029 | 0.271 |
| 48787 | RP4-639F20.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-5p | 13 | KCNJ5 | Sponge network | -0.517 | 0.02935 | -0.281 | 0.3339 | 0.271 |
| 48788 | CTD-3138B18.5 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 39 | LPP | Sponge network | 0.379 | 0.03091 | -0.459 | 0.04475 | 0.271 |
| 48789 | CTD-2619J13.9 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-628-5p | 10 | QKI | Sponge network | 0.576 | 0.0022 | -0.333 | 0.04111 | 0.271 |
| 48790 | MIR22HG |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-454-3p | 16 | ITPKB | Sponge network | -1.047 | 0 | -0.704 | 0.00106 | 0.271 |
| 48791 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 23 | SOX11 | Sponge network | 1.089 | 0 | 2.68 | 0 | 0.271 |
| 48792 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | KAT6B | Sponge network | 0.559 | 0.00026 | -0.478 | 0.00018 | 0.271 |
| 48793 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-30a-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-374b-5p | 16 | SOX11 | Sponge network | 1.4 | 0 | 2.68 | 0 | 0.271 |
| 48794 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | PAK3 | Sponge network | 0.082 | 0.55654 | -0.628 | 0.07253 | 0.271 |
| 48795 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | USP25 | Sponge network | -2.452 | 0 | -0.242 | 0.01397 | 0.271 |
| 48796 | RP4-584D14.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-940 | 21 | GAS7 | Sponge network | 1.294 | 0.0031 | -0.555 | 0.01602 | 0.271 |
| 48797 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-576-5p | 14 | CDHR3 | Sponge network | -0.147 | 0.40452 | -0.977 | 4.0E-5 | 0.271 |
| 48798 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | DKK3 | Sponge network | 0.72 | 0.0001 | -0.052 | 0.77783 | 0.271 |
| 48799 | HAND2-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | LUZP2 | Sponge network | -1.697 | 0.00889 | -0.056 | 0.89416 | 0.271 |
| 48800 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3613-5p | 14 | GPAM | Sponge network | 0.909 | 0 | 0.142 | 0.29732 | 0.271 |
| 48801 | RP11-686D22.8 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | NCAM2 | Sponge network | 0.898 | 1.0E-5 | -0.482 | 0.22565 | 0.271 |
| 48802 | RP11-1277A3.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-92a-3p | 10 | LUZP2 | Sponge network | 0.453 | 0.01839 | -0.056 | 0.89416 | 0.271 |
| 48803 | BZRAP1-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-671-5p | 17 | PPP2R5C | Sponge network | -0.465 | 0.04703 | -0.314 | 3.0E-5 | 0.271 |
| 48804 | OIP5-AS1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | PRKAR1A | Sponge network | 0.421 | 0.00084 | -0.063 | 0.50314 | 0.271 |
| 48805 | RP11-554A11.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-339-5p | 15 | ACSL6 | Sponge network | -0.583 | 0.14589 | -1.593 | 0 | 0.271 |
| 48806 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | PHC3 | Sponge network | 0.246 | 0.59912 | 0.212 | 0.0499 | 0.271 |
| 48807 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 18 | SNX29 | Sponge network | 0.349 | 0.01695 | -0.34 | 0.01371 | 0.271 |
| 48808 | RP4-614O4.11 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-766-3p | 18 | MDM4 | Sponge network | 1.042 | 0.00177 | 0.345 | 0.00762 | 0.271 |
| 48809 | HCG11 |
hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | ERCC6 | Sponge network | 0.385 | 0.10067 | 0.23 | 0.015 | 0.271 |
| 48810 | RP11-166D19.1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | RABEP1 | Sponge network | -0.576 | 0.10121 | -0.066 | 0.43142 | 0.271 |
| 48811 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-655-3p | 26 | DMD | Sponge network | 0.614 | 1.0E-5 | -1.506 | 0 | 0.271 |
| 48812 | RP11-284N8.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-93-3p;hsa-miR-940 | 11 | GRIK3 | Sponge network | -0.31 | 0.33871 | -3.208 | 0 | 0.271 |
| 48813 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ZFHX3 | Sponge network | -0.223 | 0.47816 | 0.389 | 0.01363 | 0.271 |
| 48814 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 30 | APC | Sponge network | 0.532 | 0.00505 | -0.255 | 0.04051 | 0.271 |
| 48815 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-628-5p | 14 | ACVR1 | Sponge network | -1.092 | 4.0E-5 | 0.279 | 0.00234 | 0.271 |
| 48816 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | FGF5 | Sponge network | -0.342 | 0.02288 | 0.549 | 0.28659 | 0.271 |
| 48817 | LINC00654 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-7-5p | 21 | KCNJ5 | Sponge network | 0.374 | 0.20054 | -0.281 | 0.3339 | 0.271 |
| 48818 | CTD-2002H8.2 |
hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ITGAV | Sponge network | 0.931 | 1.0E-5 | 0.337 | 0.01002 | 0.271 |
| 48819 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-421 | 12 | RBL2 | Sponge network | -4.546 | 0 | -0.282 | 0.00254 | 0.271 |
| 48820 | RP11-473I1.5 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 14 | FOXP1 | Sponge network | 0.542 | 0.00054 | 0.042 | 0.74368 | 0.271 |
| 48821 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-590-3p | 10 | DLC1 | Sponge network | 0.131 | 0.8436 | -0.382 | 0.05038 | 0.271 |
| 48822 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-940 | 18 | ZFP36L1 | Sponge network | -0.421 | 0.0114 | -0.505 | 8.0E-5 | 0.271 |
| 48823 | RP13-516M14.1 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p | 10 | DCLK1 | Sponge network | 0.389 | 0.04293 | -1.324 | 0.00014 | 0.271 |
| 48824 | LINC00641 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-589-5p | 10 | SORCS2 | Sponge network | -0.222 | 0.32445 | -0.223 | 0.4253 | 0.271 |
| 48825 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-141-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-361-3p | 10 | KAT6B | Sponge network | 0.556 | 0.00298 | -0.478 | 0.00018 | 0.271 |
| 48826 | RP11-37B2.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | ARHGEF12 | Sponge network | 1.03 | 0 | 0.09 | 0.35693 | 0.271 |
| 48827 | RP11-98I9.4 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | DDX5 | Sponge network | 0.67 | 0.00048 | -0.099 | 0.17428 | 0.271 |
| 48828 | RP11-35G9.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-940 | 23 | GAS7 | Sponge network | 0.657 | 4.0E-5 | -0.555 | 0.01602 | 0.271 |
| 48829 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 41 | MOB1B | Sponge network | -0.737 | 0.06182 | 0.197 | 0.15266 | 0.271 |
| 48830 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-582-5p;hsa-miR-655-3p | 12 | PIK3C2A | Sponge network | 0.757 | 0 | 0.061 | 0.53776 | 0.271 |
| 48831 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 25 | RAP1A | Sponge network | 0.335 | 0.20596 | -0.929 | 0 | 0.271 |
| 48832 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | MED12L | Sponge network | -0.777 | 0.1134 | -0.194 | 0.55299 | 0.271 |
| 48833 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-940 | 22 | CHD5 | Sponge network | -4.477 | 0 | -0.922 | 0.01521 | 0.271 |
| 48834 | RP11-360F5.3 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | TSC1 | Sponge network | 1.867 | 0 | 0.103 | 0.26071 | 0.271 |
| 48835 | RP11-428J1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-342-3p;hsa-miR-942-5p | 11 | FBXO31 | Sponge network | 0.538 | 0.00076 | -0.085 | 0.42389 | 0.271 |
| 48836 | SH3BP5-AS1 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 13 | INPP4A | Sponge network | 0.267 | 0.2937 | 0.07 | 0.53563 | 0.271 |
| 48837 | RP11-298J20.4 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-671-5p | 11 | GRIK3 | Sponge network | 0.002 | 0.99057 | -3.208 | 0 | 0.271 |
| 48838 | C4B-AS1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-744-3p | 27 | DST | Sponge network | 2.306 | 9.0E-5 | -0.713 | 0.00096 | 0.271 |
| 48839 | PWAR6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 32 | BHLHE41 | Sponge network | -1.575 | 6.0E-5 | 0.353 | 0.12753 | 0.271 |
| 48840 | PCA3 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 11 | ACTB | Sponge network | -0.709 | 0.18168 | -0.247 | 0.01736 | 0.271 |
| 48841 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 25 | ARID5B | Sponge network | 0.4 | 0.14611 | 0.342 | 0.01676 | 0.271 |
| 48842 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-550a-3p;hsa-miR-942-5p | 13 | PCDH17 | Sponge network | 0.082 | 0.55397 | 0.731 | 2.0E-5 | 0.271 |
| 48843 | PRKAG2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | NTRK3 | Sponge network | -0.622 | 0.02807 | -1.77 | 3.0E-5 | 0.271 |
| 48844 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | 0.385 | 0.10067 | -3.328 | 0 | 0.271 |
| 48845 | CALML3-AS1 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484 | 10 | ITGA5 | Sponge network | 0.95 | 0.15943 | 0.452 | 0.03524 | 0.271 |
| 48846 | AC010226.4 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-590-3p | 11 | MXI1 | Sponge network | -0.811 | 2.0E-5 | -0.987 | 0 | 0.271 |
| 48847 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PTPRB | Sponge network | -0.235 | 0.15142 | 0.105 | 0.47122 | 0.271 |
| 48848 | RP11-212P7.2 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 14 | INPP4A | Sponge network | 0.219 | 0.1516 | 0.07 | 0.53563 | 0.271 |
| 48849 | LINC00654 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 35 | PPP1R9A | Sponge network | 0.374 | 0.20054 | -0.903 | 0.04254 | 0.27 |
| 48850 | GUSBP11 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-497-5p | 10 | CDK6 | Sponge network | 0.856 | 0 | 0.991 | 0 | 0.27 |
| 48851 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-5p;hsa-miR-96-5p | 17 | RECK | Sponge network | 0.331 | 0.57247 | -1.043 | 0 | 0.27 |
| 48852 | RP11-67L2.2 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 41 | PPARA | Sponge network | 0.72 | 0.0001 | -0.379 | 0.00548 | 0.27 |
| 48853 | C4A-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-511-5p;hsa-miR-590-5p | 12 | EYA4 | Sponge network | 3.345 | 0 | 0.3 | 0.36876 | 0.27 |
| 48854 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | FGF5 | Sponge network | 0.064 | 0.72934 | 0.549 | 0.28659 | 0.27 |
| 48855 | LINC00476 |
hsa-miR-140-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | NOV | Sponge network | -0.615 | 0.00015 | -0.58 | 0.04676 | 0.27 |
| 48856 | AC159540.2 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | CCDC6 | Sponge network | 1.634 | 0 | 0.27 | 0.02555 | 0.27 |
| 48857 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 12 | DKK3 | Sponge network | 0.862 | 0.06213 | -0.052 | 0.77783 | 0.27 |
| 48858 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-877-5p | 22 | NRK | Sponge network | -0.355 | 0.0077 | 0.656 | 0.19217 | 0.27 |
| 48859 | RP11-53O19.3 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | DICER1 | Sponge network | 1.205 | 0 | 0.042 | 0.674 | 0.27 |
| 48860 | RP11-526I2.5 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-628-5p | 11 | SLC5A3 | Sponge network | 1.054 | 1.0E-5 | 0.493 | 5.0E-5 | 0.27 |
| 48861 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | TMEFF2 | Sponge network | 0.225 | 0.30644 | -2.426 | 0 | 0.27 |
| 48862 | RP11-1006G14.4 |
hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-590-3p | 10 | PRKAR1A | Sponge network | 0.559 | 0.00026 | -0.063 | 0.50314 | 0.27 |
| 48863 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-96-5p | 20 | ACVR1 | Sponge network | -0.976 | 0.00025 | 0.279 | 0.00234 | 0.27 |
| 48864 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 13 | BCL11A | Sponge network | 1.757 | 0 | -0.932 | 0.0063 | 0.27 |
| 48865 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | NCAM2 | Sponge network | -0.969 | 2.0E-5 | -0.482 | 0.22565 | 0.27 |
| 48866 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-375;hsa-miR-629-3p;hsa-miR-96-5p | 15 | EBF3 | Sponge network | -0.326 | 0.14314 | -0.058 | 0.81437 | 0.27 |
| 48867 | LINC00472 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 19 | OPCML | Sponge network | -0.842 | 0.01361 | -2.498 | 0 | 0.27 |
| 48868 | RP11-490M8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-33a-5p | 10 | EYA4 | Sponge network | -0.214 | 0.44248 | 0.3 | 0.36876 | 0.27 |
| 48869 | A1BG-AS1 |
hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p | 10 | ZBTB4 | Sponge network | -0.164 | 0.45106 | -0.739 | 0 | 0.27 |
| 48870 | RP11-434B12.1 |
hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | UBE2D1 | Sponge network | -0.031 | 0.89063 | 0.468 | 0 | 0.27 |
| 48871 | CD27-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-455-5p | 15 | PTGFR | Sponge network | 0.177 | 0.16681 | -0.233 | 0.45421 | 0.27 |
| 48872 | SOX2-OT |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-5p | 25 | PGR | Sponge network | -1.264 | 6.0E-5 | -1.704 | 0 | 0.27 |
| 48873 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-577 | 12 | GREM1 | Sponge network | -0.418 | 0.00068 | 0.902 | 0.01691 | 0.27 |
| 48874 | RP11-693J15.4 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320b;hsa-miR-589-5p | 11 | SLITRK3 | Sponge network | -0.72 | 0.24239 | -3.328 | 0 | 0.27 |
| 48875 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 24 | YAP1 | Sponge network | 0.219 | 0.1516 | 0.22 | 0.11836 | 0.27 |
| 48876 | AP001347.6 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-7-5p | 24 | BTG2 | Sponge network | -1.161 | 0.00591 | -1.763 | 0 | 0.27 |
| 48877 | RP11-386G11.5 |
hsa-miR-125a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-409-5p | 13 | CCDC6 | Sponge network | 2.031 | 0.00154 | 0.27 | 0.02555 | 0.27 |
| 48878 | PGM5-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-7-1-3p | 10 | EGR2 | Sponge network | -4.546 | 0 | 0.309 | 0.17079 | 0.27 |
| 48879 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 14 | SASH1 | Sponge network | 0.131 | 0.8436 | -0.502 | 0.00175 | 0.27 |
| 48880 | LINC00476 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | SEMA3E | Sponge network | -0.615 | 0.00015 | -1.717 | 0.00137 | 0.27 |
| 48881 | RP5-1198O20.4 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | SPOP | Sponge network | 0.997 | 0.00066 | -0.419 | 1.0E-5 | 0.27 |
| 48882 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | PTPN11 | Sponge network | 0.768 | 7.0E-5 | 0.431 | 2.0E-5 | 0.27 |
| 48883 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 51 | IL6ST | Sponge network | 0.61 | 0.01593 | -0.402 | 0.00676 | 0.27 |
| 48884 | AC108488.4 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577 | 12 | FMNL3 | Sponge network | 0.259 | 0.12542 | 0.555 | 4.0E-5 | 0.27 |
| 48885 | GNG12-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p | 23 | CADM1 | Sponge network | -0.793 | 7.0E-5 | -0.9 | 0.00084 | 0.27 |
| 48886 | RP11-888D10.4 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-590-3p | 10 | PICALM | Sponge network | 0.702 | 7.0E-5 | 0.086 | 0.23523 | 0.27 |
| 48887 | NNT-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | NF1 | Sponge network | 0.799 | 1.0E-5 | 0.51 | 0 | 0.27 |
| 48888 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-7-1-3p | 14 | BCL11A | Sponge network | 0.299 | 0.57722 | -0.932 | 0.0063 | 0.27 |
| 48889 | LINC00641 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | MSN | Sponge network | -0.222 | 0.32445 | -0.023 | 0.88422 | 0.27 |
| 48890 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-874-3p | 10 | ST5 | Sponge network | 0.083 | 0.66405 | -0.78 | 0 | 0.27 |
| 48891 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-664a-3p | 18 | NUAK1 | Sponge network | -2.69 | 0.0001 | 0.904 | 0 | 0.27 |
| 48892 | CKMT2-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 51 | CADM2 | Sponge network | 0.082 | 0.55654 | -3.782 | 0 | 0.27 |
| 48893 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | PDE4DIP | Sponge network | 0.421 | 0.00084 | -0.282 | 0.0257 | 0.27 |
| 48894 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 49 | FAT3 | Sponge network | -1.264 | 6.0E-5 | -1.601 | 0.00051 | 0.27 |
| 48895 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | IGSF10 | Sponge network | 0.342 | 0.03129 | -1.751 | 1.0E-5 | 0.27 |
| 48896 | RP11-678G14.2 |
hsa-miR-103a-2-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-744-3p | 13 | LPP | Sponge network | -1.765 | 0.0778 | -0.459 | 0.04475 | 0.27 |
| 48897 | FENDRR |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | RABEP1 | Sponge network | -1.281 | 0.00012 | -0.066 | 0.43142 | 0.27 |
| 48898 | C20orf166-AS1 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-532-5p;hsa-miR-582-5p | 21 | MTSS1 | Sponge network | -2.69 | 0.0001 | -0.81 | 0.00035 | 0.27 |
| 48899 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 15 | NCAM2 | Sponge network | 0.225 | 0.30644 | -0.482 | 0.22565 | 0.27 |
| 48900 | CTD-2619J13.9 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-532-3p | 12 | GAS7 | Sponge network | 0.576 | 0.0022 | -0.555 | 0.01602 | 0.27 |
| 48901 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | LIFR | Sponge network | 0.657 | 4.0E-5 | -1.794 | 0 | 0.27 |
| 48902 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-940 | 20 | IGF1 | Sponge network | 0.921 | 0.00118 | -0.204 | 0.5898 | 0.27 |
| 48903 | AC012360.6 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-26a-5p;hsa-miR-365a-3p;hsa-miR-455-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 16 | ARHGEF12 | Sponge network | 0.624 | 0.02176 | 0.09 | 0.35693 | 0.27 |
| 48904 | RP5-1139B12.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-454-3p | 13 | MED12L | Sponge network | 0.598 | 0.38481 | -0.194 | 0.55299 | 0.27 |
| 48905 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | SYNE1 | Sponge network | 0.919 | 0 | -0.738 | 0.00316 | 0.27 |
| 48906 | CTD-3064M3.3 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-7-5p | 10 | LZTS1 | Sponge network | -0.469 | 0.08721 | 1.664 | 0 | 0.27 |
| 48907 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-5p | 14 | PRKAR1A | Sponge network | 0.889 | 9.0E-5 | -0.063 | 0.50314 | 0.27 |
| 48908 | AC010226.4 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-5p | 30 | NFIB | Sponge network | -0.811 | 2.0E-5 | 0.023 | 0.90795 | 0.27 |
| 48909 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-98-5p | 10 | SCN11A | Sponge network | -0.727 | 0.04726 | -1.15 | 0.00299 | 0.27 |
| 48910 | RP11-401P9.1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-744-3p | 28 | LPP | Sponge network | 1.524 | 0.1098 | -0.459 | 0.04475 | 0.27 |
| 48911 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p | 12 | CLASP2 | Sponge network | 0.752 | 0 | -0.038 | 0.71 | 0.27 |
| 48912 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ZMYND11 | Sponge network | -0.513 | 0.04703 | -0.2 | 0.05449 | 0.27 |
| 48913 | MIR497HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | NUAK1 | Sponge network | -1.439 | 4.0E-5 | 0.904 | 0 | 0.27 |
| 48914 | AF131215.9 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-92b-3p | 15 | LUZP2 | Sponge network | -0.322 | 0.25945 | -0.056 | 0.89416 | 0.27 |
| 48915 | RP11-999E24.3 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-7-5p | 13 | IRS1 | Sponge network | -0.693 | 0.05629 | -0.026 | 0.88855 | 0.27 |
| 48916 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-942-5p | 22 | NOTCH2 | Sponge network | 1.757 | 0 | 0.138 | 0.35163 | 0.27 |
| 48917 | HOXB-AS2 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-22-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-505-5p | 10 | ETV1 | Sponge network | 1.316 | 0.01106 | -0.096 | 0.67674 | 0.27 |
| 48918 | DIO3OS |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-3p;hsa-miR-940 | 34 | TRPS1 | Sponge network | -0.773 | 0.04073 | -0.191 | 0.44109 | 0.27 |
| 48919 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-96-5p | 16 | ACVR1 | Sponge network | -1.439 | 4.0E-5 | 0.279 | 0.00234 | 0.27 |
| 48920 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | CREBBP | Sponge network | 0.401 | 0.00066 | 0.126 | 0.15186 | 0.27 |
| 48921 | ARHGEF26-AS1 |
hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | EDNRB | Sponge network | -2.46 | 0 | -0.682 | 0.00138 | 0.27 |
| 48922 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-7-5p | 17 | ATRX | Sponge network | 0.001 | 0.99753 | 0.261 | 0.04408 | 0.27 |
| 48923 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-374a-3p | 10 | MYO9A | Sponge network | 1.352 | 0 | -0.147 | 0.32373 | 0.27 |
| 48924 | RP11-720L2.4 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-940 | 11 | TRPS1 | Sponge network | -1.255 | 0.00981 | -0.191 | 0.44109 | 0.27 |
| 48925 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-98-5p | 20 | ARHGAP28 | Sponge network | -0.83 | 0 | -0.911 | 0.00013 | 0.27 |
| 48926 | AC005154.6 |
hsa-miR-125a-3p;hsa-miR-149-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | CDC27 | Sponge network | 0.848 | 0 | 0.384 | 0 | 0.27 |
| 48927 | AC127904.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | RPS6KA3 | Sponge network | 1.308 | 0 | 0.256 | 0.02419 | 0.27 |
| 48928 | RP11-379F4.7 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 15 | CBX5 | Sponge network | 1.316 | 0 | 0.165 | 0.24446 | 0.27 |
| 48929 | CTC-429P9.1 |
hsa-miR-128-3p;hsa-miR-141-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-409-3p | 11 | AFF3 | Sponge network | 0.497 | 0.01451 | -2.373 | 0 | 0.27 |
| 48930 | ZSCAN16-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-484 | 13 | ITGA5 | Sponge network | -0.342 | 0.02288 | 0.452 | 0.03524 | 0.27 |
| 48931 | CASC2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 13 | GPAM | Sponge network | 0.385 | 0.01306 | 0.142 | 0.29732 | 0.27 |
| 48932 | AC025171.1 |
hsa-let-7g-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | MEF2C | Sponge network | 0.998 | 0 | -0.499 | 0.01448 | 0.27 |
| 48933 | RP11-798M19.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 19 | CYLD | Sponge network | -0.612 | 0.00094 | -0.367 | 0.00171 | 0.27 |
| 48934 | RP11-327P2.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-664a-3p | 12 | PRUNE2 | Sponge network | -0.147 | 0.40452 | -1.733 | 0.00022 | 0.27 |
| 48935 | KCNJ2-AS1 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | CADM1 | Sponge network | -0.436 | 0.05325 | -0.9 | 0.00084 | 0.27 |
| 48936 | LINC01006 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-484 | 15 | CADM1 | Sponge network | -0.702 | 0.04814 | -0.9 | 0.00084 | 0.27 |
| 48937 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-577 | 18 | GREM1 | Sponge network | -0.899 | 6.0E-5 | 0.902 | 0.01691 | 0.27 |
| 48938 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | LRRC7 | Sponge network | 0.349 | 0.01695 | -1.341 | 0.01193 | 0.27 |
| 48939 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 10 | PDGFC | Sponge network | -0.709 | 0.18168 | -0.33 | 0.06483 | 0.27 |
| 48940 | DLEU2 |
hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 11 | CDK6 | Sponge network | 1.831 | 0 | 0.991 | 0 | 0.27 |
| 48941 | RP11-37B2.1 |
hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-625-5p;hsa-miR-664a-3p | 15 | NUAK1 | Sponge network | 1.03 | 0 | 0.904 | 0 | 0.27 |
| 48942 | FAM66C |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 19 | SUFU | Sponge network | -0.901 | 0.00019 | -0.04 | 0.65489 | 0.27 |
| 48943 | AC034220.3 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-495-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 28 | DDX6 | Sponge network | 0.309 | 0.07304 | 0.091 | 0.36428 | 0.27 |
| 48944 | RP11-678G14.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | SYT4 | Sponge network | -4.477 | 0 | -3.207 | 0 | 0.27 |
| 48945 | RP11-195B17.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-96-5p | 14 | SPRY3 | Sponge network | 0.37 | 0.27614 | 0.016 | 0.91286 | 0.27 |
| 48946 | MAGI2-AS3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 27 | CSMD1 | Sponge network | -0.129 | 0.57177 | -0.63 | 0.18881 | 0.27 |
| 48947 | RP11-403I13.7 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 17 | SFRP1 | Sponge network | 0.395 | 0.15451 | -3.566 | 0 | 0.27 |
| 48948 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | YAP1 | Sponge network | 1.302 | 7.0E-5 | 0.22 | 0.11836 | 0.27 |
| 48949 | CTD-3138B18.5 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 18 | MOB1B | Sponge network | 0.379 | 0.03091 | 0.197 | 0.15266 | 0.27 |
| 48950 | GABPB1-AS1 |
hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p | 10 | LZTS1 | Sponge network | 1.11 | 0 | 1.664 | 0 | 0.27 |
| 48951 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-25-3p;hsa-miR-320b | 11 | MYCBP2 | Sponge network | -1.092 | 4.0E-5 | -0.027 | 0.82016 | 0.27 |
| 48952 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p | 24 | BNC2 | Sponge network | 0.921 | 0.00118 | -0.491 | 0.09988 | 0.27 |
| 48953 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-940 | 32 | SNX29 | Sponge network | -0.899 | 6.0E-5 | -0.34 | 0.01371 | 0.27 |
| 48954 | RP11-531A24.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-429 | 16 | LATS2 | Sponge network | 0.75 | 0.00054 | 0.159 | 0.2304 | 0.27 |
| 48955 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-940 | 19 | PPP1R9A | Sponge network | -0.829 | 0.01343 | -0.903 | 0.04254 | 0.27 |
| 48956 | AF131215.9 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-93-3p;hsa-miR-940 | 12 | POU2F2 | Sponge network | -0.322 | 0.25945 | -0.233 | 0.32214 | 0.27 |
| 48957 | RP11-983P16.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-96-5p | 11 | LATS2 | Sponge network | 0.41 | 0.02214 | 0.159 | 0.2304 | 0.27 |
| 48958 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 15 | NF1 | Sponge network | 1.117 | 0 | 0.51 | 0 | 0.27 |
| 48959 | RP11-428J1.5 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-505-5p;hsa-miR-590-3p | 14 | MRVI1 | Sponge network | 0.538 | 0.00076 | -1.051 | 0.00022 | 0.27 |
| 48960 | SNX29P2 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | NCAM2 | Sponge network | 0.4 | 0.14611 | -0.482 | 0.22565 | 0.27 |
| 48961 | AGAP11 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-5p | 13 | KCTD8 | Sponge network | -1.645 | 0 | -3.359 | 0 | 0.27 |
| 48962 | AP001347.6 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | CREB3L2 | Sponge network | -1.161 | 0.00591 | -0.525 | 2.0E-5 | 0.27 |
| 48963 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-93-3p | 22 | PAFAH1B1 | Sponge network | -0.693 | 0.05629 | -0.429 | 0 | 0.27 |
| 48964 | RP5-1198O20.4 |
hsa-miR-1180-3p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-365a-3p;hsa-miR-429;hsa-miR-502-5p | 10 | ETS1 | Sponge network | 0.997 | 0.00066 | 0.634 | 4.0E-5 | 0.27 |
| 48965 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 30 | EPHA5 | Sponge network | 0.389 | 0.04293 | -0.957 | 0.01733 | 0.27 |
| 48966 | TINCR |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 12 | SHPRH | Sponge network | -0.664 | 0.14543 | 0.098 | 0.40994 | 0.27 |
| 48967 | AC009120.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 30 | AFF4 | Sponge network | 0.919 | 0 | -0.266 | 0.01565 | 0.27 |
| 48968 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-93-3p | 25 | EPHA3 | Sponge network | -0.371 | 0.24604 | 0.185 | 0.53588 | 0.27 |
| 48969 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935 | 29 | HBP1 | Sponge network | -0.384 | 0.40652 | -0.454 | 0 | 0.27 |
| 48970 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 36 | EPHA5 | Sponge network | 0.683 | 1.0E-5 | -0.957 | 0.01733 | 0.27 |
| 48971 | RP11-296I10.3 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-539-5p;hsa-miR-942-5p | 23 | ARHGEF12 | Sponge network | 0.592 | 0.00014 | 0.09 | 0.35693 | 0.27 |
| 48972 | RP11-244O19.1 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-625-5p | 13 | ADCY1 | Sponge network | -0.096 | 0.67958 | 0.362 | 0.16982 | 0.27 |
| 48973 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ABCA10 | Sponge network | 1.302 | 7.0E-5 | -0.571 | 0.03674 | 0.27 |
| 48974 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | MOB1B | Sponge network | 0.476 | 0.19203 | 0.197 | 0.15266 | 0.27 |
| 48975 | RP11-175O19.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-766-3p | 27 | MDM4 | Sponge network | 0.917 | 0 | 0.345 | 0.00762 | 0.27 |
| 48976 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p | 16 | ACVR1 | Sponge network | -0.777 | 0.16667 | 0.279 | 0.00234 | 0.27 |
| 48977 | RP11-46C24.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PDE4DIP | Sponge network | 0.392 | 0.0278 | -0.282 | 0.0257 | 0.27 |
| 48978 | PSMD5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p | 29 | ZFX | Sponge network | 0.569 | 0.00067 | 0.09 | 0.42563 | 0.27 |
| 48979 | AC116366.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-375;hsa-miR-577;hsa-miR-629-3p | 11 | EBF3 | Sponge network | 0.329 | 0.11122 | -0.058 | 0.81437 | 0.27 |
| 48980 | AC018890.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429 | 13 | ERCC4 | Sponge network | 1.61 | 0.00961 | 0.126 | 0.15266 | 0.27 |
| 48981 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-590-3p | 21 | ZFHX4 | Sponge network | 0.765 | 7.0E-5 | 0.018 | 0.95574 | 0.27 |
| 48982 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-874-3p | 13 | NR2C2 | Sponge network | -0.663 | 0.10887 | 0.21 | 0.03224 | 0.27 |
| 48983 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-93-5p;hsa-miR-935 | 56 | SMAD4 | Sponge network | -0.162 | 0.47687 | -0.313 | 0.00413 | 0.27 |
| 48984 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p | 18 | SPRY3 | Sponge network | 1.601 | 0 | 0.016 | 0.91286 | 0.27 |
| 48985 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-93-5p | 38 | EPHA7 | Sponge network | -0.371 | 0.24604 | -2.197 | 3.0E-5 | 0.27 |
| 48986 | RP4-605O3.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 15 | MED12L | Sponge network | 0.262 | 0.0475 | -0.194 | 0.55299 | 0.27 |
| 48987 | LINC00843 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-429 | 14 | NFIB | Sponge network | 1.106 | 0 | 0.023 | 0.90795 | 0.27 |
| 48988 | RP11-439L18.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-93-3p | 20 | BMPR2 | Sponge network | 0.224 | 0.48719 | 0.206 | 0.0635 | 0.27 |
| 48989 | LA16c-321D4.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-654-3p | 13 | AKAP11 | Sponge network | 2.459 | 5.0E-5 | 0.196 | 0.09825 | 0.27 |
| 48990 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | EBF1 | Sponge network | 1.308 | 0 | -0.384 | 0.08856 | 0.27 |
| 48991 | NEAT1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-505-3p;hsa-miR-590-3p | 16 | PTPN11 | Sponge network | 0.705 | 0.00014 | 0.431 | 2.0E-5 | 0.27 |
| 48992 | EIF3J-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | WDFY3 | Sponge network | 0.331 | 0.00509 | -0.201 | 0.0967 | 0.27 |
| 48993 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | CXXC4 | Sponge network | -0.737 | 0.06182 | -0.433 | 0.21055 | 0.27 |
| 48994 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 28 | GRIA3 | Sponge network | -0.096 | 0.67958 | -1.956 | 0 | 0.27 |
| 48995 | RP11-400K9.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-590-5p;hsa-miR-93-3p | 13 | PAFAH1B1 | Sponge network | -0.733 | 0.19469 | -0.429 | 0 | 0.27 |
| 48996 | TMEM9B-AS1 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-582-5p | 13 | ARID5B | Sponge network | 0.529 | 0.00552 | 0.342 | 0.01676 | 0.27 |
| 48997 | BCYRN1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 13 | UVRAG | Sponge network | 0.311 | 0.10344 | -0.017 | 0.83865 | 0.27 |
| 48998 | LINC00622 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZFHX4 | Sponge network | 1.169 | 0.00028 | 0.018 | 0.95574 | 0.27 |
| 48999 | PSMG3-AS1 |
hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-502-5p | 16 | PTPN14 | Sponge network | -0.125 | 0.49414 | 0.144 | 0.34595 | 0.27 |
| 49000 | CTD-2002H8.2 |
hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | CDC73 | Sponge network | 0.931 | 1.0E-5 | 0.094 | 0.20463 | 0.27 |
| 49001 | RP11-327P2.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-23a-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 16 | NRXN3 | Sponge network | -0.147 | 0.40452 | -0.893 | 0.04186 | 0.27 |
| 49002 | GUSBP11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-375;hsa-miR-495-3p;hsa-miR-539-5p | 11 | ABI2 | Sponge network | 0.856 | 0 | 0.175 | 0.14787 | 0.27 |
| 49003 | MYLK-AS1 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-484 | 18 | DDX6 | Sponge network | -1.331 | 3.0E-5 | 0.091 | 0.36428 | 0.27 |
| 49004 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | NOTCH2NL | Sponge network | -2.452 | 0 | 0.024 | 0.84307 | 0.27 |
| 49005 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CDH19 | Sponge network | -0.08 | 0.90092 | -3.434 | 0 | 0.27 |
| 49006 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 11 | GPAM | Sponge network | 0.769 | 0 | 0.142 | 0.29732 | 0.27 |
| 49007 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-148a-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | CDC73 | Sponge network | 0.702 | 7.0E-5 | 0.094 | 0.20463 | 0.27 |
| 49008 | CTD-2303H24.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | PLAG1 | Sponge network | 0.415 | 0.14657 | -0.315 | 0.26532 | 0.27 |
| 49009 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | DCN | Sponge network | 0.61 | 0.01593 | -1.288 | 0 | 0.27 |
| 49010 | BVES-AS1 |
hsa-let-7a-3p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-942-5p | 13 | PCDH17 | Sponge network | -2.62 | 0 | 0.731 | 2.0E-5 | 0.27 |
| 49011 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | ANK2 | Sponge network | 0.898 | 1.0E-5 | -1.603 | 1.0E-5 | 0.27 |
| 49012 | RP11-13K12.1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 10 | PEA15 | Sponge network | -0.154 | 0.78939 | 0.009 | 0.93318 | 0.27 |
| 49013 | CALML3-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-3p | 11 | FAM172A | Sponge network | 0.95 | 0.15943 | -0.57 | 0 | 0.27 |
| 49014 | SOX2-OT |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-30d-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-96-5p | 19 | RIMBP2 | Sponge network | -1.264 | 6.0E-5 | -1.968 | 5.0E-5 | 0.27 |
| 49015 | RP11-678G14.3 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-24-3p | 13 | DLC1 | Sponge network | -4.477 | 0 | -0.382 | 0.05038 | 0.27 |
| 49016 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-629-3p | 22 | LIFR | Sponge network | 0.253 | 0.09565 | -1.794 | 0 | 0.27 |
| 49017 | AC083843.1 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-374b-5p | 10 | CDK6 | Sponge network | 1.4 | 0 | 0.991 | 0 | 0.27 |
| 49018 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 17 | PER2 | Sponge network | -0.487 | 0.02157 | -0.396 | 0.00269 | 0.27 |
| 49019 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p | 12 | PHC3 | Sponge network | 1.206 | 0 | 0.212 | 0.0499 | 0.27 |
| 49020 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ITGAV | Sponge network | 0.727 | 3.0E-5 | 0.337 | 0.01002 | 0.27 |
| 49021 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-30b-5p | 13 | GPAM | Sponge network | 1.262 | 0 | 0.142 | 0.29732 | 0.27 |
| 49022 | RP11-758N13.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-505-5p | 15 | ASTN1 | Sponge network | 2.939 | 3.0E-5 | -2.389 | 2.0E-5 | 0.27 |
| 49023 | RP11-159D12.2 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375 | 11 | VEZT | Sponge network | 1.254 | 0 | 0.259 | 0.00296 | 0.27 |
| 49024 | RP11-46C24.7 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | LUZP2 | Sponge network | 0.392 | 0.0278 | -0.056 | 0.89416 | 0.27 |
| 49025 | FAM66C |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-940 | 56 | NFIB | Sponge network | -0.901 | 0.00019 | 0.023 | 0.90795 | 0.27 |
| 49026 | RP11-212P7.2 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-7-5p;hsa-miR-96-5p | 14 | FNBP1 | Sponge network | 0.219 | 0.1516 | -0.969 | 4.0E-5 | 0.27 |
| 49027 | CTB-113P19.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 28 | PIK3R1 | Sponge network | 0.906 | 0.31715 | -0.133 | 0.36854 | 0.27 |
| 49028 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p | 23 | ERCC4 | Sponge network | -0.405 | 0.22013 | 0.126 | 0.15266 | 0.27 |
| 49029 | CTC-429P9.3 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p | 10 | AMPH | Sponge network | 0.082 | 0.55397 | -0.916 | 5.0E-5 | 0.27 |
| 49030 | SH3RF3-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-874-3p | 11 | SORCS1 | Sponge network | 0.889 | 9.0E-5 | -3.492 | 0 | 0.27 |
| 49031 | RP11-254F7.2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-5p | 15 | PIK3CA | Sponge network | 0.176 | 0.37402 | 0.195 | 0.06908 | 0.27 |
| 49032 | CTC-378H22.2 |
hsa-miR-103a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-29b-1-5p;hsa-miR-455-5p;hsa-miR-501-5p | 11 | IL6ST | Sponge network | 0.001 | 0.99663 | -0.402 | 0.00676 | 0.27 |
| 49033 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-429 | 10 | DKK3 | Sponge network | 0.859 | 0.00331 | -0.052 | 0.77783 | 0.27 |
| 49034 | RP11-574K11.29 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326 | 12 | CBFB | Sponge network | 0.997 | 0 | 1.061 | 0 | 0.27 |
| 49035 | U91328.19 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p | 11 | KCTD8 | Sponge network | -0.303 | 0.21002 | -3.359 | 0 | 0.27 |
| 49036 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-429;hsa-miR-590-3p | 15 | USP6 | Sponge network | 1.317 | 0 | 0.188 | 0.3871 | 0.27 |
| 49037 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | CDH10 | Sponge network | -0.355 | 0.0077 | -2.397 | 0 | 0.27 |
| 49038 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | PDGFRA | Sponge network | -0.709 | 0.18168 | -0.372 | 0.0037 | 0.27 |
| 49039 | TMEM51-AS1 |
hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p | 19 | ARHGEF12 | Sponge network | 0.23 | 0.38667 | 0.09 | 0.35693 | 0.27 |
| 49040 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-96-5p | 16 | SCN9A | Sponge network | 1.2 | 0.01813 | -0.525 | 0.10593 | 0.27 |
| 49041 | RP11-434B12.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-429;hsa-miR-590-3p | 22 | DLC1 | Sponge network | -0.031 | 0.89063 | -0.382 | 0.05038 | 0.27 |
| 49042 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 24 | MYBL1 | Sponge network | -1.575 | 6.0E-5 | 0.494 | 0.02152 | 0.27 |
| 49043 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-33a-5p | 12 | ARID5B | Sponge network | -0.473 | 0.07602 | 0.342 | 0.01676 | 0.27 |
| 49044 | EIF3J-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 11 | FREM2 | Sponge network | 0.331 | 0.00509 | -0.437 | 0.46353 | 0.27 |
| 49045 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-766-3p | 28 | MAPK1 | Sponge network | 1.153 | 0 | -0.017 | 0.81465 | 0.27 |
| 49046 | LINC01006 |
hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p | 18 | MED12L | Sponge network | -0.702 | 0.04814 | -0.194 | 0.55299 | 0.27 |
| 49047 | TINCR |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-940 | 11 | FAM172A | Sponge network | -0.664 | 0.14543 | -0.57 | 0 | 0.27 |
| 49048 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-320b;hsa-miR-590-5p | 13 | FOXO3 | Sponge network | 0.983 | 0.00048 | -0.04 | 0.72028 | 0.27 |
| 49049 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 24 | CLASP2 | Sponge network | -0.513 | 0.04703 | -0.038 | 0.71 | 0.27 |
| 49050 | PDXDC2P |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-654-3p | 11 | RICTOR | Sponge network | 0.756 | 0 | 0.388 | 0.00188 | 0.27 |
| 49051 | CTB-113P19.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 19 | SNX29 | Sponge network | 0.906 | 0.31715 | -0.34 | 0.01371 | 0.27 |
| 49052 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 35 | GAS7 | Sponge network | 0.259 | 0.12542 | -0.555 | 0.01602 | 0.27 |
| 49053 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p | 25 | USP12 | Sponge network | 0.253 | 0.09565 | -0.102 | 0.40768 | 0.27 |
| 49054 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-625-3p | 19 | BMPR2 | Sponge network | 0.903 | 0 | 0.206 | 0.0635 | 0.27 |
| 49055 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | CACNA2D1 | Sponge network | 1.757 | 0 | -0.846 | 0.00675 | 0.27 |
| 49056 | LINC00094 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-5p | 18 | PTGFR | Sponge network | 0.253 | 0.09565 | -0.233 | 0.45421 | 0.27 |
| 49057 | RP11-46C24.7 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TSC1 | Sponge network | 0.392 | 0.0278 | 0.103 | 0.26071 | 0.27 |
| 49058 | RP11-531A24.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429 | 12 | ITGB3 | Sponge network | 0.75 | 0.00054 | -0.011 | 0.95751 | 0.27 |
| 49059 | AC138035.2 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326 | 12 | CCDC6 | Sponge network | 1.073 | 0 | 0.27 | 0.02555 | 0.27 |
| 49060 | RP11-774O3.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | EGFR | Sponge network | -0.487 | 0.02157 | -0.175 | 0.48451 | 0.27 |
| 49061 | WDFY3-AS2 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CTGF | Sponge network | -0.942 | 0 | 0.321 | 0.11682 | 0.27 |
| 49062 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-24-3p;hsa-miR-331-5p;hsa-miR-577 | 10 | KAT6B | Sponge network | -0.473 | 0.07602 | -0.478 | 0.00018 | 0.27 |
| 49063 | RP11-13K12.5 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | SEMA3C | Sponge network | -0.318 | 0.65847 | -0.272 | 0.31153 | 0.27 |
| 49064 | CTD-2017D11.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-940 | 11 | SIRT1 | Sponge network | 0.792 | 0.0049 | 0.109 | 0.2593 | 0.27 |
| 49065 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-455-3p | 11 | ANK3 | Sponge network | 0.331 | 0.57247 | -0.55 | 0.00086 | 0.27 |
| 49066 | JAZF1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p | 12 | MNT | Sponge network | -0.769 | 0.00035 | -0.157 | 0.06598 | 0.27 |
| 49067 | LOXL1-AS1 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 11 | PRDM5 | Sponge network | 0.487 | 0.04053 | -1.09 | 0.00014 | 0.27 |
| 49068 | AC084219.4 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30e-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 12 | KIF5B | Sponge network | 1.296 | 0.0054 | 0.362 | 0.00066 | 0.27 |
| 49069 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-500a-3p;hsa-miR-7-1-3p | 23 | ZFHX4 | Sponge network | -4.477 | 0 | 0.018 | 0.95574 | 0.27 |
| 49070 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-5p | 20 | FOXO3 | Sponge network | 0.889 | 9.0E-5 | -0.04 | 0.72028 | 0.27 |
| 49071 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-625-5p | 24 | YAP1 | Sponge network | -0.154 | 0.78939 | 0.22 | 0.11836 | 0.27 |
| 49072 | LINC00641 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | SYT4 | Sponge network | -0.222 | 0.32445 | -3.207 | 0 | 0.27 |
| 49073 | HOXB-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-576-5p | 15 | TSC1 | Sponge network | 1.316 | 0.01106 | 0.103 | 0.26071 | 0.27 |
| 49074 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CLIP1 | Sponge network | 1.302 | 7.0E-5 | -0.215 | 0.08684 | 0.27 |
| 49075 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | HERC2 | Sponge network | 0.532 | 0.00505 | 0.114 | 0.30638 | 0.27 |
| 49076 | RP11-434B12.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 10 | CELF4 | Sponge network | -0.031 | 0.89063 | -1.135 | 0.00344 | 0.27 |
| 49077 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 37 | ERBB4 | Sponge network | 0.385 | 0.10067 | -1.975 | 2.0E-5 | 0.27 |
| 49078 | HCG11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | EDA2R | Sponge network | 0.385 | 0.10067 | -0.587 | 0.01598 | 0.27 |
| 49079 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | RBMS3 | Sponge network | 0.862 | 0.06213 | -0.519 | 0.03551 | 0.27 |
| 49080 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-625-5p | 20 | CBX5 | Sponge network | 1.294 | 0 | 0.165 | 0.24446 | 0.27 |
| 49081 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-935 | 12 | DAB2 | Sponge network | -2.62 | 0 | 0.431 | 0.0113 | 0.27 |
| 49082 | RP11-400K9.3 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-664a-3p | 11 | KIF1B | Sponge network | -0.733 | 0.19469 | -0.118 | 0.32258 | 0.27 |
| 49083 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | TBX18 | Sponge network | -2.117 | 0.00161 | 0.843 | 0.02945 | 0.27 |
| 49084 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 41 | SMAD2 | Sponge network | 0.083 | 0.66405 | -0.128 | 0.12732 | 0.27 |
| 49085 | CTA-941F9.9 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27b-5p;hsa-miR-505-5p | 10 | KCNA6 | Sponge network | -0.344 | 0.49624 | -1.531 | 0.00013 | 0.27 |
| 49086 | FGD5-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | HECA | Sponge network | 0.401 | 0.00066 | -0.217 | 0.03463 | 0.27 |
| 49087 | A2M-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 33 | TRPS1 | Sponge network | -0.08 | 0.90092 | -0.191 | 0.44109 | 0.27 |
| 49088 | RP11-967K21.1 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-92a-3p | 19 | CADM1 | Sponge network | 0.056 | 0.81195 | -0.9 | 0.00084 | 0.27 |
| 49089 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | ERBB4 | Sponge network | 0.238 | 0.70544 | -1.975 | 2.0E-5 | 0.27 |
| 49090 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 22 | HECA | Sponge network | 0.174 | 0.30034 | -0.217 | 0.03463 | 0.27 |
| 49091 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-577 | 13 | GREM1 | Sponge network | 0.392 | 0.0278 | 0.902 | 0.01691 | 0.27 |
| 49092 | BCYRN1 |
hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 12 | FBXW7 | Sponge network | 0.311 | 0.10344 | -0.418 | 2.0E-5 | 0.27 |
| 49093 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-940 | 17 | LIFR | Sponge network | -4.477 | 0 | -1.794 | 0 | 0.27 |
| 49094 | RP11-244H3.1 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-590-3p | 10 | PREX2 | Sponge network | 0.659 | 3.0E-5 | -0.272 | 0.25382 | 0.27 |
| 49095 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 15 | MAP3K12 | Sponge network | -2.46 | 0 | -0.057 | 0.59369 | 0.27 |
| 49096 | CTA-204B4.2 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-590-3p | 32 | DTNA | Sponge network | 0.765 | 7.0E-5 | -1.648 | 1.0E-5 | 0.27 |
| 49097 | RP11-428J1.5 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-299-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | DTNA | Sponge network | 0.538 | 0.00076 | -1.648 | 1.0E-5 | 0.27 |
| 49098 | BCYRN1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 13 | ABCB5 | Sponge network | 0.311 | 0.10344 | -0.956 | 0.04077 | 0.27 |
| 49099 | CTD-2089N3.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-27b-5p | 10 | MCC | Sponge network | -0.314 | 0.62033 | -1.005 | 0 | 0.27 |
| 49100 | RP13-516M14.1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p | 15 | GJA1 | Sponge network | 0.389 | 0.04293 | -0.485 | 0.02179 | 0.27 |
| 49101 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-7-1-3p | 11 | CNTN5 | Sponge network | -0.49 | 0.04946 | -0.219 | 0.61957 | 0.27 |
| 49102 | CTC-523E23.11 |
hsa-miR-106a-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-224-5p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-429 | 10 | MCC | Sponge network | -0.235 | 0.35308 | -1.005 | 0 | 0.27 |
| 49103 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | SASH1 | Sponge network | -2.915 | 0.00015 | -0.502 | 0.00175 | 0.27 |
| 49104 | ARHGAP5-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-542-3p | 14 | DYNC1I1 | Sponge network | -0.644 | 0.00021 | -1.12 | 0.00107 | 0.27 |
| 49105 | AC074289.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-96-5p | 19 | IL17RD | Sponge network | -0.326 | 0.14314 | -0.019 | 0.94873 | 0.27 |
| 49106 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 36 | NOTCH2 | Sponge network | -0.323 | 0.01372 | 0.138 | 0.35163 | 0.27 |
| 49107 | FAM215B |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | BRD3 | Sponge network | 0.817 | 3.0E-5 | 0.37 | 0.00013 | 0.27 |
| 49108 | LINC00667 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | EPB41L3 | Sponge network | -0.235 | 0.15142 | -1.193 | 0 | 0.27 |
| 49109 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-7-1-3p | 21 | USP12 | Sponge network | -0.842 | 0.01361 | -0.102 | 0.40768 | 0.27 |
| 49110 | CTD-2314G24.2 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | FAM117A | Sponge network | 0.246 | 0.59912 | -1.379 | 0 | 0.27 |
| 49111 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-374b-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-708-5p | 32 | SPRY3 | Sponge network | -0.096 | 0.67958 | 0.016 | 0.91286 | 0.27 |
| 49112 | RP11-798M19.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 26 | PRDM2 | Sponge network | -0.612 | 0.00094 | -0.258 | 0.00721 | 0.27 |
| 49113 | CTD-2002H8.2 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CBX5 | Sponge network | 0.931 | 1.0E-5 | 0.165 | 0.24446 | 0.27 |
| 49114 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-3p | 15 | HDAC9 | Sponge network | -0.322 | 0.25945 | -0.755 | 0.00495 | 0.27 |
| 49115 | KCNJ2-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-3913-5p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-3p | 20 | GRIK3 | Sponge network | -0.436 | 0.05325 | -3.208 | 0 | 0.27 |
| 49116 | MAPKAPK5-AS1 |
hsa-miR-130a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 10 | CDC73 | Sponge network | 0.765 | 0 | 0.094 | 0.20463 | 0.27 |
| 49117 | RP4-791M13.3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 11 | FAT4 | Sponge network | 0.419 | 0.0319 | -0.354 | 0.14694 | 0.27 |
| 49118 | MIR17HG |
hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 11 | PAFAH1B2 | Sponge network | 1.699 | 0 | 0.318 | 0.0001 | 0.27 |
| 49119 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-577;hsa-miR-7-1-3p | 21 | CNTN1 | Sponge network | 0.389 | 0.04293 | -2.594 | 0 | 0.27 |
| 49120 | RP11-5C23.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | PTGFR | Sponge network | 0.274 | 0.07681 | -0.233 | 0.45421 | 0.27 |
| 49121 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 14 | PTGFR | Sponge network | 0.559 | 0.00026 | -0.233 | 0.45421 | 0.27 |
| 49122 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | DDX6 | Sponge network | 0.249 | 0.35902 | 0.091 | 0.36428 | 0.27 |
| 49123 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 20 | TIMP3 | Sponge network | -0.125 | 0.49414 | -0.546 | 0.02307 | 0.27 |
| 49124 | RP4-639F20.1 |
hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p | 10 | UBE4B | Sponge network | -0.517 | 0.02935 | -0.235 | 0.007 | 0.27 |
| 49125 | HCG11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | CDH19 | Sponge network | 0.385 | 0.10067 | -3.434 | 0 | 0.27 |
| 49126 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | YAP1 | Sponge network | 0.782 | 0.00016 | 0.22 | 0.11836 | 0.27 |
| 49127 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-337-3p;hsa-miR-550a-5p | 11 | HOOK3 | Sponge network | -0.176 | 0.59692 | 0.273 | 0.09957 | 0.27 |
| 49128 | AC156455.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p | 10 | DLC1 | Sponge network | 1.154 | 0.00041 | -0.382 | 0.05038 | 0.27 |
| 49129 | KB-1460A1.5 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | SH3GLB1 | Sponge network | 0.603 | 0.00127 | -0.115 | 0.14773 | 0.269 |
| 49130 | RP11-434B12.1 |
hsa-let-7d-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 13 | TLE4 | Sponge network | -0.031 | 0.89063 | -1.157 | 0 | 0.269 |
| 49131 | RP11-968O1.5 |
hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-338-3p | 10 | EYA4 | Sponge network | -0.829 | 0.01343 | 0.3 | 0.36876 | 0.269 |
| 49132 | RP11-835E18.5 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-590-3p | 11 | DMD | Sponge network | -0.462 | 0.01226 | -1.506 | 0 | 0.269 |
| 49133 | LINC00473 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-590-3p | 15 | FMNL3 | Sponge network | -1.203 | 0.03881 | 0.555 | 4.0E-5 | 0.269 |
| 49134 | FAM13A-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-942-5p | 27 | OPCML | Sponge network | -0.83 | 0 | -2.498 | 0 | 0.269 |
| 49135 | H1FX-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 14 | ARNT2 | Sponge network | -0.206 | 0.12942 | -0.332 | 0.25851 | 0.269 |
| 49136 | ADIRF-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 21 | RASSF8 | Sponge network | -0.223 | 0.47816 | -0.122 | 0.65591 | 0.269 |
| 49137 | U91328.19 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-877-5p | 32 | EGFR | Sponge network | -0.303 | 0.21002 | -0.175 | 0.48451 | 0.269 |
| 49138 | HOXB-AS1 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-539-5p | 13 | PDE4DIP | Sponge network | -0.154 | 0.53954 | -0.282 | 0.0257 | 0.269 |
| 49139 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | NOTCH2NL | Sponge network | -0.942 | 0 | 0.024 | 0.84307 | 0.269 |
| 49140 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-629-3p | 21 | ZBTB4 | Sponge network | -0.602 | 0.00331 | -0.739 | 0 | 0.269 |
| 49141 | AGAP11 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-340-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p | 46 | NFIB | Sponge network | -1.645 | 0 | 0.023 | 0.90795 | 0.269 |
| 49142 | GABPB1-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p | 28 | ZFX | Sponge network | 1.11 | 0 | 0.09 | 0.42563 | 0.269 |
| 49143 | RP11-678G14.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 11 | ITPKB | Sponge network | -4.477 | 0 | -0.704 | 0.00106 | 0.269 |
| 49144 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940;hsa-miR-96-5p | 37 | MEF2C | Sponge network | 0.537 | 0.00139 | -0.499 | 0.01448 | 0.269 |
| 49145 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p | 25 | NR2C2 | Sponge network | -0.129 | 0.57177 | 0.21 | 0.03224 | 0.269 |
| 49146 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | RSPO2 | Sponge network | -0.663 | 0.10887 | -3.038 | 0 | 0.269 |
| 49147 | MRPL23-AS1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-93-3p | 15 | ERC1 | Sponge network | -0.278 | 0.76559 | -0.108 | 0.38794 | 0.269 |
| 49148 | CTB-113P19.4 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | WWTR1 | Sponge network | 0.906 | 0.31715 | -0.345 | 0.11997 | 0.269 |
| 49149 | RP11-166D19.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935 | 27 | ACSL6 | Sponge network | -0.576 | 0.10121 | -1.593 | 0 | 0.269 |
| 49150 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FAM188A | Sponge network | -0.739 | 0.0574 | -0.44 | 0.00018 | 0.269 |
| 49151 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-744-3p | 12 | HIP1 | Sponge network | 0.556 | 0.00298 | 0.774 | 0 | 0.269 |
| 49152 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b | 10 | ITCH | Sponge network | 0.817 | 3.0E-5 | 0.084 | 0.34058 | 0.269 |
| 49153 | LINC00963 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-425-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 27 | ATRX | Sponge network | 0.523 | 9.0E-5 | 0.261 | 0.04408 | 0.269 |
| 49154 | RP11-59H7.3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-500a-5p | 10 | UBE4B | Sponge network | 2.163 | 0 | -0.235 | 0.007 | 0.269 |
| 49155 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-5p | 19 | GPC6 | Sponge network | -0.18 | 0.20343 | -0.054 | 0.81465 | 0.269 |
| 49156 | RP11-760H22.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p | 33 | RUNX1T1 | Sponge network | 0.001 | 0.99753 | -0.344 | 0.2374 | 0.269 |
| 49157 | LINC00174 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p | 10 | CLASP2 | Sponge network | 1.253 | 0 | -0.038 | 0.71 | 0.269 |
| 49158 | AC108488.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-7-1-3p | 18 | EBF1 | Sponge network | 0.259 | 0.12542 | -0.384 | 0.08856 | 0.269 |
| 49159 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 18 | PTPRD | Sponge network | -2.076 | 0.00548 | -1.005 | 0.00064 | 0.269 |
| 49160 | RAD51-AS1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-155-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-3607-3p | 10 | PLAG1 | Sponge network | 0.768 | 7.0E-5 | -0.315 | 0.26532 | 0.269 |
| 49161 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-486-5p | 28 | EPHA7 | Sponge network | -0.147 | 0.40452 | -2.197 | 3.0E-5 | 0.269 |
| 49162 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 21 | BCL6 | Sponge network | 0.494 | 0.00339 | -0.211 | 0.19489 | 0.269 |
| 49163 | RP11-819C21.1 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 23 | SYNPO2 | Sponge network | 0.537 | 0.00139 | -2.342 | 0 | 0.269 |
| 49164 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | SLITRK3 | Sponge network | 0.082 | 0.55654 | -3.328 | 0 | 0.269 |
| 49165 | CTC-297N7.9 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181d-5p;hsa-miR-590-5p;hsa-miR-744-3p | 10 | KALRN | Sponge network | -2.069 | 0 | -0.819 | 1.0E-5 | 0.269 |
| 49166 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-5p;hsa-miR-96-5p | 25 | MOB1B | Sponge network | 0.078 | 0.55803 | 0.197 | 0.15266 | 0.269 |
| 49167 | CTB-31O20.4 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | DDX3X | Sponge network | 1.619 | 0 | -0.152 | 0.07686 | 0.269 |
| 49168 | U91328.19 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 11 | UBE2QL1 | Sponge network | -0.303 | 0.21002 | -2.417 | 0 | 0.269 |
| 49169 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-93-5p | 19 | ZBTB4 | Sponge network | 0.381 | 0.25967 | -0.739 | 0 | 0.269 |
| 49170 | LOXL1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | PPP1R9A | Sponge network | 0.487 | 0.04053 | -0.903 | 0.04254 | 0.269 |
| 49171 | CTB-51J22.1 |
hsa-miR-10b-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p | 11 | TACC1 | Sponge network | -0.803 | 0.23737 | -0.362 | 0.05406 | 0.269 |
| 49172 | TPTEP1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 50 | PPM1L | Sponge network | -1.216 | 0 | -0.537 | 0.00616 | 0.269 |
| 49173 | RP11-403I13.7 |
hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-935 | 10 | PHOX2B | Sponge network | 0.395 | 0.15451 | -2.68 | 0 | 0.269 |
| 49174 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-502-5p | 23 | AR | Sponge network | 0.912 | 0.00014 | -1.554 | 0 | 0.269 |
| 49175 | HAR1A |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 11 | EBF1 | Sponge network | -0.672 | 0.19447 | -0.384 | 0.08856 | 0.269 |
| 49176 | RP11-867G23.10 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-625-5p | 12 | NUAK1 | Sponge network | -2.531 | 0 | 0.904 | 0 | 0.269 |
| 49177 | LINC00621 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-93-5p;hsa-miR-96-5p | 39 | LPP | Sponge network | 0.131 | 0.8436 | -0.459 | 0.04475 | 0.269 |
| 49178 | ADIRF-AS1 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | IGSF10 | Sponge network | -0.223 | 0.47816 | -1.751 | 1.0E-5 | 0.269 |
| 49179 | LINC00476 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | AFAP1L2 | Sponge network | -0.615 | 0.00015 | -0.284 | 0.15434 | 0.269 |
| 49180 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-96-5p | 10 | ZFP36L1 | Sponge network | -0.418 | 0.00068 | -0.505 | 8.0E-5 | 0.269 |
| 49181 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p | 21 | TCF4 | Sponge network | 1.46 | 1.0E-5 | 0.016 | 0.92271 | 0.269 |
| 49182 | RP3-402G11.26 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-361-3p;hsa-miR-500a-3p;hsa-miR-590-3p | 17 | RUNX1T1 | Sponge network | -0.254 | 0.19303 | -0.344 | 0.2374 | 0.269 |
| 49183 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-940 | 17 | IGF1 | Sponge network | 0.419 | 0.0319 | -0.204 | 0.5898 | 0.269 |
| 49184 | LINC00680 |
hsa-let-7a-3p;hsa-miR-155-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-497-5p | 11 | CBL | Sponge network | 0.884 | 0 | 0.291 | 0.01267 | 0.269 |
| 49185 | NDUFA6-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | -0.355 | 0.0077 | -1.379 | 0 | 0.269 |
| 49186 | LINC00654 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 13 | PLAG1 | Sponge network | 0.374 | 0.20054 | -0.315 | 0.26532 | 0.269 |
| 49187 | TMEM9B-AS1 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | CCDC6 | Sponge network | 0.529 | 0.00552 | 0.27 | 0.02555 | 0.269 |
| 49188 | RBPMS-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-454-3p;hsa-miR-7-1-3p | 19 | DTNA | Sponge network | 0.144 | 0.64989 | -1.648 | 1.0E-5 | 0.269 |
| 49189 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | FOXO1 | Sponge network | 0.889 | 9.0E-5 | -0.141 | 0.33874 | 0.269 |
| 49190 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-424-5p | 13 | PPM1L | Sponge network | 1.344 | 0 | -0.537 | 0.00616 | 0.269 |
| 49191 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p | 15 | BCL6 | Sponge network | 0.178 | 0.29106 | -0.211 | 0.19489 | 0.269 |
| 49192 | RP11-277P12.20 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p | 11 | PLXNC1 | Sponge network | 0.687 | 0.02753 | 0.569 | 0.00329 | 0.269 |
| 49193 | RP11-195F19.9 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | SIK1 | Sponge network | -0.726 | 0.00132 | -0.98 | 0 | 0.269 |
| 49194 | AP001347.6 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 23 | OPCML | Sponge network | -1.161 | 0.00591 | -2.498 | 0 | 0.269 |
| 49195 | RP3-402G11.26 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-330-5p;hsa-miR-505-5p;hsa-miR-590-3p | 10 | ASTN1 | Sponge network | -0.254 | 0.19303 | -2.389 | 2.0E-5 | 0.269 |
| 49196 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-424-5p;hsa-miR-708-5p | 17 | MTSS1 | Sponge network | 0.368 | 0.20093 | -0.81 | 0.00035 | 0.269 |
| 49197 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-30d-3p;hsa-miR-577 | 11 | PHC3 | Sponge network | -0.473 | 0.07602 | 0.212 | 0.0499 | 0.269 |
| 49198 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110 | 10 | USP12 | Sponge network | -1.255 | 0.00981 | -0.102 | 0.40768 | 0.269 |
| 49199 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | PDS5B | Sponge network | 0.082 | 0.55654 | 0.259 | 0.00962 | 0.269 |
| 49200 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 10 | CADM1 | Sponge network | -1.375 | 0.08642 | -0.9 | 0.00084 | 0.269 |
| 49201 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CLIP1 | Sponge network | 1.601 | 0 | -0.215 | 0.08684 | 0.269 |
| 49202 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p | 17 | ANK2 | Sponge network | 3.345 | 0 | -1.603 | 1.0E-5 | 0.269 |
| 49203 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | 0.259 | 0.12542 | -0.243 | 0.11408 | 0.269 |
| 49204 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p | 11 | CNTNAP3 | Sponge network | 0.41 | 0.02214 | -1.465 | 0.00033 | 0.269 |
| 49205 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | BNC2 | Sponge network | 0.614 | 1.0E-5 | -0.491 | 0.09988 | 0.269 |
| 49206 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-98-5p | 39 | NTRK3 | Sponge network | 0.05 | 0.95483 | -1.77 | 3.0E-5 | 0.269 |
| 49207 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 22 | THBS1 | Sponge network | -0.053 | 0.80685 | 0.741 | 0.00386 | 0.269 |
| 49208 | RP4-639F20.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-542-3p | 12 | JDP2 | Sponge network | -0.517 | 0.02935 | -0.967 | 0 | 0.269 |
| 49209 | C20orf166-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-93-5p | 22 | RNASEL | Sponge network | -2.69 | 0.0001 | -0.272 | 0.01796 | 0.269 |
| 49210 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-539-5p;hsa-miR-942-5p | 25 | DDX6 | Sponge network | -0.465 | 0.04703 | 0.091 | 0.36428 | 0.269 |
| 49211 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-450b-5p;hsa-miR-7-1-3p | 17 | AKAP11 | Sponge network | -1.092 | 4.0E-5 | 0.196 | 0.09825 | 0.269 |
| 49212 | DNAJC27-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-5p | 29 | BTG2 | Sponge network | -0.969 | 2.0E-5 | -1.763 | 0 | 0.269 |
| 49213 | RP11-798G7.7 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-375;hsa-miR-582-3p | 16 | QKI | Sponge network | 0.858 | 3.0E-5 | -0.333 | 0.04111 | 0.269 |
| 49214 | LINC00641 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 19 | FMNL3 | Sponge network | -0.222 | 0.32445 | 0.555 | 4.0E-5 | 0.269 |
| 49215 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-421;hsa-miR-505-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 22 | TCF4 | Sponge network | -4.477 | 0 | 0.016 | 0.92271 | 0.269 |
| 49216 | LINC00957 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | HOOK3 | Sponge network | -0.421 | 0.0114 | 0.273 | 0.09957 | 0.269 |
| 49217 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-5p | 14 | ABCB5 | Sponge network | -0.465 | 0.04703 | -0.956 | 0.04077 | 0.269 |
| 49218 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-940 | 14 | PBX1 | Sponge network | 0.272 | 0.44692 | -1.206 | 0 | 0.269 |
| 49219 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | BCL2 | Sponge network | -0.08 | 0.90092 | -0.899 | 0 | 0.269 |
| 49220 | FENDRR |
hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | AHCYL1 | Sponge network | -1.281 | 0.00012 | -0.479 | 0 | 0.269 |
| 49221 | MIR497HG |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | GNAQ | Sponge network | -1.439 | 4.0E-5 | -0.367 | 0.00102 | 0.269 |
| 49222 | AC005154.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ABCA10 | Sponge network | 0.848 | 0 | -0.571 | 0.03674 | 0.269 |
| 49223 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-497-5p | 12 | MYBL1 | Sponge network | 1.262 | 0 | 0.494 | 0.02152 | 0.269 |
| 49224 | CTD-2303H24.2 |
hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | ZNF382 | Sponge network | 0.415 | 0.14657 | -0.494 | 0.03831 | 0.269 |
| 49225 | TINCR |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-96-5p | 22 | MED12L | Sponge network | -0.664 | 0.14543 | -0.194 | 0.55299 | 0.269 |
| 49226 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-424-5p | 18 | CBX5 | Sponge network | 1.147 | 0 | 0.165 | 0.24446 | 0.269 |
| 49227 | RP1-59D14.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | MYBL1 | Sponge network | 1.162 | 5.0E-5 | 0.494 | 0.02152 | 0.269 |
| 49228 | RP11-464F9.20 |
hsa-let-7a-3p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-33a-3p | 12 | MOB1B | Sponge network | 0.643 | 0.00136 | 0.197 | 0.15266 | 0.269 |
| 49229 | RP11-276H19.2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-502-5p | 10 | TET1 | Sponge network | -0.472 | 0.48851 | -0.135 | 0.52138 | 0.269 |
| 49230 | LINC00473 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | RIMS2 | Sponge network | -1.203 | 0.03881 | -1.365 | 0.00208 | 0.269 |
| 49231 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 27 | ST6GAL2 | Sponge network | 0.374 | 0.20054 | -1.201 | 0.00597 | 0.269 |
| 49232 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 11 | FREM2 | Sponge network | 0.422 | 0.27021 | -0.437 | 0.46353 | 0.269 |
| 49233 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-935 | 10 | SEPT6 | Sponge network | -0.738 | 0.21233 | -0.598 | 0.00181 | 0.269 |
| 49234 | RBPMS-AS1 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-935 | 10 | FBXO32 | Sponge network | 0.144 | 0.64989 | -0.495 | 0.07561 | 0.269 |
| 49235 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 37 | PTPRD | Sponge network | 0.056 | 0.81195 | -1.005 | 0.00064 | 0.269 |
| 49236 | RP11-98I9.4 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 25 | ARHGEF12 | Sponge network | 0.67 | 0.00048 | 0.09 | 0.35693 | 0.269 |
| 49237 | BCYRN1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | WWTR1 | Sponge network | 0.311 | 0.10344 | -0.345 | 0.11997 | 0.269 |
| 49238 | RP11-458F8.4 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-338-5p | 12 | BMPR2 | Sponge network | 1.518 | 0 | 0.206 | 0.0635 | 0.269 |
| 49239 | RP11-66B24.2 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-93-3p | 10 | ST6GAL2 | Sponge network | 0.57 | 0.1866 | -1.201 | 0.00597 | 0.269 |
| 49240 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-576-5p;hsa-miR-93-3p | 18 | FOXO3 | Sponge network | 2.163 | 0 | -0.04 | 0.72028 | 0.269 |
| 49241 | RP11-20I23.13 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-505-5p;hsa-miR-511-5p | 11 | MRVI1 | Sponge network | 0.505 | 0.0014 | -1.051 | 0.00022 | 0.269 |
| 49242 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-3607-3p | 10 | APC | Sponge network | 0.706 | 0 | -0.255 | 0.04051 | 0.269 |
| 49243 | HOXB-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429 | 10 | RIMBP2 | Sponge network | -0.154 | 0.53954 | -1.968 | 5.0E-5 | 0.269 |
| 49244 | AGAP11 |
hsa-miR-106b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-93-3p | 10 | BNIP3L | Sponge network | -1.645 | 0 | -0.167 | 0.14549 | 0.269 |
| 49245 | RP11-327P2.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-625-5p;hsa-miR-629-5p | 22 | CRTC1 | Sponge network | -0.147 | 0.40452 | -0.116 | 0.28412 | 0.269 |
| 49246 | RP11-888D10.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 16 | AFF4 | Sponge network | 0.702 | 7.0E-5 | -0.266 | 0.01565 | 0.269 |
| 49247 | AC108488.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-629-3p | 18 | GPC6 | Sponge network | 0.259 | 0.12542 | -0.054 | 0.81465 | 0.269 |
| 49248 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | PTPRD | Sponge network | -0.187 | 0.23787 | -1.005 | 0.00064 | 0.269 |
| 49249 | RP11-760H22.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-629-5p;hsa-miR-7-5p | 14 | BCL2 | Sponge network | 0.001 | 0.99753 | -0.899 | 0 | 0.269 |
| 49250 | AF131215.9 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ZNF208 | Sponge network | -0.322 | 0.25945 | -0.4 | 0.22381 | 0.269 |
| 49251 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-590-3p | 15 | JAKMIP2 | Sponge network | 1.302 | 7.0E-5 | -1.388 | 0 | 0.269 |
| 49252 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CXXC4 | Sponge network | 0.083 | 0.66405 | -0.433 | 0.21055 | 0.269 |
| 49253 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | EGLN1 | Sponge network | 1.089 | 0 | -0.127 | 0.1536 | 0.269 |
| 49254 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | MYO9A | Sponge network | 0.34 | 0.00273 | -0.147 | 0.32373 | 0.269 |
| 49255 | SH3RF3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-582-5p | 14 | RYR2 | Sponge network | 0.889 | 9.0E-5 | -1.466 | 0.00031 | 0.269 |
| 49256 | UBA6-AS1 |
hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 11 | BCL9 | Sponge network | 0.312 | 0.00725 | 0.321 | 0.00267 | 0.269 |
| 49257 | RASSF8-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-3p | 18 | TLE4 | Sponge network | 0.335 | 0.20596 | -1.157 | 0 | 0.269 |
| 49258 | RP11-428J1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p | 27 | DMD | Sponge network | 0.538 | 0.00076 | -1.506 | 0 | 0.269 |
| 49259 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p | 18 | ANK2 | Sponge network | 0.921 | 0.00118 | -1.603 | 1.0E-5 | 0.269 |
| 49260 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 51 | CBX5 | Sponge network | -0.129 | 0.57177 | 0.165 | 0.24446 | 0.269 |
| 49261 | RP5-1198O20.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-550a-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 31 | TOM1L2 | Sponge network | 0.997 | 0.00066 | -0.994 | 0 | 0.269 |
| 49262 | DIO3OS |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-589-3p | 10 | ABCA10 | Sponge network | -0.773 | 0.04073 | -0.571 | 0.03674 | 0.269 |
| 49263 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM13 | Sponge network | -0.031 | 0.89063 | 0.183 | 0.06968 | 0.269 |
| 49264 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | KAT6B | Sponge network | -0.811 | 2.0E-5 | -0.478 | 0.00018 | 0.269 |
| 49265 | RP11-379F4.7 |
hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-424-5p | 10 | ZFHX3 | Sponge network | 1.316 | 0 | 0.389 | 0.01363 | 0.269 |
| 49266 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-98-5p | 11 | ST5 | Sponge network | 0.082 | 0.55397 | -0.78 | 0 | 0.269 |
| 49267 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-96-5p | 20 | HIP1 | Sponge network | 0.331 | 0.57247 | 0.774 | 0 | 0.269 |
| 49268 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-98-5p | 17 | PNRC1 | Sponge network | -0.899 | 6.0E-5 | -0.646 | 0 | 0.269 |
| 49269 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-589-3p | 29 | CBX5 | Sponge network | 0.529 | 0.00552 | 0.165 | 0.24446 | 0.269 |
| 49270 | RP11-6N17.3 |
hsa-miR-103a-2-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-455-5p | 16 | BMPR2 | Sponge network | 0.108 | 0.69556 | 0.206 | 0.0635 | 0.269 |
| 49271 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-7-1-3p | 10 | NOTCH2NL | Sponge network | -1.092 | 4.0E-5 | 0.024 | 0.84307 | 0.269 |
| 49272 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 22 | IGF1 | Sponge network | -0.322 | 0.25945 | -0.204 | 0.5898 | 0.269 |
| 49273 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p | 10 | MNT | Sponge network | -0.777 | 0.1134 | -0.157 | 0.06598 | 0.269 |
| 49274 | RP11-67L2.2 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | RBBP6 | Sponge network | 0.72 | 0.0001 | 0.163 | 0.05101 | 0.269 |
| 49275 | RP11-356J5.12 |
hsa-miR-126-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TRIM58 | Sponge network | -0.899 | 6.0E-5 | -1.692 | 0 | 0.269 |
| 49276 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-940 | 15 | NEDD4 | Sponge network | -2.095 | 0.00112 | 0.02 | 0.89834 | 0.269 |
| 49277 | AP001372.2 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-365a-3p;hsa-miR-495-3p | 13 | ARHGEF12 | Sponge network | 0.861 | 0 | 0.09 | 0.35693 | 0.269 |
| 49278 | RP11-401P9.4 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-93-5p | 41 | PPM1L | Sponge network | -0.384 | 0.40652 | -0.537 | 0.00616 | 0.269 |
| 49279 | LINC00403 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-339-5p | 11 | PDE4DIP | Sponge network | -3.586 | 0.00032 | -0.282 | 0.0257 | 0.269 |
| 49280 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-590-3p;hsa-miR-942-5p | 23 | ARHGAP28 | Sponge network | -2.915 | 0.00015 | -0.911 | 0.00013 | 0.269 |
| 49281 | RP11-720L2.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-93-5p | 17 | IL17RD | Sponge network | -1.255 | 0.00981 | -0.019 | 0.94873 | 0.269 |
| 49282 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 10 | ARID2 | Sponge network | 1.089 | 0 | 0.235 | 0.02697 | 0.269 |
| 49283 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-590-3p | 22 | PAFAH1B1 | Sponge network | -1.886 | 0 | -0.429 | 0 | 0.269 |
| 49284 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p | 36 | EGFR | Sponge network | 0.997 | 0.00066 | -0.175 | 0.48451 | 0.269 |
| 49285 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p | 13 | BCL2 | Sponge network | 0.542 | 0.00054 | -0.899 | 0 | 0.269 |
| 49286 | RP11-411K7.1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SH3GLB1 | Sponge network | 0.155 | 0.81273 | -0.115 | 0.14773 | 0.269 |
| 49287 | CTC-296K1.4 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-500a-5p | 10 | ZFHX3 | Sponge network | -2.076 | 0.00548 | 0.389 | 0.01363 | 0.269 |
| 49288 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | 0.862 | 0.06213 | -1.047 | 0.00364 | 0.269 |
| 49289 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-33a-3p | 12 | PIK3C2A | Sponge network | 0.368 | 0.20093 | 0.061 | 0.53776 | 0.269 |
| 49290 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429 | 11 | CLASP2 | Sponge network | 2.364 | 0.00134 | -0.038 | 0.71 | 0.269 |
| 49291 | FENDRR |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 28 | MXI1 | Sponge network | -1.281 | 0.00012 | -0.987 | 0 | 0.269 |
| 49292 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-532-3p | 15 | CBL | Sponge network | 0.545 | 0.05789 | 0.291 | 0.01267 | 0.269 |
| 49293 | RP11-38L15.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p | 12 | IL17RD | Sponge network | 0.344 | 0.3127 | -0.019 | 0.94873 | 0.269 |
| 49294 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-3065-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 26 | SOX11 | Sponge network | -0.576 | 0.10121 | 2.68 | 0 | 0.269 |
| 49295 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-590-3p | 24 | PIK3C2A | Sponge network | 0.101 | 0.60919 | 0.061 | 0.53776 | 0.269 |
| 49296 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CXXC4 | Sponge network | 0.194 | 0.64278 | -0.433 | 0.21055 | 0.269 |
| 49297 | AC156455.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p | 14 | RASSF8 | Sponge network | 1.154 | 0.00041 | -0.122 | 0.65591 | 0.269 |
| 49298 | RP11-67L2.2 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | RPS6KA3 | Sponge network | 0.72 | 0.0001 | 0.256 | 0.02419 | 0.269 |
| 49299 | OSER1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | RASSF8 | Sponge network | -0.13 | 0.48734 | -0.122 | 0.65591 | 0.269 |
| 49300 | RP11-822E23.8 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-940;hsa-miR-942-5p | 33 | NOTCH2 | Sponge network | -0.777 | 0.16667 | 0.138 | 0.35163 | 0.269 |
| 49301 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-590-3p | 14 | DACT1 | Sponge network | 0.082 | 0.55654 | 0.783 | 0.00072 | 0.269 |
| 49302 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-942-5p | 31 | TMEM132B | Sponge network | 1.757 | 0 | -0.83 | 0.01084 | 0.269 |
| 49303 | DNM1P35 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-590-3p | 14 | NRK | Sponge network | 0.051 | 0.83388 | 0.656 | 0.19217 | 0.269 |
| 49304 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 10 | SON | Sponge network | 1.266 | 0 | 0.168 | 0.04406 | 0.269 |
| 49305 | DNM1P35 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-744-3p | 16 | IGF2 | Sponge network | 0.051 | 0.83388 | 0.848 | 0.03842 | 0.269 |
| 49306 | RP11-25K21.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p | 14 | BCL2 | Sponge network | 0.489 | 0.25786 | -0.899 | 0 | 0.269 |
| 49307 | ZRANB2-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-935 | 24 | NFIB | Sponge network | -0.376 | 0.00337 | 0.023 | 0.90795 | 0.269 |
| 49308 | RP11-175O19.4 |
hsa-miR-125a-5p;hsa-miR-150-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-411-5p;hsa-miR-497-5p | 14 | RAD23B | Sponge network | 0.917 | 0 | 0.108 | 0.17414 | 0.269 |
| 49309 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p | 12 | RIMBP2 | Sponge network | 0.299 | 0.57722 | -1.968 | 5.0E-5 | 0.269 |
| 49310 | RP11-156P1.3 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | KCTD8 | Sponge network | 0.214 | 0.18246 | -3.359 | 0 | 0.269 |
| 49311 | PWAR6 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-5p | 23 | KCNJ5 | Sponge network | -1.575 | 6.0E-5 | -0.281 | 0.3339 | 0.269 |
| 49312 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p | 14 | SASH1 | Sponge network | 0.37 | 0.27614 | -0.502 | 0.00175 | 0.269 |
| 49313 | EIF3J-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | CXXC4 | Sponge network | 0.331 | 0.00509 | -0.433 | 0.21055 | 0.269 |
| 49314 | RP11-13K12.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | PDE4DIP | Sponge network | -0.318 | 0.65847 | -0.282 | 0.0257 | 0.269 |
| 49315 | LPP-AS2 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-3p | 13 | RASSF3 | Sponge network | -0.18 | 0.20343 | -0.226 | 0.11691 | 0.269 |
| 49316 | SERHL |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-539-5p | 13 | SETBP1 | Sponge network | -0.602 | 0.00331 | -1.302 | 1.0E-5 | 0.269 |
| 49317 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 14 | CHD1 | Sponge network | 0.91 | 0 | 0.322 | 0.00215 | 0.269 |
| 49318 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | SYNE1 | Sponge network | -0.969 | 2.0E-5 | -0.738 | 0.00316 | 0.269 |
| 49319 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p | 12 | PBRM1 | Sponge network | 0.921 | 0.00118 | -0.029 | 0.78601 | 0.269 |
| 49320 | CTD-2314G24.2 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 26 | ACSL6 | Sponge network | 0.246 | 0.59912 | -1.593 | 0 | 0.269 |
| 49321 | RP11-835E18.5 |
hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | SOX5 | Sponge network | -0.462 | 0.01226 | -1.091 | 4.0E-5 | 0.269 |
| 49322 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-576-5p | 15 | ARID5B | Sponge network | 1.044 | 2.0E-5 | 0.342 | 0.01676 | 0.269 |
| 49323 | NNT-AS1 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-625-5p | 16 | IL13RA1 | Sponge network | 0.799 | 1.0E-5 | 0.019 | 0.86949 | 0.269 |
| 49324 | RP11-263K19.4 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p | 10 | BCL9 | Sponge network | 0.859 | 0.00331 | 0.321 | 0.00267 | 0.269 |
| 49325 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 62 | KIF1B | Sponge network | -2.452 | 0 | -0.118 | 0.32258 | 0.269 |
| 49326 | TRAF3IP2-AS1 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | GABRB2 | Sponge network | -0.323 | 0.01372 | -1.841 | 0.00029 | 0.269 |
| 49327 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p | 13 | EYA4 | Sponge network | 0.792 | 0.0049 | 0.3 | 0.36876 | 0.269 |
| 49328 | LINC00621 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | CDH19 | Sponge network | 0.131 | 0.8436 | -3.434 | 0 | 0.269 |
| 49329 | TMEM161B-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-940 | 35 | MDM4 | Sponge network | -0.04 | 0.7606 | 0.345 | 0.00762 | 0.269 |
| 49330 | CTC-429P9.3 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p | 21 | CTDSPL | Sponge network | 0.082 | 0.55397 | 0.085 | 0.45121 | 0.269 |
| 49331 | HOXD-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-3607-3p | 14 | PBX1 | Sponge network | -0.208 | 0.65592 | -1.206 | 0 | 0.269 |
| 49332 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-629-5p;hsa-miR-940 | 22 | MYO9A | Sponge network | -0.773 | 0.04073 | -0.147 | 0.32373 | 0.269 |
| 49333 | LINC00667 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | -0.235 | 0.15142 | 0.321 | 0.00267 | 0.269 |
| 49334 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p | 10 | ARID5B | Sponge network | 0.997 | 0 | 0.342 | 0.01676 | 0.269 |
| 49335 | RP11-5C23.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p | 15 | PLSCR4 | Sponge network | 0.274 | 0.07681 | -0.474 | 0.03833 | 0.269 |
| 49336 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 10 | PSIP1 | Sponge network | 0.274 | 0.07681 | -0.005 | 0.96897 | 0.269 |
| 49337 | MRPL23-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p | 14 | CDH23 | Sponge network | -0.278 | 0.76559 | -0.974 | 0.00034 | 0.269 |
| 49338 | CTC-429P9.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-5p | 10 | PTPRD | Sponge network | 0.043 | 0.81628 | -1.005 | 0.00064 | 0.269 |
| 49339 | RP11-379F4.7 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-664a-3p | 16 | CCDC6 | Sponge network | 1.316 | 0 | 0.27 | 0.02555 | 0.269 |
| 49340 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | PDE4DIP | Sponge network | -0.176 | 0.59692 | -0.282 | 0.0257 | 0.269 |
| 49341 | HOXB-AS2 |
hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429 | 12 | MOB1B | Sponge network | 1.316 | 0.01106 | 0.197 | 0.15266 | 0.269 |
| 49342 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | AKAP11 | Sponge network | 0.476 | 0.19203 | 0.196 | 0.09825 | 0.269 |
| 49343 | RP11-464F9.1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-655-3p | 11 | UBE4B | Sponge network | 0.959 | 0 | -0.235 | 0.007 | 0.269 |
| 49344 | RP1-302G2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-7-5p | 12 | ZBTB4 | Sponge network | -1.483 | 9.0E-5 | -0.739 | 0 | 0.269 |
| 49345 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-511-5p | 16 | BCL6 | Sponge network | -0.777 | 0.16667 | -0.211 | 0.19489 | 0.269 |
| 49346 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-3127-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 14 | CNTNAP1 | Sponge network | -0.969 | 2.0E-5 | 0.358 | 0.13727 | 0.269 |
| 49347 | MRPL23-AS1 |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-532-3p | 19 | WDFY3 | Sponge network | -0.278 | 0.76559 | -0.201 | 0.0967 | 0.269 |
| 49348 | AC016747.3 |
hsa-miR-107;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | AFAP1L2 | Sponge network | 0.199 | 0.38015 | -0.284 | 0.15434 | 0.269 |
| 49349 | RP11-401P9.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-708-5p;hsa-miR-93-3p | 24 | TOM1L2 | Sponge network | -0.384 | 0.40652 | -0.994 | 0 | 0.269 |
| 49350 | LINC00969 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-582-3p | 16 | CBL | Sponge network | 0.683 | 1.0E-5 | 0.291 | 0.01267 | 0.269 |
| 49351 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-96-5p | 15 | CRTC3 | Sponge network | 0.197 | 0.25618 | 0.164 | 0.16786 | 0.269 |
| 49352 | EMX2OS |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | HERC1 | Sponge network | -0.777 | 0.1134 | -0.196 | 0.08544 | 0.269 |
| 49353 | FAM13A-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -0.83 | 0 | -0.704 | 0.00106 | 0.269 |
| 49354 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p | 15 | FREM2 | Sponge network | 0.056 | 0.81195 | -0.437 | 0.46353 | 0.269 |
| 49355 | RP11-37B2.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 26 | ZFX | Sponge network | 1.03 | 0 | 0.09 | 0.42563 | 0.269 |
| 49356 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 18 | GPAM | Sponge network | 0.848 | 0 | 0.142 | 0.29732 | 0.269 |
| 49357 | HAR1A |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-93-3p | 11 | WWTR1 | Sponge network | -0.672 | 0.19447 | -0.345 | 0.11997 | 0.269 |
| 49358 | TPT1-AS1 |
hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 13 | CDK6 | Sponge network | 0.903 | 0 | 0.991 | 0 | 0.269 |
| 49359 | SERTAD4-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-500a-5p | 16 | PPM1L | Sponge network | -0.932 | 0.00329 | -0.537 | 0.00616 | 0.269 |
| 49360 | RP11-2B6.2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ITGB1 | Sponge network | 0.539 | 0.03722 | 0.524 | 0.00017 | 0.269 |
| 49361 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 11 | FOXO3 | Sponge network | 0.545 | 0.05789 | -0.04 | 0.72028 | 0.269 |
| 49362 | RP11-218M22.1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 16 | ERC1 | Sponge network | 0.197 | 0.25618 | -0.108 | 0.38794 | 0.269 |
| 49363 | CTD-3126B10.4 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-590-3p | 13 | ATRX | Sponge network | 0.778 | 0.00101 | 0.261 | 0.04408 | 0.269 |
| 49364 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-589-3p | 20 | ADARB1 | Sponge network | 3.345 | 0 | -0.335 | 0.06631 | 0.269 |
| 49365 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | -2.46 | 0 | 0.783 | 0.00072 | 0.269 |
| 49366 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 13 | PEG3 | Sponge network | -0.176 | 0.59692 | -1.74 | 0 | 0.269 |
| 49367 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 40 | EPHA7 | Sponge network | 0.72 | 0.0001 | -2.197 | 3.0E-5 | 0.269 |
| 49368 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 13 | GPAM | Sponge network | 0.594 | 0.41522 | 0.142 | 0.29732 | 0.269 |
| 49369 | RP11-251G23.5 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | RB1 | Sponge network | 1.344 | 0 | 0.382 | 0.0017 | 0.269 |
| 49370 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 38 | TMEM132B | Sponge network | 0.61 | 0.01593 | -0.83 | 0.01084 | 0.269 |
| 49371 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-505-3p | 11 | ZNF292 | Sponge network | -0.303 | 0.21002 | 0.276 | 0.04148 | 0.269 |
| 49372 | RP11-384K6.6 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326 | 10 | ABL1 | Sponge network | 0.946 | 0 | -0.215 | 0.07804 | 0.269 |
| 49373 | RP11-822E23.8 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-7-5p | 24 | BTG2 | Sponge network | -0.777 | 0.16667 | -1.763 | 0 | 0.269 |
| 49374 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-96-5p | 33 | PIK3R1 | Sponge network | 0.523 | 9.0E-5 | -0.133 | 0.36854 | 0.269 |
| 49375 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-577 | 16 | GREM1 | Sponge network | 0.853 | 0.15352 | 0.902 | 0.01691 | 0.269 |
| 49376 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 17 | EBF3 | Sponge network | 0.051 | 0.83388 | -0.058 | 0.81437 | 0.269 |
| 49377 | CTD-2303H24.2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ITGB1 | Sponge network | 0.415 | 0.14657 | 0.524 | 0.00017 | 0.269 |
| 49378 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | HDAC9 | Sponge network | 0.572 | 0.01722 | -0.755 | 0.00495 | 0.269 |
| 49379 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | PIK3R1 | Sponge network | 0.4 | 0.14611 | -0.133 | 0.36854 | 0.269 |
| 49380 | OIP5-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 21 | BCL9 | Sponge network | 0.421 | 0.00084 | 0.321 | 0.00267 | 0.269 |
| 49381 | HOXB-AS2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p | 17 | FAT3 | Sponge network | 1.316 | 0.01106 | -1.601 | 0.00051 | 0.269 |
| 49382 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-93-5p | 12 | MAP3K12 | Sponge network | -0.162 | 0.47687 | -0.057 | 0.59369 | 0.269 |
| 49383 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ABCC9 | Sponge network | -0.83 | 0 | -0.466 | 0.14121 | 0.269 |
| 49384 | CKMT2-AS1 |
hsa-miR-101-5p;hsa-miR-10b-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p | 28 | UBR3 | Sponge network | 0.082 | 0.55654 | -0.021 | 0.82774 | 0.269 |
| 49385 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-940 | 20 | IGF1 | Sponge network | -0.668 | 3.0E-5 | -0.204 | 0.5898 | 0.269 |
| 49386 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-577;hsa-miR-7-1-3p | 15 | LRP1B | Sponge network | 0.419 | 0.0319 | -2.163 | 0 | 0.269 |
| 49387 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | FAT4 | Sponge network | 0.659 | 3.0E-5 | -0.354 | 0.14694 | 0.269 |
| 49388 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-590-5p;hsa-miR-671-5p | 18 | THBS1 | Sponge network | 0.078 | 0.55803 | 0.741 | 0.00386 | 0.269 |
| 49389 | DHRS4-AS1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-495-3p | 10 | GSK3B | Sponge network | 0.198 | 0.20379 | 0.266 | 0.00045 | 0.269 |
| 49390 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p | 18 | LATS1 | Sponge network | 0.95 | 0.15943 | 0.109 | 0.34208 | 0.269 |
| 49391 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ZFHX3 | Sponge network | -1.203 | 0.03881 | 0.389 | 0.01363 | 0.269 |
| 49392 | FAM13A-AS1 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | HERC1 | Sponge network | -0.83 | 0 | -0.196 | 0.08544 | 0.269 |
| 49393 | LINC00886 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p | 24 | HECA | Sponge network | -0.371 | 0.24604 | -0.217 | 0.03463 | 0.269 |
| 49394 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | FGF5 | Sponge network | 0.219 | 0.1516 | 0.549 | 0.28659 | 0.269 |
| 49395 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-424-5p;hsa-miR-582-5p | 15 | ZFHX3 | Sponge network | 1.262 | 0 | 0.389 | 0.01363 | 0.269 |
| 49396 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-942-5p | 18 | PCDH17 | Sponge network | -0.465 | 0.04703 | 0.731 | 2.0E-5 | 0.268 |
| 49397 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p | 25 | CBX5 | Sponge network | 0.799 | 1.0E-5 | 0.165 | 0.24446 | 0.268 |
| 49398 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 14 | ANK3 | Sponge network | 0.417 | 0.00445 | -0.55 | 0.00086 | 0.268 |
| 49399 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | EPHA5 | Sponge network | -0.737 | 0.10822 | -0.957 | 0.01733 | 0.268 |
| 49400 | AC127904.2 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | MED12L | Sponge network | 1.308 | 0 | -0.194 | 0.55299 | 0.268 |
| 49401 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 28 | PRKAA2 | Sponge network | 0.663 | 0.00073 | -2.462 | 0 | 0.268 |
| 49402 | RP11-66B24.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378c;hsa-miR-671-5p | 17 | RBMS3 | Sponge network | 0.57 | 0.1866 | -0.519 | 0.03551 | 0.268 |
| 49403 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-96-5p | 21 | MOB1B | Sponge network | 0.622 | 0.00015 | 0.197 | 0.15266 | 0.268 |
| 49404 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-589-3p | 18 | SCN9A | Sponge network | 0.663 | 0.00073 | -0.525 | 0.10593 | 0.268 |
| 49405 | CTD-3064M3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-625-5p;hsa-miR-93-3p | 17 | SUFU | Sponge network | -0.469 | 0.08721 | -0.04 | 0.65489 | 0.268 |
| 49406 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-93-5p;hsa-miR-942-5p | 13 | FBXO31 | Sponge network | -0.793 | 7.0E-5 | -0.085 | 0.42389 | 0.268 |
| 49407 | CTC-444N24.11 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NIN | Sponge network | 1.153 | 0 | -0.253 | 0.13794 | 0.268 |
| 49408 | RP4-669L17.10 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-539-5p | 14 | ARHGEF12 | Sponge network | 1.093 | 0 | 0.09 | 0.35693 | 0.268 |
| 49409 | RAMP2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-935 | 70 | LPP | Sponge network | -0.513 | 0.04703 | -0.459 | 0.04475 | 0.268 |
| 49410 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 26 | MOB1B | Sponge network | -0.246 | 0.35034 | 0.197 | 0.15266 | 0.268 |
| 49411 | CTD-2017D11.1 |
hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-940 | 12 | BAZ2B | Sponge network | 0.792 | 0.0049 | -0.276 | 0.02833 | 0.268 |
| 49412 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-96-5p | 32 | LPP | Sponge network | 2.306 | 9.0E-5 | -0.459 | 0.04475 | 0.268 |
| 49413 | CTD-2554C21.2 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p | 13 | LATS1 | Sponge network | -1.654 | 0.00144 | 0.109 | 0.34208 | 0.268 |
| 49414 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 27 | HECA | Sponge network | 0.064 | 0.72934 | -0.217 | 0.03463 | 0.268 |
| 49415 | OSER1-AS1 |
hsa-miR-106a-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-5p | 22 | ANK2 | Sponge network | -0.13 | 0.48734 | -1.603 | 1.0E-5 | 0.268 |
| 49416 | RP11-401P9.4 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-539-5p | 19 | RYR2 | Sponge network | -0.384 | 0.40652 | -1.466 | 0.00031 | 0.268 |
| 49417 | RP11-359B12.2 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | PTCH1 | Sponge network | 0.064 | 0.72934 | -0.1 | 0.6179 | 0.268 |
| 49418 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | FREM2 | Sponge network | -0.053 | 0.80685 | -0.437 | 0.46353 | 0.268 |
| 49419 | MAGI2-AS3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 46 | PPP1R9A | Sponge network | -0.129 | 0.57177 | -0.903 | 0.04254 | 0.268 |
| 49420 | SOX2-OT |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | KCTD8 | Sponge network | -1.264 | 6.0E-5 | -3.359 | 0 | 0.268 |
| 49421 | RP11-21A7A.2 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-589-5p | 13 | ERBB4 | Sponge network | -1.116 | 0.23686 | -1.975 | 2.0E-5 | 0.268 |
| 49422 | RP11-720L2.4 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-33a-3p | 12 | AFF1 | Sponge network | -1.255 | 0.00981 | -0.384 | 0.00053 | 0.268 |
| 49423 | FOXN3-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-93-5p | 10 | NRBP1 | Sponge network | -0.399 | 0.00917 | -0.203 | 0.01627 | 0.268 |
| 49424 | LINC00963 |
hsa-miR-101-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ATM | Sponge network | 0.523 | 9.0E-5 | 0.178 | 0.21568 | 0.268 |
| 49425 | LINC00654 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | 0.374 | 0.20054 | -0.704 | 0.00106 | 0.268 |
| 49426 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 30 | TSHZ3 | Sponge network | 0.222 | 0.29133 | -0.556 | 0.01516 | 0.268 |
| 49427 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | ERG | Sponge network | -0.899 | 6.0E-5 | 0.002 | 0.99011 | 0.268 |
| 49428 | RP11-20I23.11 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 12 | BMPR2 | Sponge network | 0.875 | 0.00019 | 0.206 | 0.0635 | 0.268 |
| 49429 | CKMT2-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-370-3p;hsa-miR-590-3p | 11 | TPO | Sponge network | 0.082 | 0.55654 | -1.87 | 2.0E-5 | 0.268 |
| 49430 | RP11-888D10.4 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 12 | PHF6 | Sponge network | 0.702 | 7.0E-5 | 0.785 | 0 | 0.268 |
| 49431 | AC159540.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | NF1 | Sponge network | 1.634 | 0 | 0.51 | 0 | 0.268 |
| 49432 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | IGSF10 | Sponge network | -0.08 | 0.90092 | -1.751 | 1.0E-5 | 0.268 |
| 49433 | GAS6-AS2 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-429 | 12 | KLF10 | Sponge network | 0.16 | 0.50785 | -0.783 | 0 | 0.268 |
| 49434 | LINC00578 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-589-3p | 20 | PRDM2 | Sponge network | 0.811 | 0.03228 | -0.258 | 0.00721 | 0.268 |
| 49435 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-629-3p | 19 | PSIP1 | Sponge network | 0.249 | 0.35902 | -0.005 | 0.96897 | 0.268 |
| 49436 | FAM13A-AS1 |
hsa-miR-142-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | RNF180 | Sponge network | -0.83 | 0 | -1.127 | 1.0E-5 | 0.268 |
| 49437 | AC034220.3 |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-628-5p | 14 | MIER3 | Sponge network | 0.309 | 0.07304 | -0.212 | 0.04957 | 0.268 |
| 49438 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p | 18 | LATS1 | Sponge network | 0.705 | 0.00014 | 0.109 | 0.34208 | 0.268 |
| 49439 | RP11-503E24.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 14 | BMPR2 | Sponge network | 2.218 | 0 | 0.206 | 0.0635 | 0.268 |
| 49440 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-484 | 18 | TGFBR3 | Sponge network | -0.829 | 0.01343 | -1.173 | 1.0E-5 | 0.268 |
| 49441 | RP11-890B15.3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 35 | PTPRT | Sponge network | 0.101 | 0.60919 | -0.907 | 0.03727 | 0.268 |
| 49442 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-942-5p;hsa-miR-96-5p | 19 | NRXN1 | Sponge network | -0.555 | 0.06156 | -3.778 | 0 | 0.268 |
| 49443 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-766-3p | 39 | AFF4 | Sponge network | 1.063 | 0 | -0.266 | 0.01565 | 0.268 |
| 49444 | RP11-798M19.6 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ZNF536 | Sponge network | -0.612 | 0.00094 | -3.115 | 0 | 0.268 |
| 49445 | FAM157A |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | CDC73 | Sponge network | 0.813 | 1.0E-5 | 0.094 | 0.20463 | 0.268 |
| 49446 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | LRP1B | Sponge network | -0.355 | 0.0077 | -2.163 | 0 | 0.268 |
| 49447 | CTD-2336O2.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 28 | ADARB1 | Sponge network | -0.141 | 0.26434 | -0.335 | 0.06631 | 0.268 |
| 49448 | LINC00863 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p | 24 | PBX1 | Sponge network | 0.189 | 0.13981 | -1.206 | 0 | 0.268 |
| 49449 | RP11-359B12.2 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | MSN | Sponge network | 0.064 | 0.72934 | -0.023 | 0.88422 | 0.268 |
| 49450 | USP46-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-362-5p | 14 | ARHGEF12 | Sponge network | 0.102 | 0.51464 | 0.09 | 0.35693 | 0.268 |
| 49451 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-628-5p | 10 | CHD2 | Sponge network | 1.163 | 0.00018 | 0.049 | 0.55371 | 0.268 |
| 49452 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | FREM2 | Sponge network | 0.222 | 0.29133 | -0.437 | 0.46353 | 0.268 |
| 49453 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-7-5p;hsa-miR-93-5p | 26 | ZBTB4 | Sponge network | 0.61 | 0.01593 | -0.739 | 0 | 0.268 |
| 49454 | RP11-180O5.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p | 11 | PIK3C2A | Sponge network | 3.607 | 0.0001 | 0.061 | 0.53776 | 0.268 |
| 49455 | RP5-1021I20.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-877-5p;hsa-miR-93-5p | 26 | GAS7 | Sponge network | -0.785 | 0.00035 | -0.555 | 0.01602 | 0.268 |
| 49456 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 28 | SNX29 | Sponge network | 1.302 | 7.0E-5 | -0.34 | 0.01371 | 0.268 |
| 49457 | FAM215B |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | NAV3 | Sponge network | 0.817 | 3.0E-5 | 0.212 | 0.47729 | 0.268 |
| 49458 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 33 | THRB | Sponge network | -0.663 | 0.10887 | -1.117 | 0.0004 | 0.268 |
| 49459 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p | 16 | BCL6 | Sponge network | -0.693 | 0.05629 | -0.211 | 0.19489 | 0.268 |
| 49460 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-582-3p | 12 | FOXO3 | Sponge network | -0.473 | 0.07602 | -0.04 | 0.72028 | 0.268 |
| 49461 | CTD-3064M3.3 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-625-5p | 14 | NUAK1 | Sponge network | -0.469 | 0.08721 | 0.904 | 0 | 0.268 |
| 49462 | RP11-35G9.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | AKAP6 | Sponge network | 0.657 | 4.0E-5 | -1.586 | 3.0E-5 | 0.268 |
| 49463 | LINC00957 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p | 11 | PNRC1 | Sponge network | -0.421 | 0.0114 | -0.646 | 0 | 0.268 |
| 49464 | RP11-411K7.1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-664a-3p | 15 | RASAL2 | Sponge network | 0.155 | 0.81273 | 0.869 | 0 | 0.268 |
| 49465 | U91328.19 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | KCNA6 | Sponge network | -0.303 | 0.21002 | -1.531 | 0.00013 | 0.268 |
| 49466 | RP11-597D13.9 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | ZDBF2 | Sponge network | 1.302 | 7.0E-5 | -0.998 | 0.00025 | 0.268 |
| 49467 | LINC00957 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 26 | CADM1 | Sponge network | -0.421 | 0.0114 | -0.9 | 0.00084 | 0.268 |
| 49468 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 19 | MOB1B | Sponge network | -0.726 | 0.00132 | 0.197 | 0.15266 | 0.268 |
| 49469 | HOXB-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-589-3p | 17 | OPCML | Sponge network | -0.154 | 0.53954 | -2.498 | 0 | 0.268 |
| 49470 | CTD-2554C21.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 11 | FREM2 | Sponge network | -1.654 | 0.00144 | -0.437 | 0.46353 | 0.268 |
| 49471 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p | 10 | LRRC4 | Sponge network | -0.405 | 0.22013 | -1.774 | 0 | 0.268 |
| 49472 | AC025171.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 32 | DST | Sponge network | 0.998 | 0 | -0.713 | 0.00096 | 0.268 |
| 49473 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 46 | ADARB1 | Sponge network | -0.727 | 0.04726 | -0.335 | 0.06631 | 0.268 |
| 49474 | RP11-819C21.1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 25 | RIMS2 | Sponge network | 0.537 | 0.00139 | -1.365 | 0.00208 | 0.268 |
| 49475 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-654-3p | 19 | DDX6 | Sponge network | 1.073 | 0.00216 | 0.091 | 0.36428 | 0.268 |
| 49476 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | CHD3 | Sponge network | 1.302 | 7.0E-5 | -0.001 | 0.99669 | 0.268 |
| 49477 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | EGFR | Sponge network | 0.998 | 0 | -0.175 | 0.48451 | 0.268 |
| 49478 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-590-3p | 24 | APC | Sponge network | 0.082 | 0.55654 | -0.255 | 0.04051 | 0.268 |
| 49479 | CTC-444N24.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | EGLN1 | Sponge network | 1.153 | 0 | -0.127 | 0.1536 | 0.268 |
| 49480 | RP11-464F9.20 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-625-5p | 10 | ABI2 | Sponge network | 0.643 | 0.00136 | 0.175 | 0.14787 | 0.268 |
| 49481 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SAMHD1 | Sponge network | 0.194 | 0.64278 | 0.072 | 0.62895 | 0.268 |
| 49482 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 25 | UNC80 | Sponge network | -0.726 | 0.00132 | -1.934 | 0 | 0.268 |
| 49483 | MIR497HG |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | IRS1 | Sponge network | -1.439 | 4.0E-5 | -0.026 | 0.88855 | 0.268 |
| 49484 | RP11-244H3.1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-503-5p | 17 | ABL1 | Sponge network | 0.659 | 3.0E-5 | -0.215 | 0.07804 | 0.268 |
| 49485 | HOXA-AS2 |
hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-744-3p;hsa-miR-940 | 11 | RHOB | Sponge network | 0.61 | 0.01593 | -1.052 | 0 | 0.268 |
| 49486 | RP11-758N13.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-375 | 11 | ABI2 | Sponge network | 2.939 | 3.0E-5 | 0.175 | 0.14787 | 0.268 |
| 49487 | RP11-384K6.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-345-5p;hsa-miR-486-5p | 18 | ADARB1 | Sponge network | 0.946 | 0 | -0.335 | 0.06631 | 0.268 |
| 49488 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 34 | BHLHE41 | Sponge network | -1.216 | 0 | 0.353 | 0.12753 | 0.268 |
| 49489 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | HDAC9 | Sponge network | -0.793 | 7.0E-5 | -0.755 | 0.00495 | 0.268 |
| 49490 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p | 14 | GIGYF2 | Sponge network | 0.064 | 0.72934 | 0.136 | 0.04403 | 0.268 |
| 49491 | AC108488.4 |
hsa-miR-107;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-224-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-577;hsa-miR-7-1-3p | 15 | GRIN2A | Sponge network | 0.259 | 0.12542 | -2.161 | 0 | 0.268 |
| 49492 | RP11-356J5.12 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 35 | AFF1 | Sponge network | -0.899 | 6.0E-5 | -0.384 | 0.00053 | 0.268 |
| 49493 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 18 | RBL2 | Sponge network | -1.216 | 0 | -0.282 | 0.00254 | 0.268 |
| 49494 | ATP1B3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-590-3p | 13 | ADARB1 | Sponge network | 1.2 | 0.01813 | -0.335 | 0.06631 | 0.268 |
| 49495 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | GNA13 | Sponge network | -0.129 | 0.57177 | 0.007 | 0.93884 | 0.268 |
| 49496 | DIO3OS |
hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 10 | PHOX2B | Sponge network | -0.773 | 0.04073 | -2.68 | 0 | 0.268 |
| 49497 | RP11-439A17.10 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 12 | NTRK2 | Sponge network | 0.051 | 0.95756 | -0.736 | 0.06495 | 0.268 |
| 49498 | FAM215B |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p | 14 | RPS6KA3 | Sponge network | 0.817 | 3.0E-5 | 0.256 | 0.02419 | 0.268 |
| 49499 | RP11-182L21.6 |
hsa-let-7g-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 11 | CBL | Sponge network | 0.353 | 0.00175 | 0.291 | 0.01267 | 0.268 |
| 49500 | AC025171.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | EBF1 | Sponge network | 0.998 | 0 | -0.384 | 0.08856 | 0.268 |
| 49501 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 37 | SPRY3 | Sponge network | 1.302 | 7.0E-5 | 0.016 | 0.91286 | 0.268 |
| 49502 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-653-5p | 18 | PDS5B | Sponge network | -0.615 | 0.00015 | 0.259 | 0.00962 | 0.268 |
| 49503 | LINC00702 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 22 | PAIP2 | Sponge network | -0.737 | 0.06182 | -0.515 | 0 | 0.268 |
| 49504 | AC116366.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-577;hsa-miR-628-5p | 10 | EPHA3 | Sponge network | 0.329 | 0.11122 | 0.185 | 0.53588 | 0.268 |
| 49505 | RP11-968O1.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p | 11 | SCN9A | Sponge network | -0.829 | 0.01343 | -0.525 | 0.10593 | 0.268 |
| 49506 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-342-3p | 11 | ZMYM2 | Sponge network | 2.163 | 0 | 0.279 | 0.01273 | 0.268 |
| 49507 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-625-3p;hsa-miR-664a-3p | 16 | FZD3 | Sponge network | 0.903 | 0 | 0.104 | 0.60316 | 0.268 |
| 49508 | EMX2OS |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | DOCK4 | Sponge network | -0.777 | 0.1134 | 0.502 | 0.00108 | 0.268 |
| 49509 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-511-5p | 23 | ZFHX3 | Sponge network | 0.622 | 0.00015 | 0.389 | 0.01363 | 0.268 |
| 49510 | FENDRR |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CSMD1 | Sponge network | -1.281 | 0.00012 | -0.63 | 0.18881 | 0.268 |
| 49511 | LIPE-AS1 |
hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-708-5p | 11 | MAT2A | Sponge network | 0.078 | 0.55803 | -0.073 | 0.36195 | 0.268 |
| 49512 | CTC-559E9.5 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 11 | CYLD | Sponge network | 0.594 | 0.00064 | -0.367 | 0.00171 | 0.268 |
| 49513 | PCA3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | CSMD1 | Sponge network | -0.709 | 0.18168 | -0.63 | 0.18881 | 0.268 |
| 49514 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-590-3p | 10 | CLIP1 | Sponge network | 0.765 | 7.0E-5 | -0.215 | 0.08684 | 0.268 |
| 49515 | RAMP2-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 24 | LRIG1 | Sponge network | -0.513 | 0.04703 | -0.537 | 0.00588 | 0.268 |
| 49516 | RP11-13K12.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-532-5p | 22 | ZMYND11 | Sponge network | -0.154 | 0.78939 | -0.2 | 0.05449 | 0.268 |
| 49517 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 30 | BCL2 | Sponge network | -0.176 | 0.59692 | -0.899 | 0 | 0.268 |
| 49518 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p | 10 | DMTF1 | Sponge network | 0.177 | 0.16681 | 0.248 | 0.02726 | 0.268 |
| 49519 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940;hsa-miR-942-5p | 32 | HLF | Sponge network | 0.72 | 0.0001 | -2.443 | 0 | 0.268 |
| 49520 | RASSF8-AS1 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | BCL9 | Sponge network | 0.335 | 0.20596 | 0.321 | 0.00267 | 0.268 |
| 49521 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | NCAM2 | Sponge network | 0.219 | 0.1516 | -0.482 | 0.22565 | 0.268 |
| 49522 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 28 | PRKCE | Sponge network | -1.575 | 6.0E-5 | -0.403 | 0.00056 | 0.268 |
| 49523 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 14 | NR2C2 | Sponge network | 0.619 | 0.00157 | 0.21 | 0.03224 | 0.268 |
| 49524 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-629-3p | 45 | CBX5 | Sponge network | -0.615 | 0.00015 | 0.165 | 0.24446 | 0.268 |
| 49525 | AC009948.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-590-3p | 13 | PKNOX1 | Sponge network | 0.532 | 0.00505 | 0.085 | 0.28917 | 0.268 |
| 49526 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 32 | MOB1B | Sponge network | -1.249 | 7.0E-5 | 0.197 | 0.15266 | 0.268 |
| 49527 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-654-3p | 20 | TP53INP1 | Sponge network | 0.267 | 0.2937 | -0.496 | 0.00064 | 0.268 |
| 49528 | AC005562.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | TET1 | Sponge network | 0.488 | 0.00084 | -0.135 | 0.52138 | 0.268 |
| 49529 | WDFY3-AS2 |
hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-7-5p | 14 | PIM1 | Sponge network | -0.942 | 0 | -1.114 | 0 | 0.268 |
| 49530 | HCG11 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | SEMA3C | Sponge network | 0.385 | 0.10067 | -0.272 | 0.31153 | 0.268 |
| 49531 | LINC00094 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-655-3p | 19 | ZMYND11 | Sponge network | 0.253 | 0.09565 | -0.2 | 0.05449 | 0.268 |
| 49532 | ZSCAN16-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NCAM2 | Sponge network | -0.342 | 0.02288 | -0.482 | 0.22565 | 0.268 |
| 49533 | RP11-48B3.4 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p | 14 | IRS1 | Sponge network | 1.757 | 0 | -0.026 | 0.88855 | 0.268 |
| 49534 | RP11-46C24.7 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-484 | 10 | LZTS1 | Sponge network | 0.392 | 0.0278 | 1.664 | 0 | 0.268 |
| 49535 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 42 | CUX1 | Sponge network | 0.219 | 0.1516 | -0.039 | 0.6705 | 0.268 |
| 49536 | LINC00672 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-423-5p;hsa-miR-590-3p | 10 | PAK3 | Sponge network | -0.227 | 0.31657 | -0.628 | 0.07253 | 0.268 |
| 49537 | PAN3-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-33a-3p | 10 | MCC | Sponge network | 0.062 | 0.55274 | -1.005 | 0 | 0.268 |
| 49538 | RP11-452L6.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-532-3p | 13 | CBL | Sponge network | 0.665 | 0.00061 | 0.291 | 0.01267 | 0.268 |
| 49539 | EIF3J-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-582-3p | 21 | PBX1 | Sponge network | 0.331 | 0.00509 | -1.206 | 0 | 0.268 |
| 49540 | RP11-65J21.3 |
hsa-let-7d-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 10 | TGFBR3 | Sponge network | -0.557 | 0.11135 | -1.173 | 1.0E-5 | 0.268 |
| 49541 | FZD10-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-935 | 13 | STAT5B | Sponge network | -0.727 | 0.04726 | -0.182 | 0.1082 | 0.268 |
| 49542 | HHIP-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 12 | EGR2 | Sponge network | -0.737 | 0.10822 | 0.309 | 0.17079 | 0.268 |
| 49543 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | LATS1 | Sponge network | 0.272 | 0.44692 | 0.109 | 0.34208 | 0.268 |
| 49544 | RP11-35G9.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | SPG20 | Sponge network | 0.657 | 4.0E-5 | -1.16 | 0 | 0.268 |
| 49545 | DNAJC3-AS1 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p | 10 | UBR3 | Sponge network | 0.753 | 0.00014 | -0.021 | 0.82774 | 0.268 |
| 49546 | RP11-127B20.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | MED12L | Sponge network | 0.411 | 0.1335 | -0.194 | 0.55299 | 0.268 |
| 49547 | FAM66C |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-92a-1-5p | 13 | PRKACA | Sponge network | -0.901 | 0.00019 | -0.255 | 0.0012 | 0.268 |
| 49548 | RP11-344B5.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p | 10 | FAT4 | Sponge network | -0.055 | 0.88482 | -0.354 | 0.14694 | 0.268 |
| 49549 | RP11-655M14.13 |
hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 11 | MLLT10 | Sponge network | 1.473 | 0.0057 | 0.105 | 0.33292 | 0.268 |
| 49550 | RAMP2-AS1 |
hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 10 | LZTS1 | Sponge network | -0.513 | 0.04703 | 1.664 | 0 | 0.268 |
| 49551 | LINC00694 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p | 12 | JAZF1 | Sponge network | -0.855 | 0.06723 | -0.377 | 0.02677 | 0.268 |
| 49552 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-874-3p;hsa-miR-98-5p | 10 | ST5 | Sponge network | 0.249 | 0.35902 | -0.78 | 0 | 0.268 |
| 49553 | NDUFA6-AS1 |
hsa-miR-17-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-23a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-589-5p | 10 | SORCS2 | Sponge network | -0.355 | 0.0077 | -0.223 | 0.4253 | 0.268 |
| 49554 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-196b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | SLC6A15 | Sponge network | 0.906 | 0.31715 | -2.373 | 2.0E-5 | 0.268 |
| 49555 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-628-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 42 | FZD3 | Sponge network | -0.405 | 0.22013 | 0.104 | 0.60316 | 0.268 |
| 49556 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | LRRC7 | Sponge network | 0.381 | 0.25967 | -1.341 | 0.01193 | 0.268 |
| 49557 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 25 | LRRC55 | Sponge network | -0.942 | 0 | -0.026 | 0.93163 | 0.268 |
| 49558 | MEG3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-940 | 19 | BAZ2B | Sponge network | 0.249 | 0.35902 | -0.276 | 0.02833 | 0.268 |
| 49559 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 25 | RICTOR | Sponge network | 1.308 | 0 | 0.388 | 0.00188 | 0.268 |
| 49560 | MIR22HG |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 10 | PTPN13 | Sponge network | -1.047 | 0 | -1.208 | 0 | 0.268 |
| 49561 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p | 14 | ABCB5 | Sponge network | 0.219 | 0.1516 | -0.956 | 0.04077 | 0.268 |
| 49562 | PART1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p | 21 | MACF1 | Sponge network | -2.925 | 0 | 0.019 | 0.90335 | 0.268 |
| 49563 | RP11-426C22.5 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 14 | BMPR2 | Sponge network | 0.749 | 0 | 0.206 | 0.0635 | 0.268 |
| 49564 | ACTA2-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | LIMCH1 | Sponge network | 0.194 | 0.64278 | -0.915 | 1.0E-5 | 0.268 |
| 49565 | FZD10-AS1 |
hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 11 | LZTS1 | Sponge network | -0.727 | 0.04726 | 1.664 | 0 | 0.268 |
| 49566 | RP11-983P16.4 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-7-1-3p | 12 | RASSF3 | Sponge network | 0.41 | 0.02214 | -0.226 | 0.11691 | 0.268 |
| 49567 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-508-3p;hsa-miR-590-3p | 11 | RIMS1 | Sponge network | -0.513 | 0.04703 | -3.143 | 0 | 0.268 |
| 49568 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | FOXO1 | Sponge network | -0.739 | 0.0574 | -0.141 | 0.33874 | 0.268 |
| 49569 | RP11-360F5.3 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 11 | RYR2 | Sponge network | 1.867 | 0 | -1.466 | 0.00031 | 0.268 |
| 49570 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-5p | 13 | TRIP11 | Sponge network | -0.567 | 0 | -0.158 | 0.06384 | 0.268 |
| 49571 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 22 | RSPO2 | Sponge network | -0.727 | 0.04726 | -3.038 | 0 | 0.268 |
| 49572 | CTC-429P9.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-500a-5p | 27 | AFF1 | Sponge network | 0.082 | 0.55397 | -0.384 | 0.00053 | 0.268 |
| 49573 | HNRNPU-AS1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-363-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TRIM13 | Sponge network | 1.601 | 0 | 0.183 | 0.06968 | 0.268 |
| 49574 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | KAT6B | Sponge network | 0.572 | 0.01722 | -0.478 | 0.00018 | 0.268 |
| 49575 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-96-5p | 15 | JAZF1 | Sponge network | 2.306 | 9.0E-5 | -0.377 | 0.02677 | 0.268 |
| 49576 | CTC-296K1.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p | 13 | ZMYND11 | Sponge network | -2.095 | 0.00112 | -0.2 | 0.05449 | 0.268 |
| 49577 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-409-3p;hsa-miR-590-3p | 14 | PHC3 | Sponge network | 1.073 | 0.00216 | 0.212 | 0.0499 | 0.268 |
| 49578 | LINC00957 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-93-3p;hsa-miR-940 | 12 | POU2F2 | Sponge network | -0.421 | 0.0114 | -0.233 | 0.32214 | 0.268 |
| 49579 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 21 | EPHA5 | Sponge network | 0.687 | 0.02753 | -0.957 | 0.01733 | 0.268 |
| 49580 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-935 | 40 | PTPRD | Sponge network | -0.405 | 0.22013 | -1.005 | 0.00064 | 0.268 |
| 49581 | LINC00476 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-877-5p | 12 | FLI1 | Sponge network | -0.615 | 0.00015 | -0.294 | 0.10905 | 0.268 |
| 49582 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 17 | EDA2R | Sponge network | 1.302 | 7.0E-5 | -0.587 | 0.01598 | 0.268 |
| 49583 | TTC25 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-940 | 10 | CRTC3 | Sponge network | -0.208 | 0.38261 | 0.164 | 0.16786 | 0.268 |
| 49584 | MRPL23-AS1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-484 | 10 | RIMS2 | Sponge network | -0.278 | 0.76559 | -1.365 | 0.00208 | 0.268 |
| 49585 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 19 | BMPR2 | Sponge network | 1.11 | 1.0E-5 | 0.206 | 0.0635 | 0.268 |
| 49586 | RP11-693J15.4 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-374b-3p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-589-5p;hsa-miR-590-5p | 16 | GRIA3 | Sponge network | -0.72 | 0.24239 | -1.956 | 0 | 0.268 |
| 49587 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 40 | TRPS1 | Sponge network | 0.494 | 0.00339 | -0.191 | 0.44109 | 0.268 |
| 49588 | RP11-13K12.1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-501-3p;hsa-miR-502-3p | 11 | RIMS1 | Sponge network | -0.154 | 0.78939 | -3.143 | 0 | 0.268 |
| 49589 | CKMT2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-942-5p | 40 | UNC80 | Sponge network | 0.082 | 0.55654 | -1.934 | 0 | 0.268 |
| 49590 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 17 | ITGAV | Sponge network | -0.896 | 0.00099 | 0.337 | 0.01002 | 0.268 |
| 49591 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-625-5p | 11 | GAS1 | Sponge network | 0.4 | 0.14611 | -1.047 | 0.00364 | 0.268 |
| 49592 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-7-1-3p | 10 | DACT1 | Sponge network | -0.13 | 0.48734 | 0.783 | 0.00072 | 0.268 |
| 49593 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-93-3p | 19 | ST6GAL2 | Sponge network | 0.174 | 0.30034 | -1.201 | 0.00597 | 0.268 |
| 49594 | RP11-517B11.7 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-664a-5p | 12 | CBFB | Sponge network | 0.706 | 0 | 1.061 | 0 | 0.268 |
| 49595 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-92a-3p | 10 | SASH1 | Sponge network | 0.453 | 0.01839 | -0.502 | 0.00175 | 0.268 |
| 49596 | LINC00672 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | RELN | Sponge network | -0.227 | 0.31657 | -2.403 | 0 | 0.268 |
| 49597 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-493-5p;hsa-miR-874-3p | 17 | NR2C2 | Sponge network | -0.384 | 0.40652 | 0.21 | 0.03224 | 0.268 |
| 49598 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-653-5p | 21 | PDS5B | Sponge network | -1.111 | 0.0003 | 0.259 | 0.00962 | 0.268 |
| 49599 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | NIN | Sponge network | 0.619 | 0.00157 | -0.253 | 0.13794 | 0.268 |
| 49600 | RP11-767N6.7 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-590-5p | 14 | CAMTA1 | Sponge network | 0.466 | 0.00155 | -0.436 | 1.0E-5 | 0.268 |
| 49601 | RP4-605O3.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p | 36 | LPP | Sponge network | 0.262 | 0.0475 | -0.459 | 0.04475 | 0.268 |
| 49602 | DNAJC27-AS1 |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p | 10 | GLIPR2 | Sponge network | -0.969 | 2.0E-5 | -0.996 | 0 | 0.268 |
| 49603 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-382-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 24 | ACSL6 | Sponge network | 0.101 | 0.60919 | -1.593 | 0 | 0.268 |
| 49604 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 65 | PTEN | Sponge network | -0.739 | 0.0574 | -0.187 | 0.07748 | 0.268 |
| 49605 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-758-3p | 11 | CDC73 | Sponge network | 0.997 | 0 | 0.094 | 0.20463 | 0.268 |
| 49606 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 19 | RBMS3 | Sponge network | 0.687 | 0.02753 | -0.519 | 0.03551 | 0.268 |
| 49607 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 10 | SOX5 | Sponge network | 0.062 | 0.55274 | -1.091 | 4.0E-5 | 0.268 |
| 49608 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p | 18 | ARHGAP29 | Sponge network | -0.487 | 0.02157 | 0.131 | 0.42953 | 0.268 |
| 49609 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | ST6GALNAC3 | Sponge network | 0.348 | 0.32912 | -0.987 | 0 | 0.268 |
| 49610 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 26 | NOTCH2 | Sponge network | 0.619 | 0.00157 | 0.138 | 0.35163 | 0.268 |
| 49611 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-98-5p | 17 | PNRC1 | Sponge network | -0.83 | 0 | -0.646 | 0 | 0.268 |
| 49612 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-589-5p | 18 | CXXC4 | Sponge network | -0.154 | 0.53954 | -0.433 | 0.21055 | 0.268 |
| 49613 | C20orf166-AS1 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-5p;hsa-miR-625-5p | 36 | CBX5 | Sponge network | -2.69 | 0.0001 | 0.165 | 0.24446 | 0.268 |
| 49614 | H1FX-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-671-5p | 11 | EHD3 | Sponge network | -0.206 | 0.12942 | -0.484 | 0.01747 | 0.268 |
| 49615 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-629-3p | 16 | RASSF2 | Sponge network | 1.122 | 0.02989 | -0.273 | 0.21961 | 0.268 |
| 49616 | AC025335.1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-339-5p;hsa-miR-664a-5p | 13 | SOX11 | Sponge network | 0.783 | 0 | 2.68 | 0 | 0.268 |
| 49617 | AP001347.6 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p | 10 | ATM | Sponge network | -1.161 | 0.00591 | 0.178 | 0.21568 | 0.268 |
| 49618 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ARHGAP28 | Sponge network | 0.374 | 0.20054 | -0.911 | 0.00013 | 0.268 |
| 49619 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-429 | 14 | SLITRK3 | Sponge network | 0.95 | 0.15943 | -3.328 | 0 | 0.268 |
| 49620 | RP4-791M13.3 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-7-5p | 10 | CRTC1 | Sponge network | 0.419 | 0.0319 | -0.116 | 0.28412 | 0.268 |
| 49621 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-424-5p | 10 | LRP1B | Sponge network | -1.054 | 0.0706 | -2.163 | 0 | 0.268 |
| 49622 | LINC00622 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PRKAB2 | Sponge network | 1.169 | 0.00028 | -0.367 | 0.01371 | 0.268 |
| 49623 | AC005562.1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | BRD3 | Sponge network | 0.488 | 0.00084 | 0.37 | 0.00013 | 0.268 |
| 49624 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | ST6GAL2 | Sponge network | 0.246 | 0.59912 | -1.201 | 0.00597 | 0.268 |
| 49625 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 31 | EPHA3 | Sponge network | 0.311 | 0.10344 | 0.185 | 0.53588 | 0.268 |
| 49626 | C4B-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 14 | FBXO32 | Sponge network | 2.306 | 9.0E-5 | -0.495 | 0.07561 | 0.268 |
| 49627 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-7-1-3p | 11 | RASSF8 | Sponge network | 0.238 | 0.70544 | -0.122 | 0.65591 | 0.268 |
| 49628 | AP001347.6 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-590-3p | 14 | FMNL3 | Sponge network | -1.161 | 0.00591 | 0.555 | 4.0E-5 | 0.268 |
| 49629 | RP11-401P9.4 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-5p | 10 | RTN4 | Sponge network | -0.384 | 0.40652 | -0.412 | 0.00014 | 0.268 |
| 49630 | RP11-263K19.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429 | 18 | DLC1 | Sponge network | 0.859 | 0.00331 | -0.382 | 0.05038 | 0.268 |
| 49631 | RP11-758N13.1 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p | 15 | EGFR | Sponge network | 2.939 | 3.0E-5 | -0.175 | 0.48451 | 0.268 |
| 49632 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | CACNA2D1 | Sponge network | -0.342 | 0.02288 | -0.846 | 0.00675 | 0.268 |
| 49633 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-1-3p | 19 | SYNE1 | Sponge network | 0.853 | 0.15352 | -0.738 | 0.00316 | 0.268 |
| 49634 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p;hsa-miR-582-5p | 11 | ZMYM2 | Sponge network | 0.419 | 0.0319 | 0.279 | 0.01273 | 0.268 |
| 49635 | LINC00957 |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | JUN | Sponge network | -0.421 | 0.0114 | -0.932 | 0 | 0.268 |
| 49636 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | HACE1 | Sponge network | 1.11 | 0 | -0.072 | 0.55239 | 0.268 |
| 49637 | GS1-124K5.11 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-629-5p | 13 | WNK2 | Sponge network | 0.494 | 0.00339 | -0.352 | 0.31066 | 0.268 |
| 49638 | DIO3OS |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-542-3p | 10 | RTN4 | Sponge network | -0.773 | 0.04073 | -0.412 | 0.00014 | 0.268 |
| 49639 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 26 | MYBL1 | Sponge network | -0.49 | 0.04946 | 0.494 | 0.02152 | 0.268 |
| 49640 | CTD-2089N3.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-378a-3p | 14 | TACC1 | Sponge network | -0.314 | 0.62033 | -0.362 | 0.05406 | 0.268 |
| 49641 | RP11-4O1.2 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 10 | CBFB | Sponge network | 1.302 | 0 | 1.061 | 0 | 0.268 |
| 49642 | RP11-554A11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-421;hsa-miR-450b-5p | 16 | DMXL1 | Sponge network | -0.583 | 0.14589 | -0.426 | 0.00056 | 0.268 |
| 49643 | NR2F1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | -0.49 | 0.04946 | 0.126 | 0.15266 | 0.268 |
| 49644 | UBA6-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 19 | MOB1B | Sponge network | 0.312 | 0.00725 | 0.197 | 0.15266 | 0.268 |
| 49645 | RP11-373D23.3 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 13 | ZFHX3 | Sponge network | -0.285 | 0.2172 | 0.389 | 0.01363 | 0.268 |
| 49646 | LINC00843 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | PRKAA1 | Sponge network | 1.106 | 0 | 0.317 | 0.0031 | 0.268 |
| 49647 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | FOXO1 | Sponge network | -0.769 | 0.00035 | -0.141 | 0.33874 | 0.268 |
| 49648 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p | 10 | PRDM5 | Sponge network | 1.302 | 7.0E-5 | -1.09 | 0.00014 | 0.268 |
| 49649 | AC016747.3 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 11 | SEPT6 | Sponge network | 0.199 | 0.38015 | -0.598 | 0.00181 | 0.268 |
| 49650 | BOLA3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-501-5p | 13 | PRKCE | Sponge network | -0.246 | 0.35034 | -0.403 | 0.00056 | 0.268 |
| 49651 | RP4-639F20.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-222-5p;hsa-miR-320b | 15 | CBL | Sponge network | -0.517 | 0.02935 | 0.291 | 0.01267 | 0.268 |
| 49652 | RP11-57H14.4 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NRG3 | Sponge network | 0.083 | 0.66405 | -1.836 | 4.0E-5 | 0.268 |
| 49653 | FGF14-AS2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-505-5p | 13 | TOM1L2 | Sponge network | -1.582 | 8.0E-5 | -0.994 | 0 | 0.268 |
| 49654 | KCNJ2-AS1 |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-590-3p | 12 | ITPKB | Sponge network | -0.436 | 0.05325 | -0.704 | 0.00106 | 0.268 |
| 49655 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-93-5p;hsa-miR-942-5p | 24 | UNC80 | Sponge network | -0.351 | 0.11358 | -1.934 | 0 | 0.268 |
| 49656 | HOXB-AS2 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-429 | 10 | PRDM2 | Sponge network | 1.316 | 0.01106 | -0.258 | 0.00721 | 0.268 |
| 49657 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-7-1-3p | 14 | MLLT10 | Sponge network | 0.411 | 0.1335 | 0.105 | 0.33292 | 0.268 |
| 49658 | XXbac-B444P24.8 |
hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | ABL2 | Sponge network | 2.264 | 0.0012 | 0.82 | 0 | 0.268 |
| 49659 | RP4-714D9.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-766-3p | 15 | MDM4 | Sponge network | 1.015 | 0 | 0.345 | 0.00762 | 0.268 |
| 49660 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLIP1 | Sponge network | 0.349 | 0.01695 | -0.215 | 0.08684 | 0.268 |
| 49661 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-671-5p;hsa-miR-874-3p | 20 | ARID5B | Sponge network | 0.637 | 0.00069 | 0.342 | 0.01676 | 0.268 |
| 49662 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 17 | PTGFR | Sponge network | 0.72 | 0.0001 | -0.233 | 0.45421 | 0.268 |
| 49663 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 26 | YAP1 | Sponge network | -1.697 | 0.00889 | 0.22 | 0.11836 | 0.268 |
| 49664 | CTB-51J22.1 |
hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-424-5p | 10 | CTDSPL | Sponge network | -0.803 | 0.23737 | 0.085 | 0.45121 | 0.268 |
| 49665 | SOX2-OT |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PTPN13 | Sponge network | -1.264 | 6.0E-5 | -1.208 | 0 | 0.268 |
| 49666 | USP3-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-511-5p;hsa-miR-589-5p | 23 | EYA4 | Sponge network | -0.162 | 0.47687 | 0.3 | 0.36876 | 0.268 |
| 49667 | MIR497HG |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-423-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 14 | CELF4 | Sponge network | -1.439 | 4.0E-5 | -1.135 | 0.00344 | 0.268 |
| 49668 | RP11-283I3.6 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-96-5p | 12 | PRKAR1A | Sponge network | 0.614 | 1.0E-5 | -0.063 | 0.50314 | 0.268 |
| 49669 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 39 | THRB | Sponge network | 0.395 | 0.15451 | -1.117 | 0.0004 | 0.268 |
| 49670 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-532-3p | 13 | CBL | Sponge network | -0.376 | 0.00337 | 0.291 | 0.01267 | 0.268 |
| 49671 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-185-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 17 | PRKAB2 | Sponge network | -4.546 | 0 | -0.367 | 0.01371 | 0.268 |
| 49672 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-376c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 43 | WDFY3 | Sponge network | 0.603 | 0.00127 | -0.201 | 0.0967 | 0.268 |
| 49673 | RP11-395G23.3 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-378c;hsa-miR-425-5p;hsa-miR-450b-5p | 16 | ERC1 | Sponge network | 0.381 | 0.25967 | -0.108 | 0.38794 | 0.268 |
| 49674 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p | 17 | DDX6 | Sponge network | 0.983 | 0.00048 | 0.091 | 0.36428 | 0.268 |
| 49675 | RP5-894A10.6 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p | 13 | CCDC6 | Sponge network | 0.697 | 9.0E-5 | 0.27 | 0.02555 | 0.268 |
| 49676 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-940 | 12 | SNX29 | Sponge network | 0.529 | 0.00552 | -0.34 | 0.01371 | 0.268 |
| 49677 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-942-5p | 39 | TMEM132B | Sponge network | -0.371 | 0.24604 | -0.83 | 0.01084 | 0.268 |
| 49678 | RP11-159D12.2 |
hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-424-5p;hsa-miR-590-3p | 12 | DOCK4 | Sponge network | 1.254 | 0 | 0.502 | 0.00108 | 0.268 |
| 49679 | HOXB-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-429 | 11 | FLI1 | Sponge network | -0.154 | 0.53954 | -0.294 | 0.10905 | 0.268 |
| 49680 | RP11-356J5.12 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-629-3p;hsa-miR-92a-3p | 15 | SPTBN4 | Sponge network | -0.899 | 6.0E-5 | -0.786 | 0.01281 | 0.268 |
| 49681 | CTD-2336O2.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-940 | 31 | GAS7 | Sponge network | -0.141 | 0.26434 | -0.555 | 0.01602 | 0.268 |
| 49682 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-96-5p | 11 | NRXN1 | Sponge network | 0.37 | 0.27614 | -3.778 | 0 | 0.268 |
| 49683 | ADAMTS9-AS2 |
hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 12 | FBXW7 | Sponge network | -2.452 | 0 | -0.418 | 2.0E-5 | 0.268 |
| 49684 | AC009120.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p | 16 | NAV3 | Sponge network | 0.919 | 0 | 0.212 | 0.47729 | 0.268 |
| 49685 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | AR | Sponge network | -0.223 | 0.47816 | -1.554 | 0 | 0.268 |
| 49686 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 22 | CSMD1 | Sponge network | -0.969 | 2.0E-5 | -0.63 | 0.18881 | 0.267 |
| 49687 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-423-5p;hsa-miR-455-5p | 15 | PNRC1 | Sponge network | -1.645 | 0 | -0.646 | 0 | 0.267 |
| 49688 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | PTPRB | Sponge network | 0.998 | 0 | 0.105 | 0.47122 | 0.267 |
| 49689 | PPP1R26-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-664a-5p | 11 | CBFB | Sponge network | 1.454 | 0 | 1.061 | 0 | 0.267 |
| 49690 | RP11-819C21.1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-629-5p;hsa-miR-96-5p | 12 | WNK2 | Sponge network | 0.537 | 0.00139 | -0.352 | 0.31066 | 0.267 |
| 49691 | RP11-20I23.13 |
hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-744-3p | 15 | PTPN14 | Sponge network | 0.505 | 0.0014 | 0.144 | 0.34595 | 0.267 |
| 49692 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | CLASP2 | Sponge network | 0.799 | 1.0E-5 | -0.038 | 0.71 | 0.267 |
| 49693 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p | 15 | MYBL1 | Sponge network | 0.959 | 0 | 0.494 | 0.02152 | 0.267 |
| 49694 | GS1-124K5.11 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p | 13 | RASAL2 | Sponge network | 0.494 | 0.00339 | 0.869 | 0 | 0.267 |
| 49695 | LINC00578 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p | 20 | ST6GALNAC3 | Sponge network | 0.811 | 0.03228 | -0.987 | 0 | 0.267 |
| 49696 | LINC00957 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-744-3p | 19 | HIP1 | Sponge network | -0.421 | 0.0114 | 0.774 | 0 | 0.267 |
| 49697 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 18 | NR2C2 | Sponge network | -0.612 | 0.00094 | 0.21 | 0.03224 | 0.267 |
| 49698 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 15 | ACSL6 | Sponge network | -1.375 | 0.08642 | -1.593 | 0 | 0.267 |
| 49699 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 35 | FZD3 | Sponge network | 0.488 | 0.00084 | 0.104 | 0.60316 | 0.267 |
| 49700 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-5p | 46 | THRB | Sponge network | 0.331 | 0.57247 | -1.117 | 0.0004 | 0.267 |
| 49701 | SNX29P2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-370-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-93-3p | 17 | RPS6KA2 | Sponge network | 0.4 | 0.14611 | -0.57 | 0.00013 | 0.267 |
| 49702 | BCYRN1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 25 | SOX11 | Sponge network | 0.311 | 0.10344 | 2.68 | 0 | 0.267 |
| 49703 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DACH2 | Sponge network | -0.129 | 0.57177 | -1.635 | 0.00034 | 0.267 |
| 49704 | RP11-720L2.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-33a-3p;hsa-miR-362-3p | 10 | CYLD | Sponge network | -1.255 | 0.00981 | -0.367 | 0.00171 | 0.267 |
| 49705 | LINC00672 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-93-3p | 22 | TOM1L2 | Sponge network | -0.227 | 0.31657 | -0.994 | 0 | 0.267 |
| 49706 | RP11-127B20.2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-628-5p | 21 | RUNX1T1 | Sponge network | 0.411 | 0.1335 | -0.344 | 0.2374 | 0.267 |
| 49707 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-7-1-3p | 11 | TLL1 | Sponge network | -0.176 | 0.59692 | -1.195 | 3.0E-5 | 0.267 |
| 49708 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-655-3p | 18 | PIK3C2A | Sponge network | 0.594 | 0.41522 | 0.061 | 0.53776 | 0.267 |
| 49709 | AC004540.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p | 13 | TMEM132B | Sponge network | -2.549 | 0.00528 | -0.83 | 0.01084 | 0.267 |
| 49710 | RP11-1277A3.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 19 | IL17RD | Sponge network | 0.453 | 0.01839 | -0.019 | 0.94873 | 0.267 |
| 49711 | CASC2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-5p | 23 | ABI2 | Sponge network | 0.385 | 0.01306 | 0.175 | 0.14787 | 0.267 |
| 49712 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | TMEM132B | Sponge network | 0.422 | 0.27021 | -0.83 | 0.01084 | 0.267 |
| 49713 | AC005618.6 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 20 | ANK2 | Sponge network | 0.862 | 0.06213 | -1.603 | 1.0E-5 | 0.267 |
| 49714 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-223-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p | 15 | MTSS1 | Sponge network | 0.178 | 0.29106 | -0.81 | 0.00035 | 0.267 |
| 49715 | LINC00963 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | DLC1 | Sponge network | 0.523 | 9.0E-5 | -0.382 | 0.05038 | 0.267 |
| 49716 | LINC01082 |
hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p | 11 | IL6ST | Sponge network | -3.322 | 8.0E-5 | -0.402 | 0.00676 | 0.267 |
| 49717 | MYLK-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-940 | 14 | BAZ2B | Sponge network | -1.331 | 3.0E-5 | -0.276 | 0.02833 | 0.267 |
| 49718 | LINC00403 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-2355-3p;hsa-miR-324-3p;hsa-miR-424-5p | 11 | CCND2 | Sponge network | -3.586 | 0.00032 | -0.267 | 0.3112 | 0.267 |
| 49719 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | SPRY3 | Sponge network | 0.91 | 0 | 0.016 | 0.91286 | 0.267 |
| 49720 | NDUFA6-AS1 |
hsa-miR-146b-5p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-589-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | SPOP | Sponge network | -0.355 | 0.0077 | -0.419 | 1.0E-5 | 0.267 |
| 49721 | HCG11 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 27 | RIMS2 | Sponge network | 0.385 | 0.10067 | -1.365 | 0.00208 | 0.267 |
| 49722 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577 | 25 | EGFR | Sponge network | 0.056 | 0.81195 | -0.175 | 0.48451 | 0.267 |
| 49723 | RP1-151F17.2 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | PPM1L | Sponge network | 0.222 | 0.29133 | -0.537 | 0.00616 | 0.267 |
| 49724 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 31 | NFIB | Sponge network | -0.358 | 0.17255 | 0.023 | 0.90795 | 0.267 |
| 49725 | AP001347.6 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-7-1-3p | 10 | EGR2 | Sponge network | -1.161 | 0.00591 | 0.309 | 0.17079 | 0.267 |
| 49726 | MRPL23-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-92a-1-5p | 12 | PRKAB2 | Sponge network | -0.278 | 0.76559 | -0.367 | 0.01371 | 0.267 |
| 49727 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-342-5p;hsa-miR-429 | 16 | ATRX | Sponge network | 0.859 | 0.00331 | 0.261 | 0.04408 | 0.267 |
| 49728 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 30 | JAZF1 | Sponge network | 0.219 | 0.1516 | -0.377 | 0.02677 | 0.267 |
| 49729 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-935 | 30 | RAP1A | Sponge network | 0.311 | 0.10344 | -0.929 | 0 | 0.267 |
| 49730 | LINC00476 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 25 | GJA1 | Sponge network | -0.615 | 0.00015 | -0.485 | 0.02179 | 0.267 |
| 49731 | RP11-46C24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-935 | 16 | ARNT2 | Sponge network | 0.392 | 0.0278 | -0.332 | 0.25851 | 0.267 |
| 49732 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577 | 16 | GREM1 | Sponge network | 0.389 | 0.04293 | 0.902 | 0.01691 | 0.267 |
| 49733 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p | 18 | MAFB | Sponge network | 1.302 | 7.0E-5 | -0.023 | 0.89865 | 0.267 |
| 49734 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 44 | NFIB | Sponge network | -1.057 | 0.00029 | 0.023 | 0.90795 | 0.267 |
| 49735 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-5p | 12 | MYBL1 | Sponge network | 1.093 | 0 | 0.494 | 0.02152 | 0.267 |
| 49736 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TRIM13 | Sponge network | 0.064 | 0.72934 | 0.183 | 0.06968 | 0.267 |
| 49737 | RP11-98I9.4 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | EPS15 | Sponge network | 0.67 | 0.00048 | -0.052 | 0.53998 | 0.267 |
| 49738 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 44 | LPP | Sponge network | -0.206 | 0.12942 | -0.459 | 0.04475 | 0.267 |
| 49739 | AC009120.5 |
hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26b-3p;hsa-miR-625-5p;hsa-miR-629-3p | 10 | CBX5 | Sponge network | 1.383 | 0 | 0.165 | 0.24446 | 0.267 |
| 49740 | CTC-296K1.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-590-5p | 15 | GNAQ | Sponge network | -2.076 | 0.00548 | -0.367 | 0.00102 | 0.267 |
| 49741 | TMEM9B-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 15 | TBL1XR1 | Sponge network | 0.529 | 0.00552 | 0.578 | 0 | 0.267 |
| 49742 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-874-3p | 12 | CRTC3 | Sponge network | 0.859 | 0.00331 | 0.164 | 0.16786 | 0.267 |
| 49743 | EMX2OS |
hsa-miR-107;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 11 | SEPT6 | Sponge network | -0.777 | 0.1134 | -0.598 | 0.00181 | 0.267 |
| 49744 | RP11-1094M14.11 |
hsa-miR-101-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-664a-5p | 10 | CBFB | Sponge network | 0.586 | 2.0E-5 | 1.061 | 0 | 0.267 |
| 49745 | BMS1P20 |
hsa-let-7e-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | COPS2 | Sponge network | 0.34 | 0.00273 | -0.178 | 0.04974 | 0.267 |
| 49746 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | PSIP1 | Sponge network | 0.335 | 0.20596 | -0.005 | 0.96897 | 0.267 |
| 49747 | AC108488.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-3613-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 42 | NTRK3 | Sponge network | 0.259 | 0.12542 | -1.77 | 3.0E-5 | 0.267 |
| 49748 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 11 | KAT6B | Sponge network | -0.739 | 0.00044 | -0.478 | 0.00018 | 0.267 |
| 49749 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | USP12 | Sponge network | 0.889 | 9.0E-5 | -0.102 | 0.40768 | 0.267 |
| 49750 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-629-5p | 18 | PCDH17 | Sponge network | -0.154 | 0.78939 | 0.731 | 2.0E-5 | 0.267 |
| 49751 | C4B-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-5p | 11 | PRKCB | Sponge network | 2.306 | 9.0E-5 | -1.313 | 2.0E-5 | 0.267 |
| 49752 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-429 | 10 | EPHA3 | Sponge network | 1.106 | 0 | 0.185 | 0.53588 | 0.267 |
| 49753 | RP11-384K6.6 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-378c;hsa-miR-628-5p | 15 | ERC1 | Sponge network | 0.946 | 0 | -0.108 | 0.38794 | 0.267 |
| 49754 | CD27-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-766-3p | 33 | IL17RD | Sponge network | 0.177 | 0.16681 | -0.019 | 0.94873 | 0.267 |
| 49755 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-29a-5p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p | 23 | TRPS1 | Sponge network | -0.555 | 0.06156 | -0.191 | 0.44109 | 0.267 |
| 49756 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p | 10 | CBFA2T3 | Sponge network | 1.344 | 0 | -1.221 | 1.0E-5 | 0.267 |
| 49757 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | KDSR | Sponge network | 0.529 | 0.00552 | 0.239 | 0.02216 | 0.267 |
| 49758 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | CDHR3 | Sponge network | -1.057 | 0.00029 | -0.977 | 4.0E-5 | 0.267 |
| 49759 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p | 15 | PIK3C2A | Sponge network | 0.556 | 0.00298 | 0.061 | 0.53776 | 0.267 |
| 49760 | U91328.19 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 20 | NAV3 | Sponge network | -0.303 | 0.21002 | 0.212 | 0.47729 | 0.267 |
| 49761 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | TET1 | Sponge network | 0.381 | 0.25967 | -0.135 | 0.52138 | 0.267 |
| 49762 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-93-5p;hsa-miR-935 | 11 | DAB2 | Sponge network | -0.162 | 0.47687 | 0.431 | 0.0113 | 0.267 |
| 49763 | LINC00969 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-625-5p | 11 | ARID2 | Sponge network | 0.683 | 1.0E-5 | 0.235 | 0.02697 | 0.267 |
| 49764 | EIF3J-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-411-5p;hsa-miR-505-3p;hsa-miR-590-3p | 19 | USP6 | Sponge network | 0.331 | 0.00509 | 0.188 | 0.3871 | 0.267 |
| 49765 | TMEM161B-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-23b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | BCLAF1 | Sponge network | -0.04 | 0.7606 | 0.211 | 0.00684 | 0.267 |
| 49766 | BZRAP1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-877-5p;hsa-miR-93-5p | 31 | IL17RD | Sponge network | -0.465 | 0.04703 | -0.019 | 0.94873 | 0.267 |
| 49767 | RP11-290F24.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-342-3p | 13 | RICTOR | Sponge network | 2.052 | 0 | 0.388 | 0.00188 | 0.267 |
| 49768 | FGD5-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-539-5p;hsa-miR-590-3p | 14 | NIN | Sponge network | 0.401 | 0.00066 | -0.253 | 0.13794 | 0.267 |
| 49769 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-532-3p;hsa-miR-940 | 15 | TRPS1 | Sponge network | 0.993 | 0 | -0.191 | 0.44109 | 0.267 |
| 49770 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 22 | KIF1B | Sponge network | 0.903 | 0 | -0.118 | 0.32258 | 0.267 |
| 49771 | FAM157B |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326 | 17 | CCDC6 | Sponge network | 0.92 | 0 | 0.27 | 0.02555 | 0.267 |
| 49772 | IQCH-AS1 |
hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | SLC5A3 | Sponge network | 0.929 | 0 | 0.493 | 5.0E-5 | 0.267 |
| 49773 | AC004893.11 |
hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-625-5p | 11 | NUAK1 | Sponge network | 1.317 | 0 | 0.904 | 0 | 0.267 |
| 49774 | RP11-274B21.10 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | MED12L | Sponge network | 1.073 | 0.00216 | -0.194 | 0.55299 | 0.267 |
| 49775 | RP11-160O5.1 |
hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-342-5p;hsa-miR-455-3p | 14 | CUX1 | Sponge network | -0.327 | 0.07353 | -0.039 | 0.6705 | 0.267 |
| 49776 | GS1-124K5.11 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-98-5p | 16 | KLF10 | Sponge network | 0.494 | 0.00339 | -0.783 | 0 | 0.267 |
| 49777 | RP5-1139B12.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p | 13 | PPP1R9A | Sponge network | 0.598 | 0.38481 | -0.903 | 0.04254 | 0.267 |
| 49778 | PSMG3-AS1 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | TPO | Sponge network | -0.125 | 0.49414 | -1.87 | 2.0E-5 | 0.267 |
| 49779 | RP11-385F5.5 |
hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326 | 10 | RICTOR | Sponge network | 1.076 | 0 | 0.388 | 0.00188 | 0.267 |
| 49780 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-940 | 26 | WDFY3 | Sponge network | 0.556 | 0.00298 | -0.201 | 0.0967 | 0.267 |
| 49781 | RP11-706C16.7 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-940 | 11 | PPP1R9A | Sponge network | 0.357 | 0.72407 | -0.903 | 0.04254 | 0.267 |
| 49782 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | NEDD4 | Sponge network | -0.769 | 0.00035 | 0.02 | 0.89834 | 0.267 |
| 49783 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-874-3p | 28 | TACC1 | Sponge network | -0.322 | 0.25945 | -0.362 | 0.05406 | 0.267 |
| 49784 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | TP53INP1 | Sponge network | -0.727 | 0.04726 | -0.496 | 0.00064 | 0.267 |
| 49785 | CALML3-AS1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p | 18 | DYNC1I1 | Sponge network | 0.95 | 0.15943 | -1.12 | 0.00107 | 0.267 |
| 49786 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-629-3p | 33 | IL6ST | Sponge network | 0.368 | 0.20093 | -0.402 | 0.00676 | 0.267 |
| 49787 | RP11-356J5.12 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | CITED2 | Sponge network | -0.899 | 6.0E-5 | -1.545 | 0 | 0.267 |
| 49788 | RP1-151F17.2 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 13 | IRS1 | Sponge network | 0.222 | 0.29133 | -0.026 | 0.88855 | 0.267 |
| 49789 | AC025171.1 |
hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 14 | USP12 | Sponge network | 0.998 | 0 | -0.102 | 0.40768 | 0.267 |
| 49790 | TRIM52-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-34a-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 11 | CNTNAP1 | Sponge network | -0.418 | 0.00068 | 0.358 | 0.13727 | 0.267 |
| 49791 | LINC00657 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | IL13RA1 | Sponge network | 0.91 | 0 | 0.019 | 0.86949 | 0.267 |
| 49792 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | PRKAA2 | Sponge network | 0.4 | 0.14611 | -2.462 | 0 | 0.267 |
| 49793 | NDUFA6-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-500b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 22 | ACSL6 | Sponge network | -0.355 | 0.0077 | -1.593 | 0 | 0.267 |
| 49794 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | NCOA1 | Sponge network | 0.225 | 0.30644 | -0.432 | 0 | 0.267 |
| 49795 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | ZMYM2 | Sponge network | 0.476 | 0.19203 | 0.279 | 0.01273 | 0.267 |
| 49796 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-577 | 15 | HIP1 | Sponge network | 0.921 | 0.00118 | 0.774 | 0 | 0.267 |
| 49797 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | FREM2 | Sponge network | -2.925 | 0 | -0.437 | 0.46353 | 0.267 |
| 49798 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | 0.537 | 0.00139 | -0.7 | 1.0E-5 | 0.267 |
| 49799 | CTD-3126B10.4 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | NR2C2 | Sponge network | 0.778 | 0.00101 | 0.21 | 0.03224 | 0.267 |
| 49800 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 15 | MYO9A | Sponge network | 1.073 | 0.00216 | -0.147 | 0.32373 | 0.267 |
| 49801 | LINC00963 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 16 | ARNT2 | Sponge network | 0.523 | 9.0E-5 | -0.332 | 0.25851 | 0.267 |
| 49802 | TMEM9B-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-766-3p | 21 | IL17RD | Sponge network | 0.529 | 0.00552 | -0.019 | 0.94873 | 0.267 |
| 49803 | CASC2 |
hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-381-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p | 22 | PPM1L | Sponge network | 0.385 | 0.01306 | -0.537 | 0.00616 | 0.267 |
| 49804 | AC093110.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p | 20 | ABI2 | Sponge network | 1.044 | 2.0E-5 | 0.175 | 0.14787 | 0.267 |
| 49805 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 23 | ARNT2 | Sponge network | -0.942 | 0 | -0.332 | 0.25851 | 0.267 |
| 49806 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | MAP3K12 | Sponge network | -0.663 | 0.10887 | -0.057 | 0.59369 | 0.267 |
| 49807 | RP11-890B15.3 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | SYT4 | Sponge network | 0.101 | 0.60919 | -3.207 | 0 | 0.267 |
| 49808 | SNHG14 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 20 | TES | Sponge network | -1.111 | 0.0003 | -0.348 | 0.00639 | 0.267 |
| 49809 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-942-5p | 32 | TMEM132B | Sponge network | -0.773 | 0.04073 | -0.83 | 0.01084 | 0.267 |
| 49810 | CALML3-AS1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-5p | 17 | PCDH10 | Sponge network | 0.95 | 0.15943 | -1.644 | 0.0078 | 0.267 |
| 49811 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-629-3p | 23 | AR | Sponge network | 1.316 | 0.01106 | -1.554 | 0 | 0.267 |
| 49812 | RP11-196G18.22 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-511-5p;hsa-miR-582-5p | 13 | ZFHX3 | Sponge network | 1.206 | 0 | 0.389 | 0.01363 | 0.267 |
| 49813 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-452-5p;hsa-miR-582-5p;hsa-miR-590-3p | 21 | FBXO32 | Sponge network | 0.274 | 0.07681 | -0.495 | 0.07561 | 0.267 |
| 49814 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 50 | PPM1A | Sponge network | -2.925 | 0 | -0.623 | 0 | 0.267 |
| 49815 | RP11-298J20.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-5p | 15 | TRPS1 | Sponge network | 0.002 | 0.99057 | -0.191 | 0.44109 | 0.267 |
| 49816 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 21 | PRKAB2 | Sponge network | 0.381 | 0.25967 | -0.367 | 0.01371 | 0.267 |
| 49817 | RP11-35G9.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ZFX | Sponge network | 0.657 | 4.0E-5 | 0.09 | 0.42563 | 0.267 |
| 49818 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-378a-5p | 12 | PDS5B | Sponge network | 0.4 | 0.14611 | 0.259 | 0.00962 | 0.267 |
| 49819 | CTD-2336O2.1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | -0.141 | 0.26434 | -1.74 | 0 | 0.267 |
| 49820 | WDFY3-AS2 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-942-5p | 30 | OPCML | Sponge network | -0.942 | 0 | -2.498 | 0 | 0.267 |
| 49821 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-542-3p;hsa-miR-590-3p | 15 | SLITRK6 | Sponge network | -0.777 | 0.1134 | -2.121 | 0 | 0.267 |
| 49822 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-484;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 17 | YAP1 | Sponge network | 0.41 | 0.02214 | 0.22 | 0.11836 | 0.267 |
| 49823 | AC109309.4 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-423-5p;hsa-miR-532-3p | 10 | CTIF | Sponge network | 0.163 | 0.87219 | -0.389 | 0.00549 | 0.267 |
| 49824 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | GIGYF2 | Sponge network | 0.986 | 0.01022 | 0.136 | 0.04403 | 0.267 |
| 49825 | RP11-395G23.3 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | RIMS2 | Sponge network | 0.381 | 0.25967 | -1.365 | 0.00208 | 0.267 |
| 49826 | H1FX-AS1 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MTSS1 | Sponge network | -0.206 | 0.12942 | -0.81 | 0.00035 | 0.267 |
| 49827 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | PCM1 | Sponge network | -0.129 | 0.57177 | 0.164 | 0.20307 | 0.267 |
| 49828 | RP11-867G23.10 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-361-3p | 15 | HIP1 | Sponge network | -2.531 | 0 | 0.774 | 0 | 0.267 |
| 49829 | CTD-2089N3.1 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p | 14 | CREB3L2 | Sponge network | -0.314 | 0.62033 | -0.525 | 2.0E-5 | 0.267 |
| 49830 | LINC00672 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-98-5p | 11 | TES | Sponge network | -0.227 | 0.31657 | -0.348 | 0.00639 | 0.267 |
| 49831 | RP11-218M22.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p | 33 | AR | Sponge network | 0.197 | 0.25618 | -1.554 | 0 | 0.267 |
| 49832 | MAGI2-AS3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | BRD3 | Sponge network | -0.129 | 0.57177 | 0.37 | 0.00013 | 0.267 |
| 49833 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | PHC3 | Sponge network | 0.272 | 0.44692 | 0.212 | 0.0499 | 0.267 |
| 49834 | AC108488.4 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-577;hsa-miR-625-5p;hsa-miR-7-1-3p | 26 | TGFBR3 | Sponge network | 0.259 | 0.12542 | -1.173 | 1.0E-5 | 0.267 |
| 49835 | RAMP2-AS1 |
hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-3614-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | -0.513 | 0.04703 | -2.417 | 0 | 0.267 |
| 49836 | RP4-758J18.10 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-331-3p | 11 | ABI2 | Sponge network | -0.091 | 0.71468 | 0.175 | 0.14787 | 0.267 |
| 49837 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TBX18 | Sponge network | -0.141 | 0.26434 | 0.843 | 0.02945 | 0.267 |
| 49838 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 15 | ACTB | Sponge network | 0.194 | 0.64278 | -0.247 | 0.01736 | 0.267 |
| 49839 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | AR | Sponge network | 0.537 | 0.00139 | -1.554 | 0 | 0.267 |
| 49840 | LINC01018 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-590-3p | 15 | STARD13 | Sponge network | -2.915 | 0.00015 | -0.187 | 0.27383 | 0.267 |
| 49841 | CTB-113P19.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | JAZF1 | Sponge network | 0.906 | 0.31715 | -0.377 | 0.02677 | 0.267 |
| 49842 | AC007879.7 |
hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-5p | 15 | RICTOR | Sponge network | 4.3 | 0 | 0.388 | 0.00188 | 0.267 |
| 49843 | LINC00473 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-874-3p | 26 | CTIF | Sponge network | -1.203 | 0.03881 | -0.389 | 0.00549 | 0.267 |
| 49844 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-93-5p | 24 | ST6GALNAC3 | Sponge network | -0.465 | 0.04703 | -0.987 | 0 | 0.267 |
| 49845 | CTD-3099C6.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-664a-5p | 21 | TRPS1 | Sponge network | 1.361 | 0 | -0.191 | 0.44109 | 0.267 |
| 49846 | JAZF1-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 46 | PTPRD | Sponge network | -0.769 | 0.00035 | -1.005 | 0.00064 | 0.267 |
| 49847 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | NCAM2 | Sponge network | 0.101 | 0.60919 | -0.482 | 0.22565 | 0.267 |
| 49848 | RP4-669P10.16 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-505-5p;hsa-miR-766-3p | 20 | PPARA | Sponge network | -0.301 | 0.06597 | -0.379 | 0.00548 | 0.267 |
| 49849 | LIPE-AS1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-539-5p;hsa-miR-582-5p | 10 | PHF3 | Sponge network | 0.078 | 0.55803 | 0.126 | 0.19652 | 0.267 |
| 49850 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 24 | NCOA1 | Sponge network | -0.125 | 0.49414 | -0.432 | 0 | 0.267 |
| 49851 | LINC00648 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PRKAB2 | Sponge network | 0.633 | 0.51678 | -0.367 | 0.01371 | 0.267 |
| 49852 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-345-5p;hsa-miR-450b-5p | 14 | CDHR3 | Sponge network | -4.546 | 0 | -0.977 | 4.0E-5 | 0.267 |
| 49853 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | MITF | Sponge network | 0.889 | 9.0E-5 | -0.769 | 0.00022 | 0.267 |
| 49854 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p | 33 | SPRY3 | Sponge network | 0.75 | 0.00054 | 0.016 | 0.91286 | 0.267 |
| 49855 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-92a-3p | 17 | UBE4B | Sponge network | -0.738 | 0.21233 | -0.235 | 0.007 | 0.267 |
| 49856 | RP11-20I23.13 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-582-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 30 | BMPR2 | Sponge network | 0.505 | 0.0014 | 0.206 | 0.0635 | 0.267 |
| 49857 | RP11-327P2.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p | 17 | PDE4DIP | Sponge network | -0.147 | 0.40452 | -0.282 | 0.0257 | 0.267 |
| 49858 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | IGF1 | Sponge network | -0.053 | 0.80685 | -0.204 | 0.5898 | 0.267 |
| 49859 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p | 23 | PIK3R1 | Sponge network | 0.657 | 4.0E-5 | -0.133 | 0.36854 | 0.267 |
| 49860 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-miR-135b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429 | 12 | ST6GAL2 | Sponge network | -0.785 | 0.00035 | -1.201 | 0.00597 | 0.267 |
| 49861 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 33 | AKAP11 | Sponge network | -0.612 | 0.00094 | 0.196 | 0.09825 | 0.267 |
| 49862 | RP13-1039J1.4 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p | 11 | HIP1 | Sponge network | 0.356 | 0.33467 | 0.774 | 0 | 0.267 |
| 49863 | AC003090.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | FMNL3 | Sponge network | -2.117 | 0.00161 | 0.555 | 4.0E-5 | 0.267 |
| 49864 | LINC00886 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-5p | 70 | LPP | Sponge network | -0.371 | 0.24604 | -0.459 | 0.04475 | 0.267 |
| 49865 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p | 11 | CHD5 | Sponge network | 0.542 | 0.00054 | -0.922 | 0.01521 | 0.267 |
| 49866 | RAD51-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-744-3p | 17 | EPS15 | Sponge network | 0.768 | 7.0E-5 | -0.052 | 0.53998 | 0.267 |
| 49867 | WDFY3-AS2 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935 | 30 | ACSL6 | Sponge network | -0.942 | 0 | -1.593 | 0 | 0.267 |
| 49868 | RP11-25K21.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-589-3p | 13 | ADARB1 | Sponge network | 0.489 | 0.25786 | -0.335 | 0.06631 | 0.267 |
| 49869 | RP11-218M22.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ARNT2 | Sponge network | 0.197 | 0.25618 | -0.332 | 0.25851 | 0.267 |
| 49870 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | THBS1 | Sponge network | -1.249 | 7.0E-5 | 0.741 | 0.00386 | 0.267 |
| 49871 | PDXDC2P |
hsa-miR-125a-3p;hsa-miR-125b-2-3p;hsa-miR-143-3p;hsa-miR-154-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p | 13 | LIMD1 | Sponge network | 0.756 | 0 | 0.103 | 0.28978 | 0.267 |
| 49872 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-143-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-7-5p | 14 | CLASP2 | Sponge network | 0.001 | 0.99753 | -0.038 | 0.71 | 0.267 |
| 49873 | RP11-379F4.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-374b-5p;hsa-miR-664a-3p | 12 | FZD3 | Sponge network | 1.316 | 0 | 0.104 | 0.60316 | 0.267 |
| 49874 | LINC00094 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429 | 11 | THSD7A | Sponge network | 0.253 | 0.09565 | 0.242 | 0.25154 | 0.267 |
| 49875 | EMX2OS |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PIK3CA | Sponge network | -0.777 | 0.1134 | 0.195 | 0.06908 | 0.267 |
| 49876 | PSMG3-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-429 | 13 | PLXNC1 | Sponge network | -0.125 | 0.49414 | 0.569 | 0.00329 | 0.267 |
| 49877 | NDUFA6-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 26 | PRKCB | Sponge network | -0.355 | 0.0077 | -1.313 | 2.0E-5 | 0.267 |
| 49878 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 12 | RBL2 | Sponge network | -0.811 | 2.0E-5 | -0.282 | 0.00254 | 0.267 |
| 49879 | RP11-203M5.7 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-429;hsa-miR-495-3p | 10 | ZFX | Sponge network | 1.684 | 1.0E-5 | 0.09 | 0.42563 | 0.267 |
| 49880 | TPTEP1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 26 | CYLD | Sponge network | -1.216 | 0 | -0.367 | 0.00171 | 0.267 |
| 49881 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940 | 41 | MEF2C | Sponge network | -1.047 | 0 | -0.499 | 0.01448 | 0.267 |
| 49882 | RP11-360F5.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | AFF4 | Sponge network | 1.867 | 0 | -0.266 | 0.01565 | 0.267 |
| 49883 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-454-3p | 21 | FOXO3 | Sponge network | 0.001 | 0.99753 | -0.04 | 0.72028 | 0.267 |
| 49884 | RP11-1277A3.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-505-3p | 12 | DIP2C | Sponge network | 0.453 | 0.01839 | -0.436 | 0.01713 | 0.267 |
| 49885 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-96-5p | 36 | PAFAH1B1 | Sponge network | 0.331 | 0.57247 | -0.429 | 0 | 0.267 |
| 49886 | RP11-428J1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-589-5p;hsa-miR-590-3p | 27 | ADARB1 | Sponge network | 0.538 | 0.00076 | -0.335 | 0.06631 | 0.267 |
| 49887 | C4A-AS1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-744-3p | 12 | ADAMTSL3 | Sponge network | 3.345 | 0 | -1.559 | 0 | 0.267 |
| 49888 | MAGI2-AS3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | CRTC1 | Sponge network | -0.129 | 0.57177 | -0.116 | 0.28412 | 0.267 |
| 49889 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | PTPRB | Sponge network | -1.349 | 0.00017 | 0.105 | 0.47122 | 0.267 |
| 49890 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | PRDM5 | Sponge network | -0.555 | 0.06156 | -1.09 | 0.00014 | 0.267 |
| 49891 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | QKI | Sponge network | 0.389 | 0.04293 | -0.333 | 0.04111 | 0.267 |
| 49892 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-671-5p | 14 | CRTC3 | Sponge network | -0.053 | 0.80685 | 0.164 | 0.16786 | 0.267 |
| 49893 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | FOXO3 | Sponge network | 1.317 | 0 | -0.04 | 0.72028 | 0.267 |
| 49894 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-629-3p | 12 | ZBTB4 | Sponge network | 1.122 | 0.02989 | -0.739 | 0 | 0.267 |
| 49895 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TBX18 | Sponge network | 0.603 | 0.00127 | 0.843 | 0.02945 | 0.267 |
| 49896 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | PAFAH1B1 | Sponge network | 0.572 | 0.01722 | -0.429 | 0 | 0.267 |
| 49897 | CKMT2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-501-5p;hsa-miR-590-3p | 11 | DCN | Sponge network | 0.082 | 0.55654 | -1.288 | 0 | 0.267 |
| 49898 | CTC-559E9.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-30d-3p;hsa-miR-324-5p | 10 | STARD13 | Sponge network | 0.594 | 0.00064 | -0.187 | 0.27383 | 0.267 |
| 49899 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 24 | MYO9A | Sponge network | -1.349 | 0.00017 | -0.147 | 0.32373 | 0.267 |
| 49900 | AC156455.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3065-3p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-484 | 19 | CBL | Sponge network | 1.154 | 0.00041 | 0.291 | 0.01267 | 0.267 |
| 49901 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-5p;hsa-miR-940 | 17 | CRTC3 | Sponge network | 0.921 | 0.00118 | 0.164 | 0.16786 | 0.267 |
| 49902 | SNHG14 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-942-5p | 16 | FBXO31 | Sponge network | -1.111 | 0.0003 | -0.085 | 0.42389 | 0.267 |
| 49903 | SNHG12 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 11 | PRKAA1 | Sponge network | 1.103 | 0 | 0.317 | 0.0031 | 0.267 |
| 49904 | NDUFA6-AS1 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 11 | TAF1 | Sponge network | -0.355 | 0.0077 | 0.39 | 3.0E-5 | 0.267 |
| 49905 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-589-5p;hsa-miR-93-5p | 13 | SLITRK3 | Sponge network | -0.13 | 0.48734 | -3.328 | 0 | 0.267 |
| 49906 | RP11-216L13.19 |
hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-497-5p;hsa-miR-664a-5p | 10 | SOX11 | Sponge network | 1.674 | 0 | 2.68 | 0 | 0.267 |
| 49907 | TTC25 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-590-5p;hsa-miR-769-5p;hsa-miR-940 | 11 | NTRK3 | Sponge network | -0.208 | 0.38261 | -1.77 | 3.0E-5 | 0.267 |
| 49908 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TBX18 | Sponge network | -0.08 | 0.90092 | 0.843 | 0.02945 | 0.267 |
| 49909 | A1BG-AS1 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-500a-3p;hsa-miR-589-3p | 12 | ZFHX4 | Sponge network | -0.164 | 0.45106 | 0.018 | 0.95574 | 0.267 |
| 49910 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MITF | Sponge network | 0.659 | 3.0E-5 | -0.769 | 0.00022 | 0.267 |
| 49911 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-629-5p | 31 | SOX5 | Sponge network | -0.147 | 0.40452 | -1.091 | 4.0E-5 | 0.267 |
| 49912 | LINC00657 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p | 13 | CBFB | Sponge network | 0.91 | 0 | 1.061 | 0 | 0.267 |
| 49913 | FGD5-AS1 |
hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | PREX2 | Sponge network | 0.401 | 0.00066 | -0.272 | 0.25382 | 0.267 |
| 49914 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 42 | CUX1 | Sponge network | 0.214 | 0.18246 | -0.039 | 0.6705 | 0.267 |
| 49915 | AGAP11 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-455-5p;hsa-miR-590-5p | 12 | THSD7A | Sponge network | -1.645 | 0 | 0.242 | 0.25154 | 0.267 |
| 49916 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-503-5p;hsa-miR-582-5p;hsa-miR-590-3p | 36 | RICTOR | Sponge network | 0.219 | 0.1516 | 0.388 | 0.00188 | 0.267 |
| 49917 | HOXB-AS3 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-550a-3p | 25 | RUNX1T1 | Sponge network | 0.343 | 0.28887 | -0.344 | 0.2374 | 0.267 |
| 49918 | RP11-400K9.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-423-5p;hsa-miR-590-5p | 13 | CCND2 | Sponge network | -0.733 | 0.19469 | -0.267 | 0.3112 | 0.267 |
| 49919 | RP11-356J5.12 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p | 17 | FAM135B | Sponge network | -0.899 | 6.0E-5 | -3.978 | 0 | 0.267 |
| 49920 | RP11-25K21.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-589-3p | 10 | PGR | Sponge network | 0.489 | 0.25786 | -1.704 | 0 | 0.267 |
| 49921 | FAM215B |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-7-1-3p | 13 | SYNE1 | Sponge network | 0.817 | 3.0E-5 | -0.738 | 0.00316 | 0.267 |
| 49922 | DNAJC27-AS1 |
hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-378a-3p;hsa-miR-7-1-3p | 11 | MLF1 | Sponge network | -0.969 | 2.0E-5 | -0.893 | 0.00727 | 0.267 |
| 49923 | CTD-2017D11.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-582-5p | 11 | LUZP2 | Sponge network | 0.792 | 0.0049 | -0.056 | 0.89416 | 0.267 |
| 49924 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-532-5p;hsa-miR-590-3p | 18 | SLITRK6 | Sponge network | -0.222 | 0.32445 | -2.121 | 0 | 0.267 |
| 49925 | OIP5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 44 | ADARB1 | Sponge network | 0.421 | 0.00084 | -0.335 | 0.06631 | 0.267 |
| 49926 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 23 | RBMS3 | Sponge network | 1.61 | 0.00961 | -0.519 | 0.03551 | 0.267 |
| 49927 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | EGFR | Sponge network | -0.285 | 0.2172 | -0.175 | 0.48451 | 0.267 |
| 49928 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-577 | 22 | GREM1 | Sponge network | 0.064 | 0.72934 | 0.902 | 0.01691 | 0.267 |
| 49929 | RP11-95D17.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-3607-3p | 14 | MOB1B | Sponge network | 0.75 | 0 | 0.197 | 0.15266 | 0.267 |
| 49930 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 66 | SMAD4 | Sponge network | -0.129 | 0.57177 | -0.313 | 0.00413 | 0.267 |
| 49931 | GNG12-AS1 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-421;hsa-miR-425-5p;hsa-miR-542-3p;hsa-miR-590-3p | 15 | RIT1 | Sponge network | -0.793 | 7.0E-5 | -0.296 | 0.00054 | 0.267 |
| 49932 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p | 16 | RET | Sponge network | -0.18 | 0.20343 | -0.833 | 0.03517 | 0.267 |
| 49933 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | EYA4 | Sponge network | 0.101 | 0.60919 | 0.3 | 0.36876 | 0.267 |
| 49934 | WDFY3-AS2 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | NR3C2 | Sponge network | -0.942 | 0 | -1.56 | 0 | 0.267 |
| 49935 | RP11-196G18.23 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-625-5p | 12 | CADM1 | Sponge network | 0.724 | 0.00039 | -0.9 | 0.00084 | 0.267 |
| 49936 | RP11-20I23.11 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | KIF1B | Sponge network | 0.875 | 0.00019 | -0.118 | 0.32258 | 0.267 |
| 49937 | RP1-151F17.2 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-708-5p | 28 | CCDC6 | Sponge network | 0.222 | 0.29133 | 0.27 | 0.02555 | 0.267 |
| 49938 | CD27-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 19 | PLSCR4 | Sponge network | 0.177 | 0.16681 | -0.474 | 0.03833 | 0.267 |
| 49939 | CALML3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p | 27 | MED12L | Sponge network | 0.95 | 0.15943 | -0.194 | 0.55299 | 0.267 |
| 49940 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | DACH2 | Sponge network | 0.335 | 0.20596 | -1.635 | 0.00034 | 0.267 |
| 49941 | RP1-59D14.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-629-3p | 19 | DST | Sponge network | 1.162 | 5.0E-5 | -0.713 | 0.00096 | 0.267 |
| 49942 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-93-3p | 16 | PAFAH1B1 | Sponge network | -0.932 | 0.00329 | -0.429 | 0 | 0.267 |
| 49943 | AC005618.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 20 | CADM1 | Sponge network | 0.862 | 0.06213 | -0.9 | 0.00084 | 0.267 |
| 49944 | RP11-774O3.3 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | OPCML | Sponge network | -0.487 | 0.02157 | -2.498 | 0 | 0.267 |
| 49945 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 13 | APC | Sponge network | 0.946 | 0 | -0.255 | 0.04051 | 0.267 |
| 49946 | LINC00472 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | ETV1 | Sponge network | -0.842 | 0.01361 | -0.096 | 0.67674 | 0.267 |
| 49947 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 38 | AR | Sponge network | -0.176 | 0.59692 | -1.554 | 0 | 0.267 |
| 49948 | OSER1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | GALNT13 | Sponge network | -0.13 | 0.48734 | -0.857 | 0.07439 | 0.267 |
| 49949 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-942-5p | 15 | CREBBP | Sponge network | -0.976 | 0.00025 | 0.126 | 0.15186 | 0.267 |
| 49950 | FZD10-AS1 |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 17 | EMP2 | Sponge network | -0.727 | 0.04726 | -0.423 | 0.00985 | 0.267 |
| 49951 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 21 | HDAC9 | Sponge network | -0.405 | 0.22013 | -0.755 | 0.00495 | 0.267 |
| 49952 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-655-3p | 12 | NF1 | Sponge network | 0.41 | 0.02214 | 0.51 | 0 | 0.267 |
| 49953 | CTC-301O7.4 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p | 12 | PTPRT | Sponge network | 0.292 | 0.43384 | -0.907 | 0.03727 | 0.267 |
| 49954 | AP001347.6 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-589-3p | 17 | MAFB | Sponge network | -1.161 | 0.00591 | -0.023 | 0.89865 | 0.267 |
| 49955 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | NEDD4 | Sponge network | -0.08 | 0.90092 | 0.02 | 0.89834 | 0.267 |
| 49956 | CTD-2336O2.1 |
hsa-miR-126-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SEMA3E | Sponge network | -0.141 | 0.26434 | -1.717 | 0.00137 | 0.267 |
| 49957 | AC093627.8 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-2110;hsa-miR-26b-5p | 14 | ZFHX4 | Sponge network | -0.787 | 0.3928 | 0.018 | 0.95574 | 0.267 |
| 49958 | CTD-2089N3.2 |
hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 12 | ZFHX3 | Sponge network | -1.375 | 0.08642 | 0.389 | 0.01363 | 0.267 |
| 49959 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 14 | LIFR | Sponge network | 0.693 | 0.00076 | -1.794 | 0 | 0.266 |
| 49960 | AC003090.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-940;hsa-miR-96-5p | 14 | POU2F2 | Sponge network | -2.117 | 0.00161 | -0.233 | 0.32214 | 0.266 |
| 49961 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 31 | CNTN1 | Sponge network | -0.83 | 0 | -2.594 | 0 | 0.266 |
| 49962 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | PDGFRA | Sponge network | 0.05 | 0.95483 | -0.372 | 0.0037 | 0.266 |
| 49963 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | ZFHX3 | Sponge network | 0.959 | 0 | 0.389 | 0.01363 | 0.266 |
| 49964 | HOXA-AS2 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-3p;hsa-miR-542-3p;hsa-miR-590-3p | 11 | USP25 | Sponge network | 0.61 | 0.01593 | -0.242 | 0.01397 | 0.266 |
| 49965 | RP11-262H14.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | MOB1B | Sponge network | -0.106 | 0.63074 | 0.197 | 0.15266 | 0.266 |
| 49966 | AC010226.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | CADM1 | Sponge network | -0.811 | 2.0E-5 | -0.9 | 0.00084 | 0.266 |
| 49967 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 14 | ABCA10 | Sponge network | -0.513 | 0.04703 | -0.571 | 0.03674 | 0.266 |
| 49968 | RP11-434B12.1 |
hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-339-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-654-3p | 10 | USP25 | Sponge network | -0.031 | 0.89063 | -0.242 | 0.01397 | 0.266 |
| 49969 | SRP14-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-361-3p;hsa-miR-486-5p;hsa-miR-766-3p;hsa-miR-93-5p | 13 | EZH1 | Sponge network | -0.061 | 0.70068 | -0.564 | 1.0E-5 | 0.266 |
| 49970 | U91328.19 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-484;hsa-miR-629-3p | 13 | ITGA5 | Sponge network | -0.303 | 0.21002 | 0.452 | 0.03524 | 0.266 |
| 49971 | RP11-347P5.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-374a-3p;hsa-miR-532-3p;hsa-miR-654-3p;hsa-miR-940 | 10 | NFIB | Sponge network | 1.364 | 0 | 0.023 | 0.90795 | 0.266 |
| 49972 | RP13-516M14.1 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | GALNT13 | Sponge network | 0.389 | 0.04293 | -0.857 | 0.07439 | 0.266 |
| 49973 | RP11-130L8.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | RELN | Sponge network | -0.663 | 0.10887 | -2.403 | 0 | 0.266 |
| 49974 | RP11-983P16.4 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-629-3p | 10 | SPTBN4 | Sponge network | 0.41 | 0.02214 | -0.786 | 0.01281 | 0.266 |
| 49975 | RP11-760H22.2 |
hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-374b-5p | 13 | SYNPO2 | Sponge network | 0.001 | 0.99753 | -2.342 | 0 | 0.266 |
| 49976 | PTOV1-AS1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 10 | GSK3B | Sponge network | 0.87 | 0 | 0.266 | 0.00045 | 0.266 |
| 49977 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p | 10 | MYO9A | Sponge network | 0.832 | 0 | -0.147 | 0.32373 | 0.266 |
| 49978 | TPTEP1 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FAM133A | Sponge network | -1.216 | 0 | 0.549 | 0.29009 | 0.266 |
| 49979 | HAR1A |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p | 11 | ERG | Sponge network | -0.672 | 0.19447 | 0.002 | 0.99011 | 0.266 |
| 49980 | KCNJ2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p;hsa-miR-671-5p | 12 | KIT | Sponge network | -0.436 | 0.05325 | -2.184 | 0 | 0.266 |
| 49981 | LINC00092 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | GALNT13 | Sponge network | -1.886 | 0 | -0.857 | 0.07439 | 0.266 |
| 49982 | MIR497HG |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 24 | RNASEL | Sponge network | -1.439 | 4.0E-5 | -0.272 | 0.01796 | 0.266 |
| 49983 | CTC-429P9.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p | 13 | GRIN2A | Sponge network | 0.497 | 0.01451 | -2.161 | 0 | 0.266 |
| 49984 | PCBP1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-5p | 14 | BCL2 | Sponge network | -0.567 | 0 | -0.899 | 0 | 0.266 |
| 49985 | AC034220.3 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p | 30 | PIK3R1 | Sponge network | 0.309 | 0.07304 | -0.133 | 0.36854 | 0.266 |
| 49986 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | IGF1 | Sponge network | 0.619 | 0.00157 | -0.204 | 0.5898 | 0.266 |
| 49987 | SNX29P2 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 24 | DIP2C | Sponge network | 0.4 | 0.14611 | -0.436 | 0.01713 | 0.266 |
| 49988 | RP13-516M14.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p | 22 | DLC1 | Sponge network | 0.389 | 0.04293 | -0.382 | 0.05038 | 0.266 |
| 49989 | RP1-122P22.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-629-3p | 11 | ASTN1 | Sponge network | 0.065 | 0.73468 | -2.389 | 2.0E-5 | 0.266 |
| 49990 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-378a-5p | 13 | PDS5B | Sponge network | -0.384 | 0.40652 | 0.259 | 0.00962 | 0.266 |
| 49991 | RP11-774O3.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p | 11 | ZYX | Sponge network | -0.487 | 0.02157 | 0.019 | 0.89763 | 0.266 |
| 49992 | CALML3-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-7-5p | 23 | SETBP1 | Sponge network | 0.95 | 0.15943 | -1.302 | 1.0E-5 | 0.266 |
| 49993 | RP11-296I10.3 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p | 10 | ITPKB | Sponge network | 0.592 | 0.00014 | -0.704 | 0.00106 | 0.266 |
| 49994 | SERTAD4-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-502-5p;hsa-miR-576-5p | 10 | TET1 | Sponge network | -0.932 | 0.00329 | -0.135 | 0.52138 | 0.266 |
| 49995 | RP11-968O1.5 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-484 | 14 | PRKCB | Sponge network | -0.829 | 0.01343 | -1.313 | 2.0E-5 | 0.266 |
| 49996 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-96-5p | 18 | SCN9A | Sponge network | 1.61 | 0.00961 | -0.525 | 0.10593 | 0.266 |
| 49997 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FREM2 | Sponge network | 1.04 | 0.00023 | -0.437 | 0.46353 | 0.266 |
| 49998 | RP11-867G23.10 |
hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-502-5p | 10 | PTCH1 | Sponge network | -2.531 | 0 | -0.1 | 0.6179 | 0.266 |
| 49999 | RP11-25K21.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p | 11 | JAZF1 | Sponge network | 0.489 | 0.25786 | -0.377 | 0.02677 | 0.266 |
| 50000 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-590-3p | 26 | NTRK2 | Sponge network | -0.555 | 0.06156 | -0.736 | 0.06495 | 0.266 |
| 50001 | LINC00403 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-589-5p | 12 | SOX5 | Sponge network | -3.586 | 0.00032 | -1.091 | 4.0E-5 | 0.266 |
| 50002 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | NEDD4 | Sponge network | -0.13 | 0.48734 | 0.02 | 0.89834 | 0.266 |
| 50003 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p | 17 | CREB3L2 | Sponge network | -1.255 | 0.00981 | -0.525 | 2.0E-5 | 0.266 |
| 50004 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-590-5p;hsa-miR-629-5p | 17 | GRIN2A | Sponge network | -0.147 | 0.40452 | -2.161 | 0 | 0.266 |
| 50005 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-539-5p | 18 | TMEM132B | Sponge network | -0.785 | 0.00035 | -0.83 | 0.01084 | 0.266 |
| 50006 | CTB-179K24.3 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-3677-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 10 | PRKAA1 | Sponge network | 0.841 | 0.04055 | 0.317 | 0.0031 | 0.266 |
| 50007 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | AFF4 | Sponge network | 0.898 | 1.0E-5 | -0.266 | 0.01565 | 0.266 |
| 50008 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | SPG20 | Sponge network | 0.898 | 1.0E-5 | -1.16 | 0 | 0.266 |
| 50009 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 44 | PRKAA2 | Sponge network | 0.349 | 0.01695 | -2.462 | 0 | 0.266 |
| 50010 | LINC00654 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 12 | FAM117A | Sponge network | 0.374 | 0.20054 | -1.379 | 0 | 0.266 |
| 50011 | RP11-834C11.4 |
hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | JAZF1 | Sponge network | -0.883 | 0.01839 | -0.377 | 0.02677 | 0.266 |
| 50012 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | CREB3L2 | Sponge network | -0.187 | 0.23787 | -0.525 | 2.0E-5 | 0.266 |
| 50013 | SH3RF3-AS1 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-486-5p | 10 | EZH1 | Sponge network | 0.889 | 9.0E-5 | -0.564 | 1.0E-5 | 0.266 |
| 50014 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-628-5p | 43 | FZD3 | Sponge network | -0.162 | 0.47687 | 0.104 | 0.60316 | 0.266 |
| 50015 | AC009948.5 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p | 23 | UBR3 | Sponge network | 0.532 | 0.00505 | -0.021 | 0.82774 | 0.266 |
| 50016 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-576-5p | 19 | EPHA5 | Sponge network | 0.594 | 0.00064 | -0.957 | 0.01733 | 0.266 |
| 50017 | TPTEP1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-3913-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p | 38 | AFF1 | Sponge network | -1.216 | 0 | -0.384 | 0.00053 | 0.266 |
| 50018 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 47 | SMAD2 | Sponge network | -0.487 | 0.02157 | -0.128 | 0.12732 | 0.266 |
| 50019 | EMX2OS |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 16 | TLE4 | Sponge network | -0.777 | 0.1134 | -1.157 | 0 | 0.266 |
| 50020 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | ZMYM2 | Sponge network | 0.556 | 0.00298 | 0.279 | 0.01273 | 0.266 |
| 50021 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 37 | ATRX | Sponge network | -0.737 | 0.06182 | 0.261 | 0.04408 | 0.266 |
| 50022 | BCYRN1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 21 | TBL1XR1 | Sponge network | 0.311 | 0.10344 | 0.578 | 0 | 0.266 |
| 50023 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | BCL11A | Sponge network | 0.064 | 0.72934 | -0.932 | 0.0063 | 0.266 |
| 50024 | CTD-3099C6.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-3p | 14 | NAV3 | Sponge network | 1.361 | 0 | 0.212 | 0.47729 | 0.266 |
| 50025 | ZNF667-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | EFNA5 | Sponge network | -0.976 | 0.00025 | -0.421 | 0.17868 | 0.266 |
| 50026 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 62 | AFF4 | Sponge network | 0.572 | 0.01722 | -0.266 | 0.01565 | 0.266 |
| 50027 | AC009120.5 |
hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26a-2-3p;hsa-miR-33a-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 13 | IL6ST | Sponge network | 1.383 | 0 | -0.402 | 0.00676 | 0.266 |
| 50028 | U91328.21 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p | 14 | EPHA5 | Sponge network | -0.735 | 0.15983 | -0.957 | 0.01733 | 0.266 |
| 50029 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-424-3p;hsa-miR-450b-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 17 | HOOK3 | Sponge network | -1.281 | 0.00012 | 0.273 | 0.09957 | 0.266 |
| 50030 | RP11-815I9.4 |
hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-24-2-5p;hsa-miR-3607-3p;hsa-miR-664a-3p | 10 | PPM1L | Sponge network | 0.537 | 0.0006 | -0.537 | 0.00616 | 0.266 |
| 50031 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 47 | IL6ST | Sponge network | 0.637 | 0.00069 | -0.402 | 0.00676 | 0.266 |
| 50032 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-93-5p | 29 | UNC80 | Sponge network | 0.299 | 0.57722 | -1.934 | 0 | 0.266 |
| 50033 | PCBP1-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10a-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-940 | 25 | CREB1 | Sponge network | -0.567 | 0 | 0.203 | 0.00537 | 0.266 |
| 50034 | GAS6-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-92a-3p | 17 | RIMBP2 | Sponge network | 0.16 | 0.50785 | -1.968 | 5.0E-5 | 0.266 |
| 50035 | AC034220.3 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 13 | PICALM | Sponge network | 0.309 | 0.07304 | 0.086 | 0.23523 | 0.266 |
| 50036 | KB-1460A1.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | TCF12 | Sponge network | 0.603 | 0.00127 | 0.174 | 0.13271 | 0.266 |
| 50037 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-92a-3p | 10 | ZNF521 | Sponge network | 0.219 | 0.1516 | -0.111 | 0.58068 | 0.266 |
| 50038 | RP11-1246C19.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-625-3p;hsa-miR-628-5p | 19 | BMPR2 | Sponge network | 1.352 | 0 | 0.206 | 0.0635 | 0.266 |
| 50039 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | WDFY3 | Sponge network | -0.053 | 0.80685 | -0.201 | 0.0967 | 0.266 |
| 50040 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-5p | 18 | ATRX | Sponge network | 0.176 | 0.37402 | 0.261 | 0.04408 | 0.266 |
| 50041 | FAM66C |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | KANK1 | Sponge network | -0.901 | 0.00019 | -0.55 | 0.00134 | 0.266 |
| 50042 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-96-5p | 23 | BCL2 | Sponge network | 0.453 | 0.01839 | -0.899 | 0 | 0.266 |
| 50043 | OSER1-AS1 |
hsa-let-7f-1-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-582-5p | 11 | PTCH1 | Sponge network | -0.13 | 0.48734 | -0.1 | 0.6179 | 0.266 |
| 50044 | DIO3OS |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-455-5p | 16 | ITPKB | Sponge network | -0.773 | 0.04073 | -0.704 | 0.00106 | 0.266 |
| 50045 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-34a-5p;hsa-miR-542-3p;hsa-miR-93-3p | 11 | BNC2 | Sponge network | -0.301 | 0.06597 | -0.491 | 0.09988 | 0.266 |
| 50046 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 16 | ZMYND11 | Sponge network | -0.246 | 0.35034 | -0.2 | 0.05449 | 0.266 |
| 50047 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-5p;hsa-miR-590-3p | 19 | SLITRK3 | Sponge network | -0.222 | 0.32445 | -3.328 | 0 | 0.266 |
| 50048 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-455-3p | 11 | EDA2R | Sponge network | 0.331 | 0.57247 | -0.587 | 0.01598 | 0.266 |
| 50049 | RP1-43E13.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-532-3p;hsa-miR-590-5p | 13 | PTPRT | Sponge network | -0.034 | 0.83228 | -0.907 | 0.03727 | 0.266 |
| 50050 | CTD-3099C6.9 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-429 | 10 | PRKAR1A | Sponge network | 1.361 | 0 | -0.063 | 0.50314 | 0.266 |
| 50051 | RP5-1021I20.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-33a-3p | 10 | SNX29 | Sponge network | -0.785 | 0.00035 | -0.34 | 0.01371 | 0.266 |
| 50052 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | FYN | Sponge network | -0.727 | 0.04726 | -0.131 | 0.37051 | 0.266 |
| 50053 | MIR22HG |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-369-3p;hsa-miR-493-3p | 10 | JAKMIP2 | Sponge network | -1.047 | 0 | -1.388 | 0 | 0.266 |
| 50054 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | PTGFR | Sponge network | 0.349 | 0.01695 | -0.233 | 0.45421 | 0.266 |
| 50055 | AC084219.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | PRKAB2 | Sponge network | 1.296 | 0.0054 | -0.367 | 0.01371 | 0.266 |
| 50056 | RP11-701H24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-93-5p | 15 | MAF | Sponge network | -2.039 | 0.03447 | -1.257 | 0 | 0.266 |
| 50057 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 11 | NR2C2 | Sponge network | -0.285 | 0.2172 | 0.21 | 0.03224 | 0.266 |
| 50058 | RP11-244H3.1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 19 | BAZ2B | Sponge network | 0.659 | 3.0E-5 | -0.276 | 0.02833 | 0.266 |
| 50059 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-98-5p | 26 | CLASP2 | Sponge network | -0.154 | 0.78939 | -0.038 | 0.71 | 0.266 |
| 50060 | LINC00622 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 15 | CBL | Sponge network | 1.169 | 0.00028 | 0.291 | 0.01267 | 0.266 |
| 50061 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | EYA4 | Sponge network | 0.603 | 0.00127 | 0.3 | 0.36876 | 0.266 |
| 50062 | RP11-158K1.3 |
hsa-miR-10b-5p;hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 17 | CSDE1 | Sponge network | 0.349 | 0.01695 | -0.493 | 0 | 0.266 |
| 50063 | AC024560.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-940 | 10 | BAZ2B | Sponge network | 2.166 | 0.00228 | -0.276 | 0.02833 | 0.266 |
| 50064 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 22 | NOTCH2 | Sponge network | 0.453 | 0.01839 | 0.138 | 0.35163 | 0.266 |
| 50065 | RP11-680C21.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-940 | 30 | NTRK2 | Sponge network | -1.424 | 0.04346 | -0.736 | 0.06495 | 0.266 |
| 50066 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-93-5p | 21 | WDFY3 | Sponge network | 0.348 | 0.32912 | -0.201 | 0.0967 | 0.266 |
| 50067 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-625-3p | 36 | FZD3 | Sponge network | 0.253 | 0.09565 | 0.104 | 0.60316 | 0.266 |
| 50068 | LINC01001 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-539-5p | 17 | KIF1B | Sponge network | 1.055 | 0 | -0.118 | 0.32258 | 0.266 |
| 50069 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429 | 18 | MYBL1 | Sponge network | 1.61 | 0.00961 | 0.494 | 0.02152 | 0.266 |
| 50070 | LINC00242 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | THRB | Sponge network | 0.745 | 0.00347 | -1.117 | 0.0004 | 0.266 |
| 50071 | CTB-113P19.4 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 17 | RIMS2 | Sponge network | 0.906 | 0.31715 | -1.365 | 0.00208 | 0.266 |
| 50072 | AC008746.12 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p | 10 | KIF1B | Sponge network | 2.279 | 0 | -0.118 | 0.32258 | 0.266 |
| 50073 | AC005154.6 |
hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p | 10 | LZTS1 | Sponge network | 0.848 | 0 | 1.664 | 0 | 0.266 |
| 50074 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-744-3p | 14 | DIP2C | Sponge network | 3.345 | 0 | -0.436 | 0.01713 | 0.266 |
| 50075 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-940 | 12 | TRIM33 | Sponge network | 0.419 | 0.0319 | 0.402 | 7.0E-5 | 0.266 |
| 50076 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p | 25 | FZD3 | Sponge network | 0.368 | 0.20093 | 0.104 | 0.60316 | 0.266 |
| 50077 | RP11-434H6.7 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-550a-5p | 15 | CCND2 | Sponge network | 0.358 | 0.14302 | -0.267 | 0.3112 | 0.266 |
| 50078 | NR2F1-AS1 |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-502-5p | 12 | CADM4 | Sponge network | -0.49 | 0.04946 | -0.635 | 0.00305 | 0.266 |
| 50079 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 21 | TRIM33 | Sponge network | 0.064 | 0.72934 | 0.402 | 7.0E-5 | 0.266 |
| 50080 | RP11-693J15.4 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | AKAP6 | Sponge network | -0.72 | 0.24239 | -1.586 | 3.0E-5 | 0.266 |
| 50081 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p | 10 | TRIM3 | Sponge network | -1.582 | 8.0E-5 | -0.577 | 0 | 0.266 |
| 50082 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | JAZF1 | Sponge network | -0.83 | 0 | -0.377 | 0.02677 | 0.266 |
| 50083 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | PTPN13 | Sponge network | 0.219 | 0.1516 | -1.208 | 0 | 0.266 |
| 50084 | MIR143HG |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | ZFX | Sponge network | -0.739 | 0.0574 | 0.09 | 0.42563 | 0.266 |
| 50085 | RP4-639F20.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-96-5p | 22 | PPM1A | Sponge network | -0.517 | 0.02935 | -0.623 | 0 | 0.266 |
| 50086 | RP11-2H8.2 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-590-3p;hsa-miR-744-3p | 10 | HIP1 | Sponge network | 1.089 | 0 | 0.774 | 0 | 0.266 |
| 50087 | CTC-297N7.9 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 17 | RASSF2 | Sponge network | -2.069 | 0 | -0.273 | 0.21961 | 0.266 |
| 50088 | LINC00672 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p | 16 | RET | Sponge network | -0.227 | 0.31657 | -0.833 | 0.03517 | 0.266 |
| 50089 | RASSF8-AS1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-508-3p;hsa-miR-7-1-3p | 11 | RASSF3 | Sponge network | 0.335 | 0.20596 | -0.226 | 0.11691 | 0.266 |
| 50090 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 11 | SEMA3E | Sponge network | 0.214 | 0.18246 | -1.717 | 0.00137 | 0.266 |
| 50091 | SNHG16 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-495-3p | 10 | DICER1 | Sponge network | 0.961 | 0 | 0.042 | 0.674 | 0.266 |
| 50092 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | PIK3C2A | Sponge network | 0.476 | 0.19203 | 0.061 | 0.53776 | 0.266 |
| 50093 | RP11-284N8.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-93-5p;hsa-miR-940 | 13 | MCC | Sponge network | -0.31 | 0.33871 | -1.005 | 0 | 0.266 |
| 50094 | RP11-967K21.1 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-589-3p | 13 | HOMER2 | Sponge network | 0.056 | 0.81195 | -2.698 | 0 | 0.266 |
| 50095 | TTC28-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p | 13 | SIRT1 | Sponge network | 0.373 | 0.00068 | 0.109 | 0.2593 | 0.266 |
| 50096 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | EBF1 | Sponge network | 0.381 | 0.25967 | -0.384 | 0.08856 | 0.266 |
| 50097 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-375;hsa-miR-590-3p | 11 | MYBL1 | Sponge network | 0.768 | 7.0E-5 | 0.494 | 0.02152 | 0.266 |
| 50098 | AC141928.1 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 11 | ADCY1 | Sponge network | 0.853 | 0.15352 | 0.362 | 0.16982 | 0.266 |
| 50099 | RP11-539I5.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | FOXO3 | Sponge network | -0.468 | 0.32587 | -0.04 | 0.72028 | 0.266 |
| 50100 | RP11-21A7A.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-455-3p | 11 | BTG2 | Sponge network | -1.116 | 0.23686 | -1.763 | 0 | 0.266 |
| 50101 | AP001347.6 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-577;hsa-miR-590-3p | 11 | PRKAR1A | Sponge network | -1.161 | 0.00591 | -0.063 | 0.50314 | 0.266 |
| 50102 | NR2F1-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-452-5p;hsa-miR-508-3p;hsa-miR-590-3p | 18 | INPP4B | Sponge network | -0.49 | 0.04946 | -0.097 | 0.60503 | 0.266 |
| 50103 | LINC00865 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 20 | PIK3CA | Sponge network | -1.249 | 7.0E-5 | 0.195 | 0.06908 | 0.266 |
| 50104 | CTD-2528L19.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | COPS2 | Sponge network | 0.953 | 0 | -0.178 | 0.04974 | 0.266 |
| 50105 | FENDRR |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-96-5p | 29 | CADM1 | Sponge network | -1.281 | 0.00012 | -0.9 | 0.00084 | 0.266 |
| 50106 | RP11-156E6.1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-497-5p | 20 | MDM4 | Sponge network | 1.266 | 0 | 0.345 | 0.00762 | 0.266 |
| 50107 | CTD-3099C6.9 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p | 11 | NRK | Sponge network | 1.361 | 0 | 0.656 | 0.19217 | 0.266 |
| 50108 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | FREM2 | Sponge network | 0.532 | 0.00505 | -0.437 | 0.46353 | 0.266 |
| 50109 | DNM3OS |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NRG3 | Sponge network | 0.572 | 0.01722 | -1.836 | 4.0E-5 | 0.266 |
| 50110 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | PPP1R9A | Sponge network | -0.031 | 0.89063 | -0.903 | 0.04254 | 0.266 |
| 50111 | LINC00621 |
hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-576-5p | 11 | ST6GAL2 | Sponge network | 0.131 | 0.8436 | -1.201 | 0.00597 | 0.266 |
| 50112 | CTD-2089N3.1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-455-5p | 10 | LRP1B | Sponge network | -0.314 | 0.62033 | -2.163 | 0 | 0.266 |
| 50113 | RP11-800A3.7 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | ZFHX4 | Sponge network | 0.18 | 0.7147 | 0.018 | 0.95574 | 0.266 |
| 50114 | AP001469.9 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-486-5p | 13 | CBFB | Sponge network | 1.096 | 0 | 1.061 | 0 | 0.266 |
| 50115 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 46 | TCF4 | Sponge network | -0.176 | 0.59692 | 0.016 | 0.92271 | 0.266 |
| 50116 | RBPMS-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-454-3p;hsa-miR-93-5p | 12 | PGR | Sponge network | 0.144 | 0.64989 | -1.704 | 0 | 0.266 |
| 50117 | HOXA-AS2 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 25 | RIMS2 | Sponge network | 0.61 | 0.01593 | -1.365 | 0.00208 | 0.266 |
| 50118 | RP11-395G23.3 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-590-3p | 16 | BRD3 | Sponge network | 0.381 | 0.25967 | 0.37 | 0.00013 | 0.266 |
| 50119 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 14 | YAP1 | Sponge network | 1.063 | 0 | 0.22 | 0.11836 | 0.266 |
| 50120 | CTD-2554C21.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-7-5p;hsa-miR-96-5p | 19 | CLASP2 | Sponge network | -1.654 | 0.00144 | -0.038 | 0.71 | 0.266 |
| 50121 | RP11-517B11.7 |
hsa-miR-10b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-493-5p;hsa-miR-590-5p | 12 | CTDSPL | Sponge network | 0.706 | 0 | 0.085 | 0.45121 | 0.266 |
| 50122 | LINC00909 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-497-5p;hsa-miR-654-3p | 10 | RICTOR | Sponge network | 0.363 | 0.00601 | 0.388 | 0.00188 | 0.266 |
| 50123 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | EYA4 | Sponge network | -0.769 | 0.00035 | 0.3 | 0.36876 | 0.266 |
| 50124 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.349 | 0.01695 | 0.943 | 0 | 0.266 |
| 50125 | CKMT2-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 14 | CELF4 | Sponge network | 0.082 | 0.55654 | -1.135 | 0.00344 | 0.266 |
| 50126 | NEAT1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 16 | PDS5B | Sponge network | 0.705 | 0.00014 | 0.259 | 0.00962 | 0.266 |
| 50127 | RP1-302G2.5 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p | 12 | LRP1B | Sponge network | -1.483 | 9.0E-5 | -2.163 | 0 | 0.266 |
| 50128 | XXyac-YX65C7_A.2 |
hsa-miR-106a-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 14 | WDFY3 | Sponge network | 0.885 | 1.0E-5 | -0.201 | 0.0967 | 0.266 |
| 50129 | RP11-1277A3.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-96-5p | 13 | SCN9A | Sponge network | 0.453 | 0.01839 | -0.525 | 0.10593 | 0.266 |
| 50130 | CTC-457L16.2 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-935 | 11 | PCDH10 | Sponge network | 1.153 | 0.00114 | -1.644 | 0.0078 | 0.266 |
| 50131 | CTA-363E19.2 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 11 | FOXO3 | Sponge network | 1.055 | 0 | -0.04 | 0.72028 | 0.266 |
| 50132 | RP11-13J8.1 |
hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-328-3p;hsa-miR-495-3p;hsa-miR-628-5p | 10 | VPS53 | Sponge network | 3.03 | 0 | 0.103 | 0.22667 | 0.266 |
| 50133 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | AFF4 | Sponge network | 0.272 | 0.44692 | -0.266 | 0.01565 | 0.266 |
| 50134 | RP11-356J5.12 |
hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-590-3p | 10 | GLIPR2 | Sponge network | -0.899 | 6.0E-5 | -0.996 | 0 | 0.266 |
| 50135 | RP11-181G12.2 |
hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 10 | TET1 | Sponge network | 0.556 | 0.00298 | -0.135 | 0.52138 | 0.266 |
| 50136 | DOCK9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-455-3p | 13 | PLSCR4 | Sponge network | -0.231 | 0.28214 | -0.474 | 0.03833 | 0.266 |
| 50137 | NDUFA6-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ZDBF2 | Sponge network | -0.355 | 0.0077 | -0.998 | 0.00025 | 0.266 |
| 50138 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 17 | KALRN | Sponge network | -2.117 | 0.00161 | -0.819 | 1.0E-5 | 0.266 |
| 50139 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p | 11 | PRKAA1 | Sponge network | 0.439 | 0.00313 | 0.317 | 0.0031 | 0.266 |
| 50140 | MRVI1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 13 | YAP1 | Sponge network | -1.092 | 4.0E-5 | 0.22 | 0.11836 | 0.266 |
| 50141 | C4A-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-628-5p | 23 | DTNA | Sponge network | 3.345 | 0 | -1.648 | 1.0E-5 | 0.266 |
| 50142 | HCG11 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 21 | GJA1 | Sponge network | 0.385 | 0.10067 | -0.485 | 0.02179 | 0.266 |
| 50143 | IQCH-AS1 |
hsa-miR-10b-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 16 | ARHGEF12 | Sponge network | 0.929 | 0 | 0.09 | 0.35693 | 0.266 |
| 50144 | RP11-77H9.2 |
hsa-let-7a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 13 | ZMYM2 | Sponge network | 1.677 | 0 | 0.279 | 0.01273 | 0.266 |
| 50145 | ZNF32-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-629-3p | 18 | KIF1B | Sponge network | 1.552 | 7.0E-5 | -0.118 | 0.32258 | 0.266 |
| 50146 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 32 | PIK3R1 | Sponge network | -1.654 | 0.00144 | -0.133 | 0.36854 | 0.266 |
| 50147 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-624-5p | 20 | LATS1 | Sponge network | 0.659 | 3.0E-5 | 0.109 | 0.34208 | 0.266 |
| 50148 | RP11-102N12.3 |
hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-7-1-3p | 13 | LIMCH1 | Sponge network | -0.351 | 0.11358 | -0.915 | 1.0E-5 | 0.266 |
| 50149 | SNHG5 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-5p;hsa-miR-664a-3p | 16 | UBE2D1 | Sponge network | 0.13 | 0.47174 | 0.468 | 0 | 0.266 |
| 50150 | RP11-6N17.3 |
hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-744-3p | 10 | EPS15 | Sponge network | 0.108 | 0.69556 | -0.052 | 0.53998 | 0.266 |
| 50151 | AC009120.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 18 | ADARB1 | Sponge network | 0.919 | 0 | -0.335 | 0.06631 | 0.266 |
| 50152 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 18 | GRIN2A | Sponge network | -0.031 | 0.89063 | -2.161 | 0 | 0.266 |
| 50153 | U91328.19 |
hsa-miR-126-3p;hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p | 14 | CXCL12 | Sponge network | -0.303 | 0.21002 | -1.421 | 0 | 0.266 |
| 50154 | AC005618.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-628-5p;hsa-miR-93-3p | 29 | ERC1 | Sponge network | 0.862 | 0.06213 | -0.108 | 0.38794 | 0.266 |
| 50155 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-374b-5p;hsa-miR-495-3p | 12 | HACE1 | Sponge network | 0.373 | 0.00068 | -0.072 | 0.55239 | 0.266 |
| 50156 | PCA3 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 21 | DOCK4 | Sponge network | -0.709 | 0.18168 | 0.502 | 0.00108 | 0.266 |
| 50157 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | PIK3C2A | Sponge network | 0.799 | 1.0E-5 | 0.061 | 0.53776 | 0.266 |
| 50158 | RP11-506M13.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | SLC5A3 | Sponge network | 0.769 | 0 | 0.493 | 5.0E-5 | 0.266 |
| 50159 | RP11-400K9.3 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-576-5p | 10 | ST6GALNAC3 | Sponge network | -0.733 | 0.19469 | -0.987 | 0 | 0.266 |
| 50160 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | PHC3 | Sponge network | -0.49 | 0.04946 | 0.212 | 0.0499 | 0.266 |
| 50161 | ADAMTS9-AS2 |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-374a-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | HOMER2 | Sponge network | -2.452 | 0 | -2.698 | 0 | 0.266 |
| 50162 | RP11-13K12.5 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 16 | PCDH17 | Sponge network | -0.318 | 0.65847 | 0.731 | 2.0E-5 | 0.266 |
| 50163 | RP11-571M6.17 |
hsa-miR-127-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29b-2-5p;hsa-miR-330-3p;hsa-miR-590-3p | 14 | KIF1B | Sponge network | 1.013 | 0 | -0.118 | 0.32258 | 0.266 |
| 50164 | LINC00473 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 32 | MAF | Sponge network | -1.203 | 0.03881 | -1.257 | 0 | 0.266 |
| 50165 | PART1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | GALNT13 | Sponge network | -2.925 | 0 | -0.857 | 0.07439 | 0.266 |
| 50166 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 10 | ABCC9 | Sponge network | 0.765 | 7.0E-5 | -0.466 | 0.14121 | 0.266 |
| 50167 | RP11-532F6.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p | 18 | RNASEL | Sponge network | -0.896 | 0.00099 | -0.272 | 0.01796 | 0.266 |
| 50168 | A1BG-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-500a-3p;hsa-miR-93-3p | 19 | RUNX1T1 | Sponge network | -0.164 | 0.45106 | -0.344 | 0.2374 | 0.266 |
| 50169 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | ZMYM2 | Sponge network | -0.323 | 0.01372 | 0.279 | 0.01273 | 0.266 |
| 50170 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484;hsa-miR-629-3p | 15 | ITGA5 | Sponge network | 0.083 | 0.66405 | 0.452 | 0.03524 | 0.266 |
| 50171 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-7-1-3p | 13 | TBX18 | Sponge network | 0.056 | 0.8494 | 0.843 | 0.02945 | 0.266 |
| 50172 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-940 | 23 | IGF1 | Sponge network | 0.389 | 0.04293 | -0.204 | 0.5898 | 0.266 |
| 50173 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-96-5p | 22 | FOXP1 | Sponge network | 0.622 | 0.00015 | 0.042 | 0.74368 | 0.266 |
| 50174 | RP11-473I1.5 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-378a-5p | 23 | DST | Sponge network | 0.542 | 0.00054 | -0.713 | 0.00096 | 0.266 |
| 50175 | JAZF1-AS1 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | RABEP1 | Sponge network | -0.769 | 0.00035 | -0.066 | 0.43142 | 0.266 |
| 50176 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | AHNAK | Sponge network | -1.057 | 0.00029 | -1.207 | 0 | 0.266 |
| 50177 | CTC-429P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-27a-3p | 11 | KCNA6 | Sponge network | 0.497 | 0.01451 | -1.531 | 0.00013 | 0.266 |
| 50178 | HAR1A |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-942-5p | 13 | NRXN1 | Sponge network | -0.672 | 0.19447 | -3.778 | 0 | 0.266 |
| 50179 | OIP5-AS1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-598-3p;hsa-miR-664a-3p | 15 | MN1 | Sponge network | 0.421 | 0.00084 | -0.358 | 0.24172 | 0.266 |
| 50180 | RASSF8-AS1 |
hsa-miR-146a-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 12 | ADCY1 | Sponge network | 0.335 | 0.20596 | 0.362 | 0.16982 | 0.266 |
| 50181 | RP11-251G23.5 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-2-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-628-5p | 20 | VPS53 | Sponge network | 1.344 | 0 | 0.103 | 0.22667 | 0.266 |
| 50182 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-628-5p | 21 | BMPR2 | Sponge network | 0.93 | 0 | 0.206 | 0.0635 | 0.266 |
| 50183 | LINC00672 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-214-5p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-590-3p | 23 | MXI1 | Sponge network | -0.227 | 0.31657 | -0.987 | 0 | 0.266 |
| 50184 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-590-5p | 22 | CBL | Sponge network | 1.04 | 0.00023 | 0.291 | 0.01267 | 0.266 |
| 50185 | ARHGEF26-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 26 | PIK3CA | Sponge network | -2.46 | 0 | 0.195 | 0.06908 | 0.266 |
| 50186 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421 | 20 | NTRK2 | Sponge network | -0.285 | 0.2172 | -0.736 | 0.06495 | 0.266 |
| 50187 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-181c-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 15 | TRIM3 | Sponge network | -1.047 | 0 | -0.577 | 0 | 0.266 |
| 50188 | ARHGEF26-AS1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ITGB1 | Sponge network | -2.46 | 0 | 0.524 | 0.00017 | 0.266 |
| 50189 | LINC00657 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p | 41 | MDM4 | Sponge network | 0.91 | 0 | 0.345 | 0.00762 | 0.266 |
| 50190 | BCYRN1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FAM133A | Sponge network | 0.311 | 0.10344 | 0.549 | 0.29009 | 0.266 |
| 50191 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 23 | PIK3C2A | Sponge network | 0.194 | 0.64278 | 0.061 | 0.53776 | 0.266 |
| 50192 | HOXB-AS2 |
hsa-miR-1266-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-576-5p | 11 | PBX1 | Sponge network | 1.316 | 0.01106 | -1.206 | 0 | 0.266 |
| 50193 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 11 | CAV1 | Sponge network | -0.513 | 0.04703 | -1.013 | 6.0E-5 | 0.266 |
| 50194 | AC004540.4 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p | 27 | LPP | Sponge network | -2.549 | 0.00528 | -0.459 | 0.04475 | 0.266 |
| 50195 | DOCK9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-500a-3p;hsa-miR-940 | 17 | HLF | Sponge network | -0.231 | 0.28214 | -2.443 | 0 | 0.266 |
| 50196 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-199b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-654-3p | 10 | PHF6 | Sponge network | 0.72 | 0.0001 | 0.785 | 0 | 0.266 |
| 50197 | CTD-2017D11.1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-484;hsa-miR-550a-5p;hsa-miR-940 | 13 | CHD5 | Sponge network | 0.792 | 0.0049 | -0.922 | 0.01521 | 0.266 |
| 50198 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-766-3p | 37 | IL6ST | Sponge network | 0.381 | 0.25967 | -0.402 | 0.00676 | 0.266 |
| 50199 | LINC00843 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | USP9X | Sponge network | 1.106 | 0 | 0.222 | 0.01689 | 0.266 |
| 50200 | CKMT2-AS1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-590-3p | 15 | TLE4 | Sponge network | 0.082 | 0.55654 | -1.157 | 0 | 0.266 |
| 50201 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | CCND2 | Sponge network | -0.031 | 0.89063 | -0.267 | 0.3112 | 0.266 |
| 50202 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 17 | LRRC55 | Sponge network | -0.08 | 0.90092 | -0.026 | 0.93163 | 0.266 |
| 50203 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | ARID5B | Sponge network | 1.073 | 0.00216 | 0.342 | 0.01676 | 0.266 |
| 50204 | CALML3-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 13 | EHD3 | Sponge network | 0.95 | 0.15943 | -0.484 | 0.01747 | 0.266 |
| 50205 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-664a-3p;hsa-miR-664a-5p | 21 | SOX11 | Sponge network | 0.614 | 1.0E-5 | 2.68 | 0 | 0.266 |
| 50206 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | RNF144A | Sponge network | -2.117 | 0.00161 | 0.594 | 0.0009 | 0.266 |
| 50207 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 14 | UBE4B | Sponge network | -0.246 | 0.35034 | -0.235 | 0.007 | 0.266 |
| 50208 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-93-5p | 31 | AFF4 | Sponge network | -0.473 | 0.07602 | -0.266 | 0.01565 | 0.266 |
| 50209 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p | 32 | SPRY3 | Sponge network | 0.61 | 0.01593 | 0.016 | 0.91286 | 0.266 |
| 50210 | KB-1410C5.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-940 | 13 | NTRK2 | Sponge network | 0.959 | 0 | -0.736 | 0.06495 | 0.266 |
| 50211 | LINC00265 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 14 | DDX3X | Sponge network | 1.07 | 0 | -0.152 | 0.07686 | 0.266 |
| 50212 | RP11-767N6.7 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3607-3p | 14 | CCDC6 | Sponge network | 0.466 | 0.00155 | 0.27 | 0.02555 | 0.266 |
| 50213 | RP11-373D23.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-335-5p;hsa-miR-7-1-3p | 10 | CDH19 | Sponge network | -0.285 | 0.2172 | -3.434 | 0 | 0.266 |
| 50214 | LINC00969 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-505-3p | 13 | PTPN11 | Sponge network | 0.683 | 1.0E-5 | 0.431 | 2.0E-5 | 0.266 |
| 50215 | U91328.19 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-625-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -0.303 | 0.21002 | -0.087 | 0.69193 | 0.266 |
| 50216 | RP11-439L18.2 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-93-3p | 13 | QKI | Sponge network | 0.224 | 0.48719 | -0.333 | 0.04111 | 0.266 |
| 50217 | AP001347.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | PRRX1 | Sponge network | -1.161 | 0.00591 | 1.316 | 0 | 0.266 |
| 50218 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-93-5p | 45 | PTEN | Sponge network | -2.69 | 0.0001 | -0.187 | 0.07748 | 0.265 |
| 50219 | CTA-204B4.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-744-3p | 14 | DIP2C | Sponge network | 0.765 | 7.0E-5 | -0.436 | 0.01713 | 0.265 |
| 50220 | LINC00854 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-628-5p;hsa-miR-940 | 13 | QKI | Sponge network | 1.18 | 0 | -0.333 | 0.04111 | 0.265 |
| 50221 | HOXA-AS2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p | 16 | BCL6 | Sponge network | 0.61 | 0.01593 | -0.211 | 0.19489 | 0.265 |
| 50222 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | PCDH8 | Sponge network | 0.194 | 0.64278 | -0.997 | 0.05366 | 0.265 |
| 50223 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | NTRK3 | Sponge network | 0.348 | 0.32912 | -1.77 | 3.0E-5 | 0.265 |
| 50224 | MYLK-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p | 12 | CSMD1 | Sponge network | -1.331 | 3.0E-5 | -0.63 | 0.18881 | 0.265 |
| 50225 | ACTA2-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-337-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | KDSR | Sponge network | 0.194 | 0.64278 | 0.239 | 0.02216 | 0.265 |
| 50226 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-93-3p | 25 | FOXO3 | Sponge network | -0.303 | 0.21002 | -0.04 | 0.72028 | 0.265 |
| 50227 | WAC-AS1 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 12 | ERCC6 | Sponge network | 0.821 | 0 | 0.23 | 0.015 | 0.265 |
| 50228 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 11 | TACC1 | Sponge network | -0.011 | 0.967 | -0.362 | 0.05406 | 0.265 |
| 50229 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-7-1-3p | 13 | DKK3 | Sponge network | 0.538 | 0.00076 | -0.052 | 0.77783 | 0.265 |
| 50230 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | PRKAB2 | Sponge network | 0.972 | 0 | -0.367 | 0.01371 | 0.265 |
| 50231 | CTD-3138B18.6 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-2-5p;hsa-miR-590-3p | 16 | KIF1B | Sponge network | 2.248 | 0 | -0.118 | 0.32258 | 0.265 |
| 50232 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 40 | ACVR2A | Sponge network | 0.101 | 0.60919 | -0.409 | 0.00039 | 0.265 |
| 50233 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 29 | SPRY3 | Sponge network | 0.683 | 1.0E-5 | 0.016 | 0.91286 | 0.265 |
| 50234 | AP000487.5 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-338-5p | 16 | RICTOR | Sponge network | 1.987 | 0 | 0.388 | 0.00188 | 0.265 |
| 50235 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | THBS1 | Sponge network | -0.811 | 2.0E-5 | 0.741 | 0.00386 | 0.265 |
| 50236 | RP11-102N12.3 |
hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-942-5p | 25 | THRB | Sponge network | -0.351 | 0.11358 | -1.117 | 0.0004 | 0.265 |
| 50237 | RP11-20I23.11 |
hsa-let-7a-3p;hsa-miR-10b-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-590-3p | 10 | ATRX | Sponge network | 0.875 | 0.00019 | 0.261 | 0.04408 | 0.265 |
| 50238 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 13 | FYN | Sponge network | -0.487 | 0.02157 | -0.131 | 0.37051 | 0.265 |
| 50239 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-339-5p;hsa-miR-369-3p | 10 | PRDM2 | Sponge network | 0.542 | 0.00054 | -0.258 | 0.00721 | 0.265 |
| 50240 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 20 | ACVR1 | Sponge network | 0.72 | 0.0001 | 0.279 | 0.00234 | 0.265 |
| 50241 | PSMG3-AS1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 22 | PCDH10 | Sponge network | -0.125 | 0.49414 | -1.644 | 0.0078 | 0.265 |
| 50242 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-582-3p | 17 | CBL | Sponge network | -0.473 | 0.07602 | 0.291 | 0.01267 | 0.265 |
| 50243 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | NR4A3 | Sponge network | -2.915 | 0.00015 | -1.385 | 0 | 0.265 |
| 50244 | LINC00578 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-576-5p | 11 | KDSR | Sponge network | 0.811 | 0.03228 | 0.239 | 0.02216 | 0.265 |
| 50245 | CTC-471J1.2 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | RASSF8 | Sponge network | 1.11 | 1.0E-5 | -0.122 | 0.65591 | 0.265 |
| 50246 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | LRRC7 | Sponge network | 0.342 | 0.03129 | -1.341 | 0.01193 | 0.265 |
| 50247 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-93-5p;hsa-miR-96-5p | 24 | IL17RD | Sponge network | 0.056 | 0.8494 | -0.019 | 0.94873 | 0.265 |
| 50248 | RP11-59H7.3 |
hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p | 11 | SIRT1 | Sponge network | 2.163 | 0 | 0.109 | 0.2593 | 0.265 |
| 50249 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p | 26 | PGR | Sponge network | 0.75 | 0.00054 | -1.704 | 0 | 0.265 |
| 50250 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 52 | TCF4 | Sponge network | 0.619 | 0.00157 | 0.016 | 0.92271 | 0.265 |
| 50251 | AC005562.1 |
hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 14 | SUFU | Sponge network | 0.488 | 0.00084 | -0.04 | 0.65489 | 0.265 |
| 50252 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p | 26 | BMPR2 | Sponge network | 1.073 | 0.00216 | 0.206 | 0.0635 | 0.265 |
| 50253 | CTC-429P9.3 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p | 13 | ABCB5 | Sponge network | 0.082 | 0.55397 | -0.956 | 0.04077 | 0.265 |
| 50254 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-7-1-3p | 12 | SON | Sponge network | 0.796 | 4.0E-5 | 0.168 | 0.04406 | 0.265 |
| 50255 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 37 | MEF2C | Sponge network | -0.355 | 0.0077 | -0.499 | 0.01448 | 0.265 |
| 50256 | LINC00641 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | RET | Sponge network | -0.222 | 0.32445 | -0.833 | 0.03517 | 0.265 |
| 50257 | RP11-1277A3.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-92a-3p | 10 | ROBO1 | Sponge network | 0.453 | 0.01839 | -0.009 | 0.96154 | 0.265 |
| 50258 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p | 27 | AFF4 | Sponge network | 1.317 | 0 | -0.266 | 0.01565 | 0.265 |
| 50259 | RP11-277P12.20 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | RASSF8 | Sponge network | 0.687 | 0.02753 | -0.122 | 0.65591 | 0.265 |
| 50260 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-27b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-378c;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p | 23 | LPP | Sponge network | -0.023 | 0.95109 | -0.459 | 0.04475 | 0.265 |
| 50261 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | FOXO1 | Sponge network | -1.575 | 6.0E-5 | -0.141 | 0.33874 | 0.265 |
| 50262 | BCYRN1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-96-5p | 53 | CADM2 | Sponge network | 0.311 | 0.10344 | -3.782 | 0 | 0.265 |
| 50263 | KB-1460A1.5 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 17 | NCAM2 | Sponge network | 0.603 | 0.00127 | -0.482 | 0.22565 | 0.265 |
| 50264 | RP11-127B20.2 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-3614-5p | 13 | JAZF1 | Sponge network | 0.411 | 0.1335 | -0.377 | 0.02677 | 0.265 |
| 50265 | RP11-981G7.6 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-7-1-3p | 10 | UBE4B | Sponge network | 0.986 | 0.01022 | -0.235 | 0.007 | 0.265 |
| 50266 | HOXB-AS3 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-1-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-452-5p;hsa-miR-625-5p | 15 | ABI2 | Sponge network | 0.343 | 0.28887 | 0.175 | 0.14787 | 0.265 |
| 50267 | MIR600HG |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 10 | ROBO1 | Sponge network | 0.594 | 0.41522 | -0.009 | 0.96154 | 0.265 |
| 50268 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-1-3p | 21 | LRP1B | Sponge network | -0.176 | 0.59692 | -2.163 | 0 | 0.265 |
| 50269 | PWAR6 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TRIM13 | Sponge network | -1.575 | 6.0E-5 | 0.183 | 0.06968 | 0.265 |
| 50270 | RP11-400N13.3 | hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-378a-3p;hsa-miR-378c | 12 | ABL2 | Sponge network | 8.612 | 0 | 0.82 | 0 | 0.265 |
| 50271 | MYLK-AS1 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-940 | 11 | SIRT1 | Sponge network | -1.331 | 3.0E-5 | 0.109 | 0.2593 | 0.265 |
| 50272 | AC093627.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-324-3p;hsa-miR-7-5p | 13 | ZBTB4 | Sponge network | -0.787 | 0.3928 | -0.739 | 0 | 0.265 |
| 50273 | LINC00654 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NRG3 | Sponge network | 0.374 | 0.20054 | -1.836 | 4.0E-5 | 0.265 |
| 50274 | RP11-701H24.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-93-5p | 18 | PTPRD | Sponge network | -2.039 | 0.03447 | -1.005 | 0.00064 | 0.265 |
| 50275 | AC034220.3 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | EPG5 | Sponge network | 0.309 | 0.07304 | 0.101 | 0.3563 | 0.265 |
| 50276 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p | 14 | BCL6 | Sponge network | 0.16 | 0.50785 | -0.211 | 0.19489 | 0.265 |
| 50277 | AC016747.3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-940 | 17 | GALNT13 | Sponge network | 0.199 | 0.38015 | -0.857 | 0.07439 | 0.265 |
| 50278 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-337-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PBRM1 | Sponge network | 0.194 | 0.64278 | -0.029 | 0.78601 | 0.265 |
| 50279 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 20 | TRIM33 | Sponge network | 0.569 | 0.00067 | 0.402 | 7.0E-5 | 0.265 |
| 50280 | RP11-182J1.1 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-7-1-3p | 18 | DDX6 | Sponge network | -0.187 | 0.23787 | 0.091 | 0.36428 | 0.265 |
| 50281 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-374b-5p;hsa-miR-495-3p | 10 | BCLAF1 | Sponge network | 0.8 | 0 | 0.211 | 0.00684 | 0.265 |
| 50282 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NCOA1 | Sponge network | 0.252 | 0.08508 | -0.432 | 0 | 0.265 |
| 50283 | GNG12-AS1 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p | 16 | IRS1 | Sponge network | -0.793 | 7.0E-5 | -0.026 | 0.88855 | 0.265 |
| 50284 | PAN3-AS1 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-33a-3p | 10 | CREB3L2 | Sponge network | 0.062 | 0.55274 | -0.525 | 2.0E-5 | 0.265 |
| 50285 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-582-3p | 16 | ATRX | Sponge network | -0.154 | 0.53954 | 0.261 | 0.04408 | 0.265 |
| 50286 | AGAP11 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p | 15 | RELN | Sponge network | -1.645 | 0 | -2.403 | 0 | 0.265 |
| 50287 | RP11-499P20.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-493-5p | 10 | ZFHX3 | Sponge network | 0.187 | 0.44121 | 0.389 | 0.01363 | 0.265 |
| 50288 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 24 | SOX11 | Sponge network | 0.959 | 0 | 2.68 | 0 | 0.265 |
| 50289 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p | 23 | ZMYM2 | Sponge network | 0.401 | 0.00066 | 0.279 | 0.01273 | 0.265 |
| 50290 | RP11-298J20.4 |
hsa-let-7g-3p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-671-5p | 11 | GAS7 | Sponge network | 0.002 | 0.99057 | -0.555 | 0.01602 | 0.265 |
| 50291 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ZNF208 | Sponge network | 0.083 | 0.66405 | -0.4 | 0.22381 | 0.265 |
| 50292 | LINC00654 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p | 17 | KANK1 | Sponge network | 0.374 | 0.20054 | -0.55 | 0.00134 | 0.265 |
| 50293 | PCBP1-AS1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-92b-3p | 12 | TRIM13 | Sponge network | -0.567 | 0 | 0.183 | 0.06968 | 0.265 |
| 50294 | SNHG16 |
hsa-let-7e-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-452-5p;hsa-miR-497-5p | 10 | COPS2 | Sponge network | 0.961 | 0 | -0.178 | 0.04974 | 0.265 |
| 50295 | RP11-890B15.3 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-590-3p | 13 | DDX3X | Sponge network | 0.101 | 0.60919 | -0.152 | 0.07686 | 0.265 |
| 50296 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-655-3p | 24 | PIK3C2A | Sponge network | 0.417 | 0.00445 | 0.061 | 0.53776 | 0.265 |
| 50297 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 50 | IL6ST | Sponge network | -0.668 | 3.0E-5 | -0.402 | 0.00676 | 0.265 |
| 50298 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-940 | 18 | PTPRT | Sponge network | 0.174 | 0.30034 | -0.907 | 0.03727 | 0.265 |
| 50299 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429 | 13 | ARNT2 | Sponge network | -0.469 | 0.08721 | -0.332 | 0.25851 | 0.265 |
| 50300 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-590-3p | 28 | EPHA7 | Sponge network | -0.322 | 0.25945 | -2.197 | 3.0E-5 | 0.265 |
| 50301 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-651-5p;hsa-miR-7-1-3p | 16 | WWTR1 | Sponge network | 0.419 | 0.0319 | -0.345 | 0.11997 | 0.265 |
| 50302 | RP11-47A8.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p | 10 | SYNE1 | Sponge network | 0.103 | 0.43169 | -0.738 | 0.00316 | 0.265 |
| 50303 | RP11-539I5.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 30 | PRKAA2 | Sponge network | -0.468 | 0.32587 | -2.462 | 0 | 0.265 |
| 50304 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 20 | TRPS1 | Sponge network | 0.673 | 0 | -0.191 | 0.44109 | 0.265 |
| 50305 | SH3BP5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | MED12L | Sponge network | 0.267 | 0.2937 | -0.194 | 0.55299 | 0.265 |
| 50306 | AC127904.2 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-3607-3p | 13 | RASAL2 | Sponge network | 1.308 | 0 | 0.869 | 0 | 0.265 |
| 50307 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | TACC1 | Sponge network | 0.619 | 0.00157 | -0.362 | 0.05406 | 0.265 |
| 50308 | LINC00473 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p | 25 | CRTC1 | Sponge network | -1.203 | 0.03881 | -0.116 | 0.28412 | 0.265 |
| 50309 | HOXA-AS2 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | CAV1 | Sponge network | 0.61 | 0.01593 | -1.013 | 6.0E-5 | 0.265 |
| 50310 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-27a-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p | 11 | PDS5B | Sponge network | 0.373 | 0.00068 | 0.259 | 0.00962 | 0.265 |
| 50311 | RP4-669P10.16 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-224-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-93-3p | 13 | SNX29 | Sponge network | -0.301 | 0.06597 | -0.34 | 0.01371 | 0.265 |
| 50312 | RP11-597D13.9 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | CAV1 | Sponge network | 1.302 | 7.0E-5 | -1.013 | 6.0E-5 | 0.265 |
| 50313 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-625-5p | 14 | ABI2 | Sponge network | 0.357 | 0.72407 | 0.175 | 0.14787 | 0.265 |
| 50314 | BVES-AS1 |
hsa-let-7a-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429 | 12 | MAFB | Sponge network | -2.62 | 0 | -0.023 | 0.89865 | 0.265 |
| 50315 | RP11-725P16.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-3p | 12 | RIMBP2 | Sponge network | 0.422 | 0.27021 | -1.968 | 5.0E-5 | 0.265 |
| 50316 | AC024560.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30a-3p | 11 | NR2C2 | Sponge network | 2.166 | 0.00228 | 0.21 | 0.03224 | 0.265 |
| 50317 | RP11-20I23.13 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 32 | RUNX1T1 | Sponge network | 0.505 | 0.0014 | -0.344 | 0.2374 | 0.265 |
| 50318 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 19 | TCF12 | Sponge network | -0.096 | 0.67958 | 0.174 | 0.13271 | 0.265 |
| 50319 | TMEM161B-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | -0.04 | 0.7606 | -0.57 | 0 | 0.265 |
| 50320 | SNHG14 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | DNMT3A | Sponge network | -1.111 | 0.0003 | 0.302 | 0.0262 | 0.265 |
| 50321 | A2M-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 11 | CITED2 | Sponge network | -0.08 | 0.90092 | -1.545 | 0 | 0.265 |
| 50322 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 32 | NCOA1 | Sponge network | -0.899 | 6.0E-5 | -0.432 | 0 | 0.265 |
| 50323 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-93-5p | 16 | TP53INP1 | Sponge network | -1.047 | 0 | -0.496 | 0.00064 | 0.265 |
| 50324 | RP11-20I23.13 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-92a-3p | 13 | NRXN3 | Sponge network | 0.505 | 0.0014 | -0.893 | 0.04186 | 0.265 |
| 50325 | ZRANB2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 13 | ARNT2 | Sponge network | -0.376 | 0.00337 | -0.332 | 0.25851 | 0.265 |
| 50326 | ADAMTS9-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-452-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | INPP4B | Sponge network | -2.452 | 0 | -0.097 | 0.60503 | 0.265 |
| 50327 | RP1-59D14.5 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-940 | 17 | WDFY3 | Sponge network | 1.162 | 5.0E-5 | -0.201 | 0.0967 | 0.265 |
| 50328 | RP11-307C18.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-188-5p;hsa-miR-26b-3p;hsa-miR-3607-3p;hsa-miR-576-5p | 10 | RICTOR | Sponge network | 1.082 | 0.00023 | 0.388 | 0.00188 | 0.265 |
| 50329 | AC025335.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-337-3p | 10 | CDC73 | Sponge network | 0.783 | 0 | 0.094 | 0.20463 | 0.265 |
| 50330 | A2M-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 24 | LRP1B | Sponge network | -0.08 | 0.90092 | -2.163 | 0 | 0.265 |
| 50331 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-590-3p | 10 | HOOK3 | Sponge network | 0.051 | 0.83388 | 0.273 | 0.09957 | 0.265 |
| 50332 | FGD5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p | 22 | DIP2C | Sponge network | 0.401 | 0.00066 | -0.436 | 0.01713 | 0.265 |
| 50333 | RP11-195F19.9 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-29a-5p | 11 | KANK1 | Sponge network | -0.726 | 0.00132 | -0.55 | 0.00134 | 0.265 |
| 50334 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | RNF144A | Sponge network | 0.986 | 0.01022 | 0.594 | 0.0009 | 0.265 |
| 50335 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 19 | HECA | Sponge network | -0.611 | 0.09607 | -0.217 | 0.03463 | 0.265 |
| 50336 | CTD-2006H14.2 |
hsa-miR-129-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | CCDC6 | Sponge network | 1.378 | 0 | 0.27 | 0.02555 | 0.265 |
| 50337 | FENDRR |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-590-3p | 12 | TMTC2 | Sponge network | -1.281 | 0.00012 | -0.215 | 0.18248 | 0.265 |
| 50338 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 14 | DAB2 | Sponge network | -0.08 | 0.90092 | 0.431 | 0.0113 | 0.265 |
| 50339 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 12 | CHD1 | Sponge network | 0.082 | 0.55654 | 0.322 | 0.00215 | 0.265 |
| 50340 | RP11-672A2.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p | 11 | TMEM132B | Sponge network | 1.344 | 0 | -0.83 | 0.01084 | 0.265 |
| 50341 | GNG12-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-744-3p | 12 | IGF2 | Sponge network | -0.793 | 7.0E-5 | 0.848 | 0.03842 | 0.265 |
| 50342 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ZFHX3 | Sponge network | 0.545 | 0.00064 | 0.389 | 0.01363 | 0.265 |
| 50343 | SH3RF3-AS1 |
hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 22 | PPM1L | Sponge network | 0.889 | 9.0E-5 | -0.537 | 0.00616 | 0.265 |
| 50344 | DIO3OS |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-625-5p;hsa-miR-877-5p;hsa-miR-940 | 32 | PTPRT | Sponge network | -0.773 | 0.04073 | -0.907 | 0.03727 | 0.265 |
| 50345 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 30 | BNC2 | Sponge network | -0.125 | 0.49414 | -0.491 | 0.09988 | 0.265 |
| 50346 | RP11-473I1.9 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 41 | TRPS1 | Sponge network | 0.683 | 1.0E-5 | -0.191 | 0.44109 | 0.265 |
| 50347 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 14 | FGF5 | Sponge network | -0.563 | 0.02545 | 0.549 | 0.28659 | 0.265 |
| 50348 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-424-5p;hsa-miR-484;hsa-miR-96-5p | 12 | CADM1 | Sponge network | -0.187 | 0.23787 | -0.9 | 0.00084 | 0.265 |
| 50349 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10a-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-337-3p;hsa-miR-34c-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-708-5p;hsa-miR-92b-3p;hsa-miR-940 | 35 | AFF4 | Sponge network | -0.567 | 0 | -0.266 | 0.01565 | 0.265 |
| 50350 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CXXC4 | Sponge network | -0.021 | 0.93598 | -0.433 | 0.21055 | 0.265 |
| 50351 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | PHC3 | Sponge network | 0.727 | 3.0E-5 | 0.212 | 0.0499 | 0.265 |
| 50352 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | LRRC7 | Sponge network | 0.219 | 0.1516 | -1.341 | 0.01193 | 0.265 |
| 50353 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-93-5p;hsa-miR-942-5p | 14 | FBXO31 | Sponge network | 0.249 | 0.35902 | -0.085 | 0.42389 | 0.265 |
| 50354 | RP11-403I13.8 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-744-3p | 12 | IGF2 | Sponge network | 0.619 | 0.00157 | 0.848 | 0.03842 | 0.265 |
| 50355 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p | 12 | TSC22D1 | Sponge network | -0.737 | 0.06182 | -0.529 | 2.0E-5 | 0.265 |
| 50356 | RP11-77H9.2 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p | 17 | KIF1B | Sponge network | 1.677 | 0 | -0.118 | 0.32258 | 0.265 |
| 50357 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p | 21 | PRKCE | Sponge network | -0.358 | 0.17255 | -0.403 | 0.00056 | 0.265 |
| 50358 | RP11-758N13.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p | 16 | IL17RD | Sponge network | 2.939 | 3.0E-5 | -0.019 | 0.94873 | 0.265 |
| 50359 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-425-5p;hsa-miR-96-5p | 15 | QKI | Sponge network | 3.21 | 0 | -0.333 | 0.04111 | 0.265 |
| 50360 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-3p | 17 | CLASP2 | Sponge network | 1.073 | 0.00216 | -0.038 | 0.71 | 0.265 |
| 50361 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 13 | SLITRK3 | Sponge network | 0.225 | 0.30644 | -3.328 | 0 | 0.265 |
| 50362 | RP11-725P16.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p | 29 | DST | Sponge network | 0.422 | 0.27021 | -0.713 | 0.00096 | 0.265 |
| 50363 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 52 | SMAD4 | Sponge network | -0.612 | 0.00094 | -0.313 | 0.00413 | 0.265 |
| 50364 | AC141928.1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-942-5p | 27 | IGFBP5 | Sponge network | 0.853 | 0.15352 | -0.806 | 0.00105 | 0.265 |
| 50365 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ATRX | Sponge network | -0.663 | 0.10887 | 0.261 | 0.04408 | 0.265 |
| 50366 | KB-1410C5.5 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-7-1-3p | 11 | TSC1 | Sponge network | 0.959 | 0 | 0.103 | 0.26071 | 0.265 |
| 50367 | EMX2OS |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p | 20 | RET | Sponge network | -0.777 | 0.1134 | -0.833 | 0.03517 | 0.265 |
| 50368 | RP11-806O11.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-376c-3p;hsa-miR-590-3p | 11 | TRIM33 | Sponge network | 0.284 | 0.52463 | 0.402 | 7.0E-5 | 0.265 |
| 50369 | SOX2-OT |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 11 | USP25 | Sponge network | -1.264 | 6.0E-5 | -0.242 | 0.01397 | 0.265 |
| 50370 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-409-3p | 16 | EPHA5 | Sponge network | 0.453 | 0.01839 | -0.957 | 0.01733 | 0.265 |
| 50371 | ADIRF-AS1 |
hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 26 | DIP2C | Sponge network | -0.223 | 0.47816 | -0.436 | 0.01713 | 0.265 |
| 50372 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 38 | TMEM132B | Sponge network | 0.385 | 0.10067 | -0.83 | 0.01084 | 0.265 |
| 50373 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 18 | PTGFR | Sponge network | -0.355 | 0.0077 | -0.233 | 0.45421 | 0.265 |
| 50374 | ADIRF-AS1 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | FGFR1 | Sponge network | -0.223 | 0.47816 | -0.509 | 0.03957 | 0.265 |
| 50375 | MRVI1-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p | 11 | MNT | Sponge network | -1.092 | 4.0E-5 | -0.157 | 0.06598 | 0.265 |
| 50376 | HAR1A |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-877-5p | 13 | ASTN1 | Sponge network | -0.672 | 0.19447 | -2.389 | 2.0E-5 | 0.265 |
| 50377 | LINC00265 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p | 14 | SLC5A3 | Sponge network | 1.07 | 0 | 0.493 | 5.0E-5 | 0.265 |
| 50378 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 11 | AKAP12 | Sponge network | 0.05 | 0.95483 | -0.932 | 0.0028 | 0.265 |
| 50379 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-577 | 10 | NALCN | Sponge network | 0.389 | 0.04293 | 1.068 | 0.00237 | 0.265 |
| 50380 | RP11-597D13.9 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-664a-3p | 14 | RASAL2 | Sponge network | 1.302 | 7.0E-5 | 0.869 | 0 | 0.265 |
| 50381 | RP11-686D22.4 |
hsa-miR-106a-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-577 | 11 | RASSF2 | Sponge network | 0.793 | 5.0E-5 | -0.273 | 0.21961 | 0.265 |
| 50382 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PHC3 | Sponge network | 1.205 | 0 | 0.212 | 0.0499 | 0.265 |
| 50383 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-628-5p | 18 | MIER3 | Sponge network | 1.254 | 0 | -0.212 | 0.04957 | 0.265 |
| 50384 | AC025171.1 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | RASSF2 | Sponge network | 0.998 | 0 | -0.273 | 0.21961 | 0.265 |
| 50385 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-98-5p | 11 | CHD3 | Sponge network | -1.216 | 0 | -0.001 | 0.99669 | 0.265 |
| 50386 | LINC00641 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 32 | CTIF | Sponge network | -0.222 | 0.32445 | -0.389 | 0.00549 | 0.265 |
| 50387 | RP11-983P16.4 |
hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-7-1-3p | 18 | PCDH9 | Sponge network | 0.41 | 0.02214 | -1.823 | 4.0E-5 | 0.265 |
| 50388 | RP11-774O3.3 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | BRD3 | Sponge network | -0.487 | 0.02157 | 0.37 | 0.00013 | 0.265 |
| 50389 | RP11-400K9.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-93-3p | 13 | UNC5C | Sponge network | -0.611 | 0.09607 | -0.178 | 0.40214 | 0.265 |
| 50390 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-590-3p | 10 | MGA | Sponge network | -0.358 | 0.17255 | 0.323 | 0.00792 | 0.265 |
| 50391 | RP11-435O5.2 |
hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 10 | PAFAH1B2 | Sponge network | 2.783 | 0 | 0.318 | 0.0001 | 0.265 |
| 50392 | LINC00265 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-2355-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-766-3p | 18 | AFF4 | Sponge network | 1.07 | 0 | -0.266 | 0.01565 | 0.265 |
| 50393 | RP11-384O8.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-429 | 10 | DUSP1 | Sponge network | -0.738 | 0.21233 | -1.754 | 0 | 0.265 |
| 50394 | HAND2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 30 | EFNA5 | Sponge network | -1.697 | 0.00889 | -0.421 | 0.17868 | 0.265 |
| 50395 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-628-5p | 13 | BCL6 | Sponge network | 0.559 | 0.00026 | -0.211 | 0.19489 | 0.265 |
| 50396 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p | 15 | LRRC7 | Sponge network | 0.921 | 0.00118 | -1.341 | 0.01193 | 0.265 |
| 50397 | AC002117.1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MED12L | Sponge network | 0.476 | 0.19203 | -0.194 | 0.55299 | 0.265 |
| 50398 | LINC01018 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 27 | NIN | Sponge network | -2.915 | 0.00015 | -0.253 | 0.13794 | 0.265 |
| 50399 | RP11-401P9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-93-5p | 27 | MAF | Sponge network | -0.384 | 0.40652 | -1.257 | 0 | 0.265 |
| 50400 | RP1-59D14.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p | 12 | NCOA1 | Sponge network | 1.162 | 5.0E-5 | -0.432 | 0 | 0.265 |
| 50401 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-590-5p | 11 | LRRC55 | Sponge network | 0.331 | 0.57247 | -0.026 | 0.93163 | 0.265 |
| 50402 | LPP-AS2 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-7-5p | 26 | IGFBP5 | Sponge network | -0.18 | 0.20343 | -0.806 | 0.00105 | 0.265 |
| 50403 | BDNF-AS |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-940;hsa-miR-96-5p | 10 | FOXO4 | Sponge network | -0.668 | 3.0E-5 | -0.516 | 2.0E-5 | 0.265 |
| 50404 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 60 | SMAD4 | Sponge network | -0.737 | 0.06182 | -0.313 | 0.00413 | 0.265 |
| 50405 | AC074289.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-935;hsa-miR-940 | 28 | GRIK3 | Sponge network | -0.326 | 0.14314 | -3.208 | 0 | 0.265 |
| 50406 | RP11-148O21.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-2355-3p;hsa-miR-361-5p | 10 | AR | Sponge network | 0.233 | 0.81029 | -1.554 | 0 | 0.265 |
| 50407 | FGF14-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 12 | PDE4DIP | Sponge network | -1.582 | 8.0E-5 | -0.282 | 0.0257 | 0.265 |
| 50408 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p | 15 | MYBL1 | Sponge network | 0.619 | 0.00157 | 0.494 | 0.02152 | 0.265 |
| 50409 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-629-3p | 15 | RBL2 | Sponge network | -2.62 | 0 | -0.282 | 0.00254 | 0.265 |
| 50410 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 16 | IL6ST | Sponge network | 0.062 | 0.55274 | -0.402 | 0.00676 | 0.265 |
| 50411 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p | 19 | ETV1 | Sponge network | 1.757 | 0 | -0.096 | 0.67674 | 0.265 |
| 50412 | SH3BP5-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | CEP170 | Sponge network | 0.267 | 0.2937 | 0.943 | 0 | 0.265 |
| 50413 | AC005562.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-625-5p | 17 | PTPN14 | Sponge network | 0.488 | 0.00084 | 0.144 | 0.34595 | 0.265 |
| 50414 | FLNB-AS1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-766-3p | 22 | MDM4 | Sponge network | 1.163 | 0.00018 | 0.345 | 0.00762 | 0.265 |
| 50415 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-582-5p | 17 | USP6 | Sponge network | 0.663 | 0.00073 | 0.188 | 0.3871 | 0.265 |
| 50416 | RP11-983P16.4 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | GABRB2 | Sponge network | 0.41 | 0.02214 | -1.841 | 0.00029 | 0.265 |
| 50417 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 11 | BRD3 | Sponge network | 1.055 | 0 | 0.37 | 0.00013 | 0.265 |
| 50418 | RP11-981G7.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-337-3p;hsa-miR-590-3p | 11 | RPS6KA3 | Sponge network | 0.986 | 0.01022 | 0.256 | 0.02419 | 0.265 |
| 50419 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NOTCH2NL | Sponge network | 0.272 | 0.44692 | 0.024 | 0.84307 | 0.265 |
| 50420 | CTD-3126B10.4 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | SON | Sponge network | 0.778 | 0.00101 | 0.168 | 0.04406 | 0.265 |
| 50421 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 35 | CBL | Sponge network | -1.697 | 0.00889 | 0.291 | 0.01267 | 0.265 |
| 50422 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-582-5p | 14 | MOB1B | Sponge network | 0.832 | 0 | 0.197 | 0.15266 | 0.265 |
| 50423 | RP11-705C15.3 |
hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-5p | 10 | PIK3R1 | Sponge network | 0.796 | 4.0E-5 | -0.133 | 0.36854 | 0.265 |
| 50424 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-532-5p;hsa-miR-660-5p | 11 | LRRC4 | Sponge network | -0.773 | 0.04073 | -1.774 | 0 | 0.265 |
| 50425 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | ZNF770 | Sponge network | 0.349 | 0.01695 | 0.206 | 0.06751 | 0.265 |
| 50426 | LPP-AS2 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-589-3p | 21 | PRDM2 | Sponge network | -0.18 | 0.20343 | -0.258 | 0.00721 | 0.265 |
| 50427 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-455-3p;hsa-miR-484 | 16 | ZFX | Sponge network | 0.453 | 0.01839 | 0.09 | 0.42563 | 0.265 |
| 50428 | MEG3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 25 | CYLD | Sponge network | 0.249 | 0.35902 | -0.367 | 0.00171 | 0.265 |
| 50429 | RP11-458F8.4 |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-27b-3p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-5p | 12 | SOX11 | Sponge network | 1.518 | 0 | 2.68 | 0 | 0.265 |
| 50430 | C4A-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-96-5p | 11 | ABI2 | Sponge network | 3.345 | 0 | 0.175 | 0.14787 | 0.265 |
| 50431 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-93-5p | 27 | TACC1 | Sponge network | 0.051 | 0.83388 | -0.362 | 0.05406 | 0.265 |
| 50432 | FZD10-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | CAV1 | Sponge network | -0.727 | 0.04726 | -1.013 | 6.0E-5 | 0.265 |
| 50433 | LA16c-321D4.2 |
hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-654-3p | 11 | DDX6 | Sponge network | 2.459 | 5.0E-5 | 0.091 | 0.36428 | 0.265 |
| 50434 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 29 | LRP1B | Sponge network | -0.303 | 0.21002 | -2.163 | 0 | 0.265 |
| 50435 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 13 | FAM188A | Sponge network | -0.487 | 0.02157 | -0.44 | 0.00018 | 0.265 |
| 50436 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-628-5p;hsa-miR-93-3p | 18 | EPHA3 | Sponge network | -0.602 | 0.00331 | 0.185 | 0.53588 | 0.265 |
| 50437 | CTD-2089N3.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 19 | HLF | Sponge network | -1.375 | 0.08642 | -2.443 | 0 | 0.265 |
| 50438 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-589-5p | 37 | ERBB4 | Sponge network | -0.154 | 0.78939 | -1.975 | 2.0E-5 | 0.265 |
| 50439 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p | 22 | CLASP2 | Sponge network | -0.322 | 0.25945 | -0.038 | 0.71 | 0.265 |
| 50440 | RP11-532F6.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-940 | 25 | ETV1 | Sponge network | -0.896 | 0.00099 | -0.096 | 0.67674 | 0.265 |
| 50441 | CTA-445C9.14 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-3607-3p | 16 | RICTOR | Sponge network | 0.718 | 4.0E-5 | 0.388 | 0.00188 | 0.265 |
| 50442 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-5p | 33 | FAT3 | Sponge network | -0.785 | 0.00035 | -1.601 | 0.00051 | 0.265 |
| 50443 | AC004893.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-590-3p | 19 | PIK3C2A | Sponge network | 1.317 | 0 | 0.061 | 0.53776 | 0.265 |
| 50444 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | CNTN1 | Sponge network | 0.657 | 4.0E-5 | -2.594 | 0 | 0.265 |
| 50445 | RP11-10N23.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-374a-3p | 13 | FZD3 | Sponge network | 1.134 | 0 | 0.104 | 0.60316 | 0.265 |
| 50446 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 21 | SCN9A | Sponge network | 0.401 | 0.00066 | -0.525 | 0.10593 | 0.265 |
| 50447 | AC141928.1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-577 | 18 | SYNPO2 | Sponge network | 0.853 | 0.15352 | -2.342 | 0 | 0.265 |
| 50448 | A2M-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | JDP2 | Sponge network | -0.08 | 0.90092 | -0.967 | 0 | 0.265 |
| 50449 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | ZNF208 | Sponge network | 0.214 | 0.18246 | -0.4 | 0.22381 | 0.265 |
| 50450 | SNHG5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-590-5p | 19 | MLLT10 | Sponge network | 0.13 | 0.47174 | 0.105 | 0.33292 | 0.265 |
| 50451 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p | 14 | LRRC55 | Sponge network | 0.494 | 0.00339 | -0.026 | 0.93163 | 0.265 |
| 50452 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-942-5p | 24 | GPC6 | Sponge network | 0.064 | 0.72934 | -0.054 | 0.81465 | 0.265 |
| 50453 | RP11-53O19.1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-375;hsa-miR-7-1-3p;hsa-miR-93-3p | 19 | DOCK4 | Sponge network | -0.405 | 0.22013 | 0.502 | 0.00108 | 0.265 |
| 50454 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | KAT6B | Sponge network | -0.08 | 0.90092 | -0.478 | 0.00018 | 0.265 |
| 50455 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | EPHA7 | Sponge network | 0.559 | 0.00026 | -2.197 | 3.0E-5 | 0.265 |
| 50456 | RP1-228H13.5 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-376c-3p;hsa-miR-654-3p | 10 | DICER1 | Sponge network | 1.397 | 0 | 0.042 | 0.674 | 0.265 |
| 50457 | EIF3J-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PHF3 | Sponge network | 0.331 | 0.00509 | 0.126 | 0.19652 | 0.265 |
| 50458 | RBPMS-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-935 | 11 | DMD | Sponge network | 0.144 | 0.64989 | -1.506 | 0 | 0.265 |
| 50459 | RP11-774O3.3 |
hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | TP63 | Sponge network | -0.487 | 0.02157 | -0.363 | 0.38191 | 0.265 |
| 50460 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p | 25 | HECA | Sponge network | -0.162 | 0.47687 | -0.217 | 0.03463 | 0.265 |
| 50461 | RP11-517B11.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-766-3p | 28 | MDM4 | Sponge network | 0.706 | 0 | 0.345 | 0.00762 | 0.265 |
| 50462 | A1BG-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-502-5p | 19 | DTNA | Sponge network | -0.164 | 0.45106 | -1.648 | 1.0E-5 | 0.265 |
| 50463 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-335-3p;hsa-miR-7-5p | 11 | FNBP1 | Sponge network | -0.285 | 0.2172 | -0.969 | 4.0E-5 | 0.265 |
| 50464 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-501-5p;hsa-miR-590-3p | 13 | EBF3 | Sponge network | 0.272 | 0.44692 | -0.058 | 0.81437 | 0.265 |
| 50465 | CTB-51J22.1 |
hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-29b-1-5p;hsa-miR-628-5p;hsa-miR-766-3p;hsa-miR-93-3p | 11 | IL6ST | Sponge network | -0.803 | 0.23737 | -0.402 | 0.00676 | 0.265 |
| 50466 | DNM3OS |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | SH3GLB1 | Sponge network | 0.572 | 0.01722 | -0.115 | 0.14773 | 0.265 |
| 50467 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940;hsa-miR-96-5p | 15 | HOOK3 | Sponge network | -0.942 | 0 | 0.273 | 0.09957 | 0.265 |
| 50468 | ERVH48-1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-744-3p | 32 | DST | Sponge network | 1.04 | 0.00023 | -0.713 | 0.00096 | 0.265 |
| 50469 | CTD-3126B10.4 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | CBX5 | Sponge network | 0.778 | 0.00101 | 0.165 | 0.24446 | 0.265 |
| 50470 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 32 | MOB1B | Sponge network | 0.61 | 0.01593 | 0.197 | 0.15266 | 0.265 |
| 50471 | RP11-819C21.1 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 17 | IL13RA1 | Sponge network | 0.537 | 0.00139 | 0.019 | 0.86949 | 0.265 |
| 50472 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | HACE1 | Sponge network | 0.415 | 0.14657 | -0.072 | 0.55239 | 0.265 |
| 50473 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 17 | TBL1XR1 | Sponge network | 0.983 | 0.00048 | 0.578 | 0 | 0.265 |
| 50474 | CTB-113P19.4 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-96-5p | 34 | GAS7 | Sponge network | 0.906 | 0.31715 | -0.555 | 0.01602 | 0.265 |
| 50475 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | PBRM1 | Sponge network | 0.415 | 0.14657 | -0.029 | 0.78601 | 0.265 |
| 50476 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 11 | GIGYF2 | Sponge network | 0.415 | 0.14657 | 0.136 | 0.04403 | 0.265 |
| 50477 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 48 | CBX5 | Sponge network | -0.49 | 0.04946 | 0.165 | 0.24446 | 0.265 |
| 50478 | BCYRN1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 20 | RPS6KA3 | Sponge network | 0.311 | 0.10344 | 0.256 | 0.02419 | 0.265 |
| 50479 | RP11-798G7.7 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-582-3p | 12 | SETBP1 | Sponge network | 0.858 | 3.0E-5 | -1.302 | 1.0E-5 | 0.265 |
| 50480 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-34a-5p | 11 | SON | Sponge network | 0.592 | 0.00014 | 0.168 | 0.04406 | 0.265 |
| 50481 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p | 12 | TSC22D1 | Sponge network | -1.575 | 6.0E-5 | -0.529 | 2.0E-5 | 0.265 |
| 50482 | CTA-363E19.2 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-495-3p | 12 | MAPK1 | Sponge network | 1.055 | 0 | -0.017 | 0.81465 | 0.265 |
| 50483 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-374b-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 37 | ATRX | Sponge network | 1.302 | 7.0E-5 | 0.261 | 0.04408 | 0.264 |
| 50484 | LPP-AS2 |
hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-330-5p | 12 | ABL1 | Sponge network | -0.18 | 0.20343 | -0.215 | 0.07804 | 0.264 |
| 50485 | RP11-894P9.1 |
hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-550a-5p | 10 | VPS53 | Sponge network | 0.993 | 0 | 0.103 | 0.22667 | 0.264 |
| 50486 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-96-5p | 16 | MED12L | Sponge network | 0.37 | 0.27614 | -0.194 | 0.55299 | 0.264 |
| 50487 | RP11-597D13.8 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p | 11 | THRB | Sponge network | 0.936 | 0.03889 | -1.117 | 0.0004 | 0.264 |
| 50488 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-502-3p;hsa-miR-576-5p | 20 | AFF4 | Sponge network | 2.163 | 0 | -0.266 | 0.01565 | 0.264 |
| 50489 | BDNF-AS |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 20 | EPHA3 | Sponge network | -0.668 | 3.0E-5 | 0.185 | 0.53588 | 0.264 |
| 50490 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-486-5p;hsa-miR-7-1-3p | 12 | PDGFC | Sponge network | -0.969 | 2.0E-5 | -0.33 | 0.06483 | 0.264 |
| 50491 | TMEM51-AS1 |
hsa-miR-106a-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-22-3p;hsa-miR-24-2-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-432-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-3p | 21 | PPM1L | Sponge network | 0.23 | 0.38667 | -0.537 | 0.00616 | 0.264 |
| 50492 | LIPE-AS1 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-5p | 12 | INPP4A | Sponge network | 0.078 | 0.55803 | 0.07 | 0.53563 | 0.264 |
| 50493 | KB-1460A1.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 30 | NTRK2 | Sponge network | 0.603 | 0.00127 | -0.736 | 0.06495 | 0.264 |
| 50494 | ZNF667-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 17 | RBL2 | Sponge network | -0.976 | 0.00025 | -0.282 | 0.00254 | 0.264 |
| 50495 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 26 | ADARB1 | Sponge network | 0.898 | 1.0E-5 | -0.335 | 0.06631 | 0.264 |
| 50496 | TPT1-AS1 |
hsa-miR-101-5p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-486-5p;hsa-miR-664a-5p | 14 | CBFB | Sponge network | 0.903 | 0 | 1.061 | 0 | 0.264 |
| 50497 | RP11-452L6.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-484 | 14 | KIF1B | Sponge network | 0.665 | 0.00061 | -0.118 | 0.32258 | 0.264 |
| 50498 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | RCAN2 | Sponge network | 2.094 | 0 | -0.33 | 0.15172 | 0.264 |
| 50499 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-93-5p | 11 | BHLHE41 | Sponge network | 0.348 | 0.32912 | 0.353 | 0.12753 | 0.264 |
| 50500 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 15 | CLASP2 | Sponge network | 1.11 | 1.0E-5 | -0.038 | 0.71 | 0.264 |
| 50501 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 24 | ST6GALNAC3 | Sponge network | 0.051 | 0.83388 | -0.987 | 0 | 0.264 |
| 50502 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | NIN | Sponge network | 0.349 | 0.01695 | -0.253 | 0.13794 | 0.264 |
| 50503 | NDUFA6-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-940 | 42 | TRPS1 | Sponge network | -0.355 | 0.0077 | -0.191 | 0.44109 | 0.264 |
| 50504 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-212-3p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | FOXP1 | Sponge network | 1.055 | 0 | 0.042 | 0.74368 | 0.264 |
| 50505 | TRAM2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-502-5p | 15 | IL17RD | Sponge network | 0.336 | 0.00739 | -0.019 | 0.94873 | 0.264 |
| 50506 | RP11-403I13.7 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | 0.395 | 0.15451 | 0.321 | 0.00267 | 0.264 |
| 50507 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | CXXC4 | Sponge network | 0.532 | 0.00505 | -0.433 | 0.21055 | 0.264 |
| 50508 | RP11-428J1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | SNX29 | Sponge network | 0.538 | 0.00076 | -0.34 | 0.01371 | 0.264 |
| 50509 | HOXA-AS3 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-590-3p | 11 | PRUNE2 | Sponge network | -0.555 | 0.06156 | -1.733 | 0.00022 | 0.264 |
| 50510 | TTC25 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-378a-5p | 10 | ADARB1 | Sponge network | -0.208 | 0.38261 | -0.335 | 0.06631 | 0.264 |
| 50511 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 25 | CADM1 | Sponge network | -0.738 | 0.21233 | -0.9 | 0.00084 | 0.264 |
| 50512 | RP11-384O8.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | CELF4 | Sponge network | -0.738 | 0.21233 | -1.135 | 0.00344 | 0.264 |
| 50513 | CKMT2-AS1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 12 | FOXO4 | Sponge network | 0.082 | 0.55654 | -0.516 | 2.0E-5 | 0.264 |
| 50514 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 44 | FAT3 | Sponge network | -0.125 | 0.49414 | -1.601 | 0.00051 | 0.264 |
| 50515 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935 | 24 | EBF1 | Sponge network | 0.267 | 0.2937 | -0.384 | 0.08856 | 0.264 |
| 50516 | LINC00672 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 26 | CREB1 | Sponge network | -0.227 | 0.31657 | 0.203 | 0.00537 | 0.264 |
| 50517 | RP11-800A3.7 |
hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-96-5p | 15 | GAS7 | Sponge network | 0.18 | 0.7147 | -0.555 | 0.01602 | 0.264 |
| 50518 | RP11-13K12.1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-365a-3p;hsa-miR-375;hsa-miR-378a-5p | 12 | CACNA1E | Sponge network | -0.154 | 0.78939 | 1.752 | 0 | 0.264 |
| 50519 | RP11-545G3.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-629-3p | 14 | CBX5 | Sponge network | 2.622 | 0.00025 | 0.165 | 0.24446 | 0.264 |
| 50520 | AC124789.1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-652-3p | 12 | QKI | Sponge network | 0.073 | 0.79848 | -0.333 | 0.04111 | 0.264 |
| 50521 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | ZFHX4 | Sponge network | -0.418 | 0.00068 | 0.018 | 0.95574 | 0.264 |
| 50522 | RP4-605O3.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 17 | CBX5 | Sponge network | 0.262 | 0.0475 | 0.165 | 0.24446 | 0.264 |
| 50523 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940 | 50 | NTRK3 | Sponge network | -0.371 | 0.24604 | -1.77 | 3.0E-5 | 0.264 |
| 50524 | RP11-401P9.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-18a-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-625-5p | 10 | TGFBR3 | Sponge network | 1.524 | 0.1098 | -1.173 | 1.0E-5 | 0.264 |
| 50525 | RP11-403I13.8 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 16 | RYR2 | Sponge network | 0.619 | 0.00157 | -1.466 | 0.00031 | 0.264 |
| 50526 | AGAP11 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -1.645 | 0 | -0.354 | 0.14694 | 0.264 |
| 50527 | RP11-77H9.2 |
hsa-let-7a-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 15 | RICTOR | Sponge network | 1.677 | 0 | 0.388 | 0.00188 | 0.264 |
| 50528 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 27 | SASH1 | Sponge network | 0.75 | 0.00054 | -0.502 | 0.00175 | 0.264 |
| 50529 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p | 11 | BCL6 | Sponge network | -0.663 | 0.10887 | -0.211 | 0.19489 | 0.264 |
| 50530 | TPTEP1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | MED12L | Sponge network | -1.216 | 0 | -0.194 | 0.55299 | 0.264 |
| 50531 | RP11-38L15.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-590-3p | 12 | ASTN1 | Sponge network | 0.344 | 0.3127 | -2.389 | 2.0E-5 | 0.264 |
| 50532 | RP11-5C23.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 12 | PIK3CA | Sponge network | 0.274 | 0.07681 | 0.195 | 0.06908 | 0.264 |
| 50533 | RP13-977J11.2 |
hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p | 13 | RASSF2 | Sponge network | 0.121 | 0.65192 | -0.273 | 0.21961 | 0.264 |
| 50534 | RP11-53O19.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-539-5p | 13 | DDX5 | Sponge network | -0.405 | 0.22013 | -0.099 | 0.17428 | 0.264 |
| 50535 | WDFY3-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p | 28 | RNASEL | Sponge network | -0.942 | 0 | -0.272 | 0.01796 | 0.264 |
| 50536 | RP4-791M13.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-212-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577 | 17 | UNC80 | Sponge network | 0.419 | 0.0319 | -1.934 | 0 | 0.264 |
| 50537 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-455-3p | 11 | KCNAB1 | Sponge network | 0.494 | 0.00339 | -0.755 | 2.0E-5 | 0.264 |
| 50538 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-629-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 60 | BMPR2 | Sponge network | -0.709 | 0.18168 | 0.206 | 0.0635 | 0.264 |
| 50539 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-93-5p | 14 | BHLHE41 | Sponge network | 0.497 | 0.01451 | 0.353 | 0.12753 | 0.264 |
| 50540 | DNM1P35 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p | 17 | EYA4 | Sponge network | 0.051 | 0.83388 | 0.3 | 0.36876 | 0.264 |
| 50541 | RP11-25K19.1 |
hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DACT1 | Sponge network | -1.057 | 0.00029 | 0.783 | 0.00072 | 0.264 |
| 50542 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 15 | ARHGAP29 | Sponge network | 0.385 | 0.10067 | 0.131 | 0.42953 | 0.264 |
| 50543 | AC005154.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | BRD3 | Sponge network | 0.848 | 0 | 0.37 | 0.00013 | 0.264 |
| 50544 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 38 | WDFY3 | Sponge network | 0.853 | 0.15352 | -0.201 | 0.0967 | 0.264 |
| 50545 | DNAJC27-AS1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | PRKCB | Sponge network | -0.969 | 2.0E-5 | -1.313 | 2.0E-5 | 0.264 |
| 50546 | AP001055.6 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-331-3p;hsa-miR-7-1-3p;hsa-miR-744-3p | 16 | PTPRT | Sponge network | 0.693 | 0.00076 | -0.907 | 0.03727 | 0.264 |
| 50547 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 29 | TSHZ3 | Sponge network | 0.392 | 0.0278 | -0.556 | 0.01516 | 0.264 |
| 50548 | RP11-927P21.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-3p | 32 | DST | Sponge network | 0.921 | 0.00118 | -0.713 | 0.00096 | 0.264 |
| 50549 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935 | 28 | PCDH10 | Sponge network | 0.75 | 0.00054 | -1.644 | 0.0078 | 0.264 |
| 50550 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484 | 13 | TRIM3 | Sponge network | 0.194 | 0.64278 | -0.577 | 0 | 0.264 |
| 50551 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3613-5p;hsa-miR-874-3p;hsa-miR-940 | 14 | CRTC3 | Sponge network | 0.817 | 3.0E-5 | 0.164 | 0.16786 | 0.264 |
| 50552 | RP11-967K21.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 21 | CSMD1 | Sponge network | 0.056 | 0.81195 | -0.63 | 0.18881 | 0.264 |
| 50553 | AC005618.6 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 60 | LPP | Sponge network | 0.862 | 0.06213 | -0.459 | 0.04475 | 0.264 |
| 50554 | RP11-195B17.1 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p | 12 | IGF1 | Sponge network | 0.37 | 0.27614 | -0.204 | 0.5898 | 0.264 |
| 50555 | LINC00672 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p | 11 | WNK2 | Sponge network | -0.227 | 0.31657 | -0.352 | 0.31066 | 0.264 |
| 50556 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-96-5p | 18 | FBXO32 | Sponge network | 0.056 | 0.8494 | -0.495 | 0.07561 | 0.264 |
| 50557 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | CBL | Sponge network | 1.122 | 0 | 0.291 | 0.01267 | 0.264 |
| 50558 | LINC00667 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | HIP1 | Sponge network | -0.235 | 0.15142 | 0.774 | 0 | 0.264 |
| 50559 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | NR2C2 | Sponge network | 1.308 | 0 | 0.21 | 0.03224 | 0.264 |
| 50560 | TINCR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-629-3p | 38 | AR | Sponge network | -0.664 | 0.14543 | -1.554 | 0 | 0.264 |
| 50561 | TMEM161B-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-21-3p;hsa-miR-214-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-590-3p | 32 | SMAD4 | Sponge network | -0.04 | 0.7606 | -0.313 | 0.00413 | 0.264 |
| 50562 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-940 | 16 | CREB1 | Sponge network | 0.453 | 0.01839 | 0.203 | 0.00537 | 0.264 |
| 50563 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-331-3p;hsa-miR-629-3p | 16 | EPHA7 | Sponge network | -0.278 | 0.76559 | -2.197 | 3.0E-5 | 0.264 |
| 50564 | MIR143HG |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 39 | CREB1 | Sponge network | -0.739 | 0.0574 | 0.203 | 0.00537 | 0.264 |
| 50565 | AC005618.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 25 | DMD | Sponge network | 0.862 | 0.06213 | -1.506 | 0 | 0.264 |
| 50566 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | HECA | Sponge network | -0.235 | 0.15142 | -0.217 | 0.03463 | 0.264 |
| 50567 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-628-5p | 16 | MAPK1 | Sponge network | 1.344 | 0 | -0.017 | 0.81465 | 0.264 |
| 50568 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110 | 17 | BNC2 | Sponge network | 0.001 | 0.99753 | -0.491 | 0.09988 | 0.264 |
| 50569 | RP11-144G6.12 |
hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 12 | UBE2D1 | Sponge network | 0.826 | 1.0E-5 | 0.468 | 0 | 0.264 |
| 50570 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-766-3p | 25 | MAPK1 | Sponge network | 0.95 | 0.15943 | -0.017 | 0.81465 | 0.264 |
| 50571 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-877-5p;hsa-miR-92a-3p | 14 | CBFA2T3 | Sponge network | 0.199 | 0.38015 | -1.221 | 1.0E-5 | 0.264 |
| 50572 | FAM13A-AS1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-625-5p | 10 | CNN1 | Sponge network | -0.83 | 0 | -2.633 | 0 | 0.264 |
| 50573 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-590-3p | 14 | CLASP2 | Sponge network | 0.439 | 0.00313 | -0.038 | 0.71 | 0.264 |
| 50574 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CACNA2D1 | Sponge network | -0.141 | 0.26434 | -0.846 | 0.00675 | 0.264 |
| 50575 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-542-3p;hsa-miR-93-3p | 18 | ST6GAL2 | Sponge network | -0.384 | 0.40652 | -1.201 | 0.00597 | 0.264 |
| 50576 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | SASH1 | Sponge network | 1.2 | 0.01813 | -0.502 | 0.00175 | 0.264 |
| 50577 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p | 29 | MTSS1 | Sponge network | 0.253 | 0.09565 | -0.81 | 0.00035 | 0.264 |
| 50578 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | NIN | Sponge network | 0.381 | 0.25967 | -0.253 | 0.13794 | 0.264 |
| 50579 | AC005562.1 |
hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 14 | SHPRH | Sponge network | 0.488 | 0.00084 | 0.098 | 0.40994 | 0.264 |
| 50580 | RP4-791M13.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-940 | 18 | ETV1 | Sponge network | 0.419 | 0.0319 | -0.096 | 0.67674 | 0.264 |
| 50581 | AC010761.8 |
hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-582-5p | 11 | MOB1B | Sponge network | 1.638 | 0 | 0.197 | 0.15266 | 0.264 |
| 50582 | GS1-124K5.11 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | ABCB5 | Sponge network | 0.494 | 0.00339 | -0.956 | 0.04077 | 0.264 |
| 50583 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-33a-3p | 10 | ABCC9 | Sponge network | 0.792 | 0.0049 | -0.466 | 0.14121 | 0.264 |
| 50584 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 15 | HERC2 | Sponge network | -1.111 | 0.0003 | 0.114 | 0.30638 | 0.264 |
| 50585 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | PRKCE | Sponge network | -0.49 | 0.04946 | -0.403 | 0.00056 | 0.264 |
| 50586 | RP11-996F15.2 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 11 | DICER1 | Sponge network | 0.983 | 0.00048 | 0.042 | 0.674 | 0.264 |
| 50587 | RP5-1085F17.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 29 | ACVR2A | Sponge network | 0.541 | 0.00125 | -0.409 | 0.00039 | 0.264 |
| 50588 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-940;hsa-miR-942-5p | 20 | HLF | Sponge network | -0.326 | 0.14314 | -2.443 | 0 | 0.264 |
| 50589 | DNAJC27-AS1 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 11 | PRDM11 | Sponge network | -0.969 | 2.0E-5 | -0.252 | 0.15511 | 0.264 |
| 50590 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 31 | CBX5 | Sponge network | -0.176 | 0.59692 | 0.165 | 0.24446 | 0.264 |
| 50591 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | NUAK1 | Sponge network | 0.537 | 0.00139 | 0.904 | 0 | 0.264 |
| 50592 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 32 | ARID5B | Sponge network | -2.452 | 0 | 0.342 | 0.01676 | 0.264 |
| 50593 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p | 16 | PRKAA1 | Sponge network | 0.705 | 0.00014 | 0.317 | 0.0031 | 0.264 |
| 50594 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 33 | TGFBR3 | Sponge network | 1.302 | 7.0E-5 | -1.173 | 1.0E-5 | 0.264 |
| 50595 | LINC00886 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-502-5p;hsa-miR-877-5p;hsa-miR-93-5p | 28 | ANK2 | Sponge network | -0.371 | 0.24604 | -1.603 | 1.0E-5 | 0.264 |
| 50596 | MALAT1 |
hsa-miR-140-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-93-3p | 10 | ZNF382 | Sponge network | 0.782 | 0.00016 | -0.494 | 0.03831 | 0.264 |
| 50597 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374a-3p;hsa-miR-495-3p | 13 | AFF4 | Sponge network | 0.757 | 0 | -0.266 | 0.01565 | 0.264 |
| 50598 | RP11-686O6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26a-2-3p;hsa-miR-577 | 13 | PRKAA2 | Sponge network | 0.453 | 0.01971 | -2.462 | 0 | 0.264 |
| 50599 | RP5-994D16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-590-3p | 13 | DLC1 | Sponge network | 0.252 | 0.08508 | -0.382 | 0.05038 | 0.264 |
| 50600 | NDUFA6-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-501-3p | 11 | ZBTB20 | Sponge network | -0.355 | 0.0077 | -0.122 | 0.57569 | 0.264 |
| 50601 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 11 | NALCN | Sponge network | 0.222 | 0.29133 | 1.068 | 0.00237 | 0.264 |
| 50602 | AC093627.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p | 12 | BHLHE41 | Sponge network | -0.787 | 0.3928 | 0.353 | 0.12753 | 0.264 |
| 50603 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-942-5p | 47 | THRB | Sponge network | 0.214 | 0.18246 | -1.117 | 0.0004 | 0.264 |
| 50604 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | RASSF2 | Sponge network | -2.915 | 0.00015 | -0.273 | 0.21961 | 0.264 |
| 50605 | LINC00641 |
hsa-miR-125a-5p;hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | ERCC6 | Sponge network | -0.222 | 0.32445 | 0.23 | 0.015 | 0.264 |
| 50606 | RP11-181G12.2 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-550a-5p | 21 | ARHGEF12 | Sponge network | 0.556 | 0.00298 | 0.09 | 0.35693 | 0.264 |
| 50607 | MIR4435-1HG |
hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-497-5p;hsa-miR-9-5p | 11 | CDK6 | Sponge network | 2.275 | 0 | 0.991 | 0 | 0.264 |
| 50608 | RP11-693J15.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p | 12 | CSMD1 | Sponge network | -0.72 | 0.24239 | -0.63 | 0.18881 | 0.264 |
| 50609 | RP11-48B3.4 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-577;hsa-miR-942-5p | 17 | IGFBP5 | Sponge network | 1.757 | 0 | -0.806 | 0.00105 | 0.264 |
| 50610 | CTC-429P9.2 |
hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-539-5p;hsa-miR-940 | 10 | LIFR | Sponge network | 0.043 | 0.81628 | -1.794 | 0 | 0.264 |
| 50611 | LINC00476 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-671-5p | 14 | SHPRH | Sponge network | -0.615 | 0.00015 | 0.098 | 0.40994 | 0.264 |
| 50612 | RP11-57H14.4 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | PICALM | Sponge network | 0.083 | 0.66405 | 0.086 | 0.23523 | 0.264 |
| 50613 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-877-5p;hsa-miR-93-5p | 47 | CCND2 | Sponge network | -0.615 | 0.00015 | -0.267 | 0.3112 | 0.264 |
| 50614 | SERHL |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-625-5p | 14 | CRTC1 | Sponge network | -0.602 | 0.00331 | -0.116 | 0.28412 | 0.264 |
| 50615 | TRAM2-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-590-3p | 17 | ZFX | Sponge network | 0.336 | 0.00739 | 0.09 | 0.42563 | 0.264 |
| 50616 | CTC-429P9.1 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-409-3p | 11 | PRKCB | Sponge network | 0.497 | 0.01451 | -1.313 | 2.0E-5 | 0.264 |
| 50617 | RASSF8-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | CREB1 | Sponge network | 0.335 | 0.20596 | 0.203 | 0.00537 | 0.264 |
| 50618 | RP11-356J5.12 |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-940 | 17 | JUN | Sponge network | -0.899 | 6.0E-5 | -0.932 | 0 | 0.264 |
| 50619 | RP5-994D16.9 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-342-3p;hsa-miR-590-3p | 10 | CCDC6 | Sponge network | 0.252 | 0.08508 | 0.27 | 0.02555 | 0.264 |
| 50620 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 10 | PCDH8 | Sponge network | -0.896 | 0.00099 | -0.997 | 0.05366 | 0.264 |
| 50621 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 32 | FOXP1 | Sponge network | 0.214 | 0.18246 | 0.042 | 0.74368 | 0.264 |
| 50622 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-590-3p | 23 | FBXO32 | Sponge network | 0.765 | 7.0E-5 | -0.495 | 0.07561 | 0.264 |
| 50623 | H1FX-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-30b-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-671-5p | 13 | CDH23 | Sponge network | -0.206 | 0.12942 | -0.974 | 0.00034 | 0.264 |
| 50624 | RP11-531A24.5 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-454-3p | 11 | IGF2 | Sponge network | 0.75 | 0.00054 | 0.848 | 0.03842 | 0.264 |
| 50625 | RP11-13K12.1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-664a-5p;hsa-miR-93-5p | 43 | PPM1L | Sponge network | -0.154 | 0.78939 | -0.537 | 0.00616 | 0.264 |
| 50626 | AC034220.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-766-3p | 10 | EZH1 | Sponge network | 0.309 | 0.07304 | -0.564 | 1.0E-5 | 0.264 |
| 50627 | AP001347.6 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | NIN | Sponge network | -1.161 | 0.00591 | -0.253 | 0.13794 | 0.264 |
| 50628 | RP1-257A7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-5p | 11 | BMPR2 | Sponge network | 0.849 | 0.00178 | 0.206 | 0.0635 | 0.264 |
| 50629 | RP11-390K5.6 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 11 | DDX3X | Sponge network | 1.148 | 2.0E-5 | -0.152 | 0.07686 | 0.264 |
| 50630 | AC159540.2 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-664a-5p | 14 | CBFB | Sponge network | 1.634 | 0 | 1.061 | 0 | 0.264 |
| 50631 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | ZFHX3 | Sponge network | 0.538 | 0.00076 | 0.389 | 0.01363 | 0.264 |
| 50632 | CTD-2139B15.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-3613-5p | 17 | CCDC6 | Sponge network | 0.286 | 0.00612 | 0.27 | 0.02555 | 0.264 |
| 50633 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p | 13 | MAP3K12 | Sponge network | -0.942 | 0 | -0.057 | 0.59369 | 0.264 |
| 50634 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 13 | CREBBP | Sponge network | -0.738 | 0.21233 | 0.126 | 0.15186 | 0.264 |
| 50635 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | FREM2 | Sponge network | 0.197 | 0.25618 | -0.437 | 0.46353 | 0.264 |
| 50636 | HAND2-AS1 |
hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-874-3p | 15 | ARID1B | Sponge network | -1.697 | 0.00889 | 0.003 | 0.97503 | 0.264 |
| 50637 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 33 | ST6GALNAC3 | Sponge network | -0.727 | 0.04726 | -0.987 | 0 | 0.264 |
| 50638 | AC007277.3 |
hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p | 10 | BRD3 | Sponge network | 2.377 | 0.00108 | 0.37 | 0.00013 | 0.264 |
| 50639 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-98-5p | 16 | MITF | Sponge network | -0.773 | 0.04073 | -0.769 | 0.00022 | 0.264 |
| 50640 | RP11-2H8.2 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p | 14 | DOCK4 | Sponge network | 1.089 | 0 | 0.502 | 0.00108 | 0.264 |
| 50641 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | KCNA6 | Sponge network | -0.351 | 0.11358 | -1.531 | 0.00013 | 0.264 |
| 50642 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | FREM2 | Sponge network | 0.199 | 0.38015 | -0.437 | 0.46353 | 0.264 |
| 50643 | C20orf166-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-93-5p | 14 | MAP3K12 | Sponge network | -2.69 | 0.0001 | -0.057 | 0.59369 | 0.264 |
| 50644 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-7-1-3p | 33 | EPHA7 | Sponge network | 0.395 | 0.15451 | -2.197 | 3.0E-5 | 0.264 |
| 50645 | C4B-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-590-5p | 12 | DMD | Sponge network | 2.306 | 9.0E-5 | -1.506 | 0 | 0.264 |
| 50646 | KB-1572G7.2 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | USP9X | Sponge network | 0.924 | 0 | 0.222 | 0.01689 | 0.264 |
| 50647 | CTC-297N7.5 |
hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-146b-3p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-382-5p | 10 | PPARA | Sponge network | -1.9 | 0 | -0.379 | 0.00548 | 0.264 |
| 50648 | RP11-894P9.1 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-532-3p;hsa-miR-940 | 19 | WDFY3 | Sponge network | 0.993 | 0 | -0.201 | 0.0967 | 0.264 |
| 50649 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 23 | THBS1 | Sponge network | 0.225 | 0.30644 | 0.741 | 0.00386 | 0.264 |
| 50650 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | ADAMTSL3 | Sponge network | 0.349 | 0.01695 | -1.559 | 0 | 0.264 |
| 50651 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-500a-5p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-7-5p | 18 | CRTC1 | Sponge network | 0.299 | 0.57722 | -0.116 | 0.28412 | 0.264 |
| 50652 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 21 | PTPN14 | Sponge network | 0.853 | 0.15352 | 0.144 | 0.34595 | 0.264 |
| 50653 | AC116366.6 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | RASSF2 | Sponge network | 0.329 | 0.11122 | -0.273 | 0.21961 | 0.264 |
| 50654 | RP11-251G23.5 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-664a-3p | 22 | KIF1B | Sponge network | 1.344 | 0 | -0.118 | 0.32258 | 0.264 |
| 50655 | CTC-296K1.3 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p;hsa-miR-744-3p | 13 | PTPN14 | Sponge network | -2.095 | 0.00112 | 0.144 | 0.34595 | 0.264 |
| 50656 | RP11-373D23.3 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | BRD3 | Sponge network | -0.285 | 0.2172 | 0.37 | 0.00013 | 0.264 |
| 50657 | CTD-2184D3.6 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NAV3 | Sponge network | 0.272 | 0.44692 | 0.212 | 0.47729 | 0.264 |
| 50658 | RP11-597D13.9 |
hsa-miR-103a-2-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | MSN | Sponge network | 1.302 | 7.0E-5 | -0.023 | 0.88422 | 0.264 |
| 50659 | RP11-800A3.7 |
hsa-miR-107;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-7-1-3p | 11 | EYA4 | Sponge network | 0.18 | 0.7147 | 0.3 | 0.36876 | 0.264 |
| 50660 | RP11-526I2.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-495-3p | 11 | NF1 | Sponge network | 1.054 | 1.0E-5 | 0.51 | 0 | 0.264 |
| 50661 | RP11-434B12.1 |
hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-2355-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p | 12 | RHOB | Sponge network | -0.031 | 0.89063 | -1.052 | 0 | 0.264 |
| 50662 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 17 | IGF1 | Sponge network | 0.381 | 0.25967 | -0.204 | 0.5898 | 0.264 |
| 50663 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-664a-5p | 18 | MAPK1 | Sponge network | 0.757 | 0 | -0.017 | 0.81465 | 0.264 |
| 50664 | CTD-2336O2.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-93-3p | 29 | RUNX1T1 | Sponge network | -0.141 | 0.26434 | -0.344 | 0.2374 | 0.264 |
| 50665 | KCNJ2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-590-3p | 14 | ZFHX3 | Sponge network | -0.436 | 0.05325 | 0.389 | 0.01363 | 0.264 |
| 50666 | RP11-686D22.8 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | MYO9A | Sponge network | 0.898 | 1.0E-5 | -0.147 | 0.32373 | 0.264 |
| 50667 | AF131215.9 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 29 | NTRK2 | Sponge network | -0.322 | 0.25945 | -0.736 | 0.06495 | 0.264 |
| 50668 | FAM66C |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 18 | FAM135B | Sponge network | -0.901 | 0.00019 | -3.978 | 0 | 0.264 |
| 50669 | LINC00403 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-589-5p | 10 | EYA4 | Sponge network | -3.586 | 0.00032 | 0.3 | 0.36876 | 0.264 |
| 50670 | RP11-53O19.1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p | 14 | PTCH1 | Sponge network | -0.405 | 0.22013 | -0.1 | 0.6179 | 0.264 |
| 50671 | LINC01018 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 44 | PPM1L | Sponge network | -2.915 | 0.00015 | -0.537 | 0.00616 | 0.264 |
| 50672 | LINC00173 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-5p | 10 | NRK | Sponge network | 0.056 | 0.8494 | 0.656 | 0.19217 | 0.264 |
| 50673 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-301a-3p | 11 | JDP2 | Sponge network | -1.654 | 0.00144 | -0.967 | 0 | 0.264 |
| 50674 | ARHGEF26-AS1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | SIAH1 | Sponge network | -2.46 | 0 | -0.057 | 0.41415 | 0.264 |
| 50675 | AF131215.9 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 22 | CADM1 | Sponge network | -0.322 | 0.25945 | -0.9 | 0.00084 | 0.264 |
| 50676 | AF131215.9 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 40 | DTNA | Sponge network | -0.322 | 0.25945 | -1.648 | 1.0E-5 | 0.264 |
| 50677 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-22-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | EPHA3 | Sponge network | 0.545 | 0.00064 | 0.185 | 0.53588 | 0.264 |
| 50678 | RP1-102E24.8 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p | 10 | ARHGEF12 | Sponge network | 1.737 | 0 | 0.09 | 0.35693 | 0.264 |
| 50679 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-874-3p | 10 | NR2C2 | Sponge network | 0.336 | 0.00739 | 0.21 | 0.03224 | 0.264 |
| 50680 | U91328.19 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-7-1-3p | 10 | WAC | Sponge network | -0.303 | 0.21002 | -0.054 | 0.43688 | 0.264 |
| 50681 | AF131215.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-29a-5p;hsa-miR-454-3p;hsa-miR-7-1-3p | 10 | RARB | Sponge network | -0.176 | 0.59692 | -0.825 | 2.0E-5 | 0.264 |
| 50682 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-532-5p | 12 | CXXC4 | Sponge network | -0.829 | 0.01343 | -0.433 | 0.21055 | 0.264 |
| 50683 | CD27-AS1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p | 24 | PGR | Sponge network | 0.177 | 0.16681 | -1.704 | 0 | 0.264 |
| 50684 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-96-5p | 15 | PCDH8 | Sponge network | -1.216 | 0 | -0.997 | 0.05366 | 0.264 |
| 50685 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 17 | KAT6B | Sponge network | 0.389 | 0.04293 | -0.478 | 0.00018 | 0.264 |
| 50686 | LINC00963 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-324-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p | 13 | FMNL3 | Sponge network | 0.523 | 9.0E-5 | 0.555 | 4.0E-5 | 0.264 |
| 50687 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-940 | 21 | IGF1 | Sponge network | 0.259 | 0.12542 | -0.204 | 0.5898 | 0.264 |
| 50688 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | KDSR | Sponge network | 0.997 | 0.00066 | 0.239 | 0.02216 | 0.264 |
| 50689 | H19 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-486-5p | 11 | EPHA3 | Sponge network | 2.439 | 0 | 0.185 | 0.53588 | 0.264 |
| 50690 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 26 | BHLHE41 | Sponge network | -0.83 | 0 | 0.353 | 0.12753 | 0.264 |
| 50691 | RAMP2-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935;hsa-miR-942-5p | 26 | OPCML | Sponge network | -0.513 | 0.04703 | -2.498 | 0 | 0.264 |
| 50692 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PTPRB | Sponge network | -1.161 | 0.00591 | 0.105 | 0.47122 | 0.264 |
| 50693 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-96-5p | 31 | HBP1 | Sponge network | 0.101 | 0.60919 | -0.454 | 0 | 0.264 |
| 50694 | LINC00680 |
hsa-let-7g-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p | 15 | CREB1 | Sponge network | 0.884 | 0 | 0.203 | 0.00537 | 0.264 |
| 50695 | MIR22HG |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-940 | 31 | SNX29 | Sponge network | -1.047 | 0 | -0.34 | 0.01371 | 0.264 |
| 50696 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-940 | 15 | GLI3 | Sponge network | -0.969 | 2.0E-5 | -0.615 | 0.01482 | 0.264 |
| 50697 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-942-5p | 14 | RB1CC1 | Sponge network | 0.194 | 0.64278 | 0.175 | 0.12559 | 0.264 |
| 50698 | FZD10-AS1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-590-3p | 16 | FMNL3 | Sponge network | -0.727 | 0.04726 | 0.555 | 4.0E-5 | 0.264 |
| 50699 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p | 16 | ARHGAP28 | Sponge network | -0.726 | 0.00132 | -0.911 | 0.00013 | 0.264 |
| 50700 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p | 16 | RARB | Sponge network | 0.051 | 0.83388 | -0.825 | 2.0E-5 | 0.264 |
| 50701 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | LATS1 | Sponge network | 0.082 | 0.55654 | 0.109 | 0.34208 | 0.264 |
| 50702 | WDFY3-AS2 |
hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 17 | BCL9 | Sponge network | -0.942 | 0 | 0.321 | 0.00267 | 0.264 |
| 50703 | PSMD5-AS1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p;hsa-miR-708-5p | 15 | MAT2A | Sponge network | 0.569 | 0.00067 | -0.073 | 0.36195 | 0.264 |
| 50704 | RP11-384O8.1 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 26 | RIMS2 | Sponge network | -0.738 | 0.21233 | -1.365 | 0.00208 | 0.264 |
| 50705 | RP4-791M13.3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-940 | 14 | SIRT1 | Sponge network | 0.419 | 0.0319 | 0.109 | 0.2593 | 0.264 |
| 50706 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-3677-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p | 13 | PRKAA1 | Sponge network | 0.946 | 0 | 0.317 | 0.0031 | 0.264 |
| 50707 | MIR143HG |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 25 | SIRT1 | Sponge network | -0.739 | 0.0574 | 0.109 | 0.2593 | 0.264 |
| 50708 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p | 20 | MYBL1 | Sponge network | 0.41 | 0.02214 | 0.494 | 0.02152 | 0.264 |
| 50709 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-96-5p | 40 | BMPR2 | Sponge network | 0.594 | 0.41522 | 0.206 | 0.0635 | 0.264 |
| 50710 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-590-3p | 25 | MOB1B | Sponge network | -0.739 | 0.00044 | 0.197 | 0.15266 | 0.264 |
| 50711 | AC016747.3 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p | 27 | CREB3L2 | Sponge network | 0.199 | 0.38015 | -0.525 | 2.0E-5 | 0.264 |
| 50712 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-96-5p | 70 | LPP | Sponge network | 0.523 | 9.0E-5 | -0.459 | 0.04475 | 0.264 |
| 50713 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 26 | PDGFRA | Sponge network | 0.299 | 0.57722 | -0.372 | 0.0037 | 0.264 |
| 50714 | RP11-1006G14.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-590-3p | 11 | LATS2 | Sponge network | 0.559 | 0.00026 | 0.159 | 0.2304 | 0.264 |
| 50715 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | YAP1 | Sponge network | 1.308 | 0 | 0.22 | 0.11836 | 0.264 |
| 50716 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 33 | RBMS3 | Sponge network | -0.223 | 0.47816 | -0.519 | 0.03551 | 0.264 |
| 50717 | RP11-356J5.12 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | RIMS2 | Sponge network | -0.899 | 6.0E-5 | -1.365 | 0.00208 | 0.264 |
| 50718 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | NR4A3 | Sponge network | -0.615 | 0.00015 | -1.385 | 0 | 0.264 |
| 50719 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-96-5p | 23 | FOXO3 | Sponge network | 0.594 | 0.41522 | -0.04 | 0.72028 | 0.264 |
| 50720 | RP11-276H19.2 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-502-5p | 17 | DST | Sponge network | -0.472 | 0.48851 | -0.713 | 0.00096 | 0.264 |
| 50721 | RP11-264B14.1 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p | 12 | CCDC6 | Sponge network | 0.557 | 0.00564 | 0.27 | 0.02555 | 0.264 |
| 50722 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p | 28 | ZFHX4 | Sponge network | 0.921 | 0.00118 | 0.018 | 0.95574 | 0.264 |
| 50723 | RP11-384O8.1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | PCDH17 | Sponge network | -0.738 | 0.21233 | 0.731 | 2.0E-5 | 0.264 |
| 50724 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 10 | EDA2R | Sponge network | 0.174 | 0.30034 | -0.587 | 0.01598 | 0.264 |
| 50725 | HCG11 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | 0.385 | 0.10067 | 0.142 | 0.04938 | 0.264 |
| 50726 | RP11-758N13.1 |
hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-375;hsa-miR-424-5p | 13 | PTPN14 | Sponge network | 2.939 | 3.0E-5 | 0.144 | 0.34595 | 0.264 |
| 50727 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 15 | GPAM | Sponge network | -0.096 | 0.67958 | 0.142 | 0.29732 | 0.264 |
| 50728 | AC006116.24 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-33a-5p | 10 | RIMBP2 | Sponge network | -0.473 | 0.07602 | -1.968 | 5.0E-5 | 0.264 |
| 50729 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-377-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | PER2 | Sponge network | 0.683 | 1.0E-5 | -0.396 | 0.00269 | 0.264 |
| 50730 | C20orf166-AS1 |
hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-577 | 14 | PIK3CA | Sponge network | -2.69 | 0.0001 | 0.195 | 0.06908 | 0.264 |
| 50731 | RP11-473I1.9 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 16 | ZMYND11 | Sponge network | 0.683 | 1.0E-5 | -0.2 | 0.05449 | 0.264 |
| 50732 | RP1-68D18.2 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-429;hsa-miR-628-5p | 10 | LPP | Sponge network | 1.987 | 0 | -0.459 | 0.04475 | 0.264 |
| 50733 | RP11-244O19.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-382-5p;hsa-miR-590-3p | 14 | INPP4B | Sponge network | -0.096 | 0.67958 | -0.097 | 0.60503 | 0.264 |
| 50734 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | ERCC4 | Sponge network | 1.302 | 7.0E-5 | 0.126 | 0.15266 | 0.264 |
| 50735 | PART1 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-508-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | RASSF3 | Sponge network | -2.925 | 0 | -0.226 | 0.11691 | 0.264 |
| 50736 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429 | 12 | HERC2 | Sponge network | 0.253 | 0.09565 | 0.114 | 0.30638 | 0.264 |
| 50737 | A2M-AS1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 16 | LUZP2 | Sponge network | -0.08 | 0.90092 | -0.056 | 0.89416 | 0.264 |
| 50738 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | APC | Sponge network | 0.274 | 0.07681 | -0.255 | 0.04051 | 0.264 |
| 50739 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PRKAB2 | Sponge network | 1.308 | 0 | -0.367 | 0.01371 | 0.264 |
| 50740 | LINC00654 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | NF1 | Sponge network | 0.374 | 0.20054 | 0.51 | 0 | 0.264 |
| 50741 | MEG3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-5p;hsa-miR-96-5p | 18 | PRKAR1A | Sponge network | 0.249 | 0.35902 | -0.063 | 0.50314 | 0.264 |
| 50742 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 14 | RARB | Sponge network | -0.726 | 0.00132 | -0.825 | 2.0E-5 | 0.264 |
| 50743 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 22 | MEF2C | Sponge network | 0.817 | 3.0E-5 | -0.499 | 0.01448 | 0.264 |
| 50744 | PTOV1-AS1 |
hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 10 | GPAM | Sponge network | 0.87 | 0 | 0.142 | 0.29732 | 0.264 |
| 50745 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | -0.125 | 0.49414 | 0.783 | 0.00072 | 0.264 |
| 50746 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p | 13 | PDZD2 | Sponge network | 0.494 | 0.00339 | -0.909 | 3.0E-5 | 0.264 |
| 50747 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-577;hsa-miR-590-3p | 13 | MAP3K12 | Sponge network | -0.896 | 0.00099 | -0.057 | 0.59369 | 0.264 |
| 50748 | LPP-AS2 |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-3127-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 21 | CTIF | Sponge network | -0.18 | 0.20343 | -0.389 | 0.00549 | 0.264 |
| 50749 | LINC00654 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 12 | TPO | Sponge network | 0.374 | 0.20054 | -1.87 | 2.0E-5 | 0.264 |
| 50750 | PCBP1-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | ATM | Sponge network | -0.567 | 0 | 0.178 | 0.21568 | 0.264 |
| 50751 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-5p | 11 | YAP1 | Sponge network | -0.473 | 0.07602 | 0.22 | 0.11836 | 0.264 |
| 50752 | ADAMTS9-AS2 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-421;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | RIT1 | Sponge network | -2.452 | 0 | -0.296 | 0.00054 | 0.264 |
| 50753 | ARHGAP5-AS1 |
hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | PRKAB2 | Sponge network | -0.644 | 0.00021 | -0.367 | 0.01371 | 0.264 |
| 50754 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-590-3p | 11 | KANK1 | Sponge network | -0.223 | 0.47816 | -0.55 | 0.00134 | 0.264 |
| 50755 | MRPL23-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-484;hsa-miR-532-3p | 15 | CBL | Sponge network | -0.278 | 0.76559 | 0.291 | 0.01267 | 0.264 |
| 50756 | RP11-46C24.7 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-484 | 17 | ABL1 | Sponge network | 0.392 | 0.0278 | -0.215 | 0.07804 | 0.264 |
| 50757 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 12 | GALNT13 | Sponge network | 0.853 | 0.15352 | -0.857 | 0.07439 | 0.264 |
| 50758 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | BMPR2 | Sponge network | 1.308 | 0 | 0.206 | 0.0635 | 0.264 |
| 50759 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-590-3p | 14 | GPAM | Sponge network | -0.355 | 0.0077 | 0.142 | 0.29732 | 0.264 |
| 50760 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-940 | 47 | WDFY3 | Sponge network | 0.082 | 0.55654 | -0.201 | 0.0967 | 0.264 |
| 50761 | MYLK-AS1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-424-5p | 14 | BRD3 | Sponge network | -1.331 | 3.0E-5 | 0.37 | 0.00013 | 0.264 |
| 50762 | CTA-204B4.2 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 11 | TAL1 | Sponge network | 0.765 | 7.0E-5 | -0.462 | 0.00574 | 0.264 |
| 50763 | ADIRF-AS1 |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ZMYND11 | Sponge network | -0.223 | 0.47816 | -0.2 | 0.05449 | 0.264 |
| 50764 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p | 25 | FOXO3 | Sponge network | 0.541 | 0.00125 | -0.04 | 0.72028 | 0.264 |
| 50765 | RP11-46C24.7 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | NIN | Sponge network | 0.392 | 0.0278 | -0.253 | 0.13794 | 0.264 |
| 50766 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p | 10 | LHX6 | Sponge network | -0.829 | 0.01343 | -0.087 | 0.69193 | 0.264 |
| 50767 | CTC-457L16.2 |
hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | PRUNE2 | Sponge network | 1.153 | 0.00114 | -1.733 | 0.00022 | 0.264 |
| 50768 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SLC6A15 | Sponge network | -0.223 | 0.47816 | -2.373 | 2.0E-5 | 0.264 |
| 50769 | RP11-967K21.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-625-5p;hsa-miR-940 | 16 | CELF4 | Sponge network | 0.056 | 0.81195 | -1.135 | 0.00344 | 0.264 |
| 50770 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 18 | TRIM33 | Sponge network | 0.401 | 0.00066 | 0.402 | 7.0E-5 | 0.264 |
| 50771 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-493-5p;hsa-miR-590-3p | 13 | MYCBP2 | Sponge network | 0.225 | 0.30644 | -0.027 | 0.82016 | 0.264 |
| 50772 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-5p | 21 | RSPO2 | Sponge network | 0.16 | 0.50785 | -3.038 | 0 | 0.263 |
| 50773 | RP11-156P1.3 |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 14 | CELF4 | Sponge network | 0.214 | 0.18246 | -1.135 | 0.00344 | 0.263 |
| 50774 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 35 | EPHA7 | Sponge network | 0.177 | 0.16681 | -2.197 | 3.0E-5 | 0.263 |
| 50775 | AC004540.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-629-3p | 14 | ESR1 | Sponge network | -2.549 | 0.00528 | -1.035 | 3.0E-5 | 0.263 |
| 50776 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | ZMYM2 | Sponge network | 0.422 | 0.27021 | 0.279 | 0.01273 | 0.263 |
| 50777 | CD27-AS1 |
hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-486-5p | 10 | DNMT3A | Sponge network | 0.177 | 0.16681 | 0.302 | 0.0262 | 0.263 |
| 50778 | LINC00667 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-589-5p | 10 | MN1 | Sponge network | -0.235 | 0.15142 | -0.358 | 0.24172 | 0.263 |
| 50779 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 13 | EGR2 | Sponge network | 0.253 | 0.09565 | 0.309 | 0.17079 | 0.263 |
| 50780 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-96-5p | 23 | ZFHX4 | Sponge network | -0.326 | 0.14314 | 0.018 | 0.95574 | 0.263 |
| 50781 | LIPE-AS1 |
hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-96-5p | 15 | PRKAR1A | Sponge network | 0.078 | 0.55803 | -0.063 | 0.50314 | 0.263 |
| 50782 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | SOX5 | Sponge network | 0.392 | 0.0278 | -1.091 | 4.0E-5 | 0.263 |
| 50783 | NEAT1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | PRKAB2 | Sponge network | 0.705 | 0.00014 | -0.367 | 0.01371 | 0.263 |
| 50784 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | EBF1 | Sponge network | 0.05 | 0.95483 | -0.384 | 0.08856 | 0.263 |
| 50785 | RP11-344B5.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p | 10 | IL17RD | Sponge network | -0.055 | 0.88482 | -0.019 | 0.94873 | 0.263 |
| 50786 | RAB30-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 10 | NF1 | Sponge network | 0.752 | 0 | 0.51 | 0 | 0.263 |
| 50787 | NEAT1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | ZNF208 | Sponge network | 0.705 | 0.00014 | -0.4 | 0.22381 | 0.263 |
| 50788 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-744-3p | 15 | HIP1 | Sponge network | -2.095 | 0.00112 | 0.774 | 0 | 0.263 |
| 50789 | SNX29P2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-378a-3p;hsa-miR-424-5p;hsa-miR-582-3p | 17 | ABL2 | Sponge network | 0.4 | 0.14611 | 0.82 | 0 | 0.263 |
| 50790 | RP11-337C18.8 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-5p | 20 | BMPR2 | Sponge network | 0.496 | 0.0011 | 0.206 | 0.0635 | 0.263 |
| 50791 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-361-5p;hsa-miR-455-3p;hsa-miR-940 | 20 | MCC | Sponge network | -1.942 | 0 | -1.005 | 0 | 0.263 |
| 50792 | RP5-1085F17.3 |
hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | UBE2D1 | Sponge network | 0.541 | 0.00125 | 0.468 | 0 | 0.263 |
| 50793 | RP11-517B11.7 |
hsa-let-7e-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-33a-3p | 12 | COPS2 | Sponge network | 0.706 | 0 | -0.178 | 0.04974 | 0.263 |
| 50794 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-590-3p | 16 | EBF3 | Sponge network | -0.053 | 0.80685 | -0.058 | 0.81437 | 0.263 |
| 50795 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577 | 10 | CNTN1 | Sponge network | 0.357 | 0.72407 | -2.594 | 0 | 0.263 |
| 50796 | AGAP11 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-590-5p | 11 | LHFP | Sponge network | -1.645 | 0 | -0.167 | 0.41371 | 0.263 |
| 50797 | RP11-806O11.1 |
hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | UBR3 | Sponge network | 0.284 | 0.52463 | -0.021 | 0.82774 | 0.263 |
| 50798 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p | 24 | ANK2 | Sponge network | 0.95 | 0.15943 | -1.603 | 1.0E-5 | 0.263 |
| 50799 | RP11-999E24.3 |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p | 11 | PAIP2 | Sponge network | -0.693 | 0.05629 | -0.515 | 0 | 0.263 |
| 50800 | OIP5-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940 | 20 | PEG3 | Sponge network | 0.421 | 0.00084 | -1.74 | 0 | 0.263 |
| 50801 | AC116366.6 |
hsa-miR-1266-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-877-5p | 10 | ANK2 | Sponge network | 0.329 | 0.11122 | -1.603 | 1.0E-5 | 0.263 |
| 50802 | HOTAIRM1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | PRRX1 | Sponge network | -0.053 | 0.80685 | 1.316 | 0 | 0.263 |
| 50803 | RP11-725P16.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 37 | MDM4 | Sponge network | 0.422 | 0.27021 | 0.345 | 0.00762 | 0.263 |
| 50804 | RP11-21A7A.2 |
hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-24-2-5p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-532-5p | 16 | PPM1L | Sponge network | -1.116 | 0.23686 | -0.537 | 0.00616 | 0.263 |
| 50805 | RP11-195F19.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-93-5p | 10 | LHX6 | Sponge network | -0.726 | 0.00132 | -0.087 | 0.69193 | 0.263 |
| 50806 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p | 20 | GREM1 | Sponge network | 0.421 | 0.00084 | 0.902 | 0.01691 | 0.263 |
| 50807 | NDUFA6-AS1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | SH3GLB1 | Sponge network | -0.355 | 0.0077 | -0.115 | 0.14773 | 0.263 |
| 50808 | AC141928.1 |
hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | PCDH10 | Sponge network | 0.853 | 0.15352 | -1.644 | 0.0078 | 0.263 |
| 50809 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | CDC73 | Sponge network | 0.768 | 7.0E-5 | 0.094 | 0.20463 | 0.263 |
| 50810 | CTC-429P9.5 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-532-5p;hsa-miR-590-3p | 20 | PPM1L | Sponge network | 0.178 | 0.29106 | -0.537 | 0.00616 | 0.263 |
| 50811 | LINC01082 |
hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p | 11 | PTPRT | Sponge network | -3.322 | 8.0E-5 | -0.907 | 0.03727 | 0.263 |
| 50812 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 23 | TGFBR2 | Sponge network | -0.615 | 0.00015 | -0.243 | 0.11408 | 0.263 |
| 50813 | CROCCP2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | RPS6KA3 | Sponge network | 0.79 | 0 | 0.256 | 0.02419 | 0.263 |
| 50814 | DNM3OS |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-940 | 19 | SIRT1 | Sponge network | 0.572 | 0.01722 | 0.109 | 0.2593 | 0.263 |
| 50815 | RP11-157J24.2 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-576-5p | 19 | DTNA | Sponge network | -1.664 | 0.05165 | -1.648 | 1.0E-5 | 0.263 |
| 50816 | HCG22 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | TRPS1 | Sponge network | 0.816 | 0.32368 | -0.191 | 0.44109 | 0.263 |
| 50817 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TBX18 | Sponge network | -0.769 | 0.00035 | 0.843 | 0.02945 | 0.263 |
| 50818 | CTB-51J22.1 |
hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 16 | SSBP2 | Sponge network | -0.803 | 0.23737 | -1.58 | 0 | 0.263 |
| 50819 | CTD-3099C6.9 |
hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-629-3p | 10 | PIK3R1 | Sponge network | 1.361 | 0 | -0.133 | 0.36854 | 0.263 |
| 50820 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 31 | CBL | Sponge network | 0.335 | 0.20596 | 0.291 | 0.01267 | 0.263 |
| 50821 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p | 10 | CNTNAP3 | Sponge network | 0.395 | 0.15451 | -1.465 | 0.00033 | 0.263 |
| 50822 | RP11-678G14.3 |
hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-484;hsa-miR-7-1-3p | 14 | RIMS2 | Sponge network | -4.477 | 0 | -1.365 | 0.00208 | 0.263 |
| 50823 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p | 16 | USP6 | Sponge network | 0.4 | 0.14611 | 0.188 | 0.3871 | 0.263 |
| 50824 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 10 | DACT1 | Sponge network | -0.72 | 0.24239 | 0.783 | 0.00072 | 0.263 |
| 50825 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 26 | SASH1 | Sponge network | -0.246 | 0.35034 | -0.502 | 0.00175 | 0.263 |
| 50826 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | ST6GALNAC3 | Sponge network | -0.777 | 0.1134 | -0.987 | 0 | 0.263 |
| 50827 | SOX2-OT |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | TMEFF2 | Sponge network | -1.264 | 6.0E-5 | -2.426 | 0 | 0.263 |
| 50828 | RP11-488L18.10 |
hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 13 | CCDC6 | Sponge network | 1.392 | 0 | 0.27 | 0.02555 | 0.263 |
| 50829 | RP11-158K1.3 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | PCDH9 | Sponge network | 0.349 | 0.01695 | -1.823 | 4.0E-5 | 0.263 |
| 50830 | RP11-158K1.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 35 | GAS7 | Sponge network | 0.349 | 0.01695 | -0.555 | 0.01602 | 0.263 |
| 50831 | DNAJC27-AS1 |
hsa-miR-141-3p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-935 | 11 | STAT5B | Sponge network | -0.969 | 2.0E-5 | -0.182 | 0.1082 | 0.263 |
| 50832 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | FYN | Sponge network | 0.853 | 0.15352 | -0.131 | 0.37051 | 0.263 |
| 50833 | H19 |
hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-454-3p | 18 | BMPR2 | Sponge network | 2.439 | 0 | 0.206 | 0.0635 | 0.263 |
| 50834 | USP3-AS1 |
hsa-miR-107;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421 | 10 | RNF103 | Sponge network | -0.162 | 0.47687 | -0.341 | 1.0E-5 | 0.263 |
| 50835 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-93-5p | 15 | CDHR3 | Sponge network | -0.13 | 0.48734 | -0.977 | 4.0E-5 | 0.263 |
| 50836 | LINC01021 |
hsa-miR-125a-3p;hsa-miR-144-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p | 12 | MDM4 | Sponge network | 4.362 | 0 | 0.345 | 0.00762 | 0.263 |
| 50837 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-5p | 34 | MEF2C | Sponge network | -0.384 | 0.40652 | -0.499 | 0.01448 | 0.263 |
| 50838 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | MED12L | Sponge network | -0.342 | 0.02288 | -0.194 | 0.55299 | 0.263 |
| 50839 | AC090587.5 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-501-5p;hsa-miR-589-3p | 11 | TACC1 | Sponge network | -0.088 | 0.65011 | -0.362 | 0.05406 | 0.263 |
| 50840 | BZRAP1-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-485-3p | 20 | ZFX | Sponge network | -0.465 | 0.04703 | 0.09 | 0.42563 | 0.263 |
| 50841 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-188-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 15 | CDHR3 | Sponge network | 0.537 | 0.00139 | -0.977 | 4.0E-5 | 0.263 |
| 50842 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p | 10 | FBXO32 | Sponge network | -0.421 | 0.12556 | -0.495 | 0.07561 | 0.263 |
| 50843 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 17 | CRTC3 | Sponge network | -0.737 | 0.10822 | 0.164 | 0.16786 | 0.263 |
| 50844 | RP4-669H2.1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-338-5p | 12 | ZFHX4 | Sponge network | 1.711 | 0.00019 | 0.018 | 0.95574 | 0.263 |
| 50845 | SNX29P2 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-33a-3p;hsa-miR-500a-3p;hsa-miR-7-1-3p | 14 | LIMCH1 | Sponge network | 0.4 | 0.14611 | -0.915 | 1.0E-5 | 0.263 |
| 50846 | RP11-116O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | DAB2 | Sponge network | 0.05 | 0.95483 | 0.431 | 0.0113 | 0.263 |
| 50847 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-338-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CXXC4 | Sponge network | -0.223 | 0.47816 | -0.433 | 0.21055 | 0.263 |
| 50848 | BZRAP1-AS1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-577;hsa-miR-582-3p | 12 | PIK3CA | Sponge network | -0.465 | 0.04703 | 0.195 | 0.06908 | 0.263 |
| 50849 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-655-3p;hsa-miR-7-1-3p | 13 | UBE4B | Sponge network | -0.031 | 0.89063 | -0.235 | 0.007 | 0.263 |
| 50850 | RP11-798G7.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-455-3p | 24 | RUNX1T1 | Sponge network | 0.858 | 3.0E-5 | -0.344 | 0.2374 | 0.263 |
| 50851 | AC116366.6 |
hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-942-5p | 10 | NRXN1 | Sponge network | 0.329 | 0.11122 | -3.778 | 0 | 0.263 |
| 50852 | LINC00863 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 36 | DST | Sponge network | 0.189 | 0.13981 | -0.713 | 0.00096 | 0.263 |
| 50853 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 48 | NFIB | Sponge network | 0.335 | 0.20596 | 0.023 | 0.90795 | 0.263 |
| 50854 | AC034220.3 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ATRX | Sponge network | 0.309 | 0.07304 | 0.261 | 0.04408 | 0.263 |
| 50855 | RP11-180O5.2 |
hsa-let-7g-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-7-1-3p | 14 | CREB1 | Sponge network | 3.607 | 0.0001 | 0.203 | 0.00537 | 0.263 |
| 50856 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-576-5p | 14 | PBX1 | Sponge network | 0.411 | 0.1335 | -1.206 | 0 | 0.263 |
| 50857 | RP11-532F6.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | ZFX | Sponge network | -0.896 | 0.00099 | 0.09 | 0.42563 | 0.263 |
| 50858 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | TSHZ3 | Sponge network | 0.603 | 0.00127 | -0.556 | 0.01516 | 0.263 |
| 50859 | TAPT1-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-582-3p | 12 | ATRX | Sponge network | 0.403 | 2.0E-5 | 0.261 | 0.04408 | 0.263 |
| 50860 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | PBRM1 | Sponge network | -1.575 | 6.0E-5 | -0.029 | 0.78601 | 0.263 |
| 50861 | CTD-2245F17.3 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-154-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-766-3p;hsa-miR-940 | 13 | CHD5 | Sponge network | -0.421 | 0.12556 | -0.922 | 0.01521 | 0.263 |
| 50862 | RP11-276H19.2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p | 10 | ADAMTSL3 | Sponge network | -0.472 | 0.48851 | -1.559 | 0 | 0.263 |
| 50863 | CD27-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-577 | 17 | NRK | Sponge network | 0.177 | 0.16681 | 0.656 | 0.19217 | 0.263 |
| 50864 | RP4-605O3.4 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 10 | BRD3 | Sponge network | 0.262 | 0.0475 | 0.37 | 0.00013 | 0.263 |
| 50865 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PBRM1 | Sponge network | 0.272 | 0.44692 | -0.029 | 0.78601 | 0.263 |
| 50866 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 25 | USP12 | Sponge network | -0.235 | 0.15142 | -0.102 | 0.40768 | 0.263 |
| 50867 | CTC-296K1.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-625-5p | 14 | CRTC1 | Sponge network | -2.095 | 0.00112 | -0.116 | 0.28412 | 0.263 |
| 50868 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 20 | RNF144A | Sponge network | -0.793 | 7.0E-5 | 0.594 | 0.0009 | 0.263 |
| 50869 | AC003090.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 29 | BHLHE41 | Sponge network | -2.117 | 0.00161 | 0.353 | 0.12753 | 0.263 |
| 50870 | RP4-605O3.4 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-590-3p | 24 | DTNA | Sponge network | 0.262 | 0.0475 | -1.648 | 1.0E-5 | 0.263 |
| 50871 | EIF3J-AS1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p | 43 | MDM4 | Sponge network | 0.331 | 0.00509 | 0.345 | 0.00762 | 0.263 |
| 50872 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | FGF5 | Sponge network | 0.225 | 0.30644 | 0.549 | 0.28659 | 0.263 |
| 50873 | EMX2OS |
hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 18 | CELF4 | Sponge network | -0.777 | 0.1134 | -1.135 | 0.00344 | 0.263 |
| 50874 | AP001347.6 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | BAZ2B | Sponge network | -1.161 | 0.00591 | -0.276 | 0.02833 | 0.263 |
| 50875 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 44 | SASH1 | Sponge network | -2.925 | 0 | -0.502 | 0.00175 | 0.263 |
| 50876 | CTD-2314G24.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-324-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-96-5p | 18 | RIMBP2 | Sponge network | 0.246 | 0.59912 | -1.968 | 5.0E-5 | 0.263 |
| 50877 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 56 | BMPR2 | Sponge network | 0.246 | 0.59912 | 0.206 | 0.0635 | 0.263 |
| 50878 | RP11-800A3.7 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-34a-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-942-5p | 12 | IGFBP5 | Sponge network | 0.18 | 0.7147 | -0.806 | 0.00105 | 0.263 |
| 50879 | LINC00702 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | PTCH1 | Sponge network | -0.737 | 0.06182 | -0.1 | 0.6179 | 0.263 |
| 50880 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-629-3p | 20 | PSIP1 | Sponge network | -1.111 | 0.0003 | -0.005 | 0.96897 | 0.263 |
| 50881 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-495-3p;hsa-miR-590-3p | 10 | MAPK1 | Sponge network | 0.545 | 0.00064 | -0.017 | 0.81465 | 0.263 |
| 50882 | PAN3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 11 | TRPS1 | Sponge network | 0.062 | 0.55274 | -0.191 | 0.44109 | 0.263 |
| 50883 | LINC00969 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-625-3p;hsa-miR-628-5p | 27 | BMPR2 | Sponge network | 0.683 | 1.0E-5 | 0.206 | 0.0635 | 0.263 |
| 50884 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | LRRC55 | Sponge network | 0.61 | 0.01593 | -0.026 | 0.93163 | 0.263 |
| 50885 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | ST6GALNAC3 | Sponge network | 0.082 | 0.55654 | -0.987 | 0 | 0.263 |
| 50886 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-425-5p | 12 | ITGB3 | Sponge network | 0.37 | 0.27614 | -0.011 | 0.95751 | 0.263 |
| 50887 | RP11-121C2.2 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p | 12 | PAWR | Sponge network | 1.147 | 0 | 0.636 | 0 | 0.263 |
| 50888 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-93-5p | 18 | MAF | Sponge network | 1.153 | 0.00114 | -1.257 | 0 | 0.263 |
| 50889 | CTD-2666L21.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-486-5p | 27 | DTNA | Sponge network | 0.368 | 0.20093 | -1.648 | 1.0E-5 | 0.263 |
| 50890 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-3p | 16 | SOX11 | Sponge network | 0.395 | 0.15451 | 2.68 | 0 | 0.263 |
| 50891 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-5p | 12 | PRKAA1 | Sponge network | 1.093 | 0 | 0.317 | 0.0031 | 0.263 |
| 50892 | AC093627.8 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p | 10 | RASSF8 | Sponge network | -0.787 | 0.3928 | -0.122 | 0.65591 | 0.263 |
| 50893 | RP11-411K7.1 |
hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | TP63 | Sponge network | 0.155 | 0.81273 | -0.363 | 0.38191 | 0.263 |
| 50894 | ADIRF-AS1 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | ANK2 | Sponge network | -0.223 | 0.47816 | -1.603 | 1.0E-5 | 0.263 |
| 50895 | A1BG-AS1 |
hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p | 10 | HIP1 | Sponge network | -0.164 | 0.45106 | 0.774 | 0 | 0.263 |
| 50896 | RP11-196G18.24 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-582-5p | 18 | CREB1 | Sponge network | 1.262 | 0 | 0.203 | 0.00537 | 0.263 |
| 50897 | RP11-130L8.1 |
hsa-miR-130b-3p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ROBO2 | Sponge network | -0.663 | 0.10887 | 0.092 | 0.78768 | 0.263 |
| 50898 | GNG12-AS1 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | PTCH1 | Sponge network | -0.793 | 7.0E-5 | -0.1 | 0.6179 | 0.263 |
| 50899 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-766-3p | 22 | IL17RD | Sponge network | -0.602 | 0.00331 | -0.019 | 0.94873 | 0.263 |
| 50900 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 34 | RICTOR | Sponge network | 0.572 | 0.01722 | 0.388 | 0.00188 | 0.263 |
| 50901 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p | 10 | TSHZ3 | Sponge network | 0.542 | 0.00054 | -0.556 | 0.01516 | 0.263 |
| 50902 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 27 | SETBP1 | Sponge network | 0.311 | 0.10344 | -1.302 | 1.0E-5 | 0.263 |
| 50903 | TUG1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-539-5p | 10 | TCF12 | Sponge network | 0.972 | 0 | 0.174 | 0.13271 | 0.263 |
| 50904 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 13 | TIMP3 | Sponge network | -0.72 | 0.24239 | -0.546 | 0.02307 | 0.263 |
| 50905 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | NUAK1 | Sponge network | -2.915 | 0.00015 | 0.904 | 0 | 0.263 |
| 50906 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p | 11 | MLLT10 | Sponge network | 0.792 | 0.0049 | 0.105 | 0.33292 | 0.263 |
| 50907 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 38 | UNC80 | Sponge network | 0.064 | 0.72934 | -1.934 | 0 | 0.263 |
| 50908 | DLGAP1-AS2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 10 | MOB1B | Sponge network | 2.406 | 0 | 0.197 | 0.15266 | 0.263 |
| 50909 | A2M-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 13 | STARD13 | Sponge network | -0.08 | 0.90092 | -0.187 | 0.27383 | 0.263 |
| 50910 | RP11-35G9.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 11 | CDH19 | Sponge network | 0.622 | 0.00015 | -3.434 | 0 | 0.263 |
| 50911 | AC002117.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | AKAP6 | Sponge network | 0.476 | 0.19203 | -1.586 | 3.0E-5 | 0.263 |
| 50912 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | WDFY3 | Sponge network | 0.419 | 0.0319 | -0.201 | 0.0967 | 0.263 |
| 50913 | RP11-6O2.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 14 | LUZP2 | Sponge network | 0.331 | 0.57247 | -0.056 | 0.89416 | 0.263 |
| 50914 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-2110;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-92a-3p | 15 | RIMBP2 | Sponge network | 0.214 | 0.18246 | -1.968 | 5.0E-5 | 0.263 |
| 50915 | TTC28-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-664a-3p;hsa-miR-664a-5p | 19 | SOX11 | Sponge network | 0.373 | 0.00068 | 2.68 | 0 | 0.263 |
| 50916 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 25 | PRKCE | Sponge network | 0.194 | 0.64278 | -0.403 | 0.00056 | 0.263 |
| 50917 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-429 | 10 | GREM1 | Sponge network | 0.274 | 0.07681 | 0.902 | 0.01691 | 0.263 |
| 50918 | AC127904.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-495-3p | 11 | VPS53 | Sponge network | 1.308 | 0 | 0.103 | 0.22667 | 0.263 |
| 50919 | RP11-983P16.4 |
hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-542-3p | 11 | THRA | Sponge network | 0.41 | 0.02214 | -0.307 | 0.05099 | 0.263 |
| 50920 | MRVI1-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-3p;hsa-miR-7-1-3p | 13 | CSMD1 | Sponge network | -1.092 | 4.0E-5 | -0.63 | 0.18881 | 0.263 |
| 50921 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-5p | 16 | LATS1 | Sponge network | 0.921 | 0.00118 | 0.109 | 0.34208 | 0.263 |
| 50922 | RP11-212P7.2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | UBE4B | Sponge network | 0.219 | 0.1516 | -0.235 | 0.007 | 0.263 |
| 50923 | AC138035.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-1976;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p | 17 | CBX5 | Sponge network | 1.073 | 0 | 0.165 | 0.24446 | 0.263 |
| 50924 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-3p | 17 | EBF3 | Sponge network | 0.859 | 0.00331 | -0.058 | 0.81437 | 0.263 |
| 50925 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SAMHD1 | Sponge network | -0.487 | 0.02157 | 0.072 | 0.62895 | 0.263 |
| 50926 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | FGF5 | Sponge network | -0.901 | 0.00019 | 0.549 | 0.28659 | 0.263 |
| 50927 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | CLASP2 | Sponge network | 0.299 | 0.57722 | -0.038 | 0.71 | 0.263 |
| 50928 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-664a-3p | 14 | PRKAA1 | Sponge network | 0.917 | 0 | 0.317 | 0.0031 | 0.263 |
| 50929 | LINC00702 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 43 | PPP1R9A | Sponge network | -0.737 | 0.06182 | -0.903 | 0.04254 | 0.263 |
| 50930 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 21 | PTPRD | Sponge network | 0.131 | 0.8436 | -1.005 | 0.00064 | 0.263 |
| 50931 | RP11-218M22.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | PDE4DIP | Sponge network | 0.197 | 0.25618 | -0.282 | 0.0257 | 0.263 |
| 50932 | GS1-124K5.11 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-451a;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-5p | 32 | BCL2 | Sponge network | 0.494 | 0.00339 | -0.899 | 0 | 0.263 |
| 50933 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-98-5p | 22 | THBS1 | Sponge network | -0.405 | 0.22013 | 0.741 | 0.00386 | 0.263 |
| 50934 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-7-1-3p | 27 | PAFAH1B1 | Sponge network | -4.546 | 0 | -0.429 | 0 | 0.263 |
| 50935 | RP11-37B2.1 |
hsa-let-7g-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p | 11 | KDM3A | Sponge network | 1.03 | 0 | 0.132 | 0.22711 | 0.263 |
| 50936 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | DMTF1 | Sponge network | 0.082 | 0.55654 | 0.248 | 0.02726 | 0.263 |
| 50937 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | HECA | Sponge network | -0.513 | 0.04703 | -0.217 | 0.03463 | 0.263 |
| 50938 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-7-1-3p | 17 | CXXC4 | Sponge network | 0.4 | 0.14611 | -0.433 | 0.21055 | 0.263 |
| 50939 | RP5-994D16.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SOX5 | Sponge network | 0.252 | 0.08508 | -1.091 | 4.0E-5 | 0.263 |
| 50940 | AC002117.1 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | PIK3R1 | Sponge network | 0.476 | 0.19203 | -0.133 | 0.36854 | 0.263 |
| 50941 | RP11-144G6.12 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p | 13 | PIK3R1 | Sponge network | 0.826 | 1.0E-5 | -0.133 | 0.36854 | 0.263 |
| 50942 | RP5-894A10.6 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-485-3p;hsa-miR-495-3p | 11 | PRKAA1 | Sponge network | 0.697 | 9.0E-5 | 0.317 | 0.0031 | 0.263 |
| 50943 | HOXD-AS2 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-3607-3p | 10 | LIMCH1 | Sponge network | -0.208 | 0.65592 | -0.915 | 1.0E-5 | 0.263 |
| 50944 | AC005062.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p | 15 | MLLT10 | Sponge network | -0.019 | 0.81504 | 0.105 | 0.33292 | 0.263 |
| 50945 | RP11-326G21.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | LRIG1 | Sponge network | -0.021 | 0.93598 | -0.537 | 0.00588 | 0.263 |
| 50946 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | ARHGAP29 | Sponge network | -1.281 | 0.00012 | 0.131 | 0.42953 | 0.263 |
| 50947 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-455-3p | 13 | CADM3 | Sponge network | 0.082 | 0.55397 | -3.663 | 0 | 0.263 |
| 50948 | RP1-151F17.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | PRRX1 | Sponge network | 0.222 | 0.29133 | 1.316 | 0 | 0.263 |
| 50949 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 24 | ARID5B | Sponge network | 0.349 | 0.01695 | 0.342 | 0.01676 | 0.263 |
| 50950 | SH3BP5-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-369-3p;hsa-miR-625-5p | 10 | ARID2 | Sponge network | 0.267 | 0.2937 | 0.235 | 0.02697 | 0.263 |
| 50951 | AC074289.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-143-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-940 | 32 | MDM4 | Sponge network | -0.326 | 0.14314 | 0.345 | 0.00762 | 0.263 |
| 50952 | LINC00094 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 25 | LRIG1 | Sponge network | 0.253 | 0.09565 | -0.537 | 0.00588 | 0.263 |
| 50953 | RP11-344B5.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b | 11 | BHLHE41 | Sponge network | -0.055 | 0.88482 | 0.353 | 0.12753 | 0.263 |
| 50954 | RP5-998N21.10 |
hsa-miR-103a-2-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-30b-3p | 10 | FAT3 | Sponge network | -0.098 | 0.76298 | -1.601 | 0.00051 | 0.263 |
| 50955 | DNM1P35 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 17 | NAV3 | Sponge network | 0.051 | 0.83388 | 0.212 | 0.47729 | 0.263 |
| 50956 | RP11-693J15.4 |
hsa-let-7g-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 15 | PCDH10 | Sponge network | -0.72 | 0.24239 | -1.644 | 0.0078 | 0.263 |
| 50957 | RP11-545G3.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p | 10 | ZFX | Sponge network | 2.622 | 0.00025 | 0.09 | 0.42563 | 0.263 |
| 50958 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p | 11 | PLSCR4 | Sponge network | -3.586 | 0.00032 | -0.474 | 0.03833 | 0.263 |
| 50959 | ADIRF-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-3p | 22 | MYBL1 | Sponge network | -0.223 | 0.47816 | 0.494 | 0.02152 | 0.263 |
| 50960 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-744-5p;hsa-miR-940 | 21 | CHD5 | Sponge network | 2.306 | 9.0E-5 | -0.922 | 0.01521 | 0.263 |
| 50961 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-942-5p;hsa-miR-96-5p | 22 | NRXN1 | Sponge network | -0.326 | 0.14314 | -3.778 | 0 | 0.263 |
| 50962 | RP11-968O1.5 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-940 | 13 | IGF1 | Sponge network | -0.829 | 0.01343 | -0.204 | 0.5898 | 0.263 |
| 50963 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | TLL1 | Sponge network | 1.302 | 7.0E-5 | -1.195 | 3.0E-5 | 0.263 |
| 50964 | AC024560.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p | 15 | USP6 | Sponge network | 2.166 | 0.00228 | 0.188 | 0.3871 | 0.263 |
| 50965 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-576-5p | 25 | TCF4 | Sponge network | 2.163 | 0 | 0.016 | 0.92271 | 0.263 |
| 50966 | AC005562.1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | TCF12 | Sponge network | 0.488 | 0.00084 | 0.174 | 0.13271 | 0.263 |
| 50967 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | ZFHX4 | Sponge network | 0.056 | 0.8494 | 0.018 | 0.95574 | 0.263 |
| 50968 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421 | 11 | PDGFC | Sponge network | -0.154 | 0.78939 | -0.33 | 0.06483 | 0.263 |
| 50969 | H1FX-AS1 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 11 | PRUNE2 | Sponge network | -0.206 | 0.12942 | -1.733 | 0.00022 | 0.263 |
| 50970 | CTB-113P19.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | LUZP2 | Sponge network | 0.906 | 0.31715 | -0.056 | 0.89416 | 0.263 |
| 50971 | TINCR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-96-5p | 32 | IL17RD | Sponge network | -0.664 | 0.14543 | -0.019 | 0.94873 | 0.263 |
| 50972 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p | 14 | NF1 | Sponge network | 0.336 | 0.00739 | 0.51 | 0 | 0.263 |
| 50973 | USP3-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-576-5p;hsa-miR-589-3p | 28 | MED12L | Sponge network | -0.162 | 0.47687 | -0.194 | 0.55299 | 0.263 |
| 50974 | CASC2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-625-3p | 19 | TMTC3 | Sponge network | 0.385 | 0.01306 | 0.123 | 0.22189 | 0.263 |
| 50975 | RP11-37B2.1 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-2355-3p;hsa-miR-339-5p;hsa-miR-495-3p | 12 | TSC1 | Sponge network | 1.03 | 0 | 0.103 | 0.26071 | 0.263 |
| 50976 | RBPMS-AS1 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-335-3p;hsa-miR-93-5p | 12 | JAZF1 | Sponge network | 0.144 | 0.64989 | -0.377 | 0.02677 | 0.263 |
| 50977 | AF131215.9 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 11 | GABRB2 | Sponge network | -0.322 | 0.25945 | -1.841 | 0.00029 | 0.263 |
| 50978 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | PRKAB2 | Sponge network | 0.997 | 0 | -0.367 | 0.01371 | 0.263 |
| 50979 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-335-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-484;hsa-miR-7-1-3p | 19 | CBL | Sponge network | 0.986 | 0.01022 | 0.291 | 0.01267 | 0.263 |
| 50980 | CTD-3092A11.2 |
hsa-miR-10b-3p;hsa-miR-10b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 15 | CTDSPL | Sponge network | 0.873 | 0 | 0.085 | 0.45121 | 0.263 |
| 50981 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | ZNF208 | Sponge network | -0.355 | 0.0077 | -0.4 | 0.22381 | 0.263 |
| 50982 | LINC00886 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 30 | BHLHE41 | Sponge network | -0.371 | 0.24604 | 0.353 | 0.12753 | 0.263 |
| 50983 | LINC00476 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | ZNF638 | Sponge network | -0.615 | 0.00015 | 0.22 | 0.01214 | 0.263 |
| 50984 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 14 | CHD2 | Sponge network | 0.4 | 0.14611 | 0.049 | 0.55371 | 0.263 |
| 50985 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-98-5p | 23 | ARHGAP28 | Sponge network | -0.222 | 0.32445 | -0.911 | 0.00013 | 0.263 |
| 50986 | RP11-774O3.3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CDH10 | Sponge network | -0.487 | 0.02157 | -2.397 | 0 | 0.263 |
| 50987 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 38 | LPP | Sponge network | 1.601 | 0 | -0.459 | 0.04475 | 0.263 |
| 50988 | RP11-1246C19.1 |
hsa-miR-127-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | CCDC6 | Sponge network | 1.352 | 0 | 0.27 | 0.02555 | 0.263 |
| 50989 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | USP12 | Sponge network | 0.67 | 0.00048 | -0.102 | 0.40768 | 0.263 |
| 50990 | HOXB-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-589-3p | 15 | SYT4 | Sponge network | -0.154 | 0.53954 | -3.207 | 0 | 0.263 |
| 50991 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 21 | MEF2C | Sponge network | -0.726 | 0.00132 | -0.499 | 0.01448 | 0.263 |
| 50992 | RP4-669P10.16 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-339-5p | 10 | CDH23 | Sponge network | -0.301 | 0.06597 | -0.974 | 0.00034 | 0.263 |
| 50993 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-590-3p | 11 | SLC6A15 | Sponge network | -0.376 | 0.00337 | -2.373 | 2.0E-5 | 0.263 |
| 50994 | AC016747.3 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | DNMT3A | Sponge network | 0.199 | 0.38015 | 0.302 | 0.0262 | 0.263 |
| 50995 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 20 | UBE4B | Sponge network | 0.494 | 0.00339 | -0.235 | 0.007 | 0.263 |
| 50996 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NR4A3 | Sponge network | -0.487 | 0.02157 | -1.385 | 0 | 0.263 |
| 50997 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | KAT6B | Sponge network | 0.539 | 0.03722 | -0.478 | 0.00018 | 0.263 |
| 50998 | SERHL |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-320b;hsa-miR-421 | 10 | SORCS1 | Sponge network | -0.602 | 0.00331 | -3.492 | 0 | 0.263 |
| 50999 | PCBP1-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p | 16 | TSC1 | Sponge network | -0.567 | 0 | 0.103 | 0.26071 | 0.263 |
| 51000 | AF131215.2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p | 37 | DST | Sponge network | -0.176 | 0.59692 | -0.713 | 0.00096 | 0.263 |
| 51001 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-330-3p;hsa-miR-7-1-3p | 12 | LRP1B | Sponge network | 0.238 | 0.70544 | -2.163 | 0 | 0.263 |
| 51002 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-455-3p;hsa-miR-625-5p | 11 | MCC | Sponge network | 0.858 | 3.0E-5 | -1.005 | 0 | 0.263 |
| 51003 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-942-5p | 14 | CREBBP | Sponge network | 0.395 | 0.15451 | 0.126 | 0.15186 | 0.263 |
| 51004 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | HOOK3 | Sponge network | 0.906 | 0.31715 | 0.273 | 0.09957 | 0.263 |
| 51005 | RP5-1021I20.1 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-200a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429 | 12 | PCDH9 | Sponge network | -0.785 | 0.00035 | -1.823 | 4.0E-5 | 0.263 |
| 51006 | MIR143HG |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 29 | CSMD1 | Sponge network | -0.739 | 0.0574 | -0.63 | 0.18881 | 0.263 |
| 51007 | SNHG5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-195-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-23b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-34c-5p;hsa-miR-495-3p | 11 | EIF1AX | Sponge network | 0.13 | 0.47174 | 0.307 | 0.011 | 0.263 |
| 51008 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 15 | TGFBR2 | Sponge network | 0.222 | 0.29133 | -0.243 | 0.11408 | 0.263 |
| 51009 | PROSER2-AS1 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-28-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-505-5p | 10 | TBL1XR1 | Sponge network | 1.825 | 0 | 0.578 | 0 | 0.263 |
| 51010 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | NEDD4 | Sponge network | 0.727 | 3.0E-5 | 0.02 | 0.89834 | 0.263 |
| 51011 | RP4-584D14.7 |
hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p;hsa-miR-577 | 18 | TACC1 | Sponge network | 1.294 | 0.0031 | -0.362 | 0.05406 | 0.263 |
| 51012 | MIR600HG |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-96-5p | 17 | LRIG1 | Sponge network | 0.594 | 0.41522 | -0.537 | 0.00588 | 0.263 |
| 51013 | MEG3 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p | 16 | TLE4 | Sponge network | 0.249 | 0.35902 | -1.157 | 0 | 0.263 |
| 51014 | RP11-57H14.4 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-590-3p | 10 | TOPORS | Sponge network | 0.083 | 0.66405 | -0.226 | 0.00524 | 0.263 |
| 51015 | OSER1-AS1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 10 | CNTNAP1 | Sponge network | -0.13 | 0.48734 | 0.358 | 0.13727 | 0.263 |
| 51016 | LINC00403 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-589-5p | 11 | PRKAA2 | Sponge network | -3.586 | 0.00032 | -2.462 | 0 | 0.263 |
| 51017 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 13 | CHD1 | Sponge network | 0.415 | 0.14657 | 0.322 | 0.00215 | 0.263 |
| 51018 | AC084219.4 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 18 | ZMYM2 | Sponge network | 1.296 | 0.0054 | 0.279 | 0.01273 | 0.263 |
| 51019 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-421 | 10 | DPYD | Sponge network | -0.583 | 0.14589 | -0.736 | 0.00076 | 0.263 |
| 51020 | AC124789.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-3127-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | SUFU | Sponge network | 0.073 | 0.79848 | -0.04 | 0.65489 | 0.263 |
| 51021 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p | 27 | LATS1 | Sponge network | -0.129 | 0.57177 | 0.109 | 0.34208 | 0.263 |
| 51022 | CTD-3099C6.9 |
hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-628-5p | 11 | ERC1 | Sponge network | 1.361 | 0 | -0.108 | 0.38794 | 0.263 |
| 51023 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-25-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | LATS2 | Sponge network | 0.178 | 0.29106 | 0.159 | 0.2304 | 0.263 |
| 51024 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-532-5p | 12 | SLITRK6 | Sponge network | 0.082 | 0.55397 | -2.121 | 0 | 0.263 |
| 51025 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-942-5p | 23 | HLF | Sponge network | -0.13 | 0.48734 | -2.443 | 0 | 0.263 |
| 51026 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-654-3p | 17 | ITCH | Sponge network | 0.349 | 0.01695 | 0.084 | 0.34058 | 0.263 |
| 51027 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | PTPRB | Sponge network | 0.219 | 0.1516 | 0.105 | 0.47122 | 0.263 |
| 51028 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 31 | DDX6 | Sponge network | -0.709 | 0.18168 | 0.091 | 0.36428 | 0.263 |
| 51029 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 23 | KIF1B | Sponge network | 0.643 | 0 | -0.118 | 0.32258 | 0.263 |
| 51030 | DOCK9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-29a-5p;hsa-miR-505-3p;hsa-miR-629-3p | 25 | AR | Sponge network | -0.231 | 0.28214 | -1.554 | 0 | 0.263 |
| 51031 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | HIVEP1 | Sponge network | 1.11 | 0 | 0.368 | 0.00064 | 0.263 |
| 51032 | AC159540.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-323a-3p;hsa-miR-340-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-629-3p | 12 | NFIB | Sponge network | 1.122 | 0 | 0.023 | 0.90795 | 0.263 |
| 51033 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | CHD2 | Sponge network | 0.392 | 0.0278 | 0.049 | 0.55371 | 0.263 |
| 51034 | MIR497HG |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 45 | PPM1L | Sponge network | -1.439 | 4.0E-5 | -0.537 | 0.00616 | 0.263 |
| 51035 | LIPE-AS1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p | 10 | GSK3B | Sponge network | 0.078 | 0.55803 | 0.266 | 0.00045 | 0.263 |
| 51036 | RP11-212P7.2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-96-5p | 27 | ASTN1 | Sponge network | 0.219 | 0.1516 | -2.389 | 2.0E-5 | 0.263 |
| 51037 | GAS6-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-550a-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 17 | CRTC1 | Sponge network | 0.16 | 0.50785 | -0.116 | 0.28412 | 0.263 |
| 51038 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p | 15 | SLITRK3 | Sponge network | -0.125 | 0.49414 | -3.328 | 0 | 0.263 |
| 51039 | RP4-758J18.10 |
hsa-miR-107;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-21-3p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-450b-5p | 17 | DTNA | Sponge network | -0.091 | 0.71468 | -1.648 | 1.0E-5 | 0.263 |
| 51040 | AC009948.5 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-362-5p;hsa-miR-505-3p;hsa-miR-589-3p | 21 | KCNJ5 | Sponge network | 0.532 | 0.00505 | -0.281 | 0.3339 | 0.263 |
| 51041 | RP11-727A23.5 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-495-3p | 10 | STAG2 | Sponge network | 1.117 | 0 | 0.252 | 0.00438 | 0.263 |
| 51042 | LINC00662 |
hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-942-5p | 10 | RB1CC1 | Sponge network | 0.213 | 0.32297 | 0.175 | 0.12559 | 0.263 |
| 51043 | HCG11 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CAV1 | Sponge network | 0.385 | 0.10067 | -1.013 | 6.0E-5 | 0.263 |
| 51044 | RP11-102N12.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 10 | CXXC4 | Sponge network | -0.351 | 0.11358 | -0.433 | 0.21055 | 0.263 |
| 51045 | PCA3 |
hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-342-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | WAC | Sponge network | -0.709 | 0.18168 | -0.054 | 0.43688 | 0.263 |
| 51046 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | HLF | Sponge network | 0.131 | 0.8436 | -2.443 | 0 | 0.263 |
| 51047 | RP11-35G9.3 |
hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ITPKB | Sponge network | 0.657 | 4.0E-5 | -0.704 | 0.00106 | 0.263 |
| 51048 | CTD-2314G24.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-423-5p;hsa-miR-7-5p;hsa-miR-96-5p | 12 | ZYX | Sponge network | 0.246 | 0.59912 | 0.019 | 0.89763 | 0.263 |
| 51049 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 36 | AFF4 | Sponge network | 0.889 | 9.0E-5 | -0.266 | 0.01565 | 0.263 |
| 51050 | RP11-723D22.3 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-652-3p;hsa-miR-940 | 10 | QKI | Sponge network | 0.562 | 0.20117 | -0.333 | 0.04111 | 0.263 |
| 51051 | AF131215.2 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-429;hsa-miR-7-1-3p | 11 | RECK | Sponge network | -0.176 | 0.59692 | -1.043 | 0 | 0.262 |
| 51052 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 14 | ITCH | Sponge network | 0.488 | 0.00084 | 0.084 | 0.34058 | 0.262 |
| 51053 | LINC00476 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | UBE2QL1 | Sponge network | -0.615 | 0.00015 | -2.417 | 0 | 0.262 |
| 51054 | LINC00843 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429 | 10 | UBE4B | Sponge network | 1.106 | 0 | -0.235 | 0.007 | 0.262 |
| 51055 | AC004893.11 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | ZFX | Sponge network | 1.317 | 0 | 0.09 | 0.42563 | 0.262 |
| 51056 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 10 | ST6GAL2 | Sponge network | 1.153 | 0.00114 | -1.201 | 0.00597 | 0.262 |
| 51057 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-1-3p | 28 | ZFHX3 | Sponge network | 0.267 | 0.2937 | 0.389 | 0.01363 | 0.262 |
| 51058 | PVT1 |
hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-20b-5p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-497-5p;hsa-miR-9-5p | 10 | CDK6 | Sponge network | 1.985 | 0 | 0.991 | 0 | 0.262 |
| 51059 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 23 | ARHGAP28 | Sponge network | -0.709 | 0.18168 | -0.911 | 0.00013 | 0.262 |
| 51060 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | ARID5B | Sponge network | 0.539 | 0.03722 | 0.342 | 0.01676 | 0.262 |
| 51061 | AGAP11 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-942-5p | 20 | COPS2 | Sponge network | -1.645 | 0 | -0.178 | 0.04974 | 0.262 |
| 51062 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 31 | BHLHE41 | Sponge network | 0.374 | 0.20054 | 0.353 | 0.12753 | 0.262 |
| 51063 | H19 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p | 14 | ERC1 | Sponge network | 2.439 | 0 | -0.108 | 0.38794 | 0.262 |
| 51064 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ADAMTSL3 | Sponge network | -0.031 | 0.89063 | -1.559 | 0 | 0.262 |
| 51065 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-93-5p | 19 | CNTNAP3 | Sponge network | -0.969 | 2.0E-5 | -1.465 | 0.00033 | 0.262 |
| 51066 | RP11-434B12.1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 11 | PICALM | Sponge network | -0.031 | 0.89063 | 0.086 | 0.23523 | 0.262 |
| 51067 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 20 | MYO9A | Sponge network | 0.199 | 0.38015 | -0.147 | 0.32373 | 0.262 |
| 51068 | AC018890.6 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-629-3p;hsa-miR-96-5p | 14 | ASTN1 | Sponge network | 1.61 | 0.00961 | -2.389 | 2.0E-5 | 0.262 |
| 51069 | RP11-760H22.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p | 10 | BRD3 | Sponge network | 0.001 | 0.99753 | 0.37 | 0.00013 | 0.262 |
| 51070 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | HIP1 | Sponge network | 0.272 | 0.44692 | 0.774 | 0 | 0.262 |
| 51071 | AC093627.8 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p | 18 | FAT3 | Sponge network | -0.787 | 0.3928 | -1.601 | 0.00051 | 0.262 |
| 51072 | AC009948.5 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 20 | PICALM | Sponge network | 0.532 | 0.00505 | 0.086 | 0.23523 | 0.262 |
| 51073 | LINC00648 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | ZFHX4 | Sponge network | 0.633 | 0.51678 | 0.018 | 0.95574 | 0.262 |
| 51074 | ZNF582-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-7-1-3p | 18 | NCOA1 | Sponge network | -1.349 | 0.00017 | -0.432 | 0 | 0.262 |
| 51075 | RP11-554A11.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-505-5p;hsa-miR-769-5p | 18 | HBP1 | Sponge network | -0.583 | 0.14589 | -0.454 | 0 | 0.262 |
| 51076 | LINC00648 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | RASSF8 | Sponge network | 0.633 | 0.51678 | -0.122 | 0.65591 | 0.262 |
| 51077 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-539-5p | 11 | ITCH | Sponge network | 0.921 | 0.00118 | 0.084 | 0.34058 | 0.262 |
| 51078 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 29 | CBL | Sponge network | -0.405 | 0.22013 | 0.291 | 0.01267 | 0.262 |
| 51079 | RP11-264I13.2 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-664a-5p | 10 | LPP | Sponge network | 0.421 | 0.01652 | -0.459 | 0.04475 | 0.262 |
| 51080 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 20 | TGFBR2 | Sponge network | -0.777 | 0.1134 | -0.243 | 0.11408 | 0.262 |
| 51081 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-629-3p | 22 | DDX6 | Sponge network | 0.368 | 0.20093 | 0.091 | 0.36428 | 0.262 |
| 51082 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-629-3p;hsa-miR-652-3p;hsa-miR-7-5p;hsa-miR-93-5p | 27 | ZBTB4 | Sponge network | 0.219 | 0.1516 | -0.739 | 0 | 0.262 |
| 51083 | LINC00173 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | DOCK4 | Sponge network | 0.056 | 0.8494 | 0.502 | 0.00108 | 0.262 |
| 51084 | CTC-296K1.4 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-629-5p | 10 | PCDH17 | Sponge network | -2.076 | 0.00548 | 0.731 | 2.0E-5 | 0.262 |
| 51085 | RP11-403I13.8 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PRUNE2 | Sponge network | 0.619 | 0.00157 | -1.733 | 0.00022 | 0.262 |
| 51086 | RP11-327P2.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-5p | 19 | TET1 | Sponge network | -0.147 | 0.40452 | -0.135 | 0.52138 | 0.262 |
| 51087 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 47 | THRB | Sponge network | 0.225 | 0.30644 | -1.117 | 0.0004 | 0.262 |
| 51088 | CTA-204B4.2 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 15 | ANK2 | Sponge network | 0.765 | 7.0E-5 | -1.603 | 1.0E-5 | 0.262 |
| 51089 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-577 | 12 | DNAJB4 | Sponge network | -0.18 | 0.20343 | -0.7 | 1.0E-5 | 0.262 |
| 51090 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 14 | MAP3K12 | Sponge network | 0.727 | 3.0E-5 | -0.057 | 0.59369 | 0.262 |
| 51091 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 36 | CHD5 | Sponge network | 0.311 | 0.10344 | -0.922 | 0.01521 | 0.262 |
| 51092 | CD27-AS1 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-769-5p | 24 | SYNPO2 | Sponge network | 0.177 | 0.16681 | -2.342 | 0 | 0.262 |
| 51093 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p;hsa-miR-93-3p | 53 | IL6ST | Sponge network | -0.773 | 0.04073 | -0.402 | 0.00676 | 0.262 |
| 51094 | RP11-554A11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-450b-5p | 14 | ARNT2 | Sponge network | -0.583 | 0.14589 | -0.332 | 0.25851 | 0.262 |
| 51095 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 31 | PRKAB2 | Sponge network | 0.246 | 0.59912 | -0.367 | 0.01371 | 0.262 |
| 51096 | ZNF582-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-96-5p | 13 | RIMBP2 | Sponge network | -1.349 | 0.00017 | -1.968 | 5.0E-5 | 0.262 |
| 51097 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577;hsa-miR-7-1-3p | 25 | TSHZ3 | Sponge network | 0.853 | 0.15352 | -0.556 | 0.01516 | 0.262 |
| 51098 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-664a-3p | 16 | ITCH | Sponge network | 0.385 | 0.10067 | 0.084 | 0.34058 | 0.262 |
| 51099 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 51 | IL6ST | Sponge network | 0.782 | 0.00016 | -0.402 | 0.00676 | 0.262 |
| 51100 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-3615;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | DMTF1 | Sponge network | -0.615 | 0.00015 | 0.248 | 0.02726 | 0.262 |
| 51101 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | FYN | Sponge network | -0.942 | 0 | -0.131 | 0.37051 | 0.262 |
| 51102 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-590-3p | 12 | EPHA3 | Sponge network | 0.545 | 0.05789 | 0.185 | 0.53588 | 0.262 |
| 51103 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 17 | HDAC9 | Sponge network | -0.176 | 0.59692 | -0.755 | 0.00495 | 0.262 |
| 51104 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-455-5p;hsa-miR-93-5p | 10 | JAK1 | Sponge network | -0.739 | 0.0574 | 0.001 | 0.98982 | 0.262 |
| 51105 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | SMARCA2 | Sponge network | -2.925 | 0 | -0.529 | 0.00014 | 0.262 |
| 51106 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | KAT6B | Sponge network | -0.942 | 0 | -0.478 | 0.00018 | 0.262 |
| 51107 | AC025171.1 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-629-3p | 15 | PTPN14 | Sponge network | 0.998 | 0 | 0.144 | 0.34595 | 0.262 |
| 51108 | HCG22 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | EBF1 | Sponge network | 0.816 | 0.32368 | -0.384 | 0.08856 | 0.262 |
| 51109 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-93-3p | 12 | NCOA1 | Sponge network | 1.46 | 1.0E-5 | -0.432 | 0 | 0.262 |
| 51110 | EIF3J-AS1 |
hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ATRX | Sponge network | 0.331 | 0.00509 | 0.261 | 0.04408 | 0.262 |
| 51111 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-455-5p;hsa-miR-542-3p | 13 | TSC22D1 | Sponge network | -1.697 | 0.00889 | -0.529 | 2.0E-5 | 0.262 |
| 51112 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-942-5p | 14 | RB1CC1 | Sponge network | 0.853 | 0.15352 | 0.175 | 0.12559 | 0.262 |
| 51113 | RP11-356J5.12 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-379-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 15 | SH3GLB1 | Sponge network | -0.899 | 6.0E-5 | -0.115 | 0.14773 | 0.262 |
| 51114 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 25 | NEDD4 | Sponge network | 0.782 | 0.00016 | 0.02 | 0.89834 | 0.262 |
| 51115 | AC016747.3 |
hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-940 | 11 | RHOB | Sponge network | 0.199 | 0.38015 | -1.052 | 0 | 0.262 |
| 51116 | LINC00476 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | SIAH1 | Sponge network | -0.615 | 0.00015 | -0.057 | 0.41415 | 0.262 |
| 51117 | LIPE-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p | 12 | CNTNAP3 | Sponge network | 0.078 | 0.55803 | -1.465 | 0.00033 | 0.262 |
| 51118 | RP11-66B24.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-3607-3p | 14 | IGF1 | Sponge network | 0.57 | 0.1866 | -0.204 | 0.5898 | 0.262 |
| 51119 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-582-5p | 15 | ARHGAP29 | Sponge network | 0.921 | 0.00118 | 0.131 | 0.42953 | 0.262 |
| 51120 | LOXL1-AS1 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | PDE4DIP | Sponge network | 0.487 | 0.04053 | -0.282 | 0.0257 | 0.262 |
| 51121 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-96-5p | 14 | CNTN1 | Sponge network | 0.37 | 0.27614 | -2.594 | 0 | 0.262 |
| 51122 | RASSF8-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | SIRT1 | Sponge network | 0.335 | 0.20596 | 0.109 | 0.2593 | 0.262 |
| 51123 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 55 | KIF1B | Sponge network | -0.976 | 0.00025 | -0.118 | 0.32258 | 0.262 |
| 51124 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-199a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-766-3p | 30 | MAPK1 | Sponge network | 0.083 | 0.66405 | -0.017 | 0.81465 | 0.262 |
| 51125 | CASC2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 20 | TRIM33 | Sponge network | 0.385 | 0.01306 | 0.402 | 7.0E-5 | 0.262 |
| 51126 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-455-3p;hsa-miR-589-5p | 10 | RARB | Sponge network | -1.942 | 0 | -0.825 | 2.0E-5 | 0.262 |
| 51127 | RP11-325K4.2 |
hsa-let-7a-3p;hsa-miR-1296-5p;hsa-miR-135b-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | MEF2C | Sponge network | 0.198 | 0.27624 | -0.499 | 0.01448 | 0.262 |
| 51128 | RP4-669P10.16 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-214-3p;hsa-miR-214-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-542-3p | 13 | PPM1A | Sponge network | -0.301 | 0.06597 | -0.623 | 0 | 0.262 |
| 51129 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 13 | PTPN13 | Sponge network | -0.355 | 0.0077 | -1.208 | 0 | 0.262 |
| 51130 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p | 14 | PLSCR4 | Sponge network | -0.555 | 0.06156 | -0.474 | 0.03833 | 0.262 |
| 51131 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-874-3p;hsa-miR-940 | 13 | CRTC3 | Sponge network | 0.946 | 0 | 0.164 | 0.16786 | 0.262 |
| 51132 | A1BG-AS1 |
hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-589-3p | 10 | MED12L | Sponge network | -0.164 | 0.45106 | -0.194 | 0.55299 | 0.262 |
| 51133 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-421;hsa-miR-484;hsa-miR-550a-5p | 10 | FAT2 | Sponge network | 0.61 | 0.01593 | -0.278 | 0.44877 | 0.262 |
| 51134 | DOCK9-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-375;hsa-miR-539-5p | 12 | ABI2 | Sponge network | -0.231 | 0.28214 | 0.175 | 0.14787 | 0.262 |
| 51135 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-629-3p | 10 | CHD2 | Sponge network | 0.368 | 0.20093 | 0.049 | 0.55371 | 0.262 |
| 51136 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-590-3p | 13 | HERC2 | Sponge network | 0.222 | 0.29133 | 0.114 | 0.30638 | 0.262 |
| 51137 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-93-5p;hsa-miR-942-5p | 32 | HLF | Sponge network | 0.253 | 0.09565 | -2.443 | 0 | 0.262 |
| 51138 | LINC00662 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | RASSF8 | Sponge network | 0.213 | 0.32297 | -0.122 | 0.65591 | 0.262 |
| 51139 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-93-5p | 19 | ARNT2 | Sponge network | -2.69 | 0.0001 | -0.332 | 0.25851 | 0.262 |
| 51140 | AC099850.1 |
hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p | 13 | MDM4 | Sponge network | 0.803 | 0 | 0.345 | 0.00762 | 0.262 |
| 51141 | AC005618.6 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-590-3p | 17 | MRVI1 | Sponge network | 0.862 | 0.06213 | -1.051 | 0.00022 | 0.262 |
| 51142 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 21 | FOXP1 | Sponge network | 0.199 | 0.52763 | 0.042 | 0.74368 | 0.262 |
| 51143 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 25 | SOX11 | Sponge network | -0.49 | 0.04946 | 2.68 | 0 | 0.262 |
| 51144 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 14 | GPAM | Sponge network | 0.95 | 0.15943 | 0.142 | 0.29732 | 0.262 |
| 51145 | HOXD-AS2 |
hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-625-5p | 16 | CHD5 | Sponge network | -0.208 | 0.65592 | -0.922 | 0.01521 | 0.262 |
| 51146 | AF131215.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-92b-3p | 15 | CBFA2T3 | Sponge network | -0.322 | 0.25945 | -1.221 | 1.0E-5 | 0.262 |
| 51147 | MIR497HG |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 17 | SIK1 | Sponge network | -1.439 | 4.0E-5 | -0.98 | 0 | 0.262 |
| 51148 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 41 | AR | Sponge network | 0.75 | 0.00054 | -1.554 | 0 | 0.262 |
| 51149 | RP4-717I23.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-582-5p | 20 | NAV3 | Sponge network | 0.637 | 0.00069 | 0.212 | 0.47729 | 0.262 |
| 51150 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 14 | EPHA7 | Sponge network | 0.411 | 0.1335 | -2.197 | 3.0E-5 | 0.262 |
| 51151 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-93-3p;hsa-miR-96-5p | 19 | EBF3 | Sponge network | 0.395 | 0.15451 | -0.058 | 0.81437 | 0.262 |
| 51152 | RP11-156P1.3 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-655-3p | 10 | SHPRH | Sponge network | 0.214 | 0.18246 | 0.098 | 0.40994 | 0.262 |
| 51153 | RP5-1198O20.4 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-590-3p | 18 | TLE4 | Sponge network | 0.997 | 0.00066 | -1.157 | 0 | 0.262 |
| 51154 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-590-3p | 13 | HERC2 | Sponge network | 0.488 | 0.00084 | 0.114 | 0.30638 | 0.262 |
| 51155 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p | 18 | ACVR1 | Sponge network | -0.612 | 0.00094 | 0.279 | 0.00234 | 0.262 |
| 51156 | RP11-244O19.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 36 | GNAQ | Sponge network | -0.096 | 0.67958 | -0.367 | 0.00102 | 0.262 |
| 51157 | ACTA2-AS1 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PTCH1 | Sponge network | 0.194 | 0.64278 | -0.1 | 0.6179 | 0.262 |
| 51158 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | EPHA7 | Sponge network | -0.351 | 0.11358 | -2.197 | 3.0E-5 | 0.262 |
| 51159 | LIPE-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-455-5p | 19 | ZMYND11 | Sponge network | 0.078 | 0.55803 | -0.2 | 0.05449 | 0.262 |
| 51160 | AC108488.4 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-625-5p | 11 | NGFR | Sponge network | 0.259 | 0.12542 | -1.271 | 0.01058 | 0.262 |
| 51161 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-409-3p;hsa-miR-629-3p;hsa-miR-940;hsa-miR-96-5p | 15 | LIFR | Sponge network | 0.453 | 0.01839 | -1.794 | 0 | 0.262 |
| 51162 | LINC00476 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 23 | KANK1 | Sponge network | -0.615 | 0.00015 | -0.55 | 0.00134 | 0.262 |
| 51163 | RP11-531A24.5 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p | 14 | FAT4 | Sponge network | 0.75 | 0.00054 | -0.354 | 0.14694 | 0.262 |
| 51164 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-96-5p | 27 | ZFHX4 | Sponge network | -0.555 | 0.06156 | 0.018 | 0.95574 | 0.262 |
| 51165 | HHIP-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 24 | BTG2 | Sponge network | -0.737 | 0.10822 | -1.763 | 0 | 0.262 |
| 51166 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p | 18 | TMEM132B | Sponge network | 0.912 | 0.00014 | -0.83 | 0.01084 | 0.262 |
| 51167 | LINC00621 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | GPC6 | Sponge network | 0.131 | 0.8436 | -0.054 | 0.81465 | 0.262 |
| 51168 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 17 | HLF | Sponge network | 0.342 | 0.03129 | -2.443 | 0 | 0.262 |
| 51169 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p | 12 | CBFA2T3 | Sponge network | -0.829 | 0.01343 | -1.221 | 1.0E-5 | 0.262 |
| 51170 | TRIM52-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p | 13 | OPCML | Sponge network | -0.418 | 0.00068 | -2.498 | 0 | 0.262 |
| 51171 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-96-5p | 17 | DAB2 | Sponge network | -0.942 | 0 | 0.431 | 0.0113 | 0.262 |
| 51172 | AC012360.6 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-654-3p | 10 | CTDSPL | Sponge network | 0.624 | 0.02176 | 0.085 | 0.45121 | 0.262 |
| 51173 | RP11-473I1.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-339-5p | 11 | NRXN3 | Sponge network | 0.542 | 0.00054 | -0.893 | 0.04186 | 0.262 |
| 51174 | CTC-444N24.11 |
hsa-miR-125a-5p;hsa-miR-150-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-411-5p;hsa-miR-497-5p | 10 | RAD23B | Sponge network | 1.153 | 0 | 0.108 | 0.17414 | 0.262 |
| 51175 | RP11-384O8.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p | 42 | BMPR2 | Sponge network | -0.738 | 0.21233 | 0.206 | 0.0635 | 0.262 |
| 51176 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p | 12 | GNA13 | Sponge network | 0.946 | 0 | 0.007 | 0.93884 | 0.262 |
| 51177 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-93-5p | 46 | ADARB1 | Sponge network | 0.61 | 0.01593 | -0.335 | 0.06631 | 0.262 |
| 51178 | WDR86-AS1 |
hsa-miR-135b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-429;hsa-miR-7-5p;hsa-miR-96-5p | 10 | SETBP1 | Sponge network | 0.348 | 0.32912 | -1.302 | 1.0E-5 | 0.262 |
| 51179 | FLNB-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 13 | ZMYM2 | Sponge network | 1.163 | 0.00018 | 0.279 | 0.01273 | 0.262 |
| 51180 | RP11-46C24.7 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-2-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 18 | DOCK4 | Sponge network | 0.392 | 0.0278 | 0.502 | 0.00108 | 0.262 |
| 51181 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-590-3p | 11 | LRRK2 | Sponge network | -0.727 | 0.04726 | -1.136 | 1.0E-5 | 0.262 |
| 51182 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-223-3p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | CXXC4 | Sponge network | 1.04 | 0.00023 | -0.433 | 0.21055 | 0.262 |
| 51183 | AC011526.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-590-3p | 13 | DIP2C | Sponge network | 0.342 | 0.03129 | -0.436 | 0.01713 | 0.262 |
| 51184 | CTC-457L16.2 |
hsa-let-7g-3p;hsa-miR-192-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-671-5p | 10 | CADM3 | Sponge network | 1.153 | 0.00114 | -3.663 | 0 | 0.262 |
| 51185 | RP11-345P4.7 |
hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | RPS6KA3 | Sponge network | 1.597 | 0 | 0.256 | 0.02419 | 0.262 |
| 51186 | BCYRN1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 18 | DYNC1I1 | Sponge network | 0.311 | 0.10344 | -1.12 | 0.00107 | 0.262 |
| 51187 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p | 17 | MYBL1 | Sponge network | 1.04 | 0.00023 | 0.494 | 0.02152 | 0.262 |
| 51188 | RP11-296I10.3 |
hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-450b-5p | 11 | PIK3R1 | Sponge network | 0.592 | 0.00014 | -0.133 | 0.36854 | 0.262 |
| 51189 | DNAJC27-AS1 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-96-5p | 11 | PRKACA | Sponge network | -0.969 | 2.0E-5 | -0.255 | 0.0012 | 0.262 |
| 51190 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | ST6GAL2 | Sponge network | -1.203 | 0.03881 | -1.201 | 0.00597 | 0.262 |
| 51191 | OIP5-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | CDYL | Sponge network | 0.421 | 0.00084 | 0.12 | 0.12425 | 0.262 |
| 51192 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-590-3p | 11 | MYCBP2 | Sponge network | 0.765 | 7.0E-5 | -0.027 | 0.82016 | 0.262 |
| 51193 | LINC00578 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p | 31 | NFIB | Sponge network | 0.811 | 0.03228 | 0.023 | 0.90795 | 0.262 |
| 51194 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-628-5p | 25 | FZD3 | Sponge network | 1.073 | 0.00216 | 0.104 | 0.60316 | 0.262 |
| 51195 | RP11-890B15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 13 | AKAP13 | Sponge network | 0.101 | 0.60919 | 0.028 | 0.7729 | 0.262 |
| 51196 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-590-3p | 14 | MYBL1 | Sponge network | 1.317 | 0 | 0.494 | 0.02152 | 0.262 |
| 51197 | CTD-2017D11.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-30b-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-378a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-628-5p | 13 | CDH23 | Sponge network | 0.792 | 0.0049 | -0.974 | 0.00034 | 0.262 |
| 51198 | SNX29P2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-149-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-5p | 18 | KCNJ5 | Sponge network | 0.4 | 0.14611 | -0.281 | 0.3339 | 0.262 |
| 51199 | LINC00702 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 28 | CSMD1 | Sponge network | -0.737 | 0.06182 | -0.63 | 0.18881 | 0.262 |
| 51200 | RP11-356J5.12 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 25 | NIN | Sponge network | -0.899 | 6.0E-5 | -0.253 | 0.13794 | 0.262 |
| 51201 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-508-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | MITF | Sponge network | 0.16 | 0.50785 | -0.769 | 0.00022 | 0.262 |
| 51202 | RP11-244O19.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-32-5p;hsa-miR-362-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | FLI1 | Sponge network | -0.096 | 0.67958 | -0.294 | 0.10905 | 0.262 |
| 51203 | FAM157C |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-342-3p | 13 | ZFX | Sponge network | 0.91 | 0 | 0.09 | 0.42563 | 0.262 |
| 51204 | RP11-59H7.3 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p | 12 | PIK3R1 | Sponge network | 2.163 | 0 | -0.133 | 0.36854 | 0.262 |
| 51205 | RP11-359E10.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-93-5p | 28 | PIK3R1 | Sponge network | 0.299 | 0.57722 | -0.133 | 0.36854 | 0.262 |
| 51206 | LINC00863 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-199b-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ARHGEF12 | Sponge network | 0.189 | 0.13981 | 0.09 | 0.35693 | 0.262 |
| 51207 | CKMT2-AS1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 10 | FAM117A | Sponge network | 0.082 | 0.55654 | -1.379 | 0 | 0.262 |
| 51208 | RP11-77H9.2 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-497-5p | 12 | CDK6 | Sponge network | 1.677 | 0 | 0.991 | 0 | 0.262 |
| 51209 | RP4-584D14.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p | 11 | DLC1 | Sponge network | 1.294 | 0.0031 | -0.382 | 0.05038 | 0.262 |
| 51210 | LINC00954 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-582-5p | 15 | NAV3 | Sponge network | 1.157 | 0.00455 | 0.212 | 0.47729 | 0.262 |
| 51211 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-508-3p | 15 | TGFBR2 | Sponge network | -0.154 | 0.53954 | -0.243 | 0.11408 | 0.262 |
| 51212 | LINC00886 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-331-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-532-5p;hsa-miR-582-5p | 10 | THSD7B | Sponge network | -0.371 | 0.24604 | 0.154 | 0.74723 | 0.262 |
| 51213 | RP11-848P1.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TET1 | Sponge network | 1.253 | 0 | -0.135 | 0.52138 | 0.262 |
| 51214 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-5p | 21 | SOX11 | Sponge network | 0.953 | 0 | 2.68 | 0 | 0.262 |
| 51215 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-5p | 19 | PTGFR | Sponge network | 0.61 | 0.01593 | -0.233 | 0.45421 | 0.262 |
| 51216 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-940 | 12 | ZFP36L1 | Sponge network | -0.465 | 0.04703 | -0.505 | 8.0E-5 | 0.262 |
| 51217 | RP11-47A8.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-625-5p | 13 | NUAK1 | Sponge network | 0.103 | 0.43169 | 0.904 | 0 | 0.262 |
| 51218 | LINC00886 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-542-3p | 22 | PDE4DIP | Sponge network | -0.371 | 0.24604 | -0.282 | 0.0257 | 0.262 |
| 51219 | PCA3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 21 | SIRT1 | Sponge network | -0.709 | 0.18168 | 0.109 | 0.2593 | 0.262 |
| 51220 | RP11-119F7.5 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-935 | 21 | GJA1 | Sponge network | 0.225 | 0.30644 | -0.485 | 0.02179 | 0.262 |
| 51221 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 14 | TGFBR2 | Sponge network | 0.225 | 0.30644 | -0.243 | 0.11408 | 0.262 |
| 51222 | AC011526.1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 10 | GALNT13 | Sponge network | 0.342 | 0.03129 | -0.857 | 0.07439 | 0.262 |
| 51223 | FENDRR |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-331-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CREM | Sponge network | -1.281 | 0.00012 | -0.345 | 0.00258 | 0.262 |
| 51224 | SOX2-OT |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | THSD7B | Sponge network | -1.264 | 6.0E-5 | 0.154 | 0.74723 | 0.262 |
| 51225 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | LATS2 | Sponge network | 0.083 | 0.66405 | 0.159 | 0.2304 | 0.262 |
| 51226 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 25 | IGF1 | Sponge network | 0.659 | 3.0E-5 | -0.204 | 0.5898 | 0.262 |
| 51227 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p | 17 | PSIP1 | Sponge network | -0.739 | 0.0574 | -0.005 | 0.96897 | 0.262 |
| 51228 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | IGSF10 | Sponge network | 0.064 | 0.72934 | -1.751 | 1.0E-5 | 0.262 |
| 51229 | RP11-497H16.9 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TSC1 | Sponge network | 0.545 | 0.00064 | 0.103 | 0.26071 | 0.262 |
| 51230 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 13 | TBX18 | Sponge network | 0.199 | 0.52763 | 0.843 | 0.02945 | 0.262 |
| 51231 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 31 | PRKAB2 | Sponge network | -1.439 | 4.0E-5 | -0.367 | 0.01371 | 0.262 |
| 51232 | BVES-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p | 19 | HECA | Sponge network | -2.62 | 0 | -0.217 | 0.03463 | 0.262 |
| 51233 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-96-5p | 12 | MED12L | Sponge network | 0.348 | 0.32912 | -0.194 | 0.55299 | 0.262 |
| 51234 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 50 | CBX5 | Sponge network | -0.739 | 0.0574 | 0.165 | 0.24446 | 0.262 |
| 51235 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-324-3p | 16 | PRKAR1A | Sponge network | -0.611 | 0.09607 | -0.063 | 0.50314 | 0.262 |
| 51236 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-628-5p;hsa-miR-664a-3p | 24 | FZD3 | Sponge network | 0.373 | 0.00068 | 0.104 | 0.60316 | 0.262 |
| 51237 | RP11-499P20.2 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-337-3p;hsa-miR-33a-5p;hsa-miR-409-3p | 12 | FOXP1 | Sponge network | 0.187 | 0.44121 | 0.042 | 0.74368 | 0.262 |
| 51238 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | CXXC4 | Sponge network | 0.559 | 0.00026 | -0.433 | 0.21055 | 0.262 |
| 51239 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | LRP1B | Sponge network | 0.249 | 0.35902 | -2.163 | 0 | 0.262 |
| 51240 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 19 | CDHR3 | Sponge network | -1.216 | 0 | -0.977 | 4.0E-5 | 0.262 |
| 51241 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-590-3p | 18 | RPS6KA3 | Sponge network | 0.101 | 0.60919 | 0.256 | 0.02419 | 0.262 |
| 51242 | RP3-402G11.26 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | NAV3 | Sponge network | -0.254 | 0.19303 | 0.212 | 0.47729 | 0.262 |
| 51243 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-486-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 22 | HDAC9 | Sponge network | 0.082 | 0.55654 | -0.755 | 0.00495 | 0.262 |
| 51244 | CTD-2126E3.4 |
hsa-let-7a-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p | 13 | RUNX1T1 | Sponge network | 0.493 | 0.61262 | -0.344 | 0.2374 | 0.262 |
| 51245 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | ERCC4 | Sponge network | 0.174 | 0.30034 | 0.126 | 0.15266 | 0.262 |
| 51246 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-577;hsa-miR-93-5p | 14 | MAP3K12 | Sponge network | 0.259 | 0.12542 | -0.057 | 0.59369 | 0.262 |
| 51247 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-590-3p | 12 | RBM5 | Sponge network | 0.421 | 0.00084 | -0.205 | 0.0129 | 0.262 |
| 51248 | LPP-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-625-5p;hsa-miR-7-5p | 21 | CRTC1 | Sponge network | -0.18 | 0.20343 | -0.116 | 0.28412 | 0.262 |
| 51249 | ZNF582-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-93-5p | 18 | ARNT2 | Sponge network | -1.349 | 0.00017 | -0.332 | 0.25851 | 0.262 |
| 51250 | RP11-403I13.8 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-625-5p | 12 | NGFR | Sponge network | 0.619 | 0.00157 | -1.271 | 0.01058 | 0.262 |
| 51251 | RP11-384O8.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | ZNF208 | Sponge network | -0.738 | 0.21233 | -0.4 | 0.22381 | 0.262 |
| 51252 | RP11-195F19.9 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 13 | ERG | Sponge network | -0.726 | 0.00132 | 0.002 | 0.99011 | 0.262 |
| 51253 | RP11-867G23.10 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-5p | 25 | PIK3R1 | Sponge network | -2.531 | 0 | -0.133 | 0.36854 | 0.262 |
| 51254 | AC074289.1 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-409-3p;hsa-miR-92a-1-5p | 15 | AKAP6 | Sponge network | -0.326 | 0.14314 | -1.586 | 3.0E-5 | 0.262 |
| 51255 | RP11-554A11.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-450b-5p | 12 | PIK3CA | Sponge network | -0.583 | 0.14589 | 0.195 | 0.06908 | 0.262 |
| 51256 | LINC00863 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | AFF1 | Sponge network | 0.189 | 0.13981 | -0.384 | 0.00053 | 0.262 |
| 51257 | RP11-890B15.3 |
hsa-miR-130b-3p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p | 10 | RASA1 | Sponge network | 0.101 | 0.60919 | -0.002 | 0.98204 | 0.262 |
| 51258 | RP11-254F7.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-5p | 12 | UNC5C | Sponge network | 0.176 | 0.37402 | -0.178 | 0.40214 | 0.262 |
| 51259 | SOX2-OT |
hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | PAIP2 | Sponge network | -1.264 | 6.0E-5 | -0.515 | 0 | 0.262 |
| 51260 | AC005562.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p | 27 | SOX11 | Sponge network | 0.488 | 0.00084 | 2.68 | 0 | 0.262 |
| 51261 | MIR600HG |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-589-3p;hsa-miR-940 | 14 | FAM172A | Sponge network | 0.594 | 0.41522 | -0.57 | 0 | 0.262 |
| 51262 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | CBX5 | Sponge network | 1.378 | 0 | 0.165 | 0.24446 | 0.262 |
| 51263 | RP11-244H3.1 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-590-3p | 17 | MRVI1 | Sponge network | 0.659 | 3.0E-5 | -1.051 | 0.00022 | 0.262 |
| 51264 | RP11-46C24.7 |
hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 10 | PAK3 | Sponge network | 0.392 | 0.0278 | -0.628 | 0.07253 | 0.262 |
| 51265 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-424-5p | 13 | ZMYM2 | Sponge network | 0.368 | 0.20093 | 0.279 | 0.01273 | 0.262 |
| 51266 | CTD-2554C21.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-339-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-96-5p | 18 | PRKCE | Sponge network | -1.654 | 0.00144 | -0.403 | 0.00056 | 0.262 |
| 51267 | CTD-2314G24.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-940 | 58 | NFIB | Sponge network | 0.246 | 0.59912 | 0.023 | 0.90795 | 0.262 |
| 51268 | AC005618.6 |
hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | MAPK10 | Sponge network | 0.862 | 0.06213 | -1.774 | 0 | 0.262 |
| 51269 | RP11-597D13.9 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-455-3p;hsa-miR-484 | 19 | ABL1 | Sponge network | 1.302 | 7.0E-5 | -0.215 | 0.07804 | 0.262 |
| 51270 | HOXB-AS2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-429 | 11 | KANK1 | Sponge network | 1.316 | 0.01106 | -0.55 | 0.00134 | 0.262 |
| 51271 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-940 | 21 | NEDD4 | Sponge network | -0.371 | 0.24604 | 0.02 | 0.89834 | 0.262 |
| 51272 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | FAT4 | Sponge network | 0.131 | 0.8436 | -0.354 | 0.14694 | 0.262 |
| 51273 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | EDA2R | Sponge network | 0.657 | 4.0E-5 | -0.587 | 0.01598 | 0.262 |
| 51274 | KB-1460A1.5 |
hsa-miR-101-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | MAP3K4 | Sponge network | 0.603 | 0.00127 | 0.058 | 0.54229 | 0.262 |
| 51275 | RP1-68D18.2 |
hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-429 | 13 | RICTOR | Sponge network | 1.987 | 0 | 0.388 | 0.00188 | 0.262 |
| 51276 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 51 | KIF1B | Sponge network | -0.612 | 0.00094 | -0.118 | 0.32258 | 0.262 |
| 51277 | CTC-490E21.10 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p | 10 | HIP1 | Sponge network | 1.46 | 1.0E-5 | 0.774 | 0 | 0.262 |
| 51278 | DNM1P35 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | ID4 | Sponge network | 0.051 | 0.83388 | -1.644 | 0 | 0.262 |
| 51279 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-629-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-96-5p | 49 | BMPR2 | Sponge network | -0.563 | 0.02545 | 0.206 | 0.0635 | 0.262 |
| 51280 | RP11-1006G14.4 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | MRVI1 | Sponge network | 0.559 | 0.00026 | -1.051 | 0.00022 | 0.262 |
| 51281 | AP001347.6 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | ADCY1 | Sponge network | -1.161 | 0.00591 | 0.362 | 0.16982 | 0.262 |
| 51282 | PWAR6 |
hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | SH3GLB1 | Sponge network | -1.575 | 6.0E-5 | -0.115 | 0.14773 | 0.262 |
| 51283 | RP11-225B17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | PRKAB2 | Sponge network | 1.055 | 0 | -0.367 | 0.01371 | 0.262 |
| 51284 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3C | Sponge network | 0.219 | 0.1516 | -0.272 | 0.31153 | 0.262 |
| 51285 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-493-5p | 10 | CUL5 | Sponge network | 1.46 | 1.0E-5 | 0.026 | 0.77301 | 0.262 |
| 51286 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 31 | ZFHX4 | Sponge network | 0.614 | 1.0E-5 | 0.018 | 0.95574 | 0.262 |
| 51287 | DNM3OS |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | TLE4 | Sponge network | 0.572 | 0.01722 | -1.157 | 0 | 0.262 |
| 51288 | RP11-890B15.3 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 15 | IRS1 | Sponge network | 0.101 | 0.60919 | -0.026 | 0.88855 | 0.262 |
| 51289 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-450b-5p | 18 | AKAP11 | Sponge network | 0.592 | 0.00014 | 0.196 | 0.09825 | 0.262 |
| 51290 | RP5-1198O20.4 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-424-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 31 | EFNA5 | Sponge network | 0.997 | 0.00066 | -0.421 | 0.17868 | 0.262 |
| 51291 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577;hsa-miR-625-5p | 26 | LPP | Sponge network | 0.357 | 0.72407 | -0.459 | 0.04475 | 0.262 |
| 51292 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 15 | ADAMTSL3 | Sponge network | 0.687 | 0.02753 | -1.559 | 0 | 0.262 |
| 51293 | RP3-402G11.26 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-590-3p | 14 | NCOA1 | Sponge network | -0.254 | 0.19303 | -0.432 | 0 | 0.262 |
| 51294 | RP4-717I23.3 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-486-5p;hsa-miR-942-5p | 12 | GLIPR1 | Sponge network | 0.637 | 0.00069 | 0.146 | 0.3276 | 0.262 |
| 51295 | SNHG14 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 27 | SUFU | Sponge network | -1.111 | 0.0003 | -0.04 | 0.65489 | 0.262 |
| 51296 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | DMTF1 | Sponge network | 0.569 | 0.00067 | 0.248 | 0.02726 | 0.262 |
| 51297 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 13 | IGSF10 | Sponge network | 0.056 | 0.8494 | -1.751 | 1.0E-5 | 0.262 |
| 51298 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | PDZRN3 | Sponge network | 0.349 | 0.01695 | -1.548 | 0 | 0.262 |
| 51299 | HCG11 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 22 | PCDH17 | Sponge network | 0.385 | 0.10067 | 0.731 | 2.0E-5 | 0.262 |
| 51300 | LINC00886 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p | 18 | SEMA3C | Sponge network | -0.371 | 0.24604 | -0.272 | 0.31153 | 0.262 |
| 51301 | DIO3OS |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-660-5p | 19 | LIMCH1 | Sponge network | -0.773 | 0.04073 | -0.915 | 1.0E-5 | 0.262 |
| 51302 | HCG11 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-744-3p | 15 | IGF2 | Sponge network | 0.385 | 0.10067 | 0.848 | 0.03842 | 0.262 |
| 51303 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 12 | MLLT10 | Sponge network | 0.274 | 0.07681 | 0.105 | 0.33292 | 0.262 |
| 51304 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-409-3p | 12 | ABCC9 | Sponge network | 0.594 | 0.00064 | -0.466 | 0.14121 | 0.262 |
| 51305 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 18 | TACC1 | Sponge network | 1.04 | 0.00023 | -0.362 | 0.05406 | 0.262 |
| 51306 | ARHGAP5-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-625-5p;hsa-miR-93-3p | 16 | TOM1L2 | Sponge network | -0.644 | 0.00021 | -0.994 | 0 | 0.262 |
| 51307 | RP11-360F5.1 |
hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-550a-3p | 10 | HECA | Sponge network | 2.364 | 0.00134 | -0.217 | 0.03463 | 0.262 |
| 51308 | ADIRF-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-671-5p | 15 | CDH23 | Sponge network | -0.223 | 0.47816 | -0.974 | 0.00034 | 0.262 |
| 51309 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | PDGFRA | Sponge network | -0.83 | 0 | -0.372 | 0.0037 | 0.262 |
| 51310 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | ZNF208 | Sponge network | 0.219 | 0.1516 | -0.4 | 0.22381 | 0.262 |
| 51311 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-542-3p | 18 | KANK1 | Sponge network | -2.69 | 0.0001 | -0.55 | 0.00134 | 0.262 |
| 51312 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-93-3p | 28 | ST6GAL2 | Sponge network | -0.615 | 0.00015 | -1.201 | 0.00597 | 0.262 |
| 51313 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p | 13 | CREB3L2 | Sponge network | 0.542 | 0.00054 | -0.525 | 2.0E-5 | 0.262 |
| 51314 | TPT1-AS1 |
hsa-miR-139-5p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | GSK3B | Sponge network | 0.903 | 0 | 0.266 | 0.00045 | 0.262 |
| 51315 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 10 | PRKAB2 | Sponge network | 0.826 | 1.0E-5 | -0.367 | 0.01371 | 0.262 |
| 51316 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-93-5p | 39 | UNC80 | Sponge network | -0.303 | 0.21002 | -1.934 | 0 | 0.262 |
| 51317 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-940 | 24 | AKAP11 | Sponge network | 0.622 | 0.00015 | 0.196 | 0.09825 | 0.262 |
| 51318 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p | 11 | GPAM | Sponge network | 0.853 | 0.15352 | 0.142 | 0.29732 | 0.262 |
| 51319 | RP11-2H8.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-590-3p | 12 | BAZ2B | Sponge network | 1.089 | 0 | -0.276 | 0.02833 | 0.262 |
| 51320 | GAS6-AS2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 11 | KDSR | Sponge network | 0.16 | 0.50785 | 0.239 | 0.02216 | 0.262 |
| 51321 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-454-3p;hsa-miR-582-5p | 15 | CXXC4 | Sponge network | -2.095 | 0.00112 | -0.433 | 0.21055 | 0.262 |
| 51322 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 43 | WDFY3 | Sponge network | -1.203 | 0.03881 | -0.201 | 0.0967 | 0.262 |
| 51323 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 54 | PTEN | Sponge network | -0.096 | 0.67958 | -0.187 | 0.07748 | 0.262 |
| 51324 | LINC00909 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p | 13 | MAT2A | Sponge network | 0.363 | 0.00601 | -0.073 | 0.36195 | 0.262 |
| 51325 | KB-1460A1.5 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | DMD | Sponge network | 0.603 | 0.00127 | -1.506 | 0 | 0.262 |
| 51326 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-93-5p | 28 | SASH1 | Sponge network | -0.777 | 0.16667 | -0.502 | 0.00175 | 0.262 |
| 51327 | PWAR6 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | CTGF | Sponge network | -1.575 | 6.0E-5 | 0.321 | 0.11682 | 0.262 |
| 51328 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-590-3p | 11 | TES | Sponge network | -0.031 | 0.89063 | -0.348 | 0.00639 | 0.262 |
| 51329 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 32 | SASH1 | Sponge network | -0.303 | 0.21002 | -0.502 | 0.00175 | 0.262 |
| 51330 | CTB-133G6.2 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-940 | 10 | MCC | Sponge network | 0.4 | 0.39299 | -1.005 | 0 | 0.262 |
| 51331 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PGR | Sponge network | 0.272 | 0.44692 | -1.704 | 0 | 0.262 |
| 51332 | LINC00621 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | PPP1R9A | Sponge network | 0.131 | 0.8436 | -0.903 | 0.04254 | 0.262 |
| 51333 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326 | 13 | TBX18 | Sponge network | 0.331 | 0.57247 | 0.843 | 0.02945 | 0.262 |
| 51334 | CTC-444N24.11 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 13 | UBE4B | Sponge network | 1.153 | 0 | -0.235 | 0.007 | 0.262 |
| 51335 | RP1-43E13.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-378a-5p;hsa-miR-590-5p | 15 | NTRK3 | Sponge network | -0.034 | 0.83228 | -1.77 | 3.0E-5 | 0.262 |
| 51336 | LINC00173 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-421 | 12 | DYNC1I1 | Sponge network | 0.056 | 0.8494 | -1.12 | 0.00107 | 0.262 |
| 51337 | RP11-458D21.1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-7-1-3p | 13 | PLSCR4 | Sponge network | 0.663 | 0.00073 | -0.474 | 0.03833 | 0.262 |
| 51338 | RAD51-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-495-3p;hsa-miR-590-3p | 15 | ABI2 | Sponge network | 0.768 | 7.0E-5 | 0.175 | 0.14787 | 0.262 |
| 51339 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | EBF1 | Sponge network | 0.272 | 0.44692 | -0.384 | 0.08856 | 0.262 |
| 51340 | RP11-327P2.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-576-5p | 21 | BRD3 | Sponge network | -0.147 | 0.40452 | 0.37 | 0.00013 | 0.262 |
| 51341 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | KIF1B | Sponge network | 0.34 | 0.00273 | -0.118 | 0.32258 | 0.262 |
| 51342 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 23 | PTPRB | Sponge network | -1.216 | 0 | 0.105 | 0.47122 | 0.262 |
| 51343 | RP11-206L10.2 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-766-3p | 10 | IL17RD | Sponge network | 0.793 | 0 | -0.019 | 0.94873 | 0.262 |
| 51344 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-577;hsa-miR-7-1-3p | 18 | CNTN1 | Sponge network | 0.419 | 0.0319 | -2.594 | 0 | 0.261 |
| 51345 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-502-5p;hsa-miR-93-3p | 20 | NTRK3 | Sponge network | 0.598 | 0.38481 | -1.77 | 3.0E-5 | 0.261 |
| 51346 | RP11-706C16.7 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-940 | 15 | WDFY3 | Sponge network | 0.357 | 0.72407 | -0.201 | 0.0967 | 0.261 |
| 51347 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | AKAP11 | Sponge network | 0.693 | 0.00076 | 0.196 | 0.09825 | 0.261 |
| 51348 | CTD-3064M3.3 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-429 | 10 | ERRFI1 | Sponge network | -0.469 | 0.08721 | -0.798 | 1.0E-5 | 0.261 |
| 51349 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 19 | EPAS1 | Sponge network | -2.46 | 0 | -0.184 | 0.14418 | 0.261 |
| 51350 | RP11-822E23.8 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p | 12 | PTCH1 | Sponge network | -0.777 | 0.16667 | -0.1 | 0.6179 | 0.261 |
| 51351 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | MTSS1 | Sponge network | -4.477 | 0 | -0.81 | 0.00035 | 0.261 |
| 51352 | LINC00662 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | MOB1B | Sponge network | 0.213 | 0.32297 | 0.197 | 0.15266 | 0.261 |
| 51353 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 45 | ERBB4 | Sponge network | -0.487 | 0.02157 | -1.975 | 2.0E-5 | 0.261 |
| 51354 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-590-3p | 11 | USP6 | Sponge network | 0.439 | 0.00313 | 0.188 | 0.3871 | 0.261 |
| 51355 | RP11-678G14.3 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-629-5p;hsa-miR-7-1-3p | 16 | RASSF8 | Sponge network | -4.477 | 0 | -0.122 | 0.65591 | 0.261 |
| 51356 | RP11-305E6.4 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-590-3p | 14 | DDX3X | Sponge network | 1.317 | 0 | -0.152 | 0.07686 | 0.261 |
| 51357 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 24 | BHLHE41 | Sponge network | 0.603 | 0.00127 | 0.353 | 0.12753 | 0.261 |
| 51358 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 50 | ERBB4 | Sponge network | -0.739 | 0.0574 | -1.975 | 2.0E-5 | 0.261 |
| 51359 | RP11-116O18.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | BHLHE41 | Sponge network | 0.05 | 0.95483 | 0.353 | 0.12753 | 0.261 |
| 51360 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 10 | PRKAB2 | Sponge network | 0.924 | 0 | -0.367 | 0.01371 | 0.261 |
| 51361 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-664a-3p;hsa-miR-671-5p | 23 | THBS1 | Sponge network | 0.95 | 0.15943 | 0.741 | 0.00386 | 0.261 |
| 51362 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 20 | SUFU | Sponge network | -0.513 | 0.04703 | -0.04 | 0.65489 | 0.261 |
| 51363 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | PPM1A | Sponge network | -1.439 | 4.0E-5 | -0.623 | 0 | 0.261 |
| 51364 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 11 | ARHGAP29 | Sponge network | 0.178 | 0.29106 | 0.131 | 0.42953 | 0.261 |
| 51365 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | JAKMIP2 | Sponge network | 0.219 | 0.1516 | -1.388 | 0 | 0.261 |
| 51366 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-582-5p;hsa-miR-590-3p | 25 | USP6 | Sponge network | 0.415 | 0.14657 | 0.188 | 0.3871 | 0.261 |
| 51367 | AC009120.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p | 33 | TCF4 | Sponge network | 0.919 | 0 | 0.016 | 0.92271 | 0.261 |
| 51368 | RP11-83A24.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | BRD3 | Sponge network | 0.417 | 0.00445 | 0.37 | 0.00013 | 0.261 |
| 51369 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | QKI | Sponge network | -0.737 | 0.10822 | -0.333 | 0.04111 | 0.261 |
| 51370 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 34 | FOXP1 | Sponge network | 0.16 | 0.50785 | 0.042 | 0.74368 | 0.261 |
| 51371 | LINC00702 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PCLO | Sponge network | -0.737 | 0.06182 | -1.843 | 0.00064 | 0.261 |
| 51372 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-708-5p | 34 | SPRY3 | Sponge network | 0.267 | 0.2937 | 0.016 | 0.91286 | 0.261 |
| 51373 | AC005618.6 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | MED12L | Sponge network | 0.862 | 0.06213 | -0.194 | 0.55299 | 0.261 |
| 51374 | AC006547.13 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-628-5p;hsa-miR-654-3p | 10 | FOXP1 | Sponge network | 1.159 | 0 | 0.042 | 0.74368 | 0.261 |
| 51375 | CTC-559E9.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p | 16 | MED12L | Sponge network | 0.594 | 0.00064 | -0.194 | 0.55299 | 0.261 |
| 51376 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-582-5p | 11 | ZNF208 | Sponge network | -0.176 | 0.59692 | -0.4 | 0.22381 | 0.261 |
| 51377 | RP11-2H8.2 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-1976;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-511-5p;hsa-miR-590-3p | 20 | WWTR1 | Sponge network | 1.089 | 0 | -0.345 | 0.11997 | 0.261 |
| 51378 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-590-3p | 13 | MYBL1 | Sponge network | 0.336 | 0.00739 | 0.494 | 0.02152 | 0.261 |
| 51379 | RP11-680C21.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p | 34 | PRKAA2 | Sponge network | -1.424 | 0.04346 | -2.462 | 0 | 0.261 |
| 51380 | AC018890.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-5p | 17 | DMD | Sponge network | 1.61 | 0.00961 | -1.506 | 0 | 0.261 |
| 51381 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p | 13 | DMTF1 | Sponge network | 0.72 | 0.0001 | 0.248 | 0.02726 | 0.261 |
| 51382 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-539-5p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | TP53INP1 | Sponge network | -0.563 | 0.02545 | -0.496 | 0.00064 | 0.261 |
| 51383 | NR2F1-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-3p | 17 | PKNOX1 | Sponge network | -0.49 | 0.04946 | 0.085 | 0.28917 | 0.261 |
| 51384 | LINC00472 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-338-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 10 | BCL9 | Sponge network | -0.842 | 0.01361 | 0.321 | 0.00267 | 0.261 |
| 51385 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-424-5p | 13 | ADAMTSL3 | Sponge network | -0.602 | 0.00331 | -1.559 | 0 | 0.261 |
| 51386 | CALML3-AS1 |
hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-5p | 12 | GLI3 | Sponge network | 0.95 | 0.15943 | -0.615 | 0.01482 | 0.261 |
| 51387 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 13 | FAM188A | Sponge network | -0.49 | 0.04946 | -0.44 | 0.00018 | 0.261 |
| 51388 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 18 | ZBTB4 | Sponge network | 0.727 | 3.0E-5 | -0.739 | 0 | 0.261 |
| 51389 | ZNF667-AS1 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-98-5p | 15 | TES | Sponge network | -0.976 | 0.00025 | -0.348 | 0.00639 | 0.261 |
| 51390 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-93-5p | 13 | TRIM3 | Sponge network | -0.612 | 0.00094 | -0.577 | 0 | 0.261 |
| 51391 | RP1-59D14.5 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 16 | DDX3X | Sponge network | 1.162 | 5.0E-5 | -0.152 | 0.07686 | 0.261 |
| 51392 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 32 | DDX6 | Sponge network | -1.697 | 0.00889 | 0.091 | 0.36428 | 0.261 |
| 51393 | FAM66C |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 26 | CYLD | Sponge network | -0.901 | 0.00019 | -0.367 | 0.00171 | 0.261 |
| 51394 | TPTEP1 |
hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-874-3p;hsa-miR-96-5p | 11 | AJAP1 | Sponge network | -1.216 | 0 | -0.561 | 0.04832 | 0.261 |
| 51395 | BCYRN1 |
hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-369-5p;hsa-miR-409-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p | 17 | MACF1 | Sponge network | 0.311 | 0.10344 | 0.019 | 0.90335 | 0.261 |
| 51396 | RP11-283I3.6 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-96-5p | 10 | ZBTB20 | Sponge network | 0.614 | 1.0E-5 | -0.122 | 0.57569 | 0.261 |
| 51397 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | KIF1B | Sponge network | 1.378 | 0 | -0.118 | 0.32258 | 0.261 |
| 51398 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-942-5p | 35 | NOTCH2 | Sponge network | -2.915 | 0.00015 | 0.138 | 0.35163 | 0.261 |
| 51399 | LINC00641 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 51 | CUX1 | Sponge network | -0.222 | 0.32445 | -0.039 | 0.6705 | 0.261 |
| 51400 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PDGFC | Sponge network | 0.064 | 0.72934 | -0.33 | 0.06483 | 0.261 |
| 51401 | RP11-296I10.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-450b-5p | 10 | PIK3CA | Sponge network | 0.592 | 0.00014 | 0.195 | 0.06908 | 0.261 |
| 51402 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-671-5p | 22 | PPP2R5C | Sponge network | 0.194 | 0.64278 | -0.314 | 3.0E-5 | 0.261 |
| 51403 | HAR1A |
hsa-miR-103a-2-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-940 | 11 | IGF1 | Sponge network | -0.672 | 0.19447 | -0.204 | 0.5898 | 0.261 |
| 51404 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | DNAJB4 | Sponge network | 0.349 | 0.01695 | -0.7 | 1.0E-5 | 0.261 |
| 51405 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-32-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 20 | WDFY3 | Sponge network | -0.557 | 0.11135 | -0.201 | 0.0967 | 0.261 |
| 51406 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-2110;hsa-miR-324-3p;hsa-miR-7-1-3p | 11 | USP12 | Sponge network | -4.546 | 0 | -0.102 | 0.40768 | 0.261 |
| 51407 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | PAFAH1B1 | Sponge network | -0.811 | 2.0E-5 | -0.429 | 0 | 0.261 |
| 51408 | RP4-614O4.11 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 18 | KIF1B | Sponge network | 1.042 | 0.00177 | -0.118 | 0.32258 | 0.261 |
| 51409 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p | 13 | FAT3 | Sponge network | 1.969 | 0.00316 | -1.601 | 0.00051 | 0.261 |
| 51410 | AC025171.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | BNC2 | Sponge network | 0.998 | 0 | -0.491 | 0.09988 | 0.261 |
| 51411 | MALAT1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-628-5p | 18 | SLC5A3 | Sponge network | 0.782 | 0.00016 | 0.493 | 5.0E-5 | 0.261 |
| 51412 | ZNF582-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-577;hsa-miR-7-1-3p | 14 | SAMHD1 | Sponge network | -1.349 | 0.00017 | 0.072 | 0.62895 | 0.261 |
| 51413 | RP11-3K16.2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p | 10 | CCDC6 | Sponge network | 0.749 | 0.17788 | 0.27 | 0.02555 | 0.261 |
| 51414 | HCG11 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TLL1 | Sponge network | 0.385 | 0.10067 | -1.195 | 3.0E-5 | 0.261 |
| 51415 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p | 14 | DDX5 | Sponge network | 0.064 | 0.72934 | -0.099 | 0.17428 | 0.261 |
| 51416 | RP11-473I1.9 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-766-3p | 25 | IL17RD | Sponge network | 0.683 | 1.0E-5 | -0.019 | 0.94873 | 0.261 |
| 51417 | RP11-418J17.1 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-495-3p;hsa-miR-497-5p | 10 | ZMYM2 | Sponge network | 0.998 | 0 | 0.279 | 0.01273 | 0.261 |
| 51418 | AC005618.6 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p | 11 | CACNA1E | Sponge network | 0.862 | 0.06213 | 1.752 | 0 | 0.261 |
| 51419 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p | 51 | SMAD4 | Sponge network | -0.793 | 7.0E-5 | -0.313 | 0.00413 | 0.261 |
| 51420 | LINC00641 |
hsa-miR-125a-5p;hsa-miR-141-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | RANBP9 | Sponge network | -0.222 | 0.32445 | -0.191 | 0.04628 | 0.261 |
| 51421 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p | 18 | HLF | Sponge network | 0.889 | 9.0E-5 | -2.443 | 0 | 0.261 |
| 51422 | LINC00672 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 11 | IRS1 | Sponge network | -0.227 | 0.31657 | -0.026 | 0.88855 | 0.261 |
| 51423 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | USP12 | Sponge network | 1.308 | 0 | -0.102 | 0.40768 | 0.261 |
| 51424 | RP5-994D16.9 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | HIP1 | Sponge network | 0.252 | 0.08508 | 0.774 | 0 | 0.261 |
| 51425 | FAM157C |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p | 13 | DDX6 | Sponge network | 0.91 | 0 | 0.091 | 0.36428 | 0.261 |
| 51426 | RP11-344B5.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-423-5p | 18 | NOTCH2 | Sponge network | -0.055 | 0.88482 | 0.138 | 0.35163 | 0.261 |
| 51427 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | ACSL6 | Sponge network | 0.178 | 0.29106 | -1.593 | 0 | 0.261 |
| 51428 | FGD5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-589-3p | 26 | IL17RD | Sponge network | 0.401 | 0.00066 | -0.019 | 0.94873 | 0.261 |
| 51429 | MIR7-3HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-532-3p | 15 | PBX1 | Sponge network | -2.688 | 0.00397 | -1.206 | 0 | 0.261 |
| 51430 | RP11-680C21.1 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | GABRB2 | Sponge network | -1.424 | 0.04346 | -1.841 | 0.00029 | 0.261 |
| 51431 | HOXB-AS3 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-324-5p;hsa-miR-532-3p | 11 | RIMBP2 | Sponge network | 0.343 | 0.28887 | -1.968 | 5.0E-5 | 0.261 |
| 51432 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | PRRX1 | Sponge network | 0.4 | 0.14611 | 1.316 | 0 | 0.261 |
| 51433 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-338-5p | 10 | MYBL1 | Sponge network | 1.11 | 1.0E-5 | 0.494 | 0.02152 | 0.261 |
| 51434 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 31 | BHLHE41 | Sponge network | -0.777 | 0.1134 | 0.353 | 0.12753 | 0.261 |
| 51435 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 47 | PAFAH1B1 | Sponge network | -0.769 | 0.00035 | -0.429 | 0 | 0.261 |
| 51436 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | ST6GALNAC3 | Sponge network | -0.726 | 0.00132 | -0.987 | 0 | 0.261 |
| 51437 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-5p | 22 | ZBTB4 | Sponge network | -0.031 | 0.89063 | -0.739 | 0 | 0.261 |
| 51438 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 32 | SASH1 | Sponge network | -0.738 | 0.21233 | -0.502 | 0.00175 | 0.261 |
| 51439 | RP11-20I23.11 |
hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-590-3p | 10 | TSC1 | Sponge network | 0.875 | 0.00019 | 0.103 | 0.26071 | 0.261 |
| 51440 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-96-5p | 16 | GAS7 | Sponge network | -0.214 | 0.44248 | -0.555 | 0.01602 | 0.261 |
| 51441 | RP11-13J8.1 |
hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-940 | 12 | CREB1 | Sponge network | 3.03 | 0 | 0.203 | 0.00537 | 0.261 |
| 51442 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | AKAP11 | Sponge network | -0.053 | 0.80685 | 0.196 | 0.09825 | 0.261 |
| 51443 | RP5-1021I20.1 |
hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429 | 15 | NTRK2 | Sponge network | -0.785 | 0.00035 | -0.736 | 0.06495 | 0.261 |
| 51444 | AC005562.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-429;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | NIN | Sponge network | 0.488 | 0.00084 | -0.253 | 0.13794 | 0.261 |
| 51445 | AC010226.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-1-5p | 21 | TRPS1 | Sponge network | -0.811 | 2.0E-5 | -0.191 | 0.44109 | 0.261 |
| 51446 | AC025171.1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 18 | PCDH17 | Sponge network | 0.998 | 0 | 0.731 | 2.0E-5 | 0.261 |
| 51447 | SERTAD4-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 14 | PRRX1 | Sponge network | -0.932 | 0.00329 | 1.316 | 0 | 0.261 |
| 51448 | WDFY3-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | ZNF208 | Sponge network | -0.942 | 0 | -0.4 | 0.22381 | 0.261 |
| 51449 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-5p | 36 | TCF4 | Sponge network | 0.921 | 0.00118 | 0.016 | 0.92271 | 0.261 |
| 51450 | CTD-2336O2.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 18 | DLC1 | Sponge network | -0.141 | 0.26434 | -0.382 | 0.05038 | 0.261 |
| 51451 | LINC00578 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-577;hsa-miR-589-3p | 15 | SYT4 | Sponge network | 0.811 | 0.03228 | -3.207 | 0 | 0.261 |
| 51452 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-421 | 10 | FLI1 | Sponge network | -0.18 | 0.20343 | -0.294 | 0.10905 | 0.261 |
| 51453 | RP11-212P7.2 |
hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-212-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p | 11 | RTN4 | Sponge network | 0.219 | 0.1516 | -0.412 | 0.00014 | 0.261 |
| 51454 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p | 16 | CLASP2 | Sponge network | 0.556 | 0.00298 | -0.038 | 0.71 | 0.261 |
| 51455 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-429;hsa-miR-93-3p | 20 | AKAP11 | Sponge network | 0.859 | 0.00331 | 0.196 | 0.09825 | 0.261 |
| 51456 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 50 | DST | Sponge network | -0.125 | 0.49414 | -0.713 | 0.00096 | 0.261 |
| 51457 | RP11-156E6.1 |
hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-497-5p | 13 | DDX3X | Sponge network | 1.266 | 0 | -0.152 | 0.07686 | 0.261 |
| 51458 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-590-3p | 13 | AKAP11 | Sponge network | 0.336 | 0.00739 | 0.196 | 0.09825 | 0.261 |
| 51459 | SNHG5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30a-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 14 | ZNF770 | Sponge network | 0.13 | 0.47174 | 0.206 | 0.06751 | 0.261 |
| 51460 | RBPMS-AS1 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p | 23 | NTRK3 | Sponge network | 0.144 | 0.64989 | -1.77 | 3.0E-5 | 0.261 |
| 51461 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-199b-5p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-484;hsa-miR-590-3p | 11 | EIF4A2 | Sponge network | 1.254 | 0 | -0.383 | 0.00046 | 0.261 |
| 51462 | A2M-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 12 | EGR2 | Sponge network | -0.08 | 0.90092 | 0.309 | 0.17079 | 0.261 |
| 51463 | RP11-20I23.11 |
hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 20 | LPP | Sponge network | 0.875 | 0.00019 | -0.459 | 0.04475 | 0.261 |
| 51464 | RP11-158K1.3 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 13 | KCNAB1 | Sponge network | 0.349 | 0.01695 | -0.755 | 2.0E-5 | 0.261 |
| 51465 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-93-3p | 11 | ERC1 | Sponge network | -0.301 | 0.06597 | -0.108 | 0.38794 | 0.261 |
| 51466 | RP11-283I3.6 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 11 | CSDE1 | Sponge network | 0.614 | 1.0E-5 | -0.493 | 0 | 0.261 |
| 51467 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p | 21 | PRKAB2 | Sponge network | 0.259 | 0.12542 | -0.367 | 0.01371 | 0.261 |
| 51468 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p | 10 | EZH1 | Sponge network | -1.054 | 0.0706 | -0.564 | 1.0E-5 | 0.261 |
| 51469 | LINC00641 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | PAK3 | Sponge network | -0.222 | 0.32445 | -0.628 | 0.07253 | 0.261 |
| 51470 | RP11-216B9.6 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-370-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 10 | TPO | Sponge network | 0.174 | 0.30034 | -1.87 | 2.0E-5 | 0.261 |
| 51471 | AC006116.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-5p;hsa-miR-454-3p | 19 | AKAP11 | Sponge network | -0.473 | 0.07602 | 0.196 | 0.09825 | 0.261 |
| 51472 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 12 | TRIM33 | Sponge network | 0.174 | 0.30034 | 0.402 | 7.0E-5 | 0.261 |
| 51473 | RP11-395A13.2 |
hsa-miR-106a-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-3607-3p | 21 | AR | Sponge network | 0.057 | 0.61225 | -1.554 | 0 | 0.261 |
| 51474 | RP13-516M14.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-505-3p | 15 | ASTN1 | Sponge network | 0.389 | 0.04293 | -2.389 | 2.0E-5 | 0.261 |
| 51475 | MRPL23-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-93-3p | 20 | RUNX1T1 | Sponge network | -0.278 | 0.76559 | -0.344 | 0.2374 | 0.261 |
| 51476 | RP11-1277A3.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-671-5p;hsa-miR-96-5p | 12 | CYLD | Sponge network | 0.453 | 0.01839 | -0.367 | 0.00171 | 0.261 |
| 51477 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 11 | PRKAB2 | Sponge network | 1.117 | 0 | -0.367 | 0.01371 | 0.261 |
| 51478 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-375;hsa-miR-455-5p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-940 | 23 | THBS1 | Sponge network | -0.465 | 0.04703 | 0.741 | 0.00386 | 0.261 |
| 51479 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NR4A3 | Sponge network | -0.342 | 0.02288 | -1.385 | 0 | 0.261 |
| 51480 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | MITF | Sponge network | -0.08 | 0.90092 | -0.769 | 0.00022 | 0.261 |
| 51481 | LIPE-AS1 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-7-5p | 15 | SH3GLB1 | Sponge network | 0.078 | 0.55803 | -0.115 | 0.14773 | 0.261 |
| 51482 | AC018890.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-5p | 17 | NTRK2 | Sponge network | 1.61 | 0.00961 | -0.736 | 0.06495 | 0.261 |
| 51483 | AC016747.3 |
hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-93-5p | 13 | PRUNE2 | Sponge network | 0.199 | 0.38015 | -1.733 | 0.00022 | 0.261 |
| 51484 | CTA-445C9.14 |
hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-582-3p | 16 | ABL2 | Sponge network | 0.718 | 4.0E-5 | 0.82 | 0 | 0.261 |
| 51485 | PCA3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | BCL9 | Sponge network | -0.709 | 0.18168 | 0.321 | 0.00267 | 0.261 |
| 51486 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-370-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-584-5p;hsa-miR-590-5p | 42 | SSBP2 | Sponge network | 0.331 | 0.57247 | -1.58 | 0 | 0.261 |
| 51487 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-654-3p;hsa-miR-7-1-3p | 27 | DDX6 | Sponge network | 0.657 | 4.0E-5 | 0.091 | 0.36428 | 0.261 |
| 51488 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | GNA13 | Sponge network | 0.572 | 0.01722 | 0.007 | 0.93884 | 0.261 |
| 51489 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-940 | 33 | NTRK2 | Sponge network | 0.862 | 0.06213 | -0.736 | 0.06495 | 0.261 |
| 51490 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-144-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-940 | 20 | NTRK3 | Sponge network | 0.959 | 0 | -1.77 | 3.0E-5 | 0.261 |
| 51491 | LINC00702 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 35 | CADM1 | Sponge network | -0.737 | 0.06182 | -0.9 | 0.00084 | 0.261 |
| 51492 | C20orf166-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-577;hsa-miR-629-5p | 45 | NFIB | Sponge network | -2.69 | 0.0001 | 0.023 | 0.90795 | 0.261 |
| 51493 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940 | 42 | AFF4 | Sponge network | 0.082 | 0.55397 | -0.266 | 0.01565 | 0.261 |
| 51494 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 11 | CREM | Sponge network | -0.513 | 0.04703 | -0.345 | 0.00258 | 0.261 |
| 51495 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-374a-5p;hsa-miR-500a-3p;hsa-miR-590-3p | 14 | CDH19 | Sponge network | -0.555 | 0.06156 | -3.434 | 0 | 0.261 |
| 51496 | CTC-487M23.5 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-93-3p | 12 | ERC1 | Sponge network | 0.912 | 0.00014 | -0.108 | 0.38794 | 0.261 |
| 51497 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | CUL5 | Sponge network | 0.8 | 0 | 0.026 | 0.77301 | 0.261 |
| 51498 | RP13-516M14.1 |
hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p | 14 | PCDH9 | Sponge network | 0.389 | 0.04293 | -1.823 | 4.0E-5 | 0.261 |
| 51499 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | PRKAB2 | Sponge network | 0.953 | 0 | -0.367 | 0.01371 | 0.261 |
| 51500 | RP11-539I5.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | PRKAB2 | Sponge network | -0.468 | 0.32587 | -0.367 | 0.01371 | 0.261 |
| 51501 | TP73-AS1 |
hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-3p;hsa-miR-26b-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-874-3p;hsa-miR-96-5p | 11 | AJAP1 | Sponge network | -0.563 | 0.02545 | -0.561 | 0.04832 | 0.261 |
| 51502 | EIF3J-AS1 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 41 | BMPR2 | Sponge network | 0.331 | 0.00509 | 0.206 | 0.0635 | 0.261 |
| 51503 | RP11-693J15.4 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-590-5p | 16 | RET | Sponge network | -0.72 | 0.24239 | -0.833 | 0.03517 | 0.261 |
| 51504 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-3p | 26 | RICTOR | Sponge network | -0.053 | 0.80685 | 0.388 | 0.00188 | 0.261 |
| 51505 | LINC00963 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 10 | HMCN1 | Sponge network | 0.523 | 9.0E-5 | 0.809 | 0.00544 | 0.261 |
| 51506 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 28 | IGF1 | Sponge network | 0.61 | 0.01593 | -0.204 | 0.5898 | 0.261 |
| 51507 | RP11-212P7.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TMEFF2 | Sponge network | 0.219 | 0.1516 | -2.426 | 0 | 0.261 |
| 51508 | AC074138.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-154-5p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-577 | 10 | PRKAA2 | Sponge network | -0.127 | 0.52761 | -2.462 | 0 | 0.261 |
| 51509 | CTC-297N7.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-34a-5p | 19 | PRKAA2 | Sponge network | -2.069 | 0 | -2.462 | 0 | 0.261 |
| 51510 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 42 | KIF1B | Sponge network | 0.569 | 0.00067 | -0.118 | 0.32258 | 0.261 |
| 51511 | RP11-73M18.8 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-328-3p | 11 | VPS53 | Sponge network | 0.832 | 0 | 0.103 | 0.22667 | 0.261 |
| 51512 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p | 17 | MLLT10 | Sponge network | 1.757 | 0 | 0.105 | 0.33292 | 0.261 |
| 51513 | CTC-297N7.9 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-590-5p | 10 | ERG | Sponge network | -2.069 | 0 | 0.002 | 0.99011 | 0.261 |
| 51514 | RP11-403I13.7 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-378c;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-7-5p;hsa-miR-93-3p | 28 | ERC1 | Sponge network | 0.395 | 0.15451 | -0.108 | 0.38794 | 0.261 |
| 51515 | RP11-835E18.5 |
hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | THRB | Sponge network | -0.462 | 0.01226 | -1.117 | 0.0004 | 0.261 |
| 51516 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-942-5p | 35 | THRB | Sponge network | 0.197 | 0.25618 | -1.117 | 0.0004 | 0.261 |
| 51517 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | TSHZ3 | Sponge network | 0.419 | 0.0319 | -0.556 | 0.01516 | 0.261 |
| 51518 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 42 | PPM1A | Sponge network | -2.117 | 0.00161 | -0.623 | 0 | 0.261 |
| 51519 | RP11-686D22.8 |
hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-590-3p | 10 | GLIPR1 | Sponge network | 0.898 | 1.0E-5 | 0.146 | 0.3276 | 0.261 |
| 51520 | HOXB-AS2 |
hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-429;hsa-miR-505-5p | 10 | MRVI1 | Sponge network | 1.316 | 0.01106 | -1.051 | 0.00022 | 0.261 |
| 51521 | FZD10-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-98-5p | 17 | KLF10 | Sponge network | -0.727 | 0.04726 | -0.783 | 0 | 0.261 |
| 51522 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | TIMP3 | Sponge network | 0.219 | 0.1516 | -0.546 | 0.02307 | 0.261 |
| 51523 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 28 | ANK2 | Sponge network | 0.311 | 0.10344 | -1.603 | 1.0E-5 | 0.261 |
| 51524 | TAPT1-AS1 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-495-3p | 14 | RICTOR | Sponge network | 0.403 | 2.0E-5 | 0.388 | 0.00188 | 0.261 |
| 51525 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | ITGAV | Sponge network | -1.111 | 0.0003 | 0.337 | 0.01002 | 0.261 |
| 51526 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | FAT4 | Sponge network | -0.668 | 3.0E-5 | -0.354 | 0.14694 | 0.261 |
| 51527 | MAGI2-AS3 |
hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 11 | PCLO | Sponge network | -0.129 | 0.57177 | -1.843 | 0.00064 | 0.261 |
| 51528 | RP11-67L2.2 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-940 | 21 | BAZ2B | Sponge network | 0.72 | 0.0001 | -0.276 | 0.02833 | 0.261 |
| 51529 | A2M-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-940 | 32 | NTRK2 | Sponge network | -0.08 | 0.90092 | -0.736 | 0.06495 | 0.261 |
| 51530 | RP11-102N12.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b | 12 | SYNM | Sponge network | -0.351 | 0.11358 | -2.309 | 1.0E-5 | 0.261 |
| 51531 | RP11-283I3.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-625-5p;hsa-miR-96-5p | 20 | JAZF1 | Sponge network | 0.614 | 1.0E-5 | -0.377 | 0.02677 | 0.261 |
| 51532 | HOXB-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-671-5p | 17 | CYLD | Sponge network | -0.154 | 0.53954 | -0.367 | 0.00171 | 0.261 |
| 51533 | RP11-981G7.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CEP170 | Sponge network | 0.986 | 0.01022 | 0.943 | 0 | 0.261 |
| 51534 | RP11-360F5.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429 | 10 | RASSF8 | Sponge network | 2.364 | 0.00134 | -0.122 | 0.65591 | 0.261 |
| 51535 | RP11-127B20.2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-539-5p | 23 | DST | Sponge network | 0.411 | 0.1335 | -0.713 | 0.00096 | 0.261 |
| 51536 | DNAJC27-AS1 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-3614-5p | 10 | RIMS1 | Sponge network | -0.969 | 2.0E-5 | -3.143 | 0 | 0.261 |
| 51537 | HHIP-AS1 |
hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | GABRB2 | Sponge network | -0.737 | 0.10822 | -1.841 | 0.00029 | 0.261 |
| 51538 | RP4-758J18.10 |
hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-450b-5p | 12 | WWTR1 | Sponge network | -0.091 | 0.71468 | -0.345 | 0.11997 | 0.261 |
| 51539 | LPP-AS2 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-577;hsa-miR-7-5p;hsa-miR-96-5p | 11 | CAV1 | Sponge network | -0.18 | 0.20343 | -1.013 | 6.0E-5 | 0.261 |
| 51540 | RP11-373D23.3 |
hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-3p;hsa-miR-424-5p | 10 | UBR3 | Sponge network | -0.285 | 0.2172 | -0.021 | 0.82774 | 0.261 |
| 51541 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-629-3p;hsa-miR-940 | 25 | LIFR | Sponge network | -0.777 | 0.16667 | -1.794 | 0 | 0.261 |
| 51542 | RP11-855A2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-629-3p | 10 | RASSF2 | Sponge network | -1.717 | 0.01522 | -0.273 | 0.21961 | 0.261 |
| 51543 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-424-5p | 10 | RET | Sponge network | -0.285 | 0.2172 | -0.833 | 0.03517 | 0.261 |
| 51544 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-624-5p | 20 | ZFX | Sponge network | -0.358 | 0.17255 | 0.09 | 0.42563 | 0.261 |
| 51545 | RP11-403I13.7 |
hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 16 | KLF10 | Sponge network | 0.395 | 0.15451 | -0.783 | 0 | 0.261 |
| 51546 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | FBXO32 | Sponge network | 0.131 | 0.8436 | -0.495 | 0.07561 | 0.261 |
| 51547 | RP13-516M14.1 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | TAL1 | Sponge network | 0.389 | 0.04293 | -0.462 | 0.00574 | 0.261 |
| 51548 | RP11-212P7.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-671-5p | 20 | CDH23 | Sponge network | 0.219 | 0.1516 | -0.974 | 0.00034 | 0.261 |
| 51549 | RP11-389C8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 18 | KCNA6 | Sponge network | 0.727 | 3.0E-5 | -1.531 | 0.00013 | 0.261 |
| 51550 | RP11-248G5.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-582-5p | 22 | BMPR2 | Sponge network | 1.294 | 0 | 0.206 | 0.0635 | 0.261 |
| 51551 | RP11-13K12.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p | 29 | PAFAH1B1 | Sponge network | -0.318 | 0.65847 | -0.429 | 0 | 0.261 |
| 51552 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p | 26 | NTRK2 | Sponge network | -0.176 | 0.59692 | -0.736 | 0.06495 | 0.261 |
| 51553 | AC011526.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | ARNT2 | Sponge network | 0.342 | 0.03129 | -0.332 | 0.25851 | 0.261 |
| 51554 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-23a-3p;hsa-miR-339-5p;hsa-miR-424-5p | 13 | TGFBR3 | Sponge network | -3.586 | 0.00032 | -1.173 | 1.0E-5 | 0.261 |
| 51555 | LINC00657 |
hsa-miR-126-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 10 | IRS1 | Sponge network | 0.91 | 0 | -0.026 | 0.88855 | 0.261 |
| 51556 | RP11-190A12.8 |
hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-654-3p | 10 | RICTOR | Sponge network | 1.521 | 0.01586 | 0.388 | 0.00188 | 0.261 |
| 51557 | RP11-359E10.1 |
hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 44 | THRB | Sponge network | 0.299 | 0.57722 | -1.117 | 0.0004 | 0.261 |
| 51558 | CTD-2184D3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | RUNX1T1 | Sponge network | 0.272 | 0.44692 | -0.344 | 0.2374 | 0.261 |
| 51559 | RP11-282O18.3 |
hsa-let-7a-3p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-374a-3p;hsa-miR-582-5p | 11 | MOB1B | Sponge network | 0.757 | 0 | 0.197 | 0.15266 | 0.261 |
| 51560 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 15 | CSRNP1 | Sponge network | -0.942 | 0 | -1.448 | 0 | 0.261 |
| 51561 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 20 | SNX29 | Sponge network | 0.225 | 0.30644 | -0.34 | 0.01371 | 0.261 |
| 51562 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 30 | IGF1 | Sponge network | 0.311 | 0.10344 | -0.204 | 0.5898 | 0.261 |
| 51563 | CTB-113P19.4 |
hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-96-5p | 21 | NAV3 | Sponge network | 0.906 | 0.31715 | 0.212 | 0.47729 | 0.261 |
| 51564 | RP11-2B6.2 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 14 | RASSF8 | Sponge network | 0.539 | 0.03722 | -0.122 | 0.65591 | 0.261 |
| 51565 | RP11-927P21.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-337-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-539-5p | 10 | KIF5B | Sponge network | 0.921 | 0.00118 | 0.362 | 0.00066 | 0.261 |
| 51566 | SNHG14 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 21 | BCL9 | Sponge network | -1.111 | 0.0003 | 0.321 | 0.00267 | 0.261 |
| 51567 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p | 20 | CREB3L2 | Sponge network | 0.178 | 0.29106 | -0.525 | 2.0E-5 | 0.261 |
| 51568 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 27 | MITF | Sponge network | -0.355 | 0.0077 | -0.769 | 0.00022 | 0.261 |
| 51569 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-326 | 21 | FAT3 | Sponge network | 0.238 | 0.70544 | -1.601 | 0.00051 | 0.261 |
| 51570 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p | 10 | TBRG1 | Sponge network | 0.194 | 0.64278 | -0.071 | 0.37324 | 0.261 |
| 51571 | CTB-133G6.2 |
hsa-let-7d-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-424-5p | 10 | KCNA6 | Sponge network | 0.4 | 0.39299 | -1.531 | 0.00013 | 0.261 |
| 51572 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 23 | JAZF1 | Sponge network | 0.311 | 0.10344 | -0.377 | 0.02677 | 0.261 |
| 51573 | AF131215.9 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-874-3p | 18 | ASTN1 | Sponge network | -0.322 | 0.25945 | -2.389 | 2.0E-5 | 0.261 |
| 51574 | CTD-3092A11.2 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-342-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | TMTC2 | Sponge network | 0.873 | 0 | -0.215 | 0.18248 | 0.261 |
| 51575 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-335-5p | 11 | DMXL1 | Sponge network | 0.796 | 4.0E-5 | -0.426 | 0.00056 | 0.261 |
| 51576 | LINC00092 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-345-5p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | CDHR3 | Sponge network | -1.886 | 0 | -0.977 | 4.0E-5 | 0.261 |
| 51577 | AC103563.8 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p | 10 | GNAQ | Sponge network | -7.551 | 0 | -0.367 | 0.00102 | 0.261 |
| 51578 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p | 12 | LATS1 | Sponge network | 0.538 | 0.00076 | 0.109 | 0.34208 | 0.261 |
| 51579 | RP13-1039J1.4 |
hsa-miR-106a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-589-3p | 11 | TACC1 | Sponge network | 0.356 | 0.33467 | -0.362 | 0.05406 | 0.261 |
| 51580 | RP11-434B12.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p | 11 | MAPK10 | Sponge network | -0.031 | 0.89063 | -1.774 | 0 | 0.261 |
| 51581 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p | 24 | SOX11 | Sponge network | 0.064 | 0.72934 | 2.68 | 0 | 0.261 |
| 51582 | AC034220.3 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-744-3p | 18 | CUL5 | Sponge network | 0.309 | 0.07304 | 0.026 | 0.77301 | 0.261 |
| 51583 | ARHGEF26-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-432-5p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 60 | PPM1L | Sponge network | -2.46 | 0 | -0.537 | 0.00616 | 0.261 |
| 51584 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-93-5p | 18 | TP53INP1 | Sponge network | -0.227 | 0.31657 | -0.496 | 0.00064 | 0.261 |
| 51585 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-92a-3p | 24 | WDFY3 | Sponge network | 0.912 | 0.00014 | -0.201 | 0.0967 | 0.261 |
| 51586 | RP11-574K11.29 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-654-3p | 10 | PIK3R1 | Sponge network | 0.997 | 0 | -0.133 | 0.36854 | 0.261 |
| 51587 | RP5-994D16.9 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | EPHA7 | Sponge network | 0.252 | 0.08508 | -2.197 | 3.0E-5 | 0.261 |
| 51588 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | FREM2 | Sponge network | -0.322 | 0.25945 | -0.437 | 0.46353 | 0.261 |
| 51589 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p | 24 | PRDM2 | Sponge network | 0.056 | 0.81195 | -0.258 | 0.00721 | 0.261 |
| 51590 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-193a-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-590-3p | 12 | NF1 | Sponge network | 0.225 | 0.30644 | 0.51 | 0 | 0.261 |
| 51591 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-7-5p | 20 | CRTC1 | Sponge network | 0.41 | 0.02214 | -0.116 | 0.28412 | 0.261 |
| 51592 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-511-5p | 14 | ZFHX3 | Sponge network | 0.93 | 0 | 0.389 | 0.01363 | 0.261 |
| 51593 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-96-5p | 11 | MITF | Sponge network | 0.37 | 0.27614 | -0.769 | 0.00022 | 0.261 |
| 51594 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-590-3p | 11 | ABCA10 | Sponge network | 0.219 | 0.1516 | -0.571 | 0.03674 | 0.261 |
| 51595 | RP11-403I13.7 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 23 | CREB1 | Sponge network | 0.395 | 0.15451 | 0.203 | 0.00537 | 0.261 |
| 51596 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p | 25 | TACC1 | Sponge network | -0.602 | 0.00331 | -0.362 | 0.05406 | 0.261 |
| 51597 | PSMD5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-874-3p | 11 | GIGYF2 | Sponge network | 0.569 | 0.00067 | 0.136 | 0.04403 | 0.261 |
| 51598 | H19 |
hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-3p | 12 | FBXO32 | Sponge network | 2.439 | 0 | -0.495 | 0.07561 | 0.261 |
| 51599 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | RUNX1T1 | Sponge network | 0.931 | 1.0E-5 | -0.344 | 0.2374 | 0.261 |
| 51600 | XXbac-B444P24.8 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-590-3p | 23 | TCF4 | Sponge network | 2.264 | 0.0012 | 0.016 | 0.92271 | 0.261 |
| 51601 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-590-3p | 15 | CREBBP | Sponge network | 0.225 | 0.30644 | 0.126 | 0.15186 | 0.261 |
| 51602 | EIF3J-AS1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p | 28 | NTRK2 | Sponge network | 0.331 | 0.00509 | -0.736 | 0.06495 | 0.261 |
| 51603 | BDNF-AS |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-96-5p | 25 | CADM1 | Sponge network | -0.668 | 3.0E-5 | -0.9 | 0.00084 | 0.261 |
| 51604 | AC004540.4 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-33a-5p | 12 | BCL2 | Sponge network | -2.549 | 0.00528 | -0.899 | 0 | 0.261 |
| 51605 | LIPE-AS1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-93-3p | 37 | TRPS1 | Sponge network | 0.078 | 0.55803 | -0.191 | 0.44109 | 0.261 |
| 51606 | AC108676.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-495-3p | 10 | ZFX | Sponge network | 4.346 | 2.0E-5 | 0.09 | 0.42563 | 0.261 |
| 51607 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-96-5p | 29 | ARID5B | Sponge network | -0.976 | 0.00025 | 0.342 | 0.01676 | 0.261 |
| 51608 | RP11-819C21.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-375;hsa-miR-429;hsa-miR-7-1-3p | 10 | VEZT | Sponge network | 0.537 | 0.00139 | 0.259 | 0.00296 | 0.261 |
| 51609 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-214-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 23 | PPM1A | Sponge network | -0.285 | 0.2172 | -0.623 | 0 | 0.261 |
| 51610 | CD27-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-450b-5p;hsa-miR-942-5p | 10 | ZBTB20 | Sponge network | 0.177 | 0.16681 | -0.122 | 0.57569 | 0.261 |
| 51611 | RP11-2H8.2 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-326;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | DDX5 | Sponge network | 1.089 | 0 | -0.099 | 0.17428 | 0.261 |
| 51612 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 21 | LRRC7 | Sponge network | 0.619 | 0.00157 | -1.341 | 0.01193 | 0.261 |
| 51613 | ADIRF-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-3p | 24 | CADM1 | Sponge network | -0.223 | 0.47816 | -0.9 | 0.00084 | 0.261 |
| 51614 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 50 | PTEN | Sponge network | 0.083 | 0.66405 | -0.187 | 0.07748 | 0.261 |
| 51615 | CTC-296K1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-671-5p | 19 | PTPRD | Sponge network | -2.095 | 0.00112 | -1.005 | 0.00064 | 0.261 |
| 51616 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 48 | THRB | Sponge network | 0.392 | 0.0278 | -1.117 | 0.0004 | 0.261 |
| 51617 | AF131215.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 15 | AKAP6 | Sponge network | -0.176 | 0.59692 | -1.586 | 3.0E-5 | 0.261 |
| 51618 | DNM3OS |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-508-3p;hsa-miR-590-3p | 11 | RIMS1 | Sponge network | 0.572 | 0.01722 | -3.143 | 0 | 0.261 |
| 51619 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p | 12 | LHX6 | Sponge network | 0.657 | 4.0E-5 | -0.087 | 0.69193 | 0.261 |
| 51620 | RP11-25K19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-493-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-744-5p | 24 | CHD5 | Sponge network | -1.057 | 0.00029 | -0.922 | 0.01521 | 0.261 |
| 51621 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-495-3p;hsa-miR-590-5p | 11 | ZMYM2 | Sponge network | 0.331 | 0.57247 | 0.279 | 0.01273 | 0.261 |
| 51622 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-378a-3p;hsa-miR-378c | 20 | TACC1 | Sponge network | 0.792 | 0.0049 | -0.362 | 0.05406 | 0.261 |
| 51623 | PCBP1-AS1 |
hsa-miR-199a-5p;hsa-miR-214-3p;hsa-miR-223-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-382-5p;hsa-miR-432-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-940 | 13 | SRGAP3 | Sponge network | -0.567 | 0 | -0.202 | 0.29297 | 0.261 |
| 51624 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | CUL5 | Sponge network | -0.899 | 6.0E-5 | 0.026 | 0.77301 | 0.261 |
| 51625 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | TMEM132B | Sponge network | 0.083 | 0.66405 | -0.83 | 0.01084 | 0.261 |
| 51626 | AC127904.2 |
hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | UBE2D1 | Sponge network | 1.308 | 0 | 0.468 | 0 | 0.261 |
| 51627 | PROSER2-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p | 10 | FOXO3 | Sponge network | 1.825 | 0 | -0.04 | 0.72028 | 0.261 |
| 51628 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-550a-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p | 45 | CCND2 | Sponge network | -0.777 | 0.16667 | -0.267 | 0.3112 | 0.261 |
| 51629 | RP4-669P10.16 |
hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-424-5p | 19 | FAT3 | Sponge network | -0.301 | 0.06597 | -1.601 | 0.00051 | 0.261 |
| 51630 | AC005618.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | ETV1 | Sponge network | 0.862 | 0.06213 | -0.096 | 0.67674 | 0.261 |
| 51631 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-301a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | TRIP11 | Sponge network | 1.2 | 0.01813 | -0.158 | 0.06384 | 0.261 |
| 51632 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | CNTNAP3 | Sponge network | 0.225 | 0.30644 | -1.465 | 0.00033 | 0.261 |
| 51633 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-874-3p | 16 | NR2C2 | Sponge network | 0.41 | 0.02214 | 0.21 | 0.03224 | 0.261 |
| 51634 | RP11-927P21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-576-5p | 17 | ZFX | Sponge network | 0.921 | 0.00118 | 0.09 | 0.42563 | 0.261 |
| 51635 | RP11-159D12.2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 11 | BCL9 | Sponge network | 1.254 | 0 | 0.321 | 0.00267 | 0.261 |
| 51636 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-654-3p | 14 | NR2C2 | Sponge network | -0.567 | 0 | 0.21 | 0.03224 | 0.261 |
| 51637 | CTB-113P19.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-590-5p | 10 | THSD7A | Sponge network | 0.906 | 0.31715 | 0.242 | 0.25154 | 0.261 |
| 51638 | LINC00641 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | PAIP2 | Sponge network | -0.222 | 0.32445 | -0.515 | 0 | 0.261 |
| 51639 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 20 | SON | Sponge network | 0.417 | 0.00445 | 0.168 | 0.04406 | 0.261 |
| 51640 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-940 | 21 | MEF2C | Sponge network | 0.921 | 0.00118 | -0.499 | 0.01448 | 0.261 |
| 51641 | LINC00886 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-421;hsa-miR-501-3p;hsa-miR-503-5p | 10 | SMARCA1 | Sponge network | -0.371 | 0.24604 | -0.684 | 0.00159 | 0.261 |
| 51642 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-362-3p | 13 | NAV3 | Sponge network | -0.829 | 0.01343 | 0.212 | 0.47729 | 0.261 |
| 51643 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 35 | AKAP11 | Sponge network | 0.249 | 0.35902 | 0.196 | 0.09825 | 0.261 |
| 51644 | LOXL1-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-362-3p;hsa-miR-590-3p | 16 | DLC1 | Sponge network | 0.487 | 0.04053 | -0.382 | 0.05038 | 0.261 |
| 51645 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-502-3p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-874-3p | 18 | ARID5B | Sponge network | 0.817 | 3.0E-5 | 0.342 | 0.01676 | 0.261 |
| 51646 | AC005154.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p | 15 | ANK3 | Sponge network | 0.848 | 0 | -0.55 | 0.00086 | 0.261 |
| 51647 | AC024560.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3614-5p;hsa-miR-539-5p | 18 | MOB1B | Sponge network | 2.166 | 0.00228 | 0.197 | 0.15266 | 0.261 |
| 51648 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-532-5p | 19 | MAF | Sponge network | -0.829 | 0.01343 | -1.257 | 0 | 0.261 |
| 51649 | RP11-283I3.6 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p | 13 | BAZ2B | Sponge network | 0.614 | 1.0E-5 | -0.276 | 0.02833 | 0.261 |
| 51650 | FGF14-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 20 | PIK3R1 | Sponge network | -1.582 | 8.0E-5 | -0.133 | 0.36854 | 0.261 |
| 51651 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 18 | EPHA5 | Sponge network | 0.817 | 3.0E-5 | -0.957 | 0.01733 | 0.261 |
| 51652 | LINC00963 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-375;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 25 | TET1 | Sponge network | 0.523 | 9.0E-5 | -0.135 | 0.52138 | 0.261 |
| 51653 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | PTPRD | Sponge network | 0.299 | 0.57722 | -1.005 | 0.00064 | 0.261 |
| 51654 | CTD-3064M3.3 |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429;hsa-miR-455-3p | 11 | JUN | Sponge network | -0.469 | 0.08721 | -0.932 | 0 | 0.261 |
| 51655 | RP11-532F6.3 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-590-3p | 13 | TMTC2 | Sponge network | -0.896 | 0.00099 | -0.215 | 0.18248 | 0.261 |
| 51656 | AC025171.1 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 26 | WDFY3 | Sponge network | 0.998 | 0 | -0.201 | 0.0967 | 0.261 |
| 51657 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | NOTCH2NL | Sponge network | -0.021 | 0.93598 | 0.024 | 0.84307 | 0.261 |
| 51658 | RP11-488C13.5 |
hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-497-5p | 10 | RICTOR | Sponge network | 0.753 | 0 | 0.388 | 0.00188 | 0.261 |
| 51659 | AF131215.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 14 | LUZP2 | Sponge network | -0.176 | 0.59692 | -0.056 | 0.89416 | 0.261 |
| 51660 | U91328.19 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 30 | ZFX | Sponge network | -0.303 | 0.21002 | 0.09 | 0.42563 | 0.26 |
| 51661 | LINC01018 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 31 | ETV1 | Sponge network | -2.915 | 0.00015 | -0.096 | 0.67674 | 0.26 |
| 51662 | LINC00863 |
hsa-let-7a-3p;hsa-miR-103a-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ADAMTSL3 | Sponge network | 0.189 | 0.13981 | -1.559 | 0 | 0.26 |
| 51663 | RP11-218M22.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 30 | DTNA | Sponge network | 0.197 | 0.25618 | -1.648 | 1.0E-5 | 0.26 |
| 51664 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-96-5p | 41 | PIK3R1 | Sponge network | 0.311 | 0.10344 | -0.133 | 0.36854 | 0.26 |
| 51665 | RP11-57H14.4 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-424-5p | 10 | USP9X | Sponge network | 0.083 | 0.66405 | 0.222 | 0.01689 | 0.26 |
| 51666 | HOXA-AS2 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TPO | Sponge network | 0.61 | 0.01593 | -1.87 | 2.0E-5 | 0.26 |
| 51667 | RP11-800A3.7 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-7-1-3p | 11 | RBMS3 | Sponge network | 0.18 | 0.7147 | -0.519 | 0.03551 | 0.26 |
| 51668 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-655-3p | 20 | LATS1 | Sponge network | -0.405 | 0.22013 | 0.109 | 0.34208 | 0.26 |
| 51669 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-590-3p | 14 | MYBL1 | Sponge network | 1.148 | 2.0E-5 | 0.494 | 0.02152 | 0.26 |
| 51670 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-133a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-28-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 15 | TBL1XR1 | Sponge network | 0.796 | 4.0E-5 | 0.578 | 0 | 0.26 |
| 51671 | MALAT1 |
hsa-let-7e-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-942-5p | 25 | COPS2 | Sponge network | 0.782 | 0.00016 | -0.178 | 0.04974 | 0.26 |
| 51672 | AC005618.6 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | DLC1 | Sponge network | 0.862 | 0.06213 | -0.382 | 0.05038 | 0.26 |
| 51673 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | ZNF292 | Sponge network | 0.194 | 0.64278 | 0.276 | 0.04148 | 0.26 |
| 51674 | RP11-67L2.2 |
hsa-miR-155-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 11 | CDH13 | Sponge network | 0.72 | 0.0001 | 0.634 | 0.00094 | 0.26 |
| 51675 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 31 | CTIF | Sponge network | 0.246 | 0.59912 | -0.389 | 0.00549 | 0.26 |
| 51676 | RP11-403I13.7 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-96-5p | 62 | LPP | Sponge network | 0.395 | 0.15451 | -0.459 | 0.04475 | 0.26 |
| 51677 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-625-5p | 15 | LHX6 | Sponge network | -0.222 | 0.32445 | -0.087 | 0.69193 | 0.26 |
| 51678 | RP11-203M5.7 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-5p | 17 | LPP | Sponge network | 1.684 | 1.0E-5 | -0.459 | 0.04475 | 0.26 |
| 51679 | LINC00672 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | BRD3 | Sponge network | -0.227 | 0.31657 | 0.37 | 0.00013 | 0.26 |
| 51680 | CTD-3064M3.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-550a-5p | 28 | CCND2 | Sponge network | -0.469 | 0.08721 | -0.267 | 0.3112 | 0.26 |
| 51681 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-505-5p | 27 | AR | Sponge network | 0.622 | 0.00015 | -1.554 | 0 | 0.26 |
| 51682 | RP11-597D13.9 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 23 | LUZP2 | Sponge network | 1.302 | 7.0E-5 | -0.056 | 0.89416 | 0.26 |
| 51683 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 33 | FOXP1 | Sponge network | -1.057 | 0.00029 | 0.042 | 0.74368 | 0.26 |
| 51684 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-589-5p | 10 | RARB | Sponge network | -0.154 | 0.53954 | -0.825 | 2.0E-5 | 0.26 |
| 51685 | SOX2-OT |
hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 13 | TRIM13 | Sponge network | -1.264 | 6.0E-5 | 0.183 | 0.06968 | 0.26 |
| 51686 | RP11-571M6.17 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-590-3p | 10 | UBR3 | Sponge network | 1.013 | 0 | -0.021 | 0.82774 | 0.26 |
| 51687 | LINC00865 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-744-3p | 26 | PTPN14 | Sponge network | -1.249 | 7.0E-5 | 0.144 | 0.34595 | 0.26 |
| 51688 | CALML3-AS1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-454-3p | 11 | THSD7A | Sponge network | 0.95 | 0.15943 | 0.242 | 0.25154 | 0.26 |
| 51689 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-93-3p | 23 | PTPN11 | Sponge network | -0.129 | 0.57177 | 0.431 | 2.0E-5 | 0.26 |
| 51690 | TINCR |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-7-5p | 21 | TET1 | Sponge network | -0.664 | 0.14543 | -0.135 | 0.52138 | 0.26 |
| 51691 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-424-5p;hsa-miR-93-5p | 20 | CHD5 | Sponge network | -0.811 | 2.0E-5 | -0.922 | 0.01521 | 0.26 |
| 51692 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 29 | NCOA1 | Sponge network | -0.563 | 0.02545 | -0.432 | 0 | 0.26 |
| 51693 | RP11-218M22.1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-590-3p | 18 | ABL2 | Sponge network | 0.197 | 0.25618 | 0.82 | 0 | 0.26 |
| 51694 | CTA-363E19.2 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-193b-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-382-5p;hsa-miR-409-3p | 13 | PPARA | Sponge network | 1.055 | 0 | -0.379 | 0.00548 | 0.26 |
| 51695 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-23a-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | AKAP11 | Sponge network | -0.021 | 0.93598 | 0.196 | 0.09825 | 0.26 |
| 51696 | RP11-57H14.4 |
hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | DNMT3A | Sponge network | 0.083 | 0.66405 | 0.302 | 0.0262 | 0.26 |
| 51697 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-5p | 22 | CHD5 | Sponge network | -0.737 | 0.10822 | -0.922 | 0.01521 | 0.26 |
| 51698 | AC003090.1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | BAZ2B | Sponge network | -2.117 | 0.00161 | -0.276 | 0.02833 | 0.26 |
| 51699 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | SOX5 | Sponge network | 0.222 | 0.29133 | -1.091 | 4.0E-5 | 0.26 |
| 51700 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 15 | DKK3 | Sponge network | 0.177 | 0.16681 | -0.052 | 0.77783 | 0.26 |
| 51701 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p | 10 | ZNF770 | Sponge network | 1.262 | 0 | 0.206 | 0.06751 | 0.26 |
| 51702 | RP11-439A17.10 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-27a-5p;hsa-miR-362-3p;hsa-miR-576-5p | 10 | ERBB4 | Sponge network | 0.051 | 0.95756 | -1.975 | 2.0E-5 | 0.26 |
| 51703 | BZRAP1-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-877-5p | 21 | CRTC1 | Sponge network | -0.465 | 0.04703 | -0.116 | 0.28412 | 0.26 |
| 51704 | WDR86-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-93-5p | 14 | AR | Sponge network | 0.348 | 0.32912 | -1.554 | 0 | 0.26 |
| 51705 | CTD-2666L21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 13 | ERCC4 | Sponge network | 0.368 | 0.20093 | 0.126 | 0.15266 | 0.26 |
| 51706 | NEAT1 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | ZNF770 | Sponge network | 0.705 | 0.00014 | 0.206 | 0.06751 | 0.26 |
| 51707 | CTD-2336O2.1 |
hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 14 | GJA1 | Sponge network | -0.141 | 0.26434 | -0.485 | 0.02179 | 0.26 |
| 51708 | CTB-50E14.4 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-2-5p | 10 | KIF1B | Sponge network | 1.449 | 0.00389 | -0.118 | 0.32258 | 0.26 |
| 51709 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-628-5p | 25 | GRIA3 | Sponge network | -0.773 | 0.04073 | -1.956 | 0 | 0.26 |
| 51710 | CTD-3092A11.2 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 10 | BCL9 | Sponge network | 0.873 | 0 | 0.321 | 0.00267 | 0.26 |
| 51711 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-590-3p | 17 | BCL6 | Sponge network | 0.083 | 0.66405 | -0.211 | 0.19489 | 0.26 |
| 51712 | RP11-736K20.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p | 19 | ETV1 | Sponge network | -0.358 | 0.17255 | -0.096 | 0.67674 | 0.26 |
| 51713 | AC016747.3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 12 | LRRC4 | Sponge network | 0.199 | 0.38015 | -1.774 | 0 | 0.26 |
| 51714 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p | 19 | SMAD2 | Sponge network | 1.147 | 0 | -0.128 | 0.12732 | 0.26 |
| 51715 | CD27-AS1 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 29 | DIP2C | Sponge network | 0.177 | 0.16681 | -0.436 | 0.01713 | 0.26 |
| 51716 | CTD-3126B10.4 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-590-3p | 14 | BMPR2 | Sponge network | 0.778 | 0.00101 | 0.206 | 0.0635 | 0.26 |
| 51717 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-590-3p | 20 | GRIA3 | Sponge network | -0.08 | 0.90092 | -1.956 | 0 | 0.26 |
| 51718 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | FGF5 | Sponge network | -1.203 | 0.03881 | 0.549 | 0.28659 | 0.26 |
| 51719 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-342-5p;hsa-miR-582-5p | 12 | ATRX | Sponge network | 0.529 | 0.00552 | 0.261 | 0.04408 | 0.26 |
| 51720 | CTD-2314G24.2 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | HERC1 | Sponge network | 0.246 | 0.59912 | -0.196 | 0.08544 | 0.26 |
| 51721 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-454-3p | 22 | FOXP1 | Sponge network | 0.001 | 0.99753 | 0.042 | 0.74368 | 0.26 |
| 51722 | SH3RF3-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-5p | 12 | KCTD8 | Sponge network | 0.889 | 9.0E-5 | -3.359 | 0 | 0.26 |
| 51723 | PART1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 14 | FYN | Sponge network | -2.925 | 0 | -0.131 | 0.37051 | 0.26 |
| 51724 | AC141928.1 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-625-5p | 12 | DCLK1 | Sponge network | 0.853 | 0.15352 | -1.324 | 0.00014 | 0.26 |
| 51725 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | PPP1R9A | Sponge network | 0.335 | 0.20596 | -0.903 | 0.04254 | 0.26 |
| 51726 | NR2F1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-503-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 33 | TOM1L2 | Sponge network | -0.49 | 0.04946 | -0.994 | 0 | 0.26 |
| 51727 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 61 | PTEN | Sponge network | -1.697 | 0.00889 | -0.187 | 0.07748 | 0.26 |
| 51728 | RP11-66B24.2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-3607-3p | 15 | MCC | Sponge network | 0.57 | 0.1866 | -1.005 | 0 | 0.26 |
| 51729 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-375 | 15 | BMPR2 | Sponge network | 1.969 | 0.00316 | 0.206 | 0.0635 | 0.26 |
| 51730 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-339-5p | 15 | HBP1 | Sponge network | -4.477 | 0 | -0.454 | 0 | 0.26 |
| 51731 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 10 | ZNF208 | Sponge network | 0.614 | 1.0E-5 | -0.4 | 0.22381 | 0.26 |
| 51732 | RP11-212P7.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 35 | PPP1R9A | Sponge network | 0.219 | 0.1516 | -0.903 | 0.04254 | 0.26 |
| 51733 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-629-5p | 26 | IGF1 | Sponge network | -1.645 | 0 | -0.204 | 0.5898 | 0.26 |
| 51734 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 48 | PTPRD | Sponge network | -0.096 | 0.67958 | -1.005 | 0.00064 | 0.26 |
| 51735 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | DAB2 | Sponge network | 0.72 | 0.0001 | 0.431 | 0.0113 | 0.26 |
| 51736 | CKMT2-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | CDYL | Sponge network | 0.082 | 0.55654 | 0.12 | 0.12425 | 0.26 |
| 51737 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-361-3p | 17 | MTSS1 | Sponge network | -0.693 | 0.05629 | -0.81 | 0.00035 | 0.26 |
| 51738 | RP11-848P1.3 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | CREB1 | Sponge network | 1.253 | 0 | 0.203 | 0.00537 | 0.26 |
| 51739 | AF131215.9 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-92b-3p;hsa-miR-940 | 39 | CADM2 | Sponge network | -0.322 | 0.25945 | -3.782 | 0 | 0.26 |
| 51740 | MEG3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | TMEFF2 | Sponge network | 0.249 | 0.35902 | -2.426 | 0 | 0.26 |
| 51741 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-590-5p | 21 | PHC3 | Sponge network | 0.176 | 0.37402 | 0.212 | 0.0499 | 0.26 |
| 51742 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-576-5p | 26 | DMXL1 | Sponge network | 0.75 | 0.00054 | -0.426 | 0.00056 | 0.26 |
| 51743 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 17 | LRRC7 | Sponge network | 0.392 | 0.0278 | -1.341 | 0.01193 | 0.26 |
| 51744 | RP11-434B12.1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | KDSR | Sponge network | -0.031 | 0.89063 | 0.239 | 0.02216 | 0.26 |
| 51745 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 27 | PHC3 | Sponge network | -0.793 | 7.0E-5 | 0.212 | 0.0499 | 0.26 |
| 51746 | PTOV1-AS1 |
hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-30e-5p | 12 | CDC73 | Sponge network | 0.87 | 0 | 0.094 | 0.20463 | 0.26 |
| 51747 | DNM1P35 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-628-5p | 16 | RIMS2 | Sponge network | 0.051 | 0.83388 | -1.365 | 0.00208 | 0.26 |
| 51748 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-766-3p | 16 | AFF4 | Sponge network | -4.463 | 0.00025 | -0.266 | 0.01565 | 0.26 |
| 51749 | OIP5-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 20 | PRKAA1 | Sponge network | 0.421 | 0.00084 | 0.317 | 0.0031 | 0.26 |
| 51750 | RP11-800A3.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-629-3p;hsa-miR-942-5p | 10 | GPC6 | Sponge network | 0.18 | 0.7147 | -0.054 | 0.81465 | 0.26 |
| 51751 | AC090587.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p | 12 | AR | Sponge network | -0.088 | 0.65011 | -1.554 | 0 | 0.26 |
| 51752 | NDUFA6-AS1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p | 17 | RPS6KA6 | Sponge network | -0.355 | 0.0077 | -2.989 | 0 | 0.26 |
| 51753 | RP11-157J24.2 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-769-5p | 12 | HBP1 | Sponge network | -1.664 | 0.05165 | -0.454 | 0 | 0.26 |
| 51754 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-93-5p | 14 | TRIM3 | Sponge network | -2.46 | 0 | -0.577 | 0 | 0.26 |
| 51755 | RP11-575A19.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-375 | 11 | ADARB1 | Sponge network | 1.969 | 0.00316 | -0.335 | 0.06631 | 0.26 |
| 51756 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-450b-5p | 16 | GAS7 | Sponge network | 0.912 | 0.00014 | -0.555 | 0.01602 | 0.26 |
| 51757 | AC093627.8 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-29b-1-5p | 16 | ASTN1 | Sponge network | -0.787 | 0.3928 | -2.389 | 2.0E-5 | 0.26 |
| 51758 | RP11-212P7.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 31 | ETV1 | Sponge network | 0.219 | 0.1516 | -0.096 | 0.67674 | 0.26 |
| 51759 | C4A-AS1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-96-5p | 17 | MEF2C | Sponge network | 3.345 | 0 | -0.499 | 0.01448 | 0.26 |
| 51760 | LINC00174 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-497-5p | 11 | MYBL1 | Sponge network | 1.253 | 0 | 0.494 | 0.02152 | 0.26 |
| 51761 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-590-5p | 26 | NTRK2 | Sponge network | 0.889 | 9.0E-5 | -0.736 | 0.06495 | 0.26 |
| 51762 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-625-3p | 37 | ERBB4 | Sponge network | -0.227 | 0.31657 | -1.975 | 2.0E-5 | 0.26 |
| 51763 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | KAT6B | Sponge network | 0.421 | 0.00084 | -0.478 | 0.00018 | 0.26 |
| 51764 | C20orf166-AS1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-577 | 20 | HIP1 | Sponge network | -2.69 | 0.0001 | 0.774 | 0 | 0.26 |
| 51765 | AC011526.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p | 11 | RARB | Sponge network | 0.342 | 0.03129 | -0.825 | 2.0E-5 | 0.26 |
| 51766 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-940 | 18 | TRIM33 | Sponge network | 0.349 | 0.01695 | 0.402 | 7.0E-5 | 0.26 |
| 51767 | RP11-130L8.1 |
hsa-miR-107;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-590-3p | 19 | ABI2 | Sponge network | -0.663 | 0.10887 | 0.175 | 0.14787 | 0.26 |
| 51768 | AC141928.1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | UBE4B | Sponge network | 0.853 | 0.15352 | -0.235 | 0.007 | 0.26 |
| 51769 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 11 | NR4A3 | Sponge network | -0.08 | 0.90092 | -1.385 | 0 | 0.26 |
| 51770 | DNAJC27-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-93-5p | 17 | EDA2R | Sponge network | -0.969 | 2.0E-5 | -0.587 | 0.01598 | 0.26 |
| 51771 | MIR22HG |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-942-5p | 11 | PICALM | Sponge network | -1.047 | 0 | 0.086 | 0.23523 | 0.26 |
| 51772 | CTD-3025N20.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-24-1-5p;hsa-miR-30a-5p;hsa-miR-590-3p | 11 | ZNF770 | Sponge network | 1.046 | 0 | 0.206 | 0.06751 | 0.26 |
| 51773 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3614-5p;hsa-miR-452-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 42 | GAS7 | Sponge network | -0.899 | 6.0E-5 | -0.555 | 0.01602 | 0.26 |
| 51774 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-625-5p | 11 | LHX6 | Sponge network | -0.342 | 0.02288 | -0.087 | 0.69193 | 0.26 |
| 51775 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1296-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-940 | 28 | LIFR | Sponge network | -0.612 | 0.00094 | -1.794 | 0 | 0.26 |
| 51776 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-96-5p | 22 | NTRK3 | Sponge network | 0.37 | 0.27614 | -1.77 | 3.0E-5 | 0.26 |
| 51777 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-98-5p | 27 | USP12 | Sponge network | 0.246 | 0.59912 | -0.102 | 0.40768 | 0.26 |
| 51778 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | KAT6B | Sponge network | -0.342 | 0.02288 | -0.478 | 0.00018 | 0.26 |
| 51779 | CD27-AS1 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | PEG3 | Sponge network | 0.177 | 0.16681 | -1.74 | 0 | 0.26 |
| 51780 | RP11-46C24.7 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-532-5p | 24 | PTPN14 | Sponge network | 0.392 | 0.0278 | 0.144 | 0.34595 | 0.26 |
| 51781 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-5p | 15 | BCL11A | Sponge network | 0.176 | 0.37402 | -0.932 | 0.0063 | 0.26 |
| 51782 | CD27-AS1 |
hsa-let-7b-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-7-1-3p | 13 | INPP4A | Sponge network | 0.177 | 0.16681 | 0.07 | 0.53563 | 0.26 |
| 51783 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-7-5p;hsa-miR-874-3p | 17 | CRTC3 | Sponge network | -0.342 | 0.02288 | 0.164 | 0.16786 | 0.26 |
| 51784 | LINC00863 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-93-5p | 24 | IL17RD | Sponge network | 0.189 | 0.13981 | -0.019 | 0.94873 | 0.26 |
| 51785 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 13 | CHD1 | Sponge network | 0.532 | 0.00505 | 0.322 | 0.00215 | 0.26 |
| 51786 | RP11-359E10.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-582-5p | 11 | ACVR1 | Sponge network | 0.299 | 0.57722 | 0.279 | 0.00234 | 0.26 |
| 51787 | RP11-403I13.8 |
hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 10 | AFAP1L2 | Sponge network | 0.619 | 0.00157 | -0.284 | 0.15434 | 0.26 |
| 51788 | RP1-122P22.2 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-942-5p | 11 | NRXN1 | Sponge network | 0.065 | 0.73468 | -3.778 | 0 | 0.26 |
| 51789 | LOXL1-AS1 |
hsa-let-7a-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | PRKAB2 | Sponge network | 0.487 | 0.04053 | -0.367 | 0.01371 | 0.26 |
| 51790 | RP11-157J24.2 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-769-5p | 12 | GRIK3 | Sponge network | -1.664 | 0.05165 | -3.208 | 0 | 0.26 |
| 51791 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-511-5p;hsa-miR-590-3p | 11 | BCL6 | Sponge network | 0.131 | 0.8436 | -0.211 | 0.19489 | 0.26 |
| 51792 | FLNB-AS1 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | DDX3X | Sponge network | 1.163 | 0.00018 | -0.152 | 0.07686 | 0.26 |
| 51793 | RP11-67L2.2 |
hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-505-3p;hsa-miR-96-5p | 12 | FNBP1 | Sponge network | 0.72 | 0.0001 | -0.969 | 4.0E-5 | 0.26 |
| 51794 | MIR600HG |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-96-5p | 21 | IL17RD | Sponge network | 0.594 | 0.41522 | -0.019 | 0.94873 | 0.26 |
| 51795 | RP1-228H13.5 |
hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-664a-5p | 13 | SOX11 | Sponge network | 1.397 | 0 | 2.68 | 0 | 0.26 |
| 51796 | RP11-20I23.11 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | CBX5 | Sponge network | 0.875 | 0.00019 | 0.165 | 0.24446 | 0.26 |
| 51797 | RP11-927P21.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p | 32 | DTNA | Sponge network | 0.921 | 0.00118 | -1.648 | 1.0E-5 | 0.26 |
| 51798 | RP11-678G14.3 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-424-5p | 11 | PTGFR | Sponge network | -4.477 | 0 | -0.233 | 0.45421 | 0.26 |
| 51799 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-590-5p | 11 | RSPO2 | Sponge network | 0.889 | 9.0E-5 | -3.038 | 0 | 0.26 |
| 51800 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-942-5p | 23 | DDX6 | Sponge network | 0.622 | 0.00015 | 0.091 | 0.36428 | 0.26 |
| 51801 | CTD-2314G24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 20 | ERCC4 | Sponge network | 0.246 | 0.59912 | 0.126 | 0.15266 | 0.26 |
| 51802 | RP11-819C21.1 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 10 | RASSF3 | Sponge network | 0.537 | 0.00139 | -0.226 | 0.11691 | 0.26 |
| 51803 | RP3-475N16.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 10 | TBL1XR1 | Sponge network | 1.056 | 0 | 0.578 | 0 | 0.26 |
| 51804 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p | 10 | NALCN | Sponge network | 1.308 | 0 | 1.068 | 0.00237 | 0.26 |
| 51805 | FENDRR |
hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-23a-3p;hsa-miR-34a-5p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | SLC2A13 | Sponge network | -1.281 | 0.00012 | -0.331 | 0.03658 | 0.26 |
| 51806 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 57 | THRB | Sponge network | -0.615 | 0.00015 | -1.117 | 0.0004 | 0.26 |
| 51807 | RP11-597D13.9 |
hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | HERC1 | Sponge network | 1.302 | 7.0E-5 | -0.196 | 0.08544 | 0.26 |
| 51808 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-98-5p | 37 | CREB3L2 | Sponge network | -0.727 | 0.04726 | -0.525 | 2.0E-5 | 0.26 |
| 51809 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-423-5p;hsa-miR-424-5p | 17 | CBFA2T3 | Sponge network | 0.299 | 0.57722 | -1.221 | 1.0E-5 | 0.26 |
| 51810 | CTD-2017D11.1 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-376c-3p | 12 | DICER1 | Sponge network | 0.792 | 0.0049 | 0.042 | 0.674 | 0.26 |
| 51811 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-96-5p | 50 | NTRK3 | Sponge network | 0.619 | 0.00157 | -1.77 | 3.0E-5 | 0.26 |
| 51812 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-590-3p | 11 | USP12 | Sponge network | -1.425 | 0.04204 | -0.102 | 0.40768 | 0.26 |
| 51813 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | PDGFRA | Sponge network | -0.125 | 0.49414 | -0.372 | 0.0037 | 0.26 |
| 51814 | RP4-669H2.1 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-3p;hsa-miR-378a-5p | 10 | CREB1 | Sponge network | 1.711 | 0.00019 | 0.203 | 0.00537 | 0.26 |
| 51815 | LINC00694 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29a-5p | 12 | RASSF2 | Sponge network | -0.855 | 0.06723 | -0.273 | 0.21961 | 0.26 |
| 51816 | KB-1460A1.5 |
hsa-miR-132-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 19 | PCDH9 | Sponge network | 0.603 | 0.00127 | -1.823 | 4.0E-5 | 0.26 |
| 51817 | CD27-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-486-5p | 10 | ZNF208 | Sponge network | 0.177 | 0.16681 | -0.4 | 0.22381 | 0.26 |
| 51818 | RP1-151F17.2 |
hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | UBE2D1 | Sponge network | 0.222 | 0.29133 | 0.468 | 0 | 0.26 |
| 51819 | RP11-195B17.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-96-5p | 16 | MEF2C | Sponge network | 0.37 | 0.27614 | -0.499 | 0.01448 | 0.26 |
| 51820 | EMX2OS |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-5p | 13 | UVRAG | Sponge network | -0.777 | 0.1134 | -0.017 | 0.83865 | 0.26 |
| 51821 | MIR22HG |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-500a-5p | 12 | SEMA3E | Sponge network | -1.047 | 0 | -1.717 | 0.00137 | 0.26 |
| 51822 | AC010226.4 |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-96-5p | 10 | PRKACA | Sponge network | -0.811 | 2.0E-5 | -0.255 | 0.0012 | 0.26 |
| 51823 | RP11-344B5.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b | 11 | RET | Sponge network | -0.055 | 0.88482 | -0.833 | 0.03517 | 0.26 |
| 51824 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-93-5p | 22 | BHLHE41 | Sponge network | -0.611 | 0.09607 | 0.353 | 0.12753 | 0.26 |
| 51825 | DOCK9-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-375;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-940 | 26 | TRPS1 | Sponge network | -0.231 | 0.28214 | -0.191 | 0.44109 | 0.26 |
| 51826 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 17 | DOCK4 | Sponge network | 0.782 | 0.00016 | 0.502 | 0.00108 | 0.26 |
| 51827 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 14 | EYA4 | Sponge network | 1.153 | 0.00114 | 0.3 | 0.36876 | 0.26 |
| 51828 | EMX2OS |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-550a-3p | 14 | CACNA1E | Sponge network | -0.777 | 0.1134 | 1.752 | 0 | 0.26 |
| 51829 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-625-5p | 12 | GAS1 | Sponge network | 0.253 | 0.09565 | -1.047 | 0.00364 | 0.26 |
| 51830 | RP5-1139B12.3 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-505-5p | 20 | CADM2 | Sponge network | 0.598 | 0.38481 | -3.782 | 0 | 0.26 |
| 51831 | SERHL |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-424-5p | 18 | CBFA2T3 | Sponge network | -0.602 | 0.00331 | -1.221 | 1.0E-5 | 0.26 |
| 51832 | RP11-13K12.5 |
hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-766-3p | 12 | ATM | Sponge network | -0.318 | 0.65847 | 0.178 | 0.21568 | 0.26 |
| 51833 | SOX2-OT |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | IL17RD | Sponge network | -1.264 | 6.0E-5 | -0.019 | 0.94873 | 0.26 |
| 51834 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 11 | ZNF292 | Sponge network | 0.385 | 0.10067 | 0.276 | 0.04148 | 0.26 |
| 51835 | MEG3 |
hsa-let-7g-5p;hsa-miR-107;hsa-miR-185-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-629-3p | 12 | SPTBN4 | Sponge network | 0.249 | 0.35902 | -0.786 | 0.01281 | 0.26 |
| 51836 | RP11-758N13.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484 | 14 | EZH1 | Sponge network | 2.939 | 3.0E-5 | -0.564 | 1.0E-5 | 0.26 |
| 51837 | CTD-2017D11.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-744-3p;hsa-miR-940 | 20 | PTPRT | Sponge network | 0.792 | 0.0049 | -0.907 | 0.03727 | 0.26 |
| 51838 | MYLK-AS1 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-484 | 30 | CBX5 | Sponge network | -1.331 | 3.0E-5 | 0.165 | 0.24446 | 0.26 |
| 51839 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-511-5p | 14 | ZFHX3 | Sponge network | 0.983 | 0.00048 | 0.389 | 0.01363 | 0.26 |
| 51840 | FGD5-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | DLG1 | Sponge network | 0.401 | 0.00066 | -0.014 | 0.8926 | 0.26 |
| 51841 | BOLA3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 13 | EPAS1 | Sponge network | -0.246 | 0.35034 | -0.184 | 0.14418 | 0.26 |
| 51842 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | MOB1B | Sponge network | -0.08 | 0.90092 | 0.197 | 0.15266 | 0.26 |
| 51843 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-5p | 18 | RNASEL | Sponge network | -0.612 | 0.00094 | -0.272 | 0.01796 | 0.26 |
| 51844 | RP11-307B6.3 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p | 10 | GNAQ | Sponge network | -4.463 | 0.00025 | -0.367 | 0.00102 | 0.26 |
| 51845 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | DDX6 | Sponge network | 1.11 | 1.0E-5 | 0.091 | 0.36428 | 0.26 |
| 51846 | LINC01018 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | JUN | Sponge network | -2.915 | 0.00015 | -0.932 | 0 | 0.26 |
| 51847 | RP11-526I2.5 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 10 | UBR3 | Sponge network | 1.054 | 1.0E-5 | -0.021 | 0.82774 | 0.26 |
| 51848 | RP11-298J20.4 |
hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 12 | PAFAH1B1 | Sponge network | 0.002 | 0.99057 | -0.429 | 0 | 0.26 |
| 51849 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 22 | SYT4 | Sponge network | 0.214 | 0.18246 | -3.207 | 0 | 0.26 |
| 51850 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-708-3p;hsa-miR-93-5p;hsa-miR-942-5p | 34 | HLF | Sponge network | -0.235 | 0.15142 | -2.443 | 0 | 0.26 |
| 51851 | RP11-5C23.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 11 | INPP4A | Sponge network | 0.274 | 0.07681 | 0.07 | 0.53563 | 0.26 |
| 51852 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 26 | NCOA1 | Sponge network | 0.249 | 0.35902 | -0.432 | 0 | 0.26 |
| 51853 | C20orf166-AS1 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-577;hsa-miR-582-5p | 22 | PHC3 | Sponge network | -2.69 | 0.0001 | 0.212 | 0.0499 | 0.26 |
| 51854 | SNHG16 |
hsa-miR-10a-5p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-495-3p | 18 | CREB1 | Sponge network | 0.961 | 0 | 0.203 | 0.00537 | 0.26 |
| 51855 | RP11-620J15.3 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | PCDH17 | Sponge network | -0.739 | 0.00044 | 0.731 | 2.0E-5 | 0.26 |
| 51856 | JAZF1-AS1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-98-5p | 15 | TES | Sponge network | -0.769 | 0.00035 | -0.348 | 0.00639 | 0.26 |
| 51857 | RP11-57H14.4 |
hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-625-5p;hsa-miR-940 | 11 | SLC12A6 | Sponge network | 0.083 | 0.66405 | -0.343 | 0.00043 | 0.26 |
| 51858 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-940 | 35 | NTRK3 | Sponge network | 0.389 | 0.04293 | -1.77 | 3.0E-5 | 0.26 |
| 51859 | MRPL23-AS1 |
hsa-miR-126-3p;hsa-miR-127-5p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p | 10 | PGR | Sponge network | -0.278 | 0.76559 | -1.704 | 0 | 0.26 |
| 51860 | RP11-307B6.3 |
hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-942-5p | 13 | OPCML | Sponge network | -4.463 | 0.00025 | -2.498 | 0 | 0.26 |
| 51861 | RP11-490M8.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-223-3p;hsa-miR-24-3p | 11 | IGFBP5 | Sponge network | -0.214 | 0.44248 | -0.806 | 0.00105 | 0.26 |
| 51862 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p | 15 | SPRY3 | Sponge network | 0.986 | 0.01022 | 0.016 | 0.91286 | 0.26 |
| 51863 | ADAMTS9-AS2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-361-3p | 13 | MNT | Sponge network | -2.452 | 0 | -0.157 | 0.06598 | 0.26 |
| 51864 | RP11-403I13.8 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | IGSF10 | Sponge network | 0.619 | 0.00157 | -1.751 | 1.0E-5 | 0.26 |
| 51865 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-93-3p;hsa-miR-940 | 33 | TRPS1 | Sponge network | 0.174 | 0.30034 | -0.191 | 0.44109 | 0.26 |
| 51866 | FAM215B |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-940 | 11 | FAM172A | Sponge network | 0.817 | 3.0E-5 | -0.57 | 0 | 0.26 |
| 51867 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-577;hsa-miR-590-3p | 19 | AFF3 | Sponge network | 0.083 | 0.66405 | -2.373 | 0 | 0.26 |
| 51868 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | GRIN2A | Sponge network | 0.349 | 0.01695 | -2.161 | 0 | 0.26 |
| 51869 | RP11-305E6.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | MYO9A | Sponge network | 1.317 | 0 | -0.147 | 0.32373 | 0.26 |
| 51870 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-671-5p | 10 | PIK3IP1 | Sponge network | 0.335 | 0.20596 | -0.187 | 0.19945 | 0.26 |
| 51871 | AC005562.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | 0.488 | 0.00084 | 0.809 | 0.00544 | 0.26 |
| 51872 | RP4-605O3.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 11 | NAV3 | Sponge network | 0.262 | 0.0475 | 0.212 | 0.47729 | 0.26 |
| 51873 | RP11-59H7.3 |
hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-93-3p | 12 | ERC1 | Sponge network | 2.163 | 0 | -0.108 | 0.38794 | 0.26 |
| 51874 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-96-5p | 15 | LATS2 | Sponge network | 0.395 | 0.15451 | 0.159 | 0.2304 | 0.26 |
| 51875 | RP11-182J1.1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-424-5p | 10 | MYBL1 | Sponge network | -0.187 | 0.23787 | 0.494 | 0.02152 | 0.26 |
| 51876 | RAD51-AS1 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-374b-5p | 10 | CDK6 | Sponge network | 0.768 | 7.0E-5 | 0.991 | 0 | 0.26 |
| 51877 | PWAR6 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 13 | FOXO4 | Sponge network | -1.575 | 6.0E-5 | -0.516 | 2.0E-5 | 0.26 |
| 51878 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-493-5p;hsa-miR-874-3p | 15 | NR2C2 | Sponge network | 0.056 | 0.8494 | 0.21 | 0.03224 | 0.26 |
| 51879 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 15 | NR2C2 | Sponge network | 0.05 | 0.95483 | 0.21 | 0.03224 | 0.26 |
| 51880 | RP11-181G12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-361-3p;hsa-miR-424-5p | 27 | RUNX1T1 | Sponge network | 0.556 | 0.00298 | -0.344 | 0.2374 | 0.26 |
| 51881 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | PAFAH1B1 | Sponge network | -2.117 | 0.00161 | -0.429 | 0 | 0.26 |
| 51882 | RP1-193H18.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-195-5p;hsa-miR-23a-3p;hsa-miR-33a-5p;hsa-miR-497-5p;hsa-miR-940 | 11 | AKAP11 | Sponge network | 0.48 | 0.05578 | 0.196 | 0.09825 | 0.26 |
| 51883 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-493-5p | 27 | KIF1B | Sponge network | -0.829 | 0.01343 | -0.118 | 0.32258 | 0.26 |
| 51884 | HOXB-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-335-5p;hsa-miR-589-3p | 21 | PPM1L | Sponge network | -0.154 | 0.53954 | -0.537 | 0.00616 | 0.26 |
| 51885 | SNHG16 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-497-5p | 15 | UBR3 | Sponge network | 0.961 | 0 | -0.021 | 0.82774 | 0.26 |
| 51886 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 14 | SON | Sponge network | 0.643 | 0 | 0.168 | 0.04406 | 0.26 |
| 51887 | LINC00865 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-744-3p | 16 | IGF2 | Sponge network | -1.249 | 7.0E-5 | 0.848 | 0.03842 | 0.26 |
| 51888 | RP11-575A19.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p | 11 | MAF | Sponge network | 1.969 | 0.00316 | -1.257 | 0 | 0.26 |
| 51889 | PCA3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | NOTCH2NL | Sponge network | -0.709 | 0.18168 | 0.024 | 0.84307 | 0.26 |
| 51890 | CTC-444N24.11 |
hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 11 | NUAK1 | Sponge network | 1.153 | 0 | 0.904 | 0 | 0.26 |
| 51891 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | DPYD | Sponge network | -1.216 | 0 | -0.736 | 0.00076 | 0.26 |
| 51892 | RP11-890B15.3 |
hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | EPB41L3 | Sponge network | 0.101 | 0.60919 | -1.193 | 0 | 0.26 |
| 51893 | RP11-473I1.5 |
hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 10 | CDC73 | Sponge network | 0.542 | 0.00054 | 0.094 | 0.20463 | 0.26 |
| 51894 | CTC-490E21.10 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-29a-5p | 11 | TSC1 | Sponge network | 1.46 | 1.0E-5 | 0.103 | 0.26071 | 0.26 |
| 51895 | RP11-967K21.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 21 | CRTC1 | Sponge network | 0.056 | 0.81195 | -0.116 | 0.28412 | 0.26 |
| 51896 | AC005062.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 20 | EPS15 | Sponge network | -0.019 | 0.81504 | -0.052 | 0.53998 | 0.26 |
| 51897 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 19 | TCF4 | Sponge network | 0.545 | 0.05789 | 0.016 | 0.92271 | 0.26 |
| 51898 | CTC-429P9.1 |
hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p | 15 | RIMS2 | Sponge network | 0.497 | 0.01451 | -1.365 | 0.00208 | 0.26 |
| 51899 | RP11-531A24.5 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-655-3p;hsa-miR-940 | 13 | DLG1 | Sponge network | 0.75 | 0.00054 | -0.014 | 0.8926 | 0.26 |
| 51900 | LINC00863 |
hsa-let-7a-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DACT1 | Sponge network | 0.189 | 0.13981 | 0.783 | 0.00072 | 0.26 |
| 51901 | RP11-597D13.9 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-455-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 14 | RASSF3 | Sponge network | 1.302 | 7.0E-5 | -0.226 | 0.11691 | 0.26 |
| 51902 | HOXA-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | HECA | Sponge network | 0.61 | 0.01593 | -0.217 | 0.03463 | 0.26 |
| 51903 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-96-5p | 17 | ARHGAP29 | Sponge network | 0.246 | 0.59912 | 0.131 | 0.42953 | 0.26 |
| 51904 | HOXA-AS3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-450b-5p;hsa-miR-532-3p | 24 | PBX1 | Sponge network | -0.555 | 0.06156 | -1.206 | 0 | 0.26 |
| 51905 | RP11-532F6.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 22 | PRDM2 | Sponge network | -0.896 | 0.00099 | -0.258 | 0.00721 | 0.26 |
| 51906 | RP11-13K12.1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-629-5p | 15 | WNK2 | Sponge network | -0.154 | 0.78939 | -0.352 | 0.31066 | 0.26 |
| 51907 | RP11-333A23.4 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | DTNA | Sponge network | -1.791 | 0.04269 | -1.648 | 1.0E-5 | 0.26 |
| 51908 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p | 25 | USP6 | Sponge network | -2.46 | 0 | 0.188 | 0.3871 | 0.26 |
| 51909 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p | 26 | FZD3 | Sponge network | 0.594 | 0.00064 | 0.104 | 0.60316 | 0.26 |
| 51910 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-362-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-935 | 28 | HBP1 | Sponge network | -0.096 | 0.67958 | -0.454 | 0 | 0.26 |
| 51911 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-940 | 40 | WDFY3 | Sponge network | -0.371 | 0.24604 | -0.201 | 0.0967 | 0.26 |
| 51912 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | CAV1 | Sponge network | -0.303 | 0.21002 | -1.013 | 6.0E-5 | 0.26 |
| 51913 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 18 | CREBBP | Sponge network | 0.782 | 0.00016 | 0.126 | 0.15186 | 0.26 |
| 51914 | RP11-384O8.1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p | 12 | EHD3 | Sponge network | -0.738 | 0.21233 | -0.484 | 0.01747 | 0.26 |
| 51915 | RP11-276H19.2 |
hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-500a-5p;hsa-miR-942-5p | 15 | THRB | Sponge network | -0.472 | 0.48851 | -1.117 | 0.0004 | 0.26 |
| 51916 | HCG11 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-590-3p | 11 | ABCA10 | Sponge network | 0.385 | 0.10067 | -0.571 | 0.03674 | 0.26 |
| 51917 | RP11-798M19.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 36 | PTPRD | Sponge network | -0.612 | 0.00094 | -1.005 | 0.00064 | 0.26 |
| 51918 | LIPE-AS1 |
hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | FOXO4 | Sponge network | 0.078 | 0.55803 | -0.516 | 2.0E-5 | 0.26 |
| 51919 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 27 | TCF4 | Sponge network | 0.238 | 0.70544 | 0.016 | 0.92271 | 0.26 |
| 51920 | MEG3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-96-5p | 21 | CBFA2T3 | Sponge network | 0.249 | 0.35902 | -1.221 | 1.0E-5 | 0.26 |
| 51921 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p | 11 | CREBBP | Sponge network | 0.657 | 4.0E-5 | 0.126 | 0.15186 | 0.26 |
| 51922 | NEAT1 |
hsa-miR-130b-3p;hsa-miR-24-3p;hsa-miR-28-5p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | DDX5 | Sponge network | 0.705 | 0.00014 | -0.099 | 0.17428 | 0.26 |
| 51923 | RP11-395G23.3 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p | 10 | RARB | Sponge network | 0.381 | 0.25967 | -0.825 | 2.0E-5 | 0.26 |
| 51924 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935;hsa-miR-96-5p | 30 | HBP1 | Sponge network | -1.281 | 0.00012 | -0.454 | 0 | 0.26 |
| 51925 | HAR1A |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-33a-3p;hsa-miR-362-3p | 10 | CACNA2D1 | Sponge network | -0.672 | 0.19447 | -0.846 | 0.00675 | 0.26 |
| 51926 | LINC00641 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 45 | PPP1R9A | Sponge network | -0.222 | 0.32445 | -0.903 | 0.04254 | 0.26 |
| 51927 | FGF14-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-96-5p | 30 | BMPR2 | Sponge network | -1.582 | 8.0E-5 | 0.206 | 0.0635 | 0.26 |
| 51928 | RP11-701H24.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 13 | TRPS1 | Sponge network | -1.425 | 0.04204 | -0.191 | 0.44109 | 0.26 |
| 51929 | GS1-124K5.11 |
hsa-miR-146b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-940 | 11 | PHOX2B | Sponge network | 0.494 | 0.00339 | -2.68 | 0 | 0.26 |
| 51930 | LINC00957 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 28 | FOXP1 | Sponge network | -0.421 | 0.0114 | 0.042 | 0.74368 | 0.26 |
| 51931 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-320b | 17 | TMTC3 | Sponge network | 0.946 | 0 | 0.123 | 0.22189 | 0.26 |
| 51932 | RP11-888D10.4 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-590-3p | 14 | ARHGEF12 | Sponge network | 0.702 | 7.0E-5 | 0.09 | 0.35693 | 0.26 |
| 51933 | GS1-124K5.11 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | ERCC6 | Sponge network | 0.494 | 0.00339 | 0.23 | 0.015 | 0.26 |
| 51934 | ARHGEF26-AS1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3613-5p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | GATA2 | Sponge network | -2.46 | 0 | -0.654 | 0.0016 | 0.26 |
| 51935 | DNAJC27-AS1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-93-3p | 11 | PEA15 | Sponge network | -0.969 | 2.0E-5 | 0.009 | 0.93318 | 0.26 |
| 51936 | RP5-1021I20.1 |
hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 10 | CSRNP1 | Sponge network | -0.785 | 0.00035 | -1.448 | 0 | 0.26 |
| 51937 | AGAP11 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-532-3p;hsa-miR-93-3p | 16 | RPS6KA2 | Sponge network | -1.645 | 0 | -0.57 | 0.00013 | 0.26 |
| 51938 | MRPL23-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-324-5p;hsa-miR-378a-5p;hsa-miR-502-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-93-3p | 28 | NTRK3 | Sponge network | -0.278 | 0.76559 | -1.77 | 3.0E-5 | 0.26 |
| 51939 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 18 | ZFHX4 | Sponge network | 1.04 | 0.00023 | 0.018 | 0.95574 | 0.26 |
| 51940 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p | 23 | FAT3 | Sponge network | 0.411 | 0.1335 | -1.601 | 0.00051 | 0.26 |
| 51941 | RP11-539I5.1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 12 | CSDE1 | Sponge network | -0.468 | 0.32587 | -0.493 | 0 | 0.26 |
| 51942 | LINC01018 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-625-5p;hsa-miR-93-5p | 14 | LHX6 | Sponge network | -2.915 | 0.00015 | -0.087 | 0.69193 | 0.26 |
| 51943 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 47 | TCF4 | Sponge network | 0.392 | 0.0278 | 0.016 | 0.92271 | 0.26 |
| 51944 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 19 | LRRC55 | Sponge network | 0.72 | 0.0001 | -0.026 | 0.93163 | 0.26 |
| 51945 | GABPB1-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | PIK3CA | Sponge network | 1.11 | 0 | 0.195 | 0.06908 | 0.26 |
| 51946 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-365a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-503-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 40 | RICTOR | Sponge network | -0.129 | 0.57177 | 0.388 | 0.00188 | 0.26 |
| 51947 | LINC00667 |
hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p | 14 | SYNM | Sponge network | -0.235 | 0.15142 | -2.309 | 1.0E-5 | 0.26 |
| 51948 | AC141928.1 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-342-3p;hsa-miR-484 | 14 | ABL1 | Sponge network | 0.853 | 0.15352 | -0.215 | 0.07804 | 0.26 |
| 51949 | LINC00954 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-429;hsa-miR-582-5p | 10 | ARID5B | Sponge network | 1.157 | 0.00455 | 0.342 | 0.01676 | 0.26 |
| 51950 | AF131215.9 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | GLIPR1 | Sponge network | -0.322 | 0.25945 | 0.146 | 0.3276 | 0.26 |
| 51951 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-424-5p;hsa-miR-550a-5p;hsa-miR-7-1-3p | 26 | CCND2 | Sponge network | 0.693 | 0.00076 | -0.267 | 0.3112 | 0.26 |
| 51952 | RP11-389C8.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 11 | CNTNAP3 | Sponge network | 0.727 | 3.0E-5 | -1.465 | 0.00033 | 0.26 |
| 51953 | LINC00449 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-342-3p | 13 | CCDC6 | Sponge network | 1.525 | 0 | 0.27 | 0.02555 | 0.26 |
| 51954 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p | 29 | KIF1B | Sponge network | 2.163 | 0 | -0.118 | 0.32258 | 0.26 |
| 51955 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-539-5p | 23 | KIF1B | Sponge network | 1.4 | 0 | -0.118 | 0.32258 | 0.26 |
| 51956 | RP11-264B14.1 |
hsa-miR-126-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p | 10 | TCF12 | Sponge network | 0.557 | 0.00564 | 0.174 | 0.13271 | 0.26 |
| 51957 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-299-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-582-5p | 15 | HDAC9 | Sponge network | 0.056 | 0.81195 | -0.755 | 0.00495 | 0.26 |
| 51958 | RP11-307B6.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-339-5p | 10 | ACSL6 | Sponge network | -4.463 | 0.00025 | -1.593 | 0 | 0.26 |
| 51959 | LINC00473 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 22 | NIN | Sponge network | -1.203 | 0.03881 | -0.253 | 0.13794 | 0.26 |
| 51960 | RP11-359E10.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-93-5p | 12 | MAP3K12 | Sponge network | 0.299 | 0.57722 | -0.057 | 0.59369 | 0.26 |
| 51961 | RP11-13K12.5 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-511-5p | 12 | BCL6 | Sponge network | -0.318 | 0.65847 | -0.211 | 0.19489 | 0.26 |
| 51962 | HOXA-AS2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p | 31 | RICTOR | Sponge network | 0.61 | 0.01593 | 0.388 | 0.00188 | 0.26 |
| 51963 | LINC00702 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | KANK1 | Sponge network | -0.737 | 0.06182 | -0.55 | 0.00134 | 0.26 |
| 51964 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-5p | 30 | ST6GALNAC3 | Sponge network | 0.253 | 0.09565 | -0.987 | 0 | 0.259 |
| 51965 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-501-5p | 12 | KIT | Sponge network | -0.602 | 0.00331 | -2.184 | 0 | 0.259 |
| 51966 | RP11-806O11.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-590-3p | 15 | PPP1R9A | Sponge network | 0.284 | 0.52463 | -0.903 | 0.04254 | 0.259 |
| 51967 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-342-5p;hsa-miR-409-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | ATRX | Sponge network | 0.439 | 0.00313 | 0.261 | 0.04408 | 0.259 |
| 51968 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-671-5p;hsa-miR-874-3p | 15 | ARID5B | Sponge network | 0.556 | 0.00298 | 0.342 | 0.01676 | 0.259 |
| 51969 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-98-5p | 11 | RSPO2 | Sponge network | 0.693 | 0.00076 | -3.038 | 0 | 0.259 |
| 51970 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-93-5p | 11 | MAP3K12 | Sponge network | 0.082 | 0.55397 | -0.057 | 0.59369 | 0.259 |
| 51971 | RP11-539I5.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-590-5p | 20 | DMD | Sponge network | -0.468 | 0.32587 | -1.506 | 0 | 0.259 |
| 51972 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | MTSS1 | Sponge network | -0.358 | 0.17255 | -0.81 | 0.00035 | 0.259 |
| 51973 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 14 | EBF1 | Sponge network | -0.726 | 0.00132 | -0.384 | 0.08856 | 0.259 |
| 51974 | LINC00641 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 22 | NCAM2 | Sponge network | -0.222 | 0.32445 | -0.482 | 0.22565 | 0.259 |
| 51975 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p | 10 | PBRM1 | Sponge network | 0.637 | 0.00069 | -0.029 | 0.78601 | 0.259 |
| 51976 | RP11-156P1.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p | 20 | CSMD1 | Sponge network | 0.214 | 0.18246 | -0.63 | 0.18881 | 0.259 |
| 51977 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | CLASP2 | Sponge network | 0.538 | 0.00076 | -0.038 | 0.71 | 0.259 |
| 51978 | AF131215.2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-671-5p | 24 | DLC1 | Sponge network | -0.176 | 0.59692 | -0.382 | 0.05038 | 0.259 |
| 51979 | RP11-263K19.4 |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-455-5p | 14 | ZMYND11 | Sponge network | 0.859 | 0.00331 | -0.2 | 0.05449 | 0.259 |
| 51980 | RP11-2B6.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-766-3p | 17 | IL17RD | Sponge network | 0.539 | 0.03722 | -0.019 | 0.94873 | 0.259 |
| 51981 | USP3-AS1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-940 | 44 | MDM4 | Sponge network | -0.162 | 0.47687 | 0.345 | 0.00762 | 0.259 |
| 51982 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-505-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 27 | AR | Sponge network | 0.238 | 0.70544 | -1.554 | 0 | 0.259 |
| 51983 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-7-5p | 33 | BCL2 | Sponge network | -0.303 | 0.21002 | -0.899 | 0 | 0.259 |
| 51984 | CTD-2184D3.6 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | ANK2 | Sponge network | 0.272 | 0.44692 | -1.603 | 1.0E-5 | 0.259 |
| 51985 | LINC00843 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-326;hsa-miR-429;hsa-miR-497-5p | 10 | SOX11 | Sponge network | 1.106 | 0 | 2.68 | 0 | 0.259 |
| 51986 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | HDAC9 | Sponge network | 0.246 | 0.59912 | -0.755 | 0.00495 | 0.259 |
| 51987 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | MYBL1 | Sponge network | 0.93 | 0 | 0.494 | 0.02152 | 0.259 |
| 51988 | SH3RF3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-576-5p;hsa-miR-7-1-3p | 12 | HECA | Sponge network | 0.889 | 9.0E-5 | -0.217 | 0.03463 | 0.259 |
| 51989 | KCNJ2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | TACC1 | Sponge network | -0.436 | 0.05325 | -0.362 | 0.05406 | 0.259 |
| 51990 | RASSF8-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 14 | CITED2 | Sponge network | 0.335 | 0.20596 | -1.545 | 0 | 0.259 |
| 51991 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-577;hsa-miR-7-1-3p | 30 | THRB | Sponge network | -0.418 | 0.00068 | -1.117 | 0.0004 | 0.259 |
| 51992 | PSMD5-AS1 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-222-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p | 12 | ERCC4 | Sponge network | 0.569 | 0.00067 | 0.126 | 0.15266 | 0.259 |
| 51993 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-330-3p | 13 | ATRX | Sponge network | 0.542 | 0.00054 | 0.261 | 0.04408 | 0.259 |
| 51994 | TRAM2-AS1 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-330-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-590-3p | 12 | BRD3 | Sponge network | 0.336 | 0.00739 | 0.37 | 0.00013 | 0.259 |
| 51995 | RP11-156P1.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | CYLD | Sponge network | 0.214 | 0.18246 | -0.367 | 0.00171 | 0.259 |
| 51996 | RP13-516M14.1 |
hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-7-1-3p | 20 | RIMS2 | Sponge network | 0.389 | 0.04293 | -1.365 | 0.00208 | 0.259 |
| 51997 | RP11-356J5.12 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 40 | ACVR2A | Sponge network | -0.899 | 6.0E-5 | -0.409 | 0.00039 | 0.259 |
| 51998 | SNX29P2 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-5p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 50 | DST | Sponge network | 0.4 | 0.14611 | -0.713 | 0.00096 | 0.259 |
| 51999 | RP11-66B24.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-425-5p;hsa-miR-502-5p;hsa-miR-550a-3p | 21 | DTNA | Sponge network | 0.57 | 0.1866 | -1.648 | 1.0E-5 | 0.259 |
| 52000 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 10 | DPYD | Sponge network | 0.194 | 0.64278 | -0.736 | 0.00076 | 0.259 |
| 52001 | CTB-113P19.5 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-369-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 10 | PCDH10 | Sponge network | 0.199 | 0.52763 | -1.644 | 0.0078 | 0.259 |
| 52002 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | LRRC4 | Sponge network | 0.083 | 0.66405 | -1.774 | 0 | 0.259 |
| 52003 | ADIRF-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 24 | DMD | Sponge network | -0.223 | 0.47816 | -1.506 | 0 | 0.259 |
| 52004 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-98-5p | 34 | THBS1 | Sponge network | -2.46 | 0 | 0.741 | 0.00386 | 0.259 |
| 52005 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-1-3p | 20 | RARB | Sponge network | 0.064 | 0.72934 | -0.825 | 2.0E-5 | 0.259 |
| 52006 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p | 12 | LRRK2 | Sponge network | 0.374 | 0.20054 | -1.136 | 1.0E-5 | 0.259 |
| 52007 | PCBP1-AS1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-655-3p | 16 | HECA | Sponge network | -0.567 | 0 | -0.217 | 0.03463 | 0.259 |
| 52008 | RP11-156P1.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-92a-3p | 20 | CADM1 | Sponge network | 0.214 | 0.18246 | -0.9 | 0.00084 | 0.259 |
| 52009 | DIO3OS |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-940 | 12 | JUN | Sponge network | -0.773 | 0.04073 | -0.932 | 0 | 0.259 |
| 52010 | HOXB-AS2 |
hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-484 | 10 | TGFBR3 | Sponge network | 1.316 | 0.01106 | -1.173 | 1.0E-5 | 0.259 |
| 52011 | RP11-758N13.1 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-455-3p;hsa-miR-505-5p;hsa-miR-532-3p | 11 | ETV1 | Sponge network | 2.939 | 3.0E-5 | -0.096 | 0.67674 | 0.259 |
| 52012 | HOXB-AS2 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-628-5p;hsa-miR-629-3p | 11 | RASSF8 | Sponge network | 1.316 | 0.01106 | -0.122 | 0.65591 | 0.259 |
| 52013 | RP11-277P12.20 |
hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-5p | 17 | PIK3R1 | Sponge network | 0.687 | 0.02753 | -0.133 | 0.36854 | 0.259 |
| 52014 | AC018890.6 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-7-1-3p | 18 | PPP1R9A | Sponge network | 1.61 | 0.00961 | -0.903 | 0.04254 | 0.259 |
| 52015 | RP11-384O8.1 |
hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-550a-3p;hsa-miR-92a-3p | 13 | RASSF3 | Sponge network | -0.738 | 0.21233 | -0.226 | 0.11691 | 0.259 |
| 52016 | AP000487.5 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-376c-3p | 10 | RASAL2 | Sponge network | 1.987 | 0 | 0.869 | 0 | 0.259 |
| 52017 | DIO3OS |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-324-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-660-5p | 16 | RIMBP2 | Sponge network | -0.773 | 0.04073 | -1.968 | 5.0E-5 | 0.259 |
| 52018 | DIO3OS |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-500a-3p;hsa-miR-542-3p;hsa-miR-625-5p | 11 | PPP2R2C | Sponge network | -0.773 | 0.04073 | -0.959 | 0.07428 | 0.259 |
| 52019 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-409-3p;hsa-miR-411-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-652-3p;hsa-miR-7-1-3p | 43 | SMAD2 | Sponge network | 0.782 | 0.00016 | -0.128 | 0.12732 | 0.259 |
| 52020 | CTD-2583A14.8 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-374a-3p | 12 | VPS53 | Sponge network | 1.134 | 0 | 0.103 | 0.22667 | 0.259 |
| 52021 | PGM5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-421;hsa-miR-450b-5p | 16 | PRKCE | Sponge network | -4.546 | 0 | -0.403 | 0.00056 | 0.259 |
| 52022 | TP73-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 34 | AFF1 | Sponge network | -0.563 | 0.02545 | -0.384 | 0.00053 | 0.259 |
| 52023 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | LRP1B | Sponge network | -0.053 | 0.80685 | -2.163 | 0 | 0.259 |
| 52024 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-484;hsa-miR-577;hsa-miR-629-3p;hsa-miR-96-5p | 20 | EBF3 | Sponge network | 0.853 | 0.15352 | -0.058 | 0.81437 | 0.259 |
| 52025 | LA16c-321D4.2 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-629-3p | 21 | KIF1B | Sponge network | 2.459 | 5.0E-5 | -0.118 | 0.32258 | 0.259 |
| 52026 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-425-5p | 14 | ATRX | Sponge network | -0.583 | 0.14589 | 0.261 | 0.04408 | 0.259 |
| 52027 | AC005618.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-590-3p | 20 | ERCC4 | Sponge network | 0.862 | 0.06213 | 0.126 | 0.15266 | 0.259 |
| 52028 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 20 | EBF3 | Sponge network | -0.031 | 0.89063 | -0.058 | 0.81437 | 0.259 |
| 52029 | HOXD-AS2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p | 13 | FAT3 | Sponge network | -0.208 | 0.65592 | -1.601 | 0.00051 | 0.259 |
| 52030 | LINC00657 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 29 | DST | Sponge network | 0.91 | 0 | -0.713 | 0.00096 | 0.259 |
| 52031 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-30c-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 14 | TBL1XR1 | Sponge network | 0.659 | 3.0E-5 | 0.578 | 0 | 0.259 |
| 52032 | AF131215.2 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-326 | 10 | CSDE1 | Sponge network | -0.176 | 0.59692 | -0.493 | 0 | 0.259 |
| 52033 | RP11-999E24.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-340-5p | 10 | LUZP2 | Sponge network | -0.693 | 0.05629 | -0.056 | 0.89416 | 0.259 |
| 52034 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-502-5p | 12 | TGFBR2 | Sponge network | -0.405 | 0.22013 | -0.243 | 0.11408 | 0.259 |
| 52035 | CTD-2666L21.1 |
hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-629-3p | 18 | RASSF2 | Sponge network | 0.368 | 0.20093 | -0.273 | 0.21961 | 0.259 |
| 52036 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-455-5p | 10 | AKAP12 | Sponge network | 0.056 | 0.8494 | -0.932 | 0.0028 | 0.259 |
| 52037 | RP11-736K20.5 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-452-5p;hsa-miR-590-3p | 27 | GNAQ | Sponge network | -0.358 | 0.17255 | -0.367 | 0.00102 | 0.259 |
| 52038 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 16 | CNTNAP3 | Sponge network | -0.342 | 0.02288 | -1.465 | 0.00033 | 0.259 |
| 52039 | LINC00094 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-589-3p | 10 | FAM172A | Sponge network | 0.253 | 0.09565 | -0.57 | 0 | 0.259 |
| 52040 | LINC00969 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 17 | PRKAA1 | Sponge network | 0.683 | 1.0E-5 | 0.317 | 0.0031 | 0.259 |
| 52041 | PGM5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 20 | HECA | Sponge network | -4.546 | 0 | -0.217 | 0.03463 | 0.259 |
| 52042 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-93-5p | 11 | EPAS1 | Sponge network | -0.785 | 0.00035 | -0.184 | 0.14418 | 0.259 |
| 52043 | MIR143HG |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 30 | TSC1 | Sponge network | -0.739 | 0.0574 | 0.103 | 0.26071 | 0.259 |
| 52044 | FAM13A-AS1 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-935;hsa-miR-98-5p | 25 | DMD | Sponge network | -0.83 | 0 | -1.506 | 0 | 0.259 |
| 52045 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 41 | SPRY3 | Sponge network | 0.311 | 0.10344 | 0.016 | 0.91286 | 0.259 |
| 52046 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | IGSF10 | Sponge network | 0.537 | 0.00139 | -1.751 | 1.0E-5 | 0.259 |
| 52047 | MEG3 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-550a-3p | 13 | CACNA1E | Sponge network | 0.249 | 0.35902 | 1.752 | 0 | 0.259 |
| 52048 | RP11-968O1.5 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p | 21 | PAFAH1B1 | Sponge network | -0.829 | 0.01343 | -0.429 | 0 | 0.259 |
| 52049 | RP11-798M19.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-338-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | BRD3 | Sponge network | -0.612 | 0.00094 | 0.37 | 0.00013 | 0.259 |
| 52050 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196b-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-96-5p | 14 | JAZF1 | Sponge network | 0.453 | 0.01839 | -0.377 | 0.02677 | 0.259 |
| 52051 | OIP5-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | ZDBF2 | Sponge network | 0.421 | 0.00084 | -0.998 | 0.00025 | 0.259 |
| 52052 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-5p | 12 | FGFR1 | Sponge network | 0.622 | 0.00015 | -0.509 | 0.03957 | 0.259 |
| 52053 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-7-1-3p | 27 | THBS1 | Sponge network | -2.915 | 0.00015 | 0.741 | 0.00386 | 0.259 |
| 52054 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-409-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 28 | HBP1 | Sponge network | -2.915 | 0.00015 | -0.454 | 0 | 0.259 |
| 52055 | CTD-2619J13.9 |
hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-29b-1-5p;hsa-miR-628-5p | 11 | IL6ST | Sponge network | 0.576 | 0.0022 | -0.402 | 0.00676 | 0.259 |
| 52056 | PSMG3-AS1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TLL1 | Sponge network | -0.125 | 0.49414 | -1.195 | 3.0E-5 | 0.259 |
| 52057 | GNG12-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-940 | 27 | THBS1 | Sponge network | -0.793 | 7.0E-5 | 0.741 | 0.00386 | 0.259 |
| 52058 | RP11-384O8.1 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-590-3p | 16 | RPS6KA6 | Sponge network | -0.738 | 0.21233 | -2.989 | 0 | 0.259 |
| 52059 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3913-5p;hsa-miR-942-5p | 13 | HLF | Sponge network | -1.054 | 0.0706 | -2.443 | 0 | 0.259 |
| 52060 | RP11-244H3.1 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | TAL1 | Sponge network | 0.659 | 3.0E-5 | -0.462 | 0.00574 | 0.259 |
| 52061 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | TCF4 | Sponge network | 0.853 | 0.15352 | 0.016 | 0.92271 | 0.259 |
| 52062 | RP5-1085F17.3 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 14 | KDSR | Sponge network | 0.541 | 0.00125 | 0.239 | 0.02216 | 0.259 |
| 52063 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-940 | 10 | SRGAP3 | Sponge network | -0.72 | 0.24239 | -0.202 | 0.29297 | 0.259 |
| 52064 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 14 | NR4A3 | Sponge network | -0.612 | 0.00094 | -1.385 | 0 | 0.259 |
| 52065 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-421;hsa-miR-629-3p | 12 | RBL2 | Sponge network | -0.602 | 0.00331 | -0.282 | 0.00254 | 0.259 |
| 52066 | HHIP-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-455-3p | 13 | EZH1 | Sponge network | -0.737 | 0.10822 | -0.564 | 1.0E-5 | 0.259 |
| 52067 | RAMP2-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 27 | PIK3R1 | Sponge network | -0.513 | 0.04703 | -0.133 | 0.36854 | 0.259 |
| 52068 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 15 | CLIP1 | Sponge network | 0.174 | 0.30034 | -0.215 | 0.08684 | 0.259 |
| 52069 | ZNF32-AS2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-629-3p | 12 | TCF12 | Sponge network | 1.552 | 7.0E-5 | 0.174 | 0.13271 | 0.259 |
| 52070 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | MXI1 | Sponge network | -0.021 | 0.93598 | -0.987 | 0 | 0.259 |
| 52071 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 24 | GRIN2A | Sponge network | 0.72 | 0.0001 | -2.161 | 0 | 0.259 |
| 52072 | RP11-999E24.3 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-3p | 14 | EFNA5 | Sponge network | -0.693 | 0.05629 | -0.421 | 0.17868 | 0.259 |
| 52073 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-96-5p | 26 | PPM1L | Sponge network | -0.517 | 0.02935 | -0.537 | 0.00616 | 0.259 |
| 52074 | CALML3-AS1 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-625-5p | 12 | IL13RA1 | Sponge network | 0.95 | 0.15943 | 0.019 | 0.86949 | 0.259 |
| 52075 | FOXN3-AS1 |
hsa-let-7f-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-30d-3p;hsa-miR-361-3p;hsa-miR-7-1-3p | 11 | THRB | Sponge network | -0.399 | 0.00917 | -1.117 | 0.0004 | 0.259 |
| 52076 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-340-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-96-5p | 33 | JAZF1 | Sponge network | 0.083 | 0.66405 | -0.377 | 0.02677 | 0.259 |
| 52077 | PART1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-324-3p;hsa-miR-331-5p;hsa-miR-429 | 12 | DUSP1 | Sponge network | -2.925 | 0 | -1.754 | 0 | 0.259 |
| 52078 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 19 | ARHGAP29 | Sponge network | 0.782 | 0.00016 | 0.131 | 0.42953 | 0.259 |
| 52079 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-7-1-3p | 17 | CBL | Sponge network | 0.545 | 0.00064 | 0.291 | 0.01267 | 0.259 |
| 52080 | RP11-693J15.4 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-590-5p | 11 | FAM135B | Sponge network | -0.72 | 0.24239 | -3.978 | 0 | 0.259 |
| 52081 | RP1-122P22.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-629-3p | 11 | EPHA7 | Sponge network | 0.065 | 0.73468 | -2.197 | 3.0E-5 | 0.259 |
| 52082 | RP11-6O2.3 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-590-5p;hsa-miR-96-5p | 30 | GNAQ | Sponge network | 0.331 | 0.57247 | -0.367 | 0.00102 | 0.259 |
| 52083 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p | 11 | RYR2 | Sponge network | -0.285 | 0.2172 | -1.466 | 0.00031 | 0.259 |
| 52084 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-582-5p | 13 | ARID5B | Sponge network | 0.792 | 0.0049 | 0.342 | 0.01676 | 0.259 |
| 52085 | RP11-130L8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 24 | NTRK2 | Sponge network | -0.663 | 0.10887 | -0.736 | 0.06495 | 0.259 |
| 52086 | RP11-119F7.5 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | DICER1 | Sponge network | 0.225 | 0.30644 | 0.042 | 0.674 | 0.259 |
| 52087 | RP11-727A23.5 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-378c;hsa-miR-484 | 11 | MED12L | Sponge network | 1.117 | 0 | -0.194 | 0.55299 | 0.259 |
| 52088 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-7-1-3p | 11 | SASH1 | Sponge network | 0.817 | 3.0E-5 | -0.502 | 0.00175 | 0.259 |
| 52089 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 51 | THRB | Sponge network | -1.203 | 0.03881 | -1.117 | 0.0004 | 0.259 |
| 52090 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | MYO9A | Sponge network | 0.197 | 0.25618 | -0.147 | 0.32373 | 0.259 |
| 52091 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 27 | PPP1R9A | Sponge network | -0.739 | 0.00044 | -0.903 | 0.04254 | 0.259 |
| 52092 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p | 20 | PCDH17 | Sponge network | 0.659 | 3.0E-5 | 0.731 | 2.0E-5 | 0.259 |
| 52093 | NR2F1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 16 | TRIM3 | Sponge network | -0.49 | 0.04946 | -0.577 | 0 | 0.259 |
| 52094 | LINC00648 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 12 | NR2C2 | Sponge network | 0.633 | 0.51678 | 0.21 | 0.03224 | 0.259 |
| 52095 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-628-5p | 16 | BCL6 | Sponge network | 0.657 | 4.0E-5 | -0.211 | 0.19489 | 0.259 |
| 52096 | TRAF3IP2-AS1 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-590-3p | 17 | FAM135B | Sponge network | -0.323 | 0.01372 | -3.978 | 0 | 0.259 |
| 52097 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-486-5p;hsa-miR-628-5p;hsa-miR-654-3p | 19 | FOXP1 | Sponge network | 0.946 | 0 | 0.042 | 0.74368 | 0.259 |
| 52098 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 16 | FOXO3 | Sponge network | 1.308 | 0 | -0.04 | 0.72028 | 0.259 |
| 52099 | RP11-774O3.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-590-3p | 12 | DAB2 | Sponge network | -0.487 | 0.02157 | 0.431 | 0.0113 | 0.259 |
| 52100 | RP11-327P2.5 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-92a-1-5p | 14 | LUZP2 | Sponge network | -0.147 | 0.40452 | -0.056 | 0.89416 | 0.259 |
| 52101 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | NRXN1 | Sponge network | -0.031 | 0.89063 | -3.778 | 0 | 0.259 |
| 52102 | AF131215.2 |
hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-378a-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 10 | ADCY1 | Sponge network | -0.176 | 0.59692 | 0.362 | 0.16982 | 0.259 |
| 52103 | CTD-3099C6.9 |
hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-3p | 12 | CCDC6 | Sponge network | 1.361 | 0 | 0.27 | 0.02555 | 0.259 |
| 52104 | CTD-2089N3.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-27a-5p;hsa-miR-590-3p;hsa-miR-629-3p | 14 | SNX29 | Sponge network | -1.375 | 0.08642 | -0.34 | 0.01371 | 0.259 |
| 52105 | ARHGEF26-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | USP25 | Sponge network | -2.46 | 0 | -0.242 | 0.01397 | 0.259 |
| 52106 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | EYA4 | Sponge network | 0.178 | 0.29106 | 0.3 | 0.36876 | 0.259 |
| 52107 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 66 | PTEN | Sponge network | -0.737 | 0.06182 | -0.187 | 0.07748 | 0.259 |
| 52108 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-539-5p | 15 | RSPO2 | Sponge network | -0.785 | 0.00035 | -3.038 | 0 | 0.259 |
| 52109 | RP13-1039J1.4 |
hsa-let-7g-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-582-5p | 15 | RUNX1T1 | Sponge network | 0.356 | 0.33467 | -0.344 | 0.2374 | 0.259 |
| 52110 | RP11-798M19.6 |
hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-128-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-3613-5p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-942-5p | 20 | EMP2 | Sponge network | -0.612 | 0.00094 | -0.423 | 0.00985 | 0.259 |
| 52111 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-576-5p | 12 | NALCN | Sponge network | -0.969 | 2.0E-5 | 1.068 | 0.00237 | 0.259 |
| 52112 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | ZFHX3 | Sponge network | -1.264 | 6.0E-5 | 0.389 | 0.01363 | 0.259 |
| 52113 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-3p | 11 | LATS1 | Sponge network | 2.094 | 0 | 0.109 | 0.34208 | 0.259 |
| 52114 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 42 | CUX1 | Sponge network | 0.72 | 0.0001 | -0.039 | 0.6705 | 0.259 |
| 52115 | CTBP1-AS2 |
hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-497-5p | 10 | SLC5A3 | Sponge network | 0.623 | 0 | 0.493 | 5.0E-5 | 0.259 |
| 52116 | AC005562.1 |
hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | PHF3 | Sponge network | 0.488 | 0.00084 | 0.126 | 0.19652 | 0.259 |
| 52117 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 17 | TBL1XR1 | Sponge network | 0.267 | 0.2937 | 0.578 | 0 | 0.259 |
| 52118 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p | 13 | GREM1 | Sponge network | 0.225 | 0.30644 | 0.902 | 0.01691 | 0.259 |
| 52119 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p | 12 | PKNOX1 | Sponge network | 0.782 | 0.00016 | 0.085 | 0.28917 | 0.259 |
| 52120 | RP3-402G11.26 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | CADM2 | Sponge network | -0.254 | 0.19303 | -3.782 | 0 | 0.259 |
| 52121 | PGM5-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-423-5p;hsa-miR-589-5p;hsa-miR-7-1-3p | 13 | SUFU | Sponge network | -4.546 | 0 | -0.04 | 0.65489 | 0.259 |
| 52122 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-96-5p | 14 | PAK3 | Sponge network | 0.219 | 0.1516 | -0.628 | 0.07253 | 0.259 |
| 52123 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-550a-3p;hsa-miR-582-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 19 | EBF1 | Sponge network | 0.614 | 1.0E-5 | -0.384 | 0.08856 | 0.259 |
| 52124 | AC141928.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-454-3p;hsa-miR-7-1-3p | 14 | HECA | Sponge network | 0.853 | 0.15352 | -0.217 | 0.03463 | 0.259 |
| 52125 | SNHG14 |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 22 | JUN | Sponge network | -1.111 | 0.0003 | -0.932 | 0 | 0.259 |
| 52126 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-214-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-542-3p;hsa-miR-577;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-93-5p | 41 | PPM1A | Sponge network | -0.227 | 0.31657 | -0.623 | 0 | 0.259 |
| 52127 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-223-3p | 12 | MEF2C | Sponge network | 1.122 | 0.02989 | -0.499 | 0.01448 | 0.259 |
| 52128 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-93-5p | 11 | EDA2R | Sponge network | -0.785 | 0.00035 | -0.587 | 0.01598 | 0.259 |
| 52129 | SOX2-OT |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-19b-3p;hsa-miR-30d-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-493-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | PRUNE2 | Sponge network | -1.264 | 6.0E-5 | -1.733 | 0.00022 | 0.259 |
| 52130 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-7-1-3p | 10 | CNTN5 | Sponge network | -0.969 | 2.0E-5 | -0.219 | 0.61957 | 0.259 |
| 52131 | RP11-296I10.3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-335-5p | 14 | PTPN14 | Sponge network | 0.592 | 0.00014 | 0.144 | 0.34595 | 0.259 |
| 52132 | FAM157B |
hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-664a-5p | 15 | SOX11 | Sponge network | 0.92 | 0 | 2.68 | 0 | 0.259 |
| 52133 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-7-1-3p | 12 | ABCC9 | Sponge network | -0.176 | 0.59692 | -0.466 | 0.14121 | 0.259 |
| 52134 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p;hsa-miR-96-5p | 50 | BMPR2 | Sponge network | 0.395 | 0.15451 | 0.206 | 0.0635 | 0.259 |
| 52135 | RP11-10N23.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-744-3p | 19 | LPP | Sponge network | 1.134 | 0 | -0.459 | 0.04475 | 0.259 |
| 52136 | RP11-686D22.8 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PRKCB | Sponge network | 0.898 | 1.0E-5 | -1.313 | 2.0E-5 | 0.259 |
| 52137 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | PDZRN3 | Sponge network | 0.72 | 0.0001 | -1.548 | 0 | 0.259 |
| 52138 | AC009948.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | MAF | Sponge network | 0.532 | 0.00505 | -1.257 | 0 | 0.259 |
| 52139 | WAC-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-511-5p;hsa-miR-590-3p | 11 | TRIM33 | Sponge network | 0.821 | 0 | 0.402 | 7.0E-5 | 0.259 |
| 52140 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 21 | BCL11A | Sponge network | -1.249 | 7.0E-5 | -0.932 | 0.0063 | 0.259 |
| 52141 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 23 | SON | Sponge network | -1.575 | 6.0E-5 | 0.168 | 0.04406 | 0.259 |
| 52142 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 15 | AKAP13 | Sponge network | -0.563 | 0.02545 | 0.028 | 0.7729 | 0.259 |
| 52143 | AP000487.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 13 | ABI2 | Sponge network | 1.987 | 0 | 0.175 | 0.14787 | 0.259 |
| 52144 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 25 | CLASP2 | Sponge network | 0.488 | 0.00084 | -0.038 | 0.71 | 0.259 |
| 52145 | TINCR |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p | 10 | DNAJB4 | Sponge network | -0.664 | 0.14543 | -0.7 | 1.0E-5 | 0.259 |
| 52146 | RP11-307C18.1 |
hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-582-3p | 11 | ABL2 | Sponge network | 1.082 | 0.00023 | 0.82 | 0 | 0.259 |
| 52147 | CTD-2184D3.6 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-502-5p | 10 | PTPN14 | Sponge network | 0.272 | 0.44692 | 0.144 | 0.34595 | 0.259 |
| 52148 | AP001055.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 21 | PTPRD | Sponge network | 0.693 | 0.00076 | -1.005 | 0.00064 | 0.259 |
| 52149 | AC141928.1 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-7-1-3p | 16 | LIMCH1 | Sponge network | 0.853 | 0.15352 | -0.915 | 1.0E-5 | 0.259 |
| 52150 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-374a-3p;hsa-miR-532-3p;hsa-miR-7-1-3p | 10 | CBL | Sponge network | -0.557 | 0.11135 | 0.291 | 0.01267 | 0.259 |
| 52151 | DNM1P35 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-590-3p | 14 | LIMCH1 | Sponge network | 0.051 | 0.83388 | -0.915 | 1.0E-5 | 0.259 |
| 52152 | TINCR |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-624-5p;hsa-miR-940;hsa-miR-942-5p | 26 | NOTCH2 | Sponge network | -0.664 | 0.14543 | 0.138 | 0.35163 | 0.259 |
| 52153 | CTD-3138B18.5 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-511-5p;hsa-miR-628-5p | 11 | TRIM33 | Sponge network | 0.379 | 0.03091 | 0.402 | 7.0E-5 | 0.259 |
| 52154 | RP11-157J24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-576-5p | 19 | RUNX1T1 | Sponge network | -1.664 | 0.05165 | -0.344 | 0.2374 | 0.259 |
| 52155 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 10 | PSIP1 | Sponge network | 1.601 | 0 | -0.005 | 0.96897 | 0.259 |
| 52156 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-628-5p | 18 | ACVR1 | Sponge network | -0.405 | 0.22013 | 0.279 | 0.00234 | 0.259 |
| 52157 | RP4-791M13.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-7-1-3p | 14 | GRIN2A | Sponge network | 0.419 | 0.0319 | -2.161 | 0 | 0.259 |
| 52158 | RP11-327P2.5 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-450b-5p;hsa-miR-590-5p;hsa-miR-664a-3p | 11 | TMEFF2 | Sponge network | -0.147 | 0.40452 | -2.426 | 0 | 0.259 |
| 52159 | AC005618.6 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-423-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 14 | ROBO1 | Sponge network | 0.862 | 0.06213 | -0.009 | 0.96154 | 0.259 |
| 52160 | RP11-747H7.3 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-376c-3p;hsa-miR-497-5p;hsa-miR-766-3p | 13 | TBL1XR1 | Sponge network | 1.379 | 0 | 0.578 | 0 | 0.259 |
| 52161 | RP11-488L18.10 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p | 10 | SLC5A3 | Sponge network | 1.392 | 0 | 0.493 | 5.0E-5 | 0.259 |
| 52162 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 25 | EPHA7 | Sponge network | 0.381 | 0.25967 | -2.197 | 3.0E-5 | 0.259 |
| 52163 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-590-3p | 24 | NOTCH2 | Sponge network | 0.178 | 0.29106 | 0.138 | 0.35163 | 0.259 |
| 52164 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p | 10 | THBS1 | Sponge network | 1.2 | 0.01813 | 0.741 | 0.00386 | 0.259 |
| 52165 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-769-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 26 | KALRN | Sponge network | -2.925 | 0 | -0.819 | 1.0E-5 | 0.259 |
| 52166 | CALML3-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-576-5p | 16 | KDSR | Sponge network | 0.95 | 0.15943 | 0.239 | 0.02216 | 0.259 |
| 52167 | CALML3-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-484 | 10 | FBXO11 | Sponge network | 0.95 | 0.15943 | 0.142 | 0.04938 | 0.259 |
| 52168 | FAM13A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-324-5p;hsa-miR-532-3p | 13 | SYNM | Sponge network | -0.83 | 0 | -2.309 | 1.0E-5 | 0.259 |
| 52169 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 35 | MOB1B | Sponge network | -0.899 | 6.0E-5 | 0.197 | 0.15266 | 0.259 |
| 52170 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-590-3p | 17 | YAP1 | Sponge network | -0.227 | 0.31657 | 0.22 | 0.11836 | 0.259 |
| 52171 | TRAF3IP2-AS1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-32-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 13 | EGR2 | Sponge network | -0.323 | 0.01372 | 0.309 | 0.17079 | 0.259 |
| 52172 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-96-5p | 12 | SCN9A | Sponge network | -0.418 | 0.00068 | -0.525 | 0.10593 | 0.259 |
| 52173 | TMEM51-AS1 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-590-3p | 10 | EGFR | Sponge network | 0.23 | 0.38667 | -0.175 | 0.48451 | 0.259 |
| 52174 | GS1-124K5.11 |
hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-550a-3p;hsa-miR-590-3p | 12 | PAK3 | Sponge network | 0.494 | 0.00339 | -0.628 | 0.07253 | 0.259 |
| 52175 | C4A-AS1 |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-628-5p;hsa-miR-96-5p | 11 | ACVR1 | Sponge network | 3.345 | 0 | 0.279 | 0.00234 | 0.259 |
| 52176 | RP11-983P16.4 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-655-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 13 | SHPRH | Sponge network | 0.41 | 0.02214 | 0.098 | 0.40994 | 0.259 |
| 52177 | FAM13A-AS1 |
hsa-miR-1296-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-758-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | CSDE1 | Sponge network | -0.83 | 0 | -0.493 | 0 | 0.259 |
| 52178 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 27 | MYBL1 | Sponge network | -1.111 | 0.0003 | 0.494 | 0.02152 | 0.259 |
| 52179 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-539-5p | 11 | CLASP2 | Sponge network | 0.924 | 0 | -0.038 | 0.71 | 0.259 |
| 52180 | RP11-389C8.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | LRIG1 | Sponge network | 0.727 | 3.0E-5 | -0.537 | 0.00588 | 0.259 |
| 52181 | PCA3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | PPP1R9A | Sponge network | -0.709 | 0.18168 | -0.903 | 0.04254 | 0.259 |
| 52182 | ZSCAN16-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-590-3p | 22 | PRDM2 | Sponge network | -0.342 | 0.02288 | -0.258 | 0.00721 | 0.259 |
| 52183 | RP11-384K6.6 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-628-5p;hsa-miR-744-3p | 37 | LPP | Sponge network | 0.946 | 0 | -0.459 | 0.04475 | 0.259 |
| 52184 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | BCL11A | Sponge network | -0.222 | 0.32445 | -0.932 | 0.0063 | 0.259 |
| 52185 | RP11-46C24.7 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p | 41 | PPM1L | Sponge network | 0.392 | 0.0278 | -0.537 | 0.00616 | 0.259 |
| 52186 | RP11-890B15.3 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-411-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TMEM30A | Sponge network | 0.101 | 0.60919 | 0.096 | 0.31031 | 0.259 |
| 52187 | RP11-116O18.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CDH19 | Sponge network | 0.05 | 0.95483 | -3.434 | 0 | 0.259 |
| 52188 | RP11-706C16.7 |
hsa-miR-1271-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577 | 11 | TACC1 | Sponge network | 0.357 | 0.72407 | -0.362 | 0.05406 | 0.259 |
| 52189 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 24 | MEF2C | Sponge network | 0.389 | 0.04293 | -0.499 | 0.01448 | 0.259 |
| 52190 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 11 | GPAM | Sponge network | 0.368 | 0.20093 | 0.142 | 0.29732 | 0.259 |
| 52191 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 14 | EPHA3 | Sponge network | 1.11 | 1.0E-5 | 0.185 | 0.53588 | 0.259 |
| 52192 | RP11-822E23.8 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3607-3p | 20 | LIMCH1 | Sponge network | -0.777 | 0.16667 | -0.915 | 1.0E-5 | 0.259 |
| 52193 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p | 15 | PIK3C2A | Sponge network | 1.11 | 1.0E-5 | 0.061 | 0.53776 | 0.259 |
| 52194 | RP11-157J24.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-550a-5p | 17 | DST | Sponge network | -1.664 | 0.05165 | -0.713 | 0.00096 | 0.259 |
| 52195 | RP11-102N12.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | TSHZ3 | Sponge network | -0.351 | 0.11358 | -0.556 | 0.01516 | 0.259 |
| 52196 | AC005618.6 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 23 | RIMS2 | Sponge network | 0.862 | 0.06213 | -1.365 | 0.00208 | 0.259 |
| 52197 | LINC00957 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 24 | HBP1 | Sponge network | -0.421 | 0.0114 | -0.454 | 0 | 0.259 |
| 52198 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p | 19 | MYBL1 | Sponge network | 1.601 | 0 | 0.494 | 0.02152 | 0.259 |
| 52199 | RP11-855A2.3 |
hsa-miR-127-5p;hsa-miR-148a-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-374b-3p;hsa-miR-590-3p | 17 | CCDC6 | Sponge network | 2.094 | 0 | 0.27 | 0.02555 | 0.259 |
| 52200 | MALAT1 |
hsa-miR-106b-5p;hsa-miR-127-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | PER2 | Sponge network | 0.782 | 0.00016 | -0.396 | 0.00269 | 0.259 |
| 52201 | CTB-113P19.4 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | LATS2 | Sponge network | 0.906 | 0.31715 | 0.159 | 0.2304 | 0.259 |
| 52202 | RP11-53O19.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-375;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 16 | KDSR | Sponge network | -0.405 | 0.22013 | 0.239 | 0.02216 | 0.259 |
| 52203 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-5p | 13 | CNTNAP3 | Sponge network | -0.773 | 0.04073 | -1.465 | 0.00033 | 0.259 |
| 52204 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 32 | ST6GAL2 | Sponge network | 0.421 | 0.00084 | -1.201 | 0.00597 | 0.259 |
| 52205 | RP11-2H8.2 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 14 | UBE2D1 | Sponge network | 1.089 | 0 | 0.468 | 0 | 0.259 |
| 52206 | RP11-2B6.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-590-3p | 15 | DMD | Sponge network | 0.539 | 0.03722 | -1.506 | 0 | 0.259 |
| 52207 | RP11-389C8.2 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3127-5p;hsa-miR-455-3p | 12 | ABL1 | Sponge network | 0.727 | 3.0E-5 | -0.215 | 0.07804 | 0.259 |
| 52208 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-935;hsa-miR-96-5p | 36 | FBXO32 | Sponge network | 0.311 | 0.10344 | -0.495 | 0.07561 | 0.259 |
| 52209 | LINC00641 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 23 | CSNK1A1 | Sponge network | -0.222 | 0.32445 | -0.19 | 0.01687 | 0.259 |
| 52210 | FAM215B |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-330-5p | 13 | TCF12 | Sponge network | 0.817 | 3.0E-5 | 0.174 | 0.13271 | 0.259 |
| 52211 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | FYN | Sponge network | 0.082 | 0.55654 | -0.131 | 0.37051 | 0.259 |
| 52212 | NNT-AS1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-363-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p | 13 | TRIM13 | Sponge network | 0.799 | 1.0E-5 | 0.183 | 0.06968 | 0.259 |
| 52213 | LINC00094 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-508-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-660-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 75 | LPP | Sponge network | 0.253 | 0.09565 | -0.459 | 0.04475 | 0.259 |
| 52214 | RP11-532F6.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p | 25 | SASH1 | Sponge network | -0.896 | 0.00099 | -0.502 | 0.00175 | 0.259 |
| 52215 | CTC-297N7.9 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-301a-3p | 20 | DTNA | Sponge network | -2.069 | 0 | -1.648 | 1.0E-5 | 0.259 |
| 52216 | EIF3J-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 19 | EPHA3 | Sponge network | 0.331 | 0.00509 | 0.185 | 0.53588 | 0.259 |
| 52217 | RP11-57H14.4 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-493-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-96-5p | 47 | CADM2 | Sponge network | 0.083 | 0.66405 | -3.782 | 0 | 0.259 |
| 52218 | CTC-490E21.10 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p | 10 | SPRY3 | Sponge network | 1.46 | 1.0E-5 | 0.016 | 0.91286 | 0.259 |
| 52219 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 15 | PTPN11 | Sponge network | 0.392 | 0.0278 | 0.431 | 2.0E-5 | 0.259 |
| 52220 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | EPHA3 | Sponge network | 0.267 | 0.2937 | 0.185 | 0.53588 | 0.259 |
| 52221 | EMX2OS |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 18 | NOTCH2NL | Sponge network | -0.777 | 0.1134 | 0.024 | 0.84307 | 0.259 |
| 52222 | RP11-434B12.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 13 | SORCS1 | Sponge network | -0.031 | 0.89063 | -3.492 | 0 | 0.259 |
| 52223 | RP11-758N13.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-455-3p | 15 | MCC | Sponge network | 2.939 | 3.0E-5 | -1.005 | 0 | 0.259 |
| 52224 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-188-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | TIMP3 | Sponge network | 0.225 | 0.30644 | -0.546 | 0.02307 | 0.259 |
| 52225 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p | 20 | LATS1 | Sponge network | -0.162 | 0.47687 | 0.109 | 0.34208 | 0.259 |
| 52226 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-486-5p | 17 | EPHA3 | Sponge network | 1.03 | 0 | 0.185 | 0.53588 | 0.259 |
| 52227 | HOXA-AS2 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-744-3p;hsa-miR-942-5p | 31 | IGFBP5 | Sponge network | 0.61 | 0.01593 | -0.806 | 0.00105 | 0.259 |
| 52228 | RP11-798M19.6 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-660-5p | 16 | IRS1 | Sponge network | -0.612 | 0.00094 | -0.026 | 0.88855 | 0.259 |
| 52229 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-93-5p | 13 | ARNT2 | Sponge network | 0.082 | 0.55397 | -0.332 | 0.25851 | 0.259 |
| 52230 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-362-3p;hsa-miR-539-5p | 11 | MGA | Sponge network | -0.405 | 0.22013 | 0.323 | 0.00792 | 0.259 |
| 52231 | RP11-359E10.1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-484;hsa-miR-500a-5p | 10 | PKNOX1 | Sponge network | 0.299 | 0.57722 | 0.085 | 0.28917 | 0.259 |
| 52232 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | EDA2R | Sponge network | -0.053 | 0.80685 | -0.587 | 0.01598 | 0.259 |
| 52233 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-590-3p | 16 | ARHGAP29 | Sponge network | 0.873 | 0 | 0.131 | 0.42953 | 0.259 |
| 52234 | RAD51-AS1 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-655-3p | 11 | HECA | Sponge network | 0.768 | 7.0E-5 | -0.217 | 0.03463 | 0.259 |
| 52235 | AF131215.9 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 20 | SYT4 | Sponge network | -0.322 | 0.25945 | -3.207 | 0 | 0.259 |
| 52236 | CTB-179K24.3 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-378a-5p;hsa-miR-940 | 15 | CREB1 | Sponge network | 0.841 | 0.04055 | 0.203 | 0.00537 | 0.259 |
| 52237 | SNX29P2 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-511-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | BTG2 | Sponge network | 0.4 | 0.14611 | -1.763 | 0 | 0.259 |
| 52238 | LINC00578 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-26b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577 | 17 | LRP1B | Sponge network | 0.811 | 0.03228 | -2.163 | 0 | 0.259 |
| 52239 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-940 | 21 | NTRK2 | Sponge network | 0.453 | 0.01839 | -0.736 | 0.06495 | 0.259 |
| 52240 | RP4-668J24.2 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-424-5p;hsa-miR-92a-1-5p | 14 | PRKAB2 | Sponge network | -1.054 | 0.0706 | -0.367 | 0.01371 | 0.259 |
| 52241 | AC108488.4 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 21 | ETV1 | Sponge network | 0.259 | 0.12542 | -0.096 | 0.67674 | 0.259 |
| 52242 | LIPE-AS1 |
hsa-miR-125a-5p;hsa-miR-141-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-582-5p | 10 | RANBP9 | Sponge network | 0.078 | 0.55803 | -0.191 | 0.04628 | 0.259 |
| 52243 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-484 | 10 | MYCBP2 | Sponge network | 0.4 | 0.14611 | -0.027 | 0.82016 | 0.259 |
| 52244 | HAND2-AS1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-340-5p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-590-3p | 10 | PKD1L2 | Sponge network | -1.697 | 0.00889 | -0.971 | 0.00438 | 0.259 |
| 52245 | LINC00654 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-96-5p | 27 | JAZF1 | Sponge network | 0.374 | 0.20054 | -0.377 | 0.02677 | 0.259 |
| 52246 | RP11-585P4.5 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 11 | PAFAH1B2 | Sponge network | 1.046 | 1.0E-5 | 0.318 | 0.0001 | 0.259 |
| 52247 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-425-5p;hsa-miR-429 | 10 | HERC2 | Sponge network | 0.249 | 0.35902 | 0.114 | 0.30638 | 0.259 |
| 52248 | AC074289.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-629-3p;hsa-miR-96-5p | 17 | RASSF8 | Sponge network | -0.326 | 0.14314 | -0.122 | 0.65591 | 0.259 |
| 52249 | RP11-160O5.1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-299-5p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-539-5p | 10 | DIP2C | Sponge network | -0.327 | 0.07353 | -0.436 | 0.01713 | 0.259 |
| 52250 | RP11-6O2.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-590-5p;hsa-miR-940 | 13 | CELF4 | Sponge network | 0.331 | 0.57247 | -1.135 | 0.00344 | 0.259 |
| 52251 | CTC-471J1.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-539-5p;hsa-miR-629-3p | 20 | DST | Sponge network | 1.11 | 1.0E-5 | -0.713 | 0.00096 | 0.259 |
| 52252 | RP11-2H8.2 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-590-3p | 14 | TMEM30A | Sponge network | 1.089 | 0 | 0.096 | 0.31031 | 0.259 |
| 52253 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-335-5p;hsa-miR-532-3p | 10 | LATS1 | Sponge network | 0.453 | 0.01839 | 0.109 | 0.34208 | 0.259 |
| 52254 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-93-5p | 12 | EDA2R | Sponge network | 0.4 | 0.14611 | -0.587 | 0.01598 | 0.259 |
| 52255 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | FSTL5 | Sponge network | 0.619 | 0.00157 | -1.3 | 0.01619 | 0.259 |
| 52256 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-942-5p | 11 | RB1CC1 | Sponge network | 0.082 | 0.55397 | 0.175 | 0.12559 | 0.259 |
| 52257 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-493-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | DDX6 | Sponge network | -0.405 | 0.22013 | 0.091 | 0.36428 | 0.259 |
| 52258 | LINC00963 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 14 | GPAM | Sponge network | 0.523 | 9.0E-5 | 0.142 | 0.29732 | 0.259 |
| 52259 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 25 | EPHA5 | Sponge network | 0.4 | 0.14611 | -0.957 | 0.01733 | 0.259 |
| 52260 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-628-5p | 17 | ACVR1 | Sponge network | 0.494 | 0.00339 | 0.279 | 0.00234 | 0.259 |
| 52261 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p | 11 | PRKAA1 | Sponge network | 0.953 | 0 | 0.317 | 0.0031 | 0.259 |
| 52262 | FAM215B |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-625-5p;hsa-miR-744-3p | 14 | PTPN14 | Sponge network | 0.817 | 3.0E-5 | 0.144 | 0.34595 | 0.259 |
| 52263 | RP11-48B3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 16 | ERCC4 | Sponge network | 1.757 | 0 | 0.126 | 0.15266 | 0.259 |
| 52264 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CTDSPL | Sponge network | -0.487 | 0.02157 | 0.085 | 0.45121 | 0.259 |
| 52265 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-628-5p | 15 | ACVR1 | Sponge network | 0.177 | 0.16681 | 0.279 | 0.00234 | 0.259 |
| 52266 | RP11-47A8.5 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 19 | WDFY3 | Sponge network | 0.103 | 0.43169 | -0.201 | 0.0967 | 0.259 |
| 52267 | RP11-181G12.2 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-486-5p | 10 | NRK | Sponge network | 0.556 | 0.00298 | 0.656 | 0.19217 | 0.259 |
| 52268 | RP11-894P9.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-940 | 16 | NOTCH2 | Sponge network | 0.993 | 0 | 0.138 | 0.35163 | 0.259 |
| 52269 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 35 | PPM1A | Sponge network | -0.611 | 0.09607 | -0.623 | 0 | 0.259 |
| 52270 | RP11-439L18.2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-454-3p | 10 | DIP2C | Sponge network | 0.224 | 0.48719 | -0.436 | 0.01713 | 0.259 |
| 52271 | RP11-20I23.13 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-671-5p;hsa-miR-92a-3p | 13 | CRTC3 | Sponge network | 0.505 | 0.0014 | 0.164 | 0.16786 | 0.259 |
| 52272 | DNM1P35 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-378a-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-671-5p | 17 | CDH23 | Sponge network | 0.051 | 0.83388 | -0.974 | 0.00034 | 0.259 |
| 52273 | LINC00957 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | DMXL1 | Sponge network | -0.421 | 0.0114 | -0.426 | 0.00056 | 0.259 |
| 52274 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 20 | YAP1 | Sponge network | 0.082 | 0.55654 | 0.22 | 0.11836 | 0.259 |
| 52275 | RP11-855A2.3 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-654-3p | 11 | PIK3R1 | Sponge network | 2.094 | 0 | -0.133 | 0.36854 | 0.259 |
| 52276 | CTD-2017D11.1 |
hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-582-5p | 15 | PIK3R1 | Sponge network | 0.792 | 0.0049 | -0.133 | 0.36854 | 0.258 |
| 52277 | A2M-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 18 | PCDH17 | Sponge network | -0.08 | 0.90092 | 0.731 | 2.0E-5 | 0.258 |
| 52278 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500b-5p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | GRIA3 | Sponge network | 0.4 | 0.14611 | -1.956 | 0 | 0.258 |
| 52279 | RP11-1246C19.1 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374a-3p;hsa-miR-424-5p | 12 | PIK3R1 | Sponge network | 1.352 | 0 | -0.133 | 0.36854 | 0.258 |
| 52280 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 10 | CHD1 | Sponge network | 0.083 | 0.66405 | 0.322 | 0.00215 | 0.258 |
| 52281 | RP13-516M14.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-7-1-3p | 17 | ADAMTSL3 | Sponge network | 0.389 | 0.04293 | -1.559 | 0 | 0.258 |
| 52282 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 13 | CREBBP | Sponge network | -2.117 | 0.00161 | 0.126 | 0.15186 | 0.258 |
| 52283 | CROCCP2 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 19 | CCDC6 | Sponge network | 0.79 | 0 | 0.27 | 0.02555 | 0.258 |
| 52284 | ARHGEF26-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 24 | TGFBR2 | Sponge network | -2.46 | 0 | -0.243 | 0.11408 | 0.258 |
| 52285 | AC108488.4 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 33 | TRPS1 | Sponge network | 0.259 | 0.12542 | -0.191 | 0.44109 | 0.258 |
| 52286 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-654-3p;hsa-miR-708-3p;hsa-miR-744-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-942-5p | 32 | HLF | Sponge network | -0.371 | 0.24604 | -2.443 | 0 | 0.258 |
| 52287 | RP11-701H24.3 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-335-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 10 | NRK | Sponge network | -1.425 | 0.04204 | 0.656 | 0.19217 | 0.258 |
| 52288 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-576-5p | 14 | SLC6A15 | Sponge network | 0.176 | 0.37402 | -2.373 | 2.0E-5 | 0.258 |
| 52289 | LINC00473 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 26 | PRKAB2 | Sponge network | -1.203 | 0.03881 | -0.367 | 0.01371 | 0.258 |
| 52290 | AC009120.6 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-484;hsa-miR-590-3p | 10 | PKNOX1 | Sponge network | 0.919 | 0 | 0.085 | 0.28917 | 0.258 |
| 52291 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | CDH19 | Sponge network | 0.619 | 0.00157 | -3.434 | 0 | 0.258 |
| 52292 | GAS6-AS2 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-301a-3p;hsa-miR-421;hsa-miR-429 | 12 | MAFB | Sponge network | 0.16 | 0.50785 | -0.023 | 0.89865 | 0.258 |
| 52293 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p | 37 | ADARB1 | Sponge network | 0.853 | 0.15352 | -0.335 | 0.06631 | 0.258 |
| 52294 | RP11-473I1.5 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-369-3p | 19 | PPARA | Sponge network | 0.542 | 0.00054 | -0.379 | 0.00548 | 0.258 |
| 52295 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.194 | 0.64278 | 0.323 | 0.00792 | 0.258 |
| 52296 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-7-1-3p | 14 | TBX18 | Sponge network | 0.853 | 0.15352 | 0.843 | 0.02945 | 0.258 |
| 52297 | AC108488.4 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3613-5p;hsa-miR-7-1-3p;hsa-miR-940 | 15 | TAL1 | Sponge network | 0.259 | 0.12542 | -0.462 | 0.00574 | 0.258 |
| 52298 | MRPL23-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p | 10 | FBXO32 | Sponge network | -0.278 | 0.76559 | -0.495 | 0.07561 | 0.258 |
| 52299 | RP11-539I5.1 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RET | Sponge network | -0.468 | 0.32587 | -0.833 | 0.03517 | 0.258 |
| 52300 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | YAP1 | Sponge network | 0.385 | 0.10067 | 0.22 | 0.11836 | 0.258 |
| 52301 | AC010226.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-5p | 19 | ETV1 | Sponge network | -0.811 | 2.0E-5 | -0.096 | 0.67674 | 0.258 |
| 52302 | BOLA3-AS1 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | PCDH17 | Sponge network | -0.246 | 0.35034 | 0.731 | 2.0E-5 | 0.258 |
| 52303 | KB-1460A1.5 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | PIK3CA | Sponge network | 0.603 | 0.00127 | 0.195 | 0.06908 | 0.258 |
| 52304 | RP11-1277A3.1 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3127-5p;hsa-miR-96-5p | 16 | TACC1 | Sponge network | 0.453 | 0.01839 | -0.362 | 0.05406 | 0.258 |
| 52305 | RP11-175O19.4 |
hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-411-5p | 13 | TMEM30A | Sponge network | 0.917 | 0 | 0.096 | 0.31031 | 0.258 |
| 52306 | RP4-669L17.10 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 19 | ZFX | Sponge network | 1.093 | 0 | 0.09 | 0.42563 | 0.258 |
| 52307 | MIR600HG |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-493-3p | 12 | PRUNE2 | Sponge network | 0.594 | 0.41522 | -1.733 | 0.00022 | 0.258 |
| 52308 | RP4-584D14.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-223-5p;hsa-miR-362-3p;hsa-miR-500a-5p | 11 | DMD | Sponge network | 1.294 | 0.0031 | -1.506 | 0 | 0.258 |
| 52309 | MEG3 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-7-1-3p | 28 | SYT4 | Sponge network | 0.249 | 0.35902 | -3.207 | 0 | 0.258 |
| 52310 | H19 |
hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-576-5p | 10 | PCDH17 | Sponge network | 2.439 | 0 | 0.731 | 2.0E-5 | 0.258 |
| 52311 | BOLA3-AS1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-539-5p | 15 | RSPO2 | Sponge network | -0.246 | 0.35034 | -3.038 | 0 | 0.258 |
| 52312 | RP11-25K19.1 |
hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | PCDH17 | Sponge network | -1.057 | 0.00029 | 0.731 | 2.0E-5 | 0.258 |
| 52313 | RP11-116O18.1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-590-3p;hsa-miR-671-5p | 18 | DLC1 | Sponge network | 0.05 | 0.95483 | -0.382 | 0.05038 | 0.258 |
| 52314 | LINC00621 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 10 | EBF3 | Sponge network | 0.131 | 0.8436 | -0.058 | 0.81437 | 0.258 |
| 52315 | RP1-122P22.2 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-501-3p;hsa-miR-502-3p | 13 | PTPRD | Sponge network | 0.065 | 0.73468 | -1.005 | 0.00064 | 0.258 |
| 52316 | TP73-AS1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-671-5p;hsa-miR-92a-3p | 12 | GATA2 | Sponge network | -0.563 | 0.02545 | -0.654 | 0.0016 | 0.258 |
| 52317 | RP11-157J24.2 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-576-5p | 10 | ZFHX4 | Sponge network | -1.664 | 0.05165 | 0.018 | 0.95574 | 0.258 |
| 52318 | LINC00403 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-339-5p;hsa-miR-424-5p | 11 | ABCB5 | Sponge network | -3.586 | 0.00032 | -0.956 | 0.04077 | 0.258 |
| 52319 | RP11-327P2.5 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-92a-1-5p;hsa-miR-93-3p;hsa-miR-935 | 33 | GRIK3 | Sponge network | -0.147 | 0.40452 | -3.208 | 0 | 0.258 |
| 52320 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 24 | CBL | Sponge network | -0.031 | 0.89063 | 0.291 | 0.01267 | 0.258 |
| 52321 | RP11-981G7.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-324-5p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-744-3p | 16 | CUL5 | Sponge network | 0.986 | 0.01022 | 0.026 | 0.77301 | 0.258 |
| 52322 | RP11-620J15.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | LUZP2 | Sponge network | -0.739 | 0.00044 | -0.056 | 0.89416 | 0.258 |
| 52323 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-429;hsa-miR-452-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 59 | PTEN | Sponge network | -1.575 | 6.0E-5 | -0.187 | 0.07748 | 0.258 |
| 52324 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 23 | RNASEL | Sponge network | -0.355 | 0.0077 | -0.272 | 0.01796 | 0.258 |
| 52325 | TINCR |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-625-5p | 15 | CSMD1 | Sponge network | -0.664 | 0.14543 | -0.63 | 0.18881 | 0.258 |
| 52326 | CTA-14H9.5 |
hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-508-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 17 | THRB | Sponge network | 0.145 | 0.53343 | -1.117 | 0.0004 | 0.258 |
| 52327 | USP3-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-452-5p;hsa-miR-495-3p;hsa-miR-508-3p;hsa-miR-576-5p | 20 | INPP4B | Sponge network | -0.162 | 0.47687 | -0.097 | 0.60503 | 0.258 |
| 52328 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 22 | PTPN11 | Sponge network | -0.739 | 0.0574 | 0.431 | 2.0E-5 | 0.258 |
| 52329 | RP11-458D21.1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-429 | 11 | DYNC1I1 | Sponge network | 0.663 | 0.00073 | -1.12 | 0.00107 | 0.258 |
| 52330 | RP11-57H14.4 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | TMEM30A | Sponge network | 0.083 | 0.66405 | 0.096 | 0.31031 | 0.258 |
| 52331 | AC002454.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p | 10 | CXXC4 | Sponge network | 1.952 | 0.02558 | -0.433 | 0.21055 | 0.258 |
| 52332 | RP11-452L6.1 |
hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-425-5p;hsa-miR-664a-5p | 11 | ADARB1 | Sponge network | 0.665 | 0.00061 | -0.335 | 0.06631 | 0.258 |
| 52333 | LOXL1-AS1 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | EPHA5 | Sponge network | 0.487 | 0.04053 | -0.957 | 0.01733 | 0.258 |
| 52334 | FENDRR |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-629-5p;hsa-miR-96-5p | 14 | WNK2 | Sponge network | -1.281 | 0.00012 | -0.352 | 0.31066 | 0.258 |
| 52335 | RP11-212P7.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | ARNT2 | Sponge network | 0.219 | 0.1516 | -0.332 | 0.25851 | 0.258 |
| 52336 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 13 | HBP1 | Sponge network | -0.376 | 0.00337 | -0.454 | 0 | 0.258 |
| 52337 | CTD-2666L21.1 |
hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-342-5p | 11 | SYNPO2 | Sponge network | 0.368 | 0.20093 | -2.342 | 0 | 0.258 |
| 52338 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-3913-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-5p | 30 | ESR1 | Sponge network | -0.83 | 0 | -1.035 | 3.0E-5 | 0.258 |
| 52339 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 33 | PRKAB2 | Sponge network | -0.777 | 0.1134 | -0.367 | 0.01371 | 0.258 |
| 52340 | CTC-297N7.9 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-877-5p | 16 | ASTN1 | Sponge network | -2.069 | 0 | -2.389 | 2.0E-5 | 0.258 |
| 52341 | LINC00886 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-5p | 15 | RPS6KA6 | Sponge network | -0.371 | 0.24604 | -2.989 | 0 | 0.258 |
| 52342 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-7-1-3p | 14 | TBX18 | Sponge network | 0.41 | 0.02214 | 0.843 | 0.02945 | 0.258 |
| 52343 | RP11-307B6.3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p | 11 | BAZ2B | Sponge network | -4.463 | 0.00025 | -0.276 | 0.02833 | 0.258 |
| 52344 | TRAF3IP2-AS1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-326;hsa-miR-3614-5p;hsa-miR-378a-5p | 11 | CACNA1E | Sponge network | -0.323 | 0.01372 | 1.752 | 0 | 0.258 |
| 52345 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-590-3p | 13 | EDA2R | Sponge network | -0.899 | 6.0E-5 | -0.587 | 0.01598 | 0.258 |
| 52346 | MIR600HG |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p | 12 | CACNA1E | Sponge network | 0.594 | 0.41522 | 1.752 | 0 | 0.258 |
| 52347 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-877-5p | 32 | CCND2 | Sponge network | -2.62 | 0 | -0.267 | 0.3112 | 0.258 |
| 52348 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-93-5p | 15 | EPAS1 | Sponge network | -0.162 | 0.47687 | -0.184 | 0.14418 | 0.258 |
| 52349 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 20 | MITF | Sponge network | 0.4 | 0.14611 | -0.769 | 0.00022 | 0.258 |
| 52350 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | EPHA7 | Sponge network | -0.235 | 0.15142 | -2.197 | 3.0E-5 | 0.258 |
| 52351 | MALAT1 |
hsa-miR-130b-3p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | RASA1 | Sponge network | 0.782 | 0.00016 | -0.002 | 0.98204 | 0.258 |
| 52352 | RP11-244O19.1 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-5p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-940 | 16 | ZFP36L1 | Sponge network | -0.096 | 0.67958 | -0.505 | 8.0E-5 | 0.258 |
| 52353 | RP11-693J15.4 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 12 | WWTR1 | Sponge network | -0.72 | 0.24239 | -0.345 | 0.11997 | 0.258 |
| 52354 | AC012360.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-92a-3p | 25 | LPP | Sponge network | 0.624 | 0.02176 | -0.459 | 0.04475 | 0.258 |
| 52355 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 21 | PCDH17 | Sponge network | -0.769 | 0.00035 | 0.731 | 2.0E-5 | 0.258 |
| 52356 | RP11-458D21.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-429;hsa-miR-495-3p;hsa-miR-7-1-3p | 16 | AKAP6 | Sponge network | 0.663 | 0.00073 | -1.586 | 3.0E-5 | 0.258 |
| 52357 | RP11-798M19.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-940 | 24 | EPS15 | Sponge network | -0.612 | 0.00094 | -0.052 | 0.53998 | 0.258 |
| 52358 | C20orf166-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-93-5p | 32 | GNAQ | Sponge network | -2.69 | 0.0001 | -0.367 | 0.00102 | 0.258 |
| 52359 | AC127904.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | HECA | Sponge network | 1.308 | 0 | -0.217 | 0.03463 | 0.258 |
| 52360 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-501-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 16 | NOTCH2NL | Sponge network | 0.335 | 0.20596 | 0.024 | 0.84307 | 0.258 |
| 52361 | RP11-890B15.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | SEMA3E | Sponge network | 0.101 | 0.60919 | -1.717 | 0.00137 | 0.258 |
| 52362 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 22 | TP53INP1 | Sponge network | -0.739 | 0.0574 | -0.496 | 0.00064 | 0.258 |
| 52363 | PRKAG2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | KIT | Sponge network | -0.622 | 0.02807 | -2.184 | 0 | 0.258 |
| 52364 | AC159540.2 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-214-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-628-5p | 11 | MAT2A | Sponge network | 1.634 | 0 | -0.073 | 0.36195 | 0.258 |
| 52365 | CTC-297N7.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p | 11 | CACNA2D1 | Sponge network | -2.069 | 0 | -0.846 | 0.00675 | 0.258 |
| 52366 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-940 | 19 | NEDD4 | Sponge network | 0.637 | 0.00069 | 0.02 | 0.89834 | 0.258 |
| 52367 | RP11-2H8.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-744-3p | 21 | EPS15 | Sponge network | 1.089 | 0 | -0.052 | 0.53998 | 0.258 |
| 52368 | LINC00863 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-671-5p | 18 | DLC1 | Sponge network | 0.189 | 0.13981 | -0.382 | 0.05038 | 0.258 |
| 52369 | DOCK9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-484 | 12 | TGFBR3 | Sponge network | -0.231 | 0.28214 | -1.173 | 1.0E-5 | 0.258 |
| 52370 | CTD-3064M3.3 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-505-5p | 11 | WNK2 | Sponge network | -0.469 | 0.08721 | -0.352 | 0.31066 | 0.258 |
| 52371 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-532-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-93-5p | 46 | NTRK3 | Sponge network | 0.051 | 0.83388 | -1.77 | 3.0E-5 | 0.258 |
| 52372 | C20orf166-AS1 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-577;hsa-miR-629-5p | 18 | PCDH17 | Sponge network | -2.69 | 0.0001 | 0.731 | 2.0E-5 | 0.258 |
| 52373 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | FYN | Sponge network | -1.216 | 0 | -0.131 | 0.37051 | 0.258 |
| 52374 | RP11-158K1.3 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 47 | DTNA | Sponge network | 0.349 | 0.01695 | -1.648 | 1.0E-5 | 0.258 |
| 52375 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-452-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-92a-3p;hsa-miR-940 | 37 | MEF2C | Sponge network | 0.056 | 0.81195 | -0.499 | 0.01448 | 0.258 |
| 52376 | RP11-212P7.2 |
hsa-miR-146b-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | HERC1 | Sponge network | 0.219 | 0.1516 | -0.196 | 0.08544 | 0.258 |
| 52377 | RP11-307B6.3 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-589-3p | 19 | CCND2 | Sponge network | -4.463 | 0.00025 | -0.267 | 0.3112 | 0.258 |
| 52378 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 18 | KIT | Sponge network | -0.355 | 0.0077 | -2.184 | 0 | 0.258 |
| 52379 | LINC01011 |
hsa-miR-125a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-497-5p | 15 | TBL1XR1 | Sponge network | 0.426 | 0.00084 | 0.578 | 0 | 0.258 |
| 52380 | HCG11 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 16 | RIMBP2 | Sponge network | 0.385 | 0.10067 | -1.968 | 5.0E-5 | 0.258 |
| 52381 | DNAJC27-AS1 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-484 | 20 | ABL1 | Sponge network | -0.969 | 2.0E-5 | -0.215 | 0.07804 | 0.258 |
| 52382 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | EPHA5 | Sponge network | 0.862 | 0.06213 | -0.957 | 0.01733 | 0.258 |
| 52383 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-10a-3p;hsa-miR-1296-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-324-3p;hsa-miR-577 | 10 | PRKAR1A | Sponge network | -0.465 | 0.04703 | -0.063 | 0.50314 | 0.258 |
| 52384 | LINC00662 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 25 | ZFHX4 | Sponge network | 0.213 | 0.32297 | 0.018 | 0.95574 | 0.258 |
| 52385 | RP11-264I13.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-26b-3p;hsa-miR-339-5p;hsa-miR-576-5p | 10 | CBX5 | Sponge network | 0.421 | 0.01652 | 0.165 | 0.24446 | 0.258 |
| 52386 | RP11-130L8.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 19 | PPP1R9A | Sponge network | -0.663 | 0.10887 | -0.903 | 0.04254 | 0.258 |
| 52387 | TPTEP1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-3613-5p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 14 | GATA2 | Sponge network | -1.216 | 0 | -0.654 | 0.0016 | 0.258 |
| 52388 | FGF14-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | PIK3CA | Sponge network | -1.582 | 8.0E-5 | 0.195 | 0.06908 | 0.258 |
| 52389 | LINC01006 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p | 13 | SETBP1 | Sponge network | -0.702 | 0.04814 | -1.302 | 1.0E-5 | 0.258 |
| 52390 | RP5-1198O20.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p | 18 | KANK1 | Sponge network | 0.997 | 0.00066 | -0.55 | 0.00134 | 0.258 |
| 52391 | RP11-359E10.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 36 | AFF4 | Sponge network | 0.299 | 0.57722 | -0.266 | 0.01565 | 0.258 |
| 52392 | DNM1P35 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-374a-5p;hsa-miR-584-5p;hsa-miR-93-3p | 14 | ST6GAL2 | Sponge network | 0.051 | 0.83388 | -1.201 | 0.00597 | 0.258 |
| 52393 | TINCR |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-93-3p | 16 | SUFU | Sponge network | -0.664 | 0.14543 | -0.04 | 0.65489 | 0.258 |
| 52394 | LINC00472 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p | 16 | KANK1 | Sponge network | -0.842 | 0.01361 | -0.55 | 0.00134 | 0.258 |
| 52395 | CTD-2303H24.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-744-3p | 48 | DST | Sponge network | 0.415 | 0.14657 | -0.713 | 0.00096 | 0.258 |
| 52396 | AC003090.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | CDHR3 | Sponge network | -2.117 | 0.00161 | -0.977 | 4.0E-5 | 0.258 |
| 52397 | RP11-2B6.2 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-7-1-3p | 10 | UBE4B | Sponge network | 0.539 | 0.03722 | -0.235 | 0.007 | 0.258 |
| 52398 | RP11-119F7.5 |
hsa-miR-1271-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | KIF5B | Sponge network | 0.225 | 0.30644 | 0.362 | 0.00066 | 0.258 |
| 52399 | RP1-151F17.2 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TRIM13 | Sponge network | 0.222 | 0.29133 | 0.183 | 0.06968 | 0.258 |
| 52400 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-450a-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 13 | CHD2 | Sponge network | -0.176 | 0.59692 | 0.049 | 0.55371 | 0.258 |
| 52401 | RP11-359B12.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | LUZP2 | Sponge network | 0.064 | 0.72934 | -0.056 | 0.89416 | 0.258 |
| 52402 | USP3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-455-5p;hsa-miR-7-5p;hsa-miR-877-5p | 32 | EGFR | Sponge network | -0.162 | 0.47687 | -0.175 | 0.48451 | 0.258 |
| 52403 | RP11-98I9.4 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TET1 | Sponge network | 0.67 | 0.00048 | -0.135 | 0.52138 | 0.258 |
| 52404 | RP11-182J1.1 |
hsa-miR-107;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 15 | FOXP1 | Sponge network | -0.187 | 0.23787 | 0.042 | 0.74368 | 0.258 |
| 52405 | RP11-83A24.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 19 | HECA | Sponge network | 0.417 | 0.00445 | -0.217 | 0.03463 | 0.258 |
| 52406 | RP11-6O2.3 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-505-5p;hsa-miR-96-5p | 11 | WNK2 | Sponge network | 0.331 | 0.57247 | -0.352 | 0.31066 | 0.258 |
| 52407 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-376c-3p;hsa-miR-590-3p | 12 | CREBBP | Sponge network | 1.063 | 0 | 0.126 | 0.15186 | 0.258 |
| 52408 | RP11-326C3.7 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p | 12 | RUNX1T1 | Sponge network | 0.295 | 0.28897 | -0.344 | 0.2374 | 0.258 |
| 52409 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-miR-101-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-664a-3p | 22 | SOX11 | Sponge network | 1.147 | 0 | 2.68 | 0 | 0.258 |
| 52410 | ATP1B3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | SPRY3 | Sponge network | 1.2 | 0.01813 | 0.016 | 0.91286 | 0.258 |
| 52411 | HNRNPU-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 13 | CBFB | Sponge network | 1.601 | 0 | 1.061 | 0 | 0.258 |
| 52412 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | FGFR1 | Sponge network | 0.603 | 0.00127 | -0.509 | 0.03957 | 0.258 |
| 52413 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 23 | NIN | Sponge network | -0.246 | 0.35034 | -0.253 | 0.13794 | 0.258 |
| 52414 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | GNA13 | Sponge network | -1.575 | 6.0E-5 | 0.007 | 0.93884 | 0.258 |
| 52415 | RP11-119F7.5 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 30 | NOTCH2 | Sponge network | 0.225 | 0.30644 | 0.138 | 0.35163 | 0.258 |
| 52416 | UBA6-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-362-5p;hsa-miR-374b-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 23 | ABI2 | Sponge network | 0.312 | 0.00725 | 0.175 | 0.14787 | 0.258 |
| 52417 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-369-3p | 10 | PPM1A | Sponge network | 0.062 | 0.55274 | -0.623 | 0 | 0.258 |
| 52418 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-339-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | PRKCE | Sponge network | -1.057 | 0.00029 | -0.403 | 0.00056 | 0.258 |
| 52419 | RP11-848P1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 11 | CACNA2D1 | Sponge network | 1.253 | 0 | -0.846 | 0.00675 | 0.258 |
| 52420 | CTD-3092A11.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429;hsa-miR-940 | 12 | CRTC3 | Sponge network | 0.873 | 0 | 0.164 | 0.16786 | 0.258 |
| 52421 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | YAP1 | Sponge network | 0.906 | 0.31715 | 0.22 | 0.11836 | 0.258 |
| 52422 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-18a-5p;hsa-miR-324-5p;hsa-miR-339-5p | 11 | PRDM2 | Sponge network | -0.301 | 0.06597 | -0.258 | 0.00721 | 0.258 |
| 52423 | GAS5 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p | 12 | CCDC6 | Sponge network | 0.728 | 0 | 0.27 | 0.02555 | 0.258 |
| 52424 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-7-1-3p | 19 | KAT6B | Sponge network | 0.614 | 1.0E-5 | -0.478 | 0.00018 | 0.258 |
| 52425 | TINCR |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-3p | 11 | BCL9 | Sponge network | -0.664 | 0.14543 | 0.321 | 0.00267 | 0.258 |
| 52426 | RP11-531A24.5 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 10 | ZDBF2 | Sponge network | 0.75 | 0.00054 | -0.998 | 0.00025 | 0.258 |
| 52427 | RP11-13J8.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-33a-5p | 11 | SOX11 | Sponge network | 3.03 | 0 | 2.68 | 0 | 0.258 |
| 52428 | SOX2-OT |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | MED12L | Sponge network | -1.264 | 6.0E-5 | -0.194 | 0.55299 | 0.258 |
| 52429 | CTA-204B4.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 11 | SETBP1 | Sponge network | 0.765 | 7.0E-5 | -1.302 | 1.0E-5 | 0.258 |
| 52430 | AC002454.1 |
hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-362-5p | 15 | IL17RD | Sponge network | 1.952 | 0.02558 | -0.019 | 0.94873 | 0.258 |
| 52431 | CTD-2314G24.2 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-96-5p;hsa-miR-98-5p | 42 | CREB3L2 | Sponge network | 0.246 | 0.59912 | -0.525 | 2.0E-5 | 0.258 |
| 52432 | RP11-283I3.6 |
hsa-let-7d-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-330-3p | 12 | GLIPR1 | Sponge network | 0.614 | 1.0E-5 | 0.146 | 0.3276 | 0.258 |
| 52433 | LINC00865 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-7-1-3p | 13 | BCL9 | Sponge network | -1.249 | 7.0E-5 | 0.321 | 0.00267 | 0.258 |
| 52434 | MIR600HG |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-655-3p | 14 | ZMYND11 | Sponge network | 0.594 | 0.41522 | -0.2 | 0.05449 | 0.258 |
| 52435 | CKMT2-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-409-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p | 13 | SEMA3E | Sponge network | 0.082 | 0.55654 | -1.717 | 0.00137 | 0.258 |
| 52436 | AGAP11 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-584-5p;hsa-miR-942-5p | 17 | ERRFI1 | Sponge network | -1.645 | 0 | -0.798 | 1.0E-5 | 0.258 |
| 52437 | C20orf166-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p | 16 | ERCC4 | Sponge network | -2.69 | 0.0001 | 0.126 | 0.15266 | 0.258 |
| 52438 | RP11-130L8.1 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 18 | LRIG1 | Sponge network | -0.663 | 0.10887 | -0.537 | 0.00588 | 0.258 |
| 52439 | RP11-672A2.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-98-5p | 12 | LRIG1 | Sponge network | 1.344 | 0 | -0.537 | 0.00588 | 0.258 |
| 52440 | EIF3J-AS1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-654-3p | 16 | MYBL1 | Sponge network | 0.331 | 0.00509 | 0.494 | 0.02152 | 0.258 |
| 52441 | RP11-244H3.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | BCL9 | Sponge network | 0.659 | 3.0E-5 | 0.321 | 0.00267 | 0.258 |
| 52442 | OIP5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | SYNE1 | Sponge network | 0.421 | 0.00084 | -0.738 | 0.00316 | 0.258 |
| 52443 | LINC00578 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-210-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-577;hsa-miR-589-3p | 14 | APC | Sponge network | 0.811 | 0.03228 | -0.255 | 0.04051 | 0.258 |
| 52444 | RASSF8-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 35 | AFF1 | Sponge network | 0.335 | 0.20596 | -0.384 | 0.00053 | 0.258 |
| 52445 | LINC00926 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-877-5p;hsa-miR-96-5p | 10 | NAV3 | Sponge network | 0.756 | 0.00739 | 0.212 | 0.47729 | 0.258 |
| 52446 | RP4-794H19.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-501-5p | 11 | RET | Sponge network | -0.825 | 1.0E-5 | -0.833 | 0.03517 | 0.258 |
| 52447 | KTN1-AS1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-374b-5p;hsa-miR-664a-3p | 11 | UBE2D1 | Sponge network | 0.927 | 0 | 0.468 | 0 | 0.258 |
| 52448 | RP11-195B17.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-96-5p | 19 | QKI | Sponge network | 0.37 | 0.27614 | -0.333 | 0.04111 | 0.258 |
| 52449 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-93-5p | 12 | FBXO31 | Sponge network | 0.259 | 0.12542 | -0.085 | 0.42389 | 0.258 |
| 52450 | DNM3OS |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-508-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | INPP4B | Sponge network | 0.572 | 0.01722 | -0.097 | 0.60503 | 0.258 |
| 52451 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | COPS2 | Sponge network | 0.673 | 0 | -0.178 | 0.04974 | 0.258 |
| 52452 | RP11-182J1.1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 11 | EPS15 | Sponge network | -0.187 | 0.23787 | -0.052 | 0.53998 | 0.258 |
| 52453 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-590-3p | 16 | FREM2 | Sponge network | 0.392 | 0.0278 | -0.437 | 0.46353 | 0.258 |
| 52454 | RP11-212P7.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 52 | CADM2 | Sponge network | 0.219 | 0.1516 | -3.782 | 0 | 0.258 |
| 52455 | AC005618.6 |
hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p | 25 | PIK3R1 | Sponge network | 0.862 | 0.06213 | -0.133 | 0.36854 | 0.258 |
| 52456 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29b-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-758-3p;hsa-miR-940 | 24 | HLF | Sponge network | 0.274 | 0.07681 | -2.443 | 0 | 0.258 |
| 52457 | MIR600HG |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-339-5p;hsa-miR-342-3p | 11 | STARD13 | Sponge network | 0.594 | 0.41522 | -0.187 | 0.27383 | 0.258 |
| 52458 | CD27-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-577 | 11 | MAP3K12 | Sponge network | 0.177 | 0.16681 | -0.057 | 0.59369 | 0.258 |
| 52459 | RP11-98I9.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-495-3p;hsa-miR-590-3p | 26 | CCDC6 | Sponge network | 0.67 | 0.00048 | 0.27 | 0.02555 | 0.258 |
| 52460 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p | 22 | SPRY3 | Sponge network | 0.727 | 3.0E-5 | 0.016 | 0.91286 | 0.258 |
| 52461 | RP11-262H14.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940;hsa-miR-96-5p | 18 | MYO9A | Sponge network | -0.121 | 0.53195 | -0.147 | 0.32373 | 0.258 |
| 52462 | RP11-161M6.2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-378a-3p | 14 | RBMS3 | Sponge network | 0.61 | 0.06611 | -0.519 | 0.03551 | 0.258 |
| 52463 | CTD-2528L19.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | KDSR | Sponge network | 0.953 | 0 | 0.239 | 0.02216 | 0.258 |
| 52464 | RP11-6O2.3 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-2355-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-590-5p;hsa-miR-940;hsa-miR-96-5p | 10 | FOXO4 | Sponge network | 0.331 | 0.57247 | -0.516 | 2.0E-5 | 0.258 |
| 52465 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-539-5p | 20 | TRPS1 | Sponge network | 0.37 | 0.27614 | -0.191 | 0.44109 | 0.258 |
| 52466 | HOTAIRM1 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-942-5p | 17 | IGFBP5 | Sponge network | -0.053 | 0.80685 | -0.806 | 0.00105 | 0.258 |
| 52467 | CTD-2303H24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 27 | DMD | Sponge network | 0.415 | 0.14657 | -1.506 | 0 | 0.258 |
| 52468 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-629-3p | 40 | CBX5 | Sponge network | -0.227 | 0.31657 | 0.165 | 0.24446 | 0.258 |
| 52469 | RP11-531A24.5 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | TAL1 | Sponge network | 0.75 | 0.00054 | -0.462 | 0.00574 | 0.258 |
| 52470 | LINC00476 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 23 | ARNT2 | Sponge network | -0.615 | 0.00015 | -0.332 | 0.25851 | 0.258 |
| 52471 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | FOXO3 | Sponge network | 0.67 | 0.00048 | -0.04 | 0.72028 | 0.258 |
| 52472 | ZSCAN16-AS1 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | ACTB | Sponge network | -0.342 | 0.02288 | -0.247 | 0.01736 | 0.258 |
| 52473 | RAMP2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-582-3p;hsa-miR-7-1-3p | 12 | PLAG1 | Sponge network | -0.513 | 0.04703 | -0.315 | 0.26532 | 0.258 |
| 52474 | FZD10-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | GALNT13 | Sponge network | -0.727 | 0.04726 | -0.857 | 0.07439 | 0.258 |
| 52475 | RP11-400K9.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 11 | AKAP13 | Sponge network | -0.611 | 0.09607 | 0.028 | 0.7729 | 0.258 |
| 52476 | DNM1P35 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-93-5p | 11 | TXNIP | Sponge network | 0.051 | 0.83388 | -1.277 | 0 | 0.258 |
| 52477 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-429 | 10 | PCDH8 | Sponge network | 0.859 | 0.00331 | -0.997 | 0.05366 | 0.258 |
| 52478 | EMX2OS |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 25 | JDP2 | Sponge network | -0.777 | 0.1134 | -0.967 | 0 | 0.258 |
| 52479 | CTD-2314G24.2 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p | 16 | PKNOX1 | Sponge network | 0.246 | 0.59912 | 0.085 | 0.28917 | 0.258 |
| 52480 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p | 17 | ADARB1 | Sponge network | 0.673 | 0 | -0.335 | 0.06631 | 0.258 |
| 52481 | RP11-835E18.5 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-154-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-576-5p | 18 | PBX1 | Sponge network | -0.462 | 0.01226 | -1.206 | 0 | 0.258 |
| 52482 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-628-5p | 15 | BCL6 | Sponge network | 0.537 | 0.00139 | -0.211 | 0.19489 | 0.258 |
| 52483 | RP11-67L2.2 |
hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CDH11 | Sponge network | 0.72 | 0.0001 | 1.427 | 0 | 0.258 |
| 52484 | RP11-298J20.4 |
hsa-miR-130b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-5p | 11 | DST | Sponge network | 0.002 | 0.99057 | -0.713 | 0.00096 | 0.258 |
| 52485 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-766-3p | 41 | IL6ST | Sponge network | 0.906 | 0.31715 | -0.402 | 0.00676 | 0.258 |
| 52486 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ZNF536 | Sponge network | 0.101 | 0.60919 | -3.115 | 0 | 0.258 |
| 52487 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 35 | AR | Sponge network | -0.322 | 0.25945 | -1.554 | 0 | 0.258 |
| 52488 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 28 | ST6GALNAC3 | Sponge network | 0.385 | 0.10067 | -0.987 | 0 | 0.258 |
| 52489 | PSMG3-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-429 | 11 | THSD7A | Sponge network | -0.125 | 0.49414 | 0.242 | 0.25154 | 0.258 |
| 52490 | RP11-277P12.20 |
hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-532-3p | 10 | MRVI1 | Sponge network | 0.687 | 0.02753 | -1.051 | 0.00022 | 0.258 |
| 52491 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-96-5p | 10 | SCN9A | Sponge network | 0.348 | 0.32912 | -0.525 | 0.10593 | 0.258 |
| 52492 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-590-3p | 10 | DACT1 | Sponge network | 0.919 | 0 | 0.783 | 0.00072 | 0.258 |
| 52493 | TINCR |
hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-589-3p | 11 | HOMER2 | Sponge network | -0.664 | 0.14543 | -2.698 | 0 | 0.258 |
| 52494 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-664a-5p | 17 | SOX11 | Sponge network | 1.361 | 0 | 2.68 | 0 | 0.258 |
| 52495 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-484;hsa-miR-98-5p | 19 | PNRC1 | Sponge network | -0.969 | 2.0E-5 | -0.646 | 0 | 0.258 |
| 52496 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | CLIP1 | Sponge network | 0.619 | 0.00157 | -0.215 | 0.08684 | 0.258 |
| 52497 | RP11-195F19.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 16 | PRRX1 | Sponge network | -0.726 | 0.00132 | 1.316 | 0 | 0.258 |
| 52498 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | RCAN2 | Sponge network | -1.057 | 0.00029 | -0.33 | 0.15172 | 0.258 |
| 52499 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-96-5p | 15 | MITF | Sponge network | 0.622 | 0.00015 | -0.769 | 0.00022 | 0.258 |
| 52500 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-423-5p;hsa-miR-450b-5p | 15 | PNRC1 | Sponge network | -0.842 | 0.01361 | -0.646 | 0 | 0.258 |
| 52501 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-940 | 16 | TRIM33 | Sponge network | 0.392 | 0.0278 | 0.402 | 7.0E-5 | 0.258 |
| 52502 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-625-3p | 22 | TMTC3 | Sponge network | 0.225 | 0.30644 | 0.123 | 0.22189 | 0.258 |
| 52503 | DNM1P35 |
hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-625-5p | 10 | NGFR | Sponge network | 0.051 | 0.83388 | -1.271 | 0.01058 | 0.258 |
| 52504 | H1FX-AS1 |
hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p | 11 | SLITRK6 | Sponge network | -0.206 | 0.12942 | -2.121 | 0 | 0.258 |
| 52505 | RP11-620J15.3 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-590-3p | 17 | DOCK4 | Sponge network | -0.739 | 0.00044 | 0.502 | 0.00108 | 0.258 |
| 52506 | C1RL-AS1 |
hsa-miR-129-5p;hsa-miR-149-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-3p | 10 | CCDC6 | Sponge network | 1.26 | 0 | 0.27 | 0.02555 | 0.258 |
| 52507 | HOXB-AS1 |
hsa-let-7f-1-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-582-3p | 11 | PIK3CA | Sponge network | -0.154 | 0.53954 | 0.195 | 0.06908 | 0.258 |
| 52508 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-455-5p;hsa-miR-93-5p | 12 | SLITRK3 | Sponge network | 0.056 | 0.8494 | -3.328 | 0 | 0.258 |
| 52509 | LINC00654 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 12 | ID4 | Sponge network | 0.374 | 0.20054 | -1.644 | 0 | 0.258 |
| 52510 | ZNF32-AS2 |
hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-339-5p;hsa-miR-342-3p | 10 | DDX3X | Sponge network | 1.552 | 7.0E-5 | -0.152 | 0.07686 | 0.258 |
| 52511 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p | 10 | GPAM | Sponge network | 0.792 | 0.0049 | 0.142 | 0.29732 | 0.258 |
| 52512 | LINC00092 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-744-5p | 11 | PRKACA | Sponge network | -1.886 | 0 | -0.255 | 0.0012 | 0.258 |
| 52513 | LINC00092 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p | 18 | SUFU | Sponge network | -1.886 | 0 | -0.04 | 0.65489 | 0.258 |
| 52514 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | YAP1 | Sponge network | -0.976 | 0.00025 | 0.22 | 0.11836 | 0.258 |
| 52515 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-493-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | NEDD4 | Sponge network | -1.216 | 0 | 0.02 | 0.89834 | 0.258 |
| 52516 | LINC00648 |
hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 11 | DNMT3A | Sponge network | 0.633 | 0.51678 | 0.302 | 0.0262 | 0.258 |
| 52517 | RP11-835E18.5 |
hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-590-3p | 10 | FGFR1 | Sponge network | -0.462 | 0.01226 | -0.509 | 0.03957 | 0.258 |
| 52518 | GHRLOS |
hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-200a-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-493-5p | 12 | LIMCH1 | Sponge network | -1.942 | 0 | -0.915 | 1.0E-5 | 0.258 |
| 52519 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p | 13 | ABCA10 | Sponge network | 1.757 | 0 | -0.571 | 0.03674 | 0.258 |
| 52520 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-874-3p | 19 | NR2C2 | Sponge network | 0.95 | 0.15943 | 0.21 | 0.03224 | 0.258 |
| 52521 | AC021224.1 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-149-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-9-5p | 10 | CCDC6 | Sponge network | 0.605 | 0 | 0.27 | 0.02555 | 0.258 |
| 52522 | LINC01001 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 15 | CBX5 | Sponge network | 1.055 | 0 | 0.165 | 0.24446 | 0.258 |
| 52523 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b | 15 | NR2C2 | Sponge network | 0.395 | 0.15451 | 0.21 | 0.03224 | 0.258 |
| 52524 | AC009948.5 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | PPM1L | Sponge network | 0.532 | 0.00505 | -0.537 | 0.00616 | 0.258 |
| 52525 | CTD-2245F17.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-335-5p;hsa-miR-505-3p;hsa-miR-7-5p | 10 | KCNJ5 | Sponge network | -0.421 | 0.12556 | -0.281 | 0.3339 | 0.258 |
| 52526 | LINC00863 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | NAV3 | Sponge network | 0.189 | 0.13981 | 0.212 | 0.47729 | 0.258 |
| 52527 | CTD-2245F17.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-5p;hsa-miR-940 | 14 | SNX29 | Sponge network | -0.421 | 0.12556 | -0.34 | 0.01371 | 0.258 |
| 52528 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3614-5p;hsa-miR-582-3p | 15 | JAZF1 | Sponge network | 0.921 | 0.00118 | -0.377 | 0.02677 | 0.258 |
| 52529 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | DACT1 | Sponge network | 0.064 | 0.72934 | 0.783 | 0.00072 | 0.258 |
| 52530 | HOTAIRM1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | FAT4 | Sponge network | -0.053 | 0.80685 | -0.354 | 0.14694 | 0.258 |
| 52531 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-424-5p | 10 | CSRNP1 | Sponge network | -4.463 | 0.00025 | -1.448 | 0 | 0.258 |
| 52532 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | LRRC7 | Sponge network | -0.811 | 2.0E-5 | -1.341 | 0.01193 | 0.258 |
| 52533 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-3p | 16 | NRK | Sponge network | -0.031 | 0.89063 | 0.656 | 0.19217 | 0.258 |
| 52534 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | CNTN1 | Sponge network | 0.349 | 0.01695 | -2.594 | 0 | 0.258 |
| 52535 | EIF3J-AS1 |
hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | NEDD4 | Sponge network | 0.331 | 0.00509 | 0.02 | 0.89834 | 0.258 |
| 52536 | AC103563.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-323a-3p | 10 | EPHA5 | Sponge network | -7.551 | 0 | -0.957 | 0.01733 | 0.258 |
| 52537 | FAM66C |
hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-625-5p;hsa-miR-7-5p | 11 | LZTS1 | Sponge network | -0.901 | 0.00019 | 1.664 | 0 | 0.258 |
| 52538 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 39 | PPP1R9A | Sponge network | -0.942 | 0 | -0.903 | 0.04254 | 0.258 |
| 52539 | HCG11 |
hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | SFRP1 | Sponge network | 0.385 | 0.10067 | -3.566 | 0 | 0.258 |
| 52540 | CD27-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-589-3p | 10 | ABCA10 | Sponge network | 0.177 | 0.16681 | -0.571 | 0.03674 | 0.258 |
| 52541 | AC004893.11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 16 | SOX11 | Sponge network | 1.317 | 0 | 2.68 | 0 | 0.258 |
| 52542 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-335-3p;hsa-miR-450b-5p | 12 | HOOK3 | Sponge network | 0.253 | 0.09565 | 0.273 | 0.09957 | 0.258 |
| 52543 | LINC00641 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-98-5p | 40 | CREB3L2 | Sponge network | -0.222 | 0.32445 | -0.525 | 2.0E-5 | 0.258 |
| 52544 | RP3-523E19.2 |
hsa-miR-130a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-505-3p;hsa-miR-539-5p | 11 | DDX6 | Sponge network | 2.887 | 0 | 0.091 | 0.36428 | 0.258 |
| 52545 | LINC00963 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 40 | CBX5 | Sponge network | 0.523 | 9.0E-5 | 0.165 | 0.24446 | 0.258 |
| 52546 | LINC00957 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-942-5p | 14 | ERRFI1 | Sponge network | -0.421 | 0.0114 | -0.798 | 1.0E-5 | 0.258 |
| 52547 | FENDRR |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-935 | 27 | RNF144A | Sponge network | -1.281 | 0.00012 | 0.594 | 0.0009 | 0.258 |
| 52548 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | MGA | Sponge network | -0.147 | 0.40452 | 0.323 | 0.00792 | 0.258 |
| 52549 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | KDSR | Sponge network | 0.683 | 1.0E-5 | 0.239 | 0.02216 | 0.258 |
| 52550 | AP001055.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 16 | TGFBR3 | Sponge network | 0.693 | 0.00076 | -1.173 | 1.0E-5 | 0.258 |
| 52551 | RP11-800A3.7 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-200c-5p;hsa-miR-338-3p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 11 | DIP2C | Sponge network | 0.18 | 0.7147 | -0.436 | 0.01713 | 0.258 |
| 52552 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | GNA13 | Sponge network | -0.031 | 0.89063 | 0.007 | 0.93884 | 0.258 |
| 52553 | RP11-597D13.9 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 12 | ABCB5 | Sponge network | 1.302 | 7.0E-5 | -0.956 | 0.04077 | 0.258 |
| 52554 | LINC00863 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 17 | CRTC1 | Sponge network | 0.189 | 0.13981 | -0.116 | 0.28412 | 0.258 |
| 52555 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 18 | EGLN1 | Sponge network | 0.826 | 1.0E-5 | -0.127 | 0.1536 | 0.258 |
| 52556 | RP11-152F13.8 |
hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p | 11 | ZFX | Sponge network | 0.843 | 0.01775 | 0.09 | 0.42563 | 0.258 |
| 52557 | AC005562.1 |
hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | INPP4B | Sponge network | 0.488 | 0.00084 | -0.097 | 0.60503 | 0.258 |
| 52558 | PCA3 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 48 | PPM1L | Sponge network | -0.709 | 0.18168 | -0.537 | 0.00616 | 0.258 |
| 52559 | AC003090.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 23 | PTPN14 | Sponge network | -2.117 | 0.00161 | 0.144 | 0.34595 | 0.258 |
| 52560 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 52 | CBX5 | Sponge network | 0.249 | 0.35902 | 0.165 | 0.24446 | 0.258 |
| 52561 | CTD-3099C6.9 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p | 16 | ZFX | Sponge network | 1.361 | 0 | 0.09 | 0.42563 | 0.258 |
| 52562 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 38 | SOX5 | Sponge network | 0.219 | 0.1516 | -1.091 | 4.0E-5 | 0.258 |
| 52563 | RP11-98I9.4 |
hsa-let-7b-3p;hsa-miR-125a-5p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30a-5p;hsa-miR-33a-5p;hsa-miR-590-3p | 12 | CHD1 | Sponge network | 0.67 | 0.00048 | 0.322 | 0.00215 | 0.258 |
| 52564 | TIPARP-AS1 |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-532-5p;hsa-miR-93-5p | 14 | PPM1L | Sponge network | -0.887 | 0 | -0.537 | 0.00616 | 0.258 |
| 52565 | AC103563.9 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-589-5p | 13 | ERBB4 | Sponge network | -6.057 | 0 | -1.975 | 2.0E-5 | 0.258 |
| 52566 | LINC00621 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 11 | LRRC7 | Sponge network | 0.131 | 0.8436 | -1.341 | 0.01193 | 0.258 |
| 52567 | RP11-157J24.2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-576-5p | 13 | BNC2 | Sponge network | -1.664 | 0.05165 | -0.491 | 0.09988 | 0.258 |
| 52568 | RP11-499P20.2 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-337-3p;hsa-miR-382-5p;hsa-miR-493-5p;hsa-miR-744-3p | 11 | CUL5 | Sponge network | 0.187 | 0.44121 | 0.026 | 0.77301 | 0.258 |
| 52569 | RP11-967K21.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-577 | 13 | FMNL3 | Sponge network | 0.056 | 0.81195 | 0.555 | 4.0E-5 | 0.258 |
| 52570 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-429 | 13 | MAP3K12 | Sponge network | 0.16 | 0.50785 | -0.057 | 0.59369 | 0.258 |
| 52571 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | FYN | Sponge network | 0.064 | 0.72934 | -0.131 | 0.37051 | 0.258 |
| 52572 | RP11-212P7.2 |
hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | TAF1 | Sponge network | 0.219 | 0.1516 | 0.39 | 3.0E-5 | 0.258 |
| 52573 | PSMG3-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-17-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 16 | WWTR1 | Sponge network | -0.125 | 0.49414 | -0.345 | 0.11997 | 0.258 |
| 52574 | RP11-404E16.1 |
hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-942-5p | 16 | THRB | Sponge network | 0.097 | 0.91311 | -1.117 | 0.0004 | 0.258 |
| 52575 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-93-5p | 12 | TGFBR2 | Sponge network | -0.785 | 0.00035 | -0.243 | 0.11408 | 0.258 |
| 52576 | FAM66C |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | ITGB1 | Sponge network | -0.901 | 0.00019 | 0.524 | 0.00017 | 0.258 |
| 52577 | RP4-605O3.4 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-370-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-93-3p | 11 | PCDH10 | Sponge network | 0.262 | 0.0475 | -1.644 | 0.0078 | 0.258 |
| 52578 | BOLA3-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-589-3p | 13 | ABCA10 | Sponge network | -0.246 | 0.35034 | -0.571 | 0.03674 | 0.258 |
| 52579 | CTC-490E21.10 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p | 10 | RPS6KA3 | Sponge network | 1.46 | 1.0E-5 | 0.256 | 0.02419 | 0.258 |
| 52580 | BDNF-AS |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-324-5p;hsa-miR-331-5p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-361-5p;hsa-miR-3613-5p;hsa-miR-378a-5p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 53 | NTRK3 | Sponge network | -0.668 | 3.0E-5 | -1.77 | 3.0E-5 | 0.258 |
| 52581 | CTC-559E9.6 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-32-5p;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p | 13 | FOXP1 | Sponge network | 0.601 | 1.0E-5 | 0.042 | 0.74368 | 0.258 |
| 52582 | RP11-195B17.1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-539-5p;hsa-miR-96-5p | 20 | AFF4 | Sponge network | 0.37 | 0.27614 | -0.266 | 0.01565 | 0.258 |
| 52583 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 24 | NCOA1 | Sponge network | 0.385 | 0.10067 | -0.432 | 0 | 0.258 |
| 52584 | LINC00672 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-382-5p;hsa-miR-590-3p | 17 | INPP4B | Sponge network | -0.227 | 0.31657 | -0.097 | 0.60503 | 0.257 |
| 52585 | TPT1-AS1 |
hsa-let-7a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-495-3p | 12 | TRIM33 | Sponge network | 0.903 | 0 | 0.402 | 7.0E-5 | 0.257 |
| 52586 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 22 | NOTCH2NL | Sponge network | -0.129 | 0.57177 | 0.024 | 0.84307 | 0.257 |
| 52587 | HOXD-AS2 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-500a-5p | 10 | ZNF208 | Sponge network | -0.208 | 0.65592 | -0.4 | 0.22381 | 0.257 |
| 52588 | RP11-479G22.8 |
hsa-miR-101-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | PKNOX1 | Sponge network | 1.308 | 0 | 0.085 | 0.28917 | 0.257 |
| 52589 | CTD-2184D3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-345-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-940 | 15 | SNX29 | Sponge network | 0.272 | 0.44692 | -0.34 | 0.01371 | 0.257 |
| 52590 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-23a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 13 | CNTN1 | Sponge network | 0.453 | 0.01839 | -2.594 | 0 | 0.257 |
| 52591 | LINC00865 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-628-5p | 21 | BCL6 | Sponge network | -1.249 | 7.0E-5 | -0.211 | 0.19489 | 0.257 |
| 52592 | GAS6-AS2 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-590-5p | 11 | PTCH1 | Sponge network | 0.16 | 0.50785 | -0.1 | 0.6179 | 0.257 |
| 52593 | BCYRN1 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 23 | SYNPO2 | Sponge network | 0.311 | 0.10344 | -2.342 | 0 | 0.257 |
| 52594 | ADAMTS9-AS2 |
hsa-miR-10b-3p;hsa-miR-16-2-3p;hsa-miR-192-5p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p | 10 | EXTL2 | Sponge network | -2.452 | 0 | 0.109 | 0.37375 | 0.257 |
| 52595 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-424-5p | 15 | CBX5 | Sponge network | 1.597 | 0 | 0.165 | 0.24446 | 0.257 |
| 52596 | RP11-2H8.2 |
hsa-let-7g-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-3607-3p | 10 | KDM3A | Sponge network | 1.089 | 0 | 0.132 | 0.22711 | 0.257 |
| 52597 | ZNF667-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-3p;hsa-miR-429 | 10 | USP25 | Sponge network | -0.976 | 0.00025 | -0.242 | 0.01397 | 0.257 |
| 52598 | RP11-196G18.23 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | MED12L | Sponge network | 0.724 | 0.00039 | -0.194 | 0.55299 | 0.257 |
| 52599 | RP11-545G3.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-629-3p | 12 | KIF1B | Sponge network | 2.622 | 0.00025 | -0.118 | 0.32258 | 0.257 |
| 52600 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | PRDM2 | Sponge network | 0.174 | 0.30034 | -0.258 | 0.00721 | 0.257 |
| 52601 | NDUFA6-AS1 |
hsa-miR-107;hsa-miR-193a-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p | 11 | SEPT6 | Sponge network | -0.355 | 0.0077 | -0.598 | 0.00181 | 0.257 |
| 52602 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-590-3p | 28 | HLF | Sponge network | -0.125 | 0.49414 | -2.443 | 0 | 0.257 |
| 52603 | TINCR |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-7-5p;hsa-miR-93-3p | 25 | ERC1 | Sponge network | -0.664 | 0.14543 | -0.108 | 0.38794 | 0.257 |
| 52604 | BDNF-AS |
hsa-let-7d-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 13 | TLE4 | Sponge network | -0.668 | 3.0E-5 | -1.157 | 0 | 0.257 |
| 52605 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 19 | ZFHX3 | Sponge network | 0.826 | 1.0E-5 | 0.389 | 0.01363 | 0.257 |
| 52606 | RP11-758N13.1 |
hsa-miR-103a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-484 | 10 | RET | Sponge network | 2.939 | 3.0E-5 | -0.833 | 0.03517 | 0.257 |
| 52607 | U91328.21 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-511-5p | 14 | CADM2 | Sponge network | -0.735 | 0.15983 | -3.782 | 0 | 0.257 |
| 52608 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | CACNA2D1 | Sponge network | 0.998 | 0 | -0.846 | 0.00675 | 0.257 |
| 52609 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | EDA2R | Sponge network | 0.219 | 0.1516 | -0.587 | 0.01598 | 0.257 |
| 52610 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 15 | LRP1B | Sponge network | 0.889 | 9.0E-5 | -2.163 | 0 | 0.257 |
| 52611 | OR2A1-AS1 |
hsa-let-7a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p | 10 | CHD2 | Sponge network | 0.542 | 0.00241 | 0.049 | 0.55371 | 0.257 |
| 52612 | SH3BP5-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.267 | 0.2937 | 1.083 | 0 | 0.257 |
| 52613 | LINC00173 |
hsa-let-7d-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-376c-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 18 | NCOA1 | Sponge network | 0.056 | 0.8494 | -0.432 | 0 | 0.257 |
| 52614 | LPP-AS2 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p | 15 | ZMYND11 | Sponge network | -0.18 | 0.20343 | -0.2 | 0.05449 | 0.257 |
| 52615 | SOX2-OT |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 31 | ABI2 | Sponge network | -1.264 | 6.0E-5 | 0.175 | 0.14787 | 0.257 |
| 52616 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-92a-1-5p | 24 | PRKAB2 | Sponge network | -0.777 | 0.16667 | -0.367 | 0.01371 | 0.257 |
| 52617 | EMX2OS |
hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-671-5p | 11 | PIK3IP1 | Sponge network | -0.777 | 0.1134 | -0.187 | 0.19945 | 0.257 |
| 52618 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-424-5p | 17 | CBFA2T3 | Sponge network | 0.082 | 0.55397 | -1.221 | 1.0E-5 | 0.257 |
| 52619 | LINC00621 |
hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | FOXO3 | Sponge network | 0.131 | 0.8436 | -0.04 | 0.72028 | 0.257 |
| 52620 | RP11-20I23.11 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | ZFHX4 | Sponge network | 0.875 | 0.00019 | 0.018 | 0.95574 | 0.257 |
| 52621 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-93-5p | 46 | PRKAA2 | Sponge network | -1.047 | 0 | -2.462 | 0 | 0.257 |
| 52622 | RP11-35G9.5 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-96-5p | 12 | LATS2 | Sponge network | 0.622 | 0.00015 | 0.159 | 0.2304 | 0.257 |
| 52623 | C20orf166-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p | 17 | KCNJ5 | Sponge network | -2.69 | 0.0001 | -0.281 | 0.3339 | 0.257 |
| 52624 | A2M-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 23 | PRKAB2 | Sponge network | -0.08 | 0.90092 | -0.367 | 0.01371 | 0.257 |
| 52625 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 17 | PTPN11 | Sponge network | -0.227 | 0.31657 | 0.431 | 2.0E-5 | 0.257 |
| 52626 | FAM13A-AS1 |
hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 10 | KCNAB1 | Sponge network | -0.83 | 0 | -0.755 | 2.0E-5 | 0.257 |
| 52627 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 19 | BCL11A | Sponge network | -0.303 | 0.21002 | -0.932 | 0.0063 | 0.257 |
| 52628 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-3614-5p;hsa-miR-484;hsa-miR-664a-3p | 15 | GREM1 | Sponge network | -0.513 | 0.04703 | 0.902 | 0.01691 | 0.257 |
| 52629 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935 | 21 | LRRC55 | Sponge network | -0.615 | 0.00015 | -0.026 | 0.93163 | 0.257 |
| 52630 | RP11-526I2.5 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-365a-3p;hsa-miR-495-3p;hsa-miR-539-5p | 12 | ARHGEF12 | Sponge network | 1.054 | 1.0E-5 | 0.09 | 0.35693 | 0.257 |
| 52631 | RP11-218M22.1 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-3607-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-660-5p | 14 | LIMCH1 | Sponge network | 0.197 | 0.25618 | -0.915 | 1.0E-5 | 0.257 |
| 52632 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-3p;hsa-miR-96-5p | 22 | EPHA3 | Sponge network | -0.235 | 0.15142 | 0.185 | 0.53588 | 0.257 |
| 52633 | U91328.19 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | FAT4 | Sponge network | -0.303 | 0.21002 | -0.354 | 0.14694 | 0.257 |
| 52634 | SERHL |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-214-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-539-5p | 23 | BTG2 | Sponge network | -0.602 | 0.00331 | -1.763 | 0 | 0.257 |
| 52635 | C4A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-744-3p;hsa-miR-96-5p | 33 | LPP | Sponge network | 3.345 | 0 | -0.459 | 0.04475 | 0.257 |
| 52636 | RP11-819C21.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 17 | LATS2 | Sponge network | 0.537 | 0.00139 | 0.159 | 0.2304 | 0.257 |
| 52637 | RP1-102E24.8 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p | 14 | MDM4 | Sponge network | 1.737 | 0 | 0.345 | 0.00762 | 0.257 |
| 52638 | PSMG3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-484;hsa-miR-590-3p | 18 | RET | Sponge network | -0.125 | 0.49414 | -0.833 | 0.03517 | 0.257 |
| 52639 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-590-5p | 11 | RNF103 | Sponge network | -0.942 | 0 | -0.341 | 1.0E-5 | 0.257 |
| 52640 | RP13-516M14.1 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-940 | 14 | MAPK10 | Sponge network | 0.389 | 0.04293 | -1.774 | 0 | 0.257 |
| 52641 | RP3-337H4.8 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-411-5p | 10 | VPS53 | Sponge network | 1.527 | 0 | 0.103 | 0.22667 | 0.257 |
| 52642 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p | 15 | ACVR1 | Sponge network | 0.176 | 0.37402 | 0.279 | 0.00234 | 0.257 |
| 52643 | RP11-181G12.2 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-671-5p | 14 | DLC1 | Sponge network | 0.556 | 0.00298 | -0.382 | 0.05038 | 0.257 |
| 52644 | AGAP11 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p | 21 | JDP2 | Sponge network | -1.645 | 0 | -0.967 | 0 | 0.257 |
| 52645 | LINC00092 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-671-5p | 15 | FOXP1 | Sponge network | -1.886 | 0 | 0.042 | 0.74368 | 0.257 |
| 52646 | RP11-2B6.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | EPS15 | Sponge network | 0.539 | 0.03722 | -0.052 | 0.53998 | 0.257 |
| 52647 | BCYRN1 |
hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | ZDBF2 | Sponge network | 0.311 | 0.10344 | -0.998 | 0.00025 | 0.257 |
| 52648 | RP11-195B17.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p | 15 | MCC | Sponge network | 0.37 | 0.27614 | -1.005 | 0 | 0.257 |
| 52649 | MRPL23-AS1 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-324-5p | 10 | SYNPO2 | Sponge network | -0.278 | 0.76559 | -2.342 | 0 | 0.257 |
| 52650 | BVES-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-375;hsa-miR-421;hsa-miR-429;hsa-miR-942-5p | 29 | NOTCH2 | Sponge network | -2.62 | 0 | 0.138 | 0.35163 | 0.257 |
| 52651 | FAM66C |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-93-3p;hsa-miR-940 | 28 | TOM1L2 | Sponge network | -0.901 | 0.00019 | -0.994 | 0 | 0.257 |
| 52652 | RP11-967K21.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-2355-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-940 | 14 | SIRT1 | Sponge network | 0.056 | 0.81195 | 0.109 | 0.2593 | 0.257 |
| 52653 | RP11-531A24.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 39 | PAFAH1B1 | Sponge network | 0.75 | 0.00054 | -0.429 | 0 | 0.257 |
| 52654 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-590-5p | 16 | PIK3C2A | Sponge network | 0.983 | 0.00048 | 0.061 | 0.53776 | 0.257 |
| 52655 | GABPB1-AS1 |
hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p | 10 | KDM3A | Sponge network | 1.11 | 0 | 0.132 | 0.22711 | 0.257 |
| 52656 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | SOX11 | Sponge network | 0.643 | 0 | 2.68 | 0 | 0.257 |
| 52657 | LIPE-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-708-5p;hsa-miR-766-3p | 15 | EZH1 | Sponge network | 0.078 | 0.55803 | -0.564 | 1.0E-5 | 0.257 |
| 52658 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-374a-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 59 | SMAD4 | Sponge network | -2.452 | 0 | -0.313 | 0.00413 | 0.257 |
| 52659 | PCA3 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-654-3p | 11 | USP25 | Sponge network | -0.709 | 0.18168 | -0.242 | 0.01397 | 0.257 |
| 52660 | LINC00957 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | TP53INP1 | Sponge network | -0.421 | 0.0114 | -0.496 | 0.00064 | 0.257 |
| 52661 | LINC00342 |
hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-181a-2-3p;hsa-miR-218-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p | 11 | PIK3C2A | Sponge network | 0.297 | 0.19933 | 0.061 | 0.53776 | 0.257 |
| 52662 | RP11-359B12.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-485-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | SAMHD1 | Sponge network | 0.064 | 0.72934 | 0.072 | 0.62895 | 0.257 |
| 52663 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 34 | MOB1B | Sponge network | 0.374 | 0.20054 | 0.197 | 0.15266 | 0.257 |
| 52664 | RP11-158K1.3 |
hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-940 | 13 | GLI3 | Sponge network | 0.349 | 0.01695 | -0.615 | 0.01482 | 0.257 |
| 52665 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-629-3p | 10 | ESR1 | Sponge network | 1.969 | 0.00316 | -1.035 | 3.0E-5 | 0.257 |
| 52666 | GS1-124K5.11 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-940 | 29 | MCC | Sponge network | 0.494 | 0.00339 | -1.005 | 0 | 0.257 |
| 52667 | RP11-597D13.9 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | PTCH1 | Sponge network | 1.302 | 7.0E-5 | -0.1 | 0.6179 | 0.257 |
| 52668 | MIR143HG |
hsa-miR-10b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-653-5p | 30 | UBR3 | Sponge network | -0.739 | 0.0574 | -0.021 | 0.82774 | 0.257 |
| 52669 | RP11-161M6.2 |
hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-96-5p | 10 | CBFA2T3 | Sponge network | 0.61 | 0.06611 | -1.221 | 1.0E-5 | 0.257 |
| 52670 | AC003090.1 |
hsa-miR-128-3p;hsa-miR-1287-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-96-5p | 19 | PAIP2 | Sponge network | -2.117 | 0.00161 | -0.515 | 0 | 0.257 |
| 52671 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 23 | ESR1 | Sponge network | 0.225 | 0.30644 | -1.035 | 3.0E-5 | 0.257 |
| 52672 | PART1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 27 | RNASEL | Sponge network | -2.925 | 0 | -0.272 | 0.01796 | 0.257 |
| 52673 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-28-5p;hsa-miR-324-5p;hsa-miR-362-3p | 12 | HACE1 | Sponge network | 0.078 | 0.55803 | -0.072 | 0.55239 | 0.257 |
| 52674 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | NEDD4 | Sponge network | 0.683 | 1.0E-5 | 0.02 | 0.89834 | 0.257 |
| 52675 | LINC00702 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 12 | USP25 | Sponge network | -0.737 | 0.06182 | -0.242 | 0.01397 | 0.257 |
| 52676 | GS1-124K5.11 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-3p | 13 | LATS2 | Sponge network | 0.494 | 0.00339 | 0.159 | 0.2304 | 0.257 |
| 52677 | HOXB-AS2 |
hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-576-5p | 19 | MDM4 | Sponge network | 1.316 | 0.01106 | 0.345 | 0.00762 | 0.257 |
| 52678 | CTA-204B4.2 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 10 | FAM172A | Sponge network | 0.765 | 7.0E-5 | -0.57 | 0 | 0.257 |
| 52679 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | BNC2 | Sponge network | -0.83 | 0 | -0.491 | 0.09988 | 0.257 |
| 52680 | RP11-693J15.4 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 16 | LRIG1 | Sponge network | -0.72 | 0.24239 | -0.537 | 0.00588 | 0.257 |
| 52681 | AC108488.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-93-5p | 10 | SLITRK3 | Sponge network | 0.259 | 0.12542 | -3.328 | 0 | 0.257 |
| 52682 | BCYRN1 |
hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-345-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-664a-5p | 10 | RANBP6 | Sponge network | 0.311 | 0.10344 | 0.175 | 0.19756 | 0.257 |
| 52683 | EMX2OS |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-455-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 35 | EGFR | Sponge network | -0.777 | 0.1134 | -0.175 | 0.48451 | 0.257 |
| 52684 | LIPE-AS1 |
hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-625-5p | 12 | BCL9 | Sponge network | 0.078 | 0.55803 | 0.321 | 0.00267 | 0.257 |
| 52685 | GUSBP11 |
hsa-let-7i-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-654-3p | 14 | DICER1 | Sponge network | 0.856 | 0 | 0.042 | 0.674 | 0.257 |
| 52686 | RP11-927P21.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-3613-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-409-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-940 | 33 | TRPS1 | Sponge network | 0.921 | 0.00118 | -0.191 | 0.44109 | 0.257 |
| 52687 | RP11-6N17.3 |
hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-21-5p;hsa-miR-26b-3p | 13 | GRIK3 | Sponge network | 0.108 | 0.69556 | -3.208 | 0 | 0.257 |
| 52688 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 30 | PRKCE | Sponge network | -0.129 | 0.57177 | -0.403 | 0.00056 | 0.257 |
| 52689 | TMEM9B-AS1 |
hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p | 13 | DICER1 | Sponge network | 0.529 | 0.00552 | 0.042 | 0.674 | 0.257 |
| 52690 | BDNF-AS |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 13 | TLL1 | Sponge network | -0.668 | 3.0E-5 | -1.195 | 3.0E-5 | 0.257 |
| 52691 | DIO3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-550a-5p;hsa-miR-940 | 11 | HOOK3 | Sponge network | -0.773 | 0.04073 | 0.273 | 0.09957 | 0.257 |
| 52692 | RP11-83A24.2 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 12 | UBE4B | Sponge network | 0.417 | 0.00445 | -0.235 | 0.007 | 0.257 |
| 52693 | RP11-705C15.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-495-3p;hsa-miR-7-1-3p | 14 | ZFX | Sponge network | 0.796 | 4.0E-5 | 0.09 | 0.42563 | 0.257 |
| 52694 | PART1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-508-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 22 | ROBO1 | Sponge network | -2.925 | 0 | -0.009 | 0.96154 | 0.257 |
| 52695 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-590-3p | 18 | KIT | Sponge network | 0.385 | 0.10067 | -2.184 | 0 | 0.257 |
| 52696 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | CNTN1 | Sponge network | 0.392 | 0.0278 | -2.594 | 0 | 0.257 |
| 52697 | U91328.19 |
hsa-miR-1271-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-589-3p | 15 | RPS6KA6 | Sponge network | -0.303 | 0.21002 | -2.989 | 0 | 0.257 |
| 52698 | LINC01018 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TAL1 | Sponge network | -2.915 | 0.00015 | -0.462 | 0.00574 | 0.257 |
| 52699 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-224-5p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-935 | 71 | SMAD4 | Sponge network | -1.111 | 0.0003 | -0.313 | 0.00413 | 0.257 |
| 52700 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TBX18 | Sponge network | -0.737 | 0.10822 | 0.843 | 0.02945 | 0.257 |
| 52701 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 42 | CADM2 | Sponge network | -0.08 | 0.90092 | -3.782 | 0 | 0.257 |
| 52702 | CROCCP2 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-337-3p | 11 | RB1 | Sponge network | 0.79 | 0 | 0.382 | 0.0017 | 0.257 |
| 52703 | RP11-157J24.2 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-576-5p | 16 | TMEM132B | Sponge network | -1.664 | 0.05165 | -0.83 | 0.01084 | 0.257 |
| 52704 | AC034220.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | USP12 | Sponge network | 0.309 | 0.07304 | -0.102 | 0.40768 | 0.257 |
| 52705 | RP11-707P17.2 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-328-3p;hsa-miR-432-5p;hsa-miR-495-3p | 11 | VPS53 | Sponge network | 1.877 | 0 | 0.103 | 0.22667 | 0.257 |
| 52706 | CTD-2017D11.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-486-5p | 12 | AKAP6 | Sponge network | 0.792 | 0.0049 | -1.586 | 3.0E-5 | 0.257 |
| 52707 | C4A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-345-5p;hsa-miR-589-3p;hsa-miR-629-3p;hsa-miR-93-3p | 22 | TRPS1 | Sponge network | 3.345 | 0 | -0.191 | 0.44109 | 0.257 |
| 52708 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-576-5p | 16 | TMTC3 | Sponge network | 2.163 | 0 | 0.123 | 0.22189 | 0.257 |
| 52709 | OR2A1-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | NR2C2 | Sponge network | 0.542 | 0.00241 | 0.21 | 0.03224 | 0.257 |
| 52710 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3614-5p;hsa-miR-375;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-940 | 41 | GAS7 | Sponge network | 0.494 | 0.00339 | -0.555 | 0.01602 | 0.257 |
| 52711 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-590-3p | 14 | MXI1 | Sponge network | -0.555 | 0.06156 | -0.987 | 0 | 0.257 |
| 52712 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-497-5p | 14 | PRKAA1 | Sponge network | 1.266 | 0 | 0.317 | 0.0031 | 0.257 |
| 52713 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-374b-5p;hsa-miR-628-5p | 16 | MIER3 | Sponge network | 0.417 | 0.00445 | -0.212 | 0.04957 | 0.257 |
| 52714 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p | 15 | ARHGAP29 | Sponge network | 0.657 | 4.0E-5 | 0.131 | 0.42953 | 0.257 |
| 52715 | RP11-403I13.7 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 38 | TRPS1 | Sponge network | 0.395 | 0.15451 | -0.191 | 0.44109 | 0.257 |
| 52716 | CALML3-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-874-3p | 22 | MXI1 | Sponge network | 0.95 | 0.15943 | -0.987 | 0 | 0.257 |
| 52717 | MYLK-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-484 | 31 | PTEN | Sponge network | -1.331 | 3.0E-5 | -0.187 | 0.07748 | 0.257 |
| 52718 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-7-1-3p | 29 | EPHA5 | Sponge network | 0.267 | 0.2937 | -0.957 | 0.01733 | 0.257 |
| 52719 | TPTEP1 |
hsa-miR-1976;hsa-miR-224-5p;hsa-miR-320b;hsa-miR-361-3p;hsa-miR-374b-3p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | PACRG | Sponge network | -1.216 | 0 | -2.248 | 0 | 0.257 |
| 52720 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p | 19 | PSIP1 | Sponge network | 0.421 | 0.00084 | -0.005 | 0.96897 | 0.257 |
| 52721 | RP11-403I13.8 |
hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-590-3p | 16 | MRVI1 | Sponge network | 0.619 | 0.00157 | -1.051 | 0.00022 | 0.257 |
| 52722 | RP4-669P10.16 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-193a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p | 16 | NOTCH2 | Sponge network | -0.301 | 0.06597 | 0.138 | 0.35163 | 0.257 |
| 52723 | RP11-532F6.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p | 12 | EPAS1 | Sponge network | -0.896 | 0.00099 | -0.184 | 0.14418 | 0.257 |
| 52724 | AC093627.10 |
hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-589-3p;hsa-miR-7-5p | 10 | KCNJ5 | Sponge network | -0.204 | 0.52643 | -0.281 | 0.3339 | 0.257 |
| 52725 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | ZNF536 | Sponge network | 0.997 | 0.00066 | -3.115 | 0 | 0.257 |
| 52726 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 36 | CCND2 | Sponge network | 0.16 | 0.50785 | -0.267 | 0.3112 | 0.257 |
| 52727 | RP11-53O19.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 33 | ZFX | Sponge network | -0.405 | 0.22013 | 0.09 | 0.42563 | 0.257 |
| 52728 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-2-3p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 28 | LATS1 | Sponge network | -1.575 | 6.0E-5 | 0.109 | 0.34208 | 0.257 |
| 52729 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-324-5p;hsa-miR-455-5p;hsa-miR-550a-5p | 18 | TSHZ3 | Sponge network | 0.859 | 0.00331 | -0.556 | 0.01516 | 0.257 |
| 52730 | RP13-977J11.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-324-5p | 13 | ESR1 | Sponge network | 0.121 | 0.65192 | -1.035 | 3.0E-5 | 0.257 |
| 52731 | SERTAD4-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-576-5p | 10 | ITGAV | Sponge network | -0.932 | 0.00329 | 0.337 | 0.01002 | 0.257 |
| 52732 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 13 | FYN | Sponge network | 0.101 | 0.60919 | -0.131 | 0.37051 | 0.257 |
| 52733 | AC106786.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p | 10 | JAZF1 | Sponge network | 1.122 | 0.02989 | -0.377 | 0.02677 | 0.257 |
| 52734 | ERVH48-1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p | 13 | BCL11A | Sponge network | 1.04 | 0.00023 | -0.932 | 0.0063 | 0.257 |
| 52735 | RP11-359B12.2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 18 | CELF4 | Sponge network | 0.064 | 0.72934 | -1.135 | 0.00344 | 0.257 |
| 52736 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-935 | 25 | RARB | Sponge network | -2.925 | 0 | -0.825 | 2.0E-5 | 0.257 |
| 52737 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | AKAP11 | Sponge network | 0.538 | 0.00076 | 0.196 | 0.09825 | 0.257 |
| 52738 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -0.663 | 0.10887 | -0.243 | 0.11408 | 0.257 |
| 52739 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-590-3p | 15 | SON | Sponge network | 0.799 | 1.0E-5 | 0.168 | 0.04406 | 0.257 |
| 52740 | LINC00865 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-7-1-3p | 11 | EGR2 | Sponge network | -1.249 | 7.0E-5 | 0.309 | 0.17079 | 0.257 |
| 52741 | RP11-996F15.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-3677-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p | 13 | PRKAA1 | Sponge network | 0.983 | 0.00048 | 0.317 | 0.0031 | 0.257 |
| 52742 | LINC00173 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 10 | DNAJB4 | Sponge network | 0.056 | 0.8494 | -0.7 | 1.0E-5 | 0.257 |
| 52743 | USP3-AS1 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-331-5p | 10 | DUSP1 | Sponge network | -0.162 | 0.47687 | -1.754 | 0 | 0.257 |
| 52744 | CTC-296K1.3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-625-5p | 10 | CADM1 | Sponge network | -2.095 | 0.00112 | -0.9 | 0.00084 | 0.257 |
| 52745 | LINC00667 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-423-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | CELF4 | Sponge network | -0.235 | 0.15142 | -1.135 | 0.00344 | 0.257 |
| 52746 | AC108488.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3613-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 36 | CADM2 | Sponge network | 0.259 | 0.12542 | -3.782 | 0 | 0.257 |
| 52747 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 32 | FOXP1 | Sponge network | -1.161 | 0.00591 | 0.042 | 0.74368 | 0.257 |
| 52748 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 12 | RNF144A | Sponge network | 0.051 | 0.83388 | 0.594 | 0.0009 | 0.257 |
| 52749 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-326 | 10 | SOX11 | Sponge network | 3.21 | 0 | 2.68 | 0 | 0.257 |
| 52750 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-550a-5p;hsa-miR-629-3p;hsa-miR-93-5p | 24 | DST | Sponge network | 1.153 | 0.00114 | -0.713 | 0.00096 | 0.257 |
| 52751 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-940 | 15 | IGF1 | Sponge network | 0.817 | 3.0E-5 | -0.204 | 0.5898 | 0.257 |
| 52752 | GAS6-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-450b-5p | 29 | JAZF1 | Sponge network | 0.16 | 0.50785 | -0.377 | 0.02677 | 0.257 |
| 52753 | LINC00094 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-378c;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 56 | DST | Sponge network | 0.253 | 0.09565 | -0.713 | 0.00096 | 0.257 |
| 52754 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p | 32 | FOXP1 | Sponge network | -0.322 | 0.25945 | 0.042 | 0.74368 | 0.257 |
| 52755 | DOCK9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-539-5p | 20 | DMD | Sponge network | -0.231 | 0.28214 | -1.506 | 0 | 0.257 |
| 52756 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-452-5p;hsa-miR-501-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 28 | RAP1A | Sponge network | 0.246 | 0.59912 | -0.929 | 0 | 0.257 |
| 52757 | RP11-999E24.3 |
hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-362-3p | 10 | GABRB2 | Sponge network | -0.693 | 0.05629 | -1.841 | 0.00029 | 0.257 |
| 52758 | LINC00926 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-92a-3p | 16 | RUNX1T1 | Sponge network | 0.756 | 0.00739 | -0.344 | 0.2374 | 0.257 |
| 52759 | RP11-2B6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 17 | SCN9A | Sponge network | 0.539 | 0.03722 | -0.525 | 0.10593 | 0.257 |
| 52760 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p | 14 | SCN9A | Sponge network | 0.946 | 0 | -0.525 | 0.10593 | 0.257 |
| 52761 | AF131215.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-484 | 10 | EZH1 | Sponge network | -0.176 | 0.59692 | -0.564 | 1.0E-5 | 0.257 |
| 52762 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-942-5p | 32 | TMEM132B | Sponge network | 0.214 | 0.18246 | -0.83 | 0.01084 | 0.257 |
| 52763 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326 | 11 | ZFX | Sponge network | 0.912 | 0.00014 | 0.09 | 0.42563 | 0.257 |
| 52764 | HAS2-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 11 | ZFHX3 | Sponge network | -0.011 | 0.967 | 0.389 | 0.01363 | 0.257 |
| 52765 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-3p;hsa-miR-369-3p;hsa-miR-7-5p | 10 | CAV1 | Sponge network | 0.253 | 0.09565 | -1.013 | 6.0E-5 | 0.257 |
| 52766 | RP11-373D23.3 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-7-1-3p | 17 | ANK2 | Sponge network | -0.285 | 0.2172 | -1.603 | 1.0E-5 | 0.257 |
| 52767 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-33a-3p;hsa-miR-940 | 10 | NEDD4 | Sponge network | -1.255 | 0.00981 | 0.02 | 0.89834 | 0.257 |
| 52768 | PART1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | CREM | Sponge network | -2.925 | 0 | -0.345 | 0.00258 | 0.257 |
| 52769 | RP11-53O19.1 |
hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-576-5p | 11 | RIT1 | Sponge network | -0.405 | 0.22013 | -0.296 | 0.00054 | 0.257 |
| 52770 | GHRLOS |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-493-5p | 21 | AR | Sponge network | -1.942 | 0 | -1.554 | 0 | 0.257 |
| 52771 | RP11-499P20.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-940 | 16 | CREB1 | Sponge network | 0.187 | 0.44121 | 0.203 | 0.00537 | 0.257 |
| 52772 | LINC00957 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 10 | ADCY1 | Sponge network | -0.421 | 0.0114 | 0.362 | 0.16982 | 0.257 |
| 52773 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 29 | PRKCE | Sponge network | -0.222 | 0.32445 | -0.403 | 0.00056 | 0.257 |
| 52774 | BCYRN1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | EYA4 | Sponge network | 0.311 | 0.10344 | 0.3 | 0.36876 | 0.257 |
| 52775 | RP11-344B5.2 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b | 11 | ZFHX3 | Sponge network | -0.055 | 0.88482 | 0.389 | 0.01363 | 0.257 |
| 52776 | PCBP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-5p | 15 | PBRM1 | Sponge network | -0.567 | 0 | -0.029 | 0.78601 | 0.257 |
| 52777 | RP4-669P10.16 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p | 12 | FAT4 | Sponge network | -0.301 | 0.06597 | -0.354 | 0.14694 | 0.257 |
| 52778 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-625-3p | 18 | BMPR2 | Sponge network | 0.832 | 0 | 0.206 | 0.0635 | 0.257 |
| 52779 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-374a-3p;hsa-miR-484;hsa-miR-532-3p | 16 | CBL | Sponge network | 0.792 | 0.0049 | 0.291 | 0.01267 | 0.257 |
| 52780 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-382-5p;hsa-miR-7-1-3p | 17 | NRXN1 | Sponge network | -0.285 | 0.2172 | -3.778 | 0 | 0.257 |
| 52781 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 10 | TBX18 | Sponge network | -0.376 | 0.00337 | 0.843 | 0.02945 | 0.257 |
| 52782 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-935 | 27 | RAP1A | Sponge network | 0.572 | 0.01722 | -0.929 | 0 | 0.257 |
| 52783 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | PIK3R1 | Sponge network | 0.219 | 0.1516 | -0.133 | 0.36854 | 0.257 |
| 52784 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-93-5p | 41 | EPHA5 | Sponge network | -1.645 | 0 | -0.957 | 0.01733 | 0.257 |
| 52785 | RP11-798M19.6 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 37 | GNAQ | Sponge network | -0.612 | 0.00094 | -0.367 | 0.00102 | 0.257 |
| 52786 | AC025171.1 |
hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 12 | RASAL2 | Sponge network | 0.998 | 0 | 0.869 | 0 | 0.257 |
| 52787 | RP11-6N17.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-342-5p;hsa-miR-455-5p | 13 | DTNA | Sponge network | 0.108 | 0.69556 | -1.648 | 1.0E-5 | 0.257 |
| 52788 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-342-3p;hsa-miR-454-3p | 10 | RB1CC1 | Sponge network | 0.001 | 0.99753 | 0.175 | 0.12559 | 0.257 |
| 52789 | LINC00667 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SEMA3E | Sponge network | -0.235 | 0.15142 | -1.717 | 0.00137 | 0.257 |
| 52790 | ERVH48-1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p;hsa-miR-624-5p | 11 | HMCN1 | Sponge network | 1.04 | 0.00023 | 0.809 | 0.00544 | 0.257 |
| 52791 | RP11-434B12.1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-582-3p;hsa-miR-590-3p | 22 | PRDM2 | Sponge network | -0.031 | 0.89063 | -0.258 | 0.00721 | 0.257 |
| 52792 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p | 41 | IL6ST | Sponge network | 1.757 | 0 | -0.402 | 0.00676 | 0.257 |
| 52793 | MEG3 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | CDH10 | Sponge network | 0.249 | 0.35902 | -2.397 | 0 | 0.257 |
| 52794 | BZRAP1-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 29 | NOTCH2 | Sponge network | -0.465 | 0.04703 | 0.138 | 0.35163 | 0.257 |
| 52795 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 33 | ST6GALNAC3 | Sponge network | 0.249 | 0.35902 | -0.987 | 0 | 0.257 |
| 52796 | RP11-890B15.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 12 | DCN | Sponge network | 0.101 | 0.60919 | -1.288 | 0 | 0.257 |
| 52797 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 14 | LRIG1 | Sponge network | 0.453 | 0.01839 | -0.537 | 0.00588 | 0.257 |
| 52798 | LINC00472 |
hsa-miR-127-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-331-3p | 16 | FAM135B | Sponge network | -0.842 | 0.01361 | -3.978 | 0 | 0.257 |
| 52799 | AC005062.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-940 | 12 | AKAP11 | Sponge network | -0.019 | 0.81504 | 0.196 | 0.09825 | 0.257 |
| 52800 | RP11-890B15.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 18 | LUZP2 | Sponge network | 0.101 | 0.60919 | -0.056 | 0.89416 | 0.257 |
| 52801 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 24 | KIF1B | Sponge network | 1.205 | 0 | -0.118 | 0.32258 | 0.257 |
| 52802 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p | 11 | TXNIP | Sponge network | 0.331 | 0.57247 | -1.277 | 0 | 0.257 |
| 52803 | OIP5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p | 56 | RUNX1T1 | Sponge network | 0.421 | 0.00084 | -0.344 | 0.2374 | 0.257 |
| 52804 | CTD-2303H24.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 26 | PTPN14 | Sponge network | 0.415 | 0.14657 | 0.144 | 0.34595 | 0.257 |
| 52805 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-3613-5p;hsa-miR-590-3p | 13 | GPAM | Sponge network | 0.222 | 0.29133 | 0.142 | 0.29732 | 0.257 |
| 52806 | LINC00578 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p | 18 | NCOA1 | Sponge network | 0.811 | 0.03228 | -0.432 | 0 | 0.257 |
| 52807 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-485-3p;hsa-miR-7-1-3p | 23 | PRKAB2 | Sponge network | -0.611 | 0.09607 | -0.367 | 0.01371 | 0.257 |
| 52808 | PSMD5-AS1 |
hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-376c-3p;hsa-miR-664a-3p | 18 | RASAL2 | Sponge network | 0.569 | 0.00067 | 0.869 | 0 | 0.257 |
| 52809 | AC009948.5 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | ADCY1 | Sponge network | 0.532 | 0.00505 | 0.362 | 0.16982 | 0.257 |
| 52810 | RP11-395G23.3 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 10 | KLF10 | Sponge network | 0.381 | 0.25967 | -0.783 | 0 | 0.257 |
| 52811 | RP11-156P1.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-664a-3p | 17 | SOX11 | Sponge network | 0.214 | 0.18246 | 2.68 | 0 | 0.257 |
| 52812 | RP13-516M14.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-7-1-3p | 14 | CDH19 | Sponge network | 0.389 | 0.04293 | -3.434 | 0 | 0.257 |
| 52813 | CTD-2184D3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 14 | ERCC4 | Sponge network | 0.272 | 0.44692 | 0.126 | 0.15266 | 0.257 |
| 52814 | ARHGAP5-AS1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-7-1-3p | 10 | GRIN2A | Sponge network | -0.644 | 0.00021 | -2.161 | 0 | 0.257 |
| 52815 | RP11-800A3.7 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 11 | SSBP2 | Sponge network | 0.18 | 0.7147 | -1.58 | 0 | 0.257 |
| 52816 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 28 | MAF | Sponge network | -0.83 | 0 | -1.257 | 0 | 0.257 |
| 52817 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 33 | ERBB4 | Sponge network | -0.421 | 0.0114 | -1.975 | 2.0E-5 | 0.257 |
| 52818 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 12 | FREM2 | Sponge network | 0.4 | 0.14611 | -0.437 | 0.46353 | 0.257 |
| 52819 | RP11-686D22.4 |
hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-582-3p;hsa-miR-590-3p | 10 | GAS7 | Sponge network | 0.793 | 5.0E-5 | -0.555 | 0.01602 | 0.257 |
| 52820 | RP11-157J24.2 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-338-3p | 14 | FBXO32 | Sponge network | -1.664 | 0.05165 | -0.495 | 0.07561 | 0.257 |
| 52821 | RP11-574K11.29 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-497-5p | 16 | SOX11 | Sponge network | 0.997 | 0 | 2.68 | 0 | 0.257 |
| 52822 | RP11-968O1.5 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-338-3p;hsa-miR-532-3p;hsa-miR-940 | 25 | GAS7 | Sponge network | -0.829 | 0.01343 | -0.555 | 0.01602 | 0.257 |
| 52823 | LINC00476 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-935 | 11 | PAX6 | Sponge network | -0.615 | 0.00015 | -0.444 | 0.04689 | 0.257 |
| 52824 | RP11-216B9.6 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-590-3p | 11 | TRIM13 | Sponge network | 0.174 | 0.30034 | 0.183 | 0.06968 | 0.257 |
| 52825 | SNX29P2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 27 | MOB1B | Sponge network | 0.4 | 0.14611 | 0.197 | 0.15266 | 0.257 |
| 52826 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-330-3p;hsa-miR-577;hsa-miR-582-5p | 11 | NOTCH2NL | Sponge network | 0.637 | 0.00069 | 0.024 | 0.84307 | 0.257 |
| 52827 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-628-5p | 32 | MAPK1 | Sponge network | 0.537 | 0.00139 | -0.017 | 0.81465 | 0.257 |
| 52828 | LINC01018 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 23 | SUFU | Sponge network | -2.915 | 0.00015 | -0.04 | 0.65489 | 0.257 |
| 52829 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 10 | DDX6 | Sponge network | -0.557 | 0.11135 | 0.091 | 0.36428 | 0.257 |
| 52830 | LINC00094 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-624-5p | 18 | SIRT1 | Sponge network | 0.253 | 0.09565 | 0.109 | 0.2593 | 0.257 |
| 52831 | HCG22 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | PRRX1 | Sponge network | 0.816 | 0.32368 | 1.316 | 0 | 0.257 |
| 52832 | RP11-403I13.5 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-7-1-3p | 17 | BRD3 | Sponge network | 1.062 | 0.00512 | 0.37 | 0.00013 | 0.257 |
| 52833 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | EBF1 | Sponge network | -0.355 | 0.0077 | -0.384 | 0.08856 | 0.257 |
| 52834 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-34a-5p;hsa-miR-429 | 28 | PRKAA2 | Sponge network | 0.859 | 0.00331 | -2.462 | 0 | 0.257 |
| 52835 | LINC00886 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-629-3p;hsa-miR-93-5p | 15 | RBL2 | Sponge network | -0.371 | 0.24604 | -0.282 | 0.00254 | 0.257 |
| 52836 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | SLITRK3 | Sponge network | 0.131 | 0.8436 | -3.328 | 0 | 0.257 |
| 52837 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-92a-3p | 11 | ZNF521 | Sponge network | 0.72 | 0.0001 | -0.111 | 0.58068 | 0.257 |
| 52838 | RP11-693J15.4 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 22 | TRPS1 | Sponge network | -0.72 | 0.24239 | -0.191 | 0.44109 | 0.257 |
| 52839 | TRIM52-AS1 |
hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-7-1-3p | 10 | DACT1 | Sponge network | -0.418 | 0.00068 | 0.783 | 0.00072 | 0.257 |
| 52840 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-324-5p;hsa-miR-582-3p;hsa-miR-651-5p | 11 | PRDM2 | Sponge network | -2.076 | 0.00548 | -0.258 | 0.00721 | 0.257 |
| 52841 | MEG3 |
hsa-miR-21-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-589-5p;hsa-miR-7-1-3p;hsa-miR-940 | 12 | SPOP | Sponge network | 0.249 | 0.35902 | -0.419 | 1.0E-5 | 0.257 |
| 52842 | RP11-359B12.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-361-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p | 18 | CDH23 | Sponge network | 0.064 | 0.72934 | -0.974 | 0.00034 | 0.257 |
| 52843 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-493-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | USP12 | Sponge network | -1.161 | 0.00591 | -0.102 | 0.40768 | 0.257 |
| 52844 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-664a-3p | 15 | ITCH | Sponge network | 0.614 | 1.0E-5 | 0.084 | 0.34058 | 0.257 |
| 52845 | EMX2OS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 29 | NCOA1 | Sponge network | -0.777 | 0.1134 | -0.432 | 0 | 0.257 |
| 52846 | AC004893.11 |
hsa-miR-139-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-495-3p | 10 | GSK3B | Sponge network | 1.317 | 0 | 0.266 | 0.00045 | 0.257 |
| 52847 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | MGA | Sponge network | 0.417 | 0.00445 | 0.323 | 0.00792 | 0.257 |
| 52848 | PWAR6 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 28 | EFNA5 | Sponge network | -1.575 | 6.0E-5 | -0.421 | 0.17868 | 0.257 |
| 52849 | RP11-66B24.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-502-5p;hsa-miR-550a-3p | 23 | TCF4 | Sponge network | 0.57 | 0.1866 | 0.016 | 0.92271 | 0.256 |
| 52850 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | BNC2 | Sponge network | 0.272 | 0.44692 | -0.491 | 0.09988 | 0.256 |
| 52851 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p | 12 | MLLT10 | Sponge network | 0.917 | 0 | 0.105 | 0.33292 | 0.256 |
| 52852 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-576-5p | 12 | PDS5B | Sponge network | 2.163 | 0 | 0.259 | 0.00962 | 0.256 |
| 52853 | HOXA-AS2 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-20a-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 12 | PREX2 | Sponge network | 0.61 | 0.01593 | -0.272 | 0.25382 | 0.256 |
| 52854 | AC005618.6 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | ZFX | Sponge network | 0.862 | 0.06213 | 0.09 | 0.42563 | 0.256 |
| 52855 | CTD-2554C21.2 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-532-3p;hsa-miR-96-5p | 27 | FOXP1 | Sponge network | -1.654 | 0.00144 | 0.042 | 0.74368 | 0.256 |
| 52856 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-376c-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 32 | EPHA5 | Sponge network | 0.603 | 0.00127 | -0.957 | 0.01733 | 0.256 |
| 52857 | HOXA-AS3 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-21-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-660-5p | 16 | LIMCH1 | Sponge network | -0.555 | 0.06156 | -0.915 | 1.0E-5 | 0.256 |
| 52858 | RP11-473I1.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-374b-5p | 11 | AKAP6 | Sponge network | 0.542 | 0.00054 | -1.586 | 3.0E-5 | 0.256 |
| 52859 | RP11-815I9.4 |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-590-5p | 12 | DST | Sponge network | 0.537 | 0.0006 | -0.713 | 0.00096 | 0.256 |
| 52860 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-629-5p | 31 | SPRY3 | Sponge network | -0.154 | 0.78939 | 0.016 | 0.91286 | 0.256 |
| 52861 | FZD10-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-500a-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | CSMD1 | Sponge network | -0.727 | 0.04726 | -0.63 | 0.18881 | 0.256 |
| 52862 | HCG22 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-7-1-3p | 16 | NIN | Sponge network | 0.816 | 0.32368 | -0.253 | 0.13794 | 0.256 |
| 52863 | RP11-401P9.4 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-3127-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 15 | RPS6KA2 | Sponge network | -0.384 | 0.40652 | -0.57 | 0.00013 | 0.256 |
| 52864 | RP5-1074L1.4 |
hsa-miR-100-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-337-3p | 11 | RB1 | Sponge network | 1.923 | 0 | 0.382 | 0.0017 | 0.256 |
| 52865 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | KAT6B | Sponge network | -1.264 | 6.0E-5 | -0.478 | 0.00018 | 0.256 |
| 52866 | OSER1-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 18 | PDE4DIP | Sponge network | -0.13 | 0.48734 | -0.282 | 0.0257 | 0.256 |
| 52867 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-708-5p | 30 | SPRY3 | Sponge network | -0.031 | 0.89063 | 0.016 | 0.91286 | 0.256 |
| 52868 | RP11-216B9.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p | 13 | SLITRK3 | Sponge network | 0.174 | 0.30034 | -3.328 | 0 | 0.256 |
| 52869 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-3p | 28 | FOXO3 | Sponge network | -0.129 | 0.57177 | -0.04 | 0.72028 | 0.256 |
| 52870 | RP11-686D22.8 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | SASH1 | Sponge network | 0.898 | 1.0E-5 | -0.502 | 0.00175 | 0.256 |
| 52871 | HOXA-AS2 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-877-5p | 27 | TET1 | Sponge network | 0.61 | 0.01593 | -0.135 | 0.52138 | 0.256 |
| 52872 | HCG11 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ERG | Sponge network | 0.385 | 0.10067 | 0.002 | 0.99011 | 0.256 |
| 52873 | RP11-458D21.1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | LUZP2 | Sponge network | 0.663 | 0.00073 | -0.056 | 0.89416 | 0.256 |
| 52874 | RP11-359E10.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-7-1-3p;hsa-miR-940 | 21 | AKAP11 | Sponge network | 0.299 | 0.57722 | 0.196 | 0.09825 | 0.256 |
| 52875 | RP11-701H24.3 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-3613-5p;hsa-miR-590-3p | 15 | NFIB | Sponge network | -1.425 | 0.04204 | 0.023 | 0.90795 | 0.256 |
| 52876 | AC034220.3 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SH3GLB1 | Sponge network | 0.309 | 0.07304 | -0.115 | 0.14773 | 0.256 |
| 52877 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 10 | PHC3 | Sponge network | 0.832 | 0 | 0.212 | 0.0499 | 0.256 |
| 52878 | LINC00476 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-542-3p;hsa-miR-590-3p | 11 | USP25 | Sponge network | -0.615 | 0.00015 | -0.242 | 0.01397 | 0.256 |
| 52879 | NR2F1-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 31 | ACSL6 | Sponge network | -0.49 | 0.04946 | -1.593 | 0 | 0.256 |
| 52880 | RP11-400K9.4 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-501-3p;hsa-miR-502-3p | 21 | CADM1 | Sponge network | -0.611 | 0.09607 | -0.9 | 0.00084 | 0.256 |
| 52881 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-429;hsa-miR-7-1-3p | 15 | DNAJB4 | Sponge network | -0.969 | 2.0E-5 | -0.7 | 1.0E-5 | 0.256 |
| 52882 | CTD-2647L4.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-376c-3p;hsa-miR-495-3p | 12 | SMAD2 | Sponge network | 0.555 | 0.00197 | -0.128 | 0.12732 | 0.256 |
| 52883 | PWAR6 |
hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | RBBP6 | Sponge network | -1.575 | 6.0E-5 | 0.163 | 0.05101 | 0.256 |
| 52884 | U91328.19 |
hsa-let-7g-3p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-7-1-3p | 11 | KCNAB1 | Sponge network | -0.303 | 0.21002 | -0.755 | 2.0E-5 | 0.256 |
| 52885 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 11 | PRKAB2 | Sponge network | 0.959 | 0 | -0.367 | 0.01371 | 0.256 |
| 52886 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-374b-5p | 15 | PIK3C2A | Sponge network | 1.117 | 0 | 0.061 | 0.53776 | 0.256 |
| 52887 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 13 | FOXP1 | Sponge network | 1.11 | 1.0E-5 | 0.042 | 0.74368 | 0.256 |
| 52888 | AC034220.3 |
hsa-let-7i-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-409-5p;hsa-miR-432-5p;hsa-miR-495-3p | 11 | STAG2 | Sponge network | 0.309 | 0.07304 | 0.252 | 0.00438 | 0.256 |
| 52889 | RP11-277P12.20 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-3613-5p;hsa-miR-375;hsa-miR-484;hsa-miR-532-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | TRPS1 | Sponge network | 0.687 | 0.02753 | -0.191 | 0.44109 | 0.256 |
| 52890 | RP11-707P17.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-154-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-495-3p;hsa-miR-654-3p | 10 | BMPR2 | Sponge network | 1.877 | 0 | 0.206 | 0.0635 | 0.256 |
| 52891 | RP11-2H8.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-3607-3p;hsa-miR-766-3p | 14 | IL17RD | Sponge network | 1.089 | 0 | -0.019 | 0.94873 | 0.256 |
| 52892 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | ST6GALNAC3 | Sponge network | 0.101 | 0.60919 | -0.987 | 0 | 0.256 |
| 52893 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p | 12 | ZNF292 | Sponge network | 0.532 | 0.00505 | 0.276 | 0.04148 | 0.256 |
| 52894 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-874-3p | 14 | MXI1 | Sponge network | 0.381 | 0.25967 | -0.987 | 0 | 0.256 |
| 52895 | RP11-150O12.3 |
hsa-let-7a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-497-5p;hsa-miR-766-3p | 10 | TBL1XR1 | Sponge network | 4.508 | 0 | 0.578 | 0 | 0.256 |
| 52896 | RP1-59D14.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-193a-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326 | 10 | AKAP6 | Sponge network | 1.162 | 5.0E-5 | -1.586 | 3.0E-5 | 0.256 |
| 52897 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-590-3p | 15 | CHD1 | Sponge network | 0.311 | 0.10344 | 0.322 | 0.00215 | 0.256 |
| 52898 | TP73-AS1 |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-542-3p | 10 | CADM4 | Sponge network | -0.563 | 0.02545 | -0.635 | 0.00305 | 0.256 |
| 52899 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-324-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-629-3p;hsa-miR-942-5p | 20 | GPC6 | Sponge network | 0.853 | 0.15352 | -0.054 | 0.81465 | 0.256 |
| 52900 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-93-5p | 25 | RASSF2 | Sponge network | 0.082 | 0.55397 | -0.273 | 0.21961 | 0.256 |
| 52901 | RP11-774O3.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | DDX5 | Sponge network | -0.487 | 0.02157 | -0.099 | 0.17428 | 0.256 |
| 52902 | PROSER2-AS1 |
hsa-let-7a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-625-5p | 10 | ZMYM2 | Sponge network | 1.825 | 0 | 0.279 | 0.01273 | 0.256 |
| 52903 | RASSF8-AS1 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-429;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-671-5p | 11 | GATA2 | Sponge network | 0.335 | 0.20596 | -0.654 | 0.0016 | 0.256 |
| 52904 | FAM215B |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-502-3p;hsa-miR-664a-5p | 19 | ADARB1 | Sponge network | 0.817 | 3.0E-5 | -0.335 | 0.06631 | 0.256 |
| 52905 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | FREM2 | Sponge network | 0.246 | 0.59912 | -0.437 | 0.46353 | 0.256 |
| 52906 | RP11-403I13.8 |
hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-584-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-96-5p | 27 | PIK3R1 | Sponge network | 0.619 | 0.00157 | -0.133 | 0.36854 | 0.256 |
| 52907 | MIR600HG |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-625-5p | 12 | CEP170 | Sponge network | 0.594 | 0.41522 | 0.943 | 0 | 0.256 |
| 52908 | RP11-254F7.2 |
hsa-miR-126-3p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-27a-3p;hsa-miR-7-5p | 10 | IRS1 | Sponge network | 0.176 | 0.37402 | -0.026 | 0.88855 | 0.256 |
| 52909 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 25 | PIK3R1 | Sponge network | -0.663 | 0.10887 | -0.133 | 0.36854 | 0.256 |
| 52910 | HOTAIRM1 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-539-5p;hsa-miR-590-3p | 16 | RYR2 | Sponge network | -0.053 | 0.80685 | -1.466 | 0.00031 | 0.256 |
| 52911 | CTD-3064M3.3 |
hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-429 | 11 | SASH1 | Sponge network | -0.469 | 0.08721 | -0.502 | 0.00175 | 0.256 |
| 52912 | CTC-429P9.1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-409-3p | 24 | PPARA | Sponge network | 0.497 | 0.01451 | -0.379 | 0.00548 | 0.256 |
| 52913 | RP11-48B3.4 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 16 | PICALM | Sponge network | 1.757 | 0 | 0.086 | 0.23523 | 0.256 |
| 52914 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 23 | ARNT2 | Sponge network | -1.216 | 0 | -0.332 | 0.25851 | 0.256 |
| 52915 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 12 | ZNF521 | Sponge network | -0.355 | 0.0077 | -0.111 | 0.58068 | 0.256 |
| 52916 | CTC-296K1.4 |
hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-590-5p | 10 | EYA4 | Sponge network | -2.076 | 0.00548 | 0.3 | 0.36876 | 0.256 |
| 52917 | CTC-487M23.5 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-450b-5p | 29 | KIF1B | Sponge network | 0.912 | 0.00014 | -0.118 | 0.32258 | 0.256 |
| 52918 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p | 25 | PHC3 | Sponge network | -0.154 | 0.78939 | 0.212 | 0.0499 | 0.256 |
| 52919 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | PRKCE | Sponge network | -0.096 | 0.67958 | -0.403 | 0.00056 | 0.256 |
| 52920 | SOX2-OT |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-96-5p | 29 | ASTN1 | Sponge network | -1.264 | 6.0E-5 | -2.389 | 2.0E-5 | 0.256 |
| 52921 | CTB-179K24.3 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-940 | 10 | BAZ2B | Sponge network | 0.841 | 0.04055 | -0.276 | 0.02833 | 0.256 |
| 52922 | CTD-2554C21.2 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-502-5p | 11 | PTCH1 | Sponge network | -1.654 | 0.00144 | -0.1 | 0.6179 | 0.256 |
| 52923 | RP11-439A17.10 |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-5p | 11 | PTPRT | Sponge network | 0.051 | 0.95756 | -0.907 | 0.03727 | 0.256 |
| 52924 | MIR4458HG |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | FAM172A | Sponge network | -0.427 | 0.0623 | -0.57 | 0 | 0.256 |
| 52925 | CTD-2666L21.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-23a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-486-5p | 10 | ROBO1 | Sponge network | 0.368 | 0.20093 | -0.009 | 0.96154 | 0.256 |
| 52926 | PART1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-542-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-942-5p | 15 | FBXO31 | Sponge network | -2.925 | 0 | -0.085 | 0.42389 | 0.256 |
| 52927 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 11 | DACH2 | Sponge network | 0.225 | 0.30644 | -1.635 | 0.00034 | 0.256 |
| 52928 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-493-5p | 12 | PDE4DIP | Sponge network | 1.46 | 1.0E-5 | -0.282 | 0.0257 | 0.256 |
| 52929 | RP11-144G6.12 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-320a | 13 | VPS53 | Sponge network | 0.826 | 1.0E-5 | 0.103 | 0.22667 | 0.256 |
| 52930 | RP5-1198O20.4 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | UVRAG | Sponge network | 0.997 | 0.00066 | -0.017 | 0.83865 | 0.256 |
| 52931 | RP11-532F6.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 15 | LUZP2 | Sponge network | -0.896 | 0.00099 | -0.056 | 0.89416 | 0.256 |
| 52932 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | TIMP3 | Sponge network | 0.559 | 0.00026 | -0.546 | 0.02307 | 0.256 |
| 52933 | RP11-13K12.5 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | ST6GALNAC3 | Sponge network | -0.318 | 0.65847 | -0.987 | 0 | 0.256 |
| 52934 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-590-3p | 15 | HDAC9 | Sponge network | -0.227 | 0.31657 | -0.755 | 0.00495 | 0.256 |
| 52935 | AF131215.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-429 | 11 | LATS2 | Sponge network | -0.176 | 0.59692 | 0.159 | 0.2304 | 0.256 |
| 52936 | RP4-605O3.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-370-3p;hsa-miR-454-3p;hsa-miR-493-3p;hsa-miR-940 | 18 | CHD5 | Sponge network | 0.262 | 0.0475 | -0.922 | 0.01521 | 0.256 |
| 52937 | CTC-471J1.2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 10 | RNF144A | Sponge network | 1.11 | 1.0E-5 | 0.594 | 0.0009 | 0.256 |
| 52938 | RP11-130L8.1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p | 15 | RET | Sponge network | -0.663 | 0.10887 | -0.833 | 0.03517 | 0.256 |
| 52939 | TMEM9B-AS1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29b-3p;hsa-miR-542-3p;hsa-miR-582-5p | 19 | BCL2 | Sponge network | 0.529 | 0.00552 | -0.899 | 0 | 0.256 |
| 52940 | MRPL23-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-378a-5p;hsa-miR-629-3p | 18 | ASTN1 | Sponge network | -0.278 | 0.76559 | -2.389 | 2.0E-5 | 0.256 |
| 52941 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-942-5p | 19 | CREBBP | Sponge network | -0.49 | 0.04946 | 0.126 | 0.15186 | 0.256 |
| 52942 | RP11-263K19.4 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-625-5p | 20 | ANK2 | Sponge network | 0.859 | 0.00331 | -1.603 | 1.0E-5 | 0.256 |
| 52943 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-96-5p | 18 | CNTN1 | Sponge network | -0.326 | 0.14314 | -2.594 | 0 | 0.256 |
| 52944 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-769-5p;hsa-miR-93-5p | 24 | HBP1 | Sponge network | -0.611 | 0.09607 | -0.454 | 0 | 0.256 |
| 52945 | TINCR |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-500a-3p;hsa-miR-940;hsa-miR-942-5p | 24 | HLF | Sponge network | -0.664 | 0.14543 | -2.443 | 0 | 0.256 |
| 52946 | RP4-798A10.7 |
hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-374b-5p | 11 | SOX11 | Sponge network | 1.757 | 0 | 2.68 | 0 | 0.256 |
| 52947 | MEG3 |
hsa-miR-194-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-423-5p;hsa-miR-532-3p;hsa-miR-940 | 11 | DIRAS1 | Sponge network | 0.249 | 0.35902 | -2.518 | 0 | 0.256 |
| 52948 | DNAJC27-AS1 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-374a-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-96-5p | 16 | RPS6KA6 | Sponge network | -0.969 | 2.0E-5 | -2.989 | 0 | 0.256 |
| 52949 | RAB11B-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-331-3p;hsa-miR-550a-5p;hsa-miR-625-5p;hsa-miR-940 | 11 | TOM1L2 | Sponge network | -0.508 | 2.0E-5 | -0.994 | 0 | 0.256 |
| 52950 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-766-3p | 30 | MAPK1 | Sponge network | 0.373 | 0.00068 | -0.017 | 0.81465 | 0.256 |
| 52951 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-18a-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-331-3p;hsa-miR-3614-5p | 14 | PHC3 | Sponge network | 0.592 | 0.00014 | 0.212 | 0.0499 | 0.256 |
| 52952 | MIR22HG |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-744-3p | 33 | DIP2C | Sponge network | -1.047 | 0 | -0.436 | 0.01713 | 0.256 |
| 52953 | RP1-151F17.2 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | KANK1 | Sponge network | 0.222 | 0.29133 | -0.55 | 0.00134 | 0.256 |
| 52954 | LINC00092 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-769-5p | 11 | KALRN | Sponge network | -1.886 | 0 | -0.819 | 1.0E-5 | 0.256 |
| 52955 | RBPMS-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-27a-5p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-7-1-3p | 20 | THRB | Sponge network | 0.144 | 0.64989 | -1.117 | 0.0004 | 0.256 |
| 52956 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PTPRB | Sponge network | 0.385 | 0.10067 | 0.105 | 0.47122 | 0.256 |
| 52957 | RP11-21A7A.2 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-942-5p | 11 | NOTCH2 | Sponge network | -1.116 | 0.23686 | 0.138 | 0.35163 | 0.256 |
| 52958 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-3p;hsa-miR-940 | 45 | NTRK3 | Sponge network | -0.141 | 0.26434 | -1.77 | 3.0E-5 | 0.256 |
| 52959 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | CXXC4 | Sponge network | -0.899 | 6.0E-5 | -0.433 | 0.21055 | 0.256 |
| 52960 | RP5-1021I20.1 |
hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-194-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-3p | 11 | CDH23 | Sponge network | -0.785 | 0.00035 | -0.974 | 0.00034 | 0.256 |
| 52961 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 38 | AKAP11 | Sponge network | -1.697 | 0.00889 | 0.196 | 0.09825 | 0.256 |
| 52962 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-625-3p;hsa-miR-7-1-3p | 19 | RAP1A | Sponge network | 0.056 | 0.8494 | -0.929 | 0 | 0.256 |
| 52963 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SMARCA2 | Sponge network | 0.349 | 0.01695 | -0.529 | 0.00014 | 0.256 |
| 52964 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p | 17 | USP6 | Sponge network | 0.176 | 0.37402 | 0.188 | 0.3871 | 0.256 |
| 52965 | CTD-2528L19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITGAV | Sponge network | 0.953 | 0 | 0.337 | 0.01002 | 0.256 |
| 52966 | RP11-61L19.3 |
hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-374b-5p;hsa-miR-497-5p | 10 | SOX11 | Sponge network | 1.411 | 0 | 2.68 | 0 | 0.256 |
| 52967 | U91328.21 |
hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-301a-3p;hsa-miR-589-5p | 10 | CXXC4 | Sponge network | -0.735 | 0.15983 | -0.433 | 0.21055 | 0.256 |
| 52968 | RP11-458D21.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 25 | SOX5 | Sponge network | 0.663 | 0.00073 | -1.091 | 4.0E-5 | 0.256 |
| 52969 | CTBP1-AS2 |
hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-497-5p | 11 | CDK6 | Sponge network | 0.623 | 0 | 0.991 | 0 | 0.256 |
| 52970 | RP11-284N8.3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-93-5p | 15 | UNC80 | Sponge network | -0.31 | 0.33871 | -1.934 | 0 | 0.256 |
| 52971 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-3p | 27 | EBF3 | Sponge network | 0.421 | 0.00084 | -0.058 | 0.81437 | 0.256 |
| 52972 | AC003090.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-96-5p | 27 | CADM1 | Sponge network | -2.117 | 0.00161 | -0.9 | 0.00084 | 0.256 |
| 52973 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 29 | EYA4 | Sponge network | -0.487 | 0.02157 | 0.3 | 0.36876 | 0.256 |
| 52974 | ACTA2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | SON | Sponge network | 0.194 | 0.64278 | 0.168 | 0.04406 | 0.256 |
| 52975 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-3614-5p;hsa-miR-454-3p;hsa-miR-628-5p | 19 | CAMTA1 | Sponge network | 0.411 | 0.1335 | -0.436 | 1.0E-5 | 0.256 |
| 52976 | RP11-327P2.5 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-625-5p | 23 | ANK2 | Sponge network | -0.147 | 0.40452 | -1.603 | 1.0E-5 | 0.256 |
| 52977 | RP11-25K21.6 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-374a-3p | 16 | MDM4 | Sponge network | 0.489 | 0.25786 | 0.345 | 0.00762 | 0.256 |
| 52978 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-5p | 28 | USP12 | Sponge network | -2.915 | 0.00015 | -0.102 | 0.40768 | 0.256 |
| 52979 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 32 | AKAP11 | Sponge network | 0.219 | 0.1516 | 0.196 | 0.09825 | 0.256 |
| 52980 | BCYRN1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-3p;hsa-miR-590-3p | 23 | PRDM2 | Sponge network | 0.311 | 0.10344 | -0.258 | 0.00721 | 0.256 |
| 52981 | GS1-124K5.11 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-3p | 14 | TRIM13 | Sponge network | 0.494 | 0.00339 | 0.183 | 0.06968 | 0.256 |
| 52982 | RP11-181G12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30e-3p | 13 | CXXC4 | Sponge network | 0.556 | 0.00298 | -0.433 | 0.21055 | 0.256 |
| 52983 | AC011526.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-320b;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-92a-3p | 10 | GRIN2A | Sponge network | 0.342 | 0.03129 | -2.161 | 0 | 0.256 |
| 52984 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-98-5p | 14 | PRKAA2 | Sponge network | 1.344 | 0 | -2.462 | 0 | 0.256 |
| 52985 | HCG11 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | GRIK3 | Sponge network | 0.385 | 0.10067 | -3.208 | 0 | 0.256 |
| 52986 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 13 | FGF5 | Sponge network | 0.95 | 0.15943 | 0.549 | 0.28659 | 0.256 |
| 52987 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 19 | NR2C2 | Sponge network | 0.335 | 0.20596 | 0.21 | 0.03224 | 0.256 |
| 52988 | RP11-464F9.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 16 | RPS6KA3 | Sponge network | 0.959 | 0 | 0.256 | 0.02419 | 0.256 |
| 52989 | RP11-119F7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | PDGFC | Sponge network | 0.225 | 0.30644 | -0.33 | 0.06483 | 0.256 |
| 52990 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-942-5p | 11 | CREBBP | Sponge network | 0.622 | 0.00015 | 0.126 | 0.15186 | 0.256 |
| 52991 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-369-3p | 33 | FZD3 | Sponge network | -0.384 | 0.40652 | 0.104 | 0.60316 | 0.256 |
| 52992 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-877-5p | 13 | CRTC3 | Sponge network | 0.594 | 0.00064 | 0.164 | 0.16786 | 0.256 |
| 52993 | RP11-531A24.5 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-940 | 11 | CELF4 | Sponge network | 0.75 | 0.00054 | -1.135 | 0.00344 | 0.256 |
| 52994 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-942-5p | 46 | TMEM132B | Sponge network | -0.615 | 0.00015 | -0.83 | 0.01084 | 0.256 |
| 52995 | CALML3-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p | 18 | ITPKB | Sponge network | 0.95 | 0.15943 | -0.704 | 0.00106 | 0.256 |
| 52996 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-590-3p | 17 | KIF1B | Sponge network | 0.545 | 0.05789 | -0.118 | 0.32258 | 0.256 |
| 52997 | RP11-395G23.3 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-26a-2-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-423-5p | 11 | ABL1 | Sponge network | 0.381 | 0.25967 | -0.215 | 0.07804 | 0.256 |
| 52998 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-409-3p;hsa-miR-493-5p | 11 | AFF3 | Sponge network | 0.368 | 0.20093 | -2.373 | 0 | 0.256 |
| 52999 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-744-3p | 13 | CUL5 | Sponge network | 0.817 | 3.0E-5 | 0.026 | 0.77301 | 0.256 |
| 53000 | AC004540.4 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-3p | 17 | ERBB4 | Sponge network | -2.549 | 0.00528 | -1.975 | 2.0E-5 | 0.256 |
| 53001 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-33a-3p | 16 | NOTCH2 | Sponge network | 0.497 | 0.01451 | 0.138 | 0.35163 | 0.256 |
| 53002 | BCYRN1 |
hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-654-3p | 10 | USP25 | Sponge network | 0.311 | 0.10344 | -0.242 | 0.01397 | 0.256 |
| 53003 | AC005562.1 |
hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-32-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | EPG5 | Sponge network | 0.488 | 0.00084 | 0.101 | 0.3563 | 0.256 |
| 53004 | RP11-855A2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-197-3p;hsa-miR-20a-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | EBF1 | Sponge network | 2.094 | 0 | -0.384 | 0.08856 | 0.256 |
| 53005 | RP11-373D23.3 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 20 | ZFHX4 | Sponge network | -0.285 | 0.2172 | 0.018 | 0.95574 | 0.256 |
| 53006 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-345-5p;hsa-miR-590-3p | 20 | ARHGAP28 | Sponge network | -0.096 | 0.67958 | -0.911 | 0.00013 | 0.256 |
| 53007 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-432-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 19 | UNC80 | Sponge network | -0.295 | 0.02604 | -1.934 | 0 | 0.256 |
| 53008 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-223-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 12 | HMCN1 | Sponge network | 0.311 | 0.10344 | 0.809 | 0.00544 | 0.256 |
| 53009 | LINC00662 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | TET1 | Sponge network | 0.213 | 0.32297 | -0.135 | 0.52138 | 0.256 |
| 53010 | RP11-678G14.3 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-7-1-3p | 10 | NAV3 | Sponge network | -4.477 | 0 | 0.212 | 0.47729 | 0.256 |
| 53011 | SOX2-OT |
hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ITPKB | Sponge network | -1.264 | 6.0E-5 | -0.704 | 0.00106 | 0.256 |
| 53012 | TTC28-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-539-5p | 13 | TCF12 | Sponge network | 0.373 | 0.00068 | 0.174 | 0.13271 | 0.256 |
| 53013 | BVES-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-154-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-452-5p | 30 | GNAQ | Sponge network | -2.62 | 0 | -0.367 | 0.00102 | 0.256 |
| 53014 | BCYRN1 |
hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | TSC1 | Sponge network | 0.311 | 0.10344 | 0.103 | 0.26071 | 0.256 |
| 53015 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 37 | NOTCH2 | Sponge network | 0.083 | 0.66405 | 0.138 | 0.35163 | 0.256 |
| 53016 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 38 | AKAP11 | Sponge network | -0.576 | 0.10121 | 0.196 | 0.09825 | 0.256 |
| 53017 | HAS2-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 15 | RASSF2 | Sponge network | -0.011 | 0.967 | -0.273 | 0.21961 | 0.256 |
| 53018 | RP11-195F19.9 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-7-5p | 13 | CRTC1 | Sponge network | -0.726 | 0.00132 | -0.116 | 0.28412 | 0.256 |
| 53019 | SH3RF3-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-30b-3p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-7-1-3p | 17 | KCNA6 | Sponge network | 0.889 | 9.0E-5 | -1.531 | 0.00013 | 0.256 |
| 53020 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-96-5p | 21 | SCN9A | Sponge network | 0.853 | 0.15352 | -0.525 | 0.10593 | 0.256 |
| 53021 | AC034220.3 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 21 | SON | Sponge network | 0.309 | 0.07304 | 0.168 | 0.04406 | 0.256 |
| 53022 | RP11-390K5.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | SOX11 | Sponge network | 1.148 | 2.0E-5 | 2.68 | 0 | 0.256 |
| 53023 | RP11-244O19.1 |
hsa-let-7b-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-378a-5p | 12 | CACNA1E | Sponge network | -0.096 | 0.67958 | 1.752 | 0 | 0.256 |
| 53024 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 30 | WDFY3 | Sponge network | 0.727 | 3.0E-5 | -0.201 | 0.0967 | 0.256 |
| 53025 | TTC28-AS1 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p | 21 | CREB1 | Sponge network | 0.373 | 0.00068 | 0.203 | 0.00537 | 0.256 |
| 53026 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-378a-3p;hsa-miR-378c | 24 | WDFY3 | Sponge network | 1.03 | 0 | -0.201 | 0.0967 | 0.256 |
| 53027 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TBX18 | Sponge network | -0.513 | 0.04703 | 0.843 | 0.02945 | 0.256 |
| 53028 | RP5-1085F17.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 14 | AKAP6 | Sponge network | 0.541 | 0.00125 | -1.586 | 3.0E-5 | 0.256 |
| 53029 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 23 | CXXC4 | Sponge network | 0.603 | 0.00127 | -0.433 | 0.21055 | 0.256 |
| 53030 | BCYRN1 |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | PCM1 | Sponge network | 0.311 | 0.10344 | 0.164 | 0.20307 | 0.256 |
| 53031 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-27a-3p;hsa-miR-29b-2-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 10 | GPAM | Sponge network | 1.378 | 0 | 0.142 | 0.29732 | 0.256 |
| 53032 | BCYRN1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-935;hsa-miR-96-5p | 31 | PCDH10 | Sponge network | 0.311 | 0.10344 | -1.644 | 0.0078 | 0.256 |
| 53033 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 10 | HIP1 | Sponge network | -0.376 | 0.00337 | 0.774 | 0 | 0.256 |
| 53034 | RP11-195F19.9 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p | 13 | HIP1 | Sponge network | -0.726 | 0.00132 | 0.774 | 0 | 0.256 |
| 53035 | H1FX-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | LRP1B | Sponge network | -0.206 | 0.12942 | -2.163 | 0 | 0.256 |
| 53036 | CTD-2303H24.2 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | TET1 | Sponge network | 0.415 | 0.14657 | -0.135 | 0.52138 | 0.256 |
| 53037 | BMS1P20 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 20 | EGLN1 | Sponge network | 0.34 | 0.00273 | -0.127 | 0.1536 | 0.256 |
| 53038 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | LRRC7 | Sponge network | 0.603 | 0.00127 | -1.341 | 0.01193 | 0.256 |
| 53039 | FAM157C |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-411-5p | 14 | VPS53 | Sponge network | 0.91 | 0 | 0.103 | 0.22667 | 0.256 |
| 53040 | HOXA-AS2 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | PCDH9 | Sponge network | 0.61 | 0.01593 | -1.823 | 4.0E-5 | 0.256 |
| 53041 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-93-3p;hsa-miR-96-5p | 21 | BMPR2 | Sponge network | 2.306 | 9.0E-5 | 0.206 | 0.0635 | 0.256 |
| 53042 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-744-3p | 18 | CUL5 | Sponge network | 0.603 | 0.00127 | 0.026 | 0.77301 | 0.256 |
| 53043 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 19 | CHD2 | Sponge network | 0.064 | 0.72934 | 0.049 | 0.55371 | 0.256 |
| 53044 | RP1-122P22.2 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-500a-3p | 10 | ERC1 | Sponge network | 0.065 | 0.73468 | -0.108 | 0.38794 | 0.256 |
| 53045 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-26a-2-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-5p;hsa-miR-766-3p | 16 | TBL1XR1 | Sponge network | 0.415 | 0.14657 | 0.578 | 0 | 0.256 |
| 53046 | RP11-611E13.2 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-497-5p | 13 | RICTOR | Sponge network | 0.822 | 0 | 0.388 | 0.00188 | 0.256 |
| 53047 | LINC00886 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-382-5p;hsa-miR-450b-5p;hsa-miR-590-5p | 11 | TMEFF2 | Sponge network | -0.371 | 0.24604 | -2.426 | 0 | 0.256 |
| 53048 | BZRAP1-AS1 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-484 | 14 | ITGA5 | Sponge network | -0.465 | 0.04703 | 0.452 | 0.03524 | 0.256 |
| 53049 | SNHG16 |
hsa-let-7a-3p;hsa-miR-125b-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-497-5p | 11 | ZMYM2 | Sponge network | 0.961 | 0 | 0.279 | 0.01273 | 0.256 |
| 53050 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-330-3p | 12 | PRKCE | Sponge network | -0.785 | 0.00035 | -0.403 | 0.00056 | 0.256 |
| 53051 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-671-5p | 37 | CAMTA1 | Sponge network | 0.311 | 0.10344 | -0.436 | 1.0E-5 | 0.256 |
| 53052 | RP11-83A24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 18 | USP6 | Sponge network | 0.417 | 0.00445 | 0.188 | 0.3871 | 0.256 |
| 53053 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ITGAV | Sponge network | 0.385 | 0.10067 | 0.337 | 0.01002 | 0.256 |
| 53054 | RP11-379F4.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p | 22 | MDM4 | Sponge network | 1.316 | 0 | 0.345 | 0.00762 | 0.256 |
| 53055 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-361-3p | 10 | MNT | Sponge network | -0.309 | 0.1145 | -0.157 | 0.06598 | 0.256 |
| 53056 | RP4-669P10.16 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-511-5p;hsa-miR-542-3p | 12 | RBMS3 | Sponge network | -0.301 | 0.06597 | -0.519 | 0.03551 | 0.256 |
| 53057 | C4B-AS1 |
hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-940 | 16 | GRIK3 | Sponge network | 2.306 | 9.0E-5 | -3.208 | 0 | 0.256 |
| 53058 | HOXB-AS3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-3p;hsa-miR-223-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-452-5p;hsa-miR-550a-3p;hsa-miR-877-5p | 13 | PGR | Sponge network | 0.343 | 0.28887 | -1.704 | 0 | 0.256 |
| 53059 | RP11-1277A3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 33 | TCF4 | Sponge network | 0.453 | 0.01839 | 0.016 | 0.92271 | 0.256 |
| 53060 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-5p | 27 | RNASEL | Sponge network | -1.216 | 0 | -0.272 | 0.01796 | 0.256 |
| 53061 | TRIM52-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-362-5p | 10 | KCNJ5 | Sponge network | -0.418 | 0.00068 | -0.281 | 0.3339 | 0.256 |
| 53062 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | PRKAB2 | Sponge network | -2.915 | 0.00015 | -0.367 | 0.01371 | 0.256 |
| 53063 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-24-2-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-7-1-3p | 13 | PCDH9 | Sponge network | 0.238 | 0.70544 | -1.823 | 4.0E-5 | 0.256 |
| 53064 | ZSCAN16-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p | 46 | DST | Sponge network | -0.342 | 0.02288 | -0.713 | 0.00096 | 0.256 |
| 53065 | HCG22 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p | 13 | HECA | Sponge network | 0.816 | 0.32368 | -0.217 | 0.03463 | 0.256 |
| 53066 | ADAMTS9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | AKAP13 | Sponge network | -2.452 | 0 | 0.028 | 0.7729 | 0.256 |
| 53067 | RP5-1139B12.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-454-3p;hsa-miR-93-3p | 16 | SSBP2 | Sponge network | 0.598 | 0.38481 | -1.58 | 0 | 0.256 |
| 53068 | RP11-686O6.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-324-5p;hsa-miR-532-3p | 15 | PBX1 | Sponge network | 0.453 | 0.01971 | -1.206 | 0 | 0.256 |
| 53069 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-16-1-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-744-3p | 16 | HLF | Sponge network | 0.368 | 0.20093 | -2.443 | 0 | 0.256 |
| 53070 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-7-1-3p | 15 | MLLT10 | Sponge network | 0.056 | 0.8494 | 0.105 | 0.33292 | 0.256 |
| 53071 | RP11-166D19.1 |
hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-24-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-542-3p | 13 | PIM1 | Sponge network | -0.576 | 0.10121 | -1.114 | 0 | 0.256 |
| 53072 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-96-5p | 19 | PIK3R1 | Sponge network | 0.622 | 0.00015 | -0.133 | 0.36854 | 0.256 |
| 53073 | C4A-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-18a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-3607-3p | 10 | ARHGAP28 | Sponge network | 3.345 | 0 | -0.911 | 0.00013 | 0.256 |
| 53074 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-629-3p | 20 | GPC6 | Sponge network | 0.331 | 0.57247 | -0.054 | 0.81465 | 0.256 |
| 53075 | RP11-20I23.13 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-582-3p;hsa-miR-671-5p | 26 | ADARB1 | Sponge network | 0.505 | 0.0014 | -0.335 | 0.06631 | 0.256 |
| 53076 | NR2F1-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | CEP170 | Sponge network | -0.49 | 0.04946 | 0.943 | 0 | 0.256 |
| 53077 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 21 | PRKAB2 | Sponge network | -0.663 | 0.10887 | -0.367 | 0.01371 | 0.256 |
| 53078 | AC007383.3 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-199b-5p;hsa-miR-22-3p;hsa-miR-24-2-5p;hsa-miR-330-5p;hsa-miR-424-5p | 12 | PPM1L | Sponge network | 0.104 | 0.48031 | -0.537 | 0.00616 | 0.256 |
| 53079 | RP11-747H7.3 |
hsa-miR-10b-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 13 | VPS53 | Sponge network | 1.379 | 0 | 0.103 | 0.22667 | 0.256 |
| 53080 | RP11-35G9.5 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p | 12 | ARNT2 | Sponge network | 0.622 | 0.00015 | -0.332 | 0.25851 | 0.256 |
| 53081 | HOXB-AS1 |
hsa-miR-128-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-429 | 10 | PAIP2 | Sponge network | -0.154 | 0.53954 | -0.515 | 0 | 0.256 |
| 53082 | AC007277.3 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-215-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-654-3p | 12 | AKAP11 | Sponge network | 2.377 | 0.00108 | 0.196 | 0.09825 | 0.256 |
| 53083 | H19 |
hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-339-5p;hsa-miR-576-5p;hsa-miR-589-3p | 17 | PTPRD | Sponge network | 2.439 | 0 | -1.005 | 0.00064 | 0.256 |
| 53084 | RP11-539I5.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | MYO9A | Sponge network | -0.468 | 0.32587 | -0.147 | 0.32373 | 0.256 |
| 53085 | RP11-440L14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-532-3p;hsa-miR-590-3p | 13 | FOXP1 | Sponge network | 0.545 | 0.05789 | 0.042 | 0.74368 | 0.256 |
| 53086 | SNHG14 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | HACE1 | Sponge network | -1.111 | 0.0003 | -0.072 | 0.55239 | 0.256 |
| 53087 | PAN3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-655-3p | 11 | PIK3C2A | Sponge network | 0.062 | 0.55274 | 0.061 | 0.53776 | 0.256 |
| 53088 | BOLA3-AS1 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-7-1-3p | 24 | LRIG1 | Sponge network | -0.246 | 0.35034 | -0.537 | 0.00588 | 0.256 |
| 53089 | RP1-122P22.2 |
hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-629-3p | 14 | ZBTB4 | Sponge network | 0.065 | 0.73468 | -0.739 | 0 | 0.256 |
| 53090 | RP4-613B23.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-671-5p | 18 | PTPRD | Sponge network | -0.309 | 0.1145 | -1.005 | 0.00064 | 0.256 |
| 53091 | RP1-43E13.2 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-150-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-532-3p | 12 | PBX1 | Sponge network | -0.034 | 0.83228 | -1.206 | 0 | 0.256 |
| 53092 | KB-1572G7.2 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 14 | SLC5A3 | Sponge network | 0.924 | 0 | 0.493 | 5.0E-5 | 0.256 |
| 53093 | RP11-262H14.4 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-7-1-3p | 12 | TSC1 | Sponge network | -0.121 | 0.53195 | 0.103 | 0.26071 | 0.256 |
| 53094 | TRIM52-AS1 |
hsa-let-7a-5p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | PRKAA2 | Sponge network | -0.418 | 0.00068 | -2.462 | 0 | 0.256 |
| 53095 | ERVH48-1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p | 11 | CEP170 | Sponge network | 1.04 | 0.00023 | 0.943 | 0 | 0.256 |
| 53096 | OIP5-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 40 | DMD | Sponge network | 0.421 | 0.00084 | -1.506 | 0 | 0.256 |
| 53097 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-190a-5p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-590-3p | 12 | MYCBP2 | Sponge network | 0.385 | 0.10067 | -0.027 | 0.82016 | 0.256 |
| 53098 | AC025171.1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | NEDD4 | Sponge network | 0.998 | 0 | 0.02 | 0.89834 | 0.256 |
| 53099 | RP11-967K21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-181a-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-93-5p | 15 | ARNT2 | Sponge network | 0.056 | 0.81195 | -0.332 | 0.25851 | 0.256 |
| 53100 | CTD-3064M3.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-455-3p | 19 | MAF | Sponge network | -0.469 | 0.08721 | -1.257 | 0 | 0.256 |
| 53101 | RBPMS-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-93-5p | 14 | RNASEL | Sponge network | 0.144 | 0.64989 | -0.272 | 0.01796 | 0.256 |
| 53102 | RP4-605O3.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-590-3p | 10 | MAP3K12 | Sponge network | 0.262 | 0.0475 | -0.057 | 0.59369 | 0.256 |
| 53103 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-425-5p | 15 | FGF5 | Sponge network | -0.384 | 0.40652 | 0.549 | 0.28659 | 0.256 |
| 53104 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10a-3p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p | 19 | MEF2C | Sponge network | 0.594 | 0.00064 | -0.499 | 0.01448 | 0.256 |
| 53105 | LINC00473 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-339-5p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 12 | PPP2R2C | Sponge network | -1.203 | 0.03881 | -0.959 | 0.07428 | 0.256 |
| 53106 | AC009948.5 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-590-3p | 14 | TLE4 | Sponge network | 0.532 | 0.00505 | -1.157 | 0 | 0.256 |
| 53107 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-93-3p | 15 | PTPN11 | Sponge network | 0.862 | 0.06213 | 0.431 | 2.0E-5 | 0.256 |
| 53108 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-3p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 55 | BMPR2 | Sponge network | -1.047 | 0 | 0.206 | 0.0635 | 0.256 |
| 53109 | U91328.19 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-539-5p;hsa-miR-93-3p | 10 | ZNF382 | Sponge network | -0.303 | 0.21002 | -0.494 | 0.03831 | 0.256 |
| 53110 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-590-3p;hsa-miR-96-5p | 12 | HOOK3 | Sponge network | 0.538 | 0.00076 | 0.273 | 0.09957 | 0.256 |
| 53111 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 13 | CITED2 | Sponge network | 0.194 | 0.64278 | -1.545 | 0 | 0.256 |
| 53112 | C20orf166-AS1 |
hsa-miR-127-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-542-3p;hsa-miR-93-5p | 14 | JUN | Sponge network | -2.69 | 0.0001 | -0.932 | 0 | 0.256 |
| 53113 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | HECA | Sponge network | -0.727 | 0.04726 | -0.217 | 0.03463 | 0.256 |
| 53114 | RP11-473I1.9 |
hsa-miR-106b-5p;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 31 | DIP2C | Sponge network | 0.683 | 1.0E-5 | -0.436 | 0.01713 | 0.256 |
| 53115 | RP4-545C24.1 |
hsa-let-7a-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-93-3p;hsa-miR-96-5p | 11 | BMPR2 | Sponge network | 0.363 | 0.02231 | 0.206 | 0.0635 | 0.256 |
| 53116 | AC003090.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 16 | UNC5C | Sponge network | -2.117 | 0.00161 | -0.178 | 0.40214 | 0.256 |
| 53117 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-590-3p | 19 | SOX5 | Sponge network | 0.765 | 7.0E-5 | -1.091 | 4.0E-5 | 0.256 |
| 53118 | LINC00672 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 21 | NIN | Sponge network | -0.227 | 0.31657 | -0.253 | 0.13794 | 0.256 |
| 53119 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429 | 22 | MYBL1 | Sponge network | 0.95 | 0.15943 | 0.494 | 0.02152 | 0.256 |
| 53120 | AC018890.6 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-338-5p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-550a-3p;hsa-miR-7-1-3p | 20 | BRD3 | Sponge network | 1.61 | 0.00961 | 0.37 | 0.00013 | 0.256 |
| 53121 | RP11-767N6.7 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-154-5p;hsa-miR-214-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-505-5p | 12 | TBL1XR1 | Sponge network | 0.466 | 0.00155 | 0.578 | 0 | 0.256 |
| 53122 | GUSBP11 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-22-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-628-5p | 10 | MAT2A | Sponge network | 0.856 | 0 | -0.073 | 0.36195 | 0.256 |
| 53123 | RP11-67L2.2 |
hsa-miR-130b-3p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-590-3p | 11 | RASA1 | Sponge network | 0.72 | 0.0001 | -0.002 | 0.98204 | 0.256 |
| 53124 | RP11-116O18.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 22 | BCL2 | Sponge network | 0.05 | 0.95483 | -0.899 | 0 | 0.256 |
| 53125 | FAM13A-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-628-5p;hsa-miR-671-5p | 13 | CDH23 | Sponge network | -0.83 | 0 | -0.974 | 0.00034 | 0.256 |
| 53126 | CTD-3064M3.3 |
hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-455-3p | 22 | PPM1L | Sponge network | -0.469 | 0.08721 | -0.537 | 0.00616 | 0.256 |
| 53127 | RP11-798G7.7 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-455-3p;hsa-miR-539-5p | 12 | ETV1 | Sponge network | 0.858 | 3.0E-5 | -0.096 | 0.67674 | 0.256 |
| 53128 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-940 | 31 | BHLHE41 | Sponge network | -0.899 | 6.0E-5 | 0.353 | 0.12753 | 0.256 |
| 53129 | RP11-218M22.1 |
hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-744-5p;hsa-miR-766-3p | 22 | CHD5 | Sponge network | 0.197 | 0.25618 | -0.922 | 0.01521 | 0.256 |
| 53130 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 38 | KIF1B | Sponge network | 1.757 | 0 | -0.118 | 0.32258 | 0.255 |
| 53131 | RP11-46C24.7 |
hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 11 | GALNT13 | Sponge network | 0.392 | 0.0278 | -0.857 | 0.07439 | 0.255 |
| 53132 | TMEM9B-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-589-3p;hsa-miR-940 | 18 | TRPS1 | Sponge network | 0.529 | 0.00552 | -0.191 | 0.44109 | 0.255 |
| 53133 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p | 12 | FGF5 | Sponge network | 0.051 | 0.83388 | 0.549 | 0.28659 | 0.255 |
| 53134 | RP11-263K19.4 |
hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-625-5p | 20 | JAZF1 | Sponge network | 0.859 | 0.00331 | -0.377 | 0.02677 | 0.255 |
| 53135 | TP73-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p | 19 | ERCC4 | Sponge network | -0.563 | 0.02545 | 0.126 | 0.15266 | 0.255 |
| 53136 | LINC00578 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-342-3p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-664a-3p | 15 | MXI1 | Sponge network | 0.811 | 0.03228 | -0.987 | 0 | 0.255 |
| 53137 | NNT-AS1 |
hsa-let-7a-3p;hsa-miR-155-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-338-5p;hsa-miR-342-3p | 10 | ZNRF3 | Sponge network | 0.799 | 1.0E-5 | 0.637 | 0.00172 | 0.255 |
| 53138 | SSSCA1-AS1 |
hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | BRD3 | Sponge network | 0.487 | 0.00018 | 0.37 | 0.00013 | 0.255 |
| 53139 | RP5-1021I20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-93-5p | 14 | IGSF10 | Sponge network | -0.785 | 0.00035 | -1.751 | 1.0E-5 | 0.255 |
| 53140 | RP11-254F7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-5p | 24 | MTSS1 | Sponge network | 0.176 | 0.37402 | -0.81 | 0.00035 | 0.255 |
| 53141 | RP11-35G9.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | FAM172A | Sponge network | 0.657 | 4.0E-5 | -0.57 | 0 | 0.255 |
| 53142 | AC127904.2 |
hsa-miR-142-5p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-374b-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | TMEM30A | Sponge network | 1.308 | 0 | 0.096 | 0.31031 | 0.255 |
| 53143 | RP11-1277A3.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-96-5p | 19 | SPRY3 | Sponge network | 0.453 | 0.01839 | 0.016 | 0.91286 | 0.255 |
| 53144 | RP11-379F4.7 |
hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p | 11 | WDFY3 | Sponge network | 1.316 | 0 | -0.201 | 0.0967 | 0.255 |
| 53145 | MIR4458HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-708-3p;hsa-miR-92a-1-5p | 22 | PRKAB2 | Sponge network | -0.427 | 0.0623 | -0.367 | 0.01371 | 0.255 |
| 53146 | RP11-395G23.3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-324-5p;hsa-miR-374a-5p;hsa-miR-590-3p | 13 | CDH19 | Sponge network | 0.381 | 0.25967 | -3.434 | 0 | 0.255 |
| 53147 | CTC-429P9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p | 20 | BHLHE41 | Sponge network | 0.178 | 0.29106 | 0.353 | 0.12753 | 0.255 |
| 53148 | RP1-39G22.7 |
hsa-miR-125b-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-374b-5p | 12 | RASAL2 | Sponge network | 0.439 | 0.00313 | 0.869 | 0 | 0.255 |
| 53149 | CD27-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p | 13 | KLF10 | Sponge network | 0.177 | 0.16681 | -0.783 | 0 | 0.255 |
| 53150 | RP11-575H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-590-3p | 10 | TMTC3 | Sponge network | -0.246 | 0.74154 | 0.123 | 0.22189 | 0.255 |
| 53151 | GAS6-AS2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940;hsa-miR-942-5p | 32 | NOTCH2 | Sponge network | 0.16 | 0.50785 | 0.138 | 0.35163 | 0.255 |
| 53152 | HCG11 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 23 | DICER1 | Sponge network | 0.385 | 0.10067 | 0.042 | 0.674 | 0.255 |
| 53153 | AP001347.6 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-362-3p | 12 | IGF2 | Sponge network | -1.161 | 0.00591 | 0.848 | 0.03842 | 0.255 |
| 53154 | RP4-758J18.10 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-450b-5p | 13 | ZFHX3 | Sponge network | -0.091 | 0.71468 | 0.389 | 0.01363 | 0.255 |
| 53155 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-660-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | EPAS1 | Sponge network | -1.697 | 0.00889 | -0.184 | 0.14418 | 0.255 |
| 53156 | AC010226.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | PRKCE | Sponge network | -0.811 | 2.0E-5 | -0.403 | 0.00056 | 0.255 |
| 53157 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-500a-5p | 18 | SASH1 | Sponge network | 0.622 | 0.00015 | -0.502 | 0.00175 | 0.255 |
| 53158 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-590-3p | 11 | LRRC4 | Sponge network | -0.227 | 0.31657 | -1.774 | 0 | 0.255 |
| 53159 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | RAP1A | Sponge network | -0.663 | 0.10887 | -0.929 | 0 | 0.255 |
| 53160 | RP1-39G22.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 11 | SON | Sponge network | 0.439 | 0.00313 | 0.168 | 0.04406 | 0.255 |
| 53161 | RAB30-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-3p | 12 | UBR3 | Sponge network | 0.752 | 0 | -0.021 | 0.82774 | 0.255 |
| 53162 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 13 | PRDM5 | Sponge network | 0.349 | 0.01695 | -1.09 | 0.00014 | 0.255 |
| 53163 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | FOXO1 | Sponge network | 0.657 | 4.0E-5 | -0.141 | 0.33874 | 0.255 |
| 53164 | RP11-395G23.3 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | PTGFR | Sponge network | 0.381 | 0.25967 | -0.233 | 0.45421 | 0.255 |
| 53165 | RP11-806O11.1 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | PIK3C2A | Sponge network | 0.284 | 0.52463 | 0.061 | 0.53776 | 0.255 |
| 53166 | RP11-127B20.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-7-1-3p | 10 | PRUNE2 | Sponge network | 0.411 | 0.1335 | -1.733 | 0.00022 | 0.255 |
| 53167 | RP11-277P12.20 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 21 | ANK2 | Sponge network | 0.687 | 0.02753 | -1.603 | 1.0E-5 | 0.255 |
| 53168 | LINC00092 |
hsa-let-7a-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-423-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-940 | 13 | ZFP36L1 | Sponge network | -1.886 | 0 | -0.505 | 8.0E-5 | 0.255 |
| 53169 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-34a-5p;hsa-miR-375;hsa-miR-532-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | BNC2 | Sponge network | 0.687 | 0.02753 | -0.491 | 0.09988 | 0.255 |
| 53170 | GUSBP11 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-193b-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-376c-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-628-5p;hsa-miR-654-3p | 20 | BMPR2 | Sponge network | 0.856 | 0 | 0.206 | 0.0635 | 0.255 |
| 53171 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-505-3p;hsa-miR-590-3p | 17 | USP6 | Sponge network | 0.919 | 0 | 0.188 | 0.3871 | 0.255 |
| 53172 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-628-5p;hsa-miR-96-5p | 40 | TCF4 | Sponge network | 0.594 | 0.41522 | 0.016 | 0.92271 | 0.255 |
| 53173 | WDR86-AS1 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-455-3p | 13 | TRPS1 | Sponge network | 0.348 | 0.32912 | -0.191 | 0.44109 | 0.255 |
| 53174 | FAM13A-AS1 |
hsa-let-7d-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 54 | RUNX1T1 | Sponge network | -0.83 | 0 | -0.344 | 0.2374 | 0.255 |
| 53175 | A2M-AS1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SEMA3E | Sponge network | -0.08 | 0.90092 | -1.717 | 0.00137 | 0.255 |
| 53176 | AP001347.6 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 42 | NFIB | Sponge network | -1.161 | 0.00591 | 0.023 | 0.90795 | 0.255 |
| 53177 | RP4-791M13.3 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-7-1-3p;hsa-miR-940 | 13 | TAL1 | Sponge network | 0.419 | 0.0319 | -0.462 | 0.00574 | 0.255 |
| 53178 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-338-5p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-500b-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 25 | SCN9A | Sponge network | -1.264 | 6.0E-5 | -0.525 | 0.10593 | 0.255 |
| 53179 | RP4-758J18.10 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-331-3p | 17 | EPHA7 | Sponge network | -0.091 | 0.71468 | -2.197 | 3.0E-5 | 0.255 |
| 53180 | RBPMS-AS1 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p | 10 | SYNM | Sponge network | 0.144 | 0.64989 | -2.309 | 1.0E-5 | 0.255 |
| 53181 | CTB-179K24.3 |
hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-324-5p | 11 | PIK3C2A | Sponge network | 0.841 | 0.04055 | 0.061 | 0.53776 | 0.255 |
| 53182 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-33a-3p | 13 | ARID5B | Sponge network | 0.368 | 0.20093 | 0.342 | 0.01676 | 0.255 |
| 53183 | RP11-276H19.2 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-338-5p;hsa-miR-500a-5p;hsa-miR-96-5p | 15 | PRKAA2 | Sponge network | -0.472 | 0.48851 | -2.462 | 0 | 0.255 |
| 53184 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 25 | SASH1 | Sponge network | 0.657 | 4.0E-5 | -0.502 | 0.00175 | 0.255 |
| 53185 | TAPT1-AS1 |
hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p | 12 | RASAL2 | Sponge network | 0.403 | 2.0E-5 | 0.869 | 0 | 0.255 |
| 53186 | RP11-13K12.1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421 | 21 | LIMCH1 | Sponge network | -0.154 | 0.78939 | -0.915 | 1.0E-5 | 0.255 |
| 53187 | FOXN3-AS1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 10 | SSBP2 | Sponge network | -0.399 | 0.00917 | -1.58 | 0 | 0.255 |
| 53188 | RP11-536O18.1 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p | 11 | DIP2C | Sponge network | -0.074 | 0.90758 | -0.436 | 0.01713 | 0.255 |
| 53189 | AF131215.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-671-5p | 18 | CYLD | Sponge network | -0.176 | 0.59692 | -0.367 | 0.00171 | 0.255 |
| 53190 | RP11-181G12.2 |
hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-331-3p;hsa-miR-486-5p | 10 | SYNM | Sponge network | 0.556 | 0.00298 | -2.309 | 1.0E-5 | 0.255 |
| 53191 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-93-3p;hsa-miR-940 | 40 | QKI | Sponge network | -0.322 | 0.25945 | -0.333 | 0.04111 | 0.255 |
| 53192 | RP11-53O19.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 15 | LATS1 | Sponge network | 1.205 | 0 | 0.109 | 0.34208 | 0.255 |
| 53193 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-935 | 29 | MTSS1 | Sponge network | -0.303 | 0.21002 | -0.81 | 0.00035 | 0.255 |
| 53194 | CTC-550B14.7 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-374a-3p | 11 | CREB1 | Sponge network | 1.161 | 0 | 0.203 | 0.00537 | 0.255 |
| 53195 | OIP5-AS1 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 13 | PAWR | Sponge network | 0.421 | 0.00084 | 0.636 | 0 | 0.255 |
| 53196 | RP11-141C7.4 |
hsa-miR-129-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-497-5p | 11 | CCDC6 | Sponge network | 2.412 | 0 | 0.27 | 0.02555 | 0.255 |
| 53197 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 19 | USP6 | Sponge network | 0.392 | 0.0278 | 0.188 | 0.3871 | 0.255 |
| 53198 | RP11-57H14.4 |
hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-625-5p | 13 | TMTC2 | Sponge network | 0.083 | 0.66405 | -0.215 | 0.18248 | 0.255 |
| 53199 | RP11-25K19.1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-590-3p | 12 | GLIPR1 | Sponge network | -1.057 | 0.00029 | 0.146 | 0.3276 | 0.255 |
| 53200 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 19 | SCN9A | Sponge network | 0.765 | 7.0E-5 | -0.525 | 0.10593 | 0.255 |
| 53201 | RP11-686D22.8 |
hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-590-3p | 12 | PCDH9 | Sponge network | 0.898 | 1.0E-5 | -1.823 | 4.0E-5 | 0.255 |
| 53202 | MIR497HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-96-5p | 26 | HBP1 | Sponge network | -1.439 | 4.0E-5 | -0.454 | 0 | 0.255 |
| 53203 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-454-3p;hsa-miR-590-3p | 27 | ESR1 | Sponge network | 0.082 | 0.55654 | -1.035 | 3.0E-5 | 0.255 |
| 53204 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-455-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-940 | 40 | NTRK2 | Sponge network | -0.769 | 0.00035 | -0.736 | 0.06495 | 0.255 |
| 53205 | ZNF32-AS2 |
hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-342-3p | 11 | CCDC6 | Sponge network | 1.552 | 7.0E-5 | 0.27 | 0.02555 | 0.255 |
| 53206 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | ABCA10 | Sponge network | 0.082 | 0.55654 | -0.571 | 0.03674 | 0.255 |
| 53207 | ADIRF-AS1 |
hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-92a-3p | 13 | CSDE1 | Sponge network | -0.223 | 0.47816 | -0.493 | 0 | 0.255 |
| 53208 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-378a-5p | 16 | PDS5B | Sponge network | 1.03 | 0 | 0.259 | 0.00962 | 0.255 |
| 53209 | RP11-774O3.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | DLG1 | Sponge network | -0.487 | 0.02157 | -0.014 | 0.8926 | 0.255 |
| 53210 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-671-5p | 32 | BMPR2 | Sponge network | 0.381 | 0.25967 | 0.206 | 0.0635 | 0.255 |
| 53211 | RP11-296I10.3 |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-335-5p;hsa-miR-450b-5p | 11 | RASSF8 | Sponge network | 0.592 | 0.00014 | -0.122 | 0.65591 | 0.255 |
| 53212 | RP11-244H3.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-409-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 30 | AFF1 | Sponge network | 0.659 | 3.0E-5 | -0.384 | 0.00053 | 0.255 |
| 53213 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-454-3p | 24 | FAT3 | Sponge network | 0.001 | 0.99753 | -1.601 | 0.00051 | 0.255 |
| 53214 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | PRKAB2 | Sponge network | -0.342 | 0.02288 | -0.367 | 0.01371 | 0.255 |
| 53215 | SOX2-OT |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-485-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-590-5p | 11 | PDZD2 | Sponge network | -1.264 | 6.0E-5 | -0.909 | 3.0E-5 | 0.255 |
| 53216 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 36 | AKAP11 | Sponge network | -0.737 | 0.06182 | 0.196 | 0.09825 | 0.255 |
| 53217 | RP11-400K9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-7-1-3p | 14 | FOXO1 | Sponge network | -0.611 | 0.09607 | -0.141 | 0.33874 | 0.255 |
| 53218 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-3p;hsa-miR-409-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 29 | ATRX | Sponge network | -0.901 | 0.00019 | 0.261 | 0.04408 | 0.255 |
| 53219 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-330-3p | 11 | GREM1 | Sponge network | 0.056 | 0.8494 | 0.902 | 0.01691 | 0.255 |
| 53220 | RP11-473I1.5 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p | 12 | SPRY3 | Sponge network | 0.542 | 0.00054 | 0.016 | 0.91286 | 0.255 |
| 53221 | RP1-228H13.5 |
hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 11 | DDX3X | Sponge network | 1.397 | 0 | -0.152 | 0.07686 | 0.255 |
| 53222 | AC018890.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | ARID5B | Sponge network | 1.61 | 0.00961 | 0.342 | 0.01676 | 0.255 |
| 53223 | RP11-458D21.1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p | 10 | CELF4 | Sponge network | 0.663 | 0.00073 | -1.135 | 0.00344 | 0.255 |
| 53224 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940;hsa-miR-96-5p | 66 | AFF4 | Sponge network | 0.064 | 0.72934 | -0.266 | 0.01565 | 0.255 |
| 53225 | HOXD-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-455-3p;hsa-miR-500a-5p | 24 | RUNX1T1 | Sponge network | -0.208 | 0.65592 | -0.344 | 0.2374 | 0.255 |
| 53226 | PCA3 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 39 | CREB3L2 | Sponge network | -0.709 | 0.18168 | -0.525 | 2.0E-5 | 0.255 |
| 53227 | RP11-384K6.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-339-5p | 13 | NAV3 | Sponge network | 0.946 | 0 | 0.212 | 0.47729 | 0.255 |
| 53228 | AC103563.9 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-577 | 10 | GRIN2A | Sponge network | -6.057 | 0 | -2.161 | 0 | 0.255 |
| 53229 | AC009948.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-96-5p | 16 | UNC5C | Sponge network | 0.532 | 0.00505 | -0.178 | 0.40214 | 0.255 |
| 53230 | RP11-359B12.2 |
hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29b-2-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 10 | GUCY1A2 | Sponge network | 0.064 | 0.72934 | 1.213 | 0 | 0.255 |
| 53231 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-365a-3p;hsa-miR-576-5p | 26 | RICTOR | Sponge network | 0.395 | 0.15451 | 0.388 | 0.00188 | 0.255 |
| 53232 | BDNF-AS |
hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-590-3p | 18 | KIT | Sponge network | -0.668 | 3.0E-5 | -2.184 | 0 | 0.255 |
| 53233 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | MLLT10 | Sponge network | 0.619 | 0.00157 | 0.105 | 0.33292 | 0.255 |
| 53234 | RP11-395G23.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-26a-2-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 12 | SLC6A15 | Sponge network | 0.381 | 0.25967 | -2.373 | 2.0E-5 | 0.255 |
| 53235 | RP11-21A7A.2 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-93-3p | 16 | CTIF | Sponge network | -1.116 | 0.23686 | -0.389 | 0.00549 | 0.255 |
| 53236 | CD27-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-505-5p;hsa-miR-532-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 42 | AR | Sponge network | 0.177 | 0.16681 | -1.554 | 0 | 0.255 |
| 53237 | LINC01018 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-345-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 34 | TRPS1 | Sponge network | -2.915 | 0.00015 | -0.191 | 0.44109 | 0.255 |
| 53238 | RP11-968O1.5 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-362-3p;hsa-miR-493-5p | 11 | ZFHX3 | Sponge network | -0.829 | 0.01343 | 0.389 | 0.01363 | 0.255 |
| 53239 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 20 | AFF3 | Sponge network | 0.374 | 0.20054 | -2.373 | 0 | 0.255 |
| 53240 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-590-5p | 15 | RCAN2 | Sponge network | -0.371 | 0.24604 | -0.33 | 0.15172 | 0.255 |
| 53241 | RP11-403I13.8 |
hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 15 | DOCK4 | Sponge network | 0.619 | 0.00157 | 0.502 | 0.00108 | 0.255 |
| 53242 | HOXA-AS2 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-942-5p | 14 | CREBBP | Sponge network | 0.61 | 0.01593 | 0.126 | 0.15186 | 0.255 |
| 53243 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-577 | 16 | DMXL1 | Sponge network | 0.921 | 0.00118 | -0.426 | 0.00056 | 0.255 |
| 53244 | GAS6-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-429;hsa-miR-92a-3p;hsa-miR-942-5p | 23 | GPC6 | Sponge network | 0.16 | 0.50785 | -0.054 | 0.81465 | 0.255 |
| 53245 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | RBMS3 | Sponge network | 0.817 | 3.0E-5 | -0.519 | 0.03551 | 0.255 |
| 53246 | RP11-83A24.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | RPS6KA3 | Sponge network | 0.417 | 0.00445 | 0.256 | 0.02419 | 0.255 |
| 53247 | CTC-559E9.5 |
hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-877-5p | 12 | GLIPR1 | Sponge network | 0.594 | 0.00064 | 0.146 | 0.3276 | 0.255 |
| 53248 | H19 |
hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-589-3p | 12 | DIP2C | Sponge network | 2.439 | 0 | -0.436 | 0.01713 | 0.255 |
| 53249 | AC016747.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 21 | MED12L | Sponge network | 0.199 | 0.38015 | -0.194 | 0.55299 | 0.255 |
| 53250 | RP11-242D8.1 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p | 11 | RB1 | Sponge network | 1.404 | 0 | 0.382 | 0.0017 | 0.255 |
| 53251 | RP11-410L14.2 |
hsa-miR-132-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-450b-5p | 12 | FAT3 | Sponge network | -0.052 | 0.76839 | -1.601 | 0.00051 | 0.255 |
| 53252 | FGD5-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-3p;hsa-miR-376c-3p;hsa-miR-625-5p | 11 | ARID2 | Sponge network | 0.401 | 0.00066 | 0.235 | 0.02697 | 0.255 |
| 53253 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-625-5p | 18 | NUAK1 | Sponge network | -0.773 | 0.04073 | 0.904 | 0 | 0.255 |
| 53254 | RP1-122P22.2 |
hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-92a-1-5p | 15 | CREB3L2 | Sponge network | 0.065 | 0.73468 | -0.525 | 2.0E-5 | 0.255 |
| 53255 | TUG1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-664a-3p | 17 | PRKAA1 | Sponge network | 0.972 | 0 | 0.317 | 0.0031 | 0.255 |
| 53256 | CD27-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-3127-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-769-5p | 21 | SETBP1 | Sponge network | 0.177 | 0.16681 | -1.302 | 1.0E-5 | 0.255 |
| 53257 | RP11-212P7.2 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-455-5p | 12 | AMPH | Sponge network | 0.219 | 0.1516 | -0.916 | 5.0E-5 | 0.255 |
| 53258 | RP11-967K21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-582-5p | 16 | ACVR1 | Sponge network | 0.056 | 0.81195 | 0.279 | 0.00234 | 0.255 |
| 53259 | RP11-196G18.24 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 13 | PHC3 | Sponge network | 1.262 | 0 | 0.212 | 0.0499 | 0.255 |
| 53260 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-93-3p | 24 | TRPS1 | Sponge network | 0.898 | 1.0E-5 | -0.191 | 0.44109 | 0.255 |
| 53261 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 40 | EPHA5 | Sponge network | 1.302 | 7.0E-5 | -0.957 | 0.01733 | 0.255 |
| 53262 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 18 | AHNAK | Sponge network | -0.727 | 0.04726 | -1.207 | 0 | 0.255 |
| 53263 | RP1-257A7.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326 | 11 | FAT3 | Sponge network | 0.849 | 0.00178 | -1.601 | 0.00051 | 0.255 |
| 53264 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-590-3p | 11 | ANK3 | Sponge network | 1.601 | 0 | -0.55 | 0.00086 | 0.255 |
| 53265 | RP11-999E24.3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 24 | ACVR2A | Sponge network | -0.693 | 0.05629 | -0.409 | 0.00039 | 0.255 |
| 53266 | RP11-428J1.5 |
hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p | 21 | DLC1 | Sponge network | 0.538 | 0.00076 | -0.382 | 0.05038 | 0.255 |
| 53267 | RP11-395G23.3 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-330-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-874-3p | 12 | SORCS1 | Sponge network | 0.381 | 0.25967 | -3.492 | 0 | 0.255 |
| 53268 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-495-3p | 14 | ZNF292 | Sponge network | -0.405 | 0.22013 | 0.276 | 0.04148 | 0.255 |
| 53269 | CTD-3138B18.5 |
hsa-let-7g-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-7-1-3p | 12 | MLLT10 | Sponge network | 0.379 | 0.03091 | 0.105 | 0.33292 | 0.255 |
| 53270 | RP11-774O3.3 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | -0.487 | 0.02157 | 0.098 | 0.40994 | 0.255 |
| 53271 | TINCR |
hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-96-5p | 13 | LRRC7 | Sponge network | -0.664 | 0.14543 | -1.341 | 0.01193 | 0.255 |
| 53272 | LINC00403 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-452-5p | 14 | GAS7 | Sponge network | -3.586 | 0.00032 | -0.555 | 0.01602 | 0.255 |
| 53273 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-501-5p | 20 | MAFB | Sponge network | -0.942 | 0 | -0.023 | 0.89865 | 0.255 |
| 53274 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-374b-5p;hsa-miR-493-5p;hsa-miR-628-5p | 20 | MIER3 | Sponge network | 0.782 | 0.00016 | -0.212 | 0.04957 | 0.255 |
| 53275 | AC005154.6 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 58 | LPP | Sponge network | 0.848 | 0 | -0.459 | 0.04475 | 0.255 |
| 53276 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-5p;hsa-miR-25-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 22 | CNTN1 | Sponge network | 0.056 | 0.8494 | -2.594 | 0 | 0.255 |
| 53277 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-935 | 11 | DAB2 | Sponge network | -0.096 | 0.67958 | 0.431 | 0.0113 | 0.255 |
| 53278 | SOX2-OT |
hsa-miR-140-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-214-3p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-421;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-652-3p;hsa-miR-940 | 14 | SLC12A6 | Sponge network | -1.264 | 6.0E-5 | -0.343 | 0.00043 | 0.255 |
| 53279 | FAM66C |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 23 | USP6 | Sponge network | -0.901 | 0.00019 | 0.188 | 0.3871 | 0.255 |
| 53280 | HAND2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 24 | ARNT2 | Sponge network | -1.697 | 0.00889 | -0.332 | 0.25851 | 0.255 |
| 53281 | RP11-767N6.7 |
hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-181a-2-3p;hsa-miR-186-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-590-5p | 12 | PIK3C2A | Sponge network | 0.466 | 0.00155 | 0.061 | 0.53776 | 0.255 |
| 53282 | RP11-244O19.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-93-5p | 20 | PRRX1 | Sponge network | -0.096 | 0.67958 | 1.316 | 0 | 0.255 |
| 53283 | DIO3OS |
hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-98-5p | 12 | KLF10 | Sponge network | -0.773 | 0.04073 | -0.783 | 0 | 0.255 |
| 53284 | AC090587.5 |
hsa-let-7g-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-501-5p | 10 | CREB3L2 | Sponge network | -0.088 | 0.65011 | -0.525 | 2.0E-5 | 0.255 |
| 53285 | LINC00622 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p | 13 | ZMYM2 | Sponge network | 1.169 | 0.00028 | 0.279 | 0.01273 | 0.255 |
| 53286 | SH3RF3-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-424-5p | 10 | ABCB5 | Sponge network | 0.889 | 9.0E-5 | -0.956 | 0.04077 | 0.255 |
| 53287 | CTC-301O7.4 |
hsa-miR-106b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-539-5p | 11 | SSBP2 | Sponge network | 0.292 | 0.43384 | -1.58 | 0 | 0.255 |
| 53288 | TRAF3IP2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-940 | 27 | BHLHE41 | Sponge network | -0.323 | 0.01372 | 0.353 | 0.12753 | 0.255 |
| 53289 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-576-5p | 12 | PTPRB | Sponge network | 0.594 | 0.00064 | 0.105 | 0.47122 | 0.255 |
| 53290 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-940 | 32 | MEF2C | Sponge network | 0.267 | 0.2937 | -0.499 | 0.01448 | 0.255 |
| 53291 | GAS6-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-651-5p | 23 | PRDM2 | Sponge network | 0.16 | 0.50785 | -0.258 | 0.00721 | 0.255 |
| 53292 | HCG11 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 31 | AFF1 | Sponge network | 0.385 | 0.10067 | -0.384 | 0.00053 | 0.255 |
| 53293 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-324-5p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p | 11 | HACE1 | Sponge network | 0.95 | 0.15943 | -0.072 | 0.55239 | 0.255 |
| 53294 | RP11-216B9.6 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p | 10 | IGF2 | Sponge network | 0.174 | 0.30034 | 0.848 | 0.03842 | 0.255 |
| 53295 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-335-5p;hsa-miR-93-3p;hsa-miR-96-5p | 16 | FOXO3 | Sponge network | 0.453 | 0.01839 | -0.04 | 0.72028 | 0.255 |
| 53296 | RP11-216B9.6 |
hsa-miR-1301-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-940 | 11 | SFRP1 | Sponge network | 0.174 | 0.30034 | -3.566 | 0 | 0.255 |
| 53297 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-590-3p | 12 | GPAM | Sponge network | 1.04 | 0.00023 | 0.142 | 0.29732 | 0.255 |
| 53298 | AF131215.2 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 15 | PLAG1 | Sponge network | -0.176 | 0.59692 | -0.315 | 0.26532 | 0.255 |
| 53299 | RP11-21A7A.2 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-500a-5p;hsa-miR-877-5p | 16 | AFF1 | Sponge network | -1.116 | 0.23686 | -0.384 | 0.00053 | 0.255 |
| 53300 | FENDRR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-93-5p | 11 | JAK1 | Sponge network | -1.281 | 0.00012 | 0.001 | 0.98982 | 0.255 |
| 53301 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-96-5p | 33 | PRKCE | Sponge network | -0.737 | 0.06182 | -0.403 | 0.00056 | 0.255 |
| 53302 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-590-3p | 18 | TMTC3 | Sponge network | 0.538 | 0.00076 | 0.123 | 0.22189 | 0.255 |
| 53303 | RP4-791M13.3 |
hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-34a-5p;hsa-miR-362-3p | 10 | IGF2 | Sponge network | 0.419 | 0.0319 | 0.848 | 0.03842 | 0.255 |
| 53304 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-589-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-629-3p | 39 | IL6ST | Sponge network | 0.594 | 0.41522 | -0.402 | 0.00676 | 0.255 |
| 53305 | LINC00621 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-766-3p | 14 | ATM | Sponge network | 0.131 | 0.8436 | 0.178 | 0.21568 | 0.255 |
| 53306 | C4B-AS1 |
hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-486-5p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-940;hsa-miR-96-5p | 10 | MYO9A | Sponge network | 2.306 | 9.0E-5 | -0.147 | 0.32373 | 0.255 |
| 53307 | RBPMS-AS1 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 16 | RBMS3 | Sponge network | 0.144 | 0.64989 | -0.519 | 0.03551 | 0.255 |
| 53308 | RP11-672A2.4 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-424-5p | 23 | KIF1B | Sponge network | 1.344 | 0 | -0.118 | 0.32258 | 0.255 |
| 53309 | RP11-620J15.3 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-330-5p;hsa-miR-532-5p;hsa-miR-590-3p | 19 | PRKCE | Sponge network | -0.739 | 0.00044 | -0.403 | 0.00056 | 0.255 |
| 53310 | HOXA-AS3 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-96-5p | 34 | NTRK3 | Sponge network | -0.555 | 0.06156 | -1.77 | 3.0E-5 | 0.255 |
| 53311 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | MEF2C | Sponge network | 0.476 | 0.19203 | -0.499 | 0.01448 | 0.255 |
| 53312 | PART1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-505-3p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 55 | CUX1 | Sponge network | -2.925 | 0 | -0.039 | 0.6705 | 0.255 |
| 53313 | SNX29P2 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 22 | SYT4 | Sponge network | 0.4 | 0.14611 | -3.207 | 0 | 0.255 |
| 53314 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | HECA | Sponge network | 0.219 | 0.1516 | -0.217 | 0.03463 | 0.255 |
| 53315 | HOXD-AS2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-342-5p | 10 | SYNPO2 | Sponge network | -0.208 | 0.65592 | -2.342 | 0 | 0.255 |
| 53316 | RP11-83A24.2 |
hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-365a-3p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 35 | ARHGEF12 | Sponge network | 0.417 | 0.00445 | 0.09 | 0.35693 | 0.255 |
| 53317 | CTA-363E19.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-223-3p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-409-3p | 20 | NFIB | Sponge network | 1.055 | 0 | 0.023 | 0.90795 | 0.255 |
| 53318 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | DACH2 | Sponge network | -0.323 | 0.01372 | -1.635 | 0.00034 | 0.255 |
| 53319 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92b-3p | 19 | BCL11A | Sponge network | -0.096 | 0.67958 | -0.932 | 0.0063 | 0.255 |
| 53320 | SRP14-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-1271-5p;hsa-miR-144-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-3614-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-590-5p | 15 | EPHA3 | Sponge network | -0.061 | 0.70068 | 0.185 | 0.53588 | 0.255 |
| 53321 | H19 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p | 10 | IGF1 | Sponge network | 2.439 | 0 | -0.204 | 0.5898 | 0.255 |
| 53322 | RP11-981G7.6 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-374a-3p;hsa-miR-590-3p | 11 | ERCC6 | Sponge network | 0.986 | 0.01022 | 0.23 | 0.015 | 0.255 |
| 53323 | RP11-2H8.2 |
hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-493-5p;hsa-miR-590-3p | 25 | SMAD2 | Sponge network | 1.089 | 0 | -0.128 | 0.12732 | 0.255 |
| 53324 | LIPE-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-582-5p;hsa-miR-93-3p | 18 | WWTR1 | Sponge network | 0.078 | 0.55803 | -0.345 | 0.11997 | 0.255 |
| 53325 | TP73-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-93-5p | 14 | TRIM3 | Sponge network | -0.563 | 0.02545 | -0.577 | 0 | 0.255 |
| 53326 | AF131215.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-628-5p;hsa-miR-93-3p | 40 | RUNX1T1 | Sponge network | -0.176 | 0.59692 | -0.344 | 0.2374 | 0.255 |
| 53327 | OIP5-AS1 |
hsa-let-7g-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-671-5p | 21 | PPP2R5C | Sponge network | 0.421 | 0.00084 | -0.314 | 3.0E-5 | 0.255 |
| 53328 | RP11-244H3.1 |
hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-3p | 23 | CCDC6 | Sponge network | 0.659 | 3.0E-5 | 0.27 | 0.02555 | 0.255 |
| 53329 | AP006216.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-26b-3p;hsa-miR-30a-3p;hsa-miR-590-3p | 10 | RICTOR | Sponge network | 0.779 | 0.00016 | 0.388 | 0.00188 | 0.255 |
| 53330 | AF131215.9 |
hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-2110;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-940 | 12 | PEG3 | Sponge network | -0.322 | 0.25945 | -1.74 | 0 | 0.255 |
| 53331 | RP11-196G18.22 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-582-5p | 22 | CREB1 | Sponge network | 1.206 | 0 | 0.203 | 0.00537 | 0.255 |
| 53332 | RP11-686D22.8 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-93-3p | 10 | SUFU | Sponge network | 0.898 | 1.0E-5 | -0.04 | 0.65489 | 0.255 |
| 53333 | AC005062.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-181a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p | 15 | USP12 | Sponge network | -0.019 | 0.81504 | -0.102 | 0.40768 | 0.255 |
| 53334 | RP11-983P16.4 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-5p;hsa-miR-744-3p | 48 | DST | Sponge network | 0.41 | 0.02214 | -0.713 | 0.00096 | 0.255 |
| 53335 | RP11-798M19.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-361-5p;hsa-miR-374a-5p;hsa-miR-382-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 21 | HDAC9 | Sponge network | -0.612 | 0.00094 | -0.755 | 0.00495 | 0.255 |
| 53336 | LINC00641 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 34 | MTSS1 | Sponge network | -0.222 | 0.32445 | -0.81 | 0.00035 | 0.255 |
| 53337 | DNM3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-5p | 23 | RNASEL | Sponge network | 0.572 | 0.01722 | -0.272 | 0.01796 | 0.255 |
| 53338 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-625-5p | 10 | LHX6 | Sponge network | 0.41 | 0.02214 | -0.087 | 0.69193 | 0.255 |
| 53339 | AC006547.13 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-940 | 15 | CREB1 | Sponge network | 1.159 | 0 | 0.203 | 0.00537 | 0.255 |
| 53340 | ZNF582-AS1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-7-5p | 22 | ERC1 | Sponge network | -1.349 | 0.00017 | -0.108 | 0.38794 | 0.255 |
| 53341 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 18 | PGR | Sponge network | 0.657 | 4.0E-5 | -1.704 | 0 | 0.255 |
| 53342 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 14 | PNRC1 | Sponge network | -0.031 | 0.89063 | -0.646 | 0 | 0.255 |
| 53343 | CTC-378H22.2 |
hsa-miR-106a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-455-5p;hsa-miR-501-5p | 10 | JAZF1 | Sponge network | 0.001 | 0.99663 | -0.377 | 0.02677 | 0.255 |
| 53344 | RP11-760H22.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-501-3p | 19 | GNAQ | Sponge network | 0.001 | 0.99753 | -0.367 | 0.00102 | 0.255 |
| 53345 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-3p | 13 | RB1CC1 | Sponge network | -0.227 | 0.31657 | 0.175 | 0.12559 | 0.255 |
| 53346 | LINC00963 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 24 | NAV3 | Sponge network | 0.523 | 9.0E-5 | 0.212 | 0.47729 | 0.255 |
| 53347 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-27b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-940 | 37 | MCC | Sponge network | 0.246 | 0.59912 | -1.005 | 0 | 0.255 |
| 53348 | CTC-559E9.5 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-423-3p | 10 | ROBO1 | Sponge network | 0.594 | 0.00064 | -0.009 | 0.96154 | 0.255 |
| 53349 | FZD10-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | MED12L | Sponge network | -0.727 | 0.04726 | -0.194 | 0.55299 | 0.255 |
| 53350 | HOXB-AS2 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-628-5p;hsa-miR-942-5p | 18 | TMEM132B | Sponge network | 1.316 | 0.01106 | -0.83 | 0.01084 | 0.255 |
| 53351 | LIPE-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-501-5p;hsa-miR-511-5p | 20 | BCL6 | Sponge network | 0.078 | 0.55803 | -0.211 | 0.19489 | 0.255 |
| 53352 | RP11-35G9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 52 | LPP | Sponge network | 0.657 | 4.0E-5 | -0.459 | 0.04475 | 0.255 |
| 53353 | RP11-490M8.1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-223-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-96-5p | 10 | EPHA3 | Sponge network | -0.214 | 0.44248 | 0.185 | 0.53588 | 0.255 |
| 53354 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-326;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-744-3p | 26 | FAT3 | Sponge network | 0.859 | 0.00331 | -1.601 | 0.00051 | 0.255 |
| 53355 | CTC-490E21.10 |
hsa-let-7g-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-93-3p | 12 | PCDH10 | Sponge network | 1.46 | 1.0E-5 | -1.644 | 0.0078 | 0.255 |
| 53356 | RP11-473I1.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | FOXO1 | Sponge network | 0.683 | 1.0E-5 | -0.141 | 0.33874 | 0.255 |
| 53357 | RP11-701H24.3 |
hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p | 15 | BMPR2 | Sponge network | -1.425 | 0.04204 | 0.206 | 0.0635 | 0.255 |
| 53358 | RP11-531A24.5 |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-576-5p | 16 | KLF10 | Sponge network | 0.75 | 0.00054 | -0.783 | 0 | 0.255 |
| 53359 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 17 | NOTCH2NL | Sponge network | 0.101 | 0.60919 | 0.024 | 0.84307 | 0.255 |
| 53360 | RP11-57H14.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-501-3p;hsa-miR-590-3p | 14 | TGFBR2 | Sponge network | 0.083 | 0.66405 | -0.243 | 0.11408 | 0.255 |
| 53361 | AC004540.4 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-484;hsa-miR-629-3p | 26 | DST | Sponge network | -2.549 | 0.00528 | -0.713 | 0.00096 | 0.255 |
| 53362 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-7-5p | 12 | PDGFC | Sponge network | 0.253 | 0.09565 | -0.33 | 0.06483 | 0.255 |
| 53363 | LINC00861 |
hsa-miR-130b-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-421;hsa-miR-93-3p;hsa-miR-940 | 11 | GRIK3 | Sponge network | 0.911 | 0.00854 | -3.208 | 0 | 0.255 |
| 53364 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-511-5p | 20 | BCL6 | Sponge network | 0.253 | 0.09565 | -0.211 | 0.19489 | 0.255 |
| 53365 | RP11-244O19.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-30b-5p;hsa-miR-342-3p;hsa-miR-382-5p;hsa-miR-485-3p;hsa-miR-539-5p | 11 | LIMD1 | Sponge network | -0.096 | 0.67958 | 0.103 | 0.28978 | 0.255 |
| 53366 | RP11-119F7.5 |
hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-502-5p;hsa-miR-590-3p | 16 | PAIP2 | Sponge network | 0.225 | 0.30644 | -0.515 | 0 | 0.255 |
| 53367 | RP11-434H6.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-409-3p | 11 | TGFBR3 | Sponge network | 0.358 | 0.14302 | -1.173 | 1.0E-5 | 0.255 |
| 53368 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-195-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p | 11 | AKAP11 | Sponge network | 0.79 | 0 | 0.196 | 0.09825 | 0.255 |
| 53369 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 41 | PPM1A | Sponge network | 0.997 | 0.00066 | -0.623 | 0 | 0.255 |
| 53370 | H19 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-589-3p | 11 | SCN9A | Sponge network | 2.439 | 0 | -0.525 | 0.10593 | 0.255 |
| 53371 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 31 | PAFAH1B1 | Sponge network | -0.031 | 0.89063 | -0.429 | 0 | 0.255 |
| 53372 | FZD10-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 38 | TGFBR3 | Sponge network | -0.727 | 0.04726 | -1.173 | 1.0E-5 | 0.255 |
| 53373 | RP11-161M6.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-93-3p;hsa-miR-96-5p | 10 | UNC5C | Sponge network | 0.61 | 0.06611 | -0.178 | 0.40214 | 0.255 |
| 53374 | CALML3-AS1 |
hsa-miR-141-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-429;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-532-3p;hsa-miR-542-3p | 16 | MRVI1 | Sponge network | 0.95 | 0.15943 | -1.051 | 0.00022 | 0.255 |
| 53375 | FAM157A |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p | 10 | PRKAA1 | Sponge network | 0.813 | 1.0E-5 | 0.317 | 0.0031 | 0.255 |
| 53376 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-485-3p;hsa-miR-93-3p;hsa-miR-940 | 24 | AKAP11 | Sponge network | -0.465 | 0.04703 | 0.196 | 0.09825 | 0.255 |
| 53377 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-299-5p;hsa-miR-376c-3p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p | 16 | TRIM33 | Sponge network | 0.603 | 0.00127 | 0.402 | 7.0E-5 | 0.255 |
| 53378 | RP11-53O19.1 |
hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-654-3p | 35 | PIK3R1 | Sponge network | -0.405 | 0.22013 | -0.133 | 0.36854 | 0.255 |
| 53379 | SOX2-OT |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | ID4 | Sponge network | -1.264 | 6.0E-5 | -1.644 | 0 | 0.255 |
| 53380 | RP11-540B6.6 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-484 | 16 | SMAD2 | Sponge network | 0.93 | 0 | -0.128 | 0.12732 | 0.255 |
| 53381 | RP11-890B15.3 |
hsa-miR-146a-5p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-582-3p;hsa-miR-7-1-3p | 10 | ADCY1 | Sponge network | 0.101 | 0.60919 | 0.362 | 0.16982 | 0.255 |
| 53382 | PWAR6 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 22 | NOTCH2NL | Sponge network | -1.575 | 6.0E-5 | 0.024 | 0.84307 | 0.255 |
| 53383 | LINC00403 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-452-5p | 13 | PPM1L | Sponge network | -3.586 | 0.00032 | -0.537 | 0.00616 | 0.255 |
| 53384 | RP11-774O3.3 |
hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CTGF | Sponge network | -0.487 | 0.02157 | 0.321 | 0.11682 | 0.255 |
| 53385 | CTC-429P9.3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p | 19 | MED12L | Sponge network | 0.082 | 0.55397 | -0.194 | 0.55299 | 0.255 |
| 53386 | KB-1410C5.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-940 | 10 | SNX29 | Sponge network | 0.959 | 0 | -0.34 | 0.01371 | 0.255 |
| 53387 | LINC01018 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7b-5p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 19 | NEDD4 | Sponge network | -2.915 | 0.00015 | 0.02 | 0.89834 | 0.255 |
| 53388 | RP11-2H8.2 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 11 | RNF144A | Sponge network | 1.089 | 0 | 0.594 | 0.0009 | 0.255 |
| 53389 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-93-5p | 25 | PTEN | Sponge network | 0.497 | 0.01451 | -0.187 | 0.07748 | 0.255 |
| 53390 | HHIP-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-5p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 17 | PDE4DIP | Sponge network | -0.737 | 0.10822 | -0.282 | 0.0257 | 0.255 |
| 53391 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-30d-3p;hsa-miR-577 | 16 | THRB | Sponge network | 0.357 | 0.72407 | -1.117 | 0.0004 | 0.255 |
| 53392 | CTD-2139B15.2 |
hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-191-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-484;hsa-miR-539-5p | 18 | DDX6 | Sponge network | 0.286 | 0.00612 | 0.091 | 0.36428 | 0.255 |
| 53393 | TPTEP1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 28 | PTPN14 | Sponge network | -1.216 | 0 | 0.144 | 0.34595 | 0.255 |
| 53394 | AC074289.1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-21-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-5p;hsa-miR-874-3p;hsa-miR-940 | 11 | SORCS1 | Sponge network | -0.326 | 0.14314 | -3.492 | 0 | 0.255 |
| 53395 | AC156455.1 |
hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-942-5p | 10 | GLIPR1 | Sponge network | 1.154 | 0.00041 | 0.146 | 0.3276 | 0.255 |
| 53396 | MEG3 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-942-5p | 28 | OPCML | Sponge network | 0.249 | 0.35902 | -2.498 | 0 | 0.255 |
| 53397 | RP4-584D14.7 |
hsa-let-7d-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-378c | 10 | ERC1 | Sponge network | 1.294 | 0.0031 | -0.108 | 0.38794 | 0.255 |
| 53398 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-96-5p | 20 | HBP1 | Sponge network | 0.906 | 0.31715 | -0.454 | 0 | 0.255 |
| 53399 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-485-3p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 42 | PPM1A | Sponge network | 0.72 | 0.0001 | -0.623 | 0 | 0.255 |
| 53400 | AP001347.6 |
hsa-miR-107;hsa-miR-1301-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | ERRFI1 | Sponge network | -1.161 | 0.00591 | -0.798 | 1.0E-5 | 0.255 |
| 53401 | FAM13A-AS1 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 19 | IGSF10 | Sponge network | -0.83 | 0 | -1.751 | 1.0E-5 | 0.255 |
| 53402 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 60 | THRB | Sponge network | 0.064 | 0.72934 | -1.117 | 0.0004 | 0.255 |
| 53403 | CTD-2303H24.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 16 | ARNT2 | Sponge network | 0.415 | 0.14657 | -0.332 | 0.25851 | 0.255 |
| 53404 | RP11-305E6.4 |
hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-582-5p;hsa-miR-590-3p | 11 | DOCK4 | Sponge network | 1.317 | 0 | 0.502 | 0.00108 | 0.255 |
| 53405 | GUSBP11 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-29a-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | RPS6KA3 | Sponge network | 0.856 | 0 | 0.256 | 0.02419 | 0.255 |
| 53406 | RP5-994D16.9 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-409-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-590-3p | 20 | DST | Sponge network | 0.252 | 0.08508 | -0.713 | 0.00096 | 0.255 |
| 53407 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 18 | ARHGAP29 | Sponge network | 0.415 | 0.14657 | 0.131 | 0.42953 | 0.255 |
| 53408 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-424-5p | 15 | LATS1 | Sponge network | 0.082 | 0.55397 | 0.109 | 0.34208 | 0.255 |
| 53409 | CTD-2314G24.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-589-5p;hsa-miR-7-1-3p | 20 | RARB | Sponge network | 0.246 | 0.59912 | -0.825 | 2.0E-5 | 0.255 |
| 53410 | AC156455.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-484;hsa-miR-502-5p | 20 | CHD5 | Sponge network | 1.154 | 0.00041 | -0.922 | 0.01521 | 0.255 |
| 53411 | MIR143HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 15 | AKAP13 | Sponge network | -0.739 | 0.0574 | 0.028 | 0.7729 | 0.255 |
| 53412 | RP11-434H6.7 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p | 10 | DMXL1 | Sponge network | 0.358 | 0.14302 | -0.426 | 0.00056 | 0.255 |
| 53413 | RP4-791M13.3 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-940 | 21 | GRIK3 | Sponge network | 0.419 | 0.0319 | -3.208 | 0 | 0.255 |
| 53414 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | AKAP11 | Sponge network | 1.308 | 0 | 0.196 | 0.09825 | 0.254 |
| 53415 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | DACH2 | Sponge network | -0.615 | 0.00015 | -1.635 | 0.00034 | 0.254 |
| 53416 | AP001347.6 |
hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-212-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-502-5p;hsa-miR-590-3p | 16 | PAIP2 | Sponge network | -1.161 | 0.00591 | -0.515 | 0 | 0.254 |
| 53417 | OIP5-AS1 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-335-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | ITPKB | Sponge network | 0.421 | 0.00084 | -0.704 | 0.00106 | 0.254 |
| 53418 | RP11-736K20.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-151a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 21 | CLASP2 | Sponge network | -0.358 | 0.17255 | -0.038 | 0.71 | 0.254 |
| 53419 | HAND2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | HECA | Sponge network | -1.697 | 0.00889 | -0.217 | 0.03463 | 0.254 |
| 53420 | RP11-999E24.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1296-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-3607-3p | 17 | LIFR | Sponge network | -0.693 | 0.05629 | -1.794 | 0 | 0.254 |
| 53421 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | MGA | Sponge network | -0.487 | 0.02157 | 0.323 | 0.00792 | 0.254 |
| 53422 | LINC00654 |
hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 21 | LIMCH1 | Sponge network | 0.374 | 0.20054 | -0.915 | 1.0E-5 | 0.254 |
| 53423 | LINC00963 |
hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-486-5p;hsa-miR-495-3p | 13 | SYNM | Sponge network | 0.523 | 9.0E-5 | -2.309 | 1.0E-5 | 0.254 |
| 53424 | RP11-25K19.1 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 16 | ZMYND11 | Sponge network | -1.057 | 0.00029 | -0.2 | 0.05449 | 0.254 |
| 53425 | RP11-400K9.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-362-5p;hsa-miR-542-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p | 28 | EGFR | Sponge network | -0.611 | 0.09607 | -0.175 | 0.48451 | 0.254 |
| 53426 | PCA3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | NUAK1 | Sponge network | -0.709 | 0.18168 | 0.904 | 0 | 0.254 |
| 53427 | RP11-127B20.2 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 12 | ANK2 | Sponge network | 0.411 | 0.1335 | -1.603 | 1.0E-5 | 0.254 |
| 53428 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-3065-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 37 | CBL | Sponge network | -0.576 | 0.10121 | 0.291 | 0.01267 | 0.254 |
| 53429 | RP11-428J1.5 |
hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-5p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | ITPKB | Sponge network | 0.538 | 0.00076 | -0.704 | 0.00106 | 0.254 |
| 53430 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-7-5p | 21 | MOB1B | Sponge network | 0.001 | 0.99753 | 0.197 | 0.15266 | 0.254 |
| 53431 | AC074289.1 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-96-5p | 19 | JAZF1 | Sponge network | -0.326 | 0.14314 | -0.377 | 0.02677 | 0.254 |
| 53432 | SNHG16 |
hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-664a-3p | 11 | UBE2D1 | Sponge network | 0.961 | 0 | 0.468 | 0 | 0.254 |
| 53433 | TINCR |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-345-5p;hsa-miR-376c-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-93-3p | 18 | IKZF2 | Sponge network | -0.664 | 0.14543 | -0.284 | 0.1942 | 0.254 |
| 53434 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 17 | RB1CC1 | Sponge network | -0.615 | 0.00015 | 0.175 | 0.12559 | 0.254 |
| 53435 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 29 | FOXO3 | Sponge network | 0.222 | 0.29133 | -0.04 | 0.72028 | 0.254 |
| 53436 | TINCR |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-624-5p | 13 | LATS1 | Sponge network | -0.664 | 0.14543 | 0.109 | 0.34208 | 0.254 |
| 53437 | TMEM161B-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-654-3p | 19 | NR2C2 | Sponge network | -0.04 | 0.7606 | 0.21 | 0.03224 | 0.254 |
| 53438 | FAM157A |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-505-5p | 15 | DDX3X | Sponge network | 0.813 | 1.0E-5 | -0.152 | 0.07686 | 0.254 |
| 53439 | RP11-148O21.2 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-671-5p | 11 | PTPRD | Sponge network | 0.233 | 0.81029 | -1.005 | 0.00064 | 0.254 |
| 53440 | RP11-35G9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-330-5p;hsa-miR-455-5p;hsa-miR-96-5p | 16 | DMXL1 | Sponge network | 0.622 | 0.00015 | -0.426 | 0.00056 | 0.254 |
| 53441 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-7-1-3p | 21 | YAP1 | Sponge network | -0.405 | 0.22013 | 0.22 | 0.11836 | 0.254 |
| 53442 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-625-5p;hsa-miR-93-5p;hsa-miR-940 | 25 | CHD5 | Sponge network | 0.056 | 0.8494 | -0.922 | 0.01521 | 0.254 |
| 53443 | HOXB-AS3 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-34a-5p | 16 | ZFHX4 | Sponge network | 0.343 | 0.28887 | 0.018 | 0.95574 | 0.254 |
| 53444 | LOXL1-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p | 19 | ABL2 | Sponge network | 0.487 | 0.04053 | 0.82 | 0 | 0.254 |
| 53445 | FAM13A-AS1 |
hsa-miR-140-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-3p;hsa-miR-323a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-98-5p | 11 | CDKN1B | Sponge network | -0.83 | 0 | -0.085 | 0.42464 | 0.254 |
| 53446 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-200a-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 20 | ZFHX3 | Sponge network | 0.197 | 0.25618 | 0.389 | 0.01363 | 0.254 |
| 53447 | LINC00909 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-664a-5p | 13 | MAPK1 | Sponge network | 0.363 | 0.00601 | -0.017 | 0.81465 | 0.254 |
| 53448 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-183-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-501-5p;hsa-miR-590-3p | 14 | BCL6 | Sponge network | 0.619 | 0.00157 | -0.211 | 0.19489 | 0.254 |
| 53449 | RP1-59D14.5 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-628-5p | 14 | SLC5A3 | Sponge network | 1.162 | 5.0E-5 | 0.493 | 5.0E-5 | 0.254 |
| 53450 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-671-5p;hsa-miR-93-5p | 34 | CHD5 | Sponge network | -0.235 | 0.15142 | -0.922 | 0.01521 | 0.254 |
| 53451 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-148a-5p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-505-3p;hsa-miR-590-3p | 12 | ZNF292 | Sponge network | 0.659 | 3.0E-5 | 0.276 | 0.04148 | 0.254 |
| 53452 | RP4-605O3.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 12 | FBXO32 | Sponge network | 0.262 | 0.0475 | -0.495 | 0.07561 | 0.254 |
| 53453 | RP11-158K1.3 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 31 | NTRK2 | Sponge network | 0.349 | 0.01695 | -0.736 | 0.06495 | 0.254 |
| 53454 | RP11-867G23.10 |
hsa-miR-107;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p | 16 | MTSS1 | Sponge network | -2.531 | 0 | -0.81 | 0.00035 | 0.254 |
| 53455 | NEAT1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-409-5p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 48 | KIF1B | Sponge network | 0.705 | 0.00014 | -0.118 | 0.32258 | 0.254 |
| 53456 | RP11-411K7.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | SLC6A15 | Sponge network | 0.155 | 0.81273 | -2.373 | 2.0E-5 | 0.254 |
| 53457 | RP11-248G5.8 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-214-5p;hsa-miR-22-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p | 12 | MAT2A | Sponge network | 1.294 | 0 | -0.073 | 0.36195 | 0.254 |
| 53458 | RP11-927P21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-940 | 26 | WDFY3 | Sponge network | 0.921 | 0.00118 | -0.201 | 0.0967 | 0.254 |
| 53459 | RP11-819C21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | PRRX1 | Sponge network | 0.537 | 0.00139 | 1.316 | 0 | 0.254 |
| 53460 | RP11-774O3.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | CDHR3 | Sponge network | -0.487 | 0.02157 | -0.977 | 4.0E-5 | 0.254 |
| 53461 | UBA6-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 27 | WDFY3 | Sponge network | 0.312 | 0.00725 | -0.201 | 0.0967 | 0.254 |
| 53462 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-3127-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | HECA | Sponge network | 1.757 | 0 | -0.217 | 0.03463 | 0.254 |
| 53463 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-331-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 62 | NTRK3 | Sponge network | 0.219 | 0.1516 | -1.77 | 3.0E-5 | 0.254 |
| 53464 | AP001055.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p | 11 | KIT | Sponge network | 0.693 | 0.00076 | -2.184 | 0 | 0.254 |
| 53465 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | ZFHX3 | Sponge network | 0.415 | 0.14657 | 0.389 | 0.01363 | 0.254 |
| 53466 | RP4-758J18.10 |
hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-450b-5p | 12 | CADM1 | Sponge network | -0.091 | 0.71468 | -0.9 | 0.00084 | 0.254 |
| 53467 | RP11-389C8.2 |
hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p | 13 | DYNC1I1 | Sponge network | 0.727 | 3.0E-5 | -1.12 | 0.00107 | 0.254 |
| 53468 | RP4-605O3.4 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | ABI2 | Sponge network | 0.262 | 0.0475 | 0.175 | 0.14787 | 0.254 |
| 53469 | SERTAD4-AS1 |
hsa-miR-1266-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-369-3p;hsa-miR-500a-5p | 14 | AFF1 | Sponge network | -0.932 | 0.00329 | -0.384 | 0.00053 | 0.254 |
| 53470 | RP11-439A17.10 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-590-5p | 10 | EGFR | Sponge network | 0.051 | 0.95756 | -0.175 | 0.48451 | 0.254 |
| 53471 | AF131215.2 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 66 | LPP | Sponge network | -0.176 | 0.59692 | -0.459 | 0.04475 | 0.254 |
| 53472 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-582-5p | 10 | ACVR1 | Sponge network | 1.03 | 0 | 0.279 | 0.00234 | 0.254 |
| 53473 | RP11-254F7.2 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-589-3p;hsa-miR-590-5p | 30 | PIK3R1 | Sponge network | 0.176 | 0.37402 | -0.133 | 0.36854 | 0.254 |
| 53474 | RP4-584D14.7 |
hsa-miR-1266-5p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-500a-5p | 12 | AFF1 | Sponge network | 1.294 | 0.0031 | -0.384 | 0.00053 | 0.254 |
| 53475 | ADIRF-AS1 |
hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-378a-3p;hsa-miR-7-5p;hsa-miR-92a-3p | 11 | FLNA | Sponge network | -0.223 | 0.47816 | -0.771 | 0.01377 | 0.254 |
| 53476 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-589-5p;hsa-miR-98-5p | 10 | ST5 | Sponge network | -0.727 | 0.04726 | -0.78 | 0 | 0.254 |
| 53477 | TPTEP1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-96-5p | 43 | PIK3R1 | Sponge network | -1.216 | 0 | -0.133 | 0.36854 | 0.254 |
| 53478 | PSMG3-AS1 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-92a-3p | 11 | ATM | Sponge network | -0.125 | 0.49414 | 0.178 | 0.21568 | 0.254 |
| 53479 | RP1-151F17.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-30c-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | PRUNE2 | Sponge network | 0.222 | 0.29133 | -1.733 | 0.00022 | 0.254 |
| 53480 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-576-5p | 18 | BNC2 | Sponge network | 1.361 | 0 | -0.491 | 0.09988 | 0.254 |
| 53481 | NEAT1 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-214-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-328-3p;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-365a-3p;hsa-miR-374a-3p;hsa-miR-374b-3p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-550a-5p;hsa-miR-628-5p;hsa-miR-758-3p | 24 | VPS53 | Sponge network | 0.705 | 0.00014 | 0.103 | 0.22667 | 0.254 |
| 53482 | RP11-244O19.1 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-424-5p;hsa-miR-590-3p | 23 | CTDSPL | Sponge network | -0.096 | 0.67958 | 0.085 | 0.45121 | 0.254 |
| 53483 | RP11-155G14.6 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p | 11 | VPS53 | Sponge network | 2.483 | 0 | 0.103 | 0.22667 | 0.254 |
| 53484 | AC005154.6 |
hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-411-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | TMEM30A | Sponge network | 0.848 | 0 | 0.096 | 0.31031 | 0.254 |
| 53485 | RP11-596D21.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-133a-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p | 12 | TBL1XR1 | Sponge network | 3.076 | 3.0E-5 | 0.578 | 0 | 0.254 |
| 53486 | RP11-473I1.9 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-30b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 22 | CYLD | Sponge network | 0.683 | 1.0E-5 | -0.367 | 0.00171 | 0.254 |
| 53487 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 34 | ST6GALNAC3 | Sponge network | -1.264 | 6.0E-5 | -0.987 | 0 | 0.254 |
| 53488 | CTC-429P9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-744-3p | 31 | PTEN | Sponge network | 0.178 | 0.29106 | -0.187 | 0.07748 | 0.254 |
| 53489 | RP11-798G7.7 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p | 12 | SCN9A | Sponge network | 0.858 | 3.0E-5 | -0.525 | 0.10593 | 0.254 |
| 53490 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | MYO9A | Sponge network | -0.141 | 0.26434 | -0.147 | 0.32373 | 0.254 |
| 53491 | RP11-404E16.1 |
hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-185-3p;hsa-miR-188-5p;hsa-miR-191-5p;hsa-miR-1976;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-29b-1-5p;hsa-miR-342-5p;hsa-miR-532-5p | 12 | CUX1 | Sponge network | 0.097 | 0.91311 | -0.039 | 0.6705 | 0.254 |
| 53492 | BVES-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-429 | 17 | TBX18 | Sponge network | -2.62 | 0 | 0.843 | 0.02945 | 0.254 |
| 53493 | RP11-212P7.2 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-374a-3p;hsa-miR-450b-5p;hsa-miR-532-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-671-5p;hsa-miR-877-5p;hsa-miR-93-5p;hsa-miR-940;hsa-miR-96-5p | 42 | GAS7 | Sponge network | 0.219 | 0.1516 | -0.555 | 0.01602 | 0.254 |
| 53494 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | TBX18 | Sponge network | 0.064 | 0.72934 | 0.843 | 0.02945 | 0.254 |
| 53495 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-424-5p;hsa-miR-493-5p;hsa-miR-497-5p;hsa-miR-590-3p | 15 | MYBL1 | Sponge network | 0.826 | 1.0E-5 | 0.494 | 0.02152 | 0.254 |
| 53496 | RP11-362F19.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-452-5p | 18 | JAZF1 | Sponge network | -0.012 | 0.98664 | -0.377 | 0.02677 | 0.254 |
| 53497 | RP11-13K12.5 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-500a-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 26 | AFF1 | Sponge network | -0.318 | 0.65847 | -0.384 | 0.00053 | 0.254 |
| 53498 | LINC00473 |
hsa-let-7f-1-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-590-3p | 11 | PTCH1 | Sponge network | -1.203 | 0.03881 | -0.1 | 0.6179 | 0.254 |
| 53499 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-3613-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-493-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | PPM1A | Sponge network | -0.976 | 0.00025 | -0.623 | 0 | 0.254 |
| 53500 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-96-5p | 27 | EBF1 | Sponge network | 0.395 | 0.15451 | -0.384 | 0.08856 | 0.254 |
| 53501 | MEG3 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-429 | 13 | DDX5 | Sponge network | 0.249 | 0.35902 | -0.099 | 0.17428 | 0.254 |
| 53502 | RP11-326G21.1 |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-590-3p | 10 | PPP2R5C | Sponge network | -0.021 | 0.93598 | -0.314 | 3.0E-5 | 0.254 |
| 53503 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-150-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-370-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-629-3p | 18 | CHD2 | Sponge network | -0.615 | 0.00015 | 0.049 | 0.55371 | 0.254 |
| 53504 | KB-1460A1.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-193a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-362-5p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | NALCN | Sponge network | 0.603 | 0.00127 | 1.068 | 0.00237 | 0.254 |
| 53505 | RP4-639F20.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p | 24 | RASSF2 | Sponge network | -0.517 | 0.02935 | -0.273 | 0.21961 | 0.254 |
| 53506 | LINC00173 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 20 | SASH1 | Sponge network | 0.056 | 0.8494 | -0.502 | 0.00175 | 0.254 |
| 53507 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-20a-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-589-5p;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 27 | EYA4 | Sponge network | -0.969 | 2.0E-5 | 0.3 | 0.36876 | 0.254 |
| 53508 | HOTAIRM1 |
hsa-let-7f-1-3p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-326;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 12 | EMP2 | Sponge network | -0.053 | 0.80685 | -0.423 | 0.00985 | 0.254 |
| 53509 | LINC00094 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-376c-3p;hsa-miR-429;hsa-miR-942-5p | 15 | CREBBP | Sponge network | 0.253 | 0.09565 | 0.126 | 0.15186 | 0.254 |
| 53510 | LINC00654 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZNF844 | Sponge network | 0.374 | 0.20054 | -0.966 | 0.00035 | 0.254 |
| 53511 | FAM66C |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 17 | TES | Sponge network | -0.901 | 0.00019 | -0.348 | 0.00639 | 0.254 |
| 53512 | C4B-AS1 |
hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-27a-3p;hsa-miR-590-5p;hsa-miR-96-5p | 10 | CADM1 | Sponge network | 2.306 | 9.0E-5 | -0.9 | 0.00084 | 0.254 |
| 53513 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-7-1-3p | 10 | DACT1 | Sponge network | 0.41 | 0.02214 | 0.783 | 0.00072 | 0.254 |
| 53514 | AGAP11 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-374b-5p;hsa-miR-576-5p | 21 | EPS15 | Sponge network | -1.645 | 0 | -0.052 | 0.53998 | 0.254 |
| 53515 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-671-5p;hsa-miR-877-5p | 23 | RBMS3 | Sponge network | 0.594 | 0.41522 | -0.519 | 0.03551 | 0.254 |
| 53516 | RP11-680C21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 27 | IL17RD | Sponge network | -1.424 | 0.04346 | -0.019 | 0.94873 | 0.254 |
| 53517 | DNM3OS |
hsa-miR-142-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374a-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TMEM30A | Sponge network | 0.572 | 0.01722 | 0.096 | 0.31031 | 0.254 |
| 53518 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 38 | CBX5 | Sponge network | 0.541 | 0.00125 | 0.165 | 0.24446 | 0.254 |
| 53519 | RP11-497H16.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-409-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | ACVR2A | Sponge network | 0.545 | 0.00064 | -0.409 | 0.00039 | 0.254 |
| 53520 | LINC00173 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-505-5p;hsa-miR-625-5p;hsa-miR-940 | 18 | TOM1L2 | Sponge network | 0.056 | 0.8494 | -0.994 | 0 | 0.254 |
| 53521 | LINC00863 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-362-5p | 17 | KCNJ5 | Sponge network | 0.189 | 0.13981 | -0.281 | 0.3339 | 0.254 |
| 53522 | TUG1 |
hsa-let-7a-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-505-3p;hsa-miR-628-5p | 11 | CHD2 | Sponge network | 0.972 | 0 | 0.049 | 0.55371 | 0.254 |
| 53523 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-33a-5p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 39 | WDFY3 | Sponge network | 0.41 | 0.02214 | -0.201 | 0.0967 | 0.254 |
| 53524 | SNHG12 |
hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-495-3p;hsa-miR-497-5p | 12 | SLC5A3 | Sponge network | 1.103 | 0 | 0.493 | 5.0E-5 | 0.254 |
| 53525 | RP11-13K12.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-455-3p | 21 | CREB1 | Sponge network | -0.154 | 0.78939 | 0.203 | 0.00537 | 0.254 |
| 53526 | CKMT2-AS1 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-375;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | CXCL12 | Sponge network | 0.082 | 0.55654 | -1.421 | 0 | 0.254 |
| 53527 | TMEM9B-AS1 |
hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-589-3p | 14 | USP12 | Sponge network | 0.529 | 0.00552 | -0.102 | 0.40768 | 0.254 |
| 53528 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-193a-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-532-3p | 18 | LATS1 | Sponge network | -0.154 | 0.78939 | 0.109 | 0.34208 | 0.254 |
| 53529 | RP11-473I1.10 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-335-5p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | DIP2C | Sponge network | 0.673 | 0 | -0.436 | 0.01713 | 0.254 |
| 53530 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374a-3p;hsa-miR-7-1-3p | 12 | MOB1B | Sponge network | -0.557 | 0.11135 | 0.197 | 0.15266 | 0.254 |
| 53531 | RP11-195B17.1 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-96-5p | 26 | CADM2 | Sponge network | 0.37 | 0.27614 | -3.782 | 0 | 0.254 |
| 53532 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 15 | PTPRB | Sponge network | 0.614 | 1.0E-5 | 0.105 | 0.47122 | 0.254 |
| 53533 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-146b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 12 | LRRC7 | Sponge network | -0.141 | 0.26434 | -1.341 | 0.01193 | 0.254 |
| 53534 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-93-5p | 10 | JAK1 | Sponge network | -0.096 | 0.67958 | 0.001 | 0.98982 | 0.254 |
| 53535 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | TSHZ3 | Sponge network | 0.538 | 0.00076 | -0.556 | 0.01516 | 0.254 |
| 53536 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | ITCH | Sponge network | 0.494 | 0.00339 | 0.084 | 0.34058 | 0.254 |
| 53537 | HOXD-AS2 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-625-5p | 12 | MCC | Sponge network | -0.208 | 0.65592 | -1.005 | 0 | 0.254 |
| 53538 | PART1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-181b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 12 | PLA2R1 | Sponge network | -2.925 | 0 | -0.265 | 0.24676 | 0.254 |
| 53539 | U91328.21 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-378c | 11 | MED12L | Sponge network | -0.735 | 0.15983 | -0.194 | 0.55299 | 0.254 |
| 53540 | LINC00094 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-7-5p | 10 | COL1A2 | Sponge network | 0.253 | 0.09565 | 1.991 | 0 | 0.254 |
| 53541 | RP4-605O3.4 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-378a-5p;hsa-miR-590-3p;hsa-miR-874-3p | 12 | ASTN1 | Sponge network | 0.262 | 0.0475 | -2.389 | 2.0E-5 | 0.254 |
| 53542 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | HIP1 | Sponge network | -0.355 | 0.0077 | 0.774 | 0 | 0.254 |
| 53543 | CTD-2002H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-192-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | AKAP6 | Sponge network | 0.931 | 1.0E-5 | -1.586 | 3.0E-5 | 0.254 |
| 53544 | RP11-359B12.2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-146b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-511-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-5p | 30 | BTG2 | Sponge network | 0.064 | 0.72934 | -1.763 | 0 | 0.254 |
| 53545 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-577;hsa-miR-590-3p | 18 | DMXL1 | Sponge network | -0.227 | 0.31657 | -0.426 | 0.00056 | 0.254 |
| 53546 | ARHGEF26-AS1 |
hsa-miR-141-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-940 | 14 | RANBP9 | Sponge network | -2.46 | 0 | -0.191 | 0.04628 | 0.254 |
| 53547 | NR2F1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-361-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-452-5p;hsa-miR-493-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p;hsa-miR-940 | 74 | AFF4 | Sponge network | -0.49 | 0.04946 | -0.266 | 0.01565 | 0.254 |
| 53548 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 47 | KIF1B | Sponge network | 0.853 | 0.15352 | -0.118 | 0.32258 | 0.254 |
| 53549 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-425-5p;hsa-miR-532-5p | 12 | SLITRK6 | Sponge network | -1.349 | 0.00017 | -2.121 | 0 | 0.254 |
| 53550 | AGAP11 |
hsa-miR-130b-3p;hsa-miR-188-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-590-5p | 10 | RASA1 | Sponge network | -1.645 | 0 | -0.002 | 0.98204 | 0.254 |
| 53551 | CTC-429P9.3 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-330-5p;hsa-miR-424-5p | 15 | PPP2R5C | Sponge network | 0.082 | 0.55397 | -0.314 | 3.0E-5 | 0.254 |
| 53552 | RP11-20I23.13 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-29b-1-5p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-744-3p | 15 | HIP1 | Sponge network | 0.505 | 0.0014 | 0.774 | 0 | 0.254 |
| 53553 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-375;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-3p | 19 | EBF3 | Sponge network | 0.61 | 0.01593 | -0.058 | 0.81437 | 0.254 |
| 53554 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-582-5p | 16 | ARHGAP29 | Sponge network | -0.176 | 0.59692 | 0.131 | 0.42953 | 0.254 |
| 53555 | HAR1A |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-671-5p | 14 | PTPRD | Sponge network | -0.672 | 0.19447 | -1.005 | 0.00064 | 0.254 |
| 53556 | RP11-597D13.9 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | HECA | Sponge network | 1.302 | 7.0E-5 | -0.217 | 0.03463 | 0.254 |
| 53557 | RP11-1006G14.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-454-3p;hsa-miR-7-5p | 17 | ZBTB4 | Sponge network | 0.559 | 0.00026 | -0.739 | 0 | 0.254 |
| 53558 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-374b-5p | 13 | MIER3 | Sponge network | 1.147 | 0 | -0.212 | 0.04957 | 0.254 |
| 53559 | RP11-474O21.5 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 21 | EPHA7 | Sponge network | -0.023 | 0.95109 | -2.197 | 3.0E-5 | 0.254 |
| 53560 | RP11-834C11.4 |
hsa-miR-128-3p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-940 | 10 | GRIK3 | Sponge network | -0.883 | 0.01839 | -3.208 | 0 | 0.254 |
| 53561 | HOXA-AS3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-96-5p | 13 | MYO9A | Sponge network | -0.555 | 0.06156 | -0.147 | 0.32373 | 0.254 |
| 53562 | RP11-678G14.3 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 43 | LPP | Sponge network | -4.477 | 0 | -0.459 | 0.04475 | 0.254 |
| 53563 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | ATRX | Sponge network | -0.053 | 0.80685 | 0.261 | 0.04408 | 0.254 |
| 53564 | RP3-402G11.26 |
hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-590-3p | 20 | CBX5 | Sponge network | -0.254 | 0.19303 | 0.165 | 0.24446 | 0.254 |
| 53565 | RP11-6N17.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-301a-3p | 13 | AR | Sponge network | 0.108 | 0.69556 | -1.554 | 0 | 0.254 |
| 53566 | MIR22HG |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-542-3p;hsa-miR-550a-3p | 26 | SYT4 | Sponge network | -1.047 | 0 | -3.207 | 0 | 0.254 |
| 53567 | RP11-212P7.2 |
hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 25 | DOCK4 | Sponge network | 0.219 | 0.1516 | 0.502 | 0.00108 | 0.254 |
| 53568 | AC074289.1 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-511-5p;hsa-miR-935;hsa-miR-96-5p | 15 | PCDH10 | Sponge network | -0.326 | 0.14314 | -1.644 | 0.0078 | 0.254 |
| 53569 | CTD-2291D10.4 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-532-3p;hsa-miR-96-5p | 12 | GAS7 | Sponge network | 3.21 | 0 | -0.555 | 0.01602 | 0.254 |
| 53570 | LINC00641 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-421;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | LIMCH1 | Sponge network | -0.222 | 0.32445 | -0.915 | 1.0E-5 | 0.254 |
| 53571 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-93-5p | 23 | ADARB1 | Sponge network | 0.131 | 0.8436 | -0.335 | 0.06631 | 0.254 |
| 53572 | RP11-395G23.3 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-942-5p | 12 | GLIPR1 | Sponge network | 0.381 | 0.25967 | 0.146 | 0.3276 | 0.254 |
| 53573 | BOLA3-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p | 10 | GPAM | Sponge network | -0.246 | 0.35034 | 0.142 | 0.29732 | 0.254 |
| 53574 | RP11-894P9.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-582-3p;hsa-miR-582-5p | 15 | RBMS3 | Sponge network | 0.993 | 0 | -0.519 | 0.03551 | 0.254 |
| 53575 | GAS6-AS2 |
hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 23 | RIMS2 | Sponge network | 0.16 | 0.50785 | -1.365 | 0.00208 | 0.254 |
| 53576 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-511-5p;hsa-miR-628-5p | 17 | BCL6 | Sponge network | 0.41 | 0.02214 | -0.211 | 0.19489 | 0.254 |
| 53577 | RP5-1085F17.3 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-590-3p | 14 | PRDM2 | Sponge network | 0.541 | 0.00125 | -0.258 | 0.00721 | 0.254 |
| 53578 | LINC00403 |
hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-424-5p | 11 | PRKAB2 | Sponge network | -3.586 | 0.00032 | -0.367 | 0.01371 | 0.254 |
| 53579 | RP11-158K1.3 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-590-3p;hsa-miR-628-5p | 11 | IRS1 | Sponge network | 0.349 | 0.01695 | -0.026 | 0.88855 | 0.254 |
| 53580 | AC106786.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-877-5p | 13 | IL17RD | Sponge network | 1.122 | 0.02989 | -0.019 | 0.94873 | 0.254 |
| 53581 | CTB-31O20.4 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29c-3p;hsa-miR-424-5p;hsa-miR-629-3p | 11 | CBX5 | Sponge network | 1.619 | 0 | 0.165 | 0.24446 | 0.254 |
| 53582 | RP11-156P1.3 |
hsa-let-7g-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-7-5p | 10 | COL1A2 | Sponge network | 0.214 | 0.18246 | 1.991 | 0 | 0.254 |
| 53583 | RP11-195B17.1 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-409-3p | 18 | AFF1 | Sponge network | 0.37 | 0.27614 | -0.384 | 0.00053 | 0.254 |
| 53584 | WDFY3-AS2 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 27 | EFNA5 | Sponge network | -0.942 | 0 | -0.421 | 0.17868 | 0.254 |
| 53585 | DNAJC27-AS1 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-194-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | CAV1 | Sponge network | -0.969 | 2.0E-5 | -1.013 | 6.0E-5 | 0.254 |
| 53586 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 35 | PRKAA2 | Sponge network | -0.322 | 0.25945 | -2.462 | 0 | 0.254 |
| 53587 | CTA-204B4.2 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 10 | IGSF10 | Sponge network | 0.765 | 7.0E-5 | -1.751 | 1.0E-5 | 0.254 |
| 53588 | WDFY3-AS2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 20 | UBE4B | Sponge network | -0.942 | 0 | -0.235 | 0.007 | 0.254 |
| 53589 | AP000487.5 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326 | 13 | NF1 | Sponge network | 1.987 | 0 | 0.51 | 0 | 0.254 |
| 53590 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p | 12 | PIK3C2A | Sponge network | 1.266 | 0 | 0.061 | 0.53776 | 0.254 |
| 53591 | RP11-298J20.4 |
hsa-let-7b-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-3607-3p | 13 | FBXO32 | Sponge network | 0.002 | 0.99057 | -0.495 | 0.07561 | 0.254 |
| 53592 | MIR143HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 29 | FOXO3 | Sponge network | -0.739 | 0.0574 | -0.04 | 0.72028 | 0.254 |
| 53593 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-629-3p;hsa-miR-940 | 22 | LIFR | Sponge network | -0.154 | 0.53954 | -1.794 | 0 | 0.254 |
| 53594 | LINC00094 |
hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-93-5p | 16 | IGSF10 | Sponge network | 0.253 | 0.09565 | -1.751 | 1.0E-5 | 0.254 |
| 53595 | GNG12-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p;hsa-miR-93-5p | 11 | DAB2 | Sponge network | -0.793 | 7.0E-5 | 0.431 | 0.0113 | 0.254 |
| 53596 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 20 | TET1 | Sponge network | -0.342 | 0.02288 | -0.135 | 0.52138 | 0.254 |
| 53597 | RP11-326G21.1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-18a-3p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-590-3p | 16 | UNC80 | Sponge network | -0.021 | 0.93598 | -1.934 | 0 | 0.254 |
| 53598 | RP11-48B3.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-590-3p | 14 | FGF5 | Sponge network | 1.757 | 0 | 0.549 | 0.28659 | 0.254 |
| 53599 | HCG18 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-655-3p | 11 | ZMYND11 | Sponge network | 1.063 | 0 | -0.2 | 0.05449 | 0.254 |
| 53600 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-625-3p | 14 | RIMBP2 | Sponge network | -0.358 | 0.17255 | -1.968 | 5.0E-5 | 0.254 |
| 53601 | RP11-686D22.8 |
hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 10 | TMEM30A | Sponge network | 0.898 | 1.0E-5 | 0.096 | 0.31031 | 0.254 |
| 53602 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-577 | 20 | MYBL1 | Sponge network | -0.303 | 0.21002 | 0.494 | 0.02152 | 0.254 |
| 53603 | SOX2-OT |
hsa-miR-128-3p;hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p | 17 | SYNM | Sponge network | -1.264 | 6.0E-5 | -2.309 | 1.0E-5 | 0.254 |
| 53604 | U91328.19 |
hsa-let-7b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-577 | 13 | AMPH | Sponge network | -0.303 | 0.21002 | -0.916 | 5.0E-5 | 0.254 |
| 53605 | RP11-216B9.6 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p | 12 | SEMA3E | Sponge network | 0.174 | 0.30034 | -1.717 | 0.00137 | 0.254 |
| 53606 | MEG3 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 16 | BCL9 | Sponge network | 0.249 | 0.35902 | 0.321 | 0.00267 | 0.254 |
| 53607 | SNHG1 |
hsa-miR-100-5p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-149-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p | 11 | RB1 | Sponge network | 1.265 | 0 | 0.382 | 0.0017 | 0.254 |
| 53608 | TRAM2-AS1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-3p | 12 | SCN9A | Sponge network | 0.336 | 0.00739 | -0.525 | 0.10593 | 0.254 |
| 53609 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 23 | BCL11A | Sponge network | -1.697 | 0.00889 | -0.932 | 0.0063 | 0.254 |
| 53610 | DNAJC27-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-93-5p;hsa-miR-935 | 23 | ARNT2 | Sponge network | -0.969 | 2.0E-5 | -0.332 | 0.25851 | 0.254 |
| 53611 | RP11-384O8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p | 15 | RBL2 | Sponge network | -0.738 | 0.21233 | -0.282 | 0.00254 | 0.254 |
| 53612 | RP11-57H14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-378a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 57 | NTRK3 | Sponge network | 0.083 | 0.66405 | -1.77 | 3.0E-5 | 0.254 |
| 53613 | JAZF1-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-421;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 36 | PPP1R9A | Sponge network | -0.769 | 0.00035 | -0.903 | 0.04254 | 0.254 |
| 53614 | RP11-307B6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-421 | 18 | FOXP1 | Sponge network | -4.463 | 0.00025 | 0.042 | 0.74368 | 0.254 |
| 53615 | RP5-994D16.9 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199a-3p;hsa-miR-29a-5p;hsa-miR-342-3p;hsa-miR-590-3p;hsa-miR-766-3p | 10 | MAPK1 | Sponge network | 0.252 | 0.08508 | -0.017 | 0.81465 | 0.254 |
| 53616 | RP11-20I23.13 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-424-5p | 12 | LRRC55 | Sponge network | 0.505 | 0.0014 | -0.026 | 0.93163 | 0.254 |
| 53617 | RP5-994D16.9 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-1976;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | EGLN1 | Sponge network | 0.252 | 0.08508 | -0.127 | 0.1536 | 0.254 |
| 53618 | RP11-127B20.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-628-5p;hsa-miR-654-3p | 16 | FBXO32 | Sponge network | 0.411 | 0.1335 | -0.495 | 0.07561 | 0.254 |
| 53619 | RP11-67L2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-425-5p;hsa-miR-486-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | FGF5 | Sponge network | 0.72 | 0.0001 | 0.549 | 0.28659 | 0.254 |
| 53620 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 29 | ST6GALNAC3 | Sponge network | -0.738 | 0.21233 | -0.987 | 0 | 0.254 |
| 53621 | WAC-AS1 |
hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-590-5p | 11 | MLLT10 | Sponge network | 0.821 | 0 | 0.105 | 0.33292 | 0.254 |
| 53622 | DIO3OS |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-744-5p;hsa-miR-940;hsa-miR-942-5p | 32 | NOTCH2 | Sponge network | -0.773 | 0.04073 | 0.138 | 0.35163 | 0.254 |
| 53623 | RP11-798M19.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 22 | ERCC4 | Sponge network | -0.612 | 0.00094 | 0.126 | 0.15266 | 0.254 |
| 53624 | MEG3 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-7-1-3p | 17 | DICER1 | Sponge network | 0.249 | 0.35902 | 0.042 | 0.674 | 0.254 |
| 53625 | RP5-1198O20.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 38 | LRP1B | Sponge network | 0.997 | 0.00066 | -2.163 | 0 | 0.254 |
| 53626 | RP11-2H8.2 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 13 | TRIM13 | Sponge network | 1.089 | 0 | 0.183 | 0.06968 | 0.254 |
| 53627 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-539-5p | 20 | PBX1 | Sponge network | 0.538 | 0.00076 | -1.206 | 0 | 0.254 |
| 53628 | MIR22HG |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-501-3p;hsa-miR-502-5p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 20 | TGFBR2 | Sponge network | -1.047 | 0 | -0.243 | 0.11408 | 0.254 |
| 53629 | HAND2-AS1 |
hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-185-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-7-5p | 16 | EHD3 | Sponge network | -1.697 | 0.00889 | -0.484 | 0.01747 | 0.254 |
| 53630 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-432-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-93-5p | 46 | UNC80 | Sponge network | -0.355 | 0.0077 | -1.934 | 0 | 0.254 |
| 53631 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-590-3p | 10 | EYA4 | Sponge network | 0.8 | 0 | 0.3 | 0.36876 | 0.254 |
| 53632 | BDNF-AS |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 11 | LHFP | Sponge network | -0.668 | 3.0E-5 | -0.167 | 0.41371 | 0.254 |
| 53633 | RP11-5C23.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-940 | 15 | EPS15 | Sponge network | 0.274 | 0.07681 | -0.052 | 0.53998 | 0.254 |
| 53634 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-98-5p | 20 | SEMA3C | Sponge network | -0.355 | 0.0077 | -0.272 | 0.31153 | 0.254 |
| 53635 | FAM13A-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-20a-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-625-5p | 15 | DCLK1 | Sponge network | -0.83 | 0 | -1.324 | 0.00014 | 0.254 |
| 53636 | CTB-51J22.1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-92a-3p | 14 | DST | Sponge network | -0.803 | 0.23737 | -0.713 | 0.00096 | 0.254 |
| 53637 | RP11-325K4.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-145-3p;hsa-miR-145-5p;hsa-miR-191-5p;hsa-miR-2355-5p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-7-1-3p | 11 | DDX6 | Sponge network | 0.178 | 0.31659 | 0.091 | 0.36428 | 0.254 |
| 53638 | BAIAP2-AS1 |
hsa-let-7g-5p;hsa-miR-140-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c | 20 | WDFY3 | Sponge network | 0.702 | 6.0E-5 | -0.201 | 0.0967 | 0.254 |
| 53639 | ACTA2-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-590-3p | 13 | MAP3K12 | Sponge network | 0.194 | 0.64278 | -0.057 | 0.59369 | 0.254 |
| 53640 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-542-3p | 19 | RARB | Sponge network | -0.773 | 0.04073 | -0.825 | 2.0E-5 | 0.254 |
| 53641 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p | 25 | LIFR | Sponge network | 0.374 | 0.20054 | -1.794 | 0 | 0.254 |
| 53642 | HOXA-AS2 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SLC6A15 | Sponge network | 0.61 | 0.01593 | -2.373 | 2.0E-5 | 0.254 |
| 53643 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 23 | PRKAB2 | Sponge network | 0.889 | 9.0E-5 | -0.367 | 0.01371 | 0.254 |
| 53644 | LINC00694 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-29b-1-5p | 10 | PDGFRA | Sponge network | -0.855 | 0.06723 | -0.372 | 0.0037 | 0.254 |
| 53645 | RP11-359B12.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-3065-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-7-1-3p | 43 | CUX1 | Sponge network | 0.064 | 0.72934 | -0.039 | 0.6705 | 0.254 |
| 53646 | RP5-1198O20.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-93-5p | 10 | JAK1 | Sponge network | 0.997 | 0.00066 | 0.001 | 0.98982 | 0.254 |
| 53647 | FZD10-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-625-5p | 10 | GAS1 | Sponge network | -0.727 | 0.04726 | -1.047 | 0.00364 | 0.254 |
| 53648 | RP11-196G18.23 |
hsa-let-7g-5p;hsa-miR-132-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-628-5p | 12 | FBXO32 | Sponge network | 0.724 | 0.00039 | -0.495 | 0.07561 | 0.254 |
| 53649 | LINC00957 |
hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-378a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | EFNA5 | Sponge network | -0.421 | 0.0114 | -0.421 | 0.17868 | 0.254 |
| 53650 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | LRRC7 | Sponge network | -1.057 | 0.00029 | -1.341 | 0.01193 | 0.254 |
| 53651 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-374a-3p;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 54 | IL6ST | Sponge network | 0.395 | 0.15451 | -0.402 | 0.00676 | 0.254 |
| 53652 | AC074289.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-935 | 19 | DMD | Sponge network | -0.326 | 0.14314 | -1.506 | 0 | 0.254 |
| 53653 | SERTAD4-AS1 |
hsa-miR-103a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-451a | 16 | BCL2 | Sponge network | -0.932 | 0.00329 | -0.899 | 0 | 0.254 |
| 53654 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-940 | 33 | IGF1 | Sponge network | -0.969 | 2.0E-5 | -0.204 | 0.5898 | 0.254 |
| 53655 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 22 | NCAM2 | Sponge network | -0.355 | 0.0077 | -0.482 | 0.22565 | 0.254 |
| 53656 | SNX29P2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 29 | TGFBR3 | Sponge network | 0.4 | 0.14611 | -1.173 | 1.0E-5 | 0.254 |
| 53657 | RP11-216B9.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | MAP3K12 | Sponge network | 0.174 | 0.30034 | -0.057 | 0.59369 | 0.254 |
| 53658 | THAP9-AS1 |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-218-5p;hsa-miR-23b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-628-5p | 12 | MIER3 | Sponge network | 1.079 | 0 | -0.212 | 0.04957 | 0.254 |
| 53659 | CTB-113P19.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-590-3p | 16 | NCOA1 | Sponge network | 0.906 | 0.31715 | -0.432 | 0 | 0.254 |
| 53660 | USP3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-625-5p | 18 | NUAK1 | Sponge network | -0.162 | 0.47687 | 0.904 | 0 | 0.254 |
| 53661 | KB-1572G7.2 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-30b-5p;hsa-miR-3607-3p;hsa-miR-495-3p | 10 | ZNF770 | Sponge network | 0.924 | 0 | 0.206 | 0.06751 | 0.254 |
| 53662 | RP11-104L21.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p | 10 | IL17RD | Sponge network | 0.527 | 0.16506 | -0.019 | 0.94873 | 0.254 |
| 53663 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577 | 11 | GREM1 | Sponge network | 0.657 | 4.0E-5 | 0.902 | 0.01691 | 0.254 |
| 53664 | CTD-2303H24.2 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-766-3p;hsa-miR-96-5p | 29 | IL17RD | Sponge network | 0.415 | 0.14657 | -0.019 | 0.94873 | 0.254 |
| 53665 | HNRNPU-AS1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-191-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | TET1 | Sponge network | 1.601 | 0 | -0.135 | 0.52138 | 0.254 |
| 53666 | WDFY3-AS2 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-98-5p | 16 | ITGA5 | Sponge network | -0.942 | 0 | 0.452 | 0.03524 | 0.254 |
| 53667 | A2M-AS1 |
hsa-let-7b-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 11 | FAM117A | Sponge network | -0.08 | 0.90092 | -1.379 | 0 | 0.254 |
| 53668 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-196a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 30 | CCND2 | Sponge network | -0.663 | 0.10887 | -0.267 | 0.3112 | 0.254 |
| 53669 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-590-3p | 11 | PRDM5 | Sponge network | 0.72 | 0.0001 | -1.09 | 0.00014 | 0.254 |
| 53670 | PWAR6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-96-5p | 39 | SPRY3 | Sponge network | -1.575 | 6.0E-5 | 0.016 | 0.91286 | 0.254 |
| 53671 | CTD-2017D11.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-33a-3p | 11 | SYNE1 | Sponge network | 0.792 | 0.0049 | -0.738 | 0.00316 | 0.254 |
| 53672 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p | 10 | BCL11A | Sponge network | -2.095 | 0.00112 | -0.932 | 0.0063 | 0.254 |
| 53673 | RP11-401P9.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-185-5p;hsa-miR-1976;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-539-5p | 14 | PBX1 | Sponge network | 1.524 | 0.1098 | -1.206 | 0 | 0.254 |
| 53674 | TPTEP1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-501-5p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 38 | PPP1R9A | Sponge network | -1.216 | 0 | -0.903 | 0.04254 | 0.254 |
| 53675 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-671-5p;hsa-miR-92a-3p | 21 | PTPRD | Sponge network | 0.453 | 0.01839 | -1.005 | 0.00064 | 0.254 |
| 53676 | GABPB1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-655-3p | 22 | PIK3C2A | Sponge network | 1.11 | 0 | 0.061 | 0.53776 | 0.254 |
| 53677 | JAZF1-AS1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | GALNT13 | Sponge network | -0.769 | 0.00035 | -0.857 | 0.07439 | 0.254 |
| 53678 | AC141928.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-486-5p | 15 | NR2C2 | Sponge network | 0.853 | 0.15352 | 0.21 | 0.03224 | 0.254 |
| 53679 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-330-3p;hsa-miR-7-1-3p | 12 | DKK3 | Sponge network | 0.395 | 0.15451 | -0.052 | 0.77783 | 0.254 |
| 53680 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-590-3p | 13 | NRK | Sponge network | 0.919 | 0 | 0.656 | 0.19217 | 0.254 |
| 53681 | RP11-822E23.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-5p;hsa-miR-378a-5p | 13 | ARHGAP29 | Sponge network | -0.777 | 0.16667 | 0.131 | 0.42953 | 0.254 |
| 53682 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-93-5p | 30 | HBP1 | Sponge network | -0.777 | 0.1134 | -0.454 | 0 | 0.253 |
| 53683 | JAZF1-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 16 | AKAP13 | Sponge network | -0.769 | 0.00035 | 0.028 | 0.7729 | 0.253 |
| 53684 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-511-5p;hsa-miR-590-3p | 20 | BCL6 | Sponge network | 0.219 | 0.1516 | -0.211 | 0.19489 | 0.253 |
| 53685 | RP11-401P9.4 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-532-5p | 10 | CXCL12 | Sponge network | -0.384 | 0.40652 | -1.421 | 0 | 0.253 |
| 53686 | CTC-487M23.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-450b-5p | 12 | JAZF1 | Sponge network | 0.912 | 0.00014 | -0.377 | 0.02677 | 0.253 |
| 53687 | LINC00863 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | NOTCH2 | Sponge network | 0.189 | 0.13981 | 0.138 | 0.35163 | 0.253 |
| 53688 | CTC-429P9.5 |
hsa-miR-125a-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-590-3p | 12 | RIMS2 | Sponge network | 0.178 | 0.29106 | -1.365 | 0.00208 | 0.253 |
| 53689 | ADIRF-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-342-3p;hsa-miR-590-3p | 10 | FBXO11 | Sponge network | -0.223 | 0.47816 | 0.142 | 0.04938 | 0.253 |
| 53690 | RP11-384O8.1 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 13 | NR3C2 | Sponge network | -0.738 | 0.21233 | -1.56 | 0 | 0.253 |
| 53691 | RP11-95D17.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-338-3p;hsa-miR-33a-5p;hsa-miR-497-5p | 12 | SOX11 | Sponge network | 0.75 | 0 | 2.68 | 0 | 0.253 |
| 53692 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p | 19 | PBX1 | Sponge network | 0.594 | 0.00064 | -1.206 | 0 | 0.253 |
| 53693 | RP11-395A13.2 |
hsa-miR-106a-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p | 16 | DST | Sponge network | 0.057 | 0.61225 | -0.713 | 0.00096 | 0.253 |
| 53694 | LINC00472 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p | 25 | HECA | Sponge network | -0.842 | 0.01361 | -0.217 | 0.03463 | 0.253 |
| 53695 | EIF3J-AS1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-409-3p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | RIMS2 | Sponge network | 0.331 | 0.00509 | -1.365 | 0.00208 | 0.253 |
| 53696 | RP11-373D23.3 |
hsa-let-7d-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-421;hsa-miR-493-3p;hsa-miR-7-1-3p | 16 | NCOA1 | Sponge network | -0.285 | 0.2172 | -0.432 | 0 | 0.253 |
| 53697 | AC108488.4 |
hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-7-1-3p | 10 | TBX18 | Sponge network | 0.259 | 0.12542 | 0.843 | 0.02945 | 0.253 |
| 53698 | ADIRF-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-671-5p | 32 | ADARB1 | Sponge network | -0.223 | 0.47816 | -0.335 | 0.06631 | 0.253 |
| 53699 | GS1-124K5.11 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-590-3p | 15 | ERG | Sponge network | 0.494 | 0.00339 | 0.002 | 0.99011 | 0.253 |
| 53700 | CD27-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-628-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 25 | RASSF8 | Sponge network | 0.177 | 0.16681 | -0.122 | 0.65591 | 0.253 |
| 53701 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7a-5p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-1-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-423-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-590-3p;hsa-miR-98-5p | 17 | PNRC1 | Sponge network | 0.246 | 0.59912 | -0.646 | 0 | 0.253 |
| 53702 | RP1-59D14.5 |
hsa-miR-125a-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-326 | 13 | DMXL1 | Sponge network | 1.162 | 5.0E-5 | -0.426 | 0.00056 | 0.253 |
| 53703 | RP5-1198O20.4 |
hsa-miR-141-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-92b-3p;hsa-miR-940 | 15 | RANBP9 | Sponge network | 0.997 | 0.00066 | -0.191 | 0.04628 | 0.253 |
| 53704 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-96-5p | 56 | TCF4 | Sponge network | 0.222 | 0.29133 | 0.016 | 0.92271 | 0.253 |
| 53705 | RP11-597D13.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-331-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-935 | 48 | PTPRD | Sponge network | 1.302 | 7.0E-5 | -1.005 | 0.00064 | 0.253 |
| 53706 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 31 | FOXO3 | Sponge network | 0.311 | 0.10344 | -0.04 | 0.72028 | 0.253 |
| 53707 | PPP1R26-AS1 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p | 10 | CREB1 | Sponge network | 1.454 | 0 | 0.203 | 0.00537 | 0.253 |
| 53708 | CTC-559E9.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-576-5p | 19 | BNC2 | Sponge network | 0.594 | 0.00064 | -0.491 | 0.09988 | 0.253 |
| 53709 | AC007566.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-30a-3p;hsa-miR-337-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p | 12 | CBL | Sponge network | 2.368 | 2.0E-5 | 0.291 | 0.01267 | 0.253 |
| 53710 | CTC-296K1.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-590-5p | 12 | HBP1 | Sponge network | -2.076 | 0.00548 | -0.454 | 0 | 0.253 |
| 53711 | AC009120.6 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-455-3p;hsa-miR-484;hsa-miR-505-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p | 32 | DST | Sponge network | 0.919 | 0 | -0.713 | 0.00096 | 0.253 |
| 53712 | ERVH48-1 |
hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 12 | CSMD1 | Sponge network | 1.04 | 0.00023 | -0.63 | 0.18881 | 0.253 |
| 53713 | AC004540.4 |
hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-484 | 12 | PRKCB | Sponge network | -2.549 | 0.00528 | -1.313 | 2.0E-5 | 0.253 |
| 53714 | TMEM9B-AS1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-589-3p | 17 | AFF1 | Sponge network | 0.529 | 0.00552 | -0.384 | 0.00053 | 0.253 |
| 53715 | RP11-196G11.4 |
hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | KIF1B | Sponge network | 0.875 | 0 | -0.118 | 0.32258 | 0.253 |
| 53716 | EMX2OS |
hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-744-5p | 12 | PRKACA | Sponge network | -0.777 | 0.1134 | -0.255 | 0.0012 | 0.253 |
| 53717 | CTC-444N24.11 |
hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-374b-5p;hsa-miR-376c-3p | 10 | PAWR | Sponge network | 1.153 | 0 | 0.636 | 0 | 0.253 |
| 53718 | CTC-296K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p | 10 | APC | Sponge network | -2.095 | 0.00112 | -0.255 | 0.04051 | 0.253 |
| 53719 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30e-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | ITGAV | Sponge network | 0.537 | 0.00139 | 0.337 | 0.01002 | 0.253 |
| 53720 | NEAT1 |
hsa-let-7a-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p | 22 | ARID5B | Sponge network | 0.705 | 0.00014 | 0.342 | 0.01676 | 0.253 |
| 53721 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 31 | RBMS3 | Sponge network | 0.267 | 0.2937 | -0.519 | 0.03551 | 0.253 |
| 53722 | RP11-531A24.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-940 | 41 | TRPS1 | Sponge network | 0.75 | 0.00054 | -0.191 | 0.44109 | 0.253 |
| 53723 | PAXIP1-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-664a-5p | 11 | CBFB | Sponge network | 0.66 | 1.0E-5 | 1.061 | 0 | 0.253 |
| 53724 | RP11-867G23.10 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-93-3p | 21 | BMPR2 | Sponge network | -2.531 | 0 | 0.206 | 0.0635 | 0.253 |
| 53725 | FAM157C |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-326 | 10 | SLC5A3 | Sponge network | 0.91 | 0 | 0.493 | 5.0E-5 | 0.253 |
| 53726 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-935 | 21 | LRRC55 | Sponge network | 0.395 | 0.15451 | -0.026 | 0.93163 | 0.253 |
| 53727 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 14 | BCL6 | Sponge network | 0.906 | 0.31715 | -0.211 | 0.19489 | 0.253 |
| 53728 | RP4-639F20.1 |
hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-942-5p | 11 | OPCML | Sponge network | -0.517 | 0.02935 | -2.498 | 0 | 0.253 |
| 53729 | TRIM52-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-34a-5p;hsa-miR-361-5p | 16 | GPC6 | Sponge network | -0.418 | 0.00068 | -0.054 | 0.81465 | 0.253 |
| 53730 | RP11-819C21.1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p;hsa-miR-940 | 27 | ETV1 | Sponge network | 0.537 | 0.00139 | -0.096 | 0.67674 | 0.253 |
| 53731 | HAR1A |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-219a-1-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320b | 13 | JAZF1 | Sponge network | -0.672 | 0.19447 | -0.377 | 0.02677 | 0.253 |
| 53732 | HCG18 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-338-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 28 | WDFY3 | Sponge network | 1.063 | 0 | -0.201 | 0.0967 | 0.253 |
| 53733 | AC127904.2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-590-3p;hsa-miR-664a-5p | 14 | SOX11 | Sponge network | 1.308 | 0 | 2.68 | 0 | 0.253 |
| 53734 | RP11-678G14.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-192-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-7-1-3p | 18 | SASH1 | Sponge network | -4.477 | 0 | -0.502 | 0.00175 | 0.253 |
| 53735 | CTD-3126B10.4 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 24 | LPP | Sponge network | 0.778 | 0.00101 | -0.459 | 0.04475 | 0.253 |
| 53736 | LINC00094 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-589-3p | 23 | SYNPO2 | Sponge network | 0.253 | 0.09565 | -2.342 | 0 | 0.253 |
| 53737 | LINC00957 |
hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | MOB1B | Sponge network | -0.421 | 0.0114 | 0.197 | 0.15266 | 0.253 |
| 53738 | AC083843.1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-374b-5p | 10 | MIER3 | Sponge network | 1.4 | 0 | -0.212 | 0.04957 | 0.253 |
| 53739 | RP11-473I1.10 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-34a-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | NAV3 | Sponge network | 0.673 | 0 | 0.212 | 0.47729 | 0.253 |
| 53740 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | BCL11A | Sponge network | 0.311 | 0.10344 | -0.932 | 0.0063 | 0.253 |
| 53741 | RP11-434B12.1 |
hsa-miR-128-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p | 13 | IRS1 | Sponge network | -0.031 | 0.89063 | -0.026 | 0.88855 | 0.253 |
| 53742 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-29b-2-5p | 11 | ABL2 | Sponge network | -0.278 | 0.76559 | 0.82 | 0 | 0.253 |
| 53743 | AF131215.2 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-7-1-3p | 11 | DNAJB4 | Sponge network | -0.176 | 0.59692 | -0.7 | 1.0E-5 | 0.253 |
| 53744 | RP11-277P12.20 |
hsa-let-7d-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p;hsa-miR-98-5p | 13 | PRRX1 | Sponge network | 0.687 | 0.02753 | 1.316 | 0 | 0.253 |
| 53745 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | SPRY3 | Sponge network | 0.959 | 0 | 0.016 | 0.91286 | 0.253 |
| 53746 | U91328.19 |
hsa-miR-142-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-502-3p;hsa-miR-7-1-3p | 11 | SHPRH | Sponge network | -0.303 | 0.21002 | 0.098 | 0.40994 | 0.253 |
| 53747 | FZD10-AS1 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-935;hsa-miR-942-5p | 28 | OPCML | Sponge network | -0.727 | 0.04726 | -2.498 | 0 | 0.253 |
| 53748 | GS1-124K5.11 |
hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-98-5p | 16 | TES | Sponge network | 0.494 | 0.00339 | -0.348 | 0.00639 | 0.253 |
| 53749 | BDNF-AS |
hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-23a-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-484;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p;hsa-miR-93-3p | 21 | CTIF | Sponge network | -0.668 | 3.0E-5 | -0.389 | 0.00549 | 0.253 |
| 53750 | RP11-53O19.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-655-3p | 18 | KANK1 | Sponge network | -0.405 | 0.22013 | -0.55 | 0.00134 | 0.253 |
| 53751 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-940 | 16 | CHD5 | Sponge network | -0.829 | 0.01343 | -0.922 | 0.01521 | 0.253 |
| 53752 | MAGI2-AS3 |
hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-940 | 21 | SIRT1 | Sponge network | -0.129 | 0.57177 | 0.109 | 0.2593 | 0.253 |
| 53753 | RP4-717I23.3 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-127-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-495-3p | 12 | NF1 | Sponge network | 0.637 | 0.00069 | 0.51 | 0 | 0.253 |
| 53754 | IQCH-AS1 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-495-3p | 21 | CREB1 | Sponge network | 0.929 | 0 | 0.203 | 0.00537 | 0.253 |
| 53755 | H19 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-629-3p | 18 | GAS7 | Sponge network | 2.439 | 0 | -0.555 | 0.01602 | 0.253 |
| 53756 | AC090587.5 |
hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-93-3p | 16 | IL6ST | Sponge network | -0.088 | 0.65011 | -0.402 | 0.00676 | 0.253 |
| 53757 | CTC-296K1.4 |
hsa-let-7b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-223-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-576-5p | 10 | ERC1 | Sponge network | -2.076 | 0.00548 | -0.108 | 0.38794 | 0.253 |
| 53758 | RP11-67L2.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | AKAP13 | Sponge network | 0.72 | 0.0001 | 0.028 | 0.7729 | 0.253 |
| 53759 | RP11-1006G14.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-550a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 11 | RIMBP2 | Sponge network | 0.559 | 0.00026 | -1.968 | 5.0E-5 | 0.253 |
| 53760 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-1296-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-421;hsa-miR-455-5p | 11 | ZNF521 | Sponge network | -1.645 | 0 | -0.111 | 0.58068 | 0.253 |
| 53761 | UBA6-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | MYBL1 | Sponge network | 0.312 | 0.00725 | 0.494 | 0.02152 | 0.253 |
| 53762 | CTD-2002H8.2 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-589-3p;hsa-miR-590-3p | 10 | SCN9A | Sponge network | 0.931 | 1.0E-5 | -0.525 | 0.10593 | 0.253 |
| 53763 | RP11-554A11.4 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-425-5p;hsa-miR-766-3p | 33 | AFF4 | Sponge network | -0.583 | 0.14589 | -0.266 | 0.01565 | 0.253 |
| 53764 | DOCK9-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-539-5p | 12 | HECA | Sponge network | -0.231 | 0.28214 | -0.217 | 0.03463 | 0.253 |
| 53765 | RP11-283I3.6 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-495-3p;hsa-miR-664a-3p | 26 | CCDC6 | Sponge network | 0.614 | 1.0E-5 | 0.27 | 0.02555 | 0.253 |
| 53766 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-1976;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | KAT6B | Sponge network | 0.476 | 0.19203 | -0.478 | 0.00018 | 0.253 |
| 53767 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-500a-5p | 10 | ABCA10 | Sponge network | 0.614 | 1.0E-5 | -0.571 | 0.03674 | 0.253 |
| 53768 | RP11-981G7.6 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TSC1 | Sponge network | 0.986 | 0.01022 | 0.103 | 0.26071 | 0.253 |
| 53769 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 33 | CBL | Sponge network | 0.374 | 0.20054 | 0.291 | 0.01267 | 0.253 |
| 53770 | CTC-429P9.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-532-5p;hsa-miR-542-3p | 15 | PTEN | Sponge network | 0.043 | 0.81628 | -0.187 | 0.07748 | 0.253 |
| 53771 | GS1-124K5.11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 11 | PCDH8 | Sponge network | 0.494 | 0.00339 | -0.997 | 0.05366 | 0.253 |
| 53772 | TPTEP1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-151a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p | 17 | FMNL3 | Sponge network | -1.216 | 0 | 0.555 | 4.0E-5 | 0.253 |
| 53773 | LINC00173 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-24-3p;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-539-5p;hsa-miR-93-5p | 15 | ACSL6 | Sponge network | 0.056 | 0.8494 | -1.593 | 0 | 0.253 |
| 53774 | FAM13A-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-3913-5p;hsa-miR-500b-5p;hsa-miR-590-3p | 14 | SLITRK6 | Sponge network | -0.83 | 0 | -2.121 | 0 | 0.253 |
| 53775 | RP11-727A23.5 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-495-3p;hsa-miR-511-5p | 24 | CREB1 | Sponge network | 1.117 | 0 | 0.203 | 0.00537 | 0.253 |
| 53776 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-362-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | DACT1 | Sponge network | -0.899 | 6.0E-5 | 0.783 | 0.00072 | 0.253 |
| 53777 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-589-3p;hsa-miR-96-5p | 20 | MTSS1 | Sponge network | 0.594 | 0.41522 | -0.81 | 0.00035 | 0.253 |
| 53778 | AC074289.1 |
hsa-miR-134-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-532-3p | 10 | SYNM | Sponge network | -0.326 | 0.14314 | -2.309 | 1.0E-5 | 0.253 |
| 53779 | RP11-395A13.2 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p | 17 | DTNA | Sponge network | 0.057 | 0.61225 | -1.648 | 1.0E-5 | 0.253 |
| 53780 | RP11-263K19.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-183-5p;hsa-miR-188-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-34a-5p;hsa-miR-429 | 20 | ZFHX4 | Sponge network | 0.859 | 0.00331 | 0.018 | 0.95574 | 0.253 |
| 53781 | LINC00092 |
hsa-miR-103a-2-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-30b-3p;hsa-miR-362-3p;hsa-miR-542-3p;hsa-miR-590-3p | 11 | EFNA5 | Sponge network | -1.886 | 0 | -0.421 | 0.17868 | 0.253 |
| 53782 | AC108488.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p | 16 | USP6 | Sponge network | 0.259 | 0.12542 | 0.188 | 0.3871 | 0.253 |
| 53783 | ERVH48-1 |
hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-3p | 19 | PIK3R1 | Sponge network | 1.04 | 0.00023 | -0.133 | 0.36854 | 0.253 |
| 53784 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-92b-3p | 27 | RASSF2 | Sponge network | -0.322 | 0.25945 | -0.273 | 0.21961 | 0.253 |
| 53785 | AC016747.3 |
hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-33a-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-584-5p;hsa-miR-590-3p | 11 | ZNF208 | Sponge network | 0.199 | 0.38015 | -0.4 | 0.22381 | 0.253 |
| 53786 | ADAMTS9-AS2 |
hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 14 | SH3GLB1 | Sponge network | -2.452 | 0 | -0.115 | 0.14773 | 0.253 |
| 53787 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b | 18 | FOXO3 | Sponge network | 0.817 | 3.0E-5 | -0.04 | 0.72028 | 0.253 |
| 53788 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-193b-5p;hsa-miR-200c-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-576-5p | 10 | PCDH17 | Sponge network | 2.163 | 0 | 0.731 | 2.0E-5 | 0.253 |
| 53789 | RP11-263K19.4 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-628-5p | 38 | DTNA | Sponge network | 0.859 | 0.00331 | -1.648 | 1.0E-5 | 0.253 |
| 53790 | SH3BP5-AS1 |
hsa-let-7g-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-7-1-3p | 12 | TRIM13 | Sponge network | 0.267 | 0.2937 | 0.183 | 0.06968 | 0.253 |
| 53791 | RP1-302G2.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p | 12 | ADARB1 | Sponge network | -1.483 | 9.0E-5 | -0.335 | 0.06631 | 0.253 |
| 53792 | ZNF667-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-7-1-3p | 13 | SAMHD1 | Sponge network | -0.976 | 0.00025 | 0.072 | 0.62895 | 0.253 |
| 53793 | RP5-1139B12.3 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-34a-5p;hsa-miR-505-5p;hsa-miR-874-3p | 13 | ASTN1 | Sponge network | 0.598 | 0.38481 | -2.389 | 2.0E-5 | 0.253 |
| 53794 | RP11-395G23.3 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 11 | LUZP2 | Sponge network | 0.381 | 0.25967 | -0.056 | 0.89416 | 0.253 |
| 53795 | RP4-639F20.1 |
hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-542-3p;hsa-miR-624-5p;hsa-miR-7-5p | 10 | SH3GLB1 | Sponge network | -0.517 | 0.02935 | -0.115 | 0.14773 | 0.253 |
| 53796 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-532-5p;hsa-miR-660-5p;hsa-miR-96-5p | 14 | LRRC4 | Sponge network | -0.969 | 2.0E-5 | -1.774 | 0 | 0.253 |
| 53797 | RP11-983P16.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-505-3p;hsa-miR-542-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 18 | LRP1B | Sponge network | 0.41 | 0.02214 | -2.163 | 0 | 0.253 |
| 53798 | RP11-95D17.1 |
hsa-miR-101-5p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-486-5p | 10 | CBFB | Sponge network | 0.75 | 0 | 1.061 | 0 | 0.253 |
| 53799 | C4B-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-629-3p;hsa-miR-96-5p | 14 | ASTN1 | Sponge network | 2.306 | 9.0E-5 | -2.389 | 2.0E-5 | 0.253 |
| 53800 | RP11-434H6.7 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-32-5p | 17 | RUNX1T1 | Sponge network | 0.358 | 0.14302 | -0.344 | 0.2374 | 0.253 |
| 53801 | RP11-497H16.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-411-5p;hsa-miR-590-3p | 14 | USP6 | Sponge network | 0.545 | 0.00064 | 0.188 | 0.3871 | 0.253 |
| 53802 | RP11-193H5.1 |
hsa-miR-125a-3p;hsa-miR-125a-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-149-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p | 10 | RB1 | Sponge network | 1.279 | 2.0E-5 | 0.382 | 0.0017 | 0.253 |
| 53803 | AP001347.6 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 29 | AFF1 | Sponge network | -1.161 | 0.00591 | -0.384 | 0.00053 | 0.253 |
| 53804 | RP5-1112D6.4 |
hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29b-2-5p;hsa-miR-30a-5p;hsa-miR-497-5p | 10 | PAFAH1B2 | Sponge network | 1.379 | 0 | 0.318 | 0.0001 | 0.253 |
| 53805 | LINC00654 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-744-3p | 24 | PTPN14 | Sponge network | 0.374 | 0.20054 | 0.144 | 0.34595 | 0.253 |
| 53806 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-877-5p | 20 | RBMS3 | Sponge network | -0.785 | 0.00035 | -0.519 | 0.03551 | 0.253 |
| 53807 | RP11-479G22.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 10 | NRXN3 | Sponge network | 1.308 | 0 | -0.893 | 0.04186 | 0.253 |
| 53808 | RAB30-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 13 | GPAM | Sponge network | 0.752 | 0 | 0.142 | 0.29732 | 0.253 |
| 53809 | RP11-403I13.7 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-5p | 13 | ABCA10 | Sponge network | 0.395 | 0.15451 | -0.571 | 0.03674 | 0.253 |
| 53810 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p | 17 | NCAM2 | Sponge network | 1.757 | 0 | -0.482 | 0.22565 | 0.253 |
| 53811 | RP11-6N17.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-192-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-331-3p | 12 | PRKAA2 | Sponge network | 0.108 | 0.69556 | -2.462 | 0 | 0.253 |
| 53812 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-550a-3p;hsa-miR-629-3p;hsa-miR-93-5p | 45 | EPHA7 | Sponge network | -1.645 | 0 | -2.197 | 3.0E-5 | 0.253 |
| 53813 | AP001347.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 36 | PTPRD | Sponge network | -1.161 | 0.00591 | -1.005 | 0.00064 | 0.253 |
| 53814 | CTD-2184D3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-30b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CLASP2 | Sponge network | 0.272 | 0.44692 | -0.038 | 0.71 | 0.253 |
| 53815 | RP11-251G23.5 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p | 10 | MAT2A | Sponge network | 1.344 | 0 | -0.073 | 0.36195 | 0.253 |
| 53816 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-590-5p | 12 | LATS1 | Sponge network | 0.796 | 4.0E-5 | 0.109 | 0.34208 | 0.253 |
| 53817 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 15 | LRRC55 | Sponge network | -0.737 | 0.10822 | -0.026 | 0.93163 | 0.253 |
| 53818 | KCNJ2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-93-3p | 16 | AKAP11 | Sponge network | -0.436 | 0.05325 | 0.196 | 0.09825 | 0.253 |
| 53819 | RP11-728F11.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-7-1-3p | 15 | EPHA5 | Sponge network | 0.238 | 0.70544 | -0.957 | 0.01733 | 0.253 |
| 53820 | RP11-464F9.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-590-3p | 15 | TMTC3 | Sponge network | 0.959 | 0 | 0.123 | 0.22189 | 0.253 |
| 53821 | OSER1-AS1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-877-5p | 17 | ASTN1 | Sponge network | -0.13 | 0.48734 | -2.389 | 2.0E-5 | 0.253 |
| 53822 | KB-1460A1.5 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-500a-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-93-3p | 44 | RUNX1T1 | Sponge network | 0.603 | 0.00127 | -0.344 | 0.2374 | 0.253 |
| 53823 | MIR600HG |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-424-5p;hsa-miR-505-5p;hsa-miR-511-5p;hsa-miR-589-3p;hsa-miR-625-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-96-5p | 50 | LPP | Sponge network | 0.594 | 0.41522 | -0.459 | 0.04475 | 0.253 |
| 53824 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-7-1-3p | 21 | CBL | Sponge network | 0.419 | 0.0319 | 0.291 | 0.01267 | 0.253 |
| 53825 | BCYRN1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-192-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-331-3p;hsa-miR-3607-3p;hsa-miR-3613-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 38 | EPHA7 | Sponge network | 0.311 | 0.10344 | -2.197 | 3.0E-5 | 0.253 |
| 53826 | RP11-1277A3.1 |
hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484 | 15 | ABL1 | Sponge network | 0.453 | 0.01839 | -0.215 | 0.07804 | 0.253 |
| 53827 | HCG11 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 40 | PTPRD | Sponge network | 0.385 | 0.10067 | -1.005 | 0.00064 | 0.253 |
| 53828 | AC106786.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-629-5p | 22 | DST | Sponge network | 1.122 | 0.02989 | -0.713 | 0.00096 | 0.253 |
| 53829 | RP11-277P12.20 |
hsa-let-7f-1-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | PIK3CA | Sponge network | 0.687 | 0.02753 | 0.195 | 0.06908 | 0.253 |
| 53830 | WDR86-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-429;hsa-miR-93-5p;hsa-miR-96-5p | 15 | PDGFRA | Sponge network | 0.348 | 0.32912 | -0.372 | 0.0037 | 0.253 |
| 53831 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p | 17 | NOTCH2 | Sponge network | -0.214 | 0.44248 | 0.138 | 0.35163 | 0.253 |
| 53832 | DNAJC27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-374a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 30 | PRKAB2 | Sponge network | -0.969 | 2.0E-5 | -0.367 | 0.01371 | 0.253 |
| 53833 | LINC00472 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 13 | NOTCH2NL | Sponge network | -0.842 | 0.01361 | 0.024 | 0.84307 | 0.253 |
| 53834 | LINC00476 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p | 25 | BRD3 | Sponge network | -0.615 | 0.00015 | 0.37 | 0.00013 | 0.253 |
| 53835 | FAM13A-AS1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-323a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-940 | 40 | MDM4 | Sponge network | -0.83 | 0 | 0.345 | 0.00762 | 0.253 |
| 53836 | OIP5-AS1 |
hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p;hsa-miR-940 | 18 | GLI3 | Sponge network | 0.421 | 0.00084 | -0.615 | 0.01482 | 0.253 |
| 53837 | RP11-968O1.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-362-3p | 27 | ADARB1 | Sponge network | -0.829 | 0.01343 | -0.335 | 0.06631 | 0.253 |
| 53838 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-125a-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | APC | Sponge network | 0.401 | 0.00066 | -0.255 | 0.04051 | 0.253 |
| 53839 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-874-3p | 26 | TACC1 | Sponge network | 0.541 | 0.00125 | -0.362 | 0.05406 | 0.253 |
| 53840 | RP11-35G9.3 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-299-5p;hsa-miR-324-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 16 | SYNPO2 | Sponge network | 0.657 | 4.0E-5 | -2.342 | 0 | 0.253 |
| 53841 | RP11-389C8.2 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-193a-3p;hsa-miR-193b-5p;hsa-miR-196b-5p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-671-5p | 10 | SORCS1 | Sponge network | 0.727 | 3.0E-5 | -3.492 | 0 | 0.253 |
| 53842 | RP11-806O11.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 25 | THRB | Sponge network | 0.284 | 0.52463 | -1.117 | 0.0004 | 0.253 |
| 53843 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-10b-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 29 | CTDSPL | Sponge network | -0.222 | 0.32445 | 0.085 | 0.45121 | 0.253 |
| 53844 | HOXB-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-429 | 14 | PHC3 | Sponge network | -0.154 | 0.53954 | 0.212 | 0.0499 | 0.253 |
| 53845 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-590-3p | 12 | CNTNAP3 | Sponge network | -0.322 | 0.25945 | -1.465 | 0.00033 | 0.253 |
| 53846 | RP11-156P1.3 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-582-5p;hsa-miR-590-3p | 15 | KAT6B | Sponge network | 0.214 | 0.18246 | -0.478 | 0.00018 | 0.253 |
| 53847 | RP11-25K19.1 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | JDP2 | Sponge network | -1.057 | 0.00029 | -0.967 | 0 | 0.253 |
| 53848 | RP11-195F19.9 |
hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-22-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 15 | OPCML | Sponge network | -0.726 | 0.00132 | -2.498 | 0 | 0.253 |
| 53849 | TTC28-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 18 | PRKAB2 | Sponge network | 0.373 | 0.00068 | -0.367 | 0.01371 | 0.253 |
| 53850 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 16 | DACH2 | Sponge network | -0.769 | 0.00035 | -1.635 | 0.00034 | 0.253 |
| 53851 | AC116366.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-27a-3p;hsa-miR-324-3p;hsa-miR-424-5p;hsa-miR-577 | 10 | ZFHX3 | Sponge network | 0.329 | 0.11122 | 0.389 | 0.01363 | 0.253 |
| 53852 | CTB-113P19.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-590-3p | 15 | PLSCR4 | Sponge network | 0.906 | 0.31715 | -0.474 | 0.03833 | 0.253 |
| 53853 | RP11-967K21.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-501-5p;hsa-miR-542-3p;hsa-miR-93-5p | 11 | TXNIP | Sponge network | 0.056 | 0.81195 | -1.277 | 0 | 0.253 |
| 53854 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-151a-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p | 13 | PTPRB | Sponge network | 0.199 | 0.38015 | 0.105 | 0.47122 | 0.253 |
| 53855 | MALAT1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 12 | LATS2 | Sponge network | 0.782 | 0.00016 | 0.159 | 0.2304 | 0.253 |
| 53856 | RP11-226L15.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-214-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-590-3p | 11 | MLLT10 | Sponge network | 0.8 | 0 | 0.105 | 0.33292 | 0.253 |
| 53857 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-502-3p;hsa-miR-671-5p | 24 | ARID5B | Sponge network | -0.154 | 0.78939 | 0.342 | 0.01676 | 0.253 |
| 53858 | ADIRF-AS1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-197-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | ZNF208 | Sponge network | -0.223 | 0.47816 | -0.4 | 0.22381 | 0.253 |
| 53859 | RP11-226L15.5 |
hsa-miR-134-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-495-3p;hsa-miR-590-3p | 12 | ERCC6 | Sponge network | 0.8 | 0 | 0.23 | 0.015 | 0.253 |
| 53860 | AC093627.8 |
hsa-miR-107;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-24-3p | 15 | DTNA | Sponge network | -0.787 | 0.3928 | -1.648 | 1.0E-5 | 0.253 |
| 53861 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 54 | IL6ST | Sponge network | 0.374 | 0.20054 | -0.402 | 0.00676 | 0.253 |
| 53862 | RP11-166D19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-625-5p;hsa-miR-629-3p;hsa-miR-7-1-3p | 48 | CBX5 | Sponge network | -0.576 | 0.10121 | 0.165 | 0.24446 | 0.253 |
| 53863 | TMEM51-AS1 |
hsa-miR-106a-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-223-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-423-5p;hsa-miR-590-3p | 14 | GNAQ | Sponge network | 0.23 | 0.38667 | -0.367 | 0.00102 | 0.253 |
| 53864 | RP1-302G2.5 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-20a-5p;hsa-miR-935 | 10 | ARNT2 | Sponge network | -1.483 | 9.0E-5 | -0.332 | 0.25851 | 0.253 |
| 53865 | RP11-119F7.5 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | UNC5C | Sponge network | 0.225 | 0.30644 | -0.178 | 0.40214 | 0.253 |
| 53866 | AC006116.24 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-93-5p | 12 | RBL2 | Sponge network | -0.473 | 0.07602 | -0.282 | 0.00254 | 0.253 |
| 53867 | CTD-2002H8.2 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200c-5p;hsa-miR-214-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | AFF1 | Sponge network | 0.931 | 1.0E-5 | -0.384 | 0.00053 | 0.253 |
| 53868 | LINC00173 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-93-5p | 22 | UNC80 | Sponge network | 0.056 | 0.8494 | -1.934 | 0 | 0.253 |
| 53869 | HAND2-AS1 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-455-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 23 | SUFU | Sponge network | -1.697 | 0.00889 | -0.04 | 0.65489 | 0.253 |
| 53870 | SERHL |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-196b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-29a-5p | 23 | BCL2 | Sponge network | -0.602 | 0.00331 | -0.899 | 0 | 0.253 |
| 53871 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-495-3p | 10 | PRKAB2 | Sponge network | 0.909 | 0 | -0.367 | 0.01371 | 0.253 |
| 53872 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-450b-5p | 13 | PTPN11 | Sponge network | 0.95 | 0.15943 | 0.431 | 2.0E-5 | 0.253 |
| 53873 | SOX2-OT |
hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | RIT1 | Sponge network | -1.264 | 6.0E-5 | -0.296 | 0.00054 | 0.253 |
| 53874 | U91328.21 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-330-3p | 14 | MCC | Sponge network | -0.735 | 0.15983 | -1.005 | 0 | 0.253 |
| 53875 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 19 | BCL11A | Sponge network | 0.41 | 0.02214 | -0.932 | 0.0063 | 0.253 |
| 53876 | RP11-35G9.3 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 19 | ABI2 | Sponge network | 0.657 | 4.0E-5 | 0.175 | 0.14787 | 0.253 |
| 53877 | RP4-605O3.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 15 | HECA | Sponge network | 0.262 | 0.0475 | -0.217 | 0.03463 | 0.253 |
| 53878 | RP11-57H14.4 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-96-5p | 22 | CYLD | Sponge network | 0.083 | 0.66405 | -0.367 | 0.00171 | 0.253 |
| 53879 | AC127904.2 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-145-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 17 | EPS15 | Sponge network | 1.308 | 0 | -0.052 | 0.53998 | 0.253 |
| 53880 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p | 15 | KANK1 | Sponge network | -0.322 | 0.25945 | -0.55 | 0.00134 | 0.253 |
| 53881 | RP11-57H14.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-935 | 19 | ARNT2 | Sponge network | 0.083 | 0.66405 | -0.332 | 0.25851 | 0.253 |
| 53882 | HOXD-AS2 |
hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-3607-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-500a-5p | 20 | WDFY3 | Sponge network | -0.208 | 0.65592 | -0.201 | 0.0967 | 0.253 |
| 53883 | RP11-517B11.7 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181a-2-3p;hsa-miR-193b-5p;hsa-miR-2355-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-5p | 17 | MYO9A | Sponge network | 0.706 | 0 | -0.147 | 0.32373 | 0.253 |
| 53884 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-132-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-425-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | FGF5 | Sponge network | 0.222 | 0.29133 | 0.549 | 0.28659 | 0.253 |
| 53885 | RP11-296I10.3 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-23a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-331-3p;hsa-miR-425-5p;hsa-miR-484;hsa-miR-539-5p;hsa-miR-942-5p | 21 | DDX6 | Sponge network | 0.592 | 0.00014 | 0.091 | 0.36428 | 0.253 |
| 53886 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 32 | MTSS1 | Sponge network | -0.709 | 0.18168 | -0.81 | 0.00035 | 0.253 |
| 53887 | RP11-65J21.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-128-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 10 | RNF144A | Sponge network | -0.557 | 0.11135 | 0.594 | 0.0009 | 0.253 |
| 53888 | CALML3-AS1 |
hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p | 15 | UBR3 | Sponge network | 0.95 | 0.15943 | -0.021 | 0.82774 | 0.253 |
| 53889 | RP11-218M22.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-942-5p | 27 | TMEM132B | Sponge network | 0.197 | 0.25618 | -0.83 | 0.01084 | 0.253 |
| 53890 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-1976;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-33a-5p | 17 | ADARB1 | Sponge network | -0.214 | 0.44248 | -0.335 | 0.06631 | 0.253 |
| 53891 | RP11-819C21.1 |
hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-24-2-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 35 | PPM1L | Sponge network | 0.537 | 0.00139 | -0.537 | 0.00616 | 0.253 |
| 53892 | AF131215.2 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-30e-5p;hsa-miR-370-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-93-3p | 15 | RPS6KA2 | Sponge network | -0.176 | 0.59692 | -0.57 | 0.00013 | 0.253 |
| 53893 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-376c-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p | 25 | ZMYM2 | Sponge network | 0.064 | 0.72934 | 0.279 | 0.01273 | 0.253 |
| 53894 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 24 | FOXO3 | Sponge network | -0.235 | 0.15142 | -0.04 | 0.72028 | 0.253 |
| 53895 | MRPL23-AS1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p | 11 | HIP1 | Sponge network | -0.278 | 0.76559 | 0.774 | 0 | 0.253 |
| 53896 | RP11-1094M14.11 |
hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-369-3p;hsa-miR-628-5p | 10 | CDC73 | Sponge network | 0.586 | 2.0E-5 | 0.094 | 0.20463 | 0.253 |
| 53897 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p;hsa-miR-495-3p;hsa-miR-582-5p;hsa-miR-625-5p | 17 | ZMYM2 | Sponge network | -0.176 | 0.59692 | 0.279 | 0.01273 | 0.253 |
| 53898 | KB-431C1.4 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-342-3p | 10 | SON | Sponge network | 0.909 | 0 | 0.168 | 0.04406 | 0.253 |
| 53899 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-493-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | FREM2 | Sponge network | 0.799 | 1.0E-5 | -0.437 | 0.46353 | 0.253 |
| 53900 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 18 | CDHR3 | Sponge network | -0.513 | 0.04703 | -0.977 | 4.0E-5 | 0.253 |
| 53901 | WDFY3-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | RBL2 | Sponge network | -0.942 | 0 | -0.282 | 0.00254 | 0.253 |
| 53902 | AC010226.4 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-590-3p;hsa-miR-7-5p | 10 | TET1 | Sponge network | -0.811 | 2.0E-5 | -0.135 | 0.52138 | 0.253 |
| 53903 | AC093627.8 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-29b-1-5p;hsa-miR-7-5p | 12 | EGFR | Sponge network | -0.787 | 0.3928 | -0.175 | 0.48451 | 0.253 |
| 53904 | AC156455.1 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-195-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-335-5p | 15 | AKAP11 | Sponge network | 1.154 | 0.00041 | 0.196 | 0.09825 | 0.253 |
| 53905 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-590-3p | 11 | MAP3K12 | Sponge network | 0.225 | 0.30644 | -0.057 | 0.59369 | 0.253 |
| 53906 | AC009948.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p | 21 | ARHGAP28 | Sponge network | 0.532 | 0.00505 | -0.911 | 0.00013 | 0.253 |
| 53907 | CTD-3064M3.3 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-629-3p | 39 | KIF1B | Sponge network | -0.469 | 0.08721 | -0.118 | 0.32258 | 0.253 |
| 53908 | NEAT1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-378c;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 22 | BRD3 | Sponge network | 0.705 | 0.00014 | 0.37 | 0.00013 | 0.253 |
| 53909 | RP11-725P16.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | SLC6A15 | Sponge network | 0.422 | 0.27021 | -2.373 | 2.0E-5 | 0.253 |
| 53910 | RP11-216B9.6 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-374a-3p;hsa-miR-375;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-940 | 30 | GAS7 | Sponge network | 0.174 | 0.30034 | -0.555 | 0.01602 | 0.253 |
| 53911 | JAZF1-AS1 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-485-3p;hsa-miR-590-3p | 15 | TMTC2 | Sponge network | -0.769 | 0.00035 | -0.215 | 0.18248 | 0.253 |
| 53912 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-340-5p;hsa-miR-34a-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-590-3p;hsa-miR-93-5p | 28 | EPHA5 | Sponge network | 0.051 | 0.83388 | -0.957 | 0.01733 | 0.253 |
| 53913 | HOTAIRM1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-424-5p;hsa-miR-590-3p | 10 | RET | Sponge network | -0.053 | 0.80685 | -0.833 | 0.03517 | 0.253 |
| 53914 | RP11-439L18.2 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-454-3p | 10 | DLC1 | Sponge network | 0.224 | 0.48719 | -0.382 | 0.05038 | 0.253 |
| 53915 | AC006116.24 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-24-3p;hsa-miR-323a-3p | 12 | MXI1 | Sponge network | -0.473 | 0.07602 | -0.987 | 0 | 0.253 |
| 53916 | HAND2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | PSIP1 | Sponge network | -1.697 | 0.00889 | -0.005 | 0.96897 | 0.253 |
| 53917 | MIR181A2HG |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-30e-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 11 | PHC3 | Sponge network | 1.822 | 1.0E-5 | 0.212 | 0.0499 | 0.253 |
| 53918 | CTC-457L16.2 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-7-1-3p;hsa-miR-940 | 10 | CREB1 | Sponge network | 1.153 | 0.00114 | 0.203 | 0.00537 | 0.253 |
| 53919 | DNM3OS |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-940 | 30 | PTPRT | Sponge network | 0.572 | 0.01722 | -0.907 | 0.03727 | 0.253 |
| 53920 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-361-3p;hsa-miR-550a-5p;hsa-miR-590-3p | 18 | CRTC1 | Sponge network | -0.227 | 0.31657 | -0.116 | 0.28412 | 0.253 |
| 53921 | AC006042.6 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-152-3p;hsa-miR-195-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | TBL1XR1 | Sponge network | 1.671 | 2.0E-5 | 0.578 | 0 | 0.253 |
| 53922 | LINC00843 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-429 | 19 | IL6ST | Sponge network | 1.106 | 0 | -0.402 | 0.00676 | 0.253 |
| 53923 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-766-3p | 30 | MAPK1 | Sponge network | 0.91 | 0 | -0.017 | 0.81465 | 0.253 |
| 53924 | MAP3K14 |
hsa-miR-106b-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | PLSCR4 | Sponge network | -0.295 | 0.02604 | -0.474 | 0.03833 | 0.253 |
| 53925 | RP11-411K7.1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 17 | RPS6KA3 | Sponge network | 0.155 | 0.81273 | 0.256 | 0.02419 | 0.253 |
| 53926 | LINC00865 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-7-1-3p | 16 | JDP2 | Sponge network | -1.249 | 7.0E-5 | -0.967 | 0 | 0.253 |
| 53927 | RP11-728F11.4 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-7-1-3p | 15 | BNC2 | Sponge network | 0.238 | 0.70544 | -0.491 | 0.09988 | 0.253 |
| 53928 | SERHL |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-382-5p;hsa-miR-421 | 10 | ZNF844 | Sponge network | -0.602 | 0.00331 | -0.966 | 0.00035 | 0.253 |
| 53929 | LINC00667 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-34a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-671-5p | 20 | KIT | Sponge network | -0.235 | 0.15142 | -2.184 | 0 | 0.253 |
| 53930 | PART1 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-338-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-651-5p | 35 | PRDM2 | Sponge network | -2.925 | 0 | -0.258 | 0.00721 | 0.253 |
| 53931 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 27 | MCC | Sponge network | 0.619 | 0.00157 | -1.005 | 0 | 0.253 |
| 53932 | CTC-297N7.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-671-5p | 11 | DLC1 | Sponge network | -2.069 | 0 | -0.382 | 0.05038 | 0.253 |
| 53933 | LINC01018 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-370-3p;hsa-miR-532-3p;hsa-miR-589-5p;hsa-miR-93-3p | 14 | RPS6KA2 | Sponge network | -2.915 | 0.00015 | -0.57 | 0.00013 | 0.253 |
| 53934 | DNM1P35 |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-369-3p;hsa-miR-374a-5p;hsa-miR-508-3p;hsa-miR-584-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-766-3p;hsa-miR-93-5p | 34 | CADM2 | Sponge network | 0.051 | 0.83388 | -3.782 | 0 | 0.253 |
| 53935 | RP11-379F4.7 |
hsa-miR-130a-3p;hsa-miR-139-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-29b-2-5p;hsa-miR-374b-5p;hsa-miR-664a-3p | 10 | UBE2D1 | Sponge network | 1.316 | 0 | 0.468 | 0 | 0.253 |
| 53936 | AC116366.6 |
hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-148b-3p;hsa-miR-152-3p;hsa-miR-182-5p;hsa-miR-200c-5p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-326;hsa-miR-421;hsa-miR-577;hsa-miR-92a-3p;hsa-miR-92b-3p | 13 | DMXL1 | Sponge network | 0.329 | 0.11122 | -0.426 | 0.00056 | 0.253 |
| 53937 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-141-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p | 20 | FBXO32 | Sponge network | -0.785 | 0.00035 | -0.495 | 0.07561 | 0.253 |
| 53938 | LINC00173 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-455-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 18 | PRRX1 | Sponge network | 0.056 | 0.8494 | 1.316 | 0 | 0.253 |
| 53939 | LINC00672 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-93-5p | 12 | LRRC55 | Sponge network | -0.227 | 0.31657 | -0.026 | 0.93163 | 0.253 |
| 53940 | RP11-554A11.9 |
hsa-let-7d-5p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-301a-3p;hsa-miR-505-5p;hsa-miR-940 | 10 | CADM2 | Sponge network | -2.279 | 0.00011 | -3.782 | 0 | 0.253 |
| 53941 | LPP-AS2 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-455-5p | 10 | THSD7A | Sponge network | -0.18 | 0.20343 | 0.242 | 0.25154 | 0.253 |
| 53942 | AF131215.9 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | PTCH1 | Sponge network | -0.322 | 0.25945 | -0.1 | 0.6179 | 0.253 |
| 53943 | HCG22 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | NAV3 | Sponge network | 0.816 | 0.32368 | 0.212 | 0.47729 | 0.253 |
| 53944 | LINC00667 |
hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-330-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 11 | DNMT3A | Sponge network | -0.235 | 0.15142 | 0.302 | 0.0262 | 0.253 |
| 53945 | RP11-13K12.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-629-3p;hsa-miR-93-5p | 15 | RBL2 | Sponge network | -0.154 | 0.78939 | -0.282 | 0.00254 | 0.253 |
| 53946 | RAD51-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-590-3p | 14 | SPRY3 | Sponge network | 0.768 | 7.0E-5 | 0.016 | 0.91286 | 0.253 |
| 53947 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-429;hsa-miR-485-3p;hsa-miR-495-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 23 | TP53INP1 | Sponge network | 0.421 | 0.00084 | -0.496 | 0.00064 | 0.253 |
| 53948 | RP11-384K6.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-338-3p | 10 | TBX18 | Sponge network | 0.946 | 0 | 0.843 | 0.02945 | 0.253 |
| 53949 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-96-5p | 11 | PCDH8 | Sponge network | -0.18 | 0.20343 | -0.997 | 0.05366 | 0.253 |
| 53950 | RP11-731F5.2 |
hsa-let-7d-5p;hsa-miR-15b-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-324-5p;hsa-miR-93-3p | 10 | RUNX1T1 | Sponge network | 0.053 | 0.95207 | -0.344 | 0.2374 | 0.253 |
| 53951 | RP11-5C23.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-193b-5p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-590-3p | 22 | ACVR2A | Sponge network | 0.274 | 0.07681 | -0.409 | 0.00039 | 0.253 |
| 53952 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-874-3p | 11 | TACC1 | Sponge network | 2.364 | 0.00134 | -0.362 | 0.05406 | 0.253 |
| 53953 | PAXIP1-AS1 |
hsa-miR-129-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-195-3p;hsa-miR-664a-5p | 10 | SOX11 | Sponge network | 0.66 | 1.0E-5 | 2.68 | 0 | 0.253 |
| 53954 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-378a-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 15 | NUAK1 | Sponge network | -0.206 | 0.12942 | 0.904 | 0 | 0.253 |
| 53955 | RP11-144G6.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | ARID5B | Sponge network | 0.826 | 1.0E-5 | 0.342 | 0.01676 | 0.253 |
| 53956 | RP11-686D22.8 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-590-3p | 17 | CREB1 | Sponge network | 0.898 | 1.0E-5 | 0.203 | 0.00537 | 0.253 |
| 53957 | CTD-3099C6.9 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p | 10 | MED12L | Sponge network | 1.361 | 0 | -0.194 | 0.55299 | 0.253 |
| 53958 | C4A-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-146a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-589-3p | 14 | KCNA6 | Sponge network | 3.345 | 0 | -1.531 | 0.00013 | 0.253 |
| 53959 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | MITF | Sponge network | 0.385 | 0.10067 | -0.769 | 0.00022 | 0.253 |
| 53960 | AF131215.2 |
hsa-miR-101-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30e-3p;hsa-miR-374b-3p;hsa-miR-576-5p | 16 | UBR3 | Sponge network | -0.176 | 0.59692 | -0.021 | 0.82774 | 0.253 |
| 53961 | A2M-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-362-5p;hsa-miR-96-5p | 15 | CBFA2T3 | Sponge network | -0.08 | 0.90092 | -1.221 | 1.0E-5 | 0.253 |
| 53962 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-590-3p | 16 | SCN9A | Sponge network | -0.125 | 0.49414 | -0.525 | 0.10593 | 0.253 |
| 53963 | LINC00963 |
hsa-let-7a-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-378c;hsa-miR-486-5p;hsa-miR-590-3p | 11 | DACT1 | Sponge network | 0.523 | 9.0E-5 | 0.783 | 0.00072 | 0.253 |
| 53964 | CALML3-AS1 |
hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-20a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-625-5p | 12 | DCLK1 | Sponge network | 0.95 | 0.15943 | -1.324 | 0.00014 | 0.253 |
| 53965 | MRPL23-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p | 15 | NTRK2 | Sponge network | -0.278 | 0.76559 | -0.736 | 0.06495 | 0.252 |
| 53966 | LOXL1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-766-3p | 21 | IL17RD | Sponge network | 0.487 | 0.04053 | -0.019 | 0.94873 | 0.252 |
| 53967 | AC024560.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-339-5p | 11 | MLLT10 | Sponge network | 2.166 | 0.00228 | 0.105 | 0.33292 | 0.252 |
| 53968 | LINC00863 |
hsa-miR-103a-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-424-5p;hsa-miR-590-3p | 14 | DYNC1I1 | Sponge network | 0.189 | 0.13981 | -1.12 | 0.00107 | 0.252 |
| 53969 | RP11-356J5.12 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 25 | CRTC1 | Sponge network | -0.899 | 6.0E-5 | -0.116 | 0.28412 | 0.252 |
| 53970 | HHIP-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-590-3p | 32 | ADARB1 | Sponge network | -0.737 | 0.10822 | -0.335 | 0.06631 | 0.252 |
| 53971 | PSMG3-AS1 |
hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 22 | LIMCH1 | Sponge network | -0.125 | 0.49414 | -0.915 | 1.0E-5 | 0.252 |
| 53972 | RP11-121C2.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-511-5p;hsa-miR-582-5p | 17 | ZFHX3 | Sponge network | 1.147 | 0 | 0.389 | 0.01363 | 0.252 |
| 53973 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 23 | PRRX1 | Sponge network | 0.614 | 1.0E-5 | 1.316 | 0 | 0.252 |
| 53974 | LINC00886 |
hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-361-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-532-3p;hsa-miR-590-5p;hsa-miR-874-3p | 10 | HSPB7 | Sponge network | -0.371 | 0.24604 | -2.268 | 1.0E-5 | 0.252 |
| 53975 | CTC-429P9.3 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-339-5p | 12 | PIK3CA | Sponge network | 0.082 | 0.55397 | 0.195 | 0.06908 | 0.252 |
| 53976 | RP11-6O2.3 |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-326;hsa-miR-340-5p;hsa-miR-452-5p;hsa-miR-590-5p | 14 | TES | Sponge network | 0.331 | 0.57247 | -0.348 | 0.00639 | 0.252 |
| 53977 | RP5-890O3.9 |
hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 10 | GPAM | Sponge network | 0.842 | 0 | 0.142 | 0.29732 | 0.252 |
| 53978 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-590-3p | 23 | MYBL1 | Sponge network | 0.219 | 0.1516 | 0.494 | 0.02152 | 0.252 |
| 53979 | RP4-758J18.10 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-330-5p | 10 | LRIG1 | Sponge network | -0.091 | 0.71468 | -0.537 | 0.00588 | 0.252 |
| 53980 | RP11-532F6.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-423-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-940 | 14 | CELF4 | Sponge network | -0.896 | 0.00099 | -1.135 | 0.00344 | 0.252 |
| 53981 | SSSCA1-AS1 |
hsa-miR-142-5p;hsa-miR-143-5p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p | 12 | RICTOR | Sponge network | 0.487 | 0.00018 | 0.388 | 0.00188 | 0.252 |
| 53982 | RP11-1006G14.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-628-5p;hsa-miR-7-1-3p;hsa-miR-766-3p;hsa-miR-93-3p | 48 | IL6ST | Sponge network | 0.559 | 0.00026 | -0.402 | 0.00676 | 0.252 |
| 53983 | RP11-116O18.1 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-505-5p;hsa-miR-590-3p | 15 | ASTN1 | Sponge network | 0.05 | 0.95483 | -2.389 | 2.0E-5 | 0.252 |
| 53984 | RP11-130L8.1 |
hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p | 14 | PCDH10 | Sponge network | -0.663 | 0.10887 | -1.644 | 0.0078 | 0.252 |
| 53985 | AC127904.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-455-3p;hsa-miR-590-3p | 17 | SPRY3 | Sponge network | 1.308 | 0 | 0.016 | 0.91286 | 0.252 |
| 53986 | CD27-AS1 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-221-3p;hsa-miR-222-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-342-5p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-486-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-628-5p;hsa-miR-7-1-3p | 48 | DTNA | Sponge network | 0.177 | 0.16681 | -1.648 | 1.0E-5 | 0.252 |
| 53987 | KCNJ2-AS1 |
hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-590-3p | 18 | PIK3R1 | Sponge network | -0.436 | 0.05325 | -0.133 | 0.36854 | 0.252 |
| 53988 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-493-5p;hsa-miR-7-1-3p | 24 | MLLT10 | Sponge network | -0.405 | 0.22013 | 0.105 | 0.33292 | 0.252 |
| 53989 | RP4-758J18.10 |
hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-330-5p;hsa-miR-450b-5p | 19 | PPM1L | Sponge network | -0.091 | 0.71468 | -0.537 | 0.00616 | 0.252 |
| 53990 | RP11-359B12.2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | ABCA10 | Sponge network | 0.064 | 0.72934 | -0.571 | 0.03674 | 0.252 |
| 53991 | DNM1P35 |
hsa-let-7a-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-7-5p | 23 | BTG2 | Sponge network | 0.051 | 0.83388 | -1.763 | 0 | 0.252 |
| 53992 | LINC00667 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-671-5p | 28 | DLC1 | Sponge network | -0.235 | 0.15142 | -0.382 | 0.05038 | 0.252 |
| 53993 | RP11-67L2.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p | 19 | LUZP2 | Sponge network | 0.72 | 0.0001 | -0.056 | 0.89416 | 0.252 |
| 53994 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | PBRM1 | Sponge network | 0.848 | 0 | -0.029 | 0.78601 | 0.252 |
| 53995 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 29 | TACC1 | Sponge network | 0.538 | 0.00076 | -0.362 | 0.05406 | 0.252 |
| 53996 | RP11-67L2.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148b-3p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-590-3p | 11 | SMARCA1 | Sponge network | 0.72 | 0.0001 | -0.684 | 0.00159 | 0.252 |
| 53997 | CTD-2314G24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-421;hsa-miR-590-3p | 11 | LRRK2 | Sponge network | 0.246 | 0.59912 | -1.136 | 1.0E-5 | 0.252 |
| 53998 | HCG22 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29b-1-5p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-671-5p;hsa-miR-7-1-3p | 16 | THBS1 | Sponge network | 0.816 | 0.32368 | 0.741 | 0.00386 | 0.252 |
| 53999 | AC025171.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | ROBO1 | Sponge network | 0.998 | 0 | -0.009 | 0.96154 | 0.252 |
| 54000 | HOTAIRM1 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-424-5p;hsa-miR-590-3p | 11 | BAZ2B | Sponge network | -0.053 | 0.80685 | -0.276 | 0.02833 | 0.252 |
| 54001 | GS1-124K5.11 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-320a;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-590-3p | 17 | RPS6KA3 | Sponge network | 0.494 | 0.00339 | 0.256 | 0.02419 | 0.252 |
| 54002 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-28-3p;hsa-miR-28-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-582-3p;hsa-miR-590-3p | 13 | FOXO3 | Sponge network | 0.643 | 0 | -0.04 | 0.72028 | 0.252 |
| 54003 | PCA3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-590-5p | 19 | KANK1 | Sponge network | -0.709 | 0.18168 | -0.55 | 0.00134 | 0.252 |
| 54004 | CTC-338M12.4 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p | 13 | TBL1XR1 | Sponge network | 0.638 | 0 | 0.578 | 0 | 0.252 |
| 54005 | NDUFA6-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200c-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | TSC1 | Sponge network | -0.355 | 0.0077 | 0.103 | 0.26071 | 0.252 |
| 54006 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-96-5p | 20 | ACVR1 | Sponge network | 0.219 | 0.1516 | 0.279 | 0.00234 | 0.252 |
| 54007 | TMEM161B-AS1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-320b;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-590-3p | 13 | CSNK1A1 | Sponge network | -0.04 | 0.7606 | -0.19 | 0.01687 | 0.252 |
| 54008 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-501-3p;hsa-miR-589-3p | 25 | UNC80 | Sponge network | -0.154 | 0.53954 | -1.934 | 0 | 0.252 |
| 54009 | RP4-717I23.3 |
hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p | 10 | IGSF10 | Sponge network | 0.637 | 0.00069 | -1.751 | 1.0E-5 | 0.252 |
| 54010 | RP11-490M8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-766-3p | 13 | CHD5 | Sponge network | -0.214 | 0.44248 | -0.922 | 0.01521 | 0.252 |
| 54011 | HOXB-AS2 |
hsa-miR-125a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-550a-5p | 10 | CCDC6 | Sponge network | 1.316 | 0.01106 | 0.27 | 0.02555 | 0.252 |
| 54012 | CTA-204B4.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 14 | PDE4DIP | Sponge network | 0.765 | 7.0E-5 | -0.282 | 0.0257 | 0.252 |
| 54013 | ACTA2-AS1 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 35 | PPP1R9A | Sponge network | 0.194 | 0.64278 | -0.903 | 0.04254 | 0.252 |
| 54014 | ZNF667-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-144-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 33 | MOB1B | Sponge network | -0.976 | 0.00025 | 0.197 | 0.15266 | 0.252 |
| 54015 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-193a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-590-3p | 10 | NF1 | Sponge network | 1.073 | 0.00216 | 0.51 | 0 | 0.252 |
| 54016 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-411-5p;hsa-miR-590-3p | 22 | USP6 | Sponge network | 0.101 | 0.60919 | 0.188 | 0.3871 | 0.252 |
| 54017 | LINC00963 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 13 | PTPN11 | Sponge network | 0.523 | 9.0E-5 | 0.431 | 2.0E-5 | 0.252 |
| 54018 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-651-5p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 50 | BMPR2 | Sponge network | 0.199 | 0.38015 | 0.206 | 0.0635 | 0.252 |
| 54019 | CTD-2006H14.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 10 | DST | Sponge network | 1.378 | 0 | -0.713 | 0.00096 | 0.252 |
| 54020 | RP4-717I23.3 |
hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-629-3p;hsa-miR-664a-5p | 31 | DST | Sponge network | 0.637 | 0.00069 | -0.713 | 0.00096 | 0.252 |
| 54021 | ARHGAP5-AS1 |
hsa-miR-128-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-29a-5p;hsa-miR-542-3p | 10 | SYNM | Sponge network | -0.644 | 0.00021 | -2.309 | 1.0E-5 | 0.252 |
| 54022 | RP11-736K20.5 |
hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-589-3p;hsa-miR-590-3p | 15 | JDP2 | Sponge network | -0.358 | 0.17255 | -0.967 | 0 | 0.252 |
| 54023 | AC010226.4 |
hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19b-1-5p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 34 | DST | Sponge network | -0.811 | 2.0E-5 | -0.713 | 0.00096 | 0.252 |
| 54024 | FZD10-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-330-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-532-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-935 | 22 | LRRC55 | Sponge network | -0.727 | 0.04726 | -0.026 | 0.93163 | 0.252 |
| 54025 | MIR600HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-589-3p;hsa-miR-625-5p | 26 | TGFBR3 | Sponge network | 0.594 | 0.41522 | -1.173 | 1.0E-5 | 0.252 |
| 54026 | CTA-204B4.2 |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-590-3p | 24 | PPP1R9A | Sponge network | 0.765 | 7.0E-5 | -0.903 | 0.04254 | 0.252 |
| 54027 | AC074289.1 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-210-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-539-5p;hsa-miR-7-5p;hsa-miR-96-5p | 22 | CLASP2 | Sponge network | -0.326 | 0.14314 | -0.038 | 0.71 | 0.252 |
| 54028 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-23a-5p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-92a-3p | 27 | GPC6 | Sponge network | -2.46 | 0 | -0.054 | 0.81465 | 0.252 |
| 54029 | CTA-14H9.5 |
hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-219a-1-3p;hsa-miR-29a-5p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 13 | EPHA5 | Sponge network | 0.145 | 0.53343 | -0.957 | 0.01733 | 0.252 |
| 54030 | RP11-403I13.7 |
hsa-miR-130b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-7-1-3p | 10 | TPO | Sponge network | 0.395 | 0.15451 | -1.87 | 2.0E-5 | 0.252 |
| 54031 | GUSBP11 |
hsa-miR-10b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-654-3p | 10 | BAZ2B | Sponge network | 0.856 | 0 | -0.276 | 0.02833 | 0.252 |
| 54032 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 53 | KIF1B | Sponge network | -0.513 | 0.04703 | -0.118 | 0.32258 | 0.252 |
| 54033 | AC093627.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p | 10 | CBFA2T3 | Sponge network | -0.787 | 0.3928 | -1.221 | 1.0E-5 | 0.252 |
| 54034 | LINC00672 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-193a-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-539-5p;hsa-miR-590-3p | 12 | PHF3 | Sponge network | -0.227 | 0.31657 | 0.126 | 0.19652 | 0.252 |
| 54035 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-134-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-154-3p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-539-5p;hsa-miR-93-3p | 35 | SSBP2 | Sponge network | -0.602 | 0.00331 | -1.58 | 0 | 0.252 |
| 54036 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-93-3p;hsa-miR-96-5p | 26 | FOXO3 | Sponge network | 0.219 | 0.1516 | -0.04 | 0.72028 | 0.252 |
| 54037 | RP11-2B6.2 |
hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-345-5p;hsa-miR-378a-5p;hsa-miR-532-3p;hsa-miR-550a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-5p | 29 | DST | Sponge network | 0.539 | 0.03722 | -0.713 | 0.00096 | 0.252 |
| 54038 | H19 |
hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-576-5p | 15 | TMTC3 | Sponge network | 2.439 | 0 | 0.123 | 0.22189 | 0.252 |
| 54039 | LINC00672 |
hsa-let-7f-1-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-590-3p | 12 | PTCH1 | Sponge network | -0.227 | 0.31657 | -0.1 | 0.6179 | 0.252 |
| 54040 | RP11-326G21.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | TMEFF2 | Sponge network | -0.021 | 0.93598 | -2.426 | 0 | 0.252 |
| 54041 | RP11-403I13.7 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 31 | DIP2C | Sponge network | 0.395 | 0.15451 | -0.436 | 0.01713 | 0.252 |
| 54042 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-18a-3p;hsa-miR-193b-3p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-19a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p | 33 | ERBB4 | Sponge network | -0.322 | 0.25945 | -1.975 | 2.0E-5 | 0.252 |
| 54043 | RP11-1277A3.1 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-190a-5p;hsa-miR-193b-5p;hsa-miR-223-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p | 13 | DMD | Sponge network | 0.453 | 0.01839 | -1.506 | 0 | 0.252 |
| 54044 | CTC-490E21.10 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p | 17 | NTRK2 | Sponge network | 1.46 | 1.0E-5 | -0.736 | 0.06495 | 0.252 |
| 54045 | MRPL23-AS1 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-339-5p | 11 | TSC1 | Sponge network | -0.278 | 0.76559 | 0.103 | 0.26071 | 0.252 |
| 54046 | RP11-384O8.1 |
hsa-miR-107;hsa-miR-128-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-194-3p;hsa-miR-21-3p;hsa-miR-30b-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-429 | 12 | ELMO1 | Sponge network | -0.738 | 0.21233 | 0.04 | 0.83147 | 0.252 |
| 54047 | LINC01023 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-3p;hsa-miR-301a-3p;hsa-miR-424-5p | 21 | AR | Sponge network | -0.45 | 0.00486 | -1.554 | 0 | 0.252 |
| 54048 | MAGI2-AS3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-508-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-93-5p | 17 | AKAP13 | Sponge network | -0.129 | 0.57177 | 0.028 | 0.7729 | 0.252 |
| 54049 | AC108676.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-539-5p | 10 | ABI2 | Sponge network | 4.346 | 2.0E-5 | 0.175 | 0.14787 | 0.252 |
| 54050 | CTD-2002H8.2 |
hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | TCF12 | Sponge network | 0.931 | 1.0E-5 | 0.174 | 0.13271 | 0.252 |
| 54051 | TPTEP1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92b-3p | 24 | BCL11A | Sponge network | -1.216 | 0 | -0.932 | 0.0063 | 0.252 |
| 54052 | CTC-487M23.5 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-2110;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-450b-5p | 11 | ASTN1 | Sponge network | 0.912 | 0.00014 | -2.389 | 2.0E-5 | 0.252 |
| 54053 | RP11-458D21.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-502-5p;hsa-miR-532-3p;hsa-miR-576-5p | 14 | RIMBP2 | Sponge network | 0.663 | 0.00073 | -1.968 | 5.0E-5 | 0.252 |
| 54054 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-454-3p;hsa-miR-455-5p | 10 | THSD7A | Sponge network | 0.853 | 0.15352 | 0.242 | 0.25154 | 0.252 |
| 54055 | RP11-384O8.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-2110;hsa-miR-324-3p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-744-5p | 17 | USP12 | Sponge network | -0.738 | 0.21233 | -0.102 | 0.40768 | 0.252 |
| 54056 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1296-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-2355-5p;hsa-miR-27a-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-629-3p;hsa-miR-93-5p | 25 | ZBTB4 | Sponge network | -0.668 | 3.0E-5 | -0.739 | 0 | 0.252 |
| 54057 | DIO3OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-660-5p | 14 | EPAS1 | Sponge network | -0.773 | 0.04073 | -0.184 | 0.14418 | 0.252 |
| 54058 | LINC01018 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-425-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 21 | CSMD1 | Sponge network | -2.915 | 0.00015 | -0.63 | 0.18881 | 0.252 |
| 54059 | C20orf166-AS1 |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-484;hsa-miR-505-5p;hsa-miR-629-5p | 14 | WNK2 | Sponge network | -2.69 | 0.0001 | -0.352 | 0.31066 | 0.252 |
| 54060 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-150-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-378a-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 12 | CHD2 | Sponge network | 0.91 | 0 | 0.049 | 0.55371 | 0.252 |
| 54061 | RP11-403I13.8 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-222-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 14 | RECK | Sponge network | 0.619 | 0.00157 | -1.043 | 0 | 0.252 |
| 54062 | RP11-98I9.4 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-30a-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-590-3p | 14 | SOX11 | Sponge network | 0.67 | 0.00048 | 2.68 | 0 | 0.252 |
| 54063 | RP11-37B2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-582-5p | 14 | USP6 | Sponge network | 1.03 | 0 | 0.188 | 0.3871 | 0.252 |
| 54064 | RP11-815I9.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-590-5p | 11 | PIK3C2A | Sponge network | 0.537 | 0.0006 | 0.061 | 0.53776 | 0.252 |
| 54065 | CTD-2336O2.1 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 11 | NCAM2 | Sponge network | -0.141 | 0.26434 | -0.482 | 0.22565 | 0.252 |
| 54066 | RP4-605O3.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-1301-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-590-3p | 13 | ANK2 | Sponge network | 0.262 | 0.0475 | -1.603 | 1.0E-5 | 0.252 |
| 54067 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-93-5p | 12 | FBXO31 | Sponge network | 0.219 | 0.1516 | -0.085 | 0.42389 | 0.252 |
| 54068 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | TBX18 | Sponge network | 0.727 | 3.0E-5 | 0.843 | 0.02945 | 0.252 |
| 54069 | NDUFA6-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-664a-5p;hsa-miR-671-5p;hsa-miR-766-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 47 | PPARA | Sponge network | -0.355 | 0.0077 | -0.379 | 0.00548 | 0.252 |
| 54070 | RBPMS-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-34a-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-935 | 12 | RARB | Sponge network | 0.144 | 0.64989 | -0.825 | 2.0E-5 | 0.252 |
| 54071 | DNM3OS |
hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-24-2-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-664a-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 40 | PPM1L | Sponge network | 0.572 | 0.01722 | -0.537 | 0.00616 | 0.252 |
| 54072 | AC004893.11 |
hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 11 | DOCK4 | Sponge network | 1.317 | 0 | 0.502 | 0.00108 | 0.252 |
| 54073 | RAMP2-AS1 |
hsa-miR-107;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-942-5p | 30 | NOTCH2 | Sponge network | -0.513 | 0.04703 | 0.138 | 0.35163 | 0.252 |
| 54074 | AC004540.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-330-5p | 10 | LRIG1 | Sponge network | -2.549 | 0.00528 | -0.537 | 0.00588 | 0.252 |
| 54075 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 16 | EPAS1 | Sponge network | 0.219 | 0.1516 | -0.184 | 0.14418 | 0.252 |
| 54076 | LINC00472 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-2110;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-550a-5p | 14 | RIMBP2 | Sponge network | -0.842 | 0.01361 | -1.968 | 5.0E-5 | 0.252 |
| 54077 | RP11-166D19.1 |
hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-155-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-625-5p | 17 | TMTC2 | Sponge network | -0.576 | 0.10121 | -0.215 | 0.18248 | 0.252 |
| 54078 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-223-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-493-5p | 17 | ZFHX3 | Sponge network | 1.044 | 2.0E-5 | 0.389 | 0.01363 | 0.252 |
| 54079 | HOXB-AS3 |
hsa-miR-107;hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200a-3p;hsa-miR-26b-5p | 10 | PCDH9 | Sponge network | 0.343 | 0.28887 | -1.823 | 4.0E-5 | 0.252 |
| 54080 | RP11-5C23.1 |
hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-3p;hsa-miR-429;hsa-miR-590-3p | 11 | RECK | Sponge network | 0.274 | 0.07681 | -1.043 | 0 | 0.252 |
| 54081 | RP11-244H3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-362-5p;hsa-miR-455-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 18 | SETBP1 | Sponge network | 0.659 | 3.0E-5 | -1.302 | 1.0E-5 | 0.252 |
| 54082 | AP001347.6 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-577;hsa-miR-590-3p | 21 | ACSL6 | Sponge network | -1.161 | 0.00591 | -1.593 | 0 | 0.252 |
| 54083 | RP11-967K21.1 |
hsa-let-7b-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-421;hsa-miR-542-3p;hsa-miR-577;hsa-miR-589-3p | 16 | SYT4 | Sponge network | 0.056 | 0.81195 | -3.207 | 0 | 0.252 |
| 54084 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-223-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | FAM188A | Sponge network | 0.541 | 0.00125 | -0.44 | 0.00018 | 0.252 |
| 54085 | RP11-728F11.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-196b-5p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-331-3p | 13 | CBFA2T3 | Sponge network | 0.238 | 0.70544 | -1.221 | 1.0E-5 | 0.252 |
| 54086 | CTD-2666L21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-144-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26a-2-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-493-5p;hsa-miR-744-3p | 25 | FAT3 | Sponge network | 0.368 | 0.20093 | -1.601 | 0.00051 | 0.252 |
| 54087 | C20orf166-AS1 |
hsa-let-7a-5p;hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-128-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-361-3p | 13 | MNT | Sponge network | -2.69 | 0.0001 | -0.157 | 0.06598 | 0.252 |
| 54088 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-382-5p;hsa-miR-3913-5p;hsa-miR-576-5p;hsa-miR-590-5p | 17 | TRIP11 | Sponge network | -1.645 | 0 | -0.158 | 0.06384 | 0.252 |
| 54089 | CTD-2666L21.1 |
hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-144-5p;hsa-miR-155-5p;hsa-miR-192-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-493-5p | 14 | PGR | Sponge network | 0.368 | 0.20093 | -1.704 | 0 | 0.252 |
| 54090 | EMX2OS |
hsa-let-7d-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-3614-5p;hsa-miR-450b-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 41 | CREB3L2 | Sponge network | -0.777 | 0.1134 | -0.525 | 2.0E-5 | 0.252 |
| 54091 | RP1-122P22.2 |
hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p | 10 | HIP1 | Sponge network | 0.065 | 0.73468 | 0.774 | 0 | 0.252 |
| 54092 | RP11-693J15.4 |
hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-335-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 10 | LUZP2 | Sponge network | -0.72 | 0.24239 | -0.056 | 0.89416 | 0.252 |
| 54093 | AC002117.1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-935 | 16 | PTPRT | Sponge network | 0.476 | 0.19203 | -0.907 | 0.03727 | 0.252 |
| 54094 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | MTSS1 | Sponge network | 0.538 | 0.00076 | -0.81 | 0.00035 | 0.252 |
| 54095 | RP11-156P1.3 |
hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-330-3p;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-942-5p | 17 | PCDH17 | Sponge network | 0.214 | 0.18246 | 0.731 | 2.0E-5 | 0.252 |
| 54096 | RP5-1198O20.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 38 | ST6GALNAC3 | Sponge network | 0.997 | 0.00066 | -0.987 | 0 | 0.252 |
| 54097 | LINC00476 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-542-3p;hsa-miR-589-5p;hsa-miR-935 | 22 | RARB | Sponge network | -0.615 | 0.00015 | -0.825 | 2.0E-5 | 0.252 |
| 54098 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-577 | 16 | GREM1 | Sponge network | 0.177 | 0.16681 | 0.902 | 0.01691 | 0.252 |
| 54099 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-484;hsa-miR-501-5p | 22 | SOX5 | Sponge network | -0.602 | 0.00331 | -1.091 | 4.0E-5 | 0.252 |
| 54100 | RP11-597D13.9 |
hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-338-3p;hsa-miR-505-3p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-664a-3p;hsa-miR-874-3p;hsa-miR-877-5p;hsa-miR-96-5p | 24 | ASTN1 | Sponge network | 1.302 | 7.0E-5 | -2.389 | 2.0E-5 | 0.252 |
| 54101 | RP4-614O4.11 |
hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-424-5p;hsa-miR-625-5p | 12 | CBX5 | Sponge network | 1.042 | 0.00177 | 0.165 | 0.24446 | 0.252 |
| 54102 | RP5-998N21.10 |
hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 11 | RUNX1T1 | Sponge network | -0.098 | 0.76298 | -0.344 | 0.2374 | 0.252 |
| 54103 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-409-3p;hsa-miR-500b-5p;hsa-miR-590-5p | 23 | SCN9A | Sponge network | -1.047 | 0 | -0.525 | 0.10593 | 0.252 |
| 54104 | PSMG3-AS1 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-181a-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-425-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 29 | PTPRT | Sponge network | -0.125 | 0.49414 | -0.907 | 0.03727 | 0.252 |
| 54105 | LINC00672 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-361-3p;hsa-miR-590-3p;hsa-miR-93-3p | 22 | FOXO3 | Sponge network | -0.227 | 0.31657 | -0.04 | 0.72028 | 0.252 |
| 54106 | RP11-479G22.8 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | EGFR | Sponge network | 1.308 | 0 | -0.175 | 0.48451 | 0.252 |
| 54107 | AC009120.6 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-369-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 14 | CUL5 | Sponge network | 0.919 | 0 | 0.026 | 0.77301 | 0.252 |
| 54108 | C4B-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-96-5p | 17 | IL17RD | Sponge network | 2.306 | 9.0E-5 | -0.019 | 0.94873 | 0.252 |
| 54109 | LINC00472 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-421;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-7-1-3p;hsa-miR-877-5p | 26 | RAP1A | Sponge network | -0.842 | 0.01361 | -0.929 | 0 | 0.252 |
| 54110 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-942-5p | 12 | RB1CC1 | Sponge network | 0.41 | 0.02214 | 0.175 | 0.12559 | 0.252 |
| 54111 | LINC00265 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 11 | GPAM | Sponge network | 1.07 | 0 | 0.142 | 0.29732 | 0.252 |
| 54112 | ERVH48-1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-299-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-590-3p | 11 | NF1 | Sponge network | 1.04 | 0.00023 | 0.51 | 0 | 0.252 |
| 54113 | RP11-226L15.5 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-495-3p;hsa-miR-590-3p | 20 | ZFX | Sponge network | 0.8 | 0 | 0.09 | 0.42563 | 0.252 |
| 54114 | RP11-473I1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-655-3p | 12 | KANK1 | Sponge network | 0.542 | 0.00054 | -0.55 | 0.00134 | 0.252 |
| 54115 | RP11-597D13.9 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-96-5p | 17 | CXCL12 | Sponge network | 1.302 | 7.0E-5 | -1.421 | 0 | 0.252 |
| 54116 | RP11-701H24.3 |
hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-15a-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-2355-3p;hsa-miR-301a-3p;hsa-miR-590-3p | 12 | PAFAH1B1 | Sponge network | -1.425 | 0.04204 | -0.429 | 0 | 0.252 |
| 54117 | RP11-760H22.2 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-199b-5p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-369-3p | 11 | SIRT1 | Sponge network | 0.001 | 0.99753 | 0.109 | 0.2593 | 0.252 |
| 54118 | RP11-535M15.1 |
hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-589-5p | 17 | ADARB1 | Sponge network | -1.14 | 0.03513 | -0.335 | 0.06631 | 0.252 |
| 54119 | CTB-113P19.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-338-5p;hsa-miR-369-3p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 21 | QKI | Sponge network | 0.199 | 0.52763 | -0.333 | 0.04111 | 0.252 |
| 54120 | RP11-411K7.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-378c;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 34 | TACC1 | Sponge network | 0.155 | 0.81273 | -0.362 | 0.05406 | 0.252 |
| 54121 | RP11-130L8.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-3607-3p;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-93-5p | 16 | LRRC55 | Sponge network | -0.663 | 0.10887 | -0.026 | 0.93163 | 0.252 |
| 54122 | AC007566.10 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p | 10 | PRKAA1 | Sponge network | 2.368 | 2.0E-5 | 0.317 | 0.0031 | 0.252 |
| 54123 | RP11-38L15.3 |
hsa-miR-103a-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-590-3p | 11 | BCL2 | Sponge network | 0.344 | 0.3127 | -0.899 | 0 | 0.252 |
| 54124 | AC016747.3 |
hsa-miR-107;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-877-5p | 11 | INPP4A | Sponge network | 0.199 | 0.38015 | 0.07 | 0.53563 | 0.252 |
| 54125 | RP11-360F5.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-376c-3p;hsa-miR-429 | 11 | WDFY3 | Sponge network | 2.364 | 0.00134 | -0.201 | 0.0967 | 0.252 |
| 54126 | CTD-2184D3.6 |
hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | BCL2 | Sponge network | 0.272 | 0.44692 | -0.899 | 0 | 0.252 |
| 54127 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 11 | PDGFC | Sponge network | 1.302 | 7.0E-5 | -0.33 | 0.06483 | 0.252 |
| 54128 | RP11-244O19.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 31 | MAF | Sponge network | -0.096 | 0.67958 | -1.257 | 0 | 0.252 |
| 54129 | BDNF-AS |
hsa-let-7g-3p;hsa-miR-132-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-3p;hsa-miR-96-5p | 21 | PCDH10 | Sponge network | -0.668 | 3.0E-5 | -1.644 | 0.0078 | 0.252 |
| 54130 | RP11-53O19.3 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-33a-3p;hsa-miR-374b-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-9-5p | 25 | CCDC6 | Sponge network | 1.205 | 0 | 0.27 | 0.02555 | 0.252 |
| 54131 | RP11-296I10.3 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-5p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-374b-3p | 16 | UBR3 | Sponge network | 0.592 | 0.00014 | -0.021 | 0.82774 | 0.252 |
| 54132 | PSMD5-AS1 |
hsa-miR-148a-3p;hsa-miR-16-5p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-370-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-493-3p;hsa-miR-625-5p;hsa-miR-766-3p | 21 | CHD5 | Sponge network | 0.569 | 0.00067 | -0.922 | 0.01521 | 0.252 |
| 54133 | RP5-1085F17.3 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-190a-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-940 | 10 | CELF4 | Sponge network | 0.541 | 0.00125 | -1.135 | 0.00344 | 0.252 |
| 54134 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-3607-3p;hsa-miR-455-5p;hsa-miR-500a-5p;hsa-miR-584-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-93-3p | 25 | ST6GAL2 | Sponge network | -0.513 | 0.04703 | -1.201 | 0.00597 | 0.252 |
| 54135 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-27a-3p;hsa-miR-29b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | FYN | Sponge network | -0.899 | 6.0E-5 | -0.131 | 0.37051 | 0.252 |
| 54136 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-96-5p | 15 | ARHGAP29 | Sponge network | 0.41 | 0.02214 | 0.131 | 0.42953 | 0.252 |
| 54137 | RP11-53O19.1 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-369-3p;hsa-miR-450b-5p | 13 | TLE4 | Sponge network | -0.405 | 0.22013 | -1.157 | 0 | 0.252 |
| 54138 | TRIM52-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-7-1-3p | 13 | DIP2C | Sponge network | -0.418 | 0.00068 | -0.436 | 0.01713 | 0.252 |
| 54139 | PCA3 |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-382-5p;hsa-miR-409-3p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-96-5p | 15 | UNC5D | Sponge network | -0.709 | 0.18168 | -3.646 | 0 | 0.252 |
| 54140 | RP11-327P2.5 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-23a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-93-3p | 25 | AKAP11 | Sponge network | -0.147 | 0.40452 | 0.196 | 0.09825 | 0.252 |
| 54141 | RP11-701H24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-222-5p;hsa-miR-576-5p | 14 | CBL | Sponge network | -2.039 | 0.03447 | 0.291 | 0.01267 | 0.252 |
| 54142 | PAXIP1-AS1 |
hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-758-3p | 13 | CDC73 | Sponge network | 0.66 | 1.0E-5 | 0.094 | 0.20463 | 0.252 |
| 54143 | RP11-927P21.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-484;hsa-miR-577;hsa-miR-582-5p;hsa-miR-589-3p;hsa-miR-590-5p | 21 | TGFBR3 | Sponge network | 0.921 | 0.00118 | -1.173 | 1.0E-5 | 0.252 |
| 54144 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | ARNT2 | Sponge network | 0.131 | 0.8436 | -0.332 | 0.25851 | 0.252 |
| 54145 | RP11-13K12.1 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-181b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 10 | PPP2R2C | Sponge network | -0.154 | 0.78939 | -0.959 | 0.07428 | 0.252 |
| 54146 | DNM1P35 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-5p;hsa-miR-93-5p | 42 | TCF4 | Sponge network | 0.051 | 0.83388 | 0.016 | 0.92271 | 0.252 |
| 54147 | AC141928.1 |
hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-29b-1-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-577;hsa-miR-7-1-3p;hsa-miR-96-5p | 16 | PIK3CA | Sponge network | 0.853 | 0.15352 | 0.195 | 0.06908 | 0.252 |
| 54148 | A2M-AS1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-590-3p | 10 | ABCA10 | Sponge network | -0.08 | 0.90092 | -0.571 | 0.03674 | 0.252 |
| 54149 | ARHGAP5-AS1 |
hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-93-3p | 29 | SSBP2 | Sponge network | -0.644 | 0.00021 | -1.58 | 0 | 0.252 |
| 54150 | FGD5-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-590-3p | 14 | GNA13 | Sponge network | 0.401 | 0.00066 | 0.007 | 0.93884 | 0.252 |
| 54151 | RP11-458D21.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-495-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 29 | ZFX | Sponge network | 0.663 | 0.00073 | 0.09 | 0.42563 | 0.252 |
| 54152 | RP11-359B12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 20 | SON | Sponge network | 0.064 | 0.72934 | 0.168 | 0.04406 | 0.252 |
| 54153 | HOTAIRM1 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-590-3p | 11 | ABCB5 | Sponge network | -0.053 | 0.80685 | -0.956 | 0.04077 | 0.252 |
| 54154 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-532-5p | 10 | LRRC4 | Sponge network | 0.082 | 0.55397 | -1.774 | 0 | 0.252 |
| 54155 | NDUFA6-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 30 | ST6GAL2 | Sponge network | -0.355 | 0.0077 | -1.201 | 0.00597 | 0.252 |
| 54156 | LINC00621 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-323a-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | RICTOR | Sponge network | 0.131 | 0.8436 | 0.388 | 0.00188 | 0.252 |
| 54157 | LINC00173 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-454-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | EPS15 | Sponge network | 0.056 | 0.8494 | -0.052 | 0.53998 | 0.252 |
| 54158 | RP11-575A19.2 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-301a-3p;hsa-miR-629-3p | 11 | RASSF2 | Sponge network | 1.969 | 0.00316 | -0.273 | 0.21961 | 0.252 |
| 54159 | AC004540.4 |
hsa-miR-1301-3p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-629-3p | 24 | IL6ST | Sponge network | -2.549 | 0.00528 | -0.402 | 0.00676 | 0.252 |
| 54160 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-132-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-744-3p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-5p | 53 | FAT3 | Sponge network | -1.047 | 0 | -1.601 | 0.00051 | 0.252 |
| 54161 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-3p | 13 | AKAP11 | Sponge network | 3.21 | 0 | 0.196 | 0.09825 | 0.252 |
| 54162 | AC141928.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | SUFU | Sponge network | 0.853 | 0.15352 | -0.04 | 0.65489 | 0.252 |
| 54163 | RAB30-AS1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-495-3p | 18 | ZFX | Sponge network | 0.752 | 0 | 0.09 | 0.42563 | 0.252 |
| 54164 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-150-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-330-3p;hsa-miR-335-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-651-5p;hsa-miR-766-3p;hsa-miR-93-5p | 54 | AFF4 | Sponge network | -0.154 | 0.78939 | -0.266 | 0.01565 | 0.252 |
| 54165 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-5p | 13 | BRD3 | Sponge network | 0.453 | 0.01839 | 0.37 | 0.00013 | 0.252 |
| 54166 | NR2F1-AS1 |
hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-188-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-484;hsa-miR-532-3p;hsa-miR-940 | 12 | FOXO4 | Sponge network | -0.49 | 0.04946 | -0.516 | 2.0E-5 | 0.252 |
| 54167 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-374b-5p;hsa-miR-628-5p | 13 | MIER3 | Sponge network | 0.537 | 0.00139 | -0.212 | 0.04957 | 0.252 |
| 54168 | SH3RF3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-1266-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-486-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-5p;hsa-miR-7-1-3p | 32 | THRB | Sponge network | 0.889 | 9.0E-5 | -1.117 | 0.0004 | 0.252 |
| 54169 | AF131215.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-590-3p | 18 | CACNA2D1 | Sponge network | -0.322 | 0.25945 | -0.846 | 0.00675 | 0.252 |
| 54170 | LINC00476 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p | 27 | CADM1 | Sponge network | -0.615 | 0.00015 | -0.9 | 0.00084 | 0.252 |
| 54171 | WDFY3-AS2 |
hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-590-3p | 10 | GLIPR2 | Sponge network | -0.942 | 0 | -0.996 | 0 | 0.252 |
| 54172 | LINC00621 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-3p;hsa-miR-299-5p;hsa-miR-29b-1-5p;hsa-miR-30e-3p;hsa-miR-486-5p;hsa-miR-590-3p | 11 | NR2C2 | Sponge network | 0.131 | 0.8436 | 0.21 | 0.03224 | 0.252 |
| 54173 | RP1-39G22.7 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-3614-5p;hsa-miR-374b-5p;hsa-miR-590-3p | 20 | ABL2 | Sponge network | 0.439 | 0.00313 | 0.82 | 0 | 0.252 |
| 54174 | AC007566.10 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-200c-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-3p;hsa-miR-582-5p | 13 | BRD3 | Sponge network | 2.368 | 2.0E-5 | 0.37 | 0.00013 | 0.252 |
| 54175 | RP11-373D23.3 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-7-1-3p | 16 | PCDH9 | Sponge network | -0.285 | 0.2172 | -1.823 | 4.0E-5 | 0.252 |
| 54176 | A2M-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-940 | 25 | BHLHE41 | Sponge network | -0.08 | 0.90092 | 0.353 | 0.12753 | 0.252 |
| 54177 | AC009120.5 |
hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-5p;hsa-miR-629-3p | 11 | DST | Sponge network | 1.383 | 0 | -0.713 | 0.00096 | 0.252 |
| 54178 | PSMG3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 24 | NRXN1 | Sponge network | -0.125 | 0.49414 | -3.778 | 0 | 0.252 |
| 54179 | CTBP1-AS2 |
hsa-miR-101-3p;hsa-miR-129-5p;hsa-miR-139-5p;hsa-miR-140-5p;hsa-miR-195-3p;hsa-miR-199b-5p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p | 14 | PHF6 | Sponge network | 0.623 | 0 | 0.785 | 0 | 0.252 |
| 54180 | FGD5-AS1 |
hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | KDSR | Sponge network | 0.401 | 0.00066 | 0.239 | 0.02216 | 0.252 |
| 54181 | WDFY3-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-2355-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 49 | PAFAH1B1 | Sponge network | -0.942 | 0 | -0.429 | 0 | 0.252 |
| 54182 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-190a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 41 | WDFY3 | Sponge network | 0.392 | 0.0278 | -0.201 | 0.0967 | 0.252 |
| 54183 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-654-3p | 21 | AKAP11 | Sponge network | 1.073 | 0.00216 | 0.196 | 0.09825 | 0.252 |
| 54184 | RP11-819C21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-940 | 23 | IGF1 | Sponge network | 0.537 | 0.00139 | -0.204 | 0.5898 | 0.252 |
| 54185 | RP11-819C21.1 |
hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-940 | 16 | TAL1 | Sponge network | 0.537 | 0.00139 | -0.462 | 0.00574 | 0.252 |
| 54186 | NEAT1 |
hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | ITGB1 | Sponge network | 0.705 | 0.00014 | 0.524 | 0.00017 | 0.252 |
| 54187 | MEG3 |
hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-361-3p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-532-5p;hsa-miR-589-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 25 | JDP2 | Sponge network | 0.249 | 0.35902 | -0.967 | 0 | 0.252 |
| 54188 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-335-5p;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-590-5p;hsa-miR-653-5p | 23 | EGLN1 | Sponge network | 0.331 | 0.57247 | -0.127 | 0.1536 | 0.252 |
| 54189 | IQCH-AS1 |
hsa-miR-100-5p;hsa-miR-130a-3p;hsa-miR-199b-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-374b-5p | 16 | CDC73 | Sponge network | 0.929 | 0 | 0.094 | 0.20463 | 0.252 |
| 54190 | RP13-225O21.2 |
hsa-miR-127-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30e-3p | 10 | KIF1B | Sponge network | 1.279 | 8.0E-5 | -0.118 | 0.32258 | 0.252 |
| 54191 | RP11-927P21.1 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-5p | 18 | NIN | Sponge network | 0.921 | 0.00118 | -0.253 | 0.13794 | 0.252 |
| 54192 | AGAP11 |
hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-423-5p;hsa-miR-455-3p | 20 | ABL1 | Sponge network | -1.645 | 0 | -0.215 | 0.07804 | 0.252 |
| 54193 | CTB-113P19.4 |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-450b-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-96-5p | 27 | BNC2 | Sponge network | 0.906 | 0.31715 | -0.491 | 0.09988 | 0.252 |
| 54194 | EMX2OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | FOXO1 | Sponge network | -0.777 | 0.1134 | -0.141 | 0.33874 | 0.252 |
| 54195 | LINC00667 |
hsa-miR-135b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-3127-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 11 | CNTNAP1 | Sponge network | -0.235 | 0.15142 | 0.358 | 0.13727 | 0.252 |
| 54196 | ARHGAP5-AS1 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-7-1-3p | 11 | BCL11A | Sponge network | -0.644 | 0.00021 | -0.932 | 0.0063 | 0.252 |
| 54197 | RP11-46C24.7 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | SEMA3E | Sponge network | 0.392 | 0.0278 | -1.717 | 0.00137 | 0.252 |
| 54198 | GAS6-AS2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-146b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-191-5p;hsa-miR-194-3p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-222-5p;hsa-miR-26a-2-3p;hsa-miR-29b-1-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-508-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 30 | CUX1 | Sponge network | 0.16 | 0.50785 | -0.039 | 0.6705 | 0.252 |
| 54199 | RP11-983P16.4 |
hsa-miR-146a-5p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 12 | ADCY1 | Sponge network | 0.41 | 0.02214 | 0.362 | 0.16982 | 0.252 |
| 54200 | HOXA-AS2 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-769-5p;hsa-miR-877-5p | 33 | EGFR | Sponge network | 0.61 | 0.01593 | -0.175 | 0.48451 | 0.252 |
| 54201 | BDNF-AS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p | 23 | SYNE1 | Sponge network | -0.668 | 3.0E-5 | -0.738 | 0.00316 | 0.252 |
| 54202 | RP11-102N12.3 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-421;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 21 | TACC1 | Sponge network | -0.351 | 0.11358 | -0.362 | 0.05406 | 0.252 |
| 54203 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-940 | 28 | IGF1 | Sponge network | 0.222 | 0.29133 | -0.204 | 0.5898 | 0.252 |
| 54204 | NUTM2A-AS1 |
hsa-let-7a-3p;hsa-miR-130a-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-24-1-5p;hsa-miR-24-2-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 16 | CDC73 | Sponge network | 0.643 | 0 | 0.094 | 0.20463 | 0.252 |
| 54205 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-324-5p;hsa-miR-424-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-3p | 23 | BHLHE41 | Sponge network | -0.739 | 0.00044 | 0.353 | 0.12753 | 0.252 |
| 54206 | RP4-791M13.3 |
hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p | 14 | ABL1 | Sponge network | 0.419 | 0.0319 | -0.215 | 0.07804 | 0.252 |
| 54207 | RP11-195F19.9 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-93-5p | 20 | PIK3R1 | Sponge network | -0.726 | 0.00132 | -0.133 | 0.36854 | 0.252 |
| 54208 | OSER1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-7-1-3p | 26 | ZFHX4 | Sponge network | -0.13 | 0.48734 | 0.018 | 0.95574 | 0.252 |
| 54209 | NNT-AS1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-200b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-374b-3p;hsa-miR-576-5p;hsa-miR-590-3p | 17 | UBR3 | Sponge network | 0.799 | 1.0E-5 | -0.021 | 0.82774 | 0.252 |
| 54210 | RP11-116O18.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 43 | TCF4 | Sponge network | 0.05 | 0.95483 | 0.016 | 0.92271 | 0.252 |
| 54211 | LINC00473 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-223-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-362-3p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-935 | 46 | NFIB | Sponge network | -1.203 | 0.03881 | 0.023 | 0.90795 | 0.252 |
| 54212 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 18 | CXXC4 | Sponge network | 0.541 | 0.00125 | -0.433 | 0.21055 | 0.252 |
| 54213 | ZSCAN16-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-590-3p | 15 | EIF4A2 | Sponge network | -0.342 | 0.02288 | -0.383 | 0.00046 | 0.252 |
| 54214 | EMX2OS |
hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-335-3p;hsa-miR-342-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-502-5p;hsa-miR-542-3p | 11 | CADM4 | Sponge network | -0.777 | 0.1134 | -0.635 | 0.00305 | 0.252 |
| 54215 | RP11-725P16.2 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | HECA | Sponge network | 0.422 | 0.27021 | -0.217 | 0.03463 | 0.252 |
| 54216 | HOXB-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-589-3p | 15 | HECA | Sponge network | -0.154 | 0.53954 | -0.217 | 0.03463 | 0.252 |
| 54217 | RP11-473I1.10 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-24-3p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-664a-5p | 12 | PPARA | Sponge network | 0.673 | 0 | -0.379 | 0.00548 | 0.252 |
| 54218 | RP11-384O8.1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p | 26 | MED12L | Sponge network | -0.738 | 0.21233 | -0.194 | 0.55299 | 0.252 |
| 54219 | EIF3J-AS1 |
hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-299-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p;hsa-miR-590-3p | 13 | CUL5 | Sponge network | 0.331 | 0.00509 | 0.026 | 0.77301 | 0.252 |
| 54220 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-484;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-9-5p | 15 | YAP1 | Sponge network | 1.254 | 0 | 0.22 | 0.11836 | 0.252 |
| 54221 | JAZF1-AS1 |
hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-197-3p;hsa-miR-212-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-423-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 19 | CSDE1 | Sponge network | -0.769 | 0.00035 | -0.493 | 0 | 0.252 |
| 54222 | AC010226.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 17 | NCOA1 | Sponge network | -0.811 | 2.0E-5 | -0.432 | 0 | 0.252 |
| 54223 | RP11-102N12.3 |
hsa-let-7d-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-7-1-3p | 12 | AKAP6 | Sponge network | -0.351 | 0.11358 | -1.586 | 3.0E-5 | 0.252 |
| 54224 | MIR497HG |
hsa-let-7b-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-590-5p;hsa-miR-625-5p | 14 | TES | Sponge network | -1.439 | 4.0E-5 | -0.348 | 0.00639 | 0.252 |
| 54225 | FAM215B |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-24-2-5p;hsa-miR-29b-3p;hsa-miR-582-5p;hsa-miR-7-1-3p | 16 | BCL2 | Sponge network | 0.817 | 3.0E-5 | -0.899 | 0 | 0.252 |
| 54226 | RP11-539I5.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-590-3p | 16 | ARNT2 | Sponge network | -0.468 | 0.32587 | -0.332 | 0.25851 | 0.252 |
| 54227 | FAM215B |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-193a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-7-1-3p | 11 | CLIP1 | Sponge network | 0.817 | 3.0E-5 | -0.215 | 0.08684 | 0.252 |
| 54228 | OIP5-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-550a-5p | 11 | FAT2 | Sponge network | 0.421 | 0.00084 | -0.278 | 0.44877 | 0.252 |
| 54229 | LINC01001 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 12 | GPAM | Sponge network | 1.055 | 0 | 0.142 | 0.29732 | 0.252 |
| 54230 | SH3BP5-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-671-5p | 31 | ADARB1 | Sponge network | 0.267 | 0.2937 | -0.335 | 0.06631 | 0.252 |
| 54231 | HOXA-AS3 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-340-5p;hsa-miR-374a-5p;hsa-miR-590-3p | 11 | DNAJB4 | Sponge network | -0.555 | 0.06156 | -0.7 | 1.0E-5 | 0.252 |
| 54232 | RP11-158K1.3 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-374b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZNF844 | Sponge network | 0.349 | 0.01695 | -0.966 | 0.00035 | 0.252 |
| 54233 | AC074289.1 |
hsa-let-7a-3p;hsa-miR-1266-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-3p;hsa-miR-194-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-96-5p | 14 | HIP1 | Sponge network | -0.326 | 0.14314 | 0.774 | 0 | 0.252 |
| 54234 | AF131215.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-374b-3p;hsa-miR-454-3p;hsa-miR-576-5p | 23 | IGF1 | Sponge network | -0.176 | 0.59692 | -0.204 | 0.5898 | 0.252 |
| 54235 | SNX29P2 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-455-3p;hsa-miR-500b-5p;hsa-miR-93-5p | 19 | ACSL6 | Sponge network | 0.4 | 0.14611 | -1.593 | 0 | 0.252 |
| 54236 | HOXB-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-338-3p;hsa-miR-429 | 17 | LIMCH1 | Sponge network | -0.154 | 0.53954 | -0.915 | 1.0E-5 | 0.252 |
| 54237 | AC034220.3 |
hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-374b-5p;hsa-miR-628-5p;hsa-miR-766-3p | 10 | KIAA1024 | Sponge network | 0.309 | 0.07304 | 1.083 | 0 | 0.252 |
| 54238 | U91328.19 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-624-5p;hsa-miR-7-1-3p | 34 | FOXP1 | Sponge network | -0.303 | 0.21002 | 0.042 | 0.74368 | 0.252 |
| 54239 | LINC00957 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-339-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-766-3p | 12 | ATM | Sponge network | -0.421 | 0.0114 | 0.178 | 0.21568 | 0.252 |
| 54240 | ADAMTS9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p | 24 | ERCC4 | Sponge network | -2.452 | 0 | 0.126 | 0.15266 | 0.252 |
| 54241 | SERHL |
hsa-let-7a-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-219a-1-3p;hsa-miR-320b | 12 | SLITRK3 | Sponge network | -0.602 | 0.00331 | -3.328 | 0 | 0.252 |
| 54242 | RP11-400K9.4 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-34a-5p;hsa-miR-93-5p | 20 | RNASEL | Sponge network | -0.611 | 0.09607 | -0.272 | 0.01796 | 0.252 |
| 54243 | RP4-758J18.10 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-5p;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p | 10 | FREM2 | Sponge network | -0.091 | 0.71468 | -0.437 | 0.46353 | 0.252 |
| 54244 | KB-1460A1.5 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-195-5p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-362-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 36 | ARHGEF12 | Sponge network | 0.603 | 0.00127 | 0.09 | 0.35693 | 0.252 |
| 54245 | RP11-67L2.2 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 21 | GJA1 | Sponge network | 0.72 | 0.0001 | -0.485 | 0.02179 | 0.252 |
| 54246 | LINC00641 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-370-3p;hsa-miR-421;hsa-miR-425-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-485-3p;hsa-miR-590-3p;hsa-miR-708-3p | 19 | ANK3 | Sponge network | -0.222 | 0.32445 | -0.55 | 0.00086 | 0.252 |
| 54247 | DOCK9-AS2 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29b-1-5p;hsa-miR-532-3p;hsa-miR-940 | 21 | WDFY3 | Sponge network | -0.231 | 0.28214 | -0.201 | 0.0967 | 0.252 |
| 54248 | RP1-151F17.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-935;hsa-miR-96-5p | 23 | HBP1 | Sponge network | 0.222 | 0.29133 | -0.454 | 0 | 0.252 |
| 54249 | LINC01006 |
hsa-let-7a-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-429;hsa-miR-671-5p | 13 | THRA | Sponge network | -0.702 | 0.04814 | -0.307 | 0.05099 | 0.252 |
| 54250 | ZRANB2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p | 16 | ATRX | Sponge network | -0.376 | 0.00337 | 0.261 | 0.04408 | 0.252 |
| 54251 | RP11-35G9.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-194-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-455-5p;hsa-miR-589-3p;hsa-miR-942-5p | 16 | UNC80 | Sponge network | 0.622 | 0.00015 | -1.934 | 0 | 0.252 |
| 54252 | SNX29P2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-455-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 14 | EPAS1 | Sponge network | 0.4 | 0.14611 | -0.184 | 0.14418 | 0.252 |
| 54253 | RP11-706C16.7 |
hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b | 19 | KIF1B | Sponge network | 0.357 | 0.72407 | -0.118 | 0.32258 | 0.251 |
| 54254 | RP11-59H7.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-502-3p;hsa-miR-576-5p | 12 | RNF144A | Sponge network | 2.163 | 0 | 0.594 | 0.0009 | 0.251 |
| 54255 | DOCK9-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-194-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-629-3p;hsa-miR-940 | 15 | SNX29 | Sponge network | -0.231 | 0.28214 | -0.34 | 0.01371 | 0.251 |
| 54256 | CTD-2184D3.6 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | NCOA1 | Sponge network | 0.272 | 0.44692 | -0.432 | 0 | 0.251 |
| 54257 | CTC-457L16.2 |
hsa-let-7a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-144-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-3607-3p;hsa-miR-589-3p;hsa-miR-7-1-3p | 11 | TIMP3 | Sponge network | 1.153 | 0.00114 | -0.546 | 0.02307 | 0.251 |
| 54258 | RP11-536O18.1 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-330-3p;hsa-miR-362-5p;hsa-miR-502-5p | 17 | DST | Sponge network | -0.074 | 0.90758 | -0.713 | 0.00096 | 0.251 |
| 54259 | KB-1460A1.5 |
hsa-miR-103a-2-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-450b-5p;hsa-miR-500a-5p;hsa-miR-539-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | RYR2 | Sponge network | 0.603 | 0.00127 | -1.466 | 0.00031 | 0.251 |
| 54260 | RP11-999E24.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-374a-5p | 17 | LRP1B | Sponge network | -0.693 | 0.05629 | -2.163 | 0 | 0.251 |
| 54261 | RP11-531A24.5 |
hsa-miR-130b-5p;hsa-miR-19b-3p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-7-1-3p | 10 | TPO | Sponge network | 0.75 | 0.00054 | -1.87 | 2.0E-5 | 0.251 |
| 54262 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-429;hsa-miR-497-5p;hsa-miR-590-3p | 16 | MYBL1 | Sponge network | 1.308 | 0 | 0.494 | 0.02152 | 0.251 |
| 54263 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | DACT1 | Sponge network | 0.349 | 0.01695 | 0.783 | 0.00072 | 0.251 |
| 54264 | RP11-283I3.6 |
hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-151a-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-96-5p | 23 | GAS7 | Sponge network | 0.614 | 1.0E-5 | -0.555 | 0.01602 | 0.251 |
| 54265 | DLG5-AS1 |
hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-222-5p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p | 15 | DTNA | Sponge network | 0.556 | 0.07377 | -1.648 | 1.0E-5 | 0.251 |
| 54266 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-215-5p;hsa-miR-2355-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-340-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-495-3p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-651-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p;hsa-miR-96-5p | 68 | BMPR2 | Sponge network | -2.46 | 0 | 0.206 | 0.0635 | 0.251 |
| 54267 | MRPL23-AS1 |
hsa-let-7g-3p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-215-5p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p | 10 | ZMYND11 | Sponge network | -0.278 | 0.76559 | -0.2 | 0.05449 | 0.251 |
| 54268 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-29a-5p;hsa-miR-502-5p;hsa-miR-590-5p | 10 | PTCH1 | Sponge network | -0.72 | 0.24239 | -0.1 | 0.6179 | 0.251 |
| 54269 | LINC00886 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-128-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-20a-3p;hsa-miR-21-5p;hsa-miR-2355-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-940 | 30 | MYO9A | Sponge network | -0.371 | 0.24604 | -0.147 | 0.32373 | 0.251 |
| 54270 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-3p;hsa-miR-375 | 21 | MYBL1 | Sponge network | -0.162 | 0.47687 | 0.494 | 0.02152 | 0.251 |
| 54271 | RP11-195F19.9 |
hsa-miR-128-3p;hsa-miR-132-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-26b-3p;hsa-miR-27a-3p | 11 | PAIP2 | Sponge network | -0.726 | 0.00132 | -0.515 | 0 | 0.251 |
| 54272 | RP11-158K1.3 |
hsa-miR-125a-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-590-3p | 23 | SYNPO2 | Sponge network | 0.349 | 0.01695 | -2.342 | 0 | 0.251 |
| 54273 | AC034220.3 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-26b-3p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-940 | 13 | DLG1 | Sponge network | 0.309 | 0.07304 | -0.014 | 0.8926 | 0.251 |
| 54274 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-155-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-429 | 10 | DMD | Sponge network | 1.316 | 0.01106 | -1.506 | 0 | 0.251 |
| 54275 | RP11-672A2.4 |
hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-191-5p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26a-2-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-424-5p | 16 | CCND2 | Sponge network | 1.344 | 0 | -0.267 | 0.3112 | 0.251 |
| 54276 | LINC00987 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-32-5p;hsa-miR-33a-5p;hsa-miR-664a-3p;hsa-miR-664a-5p | 12 | SOX11 | Sponge network | 0.857 | 0 | 2.68 | 0 | 0.251 |
| 54277 | RP11-686D22.8 |
hsa-miR-101-5p;hsa-miR-141-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-30d-3p;hsa-miR-335-5p;hsa-miR-590-3p | 13 | RICTOR | Sponge network | 0.898 | 1.0E-5 | 0.388 | 0.00188 | 0.251 |
| 54278 | CTD-2314G24.2 |
hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p | 14 | ATM | Sponge network | 0.246 | 0.59912 | 0.178 | 0.21568 | 0.251 |
| 54279 | RP11-596D21.1 |
hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-152-3p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-374b-5p | 12 | MDM4 | Sponge network | 3.076 | 3.0E-5 | 0.345 | 0.00762 | 0.251 |
| 54280 | RP11-760H22.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-374b-5p | 10 | PTPN11 | Sponge network | 0.001 | 0.99753 | 0.431 | 2.0E-5 | 0.251 |
| 54281 | RP11-66B24.2 |
hsa-let-7g-3p;hsa-miR-10a-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-502-5p | 11 | SETBP1 | Sponge network | 0.57 | 0.1866 | -1.302 | 1.0E-5 | 0.251 |
| 54282 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-30b-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-542-3p;hsa-miR-590-5p;hsa-miR-671-5p;hsa-miR-93-3p;hsa-miR-93-5p | 40 | BNC2 | Sponge network | -1.047 | 0 | -0.491 | 0.09988 | 0.251 |
| 54283 | RP11-620J15.3 |
hsa-let-7g-3p;hsa-miR-103a-2-5p;hsa-miR-1301-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-532-5p;hsa-miR-590-3p | 16 | ZMYND11 | Sponge network | -0.739 | 0.00044 | -0.2 | 0.05449 | 0.251 |
| 54284 | RP11-418J17.1 |
hsa-miR-144-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-3p;hsa-miR-29c-3p;hsa-miR-30a-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-497-5p | 13 | SLC5A3 | Sponge network | 0.998 | 0 | 0.493 | 5.0E-5 | 0.251 |
| 54285 | TUG1 |
hsa-miR-125a-5p;hsa-miR-125b-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-199b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-628-5p | 11 | MAT2A | Sponge network | 0.972 | 0 | -0.073 | 0.36195 | 0.251 |
| 54286 | RP11-144G6.12 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-582-5p;hsa-miR-590-3p | 12 | BRD3 | Sponge network | 0.826 | 1.0E-5 | 0.37 | 0.00013 | 0.251 |
| 54287 | RP11-428J1.5 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-342-3p;hsa-miR-342-5p;hsa-miR-539-5p;hsa-miR-590-3p | 15 | SYNPO2 | Sponge network | 0.538 | 0.00076 | -2.342 | 0 | 0.251 |
| 54288 | PART1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-27b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-454-3p;hsa-miR-500a-5p;hsa-miR-550a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 31 | CRTC1 | Sponge network | -2.925 | 0 | -0.116 | 0.28412 | 0.251 |
| 54289 | RP11-693J15.4 |
hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-224-5p;hsa-miR-29a-5p;hsa-miR-362-5p;hsa-miR-500b-5p;hsa-miR-590-5p | 14 | SCN9A | Sponge network | -0.72 | 0.24239 | -0.525 | 0.10593 | 0.251 |
| 54290 | RP5-1021I20.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-539-5p | 19 | NRXN1 | Sponge network | -0.785 | 0.00035 | -3.778 | 0 | 0.251 |
| 54291 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-3p | 15 | DOCK4 | Sponge network | -0.227 | 0.31657 | 0.502 | 0.00108 | 0.251 |
| 54292 | RP11-46C24.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-550a-3p;hsa-miR-590-3p | 11 | PCDH8 | Sponge network | 0.392 | 0.0278 | -0.997 | 0.05366 | 0.251 |
| 54293 | RASSF8-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | AKAP11 | Sponge network | 0.335 | 0.20596 | 0.196 | 0.09825 | 0.251 |
| 54294 | RASSF8-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-671-5p | 26 | CYLD | Sponge network | 0.335 | 0.20596 | -0.367 | 0.00171 | 0.251 |
| 54295 | AC005562.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-664a-5p | 31 | MAPK1 | Sponge network | 0.488 | 0.00084 | -0.017 | 0.81465 | 0.251 |
| 54296 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-96-5p | 13 | TACC1 | Sponge network | 3.21 | 0 | -0.362 | 0.05406 | 0.251 |
| 54297 | AC003090.1 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 13 | ITGB1 | Sponge network | -2.117 | 0.00161 | 0.524 | 0.00017 | 0.251 |
| 54298 | AC016747.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-324-3p;hsa-miR-429;hsa-miR-577;hsa-miR-590-3p;hsa-miR-93-5p | 17 | MAP3K12 | Sponge network | 0.199 | 0.38015 | -0.057 | 0.59369 | 0.251 |
| 54299 | HHIP-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | ETV1 | Sponge network | -0.737 | 0.10822 | -0.096 | 0.67674 | 0.251 |
| 54300 | RP11-159D12.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-590-3p;hsa-miR-874-3p | 20 | ARID5B | Sponge network | 1.254 | 0 | 0.342 | 0.01676 | 0.251 |
| 54301 | CTC-297N7.9 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p | 12 | DYNC1I1 | Sponge network | -2.069 | 0 | -1.12 | 0.00107 | 0.251 |
| 54302 | AP000487.5 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30c-5p;hsa-miR-326;hsa-miR-664a-5p | 13 | CBFB | Sponge network | 1.987 | 0 | 1.061 | 0 | 0.251 |
| 54303 | GABPB1-AS1 |
hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-29c-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-3614-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-629-3p | 14 | BCL9 | Sponge network | 1.11 | 0 | 0.321 | 0.00267 | 0.251 |
| 54304 | LINC00863 |
hsa-miR-103a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | RASSF8 | Sponge network | 0.189 | 0.13981 | -0.122 | 0.65591 | 0.251 |
| 54305 | MIR22HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-550a-3p;hsa-miR-92a-3p | 17 | ITGAV | Sponge network | -1.047 | 0 | 0.337 | 0.01002 | 0.251 |
| 54306 | RP11-216B9.6 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-25-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | ROBO1 | Sponge network | 0.174 | 0.30034 | -0.009 | 0.96154 | 0.251 |
| 54307 | CTA-204B4.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-590-3p | 27 | RUNX1T1 | Sponge network | 0.765 | 7.0E-5 | -0.344 | 0.2374 | 0.251 |
| 54308 | CTD-2089N3.1 |
hsa-let-7a-5p;hsa-let-7b-5p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-27b-5p | 12 | PRKAA2 | Sponge network | -0.314 | 0.62033 | -2.462 | 0 | 0.251 |
| 54309 | OIP5-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-25-3p;hsa-miR-32-5p;hsa-miR-331-5p;hsa-miR-338-3p;hsa-miR-338-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-664a-3p | 17 | AKAP12 | Sponge network | 0.421 | 0.00084 | -0.932 | 0.0028 | 0.251 |
| 54310 | CTB-179K24.3 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-424-5p;hsa-miR-497-5p | 18 | CCDC6 | Sponge network | 0.841 | 0.04055 | 0.27 | 0.02555 | 0.251 |
| 54311 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-196b-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 18 | ERG | Sponge network | 0.083 | 0.66405 | 0.002 | 0.99011 | 0.251 |
| 54312 | RP11-539I5.1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-493-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-589-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p | 32 | SMAD4 | Sponge network | -0.468 | 0.32587 | -0.313 | 0.00413 | 0.251 |
| 54313 | AC009948.5 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-590-3p;hsa-miR-7-1-3p | 19 | LIMCH1 | Sponge network | 0.532 | 0.00505 | -0.915 | 1.0E-5 | 0.251 |
| 54314 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-193a-3p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | PBRM1 | Sponge network | -0.615 | 0.00015 | -0.029 | 0.78601 | 0.251 |
| 54315 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-26a-2-3p;hsa-miR-28-3p;hsa-miR-30a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-497-5p;hsa-miR-766-3p | 17 | TBL1XR1 | Sponge network | 0.541 | 0.00125 | 0.578 | 0 | 0.251 |
| 54316 | RP11-13K12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-769-5p;hsa-miR-93-5p | 28 | HBP1 | Sponge network | -0.154 | 0.78939 | -0.454 | 0 | 0.251 |
| 54317 | CASC2 |
hsa-let-7a-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-16-2-3p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-628-5p | 11 | ANK3 | Sponge network | 0.385 | 0.01306 | -0.55 | 0.00086 | 0.251 |
| 54318 | CD27-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-154-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-324-5p;hsa-miR-338-3p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-940 | 29 | PBX1 | Sponge network | 0.177 | 0.16681 | -1.206 | 0 | 0.251 |
| 54319 | NNT-AS1 |
hsa-let-7a-3p;hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-16-2-3p;hsa-miR-29b-1-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-576-5p;hsa-miR-590-3p | 11 | HIP1 | Sponge network | 0.799 | 1.0E-5 | 0.774 | 0 | 0.251 |
| 54320 | KCNJ2-AS1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p;hsa-miR-671-5p | 15 | DLC1 | Sponge network | -0.436 | 0.05325 | -0.382 | 0.05038 | 0.251 |
| 54321 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-590-3p | 11 | RBM5 | Sponge network | 0.349 | 0.01695 | -0.205 | 0.0129 | 0.251 |
| 54322 | AC018890.6 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-96-5p | 11 | PRKAR1A | Sponge network | 1.61 | 0.00961 | -0.063 | 0.50314 | 0.251 |
| 54323 | RP11-720L2.4 |
hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-93-5p | 11 | ACSL6 | Sponge network | -1.255 | 0.00981 | -1.593 | 0 | 0.251 |
| 54324 | RP11-6O2.3 |
hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-326;hsa-miR-452-5p;hsa-miR-589-3p;hsa-miR-629-3p | 10 | RABEP1 | Sponge network | 0.331 | 0.57247 | -0.066 | 0.43142 | 0.251 |
| 54325 | HAR1A |
hsa-let-7g-5p;hsa-miR-200c-5p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-877-5p | 10 | NRK | Sponge network | -0.672 | 0.19447 | 0.656 | 0.19217 | 0.251 |
| 54326 | RP11-620J15.3 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-361-5p;hsa-miR-502-5p;hsa-miR-511-5p;hsa-miR-576-5p;hsa-miR-590-3p | 15 | MXI1 | Sponge network | -0.739 | 0.00044 | -0.987 | 0 | 0.251 |
| 54327 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-362-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-7-1-3p | 32 | RASSF2 | Sponge network | -0.342 | 0.02288 | -0.273 | 0.21961 | 0.251 |
| 54328 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-589-3p | 16 | MED12L | Sponge network | 0.811 | 0.03228 | -0.194 | 0.55299 | 0.251 |
| 54329 | BCYRN1 |
hsa-miR-125a-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-409-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-484;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-7-1-3p | 32 | RIMS2 | Sponge network | 0.311 | 0.10344 | -1.365 | 0.00208 | 0.251 |
| 54330 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-223-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | MTSS1 | Sponge network | 0.614 | 1.0E-5 | -0.81 | 0.00035 | 0.251 |
| 54331 | RP11-158K1.3 |
hsa-miR-142-3p;hsa-miR-150-5p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-29c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | SH3GLB1 | Sponge network | 0.349 | 0.01695 | -0.115 | 0.14773 | 0.251 |
| 54332 | RP11-48B3.4 |
hsa-miR-126-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-429;hsa-miR-590-3p | 10 | JAKMIP2 | Sponge network | 1.757 | 0 | -1.388 | 0 | 0.251 |
| 54333 | KB-1460A1.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 26 | ETV1 | Sponge network | 0.603 | 0.00127 | -0.096 | 0.67674 | 0.251 |
| 54334 | OIP5-AS1 |
hsa-miR-103a-2-5p;hsa-miR-134-5p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-331-3p;hsa-miR-376c-3p;hsa-miR-424-3p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-532-3p;hsa-miR-542-3p | 21 | SYNM | Sponge network | 0.421 | 0.00084 | -2.309 | 1.0E-5 | 0.251 |
| 54335 | BOLA3-AS1 |
hsa-let-7d-3p;hsa-miR-107;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-338-3p;hsa-miR-877-5p | 14 | GLIPR1 | Sponge network | -0.246 | 0.35034 | 0.146 | 0.3276 | 0.251 |
| 54336 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-10b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-942-5p | 16 | RB1CC1 | Sponge network | 0.249 | 0.35902 | 0.175 | 0.12559 | 0.251 |
| 54337 | C4B-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-27a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-96-5p | 14 | MED12L | Sponge network | 2.306 | 9.0E-5 | -0.194 | 0.55299 | 0.251 |
| 54338 | WAC-AS1 |
hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-149-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-342-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-590-5p | 13 | ATRX | Sponge network | 0.821 | 0 | 0.261 | 0.04408 | 0.251 |
| 54339 | HOXB-AS2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-200a-3p;hsa-miR-27a-5p;hsa-miR-324-5p;hsa-miR-550a-5p;hsa-miR-576-5p | 15 | TSHZ3 | Sponge network | 1.316 | 0.01106 | -0.556 | 0.01516 | 0.251 |
| 54340 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-93-5p | 27 | SASH1 | Sponge network | -0.384 | 0.40652 | -0.502 | 0.00175 | 0.251 |
| 54341 | RP11-244H3.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-149-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-532-3p;hsa-miR-590-3p | 12 | DAB2 | Sponge network | 0.659 | 3.0E-5 | 0.431 | 0.0113 | 0.251 |
| 54342 | RP1-59D14.5 |
hsa-miR-141-5p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-26b-5p;hsa-miR-424-5p;hsa-miR-874-3p | 12 | TACC1 | Sponge network | 1.162 | 5.0E-5 | -0.362 | 0.05406 | 0.251 |
| 54343 | RP1-102E24.8 |
hsa-let-7a-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-218-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-30a-3p;hsa-miR-374b-5p | 10 | PIK3C2A | Sponge network | 1.737 | 0 | 0.061 | 0.53776 | 0.251 |
| 54344 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-148b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | ZNF844 | Sponge network | 0.727 | 3.0E-5 | -0.966 | 0.00035 | 0.251 |
| 54345 | RP11-181G12.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-361-3p | 17 | NCOA1 | Sponge network | 0.556 | 0.00298 | -0.432 | 0 | 0.251 |
| 54346 | TPTEP1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-342-3p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p;hsa-miR-93-3p | 33 | NCOA1 | Sponge network | -1.216 | 0 | -0.432 | 0 | 0.251 |
| 54347 | RP11-359B12.2 |
hsa-let-7g-5p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-27a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-3607-3p | 10 | KDM3A | Sponge network | 0.064 | 0.72934 | 0.132 | 0.22711 | 0.251 |
| 54348 | RP11-384O8.1 |
hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-590-3p | 17 | FMNL3 | Sponge network | -0.738 | 0.21233 | 0.555 | 4.0E-5 | 0.251 |
| 54349 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-32-5p;hsa-miR-324-5p;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p | 32 | PRKAB2 | Sponge network | -0.563 | 0.02545 | -0.367 | 0.01371 | 0.251 |
| 54350 | LINC01018 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-590-5p | 18 | KANK1 | Sponge network | -2.915 | 0.00015 | -0.55 | 0.00134 | 0.251 |
| 54351 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-455-5p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p | 20 | PTPN11 | Sponge network | -0.737 | 0.06182 | 0.431 | 2.0E-5 | 0.251 |
| 54352 | LIPE-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-30d-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-582-5p | 13 | STARD13 | Sponge network | 0.078 | 0.55803 | -0.187 | 0.27383 | 0.251 |
| 54353 | AC084219.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-193a-3p;hsa-miR-223-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-539-5p | 15 | ABI2 | Sponge network | 1.296 | 0.0054 | 0.175 | 0.14787 | 0.251 |
| 54354 | HNRNPU-AS1 |
hsa-miR-142-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-214-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-374b-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p | 17 | TMEM30A | Sponge network | 1.601 | 0 | 0.096 | 0.31031 | 0.251 |
| 54355 | RP11-890B15.3 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-200a-3p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-590-3p | 12 | RIMS1 | Sponge network | 0.101 | 0.60919 | -3.143 | 0 | 0.251 |
| 54356 | CTD-3099C6.9 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-22-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-629-3p | 29 | KIF1B | Sponge network | 1.361 | 0 | -0.118 | 0.32258 | 0.251 |
| 54357 | RP11-798M19.6 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 29 | MAF | Sponge network | -0.612 | 0.00094 | -1.257 | 0 | 0.251 |
| 54358 | SERHL |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181a-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-539-5p;hsa-miR-625-5p;hsa-miR-628-5p | 54 | LPP | Sponge network | -0.602 | 0.00331 | -0.459 | 0.04475 | 0.251 |
| 54359 | DNM3OS |
hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-188-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-590-3p;hsa-miR-590-5p | 12 | FBXO11 | Sponge network | 0.572 | 0.01722 | 0.142 | 0.04938 | 0.251 |
| 54360 | RP11-403I13.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-186-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | LRRK2 | Sponge network | 0.619 | 0.00157 | -1.136 | 1.0E-5 | 0.251 |
| 54361 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-590-3p | 13 | RBM5 | Sponge network | -0.899 | 6.0E-5 | -0.205 | 0.0129 | 0.251 |
| 54362 | RAMP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p | 14 | LATS1 | Sponge network | -0.513 | 0.04703 | 0.109 | 0.34208 | 0.251 |
| 54363 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | GRIN2A | Sponge network | -0.141 | 0.26434 | -2.161 | 0 | 0.251 |
| 54364 | RP11-244H3.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-378a-5p;hsa-miR-582-5p;hsa-miR-590-3p | 14 | ARHGAP29 | Sponge network | 0.659 | 3.0E-5 | 0.131 | 0.42953 | 0.251 |
| 54365 | RP4-605O3.4 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-155-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-3614-5p;hsa-miR-590-3p | 10 | ABL2 | Sponge network | 0.262 | 0.0475 | 0.82 | 0 | 0.251 |
| 54366 | CKMT2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-2110;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-590-3p;hsa-miR-942-5p | 13 | CREBBP | Sponge network | 0.082 | 0.55654 | 0.126 | 0.15186 | 0.251 |
| 54367 | FZD10-AS1 |
hsa-miR-107;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-21-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-589-3p;hsa-miR-590-3p | 12 | ABCA10 | Sponge network | -0.727 | 0.04726 | -0.571 | 0.03674 | 0.251 |
| 54368 | DNM1P35 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-423-3p;hsa-miR-508-3p;hsa-miR-590-3p | 11 | ROBO1 | Sponge network | 0.051 | 0.83388 | -0.009 | 0.96154 | 0.251 |
| 54369 | FZD10-AS1 |
hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-421;hsa-miR-582-5p;hsa-miR-590-3p | 15 | STARD13 | Sponge network | -0.727 | 0.04726 | -0.187 | 0.27383 | 0.251 |
| 54370 | RP11-428J1.5 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-96-5p | 17 | SCN9A | Sponge network | 0.538 | 0.00076 | -0.525 | 0.10593 | 0.251 |
| 54371 | AC016747.3 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-144-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-590-3p;hsa-miR-940 | 10 | HOOK3 | Sponge network | 0.199 | 0.38015 | 0.273 | 0.09957 | 0.251 |
| 54372 | ADAMTS9-AS2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 21 | FOXO1 | Sponge network | -2.452 | 0 | -0.141 | 0.33874 | 0.251 |
| 54373 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-582-5p | 17 | GRIN2A | Sponge network | 0.95 | 0.15943 | -2.161 | 0 | 0.251 |
| 54374 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-374b-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-7-1-3p | 20 | UBE4B | Sponge network | -0.976 | 0.00025 | -0.235 | 0.007 | 0.251 |
| 54375 | RP11-158K1.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-192-5p;hsa-miR-1976;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-495-3p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | CUL5 | Sponge network | 0.349 | 0.01695 | 0.026 | 0.77301 | 0.251 |
| 54376 | RP11-326G21.1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-362-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 17 | MAF | Sponge network | -0.021 | 0.93598 | -1.257 | 0 | 0.251 |
| 54377 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-942-5p;hsa-miR-96-5p | 16 | PCDH17 | Sponge network | 0.41 | 0.02214 | 0.731 | 2.0E-5 | 0.251 |
| 54378 | RP11-359B12.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-200b-3p;hsa-miR-27a-3p;hsa-miR-429;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | CDYL | Sponge network | 0.064 | 0.72934 | 0.12 | 0.12425 | 0.251 |
| 54379 | RP11-672A2.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-193b-5p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-424-5p | 18 | SSBP2 | Sponge network | 1.344 | 0 | -1.58 | 0 | 0.251 |
| 54380 | LPP-AS2 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374a-5p;hsa-miR-508-3p;hsa-miR-532-3p;hsa-miR-96-5p | 12 | NR4A3 | Sponge network | -0.18 | 0.20343 | -1.385 | 0 | 0.251 |
| 54381 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-338-5p;hsa-miR-339-5p;hsa-miR-375;hsa-miR-424-5p;hsa-miR-429;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 27 | MYBL1 | Sponge network | -0.129 | 0.57177 | 0.494 | 0.02152 | 0.251 |
| 54382 | CTD-2583A14.8 |
hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-374a-3p | 11 | CREB1 | Sponge network | 1.134 | 0 | 0.203 | 0.00537 | 0.251 |
| 54383 | KB-1460A1.5 |
hsa-let-7b-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 45 | AR | Sponge network | 0.603 | 0.00127 | -1.554 | 0 | 0.251 |
| 54384 | RP11-490M8.1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-20a-5p;hsa-miR-539-5p;hsa-miR-96-5p | 13 | PRKAA2 | Sponge network | -0.214 | 0.44248 | -2.462 | 0 | 0.251 |
| 54385 | RP11-490M8.1 |
hsa-miR-1301-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-219a-1-3p;hsa-miR-223-3p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-766-3p | 19 | IL6ST | Sponge network | -0.214 | 0.44248 | -0.402 | 0.00676 | 0.251 |
| 54386 | RP11-770J1.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-577 | 16 | PRKAA2 | Sponge network | -2.011 | 0.0005 | -2.462 | 0 | 0.251 |
| 54387 | ZSCAN16-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-326;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-590-3p | 13 | DDX5 | Sponge network | -0.342 | 0.02288 | -0.099 | 0.17428 | 0.251 |
| 54388 | AP000254.8 |
hsa-miR-126-5p;hsa-miR-134-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p | 10 | CREB1 | Sponge network | 0.555 | 0.00012 | 0.203 | 0.00537 | 0.251 |
| 54389 | NEAT1 |
hsa-miR-127-3p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-345-5p;hsa-miR-425-3p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-654-3p | 15 | PER2 | Sponge network | 0.705 | 0.00014 | -0.396 | 0.00269 | 0.251 |
| 54390 | RP11-253E3.3 |
hsa-miR-129-5p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-497-5p;hsa-miR-9-5p | 14 | CCDC6 | Sponge network | 1.764 | 0 | 0.27 | 0.02555 | 0.251 |
| 54391 | LINC00173 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-330-3p;hsa-miR-369-3p;hsa-miR-421 | 10 | JAKMIP2 | Sponge network | 0.056 | 0.8494 | -1.388 | 0 | 0.251 |
| 54392 | LINC00957 |
hsa-let-7g-3p;hsa-miR-106b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-324-3p;hsa-miR-502-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | FOXO1 | Sponge network | -0.421 | 0.0114 | -0.141 | 0.33874 | 0.251 |
| 54393 | RP11-400K9.3 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-340-5p;hsa-miR-940 | 10 | ETV1 | Sponge network | -0.733 | 0.19469 | -0.096 | 0.67674 | 0.251 |
| 54394 | RP11-158K1.3 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-301a-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | SUFU | Sponge network | 0.349 | 0.01695 | -0.04 | 0.65489 | 0.251 |
| 54395 | XXbac-B444P24.8 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-20a-3p;hsa-miR-27a-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-33a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-93-3p | 23 | RUNX1T1 | Sponge network | 2.264 | 0.0012 | -0.344 | 0.2374 | 0.251 |
| 54396 | LINC00648 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | EPHA5 | Sponge network | 0.633 | 0.51678 | -0.957 | 0.01733 | 0.251 |
| 54397 | HOTAIRM1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-335-3p;hsa-miR-500a-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | TMEM132B | Sponge network | -0.053 | 0.80685 | -0.83 | 0.01084 | 0.251 |
| 54398 | RP11-212P7.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-26a-2-3p;hsa-miR-335-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-93-5p | 15 | RBL2 | Sponge network | 0.219 | 0.1516 | -0.282 | 0.00254 | 0.251 |
| 54399 | DIO3OS |
hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-455-5p;hsa-miR-582-5p | 14 | GJA1 | Sponge network | -0.773 | 0.04073 | -0.485 | 0.02179 | 0.251 |
| 54400 | RP11-728F11.4 |
hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-326;hsa-miR-331-3p;hsa-miR-501-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 19 | ABI2 | Sponge network | 0.238 | 0.70544 | 0.175 | 0.14787 | 0.251 |
| 54401 | RP11-33B1.4 |
hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-143-5p;hsa-miR-145-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | DDX3X | Sponge network | 0.826 | 0 | -0.152 | 0.07686 | 0.251 |
| 54402 | RP11-678G14.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-20a-5p;hsa-miR-30b-3p;hsa-miR-34a-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-7-1-3p | 12 | LRRC55 | Sponge network | -4.477 | 0 | -0.026 | 0.93163 | 0.251 |
| 54403 | CTC-429P9.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-330-3p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-532-5p | 20 | NUAK1 | Sponge network | 0.082 | 0.55397 | 0.904 | 0 | 0.251 |
| 54404 | RP11-274B21.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-27a-3p;hsa-miR-33a-3p;hsa-miR-409-3p;hsa-miR-590-3p | 10 | RCAN2 | Sponge network | 1.073 | 0.00216 | -0.33 | 0.15172 | 0.251 |
| 54405 | RP11-458D21.1 |
hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p | 13 | ARNT2 | Sponge network | 0.663 | 0.00073 | -0.332 | 0.25851 | 0.251 |
| 54406 | RP11-284N8.3 |
hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-577;hsa-miR-96-5p | 11 | SOX5 | Sponge network | -0.31 | 0.33871 | -1.091 | 4.0E-5 | 0.251 |
| 54407 | LINC00094 |
hsa-miR-141-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-3127-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-375;hsa-miR-429;hsa-miR-671-5p | 14 | THRA | Sponge network | 0.253 | 0.09565 | -0.307 | 0.05099 | 0.251 |
| 54408 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-450b-5p;hsa-miR-486-5p;hsa-miR-500a-5p;hsa-miR-532-3p;hsa-miR-542-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-628-5p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 48 | FOXP1 | Sponge network | -1.281 | 0.00012 | 0.042 | 0.74368 | 0.251 |
| 54409 | RP11-403I13.7 |
hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-26a-2-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-423-5p;hsa-miR-484 | 16 | ABL1 | Sponge network | 0.395 | 0.15451 | -0.215 | 0.07804 | 0.251 |
| 54410 | HOXD-AS2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-378a-3p;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-502-5p;hsa-miR-625-5p | 25 | NTRK3 | Sponge network | -0.208 | 0.65592 | -1.77 | 3.0E-5 | 0.251 |
| 54411 | SH3BP5-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-877-5p | 13 | NOTCH2NL | Sponge network | 0.267 | 0.2937 | 0.024 | 0.84307 | 0.251 |
| 54412 | CTC-429P9.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-500a-5p;hsa-miR-550a-3p;hsa-miR-93-5p;hsa-miR-98-5p | 22 | PRRX1 | Sponge network | 0.082 | 0.55397 | 1.316 | 0 | 0.251 |
| 54413 | ZNF252P-AS1 |
hsa-miR-125b-5p;hsa-miR-143-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-326;hsa-miR-664a-5p | 10 | CBFB | Sponge network | 1.076 | 0 | 1.061 | 0 | 0.251 |
| 54414 | AC018890.6 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 17 | PTPRT | Sponge network | 1.61 | 0.00961 | -0.907 | 0.03727 | 0.251 |
| 54415 | LINC00621 |
hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-30d-3p;hsa-miR-323a-3p;hsa-miR-576-5p;hsa-miR-590-3p | 10 | GJA1 | Sponge network | 0.131 | 0.8436 | -0.485 | 0.02179 | 0.251 |
| 54416 | MAGI2-AS3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-429;hsa-miR-501-5p;hsa-miR-590-3p;hsa-miR-629-3p | 18 | PSIP1 | Sponge network | -0.129 | 0.57177 | -0.005 | 0.96897 | 0.251 |
| 54417 | U91328.19 |
hsa-miR-107;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 18 | TAL1 | Sponge network | -0.303 | 0.21002 | -0.462 | 0.00574 | 0.251 |
| 54418 | MIR497HG |
hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-940 | 33 | PPP1R9A | Sponge network | -1.439 | 4.0E-5 | -0.903 | 0.04254 | 0.251 |
| 54419 | RP13-516M14.1 |
hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-26b-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-7-1-3p | 10 | LRRC55 | Sponge network | 0.389 | 0.04293 | -0.026 | 0.93163 | 0.251 |
| 54420 | RP4-758J18.10 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-150-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-331-3p;hsa-miR-450b-5p | 11 | CDHR3 | Sponge network | -0.091 | 0.71468 | -0.977 | 4.0E-5 | 0.251 |
| 54421 | LIPE-AS1 |
hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-455-5p;hsa-miR-582-5p | 16 | GJA1 | Sponge network | 0.078 | 0.55803 | -0.485 | 0.02179 | 0.251 |
| 54422 | AC005154.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-212-3p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-484;hsa-miR-493-5p;hsa-miR-577;hsa-miR-590-3p | 12 | MYCBP2 | Sponge network | 0.848 | 0 | -0.027 | 0.82016 | 0.251 |
| 54423 | RP11-6O2.3 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-15b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-539-5p;hsa-miR-629-3p | 13 | TCF12 | Sponge network | 0.331 | 0.57247 | 0.174 | 0.13271 | 0.251 |
| 54424 | RP11-532F6.3 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p | 17 | CSMD1 | Sponge network | -0.896 | 0.00099 | -0.63 | 0.18881 | 0.251 |
| 54425 | RP11-384O8.1 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-501-3p;hsa-miR-590-3p | 15 | KAT6B | Sponge network | -0.738 | 0.21233 | -0.478 | 0.00018 | 0.251 |
| 54426 | CTD-2006H14.2 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-20a-3p;hsa-miR-299-5p;hsa-miR-590-3p | 10 | NTRK2 | Sponge network | 1.378 | 0 | -0.736 | 0.06495 | 0.251 |
| 54427 | DNM1P35 |
hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-2110;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-92a-1-5p | 10 | PRKACA | Sponge network | 0.051 | 0.83388 | -0.255 | 0.0012 | 0.251 |
| 54428 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-96-5p | 14 | ARHGAP29 | Sponge network | 0.614 | 1.0E-5 | 0.131 | 0.42953 | 0.251 |
| 54429 | AC018890.6 |
hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-26b-5p;hsa-miR-338-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-7-1-3p | 12 | PCDH9 | Sponge network | 1.61 | 0.00961 | -1.823 | 4.0E-5 | 0.251 |
| 54430 | RP11-428J1.5 |
hsa-miR-106b-5p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19b-1-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | TET1 | Sponge network | 0.538 | 0.00076 | -0.135 | 0.52138 | 0.251 |
| 54431 | CTD-2006H14.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-143-5p;hsa-miR-210-3p;hsa-miR-30a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 10 | CLASP2 | Sponge network | 1.378 | 0 | -0.038 | 0.71 | 0.251 |
| 54432 | BDNF-AS |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-429;hsa-miR-484;hsa-miR-590-3p;hsa-miR-93-5p | 17 | ARNT2 | Sponge network | -0.668 | 3.0E-5 | -0.332 | 0.25851 | 0.251 |
| 54433 | CKMT2-AS1 |
hsa-miR-103a-2-5p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19b-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-362-5p;hsa-miR-365a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-495-3p;hsa-miR-501-5p;hsa-miR-505-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-942-5p | 34 | ARHGEF12 | Sponge network | 0.082 | 0.55654 | 0.09 | 0.35693 | 0.251 |
| 54434 | RP11-403I13.8 |
hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-5p | 15 | PTGFR | Sponge network | 0.619 | 0.00157 | -0.233 | 0.45421 | 0.251 |
| 54435 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-452-5p;hsa-miR-539-5p;hsa-miR-7-1-3p;hsa-miR-708-5p;hsa-miR-935 | 25 | RAP1A | Sponge network | -0.405 | 0.22013 | -0.929 | 0 | 0.251 |
| 54436 | RP11-401P9.4 |
hsa-let-7a-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-323a-3p;hsa-miR-324-3p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-342-3p;hsa-miR-511-5p;hsa-miR-708-3p;hsa-miR-874-3p | 23 | MXI1 | Sponge network | -0.384 | 0.40652 | -0.987 | 0 | 0.251 |
| 54437 | MAP3K14 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-146b-5p;hsa-miR-17-5p;hsa-miR-196a-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-98-5p | 14 | CADM2 | Sponge network | -0.295 | 0.02604 | -3.782 | 0 | 0.251 |
| 54438 | RP11-283I3.6 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-369-3p;hsa-miR-455-3p;hsa-miR-576-5p | 25 | NTRK2 | Sponge network | 0.614 | 1.0E-5 | -0.736 | 0.06495 | 0.251 |
| 54439 | RP11-983P16.4 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-132-5p;hsa-miR-141-5p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-150-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-33a-5p;hsa-miR-345-5p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-409-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-532-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-629-3p;hsa-miR-664a-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p;hsa-miR-940 | 40 | TRPS1 | Sponge network | 0.41 | 0.02214 | -0.191 | 0.44109 | 0.251 |
| 54440 | PCA3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-590-3p | 20 | USP6 | Sponge network | -0.709 | 0.18168 | 0.188 | 0.3871 | 0.251 |
| 54441 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-212-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-3065-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-508-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-93-5p | 52 | PTEN | Sponge network | 0.572 | 0.01722 | -0.187 | 0.07748 | 0.251 |
| 54442 | RP11-1277A3.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-200a-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-224-5p;hsa-miR-24-2-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-93-3p;hsa-miR-940;hsa-miR-96-5p | 23 | QKI | Sponge network | 0.453 | 0.01839 | -0.333 | 0.04111 | 0.251 |
| 54443 | RP11-156E6.1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-495-3p | 19 | MAPK1 | Sponge network | 1.266 | 0 | -0.017 | 0.81465 | 0.251 |
| 54444 | CTD-3064M3.3 |
hsa-miR-103a-2-5p;hsa-miR-128-3p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-423-5p;hsa-miR-429;hsa-miR-582-5p | 23 | GNAQ | Sponge network | -0.469 | 0.08721 | -0.367 | 0.00102 | 0.251 |
| 54445 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-200b-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 10 | STARD13 | Sponge network | -0.125 | 0.49414 | -0.187 | 0.27383 | 0.251 |
| 54446 | RP11-46C24.7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 16 | IGSF10 | Sponge network | 0.392 | 0.0278 | -1.751 | 1.0E-5 | 0.251 |
| 54447 | RP11-182J1.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 10 | PRRX1 | Sponge network | -0.187 | 0.23787 | 1.316 | 0 | 0.251 |
| 54448 | ADARB2-AS1 |
hsa-let-7g-5p;hsa-miR-146a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p | 12 | CBX5 | Sponge network | 0.876 | 0.37332 | 0.165 | 0.24446 | 0.251 |
| 54449 | RP11-284N8.3 |
hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-128-3p;hsa-miR-15b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-20a-5p;hsa-miR-324-3p;hsa-miR-335-3p;hsa-miR-493-5p;hsa-miR-93-5p;hsa-miR-96-5p | 12 | PIK3R1 | Sponge network | -0.31 | 0.33871 | -0.133 | 0.36854 | 0.251 |
| 54450 | RP11-727A23.5 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-664a-5p | 19 | SOX11 | Sponge network | 1.117 | 0 | 2.68 | 0 | 0.251 |
| 54451 | CTD-2336O2.1 |
hsa-let-7a-3p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-301a-3p;hsa-miR-33a-3p | 10 | MAFB | Sponge network | -0.141 | 0.26434 | -0.023 | 0.89865 | 0.251 |
| 54452 | RP11-479G22.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | ZFHX4 | Sponge network | 1.308 | 0 | 0.018 | 0.95574 | 0.251 |
| 54453 | CTD-2336O2.1 |
hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-423-3p;hsa-miR-450b-5p;hsa-miR-590-3p | 11 | ROBO1 | Sponge network | -0.141 | 0.26434 | -0.009 | 0.96154 | 0.251 |
| 54454 | SH3BP5-AS1 |
hsa-miR-106b-5p;hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-23a-3p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-375;hsa-miR-539-5p;hsa-miR-550a-3p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935 | 24 | DIP2C | Sponge network | 0.267 | 0.2937 | -0.436 | 0.01713 | 0.251 |
| 54455 | LINC00702 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 27 | FOXO3 | Sponge network | -0.737 | 0.06182 | -0.04 | 0.72028 | 0.251 |
| 54456 | CTD-2017D11.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-1976;hsa-miR-326;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-484;hsa-miR-744-3p | 13 | CUL5 | Sponge network | 0.792 | 0.0049 | 0.026 | 0.77301 | 0.251 |
| 54457 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-222-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 11 | FOXO3 | Sponge network | 0.476 | 0.19203 | -0.04 | 0.72028 | 0.251 |
| 54458 | RP3-402G11.26 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-148b-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-22-5p;hsa-miR-27a-3p;hsa-miR-330-5p;hsa-miR-590-3p;hsa-miR-940 | 11 | PTPRT | Sponge network | -0.254 | 0.19303 | -0.907 | 0.03727 | 0.251 |
| 54459 | AC010226.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-222-5p;hsa-miR-455-3p;hsa-miR-93-5p;hsa-miR-96-5p | 11 | EPAS1 | Sponge network | -0.811 | 2.0E-5 | -0.184 | 0.14418 | 0.251 |
| 54460 | RP11-531A24.5 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-151a-5p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-576-5p;hsa-miR-940 | 36 | NTRK2 | Sponge network | 0.75 | 0.00054 | -0.736 | 0.06495 | 0.251 |
| 54461 | LINC00657 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-23a-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-3607-3p;hsa-miR-374a-3p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-505-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 32 | FZD3 | Sponge network | 0.91 | 0 | 0.104 | 0.60316 | 0.251 |
| 54462 | CTC-444N24.11 |
hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-214-5p;hsa-miR-28-5p;hsa-miR-337-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 13 | IL13RA1 | Sponge network | 1.153 | 0 | 0.019 | 0.86949 | 0.251 |
| 54463 | RP11-277P12.20 |
hsa-miR-144-3p;hsa-miR-196b-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30d-5p;hsa-miR-323a-3p;hsa-miR-877-5p | 10 | NRK | Sponge network | 0.687 | 0.02753 | 0.656 | 0.19217 | 0.251 |
| 54464 | RP1-68D18.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-429;hsa-miR-628-5p | 10 | TCF4 | Sponge network | 1.987 | 0 | 0.016 | 0.92271 | 0.251 |
| 54465 | HOXA-AS3 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-146b-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-340-5p;hsa-miR-424-5p;hsa-miR-590-3p | 15 | DYNC1I1 | Sponge network | -0.555 | 0.06156 | -1.12 | 0.00107 | 0.251 |
| 54466 | RP11-262H14.3 |
hsa-miR-125a-3p;hsa-miR-149-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-214-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | CDC27 | Sponge network | -0.106 | 0.63074 | 0.384 | 0 | 0.251 |
| 54467 | AC009120.6 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-222-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-27a-3p;hsa-miR-28-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-335-5p;hsa-miR-576-5p;hsa-miR-590-3p | 19 | FOXO3 | Sponge network | 0.919 | 0 | -0.04 | 0.72028 | 0.251 |
| 54468 | CTC-429P9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-135b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-500a-5p | 16 | ST6GAL2 | Sponge network | 0.082 | 0.55397 | -1.201 | 0.00597 | 0.251 |
| 54469 | SNHG16 |
hsa-let-7a-3p;hsa-miR-101-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-497-5p | 14 | RICTOR | Sponge network | 0.961 | 0 | 0.388 | 0.00188 | 0.251 |
| 54470 | C4A-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-30b-3p;hsa-miR-345-5p;hsa-miR-34a-5p;hsa-miR-486-5p;hsa-miR-744-3p;hsa-miR-96-5p | 22 | FAT3 | Sponge network | 3.345 | 0 | -1.601 | 0.00051 | 0.251 |
| 54471 | CTD-3138B18.5 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-143-5p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-195-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30a-3p;hsa-miR-30b-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-7-1-3p | 34 | MDM4 | Sponge network | 0.379 | 0.03091 | 0.345 | 0.00762 | 0.251 |
| 54472 | JAZF1-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-3p;hsa-miR-301a-3p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-340-5p;hsa-miR-362-5p;hsa-miR-502-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-92a-3p;hsa-miR-93-5p;hsa-miR-96-5p | 36 | IL17RD | Sponge network | -0.769 | 0.00035 | -0.019 | 0.94873 | 0.251 |
| 54473 | CTC-487M23.5 |
hsa-let-7g-3p;hsa-miR-148a-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-24-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-92a-3p;hsa-miR-93-3p | 16 | PCDH10 | Sponge network | 0.912 | 0.00014 | -1.644 | 0.0078 | 0.251 |
| 54474 | ZNF667-AS1 |
hsa-let-7g-3p;hsa-let-7i-3p;hsa-miR-1307-3p;hsa-miR-16-5p;hsa-miR-192-3p;hsa-miR-196a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-326;hsa-miR-484;hsa-miR-7-1-3p;hsa-miR-744-5p;hsa-miR-93-3p | 13 | ACTB | Sponge network | -0.976 | 0.00025 | -0.247 | 0.01736 | 0.251 |
| 54475 | RP11-389C8.2 |
hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-200c-5p;hsa-miR-222-5p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-550a-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 28 | DTNA | Sponge network | 0.727 | 3.0E-5 | -1.648 | 1.0E-5 | 0.251 |
| 54476 | PWAR6 |
hsa-miR-10b-3p;hsa-miR-140-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-194-5p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-212-3p;hsa-miR-23a-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-629-5p;hsa-miR-653-5p | 25 | UBR3 | Sponge network | -1.575 | 6.0E-5 | -0.021 | 0.82774 | 0.251 |
| 54477 | RP4-668J24.2 |
hsa-miR-106b-5p;hsa-miR-146b-3p;hsa-miR-17-5p;hsa-miR-185-3p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-192-5p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-27a-5p;hsa-miR-3913-5p;hsa-miR-629-3p;hsa-miR-7-5p | 15 | ZBTB4 | Sponge network | -1.054 | 0.0706 | -0.739 | 0 | 0.251 |
| 54478 | RP11-83A24.2 |
hsa-let-7g-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-223-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-331-5p;hsa-miR-33a-3p;hsa-miR-409-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 25 | ACVR2A | Sponge network | 0.417 | 0.00445 | -0.409 | 0.00039 | 0.251 |
| 54479 | AF131215.9 |
hsa-miR-103a-2-5p;hsa-miR-106b-5p;hsa-miR-10a-3p;hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-151a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-19a-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-2110;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-23a-3p;hsa-miR-23a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-577;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-628-5p;hsa-miR-92b-3p | 59 | LPP | Sponge network | -0.322 | 0.25945 | -0.459 | 0.04475 | 0.251 |
| 54480 | TPTEP1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-454-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | HECA | Sponge network | -1.216 | 0 | -0.217 | 0.03463 | 0.251 |
| 54481 | LINC00622 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 10 | CEP170 | Sponge network | 1.169 | 0.00028 | 0.943 | 0 | 0.251 |
| 54482 | PAXIP1-AS1 |
hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-154-5p;hsa-miR-195-5p;hsa-miR-199a-5p;hsa-miR-199b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-495-3p;hsa-miR-664a-5p;hsa-miR-766-3p | 15 | MAPK1 | Sponge network | 0.66 | 1.0E-5 | -0.017 | 0.81465 | 0.251 |
| 54483 | MIR143HG |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-27b-5p;hsa-miR-29a-5p;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-335-3p;hsa-miR-339-5p;hsa-miR-34a-5p;hsa-miR-362-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-500b-5p;hsa-miR-539-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p;hsa-miR-935 | 33 | ACSL6 | Sponge network | -0.739 | 0.0574 | -1.593 | 0 | 0.251 |
| 54484 | MIR143HG |
hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-2110;hsa-miR-224-5p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 11 | PCM1 | Sponge network | -0.739 | 0.0574 | 0.164 | 0.20307 | 0.251 |
| 54485 | AC141928.1 |
hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-577 | 11 | CSDE1 | Sponge network | 0.853 | 0.15352 | -0.493 | 0 | 0.251 |
| 54486 | FENDRR |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 18 | IGF2 | Sponge network | -1.281 | 0.00012 | 0.848 | 0.03842 | 0.251 |
| 54487 | AC002117.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-532-3p | 11 | PBX1 | Sponge network | 0.476 | 0.19203 | -1.206 | 0 | 0.251 |
| 54488 | RP11-46C24.7 |
hsa-miR-146b-5p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-574-3p;hsa-miR-590-3p | 11 | FHL1 | Sponge network | 0.392 | 0.0278 | -2.687 | 0 | 0.251 |
| 54489 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-338-3p;hsa-miR-339-5p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-484;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 57 | KIF1B | Sponge network | 1.302 | 7.0E-5 | -0.118 | 0.32258 | 0.251 |
| 54490 | CTC-457L16.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-193b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-629-3p;hsa-miR-93-5p;hsa-miR-940 | 19 | BHLHE41 | Sponge network | 1.153 | 0.00114 | 0.353 | 0.12753 | 0.251 |
| 54491 | AC005562.1 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-335-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-651-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 24 | WWTR1 | Sponge network | 0.488 | 0.00084 | -0.345 | 0.11997 | 0.251 |
| 54492 | BVES-AS1 |
hsa-miR-125a-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-5p;hsa-miR-576-5p;hsa-miR-766-3p | 10 | ATM | Sponge network | -2.62 | 0 | 0.178 | 0.21568 | 0.251 |
| 54493 | RP11-5C23.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-21-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p | 20 | ESR1 | Sponge network | 0.274 | 0.07681 | -1.035 | 3.0E-5 | 0.251 |
| 54494 | SERHL |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-539-5p | 13 | BCL11A | Sponge network | -0.602 | 0.00331 | -0.932 | 0.0063 | 0.251 |
| 54495 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-1271-5p;hsa-miR-130a-3p;hsa-miR-134-5p;hsa-miR-144-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-323a-3p | 12 | PIK3C2A | Sponge network | 1.122 | 0 | 0.061 | 0.53776 | 0.251 |
| 54496 | USP3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-671-5p | 20 | CYLD | Sponge network | -0.162 | 0.47687 | -0.367 | 0.00171 | 0.251 |
| 54497 | AF131215.9 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-590-3p;hsa-miR-92a-1-5p | 13 | AKAP6 | Sponge network | -0.322 | 0.25945 | -1.586 | 3.0E-5 | 0.251 |
| 54498 | RP11-344B5.2 |
hsa-miR-103a-3p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29b-1-5p;hsa-miR-505-3p | 14 | IL6ST | Sponge network | -0.055 | 0.88482 | -0.402 | 0.00676 | 0.251 |
| 54499 | CTC-559E9.6 |
hsa-miR-1271-5p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-32-5p;hsa-miR-628-5p;hsa-miR-654-3p;hsa-miR-671-5p;hsa-miR-7-5p | 15 | BMPR2 | Sponge network | 0.601 | 1.0E-5 | 0.206 | 0.0635 | 0.251 |
| 54500 | CTD-2291D10.4 |
hsa-let-7a-3p;hsa-let-7d-5p;hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-96-5p | 10 | CLASP2 | Sponge network | 3.21 | 0 | -0.038 | 0.71 | 0.251 |
| 54501 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-424-5p;hsa-miR-577;hsa-miR-590-3p | 17 | SYNE1 | Sponge network | -0.322 | 0.25945 | -0.738 | 0.00316 | 0.251 |
| 54502 | DNAJC27-AS1 |
hsa-miR-1301-3p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-21-5p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-5p;hsa-miR-660-5p;hsa-miR-7-1-3p | 25 | LIMCH1 | Sponge network | -0.969 | 2.0E-5 | -0.915 | 1.0E-5 | 0.251 |
| 54503 | RP11-326G21.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-1301-3p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200c-3p;hsa-miR-26b-3p;hsa-miR-29b-1-5p;hsa-miR-590-3p | 11 | NR2C2 | Sponge network | -0.021 | 0.93598 | 0.21 | 0.03224 | 0.251 |
| 54504 | SNHG16 |
hsa-miR-10b-5p;hsa-miR-130a-3p;hsa-miR-148a-3p;hsa-miR-195-5p;hsa-miR-199b-5p;hsa-miR-214-3p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-374a-3p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-505-5p | 16 | ARHGEF12 | Sponge network | 0.961 | 0 | 0.09 | 0.35693 | 0.251 |
| 54505 | HOXA-AS2 |
hsa-let-7g-3p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-21-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-335-5p;hsa-miR-362-3p;hsa-miR-423-3p;hsa-miR-424-5p;hsa-miR-7-1-3p | 18 | UBE4B | Sponge network | 0.61 | 0.01593 | -0.235 | 0.007 | 0.251 |
| 54506 | AC116366.6 |
hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-200c-3p;hsa-miR-326;hsa-miR-3607-3p;hsa-miR-375;hsa-miR-421;hsa-miR-577 | 13 | BNC2 | Sponge network | 0.329 | 0.11122 | -0.491 | 0.09988 | 0.251 |
| 54507 | CTD-2089N3.2 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-23a-3p;hsa-miR-26b-3p;hsa-miR-590-3p | 12 | TMEM132B | Sponge network | -1.375 | 0.08642 | -0.83 | 0.01084 | 0.251 |
| 54508 | AC005618.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 13 | CLIP1 | Sponge network | 0.862 | 0.06213 | -0.215 | 0.08684 | 0.251 |
| 54509 | NDUFA6-AS1 |
hsa-miR-126-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-23a-3p;hsa-miR-29b-3p;hsa-miR-338-3p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | TLL1 | Sponge network | -0.355 | 0.0077 | -1.195 | 3.0E-5 | 0.251 |
| 54510 | RP11-73M18.8 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130a-3p;hsa-miR-148a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-5p;hsa-miR-33a-3p | 11 | CDC73 | Sponge network | 0.832 | 0 | 0.094 | 0.20463 | 0.251 |
| 54511 | FENDRR |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-190a-5p;hsa-miR-196a-5p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-331-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-539-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 22 | BCL11A | Sponge network | -1.281 | 0.00012 | -0.932 | 0.0063 | 0.251 |
| 54512 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2110;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-324-5p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-625-5p | 16 | LHX6 | Sponge network | -0.899 | 6.0E-5 | -0.087 | 0.69193 | 0.251 |
| 54513 | RP11-474O21.5 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-1976;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-30e-5p;hsa-miR-7-1-3p | 15 | NUAK1 | Sponge network | -0.023 | 0.95109 | 0.904 | 0 | 0.251 |
| 54514 | LINC00926 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-576-5p;hsa-miR-671-5p;hsa-miR-96-5p | 12 | BNC2 | Sponge network | 0.756 | 0.00739 | -0.491 | 0.09988 | 0.251 |
| 54515 | RP11-25K19.1 |
hsa-miR-128-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-330-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p | 13 | PAIP2 | Sponge network | -1.057 | 0.00029 | -0.515 | 0 | 0.251 |
| 54516 | RP11-25K19.1 |
hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-409-3p;hsa-miR-421;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 27 | SASH1 | Sponge network | -1.057 | 0.00029 | -0.502 | 0.00175 | 0.251 |
| 54517 | HOXB-AS2 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-429 | 10 | NAV3 | Sponge network | 1.316 | 0.01106 | 0.212 | 0.47729 | 0.251 |
| 54518 | RP11-6O2.3 |
hsa-miR-1271-5p;hsa-miR-1296-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-589-3p;hsa-miR-96-5p | 15 | RPS6KA6 | Sponge network | 0.331 | 0.57247 | -2.989 | 0 | 0.251 |
| 54519 | RP11-736K20.5 |
hsa-miR-1301-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-590-3p | 11 | FAM135B | Sponge network | -0.358 | 0.17255 | -3.978 | 0 | 0.251 |
| 54520 | PSMG3-AS1 |
hsa-let-7g-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-141-5p;hsa-miR-16-5p;hsa-miR-200a-3p;hsa-miR-200b-5p;hsa-miR-34a-5p;hsa-miR-3614-5p;hsa-miR-378a-5p | 10 | CACNA1E | Sponge network | -0.125 | 0.49414 | 1.752 | 0 | 0.251 |
| 54521 | SNX29P2 |
hsa-miR-18a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-32-5p;hsa-miR-324-3p;hsa-miR-671-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 10 | GATA2 | Sponge network | 0.4 | 0.14611 | -0.654 | 0.0016 | 0.251 |
| 54522 | RP11-359B12.2 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-330-3p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-532-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 24 | NUAK1 | Sponge network | 0.064 | 0.72934 | 0.904 | 0 | 0.251 |
| 54523 | RP11-575A19.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-500b-5p | 11 | SCN9A | Sponge network | 1.969 | 0.00316 | -0.525 | 0.10593 | 0.251 |
| 54524 | NDUFA6-AS1 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-222-5p;hsa-miR-25-3p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-624-5p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-940 | 38 | NOTCH2 | Sponge network | -0.355 | 0.0077 | 0.138 | 0.35163 | 0.251 |
| 54525 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-362-3p;hsa-miR-501-3p | 14 | RAP1A | Sponge network | -1.255 | 0.00981 | -0.929 | 0 | 0.251 |
| 54526 | TP73-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-15b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-222-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-500b-5p;hsa-miR-542-3p | 18 | SLITRK6 | Sponge network | -0.563 | 0.02545 | -2.121 | 0 | 0.251 |
| 54527 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-5p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-362-3p;hsa-miR-93-5p | 22 | PTEN | Sponge network | -1.255 | 0.00981 | -0.187 | 0.07748 | 0.251 |
| 54528 | NEAT1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-5p;hsa-miR-149-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3613-5p;hsa-miR-369-3p;hsa-miR-374a-3p;hsa-miR-409-3p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-493-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-629-3p;hsa-miR-654-3p;hsa-miR-7-1-3p;hsa-miR-940 | 36 | NFIB | Sponge network | 0.705 | 0.00014 | 0.023 | 0.90795 | 0.251 |
| 54529 | CTC-429P9.5 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-425-5p;hsa-miR-454-3p;hsa-miR-590-3p | 11 | CSMD1 | Sponge network | 0.178 | 0.29106 | -0.63 | 0.18881 | 0.251 |
| 54530 | RP5-994D16.9 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-29a-5p;hsa-miR-3614-5p;hsa-miR-511-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 13 | RBMS3 | Sponge network | 0.252 | 0.08508 | -0.519 | 0.03551 | 0.251 |
| 54531 | RP11-175O19.4 |
hsa-let-7a-3p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-342-3p;hsa-miR-374a-3p;hsa-miR-374b-5p | 16 | TMTC3 | Sponge network | 0.917 | 0 | 0.123 | 0.22189 | 0.251 |
| 54532 | HOXA-AS2 |
hsa-let-7g-5p;hsa-miR-10b-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-194-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-375;hsa-miR-590-3p;hsa-miR-7-1-3p | 20 | MLLT10 | Sponge network | 0.61 | 0.01593 | 0.105 | 0.33292 | 0.251 |
| 54533 | TMEM9B-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-141-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-3p;hsa-miR-2355-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-301a-3p;hsa-miR-424-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-766-3p;hsa-miR-940 | 23 | CHD5 | Sponge network | 0.529 | 0.00552 | -0.922 | 0.01521 | 0.251 |
| 54534 | LINC00654 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-20a-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-3614-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-500a-5p;hsa-miR-576-5p;hsa-miR-590-3p | 32 | RICTOR | Sponge network | 0.374 | 0.20054 | 0.388 | 0.00188 | 0.251 |
| 54535 | CTD-2303H24.2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-212-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-624-5p;hsa-miR-7-1-3p | 33 | ZFX | Sponge network | 0.415 | 0.14657 | 0.09 | 0.42563 | 0.251 |
| 54536 | RP1-151F17.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-30c-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-409-3p;hsa-miR-590-3p | 11 | SCN11A | Sponge network | 0.222 | 0.29133 | -1.15 | 0.00299 | 0.251 |
| 54537 | RP4-669P10.16 |
hsa-let-7b-5p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-146b-3p;hsa-miR-148a-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-183-5p;hsa-miR-185-3p;hsa-miR-193a-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-224-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-339-5p;hsa-miR-424-5p;hsa-miR-542-3p;hsa-miR-93-3p | 27 | RUNX1T1 | Sponge network | -0.301 | 0.06597 | -0.344 | 0.2374 | 0.251 |
| 54538 | RP11-403I13.7 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-103a-2-5p;hsa-miR-107;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-18a-3p;hsa-miR-18a-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200c-5p;hsa-miR-22-5p;hsa-miR-223-3p;hsa-miR-26b-5p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-501-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-96-5p | 31 | MEF2C | Sponge network | 0.395 | 0.15451 | -0.499 | 0.01448 | 0.251 |
| 54539 | JAZF1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-212-3p;hsa-miR-335-3p;hsa-miR-338-3p;hsa-miR-33a-3p;hsa-miR-340-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | FGF5 | Sponge network | -0.769 | 0.00035 | 0.549 | 0.28659 | 0.251 |
| 54540 | RP5-1085F17.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-337-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 12 | PBRM1 | Sponge network | 0.541 | 0.00125 | -0.029 | 0.78601 | 0.251 |
| 54541 | RP11-706C16.7 |
hsa-miR-1266-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-27a-3p;hsa-miR-625-5p | 10 | ANK2 | Sponge network | 0.357 | 0.72407 | -1.603 | 1.0E-5 | 0.25 |
| 54542 | PSMG3-AS1 |
hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-21-5p;hsa-miR-29a-5p;hsa-miR-32-5p;hsa-miR-338-3p;hsa-miR-378a-5p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-5p | 16 | PTGFR | Sponge network | -0.125 | 0.49414 | -0.233 | 0.45421 | 0.25 |
| 54543 | ADAMTS9-AS2 |
hsa-miR-107;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-324-5p;hsa-miR-361-5p;hsa-miR-421;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | KLF6 | Sponge network | -2.452 | 0 | -0.479 | 0.00015 | 0.25 |
| 54544 | RP5-1074L1.4 |
hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-149-5p;hsa-miR-150-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-342-3p;hsa-miR-374b-5p;hsa-miR-539-5p | 11 | TCF12 | Sponge network | 1.923 | 0 | 0.174 | 0.13271 | 0.25 |
| 54545 | RP11-5C23.1 |
hsa-let-7g-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-539-5p;hsa-miR-582-5p;hsa-miR-590-3p | 17 | SETBP1 | Sponge network | 0.274 | 0.07681 | -1.302 | 1.0E-5 | 0.25 |
| 54546 | CTD-2303H24.2 |
hsa-let-7d-5p;hsa-let-7i-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-154-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-192-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-369-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-654-3p;hsa-miR-7-1-3p | 20 | DICER1 | Sponge network | 0.415 | 0.14657 | 0.042 | 0.674 | 0.25 |
| 54547 | RP4-639F20.1 |
hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-301a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p;hsa-miR-500a-3p;hsa-miR-942-5p | 20 | TMEM132B | Sponge network | -0.517 | 0.02935 | -0.83 | 0.01084 | 0.25 |
| 54548 | ARHGEF26-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-128-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-199a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-215-5p;hsa-miR-320b;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-421;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | SMARCA2 | Sponge network | -2.46 | 0 | -0.529 | 0.00014 | 0.25 |
| 54549 | RP11-57H14.4 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-484 | 17 | IGF2 | Sponge network | 0.083 | 0.66405 | 0.848 | 0.03842 | 0.25 |
| 54550 | HOXA-AS3 |
hsa-miR-132-3p;hsa-miR-148b-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-34a-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-455-5p;hsa-miR-590-3p | 12 | RTN4 | Sponge network | -0.555 | 0.06156 | -0.412 | 0.00014 | 0.25 |
| 54551 | AC005562.1 |
hsa-miR-130b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-2355-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-374b-5p;hsa-miR-378a-5p;hsa-miR-576-5p;hsa-miR-625-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p;hsa-miR-96-5p | 20 | NUAK1 | Sponge network | 0.488 | 0.00084 | 0.904 | 0 | 0.25 |
| 54552 | MIR143HG |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-194-5p;hsa-miR-33a-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-486-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-1-3p | 16 | CEP170 | Sponge network | -0.739 | 0.0574 | 0.943 | 0 | 0.25 |
| 54553 | ZNF582-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7f-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-25-3p;hsa-miR-26b-5p;hsa-miR-27a-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-769-5p;hsa-miR-93-5p;hsa-miR-940 | 42 | WDFY3 | Sponge network | -1.349 | 0.00017 | -0.201 | 0.0967 | 0.25 |
| 54554 | LINC00641 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-192-5p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-330-5p;hsa-miR-361-3p;hsa-miR-500a-5p;hsa-miR-590-3p;hsa-miR-625-5p;hsa-miR-7-5p | 21 | CRTC1 | Sponge network | -0.222 | 0.32445 | -0.116 | 0.28412 | 0.25 |
| 54555 | RP11-597D13.9 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-96-5p | 34 | PDGFRA | Sponge network | 1.302 | 7.0E-5 | -0.372 | 0.0037 | 0.25 |
| 54556 | RP11-539I5.1 |
hsa-miR-106b-5p;hsa-miR-1266-5p;hsa-miR-141-3p;hsa-miR-17-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-339-5p;hsa-miR-625-5p | 10 | LHX6 | Sponge network | -0.468 | 0.32587 | -0.087 | 0.69193 | 0.25 |
| 54557 | LINC00174 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-155-5p;hsa-miR-16-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30a-3p | 13 | GPAM | Sponge network | 1.253 | 0 | 0.142 | 0.29732 | 0.25 |
| 54558 | RP11-356J5.12 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-107;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-181b-5p;hsa-miR-193a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | CLIP1 | Sponge network | -0.899 | 6.0E-5 | -0.215 | 0.08684 | 0.25 |
| 54559 | AF131215.2 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-484;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | PRKCB | Sponge network | -0.176 | 0.59692 | -1.313 | 2.0E-5 | 0.25 |
| 54560 | RP11-693J15.4 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-455-3p;hsa-miR-660-5p;hsa-miR-7-1-3p | 11 | EPAS1 | Sponge network | -0.72 | 0.24239 | -0.184 | 0.14418 | 0.25 |
| 54561 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-22-5p;hsa-miR-24-3p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-2-5p;hsa-miR-301a-3p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-3p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-342-3p;hsa-miR-345-5p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-374a-5p;hsa-miR-374b-5p;hsa-miR-450b-5p;hsa-miR-493-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-629-5p;hsa-miR-664a-3p;hsa-miR-7-1-3p | 61 | KIF1B | Sponge network | -0.405 | 0.22013 | -0.118 | 0.32258 | 0.25 |
| 54562 | CALML3-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-500a-3p;hsa-miR-503-5p;hsa-miR-505-5p;hsa-miR-542-3p;hsa-miR-625-5p | 29 | TOM1L2 | Sponge network | 0.95 | 0.15943 | -0.994 | 0 | 0.25 |
| 54563 | RP11-307B6.3 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19b-1-5p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-338-3p;hsa-miR-589-3p | 13 | KCNJ5 | Sponge network | -4.463 | 0.00025 | -0.281 | 0.3339 | 0.25 |
| 54564 | TTC28-AS1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-199a-3p;hsa-miR-199b-3p;hsa-miR-26a-2-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-370-3p;hsa-miR-655-3p | 13 | ZMYND11 | Sponge network | 0.373 | 0.00068 | -0.2 | 0.05449 | 0.25 |
| 54565 | PWAR6 |
hsa-miR-18a-3p;hsa-miR-193a-3p;hsa-miR-194-3p;hsa-miR-1976;hsa-miR-29b-2-5p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-378a-5p;hsa-miR-455-3p;hsa-miR-7-5p | 11 | LZTS1 | Sponge network | -1.575 | 6.0E-5 | 1.664 | 0 | 0.25 |
| 54566 | RP11-728F11.4 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-221-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-330-3p;hsa-miR-330-5p;hsa-miR-484;hsa-miR-500a-3p | 23 | DST | Sponge network | 0.238 | 0.70544 | -0.713 | 0.00096 | 0.25 |
| 54567 | RP4-605O3.4 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-183-5p;hsa-miR-191-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-222-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-324-5p;hsa-miR-454-3p;hsa-miR-532-3p;hsa-miR-590-3p | 17 | FOXP1 | Sponge network | 0.262 | 0.0475 | 0.042 | 0.74368 | 0.25 |
| 54568 | HOXB-AS3 |
hsa-miR-106b-5p;hsa-miR-10b-3p;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-18a-5p;hsa-miR-22-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-331-3p;hsa-miR-339-5p;hsa-miR-654-3p | 14 | CTDSPL | Sponge network | 0.343 | 0.28887 | 0.085 | 0.45121 | 0.25 |
| 54569 | RP11-130L8.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-27a-3p;hsa-miR-590-3p | 12 | MITF | Sponge network | -0.663 | 0.10887 | -0.769 | 0.00022 | 0.25 |
| 54570 | CROCCP2 |
hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-195-5p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-2-5p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-424-5p;hsa-miR-497-5p | 12 | BRD3 | Sponge network | 0.79 | 0 | 0.37 | 0.00013 | 0.25 |
| 54571 | RP11-770J1.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-142-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-34a-5p;hsa-miR-505-5p | 17 | AR | Sponge network | -2.011 | 0.0005 | -1.554 | 0 | 0.25 |
| 54572 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-30b-5p;hsa-miR-320b;hsa-miR-33a-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-655-3p | 18 | KANK1 | Sponge network | -0.235 | 0.15142 | -0.55 | 0.00134 | 0.25 |
| 54573 | WDFY3-AS2 |
hsa-miR-16-2-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-193a-3p;hsa-miR-215-5p;hsa-miR-25-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-429;hsa-miR-542-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 18 | ITGB1 | Sponge network | -0.942 | 0 | 0.524 | 0.00017 | 0.25 |
| 54574 | AC005154.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-20a-3p;hsa-miR-27b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-326;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-374b-5p;hsa-miR-497-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 24 | SOX11 | Sponge network | 0.848 | 0 | 2.68 | 0 | 0.25 |
| 54575 | RP5-1198O20.4 |
hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-30e-3p;hsa-miR-339-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-664a-3p | 14 | RASAL2 | Sponge network | 0.997 | 0.00066 | 0.869 | 0 | 0.25 |
| 54576 | CTC-296K1.4 |
hsa-miR-1266-5p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-212-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-30d-3p;hsa-miR-500a-5p | 13 | AFF1 | Sponge network | -2.076 | 0.00548 | -0.384 | 0.00053 | 0.25 |
| 54577 | CALML3-AS1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-33a-3p;hsa-miR-429;hsa-miR-450b-5p | 13 | IGSF10 | Sponge network | 0.95 | 0.15943 | -1.751 | 1.0E-5 | 0.25 |
| 54578 | RP11-2H8.2 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-144-3p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-1976;hsa-miR-200b-5p;hsa-miR-26b-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-486-5p;hsa-miR-590-3p | 16 | PRRX1 | Sponge network | 1.089 | 0 | 1.316 | 0 | 0.25 |
| 54579 | RP11-119F7.5 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-23a-3p;hsa-miR-26a-2-3p;hsa-miR-335-3p;hsa-miR-590-3p | 11 | RBL2 | Sponge network | 0.225 | 0.30644 | -0.282 | 0.00254 | 0.25 |
| 54580 | AP001055.6 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-301a-3p;hsa-miR-7-1-3p | 11 | MED12L | Sponge network | 0.693 | 0.00076 | -0.194 | 0.55299 | 0.25 |
| 54581 | RP11-705C15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-194-5p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-30b-3p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-484;hsa-miR-590-5p;hsa-miR-7-1-3p | 15 | SOX5 | Sponge network | 0.796 | 4.0E-5 | -1.091 | 4.0E-5 | 0.25 |
| 54582 | RP11-967K21.1 |
hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-301a-3p;hsa-miR-338-5p;hsa-miR-375;hsa-miR-429;hsa-miR-450b-5p;hsa-miR-532-5p;hsa-miR-625-5p | 12 | CXCL12 | Sponge network | 0.056 | 0.81195 | -1.421 | 0 | 0.25 |
| 54583 | LINC00957 |
hsa-miR-103a-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-127-5p;hsa-miR-128-3p;hsa-miR-1301-3p;hsa-miR-144-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-22-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-424-5p;hsa-miR-450b-5p;hsa-miR-455-3p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-93-5p | 33 | PIK3R1 | Sponge network | -0.421 | 0.0114 | -0.133 | 0.36854 | 0.25 |
| 54584 | SOX2-OT |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-3p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-141-5p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-15a-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-29a-5p;hsa-miR-30d-5p;hsa-miR-3127-5p;hsa-miR-320b;hsa-miR-362-3p;hsa-miR-378a-3p;hsa-miR-378c;hsa-miR-421;hsa-miR-424-5p;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-501-5p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-93-5p;hsa-miR-96-5p | 37 | TACC1 | Sponge network | -1.264 | 6.0E-5 | -0.362 | 0.05406 | 0.25 |
| 54585 | CKMT2-AS1 |
hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-3127-5p;hsa-miR-361-3p;hsa-miR-423-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-877-5p;hsa-miR-93-3p | 20 | SUFU | Sponge network | 0.082 | 0.55654 | -0.04 | 0.65489 | 0.25 |
| 54586 | SNX29P2 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-21-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-455-3p;hsa-miR-484;hsa-miR-500a-5p;hsa-miR-624-5p;hsa-miR-7-1-3p | 21 | ZFX | Sponge network | 0.4 | 0.14611 | 0.09 | 0.42563 | 0.25 |
| 54587 | RP11-263K19.4 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-146b-3p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-324-5p;hsa-miR-429;hsa-miR-455-5p;hsa-miR-628-5p;hsa-miR-93-3p | 28 | RUNX1T1 | Sponge network | 0.859 | 0.00331 | -0.344 | 0.2374 | 0.25 |
| 54588 | AC108676.1 |
hsa-miR-126-5p;hsa-miR-1271-5p;hsa-miR-142-5p;hsa-miR-154-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-495-3p;hsa-miR-628-5p;hsa-miR-654-3p | 12 | BMPR2 | Sponge network | 4.346 | 2.0E-5 | 0.206 | 0.0635 | 0.25 |
| 54589 | CALML3-AS1 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-21-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-330-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-501-5p;hsa-miR-582-5p | 15 | NOTCH2NL | Sponge network | 0.95 | 0.15943 | 0.024 | 0.84307 | 0.25 |
| 54590 | TRAM2-AS1 |
hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-136-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27b-3p;hsa-miR-28-5p;hsa-miR-299-5p;hsa-miR-374b-5p;hsa-miR-495-3p;hsa-miR-590-3p | 19 | CREB1 | Sponge network | 0.336 | 0.00739 | 0.203 | 0.00537 | 0.25 |
| 54591 | CROCCP2 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-324-5p;hsa-miR-326;hsa-miR-369-3p;hsa-miR-424-5p;hsa-miR-497-5p | 10 | PRKAB2 | Sponge network | 0.79 | 0 | -0.367 | 0.01371 | 0.25 |
| 54592 | HHIP-AS1 |
hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-182-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-223-5p;hsa-miR-224-5p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-92b-3p | 27 | GNAQ | Sponge network | -0.737 | 0.10822 | -0.367 | 0.00102 | 0.25 |
| 54593 | AC003090.1 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-744-3p | 12 | IGF2 | Sponge network | -2.117 | 0.00161 | 0.848 | 0.03842 | 0.25 |
| 54594 | RP11-434B12.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-10b-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-335-5p;hsa-miR-361-3p;hsa-miR-421;hsa-miR-429;hsa-miR-590-3p | 16 | BCL6 | Sponge network | -0.031 | 0.89063 | -0.211 | 0.19489 | 0.25 |
| 54595 | AC025171.1 |
hsa-miR-126-5p;hsa-miR-1266-5p;hsa-miR-127-5p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-20a-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-30d-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-425-5p;hsa-miR-455-3p;hsa-miR-486-5p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-942-5p | 31 | THRB | Sponge network | 0.998 | 0 | -1.117 | 0.0004 | 0.25 |
| 54596 | CTC-429P9.1 |
hsa-miR-103a-2-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-16-1-3p;hsa-miR-181b-5p;hsa-miR-196b-5p;hsa-miR-19b-1-5p;hsa-miR-221-3p;hsa-miR-27a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-511-5p | 14 | NRXN1 | Sponge network | 0.497 | 0.01451 | -3.778 | 0 | 0.25 |
| 54597 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-20a-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-3614-5p;hsa-miR-455-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-5p;hsa-miR-628-5p;hsa-miR-629-5p;hsa-miR-7-1-3p;hsa-miR-7-5p;hsa-miR-96-5p | 49 | TCF4 | Sponge network | 0.41 | 0.02214 | 0.016 | 0.92271 | 0.25 |
| 54598 | RP11-345P4.7 |
hsa-let-7a-3p;hsa-miR-129-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-27b-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-326;hsa-miR-33a-5p;hsa-miR-497-5p;hsa-miR-664a-5p | 13 | SOX11 | Sponge network | 1.597 | 0 | 2.68 | 0 | 0.25 |
| 54599 | BCYRN1 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-25-3p;hsa-miR-26a-2-3p;hsa-miR-30b-5p;hsa-miR-30d-3p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-577;hsa-miR-582-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-92a-1-5p | 22 | LUZP2 | Sponge network | 0.311 | 0.10344 | -0.056 | 0.89416 | 0.25 |
| 54600 | RP11-1006G14.4 |
hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-30d-3p;hsa-miR-539-5p;hsa-miR-576-5p;hsa-miR-590-3p | 16 | SYNPO2 | Sponge network | 0.559 | 0.00026 | -2.342 | 0 | 0.25 |
| 54601 | RP11-678G14.3 |
hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-21-5p;hsa-miR-421;hsa-miR-500a-3p;hsa-miR-7-1-3p | 13 | LIMCH1 | Sponge network | -4.477 | 0 | -0.915 | 1.0E-5 | 0.25 |
| 54602 | RP11-359E10.1 |
hsa-miR-142-3p;hsa-miR-148b-5p;hsa-miR-192-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-625-5p | 10 | PPP2R2C | Sponge network | 0.299 | 0.57722 | -0.959 | 0.07428 | 0.25 |
| 54603 | LINC00667 |
hsa-miR-106b-5p;hsa-miR-140-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-1-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-20a-5p;hsa-miR-29a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-671-5p;hsa-miR-7-1-3p;hsa-miR-93-5p | 17 | FAT4 | Sponge network | -0.235 | 0.15142 | -0.354 | 0.14694 | 0.25 |
| 54604 | RP11-212P7.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-142-3p;hsa-miR-146b-5p;hsa-miR-155-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-3614-5p;hsa-miR-362-3p;hsa-miR-455-5p;hsa-miR-505-5p;hsa-miR-590-3p;hsa-miR-93-5p;hsa-miR-96-5p | 29 | HBP1 | Sponge network | 0.219 | 0.1516 | -0.454 | 0 | 0.25 |
| 54605 | TRAF3IP2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-107;hsa-miR-141-3p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-590-3p | 11 | PCDH8 | Sponge network | -0.323 | 0.01372 | -0.997 | 0.05366 | 0.25 |
| 54606 | KCNJ2-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7i-5p;hsa-miR-132-3p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-196a-5p;hsa-miR-196b-5p;hsa-miR-1976;hsa-miR-19b-1-5p;hsa-miR-21-5p;hsa-miR-22-3p;hsa-miR-29a-5p;hsa-miR-324-3p;hsa-miR-378a-5p;hsa-miR-382-5p;hsa-miR-576-5p | 20 | PBX1 | Sponge network | -0.436 | 0.05325 | -1.206 | 0 | 0.25 |
| 54607 | RP11-479G22.8 |
hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-155-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-1976;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-200c-5p;hsa-miR-26a-2-3p;hsa-miR-342-5p;hsa-miR-429;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 15 | CUX1 | Sponge network | 1.308 | 0 | -0.039 | 0.6705 | 0.25 |
| 54608 | RP11-720L2.4 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-10b-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-16-1-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-3p;hsa-miR-33a-3p | 14 | TMEM132B | Sponge network | -1.255 | 0.00981 | -0.83 | 0.01084 | 0.25 |
| 54609 | LINC00672 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-18a-5p;hsa-miR-1976;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-30e-3p;hsa-miR-539-5p;hsa-miR-590-3p | 10 | DDX5 | Sponge network | -0.227 | 0.31657 | -0.099 | 0.17428 | 0.25 |
| 54610 | H1FX-AS1 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-335-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-629-5p;hsa-miR-93-5p | 22 | MAF | Sponge network | -0.206 | 0.12942 | -1.257 | 0 | 0.25 |
| 54611 | AC004540.4 |
hsa-let-7g-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-146a-5p;hsa-miR-15b-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-1976;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-33a-5p;hsa-miR-362-3p;hsa-miR-484;hsa-miR-629-3p | 18 | CBX5 | Sponge network | -2.549 | 0.00528 | 0.165 | 0.24446 | 0.25 |
| 54612 | AC159540.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-139-5p;hsa-miR-193b-3p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-369-3p;hsa-miR-505-3p | 10 | PTPN11 | Sponge network | 1.122 | 0 | 0.431 | 2.0E-5 | 0.25 |
| 54613 | RP11-597D13.9 |
hsa-let-7g-3p;hsa-miR-106b-5p;hsa-miR-1271-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-20a-5p;hsa-miR-223-3p;hsa-miR-301a-3p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-92a-3p;hsa-miR-96-5p | 25 | LRIG1 | Sponge network | 1.302 | 7.0E-5 | -0.537 | 0.00588 | 0.25 |
| 54614 | RP11-248G5.8 |
hsa-miR-125a-3p;hsa-miR-127-5p;hsa-miR-129-5p;hsa-miR-130a-3p;hsa-miR-142-3p;hsa-miR-148a-3p;hsa-miR-149-5p;hsa-miR-152-3p;hsa-miR-154-5p;hsa-miR-195-3p;hsa-miR-195-5p;hsa-miR-218-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-342-3p;hsa-miR-3607-3p;hsa-miR-424-5p;hsa-miR-495-3p;hsa-miR-497-5p;hsa-miR-664a-3p | 28 | CCDC6 | Sponge network | 1.294 | 0 | 0.27 | 0.02555 | 0.25 |
| 54615 | AC074138.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-200c-3p;hsa-miR-27a-5p;hsa-miR-29b-3p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-577;hsa-miR-744-3p | 11 | HLF | Sponge network | -0.127 | 0.52761 | -2.443 | 0 | 0.25 |
| 54616 | CTA-204B4.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-190a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-454-3p;hsa-miR-455-3p;hsa-miR-590-3p | 37 | TCF4 | Sponge network | 0.765 | 7.0E-5 | 0.016 | 0.92271 | 0.25 |
| 54617 | RP1-266L20.2 |
hsa-let-7g-3p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-29b-2-5p;hsa-miR-30a-3p | 10 | ABL2 | Sponge network | 0.474 | 0.00944 | 0.82 | 0 | 0.25 |
| 54618 | RP11-277P12.20 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130b-3p;hsa-miR-15b-3p;hsa-miR-186-5p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p | 12 | IGF1 | Sponge network | 0.687 | 0.02753 | -0.204 | 0.5898 | 0.25 |
| 54619 | DNM3OS |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-424-5p;hsa-miR-532-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | DMTF1 | Sponge network | 0.572 | 0.01722 | 0.248 | 0.02726 | 0.25 |
| 54620 | RP11-53O19.1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-140-5p;hsa-miR-148a-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-215-5p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-338-5p;hsa-miR-374b-5p;hsa-miR-375;hsa-miR-378a-5p | 17 | ARHGAP29 | Sponge network | -0.405 | 0.22013 | 0.131 | 0.42953 | 0.25 |
| 54621 | NNT-AS1 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-139-3p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-145-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-195-3p;hsa-miR-30a-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-326;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-590-3p | 20 | SOX11 | Sponge network | 0.799 | 1.0E-5 | 2.68 | 0 | 0.25 |
| 54622 | RP11-403I13.8 |
hsa-let-7d-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-144-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-215-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-30b-5p;hsa-miR-30e-3p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-7-1-3p | 40 | MDM4 | Sponge network | 0.619 | 0.00157 | 0.345 | 0.00762 | 0.25 |
| 54623 | RP11-6O2.3 |
hsa-miR-107;hsa-miR-1296-5p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-25-3p;hsa-miR-320b;hsa-miR-324-3p;hsa-miR-590-5p;hsa-miR-96-5p | 13 | PRKAR1A | Sponge network | 0.331 | 0.57247 | -0.063 | 0.50314 | 0.25 |
| 54624 | CTD-2303H24.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-150-5p;hsa-miR-301a-3p;hsa-miR-30e-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-450b-5p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 18 | TRIP11 | Sponge network | 0.415 | 0.14657 | -0.158 | 0.06384 | 0.25 |
| 54625 | RP4-791M13.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-18a-5p;hsa-miR-197-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-362-3p;hsa-miR-651-5p | 12 | PRDM2 | Sponge network | 0.419 | 0.0319 | -0.258 | 0.00721 | 0.25 |
| 54626 | LINC00476 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-16-5p;hsa-miR-33a-3p;hsa-miR-484;hsa-miR-590-3p | 10 | TBRG1 | Sponge network | -0.615 | 0.00015 | -0.071 | 0.37324 | 0.25 |
| 54627 | LINC00092 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-1-3p;hsa-miR-17-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-1-5p;hsa-miR-200c-5p;hsa-miR-20a-3p;hsa-miR-22-5p;hsa-miR-224-5p;hsa-miR-301a-3p;hsa-miR-3913-5p;hsa-miR-589-3p;hsa-miR-590-3p | 20 | AFF1 | Sponge network | -1.886 | 0 | -0.384 | 0.00053 | 0.25 |
| 54628 | RP11-119F7.5 |
hsa-let-7g-3p;hsa-miR-135b-5p;hsa-miR-142-5p;hsa-miR-185-3p;hsa-miR-185-5p;hsa-miR-192-3p;hsa-miR-24-3p;hsa-miR-30b-3p;hsa-miR-324-3p;hsa-miR-330-3p;hsa-miR-532-3p | 11 | CADM3 | Sponge network | 0.225 | 0.30644 | -3.663 | 0 | 0.25 |
| 54629 | SNHG14 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-210-3p;hsa-miR-23a-5p;hsa-miR-342-5p;hsa-miR-34a-5p;hsa-miR-361-3p;hsa-miR-362-3p;hsa-miR-454-3p;hsa-miR-484;hsa-miR-744-3p;hsa-miR-744-5p | 19 | IGF2 | Sponge network | -1.111 | 0.0003 | 0.848 | 0.03842 | 0.25 |
| 54630 | RP11-983P16.4 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-146b-5p;hsa-miR-16-1-3p;hsa-miR-29b-1-5p;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-342-3p;hsa-miR-362-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | NOTCH2NL | Sponge network | 0.41 | 0.02214 | 0.024 | 0.84307 | 0.25 |
| 54631 | RP5-1085F17.3 |
hsa-miR-130b-3p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-192-3p;hsa-miR-192-5p;hsa-miR-193b-3p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-20a-3p;hsa-miR-26b-5p;hsa-miR-29a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-29b-3p;hsa-miR-33a-5p;hsa-miR-342-3p;hsa-miR-34a-5p;hsa-miR-3607-3p;hsa-miR-425-5p;hsa-miR-590-3p;hsa-miR-664a-5p | 24 | ADARB1 | Sponge network | 0.541 | 0.00125 | -0.335 | 0.06631 | 0.25 |
| 54632 | RP11-216B9.6 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-5p | 17 | PRKCB | Sponge network | 0.174 | 0.30034 | -1.313 | 2.0E-5 | 0.25 |
| 54633 | RP11-531A24.5 |
hsa-miR-132-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-200a-3p;hsa-miR-215-5p;hsa-miR-24-2-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-429;hsa-miR-455-3p;hsa-miR-7-1-3p | 22 | PCDH9 | Sponge network | 0.75 | 0.00054 | -1.823 | 4.0E-5 | 0.25 |
| 54634 | HAS2-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-5p;hsa-miR-18a-3p;hsa-miR-190a-5p;hsa-miR-194-5p;hsa-miR-222-3p;hsa-miR-362-5p;hsa-miR-532-3p;hsa-miR-629-3p;hsa-miR-7-1-3p | 11 | TRPS1 | Sponge network | -0.011 | 0.967 | -0.191 | 0.44109 | 0.25 |
| 54635 | PART1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-134-5p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-144-3p;hsa-miR-16-2-3p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-24-3p;hsa-miR-29b-1-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-331-5p;hsa-miR-335-5p;hsa-miR-33a-3p;hsa-miR-361-3p;hsa-miR-450b-5p;hsa-miR-501-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p | 26 | KAT6B | Sponge network | -2.925 | 0 | -0.478 | 0.00018 | 0.25 |
| 54636 | C4A-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-182-5p;hsa-miR-19b-1-5p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-589-3p;hsa-miR-590-5p;hsa-miR-93-3p;hsa-miR-96-5p | 10 | UNC5C | Sponge network | 3.345 | 0 | -0.178 | 0.40214 | 0.25 |
| 54637 | MEG3 |
hsa-let-7g-3p;hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-148b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-200c-5p;hsa-miR-20a-5p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-223-3p;hsa-miR-24-2-5p;hsa-miR-25-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-330-5p;hsa-miR-331-3p;hsa-miR-33a-3p;hsa-miR-362-5p;hsa-miR-425-5p;hsa-miR-501-5p;hsa-miR-532-3p;hsa-miR-550a-3p;hsa-miR-576-5p;hsa-miR-590-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-744-3p;hsa-miR-935;hsa-miR-940 | 34 | PTPRT | Sponge network | 0.249 | 0.35902 | -0.907 | 0.03727 | 0.25 |
| 54638 | RP11-927P21.1 |
hsa-miR-106b-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-320b;hsa-miR-375;hsa-miR-424-5p;hsa-miR-577;hsa-miR-582-5p | 13 | DOCK4 | Sponge network | 0.921 | 0.00118 | 0.502 | 0.00108 | 0.25 |
| 54639 | RP11-736K20.5 |
hsa-miR-107;hsa-miR-1266-5p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-27a-3p;hsa-miR-330-5p | 11 | CSRNP1 | Sponge network | -0.358 | 0.17255 | -1.448 | 0 | 0.25 |
| 54640 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15b-3p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-25-3p;hsa-miR-29a-5p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-409-3p;hsa-miR-93-5p | 25 | SSBP2 | Sponge network | 0.497 | 0.01451 | -1.58 | 0 | 0.25 |
| 54641 | AC093110.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-5p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-320b;hsa-miR-326;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-493-5p | 17 | USP6 | Sponge network | 1.044 | 2.0E-5 | 0.188 | 0.3871 | 0.25 |
| 54642 | MIR600HG |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-135b-5p;hsa-miR-142-3p;hsa-miR-155-5p;hsa-miR-200c-3p;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-502-3p;hsa-miR-629-3p | 12 | PSIP1 | Sponge network | 0.594 | 0.41522 | -0.005 | 0.96897 | 0.25 |
| 54643 | HHIP-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-103a-3p;hsa-miR-107;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-16-1-3p;hsa-miR-17-3p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-330-5p;hsa-miR-33a-5p;hsa-miR-455-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 23 | EBF1 | Sponge network | -0.737 | 0.10822 | -0.384 | 0.08856 | 0.25 |
| 54644 | AC018890.6 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-338-5p;hsa-miR-33a-3p;hsa-miR-511-5p | 12 | TRIM33 | Sponge network | 1.61 | 0.00961 | 0.402 | 7.0E-5 | 0.25 |
| 54645 | RP11-53O19.1 |
hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-223-3p;hsa-miR-223-5p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-338-5p;hsa-miR-362-3p;hsa-miR-409-3p;hsa-miR-501-5p;hsa-miR-539-5p;hsa-miR-7-1-3p | 13 | GABRB2 | Sponge network | -0.405 | 0.22013 | -1.841 | 0.00029 | 0.25 |
| 54646 | RP11-403I13.7 |
hsa-miR-103a-3p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-194-3p;hsa-miR-194-5p;hsa-miR-301a-3p;hsa-miR-361-3p;hsa-miR-378a-5p;hsa-miR-423-5p;hsa-miR-589-5p;hsa-miR-625-5p;hsa-miR-7-1-3p;hsa-miR-877-5p;hsa-miR-93-3p | 19 | SUFU | Sponge network | 0.395 | 0.15451 | -0.04 | 0.65489 | 0.25 |
| 54647 | RP11-554A11.4 |
hsa-let-7g-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-186-5p;hsa-miR-192-5p;hsa-miR-20a-3p;hsa-miR-2110;hsa-miR-215-5p;hsa-miR-23a-3p;hsa-miR-33a-3p;hsa-miR-450b-5p | 19 | AKAP11 | Sponge network | -0.583 | 0.14589 | 0.196 | 0.09825 | 0.25 |
| 54648 | LINC00578 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-197-3p;hsa-miR-200b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-501-5p | 16 | CYLD | Sponge network | 0.811 | 0.03228 | -0.367 | 0.00171 | 0.25 |
| 54649 | HNRNPU-AS1 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-144-3p;hsa-miR-186-5p;hsa-miR-193b-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-323a-3p;hsa-miR-429;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 14 | GNA13 | Sponge network | 1.601 | 0 | 0.007 | 0.93884 | 0.25 |
| 54650 | LINC00648 |
hsa-miR-140-5p;hsa-miR-141-3p;hsa-miR-155-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-320a;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 14 | DYNC1I1 | Sponge network | 0.633 | 0.51678 | -1.12 | 0.00107 | 0.25 |
| 54651 | USP3-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-125a-3p;hsa-miR-140-5p;hsa-miR-141-5p;hsa-miR-151a-3p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-193a-3p;hsa-miR-320b;hsa-miR-505-5p;hsa-miR-589-5p | 13 | RBM5 | Sponge network | -0.162 | 0.47687 | -0.205 | 0.0129 | 0.25 |
| 54652 | RP11-57H14.4 |
hsa-miR-107;hsa-miR-142-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-224-5p;hsa-miR-27a-3p;hsa-miR-27a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-455-3p;hsa-miR-590-3p;hsa-miR-935 | 15 | RELN | Sponge network | 0.083 | 0.66405 | -2.403 | 0 | 0.25 |
| 54653 | RP11-356J5.12 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-20a-5p;hsa-miR-23a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-362-3p;hsa-miR-455-3p;hsa-miR-508-3p;hsa-miR-590-3p | 28 | MAF | Sponge network | -0.899 | 6.0E-5 | -1.257 | 0 | 0.25 |
| 54654 | RP11-620J15.3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-148a-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-20a-5p;hsa-miR-21-3p;hsa-miR-222-5p;hsa-miR-30d-3p;hsa-miR-361-5p;hsa-miR-589-3p | 12 | EPAS1 | Sponge network | -0.739 | 0.00044 | -0.184 | 0.14418 | 0.25 |
| 54655 | RP11-890B15.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-10b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-142-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-193b-3p;hsa-miR-200a-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-654-3p | 22 | MYBL1 | Sponge network | 0.101 | 0.60919 | 0.494 | 0.02152 | 0.25 |
| 54656 | LINC00094 |
hsa-let-7a-3p;hsa-let-7f-1-3p;hsa-miR-142-3p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-338-3p;hsa-miR-340-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-500a-3p;hsa-miR-501-3p;hsa-miR-501-5p;hsa-miR-502-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-7-5p | 22 | RNF144A | Sponge network | 0.253 | 0.09565 | 0.594 | 0.0009 | 0.25 |
| 54657 | RP11-35G9.3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-15b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-188-5p;hsa-miR-200c-3p;hsa-miR-30b-3p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-33a-3p;hsa-miR-424-5p;hsa-miR-429;hsa-miR-590-3p | 18 | LATS1 | Sponge network | 0.657 | 4.0E-5 | 0.109 | 0.34208 | 0.25 |
| 54658 | LINC00667 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-493-5p;hsa-miR-590-3p;hsa-miR-655-3p;hsa-miR-7-1-3p;hsa-miR-93-3p | 27 | NCOA1 | Sponge network | -0.235 | 0.15142 | -0.432 | 0 | 0.25 |
| 54659 | AF131215.9 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-5p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-200a-5p;hsa-miR-200c-5p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-450b-5p;hsa-miR-485-3p;hsa-miR-576-5p;hsa-miR-589-3p;hsa-miR-590-3p | 19 | HECA | Sponge network | -0.322 | 0.25945 | -0.217 | 0.03463 | 0.25 |
| 54660 | CTC-429P9.1 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-301a-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | 0.497 | 0.01451 | -0.243 | 0.11408 | 0.25 |
| 54661 | RP11-263K19.4 |
hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-150-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-192-3p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-3p;hsa-miR-26b-3p;hsa-miR-429;hsa-miR-766-3p | 18 | IL17RD | Sponge network | 0.859 | 0.00331 | -0.019 | 0.94873 | 0.25 |
| 54662 | AGAP11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-148a-5p;hsa-miR-155-5p;hsa-miR-182-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-23a-3p;hsa-miR-24-3p;hsa-miR-27a-3p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-501-5p;hsa-miR-629-3p;hsa-miR-93-3p | 21 | EBF3 | Sponge network | -1.645 | 0 | -0.058 | 0.81437 | 0.25 |
| 54663 | BZRAP1-AS1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-103a-2-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-154-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-193b-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200b-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-323a-3p;hsa-miR-335-3p;hsa-miR-33a-5p;hsa-miR-375;hsa-miR-378a-5p;hsa-miR-424-3p;hsa-miR-454-3p;hsa-miR-455-5p;hsa-miR-532-5p;hsa-miR-577;hsa-miR-582-3p;hsa-miR-671-5p;hsa-miR-7-5p;hsa-miR-877-5p;hsa-miR-93-3p | 44 | BMPR2 | Sponge network | -0.465 | 0.04703 | 0.206 | 0.0635 | 0.25 |
| 54664 | ZSCAN16-AS1 |
hsa-miR-142-5p;hsa-miR-17-5p;hsa-miR-188-5p;hsa-miR-192-5p;hsa-miR-197-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-215-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-369-3p;hsa-miR-576-5p;hsa-miR-582-5p;hsa-miR-590-3p | 16 | KLF10 | Sponge network | -0.342 | 0.02288 | -0.783 | 0 | 0.25 |
| 54665 | MALAT1 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-106b-5p;hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-130b-3p;hsa-miR-136-5p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-193b-3p;hsa-miR-212-3p;hsa-miR-222-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29b-3p;hsa-miR-30b-5p;hsa-miR-30e-5p;hsa-miR-32-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-323a-3p;hsa-miR-337-3p;hsa-miR-374b-5p;hsa-miR-429;hsa-miR-484;hsa-miR-493-5p;hsa-miR-495-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p;hsa-miR-744-3p | 39 | PTEN | Sponge network | 0.782 | 0.00016 | -0.187 | 0.07748 | 0.25 |
| 54666 | RP11-307B6.3 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-miR-130b-5p;hsa-miR-141-3p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-3p;hsa-miR-200a-3p;hsa-miR-26b-5p;hsa-miR-424-5p | 11 | ARNT2 | Sponge network | -4.463 | 0.00025 | -0.332 | 0.25851 | 0.25 |
| 54667 | CTD-2554C21.2 |
hsa-miR-16-2-3p;hsa-miR-17-3p;hsa-miR-21-5p;hsa-miR-212-3p;hsa-miR-27b-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-320b;hsa-miR-326;hsa-miR-421;hsa-miR-429 | 11 | RIT1 | Sponge network | -1.654 | 0.00144 | -0.296 | 0.00054 | 0.25 |
| 54668 | AP001347.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7g-5p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-135b-5p;hsa-miR-141-3p;hsa-miR-142-5p;hsa-miR-144-3p;hsa-miR-146a-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-192-3p;hsa-miR-194-5p;hsa-miR-1976;hsa-miR-200a-3p;hsa-miR-200b-3p;hsa-miR-21-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-340-5p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-7-1-3p | 39 | CBX5 | Sponge network | -1.161 | 0.00591 | 0.165 | 0.24446 | 0.25 |
| 54669 | RP11-216B9.6 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-193b-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-22-5p;hsa-miR-223-5p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-369-3p;hsa-miR-450b-5p;hsa-miR-539-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-625-3p;hsa-miR-940 | 23 | ETV1 | Sponge network | 0.174 | 0.30034 | -0.096 | 0.67674 | 0.25 |
| 54670 | HCG18 |
hsa-miR-125a-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-30c-5p;hsa-miR-32-5p;hsa-miR-339-5p;hsa-miR-3607-3p;hsa-miR-374b-5p;hsa-miR-582-5p;hsa-miR-590-3p | 10 | RANBP9 | Sponge network | 1.063 | 0 | -0.191 | 0.04628 | 0.25 |
| 54671 | MEG3 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-142-3p;hsa-miR-142-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-221-3p;hsa-miR-222-3p;hsa-miR-27a-3p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-320b;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-421;hsa-miR-429;hsa-miR-500a-5p;hsa-miR-577;hsa-miR-590-5p;hsa-miR-629-3p;hsa-miR-7-1-3p;hsa-miR-935;hsa-miR-940;hsa-miR-96-5p;hsa-miR-98-5p | 30 | LIFR | Sponge network | 0.249 | 0.35902 | -1.794 | 0 | 0.25 |
| 54672 | FAM13A-AS1 |
hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-3p;hsa-let-7i-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-141-3p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-181b-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-200a-5p;hsa-miR-200c-3p;hsa-miR-20a-5p;hsa-miR-24-3p;hsa-miR-335-3p;hsa-miR-361-5p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-671-5p | 26 | DLC1 | Sponge network | -0.83 | 0 | -0.382 | 0.05038 | 0.25 |
| 54673 | RP11-6O2.3 |
hsa-let-7a-3p;hsa-miR-126-5p;hsa-miR-142-5p;hsa-miR-15b-3p;hsa-miR-192-3p;hsa-miR-212-3p;hsa-miR-24-3p;hsa-miR-26a-5p;hsa-miR-26b-3p;hsa-miR-26b-5p;hsa-miR-299-5p;hsa-miR-326;hsa-miR-337-3p;hsa-miR-495-3p;hsa-miR-511-5p;hsa-miR-940 | 16 | TRIM33 | Sponge network | 0.331 | 0.57247 | 0.402 | 7.0E-5 | 0.25 |
| 54674 | AP000487.6 |
hsa-miR-129-5p;hsa-miR-142-3p;hsa-miR-149-5p;hsa-miR-154-5p;hsa-miR-218-5p;hsa-miR-26b-5p;hsa-miR-30a-3p;hsa-miR-30a-5p;hsa-miR-30e-3p;hsa-miR-33a-3p;hsa-miR-3607-3p | 11 | CCDC6 | Sponge network | 2.208 | 0 | 0.27 | 0.02555 | 0.25 |
| 54675 | RP11-359E10.1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-103a-2-5p;hsa-miR-130b-3p;hsa-miR-140-5p;hsa-miR-146b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-186-5p;hsa-miR-18a-5p;hsa-miR-222-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-26b-3p;hsa-miR-27a-3p;hsa-miR-299-5p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-330-5p;hsa-miR-7-1-3p | 21 | NCOA1 | Sponge network | 0.299 | 0.57722 | -0.432 | 0 | 0.25 |
| 54676 | BVES-AS1 |
hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-15b-3p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-200b-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-429 | 14 | CYLD | Sponge network | -2.62 | 0 | -0.367 | 0.00171 | 0.25 |
| 54677 | RP11-181G12.2 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-141-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-19b-3p;hsa-miR-22-3p;hsa-miR-223-3p;hsa-miR-2355-3p;hsa-miR-29a-5p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-424-5p;hsa-miR-505-5p | 22 | AR | Sponge network | 0.556 | 0.00298 | -1.554 | 0 | 0.25 |
| 54678 | C4A-AS1 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-140-5p;hsa-miR-146a-5p;hsa-miR-17-5p;hsa-miR-182-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-219a-1-3p;hsa-miR-23a-3p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-96-5p | 13 | JAZF1 | Sponge network | 3.345 | 0 | -0.377 | 0.02677 | 0.25 |
| 54679 | RP11-575H3.1 |
hsa-let-7g-3p;hsa-miR-126-5p;hsa-miR-130b-3p;hsa-miR-142-3p;hsa-miR-194-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-590-3p;hsa-miR-7-1-3p | 12 | TRPS1 | Sponge network | -0.246 | 0.74154 | -0.191 | 0.44109 | 0.25 |
| 54680 | RP4-669L17.10 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-126-5p;hsa-miR-142-3p;hsa-miR-148a-5p;hsa-miR-16-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-29c-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-495-3p | 12 | PRKAB2 | Sponge network | 1.093 | 0 | -0.367 | 0.01371 | 0.25 |
| 54681 | PCBP1-AS1 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-136-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-188-5p;hsa-miR-197-3p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-362-5p;hsa-miR-500a-5p;hsa-miR-590-5p;hsa-miR-671-5p | 13 | CYLD | Sponge network | -0.567 | 0 | -0.367 | 0.00171 | 0.25 |
| 54682 | HCG11 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7f-1-3p;hsa-miR-142-5p;hsa-miR-16-2-3p;hsa-miR-330-3p;hsa-miR-338-5p;hsa-miR-3607-3p;hsa-miR-450b-5p;hsa-miR-590-3p;hsa-miR-7-1-3p | 11 | PDGFC | Sponge network | 0.385 | 0.10067 | -0.33 | 0.06483 | 0.25 |
| 54683 | TAPT1-AS1 |
hsa-miR-125a-3p;hsa-miR-130a-3p;hsa-miR-143-5p;hsa-miR-150-5p;hsa-miR-152-3p;hsa-miR-16-5p;hsa-miR-195-5p;hsa-miR-22-5p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27b-3p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-337-3p;hsa-miR-33a-3p;hsa-miR-495-3p;hsa-miR-766-3p | 19 | MDM4 | Sponge network | 0.403 | 2.0E-5 | 0.345 | 0.00762 | 0.25 |
| 54684 | RP11-554A11.4 |
hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-130b-5p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-181a-5p;hsa-miR-181b-5p;hsa-miR-20a-3p;hsa-miR-23a-3p;hsa-miR-29a-5p;hsa-miR-320b | 11 | KANK1 | Sponge network | -0.583 | 0.14589 | -0.55 | 0.00134 | 0.25 |
| 54685 | MIR143HG |
hsa-let-7a-3p;hsa-miR-148a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-330-3p;hsa-miR-33a-3p;hsa-miR-532-3p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-942-5p | 10 | CD44 | Sponge network | -0.739 | 0.0574 | 0.223 | 0.30295 | 0.25 |
| 54686 | RP11-597D13.9 |
hsa-miR-1266-5p;hsa-miR-130b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-185-5p;hsa-miR-19a-3p;hsa-miR-19b-1-5p;hsa-miR-19b-3p;hsa-miR-200b-3p;hsa-miR-222-5p;hsa-miR-29a-5p;hsa-miR-29b-3p;hsa-miR-301a-3p;hsa-miR-30d-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-338-3p;hsa-miR-454-3p;hsa-miR-505-3p;hsa-miR-590-5p;hsa-miR-7-5p | 22 | KCNJ5 | Sponge network | 1.302 | 7.0E-5 | -0.281 | 0.3339 | 0.25 |
| 54687 | RP11-283I3.6 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-let-7d-5p;hsa-let-7f-1-3p;hsa-let-7g-5p;hsa-miR-141-5p;hsa-miR-144-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-192-3p;hsa-miR-200a-5p;hsa-miR-200b-5p;hsa-miR-26a-2-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-320a;hsa-miR-320b;hsa-miR-33a-3p;hsa-miR-33a-5p;hsa-miR-3607-3p;hsa-miR-369-3p;hsa-miR-374b-5p;hsa-miR-455-3p;hsa-miR-493-5p;hsa-miR-96-5p | 27 | FBXO32 | Sponge network | 0.614 | 1.0E-5 | -0.495 | 0.07561 | 0.25 |
| 54688 | RP11-193H5.1 |
hsa-miR-101-5p;hsa-miR-125a-5p;hsa-miR-130a-3p;hsa-miR-145-5p;hsa-miR-195-3p;hsa-miR-26a-5p;hsa-miR-26b-5p;hsa-miR-28-5p;hsa-miR-30a-3p;hsa-miR-30e-3p;hsa-miR-326;hsa-miR-664a-5p | 12 | CBFB | Sponge network | 1.279 | 2.0E-5 | 1.061 | 0 | 0.25 |
| 54689 | FAM66C |
hsa-let-7a-3p;hsa-miR-141-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-193b-5p;hsa-miR-19b-1-5p;hsa-miR-200a-3p;hsa-miR-200c-5p;hsa-miR-2110;hsa-miR-222-5p;hsa-miR-24-3p;hsa-miR-30d-3p;hsa-miR-33a-3p;hsa-miR-3607-3p;hsa-miR-429;hsa-miR-576-5p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-942-5p | 20 | PCDH17 | Sponge network | -0.901 | 0.00019 | 0.731 | 2.0E-5 | 0.25 |
| 54690 | BZRAP1-AS1 |
hsa-let-7d-5p;hsa-miR-125a-3p;hsa-miR-126-5p;hsa-miR-127-5p;hsa-miR-130b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-15b-3p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-182-5p;hsa-miR-183-5p;hsa-miR-186-5p;hsa-miR-191-5p;hsa-miR-200c-3p;hsa-miR-20a-3p;hsa-miR-21-3p;hsa-miR-22-5p;hsa-miR-222-5p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-30b-3p;hsa-miR-323a-3p;hsa-miR-326;hsa-miR-330-5p;hsa-miR-339-5p;hsa-miR-33a-5p;hsa-miR-34a-5p;hsa-miR-424-5p;hsa-miR-454-3p;hsa-miR-577;hsa-miR-625-5p;hsa-miR-671-5p;hsa-miR-940 | 36 | MDM4 | Sponge network | -0.465 | 0.04703 | 0.345 | 0.00762 | 0.25 |
| 54691 | CTD-2647L4.4 |
hsa-let-7a-3p;hsa-miR-125a-3p;hsa-miR-142-5p;hsa-miR-148a-3p;hsa-miR-155-5p;hsa-miR-16-2-3p;hsa-miR-26a-2-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-362-3p;hsa-miR-369-3p;hsa-miR-376c-3p;hsa-miR-590-5p | 13 | IL6ST | Sponge network | 0.555 | 0.00197 | -0.402 | 0.00676 | 0.25 |
| 54692 | MIR4458HG |
hsa-miR-106b-3p;hsa-miR-140-5p;hsa-miR-142-5p;hsa-miR-148b-3p;hsa-miR-182-5p;hsa-miR-22-3p;hsa-miR-27a-5p;hsa-miR-331-3p;hsa-miR-342-5p;hsa-miR-505-5p | 10 | WNK2 | Sponge network | -0.427 | 0.0623 | -0.352 | 0.31066 | 0.25 |
| 54693 | RP11-216B9.6 |
hsa-miR-130b-3p;hsa-miR-130b-5p;hsa-miR-148a-3p;hsa-miR-148b-3p;hsa-miR-148b-5p;hsa-miR-301a-3p;hsa-miR-320a;hsa-miR-320b;hsa-miR-330-3p;hsa-miR-532-5p;hsa-miR-628-5p;hsa-miR-629-3p | 12 | CDH23 | Sponge network | 0.174 | 0.30034 | -0.974 | 0.00034 | 0.25 |
| 54694 | BDNF-AS |
hsa-miR-130b-5p;hsa-miR-135b-5p;hsa-miR-141-5p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-15b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-185-5p;hsa-miR-18a-5p;hsa-miR-193b-3p;hsa-miR-197-3p;hsa-miR-200a-5p;hsa-miR-223-3p;hsa-miR-224-5p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-30b-3p;hsa-miR-345-5p;hsa-miR-3614-5p;hsa-miR-429;hsa-miR-501-5p;hsa-miR-502-5p;hsa-miR-629-3p;hsa-miR-942-5p | 25 | IGFBP5 | Sponge network | -0.668 | 3.0E-5 | -0.806 | 0.00105 | 0.25 |
| 54695 | RP11-389C8.2 |
hsa-let-7a-3p;hsa-let-7b-3p;hsa-miR-144-3p;hsa-miR-148a-3p;hsa-miR-16-2-3p;hsa-miR-182-5p;hsa-miR-186-5p;hsa-miR-200c-3p;hsa-miR-26a-2-3p;hsa-miR-27a-3p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-375;hsa-miR-429;hsa-miR-590-3p;hsa-miR-7-1-3p | 17 | MITF | Sponge network | 0.727 | 3.0E-5 | -0.769 | 0.00022 | 0.25 |
| 54696 | TRAF3IP2-AS1 |
hsa-let-7d-5p;hsa-let-7g-5p;hsa-let-7i-5p;hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-126-5p;hsa-miR-128-3p;hsa-miR-141-3p;hsa-miR-146a-5p;hsa-miR-146b-5p;hsa-miR-16-2-3p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-181b-5p;hsa-miR-182-5p;hsa-miR-185-5p;hsa-miR-186-5p;hsa-miR-18a-3p;hsa-miR-196b-5p;hsa-miR-197-3p;hsa-miR-1976;hsa-miR-19b-3p;hsa-miR-200a-3p;hsa-miR-20a-5p;hsa-miR-2355-3p;hsa-miR-24-3p;hsa-miR-25-3p;hsa-miR-29b-1-5p;hsa-miR-32-5p;hsa-miR-330-3p;hsa-miR-3614-5p;hsa-miR-362-5p;hsa-miR-450b-5p;hsa-miR-501-5p;hsa-miR-577;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-874-3p;hsa-miR-92a-1-5p;hsa-miR-92a-3p;hsa-miR-92b-3p | 41 | CREB3L2 | Sponge network | -0.323 | 0.01372 | -0.525 | 2.0E-5 | 0.25 |